script
stringlengths 113
767k
|
|---|
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import time
data = pd.read_csv("../input/featureselection/data.csv")
data.head()
col = data.columns
print(col)
y = data.diagnosis
list = ["Unnamed: 32", "id", "diagnosis"]
x = data.drop(list, axis=1)
x.head()
ax = sns.countplot(y, label="Count")
B, M = y.value_counts()
print("Number of Benign:", B)
print("Number of Malignant: ", M)
x.describe()
data_dia = y
data = x
data_n_2 = (data - data.mean()) / (data.std())
data = pd.concat([y, data_n_2.iloc[:, 0:10]], axis=1)
data = pd.melt(data, id_vars="diagnosis", var_name="features", value_name="value")
plt.figure(figsize=(12, 5))
sns.violinplot(
x="features", y="value", hue="diagnosis", data=data, split=True, inner="quart"
)
plt.xticks(rotation=90)
data = pd.concat([y, data_n_2.iloc[:, 10:20]], axis=1)
data = pd.melt(data, id_vars="diagnosis", var_name="features", value_name="value")
plt.figure(figsize=(12, 5))
sns.violinplot(
x="features", y="value", hue="diagnosis", data=data, split=True, inner="quart"
)
plt.xticks(rotation=60)
data = pd.concat([y, data_n_2.iloc[:, 20:31]], axis=1)
data = pd.melt(data, id_vars="diagnosis", value_name="value", var_name="features")
plt.figure(figsize=(12, 5))
sns.violinplot(
x="features", y="value", hue="diagnosis", data=data, split=True, inner="quart"
)
plt.xticks(rotation=60)
plt.figure(figsize=(12, 5))
sns.boxplot(x="features", y="value", hue="diagnosis", data=data)
plt.xticks(rotation=60)
sns.jointplot(
x.loc[:, "concavity_worst"], x.loc[:, "concave points_worst"], kind="reg", color="b"
)
sns.set(style="white")
df = x.loc[:, ["radius_worst", "perimeter_worst", "area_worst"]]
g = sns.PairGrid(df, diag_sharey=False)
g.map_lower(sns.kdeplot, cmap="Blues_d")
g.map_upper(plt.scatter)
g.map_diag(sns.kdeplot, lw=3)
sns.set(style="whitegrid", palette="muted")
data_dia = y
data = x
data_n_2 = (data - data.mean()) / (data.std())
data = pd.concat([y, data_n_2.iloc[:, 0:10]], axis=1)
data = pd.melt(data, id_vars="diagnosis", var_name="features", value_name="value")
plt.figure(figsize=(12, 12))
tic = time.time()
sns.swarmplot(x="features", y="value", hue="diagnosis", data=data)
plt.xticks(rotation=60)
data = pd.concat([y, data_n_2.iloc[:, 10:20]], axis=1)
data = pd.melt(data, id_vars="diagnosis", var_name="features", value_name="value")
plt.figure(figsize=(12, 5))
sns.swarmplot(x="features", y="value", hue="diagnosis", data=data)
plt.xticks(rotation=60)
data = pd.concat([y, data_n_2.iloc[:, 20:31]], axis=1)
data = pd.melt(data, id_vars="diagnosis", var_name="features", value_name="value")
plt.figure(figsize=(12, 10))
sns.swarmplot(x="diagnosis", y="value", hue="diagnosis", data=data)
plt.xticks(rotation=60)
f, ax = plt.subplots(figsize=(12, 12))
sns.heatmap(x.corr(), annot=True, linewidths=0.5, fmt=".1f", ax=ax)
drop_list1 = [
"perimeter_mean",
"radius_mean",
"compactness_mean",
"concave points_mean",
"radius_se",
"perimeter_se",
"radius_worst",
"perimeter_worst",
"compactness_worst",
"concave points_worst",
"compactness_se",
"concave points_se",
"texture_worst",
"area_worst",
]
x_1 = x.drop(drop_list1, axis=1)
x_1.head()
f, ax = plt.subplots(figsize=(12, 10))
sns.heatmap(x_1.corr(), annot=True, linewidths=0.5, fmt=".1f", ax=ax)
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import f1_score, confusion_matrix
from sklearn.metrics import accuracy_score
X_train, X_test, y_train, y_test = train_test_split(
x_1, y, test_size=0.3, random_state=42
)
clf_rf = RandomForestClassifier(random_state=43)
clf_rf = clf_rf.fit(X_train, y_train)
ac = accuracy_score(y_test, clf_rf.predict(X_test))
print("Accuracy is: ", ac)
cm = confusion_matrix(y_test, clf_rf.predict(X_test))
sns.heatmap(cm, annot=True, fmt="d")
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import chi2
select_feature = SelectKBest(chi2, k=5).fit(X_train, y_train)
print("Score list: ", select_feature.scores_)
print("Feature list: ", X_train.columns)
X_train_2 = select_feature.transform(X_train)
X_test_2 = select_feature.transform(X_test)
clf_rf_2 = RandomForestClassifier()
clf_rf_2 = clf_rf_2.fit(X_train_2, y_train)
ac_2 = accuracy_score(y_test, clf_rf_2.predict(X_test_2))
print("Accuracy is: ", ac_2)
cm_2 = confusion_matrix(y_test, clf_rf_2.predict(X_test_2))
sns.heatmap(cm_2, annot=True, fmt="d")
from sklearn.feature_selection import RFE
clf_rf_3 = RandomForestClassifier()
rfe = RFE(estimator=clf_rf_3, n_features_to_select=5, step=1)
rfe = rfe.fit(X_train, y_train)
print("Chosen best 5 features by rfe: ", X_train.columns[rfe.support_])
from sklearn.feature_selection import RFECV
clf_rf_4 = RandomForestClassifier()
rfecv = RFECV(estimator=clf_rf_4, step=1, cv=5, scoring="accuracy")
rfecv = rfecv.fit(X_train, y_train)
print("Optimal number of features: ", rfecv.n_features_)
print("Best features: ", X_train.columns[rfecv.support_])
import matplotlib.pyplot as plt
plt.figure()
plt.xlabel("Number of featuress selected")
plt.ylabel("Cross validation score of number of selected features")
plt.plot(range(1, len(rfecv.grid_scores_) + 1), rfecv.grid_scores_)
clf_rf_5 = RandomForestClassifier()
clf_rf_5 = clf_rf_5.fit(X_train, y_train)
importances = clf_rf_5.feature_importances_
std = np.std([tree.feature_importances_ for tree in clf_rf.estimators_], axis=0)
indices = np.argsort(importances)[::-1]
print("Feature ranking: ")
for f in range(X_train.shape[1]):
print("%d. feature %d (%f)" % (f + 1, indices[f], importances[indices[f]]))
plt.figure(1, figsize=(12, 8))
plt.title("Feature importances ")
plt.bar(
range(X_train.shape[1]),
importances[indices],
color="g",
yerr=std[indices],
align="center",
)
plt.xticks(range(X_train.shape[1]), X_train.columns[indices], rotation=60)
plt.xlim([-1, X_train.shape[1]])
X_train, X_test, y_train, y_test = train_test_split(
x, y, test_size=0.3, random_state=42
)
X_train_N = (X_train - X_train.mean()) / (X_train.max() - X_train.min())
X_test_N = (X_test - X_test.mean()) / (X_test.max() - X_test.min())
from sklearn.decomposition import PCA
pca = PCA()
pca.fit(X_train_N)
plt.figure(1, figsize=(12, 8))
plt.clf()
plt.axes([0.2, 0.2, 0.7, 0.7])
plt.plot(pca.explained_variance_ratio_, linewidth=2)
plt.axis("tight")
plt.xlabel("n_components")
plt.ylabel("explainbed_variance_ratio_")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
train = pd.read_csv("/kaggle/input/titanic/train.csv")
test = pd.read_csv("/kaggle/input/titanic/test.csv")
# ## Discriptive Analysis
train.head()
train.describe()
# Observation:
# 1. Fare is skewed as mean and 50% values are not near to each other.
train.info()
# Observation:
# 1. Age, Cabin, Embarked have null value.
# ## EDA
import plotly.express as px
import matplotlib
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_style("darkgrid")
matplotlib.rcParams["font.size"] = 14
matplotlib.rcParams["figure.figsize"] = (10, 6)
matplotlib.rcParams["figure.facecolor"] = "#00000000"
px.histogram(train, x="Survived")
# 1. 38% survived of the total people onboard survived.
sns.histplot(train, x="Pclass", hue="Survived")
# 1. It shows large amount of people traveled in 3rd class.
# 2. Survival rate of 1st class people is higher than 3rd class people.
px.histogram(train, x="Age", color="Survived")
# 1. Large number of people between age 20 - 35 age travelled.
# 2. Children and people aged between 20-40 survived more.
px.histogram(train, x="Sex", color="Survived")
# 1. Number of male survived is less than female.
train[train["Survived"] == 1].groupby("SibSp").count()["Name"] / train.groupby(
"SibSp"
).count()["Name"].sum()
train[train["Survived"] == 0].groupby("SibSp").count()["Name"] / train.groupby(
"SibSp"
).count()["Name"].sum()
# 1. 68% people have no Siblings or wife.
# 2. 23 % people without siblings or wife survived.
px.histogram(train, x="SibSp", color="Survived")
# 1. Lesser the SibSp value, more the chance of survival.
train[train["Survived"] == 1].groupby("Parch").count()["Name"] / train.groupby(
"SibSp"
).count()["Name"].sum()
train[train["Survived"] == 0].groupby("Parch").count()["Name"] / train.groupby(
"SibSp"
).count()["Name"].sum()
# 1. 75% have no parents or children and 26% survived in that.
px.histogram(train, x="Parch", color="Survived")
# 1. Lesser the value of Parch, more the chance of survival.
px.histogram(train, x="Fare", color="Survived")
# 1. Fare column have exp distribution.
px.histogram(train, x="Embarked", color="Survived")
# Large number of people embarked on S post.
# #### Summary of the obervation
# 1. 38% survived of the total people onboard survived.
# 2. Survival rate of 1st class people is higher than 3rd class people.
# 3. 68 % people have no Siblings or wife.
# 4. 23 % people without siblings or wife survived.
# 5. 75% have no parents or children and 26% survived in that.
# 6. Fare price is largely skewed and few people paid fare as high as $512.
# ## Addressing Missing Value
train.isna().sum()
train.describe(include=["O"])
# 1. Cabin is being shared by people. This gives the picture like, a family shares one cabin or alternatively cheap ticket traveller share cabins with other traveller.
# #### Assumptions
# 1. We can drop Ticket, Cabin, PassengerID cols as they dont directly contribute to Survival rate.Also Cabin col contain lot of null value.
# 2. Lets fill the age col with median of the col and Embarked with mode of the col.
def missing_value(df):
df = df.drop(["Ticket", "Cabin", "PassengerId", "Name"], axis=1)
df["Age"].fillna(df["Age"].median(), inplace=True)
df["Embarked"].fillna(df["Embarked"].mode()[0], inplace=True)
return df
train = missing_value(train)
train
# As large number of people are around 20-35 age group. We choose to fill null value of column Age with median.
train.isna().sum()
# #### Onehot Encode of Categorical Col
def categorical(df):
df["Sex"] = df["Sex"].map({"female": 1, "male": 0}).astype(int)
df["Embarked"] = df["Embarked"].map({"S": 0, "C": 1, "Q": 2})
return df
train = categorical(train)
# ## Model building
X_train = train.drop(["Survived"], axis=1)
y_train = train["Survived"]
from sklearn.linear_model import LogisticRegression
model1 = LogisticRegression(random_state=0, solver="liblinear")
model1.fit(X_train, y_train)
model1.score(X_train, y_train)
from sklearn.ensemble import RandomForestClassifier
random_forest = RandomForestClassifier(n_estimators=100)
random_forest.fit(X_train, y_train)
random_forest.score(X_train, y_train)
acc_random_forest = round(random_forest.score(X_train, y_train) * 100, 2)
acc_random_forest
# Thus Random Forest Classifier perform best.
# #### Test set Prediction
test.isna().sum()
test = missing_value(test)
test = categorical(test)
test["Fare"] = test["Fare"].fillna(test["Fare"].median())
test.isna().sum()
Y_pred = random_forest.predict(test)
test_df = pd.read_csv("/kaggle/input/titanic/test.csv")
submission = pd.DataFrame({"PassengerId": test_df["PassengerId"], "Survived": Y_pred})
submission.to_csv("submission.csv", index=False)
|
# #Assignment - Transliteration
# In this task you are required to solve the transliteration problem of names from English to Russian. Transliteration of a string means writing this string using the alphabet of another language with the preservation of pronunciation, although not always.
# ## Instructions
# To complete the assignment please do the following steps (both are requred to get the full credits):
# ### 1. Complete this notebook
# Upload a filled notebook with code (this file). You will be asked to implement a transformer-based approach for transliteration.
# You should implement your ``train`` and ``classify`` functions in this notebook in the cells below. Your model should be implemented as a special class/function in this notebook (be sure if you add any outer dependencies that everything is improted correctly and can be reproducable).
# ###2. Submit solution to the shared task
# After the implementation of models' architectures you are asked to participate in the [competition](https://competitions.codalab.org/competitions/30932) to solve **Transliteration** task using your implemented code.
# You should use your code from the previous part to train, validate, and generate predictions for the public (Practice) and private (Evaluation) test sets. It will produce predictions (`preds_translit.tsv`) for the dataset and score them if the true answers are present. You can use these scores to evaluate your model on dev set and choose the best one. Be sure to download the [dataset](https://github.com/skoltech-nlp/filimdb_evaluation/blob/master/TRANSLIT.tar.gz) and unzip it with `wget` command and run them from notebook cells.
# Upload obtained TSV file with your predictions (``preds_translit.tsv``) in ``.zip`` for the best results to both phases of the competition.
# **Important: You must indicate "DL4NLP-23" as your team name in Codalab. Without it your submission will be invalid!**
# ## Basic algorithm
# The basic algorithm is based on the following idea: for transliteration, alphabetic n-grams from one language can be transformed into another language into n-grams of the same size, using the most frequent transformation rule found according to statistics on the training sample.
# To test the implementation, download the data, unzip the datasets, predict transliteration and run the evaluation script. To do this, you need to run the following commands:
# ### Baseline code
from typing import List, Any
from random import random
import collections as col
def baseline_train(
train_source_strings: List[str], train_target_strings: List[str]
) -> Any:
"""
Trains transliretation model on the given train set represented as
parallel list of input strings and their transliteration via labels.
:param train_source_strings: a list of strings, one str per example
:param train_target_strings: a list of strings, one str per example
:return: learnt parameters, or any object you like (it will be passed to the classify function)
"""
ngram_lvl = 3
def obtain_train_dicts(train_source_strings, train_target_strings, ngram_lvl):
ngrams_dict = col.defaultdict(lambda: col.defaultdict(int))
for src_str, dst_str in zip(train_source_strings, train_target_strings):
try:
src_ngrams = [
src_str[i : i + ngram_lvl]
for i in range(len(src_str) - ngram_lvl + 1)
]
dst_ngrams = [
dst_str[i : i + ngram_lvl]
for i in range(len(dst_str) - ngram_lvl + 1)
]
except TypeError as e:
print(src_ngrams, dst_ngrams)
print(e)
raise StopIteration
for src_ngram in src_ngrams:
for dst_ngram in dst_ngrams:
ngrams_dict[src_ngram][dst_ngram] += 1
return ngrams_dict
ngrams_dict = col.defaultdict(lambda: col.defaultdict(int))
for nl in range(1, ngram_lvl + 1):
ngrams_dict.update(
obtain_train_dicts(train_source_strings, train_target_strings, nl)
)
return ngrams_dict
def baseline_classify(strings: List[str], params: Any) -> List[str]:
"""
Classify strings given previously learnt parameters.
:param strings: strings to classify
:param params: parameters received from train function
:return: list of lists of predicted transliterated strings
(for each source string -> [top_1 prediction, .., top_k prediction]
if it is possible to generate more than one, otherwise
-> [prediction])
corresponding to the given list of strings
"""
def predict_one_sample(sample, train_dict, ngram_lvl=1):
ngrams = [
sample[i : i + ngram_lvl]
for i in range(
0, (len(sample) // ngram_lvl * ngram_lvl) - ngram_lvl + 1, ngram_lvl
)
] + (
[]
if len(sample) % ngram_lvl == 0
else [sample[-(len(sample) % ngram_lvl) :]]
)
prediction = ""
for ngram in ngrams:
ngram_dict = train_dict[ngram]
if len(ngram_dict.keys()) == 0:
prediction += "?" * len(ngram)
else:
prediction += max(ngram_dict, key=lambda k: ngram_dict[k])
return prediction
ngram_lvl = 3
predictions = []
ngrams_dict = params
for string in strings:
top_1_pred = predict_one_sample(string, ngrams_dict, ngram_lvl)
predictions.append([top_1_pred])
return predictions
# ### Evaluation code
PREDS_FNAME = "preds_translit_baseline.tsv"
SCORED_PARTS = ("train", "dev", "train_small", "dev_small", "test")
TRANSLIT_PATH = "TRANSLIT"
import codecs
from pandas import read_csv
def load_dataset(data_dir_path=None, parts: List[str] = SCORED_PARTS):
part2ixy = {}
for part in parts:
path = os.path.join(data_dir_path, f"{part}.tsv")
with open(path, "r", encoding="utf-8") as rf:
# first line is a header of the corresponding columns
lines = rf.readlines()[1:]
col_count = len(lines[0].strip("\n").split("\t"))
if col_count == 2:
strings, transliterations = zip(
*list(map(lambda l: l.strip("\n").split("\t"), lines))
)
elif col_count == 1:
strings = list(map(lambda l: l.strip("\n"), lines))
transliterations = None
else:
raise ValueError("wrong amount of columns")
part2ixy[part] = (
[f"{part}/{i}" for i in range(len(strings))],
strings,
transliterations,
)
return part2ixy
def load_transliterations_only(data_dir_path=None, parts: List[str] = SCORED_PARTS):
part2iy = {}
for part in parts:
path = os.path.join(data_dir_path, f"{part}.tsv")
with open(path, "r", encoding="utf-8") as rf:
# first line is a header of the corresponding columns
lines = rf.readlines()[1:]
col_count = len(lines[0].strip("\n").split("\t"))
n_lines = len(lines)
if col_count == 2:
transliterations = [l.strip("\n").split("\t")[1] for l in lines]
elif col_count == 1:
transliterations = None
else:
raise ValueError("Wrong amount of columns")
part2iy[part] = (
[f"{part}/{i}" for i in range(n_lines)],
transliterations,
)
return part2iy
def save_preds(preds, preds_fname):
"""
Save classifier predictions in format appropriate for scoring.
"""
with codecs.open(preds_fname, "w") as outp:
for idx, preds in preds:
print(idx, *preds, sep="\t", file=outp)
print("Predictions saved to %s" % preds_fname)
def load_preds(preds_fname, top_k=1):
"""
Load classifier predictions in format appropriate for scoring.
"""
kwargs = {
"filepath_or_buffer": preds_fname,
"names": ["id", "pred"],
"sep": "\t",
}
pred_ids = list(read_csv(**kwargs, usecols=["id"])["id"])
pred_y = {
pred_id: [y]
for pred_id, y in zip(pred_ids, read_csv(**kwargs, usecols=["pred"])["pred"])
}
for y in pred_y.values():
assert len(y) == top_k
return pred_ids, pred_y
def compute_hit_k(preds, k=10):
raise NotImplementedError
def compute_mrr(preds):
raise NotImplementedError
def compute_acc_1(preds, true):
right_answers = 0
bonus = 0
for pred, y in zip(preds, true):
if pred[0] == y:
right_answers += 1
elif pred[0] != pred[0] and y == "нань":
print(
"Your test file contained empty string, skipping %f and %s"
% (pred[0], y)
)
bonus += 1 # bugfix: skip empty line in test
return right_answers / (len(preds) - bonus)
def score(preds, true):
assert len(preds) == len(
true
), "inconsistent amount of predictions and ground truth answers"
acc_1 = compute_acc_1(preds, true)
return {"acc@1": acc_1}
def score_preds(preds_path, data_dir, parts=SCORED_PARTS):
part2iy = load_transliterations_only(data_dir, parts=parts)
pred_ids, pred_dict = load_preds(preds_path)
# pred_dict = {i:y for i,y in zip(pred_ids, pred_y)}
scores = {}
for part, (true_ids, true_y) in part2iy.items():
if true_y is None:
print("no labels for %s set" % part)
continue
pred_y = [pred_dict[i] for i in true_ids]
score_values = score(pred_y, true_y)
acc_1 = score_values["acc@1"]
print("%s set accuracy@1: %.2f" % (part, acc_1))
scores[part] = score_values
return scores
# ### Train and predict results
from time import time
import numpy as np
import os
def train_and_predict(translit_path, scored_parts):
top_k = 1
part2ixy = load_dataset(translit_path, parts=scored_parts)
train_ids, train_strings, train_transliterations = part2ixy["train"]
print(
"\nTraining classifier on %d examples from train set ..." % len(train_strings)
)
st = time()
params = baseline_train(train_strings, train_transliterations)
print("Classifier trained in %.2fs" % (time() - st))
allpreds = []
for part, (ids, x, y) in part2ixy.items():
print("\nClassifying %s set with %d examples ..." % (part, len(x)))
st = time()
preds = baseline_classify(x, params)
print("%s set classified in %.2fs" % (part, time() - st))
count_of_values = list(map(len, preds))
assert np.all(np.array(count_of_values) == top_k)
# score(preds, y)
allpreds.extend(zip(ids, preds))
save_preds(allpreds, preds_fname=PREDS_FNAME)
print("\nChecking saved predictions ...")
return score_preds(
preds_path=PREDS_FNAME, data_dir=translit_path, parts=scored_parts
)
train_and_predict(TRANSLIT_PATH, SCORED_PARTS)
# ## Transformer-based approach
# To implement your algorithm, use the template code, which needs to be modified.
# First, you need to add some details in the code of the Transformer architecture, implement the methods of the class `LrScheduler`, which is responsible for updating the learning rate during training.
# Next, you need to select the hyperparameters for the model according to the proposed guide.
import torch
import torch.nn as nn
import torch.nn.functional as F
import pandas as pd
import numpy as np
import itertools as it
import collections as col
import random
import os
import copy
import json
from tqdm import tqdm
import datetime, time
import copy
import os
import pandas as pd
import torch
import torch.nn as nn
import torch.utils.data as torch_data
import itertools as it
import collections as col
import random
import Levenshtein as le
# ### Load dataset and embeddings
def load_datasets(data_dir_path, parts):
datasets = {}
for part in parts:
path = os.path.join(data_dir_path, f"{part}.tsv")
datasets[part] = pd.read_csv(path, sep="\t", na_filter=False)
print(f"Loaded {part} dataset, length: {len(datasets[part])}")
return datasets
class TextEncoder:
def __init__(self, load_dir_path=None):
self.lang_keys = ["en", "ru"]
self.directions = ["id2token", "token2id"]
self.service_token_names = {
"pad_token": "<pad>",
"start_token": "<start>",
"unk_token": "<unk>",
"end_token": "<end>",
}
service_id2token = dict(enumerate(self.service_token_names.values()))
service_token2id = {v: k for k, v in service_id2token.items()}
self.service_vocabs = dict(
zip(self.directions, [service_id2token, service_token2id])
)
if load_dir_path is None:
self.vocabs = {}
for lk in self.lang_keys:
self.vocabs[lk] = copy.deepcopy(self.service_vocabs)
else:
self.vocabs = self.load_vocabs(load_dir_path)
def load_vocabs(self, load_dir_path):
vocabs = {}
load_path = os.path.join(load_dir_path, "vocabs")
for lk in self.lang_keys:
vocabs[lk] = {}
for d in self.directions:
columns = d.split("2")
print(lk, d)
df = pd.read_csv(os.path.join(load_path, f"{lk}_{d}"))
vocabs[lk][d] = dict(zip(*[df[c] for c in columns]))
return vocabs
def save_vocabs(self, save_dir_path):
save_path = os.path.join(save_dir_path, "vocabs")
os.makedirs(save_path, exist_ok=True)
for lk in self.lang_keys:
for d in self.directions:
columns = d.split("2")
pd.DataFrame(data=self.vocabs[lk][d].items(), columns=columns).to_csv(
os.path.join(save_path, f"{lk}_{d}"), index=False, sep=","
)
def make_vocabs(self, data_df):
for lk in self.lang_keys:
tokens = col.Counter("".join(list(it.chain(*data_df[lk])))).keys()
part_id2t = dict(enumerate(tokens, start=len(self.service_token_names)))
part_t2id = {k: v for v, k in part_id2t.items()}
part_vocabs = [part_id2t, part_t2id]
for i in range(len(self.directions)):
self.vocabs[lk][self.directions[i]].update(part_vocabs[i])
self.src_vocab_size = len(self.vocabs["en"]["id2token"])
self.tgt_vocab_size = len(self.vocabs["ru"]["id2token"])
def frame(self, sample, start_token=None, end_token=None):
if start_token is None:
start_token = self.service_token_names["start_token"]
if end_token is None:
end_token = self.service_token_names["end_token"]
return [start_token] + sample + [end_token]
def token2id(self, samples, frame, lang_key):
if frame:
samples = list(map(self.frame, samples))
vocab = self.vocabs[lang_key]["token2id"]
return list(
map(
lambda s: [
vocab[t]
if t in vocab.keys()
else vocab[self.service_token_names["unk_token"]]
for t in s
],
samples,
)
)
def unframe(self, sample, start_token=None, end_token=None):
if start_token is None:
start_token = self.service_vocabs["token2id"][
self.service_token_names["start_token"]
]
if end_token is None:
end_token = self.service_vocabs["token2id"][
self.service_token_names["end_token"]
]
pad_token = self.service_vocabs["token2id"][
self.service_token_names["pad_token"]
]
return list(
it.takewhile(lambda e: e != end_token and e != pad_token, sample[1:])
)
def id2token(self, samples, unframe, lang_key):
if unframe:
samples = list(map(self.unframe, samples))
vocab = self.vocabs[lang_key]["id2token"]
return list(
map(
lambda s: [
vocab[idx]
if idx in vocab.keys()
else self.service_token_names["unk_token"]
for idx in s
],
samples,
)
)
class TranslitData(torch_data.Dataset):
def __init__(self, source_strings, target_strings, text_encoder):
super(TranslitData, self).__init__()
self.source_strings = source_strings
self.text_encoder = text_encoder
if target_strings is not None:
assert len(source_strings) == len(target_strings)
self.target_strings = target_strings
else:
self.target_strings = None
def __len__(self):
return len(self.source_strings)
def __getitem__(self, idx):
src_str = self.source_strings[idx]
encoder_input = self.text_encoder.token2id(
[list(src_str)], frame=True, lang_key="en"
)[0]
if self.target_strings is not None:
tgt_str = self.target_strings[idx]
tmp = self.text_encoder.token2id(
[list(tgt_str)], frame=True, lang_key="ru"
)[0]
decoder_input = tmp[:-1]
decoder_target = tmp[1:]
return (encoder_input, decoder_input, decoder_target)
else:
return (encoder_input,)
class BatchSampler(torch_data.BatchSampler):
def __init__(self, sampler, batch_size, drop_last, shuffle_each_epoch):
super(BatchSampler, self).__init__(sampler, batch_size, drop_last)
self.batches = []
for b in super(BatchSampler, self).__iter__():
self.batches.append(b)
self.shuffle_each_epoch = shuffle_each_epoch
if self.shuffle_each_epoch:
random.shuffle(self.batches)
self.index = 0
# print(f'Batches collected: {len(self.batches)}')
def __iter__(self):
self.index = 0
return self
def __next__(self):
if self.index == len(self.batches):
if self.shuffle_each_epoch:
random.shuffle(self.batches)
raise StopIteration
else:
batch = self.batches[self.index]
self.index += 1
return batch
def collate_fn(batch_list):
"""batch_list can store either 3 components:
encoder_inputs, decoder_inputs, decoder_targets
or single component: encoder_inputs"""
components = list(zip(*batch_list))
batch_tensors = []
for data in components:
max_len = max([len(sample) for sample in data])
# print(f'Maximum length in batch = {max_len}')
sample_tensors = [
torch.tensor(s, requires_grad=False, dtype=torch.int64) for s in data
]
batch_tensors.append(
nn.utils.rnn.pad_sequence(sample_tensors, batch_first=True, padding_value=0)
)
return tuple(batch_tensors)
def create_dataloader(
source_strings, target_strings, text_encoder, batch_size, shuffle_batches_each_epoch
):
"""target_strings parameter can be None"""
dataset = TranslitData(source_strings, target_strings, text_encoder=text_encoder)
seq_sampler = torch_data.SequentialSampler(dataset)
batch_sampler = BatchSampler(
seq_sampler,
batch_size=batch_size,
drop_last=False,
shuffle_each_epoch=shuffle_batches_each_epoch,
)
dataloader = torch_data.DataLoader(
dataset, batch_sampler=batch_sampler, collate_fn=collate_fn
)
return dataloader
# ### Metric function
def compute_metrics(predicted_strings, target_strings, metrics):
metric_values = {}
for m in metrics:
if m == "acc@1":
metric_values[m] = sum(predicted_strings == target_strings) / len(
target_strings
)
elif m == "mean_ld@1":
metric_values[m] = np.mean(
list(
map(
lambda e: le.distance(*e),
zip(predicted_strings, target_strings),
)
)
)
else:
raise ValueError(f"Unknown metric: {m}")
return metric_values
# ### Positional Encoding
# As you remember, Transformer treats an input sequence of elements as a time series. Since the Encoder inside the Transformer simultaneously processes the entire input sequence, the information about the position of the element needs to be encoded inside its embedding, since it is not identified in any other way inside the model. That is why the PositionalEncoding layer is used, which sums embeddings with a vector of the same dimension.
# Let the matrix of these vectors for each position of the time series be denoted as $PE$. Then the elements of the matrix are:
# $$ PE_{(pos,2i)} = \sin{(pos/10000^{2i/d_{model}})}$$
# $$ PE_{(pos,2i+1)} = \cos{(pos/10000^{2i/d_{model}})}$$
# where $pos$ - is the position, $i$ - index of the component of the corresponging vector, $d_{model}$ - dimension of each vector. Thus, even components represent sine values, and odd ones represent cosine values with different arguments.
# In this task you are required to implement these formulas inside the class constructor *PositionalEncoding* in the main file ``translit.py``, which you are to upload. To run the test use the following function:
# `test_positional_encoding()`
# Make sure that there is no any `AssertionError`!
#
import math
class Embedding(nn.Module):
def __init__(self, hidden_size, vocab_size):
super(Embedding, self).__init__()
self.emb_layer = nn.Embedding(vocab_size, hidden_size)
self.hidden_size = hidden_size
def forward(self, x):
return self.emb_layer(x)
class PositionalEncoding(nn.Module):
def __init__(self, hidden_size, max_len=512):
super(PositionalEncoding, self).__init__()
pe = torch.zeros(max_len, hidden_size, requires_grad=False)
position = torch.arange(0, max_len, dtype=torch.float).unsqueeze(1)
div_term = torch.exp(
torch.arange(0, hidden_size, 2).float() * (-math.log(10000.0) / hidden_size)
)
pe[:, 0::2] = torch.sin(position * div_term)
pe[:, 1::2] = torch.cos(position * div_term)
pe = pe.unsqueeze(0)
# pe shape: (1, max_len, hidden_size)
self.register_buffer("pe", pe)
def forward(self, x):
# x: shape (batch size, sequence length, hidden size)
x = x + self.pe[:, : x.size(1)]
return x
def test_positional_encoding():
pe = PositionalEncoding(max_len=3, hidden_size=4)
res_1 = torch.tensor(
[
[
[0.0000, 1.0000, 0.0000, 1.0000],
[0.8415, 0.5403, 0.0100, 0.9999],
[0.9093, -0.4161, 0.0200, 0.9998],
]
]
)
# print(pe.pe - res_1)
assert torch.all(torch.abs(pe.pe - res_1) < 1e-4).item()
print("Test is passed!")
test_positional_encoding()
# ### LayerNorm
class LayerNorm(nn.Module):
"Layer Normalization layer"
def __init__(self, hidden_size, eps=1e-6):
super(LayerNorm, self).__init__()
self.gain = nn.Parameter(torch.ones(hidden_size))
self.bias = nn.Parameter(torch.zeros(hidden_size))
self.eps = eps
def forward(self, x):
mean = x.mean(-1, keepdim=True)
std = x.std(-1, keepdim=True)
return self.gain * (x - mean) / (std + self.eps) + self.bias
# ### SublayerConnection
class SublayerConnection(nn.Module):
"""
A residual connection followed by a layer normalization.
"""
def __init__(self, hidden_size, dropout):
super(SublayerConnection, self).__init__()
self.layer_norm = LayerNorm(hidden_size)
self.dropout = nn.Dropout(dropout)
def forward(self, x, sublayer):
return self.layer_norm(x + self.dropout(sublayer(x)))
def padding_mask(x, pad_idx=0):
assert len(x.size()) >= 2
return (x != pad_idx).unsqueeze(-2)
def look_ahead_mask(size):
"Mask out the right context"
attn_shape = (1, size, size)
look_ahead_mask = np.triu(np.ones(attn_shape), k=1).astype("uint8")
return torch.from_numpy(look_ahead_mask) == 0
def compositional_mask(x, pad_idx=0):
pm = padding_mask(x, pad_idx=pad_idx)
seq_length = x.size(-1)
result_mask = pm & look_ahead_mask(seq_length).type_as(pm.data)
return result_mask
# ### FeedForward
class FeedForward(nn.Module):
def __init__(self, hidden_size, ff_hidden_size, dropout=0.1):
super(FeedForward, self).__init__()
self.pre_linear = nn.Linear(hidden_size, ff_hidden_size)
self.post_linear = nn.Linear(ff_hidden_size, hidden_size)
self.dropout = nn.Dropout(dropout)
def forward(self, x):
return self.post_linear(self.dropout(F.relu(self.pre_linear(x))))
def clone_layer(module, N):
"Produce N identical layers."
return nn.ModuleList([copy.deepcopy(module) for _ in range(N)])
# ### MultiHeadAttention
# Then you are required to implement `attention` method in the class `MultiHeadAttention`. The MultiHeadAttention layer takes as input query vectors, key and value vectors for each step of the sequence of matrices Q,K,V correspondingly. Each key vector, value vector, and query vector is obtained as a result of linear projection using one of three trained vector parameter matrices from the previous layer. This semantics can be represented in the form of formulas:
# $$
# Attention(Q, K, V)=softmax\left(\frac{Q K^{T}}{\sqrt{d_{k}}}\right) V\\
# $$
# $$
# MultiHead(Q, K, V) = Concat\left(head_1, ... , head_h\right) W^O\\
# $$
# $$
# head_i=Attention\left(Q W_i^Q, K W_i^K, V W_i^V\right)\\
# $$
# $h$ - the number of attention heads - parallel sub-layers for Scaled Dot-Product Attention on a vector of smaller dimension ($d_{k} = d_{q} = d_{v} = d_{model} / h$).
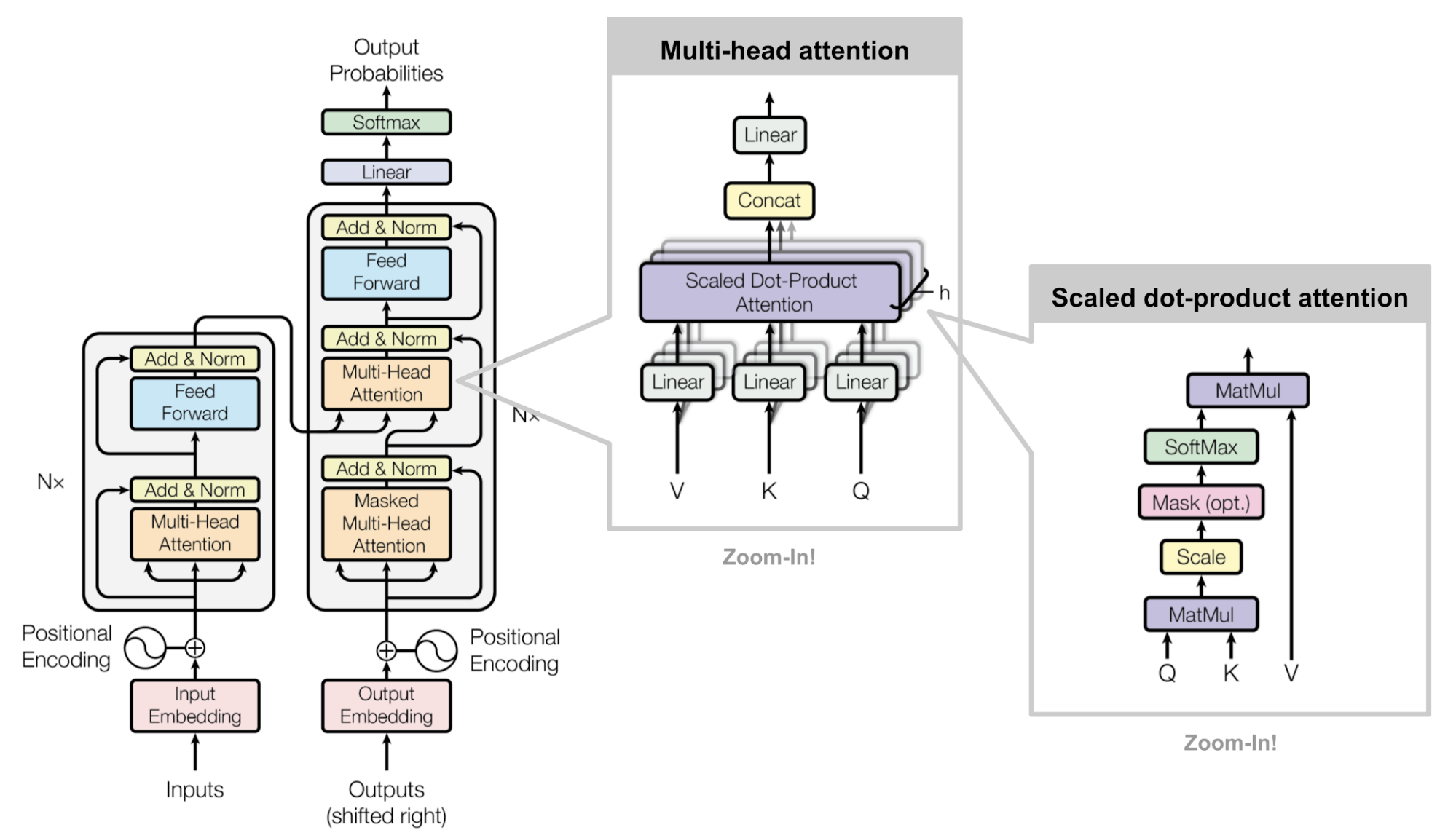
# The logic of $\texttt{MultiHeadAttention}$ is presented in the picture (from original [paper](https://arxiv.org/abs/1706.03762)):
# 
# Inside a method `attention` you are required to create a dropout layer from MultiHeadAttention class constructor. Dropout layer is to be applied directly on the attention weights - the result of softmax operation. Value of drop probability can be regulated in the train in the `model_config['dropout']['attention']`.
# The correctness of implementation can be checked with
# `test_multi_head_attention()`
#
class MultiHeadAttention(nn.Module):
def __init__(self, n_heads, hidden_size, dropout=None):
super(MultiHeadAttention, self).__init__()
assert hidden_size % n_heads == 0
self.head_hidden_size = hidden_size // n_heads
self.n_heads = n_heads
self.linears = clone_layer(nn.Linear(hidden_size, hidden_size), 4)
self.attn_weights = None
self.dropout = dropout
if self.dropout is not None:
self.dropout_layer = nn.Dropout(p=self.dropout)
def attention(self, query, key, value, mask=None):
d_k = query.size(-1)
scores = torch.matmul(query, key.transpose(-2, -1)) / math.sqrt(d_k)
if mask is not None:
scores = scores.masked_fill(mask == 0, -1e9)
attn_weights = F.softmax(scores, dim=-1)
# if self.dropout is not None:
# attn_weights = self.dropout(attn_weights)
result = torch.matmul(attn_weights, value)
return result, attn_weights
def forward(self, query, key, value, mask=None):
if mask is not None:
# Same mask applied to all h heads.
mask = mask.unsqueeze(1)
batch_size = query.size(0)
# Split vectors for different attention heads (from hidden_size => n_heads x head_hidden_size)
# and do separate linear projection, for separate trainable weights
query, key, value = [
l(x)
.view(batch_size, -1, self.n_heads, self.head_hidden_size)
.transpose(1, 2)
for l, x in zip(self.linears, (query, key, value))
]
x, self.attn_weights = self.attention(query, key, value, mask=mask)
# x shape: (batch size, number of heads, sequence length, head hidden size)
# self.attn_weights shape: (batch size, number of heads, sequence length, sequence length)
# Concatenate the output of each head
x = (
x.transpose(1, 2)
.contiguous()
.view(batch_size, -1, self.n_heads * self.head_hidden_size)
)
return self.linears[-1](x)
def test_multi_head_attention():
mha = MultiHeadAttention(n_heads=1, hidden_size=5, dropout=None)
# batch_size == 2, sequence length == 3, hidden_size == 5
# query = torch.arange(150).reshape(2, 3, 5)
query = torch.tensor(
[
[
[[0.64144618, -0.95817388, 0.37432297, 0.58427106, -0.94668716]],
[[-0.23199289, 0.66329209, -0.46507035, -0.54272512, -0.98640698]],
[[0.07546638, -0.09277002, 0.20107185, -0.97407381, -0.27713414]],
],
[
[[0.14727783, 0.4747886, 0.44992016, -0.2841419, -0.81820319]],
[[-0.72324994, 0.80643179, -0.47655449, 0.45627872, 0.60942404]],
[[0.61712569, -0.62947282, -0.95215713, -0.38721959, -0.73289725]],
],
]
)
key = torch.tensor(
[
[
[[-0.81759856, -0.60049991, -0.05923424, 0.51898901, -0.3366209]],
[[0.83957818, -0.96361722, 0.62285191, 0.93452467, 0.51219613]],
[[-0.72758847, 0.41256154, 0.00490795, 0.59892503, -0.07202049]],
],
[
[[0.72315339, -0.49896314, 0.94254637, -0.54356006, -0.04837949]],
[[0.51759322, -0.43927061, -0.59924184, 0.92241702, -0.86811696]],
[[-0.54322046, -0.92323003, -0.827746, 0.90842783, 0.88428119]],
],
]
)
value = torch.tensor(
[
[
[[-0.83895431, 0.805027, 0.22298283, -0.84849915, -0.34906026]],
[[-0.02899652, -0.17456128, -0.17535998, -0.73160314, -0.13468061]],
[[0.75234265, 0.02675947, 0.84766286, -0.5475651, -0.83319316]],
],
[
[[-0.47834413, 0.34464645, -0.41921457, 0.33867964, 0.43470836]],
[[-0.99000979, 0.10220893, -0.4932273, 0.95938905, 0.01927012]],
[[0.91607137, 0.57395644, -0.90914179, 0.97212912, 0.33078759]],
],
]
)
query = query.float().transpose(1, 2)
key = key.float().transpose(1, 2)
value = value.float().transpose(1, 2)
x, _ = torch.max(query[:, 0, :, :], axis=-1)
mask = compositional_mask(x)
mask.unsqueeze_(1)
for n, t in [("query", query), ("key", key), ("value", value), ("mask", mask)]:
print(f"Name: {n}, shape: {t.size()}")
with torch.no_grad():
output, attn_weights = mha.attention(query, key, value, mask=mask)
assert output.size() == torch.Size([2, 1, 3, 5])
assert attn_weights.size() == torch.Size([2, 1, 3, 3])
truth_output = torch.tensor(
[
[
[
[-0.8390, 0.8050, 0.2230, -0.8485, -0.3491],
[-0.6043, 0.5212, 0.1076, -0.8146, -0.2870],
[-0.0665, 0.2461, 0.3038, -0.7137, -0.4410],
]
],
[
[
[-0.4783, 0.3446, -0.4192, 0.3387, 0.4347],
[-0.7959, 0.1942, -0.4652, 0.7239, 0.1769],
[-0.3678, 0.2868, -0.5799, 0.7987, 0.2086],
]
],
]
)
truth_attn_weights = torch.tensor(
[
[
[
[1.0000, 0.0000, 0.0000],
[0.7103, 0.2897, 0.0000],
[0.3621, 0.3105, 0.3274],
]
],
[
[
[1.0000, 0.0000, 0.0000],
[0.3793, 0.6207, 0.0000],
[0.2642, 0.4803, 0.2555],
]
],
]
)
# print(torch.abs(output - truth_output))
# print(torch.abs(attn_weights - truth_attn_weights))
assert torch.all(torch.abs(output - truth_output) < 1e-4).item()
assert torch.all(torch.abs(attn_weights - truth_attn_weights) < 1e-4).item()
print("Test is passed!")
test_multi_head_attention()
# ### Encoder
class EncoderLayer(nn.Module):
"Encoder is made up of self-attn and feed forward (defined below)"
def __init__(self, hidden_size, ff_hidden_size, n_heads, dropout):
super(EncoderLayer, self).__init__()
self.self_attn = MultiHeadAttention(
n_heads, hidden_size, dropout=dropout["attention"]
)
self.feed_forward = FeedForward(
hidden_size, ff_hidden_size, dropout=dropout["relu"]
)
self.sublayers = clone_layer(
SublayerConnection(hidden_size, dropout["residual"]), 2
)
def forward(self, x, mask):
x = self.sublayers[0](x, lambda x: self.self_attn(x, x, x, mask))
return self.sublayers[1](x, self.feed_forward)
class Encoder(nn.Module):
def __init__(self, config):
super(Encoder, self).__init__()
self.embedder = Embedding(config["hidden_size"], config["src_vocab_size"])
self.positional_encoder = PositionalEncoding(
config["hidden_size"], max_len=config["max_src_seq_length"]
)
self.embedding_dropout = nn.Dropout(p=config["dropout"]["embedding"])
self.encoder_layer = EncoderLayer(
config["hidden_size"],
config["ff_hidden_size"],
config["n_heads"],
config["dropout"],
)
self.layers = clone_layer(self.encoder_layer, config["n_layers"])
self.layer_norm = LayerNorm(config["hidden_size"])
def forward(self, x, mask):
"Pass the input (and mask) through each layer in turn."
x = self.embedding_dropout(self.positional_encoder(self.embedder(x)))
for layer in self.layers:
x = layer(x, mask)
return self.layer_norm(x)
# ### Decoder
class DecoderLayer(nn.Module):
"""
Decoder is made of 3 sublayers: self attention, encoder-decoder attention
and feed forward"
"""
def __init__(self, hidden_size, ff_hidden_size, n_heads, dropout):
super(DecoderLayer, self).__init__()
self.self_attn = MultiHeadAttention(
n_heads, hidden_size, dropout=dropout["attention"]
)
self.encdec_attn = MultiHeadAttention(
n_heads, hidden_size, dropout=dropout["attention"]
)
self.feed_forward = FeedForward(
hidden_size, ff_hidden_size, dropout=dropout["relu"]
)
self.sublayers = clone_layer(
SublayerConnection(hidden_size, dropout["residual"]), 3
)
def forward(self, x, encoder_output, encoder_mask, decoder_mask):
x = self.sublayers[0](x, lambda x: self.self_attn(x, x, x, decoder_mask))
x = self.sublayers[1](
x,
lambda x: self.encdec_attn(x, encoder_output, encoder_output, encoder_mask),
)
return self.sublayers[2](x, self.feed_forward)
class Decoder(nn.Module):
def __init__(self, config):
super(Decoder, self).__init__()
self.embedder = Embedding(config["hidden_size"], config["tgt_vocab_size"])
self.positional_encoder = PositionalEncoding(
config["hidden_size"], max_len=config["max_tgt_seq_length"]
)
self.embedding_dropout = nn.Dropout(p=config["dropout"]["embedding"])
self.decoder_layer = DecoderLayer(
config["hidden_size"],
config["ff_hidden_size"],
config["n_heads"],
config["dropout"],
)
self.layers = clone_layer(self.decoder_layer, config["n_layers"])
self.layer_norm = LayerNorm(config["hidden_size"])
def forward(self, x, encoder_output, encoder_mask, decoder_mask):
x = self.embedding_dropout(self.positional_encoder(self.embedder(x)))
for layer in self.layers:
x = layer(x, encoder_output, encoder_mask, decoder_mask)
return self.layer_norm(x)
# ### Transformer
class Transformer(nn.Module):
def __init__(self, config):
super(Transformer, self).__init__()
self.config = config
self.encoder = Encoder(config)
self.decoder = Decoder(config)
self.proj = nn.Linear(config["hidden_size"], config["tgt_vocab_size"])
self.pad_idx = config["pad_idx"]
self.tgt_vocab_size = config["tgt_vocab_size"]
def encode(self, encoder_input, encoder_input_mask):
return self.encoder(encoder_input, encoder_input_mask)
def decode(
self, encoder_output, encoder_input_mask, decoder_input, decoder_input_mask
):
return self.decoder(
decoder_input, encoder_output, encoder_input_mask, decoder_input_mask
)
def linear_project(self, x):
return self.proj(x)
def forward(self, encoder_input, decoder_input):
encoder_input_mask = padding_mask(encoder_input, pad_idx=self.config["pad_idx"])
decoder_input_mask = compositional_mask(
decoder_input, pad_idx=self.config["pad_idx"]
)
encoder_output = self.encode(encoder_input, encoder_input_mask)
decoder_output = self.decode(
encoder_output, encoder_input_mask, decoder_input, decoder_input_mask
)
output_logits = self.linear_project(decoder_output)
return output_logits
def prepare_model(config):
model = Transformer(config)
for p in model.parameters():
if p.dim() > 1:
nn.init.xavier_uniform_(p)
return model
# #### LrScheduler
# The last thing you have to prepare is the class `LrScheduler`, which is in charge of learning rate updating after every step of the optimizer. You are required to fill the class constructor and the method `learning_rate`. The preferable stratagy of updating the learning rate (lr), is the following two stages:
# * "warmup" stage - lr linearly increases until the defined value during the fixed number of steps (the proportion of all training steps - the parameter `train_config['warmup\_steps\_part']` in the train function).
# * "decrease" stage - lr linearly decreases until 0 during the left training steps.
# `learning_rate()` call should return the value of lr at this step, which number is stored at self.step. The class constructor takes not only `warmup_steps_part` but the peak learning rate value `lr_peak` at the end of "warmup" stage and a string name of the strategy of learning rate scheduling. You can test other strategies if you want to with `self.type attribute`.
# Correctness check: `test_lr_scheduler()`
#
class LrScheduler:
def __init__(self, n_steps, **kwargs):
self.type = kwargs["type"]
if self.type == "warmup,decay_linear":
self.warmup_steps_part = kwargs["warmup_steps_part"]
self.lr_peak = kwargs["lr_peak"]
self.n_steps = n_steps # total number of training steps
self.warmup_steps = int(self.warmup_steps_part * n_steps)
self._lr = 0 # initialize the _lr attribute to zero
# raise NotImplementedError
else:
raise ValueError(f"Unknown type argument: {self.type}")
self._step = 0
self._lr = 0
def step(self, optimizer):
self._step += 1
lr = self.learning_rate()
for p in optimizer.param_groups:
p["lr"] = lr
def learning_rate(self, step=None):
if step is None:
step = self._step
if self.type == "warmup,decay_linear":
if step < self.warmup_steps:
self._lr = self.lr_peak * (step / self.warmup_steps)
else:
self._lr = self.lr_peak * (
(self.n_steps - step) / (self.n_steps - self.warmup_steps)
)
return self._lr
raise NotImplementedError("Unknown type of learning rate scheduling.")
def state_dict(self):
sd = copy.deepcopy(self.__dict__)
return sd
def load_state_dict(self, sd):
for k in sd.keys():
self.__setattr__(k, sd[k])
def test_lr_scheduler():
lrs_type = "warmup,decay_linear"
warmup_steps_part = 0.1
lr_peak = 3e-4
sch = LrScheduler(
100, type=lrs_type, warmup_steps_part=warmup_steps_part, lr_peak=lr_peak
)
assert sch.learning_rate(step=5) - 15e-5 < 1e-6
assert sch.learning_rate(step=10) - 3e-4 < 1e-6
assert sch.learning_rate(step=50) - 166e-6 < 1e-6
assert sch.learning_rate(step=100) - 0.0 < 1e-6
print("Test is passed!")
test_lr_scheduler()
# ### Run and translate
def format_time(elapsed):
"""
Takes a time in seconds and returns a string hh:mm:ss
"""
elapsed_rounded = int(round((elapsed)))
return str(datetime.timedelta(seconds=elapsed_rounded))
def run_epoch(data_iter, model, lr_scheduler, optimizer, device, verbose=False):
start = time.time()
local_start = start
total_tokens = 0
total_loss = 0
tokens = 0
loss_fn = nn.CrossEntropyLoss(reduction="sum", label_smoothing=0.1)
for i, batch in tqdm(enumerate(data_iter)):
encoder_input = batch[0].to(device)
decoder_input = batch[1].to(device)
decoder_target = batch[2].to(device)
logits = model(encoder_input, decoder_input)
loss = loss_fn(logits.view(-1, model.tgt_vocab_size), decoder_target.view(-1))
total_loss += loss.item()
batch_n_tokens = (decoder_target != model.pad_idx).sum().item()
total_tokens += batch_n_tokens
if optimizer is not None:
optimizer.zero_grad()
lr_scheduler.step(optimizer)
loss.backward()
optimizer.step()
tokens += batch_n_tokens
if verbose and i % 1000 == 1:
elapsed = time.time() - local_start
print(
"batch number: %d, accumulated average loss: %f, tokens per second: %f"
% (i, total_loss / total_tokens, tokens / elapsed)
)
local_start = time.time()
tokens = 0
average_loss = total_loss / total_tokens
print("** End of epoch, accumulated average loss = %f **" % average_loss)
epoch_elapsed_time = format_time(time.time() - start)
print(f"** Elapsed time: {epoch_elapsed_time}**")
return average_loss
def save_checkpoint(epoch, model, lr_scheduler, optimizer, model_dir_path):
save_path = os.path.join(model_dir_path, f"cpkt_{epoch}_epoch")
torch.save(
{
"epoch": epoch,
"model_state_dict": model.state_dict(),
"optimizer_state_dict": optimizer.state_dict(),
"lr_scheduler_state_dict": lr_scheduler.state_dict(),
},
save_path,
)
print(f"Saved checkpoint to {save_path}")
def load_model(epoch, model_dir_path):
save_path = os.path.join(model_dir_path, f"cpkt_{epoch}_epoch")
checkpoint = torch.load(save_path)
with open(
os.path.join(model_dir_path, "model_config.json"), "r", encoding="utf-8"
) as rf:
model_config = json.load(rf)
model = prepare_model(model_config)
model.load_state_dict(checkpoint["model_state_dict"])
return model
def greedy_decode(model, device, encoder_input, max_len, start_symbol):
batch_size = encoder_input.size()[0]
decoder_input = (
torch.ones(batch_size, 1)
.fill_(start_symbol)
.type_as(encoder_input.data)
.to(device)
)
for i in range(max_len):
logits = model(encoder_input, decoder_input)
_, predicted_ids = torch.max(logits, dim=-1)
next_word = predicted_ids[:, i]
# print(next_word)
rest = torch.ones(batch_size, 1).type_as(decoder_input.data)
# print(rest[:,0].size(), next_word.size())
rest[:, 0] = next_word
decoder_input = torch.cat([decoder_input, rest], dim=1).to(device)
# print(decoder_input)
return decoder_input
def generate_predictions(dataloader, max_decoding_len, text_encoder, model, device):
# print(f'Max decoding length = {max_decoding_len}')
model.eval()
predictions = []
start_token_id = text_encoder.service_vocabs["token2id"][
text_encoder.service_token_names["start_token"]
]
with torch.no_grad():
for batch in tqdm(dataloader):
encoder_input = batch[0].to(device)
prediction_tensor = greedy_decode(
model, device, encoder_input, max_decoding_len, start_token_id
)
predictions.extend(
[
"".join(e)
for e in text_encoder.id2token(
prediction_tensor.cpu().numpy(), unframe=True, lang_key="ru"
)
]
)
return np.array(predictions)
def train(source_strings, target_strings):
"""Common training cycle for final run (fixed hyperparameters,
no evaluation during training)"""
if torch.cuda.is_available():
device = torch.device("cuda")
print(f"Using GPU device: {device}")
else:
device = torch.device("cpu")
print(f"GPU is not available, using CPU device {device}")
train_df = pd.DataFrame({"en": source_strings, "ru": target_strings})
text_encoder = TextEncoder()
text_encoder.make_vocabs(train_df)
model_config = {
"src_vocab_size": text_encoder.src_vocab_size,
"tgt_vocab_size": text_encoder.tgt_vocab_size,
"max_src_seq_length": max(train_df["en"].aggregate(len))
+ 2, # including start_token and end_token
"max_tgt_seq_length": max(train_df["ru"].aggregate(len)) + 2,
"n_layers": 2,
"n_heads": 2,
"hidden_size": 128,
"ff_hidden_size": 256,
"dropout": {
"embedding": 0.15,
"attention": 0.15,
"residual": 0.15,
"relu": 0.15,
},
"pad_idx": 0,
}
model = prepare_model(model_config)
model.to(device)
train_config = {
"batch_size": 200,
"n_epochs": 200,
"lr_scheduler": {
"type": "warmup,decay_linear",
"warmup_steps_part": 0.1,
"lr_peak": 1e-3,
},
}
# Model training procedure
optimizer = torch.optim.Adam(model.parameters(), lr=0.0)
n_steps = (len(train_df) // train_config["batch_size"] + 1) * train_config[
"n_epochs"
]
lr_scheduler = LrScheduler(n_steps, **train_config["lr_scheduler"])
# prepare train data
source_strings, target_strings = zip(
*sorted(zip(source_strings, target_strings), key=lambda e: len(e[0]))
)
train_dataloader = create_dataloader(
source_strings,
target_strings,
text_encoder,
train_config["batch_size"],
shuffle_batches_each_epoch=True,
)
# training cycle
for epoch in range(1, train_config["n_epochs"] + 1):
print("\n" + "-" * 40)
print(f"Epoch: {epoch}")
print(f"Run training...")
model.train()
run_epoch(
train_dataloader,
model,
lr_scheduler,
optimizer,
device=device,
verbose=False,
)
learnable_params = {
"model": model,
"text_encoder": text_encoder,
}
return learnable_params
def classify(source_strings, learnable_params):
if torch.cuda.is_available():
device = torch.device("cuda")
print(f"Using GPU device: {device}")
else:
device = torch.device("cpu")
print(f"GPU is not available, using CPU device {device}")
model = learnable_params["model"]
text_encoder = learnable_params["text_encoder"]
batch_size = 200
dataloader = create_dataloader(
source_strings, None, text_encoder, batch_size, shuffle_batches_each_epoch=False
)
max_decoding_len = model.config["max_tgt_seq_length"]
predictions = generate_predictions(
dataloader, max_decoding_len, text_encoder, model, device
)
# return single top1 prediction for each sample
return np.expand_dims(predictions, 1)
# ### Training
PREDS_FNAME = "preds_translit.tsv"
SCORED_PARTS = ("train", "dev", "train_small", "dev_small", "test")
TRANSLIT_PATH = "TRANSLIT"
top_k = 1
part2ixy = load_dataset(TRANSLIT_PATH, parts=SCORED_PARTS)
train_ids, train_strings, train_transliterations = part2ixy["train"]
print("\nTraining classifier on %d examples from train set ..." % len(train_strings))
st = time.time()
params = train(train_strings, train_transliterations)
print("Classifier trained in %.2fs" % (time.time() - st))
allpreds = []
for part, (ids, x, y) in part2ixy.items():
print("\nClassifying %s set with %d examples ..." % (part, len(x)))
st = time.time()
preds = classify(x, params)
print("%s set classified in %.2fs" % (part, time.time() - st))
count_of_values = list(map(len, preds))
assert np.all(np.array(count_of_values) == top_k)
# score(preds, y)
allpreds.extend(zip(ids, preds))
save_preds(allpreds, preds_fname=PREDS_FNAME)
print("\nChecking saved predictions ...")
score_preds(preds_path=PREDS_FNAME, data_dir=TRANSLIT_PATH, parts=SCORED_PARTS)
# ### Hyper-parameters choice
# The model is ready. Now we need to find the optimal hyper-parameters.
# The quality of models with different hyperparameters should be monitored on dev or on dev_small samples (in order to save time, since generating transliterations is a rather time-consuming process, comparable to one training epoch).
# To generate predictions, you can use the `generate_predictions` function, to calculate the accuracy@1 metric, and then you can use the `compute_metrics` function.
# Hyper-parameters are stored in the dictionary `model_config` and `train_config` in train function. The following hyperparameters in `model_config` and `train_config` are suggested to leave unmodified:
# * n_layers $=$ 2
# * n_heads $=$ 2
# * hidden_size $=$ 128
# * fc_hidden_size $=$ 256
# * warmup_steps_part $=$ 0.1
# * batch_size $=$ 200
# You can vary the dropout value. The model has 4 types of : ***embedding dropout*** applied on embdeddings before sending to the first layer of Encoder or Decoder, ***attention*** dropout applied on the attention weights in the MultiHeadAttention layer, ***residual dropout*** applied on the output of each sublayer (MultiHeadAttention or FeedForward) in layers Encoder and Decoder and, finaly, ***relu dropout*** in used in FeedForward layer. For all 4 types it is suggested to test the same value of dropout from the list: 0.1, 0.15, 0.2.
# Also it is suggested to test several peak levels of learning rate - **lr_peak** : 5e-4, 1e-3, 2e-3.
# Note that if you are using a GPU, then training one epoch takes about 1 minute, and up to 1 GB of video memory is required. When using the CPU, the learning speed slows down by about 2 times. If there are problems with insufficient RAM / video memory, reduce the batch size, but in this case the optimal range of learning rate values will change, and it must be determined again. To train a model with batch_size $=$ 200 , it will take at least 300 epochs to achieve accuracy 0.66 on dev_small dataset.
# *Question: What are the optimal hyperpameters according to your experiments? Add plots or other descriptions here.*
# ```
# ENTER HERE YOUR ANSWER
# ```
#
import optuna
import datetime
import torch
import numpy as np
study = optuna.create_study(direction="maximize", sampler=optuna.samplers.TPESampler())
def objective(trial):
params = {
"learning_rate": trial.suggest_loguniform("learning_rate", 1e-5, 1e-1),
"optimizer": trial.suggest_categorical("optimizer", ["Adam", "RMSprop", "SGD"]),
"n_unit": trial.suggest_int("n_unit", 4, 18),
}
model = build_model(params)
accuracy = train_and_evaluate(params, model)
return accuracy
def objective(trial):
dropout_embeddings = trial.suggest_categorical(
"dropout_embeddings", [0.1, 0.15, 0.2]
)
dropout_attention = trial.suggest_categorical("dropout_attention", [0.1, 0.15, 0.2])
dropout_residual = trial.suggest_categorical("dropout_residual", [0.1, 0.15, 0.2])
dropout_relu = trial.suggest_categorical("dropout_relu", [0.1, 0.15, 0.2])
lr_peak = trial.suggest_categorical("lr_peak", [5e-4, 1e-3, 2e-3])
# train the model and return the evaluation metric for tuning
model = Model(
dropout_embeddings, dropout_attention, dropout_residual, dropout_relu, lr_peak
)
train(model)
metric = evaluate(model)
return metric
# Initialize an Optuna study
study = optuna.create_study(
direction="maximize", sampler=optuna.samplers.RandomSampler(seed=123)
)
study.optimize(objective, n_trials=100)
# Get the best hyperparameters and the best evaluation metric
best_params = study.best_params
best_metric = study.best_value
# Print the best hyperparameters and the best evaluation metric
print(f"Best hyperparameters: {best_params}")
print(f"Best evaluation metric: {best_metric}")
# ## Label smoothing
# We suggest to implement an additional regularization method - **label smoothing**. Now imagine that we have a prediction vector from probabilities at position t in the sequence of tokens for each token id from the vocabulary. CrossEntropy compares it with ground truth one-hot representation
# $$[0, ... 0, 1, 0, ..., 0].$$
# And now imagine that we are slightly "smoothed" the values in the ground truth vector and obtained
# $$[\frac{\alpha}{|V|}, ..., \frac{\alpha}{|V|}, 1(1-\alpha)+\frac{\alpha}{|V|}, \frac{\alpha}{|V|}, ... \frac{\alpha}{|V|}],$$
# where $\alpha$ - parameter from 0 to 1, $|V|$ - vocabulary size - number of components in the ground truth vector. The values of this new vector are still summed to 1. Calculate the cross-entropy of our prediction vector and the new ground truth. Now, firstly, cross-entropy will never reach 0, and secondly, the result of the error function will require the model, as usual, to return the highest probability vector compared to other components of the probability vector for the correct token in the dictionary, but at the same time not too large, because as the value of this probability approaches 1, the value of the error function increases. For research on the use of label smoothing, see the [paper](https://arxiv.org/abs/1906.02629).
#
# Accordingly, in order to embed label smoothing into the model, it is necessary to carry out the transformation described above on the ground truth vectors, as well as to implement the cross-entropy calculation, since the used `torch.nn.CrossEntropy` class is not quite suitable, since for the ground truth representation of `__call__` method takes the id of the correct token and builds a one-hot vector already inside. However, it is possible to implement what is required based on the internal implementation of this class [CrossEntropyLoss](https://pytorch.org/docs/stable/_modules/torch/nn/modules/loss.html#CrossEntropyLoss).
#
# Test different values of $\alpha$ (e.x, 0.05, 0.1, 0.2). Describe your experiments and results.
# ```
# ENTER HERE YOUR ANSWER
# ```
# ENTER HERE YOUR CODE
|
import numpy as np
import pandas as pd
import os
import cv2
labels = os.listdir("/kaggle/input/drowsiness-dataset/train")
labels
import matplotlib.pyplot as plt
plt.imshow(plt.imread("/kaggle/input/drowsiness-dataset/train/Closed/_107.jpg"))
plt.imshow(plt.imread("/kaggle/input/drowsiness-dataset/train/yawn/10.jpg"))
# # for yawn
def face_for_yawn(
direc="/kaggle/input/drowsiness-dataset/train",
face_cas_path="/kaggle/input/haarcasacades/haarcascade_frontalface_alt.xml",
):
yaw_no = []
IMG_SIZE = 224
categories = ["yawn", "no_yawn"]
for category in categories:
path_link = os.path.join(direc, category)
class_num1 = categories.index(category)
print(class_num1)
for image in os.listdir(path_link):
image_array = cv2.imread(os.path.join(path_link, image), cv2.IMREAD_COLOR)
face_cascade = cv2.CascadeClassifier(face_cas_path)
faces = face_cascade.detectMultiScale(image_array, 1.3, 5)
for x, y, w, h in faces:
img = cv2.rectangle(image_array, (x, y), (x + w, y + h), (0, 255, 0), 2)
roi_color = img[y : y + h, x : x + w]
resized_array = cv2.resize(roi_color, (IMG_SIZE, IMG_SIZE))
yaw_no.append([resized_array, class_num1])
return yaw_no
yawn_no_yawn = face_for_yawn()
# # for eyes
def get_data(dir_path="/kaggle/input/drowsiness-dataset/train"):
labels = ["Closed", "Open"]
IMG_SIZE = 224
data = []
for label in labels:
path = os.path.join(dir_path, label)
class_num = labels.index(label)
class_num += 2
print(class_num)
for img in os.listdir(path):
try:
img_array = cv2.imread(os.path.join(path, img), cv2.IMREAD_COLOR)
resized_array = cv2.resize(img_array, (IMG_SIZE, IMG_SIZE))
data.append([resized_array, class_num])
except Exception as e:
print(e)
return data
data_train = get_data()
def append_data():
yaw_no = face_for_yawn()
data = get_data()
yaw_no.extend(data)
return np.array(yaw_no)
all_data = append_data()
# # separate label and features
X = []
y = []
for feature, labelss in all_data:
X.append(feature)
y.append(labelss)
X = np.array(X)
X = X.reshape(-1, 224, 224, 3)
from sklearn.preprocessing import LabelBinarizer
label_bin = LabelBinarizer()
y = label_bin.fit_transform(y)
y = np.array(y)
from sklearn.model_selection import train_test_split
seed = 42
test_size = 0.30
X_train, X_test, y_train, y_test = train_test_split(
X, y, random_state=seed, test_size=test_size
)
# # data augmentation and model
from keras.layers import Input, Lambda, Dense, Flatten, Conv2D, MaxPooling2D, Dropout
from keras.models import Model
from keras.models import Sequential
from keras.preprocessing.image import ImageDataGenerator
import tensorflow as tf
train_generator = ImageDataGenerator(
rescale=1 / 255, zoom_range=0.2, horizontal_flip=True, rotation_range=30
)
test_generator = ImageDataGenerator(rescale=1 / 255)
train_generator = train_generator.flow(np.array(X_train), y_train, shuffle=False)
test_generator = test_generator.flow(np.array(X_test), y_test, shuffle=False)
model = Sequential()
model.add(Conv2D(256, (3, 3), activation="relu", input_shape=X_train.shape[1:]))
model.add(MaxPooling2D(2, 2))
model.add(Conv2D(128, (3, 3), activation="relu"))
model.add(MaxPooling2D(2, 2))
model.add(Conv2D(64, (3, 3), activation="relu"))
model.add(MaxPooling2D(2, 2))
model.add(Conv2D(32, (3, 3), activation="relu"))
model.add(MaxPooling2D(2, 2))
model.add(Flatten())
model.add(Dropout(0.5))
model.add(Dense(64, activation="relu"))
model.add(Dense(4, activation="softmax"))
model.compile(loss="categorical_crossentropy", metrics=["accuracy"], optimizer="adam")
model.summary()
r = model.fit(
train_generator,
epochs=50,
validation_data=test_generator,
shuffle=True,
validation_steps=len(test_generator),
)
# loss
plt.figure(figsize=(4, 2))
plt.plot(r.history["loss"], label="train loss")
plt.plot(r.history["val_loss"], label="val loss")
plt.legend()
plt.show()
# accuracies
plt.figure(figsize=(4, 2))
plt.plot(r.history["accuracy"], label="train acc")
plt.plot(r.history["val_accuracy"], label="val acc")
plt.legend()
plt.show()
model.save("drowiness_new6.h5")
# # test data
prediction = model.predict(X_test)
classes_predicted = np.argmax(prediction, axis=1)
classes_predicted
# # evaluation metrics
labels_new = ["yawn", "no_yawn", "Closed", "Open"]
from sklearn.metrics import classification_report
print(
classification_report(
np.argmax(y_test, axis=1), classes_predicted, target_names=labels_new
)
)
# # predictions
# 0-yawn, 1-no_yawn, 2-Closed, 3-Open
model = tf.keras.models.load_model("./drowiness_new6.h5")
IMG_SIZE = 224
def prepare_data(filepath, face_cas="haarcascade_frontalface_default.xml"):
image_array = cv2.imread(filepath)
face_cascade = cv2.CascadeClassifier(cv2.data.haarcascades + face_cas)
faces = face_cascade.detectMultiScale(image_array, 1.3, 5)
for x, y, w, h in faces:
img = cv2.rectangle(image_array, (x, y), (x + w, y + h), (0, 255, 0), 2)
roi_color = img[y : y + h, x : x + w]
roi_color = roi_color / 255
resized_array = cv2.resize(roi_color, (IMG_SIZE, IMG_SIZE))
return resized_array.reshape(-1, IMG_SIZE, IMG_SIZE, 3)
def prediction_function(img_path):
ready_data = prepare_data(img_path)
prediction = model.predict(ready_data)
plt.imshow(plt.imread(img_path))
if np.argmax(prediction) == 2 or np.argmax(prediction) == 0:
print("DROWSY ALERT!!!!")
else:
print("DRIVER IS ACTIVE")
prediction_function("/kaggle/input/predict-img/predict_1.jpg")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 5GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import seaborn as sns
import matplotlib.pyplot as matplot
from matplotlib import pyplot as plt
from matplotlib.pyplot import show
from sklearn import metrics
from sklearn.model_selection import train_test_split
from scipy.stats import zscore
from sklearn.neighbors import KNeighborsClassifier
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import GridSearchCV
from mlxtend.feature_selection import SequentialFeatureSelector as sfs
pd.set_option("display.max_rows", None)
pd.set_option("display.max_columns", None)
train_df = pd.read_csv("/kaggle/input/titanic/train.csv")
sub_df = pd.read_csv("/kaggle/input/titanic/gender_submission.csv")
test_df = pd.read_csv("/kaggle/input/titanic/test.csv")
train_df.head()
train_df.dtypes
test_df.head()
print(train_df.shape)
print(test_df.shape)
# The training dataset has 891 records and test_df has 418 records. The test dataset has 1 column in short since the target column is excluded
# Checking for null values
train_df.isnull().sum()
test_df.isnull().sum()
# The column Cabin has more than 70% of the records with null values in both train and test dataset
# Imputing this column will not be helpful, so deleting this column
train_df.drop("Cabin", axis=1, inplace=True)
test_df.drop("Cabin", axis=1, inplace=True)
# The age and Embarked columns has null values in training dataset which needs to be imputed.
# Similarly Age and Fare dataset has null values.
train_df[train_df["Embarked"].isnull()]
# Both the records belong to same ticket, which means they are travelling together and we can see they were in class 1.
# imputing the column with most repeated value using mode.
train_df["Embarked"].fillna(
train_df[train_df["Pclass"] == 1]["Embarked"].mode()[0], inplace=True
)
train_df.sort_values(by="Ticket")
train_df[train_df["Ticket"] == "CA. 2343"]
train_df.sort_values(by="Age")
train_df[train_df["Age"].isnull()].sort_values(by="Ticket")
train_df[(train_df["Age"].isnull()) & (train_df["Parch"] != 0)]
train_df["Age"].fillna(
train_df["Age"].median(), inplace=True
) # imputing with mdeian as there are some outlier, since only few people have travelled whose age is close to eighty. using mean will not be suitable
test_df["Age"].fillna(
test_df["Age"].median(), inplace=True
) # imputing with mdeian as there are some outlier, since only few people have travelled whose age is close to eighty. using mean will not be suitable
# We can remove the columns id and Name as it will not be helpful for our analysis
train_df.drop(["Name"], axis=1, inplace=True)
test_df.drop(["Name"], axis=1, inplace=True)
train_df.describe().transpose()
# sns.pairplot(train_df,hue='Survived')
# Univariant analysis, Bivariant and multivariant analysis
total = float(len(train_df))
ax = sns.countplot(train_df["Survived"]) # for Seaborn version 0.7 and more
for p in ax.patches:
height = p.get_height()
ax.text(
p.get_x() + p.get_width() / 2.0,
height + 3,
"{:1.2f}".format((height / total) * 100),
ha="center",
)
show()
# The graph above shows only 38.38% of people have survided
total = float(len(train_df))
ax = sns.countplot(train_df["Pclass"]) # for Seaborn version 0.7 and more
for p in ax.patches:
height = p.get_height()
ax.text(
p.get_x() + p.get_width() / 2.0,
height + 3,
"{:1.2f}".format((height / total) * 100),
ha="center",
)
show()
# The graph above shows more than 50 percentage of the people have travelled in class 3
sns.distplot(train_df["Age"])
# The distribution plot shows that the majority of the people were in the age group 20 to 40.
total = float(len(train_df))
ax = sns.countplot(train_df["Sex"]) # for Seaborn version 0.7 and more
for p in ax.patches:
height = p.get_height()
ax.text(
p.get_x() + p.get_width() / 2.0,
height + 3,
"{:1.2f}".format((height / total) * 100),
ha="center",
)
show()
# In the total passenger 64.76% were male and 35.24% people were female.
total = float(len(train_df))
ax = sns.countplot(train_df["Embarked"]) # for Seaborn version 0.7 and more
for p in ax.patches:
height = p.get_height()
ax.text(
p.get_x() + p.get_width() / 2.0,
height + 3,
"{:1.2f}".format((height / total) * 100),
ha="center",
)
show()
# Most of the traveller(72.50%) have embarked at S
total = float(len(train_df))
ax = sns.countplot(
train_df["SibSp"] + train_df["Parch"]
) # for Seaborn version 0.7 and more
for p in ax.patches:
height = p.get_height()
ax.text(
p.get_x() + p.get_width() / 2.0,
height + 3,
"{:1.2f}".format((height / total) * 100),
ha="center",
)
show()
# 60% of the people have travelled alone. This cannot be exact number as mentione in the data description children who have travelled with their nanny Parch is mentioned as 0. But we can say that a majority of the people have travelled alone.
sns.distplot(train_df["Fare"])
# The fare column shows the same prize shows something interesting. We do have fare starting from 0 to 500. The highest probability
# for the reason for the fare to be zero is they might be the crew members of the ship and the highest fare might be due to large nummber of people travelling together, as the fare displayed is at the itinerary level.
train_df.head()
sns.countplot(x="Survived", data=train_df)
total = float(len(train_df))
ax = sns.countplot(
x="Survived", hue="Pclass", data=train_df
) # for Seaborn version 0.7 and more
for p in ax.patches:
height = p.get_height()
ax.text(
p.get_x() + p.get_width() / 2.0,
height + 3,
"{:1.2f}".format((height / total) * 100),
ha="center",
)
show()
# Overall only 38.38 percentage of the travellers survived and the percentage split with respect to Pclass is shown.
p_cont1 = pd.crosstab(train_df["Pclass"], train_df["Survived"], normalize="index") * 100
p_cont1 = p_cont1.reset_index()
p_cont1.rename(columns={0: "Dead", 1: "Survived"}, inplace=True)
listv = []
for var in train_df["Pclass"].unique():
listv.append(var)
plt.figure(figsize=(20, 6))
ax1 = plt.subplot(121)
g1 = sns.countplot(x="Pclass", data=train_df, order=listv, palette="Set1")
gt = g1.twinx()
gt = sns.pointplot(
x="Pclass", y="Survived", data=p_cont1, color="black", legend=False, order=listv
)
gt.set_ylabel("% of Survived", fontsize=12)
g1.set_title("Survival Rate Passenger class wise", fontsize=14)
g1.set_ylabel("Count", fontsize=12)
plt.show()
# The above graph shows, the survival rate is much higher in the class 1 travellers and very low for class 3
p_cont1 = pd.crosstab(train_df["Sex"], train_df["Survived"], normalize="index") * 100
p_cont1 = p_cont1.reset_index()
p_cont1.rename(columns={0: "Dead", 1: "Survived"}, inplace=True)
listv = []
for var in train_df["Sex"].unique():
listv.append(var)
plt.figure(figsize=(20, 6))
ax1 = plt.subplot(121)
g1 = sns.countplot(x="Sex", data=train_df, order=listv, palette="Set1")
gt = g1.twinx()
gt = sns.pointplot(
x="Sex", y="Survived", data=p_cont1, color="black", legend=False, order=listv
)
gt.set_ylabel("% of Survived", fontsize=12)
g1.set_title("Survival Rate Genderwise", fontsize=14)
g1.set_ylabel("Count", fontsize=12)
plt.show()
# The above graph shows that the number of male passengers were almost double but more than 70% of the female passengers survived whereas only 20 % of the men survived.
p_cont1 = (
pd.crosstab(train_df["Embarked"], train_df["Survived"], normalize="index") * 100
)
p_cont1 = p_cont1.reset_index()
p_cont1.rename(columns={0: "Dead", 1: "Survived"}, inplace=True)
listv = []
for var in train_df["Embarked"].unique():
listv.append(var)
plt.figure(figsize=(20, 6))
ax1 = plt.subplot(121)
g1 = sns.countplot(x="Embarked", data=train_df, order=listv, palette="Set1")
gt = g1.twinx()
gt = sns.pointplot(
x="Embarked", y="Survived", data=p_cont1, color="black", legend=False, order=listv
)
gt.set_ylabel("% of Survived", fontsize=12)
g1.set_title("Survival Rate based on Embarked", fontsize=14)
g1.set_ylabel("Count", fontsize=12)
plt.show()
train_df[train_df["Fare"] == 0]
# Interestingly for all the passengers who have 0 as fare has embarked at S, they are all male, all are travelling alone and none of them have survived. possibillly they could be the crew members. All the travller with ticket mentioned as Line are in Class 3. They could be lower level crew member
def personclassifier(x):
age, sex = x
if age <= 12:
return "Child"
if age > 12 and age <= 19:
return "Teenager"
if age > 19 and age <= 30:
return "Young Adult"
if age > 30 and age <= 45:
return "Mid aged Adult"
if age > 45:
return "Old Adults"
def singletraveller(x):
single = x
if single == 0:
return "Yes"
else:
return "No"
train_df["Single_Traveller"] = train_df["SibSp"] + train_df["Parch"]
train_df["Single_Traveller"] = train_df["Single_Traveller"].apply(singletraveller)
test_df["Single_Traveller"] = test_df["SibSp"] + test_df["Parch"]
test_df["Single_Traveller"] = test_df["Single_Traveller"].apply(singletraveller)
train_df["Person"] = train_df[["Age", "Sex"]].apply(personclassifier, axis=1)
test_df["Person"] = test_df[["Age", "Sex"]].apply(personclassifier, axis=1)
p_cont1 = pd.crosstab(train_df["Person"], train_df["Survived"], normalize="index") * 100
p_cont1 = p_cont1.reset_index()
p_cont1.rename(columns={0: "Dead", 1: "Survived"}, inplace=True)
listv = []
for var in train_df["Person"].unique():
listv.append(var)
plt.figure(figsize=(20, 6))
ax1 = plt.subplot(121)
g1 = sns.countplot(x="Person", data=train_df, order=listv, palette="Set1")
gt = g1.twinx()
gt = sns.pointplot(
x="Person", y="Survived", data=p_cont1, color="black", legend=False, order=listv
)
gt.set_ylabel("% of Survived", fontsize=12)
g1.set_title("Survival Rate Age Group wise", fontsize=14)
g1.set_ylabel("Count", fontsize=12)
plt.show()
# The above graph clearly shows that, young adults are the majority population and the survival rate is much lesser in that group and children having the highest survival rate.
p_cont1 = (
pd.crosstab(
[train_df["Person"], train_df["Sex"]], train_df["Survived"], normalize="index"
)
* 100
)
p_cont1
p_cont1 = (
pd.crosstab(
[train_df["Person"], train_df["Sex"]], train_df["Survived"], normalize="index"
)
* 100
)
p_cont1 = p_cont1.reset_index()
p_cont1.rename(columns={0: "Dead", 1: "Survived"}, inplace=True)
listv = []
for var in train_df["Person"].unique():
listv.append(var)
plt.figure(figsize=(20, 6))
ax1 = plt.subplot(121)
g1 = sns.countplot(x="Person", hue="Sex", data=train_df, order=listv, palette="Set1")
gt = g1.twinx()
gt = sns.pointplot(
x="Person", y="Survived", data=p_cont1, color="black", legend=False, order=listv
)
gt.set_ylabel("% of Survived", fontsize=12)
g1.set_title("Survival Rate By Age Group by Gender ", fontsize=14)
g1.set_ylabel("Count", fontsize=12)
plt.show()
# In every age group the count of male is higher whereas in teenager group alone female traveller are slightly higher. Breaking the survival rate by age group by sex, children seem to have a farely same survival rate in both the gender, where as in all age group female seem to have hiher survival rate where in the teenager male having the least survival rate (less than 10%) and females in the Old Adult group have the higher survival rate with 86.66%.
p_cont1 = (
pd.crosstab(
[train_df["Person"], train_df["Pclass"]],
train_df["Survived"],
normalize="index",
)
* 100
)
p_cont1 = p_cont1.reset_index()
p_cont1.rename(columns={0: "Dead", 1: "Survived"}, inplace=True)
listv = []
for var in train_df["Person"].unique():
listv.append(var)
plt.figure(figsize=(20, 6))
ax1 = plt.subplot(121)
g1 = sns.countplot(x="Person", hue="Pclass", data=train_df, order=listv, palette="Set1")
gt = g1.twinx()
gt = sns.pointplot(
x="Person", y="Survived", data=p_cont1, color="black", legend=False, order=listv
)
gt.set_ylabel("% of Survived", fontsize=12)
g1.set_title("Survival Rate by age group by class", fontsize=14)
g1.set_ylabel("Count", fontsize=12)
plt.show()
p_cont1 = (
pd.crosstab(
[train_df["Person"], train_df["Pclass"]],
train_df["Survived"],
normalize="index",
)
* 100
)
p_cont1
# Children in class 2 have 100% survival rate and old adults in class 3 have the least survival rate with just 5.55%
test_df.isnull().sum()
test_df["Fare"].fillna(test_df["Fare"].median(), inplace=True)
p_cont1 = (
pd.crosstab(train_df["Single_Traveller"], train_df["Survived"], normalize="index")
* 100
)
p_cont1 = p_cont1.reset_index()
p_cont1.rename(columns={0: "Dead", 1: "Survived"}, inplace=True)
listv = []
for var in train_df["Single_Traveller"].unique():
listv.append(var)
plt.figure(figsize=(20, 6))
ax1 = plt.subplot(121)
g1 = sns.countplot(x="Single_Traveller", data=train_df, order=listv, palette="Set1")
gt = g1.twinx()
gt = sns.pointplot(
x="Single_Traveller",
y="Survived",
data=p_cont1,
color="black",
legend=False,
order=listv,
)
gt.set_ylabel("% of Survived", fontsize=12)
g1.set_title("Survival Rate by Single Traveller", fontsize=14)
g1.set_ylabel("Count", fontsize=12)
plt.show()
# The count of single traveller seems to be high,but only 30 percentage of the single traveller survived where as more than 50% of the traveller who travelled with someone have survived.
p_cont1 = (
pd.crosstab(
[train_df["Single_Traveller"], train_df["Sex"]],
train_df["Survived"],
normalize="index",
)
* 100
)
p_cont1 = p_cont1.reset_index()
p_cont1.rename(columns={0: "Dead", 1: "Survived"}, inplace=True)
listv = []
for var in train_df["Single_Traveller"].unique():
listv.append(var)
plt.figure(figsize=(20, 6))
ax1 = plt.subplot(121)
g1 = sns.countplot(
x="Single_Traveller", hue="Sex", data=train_df, order=listv, palette="Set1"
)
gt = g1.twinx()
gt = sns.pointplot(
x="Single_Traveller",
y="Survived",
data=p_cont1,
color="black",
legend=False,
order=listv,
)
gt.set_ylabel("% of Survived", fontsize=12)
g1.set_title("Survival Rate by Single traveller by gender", fontsize=14)
g1.set_ylabel("Count", fontsize=12)
plt.show()
# Though the number of single traveller female is very less almost 80% of them have survived, whereas only 15% of male single traveller have survived.
p_cont1 = (
pd.crosstab(
[train_df["Single_Traveller"], train_df["Sex"]],
train_df["Survived"],
normalize="index",
)
* 100
)
p_cont1
# The nummber female single traveller are very less.
p_cont1 = (
pd.crosstab(
[train_df["Pclass"], train_df["Single_Traveller"]],
train_df["Survived"],
normalize="index",
)
* 100
)
p_cont1 = p_cont1.reset_index()
p_cont1.rename(columns={0: "Dead", 1: "Survived"}, inplace=True)
listv = []
for var in train_df["Pclass"].unique():
listv.append(var)
plt.figure(figsize=(20, 6))
ax1 = plt.subplot(121)
g1 = sns.countplot(
x="Pclass", hue="Single_Traveller", data=train_df, order=listv, palette="Set1"
)
gt = g1.twinx()
gt = sns.pointplot(
x="Pclass", y="Survived", data=p_cont1, color="black", legend=False, order=listv
)
gt.set_ylabel("% of Survived", fontsize=12)
g1.set_title("Survival Rate by class by single traveller", fontsize=14)
g1.set_ylabel("Count", fontsize=12)
plt.show()
p_cont1 = (
pd.crosstab(
[train_df["Pclass"], train_df["Single_Traveller"]],
train_df["Survived"],
normalize="index",
)
* 100
)
p_cont1
# By all means single traveller has less survival rate
# Creating the model without any outlier treatment.
train_df.head()
train_df.drop("Ticket", axis=1, inplace=True)
test_df.drop("Ticket", axis=1, inplace=True)
# train_df.drop(['SibSp','Parch'],axis=1,inplace=True)
# test_df.drop(['SibSp','Parch'],axis=1,inplace=True)
col = []
for c in train_df.columns:
if train_df[c].dtypes == "object":
col.append(c)
train_df_dummies = pd.get_dummies(train_df, columns=col, drop_first=True)
corr_mat = train_df_dummies.corr()
train_df_dummies.corr()
# Getting the columns that are having multi collinearity
# Creating a dataframe with correlated column, the correlation value and the source column to which it is correlated
# Filtering only those that are correlated more than 96%
multi_col_df = pd.DataFrame(columns=["corr_col", "corr_val", "source_col"])
for i in corr_mat:
temp_df = pd.DataFrame(corr_mat[corr_mat[i] > 0.9][i])
temp_df = temp_df.reset_index()
temp_df["source_col"] = i
temp_df.columns = ["corr_col", "corr_val", "source_col"]
multi_col_df = pd.concat((multi_col_df, temp_df), axis=0)
multi_col_df
X = train_df_dummies.drop(["Survived", "PassengerId"], axis=1)
X_id = train_df_dummies["PassengerId"]
y = train_df_dummies["Survived"]
X_trainval, X_test, y_trainval, y_test = train_test_split(
X, y, test_size=0.20, random_state=1
)
X_train, X_val, y_train, y_val = train_test_split(
X_trainval, y_trainval, test_size=0.20, random_state=1
)
X_trainval_z = X_trainval.apply(zscore)
X_train_z = X_train.apply(zscore)
X_val_z = X_val.apply(zscore)
X_test_z = X_test.apply(zscore)
X_z = X.apply(zscore)
# Grid Search based on Max_features, Min_Samples_Split and Max_Depth
param_grid = [
{
"n_neighbors": list(range(1, 50)),
"algorithm": ["auto", "ball_tree", "kd_tree", "brute"],
"leaf_size": [10, 15, 20, 30],
"n_jobs": [-1],
"weights": ["uniform", "distance"],
}
]
import multiprocessing
gs = GridSearchCV(
estimator=KNeighborsClassifier(),
param_grid=param_grid,
scoring="accuracy",
n_jobs=multiprocessing.cpu_count(),
cv=3,
)
gs.fit(X_train_z, y_train)
gs.best_estimator_
gs.best_score_
knn_clfr = KNeighborsClassifier(
algorithm="auto",
leaf_size=10,
metric="minkowski",
metric_params=None,
n_jobs=-1,
n_neighbors=6,
p=2,
weights="uniform",
)
sfs1 = sfs(knn_clfr, k_features=8, forward=False, scoring="accuracy", cv=10, n_jobs=-1)
sfs1 = sfs(knn_clfr, k_features=8, forward=False, scoring="accuracy", cv=10, n_jobs=-1)
sfs1 = sfs1.fit(X_train_z.values, y_train.values)
X_train_z.head()
sfs1.get_metric_dict()
sfs1 = sfs(knn_clfr, k_features=8, forward=False, scoring="accuracy", cv=10, n_jobs=-1)
sfs1 = sfs1.fit(X_train_z.values, y_train.values)
sfs1.get_metric_dict()
columnList = list(X_train_z.columns)
feat_cols = list(sfs1.k_feature_idx_)
print(feat_cols)
sfs1 = sfs(knn_clfr, k_features=8, forward=False, scoring="accuracy", cv=10, n_jobs=-1)
sfs1 = sfs1.fit(X_train_z.values, y_train.values)
sfs1.get_metric_dict()
columnList = list(X_train_z.columns)
feat_cols = list(sfs1.k_feature_idx_)
print(feat_cols)
subsetColumnList = [columnList[i] for i in feat_cols]
print(subsetColumnList)
train_df_dummies.dtypes
train_df_dummies_knn = train_df_dummies.drop(
["Age", "Parch", "Embarked_Q", "Single_Traveller_Yes"], axis=1
)
train_df_dummies_knn.head()
X_knn = train_df_dummies_knn.drop(["PassengerId", "Survived"], axis=1)
X_id_knn = train_df_dummies_knn["PassengerId"]
y_knn = train_df_dummies_knn["Survived"]
from imblearn.combine import SMOTETomek
smk = SMOTETomek(random_state=1)
X_res, y_res = smk.fit_sample(X_knn, y_knn)
X_trainval_knn, X_test_knn, y_trainval_knn, y_test_knn = train_test_split(
X_res, y_res, test_size=0.20, random_state=1
)
X_train_knn, X_val_knn, y_train_knn, y_val_knn = train_test_split(
X_trainval_knn, y_trainval_knn, test_size=0.20, random_state=1
)
X_trainval_z_knn = X_trainval_knn.apply(zscore)
X_train_z_knn = X_train_knn.apply(zscore)
X_val_z_knn = X_val_knn.apply(zscore)
X_test_z_knn = X_test_knn.apply(zscore)
X_z_knn = X_res.apply(zscore)
import multiprocessing
gs = GridSearchCV(
estimator=KNeighborsClassifier(),
param_grid=param_grid,
scoring="accuracy",
n_jobs=multiprocessing.cpu_count(),
cv=10,
)
gs.fit(X_train_z_knn, y_train_knn)
gs.best_estimator_
gs.best_score_
knnclfr = KNeighborsClassifier(
algorithm="auto",
leaf_size=10,
metric="minkowski",
metric_params=None,
n_jobs=-1,
n_neighbors=6,
p=2,
weights="uniform",
)
knnclfr.fit(X_train_z_knn, y_train_knn)
y_predict_knn = knnclfr.predict(X_val_z_knn)
print(knnclfr.score(X_train_z_knn, y_train_knn))
print(knnclfr.score(X_val_z_knn, y_val_knn))
print(metrics.classification_report(y_val_knn, y_predict_knn))
print(metrics.confusion_matrix(y_val_knn, y_predict_knn))
knnclfr.fit(X_trainval_z_knn, y_trainval_knn)
y_predict_knn = knnclfr.predict(X_test_z_knn)
print(knnclfr.score(X_trainval_z_knn, y_trainval_knn))
print(knnclfr.score(X_test_z_knn, y_test_knn))
print(metrics.classification_report(y_test_knn, y_predict_knn))
print(metrics.confusion_matrix(y_test_knn, y_predict_knn))
knnclfr.fit(X_z_knn, y_res)
test_df.head()
test_df["Fare"].fillna(test_df["Fare"].median(), inplace=True)
col = []
for c in test_df.columns:
if test_df[c].dtypes == "object":
col.append(c)
test_df_dummies = pd.get_dummies(test_df, columns=col, drop_first=True)
test_df_dummies.head()
test_df_dummies_knn = test_df_dummies.drop(
["Age", "Parch", "Embarked_Q", "Single_Traveller_Yes"], axis=1, inplace=True
)
test_df_dummies_knn
X_test_knn = test_df_dummies.drop(["PassengerId"], axis=1)
X_test_id_knn = test_df_dummies["PassengerId"]
X_test_knn = X_test_knn.apply(zscore)
y_predict = knnclfr.predict(X_test_knn)
final_pred_df = pd.DataFrame(y_predict)
final_pred_df.columns = ["Survived"]
X_test_id_knn = pd.DataFrame(X_test_id_knn)
final_pred = X_test_id_knn.merge(final_pred_df, left_index=True, right_index=True)
final_pred.shape
final_pred.to_csv("csv_to_submit2602-2.csv", index=False)
from sklearn import preprocessing
lab_enc = preprocessing.LabelEncoder()
lab_enc = preprocessing.LabelEncoder()
col = []
for c in train_df.columns:
if train_df[c].dtypes == "object":
train_df[c] = lab_enc.fit_transform(train_df[c])
print("Column {} has been encoded".format(c))
col = []
for c in test_df.columns:
if test_df[c].dtypes == "object":
test_df[c] = lab_enc.fit_transform(test_df[c])
print("Column {} has been encoded".format(c))
X = train_df.drop(["Survived", "PassengerId"], axis=1)
X_id = train_df["PassengerId"]
y = train_df["Survived"]
from imblearn.combine import SMOTETomek
smk = SMOTETomek(random_state=1)
X_res, y_res = smk.fit_sample(X, y)
X_trainval, X_test, y_trainval, y_test = train_test_split(
X_res, y_res, test_size=0.20, random_state=1
)
X_train, X_val, y_train, y_val = train_test_split(
X_trainval, y_trainval, test_size=0.20, random_state=1
)
X_trainval_z = X_trainval.apply(zscore)
X_train_z = X_train.apply(zscore)
X_val_z = X_val.apply(zscore)
X_test_z = X_test.apply(zscore)
X_z = X_res.apply(zscore)
# Grid Search based on Max_features, Min_Samples_Split and Max_Depth
param_grid = [
{
"n_neighbors": list(range(1, 50)),
"algorithm": ["auto", "ball_tree", "kd_tree", "brute"],
"leaf_size": [10, 15, 20, 30],
"n_jobs": [-1],
"weights": ["uniform", "distance"],
}
]
import multiprocessing
gs = GridSearchCV(
estimator=KNeighborsClassifier(),
param_grid=param_grid,
scoring="accuracy",
n_jobs=multiprocessing.cpu_count(),
cv=3,
)
gs.fit(X_train_z, y_train)
gs.best_estimator_
gs.best_score_
knn_clfr = KNeighborsClassifier(
algorithm="auto",
leaf_size=10,
metric="minkowski",
metric_params=None,
n_jobs=-1,
n_neighbors=5,
p=2,
weights="uniform",
)
train_df.shape
sfs1 = sfs(knn_clfr, k_features=6, forward=True, scoring="accuracy", cv=10, n_jobs=-1)
sfs1 = sfs1.fit(X_train_z.values, y_train.values)
sfs1.get_metric_dict()
columnList = list(X_train_z.columns)
feat_cols = list(sfs1.k_feature_idx_)
print(feat_cols)
subsetColumnList = [columnList[i] for i in feat_cols]
print(subsetColumnList)
train_df.head(1)
train_df_bkp = train_df.copy()
test_df_bkp = test_df.copy()
X_knn = train_df[["Pclass", "Sex", "Age", "Fare", "Single_Traveller", "Person"]]
X_id_knn = train_df["PassengerId"]
y_knn = train_df["Survived"]
from imblearn.combine import SMOTETomek
smk = SMOTETomek(random_state=1)
X_res, y_res = smk.fit_sample(X_knn, y_knn)
X_trainval_knn, X_test_knn, y_trainval_knn, y_test_knn = train_test_split(
X_res, y_res, test_size=0.20, random_state=1
)
X_train_knn, X_val_knn, y_train_knn, y_val_knn = train_test_split(
X_trainval_knn, y_trainval_knn, test_size=0.20, random_state=1
)
X_trainval_z_knn = X_trainval_knn.apply(zscore)
X_train_z_knn = X_train_knn.apply(zscore)
X_val_z_knn = X_val_knn.apply(zscore)
X_test_z_knn = X_test_knn.apply(zscore)
X_z_knn = X_res.apply(zscore)
X_train_z_knn.shape
gs = GridSearchCV(
estimator=KNeighborsClassifier(),
param_grid=param_grid,
scoring="accuracy",
n_jobs=multiprocessing.cpu_count(),
cv=10,
)
gs.fit(X_train_z_knn, y_train_knn)
gs.best_estimator_
gs.best_score_
knn_clfr = KNeighborsClassifier(
algorithm="auto",
leaf_size=10,
metric="minkowski",
metric_params=None,
n_jobs=-1,
n_neighbors=12,
p=2,
weights="distance",
)
knn_clfr.fit(X_train_z_knn, y_train_knn)
y_predict = knn_clfr.predict(X_val_z_knn)
print(knn_clfr.score(X_train_z_knn, y_train_knn))
print(knn_clfr.score(X_val_z_knn, y_val_knn))
print(metrics.classification_report(y_val_knn, y_predict))
print(metrics.confusion_matrix(y_val_knn, y_predict))
knn_clfr.fit(X_trainval_knn, y_trainval_knn)
y_predict = knn_clfr.predict(X_test_z_knn)
print(knn_clfr.score(X_trainval_knn, y_trainval_knn))
print(knn_clfr.score(X_test_z_knn, y_test_knn))
print(metrics.classification_report(y_test_knn, y_predict))
print(metrics.confusion_matrix(y_test_knn, y_predict))
from sklearn.model_selection import RandomizedSearchCV
# Number of trees in random forest
n_estimators = [10, 50, 150, 175, 200, 250] # returns evenly spaced 10 numbers
# Number of features to consider at every split
max_features = ["auto"]
# Maximum number of levels in tree
max_depth = [
6,
7,
12,
15,
18,
22,
25,
] # returns evenly spaced numbers can be changed to any
# max_depth.append(None)
# Method of selecting samples for training each tree
bootstrap = [True, False]
subsample = [0.7, 0.8, 0.9]
# Create the random grid
random_grid = {
"n_estimators": n_estimators,
"max_features": max_features,
"max_depth": max_depth,
"bootstrap": bootstrap,
"subsample": subsample,
}
print(random_grid)
# Use the random grid to search for best hyperparameters
# Random search of parameters, using 3 fold cross validation,
# search across 100 different combinations, and use all available cores
from sklearn.model_selection import GridSearchCV
from xgboost import XGBClassifier
rf_random = GridSearchCV(
estimator=XGBClassifier(),
param_grid=random_grid,
cv=3,
verbose=0,
n_jobs=1,
return_train_score=True,
)
# Fit the random search model
rf_random.fit(X_train_z, y_train)
# currently running
rf_random.best_estimator_
rf_random.best_score_
model = XGBClassifier(
base_score=0.5,
booster=None,
bootstrap=True,
colsample_bylevel=1,
colsample_bynode=1,
colsample_bytree=1,
gamma=0,
gpu_id=-1,
importance_type="gain",
interaction_constraints=None,
learning_rate=0.300000012,
max_delta_step=0,
max_depth=7,
max_features="auto",
min_child_weight=1,
monotone_constraints=None,
n_estimators=10,
n_jobs=0,
num_parallel_tree=1,
objective="binary:logistic",
random_state=0,
reg_alpha=0,
reg_lambda=1,
scale_pos_weight=1,
subsample=0.9,
tree_method=None,
validate_parameters=False,
verbosity=None,
)
from sklearn.model_selection import KFold
from sklearn.model_selection import cross_val_score
results = cross_val_score(model, X_train_z, y_train, cv=10)
print(results)
print("Accuracy: %.3f%% (%.3f%%)" % (results.mean() * 100.0, results.std() * 100.0))
sfs1 = sfs(model, k_features=8, forward=True, scoring="accuracy", cv=10, n_jobs=-1)
sfs1 = sfs1.fit(X_train_z.values, y_train.values)
sfs1.get_metric_dict()
columnList = list(X_train_z.columns)
feat_cols = list(sfs1.k_feature_idx_)
print(feat_cols)
subsetColumnList = [columnList[i] for i in feat_cols]
print(subsetColumnList)
X = train_df_dummies[
[
"Pclass",
"Age",
"SibSp",
"Fare",
"Sex_male",
"Person_Mid aged Adult",
"Person_Teenager",
"Person_Young Adult",
]
]
x_id = train_df_dummies["PassengerId"]
y = train_df_dummies["Survived"]
from imblearn.combine import SMOTETomek
smk = SMOTETomek(random_state=1)
X_res, y_res = smk.fit_sample(X, y)
X_trainval, X_test, y_trainval, y_test = train_test_split(
X_res, y_res, test_size=0.20, random_state=1
)
X_train, X_val, y_train, y_val = train_test_split(
X_trainval, y_trainval, test_size=0.20, random_state=1
)
X_trainval_z = X_trainval.apply(zscore)
X_train_z = X_train.apply(zscore)
X_val_z = X_val.apply(zscore)
X_test_z = X_test.apply(zscore)
X_z = X_res.apply(zscore)
model.fit(X_train_z, y_train)
y_predict = model.predict(X_val_z)
print(model.score(X_train_z, y_train))
print(model.score(X_val_z, y_val))
print(metrics.classification_report(y_val, y_predict))
print(metrics.confusion_matrix(y_val, y_predict))
model.fit(X_trainval_z, y_trainval)
y_predict = model.predict(X_test)
print(model.score(X_trainval_z, y_trainval))
print(model.score(X_test_z, y_test))
print(metrics.classification_report(y_test, y_predict))
print(metrics.confusion_matrix(y_test, y_predict))
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Sales Anomaly Detection
# This notebook starts from very basic exploratory data analysis (EDA) with data visualization and analysis. Besides the visual elimination, for this example we will also use the isolation forest algorithm frok SKLearn, you can access the details from following link : https://scikit-learn.org/stable/modules/outlier_detection.html#isolation-forest
# TABLE OF CONTENTS
#
# * [1. IMPORTING LIBRARIES](#1)
#
# * [2. LOADING DATA](#2)
#
# * [3. EDA & Visualizations](#3)
#
# * [4. Anomaly Detection with Isolation Forest](#4)
#
# * [5. Result Set and Evaluation](#5)
#
# * [6. Conclusion](#6)
#
# * [7. END](#7)
# # 1. Importing Libraries
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.ensemble import IsolationForest
import warnings
warnings.filterwarnings("ignore")
#
# # 2. Loading Data
#
# Now, we can load the data set and start to analyze it.
df_original = df = pd.read_csv("/kaggle/input/retail-data-set/file_out2.csv")
df.head()
#
# # 3. EDA and Visualizations
#
df.describe()
# Let's understand the ditribution of data among columns with number of unique records in each column:
df.nunique()
df.nunique().plot.bar()
# Visually it is about 2 times for the first row (row id) to invoice id. We can say, the average number of rows per invoice is about 2. Date column is about 1/20 of total records, which means for each day we have about 20 rows or 10 invoices. Finally the total number of customers is 507, which is 1/60 of the total number of rows, so we can understand for each customer we have about 60 items sold, about 3 different dates we have invoices.
plt.scatter(range(df.shape[0]), np.sort(df["TotalSales"].values))
plt.xlabel("index")
plt.ylabel("Sales")
plt.title("Sales distribution")
sns.despine()
# for all the sales, the distribution shows the 'Total Sales' amount is close to each other, but for about 5% of the sales there are huge amounts. Also 1 point is obviously outlier in the sales. This might be the first outlier / **anomaly** we have found. Let's dig it mor:
df.sort_values("TotalSales", ascending=False).head(10)
# here, we can see the first 2 rows are very high and there is no smooth connection to the rest of the dataset. Also the quantity column shows us these two rows are impossible with unit prices. Let's create an anomaly result set and start collecting those rows to the result data set:
# we create a df_anomaly dataframe for the anomalies and starting to collect
# anomalies with 2 rows in the total sales column.
df_anomaly = df.sort_values("TotalSales", ascending=False).head(2)
df_anomaly.head()
# Let's also remove the anomalies from the original dataset:
list(df.sort_values("TotalSales", ascending=False).head(2).index)
df = df.drop(list(df.sort_values("TotalSales", ascending=False).head(2).index))
df.sort_values("TotalSales", ascending=False).head(10)
# Let's see the unit price distribution and check for anomalies:
df["UnitPrice"] = df["TotalSales"] / df["Quantity"]
df.sort_values("UnitPrice", ascending=False).head(10)
plt.scatter(range(df.shape[0]), np.sort(df["UnitPrice"].values))
plt.xlabel("index")
plt.ylabel("UnitPrice")
plt.title("UnitPrice Sales distribution")
sns.despine()
# We can also find a another sales record with a very high unit price, which might be another anomaly. Let's add this record to the anomalies also:
df_anomaly = df_anomaly.append(df.sort_values("UnitPrice", ascending=False).head(1))
df_anomaly.head()
df = df.drop(list(df.sort_values("UnitPrice", ascending=False).head(1).index))
df.head()
sns.distplot(df["TotalSales"])
plt.title("Distribution of Sales")
sns.despine()
sns.distplot(df["UnitPrice"])
plt.title("Distribution of UnitPrice")
sns.despine()
#
# # 4. Anomaly Detection with Isolation Forest
#
isolation_forest = IsolationForest(n_estimators=100)
isolation_forest.fit(df["TotalSales"].values.reshape(-1, 1))
xx = np.linspace(df["TotalSales"].min(), df["TotalSales"].max(), len(df)).reshape(-1, 1)
anomaly_score = isolation_forest.decision_function(xx)
outlier = isolation_forest.predict(xx)
plt.figure(figsize=(10, 4))
plt.plot(xx, anomaly_score, label="anomaly score")
plt.fill_between(
xx.T[0],
np.min(anomaly_score),
np.max(anomaly_score),
where=outlier == -1,
color="r",
alpha=0.4,
label="outlier region",
)
plt.legend()
plt.ylabel("anomaly score")
plt.xlabel("Sales")
plt.show()
df["AnomalyScore"] = anomaly_score
df.sort_values("AnomalyScore", ascending=False).head()
# Let's get the top 10 from the Total Sales anomalies:
df_anomaly = pd.concat(
[df_anomaly, df.sort_values("AnomalyScore", ascending=True).head(10)]
)
df_anomaly.head()
df = df.drop(list(df.sort_values("AnomalyScore", ascending=False).head(10).index))
df.head()
df[df["UnitPrice"].isna()].head()
df["UnitPrice"].isna().sum()
# There are 158 rows with NaN values in UnitPrice column. The reason is Quantity of these rows are 0. So we can get rid of these rows:
df = df.dropna()
df.head()
isolation_forest = IsolationForest(n_estimators=100)
isolation_forest.fit(df["UnitPrice"].values.reshape(-1, 1))
xx = np.linspace(df["UnitPrice"].min(), df["UnitPrice"].max(), len(df)).reshape(-1, 1)
anomaly_score = isolation_forest.decision_function(xx)
outlier = isolation_forest.predict(xx)
plt.figure(figsize=(10, 4))
plt.plot(xx, anomaly_score, label="anomaly score")
plt.fill_between(
xx.T[0],
np.min(anomaly_score),
np.max(anomaly_score),
where=outlier == -1,
color="r",
alpha=0.4,
label="outlier region",
)
plt.legend()
plt.ylabel("anomaly score")
plt.xlabel("Unit Price")
plt.show()
df["AnomalyScore"] = anomaly_score
df.sort_values("AnomalyScore", ascending=True).head()
#
# # 5. Result Set and OutputFile
#
df_anomaly = pd.concat(
[df_anomaly, df.sort_values("AnomalyScore", ascending=False).head(10)]
)
df_anomaly
df_anomaly.to_csv("anomalies.csv")
|
# <div style="padding:20px;
# color:white;
# margin:10;
# font-size:200%;
# text-align:center;
# display:fill;
# border-radius:5px;
# background-color:#c2a5ca;
# overflow:hidden;
# font-weight:700">BP & CNN Example
# # Catalogue
# 1.Data Description
# 2.Data Procession
# 3.Training(Using DNN/BP)
# 4.Training(Using CNN)
# 5.Summary
# Change the code theme
#!wget https://raw.githubusercontent.com/VvanWindring/DataAnalysis/main/CSS.css?token=GHSAT0AAAAAAB7MBASUTL7DTBW6NBHXUPYQZAQI3RA -O CSS.css -q
from IPython.core.display import HTML
with open("/kaggle/input/my-theme/CSS.css", "r") as file:
custom_css = file.read()
HTML(custom_css)
# # Part 1
# <div style="padding:20px;
# color:white;
# margin:10;
# font-size:170%;
# text-align:left;
# display:fill;
# border-radius:5px;
# background-color:#c2a5ca;
# overflow:hidden;
# font-weight:700">| 1.Data Description
#
import numpy as np
import pandas as pd
pd.set_option("display.max_rows", None)
pd.set_option("display.max_columns", None)
import matplotlib.pyplot as plt
import seaborn as sns
from scipy import stats
import math
origin = pd.read_csv("/kaggle/input/cluster-nn-data/penguins.csv")
origin.head()
# Randomly selected 10 samples from origin data for display
features = origin[origin.columns[0:4]] # =features = origin.iloc[:,0:4]
target = origin["Species"]
features.sample(10)
# summary table function
def summary(df):
print(f"data shape: {df.shape}")
summ = pd.DataFrame(df.dtypes, columns=["data type"])
summ["#missing"] = df.isnull().sum().values * 100
summ["%missing"] = df.isnull().sum().values / len(df)
summ["#unique"] = df.nunique().values
summ["skewness"] = df.skew() # 偏度系数,abs>1为严重左/右偏
desc = pd.DataFrame(df.describe(include="all").transpose())
summ["min"] = desc["min"].values
summ["max"] = desc["max"].values
summ["first value"] = df.loc[0].values
summ["second value"] = df.loc[1].values
summ["third value"] = df.loc[2].values
return summ
summary(origin)
fea = list(origin.columns.values)
n_bins = 100
histplot_hyperparams = {"kde": True, "alpha": 0.4, "stat": "percent", "bins": n_bins}
columns = fea
n_cols = 4
n_rows = math.ceil(len(columns) / n_cols)
fig, ax = plt.subplots(n_rows, n_cols, figsize=(20, n_rows * 4))
ax = ax.flatten()
for i, column in enumerate(columns):
plot_axes = [ax[i]]
sns.kdeplot(origin[column], label="origin", ax=ax[i], color="red")
# titles
ax[i].set_title(f"{column} Distribution")
ax[i].set_xlabel(None)
# remove axes to show only one at the end
plot_axes = [ax[i]]
handles = []
labels = []
for plot_ax in plot_axes:
handles += plot_ax.get_legend_handles_labels()[0]
labels += plot_ax.get_legend_handles_labels()[1]
plot_ax.legend().remove()
for i in range(i + 1, len(ax)):
ax[i].axis("off")
fig.suptitle(
f"Numerical Feature Distributions\n\n\n",
ha="center",
fontweight="bold",
fontsize=25,
)
fig.legend(
handles, labels, loc="upper center", bbox_to_anchor=(0.5, 0.96), fontsize=25, ncol=3
)
plt.tight_layout()
# describe numeric feature correlation
def df_corr(df):
plt.figure(figsize=(df.shape[1], df.shape[1]))
color = "RdYlGn"
mask = np.zeros_like(df.corr())
mask[np.triu_indices_from(mask)] = True
sns.heatmap(df.corr(), mask=mask, annot=True, linewidth=0.2, cmap=color)
df_corr(origin)
# # Part 2
# <div style="padding:20px;
# color:white;
# margin:10;
# font-size:170%;
# text-align:left;
# display:fill;
# border-radius:5px;
# background-color:#c2a5ca;
# overflow:hidden;
# font-weight:700">| 2.Data Procession
#
from imblearn.over_sampling import SMOTE
class data_procession:
def __init__(self, X=None, Y=None):
self.X = X
self.Y = Y
print("Origin X Shape:", self.X.shape)
print("Origin Y Shape:", self.Y.shape)
def process(self):
def gaussian_normalize(data):
mean = np.mean(data)
std = np.std(data)
normalized_data = (data - mean) / std
return normalized_data
# 填补缺失值
self.X = self.X.fillna(method="ffill")
print("Step1:已填补缺失值")
# 高斯归一
for col in self.X.columns:
self.X[col] = gaussian_normalize(self.X[col])
print("Step2:已将数据正态化")
# 样本数过少,采用超采样
sm = SMOTE(random_state=39)
self.X, self.Y = sm.fit_resample(self.X, self.Y)
print("Step3:已完成数据超采样")
print("Processed X Shape:", self.X.shape)
print("Processed Y Shape:", self.Y.shape)
return self.X, self.Y
X, Y = data_procession(features, target).process()
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.metrics import (
confusion_matrix,
auc,
roc_curve,
f1_score,
roc_auc_score,
accuracy_score,
)
from sklearn.model_selection import train_test_split
from sklearn.tree import DecisionTreeClassifier as DTC
from sklearn.model_selection import GridSearchCV
from sklearn.ensemble import RandomForestClassifier as RFC
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
data_train = pd.read_csv("/kaggle/input/assignment1-data/train.csv/train.csv")
data_test = pd.read_csv("/kaggle/input/assignment1-data/test.csv/test.csv")
data_train
data_train.info()
data_train.head(20)
# Check for Null values
data_train.isnull().sum()
data_train.describe()
# Checking which column range has the features
data_train.columns[1:]
data_train.columns[2:]
# Will decide if data needs modification after initial tests on models
# Initial data setup
SEED = 42
y = data_train["Bankrupt"]
X = data_train.drop(["Bankrupt", "id"], axis=1)
features = X.columns
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=SEED
)
# Makes sure that the features we are using is the same, e.g excluding both id and bankrupt columns
X_test_unlabeled = data_test[features]
# Initial Decision Tree test with default parameters
dt_test = DTC()
dt_test.fit(X_train, y_train)
dt_test_y_pred = dt_test.predict(X_test)
dt_test_roc_auc = roc_auc_score(y_test, dt_test_y_pred)
dt_test_accuracy_score = accuracy_score(y_test, dt_test_y_pred)
dt_test_f1_score = f1_score(y_test, dt_test_y_pred)
print("ROC_AUC: %f" % dt_test_roc_auc)
print("Accuracy_Score: %f" % dt_test_accuracy_score)
print("F1_Score: %f" % dt_test_f1_score)
# Initial Random Forest test with default parameters
rf_test = RFC()
rf_test.fit(X_train, y_train)
rf_test_y_pred = rf_test.predict(X_test)
rf_test_roc_auc = roc_auc_score(y_test, rf_test_y_pred)
rf_test_accuracy_score = accuracy_score(y_test, rf_test_y_pred)
rf_test_f1_score = f1_score(y_test, rf_test_y_pred)
print("ROC_AUC: %f" % rf_test_roc_auc)
print("Accuracy_Score: %f" % rf_test_accuracy_score)
print("F1_Score: %f" % rf_test_f1_score)
# ROC score and f1 scores are low. Lets try modifying the data by moving outliers using IQR
# Lets look at the data before modifying
X.describe()
# Lets use robust scaler to deal with outliers and scale the data using IQR
from sklearn.preprocessing import RobustScaler
rb = RobustScaler().fit_transform(X)
# Lets look at the data now
rb.shape
rb
# Lets tests our models now
X_train, X_test, y_train, y_test = train_test_split(
rb, y, test_size=0.2, random_state=SEED
)
# Decision tree
dt_test = DTC()
dt_test.fit(X_train, y_train)
dt_test_y_pred = dt_test.predict(X_test)
dt_test_roc_auc = roc_auc_score(y_test, dt_test_y_pred)
dt_test_accuracy_score = accuracy_score(y_test, dt_test_y_pred)
dt_test_f1_score = f1_score(y_test, dt_test_y_pred)
print("ROC_AUC: %f" % dt_test_roc_auc)
print("Accuracy_Score: %f" % dt_test_accuracy_score)
print("F1_Score: %f" % dt_test_f1_score)
# Random Forest
rf_test = RFC()
rf_test.fit(X_train, y_train)
rf_test_y_pred = rf_test.predict(X_test)
rf_test_roc_auc = roc_auc_score(y_test, rf_test_y_pred)
rf_test_accuracy_score = accuracy_score(y_test, rf_test_y_pred)
rf_test_f1_score = f1_score(y_test, rf_test_y_pred)
print("ROC_AUC: %f" % rf_test_roc_auc)
print("Accuracy_Score: %f" % rf_test_accuracy_score)
print("F1_Score: %f" % rf_test_f1_score)
# * Modifying data didn't improve the scorces, lets try adjusting the hyperperameters
# * Used grid search to find the best hyperparameters. Took a while bc of processing time.
# Grid search using decision tree estimator
params_dt = {
"max_depth": np.arange(1, 10),
"min_samples_leaf": [0.06, 0.07, 0.08, 0.09, 0.10, 0.11, 0.12, 0.14],
}
dt = DTC(criterion="gini", random_state=SEED)
grid_dt = GridSearchCV(
estimator=dt, param_grid=params_dt, scoring="roc_auc", cv=5, n_jobs=-1
)
grid_dt.fit(X_train, y_train)
best_model_dt = grid_dt.best_estimator_
best_model_dt.score(X_test, y_test)
y_pred_proba_dt = best_model_dt.predict_proba(X_test)[:, 1]
grid_dt.best_score_
roc_auc_dt = roc_auc_score(y_test, y_pred_proba_dt)
roc_auc_dt
# Grid search using random forest estimator
params_rf = {
"n_estimators": [100, 120, 150],
"max_depth": [6, 7, 8],
"min_samples_leaf": [0.0025, 0.005, 0.1],
"max_features": ["log2", "sqrt"],
}
rf = RFC(criterion="entropy")
# rf_base= RFC()
# rf_base.fit(X_train, y_train)
# rf_base_prediction = rf_base.predict_proba(X_test)[:,1]
# rf_base_prediction
# rf_base_roc_auc = roc_auc_score(y_test, rf_base_prediction)
# rf_base_roc_auc
grid_rf = GridSearchCV(
estimator=rf, param_grid=params_rf, cv=5, scoring="roc_auc", verbose=1, n_jobs=-1
)
grid_rf.fit(X_train, y_train)
best_model_rf = grid_rf.best_estimator_
grid_rf.best_params_
grid_rf.best_score_
y_pred_proba_rf = best_model_rf.predict_proba(X_test)[:, 1]
roc_auc_rf = roc_auc_score(y_test, y_pred_proba_rf)
roc_auc_rf
y_test
y_pred_proba_rf
y_pred_proba_rf
y_pred_proba_dt
fpr, tpr, thresholds = roc_curve(y_test, y_pred_proba_rf)
roc_auc_rf = auc(fpr, tpr)
plt.title("ROC Curve for RF")
plt.plot([0, 1], [0, 1], "k--")
plt.plot(fpr, tpr, label="AUC = %0.10f" % roc_auc_rf)
plt.legend(loc="center right")
plt.xlabel("False Positive Rate")
plt.ylabel("True Positive Rate")
plt.show()
fpr, tpr, thresholds = roc_curve(y_test, y_pred_proba_dt)
roc_auc_dt = auc(fpr, tpr)
plt.title("ROC Curve for DT")
plt.plot([0, 1], [0, 1], "k--")
plt.plot(fpr, tpr, label="AUC = %0.10f" % roc_auc_dt)
plt.legend(loc="center right")
plt.xlabel("False Positive Rate")
plt.ylabel("True Positive Rate")
plt.show()
# * Both models the same? Feel like I'm doing something wrong.
# * Figured it out, mistake in using a single variable :upside-down-smile:
# * Edit: its fine now
# Random Forest passed the benchmark ROC_score, so we use that to generate our file prediction
file_prediction = best_model_rf.predict_proba(X_test_unlabeled)[:, 1]
file = pd.DataFrame(file_prediction, columns=["Bankrupt"])
file.insert(0, "id", range(0, len(file)))
file.to_csv("my_submission3.csv", index=False)
file
|
# Import the general libraries
import os
import pandas as pd
import sklearn
import matplotlib.pyplot as plt
# Import all specific modules
from sklearn.decomposition import PCA
from sklearn.decomposition import IncrementalPCA
from sklearn.linear_model import LogisticRegression
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
# Verify the dataset path
os.listdir("../input/heart-disease-uci")
# Import the dataset
df_heart = pd.read_csv("../input/heart-disease-uci/heart.csv")
# Show the dataset
df_heart.head()
# Save the dataset without target row
df_heart_features = df_heart.drop(["target"], axis=1)
df_heart_features.head()
# Create a df just with target row
df_target = df_heart["target"]
df_target.head()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pathlib
p_dir = pathlib.Path("/kaggle/input/bike-sharing-demand")
train = pd.read_csv(p_dir / "train.csv")
test = pd.read_csv(p_dir / "test.csv")
submission = pd.read_csv(p_dir / "sampleSubmission.csv")
# # 1. Feature Engineering
# ## Feature Selection (피처 선택)
# ### 이상치 제거
train = train[train["weather"] != 4]
# ### 데이터 합치기
all_data = pd.concat([train, test], ignore_index=True)
all_data
# ### 파생 피처 추가
# https://steadiness-193.tistory.com/227
# ### datetime을 object에서 datetime 타입으로 변환(feat. 송석리 선생님)
# train['datetime'] = pd.to_datetime(train['datetime'])
# train['year'] = train['datetime'].dt.year
# train['month'] = train['datetime'].dt.month
# train['day'] = train['datetime'].dt.day
# train['hour'] = train['datetime'].dt.hour
# train['minute'] = train['datetime'].dt.minute
# train['second'] = train['datetime'].dt.second
# train['weekday'] = train['datetime'].dt.dayofweek
# train.info()
from datetime import datetime
all_data["datetime"] = pd.to_datetime(all_data["datetime"])
type(all_data.iloc[0]["datetime"])
all_data.iloc[0]["datetime"].date()
all_data["date"] = all_data["datetime"].dt.date
all_data.head()
all_data["year"] = all_data["datetime"].dt.year
all_data["month"] = all_data["datetime"].dt.month
all_data["hour"] = all_data["datetime"].dt.hour
all_data["weekday"] = all_data["datetime"].dt.dayofweek
all_data.sample(n=10)
# ### 필요 없는 피처 제거
drop_features = ["casual", "registered", "datetime", "date", "windspeed", "month"]
all_data.drop(drop_features, axis=1, inplace=True)
all_data
# ### 데이터 다시 나누기
X_train = all_data[~pd.isnull(all_data["count"])]
X_test = all_data[pd.isnull(all_data["count"])]
X_train.head(3)
X_test.head(3)
y = X_train["count"]
X_train = X_train.drop(["count"], axis=1)
X_test = X_test.drop(["count"], axis=1)
X_train.head(3)
# # 2. 평가지표 계산 함수 작성
import numpy as np
def rmsle(y_true, y_pred, convertExp=True):
if convertExp:
y_true = np.exp(y_true)
y_pred = np.exp(y_pred)
log_true = np.nan_to_num(np.log1p(y_true))
log_pred = np.nan_to_num(np.log1p(y_pred))
output = np.sqrt(np.mean(np.power(log_true - log_pred, 2)))
return output
# # 3. 모델 훈련
from sklearn.linear_model import LinearRegression
lr = LinearRegression()
log_y = np.log(y) # 타깃값 로그 변환
lr.fit(X_train, log_y)
# # 4. 모델 성능 검증
preds = lr.predict(X_train)
res = rmsle(log_y, preds, True)
print(f"선형 회귀의 RMSLE 값: {res:.4f}")
lr_preds_log = lr.predict(X_test) # 데스트 데이터로 예측
lr_preds = np.exp(lr_preds_log) # 지수 변환
submission["count"] = lr_preds # 예측값으로 Submission의 count값 변경
submission.to_csv("submission_lr_model.csv", index=False) # 파일로 저장
lr_preds_log
lr_preds
X_test
submission
|
os.listdir("/kaggle/input/chest-xray-pneumonia/chest_xray/train/")
import cv2
import numpy as np
import os
import tensorflow as tf
import cv2
from tensorflow import keras
from keras.models import Sequential
from keras import layers
from keras.preprocessing.image import ImageDataGenerator as IDG
data_train = "/kaggle/input/chest-xray-pneumonia/chest_xray/train"
data_val = "/kaggle/input/chest-xray-pneumonia/chest_xray/val"
# PARAMS
BATCH_SIZE = 128
IMG_SIZE = (224, 224)
train_datagen = IDG()
val_datagen = IDG()
train_gen = train_datagen.flow_from_directory(
data_train,
target_size=IMG_SIZE,
batch_size=BATCH_SIZE,
class_mode="binary",
shuffle=True,
)
valid_gen = train_datagen.flow_from_directory(
data_val,
target_size=IMG_SIZE,
batch_size=BATCH_SIZE,
class_mode="binary",
shuffle=False,
)
from sklearn.preprocessing import LabelEncoder
# Instantiate a LabelEncoder object
label_encoder = LabelEncoder()
# Fit the label encoder to the training set labels
label_encoder.fit(y_train)
# Transform the training, validation, and test set labels
y_train = label_encoder.transform(y_train)
y_val = label_encoder.transform(y_val)
y_test = label_encoder.transform(y_test)
# Print the integer-encoded labels for the first 10 images in the training set
print(y_train[:10])
# Normalization
X_train = X_train / 255.0
X_test = X_test / 255.0
X_val = X_val / 255.0
# visualize sample images
import matplotlib.pyplot as plt
# Plot some sample images from the training set
fig, axes = plt.subplots(nrows=3, ncols=3, figsize=(8, 8))
random_array = np.random.randint(low=0, high=len(X_train), size=9)
print(random_array)
for i, ax in enumerate(axes.flat):
ax.imshow(X_train[random_array[i]])
ax.set_title("Label: {}".format(y_train[random_array[i]]))
plt.tight_layout()
plt.show()
# basic statistics
print("Training set statistics:")
print("Number of images: ", len(X_train))
unique_labels, label_counts = np.unique(y_train, return_counts=True)
print(
"Class distribution: {}: {}\n{}: {}".format(
unique_labels[0], label_counts[0], unique_labels[1], label_counts[1]
)
)
print("Image shape: ", X_train[0].shape)
print("Pixel value range: [{}, {}]".format(np.min(X_train), np.max(X_train)))
print("Mean pixel value: ", np.mean(X_train))
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
from tensorflow.keras.preprocessing.image import ImageDataGenerator
# from efficientnet.tfkeras import EfficientNetB0
from tensorflow.keras.applications import EfficientNetV2B0
num_classes = 2
model = keras.Sequential(
[
layers.Conv2D(128, (3, 3), activation="relu", input_shape=(224, 224, 3)),
layers.MaxPool2D(2),
layers.Conv2D(128, (3, 3), activation="relu"),
layers.MaxPool2D(pool_size=(2, 2)),
layers.Dense(16),
layers.Flatten(),
layers.Dense(1, activation="sigmoid"),
]
)
model.compile(optimizer="adam", loss="binary_crossentropy", metrics=["accuracy"])
history = model.fit(train_gen, validation_data=valid_gen, epochs=15, batch_size=32)
base_model = EfficientNetB0(
weights="imagenet", include_top=False, input_shape=(224, 224, 3)
)
num_classes = 2
for layer in base_model.layers:
layer.trainable = False
x = base_model.output
x = tf.keras.layers.GlobalAveragePooling2D()(x)
x = tf.keras.layers.Dense(128, activation="relu")(x)
predictions = tf.keras.layers.Dense(num_classes, activation="softmax")(x)
model = tf.keras.models.Model(inputs=base_model.input, outputs=predictions)
model.compile(optimizer="adam", loss="categorical_crossentropy", metrics=["accuracy"])
history = model.fit(
X_train, y_train, batch_size=32, epochs=10, validation_data=(X_val, y_val)
)
score = model_final.evaluate(X_test, y_test, verbose=0)
print("Test loss:", score[0])
print("Test accuracy:", score[1])
|
# ## 21기 AI/Bigdata 아카데미 자연어처리 과목의 실습을 위한 견본 코드입니다.
# Kaggle 링크: https://www.kaggle.com/competitions/aibigdata21test/
# 하단의 코드 블럭을 실행하여 데이터셋을 불러옵니다.
#
# 하단의 코드에서 주석(#)을 해제하고, Colab 인스턴스에 transformers 와 datasets 라이브러리를 설치합니다. PyTorch는 Colab 인스턴스에 이미 설치되어 있습니다.
# 설치된 Transformers lib의 버전이 최소 4.11.0 이상인지 확인합니다.
import transformers
print(transformers.__version__)
# CUDA GPU가 사용 가능한지 확인합니다.
import torch
print(torch.cuda.is_available())
device = "cuda" if torch.cuda.is_available() else "cpu"
# Huggingface Dataset 레포지토리에서 GLUE-sst2 데이터를 로드합니다.
from datasets import load_dataset
train_dataset = load_dataset("glue", "sst2", split="train")
val_dataset = load_dataset("glue", "sst2", split="validation")
# train_dataset과 val_dataset의 크기를 확인합니다.
print(len(train_dataset))
print(len(val_dataset))
# train_dataset과 val_dataset의 첫 번째 샘플을 출력합니다.
print(train_dataset[0])
print(val_dataset[0])
# Casual Language Modeling 학습을 위해 일정 길이로 나눠진 Text Chunk (텍스트 조각)이 필요합니다. 이를 위해서, 데이터셋의 모든 Text를 Tokenize 한 후 하나로 합칩니다.
# 전부 다 이어붙인 Text를 미리 정해진 길이로 나눕니다. 편집 완료된 데이터의 예시는 아래와 같습니다:
# ```
# part of text 1
# ```
# or
# ```
# end of text 1 [BOS_TOKEN] beginning of text 2
# ```
# 텍스트 데이터 별로 학습시키는 것도 가능하지만, 여기서는 batch 관리를 편하게 하기 위해 일렬로 나열한 후 일정 길이 마다 잘라서 사용합니다.
# 두 경우 모두 각 Token에 대해 다음 순번의 Token을 예측하는 것을 Objective Function으로 설정합니다.
# 예시 코드에서는 '[bert-base-uncased](https://huggingface.co/bert-base-uncased)' 모델을 사용합니다.
# [Repository](https://huggingface.co/models?filter=causal-lm)에서 다양한 모델을 찾을 수 있습니다. 적절한 모델을 찾아 시도해보세요.
model_checkpoint = "bert-base-uncased"
# tokenizer와 model을 초기화 합니다.
from transformers import AutoModelForCausalLM, AutoTokenizer
tokenizer = AutoTokenizer.from_pretrained(model_checkpoint, use_fast=True)
model = AutoModelForCausalLM.from_pretrained(model_checkpoint).to(device)
# 이제 불러온 모든 Text에 대해 Tokenizer를 적용할 수 있습니다. Huggingface Datasets 라이브러리의 [`map`](https://huggingface.co/docs/datasets/package_reference/main_classes.html#datasets.Dataset.map) 메서드를 통해 데이터에 일괄적으로 전처리 함수를 적용할 수 있습니다.
# 여기서는 tokenizer로 입력된 Text를 Tokenize하는 전처리 함수를 정의합니다:
def tokenize_function(examples):
new_data = [s.replace(".", "") for s in examples["sentence"]]
return tokenizer(new_data)
# 그리고, 해당 전처리 함수를 `datasets` object의 모든 데이터에 적용합니다. `batched=True`로 설정하고 process를 4로 설정함으로써 처리 속도를 향상시킬 수 있습니다.
# 이 과정에서 에러가 발생할 경우 process 수를 줄이거나 `batched=False`로 설정해서 다시 해보세요.
# Tokenize 이후에 제출 데이터 제작을 위한 guid를 제외한 텍스트는 필요하지 않으므로, `remove_columns=['sentence','idx','label']`옵션을 통해 해당 Column을 제거합니다.
tokenized_train_dataset = train_dataset.map(
tokenize_function,
batched=True,
num_proc=1,
remove_columns=["sentence", "idx", "label"],
)
tokenized_val_dataset = val_dataset.map(
tokenize_function,
batched=True,
num_proc=1,
remove_columns=["sentence", "idx", "label"],
)
val_dataset_token_count_list = [len(t["input_ids"]) for t in tokenized_val_dataset]
len(val_dataset_token_count_list)
tokenized_train_dataset[0]
tokenized_val_dataset[0]
# Tokenize를 끝냈으면, 텍스트를 모두 하나로 이어 이를 `block_size` 만큼의 조각으로 나눕니다. 이를 위해 `batched=True`를 활성화 한 `map` method를 한번 더 활용합니다.
# `batched=True` 옵션은 입력과 출력 데이터의 개수를 다르게 지정할 수 있습니다. 이를 통해 새로운 batch dataset을 생성할 수도 있습니다.
# `block_size`를 `tokenizer.model_max_length`로 설정하는 것이 일반적이나(BERT의 경우 512), GPU 자원의 한계로 인해 이를 모두 활용하지 못할 수도 있습니다. (Colab 노트북에서는 `tokenizer.model_max_length`로 설정하면 메모리가 부족할 가능성이 매우 높습니다.)
# GPU 메모리 이슈가 발생할 경우, `block_size`를 128이나 더 작은 값으로 설정하고 다시 해보세요.
# block_size = tokenizer.model_max_length
block_size = 16
# Batch 생성 함수를 아래와 같이 작성합니다:
def group_texts(examples):
# Concatenate all texts.
concatenated_examples = {k: sum(examples[k], []) for k in examples.keys()}
total_length = len(concatenated_examples["input_ids"])
# We drop the small remainder, we could add padding if the model supported it instead of this drop, you can
# customize this part to your needs.
total_length = (total_length // block_size) * block_size
# Split by chunks of max_len.
result = {
k: [t[i : i + block_size] for i in range(0, total_length, block_size)]
for k, t in concatenated_examples.items()
}
result["labels"] = result["input_ids"].copy()
return result
# `map` 메서드의 `batch_size` 파라미터의 기본값은 1,000입니다. 즉 1,000 데이터마다 정해진 `block_size`에 맞지 않는 조그마한 데이터가 버려집니다.
# 필요에 따라 `batch_size`를 변경하는 것이 가능합니다.
#
lm_train_dataset = tokenized_train_dataset.map(
group_texts,
batched=True,
batch_size=1000,
num_proc=1,
)
lm_val_dataset = tokenized_val_dataset.map(
group_texts,
batched=True,
batch_size=1000,
num_proc=1,
)
# HyperParameter를 초기화 합니다. 여기서 값을 다양하게 바꾸거나, 다양한 세팅을 시도해 보세요.
# 하단의 코드 블럭에 명시된 파라미터를 컨트롤 하여 실험하는 것으로도 충분하긴 하나, 다른 파라미터를 도입하는 것도 가능합니다.
# 가능한 파라미터의 목록은 [API 문서](https://huggingface.co/docs/transformers/main_classes/trainer#transformers.TrainingArguments) 를 참조하실 수 있습니다.
from transformers import Trainer, TrainingArguments
model_name = model_checkpoint.split("/")[-1]
training_args = TrainingArguments(
f"{model_name}-finetuned-gluesst2",
evaluation_strategy="epoch",
learning_rate=1e-6,
weight_decay=0.1,
num_train_epochs=1,
per_device_train_batch_size=8,
report_to="none",
push_to_hub=False,
)
trainer = Trainer(
model=model,
args=training_args,
train_dataset=lm_train_dataset,
eval_dataset=lm_val_dataset,
)
import os
os.environ["WANDB_WATCH"] = "false"
trainer.train()
# 학습이 끝난 Language Model을 평가합니다.
import math
eval_results = trainer.evaluate()
print(f"Perplexity: {math.exp(eval_results['eval_loss']):.10f}")
# Kaggle System에 제출할 파일을 생성합니다.
predicted = []
from tqdm import tqdm
torch.cuda.empty_cache()
with torch.no_grad():
dataset_count = len(tokenized_val_dataset)
sentence_count = 0
idx = 1
predicted_cache = []
for datum in tqdm(tokenized_val_dataset):
input_vec = (
torch.Tensor(datum["input_ids"]).to("cuda", dtype=torch.long).unsqueeze(0)
)
output = model(input_ids=input_vec)
logits = output.logits[0]
len_vec = len(input_vec[0])
for i in range(len_vec - 1):
l = torch.softmax(logits[i], dim=-1)[datum["input_ids"][i + 1]].item()
predicted_cache.append(l)
if idx < dataset_count:
predicted_cache.append(
torch.softmax(logits[-1], dim=-1)[
tokenized_val_dataset[idx]["input_ids"][0]
].item()
)
idx += 1
predicted.append(sum(predicted_cache) / len(predicted_cache))
# label.append(tokenizer.decode(datum['input_ids'],skip_special_tokens=True))
# input_vec = torch.Tensor(datum['input_ids'][:-2]).to('cuda',dtype=torch.int32).unsqueeze(0)
# generated = model.generate(input_vec,max_length=len(input_vec[0])+1,
# no_repeat_ngram_size=2)
# predicted_word.append(tokenizer.decode(generated[0],skip_special_tokens=True))
# export generated text to .csv file
import pandas
prw_df = pandas.DataFrame({"Expected": predicted})
prw_df.to_csv("./result.csv", index_label="index")
|
isl_dir = "/kaggle/input/isl-csltr/ISL_CSLRT_Corpus/ISL_CSLRT_Corpus"
import os
import numpy as np
import pandas as pd
import torch
import tqdm.notebook as tqdm
isl_dir = "/kaggle/input/isl-csltr/ISL_CSLRT_Corpus"
csv_dir = "/kaggle/input/isl-csltr/ISL_CSLRT_Corpus/ISL_CSLRT_Corpus/corpus_csv_files"
frame_dir = (
"/kaggle/input/isl-csltr/ISL_CSLRT_Corpus/ISL_CSLRT_Corpus/Frames_Sentence_Level"
)
# for sentence in os.listdir(frame_dir):
# sentence_fdir = os.path.join(frame_dir, sentence)
# frames = []
# for annotator in os.listdir(sentence_fdir):
# annotator_fdir = os.path.join(sentence_fdir, annotator)
# for fr in os.listdir(annotator_fdir):
# if fr.endswith('jpg'):
# frames.append(fr)
device = "cuda" if torch.cuda.is_available else "cpu"
print(device)
sign_glosses = pd.read_csv(os.path.join(csv_dir, "ISL Corpus sign glosses.csv"))
corpus_details = pd.read_excel(os.path.join(csv_dir, "ISL_CSLRT_Corpus details.xlsx"))
frame_details = pd.read_excel(
os.path.join(csv_dir, "ISL_CSLRT_Corpus_frame_details.xlsx")
)
vid_files = [
os.path.join(isl_dir, corpus_details["File location"][i])
for i in range(len(corpus_details))
]
frame_files = [
os.path.join(isl_dir, frame_details["Frames path"][i]).replace("\\", "/")
for i in range(len(corpus_details))
]
from torch.utils.data import DataLoader
from torchvision.datasets import ImageFolder
from torchvision.transforms import Compose, CenterCrop, ToTensor, Normalize
from torch.utils.data import random_split
def get_frame_dataloader(frame_dir, batch_size=32):
# Define a transform to be applied to each frame
transform = Compose(
[
CenterCrop((112, 112)),
ToTensor(),
Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),
]
)
# Create an ImageFolder dataset from the list of frame paths
dataset = ImageFolder(frame_dir, transform=transform)
train_size = int(0.8 * len(dataset))
validation_size = len(dataset) - train_size
train_dataset, validation_dataset = random_split(
dataset, [train_size, validation_size]
)
# Create a DataLoader object to iterate over the dataset in batches
train_dataloader = DataLoader(train_dataset, batch_size=batch_size, shuffle=True)
val_dataloader = DataLoader(validation_dataset, batch_size=batch_size, shuffle=True)
return train_dataloader, val_dataloader
# dataloader = get_frame_dataloader(frame_dir)
def train_step(model, dataloader, criterion, optimizer, device):
model.train()
total_loss = 0.0
total = 0
total_loss, total_correct = 0.0, 0
with tqdm.tqdm(dataloader) as t:
for i, (inputs, targets) in enumerate(dataloader):
inputs, targets = inputs.to(device), targets.to(device)
optimizer.zero_grad()
outputs = model(inputs)
loss = criterion(outputs, targets)
loss.backward()
optimizer.step()
total_loss += loss.item()
total += inputs.size(0)
_, predictions = torch.max(outputs, 1)
total_correct += torch.sum(predictions == targets).item()
# print(predictions, targets, total_correct,total)
t.set_postfix({"Loss": total_loss / (i + 1), "acc": total_correct / total})
return total_loss / len(dataloader.dataset)
def evaluate(model, dataloader, criterion, device):
model.eval()
total_loss, total_correct = 0.0, 0
total = 0
with torch.no_grad():
with tqdm.tqdm(dataloader) as t:
for i, (inputs, targets) in enumerate(dataloader):
inputs, targets = inputs.to(device), targets.to(device)
outputs = model(inputs)
loss = criterion(outputs, targets)
total_loss += loss.item()
total += inputs.size(0)
_, predictions = torch.max(outputs, 1)
total_correct += torch.sum(predictions == targets).item()
t.set_postfix(
{"Loss": total_loss / (i + 1), "acc": total_correct / total}
)
avg_loss = total_loss / len(dataloader.dataset)
avg_accuracy = total_correct / len(dataloader.dataset)
return avg_loss, avg_accuracy
import torch
import torch.nn as nn
import torchvision.models as models
import torchvision.datasets as datasets
import torchvision.transforms as transforms
import torch.optim as optim
import torch.nn as nn
class MSTCN(nn.Module):
def __init__(self, input_channels=3, num_classes=97):
super(MSTCN, self).__init__()
# Define the feature extractor
self.features = nn.Sequential(
nn.Conv2d(input_channels, 16, kernel_size=3, stride=1, padding=1),
nn.ReLU(inplace=True),
nn.MaxPool2d(kernel_size=2, stride=2),
nn.Conv2d(16, 32, kernel_size=3, stride=1, padding=1),
nn.ReLU(inplace=True),
nn.MaxPool2d(kernel_size=2, stride=2),
nn.Conv2d(32, 64, kernel_size=3, stride=1, padding=1),
nn.ReLU(inplace=True),
nn.MaxPool2d(kernel_size=2, stride=2),
nn.Conv2d(64, 128, kernel_size=3, stride=1, padding=1),
nn.ReLU(inplace=True),
nn.MaxPool2d(kernel_size=2, stride=2),
)
# Define the classifier
self.classifier = nn.Sequential(
nn.Linear(128 * 7 * 7, 512),
nn.ReLU(inplace=True),
nn.Linear(512, num_classes),
)
def forward(self, x):
x = self.features(x)
x = x.view(x.size(0), -1)
x = self.classifier(x)
return x
# Load the pre-trained MSTCN model on UCF101 dataset
pretrained_model = models.video.r2plus1d_18(pretrained=True)
# Create a new instance of the MSTCN model with the same input and output dimensions as the pretrained model
lr = 0.001
input_channels = 3 # RGB video input
num_classes = 97 # number of action classes in UCF101 dataset
num_epochs = 10
mstcn_model = MSTCN(input_channels, num_classes).to(device)
# Initialize the weights of the MSTCN model with the pre-trained weights of the R(2+1)D model
mstcn_model_dict = mstcn_model.state_dict()
pretrained_dict = pretrained_model.state_dict()
for name, param in pretrained_dict.items():
if name.startswith("module."):
name = name[7:] # remove 'module.' prefix from the keys
if name in mstcn_model_dict:
mstcn_model_dict[name].copy_(param)
# Freeze the weights of the R(2+1)D layers in the MSTCN model
for name, param in mstcn_model.named_parameters():
if (
name.startswith("layers")
or name.startswith("pool_layers")
or name.startswith("fc_layers")
):
param.requires_grad = False
# Move the model to the GPU if available
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
test_loader = get_frame_dataloader(
frame_dir, 32
) # function that returns a PyTorch DataLoader object for the UCF101 test set
# Initialize the model, optimizer, and loss function
optimizer = optim.Adam(mstcn_model.parameters(), lr=lr)
criterion = nn.CrossEntropyLoss()
train_loader, val_loader = get_frame_dataloader(frame_dir, 32)
# Training loop
for epoch in range(num_epochs):
train_loss = train_step(mstcn_model, train_loader, criterion, optimizer, device)
val_loss, val_acc = evaluate(mstcn_model, val_loader, criterion, device)
print(
f"Epoch {epoch + 1}/{num_epochs} - Training Loss: {train_loss:.4f} - Validation Loss: {val_loss:.4f} - Validation Accuracy: {val_acc:.4f}"
)
from datetime import datetime
save_dir = "/kaggle/working/"
time = datetime.now().strftime("%d-%H:%M")
PATH = os.path.join(save_dir, f"16apr-{epoch}-{time}-.pt")
torch.save(model, PATH)
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
img = mpimg.imread(frames_file[0])
imgplot = plt.imshow(img)
plt.show()
import pandas as pd
pd.read_csv("ISL_CSLRT_Corpus/corpus_csv_files/ISL Corpus sign glosses.csv")
pd.read_excel("ISL_CSLRT_Corpus/corpus_csv_files/ISL_CSLRT_Corpus_word_details.xlsx")
|
import os
import numpy as np
import pandas as pd
import random
train = pd.read_csv("/kaggle/input/playground-series-s3e12/train.csv")
additional = pd.read_csv(
"/kaggle/input/kidney-stone-prediction-based-on-urine-analysis/kindey stone urine analysis.csv"
)
test = pd.read_csv("/kaggle/input/playground-series-s3e12/test.csv")
ypo = pd.read_csv("/kaggle/input/playground-series-s3e12/sample_submission.csv")
train = pd.concat([train, additional], sort=False).reset_index(drop=True)
train = train.drop_duplicates().reset_index(drop=True)
train.shape
from dataprep.eda import create_report
report = create_report(train)
report.show()
report = create_report(test)
report.show()
target = "target"
to_remove = ["id"]
X = train.drop(to_remove, axis=1)
X = X.drop(target, axis=1)
X_pred = test.drop(to_remove, axis=1)
y = train[target]
labels = list(X.columns)
for item in labels:
upper = np.quantile(X[item], 0.99)
lower = np.quantile(X[item], 0.01)
X[item] = list(
map(lambda x: upper if x > upper else (lower if x < lower else x), X[item])
)
X_pred[item] = list(
map(lambda x: upper if x > upper else (lower if x < lower else x), X_pred[item])
)
# from sklearn.preprocessing import RobustScaler
# scaler=RobustScaler()
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
scaler.fit(X)
X1 = pd.DataFrame(scaler.transform(X), columns=X.columns)
X_pred1 = pd.DataFrame(scaler.transform(X_pred), columns=X_pred.columns)
from flaml import AutoML
automl = AutoML()
from sklearn.linear_model import LogisticRegression
# from sklearn.model_selection import RepeatedStratifiedKFold
# rskf = RepeatedStratifiedKFold(n_splits=10, n_repeats=5)
automl_settings = {
"time_budget": 2500,
"metric": "roc_auc",
"task": "classification",
"estimator_list": [
"xgboost",
"xgb_limitdepth",
"lgbm",
"rf",
"extra_tree",
], # ,"extra_tree","catboost","rf","lrl1"
"ensemble": {
"final_estimator": LogisticRegression(),
"passthrough": True,
},
"eval_method": "cv",
# "split_type":rskf
"n_splits": 5,
}
automl.fit(X_train=X1, y_train=y, **automl_settings)
ypo[target] = automl.predict_proba(X_pred1)[:, 1]
ypo
ypo.to_csv("./play_12_flaml_nf_07.csv", index=False)
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
pd.set_option("display.max_columns", None)
pd.set_option("display.max_rows", None)
df = pd.read_csv("/content/smartphone_cleaned_v5.csv")
df.shape
df.head()
df.info()
df.isnull().sum()
df.head()
# brand_name
# plot a graph of top 5 brands
df["brand_name"].value_counts().head(10).plot(kind="bar")
# pie chart
df["brand_name"].value_counts().plot(kind="pie", autopct="%0.1f%%")
df["brand_name"].isnull().sum()
# model
df["model"].nunique()
# price
df["price"].describe()
sns.displot(kind="hist", data=df, x="price", kde=True)
df["price"].skew()
sns.boxplot(df["price"])
df[df["price"] > 250000]
df["price"].isnull().sum()
df["rating"].describe()
sns.displot(kind="hist", data=df, x="rating", kde=True)
df["rating"].skew()
sns.boxplot(df["rating"])
df["rating"].isnull().sum() / 980
df.head()
# has_5g
df["has_5g"].value_counts().plot(kind="pie", autopct="%0.1f%%")
# has_nfc
df["has_nfc"].value_counts().plot(kind="pie", autopct="%0.1f%%")
# has_ir_blaster
df["has_ir_blaster"].value_counts().plot(kind="pie", autopct="%0.1f%%")
df[df["has_ir_blaster"] == True]["brand_name"].value_counts()
df["processor_brand"].value_counts().plot(kind="pie", autopct="%0.1f%%")
df["num_cores"].value_counts().plot(kind="pie", autopct="%0.1f%%")
df["fast_charging_available"].value_counts().plot(kind="pie", autopct="%0.1f%%")
df["ram_capacity"].value_counts().plot(kind="pie", autopct="%0.1f%%")
df["internal_memory"].value_counts().plot(kind="pie", autopct="%0.1f%%")
df["refresh_rate"].value_counts().plot(kind="pie", autopct="%0.1f%%")
df["refresh_rate"].value_counts()
(df["num_rear_cameras"] + df["num_front_cameras"]).value_counts().plot(
kind="pie", autopct="%0.1f%%"
)
df["os"].value_counts().plot(kind="pie", autopct="%0.1f%%")
# extended_memory_available
df["extended_memory_available"].value_counts().plot(kind="pie", autopct="%0.1f%%")
df["extended_upto"].value_counts().plot(kind="pie", autopct="%0.1f%%")
def plot_graphs(column_name):
sns.displot(kind="hist", kde=True, data=df, x=column_name, label=column_name)
sns.catplot(kind="box", data=df, x=column_name)
num_columns = (
df.select_dtypes(include=["float64", "int64"])
.iloc[:, [3, 4, 6, 9, 13, 14, 16]]
.columns
)
for col in num_columns:
plot_graphs(col)
df.head()
plt.figure(figsize=(20, 10))
sns.barplot(data=df, x="brand_name", y="price")
plt.xticks(rotation="vertical")
x = df.groupby("brand_name").count()["model"]
temp_df = df[df["brand_name"].isin(x[x > 10].index)]
plt.figure(figsize=(15, 8))
sns.barplot(data=temp_df, x="brand_name", y="price")
plt.xticks(rotation="vertical")
df.head()
sns.scatterplot(data=df, x="rating", y="price")
sns.barplot(data=temp_df, x="has_5g", y="price", estimator=np.median)
sns.pointplot(data=temp_df, x="has_nfc", y="price", estimator=np.median)
sns.barplot(data=temp_df, x="has_ir_blaster", y="price", estimator=np.median)
sns.barplot(data=temp_df, x="processor_brand", y="price", estimator=np.median)
plt.xticks(rotation="vertical")
sns.barplot(data=temp_df, x="num_cores", y="price", estimator=np.median)
plt.xticks(rotation="vertical")
pd.crosstab(df["num_cores"], df["os"])
sns.scatterplot(data=df, x="processor_speed", y="price")
sns.scatterplot(data=df, x="screen_size", y="price")
df.corr()["price"]
df.isnull().sum()
df.corr()["rating"]
# knn imputer
df.shape
x_df = df.select_dtypes(include=["int64", "float64"]).drop(columns="price")
from sklearn.impute import KNNImputer
imputer = KNNImputer(n_neighbors=5)
x_df_values = imputer.fit_transform(x_df)
x_df = pd.DataFrame(x_df_values, columns=x_df.columns)
x_df["price"] = df["price"]
x_df.head()
a = x_df.corr()["price"].reset_index()
b = df.corr()["price"].reset_index()
b.merge(a, on="index")
pd.get_dummies(
df, columns=["brand_name", "processor_brand", "os"], drop_first=True
).corr()["price"]
|
# # EMNIST for ABCD
# import numpy as np # linear algebra
# import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# from cv2 import cv2
import matplotlib.pyplot as plt
from keras.models import Sequential
from keras.layers import (
Dense,
Activation,
Dropout,
Flatten,
MaxPooling2D,
Conv2D,
RandomFlip,
RandomRotation,
Rescaling,
GaussianNoise,
)
from tensorflow.keras.optimizers import Adam, RMSprop
from keras import backend as K
from subprocess import check_output
from keras.preprocessing.image import ImageDataGenerator
from keras.losses import SparseCategoricalCrossentropy
# 图像数据集的读取可以参照 https://tensorflow.google.cn/tutorials/load_data/images?hl=zh-cn 加以理解
from keras.utils import image_dataset_from_directory
K.clear_session()
n_classes = 4
batch_size = 32
img_width, img_height = 64, 64
data_dir = "/kaggle/input/myabcd/train"
trn_ds = image_dataset_from_directory(
data_dir, # 训练集目录名
subset="training",
shuffle=True,
seed=123,
validation_split=0.2,
image_size=(img_height, img_width),
batch_size=batch_size,
)
val_ds = image_dataset_from_directory(
data_dir,
validation_split=0.2,
subset="validation",
seed=123,
image_size=(img_height, img_width),
batch_size=batch_size,
)
# 训练集中查看类名
class_names = trn_ds.class_names
print(class_names)
### 下面是训练数据集中的 9 个图像。
plt.figure(figsize=(8, 8))
for images, labels in trn_ds.take(1):
for i in range(9):
ax = plt.subplot(3, 3, i + 1)
plt.imshow(images[i])
plt.title(class_names[labels[i]])
plt.axis("off")
for image_batch, labels_batch in trn_ds:
print("训练图片数据集的张量维度", image_batch.shape)
print("标签数据集的张量维度", labels_batch.shape)
break
data_augmentation = Sequential(
[
Rescaling(1.0 / 255),
RandomFlip("horizontal_and_vertical"),
RandomRotation(0.2, fill_mode="constant", fill_value=0),
GaussianNoise(0.1),
]
)
### 下面是训练数据集中的 9 个图像。
plt.figure(figsize=(8, 8))
for images, labels in trn_ds.take(1):
for i in range(9):
ax = plt.subplot(3, 3, i + 1)
augmented_image = data_augmentation(images[i])
plt.imshow(augmented_image)
plt.title(class_names[labels[i]])
plt.axis("off")
model = Sequential(
[
data_augmentation,
Conv2D(32, 3, activation="relu"),
MaxPooling2D(),
Conv2D(32, 3, activation="relu"),
MaxPooling2D(),
Conv2D(32, 3, activation="relu"),
MaxPooling2D(),
Flatten(),
Dense(128, activation="relu"),
Dense(n_classes),
]
)
model.build(input_shape=(None, img_height, img_width, 3))
model.summary()
model.compile(
optimizer="adam",
loss=SparseCategoricalCrossentropy(from_logits=True),
metrics=["accuracy"],
)
model.fit(trn_ds, validation_data=val_ds, epochs=150)
# ## 6.1 Compile the model with compile() method
# - Compilation of model can be done as follows:
model.compile(loss="categorical_crossentropy", optimizer="adam", metrics=["accuracy"])
# ### Loss function (categorical_crossentropy)
# - How far the predicted tensor is from the one-hot ground truth vector is called **loss**.
# - In this example, we use **categorical_crossentropy** as the loss function. It is the negative of the sum of the product of the target and the logarithm of the prediction.
# - There are other loss functions in Keras, such as mean_absolute_error and binary_crossentropy. The choice of the loss function is not arbitrary but should be a criterion that the model is learning.
# - For classification by category, categorical_crossentropy or mean_squared_error is a good choice after the softmax activation layer. The binary_crossentropy loss function is normally used after the sigmoid activation layer while mean_squared_error is an option for tanh output.
# ### Optimization (optimizer adam)
# - With optimization, the objective is to minimize the loss function. The idea is that if the loss is reduced to an acceptable level, the model has indirectly learned the function mapping input to output.
# - In Keras, there are several choices for optimizers. The most commonly used optimizers are; **Stochastic Gradient Descent (SGD)**, **Adaptive Moments (Adam)** and **Root Mean Squared Propagation (RMSprop)**.
# - Each optimizer features tunable parameters like learning rate, momentum, and decay.
# - Adam and RMSprop are variations of SGD with adaptive learning rates. In the proposed classifier network, Adam is used since it has the highest test accuracy.
# ### Metrics (accuracy)
# - Performance metrics are used to determine if a model has learned the underlying data distribution. The default metric in Keras is loss.
# - During training, validation, and testing, other metrics such as **accuracy** can also be included.
# - **Accuracy** is the percent, or fraction, of correct predictions based on ground truth.
# ## 6.2 Train the model with fit() method
model.fit(x_train, y_train, epochs=20, batch_size=batch_size)
# ## 6.3 Evaluating model performance with evaluate() method
loss, acc = model.evaluate(x_test, y_test, batch_size=batch_size)
print("\nTest accuracy: %.1f%%" % (100.0 * acc))
path = "/kaggle/input/testdata1/three.jpg"
img = cv2.imread(path)
img
# 通过模型预测结果并输出
path = "/kaggle/input/testdata1/three.jpg"
img = cv2.imread(path)
plt.imshow(img, cmap=plt.cm.binary)
plt.show()
img = np.array(img).astype(np.float32)
img = cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)
img = img.reshape(-1, 28 * 28)
y_pred = model.predict(img)
# print('predicted number:',np.argmax(y_pred, axis=1)[0])
y_pred
# # 7. Overfitting and Regularization
# [Back to Table of Contents](#0.1)
# - A neural network has the property to memorize the characteristics of training data. This is called **overfitting**.
# - In such a case, the network fails to generalize when subject to the test data.
# - To avoid this tendency, the model uses a regularizing layer or function. A commonly used regularizing layer is referred to as a **Dropout layer**.
# - Given a dropout rate (dropout=0.45), the **Dropout layer** randomly removes the fraction of units from participating in the next layer. For example, if the first layer has 256 units, after dropout=0.45 is applied, only (1 - 0.45) * 256 units = 140 units from layer 1 participate in layer 2.
# - The Dropout layer makes neural networks robust to unforeseen input data because the network is trained to predict correctly, even if some units are missing.
# - The dropout is not used in the output layer and it is only active during training. Moreover, dropout is not present during prediction.
# - There are regularizers that can be used other than dropouts like l1 or l2. In Keras, the bias, weight and activation output can be regularized per layer. - l1 and l2 favor smaller parameter values by adding a penalty function. Both l1 and l2 enforce the penalty using a fraction of the sum of absolute (l1) or square (l2) of parameter values.
# - So, the penalty function forces the optimizer to find parameter values that are small. Neural networks with small parameter values are more insensitive to the presence of noise from within the input data.
# - So, the l2 weight regularizer with fraction=0.001 can be implemented as:
from keras.regularizers import l2
model.add(Dense(hidden_units, kernel_regularizer=l2(0.001), input_dim=input_size))
|
import pandas as pd
import numpy as np
import nibabel as nib
import matplotlib.pyplot as plt
import tensorflow as tf
from tensorflow.keras import datasets, layers, models
import cv2
import os
from pylab import rcParams
import PIL
from PIL import Image
from sklearn.model_selection import train_test_split
import imgaug as ia
import imgaug.augmenters as iaa
import sys
import random
import warnings
import math
from keras.callbacks import Callback
from keras.losses import binary_crossentropy
from skimage.io import imread, imshow, concatenate_images
from skimage.transform import resize
from keras.models import Model, load_model
from keras.layers import Input, BatchNormalization, Activation, Dense, Dropout
from keras.layers.convolutional import Conv2D, Conv2DTranspose
from keras.layers.pooling import MaxPooling2D, GlobalMaxPool2D
from keras.layers import concatenate, add
from keras.callbacks import EarlyStopping, ModelCheckpoint
from keras.optimizers import Adam
from keras import backend as K
import joblib
import gc
raw_data = pd.read_csv("/kaggle/input/covid19-ct-scans/metadata.csv")
raw_data.head(5)
img_size = 256
def clahe_enhancer(test_img, demo):
test_img = test_img * 255
test_img = np.uint8(test_img)
test_img_flattened = test_img.flatten()
clahe = cv2.createCLAHE(clipLimit=3.0, tileGridSize=(8, 8))
clahe_image = clahe.apply(test_img)
clahe_image_flattened = clahe_image.flatten()
if demo == 1:
fig = plt.figure()
rcParams["figure.figsize"] = 10, 10
plt.subplot(2, 2, 1)
plt.imshow(test_img, cmap="bone")
plt.grid(False)
plt.title("Original CT-Scan")
plt.subplot(2, 2, 2)
plt.hist(test_img_flattened)
plt.grid(False)
plt.title("Histogram of Original CT-Scan")
plt.subplot(2, 2, 3)
plt.imshow(clahe_image, cmap="bone")
plt.grid(False)
plt.title("CLAHE Enhanced CT-Scan")
plt.subplot(2, 2, 4)
plt.hist(clahe_image_flattened)
plt.grid(False)
plt.title("Histogram of CLAHE Enhanced CT-Scan")
return clahe_image
def read_nii_demo(filepath, data, show_shape=False):
ct_scan = nib.load(filepath)
array = ct_scan.get_fdata()
array = np.rot90(np.array(array))
slices = array.shape[2]
array = array[:, :, round(slices * 0.2) : round(slices * 0.8)]
if show_shape:
print(array.shape)
array = np.reshape(
np.rollaxis(array, 2), (array.shape[2], array.shape[0], array.shape[1], 1)
)
# Min-max scaling
for img_no in range(0, array.shape[0]):
img = cv2.resize(
array[img_no], dsize=(img_size, img_size), interpolation=cv2.INTER_AREA
)
xmax, xmin = img.max(), img.min()
img = (img - xmin) / (xmax - xmin)
data.append(img)
cts = []
lungs = []
for i in range(0, 20):
if i == 0:
read_nii_demo(raw_data.loc[i, "lung_mask"], lungs, True)
read_nii_demo(raw_data.loc[i, "ct_scan"], cts, True)
else:
read_nii_demo(raw_data.loc[i, "lung_mask"], lungs)
read_nii_demo(raw_data.loc[i, "ct_scan"], cts)
new_cts = []
new_lungs = []
# # CLAHE
for img_no in range(len(lungs)):
lung_img = lungs[img_no]
lung_img[lung_img > 0] = 1
new_lungs.append(lung_img)
cts_img = cts[img_no]
cts_img = clahe_enhancer(cts_img, demo=0)
new_cts.append(cts_img)
x = 100
rcParams["figure.figsize"] = 10, 10
plt.subplot(1, 2, 1)
plt.imshow(new_cts[x], cmap="bone")
plt.grid(False)
plt.title("CLAHE Enhanced Image")
plt.subplot(1, 2, 2)
plt.imshow(new_lungs[x], cmap="bone")
plt.grid(False)
plt.title("Mask")
print(cts[x].shape, lungs[x].shape)
test_file = []
read_nii_demo(raw_data.loc[0, "ct_scan"], test_file)
test_file = np.array(test_file)
rcParams["figure.figsize"] = 10, 10
clahe_image = clahe_enhancer(test_file[100], demo=1)
# # Data augmentation
new_dim = 256
new_cts = np.array(new_cts)
new_lungs = np.array(new_lungs)
new_cts = np.uint8(new_cts)
new_lungs = np.uint8(new_lungs)
sometimes = lambda aug: iaa.Sometimes(0.5, aug)
seq = iaa.Sequential(
[
iaa.Fliplr(0.5), # horizontally flip 50% of all images
iaa.Flipud(0.2), # vertically flip 20% of all images
sometimes(
iaa.Affine(
scale={
"x": (0.8, 1.2),
"y": (0.8, 1.2),
}, # scale images to 80-120% of their size, individually per axis
translate_percent={
"x": (-0.2, 0.2),
"y": (-0.2, 0.2),
}, # translate by -20 to +20 percent (per axis)
rotate=(-30, 30), # rotate by -30 to +30 degrees
shear=(-15, 15), # shear by -15 to +15 degrees
)
),
],
random_order=True,
)
new_cts = new_cts.reshape(len(new_cts), new_dim, new_dim, 1)
new_lungs = new_lungs.reshape(len(new_lungs), new_dim, new_dim, 1)
len(new_cts) * 0.25
no_of_aug_imgs = 526
random_indices = np.random.randint(0, new_cts.shape[0], size=no_of_aug_imgs)
sample_new_cts = new_cts[random_indices]
sample_new_lungs = new_lungs[random_indices]
new_cts_aug, new_lungs_aug = seq(
images=sample_new_cts, segmentation_maps=sample_new_lungs
)
rcParams["figure.figsize"] = 60, 60
rand = np.random.randint(0, no_of_aug_imgs, size=8)
cells1 = new_cts_aug[rand]
grid_image1 = np.hstack(cells1)
plt.imshow(grid_image1, cmap="bone")
plt.grid(False)
rcParams["figure.figsize"] = 60, 60
cells1 = new_lungs_aug[rand]
grid_image1 = np.hstack(cells1)
plt.imshow(grid_image1, cmap="bone")
plt.grid(False)
print(new_cts_aug.shape, new_lungs_aug.shape)
new_cts_aug = new_cts_aug.reshape(len(new_cts_aug), new_dim, new_dim, 1)
new_lungs_aug = new_lungs_aug.reshape(len(new_lungs_aug), new_dim, new_dim, 1)
new_cts = np.concatenate((new_cts, new_cts_aug))
new_lungs = np.concatenate((new_lungs, new_lungs_aug))
new_cts = new_cts / 255
len(new_cts)
def plot_sample(array_list, color_map="nipy_spectral"):
fig = plt.figure(figsize=(10, 30))
plt.subplot(1, 2, 1)
plt.imshow(array_list[0].reshape(new_dim, new_dim), cmap="bone")
plt.title("Original Image")
plt.subplot(1, 2, 2)
plt.imshow(array_list[0].reshape(new_dim, new_dim), cmap="bone")
plt.imshow(array_list[1].reshape(new_dim, new_dim), alpha=0.5, cmap=color_map)
plt.title("Lung Mask")
plt.show()
rcParams["axes.grid"] = False
for index in [567, 127, 330, 440, 190]:
plot_sample([new_cts[index], new_lungs[index]])
# # Unet model
def dice_coeff(y_true, y_pred):
smooth = 1.0
y_true_f = K.flatten(y_true)
y_pred_f = K.flatten(y_pred)
intersection = K.sum(y_true_f * y_pred_f)
score = (2.0 * intersection + smooth) / (K.sum(y_true_f) + K.sum(y_pred_f) + smooth)
return score
def dice_loss(y_true, y_pred):
loss = 1 - dice_coeff(y_true, y_pred)
return loss
def bce_dice_loss(y_true, y_pred):
loss = 0.5 * binary_crossentropy(y_true, y_pred) + 0.5 * dice_loss(y_true, y_pred)
return loss
inputs = Input((new_dim, new_dim, 1))
c1 = Conv2D(
32, (3, 3), activation="relu", padding="same", kernel_initializer="he_normal"
)(inputs)
c1 = Conv2D(
32, (3, 3), activation="relu", padding="same", kernel_initializer="he_normal"
)(c1)
c1 = BatchNormalization()(c1)
p1 = MaxPooling2D((2, 2))(c1)
p1 = Dropout(0.25)(p1)
c2 = Conv2D(
64, (3, 3), activation="relu", padding="same", kernel_initializer="he_normal"
)(p1)
c2 = Conv2D(
64, (3, 3), activation="relu", padding="same", kernel_initializer="he_normal"
)(c2)
c2 = BatchNormalization()(c2)
p2 = MaxPooling2D((2, 2))(c2)
p2 = Dropout(0.25)(p2)
c3 = Conv2D(
128, (3, 3), activation="relu", padding="same", kernel_initializer="he_normal"
)(p2)
c3 = Conv2D(
128, (3, 3), activation="relu", padding="same", kernel_initializer="he_normal"
)(c3)
c3 = BatchNormalization()(c3)
p3 = MaxPooling2D((2, 2))(c3)
p3 = Dropout(0.25)(p3)
c4 = Conv2D(
256, (3, 3), activation="relu", padding="same", kernel_initializer="he_normal"
)(p3)
c4 = Conv2D(
256, (3, 3), activation="relu", padding="same", kernel_initializer="he_normal"
)(c4)
c4 = BatchNormalization()(c4)
p4 = MaxPooling2D(pool_size=(2, 2))(c4)
p4 = Dropout(0.25)(p4)
c5 = Conv2D(
512, (3, 3), activation="relu", padding="same", kernel_initializer="he_normal"
)(p4)
c5 = Conv2D(
512, (3, 3), activation="relu", padding="same", kernel_initializer="he_normal"
)(c5)
u6 = Conv2DTranspose(256, (2, 2), strides=(2, 2), padding="same")(c5)
u6 = concatenate([u6, c4])
u6 = BatchNormalization()(u6)
c6 = Conv2D(
256, (3, 3), activation="relu", padding="same", kernel_initializer="he_normal"
)(u6)
c6 = Conv2D(
256, (3, 3), activation="relu", padding="same", kernel_initializer="he_normal"
)(c6)
u7 = Conv2DTranspose(128, (2, 2), strides=(2, 2), padding="same")(c6)
u7 = concatenate([u7, c3])
u7 = BatchNormalization()(u7)
c7 = Conv2D(
128, (3, 3), activation="relu", padding="same", kernel_initializer="he_normal"
)(u7)
c7 = Conv2D(
128, (3, 3), activation="relu", padding="same", kernel_initializer="he_normal"
)(c7)
u8 = Conv2DTranspose(64, (2, 2), strides=(2, 2), padding="same")(c7)
u8 = concatenate([u8, c2])
u8 = BatchNormalization()(u8)
c8 = Conv2D(
64, (3, 3), activation="relu", padding="same", kernel_initializer="he_normal"
)(u8)
c8 = Conv2D(
64, (3, 3), activation="relu", padding="same", kernel_initializer="he_normal"
)(c8)
u9 = Conv2DTranspose(32, (2, 2), strides=(2, 2), padding="same")(c8)
u9 = concatenate([u9, c1], axis=3)
u9 = BatchNormalization()(u9)
c9 = Conv2D(
32, (3, 3), activation="relu", padding="same", kernel_initializer="he_normal"
)(u9)
c9 = Conv2D(
32, (3, 3), activation="relu", padding="same", kernel_initializer="he_normal"
)(c9)
outputs = Conv2D(1, (1, 1), activation="sigmoid")(c9)
model = Model(inputs=[inputs], outputs=[outputs])
model.summary()
batch_size = 32
epochs = 50
filepath_dice_coeff = "unet_covid_weights_dice_coeff.hdf5"
filepath_loss = "unet_covid_weights_val_loss.hdf5"
checkpoint_dice = ModelCheckpoint(
filepath_dice_coeff,
monitor="val_dice_coeff",
verbose=1,
save_best_only=True,
mode="max",
)
checkpoint_loss = ModelCheckpoint(
filepath_loss, monitor="val_loss", verbose=1, save_best_only=True, mode="min"
)
del new_cts_aug
del new_lungs_aug
del cts
del lungs
gc.collect()
model.compile(optimizer=Adam(lr=0.0001), loss=bce_dice_loss, metrics=[dice_coeff])
new_lungs = new_lungs.astype(np.float32)
x_train, x_valid, y_train, y_valid = train_test_split(
new_cts, new_lungs, test_size=0.2, random_state=42
)
results = model.fit(
x_train,
y_train,
batch_size=batch_size,
epochs=epochs,
validation_data=(x_valid, y_valid),
callbacks=[checkpoint_dice, checkpoint_loss],
)
del x_train
del x_valid
del y_train
del y_valid
model.load_weights(filepath_dice_coeff)
score = model.evaluate(x_valid, y_valid, batch_size=32)
print("test loss, test dice coefficient:", score)
# # Saving model
model.save_weights("unet_lung_segmentation_0.9867.h5")
model_json = model.to_json()
with open("unet_lung_segmentation_0.9867.json", "w") as json_file:
json_file.write(model_json)
gc.collect()
plt.rcParams["axes.grid"] = True
rcParams["figure.figsize"] = 7, 5
plt.xlim(0, 40)
plt.plot(results.history["dice_coeff"])
plt.plot(results.history["val_dice_coeff"])
plt.title("Dice Coefficient")
plt.ylabel("Dice coefficient")
plt.xlabel("epoch")
plt.legend(["train", "test"], loc="upper left")
plt.show()
plt.ylim(0, 2)
plt.xlim(0, 40)
plt.plot(results.history["loss"])
plt.plot(results.history["val_loss"])
plt.title("Dice Loss")
plt.ylabel("loss")
plt.xlabel("epoch")
plt.legend(["train", "test"], loc="upper left")
plt.show()
# # Comparison of normal images
def compare_actual_and_predicted(image_no):
rcParams["axes.grid"] = False
temp = model.predict(new_cts[image_no].reshape(1, new_dim, new_dim, 1))
fig = plt.figure(figsize=(15, 15))
plt.subplot(1, 3, 1)
plt.imshow(new_cts[image_no].reshape(new_dim, new_dim), cmap="bone")
plt.title("Original Image (CT)")
plt.subplot(1, 3, 2)
plt.imshow(new_lungs[image_no].reshape(new_dim, new_dim), cmap="bone")
plt.title("Actual mask")
plt.subplot(1, 3, 3)
plt.imshow(temp.reshape(new_dim, new_dim), cmap="bone")
plt.title("Predicted mask")
plt.show()
for i in [440, 269, 555, 355, 380, 90]:
compare_actual_and_predicted(i)
# # Comparison of augmented images
for i in [2001, 2200, 2355, 2380, 2290]:
compare_actual_and_predicted(i)
|
# # **6th place solution : Tabular Feb. Competition**
# It is my first time to reach this high ranking. So I am really happy!
# This is my code I used to improve my score.
# I refer to lots of discussion and codes, and It was really helpful. Thanks you for everybody who wrote it.
# And special thanks to @hiro5299834! Wrote amazing LGBM codes and fine-tuned params, it became our code baseline. [His Code](https://www.kaggle.com/hiro5299834/tps-feb-2021-with-single-lgbm-tuned)
# # **Abstract**
# I used pseudo labelling for improving score. Pseudo Labelling is kind of semi-supervised learning. I labelled all test data, and trained again with original train data and pseudo-labelled test data. [Detail](https://analyticsindiamag.com/pseudo-labelling-a-guide-to-semi-supervised-learning/)
# It is really simple trick, but score improved a lot. Original code was 0.84198 LB, but pseudo labelling code reached 0.84185 LB (0.84249 private score)
# After this, I ensembled a lot. And I can reached 0.84246 private score. But I couldn't see the public score improved, so I couldn't select that for final submission, unfortunately.
# Below is my code for training 6th place model.
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import os
import sys
import math
import time
import pickle
import psutil
import random
import numpy as np
import pandas as pd
from tqdm import tqdm
from pathlib import Path
from datetime import datetime
from contextlib import contextmanager
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.manifold import TSNE
import warnings
import optuna
from lightgbm import LGBMRegressor
from sklearn.metrics import mean_squared_error
from sklearn.compose import ColumnTransformer
from sklearn.model_selection import KFold
from sklearn.preprocessing import (
StandardScaler,
PowerTransformer,
MinMaxScaler,
QuantileTransformer,
LabelEncoder,
OneHotEncoder,
OrdinalEncoder,
)
import category_encoders as ce
plt.style.use("fivethirtyeight")
warnings.filterwarnings("ignore")
# # **Pseudo Labelling**
# Just concat well-trained submission file, test file, and train file.
original_train = pd.read_csv("../input/tabular-playground-series-feb-2021/train.csv")
test = pd.read_csv("../input/tabular-playground-series-feb-2021/test.csv")
test_target = pd.read_csv("../input/tabular-feb-submission-files/sub45_47_48_(5).csv")
test["target"] = test_target.target
train = pd.concat([original_train, test], axis=0)
train
# # **Training with LGBM**
# This code is from BIZEN's codes, but pseudo labelling always worked with any LGBM models. So you can use any fine-tuned LGBM models. Actually, I reached 0.84245 private score with single LGBM model and pseudo labelling. (0.84193 public LB)
N_FOLDS = 10
N_ESTIMATORS = 30000
SEED = 2021
BAGGING_SEED = 48
@contextmanager
def timer(name: str):
t0 = time.time()
p = psutil.Process(os.getpid())
m0 = p.memory_info()[0] / 2.0**30
try:
yield
finally:
m1 = p.memory_info()[0] / 2.0**30
delta = m1 - m0
sign = "+" if delta >= 0 else "-"
delta = math.fabs(delta)
print(
f"[{m1:.1f}GB({sign}{delta:.1f}GB): {time.time() - t0:.3f}sec] {name}",
file=sys.stderr,
)
def set_seed(seed=42):
random.seed(seed)
os.environ["PYTHONHASHSEED"] = str(seed)
np.random.seed(seed)
def score_log(df: pd.DataFrame, seed: int, num_fold: int, model_name: str, cv: float):
score_dict = {
"date": datetime.now(),
"seed": seed,
"fold": num_fold,
"model": model_name,
"cv": cv,
}
# noinspection PyTypeChecker
df = pd.concat([df, pd.DataFrame.from_dict([score_dict])])
df.to_csv(LOG_PATH / f"model_score_{model_name}.csv", index=False)
return df
with timer("Read data"):
train_df = train
test_df = pd.read_csv("../input/tabular-playground-series-feb-2021/test.csv")
sub_df = pd.read_csv(
"../input/tabular-playground-series-feb-2021/sample_submission.csv"
)
all_df = pd.concat([train_df, test_df])
all_df = all_df.sort_values("id").reset_index(drop=True)
cont_features = [f"cont{i}" for i in range(14)]
cat_features = [f"cat{i}" for i in range(10)]
all_features = cat_features + cont_features
target_feature = "target"
target = train_df[target_feature]
train_df = train_df[all_features]
test_df = test_df[all_features]
# LabelEncoder()
with timer("LabelEncoder"):
le = LabelEncoder()
for col in cat_features:
le.fit(train_df[col])
train_df[col] = le.transform(train_df[col])
test_df[col] = le.transform(test_df[col])
params = {
"random_state": SEED,
"metric": "rmse",
"n_estimators": N_ESTIMATORS,
"n_jobs": -1,
"cat_feature": [x for x in range(len(cat_features))],
"bagging_seed": SEED,
"feature_fraction_seed": SEED,
"learning_rate": 0.003899156646724397,
"max_depth": 99,
"num_leaves": 63,
"reg_alpha": 9.562925363678952,
"reg_lambda": 9.355810045480153,
"colsample_bytree": 0.2256038826485174,
"min_child_samples": 290,
"subsample_freq": 1,
"subsample": 0.8805303688019942,
"max_bin": 882,
"min_data_per_group": 127,
"cat_smooth": 96,
"cat_l2": 19,
}
oof = np.zeros(train_df.shape[0])
preds = 0
train_preds = 0
score_df = pd.DataFrame()
feature_importances = pd.DataFrame()
kf = KFold(n_splits=N_FOLDS, shuffle=True, random_state=SEED)
for fold, (train_idx, valid_idx) in enumerate(kf.split(X=train_df)):
X_train, X_valid = train_df.iloc[train_idx], train_df.iloc[valid_idx]
y_train, y_valid = target.iloc[train_idx], target.iloc[valid_idx]
with timer(f"fold {fold}: fit"):
model = LGBMRegressor(**params)
model.fit(
X_train,
y_train,
eval_set=[(X_valid, y_valid)],
eval_metric="rmse",
early_stopping_rounds=100,
verbose=0,
)
fi_tmp = pd.DataFrame()
fi_tmp["feature"] = model.feature_name_
fi_tmp["importance"] = model.feature_importances_
fi_tmp["fold"] = fold
fi_tmp["seed"] = SEED
feature_importances = feature_importances.append(fi_tmp)
oof[valid_idx] = model.predict(X_valid)
preds += model.predict(test_df) / N_FOLDS
train_preds += model.predict(train_df) / N_FOLDS
rmse = mean_squared_error(y_valid, oof[valid_idx], squared=False)
score_df = score_log(score_df, SEED, fold, "lgb", rmse)
print(f"rmse {rmse}")
rmse = mean_squared_error(target, oof, squared=False)
score_df = score_log(score_df, SEED, 999, "lgb", rmse)
print("+-" * 40)
print(f"rmse {rmse}")
pred_train_df = pd.DataFrame([])
sub_df.target = preds
sub_df.to_csv(f"pseudo_labelling_cv{rmse:.6f}.csv", index=False)
sub_df.head()
pred_train_df.target = train_preds
pred_train_df.to_csv("train_pred_v0.csv", index=False)
np.save(LOG_PATH / "train_oof", oof)
np.save(LOG_PATH / "test_preds", preds)
|
import nltk
from nltk.corpus import stopwords
nltk.download("stopwords")
from sklearn.model_selection import train_test_split
# from sklearn.preprocessing import LabelEncoder
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# stop_words = set(stopwords.words('english'))
# stop_words_removal=['not', "n't", 'against', 'no', 'nor', 'don', "don't", 'should', "should've", 'ain', 'aren', "aren't", 'couldn', "couldn't", 'didn', "didn't", 'doesn', "doesn't", 'hadn', "hadn't", 'hasn', "hasn't", 'haven', "haven't", 'isn', "isn't", 'ma', 'mightn', "mightn't", 'mustn', "mustn't", 'needn', "needn't", 'shan', "shan't", 'shouldn', "shouldn't", 'wasn', "wasn't", 'weren', "weren't", 'won', "won't", 'wouldn', "wouldn't","further"]
# for word in stop_words_removal:
# if word in stop_words:
# stop_words.remove(word)
# more_stopwords = {'USER','URL'}
# stop_words = stop_words.union(more_stopwords)
# def remove_emoji(text):
# emoji_pattern = re.compile(
# '['
# u'\U0001F600-\U0001F64F' # emoticons
# u'\U0001F300-\U0001F5FF' # symbols & pictographs
# u'\U0001F680-\U0001F6FF' # transport & map symbols
# u'\U0001F1E0-\U0001F1FF' # flags (iOS)
# u'\U00002702-\U000027B0'
# u'\U000024C2-\U0001F251'
# ']+',
# flags=re.UNICODE)
# return emoji_pattern.sub(r'', text)
# def remove_html(text):
# html = re.compile(r'^[^ ]<.*?>|&([a-z0-9]+|#[0-9]\"\'\“{1,6}|#x[0-9a-f]{1,6});[^A-Za-z0-9]+')
# return re.sub(html, '', text)
# def remove_punct(text):
# table = str.maketrans('', '', string.punctuation)
# return text.translate(table)
# import numpy as np
# def load_glove_embeddings(file_path):
# embeddings_dict = {}
# with open(file_path, 'r', encoding='utf-8') as f:
# for line in f:
# values = line.split(" ")
# word = values[0]
# embedding = np.asarray(values[1:], dtype='float32')
# embeddings_dict[word] = embedding
# return embeddings_dict
# embeddings_path = '/kaggle/input/glove840b/glove.840B.300d.txt'
# # storing embedding as a dictionary
# glove_embeddings = load_glove_embeddings(embeddings_path)
# print('Found %s word vectors.' % len(glove_embeddings))
import sys, os, re, csv, codecs, numpy as np, pandas as pd
# import matplotlib.pyplot as plt
# %matplotlib inline
import tensorflow as tf
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import LSTM, Dense
# importing Tokenizer and --Still Not final--
from tensorflow.keras.preprocessing.text import Tokenizer
from tensorflow.keras.preprocessing.sequence import pad_sequences
from keras.layers import Dense, Input, LSTM, Embedding, Dropout, Activation
from keras.layers import Bidirectional, GlobalMaxPool1D, Bidirectional
from keras.models import Model
from keras import initializers, regularizers, constraints, optimizers, layers
# import matplotlib.pyplot as plt
# %matplotlib inline
# import gensim.models.keyedvectors as word2vec
import gc
final_dataset = []
# data=pd.read_csv('/kaggle/input/twitter-emotion-analysis/compiled_dataset3.csv')
data1 = pd.read_csv("/kaggle/input/twitter-emotion-analysis/compiled_dataset1.csv")
data2 = pd.read_csv("/kaggle/input/twitter-emotion-analysis/compiled_dataset2.csv")
data3 = pd.read_csv("/kaggle/input/twitter-emotion-analysis/compiled_dataset3.csv")
final_dataset.extend([data1, data2, data3])
final_dataset = pd.concat(final_dataset).loc[:]
print(final_dataset)
import gc
del data1
gc.collect()
del data2
gc.collect()
del data3
gc.collect()
final_dataset = final_dataset[["cleaned_Text", "label"]]
# stopwords_removed
# cleaned_Text
gc.collect()
# data=data[['text','label']]
# tokenizer = Tokenizer(num_words=344944, oov_token="<OOV>")
# tokenizer.fit_on_texts(sentences)
# data['mod_text'] = data['text'].apply(lambda x: remove_emoji(x))
# data['mod_text'] = data['mod_text'].apply(lambda x: remove_html(x))
sentences = final_dataset["cleaned_Text"].astype(str)
# data['tokenized'] = data['mod_text'].apply(word_tokenize)
# data.head()
tokenizer = Tokenizer(num_words=626067, oov_token="<OOV>")
# # 626182
# 626066
tokenizer.fit_on_texts(sentences)
sequences = tokenizer.texts_to_sequences(sentences)
padded_sequences = pad_sequences(
sequences, maxlen=110, padding="post", truncating="post"
)
del sentences
gc.collect()
final_dataset = final_dataset["label"]
gc.collect()
# from sklearn.feature_extraction.text import CountVectorizer
# # Multiple documents
# # create the transform
# vectorizer = CountVectorizer()
# # tokenize and build vocab
# vectorizer.fit(sentences)
# # summarize
# print(len(vectorizer.vocabulary_))
# encoder = LabelEncoder()
# encoder.fit(data.label.to_list())
# y_val = encoder.transform(data.label.to_list())
# # y_test = encoder.transform(test_data.label.to_list())
# y_val = y_val.reshape(-1,1)
# # y_test = y_test.reshape(-1,1)
# print("y_train shape:", y_val.shape)
# # print("y_test shape:", y_test.shape)
# labels=final_dataset['label']
label_map = {
"sadness": 0,
"anger": 1,
"disgust": 2,
"fear": 3,
"joy": 4,
"surprise": 5,
"neutral": 6,
}
# Use map function to transform labels to numerical values
final_dataset = final_dataset.map(label_map)
gc.collect()
# del x_train
# gc.collect()
# del x_test
# gc.collect()
# del y_train
# gc.collect()
# del y_test
# gc.collect()
x_train, x_test, y_train, y_test = train_test_split(
padded_sequences, final_dataset, test_size=0.2, random_state=4
)
del final_dataset
gc.collect()
from keras.utils import to_categorical
# Convert target variable to one-hot encoding
y_train = to_categorical(y_train, num_classes=7)
y_test = to_categorical(y_test, num_classes=7)
word_index = tokenizer.word_index
vocab_size = len(tokenizer.word_index) + 1
print("Vocabulary Size :", vocab_size)
# del tokenizer
# gc.collect()
del sequences
gc.collect()
del padded_sequences
gc.collect()
# embedding_matrix = np.zeros((vocab_size, 300))
# for word, i in word_index.items():
# embedding_vector = glove_embeddings.get(word)
# if embedding_vector is not None:
# embedding_matrix[i] = embedding_vector
from tensorflow.keras.layers import Embedding, LSTM, Dense, Bidirectional, Dropout
from keras.models import Sequential
from keras.layers import (
Embedding,
Conv1D,
MaxPooling1D,
Bidirectional,
LSTM,
Dense,
Dropout,
)
from keras.metrics import Precision, Recall
from keras.optimizers import SGD
from keras.optimizers import RMSprop
from keras import datasets
from keras.callbacks import LearningRateScheduler
from keras.callbacks import History
from keras import losses
model = Sequential()
model.add(Embedding(vocab_size, 300, input_length=110))
# model.add(Embedding(vocab_size,300,
# weights=[embedding_matrix],
# input_length=110,
# trainable=False))
model.add(Conv1D(filters=64, kernel_size=7, padding="same", activation="relu"))
model.add(MaxPooling1D(pool_size=2))
# model.add(Bidirectional(LSTM(units=64, return_sequences=True)))
model.add(Bidirectional(LSTM(200)))
model.add(Dropout(0.2))
# model.add(Dense(128, activation='relu'))
model.add(Dense(7, activation="softmax"))
# Compile the model
model.compile(loss="categorical_crossentropy", optimizer="adam", metrics=["accuracy"])
history = model.fit(
x_train, y_train, batch_size=7000, epochs=25, validation_data=(x_test, y_test)
)
# dataset_words = set(tokenizer.word_index.keys())
# glove_words = set(glove_embeddings.keys())
# common_words = glove_words.intersection(dataset_words)
# num_common_words = len(common_words)
# print("Number of words in GloVe embeddings that are present in the dataset: ", num_common_words)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df1 = pd.read_csv("/kaggle/input/bidmc-pred/bidmc_csv/bidmc_01_Numerics.csv")
df2 = pd.read_csv("/kaggle/input/bidmc-pred/bidmc_csv/bidmc_01_Signals.csv")
df1.head()
df2.head()
print("Column Names:")
print(df1.columns.tolist())
print("Column Names:")
print(df2.columns.tolist())
df1[" PULSE"]
# Extract PPG data and time reference
pleth = df2[" PLETH"]
time_ref = df1["Time [s]"]
# Extract HR, SpO2, and RR data
hr_data = df1[" HR"]
spo2_data = df1[" SpO2"]
rr_data = df1[" RESP"]
hr_data.shape
time_ref.shape
print("Range of Heart Rate (HR):")
print("Min HR:", hr_data.min())
print("Max HR:", hr_data.max())
print("\nRange of Oxygen Saturation (SpO2):")
print("Min SpO2:", spo2_data.min())
print("Max SpO2:", spo2_data.max())
print("\nRange of Respiratory Rate (RR):")
print("Min RR:", rr_data.min())
print("Max RR:", rr_data.max()) # displaying the range
import matplotlib.pyplot as plt
ttm = df1.loc[:480, "Time [s]"]
# import matplotlib.pyplot as plt
# # Create a simple figure with 3 subplots
# plt.figure(figsize=(8, 6)) # Set the size of the figure
# # First subplot
# plt.subplot(3, 1, 1) # 3 rows, 1 column, index 1
# plt.plot([1, 2, 3, 4, 5], [2, 4, 6, 8, 10], 'r') # Plot red line
# plt.xlabel('X-axis label of Subplot 1') # Set X-axis label
# plt.ylabel('Y-axis label of Subplot 1') # Set Y-axis label
# plt.title('Subplot 1 Title') # Set subplot title
# # Second subplot
# plt.subplot(3, 1, 2) # 3 rows, 1 column, index 2
# plt.scatter([1, 2, 3, 4, 5], [2, 4, 6, 8, 10], color='g') # Plot green scatter points
# plt.xlabel('X-axis label of Subplot 2') # Set X-axis label
# plt.ylabel('Y-axis label of Subplot 2') # Set Y-axis label
# plt.title('Subplot 2 Title') # Set subplot title
# # Third subplot
# plt.subplot(3, 1, 3) # 3 rows, 1 column, index 3
# plt.bar(['A', 'B', 'C', 'D', 'E'], [2, 4, 6, 8, 10], color='b') # Plot blue bar chart
# plt.xlabel('X-axis label of Subplot 3') # Set X-axis label
# plt.ylabel('Y-axis label of Subplot 3') # Set Y-axis label
# plt.title('Subplot 3 Title') # Set subplot title
# plt.tight_layout() # Adjust spacing between subplots
# plt.show() # Display the figure
# import matplotlib.pyplot as plt
# # Create a simple figure with 2 subplots
# plt.figure(figsize=(8, 6)) # Set the size of the figure
# # First subplot
# plt.subplot(2, 1, 2) # 2 rows, 1 column, index 1
# plt.plot([1, 2, 3, 4, 5], [2, 4, 6, 8, 10], 'r') # Plot red line
# plt.xlabel('X-axis label of Subplot 1') # Set X-axis label
# plt.ylabel('Y-axis label of Subplot 1') # Set Y-axis label
# plt.title('Subplot 1 Title') # Set subplot title
# # Second subplot
# plt.subplot(2, 1, 1) # 2 rows, 1 column, index 2
# plt.scatter([1, 2, 3, 4, 5], [2, 4, 6, 8, 10], color='g') # Plot green scatter points
# plt.xlabel('X-axis label of Subplot 2') # Set X-axis label
# plt.ylabel('Y-axis label of Subplot 2') # Set Y-axis label
# plt.title('Subplot 2 Title') # Set subplot title
# plt.tight_layout() # Adjust spacing between subplots
# plt.show() # Display the figure
plt.figure(figsize=(12, 6))
plt.subplot(3, 1, 1)
plt.plot(ttm, hr_data)
plt.xlabel("Time")
plt.ylabel("Heart Rate (BPM)")
plt.title("Range of Heart Rates")
plt.subplot(3, 1, 2)
plt.plot(ttm, spo2_data)
plt.xlabel("Time")
plt.ylabel("SpO2 (%)")
plt.title("Range of Oxygen Saturation (SpO2)")
plt.subplot(3, 1, 3)
plt.plot(ttm, rr_data)
plt.xlabel("Time")
plt.ylabel("Respiratory Rate (BPM)")
plt.title("Range of Respiratory Rates")
plt.tight_layout()
plt.show()
from PyEMD import EMD
from scipy.signal import find_peaks
# Perform Empirical Mode Decomposition (EMD)
def perform_emd(signal, sampling_rate, imf_index):
emd = EMD()
emd(signal)
imf = emd.get_imfs()[imf_index]
peaks, _ = find_peaks(imf, distance=sampling_rate * 5) # Find peaks every 5 seconds
return peaks
# Estimate HR and RR using EMD
ppg_sampling_rate = 125 # PPG sampling rate
hr_imf_index = 0 # Index of the first IMF component for HR estimation
rr_imf_index = 1 # Index of the second IMF component for RR estimation
hr_peaks = perform_emd(df2, ppg_sampling_rate, hr_imf_index)
rr_peaks = perform_emd(df1, ppg_sampling_rate, rr_imf_index)
# Convert peak indices to time values
hr_times = time_reference[hr_peaks]
rr_times = time_reference[rr_peaks]
# hr_peaks = perform_emd(df1, ppg_sampling_rate, hr_imf_index, rr_imf_index)
# Estimate HR and RR using EMD
ppg_sampling_rate = 125 # PPG sampling rate
hr_imf_index = 0 # Index of the first IMF component for HR estimation
rr_imf_index = 1 # Index of the second IMF component for RR estimation
hr_peaks = perform_emd(df1, ppg_sampling_rate, hr_imf_index, rr_imf_index)
rr_peaks = perform_emd(df2, ppg_sampling_rate, hr_imf_index, rr_imf_index)
# Convert peak indices to time values
hr_times = time_reference.iloc[hr_peaks] # Use iloc to access DataFrame by index
rr_times = time_reference.iloc[rr_peaks] # Use iloc to access DataFrame by index
# Print HR and RR values every 5 seconds
print("\nEstimated Heart Rate (HR) every 5 seconds:")
for i in range(len(hr_peaks)):
print("Time:", hr_times.iloc[i], "HR:", hr_data.iloc[hr_peaks[i]])
print("\nEstimated Respiratory Rate (RR) every 5 seconds:")
for i in range(len(rr_peaks)):
print("Time:", rr_times.iloc[i], "RR:", rr_data.iloc[rr_peaks[i]])
# import numpy as np
# import pandas as pd
# import matplotlib.pyplot as plt
# from PyEMD import EMD
# from scipy.signal import find_peaks
# # Load PPG dataset
# ppg_df = pd.read_csv("https://physionet.org/files/bidmc/1.0.0/bidmc_01_signals.csv")
# numerics_df = pd.read_csv("https://physionet.org/files/bidmc/1.0.0/bidmc_01_numerics.csv")
# # Extract PPG data and time reference
# ppg_data = ppg_df['PLETH']
# time_reference = ppg_df['TIME']
# # Extract HR, SpO2, and RR data
# hr_data = numerics_df['HR']
# spo2_data = numerics_df['SPO2']
# rr_data = numerics_df['RESP']
# # Display the range of HR, SpO2, and RR
# print("Range of Heart Rate (HR):")
# print("Min HR:", hr_data.min())
# print("Max HR:", hr_data.max())
# print("\nRange of Oxygen Saturation (SpO2):")
# print("Min SpO2:", spo2_data.min())
# print("Max SpO2:", spo2_data.max())
# print("\nRange of Respiratory Rate (RR):")
# print("Min RR:", rr_data.min())
# print("Max RR:", rr_data.max())
# # Perform Empirical Mode Decomposition (EMD)
# def perform_emd(signal, sampling_rate, imf_index):
# emd = EMD()
# emd(signal)
# imf = emd.get_imfs()[imf_index]
# peaks, _ = find_peaks(imf, distance=sampling_rate*5) # Find peaks every 5 seconds
# return peaks
# # Estimate HR and RR using EMD
# ppg_sampling_rate = 125 # PPG sampling rate
# hr_imf_index = 0 # Index of the first IMF component for HR estimation
# rr_imf_index = 1 # Index of the second IMF component for RR estimation
# hr_peaks = perform_emd(ppg_data, ppg_sampling_rate, hr_imf_index)
# rr_peaks = perform_emd(ppg_data, ppg_sampling_rate, rr_imf_index)
# # Convert peak indices to time values
# hr_times = time_reference[hr_peaks]
# rr_times = time_reference[rr_peaks]
# # Print HR and RR values every 5 seconds
# print("\nEstimated Heart Rate (HR) every 5 seconds:")
# for i in range(len(hr_peaks)):
# print("Time:", hr_times.iloc[i], "HR:", hr_data.iloc[hr_peaks[i]])
# print("\nEstimated Respiratory Rate (RR) every 5 seconds:")
# for i in range(len(rr_peaks)):
# print("Time:", rr_times.iloc[i], "RR:", rr_data.iloc[rr_peaks[i]])
|
import numpy as np
import pandas as pd
import json
import os
import cv2
from matplotlib import pyplot as plt
import torch
import torch.nn as nn
import torchvision
import torchvision.transforms as transforms
from torch.utils.data import Dataset, DataLoader
from torchvision import transforms, models
import efficientnet_pytorch
from efficientnet_pytorch import EfficientNet
import warnings
warnings.filterwarnings("ignore")
from distutils.dir_util import copy_tree
# model = '../input/resnet152/'
model = "../input/efficientnet-pytorch/"
checkpoints = "/root/.cache/torch/hub/checkpoints/"
copy_tree(model, checkpoints)
BATCH = 5
EPOCHS = 1
LR = 0.0001
IM_SIZE = 512
DEVICE = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
TRAIN_DIR = "../input/cassava-leaf-disease-classification/train_images/"
TEST_DIR = "../input/cassava-leaf-disease-classification/test_images/"
labels = json.load(
open("../input/cassava-leaf-disease-classification/label_num_to_disease_map.json")
)
print(labels)
train = pd.read_csv("../input/cassava-leaf-disease-classification/train.csv")
train.head()
X_Train, Y_Train = train["image_id"].values, train["label"].values
X_Test = [name for name in (os.listdir(TEST_DIR))]
# X_Train = X_Train[0:1050]
# Y_Train = Y_Train[0:1050]
class GetData(Dataset):
def __init__(self, Dir, FNames, Labels, Transform):
self.dir = Dir
self.fnames = FNames
self.transform = Transform
self.lbs = Labels
def __len__(self):
return len(self.fnames)
def __getitem__(self, index):
image = cv2.imread(os.path.join(self.dir, self.fnames[index]))
image = cv2.cvtColor(image, cv2.COLOR_BGR2LAB)
original = image
new_color = image
lower = np.array([60, 60, 120])
upper = np.array([250, 120, 215])
mask = cv2.inRange(new_color, lower, upper)
result4 = cv2.bitwise_and(original, original, mask=mask)
result4 = cv2.cvtColor(result4, cv2.COLOR_LAB2RGB)
result4[mask == 0] = (255, 255, 255)
lower = np.array([165, 115, 185])
upper = np.array([225, 155, 210])
mask = cv2.inRange(new_color, lower, upper)
result2 = cv2.bitwise_and(original, original, mask=mask)
result2 = cv2.cvtColor(result2, cv2.COLOR_LAB2RGB)
result2[mask == 0] = (255, 255, 255)
lower = np.array([195, 105, 125])
upper = np.array([255, 125, 145])
mask = cv2.inRange(new_color, lower, upper)
result3 = cv2.bitwise_and(original, original, mask=mask)
result3 = cv2.cvtColor(result3, cv2.COLOR_LAB2RGB)
result3[mask == 0] = (255, 255, 255)
lower = np.array([115, 118, 120])
upper = np.array([217, 130, 130])
mask = cv2.inRange(new_color, lower, upper)
result5 = cv2.bitwise_and(original, original, mask=mask)
result5 = cv2.cvtColor(result5, cv2.COLOR_LAB2RGB)
result5[mask == 0] = (255, 255, 255)
result = cv2.bitwise_and(result4, result2)
result = cv2.bitwise_and(result, result3)
image = cv2.bitwise_and(result, result5)
if "train" in self.dir:
image = Transform_train(image=image)["image"]
return image, self.lbs[index]
elif "test" in self.dir:
image = Transform_test(image=image)["image"]
return image, self.fnames[index]
from albumentations import (
HorizontalFlip,
VerticalFlip,
IAAPerspective,
ShiftScaleRotate,
CLAHE,
RandomRotate90,
Transpose,
ShiftScaleRotate,
Blur,
OpticalDistortion,
GridDistortion,
HueSaturationValue,
IAAAdditiveGaussianNoise,
GaussNoise,
MotionBlur,
MedianBlur,
IAAPiecewiseAffine,
RandomResizedCrop,
IAASharpen,
IAAEmboss,
RandomBrightnessContrast,
Flip,
OneOf,
Compose,
Normalize,
Cutout,
CoarseDropout,
ShiftScaleRotate,
CenterCrop,
RandomGridShuffle,
Resize,
)
from albumentations.pytorch import ToTensorV2
Transform_train = Compose(
[
CenterCrop(IM_SIZE, IM_SIZE),
RandomGridShuffle(grid=(2, 2), p=0.5),
Transpose(p=0.5),
HorizontalFlip(p=0.5),
VerticalFlip(p=0.5),
ShiftScaleRotate(p=0.5),
# HueSaturationValue(hue_shift_limit=0.2, sat_shift_limit=0.2, val_shift_limit=0.2, p=0.5),
# RandomBrightnessContrast(brightness_limit=(-0.1,0.1), contrast_limit=(-0.1, 0.1), p=0.5),
Normalize(
mean=[0.485, 0.456, 0.406],
std=[0.229, 0.224, 0.225],
max_pixel_value=255.0,
p=1.0,
),
# CoarseDropout(p=0.5),
# Cutout(p=0.5),
ToTensorV2(p=1.0),
],
p=1.0,
)
Transform_test = Compose(
[
CenterCrop(IM_SIZE, IM_SIZE),
# Resize(IM_SIZE, IM_SIZE),
Normalize(
mean=[0.485, 0.456, 0.406],
std=[0.229, 0.224, 0.225],
max_pixel_value=255.0,
p=1.0,
),
ToTensorV2(p=1.0),
],
p=1.0,
)
model = EfficientNet.from_pretrained("efficientnet-b7", num_classes=5)
model.load_state_dict(torch.load("../input/my-model2/my_model (2)"))
model = model.to(DEVICE)
criterion = nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(model.parameters(), lr=LR)
scheduler = torch.optim.lr_scheduler.StepLR(optimizer, step_size=7, gamma=0.1)
# from sklearn.model_selection import train_test_split
# x_train_idx, x_valid_idx, y_train_idx, y_valid_idx = train_test_split(X_Train,Y_Train, test_size=0.1,shuffle =True)
# print(x_train_idx.shape, y_train_idx.shape)
# print(x_valid_idx.shape, y_valid_idx.shape)
from sklearn.model_selection import StratifiedKFold
n_splits = 5
fold = StratifiedKFold(n_splits=n_splits, shuffle=True)
for trn_idx, val_idx in fold.split(X_Train, Y_Train):
x_train_idx = X_Train[trn_idx]
y_train_idx = Y_Train[trn_idx]
x_valid_idx = X_Train[val_idx]
y_valid_idx = Y_Train[val_idx]
trainset = GetData(TRAIN_DIR, x_train_idx, y_train_idx, Transform_train)
trainloader = DataLoader(trainset, batch_size=BATCH, shuffle=True, num_workers=8)
validset = GetData(TRAIN_DIR, x_valid_idx, y_valid_idx, Transform_test)
validloader = DataLoader(validset, batch_size=BATCH, shuffle=True, num_workers=8)
for epoch in range(EPOCHS):
tr_loss = 0.0
tr_acc = 0.0
val_loss = 0.0
val_acc = 0.0
scheduler.step()
model.train()
for i, (images, labels) in enumerate(trainloader):
images = images.to(DEVICE)
labels = labels.to(DEVICE)
optimizer.zero_grad()
logits = model(images)
loss = criterion(logits, labels)
preds_class = torch.argmax(logits, 1)
loss.backward()
optimizer.step()
tr_loss += loss.detach().item()
tr_acc += (
(preds_class == labels.data).float().mean().data.cpu().numpy()
) # считаем accuracy
with torch.no_grad():
model.eval()
for i, (images, labels) in enumerate(validloader):
images = images.to(DEVICE)
labels = labels.to(DEVICE)
optimizer.zero_grad()
logits = model(images)
loss = criterion(logits, labels)
preds_class = torch.argmax(logits, 1)
val_loss += loss.detach().item()
val_acc += (
(preds_class == labels.data).float().mean().data.cpu().numpy()
) # считаем accuracy
print(
"Epoch: %d | Train_Loss: %.4f | Train_Acc: %.4f | Valid_Loss: %.4f | Valid_Acc: %.4f "
% (
epoch,
tr_loss / len(trainloader),
tr_acc / len(trainloader),
val_loss / len(validloader),
val_acc / len(validloader),
)
)
torch.save(model.state_dict(), "my_model")
testset = GetData(TEST_DIR, X_Test, None, Transform_test)
testloader = DataLoader(testset, batch_size=1, shuffle=False, num_workers=8)
s_ls = []
with torch.no_grad():
model.eval()
for image, fname in testloader:
image = image.to(DEVICE)
logits = model(image)
ps = torch.exp(logits)
_, top_class = ps.topk(1, dim=1)
for pred in top_class:
s_ls.append([fname[0], pred.item()])
sub = pd.DataFrame.from_records(s_ls, columns=["image_id", "label"])
sub.head()
sub.to_csv("submission.csv", index=False)
|
# # Big Mart Sales Prediction(Regression)
# Retail is another industry which extensively uses analytics to optimize business processes. Tasks like product placement, inventory management, customized offers, product bundling, etc. are being smartly handled using data science techniques. As the name suggests, this data comprises of transaction records of a sales store. This is a regression problem. Predict the sales of store
# ## We will explore the problem in following stages:
# ### 1.Hypothesis Generation – understanding the problem better by brainstorming possible factors that can impact the outcome
# ### 2.Data Exploration – looking at categorical and continuous feature summaries and making inferences about the data.
# ### 3.Data Cleaning – imputing missing values in the data and checking for outliers
# ### 4.Feature Engineering – modifying existing variables and creating new ones for analysis
# ### 5.Model Building – making predictive models on the data
import pandas as pd
import numpy as np
# ## Data Exploration
test = pd.read_csv("../input/big-mart-sales-prediction/test_AbJTz2l.csv")
train = pd.read_csv("../input/big-mart-sales-prediction/train_v9rqX0R.csv")
train["source"] = "train"
test["source"] = "test"
df = pd.concat([train, test], ignore_index=True)
print(train.shape, test.shape, df.shape)
# ### Missing Values
df.apply(lambda x: sum(x.isnull()))
df.describe()
df.apply(lambda x: len(x.unique()))
# Filter categorical variables
categorical_columns = [x for x in df.dtypes.index if df.dtypes[x] == "object"]
# Exclude ID cols and source:
categorical_columns = [
x
for x in categorical_columns
if x not in ["Item_Identifier", "Outlet_Identifier", "source"]
]
# Print frequency of categories
for col in categorical_columns:
print("\nFrequency of Categories for varible %s" % col)
print(df[col].value_counts())
# ## Data Cleaning
# ### Imputting Missing Values
# Determine the average weight per item:
item_avg = df.pivot_table(values="Item_Weight", index="Item_Identifier")
# Get a boolean variable specifying missing Item_Weight values
miss = df["Item_Weight"].isnull()
# Impute data and check #missing values before and after imputation to confirm
print("Orignal #missing: %d" % sum(miss))
df.loc[miss, "Item_Weight"] = df.loc[miss, "Item_Identifier"].apply(
lambda x: item_avg.loc[x]
)
print(sum(df["Item_Weight"].isnull()))
# Import mode function:
from scipy.stats import mode
# Determing the mode for each
outlet_size_mode = df.pivot_table(
values="Outlet_Size", columns="Outlet_Type", aggfunc=(lambda x: mode(x).mode[0])
)
print("Mode for each Outlet_Type:")
print(outlet_size_mode)
# Get a boolean variable specifying missing Item_Weight values
miss_bool = df["Outlet_Size"].isnull()
# Impute data and check #missing values before and after imputation to confirm
print("\nOrignal #missing: %d" % sum(miss_bool))
df.loc[miss_bool, "Outlet_Size"] = df.loc[miss_bool, "Outlet_Type"].apply(
lambda x: outlet_size_mode[x]
)
print(sum(df["Outlet_Size"].isnull()))
# ## Feature Engineering
df.pivot_table(values="Item_Outlet_Sales", index="Outlet_Type")
# Determine avg visibility of product (as some has min 0)
avg = df.pivot_table(values="Item_Visibility", index="Item_Identifier")
# impute 0 with avg visibilty of that product
miss = df["Item_Visibility"] == 0
print("Number of 0 values initially: %d" % sum(miss))
df.loc[miss, "Item_Visibility"] = df.loc[miss, "Item_Identifier"].apply(
lambda x: avg.loc[x]
)
print(sum(df["Item_Visibility"] == 0))
# Determine another variable with means ratio
df["Item_Visibility_MeanRatio"] = df.apply(
lambda x: x["Item_Visibility"] / avg.loc[x["Item_Identifier"]], axis=1
)
print(df["Item_Visibility_MeanRatio"].describe())
# Get the first two characters of ID:
df["Item_Type_Combined"] = df["Item_Identifier"].apply(lambda x: x[0:2])
# Rename them to more intuitive categories:
df["Item_Type_Combined"] = df["Item_Type_Combined"].map(
{"FD": "Food", "NC": "Non-Consumable", "DR": "Drinks"}
)
df["Item_Type_Combined"].value_counts()
# years
df["Outlet_Year"] = 2013 - df["Outlet_Establishment_Year"]
df["Outlet_Year"].describe()
# Change categories of low fat:
print("Original Categories:")
print(df["Item_Fat_Content"].value_counts())
print("\nModified Categories:")
df["Item_Fat_Content"] = df["Item_Fat_Content"].replace(
{"LF": "Low Fat", "reg": "Regular", "low fat": "Low Fat"}
)
print(df["Item_Fat_Content"].value_counts())
# Mark non-consumables as separate category in low_fat:
df.loc[df["Item_Type_Combined"] == "Non-Consumable", "Item_Fat_Content"] = "Non-Edible"
df["Item_Fat_Content"].value_counts()
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
# New variable for outlet
df["Outlet"] = le.fit_transform(df["Outlet_Identifier"])
df["Item_Fat_Content"] = le.fit_transform(df["Item_Fat_Content"].astype("bool"))
var_mod = [
"Item_Fat_Content",
"Outlet_Location_Type",
"Outlet_Size",
"Item_Type_Combined",
"Outlet_Type",
"Outlet",
]
le = LabelEncoder()
for i in var_mod:
df[i] = le.fit_transform(df[i])
# One Hot Coding:
df = pd.get_dummies(
df,
columns=[
"Item_Fat_Content",
"Outlet_Location_Type",
"Outlet_Size",
"Outlet_Type",
"Item_Type_Combined",
"Outlet",
],
)
df.dtypes
df.drop(["Item_Type", "Outlet_Establishment_Year"], axis=1, inplace=True)
train = df.loc[df["source"] == "train"]
test = df.loc[df["source"] == "test"]
test.drop(["Item_Outlet_Sales", "source"], axis=1, inplace=True)
train.to_csv("train_modified.csv", index=False)
test.to_csv("test_modified.csv", index=False)
train.drop(["source"], axis=1, inplace=True)
train.dtypes
target = "Item_Outlet_Sales"
IDcol = ["Item_Identifier", "Outlet_Identifier"]
# ## Model Building
# Mean based:
mean_sales = train["Item_Outlet_Sales"].mean()
# Define a dataframe with IDs for submission:
base1 = test[["Item_Identifier", "Outlet_Identifier"]]
base1["Item_Outlet_Sales"] = mean_sales
# Export submission file
base1.to_csv("alg0.csv", index=False)
from sklearn.model_selection import cross_val_score
from sklearn import metrics
def modelfit(alg, dtrain, dtest, predictors, target, IDcol, filename):
# Fit the algorithm on the data
alg.fit(dtrain[predictors], dtrain[target])
# Predict training set:
dtrain_predictions = alg.predict(dtrain[predictors])
# Perform cross-validation:
cv_score = cross_val_score(
alg, dtrain[predictors], dtrain[target], cv=20, scoring="neg_mean_squared_error"
)
cv_score = np.sqrt(np.abs(cv_score))
# Print model report:
print("\nModel Report")
print(
"RMSE : %.4g"
% np.sqrt(metrics.mean_squared_error(dtrain[target].values, dtrain_predictions))
)
print(
"CV Score : Mean - %.4g | Std - %.4g | Min - %.4g | Max - %.4g"
% (np.mean(cv_score), np.std(cv_score), np.min(cv_score), np.max(cv_score))
)
# Predict on testing data:
dtest[target] = alg.predict(dtest[predictors])
# Export submission file:
IDcol.append(target)
submission = pd.DataFrame({x: dtest[x] for x in IDcol})
submission.to_csv(filename, index=False)
# ## Linear Regression
from sklearn.linear_model import LinearRegression, Ridge, Lasso
predictor = [x for x in train.columns if x not in [target] + IDcol]
l = LinearRegression(normalize=True)
modelfit(l, train, test, predictor, target, IDcol, "alg1.csv")
coef1 = pd.Series(l.coef_, predictor).sort_values()
coef1.plot(kind="bar", title="Model Coefficients")
# Ridge Regression
predictors = [x for x in train.columns if x not in [target] + IDcol]
alg2 = Ridge(alpha=0.05, normalize=True)
modelfit(alg2, train, test, predictors, target, IDcol, "alg2.csv")
coef2 = pd.Series(alg2.coef_, predictors).sort_values()
coef2.plot(kind="bar", title="Model Coefficients")
# Decision Tree
from sklearn.tree import DecisionTreeRegressor
predictors = [x for x in train.columns if x not in [target] + IDcol]
alg3 = DecisionTreeRegressor(max_depth=15, min_samples_leaf=100)
modelfit(alg3, train, test, predictors, target, IDcol, "alg3.csv")
coef3 = pd.Series(alg3.feature_importances_, predictors).sort_values(ascending=False)
coef3.plot(kind="bar", title="Feature Importances")
# Decision tree with top 4 variables
predictors = ["Item_MRP", "Outlet_Type_0", "Outlet_5", "Outlet_Type_3"]
alg4 = DecisionTreeRegressor(max_depth=8, min_samples_leaf=150)
modelfit(alg4, train, test, predictors, target, IDcol, "alg4.csv")
coef4 = pd.Series(alg4.feature_importances_, predictors).sort_values(ascending=False)
coef4.plot(kind="bar", title="Feature Importances")
# Random Forest
from sklearn.ensemble import RandomForestRegressor
predictors = [x for x in train.columns if x not in [target] + IDcol]
alg5 = RandomForestRegressor(
n_estimators=200, max_depth=5, min_samples_leaf=100, n_jobs=4
)
modelfit(alg5, train, test, predictors, target, IDcol, "alg5.csv")
coef5 = pd.Series(alg5.feature_importances_, predictors).sort_values(ascending=False)
coef5.plot(kind="bar", title="Feature Importances")
|
# ### O presente documento é uma cópia desse notebook: https://www.kaggle.com/ash316/ml-from-scratch-with-iris
#
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import seaborn as sns
import matplotlib.pyplot as plt
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list the files in the input directory
from subprocess import check_output
print(check_output(["ls", "../input"]).decode("utf8"))
# Any results you write to the current directory are saved as output.
print(check_output(["ls", "../"]).decode("utf8"))
iris = pd.read_csv("../input/iris/Iris.csv") # carregando o conjunto de dados
iris.head(2) # mostra as 2 primeiras linhas do conjunto de dados
iris.info() # verificar se há alguma inconsistência no conjunto de dados
# como vemos, não há valores nulos no conjunto de dados, então os dados podem ser processados
# #### Removendo a coluna desnecessária
iris.drop("Id", axis=1, inplace=True)
# descartando a coluna Id, pois é desnecessário
# axis = 1 especifica que deve ser coluna a ser removida,
# inplace = 1 significa que as alterações devem ser refletidas no dataframe
iris.info()
# ## Algumas análises exploratórias de dados com íris
iris[
iris.Species == "Iris-setosa"
].head() # espécie de select por valor dentro do conjunto de dados
fig = iris[iris.Species == "Iris-setosa"].plot(
kind="scatter", x="SepalLengthCm", y="SepalWidthCm", color="blue", label="Setosa"
)
iris[iris.Species == "Iris-versicolor"].plot(
kind="scatter",
x="SepalLengthCm",
y="SepalWidthCm",
color="red",
label="versicolor",
ax=fig,
) # ax=fig
iris[iris.Species == "Iris-virginica"].plot(
kind="scatter",
x="SepalLengthCm",
y="SepalWidthCm",
color="brown",
label="virginica",
ax=fig,
) # ax=fig
fig.set_xlabel("Sepal Length")
fig.set_ylabel("Sepal Width")
fig.set_title("Sepal Length VS Width")
fig = plt.gcf() # retorna a figura que está em construção
fig.set_size_inches(10, 6)
plt.show()
# O gráfico acima mostra a relação entre o comprimento e a largura da 'Sepal'.
# Agora vamos verificar a relação entre o comprimento e a largura da 'Petal'.
fig = iris[iris.Species == "Iris-setosa"].plot.scatter(
x="PetalLengthCm", y="PetalWidthCm", color="orange", label="Setosa"
)
iris[iris.Species == "Iris-versicolor"].plot.scatter(
x="PetalLengthCm", y="PetalWidthCm", color="blue", label="versicolor", ax=fig
)
iris[iris.Species == "Iris-virginica"].plot.scatter(
x="PetalLengthCm", y="PetalWidthCm", color="green", label="virginica", ax=fig
)
fig.set_xlabel("Petal Length")
fig.set_ylabel("Petal Width")
fig.set_title(" Petal Length VS Width")
fig = plt.gcf()
fig.set_size_inches(10, 6)
plt.show()
# Como podemos ver, os feições pétala (Petal) estão proporcionando uma melhor divisão do cluster em relação aos feições 'Sepal'.
# Esta é uma indicação de que as pétalas podem ajudar em previsões melhores e precisas sobre o 'Sepal'.
# Verificaremos isso mais tarde.
# ### Agora vamos ver como o comprimento e a largura são distribuídos
iris.hist(edgecolor="green", linewidth=1.2)
fig = plt.gcf()
fig.set_size_inches(12, 6)
plt.show()
# ### Agora, o problema dado é um problema de classificação. Portanto, estaremos usando os algoritmos de classificação para construir um modelo.
# **Classification**: as amostras pertencem a duas ou mais classes e queremos aprender com os dados já rotulados como prever a classe de dados não rotulados
# **Regression**: se a saída desejada consistir em uma ou mais variáveis contínuas, a tarefa é chamada de regressão. Um exemplo de problema de regressão seria a previsão do comprimento de um salmão em função de sua idade e peso.
# Antes de começar, precisamos explicar algumas notações de ML.
# **attributes**-->Um atributo é uma propriedade de uma instância que pode ser usada para determinar sua classificação. No conjunto de dados a seguir, os atributos são o comprimento e largura da pétala e sépala. Também é conhecido como **Features**.
# **Target variable**, no contexto de aprendizado de máquina é a variável que é ou deveria ser a saída. Aqui, as variáveis alvo são as 3 espécies de flores.
# importar todos os pacotes necessários para usar os vários algoritmos de classificação
from sklearn.linear_model import LogisticRegression # for Logistic Regression algorithm
from sklearn.model_selection import (
train_test_split,
) # to split the dataset for training and testing
from sklearn.neighbors import KNeighborsClassifier # for K nearest neighbours
from sklearn import svm # for Support Vector Machine (SVM) Algorithm
from sklearn import metrics # for checking the model accuracy
from sklearn.tree import DecisionTreeClassifier # for using Decision Tree Algoithm
iris.shape # obter a forma do conjunto de dados
# Agora, quando treinamos qualquer algoritmo, o número de **features** e sua correlação desempenham um papel importante.
# Se houver **features** e muitos deles estiverem altamente correlacionados, treinar um algoritmo com todos os recursos reduzirá a precisão. Portanto, a seleção de recursos deve ser feita com cuidado.
# Este conjunto de dados tem poucos **features**, mas ainda veremos a correlação.
iris.corr()
plt.figure(figsize=(7, 4))
sns.heatmap(
iris.corr(), annot=True, cmap="cubehelix_r"
) # draws heatmap with input as the correlation matrix calculted by(iris.corr())
plt.show()
# **Observation--->**
# Sepal Width and Length não estão correlacionados
# Petal Width and Length são bem correlacionados
# Usaremos todos os recursos para treinar o algoritmo e verificar a precisão.
# Em seguida, usaremos 1 recurso de pétala e 1 recurso de sépala para verificar a precisão do algoritmo, pois estamos usando apenas 2 recursos que não estão correlacionados. Assim, podemos ter uma variação no conjunto de dados que pode ajudar em uma melhor precisão. Vamos verificar mais tarde.
# ### Steps To Be followed When Applying an Algorithm
# 1. Divida o conjunto de dados em conjunto de dados de treinamento e teste. O conjunto de dados de teste geralmente é menor do que o de treinamento, pois ajudará a treinar melhor o modelo.
# 2. Selecione qualquer algoritmo baseado no problema (classificação ou regressão) o que você achar que pode ser bom.
# 3. Em seguida, passe o conjunto de dados de treinamento ao algoritmo para treiná-lo. Nós usamos o **.fit()** method
# 4. Em seguida, passe os dados de teste para o algoritmo treinado para prever o resultado. Nós usamos o **.predict()** method.
# 5. Em seguida, verificamos a precisão **passando o resultado previsto e a saída real** para o modelo.
# ### Splitting The Data into Training And Testing Dataset
train, test = train_test_split(
iris, test_size=0.3
) # neste, nossos dados principais são divididos em treinar e testar
# o atributo test_size = 0.3 divide os dados em uma proporção de 70% e 30%. treinar = 70% e teste = 30%
print(train.shape)
print(test.shape)
train
# test_y e train_y são os rótulos
# test_X e train_X são os conjuntos de dados em sí
train_X = train[
["SepalLengthCm", "SepalWidthCm", "PetalLengthCm", "PetalWidthCm"]
] # taking the training data features
train_y = train.Species # output of our training data
test_X = test[
["SepalLengthCm", "SepalWidthCm", "PetalLengthCm", "PetalWidthCm"]
] # taking test data features
test_y = test.Species # output value of test data
# Vamos verificar o conjunto de dados de treinamento e teste
test_X.head(2)
train_X.head(2)
train_X.head(2)
train_y.head(2) ##output of the training data
test_X
# ### Support Vector Machine (SVM)
model = svm.SVC() # selecione o algoritmo
model.fit(
train_X, train_y
) # treinamos o algoritmo com os dados de treinamento e a saída do treinamento
prediction = model.predict(
test_X
) # agora passamos os dados de teste para o algoritmo treinado
print("The accuracy of the SVM is:", metrics.classification_report(test_y, prediction))
# print('The precision of the SVM is:',metrics.precision_score(prediction,test_y, average='macro'))
# print('The Recall of the SVM is:',metrics.recall_score(prediction,test_y, average='macro'))
# https://scikit-learn.org/stable/modules/generated/sklearn.metrics.accuracy_score.html
# https://scikit-learn.org/stable/modules/generated/sklearn.metrics.precision_score.html#sklearn.metrics.precision_score
# https://scikit-learn.org/stable/modules/generated/sklearn.metrics.recall_score.html#sklearn.metrics.recall_score
# passamos a saída prevista pelo modelo e a saída real
# SVM está dando uma precisão muito boa. Continuaremos a verificar a precisão de diferentes modelos.
# Agora seguiremos as mesmas etapas acima para treinar vários algoritmos de aprendizado de máquina.
# ### Logistic Regression
model = LogisticRegression()
model.fit(train_X, train_y)
prediction = model.predict(test_X)
print(
"The accuracy of the Logistic Regression is",
metrics.accuracy_score(test_y, prediction),
)
# ### Decision Tree
model = DecisionTreeClassifier()
model.fit(train_X, train_y)
prediction = model.predict(test_X)
print(
"The accuracy of the Decision Tree is", metrics.accuracy_score(test_y, prediction)
)
# ### K-Nearest Neighbours
model = KNeighborsClassifier(
n_neighbors=10
) # this examines 3 neighbours for putting the new data into a class
model.fit(train_X, train_y)
prediction = model.predict(test_X)
print("The accuracy of the KNN is", metrics.accuracy_score(test_y, prediction))
# ### Usamos todos os recursos da íris nos modelos acima. Agora vamos usar pétalas e sépalas separadamente
# ### Criação de dados de treinamento de pétalas e sépalas
petal = iris[["PetalLengthCm", "PetalWidthCm", "Species"]]
sepal = iris[["SepalLengthCm", "SepalWidthCm", "Species"]]
train_p, test_p = train_test_split(petal, test_size=0.3, random_state=0) # petals
train_x_p = train_p[["PetalWidthCm", "PetalLengthCm"]]
train_y_p = train_p.Species
test_x_p = test_p[["PetalWidthCm", "PetalLengthCm"]]
test_y_p = test_p.Species
train_s, test_s = train_test_split(sepal, test_size=0.3, random_state=0) # Sepal
train_x_s = train_s[["SepalWidthCm", "SepalLengthCm"]]
train_y_s = train_s.Species
test_x_s = test_s[["SepalWidthCm", "SepalLengthCm"]]
test_y_s = test_s.Species
# ### SVM
model = svm.SVC()
model.fit(train_x_p, train_y_p)
prediction = model.predict(test_x_p)
print(
"The accuracy of the SVM using Petals is:",
metrics.accuracy_score(test_y_p, prediction),
)
model = svm.SVC()
model.fit(train_x_s, train_y_s)
prediction = model.predict(test_x_s)
print(
"The accuracy of the SVM using Sepal is:",
metrics.accuracy_score(test_y_s, prediction),
)
# ### Logistic Regression
model = LogisticRegression()
model.fit(train_x_p, train_y_p)
prediction = model.predict(test_x_p)
print(
"The accuracy of the Logistic Regression using Petals is:",
metrics.accuracy_score(
test_y_p,
prediction,
),
)
model.fit(train_x_s, train_y_s)
prediction = model.predict(test_x_s)
print(
"The accuracy of the Logistic Regression using Sepals is:",
metrics.accuracy_score(test_y_s, prediction),
)
# ### Decision Tree
model = DecisionTreeClassifier()
model.fit(train_x_p, train_y_p)
prediction = model.predict(test_x_p)
print(
"The accuracy of the Decision Tree using Petals is:",
metrics.accuracy_score(test_y_p, prediction),
)
model.fit(train_x_s, train_y_s)
prediction = model.predict(test_x_s)
print(
"The accuracy of the Decision Tree using Sepals is:",
metrics.accuracy_score(test_y_s, prediction),
)
# ### K-Nearest Neighbours
model = KNeighborsClassifier(n_neighbors=3)
model.fit(train_x_p, train_y_p)
prediction = model.predict(test_x_p)
print(
"The accuracy of the KNN using Petals is:",
metrics.accuracy_score(
test_y_p,
prediction,
),
)
model.fit(train_x_s, train_y_s)
prediction = model.predict(test_x_s)
print(
"The accuracy of the KNN using Sepals is:",
metrics.accuracy_score(test_y_s, prediction),
)
# ### Observações:
# - Usar pétalas sobre sépala para treinar os dados dá uma precisão muito melhor.
# - Isso era esperado, pois vimos no mapa de calor acima, que a correlação entre a largura e o comprimento da sépala era muito baixa, enquanto a correlação entre a largura e o comprimento da pétala era muito alta.
# Assim, acabamos de implementar algumas das ferramentas de aprendizado de máquina comuns. Como o conjunto de dados é pequeno, com poucos recursos, não abordei alguns conceitos, pois seriam relevantes quando temos muitos recursos.
ls
pwd
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import re
import matplotlib.pyplot as plt
from nltk.tokenize import word_tokenize
from nltk.corpus import stopwords
from unicodedata import normalize
from sklearn.model_selection import train_test_split
from nltk.stem import SnowballStemmer
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
def csv_len(data):
words = []
letters = []
sentiments = []
tweets = []
for index, tweet in data.iterrows():
tweet_split = tweet.Tweet.split()
sentiments.append(tweet.Sentiment)
tweets.append(tweet.Tweet)
letters.append(len(tweet.Tweet))
words.append(len(tweet_split))
data["Tweet"] = tweets
data["Sentiment"] = sentiments
data["Words"] = words
data["Letters"] = letters
return data
def preprocessing(data):
tweets = []
sentiment = []
for index, tweet in data.iterrows():
words_cleaned = ""
tweet_clean = tweet.Tweet.lower()
words_cleaned = " ".join(
[
word
for word in tweet_clean.split()
if "http://" not in word
and "https://" not in word
and ".com" not in word
and ".es" not in word
and not word.startswith("@")
and not word.startswith("#")
and word != "rt"
]
)
tweet_clean = re.sub(r"\b([jh]*[aeiou]*[jh]+[aeiou]*)*\b", "", words_cleaned)
tweet_clean = re.sub(r"(.)\1{2,}", r"\1", tweet_clean)
tweet_clean = re.sub(
r"([^n\u0300-\u036f]|n(?!\u0303(?![\u0300-\u036f])))[\u0300-\u036f]+",
r"\1",
normalize("NFD", tweet_clean),
0,
re.I,
)
tweet_clean = re.sub("[^a-zA-Z]", " ", tweet_clean)
tweet_clean = re.sub("\t", " ", tweet_clean)
tweet_clean = re.sub(" +", " ", tweet_clean)
tweet_clean = re.sub("^ ", "", tweet_clean)
tweet_clean = re.sub(" $", "", tweet_clean)
tweet_clean = re.sub("\n", "", tweet_clean)
words_cleaned = ""
stemmed = ""
stop_words = set(stopwords.words("english"))
stemmer = SnowballStemmer("english")
tokens = word_tokenize(tweet_clean)
words_cleaned = [
word for word in tokens if not word in stop_words and len(word) > 2
]
stemmed = " ".join([stemmer.stem(word) for word in words_cleaned])
sentiment.append(tweet.Sentiment)
tweets.append(stemmed)
data["Tweet"] = tweets
data["Sentiment"] = sentiment
data.loc[:, ["Sentiment", "Tweet"]]
return data
c_df = pd.read_csv(
"../input/covid-19-nlp-text-classification/Corona_NLP_train.csv", encoding="latin-1"
)
c_df
header_names = [
"UserName",
"ScreenName",
"Location",
"TweetAt",
"Tweet",
"Sentiment",
"Sentiment Score",
]
training_c = pd.read_csv(
"../input/covid-19-nlp-text-classification/Corona_NLP_train.csv",
header=None,
names=header_names,
encoding="latin-1",
)
training_c = training_c.iloc[1:]
training_c = training_c.reset_index()
data_corona_train = training_c.drop(
["UserName", "ScreenName", "Location", "TweetAt", "Sentiment Score", "index"],
axis=1,
)
data_corona_train
header_names = [
"UserName",
"ScreenName",
"Location",
"TweetAt",
"Tweet",
"Sentiment",
"Sentiment Score",
]
testing_c = pd.read_csv(
"../input/covid-19-nlp-text-classification/Corona_NLP_test.csv",
header=None,
names=header_names,
)
data_corona_test = testing_c.drop(
["UserName", "ScreenName", "Location", "TweetAt", "Sentiment Score"], axis=1
)
data_corona_test
data_corona_train.Sentiment.value_counts()
data_corona_test.Sentiment.value_counts()
data_corona_train.drop(
data_corona_train[data_corona_train["Sentiment"] == "Neutral"].index, inplace=True
)
data_corona_train.drop(
data_corona_train[data_corona_train["Sentiment"] == "Sentiment"].index, inplace=True
)
# data_corona_train.drop(data_corona_train[data_corona_train['Sentiment'] == 'Extremely Positive'].index, inplace = True)
# data_corona_train.drop(data_corona_train[data_corona_train['Sentiment'] == 'Extremely Negative'].index, inplace = True)
data_corona_train["Sentiment"] = data_corona_train["Sentiment"].replace(
["Extremely Positive", "Extremely Negative"], ["Positive", "Negative"]
)
data_corona_train["Sentiment"] = data_corona_train["Sentiment"].replace(
["Positive", "Negative"], [4, 0]
)
data_corona_train
data_corona_train.Sentiment.value_counts()
data_corona_test.drop(
data_corona_test[data_corona_test["Sentiment"] == "Neutral"].index, inplace=True
)
data_corona_test.drop(
data_corona_test[data_corona_test["Sentiment"] == "Sentiment"].index, inplace=True
)
# data_corona_test.drop(data_corona_test[data_corona_test['Sentiment'] == 'Extremely Positive'].index, inplace = True)
# data_corona_test.drop(data_corona_test[data_corona_test['Sentiment'] == 'Extremely Negative'].index, inplace = True)
data_corona_test["Sentiment"] = data_corona_test["Sentiment"].replace(
["Extremely Positive", "Extremely Negative"], ["Positive", "Negative"]
)
data_corona_test["Sentiment"] = data_corona_test["Sentiment"].replace(
["Positive", "Negative"], [4, 0]
)
data_corona_test
data_corona_test.Sentiment.value_counts()
data_corona_train = csv_len(data_corona_train)
data_corona_train
train_corona_clean = preprocessing(data_corona_train)
train_corona_clean
train_corona_clean = csv_len(train_corona_clean)
train_corona_clean
train_corona_cleaned = train_corona_clean.loc[:, ["Tweet", "Sentiment"]]
data_corona_test = csv_len(data_corona_test)
data_corona_test
test_corona_clean = preprocessing(data_corona_test)
test_corona_clean
test_corona_clean = csv_len(test_corona_clean)
test_corona_clean
test_corona_cleaned = test_corona_clean.loc[:, ["Tweet", "Sentiment"]]
frames = [train_corona_cleaned, test_corona_cleaned]
result_corona = pd.concat(frames)
result_corona
result_corona = result_corona.dropna().reset_index()
result_corona
result_corona_def = result_corona.loc[:, ["Tweet", "Sentiment"]]
result_corona_def
result_corona_def.to_csv(
"corona-entity-sentiment-analysis-with-stemming.csv", index=False
)
|
# # シンプルな11位の解法【詳細】
# # very simple 11th place solution【Details】
# このNotebookではPseudo Labeling(疑似ラベリング)について説明しています。11位の解法の概要についてはこちらのNotebookを参照してください。
# 【日本語&English】TPS Feb 11th place solution
# https://www.kaggle.com/maostack/english-tps-feb-11th-place-solution
#
# This Notebook describes Pseudo Labeling, see this Notebook for an overview of the 11th solution.
# 【日本語&English】TPS Feb 11th place solution
# https://www.kaggle.com/maostack/english-tps-feb-11th-place-solution
# ## 疑似ラベリング / Pseudo Labeling
# 半教師あり学習の手法の一つ / One of the methods of semi-supervised learning
#
# 疑似ラベリングは2段階の構成になっている。
# まず、何らかの予測モデルを用意する(今回はLightGBM)。
# 第1段階では、モデルを学習させた後、普通にテストデータに対して予測を行う。その予測値をテストデータに対する疑似ラベルとする。つまり、テストデータに対する予測値を疑似的に目的変数(label・target)として扱う。
# 第2段階では、"もともとの学習データにテストデータを合体させたもの"を学習データとして用いて、テストデータに対する予測を行う。
#
# The pseudo labeling consists of two steps.
# First, prepare some kind of prediction model (LightGBM in this case).
# In the 1st stage, we train the model and make a prediction for the test data. The predicted value is used as a pseudo-label for the test data. In other words, the predicted value for the test data is treated as a pseudo target variable (label/target).
# In the 2nd stage, predictions are made for the test data using the "original training data combined with the test data" as the training data.
import os
import numpy as np
import pandas as pd
import warnings
warnings.simplefilter("ignore")
# 別のNotebookで既に前処理をしたデータをtrain, testとして読み込んでいます。
# 前処理として行ったことは、
# ・targetが外れ値の行を除外(targetが4より小さい行を除外)
# ・変数"cat6"について"G"は学習データにしか存在しない(テストデータで値がGをとるデータが存在しない)ので、cat6の値がGの行を除外
# ・カテゴリ変数に対するLabel Encoding
# ・cont列に対するRankGauss変換
# です。最後のRankGauss変換は、決定木系のモデルには影響を与えないのでしなくてもいいのですが一応しておきました。
# 除外後の学習データのデータ数は299963になりました。37行減った。テストデータの数は変わっていない。
#
# The data that has already been preprocessed in another Notebook is loaded as train and test.
# What we did as preprocessing was
# ・Exclude rows where target is an outlier (exclude rows where target is less than 4)
# ・For the variable "cat6", "G" exists only in the training data (there is no data that takes the value G in the test data), so the line with the value G in cat6 is excluded.
# ・Label Encoding for categorical variables
# ・RankGauss transform for cont columns
# The RankGauss transformation is not necessary because it does not affect the decision tree model, but I did it just in case.
# After preprocessing, the number of data in the training data is now 299963. 37 rows have been reduced. The number of test data has not changed.
train = pd.read_csv("../input/tps-feb-eda-fe/train_data.csv")
test = pd.read_csv("../input/tps-feb-eda-fe/test_data.csv")
train
test
cat_columns = [f"cat{i}" for i in range(10)]
X = train.drop(["target"], axis=1)
X_test = test
y = train.target
print(X.shape)
print(X_test.shape)
print(y.shape)
X
X_test
y
from sklearn.model_selection import KFold
import lightgbm as lgb
SEED = 8970365
kf = KFold(n_splits=5, shuffle=True, random_state=SEED)
# パラメータの値は他の方のNotebookを参考にしました。感謝します。
# The value of the parameter was taken from another person's Notebook.
# I appreciate it.
params_lgb = {
"task": "train",
"boosting_type": "gbdt",
"objective": "regression",
"metric": "rmse",
"learning_rate": 0.003899156646724397,
"num_leaves": 63,
"max_depth": 99,
"feature_fraction": 0.2256038826485174,
"bagging_fraction": 0.8805303688019942,
"min_child_samples": 290,
"reg_alpha": 9.562925363678952,
"reg_lambda": 9.355810045480153,
"max_bin": 882,
"min_data_per_group": 127,
"bagging_freq": 1,
"cat_smooth": 96,
"cat_l2": 19,
"verbosity": -1,
"bagging_seed": SEED,
"feature_fraction_seed": SEED,
"seed": SEED,
}
# まず普通にテストデータに対して予測を行う。
# First, make a prediction for the test data as usual.
# 予測値を格納するdf
# df to store the predicted value
preds_lgb = pd.DataFrame()
X[cat_columns] = X[cat_columns].astype("category")
X_test[cat_columns] = X_test[cat_columns].astype("category")
for k, (tr_id, vl_id) in enumerate(kf.split(X, y)):
print("=" * 50)
print(f" KFold{k+1}")
print("=" * 50)
X_train, X_val = X.iloc[tr_id, :], X.iloc[vl_id, :]
y_train, y_val = y.iloc[tr_id], y.iloc[vl_id]
lgb_train = lgb.Dataset(X_train, y_train)
lgb_val = lgb.Dataset(X_val, y_val)
model_lgb = lgb.train(
params=params_lgb,
train_set=lgb_train,
valid_sets=lgb_val,
num_boost_round=100000,
early_stopping_rounds=200,
verbose_eval=1000,
)
pred_lgb = model_lgb.predict(X_test, num_iteration=model_lgb.best_iteration)
pred_lgb = pd.DataFrame(pred_lgb)
# 予測値を横に連結していく
# Concatenate the predictions horizontally
preds_lgb = pd.concat([preds_lgb, pred_lgb], axis=1)
preds_lgb
# 平均を計算して、テストデータに対する疑似ラベルとする
# Calculate the mean and use it as a pseudo labels for the test data
label = preds_lgb.mean(axis=1)
label
# もともとの学習データX, yにテストデータと疑似ラベルを縦に連結する。
# これを新たな学習データとする
# Concatenate the test data and pseudo labels to the original training data X, y.
# Make this the new training data.
X = pd.concat([X, X_test], axis=0).reset_index(drop=True)
y = pd.concat([y, label], axis=0).reset_index(drop=True)
print("X.shape: ", X.shape)
print("y.shape: ", y.shape)
X
y
# 最終予測値を格納するdf
# df to store the final prediction
preds_lgb = pd.DataFrame()
X[cat_columns] = X[cat_columns].astype("category")
X_test[cat_columns] = X_test[cat_columns].astype("category")
for k, (tr_id, vl_id) in enumerate(kf.split(X, y)):
print("=" * 50)
print(f" KFold{k+1}")
print("=" * 50)
X_train, X_val = X.iloc[tr_id, :], X.iloc[vl_id, :]
y_train, y_val = y.iloc[tr_id], y.iloc[vl_id]
lgb_train = lgb.Dataset(X_train, y_train)
lgb_val = lgb.Dataset(X_val, y_val)
model_lgb = lgb.train(
params=params_lgb,
train_set=lgb_train,
valid_sets=lgb_val,
num_boost_round=100000,
early_stopping_rounds=200,
verbose_eval=1000,
)
pred_lgb = model_lgb.predict(X_test, num_iteration=model_lgb.best_iteration)
pred_lgb = pd.DataFrame(pred_lgb)
preds_lgb = pd.concat([preds_lgb, pred_lgb], axis=1)
# # Submission
submission = pd.read_csv(
"../input/tabular-playground-series-feb-2021/sample_submission.csv"
)
# 予測値の平均を計算して、最終的な予測値とする
# Calculate the average of the predictions to get the final prediction.
pred = preds_lgb.mean(axis=1)
submission.target = pred
submission.head()
submission.to_csv("submission_pseudo_lgb_5.csv", index=False)
|
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import tensorflow as tf
# tf.enable_eager_execution()
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
import unicodedata
import re
import numpy as np
import time
import string
import chart_studio.plotly
import chart_studio.plotly as py
from plotly.offline import init_notebook_mode, iplot
# %plotly.offline.init_notebook_mode(connected=True)
import plotly.graph_objs as go
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# ### As in case of any NLP task, after reading the input file, we perform the basic cleaning and preprocessing as follows:
# **The Dataset :** We need a dataset that contains English sentences and their Portuguese translations which can be freely downloaded from this [link](http://www.manythings.org/anki/). Download the file fra-eng.zip and extract it. On each line, the text file contains an English sentence and its French translation, separated by a tab.
file_path = "/kaggle/input/cheryl-fr-en/news_en_fr.txt" # please set the path according to your system
lines = open(file_path, encoding="UTF-8").read().strip().split("\n")
lines[5000:5010]
print("total number of records: ", len(lines))
exclude = set(string.punctuation) # Set of all special characters
remove_digits = str.maketrans("", "", string.digits) # Set of all digits
# ### Function to preprocess English sentence
def preprocess_eng_sentence(sent):
"""Function to preprocess English sentence"""
sent = sent.lower() # lower casing
sent = re.sub("'", "", sent) # remove the quotation marks if any
sent = "".join(ch for ch in sent if ch not in exclude)
sent = sent.translate(remove_digits) # remove the digits
sent = sent.strip()
sent = re.sub(" +", " ", sent) # remove extra spaces
sent = "<start> " + sent + " <end>" # add <start> and <end> tokens
return sent
# ### Function to preprocess Portuguese sentence
def preprocess_port_sentence(sent):
"""Function to preprocess Portuguese sentence"""
sent = re.sub("'", "", sent) # remove the quotation marks if any
sent = "".join(ch for ch in sent if ch not in exclude)
# sent = re.sub("[२३०८१५७९४६]", "", sent) # remove the digits
sent = sent.strip()
sent = re.sub(" +", " ", sent) # remove extra spaces
sent = "<start> " + sent + " <end>" # add <start> and <end> tokens
return sent
# ### Generate pairs of cleaned English and Portuguese sentences with start and end tokens added.
# Generate pairs of cleaned English and Portuguese sentences
sent_pairs = []
for line in lines:
sent_pair = []
eng = line.rstrip().split("\t")[1]
port = line.rstrip().split("\t")[0]
port = preprocess_port_sentence(port)
sent_pair.append(port)
eng = preprocess_eng_sentence(eng)
sent_pair.append(eng)
sent_pairs.append(sent_pair)
sent_pairs = [pair for pair in sent_pairs if len(pair[0].split()) <= 40]
sent_pairs = sent_pairs[:100000]
sent_pairs[5000:5010], len(sent_pairs)
# ### Create a class to map every word to an index and vice-versa for any given vocabulary.
# This class creates a word -> index mapping (e.g,. "dad" -> 5) and vice-versa
# (e.g., 5 -> "dad") for each language,
class LanguageIndex:
def __init__(self, lang):
self.lang = lang
self.word2idx = {}
self.idx2word = {}
self.vocab = set()
self.create_index()
def create_index(self):
for phrase in self.lang:
self.vocab.update(phrase.split(" "))
self.vocab = sorted(self.vocab)
self.word2idx["<pad>"] = 0
for index, word in enumerate(self.vocab):
self.word2idx[word] = index + 1
for word, index in self.word2idx.items():
self.idx2word[index] = word
def max_length(tensor):
return max(len(t) for t in tensor)
# ### Tokenization and Padding
def load_dataset(pairs, num_examples):
# pairs => already created cleaned input, output pairs
# index language using the class defined above
inp_lang = LanguageIndex(en for en, ma in pairs)
targ_lang = LanguageIndex(ma for en, ma in pairs)
# Vectorize the input and target languages
# English sentences
input_tensor = [[inp_lang.word2idx[s] for s in en.split(" ")] for en, ma in pairs]
# Marathi sentences
target_tensor = [[targ_lang.word2idx[s] for s in ma.split(" ")] for en, ma in pairs]
# Calculate max_length of input and output tensor
# Here, we'll set those to the longest sentence in the dataset
max_length_inp, max_length_tar = max_length(input_tensor), max_length(target_tensor)
# Padding the input and output tensor to the maximum length
input_tensor = tf.keras.preprocessing.sequence.pad_sequences(
input_tensor, maxlen=max_length_inp, padding="post"
)
target_tensor = tf.keras.preprocessing.sequence.pad_sequences(
target_tensor, maxlen=max_length_tar, padding="post"
)
return (
input_tensor,
target_tensor,
inp_lang,
targ_lang,
max_length_inp,
max_length_tar,
)
(
input_tensor,
target_tensor,
inp_lang,
targ_lang,
max_length_inp,
max_length_targ,
) = load_dataset(sent_pairs, len(lines))
# ### Creating training and validation sets using an 80-20 split
# Creating training and validation sets using an 80-20 split
# input_tensor_train, input_tensor_test, target_tensor_train, target_tensor_test = train_test_split(input_tensor, target_tensor, test_size=0.3, random_state = 101)
# Show length
# len(input_tensor_train), len(target_tensor_train), len(input_tensor_test), len(target_tensor_test)
# Split the data into training set, validation set, and test set
(
input_tensor_train,
input_tensor_val_test,
target_tensor_train,
target_tensor_val_test,
) = train_test_split(input_tensor, target_tensor, test_size=0.2, random_state=101)
(
input_tensor_val,
input_tensor_test,
target_tensor_val,
target_tensor_test,
) = train_test_split(
input_tensor_val_test, target_tensor_val_test, test_size=0.5, random_state=101
)
# Show length of each dataset
print("Training set length:", len(input_tensor_train))
print("Validation set length:", len(input_tensor_val))
print("Test set length:", len(input_tensor_test))
BUFFER_SIZE = len(input_tensor_train)
BATCH_SIZE = 64
N_BATCH = BUFFER_SIZE // BATCH_SIZE
embedding_dim = 256
units = 512
vocab_inp_size = len(inp_lang.word2idx)
vocab_tar_size = len(targ_lang.word2idx)
# dataset = tf.data.Dataset.from_tensor_slices((input_tensor_train, target_tensor_train)).shuffle(BUFFER_SIZE)
# dataset = dataset.batch(BATCH_SIZE, drop_remainder=True)
# 从原始数据集中选择前10000个元素,并创建一个新的数据集
dataset = tf.data.Dataset.from_tensor_slices((input_tensor_train, target_tensor_train))
# 打乱数据,并按批次输出
dataset = dataset.shuffle(BUFFER_SIZE).batch(BATCH_SIZE, drop_remainder=True)
# We'll be using GRUs instead of LSTMs as we only have to create one state and implementation would be easier.
# ### Create GRU units
def gru(units):
return tf.keras.layers.GRU(
units,
return_sequences=True,
return_state=True,
recurrent_activation="sigmoid",
recurrent_initializer="glorot_uniform",
)
# ### The next step is to define the encoder and decoder network.
# The input to the encoder will be the sentence in English and the output will be the hidden state and cell state of the GRU.
from tensorflow_probability import distributions as tfd
class Encoder(tf.keras.Model):
def __init__(self, vocab_size, embedding_dim, enc_units, batch_sz):
super(Encoder, self).__init__()
self.batch_sz = batch_sz
self.enc_units = enc_units
self.embedding = tf.keras.layers.Embedding(vocab_size, embedding_dim)
self.gru = gru(self.enc_units)
def call(self, x, hidden):
x = self.embedding(x)
output, state = self.gru(x, initial_state=hidden)
return output, state
def initialize_hidden_state(self):
return tf.zeros((self.batch_sz, self.enc_units))
# The next step is to define the decoder. The decoder will have two inputs: the hidden state and cell state from the encoder and the input sentence, which actually will be the output sentence with a token appended at the beginning.
import numpy as np
import tensorflow_probability as tfp
import tensorflow_addons as tfa
import numpy as np
import tensorflow as tf
def gumbel_softmax(o_t, temperature, eps=1e-20):
"""Sample from Gumbel(0, 1)"""
u = tf.random.uniform(tf.shape(o_t), minval=0, maxval=1)
g_t = -tf.math.log(-tf.math.log(u + eps) + eps)
gumbel_t = tf.math.add(o_t, g_t)
return tf.math.multiply(gumbel_t, temperature)
class Decoder(tf.keras.Model):
def __init__(
self,
vocab_size,
embedding_dim,
dec_units,
batch_sz,
gumbel=False,
gumbel_temp=0.5,
use_attention=True,
):
super(Decoder, self).__init__()
self.batch_sz = batch_sz
self.dec_units = dec_units
self.embedding = tf.keras.layers.Embedding(vocab_size, embedding_dim)
self.gru = gru(self.dec_units)
if gumbel:
self.fc = tf.keras.layers.Dense(
vocab_size,
activation=(lambda x: gumbel_softmax(x, temperature=gumbel_temp)),
)
else:
self.fc = tf.keras.layers.Dense(vocab_size)
self.use_attention = use_attention
# used for attention
if self.use_attention:
self.W1 = tf.keras.layers.Dense(self.dec_units)
self.W2 = tf.keras.layers.Dense(self.dec_units)
self.V = tf.keras.layers.Dense(1)
def call(self, x, hidden, enc_output, temperature=1.0):
if self.use_attention:
hidden_with_time_axis = tf.expand_dims(hidden, 1)
score = self.V(
tf.nn.tanh(self.W1(enc_output) + self.W2(hidden_with_time_axis))
)
attention_weights = tf.nn.softmax(score, axis=1)
context_vector = attention_weights * enc_output
context_vector = tf.reduce_sum(context_vector, axis=1)
x = self.embedding(x)
x = tf.concat([tf.expand_dims(context_vector, 1), x], axis=-1)
else:
x = self.embedding(x)
output, state = self.gru(x)
output = tf.reshape(output, (-1, output.shape[2]))
x = self.fc(output)
return x, state, attention_weights if self.use_attention else None
# Create encoder and decoder objects from their respective classes.
encoder = Encoder(vocab_inp_size, embedding_dim, units, BATCH_SIZE)
decoder = Decoder(
vocab_tar_size,
embedding_dim,
units,
BATCH_SIZE,
gumbel=False,
gumbel_temp=1.0,
use_attention=True,
)
# ### Define the optimizer and the loss function.
optimizer = tf.optimizers.Adam()
def loss_function(real, pred):
mask = 1 - np.equal(real, 0)
loss_ = (
tf.nn.sparse_softmax_cross_entropy_with_logits(labels=real, logits=pred) * mask
)
return tf.reduce_mean(loss_)
checkpoint_dir = "./training_checkpoints"
checkpoint_prefix = os.path.join(checkpoint_dir, "ckpt")
checkpoint = tf.train.Checkpoint(optimizer=optimizer, encoder=encoder, decoder=decoder)
# ### Training the Model
EPOCHS = 10
for epoch in range(EPOCHS):
start = time.time()
hidden = encoder.initialize_hidden_state()
total_loss = 0
for batch, (inp, targ) in enumerate(dataset):
loss = 0
with tf.GradientTape() as tape:
enc_output, enc_hidden = encoder(inp, hidden)
dec_hidden = enc_hidden
dec_input = tf.expand_dims([targ_lang.word2idx["<start>"]] * BATCH_SIZE, 1)
# Teacher forcing - feeding the target as the next input
for t in range(1, targ.shape[1]):
# passing enc_output to the decoder
predictions, dec_hidden, _ = decoder(dec_input, dec_hidden, enc_output)
loss += loss_function(targ[:, t], predictions)
# using teacher forcing
dec_input = tf.expand_dims(targ[:, t], 1)
batch_loss = loss / int(targ.shape[1])
total_loss += batch_loss
variables = encoder.variables + decoder.variables
gradients = tape.gradient(loss, variables)
optimizer.apply_gradients(zip(gradients, variables))
if batch % 100 == 0:
print(
"Epoch {} Batch {} Loss {:.4f}".format(
epoch + 1, batch, batch_loss.numpy()
)
)
# saving (checkpoint) the model every epoch
checkpoint.save(file_prefix=checkpoint_prefix)
print("Epoch {} Loss {:.4f}".format(epoch + 1, total_loss / N_BATCH))
print("Time taken for 1 epoch {} sec\n".format(time.time() - start))
# ### Restoring the latest checkpoint
# restoring the latest checkpoint in checkpoint_dir
checkpoint.restore(tf.train.latest_checkpoint(checkpoint_dir))
# Save the model
encoder.save_weights("/kaggle/working/encoder_weights.h5")
decoder.save_weights("/kaggle/working/decoder_weights.h5")
# ### Inference setup and testing:
# ### Function to predict (translate) a randomly selected test point
#
from nltk.translate.bleu_score import sentence_bleu
from nltk.translate.bleu_score import SmoothingFunction
def evaluate_sentence(sentence, max_length_inp, max_length_targ):
inputs = [inp_lang.word2idx[i] for i in sentence.split(" ")]
inputs = tf.keras.preprocessing.sequence.pad_sequences(
[inputs], maxlen=max_length_inp, padding="post"
)
inputs = tf.convert_to_tensor(inputs)
result = ""
hidden = [tf.zeros((1, units))]
enc_out, enc_hidden = encoder(inputs, hidden)
dec_hidden = enc_hidden
dec_input = tf.expand_dims([targ_lang.word2idx["<start>"]], 0)
for t in range(max_length_targ):
predictions, dec_hidden, _ = decoder(dec_input, dec_hidden, enc_out)
predicted_id = tf.argmax(predictions[0]).numpy()
result += targ_lang.idx2word[predicted_id] + " "
if targ_lang.idx2word[predicted_id] == "<end>":
return result, sentence
dec_input = tf.expand_dims([predicted_id], 0)
return result, sentence
def evaluate_bleu(reference, candidate):
smoothie = SmoothingFunction().method4
reference = [reference.split()]
candidate = candidate.split()
score = sentence_bleu(reference, candidate, smoothing_function=smoothie)
return score
def evaluate_translation(sentence):
result, input_sentence = evaluate_sentence(
sentence, max_length_inp, max_length_targ
)
return result
total_bleu_score = 0
num_examples = 20
for i in range(num_examples):
input_sentence = " ".join(
[inp_lang.idx2word[w] for w in input_tensor_test[i] if w != 0]
)
target_sentence = " ".join(
[targ_lang.idx2word[w] for w in target_tensor_test[i] if w != 0]
)
predicted_sentence = evaluate_translation(input_sentence)
# 为 predicted_sentence 加上 <start> 标记
predicted_sentence = "<start> " + predicted_sentence
bleu_score = evaluate_bleu(target_sentence, predicted_sentence)
total_bleu_score += bleu_score
print("Input: %s" % (input_sentence))
print("Target translation: {}".format(target_sentence))
print("Predicted translation: {}".format(predicted_sentence))
print("BLEU score: {:.2f}\n".format(bleu_score))
print("Average BLEU score: {:.2f}".format(total_bleu_score / num_examples))
import numpy as np
from nltk.translate.bleu_score import sentence_bleu, SmoothingFunction
def get_length_category(length):
if 0 <= length <= 5:
return 0
elif 6 <= length <= 10:
return 1
elif 11 <= length <= 15:
return 2
elif 16 <= length <= 20:
return 3
elif 21 <= length <= 25:
return 4
elif 26 <= length <= 30:
return 5
elif 31 <= length <= 35:
return 6
elif 36 <= length <= 40:
return 7
else:
return 8
num_examples = len(input_tensor_test)
weights = (0.25, 0.25, 0.25, 0.25)
smoothing = SmoothingFunction()
bleu_scores_by_length = [0] * 9
num_examples_by_length = [0] * 9
for i in range(num_examples):
input_sentence = " ".join(
[inp_lang.idx2word[w] for w in input_tensor_test[i] if w != 0]
)
target_sentence = " ".join(
[targ_lang.idx2word[w] for w in target_tensor_test[i] if w != 0]
)
predicted_sentence = evaluate_translation(input_sentence)
target_tokens = target_sentence.split()
predicted_tokens = predicted_sentence.split()
predicted_sentence = "<start> " + predicted_sentence
predicted_tokens = predicted_sentence.split()
bleu_score = sentence_bleu(
[target_tokens],
predicted_tokens,
weights=weights,
smoothing_function=SmoothingFunction().method4,
)
input_length = len(input_sentence.split())
length_category = get_length_category(input_length)
bleu_scores_by_length[length_category] += bleu_score
num_examples_by_length[length_category] += 1
for i, num_examples in enumerate(num_examples_by_length):
print(f"Number of sentence pairs for length category {i}: {num_examples}")
for i, (total_bleu_score, num_examples) in enumerate(
zip(bleu_scores_by_length, num_examples_by_length)
):
if num_examples != 0:
avg_bleu_score = total_bleu_score / num_examples
print(f"Average BLEU score for length category {i}: {avg_bleu_score:.2f}")
|
# # Project Overview
# The objective of this project was to analyze the electricity consumption per person data, which provides an estimate of power plant production minus losses, using time series analysis. The data was obtained from the World Bank and the International Energy Agency and covers the period from 1971 to 2014.
# ## Methodology
# Python was used for all the analysis steps, including data cleaning, transformation, and modeling. Initially, an Augmented Dickey-Fuller (ADF) test was performed to determine if the time series was stationary. If the time series was non-stationary, transformations and differencing were applied to make it stationary.
# # Environement Setup
# ## Importing Libraries
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
from statsmodels.graphics.gofplots import qqplot
from statsmodels.graphics.tsaplots import plot_acf, plot_pacf
from statsmodels.tsa.stattools import adfuller
from statsmodels.tsa.arima.model import ARIMA
from seaborn import distplot
warnings.filterwarnings("ignore")
sns.set()
# ## Defining Functions
def tsplot(y, ADF=True, lags=None, title=None, figsize=(15, 7), style="bmh"):
"""Plot time series, its ACF and PACF, calculate Dickey–Fullertest
y - timeseries
lags - how many lags to include in ACF, PACF calculation
"""
if title == None:
title = "Time Series Analysis Plots"
else:
title = str(title)
if not isinstance(y, pd.Series):
y = pd.Series(y)
with plt.style.context(style):
fig = plt.figure(figsize=figsize)
layout = (2, 2)
ts_ax = plt.subplot2grid(layout, (0, 0), colspan=2)
acf_ax = plt.subplot2grid(layout, (1, 0))
pacf_ax = plt.subplot2grid(layout, (1, 1))
y.plot(ax=ts_ax)
if ADF:
p_value = adfuller(y)[1]
ts_ax.set_title(title + "\n Dickey-Fuller: p_value ={0:.5f}".format(p_value))
else:
ts_ax.set_title(title)
plot_acf(y, lags=lags, ax=acf_ax)
plot_pacf(y, lags=lags, ax=pacf_ax)
plt.tight_layout()
def tsplot_resid(y, lags=None, title=None, figsize=(15, 7), style="bmh"):
"""
Plot time series, its ACF and PACF, calculate Dickey–Fuller
test
y - timeseries
lags - how many lags to include in ACF, PACF calculation
"""
if title == None:
title = "Time Series Analysis Plots"
else:
title = str(title)
if not isinstance(y, pd.Series):
y = pd.Series(y)
with plt.style.context(style):
fig = plt.figure(figsize=figsize)
layout = (3, 2)
ts_ax = plt.subplot2grid(layout, (0, 0), colspan=2)
acf_ax = plt.subplot2grid(layout, (1, 0))
pacf_ax = plt.subplot2grid(layout, (1, 1))
qqplot_ax = plt.subplot2grid(layout, (2, 0))
dist_ax = plt.subplot2grid(layout, (2, 1))
y.plot(ax=ts_ax)
ts_ax.set_title(title)
plot_acf(y, lags=lags, ax=acf_ax)
plot_pacf(y, lags=lags, ax=pacf_ax)
qqplot(y, fit=True, line="45", ax=qqplot_ax)
qqplot_ax.set_title("Normalitée")
distplot(y, ax=dist_ax)
dist_ax.set_title("Distribution")
plt.tight_layout()
# # Preprocessing
# ## Importing Data
df = pd.read_csv(
"/kaggle/input/algeria-electric-consumption-per-capita-1971-2019/Algeria electricity consumption 1971-2019.csv",
index_col=0,
)
df.head()
# ## Preprocessing the Data
df.index = pd.to_datetime(df.index, format="%Y")
df.index = df.index + pd.offsets.YearEnd(0)
df.rename(
columns={"Electric power consumption (kWh per capita)": "consumption"}, inplace=True
)
df.head()
# ## Plotting the Series
import matplotlib.pyplot as plt
# Set the style to a clean, simple format
# plt.style.use('seaborn-white')
# Create the figure object and axes
fig, ax = plt.subplots(figsize=(12, 6))
# Plot the data
df.plot(ax=ax, linewidth=2)
# Set the x and y labels and title
ax.set_xlabel("Year", fontsize=12)
ax.set_ylabel("Electricity consumption per capita", fontsize=12)
ax.set_title(
"Electricity consumption per capita 1971-2019", fontsize=16, fontweight="bold"
)
# Add a legend with a shadow effect
ax.legend(loc="upper left", shadow=True, fontsize=12)
# Remove the top and right spines
ax.spines["right"].set_visible(False)
ax.spines["top"].set_visible(False)
# Adjust the tick label font size
ax.tick_params(axis="both", which="major", labelsize=10)
# Display the plot
plt.savefig("electricity_consumption.png")
plt.show()
# # Model Selection
# ## Checking Stationarity
tsplot(df.consumption, lags=20, title="Series before integration (differencing)")
# * A p-value of 0.99898 suggests that there is very strong evidence against rejecting the null hypothesis, and that the time series is likely non-stationary.
# * This means that the series may have trends or other patterns that make it difficult to model or analyze. It may be necessary to apply transformations or differencing to make the series stationary before performing further analysis or modeling.
# ## Differencing
dff = df.diff().dropna()
dff.head()
tsplot(dff["consumption"], lags=20, title="Series after integration (diffrencing)")
# * A p-value below a significance level (e.g. 0.05) suggests that the time series is stationary, meaning that it does not exhibit any significant trend or seasonality over time.
# * No spikes are significant either in the ACF or in the PACF.
# * At first glance, an ARIMA(0,1,0) process is best for this dataset
# ## Choosing the best model
def get_best_model(TS):
AIC = []
order = []
pq_rng = range(3) # [0,1,2,3,4]
d_rng = range(1, 2) # [0,1]
for d in d_rng:
for i in pq_rng:
for j in pq_rng:
tmp_mdl = ARIMA(TS, order=(i, d, j)).fit()
AIC.append(tmp_mdl.aic)
order.append((i, d, j))
tbl = {"Ordre": order, "AIC": AIC}
rank = pd.DataFrame(tbl)
rank.sort_values(by=["AIC"], ascending=True, inplace=True)
rank.set_index("Ordre", inplace=True)
return rank
get_best_model(df)
# * According to the Akaike criterion (AIC), the best model is an ARIMA(1,1,1) with an AIC = 490.59
AR11 = ARIMA(df, order=(1, 1, 1)).fit()
AR11.summary()
# * All the parameters of the model are significant
# # Model Validation
# ## Checking Residuals
tsplot_resid(AR11.resid, lags=20, title="Residus")
# * At first glance, the residuals are the properties of White Noise
# * There is no significant peak in ACF nor PACF
# ## Engle's ARCH Test for Homoscedasticity of Errors
# * Ho: The squared residuals are a sequence of white noise - the residuals are homoscedastics.
# * H1: Squared residuals could not be fitted with a linear regression model and exhibit heteroscedasticity.
from statsmodels.stats.diagnostic import het_arch
print(
f"Critical value : {het_arch(AR11.resid)[2]:.6f}, p_value : {het_arch(AR11.resid)[3]:.6f}"
)
# A p-value of 0.082398 suggests that we cannot reject the null hypothesis at the conventional significance level of 0.05. In other words, there is not enough evidence to conclude that there is conditional heteroskedasticity present in the residuals
# ## Ljung-Box Test for Autocorrelation of Errors
# * H0: Residuals are distributed independently.
# * H1: residues are not distributed independently; they show a correlation in series.
from statsmodels.stats.diagnostic import acorr_ljungbox
LB = acorr_ljungbox(AR11.resid, lags=[10])
print(f"Critical value = {LB.iloc[0,0]:.6f}, p_value ={LB.iloc[0,1]:.6f}")
# * A p-value greater than 0.05 indicates that there is no significant autocorrelation in the residuals, and thus the null hypothesis cannot be rejected.
# * Therefore, we cannot reject the null hypothesis, and we can assume that there is no significant autocorrelation in the residuals.
# # Forecast
# ## Forecasting 2020-2024
pred = AR11.forecast(5)
years = [2020, 2021, 2022, 2023, 2024]
df_pred = pd.DataFrame({"Years": years, "Forecast": pred})
df_pred.set_index("Years", inplace=True)
df_pred
# ## Plotting Results
from statsmodels.graphics.tsaplots import plot_predict
fig, ax = plt.subplots(figsize=(15, 7))
ax = df.iloc[:].plot(ax=ax)
fig = plot_predict(AR11, "2019", "2024", ax=ax)
|
# **Llama bibliotecas del sistema**
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import imageio.v3 as iio
import matplotlib.pyplot as plt
import open3d as o3d
import os
from PIL import Image
from numpy import asarray
# Revisar: https://medium.com/yodayoda/from-depth-map-to-point-cloud-7473721d3f
# Revisar: https://betterprogramming.pub/point-cloud-computing-from-rgb-d-images-918414d57e80
def img_to_pcd_NYU(depth_path, pcd_path):
# Depth camera parameters:
FX_DEPTH = 5.8262448167737955e02
FY_DEPTH = 5.8269103270988637e02
CX_DEPTH = 3.1304475870804731e02
CY_DEPTH = 2.3844389626620386e02
depth_image = Image.open(depth_path).convert("L")
depth_image = asarray(depth_image)
# get depth resolution:
height, width = depth_image.shape
length = height * width
# compute indices:
jj = np.tile(range(width), height)
ii = np.repeat(range(height), width)
# rechape depth image
z = depth_image.reshape(length)
# compute pcd:
pcd = np.dstack(
[(ii - CX_DEPTH) * z / FX_DEPTH, (jj - CY_DEPTH) * z / FY_DEPTH, z]
).reshape((length, 3))
pcd_o3d = o3d.geometry.PointCloud() # create point cloud object
pcd_o3d.points = o3d.utility.Vector3dVector(
pcd
) # set pcd_np as the point cloud points
# Visualize:
# o3d.visualization.draw_geometries([pcd_o3d])
o3d.io.write_point_cloud(pcd_path, pcd_o3d)
# depth_path = '/kaggle/input/imagenes-jason-tres-algoritmos/imagen10_disp.jpeg'
# pcd_path = "prueba.pcd"
direccion_explorar = "/kaggle/input/resultados-nyu/depth_estimation_high_quality_NYU/"
dir_list = os.listdir(direccion_explorar)
# Genera el directorio de salida
if not (os.path.isdir("/kaggle/working/output")):
os.mkdir("/kaggle/working/output")
counter = 0
for filename in dir_list:
counter += 1
print(str(counter) + "/" + str(len(dir_list)))
# Para extraer solo aquellas imágenes que se hayan extraídas en vídeo
if filename != None and not "_" in filename and "." in filename:
depth_path = direccion_explorar + filename
pcd_path = "/kaggle/working/output/" + filename.split(".")[0] + ".pcd"
img_to_pcd_NYU(depth_path, pcd_path)
os.chdir(r"/kaggle/working/")
from IPython.display import FileLink
FileLink(r"cloud_point.tar.gz")
|
# Netflix Shows and Movies - Exploratory Data Analysis (EDA)
# First, import the packages.
import numpy as np
import seaborn as sns
import pandas as pd
import matplotlib.pyplot as plt
import matplotlib.mlab as mlab
import matplotlib
plt.style.use("ggplot")
import plotly.graph_objects as go
import plotly.express as px
matplotlib.rcParams["figure.figsize"] = (12, 8)
# Overview of Dataset Information
# Loading Data
pwd
import pandas as pd
NetFlixData = pd.read_csv("../input/netflix-shows/netflix_titles.csv")
# ### Display Data
NetFlixData.head(5)
# ### Display Last 5 values
NetFlixData.tail(5)
# Describe the Data
NetFlixData.describe()
# Information about the Data
NetFlixData.info()
# Shape of the Data
NetFlixData.shape
# Name of the Column
NetFlixData.columns
# Name of the Keys
NetFlixData.keys()
# Checking the Null values
NetFlixData.isnull().sum()
# Check Unique Values
NetFlixData.nunique()
# Check Duplicated Values
NetFlixData.duplicated().sum()
# Make a copy of Dataset
df = NetFlixData.copy()
df.shape
# Drop null values
df = df.dropna()
df.shape
# Print first 10 values
df.head(6)
# Convert Date Time format
df["date_added"] = pd.to_datetime(df["date_added"])
df["date_added"].head(3)
#
df["day_added"] = df["date_added"].dt.day
df["day_added"].head(3)
df["year_added"] = df["date_added"].dt.year
df["year_added"].head(6)
df["month_added"] = df["date_added"].dt.month
df["month_added"].head(6)
df["year_added"].astype(int).head(6)
df["day_added"].astype(int).head(6)
df["month_added"].astype(int).head(6)
df.head(6)
# Data Visualization
# sns.countplot(Netflix['type'])
ax = sns.countplot(x="type", hue="type", data=NetFlixData)
for p in ax.patches:
height = p.get_height()
ax.text(
p.get_x() + p.get_width() / 2, height + 5, "{:.0f}".format(height), ha="center"
)
fig = plt.gcf()
fig.set_size_inches(10, 5)
plt.title("Count of Movies and TV Shows on Netflix")
plt.savefig("Count of Movies and TV Shows on Netflix.png")
# Rating of shows and movies
# Replace missing values in 'rating' column with 'Unknown'
NetFlixData["rating"].fillna("Unknown", inplace=True)
# Plot countplot and rotate x-axis labels
ax = sns.countplot(x="rating", data=NetFlixData)
for p in ax.patches:
height = p.get_height()
ax.text(
p.get_x() + p.get_width() / 2, height + 5, "{:.0f}".format(height), ha="center"
)
plt.xticks(rotation=90)
# Set figure size and title
fig = plt.gcf()
fig.set_size_inches(13, 7)
plt.title("Rating")
plt.savefig("Rating.png")
# Relation Between Type and Rating
ax = sns.countplot(x="rating", hue="type", data=NetFlixData)
for p in ax.patches:
height = p.get_height()
ax.text(
p.get_x() + p.get_width() / 2, height + 5, "{:.0f}".format(height), ha="center"
)
fig = plt.gcf()
fig.set_size_inches(10, 5)
plt.title("Count of ratings for [Movies and TV] Shows on Netflix")
plt.savefig("Count of ratings for [Movies and TV] Shows on Netflix.png")
# Pie-chart for the Type: Movie and TV Shows
labels = ["Movie", "TV show"]
size = NetFlixData["type"].value_counts()
colors = plt.cm.Wistia(np.linspace(0, 1, 2))
explode = [0, 0.1]
plt.rcParams["figure.figsize"] = (5, 5)
ax = plt.pie(
size,
labels=labels,
colors=colors,
explode=explode,
shadow=True,
startangle=90,
autopct="%1.1f%%",
)
plt.title("Distribution of Type", fontsize=25)
plt.legend()
plt.show()
plt.savefig("Distribution of Type.png")
# Pie-chart for Rating
NetFlixData["rating"].value_counts().plot.pie(
autopct="%1.1f%%", shadow=False, figsize=(10, 8)
)
plt.show()
plt.savefig("rating-piechart.png")
# Group the data by 'date_added' and 'type'
grouped_data = NetFlixData.groupby(["date_added", "type"]).size().unstack()
# Plot the line chart
grouped_data.plot(kind="line", figsize=(12, 6), linewidth=2)
# Set the title and axis labels
plt.title("Number of Titles Added by Date", fontsize=16)
plt.xlabel("Date", fontsize=12)
plt.ylabel("Number of Titles", fontsize=12)
# Show the plot
plt.show()
# This code groups the data by date and type (Movie or TV Show) and creates a line chart showing the number of titles added by date. The resulting chart shows two lines, one for Movies and one for TV Shows, with each point on the line representing the number of titles added on a particular date.
# Wordcloud
from wordcloud import WordCloud
import matplotlib.pyplot as plt
plt.subplots(figsize=(25, 15))
wordcloud = WordCloud(background_color="black", width=1920, height=1080).generate(
" ".join(df.country)
)
plt.imshow(wordcloud)
plt.axis("off")
plt.savefig("country.png")
plt.show()
# Categories
plt.subplots(figsize=(25, 15))
wordcloud = WordCloud(background_color="black", width=1920, height=1080).generate(
" ".join(df.listed_in)
)
plt.imshow(wordcloud)
plt.axis("off")
plt.savefig("category.png")
plt.show()
|
# # hand written digits recognition
# accuracy 92%
# The **MNIST** dataset is a well-known dataset in the field of machine learning, it's used for testing & training algorithms and models. It consists of 70,000 images of handwritten digits (0 to 9) that are 28 pixels wide and 28 pixels height. Each image is represented as a grayscale array of 784 pixels (28x28).
# The MNIST dataset is frequently used for image classification tasks, where the goal is to train a machine learning model to correctly identify the digit represented in each image.
# Digit recognition is used in a lot of activities like identify the digits on cheques or zip postal codes ...
# and in this notebook we will build a model that will identify the hand written digits based on the mnist data set.
# # libraries
# starting by importing the required libraries, we are going to use the following :
# numpy : for manupilation vector and matrix
# pandas : for importing and working with the dataset
# matplotlib : for visulazng the dataset
# tensorflow : for building the model
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# ## importing the data set
# lets read our data frame using pandas
df = pd.read_csv("/kaggle/input/digit-recognizer/train.csv")
df_test = pd.read_csv("/kaggle/input/digit-recognizer/test.csv")
# ## inspect the dataset
# After readning the data set lets see the first 5 rows on it.
# We have the first column that contains the label for each digit and the rest contains the values(value of each pixel) of the image vector(784 pixel => (28,28))
df.head()
# the shape of the data set
# it contains 42000 image with shape of (28,28)
df.shape
# train has a label
# ## split the dataset
# Now after we read and seen the data set lets split it to a training set and testing set.
# we will take 10000 element in the trainig set and 1000 element in the testing set
test_slice = df_test[0:100]
# y_test = np.array(test_slice['label'])
# test_slice = test_slice.drop('label',axis=1)
# the training set
train_slice = df[0:10000]
y_train = np.array(train_slice["label"]) # label
X_train = np.array(train_slice.drop("label", axis=1))
# the testing set
test_slice = df[10001:11001]
y_test = np.array(test_slice["label"]) # label
x_test = test_slice.drop("label", axis=1)
# the shape of each shape
print(f"the shape of the testing set {test_slice.shape}")
print(f"the shape of the training set {train_slice.shape}")
print(f"the shape of x_train set and y_train set {y_train.shape,X_train.shape}")
print(f"the shape of x_test set and y_test set {y_test.shape,x_test.shape}")
train_slice.head()
# ## visualizing the data
fig, ax = plt.subplots(1, 10, figsize=(20, 3))
for i in range(10):
testReshaped = X_train[i].reshape(28, 28)
ax[i].imshow(testReshaped.astype("uint8"), interpolation="none", cmap="gray")
plt.show()
# ## Model
# ### import required library for building the model
import tensorflow as tf
from tensorflow.keras import Sequential
from tensorflow.keras.layers import Dense
# we will build a neural network of 3 layers, the first layer contains 35 units with relu activation, the second layer contains 30 units with relu activation and the last layer contains 10 units to predict each number with linear activation
model = Sequential(
[
tf.keras.Input(shape=(784,)),
Dense(units=512, activation="relu"),
# Dense(units=10,activation='relu'),
Dense(units=10, activation="linear"),
]
)
# for the cost/loss function we will use Sparse Categorical Cross entropy and the adam algorithm.
# train and fit the model
model.compile(
loss=tf.keras.losses.SparseCategoricalCrossentropy(from_logits=True),
optimizer=tf.keras.optimizers.Adam(learning_rate=0.001),
metrics=["accuracy"],
)
history = model.fit(X_train, y_train, epochs=20, validation_data=(x_test, y_test))
# ## predict the digit
# after training the model lets use it to predict the digit in the images
prr = np.array(
test_slice.iloc[999][1:]
) # there is 1000 element you can change between 0 999
testReshaped = prr.reshape(28, 28)
plt.imshow(testReshaped.astype("uint8"), interpolation="none", cmap="gray")
plt.title("the digit we will predict")
result = model.predict(prr.reshape(1, 784))
print(f"the digit on the image is {np.argmax(result)}")
# ## the accuracy of the model
loss, accuracy = model.evaluate(x_test, y_test, verbose=0)
print(f"the accuracy is {accuracy*100:.2f}%")
# convulution matrix and accuracy
test = [np.array(test_slice.iloc[i][1:]) for i in range(1000)]
test = np.array(test)
y_label = [test_slice.iloc[i][0] for i in range(1000)]
y_pred = [np.argmax(model.predict(test[i].reshape(1, 784))) for i in range(1000)]
from sklearn.metrics import confusion_matrix
import seaborn as sns
cm = confusion_matrix(y_pred, y_label)
sns.heatmap(cm, annot=True)
# here on the horizantal axis we have the digit we wanted to predict and on the vertical axis we have the the predicted digit and how many its predicted
# model.metrics_names
# i have imported a written digit i wrote it to predec it
from PIL import Image # fro manupilation the images
image = Image.open("/kaggle/input/itsnotadataset/2.jpeg")
# read the image
image # visualize the digit
# preprocecing
image = image.resize((28, 28))
image = image.convert("L")
imArray = np.array(image)
# predict the digit
result = model.predict(imArray.reshape(1, 784))
print(f"the number is : {np.argmax(result)}")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.preprocessing import StandardScaler, MinMaxScaler, Normalizer
from sklearn.pipeline import make_pipeline
from sklearn.model_selection import TimeSeriesSplit, train_test_split, cross_val_score
from sklearn.metrics import mean_absolute_error
from sklearn.linear_model import SGDRegressor, RidgeCV, LassoCV
sns.set_theme(style="darkgrid")
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
meat = pd.read_csv("/kaggle/input/meat-consumption/meat_consumption.csv")
print(meat.shape)
meat.head()
# # Data Analysis
meat.dtypes
data = meat.location.value_counts()
labels = data.index
plt.figure(figsize=(10, 10))
colors = sns.color_palette("pastel")[: len(labels)]
plt.pie(data.values, labels=labels, colors=colors, autopct="%.0f%%")
plt.show()
meat.indicator.value_counts()
fig, ax = plt.subplots(1, 2)
sns.countplot(ax=ax[0], x=meat.subject)
ax[0].set_title("Subject")
sns.countplot(ax=ax[1], x=meat.measure)
ax[1].set_title("Measure")
fig.tight_layout()
fig.show()
meat.frequency.value_counts()
plt.figure(figsize=(20, 6))
sns.countplot(x=meat.time)
plt.show()
sns.violinplot(y=meat.value, scale="count")
plt.show()
cat_columns = [i for i in meat.columns if i not in ["time", "value"]]
meat = pd.get_dummies(meat, columns=cat_columns)
meat.head()
sns.lineplot(
data=meat,
x="time",
y="value",
)
plt.show()
y = meat.value
X = meat.drop(columns=["value"])
X = X.sort_values(by="time")
X.head()
cat_columns = [i for i in X.columns if i != "time"]
X = pd.get_dummies(X, columns=cat_columns)
X.head()
X = X.set_index("time")
X.head()
# x_train, x_test, y_train, y_test = train_test_split(X, y, test_size=0.3)
# x_train, x_val, y_train, y_val = train_test_split(x_train, y_train, test_size=0.2)
int(len(y) * 0.6)
x_train, y_train = X[: int(len(X) * 0.6)], y[: int(len(y) * 0.6)]
x_val, y_val = (
X[int(len(X) * 0.6) : int(len(X) * 0.7)],
y[int(len(X) * 0.6) : int(len(y) * 0.7)],
)
x_test, y_test = X[int(len(X) * 0.7) :], y[int(len(y) * 0.7) :]
print(f"X_train: {x_train.shape}")
print(f"X_val: {x_val.shape}")
print(f"X_test: {x_test.shape}")
# # Basic regression
st = (StandardScaler(), MinMaxScaler(), Normalizer())
estimators = (RidgeCV(), SGDRegressor(), LassoCV())
for est in estimators:
print("*********************************************")
print(est)
for s in st:
pipe = make_pipeline(s, est)
pipe.fit(x_train, y_train)
predict_pipe = pipe.predict(x_test)
print(f" {s}: {mean_absolute_error(y_test, predict_pipe)}")
def evaluate_model(estimator):
cv = TimeSeriesSplit(n_splits=15)
scores = cross_val_score(estimator, x_val, y_val, cv=cv, n_jobs=-1)
return scores
def get_model1():
models = {}
eps_ = [0.001, 0.003, 0.006, 0.0012]
for e in eps_:
models[str(e)] = make_pipeline(StandardScaler(), LassoCV(eps=e))
return models
models1 = get_model1()
results, names = [], []
for name, model in models1.items():
scores = evaluate_model(model)
results.append(scores)
names.append(name)
print(f"{name}: {np.mean(scores):.5f} ({np.std(scores):.5f})")
plt.boxplot(results, labels=names)
plt.show()
def get_model2():
models = {}
n_ = range(25, 151, 25)
for n in n_:
models[str(n)] = make_pipeline(StandardScaler(), LassoCV(eps=0.003, n_alphas=n))
return models
models2 = get_model2()
results, names = [], []
for name, model in models2.items():
scores = evaluate_model(model)
results.append(scores)
names.append(name)
print(f"{name}: {np.mean(scores):.5f} ({np.std(scores):.5f})")
plt.boxplot(results, labels=names)
plt.show()
def get_model3():
models = {}
fit_ = [True, False]
for f in fit_:
models[str(f)] = make_pipeline(
StandardScaler(), LassoCV(eps=0.003, fit_intercept=f)
)
return models
models3 = get_model3()
results, names = [], []
for name, model in models3.items():
scores = evaluate_model(model)
results.append(scores)
names.append(name)
print(f"{name}: {np.mean(scores):.5f} ({np.std(scores):.5f})")
plt.boxplot(results, labels=names)
plt.show()
def get_model4():
models = {}
iter_ = range(500, 2501, 500)
for i in iter_:
models[str(i)] = make_pipeline(
StandardScaler(), LassoCV(eps=0.003, fit_intercept=False, max_iter=i)
)
return models
models4 = get_model4()
results, names = [], []
for name, model in models4.items():
scores = evaluate_model(model)
results.append(scores)
names.append(name)
print(f"{name}: {np.mean(scores):.5f} ({np.std(scores):.5f})")
plt.boxplot(results, labels=names)
plt.show()
def get_model5():
models = {}
split_ = range(5, 26, 5)
for s in split_:
cv_ = TimeSeriesSplit(n_splits=s)
models[str(s)] = make_pipeline(
StandardScaler(), LassoCV(eps=0.003, fit_intercept=False, cv=cv_)
)
return models
models5 = get_model5()
results, names = [], []
for name, model in models5.items():
scores = evaluate_model(model)
results.append(scores)
names.append(name)
print(f"{name}: {np.mean(scores):.5f} ({np.std(scores):.5f})")
plt.boxplot(results, labels=names)
plt.show()
def get_model6():
models = {}
tol_ = [0.00006, 0.0003, 0.0006, 0.0009, 0.0112]
for t in tol_:
models[str(t)] = make_pipeline(
StandardScaler(),
LassoCV(
eps=0.003, fit_intercept=False, cv=TimeSeriesSplit(n_splits=25), tol=t
),
)
return models
models6 = get_model6()
results, names = [], []
for name, model in models6.items():
scores = evaluate_model(model)
results.append(scores)
names.append(name)
print(f"{name}: {np.mean(scores):.5f} ({np.std(scores):.5f})")
plt.boxplot(results, labels=names)
plt.show()
model = make_pipeline(
StandardScaler(),
LassoCV(
eps=0.012, fit_intercept=False, cv=TimeSeriesSplit(n_splits=25), random_state=14
),
)
model.fit(x_train, y_train)
predict_model = model.predict(x_test)
print(f"MAE: {mean_absolute_error(y_test, predict_model)}")
# # Neural Network
from keras.models import Sequential
from keras.layers import Dense, Dropout
model_NN = Sequential(
layers=[
Dense(x_train.shape[1], input_dim=x_train.shape[1], activation="relu"),
Dense(144, activation="relu"),
Dense(72, activation="relu"),
Dropout(0.2),
Dense(16, activation="relu"),
Dense(1, activation=None),
]
)
model_NN.compile(optimizer="nadam", loss="mse", metrics=["mae"])
model_NN.summary()
model_NN.fit(
x_train,
y_train,
validation_data=(x_val, y_val),
epochs=150,
verbose=False,
batch_size=16,
)
predict_NN = model_NN.predict(x_test)
print(f"MAE: {mean_absolute_error(y_test, predict_NN)}")
|
# ## Diagnosis
# #### B = Benign Cancer
# #### M = Malignant Cancer
# ---
# ### Imports
import pandas as pd
import numpy as np
from sklearn.model_selection import train_test_split
from imblearn.over_sampling import SMOTE
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import accuracy_score, precision_score, recall_score, f1_score
# ### Data
df = pd.read_csv("/kaggle/input/cancer-data/Cancer_Data.csv")
df.head()
# ### Drop unnecessary columns
df.drop(["id", "Unnamed: 32"], axis=1, inplace=True)
# ### Descriptive information of the data
df.info()
df.describe()
# ### Checking null values
df.isna().sum()
# ### Removing outliers
# Threshold value to define the range for identifying outliers
threshold = 5
# Loop through all columns in the DataFrame, excluding the diagnosis
for column in df.loc[:, ~df.columns.isin(["diagnosis"])]:
# Mean and STD of the column
mean = df[column].mean()
std = df[column].std()
# Lower and Upper limits
lower_limit = mean - threshold * std
upper_limit = mean + threshold * std
# Remove outliers
df = df[(df[column] >= lower_limit) & (df[column] <= upper_limit)]
df.shape
# ### Swapping B and M for 0 and 1
df["diagnosis"] = df["diagnosis"].map({"B": 0, "M": 1})
# ### Split the data into X and y
X = df.drop("diagnosis", axis=1)
y = df["diagnosis"]
# ### Split X and y into Train and Test
xtrain, xtest, ytrain, ytest = train_test_split(
X, y, stratify=y, test_size=0.25, random_state=42
)
# ### Data Balancing
# Instantiating SMOTE
smt = SMOTE(random_state=0)
# Data Balancing
xtrain_res, ytrain_res = smt.fit_resample(xtrain, ytrain)
print(f"Distribution BEFORE balancing:\n{ytrain.value_counts()}")
print("=-" * 16)
print(f"Distribution AFTER balancing:\n{ytrain_res.value_counts()}")
# ### Training
rf = RandomForestClassifier(max_depth=5, random_state=0)
rf.fit(xtrain_res, ytrain_res)
# ### Predicting
pred = rf.predict(xtest)
# ### Results
accuracy = accuracy_score(ytest, pred)
precision = precision_score(ytest, pred)
recall = recall_score(ytest, pred)
f1 = f1_score(ytest, pred)
print("Accuracy:", accuracy)
print("Precision:", precision)
print("Recall:", recall)
print("F1 Score:", f1)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 5GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# ### Dataset
# NHANES datasets from 2013-2014
# https://www.kaggle.com/cdc/national-health-and-nutrition-examination-survey
# Variable Search:
# https://wwwn.cdc.gov/nchs/nhanes/search/default.aspx
# Codebook:
# https://wwwn.cdc.gov/nchs/nhanes/Search/DataPage.aspx?Component=Demographics&CycleBeginYear=2013
#
# datasets path and df names
path = [
"/kaggle/input/national-health-and-nutrition-examination-survey/demographic.csv",
"/kaggle/input/national-health-and-nutrition-examination-survey/examination.csv",
"/kaggle/input/national-health-and-nutrition-examination-survey/questionnaire.csv",
"/kaggle/input/national-health-and-nutrition-examination-survey/labs.csv",
"/kaggle/input/national-health-and-nutrition-examination-survey/diet.csv",
]
dfname = ["dm", "exam", "qs", "lab", "diet"]
# import datasets as dfs
df = {}
dfn = dict(zip(dfname, path))
df = {key: pd.read_csv(value) for key, value in dfn.items()}
dfs = df.values()
from functools import partial, reduce
inner_merge = partial(pd.merge, how="inner", on="SEQN")
# outer_merge = partial(pd.merge, how='outer')
c = reduce(inner_merge, dfs)
# check if there are duplicated SEQN
c.SEQN.duplicated().value_counts()
# show combined df
c
c.RIDRETH3
# c = pd.concat(dfs, axis=1)
# c.SEQN
# ### Exclude rows with null values or NA for MCQ160F
# The prediction target in the dataset is MCQ160F, a questionnaire question "Has a doctor or other health professional ever told you that you had a stroke?"
#
# MCQ160F (target feature): exclude null values and NA
c = c[(c.MCQ160F.notnull()) & (c.MCQ160F != 9)]
# check MCQ160F
c.MCQ160F.describe()
# target varoable counts
c.MCQ160F.value_counts()
# import code book and clean data
cbook = pd.read_excel("/kaggle/input/data-dictionary/NAHNES%202014%20Dictionary.xlsx")
cbook.dropna(inplace=True)
cbook.drop_duplicates(inplace=True)
cbook.drop(index=[767, 2116, 3984], inplace=True) # drop repeated columns
cbook["Variable Name"] = cbook["Variable Name"].apply(lambda x: x.upper())
# ### Data Cleaning
# exclude non-numeric values
d = c.select_dtypes(["number"])
# exclue columns that have over 50% NaN
d = d.dropna(thresh=0.5 * len(d), axis=1)
print(len(d.columns), "columns left")
# changing variable coding from 1, 2 to 0 (Negative), 1 (Positive)
d["MCQ160F"] = d.apply(lambda x: 1 if x.MCQ160F == 1 else 0, axis="columns")
d.MCQ160F.value_counts()
# np.unique(d.MCQ160F, return_counts=True)
# # impute missing data with mode
# from sklearn.impute import SimpleImputer
# d_imputed = pd.DataFrame(SimpleImputer().fit_transform(d), columns = d.columns)
# d_imputed
# ddd = pd.DataFrame(SimpleImputer().fit_transform(d))
# ddd.columns = d.columns
# # ddd.columns = [d.columns] ### this will result in error, beacause asigning list inside a list!!!
# ddd
# ### Feature Selection
# Reference:
# A data-driven approach to predicting diabetes and cardiovascular disease with machine learning
# https://pubmed.ncbi.nlm.nih.gov/31694707/
# X = d_imputed.loc[:, d_imputed.columns != 'MCQ160F']
# X
# y = d_imputed.MCQ160F
X = d.loc[:, d.columns != "MCQ160F"]
X
y = d.MCQ160F
# from scipy.sparse import coo_matrix
# X_sparse = coo_matrix(X)
# from sklearn.utils import resample
# X, X_sparse, y = resample(X, X_sparse, y, random_state=11)
print(X.shape)
print(y.shape)
X.shape
# for i in sorted(X.columns):
# print(i)
y.shape
y.value_counts()
# ## Feature Selection
# ### ExtraTreesClassifier
# ### Requires imputation
# from sklearn.ensemble import ExtraTreesClassifier
# import matplotlib.pyplot as plt
# model = ExtraTreesClassifier()
# model.fit(X,y)
# # print(model.feature_importances_)
# #use inbuilt class feature_importances of tree based classifiers
# #plot graph of feature importances for better visualization
# feat_importances = pd.Series(model.feature_importances_,
# index=X.columns)
# fs_ = feat_importances.nlargest(24)
# fs_.sort_values(ascending=True).plot(kind='barh', figsize=(10,8))
# plt.show()
# fs = pd.DataFrame(fs_).reset_index()
# fs.columns=['feature','score']
# features = fs.merge(cbook, how='left', left_on='feature', right_on='Variable Name')#.drop('Variable Name', axis=1)
# features
c
d
# ## Standardization
# from sklearn.preprocessing import StandardScaler
# scaler = StandardScaler()
# scaler.fit(d)
# print(scaler.mean_)
# std_d = scaler.transform(d)
# std_df = pd.DataFrame(std_d, columns=d.columns)
# std_df
# ### DownSampling
# https://elitedatascience.com/imbalanced-classes
import sklearn
sklearn.__version__
# from sklearn.utils import resample
# # # Separate majority and minority classes
# df_majority = std_df[std_df.MCQ160F==0]
# df_minority = std_df[std_df.MCQ160F==1]
# # # Downsample majority class
# df_majority_downsampled = resample(df_majority,
# replace=True, # sample without replacement
# n_samples=194, # to match minority class
# random_state=11) # reproducible results
# # # Combine minority class with downsampled majority class
# df_downsampled = pd.concat([df_majority_downsampled, df_minority])
# # # Display new class counts
# df_downsampled.balance.value_counts()
# ## Feature Selection
# ### XGBoost
from xgboost import XGBClassifier
from matplotlib import pyplot
# fit model no training data
model = XGBClassifier()
model.fit(X, y)
# # feature importance
# print(model.feature_importances_)
# # feature names
# print(model.get_booster().feature_names)
# Features selected by XGBoost
keys = list(model.get_booster().feature_names)
values = list(model.feature_importances_)
data = pd.DataFrame(data=values, index=keys, columns=["score"]).sort_values(
by="score", ascending=False
)
# Top 24 features
xgbfs_ = data[:24]
# Plot feature score
xgbfs_.sort_values(by="score").plot(kind="barh", figsize=(10, 8))
# 24 most important features
features = {}
features["age"] = "RIDAGEYR"
features["LDL cholesterol"] = "LBDLDL"
features["Chest pain"] = "CDQ001"
features["Diastolic Blood Pressure"] = "BPXDI1"
features["HDL cholesterol"] = "LBDHDD"
features["Weight"] = "BMXWT"
features["Kcal intake"] = "DR1TKCAL"
features["Close relative had heart attack"] = "MCQ300a"
features["Segmented neutrophils"] = "LBXNEPCT"
features["Senedentary activities"] = "PAD680"
features["Overnight hospitalization"] = "HUD080"
features["Systolic blood pressure"] = "BPXSY1"
features["General health condition"] = "HSD010"
features["Calcium intake"] = "DR1TCALC"
features["Ethnicity"] = "RIDRETH3"
features["Alcohol consumpion"] = "ALQ101"
features["Carbohydrate intake"] = "DR1ICARB"
features["High Blood Pressure"] = "BPQ020"
features["Fiber intake"] = "DR1IFIBE"
features["Monocyte number"] = "LBDMONO"
features["Lymphocyte number"] = "LBDLYMNO"
features["Diabetes"] = "DIQ010"
features["Eosinophils number"] = "LBDEONO"
features["Close relative had diabetes"] = "MCQ300C"
len(features)
features
xgbfs = pd.DataFrame(xgbfs_).reset_index()
xgbfs.columns = ["feature", "score"]
features = xgbfs.merge(
cbook, how="inner", left_on="feature", right_on="Variable Name"
).drop("Variable Name", axis=1)
features["label"] = [
"Need special equipment to walk",
"Age in years at screening",
"Limited in amount of work you can do",
"Have difficulty doing errands alone ?",
"Dr told to take daily low-dose aspirin?",
"Tooth Count: #9",
"How healthy is the diet",
"Grip strength (kg) hand1",
"Tooth Count: #26",
"Monocyte number ",
"Hours worked last week at all jobs",
"Require special healthcare equipment",
"HH ref person's spouse's education level",
"Tooth Count: #4",
"Tooth Count: #12",
"Salt used in preparation?",
"Ever told you had heart attack",
"HPV Type 70",
"Doctor ever told any other fractures?",
"Past yr need dental but couldn't get it",
"Combined grip strength ",
"Tooth Count: #11",
"SP ever had pain or discomfort in chest",
"Body Measures Component Status Code",
]
# Feature short names
features
features.sort_values(by="score", ascending=True).plot(
x="label", y="score", kind="barh", figsize=(10, 8)
)
# ### Test/Train
# final variables
var = features.feature.tolist()
var.append("MCQ160F")
print(var)
# final df
df_final = d.filter(var)
df_final
df_final.columns
# final variables
X = df_final.loc[:, df_final.columns != "MCQ160F"]
y = df_final.MCQ160F
X
y
print(X.shape)
print(y.shape)
type(y)
# import seaborn as sns
import matplotlib.pyplot as plt
# sns.pairplot(fdf[-10:])
df_final.iloc[:, :13].describe()
df_final.iloc[:, 12:].describe()
fdf1 = fdf.loc[:, (fdf.columns != "OCQ180") & (fdf.columns != "DR1BWATZ")]
fdf2 = fdf.filter(["OCQ180", "DR1BWATZ"])
fdf2
import seaborn as sns
import matplotlib.pyplot as plot
# sns.boxplot(fdf)
fdf1.plot.box(figsize=(30, 5))
# fdf2.iloc[:,0].plot.box(figsize=(10,10))
fdf2.iloc[:, 0].plot.box()
fdf2.iloc[:, 1].plot.box(figsize=(10, 10))
# sns.axes_style("white")
ax = plt.subplots(figsize=(20, 20))
corr = fdf.corr()
# sns.heatmap(corr, vmin=-1, vmax=1, cmap=sns.cm.rocket_r)
sns.heatmap(corr, cmap=sns.cm.rocket_r)
# ## Feature Selection
# ### GradientBoostingClassifier
# ### Requires imputation
# from sklearn.ensemble import GradientBoostingClassifier
# from sklearn.feature_selection import SelectFromModel
# # gbm = GradientBoostingClassifier()
# # selection = SelectFromModel(gbm, threshold=0.03, prefit=True)
# # selected_dataset = selection.transform(X)
# from sklearn.feature_selection import SelectFromModel
# from sklearn.ensemble import RandomForestClassifier
# embeded_rf_selector = SelectFromModel(RandomForestClassifier(n_estimators=100), max_features=24)
# embeded_rf_selector.fit(X, y)
# embeded_rf_support = embeded_rf_selector.get_support()
# embeded_rf_feature = X.loc[:,embeded_rf_support].columns.tolist()
# print(str(len(embeded_rf_feature)), 'selected features')
# embeded_rf_feature
# stroke count
# qs.MCQ160F.value_counts()
# select stroke subjects
# qs160 = qs.loc[:,['SEQN','MCQ160F','MCQ180F']]
# print(qs160)
# stroke = qs160.loc[qs160.MCQ160F == 1.0]
# stroken = stroke.SEQN.to_list()
# stroken
# exam[['BPXDI1'-'BPXDI4']]
# bp = exam.filter(regex='SEQN|BPXDI[0-9]')
# bp[bp.SEQN.isin(stroken)]
# age = dm.filter(regex='SEQN|RIDAGEYR')
# agen = age[age.SEQN.isin(stroken)]
# pd.merge(agen, qs160, how='inner', on='SEQN')
# ### Missing Value
used_df = d
from sklearn.impute import SimpleImputer
X_impu = SimpleImputer(strategy="most_frequent").fit_transform(df_final)
df_impu = SimpleImputer(strategy="most_frequent").fit_transform(df_final)
df_final = pd.DataFrame(df_impu, columns=df_final.columns)
# ### Downsampling
used_df = df_final
# Separate majority and minority classes
df_majority = used_df[used_df.MCQ160F == 0]
df_minority = used_df[used_df.MCQ160F == 1]
# Downsample majority class
df_majority_downsampled = resample(
df_majority,
replace=False, # sample without replacement
n_samples=194, # to match minority class
random_state=123,
) # reproducible results
# Combine minority class with downsampled majority class
df_downsampled = pd.concat([df_majority_downsampled, df_minority])
print(df_downsampled)
# ### Train/Test
from sklearn.model_selection import train_test_split
X = df_downsampled.loc[:, df_downsampled.columns != "MCQ160F"]
y = df_downsampled.MCQ160F
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=11)
# ### Feature Selection
from xgboost import XGBClassifier
from matplotlib import pyplot
# fit model no training data
model = XGBClassifier()
model.fit(X_train, y_train)
# # feature importance
# print(model.feature_importances_)
# # feature names
# print(model.get_booster().feature_names)
# Features selected by XGBoost
keys = list(model.get_booster().feature_names)
values = list(model.feature_importances_)
data = pd.DataFrame(data=values, index=keys, columns=["score"]).sort_values(
by="score", ascending=False
)
# Top 24 features
xgbfs_ = data[:24]
# Plot feature score
xgbfs_.sort_values(by="score").plot(kind="barh", figsize=(10, 8))
xgbfs = pd.DataFrame(xgbfs_).reset_index()
xgbfs.columns = ["feature", "score"]
features = xgbfs.merge(
cbook, how="inner", left_on="feature", right_on="Variable Name"
).drop("Variable Name", axis=1)
# final variables
var = features.feature.tolist()
print(var)
X_train = X_train.filter(var)
X_test = X_test.filter(var)
print(X_train.shape)
print(X_test.shape)
var2 = var.copy()
var2.append("MCQ160F")
print(var2)
fdf = df_downsampled.filter(var2)
import seaborn as sns
import matplotlib.pyplot as plt
# sns.boxplot(fdf)
fdf.plot.box(figsize=(30, 5))
# sns.axes_style("white")
ax = plt.subplots(figsize=(20, 20))
corr = fdf.corr()
sns.heatmap(corr, vmin=-1, vmax=1, cmap=sns.cm.rocket_r)
# ### Logistic Regression
from sklearn.linear_model import LogisticRegression
clf = LogisticRegression(max_iter=100, random_state=11).fit(X_train, y_train)
y_pred = clf.predict(X_test)
print("Accuracy Score:", clf.score(X_test, y_test))
print("Prediction:", y_pred)
from sklearn.metrics import classification_report
print(classification_report(y_test, y_pred))
# ### Random Forest
from sklearn.ensemble import RandomForestClassifier
rnd_clf = RandomForestClassifier(max_depth=5, random_state=11)
rnd_clf.fit(X_train, y_train)
y_pred = rnd_clf.predict(X_test)
print("Accuracy Score:", rnd_clf.score(X_test, y_test))
print("Prediction:", y_pred)
from sklearn.metrics import classification_report
print(classification_report(y_test, y_pred))
# ### Gradient Boosting Decision Trees
from sklearn.ensemble import GradientBoostingClassifier
gbc = GradientBoostingClassifier(learning_rate=0.1, n_estimators=20, random_state=11)
gbc.fit(X_train, y_train)
y_pred = gbc.predict(X_test)
print("Accuracy Score:", gbc.score(X_test, y_test))
print("Prediction:", y_pred)
# from sklearn.metrics import classification_report
print(classification_report(y_test, y_pred))
# ### AdaBoostClassifier
from sklearn.ensemble import AdaBoostClassifier
ada = AdaBoostClassifier(learning_rate=0.1, n_estimators=20, random_state=11)
ada.fit(X_train, y_train)
y_pred = ada.predict(X_test)
print("Accuracy Score:", ada.score(X_test, y_test))
print("Prediction:", y_pred)
# from sklearn.metrics import classification_report
print(classification_report(y_test, y_pred))
# ### SVM
from sklearn.svm import SVC
svm_clf = SVC(kernel="rbf", gamma="auto", random_state=11)
svm_clf.fit(X_train, y_train)
y_pred = svm_clf.predict(X_test)
print("Accuracy Score:", svm_clf.score(X_test, y_test))
print("Prediction:", y_pred)
# from sklearn.metrics import classification_report
print(classification_report(y_test, y_pred))
# ### xGBoost
from xgboost import XGBClassifier
# xgbc = XGBClassifier(eta=0.3, max_depth=5)
xgbc = XGBClassifier(eta=0.01)
xgbc.fit(X_train, y_train)
y_pred = xgbc.predict(X_test)
print("Accuracy Score:", svm_clf.score(X_test, y_test))
print("Prediction:", y_pred)
from sklearn.metrics import classification_report
print(classification_report(y_test, y_pred))
|
# # Assignment Instructions
# 1. Load the data containing the NBA shot log for the 2016/17 season that we used earlier
# 2. Use the same code that we used to project all shots onto a half court by defining shot[‘halfcourt_x’] and shot[‘halfcourt_y’]
# 3. Now define subsets for the following players: Kevin Durant, Dwight Howard, DeAndre Jordan and Russell Westbrook.
# 4. Create plots of their shots in the same way that we did for Steph Curry and LeBron James: copy the code we used and just change the names. Show the plots of Russell Westbrook and Kevin Durant side by side. In order to make sure that the two plots have the same ranges, for each subplot add the lines:
# - plt.xlim(500,950)
# - plt.ylim(0,500)
# 5. Create the plot of DeAndre Jordan and Dwight Howard side by side
# 6. Create the plot of Brook Lopez and Robin Lopez side by side
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
shot = pd.read_csv("/kaggle/input/nba-shotlog-16-17/NBA shotlog_16_17.csv")
pd.set_option("display.max_columns", 100)
print(shot.columns.tolist())
shot.describe()
# A simple plot of coordinates
x = shot["location_x"]
y = shot["location_y"]
plt.scatter(x, y, s=0.005, c="r", marker=".")
# The right hand half court
plt.figure(figsize=(94 / 12, 50 / 6))
plt.scatter(x, y, s=0.1, c="r", marker=".")
plt.minorticks_on()
plt.grid(which="major", linewidth=".5", color="black")
plt.grid(which="minor", linewidth=".5", color="red")
plt.xlim(933 / 2, 933)
shot["halfcourt_x"] = np.where(
shot["location_x"] < 933 / 2, 933 - shot["location_x"], shot["location_x"]
)
shot["halfcourt_y"] = np.where(
shot["location_x"] < 933 / 2, 500 - shot["location_y"], shot["location_y"]
)
shot.describe()
# all shots shown on a half court
hx = shot["halfcourt_x"]
hy = shot["halfcourt_y"]
plt.figure(figsize=(94 / 12, 50 / 6))
plt.scatter(hx, hy, s=0.01, c="r", marker=".")
plt.minorticks_on()
plt.grid(which="major", linestyle="-", linewidth=".5", color="black")
plt.grid(which="minor", linestyle=":", linewidth="0.5", color="red")
plt.title("Shots", fontsize=15)
# Comparing players
# We use a pivot table here to list players by shots
playersn = shot.groupby("shoot_player")["current_shot_outcome"].describe().reset_index()
playersn.sort_values(by="count", ascending=False)
Durant = shot[shot["shoot_player"] == "Kevin Durant"]
Howard = shot[shot["shoot_player"] == "Dwight Howard"]
Jordan = shot[shot["shoot_player"] == "DeAndre Jordan"]
Westbrook = shot[shot["shoot_player"] == "Russell Westbrook"]
hxD = Durant["halfcourt_x"]
hyD = Durant["halfcourt_y"]
colors = np.where(
Durant["current_shot_outcome"] == "SCORED",
"r",
np.where(Durant["current_shot_outcome"] == "MISSED", "b", "g"),
)
plt.figure(figsize=(94 / 12, 50 / 6))
plt.scatter(hxD, hyD, s=10, c=colors, marker=".")
plt.grid(True)
plt.title("Kevin Durant", fontsize=15)
hxH = Howard["halfcourt_x"]
hyH = Howard["halfcourt_y"]
colors = np.where(
Howard["current_shot_outcome"] == "SCORED",
"r",
np.where(Howard["current_shot_outcome"] == "MISSED", "b", "g"),
)
plt.figure(figsize=(94 / 12, 50 / 6))
plt.scatter(hxH, hyH, s=10, c=colors, marker=".")
plt.grid(True)
plt.title("Dwight Howard", fontsize=15)
hxJ = Jordan["halfcourt_x"]
hyJ = Jordan["halfcourt_y"]
colors = np.where(
Jordan["current_shot_outcome"] == "SCORED",
"r",
np.where(Jordan["current_shot_outcome"] == "MISSED", "b", "g"),
)
plt.figure(figsize=(94 / 12, 50 / 6))
plt.scatter(hxJ, hyJ, s=10, c=colors, marker=".")
plt.grid(True)
plt.title("DeAndre Jordan", fontsize=15)
hxW = Westbrook["halfcourt_x"]
hyW = Westbrook["halfcourt_y"]
colors = np.where(
Westbrook["current_shot_outcome"] == "SCORED",
"r",
np.where(Westbrook["current_shot_outcome"] == "MISSED", "b", "g"),
)
plt.figure(figsize=(94 / 12, 50 / 6))
plt.scatter(hxW, hyW, s=10, c=colors, marker=".")
plt.grid(True)
plt.title("Russel Westbrook", fontsize=15)
f = plt.figure(figsize=(94 / 6, 50 / 6))
ax = f.add_subplot(121)
colors = np.where(
Westbrook["current_shot_outcome"] == "SCORED",
"r",
np.where(Westbrook["current_shot_outcome"] == "MISSED", "b", "g"),
)
ax = plt.scatter(hxW, hyW, s=10, c=colors, marker=".")
plt.grid(True)
plt.xlim(500, 950)
plt.ylim(0, 500)
plt.title("Russel Westbrook", fontsize=15)
ax = f.add_subplot(122)
colors = np.where(
Durant["current_shot_outcome"] == "SCORED",
"r",
np.where(Durant["current_shot_outcome"] == "MISSED", "b", "g"),
)
ax = plt.scatter(hxD, hyD, s=10, c=colors, marker=".")
plt.grid(True)
plt.xlim(500, 950)
plt.ylim(0, 500)
plt.title("Kevin Durant", fontsize=15)
plt.savefig("WestbrookVDurant.png")
f = plt.figure(figsize=(94 / 6, 50 / 6))
ax = f.add_subplot(121)
colors = np.where(
Jordan["current_shot_outcome"] == "SCORED",
"r",
np.where(Jordan["current_shot_outcome"] == "MISSED", "b", "g"),
)
ax = plt.scatter(hxJ, hyJ, s=10, c=colors, marker=".")
plt.grid(True)
plt.xlim(500, 950)
plt.ylim(0, 500)
plt.title("DeAndre Jordan", fontsize=15)
ax = f.add_subplot(122)
colors = np.where(
Howard["current_shot_outcome"] == "SCORED",
"r",
np.where(Howard["current_shot_outcome"] == "MISSED", "b", "g"),
)
ax = plt.scatter(hxH, hyH, s=10, c=colors, marker=".")
plt.grid(True)
plt.xlim(500, 950)
plt.ylim(0, 500)
plt.title("Dwight Howard", fontsize=15)
BLopez = shot[shot["shoot_player"] == "Brook Lopez"]
RLopez = shot[shot["shoot_player"] == "Robin Lopez"]
hxBL = BLopez["halfcourt_x"]
hyBL = BLopez["halfcourt_y"]
hxRL = RLopez["halfcourt_x"]
hyRL = RLopez["halfcourt_y"]
f = plt.figure(figsize=(94 / 6, 50 / 6))
ax = f.add_subplot(121)
colors = np.where(
BLopez["current_shot_outcome"] == "SCORED",
"r",
np.where(BLopez["current_shot_outcome"] == "MISSED", "b", "g"),
)
ax = plt.scatter(hxBL, hyBL, s=10, c=colors, marker=".")
plt.grid(True)
plt.xlim(500, 950)
plt.ylim(0, 500)
plt.title("Brook Lopez", fontsize=15)
ax = f.add_subplot(122)
colors = np.where(
RLopez["current_shot_outcome"] == "SCORED",
"r",
np.where(RLopez["current_shot_outcome"] == "MISSED", "b", "g"),
)
ax = plt.scatter(hxRL, hyRL, s=10, c=colors, marker=".")
plt.grid(True)
plt.xlim(500, 950)
plt.ylim(0, 500)
plt.title("Robin Lopez", fontsize=15)
|
# # TP3 - Compression, Prediction, Generation: Text Entropy
# #### Francesco Saverio Pezzicoli, Guillaume Charpiat
# #### Credits: Vincenzo Schimmenti
# ### Author :
# - Benedictus Kent Rachmat
# - Diego Andres Torres Guarin
# ### Introduction
# In this TP we are interested in compressing and generating texts written in natural languages.
# Given a text of length $n$, a sequence of symbols is just a vector $(x_1, . . . , x_n)$ where each $x_i$ is a symbol i.e. $x_i = a, b, c, \dots$. We can define the alphabet of possible symbols as $\mathcal{A} = \{a_1,a_2,\dots,a_M\}$ then each $x_i$ can have $M$ values.
# In order to model the sequence of symbols we need a joint probability distribution for each symbol in the sequence, namely $p(X_1 = x_1, X_2 = x_2, \dots , X_n = x_n)$. If our alphabet had $M$ symbols, for modelling a sequence of length $n$ we would need $M^n$ probabilities. Thus some assumptions are required in order to reduce this dimensionality. In this case we will use two different models for $p$, the IID and the Markov Chain model.
# ### IID Model
# The IID model assumes:
# $$ p(X_1 = x_1, X_2 = x_2, \dots , X_n = x_n) = \prod_{i=1}^n p(X_i = x_i)$$
# i.e. that the symbols in a sequence are independent and identically distributed. With this model we need only $M$ probabilities, one for each symbol. One can generalize and use symbols not of a single character but of multiples ones. For example using 3 characters per symbol, the symbols would be of the form $aaa,aab,...,zzz$. When using $k$ characters per symbols in an alphabet of $M$ characters, the needed probabilities would be $M^k$.
# ### Markov Chain Model
# The Markov Chain model assume a limited range of dependence of the symbols. Indeed for an order $k$ Markov Chain:
# $$p(X_i | X_{i-1},X_{i-2},\dots,X_1) = p(X_i | X_{i-1},X_{i-2},\dots,X_{i-k})$$
# The meaning of the above structure is that the $i$-th symbol in the sequence depends only on the previous $k$ symbols. We add the time *invariant assumption*, meaning that the conditional probabilities do not depend on the time index $i$ i.e. $p(X_i | X_{i-1},X_{i-2},\dots,X_{i-k}) = p(X_{k+1} | X_{k},X_{k-1},\dots,X_{1})$. The most common and widely used Markov Chain is the Markov Chain of order 1:
# $$p(X_i | X_{i-1},X_{i-2},\dots,X_1) = p(X_i | X_{i-1})$$
# In this case the conditional probability $p(X_i|X_{i−1})$ can be expressed using $M^2$
# numbers. Usually this is referred to as the *transition matrix*. Given an alphabet $\mathcal{A} = \{a_1,a_2,\dots,a_M\}$ the transition matrix can be written as:
# $$ \mathbb{M}_{kl} = p(X_i = a_k| X_{i-1} = a_l) $$
# ### Entropy and Cross-Entropy
# - For the IID model of order 1 the entropy computation is straightforward:
# $$ H_{IID} = -\sum_{i=1}^M p(a_i) log p(a_i)$$
# and consequently, starting from two distributions $p,q$ fitted on two different texts, the cross-entropy:
# $$ CE_{IID} = -\sum_{i=1}^M p(a_i) log q(a_i)$$
# - For the MC model of order 1 the entropy is defined as follows:
# $$ H_{MC} = - \sum_{kl} \pi(a_k) p(X_i = a_k| X_{i-1} = a_l) log \left(p(X_i = a_k| X_{i-1} = a_l)\right)= - \sum_{kl} \pi_k\mathbb{M}_{kl} log \mathbb{M}_{kl}$$
# where $\pi$ is the stationary distribution of the Markov Chain i.e. $\pi_k = \mathbb{M}_{kl} \pi_l$. The code to compute the stationary distribution is already given.
# The cross-entropy:
# $$ CE_{IID} = - \sum_{kl} \pi_k\mathbb{M}_{kl} log \mathbb{M'}_{kl}$$
# with $\mathbb{M}$ and $\mathbb{M'}$ are fitted on two different texts.
# ### Theoretical Questions:
# 1) Interpret the time invariant assumption associated to our Markov chains in the contex of text generation.
# **Answer:**
# In the context of text generation, the time invariant assumption means that the generated text won't change depending on how further in the text we are in. For example, if we work with a Markov chain of order 3 to generate text, and the last three words are "I like eating", the probability distribution of following words will be the same, no matter if this is the first sentence or the eleventh. At first this assumption may seem a bit unsettling, because the some sentences are more or less likely depending on what has been said before. However, we need to keep in mind that this effect is due to the previously generated text, and not simply the index of the current position.
# 2) How can we rewrite a Markov chain of higher order as a Markov chain of order 1?
# **Answer:**
# We can group the chain of symbols in a sort of sliding window. For example, if we are working with a Markov chain of order 2 and are given the sequence *A,B,A,C,D*, it is possible to rewrite it as *AB, BA, AC, CD*. In this scenario, the new vocabulary will be composed of all the possible pairs of symbols $M^{2}$, and the new transition matrix will have $M^{4}$ entries. However, note that not all of these entries will be nonzero, since there would be impossible transitions. For instance coming from *AB*, the following pair of symbols will need to have *B* as its first symbol, so to keep the consistency of the original chain.
# As a concrete example, consider the following transition matrix
# | /// | /// | /// |
# | ----------- | ----------- | ----------- |
# | P(A\|AB) = 0.6 | P(B\|AB) = 0.3 | P(C\|AB) = 0.1 |
# | P(A\|BC) = 0.2 | P(B\|BC) = 0.5 |P(C\|BC) = 0.3 |
# |P(A\|CA) = 0.4| P(B\|CA) = 0.1| P(C\|CA) = 0.5 |
# We can rewrite this as a first-order Markov chain by concatenating the previous state with the current state. So, our new state space will consist of {AA, AB, AC, BA, BB, BC, CA, CB, CC}. The transition probabilities for the state AB will be:
# - P(AA|AB) = 0
# - P(AB|AB) = 0
# - P(AC|AB) = 0
# - P(BA|AB) = 0.6
# - P(BB|AB) = 0.3
# - P(BC|AB) = 0.1
# - P(CA|AB) = 0
# - P(CB|AB) = 0
# - P(CC|AB) = 0
# We can generalize this procedure for Markov chains of order higher than 2.
# 3) Given a probability distribution over symbols, how to use it for generating sentences?
#
# **Answer:**
#
# If we are given the probability distribution of all sequences of arbitrary number of symbols, say
# $$ p(X_1, X_2, \dots , X_n) $$
# We can compute the conditional probability of the new symbol given the ones already generated as:
# $$p(X_{n+1} | X_1, X_2, \dots , X_n) = \frac{p(X_1, X_2, \dots , X_n, X_{n+1})}{p(X_1, X_2, \dots , X_n)}$$
# Then, we can start with a randomly chosen symbol (either by hand or by the single symbol frequencies), and sample the next one each time using these conditional probabilities.
# ### Practical questions
# In order to construct our IID and Markov Chain models we need some text. Our source will be a set of classical novels available at: https://www.lri.fr/~gcharpia/informationtheory/TP2_texts.zip
# We will use the symbols in each text to learn the probabilities of each model. The alphabet we suggest for the characters to use is string.printable which is made of $\sim 100$ characters. (see below)
# For both models, perform the following steps:
# 1) For different orders of dependencies, train the model on a novel and compute the associated entropy. What do you observe as the order increases? Explain your observations.
# 2) Use the other novels as test sets and compute the cross-entropy for each model trained previously. How to handle symbols (or sequences of symbols) not seen in the training set?
# 3) For each order of dependencies, compare the cross-entropy with the entropy. Explain and interpret the differences.
# 4) Choose the order of dependencies with the lowest cross-entropy and generate some sentences.
# 5) Train one model per novel and use the KL divergence in order to cluster the novels.
# Hints :
# - In the MC case limit yourself to order $2$ (the computation can become quite expensive). If you have $ M \sim 100$ characters, for order $1$ you will need a $\sim 100 \times 100$ matrix, for order $2$ a $\sim 10^4 \times 10^4$ matrix.
# - For the second order MC model you need to compute: $p(X_{i+1},X_{i}|X_{i},X_{i-1})$
# - It is possible to implement efficiently the two models with dictionaries inPython. For the IID model, a key of the dictionary is simply a symbol and the value is the number of occurrences of the symbol in the text. For a Markov chain, a key of the dictionary is also a symbol, but the value is a vector that contains the number of occurrences of each character of the alphabet. Notice that a symbol may consist of one or several characters. Note also that there is no need to explicitly consider all possible symbols; the ones that are observed in the training set are sufficient.
# - A low probability can be assigned to symbols not observed in the training-set.
# #### Computing stationary distribution
# Here we provide you two version of the function to compute the stationary distirbution of a markov chain and show a small example
# direct way to find pi (can be slow)
import math
import string
from collections import Counter
from itertools import product
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import seaborn as sns
def Compute_stationary_distribution(P_kl):
## P_kl must be the transition matrix from state l to state k!
evals, evecs = np.linalg.eig(P_kl)
evec1 = evecs[:, np.isclose(evals, 1)]
evec1 = evec1[:, 0]
pi = evec1 / evec1.sum()
pi = pi.real # stationary probability
return pi
# iteative way (should be faster)
def Compute_stationary_distribution_it(P_kl, n_it):
pi = np.random.uniform(size=P_kl.shape[0]) # initial state, can be a random one!
pi /= pi.sum()
# print(pi,pi.sum())
for t in range(n_it):
pi = np.matmul(P_kl, pi)
return pi
##simple example of computation of stationary distribution
n_it = 1000 ##remind to check that n_it is enough to reach convergence
P_kl = np.array([[0.7, 0.5], [0.3, 0.5]])
Compute_stationary_distribution_it(P_kl, n_it)
Compute_stationary_distribution(P_kl)
# #### Defining the Alphabet
# Example of uploading a text and filtering out characters which are not in the chosen alphabet
def import_text(file_name):
lines = []
with open(f"/kaggle/input/ittexts/texts/{file_name}.txt", encoding="UTF8") as f:
lines = f.readlines()
text = "\n".join(lines)
printable = set(string.printable)
text = "".join(filter(lambda x: x in printable, text))
return text
# #### IID - MODEL
class IIDModel:
"""An interface for the text model"""
VOCAB = sorted(string.printable)
def __init__(self, order=1):
self.order = order
self.counts = Counter("".join(x) for x in product(*[self.VOCAB] * order))
self.prob_distribution = None
def process(self, text):
for i in range(0, len(text), self.order):
if text[i : i + self.order] not in self.counts:
continue
self.counts[text[i : i + self.order]] += 1
self.prob_distribution = np.array(
[self.counts[w] for w in sorted(self.counts.keys())]
)
self.prob_distribution = self.prob_distribution / self.prob_distribution.sum()
return self
def getEntropy(self):
return -np.sum(self.prob_distribution * np.log2(self.prob_distribution))
def getCrossEntropy(self, text):
if isinstance(text, str):
other_model = IIDModel(self.order).process(text)
elif isinstance(text, IIDModel):
other_model = text
else:
raise ValueError("text must be a string or a IIDModel")
return -np.sum(self.prob_distribution * np.log2(other_model.prob_distribution))
def generate(self, length):
chars = np.random.choice(
sorted(self.counts.keys()), p=self.prob_distribution, size=length
)
return "".join(chars)
##clustering texts
def KL_divergence(dist1, dist2):
return np.sum(dist1 * np.log(dist1 / dist2))
# #### MARKOV CHAIN - MODEL
class MarkovModel:
"""An interface for the text model"""
VOCAB = sorted(string.printable)
def __init__(self, order=2):
self.order = order
self.transition_matrix = None
self.stationary = None
self.counts, self.priors = self.initialize_counts(order)
@classmethod
def initialize_counts(cls, order):
counts = dict()
priors = list()
for characters in product(*[cls.VOCAB] * order):
counts["".join(characters)] = {next_char: 1 for next_char in cls.VOCAB}
priors.append("".join(characters))
return counts, priors
def process(self, text):
for i in range(len(text) - self.order):
key = text[i : i + self.order]
value = text[i + self.order]
if key in self.counts:
if value in self.counts[key]:
self.counts[key][value] += 1
return self
def build_transition_matrix(self):
self.transition_matrix = np.zeros((len(self.priors), len(self.priors)))
for i, start in enumerate(self.priors):
for next_char, count in self.counts[start].items():
idx = self.priors.index(start[1:] + next_char)
self.transition_matrix[i, idx] += count
self.transition_matrix = self.transition_matrix / self.transition_matrix.sum(
axis=1, keepdims=True
)
self.stationary = Compute_stationary_distribution_it(
self.transition_matrix.T, 300
) ## iterations
return self
def getEntropy(self):
entropy = self.getCrossEntropy(self)
return entropy
def getCrossEntropy(self, text):
if self.transition_matrix is None:
self.build_transition_matrix()
if isinstance(text, str):
other_model = (
MarkovModel(self.order).process(text).build_transition_matrix()
)
elif isinstance(text, MarkovModel):
other_model = text
if other_model.transition_matrix is None:
other_model.build_transition_matrix()
entropy = 0
for k in range(len(self.stationary)):
for l in range(len(self.stationary)):
if other_model.transition_matrix[l, k] == 0:
continue
entropy -= (
self.stationary[k]
* self.transition_matrix[l, k]
* np.log2(other_model.transition_matrix[l, k])
)
return entropy
def generate(self, length):
initial = np.random.choice(list(self.counts.keys()))
text = initial
for i in range(length - self.order):
key = text[i : i + self.order]
if key in self.counts:
values, counts = zip(*self.counts[key].items())
probs = np.array(counts) / np.sum(counts)
char = np.random.choice(values, p=probs)
text += char
else:
print("Key not found")[1, 2, 3]
break
return text
# ## Questions
texts = ["Dostoevsky", "Alighieri", "Goethe", "Hamlet"]
models = []
for text_name in texts:
text = import_text(text_name)
for order in [1, 2, 3]:
iid_model = IIDModel(order).process(text)
models.append(
{"model": iid_model, "order": order, "text": text_name, "model_type": "iid"}
)
for order in [1, 2]:
markov_model = MarkovModel(order).process(text)
models.append(
{
"model": markov_model,
"order": order,
"text": text_name,
"model_type": "markov",
}
)
# 1) For different orders of dependencies, train the model on a novel and compute the associated entropy. What do you observe as the order increases? Explain your observations.
#
models = pd.DataFrame.from_records(models)
results = []
for ix, row in models[["order", "model_type"]].drop_duplicates().iterrows():
sub_df = models.loc[
(models["order"] == row.order) & (models["model_type"] == row.model_type),
["model", "text"],
]
for (model1, text1), (model2, text2) in product(sub_df.values, sub_df.values):
entropy = model1.getCrossEntropy(model2)
results.append(
{
"model_type": row.model_type,
"order": row.order,
"text_1": text1,
"text_2": text2,
"entropy": entropy,
}
)
results_df = pd.DataFrame.from_records(results)
plot_data = results_df[
(results_df["model_type"] == "iid") & (results_df["text_1"] == results_df["text_2"])
]
sns.lineplot(x="order", y="entropy", hue="text_1", data=plot_data)
plt.title("Entropy of IID model")
plot_data = results_df[
(results_df["model_type"] == "markov")
& (results_df["text_1"] == results_df["text_2"])
]
sns.barplot(hue="text_1", x="order", y="entropy", data=plot_data)
plt.title("Entropy of Markov chain model")
# **Answer**: The behavior of the entropy changes depending on the type of model. In the IID model, the entropy increases with the order, going from around 4 bits to almost 20 bits. We can associate this growth with the increase in the size of the vocabulary and the possibilities for the next character. As for the Markov chain model, the entropy decreases from around 10 bits in order 1 to close to 7 bits in order 2. The decrease in entropy can be understood by considering the way Markov chains work. By increasing the order, the model learns more from the data (since given 2 characters there is more information to try to predict the next one) and hence the probability distribution on the next character is more narrow, decreasing the entropy.
# 2) Use the other novels as test sets and compute the cross-entropy for each model trained previously. How to handle symbols (or sequences of symbols) not seen in the training set?
#
plot_dict = {}
for ix, row in results_df[["order", "model_type"]].drop_duplicates().iterrows():
sub_df = results_df.loc[
(results_df["order"] == row.order)
& (results_df["model_type"] == row.model_type),
["text_1", "text_2", "entropy"],
]
heatmap = sub_df.pivot(index="text_1", columns="text_2", values="entropy")
plot_dict[(row.model_type, row.order)] = heatmap
## IID model
fig, axes = plt.subplots(3, 1, figsize=(3, 9), sharex=True)
for i, ax in enumerate(axes):
sns.heatmap(plot_dict[("iid", i + 1)], annot=True, ax=ax)
## MC model
fig, axes = plt.subplots(2, 1, figsize=(3, 6), sharex=True)
for i, ax in enumerate(axes):
sns.heatmap(plot_dict[("markov", i + 1)], annot=True, ax=ax)
# **Answer**: We simply initialized the count with all the possibilites to 1 so we would never get zero probabilities that will make the cross entropy diverge.
# 3) For each order of dependencies, compare the cross-entropy with the entropy. Explain and interpret the differences.
# **Answer:** For the iid model we found that the differences between the entropy and the cross entropy decreases as the order increases. For instance, in the order 1 model, we encountered that the entropy of the *Alighieri* text was 4.2 bits and the cross entropy of this text with the others was more than 5.2 bit, which represents an increase of almost 25%. On the contrary, for the order 3 model, the increase was barely noticeable, 1 bit, or around 5% of the value of the entropy. This tendency is reverted in the Markov chain model, where the cross entropy increases with an increasing order. Again, we can explain this because with higher order the Markov chain is more deterministic and different texts will have more different distribution, increasing the cross entropy.
# 4) Choose the order of dependencies with the lowest cross-entropy and generate some sentences.
IID_LOWEST_ENTROPY = ("Alighieri", 1)
MARKOV_LOWEST_ENTROPY = ("Goethe", 2)
## IID MODEL
iid_model = models.loc[
(models["model_type"] == "iid")
& (models["text"] == IID_LOWEST_ENTROPY[0])
& (models["order"] == IID_LOWEST_ENTROPY[1]),
"model",
].values[0]
print(iid_model.generate(200))
## MC MODEL
mc_model = models.loc[
(models["model_type"] == "markov")
& (models["text"] == MARKOV_LOWEST_ENTROPY[0])
& (models["order"] == MARKOV_LOWEST_ENTROPY[1]),
"model",
].values[0]
print(mc_model.generate(200))
# 5) Train one model per novel and use the KL divergence in order to cluster the novels.
from sklearn.cluster import AgglomerativeClustering
from sklearn.metrics import pairwise_distances
from scipy.cluster.hierarchy import dendrogram
def KL_distance(dist1, dist2):
return 0.5 * KL_divergence(dist1, dist2) + 0.5 * KL_divergence(dist2, dist1)
def KL_distance_affinity(X):
return pairwise_distances(X, metric=KL_distance)
my_models = models[(models["model_type"] == "iid") & (models["order"] == 1)]
my_models
distributions = np.array(
my_models["model"].apply(lambda x: x.prob_distribution.tolist()).tolist()
)
cluster = AgglomerativeClustering(
distance_threshold=0,
n_clusters=None,
affinity=KL_distance_affinity,
linkage="single",
)
cluster.fit(distributions)
def plot_dendrogram(model, **kwargs):
# Create linkage matrix and then plot the dendrogram
# create the counts of samples under each node
counts = np.zeros(model.children_.shape[0])
n_samples = len(model.labels_)
for i, merge in enumerate(model.children_):
current_count = 0
for child_idx in merge:
if child_idx < n_samples:
current_count += 1 # leaf node
else:
current_count += counts[child_idx - n_samples]
counts[i] = current_count
linkage_matrix = np.column_stack(
[model.children_, model.distances_, counts]
).astype(float)
# Plot the corresponding dendrogram
dendrogram(linkage_matrix, **kwargs)
plot_dendrogram(cluster, truncate_mode="level", p=3)
|
# ## Impoprt dependencies and dataset
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
from sklearn.datasets import load_iris
iris = load_iris()
dir(iris)
df = pd.DataFrame(iris.data, columns=iris.feature_names)
df.head()
df.shape
df["target"] = iris.target
df.head()
df.isnull().sum()
# ## Visualize the dataset
df1 = df[df.target == 0]
df2 = df[df.target == 1]
df3 = df[df.target == 2]
plt.xlabel("sepal length (cm)")
plt.ylabel("sepal width (cm)")
plt.scatter(
df1["sepal length (cm)"], df1["sepal width (cm)"], color="black", marker="."
)
plt.scatter(df2["sepal length (cm)"], df2["sepal width (cm)"], color="red", marker=".")
plt.scatter(
df3["sepal length (cm)"], df3["sepal width (cm)"], color="green", marker="."
)
plt.xlabel("petal length (cm)")
plt.ylabel("petal width (cm)")
plt.scatter(
df1["petal length (cm)"], df1["petal width (cm)"], color="black", marker="+"
)
plt.scatter(df2["petal length (cm)"], df2["petal width (cm)"], color="red", marker="+")
plt.scatter(
df3["petal length (cm)"], df3["petal width (cm)"], color="green", marker="+"
)
# ## Split dataset for training and testing
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(
df.drop(["target"], axis="columns"), df.target, test_size=0.2, random_state=1
)
x_train.shape
x_test.shape
# # Using KNN
from sklearn.neighbors import KNeighborsClassifier
knn = KNeighborsClassifier(n_neighbors=3)
knn.fit(x_train, y_train)
knn.score(x_test, y_test)
from sklearn.metrics import confusion_matrix
y_pred = knn.predict(x_test)
cm = confusion_matrix(y_test, y_pred)
cm
import seaborn as sn
sn.heatmap(cm, annot=True)
plt.xlabel("Predicted")
plt.ylabel("True value")
from sklearn.metrics import classification_report
print(classification_report(y_test, y_pred))
# # Try with the digits dataset
from sklearn.datasets import load_digits
digits = load_digits()
dir(digits)
data = pd.DataFrame(digits.data)
target = pd.DataFrame(digits.target)
data.shape
target.shape
plt.gray()
plt.matshow(digits.images[0])
xtrain, xtest, ytrain, ytest = train_test_split(
data, target, test_size=0.2, random_state=1
)
d_knn = KNeighborsClassifier(n_neighbors=10)
d_knn.fit(xtrain, ytrain)
d_knn.score(xtest, ytest)
ypred = d_knn.predict(xtest)
dcm = confusion_matrix(ytest, ypred)
sn.heatmap(dcm, annot=True)
plt.xlabel("Predicted")
plt.ylabel("True value")
print(classification_report(ytest, ypred))
|
import tensorflow as tf
from tensorflow import keras
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
# import seaborn as sns
import cv2
from keras.models import Sequential
from keras.layers import Dense, Dropout, Flatten, Conv2D, MaxPool2D
from keras.callbacks import (
EarlyStopping,
ReduceLROnPlateau,
TensorBoard,
ModelCheckpoint,
)
from glob import glob
import os
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
train_dir = "/kaggle/working/train"
for dirname, _, filenames in os.walk("/kaggle/working"):
for filename in filenames:
print(os.path.join(dirname, filename))
# load data
filenames = glob("/kaggle/working/train/*.jpg")
labels = [1 if "dog" in pic else 0 for pic in os.listdir("/kaggle/working/train")]
# os.listdir trả về một list các file trong thư mục (trả về theo tên file)
# glob trả về một list các file trong thư mục theo định dạng
print(filenames[:5])
print(labels[:5])
df = pd.DataFrame({"filename": filenames, "label": labels})
df.head()
df["label"].value_counts().plot.bar()
df["label"] = df["label"].replace({0: "cat", 1: "dog"})
df.head()
# train test split
from sklearn.model_selection import train_test_split
train_df, validate_df = train_test_split(df, test_size=0.20, random_state=42)
train_df = train_df.reset_index(drop=True)
validate_df = validate_df.reset_index(drop=True)
# reset_index để reset lại index của dataframe
print(train_df.shape)
print(validate_df.shape)
# read images test
filenames_test = os.listdir("/kaggle/working/test1")
test_df = pd.DataFrame({"filename": filenames_test})
print(test_df.shape)
test_df.head()
# img processing
from keras.preprocessing.image import ImageDataGenerator
IMG_SIZE = (150, 150)
BATCH_SIZE = 32
datagen = ImageDataGenerator(rescale=1.0 / 255)
# rescale = 1./255: chuyển đổi các giá trị pixel từ 0-255 thành 0-1
train_generator = datagen.flow_from_dataframe(
dataframe=train_df,
# directory='../dataset/train', # thư mục chứa ảnh
x_col="filename",
y_col="label",
class_mode="binary", # binary: 2 lớp, categorical: nhiều lớp
target_size=IMG_SIZE,
batch_size=BATCH_SIZE,
)
# flow_from_dataframe: tạo ra một generator từ dataframe
validation_generator = datagen.flow_from_dataframe(
dataframe=validate_df,
# directory='../dataset/train',
x_col="filename",
y_col="label",
class_mode="binary",
target_size=IMG_SIZE,
batch_size=BATCH_SIZE,
)
test_generator = datagen.flow_from_dataframe(
dataframe=test_df,
directory="/kaggle/working/test1",
x_col="filename",
y_col=None,
class_mode=None,
target_size=IMG_SIZE,
batch_size=BATCH_SIZE,
shuffle=False,
)
print(train_generator.class_indices)
# trả về một dict chứa các class và index của class đó
images = train_generator.next()[:10]
# plot 9 original training images
plt.figure(figsize=(10, 10))
for i in range(9):
plt.subplot(3, 3, i + 1)
plt.imshow(images[0][i])
plt.axis("off")
plt.tight_layout()
plt.show()
from keras.applications import VGG16
conv_base = VGG16(weights="imagenet", include_top=False, input_shape=(150, 150, 3))
# weights: 'imagenet' sử dụng các trọng số được học trên tập dữ liệu ImageNet
# include_top: False: không sử dụng các lớp fully-connected ở cuối mạng
# input_shape: kích thước ảnh đầu vào
# Build model
model = Sequential()
model.add(
Conv2D(
filters=32,
kernel_size=(3, 3),
input_shape=(150, 150, 3),
activation="relu",
padding="same",
)
)
model.add(MaxPool2D(pool_size=(2, 2)))
model.add(Conv2D(filters=64, kernel_size=(3, 3), activation="relu", padding="same"))
model.add(MaxPool2D(pool_size=(2, 2)))
model.add(Conv2D(filters=128, kernel_size=(3, 3), activation="relu", padding="same"))
model.add(MaxPool2D(pool_size=(2, 2)))
# Flatten
model.add(Flatten())
model.add(Dense(32, activation="relu"))
model.add(Dropout(0.5))
model.add(Dense(1, activation="sigmoid"))
model.summary()
# fit model
model.compile(loss="binary_crossentropy", optimizer="adam", metrics=["accuracy"])
callbacks = [
EarlyStopping(
monitor="val_loss", patience=5, verbose=1, mode="min"
), # mode in {'auto', 'min', 'max'}, patience: sau bao nhiêu epoch không có sự cải thiện thì dừng lại, mode='min': chỉ dừng lại khi loss giảm dần, mode='max': chỉ dừng lại khi loss tăng dần
ReduceLROnPlateau(
monitor="val_loss", factor=0.1, patience=3, verbose=1, mode="min"
), # factor: giảm learning rate bằng factor, patience: số lần không có sự cải thiện nào thì dừng lại, verbose: thông báo
]
history = model.fit(
train_generator,
epochs=100,
batch_size=BATCH_SIZE,
validation_data=validation_generator, # dùng để đánh giá mô hình
validation_steps=validate_df.shape[0]
// BATCH_SIZE, # số lượng ảnh dùng để đánh giá mô hình
steps_per_epoch=train_df.shape[0] // BATCH_SIZE, # số lượng ảnh dùng để train
verbose=1, # 0: không hiển thị, 1: hiển thị tiến trình, 2: hiển thị tiến trình chi tiết\
callbacks=callbacks,
)
# plot performance
pd.DataFrame(history.history).plot(figsize=(8, 5))
# predict
test_generator.reset()
pred = model.predict(test_generator, verbose=1)
print(pred[:10])
pred = pred > 0.5
pred_label = ["dog" if p == 1 else "cat" for p in pred]
print(pred_label[:10])
# plot 9 images with their predicted and true labels
plt.figure(figsize=(10, 10))
for i in range(9):
plt.subplot(3, 3, i + 1)
plt.imshow(images[0][i])
plt.title("Predicted: " + pred_label[i])
plt.axis("off")
plt.tight_layout()
# evaluate
model.evaluate(validation_generator)
model.save("model_first.h5")
# vgg16 + model
for layer in conv_base.layers:
layer.trainable = False
model_vgg = Sequential()
model_vgg.add(conv_base)
model_vgg.add(Flatten())
model_vgg.add(Dense(32, activation="relu"))
model_vgg.add(Dropout(0.5))
model_vgg.add(Dense(1, activation="sigmoid"))
model_vgg.summary()
model_vgg.compile(loss="binary_crossentropy", optimizer="adam", metrics="accuracy")
callbacks = [
EarlyStopping(
monitor="val_loss", patience=5, verbose=1, mode="min"
), # mode in {'auto', 'min', 'max'}, patience: sau bao nhiêu epoch không có sự cải thiện thì dừng lại, mode='min': chỉ dừng lại khi loss giảm dần, mode='max': chỉ dừng lại khi loss tăng dần
ReduceLROnPlateau(
monitor="val_loss", factor=0.1, patience=3, verbose=1, mode="min"
), # factor: giảm learning rate bằng factor, patience: số lần không có sự cải thiện nào thì dừng lại, verbose: thông báo
]
history_vgg = model_vgg.fit(
train_generator,
epochs=100,
batch_size=BATCH_SIZE,
validation_data=validation_generator, # dùng để đánh giá mô hình
validation_steps=validate_df.shape[0]
// BATCH_SIZE, # số lượng ảnh dùng để đánh giá mô hình
steps_per_epoch=train_df.shape[0] // BATCH_SIZE, # số lượng ảnh dùng để train
verbose=1, # 0: không hiển thị, 1: hiển thị tiến trình, 2: hiển thị tiến trình chi tiết
callbacks=callbacks,
)
# plot performance
pd.DataFrame(history_vgg.history).plot(figsize=(8, 5))
# predict
test_generator.reset()
pred = model_vgg.predict(test_generator, verbose=1)
print(pred[:10])
pred = pred > 0.5
pred_label = ["dog" if p == 1 else "cat" for p in pred]
print(pred_label[:10])
# plot 9 images with their predicted and true labels
plt.figure(figsize=(10, 10))
for i in range(9):
plt.subplot(3, 3, i + 1)
plt.imshow(images[0][i])
plt.title("Predicted: " + pred_label[i])
plt.axis("off")
plt.tight_layout()
model_vgg.save("model_second_vgg.h5")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import numpy as np
import seaborn as sns
import pandas as pd
import matplotlib.pyplot as plt
import sklearn
file_path = "/kaggle/input/house-prices-advanced-regression-techniques/train.csv"
file = pd.read_csv(file_path)
file.head()
file.columns
null_val = file.isnull()
null_count = null_val.sum()
columns = null_count.sort_values(ascending=False)
file.set_index("Id", inplace=True)
columns.index
file[
["PoolQC", "MiscFeature", "Alley", "Fence", "FireplaceQu", "LotFrontage"]
].isnull().sum()
file.drop_duplicates(inplace=True)
# these 4 columns have high missing values(around 1400 values are missing out of 1100) so they were deleted
file.drop(columns=["PoolQC", "MiscFeature", "Alley", "Fence"], inplace=True)
column = file.isnull().sum().sort_values(ascending=False)
file.info()
file[
[
"FireplaceQu",
"LotFrontage",
"GarageYrBlt",
"GarageCond",
"GarageType",
"GarageFinish",
"GarageQual",
"BsmtFinType2",
"BsmtExposure",
"BsmtQual",
"BsmtCond",
"BsmtFinType1",
"MasVnrArea",
"MasVnrType",
"Electrical",
]
].isnull().sum()
file["FireplaceQu"].describe()
# lot frontage should be of integer or float type but some values are in string form so
# Replace all string values with NaN
file["LotFrontage"] = file["LotFrontage"].replace(
to_replace=r"^.*$", value=np.nan, regex=True
)
file.LotFrontage.astype("float64")
file.describe()
# missing values in lotfrontage replaced by median
file.LotFrontage.fillna(file.LotFrontage.median(), inplace=True)
file.isnull().sum().sort_values(ascending=False)
file.FireplaceQu.fillna(file.FireplaceQu.mode()[0], inplace=True)
file.isnull().sum().sort_values(ascending=False)
file.GarageYrBlt.fillna(file.GarageYrBlt.median(), inplace=True)
file.isnull().sum().sort_values(ascending=False)
file.fillna(file.mode(), inplace=True)
file.isnull().sum()
file.to_csv("train cleaned.csv")
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import plotly.express as px
from xgboost import XGBRegressor
from lightgbm import LGBMRegressor
from catboost import CatBoostRegressor
from sklearn.linear_model import HuberRegressor, TheilSenRegressor
from sklearn.linear_model import LinearRegression, Ridge, Lasso, ElasticNet
from sklearn.metrics import (
mean_squared_error,
mean_absolute_error,
r2_score,
make_scorer,
)
from sklearn.model_selection import train_test_split, cross_val_score, GridSearchCV
from sklearn.preprocessing import LabelEncoder, StandardScaler, RobustScaler
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import (
accuracy_score,
roc_auc_score,
confusion_matrix,
classification_report,
)
from sklearn.model_selection import train_test_split, cross_validate
from sklearn.neighbors import KNeighborsClassifier
from sklearn.model_selection import GridSearchCV, cross_validate
from sklearn.preprocessing import StandardScaler
from sklearn.tree import DecisionTreeRegressor
from sklearn.ensemble import RandomForestRegressor, GradientBoostingRegressor
from sklearn.svm import SVR
from sklearn.experimental import enable_iterative_imputer
from sklearn.impute import IterativeImputer
from sklearn.neighbors import KNeighborsRegressor
pd.set_option("display.max_rows", None)
pd.reset_option("display.max_rows")
import warnings
warnings.filterwarnings("ignore")
pd.set_option("display.max_columns", None)
pd.set_option("display.width", 500)
# # Import Data
#
df = pd.read_csv(
"/kaggle/input/prediction-of-surgery-duration/train.csv", encoding="Latin-1"
)
df.head()
# # Data Overwiev
# Check Dataset:
def check_data(dataframe, head=5):
print(20 * "-" + "Information".center(20) + 20 * "-")
print(dataframe.info())
print(20 * "-" + "Data Shape".center(20) + 20 * "-")
print(dataframe.shape)
print("\n" + 20 * "-" + "The First 5 Data".center(20) + 20 * "-")
print(dataframe.head())
print("\n" + 20 * "-" + "The Last 5 Data".center(20) + 20 * "-")
print(dataframe.tail())
print("\n" + 20 * "-" + "Missing Values".center(20) + 20 * "-")
print(dataframe.isnull().sum())
print("\n" + 40 * "-" + "Describe the Data".center(40) + 40 * "-")
print(dataframe.describe().T)
check_data(df)
df.describe().transpose()
set_ICD10_unique = set()
list_len_ICD10 = []
list_spell_data = []
def DiagnosticICD10Code(values):
"""
This function takes in a string of ICD-10 codes separated by commas, and returns the same string as well
as the number of codes and a list of the first letter of each code in separate columns.
"""
try:
splited_value = values.split("'")
except AttributeError:
list_len_ICD10.append(np.nan)
list_spell_data.append(np.nan)
return values
return_str = ""
temp_list = []
for index in range(1, len(splited_value), 2):
val = splited_value[index]
return_str += val + ","
set_ICD10_unique.add(val[0])
temp_list.append(val[0])
list_len_ICD10.append(len(temp_list))
list_spell_data.append(temp_list)
return return_str, len(temp_list), temp_list
df[["DiagnosticICD10Code", "Lenght_ICD10_Code", "ICD10_Code_Spell"]] = (
df["DiagnosticICD10Code"].apply(DiagnosticICD10Code).apply(pd.Series)
)
set_SurgeyGroup_unique = set(["A1", "A2", "A3", "B", "C", "D", "E"])
def SurgeryGroup(values):
# Again we build the Null value trap to throw these back
try:
values.split(",")
# Don't forget the null values
except AttributeError:
return np.nan
# Quick fix for our str input
values = values[1:-1]
# Check if it is include 0 or not
start_index = values.find("0")
if start_index + 1:
values = values[start_index + 3 : -1]
return_str = ""
# If its not we can change the our str data more usefull form
for val in values.split(","):
val = val.strip(" ").strip("'")
return_str += val + ","
return return_str
SurgeryGroup(df)
df.replace("{0}", np.nan, inplace=True)
df.info()
df["SurgeryGroup"] = df["SurgeryGroup"].apply(SurgeryGroup)
df.replace(",", np.nan, inplace=True)
df.head()
df.groupby("AnesthesiaType")["ElapsedTime(second)"].mean()
import matplotlib.pyplot as plt
anesthesia_type_mean_time = df.groupby("AnesthesiaType")["ElapsedTime(second)"].mean()
plt.plot(anesthesia_type_mean_time.index, anesthesia_type_mean_time.values)
plt.xticks(rotation=90)
plt.xlabel("Anesthesia Type")
plt.ylabel("Elapsed Time (seconds)")
plt.title("Mean Elapsed Time by Anesthesia Type")
plt.show()
list_Index = ["A1", "A2", "A3", "B", "C", "D", "E"]
def SurgeryGroup_OneHotEncoder(data_fream):
# Let's create the our storage lists
list_New_columns = [[], [], [], [], [], [], []]
# Go throug the all the values on our columns
for val in data_fream.SurgeryGroup.values:
# Split the our values create the list to easy check
val = str(val).split(",")
# Fast tour on our list
for index in range(len(list_Index)):
# Check the is value NaN or not
if "nan" in val:
list_New_columns[index].append(np.nan)
# Quick check the we have the value in our data point
elif list_Index[index] in val:
list_New_columns[index].append(1)
else:
list_New_columns[index].append(0)
return list_New_columns
new_col_list = SurgeryGroup_OneHotEncoder(df)
# Little for loop go trough our list
for index in range(len(list_Index)):
df[list_Index[index]] = new_col_list[index]
df.head()
# # Label Encoding
labelencoder = LabelEncoder()
for i in [
"A",
"B",
"C",
"D",
"E",
"F",
"G",
"H",
"I",
"J",
"K",
"L",
"M",
"N",
"O",
"P",
"Q",
"R",
"S",
"U",
"V",
"Z",
]:
df[i] = df["ICD10_Code_Spell"].apply(
lambda x: sum([1 for code in " ".join(str(x)).split() if code.startswith(i)])
)
print(i, df[i].sum())
for i in [
"A",
"B",
"C",
"D",
"E",
"F",
"G",
"H",
"I",
"J",
"K",
"L",
"M",
"N",
"O",
"P",
"Q",
"R",
"S",
"U",
"V",
"Z",
]:
df[i] = df[i].apply(lambda x: 1 if x > 1 else x)
df.head()
df["AnesthesiaType"] = labelencoder.fit_transform(df["AnesthesiaType"])
df["Service"] = labelencoder.fit_transform(df["Service"])
df["SurgeryNameEncoded"] = labelencoder.fit_transform(df["SurgeryName"])
df.head()
# # Binary Coding
def label_encoder(dataframe, binary_col):
labelencoder = LabelEncoder()
dataframe[binary_col] = labelencoder.fit_transform(dataframe[binary_col])
return dataframe
label_encoder(df, "Sex")
def ICD10_Code_Laplace(dataFrame, s):
# Return list storage for our function
return_list_col = []
# Quick go trough the values
for val in dataFrame.DiagnosticICD10Code.values:
# Assign the our sum variable
res_val = 0
# Little "try except" block for our NaN values
try:
list_val = val.split(",")[:-1]
except AttributeError:
res_val = np.nan
list_val = []
# Quick go trough of in our data point values
for str_val in list_val:
# Determine the our frequency values with little math
val = ord(str_val[0:1]) - 64
freq = val / 4
freq_2 = int(str_val[1:3]) / 20
# Little calculation
res_val += (s + freq) / ((s + freq) ** 2 + freq_2**2)
return_list_col.append(res_val)
return return_list_col
def SurgeryGroup_Laplace(dataFrame, s):
# Return list storage for our function
return_list_col_SurgeryGroup = []
# Quick go trough the values
for val in dataFrame.SurgeryGroup.values:
# Assign the our sum variable
res_val = 0
# Little "try except" block for our NaN values
try:
list_val = val.split(",")[:-1]
except AttributeError:
res_val = np.nan
list_val = []
# Quick go trough of in our data point values
for str_val in list_val:
# Determine the our frequency values with little math
val = ord(str_val[0:1]) - 64
freq = val / 4
if len(str_val) > 2:
freq_2 = int(str_val[1:3]) / 20
else:
freq_2 = 0
# Little calculation
res_val += (s + freq) / ((s + freq) ** 2 + freq_2**2)
return_list_col_SurgeryGroup.append(res_val)
return return_list_col_SurgeryGroup
df["ICD10_Code_Laplace"] = ICD10_Code_Laplace(df, 1)
df["SurgeryGroup_Laplace"] = SurgeryGroup_Laplace(df, 1)
df.drop(["DiagnosticICD10Code", "SurgeryGroup", "SurgeryName"], axis=1, inplace=True)
df = df.drop("ICD10_Code_Spell", axis=1)
df.head()
df.info()
# İteratif imputasyon modeli
# NaN değerleri iteratif olarak doldur
imp = IterativeImputer(KNeighborsRegressor(), max_iter=10, random_state=0)
df_imputed_array = imp.fit_transform(df)
df_imputed = pd.DataFrame(data=df_imputed_array, columns=df.columns)
# Let's scaling with RobustScaler
df_imputed["Age"] = RobustScaler().fit_transform(df_imputed[["Age"]])
# target and independent variables:
y = df_imputed["ElapsedTime(second)"]
X = df_imputed.drop(["ElapsedTime(second)", "ID"], axis=1)
X_train, X_test, y_train, y_test = train_test_split(
X, y, random_state=15, test_size=0.2, shuffle=True
)
print(f"The shape of X_train is --> {(X_train.shape)}")
print(f"The shape of X_test is --> {(X_test.shape)}")
print(f"The shape of y_train is --> {(y_train.shape)}")
print(f"The shape of y_test is --> {(y_test.shape)}")
def evaluate_models(X_train, X_test, y_train, y_test):
models = [
LinearRegression(),
Ridge(),
Lasso(),
ElasticNet(),
DecisionTreeRegressor(),
RandomForestRegressor(),
GradientBoostingRegressor(),
XGBRegressor(),
LGBMRegressor(),
CatBoostRegressor(verbose=False),
]
for model in models:
model_name = type(model).__name__
model.fit(X_train, y_train)
y_train_pred = model.predict(X_train)
y_test_pred = model.predict(X_test)
train_rmse = mean_squared_error(y_train, y_train_pred, squared=False)
test_rmse = mean_squared_error(y_test, y_test_pred, squared=False)
train_r2 = r2_score(y_train, y_train_pred)
test_r2 = r2_score(y_test, y_test_pred)
print(f"{model_name}:")
print(f"Train RMSE: {train_rmse:.2f}")
print(f"Test RMSE: {test_rmse:.2f}")
print(f"Train R^2: {train_r2:.2f}")
print(f"Test R^2: {test_r2:.2f}")
print("-------------------------------------------------------")
evaluate_models(X_train, X_test, y_train, y_test)
df_test = pd.read_csv(
"/kaggle/input/prediction-of-surgery-duration/test.csv", encoding="Latin-1"
)
df_test[["DiagnosticICD10Code", "Lenght_ICD10_Code", "ICD10_Code_Spell"]] = (
df_test["DiagnosticICD10Code"].apply(DiagnosticICD10Code).apply(pd.Series)
)
SurgeryGroup(df_test)
df_test.replace("{0}", np.nan, inplace=True)
df_test.info()
df_test["SurgeryGroup"] = df_test["SurgeryGroup"].apply(SurgeryGroup)
df_test.replace(",", np.nan, inplace=True)
new_col_list = SurgeryGroup_OneHotEncoder(df_test)
# Little for loop go trough our list
for index in range(len(list_Index)):
df_test[list_Index[index]] = new_col_list[index]
df_test["AnesthesiaType"] = labelencoder.fit_transform(df_test["AnesthesiaType"])
df_test["Service"] = labelencoder.fit_transform(df_test["Service"])
df_test["SurgeryNameEncoded"] = labelencoder.fit_transform(df_test["SurgeryName"])
label_encoder(df_test, "Sex")
df_test["ICD10_Code_Laplace"] = ICD10_Code_Laplace(df_test, 1)
df_test["SurgeryGroup_Laplace"] = SurgeryGroup_Laplace(df_test, 1)
for i in [
"A",
"B",
"C",
"D",
"E",
"F",
"G",
"H",
"I",
"J",
"K",
"L",
"M",
"N",
"O",
"P",
"Q",
"R",
"S",
"U",
"V",
"Z",
]:
df_test[i] = df_test["ICD10_Code_Spell"].apply(
lambda x: sum([1 for code in " ".join(str(x)).split() if code.startswith(i)])
)
print(i, df_test[i].sum())
for i in [
"A",
"B",
"C",
"D",
"E",
"F",
"G",
"H",
"I",
"J",
"K",
"L",
"M",
"N",
"O",
"P",
"Q",
"R",
"S",
"U",
"V",
"Z",
]:
df_test[i] = df_test[i].apply(lambda x: 1 if x > 1 else x)
df_test.drop(
["DiagnosticICD10Code", "SurgeryGroup", "SurgeryName", "ICD10_Code_Spell"],
axis=1,
inplace=True,
)
# İteratif imputasyon modeli
# NaN değerleri iteratif olarak doldur
imp = IterativeImputer(KNeighborsRegressor(), max_iter=10, random_state=0)
df_imputed_array_test = imp.fit_transform(df_test)
df_imputed_test = pd.DataFrame(data=df_imputed_array_test, columns=df_test.columns)
# Let's scaling with RobustScaler
df_imputed_test["Age"] = RobustScaler().fit_transform(df_imputed_test[["Age"]])
df_imputed_test.head()
df_imputed.head()
# df_test1 ve df_train veri çerçevelerindeki sütun sayılarını bulun
print("df_test1 sütun sayısı:", df_imputed_test.shape[1])
print("df_train sütun sayısı:", df_imputed.shape[1])
df_imputed_test["ElapsedTime(second)"] = 0
# Özelliklerin ve hedef değişkenin ayrılması
X_train = df_imputed.drop(["ElapsedTime(second)", "ID"], axis=1)
y_train = df_imputed["ElapsedTime(second)"]
X_test = df_imputed_test.drop(["ID"], axis=1)
y_test = df_imputed_test["ElapsedTime(second)"]
X_train
df_imputed_test.isnull().sum()
model = CatBoostRegressor(verbose=False, random_state=42)
model.fit(X_train, y_train)
model.get_all_params()
params = {"learning_rate": [0.01], "depth": [7], "l2_leaf_reg": [1], "subsample": [1.0]}
grid = GridSearchCV(estimator=model, param_grid=params, cv=5, n_jobs=-1)
grid.fit(X_train, y_train)
print(grid.best_params_)
best_model = CatBoostRegressor(**grid.best_params_, verbose=False, random_state=42)
best_model.fit(X_train, y_train)
y_pred = best_model.predict(X_test)
mse = mean_squared_error(y_test, y_pred)
rmse = np.sqrt(mse)
print(f"RMSE: {rmse}")
df_imputed_test["ElapsedTime(second)"] = model.predict(X_test)
df_imputed_test["ElapsedTime(second)"]
df_imputed_test["ID"] = df_imputed_test["ID"].astype("Int32")
# özellik önemlerini bir dizi olarak al
importances = model.feature_importances_
# özelliklerin isimlerini bir dizi olarak al
feature_names = X_train.columns
# özellik isimleri ve önemleri içeren bir veri çerçevesi oluştur
feature_importances = pd.DataFrame(
{"feature": feature_names, "importance": importances}
)
# Önemliliğe göre sırala
feature_importances = feature_importances.sort_values(by="importance", ascending=False)
# Veri çerçevesini kullanarak bir bar grafik oluştur
plt.figure(figsize=(10, 8))
plt.bar(x=feature_importances["feature"], height=feature_importances["importance"])
plt.xticks(rotation=90, fontsize=5)
plt.title("Random Forest Feature Importance")
plt.xlabel("Feature")
plt.ylabel("Importance")
plt.show()
# submission.csv dosyasının oluşturulması
submission = pd.DataFrame(
{
"ID": df_imputed_test["ID"],
"ElapsedTime(second)": df_imputed_test["ElapsedTime(second)"],
}
)
submission.to_csv("submission.csv", index=False)
|
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# # Import Dependecies
import pandas as pd
import numpy as np
from sklearn.model_selection import TimeSeriesSplit
from sklearn.metrics import mean_absolute_percentage_error
import matplotlib.pyplot as plt
import seaborn as sns
import os
import xgboost
from xgboost import XGBRegressor
from lightgbm import LGBMClassifier
from sklearn.metrics import mean_squared_error
color_pal = sns.color_palette()
plt.style.use("fivethirtyeight")
df = pd.read_csv("/kaggle/input/gdz-elektrik-datathon-2023/train.csv")
med_df = pd.read_csv("/kaggle/input/gdz-elektrik-datathon-2023/med.csv")
df_sub = pd.read_csv("/kaggle/input/gdz-elektrik-datathon-2023/sample_submission.csv")
df = df.set_index("Tarih")
df_sub = df_sub.set_index("Tarih")
df.index = pd.to_datetime(df.index)
df_sub.index = pd.to_datetime(df_sub.index)
df.plot(style=".", figsize=(15, 5), color=color_pal[0], title="Dağıtılan Enerji (MWh)")
plt.show()
tscv = TimeSeriesSplit(n_splits=5)
fig, ax = plt.subplots(5, 1, figsize=(15, 12))
fig.subplots_adjust(bottom=0.2)
for i, (train_index, test_index) in enumerate(tscv.split(df)):
train_part = df.iloc[train_index]
test_part = df.iloc[test_index]
print(f"Fold {i}:")
print(f" Train: index={train_index}")
print(f" Test: index={test_index}")
train_part.plot(
ax=ax[i], label="Training Set", title="Time Series Train/Test Split"
)
test_part.plot(ax=ax[i], label="Test Set")
plt.tight_layout()
plt.show()
# # Feature Engineering
def get_time_features(df):
df = df.copy()
df["hour"] = df.index.hour
df["dayofweek"] = df.index.dayofweek.astype("int32")
df["quarter"] = df.index.quarter
df["month"] = df.index.month
df["year"] = df.index.year
df["dayofyear"] = df.index.dayofyear
df["dayofmonth"] = df.index.day
df["weekofyear"] = df.index.isocalendar().week.astype("int32")
return df
def get_lags(df):
df["6_hrs_lag"] = df["Dağıtılan Enerji (MWh)"].shift(6)
df["12_hrs_lag"] = df["Dağıtılan Enerji (MWh)"].shift(12)
df["24_hrs_lag"] = df["Dağıtılan Enerji (MWh)"].shift(24)
return df
def get_mean(df):
df["6_hrs_mean"] = df["Dağıtılan Enerji (MWh)"].rolling(window=6).mean()
df["12_hrs_mean"] = df["Dağıtılan Enerji (MWh)"].rolling(window=12).mean()
df["24_hrs_mean"] = df["Dağıtılan Enerji (MWh)"].rolling(window=24).mean()
return df
def get_std(df):
df["6_hrs_std"] = df["Dağıtılan Enerji (MWh)"].rolling(window=6).std()
df["12_hrs_std"] = df["Dağıtılan Enerji (MWh)"].rolling(window=12).std()
df["24_hrs_std"] = df["Dağıtılan Enerji (MWh)"].rolling(window=24).std()
return df
def get_max(df):
df["6_hrs_max"] = df["Dağıtılan Enerji (MWh)"].rolling(window=6).max()
df["12_hrs_max"] = df["Dağıtılan Enerji (MWh)"].rolling(window=12).max()
df["24_hrs_max"] = df["Dağıtılan Enerji (MWh)"].rolling(window=24).max()
return df
def get_min(df):
df["6_hrs_min"] = df["Dağıtılan Enerji (MWh)"].rolling(window=6).max()
df["12_hrs_min"] = df["Dağıtılan Enerji (MWh)"].rolling(window=12).max()
df["24_hrs_min"] = df["Dağıtılan Enerji (MWh)"].rolling(window=24).max()
return df
def get_med_day(df):
pass
df_all = get_time_features(df)
df_all = get_lags(df_all)
df_all = get_mean(df_all)
df_all = get_std(df_all)
df_all = get_max(df_all)
df_all = get_min(df_all)
df_all = df_all.fillna(0)
df_all.head()
# # Parameters Optimization
# # Model Training
tss = TimeSeriesSplit(n_splits=5)
df_all = df_all.sort_index()
preds = []
scores = []
for train_idx, val_idx in tss.split(df_all):
train = df_all.iloc[train_idx]
test = df_all.iloc[val_idx]
FEATURES = [
"hour",
"6_hrs_max",
"24_hrs_lag",
"dayofweek",
"6_hrs_std",
"6_hrs_mean",
"12_hrs_std",
"12_hrs_lag",
"dayofyear",
]
# ['hour', 'dayofweek', 'quarter', 'month',
# 'year', 'dayofyear', 'dayofmonth', 'weekofyear', '6_hrs_lag',
# '12_hrs_lag', '24_hrs_lag', '6_hrs_mean', '12_hrs_mean', '24_hrs_mean',
# '6_hrs_std', '12_hrs_std', '24_hrs_std', '6_hrs_max', '12_hrs_max',
# '24_hrs_max', '6_hrs_min', '12_hrs_min', '24_hrs_min']
TARGET = "Dağıtılan Enerji (MWh)"
X_train = train[FEATURES]
y_train = train[TARGET]
X_test = test[FEATURES]
y_test = test[TARGET]
reg = xgb.XGBRegressor(
base_score=0.5,
booster="gbtree",
n_estimators=1000,
early_stopping_rounds=50,
objective="reg:squarederror",
max_depth=3,
learning_rate=0.01,
)
reg.fit(
X_train, y_train, eval_set=[(X_train, y_train), (X_test, y_test)], verbose=100
)
y_pred = reg.predict(X_test)
preds.append(y_pred)
score = mean_absolute_percentage_error(y_test, y_pred)
scores.append(score)
print(f"Score across folds {np.mean(scores):0.4f}")
print(f"Fold scores:{scores}")
xgboost.plot_importance(reg)
plt.figure(figsize=(16, 12))
plt.show()
df_sub
df_sub["isFuture"] = True
df["isFuture"] = False
df_and_future = pd.concat([df, df_sub])
df_and_future = get_time_features(df_and_future)
df_and_future = get_lags(df_and_future)
df_and_future = get_mean(df_and_future)
df_and_future = get_std(df_and_future)
df_and_future = get_max(df_and_future)
df_and_future = get_min(df_and_future)
df_and_future = df_and_future.fillna(0)
df_and_future.head()
future_w_features = df_and_future.query("isFuture").copy()
future_w_features["pred"] = reg.predict(future_w_features[FEATURES])
future_w_features["pred"].plot(
figsize=(10, 5), color=color_pal[4], ms=1, lw=1, title="Future Predictions"
)
plt.show()
sample_submission_df = (
future_w_features["pred"]
.reset_index()
.rename(columns={"pred": "Dağıtılan Enerji (MWh)"})
)
sample_submission_df.to_csv("sample_submission.csv", index=False)
sample_submission_df
df_sub.drop(["isFuture"], axis=1).reset_index()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import numpy as np
import pandas as pd
from sklearn.preprocessing import MinMaxScaler
from sklearn.model_selection import train_test_split
from keras.models import Sequential
from keras.layers import LSTM, Dense
from keras.callbacks import EarlyStopping
import matplotlib.pyplot as plt
# 读入数据
df = pd.read_csv("/kaggle/input/stockprice1/chinadianxing1.csv")
x = df.columns.difference(["Date", "close"])
# 将数据拆分成特征和标签
X = df[x].values
y = df[["close"]].values
# 将数据集拆分为训练集和测试集
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=42
)
# 归一化数据
scaler_X = MinMaxScaler()
scaler_y = MinMaxScaler()
X_train = scaler_X.fit_transform(X_train)
X_test = scaler_X.transform(X_test)
y_train = scaler_y.fit_transform(y_train)
y_test = scaler_y.transform(y_test)
# 准备数据成 LSTM 模型所需的格式
X_train = np.reshape(X_train, (X_train.shape[0], 1, X_train.shape[1]))
X_test = np.reshape(X_test, (X_test.shape[0], 1, X_test.shape[1]))
# 建立 LSTM 模型
model = Sequential()
model.add(LSTM(units=50, input_shape=(X_train.shape[1], X_train.shape[2])))
model.add(Dense(units=1))
model.compile(loss="mean_squared_error", optimizer="adam")
# 训练模型
early_stop = EarlyStopping(monitor="val_loss", patience=3)
history = model.fit(
X_train,
y_train,
epochs=30,
batch_size=32,
validation_data=(X_test, y_test),
callbacks=[early_stop],
verbose=2,
)
# 使用模型进行预测
y_pred = model.predict(X_test)
# 反归一化预测结果和真实结果
y_pred = scaler_y.inverse_transform(y_pred)
y_test = scaler_y.inverse_transform(y_test)
# 画出 true-prediction 图
plt.plot(y_test, label="True")
plt.plot(y_pred, label="Prediction")
plt.legend()
plt.show()
# 画出 loss-epoch 图
plt.plot(history.history["loss"], label="Training Loss")
plt.plot(history.history["val_loss"], label="Validation Loss")
plt.title("Training and Validation Loss")
plt.xlabel("Epochs")
plt.ylabel("Loss")
plt.legend()
plt.show()
from keras.layers import GRU
# 建立 GRU 模型
model_gru = Sequential()
model_gru.add(GRU(units=50, input_shape=(X_train.shape[1], X_train.shape[2])))
model_gru.add(Dense(units=1))
model_gru.compile(loss="mean_squared_error", optimizer="adam")
# 训练 GRU 模型
history_gru = model_gru.fit(
X_train,
y_train,
epochs=30,
batch_size=32,
validation_data=(X_test, y_test),
callbacks=[early_stop],
verbose=2,
)
# 使用 GRU 模型进行预测
y_pred_gru = model_gru.predict(X_test)
# 反归一化预测结果
y_pred_gru = scaler_y.inverse_transform(y_pred_gru)
from keras.layers import Dropout
# 建立改进的 LSTM 模型
improved_model = Sequential()
improved_model.add(
LSTM(
units=50,
return_sequences=True,
input_shape=(X_train.shape[1], X_train.shape[2]),
)
)
improved_model.add(Dropout(0.2))
improved_model.add(LSTM(units=50, return_sequences=True))
improved_model.add(Dropout(0.2))
improved_model.add(LSTM(units=50))
improved_model.add(Dropout(0.2))
improved_model.add(Dense(units=1))
improved_model.compile(loss="mean_squared_error", optimizer="adam")
history_improved = improved_model.fit(
X_train,
y_train,
epochs=30,
batch_size=32,
validation_data=(X_test, y_test),
callbacks=[early_stop],
verbose=2,
)
y_pred_improved = improved_model.predict(X_test)
y_pred_improved = scaler_y.inverse_transform(y_pred_improved)
from tcn import TCN
# 建立 TCN 模型
tcn_model = Sequential()
tcn_model.add(
TCN(
input_shape=(X_train.shape[1], X_train.shape[2]),
nb_filters=32,
kernel_size=2,
nb_stacks=1,
dilations=[1, 2, 4, 8],
activation="relu",
)
)
tcn_model.add(Dropout(0.2))
tcn_model.add(Dense(units=1))
tcn_model.compile(loss="mean_squared_error", optimizer="adam")
history_tcn = tcn_model.fit(
X_train,
y_train,
epochs=100,
batch_size=32,
validation_data=(X_test, y_test),
callbacks=[early_stop],
verbose=2,
)
y_pred_tcn = tcn_model.predict(X_test)
y_pred_tcn = scaler_y.inverse_transform(y_pred_tcn)
plt.plot(y_test, label="True")
plt.plot(y_pred, label="LSTM Prediction")
plt.plot(y_pred_improved, label="Improved LSTM Prediction")
plt.plot(y_pred_gru, label="GRU Prediction")
plt.plot(y_pred_tcn, label="TCN Prediction")
plt.legend()
plt.show()
from sklearn.metrics import mean_squared_error
from sklearn.metrics import mean_absolute_error
# 计算 LSTM 的 MSE 和 RMSEmean_absolute_error
mae_lstm = mean_absolute_error(y_test, y_pred)
mse_lstm = mean_squared_error(y_test, y_pred)
rmse_lstm = np.sqrt(mse_lstm)
# 计算 GRU 的 MSE 和 RMSE
mae_gru = mean_absolute_error(y_test, y_pred_gru)
mse_gru = mean_squared_error(y_test, y_pred_gru)
rmse_gru = np.sqrt(mse_gru)
mae_improve = mean_absolute_error(y_test, y_pred_improved)
mse_improve = mean_squared_error(y_test, y_pred_improved)
rmse_improve = np.sqrt(mse_improve)
mae_tcn = mean_absolute_error(y_test, y_pred_tcn)
mse_tcn = mean_squared_error(y_test, y_pred_tcn)
rmse_tcn = np.sqrt(mse_tcn)
print("LSTM,GRU,IMPROVE_LSTM,TCN MAE")
print(mae_lstm, mae_gru, mae_improve, mae_tcn)
print("LSTM,GRU,IMPROVE_LSTM,TCN MSE")
print(mse_lstm, mse_gru, mse_improve, mse_tcn)
print("LSTM,GRU,IMPROVE_LSTM,TCN RMSE")
print(rmse_lstm, rmse_gru, rmse_improve, rmse_tcn)
# 创建指标列表
metrics = ["MSE", "RMSE", "MAE"]
lstm_values = [mse_lstm, rmse_lstm, mae_lstm]
improved_lstm_values = [mse_improve, rmse_improve, mae_improve]
gru_values = [mse_gru, rmse_gru, mae_gru]
tcn_values = [mse_tcn, rmse_tcn, mae_tcn]
# 设置绘图参数
x = np.arange(len(metrics))
width = 0.25
# 绘制条形图
fig, ax = plt.subplots()
rects1 = ax.bar(x - width, lstm_values, width, label="LSTM")
rects2 = ax.bar(x, improved_lstm_values, width, label="Improved LSTM")
rects3 = ax.bar(x + width, gru_values, width, label="GRU")
rects4 = ax.bar(x + 2 * width, tcn_values, width, label="TCN")
# 设置坐标轴和图例
ax.set_ylabel("Error")
ax.set_title("Error Comparison between LSTM, Improved LSTM, and GRU")
ax.set_xticks(x)
ax.set_xticklabels(metrics)
ax.legend()
# 定义显示数值的函数
def autolabel(rects):
for rect in rects:
height = rect.get_height()
ax.annotate(
f"{height:.4f}",
xy=(rect.get_x() + rect.get_width() / 2, height),
xytext=(0, 3),
textcoords="offset points",
ha="center",
va="bottom",
)
# 显示数值
autolabel(rects1)
autolabel(rects2)
autolabel(rects3)
autolabel(rects4)
# 设置y轴格式
ax.yaxis.set_major_formatter(mticker.FormatStrFormatter("%.4f"))
fig.tight_layout()
plt.show()
|
# # **Introduction**
# The FAOSTAT Temperature Change domain disseminates statistics of mean surface temperature change by country, with annual updates. The current dissemination covers the period 1961–2019. Statistics are available for monthly, seasonal and annual mean temperature anomalies, i.e., temperature change with respect to a baseline climatology, corresponding to the period 1951–1980. The standard deviation of the temperature change of the baseline methodology is also available. Data are based on the publicly available GISTEMP data, the Global Surface Temperature Change data distributed by the National Aeronautics and Space Administration Goddard Institute for Space Studies (NASA-GISS).
# libraries used
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
from matplotlib import pyplot as plt
import seaborn as sns
from sklearn.linear_model import LinearRegression
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import PolynomialFeatures
from sklearn.pipeline import Pipeline
from sklearn import metrics
# Below is the data present in the file.
Etemp_path = (
"../input/temperature-change/Environment_Temperature_change_E_All_Data_NOFLAG.csv"
)
Code_path = "../input/temperature-change/FAOSTAT_data_11-24-2020.csv"
data = pd.read_csv(Etemp_path, encoding="latin-1")
data2 = pd.read_csv(Code_path)
data
plt.figure(figsize=(20, 8))
sns.heatmap(data.isnull(), yticklabels=False, cbar=False, cmap="viridis")
plt.show()
# Due to world politics being in contant flux many countries have come into existence or have gone the way of the dodo. This means our dataset is incomplete as shown by the above plot where the bright lines show null/nan values in our dataset. A simple thing we do to remedy this is to just remove the rows which contain nan values.
data = data.dropna()
# Next let us look at the second file.
data2
# The second dataset just contains codes used for different countries and groups. This set is not important for our study.
# As we can see the main dataset contains the temperature change and standard deviation for countries around the world from 1961 to 2019.
# For the purposes of this investigation we:
# * delete columns which contain the various codes as they wont be nessessary.
# * rename the Area column to country
# * keep the temperature change and standard deviation
# * keep only the 12 months and get rid of the three month grouping
data = data.rename(columns={"Area": "Country"})
# data=data[data['Element']=='Temperature change']
data = data.drop(columns=["Area Code", "Months Code", "Element Code", "Unit"])
TempC = data.loc[
data.Months.isin(
[
"January",
"February",
"March",
"April",
"May",
"June",
"July",
"August",
"September",
"October",
"November",
"December",
]
)
]
# After getting rid of the extra columns that are not needed the dataset now looks like the table below.
TempC.head()
# Lets look at how many countries we have in the data set.
TempC.Country.unique()
# This is obviously a lot, the dataset also contains some groupings of countries such as EU, Least Developed Countries, etc. Lets just use one country for now to see what data can be extrated and understood
# # **Afghanistan as a case study**
# Let use start the study by looking at what data in present for each country and what information can be extracted from it. The table below show the data present for just Afghanistan.
Afg = TempC.loc[TempC.Country == "Afghanistan"]
Afg
# Lets make a simple plot to see how the temperature varies over the year.
plt.figure(figsize=(15, 10))
sns.lineplot(
x=Afg.Months.loc[Afg.Element == "Temperature change"],
y=Afg.Y1961.loc[Afg.Element == "Temperature change"],
label="Y1961",
)
sns.lineplot(
x=Afg.Months.loc[Afg.Element == "Temperature change"],
y=Afg.Y1971.loc[Afg.Element == "Temperature change"],
label="Y1971",
)
sns.lineplot(
x=Afg.Months.loc[Afg.Element == "Temperature change"],
y=Afg.Y1981.loc[Afg.Element == "Temperature change"],
label="Y1981",
)
sns.lineplot(
x=Afg.Months.loc[Afg.Element == "Temperature change"],
y=Afg.Y1991.loc[Afg.Element == "Temperature change"],
label="Y1991",
)
sns.lineplot(
x=Afg.Months.loc[Afg.Element == "Temperature change"],
y=Afg.Y2001.loc[Afg.Element == "Temperature change"],
label="Y2001",
)
plt.xlabel("Months")
plt.ylabel("Temperature change (C)")
plt.title("Temperature Change in Afganistan")
plt.show()
# We reshape the dataset so instead of a column for each year we now have all the years in one column. Although this makes the dataset have more rows it makes maipulation a bit simpler (for me anyway).
Afg = Afg.melt(
id_vars=["Country", "Months", "Element"], var_name="Year", value_name="TempC"
)
Afg["Year"] = Afg["Year"].str[1:].astype("str")
Afg.info()
# Lets replot the temperature change over the year and the standard deviation provided for each month.
plt.figure(figsize=(15, 15))
plt.subplot(211)
for i in Afg.Year.unique():
plt.plot(
Afg.Months.loc[Afg.Year == str(i)].loc[Afg.Element == "Temperature change"],
Afg.TempC.loc[Afg.Year == str(i)].loc[Afg.Element == "Temperature change"],
linewidth=0.5,
)
plt.plot(
Afg.Months.unique(),
Afg.loc[Afg.Element == "Temperature change"].groupby(["Months"]).mean(),
"r",
linewidth=2.0,
label="Average",
)
plt.xlabel(
"Months",
)
plt.xticks(rotation=45)
plt.ylabel("Temperature change")
plt.title("Temperature Change in Afganistan")
plt.legend()
plt.subplot(212)
plt.plot(
Afg.Months.loc[Afg.Year == "1961"].loc[Afg.Element == "Standard Deviation"],
Afg.TempC.loc[Afg.Year == "1961"].loc[Afg.Element == "Standard Deviation"],
)
plt.xlabel("Year")
plt.xticks(rotation=45)
plt.ylabel("Standard Deviation")
plt.title("Standard Deviation of Temperature Change in Afganistan")
plt.subplots_adjust(hspace=0.3)
plt.show()
# It looks like the month of July has the smallest devation while the winter months of December January and Febuary have a lage spread with febuary having a highest. The Standard Deviation is calculated by looking at the temperature for that month in a country from 1961 to 2019. So each country has their own standerd deviation of temperature for the different months.
# Next lets look at the how the data is spread over the different years and how the mean temperature changes.
plt.figure(figsize=(15, 10))
plt.scatter(
Afg["Year"].loc[Afg.Element == "Temperature change"],
Afg["TempC"].loc[Afg.Element == "Temperature change"],
)
plt.plot(
Afg.loc[Afg.Element == "Temperature change"].groupby(["Year"]).mean(),
"r",
label="Average",
)
plt.axhline(y=0.0, color="k", linestyle="-")
plt.xlabel("Year")
plt.xticks(np.linspace(0, 58, 20), rotation=45)
plt.ylabel("Temperature change")
plt.legend()
plt.title("Temperature Change in Afganistan")
plt.show()
# We can also look at the histogram of the temperature changes
plt.figure(figsize=(15, 10))
sns.histplot(
Afg.TempC.loc[Afg.Element == "Temperature change"], kde=True, stat="density"
)
plt.axvline(x=0.0, color="b", linestyle="-")
plt.xlabel("Temperature change")
plt.title("Temperature Change in Afganistan")
plt.show()
# Clearly a majority of Afganistan's temperature is higher then the world baseline corresponding to the period 1951–1980. Even the average temperature change for Afganistan is rising as the years progress.
# # **World Temperature**
# Obviously the above study can be done to every contry and area in the dataset but this will just be repeting the above graphs for more then 200 times. We need to try and summerise the data and present it in a more digestable way. We create a similar dataset to what was used above for the world datset.
TempC = TempC.melt(
id_vars=["Country", "Months", "Element"], var_name="Year", value_name="TempC"
)
TempC["Year"] = TempC["Year"].str[1:].astype("str")
TempC
# To make sure country groupings such as EU or Africa don't skew our calculations we remove them for the world data list. So we are just left with individual countires. We can keep the regions data in the different dataset in case we want to use it later.
regions = TempC[
TempC.Country.isin(
[
"World",
"Africa",
"Eastern Africa",
"Middle Africa",
"Northern Africa",
"Southern Africa",
"Western Africa",
"Americas",
"Northern America",
"Central America",
"Caribbean",
"South America",
"Asia",
"Central Asia",
"Eastern Asia",
"Southern Asia",
"South-Eastern Asia",
"Western Asia",
"Europe",
"Eastern Europe",
"Northern Europe",
"Southern Europe",
"Western Europe",
"Oceania",
"Australia and New Zealand",
"Melanesia",
"Micronesia",
"Polynesia",
"European Union",
"Least Developed Countries",
"Land Locked Developing Countries",
"Small Island Developing States",
"Low Income Food Deficit Countries",
"Net Food Importing Developing Countries",
"Annex I countries",
"Non-Annex I countries",
"OECD",
]
)
]
TempC = TempC[
~TempC.Country.isin(
[
"World",
"Africa",
"Eastern Africa",
"Middle Africa",
"Northern Africa",
"Southern Africa",
"Western Africa",
"Americas",
"Northern America",
"Central America",
"Caribbean",
"South America",
"Asia",
"Central Asia",
"Eastern Asia",
"Southern Asia",
"South-Eastern Asia",
"Western Asia",
"Europe",
"Eastern Europe",
"Northern Europe",
"Southern Europe",
"Western Europe",
"Oceania",
"Australia and New Zealand",
"Melanesia",
"Micronesia",
"Polynesia",
"European Union",
"Least Developed Countries",
"Land Locked Developing Countries",
"Small Island Developing States",
"Low Income Food Deficit Countries",
"Net Food Importing Developing Countries",
"Annex I countries",
"Non-Annex I countries",
"OECD",
]
)
]
TempC
# Now we can look at the distribution data. Lets first look at a histogram for temperature change.
plt.figure(figsize=(15, 10))
sns.histplot(
TempC.TempC.loc[TempC.Element == "Temperature change"], kde=True, stat="density"
)
plt.axvline(x=0.0, color="b", linestyle="-")
plt.xlabel("Temperature change")
plt.title("Temperature Change distribution of the World")
plt.xlim(-5, 5)
plt.show()
# Let us calculate some averages that we can easily use for our plots.
# Average for the whole world
AvgT = (
TempC.loc[TempC.Element == "Temperature change"]
.groupby(["Year"], as_index=False)
.mean()
)
# Average for every country
AvgTC = (
TempC.loc[TempC.Element == "Temperature change"]
.groupby(["Country", "Year"], as_index=False)
.mean()
)
# We can also do a scatter plot, like before, for different years for all the countries and plot the world average.
plt.figure(figsize=(15, 10))
plt.scatter(
TempC["Year"].loc[TempC.Element == "Temperature change"],
TempC["TempC"].loc[TempC.Element == "Temperature change"],
)
plt.plot(AvgT.Year, AvgT.TempC, "r", label="Average")
plt.axhline(y=0.0, color="k", linestyle="-")
plt.xlabel("Year")
plt.xticks(np.linspace(0, 58, 20), rotation=45)
plt.ylabel("Temperature change")
plt.legend()
plt.title("Temperature Change of the World")
plt.show()
# Finally we can plot the temperatures for each country and plot the world average on top.
plt.figure(figsize=(15, 10))
for i in AvgTC.Country.unique():
plt.plot(
AvgTC.Year.loc[AvgTC.Country == str(i)],
AvgTC.TempC.loc[AvgTC.Country == str(i)],
linewidth=0.5,
)
plt.plot(AvgT.Year, AvgT.TempC, "r", linewidth=2.0)
plt.axhline(y=0.0, color="k", linestyle="-")
plt.xlabel("Year")
plt.xticks(np.linspace(0, 58, 20), rotation=45)
plt.ylabel("Average Temperature change")
plt.title("Average Temperature Change of the World")
plt.show()
# The plot clearly shows how the temperature of the world is rising when compared to the basline corresponding to the period 1951–1980. A lot of work needs to be clearly done to get the temperature in control. In the next part we will try and create a machine learning model to predict how the temperatures will change in the future.
# # Test-Train Split
# Before we can make a prediction we need to train our model. We will split the data into a test and train dataset. This can be used to verify the predictability of our model.
MonthV = {
"January": "1",
"February": "2",
"March": "3",
"April": "4",
"May": "5",
"June": "6",
"July": "7",
"August": "8",
"September": "9",
"October": "10",
"November": "11",
"December": "12",
}
TempC = TempC.replace(MonthV)
TempC.head()
y = TempC["TempC"].loc[TempC.Element == "Temperature change"]
X = TempC.drop(columns=["TempC", "Country", "Months", "Element"]).loc[
TempC.Element == "Temperature change"
]
X_train, X_valid, y_train, y_valid = train_test_split(
X, y, train_size=0.8, random_state=42
)
# # Regression
# One of the simplest models we can use on the system is regression. Below we use both a linear model and polynomial models to predict world the temperatures in the future.
# ## Simple Linear Regression
# We use the test-train data to train the model and compare the predictions to actual data.
LR = LinearRegression()
LR.fit(X_train, y_train)
LRpreds = LR.predict(X_valid)
print("RMSE:", np.sqrt(metrics.mean_squared_error(y_valid, LRpreds)))
plt.figure(figsize=(15, 8))
plt.plot(y_valid - LRpreds, "o")
plt.axhline(y=0.0, color="k", linestyle="-")
plt.ylabel("Actual value - Predicited value")
plt.show()
# We are happy with what the model is prediciting so we use the whole dataset to train a model.
# Fit the model to the training data
LR.fit(X, y)
# We now create artifical data that we can use to test what the model predicts for the future.
# Creating prediction data
LR_test = pd.DataFrame({"Year": np.random.randint(1980, 2060, size=1000)})
LR_test = LR_test.sort_values(by=["Year"]).reset_index(drop=True).astype(str)
# T_test=pd.DataFrame(np.arange(2020, 2046),columns=['Year']).astype(str)
# Generate test predictions
preds_test = LR.predict(LR_test)
LR_test["TempC"] = pd.Series(preds_test, index=LR_test.index)
# ## Polynomial regression
# As seen in different plots the data is clearly not linear so let us experiment with some nonlinear features to see if we can get a more accirate prediction.
PR2_mod = Pipeline(
[
("poly", PolynomialFeatures(degree=2)),
("linear", LinearRegression(fit_intercept=False)),
]
)
PR3_mod = Pipeline(
[
("poly", PolynomialFeatures(degree=5)),
("linear", LinearRegression(fit_intercept=False)),
]
)
# Fit the model to the training data
PR2_mod.fit(X, y)
PR3_mod.fit(X, y)
# Creating prediction data
PR2_test = pd.DataFrame({"Year": np.random.randint(1980, 2060, size=1000)})
PR2_test = PR2_test.sort_values(by=["Year"]).reset_index(drop=True).astype(str)
PR3_test = pd.DataFrame({"Year": np.random.randint(1980, 2060, size=1000)})
PR3_test = PR3_test.sort_values(by=["Year"]).reset_index(drop=True).astype(str)
# Generate test predictions
pred2_test = PR2_mod.predict(PR2_test)
pred3_test = PR3_mod.predict(PR3_test)
PR2_test["TempC"] = pd.Series(pred2_test, index=PR2_test.index)
PR3_test["TempC"] = pd.Series(pred3_test, index=PR3_test.index)
# # Plotting Results
# Lets plot the reuslts for the linear and polynomial models side-by-side to see how they fair.
plt.figure(figsize=(15, 10))
for i in AvgTC.Country.unique():
plt.plot(
AvgTC.Year.loc[AvgTC.Country == str(i)],
AvgTC.TempC.loc[AvgTC.Country == str(i)],
linewidth=0.5,
)
plt.plot(AvgT.Year, AvgT.TempC, "r", linewidth=2.0)
plt.plot(
LR_test.Year.unique(),
LR_test.groupby("Year").mean(),
"b",
linewidth=2.0,
label="Linear Model",
)
plt.plot(
PR2_test.Year.unique(),
PR2_test.groupby("Year").mean(),
"g",
linewidth=2.0,
label="Poly-2 Model",
)
plt.plot(
PR3_test.Year.unique(),
PR3_test.groupby("Year").mean(),
"c",
linewidth=2.0,
label="Poly-5 Model",
)
plt.axhline(y=0.0, color="k", linestyle="-")
plt.xticks(np.linspace(0, 100, 40), rotation=45)
plt.xlabel("Year")
plt.ylabel("Average Temperature change")
plt.title("Average Temperature Change of the World")
plt.legend()
plt.show()
|
# **What have done here:**
# Predicted Next 7 days data using Support Vector Machine, Linear Regression, Deep Neural Network.
# Import Libraries
import pandas as pd
import math
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import sklearn
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
from sklearn import preprocessing
from sklearn.model_selection import cross_validate
from sklearn.model_selection import train_test_split
from sklearn.svm import SVR
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import LSTM, Dense, Dropout
from sklearn.linear_model import LinearRegression
# Data loading
data = pd.read_csv(
"/kaggle/input/stock-price-forecast/ISCTR.IS (1).csv", parse_dates=True, index_col=0
)
data = data.dropna()
data["High_Low_per"] = (data["High"] - data["Close"]) / data["Close"] * 100
data["Per_change"] = (data["Open"] - data["Open"]) / data["Close"] * 100
data = data[["Adj Close", "High_Low_per", "Per_change", "Volume"]]
label_col = "Adj Close"
ceil_data = int(math.ceil(0.002 * len(data)))
data["label"] = data[label_col].shift(-ceil_data)
X = data.drop(["label"], axis=1).values
X = preprocessing.scale(X)
X = X[:-ceil_data]
# Predict on next days
forecast = X[-ceil_data - 7 : -ceil_data]
y = data["label"].values
# Drop rows with missing values
data.dropna(inplace=True)
# Convert label column to numpy array
y = data["label"].values
# Split the data into training and testing sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
# # **Support Vector Machine**
svr = SVR(kernel="rbf", C=1000, gamma=0.1)
svr.fit(X_train, y_train)
# Predict the test data
prediction_svm = svr.predict(X_test)
# Predict the next 7 days of data
svm_next7days = svr.predict(forecast)
print("Predictions on Test Data:", prediction_svm)
# Prediction for next 7 days
print("Predictions for Next 7 Days:", svm_next7days)
# # **Linear Regression**
lr = LinearRegression()
lr.fit(X_train, y_train)
lr_prediction = lr.predict(X_test)
# Predict the next 7 days of data
lr_next7days = lr.predict(forecast)
print("Predictions on Test Data:", lr_prediction)
# Prediction for next 7 days
print("Predictions for Next 7 Days:", lr_next7days)
# # **Deep Neural Network**
# Model Building
model = Sequential()
model.add(LSTM(units=50, return_sequences=True, input_shape=(X_train.shape[1], 1)))
model.add(Dropout(0.2))
model.add(LSTM(units=50, return_sequences=True))
model.add(Dropout(0.2))
model.add(LSTM(units=50))
model.add(Dropout(0.2))
model.add(Dense(units=1))
# Compile the model
model.compile(optimizer="adam", loss="mean_squared_error")
# Train the model
model.fit(X_train, y_train, epochs=40, batch_size=30, validation_data=(X_test, y_test))
dnn_prediction = model.predict(X_test)
# Predict for the next 7 days data
dnn_next7days = model.predict(forecast)
print("Predictions on Test Data:", dnn_prediction)
# Prediction for next 7 days
print("Predictions for Next 7 Days:", dnn_next7days)
# # **Visualization**
# Visualizing the comparision of the days
# Create a list of days
days = [1, 2, 3, 4, 5, 6, 7]
# Plot the data
plt.plot(days, svm_next7days, color="blue", label="SVM")
plt.plot(days, lr_next7days, color="red", label="Linear Regression")
plt.plot(days, dnn_next7days, color="green", label="DNN")
# Add labels and title
plt.xlabel("Days")
plt.ylabel("Predicted Stock Price")
plt.title("Stock Price Prediction for Next 7 Days")
# Add legend
plt.legend()
# Show the plot
plt.show()
# Define the predicted values for SVM, Linear Regression, and DNN
svm_next7days = [
12.25693225,
10.1123252,
10.7488707,
11.30425637,
10.27959834,
11.24091329,
9.13999752,
]
lr_next7days = [
12.11145881,
11.59601589,
11.26758707,
10.94751006,
10.76198022,
10.94537396,
9.76628752,
]
dnn_next7days = [
11.5351515,
11.206194,
11.2260275,
11.203941,
11.088118,
11.192676,
10.310493,
]
# Create a pandas dataframe with the predicted values
data = {
"Days": [1, 2, 3, 4, 5, 6, 7],
"SVM": svm_next7days,
"Linear Regression": lr_next7days,
"DNN": dnn_next7days,
}
df = pd.DataFrame(data)
# Reshape the data to create a "long" format for the barplot
df = df.melt(id_vars=["Days"], var_name="Algorithm", value_name="Predicted Stock Price")
# Create the barplot using Seaborn
sns.set(style="whitegrid")
ax = sns.barplot(x="Days", y="Predicted Stock Price", hue="Algorithm", data=df)
# Adjust the position of the legend
ax.legend(loc="upper left", bbox_to_anchor=(1.02, 1))
# Add a title
ax.set_title("Stock Price Prediction for Next 7 Days")
# Show the plot
plt.show()
|
#!pip install https://download.pytorch.org/whl/cu101/torch-1.10.0-cp38-cp38-linux_x86_64.whl
#!pip uninstall torch -y
#!pip3 install https://download.pytorch.org/whl/cu100/torch-1.10.0-cp36-cp36m-linux_x86_64.whl
# import torch
# print(torch.__version__)
import torch
import torch.nn.functional as F
import matplotlib.pyplot as plt
from torch import nn
from torch import Tensor
from PIL import Image
from torchvision.transforms import Compose, Resize, ToTensor
from torchvision.datasets import ImageFolder
from torch.utils.data import DataLoader
from torch.utils.data import random_split
from einops import rearrange, reduce
from einops.layers.torch import Rearrange, Reduce
transforms = Compose([Resize((224, 224)), ToTensor()])
training_data = ImageFolder(
root="/kaggle/input/100-bird-species/train", transform=transforms
)
test_data = ImageFolder(
root="/kaggle/input/100-bird-species/test", transform=transforms
)
train_set, test_set = random_split(
training_data, (int(len(training_data) * 0.7) + 1, int(len(training_data) * 0.3))
)
train_dataloader = DataLoader(train_set, batch_size=64, shuffle=True)
test_dataloader = DataLoader(test_set, batch_size=64, shuffle=True)
print(f"Training data size: {train_set}")
clases_list = training_data.classes
clases = {}
cont = 0
for i in clases_list:
clases[cont] = i
cont += 1
print(clases)
train_features, train_labels = training_data.__getitem__(0)
print(f"Tamaño de cada imagen: {train_features.size()}")
figure = plt.figure(figsize=(8, 8))
cols, rows = 3, 3
for i in range(1, cols * rows + 1):
sample_idx = torch.randint(len(training_data), size=(1,)).item()
img, label = training_data[sample_idx]
figure.add_subplot(rows, cols, i)
plt.title(clases[label])
plt.axis("off")
plt.imshow(img[1][:][:], cmap="gray")
plt.show()
for X, y in train_dataloader:
print(f"Shape of X [N, C, H, W]: {X.shape}")
print(f"Shape of y: {y.shape} {y.dtype}")
break
device = "cuda" if torch.cuda.is_available() else "cpu"
print(f"Using {device} device")
class PatchEmbedding(nn.Module):
def __init__(
self,
img_size: int = 224,
channels: int = 3,
section: int = 16,
output_net: int = 768,
):
super().__init__()
self.positions = nn.Parameter(
torch.randn((img_size // section) ** 2, output_net)
)
self.pos_drop = nn.Dropout(p=0.1)
self.network = nn.Sequential(
Rearrange("b c (h s1) (w s2) -> b (h w) (s1 s2 c)", s1=section, s2=section),
nn.Linear(section * section * channels, output_net),
)
def forward(self, images):
images = self.network(images)
images = self.pos_drop(images + self.positions)
return images
class MultiHeadAttention(nn.Module):
def __init__(self, output_net: int = 512, num_heads: int = 8):
super().__init__()
self.output_net = output_net
self.num_heads = num_heads
self.keys = nn.Linear(output_net, output_net)
self.queries = nn.Linear(output_net, output_net)
self.values = nn.Linear(output_net, output_net)
self.network = nn.Linear(output_net, output_net)
def forward(self, images):
# Creamos las matrices queries, keys y values haciendo una subdivision de las obtenidas de la red
queries = rearrange(
self.queries(images), "b n (h d) -> b h n d", h=self.num_heads
)
keys = rearrange(self.keys(images), "b n (h d) -> b h n d", h=self.num_heads)
values = rearrange(
self.values(images), "b n (h d) -> b h n d", h=self.num_heads
)
atencion = torch.einsum("bhqd, bhkd -> bhqk", queries, keys)
tamano = self.output_net ** (1 / 2)
atencion = atencion / tamano
att = F.softmax(atencion, dim=-1)
out = torch.einsum("bhal, bhlv -> bhav ", att, values)
out = rearrange(out, "b h n d -> b n (h d)")
out = self.network(out)
return out
class FeedForward(nn.Module):
def __init__(self, output: int = 768):
super().__init__()
self.network = nn.Sequential(
nn.Linear(output, 4 * output),
nn.GELU(),
nn.Linear(output * 4, output),
nn.Dropout(0.1),
)
def forward(self, images):
images = self.network(images)
return images
class ResidualConection(nn.Module):
def __init__(self, fn):
super().__init__()
self.fn = fn
def forward(self, images):
res = images
images = self.fn(images)
images += res
return images
output = 768
capa0 = ResidualConection(
nn.Sequential(nn.LayerNorm(output), MultiHeadAttention(output), nn.Dropout(0.1))
)
capa1 = ResidualConection(
nn.Sequential(nn.LayerNorm(output), FeedForward(output), nn.Dropout(0.1)),
)
class TransformerEncoder(nn.Module):
def __init__(self):
super().__init__()
self.network = nn.Sequential(capa0, capa1)
def forward(self, images):
images = self.network(images)
return images
class Transformer(nn.Module):
def __init__(self):
super().__init__()
self.network = nn.Sequential(
TransformerEncoder(),
TransformerEncoder(),
TransformerEncoder(),
TransformerEncoder(),
TransformerEncoder(),
TransformerEncoder(),
TransformerEncoder(),
TransformerEncoder(),
)
def forward(self, images):
images = self.network(images)
return images
class MLPHead(nn.Module):
def __init__(self, output: int = 768, n_classes: int = 400):
super().__init__()
self.network = nn.Sequential(
Reduce("b n e -> b e", reduction="mean"),
nn.LayerNorm(output),
nn.Linear(output, n_classes),
)
def forward(self, images):
images = self.network(images)
return images
class ViT(nn.Module):
def __init__(self):
super().__init__()
self.network = nn.Sequential(PatchEmbedding(), Transformer(), MLPHead())
def forward(self, images):
images = self.network(images)
return images
model = ViT()
model = torch.load("/kaggle/input/models/model116pth")
model.to(device)
loss_fn = nn.CrossEntropyLoss()
optimizer = torch.optim.SGD(model.parameters(), lr=3e-4)
def train(train_dataloader, model, loss_fn, optimizer):
size = len(train_dataloader.dataset)
model.train()
for batch, (X, y) in enumerate(train_dataloader):
X, y = X.to(device), y.to(device)
# Compute prediction error
pred = model(X)
loss = loss_fn(pred, y)
# Backpropagation
optimizer.zero_grad()
loss.backward()
optimizer.step()
if batch % 100 == 0:
loss, current = loss.item(), batch * len(X)
print(f"loss: {loss:>7f} [{current:>5d}/{size:>5d}]")
def test(dataloader, model, loss_fn):
size = len(dataloader.dataset)
num_batches = len(dataloader)
model.eval()
test_loss, correct = 0, 0
with torch.no_grad():
for X, y in dataloader:
X, y = X.to(device), y.to(device)
pred = model(X)
test_loss += loss_fn(pred, y).item()
correct += (pred.argmax(1) == y).type(torch.float).sum().item()
test_loss /= num_batches
correct /= size
print(
f"Test Error: \n Accuracy: {(100*correct):>0.1f}%, Avg loss: {test_loss:>8f} \n"
)
epochs = 3
for t in range(epochs):
print(f"Epoch {t+1}\n-------------------------------")
train(train_dataloader, model, loss_fn, optimizer)
test(test_dataloader, model, loss_fn)
print("Done!")
torch.save(model, "model116pth")
print("Model saved")
# define la ruta de la imagen que deseas cargar
ruta_imagen = "/kaggle/input/100-bird-species/valid/ABBOTTS BABBLER/1.jpg"
# carga la imagen utilizando la biblioteca PIL
imagen = Image.open(ruta_imagen).convert("RGB")
# aplica las transformaciones necesarias para la entrada de la red neuronal
imagen = transforms(imagen).unsqueeze(0)
# muestra la imagen cargada
plt.imshow(imagen.squeeze().permute(1, 2, 0))
# realiza la predicción de la imagen utilizando el modelo
with torch.no_grad():
imagen = imagen.to(device)
pred = model(imagen)
_, predicted_label = torch.max(pred, dim=1)
nombre_archivo = ruta_imagen.split("/")[-2]
# Reemplazar los guiones bajos con espacios para obtener el nombre del ave
nombre_ave = nombre_archivo.replace("_", " ")
print("Etiqueta verdadera: ", nombre_ave)
print("Etiqueta predicha: ", clases_list[predicted_label.item()])
|
# # Описание проекта
# Вы работаете специалистом по обработке данных в глобальной финансовой компании. На протяжении многих лет компания собирала основные банковские реквизиты и собрала много информации, связанной с кредитом. Руководство хочет создать интеллектуальную систему для разделения сотрудников по категориям кредитных баллов, чтобы сократить ручные усилия.
# # Цель и задачи проекта:
# ## Цель - предсказание кредитного рейтинга клиентов компании.
# ## Задачи:
# 1. Очистка и подготовка датасета к моделированию.
# 2. Отбор признаков, значимых для построения модели.
# 3. Учитывая информацию, связанную с кредитом человека, создать модель машинного обучения, которая может классифицировать кредитный рейтинг.
# Метрика В качестве метрики качества модели использовалась Accuracy - доля правильных ответов алгоритма.
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
import seaborn as sns
import plotly.express as px
from sklearn.linear_model import LogisticRegression
from sklearn.neighbors import KNeighborsClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.ensemble import AdaBoostClassifier
from sklearn.svm import SVC
from sklearn import ensemble
from sklearn import model_selection
from sklearn import metrics
from sklearn import neighbors
from sklearn import tree
from sklearn.metrics import (
confusion_matrix,
roc_auc_score,
accuracy_score,
f1_score,
recall_score,
precision_score,
)
from sklearn.metrics import classification_report
from sklearn.model_selection import cross_validate
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
name = os.path.join(dirname, filename)
print(name)
if "train.csv" in name:
train_file_name = name
elif "test.csv" in name:
test_file_name = name
print("train data :", train_file_name)
print("test data :", test_file_name)
# # 1. Предварительный анализ набора данных
# прочитаем training и test data
train_data = pd.read_csv(train_file_name)
test_data = pd.read_csv(test_file_name)
# Описание данных:
# 0. ID: Уникальный идентификатор записи
# 1. Customer_ID: Уникальный идентификатор клиента
# 2. Month: Месяц в году
# 3. Name: Имя клиента
# 4. Age: Возраст клиента
# 5. SSN: Номер социального страхования данного лица
# 6. Occupation: Род занятий клиента
# 7. Annual_Income: Годовой доход лица
# 8. Monthly_Inhand_Salary: Ежемесячная заработная плата клиента
# 9. Num_Bank_Accounts: Количество банковских счетов данного лица
# 10. Num_Credit_Card: Количество кредитных карт, имеющихся у данного лица
# 11. Interest_Rate: Процентная ставка по кредитной карте данного лица
# 12. Num_of_Loan: Количество кредитов, взятых лицом в банке
# 13. Type_of_Loan: Виды кредитов, взятых лицом в банке
# 14. Delay_from_due_date: Среднее количество дней, просроченных лицом с момента оплаты
# 15. Num_of_Delayed_Payment: Количество платежей, задержанных данным лицом
# 16. Changed_Credit_Card: Процентное изменение лимита кредитной карты данного лица
# 17. Num_Credit_Inquiries: Количество запросов по кредитной карте, сделанных данным лицом
# 18. Credit_Mix: Классификация кредитного портфеля клиента
# 19. Outstanding_Debt: Непогашенный баланс клиента
# 20. Credit_Utilization_Ratio: Коэффициент использования кредита по кредитной карте клиента
# 21. Credit_History_Age: Возраст кредитной истории лица
# 22. Payment_of_Min_Amount: Да, если лицо оплатило только минимальную сумму, подлежащую выплате, в противном случае нет
# 23. Total_EMI_per_month: Общий EMI человека в месяц
# 24. Amount_invested_monthly: Ежемесячная сумма, инвестируемая лицом
# 25. Payment_Behaviour: Платежное поведение лица
# 26. Monthly_Balance: Ежемесячный остаток, оставшийся на счете данного лица
# 27. Credit_Score: Кредитный рейтинг клиента
# посмотрим на данные
train_data.head(15)
test_data.head(15)
# Видно, что есть странные значения в различных колонках.
# метод info() покажет пропуски в данных и их тип
train_data.info()
cols = train_data.columns
# желтый - пропущенные данные, синий - не пропущенные
colours = ["#000099", "#ffff00"]
sns.heatmap(train_data[cols].isnull(), cmap=sns.color_palette(colours))
# Пропуски в процентах
train_data.isnull().sum() / len(train_data) * 100
# Во-первых, можно отметить, что в колонках ```Age```, ```Occupation```, ```Payment_Behaviour``` есть странные значения.
# Во-вторых, в колонках ```Name```, ```Monthly_Inhand_Salary```, ```Type_of_Loan```, ```Num_of_Delayed_Payment```, ```Num_Credit_Inquiries```, ```Credit_Hitory_Age```, ```Amount_invested_monthly```, и ```Monthly_Balance``` еть нулевые значения.
# # 2. Трансформирование и чистка данных
# удалим ID, Name и SSN (Customer_ID необходим для удаления дубликатов)
train_data = train_data.drop(columns=["ID", "Name", "SSN"])
test_data = test_data.drop(columns=["ID", "Name", "SSN"])
train_data[train_data["Customer_ID"] == "CUS_0xd40"]
# При выборе данных с определенным Customer_ID, можно заметить, что на каждого клиента в train_data приходится по 8 строк, а в test_data по 4 строки, это будем использовать позднее. Также отмечу, что есть различные пропуски в колонках, которые можно убрать путем соспоставления по Customer_ID. Есть данные, которые не меняются для одного и того же клиента, а есть данные, которые отличаются от месяца к месяцу.
# удаляем '_' в данных
# fixed /var : заменим ошибочные значения (используя замену значениями, указывающими на одного и того же клиента)
# int_fixed : целочисленное значение, имеет одинаковое значение для каждого клиента
# int_var : целочисленное значение, имеет разные значения для каждого клиента
int_fixed = [
"Age",
"Num_Bank_Accounts",
"Num_Credit_Card",
"Interest_Rate",
"Num_of_Loan",
]
int_var = ["Delay_from_due_date", "Num_of_Delayed_Payment"]
def convert_to_int(string):
try:
return int(str(string).replace("_", ""))
except:
return "nan"
# удалим '_' чтобы конвертировать в int
for data in [train_data, test_data]:
int_col_with_str = int_fixed + int_var
for int_col in int_col_with_str:
data[int_col] = data[int_col].apply(lambda x: convert_to_int(x))
# float_fixed : значение с плавающей точкой, имеет одинаковое значение для каждого клиента
# float_var : значение с плавающей точкой, имеет разные значения для каждого клиента
float_fixed = [
"Annual_Income",
"Monthly_Inhand_Salary",
"Num_Credit_Inquiries",
"Outstanding_Debt",
"Total_EMI_per_month",
]
float_var = [
"Credit_Utilization_Ratio",
"Amount_invested_monthly",
"Monthly_Balance",
"Changed_Credit_Limit",
]
def convert_to_float(string):
try:
return float(str(string).replace("_", ""))
except:
return "nan"
# удалим '_' чтобы конвертировать в float
for data in [train_data, test_data]:
float_col_with_str = float_fixed + float_var
for float_col in float_col_with_str:
data[float_col] = data[float_col].apply(lambda x: convert_to_float(x))
# Credit_History_Age преобразуем в месяцы
def convert_credit_history(credit_history_age):
if str(credit_history_age) == "nan":
return "nan"
else:
years = int(credit_history_age.split(" ")[0])
months = int(credit_history_age.split(" ")[3])
return 12 * years + months
# Проверю, какие значения есть в столбце 'Payment_Behaviour'
train_data["Payment_Behaviour"].value_counts()
# Разделяю Payment_Behaviour на 'spent' и 'value'
def convert_payment_behaviour(behaviour, split_name):
try:
if split_name == "spent":
return behaviour.split("_")[0]
elif split_name == "value":
return behaviour.split("_")[2]
else:
return "nan"
except:
return "nan"
# Проверю, какие значения есть в столбце 'Type_of_Loan'
train_data["Type_of_Loan"].value_counts()
# Type_of_Loan: если включает = 1 и не включает = 0 в определенный тип займа
def convert_type_of_loan(original_text, loan_type):
if original_text == "":
return "nan"
try:
loans = original_text.split(", ")
if loan_type in loans:
return 1
else:
return 0
except:
return "nan"
# найду все виды кредитов, такие как 'Credit-Builder Loan', 'Personal Loan', ...
merged_data = pd.concat([train_data, test_data])
loan_type_column = merged_data["Type_of_Loan"]
loan_type_all = []
for i in range(len(merged_data)):
try:
loan_types = loan_type_column.iloc[i].split(", ")
for loan_type in loan_types:
if len(loan_type) >= 5 and loan_type[:4] == "and ":
loan_type_all.append(loan_type[4:])
else:
loan_type_all.append(loan_type)
except:
pass
# не содержит 'Not Specified' тип займа
loan_type_all = list(set(loan_type_all) - set(["Not Specified"]))
print(loan_type_all)
# конвертируем колонки, для которых были написаны функции для конвертации
for data in [train_data, test_data]:
data["Credit_History_Age"] = data["Credit_History_Age"].apply(
lambda x: convert_credit_history(x)
)
data["Payment_Behaviour_Spent"] = data["Payment_Behaviour"].apply(
lambda x: convert_payment_behaviour(x, "spent")
)
data["Payment_Behaviour_Value"] = data["Payment_Behaviour"].apply(
lambda x: convert_payment_behaviour(x, "value")
)
for loan_type in loan_type_all:
data["Loan_Type_" + loan_type.replace(" ", "_")] = data["Type_of_Loan"].apply(
lambda x: convert_type_of_loan(x, loan_type)
)
# удаляю повторяющиеся колонки
train_data = train_data.drop(columns=["Payment_Behaviour", "Type_of_Loan"])
test_data = test_data.drop(columns=["Payment_Behaviour", "Type_of_Loan"])
# преобразуем месяцы из строкового типа в float, причем, чтобы их значения были в пределах от 0 до 1
def map_month(month_str):
months = [
"January",
"February",
"March",
"April",
"May",
"June",
"July",
"August",
"September",
"October",
"November",
"December",
]
return months.index(month_str) / 11
for data in [train_data, test_data]:
data["Month"] = data["Month"].apply(lambda x: map_month(x))
# проверяем
train_data["Month"].value_counts()
# заменила значения ошибок для столбцов, фиксированных для клиента
fixed_numeric_columns = int_fixed + float_fixed
nRowsTrain = len(train_data)
nRowsTest = len(test_data)
for col in fixed_numeric_columns:
print(
"current processing column : " + col
) # чтобы было видно, какой из столбцов обработался
# заменила на наиболее частое значение для того же клиента (всего 8 (train), 4 (test) значения для каждого клиента)
for i in range(nRowsTrain // 8):
column = train_data.loc[
train_data["Customer_ID"] == train_data["Customer_ID"].iloc[i * 8]
][col]
most_frequent_values = column.dropna().mode()
if len(most_frequent_values) > 0:
train_data[col].iloc[8 * i : 8 * (i + 1)] = most_frequent_values[0]
for i in range(nRowsTest // 4):
column = test_data.loc[
test_data["Customer_ID"] == test_data["Customer_ID"].iloc[i * 4]
][col]
most_frequent_values = column.dropna().mode()
if len(most_frequent_values) > 0:
test_data[col].iloc[4 * i : 4 * (i + 1)] = most_frequent_values[0]
# признаки Monthly_Inhand_Salary, Num_Credit_Inquiries, Amount_invested_monthly из NaN в str(NaN), чтобы потом заменить
for col in [
"Monthly_Inhand_Salary",
"Num_Credit_Inquiries",
"Amount_invested_monthly",
"Monthly_Balance",
]:
train_data[col] = train_data[col].apply(
lambda x: x if (pd.notnull(x) and convert_to_float(x) != "nan") else "NaN_float"
)
test_data[col] = test_data[col].apply(
lambda x: x if (pd.notnull(x) and convert_to_float(x) != "nan") else "NaN_float"
)
# словарь с нулевыми значениями для замены
null_values = {
"Occupation": "_______",
"Monthly_Inhand_Salary": "NaN_float",
"Num_Credit_Inquiries": "NaN_float",
"Amount_invested_monthly": "NaN_float",
"Monthly_Balance": "NaN_float",
"Loan_Type_Mortgage_Loan": "nan",
"Loan_Type_Auto_Loan": "nan",
"Loan_Type_Student_Loan": "nan",
"Loan_Type_Payday_Loan": "nan",
"Loan_Type_Debt_Consolidation_Loan": "nan",
"Loan_Type_Home_Equity_Loan": "nan",
"Loan_Type_Personal_Loan": "nan",
"Loan_Type_Credit-Builder_Loan": "nan",
"Num_of_Delayed_Payment": "nan",
"Credit_History_Age": "nan",
"Changed_Credit_Limit": "nan",
"Payment_Behaviour_Value": "nan",
"Credit_Mix": "_",
"Payment_Behaviour_Spent": "!@9#%8",
}
for null_value_col in [
"Occupation",
"Monthly_Inhand_Salary",
"Num_Credit_Inquiries",
"Credit_Mix",
"Loan_Type_Mortgage_Loan",
"Loan_Type_Auto_Loan",
"Loan_Type_Student_Loan",
"Loan_Type_Payday_Loan",
"Loan_Type_Debt_Consolidation_Loan",
"Loan_Type_Home_Equity_Loan",
"Loan_Type_Personal_Loan",
"Loan_Type_Credit-Builder_Loan",
]:
print("current processing column : " + null_value_col)
# замените на наиболее частое значение для того же клиента (всего 8 (train), 4 (test) значения для каждого клиента)
for i in range(nRowsTrain // 8):
column = train_data.loc[
train_data["Customer_ID"] == train_data["Customer_ID"].iloc[i * 8]
][null_value_col]
mode_values = column.loc[column != null_values[null_value_col]].mode()
if len(mode_values) > 0:
most_frequent = mode_values[0]
train_data[null_value_col].iloc[8 * i : 8 * (i + 1)] = most_frequent
for i in range(nRowsTest // 4):
column = test_data.loc[
test_data["Customer_ID"] == test_data["Customer_ID"].iloc[i * 4]
][null_value_col]
mode_values = column.loc[column != null_values[null_value_col]].mode()
if len(mode_values) > 0:
most_frequent = mode_values[0]
test_data[null_value_col].iloc[4 * i : 4 * (i + 1)] = most_frequent
# Поскольку столбцы были обработаны, столбец Customer_ID можно удалить
train_data = train_data.drop(columns=["Customer_ID"])
test_data = test_data.drop(columns=["Customer_ID"])
# заполнила медианой для того же клиента
def replace_with_median(
value, idx, data_arr, rows_per_customer, null_value, is_round=False
):
# если совпадает с null_value, то значение обрабатывается как есть
if value != null_value:
return value
start_idx = (idx // rows_per_customer) * rows_per_customer
end_idx = (idx // rows_per_customer + 1) * rows_per_customer
data_range = data_arr[start_idx:end_idx]
values = []
fraction = -1
for data_value in data_range:
if data_value != null_value:
values.append(float(data_value))
fraction = float(data_value) % 1.0
if len(values) == 0:
return null_value
else:
result = np.median(values)
if is_round:
return result if abs(result % 1.0 - fraction) < 0.25 else result + 0.5
else:
return result
# заполнила значение другим значением, указывающим на некоторую информацию о клиенте
# даже если значение столбца не полностью совпадает с конкретным клиентом
# Amount_invested_monthly, Monthly_Balance -> медиана (от того же клиента)
# Changed_Credit_Limit, Num_of_Delayed_Payment -> округленная медиана с той же дробной частью (от того же клиента)
# НЕ использую среднее значение из-за экстремальных выбросов в некоторых столбцах
for null_value_col in [
"Amount_invested_monthly",
"Num_of_Delayed_Payment",
"Changed_Credit_Limit",
"Monthly_Balance",
]:
print("current processing column : " + null_value_col)
train_data_np = []
test_data_np = []
rounded = null_value_col in ["Num_of_Delayed_Payment", "Changed_Credit_Limit"]
for i in range(nRowsTrain):
train_data_np.append(
replace_with_median(
train_data[null_value_col].iloc[i],
i,
train_data[null_value_col],
8,
null_values[null_value_col],
rounded,
)
)
for i in range(nRowsTest):
test_data_np.append(
replace_with_median(
test_data[null_value_col].iloc[i],
i,
test_data[null_value_col],
4,
null_values[null_value_col],
rounded,
)
)
train_data[null_value_col] = pd.Series(train_data_np)
test_data[null_value_col] = pd.Series(test_data_np)
# Credit_History_Age : Для каждого клиента заполнила нулевые значения, используя разные записи
# month count column Функция для заполнения
def fill_month_count_column(value, idx, data_arr, rows_per_customer, null_value):
# если совпадает с null_value, то значение обрабатывается как есть
if value != null_value:
return value
start_idx = (idx // rows_per_customer) * rows_per_customer
end_idx = (idx // rows_per_customer + 1) * rows_per_customer
data_range = data_arr[start_idx:end_idx]
# Об одном и том же клиенте [300, 301, 302, 303, 304, 305, 306, 307]
# Как показано, каждый раз, когда индекс увеличивается на 1, значение также увеличивается на 1
first_valid_value = None
for value_idx in range(rows_per_customer):
if data_arr[value_idx] != null_value:
first_valid_value = [value_idx - start_idx, data_arr[value_idx]]
break
if first_valid_value == None:
return null_value
else:
return first_valid_value[1] + (idx % rows_per_customer)
# Credit_History_Age Заполнила нулевые значения разными записями для одного и того же клиента по столбцу
train_data_np = []
test_data_np = []
col = "Credit_History_Age"
for i in range(nRowsTrain):
train_data_np.append(
fill_month_count_column(
train_data[col].iloc[i], i, train_data[col], 8, null_values[col]
)
)
for i in range(nRowsTest):
test_data_np.append(
fill_month_count_column(
test_data[col].iloc[i], i, test_data[col], 4, null_values[col]
)
)
train_data[col] = pd.Series(train_data_np)
test_data[col] = pd.Series(test_data_np)
# Payment_Behaviour_Spent, Payment_Behaviour_Value Чтобы обработать столбцы,
# сначала определила количество высоких и низких значений в данных train и test
train_pb_spent = train_data["Payment_Behaviour_Spent"].value_counts()
test_pb_spent = test_data["Payment_Behaviour_Spent"].value_counts()
train_pb_value = train_data["Payment_Behaviour_Value"].value_counts()
test_pb_value = test_data["Payment_Behaviour_Value"].value_counts()
# Результат : Расходы: Низкие > высокие в порядке убывания (train и для test то же самое)
# Значение: Маленький > Средний > Большой (train и для test то же самое)
# Payment_Behaviour_Spent: Если при записи одного и того же клиента наблюдается более низкий уровень или та же частота возникновения,
# он рассматривается как низкий, если более высокий, он рассматривается как высокий
# Payment_Behaviour_Value : Обрабатывается как наибольшее количество записей одного и того же клиента,
# но в порядке малого, среднего и крупного, если есть элементы с одинаковой частотой появления.
# categorical column Функция для заполнения
def fill_categorical_column(
value, idx, data_arr, rows_per_customer, null_value, pb_count
):
if value != null_value:
return value
start_idx = (idx // rows_per_customer) * rows_per_customer
end_idx = (idx // rows_per_customer + 1) * rows_per_customer
data_range = data_arr[start_idx:end_idx]
# pb_count Копировать
# { значение: [Количество появлений, количество появлений одного и того же клиента],... В формате }
pb_count_copied = pb_count.copy()
for data_value in data_range:
pb_count_copied[data_value][1] += 1
# Приоритет: Одинаковые появления клиентов > Сортировка по количеству появлений
is_all_null = True
pb_count_list_customer = []
for cnt_key, cnt_value in pb_count_copied.items():
pb_count_list_customer.append([cnt_key, cnt_value[0], cnt_value[1]])
if cnt_key != null_value and cnt_value[1] > 0:
is_all_null = False
pb_count_list_customer.sort(
key=lambda x: x[1], reverse=True
) # Количество появлений
pb_count_list_customer.sort(
key=lambda x: x[2], reverse=True
) # Такое же количество посещений клиентов
if is_all_null:
return null_value
else:
return pb_count_list_customer[0][
0
] # Значения, которые чаще всего отображаются для этого клиента на основе отсортированных приоритетов
# Payment_Behaviour_Spent Заполнила нулевые значения разными записями одного и того же клиента для столбца
cols_to_pb_count = {
"Payment_Behaviour_Spent": train_pb_spent,
"Payment_Behaviour_Value": train_pb_value,
}
for col, pb_count in cols_to_pb_count.items():
train_data_np = []
test_data_np = []
# pb_count { значение: [Количество появлений, 0],... В формате }
pb_count_dict = {}
for val, cnt in pb_count.items():
pb_count_dict[val] = [cnt, 0]
print("count of column " + col + ": ", pb_count_dict)
# null Значения заполнения
for i in range(nRowsTrain):
train_data_np.append(
fill_categorical_column(
train_data[col].iloc[i],
i,
train_data[col],
8,
null_values[col],
pb_count_dict,
)
)
for i in range(nRowsTest):
test_data_np.append(
fill_categorical_column(
test_data[col].iloc[i],
i,
test_data[col],
4,
null_values[col],
pb_count_dict,
)
)
train_data[col] = pd.Series(train_data_np)
test_data[col] = pd.Series(test_data_np)
# Вычислим среднее значение, чтобы определить, будут ли все нулевые значения ('nan')
# в каждом типе кредита рассматриваться как 0 или 1
nullable_columns = null_values.keys()
loan_type_columns = []
for col in nullable_columns:
if len(col) >= 10 and col[:10] == "Loan_Type_":
loan_type_columns.append(col)
for col in loan_type_columns:
arr = np.array(train_data[col])
print("(train) mean of [" + col + "] :", arr[arr != null_values[col]].mean())
print("")
for col in loan_type_columns:
arr = np.array(test_data[col])
print("(test) mean of [" + col + "] :", arr[arr != null_values[col]].mean())
# Результат выполнения составляет примерно от 0,28 до 0,29, поэтому все обрабатывается как 0
# Рассматриваем все нулевые значения каждого типа кредита как нулевые
for col in loan_type_columns:
train_data[col] = train_data[col].apply(lambda x: x if x != "nan" else 0)
test_data[col] = test_data[col].apply(lambda x: x if x != "nan" else 0)
# Monthly_Inhand_Salary, Monthly_Balance, Num_of_Delayed_Payment, Num_Credit_Inquiries, Credit_Mix
# осуществим процесс, который позволит обработать null value
# Monthly_Inhand_Salary, Monthly_Balance -> Применила медиану ко всем данным
# Num_of_Delayed_Payment, Num_Credit_Inquiries -> Применила медиану всех данных (дробь = 0,5 дня, округленная до целого числа)
# Credit_Mix -> Заполним Low все нули
for median_col in [
"Monthly_Inhand_Salary",
"Monthly_Balance",
"Num_of_Delayed_Payment",
"Num_Credit_Inquiries",
]:
arr_train = np.array(train_data[median_col])
arr_test = np.array(test_data[median_col])
median_train = np.median(arr_train[arr_train != null_values[median_col]])
median_test = np.median(arr_test[arr_test != null_values[median_col]])
median_all = (median_train * nRowsTrain + median_test * nRowsTest) / (
nRowsTrain + nRowsTest
)
# fraction = 0.5 Округлено до целого числа по времени суток
if median_col in ["Num_of_Delayed_Payment", "Num_Credit_Inquiries"]:
median_all = (
median_all + 0.5 if abs(median_all % 1.0 - 0.5) < 0.25 else median_all
)
print("median of [" + median_col + "] :", median_all)
for data in [train_data, test_data]:
data[median_col] = data[median_col].apply(
lambda x: median_all if x == null_values[median_col] else x
)
for data in [train_data, test_data]:
data["Credit_Mix"] = data["Credit_Mix"].apply(
lambda x: "Low" if x == null_values["Credit_Mix"] else x
)
# просмотрим на training data, отметим, что больше пропущенных значений нет
train_data.head(n=20)
# В результате очиски данных были заменены ошибочные данные в соотвествии с id клиента, а также были заполнены различные нулевые значения также в соответствии с id клиента.
# # 3. Визуализация
age_count = train_data["Age"].value_counts(dropna=False)
sns.set(rc={"figure.figsize": (20, 10)})
sns.barplot(x=age_count.index, y=age_count.values)
plt.title("Bar graph showing the value counts of the column - Age", fontsize=16)
plt.ylabel("Count", fontsize=14)
plt.xlabel("Age", fontsize=14)
plt.xticks(rotation=45)
plt.show()
# По данному графику можно определить, что большинство клиентов находятся в возрастном диапазоне от 18 до 45 лет.
Occupation = train_data["Occupation"].value_counts().tolist()
labels = train_data["Occupation"].values.tolist()
labels = list(set(labels))
plt.figure(figsize=(15, 15))
plt.pie(Occupation, labels=labels, autopct="%1.2f%%")
plt.show()
# По данному же графику делаю вывод, клиенты достаточно равномерно распределены по всем профессиям.
num_of_loan_count = train_data["Num_of_Loan"].value_counts(dropna=False)
sns.set(rc={"figure.figsize": (20, 10)})
sns.barplot(x=num_of_loan_count.index, y=num_of_loan_count.values)
plt.title("Bar graph showing the value counts of the column - Num_of_Loan", fontsize=16)
plt.ylabel("Count", fontsize=14)
plt.xlabel("Num_of_Loan", fontsize=14)
plt.xticks(rotation=45)
plt.show()
# График распределения количества займов говорит о том, что большинство клиентов берут от 2 до 4 займов.
plt.figure(figsize=(20, 20))
sns.histplot(data=train_data, x="Annual_Income", kde=True, stat="probability")
plt.show()
# Распределение годового дохода не похоже на нормальное распределение, данную информацию буду использовать, когда буду обрабатывать данные для загрузки в модель.
# Также распределение является экспоненциальным и тримодальным, пик приходится примерно на 25000, а также на 30000 и 60000.
plt.figure(figsize=(10, 10))
sns.histplot(
data=train_data,
x="Age",
hue="Credit_Score",
kde=True,
stat="probability",
palette="husl",
)
plt.show()
# По данному графику делаем вывод, что клиенты с плохим кредитным рейтингом в основном молодого или среднего возраста, а большинство же клиентов имеют стандартный кредитный рейтинг.
plt.figure(figsize=(20, 20))
sns.histplot(data=train_data, x="Monthly_Inhand_Salary", kde=True, stat="probability")
plt.show()
# По данному графику делаем вывод, что данные распределены не нормально.
plt.figure(figsize=(20, 20))
sns.boxplot(data=train_data, x="Occupation", y="Annual_Income")
plt.show()
# По данной диаграмме можно видеть, что годовой доход для разных родов деятельности не сильно различается.
plt.figure(figsize=(20, 20))
sns.boxplot(data=train_data, x="Occupation", y="Monthly_Inhand_Salary")
plt.show()
# Месячная заработная плата также не сильно различается для разных родов деятельности.
plt.figure(figsize=(20, 20))
sns.boxplot(data=train_data, x="Credit_Score", y="Annual_Income")
plt.show()
# По данному графику можно сделать достаточно очевидный вывод: чем больше годовой доход, тем лучше кредитный рейтинг.
plt.figure(figsize=(20, 20), dpi=200)
sns.boxplot(data=train_data, x="Credit_Score", y="Total_EMI_per_month")
plt.show()
# Таким образом, общий EMI в месяц не влияет на кредитный рейтинг.
plt.figure(figsize=(20, 20))
sns.boxplot(data=train_data, x="Num_Bank_Accounts", y="Monthly_Inhand_Salary")
plt.show()
# А вот по данной диаграмме видно, что чем меньше месячная заработная плата, тем больше число банковских аккаунтов.
plt.figure(figsize=(20, 20))
sns.countplot(data=train_data, x="Num_Credit_Card", hue="Credit_Score")
# По данной диаграмме видно, что клиенты с плохим кредитным рейтингом имеют кредитных карт больше, чем клиенты с хорошим и стандартными кредитными рейтингами.
plt.figure(figsize=(20, 20))
sns.countplot(data=train_data, x="Num_Credit_Inquiries", hue="Credit_Score")
# По данному графику понятно, что клиенты с плохим кредитным рейтингом имееют больше запросов на кредитные карты по сравнению с клиентами с хорошим и стандартным кредитными рейтингами.
plt.figure(figsize=(20, 20))
sns.countplot(data=train_data, x="Num_of_Loan", hue="Credit_Score")
# По числу заемов можно сказать, что клиенты с хорошим кредитным рейтингом берут небольшое количество заемов по сравнению с остальными клиентами.
plt.figure(figsize=(10, 6))
sns.histplot(x=train_data["Credit_History_Age"], kde=True)
plt.title("Credit_History_Age distribution")
# Опять же, распределение не является нормальным. Большой пик приходится на 260-270 количнство месяцев, это примерно 21-22 года.
plt.figure(figsize=(10, 6))
sns.histplot(x=train_data["Outstanding_Debt"])
plt.title("Outstanding debit distribution")
# Распределение не нормально, пик приходится на 1500, далее следует сильный спад.
plt.figure(figsize=(20, 20))
sns.countplot(data=train_data, x="Occupation", hue="Credit_Score")
# Опять же, по данному графику можно предположить, что род деятельности не сильно влияет на кредитный рейтинг.
label = train_data.Credit_Mix.value_counts().index
label_count = train_data.Credit_Mix.value_counts().values
plt.pie(
data=train_data,
x=label_count,
labels=label,
autopct="%1.1f%%",
shadow=True,
radius=1,
)
plt.show()
# По данной диаграмме видно, что большинство клиентов имеют стандартный кредитный портфель.
mask = train_data[train_data["Amount_invested_monthly"] < 1000]
plt.figure(figsize=(20, 10))
sns.histplot(
data=mask,
x="Amount_invested_monthly",
kde=True,
hue="Credit_Score",
stat="probability",
)
plt.xticks(np.arange(0, 500, 25))
plt.show()
# Из приведенного выше графика ясно, что большинство клиентов, которые инвестируют, имеют стандартный кредитный рейтинг.
pivot_data = train_data.pivot_table(
index="Credit_Score", values="Annual_Income", columns="Num_of_Loan", aggfunc="mean"
)
fig, ax = plt.subplots(figsize=(20, 5))
sns.heatmap(data=pivot_data, annot=True, fmt=".1f", cmap="coolwarm", ax=ax)
ax.set_title(
"Distribution of the average annual income of the client depending on the number of loans"
)
ax.set_ylabel("Credit_Score")
ax.set_xlabel("Num_of_Loan")
# А вот по данной тепловой карте понятно, что клипенты с высоким годовым доходом предпочитают брать небольшое число заемов, что достаточно понятно.
pivot_data_bank = train_data.pivot_table(
index="Credit_Score",
values="Annual_Income",
columns="Num_Bank_Accounts",
aggfunc="mean",
)
fig, ax = plt.subplots(figsize=(20, 5))
sns.heatmap(data=pivot_data_bank, annot=True, fmt=".1f", cmap="coolwarm", ax=ax)
ax.set_title(
"Distribution of the average annual income of the client depending on the number of bank accounts"
)
# По данной диаграмме также отмечу, что клиенты с низким годовым доходом имею много банковских аккаунтов.
pivot_data_occupation = train_data.pivot_table(
index="Credit_Score", values="Annual_Income", columns="Occupation", aggfunc="mean"
)
fig, ax = plt.subplots(figsize=(20, 5))
sns.heatmap(data=pivot_data_occupation, annot=True, fmt=".1f", cmap="coolwarm", ax=ax)
ax.set_title(
"Distribution of the average annual income of the client depending on the occupation"
)
# Очевидно, что люди любой профессии со средним годовым доходом более 60000 долларов имеют хороший кредитный рейтинг
# Люди со средним годовым доходом 50000 имеют стандартный кредитный рейтинг
# Люди со средним годовым доходом в 40000 долларов имеют плохой кредитный рейтинг
plt.figure(figsize=(20, 20))
sns.heatmap(train_data.corr(), annot=True)
plt.tight_layout()
# По данной тепловой диаграмме видно, что признаки 'Annual_Income' и 'Monthly_Inhand_Salary' имеют коэффициент корреляции 1, этот факт может привести к утечке данных. Также важно отметить, что такие признаки как 'Occupation' и 'Month' не влияют на значение кредитного рейтинга клиента. Многие числовые данные не подчиняются нормальному распределению и имеют большой разброс значений, поэтому делаю вывод, что такие данные требуют обработки в следующем разделе.
# # 4. Преобразование данных
# Сделаем копии датасетов, чтобы преоразовать их для передачи в модель
copy_train = train_data.copy()
copy_test = test_data.copy()
# удаляем месяц, так как он слабо влияет на кредитный рейтинг
del copy_train["Month"]
del copy_test["Month"]
# На мой взгляд, род деятельности не сильно коррелирует с остальными данными, этот признак можно удалить
del copy_train["Occupation"]
del copy_test["Occupation"]
# закодируем признаки 'Credit_Score' и 'Credit_Mix'
def cod_score(x):
if x == "Good":
return 1
elif x == "Standard":
return 0
else:
return -1
copy_train["Credit_Score"] = copy_train["Credit_Score"].apply(cod_score)
copy_train["Credit_Mix"] = copy_train["Credit_Mix"].apply(cod_score)
copy_test["Credit_Mix"] = copy_test["Credit_Mix"].apply(cod_score)
# также закодируем следующие признаки однократным кодированием
cols_to_onehot = [
"Payment_of_Min_Amount",
"Payment_Behaviour_Spent",
"Payment_Behaviour_Value",
]
for data in [copy_train, copy_test]:
for col in cols_to_onehot:
unique_values = data[col].unique()
for uniq in unique_values:
data[col + "_" + uniq] = data[col].apply(
lambda x: 1.0 if x == uniq else 0.0
)
copy_train = copy_train.drop(columns=cols_to_onehot)
copy_test = copy_test.drop(columns=cols_to_onehot)
# удаляем 'Monthly_Inhand_Salary', чтобы избежать утечки данных
del copy_train["Monthly_Inhand_Salary"]
del copy_test["Monthly_Inhand_Salary"]
# проверяем тепловую диаграмму
plt.figure(figsize=(20, 20))
sns.heatmap(copy_train.corr(), annot=True)
plt.tight_layout()
# используем логарифмическое преобразование для следующих признаков, чтобы их распределение приблизилось к нормальному
cols_to_log = ["Annual_Income", "Total_EMI_per_month", "Amount_invested_monthly"]
train_mean = {}
train_std = {}
for col in cols_to_log:
train_mean[col] = data[col].mean()
train_std[col] = data[col].std()
for data in [copy_train, copy_test]:
for col in cols_to_log:
data[col] = data[col].apply(lambda x: np.log(x + 1.0))
data[col] = data[col].apply(lambda x: (x - train_mean[col]) / train_std[col])
plt.figure(figsize=(20, 10))
sns.histplot(
data=copy_train,
x="Amount_invested_monthly",
kde=True,
hue="Credit_Score",
stat="probability",
)
plt.show()
# Действительно, распределение признака больше напоминает нормальное.
plt.figure(figsize=(20, 10))
sns.histplot(
data=copy_train, x="Annual_Income", kde=True, hue="Credit_Score", stat="probability"
)
plt.show()
# Годовой доход сохранил три пика даже после логарифмического преобразования.
# Нормализуем остальные признаки
names_to_norm = [
"Age",
"Num_Bank_Accounts",
"Num_Credit_Card",
"Interest_Rate",
"Num_of_Loan",
"Delay_from_due_date",
"Num_of_Delayed_Payment",
"Changed_Credit_Limit",
"Num_Credit_Inquiries",
"Outstanding_Debt",
"Credit_History_Age",
"Monthly_Balance",
]
train_mean = {}
train_std = {}
for col in names_to_norm:
train_mean[col] = data[col].mean()
train_std[col] = data[col].std()
for data in [copy_train, copy_test]:
for col in names_to_norm:
data[col] = data[col].apply(lambda x: (x - train_mean[col]) / train_std[col])
# проверяем тепловую диаграмму
plt.figure(figsize=(20, 20))
sns.heatmap(copy_train.corr(), annot=True, cmap="YlGnBu")
plt.tight_layout()
# После нормализации и логарифмирования данных можно отметить, что распределение числовых признаков стало больше напоминать нормальное, нечисловые признаки были закодированы для передачи в модель.
# # 5. Обучение модели
# Определяем целевой признак, т.е. "Credit_Score". Разделяем тренинговый датасет: 75% на обучение, 25% на валидацию
y = copy_train["Credit_Score"]
X = copy_train
del copy_train["Credit_Score"]
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.25, shuffle=True, random_state=42
)
# ## 5.1. Логистическая регрессия
# Для начала я применяю самую простую модель со стандартными параметрами.
log_reg = LogisticRegression(random_state=42)
log_reg = log_reg.fit(X_train, y_train)
y_pred = log_reg.predict(X_test)
print(classification_report(y_test, y_pred))
# Резултьтат слабый, логистическая регрессия слабо предсказывает "Credit_Score".
# ## 5.2. Ближайшие соседи
knn = KNeighborsClassifier()
knn = knn.fit(X_train, y_train)
y_pred = knn.predict(X_test)
print(classification_report(y_test, y_pred))
# Результат также слабый, использовала стандартные параметры.
# ## 5.3. Деревья решений
clf = DecisionTreeClassifier(random_state=42)
clf = clf.fit(X_train, y_train)
y_pred = clf.predict(X_test)
print(classification_report(y_test, y_pred))
# Уже несколько лучший результат, использовала стандартные параметры.
# ## 5.4. Леса рандомизированных деревьев (Рандомный лес)
rfc = RandomForestClassifier()
rfc = rfc.fit(X_train, y_train)
y_pred = rfc.predict(X_test)
print(classification_report(y_test, y_pred))
# вывводим самые важные признаки для модели
fi = pd.DataFrame(
rfc.feature_importances_, index=rfc.feature_names_in_, columns=["importance"]
).sort_values(["importance"], ascending=False)
fi
# также отражаем их на графике
ax = fi.sort_values("importance").plot(
kind="barh", title="Features Importance", figsize=(10, 10)
)
ax.set_ylabel("Features")
plt.show()
# Получается, что самый важный признак - Outstanding_Deb, что достаточно логично. Важное значение также имеют: Interest_Rate, Gredit_Mix и Credit_History_Age.
# ## 5.5. Наивные методы Байеса
clf = GaussianNB()
clf = clf.fit(X_train, y_train)
y_pred = clf.predict(X_test)
print(classification_report(y_test, y_pred))
# Результат слабый. Дааная модель достаточно посредственно предсказывает результаты.
# ## 5.6. AdaBoost
clf = AdaBoostClassifier(n_estimators=120, random_state=42, learning_rate=0.5)
clf = clf.fit(X_train, y_train)
y_pred = clf.predict(X_test)
print(classification_report(y_test, y_pred))
# Значение метрик не очень большое, тут я не стала оставлять стандартные параметры, так как они показывали плохой результат.
# ## 5.7. GridSearchCV
# В данном разделе я использовала три модели с различными параметрами, чтобы найти наилучший результат.
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.25, shuffle=True, random_state=42
)
param_grid = {
"max_depth": range(5, 15),
"criterion": ["gini", "entropy"],
"n_estimators": [100, 200, 500],
}
base_estimator = ensemble.RandomForestClassifier(random_state=42)
model = model_selection.GridSearchCV(base_estimator, param_grid, cv=5, n_jobs=-1)
model.fit(X_train, y_train)
print(model.best_estimator_)
y_train_pred = model.predict(X_train)
print(
"Accuracy на тренировочной выборке: {:.3f}".format(
metrics.accuracy_score(y_train, y_train_pred)
)
)
y_test_pred = model.predict(X_test)
print(
"Accuracy на тестовой выборке: {:.3f}".format(
metrics.accuracy_score(y_test, y_test_pred)
)
)
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.25, shuffle=True, random_state=42
)
param_grid = {"max_depth": range(3, 15), "criterion": ["gini", "entropy"]}
base_estimator = tree.DecisionTreeClassifier(random_state=42)
model = model_selection.GridSearchCV(base_estimator, param_grid, cv=5, n_jobs=-1)
model.fit(X_train, y_train)
print(model.best_estimator_)
y_train_pred = model.predict(X_train)
print(
"Accuracy на тренировочной выборке: {:.3f}".format(
metrics.accuracy_score(y_train, y_train_pred)
)
)
y_test_pred = model.predict(X_test)
print(
"Accuracy на тестовой выборке: {:.3f}".format(
metrics.accuracy_score(y_test, y_test_pred)
)
)
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.25, shuffle=True, random_state=42
)
param_grid = {
"n_neighbors": range(3, 15),
"algorithm": ["auto", "ball_tree", "kd_tree", "brute"],
}
base_estimator = neighbors.KNeighborsClassifier()
model = model_selection.GridSearchCV(base_estimator, param_grid, cv=5, n_jobs=-1)
model.fit(X_train, y_train)
print(model.best_estimator_)
y_train_pred = model.predict(X_train)
print(
"Accuracy на тренировочной выборке: {:.3f}".format(
metrics.accuracy_score(y_train, y_train_pred)
)
)
y_test_pred = model.predict(X_test)
print(
"Accuracy на тестовой выборке: {:.3f}".format(
metrics.accuracy_score(y_test, y_test_pred)
)
)
# В результе можно отметить, что стандартные методы с параметрами по умолчанию имеют значения метрики Accuracy ниже, чем
# результаты, полученные с помощью поиска по сетке GridSearchCV. А лучший результат на этом датасете показывает рандомный лес со следующими параметрами max_depth=14, n_estimators=500.
# А именно:
# RandomForestClassifier(max_depth=14, n_estimators=500, random_state=42)
# Accuracy на тренировочной выборке: 0.822
# Accuracy на тестовой выборке: 0.758
# # 6. Optuna
# ## 6.1 RandomForestClassifier
# Настрою оптимизацию гиперпараметров для алгоритма случайного леса.
import optuna
import catboost as cb
def optuna_rf(trial):
# задаем пространства поиска гиперпараметров
n_estimators = trial.suggest_int("n_estimators", 100, 200, 1)
max_depth = trial.suggest_int("max_depth", 10, 30, 1)
min_samples_leaf = trial.suggest_int("min_samples_leaf", 2, 10, 1)
# создаем модель
model = ensemble.RandomForestClassifier(
n_estimators=n_estimators,
max_depth=max_depth,
min_samples_leaf=min_samples_leaf,
random_state=42,
)
# обучаем модель
model.fit(X_train, y_train)
score = metrics.accuracy_score(y_train, model.predict(X_train))
return score
# cоздаем объект исследования
# необходимо максимизировать метрику direction="maximize"
study = optuna.create_study(study_name="RandomForestClassifier", direction="maximize")
# ищем лучшую комбинацию гиперпараметров n_trials раз
study.optimize(optuna_rf, n_trials=20)
# выводим результаты на обучающей выборке
print("Наилучшие значения гиперпараметров {}".format(study.best_params))
print("accuracy на обучающем наборе: {:.2f}".format(study.best_value))
# рассчитаем точность для тестовой выборки
model = ensemble.RandomForestClassifier(**study.best_params, random_state=42)
model.fit(X_train, y_train)
y_train_pred = model.predict(X_train)
print("accuracy на тестовом наборе: {:.2f}".format(model.score(X_test, y_test)))
study.optimize(optuna_rf, n_trials=10)
optuna.visualization.plot_optimization_history(study, target_name="accuracy_score")
# График важности гиперпараметров - помогает понять, какие гиперпараметры вносят больший вклад в минимизацию/максимизацию метрики.
optuna.visualization.plot_param_importances(study, target_name="accuracy_score")
# Из этого графика можно сделать вывод, что стоит обратить большее внимание на настройку гиперпараметра max_depth
optuna.visualization.plot_contour(
study, params=["max_depth", "n_estimators"], target_name="accuracy_score"
)
# Точки с белым фоном это и есть лучшии комбинации n_estimator, max_depth.
# ## 6.2 CatBoostClassifier
# Настрою оптимизацию CatBoostClassifier, для это я повторю обработку исходного датасета, но по другому закодирую целевой признак.
copy_train_cb = train_data.copy()
del copy_train_cb["Month"]
del copy_train_cb["Occupation"]
del copy_train_cb["Monthly_Inhand_Salary"]
cols_to_onehot = [
"Payment_of_Min_Amount",
"Payment_Behaviour_Spent",
"Payment_Behaviour_Value",
]
for data in [copy_train_cb]:
for col in cols_to_onehot:
unique_values = data[col].unique()
for uniq in unique_values:
data[col + "_" + uniq] = data[col].apply(
lambda x: 1.0 if x == uniq else 0.0
)
copy_train_cb = copy_train_cb.drop(columns=cols_to_onehot)
# закодируем признаки 'Credit_Score' и 'Credit_Mix'
def cod_score(x):
if x == "Good":
return 1
elif x == "Standard":
return 1
else:
return 0
copy_train_cb["Credit_Score"] = copy_train_cb["Credit_Score"].apply(cod_score)
copy_train_cb["Credit_Mix"] = copy_train_cb["Credit_Mix"].apply(cod_score)
cols_to_log = ["Annual_Income", "Total_EMI_per_month", "Amount_invested_monthly"]
train_mean = {}
train_std = {}
for col in cols_to_log:
train_mean[col] = data[col].mean()
train_std[col] = data[col].std()
for data in [copy_train_cb]:
for col in cols_to_log:
data[col] = data[col].apply(lambda x: np.log(x + 1.0))
data[col] = data[col].apply(lambda x: (x - train_mean[col]) / train_std[col])
# Нормализуем остальные признаки
names_to_norm = [
"Age",
"Num_Bank_Accounts",
"Num_Credit_Card",
"Interest_Rate",
"Num_of_Loan",
"Delay_from_due_date",
"Num_of_Delayed_Payment",
"Changed_Credit_Limit",
"Num_Credit_Inquiries",
"Outstanding_Debt",
"Credit_History_Age",
"Monthly_Balance",
]
train_mean = {}
train_std = {}
for col in names_to_norm:
train_mean[col] = data[col].mean()
train_std[col] = data[col].std()
for data in [copy_train_cb]:
for col in names_to_norm:
data[col] = data[col].apply(lambda x: (x - train_mean[col]) / train_std[col])
y = copy_train_cb["Credit_Score"]
X = copy_train_cb
del copy_train_cb["Credit_Score"]
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.25, shuffle=True, random_state=42
)
from catboost import CatBoostClassifier, Pool
def optuna_cb(trial):
# задаем пространства поиска гиперпараметров
iterations = trial.suggest_int("iterations", 2, 5, 1)
depth = trial.suggest_int("depth", 1, 5, 1)
learning_rate = trial.suggest_float("learning_rate", 0.1, 1)
# создаем модель
model = CatBoostClassifier(
iterations=iterations,
depth=depth,
learning_rate=learning_rate,
loss_function="Logloss",
verbose=True,
)
# обучаем модель
model.fit(X_train, y_train)
score = metrics.accuracy_score(y_train, model.predict(X_train))
return score
# cоздаем объект исследования
# можем напрямую указать, что нам необходимо максимизировать метрику direction="maximize"
study = optuna.create_study(study_name="CatBoostClassifier", direction="maximize")
# ищем лучшую комбинацию гиперпараметров n_trials раз
study.optimize(optuna_rf, n_trials=20)
# выводим результаты на обучающей выборке
print("Наилучшие значения гиперпараметров {}".format(study.best_params))
print("accuracy на обучающем наборе: {:.2f}".format(study.best_value))
# рассчитаем точность для тестовой выборки
model = CatBoostClassifier(**study.best_params, random_state=42)
model.fit(X_train, y_train)
y_train_pred = model.predict(X_train)
print("accuracy на тестовом наборе: {:.2f}".format(model.score(X_test, y_test)))
study.optimize(optuna_cb, n_trials=10)
optuna.visualization.plot_optimization_history(study, target_name="accuracy_score")
optuna.visualization.plot_param_importances(study, target_name="accuracy_score")
# По данному графику можно сделать вывод, что параметр deth имеет наибольшее значение.
optuna.visualization.plot_contour(
study, params=["depth", "learning_rate"], target_name="accuracy_score"
)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
netflix = pd.read_csv("/kaggle/input/netflix-shows/netflix_titles.csv")
netflix.head()
netflix.loc[0, "cast"]
netflix.cast.describe()
netflix.director.describe()
# how many movies/tv shows has every director
netflix.groupby("director").show_id.count().idxmax()
netflix.cast = netflix.cast.fillna("Unknown")
netflix.loc[netflix.cast.isnull()]
show_actor = {"show_id": [], "actor": []}
for cast, show_id in zip(netflix.cast.values, netflix.show_id.values):
actors = cast.lower().split(", ")
show_actor["show_id"] += [show_id] * len(actors)
show_actor["actor"] += actors
show_actor = pd.DataFrame(show_actor)
show_actor.head()
show_actor.to_csv("show_actor.csv")
|
# # **Minor Project**
# ## Credit Card Fraud **Detection**
#
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import matplotlib.pyplot as plt
import numpy as np
import itertools
import calendar
from scipy.stats import norm, skew
import random
from sklearn.metrics import roc_auc_score
from sklearn.metrics import roc_curve
from sklearn.model_selection import train_test_split
from sklearn.utils import (
resample,
) # Correcting the imbalance discovered by using resample
from sklearn.linear_model import LogisticRegression
from sklearn.experimental import enable_halving_search_cv
from sklearn.metrics import (
accuracy_score,
mean_absolute_error,
mean_squared_error,
confusion_matrix,
median_absolute_error,
classification_report,
f1_score,
recall_score,
precision_score,
)
from sklearn.tree import DecisionTreeClassifier
from sklearn.preprocessing import StandardScaler
from sklearn.metrics import RocCurveDisplay
from sklearn.model_selection import HalvingRandomSearchCV, RandomizedSearchCV
# Checking p-values and Variation Inflation Factor
import statsmodels.api as sm
from statsmodels.stats.outliers_influence import variance_inflation_factor
# ## Loading the Dataset
def plot_confusion_matrix(
cm, target_names, title="Confusion matrix", cmap=None, normalize=True
):
"""
given a sklearn confusion matrix (cm), make a nice plot
Arguments
---------
cm: confusion matrix from sklearn.metrics.confusion_matrix
target_names: given classification classes such as [0, 1, 2]
the class names, for example: ['high', 'medium', 'low']
title: the text to display at the top of the matrix
cmap: the gradient of the values displayed from matplotlib.pyplot.cm
see http://matplotlib.org/examples/color/colormaps_reference.html
plt.get_cmap('jet') or plt.cm.Blues
normalize: If False, plot the raw numbers
If True, plot the proportions
Usage
-----
plot_confusion_matrix(cm = cm, # confusion matrix created by
# sklearn.metrics.confusion_matrix
normalize = True, # show proportions
target_names = y_labels_vals, # list of names of the classes
title = best_estimator_name) # title of graph
Citiation
---------
http://scikit-learn.org/stable/auto_examples/model_selection/plot_confusion_matrix.html
"""
import matplotlib.pyplot as plt
import numpy as np
import itertools
accuracy = np.trace(cm) / float(np.sum(cm))
misclass = 1 - accuracy
if cmap is None:
cmap = plt.get_cmap("Blues")
plt.figure(figsize=(8, 6))
plt.imshow(cm, interpolation="nearest", cmap=cmap)
plt.title(title)
plt.colorbar()
if target_names is not None:
tick_marks = np.arange(len(target_names))
plt.xticks(tick_marks, target_names, rotation=45)
plt.yticks(tick_marks, target_names)
if normalize:
cm = cm.astype("float") / cm.sum(axis=1)[:, np.newaxis]
thresh = cm.max() / 1.5 if normalize else cm.max() / 2
for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):
if normalize:
plt.text(
j,
i,
"{:0.4f}".format(cm[i, j]),
horizontalalignment="center",
color="white" if cm[i, j] > thresh else "black",
)
else:
plt.text(
j,
i,
"{:,}".format(cm[i, j]),
horizontalalignment="center",
color="white" if cm[i, j] > thresh else "black",
)
plt.tight_layout()
plt.ylabel("True label")
plt.xlabel(
"Predicted label\naccuracy={:0.4f}; misclass={:0.4f}".format(accuracy, misclass)
)
plt.show()
train = pd.read_csv("/kaggle/input/fraud-detection/fraudTrain.csv")
test = pd.read_csv("/kaggle/input/fraud-detection/fraudTrain.csv")
train.head()
# test.head()
train.columns
# This prints the shape of dataset
print("fraudTrain.csv Shape : ", test.shape)
print("fraudTest.csv Shape : ", train.shape)
# ## Data Cleaning
test.isnull().sum()
train.isnull().sum()
test.info()
train.info()
# Converting dob, trans_date_trans_time column in both test & train to datetime data type and creating new 'trans_date' column -
train["trans_date_trans_time"] = pd.to_datetime(train["trans_date_trans_time"])
train["trans_date"] = train["trans_date_trans_time"].dt.strftime("%Y-%m-%d")
train["trans_date"] = pd.to_datetime(train["trans_date"])
train["dob"] = pd.to_datetime(train["dob"])
test["trans_date_trans_time"] = pd.to_datetime(test["trans_date_trans_time"])
test["trans_date"] = test["trans_date_trans_time"].dt.strftime("%Y-%m-%d")
test["trans_date"] = pd.to_datetime(test["trans_date"])
test["dob"] = pd.to_datetime(test["dob"])
test.trans_date.head(), test.dob.head(), train.trans_date.head(), train.dob.head()
train.drop("Unnamed: 0", axis=1, inplace=True)
test.drop("Unnamed: 0", axis=1, inplace=True)
train.head()
# **EDA, Feature Engineering**
# Categorical Variable Analysis
#
total = pd.concat([test, train])
total.info()
total["is_fraud_cat"] = total.is_fraud.apply(lambda x: "T" if x == 1 else "F")
total["is_fraud_cat"].astype("object")
totalcat = total.select_dtypes(include=["object"])
total[totalcat.columns]
plt.figure(figsize=(10, 5))
sns.countplot(x=total[total["is_fraud_cat"] == "T"].category)
plt.xticks(rotation=45)
plt.show()
# Most frauds occured in categories of shopping_net and grocery_pos
#
sns.countplot(x=total[total["is_fraud_cat"] == "T"].gender)
plt.xticks(rotation=45)
plt.show()
# Although more cases of fraud happened with female customers, the number is almost the same for both Males and Females
fig, ax = plt.subplots(figsize=(120, 60))
plt.rcParams.update({"font.size": 60})
sns.countplot(x=total[total["is_fraud_cat"] == "T"].state)
plt.xticks(rotation=45)
for p, label in zip(ax.patches, total["state"].value_counts().index):
ax.annotate(label, (p.get_x(), p.get_height() + 0.15))
plt.title("Number of Credit Card Frauds by State")
plt.show()
# States OH, TX and LA report the most number of credit card frauds
def randomcolor():
r = random.random()
b = random.random()
g = random.random()
rgb = [r, g, b]
return rgb
plt.rcParams.update({"font.size": 20})
total[total["is_fraud_cat"] == "T"]["city"].value_counts(
sort=True, ascending=False
).head(10).plot(kind="bar", color=randomcolor())
plt.title("Number of Credit Card Frauds by City")
plt.show()
# Dallas,Houston and Birmingham report the most frauds city wise.
total[total["is_fraud_cat"] == "T"]["job"].value_counts(
sort=True, ascending=False
).head(10).plot(kind="bar", color=randomcolor())
plt.title("Number of Credit Card Frauds by Job")
plt.show()
# Most frauds occured in jobs of quantity surveyor followed by naval architect and materials engineer
# **Numerical Variable Analysis**
# checking the spread & skewness of all numerical variables
del total["is_fraud_cat"]
# finding numerical columns
testnum = test.select_dtypes(include=np.number)
test[testnum.columns]
total.isnull().sum()
total[testnum.columns].info()
plt.rcParams.update({"font.size": 10})
skewness = str(skew(total["amt"]))
sns.distplot(total["amt"], fit=norm, color=randomcolor())
plt.title("Skewness of amt" + " = " + skewness)
plt.show()
# Most transactions are limited within a very small bracket, although transactions of large sums are very much present, which are likely to be fraud transactions.
skewness = str(skew(total["city_pop"]))
sns.distplot(total["city_pop"], fit=norm, color=randomcolor())
plt.title("Skewness of population" + " = " + skewness)
plt.show()
sns.distplot(total["is_fraud"], fit=norm, color=randomcolor())
plt.title("Distribution of is_fraud")
plt.show()
# Certain numerical columns are not needed for modeling and hence can be removed.
# Moreover, a lot of the data is highly skewed.
# one insight is that proportion of non fraud transactions are much much larger than fraud transactions so we are looking at an imbalanced dataset.
total.drop(
["cc_num", "merchant", "first", "last", "street", "zip", "trans_num", "unix_time"],
axis=1,
inplace=True,
)
total.info()
plt.figure(figsize=(8, 5))
ax = sns.countplot(x="is_fraud", data=total, color=randomcolor())
for p in ax.patches:
ax.annotate(
"{:.1f}".format(p.get_height()), (p.get_x() + 0.25, p.get_height() + 0.01)
)
plt.show()
# only around 10,000 entries represent fraud transactions out of nearly 1.8 million entries, hence we are looking at an imbalanced dataset.
# creating age variable from transaction dates and DOB.
total["age"] = total["trans_date"] - total["dob"]
total["age"] = total["age"].astype("timedelta64[Y]")
total["age"].head()
total.info()
fraud = total[total["is_fraud"] == 1]
fig, ax = plt.subplots()
ax.hist(fraud.age, edgecolor="black", bins=5, color=randomcolor())
plt.title("Number of Credit Card Frauds by Age Groups")
plt.show()
# creating transacation month and transaction year columns
total["trans_month"] = pd.DatetimeIndex(total["trans_date"]).month
total["trans_year"] = pd.DatetimeIndex(total["trans_date"]).year
total["Month_name"] = total["trans_month"].apply(lambda x: calendar.month_abbr[x])
sns.countplot(x=total[total["is_fraud"] == 1]["Month_name"], color=randomcolor())
plt.title("Number of Credit Card Frauds by month")
plt.show()
del total["Month_name"]
sns.countplot(x=total[total["is_fraud"] == 1]["gender"], color=randomcolor())
plt.title("Number of Credit Card Frauds by Gender")
plt.show()
sns.countplot(x=total[total["is_fraud"] == 1]["trans_year"], color=randomcolor())
plt.title("Number of Credit Card Frauds by year")
plt.show()
# Finding distance from customer location to merchant location in degrees latitude and degrees longitude
total["latitudinal_distance"] = abs(round(total["merch_lat"] - total["lat"], 3))
total["longitudinal_distance"] = abs(round(total["merch_long"] - total["long"], 3))
fraud = total[total["is_fraud"] == 1]
fig, ax = plt.subplots()
ax.hist(fraud.latitudinal_distance, edgecolor="black", bins=5, color=randomcolor())
plt.title("Number of Credit Card Frauds by latitudinal distance")
plt.show()
fig, ax = plt.subplots()
ax.hist(fraud.longitudinal_distance, edgecolor="black", bins=5, color=randomcolor())
plt.title("Number of Credit Card Frauds by longitudinal distance")
plt.show()
total.info()
# Changing gender values to binary values
total.gender.value_counts()
total.gender = total.gender.apply(lambda x: 1 if x == "M" else 0)
total.gender.value_counts()
# Dropping final set of variables not useful for model building
drop_cols = [
"trans_date_trans_time",
"city",
"lat",
"long",
"job",
"dob",
"merch_lat",
"merch_long",
"trans_date",
"state",
]
total = total.drop(drop_cols, axis=1)
total.info()
total = pd.get_dummies(total, columns=["category"], drop_first=True)
total.info()
total.head()
# # Model Building
#
# Creating two different dataframe of majority and minority class
df_majority = total[(total["is_fraud"] == 0)]
df_minority = total[(total["is_fraud"] == 1)]
df_majority.shape, df_minority.shape
# Upsampling minority class
df_minority_upsampled = resample(
df_minority,
replace=True, # sample with replacement
n_samples=1842743, # to match majority class
random_state=42,
) # reproducible results
df_minority_upsampled.shape
# Combining majority class with upsampled minority class
total_upsampled = pd.concat([df_minority_upsampled, df_majority])
total_upsampled.shape
x_cols = list(total_upsampled.columns)
x_cols.remove("is_fraud")
x_cols
X = total_upsampled[x_cols]
Y = total_upsampled["is_fraud"]
X.info()
X_train, X_test, Y_train, Y_test = train_test_split(
X, Y, test_size=0.3, random_state=42
)
# Scaling the X-Variable
sc = StandardScaler()
X_train_std = sc.fit_transform(X_train)
X_test_std = sc.fit_transform(X_test)
# Feature Importances
logit_model = LogisticRegression(solver="liblinear")
logit_model.fit(X_train_std, Y_train)
feature = pd.DataFrame()
feature["column"] = X_train.columns
feature["importance"] = logit_model.coef_[0]
feature.sort_values("importance", ascending=False, inplace=True)
feature
# ## Logistic Regression
#
Logit1 = LogisticRegression(solver="liblinear")
Logit1.fit(X_train_std, Y_train)
print(
"Score of the model with X-train and Y-train is : ",
str(round(Logit1.score(X_train, Y_train) * 100, 2)),
"%",
)
print(
"Score of the model with X-test and Y-test is : ",
str(round(Logit1.score(X_test, Y_test) * 100, 2)),
"%",
)
Y_pred = Logit1.predict(X_test_std)
print(" Mean absolute error is ", (mean_absolute_error(Y_test, Y_pred)))
print(" Mean squared error is ", mean_squared_error(Y_test, Y_pred))
print(" Median absolute error is ", median_absolute_error(Y_test, Y_pred))
print("Accuracy is ", round(accuracy_score(Y_test, Y_pred) * 100, 2), "%")
print("F1 score: ", round(f1_score(Y_test, Y_pred, average="weighted") * 100, 2), "%")
plot_confusion_matrix(
confusion_matrix(Y_test, Y_pred), ["Fraud", "Not Fraud"], normalize=False
)
plot_confusion_matrix(
confusion_matrix(Y_test, Y_pred), ["Fraud", "Not Fraud"], normalize=True
)
# Fine Tuning
X_train_new = X_train[[x for x in feature[feature["importance"] > 0].column]]
X_test_new = X_test[[x for x in feature[feature["importance"] > 0].column]]
X_train_sm = sm.add_constant(X_train_new)
logm = sm.GLM(Y_train, X_train_sm, family=sm.families.Binomial())
res = logm.fit()
res.summary()
vif = pd.DataFrame()
vif["Features"] = X_train_new.columns
vif["VIF"] = [
variance_inflation_factor(X_train_new.values, i)
for i in range(X_train_new.shape[1])
]
vif["VIF"] = round(vif["VIF"], 2)
vif = vif.sort_values(by="VIF", ascending=False)
vif
# Since all the columns have VIF<5, we'll continue with all columns
# x_train_vif_adj = X_train_new[[x for x in list(vif[vif['VIF']<=5]['Features'])]]
x_train_vif_adj = X_train_new
# x_test_vif_adj = X_test_new[[x for x in list(vif[vif['VIF']<=5]['Features'])]]
x_test_vif_adj = X_test_new
# Scaling the new test and train sets
sc = StandardScaler()
X_train_vif_adj_std = sc.fit_transform(x_train_vif_adj)
X_test_vif_adj_std = sc.fit_transform(x_test_vif_adj)
# Training a new Logistic Regression Model to reflect the changes:-
Logit2 = LogisticRegression(solver="liblinear")
Logit2.fit(X_train_vif_adj_std, Y_train)
print(
"Score of the model with X-train and Y-train is : ",
str(round(Logit2.score(X_train_vif_adj_std, Y_train) * 100, 2)),
"%",
)
print(
"Score of the model with X-test and Y-test is : ",
str(round(Logit2.score(X_test_vif_adj_std, Y_test) * 100, 2)),
"%",
)
Y_pred = Logit2.predict(X_test_vif_adj_std)
print(" Mean absolute error is ", (mean_absolute_error(Y_test, Y_pred)))
print(" Mean squared error is ", mean_squared_error(Y_test, Y_pred))
print(" Median absolute error is ", median_absolute_error(Y_test, Y_pred))
print("Accuracy is ", round(accuracy_score(Y_test, Y_pred) * 100, 2), "%")
print("F1 score: ", round(f1_score(Y_test, Y_pred, average="weighted") * 100, 2), "%")
plot_confusion_matrix(
confusion_matrix(Y_test, Y_pred), ["Fraud", "Not Fraud"], normalize=False
)
plot_confusion_matrix(
confusion_matrix(Y_test, Y_pred), ["Fraud", "Not Fraud"], normalize=True
)
matrix = classification_report(Y_test, Y_pred, labels=[1, 0])
print("Classification report : \n", matrix)
RocCurveDisplay.from_estimator(Logit2, X_test_vif_adj_std, Y_test)
plt.show()
# ## Decision Tree
#
dtc = DecisionTreeClassifier()
dtc.fit(X_train, Y_train)
# Feature Importances using Decision Tree
importance = dtc.feature_importances_
for i, v in enumerate(importance):
print(X_train.columns[int(i)], "- ", v)
plt.bar([x for x in range(len(importance))], importance)
plt.show()
print("Score the X-train with Y-train is : ", dtc.score(X_train, Y_train))
print("Score the X-test with Y-test is : ", dtc.score(X_test, Y_test))
Y_pred = dtc.predict(X_test)
print(" Mean absolute error is ", mean_absolute_error(Y_test, Y_pred))
print(" Mean squared error is ", mean_squared_error(Y_test, Y_pred))
print(" Median absolute error is ", median_absolute_error(Y_test, Y_pred))
print("Accuracy score ", accuracy_score(Y_test, Y_pred))
print("F1 score: ", round(f1_score(Y_test, Y_pred, average="weighted") * 100, 2), "%")
plot_confusion_matrix(
confusion_matrix(Y_test, Y_pred), ["Fraud", "Not Fraud"], normalize=False
)
plot_confusion_matrix(
confusion_matrix(Y_test, Y_pred), ["Fraud", "Not Fraud"], normalize=True
)
# Perfect score on training set indicates overfitting. Using hyperparameter tuning.
# ### Hyperparameter Tuning
# Normal Randomised Search takes too much time to execute on a dataset this large.
"""dtc1 = DecisionTreeClassifier()
params_dtc = {
"splitter":["best"],
'max_depth': [10, 20, 50, 100, 200],
'min_samples_leaf': [10, 20, 50, 100, 200],
'min_samples_split' : [10, 20, 50, 100, 200],
'criterion': ["gini", "entropy"]
}
random_search=RandomizedSearchCV(estimator=dtc1,param_distributions = params_dtc, scoring = 'f1',cv=5,n_iter=100)
random_search.fit(X_train,Y_train)"""
# Since dataset is very large(close to 1.8 million rows originally and even more after treating for unbalanced condition), we will use halving randomized search cross validation which is an experimental variant of the randomised search, much faster compared to either randomised search or grid search cross validation.
dtc1 = DecisionTreeClassifier()
params_dtc = {
"max_depth": [10, 20, 50, 100, 200],
"min_samples_leaf": [10, 20, 50, 100, 200],
"min_samples_split": [10, 20, 50, 100, 200],
"criterion": ["gini", "entropy"],
}
halving_random_search = HalvingRandomSearchCV(
estimator=dtc1, param_distributions=params_dtc, cv=5
)
halving_random_search.fit(X_train, Y_train)
print(halving_random_search.best_params_)
print(halving_random_search.best_params_)
dtc2 = DecisionTreeClassifier(
min_samples_split=100, min_samples_leaf=20, max_depth=200, criterion="gini"
)
dtc2.fit(X_train, Y_train)
print("Score the X-train with Y-train is : ", dtc2.score(X_train, Y_train))
print("Score the X-test with Y-test is : ", dtc2.score(X_test, Y_test))
Y_pred = dtc2.predict(X_test)
print(" Mean absolute error is ", mean_absolute_error(Y_test, Y_pred))
print(" Mean squared error is ", mean_squared_error(Y_test, Y_pred))
print(" Median absolute error is ", median_absolute_error(Y_test, Y_pred))
print("Accuracy score ", accuracy_score(Y_test, Y_pred))
print("F1 score: ", round(f1_score(Y_test, Y_pred, average="weighted") * 100, 2), "%")
plot_confusion_matrix(
confusion_matrix(Y_test, Y_pred), ["Fraud", "Not Fraud"], normalize=False
)
plot_confusion_matrix(
confusion_matrix(Y_test, Y_pred), ["Fraud", "Not Fraud"], normalize=True
)
matrix = classification_report(Y_test, Y_pred, labels=[1, 0])
print("Classification report : \n", matrix)
RocCurveDisplay.from_estimator(dtc2, X_test, Y_test)
plt.show()
# In accordance with the confusion matrix, the roc curve is almost perfect.
# # ADA Boost
from sklearn.ensemble import AdaBoostClassifier
from sklearn.ensemble import GradientBoostingClassifier
from xgboost import XGBClassifier
# Create adaboost classifer object
dtc = DecisionTreeClassifier()
abc = AdaBoostClassifier(
n_estimators=100, base_estimator=dtc, learning_rate=1.0, random_state=0
)
# Train Adaboost Classifer
abc.fit(X_train, Y_train)
# Predict the response for test dataset
y_pred_lg = abc.predict(X_test)
precision_score_ab = precision_score(Y_test, y_pred_lg)
accuracy_score_ab = accuracy_score(Y_test, y_pred_lg)
print("The precision score is : ", round(precision_score_ab * 100, 2), "%")
print("The accuracy score is : ", round(accuracy_score_ab * 100, 2), "%")
print("\nClassification Report TEST:\n", classification_report(Y_test, y_pred_lg))
plot_confusion_matrix(
confusion_matrix(Y_test, y_pred_lg), ["Fraud", "Not Fraud"], normalize=False
)
plot_confusion_matrix(
confusion_matrix(Y_test, y_pred_lg), ["Fraud", "Not Fraud"], normalize=True
)
RocCurveDisplay.from_estimator(abc, X_test, Y_test)
plt.show()
matrix = classification_report(Y_test, y_pred_lg, labels=[1, 0])
print("Classification report : \n", matrix)
# # Auto-Sklearn
import autosklearn.classification
automl = autosklearn.classification.AutoSklearnClassifier(
time_left_for_this_task=9000,
memory_limit=15400,
)
automl.fit(X_train, Y_train)
y_hat = automl.predict(X_test)
print("Accuracy score", accuracy_score(Y_test, y_hat))
matrix = confusion_matrix(Y_test, y_hat, labels=[1, 0])
print("Confusion matrix : \n", matrix)
tp, fn, fp, tn = confusion_matrix(Y_test, y_hat, labels=[1, 0]).reshape(-1)
print("Outcome values : \n", tp, fn, fp, tn)
matrix = classification_report(Y_test, y_hat, labels=[1, 0])
print("Classification report : \n", matrix)
a_sklrn_df = pd.DataFrame.from_dict(automl.cv_results_)
a_sklrn_df = a_sklrn_df.sort_values(by=["mean_test_score"], ascending=False)
a_sklrn_df.to_csv("autosklearn.csv", index=True)
a_sklrn_df
precision_score_ab = precision_score(Y_test, y_hat)
accuracy_score_ab = accuracy_score(Y_test, y_hat)
print("The precision score is : ", round(precision_score_ab * 100, 2), "%")
print("The accuracy score is : ", round(accuracy_score_ab * 100, 2), "%")
print("\nClassification Report TEST:\n", classification_report(Y_test, y_hat))
plot_confusion_matrix(
confusion_matrix(Y_test, y_hat), ["Fraud", "Not Fraud"], normalize=False
)
plot_confusion_matrix(
confusion_matrix(Y_test, y_hat), ["Fraud", "Not Fraud"], normalize=True
)
# # KNN
# Downsampling majority class
df_majority_downsampled = resample(
df_majority,
replace=False, # sample without replacement
n_samples=len(df_minority), # to match minority class
random_state=42,
) # reproducible results
df_majority_downsampled.shape
# Combining downsampled majority class with minority class
total_downsampled = pd.concat([df_majority_downsampled, df_minority])
total_downsampled.shape
# Splitting into training and testing sets
x_cols = list(total_downsampled.columns)
x_cols.remove("is_fraud")
X = total_downsampled[x_cols]
Y = total_downsampled["is_fraud"]
X_train, X_test, Y_train, Y_test = train_test_split(
X, Y, test_size=0.3, random_state=42
)
X_train.shape
from sklearn.neighbors import KNeighborsClassifier
# Setup arrays to store training and test accuracies
neighbors = np.arange(1, 21)
# print(neighbors)
train_accuracy = np.empty(len(neighbors))
test_accuracy = np.empty(len(neighbors))
for i, k in enumerate(neighbors):
# Setup a knn classifier with k neighbors
knn = KNeighborsClassifier(n_neighbors=k)
# Fit the model
knn.fit(X_train, Y_train)
# Compute accuracy on the training set
train_accuracy[i] = knn.score(X_train, Y_train)
# Compute accuracy on the test set
test_accuracy[i] = knn.score(X_test, Y_test)
# Generate plot
plt.title("k-NN Varying number of neighbors")
plt.plot(neighbors, test_accuracy, label="Testing Accuracy")
plt.plot(neighbors, train_accuracy, label="Training accuracy")
plt.legend()
plt.xlabel("Number of neighbors")
plt.ylabel("Accuracy")
plt.show()
# Setup a knn classifier with k neighbors
knn = KNeighborsClassifier(n_neighbors=1)
# Fit the model
knn.fit(X_train, Y_train)
# Get accuracy. Note: In case of classification algorithms score method represents accuracy.
knn.score(X_test, Y_test)
# let us get the predictions using the classifier we had fit above
y_pred = knn.predict(X_test)
y_pred_proba = knn.predict_proba(X_test)[:, 1]
fpr, tpr, thresholds = roc_curve(Y_test, y_pred_proba)
plt.plot([0, 1], [0, 1], "k--")
plt.plot(fpr, tpr, label="Knn")
plt.xlabel("fpr")
plt.ylabel("tpr")
plt.title("Knn(n_neighbors=1) ROC curve")
plt.show()
# Area under ROC curve
roc_auc_score(Y_test, y_pred_proba)
precision_score_ab = precision_score(Y_test, y_pred)
accuracy_score_ab = accuracy_score(Y_test, y_pred)
print("The precision score is : ", round(precision_score_ab * 100, 2), "%")
print("The accuracy score is : ", round(accuracy_score_ab * 100, 2), "%")
print("\nClassification Report TEST:\n", classification_report(Y_test, y_pred))
plot_confusion_matrix(
confusion_matrix(Y_test, y_pred), ["Fraud", "Not Fraud"], normalize=False
)
plot_confusion_matrix(
confusion_matrix(Y_test, y_pred), ["Fraud", "Not Fraud"], normalize=True
)
# # XG Boost
from xgboost import XGBRegressor
xgb = XGBRegressor(n_estimators=500)
xgb.fit(
X_train,
Y_train,
early_stopping_rounds=5,
eval_set=[(X_test, Y_test)],
verbose=False,
)
Y_pred = xgb.predict(X_test)
Y_pred = [0 if i <= 0.5 else 1 for i in Y_pred]
precision_score_ab = precision_score(Y_test, Y_pred)
accuracy_score_ab = accuracy_score(Y_test, Y_pred)
print("The precision score is : ", round(precision_score_ab * 100, 2), "%")
print("The accuracy score is : ", round(accuracy_score_ab * 100, 2), "%")
print("\nClassification Report TEST:\n", classification_report(Y_test, Y_pred))
plot_confusion_matrix(
confusion_matrix(Y_test, Y_pred), ["Fraud", "Not Fraud"], normalize=False
)
plot_confusion_matrix(
confusion_matrix(Y_test, Y_pred), ["Fraud", "Not Fraud"], normalize=True
)
# # Gradient Boost
from sklearn.ensemble import GradientBoostingClassifier
gb = GradientBoostingClassifier(
learning_rate=0.11042628136263043,
max_depth=None,
max_leaf_nodes=30,
min_samples_leaf=22,
)
gb.fit(X_train, Y_train)
y_pred = gb.predict(X_test)
plot_confusion_matrix(
confusion_matrix(Y_test, y_pred), ["Fraud", "Not Fraud"], normalize=True
)
plot_confusion_matrix(
confusion_matrix(Y_test, y_pred), ["Fraud", "Not Fraud"], normalize=False
)
precision_score_ab = precision_score(Y_test, y_pred)
accuracy_score_ab = accuracy_score(Y_test, y_pred)
print("The precision score is : ", round(precision_score_ab * 100, 2), "%")
print("The accuracy score is : ", round(accuracy_score_ab * 100, 2), "%")
print("\nClassification Report TEST:\n", classification_report(Y_test, y_pred))
RocCurveDisplay.from_estimator(gb, X_test, Y_test)
plt.show()
# # Neural Network
import tensorflow as tf
from keras.models import Sequential
from keras.layers import Dense, Dropout, Input
from tensorflow.keras.callbacks import ReduceLROnPlateau, EarlyStopping
from keras.utils.vis_utils import plot_model
model = Sequential()
model.add(Input(shape=(X_train.shape[1],)))
model.add(
Dense(
16,
activation="LeakyReLU",
)
)
model.add(
Dense(
8,
activation="relu",
)
)
model.add(
Dense(
4,
activation="LeakyReLU",
)
)
model.add(
Dense(
2,
activation="LeakyReLU",
)
)
model.add(
Dense(
1,
activation="sigmoid",
)
)
model.summary()
model.compile(loss="binary_crossentropy", optimizer="adam", metrics=["accuracy"])
history = model.fit(
X_train, Y_train, epochs=25, batch_size=512, validation_split=0.2, shuffle=True
)
# Evaluate the model
y_pred = model.predict(X_test)
loss, accuracy = model.evaluate(X_test, Y_test)
print("Test loss:", loss)
print("Test accuracy:", accuracy)
y_pred = [0 if i <= 0.5 else 1 for i in Y_pred]
plot_confusion_matrix(
confusion_matrix(Y_test, y_pred), ["Fraud", "Not Fraud"], normalize=False
)
plot_confusion_matrix(
confusion_matrix(Y_test, y_pred), ["Fraud", "Not Fraud"], normalize=True
)
# Assuming history is the object returned by model.fit()
def plot_history(history):
plt.figure(figsize=(12, 4))
# Plot training & validation accuracy values
plt.subplot(1, 2, 1)
plt.plot(history.history["accuracy"])
plt.plot(history.history["val_accuracy"])
plt.title("Model accuracy")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(["Train", "Validation"], loc="upper left")
# Plot training & validation loss values
plt.subplot(1, 2, 2)
plt.plot(history.history["loss"])
plt.plot(history.history["val_loss"])
plt.title("Model loss")
plt.ylabel("Loss")
plt.xlabel("Epoch")
plt.legend(["Train", "Validation"], loc="upper right")
plt.show()
plot_history(history)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Importing core libraries
import numpy as np
import pandas as pd
from time import time
import pprint
import joblib
from functools import partial
# Suppressing warnings because of skopt verbosity
import warnings
warnings.filterwarnings("ignore")
# Classifiers
import lightgbm as lgb
# Model selection
from sklearn.model_selection import KFold
# Metrics
from sklearn.metrics import mean_squared_error
from sklearn.metrics import make_scorer
# Skopt functions
from skopt import BayesSearchCV
from skopt.callbacks import DeadlineStopper, DeltaYStopper
from skopt.space import Real, Categorical, Integer
# Loading data
X = pd.read_csv("../input/playground-series-s3e12/train.csv")
X_test = pd.read_csv("../input/playground-series-s3e12/test.csv")
# Preparing data as a tabular matrix
y = X.target
X = X.set_index("id").drop("target", axis="columns")
X_test = X_test.set_index("id")
# Reporting util for different optimizers
def report_perf(optimizer, X, y, title="model", callbacks=None):
"""
A wrapper for measuring time and performance of optimizers
optimizer = a sklearn or a skopt optimizer
X = the training set
y = our target
title = a string label for the experiment
"""
start = time()
if callbacks is not None:
optimizer.fit(X, y, callback=callbacks)
else:
optimizer.fit(X, y)
d = pd.DataFrame(optimizer.cv_results_)
best_score = optimizer.best_score_
best_score_std = d.iloc[optimizer.best_index_].std_test_score
best_params = optimizer.best_params_
print(
(
title
+ " took @.2f seconds, candidates checked: %d, best CV score: %.3f"
+ "\u00B1"
+ " %.3f"
)
% (
time() - start,
len(optimizer.cv_results_["params"]),
best_score,
best_score_std,
)
)
print("Best parameters:")
pprint.pprint(best_params)
print()
return best_params
# Setting the scoring function
scoring = make_scorer(
partial(mean_squared_error, squared=False), greater_is_better=False
)
# Setting the validation strategy
kf = KFold(n_splits=5, shuffle=True, random_state=0)
# Setting the basic regressor
reg = lgb.LGBMRegressor(
boosting_type="gbdt",
metric="rmse",
objective="regression",
n_jobs=1,
verbose=-1,
random_state=0,
)
# Setting the search space
search_spaces = {
# Boosting learning rate
"learning_rate": Real(0.01, 1.0, "log-uniform"),
# Number of boosted trees to fit
"n_estimators": Integer(30, 5000),
# Maximum tree leaves for base learners
"num_leaves": Integer(2, 512),
# Maximum tree depth for base learners
"max_depth": Integer(-1, 256),
# Minimal number of data in one leaf
"min_child_samples": Integer(1, 256),
# Max number of bins buckets
"max_bin": Integer(100, 1000),
# Subsample ratio of the training instance
"subsample": Real(0.01, 1.0, "uniform"),
# Frequency of subsample
"subsample_freq": Integer(0, 10),
# Subsample ratio of columns
"colsample_bytree": Real(0.01, 10.0, "uniform"),
# Minimum sum of instance weight
"min_child_weight": Real(0.01, 10.0, "uniform"),
# L2 regularization
"reg_lambda": Real(1e-9, 100.0, "log-uniform"),
# L1 regularization
"reg_alpha": Real(1e-9, 100.0, "log-uniform"),
}
# Wrapping everything up into the Bayesian optimizer
opt = BayesSearchCV(
estimator=reg,
search_spaces=search_spaces,
scoring=scoring,
cv=kf,
n_iter=100, # max number of trials
n_jobs=-1, # number of jobs
iid=False,
# if not iid it optimizes on the cv score
return_train_score=False,
refit=False,
# Gaussian Processes (GP)
optimizer_kwargs={"base_estimator": "GP"},
# random state for replicability
random_state=0,
)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
assignees = pd.read_stata("./Assignees_1975-2010.dta")
citation1 = pd.read_stata("./Citations_1975-1999.dta")
citation2_1 = pd.read_stata("./Citations_2000-2010 part 1.dta")
citation2_2 = pd.read_stata("./Citations_2000-2010 part 2.dta")
patents = pd.read_stata("./Patents_1975-2010.dta")
inventors1 = pd.read_stata("./Inventors_1975-2010 part 1.dta")
# inventors2 = pd.read_stata('./Inventors_1975-2010 part 2.dta')
class_subclass = pd.read_stata("./Class-Subclass_1975-2010.dta")
assignees.head()
citation1.head()
citation1["category"].isnull().sum() / len(citation1)
|
import numpy as np
import pandas as pd
import tensorflow as tf
df = pd.read_csv("/kaggle/input/crop-production-data/cpdata.csv")
df["label"].unique().shape
class_names = list(df["label"].unique())
X = df.drop(["ph", "label"], axis=1)
y = df["label"].apply(class_names.index)
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, shuffle=True, random_state=0
)
y_train
model = aggtf.keras.Sequential(
[
tf.keras.layers.Dense(32, activation="relu"),
tf.keras.layers.Dense(64, activation="relu"),
tf.keras.layers.Dense(31),
]
)
model.compile(
optimizer="adam",
loss=tf.keras.losses.CategoricalCrossentropy(from_logits=True),
metrics=["accuracy"],
)
model.fit(X_train, y_train, epochs=300)
model.evaluate(test_ds)
model.save("/kaggle/working/my_saved_model")
|
# # 1. Introduction
# Name: Tomasz Abels and Jack Chen
# Username: JackChenXJ
# Score:
# Leaderbord rank:
# # 2. Data
# ### 2.1 Dataset
# In this section, we load and explore the dataset.
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import dask.dataframe
import os
print(os.listdir("../input/LANL-Earthquake-Prediction"))
train = pd.read_csv(
"../input/LANL-Earthquake-Prediction/train.csv",
dtype={"acoustic_data": np.int16, "time_to_failure": np.float64},
)
print(train.shape)
acoustic_data = train["acoustic_data"].values[::100]
time_data = train["time_to_failure"].values[::100]
print(acoustic_data.shape)
print(time_data.shape)
fig, ax1 = plt.subplots(figsize=(16, 8))
plt.title("Trends of acoustic_data and time_to_failure. 2% of data (sampled)")
plt.plot(acoustic_data, color="b")
ax1.set_ylabel("acoustic_data", color="b")
plt.legend(["acoustic_data"])
ax2 = ax1.twinx()
plt.plot(time_data, color="g")
ax2.set_ylabel("time_to_failure", color="g")
plt.legend(["time_to_failure"], loc=(0.875, 0.9))
test1 = pd.read_csv(
"/kaggle/input/LANL-Earthquake-Prediction/test/seg_00030f.csv",
dtype={"acoustic_data": np.int16, "time_to_failure": np.float64},
)
test2 = pd.read_csv(
"/kaggle/input/LANL-Earthquake-Prediction/test/seg_0012b5.csv",
dtype={"acoustic_data": np.int16, "time_to_failure": np.float64},
)
test3 = pd.read_csv(
"/kaggle/input/LANL-Earthquake-Prediction/test/seg_00184e.csv",
dtype={"acoustic_data": np.int16, "time_to_failure": np.float64},
)
# Initialize the subplots
fig, ax = plt.subplots(3, 1, figsize=(16, 12))
# Plot the time domain, label the graph and limit the axis
ax[0].plot(test1, color="b")
ax[0].legend(["acoustic_data"])
ax[0].set_ylabel("acoustic_data")
ax[1].plot(test2, color="b")
ax[1].legend(["acoustic_data"])
ax[1].set_ylabel("acoustic_data")
ax[2].plot(test3, color="b")
ax[2].legend(["acoustic_data"])
ax[2].set_ylabel("acoustic_data")
# ### 2.1.1 Train-test split
# In the above below, we split the train data into a test and a train set. Set a value for the `test_size` yourself. Argue why the test value can not be too small or too large. You can also use k-fold cross validation.
# Secondly, we have set the `random_state` to 102. Can you think of a reason why we set a `random_state` at all?
# ### 2.2 Data Exploration
# Explore the features and target variables of the dataset. Think about making some scatter plots, box plots, histograms or printing the data, but feel free to choose any method that suits you.
# What do you think is the right performance
# metric to use for this dataset? Clearly explain which performance metric you
# choose and why.
# Algorithmic bias can be a real problem in Machine Learning. So based on this,
# should we use the Race and the Sex features in our machine learning algorithm? Explain what you believe.
# Code from Basic Feature Benchmark by INVERSION, https://www.kaggle.com/code/inversion/basic-feature-benchmark
# Code from Earthquakes FE. More features and samples by ANDREW LUKYANENKO, https://www.kaggle.com/code/artgor/earthquakes-fe-more-features-and-samples
from tqdm import tqdm
from scipy.signal import hilbert, convolve, hann
from scipy import stats
rows = 150000
segments = int(np.floor(train.shape[0] / rows))
X_train = pd.DataFrame(
index=range(segments), dtype=float, columns=["ave", "std", "max", "min"]
)
y_train = pd.DataFrame(index=range(segments), dtype=float, columns=["time_to_failure"])
def add_trend_feature(arr, abs_values=False):
idx = np.array(range(len(arr)))
if abs_values:
arr = np.abs(arr)
lr = LinearRegression()
lr.fit(idx.reshape(-1, 1), arr)
return lr.coef_[0]
def classic_sta_lta(x, length_sta, length_lta):
sta = np.cumsum(x**2)
# Convert to float
sta = np.require(sta, dtype=float)
# Copy for LTA
lta = sta.copy()
# Compute the STA and the LTA
sta[length_sta:] = sta[length_sta:] - sta[:-length_sta]
sta /= length_sta
lta[length_lta:] = lta[length_lta:] - lta[:-length_lta]
lta /= length_lta
# Pad zeros
sta[: length_lta - 1] = 0
# Avoid division by zero by setting zero values to tiny float
dtiny = np.finfo(0.0).tiny
idx = lta < dtiny
lta[idx] = dtiny
return sta / lta
def calc_change_rate(x):
change = (np.diff(x) / x[:-1]).values
change = change[np.nonzero(change)[0]]
change = change[~np.isnan(change)]
change = change[change != -np.inf]
change = change[change != np.inf]
return np.mean(change)
for segment in tqdm(range(segments)):
seg = train.iloc[segment * rows : segment * rows + rows]
x = pd.Series(seg["acoustic_data"].values)
y = seg["time_to_failure"].values[-1]
y_train.loc[segment, "time_to_failure"] = y
X_train.loc[segment, "ave"] = x.mean()
X_train.loc[segment, "std"] = x.std()
X_train.loc[segment, "max"] = x.max()
X_train.loc[segment, "min"] = x.min()
X_train.loc[segment, "mean_change_abs"] = np.mean(np.diff(x))
X_train.loc[segment, "mean_change_rate"] = calc_change_rate(x)
X_train.loc[segment, "abs_max"] = np.abs(x).max()
X_train.loc[segment, "abs_max"] = np.abs(x).min()
X_train.loc[segment, "std_first_50000"] = x[:50000].std()
X_train.loc[segment, "std_last_50000"] = x[-50000:].std()
X_train.loc[segment, "std_first_10000"] = x[:10000].std()
X_train.loc[segment, "std_last_10000"] = x[-10000:].std()
X_train.loc[segment, "avg_first_50000"] = x[:50000].mean()
X_train.loc[segment, "avg_last_50000"] = x[-50000:].mean()
X_train.loc[segment, "avg_first_10000"] = x[:10000].mean()
X_train.loc[segment, "avg_last_10000"] = x[-10000:].mean()
X_train.loc[segment, "min_first_50000"] = x[:50000].min()
X_train.loc[segment, "min_last_50000"] = x[-50000:].min()
X_train.loc[segment, "min_first_10000"] = x[:10000].min()
X_train.loc[segment, "min_last_10000"] = x[-10000:].min()
X_train.loc[segment, "max_first_50000"] = x[:50000].max()
X_train.loc[segment, "max_last_50000"] = x[-50000:].max()
X_train.loc[segment, "max_first_10000"] = x[:10000].max()
X_train.loc[segment, "max_last_10000"] = x[-10000:].max()
X_train.loc[segment, "max_to_min"] = x.max() / np.abs(x.min())
X_train.loc[segment, "max_to_min_diff"] = x.max() - np.abs(x.min())
X_train.loc[segment, "count_big"] = len(x[np.abs(x) > 500])
X_train.loc[segment, "sum"] = x.sum()
X_train.loc[segment, "mean_change_rate_first_50000"] = calc_change_rate(x[:50000])
X_train.loc[segment, "mean_change_rate_last_50000"] = calc_change_rate(x[-50000:])
X_train.loc[segment, "mean_change_rate_first_10000"] = calc_change_rate(x[:10000])
X_train.loc[segment, "mean_change_rate_last_10000"] = calc_change_rate(x[-10000:])
X_train.loc[segment, "q95"] = np.quantile(x, 0.95)
X_train.loc[segment, "q99"] = np.quantile(x, 0.99)
X_train.loc[segment, "q05"] = np.quantile(x, 0.05)
X_train.loc[segment, "q01"] = np.quantile(x, 0.01)
X_train.loc[segment, "abs_q95"] = np.quantile(np.abs(x), 0.95)
X_train.loc[segment, "abs_q99"] = np.quantile(np.abs(x), 0.99)
X_train.loc[segment, "abs_q05"] = np.quantile(np.abs(x), 0.05)
X_train.loc[segment, "abs_q01"] = np.quantile(np.abs(x), 0.01)
X_train.loc[segment, "abs_mean"] = np.abs(x).mean()
X_train.loc[segment, "abs_std"] = np.abs(x).std()
X_train.loc[segment, "mad"] = x.mad()
X_train.loc[segment, "kurt"] = x.kurtosis()
X_train.loc[segment, "skew"] = x.skew()
X_train.loc[segment, "med"] = x.median()
X_train.loc[segment, "Hilbert_mean"] = np.abs(hilbert(x)).mean()
X_train.loc[segment, "Hann_window_mean"] = (
convolve(x, hann(150), mode="same") / sum(hann(150))
).mean()
X_train.loc[segment, "Moving_average_700_mean"] = (
x.rolling(window=700).mean().mean(skipna=True)
)
ewma = pd.Series.ewm
X_train.loc[segment, "exp_Moving_average_300_mean"] = (
ewma(x, span=300).mean()
).mean(skipna=True)
X_train.loc[segment, "exp_Moving_average_3000_mean"] = (
ewma(x, span=3000).mean().mean(skipna=True)
)
X_train.loc[segment, "exp_Moving_average_30000_mean"] = (
ewma(x, span=30000).mean().mean(skipna=True)
)
no_of_std = 3
X_train.loc[segment, "MA_700MA_std_mean"] = x.rolling(window=700).std().mean()
X_train.loc[segment, "MA_700MA_BB_high_mean"] = (
X_train.loc[segment, "Moving_average_700_mean"]
+ no_of_std * X_train.loc[segment, "MA_700MA_std_mean"]
).mean()
X_train.loc[segment, "MA_700MA_BB_low_mean"] = (
X_train.loc[segment, "Moving_average_700_mean"]
- no_of_std * X_train.loc[segment, "MA_700MA_std_mean"]
).mean()
X_train.loc[segment, "MA_400MA_std_mean"] = x.rolling(window=400).std().mean()
X_train.loc[segment, "MA_400MA_BB_high_mean"] = (
X_train.loc[segment, "Moving_average_700_mean"]
+ no_of_std * X_train.loc[segment, "MA_400MA_std_mean"]
).mean()
X_train.loc[segment, "MA_400MA_BB_low_mean"] = (
X_train.loc[segment, "Moving_average_700_mean"]
- no_of_std * X_train.loc[segment, "MA_400MA_std_mean"]
).mean()
X_train.loc[segment, "MA_1000MA_std_mean"] = x.rolling(window=1000).std().mean()
X_train.drop("Moving_average_700_mean", axis=1, inplace=True)
X_train.loc[segment, "iqr"] = np.subtract(*np.percentile(x, [75, 25]))
X_train.loc[segment, "q999"] = np.quantile(x, 0.999)
X_train.loc[segment, "q001"] = np.quantile(x, 0.001)
X_train.loc[segment, "ave10"] = stats.trim_mean(x, 0.1)
print(X_train.shape)
print(y_train.shape)
print(X_train)
# standard normalize the data
from sklearn import preprocessing
scaler = preprocessing.StandardScaler().fit(X_train)
X_train = scaler.transform(X_train)
# train pca
from sklearn.decomposition import PCA
pca = PCA(
n_components=20
) # doing pca and keeping only n_components, shows the first 20 components
pca = pca.fit(
X_train
) # the correct dimension of X for sklearn is P*N (samples*features)
X_pca_skl = pca.transform(X_train)
# perform pca on features
plt.bar(range(0, 20), pca.explained_variance_ratio_, label="individual var")
plt.step(
range(0, 20), np.cumsum(pca.explained_variance_ratio_), "r", label="cumulative var"
)
plt.xlabel("Principal component index")
plt.ylabel("explained variance ratio %")
plt.legend()
# fig, ax1 = plt.subplots(figsize=(16, 8))
plt.title("Trends of acoustic_data and time_to_failure. 2% of data (sampled)")
plt.plot(X_pca_skl[:, 0], color="b")
ax1.set_ylabel("acoustic_data", color="b")
plt.legend(["acoustic_data"])
# ### 2.3 Data Preparation
# This dataset hasn’t been cleaned yet. Meaning that some attributes (features) are in numerical format and some are in categorial format. Moreover, there are missing values as well. However, all Scikit-learn’s implementations of these algorithms expect numerical features. Check for all features if they are in categorial and use a method to transform them to numerical values. For the numerical data, handle the missing data and normalize the data.
# Note that you are only allowed to use training data for preprocessing but you then need to perform similar changes on test data too.
# You can use [pipelining](https://scikit-learn.org/stable/modules/generated/sklearn.pipeline.Pipeline.html) to help with the preprocessing.
from sklearn.model_selection import train_test_split
# Randomize the set and split it into a training and test set
x_train, x_test, y_train, y_test = train_test_split(
X_pca_skl, y_train, test_size=0.3, shuffle=True, random_state=102
)
# standard normalize the data
from sklearn import preprocessing
scaler = preprocessing.StandardScaler().fit(x_train)
x_train_norm = scaler.transform(x_train)
x_test_norm = scaler.transform(x_test)
# convert y values to categorical values
y_train = np.ravel(y_train)
y_test = np.ravel(y_test)
lab = preprocessing.LabelEncoder()
y_train_t = lab.fit_transform(y_train)
y_test_t = lab.fit_transform(y_test)
# ## 3. Training and Results
# Briefly introduce the classification algorithms you choose.
# Present your final confusion matrices (2 by 2) and balanced accuracies for both test and training data for all classifiers. Analyse the performance on test and training in terms of bias and variance. Give one advantage and one drawback of the method you use.
print(x_train_norm.shape)
print(x_test_norm.shape)
print(y_train_t.shape)
print(y_test_t.shape)
# from sklearn.datasets import make_classification
from sklearn.ensemble import RandomForestClassifier
from sklearn import datasets, svm
from sklearn.linear_model import LogisticRegression
from matplotlib import pyplot
# define the model
model = RandomForestClassifier()
# fit/train the model on all features
model.fit(x_train_norm, y_train_t)
print(model.score(x_test_norm, y_test_t))
# get feature importance
importance = model.feature_importances_
# get feature importance
importance = model.feature_importances_
# plot feature importance
pyplot.bar([x for x in range(len(importance))], importance)
pyplot.show()
# Predict
y_test_pred = model.predict(x_test_norm)
y_train_pred = model.predict(x_train_norm)
# Accuracy for training and test set
score1 = model.score(x_train_norm, y_train_t)
score2 = model.score(x_test_norm, y_test_t)
print(score1)
print(score2)
print(y_train_pred)
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn import metrics
from sklearn.linear_model import LogisticRegression
# creates a confusion matrix for the training set
cm = metrics.confusion_matrix(y_train_t, y_train_pred)
plt.figure(figsize=(4, 4))
sns.heatmap(cm, annot=True, fmt=".0f", linewidths=0.5, square=True, cmap="Blues_r")
plt.ylabel("Actual label")
plt.xlabel("Predicted label")
all_sample_title = "Accuracy Train Score: {0}".format(score1) # accuracy score
plt.title(all_sample_title, size=10)
# creates a confusion matrix for the test set
cm2 = metrics.confusion_matrix(y_test_t, y_test_pred)
plt.figure(figsize=(4, 4))
sns.heatmap(cm2, annot=True, fmt=".0f", linewidths=0.5, square=True, cmap="Blues_r")
plt.ylabel("Actual label")
plt.xlabel("Predicted label")
all_sample_title = "Accuracy Test Score: {0}".format(score2) # accuracy score
plt.title(all_sample_title, size=10)
# ## 4. Discussion and Conclusion
# Discuss all the choices you made during the process and your final conclusions. Highlight the strong points of your approach, discuss its shortcomings and suggest some future approaches that may improve it. Please be self critical here. The assignment is not about achieving a state of the art performance, but about showing what you have learned the concepts during the course.
# my_submission = pd.DataFrame({'Id': test.Id, 'SalePrice': predicted_data})
# you could use any filename. We choose submission here
# my_submission.to_csv('submission.csv', index=False)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Hello! This is a test datast from the wonderful people at IBM. It is my first solo project here on Kaggle that is completed. The dataset is trying to determine some of the causes of attrition among employees or what could cause them to leave. Let's dive in.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import accuracy_score
# Bringing in all of our necessary packages!
data = pd.read_csv(
"/kaggle/input/ibm-hr-analytics-attrition-dataset/WA_Fn-UseC_-HR-Employee-Attrition.csv"
)
data.head()
# Reading in our data and taking our first look at it.
data.info()
# Looking at the types for each column.
data.describe()
# Quick stats for the columns where it applies.
# Checking for NA's in the data.
missing_data = data.isnull().sum()
missing_data
# Making sure they are no null or NA values.
# Beginning the EDA. I want to see if there are any obvious patterns that could tell me if an employee would leave or not.
sns.countplot("Gender", hue="Attrition", data=data)
sns.countplot("MaritalStatus", hue="Attrition", data=data)
sns.countplot("NumCompaniesWorked", hue="Attrition", data=data)
sns.countplot("StockOptionLevel", hue="Attrition", data=data)
plt.figure(figsize=(12, 12))
sns.countplot("DistanceFromHome", hue="Attrition", data=data)
plt.xticks(rotation=45)
plt.show()
# Checking variables that could be important
sns.relplot(x="YearsAtCompany", y="JobSatisfaction", hue="Attrition", data=data)
# Continuing comparing variables I think are important
sns.countplot(x="YearsSinceLastPromotion", hue="Attrition", data=data)
sns.countplot("EducationField", hue="Attrition", data=data)
plt.xticks(rotation=30)
plt.show()
sns.countplot("PercentSalaryHike", hue="Attrition", data=data)
# Checking for collinearity by using a correlation matrix.
correlation_heatmap = data.drop(["Attrition"], axis=1).corr()
plt.figure(figsize=(12, 12))
sns.heatmap(correlation_heatmap, cmap="Blues")
plt.show()
# Looking at the ratio
data["Attrition"].value_counts()
# I want to see how the target, in this case Attrition is split. It looks like we have a lot more people who are not leaving which is good for this made up company but we could run into the problem of bias in the data.
data.Attrition.replace({"Yes": 1, "No": 0}, inplace=True)
data.BusinessTravel.replace(
{"Non-Travel": 0, "Travel_Rarely": 1, "Travel_Frequently": 2}, inplace=True
)
data.Department.replace(
{
"Sales": 0,
"Research & Development": 1,
"Human Resources": 2,
},
inplace=True,
)
data.Gender.replace({"Female": 0, "Male": 1}, inplace=True)
data.OverTime.replace({"No": 0, "Yes": 1}, inplace=True)
data.EducationField.replace(
{
"Life Sciences": 0,
"Medical": 1,
"Marketing": 2,
"Technical Degree": 3,
"Human Resources": 4,
"Other": 5,
},
inplace=True,
)
data.JobRole.replace(
{
"Sales Executive": 0,
"Research Scientist": 1,
"Laboratory Technician": 2,
"Manufacturing Director": 3,
"Healthcare Representative": 4,
"Manager": 5,
"Sales Representative": 6,
"Research Director": 7,
"Human Resources": 8,
},
inplace=True,
)
data.MaritalStatus.replace({"Single": 0, "Married": 1, "Divorced": 2}, inplace=True)
num_cols = [
"Age",
"DailyRate",
"DistanceFromHome",
"Education",
"HourlyRate",
"EnvironmentSatisfaction",
"JobInvolvement",
"JobLevel",
"JobSatisfaction",
"MonthlyIncome",
"MonthlyRate",
"NumCompaniesWorked",
"PercentSalaryHike",
"PerformanceRating",
"RelationshipSatisfaction",
"StockOptionLevel",
"WorkLifeBalance",
"YearsatCompany",
"YearsinCurrentRole",
"YearsSinceLastPromotion",
"YearswithCurrManager",
]
num_cols
cat_cols = [
"Attrition",
"BusinessTravel",
"Department",
"EducationField",
"Gender",
"JobRole",
"MartialStatus",
"OverTime",
]
cat_cols
# I want to organize the columns to see if which are categorical against which are numeric. I changed some of the numeric to categorical to make it easy for our model.
df1 = data.drop(["EmployeeCount", "EmployeeNumber", "StandardHours", "Over18"], axis=1)
df2 = pd.get_dummies(df1)
df2.head()
X = df2.drop(columns=["Attrition"])
y = df2["Attrition"]
# After our data has been changed to my satisfaction, I am splitting the data one last time into what will become our test and train sets. The X is all of the variables that I want to use to see how they affect attrition rate within the company. The y is the dependent variable I want to observe from the model.
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=137
)
# Splitting into testing and training. Random state for the sake of reproducibility.
rf = RandomForestClassifier()
rf.fit(X_train, y_train)
rf_predictions = rf.predict(X_test)
print("Accurary Score: {}".format(accuracy_score(y_test, rf_predictions)))
# The accuracy score! Not bad for the model.
from sklearn.ensemble import GradientBoostingClassifier
GBCmodel = GradientBoostingClassifier(n_estimators=50, max_depth=4, random_state=137)
GBCmodel.fit(X_train, y_train)
print("GBCmodel Training Score is : ", GBCmodel.score(X_train, y_train))
print("GBCmodel Test Score is : ", GBCmodel.score(X_test, y_test))
y_pred = GBCmodel.predict(X_test)
# Using GradientBoosting, which is commonly used in decision trees and by extension, randomforests.
from sklearn.metrics import confusion_matrix
CM = confusion_matrix(y_test, y_pred)
print("Confusion Matrix is : \n", CM)
sns.heatmap(CM, center=True, color="rgb")
plt.show()
# Checking our confusion matrix. As we can see, our model successfully classified people who did not leave the company.
rf.feature_importances_
from sklearn.inspection import permutation_importance
col_name = list(X.columns)
plt.figure(figsize=(12, 12))
plt.barh(col_name, rf.feature_importances_)
plt.show()
|
# # Customer Churn Prediction With EDA and Model Building
# > In this notebook we will be using plotly and seaborn for EDA. We wiill also apply multiple classification model for better prediction
# importing required libraries
import pandas as pd
import numpy as np
import seaborn as sns
import plotly.express as px
import missingno as msno
import warnings
warnings.filterwarnings("ignore")
# ----------------- Importing Machine Learnig Libraries---------------#
from sklearn.metrics import classification_report
from sklearn.preprocessing import StandardScaler
from sklearn.preprocessing import LabelEncoder
from sklearn.linear_model import LogisticRegression
from sklearn.neighbors import KNeighborsClassifier
from sklearn.svm import LinearSVC
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.ensemble import AdaBoostClassifier
from sklearn.ensemble import GradientBoostingClassifier
from sklearn.ensemble import VotingClassifier
from xgboost import XGBClassifier
# # Reading and Visualizing data
data = pd.read_csv(
"/kaggle/input/telco-customer-churn/WA_Fn-UseC_-Telco-Customer-Churn.csv"
)
data.head()
data.info()
# # Performing EDA to get insights from data
msno.matrix(data)
# Looking into data information
data.info()
# converting ToralCharges string(object) format into numerical format
data["TotalCharges"] = pd.to_numeric(data["TotalCharges"], errors="coerce")
# printing unique value of categorical data
for col in data.columns: # looping through column names
if data[col].dtype == "object": # checking for object type data
unique_values = pd.unique(
data[col]
) # storing unique values of features in a array
print(
f"{col} : {unique_values}\n"
) # printing formated string with column name and it's values
px.histogram(
data,
x="Churn",
color="Dependents",
barmode="group",
title="Churn with Dependent People",
)
# ploating graph for Online Security with respect to churn
px.histogram(
data,
x="Churn",
color="OnlineSecurity",
barmode="group",
title="Churn vs Online Security",
)
# 🔑 People with no online Security leave the company most. Company should focus on the people with no security more to make them stay
# Ploting Tenure Distribution with Box plot
px.box(data, x="Churn", y="tenure")
# 🔑 From the above box plot we can understand the folloowing:
# * Customer with tenure lower then 10 months are more likely to leave the company
# * Cutomer with tenure more then 15 months are more likely to stay with company
# Telicom company should focus more on short term nures to make them stay for longer business
# ploating donut graph for gender
px.pie(data, names="gender", hole=0.45, title="Gender Distribution")
# >our data is perfectly balanced between male and female
# visualizing Churn
px.pie(data, names="Churn", hole=0.45, title="Churn Distribution")
# > We have less data in **Yes** category as compaired to **NO**, which can cause model to over fit for NO and result in less f1 score
px.histogram(data, x="Churn", color="InternetService", barmode="group")
# 🔑 People With Optical fiber have a lot more tendency to leave the company as compared to others. Maybe they are unhappy with the optical service. Company should look into this.
px.histogram(
data, x="Churn", color="gender", barmode="group", title="Churn with gender"
)
# > From the above graph we can see that gender have no role in churn, both male and female play similar role in churn
data.columns
px.histogram(data, x="MonthlyCharges", nbins=50)
px.histogram(data, x="tenure", color="Churn")
# # Preparing Data fot Modeling
# droping Useless columns
data.drop(["customerID", "gender"], inplace=True, axis=1)
# creating a function to convert all categorical values into numerical values
def obj_to_num(df):
le = LabelEncoder() # using LabelEncoder from sklearn
for col in df.columns: # using for loop to iterate through all features
if (
df[col].dtype == "object"
): # if a column is 'object' dtype using LabelEncoder on them
df[col] = le.fit_transform(df[col])
return df # returning numerical values
# converting data into numerical data
numeric_data = obj_to_num(data)
# ploting Coorelations for features
px.imshow(
numeric_data.corr(), title="Correlations among features", height=700, width=700
)
data[["MonthlyCharges", "TotalCharges"]]
# Scaling features
scaler = StandardScaler()
scaler.fit(data[["MonthlyCharges", "TotalCharges", "tenure"]])
data[["MonthlyCharges", "TotalCharges", "tenure"]] = scaler.transform(
data[["MonthlyCharges", "TotalCharges", "tenure"]]
)
data[["MonthlyCharges", "TotalCharges"]]
# droping null values if any present
data.dropna(inplace=True)
# checking data for info once again
data.info()
# saperating dependent variable from independent variables
x = data.drop("Churn", axis=1)
y = data["Churn"]
# splitting data into training and testing data
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.25, random_state=47
)
# # Creating ML Models
# > Here we will go through some different type of ML Models/algorithms to find the best one to take in for ensembeling
# ## Logistic Regression
# creating machine learning Logistic Regression model named "lr"
lr = LogisticRegression()
# fitting data in our model
lr.fit(x_train, y_train)
# predicating x_test values which we saperated during train_test_split()
pred = lr.predict(x_test)
pred = lr.predict(x_test)
print(classification_report(y_test, pred))
# ## K-Neighbours Classifier (KNN)
knn = KNeighborsClassifier(n_neighbors=10)
knn.fit(x_train, y_train)
pred = knn.predict(x_test)
print(classification_report(y_test, pred))
# ## LinearSVC (Support Vector Machine)
svm = LinearSVC()
svm.fit(x_train, y_train)
pred = svm.predict(x_test)
print(classification_report(y_test, pred))
# ## Decesion Tree Classifier
dt = DecisionTreeClassifier(max_depth=5)
dt.fit(x_train, y_train)
pred = dt.predict(x_test)
print(classification_report(y_test, pred))
# ## Random Forest Classifier
rt = RandomForestClassifier(max_depth=5)
rt.fit(x_train, y_train)
pred = rt.predict(x_test)
print(classification_report(y_test, pred))
# ## Adaptive Boosting Classifier
abc = AdaBoostClassifier()
abc.fit(x_train, y_train)
pred = abc.predict(x_test)
print(classification_report(y_test, pred))
# # Gradient Boosting Classifier
gbc = GradientBoostingClassifier()
gbc.fit(x_train, y_train)
pred = gbc.predict(x_test)
print(classification_report(y_test, pred))
# ## XGBoost Classifier
xgb = XGBClassifier()
xgb.fit(x_train, y_train)
pred = xgb.predict(x_test)
print(classification_report(y_test, pred))
# # Ensemble top 3 Machine Learning Models
xgb = XGBClassifier()
gbc = GradientBoostingClassifier()
abc = AdaBoostClassifier()
# using Voting Classifier for Ensembling(you can use stacking as well)
ensemble = VotingClassifier(
estimators=[("lr", lr), ("gbc", gbc), ("abc", abc)], voting="soft"
)
ensemble.fit(x_train, y_train)
predictions = ensemble.predict(x_test)
print("Final Accuracy Score ")
print(classification_report(y_test, predictions))
|
a = 2
print(a)
a = 3
print(a)
c = 2 * 3**3 / 2
print(c)
print("The value of a and b is", a, c, "Thank you")
# **Swapping 2 variables**
# ****
a = 2
b = 3
c = a
a = b
b = c
print(a, b) # Swapping 2 variables....
|
# This is the submission for milestone 2 for Group 38 (Lee Soon Yoong, Ma Tsz Kiu, and Mao Yu Qi).
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
from sklearn.linear_model import LinearRegression
from sklearn.model_selection import cross_val_score
from sklearn import preprocessing
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import f_regression
from sklearn.ensemble import RandomForestRegressor
from sklearn.decomposition import PCA
from sklearn.model_selection import ShuffleSplit
import xgboost as xgb
from sklearn.model_selection import GridSearchCV
from sklearn.tree import DecisionTreeRegressor
from sklearn.ensemble import GradientBoostingRegressor
from sklearn.ensemble import ExtraTreesRegressor
# ### 1. Importing the data
df = pd.read_csv("/kaggle/input/us-college-completion-rate-analysis/train.csv")
df
# Drop index column
df = df.drop("Unnamed: 0", axis=1)
# ### 2.1 Data Exploration & Preprocessing - Check for null data
df.isnull().sum()
# There are no null values in the dataset, so we do not need to handle null values.
# ### 2.2 Data Exploration & Preprocessing - Normalize the data
x_cols = [x for x in df.columns if (x != "Completion_rate")]
x_cols
numerical_cols = [
"Tuition_in_state",
"Tuition_out_state",
"Faculty_salary",
"Pell_grant_rate",
"SAT_average",
"ACT_50thPercentile",
]
for x in numerical_cols:
df[x] = preprocessing.StandardScaler().fit_transform(np.array(df[x]).reshape(-1, 1))
df
# ### 2.3 Data Exploration & Preprocessing - Correlation matrix
corr = df.corr()
corr.style.background_gradient(cmap="RdYlBu")
# We can see that for completion rate, variables that are highly correlated with it are SAT_average, ACT_50thPercentile, Parents_highsch.
# ### 2.4 Data Exploration & Preprocessing - PCA
pca = PCA()
pca.fit(df[x_cols])
plt.plot(np.cumsum(pca.explained_variance_ratio_))
plt.xlabel("number of components")
plt.ylabel("cumulative explained variance")
# ### 2.5 Data Exploration & Preprocessing - Scatter plots
fig, axs = plt.subplots(nrows=3, ncols=5, figsize=(20, 12))
axs = axs.flatten()
for i in range(len(x_cols)):
sns.scatterplot(data=df, x=x_cols[i], y="Completion_rate", ax=axs[i])
axs[-1].set_visible(False)
axs[-2].set_visible(False)
plt.tight_layout()
# ### 2.6 Data Exploration & Preprocessing - Histogram plots
fig, axs = plt.subplots(nrows=3, ncols=5, figsize=(20, 12))
axs = axs.flatten()
for i in range(len(df.columns)):
sns.histplot(data=df, x=df[df.columns[i]], ax=axs[i])
axs[-1].set_visible(False)
plt.tight_layout()
# ### 3. Feature Selection
x = df.drop(columns=["Completion_rate"])
y = df["Completion_rate"]
best_var = SelectKBest(f_regression, k=10)
best_var.fit(x, y)
selected_features = x.columns[best_var.get_support()].tolist()
print(selected_features)
# ### 4. Model Fitting
x = df[selected_features].values
y = df["Completion_rate"]
x, y
# ### 4.1 Model Fitting - Simple regression model
linear_model = LinearRegression().fit(x, y)
r_sq = linear_model.score(x, y)
print(r_sq)
# Cross validation
cv = ShuffleSplit(n_splits=5, test_size=0.3, random_state=0)
print("R2:", cross_val_score(linear_model, x, y, scoring="r2", cv=cv))
print(
"MSE:", cross_val_score(linear_model, x, y, scoring="neg_mean_squared_error", cv=cv)
)
# ### 4.2 Model Fitting - Random forest model
param_grid = {"max_depth": [15, 20], "n_estimators": [100, 200]}
regr = RandomForestRegressor()
grid_search = GridSearchCV(estimator=regr, param_grid=param_grid, cv=5)
grid_search.fit(x, y)
best_pars = grid_search.best_params_
print("Best score:", grid_search.best_score_)
print("Best parameters:", best_pars)
regr_final_model = RandomForestRegressor(**best_pars)
regr_final_model.fit(x, y)
r_sq = regr_final_model.score(x, y)
print(r_sq)
# Cross validation
cv = ShuffleSplit(n_splits=5, test_size=0.3, random_state=0)
print("R2:", cross_val_score(regr_final_model, x, y, scoring="r2", cv=cv))
print(
"MSE:",
cross_val_score(regr_final_model, x, y, scoring="neg_mean_squared_error", cv=cv),
)
print(cross_val_score(regr_final_model, x, y, cv=cv).mean())
# For k = 12,
# Best score: 0.8529040010500083
# Best parameters: {'max_depth': 15, 'n_estimators': 100}
# 0.9788860324131724
# R2: [0.83195388 0.86202789 0.85862123 0.85882481 0.85746829]
# MSE: [-0.00527411 -0.00459544 -0.00471425 -0.00405235 -0.00426113]
# 0.8532691502238553
# For k = 10,
# Best score: 0.8577984281332242
# Best parameters: {'max_depth': 20, 'n_estimators': 200}
# 0.9811670294363567
# R2: [0.83462858 0.8629031 0.86093038 0.86400979 0.86409165]
# MSE: [-0.00510008 -0.00444626 -0.00451405 -0.00392749 -0.00422858]
# 0.857600415571915
# ### 4.3 Model Fitting - Decision Tree
param_grid = {"max_depth": [7, 10], "min_samples_leaf": [1, 2]}
dt = DecisionTreeRegressor()
grid_search = GridSearchCV(estimator=dt, param_grid=param_grid, cv=5)
grid_search.fit(x, y)
best_pars = grid_search.best_params_
print("Best score:", grid_search.best_score_)
print("Best parameters:", best_pars)
dt_final_model = DecisionTreeRegressor(**best_pars)
dt_final_model.fit(x, y)
r_sq = dt_final_model.score(x, y)
print(r_sq)
# Cross validation
cv = ShuffleSplit(n_splits=5, test_size=0.3, random_state=0)
print("R2:", cross_val_score(dt_final_model, x, y, scoring="r2", cv=cv))
print(
"MSE:",
cross_val_score(dt_final_model, x, y, scoring="neg_mean_squared_error", cv=cv),
)
print(cross_val_score(dt_final_model, x, y, cv=cv).mean())
# ### 4.4 Model Fitting - XGBoost
param_grid = {
"max_depth": [7, 10],
"n_estimators": [200, 300],
}
xgb_model = xgb.XGBRegressor()
grid_search = GridSearchCV(estimator=xgb_model, param_grid=param_grid, cv=5)
grid_search.fit(x, y)
best_pars = grid_search.best_params_
print("Best score:", grid_search.best_score_)
print("Best parameters:", best_pars)
xgb_final_model = xgb.XGBRegressor(**best_pars)
xgb_final_model.fit(x, y)
r_sq = xgb_final_model.score(x, y)
print(r_sq)
# Cross validation
cv = ShuffleSplit(n_splits=5, test_size=0.3, random_state=0)
print("R2:", cross_val_score(xgb_final_model, x, y, scoring="r2", cv=cv))
print(
"MSE:",
cross_val_score(xgb_final_model, x, y, scoring="neg_mean_squared_error", cv=cv),
)
print(cross_val_score(xgb_final_model, x, y, cv=cv).mean())
# ### 4.5 Gradient boosting
param_grid = {
"max_depth": [7, 10],
"n_estimators": [100, 200],
}
grad = GradientBoostingRegressor()
grid_search = GridSearchCV(estimator=grad, param_grid=param_grid, cv=5)
grid_search.fit(x, y)
best_pars = grid_search.best_params_
print("Best score:", grid_search.best_score_)
print("Best parameters:", best_pars)
grad_final_model = GradientBoostingRegressor(**best_pars)
grad_final_model.fit(x, y)
r_sq = grad_final_model.score(x, y)
print(r_sq)
# Cross validation
cv = ShuffleSplit(n_splits=5, test_size=0.3, random_state=0)
print("R2:", cross_val_score(grad_final_model, x, y, scoring="r2", cv=cv))
print(
"MSE:",
cross_val_score(grad_final_model, x, y, scoring="neg_mean_squared_error", cv=cv),
)
print(cross_val_score(grad_final_model, x, y, cv=cv).mean())
# ### 4.6 Extra Trees Regressor
param_grid = {
"max_depth": [10, 20],
"n_estimators": [100, 200],
}
ext = ExtraTreesRegressor()
grid_search = GridSearchCV(estimator=ext, param_grid=param_grid, cv=5)
grid_search.fit(x, y)
best_pars = grid_search.best_params_
print("Best score:", grid_search.best_score_)
print("Best parameters:", best_pars)
ext_final_model = ExtraTreesRegressor(**best_pars)
ext_final_model.fit(x, y)
r_sq = ext_final_model.score(x, y)
print(r_sq)
# Cross validation
cv = ShuffleSplit(n_splits=5, test_size=0.3, random_state=0)
print("R2:", cross_val_score(ext_final_model, x, y, scoring="r2", cv=cv))
print(
"MSE:",
cross_val_score(ext_final_model, x, y, scoring="neg_mean_squared_error", cv=cv),
)
print(cross_val_score(ext_final_model, x, y, cv=cv).mean())
# ### 4.7 Stacking models
"""
models = [regr_final_model, xgb_final_model, grad_final_model, ext_final_model]
train_preds = []
for i in models:
train_preds.append(i.predict(x))
meta_train_preds = np.column_stack(train_preds)
meta_model = LinearRegression()
meta_model.fit(meta_train_preds, y)
test_preds =[]
for i in models:
test_preds.append(i.predict(x_pred))
meta_test_preds = np.column_stack(test_preds)
meta_preds = meta_model.predict(meta_test_preds)
"""
test = pd.read_csv("/kaggle/input/us-college-completion-rate-analysis/x_test.csv")
for x in numerical_cols:
test[x] = preprocessing.StandardScaler().fit_transform(
np.array(test[x]).reshape(-1, 1)
)
x_pred = test[selected_features].values
y_pred = ext_final_model.predict(x_pred)
print(y_pred)
test[selected_features]
# ### 5. Submission
submission = pd.DataFrame.from_dict({"Completion_rate": y_pred})
submission
submission.to_csv("submission.csv", index=True, index_label="id")
|
import sys
import os
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.model_selection import cross_validate
from sklearn.pipeline import Pipeline
from sklearn.compose import ColumnTransformer
from sklearn.impute import SimpleImputer
from sklearn.preprocessing import (
OneHotEncoder,
OrdinalEncoder,
MinMaxScaler,
StandardScaler,
)
import warnings
warnings.filterwarnings("ignore")
print("python==", sys.version)
print("pandas==", pd.__version__)
print("numpy==", np.__version__)
print("seaborn==", sns.__version__)
print(os.listdir("../input/titanic"))
df_train = pd.read_csv("../input/titanic/train.csv")
df_test = pd.read_csv("../input/titanic/test.csv")
print("train shape:", df_train.shape)
print("test shape:", df_test.shape)
df_train.head()
df_train.info()
df_test.info()
# check for duplicates
df_train["PassengerId"].nunique() == df_train.shape[0]
sns.countplot(data=df_train, x="Survived")
plt.show()
all_results = []
scoring = ["accuracy", "f1", "precision", "recall"]
# ## Preprocessing pipeline
numeric_cols = ["Age", "SibSp", "Parch", "Fare"]
categorical_cols = ["Sex", "Embarked"]
ordinal_cols = ["Pclass"]
ignore_cols = ["PassengerId", "Name", "Cabin"]
X_train = df_train.drop(columns=["Survived"] + ignore_cols)
y_train = df_train["Survived"]
X_test = df_test.drop(columns=ignore_cols)
cat_pipe = Pipeline(
[
("cat_impute", SimpleImputer(strategy="most_frequent")),
("cat_one_hot_encode", OneHotEncoder()),
]
)
num_pipe = Pipeline(
[
("num_impute", SimpleImputer(strategy="mean")),
]
)
ord_pipe = Pipeline(
[
("ord_impute", SimpleImputer(strategy="most_frequent")),
("ord_ordinal_encoder", OrdinalEncoder()),
]
)
preprocess = ColumnTransformer(
[
("cat_pipe", cat_pipe, categorical_cols),
("num_pipe", num_pipe, numeric_cols),
("ord_pipe", ord_pipe, ordinal_cols),
]
)
# ## Functions
def get_one_score_from_cv(cv_result: dict) -> dict:
"""
Params:
@cv_result dict: results from sklearn cross_validate
Return:
Aggregated cross validation results
"""
better_name = cv_result["estimator"][0].steps[-1][0]
new_cv_result = {}
for score, vals in cv_result.items():
if isinstance(vals, np.ndarray):
new_cv_result[score] = np.mean(vals)
else:
new_cv_result[score] = vals[0]
new_cv_result["better_name"] = better_name
return new_cv_result
# ## Models
# ### Dummy classifier
from sklearn.dummy import DummyClassifier
# all this preproces is not needed for dummy model
pred_pipe = Pipeline(
[
("preprocess", preprocess),
("dummy", DummyClassifier(strategy="most_frequent")),
]
)
cur_result = cross_validate(
pred_pipe,
X_train,
y_train,
cv=3,
scoring=scoring,
return_estimator=True,
return_train_score=True,
)
all_results.append(get_one_score_from_cv(cur_result))
pd.DataFrame(all_results)
# Predicting the most frequent label (0 - not survived) gives an accuracy of 61%. Precision and recall values are zero because these metrics are not present for the positive label (1 - survived). TP (true positives) are 0.
# ### LGBM
from lightgbm import LGBMClassifier
pred_pipe = Pipeline(
[
("preprocess", preprocess),
("lgbm", LGBMClassifier()),
]
)
cur_result = cross_validate(
pred_pipe,
X_train,
y_train,
cv=3,
scoring=scoring,
return_estimator=True,
return_train_score=True,
)
all_results.append(get_one_score_from_cv(cur_result))
pd.DataFrame(all_results).sort_values(by=["test_accuracy"], ascending=False)
# LGBM (light gradient boosting) is a tree based model. The model has a tendecy to overfit. This can be observed by comparing the test_accuracy (cross validated): .8 with the train_accuracy: .94. The large difference in train and test indicates over-fitting.
# ### XGBOOST
from xgboost import XGBClassifier
pred_pipe = Pipeline(
[
("preprocess", preprocess),
("xgb", XGBClassifier()),
]
)
cur_result = cross_validate(
pred_pipe,
X_train,
y_train,
cv=3,
scoring=scoring,
return_estimator=True,
return_train_score=True,
)
all_results.append(get_one_score_from_cv(cur_result))
pd.DataFrame(all_results).sort_values(by=["test_accuracy"], ascending=False)
# Similar to LGBM we observe over-fitting.
# ### Naive Bayes
from sklearn.naive_bayes import GaussianNB
pred_pipe = Pipeline(
[
("preprocess", preprocess),
("gauss_nb", GaussianNB()),
]
)
cur_result = cross_validate(
pred_pipe,
X_train,
y_train,
cv=3,
scoring=scoring,
return_estimator=True,
return_train_score=True,
)
all_results.append(get_one_score_from_cv(cur_result))
pd.DataFrame(all_results).sort_values(by=["test_accuracy"], ascending=False)
# NB is not over-fitting but it is not nearly as performant as the tree based models.
# ## Hyperparameter tuning
from sklearn.model_selection import GridSearchCV
pred_pipe = Pipeline(
[
("preprocess", preprocess),
(
"tuned_lgbm",
LGBMClassifier(
verbose=-1,
),
),
]
)
# parameters recomended to tune from offical lgbm documentation to deal with overfitting.
# https://lightgbm.readthedocs.io/en/latest/Parameters-Tuning.html
param_grid = {
"tuned_lgbm__max_bin": [100, 50],
"tuned_lgbm__num_leaves": [20, 10],
"tuned_lgbm__feature_fraction": [0.7, 0.5],
"tuned_lgbm__lambda_l2": [0.01, 0.1],
"tuned_lgbm__max_depth": [20, 10, 5],
"tuned_lgbm__extra_trees": [True, False],
"tuned_lgbm__path_smooth": [0.1, 1, 10],
}
lgbm_gcv = GridSearchCV(
estimator=pred_pipe,
param_grid=param_grid,
scoring="accuracy",
cv=3,
return_train_score=True,
)
lgbm_gcv.fit(X_train, y_train)
# We are happy with the performance of LGBM. So lets grab the params that give us the best generalization performance.
df_cv_result = pd.DataFrame(lgbm_gcv.cv_results_)
df_cv_result["test_train_delta"] = abs(
df_cv_result["mean_train_score"] - df_cv_result["mean_test_score"]
)
best_params = (
df_cv_result.sort_values(
by=["test_train_delta"],
ascending=True,
)
.reset_index()
.loc[0, "params"]
)
best_params
# # This is one way we can get the best model
# best_model = pd.DataFrame(all_results).sort_values(by=["test_accuracy"], ascending=False).reset_index().head(1)["estimator"][0]
best_model = Pipeline(
[
("preprocess", preprocess),
(
"tuned_lgbm",
LGBMClassifier(
extra_trees=True,
feature_fraction=0.5,
lambda_l2=0.1,
max_bin=100,
max_depth=5,
num_leaves=20,
path_smooth=1,
),
),
]
)
best_model.fit(X_train, y_train)
y_pred = best_model.predict(X_test)
df_test["Survived"] = y_pred
df_result = df_test[["PassengerId", "Survived"]]
df_result
df_result.to_csv("titanic_submission.csv", index=False)
|
# 
# Pandas is a popular open-source data manipulation and analysis library for Python. It provides powerful data structures such as DataFrame and Series for efficient data handling, cleaning, transformation, and analysis. Pandas is widely used in data science, machine learning, and data analysis tasks due to its flexibility and ease of use. It offers a wide range of data manipulation functionalities, including data indexing, filtering, sorting, merging, reshaping, and aggregation. Pandas also provides powerful data visualization capabilities through integration with other libraries such as Matplotlib and Seaborn. With its rich set of functions and methods, Pandas simplifies the process of working with structured and tabular data in Python, making it a popular choice for data analysis tasks.
# # Basic of pandas
# Pandas: Python library for data manipulation, analysis, and visualization; provides DataFrame and Series data structures for handling tabular data efficiently.
## Importing nasseries libraries
import pandas as pd
import numpy as np
import warnings
warnings.filterwarnings("ignore")
# ### Create dictionary
# Python dictionary: key-value pairs, mutable, unordered, efficient, hash table, iterable, lookup, insertion, deletion, curly braces.
data = {
"Name": ["a", "b", "c", "d"],
"Salary": [1200, 1500, 1600, 1440],
"Digination": ["DS", "DA", "DE", "HR"],
}
data
# ### Convert dict to datadrame
# Pandas DataFrame: Tabular data, rows/columns, flexible, powerful, data manipulation, indexing, analysis, Python library, DataFrame object, data structure.
df = pd.DataFrame(data)
df
df.head(
2
) # pandas head(): DataFrame preview, top rows, quick data inspection, initial view, summary.
df.tail(
2
) # pandas tail(): DataFrame preview, bottom rows, quick data inspection, summary view, last records.
# pandas shape: DataFrame size, rows and columns, data dimensions, shape attribute, tuple.
df.shape
type(
df
) # pandas type(): DataFrame data type, data structure, Python data types, dtype, data representation.
# ### Select statement
# pandas select: DataFrame column selection, data extraction, column filtering, subsetting data, indexing.
# Select a single column
df["Name"]
## select maltiple columns
df[["Name", "Salary"]]
## select a single row
df[df["Name"] == "b"]
## how mani persons salary is > 1500
df[df["Salary"] > 1500]
# how mani persons salary is > 1500
df[df["Salary"] < 1500]
## how mani persons salary is > 1500
df[df["Salary"] >= 1500]
# particalar select row data not showing using this commant !=
df[df["Digination"] != "DS"]
# # pandas loc function used some operators:
# 1. Label-based data selection, DataFrame filtering, row/column indexing, data retrieval, locatio
# 2. Pandas: Supports +, -, *, /, >, <, ==, !=, &, |, ~ operators for data manipulation.
df.head(2)
df.loc[df["Name"] == "a"] # find the a person row
df.loc[df["Salary"] == 1200, "Salary"] ## select the particaler columns values
df["Salary"][0] # same option find the same result
df.loc[df["Salary"] > 1500] ## find the salary columns grether then 1500 salary
df.loc[
df["Salary"] > 1500, "Salary"
] ## find the salary columns grether then 1500 and show only salary values
# "!=" in pandas: Comparison operator for not equal to in DataFrame filtering, data selection, and conditional operations.
df.loc[df["Name"] != "a"]
# ### iloc function
# pandas iloc: Integer-based data selection, DataFrame indexing, row/column filtering, data retrieval, position.
df.iloc[:1] ## select the first index using iloc funtion
df.iloc[3:] ## select the 3 index using iloc funtion
df.iloc[:2]
df.iloc[:, :1] ## select the first columns
df.iloc[:, -1:] # select the second column
df.iloc[:, 1:2] # select the last column
# ### Update
# pandas update: DataFrame value update, inplace modification, data modification, data synchronization.
df.head(2)
## how to update 1200 to 15000
df.loc[df["Salary"] == 1200, "Salary"] = 15000
df
## 1 option : change the person name like a to abcd
df.loc[df["Name"] == "a", "Name"] = "Abcd"
df
## 2 option: also used this second way
df["Digination"][1] = "Data_Analyst"
df
## 3 option chage update values name whatever you wand
df["Digination"] = df["Digination"].replace({"DS": "Data_Science"})
df
# this is 4 option update values
df.replace({"DE": "Data_engineer"})
# ### Delete in DataFrame
# pandas delete: DataFrame data removal, row/column deletion, data manipulation, data modification.
df.head(2)
# drop the 0 , 2 index data usesing the axis=0 it used the select of the row
df.drop([0, 2], axis=0)
# drop the columns apply the axis=1
df.drop(columns="Name", axis=1)
# drop the maltiple columns
df.drop(columns=["Name", "Salary"], axis=1)
# ### Joing Data
# pandas join: DataFrame merging, combining data, concatenating, data integration, data consolidation.
# create the random data1
df_1 = pd.DataFrame(np.random.rand(50).reshape(10, 5), columns=list("ABCDE"))
df_1
# create datafram 2
df_2 = pd.DataFrame(np.random.rand(25).reshape(5, 5), columns=list("ABCDE"))
df_2
# check the df1 data shape
df_1.shape
# check the df2 data shape
df_2.shape
# ## concat
# "concat" in pandas: Function to concatenate and stack DataFrames along specified axis for data combination.
# marge the new dataset row wise
pd.concat([df_1, df_2], axis=0)
# marge the dataset column wise
pd.concat([df_1, df_2], axis=1)
# ## Merge funtion in pandas
# pandas merge: DataFrame joining, data merging, combining data, database-style merging, data integration.
# creathe df1 dataset and df2 dataset
df1 = pd.DataFrame({"Name": ["a", "b", "c", "d"], "Salary": [1000, 2000, 3000, 5000]})
df2 = pd.DataFrame({"Name": ["a", "b", "c", "d"], "Salary": [5500, 6500, 7500, 8500]})
df1
df2 # read_dataset
## first mathod joind the dataset like left to right columns wise
marge_by_name = df1.merge(df2, left_on="Name", right_on="Name")
marge_by_name
## second method joind the dataset like left_index to right_index wise
marge_index_wise = df1.merge(df2, right_index=True, left_index=True)
marge_index_wise
## 3 method joind the dataset inner join wise
marge_inner = df1.merge(df2, how="inner", on="Name")
marge_inner
## marge and change the header name used suffixes parmater
marge = df1.merge(df2, left_on="Name", right_on="Name", suffixes=("A", "B"))
marge
## save csv file
marge.to_csv("Employee_salary")
# ### pandas read_csv():
# DataFrame CSV file reading, data loading, data ingestion, data import.
## read the dataset
read_data = pd.read_csv(r"C:\Users\Dell\Downloads\sales data\Superstore_sales.csv")
read_data.head()
read_data.columns # pandas columns: DataFrame column names, data variables, data features, data attributes.
# pandas nunique: Count of unique values in DataFrame, distinct values, data cardinality, data uniqueness.
read_data.nunique()
# pandas drop: DataFrame data removal, row/column deletion, data dropping, data exclusion.
final_df = read_data.drop(
columns=["Row ID", "Customer ID", "Country", "Order ID"], axis=1
)
final_df.head(2)
# check the dataset shape like columns and row
final_df.shape
# check the dataset size
final_df.size
# # Handling missing values in pandas
# 1. can be done using methods like dropna(), fillna(), interpolate(), ffill(), bfill(), fillna with groupby, dropna with thresholds, or advanced imputation techniques, depending on data and analysis requirements.
# 2. set the columns of like (min,mode,median)
# pandas isna: DataFrame missing data detection, null/NaN values, data validation, data quality.
final_df.isna().sum()
# pandas mean, median, mode, max, min: DataFrame statistical measures for central tendency, data distribution, and data summary statistics, used for data analysis and summary.
final_df["City"].mode()[
0
] # pandas mode: DataFrame most frequent value(s), data mode, data occurrence count, data analysis.
final_df["City"] = final_df["City"].fillna(final_df["City"].mode()[0])
# pandas median: DataFrame middle value, data median, data central tendency, data analysis.
final_df["Postal Code"].median()
final_df["Postal Code"] = final_df["Postal Code"].fillna(
final_df["Postal Code"].median()
)
# pandas mode: DataFrame most frequent value(s), data mode, data occurrence count, data analysis.
final_df["Category"].mode()[0]
final_df["Category"] = final_df["Category"].fillna(final_df["Category"].mode()[0])
# pandas median: DataFrame middle value, data median, data central tendency, data analysis.
final_df["Sales"].median()
final_df["Sales"] = final_df["Sales"].fillna(final_df["Sales"].median())
## after fill missing values show
final_df.isna().sum()
final_df.info() # pandas info(): DataFrame summary, data information, data overview, data statistics, data details.
# astype function used change dtypes like (float to int ),(str to bool)
final_df["Postal Code"] = final_df["Postal Code"].astype(int)
## chage the dtype then showing int
final_df.dtypes
# ## pandas describe():
# DataFrame statistical summary, data distribution, data summary statistics, data insights.
final_df.describe()
# ### Find the insight of the dataset using same funtion like (groupby,pivot,value_counts)
final_df.head(2)
## check the columns of dataset
final_df.columns
## check the total sales
final_df["Sales"].sum()
## check the maximum sales
final_df["Sales"].max()
# ## value_counts():
# DataFrame value frequency, data occurrence count, data distribution analysis, categorical data analysis.
## check the region columns values of counts
final_df["Region"].value_counts()
# ## groupby:
# DataFrame grouping, data aggregation, data grouping by keys, data summarization.
## cehck the sales by region wise
final_df["Sales"].groupby(final_df["Region"]).sum()
# check the top ten state highest sales
final_df["Sales"].groupby(final_df["State"]).sum().sort_values(ascending=False).head(10)
# check the top ten state lowest sales
final_df["Sales"].groupby(final_df["State"]).sum().sort_values(ascending=False).tail(10)
## check the segment wise sales
pd.pivot_table(
data=final_df, values=["Sales"], index=["Segment"], aggfunc=max
).reset_index()
# ## pivot_table:
# DataFrame pivoting, data reshaping, data aggregation, multi-dimensional data analysis.
## check the categries wise sales
pd.pivot_table(
data=final_df, values=["Sales"], index=["Category"], aggfunc=max
).reset_index()
## cehck the catogory and region wise sales
pd.pivot_table(
data=final_df, values=["Sales"], index=["Category", "Region"], aggfunc=sum
)
# ### Lambda finction in pandas:
# Anonymous function, shorthand for data transformation, data manipulation, data filtering.
final_df.head(2)
## create new column for increasing 10 percent sales
final_df = final_df.assign(ten_percen_increage_sales=lambda x: (x["Sales"] * 110 / 100))
final_df.head(2)
final_df.drop(["Percentage", "ten_percen_increage"], axis=1, inplace=True)
## create new columns and fint rate of sales columns and percentage columns
final_df.assign(
Rate=lambda x: (x["Sales"] / x["ten_percen_increage_sales"] * 100)
).head(2)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv(
"/kaggle/input/international-rugby-union-results-from-18712022/results.csv"
)
df
df.info()
# # **Number of different competitions**
len(df["competition"].unique())
df["competition"].unique()
# ## **Since the name of the competion starts with the year, I can remove the first 5 characters if the 1st character starts with a 1 or a 2**
# Remove first 5 characters if first character is '1' or '2'
df["competition"] = df["competition"].astype(str)
df["competition"] = df["competition"].apply(
lambda x: x[5:] if x.startswith(("1", "2")) else x
)
len(df["competition"].unique())
# ## **By removing the years I was able to get the amount of competitions down from 788 to 274**
df["competition"].unique()
# ## **Lets now remove the first 3 characters for those that are dipalying the year in 2 character format as apposed to the 4 character format**
# Remove first 3 characters if first character is from 0 to 8
df["competition"] = df["competition"].apply(
lambda x: x[2:]
if x.startswith(("0", "1", "2", "3", "4", "5", "6", "7", "8"))
else x
)
len(df["competition"].unique())
# ## **Trim all competitions names to remove all whitespaces**
df["competition"] = df["competition"].apply(
lambda x: str.strip(x) if x.startswith(" ") else x
)
len(df["competition"].unique())
df["competition"].unique()[-5:]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Installations
# ### Imports
import matplotlib.pyplot as plt
import seaborn as sns
import tabulate as tbl
import geopandas as gp
import pycountry as pyc
from collections import Counter
# Data set
data = pd.read_csv("/kaggle/input/migration-nz/migration_nz.csv", header=0)
# View
data.head(1)
# Color palette
palette = sns.color_palette("icefire", as_cmap=True)
# Expand the color palette above
# Special palette for denoting loss values
loss_colors = [sns.color_palette("flare")[1], sns.color_palette("viridis")[4]]
# # Data Exploration
# Data set contents:
# - 5 columns
# - 86,526 records
# Size
print(f"There are {data.shape[0]} rows across {data.shape[1]} columns.")
# Checking dtype and missing values
data.info()
# For each object dtype, return the unique values
for cols in data.columns:
if data[cols].dtype == "object" or data[cols].dtype == "int64":
print(f"{cols}: {data[cols].nunique()}")
# Unique Values - Measure
print(f"Measure unique values:\n{data.Measure.value_counts()}")
# Unique Values - Year
print(f"Year unique values:\n{data.Year.value_counts()}")
# Unique Values - Country
print(f"Country unique values:\n{data.Country.value_counts()}")
# Summary of Value
data.Value.describe()
# ## Visualizations
# Visualizations will be most effective when looking at the intersections of categorical variables. Example: Arrivals by year.
# ### Measure across Years
# Within Measure, there are three categories:
# - Arrivals, for individuals arriving to New Zealand.
# - Departures, for individuals leaving New Zealand.
# - Net, the difference between arrivals and departures.
# There is an interesting question to ask about the Net migration per year in the timeframe of 1979 thru 2016.
# Filtered dataset
new_data = data[
(data["Citizenship"] == "Total All Citizenships")
& (data["Country"] != "All countries")
]
# Reset the index
new_data.reset_index(inplace=True)
# Check
new_data.head(10)
# Years
years = [*new_data.Year.unique()]
years = sorted(
years, reverse=False
) # Do this step to ensure years are correctly sorted in ascending order
# Measures
measures = [*new_data.Measure.unique()]
# Function to calculate measure sum based on year
def measurePerYear(measure, year) -> int:
"""
measure is "Arrivals", "Departures", or "Net"
year in any int64 from 1979 thru 2016 that appears in years
"""
# Return
return int(
new_data["Value"]
.loc[(new_data["Measure"] == measure) & (new_data["Year"] == year)]
.sum()
)
# When calculating arrivals, departures, and net migrations, do not use the all locations rows. Also, focus only on the Total All Citizenships rows.
### Measures per year
# Lists
arrivals_per_year = []
departures_per_year = []
net_per_year = []
# Iterate to calculate
for y in years:
arrivals_per_year.append(measurePerYear("Arrivals", y))
departures_per_year.append(measurePerYear("Departures", y))
net_per_year.append(measurePerYear("Net", y))
### Arrivals vs. Departures vs. Net
# Make subplots
fig, (ax0, ax1) = plt.subplots(nrows=2, figsize=(18, 18), sharex=True)
# Plots
sns.lineplot(
data=[arrivals_per_year, departures_per_year],
marker="o",
linestyle="dashed",
ax=ax0,
)
# Set the line chart title
ax0.set_title("Arrivals vs. Departures")
# Fix the legend
ax0.legend(title="Migration Type", labels=["Arrivals", "Departures"])
# Centered barplot
sns.barplot(
x=years,
y=net_per_year,
palette=[loss_colors[0] if y < 0 else loss_colors[1] for y in net_per_year],
ax=ax1,
)
# Center the axis
ax1.axhline(0, color="black")
# Set the title for the barplot
ax1.set_title("Net Migration")
# View
fig.show()
### Create a table for plain language
# Compile necessary columsn into a dataframe
yearly_migration_summary = pd.DataFrame(
list(zip(years, arrivals_per_year, departures_per_year, net_per_year)),
columns=["year", "arrivals_tot", "departures_tot", "net_tot"],
)
# Table
print(
tbl.tabulate(
yearly_migration_summary,
headers=["Years", "Total Arrivals", "Total Departures", "Total Net"],
tablefmt="prettytable",
)
)
### Histograms of arrivals, departures, and net migration
# Make subplots
fig, (ax0, ax1, ax2) = plt.subplots(ncols=3, figsize=(30, 10))
# Arrivals
sns.histplot(data=yearly_migration_summary, x="arrivals_tot", kde=True, ax=ax0)
ax0.set_title("Arrivals Histogram")
# Departures
sns.histplot(data=yearly_migration_summary, x="departures_tot", kde=True, ax=ax1)
ax1.set_title("Departures Histogram")
# Net
sns.histplot(data=yearly_migration_summary, x="net_tot", kde=True, ax=ax2)
ax2.set_title("Net Migration Histogram")
# View
fig.show()
# Looking at the charts above, the Arrivals histogram shows some bi-modality. There may be world events at play during the timeframes where those spikes in arrivals occurred, such as economic distress or civil unrest, causing an influx of immigrants. On the other hand, departures from New Zealand seem to be near normally distributed. Net Migration, which is determined from Arrivals and Departures also seems somewhat normally distributed. However, because it is impacted by the two other distributions, any extreme occurrences (like bimodality), would influence the shape of the distribution.
# ### Countries
# There are a lot of countries each year serving as the point of departure for migrants into New Zealand. To make the analysis easier, I will group those countries according to regions. Example:
# - Paraguay in a category 'South America'
# - Croatia, Hungary, Greece in a category 'Europe'
# However, this part of the exploration will start with some overall calculations across 1979 thru 2016.
### Create the list of countries
countries = [*new_data.Country.unique()]
# Function to sum the amount of arrivals/departures from/to a particular country regardless of year
def countryMovementSum(measure, country) -> int:
"""
measure is either "Arrivals" or "Departure."
country is one of the str values from countries list.
Will return an int64
"""
return int(
new_data["Value"]
.loc[(new_data["Measure"] == measure) & (new_data["Country"] == country)]
.sum()
)
# Calculate arrival and departure sums per country regardless of year
arrivals_per_country = []
departures_per_country = []
# Arrivals
for c in countries:
arrivals_per_country.append(countryMovementSum("Arrivals", c))
departures_per_country.append(countryMovementSum("Departures", c))
# Create a dataframe of just this information
country_movement_df = pd.DataFrame(
list(zip(countries, arrivals_per_country, departures_per_country)),
columns=["countries", "arrivals_tot", "departures_tot"],
)
# Remove Not stated and All countries
clean_movement_df = country_movement_df[0:251]
### Continents/Regions
# STEP 1: Add relevant geopandas data into clean_movement_df
# Read in the world data
world = gp.read_file(gp.datasets.get_path("naturalearth_lowres")) # Low res earth file
# View
world.head(3)
# In the current data set, some of the existing country names are actually continent names, such as Oceania. That's fine, there will simply be repetition between the Country and Region column. In summary, the columns take from world and added to the clean_movement_df will be:
# - continent
# - geometry
# STEP 2: Find the intersection between world.name and clean_movement_df
country_name_set = [
i for j in clean_movement_df.countries for i in world.name if i == j
]
# STEP 3: Build the continent column in clean_movement_df
continents = []
# Iterate
for c in clean_movement_df.countries:
# Check whether the country name is in the country name set
if c not in country_name_set:
continents.append(c)
# When c is in country name set, retrieve the continent name
else:
continents.append(world.continent.loc[world.name == c].values[0])
# Add to the dataframe
clean_movement_df["continent"] = continents
# Check
clean_movement_df.head(3)
# STEP 4: Return the list of continent values where continent == countries
continent_clean_up = [
i
for j in clean_movement_df.continent
for i in clean_movement_df.countries
if i == j
]
continent_clean_set = set(continent_clean_up)
# Create the continent listing
world_continent_list = [*world.continent.unique()]
# Remove any items already in world_continent_list
for c in world_continent_list:
# Check if the value in world_continent_list
if c in continent_clean_set:
# Remove that item
continent_clean_set.remove(c)
else:
continue
# View
print(continent_clean_set)
# Create the list of locations for each item in the set
# NOTE - this would be easier to do in a spreadsheet, but I like being tedious, so here we are.
continent_clean_updater = {
"USSR": "Europe",
"Macau": "Asia",
"San Marino": "Europe",
"Bermuda": "North America",
"Guam": "Oceania",
"Nauru": "Oceania",
"Czechoslovakia": "Europe",
"Equatorial Guinea": "Africa",
"Solomon Islands": "Oceania",
"Western Sahara": "Africa",
"South Georgia and the South Sandwich Islands": "South America",
"Mauritius": "Africa",
"St Lucia": "North America",
"French Southern Territories": "Antarctica",
"UK": "Europe",
"St Helena": "Africa",
"Norfolk Island": "Oceania",
"Andorra": "Europe",
"Bosnia and Herzegovina": "Europe",
"Guadeloupe": "North America",
"Bahrain": "Asia",
"Wallis and Futuna": "Oceania",
"Antigua and Barbuda": "North America",
"Democrative Republic of the Congo": "Africa",
"Liechtenstein": "Europe",
"St Pierre and Miquelon": "North America",
"Dominican Republic": "North America",
"Micronesia": "Oceania",
"Hong Kong": "Asia",
"Cayman Islands": "North America",
"Cape Verde": "Africa",
"Pitcairn Island": "Oceania",
"Cook Islands": "Oceania",
"Turks and Caicos": "North America",
"Niue": "Oceania",
"Gibraltar": "Europe",
"French Polynesia": "Oceania",
"Christmas Island": "Asia",
"Seychelles": "Africa",
"Mayotte": "Africa",
"Malta": "Europe",
"Curacao": "South America",
"US Virgin Islands": "North America",
"Dominica": "North America",
"US Minor Outlying Islands": "North America",
"Grenada": "North America",
"Yugoslavia/Serbia and Montenegro": "Europe",
"Cocos Islands": "Asia",
"Brunei Darussalam": "Asia",
"Reunion": "Africa",
"Comoros": "Africa",
"South Sudan": "Africa",
"Central African Republic": "Africa",
"Kiribati": "Oceania",
"American Samoa": "Oceania",
"Marshall Islands": "Oceania",
"Palau": "Oceania",
"Sao Tome and Principe": "Africa",
"Vatican City": "Europe",
"Falkland Islands": "South America",
"Monaco": "Europe",
"St Maarten": "North America",
"Cote d'Ivoire": "Africa",
"British Virgin Islands": "North America",
"Northern Mariana Islands": "Oceania",
"St Kitts and Nevis": "North America",
"Singapore": "Asia",
"USA": "North America",
"British Indian Ocean Territory": "Asia",
"St Vincent and the Grenadines": "North America",
"Netherlands Antilles": "South America",
"Anguilla": "North America",
"Martinique": "North America",
"French Guiana": "South America",
"Tokelau": "Oceania",
"Maldives": "Asia",
"Aruba": "South America",
"Tonga": "Oceania",
"Swaziland": "Africa",
"Barbados": "North America",
"Samoa": "Oceania",
"UAE": "Asia",
"Faeroe Islands": "Europe",
"Montserrat": "North America",
"East Germany": "Europe",
"South Yemen": "Asia",
"Tuvalu": "Oceania",
"Democratic Republic of the Congo": "Africa",
}
# For each country listed, tie to a continent
# If no matching value leave unchanged
for i in range(0, clean_movement_df.shape[0]):
# If the country is in the dictionary
if clean_movement_df.countries[i] in continent_clean_updater.keys():
# Update the continent value
clean_movement_df.continent[i] = continent_clean_updater[
clean_movement_df.countries[i]
]
# Not in the dictionary
else:
# Do nothing
continue
# Function to calculate measure sum
def continentCounts(measure, continent) -> int:
# If arrivals
if measure.lower() == "arrivals":
# Return
return int(
clean_movement_df["arrivals_tot"]
.loc[clean_movement_df["continent"] == continent]
.sum()
)
elif measure.lower() == "departures":
# Return
return int(
clean_movement_df["departures_tot"]
.loc[clean_movement_df["continent"] == continent]
.sum()
)
elif measure.lower() == "net":
# Return
return int(
clean_movement_df["arrivals_tot"]
.loc[clean_movement_df["continent"] == continent]
.sum()
) - int(
clean_movement_df["departures_tot"]
.loc[clean_movement_df["continent"] == continent]
.sum()
)
else:
print("Measure not found. Try entering arrivals, departures, or net.")
# Continents counts
arrivals_by_continent = []
departures_by_continent = []
net_by_continent = []
### Create the list of continents
continent_names = sorted([*clean_movement_df.continent.unique()])
for c in continent_names:
arrivals_by_continent.append(continentCounts("arrivals", c))
departures_by_continent.append(continentCounts("departures", c))
net_by_continent.append(continentCounts("net", c))
### Arrivals vs. Departures vs. Net by Continent
# Make subplots
fig, (ax0, ax1) = plt.subplots(nrows=2, figsize=(18, 12), sharex=True)
# Plots
sns.lineplot(
data=[arrivals_by_continent, departures_by_continent],
marker="o",
linestyle="dashed",
ax=ax0,
)
# Set the line chart title
ax0.set_title("Arrivals vs. Departures by Continent (1979 - 2016)")
# Fix the legend
ax0.legend(title="Migration Type", labels=["Arrivals", "Departures"])
# Centered barplot
sns.barplot(
x=continent_names,
y=net_by_continent,
palette=[loss_colors[0] if y < 0 else loss_colors[1] for y in net_by_continent],
ax=ax1,
)
# Center the axis
ax1.axhline(0, color="black")
# Set the title for the barplot
ax1.set_title("Net Migration by Continent (1979 - 2016)")
# View
fig.show()
### Continents by Year
continents_by_year = []
# iterate through data.Country
for c in range(0, new_data.shape[0]):
if new_data.Country[c] in continent_clean_updater.keys():
continents_by_year.append(continent_clean_updater[new_data.Country[c]])
elif new_data.Country[c] in country_name_set:
continents_by_year.append(
world.continent.loc[world.name == new_data.Country[c]].values[0]
)
else:
continents_by_year.append(new_data.Country[c])
# Add to the dataframe
new_data["Continent"] = continents_by_year
# Function to sum the amount of arrivals/departures from/to a particular country regardless of year
def continentYearlySum(measure, continent, year) -> int:
"""
measure is either "Arrivals" or "Departure."
continent is one of the str values from continent list.
Will return an int64
"""
return int(
new_data["Value"]
.loc[
(new_data["Measure"] == measure)
& (new_data["Continent"] == continent)
& (new_data["Year"] == y)
]
.sum()
)
# Continent counts by year
arr_cont_yearly = {}
dep_cont_yearly = {}
# Iterate through continent names
for c in continent_names:
# Set the continent as the key
arr_count_list = []
dep_count_list = []
# Bulid the yearly list
for y in years:
# Arrivals
arr_count_list.append(continentYearlySum("Arrivals", c, y))
# Departures
dep_count_list.append(continentYearlySum("Departures", c, y))
# Append to the dictionary
arr_cont_yearly[c] = arr_count_list
#
dep_cont_yearly[c] = dep_count_list
# Net
net_cont_yearly = {}
# Same set of keys
for c in continent_names:
net_cont_yearly[c] = np.subtract(arr_cont_yearly[c], dep_cont_yearly[c])
# Check
net_cont_yearly.keys()
# Add for 'Not stated'
arr_not_stated = []
dep_not_stated = []
# Iterate
for y in years:
arr_not_stated.append(continentYearlySum("Arrivals", "Not stated", y))
dep_not_stated.append(continentYearlySum("Departures", "Not stated", y))
# Net
net_not_stated = [np.subtract(arr_not_stated, dep_not_stated)]
# Update dictionaries
arr_cont_yearly["Not stated"] = arr_not_stated
dep_cont_yearly["Not stated"] = dep_not_stated
net_cont_yearly["Not stated"] = net_not_stated
### Arrivals vs. Departures vs. Net by Continent
# Make subplots
fig, (ax0, ax1) = plt.subplots(nrows=2, figsize=(18, 12))
# Plots
sns.lineplot(
data=[
arr_cont_yearly["Africa"],
arr_cont_yearly["Africa and the Middle East"],
arr_cont_yearly["Americas"],
arr_cont_yearly["Antarctica"],
arr_cont_yearly["Asia"],
arr_cont_yearly["Europe"],
arr_cont_yearly["North America"],
arr_cont_yearly["Oceania"],
arr_cont_yearly["South America"],
arr_cont_yearly["Not stated"],
],
marker="o",
linestyle="dashed",
ax=ax0,
)
# Set the line chart title
ax0.set_title("Yearly Arrivals by Continent")
# Tick marks
ax0.set_xticks(range(len(years)))
ax0.set_xticklabels(years, rotation=45)
# Fix the legend
ax0.legend(title="Continent", labels=arr_cont_yearly.keys())
sns.lineplot(
data=[
dep_cont_yearly["Africa"],
dep_cont_yearly["Africa and the Middle East"],
dep_cont_yearly["Americas"],
dep_cont_yearly["Antarctica"],
dep_cont_yearly["Asia"],
dep_cont_yearly["Europe"],
dep_cont_yearly["North America"],
dep_cont_yearly["Oceania"],
dep_cont_yearly["South America"],
dep_cont_yearly["Not stated"],
],
marker="o",
linestyle="dashed",
ax=ax1,
)
# Set the title
ax1.set_title("Yearly Departures by Continent")
# X tick labeles
ax1.set_xticks(range(len(years)))
ax1.set_xticklabels(years, rotation=45)
# Fix the legend
ax1.legend(title="Continent", labels=dep_cont_yearly.keys())
# View
fig.show()
# ## CUSUM Analysis - Arrivals and Departures
# CUSUM analysis is a method of detecting notable changes in data. It would be interesting to see if year-over-year any noteworthy changes in arrivals to or departures from New Zealand occurred.
# - Yearly arrivals, irrespective of country, are found in arrivals_per_year
# - Yearly departures, irrespective of country, are found in departures_per_year
# $X_{t}$ values, those we are using for change detection, are from the two lists above.
# $\mu$ will be defined based on the average values of each list.
# $\theta$ will act as the threshold.
# $c$ will be the dampening constant to keep our judgments from getting too big or too small.
# Note that this won't be very accurate to start, as I'm not accustomed to dealing with immigration data.
# ### Arrivals
### NOTE: X_t values are in arrivals_per_year
# Set mu to average of X_t
mu_arr = np.mean(arrivals_per_year)
# Threshold - usually 4 or 5 times the standard deviation
theta_arr = 5 * np.std(arrivals_per_year)
# C - usually 1 standard deviation, or half a standard deviation
c_arr = np.std(arrivals_per_year)
# Display our starting values
print(f"Mu-arrivals: {mu_arr} | Threshold: {theta_arr} | C: {c_arr}")
# Use a 0 start method
inc_arr_st_vals = [0] * len(arrivals_per_year)
dec_arr_st_vals = [0] * len(arrivals_per_year)
# Iterate through the arrivals values
for a in range(1, len(arrivals_per_year)):
inc_arr_st_vals[a] = max(
0, inc_arr_st_vals[a - 1] + (arrivals_per_year[a] - mu_arr - c_arr)
)
dec_arr_st_vals[a] = max(
0, dec_arr_st_vals[a - 1] + (mu_arr - arrivals_per_year[a] - c_arr)
)
# Build a dataframe
arr_yearly_cusum = pd.DataFrame(
list(zip(years, inc_arr_st_vals, dec_arr_st_vals)),
columns=["year", "increase_detection", "decrease_detection"],
)
# Classify as increase, decrease, or no change
arr_chg_type = []
# Iterate
for a in range(0, len(arr_yearly_cusum.year)):
# Increase
if arr_yearly_cusum.increase_detection[a] > arr_yearly_cusum.decrease_detection[a]:
arr_chg_type.append("increase")
# Decrease
elif (
arr_yearly_cusum.decrease_detection[a] > arr_yearly_cusum.increase_detection[a]
):
arr_chg_type.append("decrease")
# No change
else:
arr_chg_type.append("no change")
# Add to dataframe
arr_yearly_cusum["change_type"] = arr_chg_type
# Display the change type per year
# Table
print(
tbl.tabulate(
arr_yearly_cusum[["year", "change_type"]],
headers=["Years", "Change Type"],
tablefmt="prettytable",
)
)
# Arrivals only chart
arr_cusum = sns.lineplot(
data=[arrivals_per_year], marker="o", linestyle="solid", legend=False
)
# Tickmarks
arr_cusum.set_xticks(range(len(years)))
arr_cusum.set_xticklabels(years, rotation=45)
# Title
arr_cusum.set_title("Arrivals per Year (1979 - 2016)")
# Add mu_arr line
plt.axhline(mu_arr, c="black", ls="-")
# Add threshold lines
plt.axhline(theta_arr, c="red", ls="--")
plt.axhline(mu_arr - (theta_arr - mu_arr), c="red", ls="--")
# Display
plt.figure(figsize=(18, 12))
plt.show()
# Counter
arr_chg_type_counter = Counter(arr_yearly_cusum.change_type)
# Breakdown of change types
plt.pie(arr_chg_type_counter.values(), labels=arr_chg_type_counter.keys())
# Title
plt.title("Arrivals - Breakdown of Change Types")
# displaying chart
plt.show()
# ### Departures
### NOTE: X_t values are in departures_per_year
# Set mu to average of X_t
mu_dep = np.mean(departures_per_year)
# Threshold - usually 4 or 5 times the standard deviation
theta_dep = 5 * np.std(departures_per_year)
# C - usually 1 standard deviation, or half a standard deviation
c_dep = np.std(departures_per_year)
# Display our starting values
print(f"Mu: {mu_dep} | Threshold: {theta_dep} | C: {c_dep}")
# Use a 0 start method
inc_dep_st_vals = [0] * len(departures_per_year)
dec_dep_st_vals = [0] * len(departures_per_year)
# Iterate through the arrivals values
for a in range(1, len(departures_per_year)):
inc_dep_st_vals[a] = max(
0, inc_dep_st_vals[a - 1] + (departures_per_year[a] - mu_dep - c_dep)
)
dec_dep_st_vals[a] = max(
0, dec_dep_st_vals[a - 1] + (mu_dep - departures_per_year[a] - c_dep)
)
# Build a dataframe
dep_yearly_cusum = pd.DataFrame(
list(zip(years, inc_dep_st_vals, dec_dep_st_vals)),
columns=["year", "increase_detection", "decrease_detection"],
)
# Classify as increase, decrease, or no change
dep_chg_type = []
# Iterate
for a in range(0, len(dep_yearly_cusum.year)):
# Increase
if dep_yearly_cusum.increase_detection[a] > dep_yearly_cusum.decrease_detection[a]:
dep_chg_type.append("increase")
# Decrease
elif (
dep_yearly_cusum.decrease_detection[a] > dep_yearly_cusum.increase_detection[a]
):
dep_chg_type.append("decrease")
# No change
else:
dep_chg_type.append("no change")
# Add to dataframe
dep_yearly_cusum["change_type"] = dep_chg_type
# Display the change type per year
# Table
print(
tbl.tabulate(
dep_yearly_cusum[["year", "change_type"]],
headers=["Years", "Change Type"],
tablefmt="prettytable",
)
)
# Departures only chart
dep_cusum = sns.lineplot(
data=[departures_per_year], marker="o", linestyle="solid", legend=False
)
# Tickmarks
dep_cusum.set_xticks(range(len(years)))
dep_cusum.set_xticklabels(years, rotation=45)
# Title
dep_cusum.set_title("Departures per Year (1979 - 2016)")
# Add mu_arr line
plt.axhline(mu_dep, c="black", ls="-")
# Add threshold lines
plt.axhline(theta_dep, c="red", ls="--")
plt.axhline(mu_dep - (theta_dep - mu_dep), c="red", ls="--")
# Display
plt.figure(figsize=(18, 12))
plt.show()
# Counter
dep_chg_type_counter = Counter(dep_yearly_cusum.change_type)
# Breakdown of change types
plt.pie(dep_chg_type_counter.values(), labels=dep_chg_type_counter.keys())
# Title
plt.title("Departures - Breakdown of Change Types")
# displaying chart
plt.show()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df_sales_lv1 = pd.read_csv("/kaggle/input/m5-data/m5/processed/sales_lv1.csv")
df_sales_lv2 = pd.read_csv("/kaggle/input/m5-data/m5/processed/sales_lv2.csv")
df_sales_lv3 = pd.read_csv("/kaggle/input/m5-data/m5/processed/sales_lv3.csv")
df_sales_lv4 = pd.read_csv("/kaggle/input/m5-data/m5/processed/sales_lv4.csv")
df_sales_lv5 = pd.read_csv("/kaggle/input/m5-data/m5/processed/sales_lv5.csv")
df_sales_lv6 = pd.read_csv("/kaggle/input/m5-data/m5/processed/sales_lv6.csv")
df_sales_lv7 = pd.read_csv("/kaggle/input/m5-data/m5/processed/sales_lv7.csv")
df_sales_lv8 = pd.read_csv("/kaggle/input/m5-data/m5/processed/sales_lv8.csv")
df_sales_lv9 = pd.read_csv("/kaggle/input/m5-data/m5/processed/sales_lv9.csv")
df_sales_lv1["unique_id"] = "Total"
df_sales_lv2 = df_sales_lv2.rename(columns={"state_id": "unique_id"})
df_sales_lv3 = df_sales_lv3.rename(columns={"store_id": "unique_id"})
df_sales_lv4 = df_sales_lv4.rename(columns={"cat_id": "unique_id"})
df_sales_lv5 = df_sales_lv5.rename(columns={"dept_id": "unique_id"})
df_sales_lv6["unique_id"] = df_sales_lv6.state_id + "/" + df_sales_lv6.cat_id
df_sales_lv6 = df_sales_lv6.drop(["state_id", "cat_id"], axis=1)
df_sales_lv7["unique_id"] = df_sales_lv7.state_id + "/" + df_sales_lv7.dept_id
df_sales_lv7 = df_sales_lv7.drop(["state_id", "dept_id"], axis=1)
df_sales_lv8["unique_id"] = df_sales_lv8.store_id + "/" + df_sales_lv8.cat_id
df_sales_lv8 = df_sales_lv8.drop(["store_id", "cat_id"], axis=1)
df_sales_lv9["unique_id"] = df_sales_lv9.store_id + "/" + df_sales_lv9.dept_id
df_sales_lv9 = df_sales_lv9.drop(["store_id", "dept_id"], axis=1)
Y_df = (
df_sales_lv1.append(df_sales_lv2)
.append(df_sales_lv3)
.append(df_sales_lv4)
.append(df_sales_lv5)
.append(df_sales_lv6)
.append(df_sales_lv7)
.append(df_sales_lv8)
.append(df_sales_lv9)
)
Y_df.ds = pd.to_datetime(Y_df.ds)
date_last = np.max(Y_df["ds"])
date_cutoff = date_last - pd.Timedelta(days=28)
Y_test_df = Y_df[Y_df["ds"] > date_cutoff]
Y_train_df = Y_df[Y_df["ds"] <= date_cutoff]
df_tsfresh_lv3 = pd.read_csv(
"/kaggle/input/m5-data/m5/base_models/tsfresh_forecast_lv3.csv"
)
df_tsfresh_lv8 = pd.read_csv(
"/kaggle/input/m5-data/m5/base_models/tsfresh_forecast_lv8.csv"
)
df_tsfresh_lv9 = pd.read_csv(
"/kaggle/input/m5-data/m5/base_models/tsfresh_forecast_lv9.csv"
)
df_tsfresh_lv3 = df_tsfresh_lv3.drop(["y"], axis=1)
df_tsfresh_lv3 = df_tsfresh_lv3.rename(columns={"store_id": "unique_id", "y_pred": "y"})
df_tsfresh_lv8 = df_tsfresh_lv8.drop(["y"], axis=1)
df_tsfresh_lv8["unique_id"] = df_tsfresh_lv8.store_id + "/" + df_tsfresh_lv8.cat_id
df_tsfresh_lv8 = df_tsfresh_lv8.rename(columns={"y_pred": "y"})
df_tsfresh_lv8 = df_tsfresh_lv8.drop(["store_id", "cat_id"], axis=1)
df_tsfresh_lv9 = df_tsfresh_lv9.drop(["y"], axis=1)
df_tsfresh_lv9["unique_id"] = df_tsfresh_lv9.store_id + "/" + df_tsfresh_lv9.dept_id
df_tsfresh_lv9 = df_tsfresh_lv9.rename(columns={"y_pred": "y"})
df_tsfresh_lv9 = df_tsfresh_lv9.drop(["store_id", "dept_id"], axis=1)
df_first_lv1 = pd.read_csv("/kaggle/input/m5-data/m5/submissions/m5_1st_eval_lv1.csv")
df_first_lv2 = pd.read_csv("/kaggle/input/m5-data/m5/submissions/m5_1st_eval_lv2.csv")
df_first_lv3 = pd.read_csv("/kaggle/input/m5-data/m5/submissions/m5_1st_eval_lv3.csv")
df_first_lv4 = pd.read_csv("/kaggle/input/m5-data/m5/submissions/m5_1st_eval_lv4.csv")
df_first_lv5 = pd.read_csv("/kaggle/input/m5-data/m5/submissions/m5_1st_eval_lv5.csv")
df_first_lv6 = pd.read_csv("/kaggle/input/m5-data/m5/submissions/m5_1st_eval_lv6.csv")
df_first_lv7 = pd.read_csv("/kaggle/input/m5-data/m5/submissions/m5_1st_eval_lv7.csv")
df_first_lv8 = pd.read_csv("/kaggle/input/m5-data/m5/submissions/m5_1st_eval_lv8.csv")
df_first_lv9 = pd.read_csv("/kaggle/input/m5-data/m5/submissions/m5_1st_eval_lv9.csv")
df_first_lv1["unique_id"] = "Total"
df_first_lv2 = df_first_lv2.rename(columns={"state_id": "unique_id"})
df_first_lv3 = df_first_lv3.rename(columns={"store_id": "unique_id"})
df_first_lv4 = df_first_lv4.rename(columns={"cat_id": "unique_id"})
df_first_lv5 = df_first_lv5.rename(columns={"dept_id": "unique_id"})
df_first_lv6["unique_id"] = df_first_lv6.state_id + "/" + df_first_lv6.cat_id
df_first_lv6 = df_first_lv6.drop(["state_id", "cat_id"], axis=1)
df_first_lv7["unique_id"] = df_first_lv7.state_id + "/" + df_first_lv7.dept_id
df_first_lv7 = df_first_lv7.drop(["state_id", "dept_id"], axis=1)
df_first_lv8["unique_id"] = df_first_lv8.store_id + "/" + df_first_lv8.cat_id
df_first_lv8 = df_first_lv8.drop(["store_id", "cat_id"], axis=1)
df_first_lv9["unique_id"] = df_first_lv9.store_id + "/" + df_first_lv9.dept_id
df_first_lv9 = df_first_lv9.drop(["store_id", "dept_id"], axis=1)
Y_hat_df = (
df_first_lv1.append(df_first_lv2)
.append(df_tsfresh_lv3)
.append(df_first_lv4)
.append(df_first_lv5)
.append(df_first_lv6)
.append(df_first_lv7)
.append(df_tsfresh_lv8)
.append(df_tsfresh_lv9)
)
categorical_exog = pd.read_csv(
"/kaggle/input/m5-data/m5/processed/categorical_exog.csv"
)
categorical_exog = categorical_exog[["dept_id", "cat_id", "store_id", "state_id"]]
categorical_exog = categorical_exog.drop_duplicates()
categorical_exog = categorical_exog.reset_index(drop=True)
state_cat = list(set(categorical_exog.state_id + "/" + categorical_exog.cat_id))
state_dept = list(set(categorical_exog.state_id + "/" + categorical_exog.dept_id))
store_cat = list(set(categorical_exog.store_id + "/" + categorical_exog.cat_id))
store_dept = list(set(categorical_exog.store_id + "/" + categorical_exog.dept_id))
dept_ids = list(categorical_exog.dept_id.unique())
cat_ids = list(categorical_exog.cat_id.unique())
store_ids = list(categorical_exog.store_id.unique())
state_ids = list(categorical_exog.state_id.unique())
total = ["Total"]
S_df = pd.DataFrame(
index=total
+ state_ids
+ store_ids
+ cat_ids
+ dept_ids
+ state_cat
+ state_dept
+ store_cat
+ store_dept,
columns=state_ids + store_ids + cat_ids + dept_ids,
)
for col in S_df.columns:
for row in S_df.index:
if col in row:
S_df.loc[row][col] = 1
else:
S_df.loc[row][col] = 0
if row == "Total":
S_df.loc[row][col] = 1
tags = {
"Total": total,
"State": state_ids,
"Store": store_ids,
"Cat": cat_ids,
"Dept": dept_ids,
"State/Cat": state_cat,
"State/Dept": state_dept,
"Store/Cat": store_cat,
"Store/Dept": store_dept,
}
S_df
from hierarchicalforecast.core import HierarchicalReconciliation
from hierarchicalforecast.methods import BottomUp, TopDown, MiddleOut
reconcilers = [
BottomUp(),
TopDown(method="forecast_proportions"),
]
hrec = HierarchicalReconciliation(reconcilers=reconcilers)
Y_rec_df = hrec.reconcile(Y_hat_df=Y_hat_df, Y_df=Y_train_df, S=S_df, tags=tags)
Y_rec_df
from hierarchicalforecast.evaluation import HierarchicalEvaluation
def mse(y, y_hat):
return np.mean((y - y_hat) ** 2)
evaluator = HierarchicalEvaluation(evaluators=[mse])
evaluation = evaluator.evaluate(
Y_hat_df=Y_rec_df, Y_test_df=Y_test_df, tags=tags, benchmark="Naive"
)
evaluation.filter(like="ARIMA", axis=1).T
|
import torch, os
import numpy as np
import pandas as pd
from transformers import (
AutoTokenizer,
AutoModelForSequenceClassification,
Trainer,
TrainingArguments,
)
from sklearn.metrics import (
classification_report,
f1_score,
precision_score,
recall_score,
accuracy_score,
roc_auc_score,
)
from sklearn.model_selection import train_test_split
if torch.cuda.is_available():
device = torch.device("cuda")
else:
print("No GPU available")
# # Hyperparameters
SEED_VAL = 42
MAX_LENGTH = 64
LR = 4e-5
EPS = 1e-8
BATCH_SIZE = 16
EPOCHS = 4
MODEL = "xlm-roberta-base"
# # Data
df = pd.read_csv("/kaggle/input/pold-dataset/dataset.csv")
train, test = train_test_split(
df, test_size=0.2, stratify=df.label, random_state=SEED_VAL
)
test, val = train_test_split(
test, test_size=0.5, stratify=test.label, random_state=SEED_VAL
)
train_text = list(train.text)
train_labels = list(train.label)
val_text = list(val.text)
val_labels = list(val.label)
test_text = list(test.text)
test_labels = list(test.label)
print("Total: ", len(df))
print("Train: ", len(train))
print("Val: ", len(val))
print("Test: ", len(test))
# # Tokenization
tokenizer = AutoTokenizer.from_pretrained(MODEL)
train_encodings = tokenizer(
train_text, truncation=True, padding="max_length", max_length=MAX_LENGTH
)
val_encodings = tokenizer(
val_text, truncation=True, padding="max_length", max_length=MAX_LENGTH
)
test_encodings = tokenizer(
test_text, truncation=True, padding="max_length", max_length=MAX_LENGTH
)
# # Dataset
class MyDataset(torch.utils.data.Dataset):
def __init__(self, encodings, labels):
self.encodings = encodings
self.labels = labels
def __getitem__(self, idx):
item = {key: torch.tensor(val[idx]) for key, val in self.encodings.items()}
item["labels"] = torch.tensor(self.labels[idx])
return item
def __len__(self):
return len(self.labels)
train_dataset = MyDataset(train_encodings, train_labels)
val_dataset = MyDataset(val_encodings, val_labels)
test_dataset = MyDataset(test_encodings, test_labels)
# # Finetuning
training_args = TrainingArguments(
num_train_epochs=EPOCHS,
per_device_train_batch_size=BATCH_SIZE,
per_device_eval_batch_size=BATCH_SIZE,
warmup_steps=10,
logging_steps=100,
adam_epsilon=EPS,
report_to="none",
output_dir="./results",
logging_dir="./logs",
)
model = AutoModelForSequenceClassification.from_pretrained(MODEL)
trainer = Trainer(
model=model,
args=training_args,
train_dataset=train_dataset,
eval_dataset=val_dataset,
)
trainer.train()
# # Testing/Predictions
preds_raw, test_labels, _ = trainer.predict(test_dataset)
preds = np.argmax(preds_raw, axis=-1)
# # Results
pre = round(precision_score(test_labels, preds, average="macro") * 100, 2)
rec = round(recall_score(test_labels, preds, average="macro") * 100, 2)
f1 = round(f1_score(test_labels, preds, average="macro") * 100, 2)
acc = round(accuracy_score(test_labels, preds) * 100, 2)
print("Pre\tRec\tF1\tAcc")
print(f"{pre}\t{rec}\t{f1}\t{acc}")
# # Save the Model
trainer.save_model("model")
|
s = set()
type(s)
s = {"india": 75, "brazil": 56, "germany": 47, "japan": 23}
type(s)
s = {"india", "brazil", "germany", "japan"}
type(s)
# **Intersection**
A = {1, 2, 3, 4}
B = {3, 4, 5, 6}
C = A.intersection(B)
E = B.intersection(A) # THESE ARE SEMMETRIC OPERTION
C
A = {1, 2, 3, 4}
B = {3, 4, 5, 6}
C = A & B # INTERSECTION OPERTION
C
# **Union**
D = A.union(B)
F = B.union(A) # THESE ARE SEMMETRIC OPERTION
F
D = A | B # UNION OPERTION
D
# **Difference**
P = A - B
P
G = A.difference(B)
G
K = B - A
K
# **Symmetric Difference**
V = A ^ B
V
Sym_def = A.symmetric_difference(B)
Sym_def
Sym_def = B.symmetric_difference(A)
Sym_def
# **Other methods in set**
A
A.add(99)
A
A.pop()
A.remove(56)
B = {3, 4, 5}
A.update(B)
A
N = A.discard(70)
print(N)
A.clear()
A
# ***ASSIGNMENT*****
# **Check whether a string is a palangram or not** ( palangram is a sentence that contains every letter in english alphabet eg: **The quick brown fox jumps over the lazy dog**
a = "The quick brown fox jumps over the lazy dog"
b = set(a)
b
a = "The quick brown fox jumps over the lazy dog"
b = set(a)
count = 0
for i in b:
count = count + 1
print(count)
a = "The quick brown fox jumps over the lazy dog"
b = a.lower()
c = set(b)
count = 0
for i in c:
if i.isalpha() == True:
count = count + 1
if count == 26:
print("The string is palangram")
else:
print("The string is not palangram")
# **Program to check if a set is a subset or not**
A = {3, 4, 5, 6, 7}
B = {4, 5, 3}
count = 0
for i in B:
for j in A:
if i == j:
count = count + 1
if count == len(B):
print("B is the subset of A")
else:
print("B is not subset of A")
|
import cv2
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.image as img
image = cv2.imread("/kaggle/input/m-g-odev1/mevlana.jpg")
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
kernel = np.array([[2, 0, -1], [1, 0, -2], [1, 0, -1]])
img = cv2.filter2D(image, -1, kernel) # çıkış boyutunun aynı olması (-1)
fig, ax = plt.subplots(1, 2, figsize=(10, 6))
ax[0].imshow(image) # normal image
ax[1].imshow(img) # filter image
image = cv2.imread("/kaggle/input/m-g-odev1/mevlana.jpg")
image = image[400:500, 100:200, :]
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
kernel = np.array([[1, 2, 1], [0, 0, 0], [-1, -2, -1]])
kernel2 = np.array([[1, 0, -1], [2, 0, -2], [1, 0, -1]])
img = cv2.filter2D(image, -1, kernel)
img2 = cv2.filter2D(image, -1, kernel2)
fig, ax = plt.subplots(1, 3, figsize=(10, 6))
ax[0].imshow(image) # normal image
ax[1].imshow(img) # filter image yatay kernel geçişleri (sobel)
ax[2].imshow(img2) # filter image dikey kenel geçişleri (sobel)
image = cv2.imread("/kaggle/input/m-g-odev1/mevlana.jpg")
image = image[400:500, 100:200, :]
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
kernel = np.array([[3, 10, 3], [0, 0, 0], [-3, -10, -3]])
kernel2 = np.array([[3, 0, -3], [10, 0, -10], [3, 0, -3]])
img = cv2.filter2D(image, -1, kernel)
img2 = cv2.filter2D(image, -1, kernel2)
fig, ax = plt.subplots(1, 3, figsize=(10, 6))
ax[0].imshow(image) # normal image
ax[1].imshow(img) # filter image yatay kernel geçişleri (scharr)
ax[2].imshow(img2) # filter image dikey kenel geçişleri (scharr)
from skimage import color
img = cv2.imread("/kaggle/input/m-g-odev1/mevlana.jpg")
image = color.rgb2gray(img)
fig, ax = plt.subplots(1, figsize=(12, 8))
plt.imshow(image, cmap="gray")
kernel = np.ones((3, 3), np.float32) / 16
img = cv2.filter2D(image, -1, kernel)
fig, ax = plt.subplots(1, 2, figsize=(10, 6))
ax[0].imshow(image, cmap="gray")
ax[1].imshow(img, cmap="gray")
img = cv2.imread("/kaggle/input/m-g-odev1/mevlana.jpg")
grayimg = cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)
plt.imshow(
grayimg, cmap="gray"
) # cmap has been used as matplotlib uses some default colormap to plot grayscale images
plt.xticks([]) # To get rid of the x-ticks and y-ticks on the image axis
plt.yticks([])
print("New Image Shape", grayimg.shape)
# Finding optimal threshold
from skimage.filters import threshold_otsu
thresh_val = threshold_otsu(grayimg)
print("The optimal seperation value is", thresh_val)
thresh = 120 # set a random thresh value
binary_high = grayimg > thresh_val
binary_low = grayimg <= thresh_val
plt.imshow(binary_high)
plt.imshow(binary_low)
from skimage.filters import try_all_threshold
fig, ax = try_all_threshold(grayimg, verbose=False)
from skimage.filters import threshold_otsu
thresh = threshold_otsu(grayimg)
text_binary_otsu = grayimg > thresh
plt.imshow(text_binary_otsu)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
data = pd.read_csv("/kaggle/input/hotel-booking-demand/hotel_bookings.csv")
data
data.shape
data.describe()
data.info()
data.isnull().sum()
data["agent"].fillna("agent not avvailable", inplace=True)
data["company"].fillna("company not available", inplace=True)
data.dropna(subset=["country", "children"], inplace=True)
data.isnull().sum()
import matplotlib.pyplot as plt
import seaborn as sns
plt.figure(figsize=(5, 4))
plt.title("Reservation status count")
plt.bar(["Not canceled", "canceled"], data["is_canceled"].value_counts())
plt.show()
plt.figure(figsize=(8, 4))
ax1 = sns.countplot(x="hotel", hue="is_canceled", data=data, palette="Blues")
legend_labels, _ = ax1.get_legend_handles_labels()
plt.title("Reservation status in different hotels", size=20)
plt.xlabel("hotel")
plt.ylabel("number of reservations")
cancelled_data = data[data["is_canceled"] == 1]
top_10_country = cancelled_data["country"].value_counts()[:10]
plt.figure(figsize=(8, 8))
plt.title("Top 10 countries with reservation canceled")
plt.pie(top_10_country, autopct="%.2f", labels=top_10_country.index)
plt.show()
|
# #### The goal of this notebook is use this very small dataset to quickly and simply get a model training using PyTorch.
# [PyTorch: Training a Classifier](https://pytorch.org/tutorials/beginner/blitz/cifar10_tutorial.html)
# [PyTorch: Fine Tuning a Model](https://pytorch.org/tutorials/beginner/finetuning_torchvision_models_tutorial.html)
# ****
# import the relevant libraries
import numpy as np
from pathlib import Path
import random
import torch
import torchvision
from torchvision import transforms
import tqdm
data_dir = Path(
"/kaggle/input/brain-mri-images-for-brain-tumor-detection/brain_tumor_dataset"
)
list(data_dir.iterdir())
# ### Dataset Normalization
# The images in this dataset are MRI scans of brains and are therefore very different to the images in the used in the pretrained classification models from PyTorch, which use the IMAGENET dataset. For this reason I want to calculate the mean and standard deviation of the MRI scan dataset so that it can be normalised properly.
# *Thanks to [Binary Study](https://www.binarystudy.com/2022/04/how-to-normalize-image-dataset-inpytorch.html) for the guidance*
# calulating mean and std of this image dataset
def calculate_dataset_mean(dataloader):
images, labels = next(iter(dataloader))
# shape of images = [batch,channel,width,height]
return images.mean([0, 2, 3])
def calculate_dataset_std(dataloader):
images, labels = next(iter(dataloader))
# shape of images = [batch,channel,width,height]
return images.std([0, 2, 3])
raw_dataset_transforms = transforms.Compose(
[transforms.Resize(255), transforms.CenterCrop(225), transforms.ToTensor()]
)
raw_dataset = torchvision.datasets.ImageFolder(
root=str(data_dir), transform=raw_dataset_transforms
)
raw_dataloader = torch.utils.data.DataLoader(raw_dataset, batch_size=len(raw_dataset))
print(
f"mean = {calculate_dataset_mean(raw_dataloader)} and std = {calculate_dataset_std(raw_dataloader)}"
)
# ### Creating the dataloaders
# * Compose transformations for the training and validation datasets
# * Create datasets with the appropicate transformations applied
# * Create dataloaders using the datasets
# Dataset Configurations
CLASSES = ["no", "yes"]
NUMBER_OF_CLASSES = len(CLASSES)
SHUFFLE = True
VALIDATION_SIZE = 0.2
RESIZE = 64
# Compose image transformations for the training and validation datasets
normalize = transforms.Normalize(
mean=calculate_dataset_mean(raw_dataloader),
std=calculate_dataset_std(raw_dataloader),
)
training_transform = transforms.Compose(
[
transforms.RandomRotation(30),
transforms.Resize(RESIZE),
transforms.CenterCrop(RESIZE),
transforms.RandomHorizontalFlip(),
transforms.ToTensor(),
normalize,
]
)
validation_transform = transforms.Compose(
[
transforms.Resize(RESIZE),
transforms.CenterCrop(RESIZE),
transforms.ToTensor(),
normalize,
]
)
# Create the datasets and apply the relavant transformations.
# At this point both datasets contain all the data from the data directory.
# I will split the data later using a data sampler.
training_dataset = torchvision.datasets.ImageFolder(
root=str(data_dir),
transform=training_transform,
)
validation_dataset = torchvision.datasets.ImageFolder(
root=str(data_dir), transform=validation_transform
)
# Check dataset normalization.
# Mean should be close to 0 and std should be close to 1 after normalization
normalized_dataloader = torch.utils.data.DataLoader(
validation_dataset,
batch_size=len(validation_dataset),
)
print(
f"mean = {calculate_dataset_mean(normalized_dataloader)}",
f"std = {calculate_dataset_std(normalized_dataloader)}",
)
# Use samplers to split the data between the training and validation.
split = int(np.floor(len(training_dataset) * VALIDATION_SIZE))
indices = list(range(len(training_dataset)))
if SHUFFLE:
random.shuffle(indices)
validation_indices, training_indices = indices[:split], indices[split:]
training_sampler = torch.utils.data.sampler.SubsetRandomSampler(training_indices)
validation_sampler = torch.utils.data.sampler.SubsetRandomSampler(validation_indices)
# Dataloader Configurations
BATCH_SIZE = 4
NUMBER_OF_WORKERS = 2
PIN_MEMORY = False
# Create the dataloaders
training_dataloader = torch.utils.data.DataLoader(
training_dataset,
batch_size=BATCH_SIZE,
sampler=training_sampler,
num_workers=NUMBER_OF_WORKERS,
pin_memory=PIN_MEMORY,
)
validation_dataloader = torch.utils.data.DataLoader(
training_dataset,
batch_size=BATCH_SIZE,
sampler=validation_sampler,
num_workers=NUMBER_OF_WORKERS,
pin_memory=PIN_MEMORY,
)
# #### Lets have a look at the data :)
import matplotlib.pyplot as plt
# display images
for images, labels in training_dataloader:
fig = plt.figure(figsize=(14, 7))
for i in range(BATCH_SIZE):
ax = fig.add_subplot(2, 4, i + 1, xticks=[], yticks=[])
ax.set_xlabel(f"cancer = {CLASSES[labels[i]]}")
image = images[i][0, :, :]
plt.imshow(image)
break
# ### Training
# Training Configurations
MODEL_NAME = "resnet18"
WEIGHTS = "DEFAULT"
LEARNING_RATE = 0.0001
MOMENTUM = 0.9
NUMBER_OF_EPOCHS = 10
MODEL_SAVE_PATH = "model.pt"
# Initialize a pretrained model
model = torchvision.models.get_model(MODEL_NAME, weights=WEIGHTS)
model.fc = torch.nn.Linear(512, 2)
# Choose a loss function and an optimazation function
criterion = torch.nn.CrossEntropyLoss()
optimizer = torch.optim.SGD(model.parameters(), lr=LEARNING_RATE, momentum=MOMENTUM)
# #### Now lets train the model
# somewhere to store the training process stats
training_loss_history = []
validation_loss_history = []
validation_accuracy_history = []
# Training and validation loop
for epoch in range(NUMBER_OF_EPOCHS):
model.train()
training_step_loss = []
print(f"Epoch {epoch + 1}/{NUMBER_OF_EPOCHS}")
for data in tqdm.tqdm(training_dataloader, desc="training"):
features, labels = data
# zero the parameter gradients
optimizer.zero_grad()
outputs = model(features)
training_loss = criterion(outputs, labels)
training_loss.backward()
optimizer.step()
training_step_loss.append(training_loss.item())
training_epoch_loss = sum(training_step_loss) / len(training_step_loss)
training_loss_history.append(training_epoch_loss)
model.eval()
validation_step_loss = []
correct_predictions = 0
for data in tqdm.tqdm(validation_dataloader, desc="validating"):
features, labels = data
outputs = model(features)
correct_predictions += torch.sum(torch.argmax(outputs, axis=1) == labels)
validation_loss = criterion(outputs, labels)
validation_step_loss.append(validation_loss.item())
validation_epoch_loss = sum(validation_step_loss) / len(validation_step_loss)
validation_loss_history.append(validation_epoch_loss)
validation_epoch_accuracy = correct_predictions / (
len(validation_dataloader) * BATCH_SIZE
)
print(
f"Training Loss: {training_epoch_loss:.4f},"
f"Validation Loss: {validation_epoch_loss:.4f},"
f"Validation Acc: {validation_epoch_accuracy:.4f}"
)
# save model
if epoch == 0 or validation_epoch_accuracy > max(validation_accuracy_history):
print("Validation loss improved, saving checkpoint.")
torch.save(
{
"epoch": epoch,
"model_state_dict": model.state_dict(),
"optimizer_state_dict": optimizer.state_dict(),
"loss": validation_epoch_loss,
},
MODEL_SAVE_PATH,
)
print("Checkpoint saved")
validation_accuracy_history.append(validation_epoch_accuracy)
print("Finished Training")
# #### Plot the training and validation loss
plt.plot(training_loss_history, label="training_loss")
plt.plot(validation_loss_history, label="validation_loss")
plt.legend()
plt.show
# #### Plot the validation accuracy
plt.plot(validation_accuracy_history, label="validation accuracy")
plt.legend()
plt.show
|
import pandas as pd # 导入csv文件的库
import numpy as np # 进行矩阵运算的库
import matplotlib.pyplot as plt # 作图的库
import torch # 一个深度学习的库Pytorch
import torch.nn as nn # neural network,神经网络
from torch.autograd import Variable # 从自动求导中引入变量
import torch.optim as optim # 一个实现了各种优化算法的库
import torch.nn.functional as F # 神经网络函数库
import os # 与操作系统交互,处理文件和目录、管理进程、获取环境变量
from PIL import Image, ImageOps, ImageFilter, ImageEnhance # PIL是图像处理库
import torchvision.transforms as transforms # 图像、视频、文本增强和预处理的库
import warnings # 避免一些可以忽略的报错
warnings.filterwarnings("ignore") # filterwarnings()方法是用于设置警告过滤器的方法,它可以控制警告信息的输出方式和级别。
def getPhoto(path_photo):
files_list = os.listdir(path_photo)
return files_list
data = []
label = []
# 手的数字是i
for i in range(10):
path = (
"/kaggle/input/sign-language-digits-dataset/Sign-Language-Digits-Datase/train/A"
+ str(i)
+ "/"
)
list = getPhoto(path)
for j in range(len(list)):
full_path = path + list[j]
data.append(full_path)
label.append(i)
data = np.array(data)
label = np.array(label)
num = [i for i in range(len(data))]
np.random.shuffle(num)
data = data[num]
label = label[num]
train_X = np.array(data[: 8 * len(data) // 10])
train_y = np.array(label[: 8 * len(data) // 10])
test_X = np.array(data[8 * len(data) // 10 :])
test_y = np.array(label[8 * len(data) // 10 :])
np.sum(train_y == 0), np.sum(train_y == 1), np.sum(train_y == 2), np.sum(
train_y == 3
), np.sum(train_y == 4), np.sum(train_y == 5), np.sum(train_y == 6), np.sum(
train_y == 7
), np.sum(
train_y == 8
), np.sum(
train_y == 9
),
# 定义正则化的标准,3个颜色通道的均值和方差,这是从ImageNet数据集上得出的数值
normalize = transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225])
# 均值和方差的数值是从ImageNet得到的
transform = transforms.Compose(
[
transforms.RandomRotation(30), # 在[-30,30]的范围内随机旋转一个角度
transforms.RandomResizedCrop(
size=224,
scale=(0.8, 1.2),
interpolation=transforms.functional.InterpolationMode.BILINEAR,
), # 先将图像随机进行缩放操作,然后再将图像变成128*128的图像
transforms.RandomHorizontalFlip(), # 随机水平翻转
transforms.ColorJitter(brightness=0.5, contrast=0.5, saturation=0.5), # 随机颜色变换
transforms.ToTensor(), # 转换为张量
normalize, # 图像的标准化操作
]
)
X_train = []
y_train = []
for epoch in range(4):
for i in range(len(train_X)):
img = Image.open(train_X[i])
img = np.array(transform(img))
X_train.append(img)
y_train.append(train_y[i])
print(epoch)
X_test = []
y_test = []
for epoch in range(4):
for i in range(len(test_X)):
img = Image.open(test_X[i])
img = np.array(transform(img))
X_test.append(img)
y_test.append(test_y[i])
print(epoch)
X_train = np.array(X_train)
y_train = np.array(y_train)
X_test = np.array(X_test)
y_test = np.array(y_test)
y_test
class VGG(nn.Module):
# 初始化
def __init__(self):
# 继承父类的所有方法
super(VGG, self).__init__()
# 构建序列化的神经网络,将网络层按照传入的顺序组合起来
self.model = nn.Sequential()
# 第一层卷积神经网络
# 传入3*224*224
self.model.add_module("conv1_1", nn.Conv2d(3, 64, 3, 1, 1, bias=False))
# 变成64*224*224
self.model.add_module("relu1_1", nn.ReLU())
# self.model.add_module('conv1_2',nn.Conv2d(64,64,3,1,1,bias=False))
# self.model.add_module('relu1_2',nn.ReLU())
# 变成64*224*224
# 最大池化第一层,2*2的池化核,然后步长为2,周围加上0层0
self.model.add_module("maxpool1", nn.MaxPool2d(2, 2, 0))
# 此操作过后会变成64*112*112的图像
self.model.add_module("batch1", nn.BatchNorm2d(64))
# 第二层卷积神经网络
# 传入64*112*112
self.model.add_module("conv2_1", nn.Conv2d(64, 128, 3, 1, 1, bias=False))
# 变成128*112*112
self.model.add_module("relu2_1", nn.ReLU())
# self.model.add_module('conv2_2',nn.Conv2d(128,128,3,1,1,bias=False))
# self.model.add_module('relu2_2',nn.ReLU())
# 变成128*112*112
# 最大池化第二层,2*2的池化核,然后步长为2,周围加上0层0
self.model.add_module("maxpool2", nn.MaxPool2d(2, 2, 0))
# 变成128*56*56
self.model.add_module("batch2", nn.BatchNorm2d(128))
# 第三层卷积神经网络
# 传入128*56*56
self.model.add_module("conv3_1", nn.Conv2d(128, 256, 3, 1, 1, bias=False))
# 变成256*56*56
self.model.add_module("relu3_1", nn.ReLU())
# self.model.add_module('conv3_2',nn.Conv2d(256,256,3,1,1,bias=False))
# self.model.add_module('relu3_2',nn.ReLU())
# self.model.add_module('conv3_3',nn.Conv2d(256,256,3,1,1,bias=False))
# self.model.add_module('relu3_3',nn.ReLU())
# 最大池化第三层,2*2的池化核,然后步长为2,周围加上0层0
self.model.add_module("maxpool3", nn.MaxPool2d(2, 2, 0))
# 变成256*28*28
self.model.add_module("batch3", nn.BatchNorm2d(256))
# 第四层卷积神经网络
# 传入256*28*28
self.model.add_module("conv4_1", nn.Conv2d(256, 512, 3, 1, 1, bias=False))
# 变成512*28*28
self.model.add_module("relu4_1", nn.ReLU())
# self.model.add_module('conv4_2',nn.Conv2d(512,512,3,1,1,bias=False))
# self.model.add_module('relu4_2',nn.ReLU())
# self.model.add_module('conv4_3',nn.Conv2d(512,512,3,1,1,bias=False))
# self.model.add_module('relu4_3',nn.ReLU())
# 最大池化第四层,2*2的池化核,然后步长为2,周围加上0层0
self.model.add_module("maxpool4", nn.MaxPool2d(2, 2, 0))
# 变成512*14*14
self.model.add_module("batch4", nn.BatchNorm2d(512))
# 第五层卷积神经网络
# 传入512*14*14
self.model.add_module("conv5_1", nn.Conv2d(512, 512, 3, 1, 1, bias=False))
# 变成512*14*14
self.model.add_module("relu5_1", nn.ReLU())
# self.model.add_module('conv5_2',nn.Conv2d(512,512,3,1,1,bias=False))
# self.model.add_module('relu5_2',nn.ReLU())
# self.model.add_module('conv5_3',nn.Conv2d(512,512,3,1,1,bias=False))
# self.model.add_module('relu5_3',nn.ReLU())
# 最大池化第四层,2*2的池化核,然后步长为2,周围加上0层0
self.model.add_module("maxpool5", nn.MaxPool2d(2, 2, 0))
# 变成512*7*7
self.model.add_module("batch5", nn.BatchNorm2d(512))
# 全连接网络层(这里没有照搬VGG)
self.model.add_module("linear1", nn.Linear(512 * 7 * 7, 4096))
self.model.add_module("relu6_1", nn.ReLU())
self.model.add_module("dropout1", nn.Dropout(0.5))
self.model.add_module("linear2", nn.Linear(4096, 1000))
self.model.add_module("relu6_2", nn.ReLU())
self.model.add_module("dropout2", nn.Dropout(0.5))
self.model.add_module("linear3", nn.Linear(1000, 10))
# 前向传播
def forward(self, input):
# 传入数据
output = input
# 按照神经网络的顺序去跑一遍
for name, module in self.model.named_children():
# 如果这一层是全连接层,那就先展平,得到一维的32*16的向量
if name == "linear1":
output = output.view(-1, 512 * 7 * 7)
output = module(output)
return F.softmax(output, dim=1)
def weight_init(m):
# 获取对象所属的类的名称
class_name = m.__class__.__name__
# 当对象的name中出现"conv",也就是卷积操作
if class_name.find("conv") != -1:
# 对卷积核按照正态分布的均值和标准差随机初始化
m.weight.data.normal_(0, 0.02)
# 初始化神经网络
netC = VGG()
netC.apply(weight_init)
print(netC)
# 优化器
optimizer = optim.Adam(netC.parameters(), lr=0.0002, betas=(0.5, 0.999))
# 损失函数
criterion = nn.NLLLoss() # 负对数似然损失函数,也是交叉熵损失函数的一种
# 训练周期为5次
num_epochs = 5
for epoch in range(num_epochs):
num = [i for i in range(len(X_train))]
np.random.shuffle(num)
X_train = X_train[num]
y_train = y_train[num]
for i in range(10):
image = []
label = []
for j in range(300):
image.append(X_train[500 * i + j].reshape((3, 224, 224)))
label.append(y_train[500 * i + j])
# 将数据转换成可以处理的张量格式
image = torch.Tensor(image) # .to(device)
label = torch.Tensor(label).long() # .to(device)
# 训练
netC.train()
# 将数据放进去训练
output = netC(image) # .to(device)
# 计算每次的损失函数
error = criterion(output, label) # .to(device)
# 反向传播
error.backward()
print(i)
# 优化器进行优化(梯度下降,降低误差)
optimizer.step()
# 将梯度清空
optimizer.zero_grad()
print(epoch, error)
pred_y = []
for i in range(len(X_train)):
pred = netC(torch.Tensor(X_train[i].reshape((1, 3, 224, 224))))[0] # .to(device)
pred = np.argmax(pred.detach().cpu().numpy())
pred_y.append(pred)
if i % 500 == 0:
print(i)
print("训练集的准确率:", np.sum(pred_y == y_train) / len(y_train), "训练集总数为:", len(y_train))
pred_y = []
for i in range(len(X_test)):
pred = netC(torch.Tensor(X_test[i].reshape((1, 3, 224, 224))))[0] # .to(device)
pred = np.argmax(pred.detach().cpu().numpy())
pred_y.append(pred)
if i % 500 == 0:
print(i)
print("测试集的准确率:", np.sum(pred_y == y_test) / len(y_test), "测试集总数为:", len(y_test))
torch.save(netC, "sign_language.pth")
|
# Tyler VanderMate
# Assignment 1
# TODO
# 1. You must load the data from the provided CSV files. X
# 2. You must check for missing values within the training data. X
# 3. If the training data contains missing values, you must describe and implement an approach to handle those missing values. X
# 4. You must check for outliers within the training data.
# 5. If the training data contains outliers, you must describe and implement an approach to handle those outliers.
# 6. You must determine whether or not you will implement normalization or standardization, and explain your decision. X
# 7. You must build and train a decision tree model on the training data. X
# 8. You must report the best ROC AUC score, F1 score, and accuracy score that you were able to obtain for your decision tree model. X
# 9. You must build and train a random forest model on the training data. X
# 10. You must report the best ROC AUC score, F1 score, and accuracy score that you were able to obtain for your random forest model. X
# 11. You must select the best model that you are able to generate and use that model to predict the target vector for the test data. X
# 12. Your notebook must be saved with the output enabled so that we can see the results of each cell after it has been run.
# Failure to adhere to this criterion will result in a 0 for this portion of your score. X
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import seaborn as sns # plotting
from scipy import stats
from sklearn import datasets # Basically ripped off of Datacamp
from sklearn.model_selection import train_test_split
from sklearn.tree import DecisionTreeClassifier
from sklearn.metrics import accuracy_score
from sklearn.metrics import f1_score # From Dr. H in Discord
from sklearn.metrics import roc_auc_score
from sklearn.ensemble import RandomForestClassifier as rfc
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Uncomment for help
# rfc
# ?roc_auc_score
# ?f1_score
# accuracy_score
# ?DecisionTreeClassifier
# ?train_test_split
# ?datasets
# ?sns
# ?pd
# ?np
# 1. You must load the data from the provided CSV files
train_set = pd.read_csv("../input/cap-4611-spring-21-assignment-1/train.csv")
test_set = pd.read_csv("../input/cap-4611-spring-21-assignment-1/test.csv")
train_set.drop("id", axis=1, inplace=True)
test_set.drop("id", axis=1, inplace=True)
y_train = train_set["Bankrupt"]
X_train = train_set.drop("Bankrupt", axis=1)
print(y_train.shape)
print(X_train.shape)
train_set.shape
# 2. You must check for missing values within the training data.
train_set.info()
test_set.info()
# 3. If the training data contains missing values, you must describe and implement an approach to handle those missing values.
# Looks like we aren't missing any values in the entire dataset since there are no non-null values, so we won't have to do 3 here.
# There are also no duplicate values.
print("There are missing values: " + str(X_train.isna().sum().any()))
print("Total duplicate values: " + str(X_train.duplicated().sum()))
# Look at ALL the features with eyeballs
with pd.option_context("display.max_columns", None):
display(train_set.describe())
# 4. You must check for outliers within the training data.
# 5. If the training data contains outliers, you must describe and implement an approach to handle those outliers.
# If any values for and feature fall beyond 3 standard deviations, we can *think* about removing them.
# scipy.stats.zscore : Compute the z score of each value in the sample, relative to the sample mean and standard deviation.
# Returns
# zscorearray_like
# The z-scores, standardized by mean and standard deviation of input array a.
for column in X_train:
print(column)
print(np.where(stats.zscore(X_train.loc[:, column]) > 3))
# It looks like we have a few outliers within the dataset, however, this is expected with a large dataset.
# Since this is in a business context, larger more established businesses who own a huge share of their respective market are expected to be within the top 99.7%. Since .03% of 3409 = 10.227, we should expect about 10 companies to stand out within each feature. This is *roughly* true in our dataset.
# Therefore, we shouldn't remove outliers yet if at all.
# 6. You must determine whether or not you will implement normalization or standardization, and explain your decision.
# Looks like not all the values are normalized; however, because we are using decision trees and random forest models, scaling our features down to a fixed range will not have much of an affect on our final model. Therefore, in this case, there is no need to normalize the data.
#
print("Normalized feature mean example:" + str(train_set.describe().iloc[1, 1]))
print()
print("Non-normalized feature means:")
print("feature 11: " + str(train_set.describe().iloc[1, 11]))
print("feature 12: " + str(train_set.describe().iloc[1, 12]))
print("feature 14: " + str(train_set.describe().iloc[1, 14]))
print("feature 21: " + str(train_set.describe().iloc[1, 21]))
print("feature 29: " + str(train_set.describe().iloc[1, 29]))
# all features samples
with pd.option_context("display.max_rows", 11, "display.max_columns", None):
display(train_set)
sns.countplot(data=train_set, x="Bankrupt")
sns.countplot(
data=train_set, x="one if total liabilities exceeds total assets zero otherwise"
)
sns.countplot(
data=train_set,
x="one if net income was negative for the last two year zero otherwise",
)
print(X_train.keys())
# It looks like some of the keys have spaces in them; that's annoying.
# 7. You must build and train a decision tree model on the training data.
dt = DecisionTreeClassifier(max_depth=6, random_state=1)
dt.fit(X_train, y_train)
y_pred = dt.predict_proba(X_train)
tree_output = pd.DataFrame({"Bankrupt": y_pred[:, 1]})
tree_output.to_csv("tree_pred.csv", index=True, index_label="Id")
# 8. You must report the best ROC AUC score, F1 score, and accuracy score that you were able to obtain for your decision tree model.
# **ROC, AUC, abd f1 scores for the Decision Tree**
decision_tree_roc_auc = roc_auc_score(y_train, dt.predict(X_train))
decision_tree_f1 = f1_score(y_train, dt.predict(X_train))
decision_tree_accuracy = accuracy_score(y_train, dt.predict(X_train))
print("ROC_AUC: " + str(decision_scores))
print("f1: " + str(decision_tree_f1))
print("accuracy: " + str(decision_tree_accuracy))
# 9. You must build and train a random forest model on the training data.
# After doing a few of these, it looks like 6 is the magic number of the depth with a lucky roll of 69 as the seed
# The random forest model defaults to using the gini impurity because it's faster, however to get a more accurate model in this case, we should instead branch based on information gain using entropy. Also, we get a slightly better score when we use entropy when predicting about the testing data on Kaggle.
model = rfc(max_depth=6, criterion="entropy", random_state=69)
model.fit(X_train, y_train)
y_pred = model.predict_proba(test_set)
forest_output = pd.DataFrame({"Bankrupt": y_pred[:, 1]})
forest_output.to_csv("forest_entropy_69_6.csv", index=True, index_label="Id")
# 10. You must report the best ROC AUC score, F1 score, and accuracy score that you were able to obtain for your random forest model.
random_forest_roc_auc = roc_auc_score(y_train, model.predict(X_train))
random_forest_f1 = f1_score(y_train, model.predict(X_train))
random_forest_accuracy = accuracy_score(y_train, model.predict(X_train))
print("ROC_AUC: " + str(random_forest_roc_auc))
print("f1: " + str(random_forest_f1))
print("accuracy: " + str(random_forest_accuracy))
|
import tensorflow as tf
from tensorflow.keras.preprocessing.image import ImageDataGenerator
import numpy as np
import pandas as pd
from PIL import Image
import matplotlib.pyplot as plt
import cv2
import os
from tqdm import tqdm
from tensorflow.keras.models import load_model
model = load_model("../input/unetresnet34trainedmodel/unet-resnet34.h5", compile=False)
# https://www.kaggle.com/titericz/building-and-visualizing-masks
# https://www.kaggle.com/paulorzp/rle-functions-run-lenght-encode-decode
# defining function for converting EncodedPixels(rle: run length encoding) to mask
def rle2mask(rle_string, img_shape=(256, 1600)):
"""
input: EncodedPixels (run-length-encoded) string & image shape:-(width,height)
output: mask in numpy.ndarray format with shape (256,1600)
"""
rle_array = np.array([int(s) for s in rle_string.split()])
starts_array = rle_array[::2] - 1
lengths_array = rle_array[1::2]
mask_array = np.zeros(img_shape[0] * img_shape[1], dtype=np.uint8)
# print(starts_array,lengths_array)
for i in range(len(starts_array)):
mask_array[starts_array[i] : starts_array[i] + lengths_array[i]] = 1
# order='F' because encoded pixels are numbered from top to bottom, then left to right
return mask_array.reshape(img_shape, order="F")
# defining function for converting given mask to EncodedPixels(rle: run length encoding)
def mask2rle(mask_array):
"""
input: mask in numpy.ndarray format
output: EncodedPixels (run-length-encoded) string
"""
mask_array = mask_array.T.flatten()
mask_array = np.concatenate([[0], mask_array, [0]])
rle_array = np.where(mask_array[1:] != mask_array[:-1])[0] + 1
rle_array[1::2] -= rle_array[::2]
rle_string = " ".join(map(str, rle_array))
return rle_string
# defining function for calculation of metric dice coefficient
def dice_coefficient(y_true, y_pred):
y_true_f = tf.reshape(y_true, [-1])
y_pred_f = tf.reshape(y_pred, [-1])
intersection = tf.math.reduce_sum(y_true_f * y_pred_f)
smoothing_const = 1e-9
return (2.0 * intersection + smoothing_const) / (
tf.math.reduce_sum(y_true_f) + tf.math.reduce_sum(y_pred_f) + smoothing_const
)
class PredictDataGenerator(tf.keras.utils.Sequence):
def __init__(
self,
dataframe,
list_idcs,
batch_size=32,
):
self.batch_size = batch_size
self.df = dataframe
self.list_idcs = list_idcs
self.indices = self.df.index.tolist()
self.rem = len(self.list_idcs) % (self.batch_size)
self.on_epoch_end()
def __len__(self):
return len(self.list_idcs) // (self.batch_size)
# if (self.rem) == 0:
# return len(self.list_idcs) // (self.batch_size)
# else:
# return (len(self.list_idcs) // (self.batch_size) )+1
def __getitem__(self, index):
index = self.indices[index * self.batch_size : (index + 1) * self.batch_size]
# if ((index + 1) * self.batch_size) < len(self.list_idcs):
# index = self.indices[index * self.batch_size:(index + 1) * self.batch_size]
# else:
# index = self.indices[index * self.batch_size: (index * self.batch_size)+ self.rem]
batch = [self.list_idcs[k] for k in index]
X = self.__get_data(batch)
return X
def on_epoch_end(self):
self.index = np.arange(len(self.indices))
def __get_data(self, batch):
X = np.empty(
(self.batch_size, 256, 1600, 3), dtype=np.float32
) # image place-holders
for i, id in enumerate(batch):
img = Image.open(
"../input/severstal-steel-defect-detection/test_images/"
+ str(self.df["ImageId"].loc[id])
)
X[i,] = img # input image
return X
test_img_IDs = list(os.listdir("../input/severstal-steel-defect-detection/test_images"))
test_imgsIds_df = pd.DataFrame({"ImageId": test_img_IDs})
print(len(test_imgsIds_df))
test_imgsIds_df.head()
SubmissionDf = pd.DataFrame(columns=["ImageId", "EncodedPixels", "ClassId"])
for i in range(0, len(test_imgsIds_df), 320):
batch_idcs = list(
range(i, min(test_imgsIds_df.shape[0], i + 320))
) # .iloc[batch_idcs]
if len(batch_idcs) == 320:
test_subbatch = PredictDataGenerator(
dataframe=test_imgsIds_df, list_idcs=batch_idcs
)
else:
test_subbatch = PredictDataGenerator(
dataframe=test_imgsIds_df, list_idcs=batch_idcs, batch_size=len(batch_idcs)
)
# print(len(test_subbatch))
subbatch_pred_masks = model.predict(test_subbatch)
# print(len(subbatch_pred_masks))
# break
for j, idx in tqdm(enumerate(batch_idcs)):
filename = test_imgsIds_df["ImageId"].iloc[idx]
rle1 = mask2rle(subbatch_pred_masks[j, :, :, 0].round().astype(int))
rle2 = mask2rle(subbatch_pred_masks[j, :, :, 1].round().astype(int))
rle3 = mask2rle(subbatch_pred_masks[j, :, :, 2].round().astype(int))
rle4 = mask2rle(subbatch_pred_masks[j, :, :, 3].round().astype(int))
df = pd.DataFrame(
{
"ImageId": [filename] * 4,
"EncodedPixels": [rle1, rle2, rle3, rle4],
"ClassId": ["1", "2", "3", "4"],
}
)
SubmissionDf = SubmissionDf.append(df, ignore_index=True)
# print(SubmissionDf.head())
# print(SubmissionDf.shape)
# break
# break
SubmissionDf.sort_values(by=["ImageId", "ClassId"], inplace=True)
print(SubmissionDf.shape)
SubmissionDf.head(10)
SubmissionDf["ImageId_ClassId"] = (
SubmissionDf["ImageId"] + "_" + SubmissionDf["ClassId"]
)
SubmissionDf
SubmissionDf[["ImageId_ClassId", "EncodedPixels"]].to_csv("submission.csv", index=False)
|
import pandas as pd
import numpy as np
df = pd.read_csv("/kaggle/input/titanic/train.csv")
df.keys()
num = [x for x in df.columns if df.dtypes[x] in ("float", "int64")]
cat = [x for x in df.columns if df.dtypes[x] == "object"]
target = "Survived"
num.remove(target)
from sklearn.model_selection import train_test_split
train, test = train_test_split(df, test_size=0.2)
# # Explore
train.corr()[target].sort_values(ascending=False)
import seaborn as sns
cor = train.corr()
np.fill_diagonal(cor.values, 0)
sns.heatmap(cor, annot=True, cmap="RdBu", center=0)
# # Clean
train.isnull().any()
# # Pipeline
from sklearn.base import BaseEstimator, TransformerMixin
class DataSelect(BaseEstimator, TransformerMixin):
def __init__(self, attributes):
self.attributes = attributes
def fit(self, X, y=None):
return self
def transform(self, X):
return X[self.attributes]
from sklearn.pipeline import Pipeline
from sklearn.impute import SimpleImputer
num_pip = Pipeline(
[("dataselect", DataSelect(num)), ("impute", SimpleImputer(strategy="median"))]
)
from sklearn.preprocessing import OneHotEncoder
cat_pip = Pipeline([("dataselect", DataSelect(["Sex"])), ("enc", OneHotEncoder())])
from sklearn.pipeline import FeatureUnion
pipe = FeatureUnion([("numeric", num_pip), ("categorical", cat_pip)])
# # Machine Models
pipe.fit(train)
train_pre = pipe.transform(train).toarray()
test_pre = pipe.transform(test).toarray()
from sklearn.naive_bayes import GaussianNB
m_gaus = GaussianNB()
m_gaus.fit(train_pre, train[target])
# # Cross Validation
from sklearn.model_selection import cross_val_score
cross_val_score(m_gaus, train_pre, train[target], cv=3).mean()
cross_val_score(m_gaus, test_pre, test[target], cv=3).mean()
# # Submission
sample = pd.read_csv("/kaggle/input/titanic/test.csv")
sample_pre = pipe.transform(sample).toarray()
submission = pd.DataFrame(
{"PassengerId": sample["PassengerId"], "Survived": m_gaus.predict(sample_pre)}
)
submission.to_csv("submission.csv", index=False)
|
import pandas as pd
import numpy as np
from random import gauss
from pandas.plotting import autocorrelation_plot
import warnings
import itertools
from random import random
import statsmodels.formula.api as smf
import statsmodels.api as sm
from statsmodels.graphics.tsaplots import plot_acf, plot_pacf
from statsmodels.tsa.ar_model import AR
from statsmodels.tsa.seasonal import seasonal_decompose
from statsmodels.tsa.stattools import adfuller
import matplotlib.pyplot as plt
plt.style.use("fivethirtyeight")
# import matplotlib as mpl
import seaborn as sns
# general settings
class CFG:
data_folder = "../input/tsdata-1/"
img_dim1 = 20
img_dim2 = 10
# adjust the parameters for displayed figures
plt.rcParams.update({"figure.figsize": (CFG.img_dim1, CFG.img_dim2)})
#
# List of Contents
#
# * [Groundwork](#section-one)
# * [Patterns](#section-two)
# * [Dependence](#section-three)
# * [Stationarity](#section-four)
# [Table of Contents](#0.1)
# # **1. Introduction to Time-Series Analysis**
# - A **time-series** data is a series of data points or observations recorded at different or regular time intervals. In general, a time series is a sequence of data points taken at equally spaced time intervals. The frequency of recorded data points may be hourly, daily, weekly, monthly, quarterly or annually.
# - **Time-Series Forecasting** is the process of using a statistical model to predict future values of a time-series based on past results.
# - A time series analysis encompasses statistical methods for analyzing time series data. These methods enable us to extract meaningful statistics, patterns and other characteristics of the data. Time series are visualized with the help of line charts. So, time series analysis involves understanding inherent aspects of the time series data so that we can create meaningful and accurate forecasts.
# - Applications of time series are used in statistics, finance or business applications. A very common example of time series data is the daily closing value of the stock index like NASDAQ or Dow Jones. Other common applications of time series are sales and demand forecasting, weather forecasting, econometrics, signal processing, pattern recognition and earthquake prediction.
# ### **Components of a Time-Series**
# - **Trend** - The trend shows a general direction of the time series data over a long period of time. A trend can be increasing(upward), decreasing(downward), or horizontal(stationary).
# - **Seasonality** - The seasonality component exhibits a trend that repeats with respect to timing, direction, and magnitude. Some examples include an increase in water consumption in summer due to hot weather conditions.
# - **Cyclical Component** - These are the trends with no set repetition over a particular period of time. A cycle refers to the period of ups and downs, booms and slums of a time series, mostly observed in business cycles. These cycles do not exhibit a seasonal variation but generally occur over a time period of 3 to 12 years depending on the nature of the time series.
# - **Irregular Variation** - These are the fluctuations in the time series data which become evident when trend and cyclical variations are removed. These variations are unpredictable, erratic, and may or may not be random.
# - **ETS Decomposition** - ETS Decomposition is used to separate different components of a time series. The term ETS stands for Error, Trend and Seasonality.
# Groundwork
# [Table of Contents](#0.1)
# Time series is any sequence you record over time and applications are everywhere.
# More formally, time series data is a sequence of data points (or observations) recorded at different time intervals - those intervals are frequently, but not always, regular (hourly, daily, weekly, monthly, quarterly etc):
# \begin{equation}
# \{X_t\} \quad t= 1,\ldots,T
# \end{equation}
# A strict formulation would be that a time series (discrete) realization of a (continuous) stochastic process generating the data and the underlying reason why we can infer from the former about the latter is the Kolmogorov extension theorem. The proper mathematical treatment of this theory is way beyond the scope of this notebook, so a mathematically inclinded reader is advised to look up those terms and then follow the references.
# Phenomena measured over time are everywhere, so a natural question is: what can we do with time series? Some of the more popular applications / reasons to bother are:
# * interpretation: we want to be able to make sense of diverse phenomena and capture the nature of the underlying dynamics
# * modelling: understanding inherent aspects of the time series data so that we can create meaningful and accurate forecasts.
# * **forecasting / prediction**: we want to know something about the future :-)
# * filtering / smoothing: we want to get a better understanding of the process based on partially / fully observed sample
# * simulation: in certain applications calculating e.g. high quantiles of a distribution is only possible with simulation, because there is not enough historical data
# # **2. Types of data**
# [Table of Contents](#0.1)
# As stated above, the time series analysis is the statistical analysis of the time series data. A time series data means that data is recorded at different time periods or intervals. The time series data may be of three types:-
# 1 **Time series data** - The observations of the values of a variable recorded at different points in time is called time series data.
# 2 **Cross sectional data** - It is the data of one or more variables recorded at the same point in time.
# 3 **Pooled data**- It is the combination of time series data and cross sectional data.
# # **3. Time Series terminology**
# [Table of Contents](#0.1)
# There are various terms and concepts in time series that we should know. These are as follows:-
# 1 **Dependence**- It refers to the association of two observations of the same variable at prior time periods.
# 2 **Stationarity**- It shows the mean value of the series that remains constant over the time period. If past effects accumulate and the values increase towards infinity then stationarity is not met.
# 3 **Differencing**- Differencing is used to make the series stationary and to control the auto-correlations. There may be some cases in time series analyses where we do not require differencing and over-differenced series can produce wrong estimates.
# 4 **Specification** - It may involve the testing of the linear or non-linear relationships of dependent variables by using time series models such as ARIMA models.
# 5 **Exponential Smoothing** - Exponential smoothing in time series analysis predicts the one next period value based on the past and current value. It involves averaging of data such that the non-systematic components of each individual case or observation cancel out each other. The exponential smoothing method is used to predict the short term prediction.
# 6 **Curve fitting** - Curve fitting regression in time series analysis is used when data is in a non-linear relationship.
# 7 **ARIMA** - ARIMA stands for Auto Regressive Integrated Moving Average.
# Patterns
# The first we can do to identify patterns in a time series is separate it into components with easily understandable characteristics:
# \begin{equation}
# X_t = T_t + S_t + C_t + I_t \quad
# \end{equation}
# where:
# * $T_t$: the trend shows a general direction of the time series data over a long period of time. It represents a long-term progression of the series (secular variation)
# * $S_t$: the seasonal component with fixed and known period. It is observed when there is a distinct repeated pattern observed between regular intervals due to seasonal factors: annual, monthly or weekly. Obvious examples include daily power consumption patterns or annual sales of seasonal goods.
# * $C_t$: (optional) cyclical component is a repetitive pattern which does not occur at fixed intervals - usually observed in an economic context like business cycles.
# * $I_t$: the irregular component (residuals ) consists of the fluctuations in the time series that are observed after removing trend and seasonal / cyclical variations.
# It is worth pointing out that an alternative to using a multiplicative decomposition is to first transform the data until the variation in the series appears to be stable over time, then use an additive decomposition. When a log transformation has been used, this is equivalent to using a multiplicative decomposition because
# \begin{equation}
# X_t = T_t * S_t * I_t
# \end{equation}
# is equivalent to
# \begin{equation}
# log X_t = log T_t + log S_t + log I_t
# \end{equation}
# A popular implementation for calculating the fundamental decomposition can be used via the statsmodels package:
# Airline Passenger data - between 1949 and 1960, first compiled by Box and Jenkins. We will use this dataset to demonstrate in practice what kind of information can be obtained using seasonal decomposition.
#
air_df = pd.read_csv(CFG.data_folder + "passengers.csv")
def plot_df(df, x, y, title="", xlabel="Date", ylabel="Number of Passengers", dpi=100):
plt.figure(figsize=(15, 4), dpi=dpi)
plt.plot(x, y, color="tab:red")
plt.gca().set(title=title, xlabel=xlabel, ylabel=ylabel)
plt.show()
air_df["date"] = pd.to_datetime(air_df["date"])
air_df.columns = ["Date", "Number of Passengers"]
plot_df(
air_df,
x=air_df["Date"],
y=air_df["Number of Passengers"],
title="Number of US Airline passengers from 1949 to 1960",
)
#
# [Table of Contents](#0.1)
# - Any time series visualization may consist of the following components: **Base Level + Trend + Seasonality + Error**.
# ### **Trend**
# - A **trend** is observed when there is an increasing or decreasing slope observed in the time series.
# ### **Seasonality**
# - A **seasonality** is observed when there is a distinct repeated pattern observed between regular intervals due to seasonal factors. It could be because of the month of the year, the day of the month, weekdays or even time of the day.
# However, It is not mandatory that all time series must have a trend and/or seasonality. A time series may not have a distinct trend but have a seasonality and vice-versa.
# ### **Cyclic behaviour**
# - Another important thing to consider is the **cyclic behaviour**. It happens when the rise and fall pattern in the series does not happen in fixed calendar-based intervals. We should not confuse 'cyclic' effect with 'seasonal' effect.
# - If the patterns are not of fixed calendar based frequencies, then it is cyclic. Because, unlike the seasonality, cyclic effects are typically influenced by the business and other socio-economic factors.
from statsmodels.tsa.seasonal import seasonal_decompose
from dateutil.parser import parse
# Multiplicative Decomposition
multiplicative_decomposition = seasonal_decompose(
air_df["Number of Passengers"], model="multiplicative", period=12
)
# Additive Decomposition
additive_decomposition = seasonal_decompose(
air_df["Number of Passengers"], model="additive", period=12
)
# Plot
plt.rcParams.update({"figure.figsize": (16, 12)})
additive_decomposition.plot().suptitle("Additive Decomposition", fontsize=16)
plt.tight_layout(rect=[0, 0.03, 1, 0.95])
plt.show()
#
#
# If we look at the residuals of the additive decomposition closely, it has some pattern left over
multiplicative_decomposition.plot().suptitle(
"Multiplicative Decomposition", fontsize=16
)
plt.tight_layout(rect=[0, 0.03, 1, 0.95])
plt.show()
#
#
# Not much of a qualitative change in trend and seasonality components, but the residuals looks much more stable around a constant level - such phenomenon does not of course imply stationarity by itself, but at least a clear signal in the opposite direction is not there anymore.
# # **7. Additive and Multiplicative Time Series**
# [Table of Contents](#0.1)
# - We may have different combinations of trends and seasonality. Depending on the nature of the trends and seasonality, a time series can be modeled as an additive or multiplicative time series. Each observation in the series can be expressed as either a sum or a product of the components.
# ### **Additive time series:**
# Value = Base Level + Trend + Seasonality + Error
# ### **Multiplicative Time Series:**
# Value = Base Level x Trend x Seasonality x Error
# # **9. Stationary and Non-Stationary Time Series**
# [Table of Contents](#0.1)
# - Now, we wil discuss **Stationary and Non-Stationary Time Series**. **Stationarity** is a property of a time series. A stationary series is one where the values of the series is not a function of time. So, the values are independent of time.
# - Hence the statistical properties of the series like mean, variance and autocorrelation are constant over time. Autocorrelation of the series is nothing but the correlation of the series with its previous values.
# - A stationary time series is independent of seasonal effects as well.
# - Now, we will plot some examples of stationary and non-stationary time series for clarity.
# We can covert any non-stationary time series into a stationary one by applying a suitable transformation.
# Mostly statistical forecasting methods are designed to work on a stationary time series.
# The first step in the forecasting process is typically to do some transformation to convert a non-stationary series to stationary.
# # **10. How to make a time series stationary?**
# [Table of Contents](#0.1)
# - We can apply some sort of transformation to make the time-series stationary. These transformation may include:
# 1. Differencing the Series (once or more)
# 2. Take the log of the series
# 3. Take the nth root of the series
# 4. Combination of the above
# - The most commonly used and convenient method to stationarize the series is by differencing the series at least once until it becomes approximately stationary.
# ## **Wait, But what's differencing???**
# [Table of Contents](#0.1)
# - If Y_t is the value at time t, then the first difference of Y = Yt – Yt-1.
# - In simpler terms, differencing the series is nothing but subtracting the next value by the current value.
# - If the first difference doesn’t make a series stationary, we can go for the second differencing and so on.
# - For example, consider the following series: [1, 5, 2, 12, 20]
# - First differencing gives: [5-1, 2-5, 12-2, 20-12] = [4, -3, 10, 8]
# - Second differencing gives: [-3-4, -10-3, 8-10] = [-7, -13, -2]
# # **11. How to test for stationarity?**
# [Table of Contents](#0.1)
# - The stationarity of a series can be checked by looking at the plot of the series.
# - Another method is to split the series into 2 or more contiguous parts and computing the summary statistics like the mean, variance and the autocorrelation. If the stats are quite different, then the series is not likely to be stationary.
# - There are several quantitative methods we can use to determine if a given series is stationary or not. This can be done using statistical tests called [Unit Root Tests](https://en.wikipedia.org/wiki/Unit_root). This test checks if a time series is non-stationary and possess a unit root.
# - There are multiple implementations of Unit Root tests like:
# **1. Augmented Dickey Fuller test (ADF Test)**
# **2. Kwiatkowski-Phillips-Schmidt-Shin – KPSS test (trend stationary)**
# **3. Philips Perron test (PP Test)**
#
X = air_df["Number of Passengers"].values
split = int(len(X) / 2)
X1, X2 = X[0:split], X[split:]
mean1, mean2 = X1.mean(), X2.mean()
var1, var2 = X1.var(), X2.var()
print("\n\n")
print("Mean:")
print("Chunk1: %.2f vs Chunk2: %.2f" % (mean1, mean2))
print("\n\n")
print("Variance:")
print("Chunk1: %.2f vs Chunk2: %.2f" % (var1, var2))
#
# The values are clearly very different across the two data subsets, which strongly suggests non-stationarity.
#
# However, visual inspection is not what one would could a rigorous criterion - so let's define things in a formal manner.
# ADF test is a unit root test. It determines how strongly a time series is defined by a trend.
# - Null Hypothesis (H0): Null hypothesis of the test is that the time series can be represented by a unit root that is not stationary.
# - Alternative Hypothesis (H1): Alternative Hypothesis of the test is that the time series is stationary.
# Interpretation of p value:
# - above $\alpha$: Accepts the Null Hypothesis (H0), the data has a unit root and is non-stationary.
# - below $\alpha$ : Rejects the Null Hypothesis (H0), the data is stationary.
#
decomposition = seasonal_decompose(X, model="additive", period=12)
decomposition.plot().suptitle("Multiplicative Decomposition", fontsize=16)
plt.tight_layout(rect=[0, 0.03, 1, 0.95])
plt.show()
result = adfuller(X)
print("ADF Statistic: %f" % result[0])
print("p-value: %f" % result[1])
print("Critical Values:")
for key, value in result[4].items():
print("\t%s: %.3f" % (key, value))
# Skip the start of the series: adfuller does not handle missing values which appear for values within the first full period
result = adfuller(decomposition.trend[10:-10])
print("ADF Statistic: %f" % result[0])
print("p-value: %f" % result[1])
print("\n")
print("p-value is not less than 0.05, so its non-stationary")
result = adfuller(decomposition.seasonal[10:-10])
print("ADF Statistic: %f" % result[0])
print("p-value: %f" % result[1])
print("\n")
print("p-value is less than 0.05")
print("Passed ✅")
result = adfuller(decomposition.resid[10:-10])
print("ADF Statistic: %f" % result[0])
print("p-value: %f" % result[1])
print("\n")
print("p-value is less than 0.05")
print("Passed ✅")
air_df["passengers2"] = np.log(air_df["Number of Passengers"])
air_df.passengers2.plot()
plt.show()
#
# Applying a logarithm does not remove the trend, but it does seem to stabilize the amplitude (periodic variations have comparable magnitude now). How does that translate into ADF results?
#
result = adfuller(air_df.passengers2)
print("p-value: %f" % result[1])
print("\n")
print("Nope 😅, p-value is still not less than 0.05, so its non-stationary")
print("\n")
print("But on the bright side, the p-value has dropped from 0.994532 to 0.422367 🔥")
air_df["passengers3"] = air_df["Number of Passengers"].diff()
air_df.passengers3.plot()
plt.show()
#
# As expected, differentiation removes the trend (oscillations happen around a fixed level), but variations amplitude is magnified.
result = adfuller(air_df.passengers3[10:])
print("ADF Statistic: %f" % result[0])
print("p-value: %f" % result[1])
print("\n")
print("Nope 😅, p-value is still not less than 0.05, so its non-stationary")
print("\n")
print("But on the bright side, the p-value has dropped from 0.422367 to 0.054094🔥")
#
# We continue moving in the right direction - what happens if we combine the two transformations?
#
air_df["passengers4"] = air_df["Number of Passengers"].apply(np.log).diff()
air_df.passengers4.plot()
plt.show()
result = adfuller(air_df.passengers4[10:])
print("ADF Statistic: %f" % result[0])
print("p-value: %f" % result[1])
print("\nOMG Finally 🙏🏻, p-value is less than 0.05, Now we are stationary")
#
# So after applying logarithm (to stabilize the variance) and differentiation (to remove the trend), we have transformed our series to one that can be plausibly treated as stationary. We can verify that intuition by examining ACF and PACF
#
plot_acf(air_df["passengers4"][10:], lags=10)
plt.show()
plot_pacf(air_df["passengers4"][10:], lags=10)
plt.show()
# As mentioned above, exponential moving average (EMA) assigns exponentially decreasing weights over time. The functions as a low-pass filter that removes high-frequency noise (and can be formulated as a special case of a more general problem of recursive filtering). Exponential smoothing models discussed in this module can be formulated as a special case of a general framework of state space models - those are discussed in part 4 of this series (there will be a link to the notebook once I've written it ;-)
# A complete taxonomy of ETS (error, trend, seasonality) models can be found in Hyndman et al "Forecasting with Exponential Smoothing: The State Space Approach" - below we discuss the three most popular models from that class.
# # Popular methods
# ## EWMA
# Simple Exponential Smoothing (Brown method) is defined by the relationship:
# \begin{equation}
# S_t = \alpha X_t + (1-\alpha) S_{t-1} \quad \text{where} \quad \alpha \in (0,1)
# \end{equation}
# or equivalently:
# \begin{equation}
# S_t = S_{t-1} + \alpha (X_t - S_{t-1})
# \end{equation}
# Few observations around that definition:
# * the smoothed series is a simple weighted average of the past and the present
# * interpretation of smoothing factor $\alpha$: recency vs smoothing (see below). It defines how quickly we will "forget" the last available true observation.
# * $\alpha$ is selected on the basis of expert judgement or estimated (with MSE); statsmodels does the estimation by default
# * by its very nature, smoothing needs some time to catch up with the dynamics of your time series. A rule of thumb for a reasonable sample size is that you need $\frac{3}{\alpha}$ observations.
# * Exponentiality is hidden in the recursiveness of the function -- we multiply by $(1−\alpha)$ each time, which already contains a multiplication by the same factor of previous model values.
# * the method is suitable for forecasting data with no clear trend or seasonal pattern
# With the setup of the above equation, we have the following form of a long term forecast:
# \begin{equation}
# \hat{X}_{t+h} = S_t
# \end{equation}
# which means simply that out of sample, our forecast is equal to the most recent value of the smoothed series.
# It's an old cliche that a picture is worth a thousand words - so the three pictures below should give you a truly excellent intuition ;-) on how single exponential smoothing works.
#
from statsmodels.tsa.holtwinters import SimpleExpSmoothing, ExponentialSmoothing, Holt
for alpha_sm in [0.2, 0.5, 0.9]:
df = pd.read_csv(CFG.data_folder + "exp1.csv", header=None)
df.columns = ["series"]
df.plot.line()
fit1 = SimpleExpSmoothing(df).fit(smoothing_level=alpha_sm, optimized=False)
fcast1 = fit1.forecast(12).rename("alpha = " + str(alpha_sm))
fcast1.plot(marker="o", color="red", legend=True)
fit1.fittedvalues.plot(color="red")
plt.show()
# As you can see from the graphs above, the for small values of the smoothing constant $\alpha$ most of the variation has been removed and we have a series following just the general trend; on the other hand, high value of the $\alpha$ parameter results in hardly any smoothing at all and the new series follows the original very closely (albeit with a delay, which is obvious given the relationship between raw and smoothed values).
# **Pro tip**: anytime you are using exponential smoothing that you did not write yourself, double-check the parametrization - does small $\alpha$ mean heavy smoothing or hardly any at all? The idea that the coefficient closer to 1 means less smoothing is merely a convention.
# What happens if we apply the method to the passengers dataset, first introduced in [part 1](https://www.kaggle.com/konradb/practical-time-series-part-1-the-basics) ?
# ## Double Exponential Smoothing
# Moving towards double exponential smoothing is akin to taking one more component in the seasonal decomposition: we began with level only, and we take into account the trend as well. We have learnt to predict intercept with our previous methods; now, we will apply the same exponential smoothing to the trend by assuming that the future direction of the time series changes depends on the previous weighted changes. Double exponential smoothing, a.k.a. the Holt method is defined by the relationship:
# \begin{equation}
# S_t = \alpha X_t + (1 - \alpha) (S_{t-1} + b_{t-1})\\
# b_t = \beta (S_t - S_{t-1}) + (1- \beta) b_{t-1}
# \end{equation}
# where $S_1 = X_1$, $\quad b_1 = X_1 - X_0$ and $\alpha, \beta \in (0,1)$
# The first equation describes the intercept, which depends on the current value of the series. The second term is now split into previous values of the level and of the trend. The second function describes the trend, which depends on the level changes at the current step and on the previous value of the trend. Complete prediction is composed of the sum of level and trend and the difference with simple exponential smoothing is that we need a second parameter to smooth the trend - as before, those can be set based on expert judgement or estimated (jointly) from the data.
# The forecast $h$ steps ahead is defined by
# \begin{equation}
# \hat{X}_{t+h} = S_t + h b_t
# \end{equation}
# The forecast function is no longer flat but trending: $h$-step-ahead forecast is equal to the last estimated level plus $h$ times the last estimated trend value.
# Let's compare the performance of the two methods on the passenger dataset:
#
alpha = 0.5
df = pd.read_csv(CFG.data_folder + "passengers.csv", usecols=["passengers"])
df.plot.line()
fit1 = SimpleExpSmoothing(df).fit(smoothing_level=alpha, optimized=False)
fcast1 = fit1.forecast(12).rename(r"$\alpha=0.5$")
fcast1.plot(marker="o", color="red", legend=True)
fit1.fittedvalues.plot(color="red")
plt.show()
df.plot.line()
fit1 = Holt(df).fit(smoothing_level=0.5, smoothing_slope=0.5, optimized=False)
fcast1 = fit1.forecast(12).rename("Holt's linear trend")
fit1.fittedvalues.plot(color="red")
fcast1.plot(color="red", legend=True)
plt.show()
#
# It seems like we are moving in the right direction - the forecast going forward is not constant, but follows a trend. However, it is simply an extrapolation of the most recent (smoothed) trend in the data which means we can expect the forecast to turn negative shortly. This is suspicious in general, and clearly renders the forecast unusable in the domain context.
#
# ## Triple Exponential Smoothing
# If it worked once, maybe it can work twice? A natural extension is to introduce a smoothed seasonal component: triple exponential smoothing, a.k.a. Holt-Winters, is defined by:
# \begin{equation}
# S_t = \alpha (X_t - c_{t - L}) + (1 - \alpha) (S_{t-1} + b_{t-1}) \\
# \end{equation}
# \begin{equation}
# b_t = \beta (S_t - S_{t-1}) + (1- \beta) b_{t-1} \\
# \end{equation}
# \begin{equation}
# c_t = \gamma (X_t - S_{t-1} - b_{t-1})+ (1 - \gamma) c_{t-L}
# \end{equation}
# with $\alpha, \beta, \gamma \in (0,1)$.
# The most important addition is the seasonal component to explain repeated variations around intercept and trend, and it will be specified by the period. For each observation in the season, there is a separate component; for example, if the length of the season is 7 days (a weekly seasonality), we will have 7 seasonal components, one for each day of the week. An obvious, yet worth repeating caveat: it makes sense to estimate seasonality with period $L$ only if your sample size is bigger than $2L$.
# The forecast $h$ steps ahead is defined by
# \begin{equation}
# \hat{X}_{t+h} = S_t + h b_t + c_{(t-L + h) \;\; mod \;\; L }
# \end{equation}
#
alpha = 0.5
df = pd.read_csv(CFG.data_folder + "passengers.csv", usecols=["passengers"])
df.plot.line()
fit1 = ExponentialSmoothing(df, seasonal_periods=12, trend="add", seasonal="add")
fit1 = fit1.fit(smoothing_level=0.5) # ,use_boxcox=True)
fit1.fittedvalues.plot(color="red")
fit1.forecast(12).rename("Holt-Winters smoothing").plot(color="red", legend=True)
plt.ylim(0, 800)
plt.show()
from statsmodels.graphics.tsaplots import plot_pacf
plot_pacf(air_df["Number of Passengers"].diff().dropna())
#
# Here we can see that the first lag is significantly out of the limit and the second one is also out of the significant limit but it is not that far so we can select the order of the p as 1.
#
plot_acf(air_df["Number of Passengers"].diff().dropna())
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Imports
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split, StratifiedKFold
from sklearn.linear_model import LogisticRegression
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from xgboost import XGBClassifier
from sklearn import metrics
from imblearn.over_sampling import RandomOverSampler
from sklearn.model_selection import GridSearchCV
from catboost import CatBoostClassifier
from lightgbm import LGBMClassifier
# # Dataset
df = pd.read_csv("/kaggle/input/playground-series-s3e12/train.csv")
df_test = pd.read_csv("/kaggle/input/playground-series-s3e12/test.csv")
# # Understanding the data
df.head()
df.shape
df.info()
df.describe().T
df.columns
df.isnull().sum()
df.duplicated().sum()
df.value_counts("target")
sns.heatmap(df.corr(), annot=True)
df.drop(columns="id", inplace=True)
df[["gravity", "ph", "osmo", "cond", "urea", "calc"]].agg(["skew", "kurtosis"]).T
sns.histplot(df["gravity"])
sns.histplot(df["ph"])
sns.histplot(df["osmo"])
sns.histplot(df["cond"])
sns.histplot(df["urea"])
sns.histplot(df["calc"])
df["calc/ph"] = df["calc"] / df["ph"]
df["osmo*calc"] = df["osmo"] * df["calc"]
df["urea*calc"] = df["urea"] * df["calc"]
X = df.drop(columns=["target", "ph", "cond", "gravity"], axis=1)
y = df["target"]
ROS = RandomOverSampler(sampling_strategy="minority")
X_balanced, y_balanced = ROS.fit_resample(X, y)
ss = StandardScaler()
X_std = ss.fit_transform(X_balanced)
X_train, X_val, y_train, y_val = train_test_split(
X_std, y_balanced, test_size=0.2, random_state=42
)
lr = LogisticRegression(penalty="l2", C=0.01, max_iter=10000)
lr.fit(X_train, y_train)
y1_pred = lr.predict(X_val)
df_check1 = pd.DataFrame({"Predicted": y1_pred, "Actual": y_val})
df_check1.sample(5)
print(metrics.classification_report(y_val, y1_pred))
print(metrics.roc_auc_score(y_val, y1_pred))
dtc = DecisionTreeClassifier(max_depth=100)
dtc.fit(X_train, y_train)
y2_pred = dtc.predict(X_val)
df_check2 = pd.DataFrame({"Actual": y_val, "Predicted": y2_pred})
df_check2.sample(5)
print(metrics.classification_report(y_val, y2_pred))
print(metrics.roc_auc_score(y_val, y2_pred))
rfc = RandomForestClassifier(n_estimators=200, max_depth=7, random_state=4)
rfc.fit(X_train, y_train)
y3_pred = rfc.predict(X_val)
df_check3 = pd.DataFrame({"Actual": y_val, "Predicted": y3_pred})
df_check3.sample(5)
print(metrics.classification_report(y_val, y3_pred))
print(metrics.roc_auc_score(y_val, y3_pred))
xgb = XGBClassifier(
colsample_bytree=1,
max_depth=2,
learning_rate=0.02,
min_child_weight=1,
gamma=0.0005,
reg_alpha=1,
reg_lambda=0.5,
)
xgb.fit(X_train, y_train)
y4_pred = xgb.predict(X_val)
df_check4 = pd.DataFrame({"Actual": y_val, "Predicted": y4_pred})
df_check4.sample(5)
print(metrics.classification_report(y_val, y4_pred))
print(metrics.roc_auc_score(y_val, y4_pred))
cbc = CatBoostClassifier(learning_rate=0.1, depth=3)
cbc.fit(X_train, y_train)
y5_pred = cbc.predict(X_val)
df_check5 = pd.DataFrame({"Actual": y_val, "Predicted": y5_pred})
df_check5.sample(5)
print(metrics.classification_report(y_val, y5_pred))
print(metrics.roc_auc_score(y_val, y5_pred))
lgbm = LGBMClassifier(learning_rate=0.1)
lgbm.fit(X_train, y_train)
y6_pred = lgbm.predict(X_val)
df_check6 = pd.DataFrame({"Actual": y_val, "Predicted": y6_pred})
df_check6.sample(5)
print(metrics.classification_report(y_val, y6_pred))
print(metrics.roc_auc_score(y_val, y6_pred))
df_test1 = df_test.copy()
df_test["calc/ph"] = df_test["calc"] / df_test["ph"]
df_test["osmo*calc"] = df_test["osmo"] * df_test["calc"]
df_test["urea*calc"] = df_test["urea"] * df_test["calc"]
X_test = df_test.drop(columns=["ph", "cond", "gravity"], axis=1)
X_test = ss.fit_transform(X_test)
final_prediction = cbc.predict(X_test)
final_test = pd.DataFrame({"id": df_test1["id"], "target": final_prediction})
final_test
final_test.set_index("id", inplace=True)
final_test.to_csv("submission.csv")
final_test.head()
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.tree import DecisionTreeClassifier
df = pd.read_csv("/kaggle/input/suv-nanze/suv.csv")
df.drop("User ID", axis=1, inplace=True)
df.head(5)
df.Gender = pd.get_dummies(df.Gender, drop_first=True)
X = df.to_numpy()
np.random.seed = 0
X = X[np.random.permutation(X.shape[0])]
y = X[:, -1]
X = X[:, :-1]
split = int(X.shape[0] * 0.8)
X_train = X[:split]
y_train = y[:split]
X_test = X[split:]
y_test = y[split:]
"""
the parameters > criterion > gini, entropy > gini is quicker because it removes the log
splitter > where to split the tree > best or random > in some cases random is better
max_depth
min_samples_split
"""
clf = DecisionTreeClassifier()
clf.fit(X_train, y_train)
print("max depth : ", clf.tree_.max_depth)
print("train accuracy : ", clf.score(X_train, y_train))
print("test accuracy : ", clf.score(X_test, y_test))
train_scores = []
test_scores = []
for d in range(1, 14):
clf = DecisionTreeClassifier(max_depth=d)
clf.fit(X_train, y_train)
train_scores.append(clf.score(X_train, y_train))
test_scores.append(clf.score(X_test, y_test))
plt.plot(np.arange(1, 14, 1), np.array(train_scores), "b-")
plt.plot(np.arange(1, 14, 1), np.array(test_scores), "r-")
clf = DecisionTreeClassifier(max_depth=2, criterion="entropy")
clf.fit(X_train, y_train)
print("max depth : ", clf.tree_.max_depth)
print("train accuracy : ", clf.score(X_train, y_train))
print("test accuracy : ", clf.score(X_test, y_test))
# for finding the best depth > it recommended to train with k fold cross validation too
plt.plot(X[y == 0, 1], X[y == 0, 2], "ro")
plt.plot(X[y == 1, 1], X[y == 1, 2], "bo")
# it is separable with two broken lines
# # Visualization
from sklearn.tree import export_text
print(export_text(clf, feature_names=["Gender", "Age", "EstimatedSalary"]))
# age is repeated two times
plt.plot(
df[df["Purchased"] == 1]["Age"],
df[df["Purchased"] == 1]["EstimatedSalary"],
"bo",
label="1",
)
plt.plot(
df[df["Purchased"] == 0]["Age"],
df[df["Purchased"] == 0]["EstimatedSalary"],
"rx",
label="0",
)
plt.vlines(42.5, 0, 160000)
plt.hlines(89500, 16, 42.5)
plt.xlabel("Age")
plt.ylabel("EstimatedSalary")
plt.legend()
from sklearn.tree import export_graphviz
# output format > .dot
# class_names > value list in each node > #False , #True
# class 0 > not perchased > false
export_graphviz(
clf,
out_file="DT.dot",
feature_names=["Gender", "Age", "EstimatedSalary"],
filled=True,
class_names=["False", "True"],
)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
#
# # Introduction
# #### Hi! This is my first kernel and project. I benefited from many sources and learned a lot while preparing this kernel. I will be waiting for your feedback :). I will publish new versions with what I learned with your advice and feedback.
# ### **Import The Required Libraries**
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
# ### **Load Dataset**
data = pd.read_csv("../input/students-performance-in-exams/StudentsPerformance.csv")
# ### **Copy Real Dataset**
df = data.copy()
# ### **Quick Check (There seems to be no missing value)**
df.info()
# ### **First look**
df.head()
# ### **Rename columns names for easier reading**
df.rename(
inplace=True,
columns={
"race/ethnicity": "race_ethnicity",
"parental level of education": "education_level",
"test preparation course": "prep_course",
"math score": "math_score",
"reading score": "reading_score",
"writing score": "writing_score",
},
)
# ### **Create a new column called "average_score" to see the scores together.**
df["average_score"] = df[["math_score", "reading_score", "writing_score"]].mean(axis=1)
# ### Describe our dataset and understand basic information
df.describe().T
# ### Let us create a new column called "grade" and assign the grades
def Grade(AverageScore):
if AverageScore >= 80:
return "A"
if AverageScore >= 70:
return "B"
if AverageScore >= 60:
return "C"
if AverageScore >= 50:
return "D"
if AverageScore >= 40:
return "E"
else:
return "F"
df["grade"] = df.apply(lambda x: Grade(x["average_score"]), axis=1)
# ### Let's create a simple pie chart to see the grade distributions.
plt.figure(figsize=(7, 7))
plt.pie(
df["grade"].value_counts().values,
labels=df["grade"].value_counts().index,
autopct="%1.1f%%",
shadow=True,
explode=[0, 0, 0.1, 0, 0, 0],
)
plt.title("Grade Pie Chart", color="Black", fontsize=20)
plt.show()
# ### Take a look at Score Heatmap
plt.figure(figsize=(7, 7))
plt.title("Score Heatmap", color="Black", fontsize=20, pad=40)
sns.heatmap(df.corr(), annot=True, linewidths=0.5)
# #
# #
# # What I want to know
# #
# ### **Q1**: What is the effect of gender and education level on average score?
sns.catplot(
data=df, x="gender", y="average_score", hue="education_level", kind="bar", height=5
)
# ### -----------------------------------------------------------------------------------------
# ### Q2: What is the effect of gender and preparation course on average score?
sns.catplot(
data=df, x="gender", y="average_score", hue="prep_course", kind="bar", height=5
)
# ### -----------------------------------------------------------------------------------------
# ### Q3: Which group is the most successful? (on average_score)
sns.barplot(x="race_ethnicity", y="average_score", data=df)
df.groupby("race_ethnicity")["average_score"].mean()
# ### **"group E"** is the most successful group. However, why?. I make some research.
df.groupby("race_ethnicity")["education_level"].describe()
df.groupby("race_ethnicity")["prep_course"].describe()
df.groupby("race_ethnicity")["grade"].describe()
(
sns.FacetGrid(df, hue="race_ethnicity", height=5, xlim=(0, 100))
.map(sns.kdeplot, "average_score")
.add_legend()
)
sns.catplot(x="race_ethnicity", y="average_score", hue="grade", kind="point", data=df)
# ### Finally, I want to show you the last graph. I believe this graph will give us important assumptions about scores.
sns.displot(df)
|
# # 催收评分卡项目
# ## 环境处理
# - [依赖的外部环境](#env) 其中主要封装了一些模型的训练和模型效果评估的代码, 以及依赖包的导入
# - model_trains 模型训练
# - model_estimate 模型效果评估
# - start_load 依赖包的导入和一些工具方法的定义
#
# ## [加载数据](#load_data)
# - [列名替换](#将列名进行映射)这里为了方便理解用中文列名进行了替换
# - [切分数据集和验证集](#切分数据集和验证集)
# - [查看样本分布](#查看样本分布)
# ## [缺失值处理d](#缺失值处理)
# ## 模型训练
# ### [1辅助模型](#辅助模型)
# - [特征重要性排序](#重要性排序)
# - [IV值排序](#iv值排序)
# ## [分箱](#split_box)
# - [特征类型划分](#特征分类)
# ## 环境处理
# ```shell
# !mkdir -p libs/utils
# !touch libs/__init__.py libs/utils/__init__.py
# !wget -O libs/utils/model_trains.py https://gitee.com/mill_teacher/machine_learn/raw/master/card/libs/utils/model_trains.py
# !wget -O libs/utils/model_estimate.py https://gitee.com/mill_teacher/machine_learn/raw/master/card/libs/utils/model_estimate.py
# !wget -O start_load.py https://gitee.com/mill_teacher/machine_learn/raw/master/card/script/%E8%AF%84%E5%88%86%E5%8D%A1%E4%BB%A3%E7%A0%81/start_load.py
# !pip install toad==0.0.61
# ```
#
# ## 加载数据
data_path = os.path.join("../input/give-me-some-credit-dataset", "cs-training.csv")
data = pd.read_csv(data_path, index_col=0)
data.shape
import toad
#
# ### 将列名进行映射
# field|describe|type
# ---|---|---
# SeriousDlqin2yrs|逾期90天以上|Y/N
# RevolvingUtilizationOfUnsecuredLines|信用卡和个人信用额度的总余额(不动产和汽车贷款等无分期付款债务除外)除以信用额度之和|percentage
# age|年龄|integer
# NumberOfTime30-59DaysPastDueNotWorse|借款人逾期30-59天的次数,但在过去2年中没有恶化。|integer
# DebtRatio|负债率: 每月还债,赡养费,生活费除以每月总收入|percentage
# MonthlyIncome|实际月收入|
# NumberOfOpenCreditLinesAndLoans|未结贷款(分期付款,如汽车贷款或抵押贷款)和信贷额度(如信用卡)的数量|integer
# NumberOfTimes90DaysLate|借款人逾期90天或以上的次数|integer
# NumberRealEstateLoansOrLines|抵押贷款和房地产贷款的数量,包括房屋净值信贷额度|integer
# NumberOfTime60-89DaysPastDueNotWorse|借款人逾期60-89天的次数,但在过去2年中没有恶化。|integer
# NumberOfDependents|家庭中不包括自己的受抚养人人数(配偶、子女等)|integer
column_map = {
"SeriousDlqin2yrs": "target",
"RevolvingUtilizationOfUnsecuredLines": "信用额度使用率",
"age": "年龄",
"NumberOfTime30-59DaysPastDueNotWorse": "逾期30-59天的次数",
"DebtRatio": "负债率",
"MonthlyIncome": "实际月收入",
"NumberOfOpenCreditLinesAndLoans": "未结贷款的数量",
"NumberOfTimes90DaysLate": "连续逾期90天以上的次数",
"NumberRealEstateLoansOrLines": "抵押贷款笔数",
"NumberOfTime60-89DaysPastDueNotWorse": "连续逾期60~90天的次数",
"NumberOfDependents": "家庭人口数",
}
data = data.rename(columns=column_map)
data.describe().T
#
# ### 切分数据集和验证集
# 将数据分为
# - 训练集
# - 测试集
# - 验证集 正常情况下,验证集从时间外样本上获取
# #### 切分数据集和验证集
train_data, oot_data = train_test_split(data, stratify=data["target"], random_state=47)
train_data.shape, oot_data.shape
# #### 切分训练集和测试集
train_data, test_data = train_test_split(
train_data, stratify=train_data["target"], random_state=47
)
train_data.shape, test_data.shape
# ### 打上类型标记
# > 所有的数据都在一起进行处理,如缺失值填补和异常值处理。但是标注好类型,方便分开
train_data["type"] = "train"
oot_data["type"] = "oot"
test_data["type"] = "test"
data = pd.concat([train_data, oot_data, test_data])
data.shape
#
# ## 查看样本分布
# > 可以发现坏样本率在6.6%
samples_rate = (
data.groupby(["type", "target"])
.agg({"年龄": "count"})
.reset_index()
.rename(columns={"年龄": "count"})
) # 按类型的好坏样本分布
samples_total = (
data["type"]
.value_counts()
.reset_index()
.rename(columns={"index": "type", "type": "total"})
) # 按类型总客户数
samples_cal_pd = pd.merge(samples_rate, samples_total, on="type")
samples_cal_pd["rate"] = samples_cal_pd["count"] / samples_cal_pd["total"] # 计算好坏客户占比
samples_cal_pd
#
# ## 缺失值处理
# 一般的处理方式如下:
# 1. 缺失值超过90%直接删除
# 2. 缺失值超过50%将缺失值单独作为一类
# 3. 通过分箱解决
# 4. 填充固定值,这个常用,因为缺失值往往有一定的意义
# 5. 填充中位数
# 6. knn或者随机森林填充
# 本案中`实际月收入`和`家庭人口数` 的缺失值都没有实际的含义。缺失率也不多,应该使用插补填充。
missing_data = (
pd.DataFrame(data.isnull().sum())
.reset_index()
.rename(columns={"index": "column", 0: "count"})
)
missing_data_sortd = missing_data.sort_values("count", ascending=False)
missing_data_sortd["missing_rate"] = missing_data_sortd["count"] / data.shape[0]
missing_data_sortd
# ### 尝试使用KNN填补
# 因为内存消耗太大无法计算
# ```python
# from sklearn.impute import KNNImputer
# train_columns = set(data.columns)-{'target','type'}
# imputer = KNNImputer(n_neighbors=5)
# data.loc[:,train_columns] = imputer.fit_transform(data.loc[:,train_columns])
# ```
# ### 尝试使用xgboost填充
# 也是内存使用太大,无法计算
# ```python
# train_columns = list(set(data.columns)-{'target','type','实际月收入'})
# valide_data = data.loc[data['实际月收入'].isnull(),:]
# train_data = data.loc[data['实际月收入'].notnull(),:]
# x_train,x_test,y_train,y_test = train_test_split(train_data[train_columns].values,train_data['实际月收入'].values,random_state=37)
# xgb_model(x_train,y_train,x_test,y_test,estimators=100)
# ```
# ### 这里先使用均值填充
from sklearn.impute import SimpleImputer
imputer = SimpleImputer()
train_columns = set(data.columns) - {"target", "type"}
data.loc[:, train_columns] = imputer.fit_transform(data.loc[:, train_columns])
missing = data.isnull().sum()
missing[missing > 0]
#
# ## 异常值处理
# 查看数据的分布情况
plt.rcParams["font.sans-serif"] = ["Droid Sans Fallback"]
plt.rcParams["axes.unicode_minus"] = False
plt.rcParams["font.family"] = ["Times New Roman"]
plt.rcParams.update({"font.size": 8})
number_features = data[train_columns].select_dtypes(["float", "int"]).columns
plot_distplot(data[number_features])
# ### 箱线图查看长尾情况
plot_box(data[number_features])
#
# ## 辅助模型
X_train, X_test, y_train, y_test = train_test_split(data[train_columns], data["target"])
xgb_model_obj, xgb_test_pred = xgb_model(X_train, y_train, X_test, y_test)
#
# ### xgboost重要性排序
from xgboost import plot_importance
_, ax = plt.subplots(figsize=(12, 8))
plot_importance(xgb_model_obj, ax=ax)
# ### 用列表显示特征重要性
importants = xgb_model_obj.get_booster().get_score()
import_features = (
pd.DataFrame(importants, index=["import"])
.T.reset_index()
.rename(columns={"index": "name"})
)
import_features.sort_values("import", ascending=False)
#
# ### iv值排序
toad.quality(data.drop("type", axis=1), cpu_cores=1, iv_only=True)
#
# ## 分箱
# ### 初步筛选
# 在特征分箱之前,先行进行一次筛选,目的是减少分箱的工作
selected, drop_list = toad.select(
data, return_drop=True, iv=0.03, corr=1, exclude=["type"]
)
drop_list
#
# ## 将特征分为
# - 二值型
# - 类别型
# - 数值型
# 其中只有数值型需要参与分箱,本案中没有类别型,全部都是数值型
pd.DataFrame(data.drop(["target", "type"], axis=1).nunique().sort_values())
# #### 样本划分
# 这里只采用训练集的数据进行分箱,而用测试集和验证集来进行验证
train_data = data.loc[data["type"] == "train", :]
oot_data = data.loc[data["type"] == "oot", :]
test_data = data.loc[data["type"] == "test", :]
# ### 自动卡方分箱
# 使用卡方分箱,`min_samples`参数可以设置最小一箱的样本量。 这里为了代码重跑,做了缓存判断
import pickle
MODEL_PATH = "./combiner_model_v1.pkl"
if os.path.exists(MODEL_PATH):
with open(MODEL_PATH, "rb") as f:
combiner = pickle.load(f)
else:
combiner = toad.transform.Combiner()
print("start fit...")
combiner.fit(
train_data[train_columns], train_data["target"], method="chi", min_samples=0.05
)
print("end fit...")
with open(MODEL_PATH, "wb") as f:
pickle.dump(combiner, f)
# ### 分箱的分隔
bin = combiner.export()
bin
def transform(data):
"""
对数据进行分箱
"""
data_number = data.copy() # copy一份数据,因为需要多次操作
data_number.loc[:, train_columns] = combiner.transform(data[train_columns])
return data_number
data_number = transform(data)
def bin_badrate_plot(data, col, t="type", target="target"):
"""
画图查看分箱情况
"""
badrate_plot(data, x=t, target=target, by=col)
data_train = data.loc[data["type"] == "train", :]
bin_plot(data_train, x=col, annotate_format=".2f")
bin_bg_plot(data_train, col)
# #### 查看不稳定的特征
list(filter(lambda x: not is_stable(data_number, x), number_features))
# #### 查看没有单调性的特征
list(filter(lambda x: not is_monotonic(data_number, x), number_features))
bin["实际月收入"] # 查看分箱区隔
ajd_bin = {
"抵押贷款笔数": [
1.0,
],
"负债率": [
0.020232659,
0.406828251,
0.50950794,
],
"未结贷款的数量": [
3.0,
5.0,
],
"实际月收入": [4839.0, 7542.0],
}
combiner.set_rules(ajd_bin)
data_number = transform(data)
bin_badrate_plot(data_number, "实际月收入")
features_count_lst = []
for i in set(data_number.columns) - {"target", "type"}:
tmp = data_number[i].value_counts().reset_index().rename(columns={i: "count"})
tmp["feature"] = i
features_count_lst.append(tmp)
box_score_pd = pd.concat(features_count_lst)
t = toad.transform.WOETransformer()
data_number.loc[:, train_columns] = t.fit_transform(
data_number[train_columns], data_number["target"]
)
woe_map = t.export()
data_selected, drop_lst = toad.selection.select(
data_number[train_columns],
data_number["target"],
empty=0.6,
iv=0.002,
corr=0.7,
return_drop=True,
)
drop_lst
test_data.head()
train_data = data_number.loc[data["type"] == "train", :]
oot_data = data_number.loc[data["type"] == "oot", :]
test_data = data_number.loc[data["type"] == "test", :]
# model, val_pred = xgb_model(train_data[train_columns],train_data['target'],test_data[train_columns],test_data['target'])
y_test_pred, y_train_pred, lr_model_obj = lr_model(
train_data[train_columns],
train_data["target"],
test_data[train_columns],
test_data["target"],
C=0.1,
)
# ## 参数搜索
param_gric = [
{
"penalty": ["l2"],
"solver": ["newton-cg", "lbfgs", "liblinear", "sag", "saga"],
"C": np.arange(0.5, 1.5, 0.1),
}
]
grid_search = GridSearchCV(lr_model_obj, param_gric, n_jobs=1, verbose=1)
grid_search.fit(train_data[train_columns], train_data["target"])
grid_search.best_params_
y_test_pred, y_train_pred, lr_model_obj = lr_model(
train_data[train_columns],
train_data["target"],
test_data[train_columns],
test_data["target"],
**grid_search.best_params_
)
import pickle
LR_MODEL_PATH = "./lr_model.plk"
with open(LR_MODEL_PATH, "wb") as f:
pickle.dump(lr_model_obj, f)
pred_train = lr_model_validation(
lr_model_obj, oot_data[train_columns], oot_data["target"]
)
toad.metrics.PSI(y_train_pred, pred_train)
toad.metrics.PSI(y_train_pred, y_test_pred)
toad.metrics.PSI(y_test_pred, pred_train)
PDO = (1000 - 0) / np.log2(9999 / (1 / 9999))
B = PDO / np.log(2)
A = 0 - B * np.log(1 / 9999)
# ## 计算总分
score_pd = pd.DataFrame(
{
"score": pd.Series(y_test_pred).apply(get_score_with_model, args=(A, B)).values,
"possi": y_test_pred,
"real": test_data["target"].values,
}
)
score_pd.head()
box_score_pd["woe"] = box_score_pd.apply(
lambda row: woe_map.get(row["feature"]).get(row["index"]), axis=1
)
def get_combine_max(row):
"""
根据分箱获取每一个变量的每一个分箱的值范围上限
"""
tmp = bin.get(row["feature"])
if tmp == None:
return row["index"]
idx = int(row["index"])
if len(tmp) <= idx:
return np.inf
return tmp[idx]
def get_combine_min(row):
"""
根据分箱获取每一个变量的每一个分箱的值范围下限
"""
tmp = bin.get(row["feature"])
if tmp == None:
return row["index"]
idx = row["index"] - 1
if idx < 0:
return -np.inf
return tmp[int(idx)]
bin = combiner.export()
box_score_pd["min"] = box_score_pd.apply(get_combine_min, axis=1)
box_score_pd["max"] = box_score_pd.apply(get_combine_max, axis=1)
box_score_pd = box_score_pd.sort_values("feature", "index")
feature_cnt = len(train_columns)
a = lr_model_obj.intercept_
b_dict = dict(zip(list(X_train.columns), lr_model_obj.coef_.tolist()[0])) # 系数
box_score_pd["a"] = a[0]
box_score_pd["b"] = box_score_pd["feature"].apply(lambda x: b_dict.get(x))
def cal_score(row, A, B):
"""
计算打分结果, 一定要检查feature_cnt, 搞了一下午这个值都搞错了
"""
woe, b, a = row[["woe", "b", "a"]]
return A / feature_cnt - B * (woe * b + a / feature_cnt)
box_score_pd["score"] = box_score_pd.apply(cal_score, axis=1, args=(A, B))
from IPython.display import HTML
HTML(box_score_pd.to_html())
# ## 模型效果评估
# ### KS值
from libs.utils.model_estimate import model_monotony, calculate_ks
ks_value, probability, crossdens = calculate_ks(y_test_pred, test_data["target"])
ks_value
# ## 查看效果评估报告
from sklearn.metrics import classification_report
rs = classification_report(
y_pred=np.where(y_test_pred >= probability, 1, 0), y_true=test_data["target"]
)
print(rs)
from sklearn.metrics import confusion_matrix, recall_score
pd.DataFrame(
confusion_matrix(
test_data["target"], y_pred=np.where(y_test_pred >= probability, 1, 0)
)
)
recall_score(test_data["target"], y_pred=np.where(y_test_pred >= probability, 1, 0))
ks_score = get_score_with_model(probability, A, B)
ks_score, ks_value, probability
ks_bucket_pd = cal_lift(y_test_pred, test_data["target"], A, B, ks_score)
ks_bucket_pd.applymap(lambda x: round(x, 2))
oot_ks_bucket_pd = cal_lift(pred_train, oot_data["target"], A, B, ks_score)
oot_ks_bucket_pd.applymap(lambda x: round(x, 2))
# ## 入模特征
train_cols_ivs = toad.quality(
data_number[list(train_columns) + ["target"]], iv_only=True
)
train_cols_ivs
# ## 找几个样例客户
test_data["score"] = (
pd.Series(y_test_pred).apply(get_score_with_model, args=(A, B)).values
)
good_sample_idx = (
test_data.sort_values("score", ascending=False).head(2).index
) # 好客户的索引
good_sample = data.loc[good_sample_idx, list(train_columns) + ["target"]].T # 查看好样本
good_sample
bad_sample_idx = test_data.sort_values("score", ascending=True).head(2).index # 坏客户的索引
bad_sample = data.loc[bad_sample_idx, list(train_columns) + ["target"]].T # 坏客户样本
bad_sample
def cal_score_split(row):
score_lst = list(
map(
lambda idx: box_score_pd.loc[
(box_score_pd["feature"] == idx[0]) & (box_score_pd["index"] == idx[1]),
"score",
].values[0],
zip(row.index, row),
)
)
return pd.Series(score_lst, index=row.index) # index对齐,方便做concat
def get_sample_detail(sample):
data_train = combiner.transform(sample.T) # 分箱
# result_type='expand' 参数表示将like-array扩展成pd,这里其实没有必要设置,因为返回的是series
score_pd = (
data_train[train_columns].apply(cal_score_split, axis=1, result_type="expand").T
)
return pd.concat([score_pd, sample], axis=1)
good_sample_pd = get_sample_detail(good_sample).applymap(lambda x: round(x, 2)) # 好样本
bad_sample_pd = get_sample_detail(bad_sample).applymap(lambda x: round(x, 2)) # 坏样本
tpl_samples_pd = pd.concat(
[good_sample_pd, bad_sample_pd, train_cols_ivs], axis=1
) # 合并
tpl_samples_pd.drop(["gini", "entropy"], axis=1, inplace=True) # 删除空字段
tpl_samples_pd.sort_values("iv", ascending=False) # 找的坏样本中,还有个误杀-_-||
# ## 输出依赖的版本号
import xgboost as xgb
import sklearn as sk
for m in {
inspect,
math,
np,
sk,
os,
pd,
pickle,
plt,
re,
relativedelta,
sns,
sys,
toad,
warnings,
xgb,
}:
try:
print("{}--{}".format(m.__name__, m.__version__))
except:
pass
# ## 评分推导
from sympy import *
import sympy as sy
init_session(use_latex=True)
A, B, Odds, PDO, Score, p = symbols("A,B,Odds,PDO,Score,p")
# - Odds 好坏比或者坏好比
# - PDO 好坏比增加一倍的时候,分数增加PDO
Eq(Score, A + B * log(Odds)) # 基本公式
Eq(PDO + Score, A + B * log(2 * Odds)) # 好坏比增加一倍的时候,分数增加PDO
# #### 定义Odds,算出B的值
# 设Odds为坏好比,Odds=9999/1的时候客户最坏, Odds=1/9999的时候客户最好
Eq(0, A + B * log(Odds)).subs({Odds: (9999 / 1)}) # 最坏的时候分数为0
Eq(1000, A + B * log(Odds)).subs({Odds: (1 / 9999)}) # 最好的时候分数为1000
exp1 = Eq((A + B * log(1 / 9999)) - (A + B * log(9999 / 1)), 1000) # 上两个等式化简后可以算出B的值
exp1
BVal = solve(exp1)[0] # 算出B的值
BVal
# #### 根据B算出A的值
# 根据下面任意一种情况算出的A值都是一样的
exp2 = Eq(Score, A + B * log(Odds)).subs(
{Score: 0, Odds: 9999 / 1, B: BVal}
) # 0分对应的好坏比
exp2
Eq(Score, A + B * log(Odds)).subs({Score: 1000, Odds: 1 / 9999, B: BVal}) # 1000分对应的好坏比
AVal = solve(exp2)[0]
AVal
# #### 计算出PDO的值
# 实际上PDO和B是对应的值,只要求出B就能给出PDO
exp3 = Eq(PDO, (A + B * log(2 * Odds)) - (A + B * log(Odds)))
exp3
PDOVal = solve(exp3.subs({B: BVal}))[0][PDO]
PDOVal
# ### 逻辑回归计算变量分推理
# 到这里,实际上能算出总分来了。但是根据每一个特征算分,还需要推理一下
Eq(p, 1 / (1 + E ** -g(x))) # 逻辑回归的激活函数sigmoid函数
# 假设P为好客户概率,1-P为坏客户概率,好坏比Odds为:
# $Odds = \frac{p}{1 - p}$
exp3 = Eq(Score, A + B * log(Odds)).subs({Odds: (p / (1 - p))})
exp3
exp4 = exp3.subs({p: 1 / (1 + E ** -g(x))}) # 把P的sigmoid定义带入
exp4
simplify(exp4) # 化简后
Eq(Score, A + B * g(x)) # log和e抵消
|
# # Lab Report 06: Regression, feature selection and PCA
# **Mohammad Adeel (033-19-0020)**
# **Loading Bostin Housing Price Dataset from sklearn**
from sklearn import datasets
boston = datasets.load_boston()
boston.keys()
# **Understanding the dataset**
print(boston.DESCR)
# **Printing out the features**
print(boston.data)
# **Printing out the targets (Price in thoussand dollars)**
print(boston.target)
# **Checking the shape of the dataset**
boston.data.shape
# **Seperating features and labels:**
X = boston.data
y = boston.target
X.shape, y.shape
# **Converting data from arrays to give it a better look**
import pandas as pd
df_boston = pd.DataFrame(X, columns=boston.feature_names)
df_boston["Price"] = boston.target
df_boston.head()
# **Seperatin the features and labels in DataFrame:**
X, y = df_boston.loc[:, df_boston.columns != "Price"], df_boston.Price
X.shape, y.shape
# **Splitting into train and test dataset**
from sklearn.model_selection import train_test_split
Xtrain, Xtest, ytrain, ytest = train_test_split(X, y, test_size=0.25, shuffle=True)
Xtrain.shape, Xtest.shape, ytrain.shape, ytest.shape
# **Linear Regression:**
# **Initializing the model, Fitting and calculating testing accuracy using Linear Regression:**
from sklearn.linear_model import LinearRegression
LReg = LinearRegression()
LReg.fit(Xtrain, ytrain)
LReg.score(Xtest, ytest)
# **Visualization of Linear Regression model:**
import pandas as pd
dXtest = pd.DataFrame(Xtest, columns=boston.feature_names)
dXtrain = pd.DataFrame(Xtrain, columns=boston.feature_names)
dXtest = dXtest[["LSTAT"]]
dXtrain = dXtrain[["LSTAT"]]
# **Plotting the selected feature value distribution along with price of the house and model line:**
LReg.fit(dXtrain, ytrain)
import matplotlib.pyplot as plt
plt.xlabel("Feature Value")
plt.ylabel("Price of House")
plt.title("Linear Regression Visulization on single feature")
plt.scatter(dXtest, ytest, color="black", label="Original Data Distribution")
plt.plot(
dXtest,
LReg.predict(dXtest),
color="blue",
linewidth=3,
label="Linear Refression Line trying to fit on data",
)
plt.legend(loc="upper right")
# **Feature Importance:**
# **Making slots of features from the dataset of both train and test**
slot_1_Xtr = Xtrain[["CRIM", "ZN", "INDUS"]]
slot_2_Xtr = Xtrain[["CHAS", "NOX", "RM"]]
slot_3_Xtr = Xtrain[["AGE", "DIS", "RAD"]]
slot_4_Xtr = Xtrain[["TAX", "PTRATIO", "B", "LSTAT"]]
slot_1_Xts = Xtest[["CRIM", "ZN", "INDUS"]]
slot_2_Xts = Xtest[["CHAS", "NOX", "RM"]]
slot_3_Xts = Xtest[["AGE", "DIS", "RAD"]]
slot_4_Xts = Xtest[["TAX", "PTRATIO", "B", "LSTAT"]]
# **Fitting and evaluating on 1st slot:**
from sklearn.linear_model import LinearRegression
LReg = LinearRegression()
LReg.fit(slot_1_Xtr, ytrain)
LReg.score(slot_1_Xts, ytest)
# **Fitting and evaluating on second slot**
from sklearn.linear_model import LinearRegression
LReg = LinearRegression()
LReg.fit(slot_2_Xtr, ytrain)
LReg.score(slot_2_Xts, ytest)
# **Fitting and evaluating on the 3rd slot**
from sklearn.linear_model import LinearRegression
LReg = LinearRegression()
LReg.fit(slot_3_Xtr, ytrain)
LReg.score(slot_3_Xts, ytest)
# **Fitting and evaluating on the 4th slot**
from sklearn.linear_model import LinearRegression
LReg = LinearRegression()
LReg.fit(slot_4_Xtr, ytrain)
LReg.score(slot_4_Xts, ytest)
# **Calculating error metrics for slot 1 using linear regression**
from sklearn.metrics import (
mean_absolute_error,
explained_variance_score,
mean_squared_error,
)
LReg1 = LinearRegression()
LReg1.fit(slot_1_Xtr, ytrain)
print(
"Mean Absolute error of slot 1:",
mean_absolute_error(ytest, LReg1.predict(slot_1_Xts)),
)
print(
"Mean Sqaured error of slot 1:",
mean_squared_error(ytest, LReg1.predict(slot_1_Xts)),
)
print(
"Explained Variance Score of slot 1:",
mean_absolute_error(ytest, LReg1.predict(slot_1_Xts)),
)
# **For Slot 2**
from sklearn.metrics import (
mean_absolute_error,
explained_variance_score,
mean_squared_error,
)
LReg1 = LinearRegression()
LReg1.fit(slot_2_Xtr, ytrain)
print(
"Mean Absolute error of slot 2:",
mean_absolute_error(ytest, LReg1.predict(slot_2_Xts)),
)
print(
"Mean Sqaured error of slot 2:",
mean_squared_error(ytest, LReg1.predict(slot_2_Xts)),
)
print(
"Explained Variance Score of slot 2:",
mean_absolute_error(ytest, LReg1.predict(slot_2_Xts)),
)
# **Slot 3**
from sklearn.metrics import (
mean_absolute_error,
explained_variance_score,
mean_squared_error,
)
LReg1 = LinearRegression()
LReg1.fit(slot_3_Xtr, ytrain)
print(
"Mean Absolute error of slot 3:",
mean_absolute_error(ytest, LReg1.predict(slot_3_Xts)),
)
print(
"Mean Sqaured error of slot 3:",
mean_squared_error(ytest, LReg1.predict(slot_3_Xts)),
)
print(
"Explained Variance Score of slot 3:",
mean_absolute_error(ytest, LReg1.predict(slot_3_Xts)),
)
# **Slot 4**
from sklearn.metrics import (
mean_absolute_error,
explained_variance_score,
mean_squared_error,
)
LReg1 = LinearRegression()
LReg1.fit(slot_4_Xtr, ytrain)
print(
"Mean Absolute error of slot 4:",
mean_absolute_error(ytest, LReg1.predict(slot_4_Xts)),
)
print(
"Mean Sqaured error of slot 4:",
mean_squared_error(ytest, LReg1.predict(slot_4_Xts)),
)
print(
"Explained Variance Score of slot 4:",
mean_absolute_error(ytest, LReg1.predict(slot_4_Xts)),
)
# **Calculating all metrics for test dataset containing all features:**
from sklearn.metrics import (
mean_absolute_error,
explained_variance_score,
mean_squared_error,
)
LReg1 = LinearRegression()
LReg1.fit(Xtrain, ytrain)
print("Mean Absolute error:", mean_absolute_error(ytest, LReg1.predict(Xtest)))
print("Mean Sqaured error:", mean_squared_error(ytest, LReg1.predict(Xtest)))
print("Explained Variance Score:", mean_absolute_error(ytest, LReg1.predict(Xtest)))
# **Principal Components Analysis (PCA)**
# **Analyzing work of PCA with sklearn:**
# **Accuracy without PCA with all 13 features**
from sklearn.linear_model import LinearRegression
LReg = LinearRegression()
LReg.fit(Xtrain, ytrain)
LReg.score(Xtest, ytest)
# **Calculating Variance by top 5 components**
from sklearn.decomposition import PCA
pca = PCA(n_components=5)
pca.fit(X)
pca.explained_variance_ratio_
# **Transformed components values by PCA**
from sklearn.decomposition import PCA
pca = PCA(n_components=11)
arraystr = pca.fit_transform(Xtrain)
arraysts = pca.transform(Xtest)
arraystr
# **Shape of the transformed data:**
from sklearn.decomposition import PCA
pca = PCA(n_components=11)
arraystr = pca.fit_transform(Xtrain)
arraysts = pca.transform(Xtest)
arraystr.shape, arraysts.shape
# **Accuracy with 11 features extracted by PCA**
from sklearn.decomposition import PCA
pca = PCA(n_components=11)
arraystr = pca.fit_transform(Xtrain)
arraysts = pca.transform(Xtest)
LReg.fit(arraystr, ytrain)
LReg.score(arraysts, ytest)
# **Fitting the model, applying permutation importance on data along with model and printing them out in ascending order:**
from sklearn.linear_model import LinearRegression
LReg = LinearRegression()
LReg.fit(X, y)
from sklearn.inspection import permutation_importance
results = permutation_importance(LReg, X, y, n_repeats=10, random_state=0)
import pandas as pd
for i in results.importances_mean.argsort():
print("Feature", X.columns[i], ": has Importance", results.importances_mean[i])
# **Plotting the features along with its corresponding importance using bar plot**
import matplotlib.pyplot as plt
feature = []
importances = []
for i in results.importances_mean.argsort():
feature.append(i)
importances.append(results.importances_mean[i])
plt.xlabel("Feature")
plt.ylabel("Importance")
plt.title("Plot of importance of corrosponding feature")
plt.bar(range(len(importances)), importances, align="center")
plt.xticks(range(len(feature)), X.columns[feature], rotation="vertical")
# # Exercise
# **Load and define briefly the diabetes dataset available in sklearn.**
from sklearn.datasets import load_diabetes
# Load the diabetes dataset
diabetes_dataset = load_diabetes()
# Print a brief description of the dataset
print(diabetes_dataset.DESCR)
# **Write brief summary about Diabetes Dataset**
# The load_diabetes() function from sklearn.datasets module loads the diabetes dataset and returns an object that contains the dataset's features, target values, and metadata.
# The DESCR attribute of the diabetes_dataset object contains a brief description of the dataset, which includes the following information:
# Number of instances: 442
# Number of attributes: 10 (age, sex, body mass index, average blood pressure, and six blood serum measurements)
# Target variable: a quantitative measure of disease progression one year after baseline
# Features: All features have been mean-centered and scaled by the standard deviation multiplied by n_samples (n_samples is the number of instances).
# **Print, Check and define keys and shape of diabetes dataset.**
from sklearn.datasets import load_diabetes
# Load the diabetes dataset
diabetes_dataset = load_diabetes()
# Print the keys of the diabetes dataset
print(diabetes_dataset.keys())
# Check the shape of the data and target arrays
print("Data shape:", diabetes_dataset.data.shape)
print("Target shape:", diabetes_dataset.target.shape)
from sklearn.datasets import load_diabetes
# Load the diabetes dataset
diabetes_dataset = load_diabetes()
# **Keys**
# Print the keys of the diabetes dataset
print(diabetes_dataset.keys())
# **Data Shape**
# Check the shape of the data and target arrays
print("Data shape:", diabetes_dataset.data.shape)
# **Target Shape**
print("Target shape:", diabetes_dataset.target.shape)
# **Split the dataset into train and test**
from sklearn.datasets import load_diabetes
from sklearn.model_selection import train_test_split
# Load the diabetes dataset
diabetes_dataset = load_diabetes()
# Split the dataset into training and testing subsets
X_train, X_test, y_train, y_test = train_test_split(
diabetes_dataset.data, diabetes_dataset.target, test_size=0.2, random_state=42
)
# Print the shapes of the training and testing subsets
print("X_train shape:", X_train.shape)
print("y_train shape:", y_train.shape)
print("X_test shape:", X_test.shape)
print("y_test shape:", y_test.shape)
# **Fit the model on test data and compute accuracy using Linear Regression**
from sklearn.datasets import load_diabetes
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_squared_error, r2_score
# Load the diabetes dataset
diabetes_dataset = load_diabetes()
# Split the dataset into training and testing subsets
X_train, X_test, y_train, y_test = train_test_split(
diabetes_dataset.data, diabetes_dataset.target, test_size=0.2, random_state=42
)
# Create a linear regression model object
linear_regression = LinearRegression()
# Train the model on the training data
linear_regression.fit(X_train, y_train)
# Make predictions on the testing data
y_pred = linear_regression.predict(X_test)
# Compute the mean squared error and r2 score of the model
# **Make slots of features on data set and evaluate using regression metrics.**
from sklearn.datasets import load_diabetes
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_squared_error, r2_score
# Load the diabetes dataset
diabetes_dataset = load_diabetes()
# Use only one feature (column 2) from the dataset
X = diabetes_dataset.data[:, 2].reshape(-1, 1)
# Split the dataset into training and testing subsets
X_train, X_test, y_train, y_test = train_test_split(
X, diabetes_dataset.target, test_size=0.2, random_state=42
)
# Create a linear regression model object
linear_regression = LinearRegression()
# Train the model on the training data
linear_regression.fit(X_train, y_train)
# Make predictions on the testing data
y_pred = linear_regression.predict(X_test)
# Compute the mean squared error and r2 score of the model
mse = mean_squared_error(y_test, y_pred)
r2 = r2_score(y_test, y_pred)
# Print the mean squared error and r2 score
print("Mean Squared Error:", mse)
print("R2 Score:", r2)
# **Apply PCA and analyze the results with results without PCA.**
from sklearn.datasets import load_diabetes
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.decomposition import PCA
from sklearn.metrics import mean_squared_error, r2_score
# Load the diabetes dataset
diabetes_dataset = load_diabetes()
# Split the dataset into training and testing subsets
X_train, X_test, y_train, y_test = train_test_split(
diabetes_dataset.data, diabetes_dataset.target, test_size=0.2, random_state=42
)
# Fit a PCA model to the training data
pca = PCA(n_components=3)
pca.fit(X_train)
# Transform the training and testing data using the PCA model
X_train_pca = pca.transform(X_train)
X_test_pca = pca.transform(X_test)
# Create a linear regression model object
linear_regression = LinearRegression()
# Train the model on the training data without PCA
linear_regression.fit(X_train, y_train)
y_pred = linear_regression.predict(X_test)
mse = mean_squared_error(y_test, y_pred)
r2 = r2_score(y_test, y_pred)
print("Without PCA:")
print("Mean Squared Error:", mse)
print("R2 Score:", r2)
# Train the model on the training data with PCA
linear_regression.fit(X_train_pca, y_train)
y_pred_pca = linear_regression.predict(X_test_pca)
mse_pca = mean_squared_error(y_test, y_pred_pca)
r2_pca = r2_score(y_test, y_pred_pca)
print("With PCA:")
print("Mean Squared Error:", mse_pca)
print("R2 Score:", r2_pca)
# **Apply permutation importance function and plot feature importances.**
from sklearn.inspection import permutation_importance
import matplotlib.pyplot as plt
# Fit a linear regression model to the training data
linear_regression = LinearRegression()
linear_regression.fit(X_train, y_train)
# Compute feature importances using permutation importance
results = permutation_importance(
linear_regression, X_test, y_test, n_repeats=10, random_state=42
)
# Get the sorted indices of feature importances
importance_indices = results.importances_mean.argsort()
# Plot the feature importances
plt.barh(range(X_test.shape[1]), results.importances_mean[importance_indices])
plt.xlabel("Permutation Importance")
plt.ylabel("Features")
plt.title("Feature Importance Using Permutation Importance")
plt.show()
|
# # **第一部分:程序前期准备
# # 第一次使用的话请全部运行一次
# # 后续如果选择了presistent不删除文件的话, 无须再运行,可以直接跳到第二部分**
# 点击左侧按钮运行, 选择Persistence->Variables and Files之后, 下次使用, 无须再重复运行
# 选择Persistence->Variables and Files之后, 下次使用, 无须再重复运行
#!pip install llvmlite0.31 --ignore-installed
# 选择Persistence->Variables and Files之后, 下次使用, 无须再重复运行
# 选择Persistence->Variables and Files之后, 下次使用, 无须再重复运行
# # 第二部分:合成视频
# # 把准备好的视频和音频文件上传到input里面, 预先把文件名称改好 (又或者更改代码上写好的文件名称), 然后运行就行
# # 方法:
# # 1. 首先在右侧的功能栏目, 点击上传的按钮
# # 2. 填写文件夹的名称
# # 3. 点击复制路径
# # 4. 把音频和视频的路径贴到代码上
# # 5. 运行
# 注意检查文件夹和文件名称是否正确, 要和上传的文件一致
# 查看生成的文件地址和名称
# 把文件从results文件夹中移出来, 在working目录的底下, 点击下载
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import sympy
import random
from sympy import *
from math import *
import hashlib
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# **Códigos complementarios**
####### Sample in ball #########
def sampleinball(tau, a):
c = np.zeros(a, int)
for i in range(a - tau, a):
j = random.randint(0, i)
s = random.randint(0, 1)
c[i] = c[j]
c[j] = (-1) ** s
return c
def modpm(r, alpha):
a = int((alpha - 1) / 2) + 1
b = int(r / 2 + 1)
if alpha % 2 == 0:
r1 = r % alpha
if r1 in range(0, b):
r1 = r1
else:
r1 = r1 - alpha
else:
r1 = int(r % alpha)
if r1 in range(0, a):
r1 = r1
else:
r1 = r1 - alpha
return r1
def decompose(r, alpha, q):
r = r % q
r0 = modpm(r, alpha)
if r - r0 == q - 1:
r1 = 0
r0 = r0 - 1
else:
r1 = int((r - r0) / alpha)
return (r1, r0)
def HighBits(r, alpha, q):
(r1, r0) = decompose(r, alpha, q)
return r1
def LowBits(r, alpha, q):
(r1, r0) = decompose(r, alpha, q)
return r0
def norma_infinito_matriz(matriz):
return max(np.linalg.norm(matriz, ord=np.inf, axis=1))
def hashear(texto, w_1):
hash_obj = hashlib.sha256(texto.encode("utf-8"))
hash_result = int.from_bytes(hash_obj.digest(), byteorder="big") % (10**w_1)
return hash_result
# **A partir de ahora presentamos la KeyGen**
def keygen(n, m, q):
A = np.random.randint(
1000000000000000000, size=(n, m)
) ##Puesto que queremos una matriz 4x3
A = A % q
s1 = (np.random.randint(1000000000000000000, size=(m, 1))) % 73
s2 = (np.random.randint(1000000000000000000, size=(n, 1))) % 119
t = (np.matmul(A, s1) + s2) % q
pk = (t, A)
sk = (t, A, s1, s2)
return pk, sk
# **El algoritmo que genera la firma**
##### Creación de firma ######
def sign(sk, M):
t = sk[0]
A = sk[1]
s1 = sk[2]
s2 = sk[3]
gamma1 = 104659
gamma2 = int(q / 3)
[n, m] = A.shape
y = (np.random.randint(1000000000000000000, size=(n, 1))) % gamma1
Ay = int(norma_infinito_matriz(np.matmul(A, y)))
w_1 = HighBits(Ay, 2 * gamma2, q)
c = hashear(M, abs(w_1))
z = y + c * s1
sigma = (z, c)
##Calculamos beta para la parte final, necesitamos los máximos
a = int(norma_infinito_matriz((c * s1)))
b = int(norma_infinito_matriz((c * s2)))
beta = int(max(a, b))
if norma_infinito_matriz(z) >= (gamma1 - beta) and LowBits(
Ay - b, 2 * gamma2, q
) >= (gamma2 - beta):
sign(sk, M)
else:
pass
return sigma
# **Algoritmo de verificación**
######## Verificación de firma #########
def verify(pk, M, sigma):
t = pk[0]
A = pk[1]
s1 = (sk[2],)
s2 = sk[3]
z = sigma[0]
c = sigma[1]
gamma1 = 104659
gamma2 = int(q / 3)
gamma2 = int(q / 3)
Az = norma_infinito_matriz(np.matmul(A, z))
ct = norma_infinito_matriz(c * t)
w1 = HighBits(Az - ct, 2 * gamma2, q)
a = int(norma_infinito_matriz((c * s1)))
b = int(norma_infinito_matriz((c * s2)))
beta = int(max(a, b))
if norma_infinito_matriz(z) < (gamma1 - beta) and c == hashear(M, w1):
return True
else:
return False
##Definimos un n aleatorio para el número de filas y columnas
n = np.random.randint(2, 30)
q = 2**23 - 2**13 + 1
##Creamos las claves públicas y privadas
pk = keygen(n, n, q)[0]
sk = keygen(n, n, q)[1]
pk, sk
##Creamos la firma
sigma = sign(sk, "Mensaje")
sigma
verify(pk, "Mensaje", sigma)
|
import numpy as np
import rawpy
import math
import cv2
from skimage.metrics import structural_similarity as ssim
from matplotlib import pyplot as plt
def mse(imageA, imageB):
err = np.sum((imageA.astype("float") - imageB.astype("float")) ** 2)
err /= float(imageA.shape[0] * imageA.shape[1])
return err
def compare(img1, img2):
img1 = cv2.cvtColor(img1, cv2.COLOR_RGB2GRAY)
img2 = cv2.cvtColor(img2, cv2.COLOR_RGB2GRAY)
MSE_val = mse(img1, img2)
SSIM_val = ssim(img1, img2) * 100
RMSE_val = math.sqrt(MSE_val)
if RMSE_val != 0:
PSNR_val = 20 * math.log10(255 / RMSE_val)
RMSE_val = 255 / pow(10.0, (PSNR_val / 20.0))
else:
PSNR_val = 100
return MSE_val, SSIM_val, RMSE_val, PSNR_val
def size_compare(raw, compressed, format_name):
decimg = cv2.imdecode(compressed, 1)
raw_size = raw.size * raw.itemsize
compressed_size = compressed.size * compressed.itemsize
print(
"raw:",
raw_size,
format_name + ":",
compressed_size,
"compression:",
raw_size / compressed_size,
)
print(
"MSE:{0[0]:.2f} SSIM:{0[1]:.2f} RMSE:{0[2]:.2f} PSNR:{0[3]:.2f}".format(
compare(raw, decimg)
)
)
fig, ax = plt.subplots(nrows=1, ncols=2, figsize=(10, 10))
ax[0].imshow(decimg)
ax[1].imshow(raw)
raw = rawpy.imread("/kaggle/input/raw-imgs/building4.CR2")
rgb = raw.postprocess(use_camera_wb=True)
raw_size = rgb.size * rgb.itemsize
quality = 35
print(quality)
encode_param = [int(cv2.IMWRITE_JPEG_QUALITY), quality]
result, jpg_img = cv2.imencode(".jpg", rgb, encode_param)
size_compare(rgb, jpg_img, "jpg")
plt.show()
result, bmp_img = cv2.imencode(".bmp", rgb)
size_compare(rgb, bmp_img, "bmp")
result, png_img = cv2.imencode(".png", rgb)
size_compare(rgb, png_img, "png")
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import re
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
df11 = pd.read_csv(
"/kaggle/input/the-world-university-rankings-2011-2023/2011_rankings.csv"
)
df11.info()
df11["location"].value_counts()
df11["scores_overall"].plot(kind="box", vert=False)
df11.describe()
df11["aliases"]
df23 = pd.read_csv(
"/kaggle/input/the-world-university-rankings-2011-2023/2023_rankings.csv"
)
df23.info()
subdf23 = df23[
[
"name",
"stats_number_students",
"stats_student_staff_ratio",
"stats_pc_intl_students",
"stats_female_male_ratio",
]
]
subdf23.head()
# load the datasets of top university
df22 = pd.read_csv(
"/kaggle/input/the-world-university-rankings-2011-2023/2022_rankings.csv"
)
df21 = pd.read_csv(
"/kaggle/input/the-world-university-rankings-2011-2023/2021_rankings.csv"
)
df20 = pd.read_csv(
"/kaggle/input/the-world-university-rankings-2011-2023/2020_rankings.csv"
)
df19 = pd.read_csv(
"/kaggle/input/the-world-university-rankings-2011-2023/2019_rankings.csv"
)
df22.info()
df23["location"].value_counts()[:10]
# ### Viz ideas
# 1. Top Uni rankings over a period of past 5 years
# 2. Top Uni with rankings change in over past 5 years in all measures
# 3. radar chart of of scores by individual uni.
# ### 1. Top university ranking over past 5 years
def ranking_by_year(df, year):
rank_df = df[["name", "rank"]][:10]
rank_df["year"] = year
return rank_df
rank_19df = ranking_by_year(df19, 2019)
rank_20df = ranking_by_year(df20, 2020)
rank_21df = ranking_by_year(df21, 2021)
rank_22df = ranking_by_year(df22, 2022)
rank_23df = ranking_by_year(df23, 2023)
combined_df = pd.concat([rank_19df, rank_20df, rank_21df, rank_22df, rank_23df])
combined_df["rank"] = combined_df["rank"].apply(
lambda x: "".join([re.sub("^=", "", s) for s in x])
)
combined_df
# plot the the rankings of each over past 5 years
plt.figure(figsize=(10, 7))
sns.lineplot(x="year", y="rank", hue="name", data=combined_df)
plt.show()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# IMPORT ADDITIONAL LIBRARIES
import matplotlib.pyplot as plt
# CLEAN UP AND REARRANGE DATA
dataset = pd.read_csv("/kaggle/input/real-estate-price-prediction/Real estate.csv")
dataset = dataset.set_index("No")
dataset.rename(
columns={
"X1 transaction date": "transaction date",
"X2 house age": "house age",
"X3 distance to the nearest MRT station": "distance to the nearest MRT station",
"X4 number of convenience stores": "number of convenience stores",
"X5 latitude": "latitude",
"X6 longitude": "longitude",
"Y house price of unit area": "house price of unit area",
},
inplace=True,
)
dataset.head()
# DESCRIBE AND SUMMARISE DATA
dataset.describe()
dataset.info()
dataset.hist(bins=50, figsize=(20, 15))
plt.show()
# SPLIT DATA INTO TEST AND TRAINING SET
# Create a stratiffied shuffle of data to evenly shuffle the data
dataset["distance_cat"] = pd.cut(
dataset["distance to the nearest MRT station"],
bins=[0, 200, 500, 1000, 3000, np.inf],
labels=[1, 2, 3, 4, 5],
)
dataset["distance_cat"].hist()
# Split the Data
from sklearn.model_selection import StratifiedShuffleSplit
split = StratifiedShuffleSplit(n_splits=1, test_size=0.2, random_state=42)
for train_index, test_index in split.split(dataset, dataset["distance_cat"]):
strat_train_set = dataset.reindex(train_index)
strat_test_set = dataset.reindex(test_index)
# Drop possible NaN values
strat_train_set.dropna(axis=0, inplace=True)
# Drop the shuffle column
for set_ in (strat_train_set, strat_test_set):
set_.drop("distance_cat", axis=1, inplace=True)
# Check for correlation in the Data
corr_matrix = dataset.corr()
corr_matrix["house price of unit area"]
# Prepare the data for Machine Learning Algorithm
# Create inputs and outputs
dataset = strat_train_set.drop("house price of unit area", axis=1)
dataset_labels = strat_train_set["house price of unit area"].copy()
# Perform feature Scaling on the Data and keep scaled data in a pipeline
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import StandardScaler
num_pipeline = Pipeline([("std_scaler", StandardScaler())])
prepared_data = num_pipeline.fit_transform(dataset)
# Perform Random forest Regression on data
from sklearn.ensemble import RandomForestRegressor
forest_reg = RandomForestRegressor()
forest_reg.fit(prepared_data, dataset_labels)
# Evaluate algorithm performance
from sklearn.metrics import mean_squared_error
from sklearn.model_selection import cross_val_score
dataset_predictions = forest_reg.predict(prepared_data)
forest_mse = mean_squared_error(dataset_labels, dataset_predictions)
forest_rmse = np.sqrt(forest_mse)
forest_rmse
def display_scores(scores):
print("Scores:", scores)
print("Mean:", scores.mean())
print("Standard deviation:", scores.std())
forest_scores = cross_val_score(
forest_reg, prepared_data, dataset_labels, scoring="neg_mean_squared_error", cv=10
)
forest_rmse_scores = np.sqrt(-forest_scores)
display_scores(forest_rmse_scores)
# Perform Grid Search to determine best parameters to use
from sklearn.model_selection import GridSearchCV
param_grid = {"n_estimators": [3, 10, 30], "max_features": [2, 4, 6, 8]}
forest_reg = RandomForestRegressor()
grid_search = GridSearchCV(
forest_reg,
param_grid,
cv=5,
scoring="neg_mean_squared_error",
return_train_score=True,
verbose=3,
)
grid_search.fit(prepared_data, dataset_labels)
# Test on test Data
final_model = grid_search.best_estimator_
X_test = strat_test_set.drop("house price of unit area", axis=1)
y_test = strat_test_set["house price of unit area"].copy()
X_test_prepared = num_pipeline.transform(X_test)
final_prediction = final_model.predict(X_test_prepared)
final_mse = mean_squared_error(y_test, final_prediction)
final_rmse = np.sqrt(final_mse)
final_rmse
# View Confidence Interval
from scipy import stats
confidence = 0.95
squared_errors = (final_prediction - y_test) ** 2
np.sqrt(
stats.t.interval(
confidence,
len(squared_errors) - 1,
loc=squared_errors.mean(),
scale=stats.sem(squared_errors),
)
)
|
# # Task for Today
# ***
# ## Fertilizer Type Prediction
#
# Given *data about different fertilizers*, let's try to predict the **type of fertilizer** for a given record.
#
# We will use a random forest classifier to make our predictions.
# # Getting Started
import numpy as np
import pandas as pd
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import OneHotEncoder, StandardScaler
from sklearn.compose import ColumnTransformer
from sklearn.pipeline import Pipeline
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import classification_report
data = pd.read_csv("../input/fertilizer-prediction/Fertilizer Prediction.csv")
data
data.info()
# # Preprocessing Pipeline
y = data["Fertilizer Name"].copy()
X = data.drop("Fertilizer Name", axis=1).copy()
X_train, X_test, y_train, y_test = train_test_split(
X, y, train_size=0.7, shuffle=True, random_state=1
)
nominal_transformer = Pipeline(steps=[("onehot", OneHotEncoder(sparse=False))])
preprocessor = ColumnTransformer(
transformers=[("nominal", nominal_transformer, ["Soil Type", "Crop Type"])],
remainder="passthrough",
)
model = Pipeline(
steps=[
("preprocessor", preprocessor),
("scaler", StandardScaler()),
("classifier", RandomForestClassifier()),
]
)
# # Training
model.fit(X_train, y_train)
# # Results
print("Test Accuracy: {:.2f}%".format(model.score(X_test, y_test) * 100))
y_pred = model.predict(X_test)
clr = classification_report(y_test, y_pred)
print("Classification Report:\n----------------------\n", clr)
|
# # Apple Stock Price Using Support Vector Regression
# ## Table of Content
# ### 1. What is Support Vector Regression?
# ### 2. Importing Libraries
# ### 3. Loading Dataset
# ### 4. Data PreProcessing
# ### 5. EDA
# ### 6. Data Splitting
# ### 7. Model Training
# ### 8. Model Evaluation
# ### 9. Conclusion
# ## 1. What is Support Vector Regression?
# ### Support Vector Regression (SVR) is a type of regression analysis that uses Support Vector Machines (SVMs) to model the relationship between input variables and output variables. SVR is a powerful machine learning algorithm that is used for solving regression problems.
# ### The main idea behind SVR is to find a hyperplane that maximizes the margin between the predicted output and the actual output, while also minimizing the number of points that fall outside the margin. The hyperplane is found by minimizing a cost function that penalizes errors in the predictions.
# ### SVR is widely used in fields such as finance, economics, and engineering for predicting numerical values, such as stock prices, housing prices, and product demand.
# 
# ## 2. Importing Libraries
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import plotly.express as px
import seaborn as sns
# ## 3. Loading Dataset
aapl_stock = pd.read_csv("/kaggle/input/apple-stock-price-from-19802021/AAPL.csv")
aapl_stock.head()
aapl_stock.info()
# ## 4. Data Preprocessing
aapl_stock.isnull().sum()
# ### Here, In this Dataset No null or missing values.
# remove the unnecessary features
del aapl_stock["Adj Close"]
aapl_stock.head()
aapl_stock.shape
X = aapl_stock[["Open", "High", "Low"]]
Y = aapl_stock["Close"]
# ## 5. EDA
plt.figure(figsize=(10, 4))
sns.scatterplot(aapl_stock["High"])
plt.title("Apple Stocks from 2021 to 2022")
plt.ylabel("Stocks")
plt.xlabel("Months")
plt.xticks(rotation=90)
plt.show()
sns.heatmap(aapl_stock.corr(), annot=True)
# ## 6. Data Splitting
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(X, Y, test_size=0.3)
print("x_train:", x_train.shape)
print("x_test: ", x_test.shape)
print("y_train:", y_train.shape)
print("y_test: ", y_test.shape)
# ## 7. Model Training (SVR)
from sklearn.svm import SVR
model = SVR()
model.fit(x_train, y_train)
model.score(x_test, y_test)
predict = model.predict(x_test)
predict
# ## 8. Model Evaluation
# ### 1. Mean Absolute Error
from sklearn.metrics import mean_absolute_error
print(mean_absolute_error(predict, y_test))
# ### 2. Mean Squared Error
from sklearn.metrics import mean_squared_error
print(mean_squared_error(y_test, predict))
# ### 3. Mean Squared Log Error
from sklearn.metrics import mean_squared_log_error
mean_squared_log_error(y_test, predict, squared=False)
# ### 4. R2 Squared
from sklearn.metrics import r2_score
r2 = r2_score(y_test, predict)
print(r2)
# ### 5. Adjusted R Squared
n = 60
k = 10
r2 = r2_score(y_test, predict) # r2 = r square score
adj_r2_score = 1 - ((1 - r2) * (n - 1) / (n - k - 1))
print(adj_r2_score)
|
# Import libraries, packages, modules, functions, etc...
import numpy as np
import cv2
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import (
Activation,
Dense,
Flatten,
BatchNormalization,
Conv2D,
MaxPool2D,
Dropout,
)
from tensorflow.keras.optimizers import Adam
from tensorflow.keras.metrics import categorical_crossentropy
from tensorflow.keras.preprocessing.image import ImageDataGenerator
from sklearn.metrics import confusion_matrix
import itertools
import os
import shutil
import random
import glob
import matplotlib.pyplot as plt
from sklearn.preprocessing import MinMaxScaler
import warnings
from sklearn.utils import shuffle
from sklearn.model_selection import train_test_split
import skimage
from skimage.restoration import (
denoise_nl_means,
estimate_sigma,
denoise_tv_chambolle,
denoise_bilateral,
denoise_wavelet,
unsupervised_wiener,
)
from scipy.signal import convolve2d as conv2
from skimage.filters import median
import copy
from tqdm import tqdm
from scipy import ndimage as nd
import multiprocessing
from skimage.metrics import peak_signal_noise_ratio
from skimage.metrics import mean_squared_error
from skimage.metrics import structural_similarity as ssim
from itertools import repeat
import zipfile
import os
from IPython.display import FileLink
from sklearn.svm import SVC
from keras.callbacks import EarlyStopping
import pandas as pd
from sklearn.cluster import KMeans
from sklearn import metrics
from sklearn.cluster import MiniBatchKMeans
from sklearn.metrics import accuracy_score
from tensorflow.keras.preprocessing import image
from skimage.morphology import disk
from tensorflow.keras.optimizers import SGD
warnings.simplefilter(action="ignore", category=FutureWarning)
import natsort
from medpy.filter.smoothing import anisotropic_diffusion
# # **Augmentation**
# rotation_range: Int. Degree range for random rotations.
# width_shift_range: Float, 1-D array-like or int(measured as a fraction of the width of the image.)
# height_shift_range: Float, 1-D array-like or int(measured as a fraction of the width of the image.)
# shear_range: Float. Shear Intensity (Shear angle in counter-clockwise direction in degrees)
# zoom_range: Float or [lower, upper]. Range for random zoom. If a float, [lower, upper] = [1-zoom_range, 1+zoom_range].
# channel_shift_range: Float. Range for random channel shifts.
# horizontal_flip: Boolean. Randomly flip inputs horizontally.
gen = ImageDataGenerator(
rotation_range=90,
width_shift_range=0.1,
height_shift_range=0.1,
shear_range=0.15,
zoom_range=0.2,
channel_shift_range=10.0,
horizontal_flip=True,
vertical_flip=True,
featurewise_center=True,
featurewise_std_normalization=True,
)
def Augmentation(path_from, path_to):
ind = 1
for chosen_image in os.listdir(path_from):
# Choosing a imagefrom the directory and setting its dimensions
if (
chosen_image.endswith(".png")
or chosen_image.endswith(".jpg")
or chosen_image.endswith(".jpeg")
):
image_path1 = path_from + chosen_image
images = image.load_img(image_path1, target_size=(128, 128, 3))
images = image.img_to_array(images)
images = np.expand_dims(images, axis=0)
# Saving the images to the directory
# Next, we'll generate batches of augmented images from the original image.
# The flow() function takes numpy data and generates batches of augmented data.
for x, val in zip(
gen.flow(
images, # image we chose
save_to_dir=path_to, # Directory to save the data
save_prefix="x", # it will save the images as 'x_0_912' some number for every new augmented image
save_format="png",
),
range(5),
): # here we define a range because we want 10 augmented images
pass
# Renaming the augmentated images and masks saved to the format we require
for i in os.listdir(path_to):
image_path = path_to + i
new_path = path_to + "KS" + str(ind) + ".png"
ind += 1
os.rename(image_path, new_path)
count_value = 1
def Rename(path_to, name_val):
global count_value
for i in os.listdir(path_to):
image_path = path_to + i
new_path = path_to + name_val + str(count_value) + ".png"
count_value += 1
os.rename(image_path, new_path)
count_value = 1
# Rename the Images
Rename("/kaggle/working/Aug_Kidney_Stone/", "KS")
Rename("/kaggle/working/Aug_Normal/", "N")
# Augmentation for Kidney Stones and Normal Kidney Data
Augmentation(
"/kaggle/input/kidneystone/CT_SCAN/Kidney_stone/",
"/kaggle/working/Aug_Kidney_Stone/",
)
Augmentation("/kaggle/input/kidneystone/CT_SCAN/Normal/", "/kaggle/working/Aug_Normal/")
# Total Number of Images after augmentation
print(
"Number of kidney stone images",
len(os.listdir("/kaggle/working/Aug_Kidney_Stone/")),
)
print("Number of Normal kidney images", len(os.listdir("/kaggle/working/Aug_Normal/")))
# Display Images in a sorted manner
path = "/kaggle/working/Aug_Kidney_Stone/"
imglist_KS = natsort.natsorted(os.listdir(path))
path = "/kaggle/working/Aug_Normal/"
imglist_Normal = natsort.natsorted(os.listdir(path))
# Zip files and working kaggle directly for data accumulation
# The shutil module offers a number of high-level operations on files and collections of files. In particular, functions are provided which support file copying and removal.
# import shutil
# The shutil. rmtree() function is used to delete an entire directory
# shutil.rmtree("/kaggle/working/")
# Create a zip file for file or folder in the directory
def zip_dir(directory=os.curdir, file_name="directory.zip"):
"""
zip all the files in a directory
Parameters
_____
directory: str
directory needs to be zipped, default is current working directory
file_name: str
the name of the zipped file (including .zip), default is 'directory.zip'
Returns
_____
Creates a hyperlink, which can be used to download the zip file)
"""
os.chdir(directory)
zip_ref = zipfile.ZipFile(file_name, mode="w")
for folder, _, files in os.walk(directory):
for file in files:
if file_name in file:
pass
else:
zip_ref.write(os.path.join(folder, file))
return FileLink(file_name)
# Create a zip file for file or folder in the directory
zip_dir()
# Creating a directory
# makedirs creates all directories that are part of path and do not yet exist. mkdir creates only the rightmost directory of path
os.mkdir("/kaggle/input/kidneystone/CT_SCAN/Kidney_stone/Aug_KS/")
os.makedirs("/kaggle/working/Aug_Kidney_Stone")
os.makedirs("/kaggle/working/Aug_Normal")
# Copy all the contents from one directory to another directory
# # **Data Accumulation**
# Get the complete data of kidney stone images
def Get_data(dir, catagories, data):
for category in catagories:
path = os.path.join(dir, category)
class_number = catagories.index(category)
for img in os.listdir(path):
try:
img_array = cv2.imread(os.path.join(path, img))
new_image = cv2.resize(img_array, (128, 128))
data.append([new_image, class_number])
except Exception as e:
pass
data = []
# Augmented Dataset
Get_data(
"/kaggle/input/combined-aug-ks/KS_Detection", ["Combined_N", "Combined_KS"], data
)
# Original Dataset
Get_data("/kaggle/input/kidneystone/CT_SCAN", ["Kidney_stone", "Normal"], data)
# Categories of Kidney Stone images
catagories = ["Kidney_stone", "Normal"]
print(catagories.index("Kidney_stone"))
print(catagories.index("Normal"))
print(
"Number of kidney stone images",
len(os.listdir("/kaggle/input/kidneystone/CT_SCAN/Kidney_stone")),
)
print(
"Number of Normal kidney images",
len(os.listdir("/kaggle/input/kidneystone/CT_SCAN/Normal")),
)
print(data[143][0].shape)
plt.imshow(data[143][0])
plt.show()
# Extract Features and Labels from the data
Features = []
Labels = []
for features, labels in data:
Features.append(features)
Labels.append(labels)
Features = np.array(Features)
Labels = np.array(Labels)
# Shape of Features and Labels
print(Features.shape)
print(Labels.shape)
# # **Preprocessing**
# **Denoising**
def Processed(Processed_images):
img_per_row = 4
fig, ax = plt.subplots(
nrows=2,
ncols=img_per_row,
figsize=(10, 10),
subplot_kw=dict(xticks=[], yticks=[]),
)
for row in [0, 1]:
for col in range(img_per_row):
if row * img_per_row + col == 7:
break
if (
Processed_images[row * img_per_row + col][1] == "Bilateral_Denoised"
or Processed_images[row * img_per_row + col][1] == "Wavelet_Denoised"
or Processed_images[row * img_per_row + col][1] == "Non_Local_Means"
):
ax[row, col].imshow(
Processed_images[row * img_per_row + col][0].astype("float64"),
cmap="gray",
)
else:
ax[row, col].imshow(
Processed_images[row * img_per_row + col][0].astype("uint8"),
cmap="gray",
)
ax[row, col].set_title(Processed_images[row * img_per_row + col][1])
plt.axis("off")
plt.show()
# Filters
def AnisotropicFilter_wholedataset(noised_dataset, niter, kappa, gamma, option, size):
anisotropic_dataset = copy.deepcopy(noised_dataset)
for i in tqdm(range(anisotropic_dataset.shape[0])):
anisotropic_dataset[i] = anisotropic_diffusion(
noised_dataset[i, :, :, :],
niter=niter,
kappa=kappa,
gamma=gamma,
option=option,
).reshape(size, size, 1)
return anisotropic_dataset
def median_wholedataset(noised_dataset, filtersize, size):
median_wholedata = copy.deepcopy(noised_dataset)
for i in tqdm(range(noised_dataset.shape[0])):
median_wholedata[i] = median(
noised_dataset[i, :, :, :][:, :, 0], np.ones((filtersize, filtersize))
).reshape(size, size, 1)
return median_wholedata
def wavelet_wholedataset(noised_dataset, sigma, wavelet_levels, size):
wavelet_wholedata = copy.deepcopy(noised_dataset)
for i in tqdm(range(noised_dataset.shape[0])):
wavelet_wholedata[i] = denoise_wavelet(
noised_dataset[i, :, :, :],
sigma=sigma,
channel_axis=-1,
wavelet_levels=wavelet_levels,
rescale_sigma=True,
).reshape(size, size, 1)
return wavelet_wholedata
def BilateralFilter_wholedataset(
noised_dataset, sigma_color, sigma_spatial, channel_axis, size
):
bilateral_dataset = copy.deepcopy(noised_dataset)
for i in tqdm(range(bilateral_dataset.shape[0])):
bilateral_dataset[i] = denoise_bilateral(
noised_dataset[i, :, :, :],
sigma_color=sigma_color,
sigma_spatial=sigma_spatial,
channel_axis=channel_axis,
).reshape(size, size, 1)
return bilateral_dataset
def GaussianFilter_wholedataset(noised_dataset, sigma):
gaussian_dataset = copy.deepcopy(noised_dataset)
for i in tqdm(range(gaussian_dataset.shape[0])):
gaussian_dataset[i] = nd.gaussian_filter(
tuple(noised_dataset[i, :, :, :]), sigma=sigma
)
return gaussian_dataset
def non_local_mean(image):
sigma_est = np.mean(estimate_sigma(image, channel_axis=-1))
patch_kw = dict(
patch_size=5, # 5x5 patches
patch_distance=6, # 13x13 search area
channel_axis=-1,
)
denoise_fast = denoise_nl_means(
image, h=0.6 * sigma_est, sigma=sigma_est, fast_mode=True, **patch_kw
)
return denoise_fast
# Apply bilateral filter with d = 15
# sigmaColor = sigmaSpace = 75
def cv2bilateralFilter(image, d, sigmaColor, sigmaSpace):
bilateral = cv2.bilateralFilter(image, d, sigmaColor, sigmaSpace)
bilateral = bilateral.astype("uint8")
return bilateral
# Execution
# ani = AnisotropicFilter_wholedataset(noised_dataset, 50, 20, 0.2, 1)
# bi = BilateralFilter_wholedataset(noised_dataset, 15, -1)
# gauss = GaussianFilter_wholedataset(noised_dataset, 2)
# median_img = median(Features[0, :, :, :][:,:,0], disk(3), mode = 'constant', cval = 0.0).reshape(128, 128, 1)
# wavelet_img = denoise_wavelet(Features[0, :, :, :], sigma = 0.12, channel_axis = -1,convert2ycbcr=True, method='BayesShrink', mode='soft', rescale_sigma=True).reshape(128, 128, 3)
# bilateral_image = denoise_bilateral(Features[0], sigma_spatial = 10, channel_axis = -1)
# anisotropic_image = anisotropic_diffusion(Features[700,:,:,:][:,:,0], niter = 100, kappa = 10, gamma = 0.02, option=1)
# anisotropic_image = anisotropic_diffusion(Features[700,:,:,:][:,:,0], niter = 50, kappa = 10, gamma = 0.02, option=1)
# anisotropic_image = anisotropic_diffusion(Features[60,:,:,:][:,:,0], niter = 50, kappa = 5, gamma = 0.005, option=1)
# anisotropic_image = anisotropic_diffusion(Features[600,:,:,:][:,:,0], niter = 50, kappa = 5, gamma = 0.000001, option=1)
# anisotropic_image = anisotropic_image.astype('uint8')
# Metrics
def psnr_wholedataset(dataset_original, dataset_denoised):
sumpsnr = 0
avgpsnr = 0
for i in tqdm(range(dataset_original.shape[0])):
true_min, true_max = np.min(dataset_original[i, :, :, :]), np.max(
dataset_original[i, :, :, :]
)
dataRange = abs(true_min) + abs(true_max)
psnr = peak_signal_noise_ratio(
dataset_original[i], dataset_denoised[i], data_range=dataRange
)
sumpsnr += psnr
avgpsnr = sumpsnr / dataset_original.shape[0]
return avgpsnr
# psnr_wholedataset(data, noised_dataset)
# Execution
# true_min, true_max = np.min(Features[0]), np.max(Features[0])
# dataRange = abs(true_min) + abs(true_max)
# print("PSNR: ", peak_signal_noise_ratio(Features[143, :, :, :][:,:,0], anisotropic_image, data_range = dataRange))
# print("MSE: ", mean_squared_error(Features[143,:,:,:][:,:,0], anisotropic_image))
# print("SSIM: ", ssim(Features[143, :, :, :][:,:,0], anisotropic_image, multichannel=True, gaussian_weights = True, sigma=1.5, use_sample_covariance=False))
# plt.imshow(np.hstack((wavelet_img, Features[143, :, :, :].reshape(128,128,3))), cmap = "gray")
true_min, true_max = np.min(Features[143]), np.max(Features[143])
dataRange = abs(true_min) + abs(true_max)
bilateral = cv2bilateralFilter(Features[143, :, :, :], 15, 75, 75)
print(
"PSNR: ",
peak_signal_noise_ratio(Features[143, :, :, :], bilateral, data_range=dataRange),
)
print("MSE: ", mean_squared_error(Features[143, :, :, :], bilateral))
print(
"SSIM: ",
ssim(
Features[143, :, :, :],
bilateral,
multichannel=True,
gaussian_weights=True,
sigma=1.5,
use_sample_covariance=False,
),
)
anisotropic_image = anisotropic_diffusion(
Features[143, :, :, :][:, :, 0], niter=50, kappa=5, gamma=0.005, option=1
)
bilateral_image = denoise_bilateral(Features[143], sigma_spatial=10, channel_axis=-1)
wavelet_img = denoise_wavelet(
Features[143, :, :, :],
sigma=0.12,
channel_axis=-1,
convert2ycbcr=True,
method="BayesShrink",
mode="soft",
rescale_sigma=True,
).reshape(128, 128, 3)
median_img = median(
Features[143, :, :, :][:, :, 0], disk(3), mode="constant", cval=0.0
).reshape(128, 128, 1)
guassian_img = nd.gaussian_filter(tuple(Features[143, :, :, :]), sigma=2)
non_local_mean_img = non_local_mean(Features[143])
Processed_images = [
[Features[143, :, :, :], "Original_Image"],
[anisotropic_image, "Anisotropic_Denoised"],
[bilateral_image, "Bilateral_Denoised"],
[wavelet_img, "Wavelet_Denoised"],
[median_img, "Median_Denoised"],
[guassian_img, "Gaussian_Denoised"],
[non_local_mean_img, "Non_Local_Means"],
]
Processed(Processed_images)
# Denoising the given data with anisotropic Filter
true_min, true_max = np.min(Features[0]), np.max(Features[0])
dataRange = abs(true_min) + abs(true_max)
anis_denoised = []
for i in range(len(Features)):
anisotropic_image = anisotropic_diffusion(
Features[i, :, :, :], niter=100, kappa=10, gamma=0.02, option=1
)
anis_denoised.append(anisotropic_image)
anis_denoised = np.array(anis_denoised)
# **Cropping, Thersholding(or masking), Edge Detection, Morphological Analysis**
def crop(image_to_be_cropped):
height, width = image_to_be_cropped.shape[:2]
start_row, start_col = int(height * 0.24), int(width * 0.24)
end_row, end_col = int(height * 0.78), int(width * 0.78)
cropped_image = image_to_be_cropped[start_row:end_row, start_col:end_col]
return cropped_image
def threshold(img, thresh1=150):
return ((img > thresh1) * 255).astype("uint8")
def edge_detection(Image):
img = cv2.Canny(Image, 100, 200)
return img
# To see the pixel values division in the images
def hist_plot(Image):
plt.hist(Image.flat, bins=100, range=(0, 255))
plt.show()
# Convert BGR image to Gray Scale image
gray_image = cv2.cvtColor(data[143][0], cv2.COLOR_BGR2GRAY)
# expanding dimensions from 128,128 to 128,128,1
gray_image = np.expand_dims(gray_image, axis=-1)
plt.imshow(gray_image, cmap="gray")
plt.show()
# Crop a Image
cropped_image = crop(Features[143, :, :, :])
plt.imshow(cropped_image)
plt.show()
# Edge Detection
img = edge_detection(gray_image)
plt.imshow(img, cmap="gray")
plt.show()
# Threshold masking using OTSU method
# Automatic Thresholding
# Otsu's method looks at every possible value for the threshold between background and foreground, calculates the variance within each of the two clusters, and selects the value for which the weighted sum of these variances is the least.
re, th = cv2.threshold(gray_image, 0, 255, cv2.THRESH_BINARY + cv2.THRESH_OTSU)
plt.imshow(th, cmap="gray")
plt.show()
# See the pixel values division in the images
hist_plot(gray_image)
# Manual Thresholding on the basis of pixel divisions
img = threshold(gray_image)
plt.imshow(img, cmap="gray")
plt.show()
# Mask of Image
lower_yellow = np.array([35, 255, 255])
upper_yellow = np.array([25, 50, 70])
lower_blue = np.array([110, 50, 50])
upper_blue = np.array([130, 255, 255])
lower_white = np.array([180, 18, 255])
upper_white = np.array([0, 0, 231])
lower_black = np.array([180, 255, 30])
upper_black = np.array([0, 0, 0])
bgr_image = cv2.cvtColor(Features[143, :, :][:, :, 0], cv2.COLOR_GRAY2BGR)
hsv = cv2.cvtColor(bgr_image, cv2.COLOR_BGR2HSV)
mask = cv2.inRange(hsv, lower_black, upper_black)
detected_output = cv2.bitwise_and(hsv, hsv, mask=mask)
plt.imshow(detected_output, cmap="gray")
plt.show()
# Crop all the input images
Cropped_Features = []
for i in range(len(Features)):
cropped_image = crop(Features[i, :, :, :])
Cropped_Features.append(cropped_image)
Cropped_Features = np.array(Cropped_Features)
# Denoise all the input images with Bilateral Filte
# On original images
bil_de = []
for i in range(len(Features)):
bilateral_de = cv2.bilateralFilter(Features[i], 15, 75, 75)
bil_de.append(bilateral_de)
bil_de = np.array(bil_de)
# On cropped images
bilateral_denoised = []
for i in range(len(Cropped_Features)):
bilateral = cv2.bilateralFilter(Cropped_Features[i], 15, 75, 75)
bilateral_denoised.append(bilateral)
bilateral_denoised = np.array(bilateral_denoised)
# Threshold masking on all the input images
# Original
thresholded = []
for i in range(len(bilateral_denoised)):
temp = threshold(bilateral_denoised[i])
thresholded.append(temp)
thresholded = np.array(thresholded)
# Cropped
thresholded = []
for i in range(len(Cropped_Features)):
temp = threshold(Cropped_Features[i])
thresholded.append(temp)
thresholded = np.array(thresholded)
# # **Splitting of Data**
Xtrain, Xtest, Ytrain, Ytest = train_test_split(
Features, Labels, test_size=0.30, random_state=80, shuffle=True
)
print(len(Xtrain), len(Xtest), len(Ytrain), len(Ytest))
# Converting the list to a numpy array as a requirement for the input in fit function.
Xtrain = np.array(Xtrain)
Xtest = np.array(Xtest)
Ytrain = np.array(Ytrain)
Ytest = np.array(Ytest)
# # **Different types of Model Training and Testing**
# **Convolutional Neural Network(CNN)**
# Model 1
model1 = Sequential(
[
(Conv2D(128, 3, 3, input_shape=(128, 128, 3))),
Activation("relu"),
MaxPool2D(pool_size=(2, 2)),
Conv2D(128, 3, 3),
Activation("relu"),
MaxPool2D(pool_size=(2, 2)),
Flatten(),
Dense(128),
Activation("relu"),
Dropout(0.4),
Dense(1),
Activation("sigmoid"),
]
)
# Model 2
model2 = Sequential(
[
Conv2D(
filters=32,
kernel_size=(3, 3),
activation="relu",
padding="same",
input_shape=(128, 77, 3),
),
MaxPool2D(pool_size=(2, 2), strides=2),
Conv2D(filters=64, kernel_size=(3, 3), activation="relu", padding="same"),
MaxPool2D(pool_size=(2, 2), strides=2),
Flatten(),
Dense(units=1, activation="softmax"),
]
)
# Model 3
model3 = Sequential()
model3.add(
Conv2D(32, (3, 3), activation="relu", input_shape=(128, 128, 3), padding="same")
)
model3.add(MaxPool2D(2))
model3.add(Dropout(0.2))
model3.add(Conv2D(32, (3, 3), activation="relu", padding="same"))
model3.add(MaxPool2D(pool_size=(2, 2), strides=2))
model3.add(Dropout(0.4))
model3.add(Conv2D(64, (3, 3), activation="relu", padding="same"))
model3.add(MaxPool2D(pool_size=(2, 2), strides=2))
model3.add(Dropout(0.5))
model3.add(Conv2D(64, (3, 3), activation="relu", padding="same"))
model3.add(MaxPool2D(pool_size=(2, 2), strides=2))
model3.add(Conv2D(128, (3, 3), activation="relu", padding="same"))
model3.add(MaxPool2D(pool_size=(2, 2), strides=2))
model3.add(Conv2D(128, (3, 3), activation="relu", padding="same"))
model3.add(MaxPool2D(pool_size=(2, 2), strides=2))
model3.add(Dropout(0.6))
model3.add(Flatten())
model3.add(Dense(512, activation="relu"))
model3.add(Dense(1, activation="sigmoid"))
model1.summary()
model2.summary()
model3.summary()
# compile function configures the Sequential model for training.
# Optimizers are algorithms or methods used to change the attributes of the neural network such as weights and learning rate to reduce the losses. Optimizers are used to solve optimization problems by minimizing the function.
# Crossentropy loss function when there are two or more label classes.
# We expect labels to be provided as integers.
# model.compile(optimizer = Adam(learning_rate=0.001), loss='binary_crossentropy',metrics=['accuracy'])
# model.compile(optimizer='rmsprop', loss='categorical_crossentropy',metrics=['accuracy'])
sgd = SGD(learning_rate=0.001, decay=1e-6, momentum=0.9, nesterov=True)
model3.compile(loss="binary_crossentropy", optimizer=sgd, metrics=["accuracy"])
# verbose=2 just specifies how much output to the console we want to see during each epoch of training. The verbosity levels range from 0 to 2, so we're getting the most verbose output.
# model.fit(Xtrain, Ytrain, epochs = 50, batch_size = 20, verbose = 2)
# Define a callback for early stopping
early_stopping = EarlyStopping(monitor="val_loss", patience=10)
# Train the model
history = model3.fit(
Xtrain, Ytrain, epochs=200, batch_size=20, callbacks=[early_stopping]
)
# To this function, we pass in the test samples x, specify a batch_size, and specify which level of verbosity we want from log messages during prediction generation. The output from the predictions won't be relevant for us, so we're setting verbose=0 for no output.
# Note that, unlike with training and validation sets, we do not pass the labels of the test set to the model during the inference stage.
predictions = model3.predict(x=Xtest, batch_size=20, verbose=0)
Accuracy = model3.evaluate(Xtest, Ytest, verbose=0)
print("Accuracy: ", Accuracy[1] * 100)
# **Support Vector Machine (SVM)**
Xtrain.shape
Xtest.shape
Xtrain_SVC = Xtrain.reshape(1126, 128 * 128 * 3)
Xtest_SVC = Xtest.reshape(483, 128 * 128 * 3)
model_SVC = SVC(C=1, kernel="poly", gamma="auto")
model_SVC.fit(Xtrain_SVC, Ytrain)
prediction = model_SVC.predict(Xtest_SVC)
accuracy = model_SVC.score(Xtest_SVC, Ytest)
train_accuracy = model_SVC.score(Xtrain_SVC, Ytrain)
print("Train_Accuracy", train_accuracy * 100)
print("Test_Accuracy ", accuracy * 100)
# **Confusion Matrix**
# cm = confusion_matrix(y_true = Ytest, y_pred = np.argmax(predictions,axis=-1))
cm = confusion_matrix(Ytest, (predictions > 0.75) * 1)
# Function for plotting the confusion matrix
def plot_confusion_matrix(
cm, classes, normalize=False, title="Confusion matrix", cmap=plt.cm.Blues
):
"""
This function prints and plots the confusion matrix.
Normalization can be applied by setting `normalize=True`.
"""
plt.imshow(cm, interpolation="nearest", cmap=cmap)
plt.title(title)
plt.colorbar()
tick_marks = np.arange(len(classes))
plt.xticks(tick_marks, classes, rotation=45)
plt.yticks(tick_marks, classes)
if normalize:
cm = cm.astype("float") / cm.sum(axis=1)[:, np.newaxis]
print("Normalized confusion matrix")
else:
print("Confusion matrix, without normalization")
print(cm)
thresh = cm.max() / 2.0
for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):
plt.text(
j,
i,
cm[i, j],
horizontalalignment="center",
color="white" if cm[i, j] > thresh else "black",
)
plt.tight_layout()
plt.ylabel("True label")
plt.xlabel("Predicted label")
cm_plot_labels = ["Normal", "Kidnet_Stone"]
plot_confusion_matrix(cm=cm, classes=cm_plot_labels, title="Confusion Matrix")
def image_prediction(img):
plt.imshow(img)
img1 = img.reshape(1, 128, 128, 3)
predict = model3.predict(img1)
if (predict > 0.75) * 1:
print("The condition is normal and stable.")
else:
print("Person is having kidney stone(s)")
image_prediction(Xtest[143])
image_prediction(Xtest[50])
# **KMeans**
temp = Xtrain
temp = temp.reshape(1126, 128 * 128 * 3)
kmeans_model = KMeans(n_clusters=2, max_iter=100).fit(temp)
tempTest = Xtest
tempTest = tempTest.reshape(483, 128 * 128 * 3)
predictions = kmeans_model.predict(tempTest)
tempY = Ytest.reshape(483, 1)
print(accuracy_score(Ytest, predictions))
predictionsTrain = kmeans_model.predict(temp)
print(accuracy_score(Ytrain, predictionsTrain))
def image_prediction_KMeans(img):
img = img.reshape(128, 128, 3)
plt.imshow(img)
img1 = img.reshape(1, 128 * 128 * 3)
predict = kmeans_model.predict(img1)
print(predict)
if predict:
print("Person is having kidney stone(s)")
else:
print("The condition is normal and stable.")
path_2 = "/kaggle/input/kidneystone/CT_SCAN/Normal/N1.png"
image_array_2 = cv2.imread(path_2)
new_image_2 = cv2.resize(image_array_2, (128, 128))
plt.imshow(new_image_2)
plt.show()
image_prediction_KMeans(new_image_2)
path = "/kaggle/input/kidneystone/CT_SCAN/Kidney_stone/KS1.png"
image_array = cv2.imread(path)
new_image_1 = cv2.resize(image_array, (128, 128))
plt.imshow(new_image_1)
plt.show()
image_prediction_KMeans(new_image_1)
# Save CNN Model
model3.save("kidney_stones_model.h5")
|
# Imported libraries
#
import os
import glob
import random
import shutil
import warnings
import json
import itertools
import numpy as np
import pandas as pd
from collections import Counter
import seaborn as sns
import plotly.express as px
import matplotlib.pyplot as plt
import seaborn as sns
import cv2
import keras
from keras.preprocessing.image import ImageDataGenerator
import tensorflow as tf
from PIL import Image
from glob import glob
from sklearn.model_selection import train_test_split
from keras.models import Sequential
from keras.layers import (
GlobalAveragePooling2D,
Flatten,
Dense,
Dropout,
BatchNormalization,
)
from keras.optimizers import RMSprop, Adam
from keras.callbacks import EarlyStopping, ModelCheckpoint, ReduceLROnPlateau
from tensorflow.keras.applications import EfficientNetB3
import os
work_dir = "../input/cassava-leaf-disease-classification/"
os.listdir(work_dir)
train_path = "/kaggle/input/cassava-leaf-disease-classification/train_images/"
# csv_path = '/kaggle/input/cassava-leaf-disease-classification/train.csv'
# plt.figure(figsize=(8, 4))
# sns.countplot(y="class_name", data=traindf);
# Check for GPU
print("Num GPUs Available: ", len(tf.config.experimental.list_physical_devices("GPU")))
with tf.device("/GPU:0"):
print("Yes, there is GPU")
tf.debugging.set_log_device_placement(True)
# Seed the dataset
# Lets set all random seeds
def seed_everything(seed=0):
random.seed(seed)
np.random.seed(seed)
tf.random.set_seed(seed)
os.environ["PYTHONHASHSEED"] = str(seed)
os.environ["TF_DETERMINISTIC_OPS"] = "1"
seed = 66
seed_everything(seed)
warnings.filterwarnings("ignore")
print("here")
# Read the data
data = pd.read_csv(work_dir + "train.csv")
print(data["label"].value_counts()) # Checking the frequencies of the labels
# Importing the json file with labels
with open(work_dir + "label_num_to_disease_map.json") as f:
real_labels = json.load(f)
real_labels = {int(k): v for k, v in real_labels.items()}
# Defining the working dataset
data["class_name"] = data["label"].map(real_labels)
real_labels
df_train = pd.read_csv("../input/cassava-leaf-disease-classification/train.csv")
df_train["class_name"] = df_train["label"].map(real_labels)
df_train
print("The number of train images is :", df_train.shape[0])
plt.figure(figsize=(8, 4))
sns.countplot(y="class_name", data=df_train)
def visualize_batch(image_ids, labels):
plt.figure(figsize=(16, 12))
for ind, (image_id, label) in enumerate(zip(image_ids, labels)):
plt.subplot(4, 4, ind + 1)
image = cv2.imread(os.path.join(work_dir, "train_images", image_id))
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
plt.imshow(image)
# plt.title(f"Class: {label}", fontsize=12)
plt.axis("off")
plt.show()
tmp_df = df_train.sample(16)
image_ids = tmp_df["image_id"].values
labels = tmp_df["class_name"].values
visualize_batch(image_ids, labels)
# Split the dataset
# generate train and test sets
train, test = train_test_split(
data, test_size=0.05, random_state=42, stratify=data["class_name"]
)
print("here")
# Initialise hyperparameters
IMG_SIZE = 256
size = (IMG_SIZE, IMG_SIZE)
n_CLASS = 5
BATCH_SIZE = 10
print("here")
# Data preprocessing
datagen_train = ImageDataGenerator(
preprocessing_function=tf.keras.applications.efficientnet.preprocess_input,
rotation_range=40,
width_shift_range=0.2,
height_shift_range=0.2,
shear_range=0.2,
zoom_range=0.2,
horizontal_flip=True,
vertical_flip=True,
fill_mode="nearest",
)
datagen_val = ImageDataGenerator(
preprocessing_function=tf.keras.applications.efficientnet.preprocess_input,
)
print("here")
# training & validation initialisation
train_set = datagen_train.flow_from_dataframe(
train,
directory=train_path,
seed=42,
x_col="image_id",
y_col="class_name",
target_size=size,
class_mode="categorical",
interpolation="nearest",
shuffle=True,
batch_size=BATCH_SIZE,
)
test_set = datagen_val.flow_from_dataframe(
test,
directory=train_path,
seed=42,
x_col="image_id",
y_col="class_name",
target_size=size,
class_mode="categorical",
interpolation="nearest",
shuffle=True,
batch_size=BATCH_SIZE,
)
print("here")
# Building the model
def create_model():
model = Sequential()
# initialize the model with input shape
model.add(
EfficientNetB3(
input_shape=(IMG_SIZE, IMG_SIZE, 3),
include_top=False,
weights="imagenet",
drop_connect_rate=0.6,
)
)
model.add(BatchNormalization())
model.add(GlobalAveragePooling2D())
model.add(Flatten())
model.add(
Dense(
32,
activation="relu",
bias_regularizer=tf.keras.regularizers.L1L2(l1=0.01, l2=0.001),
)
)
model.add(Dropout(0.5))
model.add(Dense(10, activation="softmax"))
# model.add(Dropout(0.5))
model.add(
Dense(
10,
activation="softmax",
)
)
model.add(
Dense(
5,
activation="relu",
)
)
return model
leaf_model = create_model()
leaf_model.summary()
print("here")
EPOCHS = 5
STEP_SIZE_TRAIN = train_set.n // train_set.batch_size
STEP_SIZE_TEST = test_set.n // test_set.batch_size
def model_fit():
leaf_model = create_model()
# Loss function
# https://www.tensorflow.org/api_docs/python/tf/keras/losses/CategoricalCrossentropy
loss = tf.keras.losses.CategoricalCrossentropy(
from_logits=False, label_smoothing=0.0001, name="categorical_crossentropy"
)
# Compile the model
leaf_model.compile(
optimizer=RMSprop(learning_rate=1e-3),
loss=loss, #'categorical_crossentropy'
metrics=["categorical_accuracy"],
)
# Stop training when the val_loss has stopped decreasing for 3 epochs.
# https://www.tensorflow.org/api_docs/python/tf/keras/callbacks/EarlyStopping
es = EarlyStopping(
monitor="val_loss",
mode="min",
patience=3,
restore_best_weights=True,
verbose=1,
)
# Save the model with the minimum validation loss
# https://www.tensorflow.org/api_docs/python/tf/keras/callbacks/ModelCheckpoint
checkpoint_cb = ModelCheckpoint(
"Cassava_best_model.h5",
save_best_only=True,
monitor="val_loss",
mode="min",
)
# Reduce learning rate once learning stagnates
# https://www.tensorflow.org/api_docs/python/tf/keras/callbacks/ReduceLROnPlateau
reduce_lr = ReduceLROnPlateau(
monitor="val_loss",
factor=0.2,
patience=2,
min_lr=1e-6,
mode="min",
verbose=1,
)
# Fit the model
history = leaf_model.fit(
train_set,
validation_data=test_set,
epochs=EPOCHS,
batch_size=BATCH_SIZE,
steps_per_epoch=STEP_SIZE_TRAIN,
validation_steps=STEP_SIZE_TEST,
callbacks=[es, checkpoint_cb, reduce_lr],
)
# Save the model
leaf_model.save("Cassava_model" + ".h5")
return history
print("here")
sess = tf.compat.v1.Session(config=tf.compat.v1.ConfigProto(log_device_placement=True))
from tensorflow.compat.v1.keras import backend as K
K.set_session(sess)
try:
final_model = keras.models.load_model("Cassava_model.h5")
except Exception as e:
with tf.device("/GPU:0"):
results = model_fit()
print("Train Categorical Accuracy: ", max(results.history["categorical_accuracy"]))
print(
"Test Categorical Accuracy: ", max(results.history["val_categorical_accuracy"])
)
# draw the results & try to fix the bug of an empty plot
import plotly.graph_objects as go
from plotly.offline import init_notebook_mode, iplot
init_notebook_mode(connected=True)
def trai_test_plot(acc, test_acc, loss, test_loss):
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(15, 10))
fig.suptitle("Model's metrics comparisson", fontsize=20)
ax1.plot(range(1, len(acc) + 1), acc)
ax1.plot(range(1, len(test_acc) + 1), test_acc)
ax1.set_title("History of Accuracy", fontsize=15)
ax1.set_xlabel("Epochs", fontsize=15)
ax1.set_ylabel("Accuracy", fontsize=15)
ax1.legend(["training", "validation"])
ax2.plot(range(1, len(loss) + 1), loss)
ax2.plot(range(1, len(test_loss) + 1), test_loss)
ax2.set_title("History of Loss", fontsize=15)
ax2.set_xlabel("Epochs", fontsize=15)
ax2.set_ylabel("Loss", fontsize=15)
ax2.legend(["training", "validation"])
plt.show()
trai_test_plot(
results.history["categorical_accuracy"],
results.history["val_categorical_accuracy"],
results.history["loss"],
results.history["val_loss"],
)
# **results**
STEP_SIZE = test_set.n // test_set.batch_size
predict = model.predict(test_set, STEP_SIZE)
print(predict)
class_prob = model.predict_classes(test_set)
top_values_index = sorted(range(len(class_prob)), key=lambda i: class_prob[i])[-5:]
top_values = [class_prob[i] for i in np.argsort(class_prob)[-5:]]
print(top_values)
input_dir = os.path.join("..", "input")
output_dir = os.path.join("..", "output")
dataset_dir = os.path.join(input_dir, "cassava-leaf-disease-classification")
train_dir = os.path.join(dataset_dir, "train_images")
def get_batch(dataframe, start, batch_size):
image_array = []
label_array = []
end_img = start + batch_size
if end_img > len(dataframe):
end_img = len(dataframe)
for idx in range(start, end_img):
n = idx
im, label = get_image_from_number(n, dataframe)
im = image_reshape(im, (224, 224)) / 255.0
image_array.append(im)
label_array.append(label)
label_array = encode_label(label_array)
return np.array(image_array), np.array(label_array)
def get_image_from_number(num, data):
fname, label = data.iloc[num, :]
fname = fname + ".jpg"
f1 = fname[0]
f2 = fname[1]
f3 = fname[2]
path = os.path.join(f1, f2, f3, fname)
im = cv2.imread(
os.path.join(
r"/kaggle/input/cassava-leaf-disease-classification/train_images", path
)
)
return im, label
batch_size = 10
errors = 0
good_preds = []
bad_preds = []
for it in range(int(np.ceil(len(test_set) / batch_size))):
X_train, y_train = get_batch(test_set, it * batch_size, batch_size)
result = model.predict(work_dir)
cla = np.argmax(result, axis=1)
for idx, res in enumerate(result):
if cla[idx] != y_train[idx]:
errors = errors + 1
bad_preds.append([batch_size * it + idx, cla[idx], res[cla[idx]]])
else:
good_preds.append([batch_size * it + idx, cla[idx], res[cla[idx]]])
print(
"Total errors: ",
errors,
"out of",
len(validate),
"\nAccuracy:",
np.round(100 * (len(validate) - errors) / len(validate), 2),
"%",
)
good_preds = np.array(good_preds)
good_preds = np.array(sorted(good_preds, key=lambda x: x[2], reverse=True))
print("5 images where classification went well:")
fig = plt.figure(figsize=(16, 16))
for i in range(1, 6):
n = int(good_preds[i, 0])
img, lbl = get_image_from_number(n, validate)
img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)
fig.add_subplot(1, 5, i)
plt.imshow(img)
lbl2 = np.array(int(good_preds[i, 1])).reshape(1, 1)
sample_cnt = int(temp[int(encode_label(np.array([lbl]))) - 1][1])
plt.title(
"Label: "
+ str(lbl)
+ "\nClassified as: "
+ str(decode_label(lbl2))
+ "\nSamples in class "
+ str(lbl)
+ ": "
+ str(sample_cnt)
)
plt.axis("off")
plt.show()
val_filenames = test_set.filenames
label_map = test_set.class_indices
# label_categories = to_categorical(np.asarray(labels))
cla = np.argmax(predict, axis=-1)
label_map = list(map(int, label_map.keys()))
val_label = test_set.labels
for idx, res in enumerate(predict):
# print("image_id: ", val_filenames[idx], ", class predict: ", label_map[cla[idx]], "class: ", label_map[val_label[idx]])
if label_map[cla[idx]] != label_map[val_label[idx]]:
bad_preds.append(
[
val_filenames[idx],
label_map[cla[idx]],
label_map[val_label[idx]],
res[cla[idx]],
]
)
else:
good_preds.append(
[
val_filenames[idx],
label_map[cla[idx]],
label_map[val_label[idx]],
res[cla[idx]],
]
)
print(
"wrong predictions: ",
len(bad_preds),
" right predictions: ",
len(good_preds),
" acc: ",
np.round(100 * (len(predict) - len(bad_preds)) / len(predict), 2),
)
sorted_preds = np.array(sorted(predict, key=lambda x: x[2], reverse=True))
print(sorted_preds[:5])
print(b)
rows = 2
# os.chdir('/home/brian/Desktop/cats/')
# files = os.listdir('/home/brian/Desktop/cats/')
import PIL
from PIL import Image
for num, x in enumerate(good_preds):
img = PIL.Image.fromarray(x)
plt.subplot(rows, 6, num + 1)
plt.title(x.split(".")[0])
plt.axis("off")
plt.imshow(img)
good_preds = []
bad_preds = []
val_filenames = test_set.filenames
label_map = test_set.class_indices
# label_categories = to_categorical(np.asarray(labels))
cla = np.argmax(predict, axis=-1)
label_map = list(map(int, label_map.keys()))
val_label = test_set.labels
for idx, res in enumerate(predict):
# print("image_id: ", val_filenames[idx], ", class predict: ", label_map[cla[idx]], "class: ", label_map[val_label[idx]])
if label_map[cla[idx]] != label_map[val_label[idx]]:
bad_preds.append(
[
val_filenames[idx],
label_map[cla[idx]],
label_map[val_label[idx]],
res[cla[idx]],
]
)
else:
good_preds.append(
[
val_filenames[idx],
label_map[cla[idx]],
label_map[val_label[idx]],
res[cla[idx]],
]
)
print(
"wrong predictions: ",
len(bad_preds),
" right predictions: ",
len(good_preds),
" acc: ",
np.round(100 * (len(predict) - len(bad_preds)) / len(predict), 2),
)
# Convert
sub = pd.DataFrame({"image_id": test_images, "label": predictions})
display(sub)
sub.to_csv("submission.csv", index=False)
|
# Written by Noah Halgren
# CAP 4611
# Assignment 1
# **-------- Importing --------**
# Importing Libraries
# Importing libraries and set constants
import numpy as np
import pandas as pd
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import classification_report, confusion_matrix, roc_auc_score
from sklearn.model_selection import RandomizedSearchCV
from sklearn.model_selection import train_test_split
# Importing Data
# Importing CSV file
train_data = pd.read_csv("../input/cap-4611-spring-21-assignment-1/train.csv").drop(
columns=["id"]
)
test_data = pd.read_csv("../input/cap-4611-spring-21-assignment-1/test.csv").drop(
columns=["id"]
)
# Display Training and Testing Data
train_data
test_data
# **-------- Data Cleaning --------**
# Handling Outliers
# This is where i would handel outliers, but after inspecting this data set, it seems there aren't any outliers to remove
# If i did have a data set with outliers, i would find and fix them by standardizing the feature using something like a z-score
# and i would replace any outliers (2-3 standard deviations away) with either the mean, min, or max.
# Handling Missing Values
# Go through each col in training data
for col in train_data:
numZero = 0
# This loop counts the number of 0's in each col and saves it to numZero
for i in train_data[col].values:
if i == 0:
numZero = numZero + 1
# Make sure we're not messing with target feature
if col != "Bankrupt":
# If we are only missing 10% or less of values, we replace with the mean of that col
if (numZero / len(train_data.index)) > 0.10:
train_data = train_data.drop(col, axis=1)
print(train_data.shape)
# Applying Feature Removal to Test Set
# Remove all features where every value is the same
train_data = train_data.drop(
[e for e in train_data.columns if train_data[e].nunique() == 1], axis=1
)
# Remove those same features in out test data
common_cols = [col for col in set(test_data.columns).intersection(train_data.columns)]
test_data = test_data[common_cols]
# Check that both dataframes have the same number of features
print(train_data.shape)
print(test_data.shape)
# Standardization / Normalization
# I decided against implementing standardization/normalization
# Display Training and Testing Data
test_data
train_data
# Seperating Target Feature from Training Data
# Creating seperated dataframes for features and target
x = train_data
y = train_data["Bankrupt"]
# Drop target feature and index column from dataframes
x = x.drop("Bankrupt", axis=1)
# Partition Dataset into Test and Train Sections
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.20, random_state=420
)
# **-------- Decission Tree --------**
# Hyperparameter Tuning Decission Tree
# Create a list of parameters and possible values
tree_params = {
"criterion": ["gini", "entropy"],
"max_depth": range(2, 10),
"min_samples_split": range(10, 31),
"min_samples_leaf": range(1, 100),
"max_features": range(1, 90),
}
# Create test decission tree
tree = DecisionTreeClassifier()
tree_cv = RandomizedSearchCV(tree, tree_params, cv=5, scoring="roc_auc")
# Fit it to the data
tree_cv.fit(x_train, y_train)
# Print results
print(tree_cv.best_params_)
print(tree_cv.best_score_)
dt = tree_cv.best_estimator_
# Running / Evaluating Decision Tree
dt.fit(x_train, y_train)
y_pred = dt.predict_proba(x_test)
y_pred = y_pred[:, 1]
print(roc_auc_score(y_test, y_pred))
y_pred = dt.predict_proba(test_data)
y_pred = y_pred[:, 1]
d = {"Bankrupt": y_pred}
df = pd.DataFrame(data=d)
df.to_csv("submission_dt.csv", index_label="id")
# **-------- Random Forest Model --------**
# Hyperparameter Tuning Random Forest
#
# Create a list of parameters and possible values
forest_params = {
"n_estimators": range(2, 200),
"criterion": ["gini", "entropy"],
"max_depth": range(2, 10),
"class_weight": ["balanced", "balanced_subsample", None],
"max_samples": range(5, 2000),
}
# Create test random forest
forest = RandomForestClassifier()
forest_cv = RandomizedSearchCV(forest, forest_params, cv=5, scoring="roc_auc")
# Fit it to the data
forest_cv.fit(x_train, y_train)
# Print results
print(forest_cv.best_params_)
print(forest_cv.best_score_)
rf = forest_cv.best_estimator_
# Running / Evaluating Random Forest Model
rf.fit(x_train, y_train)
y_pred_rf = rf.predict_proba(x_test)
y_pred_rf = y_pred_rf[:, 1]
print(roc_auc_score(y_test, y_pred_rf))
y_pred_rf = rf.predict_proba(test_data)
y_pred_rf = y_pred_rf[:, 1]
d = {"Bankrupt": y_pred_rf}
df = pd.DataFrame(data=d)
df.to_csv("submission_rf.csv", index_label="id")
|
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import seaborn as sns
import os
from keras.preprocessing.image import load_img, img_to_array
from keras.preprocessing.image import ImageDataGenerator
from keras.layers import (
Dense,
Input,
Dropout,
GlobalAveragePooling2D,
Flatten,
Conv2D,
BatchNormalization,
Activation,
MaxPooling2D,
)
from keras.models import Model, Sequential
from tensorflow.keras.optimizers import Adam, SGD, RMSprop
import tensorflow as tf
gpus = tf.config.list_logical_devices("GPU")
stg = tf.distribute.MirroredStrategy(gpus)
picture_size = 244
folder_path = "/kaggle/input/emotionv3/facemo/images/"
from PIL import Image
import cv2
import os
folder_dir = "/kaggle/input/emotionv3/facemo/images/train"
SIZE = 244
DOWNSAMPLE_RATIO = 4
for folder in os.listdir(folder_dir):
for file in os.listdir(os.path.join(folder_dir, folder)):
if file.endswith("jpg"):
image_path = os.path.join(folder_dir, folder, file)
img = cv2.imread(image_path)
img_resized = cv2.resize(img, (SIZE, SIZE))
cv2.imwrite(image_path, img_resized)
else:
continue
batch_size = 32
datagen_train = ImageDataGenerator()
datagen_val = ImageDataGenerator()
train_set = datagen_train.flow_from_directory(
folder_path + "train",
target_size=(picture_size, picture_size),
batch_size=batch_size,
class_mode="categorical",
shuffle=True,
)
test_set = datagen_val.flow_from_directory(
folder_path + "validation",
target_size=(picture_size, picture_size),
batch_size=batch_size,
class_mode="categorical",
shuffle=True,
)
from keras.applications.vgg16 import VGG16
from keras.models import Model
from keras import layers
from keras.models import Sequential
from keras.layers import (
Conv2D,
MaxPooling2D,
Dropout,
Flatten,
Dense,
Activation,
GlobalMaxPooling2D,
)
from keras import applications
from keras.preprocessing.image import ImageDataGenerator
from tensorflow.keras.optimizers import Adam, SGD, RMSprop
from keras.layers import BatchNormalization
image_size = 244
input_shape = (image_size, image_size, 3)
epochs = 5
batch_size = 16
from tensorflow.keras.optimizers import Adam, SGD, RMSprop
no_of_classes = 5
with stg.scope():
pre_trained_model = VGG16(
input_shape=input_shape,
include_top=False,
weights="/kaggle/input/vgg16v1/vgg16_weights_tf_dim_ordering_tf_kernels_notop.h5",
)
for layer in pre_trained_model.layers:
layer.trainable = False
last_layer = pre_trained_model.get_layer("block1_conv1")
last_output = last_layer.output
x = Conv2D(128, (3, 3))(last_output)
vgm = Model(pre_trained_model.input, x)
model = Sequential()
model.add(vgm)
# 1st CNN layer
model.add(Conv2D(64, (3, 3)))
model.add(Activation("relu"))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
# 2nd CNN layer
model.add(Conv2D(128, (5, 5), padding="same"))
model.add(BatchNormalization())
model.add(Activation("relu"))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
# 3rd CNN layer
model.add(Conv2D(512, (3, 3), padding="same"))
model.add(BatchNormalization())
model.add(Activation("relu"))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
# 4th CNN layer
model.add(Conv2D(512, (3, 3), padding="same"))
model.add(BatchNormalization())
model.add(Activation("relu"))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Flatten())
# Fully connected 1st layer
model.add(Dense(256))
model.add(BatchNormalization())
model.add(Activation("relu"))
model.add(Dropout(0.25))
# Fully connected layer 2nd layer
model.add(Dense(512))
model.add(BatchNormalization())
model.add(Activation("relu"))
model.add(Dropout(0.25))
# Fully connected layer 3rd layer
model.add(Dense(512))
model.add(BatchNormalization())
model.add(Activation("relu"))
model.add(Dropout(0.22))
model.add(Dense(no_of_classes, activation="softmax"))
opt = Adam(learning_rate=0.0001)
model.compile(optimizer=opt, loss="categorical_crossentropy", metrics=["accuracy"])
model.summary()
from tensorflow.keras.optimizers import RMSprop, SGD, Adam
from tensorflow.keras.callbacks import ModelCheckpoint, EarlyStopping, ReduceLROnPlateau
checkpoint = ModelCheckpoint(
"./model.h5", monitor="val_acc", verbose=1, save_best_only=True, mode="max"
)
early_stopping = EarlyStopping(
monitor="val_loss", min_delta=0, patience=3, verbose=1, restore_best_weights=True
)
reduce_learningrate = ReduceLROnPlateau(
monitor="val_loss", factor=0.2, patience=3, verbose=1, min_delta=0.0001
)
callbacks_list = [early_stopping, checkpoint, reduce_learningrate]
epochs = 4
model.compile(
loss="categorical_crossentropy",
optimizer=Adam(learning_rate=0.001),
metrics=["accuracy"],
)
history = model.fit(
train_set,
steps_per_epoch=train_set.n // train_set.batch_size,
epochs=epochs,
validation_data=test_set,
validation_steps=test_set.n // test_set.batch_size,
callbacks=callbacks_list,
)
test_loss, test_acc = model.evaluate(test_set)
print("Test accuracy:", test_acc)
model.save("/kaggle/working/my_model_emo.h5")
import numpy as np
import matplotlib.pyplot as plt
e = ["Angry", "Happy", "Neutral", "Sad", "Surprise"]
for i in range(len(e)):
print(i, "=", e[i])
test_batch = test_set.next()
images = test_batch[0][:10]
true_labels = test_batch[1][:10]
pred_probs = model.predict(images)
pred_labels = np.argmax(pred_probs, axis=1)
fig, axs = plt.subplots(2, 5, figsize=(15, 7))
axs = axs.flatten()
for i in range(len(images)):
img = (images[i] * 255).astype(np.uint8)
axs[i].imshow(img)
axs[i].set_title(f"True: {true_labels[i]}, Pred: {pred_labels[i]}")
axs[i].axis("off")
plt.show()
|
# ---
# # Stage-2 of Tree Counting Image Regression
# ---
# ---
# #### Daniel Bruintjies
# #### March 2022
# ___
# ### ZINDI COMPETITION
# ### 3RD Place Solution
# #### METRIC: RMSE
# #### Public LB: 2.1079
# #### PRIVATE LB: 1.595
# ___
# # Digital Africa Plantation Counting Challenge
# ---
# #### The objective of this challenge is to create a semi-supervised machine learning algorithm to count the number of palm oil trees in an image. This will aid farmers to determine the number of trees on their plot and estimated crop yield. The semi supervised nature of this solution will allow this solution to be applied to other plantations such as banana palms.
# https://zindi.africa/competitions/digital-africa-plantation-counting-challenge
# ### Stage-1 of Tree Counting Image Regression can be found here:
# https://www.kaggle.com/code/danielbruintjies/stage-1-of-tree-counting-image-regression
# # Imports
from typing import Callable
from typing import Dict
from typing import Optional
from typing import Tuple
from pathlib import Path
import os
import math
import io
import gc
gc.enable()
import time
import random
from numpy.random import seed
from PIL import Image, ImageEnhance
import cv2
import seaborn as sns
import matplotlib
matplotlib.use("agg")
import matplotlib.image as mpimg
import matplotlib.pyplot as plt
import shutil
import warnings
warnings.filterwarnings("ignore")
import numpy as np
import pandas as pd
import torch
import torch.nn as nn
import torch.nn.functional as F
from torch import optim
from torch.utils.data import DataLoader
from torch.utils.data import Dataset
import albumentations as A
from albumentations.pytorch import ToTensorV2
from torch.optim.lr_scheduler import ReduceLROnPlateau
from torch.optim import lr_scheduler
import timm
import torchvision.models as models
from tqdm.notebook import tqdm
from sklearn.model_selection import StratifiedKFold, KFold, StratifiedShuffleSplit
from sklearn.preprocessing import normalize
from sklearn.preprocessing import StandardScaler, RobustScaler
from sklearn.metrics import mean_squared_error as MSE
from sklearn.metrics import mean_squared_error
import lightgbm as lgb
from lightgbm import LGBMRegressor
import pickle
def rmse(actual, predicted):
mse = MSE(actual, predicted)
return math.sqrt(mse)
def set_seed(seed=42):
"""Sets the seed of the entire notebook so results are the same every time we run.
This is for REPRODUCIBILITY."""
np.random.seed(seed)
random.seed(seed)
torch.manual_seed(seed)
torch.cuda.manual_seed(seed)
# When running on the CuDNN backend, two further options must be set
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = True
# Set a fixed value for the hash seed
os.environ["PYTHONHASHSEED"] = str(seed)
os.environ["PL_GLOBAL_SEED"] = str(seed)
set_seed(42)
# # Paths & Settings
"""
"data_path" should follow this structure:
->digital-africa-plantation-counting-challenge
->>images
SampleSubmission.csv
Test.csv
Train.csv
"""
data_path = "/kaggle/input/digital-africa-plantation-counting-challenge/"
"""
Change val_df_preds_path to the path of the saved val_df_preds.csv
from Stage-1 of Tree Counting Notebook
"""
val_df_preds_path = "/kaggle/input/dapcc-stage1-val-df-preds/val_df_preds.csv"
"""
"OUTPUT_DIR" should be changed to just "Path("/")" if not used in Kaggle env
"""
OUTPUT_DIR = Path("/") / "kaggle" / "working"
IMG_DIR = data_path + "images/"
TRAIN_CSV_PATH = data_path + "Train.csv"
TEST_CSV_PATH = data_path + "Test.csv"
SAMPLE_SUBMISSION_CSV_PATH = data_path + "SampleSubmission.csv"
TRAIN_CSV_ENCODED_FOLDED_PATH = OUTPUT_DIR / "Train_ENCODED.csv"
TEST_CSV_ENCODED_FOLDED_PATH = OUTPUT_DIR / "Test_ENCODED.csv"
CHECKPOINTS_DIR = OUTPUT_DIR / "checkpoints"
SUBMISSION_CSV_PATH = OUTPUT_DIR / "submission.csv"
# ### Make sure to connect to the GPU P100 accelerator in Notebook options -->
"""
N.B. Do not change the CFG config class if you wish to reproduce my score.
If, however, you have run Stage-1 of Tree Counting Notebook and have the val_df_preds.csv file,
then feel free to set FIND_BADLABELS = True and alter the neccessary code as described in the
Paths & Settings section and the FIND BADLABELS section and then my score will still be reproducible.
If you want to visualise the cool gradcam heatmaps, youll need a pretrained convnext model,
so run this notebook, get the trained model, create a dataset and add the
models path in model_dir in the gradcam section!
"""
class CFG:
TRAIN = True
INFER = True
VIS_GRADCAM = False
CALC_GRADCAM = False
USE_GRADCAM = False
PRED_VAL = True
FIND_BADLABELS = False
REMOVE_BADLABELS = True
if torch.cuda.is_available():
device = "cuda"
print(torch.cuda.get_device_name())
else:
device = "cpu"
print(f"device : {device}")
# # Prepare DataFrames
def get_image_path(id_: str, dir_: str) -> str:
return dir_ + id_
def crop_and_resize(pil_img, imsize):
"""Crops square from center of image and resizes to (img_size, img_size)"""
w, h = pil_img.size
crop_size = min(w, h)
crop = pil_img.crop(
(
(w - crop_size) // 2,
(h - crop_size) // 2,
(w + crop_size) // 2,
(h + crop_size) // 2,
)
)
return crop.resize((imsize, imsize))
def load_image(img_path, imsize):
img = Image.open(img_path)
img.draft("RGB", (imsize, imsize))
img = crop_and_resize(img, imsize)
return np.array(img)
"""
Reading of Files
"""
train_df = pd.read_csv(TRAIN_CSV_PATH)
test_df = pd.read_csv(TEST_CSV_PATH)
sample_submission = pd.read_csv(SAMPLE_SUBMISSION_CSV_PATH)
for df in [train_df, test_df]:
df.rename(columns={"ImageId": "image"}, inplace=True)
train_df["image_path"] = train_df["image"].apply(get_image_path, dir_=IMG_DIR)
test_df["image_path"] = test_df["image"].apply(get_image_path, dir_=IMG_DIR)
"""
Creation of Train/Val splits
"""
low_counts = train_df["Target"].value_counts().iloc[-4:].index
low_counts_df = pd.DataFrame([])
low_counts_df = low_counts_df.append(train_df[train_df["Target"].isin(low_counts)])
train_df = train_df[~train_df["Target"].isin(low_counts)].reset_index(drop=True)
N_SPLITS = 4
FOLDS = list(range(N_SPLITS))
kf = StratifiedShuffleSplit(n_splits=N_SPLITS, test_size=250, random_state=5)
for fold, (_, val_) in enumerate(kf.split(X=train_df, y=train_df["Target"])):
train_df.loc[val_, f"kfold_{fold}"] = 1
train_df = train_df.append(low_counts_df).reset_index(drop=True)
test_df["Target"] = 1000
train_df.to_csv(TRAIN_CSV_ENCODED_FOLDED_PATH, index=False)
test_df.to_csv(TEST_CSV_ENCODED_FOLDED_PATH, index=False)
display(train_df.head(), train_df.shape)
display(test_df.head(), test_df.shape)
display(sample_submission.head(), sample_submission.shape)
"""
Finding Images that are completely black
"""
for i, row in tqdm(train_df.iterrows()):
train_df.loc[i, "img_mean"] = (
load_image(train_df.loc[i, "image_path"], 256) / 255
).mean()
for i, row in tqdm(test_df.iterrows()):
test_df.loc[i, "img_mean"] = (
load_image(test_df.loc[i, "image_path"], 256) / 255
).mean()
"""
Our Train/Val shuffle split method will yield some samples that occur across different validation sets.
"""
num_training_samples = []
num_validation_samples = []
unique_val_samples = []
for k in FOLDS:
num_validation_samples.append(train_df[train_df[f"kfold_{k}"] == 1].shape[0])
unique_val_samples += train_df[train_df[f"kfold_{k}"] == 1]["image"].tolist()
num_training_samples.append(
train_df.shape[0] - train_df[train_df[f"kfold_{k}"] == 1].shape[0]
)
print("FOLDS:\t\t\t\t", FOLDS)
print("num_training_samples:\t\t", num_training_samples)
print("num_validation_samples:\t\t", num_validation_samples)
print("total_unique_validation_samples:", len(set(unique_val_samples)))
"""
From stage 1 found that the Train/Val split with Fold 1 has the best
train/val RMSE score and convergence, therefore I select only this split to be run when training.
"""
FOLDS = [1]
RUN_FOLDS = 1000
"""
Visualize the value_counts of the target count from the train and validation stratified shuffle
"""
USE_FOLD = 1
for k in [USE_FOLD]:
df = train_df[train_df[f"kfold_{k}"] == 1]
display(df["Target"].value_counts().sort_index())
df2 = train_df[train_df[f"kfold_{k}"] != 1]
display(df2["Target"].value_counts().sort_index())
plt.figure(figsize=(3, 2))
df["Target"].hist()
plt.show()
plt.figure(figsize=(3, 2))
df2["Target"].hist()
plt.show()
def image_gen(train, tree_counts=[0, 49]):
"""
Generator funtion to visualise the images in dataset given a pandas df [train]
and range of targets [tree_counts] to iterate over.
"""
for l in range(tree_counts[0], tree_counts[1]):
df = train[train["Target"] == l]
n = 0
while n < len(df):
plt.figure(figsize=(25, 15), dpi=50)
info = []
for i, (im, p) in enumerate(
zip(df["image"].values[n : n + 15], df["image_path"].values[n : n + 15])
):
ax = plt.subplot(3, 5, i + 1)
img = load_image(p, 256) / 255
info.append([im, img.mean()])
plt.imshow(img)
plt.title(f"{im}")
n += 15
print(f"TARGET = {l}")
print(info)
print(f"{n}/{len(df)}")
plt.show()
yield
"""
Initialise the image generator function
Set tree_counts to a range you want to visualise
"""
USE_FOLD = 1
train_image_generator = image_gen(
train_df[train_df[f"kfold_{USE_FOLD}"] != 1], tree_counts=[1, 49]
)
val_image_generator = image_gen(
train_df[train_df[f"kfold_{USE_FOLD}"] == 1], tree_counts=[1, 49]
)
"""
Visualize the images in train split by continuosly running this cell
"""
SHOW = True
if SHOW:
next(train_image_generator)
"""
Visualize the images in val split by continuosly running this cell
"""
SHOW = True
if SHOW:
next(val_image_generator)
"""
If you have predictions for the test set in the submission format,
then load it into pred_test_df_path
"""
pred_test_df_path = "/kaggle/input/dapcc-models/sub_allfolds.csv"
SHOW_TEST = False
if SHOW_TEST:
pred_test_df = pd.read_csv(pred_test_df_path)
pred_test_df["image"] = pred_test_df["ImageId"]
pred_test_df["image_path"] = pred_test_df["image"].apply(
get_image_path, dir_=IMG_DIR
)
pred_test_df["Target"] = np.round(pred_test_df["Target"], 0)
"""
Initialise the image generator function
Set tree_counts to a range you want to visualise
"""
test_image_generator = image_gen(pred_test_df, tree_counts=[6, 49])
"""
Visualize the images in testset by continuosly running this cell
"""
if SHOW_TEST:
next(test_image_generator)
"""
Finding Images that are completely black in the testset
"""
test_df[test_df["img_mean"] == 0]
"""
Finding Images that are completely black in the trainset
"""
train_df[train_df["img_mean"] == 0]
# ### Find all completely black images for use to downsample later
"""
Select x images that are completely black in the trainset for use later
to omit from training split of fold 1 [k==1]
"""
x = 200
k = 1
df2 = train_df[train_df[f"kfold_{k}"] != 1]
sample_zero_images = (
df2[df2["img_mean"] == 0].sample(x, random_state=5)["image"].tolist()
)
# ### FIND BADLABELS
"""
Select validation images from Stage 1 with absolute error greater than a threshold (3)
to mark as badly labelled images for use later to omit from validation set.
"""
if CFG.FIND_BADLABELS:
val_df_preds = pd.read_csv(val_df_preds_path)
val_df_preds.loc[val_df_preds["pred"] < 0, "pred"] = 0
val_df_preds = (
val_df_preds.groupby(["image", "image_path", "Target"])
.agg({"pred": "mean"})
.reset_index()
)
val_df_preds["abs_error"] = abs(val_df_preds["pred"] - val_df_preds["Target"])
display(val_df_preds)
abs_error_THRESH = 3
badlabels_df = val_df_preds[val_df_preds["abs_error"] > abs_error_THRESH]
badlabels = badlabels_df["image"].tolist()
"""
Save badly labelled images so dont need to run FIND_BADLABELS again.
Or comment out if you ran FIND_BADLABELS
"""
"""
Visualise sample image given id
"""
sampleid = "Id_063httgrzd.png"
sample_df = train_df[train_df["image"] == sampleid]
sampleid_path = sample_df["image_path"].values[0]
sampleid_path_target = sample_df["Target"].values[0]
print(sampleid_path_target)
img = load_image(sampleid_path, 256) / 255
plt.figure(figsize=(5, 5))
plt.imshow(img)
# # Dataset Setup
class df_to_datatset(Dataset):
def __init__(
self,
df,
kfold=None,
is_train=True,
is_inference=False,
transform=None,
imsize=256,
my_shuffle=False,
resize=None,
remove_ids=None,
return_ids=False,
return_gradcam=False,
):
self.transform = transform
self.is_inference = is_inference
self.imsize = imsize
self.resize = resize
self.return_ids = return_ids
self.return_gradcam = return_gradcam
if is_inference:
self.df = df
else:
if is_train:
self.df = df[df[f"kfold_{kfold}"] != 1].reset_index(drop=True)
print("NUM TRAINING SAMPLES: ", self.df.shape[0])
if remove_ids != None:
train_remove_ids = remove_ids[0]
if train_remove_ids != None:
self.df = self.df[
~self.df["image"].isin(train_remove_ids)
].reset_index(drop=True)
print("NUM TRAINING SAMPLES: ", self.df.shape[0])
else:
self.df = df[df[f"kfold_{kfold}"] == 1].reset_index(drop=True)
print("NUM VALIDATION SAMPLES: ", self.df.shape[0])
if remove_ids != None:
val_remove_ids = remove_ids[1]
if val_remove_ids != None:
self.df = self.df[
~self.df["image"].isin(val_remove_ids)
].reset_index(drop=True)
print("NUM VALIDATION SAMPLES: ", self.df.shape[0])
def __len__(self):
return self.df.shape[0]
def __getitem__(self, index):
image_id = self.df["image"][index]
image_path = self.df["image_path"][index]
if self.return_gradcam:
gradcam_path = self.df["gradcam_path_fold1"][index]
if not self.is_inference:
label = self.df["Target"][index]
if self.resize == None:
image = load_image(image_path, self.imsize)
if self.return_gradcam:
gradcam = load_image(gradcam_path, self.imsize)
else:
image = Image.open(image_path)
image.draft("RGB", (self.resize, self.resize))
image = crop_and_resize(image, self.resize)
image = crop_and_resize(image, self.imsize)
image = np.array(image)
if self.return_gradcam:
gradcam = Image.open(gradcam_path)
gradcam.draft("RGB", (self.resize, self.resize))
gradcam = crop_and_resize(gradcam, self.resize)
gradcam = crop_and_resize(gradcam, self.imsize)
gradcam = np.array(gradcam)
if self.transform is not None:
augmentations = self.transform(image=image)
image = augmentations["image"]
if self.return_gradcam:
augmentations = self.transform(image=gradcam[:, :, :3])
gradcam = augmentations["image"]
if not self.is_inference:
if self.return_gradcam:
return image, gradcam, torch.as_tensor(label)
return image, torch.as_tensor(label)
if self.return_ids:
return image, image_id
if self.return_gradcam:
return image, gradcam
return image
# # Training Setup
def check_acc(loader, model, return_preds=False, return_gradcam=False):
all_preds, all_groud_truth = np.array([]), np.array([])
model.eval()
with torch.no_grad():
for batch in tqdm(loader):
x, y = batch
x = x.to("cuda").to(torch.float32)
y = y.to(torch.float).unsqueeze(1)
all_preds = np.append(all_preds, (model(x).cpu()))
all_groud_truth = np.append(all_groud_truth, (y.cpu()))
loss = rmse(all_groud_truth, all_preds)
if return_preds:
return all_groud_truth, all_preds
return loss
def save_checkpoint(checkpoint, filename):
torch.save(checkpoint, filename)
print(f"\n--> Saved checkpoint: {filename.split('.')[0]}")
def load_checkpoint(filename, model):
model.load_state_dict(torch.load(filename)["state_dict"])
return model
def start_training(
train_df,
train_transform,
val_transform,
imsize,
batch_size,
loss_fn,
model,
opt,
model_filename,
kfold=0,
patience=4,
num_epochs=1000,
my_shuffle=False,
remove_ids=None,
return_gradcam=False,
use_amp=False,
):
"""
remove_ids should be a tuple of (ids_to_remove_from_training, ids_to_remove_from_validation)
use_amp does not seem to work with my scheduler ReduceLROnPlateau and should remain False
return_gradcam and my_shuffle should remain false because no longer in my pipeline
"""
scheduler = ReduceLROnPlateau(
opt, mode="min", factor=0.7, patience=1, verbose=True, min_lr=1e-6
)
scaler = torch.cuda.amp.GradScaler(enabled=use_amp)
def train_fn(
loader,
model,
opt,
loss_fn,
scheduler=None,
use_amp=False,
return_gradcam=False,
scaler=None,
):
model.train()
all_preds, all_groud_truth = np.array([]), np.array([])
for batch in tqdm(loader):
x, y = batch
x = x.to("cuda").to(torch.float32)
y = y.to(torch.float).unsqueeze(1).to("cuda")
with torch.cuda.amp.autocast(enabled=use_amp):
preds = model(x).to(torch.float)
loss = loss_fn(preds, y)
scaler.scale(loss).backward()
scaler.step(opt)
scaler.update()
opt.zero_grad()
all_groud_truth = np.append(all_groud_truth, (y.detach().cpu()))
all_preds = np.append(all_preds, (preds.detach().cpu()))
all_preds[all_preds < 0] = 0
loss = rmse(all_groud_truth, all_preds)
return loss
train_ds = df_to_datatset(
train_df,
kfold=kfold,
imsize=imsize,
is_train=True,
is_inference=False,
transform=train_transform,
remove_ids=remove_ids,
return_gradcam=return_gradcam,
)
train_loader = DataLoader(train_ds, batch_size=batch_size, shuffle=True)
val_ds = df_to_datatset(
train_df,
kfold=kfold,
imsize=imsize,
is_train=False,
is_inference=False,
transform=val_transform,
remove_ids=remove_ids,
return_gradcam=return_gradcam,
)
val_loader = DataLoader(val_ds, batch_size=batch_size, shuffle=False)
best_val_rmse = 1000000
epochs_without_improvement = 0
for epoch in range(num_epochs):
print("\n------------------------------- Epoch: " + str(epoch))
train_rmse = train_fn(
train_loader,
model,
opt,
loss_fn,
return_gradcam=return_gradcam,
use_amp=use_amp,
scaler=scaler,
)
val_rmse = check_acc(val_loader, model, return_gradcam=return_gradcam)
print(f"TRAIN RMSE: {np.round(train_rmse,5)}")
print(f"VAL RMSE: {np.round(val_rmse,5)}")
if val_rmse < best_val_rmse:
best_val_rmse = val_rmse
epochs_without_improvement = 0
checkpoint = {
"state_dict": model.state_dict(),
"optimizer": opt.state_dict(),
}
save_checkpoint(checkpoint=checkpoint, filename=model_filename)
else:
epochs_without_improvement += 1
if epochs_without_improvement == patience:
break
scheduler.step(val_rmse)
model = load_checkpoint(filename=model_filename, model=model)
groud_truth, preds = check_acc(
val_loader, model, return_preds=True, return_gradcam=return_gradcam
)
val_rmse = rmse(groud_truth, preds)
print(f"BEST VAL RMSE: {np.round(val_rmse,5)}")
return groud_truth, preds
def start_inference(
model,
test_df,
val_transform,
imsize,
batch_size,
pred_name,
resize=None,
return_gradcam=False,
):
"""
Standard inference function
"""
def inference_func(loader, model, return_gradcam=False):
model.eval()
all_preds = np.array([])
with torch.no_grad():
for batch in tqdm(loader):
x = batch
x = x.to("cuda").to(torch.float32)
all_preds = np.append(all_preds, model(x).cpu())
all_preds[all_preds < 0] = 0
return all_preds
test_ds = df_to_datatset(
test_df,
transform=val_transform,
imsize=imsize,
is_train=False,
is_inference=True,
resize=resize,
return_gradcam=return_gradcam,
)
test_loader = DataLoader(test_ds, batch_size=batch_size, shuffle=False)
all_preds = inference_func(test_loader, model, return_gradcam=return_gradcam)
test_df["Target"] = all_preds
test_df["ImageId"] = test_df["image"]
test_df[["ImageId", "Target"]].to_csv(f"sub_{pred_name}.csv", index=False)
return all_preds
# # Model Setup
"""
My palm tree counter model
"""
class SelfAttention(nn.Module):
def __init__(
self, in_channels, key_channels=None, value_channels=None, out_channels=None
):
super(SelfAttention, self).__init__()
self.key_channels = (
key_channels if key_channels is not None else in_channels // 8
)
self.value_channels = (
value_channels if value_channels is not None else in_channels // 2
)
self.out_channels = out_channels if out_channels is not None else in_channels
self.query_conv = nn.Conv2d(in_channels, self.key_channels, kernel_size=1)
self.key_conv = nn.Conv2d(in_channels, self.key_channels, kernel_size=1)
self.value_conv = nn.Conv2d(in_channels, self.value_channels, kernel_size=1)
self.gamma = nn.Parameter(torch.zeros(1))
self.out_conv = nn.Conv2d(self.value_channels, self.out_channels, kernel_size=1)
self.bn = nn.BatchNorm2d(self.out_channels)
self.relu = nn.ReLU(inplace=True)
def forward(self, x):
batch_size, channels, height, width = x.size()
query = self.query_conv(x).view(batch_size, self.key_channels, -1)
key = self.key_conv(x).view(batch_size, self.key_channels, -1)
value = self.value_conv(x).view(batch_size, self.value_channels, -1)
energy = torch.bmm(query.transpose(1, 2), key)
attention = F.softmax(energy, dim=-1)
out = torch.bmm(value, attention.transpose(1, 2))
out = out.view(batch_size, self.value_channels, height, width)
out = self.out_conv(out)
out = self.bn(out)
out = self.relu(out)
out = self.gamma * out + x
return out
class PalmTreeCounter(nn.Module):
def __init__(self):
super(PalmTreeCounter, self).__init__()
self.model = timm.create_model("convnext_base", pretrained=True)
self.attention1 = SelfAttention(in_channels=1024)
self.attention2 = SelfAttention(in_channels=1024)
self.fc = nn.Linear(1024, 1)
def forward(self, image):
x = self.model.forward_features(image)
x = self.attention1(x)
x = self.attention2(x)
x = x.mean(dim=[2, 3])
x = self.fc(x)
return x
# # Grad-CAM
"""
A setup I created when trying to make use of gradcam images in my pipeline,
however they only helped visualise the locations of the trees in the image for analysis and
using this information to mask areas of image or add raw gradcam image as input to model with original
did not prove useful.
Having this function created now is pretty cool though as it can be used during postpressing
with some additional steps to create an object detection visualisation from a pure image regression model.
"""
class GradCAM:
def __init__(self, model, target_layer):
self.model = model
self.target_layer = target_layer
self.feature_maps = None
self.gradient = None
self.hooks = []
self.register_hooks()
def register_hooks(self):
def hook_fn(module, input, output):
self.feature_maps = output.detach()
def backward_hook_fn(module, grad_out, grad_in):
self.gradient = grad_out[0].detach()
for name, module in self.model.named_modules():
if name == self.target_layer:
self.hooks.append(module.register_forward_hook(hook_fn))
self.hooks.append(module.register_backward_hook(backward_hook_fn))
def remove_hooks(self):
for hook in self.hooks:
hook.remove()
def generate_heatmap(self, input_image, return_pred=False):
self.model.zero_grad()
output = self.model(input_image)
pred = output.detach().cpu().numpy()[0]
loss = output[0, 0]
loss.backward(retain_graph=True)
alpha = self.gradient.mean(dim=(2, 3), keepdim=True)
heatmap = (alpha * self.feature_maps).sum(dim=1, keepdim=True)
heatmap = torch.relu(heatmap)
heatmap /= torch.max(heatmap)
heatmap = heatmap.squeeze().cpu().numpy()
heatmap = cv2.resize(heatmap, (input_image.shape[2], input_image.shape[3]))
if return_pred:
return heatmap, pred
return heatmap, None
def generate_batch_heatmap(self, input_images, return_pred=False):
heatmaps = []
preds = []
for input_image in input_images:
# Generate heatmap
heatmap, pred = self.generate_heatmap(
input_image.unsqueeze(0), return_pred=return_pred
)
heatmaps.append(heatmap)
preds.append(pred)
if return_pred:
return heatmaps, preds
else:
return heatmaps
def generate_batch_mask(self, input_images):
masked_imgs = []
for input_image in input_images:
# Generate heatmap
heatmap = self.generate_heatmap(input_image.unsqueeze(0))
# Threshold density map to create binary mask
threshold = 0.001
_, binary_map = cv2.threshold(heatmap, threshold, 255, cv2.THRESH_BINARY)
# Dilate binary map to fill in gaps between trees
kernel = np.ones((5, 5), np.uint8)
dilated_map = cv2.dilate(binary_map, kernel, iterations=2)
# Apply mask to input image
masked_image = input_image.cpu().numpy().transpose((1, 2, 0)).copy()
masked_image[dilated_map == 0] = 0
masked_imgs.append(masked_image)
# masked_imgs_tensor = torch.tensor(masked_imgs).cuda()
return masked_imgs
"""
After training the model on the data save them and enter the path to it here
"""
model_dir = "/kaggle/input/models-stage-2-of-tree-counting/"
IMG_SIZE = 256
"""
Set CALC_GRADCAM to true in CFG to create the gradcams for the entire train and test dataset
"""
if CFG.CALC_GRADCAM:
# define target layer for Grad-CAM
target_layer = "model.stages.2.blocks.26"
FOLDS = [1]
RUN_FOLDS = 1000
all_preds_df, test_all_preds_df = [], []
for VAL_FOLD in FOLDS[:RUN_FOLDS]:
model = PalmTreeCounter().to(device)
model_filename = f"{model_dir}TreeCounterModel_fold_{VAL_FOLD}.pth.tar"
model = load_checkpoint(filename=model_filename, model=model)
model.to(device)
model.eval()
normalize = A.Normalize(
mean=(0.485, 0.456, 0.406), std=(0.229, 0.224, 0.225), max_pixel_value=255
)
val_transform = A.Compose([normalize, ToTensorV2()])
USE_DF = train_df.copy()
img_ds = df_to_datatset(
USE_DF,
kfold=VAL_FOLD,
imsize=IMG_SIZE,
is_train=False,
is_inference=True,
transform=val_transform,
return_ids=True,
)
img_loader = DataLoader(img_ds, batch_size=8, shuffle=False)
saved_heatmaps_folder = f"heatmap_imgs_fold_{VAL_FOLD}"
try:
shutil.rmtree(saved_heatmaps_folder)
except:
pass
os.mkdir(saved_heatmaps_folder)
gradcam = GradCAM(model, target_layer)
# register hooks
gradcam.register_hooks()
preds_df = pd.DataFrame([])
for imgs, img_ids in tqdm(img_loader):
imgs = imgs.to(device)
# generate heatmap
heatmap_images, preds = gradcam.generate_batch_heatmap(
imgs, return_pred=True
)
# save heatmaps
for heatmap_image, pred, image_id in zip(heatmap_images, preds, img_ids):
plt.imsave(saved_heatmaps_folder + "/" + image_id, heatmap_image)
tmp_df = pd.DataFrame([])
tmp_df[f"pred_fold_{VAL_FOLD}"] = pred
tmp_df["image"] = image_id
preds_df = preds_df.append(tmp_df)
all_preds_df.append(preds_df)
gradcam.remove_hooks()
torch.cuda.empty_cache()
gc.collect()
del heatmap_images, preds, img_loader, gradcam
torch.cuda.empty_cache()
gc.collect()
USE_DF = test_df.copy()
img_ds = df_to_datatset(
USE_DF,
kfold=VAL_FOLD,
imsize=IMG_SIZE,
is_train=False,
is_inference=True,
transform=val_transform,
return_ids=True,
)
img_loader = DataLoader(img_ds, batch_size=8, shuffle=False)
saved_heatmaps_folder = f"test_heatmap_imgs_fold_{VAL_FOLD}"
try:
shutil.rmtree(saved_heatmaps_folder)
except:
pass
os.mkdir(saved_heatmaps_folder)
# define target layer for Grad-CAM
target_layer = "model.stages.2.blocks.26"
gradcam = GradCAM(model, target_layer)
# register hooks
gradcam.register_hooks()
preds_df = pd.DataFrame([])
for imgs, img_ids in tqdm(img_loader):
imgs = imgs.to(device)
# generate heatmap
heatmap_images, preds = gradcam.generate_batch_heatmap(
imgs, return_pred=True
)
# save heatmaps
for heatmap_image, pred, image_id in zip(heatmap_images, preds, img_ids):
plt.imsave(saved_heatmaps_folder + "/" + image_id, heatmap_image)
tmp_df = pd.DataFrame([])
tmp_df[f"pred_fold_{VAL_FOLD}"] = pred
tmp_df["image"] = image_id
preds_df = preds_df.append(tmp_df)
test_all_preds_df.append(preds_df)
gradcam.remove_hooks()
torch.cuda.empty_cache()
gc.collect()
del heatmap_images, preds, img_loader, gradcam
torch.cuda.empty_cache()
gc.collect()
df = pd.concat(all_preds_df, axis=1)
df = df.loc[:, ~df.columns.duplicated()]
df = df[sorted(df.columns)]
df = df.merge(train_df[["image", "Target"]], on="image", how="left")
df.to_csv("pred_train_df_dapcc.csv", index=False)
display(df)
df = pd.concat(test_all_preds_df, axis=1)
df = df.loc[:, ~df.columns.duplicated()]
df = df[sorted(df.columns)]
df = df.merge(test_df[["image", "Target"]], on="image", how="left")
df.to_csv("pred_test_df_dapcc.csv", index=False)
display(df)
torch.cuda.empty_cache()
gc.collect()
"""
Look at the names [name] of the various target_layers to choose from when creating gradcam
"""
if CFG.VIS_GRADCAM and CFG.USE_GRADCAM:
VAL_FOLD = 1
model = PalmTreeCounter().to(device)
model_filename = f"{model_dir}TreeCounterModel_fold_{VAL_FOLD}.pth.tar"
model = load_checkpoint(filename=model_filename, model=model)
for name, module in model.named_modules():
print(name)
# define target layer for Grad-CAM
target_layer = "model.stages.2.blocks.26"
if CFG.VIS_GRADCAM and CFG.USE_GRADCAM:
r = 114
t = 12
# Sample an Image with target == t
sample = train_df[train_df["Target"] == t].sample(1, random_state=r)
sampleid_path = sample["image_path"].values[0]
sampleid_target = sample["Target"].values[0]
img = load_image(sampleid_path, IMG_SIZE)
normalize = A.Normalize(
mean=(0.485, 0.456, 0.406), std=(0.229, 0.224, 0.225), max_pixel_value=255
)
val_transform = A.Compose(
[
normalize,
ToTensorV2(),
]
)
input_image = val_transform(image=img)["image"].unsqueeze(0).to(device)
model.to(device)
model.eval()
gradcam = GradCAM(model, target_layer)
# register hooks
gradcam.register_hooks()
# generate heatmap
heatmap, count = gradcam.generate_heatmap(input_image, return_pred=True)
print("Predicted:", count, "\nTarget:", sampleid_target)
gradcam.remove_hooks()
# Threshold density map to create binary mask
threshold = 0.5
_, binary_map = cv2.threshold(heatmap, threshold, 255, cv2.THRESH_BINARY)
# Dilate binary map to fill in gaps between trees
kernel = np.ones((5, 5), np.uint8)
dilated_map = cv2.dilate(binary_map, kernel, iterations=2)
# Apply mask to input image
masked_image = img.copy()
masked_image[dilated_map == 0] = 0
# Display original and transformed images
fig, axs = plt.subplots(1, 3, figsize=(10, 5))
axs[0].imshow(cv2.cvtColor(img, cv2.COLOR_BGR2RGB))
axs[0].set_title("Original")
axs[1].imshow(heatmap, cmap="jet")
axs[1].set_title("Heatmap")
axs[2].imshow(masked_image)
axs[2].set_title("Masked")
plt.show()
# 
# define target layer for Grad-CAM
target_layer = "model.stages.2.blocks.26"
if CFG.VIS_GRADCAM and CFG.USE_GRADCAM:
r = 17
t = 8
VAL_FOLD = 1
model = PalmTreeCounter().to(device)
model_filename = f"{model_dir}TreeCounterModel_fold_{VAL_FOLD}.pth.tar"
model = load_checkpoint(filename=model_filename, model=model)
sample = train_df[train_df[f"kfold_{VAL_FOLD}"] == 1]
sample = sample[sample["Target"] == t].sample(4, random_state=r, replace=False)
orig_images = []
input_images = []
targets = []
for i in range(4):
sampleid_path = sample["image_path"].values[i]
sampleid_target = sample["Target"].values[i]
img = load_image(sampleid_path, IMG_SIZE)
input_image = val_transform(image=img)["image"].to(device)
orig_images.append(img)
input_images.append(input_image)
targets.append(sampleid_target)
input_images = torch.stack(input_images, dim=0).to(device)
model.to(device)
model.eval()
counts = model(input_images).cpu().detach().numpy()
print("Predicted\t:", list(counts.ravel()), "\nTarget\t\t:", targets)
gradcam = GradCAM(model, target_layer)
# register hooks
gradcam.register_hooks()
# generate heatmap
heatmap_images, preds = gradcam.generate_batch_heatmap(
input_images, return_pred=True
)
gradcam.remove_hooks()
# Display original and transformed images
fig, axs = plt.subplots(1, 4, figsize=(12, 4))
for i in range(4):
axs[i].imshow(heatmap_images[i], cmap="jet")
axs[i].set_title(f"{targets[i]} - {counts[i]}")
plt.show()
# Display original and transformed images
fig, axs = plt.subplots(1, 4, figsize=(12, 4))
for i in range(4):
axs[i].imshow(orig_images[i])
axs[i].set_title(f"{targets[i]} - {counts[i]}")
plt.show()
def heatmap_to_pointmap(heatmap, threshold=0.5):
# Threshold the heatmap to identify regions with high activation values
thresholded = (heatmap > threshold).astype(np.uint8)
# Find connected components and their centroids in the thresholded heatmap
num_components, labels, stats, centroids = cv2.connectedComponentsWithStats(
thresholded
)
# Convert centroids to points in original image space
points = []
for i in range(1, num_components):
x, y = centroids[i]
points.append((int(x), int(y)))
return points
if CFG.VIS_GRADCAM and CFG.USE_GRADCAM:
i = 1
gradcam_img = heatmap_images[i].copy()
points = heatmap_to_pointmap(gradcam_img, threshold=0.1)
print(len(points))
# Plot the image with the points as squares
fig, ax = plt.subplots(figsize=(4, 4))
ax.imshow(orig_images[i])
for x, y in points:
rect = plt.Rectangle(
(x - 5, y - 5), 10, 10, linewidth=1, edgecolor="r", facecolor="none"
)
ax.add_patch(rect)
plt.show()
# 
if CFG.VIS_GRADCAM and CFG.USE_GRADCAM:
plt.figure(figsize=(4, 4))
# Convert the image to binary based on a threshold value
binary_img = (gradcam_img > 0.1).astype(np.uint8)
# Get connected components and their stats
num_labels, labels, stats, centroids = cv2.connectedComponentsWithStats(binary_img)
gradcam_img_lines = gradcam_img.copy()
# Loop over the centroids and draw lines between connected components
for i in range(1, num_labels):
for j in range(i + 1, num_labels):
# Calculate distance between centroids
dist = np.sqrt(
(centroids[i][0] - centroids[j][0]) ** 2
+ (centroids[i][1] - centroids[j][1]) ** 2
)
# Draw line between centroids if distance is below threshold
if dist < 384:
cv2.line(
gradcam_img_lines,
(int(centroids[i][0]), int(centroids[i][1])),
(int(centroids[j][0]), int(centroids[j][1])),
(255, 0, 0),
2,
)
# Display the GradCAM image with lines drawn
plt.imshow(gradcam_img_lines)
plt.show()
"""
Prepare the train/test files to work with the saved gradcam images
"""
if CFG.USE_GRADCAM:
grad_cam_dir_root = "/kaggle/input/gradcam-heatmaps/"
train = pd.read_csv(grad_cam_dir_root + "pred_train_df_dapcc.csv")
test = pd.read_csv(grad_cam_dir_root + "pred_test_df_dapcc.csv")
pred_cols = [c for c in train.columns if "pred" in c]
for col in pred_cols:
train.loc[train[col] < 0, col] = 0
test.loc[test[col] < 0, col] = 0
train["image_path"] = train["image"].apply(get_image_path, dir_=IMG_DIR)
test["image_path"] = test["image"].apply(get_image_path, dir_=IMG_DIR)
for VAL_FOLD in FOLDS:
grad_cam_img_path = grad_cam_dir_root + f"heatmap_imgs_fold_{VAL_FOLD}/"
train[f"gradcam_path_fold{VAL_FOLD}"] = train["image"].apply(
get_image_path, dir_=grad_cam_img_path
)
grad_cam_img_path = grad_cam_dir_root + f"test_heatmap_imgs_fold_{VAL_FOLD}/"
test[f"gradcam_path_fold{VAL_FOLD}"] = test["image"].apply(
get_image_path, dir_=grad_cam_img_path
)
display("TRAIN", train, "TEST", test)
if CFG.USE_GRADCAM:
train_df = train_df.merge(
train[["image", "gradcam_path_fold1"]], how="left", on="image"
)
test_df = test_df.merge(
test[["image", "gradcam_path_fold1"]], how="left", on="image"
)
if CFG.USE_GRADCAM:
IMG_SIZE = 256
r = 5
sample = train[train["Target"] == 3].copy()
sample = sample.sample(1, random_state=r, replace=False)
orig_images = []
heatmap_images = []
targets = []
counts = []
i = 1
sampleid_path = sample["image_path"].values[0]
sampleid_target = sample["Target"].values[0]
count = sample[f"pred_fold_{i}"].values[0]
img = load_image(sampleid_path, IMG_SIZE)
gradcam_path = sample[f"gradcam_path_fold{i}"].values[0]
heatmap_image = load_image(gradcam_path, IMG_SIZE)
# Display original and transformed images
fig, axs = plt.subplots(1, 2, figsize=(8, 5))
axs[0].imshow(img)
axs[0].set_title("Original " + f"{sampleid_target} - {count}")
axs[1].imshow(heatmap_image[:, :, :3], cmap="jet")
axs[1].set_title("Heatmap " + f"{sampleid_target} - {count}")
plt.show()
class PalmTreeCounter(nn.Module):
def __init__(self):
super(PalmTreeCounter, self).__init__()
self.model = timm.create_model("convnext_base", pretrained=True)
self.attention1 = SelfAttention(in_channels=1024)
self.attention2 = SelfAttention(in_channels=1024)
self.fc = nn.Linear(1024, 1)
def forward(self, image):
x = self.model.forward_features(image)
x = self.attention1(x)
x = self.attention2(x)
x = x.mean(dim=[2, 3])
x = self.fc(x)
return x
"""
input_tensor = torch.rand(1, 3, 256, 256).to(device)
model_ = PalmTreeCounter().to(device)
print(model_(input_tensor).shape)
del model_,input_tensor
torch.cuda.empty_cache()
gc.collect()
"""
torch.cuda.empty_cache()
gc.collect()
# timm.list_models()
# # Training
"""
Setting all the training parameters
"""
USE_THIS_DF = train_df.copy()
IMG_SIZE = 256
BATCH_SIZE = 40
PAITENCE = 8
RUN_FOLDS = 1000
NUM_EPOCHS = 100
LR = 7e-4
DEBUG = False
if DEBUG:
BATCH_SIZE = 4
NUM_VAL_SAMPLES = 50
NUM_TRAIN_SAMPLES = 50
IMG_SIZE = 256
NUM_EPOCHS = 4
RUN_FOLDS = 1
PAITENCE = 4
df = pd.DataFrame([])
for VAL_FOLD in FOLDS:
df = df.append(
train_df[train_df[f"kfold_{VAL_FOLD}"] == 1].sample(
NUM_VAL_SAMPLES, random_state=1
)
)
df = df.append(
train_df[train_df[f"kfold_{VAL_FOLD}"] != 1].sample(
NUM_TRAIN_SAMPLES, random_state=1
)
)
USE_THIS_DF = df.reset_index(drop=True).copy()
display(USE_THIS_DF.head(5), USE_THIS_DF.shape)
plt.figure(figsize=(4, 2))
USE_THIS_DF["Target"].hist()
plt.show()
normalize = A.Normalize(
mean=(0.485, 0.456, 0.406), std=(0.229, 0.224, 0.225), max_pixel_value=255
)
train_transforms = [
A.Sharpen(p=0.2),
A.OneOf(
[
A.RandomFog(
fog_coef_lower=0.1,
fog_coef_upper=0.5,
alpha_coef=0.15,
always_apply=False,
p=0.3,
),
A.Blur(p=0.2),
],
p=1,
),
A.RandomRotate90(p=0.5),
A.HorizontalFlip(p=0.5),
A.VerticalFlip(p=0.5),
A.CoarseDropout(
max_holes=22,
max_height=15,
max_width=15,
min_holes=16,
min_height=7,
min_width=7,
p=0.4,
),
normalize,
ToTensorV2(),
]
train_transform = A.Compose(train_transforms)
val_transform = A.Compose(
[
normalize,
ToTensorV2(),
]
)
vis_transform = A.Compose(train_transforms[-7:-1])
torch.cuda.empty_cache()
gc.collect()
"""
Visualising the augmentaions
"""
t = 8
r = 8
sample_df = train_df[train_df["Target"] == t].sample(1, random_state=r)
sampleid_path = sample_df["image_path"].values[0]
sampleid_path_target = sample_df["Target"].values[0]
print(sampleid_path_target)
img = load_image(sampleid_path, 256)
# Apply transformations
transformed = vis_transform(image=img)["image"]
if CFG.USE_GRADCAM:
sampleid_path_gcam = sample_df["gradcam_path_fold1"].values[0]
gradcam_img = load_image(sampleid_path_gcam, 256)
# Display original and transformed images
fig, axs = plt.subplots(1, 3, figsize=(9, 3))
axs[0].imshow(img)
axs[0].set_title("Original")
axs[1].imshow(transformed)
axs[1].set_title("Transformed")
axs[2].imshow(gradcam_img, cmap="jet")
axs[2].set_title("Grad-CAM")
plt.tight_layout()
plt.show()
else:
# Display original and transformed images
fig, axs = plt.subplots(1, 2, figsize=(6, 3))
axs[0].imshow(img)
axs[0].set_title("Original")
axs[1].imshow(transformed)
axs[1].set_title("Transformed")
plt.tight_layout()
plt.show()
set_seed(42)
FOLDS = [1]
"""
Start Training
"""
if CFG.TRAIN:
val_df_preds = pd.DataFrame([])
for VAL_FOLD in FOLDS[:RUN_FOLDS]:
if DEBUG:
print("==!!== IN DEBUB MODE ==!!==")
print("\n", "*" * 50, f"\nTRAINING WITH FOLD !={VAL_FOLD}\n", "*" * 50, "\n")
model = PalmTreeCounter().to(device)
opt = optim.AdamW(model.parameters(), lr=LR)
loss_fn = nn.SmoothL1Loss().to(device)
model_filename = f"TreeCounterModel_fold_{VAL_FOLD}.pth.tar"
remove_ids = None
if CFG.REMOVE_BADLABELS:
if VAL_FOLD == 1:
remove_ids = sample_zero_images, badlabels
groud_truth, preds = start_training(
kfold=VAL_FOLD,
train_df=USE_THIS_DF,
train_transform=train_transform,
val_transform=val_transform,
loss_fn=loss_fn,
model=model,
opt=opt,
patience=PAITENCE,
imsize=IMG_SIZE,
batch_size=BATCH_SIZE,
num_epochs=NUM_EPOCHS,
model_filename=model_filename,
remove_ids=remove_ids,
return_gradcam=False,
use_amp=False,
my_shuffle=False,
)
val_df = USE_THIS_DF[USE_THIS_DF[f"kfold_{VAL_FOLD}"] == 1].reset_index(
drop=True
)
if remove_ids != None:
val_remove_ids = remove_ids[1]
if val_remove_ids != None:
val_df = val_df[~val_df["image"].isin(val_remove_ids)].reset_index(
drop=True
)
val_df["pred"] = preds
val_df["groud_truth"] = groud_truth
val_df_preds = val_df_preds.append(val_df)
val_df_preds = (
val_df_preds.groupby("image")
.agg({"pred": "mean", "Target": "mean"})
.reset_index()
)
all_groud_truth = val_df_preds["Target"].tolist()
all_preds = val_df_preds["pred"].tolist()
oof_rmse = rmse(all_groud_truth, all_preds)
print("\nOOF RMSE: ", oof_rmse)
def start_inference(
model,
test_df,
val_transform,
imsize,
batch_size,
pred_name,
resize=None,
return_gradcam=False,
tta=False,
test_transforms=None,
save=True,
):
"""
Inference function that works with Test-Time-Augmentation.
The first TTA is always the original image and predictions on it is saved if save,
as puresub_{pred_name}.csv
"""
def inference_func(loader, model, return_gradcam=False, tt=None, tta=False):
model.eval()
all_preds = np.array([])
with torch.no_grad():
for batch in tqdm(loader):
x = batch
if tta:
if type(tt) != list:
augs = [tt] + [normalize, ToTensorV2()]
else:
augs = tt + [normalize, ToTensorV2()]
if None in augs:
augs.remove(None)
test_transform = A.Compose(augs)
x = x.numpy()
augmented_xs = []
for img in x:
augmented_xs.append(test_transform(image=img)["image"])
augmented_xs = torch.stack(augmented_xs)
preds = (
model(augmented_xs.to("cuda").to(torch.float32)).cpu().numpy()
)
all_preds = np.append(all_preds, preds)
else:
x = x.to("cuda").to(torch.float32)
preds = model(x).cpu()
all_preds = np.append(all_preds, preds)
all_preds[all_preds < 0] = 0
return all_preds
if tta:
val_transform = None
test_ds = df_to_datatset(
test_df,
transform=val_transform,
imsize=imsize,
is_train=False,
is_inference=True,
resize=resize,
return_gradcam=return_gradcam,
)
test_loader = DataLoader(test_ds, batch_size=batch_size, shuffle=False)
all_preds = []
if tta:
num_orig = 1
test_transforms = [None] * num_orig + test_transforms
else:
test_transforms = [None]
for tt in test_transforms:
preds = inference_func(
test_loader, model, return_gradcam=return_gradcam, tt=tt, tta=tta
)
all_preds.append(preds)
for p in all_preds:
print(p[:10])
all_preds_mean = np.mean(all_preds, axis=0)
if save:
test_df["Target"] = all_preds_mean
test_df["ImageId"] = test_df["image"]
test_df[["ImageId", "Target"]].to_csv(f"sub_{pred_name}.csv", index=False)
if tta:
test_df["Target"] = all_preds[0]
test_df["ImageId"] = test_df["image"]
test_df[["ImageId", "Target"]].to_csv(
f"puresub_{pred_name}.csv", index=False
)
return all_preds_mean
"""
Run Test Time Augmentation on the Validation Set to find the ones that
decrease the validation RMSE the most
"""
class CropBorders:
def __init__(self, border_size):
self.border_size = border_size
def __call__(self, image, cols, rows):
img = image
height, width = img.shape[:2]
new_height, new_width = (
height - 2 * self.border_size,
width - 2 * self.border_size,
)
cropped_img = img[
self.border_size : height - self.border_size,
self.border_size : width - self.border_size,
]
padded_img = np.zeros((height, width, img.shape[2]), dtype=img.dtype)
padded_img[
self.border_size : new_height + self.border_size,
self.border_size : new_width + self.border_size,
] = cropped_img
return padded_img
RESIZE = None
BATCH_SIZE = 32
TEST_TIME_AUGMENTS = True
if TEST_TIME_AUGMENTS:
# Define augments to use.
test_transforms = [
A.Lambda(image=CropBorders(border_size=2), p=1),
A.Lambda(image=CropBorders(border_size=4), p=1),
A.Sharpen(p=1),
A.RandomFog(
fog_coef_lower=0.1,
fog_coef_upper=0.5,
alpha_coef=0.15,
always_apply=False,
p=1,
),
A.RandomRotate90(p=1),
A.HorizontalFlip(p=1),
A.VerticalFlip(p=1),
]
test_transforms = test_transforms[:]
else:
test_transforms = None
if CFG.PRED_VAL:
val_df_preds = pd.DataFrame([])
for VAL_FOLD in FOLDS:
# if using model trained in diffent session, change "model_dir" to the
# respective path of the model
model_dir = ""
model_filename = f"{model_dir}TreeCounterModel_fold_{VAL_FOLD}.pth.tar"
model = PalmTreeCounter().to(device)
model = load_checkpoint(filename=model_filename, model=model)
pred_name = f"val_df_preds_fold{VAL_FOLD}"
remove_ids = None
if CFG.REMOVE_BADLABELS:
if VAL_FOLD == 1:
remove_ids = None, badlabels
val_df = USE_THIS_DF[USE_THIS_DF[f"kfold_{VAL_FOLD}"] == 1].reset_index(
drop=True
)
if remove_ids != None:
val_remove_ids = remove_ids[1]
if val_remove_ids != None:
val_df = val_df[~val_df["image"].isin(val_remove_ids)].reset_index(
drop=True
)
preds = start_inference(
model=model,
test_df=val_df,
val_transform=val_transform,
imsize=IMG_SIZE,
resize=RESIZE,
batch_size=BATCH_SIZE,
pred_name=pred_name,
return_gradcam=False,
tta=TEST_TIME_AUGMENTS,
test_transforms=test_transforms,
save=False,
)
groud_truth = val_df["Target"].values
val_rmse = rmse(groud_truth, preds)
print(f"BEST VAL RMSE: {np.round(val_rmse,5)}")
val_df["pred"] = preds
val_df["groud_truth"] = groud_truth
val_df_preds = val_df_preds.append(val_df)
val_df_preds = val_df_preds.reset_index(drop=True)
val_df_preds.to_csv(f"val_df_preds.csv", index=False)
# BEST VAL RMSE: 1.03187
# # Inference
"""
Run Inference on the testset for submission
"""
if CFG.INFER and not DEBUG and CFG.TRAIN:
all_preds = []
best_trainall_preds = []
for VAL_FOLD in FOLDS[:RUN_FOLDS]:
print(
"\n",
"*" * 50,
f"\nRUNNING INFERENCE WITH FOLD !={VAL_FOLD}\n",
"*" * 50,
"\n",
)
model_filename = f"TreeCounterModel_fold_{VAL_FOLD}.pth.tar"
model = PalmTreeCounter().to(device)
pred_name = f"fold_{VAL_FOLD}"
model = load_checkpoint(filename=model_filename, model=model)
preds = start_inference(
model=model,
test_df=test_df,
val_transform=val_transform,
imsize=IMG_SIZE,
resize=RESIZE,
batch_size=BATCH_SIZE,
pred_name=pred_name,
return_gradcam=False,
tta=TEST_TIME_AUGMENTS, # <-- Using the TTA
test_transforms=test_transforms,
save=True,
) # <-- Saving submission preds of each fold
all_preds.append(preds)
if CFG.INFER and not DEBUG and not CFG.TRAIN:
all_preds = []
for VAL_FOLD in FOLDS[:RUN_FOLDS]:
print(
"\n",
"*" * 50,
f"\nRUNNING INFERENCE WITH FOLD !={VAL_FOLD}\n",
"*" * 50,
"\n",
)
# if using model trained in diffent session, change "model_dir" to the
# respective path of the model
model_dir = "/kaggle/input/models-stage-2-of-tree-counting/"
model_filename = f"{model_dir}TreeCounterModel_fold_{VAL_FOLD}.pth.tar"
model = PalmTreeCounter().to(device)
model = load_checkpoint(filename=model_filename, model=model)
pred_name = f"fold_{VAL_FOLD}"
preds = start_inference(
model=model,
test_df=test_df,
val_transform=val_transform,
imsize=IMG_SIZE,
resize=RESIZE,
batch_size=BATCH_SIZE,
pred_name=pred_name,
return_gradcam=False,
tta=TEST_TIME_AUGMENTS,
test_transforms=test_transforms,
save=True,
)
all_preds.append(preds)
"""
Create submission
"""
if CFG.INFER and not DEBUG:
sub_df = test_df.copy()
sub_df["ImageId"] = sub_df["image"]
sub_df = sub_df[["ImageId", "Target"]]
mean_all_preds = np.mean(all_preds, 0)
sub_df["Target"] = mean_all_preds
sub_df.to_csv(f"sub_allfolds.csv", index=False)
display(sub_df)
|
import pandas as pd
import numpy as np
from tqdm import tqdm
from collections import defaultdict
import seaborn as sns
import matplotlib.pyplot as plt
import numpy as np
import matplotlib
tqdm.pandas()
train = pd.read_csv(r"../input/quora-insincere-questions-classification/train.csv")
test = pd.read_csv(r"../input/quora-insincere-questions-classification/test.csv")
train.head()
print(train.shape)
print(test.shape)
train.isna().sum()
train.groupby("target").count()["qid"]
y = [
train.groupby("target").count()["qid"][0],
train.groupby("target").count()["qid"][1],
]
x = [0, 1]
plt.bar(x, y, tick_label=[0, 1])
for i, num in enumerate(y):
plt.annotate(num, (x[i], y[i]))
plt.xlabel("target")
plt.ylabel("count")
plt.show()
from wordcloud import WordCloud, STOPWORDS
def plot_wordcloud(text, title):
stopwords = set(STOPWORDS) | {"one", "br", "Po", "th", "sayi", "fo", "Unknown"}
wordcloud = WordCloud(
background_color="white",
stopwords=stopwords,
max_words=200,
max_font_size=100,
random_state=42,
width=800,
height=400,
)
wordcloud.generate(str(text))
plt.figure(figsize=(10, 8))
plt.imshow(wordcloud)
plt.title(
title, fontdict={"size": 20, "color": "black", "verticalalignment": "bottom"}
)
plt.axis("off")
plt.tight_layout()
sincere_data = train[train["target"] == 0].copy()
insincere_data = train[train["target"] == 1].copy()
plot_wordcloud(sincere_data["question_text"], title="Word Cloud of Sincere Questions")
plot_wordcloud(
insincere_data["question_text"], title="Word Cloud of Insincere Questions"
)
def generate_ngrams(text, n_gram=1):
token = [
token
for token in text.lower().split(" ")
if token != ""
if token not in STOPWORDS
]
ngrams = zip(*[token[i:] for i in range(n_gram)])
return [" ".join(ngram) for ngram in ngrams]
def generate_freq_dict(text, ngram, network=False):
freq_dict = defaultdict(int)
for line in text:
for word in generate_ngrams(line, ngram):
freq_dict[word] += 1
fd_sorted = pd.DataFrame(sorted(freq_dict.items(), key=lambda x: x[1])[::-1])
fd_sorted.columns = ["word", "wordcount"]
if network:
return fd_sorted[:200]
else:
return fd_sorted.head(20)
def drawhist(sincere_text, insincere_text, ngram, first_title, second_title):
fd_data_s = generate_freq_dict(sincere_text, ngram)
fd_data_is = generate_freq_dict(insincere_text, ngram)
fig, axes = plt.subplots(1, 2, figsize=(25, 18))
for fd_data, ax, title in zip(
[fd_data_s, fd_data_is], axes, [first_title, second_title]
):
x = list(fd_data["word"])
y = list(fd_data["wordcount"])
ax.barh(range(len(x)), y[::-1], height=0.7, color="steelblue", alpha=0.8)
ax.set_yticks(range(len(x)))
ax.set_yticklabels(x[::-1])
ax.set_title(title, fontsize=15)
ax.tick_params(axis="both", labelsize=15)
plt.show()
first_title = "Words' frequency of sincere questions"
second_title = "Words' frequency of insincere questions"
drawhist(
sincere_data["question_text"],
insincere_data["question_text"],
1,
first_title,
second_title,
)
first_title = "Words' frequency of train data"
second_title = "Words' frequency of test data"
drawhist(train["question_text"], test["question_text"], 1, first_title, second_title)
first_title = "Bigrams' frequency of sincere questions"
second_title = "Bigrams' frequency of insincere questions"
drawhist(
sincere_data["question_text"],
insincere_data["question_text"],
2,
first_title,
second_title,
)
first_title = "Trigrams' frequency of sincere questions"
second_title = "Trigrams' frequency of insincere questions"
drawhist(
sincere_data["question_text"],
insincere_data["question_text"],
3,
first_title,
second_title,
)
import networkx as nx
def draw_network(data):
draw_data = generate_freq_dict(data, 2, network=True)["word"]
plt.figure(figsize=(20, 20))
network = [i.split(" ") for i in draw_data]
G = nx.Graph()
for i in network:
G.add_edge(i[0], i[1])
nx.draw_networkx(
G,
pos=nx.spring_layout(G),
node_size=20,
with_labels=True,
font_size=12,
width=1,
style="dashed",
)
plt.show()
draw_network(sincere_data["question_text"])
draw_network(insincere_data["question_text"])
train["length"] = train["question_text"].progress_apply(len)
train.head()
import string
import re
import spacy
from spacy.lang.en.stop_words import STOP_WORDS
from spacy.lang.en import English
regex = re.compile("[" + re.escape(string.punctuation) + "0-9\\r\\t\\n]")
stopwords = list(STOP_WORDS)
parser = English()
def filter_stopwords_punctuation(line):
words = parser(line)
filter_stop_punc = [
word.lemma_.lower().strip() if word.lemma_ != "-PRON-" else word.lower_
for word in words
]
filter_stop_punc = [
word
for word in filter_stop_punc
if word not in stopwords and not regex.match(word)
]
return (" ".join(filter_stop_punc), len(words), len(filter_stop_punc))
train["filter_stop_punc"], train["origin_word_count"], train["filter_word_count"] = zip(
*train["question_text"].progress_apply(filter_stopwords_punctuation)
)
test["length"] = test["question_text"].progress_apply(len)
test["filter_stop_punc"], test["origin_word_count"], test["filter_word_count"] = zip(
*test["question_text"].progress_apply(filter_stopwords_punctuation)
)
import matplotlib.pyplot as plt
import seaborn as sns
sincere = train[train["target"] == 0].copy()
insincere = train[train["target"] == 1].copy()
sincere.head()
f, (ax1, ax2) = plt.subplots(2, 1, figsize=(12, 6))
sns.boxplot(data=sincere, x="length", ax=ax1).set_title("Sincere Questions")
sns.boxplot(data=insincere, x="length", ax=ax2).set_title("Insincere Questions")
plt.show()
f, (ax1, ax2) = plt.subplots(1, 2, figsize=(12, 6))
sns.histplot(data=sincere, x="length", kde=True, ax=ax1).set_title("Sincere Questions")
ax1.set_xlim(0, 200)
sns.histplot(data=insincere, x="length", kde=True, ax=ax2).set_title(
"Insincere Questions"
)
ax2.set_xlim(0, 200)
plt.show()
f, (ax1, ax2) = plt.subplots(1, 2, figsize=(12, 6))
sns.histplot(data=sincere, x="origin_word_count", kde=True, ax=ax1).set_title(
"Sincere Questions"
)
sns.histplot(data=insincere, x="origin_word_count", kde=True, ax=ax2).set_title(
"Insincere Questions"
)
plt.show()
f, (ax1, ax2) = plt.subplots(1, 2, figsize=(12, 6))
sns.histplot(data=sincere, x="filter_word_count", kde=True, ax=ax1).set_title(
"Sincere Questions"
)
sns.histplot(data=insincere, x="filter_word_count", kde=True, ax=ax2).set_title(
"Insincere Questions"
)
plt.show()
f, (ax1, ax2) = plt.subplots(2, 1, figsize=(12, 6), sharex=True)
sns.boxplot(data=sincere, x="origin_word_count", ax=ax1).set_title("Sincere Questions")
sns.boxplot(data=insincere, x="origin_word_count", ax=ax2).set_title(
"Insincere Questions"
)
plt.show()
f, (ax1, ax2) = plt.subplots(1, 2, figsize=(12, 6))
sns.histplot(
sincere["filter_word_count"] / sincere["origin_word_count"], kde=True, ax=ax1
).set_title("Sincere Questions")
sns.histplot(
insincere["filter_word_count"] / insincere["origin_word_count"], kde=True, ax=ax2
).set_title("Insincere Questions")
plt.show()
f, (ax1, ax2) = plt.subplots(2, 1, figsize=(12, 6), sharex=True)
sns.boxplot(x=sincere["filter_word_count"], ax=ax1).set_title("Sincere Questions")
sns.boxplot(x=insincere["filter_word_count"], ax=ax2).set_title("Insincere Questions")
plt.show()
f, (ax1, ax2) = plt.subplots(2, 1, figsize=(12, 6), sharex=True)
sns.boxplot(
x=sincere["filter_word_count"] / sincere["origin_word_count"], ax=ax1
).set_title("Sincere Questions")
sns.boxplot(
x=insincere["filter_word_count"] / insincere["origin_word_count"], ax=ax2
).set_title("Insincere Questions")
plt.show()
sincere["origin_word_count"].describe()[1:]
insincere["origin_word_count"].describe()[1:]
sincere["filter_word_count"].describe()[1:]
insincere["filter_word_count"].describe()[1:]
traindata = train.copy()
testdata = test.copy()
label = traindata["target"].to_numpy()
from sklearn.metrics import f1_score, accuracy_score, precision_score, recall_score
from sklearn.model_selection import KFold
from sklearn.feature_extraction.text import TfidfVectorizer, CountVectorizer
# tf_vectorizer=TfidfVectorizer()
# tf_vectorizer.fit(traindata.append(testdata).question_text)
cv_vectorizer = CountVectorizer()
cv_vectorizer.fit(traindata.append(testdata).question_text)
# train_vec_tf = tf_vectorizer.transform(traindata.question_text)
# test_vec_tf = tf_vectorizer.transform(testdata.question_text)
train_vec_cv = cv_vectorizer.transform(traindata.question_text)
test_vec_cv = cv_vectorizer.transform(testdata.question_text)
from sklearn.cluster import KMeans
sincere_cv = cv_vectorizer.transform(sincere.question_text)
kmeans = KMeans()
sincere["cluster"] = kmeans.fit(sincere_cv).labels_
sincere.groupby("cluster").count()
insincere_cv = cv_vectorizer.transform(insincere.question_text)
kmeans = KMeans()
insincere["cluster"] = kmeans.fit(insincere_cv).labels_
insincere.groupby("cluster").count()
from sklearn.naive_bayes import MultinomialNB, ComplementNB, BernoulliNB
def split_and_train(train_data, label_data, test_data, nsplit):
kfold = KFold(n_splits=nsplit, shuffle=True, random_state=54142)
train_prob_preds = np.zeros(train_data.shape[0])
test_predict = np.zeros(test_data.shape[0])
for train_idx, valid_idx in tqdm(kfold.split(train_data), total=nsplit):
classifier = MultinomialNB()
x_train, x_valid = train_data[train_idx], train_data[valid_idx]
y_train, y_valid = label_data[train_idx], label_data[valid_idx]
classifier.fit(x_train, y_train)
train_prob_preds[valid_idx] = classifier.predict_proba(x_valid)[:, 1]
test_predict += (1 / nsplit) * classifier.predict_proba(test_data)[:, 1]
return train_prob_preds, test_predict
train_prob_preds_cv, test_prob_preds_cv = split_and_train(
train_vec_cv, label, test_vec_cv, 5
)
print("******* cv *************")
pred_train = (train_prob_preds_cv > 0.27).astype(np.int)
print(f"acc: {accuracy_score(label, pred_train)}")
print(f"precistion: {precision_score(label, pred_train)}")
print(f"f1 score: {f1_score(label, pred_train)}")
print(f"recall: {recall_score(label, pred_train)}")
import scipy.sparse
train_vec_cv_t = scipy.sparse.hstack(
(train_vec_cv, traindata[["length", "origin_word_count", "filter_word_count"]])
).tocsr()
test_vec_cv_t = scipy.sparse.hstack(
(test_vec_cv, testdata[["length", "origin_word_count", "filter_word_count"]])
).tocsr()
train_prob_preds_cv, test_prob_preds_cv = split_and_train(
train_vec_cv_t, label, test_vec_cv_t, 5
)
print("******* cv *************")
pred_train = (train_prob_preds_cv > 0.65).astype(np.int)
print(f"acc: {accuracy_score(label, pred_train)}")
print(f"precistion: {precision_score(label, pred_train)}")
print(f"f1 score: {f1_score(label, pred_train)}")
print(f"recall: {recall_score(label, pred_train)}")
submission = pd.DataFrame.from_dict({"qid": test["qid"]})
submission["prediction"] = (test_prob_preds_cv > 0.6).astype(np.int)
submission.to_csv("submission.csv", index=False)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
from sklearn.decomposition import PCA
from sklearn.cluster import KMeans
from sklearn.preprocessing import MinMaxScaler, OneHotEncoder, StandardScaler
from sklearn.decomposition import FastICA
# from sklearn.random_projection import SparseRandomProjection
from sklearn.feature_selection import SelectFromModel
from sklearn.metrics import silhouette_score
from sklearn.random_projection import GaussianRandomProjection
from sklearn.metrics import classification_report, confusion_matrix
from sklearn.metrics import (
roc_curve,
roc_auc_score,
precision_score,
recall_score,
f1_score,
)
from sklearn.metrics import make_scorer, accuracy_score
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
df = pd.read_csv("/kaggle/input/hungpn5-lead-5-dy/lead5day.csv")
df.info()
list_col_2 = [
"NO_PHAI_TRA",
"VCSH",
"DOANH_THU_THUAN",
"LNST",
"TIEN_VA_TUONG_DUONG_TIEN",
"CHI_PHI_LAI_VAY",
"NGAY_DKTD",
"NGAY_TL",
"TEN_TINH",
"NNKD_CHINH",
"VON",
"SO_LUONG_LAO_DONG",
"LOAI_HINH_DN",
"TÀI SẢN",
"I# Tiền và các khoản tương đương tiền",
"IV# Hàng tồn kho",
"- giá trị hao mòn lũy kế",
"TỔNG CỘNG TÀI SẢN ",
"3# Thuế và các khoản phải nộp Nhà nước",
"TỔNG CỘNG NGUỒN VỐN ",
"1# Doanh thu bán hàng và cung cấp dịch vụ",
"3# Doanh thu thuần về bán hàng và cung cấp dịch vụ ",
"4# Giá vốn hàng bán",
"5# Lợi nhuận gộp về bán hàng và cung cấp dịch vụ ",
"6# Doanh thu hoạt động tài chính",
"7# Chi phí tài chính",
"- Trong đó: Chi phí lãi vay",
"I# Lưu chuyển tiền từ hoạt động kinh doanh",
"Lưu chuyển tiền thuần từ hoạt động kinh doanh",
"II# Lưu chuyển tiền từ hoạt động đầu tư",
"III# Lưu chuyển tiền từ hoạt động tài chính",
"3#Tiền thu từ đi vay",
"Lưu chuyển tiền thuần từ hoạt động tài chính",
"Lưu chuyển tiền thuần trong kỳ ",
"1# Lợi nhuận trước thuế",
]
data = df[list_col_2]
data.info()
data.corr()
# Correlation Plot
plt.figure(figsize=(14, 14))
sns.set(font_scale=0.6)
sns.heatmap(data.corr(), cmap="GnBu_r", annot=True, square=True, linewidths=0.5)
plt.title("Variable Correlation")
def label_race(row):
if row["CHI_PHI_LAI_VAY"] > 0:
return 1
else:
return 0
data["LOAN"] = data.apply(lambda row: label_race(row), axis=1)
|
# # Importing Required Packages
import numpy as np
import seaborn as sns
from matplotlib import pyplot as plt
import pandas as pd
# # Simulation
# data=
P_psi = 1 * 14.6959
# Seperator K Values
#
def val_K(P): # write a function to find k values
K = None
if P >= 1 and P < 15:
K = 0.1821 + 0.00 * P + 0.0460 * np.log(P)
elif P <= 15 and P < 40:
K = 0.35
else:
K = 0.430 - 0.023 * np.log(P)
return K
# Determine constant Terminal Vapor velocity
#
rL, rV = 0.895810189026424 * 1e3, 0.00189077530801796 * 1e3
UT = val_K(14.6959) * np.sqrt((rL - rV) / rL)
UT
# Determine constant Design Vapor velocity
UV = 0.75 * UT
UV
# Calculate Vapor disengagement diameter.
MV = 4910.6109721109
QV = (MV / 3600) / rV
QV
Dv = np.sqrt((4 * QV) / (np.pi * UV))
Dv
# Calculate Liquid volumetric flow rate
ML = 12281.7825333688 / 3600
QL = ML / rL
QL
# **Calculate liquid holdup and surge times**
# 1. Feed to column.
# 2. Feed to other drum or tankage with pump or exchanger.
# 3. Feed to other drum or tankage without pump.
# 4. Feed to fired heater.
def L_HT(n):
time = None
if n == 1:
time = 5 * 60
elif n == 2:
time = 5 * 60
elif n == 3:
time = 2 * 60
elif n == 4:
time = 10 * 60
return time
def S_T(n):
time = None
if n == 1:
time = 3 * 60
elif n == 2:
time = 2 * 60
elif n == 3:
time = 1 * 60
elif n == 4:
time = 3 * 60
return time
# Calculate Holdup volume
VH = L_HT(1) * QL
VH
# Calculate Surge volume
VS = S_T(1) * QL
VS
Dv_inch = Dv * 39.3701
Dv_inch
def H_LLL(D, P):
D = D / 12
HLLL = None # D=Dv_inch
if D <= 8 and P < 300:
HLLL = 15 * 0.0254
elif D <= 8 and P > 300:
HLLL = 6 * 0.0254
elif D > 8 and D <= 16:
HLLL = 6 * 0.0254
return HLLL
print(H_LLL(Dv_inch, P_psi))
# Calculate the height from low liquid level to normal liquid level
H_H = VH / ((np.pi / 4) * (Dv**2))
H_H
# Calculate the height from normal liquid level to high liquid level
H_S = VS / ((np.pi / 4) * (Dv**2))
H_S
# Calculate the height froin high liquid level to the centerlihe of the inlet nozzle:
rM = 0.0065849750620396 * 1000
MM = 17192.3935054797 / 3600
Qm = MM / rM
dn = np.sqrt((4 * Qm) / ((60 * np.pi) / rM))
H_LIN = 12 * 0.0254
|
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Load the data into a Pandas dataframe
data = pd.read_csv(
"/kaggle/input/employee-names-salaries-and-position-titles/Current_Employee_Names__Salaries__and_Position_Titles.csv"
)
data.head()
# Filter the data to only include employees with a "Salary" pay frequency
salary_data = data[data["Salary or Hourly"] == "SALARY"]
# Create a histogram of the annual salary rates
sns.histplot(data=salary_data, x="Annual Salary", bins=30, kde=True)
plt.show()
# Filter the data to only include employees with an "Hourly" pay frequency
hourly_data = data[data["Salary or Hourly"] == "HOURLY"]
# Create a histogram of the hourly rates
sns.histplot(data=hourly_data, x="Hourly Rate", bins=30, kde=True)
plt.show()
# Count the number of employees in each Full or Part-Time category
count_data = data["Full or Part-Time"].value_counts()
# Create a bar plot of the Full or Part-Time counts
sns.barplot(x=count_data.index, y=count_data.values)
# Add labels to the plot
plt.title("Number of Employees by Full or Part-Time Status")
plt.xlabel("Full or Part-Time Status")
plt.ylabel("Number of Employees")
# Display the plot
plt.show()
# Count the number of employees in each Salary or Hourly category
count_data = data["Salary or Hourly"].value_counts()
# Create a pie chart of the Salary or Hourly counts
plt.figure(figsize=(7, 7))
plt.pie(count_data.values, labels=count_data.index, autopct="%1.1f%%")
plt.title("Salary or Hourly Distribution")
plt.show()
# Filter data to only include hourly employees
hourly_data = data[data["Salary or Hourly"] == "HOURLY"]
# Create a histogram of Typical Hours column
sns.histplot(hourly_data, x="Typical Hours", bins=20)
plt.show()
# Count the number of employees in each department
count_data = data["Department"].value_counts()
# Create a horizontal bar chart of the department counts
plt.figure(figsize=(10, 6))
sns.barplot(x=count_data.values, y=count_data.index)
plt.title("Employee Count by Department")
plt.xlabel("Count")
plt.ylabel("Department")
plt.show()
# Filter data to only include SALARY employees
salary_data = data[data["Salary or Hourly"] == "SALARY"]
# Calculate the average Annual Salary for each department
avg_salary_by_dept = (
salary_data.groupby("Department")["Annual Salary"]
.mean()
.sort_values(ascending=False)
)
# Create a horizontal bar chart of the average salary by department
plt.figure(figsize=(10, 6))
sns.barplot(x=avg_salary_by_dept.values, y=avg_salary_by_dept.index)
plt.title("Average Salary by Department (SALARY Employees Only)")
plt.xlabel("Average Salary ($)")
plt.ylabel("Department")
plt.show()
# Sort the data by Annual Salary and take the top 10 records
top_salaries = salary_data.sort_values("Annual Salary", ascending=False).head(10)
# Create a bar chart of the top 10 salaries by job title
plt.figure(figsize=(10, 6))
sns.barplot(x="Annual Salary", y="Job Titles", data=top_salaries)
plt.title("Top 10 Annual Salaries (SALARY Employees Only)")
plt.xlabel("Annual Salary ($)")
plt.ylabel("Job Title")
plt.show()
# Sort the data by Annual Salary and take the bottom records
bottom_salaries = salary_data.sort_values("Annual Salary", ascending=True).head(50)
# Create a horizontal bar chart of the bottom salaries by job title
plt.figure(figsize=(10, 6))
sns.barplot(x="Annual Salary", y="Job Titles", data=bottom_salaries)
plt.title("Bottom Annual Salaries (SALARY Employees Only)")
plt.xlabel("Annual Salary ($)")
plt.ylabel("Job Title")
plt.show()
# Filter data to only include HOURLY employees
hourly_data = data[data["Salary or Hourly"] == "HOURLY"]
# Calculate the average Hourly Rate for each department
avg_hourly_rate_by_dept = (
hourly_data.groupby("Department")["Hourly Rate"].mean().sort_values(ascending=False)
)
# Create a horizontal bar chart of the average hourly rate by department
plt.figure(figsize=(10, 6))
sns.barplot(x=avg_hourly_rate_by_dept.values, y=avg_hourly_rate_by_dept.index)
plt.title("Average Hourly Rate by Department (HOURLY Employees Only)")
plt.xlabel("Average Hourly Rate ($)")
plt.ylabel("Department")
plt.show()
# Sort the data by Hourly Rate and take the top records
top_hourly_rates = hourly_data.sort_values("Hourly Rate", ascending=False).head(50)
# Create a horizontal bar chart of the top hourly rates by job title
plt.figure(figsize=(10, 6))
sns.barplot(x="Hourly Rate", y="Job Titles", data=top_hourly_rates)
plt.title("Top Hourly Rates (HOURLY Employees Only)")
plt.xlabel("Hourly Rate ($)")
plt.ylabel("Job Title")
plt.show()
# Sort the data by Hourly Rate and take the bottom records
bottom_hourly_rates = hourly_data.sort_values("Hourly Rate", ascending=True).head(50)
# Create a horizontal bar chart of the bottom hourly rates by job title
plt.figure(figsize=(10, 6))
sns.barplot(x="Hourly Rate", y="Job Titles", data=bottom_hourly_rates)
plt.title("Bottom Hourly Rates (HOURLY Employees Only)")
plt.xlabel("Hourly Rate ($)")
plt.ylabel("Job Title")
plt.show()
|
# # The Best Cities In The World For A Workation
# ---
# * author: Kornkanok Klinsumalee
# * e-mail: [email protected]
# * [Chulalongkorn Business School, Thailand](https://www.cbs.chula.ac.th/en/home/)
# ---
# * Source : https://www.holidu.co.uk/magazine/the-best-cities-for-a-workation
# * Data source : https://www.kaggle.com/datasets/prasertk/the-best-cities-for-a-workation
# * Image source : https://holidu-magazine.s3.eu-west-1.amazonaws.com/wp-content/uploads/20210819124230/mikey-harris-kw0z6RyvC0s-unsplash-min-scaled.jpg
# ---
# คุณอาจเบื่อการทำงานจากบ้านหรือร้านกาแฟใกล้บ้านของคุณหรือไม่? ลองคิดถึงเดินทางไปเมืองอื่น ๆ เพื่อสัมผัสประสบการณ์การทำงานร่วมพร้อมกับการพักผ่อนในที่ต่าง ๆ โดยการทำงานร่วมกับการเดินทางนี้เรียกว่า "workation" ซึ่งเป็นการเดินทางเพื่อทำงานโดยรวมกับการผ่อนคลายในสถานที่ใหม่ ๆ พร้อมสำรวจประสบการณ์ใหม่ ๆ ในสถานที่นั้น ๆ โดยยังคงทำงานปรกติตามกำหนดเวลาทุกวัน
# ดังนั้นเราจึงรวบรวมดัชนี 'Workation Index' ในการจัดอันดับเมืองที่ดีที่สุดทั่วโลกในการทำงาน
# ---
# # Import libraries
import sys
import numpy as np
import pandas as pd
import matplotlib
import matplotlib.pyplot as plt
import seaborn as sns
print(f"python version: {sys.version}")
print(f"pandas version: {pd.__version__}")
print(f"matplotlib version: {matplotlib.__version__}")
print(f"seaborn version: {sns.__version__}")
#
#
# Import module
# Description
#
#
# Pandas
# This module is a Python library for data manipulation and analysis.
#
#
# Seaborn
# This module is a Python library for statistical data visualization.
#
#
# Numpy
# This module is a Python library for numerical computing.
#
#
# Matplotlib
# This module is a Python library for creating static, interactive, and animated visualizations.
#
#
# ipywidgets
# This module is a Python library for creating interactive user interfaces in Jupyter notebooks.
#
# ---
# # Read Data
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
path = os.path.join(dirname, filename)
df = pd.read_csv(path)
df.head()
# ---
# # Exploratory Data Analysis
df.info()
df.describe()
df.columns
df.corr()
# We will utilize numeric and float data types as the features for Create Cluster Model.
col = [
"Remote connection: Average WiFi speed (Mbps per second)",
"Co-working spaces: Number of co-working spaces",
"Caffeine: Average price of buying a coffee",
"Travel: Average price of taxi (per km)",
"After-work drinks: Average price for 2 beers in a bar",
"Accommodation: Average price of 1 bedroom apartment per month",
"Food: Average cost of a meal at a local, mid-level restaurant",
"Climate: Average number of sunshine hours",
"Tourist attractions: Number of ‘Things to do’ on Tripadvisor",
"Instagramability: Number of photos with #",
]
# ---
# ## Plot the pair plot
sns.pairplot(df, vars=col, plot_kws={"alpha": 0.4})
# ---
# ## Plot the histogram
# > Plot การกระจายตัวข้อมูลแต่ละ columns เพื่อดูว่าข้อมูลในแต่ละ columns มีลักษณะเป็นอย่างไร
# > จะเห็นได้ว่า ข้อมูลส่วนใหญ่ **เบ้ขวา**
df[col].hist(layout=(2, 5), figsize=(5 * 10, 15))
# ---
# # Data Preprocessing
# > จากกราฟด้านบนจะเห็นว่า ข้อมูลมีการเบ้ และไม่พร้อมสำหรับการใช้งาน
# > จึงต้องเตรียมข้อมูลให้อยู่ในรูปแบบเดียวกัน
from sklearn import preprocessing
from sklearn.cluster import KMeans
from sklearn.metrics import silhouette_samples, silhouette_score
from yellowbrick.cluster import SilhouetteVisualizer
pt = preprocessing.PowerTransformer(
method="yeo-johnson", standardize=True
) # support only positive value
mat = pt.fit_transform(df[col])
mat[:5].round(4)
df_model = pd.DataFrame(mat, columns=col)
df_model.head()
# ## After Data Preprocessing
# > จากการปรับปรุงข้อมูล จะทำให้เห็นว่า กราฟจะอยู่ในรูป Normal (ระฆังคว่ำ)
df_model[col].hist(
layout=(2, 5),
figsize=(5 * 10, 15),
color="green",
)
# ---
# # Optimal Number of Clusters
# จาก Elbow Medthod และ Silhouette Medthod จำแสดงให้เห็นว่า optimal number of clusters = 2
#
def elbow_plot(X, from_k=2, to_k=5):
"""
plot elbow chart to help determining optimal number of clusters
"""
ssd = []
for k in range(from_k, to_k + 1):
m = KMeans(n_clusters=k)
m.fit(X)
ssd.append([k, m.inertia_])
dd = pd.DataFrame(ssd, columns=["k", "ssd"])
dd["pct_chg"] = dd["ssd"].pct_change() * 100
plt.plot(dd["k"], dd["ssd"], linestyle="--", marker="o")
for index, row in dd.iterrows():
plt.text(
row["k"] + 0.02, row["ssd"] + 0.02, f'{row["pct_chg"]:.2f}', fontsize=12
)
def sil_score(X, from_k=2, to_k=6):
"""
calculate silhouette score for k clusters
"""
sils = []
for k in range(from_k, to_k + 1):
m = KMeans(n_clusters=k)
m.fit(X)
silhouette_avg = silhouette_score(X, m.labels_).round(4)
sils.append([silhouette_avg, k])
return sils
X = df_model[col]
elbow_plot(X, from_k=1, to_k=10)
ss = sil_score(X, 2, 10)
print(f"scores = {ss}")
print(f"optimal number of clusters = {max(ss)[1]}")
# # Cluster Model
# > จาก medthod ด้านบน จะพบว่า optimal number of cluster = 2
model = KMeans(n_clusters=2)
model
X = df_model[col]
model.fit(X)
model.labels_
df["cluster"] = model.labels_
df.head(10)
# ---
# # Data Visualization
# > Violinplot เพื่อดูพฤติกรรมของข้อมูล ในแต่ละ cluster
fig, ax = plt.subplots(nrows=2, ncols=5, figsize=(20, 9), sharex=True)
ax = ax.ravel()
for i, col in enumerate(col):
sns.violinplot(x="cluster", y=col, data=df, ax=ax[i])
df[df["Country"] == "Italy"]
|
# # **How to become rich without earning money ?**
# > ## *By contributing in such competition..*
import numpy as np
import pandas as pd
from scipy.stats import linregress
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
sample_submission = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/example_test_files/sample_submission.csv"
)
test = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/example_test_files/test.csv"
)
test_peptides = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/example_test_files/test_peptides.csv"
)
test_proteins = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/example_test_files/test_proteins.csv"
)
supplemental_clinical_data = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/supplemental_clinical_data.csv"
)
train_clinical_data = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/train_clinical_data.csv"
)
train_peptides = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/train_peptides.csv"
)
train_proteins = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/train_proteins.csv"
)
sample_submission
test
test_peptides["group_key"]
test_proteins
test_proteins[test_proteins["visit_id"] == "3342_0"]
supplemental_clinical_data
supplemental_clinical_data[supplemental_clinical_data["visit_id"] == "3342_0"]
# # Feature engineering
# ### Here we create a new dataframe where UPDRSs slopes are stored for each patient
# ### When the slope is negative this means that the UPDRS is going downward along visits, the patient can be considered as being in a good process of improvement
train_clinical_data
def calculate_slope(group):
cols = ["updrs_1", "updrs_2", "updrs_3", "updrs_4"]
slopes = []
for col in cols:
res = linregress(group["visit_month"], group[col])
slopes.append(res.slope)
return pd.Series(slopes, index=[c for c in cols])
slope_by_id = train_clinical_data.groupby("patient_id", as_index=False).apply(
calculate_slope
)
slope_by_id.head(20)
slope_by_id
# # Classes engineering
slope_by_id["updrs_1_good"] = [int(x <= 0) for x in slope_by_id["updrs_1"]]
slope_by_id["updrs_2_good"] = [int(x <= 0) for x in slope_by_id["updrs_2"]]
slope_by_id["updrs_3_good"] = [int(x <= 0) for x in slope_by_id["updrs_3"]]
slope_by_id["updrs_4_good"] = [int(x <= 0) for x in slope_by_id["updrs_4"]]
slope_by_id["overall_score"] = (
slope_by_id["updrs_1_good"]
+ slope_by_id["updrs_2_good"]
+ slope_by_id["updrs_3_good"]
+ slope_by_id["updrs_4_good"]
)
slope_by_id
# #### Taking the four scores we have 2 x 2 x 2 x 2 = 16 combinations of possible classes,
# #### Following are notation examples used for each class
# #### class_0_0_0_0 : is the class of patients with any improvements over time has been observed
# #### class_1_0_0_0 : is the class of patients recorded a good updrs_1 value over time
# #### class_1_1_0_0 : is the class of patients recorded a good value on both updrs_1 and updrs_2 ratios
# #### class_0_1_1_0 : is patients class with improvement on both updrs_2 and updrs_3 ratios
# #### class_1_1_1_1 : This class is of patient making improvement on all the four ratios
slope_by_id["class_0_0_0_0"] = [
int((w == 0) and (x == 0) and (y == 0) and (z == 0))
for (w, x, y, z) in zip(
slope_by_id["updrs_1_good"],
slope_by_id["updrs_2_good"],
slope_by_id["updrs_3_good"],
slope_by_id["updrs_4_good"],
)
]
slope_by_id["class_1_0_0_0"] = [
int((w == 1) and (x == 0) and (y == 0) and (z == 0))
for (w, x, y, z) in zip(
slope_by_id["updrs_1_good"],
slope_by_id["updrs_2_good"],
slope_by_id["updrs_3_good"],
slope_by_id["updrs_4_good"],
)
]
slope_by_id["class_0_1_0_0"] = [
int((w == 0) and (x == 1) and (y == 0) and (z == 0))
for (w, x, y, z) in zip(
slope_by_id["updrs_1_good"],
slope_by_id["updrs_2_good"],
slope_by_id["updrs_3_good"],
slope_by_id["updrs_4_good"],
)
]
slope_by_id["class_0_0_1_0"] = [
int((w == 0) and (x == 0) and (y == 1) and (z == 0))
for (w, x, y, z) in zip(
slope_by_id["updrs_1_good"],
slope_by_id["updrs_2_good"],
slope_by_id["updrs_3_good"],
slope_by_id["updrs_4_good"],
)
]
slope_by_id["class_0_0_0_1"] = [
int((w == 0) and (x == 0) and (y == 0) and (z == 1))
for (w, x, y, z) in zip(
slope_by_id["updrs_1_good"],
slope_by_id["updrs_2_good"],
slope_by_id["updrs_3_good"],
slope_by_id["updrs_4_good"],
)
]
slope_by_id["class_1_1_0_0"] = [
int((w == 1) and (x == 1) and (y == 0) and (z == 0))
for (w, x, y, z) in zip(
slope_by_id["updrs_1_good"],
slope_by_id["updrs_2_good"],
slope_by_id["updrs_3_good"],
slope_by_id["updrs_4_good"],
)
]
slope_by_id["class_1_0_1_0"] = [
int((w == 1) and (x == 0) and (y == 1) and (z == 0))
for (w, x, y, z) in zip(
slope_by_id["updrs_1_good"],
slope_by_id["updrs_2_good"],
slope_by_id["updrs_3_good"],
slope_by_id["updrs_4_good"],
)
]
slope_by_id["class_1_0_0_1"] = [
int((w == 1) and (x == 0) and (y == 0) and (z == 1))
for (w, x, y, z) in zip(
slope_by_id["updrs_1_good"],
slope_by_id["updrs_2_good"],
slope_by_id["updrs_3_good"],
slope_by_id["updrs_4_good"],
)
]
slope_by_id["class_1_1_1_0"] = [
int((w == 1) and (x == 1) and (y == 1) and (z == 0))
for (w, x, y, z) in zip(
slope_by_id["updrs_1_good"],
slope_by_id["updrs_2_good"],
slope_by_id["updrs_3_good"],
slope_by_id["updrs_4_good"],
)
]
slope_by_id["class_1_1_0_1"] = [
int((w == 1) and (x == 1) and (y == 0) and (z == 1))
for (w, x, y, z) in zip(
slope_by_id["updrs_1_good"],
slope_by_id["updrs_2_good"],
slope_by_id["updrs_3_good"],
slope_by_id["updrs_4_good"],
)
]
slope_by_id["class_0_1_1_0"] = [
int((w == 0) and (x == 1) and (y == 1) and (z == 0))
for (w, x, y, z) in zip(
slope_by_id["updrs_1_good"],
slope_by_id["updrs_2_good"],
slope_by_id["updrs_3_good"],
slope_by_id["updrs_4_good"],
)
]
slope_by_id["class_0_1_0_1"] = [
int((w == 0) and (x == 1) and (y == 0) and (z == 1))
for (w, x, y, z) in zip(
slope_by_id["updrs_1_good"],
slope_by_id["updrs_2_good"],
slope_by_id["updrs_3_good"],
slope_by_id["updrs_4_good"],
)
]
slope_by_id["class_1_0_0_1"] = [
int((w == 1) and (x == 0) and (y == 0) and (z == 1))
for (w, x, y, z) in zip(
slope_by_id["updrs_1_good"],
slope_by_id["updrs_2_good"],
slope_by_id["updrs_3_good"],
slope_by_id["updrs_4_good"],
)
]
slope_by_id["class_1_0_1_0"] = [
int((w == 1) and (x == 0) and (y == 1) and (z == 0))
for (w, x, y, z) in zip(
slope_by_id["updrs_1_good"],
slope_by_id["updrs_2_good"],
slope_by_id["updrs_3_good"],
slope_by_id["updrs_4_good"],
)
]
slope_by_id["class_0_1_1_1"] = [
int((w == 0) and (x == 1) and (y == 1) and (z == 1))
for (w, x, y, z) in zip(
slope_by_id["updrs_1_good"],
slope_by_id["updrs_2_good"],
slope_by_id["updrs_3_good"],
slope_by_id["updrs_4_good"],
)
]
slope_by_id["class_1_1_1_1"] = [
int((w == 1) and (x == 1) and (y == 1) and (z == 1))
for (w, x, y, z) in zip(
slope_by_id["updrs_1_good"],
slope_by_id["updrs_2_good"],
slope_by_id["updrs_3_good"],
slope_by_id["updrs_4_good"],
)
]
slope_by_id
slope_by_id_sum = slope_by_id.sum()
slope_by_id_sum
len(train_clinical_data["patient_id"].unique())
train_peptides
train_peptides[
(train_peptides["UniProt"] == "O00533")
& (train_peptides["Peptide"] != "GNPEPTFSWTK")
]
len(train_peptides["patient_id"].unique())
train_peptides["UniProt"].unique()
# ### There is 968 Peptide
len(train_peptides["Peptide"].unique())
# ### And 227 UniProt
len(set(train_peptides["UniProt"]))
g_train_peptides = train_peptides.groupby(
["patient_id", "Peptide"], as_index=False
).agg({"PeptideAbundance": "mean"})
g_train_peptides
train_proteins
g_train_proteins = train_proteins.groupby(
["patient_id", "UniProt"], as_index=False
).agg({"NPX": "mean"})
g_train_proteins = g_train_proteins.rename(columns={"NPX": "NPX_mean"})
g_train_proteins
# 54922
new_train = g_train_proteins.merge(slope_by_id, on="patient_id")
new_train
sample_submission
sample_submission["patient_id"] = [
int(x[0]) for x in sample_submission["prediction_id"].str.split("_")
]
sample_submission["updrs"] = [
int(x[3]) for x in sample_submission["prediction_id"].str.split("_")
]
sample_submission["patient_id"].unique()
sample_submission
slope_by_id[slope_by_id["updrs_1"] > 0]
updrs_1_bad = slope_by_id[slope_by_id["updrs_1"] > 0]["updrs_1"].mean()
updrs_1_good = slope_by_id[slope_by_id["updrs_1"] <= 0]["updrs_1"].mean()
updrs_2_bad = slope_by_id[slope_by_id["updrs_2"] > 0]["updrs_2"].mean()
updrs_2_good = slope_by_id[slope_by_id["updrs_2"] <= 0]["updrs_2"].mean()
updrs_3_bad = slope_by_id[slope_by_id["updrs_3"] > 0]["updrs_3"].mean()
updrs_3_good = slope_by_id[slope_by_id["updrs_3"] <= 0]["updrs_3"].mean()
updrs_4_bad = slope_by_id[slope_by_id["updrs_4"] > 0]["updrs_4"].mean()
updrs_4_good = slope_by_id[slope_by_id["updrs_4"] <= 0]["updrs_4"].mean()
for score in [
updrs_1_bad,
updrs_1_good,
updrs_2_bad,
updrs_2_good,
updrs_3_bad,
updrs_3_good,
updrs_4_bad,
updrs_4_good,
]:
print(score)
my_dict_bad = {1: 0.0715, 2: 0.08482, 3: 0.1685, 4: 0.09723}
my_dict_good = {1: -0.0410, 2: -0.0194, 3: -0.0933, 4: -0.0251}
sample_submission_3342 = sample_submission[sample_submission["patient_id"] == 3342]
sample_submission_3342["rating"] = sample_submission_3342["updrs"].replace(my_dict_bad)
sample_submission_50423 = sample_submission[sample_submission["patient_id"] == 50423]
sample_submission_50423["rating"] = sample_submission_50423["updrs"].replace(
my_dict_good
)
sample_submission = pd.concat([sample_submission_3342, sample_submission_50423])
sample_submission
sample_submission["prediction_id"].unique()
test
test_peptides
sample_submission = sample_submission.drop(columns=["patient_id", "updrs"])
sample_submission
sample_prediction_df = sample_submission.copy()
# sample_submission.to_csv("submission.csv", index = False)
# ## **Submitting predictions**
import amp_pd_peptide
env = amp_pd_peptide.make_env() # initialize the environment
iter_test = env.iter_test() # an iterator which loops over the test files
for test, test_peptides, test_proteins, sample_submission in iter_test:
sample_prediction_df["rating"] = sample_submission[
"rating"
] # make your predictions here
env.predict(sample_prediction_df) # register your predictions
|
import os
import numpy as np
import pandas as pd
import warnings
import seaborn as sns
import matplotlib.pyplot as plt
import plotly.express as px
warnings.filterwarnings("ignore")
pd.set_option("display.max_rows", None)
from sklearn import preprocessing
import matplotlib
matplotlib.style.use("ggplot")
from sklearn.preprocessing import LabelEncoder
import plotly.graph_objects as go
import plotly.offline as po
from plotly.offline import download_plotlyjs, init_notebook_mode, plot, iplot
import matplotlib.pyplot as plt
import dash
import plotly.express as px
import random
import plotly.figure_factory as ff
from plotly import tools
from plotly.subplots import make_subplots
from plotly.offline import iplot
import warnings
warnings.filterwarnings("ignore")
pd.set_option("display.max_rows", None)
data = pd.read_csv(
"https://archive.ics.uci.edu/ml/machine-learning-databases/adult/adult.data",
header=None,
)
data.columns = [
"age",
"workclass",
"fnlwgt",
"education",
"education-num",
"marital-status",
"occupation",
"relationship",
"race",
"sex",
"capital-gain",
"capital-loss",
"hours-per-week",
"native-country",
"class",
]
data = data.drop(["fnlwgt", "education-num"], axis=1)
data = data.replace(" ?", np.nan)
data.sample(10)
# ## Check Null Values
data.isna().sum()
data = data.dropna()
data = data.drop_duplicates()
# ## Check datatype - convert object into string
data.dtypes
data = data.rename(
columns={
"marital-status": "maritalstatus",
"capital-gain": "capitalgain",
"capital-loss": "capitalloss",
"hours-per-week": "hoursperweek",
"native-country": "nativecountry",
}
)
data = data.rename(columns={"class": "label"})
data.columns
numcols = []
stringcols = []
for col in data.columns:
if data[col].dtype == "object":
data[col] = data[col].astype("string")
if col != "label":
stringcols.append(col)
else:
numcols.append(col)
stringcols
numcols
# # Visualization
fig = go.Figure(
data=[
go.Table(
header=dict(
values=list(data.columns), # Header values
line_color="black", # Line Color of header
fill_color="orange", # background color of header
align="center", # Align header at center
height=40, # Height of Header
font=dict(color="white", size=10), # Font size & color of header text
),
cells=dict(
values=[
data.age,
data.workclass,
data.education,
data.maritalstatus,
data.occupation,
data.relationship,
data.race,
data.sex,
data.capitalgain,
data.capitalloss,
data.hoursperweek,
data.nativecountry,
data.label,
],
line_color="darkgrey", # Line color of the cell
fill_color="lightcyan", # Color of the cell
align="left", # Align text to left in cell
),
)
]
)
fig.update_layout(width=1500, height=500)
fig.show()
# # Categorical data analysis
def plot_barchart(fig, col, row, column):
fig.add_trace(
go.Bar(
y=data[col].value_counts().values,
x=data[col].value_counts().index,
text=data[col].value_counts().values,
textfont=dict(size=15),
textposition="outside",
showlegend=False,
marker=dict(color=colors, line_color="black", line_width=3),
),
row=row,
col=column,
)
return fig
def plot_piechart(fig, col, row, column):
fig.add_trace(
(
go.Pie(
labels=data[col].value_counts().keys(),
values=data[col].value_counts().values,
textfont=dict(size=16),
hole=0.5,
marker=dict(colors=colors, line_color="black", line_width=2),
textinfo="label+percent",
hoverinfo="label",
)
),
row=row,
col=column,
)
return fig
colors = px.colors.cyclical.Phase
def plot_graph_bar(col):
fig = make_subplots(rows=1, cols=1, subplot_titles="", specs=[[{"type": "xy"}]])
fig = plot_barchart(fig, col, 1, 1)
fig.update_yaxes(range=[0, max(data[col].value_counts())])
# Changing plot & figure background
fig.update_layout(
paper_bgcolor="#FFFDE7",
plot_bgcolor="#FFFDE7",
title=dict(text=col.title() + " Distribution", x=0.5, y=0.95),
title_font_size=30,
)
iplot(fig)
def plot_graphs(col):
fig = make_subplots(
rows=1,
cols=2,
subplot_titles=("Countplot", "Percentages"),
specs=[[{"type": "xy"}, {"type": "domain"}]],
)
fig = plot_barchart(fig, col, 1, 1)
fig = plot_piechart(fig, col, 1, 2)
fig.update_yaxes(range=[0, max(data[col].value_counts())])
# Changing plot & figure background
fig.update_layout(
paper_bgcolor="#FFFDE7",
plot_bgcolor="#FFFDE7",
title=dict(text=col.title() + " Distribution", x=0.5, y=0.95),
title_font_size=30,
)
iplot(fig)
for col in stringcols:
if col not in ["nativecountry", "education"]:
plot_graphs(col)
else:
plot_graph_bar(col)
plot_graphs("label")
colors = px.colors.cyclical.Twilight
sns.set_theme(
rc={
"figure.dpi": 500,
"axes.labelsize": 6,
"axes.facecolor": "#FFFDE7",
"grid.color": "#fffdfa",
"figure.facecolor": "#FFFDE7",
},
font_scale=0.55,
)
fig, ax = plt.subplots(7, 1, figsize=(12, 22))
for indx, (column, axes) in list(enumerate(list(zip(stringcols, ax.flatten())))):
if column == "nativecountry":
continue
if column not in "label":
sns.countplot(
ax=axes, x=data[column], hue=data["label"], palette=colors, alpha=1
)
else:
[axes.set_visible(False) for axes in ax.flatten()[indx + 1 :]]
axes_legend = ax.flatten()
axes_legend[1].legend(title="label", loc="upper right")
axes_legend[2].legend(title="label", loc="upper right")
plt.show()
# ## Lets process the some of the columns.
# ### 1. Label Distribution
# Here we got two label categories, 1) Income with >50K 2) Income with <= 50 K
# But classes are imbalanced, so we need to take care about class weight in future while training
# ### 2. Native country distribution
# Here we got so many instances of country with label "United states" while very less for other countries,
# Thus we will replace other all countries with label "Other"
data["nativecountry"].value_counts()
data["nativecountry"].where(
data["nativecountry"] == " United-States", "Other", inplace=True
)
data["nativecountry"].value_counts()
plot_graph_bar("nativecountry")
# ### 3. Education distribution
# Here we got 16 diff categories lets try to reduce the possible categories.
fig, ax = plt.subplots(1, 1, figsize=(12, 4))
sns.countplot(ax=ax, x=data["education"], hue=data["label"], palette=colors, alpha=1)
axes_legend[1].legend(title="label", loc="upper right")
axes_legend[2].legend(title="label", loc="upper right")
plt.show()
# From graph we can combine four categories into one:
# 11th, 9th, 7th-8th, 5th-6th, 10th, preschool, 12th and 1st - 4th into one - Basic
data["education"] = data["education"].str.strip()
data["education"] = data["education"].where(
(data["education"] != "11th")
& (data["education"] != "9th")
& (data["education"] != "7th-8th")
& (data["education"] != "5th-6th")
& (data["education"] != "10th")
& (data["education"] != "Preschool")
& (data["education"] != "12th")
& (data["education"] != "1st-4th"),
"Basic",
)
data["education"] = data["education"].where(
(data["education"] != "Doctorate") & (data["education"] != "Prof-school"), "Higher"
)
data["education"].value_counts()
fig, ax = plt.subplots(1, 1, figsize=(12, 4))
sns.countplot(ax=ax, x=data["education"], hue=data["label"], palette=colors, alpha=1)
axes_legend[1].legend(title="label", loc="upper right")
axes_legend[2].legend(title="label", loc="upper right")
plt.show()
# ### 4. Marital status distribution
# Here we got 7 diff categories lets try to reduce the possible categories.
fig, ax = plt.subplots(1, 1, figsize=(12, 4))
sns.countplot(
ax=ax, x=data["maritalstatus"], hue=data["label"], palette=colors, alpha=1
)
axes_legend[1].legend(title="label", loc="upper right")
axes_legend[2].legend(title="label", loc="upper right")
plt.show()
data["maritalstatus"] = data["maritalstatus"].str.strip()
data["maritalstatus"].value_counts()
# Lets combine last 4 categories as all of them have incom <= 50K
data["maritalstatus"] = data["maritalstatus"].where(
(data["maritalstatus"] != "Widowed")
& (data["maritalstatus"] != "Married-spouse-absent")
& (data["maritalstatus"] != "Married-AF-spouse")
& (data["maritalstatus"] != "Separated"),
"Other",
)
data["maritalstatus"].value_counts()
fig, ax = plt.subplots(1, 1, figsize=(12, 4))
sns.countplot(
ax=ax, x=data["maritalstatus"], hue=data["label"], palette=colors, alpha=1
)
axes_legend[1].legend(title="label", loc="upper right")
axes_legend[2].legend(title="label", loc="upper right")
plt.show()
# ### 5. Relationship distribution
# Lets combine some categories.
fig, ax = plt.subplots(1, 1, figsize=(12, 4))
sns.countplot(ax=ax, x=data["relationship"], hue=data["label"], palette=colors, alpha=1)
axes_legend[1].legend(title="label", loc="upper right")
axes_legend[2].legend(title="label", loc="upper right")
plt.show()
data["relationship"] = data["relationship"].str.strip()
data["relationship"] = data["relationship"].where(
(data["relationship"] != "Other-relative") & (data["relationship"] != "Own-child"),
"Other",
)
data["relationship"].value_counts()
# ## Encoding target class using label encoder
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
data["label"] = le.fit_transform(data["label"])
data["label"].value_counts()
# # Numerical data analysis
import plotly.graph_objs as go
import plotly.io as pio
import pandas as pd
import numpy as np
import seaborn as sns
def corrMat(df, id=False):
corr_mat = df.corr().round(2)
mask = np.triu(np.ones_like(corr_mat, dtype=bool))
mask = mask[1:, :-1]
corr = corr_mat.iloc[1:, :-1].copy()
annot = corr.astype(str)
annot[annot == "1.0"] = ""
fig = go.Figure(
data=go.Heatmap(
z=corr.values,
x=corr.columns,
y=corr.index,
xgap=1,
ygap=1,
colorscale="RdPu_r",
zmin=-0.3,
zmax=0.3,
hoverongaps=False,
hovertemplate="Feature A: %{y}<br>Feature B: %{x}<br>Correlation: %{z:.2f}<extra></extra>",
showscale=False,
name="",
customdata=annot.values.tolist(),
meta=corr_mat,
text=corr.values.round(2),
)
)
fig.update_layout(
title={
"text": "Shifted Linear Correlation Matrix",
"y": 0.95,
"x": 0.5,
"xanchor": "center",
"yanchor": "top",
},
xaxis_tickangle=-45,
xaxis_showgrid=False,
yaxis_showgrid=False,
yaxis_autorange="reversed",
yaxis_tickmode="linear",
yaxis_tick0=-0.5,
yaxis_dtick=1,
width=450,
height=450,
margin=dict(l=70, r=20, b=60, t=80),
)
return fig
corrMat(data)
# Corelation matric suggest that age, capital gain and hours per week columns directly affects the label column, lets see how they are related.
# Plot a subset of variables
g = sns.PairGrid(
data, hue="label", vars=["age", "capitalgain", "capitalloss"], height=3, aspect=1
)
g = g.map_offdiag(plt.scatter, edgecolor="w", s=15)
g = g.map_diag(plt.hist, edgecolor="w", linewidth=1)
g = g.add_legend()
plt.show()
# ## Standardization and Normalization
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
for col in numcols:
data[col] = scaler.fit_transform(data[[col]])
data.head()
plt.figure(figsize=(16, 11))
sns.set(
rc={
"axes.facecolor": "#b0deff",
"axes.grid": False,
"xtick.labelsize": 15,
"ytick.labelsize": 15,
"axes.labelsize": 20,
"figure.figsize": (6.0, 5.0),
}
)
params = dict(data=data, x=data.sex, y=data.hoursperweek, hue=data.label, dodge=True)
sns.stripplot(
**params,
size=7,
jitter=0.10,
palette=["#33FF66", "#FF6600"],
edgecolor="black",
linewidth=1
)
sns.boxplot(**params, palette=["#BDBDBD", "#E0E0E0"], linewidth=6)
plt.show()
# ## Outlier detection
from scipy import stats
for col in numcols:
z = stats.zscore(data[col])
outliers = (z > 3) | (z < -3)
df = data[outliers]
print(col, df.shape[0])
# capital gain and age's outlier are small so we can remove it
for col in ["age", "capitalgain", "hoursperweek"]:
z = stats.zscore(data[col])
outliers = (z > 3) | (z < -3)
data = data.drop(data[outliers].index)
# # Train Test Split and encoding
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
data.drop("label", axis=1),
data["label"],
test_size=0.3,
stratify=data["label"],
random_state=42,
)
from category_encoders.target_encoder import TargetEncoder
encoder = TargetEncoder(cols=stringcols)
X_train = encoder.fit_transform(X_train, y_train)
X_test = encoder.transform(X_test)
X_train.head()
X_test.head()
# Now data is ready to feed in ML models
# # Decision tree classifier
y_train.value_counts()
weights = {0: 1, 1: 3}
from sklearn import tree
from sklearn.metrics import classification_report, accuracy_score
def line_plot(X, Y, SecondY):
fig = go.Figure()
fig.add_trace(go.Scatter(x=X, y=Y, mode="lines+markers", name="Training Accuracy"))
fig.add_trace(
go.Scatter(x=X, y=SecondY, mode="lines+markers", name="Test Accuracy")
)
fig.update_layout(
xaxis_title="Number of depth",
yaxis_title="Accuracy",
legend=dict(
x=0,
y=1,
traceorder="normal",
font=dict(family="sans-serif", size=12, color="black"),
bgcolor="WhiteSmoke",
bordercolor="Black",
borderwidth=1,
),
width=700,
height=500,
margin=dict(l=50, r=50, b=50, t=50, pad=4),
)
fig.show()
def confusion_plot(cm):
fig = go.Figure(
data=go.Heatmap(
z=cm, x=["0", "1"], y=["0", "1"], colorscale="Reds", showscale=False
)
)
# Add label counts as annotations
annotations = []
for i, row in enumerate(cm):
for j, val in enumerate(row):
annotations.append(
go.layout.Annotation(
x=j,
y=i,
text=str(val),
font=dict(color="black", size=14),
showarrow=False,
)
)
fig.update_layout(
title="Confusion Matrix",
xaxis_title="Predicted label",
yaxis_title="True label",
annotations=annotations,
width=500,
height=500,
)
fig.show()
import plotly.graph_objs as go
trainAcc = []
testAcc = []
depthArray = [1, 5, 8, 10, 12, 15, 20, 25, 30]
for depth in depthArray:
clf = tree.DecisionTreeClassifier(
criterion="entropy", max_depth=depth, class_weight=weights, random_state=42
)
clf = clf.fit(X_train, y_train)
Y_predTrain = clf.predict(X_train)
Y_predTest = clf.predict(X_test)
trainAcc.append(accuracy_score(y_train, Y_predTrain))
testAcc.append(accuracy_score(y_test, Y_predTest))
line_plot(depthArray, trainAcc, testAcc)
clf = tree.DecisionTreeClassifier(
criterion="entropy", max_depth=10, class_weight=weights, random_state=42
)
clf = clf.fit(X_train, y_train)
Y_predTest = clf.predict(X_test)
print(classification_report(y_test, Y_predTest, target_names=["<=50k", ">50k"]))
import plotly.graph_objects as go
from sklearn.metrics import confusion_matrix
# Compute confusion matrix
cm = confusion_matrix(y_test, clf.predict(X_test))
# Create heatmap using Plotly
confusion_plot(cm)
# # Ensemble Techniques
import plotly.graph_objs as go
from sklearn import ensemble
n_trees = range(1, 20, 2)
trainAcc = []
testAcc = []
for n in n_trees:
clf = ensemble.RandomForestClassifier(
n_estimators=n, random_state=42, class_weight=weights
)
clf.fit(X_train, y_train)
Y_predTrain = clf.predict(X_train)
Y_predTest = clf.predict(X_test)
trainAcc.append(accuracy_score(y_train, Y_predTrain))
testAcc.append(accuracy_score(y_test, Y_predTest))
line_plot(list(n_trees), trainAcc, testAcc)
clf = ensemble.RandomForestClassifier(
n_estimators=7, random_state=42, class_weight=weights
)
clf.fit(X_train, y_train)
Y_predTest = clf.predict(X_test)
print(classification_report(y_test, Y_predTest, target_names=["<=50k", ">50k"]))
cm = confusion_matrix(y_test, Y_predTest)
confusion_plot(cm)
# # SVC
from sklearn.svm import SVC
from sklearn.model_selection import GridSearchCV
model = SVC(random_state=42, class_weight=weights)
param_grid = {"C": [1, 10], "gamma": [1, 0.1], "kernel": ["rbf"]}
grid = GridSearchCV(model, param_grid, cv=5)
# fitting the model for grid search
grid.fit(X_train, y_train)
grid.best_params_
new_grid = SVC(C=10, kernel="rbf", gamma=1, random_state=42, class_weight=weights)
new_grid.fit(X_train, y_train)
y_pred = new_grid.predict(X_test)
print(classification_report(y_test, y_pred, target_names=["<=50K", ">50K"]))
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# **Introduction**
# There is no perfect way of replacing the null values, As it depends totally on the data and the analyst whi is cleaning the data. There are many operations in order to replace or fill the null values contained in a dataframe. However, the most common method is replacing the null values with the mean of that series. Let's try the same on this dataset
# **Importing the Dataset******
df = pd.read_csv(
"../input/2022-ipl-auction-dataset/ipl_2022_dataset.csv", index_col="Unnamed: 0"
)
df
df.info()
df.isna().sum()
# Note:
# We have 396 null values in column COST IN ₹ (CR.), however it is not suggested to use mean in order to fill the null values when the number is quite huge. But, just for the sake of learning let me teach you how to do it!
# Calculating the mean of the series by skipping null values
mean = df["COST IN ₹ (CR.)"].mean(skipna=True)
mean
# Using the mean to fill the null values in the same series
df["COST IN ₹ (CR.)"] = df["COST IN ₹ (CR.)"].fillna(mean)
df.isna().sum()
df
# Checking the occurrence of values using value_counts()
df["COST IN ₹ (CR.)"].value_counts(dropna=False)
# **Filling a string in 2021 Squad Series**
# We have seen how to fill a float value inplace of null values, Now let's see if we could do the same for string values
df["2021 Squad"] = df["2021 Squad"].fillna("No Team")
df.isnull().sum()
df["2021 Squad"].value_counts()
|
import os
import shutil
import random
from tensorflow.keras.applications import Xception
from tensorflow.keras.applications.xception import preprocess_input
from tensorflow.keras.layers import Dense, Flatten
from tensorflow.keras.models import Model
import numpy as np
import matplotlib.pyplot as plt
from tensorflow.keras.preprocessing import image
from tensorflow.keras.preprocessing.image import ImageDataGenerator
subdirectories = list(os.listdir("/kaggle/input/jkltgutg/Retail_Product"))
subdirectories
# Define the initial path
initial_path = "/kaggle/input/jkltgutg/Retail_Product"
# Define the new top-level directory names
directories = ["train", "val", "test"]
# Define the subdirectory names for each top-level directory
# Define the proportions for train, validation, and test sets
props = [0.7, 0.2, 0.1]
# Loop over the top-level directories and subdirectories, and create the corresponding directories
for directory in directories:
for subdirectory in subdirectories:
path = os.path.join(initial_path, subdirectory)
files = os.listdir(path)
random.shuffle(files)
total = len(files)
train_end = int(total * props[0])
val_end = train_end + int(total * props[1])
if directory == "train":
new_files = files[:train_end]
elif directory == "val":
new_files = files[train_end:val_end]
else:
new_files = files[val_end:]
new_path = os.path.join(directory, subdirectory)
os.makedirs(new_path, exist_ok=True)
for file in new_files:
old_file_path = os.path.join(path, file)
new_file_path = os.path.join(new_path, file)
shutil.copy(old_file_path, new_file_path)
# Define the directories for the train, validation, and test sets
train_dir = "train"
val_dir = "val"
# Define the image dimensions and batch size
img_height = 224
img_width = 224
batch_size = 32
# Define the data generators for the train, validation, and test sets
train_datagen = ImageDataGenerator(
preprocessing_function=preprocess_input,
rotation_range=20,
width_shift_range=0.2,
height_shift_range=0.2,
shear_range=0.2,
zoom_range=0.2,
horizontal_flip=True,
)
train_generator = train_datagen.flow_from_directory(
train_dir,
target_size=(img_height, img_width),
batch_size=batch_size,
class_mode="categorical",
color_mode="rgb",
)
val_datagen = ImageDataGenerator(preprocessing_function=preprocess_input)
val_generator = val_datagen.flow_from_directory(
val_dir,
target_size=(img_height, img_width),
batch_size=batch_size,
class_mode="categorical",
color_mode="rgb",
)
# Load the pre-trained Xception model without the top classification layer
base_model = Xception(
weights="imagenet", include_top=False, input_shape=(img_height, img_width, 3)
)
# Freeze the pre-trained layers so they are not updated during training
for layer in base_model.layers:
layer.trainable = False
# Add your own classification layers on top of the pre-trained model
x = base_model.output
x = Flatten()(x)
x = Dense(256, activation="relu")(x)
predictions = Dense(25, activation="softmax")(x)
# Create the full model with both the pre-trained and new classification layers
model = Model(inputs=base_model.input, outputs=predictions)
# Compile the model
model.compile(optimizer="adam", loss="categorical_crossentropy", metrics=["accuracy"])
num_epochs = 50
history = model.fit(
train_generator,
steps_per_epoch=train_generator.samples // batch_size,
epochs=num_epochs,
validation_data=val_generator,
validation_steps=val_generator.samples // batch_size,
)
|
# Importing the libraries
import tensorflow as tf
from tensorflow.keras import layers, losses
from tensorflow.keras.models import Model
from tensorflow.keras import mixed_precision
from sklearn.metrics import accuracy_score, confusion_matrix
import numpy as np
import matplotlib.pyplot as plt
AUTOTUNE = tf.data.AUTOTUNE
ACTIVATION = "sigmoid"
EPOCHS = 10000
ERROR = "mean_squared_error"
BATCH_SIZE = 64
NUM_CLASSES = 5
mixed_precision.set_global_policy("mixed_float16")
from tensorflow.python.ops.numpy_ops import np_config
np_config.enable_numpy_behavior()
class ModelCallBack(tf.keras.callbacks.Callback):
def on_train_begin(self, logs={}):
self.avg_error = []
self.prev_error = 0
self.current_error = 0
self.end_epoch = 0
def on_epoch_end(self, epoch, logs=None):
# print(logs)
if epoch == 0:
self.prev_error = logs["sparse_categorical_crossentropy"]
self.current_error = logs["sparse_categorical_crossentropy"]
else:
self.current_error = logs["sparse_categorical_crossentropy"]
if abs(self.current_error - self.prev_error) < 1e-4 or (
tf.math.is_nan(self.prev_error)
):
# self.end_epoch = epoch
self.model.stop_training = True
else:
self.prev_error = self.current_error
# print(logs["sparse_categorical_crossentropy"])
print("Epoch", epoch, "done | loss:", logs["loss"])
self.end_epoch = epoch
self.avg_error.append(self.prev_error)
model_callback = ModelCallBack()
train_ds = tf.keras.utils.image_dataset_from_directory(
"/kaggle/input/group17/Group_17/train",
labels="inferred",
label_mode="int",
class_names=["0", "1", "3", "4", "5"],
color_mode="grayscale",
batch_size=BATCH_SIZE,
image_size=(28, 28),
shuffle=False,
seed=None,
validation_split=None,
# subset= "training",
)
# flatten the images
# train_ds = train_ds.map(lambda x, y: (tf.reshape(x, (28, 28)), y), num_parallel_calls=AUTOTUNE)
val_ds = tf.keras.utils.image_dataset_from_directory(
"/kaggle/input/group17/Group_17/val",
labels="inferred",
label_mode="int",
class_names=["0", "1", "3", "4", "5"],
color_mode="grayscale",
batch_size=BATCH_SIZE,
image_size=(28, 28),
shuffle=False,
seed=None,
validation_split=None,
# subset= "validation",
)
test_ds = tf.keras.utils.image_dataset_from_directory(
"/kaggle/input/group17/Group_17/test",
labels="inferred",
label_mode="int",
class_names=["0", "1", "3", "4", "5"],
color_mode="grayscale",
batch_size=BATCH_SIZE,
image_size=(28, 28),
shuffle=False,
seed=None,
validation_split=None,
# subset= "validation",
)
normalization_layer = layers.Rescaling(1.0 / 255)
normalized_train_ds = train_ds.map(
lambda x, y: (normalization_layer(x), y), num_parallel_calls=AUTOTUNE
)
normalized_val_ds = val_ds.map(
lambda x, y: (normalization_layer(x), y), num_parallel_calls=AUTOTUNE
)
normalized_test_ds = test_ds.map(
lambda x, y: (normalization_layer(x), y), num_parallel_calls=AUTOTUNE
)
train_ds = normalized_train_ds.cache().prefetch(buffer_size=AUTOTUNE)
val_ds = normalized_val_ds.cache().prefetch(buffer_size=AUTOTUNE)
test_ds = normalized_test_ds.cache().prefetch(buffer_size=AUTOTUNE)
class Autoencoder_1(Model):
def __init__(self, bottleneck):
super(Autoencoder_1, self).__init__()
self.bottleneck = bottleneck
self.encoder = tf.keras.Sequential(
[
layers.Flatten(),
layers.Dense(self.bottleneck, activation=ACTIVATION),
]
)
self.decoder = tf.keras.Sequential(
[layers.Dense(784, activation=ACTIVATION), layers.Reshape((28, 28))]
)
self.avg_error = []
self.end_epoch = 0
def call(self, x):
encoded = self.encoder(x)
decoded = self.decoder(encoded)
return decoded
class Autoencoder_3(Model):
def __init__(self, bottleneck):
super(Autoencoder_3, self).__init__()
self.bottleneck = bottleneck
self.encoder = tf.keras.Sequential(
[
layers.Flatten(),
layers.Dense(400, activation=ACTIVATION),
layers.Dense(self.bottleneck, activation=ACTIVATION),
]
)
self.decoder = tf.keras.Sequential(
[
layers.Dense(400, activation=ACTIVATION),
layers.Dense(784, activation=ACTIVATION),
layers.Reshape((28, 28)),
]
)
def call(self, x):
encoded = self.encoder(x)
decoded = self.decoder(encoded)
return decoded
for data, label in train_ds.take(1):
print(data.shape)
train_ds
def run_autoencoder(autoencoder, name):
model_callback = ModelCallBack()
autoencoder.compile(
optimizer="adam", loss="mean_squared_error", metrics=["mean_squared_error"]
)
autoencoder.fit(
train_ds_numpy,
train_ds_numpy,
batch_size=1,
epochs=EPOCHS,
shuffle=True,
# validation_data=val_ds,
callbacks=[model_callback],
verbose=0,
)
autoencoder.avg_error = model_callback.avg_error
autoencoder.end_epoch = model_callback.end_epoch
# Take one image, from each class, from the training, validation, and test set.
# Give their reconstructed images for each of the architectures (along with original images).
plt.figure(figsize=(20, 4))
for images, labels in train_ds.take(1):
# display original
ax = plt.subplot(2, 3, 1)
plt.imshow(images[0].numpy().reshape(28, 28), cmap="gray")
# print(images[0].numpy().reshape(28, 28))
plt.title("Original(training)")
ax.get_xaxis().set_visible(False)
ax.get_yaxis().set_visible(False)
# display reconstruction
ax = plt.subplot(2, 3, 4)
enc = autoencoder.encoder(images)
dec = autoencoder.decoder(enc)
plt.imshow(dec.numpy().reshape(28, 28), cmap="gray")
# print(dec.numpy().reshape(28, 28))
plt.title("Reconstructed(training)")
ax.get_xaxis().set_visible(False)
ax.get_yaxis().set_visible(False)
# validation
for images, labels in val_ds.take(1):
# display original
ax = plt.subplot(2, 3, 2)
plt.imshow(images[0].numpy().reshape(28, 28), cmap="gray")
plt.title("Original(validation)")
ax.get_xaxis().set_visible(False)
ax.get_yaxis().set_visible(False)
# display reconstruction
ax = plt.subplot(2, 3, 5)
enc = autoencoder.encoder(images)
dec = autoencoder.decoder(enc)
plt.imshow(dec.numpy().reshape(28, 28), cmap="gray")
plt.title("Reconstructed(validation)")
ax.get_xaxis().set_visible(False)
ax.get_yaxis().set_visible(False)
# test
for images, labels in test_ds.take(1):
# display original
ax = plt.subplot(2, 3, 3)
plt.imshow(images[0].numpy().reshape(28, 28), cmap="gray")
plt.title("Original(test)")
ax.get_xaxis().set_visible(False)
ax.get_yaxis().set_visible(False)
# display reconstruction
ax = plt.subplot(2, 3, 6)
enc = autoencoder.encoder(images)
dec = autoencoder.decoder(enc)
plt.imshow(dec.numpy().reshape(28, 28), cmap="gray")
plt.title("Reconstructed(test)")
ax.get_xaxis().set_visible(False)
ax.get_yaxis().set_visible(False)
plt.show()
# Observe the average reconstruction errors for the training, validation, and test data.
# Average reconstruction error is computed after the model is trained.
train_loss = autoencoder.evaluate(train_ds_numpy, train_ds_numpy, batch_size=1)
val_loss = autoencoder.evaluate(val_ds_numpy, val_ds_numpy, batch_size=1)
test_loss = autoencoder.evaluate(test_ds_numpy, test_ds_numpy, batch_size=1)
print("Average reconstruction error for training data: ", train_loss[1])
print("Average reconstruction error for validation data: ", val_loss[1])
print("Average reconstruction error for test data: ", test_loss[1])
autoencoder.save(name, save_format="tf")
# run_autoencoder(Autoencoder_1(32),"1_32")
# convert training data to reduced dimension using Autoencoders
autoencoder_1_32 = tf.keras.models.load_model(
"/kaggle/input/model-zip/1_32", compile=False
)
train_ds_1_32 = train_ds.map(lambda x, y: (autoencoder_1_32.encoder(x), y))
val_ds_1_32 = val_ds.map(lambda x, y: (autoencoder_1_32.encoder(x), y))
test_ds_1_32 = test_ds.map(lambda x, y: (autoencoder_1_32.encoder(x), y))
autoencoder_1_64 = tf.keras.models.load_model(
"/kaggle/input/model-zip/1_64", compile=False
)
train_ds_1_64 = train_ds.map(lambda x, y: (autoencoder_1_64.encoder(x), y))
val_ds_1_64 = val_ds.map(lambda x, y: (autoencoder_1_64.encoder(x), y))
test_ds_1_64 = test_ds.map(lambda x, y: (autoencoder_1_64.encoder(x), y))
autoencoder_1_128 = tf.keras.models.load_model(
"/kaggle/input/model-zip/1_128", compile=False
)
train_ds_1_128 = train_ds.map(lambda x, y: (autoencoder_1_128.encoder(x), y))
val_ds_1_128 = val_ds.map(lambda x, y: (autoencoder_1_128.encoder(x), y))
test_ds_1_128 = test_ds.map(lambda x, y: (autoencoder_1_128.encoder(x), y))
autoencoder_1_256 = tf.keras.models.load_model(
"/kaggle/input/model-zip/1_256", compile=False
)
train_ds_1_256 = train_ds.map(lambda x, y: (autoencoder_1_256.encoder(x), y))
val_ds_1_256 = val_ds.map(lambda x, y: (autoencoder_1_256.encoder(x), y))
test_ds_1_256 = test_ds.map(lambda x, y: (autoencoder_1_256.encoder(x), y))
autoencoder_3_32 = tf.keras.models.load_model(
"/kaggle/input/model-zip/3_32", compile=False
)
train_ds_3_32 = train_ds.map(lambda x, y: (autoencoder_3_32.encoder(x), y))
val_ds_3_32 = val_ds.map(lambda x, y: (autoencoder_3_32.encoder(x), y))
test_ds_3_32 = test_ds.map(lambda x, y: (autoencoder_3_32.encoder(x), y))
autoencoder_3_64 = tf.keras.models.load_model(
"/kaggle/input/model-zip/3_64", compile=False
)
train_ds_3_64 = train_ds.map(lambda x, y: (autoencoder_3_64.encoder(x), y))
val_ds_3_64 = val_ds.map(lambda x, y: (autoencoder_3_64.encoder(x), y))
test_ds_3_64 = test_ds.map(lambda x, y: (autoencoder_3_64.encoder(x), y))
autoencoder_3_128 = tf.keras.models.load_model(
"/kaggle/input/model-zip/3_128", compile=False
)
train_ds_3_128 = train_ds.map(lambda x, y: (autoencoder_3_128.encoder(x), y))
val_ds_3_128 = val_ds.map(lambda x, y: (autoencoder_3_128.encoder(x), y))
test_ds_3_128 = test_ds.map(lambda x, y: (autoencoder_3_128.encoder(x), y))
autoencoder_3_256 = tf.keras.models.load_model(
"/kaggle/input/model-zip/3_256", compile=False
)
train_ds_3_256 = train_ds.map(lambda x, y: (autoencoder_3_256.encoder(x), y))
val_ds_3_256 = val_ds.map(lambda x, y: (autoencoder_3_256.encoder(x), y))
test_ds_3_256 = test_ds.map(lambda x, y: (autoencoder_3_256.encoder(x), y))
for data, label in train_ds_1_32.take(1):
print(data.shape)
# build a classifier with 1 hidden layer
arch_1 = [32]
arch_2 = [128]
# build a classifier with 2 hidden layers
arch_3 = [128, 32]
# build a classifier with 3 hidden layers
arch_4 = [128, 64, 16]
def create_classifier(arch, input_shape):
model = tf.keras.Sequential()
model.add(tf.keras.layers.InputLayer(input_shape=input_shape))
for i in arch:
model.add(tf.keras.layers.Dense(i, activation="relu"))
model.add(tf.keras.layers.Dense(NUM_CLASSES, activation="softmax"))
model.compile(
optimizer="adam",
loss="sparse_categorical_crossentropy",
metrics=["accuracy", "sparse_categorical_crossentropy"],
jit_compile=True,
)
return model
input_32 = (32,)
input_64 = (64,)
input_128 = (128,)
input_256 = (256,)
def Q3(): # 1 layer autoencoder
def val_acc_calc(ds, model):
y_pred = model.predict(ds)
y_pred = np.argmax(y_pred, axis=1)
y_true = []
for _, y in ds:
for i in y.numpy():
y_true.append(i)
return accuracy_score(y_true, y_pred)
# classification accuracy on validation set for different FCNN (for all reduced dimensions)
val_acc_32 = []
val_acc_64 = []
val_acc_128 = []
val_acc_256 = []
def run_on_all_arch():
model_tuple = (arch_1, arch_2, arch_3, arch_4)
for arch in model_tuple:
# 32
model = create_classifier(arch, input_32)
model.fit(
train_ds_1_32,
epochs=EPOCHS,
validation_data=val_ds_1_32,
callbacks=[model_callback],
verbose=0,
)
val_acc_32.append(val_acc_calc(val_ds_1_32, model))
# 64
model = create_classifier(arch, input_64)
model.fit(
train_ds_1_64,
epochs=EPOCHS,
validation_data=val_ds_1_64,
callbacks=[model_callback],
verbose=0,
)
val_acc_64.append(val_acc_calc(val_ds_1_64, model))
# 128
model = create_classifier(arch, input_128)
model.fit(
train_ds_1_128,
epochs=EPOCHS,
validation_data=val_ds_1_128,
callbacks=[model_callback],
verbose=0,
)
val_acc_128.append(val_acc_calc(val_ds_1_128, model))
# 256
model = create_classifier(arch, input_256)
model.fit(
train_ds_1_256,
epochs=EPOCHS,
validation_data=val_ds_1_256,
callbacks=[model_callback],
verbose=0,
)
val_acc_256.append(val_acc_calc(val_ds_1_256, model))
# print the results
print("order of architectures: ", model_tuple)
print(
"accuracy on validation set for different FCNN (for all reduced dimensions):"
)
print("32: ", val_acc_32)
print("64: ", val_acc_64)
print("128: ", val_acc_128)
print("256: ", val_acc_256)
# find best architecture for each dimension
print("for 32: ", model_tuple[np.argmax(val_acc_32)])
print("for 64: ", model_tuple[np.argmax(val_acc_64)])
print("for 128: ", model_tuple[np.argmax(val_acc_128)])
print("for 256: ", model_tuple[np.argmax(val_acc_256)])
def plot_confusion_matrix(cm):
matrix2 = np.zeros((5, 5))
for i in range(5):
for j in range(5):
matrix2[i][j] = cm[i][j] / np.sum(cm[i])
plt.imshow(matrix2, cmap=plt.cm.Blues)
for i in range(5):
for j in range(5):
if matrix2[i][j] > 0.5:
plt.text(
j,
i,
round(matrix[i][j], 2),
horizontalalignment="center",
color="white",
)
else:
plt.text(
j,
i,
round(matrix[i][j], 2),
horizontalalignment="center",
color="black",
)
plt.title("Confusion Matrix")
plt.ylabel("True label")
plt.xlabel("Predicted label")
label = ["0", "1", "3", "4", "5"]
plt.xticks(np.arange(5), label)
plt.yticks(np.arange(5), label)
plt.show()
# test accuracy and confusion matrix for best architecture for each dimension
test_acc = []
# 32
model = create_classifier(model_tuple[np.argmax(val_acc_32)], input_32)
model.fit(
train_ds_1_32,
epochs=EPOCHS,
validation_data=val_ds_1_32,
callbacks=[model_callback],
verbose=0,
)
y_pred = model.predict(test_ds_1_32)
y_pred = np.argmax(y_pred, axis=1)
y_true = []
for _, y in test_ds_1_32:
for i in y.numpy():
y_true.append(i)
print("test accuracy for 32: ", test_acc.append(accuracy_score(y_true, y_pred)))
print("confusion matrix for 32: ")
plot_confusion_matrix(confusion_matrix(y_true, y_pred))
# 64
model = create_classifier(model_tuple[np.argmax(val_acc_64)], input_64)
model.fit(
train_ds_1_64,
epochs=EPOCHS,
validation_data=val_ds_1_64,
callbacks=[model_callback],
verbose=0,
)
y_pred = model.predict(test_ds_1_64)
y_pred = np.argmax(y_pred, axis=1)
y_true = []
for _, y in test_ds_1_64:
for i in y.numpy():
y_true.append(i)
print("test accuracy for 64: ", test_acc.append(accuracy_score(y_true, y_pred)))
print("confusion matrix for 64: ")
plot_confusion_matrix(confusion_matrix(y_true, y_pred))
# 128
model = create_classifier(model_tuple[np.argmax(val_acc_128)], input_128)
model.fit(
train_ds_1_128,
epochs=EPOCHS,
validation_data=val_ds_1_128,
callbacks=[model_callback],
verbose=0,
)
y_pred = model.predict(test_ds_1_128)
y_pred = np.argmax(y_pred, axis=1)
y_true = []
for _, y in test_ds_1_128:
for i in y.numpy():
y_true.append(i)
print(
"test accuracy for 128: ", test_acc.append(accuracy_score(y_true, y_pred))
)
print("confusion matrix for 128: ")
plot_confusion_matrix(confusion_matrix(y_true, y_pred))
# 256
model = create_classifier(model_tuple[np.argmax(val_acc_256)], input_256)
model.fit(
train_ds_1_256,
epochs=EPOCHS,
validation_data=val_ds_1_256,
callbacks=[model_callback],
verbose=0,
)
y_pred = model.predict(test_ds_1_256)
y_pred = np.argmax(y_pred, axis=1)
y_true = []
for _, y in test_ds_1_256:
for i in y.numpy():
y_true.append(i)
print(
"test accuracy for 256: ", test_acc.append(accuracy_score(y_true, y_pred))
)
print("confusion matrix for 256: ")
plot_confusion_matrix(confusion_matrix(y_true, y_pred))
print("test accuracy: ", test_acc)
dim_list = [32, 64, 128, 256]
print("best dimension: ", dim_list[np.argmax(test_acc)])
run_on_all_arch()
Q3()
# Q4 same as Q3 but for 3 layer encoder
def Q4():
print("Q4")
def val_acc_calc(ds, model):
y_pred = model.predict(ds)
y_pred = np.argmax(y_pred, axis=1)
y_true = []
for _, y in ds:
for i in y.numpy():
y_true.append(i)
return accuracy_score(y_true, y_pred)
# classification accuracy on validation set for different FCNN (for all reduced dimensions)
val_acc_32 = []
val_acc_64 = []
val_acc_128 = []
val_acc_256 = []
def run_on_all_arch_2():
model_tuple = (arch_1, arch_2, arch_3, arch_4)
for arch in model_tuple:
# 32
model = create_classifier(arch, input_32)
model.fit(
train_ds_3_32,
epochs=EPOCHS,
validation_data=val_ds_3_32,
callbacks=[model_callback],
verbose=0,
)
val_acc_32.append(val_acc_calc(val_ds_1_32, model))
# 64
model = create_classifier(arch, input_64)
model.fit(
train_ds_3_64,
epochs=EPOCHS,
validation_data=val_ds_3_64,
callbacks=[model_callback],
verbose=0,
)
val_acc_64.append(val_acc_calc(val_ds_1_64, model))
# 128
model = create_classifier(arch, input_128)
model.fit(
train_ds_3_128,
epochs=EPOCHS,
validation_data=val_ds_3_128,
callbacks=[model_callback],
verbose=0,
)
val_acc_128.append(val_acc_calc(val_ds_1_128, model))
# 256
model = create_classifier(arch, input_256)
model.fit(
train_ds_3_256,
epochs=EPOCHS,
validation_data=val_ds_3_256,
callbacks=[model_callback],
verbose=0,
)
val_acc_256.append(val_acc_calc(val_ds_1_256, model))
# print the results
print("order of architectures: ", model_tuple)
print(
"accuracy on validation set for different FCNN (for all reduced dimensions):"
)
print("32: ", val_acc_32)
print("64: ", val_acc_64)
print("128: ", val_acc_128)
print("256: ", val_acc_256)
# find best architecture for each dimension
print("for 32: ", model_tuple[np.argmax(val_acc_32)])
print("for 64: ", model_tuple[np.argmax(val_acc_64)])
print("for 128: ", model_tuple[np.argmax(val_acc_128)])
print("for 256: ", model_tuple[np.argmax(val_acc_256)])
def plot_confusion_matrix(cm):
matrix2 = np.zeros((5, 5))
for i in range(5):
for j in range(5):
matrix2[i][j] = cm[i][j] / np.sum(cm[i])
plt.imshow(matrix2, cmap=plt.cm.Blues)
for i in range(5):
for j in range(5):
if matrix2[i][j] > 0.5:
plt.text(
j,
i,
round(matrix[i][j], 2),
horizontalalignment="center",
color="white",
)
else:
plt.text(
j,
i,
round(matrix[i][j], 2),
horizontalalignment="center",
color="black",
)
plt.title("Confusion Matrix")
plt.ylabel("True label")
plt.xlabel("Predicted label")
label = ["0", "1", "3", "4", "5"]
plt.xticks(np.arange(5), label)
plt.yticks(np.arange(5), label)
plt.show()
# test accuracy and confusion matrix for best architecture for each dimension
test_acc = []
# 32
model = create_classifier(model_tuple[np.argmax(val_acc_32)], input_32)
model.fit(
train_ds_1_32,
epochs=EPOCHS,
validation_data=val_ds_1_32,
callbacks=[model_callback],
verbose=0,
)
y_pred = model.predict(test_ds_1_32)
y_pred = np.argmax(y_pred, axis=1)
y_true = []
for _, y in test_ds_1_32:
for i in y.numpy():
y_true.append(i)
print("test accuracy for 32: ", test_acc.append(accuracy_score(y_true, y_pred)))
print("confusion matrix for 32: ")
plot_confusion_matrix(confusion_matrix(y_true, y_pred))
# 64
model = create_classifier(model_tuple[np.argmax(val_acc_64)], input_64)
model.fit(
train_ds_1_64,
epochs=EPOCHS,
validation_data=val_ds_1_64,
callbacks=[model_callback],
verbose=0,
)
y_pred = model.predict(test_ds_1_64)
y_pred = np.argmax(y_pred, axis=1)
y_true = []
for _, y in test_ds_1_64:
for i in y.numpy():
y_true.append(i)
print("test accuracy for 64: ", test_acc.append(accuracy_score(y_true, y_pred)))
print("confusion matrix for 64: ")
plot_confusion_matrix(confusion_matrix(y_true, y_pred))
# 128
model = create_classifier(model_tuple[np.argmax(val_acc_128)], input_128)
model.fit(
train_ds_1_128,
epochs=EPOCHS,
validation_data=val_ds_1_128,
callbacks=[model_callback],
verbose=0,
)
y_pred = model.predict(test_ds_1_128)
y_pred = np.argmax(y_pred, axis=1)
y_true = []
for _, y in test_ds_1_128:
for i in y.numpy():
y_true.append(i)
print(
"test accuracy for 128: ", test_acc.append(accuracy_score(y_true, y_pred))
)
print("confusion matrix for 128: ")
plot_confusion_matrix(confusion_matrix(y_true, y_pred))
# 256
model = create_classifier(model_tuple[np.argmax(val_acc_256)], input_256)
model.fit(
train_ds_1_256,
epochs=EPOCHS,
validation_data=val_ds_1_256,
callbacks=[model_callback],
verbose=0,
)
y_pred = model.predict(test_ds_1_256)
y_pred = np.argmax(y_pred, axis=1)
y_true = []
for _, y in test_ds_1_256:
for i in y.numpy():
y_true.append(i)
print(
"test accuracy for 256: ", test_acc.append(accuracy_score(y_true, y_pred))
)
print("confusion matrix for 256: ")
plot_confusion_matrix(confusion_matrix(y_true, y_pred))
print("test accuracy: ", test_acc)
dim_list = [32, 64, 128, 256]
print("best dimension: ", dim_list[np.argmax(test_acc)])
run_on_all_arch_2()
Q4()
# #Build denoising autoencoders with one hidden layer with 20% noise and 40% noise.
# #noise adding Class
# #adds noise probabilistically to the input
# #noise is sampled from a normal distribution with mean 0 and standard deviation 1
# #this Class is active only during training
# class AddNoise(layers.Layer):
# def __init__(self,noise_prob):
# super(AddNoise, self).__init__()
# self.noise_prob = noise_prob
# def call(self, x, training=True):
# if training:
# #add noise probabilistically
# noise = tf.random.normal(shape=tf.shape(x), mean=0.0, stddev=1.0, dtype=tf.float32)
# x = tf.where(tf.random.uniform(shape=tf.shape(x)) < self.noise_prob, x + noise, x)#x+noise when random.uniform < noise_prob, else x
# return x
# else:
# return x
# BEST_NEURONS = 128
# #denoising autoencoder with one hidden layer
# denoising_autoencoder_20 = tf.keras.Sequential([
# layers.Flatten(),
# AddNoise(0.2),
# layers.Dense(BEST_NEURONS, activation=ACTIVATION),
# layers.Dense(784, activation=ACTIVATION),
# layers.Reshape((28, 28))
# ])
# denoising_autoencoder_40 = tf.keras.Sequential([
# layers.Flatten(),
# AddNoise(0.4),
# layers.Dense(BEST_NEURONS, activation=ACTIVATION),
# layers.Dense(784, activation=ACTIVATION),
# layers.Reshape((28, 28))
# ])
# run_autoencoder(denoising_autoencoder_20,"denoising_autoencoder_20")
# run_autoencoder(denoising_autoencoder_40,"denoising_autoencoder_40")
# #convert training data to reduced dimension using denoising_autoencoder_20
# denoising_autoencoder_20 = tf.keras.models.load_model("denoising_autoencoder_20")
# train_ds_denoising_20 = denoising_autoencoder_20.encoder(train_ds)
# #convert training data to reduced dimension using denoising_autoencoder_40
# denoising_autoencoder_40 = tf.keras.models.load_model("denoising_autoencoder_40")
# train_ds_denoising_40 = denoising_autoencoder_40.encoder(train_ds)
# #plot weights of denoising_autoencoder
# def plot_weights(denoising_autoencoder):
# weights = denoising_autoencoder.layers[2].get_weights()[0] #bottle neck layer
# weights = weights.reshape(28, 28, BEST_NEURONS)
# plt.figure(figsize=(20, 4))
# for i in range(BEST_NEURONS):
# ax = plt.subplot(2, BEST_NEURONS / 2, i + 1)
# plt.imshow(weights[:, :, i], cmap='gray')
# ax.get_xaxis().set_visible(False)
# ax.get_yaxis().set_visible(False)
# plt.show()
# #plot weights of autoencoder(non denoising)
# def plot_weights_autoencoder(autoencoder):
# weights = autoencoder.layers[1].get_weights()[0] #bottle neck layer
# weights = weights.reshape(28, 28, BEST_NEURONS)
# plt.figure(figsize=(20, 4))
# for i in range(BEST_NEURONS):
# ax = plt.subplot(2, BEST_NEURONS / 2, i + 1)
# plt.imshow(weights[:, :, i], cmap='gray')
# ax.get_xaxis().set_visible(False)
# ax.get_yaxis().set_visible(False)
# plt.show()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
train = pd.read_csv("/kaggle/input/titanic/train.csv")
train.head()
test = pd.read_csv("/kaggle/input/titanic/test.csv")
test.head()
df_sub = pd.read_csv("/kaggle/input/titanic/gender_submission.csv")
df_sub.head()
train.describe()
train.isnull().sum()
train.info()
train.drop(columns=["PassengerId", "Name", "Ticket", "Cabin"], inplace=True)
test.drop(columns=["PassengerId", "Name", "Ticket", "Cabin"], inplace=True)
test.head()
test.isnull().sum()
train.head()
X = train.drop(columns=["Survived"])
Y = train["Survived"]
X.shape, Y.shape
X.head()
Y.head()
from sklearn.model_selection import train_test_split
X_train, X_test, Y_train, Y_test = train_test_split(
X, Y, test_size=0.2, random_state=42
)
X_train.shape, Y_train.shape
X_train["Embarked"].value_counts()
test["Embarked"].value_counts()
from sklearn.impute import SimpleImputer
si_age = SimpleImputer()
si_embarked = SimpleImputer(strategy="most_frequent")
X_train_age = si_age.fit_transform(X_train[["Age"]])
X_train_embarked = si_embarked.fit_transform(X_train[["Embarked"]])
test_age = si_age.fit_transform(test[["Age"]])
test_fare = si_embarked.fit_transform(test[["Fare"]])
test_embarked = si_embarked.fit_transform(test[["Embarked"]])
X_test_age = si_age.transform(X_test[["Age"]])
X_test_embarked = si_embarked.transform(X_test[["Embarked"]])
from sklearn.preprocessing import OneHotEncoder
one = OneHotEncoder(sparse=False, handle_unknown="ignore")
X_train_sex = one.fit_transform(X_train[["Sex"]])
X_test_sex = one.transform(X_test[["Sex"]])
test_sex = one.fit_transform(test[["Sex"]])
test_embarked = one.fit_transform(test[["Embarked"]])
X_train_embarked = one.fit_transform(X_train[["Embarked"]])
X_test_embarked = one.transform(X_test[["Embarked"]])
test_embarked
X_train_rem = X_train.drop(columns=["Sex", "Age", "Embarked"])
X_test_rem = X_test.drop(columns=["Sex", "Age", "Embarked"])
test_rem = test.drop(columns=["Sex", "Age", "Fare", "Embarked"])
X_train_rem.head()
test_rem.head()
X_train_transformed = np.concatenate(
(X_train_age, X_train_sex, X_train_embarked, X_train_rem), axis=1
)
X_test_transformed = np.concatenate(
(X_test_age, X_test_sex, X_test_embarked, X_test_rem), axis=1
)
test_transformed = np.concatenate(
(test_age, test_sex, test_embarked, test_rem, test_fare), axis=1
)
test_transformed.shape, X_train_transformed.shape
df2 = pd.DataFrame(test_transformed)
df2
df1 = pd.DataFrame(X_train_transformed)
df1
X_train_transformed.shape, X_test_transformed.shape
from sklearn.tree import DecisionTreeClassifier
clf = DecisionTreeClassifier()
clf.fit(X_train_transformed, Y_train)
clf.score(X_train_transformed, Y_train)
y_pred = clf.predict(X_test_transformed)
y_pred
from sklearn.metrics import accuracy_score
accuracy_score(Y_test, y_pred)
df_sub.shape, y_pred.shape
test.head()
test_pred = clf.predict(test_transformed)
df_sub["target"] = y_pred
df_sub
df_sub.to_csv("submission.csv", index=False)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import seaborn as sns
import warnings
import matplotlib.pyplot as plt
from sklearn.preprocessing import LabelEncoder, MinMaxScaler
from sklearn import preprocessing, neighbors
from sklearn.model_selection import StratifiedKFold
from sklearn.model_selection import cross_validate
from sklearn.linear_model import LinearRegression, LogisticRegression
from sklearn.cluster import KMeans
from sklearn.ensemble import (
RandomForestRegressor,
ExtraTreesRegressor,
BaggingRegressor,
AdaBoostRegressor,
)
from sklearn.model_selection import cross_val_predict
from sklearn.metrics import roc_auc_score
from sklearn.tree import DecisionTreeClassifier
from sklearn.model_selection import GridSearchCV
from pandas.api.types import is_numeric_dtype
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
warnings.simplefilter(action="ignore", category=FutureWarning)
warnings.filterwarnings("ignore")
pd.set_option("display.max_rows", 500)
df = pd.read_csv(
"/kaggle/input/cost-prediction-for-logistic-company-2023w-aml1413/train.csv",
parse_dates=["date"],
)
df_test = pd.read_csv(
"/kaggle/input/cost-prediction-for-logistic-company-2023w-aml1413/test.csv",
parse_dates=["date"],
)
df
df.shape
df.info()
df.isna().sum()
df["type"].unique()
df["packageType"].unique()
df["day"] = df["date"].dt.day
df["month"] = df["date"].dt.month
df["year"] = df["date"].dt.year
df.drop("date", axis=1, inplace=True)
df["year"].value_counts()
df.fillna("Unknown", inplace=True)
diff_categ_count = df["exWeatherTag"].value_counts()
group_table = df.groupby(["exWeatherTag", "month"]).size().astype(float)
for categ in group_table.index.levels[0]:
for income in group_table[categ].index:
group_table[categ][income] = group_table[categ][income]
group_table.unstack().plot(kind="bar", stacked=True)
plt.ylabel("Percentage of Subscription Rate")
plt.title("Subscription Rate by Job Type")
print(group_table)
df[df["month"] == 12]["exWeatherTag"].unique()
df[df["month"] == 6]["exWeatherTag"].unique()
df[df["month"] == 6]["exWeatherTag"].replace("Unknown", "heat")
df.distance.value_counts().plot(kind="bar")
plt.xlabel("Distance")
plt.ylabel("Count")
sns.boxplot(data=df, x="weight")
sns.boxplot(data=df, x="cost")
df.drop(columns="trip", axis=1, inplace=True)
sns.countplot(x=df["originLocation"])
df["originLocation"] = df["originLocation"].str.replace(r"S", "").astype(int)
plt.scatter(df["originLocation"], df["cost"], c=df["weight"])
plt.legend(labels=df["weight"])
df[df["weight"] > 1000]
sns.displot(df["weight"][(df["cost"] < 200) & (df["cost"] > 100)])
df.drop(index=df.loc[(df["cost"] < 200) & (df["cost"] > 100)].index, inplace=True)
df.shape
# df_cost_weight = df[df['weight'] <= 250]
# df_cost_weight = df_cost_weight[df_cost_weight['cost'] > 150]
# index_to_delete = df_cost_weight.index.values.tolist()
# df.drop(index=index_to_delete,inplace=True)
df.shape
sns.countplot(x=df["destinationLocation"])
plt.scatter(df["destinationLocation"], df["cost"], c=df["weight"])
sns.countplot(x=df["carrier"])
df["carrier_numeric"] = df["carrier"].apply(
lambda x: 0 if x == "A" else (1 if x == "B" else (2 if x == "C" else 3))
)
plt.scatter(df["carrier"], df["distance"], c=df["cost"], alpha=1)
# df['distance_segment'] = df['distance'].apply(lambda x: 0 if x > 0 and x < 1000 else (1 if x >= 1000 and x < 2000 else (2 if x>= 2000 and x < 3000 else 3)))
# df_ca_d0 = df[(df['carrier'] == 'A') & (df['distance_segment'] == 0)]
# df_cb_d1= df[(df['carrier'] == 'B') & (df['distance_segment'] == 1)]
# df_cc_d2 = df[(df['carrier'] == 'C') & (df['distance_segment'] == 2)]
# df_cd_d3 = df[(df['carrier'] == 'D') & (df['distance_segment'] == 3)]
# # df_distance_segment_0 = df[df['distance_segment'] == 0]
# # df_distance_segment_1 = df[df['distance_segment'] == 1]
# # df_distance_segment_2 = df[df['distance_segment'] == 2]
# # df_distance_segment_3 = df[df['distance_segment'] == 3]
# plt.scatter(df_ca_d0,df_ca_d0,alpha=0.5)
# plt.scatter(df_cb_d1,df_ca_d0,alpha=0.5)
# plt.scatter(df_cc_d0,df_ca_d0,alpha=0.02)
# plt.scatter(df_cd_d0,df_ca_d0,alpha=0.02)
df_carrier_a = df[df["carrier"] == "A"]
df_carrier_b = df[df["carrier"] == "B"]
df_carrier_c = df[df["carrier"] == "C"]
df_carrier_d = df[df["carrier"] == "D"]
plt.scatter(df_carrier_a["distance"], df_carrier_a["cost"], alpha=0.5)
plt.scatter(df_carrier_b["distance"], df_carrier_b["cost"], alpha=0.5)
plt.scatter(df_carrier_c["distance"], df_carrier_c["cost"], alpha=0.5)
plt.scatter(df_carrier_d["distance"], df_carrier_d["cost"], alpha=0.5)
plt.scatter(df["carrier"], df["weight"], c=df["cost"], alpha=0.6, s=10)
plt.scatter(df_carrier_a["weight"], df_carrier_a["cost"], alpha=0.5)
plt.scatter(df_carrier_b["weight"], df_carrier_b["cost"], alpha=0.5)
plt.scatter(df_carrier_c["weight"], df_carrier_c["cost"], alpha=0.02)
plt.scatter(df_carrier_d["weight"], df_carrier_d["cost"], alpha=0.02)
sns.displot(df["distance"])
df
df["day"].value_counts().sort_index()
sns.countplot(x=df["day"])
df["dayPart"].value_counts()
sns.countplot(x=df["dayPart"])
categorical_col = []
for i in df.columns:
if (df[i].dtypes) == "object":
categorical_col.append(i)
le = LabelEncoder()
df[categorical_col] = df[categorical_col].apply(le.fit_transform)
df
df[categorical_col] = df[categorical_col].astype(int)
corr = df.corr()
plt.figure(figsize=(12, 8))
sns.heatmap(corr, cmap="Reds", annot=True)
plt.show()
df.drop(columns=["carrier_numeric"], axis=1, inplace=True)
scaler = MinMaxScaler()
df[["distance", "weight"]] = scaler.fit_transform(df[["distance", "weight"]])
df
type(df.columns)
columns = list(df.columns)
columns.remove("cost")
X = df[columns]
Y = df["cost"].astype(int)
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, Y, test_size=0.33, random_state=42
)
linreg = LinearRegression()
linreg.fit(X_train, y_train)
print(
"Accuracy of LinearRegression on training set: {:.2f}".format(
linreg.score(X_train, y_train)
)
)
print(
"Accuracy of LinearRegression on test set: {:.2f}".format(
linreg.score(X_test, y_test)
)
)
from sklearn.model_selection import cross_val_score
scores = cross_val_score(linreg, X_train, y_train, cv=5)
scores
RFReg = RandomForestRegressor(n_estimators=500, random_state=1, n_jobs=-1)
param_grid = {
"max_features": ["auto", "sqrt", "log2"],
"min_samples_split": np.linspace(0.1, 1.0, 10),
"max_depth": [x for x in range(1, 20)],
}
from sklearn.model_selection import RandomizedSearchCV
rfr = RandomizedSearchCV(
estimator=RFReg, param_distributions=param_grid, n_jobs=-1, cv=10, n_iter=50
)
rfr.fit(X_train, y_train)
print(
"Accuracy of Random Forest classifier on training set: {:.2f}".format(
rfr.score(X_train, y_train)
)
)
print(
"Accuracy of Random Forest classifier on test set: {:.2f}".format(
rfr.score(X_test, y_test)
)
)
df_test
df_test["day"] = df_test["date"].dt.day
df_test["month"] = df_test["date"].dt.month
df_test["year"] = df_test["date"].dt.year
df_test.drop(columns=["date"], inplace=True)
df_test.fillna("Unknown", inplace=True)
df_test.info()
categorical_col = []
for i in df_test.columns:
if (df_test[i].dtypes) == "object" and (i != "trip"):
categorical_col.append(i)
le = LabelEncoder()
df_test[categorical_col] = df_test[categorical_col].apply(le.fit_transform)
df_test
df_test[["distance", "weight"]] = scaler.fit_transform(df_test[["distance", "weight"]])
pred = rfr.predict(df_test.drop(columns=["trip"]))
len(pred)
output = pd.DataFrame()
output["trip"] = df_test["trip"]
output["cost"] = pred
output.to_csv("submission.csv", index=False)
print("Your submission was successfully saved!")
# # Conclusion
# This For better results down-sampling has to be done
# criterion = ['gini', 'entropy']
# max_depth = [1, 2, 3, 4, 5]
# min_samples_split = list(range(2,10))
# min_sampes_leaf = [(i + 1) / 10 for i in range(5)]
# max_leaf_nodes = list(range(2, 35))
# max_features = ['sqrt','log2',None]
# parameter_distribution = {'criterion' : criterion, 'max_depth' : max_depth,
# 'min_samples_split' : min_samples_split, 'min_samples_leaf' : min_sampes_leaf,
# 'max_leaf_nodes' : max_leaf_nodes, 'max_features' : max_features}
# dt = GridSearchCV(DecisionTreeClassifier(), parameter_distribution, cv=5, scoring='roc_auc', refit=True)
# dt.fit(X_train,y_train)
# # dt.score(X_train,y_train)
# best_estimators_dt = dt.best_estimator_
# best_paras_dt = dt.best_params_
# print(best_estimators_dt)
# print(best_paras_dt)
# best_estimators_dt.score(X_train,y_train)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Slicing Strings - Dilimleme
# Dilim sözdizimini kullanarak bir dizi karakter döndürebilirsiniz.
# Dizenin bir bölümünü döndürmek için başlangıç dizinini ve bitiş dizinini iki nokta üst üste ile ayırarak belirtin.
# Karakterleri 2. konumdan 5. konuma getirin (dahil değildir):
b = "fatih, yorgun !"
print(b[1:6])
# Not: İlk karakterin indeksi 0'dır.
# # Slice From the Start
# Başlangıç dizinini dışarıda bırakırsanız, aralık ilk karakterden başlar:
# Karakterleri baştan 5. konuma getirin (dahil değildir):
b = "fatih! ince!"
print(b[:5])
# # Slice To the End
# Son dizini dışarıda bırakarak, aralık sonuna gider:
# karakterleri 2. konumdan ve sonuna kadar alın:
b = "dunya, selam! fatih ince "
print(b[14:19])
# # Negative Indexing - Negatif İndeksleme
# Dilimi dizenin sonundan başlatmak için negatif dizinler kullanın:
b = "fati, ince!"
print(b[-5:-2])
# # Modify Strings - Dizeleri Değiştir
# # Upper Case - Büyük Harf
# Upper() yöntemi, dizeyi büyük harfle döndürür:
a = "baba, fatih!"
print(a.upper())
# BABA FATİH !
# # Lower Case - Küçük Harf
# # lower() yöntemi, dizeyi küçük harfle döndürür:
a = "Hello, World! FATİH İNCE"
print(a.lower())
# fatih ince
# # Remove Spaces - Boşlukları Kaldırma
# strip() fonksiyonu, baştaki veya sondaki tüm boşlukları kaldırır:
a = " Fatih ince! "
print(a.strip()) # returns "Hello, World!"
# fatih ince!
# # Replace String
# replace() yöntemi, bir dizeyi başka bir dizeyle değiştirir:
a = "Hello, ali"
print(a.replace("değişecek_olan", "yerine_gelmesini_istediğimiz"))
# # Split String - Bölünmüş Dize
# split() yöntemi, belirtilen ayırıcı arasındaki metnin liste öğeleri haline geldiği bir liste döndürür.
# split kelimeleri ayırır
a = "Hello, World!, ince"
print(a.split(",")) # returns ['Hello', ' World!']
# # String Concatenation - Dize Birleştirme
# İki diziyi birleştirmek veya birleştirmek için + operatörünü kullanabilirsiniz.
# Merge variable a with variable b into variable c:
a = "gs"
b = "cahmp"
c = a + b
print(c)
# gs,cahmp
# # Dize Biçimi
# Python Değişkenleri bölümünde öğrendiğimiz gibi, dizileri ve sayıları şu şekilde birleştiremeyiz:
age = 20
txt = "My name is fatih, I am " + age
print(txt)
# Ancak, format() yöntemini kullanarak dizeleri ve sayıları birleştirebiliriz!
# format() yöntemi iletilen bağımsız değişkenleri alır, biçimlendirir ve {} yer tutucularının bulunduğu dizeye yerleştirir:
# Dizelere sayı eklemek için format() yöntemini kullanın:
age = 20
txt = "My name is fatih, and I am {}"
print(txt.format(age))
# My name is fatih, and I am 20
# format() yöntemi sınırsız sayıda bağımsız değişken alır ve ilgili yer tutuculara yerleştirilir:
quantity = 5
itemno = 666
price = 54.95
myorder = "I want {} pieces of item {} for {} dollars."
print(myorder.format(quantity, itemno, price))
# I want 5 pieces of item 666 for 54.95 dollars.
# Bağımsız değişkenlerin doğru yer tutuculara yerleştirildiğinden emin olmak için dizin numaralarını {0} kullanabilirsiniz:
quantity = 3
itemno = 567
price = 49.95
myorder = "I want to pay {0} dollars for {1} pieces of item {2}."
print(myorder.format(quantity, itemno, price))
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.