script
stringlengths 113
767k
|
|---|
import bq_helper
import matplotlib.pyplot as plt
import matplotlib.cm as cm
stackOverflow = bq_helper.BigQueryHelper(
active_project="bigquery-public-data", dataset_name="stackoverflow"
)
query = """
SELECT users.display_name as `Display Name`, COUNT(users.id) as Count
FROM bigquery-public-data.stackoverflow.users AS users
INNER JOIN bigquery-public-data.stackoverflow.comments AS comments
ON users.id = comments.user_id
WHERE users.id > 0
GROUP BY users.display_name
ORDER BY count DESC
LIMIT 25;
"""
top_users = stackOverflow.query_to_pandas_safe(query, max_gb_scanned=2)
top_users = top_users.sort_values(["Count"], ascending=True)
top_users
_, ax = plt.subplots(figsize=(22, 10))
top_users.plot(
x="Display Name",
y="Count",
kind="barh",
ax=ax,
color=cm.viridis_r(top_users.Count / float(max(top_users.Count))),
)
ax.spines["top"].set_visible(False)
ax.spines["right"].set_visible(False)
ax.spines["bottom"].set_visible(False)
ax.set_title("Top 25 Users With The Most Comments", fontsize=20)
for i, v in enumerate(top_users.Count):
ax.text(v + 750, i - 0.15, str(v), color="black", fontsize=12)
ax.xaxis.set_visible(False)
ax.yaxis.get_label().set_visible(False)
ax.tick_params(axis="y", labelsize=14)
ax.get_legend().remove()
|
#
# - Learn about the prices of all cars and the changes made to all car brands
# - The automobile industry has seen a dramatic shift in pricing over the past decade. With the introduction of new technologies, increased competition, and changing consumer preferences, car prices have been on the rise. This dataset will explore the changes in car prices over time, looking at factors such as inflation, technological advances, and consumer demand.
# - you will understand collecting, organizing, analyzing, and interpreting my data
# Table of Contents
# 1. [import library](#dataA)
# 2. [Data Exploration](#dataB)
# 3. [describe data](#dataC)
# 4. [feature engineering](#dataD)
# 4.1. [handling data types](#dataD1)
# 4.2. [Exploratory Data Analysis (EDA)](#dataD2)
# 4.3. [performing a data filtering operation](#dataD3)
#
#
# 5. [Conclusion](#dataE)
# 1. import library
#
# import necessary library for work
import numpy as np
import pandas as pd
import pandas_profiling as pp
import matplotlib.pyplot as plt
import seaborn as sns
import skimpy
#
# 2. Data Exploration
#
# read data that we will work about it
df = pd.read_csv("/kaggle/input/car-price/car_price.csv", index_col=False)
my_data = df.copy()
my_data
#
# 3. describe data
# - overview about data to know some of properties as Dataset statistics name of columns and number of columns , interaction , correlations , missing values and sample about data
over_view = pp.ProfileReport(my_data)
over_view
df = pd.read_csv(r"/kaggle/input/car-price/car_price.csv") # describe data
report = skimpy.skim(df)
print(report)
my_data.isnull().count() # to know missing value
my_data.head(3) # show some of data to know info about the name of columns
my_data.columns # my data content of 5 columns
#
# 4. feature engineering
#
my_data.drop(
"Unnamed: 0", axis=1, inplace=True
) # drop the columns which named "Unnamed" from columns
new_col = [
"cars_names",
"old_price",
"price_change",
"new_price",
"change_date",
] # list of new name
my_data.columns = new_col
my_data.head(5)
my_data.dtypes # show the data type of my columns to know how will work with it
my_data[
"change_date"
] # this column should have date this mean it's dtype should be datetime
#
# 4.1. handling data types
#
my_data["change_date"] = pd.to_datetime(my_data["change_date"])
my_data["change_date"]
my_data.dtypes
my_data[
"old_price"
] # this column content is mix of string and numerical we will change dtype to number
# - create function to convert columns which contan of mix values int,str , take name of column and the string which you want to delete and replace the string with "" and replace "," with "" to ease the convert to int
def to_int(column):
# my_data[column] = my_data[column].astype(str).replace(string , "")
my_data[column] = my_data[column].astype(str).replace(r"\D", "", regex=True)
for i in range(len(my_data[column])):
my_data[column][i] = my_data[column][i].replace(",", "")
my_data.dtypes
to_int("old_price")
# my_data_copy["old_price"] = my_data_copy["old_price"].astype(int)
my_data.loc[:, "old_price"] = my_data["old_price"].astype(int)
my_data["old_price"]
my_data.head(3)
my_data[
"price_change"
] # this column contains of 3 different string are "trending_up" , "trending_down" , "EGP" , ","
# my_data["price_change"] = my_data["price_change"].str.replace("EGP" , " ")
# li_str = ["trending_up" , "trending_down"]
# for i in li_str:
to_int("price_change")
# my_data.iloc[:, "price_change"] = my_data.iloc[:, "price_change"].astype(int)
my_data["price_change"] = my_data["price_change"].astype(int)
my_data["price_change"]
my_data.head(4)
to_int("new_price")
my_data["new_price"] = my_data["new_price"].astype(int)
my_data
my_data["cars_names"] = my_data["cars_names"].astype("string")
my_data["cars_names"].describe()
my_data.dtypes
#
# 4.2. Exploratory Data Analysis (EDA)
#
plt.figure(figsize=(20, 10))
sns.set_style("darkgrid")
sns.pairplot(my_data)
plt.show()
plt.figure(figsize=(10, 10))
sns.set(
rc={
"axes.facecolor": "#2b4969",
"axes.grid": False,
"xtick.labelsize": 14,
"ytick.labelsize": 14,
}
)
# sns.set_style("dark")
sns.scatterplot(
x=my_data["old_price"], y=my_data["new_price"], color="#FF0000", markers="."
)
plt.show()
my_data["cars_names"].duplicated().sum()
my_data["cars_names"].nunique()
car_brand = []
for i in range(len(my_data["cars_names"])):
car_brand.append(my_data["cars_names"][i].split()[0])
my_data["car_brand"] = car_brand
my_data
my_data["car_brand"].nunique()
my_data["car_brand"].value_counts()
# Knowing some statistical information for each company or mark,
# such as the average, the largest value, the smallest value, and the sum of values in relation to the price increase column
info = pd.DataFrame(
my_data.groupby("car_brand").price_change.agg(
["count", "mean", "min", "max", "sum"]
)
)
info
# restore the index name, and move the index back to a column
info.reset_index(inplace=True)
# info.drop("level_0" , axis=1 , inplace=True)
info
# the name brand of max change in price change and the sum of its changes
pd.DataFrame(info[["car_brand", "sum"]][info["sum"] == info["sum"].max()])
plt.figure(figsize=(20, 20))
plt.title("Mercedes")
sns.set(
rc={
"axes.facecolor": "#414547",
"axes.grid": False,
"xtick.labelsize": 14,
"ytick.labelsize": 14,
}
)
sns.pointplot(
data=my_data,
x=my_data["old_price"][my_data["car_brand"] == "Mercedes"],
y=my_data["price_change"][my_data["car_brand"] == "Mercedes"],
)
plt.show()
# the name brand of min change in price change and the sum of its changes
pd.DataFrame(info[["car_brand", "sum"]][info["sum"] == info["sum"].min()])
plt.figure(figsize=(20, 20))
plt.title("Porsche")
sns.set(
rc={
"axes.facecolor": "#132c47",
"axes.grid": False,
"xtick.labelsize": 14,
"ytick.labelsize": 14,
}
)
sns.pointplot(
x=my_data["old_price"][my_data["car_brand"] == "Porsche"],
y=my_data["price_change"][my_data["car_brand"] == "Porsche"],
)
plt.show()
#
# 4.3. performing a data filtering operation
#
my_data[my_data["car_brand"] == "Mercedes"]
# the mean , min , max values for brand who is the max change in price change
info_mercedes = pd.DataFrame(
my_data["price_change"][my_data["car_brand"] == "Mercedes"]
)
pd.DataFrame(info_mercedes.agg(["mean", "min", "max"]).astype(int))
# the mean , min , max values for brand who is the min change in price change
info_Porsche = pd.DataFrame(my_data["price_change"][my_data["car_brand"] == "Porsche"])
pd.DataFrame(info_Porsche.agg(["mean", "min", "max"]).astype(int))
my_data[my_data["car_brand"] == "Porsche"]
# The sum of the changes that occurred to the Mercedes company,
# which is the owner of the largest increase in car prices among the companies,
# starting from [2/2/2021] to [17/1/2023] for each day of the change,
# whether with an increase or decrease, and their number
df_mercedes = pd.DataFrame(
my_data.groupby(
my_data["change_date"][my_data["car_brand"] == "Mercedes"]
).price_change.agg(["sum", "count"])
)
df_mercedes.reset_index(inplace=True)
df_mercedes
plt.figure(figsize=(10, 8))
sns.scatterplot(x=df_mercedes["change_date"], y=df_mercedes["sum"], color="#FF0000")
plt.show()
# The sum of the changes that occurred to the Porsche company,
# which is the owner of the smallest increase in car prices among the companies,
# starting from [14/10/2019] to [18/11/2020] for each day of the change,
# whether with an increase or decrease, and their number
df_porshe = pd.DataFrame(
my_data.groupby(
my_data["change_date"][my_data["car_brand"] == "Porsche"]
).price_change.agg(["sum", "count"])
)
df_porshe.reset_index(inplace=True)
df_porshe
plt.figure(figsize=(8, 6))
sns.scatterplot(x=df_porshe["change_date"], y=df_porshe["sum"], color="#ed0915")
plt.show()
my_data["cars_names"][my_data["cars_names"].duplicated()]
my_data["cars_names"].nunique()
info_names = pd.DataFrame(my_data.groupby("cars_names").price_change.agg("count"))
info_top30 = pd.DataFrame(
info_names["price_change"].nlargest(30)
) # to know 30 top of changes
info_top30.reset_index(inplace=True)
info_top30
plt.figure(figsize=(20, 10))
sns.set(
rc={
"axes.facecolor": "#283747",
"axes.grid": False,
"xtick.labelsize": 14,
"ytick.labelsize": 14,
}
)
colors = [
"#8f285b",
"#4b78b8",
"#aecd6e",
"#d47f3f",
"#8e2991",
"#7a3f3e",
"#418274",
"#b23b47",
"#ffb00d",
"#4b4b4b",
"#d6adad",
"#3978b8",
"#e6f088",
"#d46b6b",
"#a8c6e5",
]
plt.bar(info_top30["cars_names"][0:15], info_top30["price_change"][0:15], color=colors)
plt.show()
plt.figure(figsize=(10, 7))
sns.set(
rc={
"axes.facecolor": "#283747",
"axes.grid": False,
"xtick.labelsize": 14,
"ytick.labelsize": 14,
}
)
sns.barplot(data=info_top30, x=info_top30["cars_names"], y=my_data["old_price"])
plt.show()
# The total change for each type of car brands represents the owners of the top 30 changes in their prices
# for 16 times duplicated
list = []
for i in range(30):
list.append(
my_data["price_change"][
my_data["cars_names"] == info_top30["cars_names"][i]
].sum()
)
changes_top30 = pd.DataFrame(list, columns=["sum_changes_top30"])
changes_top30
plt.figure(figsize=(20, 10))
sns.set(
rc={
"axes.facecolor": "#283747",
"axes.grid": False,
"xtick.labelsize": 14,
"ytick.labelsize": 14,
}
)
sns.scatterplot(x=info_top30["cars_names"], y=changes_top30["sum_changes_top30"])
plt.show()
plt.figure(figsize=(20, 15))
sns.set(
rc={
"axes.facecolor": "#283747",
"axes.grid": False,
"xtick.labelsize": 14,
"ytick.labelsize": 14,
}
)
sns.barplot(
x=info_top30["cars_names"][0:11], y=changes_top30["sum_changes_top30"][0:11]
)
plt.show()
df_old = pd.DataFrame(my_data["old_price"].describe().astype(int))
df_old.reset_index(inplace=True)
df_old
plt.bar(df_old["index"], df_old["old_price"])
plt.show()
df_new = pd.DataFrame(my_data["new_price"].describe().astype(int))
df_new.reset_index(inplace=True)
df_new
plt.scatter(df_new["index"], df_new["new_price"], color="#c22961")
plt.show()
|
# #COVID-19 vaccine doses administered per 100 people
# Total number of vaccination doses administered per 100 people in the total population. This is counted as a single dose, and may not equal the total number of people vaccinated, depending on the specific dose regime (e.g. people receive multiple doses).
# https://data.humdata.org/dataset/covid-19-vaccinations
# youtube.com
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
from sklearn.preprocessing import StandardScaler
from sklearn.cluster import KMeans, AffinityPropagation
import matplotlib.pyplot as plt
import seaborn as sns
import plotly as py
import plotly.graph_objs as go
import plotly.express as px
py.offline.init_notebook_mode(connected=True)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
nRowsRead = 1000 # specify 'None' if want to read whole file
df = pd.read_csv(
"../input/cusersmarildownloadsvaccinatedcsv/vaccinated.csv",
delimiter=";",
encoding="utf8",
nrows=nRowsRead,
)
df.dataframeName = "vaccinated.csv"
nRow, nCol = df.shape
print(f"There are {nRow} rows and {nCol} columns")
df.head()
# #Dataprep
# Acknowlegdements
# SFU Data Science Research Group - SIMON FRASER UNIVERSITY
# https://sfu-db.github.io/dataprep/user_guide/eda/plot.html
# https://dataprep.ai/
# https://www.cs.sfu.ca/~jnwang/
# https://www.cs.sfu.ca/~jnwang/ppt/DataPrep-Overview-Databricks.pdf
from dataprep.eda import plot, plot_correlation, create_report, plot_missing
plot_missing(df)
plot(df)
# API Correlation
plot_correlation(df)
brz = df[(df["location"] == "Brazil")].reset_index(drop=True)
brz.head()
# Code by Joshua Swords https://www.kaggle.com/joshuaswords/data-visualization-clustering-mall-data
# Preparing to cluster: What about the distribution of our data?
import matplotlib.gridspec as gridspec
# Create 2x2 sub plots
gs = gridspec.GridSpec(1, 3)
fig = plt.figure(figsize=(15, 3))
# Title and sub-title
fig.text(
0.09,
1.09,
"Numeric variable distribution",
fontsize=15,
fontweight="bold",
fontfamily="serif",
)
fig.text(
0.09,
1,
"Our data appears to be relatively normal, therefore we will not transform it.",
fontsize=12,
fontweight="light",
fontfamily="serif",
)
# Grid spec layout
ax1 = fig.add_subplot(gs[0, 0]) # row 0, col 0
ax1 = sns.distplot(
df["people_vaccinated"],
color="#244747",
hist_kws=dict(edgecolor="white", linewidth=1, alpha=0.8),
)
ax2 = fig.add_subplot(gs[0, 1]) # row 0, col 1
ax2 = sns.distplot(
df["people_fully_vaccinated"],
color="#244747",
hist_kws=dict(edgecolor="white", linewidth=1, alpha=0.8),
)
ax3 = fig.add_subplot(gs[0, 2]) # row 1, span all columns
ax3 = sns.distplot(
df["daily_vaccinations_per_million"],
color="#244747",
hist_kws=dict(edgecolor="white", linewidth=1, alpha=0.8),
)
sns.despine()
plt.subplots_adjust(
left=None, bottom=None, right=None, top=None, wspace=0.3, hspace=None
) # useful for adjusting space between subplots
fig = px.bar(
df,
x="date",
y="people_vaccinated",
color_discrete_sequence=["blue"],
title="Covid Vaccine Administered",
text="location",
)
fig.show()
fig = px.bar(
brz,
x="date",
y="people_vaccinated",
color_discrete_sequence=["darkolivegreen"],
title="Covid Vaccine Administered in Brazil",
text="people_fully_vaccinated",
)
fig.show()
fig = px.parallel_categories(
df, color="people_vaccinated", color_continuous_scale=px.colors.sequential.OrRd
)
fig.show()
fig = px.line(
df,
x="date",
y="people_fully_vaccinated",
color_discrete_sequence=["darksalmon"],
title="Covid-19 Doses Administered",
)
fig.show()
fig = px.scatter(
df,
x="date",
y="people_fully_vaccinated",
color_discrete_sequence=["#4257f5"],
title="Covid Doses Administered",
)
fig.show()
# Code by Olga Belitskaya https://www.kaggle.com/olgabelitskaya/sequential-data/comments
from IPython.display import display, HTML
c1, c2, f1, f2, fs1, fs2 = "#eb3434", "#eb3446", "Akronim", "Smokum", 30, 15
def dhtml(string, fontcolor=c1, font=f1, fontsize=fs1):
display(
HTML(
"""<style>
@import 'https://fonts.googleapis.com/css?family="""
+ font
+ """&effect=3d-float';</style>
<h1 class='font-effect-3d-float' style='font-family:"""
+ font
+ """; color:"""
+ fontcolor
+ """; font-size:"""
+ str(fontsize)
+ """px;'>%s</h1>""" % string
)
)
dhtml("Be patient. Marília Prata, @mpwolke was Here.")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# The thing should take around 10gb of gpu memory when being trained
device = "cuda"
model_checkpoint = "bert-large-uncased"
# ## Exploring the data
import pandas as pd
raw_train_df = pd.read_csv("/kaggle/input/nlp-getting-started/train.csv")
raw_test_df = pd.read_csv("/kaggle/input/nlp-getting-started/test.csv")
len(raw_train_df)
raw_train_df.head()
raw_test_df.head()
duplicates_df = raw_train_df[raw_train_df.text.duplicated(keep=False)].sort_values(
"text"
)
duplicates_df.head()
# ## Clean the data
# As we've seen, there are dosens of duplicated tweets. Moreover, their labes are sometimes contradictory.
duplicates_df[:20]
duplicated_tweets = list(duplicates_df.text)
len(duplicated_tweets)
# Now search for contradictions
contradictory_tweets = set()
for tweet in duplicated_tweets:
if len(set(duplicates_df[duplicates_df["text"] == tweet].target)) > 1:
contradictory_tweets.add(tweet)
contradictory_tweets = list(contradictory_tweets)
print(len(contradictory_tweets), type(contradictory_tweets))
# We're going to label those manually
for i, tweet in enumerate(contradictory_tweets):
print(i, tweet, "\n")
no_duplicates_train_df = raw_train_df.drop_duplicates("text")
print(len(no_duplicates_train_df))
no_duplicates_train_df.loc[
no_duplicates_train_df["text"] == contradictory_tweets[0], "target"
] = 0
no_duplicates_train_df.loc[
no_duplicates_train_df["text"] == contradictory_tweets[1], "target"
] = 0
no_duplicates_train_df.loc[
no_duplicates_train_df["text"] == contradictory_tweets[2], "target"
] = 0
no_duplicates_train_df.loc[
no_duplicates_train_df["text"] == contradictory_tweets[3], "target"
] = 0
no_duplicates_train_df.loc[
no_duplicates_train_df["text"] == contradictory_tweets[4], "target"
] = 0
no_duplicates_train_df.loc[
no_duplicates_train_df["text"] == contradictory_tweets[5], "target"
] = 0
no_duplicates_train_df.loc[
no_duplicates_train_df["text"] == contradictory_tweets[6], "target"
] = 0
no_duplicates_train_df.loc[
no_duplicates_train_df["text"] == contradictory_tweets[7], "target"
] = 0
no_duplicates_train_df.loc[
no_duplicates_train_df["text"] == contradictory_tweets[8], "target"
] = 0
no_duplicates_train_df.loc[
no_duplicates_train_df["text"] == contradictory_tweets[9], "target"
] = 1
no_duplicates_train_df.loc[
no_duplicates_train_df["text"] == contradictory_tweets[10], "target"
] = 0
no_duplicates_train_df.loc[
no_duplicates_train_df["text"] == contradictory_tweets[11], "target"
] = 1
no_duplicates_train_df.loc[
no_duplicates_train_df["text"] == contradictory_tweets[12], "target"
] = 0
no_duplicates_train_df.loc[
no_duplicates_train_df["text"] == contradictory_tweets[13], "target"
] = 0
no_duplicates_train_df.loc[
no_duplicates_train_df["text"] == contradictory_tweets[14], "target"
] = 0
no_duplicates_train_df.loc[
no_duplicates_train_df["text"] == contradictory_tweets[15], "target"
] = 0
no_duplicates_train_df.loc[
no_duplicates_train_df["text"] == contradictory_tweets[16], "target"
] = 0
no_duplicates_train_df.loc[
no_duplicates_train_df["text"] == contradictory_tweets[17], "target"
] = 1
no_duplicates_train_df.loc[
no_duplicates_train_df["text"] == contradictory_tweets[12], "target"
]
# ## Create the Datasets
import torch
from torch.utils.data import Dataset
from transformers import AutoTokenizer, AutoModel
from tqdm import tqdm
from copy import deepcopy
import random
tokenizer = AutoTokenizer.from_pretrained(model_checkpoint)
class LabeledDisasterTweetsDataset(Dataset):
def __init__(
self,
df,
tokenizer,
max_tweet_len=180,
tweet_column="text",
location_column="location",
keyword_column="keyword",
target_column="target",
device="cpu",
text_prep_f=None,
):
self._data = []
for index, row in tqdm(df.iterrows(), total=len(df), ncols=70):
text = (
row[tweet_column]
if text_prep_f == None
else text_prep_f(
row[tweet_column], row[keyword_column], row[location_column]
)
)
tokenized = tokenizer(
text,
padding="max_length",
truncation=True,
max_length=256,
return_tensors="pt",
)
self._data.append(
(
(
tokenized["input_ids"][0].to(device),
tokenized["attention_mask"][0].to(device),
),
torch.tensor(row[target_column]).to(device),
)
)
print("NOTE: you can safely delete the dataframe")
def __len__(self):
return len(self._data)
def __getitem__(self, i):
"""return: ((input_ids, attention_mask), target)"""
return self._data[i]
def train_valid_split(self, train_fraction=0.9, shuffle=True):
num_train_examples = int(len(self) * train_fraction)
train_dataset = deepcopy(self)
if shuffle:
random.shuffle(train_dataset._data)
valid_dataset = deepcopy(train_dataset)
train_dataset._data = train_dataset._data[:num_train_examples]
valid_dataset._data = valid_dataset._data[num_train_examples:]
print(
"NOTE: you can safely delete parent dataset. The train|valid split is {}|{}.".format(
len(train_dataset), len(valid_dataset)
)
)
return train_dataset, valid_dataset
class UnlabeledDisasterTweetsDataset(Dataset):
def __init__(
self,
df,
tokenizer,
max_tweet_len=180,
tweet_column="text",
location_column="location",
keyword_column="keyword",
id_column="id",
device="cpu",
text_prep_f=None,
):
self._data = []
for index, row in tqdm(df.iterrows(), total=len(df), ncols=70):
text = (
row[tweet_column]
if text_prep_f == None
else text_prep_f(
row[tweet_column], row[keyword_column], row[location_column]
)
)
tokenized = tokenizer(
text,
padding="max_length",
truncation=True,
max_length=256,
return_tensors="pt",
)
self._data.append(
(
torch.tensor(row[id_column]).to(device),
(
tokenized["input_ids"][0].to(device),
tokenized["attention_mask"][0].to(device),
),
)
)
print("NOTE: you can safely delete the dataframe")
def __len__(self):
return len(self._data)
def __getitem__(self, i):
"""return: (id, (input_ids, attention_mask))"""
return self._data[i]
def prep_fn(text, keywords, location):
return text + " Location: " + str(location) + ". Keywords: " + str(keywords) + "."
dataset = LabeledDisasterTweetsDataset(
no_duplicates_train_df, tokenizer, device="cuda", text_prep_f=prep_fn
)
train_dataset, valid_dataset = dataset.train_valid_split()
eval_dataset = UnlabeledDisasterTweetsDataset(
raw_test_df, tokenizer, device="cuda", text_prep_f=prep_fn
)
tokenizer.decode(train_dataset[0][0][0])
tokenizer.decode(eval_dataset[0][1][0])
# ## Defining the model
class BertClassifier(torch.nn.Module):
def __init__(
self, n_classes=2, head_dropout=0.2, model_checkpoint=model_checkpoint
):
super().__init__()
self.Base = AutoModel.from_pretrained(model_checkpoint)
self.Head = torch.nn.Sequential(
torch.nn.Dropout(head_dropout),
torch.nn.Linear(1024, 1024),
torch.nn.Dropout(head_dropout),
torch.nn.Linear(1024, n_classes), # projection
)
def forward(self, input_ids, attention_mask=None, *argv):
res = self.Base.forward(
input_ids=input_ids, attention_mask=attention_mask, return_dict=False
)
res = res[0]
res = res[:, 0, :] # encoding for <s> token
res = self.Head(res)
return res
def parameters_num(self):
return sum(p.numel() for p in self.parameters())
model = BertClassifier(head_dropout=0.5)
model.forward(torch.tensor([1, 2, 33, 2])[None])
model.to(device)
model.forward(torch.tensor([[1]], device=device))
print("Our Roberta has {:,d} parameters...".format(model.parameters_num()))
# ## Fine-tuning
from torch.utils.data import DataLoader
batch_size = 4
train_dataloader = DataLoader(train_dataset, batch_size=batch_size, shuffle=True)
valid_dataloader = DataLoader(valid_dataset, batch_size=batch_size, shuffle=False)
from sklearn.metrics import accuracy_score, precision_score, recall_score, f1_score
def train(
model,
train_dataloader,
valid_dataloader,
steps,
optimizer,
blind_steps=None,
loss_fn=torch.nn.BCELoss(),
main_metric=("f1", f1_score),
additional_metrics=[],
filepath="model_sd.pt",
load_best=True,
scheduler=None,
losses_dict=None,
):
if blind_steps == None:
blind_steps = len(train_dataloader) // 4
def evaluate(): # the first score returned is the main
model.eval()
y_trues = []
y_hats = []
loss = 0
k = 0
with torch.no_grad():
for batch in valid_dataloader:
(ids, mask), y_true = batch
hots = torch.nn.functional.one_hot(y_true, 2).to(device, torch.float)
y_hat = torch.softmax(
model.forward(input_ids=ids, attention_mask=mask), dim=-1
)
loss += float(loss_fn(y_hat, hots))
k += 1
for i in range(y_true.shape[0]):
y_trues.append(int(y_true[i]))
y_hats.append(1 if y_hat[i][0] < y_hat[i][1] else 0)
scores = [(main_metric[0], main_metric[1](y_trues, y_hats))]
for metric in additional_metrics:
scores.append((metric[0], metric[1](y_trues, y_hats)))
model.train()
return scores + [("valid_loss", loss / k)]
def render_scores(scores, step, best=None):
print("{:05d} steps".format(step), end=" ")
for score in scores:
print("| {}: {:.3f}".format(*score), end=" ")
if best != None:
print("| best_score: {:.3f}".format(best))
# initial scores
scores = evaluate()
render_scores(scores, 0)
best_score = scores[0][1]
torch.save(model.state_dict(), filepath)
# logs
if losses_dict != None:
losses_dict["train_loss"] = []
losses_dict["valid_loss"] = []
losses_dict[main_metric[0]] = []
epoch_loss = 0
k = 0
train_iter = iter(train_dataloader)
model.train()
for step in tqdm(range(steps)):
# retrieving a batch
try:
batch = next(train_iter)
except:
train_iter = iter(train_dataloader)
batch = next(train_iter)
(ids, mask), y_true = batch
# prediction
y_hat = torch.softmax(model.forward(input_ids=ids, attention_mask=mask), dim=-1)
hots = torch.nn.functional.one_hot(y_true, 2).to(device, torch.float)
loss = loss_fn(y_hat, hots)
# backprop
optimizer.zero_grad()
loss.backward()
optimizer.step()
if scheduler != None:
scheduler.step()
epoch_loss += float(loss)
k += 1
# evaluation
if (step + 1) % blind_steps == 0:
scores = evaluate() + [("train_loss", epoch_loss / k)]
if losses_dict != None:
losses_dict["valid_loss"].append(float(scores[-2][1]))
losses_dict["train_loss"].append(float(scores[-1][1]))
losses_dict[main_metric[0]].append(float(scores[0][1]))
if scores[0][1] > best_score:
best_score = scores[0][1]
torch.save(model.state_dict(), filepath)
render_scores(scores, step + 1, best=best_score)
epoch_loss = 0
k = 0
if load_best:
model.load_state_dict(torch.load(filepath))
optimizer = torch.optim.Adam(model.parameters(), lr=5e-6, weight_decay=2e-3)
scheduler = torch.optim.lr_scheduler.LinearLR(optimizer, total_iters=500)
logs_dict = {}
train(
model,
train_dataloader,
valid_dataloader,
7000,
optimizer,
blind_steps=125,
additional_metrics=[
("precision", precision_score),
("recall", recall_score),
("accuracy", accuracy_score),
],
losses_dict=logs_dict,
scheduler=scheduler,
)
import numpy as np
import seaborn as sns
sns.set_theme(style="whitegrid")
sns.lineplot(
data=pd.DataFrame(
logs_dict, [(i + 1) * 125 for i in range(len(logs_dict["train_loss"]))]
),
palette="Set2",
linewidth=2.5,
)
# ## Evaluate
model.load_state_dict(torch.load("model_sd.pt"))
def evaluate(
model,
valid_dataloader,
metrics=[
("f1", f1_score),
("precision", precision_score),
("recall", recall_score),
("accuracy", accuracy_score),
],
):
model.eval()
y_trues = []
y_hats = []
with torch.no_grad():
for batch in valid_dataloader:
(ids, mask), y_true = batch
y_hat = torch.softmax(
model.forward(input_ids=ids, attention_mask=mask), dim=-1
)
for i in range(y_true.shape[0]):
y_trues.append(int(y_true[i]))
y_hats.append(1 if y_hat[i][0] < y_hat[i][1] else 0)
scores = []
for metric in metrics:
scores.append((metric[0], metric[1](y_trues, y_hats)))
return scores
scores = evaluate(model, valid_dataloader)
print(scores)
def classify(tweet):
ids = torch.tensor(tokenizer(tweet)["input_ids"])[None].to(device)
return model.forward(input_ids=ids)
classify("Over 2 hundred families have become victims of my cookie-shop iniciative")
classify(
"These events are terrible. The God will make those terorists suffer. #karma #teror"
)
# ## Make predictions
predictions_df = pd.DataFrame()
for i, (ids, mask) in tqdm(eval_dataset):
pred = model(input_ids=ids[None], attention_mask=mask[None])[0]
y_hat = 1 if pred[0] < pred[1] else 0
r = [int(i), y_hat]
predictions_df = pd.concat(
[predictions_df, pd.DataFrame(np.array(r)[None, :], columns=["id", "target"])]
)
predictions_df.target = predictions_df.target.astype(int)
predictions_df.id = predictions_df.id.astype(int)
predictions_df.head(20)
predictions_df.to_csv("submission.csv", index=False)
predictions_df.to_csv(str(scores[0][1]) + ".csv", index=False)
print("Done!")
|
# In this notebook, I'll build a **Support Vector Machine** for classification using **scikit-learn** and the **Radial Basis Function (RBF) Kernel**. My training dataset contains continous and categorical data from the UCI Machine Learning Repository to predict *whether or not a perso will default on their credit card*.
# SVM are one of the best machine learning methods when getting the correct answer is a higher priority than understanding why you get the correct answer. They work really well with relatively small datasets and they tend to work well "out of the box". In other words, they do not require much optimization.
# ## 1. Load Libraries
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.colors as colors
from sklearn.utils import resample # downsample the data
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import scale # scale and center data
from sklearn.svm import SVC # SVM for classification
from sklearn.model_selection import GridSearchCV # cross validation
from sklearn.metrics import confusion_matrix
from sklearn.metrics import plot_confusion_matrix
from sklearn.decomposition import PCA # to perform PCA to plot the data
# ## 2. Importing Data
df = pd.read_csv(
"/kaggle/input/default-on-their-credit-card/default of credit card clients.csv",
header=1,
)
df.head()
# We see a bunch of columns for the variables collected for each customer. The columns are...
# * **ID**: The ID number assigned to each customer
# * **LIMIT_BAL**: Credit limit
# * **SEX**: Gender (1 = male; 2 = female).
# * **EDUCATION**: Education (1 = graduate school; 2 = university; 3 = high school; 4 = others).
# * **MARRIAGE**: Marital status (1 = married; 2 = single; 3 = others).
# * **AGE**: Age (year).
# * **PAY_**: When the last 6 bills were payed
# * **BLL_AMT**: What the last 6 bills were
# * **PAY_AMT**: How much the last payments were
# * **default payment next month**
# I'll change the last column to **DEFAULT**.
df.rename({"default payment next month": "DEFAULT"}, axis="columns", inplace=True)
df.head()
# Drop ID, cause it was randomly assigned
df.drop("ID", axis=1, inplace=True)
df.head()
# ## 3. Missing Data
# ### 3.1 Identifying Missing Data
df.dtypes
# All of the columns is int64, which is good, since it tells us that they did not mix letters and numbers. In other words, there are no **NA** values. But **SEX**, **EDUCATION**, **MARRIAGE**,**PAY_** and **DEFAULT** should be **category** type.
# Make sure every category column contains the categpries which it should have and no more else.
df["SEX"].unique() # good
df["EDUCATION"].unique()
# EDUCATION only allowed numbers: 1,2,3,4. But it contains 0, 5 and 6. It is possible that 0 means missing data, and 5 and 6 represent categories not mentioned in the specification, but that is just a guess.
df["MARRIAGE"].unique() # 0 may represents missing data
# ### 3.2 Dealing with Missing Data
# Because scikit-learn's support vector machines do not support missing values, I need to figure out what to do with the 0s in the dataset. I can either delete these or impute them.
len(df.loc[(df["EDUCATION"] == 0) | (df["MARRIAGE"] == 0)])
# 68 rows have missing value.
len(df)
# So 68 of the 30000 rows, less than 1%, contains missing values. Since we have enough data for SVM when we drop these missing value rather than impute them. So I do this by selecting all of the rows that do not contain 0 in these two columns.
df_no_missing = df.loc[(df["EDUCATION"] != 0) & (df["MARRIAGE"] != 0)]
len(df_no_missing)
# Verify
df_no_missing["EDUCATION"].unique()
# Verify
df_no_missing["MARRIAGE"].unique()
# ## 4. Downsample the Data
# SVMs are great with small dataset, and this dataset will take a long time to optimize with Cross Validation. So I'll downsample both categories, customers who did and did not default, to 1000 each.
df_no_default = df_no_missing[df_no_missing["DEFAULT"] == 0]
df_default = df_no_missing[df_no_missing["DEFAULT"] == 1]
# Downsample the dataset that did not default
df_no_default_downsampled = resample(df_no_default, replace=False, n_samples=1000)
len(df_no_default_downsampled)
# Downsample the dataset that defaulted
df_default_downsampled = resample(df_default, replace=False, n_samples=1000)
len(df_default_downsampled)
# Merge them back into a single dataframe
df_downsample = pd.concat([df_no_default_downsampled, df_default_downsampled])
len(df_downsample)
# ## 5. Format the Data
# ### 5.1 Split the data into Dependent and Independent Variables
X = df_downsample.drop("DEFAULT", axis=1).copy()
# Or: X = df_no_missing.iloc[:,:-1].copy
X.head()
y = df_downsample["DEFAULT"].copy()
y.head()
# ### 5.2 One-Hot coding
X_encoded = pd.get_dummies(
X,
columns=[
"SEX",
"EDUCATION",
"MARRIAGE",
"PAY_0",
"PAY_2",
"PAY_3",
"PAY_4",
"PAY_5",
"PAY_6",
],
)
X_encoded
# ### 5.3 Centering and Scaling
# I split the data into training and testing datasets and then scale them separatly to avoid Data Leakage. Data Leakage occurs when informantion about the training dataset currupts or influences the testing dataset.
X_train, X_test, y_train, y_test = train_test_split(X_encoded, y)
X_train_scaled = scale(X_train)
X_test_scaled = scale(X_test)
# ## 6. Build a Preliminary Support Vector Machine
clf_svm = SVC()
clf_svm.fit(X_train_scaled, y_train)
plot_confusion_matrix(
clf_svm,
X_test_scaled,
y_test,
values_format="d",
display_labels=["Did not default", "Defaulted"],
)
# In this confusion matrix, we see that of the 250 people that did not default, 198(79.2%) were correctly classified. And of the 250 people that defaulted, 152(60.8%) were correctly calssified. So the SVM was not awesome. And I'll try to improve predictions using Cross validation to optimize the parameters.
# ## 7. Optimize Parameters with CV
param_grid = [
{
"C": [0.5, 1, 10, 100],
"gamma": ["scale", 1, 0.1, 0.01, 0.001, 0.0001],
"kernel": ["rbf"],
},
]
optimal_params = GridSearchCV(SVC(), param_grid, cv=5, scoring="accuracy", verbose=0)
optimal_params.fit(X_train_scaled, y_train)
print(optimal_params.best_params_)
# So the ideal value for C is 100, and the ideal value for gamma is 0.001.
# ## 8. Building, Evaluating, Drawing the Interpreting the Final Support Vector machine.
clf_svm = SVC(C=100, gamma=0.001)
clf_svm.fit(X_train_scaled, y_train)
plot_confusion_matrix(
clf_svm,
X_test_scaled,
y_test,
values_format="d",
display_labels=["Did not default", "Defaulted"],
)
# The results from the optimized SVM do not better than before. In other words, the SVM was pretty good straight out of the box without much optimization. This makes SVMs a great, quick and dirty method for relatively small datasets.
len(df_downsample.columns)
pca = PCA()
X_train_pca = pca.fit_transform(X_train_scaled)
per_var = np.round(pca.explained_variance_ratio_ * 100, decimals=1)
labels = [str(x) for x in range(1, len(per_var) + 1)]
plt.bar(x=range(1, len(per_var) + 1), height=per_var)
plt.tick_params(axis="x", which="both", bottom=False, top=False, labelbottom=False)
plt.ylabel("Percentage of Explained Variance")
plt.xlabel("Principal Components")
plt.title("Scree Plot")
plt.show()
# The Scree Plot shows that PC1 accounts for a relatively large amount of variation in the raw data, this means that it will be a good candidate for the x-axis in the 2-dimensional graph. However, PC2 is not much different with PC3 or PC4, which doesn't bode well for dimension reduction.
train_pc1_coords = X_train_pca[:, 0]
train_pc2_coords = X_train_pca[:, 1]
pca_train_scaled = scale(np.column_stack((train_pc1_coords, train_pc2_coords)))
param_grid = [
{
"C": [1, 10, 100, 1000],
"gamma": ["scale", 1, 0.1, 0.01, 0.001, 0.0001],
"kernel": ["rbf"],
},
]
optimal_params = GridSearchCV(SVC(), param_grid, cv=5, scoring="accuracy", verbose=0)
optimal_params.fit(pca_train_scaled, y_train)
print(optimal_params.best_params_)
clf_svm = SVC(C=10, gamma=0.1)
clf_svm.fit(pca_train_scaled, y_train)
X_test_pca = pca.transform(X_train_scaled)
test_pc1_coords = X_test_pca[:, 0]
test_pc2_coords = X_test_pca[:, 1]
x_min = test_pc1_coords.min() - 1
x_max = test_pc1_coords.max() + 1
y_min = test_pc2_coords.min() - 1
y_max = test_pc2_coords.max() + 1
xx, yy = np.meshgrid(
np.arange(start=x_min, stop=x_max, step=0.1),
np.arange(start=y_min, stop=y_max, step=0.1),
)
Z = clf_svm.predict(np.column_stack((xx.ravel(), yy.ravel())))
Z = Z.reshape(xx.shape)
fig, ax = plt.subplots(figsize=(10, 10))
ax.contourf(xx, yy, Z, alpha=0.1)
cmap = colors.ListedColormap(["#e41a1c", "#4daf4a"])
scatter = ax.scatter(
test_pc1_coords,
test_pc2_coords,
c=y_train,
cmap=cmap,
s=100,
edgecolors="k",
alpha=0.7,
)
legend = ax.legend(
scatter.legend_elements()[0], scatter.legend_elements()[1], loc="upper right"
)
legend.get_texts()[0].set_text("No Default")
legend.get_texts()[1].set_text("Yes Default")
ax.set_ylabel("PC2")
ax.set_xlabel("PC1")
ax.set_title("Decision surface using the PCA transformed features")
plt.show()
|
# # 📋 Variable description
# ---
# **Customer_id** = unique customer id
# **Age** = customer's age
# **Gender** = 0: Male, 1: Female
# **Revenue_Total** = total sales by customer
# **N_Purchases** = number of purchases to date
# **Purchase_DATE** = date latest purchase, dd.mm.yy
# **Purchase_VALUE** = latest purchase in €
# **Pay_Method** = 0: Digital Wallets, 1: Card, 2: PayPal, 3: Other
# **Time_Spent** = time spent (in sec) on website
# **Browser** = 0: Chrome, 1: Safari, 2: Edge, 3: Other
# **Newsletter** = 0: not subscribed, 1: subscribed
# **Voucher** = 0: not used, 1: used
# # 📚 Imports
# ---
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import warnings
warnings.filterwarnings("ignore")
# # 📖 Data
# ---
df = pd.read_csv(
"/kaggle/input/online-shop-customer-sales-data/Online Shop Customer Sales Data.csv"
)
df.head()
df.info()
df.describe().T
# Checking null values
df.isna().sum()
# Random sampling
proportion_sample = 0.1
# Calculate the sample size based on the proportion
sample_size = int(len(df) * proportion_sample)
# Perform random sampling based on the sample size
sample = df.sample(n=sample_size, random_state=0)
# Display the sample
print(sample)
# Drop Customer_id
df.drop("Customer_id", axis=1, inplace=True)
# Splitting the date into month and year
df["Month"] = df["Purchase_DATE"].str.split(".").str[1]
df["Year"] = df["Purchase_DATE"].str.split(".").str[2]
df["Year"] = df["Year"].astype(int)
df["Month"] = df["Month"].astype(int)
# Drop Purchase_DATE
df.drop("Purchase_DATE", axis=1, inplace=True)
df.head()
# # 🔎 Identifying and excluding outliers 🗑
# ---
# Create the boxplot
sns.boxplot(data=df[["Revenue_Total", "Purchase_VALUE"]], palette="cool")
# Set the title/labels
plt.title("Boxplot of Revenue Total and Purchase Value")
plt.xlabel("Features")
plt.ylabel("Values")
plt.show()
# I will use **z-score** to exclude outliers.
# The z-score is calculated by the difference between the specific value and the mean of the data set, divided by the standard deviation of the data set. The formula for calculating the z-score is as follows:
# z = (x - μ) / σ
# **where:**
# **x** is the specific value
# **μ** is the mean of the data set
# **σ** is the standard deviation of the data set
# Calculate the mean and std
mean = np.mean(df["Purchase_VALUE"])
std = np.std(df["Purchase_VALUE"])
# Calculate the Z-score
z_scores = (df["Purchase_VALUE"] - mean) / std
# Set a Z-score threshold
threshold = 3
# Identify the indices of the outliers based on the Z-score threshold
outlier_indices = np.where(np.abs(z_scores) > threshold)
# Drop the outliers
df.drop(df.index[outlier_indices], inplace=True)
# # 📊 Visualization
# ---
# # Distribution of variables
# ---
# Set the background color and figsize
plt.style.use("dark_background")
plt.figure(figsize=(15, 28))
# Adjust subplot vertical spacing
plt.subplots_adjust(hspace=0.2)
# 6x2 subplot figure
fig, axs = plt.subplots(6, 2, figsize=(15, 28))
i = 1
# Loop through dataframe columns
for feature in df.columns:
# Create a subplot
plt.subplot(6, 2, i)
# Create histogram
sns.histplot(data=df, x=feature, kde=True, palette="cool")
i += 1
# Show the plot
plt.show()
# ## ♂️ Gender Distribution ♀️
# ---
# Set the background color and figsize
plt.style.use("dark_background")
plt.figure(figsize=(10, 6))
# Gender value count
gender_counts = df["Gender"].value_counts()
# Create the pie chart
fig, ax = plt.subplots()
ax.pie(
gender_counts,
labels=gender_counts.index,
autopct="%1.1f%%",
startangle=90,
colors=["#007fff", "#FF40B4"],
)
# Add title and show
ax.set_title("Gender Distribution")
plt.show()
# ## 👶 Age by Gender 👴
# ---
# Set the background color and figsize
plt.style.use("dark_background")
plt.figure(figsize=(10, 6))
# Create histogram
sns.histplot(x="Age", hue="Gender", multiple="stack", bins=50, data=df, palette="cool")
# Customize the title and labels
plt.title("Distribution of Gender")
plt.xlabel("Age")
plt.ylabel("Count")
# Display the plot
plt.show()
# ## Revenue Total by Gender
# ---
# Set the background color and figsize
plt.style.use("dark_background")
plt.figure(figsize=(10, 6))
# Bar plot
sns.barplot(x="Gender", y="Revenue_Total", data=df, palette="cool")
# Labels/Title
plt.xlabel("Gender")
plt.ylabel("Average of Revenue Total")
plt.title("Revenue Total by Gender")
plt.show()
# ## 💵 Pay Method Distribution
# ---
# Set the background color and figsize
plt.style.use("dark_background")
plt.figure(figsize=(10, 6))
# Gender value count
pay_method_counts = df["Pay_Method"].value_counts()
# Set colors
colors = ["#07d5f5", "#04a4bd", "#d102d1", "#ff21ff"]
# Create the pie chart
fig, ax = plt.subplots()
ax.pie(
pay_method_counts,
labels=pay_method_counts.index,
autopct="%1.1f%%",
startangle=90,
colors=colors,
)
# Add title and show
ax.set_title("Pay Method Distribution")
plt.show()
# ## 💻 Browser Distribution
# ---
# Browser value count
browser_count = df["Browser"].value_counts()
# Set colors
colors = ["#07d5f5", "#04a4bd", "#d102d1", "#ff21ff"]
# Create the pie chart
fig, ax = plt.subplots()
ax.pie(
browser_count,
labels=browser_count.index,
autopct="%1.1f%%",
startangle=90,
colors=colors,
)
# Add title and show
ax.set_title("Browser Distribution")
plt.show()
# ## 📰 Newsletter Distribution
# ---
# Newsletter value count
newsletter_count = df["Newsletter"].value_counts()
# Set colors
colors = ["#07d5f5", "#d102d1"]
# Create the pie chart
fig, ax = plt.subplots()
ax.pie(
newsletter_count,
labels=newsletter_count.index,
autopct="%1.1f%%",
startangle=90,
colors=colors,
)
# Add title and show
ax.set_title("Newsletter Distribution")
plt.show()
# ## ✔️ Voucher Distribution
# ---
# Vouncher value count
voucher_count = df["Voucher"].value_counts()
# Set colors
colors = ["#07d5f5", "#d102d1"]
# Create the pie chart
fig, ax = plt.subplots()
ax.pie(
voucher_count,
labels=voucher_count.index,
autopct="%1.1f%%",
startangle=90,
colors=colors,
)
# Add title and show
ax.set_title("Voucher Distribution")
plt.show()
# ## Purchase Value by Payment Method
# ---
# Set the background color and figsize
plt.style.use("dark_background")
plt.figure(figsize=(10, 6))
# Barplot
sns.barplot(x="Pay_Method", y="Purchase_VALUE", data=df, palette="cool")
# Labels/Title
plt.xlabel("Payment Method")
plt.ylabel("Purchase Value")
plt.title("Purchase Value by Payment Method")
plt.show()
# ## Revenue Total by Number of Purchases
# ---
# Set the background color and figsize
plt.style.use("dark_background")
plt.figure(figsize=(10, 6))
# Bar plot
sns.barplot(x="N_Purchases", y="Revenue_Total", data=df, palette="cool")
# Labels/Title
plt.xlabel("N_Purchases")
plt.ylabel("Revenue Total")
plt.title("Revenue Total by Number of Purchases")
plt.show()
# ## Revenue Total by Browser
# ---
# Set the background color and figsize
plt.style.use("dark_background")
plt.figure(figsize=(10, 6))
# Bar plot
sns.barplot(x="Browser", y="Revenue_Total", data=df, palette="cool")
# Labels/Title
plt.xlabel("Browser")
plt.ylabel("Revenue Total")
plt.title("Revenue Total by Browser")
plt.show()
# ## Revenue Total by Newsletter
# ---
# Set the background color and figsize
plt.style.use("dark_background")
plt.figure(figsize=(10, 6))
# Bar plot
sns.barplot(x="Newsletter", y="Revenue_Total", data=df, palette="cool")
# Labels/Title
plt.xlabel("Newsletter")
plt.ylabel("Revenue Total")
plt.title("Revenue Total by Newsletter")
plt.show()
# ## Revenue Total by Voucher
# ---
# Set the background color and figsize
plt.style.use("dark_background")
plt.figure(figsize=(10, 6))
# Bar plot
sns.barplot(x="Voucher", y="Revenue_Total", data=df, palette="cool")
# Labels/Title
plt.xlabel("Voucher")
plt.ylabel("Revenue Total")
plt.title("Revenue Total by Voucher")
plt.show()
# ## Time Spent by Browser
# ---
# Set the background color and figsize
plt.style.use("dark_background")
plt.figure(figsize=(10, 6))
# Bar plot
sns.barplot(x="Browser", y="Time_Spent", data=df, palette="cool")
# Labels/Title
plt.xlabel("Browser")
plt.ylabel("Time Spent")
plt.title("Time Spent by Browser")
plt.show()
# ## Time Spent by Voucher
# ---
# Set the background color and figsize
plt.style.use("dark_background")
plt.figure(figsize=(10, 6))
# Bar plot
sns.barplot(x="Voucher", y="Time_Spent", data=df, palette="cool")
# Labels/Title
plt.xlabel("Voucher")
plt.ylabel("Time Spent Total")
plt.title("Time Spent by Voucher")
plt.show()
# ## Time Spent by Newsletter
# ---
# Set the background color and figsize
plt.style.use("dark_background")
plt.figure(figsize=(10, 6))
# Bar plot
sns.barplot(x="Newsletter", y="Time_Spent", data=df, palette="cool")
# Labels/Title
plt.xlabel("Newsletter")
plt.ylabel("Time Spent")
plt.title("Time Spent by Newsletter")
plt.show()
# ## Distribution of Purchase Value by Gender
# ---
# Set the background color and figsize
plt.style.use("dark_background")
plt.figure(figsize=(10, 6))
# Create the Violin plot
sns.violinplot(x="Gender", y="Purchase_VALUE", data=df, palette="cool")
# Set the axis and title
plt.title("Distribution of Purchase Value by Gender")
plt.xlabel("Gender")
plt.ylabel("Purchase Value")
plt.show()
# ## Distribution of Revenue Total by Gender
# ---
# Set the background color and figsize
plt.style.use("dark_background")
plt.figure(figsize=(10, 6))
# Create the Violin plot
sns.violinplot(x="Gender", y="Revenue_Total", data=df, palette="cool")
# Set the axis and title
plt.title("Distribution of Revenue Total by Gender")
plt.xlabel("Gender")
plt.ylabel("Revenue Total")
plt.show()
# ## Purchase Value by Gender
# ---
# Set the background color and figsize
plt.style.use("dark_background")
plt.figure(figsize=(10, 6))
# Bar plot
sns.barplot(x="Age", y="Purchase_VALUE", data=df, palette="cool")
# Labels/Title
plt.xlabel("Age")
plt.ylabel("Purchase Value")
plt.title("Purchase by Gender")
plt.show()
# ## Revenue Total by Gender
# ---
# Set the background color and figsize
plt.style.use("dark_background")
plt.figure(figsize=(10, 6))
# Bar plot
sns.barplot(x="Age", y="Revenue_Total", data=df, palette="cool")
# Labels/Title
plt.xlabel("Age")
plt.ylabel("Revenue Total")
plt.title("Revenue Total by Gender")
plt.show()
# ## Monthly Revenue Analysis
# ---
# Set the background color and figsize
plt.style.use("dark_background")
plt.figure(figsize=(10, 6))
# Grouping
grouped_df = df.groupby("Month").sum()[["Revenue_Total", "N_Purchases"]]
# Sorting the dataframe by 'Revenue_Total'
sorted_df = grouped_df.sort_values(by="Revenue_Total", ascending=False)
# Creating a bar chart
sns.barplot(x=sorted_df.index, y="Revenue_Total", data=sorted_df, palette="cool")
# Set labels/title
plt.xlabel("Month")
plt.ylabel("Number of purchases")
plt.title("Monthly Revenue Analysis")
plt.show()
# ## Revenue Total vs Time Spent
# ---
# Set the background color and figsize
plt.style.use("dark_background")
plt.figure(figsize=(10, 6))
# Set line color
color = "#FF40B4"
# Create the scatter plot
plt.scatter(df["Revenue_Total"], df["Time_Spent"], color=color, alpha=0.1)
# Set the axis labels
plt.xlabel("Revenue_Total")
plt.ylabel("Time_Spent")
# Set the title
plt.title("Revenue Total vs TimeSpent")
plt.show()
# ## Purshase Value vs Time Spent
# ---
# Set the background color and figsize
plt.style.use("dark_background")
plt.figure(figsize=(10, 6))
# Set line color
color = "#FF40B4"
# Create the scatter plot
plt.scatter(df["Purchase_VALUE"], df["Time_Spent"], color=color, alpha=0.1)
# Set the axis labels
plt.xlabel("Purshase_VALUE")
plt.ylabel("Time_Spent")
# Set the title
plt.title("Purshase Value vs Time Spent")
plt.show()
# ## Purchase Value by Month
# ---
# Set the background color and figsize
plt.style.use("dark_background")
plt.figure(figsize=(10, 6))
# Set line color
line_color = "#FF00B4"
# Group the data by 'Month' and the sum of 'Purchase_VALUE'
monthly_purchase_value = df.groupby("Month")["Purchase_VALUE"].sum()
# Create a line plot
plt.plot(monthly_purchase_value.index, monthly_purchase_value.values, color=line_color)
# Set the axis labels
plt.xlabel("Month")
plt.ylabel("Purchase_VALUE")
# Set the grid color
plt.grid(color="lightgray", alpha=0.3)
# Set the title
plt.title("Purchase Value by Month")
plt.show()
# ## Revenue Total by Month
# ---
# Set the background color and figsize
plt.style.use("dark_background")
plt.figure(figsize=(10, 6))
# Set line color
line_color = "#FF00B4"
# Group the data by 'Month' and the sum of 'Revenue_Total'
monthly_revenue_total = df.groupby("Month")["Revenue_Total"].sum()
# Create a line plot
plt.plot(monthly_revenue_total.index, monthly_revenue_total.values, color=line_color)
# Set the axis labels
plt.xlabel("Month")
plt.ylabel("Revenue_Total")
# Set the grid color
plt.grid(color="lightgray", alpha=0.3)
# Set the title
plt.title("Revenue Total by Month")
plt.show()
# ## Time Spent by Month
# ---
# Set the background color and figsize
plt.style.use("dark_background")
plt.figure(figsize=(10, 6))
# Set line color
line_color = "#FF00B4"
# Group the data by 'Month' and the sum of 'Purchase_VALUE'
monthly_purchase_value = df.groupby("Month")["Time_Spent"].sum()
# Create a line plot
plt.plot(monthly_purchase_value.index, monthly_purchase_value.values, color=line_color)
# Set the axis labels
plt.xlabel("Month")
plt.ylabel("Time_Spent")
# Set the grid color
plt.grid(color="lightgray", alpha=0.3)
# Set the title
plt.title("Time Spent by Month")
plt.show()
|
# **Load the important required libraries**
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import warnings
warnings.filterwarnings("ignore")
# **Let's Load the dataset**
fifa18 = pd.read_csv("../input/fifa-worldcup-2018/2018_worldcup_v3.csv")
# ### **Data Analysis On Fifa World Cup 2018 Data Set**
# **Checking first 5 and last 5 records from the datasets**
fifa18.head(5)
fifa18.tail(5)
# **Let's check the duplicate data in data set**
fifa18.duplicated().sum()
fifa18.shape
fifa18.info()
# **So, there 64 records in 14 columns. Also, there are no null records as well as duplicate values.**
# **Let's extract hour from datetime and add it to the new column.**
fifa18["Hour"] = fifa18.Datetime.apply(lambda x: x.split(" - ")[1])
fifa18.Datetime = fifa18.Datetime.apply(lambda x: x.split(" - ")[0])
fifa18.head()
# **Let's add total goals from home and away goals.**
fifa18["Total_Goals"] = fifa18["Home Team Goals"] + fifa18["Away Team Goals"]
fifa18.head()
fifa18.rename(
columns={
"Home Team Name": "Home_Team",
"Away Team Name": "Away_Team",
"Home Team Goals": "Home_Team_Goals",
"Away Team Goals": "Away_Team_Goals",
},
inplace=True,
)
fifa18.head()
# ### **Exploratory Data Analysis - EDA**
fifa18["City"].value_counts().sort_index()
plt.figure(figsize=(10, 5))
plt.title("Number of Matches held in each Russian City", fontsize=14)
plt.xlabel("City", fontsize=12)
plt.ylabel("Count", fontsize=12)
plt.xticks(rotation=90, fontsize=12)
plt.yticks(fontsize=12)
sns.countplot(x="City", data=fifa18, palette="rocket")
# **From above table and plot, we can observe that most number of the matches are held in Moscow.**
fifa18["Hour"].value_counts().sort_index()
plt.figure(figsize=(10, 5))
plt.title("Number of Matches held in each Hour", fontsize=14)
plt.xlabel("Hour", fontsize=12)
plt.ylabel("Count", fontsize=12)
plt.xticks(rotation=90, fontsize=12)
plt.yticks(fontsize=12)
sns.countplot(x="Hour", data=fifa18, palette="mako")
# **From above table and plot, we can observe that most number of the matches are held in during 21:00 Hour time.**
fifa18["Stadium"].value_counts().sort_index()
plt.figure(figsize=(10, 5))
plt.title("Number of Matches held in each Stadium", fontsize=14)
plt.xlabel("Stadiums", fontsize=12)
plt.ylabel("Count", fontsize=12)
plt.xticks(rotation=90, fontsize=12)
plt.yticks(fontsize=12)
sns.countplot(x="Stadium", data=fifa18, palette="Greens_d")
# **From above table and plot, we can observe that most number of the matches are held in Luzhniki Stadium and Saint Petersburg Stadium with 7 no of matches each.**
goals_by_day = fifa18.groupby("Datetime").sum().Total_Goals.to_frame().reset_index()
goals_by_day.columns = ["Datetime", "Total Goals By Day"]
goals_by_day = goals_by_day.sort_values("Datetime", ascending=False)
goals_by_day
plt.figure(figsize=(12, 8))
sns.barplot(
y=goals_by_day["Datetime"],
x=goals_by_day["Total Goals By Day"],
palette="twilight",
orient="h",
)
plt.title("No of goals scored each day", fontsize=15)
plt.xlabel("Goals")
plt.ylabel("Date")
plt.xticks(fontsize=12)
plt.yticks(fontsize=12)
# **From above table and plot, we can observe that most number of goals scored on 24th June 2018 where as least number of goals scored on 10th July 2018.**
# **Also, there is difference in group stages, QF, SF stage. So we will also have to look on no of matches held on each day.**
fifa18["Datetime"].value_counts().sort_index()
plt.figure(figsize=(10, 5))
plt.title("Number of Matches held in each Day", fontsize=14)
plt.xlabel("Days", fontsize=12)
plt.ylabel("Count", fontsize=12)
plt.xticks(rotation=90, fontsize=12)
plt.yticks(fontsize=12)
sns.countplot(x="Datetime", data=fifa18, palette="cool_d")
# **From above table and plot, we can observe that there were 4 days when 4 matches were held in one single day**
# **Now, let'swork on total team goals, away and home.**
goals_by_home = (
fifa18.groupby("Home_Team")
.sum()[["Home_Team_Goals", "Away_Team_Goals"]]
.reset_index()
)
goals_by_away = (
fifa18.groupby("Away_Team")
.sum()[["Home_Team_Goals", "Away_Team_Goals"]]
.reset_index()
)
goals_total = pd.concat([goals_by_home, goals_by_away], axis=1)
goals_total.columns = [
"Home_Team",
"Home_Scored",
"Home_Conceded",
"Away_Team",
"Away_Conceded",
"Away_Scored",
]
goals_total["Scored"] = goals_total.Home_Scored + goals_total.Away_Scored
goals_total["Conceded"] = goals_total.Home_Conceded + goals_total.Away_Conceded
goals_total = goals_total.drop(
["Home_Scored", "Home_Conceded", "Away_Team", "Away_Scored", "Away_Conceded"],
axis=1,
)
goals_total
goals_total["Goal_Diff"] = goals_total.Scored - goals_total.Conceded
goals_total = goals_total.rename(columns={"Home_Team": "Team_Name"})
goals_total
goals_total = goals_total.sort_values("Scored", ascending=False)
plt.figure(figsize=(12, 8))
sns.barplot(x=goals_total["Team_Name"], y=goals_total["Scored"], palette="coolwarm")
plt.title("No of goals scored by each teams", fontsize=15)
plt.xlabel("Teams")
plt.ylabel("No of Goals")
plt.xticks(rotation=90, fontsize=12)
plt.yticks(fontsize=12)
goals_total = goals_total.sort_values("Conceded", ascending=False)
plt.figure(figsize=(12, 8))
sns.barplot(x=goals_total["Team_Name"], y=goals_total["Conceded"], palette="coolwarm")
plt.title("No of goals conceded by each teams", fontsize=15)
plt.xlabel("Teams")
plt.ylabel("No of Goals")
plt.xticks(rotation=90, fontsize=12)
plt.yticks(fontsize=12)
plt.figure(figsize=(12, 8))
sns.barplot(x=fifa18["City"], y=fifa18["Home_Team_Goals"], palette="inferno")
plt.title("No of goals scored by Home Team in each city", fontsize=15)
plt.xlabel("City")
plt.ylabel("No of Goals")
plt.xticks(rotation=90, fontsize=12)
plt.yticks(fontsize=12)
plt.figure(figsize=(12, 8))
sns.barplot(x=fifa18["City"], y=fifa18["Away_Team_Goals"], palette="inferno")
plt.title("No of goals scored by aw away team in each city", fontsize=15)
plt.xlabel("City")
plt.ylabel("No of Goals")
plt.xticks(rotation=90, fontsize=12)
plt.yticks(fontsize=12)
plt.figure(figsize=(12, 8))
sns.barplot(x=fifa18["Stadium"], y=fifa18["Home_Team_Goals"], palette="flare")
plt.title("No of goals scored by Home Team in each stadium", fontsize=15)
plt.xlabel("City")
plt.ylabel("No of Goals")
plt.xticks(rotation=90, fontsize=12)
plt.yticks(fontsize=12)
plt.figure(figsize=(12, 8))
sns.barplot(x=fifa18["Stadium"], y=fifa18["Away_Team_Goals"], palette="flare")
plt.title("No of goals scored by Away Team in each stadium", fontsize=15)
plt.xlabel("City")
plt.ylabel("No of Goals")
plt.xticks(rotation=90, fontsize=12)
plt.yticks(fontsize=12)
|
import tensorflow as tf
from tensorflow.keras import models, layers
import matplotlib.pyplot as plt
import numpy as np
import pathlib
import os
os.environ["TF_CPP_MIN_LOG_LEVEL"] = "2" # to disable all debugging logs
# Global initialization of some imp variables
IMAGE_SIZE = 256
BATCH_SIZE = 32
CHANNELS = 3
EPOCHS = 50
train = tf.keras.preprocessing.image_dataset_from_directory(
"/kaggle/input/tomato/Tomato_images/train",
shuffle=True,
image_size=(IMAGE_SIZE, IMAGE_SIZE),
batch_size=BATCH_SIZE,
)
val = tf.keras.preprocessing.image_dataset_from_directory(
"/kaggle/input/tomato/Tomato_images/val",
shuffle=True,
image_size=(IMAGE_SIZE, IMAGE_SIZE),
batch_size=BATCH_SIZE,
)
test = tf.keras.preprocessing.image_dataset_from_directory(
"/kaggle/input/tomato/Tomato_images/test",
shuffle=True,
image_size=(IMAGE_SIZE, IMAGE_SIZE),
batch_size=BATCH_SIZE,
)
class_names = train.class_names
class_names
# Number of Batches = (total number of files belonging to all classes / Batch_Size)
len(train)
# prints Elements in dataset: here 1st element is image and 2nd index of that image.
print(train)
for image_batch, labels_batch in train.take(1):
print(image_batch.shape)
print(labels_batch.numpy())
plt.figure(figsize=(20, 10))
for image_batch, labels_batch in train.take(1):
for i in range(15):
ax = plt.subplot(3, 5, i + 1)
plt.imshow(image_batch[i].numpy().astype("uint8"))
plt.title(class_names[labels_batch[i]])
plt.axis("off")
train.take(41)
train = train.cache().shuffle(1000).prefetch(buffer_size=tf.data.AUTOTUNE)
val = val.cache().shuffle(1000).prefetch(buffer_size=tf.data.AUTOTUNE)
test = test.cache().shuffle(1000).prefetch(buffer_size=tf.data.AUTOTUNE)
resize_and_rescale = tf.keras.Sequential(
[
layers.experimental.preprocessing.Resizing(IMAGE_SIZE, IMAGE_SIZE),
layers.experimental.preprocessing.Rescaling(1.0 / 255),
]
)
data_augmentation = tf.keras.Sequential(
[
layers.experimental.preprocessing.RandomFlip("horizontal_and_vertical"),
layers.experimental.preprocessing.RandomRotation(0.2),
]
)
input_shape = (BATCH_SIZE, IMAGE_SIZE, IMAGE_SIZE, CHANNELS)
n_classes = 3
model = models.Sequential(
[
resize_and_rescale,
data_augmentation,
layers.Conv2D(
32, kernel_size=(3, 3), activation="relu", input_shape=input_shape
),
layers.MaxPooling2D((2, 2)),
layers.Conv2D(64, kernel_size=(3, 3), activation="relu"),
layers.MaxPooling2D((2, 2)),
layers.Conv2D(64, kernel_size=(3, 3), activation="relu"),
layers.MaxPooling2D((2, 2)),
layers.Conv2D(64, (3, 3), activation="relu"),
layers.MaxPooling2D((2, 2)),
layers.Conv2D(64, (3, 3), activation="relu"),
layers.MaxPooling2D((2, 2)),
layers.Conv2D(32, (3, 3), activation="relu"),
layers.MaxPooling2D((2, 2)),
layers.Flatten(),
layers.Dense(32, activation="relu"),
layers.Dense(n_classes, activation="softmax"),
]
)
model.build(input_shape=input_shape)
model.summary()
model.compile(
optimizer="adam",
loss=tf.keras.losses.SparseCategoricalCrossentropy(from_logits=False),
metrics=["accuracy"],
)
history = model.fit(
train, batch_size=BATCH_SIZE, validation_data=val, verbose=1, epochs=50
)
scores = model.evaluate(test)
scores
# # VGG16
from tensorflow.keras.applications.vgg16 import VGG16
input_shape = (IMAGE_SIZE, IMAGE_SIZE, CHANNELS)
vgg16 = VGG16(input_shape=input_shape, weights="imagenet", include_top=False)
VGG_16 = models.Sequential(
[
resize_and_rescale,
data_augmentation,
vgg16,
layers.Flatten(),
# layers.Dense(64, activation='relu'),
layers.Dense(n_classes, activation="softmax"),
]
)
VGG_16.compile(
optimizer="adam",
loss=tf.keras.losses.SparseCategoricalCrossentropy(from_logits=False),
metrics=["accuracy"],
)
history = VGG_16.fit(
train, batch_size=BATCH_SIZE, validation_data=val, verbose=1, epochs=10
)
scores = VGG_16.evaluate(test)
scores
# # VGG19
from tensorflow.keras.applications.vgg19 import VGG19
input_shape = (IMAGE_SIZE, IMAGE_SIZE, CHANNELS)
vgg19 = VGG19(input_shape=input_shape, weights="imagenet", include_top=False)
VGG_19 = models.Sequential(
[
resize_and_rescale,
data_augmentation,
vgg19,
layers.Flatten(),
# layers.Dense(64, activation='relu'),
layers.Dense(n_classes, activation="softmax"),
]
)
VGG_19.compile(
optimizer="adam",
loss=tf.keras.losses.SparseCategoricalCrossentropy(from_logits=False),
metrics=["accuracy"],
)
history = VGG_19.fit(
train, batch_size=BATCH_SIZE, validation_data=val, verbose=1, epochs=10
)
scores = VGG_19.evaluate(test)
scores
# # VGG21
VGG_21 = models.Sequential(
[
resize_and_rescale,
data_augmentation,
vgg19,
layers.Conv2D(512, kernel_size=(3, 3), activation="relu"),
layers.MaxPooling2D(pool_size=(2, 2), strides=(2, 2)),
layers.Flatten(),
layers.Dense(64, activation="relu"),
layers.Dense(n_classes, activation="softmax"),
]
)
VGG_21.compile(
optimizer="adam",
loss=tf.keras.losses.SparseCategoricalCrossentropy(from_logits=False),
metrics=["accuracy"],
)
history = VGG_21.fit(
train, batch_size=BATCH_SIZE, validation_data=val, verbose=1, epochs=10
)
VGG_21.summary()
scores = VGG_21.evaluate(test)
scores
train_loss = history.history["loss"]
train_acc = history.history["accuracy"]
val_loss = history.history["val_loss"]
val_acc = history.history["val_accuracy"]
# graphs for accuracy and loss of training and validation data
plt.figure(figsize=(15, 15))
plt.subplot(2, 3, 1)
plt.plot(range(10), train_acc, label="Training Accuracy")
plt.plot(range(10), val_acc, label="Validation Accuracy")
plt.legend(loc="lower right")
plt.title("Training and Validation Accuracy")
plt.subplot(2, 3, 2)
plt.plot(range(10), train_loss, label="Training Loss")
plt.plot(range(10), val_loss, label="Validation Loss")
plt.legend(loc="upper right")
plt.title("Training and Validation Loss")
# # VGG14
input_shape = (BATCH_SIZE, IMAGE_SIZE, IMAGE_SIZE, CHANNELS)
n_classes = 3
VGG_14 = models.Sequential(
[
resize_and_rescale,
data_augmentation,
# Block1
layers.Conv2D(
64,
kernel_size=(3, 3),
activation="relu",
padding="same",
name="block1_conv1",
),
layers.Conv2D(
64,
kernel_size=(3, 3),
activation="relu",
padding="same",
name="block1_conv2",
),
layers.MaxPooling2D(pool_size=(2, 2), strides=(2, 2), name="block1_pool"),
# Block2
layers.Conv2D(
128,
kernel_size=(3, 3),
activation="relu",
padding="same",
name="block2_conv1",
),
layers.Conv2D(
128,
kernel_size=(3, 3),
activation="relu",
padding="same",
name="block2_conv2",
),
layers.MaxPooling2D(pool_size=(2, 2), strides=(2, 2), name="block2_pool"),
# Block3
layers.Conv2D(
256,
kernel_size=(3, 3),
activation="relu",
padding="same",
name="block3_conv1",
),
layers.Conv2D(
256,
kernel_size=(3, 3),
activation="relu",
padding="same",
name="block3_conv2",
),
layers.Conv2D(
256,
kernel_size=(3, 3),
activation="relu",
padding="same",
name="block3_conv3",
),
layers.MaxPooling2D(pool_size=(2, 2), strides=(2, 2), name="block3_pool"),
# Block4
layers.Conv2D(
512,
kernel_size=(3, 3),
activation="relu",
padding="same",
name="block4_conv1",
),
layers.Conv2D(
512,
kernel_size=(3, 3),
activation="relu",
padding="same",
name="block4_conv2",
),
layers.Conv2D(
512,
kernel_size=(3, 3),
activation="relu",
padding="same",
name="block4_conv3",
),
layers.MaxPooling2D(pool_size=(2, 2), strides=(2, 2), name="block4_pool"),
layers.Flatten(),
layers.Dense(64, activation="relu"),
layers.Dense(n_classes, activation="softmax"),
]
)
VGG_14.build(input_shape=input_shape)
VGG_14.summary()
VGG_14.compile(
optimizer="adam",
loss=tf.keras.losses.SparseCategoricalCrossentropy(from_logits=False),
metrics=["accuracy"],
)
history = VGG_14.fit(
train, batch_size=BATCH_SIZE, validation_data=val, verbose=1, epochs=10
)
scores = VGG_14.evaluate(test)
scores
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
data = pd.read_csv("../input/resume-dataset/UpdatedResumeDataSet.csv")
data.head()
# To identify the number of unique categories
print(data["Category"].unique())
print("")
print("Number of datapoints in each categories")
print(data["Category"].value_counts())
# Visualization of various categories
data["Category"].value_counts(sort=True).nlargest(25).plot.bar()
# # remove empty documents from the dataframe
# empty_doc = []
# for i in range(len(data)):
# if(len(data['Resume'][i]) <=10):
# empty_doc.append(i)
# print(empty_doc)
# data = data.drop(empty_doc)
# We now need to clean the text in the "Resume" column. So some standard data preprocessing steps are followed as below.
import re
import string
import nltk
nltk.download("stopwords")
stopwords = nltk.corpus.stopwords.words("english")
def clean_text(text):
# convert text to lowercase
text = text.lower()
# remove any numeric characters
text = "".join([word for word in text if not word.isdigit()])
# text = [word for word in text if re.search("\d", word)== None]
# remove URLs
text = re.sub("http\S+\s*", " ", text)
# remove RT and cc
text = re.sub("RT|cc", " ", text)
# remove hashtags
text = re.sub("#\S+", "", text)
# remove mentions
text = re.sub("@\S+", " ", text)
# punctuations removal
text = "".join([word for word in text if word not in string.punctuation])
text = re.sub("\W", " ", str(text))
# stopwords removal
ext = [word for word in text.split() if word not in stopwords]
# replace consecutive non-ASCII characters with a space
text = re.sub(r"[^\x00-\x7f]", r" ", text)
# extra whitespace removal
text = re.sub("\s+", " ", text)
return text
data["cleaned_text"] = data["Resume"].apply(lambda x: clean_text(x))
data
data["cleaned_text"][10]
sent_lens = []
for i in data.cleaned_text:
length = len(i.split())
sent_lens.append(length)
print(len(sent_lens))
print(max(sent_lens))
data["Resume"][100]
data["cleaned_text"][100]
from sklearn.preprocessing import LabelEncoder
labelencoder = LabelEncoder()
data["Category_N"] = labelencoder.fit_transform(data["Category"])
# print(type(labels))
data.tail()
from sklearn.feature_extraction.text import TfidfVectorizer
Text = data["cleaned_text"].values
op_labels = data["Category_N"].values
word_vectorizer = TfidfVectorizer(max_features=1500)
word_vectorizer.fit(Text)
features = word_vectorizer.transform(Text)
word_vectorizer.get_feature_names()
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
features, op_labels, random_state=0, test_size=0.2
)
from sklearn.multiclass import OneVsRestClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn import metrics
clf = OneVsRestClassifier(KNeighborsClassifier())
clf.fit(X_train, y_train)
prediction = clf.predict(X_test)
print(
"Accuracy of KNeighbors Classifier on training set: {:.2f}".format(
clf.score(X_train, y_train)
)
)
print(
"Accuracy of KNeighbors Classifier on test set: {:.2f}".format(
clf.score(X_test, y_test)
)
)
print(
"\n Classification report for classifier %s:\n%s\n"
% (clf, metrics.classification_report(y_test, prediction))
)
|
# 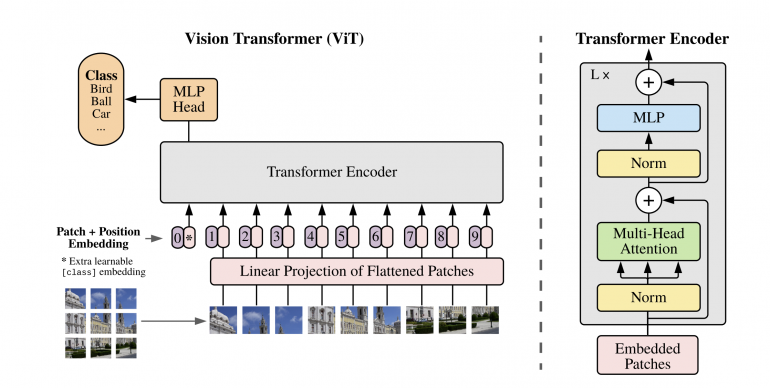
# We split an image into fixed-size patches, linearly embed each of them, add position embeddings, and feed the resulting sequence of vectors to a standard Transformer encoder. In order to perform classification, we use the standard approach of adding an extra learnable “classification token” to the sequence.
# A major challenge of applying Transformers without CNN to images is applying Self-Attention between pixels. ViT has overcome this problem by segmenting images into small patches (like 16x16 as implemented in this notebook).
# Vision Transformer (ViT) attains excellent results compared to state-of-the-art convolutional networks while requiring substantially fewer computational resources to train.
# # Install and Import Libraries
# Python library used for working with arrays.
import numpy as np
# Python library to interact with the file system.
import os
# Software library written for data manipulation and analysis.
import pandas as pd
# fastai library for computer vision tasks
from fastai.vision.all import *
from fastai.metrics import *
# Developing and training neural network based deep learning models.
import torch
# Vision Transformer
from vit_pytorch.efficient import ViT
# Nystromformer
from nystrom_attention import Nystromformer
dataset_path = Path("../input/ranzcr-clip-catheter-line-classification")
os.listdir(dataset_path)
train_df = pd.read_csv(dataset_path / "train.csv")
train_df.head()
train_df["path"] = train_df["StudyInstanceUID"].map(
lambda x: str(dataset_path / "train" / x) + ".jpg"
)
train_df = train_df.drop(columns=["StudyInstanceUID"])
train_df.head(10)
# # Dataset
# Transforms we need to do for each image in the dataset (ex: resizing).
item_tfms = RandomResizedCrop(224, min_scale=0.75, ratio=(1.0, 1.0))
# Transforms that can take place on a batch of images (ex: many augmentations).
batch_tfms = [
*aug_transforms(size=224, max_warp=0),
Normalize.from_stats(*imagenet_stats),
]
label_names = list(train_df.columns[:11])
data = DataBlock(
blocks=(
ImageBlock,
MultiCategoryBlock(encoded=True, vocab=label_names),
), # multi-label target
splitter=RandomSplitter(
seed=42
), # split data into training and validation subsets.
get_x=ColReader(12), # obtain the input images.
get_y=ColReader(list(range(11))), # obtain the targets.
item_tfms=item_tfms,
batch_tfms=batch_tfms,
)
dls = data.dataloaders(train_df, bs=16)
# We can call show_batch() to see what a sample of a batch looks like.
dls.show_batch()
# # Model Definition
# ## *Nystromformer*
# 
# * The proposed architecture of efficient self-attention via Nystrom approximation.
# * Each box represents an input, output, or intermediate matrix.
# * The variable name and the size of the matrix are inside box. × denotes matrix multiplication, and + denotes matrix addition.
# * The orange colored boxes are those matrices used in the Nystrom approximation. The green boxes are the skip connection added in parrallel to the approximation.
# * The dashed bounding box illustrates the three matrices of Nystroom approximate softmax matrix in self-attention.
efficient_transformer = Nystromformer(
# Last dimension of output tensor after linear transformation nn.Linear(..., dim).
dim=128,
# Number of Transformer blocks.
depth=6,
# Number of heads in Multi-head Attention layer.
heads=8,
# # number of landmarks
num_landmarks=256,
)
model = ViT(
# Last dimension of output tensor after linear transformation nn.Linear(..., dim).
dim=128,
# #If you have rectangular images, make sure your image size is the maximum of the width and height
image_size=224,
# n = (image_size // patch_size) ** 2 and n must be greater than 16.
patch_size=16,
# Number of classes to classify.
num_classes=11,
# plugin your own sparse attention transformer (Linformer/Reformer/Nystromformer)
transformer=efficient_transformer,
)
# Group together some dls, a model, and metrics to handle training
learn = Learner(
dls, model, metrics=[accuracy_multi]
) # Compute accuracy when input and target are the same size.
# Choosing a good learning rate
learn.lr_find()
# We can use the fine_tune function to train a model with this given learning rate
learn.fine_tune(1, base_lr=0.0003311311302240938)
# # Inference
sample_df = pd.read_csv(dataset_path / "sample_submission.csv")
sample_df.head()
_sample_df = sample_df.copy()
_sample_df["PatientID"] = "None"
_sample_df["path"] = _sample_df["StudyInstanceUID"].map(
lambda x: str(dataset_path / "test" / x) + ".jpg"
)
_sample_df = _sample_df.drop(columns=["StudyInstanceUID"])
test_dl = dls.test_dl(_sample_df)
test_dl.show_batch()
# ## *Test Time Augmentation (TTA)*
# 
# Similar to what Data Augmentation is doing to the training set, the purpose of Test Time Augmentation is to perform random modifications to the test images. Thus, instead of showing the regular, “clean” images, only once to the trained model, we will show it the augmented images several times. We will then average the predictions of each corresponding image and take that as our final guess.
# The reason why it works is that, by averaging our predictions, on randomly modified images, we are also averaging the errors. The error can be big in a single vector, leading to a wrong answer, but when averaged, only the correct answer stand out.
# Return predictions on the ds_idx dataset or dl using Test Time Augmentation
preds, _ = learn.tta(dl=test_dl, n=5)
# # Submission
submission_df = sample_df
for i in range(len(submission_df)):
for j in range(len(label_names)):
submission_df.iloc[i, j + 1] = preds[i][j].numpy().astype(np.float32)
submission_df.head(10)
submission_df.to_csv(f"submission.csv", index=False)
|
# Project: Photo to Monet
# Understanding of CycleGAN
# <img src = "https://img.wikioo.org/ADC/Art-ImgScreen-1.nsf/O/A-9GEHSH/$FILE/Claude-monet-le-bassin-aux-nympheas-reflets-verts.Jpg"
# width = "1000" height = "400"/>
# OVERVIEW
# A GAN consists of at least two neural networks: a generator model and a discriminator model. The generator is a neural network that creates the images. For our competition, you should generate images in the style of Monet. This generator is trained using a discriminator.
# The two models will work against each other, with the generator trying to trick the discriminator, and the discriminator trying to accurately classify the real vs. generated images.
# Your task is to build a GAN that generates 7,000 to 10,000 Monet-style images.
# Table of Contents
# Basic idea of CycleGAN1. CycleGAN compnonets2. Basic schema of CycleGAN training3. CycleGAN Generators Losses4. CycleGAN Discrminators training5. CycleGAN Discrminators LossesData descriptionImport modulesConstants and VariablesConstants and VariablesAuxilary functions and classesVisualLoading dataNeural NetoworkLoad DataCreate datasetCreate DataloaderCheck input imagesBuild CycleGAN ModelGeneratorConvolutional LayerTranspose Convolutional LayerResidual blockGeneratorDiscriminatorCycleGANTrainLearning rateInitialize modelTrain modelTrain resultsPlot lossesModel predictionsSave resultsSave modelSave predictions
# ##
# Basic idea of CycleGAN
# - With CycleGAN we want to get such Generator that will be able to generate Monet-style image from Photo input
# - In CycleGAN we dont have ground thruth output for generators.
# - Instead of that, couple of Generators and Dsicrimiantors will compete with each other in order to produce the most accurate Generator
# #### 1. CycleGAN compnonets
# - In CycleGAN we have:
# 1. Generator Photo - generates photo-like images from input
# 2. Generator Monet - generates Monet-like images from input
#
# 3. Discriminator photo - it should predict if input photo image was generated - then output '0'
# or it is real - in that case output '1'
#
# 4. Discriminator Monet - it should predict if input Monet image was generated - then output '0'
# or it is real - in that case output '1'
#
# #### 2. Basic schema of CycleGAN training
# - There are two branches in trianing - for photo and Monet image, which basically mirror each other
# - Training iteraion of photo branch:
# 1. Starting with 'real photo' with 'Generator Monet' we get generated Monet image
#
# 2. Given generated Monet image with 'Generator Photo' we can once again produce photo image and called it 'cycle photo'
#
# 3. Ideally 'real photo' and 'cycle photo' should be the same and that brings as to the first loss function which estimates the difference between them - 'Cycle Loss'
#
# 4. Next thing we need to consider is that if 'Generator Photo' will get photo image as an input it should generate the same photo. Such examine is called 'Identity check' and estimating the difference between those input and output from generator we can evaluate 'Identity Loss'
#
# 5. Last but not least, Generator should produce such a good image that it would fool Discriminator into consider generated image as real. So if we take 'generated Monet' after 'Generator Monet' as an input to 'Monet Discriminator' we want to get the output value as close to '1' as possible, which will mean that Discriminator is fooled. Estimating the difference between output value from Discriminator and '1' we will calculate 'Adversarial Loss'
#
# - The same steps with mirrored Generators and Discriminators are made for Monet image as an input
# 
# #### 3. CycleGAN Generators Losses
# - Total generator loss is a sum of 'Identity', 'Adversarial' and 'Cycle'
# - It is also reasonable to add weights to each of them
# 
# #### 4. CycleGAN Discrminators training
# - In order for Dicriminator not to be easily fooled by Generator, it should also be trained
# - The main purpose of discriminator is to correctly identify if input image was generated or it is real. With taht in mind training process is cinstructed pretty straignt forward:
# 1. Take a generated or real image as in input to discriminator
#
# 2. The produced output should be '0' in case of generated input and '1' in the other case
#
# 3. Estimate the difference between output and target value ('1' or '0') to alculate 'Disc Loss'
# - Monet Discriminator is trained in the same manner as Photo Discriminator
# 
# #### 5. CycleGAN Discrminators Losses
# - Total Discriminator loss is calculated as arithmetic mean between Loss with generated image in the input and real image in the input
# 
# ##
# Data description
# The dataset contains four directories: monet_tfrec, photo_tfrec, monet_jpg, and photo_jpg. The monet_tfrec and monet_jpg directories contain the same painting images, and the photo_tfrec and photo_jpg directories contain the same photos.
# We recommend using TFRecords as a Getting Started competition is a great way to become more familiar with a new data format, but JPEG images have also been provided.
# The monet directories contain Monet paintings. Use these images to train your model.
# The photo directories contain photos. Add Monet-style to these images and submit your generated jpeg images as a zip file. Other photos outside of this dataset can be transformed but keep your submission file limited to 10,000 images.
# Note: Monet-style art can be created from scratch using other GAN architectures like DCGAN. The submitted image files do not necessarily have to be transformed photos.
# - Files:
# - monet_jpg - 300 Monet paintings sized 256x256 in JPEG format
# - monet_tfrec - 300 Monet paintings sized 256x256 in TFRecord format
# - photo_jpg - 7028 photos sized 256x256 in JPEG format
# - photo_tfrec - 7028 photos sized 256x256 in TFRecord format
# ##
# Import modules
import os
import shutil
import time
import pandas as pd
import numpy as np
import itertools
import matplotlib.pyplot as plt
from PIL import Image
import torch
import torch.nn as nn
import torch.nn.functional as F
import torch.nn.init as init
from torch.nn import Conv2d, LeakyReLU, InstanceNorm2d
from torch.utils.data import Dataset, DataLoader
from torchvision import transforms as T
import warnings
warnings.filterwarnings("ignore")
# ##
# Constants and Variables
MONET_DIR = "../input/gan-getting-started/monet_jpg/"
PHOTO_DIR = "../input/gan-getting-started/photo_jpg/"
SAVE_PATH = "../images/"
RADNOM_STATE = 12345
IMAGE_SIZE = [256, 256]
BATCH_SIZE = 1
device = "cuda" if torch.cuda.is_available() else "cpu"
device
# ##
# Auxilary functions and classes
# ### Visual
# - Unnormalize image
def unnormalize(img, mean=[0.5, 0.5, 0.5], std=[0.5, 0.5, 0.5]):
# To normalize: X_norm = (X - mean) / std
# To unnormalize: X = (X_norm * std) + mean
for t, m, s in zip(img, mean, std):
t.mul_(s).add_(s)
return img
# - Show grid of images from dataloader
def show_images(data):
fig, ax = plt.subplots(2, 4, figsize=(10, 6))
it = iter(data)
for i in range(4):
p, m = next(it)
ax[0][i].imshow(unnormalize(p[0]).permute(1, 2, 0))
ax[1][i].imshow(unnormalize(m[0]).permute(1, 2, 0))
ax[0][i].set_title("Photo")
ax[1][i].set_title("Monet")
plt.show()
# - Visualize training losses per epoch
def plot_train_loss(log):
df = pd.read_csv(log)
fig, ax = plt.subplots(nrows=2, ncols=1, figsize=(7, 6))
df.plot(
x="epoch",
y="loss_gen_total_avg",
kind="line",
ax=ax[0],
color="b",
label="Total",
)
df.plot(
x="epoch",
y="loss_gen_photo_avg",
kind="line",
ax=ax[0],
color="r",
label="Photo Generator",
)
df.plot(
x="epoch",
y="loss_gen_monet_avg",
kind="line",
ax=ax[0],
color="g",
label="Monet Generator",
)
df.plot(
x="epoch",
y="loss_disc_total_avg",
kind="line",
ax=ax[1],
color="b",
label="Total",
)
df.plot(
x="epoch",
y="loss_disc_photo_avg",
kind="line",
ax=ax[1],
color="r",
label="Photo Discriminator",
)
df.plot(
x="epoch",
y="loss_disc_monet_avg",
kind="line",
ax=ax[1],
color="g",
label="Monet Discriminator",
)
ax[0].set_title("Generator Losses")
ax[1].set_title("Discriminator Losses")
ax[0].set_ylabel("Loss")
ax[1].set_ylabel("Loss")
plt.tight_layout()
plt.show()
# ### Loading data
# - Custom dataset class for training
class ImageDatasetCV(Dataset):
def __init__(self, monet_dir, photo_dir, size=(256, 256), normalize=True):
super().__init__()
self.monet_dir = None
if monet_dir:
self.monet_dir = monet_dir
self.monet_idx = dict()
for i, filename in enumerate(os.listdir(self.monet_dir)):
self.monet_idx[i] = filename
self.photo_dir = photo_dir
self.photo_idx = dict()
for i, filename in enumerate(os.listdir(self.photo_dir)):
self.photo_idx[i] = filename
if normalize:
self.transforms = T.Compose(
[
T.Resize(size),
T.ToTensor(),
T.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5)),
]
)
else:
self.transforms = T.Compose([T.Resize(size), T.ToTensor()])
def __getitem__(self, idx):
random_idx = idx
if self.monet_dir:
random_idx = int(np.random.uniform(0, len(self.monet_idx.keys())))
monet_path = os.path.join(self.monet_dir, self.monet_idx[random_idx])
monet_img = self.transforms(Image.open(monet_path))
photo_path = os.path.join(self.photo_dir, self.photo_idx[random_idx])
photo_img = self.transforms(Image.open(photo_path))
if self.monet_dir:
return photo_img, monet_img
else:
return photo_img
def __len__(self):
if self.monet_dir:
return min(len(self.monet_idx.keys()), len(self.photo_idx.keys()))
else:
return len(self.photo_idx.keys())
# ### Neural Netowork
# - Save model state
def save_checkpoint(state, save_path):
torch.save(state, save_path)
# - Load model state
def load_checkpoint(ckpt_path, map_location=None):
ckpt = torch.load(ckpt_path, map_location=map_location)
print(" [*] Loading checkpoint from %s succeed!" % ckpt_path)
return ckpt
# ##
# Load Data
# ### Create dataset
img_dataset = ImageDatasetCV(monet_dir=MONET_DIR, photo_dir=PHOTO_DIR)
# ### Create Dataloader
img_dataloader = DataLoader(img_dataset, batch_size=BATCH_SIZE, pin_memory=True)
# Enable pin_memory to automatically put the fetched data Tensors
# in pinned memory, and thus enables faster data transfer
# to CUDA-enabled GPUs
# ### Check input images
show_images(img_dataloader)
# ##
# Build CycleGAN Model
# ### Generator
# #### Convolutional Layer
def Convlayer(
in_ch,
out_ch,
kernel_size=3,
stride=2,
use_leaky=True,
use_inst_norm=True,
use_pad=True,
):
# Convolution
if use_pad:
conv = nn.Conv2d(in_ch, out_ch, kernel_size, stride, 1, bias=True)
else:
conv = nn.Conv2d(in_ch, out_ch, kernel_size, stride, 0, bias=True)
# Activation Function
if use_leaky:
actv = nn.LeakyReLU(negative_slope=0.2, inplace=True)
else:
actv = nn.GELU()
# Normalization
if use_inst_norm:
norm = nn.InstanceNorm2d(out_ch)
else:
norm = nn.BatchNorm2d(out_ch)
return nn.Sequential(conv, norm, actv)
# #### Transpose Convolutional Layer
def Upsample(in_ch, out_ch, use_dropout=False, dropout_ratio=0.5):
# Transposed Convolution
convtrans = nn.ConvTranspose2d(
in_ch, out_ch, 3, stride=2, padding=1, output_padding=1
)
# Normalization
norm = nn.InstanceNorm2d(out_ch)
# Activatin Function
actv = nn.GELU()
if use_dropout:
# Dropout layer
drop = nn.Dropout(dropout_ratio)
return nn.Sequential(convtrans, norm, drop, actv)
else:
return nn.Sequential(convtrans, norm, actv)
# #### Residual block
class Resblock(nn.Module):
def __init__(self, in_features, use_dropout=False, dropout_ratio=0.5):
super().__init__()
layers = list()
# Padding
layers.append(nn.ReflectionPad2d(1))
# Convolution layer
layers.append(
Convlayer(in_features, in_features, 3, 1, use_leaky=False, use_pad=False)
)
# Dropout
if use_dropout:
layers.append(nn.Dropout(dropout_ratio))
# Padding
layers.append(nn.ReflectionPad2d(1))
# Convolution
layers.append(nn.Conv2d(in_features, in_features, 3, 1, padding=0, bias=True))
# Normalization
layers.append(nn.InstanceNorm2d(in_features))
self.res = nn.Sequential(*layers)
def forward(self, x):
return x + self.res(x)
# #### Generator
class Generator(nn.Module):
def __init__(self, in_ch, out_ch, num_res_blocks=6):
super().__init__()
model = list()
# Padding layer
model.append(nn.ReflectionPad2d(3))
# Convolution input_channels -> 64
model.append(
Convlayer(
in_ch=in_ch,
out_ch=64,
kernel_size=7,
stride=1,
use_leaky=False,
use_inst_norm=True,
use_pad=False,
)
)
# Convolution 64 -> 128
model.append(
Convlayer(
in_ch=64,
out_ch=128,
kernel_size=3,
stride=2,
use_leaky=False,
use_inst_norm=True,
use_pad=True,
)
)
# Convolution 128 -> 256
model.append(
Convlayer(
in_ch=128,
out_ch=256,
kernel_size=3,
stride=2,
use_leaky=False,
use_inst_norm=True,
use_pad=True,
)
)
# Residual Block
for _ in range(num_res_blocks):
model.append(Resblock(in_features=256))
# Transposed convolution 256 -> 128
model.append(Upsample(in_ch=256, out_ch=128))
# Transposed convolution 128 -> 256
model.append(Upsample(in_ch=128, out_ch=64))
# Padding Layer
model.append(nn.ReflectionPad2d(3))
# Convolutional layer
model.append(
nn.Conv2d(in_channels=64, out_channels=out_ch, kernel_size=7, padding=0)
)
# Activation function Tanh
model.append(nn.Tanh())
self.gen = nn.Sequential(*model)
def forward(self, x):
return self.gen(x)
# ### Discriminator
class Discriminator(nn.Module):
def __init__(self, in_ch, num_layers=4):
super().__init__()
model = list()
# Convolution in_channels -> 64
model.append(
nn.Conv2d(
in_channels=in_ch, out_channels=64, kernel_size=4, stride=2, padding=1
)
)
# Convolutions i=1: 64 -> 64
# i=2: 64 -> 128
# i=3: 128 -> 256
# i=4: 256 -> 512
for i in range(1, num_layers):
in_chs = 64 * 2 ** (i - 1)
out_chs = in_chs * 2
if i == num_layers - 1:
model.append(Convlayer(in_chs, out_chs, kernel_size=4, stride=1))
else:
model.append(Convlayer(in_chs, out_chs, kernel_size=4, stride=2))
# Convolution 512 -> 1
model.append(
nn.Conv2d(
in_channels=512, out_channels=1, kernel_size=4, stride=1, padding=1
)
)
self.disc = nn.Sequential(*model)
def forward(self, x):
return self.disc(x)
# ### CycleGAN
class CycleGAN(object):
def __init__(
self,
in_ch,
out_ch,
epochs,
device,
start_lr=0.01,
lmbda=10,
idt_coef=0.5,
decay_epoch=0,
):
# Regularization coefficients
self.lmbda = lmbda
self.idt_coef = idt_coef
# Set device
self.device = device
# Generator Monet -> Photo
self.gen_mtp = Generator(in_ch, out_ch)
# Generator Photo -> Monet
self.gen_ptm = Generator(in_ch, out_ch)
# discriminator for Monet-generated images
self.disc_m = Discriminator(in_ch)
# discriminator for Photo-generated images
self.disc_p = Discriminator(in_ch)
# Initialize model weights
self.init_models()
# Optimizator for generators
self.adam_gen = torch.optim.Adam(
itertools.chain(self.gen_mtp.parameters(), self.gen_ptm.parameters()),
lr=start_lr,
betas=(0.5, 0.999),
)
# Optimizator for discriminator
self.adam_disc = torch.optim.Adam(
itertools.chain(self.disc_m.parameters(), self.disc_p.parameters()),
lr=start_lr,
betas=(0.5, 0.999),
)
# Set number of epochs and start of learning rate decay
self.epochs = epochs
# Set decay epoch
self.decay_epoch = decay_epoch if decay_epoch > 0 else int(self.epochs / 2)
# Set rule for learning step decay
lambda_decay = (
lambda epoch: start_lr / (epoch - self.decay_epoch)
if epoch > self.decay_epoch
else start_lr
)
# Define scheduler for generator and discriminator
self.scheduler_gen = torch.optim.lr_scheduler.LambdaLR(
self.adam_gen, lr_lambda=lambda_decay
)
self.scheduler_disc = torch.optim.lr_scheduler.LambdaLR(
self.adam_disc, lr_lambda=lambda_decay
)
# Initialize weights
def init_weights(self, net, gain=0.02):
def init_func(m):
# Name of the class
classname = m.__class__.__name__
# If class has attribute "weight" (to initialize) and
# has either convolutional layer or linear
if hasattr(m, "weight") and (
classname.find("Conv") != -1 or classname.find("Linear") != -1
):
# Initialize weights with values drawn from normal distribution N(mean, std)
init.normal_(m.weight.data, mean=0.0, std=gain)
# Set bias value with constant val
if hasattr(m, "bias") and m.bias is not None:
init.constant_(m.bias.data, val=0.0)
# Initialize BatchNorm weights
elif classname.find("BatchNorm2d") != -1:
init.normal_(m.weight.data, mean=1.0, std=gain)
init.constant_(m.bias.data, val=0.0)
# Apply weight initialization to every submodule of model
net.apply(init_func)
# Initialize models
def init_models(self):
# Initialize weights
self.init_weights(self.gen_mtp)
self.init_weights(self.gen_ptm)
self.init_weights(self.disc_m)
self.init_weights(self.disc_p)
# Set device for models
self.gen_mtp = self.gen_mtp.to(self.device)
self.gen_ptm = self.gen_ptm.to(self.device)
self.disc_m = self.disc_m.to(self.device)
self.disc_p = self.disc_p.to(self.device)
# Enable/ disable gradients for model parameters
def param_require_grad(self, models, requires_grad=True):
for model in models:
for param in model.parameters():
param.requires_grad = requires_grad
# Cycle generation: x -> y_gen -> x_cycle
def cycle_gen(self, x, G_x_to_y, G_y_to_x):
y_gen = G_x_to_y(x)
x_cycle = G_y_to_x(y_gen)
return y_gen, x_cycle
# Define BCE logistic loss
def mse_loss(self, x, target):
if target == 1:
return nn.MSELoss()(x, torch.ones(x.size()).to(self.device))
else:
return nn.MSELoss()(x, torch.zeros(x.size()).to(self.device))
# Define Generator Loss
def loss_gen(self, idt, real, cycle, disc):
# Identity Losses:
loss_idt = nn.L1Loss()(idt, real) * self.lmbda * self.idt_coef
# Cycle Losses:
loss_cycle = nn.L1Loss()(cycle, real) * self.lmbda
# Adversarial Losses:
loss_adv = self.mse_loss(disc, target=1)
# Total Generator loss:
loss_gen = loss_cycle + loss_adv + loss_idt
return loss_gen
# Discriminator Loss
# Ideal Discriminator will classify real image as 1 and fake as 0
def loss_disc(self, real, gen):
loss_real = self.mse_loss(real, target=1)
loss_gen = self.mse_loss(gen, target=0)
return (loss_real + loss_gen) / 2
# Train
def train(self, img_dl):
history = []
dataset_len = img_dl.__len__()
print_header = True
for epoch in range(self.epochs):
# Start measuring time for epoch
start_time = time.time()
# Nulify average losses for an epoch
loss_gen_photo_avg = 0.0
loss_gen_monet_avg = 0.0
loss_gen_total_avg = 0.0
loss_disc_photo_avg = 0.0
loss_disc_monet_avg = 0.0
loss_disc_total_avg = 0.0
# Iterate through dataloader with images
for i, (photo_real, monet_real) in enumerate(img_dl):
photo_real, monet_real = photo_real.to(device), monet_real.to(device)
# Disable gradients for discriminators during generator training
self.param_require_grad([self.disc_m, self.disc_p], requires_grad=False)
# Set gradients for generators to zero at the start of the training pass
self.adam_gen.zero_grad()
# =======================================
# FORWARD PASS THROUGH GENERATOR
# ----------- Cycle photo ---------------
monet_gen, photo_cycle = self.cycle_gen(
photo_real, self.gen_ptm, self.gen_mtp
)
# ----------- Cycle Monet ---------------
photo_gen, monet_cycle = self.cycle_gen(
monet_real, self.gen_mtp, self.gen_ptm
)
# ----------- Generate itself ---------------
# Real Monet -> Identical Monet
monet_idt = self.gen_ptm(monet_real)
# Real photo -> Identical photo
photo_idt = self.gen_mtp(photo_real)
# =======================================
# DISCRIMINATOR PRECTION ON GENERATED IMAGES
# Discriminator M: Check generated Monet
monet_disc = self.disc_m(monet_gen)
# Discriminator P: Check generated photo
photo_disc = self.disc_p(photo_gen)
# =======================================
# CALCULATE LOSSES FOR GENERATORS
# Generator Losses
loss_gen_photo = self.loss_gen(
photo_idt, photo_real, photo_cycle, photo_disc
)
loss_gen_monet = self.loss_gen(
monet_idt, monet_real, monet_cycle, monet_disc
)
# Total Generator loss:
loss_gen_total = loss_gen_photo + loss_gen_monet
# Update average Generator loss:
loss_gen_photo_avg += loss_gen_photo.item()
loss_gen_monet_avg += loss_gen_monet.item()
loss_gen_total_avg += loss_gen_total.item()
# =======================================
# GENERATOR BACKWARD PASS
# Propagate loss backward
loss_gen_total.backward()
# Make step with optimizer
self.adam_gen.step()
# =======================================
# FORWARD PASS THROUGH DISCRIMINATORS
# Enable gradients for discriminators during discriminator trainig
self.param_require_grad([self.disc_m, self.disc_p], requires_grad=True)
# Set zero gradients
self.adam_disc.zero_grad()
# discriminator M: Predictions on real and generated Monet:
monet_disc_real = self.disc_m(monet_real)
monet_disc_gen = self.disc_m(monet_gen.detach())
# discriminator P: Predictions on real and generated photo:
photo_disc_real = self.disc_p(photo_real)
photo_disc_gen = self.disc_p(photo_gen.detach())
# =======================================
# CALCULATE LOSSES FOR DISCRIMINATORS
# Discriminator losses
loss_disc_photo = self.loss_disc(photo_disc_real, photo_disc_gen)
loss_disc_monet = self.loss_disc(monet_disc_real, monet_disc_gen)
# Total discriminator loss:
loss_disc_total = loss_disc_photo + loss_disc_monet
# =======================================
# DISCRIMINATOR BACKWARD PASS
# Propagate losses backward
loss_disc_total.backward()
# Make step with optimizer
self.adam_disc.step()
# =======================================
# Update average Discriminator loss
loss_disc_photo_avg += loss_disc_photo.item()
loss_disc_monet_avg += loss_disc_monet.item()
loss_disc_total_avg += loss_disc_total.item()
# Calculate average losses per epoch
loss_gen_photo_avg /= dataset_len
loss_gen_monet_avg /= dataset_len
loss_gen_total_avg /= dataset_len
loss_disc_photo_avg /= dataset_len
loss_disc_monet_avg /= dataset_len
loss_disc_total_avg /= dataset_len
# Estimate training time per epoch
time_req = time.time() - start_time
# Expand training history
history.append(
[
epoch,
loss_gen_photo_avg,
loss_gen_monet_avg,
loss_gen_total_avg,
loss_disc_photo_avg,
loss_disc_monet_avg,
loss_disc_total_avg,
]
)
# Print statistics
if print_header:
print(
"EPOCH | LOSS: Gen photo | Gen Monet | Disc photo | Disc Monet | TIME MIN"
)
print_header = False
print(
"------+-----------------+-----------+------------+------------+----------"
)
print(
" {:3} | {:>8.2f} | {:>8.2f} | {:>8.2f} | {:>8.2f} | {:4.0f}".format(
epoch + 1,
loss_gen_photo_avg,
loss_gen_monet_avg,
loss_disc_photo_avg,
loss_disc_monet_avg,
time_req // 60,
)
)
# Step learning rate scheduler
self.scheduler_gen.step()
self.scheduler_disc.step()
# Save training history
history = pd.DataFrame(
history,
columns=[
"epoch",
"loss_gen_photo_avg",
"loss_gen_monet_avg",
"loss_gen_total_avg",
"loss_disc_photo_avg",
"loss_disc_monet_avg",
"loss_disc_total_avg",
],
)
history.to_csv("history.csv", index=False)
def predict(self, image):
with torch.no_grad():
self.gen_ptm.eval()
image = image.to(self.device)
monet_gen = self.gen_ptm(image)
return monet_gen
# ##
# Train
# ### Initialize model
gan = CycleGAN(3, 3, epochs=30, device=device)
# ### Train model
gan.train(img_dataloader)
# ##
# Train results
# ### Plot losses
plot_train_loss(log="history.csv")
# ### Model predictions
# - Define dataset and dataloader for photos only
photo_dataset = ImageDatasetCV(monet_dir=None, photo_dir=PHOTO_DIR)
photo_dataloader = DataLoader(photo_dataset, batch_size=BATCH_SIZE)
# - View predicted results
with torch.no_grad():
j = 0
for photo in photo_dataloader:
photo = photo.to(device)
monet_gen = gan.gen_ptm(photo)
fig, ax = plt.subplots(nrows=1, ncols=2, figsize=(10, 4))
ax[0].imshow(unnormalize(photo[0].cpu()).permute(1, 2, 0))
ax[1].imshow(unnormalize(monet_gen[0].cpu().detach()).permute(1, 2, 0))
ax[0].set_title("Photo")
ax[1].set_title("Monetesque photo")
plt.show()
j += 1
if j > 3:
break
test1 = "/kaggle/input/part-2-4/Part 2-4"
photo_dataset1 = ImageDatasetCV(monet_dir=None, photo_dir=test1)
photo_dataloader1 = DataLoader(photo_dataset1, batch_size=BATCH_SIZE)
for photo in photo_dataloader1:
photo = photo.to(device)
monet_gen = gan.gen_ptm(photo)
fig, ax = plt.subplots(nrows=1, ncols=2, figsize=(10, 4))
ax[0].imshow(unnormalize(photo[0].cpu()).permute(1, 2, 0))
ax[1].imshow(unnormalize(monet_gen[0].cpu().detach()).permute(1, 2, 0))
ax[0].set_title("Photo")
ax[1].set_title("Monetesque photo")
plt.show()
test2 = "/kaggle/input/part-2-5/Part 2-5"
photo_dataset2 = ImageDatasetCV(monet_dir=None, photo_dir=test2)
photo_dataloader2 = DataLoader(photo_dataset2, batch_size=BATCH_SIZE)
for photo in photo_dataloader2:
photo = photo.to(device)
monet_gen = gan.gen_ptm(photo)
fig, ax = plt.subplots(nrows=1, ncols=2, figsize=(10, 4))
ax[0].imshow(unnormalize(photo[0].cpu()).permute(1, 2, 0))
ax[1].imshow(unnormalize(monet_gen[0].cpu().detach()).permute(1, 2, 0))
ax[0].set_title("Photo")
ax[1].set_title("Monetesque photo")
plt.show()
# ## Save results
# ### Save model
save_dict = {
"epoch": 0,
"gen_mtp": gan.gen_mtp.state_dict(),
"gen_ptm": gan.gen_ptm.state_dict(),
"desc_m": gan.disc_m.state_dict(),
"desc_p": gan.disc_p.state_dict(),
"optimizer_gen": gan.adam_gen.state_dict(),
"optimizer_desc": gan.adam_disc.state_dict(),
}
save_checkpoint(save_dict, "model.ckpt")
# ##
# Save results
# - Get prediction for all photos
with torch.no_grad():
for i, photo in enumerate(photo_dataloader):
monet_gen = gan.predict(photo)
monet_gen = T.ToPILImage()(unnormalize(monet_gen[0].cpu().detach())).convert(
"RGB"
)
monet_gen.save(SAVE_PATH + str(i + 1) + ".jpg")
# - Create ZIP archive
shutil.make_archive("/kaggle/working/images", "zip", "/kaggle/images")
|
import numpy as np
import pandas as pd
# hyperparameter optimization is done to get best parameters for ML algorithms
df = pd.read_csv("/kaggle/input/churn-modelling/Churn_Modelling.csv")
df.head()
# dataset is about customer will stay in the bank in future or not based on current offers and policies of bank for that particular customer. This data set contains details of a bank's customers and the target variable is a binary variable reflecting the fact whether the customer left the bank (closed his account) or he continues to be a customer.So we can do work on this data so he stays in the particular bank
df.info()
import matplotlib.pyplot as plt
import seaborn as sns
corrmat = df.corr()
top_corr_features = corrmat.index
plt.figure(figsize=(20, 20))
sns.heatmap(df[top_corr_features].corr(), annot=True)
# independent features and dependent features
x = df.iloc[:, 3:13]
y = df.iloc[:, 13]
# taking care of categorical features
geo = pd.get_dummies(x["Geography"], drop_first=True)
# we do drop_first = true coz we can get info from remaining columns only
gender = pd.get_dummies(x["Gender"], drop_first=True)
print(geo)
print(gender)
# now we can drop those columns as later we will append the above columns in our dataset
x.head()
# we dropped geography and gender from dataset
x = pd.concat([x, geo, gender], axis=1)
# axis =1 means column wise
x.head()
# now we have all features with us in optimised version , we can start working on model buidling
# hyperparameters
# these parameters are present in xgboost classifier
params = {
"learning_rate": [0.05, 0.10, 0.15, 0.20, 0.25, 0.30],
# don't lower learning rate value than 0.05 , otherwise leads to overfitting case
"max_depth": [3, 4, 5, 6, 8, 10, 12, 15],
"min_child_weight": [1, 3, 5, 7],
"gamma": [0.0, 0.1, 0.2, 0.3, 0.4],
"colsample_bytree": [0.3, 0.4, 0.5, 0.7],
}
from sklearn.model_selection import RandomizedSearchCV, GridSearchCV
import xgboost
# in xgboost there are many parameter and to get best value for any parameter we need to perform hyperparameter optimization using RandomizedSearchCV and GridSearchCV
# creating timer function to find out how much time it takes to perform RandomizedSearchCV
def timer(start_time=None):
if not start_time:
start_time = datetime.now()
return start_time
elif start_time:
thour, temp_sec = divmod((datetime.now() - start_time).total_seconds(), 3600)
tmin, tsec = divmod(temp_sec, 60)
print(
"\n Time taken: %i hours %i minutes and %s seconds."
% (thour, tmin, round(tsec, 2))
)
classifier = xgboost.XGBClassifier()
random_search = RandomizedSearchCV(
classifier,
param_distributions=params,
n_iter=5,
scoring="roc_auc",
n_jobs=-1,
cv=5,
verbose=3,
)
# cv=cross validation, jobs makes sure it uses all the code present
# verbose gives message about fitting
from datetime import datetime
start_time = timer(None) # timing starts from this point for "start_time" variable
random_search.fit(x, y)
timer(start_time) # timing ends here for "start_time" variable
random_search.best_estimator_
# this gives the best parameters selected using RandomizedSearchCV
|
import pandas as pd
train = pd.read_excel(
"/kaggle/input/d/muratkokludataset/rice-msc-dataset/Rice_MSC_Dataset/Rice_MSC_Dataset.xlsx"
)
train.head()
from sklearn.metrics import mean_absolute_percentage_error
train.info()
train.drop_duplicates(keep=False, inplace=True)
train.shape
train.CLASS.value_counts()
from sklearn.model_selection import train_test_split
train, test = train_test_split(
train, test_size=0.2, random_state=42, shuffle=True, stratify=train.CLASS
)
train.shape, test.shape
train.isnull().sum().sort_values(ascending=False)[:10]
test.isnull().sum().sort_values(ascending=False)[:10]
pd.options.display.max_columns = None
train[train.isna().any(axis=1)]
test[test.isna().any(axis=1)]
train[train.CLASS == "Ipsala"].isnull().sum().sort_values(ascending=False)[:5]
train[train.CLASS == "Ipsala"][
["skewB", "skewCb", "kurtosisB", "kurtosisCb"]
].describe()
train["skewB"] = train["skewB"].fillna(train[train.CLASS == "Ipsala"]["skewB"].median())
train["skewCb"] = train["skewCb"].fillna(
train[train.CLASS == "Ipsala"]["skewCb"].median()
)
train["kurtosisB"] = train["kurtosisB"].fillna(
train[train.CLASS == "Ipsala"]["kurtosisB"].median()
)
train["kurtosisCb"] = train["kurtosisCb"].fillna(
train[train.CLASS == "Ipsala"]["kurtosisCb"].median()
)
train[train.CLASS == "Ipsala"].isnull().sum().sort_values(ascending=False)[:5]
test["skewB"] = test["skewB"].fillna(train[train.CLASS == "Ipsala"]["skewB"].median())
test["kurtosisB"] = test["kurtosisB"].fillna(
train[train.CLASS == "Ipsala"]["kurtosisB"].median()
)
test[test.CLASS == "Ipsala"].isnull().sum().sort_values(ascending=False)[:5]
train[train.CLASS == "Jasmine"].isnull().sum().sort_values(ascending=False)[:5]
train["kurtosisCr"] = train["kurtosisCr"].fillna(
train[train.CLASS == "Jasmine"]["skewB"].median()
)
train["skewCr"] = train["skewCr"].fillna(
train[train.CLASS == "Jasmine"]["skewCb"].median()
)
train[train.CLASS == "Jasmine"].isnull().sum().sort_values(ascending=False)[:5]
train[train.isna().any(axis=1)]
test[test.isna().any(axis=1)]
from flaml import AutoML
automl = AutoML()
y = train.pop("CLASS")
X = train
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=42, shuffle=True, stratify=y
)
automl.fit(
X_train, y_train, task="classification", metric="roc_auc_ovr", time_budget=900
)
print("Best ML leaner:", automl.best_estimator)
print("Best hyperparmeter config:", automl.best_config)
print("Best roc_auc_ovr on validation data: {0:.4g}".format(1 - automl.best_loss))
print("Training duration of best run: {0:.4g} s".format(automl.best_config_train_time))
from sklearn.metrics import classification_report
print(classification_report(y_train, automl.predict(X_train)))
print(classification_report(y_test, automl.predict(X_test)))
test_ = test.drop("CLASS", axis=1)
test_.head()
y_pred = automl.predict(test_)
y_pred[:5]
df = pd.DataFrame(y_pred, columns=["CLASS"])
df.head()
print(classification_report(test.CLASS, df.CLASS))
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
netflix = pd.read_csv("/kaggle/input/netflix-shows/netflix_titles.csv")
netflix.head(6)
netflix.isnull().sum() / len(netflix) * 100
netflix["country"] = netflix["country"].fillna(netflix["country"].mode()[0])
netflix.head()
netflix["count"] = 1
netflix["country"] = netflix["country"].apply(lambda x: x.split(",")[0])
data = netflix.groupby("country")["count"].sum().sort_values(ascending=False)
df = data[:10]
# countries producing the max shows
colors = ["#f5f5f1" for _ in range(10)]
colors[0] = colors[1] = colors[2] = "#b20710"
fig, ax = plt.subplots(figsize=(10, 6))
plt.bar(df.index, df.values, color=colors, edgecolor="#f5f5f1")
spine = ["top", "right", "left"]
ax.spines[spine].set_visible(False)
fig.text(0.15, 0.9, "Top 10 countries on netflix ")
fig.text(1, 0.8, "Insights")
fig.text(1, 0.75, "most number of content producers are USA ,India and UK")
ax.grid(axis="y")
color_map = ["#f5f5f1" for _ in range(10)]
color_map
color_map = ["#f5f5f1"] * 10
color_map
netflix.dtypes
netflix.head()
netflix["date_added"].isna().sum()
netflix.dropna().reset_index = True
netflix.head()
netflix["date_added"] = pd.to_datetime(netflix["date_added"])
netflix.dropna(inplace=True)
netflix.isna().sum()
netflix["year_added"] = netflix["date_added"].dt.year
netflix.head()
netflix["count"] = 1
netflix.groupby(["year_added"])["count"].sum()
lineplot
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
# for dirname, _, filenames in os.walk('/kaggle/input'):
# for filename in filenames:
# print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib as mpl
import matplotlib.pyplot as plt
from scipy.stats import norm
mpl.rcParams["font.sans-serif"] = ["SimHei"] # 中文显示解决方案
plt.rcParams["axes.unicode_minus"] = False # 符号显示解决方案
plt.rc("font", family="SimHei", size="15") # 解决标签中文乱码问题
online_date = pd.read_csv("../input/ecommerce-data/data.csv", encoding="ISO-8859-1")
online_date.info()
online_date.head(3)
# 修改InvoiceDate和CustomerID的数据类型
online_date["InvoiceDate"] = online_date["InvoiceDate"].astype("datetime64")
online_date["CustomerID"] = online_date["CustomerID"].astype("object")
# 统计每列的缺失值个数
np.sum(online_date.isnull(), axis=0)
# 统计每列的缺失率
online_date.apply(lambda x: sum(x.isnull()) / len(x), axis=0)
# 将缺失值删除赋值给新变量
df1 = online_date.dropna(how="any")
df1.info()
df1.head(3)
# 将时间的日期提取出来
df1["InvoiceDate"] = df1["InvoiceDate"].dt.date
df1.info()
# 修改InvoiceDate的数据类型
df1["InvoiceDate"] = df1["InvoiceDate"].astype("datetime64")
df1.head(3)
# 计算每位客户消费总金额
df1["Price"] = df1["Quantity"] * df1["UnitPrice"]
df1.head(3)
# 将订单数据大于0的筛选出来,根据国家进行分组求和,进行排名,取前十名,赋值给新变量
quantity_first_10 = (
df1[df1["Quantity"] > 0]
.groupby("Country")
.sum()["Quantity"]
.sort_values(ascending=False)
.head(10)
)
# 将索引变成列名
quantity_first_10 = quantity_first_10.reset_index()
# 销量前十的国家
plt.figure(
figsize=(8, 6), dpi=100, facecolor="white"
) # 设置画布,figsize=(5,4)是画布大小,dpi=100是画布分辨率,facecolor='white'是画布颜色
sns.set(
style="darkgrid", context="notebook", font_scale=1.2
) # style='darkgrid'是主题样式,context='notebook',控制图片缩放,font_scale=1.2控制坐标轴缩放
sns.barplot(
x=quantity_first_10.Country,
y=quantity_first_10.Quantity,
data=quantity_first_10,
color="g",
orient="vertical",
) # orient='vertical'柱状图的朝向
plt.title("TOP10", pad=10, fontdict={"fontsize": 15}) # 标题名称,距离,字体大小
plt.xlabel("Country", labelpad=15, fontdict={"fontsize": 15}) # x抽的名称,距离,字体大小
plt.ylabel("Quantity", labelpad=15, fontdict={"fontsize": 15}) # y抽的名称,距离,字体大小
plt.xticks(rotation=65) # rotation=1,将X轴标签标签旋转角度
mpl.rcParams["font.sans-serif"] = ["SimHei"] # 中文显示解决方案
plt.rcParams["axes.unicode_minus"] = False # 符号显示解决方案
plt.rc("font", family="SimHei", size="15") # 解决标签中文乱码问题
# 将订单数据大于0的筛选出来,根据国家进行分组求和,进行排名,取前十名,赋值给新变量
Price_first_10 = (
df1[df1["Quantity"] > 0]
.groupby("Country")
.sum()["Price"]
.sort_values(ascending=False)
.head(10)
)
# 将索引变成列名
Price_first_10 = Price_first_10.reset_index()
Price_first_10
# 销量前十的国家
plt.figure(
figsize=(8, 6), dpi=100, facecolor="white"
) # 设置画布,figsize=(5,4)是画布大小,dpi=100是画布分辨率,facecolor='white'是画布颜色
sns.set(
style="darkgrid", context="notebook", font_scale=1.2
) # style='darkgrid'是主题样式,context='notebook',控制图片缩放,font_scale=1.2控制坐标轴缩放
sns.barplot(
x=Price_first_10.Country,
y=Price_first_10.Price,
data=Price_first_10,
color="g",
orient="vertical",
) # orient='vertical'柱状图的朝向
plt.title("TOP10", pad=10, fontdict={"fontsize": 15}) # 标题名称,距离,字体大小
plt.xlabel("Country", labelpad=15, fontdict={"fontsize": 15}) # x抽的名称,距离,字体大小
plt.ylabel("Price", labelpad=15, fontdict={"fontsize": 15}) # y抽的名称,距离,字体大小
plt.xticks(rotation=65) # rotation=1,将X轴标签标签旋转角度
mpl.rcParams["font.sans-serif"] = ["SimHei"] # 中文显示解决方案
plt.rcParams["axes.unicode_minus"] = False # 符号显示解决方案
plt.rc("font", family="SimHei", size="15") # 解决标签中文乱码问题
# 将日期中的年和月分别提取出来
df1["year"] = df1["InvoiceDate"].dt.year
df1["month"] = df1["InvoiceDate"].dt.month
# 修改数据类型
df1["year"] = df1["year"].astype("object")
df1["month"] = df1["month"].astype("object")
df1.info()
df1.head()
# 按时间进行分组分别计算各字段不同的指标
Quantity_date = (
df1[df1["Quantity"] > 0]
.groupby(["year", "month"])
.agg(
{
"Quantity": np.sum,
"UnitPrice": np.mean,
"Price": np.sum,
"InvoiceNo": "count",
}
)
)
Quantity_date
# 统计每月的消费人数,注意去重
customer_date = (
df1[df1["Quantity"] > 0].groupby(["year", "month"])["CustomerID"].nunique()
)
customer_date
customer_date = customer_date.reset_index()
customer_date
customer_date = customer_date.groupby(["year", "month"]).sum()
customer_date
# 查看CustomerID去重以后的个数
df1["CustomerID"].nunique()
df1.head()
# 计算客单价
df1[df1["Quantity"] > 0]["Price"].sum() / df1[df1["Quantity"] > 0]["InvoiceNo"].count()
# 统计每个顾客最近一次消费时间、消费次数,购买数量和消费总金额
customer = (
df1[df1["Quantity"] > 0]
.groupby("CustomerID")
.agg(
{
"InvoiceDate": np.max,
"InvoiceNo": "nunique",
"Quantity": np.sum,
"Price": np.sum,
}
)
)
customer
customer.describe()
# - 用户平均消费次数为4,平均消费数量为1194,平均消费金额为2053
# - 但用户消费数量和消费金额的平均数远远大于中位数,标准差也比较大,说明数据受到极大值干扰,小部分用户购买了大部分商品
# 将最大值排除然后用户消费数量和消费金额的关系
plt.figure(
figsize=(6, 5), dpi=100, facecolor="white"
) # 设置画布,figsize=(5,4)是画布大小,dpi=100是画布分辨率,facecolor='white'是画布颜色
sns.scatterplot(
x="Quantity", # x轴的值
y="Price", # y轴的值
data=customer[customer["Quantity"] < 197491], # 数据来源
marker="o", # 散点的形状,
s=20,
) # 散点的大小
plt.xlabel("Quantity", labelpad=15, fontdict={"fontsize": 20}) # x抽的名称,距离,字体大小
plt.ylabel("Price", labelpad=15, fontdict={"fontsize": 20}) # y抽的名称,距离,字体大小
plt.title("relationship", pad=18, fontdict={"fontsize": 20}) # 标题名称,距离,字体大小
# 从上图来看用户消费金额与消费数量呈一定的线性关系
# 查看各变量之间的相关系数
customer.corr()
# 计算两个变量之间的相关系数
customer.Quantity.corr(customer.Price)
# 将用户按金额进行20等分
customer.Price.plot.hist(bins=20)
# - 从直方图可知,用户消费金额呈现集中趋势,大部分用户消费金额都比较小,小部分极大值影响数据分布,可以使用过滤排除异常
# 使用切比雪夫定理将极值剔除后按用户购买数量进行20等分
customer[customer.Quantity < 26472]["Quantity"].plot.hist(bins=20)
# 按消费金额进行升序排列
customer.sort_values("Price")
# 按订单金额进行升序排序,然后对InvoiceNo Quantity Price进行滚动求和
customer.sort_values("Price")[["InvoiceNo", "Quantity", "Price"]].cumsum()
# 按订单金额进行升序排序,使用apply()对InvoiceNo Quantity Price进行滚动求和计算当前数值占总数的比例
customer_cumsum = customer.sort_values("Price")[
["InvoiceNo", "Quantity", "Price"]
].apply(lambda x: x.cumsum() / x.sum())
customer_cumsum.tail()
# 消费金额占比图
customer_cumsum.reset_index().Price.plot()
# - 根据上图可知,大约3000个用户仅贡献了20%的消费额度,1339个用户贡献了80%的额度
# - 大约4000个用户贡献40%的消费额度,339个用户贡献了60%的消费额度
# 将消费金额占比60%以上的用户ID筛选出来
customer_cumsum[customer_cumsum.Price > 0.6].index
df1[df1["Quantity"] > 0].groupby("CustomerID").agg(
{"InvoiceDate": np.max, "InvoiceNo": "nunique", "Price": np.sum}
)
# 统计每个顾客最近一次消费时间、消费次数,购买数量和消费总金额
user = df1[df1["Quantity"] > 0].groupby("CustomerID")
# 统计每天首购的客户
user.min().InvoiceDate.value_counts().reset_index()
# 统计每天首购的客户然后可视化
user.min().InvoiceDate.value_counts().plot()
# - 用户第一次购买集中在2011年前,因为该数据集时间最早为2010年12月
# - 其他月份首购客户数量基本一致,波动较小
# 统计客户最后一次消费的时间占比然后可视化
user.max().InvoiceDate.value_counts().plot()
# 统计客户第一次和最后一次的消费时间
user_life = user.InvoiceDate.agg(["min", "max"])
user_life
# 统计有多少客户只消费了一次
(user_life["min"] == user_life["max"]).value_counts()
# - 大约有35%用户只消费了一次
# 统计每个顾客最近一次消费时间、消费次数,购买数量和消费总金额
rfm = (
df1[df1["Quantity"] > 0]
.groupby("CustomerID")
.agg({"InvoiceDate": np.max, "InvoiceNo": "nunique", "Price": np.sum})
)
# 计算最后一次消费时间距离现在有多远
rfm["R"] = -(rfm.InvoiceDate - rfm.InvoiceDate.max()) / np.timedelta64(1, "D")
# 对列名进行重命名
rfm.rename(columns={"InvoiceNo": "F", "Price": "M"}, inplace=True)
rfm.head()
def rfm_func(x):
level = x.apply(lambda x: "1" if x >= 0 else "0")
label = level.R + level.F + level.M
d = {
"111": "重要价值客户",
"011": "重要保持客户",
"101": "重要挽留客户",
"001": "重要发展客户",
"110": "一般价值客户",
"010": "一般保持客户",
"100": "一般挽留客户",
"000": "一般发展客户",
}
result = d[label]
return result
rfm["label"] = (
rfm[["R", "F", "M"]].apply(lambda x: x - x.mean()).apply(rfm_func, axis=1)
)
rfm.head()
rfm.groupby("label").sum()
|
# PROBLEM:
# The task at hand is to build a Hindi to English machine translation system from scratch using only the given data. We are allowed to use non-contextualised word embeddings.
# I have chosen FastText embeddings since Hindi is a morphologically rich language. However, the default FastText vectors appeared deficient on inspection so I downloaded FastText embeddings trained by independent researchers on a much larger dataset.
# Unlike
# ARCHITECTURE
# 1. Hindi embedding layer: Converts a given text string into a tensor of stacked embedding vectors. I plan for this to be non-trainable to enable faster training.
# 2.
# The ENglish embeddings were initialised using their GloVe word vectors.
# Approach 1:
# Build Hindi vocabulary and initialise Hindi embedding layer with those words, train it further
# Approach 2:
# Use Fasttext directly
# REFERENCES:
# 1. https://www.youtube.com/watch?v=wzfWHP6SXxY
# 2.
# from google.colab import drive
# drive.mount("/content/gdrive")
import gensim
import os
import spacy
import re
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
from tqdm.auto import tqdm
import torch
import torch.nn as nn
import torch.nn.functional as F
from torch.utils.data import TensorDataset, DataLoader, Dataset
from torch.nn.utils.rnn import pad_sequence
from torch.nn import Transformer
from torch import Tensor
import math
import random
from torch import optim
import torchtext.vocab as vocab
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
df = pd.read_csv(
"/content/gdrive/My Drive/input_data/eng_Hindi_data_train.csv", header=None
)
# Exploratory analysis of train data
df.head()
from collections import Counter
def process_chunk(chunk):
all_text = " ".join(chunk.iloc[:, 1])
words = all_text.split()
word_counts = Counter(words)
return word_counts
file_path = "/content/gdrive/My Drive/input_data/eng_Hindi_data_train.csv"
chunk_size = 10000
all_word_counts = Counter()
for chunk in pd.read_csv(file_path, chunksize=chunk_size):
chunk_word_counts = process_chunk(chunk)
all_word_counts.update(chunk_word_counts)
most_common_words = all_word_counts.most_common(10)
print(most_common_words)
len(all_word_counts)
def process_chunk(chunk):
all_text = " ".join(chunk.iloc[:, 0])
words = all_text.split()
word_counts = Counter(words)
return word_counts
file_path = "/content/gdrive/My Drive/input_data/eng_Hindi_data_train.csv"
chunk_size = 10000
all_word_counts = Counter()
for chunk in pd.read_csv(file_path, chunksize=chunk_size):
chunk_word_counts = process_chunk(chunk)
all_word_counts.update(chunk_word_counts)
most_common_words = all_word_counts.most_common(10)
print(most_common_words)
lengths = [len(sentence.split()) for sentence in df.iloc[:, 0]]
plt.hist(lengths, bins=20, range=(0, 140))
plt.grid()
# LOADING HINDI EMBEDDINGS
# Fasttext
# Non contextual embeddings from:
# https://www.cse.iitb.ac.in/~pb/papers/sltu-ccurl20-il-we.pdf
# TO RUN FOR INFERENCE
from gensim.models import FastText
model_path = "/kaggle/input/d/arqamp/fasttext/fasttext/hi-d50-m2-fasttext.model"
embed = FastText.load(model_path)
torch.tensor(embed.wv.get_vector("कहाँ")).to(device)
# PREPROCESSING FOR ENGLISH:
# 1. Creating vocabulary
# 2. Function for converting sentences to lists of vocabulary indices
# 3. Creating embedding matrix corresponding to vocabulary
max_len = 64
from spacy.lang.en import English
nlp = English()
tokenizer = nlp.tokenizer
en_list = df.iloc[:, 0].tolist()
len(en_list)
# TO RUN FOR INFERENCE AND TRAINING
class Vocabulary:
def __init__(self, sentences):
self.no_tokens = 3
# Dictionaries
self.index_to_token = {
0: "<s>",
1: "</s>",
2: "<pad>",
} # Key: index, Value: token
self.token_to_index = {} # Key: token, Value: index
self.frequency = {} # Key: index, Value: frequency
for sentence in sentences:
self.process_sentence(sentence)
# Method to add a single token to vocabulary
def add_token(self, token):
if token not in self.token_to_index:
self.token_to_index[token] = self.no_tokens
self.frequency[token] = 1
self.index_to_token[self.no_tokens] = token
self.no_tokens += 1
else:
self.frequency[token] += 1
# Method for processing sentences
def process_sentence(self, sentence):
for token in tokenizer(sentence.lower()):
self.add_token(token.text)
sentences = ["This is a sentence.", "This is another sentence."]
test_vocab = Vocabulary(sentences)
# Print vocabulary stats
print("Vocabulary:", test_vocab.index_to_token)
print("Vocabulary size:", test_vocab.no_tokens)
print("Token frequencies:", test_vocab.frequency)
# en_vocab = Vocabulary(en_list)
# print('Vocabulary size:', en_vocab.no_tokens)
# torch.save(en_vocab, "/content/gdrive/My Drive/vocab.pth")
# TO RUN FOR INFERENCE & TRAINING
en_vocab = torch.load("/kaggle/input/en-vocab/vocab.pth", map_location=device)
# Creating embedding matrix corresponding to vocabulary
# Define the vocabulary size and the embedding size
vocab_size = en_vocab.no_tokens
embedding_size = 50
# Load GloVe embeddings
glove = vocab.GloVe(name="6B", dim=embedding_size)
# Create an embedding matrix with the pre-trained embeddings
# embedding_matrix = torch.zeros((vocab_size, embedding_size))
# for i in range(vocab_size):
# if glove.stoi.get( en_vocab.index_to_token[i]) is not None:
# embedding_matrix[i] = glove.vectors[glove.stoi[en_vocab.index_to_token[i]]]
## URGENT: Maybe randomly initialise start and stop token representations (learn it?)
# embedding_matrix.shape
# torch.save(embedding_matrix, "/content/gdrive/My Drive/emb_mtx.pth")
# TO RUN FOR TRAINING & INFERENCE
embedding_matrix = torch.load("/kaggle/input/emb-mtx/emb_mtx.pth", map_location=device)
# check whether emb mtx is in in correct order
glove.vectors[glove.stoi["how"]]
embedding_matrix[en_vocab.token_to_index["how"]]
# Sentence to indexlists conversion for dataloader
def sentence_to_indices(sentence):
tok = tokenizer(sentence.lower())
if len(tok) >= max_len - 2:
tok = tok[1 : max_len - 1]
output = [0]
output.extend([en_vocab.token_to_index[i.text] for i in tok])
output.append(1)
return output
sentence_to_indices("how are you")
indices_tensors_list = [torch.tensor(sentence_to_indices(s)) for s in en_list]
padded_tensors = pad_sequence(indices_tensors_list, batch_first=True, padding_value=2)
padded_tensors.shape
# TO RUN FOR INFERENCE
def indices_to_sentence(indices):
output = " ".join([en_vocab.index_to_token[i] for i in indices])
return output
#
# PREPROCESS HINDI DATA
# TO RUN FOR INFERENCE
# Simple rule based tokeniser for Hindi
def hindi_tokenize(text):
patterns = [r",|\-", r"\s+", r"[^\w\s]", r"\d+", r"[\u0900-\u097F]+"]
token_regex = "|".join(patterns)
return [token for token in re.findall(token_regex, text) if not token.isspace()]
# Example usage
text = "चलो देखें इसमें कितना डैम है, है की नहीं - अब तो पता चल ही जाएगा "
hindi_tokenize(text)
# TO RUN FOR INFERENCE
# positional embedding
def positional_embedding(pos, d_model=50):
# pos: the position of the word in the sentence
# d_model: the dimension of the FastText embeddings
pe = torch.zeros(d_model)
for i in range(0, d_model, 2):
pe[i] = np.sin(pos / (10000 ** ((2 * i) / d_model)))
pe[i + 1] = np.cos(pos / (10000 ** ((2 * (i + 1)) / d_model)))
return pe
# TO RUN FOR INFERENCE
# no padding, for normal input
def embed_hindi_ft(sentence):
tok = hindi_tokenize(sentence) # tokenization
tok1 = ["<s>"] # SOS
tok1.extend(tok)
tok1.append("</s>") # EOS
output = []
for i in range(len(tok1)):
pos_word_embedding = torch.tensor(
embed.wv.get_vector(tok1[i])
) + positional_embedding(i)
output.append(pos_word_embedding)
return torch.stack(output).to(device)
# with padding for training data
def embed_hindi_ft_pad(sentence):
tok = hindi_tokenize(sentence) # tokenization
if len(tok) >= max_len - 2:
tok = tok[1 : max_len - 1]
tok1 = ["<s>"] # SOS
tok1.extend(tok)
tok1.append("</s>") # EOS
for i in range(max_len - len(tok1)):
tok1.append("<pad>")
output = []
for i in range(len(tok1)):
pos_word_embedding = torch.tensor(
embed.wv.get_vector(tok1[i])
) + positional_embedding(i)
output.append(pos_word_embedding)
return torch.stack(output)
embed_hindi_ft_pad("देखते हैं कितना सही है,").shape
# hi_list = df.iloc[:, 1].tolist()
# embed_list = []
# for i in tqdm(hi_list):
# embed_list.append(embed_hindi_ft_pad(i))
# len(embed_list)
# embed_list[0].shape
# torch.save(embed_list, "/content/gdrive/My Drive/embed_list")
# embed_tensor = torch.stack(embed_list)
# embed_tensor.shape
# dataset = TensorDataset(embed_tensor, padded_tensors)
# create DataLoader from dataset
# batch_size = 32 # set batch size
# dataloader = DataLoader(dataset, batch_size=batch_size, shuffle=True)
# torch.save(dataloader, "/content/gdrive/My Drive/NMT_train_loader")
# TO RUN FOR TRAINING
dataloader = torch.load(
"/kaggle/input/nmt-train-loader/NMT_train_loader", map_location=device
)
for i, batch in enumerate(dataloader):
# Move tensors to device
batch[0].to(device)
batch[1].to(device)
# MODEL
class Encoder(nn.Module):
def __init__(self, hidden_size=512, num_layers=4, embedding_dim=50):
super().__init__()
self.hidden_size = hidden_size
self.num_layers = num_layers
self.rnn = nn.LSTM(embedding_dim, hidden_size, num_layers, batch_first=True)
def forward(self, input, batched=True):
if not batched:
embedded = embed_hindi_ft(input).unsqueeze(0).to(device)
else:
embedded = input[0].to(device)
if batched:
# h0 = self.hidden
h0 = torch.zeros(self.num_layers, 32, self.hidden_size).to(device)
c0 = torch.zeros(self.num_layers, 32, self.hidden_size).to(device)
else:
# h0 = torch.zeros(self.num_layers, self.hidden_size).unsqueeze(1).to(device)
h0 = torch.zeros(self.num_layers, 1, self.hidden_size).to(device)
c0 = torch.zeros(self.num_layers, 1, self.hidden_size).to(device)
output, (hn, cn) = self.rnn(embedded.to(device), (h0.to(device), c0.to(device)))
return hn, cn
h0, c0 = enc(batch)
# Decoder debugging
x = torch.zeros((32, 1), dtype=torch.int64, device=device)
x.shape
# x = torch.zeros(32, dtype = torch.int64, device = device)
# x = x.unsqueeze(0)
prev_output_embed = embedding(x)
prev_output_embed.shape
dec = Decoder().to(device)
dec(torch.zeros((32, 1), dtype=torch.int64, device=device), h0, c0)[0].shape
NMT = NMTModel(enc, dec)
output = NMT(batch)
target = batch[1]
print(output.shape)
print(target.shape)
def train(no_epochs, model, criterion, optimizer, dataloader):
for i in range(no_epochs):
j = 0
epoch_loss = 0
iterator = tqdm(dataloader, desc="training")
for batch in iterator:
j += 1
output = model(batch)
target = batch[1]
output = output[1:].reshape(-1, output.shape[2]).float()
target = target[1:].reshape(-1).float()
optimizer.zero_grad()
loss = criterion(output, target.long())
epoch_loss += loss.item()
loss.backward()
torch.nn.utils.clip_grad_norm_(model.parameters(), max_norm=1)
optimizer.step()
iterator.set_postfix(loss=epoch_loss / j)
encoder1 = Encoder().to(device)
decoder1 = Decoder().to(device)
model1 = NMTModel(encoder1, decoder1).to(device)
optimizer = optim.Adam(model1.parameters())
criterion = nn.CrossEntropyLoss()
train(12, model1, criterion, optimizer, dataloader)
torch.save(model1, "model1.pt")
torch.save(model1.state_dict, "model_state.pth")
torch.save(optimizer.state_dict, "opt_state.pth")
criterion = nn.CrossEntropyLoss()
criterion(output, target.long())
enc("क्या हुआ छूटी", batched=False)[0].shape
embedding = nn.Embedding.from_pretrained(embedding_matrix)
prev_output_embed.shape
c0.to(device)
dec(torch.tensor(0).to(device), h0, c0, batched=False)
NMT = NMTModel(enc, dec)
class Decoder(nn.Module):
def __init__(
self,
hidden_size=512,
num_layers=4,
output_size=en_vocab.no_tokens,
tgt_embedding_matrix=embedding_matrix,
):
super().__init__()
self.hidden_size = hidden_size
self.output_size = output_size
self.num_layers = num_layers
self.embedding = nn.Embedding.from_pretrained(tgt_embedding_matrix)
self.rnn = nn.LSTM(50, hidden_size, num_layers, batch_first=True)
self.out = nn.Linear(hidden_size, output_size)
def forward(self, x, hn, cn, batched=True):
if batched:
# x.shape: 32, 1
prev_output_embed = self.embedding(x).to(device) # (32, 1, embedding_size)
output, (hn, cn) = self.rnn(
prev_output_embed, (hn.to(device), cn.to(device))
)
# output shape 32, 1, 512
probabilities = self.out(output).to(device)
# probabilities shape 32, 1, 26k
return probabilities, hn, cn
else:
x = x.unsqueeze(0)
prev_embed = self.embedding(x).unsqueeze(1).to(device)
output, (hn, cn) = self.rnn(prev_embed, (hn.to(device), cn.to(device)))
predictions = self.out(output).to(device)
return predictions, hn, cn
NMT = NMTModel(enc, dec)
x = torch.zeros((32, 1), dtype=torch.int64, device=device)
probs, hn, cn = dec(x, h0, c0, batched=True)
guess = torch.argmax(probs, dim=2).to(device)
guess.shape
tgtseq = batch[1]
tgtseq.shape
tgtseq[:, 1].unsqueeze(1).shape
class NMTModel(nn.Module):
def __init__(self, encoder, decoder):
super().__init__()
self.encoder = encoder
self.decoder = decoder
def forward(self, input, tgt_vocab=en_vocab, tf_ratio=0.5, batched=True):
batch_size = 32
tgt_vocab_size = tgt_vocab.no_tokens
if batched:
tgt_seq = input[1]
# tgt_len = -1
# for i in tgt_seq:
# tgt_len = tgt_len +1
outputs = torch.zeros(
(32, 64, tgt_vocab_size), dtype=torch.float32, device=device
)
hn, cn = self.encoder(input, batched=True)
x = torch.zeros((32, 1), dtype=torch.int64, device=device)
for t in range(1, 64):
# print(x.shape) 32, 1
probs, hn, cn = self.decoder(x, hn, cn, batched=True)
# probabilities.shape: 32, 1, 24260
guess = torch.argmax(probs, dim=2).to(device)
# 32, 1
teacher_force = tgt_seq[:, t]
teacher_force = teacher_force.unsqueeze(1)
x = teacher_force if random.random() < tf_ratio else guess.to(device)
outputs.index_copy_(1, torch.tensor([t], device=device), probs)
return outputs
# return outputs.squeeze(1)
else:
outputs = [0]
x = torch.tensor(0).to(device)
hn, cn = self.encoder(input, batched=False)
# hn = hn.squeeze(1)
# cn = cn.squeeze(1)
for t in range(1, 64):
probs, hn, cn = self.decoder(x, hn, cn, batched=False)
guess = torch.argmax(probs)
x = guess.to(device)
outputs.append(x.item())
if guess.item() == 1:
break
return indices_to_sentence(outputs)
num_epochs = 20
lr = 0.001
enc = Encoder().to(device)
dec = Decoder().to(device)
model = NMTModel(enc, dec).to(device)
model("क्या हुआ जहां लादी छुट्टी", batched=False)
def train(model, dataloader, optimizer, criterion, epochs):
for epoch in range(epochs):
epoch_loss = 0.0
model.train()
for i, batch in enumerate(tqdm(dataloader)):
src = batch
tgt = batch[1]
optimizer.zero_grad()
try:
output = model(src)
except:
continue
output = output[1:].reshape(-1, output.shape[2])
tgt = tgt[1:].reshape(-1)
loss = criterion(output, tgt)
loss.backward()
optimizer.step()
epoch_loss += loss.item()
if (i + 1) % 100 == 0:
print(
"Epoch [{}/{}], Step [{}/{}], Loss: {:.4f}".format(
epoch + 1, epochs, i + 1, len(dataloader), loss.item()
)
)
print(
"Epoch [{}/{}], Loss: {:.4f}".format(
epoch + 1, epochs, epoch_loss / len(dataloader)
)
)
epochs = 10
criterion = nn.CrossEntropyLoss()
optimizer = optim.Adam(model.parameters(), lr=0.001)
# train(model, dataloader, optimizer, criterion, epochs)
j = 0
for batch in dataloader:
print(model(batch))
j = j + 1
# print(batch[1])
if j == 1:
break
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from tqdm.notebook import tqdm
plt.style.use("seaborn-colorblind")
BASE_DIR = "../input/asl-signs/"
train = pd.read_csv(f"{BASE_DIR}/train.csv")
train.head()
fig, ax = plt.subplots(figsize=(8, 8))
train["sign"].value_counts().head(50).sort_values(ascending=True).plot(
kind="barh", ax=ax, title="Top 50 Signs in Training Dataset"
)
ax.set_xlabel("Number of Training Examples")
plt.show()
fig, ax = plt.subplots(figsize=(8, 8))
train["sign"].value_counts().tail(50).sort_values(ascending=True).plot(
kind="barh", ax=ax, title="Bottom 50 Signs in Training Dataset"
)
ax.set_xlabel("Number of Training Examples")
plt.show()
example_fn = train.query('sign == "listen"')["path"].values[0]
example_landmark = pd.read_parquet(f"{BASE_DIR}/{example_fn}")
example_landmark.head()
unique_frames = example_landmark["frame"].nunique()
unique_types = example_landmark["type"].nunique()
types_in_video = example_landmark["type"].unique()
print(
f"The file has {unique_frames} unique frames and {unique_types} unique types: {types_in_video}"
)
listen_files = train.query('sign == "listen"')["path"].values
for i, f in enumerate(listen_files):
example_landmark = pd.read_parquet(f"{BASE_DIR}/{f}")
unique_frames = example_landmark["frame"].nunique()
unique_types = example_landmark["type"].nunique()
types_in_video = example_landmark["type"].unique()
print(
f"The file has {unique_frames} unique frames and {unique_types} unique types: {types_in_video}"
)
if i == 20:
break
for i, d in tqdm(train.iterrows(), total=len(train)):
file_path = d["path"]
example_landmark = pd.read_parquet(f"{BASE_DIR}/{file_path}")
meta = example_landmark["type"].value_counts().to_dict()
meta["frames"] = example_landmark["frame"].nunique()
break
example_landmark.agg(
{
"x": ["min", "max", "mean"],
"y": ["min", "max", "mean"],
"z": ["min", "max", "mean"],
}
).unstack().to_dict()
|
# 
# ## Import Libraries
# pip install opencv-python
# !sudo apt-get install libgtk2.0-dev
# !sudo apt-get install pkg-config
import cv2
import pandas as pd
import numpy as np
from ultralytics import YOLO
import torch
import supervision as sv
from datetime import timedelta
# ## Load YOLO model
model = YOLO("yolov8s.pt")
#
# This line of code initializes an instance of the YOLO class using the 'yolov8s.pt' pre-trained model.
# ## Check GPU is working or not
import torch
print(
f"Using torch {torch.__version__} ({torch.cuda.get_device_properties(0).name if torch.cuda.is_available() else 'CPU'})"
)
# ## Open the video file
cap = cv2.VideoCapture("/kaggle/input/parking-area/parking_area/park.mp4")
# ## Video Information Extraction
def get_video_info(video_path):
# Extracting information about the video
video_info = sv.VideoInfo.from_video_path(video_path)
width, height, fps, total_frames = (
video_info.width,
video_info.height,
video_info.fps,
video_info.total_frames,
)
# Calculating the length of the video by dividing the total number of frames by the frame rate and rounding to the nearest second
video_length = timedelta(seconds=round(total_frames / fps))
# Print out the video resolution, fps, and length u
print(f"\033[1mVideo Resolution:\033[0m ({width}, {height})")
print(f"\033[1mFPS:\033[0m {fps}")
print(f"\033[1mLength:\033[0m {video_length}")
# Extracting information of the test video
get_video_info("/kaggle/input/parking-area/parking_area/park.mp4")
# ## Open the COCO dataset file
my_file = open("/kaggle/input/parking-area/parking_area/coco.txt", "r")
data = my_file.read()
class_list = data.split("\n")
# print(class_list)
count = 0
# ## Object Detection in a Region of Interest using YOLO Model
# Define the area of interest polygon (top-left, bottom-left, bottom-right, top-right)
area = [(381, 104), (342, 125), (599, 192), (597, 151)]
# Define the codec and create VideoWriter object
fourcc = cv2.VideoWriter_fourcc(*"mp4v")
out = cv2.VideoWriter("/kaggle/working/output.mp4", fourcc, 30.0, (1020, 500))
# Loop over the frames of the video
while True:
ret, frame = cap.read()
if not ret:
break
count += 1
# Process every third frame to reduce processing load
if count % 3 != 0:
continue
# Skip frames with no data
if frame is None or frame.shape[0] == 0 or frame.shape[1] == 0:
continue
# Resize the frame for faster processing
frame = cv2.resize(frame, (1020, 500))
# Predict the objects in the frame with YOLO model
results = model.predict(frame)
a = results[0].boxes.data
px = pd.DataFrame(a.cpu().numpy()).astype("float")
list = []
# Loop over the detected objects
for index, row in px.iterrows():
x1 = int(row[0])
y1 = int(row[1])
x2 = int(row[2])
y2 = int(row[3])
d = int(row[5])
c = class_list[d]
# Check if the detected object is a car and is inside the area of interest
if "car" in c:
cx = int(x1 + x2) // 2
cy = int(y1 + y2) // 2
results = cv2.pointPolygonTest(np.array(area, np.int32), ((cx, cy)), False)
if results >= 0:
# Draw a rectangle around the car, a circle on its centroid, and its class name
cv2.rectangle(frame, (x1, y1), (x2, y2), (0, 255, 0), 2)
cv2.circle(frame, (cx, cy), 4, (255, 0, 255), -1)
cv2.putText(
frame,
str(c),
(x1, y1),
cv2.FONT_HERSHEY_COMPLEX,
0.5,
(255, 0, 0),
1,
)
list.append([c])
# Draw the area of interest polygon and the number of detected cars in the frame
cv2.polylines(frame, [np.array(area, np.int32)], True, (255, 0, 2), 2)
k = len(list)
cv2.putText(frame, str(k), (50, 60), cv2.FONT_HERSHEY_PLAIN, 5, (255, 0, 0), 3)
cv2.imshow("RGB", frame)
out.write(frame)
# Exit the loop when 'ESC' key is pressed
if cv2.waitKey(1) & 0xFF == 27:
break
# Release the video file and close all windows
cap.release()
out.release()
cv2.destroyAllWindows()
# ## output
import cv2
import matplotlib.pyplot as plt
from matplotlib import animation, rcParams
from IPython.display import HTML
# Set the embed limit to 50 MB
rcParams["animation.embed_limit"] = 50.0
# Read the video file and extract its frames
video_path = "/kaggle/input/output/output.mp4"
video = cv2.VideoCapture(video_path)
frames = []
while video.isOpened():
ret, frame = video.read()
if not ret:
break
frames.append(frame)
video.release()
# Create the animation function
def create_animation(ims):
fig = plt.figure(figsize=(10, 6))
plt.axis("off")
im = plt.imshow(ims[0])
plt.close()
def animate_func(i):
im.set_array(cv2.cvtColor(ims[i], cv2.COLOR_BGR2RGB))
return [im]
return animation.FuncAnimation(
fig, animate_func, frames=len(ims), interval=1000 // 30
)
# Create the animation object
animation_obj = create_animation(frames)
# Display the animation in the notebook
HTML(animation_obj.to_jshtml())
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import numpy as np
from transformers import TFAutoModel, AutoTokenizer
from tensorflow.keras.layers import Input, Dense
from tensorflow.keras.optimizers import Adam
from tensorflow.keras.models import Model
from sklearn.metrics import accuracy_score, f1_score
from sklearn.model_selection import train_test_split
from tqdm import tqdm
# 读取数据集
data = pd.read_csv("/kaggle/input/yelpzip/yelpdata7.csv")
# 对其他特征进行缺失值处理
other_features = (
data[["user_id", "product_id", "product_id", "rating", "max_daily_count"]]
.fillna(0)
.values
)
scaled_features = (other_features - other_features.mean(axis=0)) / other_features.std(
axis=0
)
tokenizer = AutoTokenizer.from_pretrained("bert-base-uncased")
def encode_text(text, max_length):
encoded = tokenizer.encode_plus(
text,
add_special_tokens=True,
max_length=max_length,
padding="max_length",
return_attention_mask=True,
return_token_type_ids=False,
return_tensors="tf",
)
return encoded["input_ids"], encoded["attention_mask"]
max_length = 512 # 设置固定的序列长度
input_ids = []
attention_masks = []
for text in tqdm(data["user_content"].values, desc="Encoding Text"):
ids, masks = encode_text(text, max_length)
input_ids.append(ids)
attention_masks.append(masks)
input_ids = tf.keras.preprocessing.sequence.pad_sequences(
input_ids,
maxlen=max_length,
dtype="tf.int32",
padding="post",
truncating="post",
value=0,
)
attention_masks = tf.keras.preprocessing.sequence.pad_sequences(
attention_masks,
maxlen=max_length,
dtype="tf.int32",
padding="post",
truncating="post",
value=0,
)
input_ids = np.array(input_ids, dtype=np.int32)
attention_masks = np.array(attention_masks, dtype=np.int32)
# 将数据集分成训练集和测试集
(
train_input_ids,
test_input_ids,
train_attention_masks,
test_attention_masks,
train_labels,
test_labels,
) = train_test_split(
input_ids, attention_masks, data["label"].values, test_size=0.2, random_state=42
)
# 构建模型
bert_model = TFAutoModel.from_pretrained("bert-base-uncased")
input_ids_input = Input(shape=(512,), dtype="int32", name="input_ids")
attention_masks_input = Input(shape=(512,), dtype="int32", name="attention_masks")
output = bert_model(
{"input_ids": input_ids_input, "attention_mask": attention_masks_input}
)[1]
output = Dense(64, activation="relu")(output)
output = Dense(1, activation="sigmoid")(output)
model = Model(inputs=[input_ids_input, attention_masks_input], outputs=output)
model.compile(optimizer=Adam(lr=1e-5), loss="binary_crossentropy")
# 训练模型
history = model.fit(
[train_input_ids, train_attention_masks],
train_labels,
batch_size=16,
epochs=10,
validation_split=0.2,
shuffle=True,
)
# 使用模型预测标签
test_predictions = model.predict([test_input_ids, test_attention_masks])
test_predictions = (test_predictions > 0.5).astype(int)
# 计算模型评估指标
accuracy = accuracy_score(test_labels, test_predictions)
f1 = f1_score(test_labels, test_predictions)
print(f"Accuracy: {accuracy}")
print(f"F1 Score: {f1}")
|
# # Importing python libraies
import pandas as pd
import numpy as np
import seaborn as sns
# # Loading Dataset
dataset = pd.read_csv("/kaggle/input/earthquake-dataset/earthquake_data.csv")
dataset
# # Analysing the dataset
dataset.info()
del dataset["title"]
del dataset["location"]
del dataset["country"]
del dataset["continent"]
dataset
# # Checking for NullValues
dataset.isnull().sum()
dataset["alert"].value_counts()
# # Handling Missing values
dataset["alert"] = dataset["alert"].fillna("red")
dataset.isnull().sum()
dataset
# # Changing datatype of datetime column
dataset["date_time"] = pd.to_datetime(dataset["date_time"])
dataset.info()
dataset["date_time"] = pd.DatetimeIndex(dataset["date_time"]).month
# dataset.describe()
dataset.describe(include=["object"])
dataset.magType.value_counts()
# # Label Encoding
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
alert_le = LabelEncoder()
magtype_le = LabelEncoder()
net_le = LabelEncoder()
dataset["alert"] = alert_le.fit_transform(dataset["alert"])
dataset["magType"] = magtype_le.fit_transform(dataset["magType"])
dataset["net"] = net_le.fit_transform(dataset["net"])
dataset
dataset.corr()
dataset.hist()
# # Slicing the dataset
x = dataset.iloc[:, [1, 2, 3, 4, 6, 7, 8, 9, 10, 11, 12, 13, 14]]
y = dataset.iloc[:, [5]]
# # Balance data using Imbalancing technique
dataset["tsunami"].value_counts()
from imblearn.over_sampling import SMOTE
s = SMOTE()
x_data, y_data = s.fit_resample(x, y)
from collections import Counter
print(Counter(y_data))
# # Feature Scaling
from sklearn.preprocessing import StandardScaler
ss = StandardScaler()
x_scaled = ss.fit_transform(x_data)
x_scaled
# # Splitting into Train and Test Data
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(
x_scaled, y_data, random_state=11, test_size=0.2
)
# # Creating model
from sklearn.linear_model import LogisticRegression
l1 = LogisticRegression()
l1.fit(x_train, y_train)
y_pred = l1.predict(x_test)
y_pred
from sklearn.metrics import accuracy_score
ac = accuracy_score(y_test, y_pred) * 100
ac
from sklearn.svm import SVC
SVM = SVC(kernel="linear", random_state=2)
SVM.fit(x_train, y_train)
y_pred1 = SVM.predict(x_test)
y_pred1
from sklearn.metrics import accuracy_score
ac1 = accuracy_score(y_test, y_pred1) * 100
ac1
from sklearn.naive_bayes import GaussianNB
nb = GaussianNB()
nb.fit(x_train, y_train)
y_pred2 = nb.predict(x_test)
y_pred2
from sklearn.metrics import accuracy_score
ac2 = accuracy_score(y_test, y_pred2) * 100
ac2
# # Ensemble technique
from sklearn.ensemble import VotingClassifier
bc = VotingClassifier(
estimators=[("logisticRegression", l1), ("svm", SVM), ("naivebayes", nb)]
)
bc.fit(x_train, y_train)
y_pred3 = bc.predict(x_test)
y_pred3
from sklearn.metrics import accuracy_score
ac3 = accuracy_score(y_test, y_pred3) * 100
ac3
# # Cross validation using KFold technique
from sklearn.model_selection import KFold
kf = KFold()
kf.split(x_train, y_train)
kf
from sklearn.model_selection import cross_val_predict
cross_pred = cross_val_predict(bc, x_test, y_test, cv=kf)
cross_pred
from sklearn.model_selection import cross_val_score
cross_score = cross_val_score(bc, x_train, y_train, cv=kf)
cross_score
ac5 = cross_score.mean() * 100
ac5
import pickle
filename = "magtype_le.pickle"
# save model
pickle.dump(magtype_le, open(filename, "wb"))
|
import random
# # Step One: Confirm the probability of winning
# The contestant is given the choice of three doors. One of these doors contains the prize. The contestant chooses one door at random. The assumption would be that the odds that the contestant will choose correctly will be one in three. This is confirmed below.
won = 0
for count in range(1000000):
door_selected = random.randint(1, 3)
door_winning = random.randint(1, 3)
if door_selected == door_winning:
won += 1
print(round((won / count), 2))
# # Step Two: Confirm the probability of winning without switching
# After the initial selection there are only two possibilities. Either the contestant has chosen the correct door, or the winning door in one of the remaining two doors. If the winning door is one of the remaining doors then one of these door is picked at random and shown. Otherwise, the non-winning door is shown. The probability of the contestant choosing the winning door remains unchanged at about one in three.
won = 0
for count in range(1000000):
door_selected = random.randint(1, 3)
door_winning = random.randint(1, 3)
door_shown = 0
if door_selected == door_winning:
if door_selected == 1:
if random.randint(1, 2) == 1:
door_shown = 2
else:
door_shown = 3
elif door_selected == 2:
if random.randint(1, 2) == 1:
door_shown = 1
else:
door_shown = 3
elif door_selected == 3:
if random.randint(1, 2) == 1:
door_shown = 1
else:
door_shown = 2
else:
if door_selected == 1:
if door_winning == 2:
door_shown = 3
else:
door_shown = 2
elif door_selected == 2:
if door_winning == 1:
door_shown = 3
else:
door_shown = 1
elif door_selected == 3:
if door_winning == 2:
door_shown = 3
else:
door_shown = 2
if door_selected == door_winning:
won += 1
print(round((won / count), 2))
# # Step Three: Confirm the probability of winning with switching
# It would appear that the probability of the contestant winning doubles if the selected door is switched to the other door. Therefore, it is in the contestant's advantage to switch their choice.
won = 0
for count in range(1000000):
door_first_selected = random.randint(1, 3)
door_winning = random.randint(1, 3)
door_shown = 0
if door_first_selected == door_winning:
if door_first_selected == 1:
if random.randint(1, 2) == 1:
door_shown = 2
else:
door_shown = 3
elif door_first_selected == 2:
if random.randint(1, 2) == 1:
door_shown = 1
else:
door_shown = 3
elif door_first_selected == 3:
if random.randint(1, 2) == 1:
door_shown = 1
else:
door_shown = 2
else:
if door_first_selected == 1:
if door_winning == 2:
door_shown = 3
else:
door_shown = 2
elif door_first_selected == 2:
if door_winning == 1:
door_shown = 3
else:
door_shown = 1
elif door_first_selected == 3:
if door_winning == 2:
door_shown = 3
else:
door_shown = 2
if door_first_selected != door_winning:
won += 1
print(round((won / count), 2))
|
# Product reviews are becoming more important with the evolution of traditional brick and mortar retail stores to online shopping.
# Consumers are posting reviews directly on product pages in real time. With the vast amount of consumer reviews, this creates an opportunity to see how the market reacts to a specific product.
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import seaborn as sns
import math
import warnings
warnings.filterwarnings("ignore") # Hides warning
warnings.filterwarnings("ignore", category=DeprecationWarning)
warnings.filterwarnings("ignore", category=UserWarning)
sns.set_style("whitegrid") # Plotting style
np.random.seed(42) # seeding random number generator
df = pd.read_csv("../input/amazon-product-reviews/amazon.csv")
df.head()
# # Describing the Dataset
data = df.copy()
data.describe()
data.info()
data["asins"].unique()
asins_unique = len(data["asins"].unique())
print("Number of Unique ASINs: " + str(asins_unique))
# # Visualizing the distributions of numerical variables:
data["reviews.numHelpful"].hist(figsize=(20, 5))
plt.show()
data["reviews.id"].hist(figsize=(20, 5))
plt.show()
plt.figure(figsize=(20, 5))
ax1 = sns.countplot(x="reviews.rating", data=data)
for p in ax1.patches:
ax1.annotate(str(p.get_height()), (p.get_x() * 1.01, p.get_height() * 1.01))
plt.show()
# # Split the data into Train and Test
from sklearn.model_selection import StratifiedShuffleSplit
print("Before : {}".format(len(data)))
dataAfter = data.dropna(subset=["reviews.rating"])
# Removes all NAN in reviews.rating
print("After : {}".format(len(dataAfter)))
dataAfter["reviews.rating"] = dataAfter["reviews.rating"].astype(int)
split = StratifiedShuffleSplit(n_splits=10, test_size=0.2)
for train_index, test_index in split.split(dataAfter, dataAfter["reviews.rating"]):
strat_train = dataAfter.reindex(train_index)
strat_test = dataAfter.reindex(test_index)
print(len(strat_train))
print(len(strat_test))
round((strat_test["reviews.rating"].value_counts() * 100 / len(strat_test)), 2)
# # Data Exploration
reviews = strat_train.copy()
reviews.head()
len(reviews["name"].unique())
len(reviews["asins"].unique())
reviews.info()
reviews.groupby("asins")["name"].unique()
different_names = reviews[reviews["asins"] == "B00L9EPT8O,B01E6AO69U"]["name"].unique()
for name in different_names:
print(name)
reviews[reviews["asins"] == "B00L9EPT8O,B01E6AO69U"]["name"].value_counts()
fig = plt.figure(figsize=(16, 10))
ax1 = plt.subplot(211)
ax2 = plt.subplot(212, sharex=ax1)
reviews["asins"].value_counts().plot(kind="bar", ax=ax1, title="ASIN Frequency")
np.log10(reviews["asins"].value_counts()).plot(
kind="bar", ax=ax2, title="ASIN Frequency (Log10 Adjusted)"
)
for p in ax1.patches:
ax1.annotate(str(p.get_height()), (p.get_x() * 1.01, p.get_height() * 1.01))
for p in ax2.patches:
ax2.annotate(
str(round((p.get_height()), 2)), (p.get_x() * 1.01, p.get_height() * 1.01)
)
plt.show()
# # Entire training dataset average rating
reviews["reviews.rating"].mean()
asins_count_ix = reviews["asins"].value_counts().index
fig = plt.figure(figsize=(16, 5))
ax = reviews["asins"].value_counts().plot(kind="bar", title="ASIN Frequency")
for p in ax.patches:
ax.annotate(str(p.get_height()), (p.get_x() * 1.01, p.get_height() * 1.01))
plt.xticks(rotation=90)
plt.show()
fig = plt.figure(figsize=(16, 5))
sns.pointplot(x="asins", y="reviews.rating", order=asins_count_ix, data=reviews)
plt.xticks(rotation=90)
plt.show()
# # Sentiment Analysis
def sentiments(rating):
if (rating == 5) or (rating == 4):
return "Positive"
elif rating == 3:
return "Neutral"
elif (rating == 2) or (rating == 1):
return "Negative"
# Add sentiments to the data
strat_train["Sentiment"] = strat_train["reviews.rating"].apply(sentiments)
strat_test["Sentiment"] = strat_test["reviews.rating"].apply(sentiments)
print(strat_train["Sentiment"][:15])
round((strat_train["Sentiment"].value_counts() * 100 / len(strat_train)), 2)
fig = plt.figure(figsize=(16, 5))
ax = (
strat_train["Sentiment"]
.value_counts()
.plot(kind="bar", title="Train Data Sentimental Data")
)
for p in ax.patches:
ax.annotate(str(p.get_height()), (p.get_x() * 1.01, p.get_height() * 1.01))
plt.show()
round((strat_test["Sentiment"].value_counts() * 100 / len(strat_test)), 2)
fig = plt.figure(figsize=(16, 5))
ax = (
strat_test["Sentiment"]
.value_counts()
.plot(kind="bar", title="Test Data Sentimental Data")
)
for p in ax.patches:
ax.annotate(str(p.get_height()), (p.get_x() * 1.01, p.get_height() * 1.01))
plt.show()
|
from tqdm import tqdm_notebook, tnrange
from tqdm.auto import tqdm
tqdm.pandas(desc="Progress")
import re
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import nltk
nltk.download("omw-1.4")
nltk.download("wordnet")
# use only while uploading and running notebook for the first time
from nltk.stem import WordNetLemmatizer
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
from nltk.corpus import stopwords
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# columns for sentiment 140
DATASET_COLUMNS = ["sentiment", "ids", "date", "flag", "user", "text"]
DATASET_ENCODING = "ISO-8859-1"
dataset1 = pd.read_csv(
"/kaggle/input/sentiment-dataset-with-1-million-tweets/dataset.csv"
)
dataset2 = pd.read_csv("/kaggle/input/combined-twitter-dataset-for-training/result.csv")
dataset3 = pd.read_csv(
"/kaggle/input/sentiment140/training.1600000.processed.noemoticon.csv",
encoding=DATASET_ENCODING,
names=DATASET_COLUMNS,
)
dataset1 = dataset1[dataset1["Language"] == "en"]
dataset1 = dataset1[["Text"]].dropna()
dataset2 = dataset2[["Text"]].dropna()
dataset3 = dataset3[["text"]].dropna()
print(dataset1)
print(dataset2)
print(dataset3)
# Defining dictionary containing all emojis with their meanings.
emojis = {
":)": "smile",
":-)": "smile",
";d": "wink",
":-E": "vampire",
":(": "sad",
":-(": "sad",
":-<": "sad",
":P": "raspberry",
":O": "surprised",
":-@": "shocked",
":@": "shocked",
":-$": "confused",
":\\": "annoyed",
":#": "mute",
":X": "mute",
":^)": "smile",
":-&": "confused",
"$_$": "greedy",
"@@": "eyeroll",
":-!": "confused",
":-D": "smile",
":-0": "yell",
"O.o": "confused",
"<(-_-)>": "robot",
"d[-_-]b": "dj",
":'-)": "sadsmile",
";)": "wink",
";-)": "wink",
"O:-)": "angel",
"O*-)": "angel",
"(:-D": "gossip",
"=^.^=": "cat",
}
stop_words = set(stopwords.words("english"))
stop_words_removal = [
"not",
"n't",
"against",
"no",
"nor",
"don",
"don't",
"should",
"should've",
"ain",
"aren",
"aren't",
"couldn",
"couldn't",
"didn",
"didn't",
"doesn",
"doesn't",
"hadn",
"hadn't",
"hasn",
"hasn't",
"haven",
"haven't",
"isn",
"isn't",
"ma",
"mightn",
"mightn't",
"mustn",
"mustn't",
"needn",
"needn't",
"shan",
"shan't",
"shouldn",
"shouldn't",
"wasn",
"wasn't",
"weren",
"weren't",
"won",
"won't",
"wouldn",
"wouldn't",
"further",
]
for word in stop_words_removal:
if word in stop_words:
stop_words.remove(word)
more_stopwords = {"USER", "URL"}
stop_words = stop_words.union(more_stopwords)
# abbrevation and some important punctuations:
def replace_words(text, replace_dict):
regex = re.compile(r"\b(" + "|".join(replace_dict.keys()) + r")\b")
return regex.sub(
lambda x: replace_dict[x.group()] if x.group() in replace_dict else x.group(),
text,
)
dataset1["Text"] = dataset1["Text"].apply(lambda x: replace_words(x, all_abb_others))
print(dataset1)
dataset1["Text"] = dataset1["Text"].progress_apply(lambda x: x.lower())
dataset1["Text"] = dataset1["Text"].apply(lambda x: x.lower())
dataset2["Text"] = dataset2["Text"].progress_apply(lambda x: x.lower())
dataset2["Text"] = dataset2["Text"].apply(lambda x: x.lower())
dataset2["Text"] = dataset2["Text"].progress_apply(lambda x: x.lower())
dataset3["text"] = dataset3["text"].apply(lambda x: x.lower())
dataset1["Text"] = dataset1["Text"].progress_apply(
lambda x: replace_words(x, all_abb_others)
)
dataset1["Text"] = dataset1["Text"].apply(lambda x: replace_words(x, all_abb_others))
dataset2["Text"] = dataset2["Text"].progress_apply(
lambda x: replace_words(x, all_abb_others)
)
dataset2["Text"] = dataset2["Text"].apply(lambda x: replace_words(x, all_abb_others))
dataset2["Text"] = dataset2["Text"].progress_apply(
lambda x: replace_words(x, all_abb_others)
)
dataset3["text"] = dataset3["text"].apply(lambda x: replace_words(x, all_abb_others))
def remove_emoji(text):
emoji_pattern = re.compile(
"["
"\U0001F600-\U0001F64F" # emoticons
"\U0001F300-\U0001F5FF" # symbols & pictographs
"\U0001F680-\U0001F6FF" # transport & map symbols
"\U0001F1E0-\U0001F1FF" # flags (iOS)
"\U00002702-\U000027B0"
"\U000024C2-\U0001F251"
"]+",
flags=re.UNICODE,
)
return emoji_pattern.sub(r"", text)
def remove_html(text):
html = re.compile(
r"^[^ ]<.*?>|&([a-z0-9]+|#[0-9]\"\'\“{1,6}|#x[0-9a-f]{1,6});[^A-Za-z0-9]+"
)
return re.sub(html, "", text)
dataset1["Text"] = dataset1["Text"].apply(lambda x: remove_emoji(x))
dataset1["Text"] = dataset1["Text"].apply(lambda x: remove_html(x))
dataset2["Text"] = dataset2["Text"].apply(lambda x: remove_emoji(x))
dataset2["Text"] = dataset2["Text"].apply(lambda x: remove_html(x))
dataset3["text"] = dataset3["text"].apply(lambda x: remove_emoji(x))
dataset3["text"] = dataset3["text"].apply(lambda x: remove_html(x))
def preprocess(textdata):
processedText = []
# Create Lemmatizer and Stemmer.
wordLemm = WordNetLemmatizer()
# Defining regex patterns.
urlPattern = r"((http://)[^ ]*|(https://)[^ ]*|( www\.)[^ ]*)"
userPattern = "@[^\s]+"
alphaPattern = "[^a-zA-Z0-9]"
sequencePattern = r"(.)\1\1+"
seqReplacePattern = r"\1\1"
for tweet in textdata:
# tweet = tweet.lower()
# Replace all URls with 'URL'
tweet = re.sub(urlPattern, " URL", tweet)
# Replace all emojis.
for emoji in emojis.keys():
tweet = tweet.replace(emoji, "EMOJI" + emojis[emoji])
# Replace @USERNAME to 'USER'.
tweet = re.sub(userPattern, " USER", tweet)
# Replace all non alphabets.
tweet = re.sub(alphaPattern, " ", tweet)
# Replace 3 or more consecutive letters by 2 letter.
tweet = re.sub(sequencePattern, seqReplacePattern, tweet)
tweetwords = ""
for word in tweet.split():
if len(word) > 1:
# Lemmatizing the word.
word = wordLemm.lemmatize(word)
tweetwords += word + " "
processedText.append(tweetwords)
return processedText
def stop_words_removal(textdata):
processedText = []
# Create Lemmatizer and Stemmer.
for tweet in textdata:
tweetwords = ""
for word in tweet.split():
# Checking if the word is a stopword.
if word not in stop_words:
tweetwords += word + " "
processedText.append(tweetwords)
return processedText
text1 = list(dataset1["Text"])
text2 = list(dataset2["Text"])
text3 = list(dataset3["text"])
import time
t = time.time()
processedtext1 = preprocess(text1)
stopwordremoved1 = stop_words_removal(processedtext1)
processedtext2 = preprocess(text2)
stopwordremoved2 = stop_words_removal(processedtext2)
processedtext3 = preprocess(text3)
stopwordremoved3 = stop_words_removal(processedtext3)
print(f"Total Time taken for Preprocessing: {round(time.time()-t)}")
# print(round(time.time()-t))
df1 = pd.DataFrame(
{
"cleaned_Text": processedtext1,
"orignal_text": text1,
"stopwords_removed": stopwordremoved1,
}
).dropna()
df2 = pd.DataFrame(
{
"cleaned_Text": processedtext2,
"orignal_text": text2,
"stopwords_removed": stopwordremoved2,
}
).dropna()
df3 = pd.DataFrame(
{
"cleaned_Text": processedtext3,
"orignal_text": text3,
"stopwords_removed": stopwordremoved3,
}
).dropna()
df1.to_csv("export1.csv", index=None)
df2.to_csv("export2.csv", index=None)
df3.to_csv("export3.csv", index=None)
x1 = pd.read_csv("export1.csv").dropna()
print(x1)
x2 = pd.read_csv("export2.csv").dropna()
print(x2)
x3 = pd.read_csv("export3.csv").dropna()
print(x3)
x1.to_csv("export1.csv", index=None)
x2.to_csv("export2.csv", index=None)
x3.to_csv("export3.csv", index=None)
# for some reason it seems to not drop na tuples at first if anyone knows why please do tell
x1 = pd.read_csv("export1.csv").dropna()
print(x1)
x2 = pd.read_csv("export2.csv").dropna()
print(x2)
x3 = pd.read_csv("export3.csv").dropna()
print(x3)
|
# # Python Booleans - Mantıksal Operatörler
# * Mantıksal operatörler iki değerden oluşur. True - False True: doğru False: Yanlış
# ## Boolean Values
# * Programlamada genellikle bir ifadenin Doğru mu yoksa Yanlış mı olduğunu bilmeniz gerekir.
# * Python'da herhangi bir ifadeyi değerlendirebilir ve True veya False olmak üzere iki yanıttan birini alabilirsiniz.
# * İki değeri karşılaştırdığınızda, ifade değerlendirilir ve Python, Boole yanıtını döndürür:
print(180 > 120)
print(500 == 250)
print(180 < 120)
# * if ifadesinde bir koşul çalıştırdığınızda, Python True veya False değerini döndürür:
z = 250
p = 99
if p > z:
print("p büyüktür z'den")
else:
print("p büyük değildir z'den")
# # Değerleri ve Değişkenleri Değerlendirme
# * bool() işlevi, herhangi bir değeri değerlendirmenize ve karşılığında True veya False vermenize izin verir,
# * Örneğin: Bir diziyi ve bir sayıyı değerlendirin:
print(bool("Selam"))
print(bool(5))
b = "Selam"
k = 8
print(bool(b))
print(bool(k))
# # Çoğu Değer Doğrudur
# * Bir tür içeriğe sahipse, hemen hemen her değer True olarak değerlendirilir.
# * Boş diziler dışında tüm diziler True'dur.
# * 0 dışında herhangi bir sayı True'dur.
# * Boş olanlar dışında tüm liste, demet, küme ve sözlük True'dur.
bool("tdk")
bool(13579)
bool(["CHARGER", "MUSTANG", "AMG"])
# # Bazı Değerler Yanlış
# * Aslında, (), [], {}, "", 0 sayısı ve Yok değeri gibi boş değerler dışında False olarak değerlendirilen çok fazla değer yoktur. Ve elbette False değeri False olarak değerlendirilir.
# * Aşağıdaki örnekler False olarak çıktı verecektir
bool(False)
bool(None)
bool(0)
bool("")
bool(())
bool([])
bool({})
# # Fonksiyonlar bir Boole Döndürebilir
# * Bir Boole Değeri döndüren fonksiyonlar oluşturabilirsiniz:
def myFunction():
return True
print(myFunction())
# * Bir işlevin Boole yanıtına göre kod çalıştırabilirsiniz:
# * Örnek "EVET!" Yazdır işlev True döndürürse, aksi takdirde "NO!" yazdırın
def myFunction():
return True
if myFunction():
print("Ja!")
else:
print("Nein!")
# * Python ayrıca, bir nesnenin belirli bir veri türünde olup olmadığını belirlemek için kullanılabilen isinstance() fonksiyonu gibi bir boolean değeri döndüren birçok yerleşik işleve sahiptir:
# * Örnek Bir nesnenin tamsayı olup olmadığını kontrol edin
t = 456
print(isinstance(t, int))
tf = 456.5
print(isinstance(tf, float))
A = "Kumsalda Ateş Harikadır."
print(isinstance(A, str))
# * Aşağıdaki boolean hangi değeri çıktı verir.
print(18 > 5)
print(18 == 5)
print(18 < 5)
print(bool("tdk"))
print(bool(0))
# # Python Operatörleri
# * Operatörler, değişkenler ve değerler üzerinde işlem yapmak için kullanılır.
# * Aşağıdaki örnekte, iki değeri bir araya getirmek için + operatörünü kullanıyoruz:
print(18 + 5)
# #### Python, operatörleri aşağıdaki gruplara ayırır:
# * Arithmetic operators
# * Assignment operators
# * Comparison operators
# * Logical operators
# * Identity operators
# * Membership operators
# * Bitwise operators
# ## Python Arithmetic Operators
# * Aritmetik opetaörler, yaygın matematiksel işlemleri gerçekleştirmek için sayısal değerlerle birlikte kullanılır:
# * Addition(toplama) x + y
# * Subtraction(çıkarma) x - y
# * Multiplication(çarpma) x * y
# * Division(bölme) x / y
# * Modulus(kalan) x % y
# * Exponentiation(üs alma) x ** y
# * Floor division(taban fonksiyon) x // y
# addition - toplama
x = 8
y = 14
print(x + y)
# Subtraction - çıkarma
x = 50
y = 46
print(x - y)
# Multiplication - Çarpma
x = 8
y = 8
print(x * y)
# Division - bölme
x = 256
y = 4
print(x / y)
# modulus - modüller
x = 17456
y = 562
print(x % y)
# Exponentiation - üst alma
x = 16
y = 5
print(x**y) # Diğer Şekli 2*2*2*2*2
# Floor division - taban fonksiyonu
x = 864
y = 32
print(x // y)
# kat bölümü // sonucu en yakın tam sayıya yuvarlar
# # Python Atama Operatörleri
# * Atama işleçleri, değişkenlere değer atamak için kullanılır:
# = EŞİTTİR
x = 8
x
# += ARTI EŞİTTİR
x = 8
x += 5
print(x)
# -= EKSİ EŞİTTİR
x = 8
x -= 5
print(x)
# *= ÇARPI EŞİTTİR
x = 6
x *= 8
print(x)
# /=
x = 9
x /= 8
print(x)
# %= bölümden kalan sayıyı verir
x = 78
x %= 20
print(x)
# //= kaç defa bölündüğü
x = 560
x //= 145
print(x)
# **= sayının üssünü alır
x = 90
x **= 5
print(x)
# # Python Karşılaştırma Operatörleri
# * Karşılaştırma işleçleri iki değeri karşılaştırmak için kullanılır:
# == Equal - Eşit mi
x = 78
y = 56
print(x == y)
# 5, 3'e eşit olmadığı için False döndürür
# != Not equal - Eşit değil
x = 78
y = 6
print(x != y)
# 5, 3'e eşit olmadığı için True döndürür
# > Greater than , büyüktür
x = 18
y = 6
print(x > y)
# 5, 3'ten büyük olduğu için True döndürür
# < Less than , küçüktür
x = 96
y = 87
print(x < y)
# 5, 3'ten küçük olmadığı için False döndürür
# >= Greater than or equal to , Büyük eşittir
x = 8
y = 6
print(x >= y)
# 8, 6'dan büyük veya eşit olduğuğ için True Döndürür
# Less than or equal to , küçük eşittir
x = 8
y = 6
print(x <= y)
# 8, 6'dan küçük veya eşit olmadığı için False Döndürür.
# # Python Mantıksal Operatörler
# * Mantıksal işleçler, koşullu ifadeleri birleştirmek için kullanılır: "and, or, not"
# and, Her iki ifade de doğruysa True döndürür
x = 8
print(x > 2 and x < 12)
# 8, 2'den büyük 12'den küçük ise True Döndürür.
# or İfadelerden biri doğruysa True döndürür
x = 8
print(x > 2 or x < 6)
# #, koşullardan biri doğru olduğu için True döndürür (8, 2'den büyüktür, ancak 8, 6'dan küçük değildir)
# not Sonucu tersine çevirin, sonuç doğruysa False döndürür
x = 8
print(not (x > 2 and x < 12))
# sonucu tersine çevirmek için kullanılır. Norlamde true çıktısı verecekti
# # Python Kimlik Operatörleri
# * Kimlik fonksiyonları, nesneleri eşit olup olmadıklarını değil, aslında aynı nesne olup olmadıklarını ve aynı bellek konumuna sahip olup olmadıklarını karşılaştırmak için kullanılır:
# is
# Her iki değişken de aynı nesneyse True döndürür
t = ["Thy", "Pegasus"]
d = ["Thy", "Pegasus"]
k = t
print(t is k)
# True değerini döndürür çünkü z, x ile aynı nesnedir
print(t is d)
# aynı içeriğe sahip olsalar bile x, y ile aynı nesne olmadığı için False döndürür
print(t == d)
# "is" ve "==" arasındaki farkı göstermek için: x eşittir y olduğu için bu karşılaştırma True değerini döndürür
# is not
# Her iki değişken de aynı nesne değilse True döndürür
t = ["Thy", "Pegasus"]
d = ["Thy", "Pegasus"]
k = t
print(t is not k)
# z, x ile aynı nesne olduğu için False döndürür
print(t is not d)
# True döndürür çünkü x, aynı içeriğe sahip olsalar bile y ile aynı nesne değildir
print(t != d)
# "is not" ve "!=" arasındaki farkı göstermek için: x eşittir y olduğu için bu karşılaştırma False döndürür
|
# ## Executive Summary
# This's S03E01 of Kaggle playground series
# ### Objective:
# To predict median house value of California areas
# ### List of independent variables
# - MedInc: Median Income of neighborhood
# - HouseAge: How old the house is
# - AveRooms: Number of Rooms
# - AveBedrms: Number of Bed rooms
# - Population: Population of the neighborhood
# - AveOccup: Number of person in a family
# - Latitude/Longtitude: Co-ordinate of the areas
# - Target: MedianHouseVal
# ### Approach
# Inspiration comes from these 2 notebooks/authors:
# https://www.kaggle.com/code/phongnguyen1/feature-engineering-with-coordinates
# https://www.kaggle.com/code/ravi20076/playgrounds3e1-eda-model-pipeline
# From own's understanding, the most important factor in real estate is: location, location, location.
# Therefore, we will heavily utilize co-ordinates independent variables by:
# - Calculating distance to big cities
# - Distance to coastline
# ### Functions to write:
# - Baseline model with xgboost & all features
# - Distance_to_big_cities
# - Distance_to_coastline
# - run_xgboost/lightgbm with all features
import pandas as pd
import numpy as np
from sklearn.datasets import fetch_california_housing
from sklearn.model_selection import KFold, train_test_split
from sklearn.metrics import mean_squared_error as mse
from sklearn.preprocessing import LabelEncoder
from sklearn.cluster import KMeans
from xgboost import XGBRegressor
from lightgbm import LGBMRegressor
from folium import Map
from folium.plugins import HeatMap
from sklearn.metrics.pairwise import haversine_distances
from math import radians
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_style("whitegrid")
# Load data
df_train = pd.read_csv("/kaggle/input/playground-series-s3e1/train.csv")
df_test = pd.read_csv("/kaggle/input/playground-series-s3e1/test.csv")
# Check missing values
df_train.isna().sum()
# ## EDA
# Distribution
cols = df_train.columns[1:10]
# Create a grid of hisplots
fig, axes = plt.subplots(3, 3, figsize=(12, 12))
fig.tight_layout(pad=5)
for i, ax in enumerate(axes.flat):
col = cols[i]
sns.histplot(data=df_train, x=col, kde=True, ax=ax)
ax.set_title(f"{col}")
plt.show()
# Correlation Heatmap
matrix = df_train[cols].corr().round(2)
mask = np.triu(np.ones_like(matrix, dtype=bool))
plt.subplots(figsize=(15, 7))
sns.heatmap(matrix, annot=True, mask=mask, square=True)
# **Observation**
# The two most correlated independent variables with target are MedInc and AveRooms.
# Of course, longtitude & latitude doesn't show any correlation but let's draw some maps.
# Draw a map to describe median house value according to location
import folium
import branca
inferno_colors = [
(0, 0, 4),
(40, 11, 84),
(101, 21, 110),
(159, 42, 99),
(212, 72, 66),
(245, 125, 21),
(250, 193, 39),
(252, 255, 164),
]
map = Map(
df_train[["Latitude", "Longitude"]].mean(axis=0),
zoom_start=6,
zoom_control=False,
dragging=False,
scrollWheelZoom=False,
boxZoom=False,
touchZoom=False,
)
lat = list(df_train["Latitude"])
lon = list(df_train["Longitude"])
targets = list(df_train["MedHouseVal"])
cmap = branca.colormap.LinearColormap(
inferno_colors, vmin=min(targets), vmax=max(targets)
)
for loc, target in zip(zip(lat, lon), targets):
folium.Circle(
location=loc,
radius=target,
fill=True,
color=cmap(target),
fill_opacity=0.2,
weight=0,
).add_to(map)
map.add_child(cmap)
display(map)
# **Observation**
# The closer to big cities & coastline, the higher the median house value.
# ## Feature Engineering
# We are gonna perform 3 tricks: distance to large cities, distance to coastline & geo-clustering
# Def distance_to_big_cities & distance_to_coastline using harversine_distance
Sacramento = (38.576931, -121.494949)
SanFrancisco = (37.780080, -122.420160)
SanJose = (37.334789, -121.888138)
LosAngeles = (34.052235, -118.243683)
SanDiego = (32.715759, -117.163818)
coast = np.array(
[
[32.664472968971786, -117.16139777220666],
[33.20647603453836, -117.38308931734736],
[33.77719697387153, -118.20238415808473],
[34.46343131623148, -120.01447157053916],
[35.42731619324845, -120.8819602254066],
[35.9284107340049, -121.48920228383551],
[36.982737132545495, -122.028973002425],
[37.61147966825591, -122.49163361836126],
[38.3559871217218, -123.06032062543764],
[39.79260770260524, -123.82178288918176],
[40.799744611668416, -124.18805587680554],
[41.75588735544064, -124.19769463963775],
]
)
def compute_distance(loc1, loc2):
loc1_rad = np.radians(loc1)
loc2_rad = np.radians(loc2)
distance_rad = haversine_distances([loc1_rad, loc2_rad])
distance_km = (distance_rad * 6371)[0][1]
return distance_km
# Add distance_to_cities features by applying compute_to_distance function on df with big cities co-ordinates
def add_distance_to_nearest_cities(df):
df["to_Sacramento"] = df.apply(
lambda x: compute_distance((x.Latitude, x.Longitude), Sacramento), axis=1
)
df["to_SanFrancisco"] = df.apply(
lambda x: compute_distance((x.Latitude, x.Longitude), SanFrancisco), axis=1
)
df["to_SanJose"] = df.apply(
lambda x: compute_distance((x.Latitude, x.Longitude), SanJose), axis=1
)
df["to_LosAngeles"] = df.apply(
lambda x: compute_distance((x.Latitude, x.Longitude), LosAngeles), axis=1
)
df["to_SanDiego"] = df.apply(
lambda x: compute_distance((x.Latitude, x.Longitude), SanDiego), axis=1
)
# Add distance_to_coastline features by applying compute_to_distance function on df & coastline co-ordinates
def add_distance_to_coastlines(df):
for i, coast_line in enumerate(coast):
df[f"coast_{i}"] = df.apply(
lambda x: compute_distance((x.Latitude, x.Longitude), coast_line), axis=1
)
# Define run_xgboost function
N_SPLITS = 10
XGB_PARAMS = {
"n_estimators": 50000,
"max_depth": 9,
"learning_rate": 0.01,
"colsample_bytree": 0.66,
"subsample": 0.76,
"min_child_weight": 22,
"reg_lambda": 16,
"early_stopping_rounds": 1000,
"tree_method": "gpu_hist",
"eval_metric": "rmse",
"seed": 0,
}
def run_xgboost(features, params=None, return_models=False, verbose=0):
X_train, y_train = df_train[features].values, df_train[targets].values
cv = KFold(n_splits=N_SPLITS, shuffle=True, random_state=0)
oof = np.zeros(len(X_train))
models = []
for fold, (idx_tr, idx_vl) in enumerate(cv.split(X_train)):
X_tr, y_tr = X_train[idx_tr], y_train[idx_tr]
X_vl, y_vl = X_train[idx_vl], y_train[idx_vl]
model = XGBRegressor(**params)
eval_set = [(X_vl, y_vl)]
model.fit(X_tr, y_tr, eval_set=eval_set, verbose=verbose)
oof[idx_vl] = model.predict(X_vl)
models.append(model)
r = mse(y_vl, oof[idx_vl], squared=False)
print(f"Fold {fold} rmse {r:.4}")
print(f"OOF mse: {mse(y_train, oof, squared=False): .4}")
if return_models:
return models
# Baseline model with original features
features = list(df_train.columns[1:9])
targets = "MedHouseVal"
run_xgboost(features, params=XGB_PARAMS, return_models=False)
# Add distance to cities feature and run xg_boost
add_distance_to_nearest_cities(df_train)
city_features = list(c for c in df_train.columns if c.startswith("to_"))
run_xgboost(features + city_features, params=XGB_PARAMS, return_models=False)
# **Observation**
# A bit improvement. Let's see if adding coastline_feature would help. Then we add all features
# Add distance to coastline feature & run xg_boost
add_distance_to_coastlines(df_train)
coastline_features = list(c for c in df_train.columns if c.startswith("coast_"))
run_xgboost(features + coastline_features, params=XGB_PARAMS, return_models=False)
# **Observation**
# This one has big improvement. Now we combine all
xgb_models = run_xgboost(
features + coastline_features + city_features, params=XGB_PARAMS, return_models=True
)
# **Observation**
# We see a gradual improvement from original features (rmse:0.5577) ---> all features (incl. city_features & coastline_features) (rmse:0.5552).
# So we'll use xgb_models with all features to predict for df_test and make submission
# Add coastline_feature & city_feature to df_test
add_distance_to_nearest_cities(df_test)
add_distance_to_coastlines(df_test)
def submit(models, name):
y_preds = np.mean(
[
m.predict(df_test[features + coastline_features + city_features].values)
for m in models
],
axis=0,
)
submission = pd.DataFrame(data={"id": df_test.index, target: y_preds})
submission.loc[submission[target].gt(5), target] = 5
submission.to_csv(f"submission_{name}.csv", index=None)
submit(xgb_models, "xgb")
df_xgb = pd.read_csv("/kaggle/working/submission_xgb.csv", index_col=0)
target = "MedHouseVal"
df = df_xgb
df.to_csv("submission.csv")
|
# # My CycleGAN is somewhat of a painter itself
# This notebook is part of the Machine Learning in Practice Course 2021 at Radboud University. With it, we are participating in the "I am somewhat of a painter myself" challenge.
# **Objective of the challenge**: Build a GAN that generates 7,000 to 10,000 Monet-style images
# ## Outline of this notebook
# 0. Imports
# 1. Exploratory Data Analysis (EDA) (short version)
# 2. Data Loading
# 3. Data Augmentation
# 4. The model
# 1. Generator
# 2. Discriminator
# 3. CycleGan
# 5. Model Training
# 6. Results
# 7. References
# 8. Exhaustive EDA
# ## Imports
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
import tensorflow_addons as tfa
from kaggle_datasets import KaggleDatasets
import matplotlib.pyplot as plt
import numpy as np
import os, random, json, PIL, shutil, re, imageio, glob
try:
tpu = tf.distribute.cluster_resolver.TPUClusterResolver()
print("Device:", tpu.master())
tf.config.experimental_connect_to_cluster(tpu)
tf.tpu.experimental.initialize_tpu_system(tpu)
strategy = tf.distribute.experimental.TPUStrategy(tpu)
except:
strategy = tf.distribute.get_strategy()
print("Number of replicas:", strategy.num_replicas_in_sync)
AUTOTUNE = tf.data.experimental.AUTOTUNE
print(tf.__version__)
#
# ## Exploratory Data Analysis
# ## Visualization of a few images
# Let's have a first look at the data we got. All the images for the challenge are of the size 256x256 and sorted by type already
BASE_PATH = "../input/gan-getting-started"
MONET_PATH = os.path.join(BASE_PATH, "monet_jpg")
PHOTO_PATH = os.path.join(BASE_PATH, "photo_jpg")
import cv2
import math
import random
def load_images(paths):
images = []
for img in paths:
try:
img = cv2.imread(img)
img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)
except:
print("Could not load {}".format(img))
images.append(img)
return images
def show_folder_info(path):
d_image_sizes = {}
for image_name in os.listdir(path):
image = cv2.imread(os.path.join(path, image_name))
d_image_sizes[image.shape] = d_image_sizes.get(image.shape, 0) + 1
for size, count in d_image_sizes.items():
print(f"shape: {size}\tcount: {count}")
def visualize_images(images):
plt.figure(figsize=(16, 16))
w = int(len(images) ** 0.5)
h = math.ceil(len(images) / w)
for idx, image in enumerate(images):
plt.subplot(h, w, idx + 1)
plt.imshow(image)
plt.axis("off")
plt.show()
MONET_IMAGES = [os.path.join(MONET_PATH, file) for file in os.listdir(MONET_PATH)]
monet_images = load_images(MONET_IMAGES)
PHOTO_IMAGES = [os.path.join(PHOTO_PATH, file) for file in os.listdir(PHOTO_PATH)]
photo_images = load_images(PHOTO_IMAGES)
# ### Monet Styled images
visualize_images(random.sample(monet_images, 15))
# ### Photos
visualize_images(random.sample(photo_images, 15))
# *Visual Inspection*
# Comparing the Monet images and the photos we can obviously see which are paintings and which are photos. The photos have way smoother transitions, the monet images "jump" in colours. Monet was a impressionist painter, an art style which is characterized by a small but still visible brush strokes.
# So our final images should also have this characteristic but showing the content of the content images.
# Further, the colours of photos are more natural than the colours of the mone style images. This is not too surprising, but we want to keep this in mind when judging the final output of our GAN
# ### Data Stats:
print("Monet Picture Overview")
show_folder_info(MONET_PATH)
print("Content Picture Overview")
show_folder_info(PHOTO_PATH)
|
import numpy as np
import pandas as pd
data = pd.read_csv("../input/digit-recognizer/train.csv")
data.head()
labels = data[data.columns[0]]
pixels = data.columns[1:]
import numpy as np
a = np.array(data.loc[:, pixels])
print(a.shape)
test = np.reshape(a, (a.shape[0], 28, 28, 1))
print(test.shape)
test = test / 256
import matplotlib.pyplot as plt
plt.imshow(test[1]), print(labels[1])
from keras.layers import (
Input,
Lambda,
Conv1D,
Conv2D,
BatchNormalization,
LeakyReLU,
ZeroPadding2D,
UpSampling2D,
)
from keras.layers import (
Conv2D,
MaxPool2D,
Dropout,
Dense,
Lambda,
Input,
concatenate,
GlobalAveragePooling2D,
AveragePooling2D,
Flatten,
)
import tensorflow as tf
from keras.models import load_model, Model
epoch = 8
batch_size = 64
y = list(labels)
len_y = len(y)
len_y
y_onehot = np.zeros((len_y, 10))
y_onehot.shape
for i in range(len_y):
y_onehot[i, y[i]] = 1
y[0:5]
y_onehot
x = test
y = y_onehot
model = tf.keras.Sequential()
model.add(
Conv2D(filters=16, kernel_size=(3, 3), padding="Same", input_shape=(28, 28, 1))
)
model.add(LeakyReLU(alpha=0.05))
model.add(MaxPool2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Conv2D(filters=32, kernel_size=(3, 3), padding="Same"))
model.add(LeakyReLU(alpha=0.05))
model.add(MaxPool2D(pool_size=(2, 2)))
model.add(Conv2D(filters=64, kernel_size=(3, 3), padding="Same"))
model.add(LeakyReLU(alpha=0.05))
model.add(MaxPool2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Conv2D(filters=32, kernel_size=(3, 3), padding="Same"))
model.add(LeakyReLU(alpha=0.05))
model.add(MaxPool2D(pool_size=(2, 2), strides=(2, 2)))
model.add(Flatten())
model.add(Dense(256))
model.add(LeakyReLU(alpha=0.05))
model.add(Dropout(0.5))
model.add(Dense(10, activation="softmax"))
model.summary()
opt = tf.keras.optimizers.Adam(learning_rate=0.008)
model.compile(optimizer=opt, loss="categorical_crossentropy", metrics=["accuracy"])
history = model.fit(
x, y, validation_split=0.2, batch_size=128, epochs=epoch, shuffle=True
)
import matplotlib.pyplot as plt
train_loss = history.history["loss"]
val_loss = history.history["val_loss"]
xc = range(8)
plt.figure()
plt.plot(xc, train_loss)
plt.plot(xc, val_loss)
|
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.preprocessing import OrdinalEncoder
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from tensorflow import keras
from tensorflow.keras.utils import to_categorical
from tensorflow.keras.optimizers import Adam
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import (
BatchNormalization,
Conv2D,
MaxPooling2D,
Dense,
Dropout,
Flatten,
Activation,
)
from keras.callbacks import Callback
# # Importing tools that we needed to predict with best accuracy is important
# 1. We have different methods to predict the prices
# 2. We can compare them and find the best score
df_train = pd.read_csv(
"/kaggle/input/house-prices-advanced-regression-techniques/train.csv"
)
df_train
# # Learning a data
# I am going to be fimiliar with columns and types of the data
df_train.describe()
df_train.info()
# # NaN values or missed datas
# I checked how many missed information is in every column.
#
print(df_train.isna().sum())
len(df_train.columns)
# # Fill the information that has been omitted
# **ffill** fills the column with information taken from a former row from up to down
# **bfill** is the opposite of ffill
# 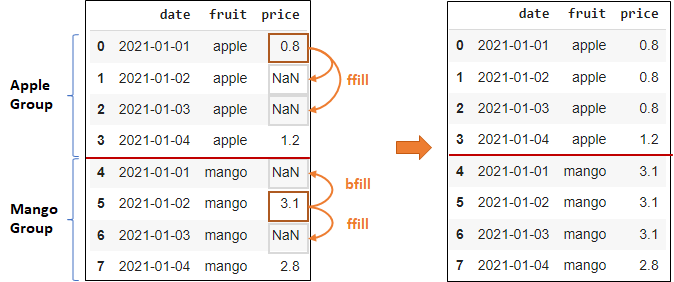
#
df_train.isna().sum()
df_train.isnull()
# # Visualisation
# * Using heatmap we can see how columns correlated with each others
# * matplotlib can show the range of a specific column especially SalesPrice that we predict
plt.figure(figsize=(25, 16))
sns.heatmap(df_train.corr(), annot=True)
plt.show()
x = df_train.SalePrice.values
plt.plot(x, ".", color="g")
# # Erasing unwanted data
# * I deleted the data that affects negatively to the model's accuracy.
df_train = df_train[df_train["SalePrice"] < 700000]
x = df_train.SalePrice.values
plt.plot(x, ".", color="g")
sns.histplot(df_train.SalePrice)
print("Skewness: %f" % df_train["SalePrice"].skew())
print("Kurtosis: %f" % df_train["SalePrice"].kurt())
sns.set()
cols = [
"SalePrice",
"OverallQual",
"GrLivArea",
"GarageCars",
"TotalBsmtSF",
"FullBath",
"YearBuilt",
]
sns.pairplot(df_train[cols], height=2.5)
plt.show()
# # Missing data information
# * We need to clean the missing datas that we have
# * If the percentage of NaN values in each column is so high , those datas can be deleted
full = df_train.isnull().sum().sort_values(ascending=False)
percent = (df_train.isnull().sum() / df_train.isnull().count()).sort_values(
ascending=False
)
nan_data = pd.concat([full, percent], axis=1, keys=["Total", "Percent"])
nan_data.head(20)
# deleting NaN value existed rows
df_train = df_train.drop((nan_data[nan_data["Total"] > 100]).index, 1)
df_train = df_train.fillna(method="bfill")
if df_train.isna().sum().max() > 0:
df_train = df_train.fillna(method="ffill")
df_train.isna().sum().max()
# # Ready data
# * Now I can train and check the clean data
df_train.info()
# # Encoding
# * It is important to check data to choose an appropriate encoder.
# * I have used ordinal encoder because we have no more than 20 classes for each column.
encode = OrdinalEncoder()
obj = [
"MSZoning",
"Street",
"LotShape",
"LandContour",
"Utilities",
"LotConfig",
"LandSlope",
"Neighborhood",
"Condition1",
"Condition2",
"BldgType",
"HouseStyle",
"RoofStyle",
"RoofMatl",
"Exterior1st",
"Exterior2nd",
"ExterQual",
"ExterCond",
"Foundation",
"Heating",
"HeatingQC",
"CentralAir",
"Electrical",
"KitchenQual",
"Functional",
"PavedDrive",
"SaleType",
"SaleCondition",
"MasVnrType",
"BsmtQual",
"BsmtExposure",
"BsmtFinType1",
"GarageType",
"BsmtFinType2",
"GarageFinish",
"GarageQual",
"GarageQual",
"GarageCond",
"BsmtCond",
]
df_train[obj] = encode.fit_transform(df_train[obj])
df_train.info()
x_train = df_train.drop("SalePrice", axis=1).values
y_train = df_train["SalePrice"].values
x_train, a_test, y_train, b_test = train_test_split(
x_train, y_train, test_size=0.15, random_state=42
)
# # Test data cleaning
# * the same cleaning process occurs here
df_test = pd.read_csv(
"/kaggle/input/house-prices-advanced-regression-techniques/test.csv"
)
df_test
df_test.isna().sum().max()
full = df_test.isnull().sum().sort_values(ascending=False)
percent = (df_test.isnull().sum() / df_test.isnull().count()).sort_values(
ascending=False
)
nan_data = pd.concat([full, percent], axis=1, keys=["Overall", "Percent"])
nan_data
df_test = df_test.fillna(method="bfill")
df_test = df_test.fillna(method="ffill")
df_test.isna().sum().max()
encode = OrdinalEncoder()
column = list(df_test.columns)
obj = []
v = []
for i in column:
if type(df_test[i].values[1]) == str:
obj.append(i)
df_test[obj] = encode.fit_transform(df_test[obj])
df_test.info()
ID = df_test["Id"].values
ID
df_test = df_test[
[
"Id",
"MSSubClass",
"MSZoning",
"LotArea",
"Street",
"LotShape",
"LandContour",
"Utilities",
"LotConfig",
"LandSlope",
"Neighborhood",
"Condition1",
"Condition2",
"BldgType",
"HouseStyle",
"OverallQual",
"OverallCond",
"YearBuilt",
"YearRemodAdd",
"RoofStyle",
"RoofMatl",
"Exterior1st",
"Exterior2nd",
"MasVnrType",
"MasVnrArea",
"ExterQual",
"ExterCond",
"Foundation",
"BsmtQual",
"BsmtCond",
"BsmtExposure",
"BsmtFinType1",
"BsmtFinSF1",
"BsmtFinType2",
"BsmtFinSF2",
"BsmtUnfSF",
"TotalBsmtSF",
"Heating",
"HeatingQC",
"CentralAir",
"Electrical",
"1stFlrSF",
"2ndFlrSF",
"LowQualFinSF",
"GrLivArea",
"BsmtFullBath",
"BsmtHalfBath",
"FullBath",
"HalfBath",
"BedroomAbvGr",
"KitchenAbvGr",
"KitchenQual",
"TotRmsAbvGrd",
"Functional",
"Fireplaces",
"GarageType",
"GarageYrBlt",
"GarageFinish",
"GarageCars",
"GarageArea",
"GarageQual",
"GarageCond",
"PavedDrive",
"WoodDeckSF",
"OpenPorchSF",
"EnclosedPorch",
"3SsnPorch",
"ScreenPorch",
"PoolArea",
"MiscVal",
"MoSold",
"YrSold",
"SaleType",
"SaleCondition",
]
]
x_test = df_test.values
df_test.info()
x_test.shape
# # Now we can train and predict the prices
# * I prefer to Linear model , if you want you can use metrics
Model = Sequential(
[
Dense(64, input_shape=[74], activation="relu"),
Dense(64, activation="relu"),
Dense(32, activation="relu"),
Dense(32, activation="relu"),
Dense(16, activation="relu"),
Dense(1, activation="linear"),
]
)
class myCallback(Callback):
def on_epoch_end(self, epoch, logs={}):
if logs.get("loss") < 0.01:
print("\n accuracy reached the highest point,cancelling session")
self.model.stop_training = True
callback = myCallback()
num_epoch = 1000
Model.compile(optimizer="rmsprop", loss="mse", metrics=["mae"])
hist = Model.fit(x_train, y_train, epochs=num_epoch, batch_size=16, callbacks=callback)
Model.evaluate(a_test, b_test)
model = LinearRegression()
model.fit(x_train, y_train)
predict = model.predict(x_test)
prediction = Model.predict(x_test)
prediction = prediction.reshape(-1)
# # Results
# *My model score works up to 85 %
submission = pd.DataFrame({"Id": ID, "SalePrice": prediction})
submission
submission.to_csv("submission.csv", index=False)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import numpy as np
import tensorflow as tf
import optuna
import matplotlib.pyplot as plt
x = np.linspace(-2, 2, 1000)
plt.plot(x, 2 * x**4 - 5 * x**2 + x)
# ### 使用Optuna超参数调整
# 普通的多项式x最优值预测也是可以的,不一定非要模型里面的超参数
def objective(trial):
x = trial.suggest_uniform("x", -2, 2)
return 2 * x**4 - 5 * x**2 + x
study = optuna.create_study()
study.optimize(objective, n_trials=30)
study.best_value, study.best_params
# ### 结果可视化
# b代表蓝色
plt.plot(x, 2 * x**4 - 5 * x**2 + x, "b")
# ro 代表红色圆圈
plt.plot(study.best_params["x"], study.best_value, "ro")
plt.legend(["objective function f(x)", "minimized solution"])
|
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import sklearn.linear_model
from sklearn.datasets import fetch_openml
mnist = fetch_openml("mnist_784", version=1, as_frame=False)
mnist.keys()
x, y = mnist["data"], mnist["target"]
y.shape
x[0].shape
plt.imshow(x[0].reshape(28, 28))
x_train, x_test, y_train, y_test = x[:60000], x[60000:], y[:60000], y[60000:]
y_test
y_train_5 = list(map(lambda x: True if (x == "5") else False, y_train))
y_test_5 = list(map(lambda x: True if (x == "5") else False, y_test))
from sklearn.linear_model import SGDClassifier
sgd_clf = SGDClassifier(random_state=42)
sgd_clf.fit(x_train, y_train_5)
some = x[0]
y[0]
sgd_clf.predict([some])
from sklearn.model_selection import cross_val_score
cross_val_score(sgd_clf, x_train, y_train_5, cv=5, scoring="accuracy")
from sklearn.base import BaseEstimator
class asla_5_olmayanlar(BaseEstimator):
def fit(self, x, y=None):
return self
def predict(self, x):
return np.zeros((len(x), 1), dtype=bool)
asla_5 = asla_5_olmayanlar()
cross_val_score(asla_5, x_train, y_train_5, cv=5, scoring="accuracy")
from sklearn.model_selection import cross_val_predict
y_train_pred = cross_val_predict(sgd_clf, x_train, y_train_5, cv=5)
from sklearn.metrics import confusion_matrix
confusion_matrix(y_train_5, y_train_pred)
perfect_Res = y_train_5
confusion_matrix(y_train_5, perfect_Res)
from sklearn.metrics import precision_score, recall_score, f1_score
precision_score(y_train_5, y_train_pred)
recall_score(y_train_5, y_train_pred)
f1_score(y_train_5, y_train_pred)
|
# Welcome!
# This notebook contains a mixture of Pandas and PySpark code to perform data analysis and anomaly detection on the given dataset.
# ## **Features**
# ___
# - Uses both Pandas and PySpark for data processing and analysis.
# - Uses caching to take advantage of the limited memory situation.
# - [Analyzes missing data in all the columns.](#1)
# - [Which columns containig object data type can potentially be used as categorical columns.](#2)
# - [Only event `notebook_click` has `page` info, checks if that is true](#3)
# - [Shows which events are more likely to occur in different levels of the game](#4)
# - [Provides anomaly detection for the given dataset in the `text` column.](#5)
# - [Provides pattern detection for the `text` column](#6)
# ## **Data Analysis**
# ___
# Data analysis is performed using Pandas and PySpark. PySpark is used to perform distributed computing on large datasets, while Pandas is used to perform data manipulation and exploration.
# ## **Anomaly Detection & Pattern Analysis of Text data.**
# * Anomaly detection is performed by identifying `text` anomalies in the dataset.
# * Rows containing text containing 7, 8, or 10 splits are flagged as anomalies.
# Quick look at the columns (paraphasing the general understandings only)
# ___
# * **session_id:** A unique identifier for each gameplay session
# * **index:** A number indicating the order in which each event occurred within a session
# * **elapsed_time:** How much time has passed in the session (in milliseconds) when each event was recorded
# * **event_name:** A description of the type of event that occurred (e.g. click, hover, etc.)
# * **name:** A more specific description of the event that occurred (e.g. which button was clicked)
# * **level:** The level of the game where the event occurred (ranging from 0 to 22)
# * **page:** The page number of the event (only for notebook-related events)
# * **room_coor_x, room_coor_y:** The coordinates of the player's click within the in-game room (only for click events)
# * **screen_coor_x, screen_coor_y:** The coordinates of the player's click within the player's screen (only for click events)
# * **hover_duration:** How long the player hovered over an object (in milliseconds)
# * **text:** The text that the player saw during the event
# * **fqid:** A unique identifier for each event
# * **room_fqid:** A unique identifier for the room where the event occurred
# * **text_fqid:** A unique identifier for the text that the player saw during the event
# * **fullscreen:** Whether the player was in fullscreen mode
#
# * **hq:** Whether the game was in high-quality
#
# * **music:** Whether the game music was on or off
#
# * **level_group:** Which group of levels the event belongs to (0-4, 5-12, or 13-22)
# *This notebook showcases the use of both Pandas and PySpark for data analysis and anomaly detection. The code is written in a clear and concise manner, making it easy to follow along and understand the steps involved in the analysis.*
# ___
# ### **Import Pandas**
import pandas as pd
# ### **Installing PySpark**
# ### Loading the dataset
from pyspark.sql import SparkSession
# create a SparkSession object
spark = SparkSession.builder.appName("predict-student-performance").getOrCreate()
# reading the data & caching it using `persist()`
train = spark.read.csv(
"/kaggle/input/predict-student-performance-from-game-play/train.csv",
header=True,
inferSchema=True,
)
# inspect infered schema
train.printSchema()
# ### Check first five rows of the training data
# ___
train_head = train.limit(5).toPandas()
train_head
# ### **The Dataset contains following Columns and Datatypes**
# ___
train_head.dtypes.to_frame(name="Data Types")
# Missing Data Inspection
# ___
from pyspark.sql.functions import col, sum
# Count the number of missing values in each column
missing_value_counts = train.agg(
*[sum(col(c).isNull().cast("int")).alias(c) for c in train.columns]
)
missing_value_counts = missing_value_counts.toPandas()
missing_value_counts
# ### Total Rows of Data
total_rows = train.select(
"session_id"
).count() # for faster processing we are usning one column
print(f"Total rows:{total_rows}")
# In this list comprehension we store data
column_with_missing_vals, data_type, missing_value_percentage = zip(
*[
(
col,
train_head[col].dtype,
round(((missing_value_counts[col][0]) / total_rows) * 100, 2),
)
for col in train.columns
if missing_value_counts[col][0] > 0
]
)
print(f"{len(column_with_missing_vals)} Features contain missing values")
# ### Plotting the Missing value percentage in each column
import matplotlib.pyplot as plt
num_rows = 2
num_cols = 7
# Create subplots with the specified number of rows and columns
fig, axes = plt.subplots(num_rows, num_cols, figsize=(20, 10))
# Flatten the axes array for easy iteration
axes = axes.flatten()
# Plot each column with missing values as a pie chart
for i, col in enumerate(column_with_missing_vals):
# Calculate the percentage of missing values
missing_percent = missing_value_percentage[i]
present_percent = 100 - missing_percent
# Create a pie chart with missing and present values
axes[i].pie(
[missing_percent, present_percent],
autopct="%1.1f%%",
startangle=90,
colors=["#CE3E3E", "#7CB342"],
)
axes[i].set_title(f"{col}[{data_type[i]}]")
# Remove unused subplots and adjust spacing
# for i in range(len(column_with_missing_vals), num_rows*num_cols):
# fig.delaxes(axes[i])
fig.tight_layout()
fig.legend(["Missing", "Present"], loc="upper right")
# Set the plot title
fig.suptitle("Percentage of Missing Values in Columns", fontweight="bold", fontsize=16)
# Show the plot
plt.show()
# Which of the Object type columns could be categorical? 🤔
# ___
# from pyspark_dist_explore import hist
from pyspark.sql.types import StringType
# here we are going to make use of the 'train_head' to extract the datatypes easily 😜
cols = train_head.select_dtypes(include="object").columns.tolist()
vals = [train.select(col).distinct().count() for col in cols]
unique_vals = pd.DataFrame({"cols": cols, "unique_vals": vals})
# Sort DataFrame based on unique_vals column
unique_vals = unique_vals.sort_values(by="unique_vals", ascending=True).reset_index(
drop=True
)
plt.bar("cols", "unique_vals", data=unique_vals)
plt.xticks(rotation=90)
plt.title("Unique value count for object Data Types")
plt.xlabel("Column Names")
plt.ylabel("Counts")
plt.show()
# `level_group`,`event_name` and `name` should be categorized
# Only event 'notebook_click' has 'page' info. Let's check if there are any outliers.
# ___
# event_name` vs page
from pyspark.sql.functions import col
unique_event_names = (
train.select(col("event_name")).distinct().rdd.flatMap(lambda x: x).collect()
)
print("\n".join(unique_event_names))
from pyspark.sql.functions import col, isnull
event_vs_page = train.select("event_name", "page").filter(
col("event_name") == "notebook_click"
)
event_vs_page.cache() # caching the dataframe
from pyspark.sql.functions import min, max
# Find the maximum value in the "page" column
max_value = event_vs_page.agg(max("page")).take(1)[0][0]
min_value = event_vs_page.agg(min("page")).take(1)[0][0]
# Print the maximum value
print("Max value in page: ", max_value)
print("Min value in page: ", min_value)
event_vs_page.count() == (total_rows - missing_value_counts["page"][0])
event_vs_page.unpersist() # remove it from cache
# **Conclusion:**
# ___
# * `page` column only contains values for `notebook_click` events
# Which events are more likely to occur in different levels of the game?
# ___
level_vs_event = train.groupBy("event_name").pivot("level").agg({"event_name": "count"})
level_vs_event.cache()
level_vs_event = level_vs_event.toPandas()
level_vs_event
import matplotlib.pyplot as plt
# Generate the subplots with 4 rows and 6 columns
fig, axs = plt.subplots(nrows=4, ncols=6, figsize=(40, 20))
# Flatten the axs array so that we can iterate over it more easily
axs = axs.flatten()
# Loop over each column in the level_vs_event DataFrame and create a boxplot
for i, col in enumerate(level_vs_event.columns[1:]):
if i > 22:
break
else:
ax = axs[i]
# Create the plot for the current column
level_vs_event.plot(x="event_name", y=col, kind="bar", ax=ax, legend=False)
# Set the title for the current axis
ax.set_title(f"Level: {col}")
# Adjust the layout of the subplots
plt.suptitle(
"Which events are more likely to occur in different levels?\n",
fontsize=24,
fontweight="bold",
)
plt.subplots_adjust(wspace=0.3, hspace=0.5)
plt.tight_layout()
# Show the figure
plt.show()
# Are there any Inconsistent Data in the text column?
# ___
#
from pyspark.sql.functions import monotonically_increasing_id
# Selecting the columns we need
sess_idx_txt = train.select("session_id", "index", "text")
# Adding a row number to track anomalies
sess_idx_txt = sess_idx_txt.withColumn("row_index", monotonically_increasing_id())
# Let's put the row_index column at front
sess_idx_txt = sess_idx_txt.select("row_index", *sess_idx_txt.columns[:-1])
sess_idx_txt.cache()
sess_idx_txt.orderBy("session_id", "index").show(5)
# ### Let's try and split the texts by '.' and see if we can detect some anomaly.
from pyspark.sql.functions import length, size, split, monotonically_increasing_id
# Let's just split the texts with '.' and see what pops up, going on a blind intuition here.
leng = sess_idx_txt.withColumn("text_splits", size(split(sess_idx_txt["text"], "\.")))
sess_idx_txt.unpersist()
leng.show(5)
# Let's check the min and max values in the 'text_splits' column, because... erm... I have no idea what awaits.
max_value = leng.agg(max("text_splits")).take(1)[0][0]
min_value = leng.agg(min("text_splits")).take(1)[0][0]
# Print the minimum and maximum values
print("Max value in page: ", max_value)
print("Min value in page: ", min_value)
from tabulate import tabulate
# Assuming that leng is a dataframe with a 'text_splits' column
unique_values = leng.select("text_splits").distinct().collect()
# Convert the list of Row objects to a list of values
unique_values = [row[0] for row in unique_values]
# let's sort out the unique_values
unique_values = sorted(unique_values)
# just being fancy here
print("Number of text splits:")
print(tabulate([unique_values], tablefmt="psql", stralign="center"))
# ##### Interesting! the split was successful I'd say. But we aren't finished here
# ##### Let's check out some of the sample texts that has the available split by '.'
for j in unique_values:
print(f"\033[34;4mFor \033[0m{j} \033[34;4m text split(s)\033[0m".center(100))
for i in (
leng.select(leng.row_index, leng.text).filter(leng.text_splits == j).head(5)
):
# Printing the row where the anomaly exists
print(
"\n\033[31mRow Number:\033[0m ", i.row_index
) # Those numbers are ANSI escape codes for fancy outputs in color.
# printing the text containing anomaly
print("\n\033[32mText:\033[35m ", i.text)
# printing the data type
print("\033[33m\nText Data Type: ", type(i.text))
print(
"\033[32m------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------\033[0m"
)
# #### Looks like unique values with 7, 8, 10 length has some unrecognizable values
# Checking the heads
for j in [7, 8, 10]:
print(f"\033[34;4mFor \033[0m{j} \033[34;4m text split(s)\033[0m".center(100))
for i in (
leng.select(leng.row_index, leng.text).filter(leng.text_splits == j).head(5)
):
# Printing the row where the anomaly exists
print(
"\033[31mAnomaly Detected! at row:\033[0m ", i.row_index
) # Those numbers are ANSI escape codes for fancy outputs in color.
# printing the text containing anomaly
print("\n\033[32mText:\033[35m ", i.text)
# printing the data type
print("\033[33m\nText Data Type: ", type(i.text))
print(
"\033[32m------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------\033[0m"
)
# Checking the tails
for j in [7, 8, 10]:
print(f"\033[34;4mFor \033[0m{j} \033[34;4m text split(s)\033[0m".center(100))
for i in (
leng.select(leng.row_index, leng.text).filter(leng.text_splits == j).tail(5)
):
# Printing the row where the anomaly exists
print(
"\033[31mAnomaly Detected! at row:\033[0m ", i.row_index
) # Those numbers are ANSI escape codes for fancy outputs in color.
# printing the text containing anomaly
print("\n\033[32mText:\033[35m ", i.text)
# printing the data type
print("\033[33m\nText Data Type: ", type(i.text))
print(
"\033[32m------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------\033[0m"
)
rows_with_text_anomaly = leng.filter(leng.text_splits.isin([7, 8, 10]))
rows_with_text_anomaly.show()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
train = pd.read_csv("../input/nlp-getting-started/train.csv")
test = pd.read_csv("../input/nlp-getting-started/test.csv")
print(train.head(), "\n")
print(train.info(), "\n")
print(test.head(), "\n"),
print(test.info())
print(train.isnull().sum(), "\n")
print(test.isnull().sum())
train = train.drop(["keyword", "location"], axis=1)
test = test.drop(["keyword", "location"], axis=1)
print(train.head(), "\n")
print(test.head())
import matplotlib.pyplot as plt
label_count = train["target"].value_counts()
labels = ["Negative", "Positive"]
plt.pie(label_count, labels=labels, autopct="%1.1f%%")
plt.show()
print(label_count[0] / label_count[1])
from tifffile import imread
from wordcloud import WordCloud
negative_cases = "".join(train[train["target"] == 0]["text"].values)
positive_cases = "".join(train[train["target"] == 1]["text"].values)
fig, axs = plt.subplots(1, 2, figsize=(10, 4))
wc1 = WordCloud(background_color="white").generate(negative_cases)
wc2 = WordCloud(background_color="white").generate(positive_cases)
axs[0].imshow(wc1, interpolation="bilinear")
axs[0].set_title("Negative")
axs[1].imshow(wc2, interpolation="bilinear")
axs[1].set_title("Positive")
train["text"] = train["text"].str.replace(r"t|https|co|Û_", "", regex=True)
test["text"] = test["text"].str.replace(r"t|https|co|Û_", "", regex=True)
negative_cases = "".join(train[train["target"] == 0]["text"].values)
positive_cases = "".join(train[train["target"] == 1]["text"].values)
fig, axs = plt.subplots(1, 2, figsize=(10, 4))
wc1 = WordCloud(background_color="white").generate(negative_cases)
wc2 = WordCloud(background_color="white").generate(positive_cases)
axs[0].imshow(wc1, interpolation="bilinear")
axs[0].set_title("Negative")
axs[1].imshow(wc2, interpolation="bilinear")
axs[1].set_title("Positive")
plt.show()
sentences_train = train["text"].values
sentences_test = test["text"].values
print(np.mean([len(text) for text in train["text"]]))
print(train["text"][7])
print(len(train["text"][7]))
import keras
from keras.models import Sequential
from keras.layers import (
Dense,
Conv2D,
MaxPool2D,
Flatten,
BatchNormalization,
Activation,
LSTM,
CuDNNLSTM,
Embedding,
Dropout,
)
from keras.preprocessing.image import ImageDataGenerator
from tensorflow.keras.preprocessing.text import Tokenizer
from tensorflow.keras.preprocessing.sequence import pad_sequences
from keras.optimizers import Adam
tokenizer = Tokenizer()
tokenizer.fit_on_texts(sentences_train)
sequences_train = tokenizer.texts_to_sequences(sentences_train)
# This is used for Embedding layer afterwards
num_words = len(tokenizer.word_index)
print(num_words)
maxlen_tokens = 20
# The reason that the maxlength is different than the true max length is that the texts are tokenized
padded_sequences_train = pad_sequences(
sequences_train, maxlen=maxlen_tokens, padding="post", truncating="post"
)
padded_sequences_train[0]
num_validation_samples = round(0.2 * len(padded_sequences_train))
seq_train = padded_sequences_train[num_validation_samples:]
labels_train = train["target"].values[num_validation_samples:]
seq_validate = padded_sequences_train[:num_validation_samples]
labels_validate = train["target"].values[:num_validation_samples]
print(len(seq_train))
print(len(seq_validate))
print(seq_train[0])
print(seq_train.shape)
# ## Model Building
model = Sequential()
model.add(Embedding(num_words, 32, input_length=maxlen_tokens))
model.add(CuDNNLSTM(128, return_sequences=True))
model.add(CuDNNLSTM(128))
model.add(Dense(units=256, activation="relu"))
model.add(Dense(units=1, activation="sigmoid"))
model.build(input_shape=(None, maxlen_tokens))
model.summary()
optimizers = Adam(learning_rate=0.00001, decay=0.000001)
model.compile(optimizer=optimizers, loss="binary_crossentropy", metrics=["accuracy"])
history = model.fit(
seq_train, labels_train, validation_data=(seq_validate, labels_validate), epochs=40
)
plt.plot(history.history["accuracy"])
plt.plot(history.history["val_accuracy"])
plt.plot(history.history["loss"])
plt.plot(history.history["val_loss"])
plt.title("Model Evaluation")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(["Accuracy", "Validation Accuracy", "Loss", "Validation Loss"])
plt.show()
model_1 = Sequential()
model_1.add(Embedding(num_words, 32, input_length=maxlen_tokens))
model_1.add(CuDNNLSTM(128, return_sequences=True))
model_1.add(Dropout(0.2))
model_1.add(CuDNNLSTM(128))
model_1.add(Dropout(0.2))
model_1.add(Dense(units=256, activation="relu"))
model_1.add(Dense(units=1, activation="sigmoid"))
model_1.build(input_shape=(None, maxlen_tokens))
model_1.summary()
optimizer_2 = Adam(learning_rate=0.00001, decay=0.000001)
model_1.compile(optimizer=optimizer_2, loss="binary_crossentropy", metrics=["accuracy"])
history_2 = model_1.fit(seq_train, labels_train, epochs=10)
plt.plot(history_2.history["accuracy"])
plt.plot(history_2.history["loss"])
plt.title("Model Evaluation")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(["Accuracy", "Validation Accuracy", "Loss", "Validation Loss"])
model_final = Sequential()
model_final.add(Embedding(num_words, 32, input_length=maxlen_tokens))
model_final.add(CuDNNLSTM(64, return_sequences=True))
model.add(BatchNormalization())
model_final.add(CuDNNLSTM(128))
model.add(BatchNormalization())
model_final.add(Dense(units=256, activation="relu"))
model.add(BatchNormalization())
model_final.add(Dense(units=1, activation="sigmoid"))
model_final.build(input_shape=(None, maxlen_tokens))
model_final.summary()
opt_final = Adam(learning_rate=0.00001, decay=0.000001)
model_final.compile(
optimizer=opt_final, loss="binary_crossentropy", metrics=["accuracy"]
)
hist_final = model_final.fit(seq_train, labels_train, epochs=15)
plt.plot(hist_final.history["accuracy"])
plt.plot(hist_final.history["loss"])
plt.title("Model Evaluation")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(["Accuracy", "Validation Accuracy", "Loss", "Validation Loss"])
plt.show()
sequences_test = tokenizer.texts_to_sequences(sentences_test)
padded_sequences_test = pad_sequences(
sequences_test, maxlen=maxlen_tokens, padding="post", truncating="post"
)
padded_sequences_test[7]
predictions = model_final.predict(padded_sequences_test, verbose=1)
print(predictions)
y_pred = model_final.predict(seq_validate, verbose=1)
import pandas as pd
print(labels_validate)
ypred = pd.DataFrame(y_pred)
ypred.iloc[y_pred < 0.5] = 0
ypred.iloc[y_pred >= 0.5] = 1
ypredf = ypred.astype(int, copy=True)
print(ypredf)
from sklearn.metrics import ConfusionMatrixDisplay
from sklearn.metrics import f1_score
F1 = f1_score(labels_validate, ypredf)
print(F1)
print(predictions)
pred = np.transpose(predictions)[0]
print(pred)
submission_df = pd.DataFrame()
submission_df["id"] = test["id"]
submission_df["target"] = list(map(lambda x: 0 if x < 0.5 else 1, pred))
print(submission_df.head())
submission_df["target"].value_counts()
submission_df.to_csv("submission.csv", index=False)
|
# # Import Libraries
from mlxtend.plotting import plot_decision_regions
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
sns.set()
import warnings
warnings.filterwarnings("ignore")
# # Basic Data Science and ML Pipeline
# Loading the dataset
diabetes = pd.read_csv("../input/pima-indians-diabetes-database/diabetes.csv")
# Print the first 10 rows of the dataframe.
diabetes.head(5)
# # EDA and statistical analysis
# gives information about the data types,columns, null value counts, memory usage for all the features
diabetes.info(verbose=True)
# **DataFrame.describe(**) method generates descriptive statistics that summarize the central tendency, dispersion and shape of a dataset’s distribution, excluding NaN values. This method tells us a lot of things about a dataset. One important thing is that the describe() method deals only with numeric values. It doesn't work with any categorical values. So if there are any categorical values in a column the describe() method will ignore it and display summary for the other columns unless parameter include="all" is passed.
# Now, let's understand the statistics that are generated by the describe() method:
# Count tells us the number of NoN-empty rows in a feature.
# Mean tells us the mean value of that feature.
# Std tells us the Standard Deviation Value of that feature.
# Min tells us the minimum value of that feature.
# 25%, 50%, and 75% are the percentile/quartile of each features. This quartile information helps us to detect Outliers.
# Max tells us the maximum value of that feature.
diabetes.describe()
diabetes.describe().T
diabetes_copy = diabetes.copy(deep=True)
diabetes_copy[
["Glucose", "BloodPressure", "SkinThickness", "Insulin", "BMI"]
] = diabetes_copy[
["Glucose", "BloodPressure", "SkinThickness", "Insulin", "BMI"]
].replace(
0, np.NaN
)
# showing the count of Nans
print(diabetes_copy.isnull().sum())
a = diabetes.hist(figsize=(20, 20))
diabetes_copy["Glucose"].fillna(diabetes_copy["Glucose"].mean(), inplace=True)
diabetes_copy["BloodPressure"].fillna(
diabetes_copy["BloodPressure"].mean(), inplace=True
)
diabetes_copy["SkinThickness"].fillna(
diabetes_copy["SkinThickness"].median(), inplace=True
)
diabetes_copy["Insulin"].fillna(diabetes_copy["Insulin"].median(), inplace=True)
diabetes_copy["BMI"].fillna(diabetes_copy["BMI"].median(), inplace=True)
p = diabetes_copy.hist(figsize=(20, 20))
## observing the shape of the data
diabetes.shape
diabetes.dtypes.value_counts()
print(diabetes.dtypes)
## null count analysis
import missingno as msno
a = msno.bar(diabetes)
## checking the balance of the data by plotting the count of outcomes by their value
color_wheel = {1: "#0392cf", 2: "#7bc043"}
colors = diabetes["Outcome"].map(lambda x: color_wheel.get(x + 1))
print(diabetes.Outcome.value_counts())
a = diabetes.Outcome.value_counts().plot(kind="bar")
from pandas.plotting import scatter_matrix
p = scatter_matrix(diabetes, figsize=(25, 25))
a = sns.pairplot(diabetes_copy, hue="Outcome")
plt.figure(figsize=(12, 10)) # on this line I just set the size of figure to 12 by 10.
p = sns.heatmap(
diabetes.corr(), annot=True, cmap="RdYlGn"
) # seaborn has very simple solution for heatmap
plt.figure(figsize=(12, 10)) # on this line I just set the size of figure to 12 by 10.
p = sns.heatmap(
diabetes_copy.corr(), annot=True, cmap="RdYlGn"
) # seaborn has very simple solution for heatmap
from sklearn.preprocessing import StandardScaler
sc_X = StandardScaler()
X = pd.DataFrame(
sc_X.fit_transform(
diabetes_copy.drop(["Outcome"], axis=1),
),
columns=[
"Pregnancies",
"Glucose",
"BloodPressure",
"SkinThickness",
"Insulin",
"BMI",
"DiabetesPedigreeFunction",
"Age",
],
)
X.head(5)
# X = diabetes.drop("Outcome",axis = 1)
y = diabetes_copy.Outcome
y.head(5)
# importing train_test_split
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=1 / 3, random_state=42, stratify=y
)
from sklearn.neighbors import KNeighborsClassifier
test_scores = []
train_scores = []
for i in range(1, 15):
knn = KNeighborsClassifier(i)
knn.fit(X_train, y_train)
train_scores.append(knn.score(X_train, y_train))
test_scores.append(knn.score(X_test, y_test))
## score that comes from testing on the same datapoints that were used for training
max_train_score = max(train_scores)
train_scores_ind = [i for i, v in enumerate(train_scores) if v == max_train_score]
print(
"Max train score {} % and k = {}".format(
max_train_score * 100, list(map(lambda x: x + 1, train_scores_ind))
)
)
## score that comes from testing on the datapoints that were split in the beginning to be used for testing solely
max_test_score = max(test_scores)
test_scores_ind = [i for i, v in enumerate(test_scores) if v == max_test_score]
print(
"Max test score {} % and k = {}".format(
max_test_score * 100, list(map(lambda x: x + 1, test_scores_ind))
)
)
plt.figure(figsize=(12, 5))
p = sns.lineplot(range(1, 15), train_scores, marker="*", label="Train Score")
p = sns.lineplot(range(1, 15), test_scores, marker="o", label="Test Score")
# Setup a knn classifier with k neighbors
knn = KNeighborsClassifier(11)
knn.fit(X_train, y_train)
knn.score(X_test, y_test)
value = 20000
width = 20000
plot_decision_regions(
X.values,
y.values,
clf=knn,
legend=2,
filler_feature_values={2: value, 3: value, 4: value, 5: value, 6: value, 7: value},
filler_feature_ranges={2: width, 3: width, 4: width, 5: width, 6: width, 7: width},
X_highlight=X_test.values,
)
plt.title("KNN with Diabetes Data")
plt.show()
|
# # Playground Series - Season 3, Episode 12
# With the same goal to give the Kaggle community a variety of fairly light-weight challenges that can be used to learn and sharpen skills in different aspects of machine learning and data science, we will continue launching the Tabular Tuesday in April every Tuesday 00:00 UTC, with each competition running for 2 weeks. Again, these will be fairly light-weight datasets that are synthetically generated from real-world data, and will provide an opportunity to quickly iterate through various model and feature engineering ideas, create visualizations, etc..
# # Libraries and settings
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.cluster import KMeans
from sklearn.preprocessing import MinMaxScaler
from sklearn.decomposition import PCA
from sklearn.ensemble import RandomForestClassifier, ExtraTreesClassifier
from sklearn.model_selection import RepeatedStratifiedKFold, cross_val_score, KFold
from sklearn.metrics import roc_auc_score
import os
import random
import xgboost as xgb
import lightgbm as lgb
from catboost import CatBoostClassifier
import optuna
import missingno as msno
sns.set_palette("muted")
seed = 42
np.random.seed(seed)
random.seed(seed)
os.environ["PYTHONHASHSEED"] = str(seed)
test = pd.read_csv("/kaggle/input/playground-series-s3e12/test.csv")
sample_submission = pd.read_csv(
"/kaggle/input/playground-series-s3e12/sample_submission.csv"
)
# ## Understanding the dataset
df = pd.read_csv("/kaggle/input/playground-series-s3e12/train.csv").drop("id", axis=1)
df
msno.matrix(
df,
figsize=(12, 8),
)
fig = plt.figure(figsize=(10, 6))
ax = sns.countplot(x=df["target"], linewidth=1, edgecolor="black", alpha=0.8)
for i in ax.containers:
ax.bar_label(
i,
)
fig.suptitle("Target variable distribution\n", fontsize=24)
ax.set_ylabel("")
plt.yticks([])
sns.despine()
plt.show()
# ### Findings
# - The number of available samples is very low
# - All the features are numeric
# - The target is binary 0 - 1
# - The target is unbalanced
# - There are no missing values
def oversample_minority_class(df, target_col):
"""Oversamples the minority class in a given DataFrame based on a target column."""
# Determine the value counts of the target column
target_counts = df[target_col].value_counts()
# Find the minority and majority classes
minority_class = target_counts.idxmin()
majority_class = target_counts.idxmax()
diff = target_counts[majority_class] - target_counts[minority_class]
if diff == 0:
return df
minority_df = df[df[target_col] == minority_class]
replacement = False
if len(minority_df) < diff:
replacement = True
balanced_df = pd.concat([df, minority_df.sample(n=diff, replace=replacement)])
return balanced_df
# ## Exploring and transforming the data
# ### Oversampling to balance the target variable
df = oversample_minority_class(df, "target")
fig = plt.figure(figsize=(10, 6))
ax = sns.countplot(x=df["target"], linewidth=1, edgecolor="black", alpha=0.8)
for i in ax.containers:
ax.bar_label(
i,
)
fig.suptitle("Target distribution after oversampling\n", fontsize=24)
ax.set_ylabel("")
plt.yticks([])
sns.despine()
plt.show()
fig = plt.figure(figsize=(10, 6))
mask = np.triu(np.ones_like(df.corr(), dtype=bool))
cmap = sns.diverging_palette(250, 15, s=75, l=40, n=9, as_cmap=True)
ax = sns.heatmap(df.corr(), annot=True, mask=mask, cmap=cmap)
fig.suptitle("Features correlation\n", fontsize=24)
plt.show()
# - The correlations are not very promising
# - Calc has a good correlation with the target
# - The other features are correlated to each other especially osmo
fig, ax = plt.subplots(nrows=3, ncols=2, figsize=(10, 10))
ax = ax.flatten()
for i, col in enumerate(["gravity", "ph", "osmo", "cond", "urea", "calc"]):
sns.kdeplot(data=df, x=col, ax=ax[i], fill=True, legend=False, hue="target")
ax[i].set_title(f"{col} Distribution")
ax[i].set_xlabel(None)
ax[i].set_ylabel(None)
fig.suptitle("Distribution of Features per Class\n", fontsize=24)
fig.legend(["Crystal (1)", "No Crystal (0)"])
plt.tight_layout()
plt.show()
# - Its is possible to observe that for each variable there are certain threshoulds where the class probability shifts.
# - Calc is the most promising feature
# - PH seems be useless from a univariant point of view
# - The other variables may be useful
# - We made a dummy predictor using only calc to estimate the target, the predictor got a score of .83 in the test set.
# ## KMEANS clustering
# Unfortunately we only have a few samples available and so we need to be careful when selection the amount of training features:
# If we selected so many variables we will fall into the curse of dimensionality which is caused by the relative lack of samples in comparison the amount of features.
# We choose to train our model with only features, one will calc and we'll create another categorical variable by clustering the remaining features.
# The idea behinde clustering is to understand and capture multivariant behaviors into a single 'cluster' variable.
# We choose to leave 'calc' out of the clustering in order to avoid redundancy.
cluster_cols = ["gravity", "ph", "osmo", "cond", "urea"]
scaler = MinMaxScaler()
df_norm = scaler.fit_transform(df[cluster_cols])
df_norm = pd.DataFrame(df_norm, columns=cluster_cols)
test_norm = scaler.transform(test[cluster_cols])
test_norm = pd.DataFrame(test_norm, columns=cluster_cols)
kmeans = KMeans(n_clusters=3, random_state=0)
df_norm["cluster"] = kmeans.fit_predict(df_norm)
df["cluster"] = df_norm["cluster"]
test["cluster"] = kmeans.predict(test_norm)
pca = PCA(n_components=2)
principal_components = pca.fit_transform(df_norm[cluster_cols])
fig = plt.figure(figsize=(10, 6))
ax = sns.scatterplot(
x=principal_components[:, 0],
y=principal_components[:, 1],
hue=df["cluster"],
style=df["target"],
s=100,
alpha=0.9,
)
fig.suptitle("Cluster vs Target\n", fontsize=24)
plt.xlabel("Principal Component 1")
plt.ylabel("Principal Component 2")
plt.tight_layout()
plt.show()
cluster_probs = df.groupby("cluster").agg(prob=("target", "mean"))
centroids = pd.DataFrame(
columns=["gravity", "ph", "osmo", "cond", "urea"],
data=scaler.inverse_transform(kmeans.cluster_centers_),
)
cluster_info = pd.concat([cluster_probs, centroids], axis=1)
cluster_info.index.name = "cluster"
cluster_info
df["kmeans_feature"] = df["cluster"].replace(
cluster_probs.to_dict()["prob"]
) + np.random.normal(loc=0, scale=0.05, size=len(df))
test["kmeans_feature"] = test["cluster"].replace(
cluster_probs.to_dict()["prob"]
) + np.random.normal(loc=0, scale=0.05, size=len(test))
# I did my best to make this graph readble so we can get some insights from the clusters:
# - When the features (except for ph) have low values the target is more likely to be 0 (about 75% prob)
# - When the values are close to the middle of the range the positive target change slightly increases (65%)
# - When the values are high the probabilty is approximately equal for both classes.
# We created two new features out of the clustering model:
# - cluster: a categorical variable representing the cluster label
# - kmeans_feature: a numerical variable that defined by the sum of the class 1 probability for the respective cluster + some white noise.
# We avoided using techniques like one hot encoding because as mentioned before, we decide not to increase the dimensionality of the data.
# ## Modeling
# We have evaluate the CatBoostClassifier within three different feature combinations:
# - calc + kmenas_feature
# - calc + cond
# - calc + cluster(using the cat_feature resource of the model)
def evaluate_model(model, X, y, cat_features=[]):
# Define the cross-validation method
cv = RepeatedStratifiedKFold(n_splits=4, n_repeats=100, random_state=seed)
# Define the metric to optimize
scoring = "roc_auc"
if len(cat_features) > 0:
# Evaluate the model using cross-validation
scores = cross_val_score(
model,
X,
y,
scoring=scoring,
cv=cv,
n_jobs=-1,
fit_params={"cat_features": cat_features},
)
else:
scores = cross_val_score(model, X, y, scoring=scoring, cv=cv, n_jobs=-1)
# Calculate the mean and standard deviation of the scores
mean_score, std_score = np.mean(scores), np.std(scores)
# Print the mean and standard deviation of the scores
print(f"Mean ROC AUC: {mean_score:.3f} (std: {std_score:.3f})")
evaluate_model(
model=CatBoostClassifier(random_state=seed, verbose=0),
X=df[["calc", "kmeans_feature"]],
y=df["target"],
)
evaluate_model(
model=CatBoostClassifier(random_state=seed, verbose=0),
X=df[["calc", "cond"]],
y=df["target"],
)
evaluate_model(
model=CatBoostClassifier(random_state=seed, verbose=0),
X=df[["calc", "cluster"]],
y=df["target"],
cat_features=["cluster"],
)
# We managed to score about .81 on using calc+cond and calc+cluster.
# **The test score was .858.**
# We tried to tune the model but it appear that the default CatBoost parameters are more apropriated.
# ## Submission
model = CatBoostClassifier(random_state=seed, verbose=0)
model.fit(df[["calc", "cluster"]], df["target"], cat_features=["cluster"])
sample_submission["target"] = model.predict_proba(test[["calc", "cluster"]])[:, 1]
sample_submission.to_csv("submission.csv", index=False)
# ## Dummy submission (public roc = .83)
# This is the baseline dummy we mentioned before. We used it as the basline result.
#
target = (test["calc"] >= 4).astype(int)
sample_submission["target"] = target
# sample_submission.to_csv('submission.csv', index=False)
|
import tensorflow as tf
import tensorflow_decision_forests as tfdf
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
train_file_path = "/kaggle/input/house-prices-advanced-regression-techniques/train.csv"
dataset_df = pd.read_csv(train_file_path)
print("Full train dataset shape is {}".format(dataset_df.shape))
print(dataset_df["SalePrice"].describe())
plt.figure(figsize=(9, 8))
sns.distplot(dataset_df["SalePrice"], color="g", bins=100, hist_kws={"alpha": 0.4})
list(set(dataset_df.dtypes.tolist()))
df_num = dataset_df.select_dtypes(include=["float64", "int64"])
df_num.head()
df_num.hist(figsize=(16, 20), bins=50, xlabelsize=8, ylabelsize=8)
print(df_num)
# 2.1 Train-test split
# In the above below, we split the train data into a test and a train set. Set a value for the test_size yourself. Argue why the test value can not be too small or too large. You can also use k-fold cross validation. Secondly, we have set the random_state to 102. Can you think of a reason why we set a random_state at all?
#
YearBuilt = np.array(df_num.iloc[:, 6])
YearBuilt = YearBuilt.reshape((len(YearBuilt), 1))
YearRemodAdd = np.array(df_num.iloc[:, 7]).reshape((len(YearBuilt), 1))
SalePrice = np.array(df_num.iloc[:, 37]).reshape((len(YearBuilt), 1))
print(SalePrice)
X = np.concatenate((YearBuilt, YearRemodAdd), axis=1).T
print(X.shape)
from sklearn.decomposition import PCA
# train pca
pca = PCA(n_components=2)
pca = pca.fit(X.T)
X_pca_skl = pca.transform(X.T)
def compute_pcs(X, lam):
# correlation matrix
P = float(X.shape[1])
Cov = 1 / P * np.dot(X, X.T) + lam * np.eye(X.shape[0])
# use numpy function to compute eigenvalues /vectors
D, V = np.linalg.eigh(Cov)
return D, V
# Find eigen values D and eigen vectors V
D, V = compute_pcs(X, lam=10 ** (-7))
# Rearrange from largest eigwn value to smallest
V = V[:, range(X.shape[0] - 1, -1, -1)]
D = D[range(X.shape[0] - 1, -1, -1)]
X_pca = np.dot(V.T, X)
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X_pca.T, SalePrice, test_size=0.3, shuffle=True, random_state=42
)
from sklearn.linear_model import LinearRegression
# define the model
# model = LogisticRegression()
# define the model
model = LinearRegression()
# fit the model
model.fit(X_train, y_train)
predictions = model.predict(X_test)
print(predictions)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv("/kaggle/input/unridd-intrusion-detection-dataset/UNR-IDD.csv")
df.head(5)
df.drop(columns={"is_valid", "Port Number", "Switch ID", "Binary Label"}, inplace=True)
df.info()
df["Label"].value_counts()
# # Select Normal and Portscan's label
df = df[(df["Label"] == "Normal") | (df["Label"] == "PortScan")]
# * Features selection
X = df.iloc[:, :-1]
from sklearn.feature_selection import VarianceThreshold
# Normalize data
normalized_df = X / X.mean()
# Init, fit VT
vt = VarianceThreshold(threshold=0.002)
_ = vt.fit(normalized_df)
# Get a boolean mask
mask = vt.get_support()
# Subset the data
X_reduced = X.loc[:, mask]
X_reduced.shape
def identify_correlated(df, threshold):
"""
A function to identify highly correlated features.
"""
# Compute correlation matrix with absolute values
matrix = df.corr().abs()
# Create a boolean mask
mask = np.triu(np.ones_like(matrix, dtype=bool))
# Subset the matrix
reduced_matrix = matrix.mask(mask)
# Find cols that meet the threshold
to_drop = [c for c in reduced_matrix.columns if any(reduced_matrix[c] > threshold)]
return to_drop
to_drop = identify_correlated(X_reduced, threshold=0.9)
len(to_drop)
df.drop(to_drop, axis=1, inplace=True)
# # Split X_train/test & y_train/test and Encoding y
from sklearn.preprocessing import LabelEncoder
# Chuyển đổi nhãn thành các số nguyên
le = LabelEncoder()
df["Label"] = le.fit_transform(df["Label"])
y = df["Label"]
from sklearn.model_selection import train_test_split
np.random.seed(42)
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=1121218
)
from sklearn.preprocessing import MinMaxScaler
scaler = MinMaxScaler()
X = scaler.fit_transform(X)
X
# # Decision Tree Model
from sklearn.tree import DecisionTreeClassifier
# Define model
model = DecisionTreeClassifier()
# Fit model
model.fit(X_train, y_train)
from sklearn.metrics import confusion_matrix
y_pred = model.predict(X_test)
confusion_matrix(y_test, y_pred)
from sklearn.metrics import f1_score
df_f1 = f1_score(y_test, y_pred, average="macro")
df_f1
from sklearn.metrics import accuracy_score
dt_accurracy = accuracy_score(y_test, y_pred)
dt_accurracy
from sklearn.metrics import classification_report
print(classification_report(y_test, y_pred))
# # Random Forest Model
from sklearn.ensemble import RandomForestClassifier
RFC = RandomForestClassifier(max_depth=10, random_state=0).fit(X_train, y_train)
y_pred_RFC = RFC.predict(X_test)
confusion_matrix(y_test, y_pred_RFC)
df_f1_DTC = f1_score(y_test, y_pred_RFC, average="macro")
df_f1_DTC
dt_accurracy_RFC = accuracy_score(y_test, y_pred_RFC)
dt_accurracy_RFC
print(classification_report(y_test, y_pred_RFC))
# # LGBM Model
import lightgbm as lgb
clf = lgb.LGBMClassifier()
clf.fit(X_train, y_train)
y_pred_LGBM = clf.predict(X_test)
confusion_matrix(y_test, y_pred_LGBM)
df_f1_LGBM = f1_score(y_test, y_pred_LGBM, average="macro")
df_f1_LGBM
dt_accurracy_LGBM = accuracy_score(y_test, y_pred_LGBM)
dt_accurracy_LGBM
print(classification_report(y_test, y_pred_LGBM))
# # XG Boost
from xgboost import XGBClassifier
XC = XGBClassifier().fit(X_train, y_train)
y_pred_XC = XC.predict(X_test)
confusion_matrix(y_test, y_pred_XC)
df_f1_XC = f1_score(y_test, y_pred_XC, average="macro")
df_f1_XC
dt_accurracy_XC = accuracy_score(y_test, y_pred_XC)
dt_accurracy_XC
print(classification_report(y_test, y_pred_XC))
# # CNN Model
import keras
from keras.layers import (
Conv2D,
Conv1D,
MaxPooling2D,
MaxPooling1D,
Flatten,
BatchNormalization,
Dense,
Dropout,
GlobalMaxPooling1D,
)
from keras.utils.np_utils import to_categorical
from keras.models import Sequential
# check for encoded labels
df["Label"].value_counts()
# make 3 seperate datasets for 3 feature labels
data_1 = df[df["Label"] == 0]
data_2 = df[df["Label"] == 1]
# make normal feature
y_1 = np.zeros(data_1.shape[0])
y_normal = pd.DataFrame(y_1)
# make portscan feature
y_2 = np.ones(data_2.shape[0])
y_pc = pd.DataFrame(y_2)
# merging the original dataframe
X = pd.concat([data_1, data_2], sort=True)
y = pd.concat([y_normal, y_pc], sort=True)
print(X.shape)
print(y.shape)
from sklearn.utils import resample
data_1_resample = resample(data_1, n_samples=5000, random_state=123, replace=True)
data_2_resample = resample(data_2, n_samples=5000, random_state=123, replace=True)
train_dataset = pd.concat([data_1, data_2])
train_dataset.head(2)
train_dataset["Label"].unique()
test_dataset = train_dataset.sample(frac=0.2)
target_train = train_dataset["Label"]
target_test = test_dataset["Label"]
target_train.unique(), target_test.unique()
y_train = to_categorical(target_train, num_classes=2)
y_test = to_categorical(target_test, num_classes=2)
train_dataset
from sklearn.preprocessing import MinMaxScaler
scaler = MinMaxScaler()
train_dataset = scaler.fit_transform(train_dataset, target_train)
test_dataset = scaler.fit_transform(test_dataset, target_test)
# making train & test splits
X_train = train_dataset[:, :-1]
X_test = test_dataset[:, :-1]
X_test
# reshape the data for CNN
X_train = X_train.reshape(len(X_train), X_train.shape[1], 1)
X_test = X_test.reshape(len(X_test), X_test.shape[1], 1)
X_train.shape, X_test.shape
# making the deep learning function
def model():
model = Sequential()
model.add(
Conv1D(
filters=64,
kernel_size=6,
activation="relu",
padding="same",
input_shape=(24, 1),
)
)
model.add(BatchNormalization())
# adding a pooling layer
model.add(MaxPooling1D(pool_size=(3), strides=2, padding="same"))
model.add(Conv1D(filters=64, kernel_size=6, activation="relu", padding="same"))
model.add(BatchNormalization())
model.add(Dropout(0.3))
model.add(MaxPooling1D(pool_size=(3), strides=2, padding="same"))
model.add(Conv1D(filters=64, kernel_size=6, activation="relu", padding="same"))
model.add(BatchNormalization())
model.add(MaxPooling1D(pool_size=(3), strides=2, padding="same"))
model.add(Dropout(0.3))
model.add(Flatten())
model.add(Dense(64, activation="relu"))
model.add(Dense(64, activation="relu"))
model.add(Dense(2, activation="softmax"))
model.compile(
loss="categorical_crossentropy", optimizer="adam", metrics=["accuracy"]
)
return model
model = model()
model.summary()
from keras.callbacks import CSVLogger, ModelCheckpoint
logger = CSVLogger("logs.csv", append=True)
his = model.fit(
X_train, y_train, epochs=10, batch_size=8, validation_split=0.3, callbacks=[logger]
)
# check the model performance on test data
cnn_scores = model.evaluate(X_test, y_test)
print("%s: %.2f%%" % (model.metrics_names[1], cnn_scores[1] * 100))
model.save("model.h5")
print("Saved model to disk")
# check history of model
history = his.history
history.keys()
import matplotlib.pyplot as plt
import seaborn as sns
epochs = range(1, len(history["loss"]) + 1)
acc = history["accuracy"]
loss = history["loss"]
val_acc = history["val_accuracy"]
val_loss = history["val_loss"]
# visualize training and val accuracy
plt.figure(figsize=(20, 10))
plt.title("Training and Validation Accuracy (CNN)")
plt.xlabel("Epochs")
plt.ylabel("Accuracy")
plt.plot(epochs, acc, label="accuracy")
plt.plot(epochs, val_acc, label="val_acc")
plt.legend()
# visualize train and val loss
plt.figure(figsize=(20, 10))
plt.title("Training and Validation Loss(CNN)")
plt.xlabel("Epochs")
plt.ylabel("Loss")
plt.plot(epochs, loss, label="loss", color="g")
plt.plot(epochs, val_loss, label="val_loss", color="r")
plt.legend()
from sklearn.metrics import (
accuracy_score,
confusion_matrix,
precision_recall_fscore_support,
roc_curve,
auc,
)
import tensorflow as tf
y_pred = model.predict(X_test)
y_pred_cm = np.argmax(y_pred, axis=1)
y_test_cm = np.argmax(y_test, axis=1)
cm = confusion_matrix(y_test_cm, y_pred_cm)
cm = confusion_matrix(y_test_cm, y_pred_cm)
group_counts = ["{0:0.0f}".format(value) for value in cm.flatten()]
group_percentages = ["{0:.2%}".format(value) for value in cm.flatten() / np.sum(cm)]
labels = [f"{v1}\n{v2}" for v1, v2 in zip(group_counts, group_percentages)]
labels = np.asarray(labels).reshape(2, 2)
label = ["normal", "portscan"]
plt.figure(figsize=(9, 9))
sns.heatmap(
cm,
xticklabels=label,
yticklabels=label,
annot=labels,
fmt="",
cmap="Blues",
vmin=0.2,
)
plt.title("Confusion Matrix for" + " CNN" + " model")
plt.ylabel("True Class")
plt.xlabel("Predicted Class")
plt.savefig("./" + "CNN" + "_CM.png")
plt.show()
cnn_f1 = f1_score(y_test_cm, y_pred_cm, average="macro")
cnn_avg_score = sum(cnn_scores) / len(cnn_scores)
import matplotlib
import matplotlib.pyplot as plt
import numpy as np
labels = ["Decision Tree", "Random\nForest(RF)", "LGBM", "XGBoost", "CNN"]
f1_scores = np.array([df_f1, df_f1_DTC, df_f1_LGBM, df_f1_XC, cnn_f1]).round(decimals=2)
accu = np.array(
[dt_accurracy, dt_accurracy_RFC, dt_accurracy_LGBM, dt_accurracy_XC, cnn_avg_score]
).round(decimals=2)
x = np.arange(len(labels)) # the label locations
width = 0.3 # the width of the bars
fig, ax = plt.subplots()
rects1 = ax.bar(x - width / 2, f1_scores, width, label="f1 score")
rects2 = ax.bar(x + width / 2, accu, width, label="accuracy")
# Add some text for labels, title and custom x-axis tick labels, etc.
ax.set_title("Performance comparison of multi class classification algorithms")
ax.set_xticks(x)
ax.set_xticklabels(labels)
ax.legend()
def autolabel(rects):
"""Attach a text label above each bar in *rects*, displaying its height."""
for rect in rects:
height = rect.get_height()
ax.annotate(
"{}".format(height),
xy=(rect.get_x() + rect.get_width() / 2, height),
xytext=(0, 3), # 3 points vertical offset
textcoords="offset points",
ha="center",
va="bottom",
)
autolabel(rects1)
autolabel(rects2)
fig.tight_layout()
plt.show()
|
import pandas as pd
inpath = "/kaggle/input/fathomnet-2023-first-glance/"
train = pd.read_csv(inpath + "train_with_labels.csv")
test = pd.read_csv(inpath + "eval_images.csv")
List = train["categories"].tolist()
most_freq = max(set(List), key=List.count)
most_freq = most_freq.replace(".0", "")
print(most_freq)
test["id"] = test["file_name"].str[:-4]
test["categories"] = most_freq
test["osd"] = 0.1
test[["id", "categories", "osd"]].to_csv("submission.csv", index=False)
|
# # シンプルな11位の解法【概要】
# # very simple 11th place solution【Overview】
# このNotebookではsubmission ensemble について説明しています。pseudo labelingについてはこちらのNotebookを参照してください。
# 【日本語&English】TPS Feb Pseudo Labeling (11th place)
# https://www.kaggle.com/maostack/english-tps-feb-pseudo-labeling-11th-place
#
# This Notebook describes submission ensemble; see this Notebook for pseudo labeling.
# 【日本語&English】TPS Feb Pseudo Labeling (11th place)
# https://www.kaggle.com/maostack/english-tps-feb-pseudo-labeling-11th-place
# 私の解法はとてもシンプルで、主に2つのテクニックしか使っていません。
# したがって、初心者の方にも真似しやすいと思います。
# 私が使ったのはPseudo LabelingとSubmission Ensembleです。
# 用いたモデルはLightGBMのみです。
#
# Submission Ensembleでは、まず、これまでに提出したsubmissionの中で、
# public leaderboardにおいてスコアが高い順に4つ選び、
# submissionのtargetを平均した(足して4で割った)だけです。
# 選んだ4つのsubmissionはすべてpseudo labelingを用いて予測したものです。
#
# My solution is very simple. I used only 2 main techniques.
# Therefore, I think it is easy for beginners to understand.
# I used Pseudo Labeling and Submission Ensemble.
# The only model I used is LightGBM.
#
# At the Submission Ensemble stage,
# I selected top 4 (at public leaderboard) predictions I submitted so far.
# Then, I just calculated the average of each predictions(added them together and divided by 4).
# All 4 selected submissions were predicted using pseudo labeling.
# public score / private score / submission file
# 0.84192 / 0.84253 / submission_pseudo_lgb
# 0.84195 / 0.84253 / submission_pseudo_lgb_4
# 0.84196 / 0.84252 / submission_pseudo_lgb_5
# 0.84196 / 0.84260 / pseudo_lgb_1
import os
import numpy as np
import pandas as pd
pred1 = pd.read_csv("../input/tps-feb-submission-ensemble/submission_pseudo_lgb.csv")
pred2 = pd.read_csv("../input/tps-feb-submission-ensemble/submission_pseudo_lgb_4.csv")
pred3 = pd.read_csv("../input/tps-feb-submission-ensemble/submission_pseudo_lgb_5.csv")
pred4 = pd.read_csv("../input/tps-feb-submission-ensemble/pseudo_lgb_1.csv")
pred3
# 各提出ファイルのtarget部分の平均をとる.
# calculate average
pred = (pred1.target + pred2.target + pred3.target + pred4.target) / 4
pred
submission = pd.read_csv(
"../input/tabular-playground-series-feb-2021/sample_submission.csv"
)
submission.target = pred
submission
submission.to_csv("ensemble.csv", index=False)
|
import matplotlib.pyplot as plt
import seaborn as sns
import pandas as pd
df = pd.read_csv("/kaggle/input/house-prices-advanced-regression-techniques/train.csv")
df.shape
df.head()
df.describe().style.background_gradient()
# # Clustering
# # Missing percent
# Those feature with too much missing values, for example the missing percentage is greater than 70%, should be consider to be removed.
feature_names = df.columns.tolist()
na_series = df[feature_names].isnull().sum()
missing_percent = na_series.apply(lambda x: x * 1.0 / df.shape[0])
missing_percent[missing_percent != 0.0].sort_values().plot.barh(
x="Features", y="Percent"
)
# # Imputation
# The chart below depict the number of unique values of each feature. For imputation, if a feature with less unique count may be more relevant with imputing with its mode value. On the other hand, a feature with high unique count may be better to impute with mean value.
num_vars = df.select_dtypes(["int64", "float64"]).columns.tolist()
num_vars.remove("Id")
col_name_list = list()
unique_count = list()
for col_name in num_vars:
col_name_list.append(col_name)
unique_count.append(len(df[col_name].unique()))
unique_count_series = pd.Series(unique_count, index=col_name_list).sort_values(
ascending=True
)
unique_count_series.plot.barh(x="Feature", y="Unique count")
# # Distribution
# ## Numeric distribution
fig = plt.figure(figsize=(28, 160))
num_vars = df.select_dtypes(["int64", "float64"]).columns.tolist()
num_vars.remove("Id")
for index, col in enumerate(num_vars):
plt.subplot(26, 2, index + 1)
sns.histplot(x=col, data=df)
plt.ylabel("COUNT", size=25)
plt.xlabel(col, fontsize=25)
plt.xticks(size=20, rotation=45)
plt.yticks(size=20)
fig.tight_layout(pad=1.0)
# The distribution of 'SalePrice' is skew to the left. For the linear model like linear regression, this may have a negative effect on the predictions. You can learn more in a post from scikit-learn https://scikit-learn.org/stable/auto_examples/compose/plot_transformed_target.html. Therefore, logarithm may be used to transform the target variable so that it follow a normal distribution as you can see in the chart below. Below are the distribtion of 'SalePrice' before and after it is applied logarithm.
fig = plt.figure(figsize=(20, 5))
sns.histplot(x="SalePrice", data=df, bins=300, kde=True)
tran_y = df.SalePrice.apply(np.log)
# tran_y.hist(bins=200)
fig = plt.figure(figsize=(20, 5))
sns.histplot(data=tran_y, bins=300, kde=True)
# ## Categorical distribution
fig = plt.figure(figsize=(26, 160))
cat_vars = df.select_dtypes(["object"]).columns.tolist()
for index, col in enumerate(cat_vars):
plt.subplot(25, 2, index + 1)
sns.countplot(x=col, data=df)
plt.ylabel("COUNT", size=25)
plt.xlabel(col, fontsize=25)
plt.xticks(size=20, rotation=45)
plt.yticks(size=20)
fig.tight_layout(pad=1.0)
# # Mutual information
# This methods provide the importance of a variable. In the chart below, the more higher number, the more crucial a feature. Learn more from this link to scikit-learn https://scikit-learn.org/stable/modules/feature_selection.html#univariate-feature-selection.
import numpy as np
import matplotlib.pyplot as plt
from sklearn.feature_selection import mutual_info_regression
# Get not null train categorical features
categorical_features = df.select_dtypes("object").columns.tolist()
na_series = df[categorical_features].isnull().sum() > 0
categorical_missing_features = na_series[na_series == True].index.tolist()
used_cate_features = list(
set(categorical_features).difference(set(categorical_missing_features))
)
# Get not null train numerical features
num_features = df.select_dtypes(["int64", "float64"]).columns.tolist()
na_series = df[num_features].isnull().sum() > 0
num_missing_features = na_series[na_series == True].index.tolist()
used_num_features = list(set(num_features).difference(set(num_missing_features)))
used_features = used_cate_features + used_num_features
used_features.remove("SalePrice")
X = df[used_features]
y = df["SalePrice"]
from sklearn.preprocessing import OrdinalEncoder
enc = OrdinalEncoder()
X_encoded = enc.fit_transform(X)
scores = mutual_info_regression(X_encoded, y)
scores = pd.Series(scores, name="MI Scores", index=X.columns)
scores = scores.sort_values(ascending=False)
width = np.arange(len(scores))
ticks = list(scores.index)
fig = plt.figure(figsize=(15, 15))
plt.barh(width, scores)
plt.yticks(width, ticks, size=10)
plt.xticks(size=10)
plt.title("Mutual Information Scores", size=15)
plt.ylabel("Features", size=15)
# # Correlation
import matplotlib.pyplot as plt
import seaborn as sns
# Get not null train categorical features
categorical_features = df.select_dtypes("object").columns.tolist()
na_series = df[categorical_features].isnull().sum() > 0
categorical_missing_features = na_series[na_series == True].index.tolist()
used_cate_features = list(
set(categorical_features).difference(set(categorical_missing_features))
)
# Get not null train numerical features
num_features = df.select_dtypes(["int64", "float64"]).columns.tolist()
na_series = df[num_features].isnull().sum() > 0
num_missing_features = na_series[na_series == True].index.tolist()
used_num_features = list(set(num_features).difference(set(num_missing_features)))
used_features = used_cate_features + used_num_features
for f in ["GarageArea", "TotRmsAbvGrd", "BsmtFullBath", "TotalBsmtSF"]:
used_features.remove(f)
sns.set(rc={"figure.figsize": (10, 10)})
axis_corr = sns.heatmap(
df[used_num_features].corr(),
vmin=-1,
vmax=1,
center=0,
cmap=sns.diverging_palette(50, 500, n=500),
square=True,
)
plt.show()
|
import numpy as np
import pandas as pd
from sklearn.linear_model import LogisticRegression, Ridge
from sklearn.feature_selection import SelectFromModel
df = pd.read_csv("/kaggle/input/cleaning-after-corr/Cleaning_after_corr.csv")
label = df["Label"]
df.head()
df.drop(["Label", "Unnamed: 0"], inplace=True, axis="columns")
df.head()
sel_ = SelectFromModel(LogisticRegression(solver="saga", penalty="l2", C=1))
sel_.fit(df, label)
non_imp = sel_.get_support()
non_imp
non_imp_index = []
for i in range(len(non_imp)):
if non_imp[i] == False:
non_imp_index.append(i)
print(non_imp_index)
len(non_imp_index)
df_final = df.drop(df.columns[non_imp_index], axis="columns")
df_final.head()
from sklearn.metrics import classification_report
class elm_model:
def __init__(
self, hidden_units, activation_function, x, weight, bias, distribution
):
self.hidden_units = hidden_units
self.activation_function = activation_function
self.m, self.n = x.shape
self.mu, self.sigma = 0, 0.01
self.beta = 0
if distribution == "normal":
self.weight = np.matrix(
np.random.normal(self.mu, self.sigma, (self.hidden_units, self.n))
)
self.bias = np.matrix(
np.random.normal(self.mu, self.sigma, (1, self.hidden_units))
)
elif distribution == "logistic":
self.weight = np.matrix(
np.random.logistic(self.mu, self.sigma, (self.hidden_units, self.n))
)
self.bias = np.matrix(
np.random.logistic(self.mu, self.sigma, (1, self.hidden_units))
)
else:
self.weight = np.matrix(
np.random.uniform(weight[0], weight[1], (self.hidden_units, self.n))
)
self.bias = np.matrix(
np.random.uniform(bias[0], bias[1], (1, self.hidden_units))
)
def sigmoid(self, z):
return 1 / (1 + np.exp(-1 * z))
def swish(self, z):
return z / (1 + np.exp(-1 * z))
def bipolar_sigmoid(self, z):
return (1 - np.exp(-1 * z)) / (1 + np.exp(-1 * z))
def relu(self, z):
z[z < 0] = 0
return z
def linear(self, z):
return z
def arc_tan(self, z):
return np.arctan(z)
def train(self, x, y):
x = np.matrix(x)
y = pd.get_dummies(y)
y = np.matrix(y)
self.H = np.dot(x, self.weight.T) + self.bias
if self.activation_function == "sigmoid":
self.H = self.sigmoid(self.H)
elif self.activation_function == "swish":
self.H = self.swish(self.H)
elif self.activation_function == "arc_tan":
self.H = self.arc_tan(self.H)
elif self.activation_function == "bipolar_sigmoid":
self.H = self.bipolar_sigmoid(self.H)
elif self.activation_function == "relu":
self.H = self.relu(self.H)
elif self.activation_function == "linear":
self.H = self.linear(self.H)
H_plus = np.dot(np.linalg.inv(np.dot(self.H.T, self.H)), self.H.T)
self.beta = np.dot(H_plus, y)
return
def predict(self, x):
y_pred = self.predict_proba(x)
y_pred = y_pred.T[0]
y_pred[y_pred > 0.5] = 0
y_pred[y_pred != 0] = 1
return y_pred
def softmax(self, x):
return np.exp(x) / np.sum(np.exp(x), axis=1)
def predict_proba(self, x):
x = np.matrix(x)
self.H = np.dot(x, self.weight.T) + self.bias
if self.activation_function == "sigmoid":
self.H = self.sigmoid(self.H)
elif self.activation_function == "swish":
self.H = self.swish(self.H)
elif self.activation_function == "arc_tan":
self.H = self.arc_tan(self.H)
elif self.activation_function == "bipolar_sigmoid":
self.H = self.bipolar_sigmoid(self.H)
elif self.activation_function == "relu":
self.H = self.relu(self.H)
elif self.activation_function == "linear":
self.H = self.linear(self.H)
y_pred = np.dot(self.H, self.beta)
return np.array(self.softmax(y_pred).tolist())
def score(self, x, y):
y_pred = self.predict(x)
print(classification_report(y, y_pred, digits=5))
return
from sklearn.model_selection import train_test_split
# np.set_printoptions(formatter={'float_kind':'{:f}'.format})
x_train, x_test, y_train, y_test = train_test_split(
df_final, label, test_size=0.2, random_state=42, stratify=label
)
# weight = [[-0.5, 0.5], [-1,1], [0,1]]
# bias = [0,1.]
# activation_fun = ['linear', 'relu','arc_tan', 'sigmoid','bipolar_sigmoid','swish']
# x_neurons = [10, 20, 30, 40, 50, 60, 70, 80, 90, 100, 110, 120, 150, 200, 250, 300, 350, 400, 450, 500, 600, 700, 800, 900,1000]
# for activation in activation_fun:
# print(activation)
# for i in weight:
# print('Weight: ', i)
# for neu in x_neurons:
# print('Neurons: ', neu)
# elm = elm_model(neu, activation, x_train, i, bias, 'normal')
# elm.train(x_train, y_train)
# # y_pred = elm.predict(x_test)
# elm.score(x_test, y_test)
# print()
from sklearn.metrics import accuracy_score
from sklearn.metrics import precision_score
from sklearn.metrics import recall_score
from sklearn.metrics import f1_score
from sklearn.metrics import roc_auc_score
import matplotlib.pyplot as plt
def graphing(activation_fun, distribution):
weight = [[-0.5, 0.5], [-1, 1], [0, 1]]
bias = [0, 1.0]
x_neurons = [
10,
20,
30,
40,
50,
60,
70,
80,
90,
100,
110,
120,
150,
200,
250,
300,
350,
400,
450,
500,
600,
700,
800,
900,
1000,
]
print(activation_fun)
for w in weight:
accuracy = []
precision = []
recall = []
f1score = []
aucscore = []
print("Weight: ", w)
for neu in x_neurons:
elm = elm_model(neu, activation_fun, x_train, w, bias, distribution)
elm.train(x_train, y_train)
y_pred = elm.predict(x_test)
# elm.score(x_test, y_test)
acc = accuracy_score(y_test, y_pred) * 100
prec = precision_score(y_test, y_pred) * 100
rec = recall_score(y_test, y_pred) * 100
f1 = f1_score(y_test, y_pred) * 100
auc = roc_auc_score(y_test, y_pred) * 100
# print(acc)
accuracy.append(acc)
precision.append(prec)
recall.append(rec)
f1score.append(f1)
aucscore.append(auc)
plt.plot(x_neurons, accuracy, label="Weights: " + str(w), color="red")
plt.xlabel("Number of hidden neurons")
plt.title("Accuracy")
plt.legend()
plt.show()
plt.plot(x_neurons, precision, label="Weights: " + str(w), color="green")
plt.xlabel("Number of hidden neurons")
plt.title("Precision")
plt.legend()
plt.show()
plt.plot(x_neurons, recall, label="Weights: " + str(w), color="orange")
plt.xlabel("Number of hidden neurons")
plt.title("Recall")
plt.legend()
plt.show()
plt.plot(x_neurons, f1score, label="Weights: " + str(w), color="purple")
plt.xlabel("Number of hidden neurons")
plt.title("F1 score")
plt.legend()
plt.show()
plt.plot(x_neurons, aucscore, label="Weights: " + str(w), color="blue")
plt.xlabel("Number of hidden neurons")
plt.title("AUC")
plt.legend()
plt.show()
graphing("linear", "normal")
graphing("relu", "normal")
graphing("arc_tan", "normal")
graphing("sigmoid", "normal")
graphing("bipolar_sigmoid", "normal")
graphing("swish", "normal")
graphing("linear", "logistic")
graphing("relu", "logistic")
graphing("arc_tan", "logistic")
graphing("sigmoid", "logistic")
graphing("bipolar_sigmoid", "logistic")
graphing("swish", "logistic")
graphing("linear", "uniform")
graphing("relu", "uniform")
graphing("arc_tan", "uniform")
graphing("sigmoid", "uniform")
graphing("bipolar_sigmoid", "uniform")
graphing("swish", "uniform")
weight = [[-0.5, 0.5], [-1, 1], [0, 1]]
bias = [0, 1.0]
activation_fun = ["linear", "relu", "arc_tan", "sigmoid", "bipolar_sigmoid", "swish"]
for activation in activation_fun:
print(activation)
for i in weight:
print("Weight: ", i)
elm = elm_model(70, activation, x_train, i, bias, "logistic")
elm.train(x_train, y_train)
# y_pred = elm.predict(x_test)
elm.score(x_test, y_test)
print()
weight = [-0.5, 0.5]
bias = [0, 1.0]
activation_fun = ["linear", "relu", "arc_tan", "sigmoid", "bipolar_sigmoid", "swish"]
for activation in activation_fun:
print(activation)
elm = elm_model(70, activation, x_train, weight, bias, "uniform")
elm.train(x_train, y_train)
# y_pred = elm.predict(x_test)
elm.score(x_test, y_test)
print()
elm = elm_model(70, "arc_tan", x_train, weight, bias, "normal")
elm.train(x_train, y_train)
from lime.lime_tabular import LimeTabularExplainer
class_names = ["ATTACK", "BENIGN"]
explainer = LimeTabularExplainer(
x_train.values,
feature_names=df_final.columns,
class_names=class_names,
mode="classification",
)
def cf(d):
# print(d)
x = elm.predict_proba(d)
# print(x)
return x
print(x_test.iloc[2])
np.set_printoptions(formatter={"float_kind": "{:f}".format})
y_test.iloc[500]
explaination = explainer.explain_instance(
x_test.iloc[500], elm.predict_proba, num_features=12
)
explaination.show_in_notebook(show_all=False)
|
# # New York City: Open Data
# Looking at 311 Service Calls relted to Noise Complaints and Correlation with DOB Permits
# ### Datasets
# **[311 Service Requests from 2010 to Present](https://data.cityofnewyork.us/Social-Services/311-Service-Requests-from-2010-to-Present/erm2-nwe9)**
# * 32.8M Rows and 49 Columns. Updated everyday.
# * The CSV data file is too large to download and analyze, thus, sodapy was used to filter and load files relevant to the project.
# * Filter to rows with noise complaints on and after 2018 (last 5 years).
# 
# **[DOB Permit Issuance](https://data.cityofnewyork.us/Housing-Development/DOB-Permit-Issuance/ipu4-2q9a)**
# * 3.94M Rows and 60 Columns
# * Filter to rows with Job start date on and after 2018 (last 5 years).
# 
#
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from sodapy import Socrata
import json, requests
import os
import geopandas as gpd
socrata_domain = "data.cityofnewyork.us"
socrata_dataset_identifier_311 = "erm2-nwe9"
rows_311 = 32749225
socrata_dataset_identifier_DOB = "ipu4-2q9a"
rows_dob = 3937674
# Reference: https://github.com/xmunoz/sodapy/blob/master/examples/soql_queries.ipynb
from kaggle_secrets import UserSecretsClient
user_secrets = UserSecretsClient()
secret_value_0 = user_secrets.get_secret("socrata_token")
client = Socrata(socrata_domain, socrata_token)
client.timeout = 1000
# raw_data = pd.read_json(query)
metadata_311 = client.get_metadata(socrata_dataset_identifier_311)
[x["name"] for x in metadata_311["columns"]]
loop_size = 1000000
num_loops = round(rows_311 / loop_size)
raw_data = []
id_str = "(complaint_type like 'Noise%') AND (created_date > '2018-01-01T00:00:00')"
for i in range(num_loops):
results = client.get(
socrata_dataset_identifier_311,
select="created_date, closed_date, agency, \
complaint_type, incident_zip, location_type, status, \
city, borough, latitude, longitude",
where=id_str,
order="created_date ASC",
limit=loop_size,
offset=loop_size * i,
)
print("\n> Loop number: {}".format(i))
raw_data.extend(results)
df_noise = pd.DataFrame.from_records(raw_data)
df_noise.to_csv("data_noise1.csv")
df_noise.shape
metadata_DOB = client.get_metadata(socrata_dataset_identifier_DOB)
[x["name"] for x in metadata_DOB["columns"]]
loop_size = 1000000
num_loops_dob = round(rows_dob / loop_size)
raw_data_dob = []
# Could not filter or order through date columns because the data is stored in text instead of datetime format.
id_str_dob = "job_start_date > '2018-01-01'"
for i in range(num_loops_dob):
results_dob = client.get(
socrata_dataset_identifier_DOB,
select="borough, bin__, zip_code, \
job__, job_type, work_type, permit_status, filing_status,\
permit_type, bldg_type, residential,filing_date, issuance_date, \
expiration_date, job_start_date ,gis_latitude, gis_longitude",
# where= id_str_dob,
# order= 'filing_date ASC',
limit=loop_size,
offset=loop_size * i,
)
print("\n> Loop number: {}".format(i))
raw_data_dob.extend(results_dob)
df_dob = pd.DataFrame.from_records(raw_data_dob)
df_dob.to_csv("data_dob.csv")
df_dob
|
# Thank you for reading this notebook. I'm new to Kaggle and machine-learing algorithms, and this competition is the second one for me after TPS-January. In this notebook I wrote down the basic flows I used in this competition. I didn't use any special techniques, but used GBDT modules commonly-used in Kaggle. I'm glad if this notebook would help other biginners.
# **0. Import modules and dataset**
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import optuna
from tqdm.notebook import tqdm
from sklearn.cluster import KMeans
from sklearn.mixture import GaussianMixture
from sklearn.metrics import mean_squared_error
from sklearn.preprocessing import LabelEncoder
from sklearn.model_selection import train_test_split, KFold
from lightgbm import LGBMRegressor, plot_importance
train = pd.read_csv("/kaggle/input/tabular-playground-series-feb-2021/train.csv")
test = pd.read_csv("/kaggle/input/tabular-playground-series-feb-2021/test.csv")
cont_features = [f for f in train.columns.tolist() if f.startswith("cont")]
cat_features = [f for f in train.columns.tolist() if f.startswith("cat")]
features = cat_features + cont_features
data = train[features]
target = train["target"]
all_data = pd.concat([data, test])
# # 1. Feature Engineering
# I did a slight feature-engineering.
# Histograms of the cont features show multiple components. For instance, the cont1 has 7 discrete peaks as shown below. I thought these characteristics could be used as an additional feature.
# So, I tried `sklearn.mixture.GaussianMixture` to devide into several groups [Ref: [Notebooks of TPS-Jan. by Dave E](https://www.kaggle.com/davidedwards1/jan21-tabplayground-nn-final-fewer-features)].
# See also https://scikit-learn.org/stable/modules/mixture.html#gmm for Gaussian Mixture Models.
# The scatter plots below show the cont-feature values and target, with the results of GMM.
# The bottom histgrams also show the results of GMM.
fig, ax = plt.subplots(5, 3, figsize=(14, 24))
for i, feature in enumerate(cont_features):
plt.subplot(5, 3, i + 1)
sns.histplot(all_data[feature][::100], color="blue", kde=True, bins=100)
plt.xlabel(feature, fontsize=9)
plt.show()
inits = [
[0.3, 0.5, 0.7, 0.9],
[0.039, 0.093, 0.24, 0.29, 0.35, 0.42, 0.49, 0.56, 0.62, 0.66, 0.76],
[0.176, 0.322, 0.416, 0.495, 0.548, 0.618, 0.707, 0.937],
[0.2, 0.35, 0.44, 0.59, 0.75, 0.83],
[0.28, 0.31, 0.42, 0.5, 0.74, 0.85],
[0.25, 0.38, 0.43, 0.58, 0.75, 0.9],
[0.34, 0.48, 0.7, 0.88],
[0.25, 0.29, 0.35, 0.48, 0.61, 0.68, 0.78, 0.9],
[0.11, 0.2, 0.3, 0.35, 0.45, 0.6, 0.76, 0.9],
[0.22, 0.32, 0.38, 0.44, 0.53, 0.63, 0.71, 0.81, 0.87],
[0.19, 0.27, 0.37, 0.46, 0.56, 0.61, 0.71, 0.86],
[0.23, 0.35, 0.52, 0.7, 0.84],
[0.27, 0.32, 0.35, 0.49, 0.63, 0.7, 0.79, 0.88],
[0.22, 0.29, 0.35, 0.4, 0.47, 0.58, 0.68, 0.72, 0.8],
]
gmms = []
for feature, init in zip(cont_features, inits):
X_ = np.array(all_data[feature].tolist()).reshape(-1, 1)
means_init = np.array(init)[:, None]
gmm_ = GaussianMixture(
n_components=len(init), means_init=means_init, random_state=0
).fit(X_)
gmms.append(gmm_)
preds = gmm_.predict(X_)
all_data[f"{feature}_gmm"] = preds
train[f"{feature}_gmm"] = preds[: len(train)]
test[f"{feature}_gmm"] = preds[len(train) :]
fig, ax = plt.subplots(5, 3, figsize=(24, 30))
for i, feature in enumerate(cont_features):
plt.subplot(5, 3, i + 1)
sns.scatterplot(
x=feature, y="target", data=train[::150], hue=f"{feature}_gmm", palette="muted"
)
plt.xlabel(feature, fontsize=9)
plt.show()
fig, ax = plt.subplots(5, 3, figsize=(24, 30))
for i, feature in enumerate(cont_features):
plt.subplot(5, 3, i + 1)
sns.histplot(
x=feature,
data=train[::150],
hue=f"{feature}_gmm",
kde=True,
bins=100,
palette="muted",
)
plt.xlabel(feature, fontsize=9)
plt.show()
# I calculated the standard deviations for each group and added as new features.
for feature in cont_features:
mu = all_data.groupby(f"{feature}_gmm")[feature].transform("mean")
sigma = all_data.groupby(f"{feature}_gmm")[feature].transform("std")
train[f"{feature}_gmm_dev"] = (train[feature] - mu[: len(train)]) / sigma[
: len(train)
]
test[f"{feature}_gmm_dev"] = (test[feature] - mu[len(train) :]) / sigma[
len(train) :
]
# For categorical features, I used label-encoding (`sklearn.preprocessing.LabelEncoder`).
for feature in cat_features:
le = LabelEncoder()
le.fit(train[feature])
train[feature] = le.transform(train[feature])
test[feature] = le.transform(test[feature])
features = [col for col in train.columns.to_list() if col not in ["id", "target"]]
test[features].head().T
# # Hyperparameter Tuning
def objective(trial, data=train[features], target=target):
train_x, test_x, train_y, test_y = train_test_split(
data, target, test_size=0.2, random_state=41
)
param = {
"metric": "rmse",
"random_state": 41,
"n_estimators": 20000,
"learning_rate": 0.01,
"reg_alpha": trial.suggest_loguniform("reg_alpha", 1e-2, 100.0),
"reg_lambda": trial.suggest_loguniform("reg_lambda", 1e-3, 10.0),
"colsample_bytree": trial.suggest_uniform("colsample_bytree", 0.01, 1.0),
"subsample": trial.suggest_categorical(
"subsample", [0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 1.0]
),
"subsample_freq": trial.suggest_int("subsample_freq", 1, 20),
"max_depth": trial.suggest_categorical("max_depth", [-1, 30, 100, 300]),
"num_leaves": trial.suggest_int("num_leaves", 2, 500),
"min_child_samples": trial.suggest_int("min_child_samples", 1, 200),
"min_child_weight": trial.suggest_loguniform("min_child_weight", 1e-3, 10),
"cat_smooth": trial.suggest_int("cat_smooth", 1, 100),
}
model = LGBMRegressor(**param)
model.fit(
train_x,
train_y,
eval_set=[(test_x, test_y)],
early_stopping_rounds=100,
verbose=False,
)
preds = model.predict(test_x)
rmse = mean_squared_error(test_y, preds, squared=False)
return rmse
study = optuna.create_study(direction="minimize")
study.optimize(objective, n_trials=50)
print("Best trial:", study.best_params)
study.trials_dataframe()
optuna.visualization.plot_param_importances(study)
optuna.visualization.plot_parallel_coordinate(study)
optuna.visualization.plot_optimization_history(study)
# ttt
# ```python
# NUM_FOLDS = 10
# seed_list = [0,1,2]
# test_pred = np.zeros(len(test))
# val_pred = np.zeros(len(train))
# for seed in tqdm(seed_list):
# tmp_test_pred = np.zeros(len(test))
# tmp_val_pred = np.zeros(len(train))
# kf = KFold(n_splits=NUM_FOLDS, shuffle=True, random_state=seed)
# for f, (train_idx, val_idx) in tqdm(enumerate(kf.split(train[features], target))):
# print("*" * 20)
# print(f"Seed-#{seed}; Fold-#{f}")
# train_x, val_x = train.iloc[train_idx][features], train.iloc[val_idx][features]
# train_y, val_y = target[train_idx], target[val_idx]
# model = LGBMRegressor(metric = 'rmse',
# random_state=seed,
# learning_rate = 0.002,
# n_estimators = 20000,
# **study.best_params)
# model.fit(train_x,train_y,eval_set=[(val_x,val_y)],early_stopping_rounds=100,verbose=5000)
#
# temp_oof = model.predict(val_x)
# temp_test = model.predict(test[features])
# tmp_test_pred += temp_test
# tmp_val_pred[val_idx] = temp_oof
# print(mean_squared_error(temp_oof, val_y, squared=False))
#
# print("*" * 20)
# print(f"Seed-#{seed}\n{mean_squared_error(tmp_val_pred, target, squared=False)}")
# val_pred += tmp_val_pred
# test_pred += tmp_test_pred / NUM_FOLDS
# val_pred /= len(seed_list)
# test_pred /= len(seed_list)
# print("*" * 20)
# print(mean_squared_error(val_pred, target, squared=False))
# ```
# # Ensemble
lgbm_1 = pd.read_csv("../input/tps2-submissions/submission1.csv")
lgbm_2 = pd.read_csv("../input/tps2-submissions/submission2.csv")
xgb_1 = pd.read_csv("../input/tps2-submissions/submission3.csv")
cat_1 = pd.read_csv("../input/tps2-submissions/submission4.csv")
models = [lgbm_1, lgbm_2, xgb_1, cat_1]
weights = [10.0, 5.0, 2.0, 1.0]
sample_submission.target = 0
for model, weight in zip(models, weights):
sample_submission.target += weight * model.target / sum(weights)
sample_submission.to_csv("sub-ensemble.csv", index=False)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input/sf-booking/"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# День добрый!
# Данный ноутбук я планирвал вести на двух языках на русском для курса, и на англиском (google translate),
# чтобы привыкать это делать на языке которым сопровождается большая часть документации в программировании.
# Это финальная версия ноутбука будет восновном на русском.
# В предыдущих, промежуточных версиях может быть смешаный язык, не везде будут имется комментарии, а местами их будет слишком много.
# _Спасибо за понимание и приятного просмотра._
# ___
# Описание
# Представьте, что вы работаете датасаентистом в компании Booking. Одна из проблем компании — это нечестные отели, которые накручивают себе рейтинг. Одним из способов нахождения таких отелей является построение модели, которая предсказывает рейтинг отеля. Если предсказания модели сильно отличаются от фактического результата, то, возможно, отель играет нечестно, и его стоит проверить.
# Вам поставлена задача создать такую модель. Готовы приступить?
# __Условия соревнования:__
# * Данное соревнование является бессрочным и доступно для всех потоков.
# * Срок выполнения соревнования устанавливается индивидуально в каждом потоке.
# * Тестовая выборка представлена в LeaderBoard целиком.
# * Делаем реальный ML продукт, который потом сможет нормально работать на новых данных.
# Определение качества
# * Метрика качества Результаты оцениваются по метрике MAPE (см. 3_Машинное обучение для самых маленьких)
# * Файл представления
# Для каждого **id** отеля в наборе тестовых данных вы должны предсказать рейтинг отеля для **reviewer_score** переменной.
# * Файл должен содержать заголовок и иметь следующий формат: ``` reviewer_score,id 1,1 ```
# Описание Датасета
# **Файлы для соревнования**
# * hotels_train.csv - набор данных для обучения
# * hotels_test.csv - набор данных для оценки качества
# * submission.csv - файл сабмишна в нужном формате
# **Признаки**
# * hotel_address - адрес отеля
# * review_date - дата, когда рецензент разместил соответствующий отзыв.
# * average_score - средний балл отеля, рассчитанный на основе последнего комментария за последний год
# * hotel_name - название отеля
# * reviewer_nationality - национальность рецензента
# * negative_review - отрицательный отзыв, который рецензент дал отелю.
# * review_total_negative_word_counts - общее количество слов в отрицательном отзыв
# * positive_review - положительный отзыв, который рецензент дал отелю
# * review_total_positive_word_counts - общее количество слов в положительном отзыве
# * reviewer_score - оценка, которую рецензент поставил отелю на основе своего опыта
# * total_number_of_reviews_reviewer_has_given - количество отзывов, которые рецензенты дали в прошлом
# * total_number_of_reviews - общее количество действительных отзывов об отеле
# * tags - теги, которые рецензент дал отелю.
# * days_since_review - продолжительность между датой проверки и датой очистки
# * additional_number_of_scoring - есть также некоторые гости, которые просто поставили оценку сервису, а не оставили отзыв. Это число указывает, сколько там действительных оценок без проверки.
# * lat - широта отеля
# * lng - долгота отеля
# ___
# Изначально у меня был план действий как в курсе, но как показала практика, некоторые элементы могу повторяться или менятся местами.
# А для того, чтобы видеть динамику улучшения мне приходилось создавать модель несколько раз.
# По этому я просто опишу посделовательность моих действий и конечный результат этих действий.
# ___
# **Выполненые действия:**
# 1. Исследование данных из датасета
# 2. Преобразование признаков и отчистка датасета
# 1. Исследование данных из датасета
# ___
# Исследование данных я провел двумя способами:
# 1. Вручную пересматривал все данные, анализировал, решал что делать с ними дальше.(на протяжении всего проекта)
# В результате данной работы я создал несколько функций, которые будут повторяться и я постараюсь их оптимизирвать их использование, а так же описать их работу.
# 2. И еще использоватл готовое решение для анализа данных метод ___ProfileReport___ из библиотеки _ydata_profiling_ этот способ будет использован при финальной версии. Жалко, что про него вспомнил только перечитывая курс в посиках подсказок.
# *(на момент создания ноутбука установить можно коммандой прям в коде !pip install -U ydata-profiling)*
# Давайте подключим все необходмые для работы модули и библиотеки, чтобы не вписывать их позже, это будет более удобно, чем прописывать их потом.
# Подключим датасет, создадим отчет с помощью ___ProfileReport___ и разберем его.
# Так же при попытке получить ответ на 100% данных я столкнулся с ситуацией когда столбцы не совмпали.По этому я сразу создам объедененный датасет который буду преобразовывать, а потом в самом конце разрежу его обратно.
# Только мне нужно придумать еще какую-нибудь метку для каждого датасета пусть так и завется ***mark***
#!pip install ydata_profiling
import plotly.express as px
from sklearn.ensemble import RandomForestRegressor
from sklearn.model_selection import train_test_split
from sklearn import metrics
import re
import random
from sklearn.feature_selection import chi2
from sklearn.feature_selection import f_classif # anova
from ydata_profiling import ProfileReport
import category_encoders as ce
# Load datasets from kaggle uncomment in kaggle
hotels_test = pd.read_csv("/kaggle/input/sf-booking/hotels_test.csv")
hotels_train = pd.read_csv("/kaggle/input/sf-booking/hotels_train.csv")
submission = pd.read_csv("/kaggle/input/sf-booking/submission.csv")
# Load datasets from local
# hotels_test = pd.read_csv('../Block-3-PROJECT-3_EDA_and_Feature_Engineering__Kaggle/data/kaggle/hotels_test.csv')
# hotels_train = pd.read_csv('../Block-3-PROJECT-3_EDA_and_Feature_Engineering__Kaggle/data/kaggle/hotels_train.csv')
# submission = pd.read_csv('../Block-3-PROJECT-3_EDA_and_Feature_Engineering__Kaggle/data/kaggle/submission.csv')
# рабочая копия датасета
df_hotels_trn = hotels_train.copy()
df_hotels_trn.info()
df_hotels_tst = hotels_test.copy()
df_hotels_tst.info()
def combine_func():
df_hotels_trn["mark"] = 1
df_hotels_tst["mark"] = 2
# добавим недостаящего признака в тестовый датасет Он может быть нулем т.к. после преобразований разделим датасеты обратно и мы все равно его удалим.
# Пробовал сделать np.nan но при проверке весов датасет не прошел обработку пришлось заменить на 0
df_hotels_tst["reviewer_score"] = 0
combine_df = pd.concat([df_hotels_tst, df_hotels_trn], sort=False).reset_index(
drop=True
)
return combine_df
combine_hotel_df = combine_func()
profile = ProfileReport(hotels_train, title="Pandas Profiling Report")
# display(profile)
# * Признаки ***hotel_address*** и ***hotel_name*** количественно совпадают и один из них точно не нужен. Сликом много различных значений.
# При самостоятельном разборе полезным оказался только ***hotel_address*** из него мы доставли название городов, а ***hotel_name*** можно будет сразу удалить.
# Но думаю это возможно было бы сделать и из координат ***lng***, ***lat***
# * Признак ***additional_number_of_scoring*** вначале я убрал этот признак, думал что он не поможет в вычислениях т.к. он ни как не связан с комментариями.
# Но сейчас я задумался о том насколько это бесполезный попробую его еще использовать. При просмотре через *ProfileReport* я увидил положительную связь между целевым признаком ***reviewer_score***.
# * Признак ***review_date*** из него мы можем получить нормальную дату, но забегу вперед даже созданные признаки, ***year*** и ***day*** не сильно помогли, месяц же оказал влияние на результаты. Что говорит о возможной сезонности (к примеру зима, лето если это курортные отели) По этому месяц я оставил.
# * Признак ***average_score*** это средняя оцена отеля в целом. И имеет положительную связь с целевым признаком ***reviewer_score***.
# * Признак ***reviewer_nationality*** национальность гостя, как видно из отчета 47% это англечане, ~7% американцы, ~4% австралийцы, ~3% процента ирланды, арабы итд.
# Я выделил всех у кого менее 3-х процентов в группу 'Other'
# * ***negative_review*** около 30% значений в этом признаке не являются негативными комментариями, это видно в отчете, и те же результаты я получил при ручной обработке этого признака. Далее из ***negative_review*** я создам новый признак ***presence_negative_review***, в старых версиях название было ***cleaned_negative_review***.
# так же *ProfileReport* не дал приемлемого приимущества при разборе слов. по этому нет смысла хранить данный признак в датасете. _(К тому же в курсе было правило, что если данные повреждены на 30 и более процентов можно отказаться от данного признака, почему я не вспомнил это правило сразу.)_
# хотя если использовать какие-либо сторонние библиотеки можно будет собрать еще какую нибудь вижимку, но я этого не сделал.
# * ***review_total_negative_word_counts*** т.к. 30% значений в негативном признаке были неприемлемы для работы этот производный признак так же не сильно функциональный. Оставим его посмотрим на вес и решим что делать.
# * ***total_number_of_reviews*** Ниже мы будем еще проверять силу признаков и возможно откажемся от этого признака.
# * ***positive_review*** аналагично негативным отзывам много значений малоинформативных, но общий процент пропусков менее 30%, это разрешает его оставить при желании, но можно и убрать для баланса.
# * ***review_total_positive_word_counts*** аналично ***review_total_negative_word_counts***
# * ***tags*** На первый взгляд информативный признак в нем содержится много ифномрации от типа клиента(***guest_type***), до количества ночей (***stayed_night***) которое он провел в отеле, а так же классы комнат(***rooms_type***) и какой тип путешествия был у клиента отдых или рабочая(***trip_type***).
# Но я почитал правила создания среднего бала для отелей в правилх booking.com, а по условию мы работаем в этой компании, по этому не вижу запрета на просмотр.
# В общем прямого влияния на среднюю оценку отеля tag-и не оказывают. И нужны лишь для сведений другим пользователям.
# Но мы все равно выделим несколько компонентов. Например люди которые провели несколько ночей в отеле скорее всего поставят оценку отелю, нежели те кто провел всего одну ночь. По этому мы создадим такой признак и еще несколько.
# Класс комнат оказался не сильно важным мы увидим.
# * ***total_number_of_reviews_reviewer_has_given*** Ниже мы будем еще проверять силу признаков и откажемся от этого признака.
# * ***days_since_review*** -- пропущенный мною признак который возможно оказыват большее влияние, чем я думал.
# В правилах Booking есть максимальное время (количество месяцев 36) после которого оценки не влияют. Возможно этот признак будет более влиятельным.
# * ***lat*** и ***lng*** координаты отеля, но т.к. мы знаем города где находится отель я думаю можно от координат отказаться. Ну или использовать их для подтверждения.
# Все с информацией о датасете закончили местами забегая вперед.
# 2. Преобразование признаков и отчистка датасета
# ____
# Первоначально я оставлял координаты и добавлял пропущенные значения
# Вот таким образом
# ```
# df_hotels_trn['lat'] = df_hotels_trn['lat'].fillna(df_hotels_trn['lat'].mean())
# df_hotels_trn['lng'] = df_hotels_trn['lng'].fillna(df_hotels_trn['lng'].mean())
# ```
# Но в финальной версии мы отказываемся от этих признаков по этому давайте сразу удалим их.
# Но чтобы не делать это потом по 100 раз если вдруг нам понадобится обернем в функцию (описание в самой функции)
# У нас два датасета тестовый и тренировочный.
# Моей ошибкой было не создать таких функция, т.к. я много много раз делал одно и тоже.
# Мы не станем удалять все сразу сейчас, мы будем собирать то, что хотим удалить в переменную del_list и потом исплоьзовать ее вместе с функцией на удаление
del_list = ["lng", "lat", "hotel_name"]
def delete_futures(df):
"""
Принимает датасет
Фунция будет удалять столбцы взятые из переменной del_list
По этому нам просто нужно следить за перменной del_list
Вовзращает датасет без удаленных столбцов.
"""
df = df.drop(del_list, axis=1)
return df
# Хотя из отчета мы видили кореляцию по неприрывным признакам,
# давайте сразу создадим данную фунцию, чтобы не вызывать профайл постоянно.
#
def show_correlation(dataframe):
"""
Функция рисует Матрицу корреляции датасета.
"""
df_graph = round(dataframe.corr(), 2)
fig = px.imshow(df_graph, text_auto=True)
fig.show()
show_correlation(df_hotels_trn)
# Мы видим как и до этого кореляцию с признаком ***total_number_of_reviews*** мы его хотели убрать т.к. этот признак имеет слабый вес, но пока не тронем. чтобы это показать.
# additional_number_of_scoring уже имеет числовой признак и пока не требует преобзований.
# Давайте пока преобразуем review_date в дату, и создадим на его основе 3 признака ***day***, ***month***, ***year***
# Обернем данное действие в функцию, чтобы не делать это действие по 100 раз
# после чего добавим его на удаление больше он нам не нужен. добавим так же функцию
def create_YMD_from_data(df):
df["month"] = pd.to_datetime(df["review_date"]).dt.month
df["year"] = pd.to_datetime(df["review_date"]).dt.year
df["day"] = pd.to_datetime(df["review_date"]).dt.day
if "review_date" not in del_list:
del_list.append("review_date")
return df
combine_hotel_df = create_YMD_from_data(combine_hotel_df)
# df_hotels_tst = create_YMD_from_data(df_hotels_tst)
# Займемся извлечением признаков из reviewer_nationality
# Возмьу уже созданную мною функцию, и добавлю сразу возможность передать туда dataset, чтобы разом все изменения сделать в конце
def transformation_reviewer_nationality(df):
"""
Принимает на вход dataset
Перезаписывает признак reviewer_nationality
А затем кодирует его, после чего готовм признак к удалению за ненадобностью
Возвращает измененный и дополненый dataset
"""
def extract_country_from_rn(row):
"""
сортеруем значения оставляя лишь 4 основных остальные в 'Other'
"""
list_of_country = [
"United Kingdom",
"United States of America",
"Australia",
"Ireland",
]
for element in list_of_country:
if element in row:
return element
return "Other"
df["reviewer_nationality"] = df["reviewer_nationality"].apply(
lambda x: extract_country_from_rn(x)
)
bin_encoder = ce.BinaryEncoder(cols=["reviewer_nationality"])
type_bin = bin_encoder.fit_transform(df["reviewer_nationality"])
df = pd.concat([df, type_bin], axis=1)
del_list.append("reviewer_nationality")
return df
combine_hotel_df = transformation_reviewer_nationality(combine_hotel_df)
# df_hotels_tst = transformation_reviewer_nationality(df_hotels_tst)
# Следующие два признака
# * ***positive_review***
# * ***negative_review***
# Давайте писать функции для обработки этой парочки. В результате мы получим другую пару
# * ***presence_negative_review***
# * ***presence_positive_review***
# Для позитивного комментария функция похожа, но набор слов по которым мы будем собирать признак немного отличается.
# Так как не все так прямоленейно как с негативными отзывами попадаются много значений, которые не смотря на длинну строки очень даже информативный пример ниже
# * *No Positive* -- это не является положительным отзывом по этому можно присвоить "0"
# * *Location* -- Человеку понравилась локация (место) можно считать позитивным "1"
# * *Everything* -- Все понравилось ставим "1"
# * *location* -- как во втором примере ставим "1"
# * *Nothing* -- человеку ни чего не понравилось, почти негативный отзыв даже ставим "0"
def create_presence_negative_review(df):
"""
Я не стал глубоко разбираться насколько отзыв сильно негативный или позитивный, но отобрал неправильные значения которые точно не являются целевым отзывом.
Если нам попадается такой комментарий, из созданного списка я считаю что комментария небыло.
Если такого значения нет, я считаю это полноценным комментарием. Грубо, но приемлемо если посмотреть отчет от ProfileReport то большая часть этих слов найдется.
Так же я смотрю, чтобы комментарий был длинне 15 символов, чтобы я не убрал случайно полноценные комментарии где встречаются такие сочетания слов.
И убираем этоти признаки, обольше они нам не нужены
"""
# Эти короткие значения в отзывах я в ручную отобрал они составляют 30% в колонке negative_review.
false_netgative = [
"No Negative",
" Nothing",
" Nothing ",
" nothing",
" N A",
" None",
" ",
" N a",
" All good",
" nothing ",
" No complaints",
" Nil",
" n a",
" None ",
" All good ",
" NA",
" No",
" NOTHING",
" Na",
" Non",
" All was good",
]
false_positive = ["No Positive", " Nothing", " Nothing ", " nothing", " "]
def rewiew_clean_with_list(review_string, false_list):
if review_string in false_list:
return 0
else:
return 1
df["presence_negative_review"] = df["negative_review"].apply(
lambda x: rewiew_clean_with_list(x, false_netgative) if len(x) < 15 else 1
)
df["presence_positive_review"] = df["positive_review"].apply(
lambda x: rewiew_clean_with_list(x, false_positive) if len(x) < 15 else 1
)
del_list.append("negative_review")
del_list.append("positive_review")
return df
combine_hotel_df = create_presence_negative_review(combine_hotel_df)
# df_hotels_tst = create_presence_negative_review(df_hotels_tst)
# Подготовим признаки
# review_total_negative_word_counts,
# review_total_positive_word_counts
# к удалению
# Теперь преобразуем tags в несколько новых признаков. И так же сделаем для этого функцию.
def pars_tag_future(df):
"""
Функция принимает датасет и работает со столбцом 'tags',
rev_func() первоначально выделяет из строки отдельные тэги и собирает их в список.
Каждый элемент которого по прежнему является строкой.
extract_rooms_type() функция по ключевому слову создает признак. какие ключевые слова использовать мы поняли из анализа ProfileReport
(stayed, trip, room), а так же тип гостя guest_type
Признаки сразу имеют приемлемый для обучения вид
Так же подготавливается к удалению признак tag
"""
def rev_func(tags):
"""
функция принимает строку и создает список используе (', ') как разделитель.
# Разделяем тэги на отдельные элементы
"""
tags = tags[2:-2]
result = tags.strip().split(" ', ' ")
return result
def extract_tag_type(tags, word):
"""
Фукнция по ключевым словам определяет с каким списком будет работат, и в резульате получем тэг с которым будем работать.
Сразу кодирует значение по количеству элементво в списке.
"""
if word == "room":
list_of_types = ["standard", "superior", "double", "deluxe", "classic"]
array_data = [0, 0, 0, 0, 0, 0, 0]
no_data = [0, 0, 0, 0, 0, 0, 1]
other_n = 5
elif word == "trip":
list_of_types = ["leisure", "business"]
# [leisure,buisnes,other,no trip]
array_data = [0, 0, 0, 0]
no_data = [0, 0, 0, 1]
other_n = 2
elif word == "stayed":
list_of_types = ["1", "2", "3", "4"]
array_data = [0, 0, 0, 0, 0, 0]
no_data = [0, 0, 0, 0, 0, 1]
other_n = 4
else:
word == ""
list_of_types = ["couple", "solo", "family"]
# [couple,solo,family,other,not gtype]
array_data = [0, 0, 0, 0, 0]
no_data = [0, 0, 0, 0, 1]
other_n = 3
listed_tags = rev_func(tags)
for num_type_room in range(len(list_of_types)):
for element in range(len(listed_tags)):
if word in listed_tags[element].lower():
if (
list_of_types[num_type_room].lower()
== listed_tags[element].lower().split(" ")[0]
):
array_data[num_type_room] = 1
else:
array_data[other_n] = 1
elif word not in listed_tags[element].lower():
array_data[other_n + 1] = 1
for i in range(len(array_data)):
# print(array_data)
if array_data == no_data:
return 0
elif array_data[i] == 1:
return i + 1
df["room_type"] = df["tags"].apply(lambda x: extract_tag_type(x, "room"))
df["trip_type"] = df["tags"].apply(lambda x: extract_tag_type(x, "trip"))
df["stayed_type"] = df["tags"].apply(lambda x: extract_tag_type(x, "stayed"))
df["guest_type"] = df["tags"].apply(lambda x: extract_tag_type(x, ""))
del_list.append("tags")
return df
combine_hotel_df = pars_tag_future(combine_hotel_df)
# df_hotels_tst = pars_tag_future(df_hotels_tst)
# display(df_hotels_trn.info())
display(
combine_hotel_df[["room_type", "trip_type", "stayed_type", "guest_type"]].describe()
)
print(del_list)
# Я просмотрел несколько рандомных наборов строк и пока вижу такую последовательность.
# страна пишется в конце адреса, в то время как город бывает предпаследним и 5 с конца для Великобритании (благодаря названию страны United Kingdom) , в начале бывает попадаются цифры бывает нет. По этому придется делать парсинг через конце строки.
# В первоначальной функции я получал и страну и город, но сейчас мы будем получать только город.(функция оставлю прежней, если понадобится взять страну вместо города)
# Еще нужно закодировать города тут подоходит метод однократного кодирования. Возьму пример прям из обучающего курса он подходит.
def extract_city_from_addres(df):
import sys
"""
Функция принимает датасет работает с признаком hotel_address и извлекает город из адеса.
может так же извлекать отдельно страну(country=True) для этого нужно выставить параметр
"""
def pars_hotel_address(address, country=False, city=False):
if (country is False) and (city is False):
city = True
if (country is True) and (city is True):
raise ("Please select one option or city or country as True")
else:
listed_address = address.split(" ")
if listed_address[-1] == "Kingdom":
if country is True:
return "United Kingdom"
if city is True:
return listed_address[-5]
else:
if country is True:
return listed_address[-1]
if city is True:
return listed_address[-2]
df["city"] = df["hotel_address"].apply(lambda x: pars_hotel_address(x, city=True))
encoder = ce.OneHotEncoder(
cols=["city"], use_cat_names=True
) # указываем столбец для кодирования
type_bin = encoder.fit_transform(df["city"])
df = pd.concat([df, type_bin], axis=1)
# Подготовка к удалению
del_list.append("hotel_address")
del_list.append("city")
return df
combine_hotel_df = extract_city_from_addres(combine_hotel_df)
# df_hotels_tst = extract_city_from_addres(df_hotels_tst)
combine_hotel_df.info()
print(del_list)
# days_since_review МЫ чуть не пропустили этот признак снова
def extract_days_num(df):
def transform_days_since_review(days):
number = re.findall(r"[\d]+", days)
return int(number[0])
df["num_days_since_review"] = df["days_since_review"].apply(
lambda x: transform_days_since_review(x)
)
del_list.append("days_since_review")
return df
combine_hotel_df = extract_days_num(combine_hotel_df)
# df_hotels_tst = extract_days_num(df_hotels_tst)
# Давайте тогда определим веса у всех признаков и попробуем улучшить наши показатели.
# Вначале посмотрим на категориальные признаки, затем на числовые(непрерывные) признаки
# А так же глянем на корреляции между признаками
# Давайте я временно создам копию рабочего датасета удалим от туда заготовленные признаки.
tmp_df = combine_hotel_df.copy()
tmp_df = delete_futures(tmp_df)
def preapre_to_test(df):
"""
Функция принимает датасет и подгатавливает его к передаче тест-предсказание
создает:
Dataset X -- это датасет без целевого признака (примеры для обработки)
Dataset y -- это целевой признка который нам известен (ответы к примерам)
Dataset X_train -- 75% Данных для тренировки модели
Dataset X_test -- 25% Для проверки модели на знание материала.
Dataset y_train -- 75% тренировочных ответов (для обучения)
Dataset y_test -- 25% тестовый ответов с которыми мы потом сравниваем ответы модели.
Возвращает все перечисленные датасеты
"""
X = df.drop(["reviewer_score"], axis=1)
y = df["reviewer_score"]
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.25, random_state=17
)
return X, y, X_train, X_test, y_train, y_test
# категориальные признаки
cat_cols = [
"average_score",
"presence_negative_review",
"presence_positive_review",
"month",
"year",
"day",
"room_type",
"stayed_type",
"trip_type",
"guest_type",
"city_London",
"city_Paris",
"city_Amsterdam",
"city_Milan",
"city_Vienna",
"city_Barcelona",
"reviewer_nationality_0",
"reviewer_nationality_1",
"reviewer_nationality_2",
]
# Увеличиваем все значения на одну велечину,т.к. функция не принимает отрицательных значений
# minimal = df_hotels_trn['lng'].min()
# tmp_df['lng'] = df_hotels_trn['lng'] + abs(minimal)
X, y, X_train, X_test, y_train, y_test = preapre_to_test(tmp_df)
y = y.astype("int")
def imp_cat_():
imp_cat = pd.Series(chi2(X[cat_cols], y)[0], index=cat_cols)
imp_cat.sort_values(inplace=True)
imp_cat.plot(kind="barh")
imp_cat_()
# Как мы видим больший вес имеют наличие негативных отзывов, долгота (видимо от теплоты), наличие позитивных отзывов, средняя отценка, две национальности потом уже остальные показатели.
# Но мы смело можем убрать (год и день) Добавим эти признаки сразу в список удаления
del_list.append("year")
del_list.append("day")
print(del_list)
# Мы не зря делали эти признаки, мы узнали что они не имеют веса
from sklearn.feature_selection import f_classif # anova
num_cols = [
"review_total_negative_word_counts",
"total_number_of_reviews",
"review_total_positive_word_counts",
"total_number_of_reviews_reviewer_has_given",
"additional_number_of_scoring",
]
def imp_num_():
imp_num = pd.Series(f_classif(X[num_cols], y)[0], index=num_cols)
imp_num.sort_values(inplace=True)
imp_num.plot(kind="barh")
imp_num_()
# По результату видно, что три признака очень слабы total_number_of_reviews, total_number_of_reviews_reviewer_has_given, additional_number_of_scoring
# Все же этот признак так же не сильно подходит и остался кандидатом на удаление (но мне интересно сильно ли он повлияет на результат рачетов по этому мы его удалим паследним)
# del_list.append('review_total_negative_word_counts')
# del_list.append('review_total_positive_word_counts')
del_list.append("total_number_of_reviews_reviewer_has_given")
del_list.append("total_number_of_reviews")
del_list.append("additional_number_of_scoring")
del_list.append("room_type")
del_list.append("review_total_positive_word_counts")
# del_list.append('num_days_since_review')
# Настало время провести обучение модели. Первый тест на тренировочных данных разделенных на 75/25
# Потом второе обучение будет на тренировочные данные против тестовых
combine_hotel_df = delete_futures(combine_hotel_df)
# df_hotels_trn = delete_futures(df_hotels_trn)
combine_hotel_df.info()
# df_hotels_trn.info()
# Разделим датасеты обратно
df_hotels_trn = combine_hotel_df[combine_hotel_df["mark"] == 1]
df_hotels_tst = combine_hotel_df[combine_hotel_df["mark"] == 2]
display(df_hotels_trn.info())
display(df_hotels_tst.info())
del_list = ["mark"]
df_hotels_tst = delete_futures(df_hotels_tst)
df_hotels_tst = df_hotels_tst.drop(["reviewer_score"], axis=1)
df_hotels_trn = delete_futures(df_hotels_trn)
display(df_hotels_trn.info())
display(df_hotels_tst.info())
X, y, X_train, X_test, y_train, y_test = preapre_to_test(df_hotels_trn)
# df_hotels_trn будет у нас выступать как X_train Без целевой переменной, а df['reviewer_score'] как y_train
def create_model(X_train, y_train, test_data):
"""
Функция принимает тренировочные данные и обучает модель.
в этой версии я убрал # submission_test = regr.predict(submissoin_df)
Поняв, что перед проверкой я могу обучить модель на 100% тренировочных данных.
Так будет больше примеров, и лучший результат наверное, вот и проверим.
"""
regr = RandomForestRegressor(n_estimators=100)
regr.fit(X_train, y_train)
y_pred = regr.predict(test_data)
return y_pred
# y_pred = create_model(X_train, y_train, X_test)
# print('train_test_MAPE:', metrics.mean_absolute_percentage_error(y_test, y_pred))
# print(X.info(), y.info(), df_hotels_tst.info())
submission_pred = create_model(X, y, df_hotels_tst)
print(
"test_MAPE:",
metrics.mean_absolute_percentage_error(
submission["reviewer_score"], submission_pred
),
)
print("END")
# Резульат хуже чем в наших предыдущих проверках что очень странно
# Попробуем использовать LightAutoML В первую очередь для проверки
# Т.к. это непредвиденное решение, придется все необходимые библиотеки и модули подргузить по ходу дела
# Структуру подачи переменных мы возьмем из уроков skillfactory
# N_THREADS - количество виртуальных ЦП для создания модели LightAutoML у меня как раз 4
# N_FOLDS - количество сгибов во внутреннем CV LightAutoML 5 как в уроке
# RANDOM_STATE - random seed будет такой как у меня в ноутбуке "17"
# TEST_SIZE - размет тестовой части данных 25%
# TIMEOUT - Время на тест. Мы не торопимся по этому 15 минут будет достаточно. При необохдимости увеличу 900
# TARGET_NAME - целевой признак 'reviewer_score'
from sklearn.metrics import mean_absolute_error
from sklearn.model_selection import train_test_split
import torch
# LightAutoML presets, task and report generation
from lightautoml.automl.presets.tabular_presets import TabularAutoML
from lightautoml.tasks import Task
N_THREADS = 4
N_FOLDS = 5
RANDOM_STATE = 17
TEST_SIZE = 0.2
TIMEOUT = 900
TARGET_NAME = "reviewer_score"
np.random.seed(RANDOM_STATE)
torch.set_num_threads(N_THREADS)
task = Task("reg", loss="mae", metric="mae")
roles = {"target": TARGET_NAME, "drop": ["row_ID"]}
automl = TabularAutoML(
task=task,
timeout=TIMEOUT,
cpu_limit=N_THREADS,
reader_params={"n_jobs": N_THREADS, "cv": N_FOLDS, "random_state": RANDOM_STATE},
)
# Тренируем модель
oof_pred = automl.fit_predict(df_hotels_trn, roles=roles, verbose=1)
# Делаем предсказание
te_pred = automl.predict(df_hotels_tst)
print(
"lama_test_MAPE:",
metrics.mean_absolute_percentage_error(
submission["reviewer_score"], pd.DataFrame(te_pred.data[:, 0])
),
)
print(
"rndf_test_MAPE:",
metrics.mean_absolute_percentage_error(
submission["reviewer_score"], submission_pred
),
)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import warnings
warnings.filterwarnings("ignore")
df_sample = pd.read_csv("/kaggle/input/playground-series-s3e12/sample_submission.csv")
df_train = pd.read_csv("/kaggle/input/playground-series-s3e12/train.csv")
df_test = pd.read_csv("/kaggle/input/playground-series-s3e12/test.csv")
df_test
print("Rows X Columns of df_train : ", df_train.shape)
df_train
df_train["target"].value_counts()
# Not highly imbalance, we can take it as almost balanced dataset with respect to the target variable
# Data type for each column in the df_train
df_train.info()
# Description for the column in the dataframe
df_train.describe()
# Missing values
for item in df_train.columns:
print("Item :", item, ".\t\t Count of null :", df_train[item].isnull().sum())
# ## UNIVARIATE ANALYSIS
# Plotting the graph for each column
import matplotlib.pyplot as plt
# print(df_train.columns)
for item in df_train.columns:
if item == "id" or item == "target":
continue
else:
plt.figure
plt.plot(df_train[item])
print("Histogram plot for :", item)
plt.show()
# Plotting the graph for each column
import matplotlib.pyplot as plt
# print(df_train.columns)
for item in df_train.columns:
if item == "id" or item == "target":
continue
else:
plt.figure
plt.hist(df_train[item])
print("Histogram plot for :", item)
plt.show()
# ## BIVARIATE ANALYSIS
df_train.columns
# Plotting few
df_train.plot(x="gravity", y="ph", kind="scatter")
# Heatmap
import seaborn as sb
corr = df_train.corr()
sb.heatmap(corr, cmap="Blues", annot=True)
df_train.head()
df_test.head()
# Trying removing the osmo column
df_train = df_train.drop(columns="osmo", axis=0)
df_test = df_test.drop(columns="osmo", axis=0)
df_train = df_train.drop(columns="urea", axis=0)
df_test = df_test.drop(columns="urea", axis=0)
df_train.head()
df_test.head()
# Splitting training set into X and Y
X = df_train.iloc[:, 1:-1]
y = df_train.iloc[:, -1]
# # Splitting train and test set for trianing and validation
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.1)
# print(X_train.shape)
# print(y_train.shape)
# print(X_val.shape)
# print(y_val.shape)
# Training a linear model to check for baseline accuracy
# from sklearn.model_selection import GridSearchCV
# # example of grid searching key hyperparametres for logistic regression
# from sklearn.datasets import make_blobs
# from sklearn.model_selection import RepeatedStratifiedKFold
# solvers = ['newton-cg', 'lbfgs', 'liblinear']
# penalty = ['l2']
# c_values = [100, 10, 1.0, 0.1, 0.01]
# # Define grid search
# model = LogisticRegression()
# grid = dict(solver=solvers,penalty=penalty,C=c_values)
# cv = RepeatedStratifiedKFold(n_splits=10, n_repeats=3, random_state=1)
# grid_search = GridSearchCV(estimator=model, param_grid=grid, n_jobs=-1, cv=cv, scoring='accuracy',error_score=0)
# Find out the best parameters for the model using grid search and Stratified CV
# # Summarize results
# grid_result = grid_search.fit(X, y) # Caluclate the result
# print("Best: %f using %s" % (grid_result.best_score_, grid_result.best_params_))
# means = grid_result.cv_results_['mean_test_score']
# stds = grid_result.cv_results_['std_test_score']
# params = grid_result.cv_results_['params']
# for mean, stdev, param in zip(means, stds, params):
# print("%f (%f) with: %r" % (mean, stdev, param))
from sklearn.linear_model import LogisticRegression
clf = LogisticRegression(solver="newton-cg", penalty="l2", C=100, random_state=0)
clf.fit(X_train, y_train)
y_hat = clf.predict(X_test)
# print(" y_test shape :",y_val.shape)
from sklearn.metrics import accuracy_score
print("Accuracy Score :", accuracy_score(y_test, y_hat, normalize=True))
clf.coef_
X_train
# df_test_final.head()
df_test_sub = df_test.copy()
# df_test_sub = df_test_sub.drop(columns = 'osmo', axis = 0)
df_test_final = df_test.iloc[:, 1:]
df_test_pred = clf.predict(df_test_final)
df_test_sub["target"] = df_test_pred.tolist()
df_test_sub = df_test_sub[["id", "target"]]
df_test_sub.to_csv("submission.csv", index=False)
# df_test_sub
|
# # Cancer Analysis Using Decision Tree Classifier
# ## Table of Content
# ### 1. What is Decision Tree Classifier?
# ### 2. Importing Libraries
# ### 3. Loading Dataset
# ### 4. Data PreProcessing
# ### 5. Feature Scaling
# ### 6. EDA
# ### 7. Data Splitting
# ### 8. Model Selection and Training
# ### 9. Model Evaluation
# ### 10. AUC - ROC Curve
# ### 11. Conclusion
# ## 1. What is Decision Tree Classifier?
# ### A decision tree classifier is a machine learning algorithm that uses a tree-like model of decisions and their possible consequences to classify input data into different categories or predict outcomes.The decision tree is a graphical representation of all possible outcomes, which starts with a single node called the root node, and then branches out to various sub-nodes representing different possible choices or decisions.
# ### In a decision tree classifier, the input data is fed into the model, and the algorithm iteratively selects the best features to split the data at each node to create the most accurate classification model. The decision tree classifier is commonly used in many applications such as in finance, healthcare, and marketing, and is a popular algorithm for data mining and machine learning tasks.
# ## Entropy:-
# ### Entropy measures the impurity in the given dataset. In Physics and Mathematics, entropy is referred to as the randomness or uncertainty of a random variable X. In information theory, it refers to the impurity in a group of examples. Information gain is the decrease in entropy. Information gain computes the difference between entropy before split and average entropy after split of the dataset based on given attribute values.
# ### Entropy is represented by the following formula:-
# 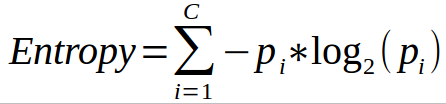
# ### Here, c is the number of classes and pi is the probability associated with the ith class.
# ### ID3 (Iterative Dichotomiser) Decision Tree algorithm uses entropy to calculate information gain. So, by calculating decrease in entropy measure of each attribute we can calculate their information gain. The attribute with the highest information gain is chosen as the splitting attribute at the node.
# ## 2. Importing Libraries
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import plotly.express as px
# ## 3. Loading Dataset
cancer_data = pd.read_csv("/kaggle/input/cancer-data/Cancer_Data.csv")
cancer_data.head()
cancer_data.info()
# ## 4. Data Preprocessing
cancer_data.shape
cancer_data.isnull().sum()
# removing the last column which only contains NaN values and it's not a dependent feature.
cancer_data = cancer_data.drop(["Unnamed: 32"], axis=1)
cancer_data.columns
cancer_data.isnull().sum()
cancer_data["diagnosis"].unique()
# ### Note: M = 1, B = 0
from sklearn import preprocessing
label_encoder = preprocessing.LabelEncoder()
cancer_data["diagnosis"] = label_encoder.fit_transform(cancer_data["diagnosis"])
cancer_data.head()
cancer_data["diagnosis"].unique()
cancer_data.describe()
# ## 5. Feature Scaling
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
cancer_data[["diagnosis", "smoothness_mean", "texture_mean"]],
cancer_data.diagnosis,
test_size=0.3,
random_state=0,
)
X_train.shape, X_test.shape
from sklearn.preprocessing import RobustScaler
scaler = RobustScaler()
X_train_scaled_ro = scaler.fit_transform(X_train)
X_test_scaled_ro = scaler.transform(X_test)
# Feature Scaling Using Robust Scaler.
print(
"mean value of diagnosis, smoothness_mean, texture_mean features: ",
X_train_scaled_ro.mean(axis=0),
)
print(
"std value of diagnosis, smoothness_mean, texture_mean features: ",
X_test_scaled_ro.std(axis=0),
)
# the distribution of the transformed variable Age
plt.hist(X_train_scaled_ro[:, 1], bins=8)
# the distribution of the transformed variable Fare
plt.hist(X_train_scaled_ro[:, 2], bins=20)
# ## 6. EDA
plt.figure(figsize=(8, 4))
sns.countplot(x=cancer_data["diagnosis"], palette="RdBu")
benign, malignant = cancer_data["diagnosis"].value_counts()
print("Number of cells labeled Benign : ", benign)
print("Number of cells labeled Malignant : ", malignant)
print("")
print("% of cells labeled Benign", round(benign / len(cancer_data) * 100, 2), "%")
print("% of cells labeled Malignant", round(malignant / len(cancer_data) * 100, 2), "%")
plt.show()
# Note: M = 1, B = 0
fig, ax = plt.subplots(figsize=(20, 20))
sns.heatmap(cancer_data.corr(), annot=True, linewidths=0.5, fmt=".1f", ax=ax)
plt.show()
sns.jointplot(
x=cancer_data.loc[:, "smoothness_mean"],
y=cancer_data.loc[:, "concave points_worst"],
kind="reg",
color="#ce1414",
)
plt.show()
# ## 7. Data Splitting
cancer_data.columns
X = cancer_data[
[
"id",
"radius_mean",
"texture_mean",
"perimeter_mean",
"area_mean",
"smoothness_mean",
"compactness_mean",
"concavity_mean",
"concave points_mean",
"symmetry_mean",
"fractal_dimension_mean",
"radius_se",
"texture_se",
"perimeter_se",
"area_se",
"smoothness_se",
"compactness_se",
"concavity_se",
"concave points_se",
"symmetry_se",
"fractal_dimension_se",
"radius_worst",
"texture_worst",
"perimeter_worst",
"area_worst",
"smoothness_worst",
"compactness_worst",
"concavity_worst",
"concave points_worst",
"symmetry_worst",
"fractal_dimension_worst",
]
]
Y = cancer_data["diagnosis"]
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(X, Y, test_size=0.3)
print("x_train: ", x_train.shape)
print("x_test: ", x_test.shape)
print("y_train: ", y_train.shape)
print("y_test: ", y_test.shape)
# ## 8. Model Selection and Training
from sklearn import tree
clf = tree.DecisionTreeClassifier(criterion="entropy")
clf.fit(x_train, y_train)
predict = clf.predict(x_test)
predict
clf.score(x_test, y_test)
# ## 9. Model Evaluation
from sklearn.metrics import accuracy_score
print("Accuracy Score: ", accuracy_score(y_test, predict))
from sklearn.metrics import precision_score
print("Precision Score: ", precision_score(y_test, predict))
from sklearn.metrics import recall_score
print("Recall Score: ", recall_score(y_test, predict))
from sklearn.metrics import f1_score
print("F1 Score: ", f1_score(y_test, predict))
from sklearn.metrics import classification_report
print(classification_report(y_test, predict))
from sklearn.metrics import confusion_matrix
cm = confusion_matrix(y_test, predict)
cm
# visualize confusion metrix
ax = sns.heatmap(cm, annot=True, cmap="Blues")
ax.set_title("Confusion Matrix with labels\n\n")
ax.set_xlabel("\nPredicted Values")
ax.set_ylabel("Actual Values ")
plt.show()
# ## 10. AUC-ROC Curve
# import metrics library from sklearn
from sklearn import metrics
y_pred_proba = clf.predict_proba(x_test)[::, 1]
fpr, tpr, _ = metrics.roc_curve(y_test, y_pred_proba)
# plot the ROC curve
plt.plot(fpr, tpr)
plt.ylabel("True Positive Rate")
plt.xlabel("False Positive Rate")
plt.show()
y_pred_proba = clf.predict_proba(x_test)[::, 1]
fpr, tpr, _ = metrics.roc_curve(y_test, y_pred_proba)
auc = metrics.roc_auc_score(y_test, y_pred_proba)
# plot the ROC curve
plt.plot(fpr, tpr, label="AUC = " + str(auc))
plt.ylabel("True Positive Rate")
plt.xlabel("False Positive Rate")
plt.legend(loc=4)
plt.show()
|
# Census data on income needs to be unpivoted so it can be used in a data tool such as Tableau, pivot tables, etc.
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Load data set
path_str = "../input/incomecsv/income.csv"
df = pd.read_csv(path_str)
df.head(10)
# Get rid of rows with duplicate year data
df.drop_duplicates(subset=["Racial_category", "Year"], keep="first", inplace=True)
df.duplicated(subset=["Racial_category", "Year"], keep="first")
# Create 3 summary categories (over 200 already exists)
df["under_50"] = (
df["Under_15"]
+ df["15_to_less_than_25"]
+ df["25_to_less_than_35"]
+ df["35_to_less_than_49"]
)
df["between_50_and_200"] = (
df["50_to_less_than_75"]
+ df["75_to_less_than_100"]
+ df["100_to_less_than_150"]
+ df["150_to_less_than_200"]
)
df.head()
# use melt function to unpivot columns
df_unpivoted = df.melt(
id_vars=["Racial_category", "Year", "median income", "mean income"],
var_name="income_group",
value_name="percentage",
)
df_unpivoted.head()
# Create dataframe to hold distinct income group names to map user-friendly descriptions.
df_income_group = pd.DataFrame(df_unpivoted["income_group"].unique())
df_income_group
# Create user-friendly names for income groupings
user_friendly_income_names = [
"Under 15,000",
"between 15,000 and 24,999",
"between 25,000 and 34,999",
"between 35,000 and 49,999",
"between 50,000 and 74,999",
"between 75,000 and 99,999",
"between 100,000 and 149,999",
"between 150,000 and 199,999",
"200,000 and above",
"Under 50,000",
"Between 50,000 and 200,000",
]
frames = [df_income_group, pd.DataFrame(user_friendly_income_names)]
map_income_name = pd.concat(frames, axis=1)
# name columns
map_income_name.columns = ["name", "alias"]
map_income_name.head(11)
# now join mapping to main data table
frames1 = [df_unpivoted, map_income_name]
df1 = pd.merge(df_unpivoted, map_income_name, left_on="income_group", right_on="name")
df1.head()
# rename alias column
df1.rename(
columns={"alias": "income_category", "percentage": "income_category_percentage"},
inplace=True,
)
df1.head()
# remove extra columns
df1.pop("income_group")
df1.pop("name")
df1.head()
# export to file for downloading
df1.to_csv(r"/kaggle/working/df_unpivoted.csv")
|
# # INTRODUCTION
# * **MDS-UPDRS** - Movement Disorder Society UPDRS revision.
# - Revised by removing all `yes` and `no` portions of the original questionaire.
# - Currently consists of `0-4` rating scale to maintain consistency.
# **MDS-UPDRS consists of 4 parts:**
# - Part I: Non-Motor Experiences of Daily Living
# - Part II: motor Experiences of Daily Living
# - Part III: Motor Examination
# - Part IV: Motor Complications
# **MDS-UPDRS staging scales:**
# - Modified Hoehn and Yahr Staging
# - Modeified Schwab and England Activities on Daily Living Scale
# # EXPLORATORY DATA ANALYSIS
# EDA will cover the following data:
# 1. Clinical Data
# 2. Peptide Data
# 3. Protein Data
# **Imports:**
# Imports
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn import model_selection
from sklearn import metrics
# 垃圾处理器
import gc
# **Read in DataFrames:**
df_train_clin = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/train_clinical_data.csv"
)
df_train_pept = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/train_peptides.csv"
)
df_train_prot = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/train_proteins.csv"
)
# ## Data Summary
# ### Clincal Data
df_train_clin.head(10)
print(
f'Unique Clinical Data patient #: {df_train_clin["patient_id"].nunique()}'
) # 查看病人个数
print("----------------------------------------------------------")
print(f"Null Values Found in Clinical Data:") # 临床数据中的空值
for col in df_train_clin.columns:
print(f"Null values found in {col}: {df_train_clin[col].isna().sum()}")
print("")
# ## 注释
# 上述代码分析了**临床数据**
# 一共有 **248** 位病人
# 缺失值分析:
# Null values found in updrs_1: **1**
# Null values found in updrs_2: **2**
# Null values found in updrs_3: **25**
# Null values found in updrs_4: **1038**
# Null values found in upd23b_clinical_state_on_medication: **1327**
# ### Peptide Data
df_train_pept.head(10)
print(f'Unique Peptide Data patient #: {df_train_pept["patient_id"].nunique()}') # 病人
print(f'Unique Peptides #: {df_train_pept["Peptide"].nunique()}') # 肽的种类
print("----------------------------------------------------------")
print(f"Null Values Found in Peptide Data:") # 肽的缺失值
for col in df_train_pept.columns:
print(f"Null values found in {col}: {df_train_pept[col].isna().sum()}")
print("")
# ## 注释
# 代码分析了 **肽** 的数据
# 仍然是 **248** 位病人
# 一共有 **968** 种肽
# 没啥缺失值 😁
# ### Protein Data
df_train_prot.head(10)
print(f'Unique Protein Data patient #: {df_train_prot["patient_id"].nunique()}')
print(f'Unique Proteins #: {df_train_prot["UniProt"].nunique()}')
print("----------------------------------------------------------")
print(f"Null Values Found in Protein Data:")
for col in df_train_prot.columns:
print(f"Null values found in {col}: {df_train_prot[col].isna().sum()}")
print("")
# ## 注释
# 还是这 **248** 位病人
# 蛋白质有 **227** 种
# 没啥缺失值 😁
# Null Values Found in Protein Data:
# Null values found in visit_id: 0
# Null values found in visit_month: 0
# Null values found in patient_id: 0
# Null values found in UniProt: 0
# Null values found in NPX: 0
# ## MDS-UPDRS SCORE ANALYSIS
# ### UPDRS_1-4 Score Analysis
# plt.figure(figsize = (10, 3))
# sns.histplot(x=df_train_clin['updrs_1'], stat='count', kde = True)
# plt.xlabel('UPDRS 1')
# plt.ylabel('COUNT')
# plt.title('UPDRS 1 SCORE (total)')
# plt.show()
# # 看看其他的分数
# plt.figure(figsize = (10, 3))
# sns.histplot(x=df_train_clin['updrs_2'], stat='count', kde = True)
# plt.xlabel('UPDRS 2')
# plt.ylabel('COUNT')
# plt.title('UPDRS 2 SCORE (total)')
# plt.show()
# plt.figure(figsize = (10, 3))
# sns.histplot(x=df_train_clin['updrs_3'], stat='count', kde = True)
# plt.xlabel('UPDRS 3')
# plt.ylabel('COUNT')
# plt.title('UPDRS 3 SCORE (total)')
# plt.show()
# plt.figure(figsize = (10, 3))
# sns.histplot(x=df_train_clin['updrs_4'], stat='count', kde = True)
# plt.xlabel('UPDRS 4')
# plt.ylabel('COUNT')
# plt.title('UPDRS 4 SCORE (total)')
# plt.show()
# **Continuing EDA Soon**
# ## Target Preperation
# ## 注释
# 构造分数 也就是 要预测的值 **y** 啦
# **有一些问题**
# 1. 数据使用率低
# 2. 构造的数据中有 [0 0 0 0] 我觉得不对 😂
# 总体感觉挺好的 👍👍👍👍
# Create Targets
patients = {}
for e in range(1, 5): # 1 2 3 4
for m in [0, 6, 12, 24]:
df_train_clin[f"updrs_{e}_plus_{m}_months"] = 0
"""
为 df_train_clin 增加了 4 * 4 = 16 列
分别对应要预测的 16 个值 初始化为了 0 好奇后续是怎么赋值的 🧐
updrs_1_plus_0_months
updrs_1_plus_6_months
updrs_1_plus_12_months
updrs_1_plus_24_months
updrs_2_plus_0_months
......
......
updrs_4_plus_12_months
updrs_4_plus_24_months
"""
for patient in df_train_clin.patient_id.unique():
# 开始处理每个人的数据 👏
temp = df_train_clin[df_train_clin.patient_id == patient] # 挑出这个人的看病数据
# debug
# print("👏👏👏👏temp:\n", temp)
month_list = []
month_windows = [0, 6, 12, 24]
for month in temp.visit_month.values:
month_list.append(
[month, month + 6, month + 12, month + 24]
) # 每一个访问的月份都对应 4 个月?
# debug
# print("👏👏👏👏month:\n", month_list)
for month in range(len(month_list)):
for x in range(1, 5): # 找出每四个月对应的 1 2 3 4 分数
arr = (
temp[temp.visit_month.isin(month_list[month])][f"updrs_{x}"]
.fillna(0)
.to_list()
) # 一种分数 四个月 的 list 👏👏👏👏👏👏👏👏👏
# debug
# print("👏👏👏👏arr:\n", arr)
if len(arr) == 4: # 如果4 个月的分数都找到了
# if len(arr) == 4 and not sum(arr) == 0: # 如果4 个月的分数都找到了
# debug
# print("add", arr)
for e, i in enumerate(arr):
m = month_list[month][0] # 起始月份 m
temp.loc[
temp.visit_month == m,
[f"updrs_{x}_plus_{month_windows[e]}_months"],
] = i # 对应分数
else:
# debug
# print("delete", arr)
temp = temp[~temp.visit_month.isin(month_list[month])] # 应该是删掉这条数据
patients[patient] = temp # 一位病人的数据就处理好了
# debug
# break
# 看看 patients 张啥样 👀
print(len(patients)) # 248 位病人
patients[55] # 这是一个 dataframe
formatted_clin = (
pd.concat(patients.values(), ignore_index=True).set_index("visit_id").iloc[:, 7:]
) # visit_id 区分不同的访问
print(formatted_clin.shape) # 数据集大小 (954, 16)
formatted_clin.iloc[953, :]
# ## Feature Preperation
protfeatures = df_train_prot.pivot(index="visit_id", columns="UniProt", values="NPX")
print(protfeatures.shape) # (1113, 227) 一共 1113 条看病数据 每次 227 种蛋白质
protfeatures.head()
# 加入 肽的数据
peptfeatures = df_train_pept.pivot(
index="visit_id", columns="Peptide", values="PeptideAbundance"
)
print(peptfeatures.shape) # (1113, 968) 一共 1113 条看病数据 每次 968 种肽
peptfeatures.head(10)
# **Peptide features coming soon**
# df1 = protfeatures.merge(formatted_clin, left_index=True, right_index=True, how='right') # 蛋白质 和 分数 merge 到一起
# df2 = peptfeatures.merge(df1, left_index=True, right_index=True, how='right') # 肽 和 蛋白质 和 分数 merge 到一起
# df3 = df2.fillna(0)
# print(f'\n protfeatures NA values: {df3[protfeatures.columns].isna().sum().sum()/(len(df3)*len(protfeatures.columns)):.2%}') # 蛋白质缺失率 53.64%
# print(f'\n peptfeatures NA values: {df3[peptfeatures.columns].isna().sum().sum()/(len(df3)*len(peptfeatures.columns)):.2%}') # 肽缺失率 53.64%
# df3['visit_month'] = df3.reset_index().visit_id.str.split('_').apply(lambda x: int(x[1])).values
# print(df3.shape)
# df3.head(10)
# df = peptfeatures.merge(formatted_clin, left_index=True, right_index=True, how='right').fillna(0) # 肽 和 分数 merge 到一起
# print(f'\n peptfeatures NA values: {df[peptfeatures.columns].isna().sum().sum()/(len(df)*len(peptfeatures.columns)):.2%}') # 肽缺失率 53.64%
# df['visit_month'] = df.reset_index().visit_id.str.split('_').apply(lambda x: int(x[1])).values
# print(df.shape)
# df.head(10)
df = protfeatures.merge(
formatted_clin, left_index=True, right_index=True, how="right"
).fillna(
0
) # 蛋白质 和 分数 merge 到一起
print(
f"\n protfeatures NA values: {df[protfeatures.columns].isna().sum().sum()/(len(df)*len(protfeatures.columns)):.2%}"
) # 蛋白质缺失率 53.64%
df["visit_month"] = (
df.reset_index().visit_id.str.split("_").apply(lambda x: int(x[1])).values
)
print(df.shape)
df.head(10)
# visit_month_list = df3.reset_index().visit_id.str.split('_').apply(lambda x: int(x[1])).unique().tolist() # 有哪几个月 [0, 6, 12, 18, 24, 30, 36]
# peptide_protein_list = peptfeatures.columns.to_list() + protfeatures.columns.to_list() # 肽 和 蛋白质种类
# len(peptide_protein_list) # 1195 = 277 + 968
# visit_month_list = df.reset_index().visit_id.str.split('_').apply(lambda x: int(x[1])).unique().tolist() # 有哪几个月 [0, 6, 12, 18, 24, 30, 36]
# peptide_list = peptfeatures.columns.to_list() # 肽
# len(peptide_list) # 968
visit_month_list = (
df.reset_index()
.visit_id.str.split("_")
.apply(lambda x: int(x[1]))
.unique()
.tolist()
) # 有哪几个月 [0, 6, 12, 18, 24, 30, 36]
protein_list = protfeatures.columns.to_list() # 肽 和 蛋白质种类
len(protein_list) # 277
# X = df3[peptide_protein_list + ["visit_month"]] # visit_month 也作为了一种影响因素
# # print("👏👏👏👏👏👏X", X.head(8))
# y = df3[formatted_clin.columns]
# # print("👏👏👏👏👏👏y", y.head(8))
# print('\nX and y shapes:') # 看样子整挺好 😁👍👍👍👍
# X.shape, y.shape
# X = df[peptide_list + ["visit_month"]] # visit_month 也作为了一种影响因素
# # print("👏👏👏👏👏👏X", X.head(8))
# y = df[formatted_clin.columns]
# # print("👏👏👏👏👏👏y", y.head(8))
# print('\nX and y shapes:') # 看样子整挺好 😁👍👍👍👍
# X.shape, y.shape
X = df[protein_list + ["visit_month"]] # visit_month 也作为了一种影响因素
# print("👏👏👏👏👏👏X", X.head(8))
y = df[formatted_clin.columns]
# print("👏👏👏👏👏👏y", y.head(8))
print("\nX and y shapes:") # 看样子整挺好 😁👍👍👍👍
X.shape, y.shape
# # **上述数据处理部分总结**
# > 个人感觉这篇文章不错 👍👍👍👍👍
# 构造的X和Y很值得我们学习,也有一些不足我们可以改进
from sklearn.preprocessing import (
OrdinalEncoder,
StandardScaler,
) # OrdinalEncoder 顺序编码器 StandardScaler 数据归一化
from sklearn.compose import ColumnTransformer, make_column_selector # 特征变换方法
from sklearn.pipeline import make_pipeline
from sklearn.experimental import enable_iterative_imputer
from sklearn.impute import IterativeImputer
X.visit_month = X.visit_month.astype("float")
y = y.astype("float")
feature_trans = ColumnTransformer(
[
(
"numerical",
# make_pipeline(IterativeImputer(), StandardScaler()),
make_pipeline(StandardScaler()),
make_column_selector(dtype_include="number"),
),
]
)
X_transformed = feature_trans.fit_transform(X)
print(X_transformed.shape)
# # sMAPE Function
import tensorflow.keras.backend as K
def smape_loss(y_true, y_pred):
epsilon = 0.1
numer = K.abs(y_pred - y_true)
denom = K.maximum(K.abs(y_true) + K.abs(y_pred) + epsilon, 0.5 + epsilon)
smape = numer / (denom / 2)
smape = tf.where(tf.math.is_nan(smape), tf.zeros_like(smape), smape)
return smape
def calculate_smape(y_true, y_pred):
y_true, y_pred = np.array(y_true), np.array(y_pred)
numer = np.round(np.abs(y_pred - y_true), 0)
denom = np.round(np.abs(y_true) + np.abs(y_pred), 0)
return 1 / len(y_true) * np.sum(np.nan_to_num(numer / (denom / 2))) * 100
# # TensorFlow Model
import tensorflow as tf
from tensorflow.keras.layers import Dense
from tensorflow.keras.layers import Dropout
input_shape = [X.shape[1]]
model = tf.keras.Sequential() # 多层感知机模型
model.add(Dense(256, input_shape=input_shape, activation="relu"))
model.add(Dropout(0.4))
model.add(Dense(128, activation="relu"))
model.add(Dropout(0.4))
model.add(Dense(64, activation="relu"))
model.add(Dropout(0.4))
model.add(Dense(32, activation="relu"))
model.add(Dropout(0.4))
model.add(Dense(y.shape[1]))
model.summary()
from keras.utils.vis_utils import plot_model
model.compile(optimizer="adam", loss=smape_loss)
history = model.fit(
X_transformed,
y,
epochs=150, # 原本 150 得分 58.1 200 得分 59.4 100 得分 58.5
verbose=False,
validation_split=0.2,
)
pd.DataFrame(history.history).plot()
# # Submission Preperation
test_proteins = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/example_test_files/test_proteins.csv"
)
test_peptides = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/example_test_files/test_peptides.csv"
)
sample_submission = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/example_test_files/sample_submission.csv"
)
clinical = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/example_test_files/test.csv"
)
def prepare_features(clinical, peptide, proteins):
"""Format features"""
# X_proteins = proteins.pivot(index='visit_id',columns='UniProt', values='NPX')
# X_peptide = peptide.pivot(index='visit_id', columns='Peptide', values='PeptideAbundance')
# X_submission = X_peptide.merge(X_proteins, left_index=True, right_index=True, how='right')
# X_submission = peptide.pivot(index='visit_id', columns='Peptide', values='PeptideAbundance')
X_submission = proteins.pivot(index="visit_id", columns="UniProt", values="NPX")
# print(X_submission.shape)
missing_visits = np.setdiff1d(
np.array(clinical.visit_id.unique()), np.array(X_submission.index)
).tolist()
missing_peptide_protein_list = np.setdiff1d(
np.array(protein_list), np.array(X_submission.columns)
).tolist()
for mv in missing_visits:
X_submission.loc[mv] = np.NaN
for mp in missing_peptide_protein_list:
X_submission.loc[:, mp] = np.NaN
X_submission = X_submission[protein_list]
X_submission["visit_month"] = (
X_submission.reset_index()
.visit_id.str.split("_")
.apply(lambda x: int(x[1]))
.values
)
X_submission.visit_month = X_submission.visit_month.astype("float")
X_submission = X_submission.fillna(0)
return X_submission
def get_predictions(features, model):
"""Make predictions from prepared features"""
X_submission_trans = feature_trans.transform(features)
# Predict and format ratings
pred_submission = np.around(np.abs(model.predict(X_submission_trans)), 0)
# pred_submission = np.abs(model.predict(X_submission_trans))
pred_submission = pd.DataFrame(
pred_submission, index=features.index, columns=y.columns
)
return pred_submission
def format_predictions(predictions, sample_submission):
"""Format predictions for submission"""
pred_submission = pd.DataFrame(predictions.stack())
# Map predictions to sample_submission
pred_submission.index = pred_submission.index.map("_".join)
pred_submission.columns = ["rating"]
sample_submission.rating = sample_submission.prediction_id.map(
pred_submission.rating
)
return sample_submission.fillna(0)
test_features = prepare_features(clinical, test_peptides, test_proteins)
test_predictions = get_predictions(test_features, model)
test_submission = format_predictions(test_predictions, sample_submission)
test_submission
# # Submission
import sys
sys.path.append("/kaggle/input/amp-parkinsons-disease-progression-prediction")
import amp_pd_peptide
env = amp_pd_peptide.make_env() # environment provided by competition
iter_test = env.iter_test()
for test, test_peptides, test_proteins, sample_submission in iter_test:
sub_features = prepare_features(test, test_peptides, test_proteins)
sub_predictions = get_predictions(sub_features, model)
submission = format_predictions(sub_predictions, sample_submission)
env.predict(submission)
pd.read_csv("/kaggle/working/submission.csv")
|
import numpy as np
import pandas as pd
df = pd.read_csv(
"/kaggle/input/bookcrossing-dataset/Books Data with Category Language and Summary/Preprocessed_data.csv"
)
df.head()
df.shape
# We can drop all the unnecessary columns now. {ALL THE columns name}. But before dropping book title, we need some way to access each book. So, lets create a book id for each book.
#
df["book_id"] = pd.factorize(df["book_title"])[0]
# As random forest is a ML model, it cannot accomodate large amounts of data/it will be so slow. So, lets take the first 2000 columns and proceed.
new_df = df.head(5000)
# Now, let's drop all the unnecessary columns.
new_df = new_df.drop(
[
"Unnamed: 0",
"location",
"book_title",
"isbn",
"img_s",
"img_m",
"img_l",
"Summary",
"city",
"state",
],
axis=1,
)
# Next, we can remove noise in our data. Books which are read by very little number of people will not be of much use to us. So, lets drop all the books that are read by 20 or less than 20 people.
book_counts = new_df[
"book_id"
].value_counts() # Returns bookid: frequency , dictionary type object
popular_books = []
threshold = 20
for item in book_counts.items():
# items are tuples with 2 values. item=(key,value)
# Here key is book_id and value is it's frequency , ie count.
book_id = item[0]
count = item[1]
if count >= threshold:
popular_books.append(book_id)
# filter new_df to include only rows with book IDs in the popular_books list
new_df = new_df[new_df["book_id"].isin(popular_books)]
# Now let's do a similar thing for countries. But, instead of dropping the values below the threshold. Let's put that into a separate column. Basically, we are going to take top 10 countries and one-hot encode it and put the rest in a separate column called 'others'.
country_counts = new_df["country"].value_counts()
top_countries = country_counts.keys()[:10]
top_countries = top_countries.to_list()
index = new_df.index.tolist()
length = len(new_df)
j = 0
for i in index:
if new_df.loc[i, "country"] not in top_countries:
new_df.loc[i, "country"] = "other"
country = pd.get_dummies(new_df["country"])
new_df = pd.concat([new_df, country], axis=1)
new_df = new_df.drop("country", axis=1)
# Now, next we said we are going to use random forest regressor. Can we give categorical or string values to it?
# No , we can't. We can only give numerical values. So, lets one-hot encode it. Before that, lets find which columns we should one-hot encode.
new_df.info()
# - book_author
# - publisher
# - Language
# - Category
# These are all categorical columns. Lets one hot encode them. But before that, can we actually one hot encode them?( number of columns constraint)
print(" Number of unique Languages: ", new_df["Language"].nunique())
print(" Number of unique Publishers: ", new_df["publisher"].nunique())
print(" Number of unique Categories: ", new_df["Category"].nunique())
print(" Number of unique Authors: ", new_df["book_author"].nunique())
# Only languages are one hot encodable.
# So basically for other columns ,we can either
# - 1) Label encode , but it just gives them a number and our model might think there is a correlation
# - 2) Drop , we lose valuable info
# Lets do both and try which works better.
# One hot encoding Language
Languages = pd.get_dummies(new_df["Language"])
new_df = pd.concat([new_df, Languages], axis=1)
new_df = new_df.drop("Language", axis=1)
# Let's do the first method and see.
new_df["book_author"], authors = pd.factorize(new_df["book_author"])
new_df["publisher"], publishers = pd.factorize(new_df["publisher"])
new_df["Category"], categories = pd.factorize(new_df["Category"])
# Now let's drop, which is the second method. And put it in separate dataframe. So, that it will be easier to compare.
dropDF = new_df.drop(columns=["book_author", "publisher", "Category"], axis=1)
from sklearn.ensemble import RandomForestRegressor
from sklearn.model_selection import train_test_split
# Let's split our dataset. Why do we do that?
train_data = new_df.sample(frac=0.8, random_state=1)
test_data = new_df.drop(train_data.index)
# train_data =dropDF.sample(frac=0.8, random_state=1)
# test_data = dropDF.drop(train_data.index)
# Let's define X and y
X_train = train_data.drop(["rating"], axis=1)
y_train = train_data["rating"]
X_test = test_data.drop(["rating"], axis=1)
y_test = test_data["rating"]
# Let's build our model and predict.
rf = RandomForestRegressor()
rf.fit(X_train, y_train)
y_pred = rf.predict(X_test)
# Let's find the error.
from sklearn.metrics import mean_squared_error
mse = mean_squared_error(y_test, rf.predict(X_test))
print(mse)
# Now we have made our model predict the ratinga user might give for each book. But what we need as an endproduct?? What are all the books a user might read right?
# For that, lets first sort the rating.
rating_index = y_pred.argsort()[::-1]
sorted_rating = y_pred[rating_index]
# Now lets print out the top 10 ratings and print the book title for the respective book title.
top_recommendations = test_data.iloc[rating_index[:10]]["book_id"]
book_titles = df[df["book_id"].isin(top_recommendations)]["book_title"][:10]
for book_title in book_titles:
print(book_title)
# Can you see what's the mistake?
top_recommendations = test_data.iloc[rating_index[:10]]["book_id"]
book_titles = df[df["book_id"].isin(top_recommendations)]["book_title"].unique()[:10]
for book_title in book_titles:
print(book_title)
|
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import numpy as np
df = pd.read_csv("../input/hr-analytics-job-change-of-data-scientists/aug_train.csv")
null_df = (
df.isnull().sum().to_frame("null_count").rename_axis("column_name").reset_index()
)
null_df.head()
null_df["null_perc"] = null_df["null_count"] * 100 / 19158
null_df
gender_df = df.groupby("gender").agg(["count", "sum"])["target"].reset_index()
bar1 = np.arange(len(gender_df["gender"]))
bar2 = [i + 0.4 for i in bar1]
plt.bar(bar1, gender_df["count"], 0.5, label="total")
plt.bar(bar2, gender_df["sum"], 0.5, label="willing to join")
plt.xticks(bar1, gender_df["gender"])
plt.legend()
plt.show()
gender_df["leaving_perc"] = gender_df["sum"] * 100 / gender_df["count"]
gender_df
education_df = (
df.groupby("education_level").agg(["count", "sum"])["target"].reset_index()
)
bar1 = np.arange(len(education_df["education_level"]))
bar2 = [i + 0.2 for i in bar1]
plt.bar(bar1, education_df["count"], 0.2, label="total")
plt.bar(bar2, education_df["sum"], 0.2, label="willing to join")
plt.xticks(bar1, education_df["education_level"])
plt.legend()
plt.show()
education_df["leaving_perc"] = education_df["sum"] * 100 / education_df["count"]
education_df.sort_values(by="leaving_perc", ascending=False)
ax = sns.boxplot(x="gender", y="training_hours", data=df)
ax.set_xticklabels(ax.get_xticklabels(), rotation=90)
experience = df[df["target"] == 1.0]
sns.countplot(
x="experience",
hue="gender",
data=experience,
order=experience["experience"].value_counts().sort_values(ascending=False).index,
)
# Show the plot
plt.title("people willing to leave company")
plt.ylabel("count of ppl")
plt.show()
company_type_df = df[df["target"] == 1.0]
sns.countplot(
x="company_type",
hue="gender",
data=company_type_df,
order=company_type_df["company_type"]
.value_counts()
.sort_values(ascending=False)
.index,
)
# Show the plot
plt.title("people willing to leave company")
plt.ylabel("count of ppl")
plt.xticks(rotation=90)
plt.show()
sns.heatmap(df.corr(), vmin=-1, vmax=1, annot=True)
df.drop(["enrollee_id"], axis=1, inplace=True)
sns.pairplot(df, hue="target", size=3)
plt.show()
|
# submission_pseudo_lgb 0.84192
# submission_pseudo_lgb_4 0.84195
# submission_pseudo_lgb_5 0.84196
# pseudo_lgb_1 0.84196
import os
import numpy as np
import pandas as pd
pred1 = pd.read_csv("../input/tps-feb-submission-ensemble/submission_pseudo_lgb.csv")
pred2 = pd.read_csv("../input/tps-feb-submission-ensemble/submission_pseudo_lgb_4.csv")
pred3 = pd.read_csv("../input/tps-feb-submission-ensemble/submission_pseudo_lgb_5.csv")
pred4 = pd.read_csv("../input/tps-feb-submission-ensemble/pseudo_lgb_1.csv")
pred = (pred1.target * 4 + pred2.target * 3 + pred3.target * 2 + pred4.target * 1) / 10
pred
submission = pd.read_csv(
"../input/tabular-playground-series-feb-2021/sample_submission.csv"
)
submission.target = pred
submission
submission.to_csv("ensemble.csv", index=False)
|
# # Wine Quality Test Using Support Vector Classifier
# ## Table of Content
# ### 1. What is Support Vector Machine?
# ### 2. Importing Libraries
# ### 3. Uploading Dataset
# ### 4. Data PreProcessing
# ### 5. Feature Scaling
# ### 6. EDA
# ### 7. Data Splitting
# ### 8. Model Selection and Training
# ### 9. Model Evaluation
# ### 10. Conclusion
# ## 1. What is Support Vector Machine?
# ### Support Vector Machine or SVM is one of the most popular Supervised Learning algorithms, which is used for Classification as well as Regression problems. However, primarily, it is used for Classification problems in Machine Learning.
#
# ### The goal of the SVM algorithm is to create the best line or decision boundary that can segregate n-dimensional space into classes so that we can easily put the new data point in the correct category in the future. This best decision boundary is called a hyperplane.
#
# ### There are two types of SVM:-
# 1. Linear SVM: Linear SVM is used for linearly separable data, which means if a dataset can be classified into two classes by using a single straight line, then such data is termed as linearly separable data, and classifier is used called as Linear SVM classifier.
#
# 2. Non-Linear SVM: Non-Linear SVM is used for non-linearly separated data, which means if a dataset cannot be classified by using a straight line, then such data is termed as non-linear data and classifier used is called as Non-linear SVM classifier.
# ## 2. Import Libraries
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import plotly.express as px
# ## 3. Uploading DataSet
WineQT_data = pd.read_csv("/kaggle/input/wine-quality-test.csv")
WineQT_data.head()
WineQT_data.info()
# ## 4. Data Preprocessing
WineQT_data.isnull().sum()
# #### Here, In this Data no null or missing values.
WineQT_data.duplicated().sum()
WineQT_data["alcohol"].unique()
WineQT_data["quality"].unique()
# remove unnecessary features
del WineQT_data["Id"]
WineQT_data.describe()
# ## 5. Feature Scaling
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
WineQT_data[["alcohol", "quality", "density"]],
WineQT_data.alcohol,
test_size=0.3,
random_state=0,
)
X_train.shape, X_test.shape
# feature scaling using min-max scaler
from sklearn.preprocessing import MinMaxScaler
scaler = MinMaxScaler()
X_train_scaled_minmax = scaler.fit_transform(X_train)
X_test_scaled_minmax = scaler.transform(X_test)
print(
"mean value of Alcohol, Quality and Density features: ",
X_train_scaled_minmax.mean(axis=0),
)
print(
"std value of Alcohol, Quality and Density features: ",
X_test_scaled_minmax.std(axis=0),
)
# plot the histogram of x_train min-max scale
plt.hist(X_train_scaled_minmax[:, 1], bins=20)
plt.hist(X_train_scaled_minmax[:, 2], bins=20)
from sklearn.preprocessing import MinMaxScaler
scaler = MinMaxScaler(feature_range=(0, 1))
df = scaler.fit_transform(WineQT_data)
df = pd.DataFrame(df, columns=WineQT_data.columns)
print(df.head())
# ## 6. EDA
f, ax = plt.subplots(1, 1)
sns.set(style="darkgrid")
sns.histplot(data=WineQT_data, x="density", color="g", bins=10, kde=True, legend=False)
# ### In this histogram plot analysis of density of Wine.
fig = plt.figure(figsize=(8, 4))
sns.barplot(x="quality", y="alcohol", data=WineQT_data)
plt.figure(figsize=(16, 10))
ax = sns.heatmap(WineQT_data.corr(), annot=True)
plt.show()
# ## 7. Data splitting
WineQT_data.columns
X = WineQT_data[
[
"fixed acidity",
"volatile acidity",
"citric acid",
"residual sugar",
"chlorides",
"free sulfur dioxide",
"total sulfur dioxide",
"density",
"pH",
"sulphates",
"alcohol",
]
]
Y = WineQT_data[["quality"]]
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(X, Y, test_size=0.3)
# ## 8. Model Selection and Training
from sklearn.svm import SVC
model = SVC()
model.fit(x_train, y_train)
model.score(x_test, y_test)
predict = model.predict(x_test)
predict
# ## 9. Model Evaluation
from sklearn.metrics import accuracy_score
accuracy_score(y_test, predict)
from sklearn.metrics import classification_report
print(classification_report(y_test, predict))
# ## Confusion Matrix
from sklearn.metrics import confusion_matrix
cm = confusion_matrix(y_test, predict)
cm
import seaborn as sns
ax = sns.heatmap(cm, annot=True, cmap="Blues")
ax.set_title("Confusion Matrix with labels\n\n")
ax.set_xlabel("\nPredicted Values")
ax.set_ylabel("Actual Values ")
plt.show()
|
# # **Predict the Colombo Stock Exchange Prices using Recurrent Neural Network (RNN) and Long Short-Term Memory (LSTM).**
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import math
import seaborn as sns
from tensorflow.keras import Sequential
from tensorflow.keras.layers import Dense, LSTM, Dropout
data = pd.read_csv("../input/colombo-stock-exchange/CSE.csv", date_parser=True)
data.isnull().sum()
data = data.dropna()
# here we are printing shape of data
data.shape
plt.figure(figsize=(15, 7))
sns.lineplot(data=data[["Open", "High", "Low", "Close"]], linewidth=2)
plt.grid(True)
plt.show()
# here we are Visualising the closing price history
plt.figure(figsize=(14, 5))
plt.title("Close Price History")
plt.plot(data["Close"])
plt.xlabel("Date")
plt.ylabel("Close Price USD ($)")
plt.show()
# Create a new data frame with only the closing price and convert it to an array. Then create a variable to store the length of the training data set. I want the training data set to contain about 80% of the data.
# Creating a new dataframe with only the 'Close' column
df = data.filter(["Close"])
# Converting the dataframe to a numpy array
dataset = df.values
# Get /Compute the number of rows to train the model on
training_data_len = math.ceil(len(dataset) * 0.8)
training_data_len
# Now scale the data set to be values between 0 and 1 inclusive, I do this because it is generally good practice to scale your data before giving it to the neural network.
# here we are Scaling the all of the data to be values between 0 and 1
from sklearn.preprocessing import MinMaxScaler
scaler = MinMaxScaler(feature_range=(0, 1))
scaled_data = scaler.fit_transform(dataset)
scaled_data
# Creating the scaled training data set
train_data = scaled_data[0:training_data_len, :]
# Spliting the data into x_train and y_train data sets
x_train = []
y_train = []
for i in range(60, len(train_data)):
x_train.append(train_data[i - 60 : i, 0])
y_train.append(train_data[i, 0])
if i <= 61:
print(x_train)
print(y_train)
print()
# Here we are Converting x_train and y_train to numpy arrays
x_train, y_train = np.array(x_train), np.array(y_train)
# Here we are reshaping the data into the shape accepted by the LSTM
x_train = np.reshape(x_train, (x_train.shape[0], x_train.shape[1], 1))
# now we are Building the LSTM network model
model = Sequential()
model.add(LSTM(units=50, return_sequences=True, input_shape=(x_train.shape[1], 1)))
model.add(LSTM(units=50, return_sequences=False))
model.add(Dense(units=25))
model.add(Dense(units=1))
# here we are Compiling the model
model.compile(optimizer="adam", loss="mean_squared_error")
# here we are training the model
model.fit(x_train, y_train, batch_size=1, epochs=1)
# here we are testing data set
test_data = scaled_data[training_data_len - 60 :, :]
# Creating the x_test and y_test data sets
x_test = []
y_test = dataset[
training_data_len:, :
] # Get all of the rows from index 1603 to the rest and all of the columns (in this case it's only column 'Close'), so 2003 - 1603 = 400 rows of data
for i in range(60, len(test_data)):
x_test.append(test_data[i - 60 : i, 0])
# here we are converting x_test to a numpy array
x_test = np.array(x_test)
# here we are reshaping the data into the shape accepted by the LSTM
x_test = np.reshape(x_test, (x_test.shape[0], x_test.shape[1], 1))
# now we are getting the models predicted price values
predictions = model.predict(x_test)
predictions = scaler.inverse_transform(predictions) # Undo scaling
# here we are calculaing the value of RMSE
rmse = np.sqrt(np.mean(((predictions - y_test) ** 2)))
rmse
# Plot/Create the data for the graph
train = data[:training_data_len]
valid = data[training_data_len:]
valid["Predictions"] = predictions
# Visualize the data
plt.figure(figsize=(16, 8))
plt.title("Model")
plt.xlabel("Date")
plt.ylabel("Close Price USD ($)")
plt.plot(train["Close"])
plt.plot(valid[["Close", "Predictions"]])
plt.legend(["Train", "Val", "Predictions"], loc="lower right")
plt.show()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import matplotlib.pyplot as plt
import seaborn as sns
import tensorflow
import keras
from keras.preprocessing.text import Tokenizer
from keras.preprocessing.sequence import pad_sequences
from sklearn.model_selection import train_test_split
from keras.layers import Input, Embedding, LSTM, Conv1D, GlobalMaxPooling1D, Dense
from keras.models import Model
from sklearn.preprocessing import LabelEncoder
from keras.utils import to_categorical
df = pd.read_csv("/kaggle/input/netflix-shows/netflix_titles.csv")
df.head()
df.isna().sum()
df["Genre"] = df["listed_in"].apply(lambda x: x.split(",")[0])
df["Genre"]
plt.figure(figsize=(10, 6))
sns.countplot(df["Genre"])
plt.xticks(rotation=90)
plt.show()
genre = [
"Dramas",
"Comedies",
"Documentaries",
"Action & Adventure",
"International TV Shows",
"Children & Family Movies",
"Stand-Up Comedy",
"Crime TV Shows",
"Horror Movies",
"Kids' TV",
]
len(df[df.Genre.isin(genre)]) / len(df)
df["len_desc"] = df["description"].apply(lambda x: len(x.split(" ")))
sns.displot(df["len_desc"])
data = df[df.Genre.isin(genre)][["Genre", "description"]]
data
tokenizer = Tokenizer()
tokenizer.fit_on_texts(data["description"])
sequences = tokenizer.texts_to_sequences(data["description"])
padded_seq = pad_sequences(sequences, maxlen=30, padding="post")
vocab_size = len(tokenizer.word_index)
le = LabelEncoder()
labels = le.fit_transform(data["Genre"])
labels = to_categorical(labels)
le.classes_
X_train, X_test, y_train, y_test = train_test_split(
padded_seq, labels, test_size=0.3, stratify=labels
)
inputs = Input(shape=(30,))
embedding = Embedding(vocab_size + 1, 64)(inputs)
lstm = LSTM(30, return_sequences=True)(embedding)
cnn = Conv1D(200, 5, activation="relu")(lstm)
maxpool = GlobalMaxPooling1D()(cnn)
outputs = Dense(len(le.classes_), activation="softmax")(maxpool)
model = Model(inputs, outputs)
model.summary()
model.compile(optimizer="adam", loss="categorical_crossentropy", metrics=["accuracy"])
model.fit(X_train, y_train, epochs=10, batch_size=32, validation_split=0.2)
[np.argmax(model.predict(X_test)[i]) for i in range(len(X_test))]
|
import os
import sys
import cv2
import json
import glob
import numpy as np
import pandas as pd
from tqdm.auto import tqdm
import matplotlib.pyplot as plt
from random import shuffle
import wandb
import torch
import torch.nn as nn
from torch.utils.data import DataLoader, Dataset
from torch.cuda.amp import GradScaler, autocast
from transformers import (
AutoProcessor,
Pix2StructConfig,
Pix2StructForConditionalGeneration,
get_linear_schedule_with_warmup,
)
import albumentations as A
from albumentations.pytorch import ToTensorV2
import warnings
warnings.simplefilter("ignore")
Config = {
"IMAGE_DIR": "/kaggle/input/benetech-making-graphs-accessible/train/images/",
"MAX_PATCHES": 1024,
"MODEL_NAME": "ybelkada/pix2struct-base",
"IMG_SIZE": (224, 224),
"MAX_LEN": 256,
"LR": 3e-5,
"NB_EPOCHS": 2,
"TRAIN_BS": 2,
"VALID_BS": 2,
"ALL_SAMPLES": 1000000,
"_wandb_kernel": "tanaym",
}
# ### About W&B:
# WandB is a developer tool for companies turn deep learning research projects into deployed software by helping teams track their models, visualize model performance and easily automate training and improving models.
# We will use their tools to log hyperparameters and output metrics from your runs, then visualize and compare results and quickly share findings with your colleagues.
# To login to W&B, you can use below snippet.
# ```python
# from kaggle_secrets import UserSecretsClient
# user_secrets = UserSecretsClient()
# wb_key = user_secrets.get_secret("WANDB_API_KEY")
# wandb.login(key=wb_key)
# ```
# Make sure you have your W&B key stored as `WANDB_API_KEY` under Add-ons -> Secrets
# You can view [this](https://www.kaggle.com/ayuraj/experiment-tracking-with-weights-and-biases) notebook to learn more about W&B tracking.
# If you don't want to login to W&B, the kernel will still work and log everything to W&B in anonymous mode.
def wandb_log(**kwargs):
for k, v in kwargs.items():
wandb.log({k: v})
# Start W&B logging
# W&B Login
from kaggle_secrets import UserSecretsClient
user_secrets = UserSecretsClient()
wb_key = user_secrets.get_secret("WANDB_API_KEY")
wandb.login(key=wb_key)
run = wandb.init(
project="pytorch",
config=Config,
group="multi_modal",
job_type="train",
)
# Let's add chart types as special tokens and a special BOS token
BOS_TOKEN = "<|BOS|>"
X_START = "<x_start>"
X_END = "<x_end>"
Y_START = "<y_start>"
Y_END = "<y_end>"
new_tokens = [
"<line>",
"<vertical_bar>",
"<scatter>",
"<dot>",
"<horizontal_bar>",
X_START,
X_END,
Y_START,
Y_END,
BOS_TOKEN,
]
def augments():
return A.Compose(
[
A.Resize(width=Config["IMG_SIZE"][0], height=Config["IMG_SIZE"][1]),
A.Normalize(
mean=[0, 0, 0],
std=[1, 1, 1],
max_pixel_value=255,
),
ToTensorV2(),
]
)
class BeneTechDataset(Dataset):
def __init__(self, dataset, processor, augments=None):
self.dataset = dataset
self.processor = processor
self.augments = augments
def __len__(self):
return len(self.dataset)
def __getitem__(self, idx):
item = self.dataset[idx]
image = cv2.imread(item["image"])
if self.augments:
image = self.augments(image=image)["image"]
encoding = self.processor(
images=image,
return_tensors="pt",
add_special_tokens=True,
max_patches=Config["MAX_PATCHES"],
)
encoding = {k: v.squeeze() for k, v in encoding.items()}
encoding["text"] = item["label"]
return encoding
def get_model(extra_tokens=new_tokens):
processor = AutoProcessor.from_pretrained(Config["MODEL_NAME"])
model = Pix2StructForConditionalGeneration.from_pretrained(Config["MODEL_NAME"])
processor.image_processor.size = {
"height": Config["IMG_SIZE"][0],
"width": Config["IMG_SIZE"][1],
}
processor.tokenizer.add_tokens(extra_tokens)
model.resize_token_embeddings(len(processor.tokenizer))
return processor, model
def collator(batch):
new_batch = {"flattened_patches": [], "attention_mask": []}
texts = [item["text"] for item in batch]
text_inputs = processor(
text=texts,
padding="max_length",
truncation=True,
return_tensors="pt",
add_special_tokens=True,
max_length=Config["MAX_LEN"],
)
new_batch["labels"] = text_inputs.input_ids
for item in batch:
new_batch["flattened_patches"].append(item["flattened_patches"])
new_batch["attention_mask"].append(item["attention_mask"])
new_batch["flattened_patches"] = torch.stack(new_batch["flattened_patches"])
new_batch["attention_mask"] = torch.stack(new_batch["attention_mask"])
return new_batch
def train_one_epoch(model, processor, train_loader, optimizer, scaler):
model.train()
avg_loss = 0
with autocast():
prog_bar = tqdm(enumerate(train_loader), total=len(train_loader))
for idx, batch in prog_bar:
labels = batch.pop("labels").to("cuda")
flattened_patches = batch.pop("flattened_patches").to("cuda")
attention_mask = batch.pop("attention_mask").to("cuda")
outputs = model(
flattened_patches=flattened_patches,
attention_mask=attention_mask,
labels=labels,
)
loss = outputs.loss
scaler.scale(loss).backward()
scaler.step(optimizer)
scaler.update()
optimizer.zero_grad(set_to_none=True)
prog_bar.set_description(f"loss: {loss.item():.4f}")
wandb_log(train_step_loss=loss.item())
avg_loss += loss.item()
avg_loss = avg_loss / len(train_loader)
print(f"Average training loss: {avg_loss:.4f}")
wandb_log(train_loss=avg_loss)
return avg_loss
@torch.no_grad()
def valid_one_epoch(model, processor, valid_loader):
model.eval()
avg_loss = 0
prog_bar = tqdm(enumerate(valid_loader), total=len(valid_loader))
for idx, batch in prog_bar:
labels = batch.pop("labels").to("cuda")
flattened_patches = batch.pop("flattened_patches").to("cuda")
attention_mask = batch.pop("attention_mask").to("cuda")
outputs = model(
flattened_patches=flattened_patches,
attention_mask=attention_mask,
labels=labels,
)
loss = outputs.loss
prog_bar.set_description(f"loss: {loss.item():.4f}")
wandb_log(val_step_loss=loss.item())
avg_loss += loss.item()
avg_loss = avg_loss / len(valid_loader)
print(f"Average validation loss: {avg_loss:.4f}")
wandb_log(val_loss=avg_loss)
return avg_loss
def fit(model, processor, train_loader, valid_loader, optimizer, scaler):
best_val_loss = int(1e5)
for epoch in range(Config["NB_EPOCHS"]):
print(f"{'='*20} Epoch: {epoch+1} / {Config['NB_EPOCHS']} {'='*20}")
_ = train_one_epoch(model, processor, train_loader, optimizer, scaler)
val_avg_loss = valid_one_epoch(model, processor, valid_loader)
if val_avg_loss < best_val_loss:
best_val_loss = val_avg_loss
print(f"Saving best model so far with loss: {best_val_loss:.4f}")
torch.save(model.state_dict(), f"pix2struct_base_benetech.pt")
print(f"Best model with val_loss: {best_val_loss:.4f}")
# Training cell
if __name__ == "__main__":
# Read the processed JSON file
with open("/kaggle/input/benetech-processed-data-json/data.json", "r") as fl:
dataset = json.load(fl)["data"]
# Shuffle the dataset and select only a few thousand examples (for faster training)
shuffle(dataset)
dataset = dataset[: Config["ALL_SAMPLES"]]
# We are splitting the data naively for now
split = 0.90
train_samples = int(len(dataset) * split)
train_ds = dataset[: train_samples + 1]
valid_ds = dataset[train_samples:]
processor, model = get_model()
model.to("cuda")
wandb.watch(model)
optimizer = torch.optim.Adam(params=model.parameters(), lr=Config["LR"])
# Load the data into Datasets and then make DataLoaders for training
train_dataset = BeneTechDataset(train_ds, processor, augments=augments())
train_dataloader = DataLoader(
train_dataset, shuffle=True, batch_size=Config["TRAIN_BS"], collate_fn=collator
)
valid_dataset = BeneTechDataset(valid_ds, processor, augments=augments())
valid_dataloader = DataLoader(
valid_dataset, shuffle=False, batch_size=Config["VALID_BS"], collate_fn=collator
)
nb_train_steps = int(train_samples / Config["TRAIN_BS"] * Config["NB_EPOCHS"])
# Print out the data sizes we are training on
print(f"Training on {len(train_ds)} samples, Validating on {len(valid_ds)} samples")
# Train the model now
fit(
model=model,
processor=processor,
train_loader=train_dataloader,
valid_loader=valid_dataloader,
optimizer=optimizer,
scaler=GradScaler(),
)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files |nder the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import os
from pathlib import Path
from tokenizers import normalizers
from tokenizers.normalizers import NFD, Lowercase, StripAccents
from tokenizers import pre_tokenizers
from tokenizers.pre_tokenizers import Digits, Whitespace, ByteLevel, Punctuation
from tokenizers.processors import TemplateProcessing
from tokenizers import Tokenizer, models, trainers
from transformers import AutoTokenizer, PreTrainedTokenizerFast
from transformers.models.gpt2.tokenization_gpt2 import bytes_to_unicode
from tqdm.auto import tqdm
from datasets import load_dataset, Dataset
from sklearn.metrics import f1_score, precision_score, recall_score, accuracy_score
import torch
from transformers import BertTokenizerFast, DataCollatorForLanguageModeling
from transformers import BertConfig, BertForMaskedLM
from transformers import LineByLineTextDataset
from transformers import Trainer, TrainingArguments, EarlyStoppingCallback
from tokenizers import Tokenizer
import pandas as pd
os.environ["WANDB_DISABLED"] = str("true")
os.environ["TOKENIZERS_PARALLELISM"] = str("true")
# ## Split data
def clean_text(examples):
return {"text": examples["text"].split("\t")[1].split(":")[-1].strip()}
dataset = load_dataset(
"text",
data_files="/kaggle/input/3-million-german-sentences/deu_news_2015_3M-sentences.txt",
split="train",
streaming=False,
)
dataset = dataset.map(clean_text)
dataset = dataset.train_test_split(test_size=0.1)
dataset["train"].save_to_disk("/kaggle/working/train")
dataset["test"].save_to_disk("/kaggle/working/eval")
# ## Data
train_dataset = Dataset.from_file("/kaggle/working/train/dataset.arrow")
eval_dataset = Dataset.from_file("/kaggle/working/eval/dataset.arrow")
# ## Tokenizer
vocab_size = 32768
batch_size = 10
length = len(train_dataset) // batch_size
def batch_iterator(batch_size=100):
for _ in tqdm(range(0, length, batch_size)):
yield [next(iter(train_dataset))["text"] for _ in range(batch_size)]
byte_to_unicode_map = bytes_to_unicode()
unicode_to_byte_map = dict((v, k) for k, v in byte_to_unicode_map.items())
base_vocab = list(unicode_to_byte_map.keys())
print(f"Size of our base vocabulary: {len(base_vocab)}")
print(f"First element: `{base_vocab[0]}`, last element: `{base_vocab[-1]}`")
tokenizer = Tokenizer(models.BPE(unk_token="<unk>"))
normalizer = normalizers.Sequence(
[
NFD(),
Lowercase(),
# StripAccents()
]
)
pre_tokenizer = pre_tokenizers.Sequence(
[ByteLevel(add_prefix_space=False), Whitespace()]
)
tokenizer.normalizer = normalizer
tokenizer.pre_tokenizer = pre_tokenizer
print(
tokenizer.normalizer.normalize_str("""Dies ist ein Beispielsatz in Deutsch Äoüß""")
)
trainer = trainers.BpeTrainer(
vocab_size=vocab_size,
initial_alphabet=base_vocab,
special_tokens=["[UNK]", "[CLS]", "[SEP]", "[PAD]", "[MASK]"],
)
tokenizer.train_from_iterator(batch_iterator(), trainer=trainer)
encoding = tokenizer.encode("Héllò hôw1 are äöü? 1234.12 1,2 12 euro po12")
print(encoding.tokens)
eval_dataset["text"][1].split("\t")[1].split(":")[-1].strip()
words = pd.read_csv(
"/kaggle/input/3-million-german-sentences/deu_news_2015_3M-words.csv", dtype=str
)
words.rename(columns={"!": "word", "53658": "count"}, inplace=True)
words = words[["word", "count"]]
words.head()
many_tokens = []
for word in tqdm(words["word"].tolist()):
if not isinstance(word, str):
continue
enc = tokenizer.encode(word)
if len(enc) > 2: # ignore cls and sep
many_tokens.append(word)
many_tokens[0]
tokenizer.encode("wieso").ids
tokenizer.save(f"/kaggle/working/tokenizer.json")
# ## Model Training
model_path = "/kaggle/working/model"
use_mlm = True
mlm_probability = 0.2 # still keeping the 80 - 10 - 10 rule
batch_size = 16
seed, buffer_size = 42, 10_000
max_length = 512
block_size = 512
max_position_embeddings = 512
hidden_size = 768
num_hidden_layers = 2 # 12
num_attention_heads = 2 # 12
intermediate_size = 3072
drop_out = 0.1
config = BertConfig(
attention_window=[block_size] * num_attention_heads,
# mask_token_id = 4,
bos_token_id=1,
sep_token_id=2,
# pad_token_id = 3,
eos_token_id=2,
max_position_embeddings=max_position_embeddings,
hidden_size=hidden_size,
num_hidden_layers=num_hidden_layers,
num_attention_heads=num_attention_heads,
intermediate_size=intermediate_size,
hidden_act="gelu",
hidden_dropout_prob=drop_out,
attention_probs_dropout_prob=drop_out,
type_vocab_size=2,
initializer_range=0.02,
layer_norm_eps=1e-12,
vocab_size=vocab_size,
use_cache=True,
classifier_dropout=None,
onnx_export=False,
)
# train_dataset = load_dataset('parquet', data_files=f'../corpus/lufthansa.snap.parquet',
# streaming=True,
# split="train"
# )
# eval_dataset = load_dataset('parquet', data_files=f'../corpus/lufthansa.snap.parquet',
# streaming=True,
# split="train"
# )
def encode(examples, max_length, tokenizer):
return tokenizer.batch_encode_plus(
examples["text"],
padding=True,
truncation=True,
max_length=max_length,
# return_special_tokens_mask=True,
# return_tensors="pt"
)
tk_tokenizer = Tokenizer.from_file(f"/kaggle/working/tokenizer.json")
tokenizer = PreTrainedTokenizerFast(tokenizer_object=tk_tokenizer)
tokenizer.add_special_tokens(
{
"pad_token": "[PAD]",
"unk_token": "[UNK]",
"sep_token": "[SEP]",
"cls_token": "[CLS]",
"bos_token": "[CLS]",
"eos_token": "[SEP]",
"mask_token": "[MASK]",
}
)
train_dataset = train_dataset.map(
encode,
remove_columns=["text"],
batched=True,
batch_size=batch_size,
fn_kwargs={"max_length": max_length, "tokenizer": tokenizer},
)
eval_dataset = eval_dataset.map(
encode,
remove_columns=["text"],
batched=True,
batch_size=batch_size,
fn_kwargs={"max_length": max_length, "tokenizer": tokenizer},
)
train_dataset = train_dataset.with_format("torch")
eval_dataset = eval_dataset.with_format("torch")
data_collator = DataCollatorForLanguageModeling(
tokenizer=tokenizer, mlm=use_mlm, mlm_probability=mlm_probability
)
model = BertForMaskedLM(config=config)
print(f"n of parameters: {model.num_parameters()}")
# comp_model = torch.compile(model)
# print(f"n of parameters: {comp_model.num_parameters()}")
# ### Trainer config
early_stopping = EarlyStoppingCallback(
early_stopping_patience=3, early_stopping_threshold=0.02
)
callbacks = [early_stopping]
learning_rate = 1e-4 # bert
weight_decay = 1e-2 # bert
lr_scheduler_type = "linear"
num_train_epochs = 20 # 5 but training set is small
train_batch_size = 32
eval_batch_size = 32
gradient_accumulation_steps = 2
eval_accumulation_steps = 2
warmup_steps = 1_000
adam_beta1 = 0.9 # bert
adam_beta2 = 0.999 # bert
adam_epsilon = 1e-8 # bert
max_grad_norm = 1.0 # bert
max_steps = 1_000_000
training_args = TrainingArguments(
output_dir=model_path,
overwrite_output_dir=True,
learning_rate=learning_rate,
weight_decay=weight_decay,
lr_scheduler_type=lr_scheduler_type,
num_train_epochs=num_train_epochs,
adam_beta1=adam_beta1,
adam_beta2=adam_beta2,
adam_epsilon=adam_epsilon,
max_grad_norm=max_grad_norm,
evaluation_strategy="steps",
eval_steps=1_000,
max_steps=max_steps,
per_device_train_batch_size=train_batch_size, # depends on memory
per_device_eval_batch_size=eval_batch_size,
gradient_accumulation_steps=gradient_accumulation_steps,
# eval_accumulation_steps=eval_accumulation_steps,
save_strategy="steps",
save_steps=1_000,
save_total_limit=3,
prediction_loss_only=False,
report_to="tensorboard",
log_level="warning",
logging_strategy="steps",
# fp16 = True,
# fp16_full_eval=True,
load_best_model_at_end=True,
metric_for_best_model="loss",
greater_is_better=False,
push_to_hub=False,
dataloader_pin_memory=True,
)
def compute_metrics(eval_pred):
logits, labels = eval_pred
predictions = np.argmax(logits, axis=-1)
accuracy = accuracy_score(y_true=labels, y_pred=predictions)
recall = recall_score(y_true=labels, y_pred=predictions, average="weighted")
precision = precision_score(y_true=labels, y_pred=predictions, average="weighted")
f1 = f1_score(y_true=labels, y_pred=predictions, average="weighted")
return {"accuracy": accuracy, "precision": precision, "recall": recall, "f1": f1}
trainer = Trainer(
model=model,
args=training_args,
data_collator=data_collator,
train_dataset=train_dataset,
eval_dataset=eval_dataset,
# compute_metrics=compute_metrics,
tokenizer=tokenizer,
callbacks=callbacks,
)
trainer.train()
|
# ## Keşifçi Veri Analizi | Becerileri Pekiştirme
# Aşağıda ihtiyacımız doğrultusunda kullanacağımız kütüphaneleri yükleyelim.
import numpy as np
import seaborn as sns
import pandas as pd
# Veri çerçevemizi bulunduğumuz dizinden yükleyelim ve bir veri çerçevesi haline getirerek df değişkenine atayalım. (pd.read_csv(...csv))
df = pd.read_csv("/kaggle/input/iris-flower-dataset/IRIS.csv")
# Veri çerçevesinin ilk 5 gözlemini görüntüleyelim.
print(df.head(5))
# Veri çerçevesinin kaç öznitelik ve kaç gözlemden oluştuğunu görüntüleyelim.
print("Öznitelik sayısı:", len(df.columns))
print("Gözlem sayısı:", len(df))
# Veri çerçevesindeki değişkenlerin hangi tipte olduğunu ve bellek kullanımını görüntüleyelim.
print(df.info())
# Veri çerçevesindeki sayısal değişkenler için temel istatistik değerlerini görüntüleyelim.
# Standart sapma ve ortalama değerlerden çıkarımda bulunarak hangi değişkenlerin ne kadar varyansa sahip olduğu hakkında fikir yürütelim.
print(df.describe())
# Veri çerçevesinde hangi öznitelikte kaç adet eksik değer olduğunu gözlemleyelim.
print(df.isnull().sum())
# Sayısal değişkenler arasında korelasyon olup olmadığını göstermek için korelasyon matrisi çizdirelim. Korelasyon katsayıları hakkında fikir yürütelim.
# En güçlü pozitif ilişki hangi iki değişken arasındadır?
korelasyon = df.corr()
sns.heatmap(korelasyon, annot=True, cmap="coolwarm")
# Burada en yüksek pozitif korelasyon katsayısı 0.97 ile "yas" ve "maas" arasındadır. Bu, yaş ve maaş arasında çok güçlü bir pozitif ilişki olduğunu gösterir.
# Korelasyon katsayılarını daha iyi okuyabilmek için ısı haritası çizdirelim.
korelasyonMatrisi = df.corr()
sns.heatmap(korelasyonMatrisi, annot=True, cmap="coolwarm")
# Veri çerçevemizin hedef değişkeninin "variety" benzersiz değerlerini görüntüleyelim.
print(
df["species"].unique()
) # veriet değişkenini bulamadım o yuzden speciesin benzersiz değerlerini görüntüledim.
print(df.columns) # burada değişkenlere baktım
# Veri çerçevemizin hedef değişkeninin "variety" benzersiz kaç adet değer içerdiğini görüntüleyelim.
print(df["species"].nunique())
# Veri çerçevesindeki sepal.width ve sepal.length değişkenlerinin sürekli olduğunu görüyoruz. Bu iki sürekli veriyi görselleştirmek için önce scatterplot kullanalım.
sns.scatterplot(x="sepal_length", y="sepal_width", data=df)
# Aynı iki veriyi daha farklı bir açıdan frekanslarıyla incelemek için jointplot kullanarak görselleştirelim.
sns.jointplot(x="sepal_length", y="sepal_width", data=df, kind="hex")
# Aynı iki veriyi scatterplot ile tekrardan görselleştirelim fakat bu sefer "variety" parametresi ile hedef değişkenine göre kırdıralım.
# 3 farklı renk arasında sepal değişkenleriyle bir kümeleme yapılabilir mi? Ne kadar ayırt edilebilir bunun üzerine düşünelim.
sns.scatterplot(x="sepal_length", y="sepal_width", hue="species", data=df)
# value_counts() fonksiyonu ile veri çerçevemizin ne kadar dengeli dağıldığını sorgulayalım.
class_counts = df["species"].value_counts()
print(class_counts)
# Keman grafiği çizdirerek sepal.width değişkeninin dağılımını inceleyin.
# Söz konusu dağılım bizim için ne ifade ediyor, normal bir dağılım olduğunu söyleyebilir miyiz?
sns.kdeplot(data=df, x="sepal_width")
# Daha iyi anlayabilmek için sepal.width üzerine bir distplot çizdirelim.
sns.displot(df["sepal_width"]) # distplot yerine displot kullanıldı
# Üç çiçek türü için üç farklı keman grafiğini sepal.length değişkeninin dağılımı üzerine tek bir satır ile görselleştirelim.
ax = sns.violinplot(x="species", y="sepal_length", data=df)
|
# # Features
# #### enrollee_id : Unique ID for candidate
# #### city: City code
# #### city_ development _index : Developement index of the city (scaled)
# #### gender: Gender of candidate
# #### relevent_experience: Relevant experience of candidate
# #### enrolled_university: Type of University course enrolled if any
# #### education_level: Education level of candidate
# #### major_discipline :Education major discipline of candidate
# #### experience: Candidate total experience in years
# #### company_size: No of employees in current employer's company
# #### company_type : Type of current employer
# #### last_new_job: Difference in years between previous job and current job
# #### training_hours: training hours completed
# #### target: 0 – Not looking for job change, 1 – Looking for a job change
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import plotly.express as px
from sklearn.metrics import accuracy_score, f1_score, recall_score
import warnings
warnings.filterwarnings("ignore")
df_train = pd.read_csv(
"/kaggle/input/hr-analytics-job-change-of-data-scientists/aug_train.csv"
)
df_test = pd.read_csv(
"/kaggle/input/hr-analytics-job-change-of-data-scientists/aug_test.csv"
)
df_train.head()
df_test.head()
df_train.info()
plt.style.use("Solarize_Light2")
# # EDA
dd = df_train.drop("enrollee_id", axis=1)
dd.hist(bins=20, figsize=(10, 5))
plt.style.use("fast")
plt.style.context("fast")
plt.show()
# # The Outliers
dd.plot(kind="box", subplots=True, figsize=(12, 4), layout=(1, 4))
# ## The training hours has outliers
sns.boxplot(x=df_train.training_hours)
plt.scatter(df_train.training_hours, df_train.enrollee_id)
plt.xlabel("Training hours")
plt.ylabel("Enrollee id")
out_liear = df_train[df_train.training_hours > 300]
plt.scatter(out_liear.training_hours, out_liear.enrollee_id)
plt.xlabel("Training hours")
plt.ylabel("Enrollee id")
round(
out_liear.training_hours.value_counts().sum()
/ df_train.training_hours.value_counts().sum(),
4,
)
# # What the average training hours of candiantes who want job change?
df_train.training_hours.plot.hist()
plt.xlabel("Training hours")
plt.ylabel("Frequency")
t_h = pd.DataFrame(df_train.training_hours[df_train.target == 1].describe())
t_h
# ## The average training hours for candidates the looking for job change is 63 hours
# # What is the percentage of gender that looking for a job change ?
plt.pie(
df_train.gender.value_counts(),
labels=df_train.gender.value_counts().index,
autopct="%.0f%%",
)
plt.legend(title="Segments")
plt.show()
plt.pie(
df_train[df_train["target"] == 1].gender.value_counts(),
labels=df_train[df_train["target"] == 1].gender.value_counts().index,
autopct="%.0f%%",
)
plt.legend(title="Segments")
plt.show()
# ## Most of our candidate are males by 90% and 8% are female and who looking for a job change are 89% male and 10% female. So this explains you the percentage of male is very high.
sns.kdeplot(
data=df_train, x="training_hours", hue="target", fill=True, palette=["red", "black"]
)
# #
plt.figure(figsize=(10, 6))
ax = sns.countplot(x="relevent_experience", data=df_train[df_train["target"] == 1])
plt.xlabel("Relevant Experience")
plt.ylabel("Count")
total = len(df_train[df_train["target"] == 1])
for p in ax.patches:
percentage = "{:.1f}%".format(100 * p.get_height() / total)
x = p.get_x() + p.get_width() / 2 - 0.1
y = p.get_height() + 10
ax.annotate(percentage, (x, y), size=12)
plt.show()
# ## Most of candidates who are looking for a job change are trained about 50 hours and the number candidates decrease as the number of hours increase
# # What is the number of employees in current employer's company
plt.figure(figsize=(10, 5))
ax = sns.countplot(x="company_size", data=df_train.sort_values(by="enrollee_id"))
plt.xlabel("Company Size")
plt.ylabel("Count")
total = len(df_train)
# to show the percentage
for p in ax.patches:
percentage = "{:.1f}%".format(100 * p.get_height() / total)
x = p.get_x() + p.get_width() / 3 - 0.1
y = p.get_height() + 12
ax.annotate(percentage, (x, y), size=12)
plt.show()
# ## Most companies in our dataset have 50-99 employees in thier companies by 16.1%, then 13.4% have 100-500 and 10.5% have more than 10,000 employees
# # What is the number of employees in current employer's company that looking for a job change ?
#
plt.figure(figsize=(10, 6))
ax = sns.countplot(x="company_size", data=df_train[df_train["target"] == 1])
plt.xlabel("Company Size")
plt.ylabel("Count")
total = len(df_train[df_train["target"] == 1])
for p in ax.patches:
percentage = "{:.1f}%".format(100 * p.get_height() / total)
x = p.get_x() + p.get_width() / 2 - 0.1
y = p.get_height() + 10
ax.annotate(percentage, (x, y), size=12)
plt.show()
# ## Most employees that looking for a job change are in company that have 50-99 employees by 11.4%, then 8.7% that have 100-500 employees and 8.1% have more than 10,000 employees
# # What is the education level of candidate ?
plt.pie(
df_train.education_level.value_counts(),
labels=df_train.education_level.value_counts().index,
autopct="%.0f%%",
)
plt.legend(title="Segments")
plt.show()
# ## Most candidates are graduate by 62% and 23% have masters
# # What is the education level of candidates who are looking for a job change ?
plt.figure(figsize=(10, 6))
ax = sns.countplot(
x="education_level",
data=df_train[df_train["target"] == 1].sort_values(by="education_level"),
)
plt.xlabel("Education Level")
plt.ylabel("Count")
total = len(df_train[df_train["target"] == 1])
for p in ax.patches:
percentage = "{:.1f}%".format(100 * p.get_height() / total)
x = p.get_x() + p.get_width() / 2 - 0.1
y = p.get_height() + 10
ax.annotate(percentage, (x, y), size=12)
plt.savefig("education level of candidates.png")
plt.show()
# ## 67% of the candidate who looking for a job change are graduated and 19.6% have master
# # What is the major discipline of candidates who are looking for a job change ?
plt.figure(figsize=(10, 4))
plt.figure(figsize=(10, 6))
ax = sns.countplot(x="major_discipline", data=df_train[df_train["target"] == 1])
plt.xlabel("Major Discipline")
plt.ylabel("Count")
total = len(df_train[df_train["target"] == 1])
for p in ax.patches:
percentage = "{:.1f}%".format(100 * p.get_height() / total)
x = p.get_x() + p.get_width() / 2 - 0.1
y = p.get_height() + 10
ax.annotate(percentage, (x, y), size=12)
plt.show()
# # What is the experience of the candidates who are looking for a job change ?
df_train.experience.dtype
df_train.experience.value_counts
df2 = df_train.copy()
experience_map = {
"<1": "0-5",
"1": "0-5",
"2": "0-5",
"3": "0-5",
"4": "0-5",
"5": "0-5",
"6": "6-10",
"7": "6-10",
"8": "6-10",
"9": "6-10",
"10": "6-10",
"11": "11-15",
"12": "11-15",
"13": "11-15",
"14": "11-15",
"15": "11-15",
"16": "16-20",
"17": "16-20",
"18": "16-20",
"19": "16-20",
"20": "16-20",
">20": ">20",
}
df2.loc[:, "experience"] = df2["experience"].map(experience_map)
pd.DataFrame(df2.experience.value_counts())
plt.figure(figsize=(10, 6))
ax = sns.countplot(x="experience", data=df2[df2.target == 1])
plt.xlabel("Experience")
plt.ylabel("Count")
total = len(df2[df2.target == 1])
for p in ax.patches:
percentage = "{:.1f}%".format(100 * p.get_height() / total)
x = p.get_x() + p.get_width() / 2 - 0.1
y = p.get_height() + 10
ax.annotate(percentage, (x, y), size=12)
plt.show()
plt.figure(figsize=(10, 5))
ax = sns.countplot(
x="experience",
data=df2[df2.target == 1].sort_values(by="experience"),
hue=df2.gender[df2.target == 1],
)
plt.xlabel("Experience")
plt.ylabel("Count")
total = len(df2[df2.target == 1])
for p in ax.patches:
percentage = "{:.1f}%".format(100 * p.get_height() / total)
x = p.get_x() + p.get_width() / 2 - 0.1
y = p.get_height() + 10
ax.annotate(percentage, (x, y), size=12)
plt.savefig("Agent_real_estat.png")
plt.show()
# ## Many candidates who are looking for a job change have experience from 0 to 5 and they normally males
# # What is the level of the city development index of the candidates who are looking for a job change ?
df_train.city_development_index.describe()
interval = [0.3, 0.5, 0.7, 0.9, 1]
names = [
"Low development",
"Moderate development",
"High development",
"Very high development",
]
df2["city_development_index_scale"] = pd.cut(
df2.city_development_index, bins=interval, labels=names
)
df2.city_development_index_scale.value_counts()
plt.pie(
df2.city_development_index_scale.value_counts(),
labels=df2.city_development_index_scale.value_counts().index,
autopct="%.0f%%",
)
plt.show()
# ## Most candidates are from very high development and high development cities by 50% and 26% respectivily
plt.pie(
df2.city_development_index_scale[df2["target"] == 1].value_counts(),
labels=df2.city_development_index_scale.value_counts().index,
autopct="%.0f%%",
)
plt.show()
sns.kdeplot(
data=df2,
x="city_development_index",
hue="target",
fill=True,
palette=["red", "black"],
)
pd.DataFrame(df2.city_development_index_scale[df2["target"] == 1].describe())
# ## Most candidates who are looking for a job change are from Moderate development then very high development
# # What is the last new job for the candidates who are looking for a job change?
sns.displot(df_train.last_new_job[df_train["target"] == 1], kde=True)
plt.figure(figsize=(10, 6))
ax = sns.countplot(x="last_new_job", data=df_train[df_train["target"] == 1])
plt.xlabel("Last New Job")
plt.ylabel("Percentage")
total = len(df_train[df_train["target"] == 1])
for p in ax.patches:
percentage = "{:.1f}%".format(100 * p.get_height() / total)
x = p.get_x() + p.get_width() / 2 - 0.1
y = p.get_height() + 10
ax.annotate(percentage, (x, y), size=12)
plt.show()
plt.figure(figsize=(9, 6))
ax = sns.countplot(x="company_type", data=df_train[df_train["target"] == 1])
plt.xlabel("company type")
plt.ylabel("Percentage")
total = len(df_train[df_train["target"] == 1])
for p in ax.patches:
percentage = "{:.1f}%".format(100 * p.get_height() / total)
x = p.get_x() + p.get_width() / 2 - 0.1
y = p.get_height() + 10
ax.annotate(percentage, (x, y), size=12)
plt.savefig("company_type.png")
plt.show()
# # Concatenate the train and test data to preprocessing them
# Make the target of the test data equal -1 so we can separate them after the preprocessing
df_test["target"] = -1
df_concat = pd.concat([df_train, df_test], axis=0).reset_index(drop=True)
df_concat.target.value_counts()
# # Handing the missing values
df_concat.info()
pd.DataFrame(df_concat.isna().sum() / len(df_train))
sns.heatmap(df_train.isnull(), cmap="plasma")
plt.savefig("missingvales.png")
# cat_columns=[column for column in df_concat.columns if df_concat[column].dtype=="object"]
# for column in cat_columns:
# if df_concat[column].isnull().sum()>0:
# df_concat[column].replace(np.nan,df_concat[column].mode()[0],inplace=True)
# pd.DataFrame(df_concat.isnull().sum())
df_concat.dropna(
subset=["enrolled_university", "education_level", "experience", "last_new_job"],
inplace=True,
)
pd.DataFrame(df_concat.isna().sum() / len(df_train) * 100)
df_before = df_concat.copy()
fill_list = df_concat["major_discipline"].dropna()
df_concat["major_discipline"] = df_concat["major_discipline"].fillna(
pd.Series(np.random.choice(fill_list, size=len(df_concat.index)))
)
sns.kdeplot(x=df_concat["major_discipline"].value_counts())
sns.kdeplot(x=df_before["major_discipline"].value_counts(), color="g")
fill_list = df_concat["gender"].dropna()
df_concat["gender"] = df_concat["gender"].fillna(
pd.Series(np.random.choice(fill_list, size=len(df_concat.index)))
)
sns.kdeplot(x=df_concat["gender"].value_counts())
sns.kdeplot(x=df_before["gender"].value_counts(), color="g")
# df_before['gender'].value_counts().plot(kind = 'kde')
df_before.company_size.value_counts()
fill_list = df_concat["company_size"].dropna()
df_concat["company_size"] = df_concat["company_size"].fillna(
pd.Series(np.random.choice(fill_list, size=len(df_concat.index)))
)
sns.kdeplot(x=df_concat["company_size"].value_counts())
sns.kdeplot(x=df_before["company_size"].value_counts(), color="g")
df_concat.company_type.value_counts()
fill_list = df_concat["company_type"].dropna()
df_concat["company_type"] = df_concat["company_type"].fillna(
pd.Series(np.random.choice(fill_list, size=len(df_concat.index)))
)
sns.kdeplot(x=df_concat["company_type"].value_counts())
sns.kdeplot(x=df_before["company_type"].value_counts(), color="g")
df_concat.isna().sum()
# # Label Encoding
data = df_concat.copy()
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
obj_list = data.select_dtypes(include="object").columns
for obj in obj_list:
data[obj] = le.fit_transform(data[obj].astype(str))
data.head().T
data.info()
# # The Correlation of the data
correlation = data.corr()
plt.figure(figsize=(20, 7))
sns.heatmap(correlation, annot=True)
plt.savefig("corr.png")
# # Separate train and test data
# separate the df_train from df_test
train_data = data[data["target"] != -1].reset_index(drop=True)
test_data = data[data["target"] == -1].reset_index(drop=True)
# then drop target of -1
test_data = test_data.drop(columns=["enrollee_id", "target", "city"], axis=1)
test_data.head(1)
# y.value_counts()
# # Handling Imbalance data
sns.countplot(x=train_data["target"])
ax = sns.countplot(x="target", data=train_data)
plt.xlabel("target")
plt.ylabel("Count")
total = len(train_data)
for p in ax.patches:
percentage = "{:.1f}%".format(100 * p.get_height() / total)
x = p.get_x() + p.get_width() / 2 - 0.1
y = p.get_height() + 10
ax.annotate(percentage, (x, y), size=12)
plt.show()
# #
x = train_data.drop(["enrollee_id", "target", "city"], axis=1)
y = train_data["target"]
from imblearn.over_sampling import SMOTE
smote = SMOTE(sampling_strategy="minority")
x_sm, y_sm = smote.fit_resample(x, y)
y_sm.value_counts()
sns.countplot(x=y_sm)
# # Train & Test split
from sklearn.model_selection import train_test_split, cross_val_score
x_train, x_test, y_train, y_test = train_test_split(
x_sm, y_sm, test_size=0.25, random_state=42, stratify=y_sm
)
print(y_train.value_counts())
print(y_test.value_counts())
# # Scaling the data
from sklearn.preprocessing import StandardScaler
sc = StandardScaler()
x_train = sc.fit_transform(x_train)
x_test = sc.fit_transform(x_test)
# # Logistic Regression model
#
from sklearn.linear_model import LogisticRegression
lr = LogisticRegression(penalty="l2", C=0.5)
# # Cross Validation
from sklearn.model_selection import KFold, cross_val_score
K_fold = KFold(n_splits=10, shuffle=True, random_state=42)
scoring = "accuracy"
score = cross_val_score(lr, x, y, cv=K_fold, scoring=scoring)
print(score)
round(np.mean(score) * 100, 2)
lr.fit(x_train, y_train)
print("Train Score =>", lr.score(x_train, y_train))
print("Test Score =>", lr.score(x_test, y_test))
y_pred = lr.predict(x_test)
print("F1 Score =>", f1_score(y_test, y_pred))
print("Recall Score =>", recall_score(y_test, y_pred))
print("Accuracy Score =>", accuracy_score(y_test, y_pred))
# # Evaluation of Logistic Regression model
from sklearn.metrics import (
confusion_matrix,
classification_report,
accuracy_score,
f1_score,
)
ev = confusion_matrix(y_test, lr.predict(x_test))
ev
from mlxtend.plotting import plot_confusion_matrix
plot_confusion_matrix(
ev, class_names=["Not looking for job change", "Looking for a job change"]
)
print(classification_report(y_test, lr.predict(x_test)))
# # Decision tree model
from sklearn.tree import DecisionTreeClassifier
x_train, x_test, y_train, y_test = train_test_split(
x_sm, y_sm, test_size=0.25, random_state=42, stratify=y_sm
)
sc = StandardScaler()
x_train = sc.fit_transform(x_train)
x_test = sc.transform(x_test)
dt = DecisionTreeClassifier(max_depth=7)
dt.fit(x_train, y_train)
print(dt.score(x_train, y_train))
print(dt.score(x_test, y_test))
ev2 = confusion_matrix(y_test, dt.predict(x_test))
ev2
plot_confusion_matrix(
ev2, class_names=["Not looking for job change", "Looking for a job change"]
)
print(classification_report(y_test, dt.predict(x_test)))
# # Grid Search for decision tree
from sklearn.model_selection import GridSearchCV
x_train, x_test, y_train, y_test = train_test_split(
x_sm, y_sm, test_size=0.25, random_state=42, stratify=y_sm
)
param_grid = {"max_depth": [3, 5, 7, 8, 9, 10], "max_features": [3, 5, 7, 8, 9, 10]}
dt = DecisionTreeClassifier()
grid_search = GridSearchCV(estimator=dt, param_grid=param_grid, cv=5)
grid_search.fit(x_train, y_train)
print("Best: %f using %s" % (grid_search.best_score_, grid_search.best_params_))
ev3 = confusion_matrix(y_test, grid_search.predict(x_test))
ev3
plot_confusion_matrix(
ev3, class_names=["Not looking for job change", "Looking for a job change"]
)
print(classification_report(y_test, grid_search.predict(x_test)))
# # Random forest classifier use grid search
from sklearn.ensemble import GradientBoostingClassifier, RandomForestClassifier
pram_grid = {
"max_depth": [4, 5, 6, 7],
"max_features": [3, 4, 5, 7],
"n_estimators": [200, 100, 20, 40],
}
clf = RandomForestClassifier()
grid = GridSearchCV(estimator=clf, param_grid=pram_grid, cv=10)
grid_result = grid.fit(x_train, y_train)
print("Best: %f using %s" % (grid_result.best_score_, grid_result.best_params_))
ev4 = confusion_matrix(y_test, grid_search.predict(x_test))
ev4
plot_confusion_matrix(
ev4, class_names=["Not looking for job change", "Looking for a job change"]
)
print(classification_report(y_test, grid_result.predict(x_test)))
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Context
# * Cardiovascular diseases (CVDs) are the number 1 cause of death globally, taking an estimated 17.9 million lives each year, which accounts for 31% of all deaths worldwide. Four out of 5CVD deaths are due to heart attacks and strokes, and one-third of these deaths occur prematurely in people under 70 years of age. Heart failure is a common event caused by CVDs and this dataset contains 11 features that can be used to predict a possible heart disease.
# * People with cardiovascular disease or who are at high cardiovascular risk (due to the presence of one or more risk factors such as hypertension, diabetes, hyperlipidaemia or already established disease) need early detection and management wherein a machine learning model can be of great help.
# # Data input
df = pd.read_csv("/kaggle/input/heart-failure-prediction/heart.csv")
df.head() # data view
# # Attribute Information
# * Age: age of the patient [years]
# * Sex: sex of the patient [M: Male, F: Female]
# * ChestPainType: chest pain type [TA: Typical Angina, ATA: Atypical Angina, NAP: Non-Anginal Pain, ASY: Asymptomatic]
# * RestingBP: resting blood pressure [mm Hg]
# * Cholesterol: serum cholesterol [mm/dl]
# * FastingBS: fasting blood sugar [1: if FastingBS > 120 mg/dl, 0: otherwise]
# * RestingECG: resting electrocardiogram results [Normal: Normal, ST: having ST-T wave abnormality (T wave inversions and/or ST elevation or depression of > 0.05 mV), LVH: showing probable or definite left ventricular hypertrophy by Estes' criteria]
# * MaxHR: maximum heart rate achieved [Numeric value between 60 and 202]
# * ExerciseAngina: exercise-induced angina [Y: Yes, N: No]
# * Oldpeak: oldpeak = ST [Numeric value measured in depression]
# * ST_Slope: the slope of the peak exercise ST segment [Up: upsloping, Flat: flat, Down: downsloping]
# * HeartDisease: output class [1: heart disease, 0: Normal]
df.shape # the shape of data is 918 rows and 12 columns
df.dtypes # the type of data
df.nunique() # every columns unique value number
df.ChestPainType.unique() # view unique value in ChestPainType
df.isnull().sum() # judge null value
# change to categorical number
# Sex
df.Sex.replace({"M": 1, "F": 0}, inplace=True)
# ChestPainType
df.ChestPainType.replace({"TA": 0, "ATA": 1, "NAP": 2, "ASY": 3}, inplace=True)
# RestingECG
df.RestingECG.replace({"Normal": 0, "ST": 1, "LVH": 2}, inplace=True)
# ExerciseAngina
df.ExerciseAngina.replace({"N": 0, "Y": 1}, inplace=True)
# ST_Slope
df.ST_Slope.replace({"Up": 0, "Flat": 1, "Down": 2}, inplace=True)
df.head()
# define categorical and continuous feature
cat_col = [
"Sex",
"ChestPainType",
"RestingECG",
"ExerciseAngina",
"ST_Slope",
] # categorical features
con_col = [
"Age",
"RestingBP",
"Cholesterol",
"FastingBS",
"MaxHR",
"Oldpeak",
"HeartDisease",
] # continuous features
round(df[con_col].describe(), 2) # two decimal
|
# # Python Booleans - Mantıksal Operatörler
# Mantıksal operatörler iki değerden oluşur. True - False
# True: doğru
# False: Yanlış
# ## Boolean Values
# Programlamada genellikle bir ifadenin Doğru mu yoksa Yanlış mı olduğunu bilmeniz gerekir.
# Python'da herhangi bir ifadeyi değerlendirebilir ve True veya False olmak üzere iki yanıttan birini alabilirsiniz.
# İki değeri karşılaştırdığınızda, ifade değerlendirilir ve Python, Boole yanıtını döndürür:
print(4 > 1)
print(4 == 1)
print(4 < 1)
# if ifadesinde bir koşul çalıştırdığınızda, Python True veya False değerini döndürür:
# Koşulun Doğru veya Yanlış olmasına bağlı olarak bir mesaj yazdıralım.
number1 = 411
number2 = 15
if b > a:
print("Number2, Number1'dan büyüktür")
else:
print("Number2, Number1'dan büyük değildir")
# # Değerleri ve Değişkenleri Değerlendirme
# bool() işlevi, herhangi bir değeri değerlendirmenize ve karşılığında True veya False vermenize izin verir,
# Örneğin:
# Bir diziyi ve bir sayıyı değerlendirin:
print(bool("Python"))
print(bool(41))
# iki değişkeni değerlendirme
word = "Python"
number = 41
print(bool(word))
print(bool(number))
# # Çoğu Değer Doğrudur
# Bir tür içeriğe sahipse, hemen hemen her değer True olarak değerlendirilir.
# Boş diziler dışında tüm diziler True'dur.
# 0 dışında herhangi bir sayı True'dur.
# Boş olanlar dışında tüm liste, demet, küme ve sözlük True'dur.
# Aşağıdakiler True olarak çıktı verir
bool("Kelime")
bool(411)
bool(["hava", "su", "toprak"])
# # Bazı Değerler Yanlış
# Aslında, (), [], {}, "", 0 sayısı ve Yok değeri gibi boş değerler dışında False olarak değerlendirilen çok fazla değer yoktur. Ve elbette False değeri False olarak değerlendirilir.
# Aşağıdaki örnekler False olarak çıktı verecektir
bool(False)
bool(None)
bool(0)
bool("")
bool(())
bool([])
bool({})
# # Fonksiyonlar bir Boole Döndürebilir
# Bir Boole Değeri döndüren fonksiyonlar oluşturabilirsiniz:
def giveTrue():
return True
print(giveTrue())
# Bir işlevin Boole yanıtına göre kod çalıştırabilirsiniz:
# Örnek
# "DOĞRU!" Yazdır işlev True döndürürse, aksi takdirde "YANLIŞ!" yazdırın:
def giveTrue():
return True
if giveTrue():
print("DOĞRU!")
else:
print("YANLIŞ!")
# Python ayrıca, bir nesnenin belirli bir veri türünde olup olmadığını belirlemek için kullanılabilen isinstance() fonksiyonu gibi bir boolean değeri döndüren birçok yerleşik işleve sahiptir:
# Örnek
# Bir nesnenin tamsayı olup olmadığını kontrol edin:
number = 411
print(isinstance(number, int))
word = "Hava bugün yağmurlu"
print(isinstance(word, str))
# Aşağıdaki boolean hangi değeri çıktı verir.
print(4 > 4)
print(4 == 4)
print(4 < 4)
print(bool("word"))
print(bool(0))
# # Python Operatörleri
# Operatörler, değişkenler ve değerler üzerinde işlem yapmak için kullanılır.
# Aşağıdaki örnekte, iki değeri bir araya getirmek için + operatörünü kullanıyoruz:
print(4 + 6)
# Python, operatörleri aşağıdaki gruplara ayırır:
# Arithmetic operators
# Assignment operators
# Comparison operators
# Logical operators
# Identity operators
# Membership operators
# Bitwise operators
# # Python Arithmetic Operators
# Aritmetik opetaörler, yaygın matematiksel işlemleri gerçekleştirmek için sayısal değerlerle birlikte kullanılır:
# Name Example Try it
# + Addition x + y
# - Subtraction x - y
# * Multiplication x * y
# / Division x / y
# % Modulus x % y
# ** Exponentiation x ** y
# // Floor division x // y
# addition - toplama
number1 = 4
number2 = 6
print(number1 + number2)
# Subtraction - çıkarma
number1 = 4
number2 = 6
print(number1 - number2)
# Multiplication
number1 = 4
number2 = 6
print(number1 * number2)
# Division - bölme
number1 = 20
number2 = 2
print(number1 / number2)
# modulus - modüller
number1 = 18
number2 = 3
print(number1 % number2)
# Exponentiation - üst alma
number1 = 5
number2 = 3
print(number1**number2) # same as 2*2*2*2*2
# Floor division - taban fonksiyonu
number1 = 21
number2 = 4
print(number1 // number2)
# kat bölümü // sonucu en yakın tam sayıya yuvarlar
# # Python Atama Operatörleri
# Atama işleçleri, değişkenlere değer atamak için kullanılır:
# = EŞİTTİR
number = 14
number
# += ARTI EŞİTTİR
number = 14
number += 6
print(number)
# -= EKSİ EŞİTTİR
number = 14
number -= 4
print(number)
# *= ÇARPI EŞİTTİR
number = 14
number *= 2
print(number)
# /=
number = 8
number /= 6
print(number)
# %= bölümden kalan sayıyı verir
number = 28
number %= 9
print(number)
# //= kaç defa bölündüğü
number = 26
number //= 5
print(number)
# **= sayının üssünü alır
number = 6
number **= 3
print(number)
# # Python Karşılaştırma Operatörleri
# Karşılaştırma işleçleri iki değeri karşılaştırmak için kullanılır:
# == Equal - Eşit mi
number1 = 9
number2 = 6
print(number1 == number2)
# 9, 6'ya eşit olmadığı için False döndürür
# != Not equal - Eşit değil
number1 = 9
number2 = 6
print(number1 != number2)
# 9, 6'ya eşit olmadığı için True döndürür
# > Greater than , büyüktür
number1 = 9
number2 = 6
print(number1 > number2)
# 9, 6'dan büyük olduğu için True döndürür
# < Less than , küçüktür
number1 = 9
number2 = 6
print(number1 < number2)
# 9, 6'dan küçük olmadığı için False döndürür
# >= Greater than or equal to , Büyük eşittir
number1 = 9
number2 = 6
print(number1 >= number2)
# returns True because 9 is greater, or equal, to 6
# Less than or equal to , küçük eşittir
number1 = 9
number2 = 6
print(number1 <= number2)
# returns False because 9 is neither less than or equal to 6
# # Python Mantıksal Operatörler
# Mantıksal işleçler, koşullu ifadeleri birleştirmek için kullanılır: "and, or, not"
# and, Her iki ifade de doğruysa True döndürür
number1 = 11
print(number1 > 10 and number1 < 20)
# returns True because 5 is greater than 3 AND 5 is less than 10
# or İfadelerden biri doğruysa True döndürür
number1 = 11
print(number1 > 10 or number1 < 2)
# #, koşullardan biri doğru olduğu için True döndürür (5, 3'ten büyüktür, ancak 5, 4'ten küçük değildir)
# not Sonucu tersine çevirin, sonuç doğruysa False döndürür
number1 = 11
print(not (number1 > 10 and number1 < 20))
# sonucu tersine çevirmek için kullanılır. Norlamde true çıktısı verecekti
# # Python Kimlik Operatörleri
# Kimlik fonksiyonları, nesneleri eşit olup olmadıklarını değil, aslında aynı nesne olup olmadıklarını ve aynı bellek konumuna sahip olup olmadıklarını karşılaştırmak için kullanılır:
# is
# Her iki değişken de aynı nesneyse True döndürür
array1 = ["hava", "su"]
array2 = ["hava", "su"]
array3 = array1
print(array1 is array3)
# True değerini döndürür çünkü z, x ile aynı nesnedir
print(array1 is array2)
# aynı içeriğe sahip olsalar bile x, y ile aynı nesne olmadığı için False döndürür
print(array1 == array2)
# "is" ve "==" arasındaki farkı göstermek için: x eşittir y olduğu için bu karşılaştırma True değerini döndürür
# is not
# Her iki değişken de aynı nesne değilse True döndürür
# is not
# Her iki değişken de aynı nesne değilse True döndürür
array1 = ["hava", "su"]
array2 = ["hava", "su"]
array3 = array1
print(array1 is not array3)
# z, x ile aynı nesne olduğu için False döndürür
print(array1 is not array2)
# True döndürür çünkü x, aynı içeriğe sahip olsalar bile y ile aynı nesne değildir
print(array1 != array2)
# "is not" ve "!=" arasındaki farkı göstermek için: x eşittir y olduğu için bu karşılaştırma False döndürür
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# # Reading in the data
# The following code reads in the data.
# The first two lines will read in a single file in order to test code.
# Once the code works, use the remaining lines to get all of the labratory data.
df = []
# path1 = '/kaggle/input/tlvmc-parkinsons-freezing-gait-prediction/train/tdcsfog/08d6702e8a.csv'
# df = pd.read_csv(path1)
for dirname, _, filenames in os.walk(
"/kaggle/input/tlvmc-parkinsons-freezing-gait-prediction/train/tdcsfog"
):
for filename in filenames:
identifier = filename[:-4]
tdcs_inst = pd.read_csv(os.path.join(dirname, filename))
tdcs_inst = tdcs_inst.assign(id=identifier)
df.append(tdcs_inst)
df = pd.concat(df)
df.info()
# Getting a general view of the data
df.describe()
df.head()
df
# # Recode to a categorical variable
# Now we need to code the A-Kinetic, Turning, Walking, and StartHesitation into a single variable
df["state"] = "Error"
df.describe(include="all")
df2 = df.groupby("Turn").describe(include="all")
print(df2)
df.loc[df["Turn"] == 1, "state"] = "Turn"
df.loc[df["StartHesitation"] == 1, "state"] = "Hesitate"
df.loc[df["Walking"] == 1, "state"] = "Walk"
df.loc[df["state"] == "Error", "state"] = "AKinetic"
print(df["state"].unique())
df.describe(include="all")
# # Analysis!
# Finally we need to look at our data.
# The null hypothesis is that the distributions for the acceleration data are the same regardless of the state of the patient.
# In order to test this, we can look at a number of different atributes. The most common would be to do a test of "center."
# Normally, this would be ANOVA, but that has some assumptions. We will consider these first.
#
import scipy.stats as stats
import matplotlib.pyplot as plt
# ## The AccV variable
# Testing normality of the AccV variable. We produce normality plots for the data stratified by the level of state that the patient is in.
Unique_States = df["state"].unique()
for State in Unique_States:
stats.probplot(df[df["state"] == State]["AccV"], dist="norm", plot=plt)
plt.title("Probability Plot - " + State)
plt.show()
# Oh... that's not good. These are all very fat tailed distributions. The patterns look similar, so it seems like
# it may be reasonable to test the "center" of the data still, but we should look at a univariate histogram or density plot of each to see what we
# are looking at.
fig, axes = plt.subplots(2, 1, figsize=(22, 14))
ax = axes.ravel()
# ['AKinetic' 'Turn' 'Hesitate' 'Walk']
df.loc[df.state == "AKinetic"].AccV.plot(
kind="hist",
ax=ax[0],
alpha=0.5,
bins=50,
sharex=True,
label="A-Kinetic",
edgecolor="black",
linewidth=3,
)
df.loc[df.state == "Turn"].AccV.plot(
kind="hist",
ax=ax[0],
alpha=0.5,
bins=50,
sharex=True,
label="Turn",
edgecolor="blue",
linewidth=3,
)
df.loc[df.state == "Hesitate"].AccV.plot(
kind="hist",
ax=ax[0],
alpha=0.5,
bins=50,
sharex=True,
label="Hesitate",
edgecolor="red",
linewidth=3,
)
df.loc[df.state == "Walk"].AccV.plot(
kind="hist",
ax=ax[0],
alpha=0.5,
bins=50,
sharex=True,
label="Walk",
edgecolor="green",
linewidth=3,
)
fig.suptitle("Histogram and Density plot of AccV by state", fontsize=20)
df.loc[df.state == "AKinetic"].AccV.plot(
kind="density", ax=ax[1], alpha=0.5, label="A-Kinetic", linewidth=3
)
df.loc[df.state == "Turn"].AccV.plot(
kind="density", ax=ax[1], alpha=0.5, label="Turn", linewidth=3
)
df.loc[df.state == "Hesitate"].AccV.plot(
kind="density", ax=ax[1], alpha=0.5, label="Hesitate", linewidth=3
)
df.loc[df.state == "Walk"].AccV.plot(
kind="density", ax=ax[1], alpha=0.5, label="Walk", linewidth=3
)
ax[0].legend(fontsize=18)
ax[0].set_xlim(-16, 4)
ax[0].set_title("Histogram of AccV based on state")
ax[0].tick_params(axis="both", which="major", labelsize=14)
ax[0].tick_params(axis="both", which="minor", labelsize=14)
ax[1].legend(fontsize=18)
ax[1].set_xlim(-16, 4)
ax[1].set_title("Density of AccV based on state")
ax[1].tick_params(axis="both", which="major", labelsize=14)
ax[1].tick_params(axis="both", which="minor", labelsize=14)
boxplot = df.boxplot(column=["AccV"], by="state")
# Wow,okay, so now what? The distributions of this data appear to be somewhat unimodal (for the most part) and pretty symmetric in general which means that the mean is probably a representative measure of center. That means we can probably apply one way ANOVA to see of the centers are all reasonably similar; however, the number of outliers is pretty significant and the spread of the data within group is very high when compared to the between group spread. This indicates that we probably don't expect to be able to see a difference in means.
# In general, we can see that there is likely a difference in spread; however, tests of spread are highly sensitive to outliers and won't be a good measure here. Furthermore, it appears that skewness differs between datasets (skewed left, right, and symmetric.) I think the best way of testing this is the K-S (Kolmogorov-Smirnov test) which is a non-parametric test of distributional heterogeneity. Admittedly, this can only test two distributional differences, not 4, so we would have to run pairwise tests. In that case, 6 tests. Given that there are 3 univariate continuous variables we will consider, that's 18 hypothesis tests so by taking the bonferroni post-hoc adjustment of 0.05/18 = 0.00277 will be our critical p-value if we run all of these tests. Realistically, there is probably a better approach.
# 1.) For each of these distributions, look for those that appear to be the closest on the density plots.
# 2.) Apply the K-S test on these. If the p-value is very small, we can infer that these are from different distributions.
# 3.) Based on logic, and without running the tests, we can assume that the others will also be different and thus we can start looking for statistics that can characterize these differences effectivelly.
#
AccV_AKin = df.loc[df["state"] == "AKinetic", "AccV"]
AccV_Walk = df.loc[df["state"] == "Walk", "AccV"]
AccV_Turn = df.loc[df["state"] == "Turn", "AccV"]
AccV_Hesitate = df.loc[df["state"] == "Hesitate", "AccV"]
# function to calculate Cohen's d for independent samples
def cohend(d1, d2):
# calculate the size of samples
n1, n2 = len(d1), len(d2)
# calculate the variance of the samples
s1, s2 = np.var(d1, ddof=1), np.var(d2, ddof=1)
# calculate the pooled standard deviation
s = np.sqrt(((n1 - 1) * s1 + (n2 - 1) * s2) / (n1 + n2 - 2))
# calculate the means of the samples
u1, u2 = np.mean(d1), np.mean(d2)
# calculate the effect size
return (u1 - u2) / s
print(stats.ks_2samp(AccV_AKin, AccV_Walk))
print(cohend(AccV_AKin, AccV_Walk))
print(stats.ks_2samp(AccV_AKin, AccV_Turn))
print(cohend(AccV_AKin, AccV_Turn))
print(stats.ks_2samp(AccV_AKin, AccV_Hesitate))
print(cohend(AccV_AKin, AccV_Hesitate))
print(stats.ks_2samp(AccV_Hesitate, AccV_Walk))
print(cohend(AccV_Hesitate, AccV_Walk))
print(stats.ks_2samp(AccV_Hesitate, AccV_Turn))
print(cohend(AccV_Hesitate, AccV_Turn))
print(stats.ks_2samp(AccV_Walk, AccV_Turn))
print(cohend(AccV_Walk, AccV_Turn))
# ## The AccML variable
Unique_States = df["state"].unique()
for State in Unique_States:
stats.probplot(df[df["state"] == State]["AccML"], dist="norm", plot=plt)
plt.title("Probability Plot - " + State)
plt.show()
fig, axes = plt.subplots(2, 1, figsize=(22, 14))
ax = axes.ravel()
# ['AKinetic' 'Turn' 'Hesitate' 'Walk']
df.loc[df.state == "AKinetic"].AccML.plot(
kind="hist",
ax=ax[0],
alpha=0.5,
bins=50,
sharex=True,
label="A-Kinetic",
edgecolor="black",
linewidth=3,
)
df.loc[df.state == "Turn"].AccML.plot(
kind="hist",
ax=ax[0],
alpha=0.5,
bins=50,
sharex=True,
label="Turn",
edgecolor="blue",
linewidth=3,
)
df.loc[df.state == "Hesitate"].AccML.plot(
kind="hist",
ax=ax[0],
alpha=0.5,
bins=50,
sharex=True,
label="Hesitate",
edgecolor="red",
linewidth=3,
)
df.loc[df.state == "Walk"].AccML.plot(
kind="hist",
ax=ax[0],
alpha=0.5,
bins=50,
sharex=True,
label="Walk",
edgecolor="green",
linewidth=3,
)
fig.suptitle("Histogram and Density plot of AccML by state", fontsize=20)
df.loc[df.state == "AKinetic"].AccML.plot(
kind="density", ax=ax[1], alpha=0.5, label="A-Kinetic", linewidth=3
)
df.loc[df.state == "Turn"].AccML.plot(
kind="density", ax=ax[1], alpha=0.5, label="Turn", linewidth=3
)
df.loc[df.state == "Hesitate"].AccML.plot(
kind="density", ax=ax[1], alpha=0.5, label="Hesitate", linewidth=3
)
df.loc[df.state == "Walk"].AccML.plot(
kind="density", ax=ax[1], alpha=0.5, label="Walk", linewidth=3
)
ax[0].legend(fontsize=18)
ax[0].set_xlim(-8, 8)
ax[0].set_title("Histogram of AccML based on state")
ax[0].tick_params(axis="both", which="major", labelsize=14)
ax[0].tick_params(axis="both", which="minor", labelsize=14)
ax[1].legend(fontsize=18)
ax[1].set_xlim(-8, 8)
ax[1].set_title("Density of AccML based on state")
ax[1].tick_params(axis="both", which="major", labelsize=14)
ax[1].tick_params(axis="both", which="minor", labelsize=14)
boxplot = df.boxplot(column=["AccML"], by="state")
AccML_AKin = df.loc[df["state"] == "AKinetic", "AccML"]
AccML_Walk = df.loc[df["state"] == "Walk", "AccML"]
AccML_Turn = df.loc[df["state"] == "Turn", "AccML"]
AccML_Hesitate = df.loc[df["state"] == "Hesitate", "AccML"]
print(stats.ks_2samp(AccML_AKin, AccML_Walk))
print(cohend(AccML_AKin, AccML_Walk))
print(stats.ks_2samp(AccML_AKin, AccML_Turn))
print(cohend(AccML_AKin, AccML_Hesitate))
print(stats.ks_2samp(AccML_AKin, AccML_Hesitate))
print(cohend(AccML_AKin, AccML_Hesitate))
print(stats.ks_2samp(AccML_Walk, AccML_Turn))
print(cohend(AccML_Walk, AccML_Turn))
print(stats.ks_2samp(AccML_Walk, AccML_Hesitate))
print(cohend(AccML_Walk, AccML_Hesitate))
print(stats.ks_2samp(AccML_Turn, AccML_Hesitate))
print(cohend(AccML_Turn, AccML_Hesitate))
# ## The AccAP variable
Unique_States = df["state"].unique()
for State in Unique_States:
stats.probplot(df[df["state"] == State]["AccAP"], dist="norm", plot=plt)
plt.title("Probability Plot - " + State)
plt.show()
fig, axes = plt.subplots(2, 1, figsize=(22, 14))
ax = axes.ravel()
# ['AKinetic' 'Turn' 'Hesitate' 'Walk']
df.loc[df.state == "AKinetic"].AccAP.plot(
kind="hist",
ax=ax[0],
alpha=0.5,
bins=50,
sharex=True,
label="A-Kinetic",
edgecolor="black",
linewidth=3,
)
df.loc[df.state == "Turn"].AccAP.plot(
kind="hist",
ax=ax[0],
alpha=0.5,
bins=50,
sharex=True,
label="Turn",
edgecolor="blue",
linewidth=3,
)
df.loc[df.state == "Hesitate"].AccAP.plot(
kind="hist",
ax=ax[0],
alpha=0.5,
bins=50,
sharex=True,
label="Hesitate",
edgecolor="red",
linewidth=3,
)
df.loc[df.state == "Walk"].AccAP.plot(
kind="hist",
ax=ax[0],
alpha=0.5,
bins=50,
sharex=True,
label="Walk",
edgecolor="green",
linewidth=3,
)
fig.suptitle("Histogram and Density plot of AccAP by state", fontsize=20)
df.loc[df.state == "AKinetic"].AccAP.plot(
kind="density", ax=ax[1], alpha=0.5, label="A-Kinetic", linewidth=3
)
df.loc[df.state == "Turn"].AccAP.plot(
kind="density", ax=ax[1], alpha=0.5, label="Turn", linewidth=3
)
df.loc[df.state == "Hesitate"].AccAP.plot(
kind="density", ax=ax[1], alpha=0.5, label="Hesitate", linewidth=3
)
df.loc[df.state == "Walk"].AccAP.plot(
kind="density", ax=ax[1], alpha=0.5, label="Walk", linewidth=3
)
ax[0].legend(fontsize=18)
ax[0].set_xlim(-8, 8)
ax[0].set_title("Histogram of AccAP based on state")
ax[0].tick_params(axis="both", which="major", labelsize=14)
ax[0].tick_params(axis="both", which="minor", labelsize=14)
ax[1].legend(fontsize=18)
ax[1].set_xlim(-8, 8)
ax[1].set_title("Density of AccAP based on state")
ax[1].tick_params(axis="both", which="major", labelsize=14)
ax[1].tick_params(axis="both", which="minor", labelsize=14)
AccAP_AKin = df.loc[df["state"] == "AKinetic", "AccAP"]
AccAP_Walk = df.loc[df["state"] == "Walk", "AccAP"]
AccAP_Turn = df.loc[df["state"] == "Turn", "AccAP"]
AccAP_Hesitate = df.loc[df["state"] == "Hesitate", "AccAP"]
boxplot = df.boxplot(column=["AccAP"], by="state")
print(stats.ks_2samp(AccAP_AKin, AccAP_Walk))
print(cohend(AccAP_AKin, AccAP_Walk))
print(stats.ks_2samp(AccAP_AKin, AccAP_Turn))
print(cohend(AccAP_AKin, AccAP_Turn))
print(stats.ks_2samp(AccAP_AKin, AccAP_Hesitate))
print(cohend(AccAP_AKin, AccAP_Hesitate))
print(stats.ks_2samp(AccAP_Walk, AccAP_Turn))
print(cohend(AccAP_Walk, AccAP_Turn))
print(stats.ks_2samp(AccAP_Walk, AccAP_Hesitate))
print(cohend(AccAP_Walk, AccAP_Hesitate))
print(stats.ks_2samp(AccAP_Turn, AccAP_Hesitate))
print(cohend(AccAP_Turn, AccAP_Hesitate))
pd.set_option("display.max_rows", 999)
df.groupby("state")[["AccV", "AccML", "AccAP"]].describe().unstack()
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import pandas_datareader as pdr
import torch
import torch.nn as nn
key = "29d36f088ce550d9ac084e401f752e983045e0b5"
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
df = pdr.get_data_tiingo("AAPL", api_key=key)
df.to_csv("AAPL.csv")
df1 = pd.read_csv("AAPL.csv")
# convert datetime column to datetime dtype
df1["date"] = pd.to_datetime(df1["date"])
# extract date from datetime column
df1["date1"] = df1["date"].dt.date
df1
# # Preprocessing the Data - Making It Daily Frequency Based and Filling the Missing Values
new_df = pd.DataFrame(df1[["date1", "high"]])
new_df["Date"] = pd.to_datetime(new_df["date1"])
new_df = new_df.drop(columns="date1")
new_df = new_df.set_index("Date")
new_freq = "D"
new_df = new_df.resample(new_freq).asfreq()
new_df["high"] = new_df["high"].fillna(method="ffill").fillna(method="bfill")
new_df
# # Trend Analysis - Plotting Time Series Data
new_df.plot(figsize=(12, 6))
plt.xticks(rotation=90)
plt.show()
# # Decomposing the Various Components of the above Plotted Graph
from statsmodels.tsa.seasonal import seasonal_decompose
results = seasonal_decompose(new_df["high"])
results.plot()
# # Splitting Data into Training and Testing set
len(new_df)
train = new_df.iloc[:1274]
test = new_df.iloc[1274:]
from sklearn.preprocessing import MinMaxScaler
scaler = MinMaxScaler()
train_t = scaler.fit_transform(train)
test_t = scaler.transform(test)
train_t[:5], test_t[:5]
# # Making Input Data Batches
input_size = 30
output_size = 1
def create_batches(data, input_size, output_size, batch_size):
x = []
y = []
for i in range(len(data) - input_size - output_size + 1):
x.append(data[i : i + input_size, :])
y.append(data[i + input_size : i + input_size + output_size, :])
x = np.array(x)
y = np.array(y)
num_batches = len(x) // batch_size
x = x[: num_batches * batch_size, :, :]
y = y[: num_batches * batch_size, :, :]
return x, y
# Create batches of training data
x_train1, y_train1 = create_batches(train_t, input_size, output_size, batch_size=32)
# Create batches of testing data
x_test1, y_test1 = create_batches(test_t, input_size, output_size, batch_size=32)
x_train1.dtype
y_train1
# # Creating LSTM Model
class StockLSTM(nn.Module):
def __init__(self):
super(StockLSTM, self).__init__()
self.lstm = nn.LSTM(input_size=1, hidden_size=128, num_layers=2)
self.act1 = nn.ReLU()
self.linear = nn.Linear(in_features=128, out_features=1)
self.act2 = nn.ReLU()
def forward(self, x):
x, y = self.lstm(x)
x = self.act1(x)
x = self.linear(x)
output = self.act2(x)
return output
model = StockLSTM()
model.state_dict()
# # Defining the Loss Function and the Optimizer for the Model
loss_fn = nn.MSELoss()
optimizer = torch.optim.Adam(params=model.parameters(), lr=0.01)
X = torch.from_numpy(x_train1).to(torch.float32)
X
# # Creating the Training and Testing Loop
epochs = 10
train_loss = []
test_loss = []
epoch_count = []
for epoch in range(epochs):
# Training Loop
model.train()
optimizer.zero_grad()
y_pred = model(X)
loss = loss_fn(
y_pred.to(torch.float32), torch.from_numpy(y_train1).to(torch.float32)
)
train_loss.append(loss)
loss.backward()
optimizer.step()
# Testing Loop
model.eval()
y_pred_t = model(torch.from_numpy(x_test1).to(torch.float32))
loss1 = loss_fn(
y_pred_t.to(torch.float32), torch.from_numpy(y_test1).to(torch.float32)
)
test_loss.append(loss1)
if epoch % 1 == 0:
epoch_count.append(epoch)
train_loss.append(loss.detach().numpy())
test_loss.append(loss1.detach().numpy())
print(f"Epoch: {epoch} | Train Loss: {loss} | Test Loss: {loss1} ")
print("\n")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import warnings
warnings.filterwarnings("ignore")
from sklearn.feature_extraction.text import TfidfVectorizer
import matplotlib.pyplot as plt
from wordcloud import WordCloud
from sklearn.model_selection import train_test_split
import nltk
nltk.download("punkt")
nltk.download("stopwords")
from nltk.corpus import stopwords
import re
from bs4 import BeautifulSoup
# import contractions
from nltk.stem.wordnet import WordNetLemmatizer
data = pd.read_csv("/kaggle/input/amareview/AmazonReview.csv")
print(data.shape)
data.head()
data.dropna(inplace=True)
data.loc[data["Sentiment"] <= 3, "Sentiment"] = 0
data.loc[data["Sentiment"] > 3, "Sentiment"] = 1
stp_words = stopwords.words("english")
def clean_review(review):
pattern = "#[\w]*"
re.sub(pattern, "", review)
review = review.replace("<br />", "")
review = review.replace("<br/>", "")
review = review.replace("<br>", "")
review = review.replace("</br>", "")
cleanreview = " ".join(word for word in review.split() if word not in stp_words)
return cleanreview
data["Review"] = data["Review"].apply(clean_review)
print(data["Sentiment"].value_counts())
consolidated = " ".join(
word for word in data["Review"][data["Sentiment"] == 0].astype(str)
)
wordCloud = WordCloud(width=1600, height=800, random_state=21, max_font_size=110)
plt.figure(figsize=(15, 10))
plt.imshow(wordCloud.generate(consolidated), interpolation="bilinear")
plt.axis("off")
plt.show()
consolidated = " ".join(
word for word in data["Review"][data["Sentiment"] == 1].astype(str)
)
wordCloud = WordCloud(width=1600, height=800, random_state=21, max_font_size=110)
plt.figure(figsize=(15, 10))
plt.imshow(wordCloud.generate(consolidated), interpolation="bilinear")
plt.axis("off")
plt.show()
cv = TfidfVectorizer(max_features=2500)
X = cv.fit_transform(data["Review"]).toarray()
x_train, x_test, y_train, y_test = train_test_split(
X, data["Sentiment"], test_size=0.25, random_state=42, stratify=data["Sentiment"]
)
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import accuracy_score
model = LogisticRegression()
model.fit(x_train, y_train)
pred = model.predict(x_test)
print(accuracy_score(y_test, pred))
from sklearn import metrics
cm = metrics.confusion_matrix(y_test, pred)
cm_display = metrics.ConfusionMatrixDisplay(
confusion_matrix=cm, display_labels=[False, True]
)
cm_display.plot()
plt.show()
a = "THis is a bad product"
k = cv.transform([a]).toarray()
# print(cv.transform([a]).toarray())
model.predict(k)
|
# Import the libraries that will be used throughout the notebook.
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import seaborn as sns
import matplotlib.pyplot as plt
import sklearn.metrics as metrics
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Set pd to display fully so I can investigate the full datasets.
pd.set_option("display.max_rows", None)
pd.set_option("display.max_columns", None)
# Loading data into notebook, and drop id column from training and test data, to not mess up the training.
train_data = pd.read_csv("/kaggle/input/cap-4611-spring-21-assignment-1/train.csv")
test_data = pd.read_csv("/kaggle/input/cap-4611-spring-21-assignment-1/test.csv")
test_ids = test_data["id"]
train_data.drop(["id"], axis=1, inplace=True)
test_data.drop(["id"], axis=1, inplace=True)
# Remove spaces on the beginning of the column names to make it easier to reference columns
for column in train_data:
if column.startswith(" "):
train_data.rename(columns={column: column[1:]}, inplace=True)
for column in test_data:
if column.startswith(" "):
test_data.rename(columns={column: column[1:]}, inplace=True)
# I checked for missing values by first looking over the datatypes and non-null count through the .info() function. No values in the dataset appeared to be null, and all data types were numbers, so I figured there weren't any non-number values representing missing or invalid data.
train_data.info()
# Next, I checked for missing data using .describe(), to see if any number values appeared to not make sense for what it's suppose to represent. From this, I did not notice anything that seemed out of place, so I decided that there were no missing values to deal with for this dataset.
train_data.describe()
# I tried then making a boxplot of the whole dataset. This led to noticeable outliers for the following columns that seemed out of place:
# - fix assets to assets
# - revenue per person
# - Revenue Per Share (Yuan)
# - Quick asset /current liabilities
# There were others that were outside the whiskers, but seemed like important data to keep or were multiple values outside the whiskers.
fig = plt.figure(figsize=(40, 6))
# Creating plot
plt.boxplot(train_data)
# show plot
plt.show()
# I looked at the data directly of the four columns that appeared to have outliers according to the box plot, and sorted by ascending so the outliers stand out.
train_data["Quick asset /current liabilities"].sort_values(ascending=True)
# train_data['Revenue Per Share (Yuan)'].sort_values(ascending=True)
# train_data['revenue per person'].sort_values(ascending=True)#
# train_data['fix assets to assets'].sort_values(ascending=True)#
# I grabbed the indexes of the outliers from the printed data, and replaced their values with the upper whisker. I used the upper whisker on all of them as they each were much higher than the rest of the values in their columns.
# To calculate the whiskers, I created the functions get_upper_whisker and get_lower_whisker to calculate the upper or lower whisker of a column x's values, using the 75% and 25% quartiles, and calculated IQR.
# I went with replacing the outlier values with the upper or lower whisker value because it seemed better in terms of preserving the data, rather than completely deleting the value or row containing it.
def get_upper_whisker(x):
upper_quartile = x.quantile(0.75)
lower_quartile = x.quantile(0.25)
iqr = upper_quartile - lower_quartile
upper_whisker = upper_quartile + 1.5 * iqr
return upper_whisker
def get_lower_whisker(x):
upper_quartile = x.quantile(0.75)
lower_quartile = x.quantile(0.25)
iqr = upper_quartile - lower_quartile
lower_whisker = lower_quartile - 1.5 * iqr
return lower_whisker
train_data.loc[3002, "fix assets to assets"] = get_upper_whisker(
train_data["fix assets to assets"]
)
train_data.loc[2416, "revenue per person"] = get_upper_whisker(
train_data["revenue per person"]
)
train_data.loc[[1663, 2416, 2287], "Revenue Per Share (Yuan)"] = get_upper_whisker(
train_data["Revenue Per Share (Yuan)"]
)
train_data.loc[
[416, 729, 2287], "Quick asset /current liabilities"
] = get_upper_whisker(train_data["Quick asset /current liabilities"])
# Remove *Bankrupt* column from training data, so it wouldn't be included when fitting the model.
y = train_data["Bankrupt"]
train_data.drop(["Bankrupt"], axis=1, inplace=True)
# To garner more un-biased info when fitting the model and looking at predictions, I split the training data into two datasets, one for training/fitting the model, and one for validation and predictions of that fitted model. I did this as the prediction results I was getting with the training data without splitting seemed overfitted, so splitting the data helped tweak the hyperparameters of the model better.
from sklearn.model_selection import train_test_split
X_train, X_valid, y_train, y_valid = train_test_split(train_data, y, test_size=0.33)
X_test = test_data
# Next, I decided to implement normalization using MinMaxScaler from the sklearn library. I decided to use normalization as the values in the data for some of the columns seemed to vary in scale from one another, and normalizing the values might help when fitting the model with tree based models.
from sklearn.preprocessing import MinMaxScaler
# fit scaler on training data
norm = MinMaxScaler().fit(X_train)
# transform training data
X_train = norm.transform(X_train)
# transform validation data
X_valid = norm.transform(X_valid)
# transform testing dataabs
X_test = norm.transform(X_test)
# Highest DecisionTree scores:
# F1 Score: 0.25316455696202533
# Accuracy Score: 0.9476021314387212
# ROC AUC Score: 0.6439387244071291
from sklearn.tree import DecisionTreeClassifier
model = DecisionTreeClassifier(criterion="entropy", max_depth=12)
model.fit(X_train, y_train)
pred_valid = model.predict(X_valid)
f1 = metrics.f1_score(y_valid, pred_valid)
accuracy = metrics.accuracy_score(y_valid, pred_valid)
auc = metrics.roc_auc_score(y_valid, pred_valid)
print("F1 Score: ", f1)
print("Accuracy Score: ", accuracy)
print("ROC AUC Score: ", auc)
test_predictions = model.predict_proba(X_test)
print(np.round(test_predictions[:, 1], 2))
output = pd.DataFrame({"id": test_ids, "Bankrupt": np.round(test_predictions[:, 1], 2)})
output.to_csv("assignment1.csv", index=False)
print("Your submission was successfully saved!")
# Highest RandomForest score:
# F1 Score: 0.4423076923076923
# Accuracy Score: 0.9484902309058615
# ROC AUC Score: 0.8481366917071734
from sklearn.ensemble import RandomForestClassifier
model = RandomForestClassifier(
n_estimators=130,
max_depth=6,
class_weight="balanced",
min_samples_leaf=5,
max_features="log2",
)
model.fit(X_train, y_train)
pred_valid = model.predict(X_valid)
f1 = metrics.f1_score(y_valid, pred_valid)
accuracy = metrics.accuracy_score(y_valid, pred_valid)
auc = metrics.roc_auc_score(y_valid, pred_valid)
print("F1 Score: ", f1)
print("Accuracy Score: ", accuracy)
print("ROC AUC Score: ", auc)
test_predictions = model.predict_proba(X_test)
print(np.round(test_predictions[:, 1], 2))
output = pd.DataFrame({"id": test_ids, "Bankrupt": np.round(test_predictions[:, 1], 2)})
output.to_csv("assignment1.csv", index=False)
print("Your submission was successfully saved!")
|
# # Notebook Overview
# In this notebook, we explore how the timing of each group of levels affects the players' final score. We analyse the correlation between the average passing time of the levels and see how this affects the final score. The basic hypothesis is that the faster a player completes groups of levels, the higher their final score will be.
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import scipy.stats as stats
import plotly.express as px
# # Imports
# ## Loading Students Logs
use_columns = ["session_id", "index", "elapsed_time", "level_group", "event_name"]
dtypes = {
"session_id": "category",
"elapsed_time": np.int32,
"event_name": "category",
"name": "category",
"level": np.uint8,
"page": "category",
"room_coor_x": np.float32,
"room_coor_y": np.float32,
"screen_coor_x": np.float32,
"screen_coor_y": np.float32,
"hover_duration": np.float32,
"text": "category",
"fqid": "category",
"room_fqid": "category",
"text_fqid": "category",
"fullscreen": "category",
"hq": "category",
"music": "category",
"level_group": "category",
}
df = pd.read_csv(
"/kaggle/input/predict-student-performance-from-game-play/train.csv",
dtype=dtypes,
usecols=use_columns,
)
# # Loading Data
# ## Loading Students Answers
labels = pd.read_csv(
"/kaggle/input/predict-student-performance-from-game-play/train_labels.csv"
)
labels[["session_id", "question"]] = labels["session_id"].str.split("_", expand=True)
# Needed for proper sorting
labels["question"] = labels["question"].str.slice(1)
labels["question"] = labels["question"].astype(str)
labels.head()
# ## Evaluating Students Score
pivoted_questions = labels.pivot(
columns="question", values="correct", index="session_id"
)
students_score = pivoted_questions.iloc[:, 0:18].sum(axis=1)
students_score = students_score.rename("student_score")
students_score.head()
# # Data Extraction
q_inf = df[["session_id", "elapsed_time", "level_group", "event_name"]].copy()
q_inf.head()
# Find how much time each user spends on each group of levels
a = (
q_inf.groupby(["level_group", "session_id"])["elapsed_time"].max()
- q_inf.groupby(["level_group", "session_id"])["elapsed_time"].min()
)
a.reset_index()
# Split into three separate groups and look at each
a.groupby(["level_group"]).mean()
a_1 = a.reset_index()[a.reset_index()["level_group"] == "0-4"]
a_1
a_1["elapsed_time"] = np.log(a_1["elapsed_time"] + 2)
mean_value = a_1["elapsed_time"].mean()
plt.axvline(x=mean_value, color="r", linestyle="--")
sns.histplot(data=a_1, x="elapsed_time", kde=True)
a_2 = a.reset_index()[a.reset_index()["level_group"] == "5-12"]
a_2
a_2["elapsed_time"] = np.log(a_2["elapsed_time"] + 2)
mean_value = a_2["elapsed_time"].mean()
plt.axvline(x=mean_value, color="r", linestyle="--")
sns.histplot(data=a_2, x="elapsed_time", kde=True)
a_3 = a.reset_index()[a.reset_index()["level_group"] == "13-22"]
a_3
a_3["elapsed_time"] = np.log(a_3["elapsed_time"] + 2)
mean_value = a_3["elapsed_time"].mean()
plt.axvline(x=mean_value, color="r", linestyle="--")
sns.histplot(data=a_3, x="elapsed_time", kde=True)
# ## Dependence of the final score on time
labels.groupby(["session_id"]).sum()
score = labels.groupby(["session_id"]).sum()
a_2_join = pd.merge(a_2, score, on="session_id")
a_2_join
a_1_join = pd.merge(a_1, score, on="session_id")
a_1_join
a_3_join = pd.merge(a_3, score, on="session_id")
a_3_join
mean_time_score3 = a_3_join.groupby(["correct"]).mean()
mean_time_score3
# ## 13-22
sns.lineplot(x=mean_time_score3.index, y=mean_time_score3.elapsed_time)
mean_time_score2 = a_2_join.groupby(["correct"]).mean()
mean_time_score2
# ## 5-12
sns.lineplot(x=mean_time_score2.index, y=mean_time_score2.elapsed_time)
mean_time_score1 = a_1_join.groupby(["correct"]).mean()
mean_time_score1
# ## 0-4
sns.lineplot(x=mean_time_score1.index, y=mean_time_score1.elapsed_time)
|
# # CLASIFICACIÓN DE RAZAS DE PERROS
# *
# # **Integrantes:**
# 1. Hollman Esteban González Suárez 2172002
# 2. Kevin Alonso Luna Bustos 2172022
# 3. Jhoann Sebastian Martinez Oviedo 2171995
# # Contexto
# El conjunto de datos de Stanford Dogs contiene imágenes de 120 razas de perros de todo el mundo. Este conjunto de datos se ha creado utilizando imágenes y anotaciones de ImageNet para la tarea de categorización detallada de imágenes. Originalmente se recopiló para la categorización de imágenes de grano fino, un problema desafiante ya que ciertas razas de perros tienen características casi idénticas o difieren en color y edad.
# # Contenido
# Número de categorías: 120
# Número de imágenes: 20.580
# Anotaciones: etiquetas de clase, cuadros delimitadores
# # Agradecimientos
# La fuente de datos original se encuentra en http://vision.stanford.edu/aditya86/ImageNetDogs/ y contiene información adicional sobre las divisiones de entrenamiento / prueba y los resultados de referencia.
# # DATASET Usado
# Se uso la versión del dataset publicado por Dog-breed-identification en la plataforma de Kaggle.
# # **Librerías a importar**
import matplotlib.pyplot as plt
import os, shutil, math, scipy, cv2
from tensorflow import keras
import tensorflow as tf
import seaborn as sns
import pandas as pd
import random as rn
import numpy as np
from sklearn.utils import shuffle
from sklearn.metrics import classification_report
from sklearn.model_selection import train_test_split
from PIL import Image
from PIL import Image as pil_image
from PIL import ImageDraw
from tqdm import tqdm
from skimage.io import imread
from IPython.display import SVG
from scipy import misc, ndimage
from scipy.ndimage.interpolation import zoom
from keras.utils.np_utils import to_categorical
from keras import layers
from keras.applications.vgg16 import VGG16, preprocess_input
from keras.models import Sequential, Input, Model
from keras.layers import (
Dense,
Flatten,
Dropout,
Concatenate,
GlobalAveragePooling2D,
Lambda,
ZeroPadding2D,
)
from keras.preprocessing.image import ImageDataGenerator
from keras.optimizers import Adam, SGD
# # **DATASET**
# Dirección de los ficheros
labels = pd.read_csv("/kaggle/input/dog-breed-identification/labels.csv")
train_dir = "/kaggle/input/dog-breed-identification/train"
test_dir = "/kaggle/input/dog-breed-identification/test"
# Razas de perros
dogs_breeds = [
"scottish_deerhound",
"maltese_dog",
"entlebucher",
"pomeranian",
"labrador_retriever",
"basenji",
"airedale",
"leonberg",
"blenheim_spaniel",
"siberian_husky",
]
D = []
Z = []
imgsize = 150
def label_assignment(img):
img = img.rsplit(".", 1)[0]
label = "".join(labels[labels.id == img].breed.unique())
return label
def resize_dataset(data_dir):
for img in tqdm(os.listdir(data_dir)):
label = label_assignment(img)
if label in dogs_breeds:
path = os.path.join(data_dir, img)
img = cv2.imread(path, cv2.IMREAD_COLOR)
img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)
img = cv2.resize(img, (imgsize, imgsize))
D.append(np.array(img))
Z.append(str(label))
resize_dataset(train_dir)
resize_dataset(test_dir)
df = pd.DataFrame()
df["breed"] = Z
print(df["breed"].value_counts())
print("\n#Total Load Image: " + str(len(df.breed)))
# # Cantidad de Imagenes de Cada Raza Seleccionada
# - scottish_deerhound
# - maltese_dog
# - entlebucher
# - pomeranian
# - labrador_retriever
# - basenji
# - airedale
# - leonberg
# - blenheim_spaniel
# - siberian_husky
# function to show bar length
def barw(ax):
for p in ax.patches:
val = p.get_width() # height of the bar
x = p.get_x() + p.get_width() # x-position
y = p.get_y() + p.get_height() / 2 # y-position
ax.annotate(round(val, 2), (x, y))
# finding top dog brands
plt.figure(figsize=(15, 4))
lab = labels[labels["breed"].isin(dogs_breeds)].breed
ax0 = sns.countplot(y=lab, order=lab.value_counts().index)
barw(ax0)
plt.show()
# Se crean dos diccionarios para pasar las clases de forma numerica a texto.
class_dogs_breeds = {
"scottish_deerhound": 0,
"maltese_dog": 1,
"entlebucher": 2,
"pomeranian": 3,
"labrador_retriever": 4,
"basenji": 5,
"airedale": 6,
"leonberg": 7,
"blenheim_spaniel": 8,
"siberian_husky": 9,
}
class_dogs_breeds2 = {
0: "scottish_deerhound",
1: "maltese_dog",
2: "entlebucher",
3: "pomeranian",
4: "labrador_retriever",
5: "basenji",
6: "airedale",
7: "leonberg",
8: "blenheim_spaniel",
9: "siberian_husky",
}
T = [class_dogs_breeds[item] for item in Z]
Y = to_categorical(T, 10)
X = np.array(D)
X = X / 255
x_train, x_test, y_train, y_test = train_test_split(
X, Y, test_size=0.2, random_state=42
)
# # Imagenes Aleatoria de las Razas de Perros
fig, ax = plt.subplots(5, 2)
fig.set_size_inches(20, 20)
for i in range(5):
for j in range(2):
l = rn.randint(0, len(Z))
ax[i, j].imshow(X[l])
ax[i, j].set_title("Dog: " + Z[l])
plt.tight_layout()
# # MODELO VGG16 (RED NEURONAL)
# 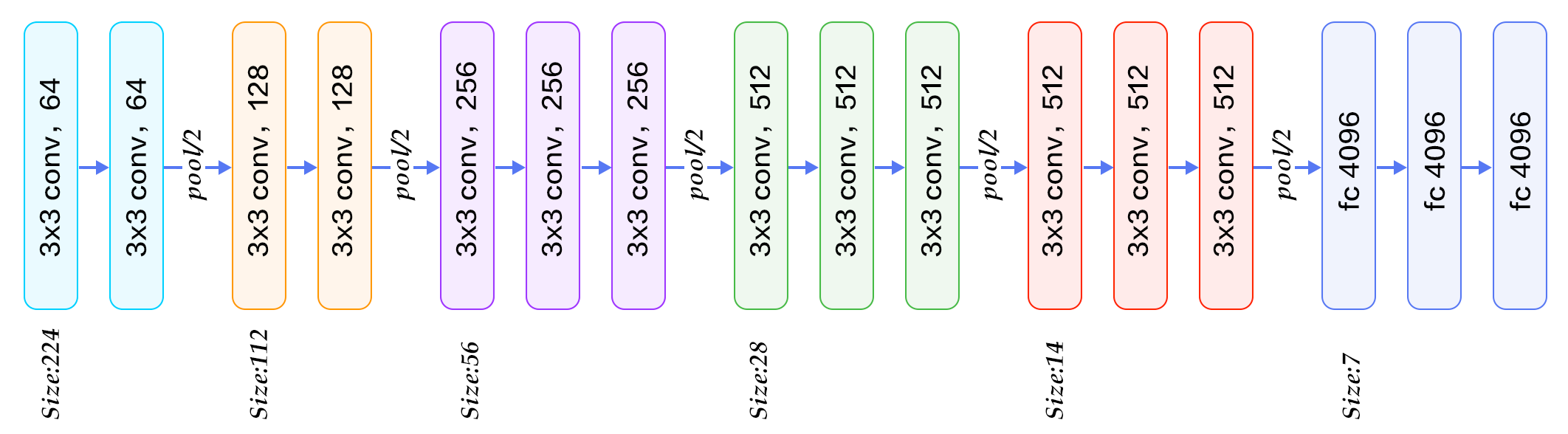*
# 
base_model = VGG16(
include_top=False, input_shape=(imgsize, imgsize, 3), weights="imagenet"
)
for layer in base_model.layers:
layer.trainable = False
for layer in base_model.layers:
print(layer, layer.trainable)
model = Sequential()
model.add(base_model)
model.add(GlobalAveragePooling2D())
model.add(Dropout(0.5))
model.add(Dense(10, activation="softmax"))
model.summary()
path = os.path.join(
"../input/dog-breed-identification/test/00102ee9d8eb90812350685311fe5890.jpg"
)
img = cv2.imread(path, cv2.IMREAD_COLOR)
img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)
img = cv2.resize(img, (imgsize, imgsize))
x = img_to_array(img)
x = x.reshape((1,) + x.shape)
datagen1 = ImageDataGenerator(rotation_range=30, fill_mode="nearest")
datagen2 = ImageDataGenerator(width_shift_range=0.2, height_shift_range=0.2)
datagen3 = ImageDataGenerator(horizontal_flip=True, vertical_flip=False)
datagen4 = ImageDataGenerator(zoom_range=0.3)
datagen5 = ImageDataGenerator(
rotation_range=10,
zoom_range=0.1,
width_shift_range=0.2,
height_shift_range=0.2,
horizontal_flip=True,
vertical_flip=False,
)
aug_iter1 = datagen1.flow(x, batch_size=1)
aug_iter2 = datagen2.flow(x, batch_size=1)
aug_iter3 = datagen3.flow(x, batch_size=1)
aug_iter4 = datagen4.flow(x, batch_size=1)
aug_iter5 = datagen5.flow(x, batch_size=1)
image1 = next(aug_iter1)[0].astype("uint8")
image2 = next(aug_iter2)[0].astype("uint8")
image3 = next(aug_iter3)[0].astype("uint8")
image4 = next(aug_iter4)[0].astype("uint8")
image5 = next(aug_iter5)[0].astype("uint8")
# plot image
fig, ax = plt.subplots(2, 3)
fig.set_size_inches(10, 10)
ax[0, 0].imshow(img)
ax[0, 1].imshow(image5)
ax[0, 2].imshow(image1)
ax[1, 0].imshow(image2)
ax[1, 1].imshow(image3)
ax[1, 2].imshow(image4)
ax[0, 0].set_title("ORIGINAL IMAGE")
ax[0, 1].set_title("DISTORTED")
ax[0, 2].set_title("ROTATION")
ax[1, 0].set_title("WIDTH AND HEIGHT SHIFT")
ax[1, 1].set_title("HORIZONTAL FLIP")
ax[1, 2].set_title("ZOOM RANGE")
plt.tight_layout()
model.compile(loss="categorical_crossentropy", optimizer="adam", metrics=["accuracy"])
history = model.fit_generator(
augs_gen.flow(x_train, y_train, batch_size=16),
validation_data=(x_test, y_test),
epochs=20,
)
plt.plot(history.history["accuracy"], linestyle="-.", color="g")
plt.plot(history.history["val_accuracy"], linestyle="--", color="y")
plt.title("ACCURACY MODEL")
plt.ylabel("ACCURACY")
plt.xlabel("EPOCH")
plt.legend(["TEST", "TRAIN"], loc="upper left")
plt.show()
plt.plot(history.history["loss"], linestyle="-.", color="g")
plt.plot(history.history["val_loss"], linestyle="--", color="y")
plt.title("LOSS MODEL")
plt.ylabel("LOSS")
plt.xlabel("EPOCH")
plt.legend(["TEST", "TRAIN"], loc="upper left")
plt.show()
list_img = np.random.randint(0, x_test.shape[0] - 1, size=10)
for i, img_test in enumerate(x_test[list_img]):
x = np.expand_dims(img_test, axis=0)
predictions = model.predict(x)
print("valor predicho:", np.argmax(predictions))
print("raza predicha: ", class_dogs_breeds2[np.argmax(predictions)])
print("max prob: ", np.max(predictions))
print("ground truth: ", np.argmax(y_test[list_img[i]]))
print("raza ground truth: ", class_dogs_breeds2[np.argmax(y_test[list_img[i]])])
plt.imshow(img_test)
plt.show()
from sklearn.ensemble import RandomForestClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.tree import DecisionTreeClassifier
from sklearn.model_selection import KFold, cross_val_score
from sklearn.metrics import *
from sklearn.decomposition import PCA
from sklearn.pipeline import Pipeline
from sklearn.svm import SVC
X1 = [X[i].flatten() for i in range(X.shape[0])]
s = cross_val_score(
GaussianNB(), X1, T, cv=KFold(5, shuffle=True), scoring=make_scorer(accuracy_score)
)
print("accuracy %.3f (+/- %.5f)" % (np.mean(s), np.std(s)))
s = cross_val_score(
RandomForestClassifier(),
X1,
T,
cv=KFold(5, shuffle=True),
scoring=make_scorer(accuracy_score),
)
print("accuracy %.3f (+/- %.5f)" % (np.mean(s), np.std(s)))
p = Pipeline((("pca", PCA(n_components=70)), ("classifier", SVC(kernel="rbf"))))
s = cross_val_score(
p, X1, T, cv=KFold(5, shuffle=True), scoring=make_scorer(accuracy_score)
)
print("accuracy %.3f (+/- %.5f)" % (np.mean(s), np.std(s)))
p = Pipeline((("pca", PCA(n_components=80)), ("classifier", RandomForestClassifier())))
s = cross_val_score(
p, X1, T, cv=KFold(5, shuffle=True), scoring=make_scorer(accuracy_score)
)
print("accuracy %.3f (+/- %.5f)" % (np.mean(s), np.std(s)))
p = Pipeline(
(
("pca", PCA(n_components=40)),
("classifier", DecisionTreeClassifier(max_depth=10)),
)
)
s = cross_val_score(
p, X1, T, cv=KFold(5, shuffle=True), scoring=make_scorer(accuracy_score)
)
print("accuracy %.3f (+/- %.5f)" % (np.mean(s), np.std(s)))
|
# !pip install opencv-python-headless
# !pip install numpy
# !pip install tqdm
# !pip install torch -f https://download.pytorch.org/whl/torch_stable.html
# !pip install natsort
# !pip install typing
# !pip install torchvision
# !pip install scipy
# # test.py
# !pip install sudo
# !pip install torchmetrics[image]
# # Train_SRGAN.py
# !pip install piq
# # !python --version
# import piq
# import torch
# x = torch.rand(3, 3, 256, 256, requires_grad=True)
# y = torch.rand(3, 3, 256, 256)
# loss = piq.information_weighted_ssim(x,y)
# # output = loss(x, y)
# # output
# print(loss)
# # output.backward()
# # Copyright 2022 Dakewe Biotech Corporation. All Rights Reserved.
# # Licensed under the Apache License, Version 2.0 (the "License");
# # you may not use this file except in compliance with the License.
# # You may obtain a copy of the License at
# #
# # http://www.apache.org/licenses/LICENSE-2.0
# #
# # Unless required by applicable law or agreed to in writing, software
# # distributed under the License is distributed on an "AS IS" BASIS,
# # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# # See the License for the specific language governing permissions and
# # limitations under the License.
# # ==============================================================================
# import os
# os.chdir("/kaggle/input/d/sahilchawla7/srganimplementation/")
# import time
# import torch
# from torch import nn
# from torch import optim
# from torch.optim import lr_scheduler
# from torch.utils.data import DataLoader
# from torch.utils.tensorboard import SummaryWriter
# import piq
# import model
# import srgan_config
# from dataset import CUDAPrefetcher, TrainValidImageDataset, TestImageDataset
# import image_quality_assessment
# from myUtils import load_state_dict, make_directory, save_checkpoint, AverageMeter, ProgressMeter
# # reset our working directory
# os.chdir("/kaggle/working/")
# model_names = sorted(
# name for name in model.__dict__ if
# name.islower() and not name.startswith("__") and callable(model.__dict__[name]))
# srgan_config.train_gt_images_dir = "/kaggle/input/superresolution-datasetpreprocessing/div2k-dataset/SRGAN/train/"
# # srgan_config.train_gt_images_dir = "/kaggle/input/div2ksmall/"
# srgan_config.test_gt_images_dir = "/kaggle/input/set5dataset/Set5/GTmod12/"
# srgan_config.test_lr_images_dir = f"/kaggle/input/set5dataset/Set5/LRbicx{srgan_config.upscale_factor}/"
# srgan_config.pretrained_g_model_weights_path = "/kaggle/input/srresnetbestepoch4/g_epoch_4.pth.tar"
# srgan_config.epochs = 4
# def main():
# # Initialize the number of training epochs
# start_epoch = 0
# # Initialize training to generate network evaluation indicators
# best_psnr = 0.0
# best_ssim = 0.0
# train_prefetcher, test_prefetcher = load_dataset()
# print("Load all datasets successfully.")
# d_model, g_model = build_model()
# print(f"Build `{srgan_config.g_arch_name}` model successfully.")
# pixel_criterion, content_criterion, adversarial_criterion = define_loss()
# print("Define all loss functions successfully.")
# d_optimizer, g_optimizer = define_optimizer(d_model, g_model)
# print("Define all optimizer functions successfully.")
# d_scheduler, g_scheduler = define_scheduler(d_optimizer, g_optimizer)
# print("Define all optimizer scheduler functions successfully.")
# print("Check whether to load pretrained d model weights...")
# if srgan_config.pretrained_d_model_weights_path:
# d_model = load_state_dict(d_model, srgan_config.pretrained_d_model_weights_path)
# print(f"Loaded `{srgan_config.pretrained_d_model_weights_path}` pretrained model weights successfully.")
# else:
# print("Pretrained d model weights not found.")
# print("Check whether to load pretrained g model weights...")
# if srgan_config.pretrained_g_model_weights_path:
# g_model = load_state_dict(g_model, srgan_config.pretrained_g_model_weights_path)
# print(f"Loaded `{srgan_config.pretrained_g_model_weights_path}` pretrained model weights successfully.")
# else:
# print("Pretrained g model weights not found.")
# print("Check whether the pretrained d model is restored...")
# if srgan_config.resume_d_model_weights_path:
# d_model, _, start_epoch, best_psnr, best_ssim, optimizer, scheduler = load_state_dict(
# d_model,
# srgan_config.resume_d_model_weights_path,
# optimizer=d_optimizer,
# scheduler=d_scheduler,
# load_mode="resume")
# print("Loaded pretrained model weights.")
# else:
# print("Resume training d model not found. Start training from scratch.")
# print("Check whether the pretrained g model is restored...")
# if srgan_config.resume_g_model_weights_path:
# g_model, _, start_epoch, best_psnr, best_ssim, optimizer, scheduler = load_state_dict(
# g_model,
# srgan_config.resume_g_model_weights_path,
# optimizer=g_optimizer,
# scheduler=g_scheduler,
# load_mode="resume")
# print("Loaded pretrained model weights.")
# else:
# print("Resume training g model not found. Start training from scratch.")
# # Create a experiment results
# samples_dir = os.path.join("samples", srgan_config.exp_name)
# results_dir = os.path.join("results", srgan_config.exp_name)
# make_directory(samples_dir)
# make_directory(results_dir)
# # Create training process log file
# writer = SummaryWriter(os.path.join("samples", "logs", srgan_config.exp_name))
# # Create an IQA evaluation model
# psnr_model = image_quality_assessment.PSNR(srgan_config.upscale_factor, srgan_config.only_test_y_channel)
# ssim_model = image_quality_assessment.SSIM(srgan_config.upscale_factor, srgan_config.only_test_y_channel)
# # Transfer the IQA model to the specified device
# psnr_model = psnr_model.to(device=srgan_config.device)
# ssim_model = ssim_model.to(device=srgan_config.device)
# for epoch in range(start_epoch, srgan_config.epochs):
# train(d_model,
# g_model,
# train_prefetcher,
# pixel_criterion,
# content_criterion,
# adversarial_criterion,
# d_optimizer,
# g_optimizer,
# epoch,
# writer)
# psnr, ssim = validate(g_model,
# test_prefetcher,
# epoch,
# writer,
# psnr_model,
# ssim_model,
# "Test")
# print("\n")
# # Update LR
# d_scheduler.step()
# g_scheduler.step()
# # Automatically save the model with the highest index
# is_best = psnr > best_psnr and ssim > best_ssim
# is_last = (epoch + 1) == srgan_config.epochs
# best_psnr = max(psnr, best_psnr)
# best_ssim = max(ssim, best_ssim)
# save_checkpoint({"epoch": epoch + 1,
# "best_psnr": best_psnr,
# "best_ssim": best_ssim,
# "state_dict": d_model.state_dict(),
# "optimizer": d_optimizer.state_dict(),
# "scheduler": d_scheduler.state_dict()},
# f"d_epoch_{epoch + 1}.pth.tar",
# samples_dir,
# results_dir,
# "d_best.pth.tar",
# "d_last.pth.tar",
# is_best,
# is_last)
# save_checkpoint({"epoch": epoch + 1,
# "best_psnr": best_psnr,
# "best_ssim": best_ssim,
# "state_dict": g_model.state_dict(),
# "optimizer": g_optimizer.state_dict(),
# "scheduler": g_scheduler.state_dict()},
# f"g_epoch_{epoch + 1}.pth.tar",
# samples_dir,
# results_dir,
# "g_best.pth.tar",
# "g_last.pth.tar",
# is_best,
# is_last)
# def load_dataset() -> [CUDAPrefetcher, CUDAPrefetcher]:
# # Load train, test and valid datasets
# train_datasets = TrainValidImageDataset(srgan_config.train_gt_images_dir,
# srgan_config.gt_image_size,
# srgan_config.upscale_factor,
# "Train")
# test_datasets = TestImageDataset(srgan_config.test_gt_images_dir, srgan_config.test_lr_images_dir)
# # Generator all dataloader
# train_dataloader = DataLoader(train_datasets,
# batch_size=srgan_config.batch_size,
# shuffle=True,
# num_workers=srgan_config.num_workers,
# pin_memory=True,
# drop_last=True,
# persistent_workers=True)
# test_dataloader = DataLoader(test_datasets,
# batch_size=1,
# shuffle=False,
# num_workers=1,
# pin_memory=True,
# drop_last=False,
# persistent_workers=True)
# # Place all data on the preprocessing data loader
# train_prefetcher = CUDAPrefetcher(train_dataloader, srgan_config.device)
# test_prefetcher = CUDAPrefetcher(test_dataloader, srgan_config.device)
# return train_prefetcher, test_prefetcher
# def build_model() -> [nn.Module, nn.Module, nn.Module]:
# d_model = model.__dict__[srgan_config.d_arch_name]()
# g_model = model.__dict__[srgan_config.g_arch_name](in_channels=srgan_config.in_channels,
# out_channels=srgan_config.out_channels,
# channels=srgan_config.channels,
# num_rcb=srgan_config.num_rcb)
# d_model = d_model.to(device=srgan_config.device)
# g_model = g_model.to(device=srgan_config.device)
# return d_model, g_model
# def define_loss() -> [nn.MSELoss, model.content_loss, nn.BCEWithLogitsLoss]:
# pixel_criterion = nn.MSELoss()
# content_criterion = model.content_loss(feature_model_extractor_node=srgan_config.feature_model_extractor_node,
# feature_model_normalize_mean=srgan_config.feature_model_normalize_mean,
# feature_model_normalize_std=srgan_config.feature_model_normalize_std)
# adversarial_criterion = nn.BCEWithLogitsLoss()
# # Transfer to CUDA
# pixel_criterion = pixel_criterion.to(device=srgan_config.device)
# content_criterion = content_criterion.to(device=srgan_config.device)
# adversarial_criterion = adversarial_criterion.to(device=srgan_config.device)
# return pixel_criterion, content_criterion, adversarial_criterion
# def define_optimizer(d_model, g_model) -> [optim.Adam, optim.Adam]:
# d_optimizer = optim.Adam(d_model.parameters(),
# srgan_config.model_lr,
# srgan_config.model_betas,
# srgan_config.model_eps,
# srgan_config.model_weight_decay)
# g_optimizer = optim.Adam(g_model.parameters(),
# srgan_config.model_lr,
# srgan_config.model_betas,
# srgan_config.model_eps,
# srgan_config.model_weight_decay)
# return d_optimizer, g_optimizer
# def define_scheduler(
# d_optimizer: optim.Adam,
# g_optimizer: optim.Adam
# ) -> [lr_scheduler.StepLR, lr_scheduler.StepLR]:
# d_scheduler = lr_scheduler.StepLR(d_optimizer,
# srgan_config.lr_scheduler_step_size,
# srgan_config.lr_scheduler_gamma)
# g_scheduler = lr_scheduler.StepLR(g_optimizer,
# srgan_config.lr_scheduler_step_size,
# srgan_config.lr_scheduler_gamma)
# return d_scheduler, g_scheduler
# def train(
# d_model: nn.Module,
# g_model: nn.Module,
# train_prefetcher: CUDAPrefetcher,
# pixel_criterion: nn.MSELoss,
# content_criterion: model.content_loss,
# adversarial_criterion: nn.BCEWithLogitsLoss,
# d_optimizer: optim.Adam,
# g_optimizer: optim.Adam,
# epoch: int,
# writer: SummaryWriter
# ) -> None:
# # Calculate how many batches of data are in each Epoch
# batches = len(train_prefetcher)
# # Print information of progress bar during training
# batch_time = AverageMeter("Time", ":6.3f")
# data_time = AverageMeter("Data", ":6.3f")
# pixel_losses = AverageMeter("Pixel loss", ":6.6f")
# content_losses = AverageMeter("Content loss", ":6.6f")
# adversarial_losses = AverageMeter("Adversarial loss", ":6.6f")
# gmsd_losses = AverageMeter("GMSD Loss",":6.6f")
# d_gt_probabilities = AverageMeter("D(GT)", ":6.3f")
# d_sr_probabilities = AverageMeter("D(SR)", ":6.3f")
# progress = ProgressMeter(batches,
# [batch_time, data_time,
# pixel_losses, content_losses, adversarial_losses,gmsd_losses,
# d_gt_probabilities, d_sr_probabilities],
# prefix=f"Epoch: [{epoch + 1}]")
# # Put the adversarial network model in training mode
# d_model.train()
# g_model.train()
# # Initialize the number of data batches to print logs on the terminal
# batch_index = 0
# # Initialize the data loader and load the first batch of data
# train_prefetcher.reset()
# batch_data = train_prefetcher.next()
# # Get the initialization training time
# end = time.time()
# while batch_data is not None:
# # Calculate the time it takes to load a batch of data
# data_time.update(time.time() - end)
# # Transfer in-memory data to CUDA devices to speed up training
# gt = batch_data["gt"].to(device=srgan_config.device, non_blocking=True)
# lr = batch_data["lr"].to(device=srgan_config.device, non_blocking=True)
# # Set the real sample label to 1, and the false sample label to 0
# batch_size, _, height, width = gt.shape
# real_label = torch.full([batch_size, 1], 1.0, dtype=gt.dtype, device=srgan_config.device)
# fake_label = torch.full([batch_size, 1], 0.0, dtype=gt.dtype, device=srgan_config.device)
# # Start training the discriminator model
# # During discriminator model training, enable discriminator model backpropagation
# for d_parameters in d_model.parameters():
# d_parameters.requires_grad = True
# # Initialize the discriminator model gradients
# d_model.zero_grad(set_to_none=True)
# # Calculate the classification score of the discriminator model for real samples
# gt_output = d_model(gt)
# d_loss_gt = adversarial_criterion(gt_output, real_label)
# # Call the gradient scaling function in the mixed precision API to
# # back-propagate the gradient information of the fake samples
# d_loss_gt.backward(retain_graph=True)
# # Calculate the classification score of the discriminator model for fake samples
# # Use the generator model to generate fake samples
# sr = g_model(lr)
# sr_output = d_model(sr.detach().clone())
# d_loss_sr = adversarial_criterion(sr_output, fake_label)
# # Call the gradient scaling function in the mixed precision API to
# # back-propagate the gradient information of the fake samples
# d_loss_sr.backward()
# # Calculate the total discriminator loss value
# d_loss = d_loss_gt + d_loss_sr
# # Improve the discriminator model's ability to classify real and fake samples
# d_optimizer.step()
# # Finish training the discriminator model
# # Start training the generator model
# # During generator training, turn off discriminator backpropagation
# for d_parameters in d_model.parameters():
# d_parameters.requires_grad = False
# # Initialize generator model gradients
# g_model.zero_grad(set_to_none=True)
# # Calculate the perceptual loss of the generator, mainly including pixel loss, feature loss and adversarial loss
# # srgan_config.pixel_weight = 0.01
# srgan_config.adversarial_weight = 0.01
# pixel_loss = srgan_config.pixel_weight * pixel_criterion(sr, gt)
# content_loss = srgan_config.content_weight * content_criterion(sr, gt)
# adversarial_loss = srgan_config.adversarial_weight * adversarial_criterion(d_model(sr), real_label)
# gmsd_loss_criterion = piq.GMSDLoss()
# gmsd_loss = 0
# sr_copy = sr.detach().clone()
# gt_copy = gt.detach().clone()
# # fsim_loss_criterion=piq.fsim()
# fsim_loss=0
# sr_copy2 = sr.detach().clone()
# gt_copy2 = gt.detach().clone()
# try:
# gmsd_loss = gmsd_loss_criterion(sr_copy,gt_copy)
# fsim_loss = piq.fsim(sr_copy2,gt_copy2)
# except:
# print("inside except")
# gmsd_loss = 0
# fsim_loss = 0
# pass
# # print(fsim_loss)
# # Calculate the generator total loss value
# g_loss = pixel_loss + content_loss + adversarial_loss + gmsd_loss #+ (1-fsim_loss)
# # Call the gradient scaling function in the mixed precision API to
# # back-propagate the gradient information of the fake samples
# g_loss.backward()
# # Encourage the generator to generate higher quality fake samples, making it easier to fool the discriminator
# g_optimizer.step()
# # Finish training the generator model
# # Calculate the score of the discriminator on real samples and fake samples,
# # the score of real samples is close to 1, and the score of fake samples is close to 0
# d_gt_probability = torch.sigmoid_(torch.mean(gt_output.detach()))
# d_sr_probability = torch.sigmoid_(torch.mean(sr_output.detach()))
# # Statistical accuracy and loss value for terminal data output
# pixel_losses.update(pixel_loss.item(), lr.size(0))
# content_losses.update(content_loss.item(), lr.size(0))
# adversarial_losses.update(adversarial_loss.item(), lr.size(0))
# gmsd_losses.update(gmsd_loss.item(),lr.size(0))
# d_gt_probabilities.update(d_gt_probability.item(), lr.size(0))
# d_sr_probabilities.update(d_sr_probability.item(), lr.size(0))
# # Calculate the time it takes to fully train a batch of data
# batch_time.update(time.time() - end)
# end = time.time()
# # Write the data during training to the training log file
# if batch_index % srgan_config.train_print_frequency == 0:
# iters = batch_index + epoch * batches + 1
# writer.add_scalar("Train/D_Loss", d_loss.item(), iters)
# writer.add_scalar("Train/G_Loss", g_loss.item(), iters)
# writer.add_scalar("Train/Pixel_Loss", pixel_loss.item(), iters)
# writer.add_scalar("Train/Content_Loss", content_loss.item(), iters)
# writer.add_scalar("Train/Adversarial_Loss", adversarial_loss.item(), iters)
# writer.add_scalar("Train/GMSD_Loss",gmsd_loss.item(),iters)
# writer.add_scalar("Train/D(GT)_Probability", d_gt_probability.item(), iters)
# writer.add_scalar("Train/D(SR)_Probability", d_sr_probability.item(), iters)
# progress.display(batch_index + 1)
# # Preload the next batch of data
# batch_data = train_prefetcher.next()
# # After training a batch of data, add 1 to the number of data batches to ensure that the
# # terminal print data normally
# batch_index += 1
# def validate(
# g_model: nn.Module,
# data_prefetcher: CUDAPrefetcher,
# epoch: int,
# writer: SummaryWriter,
# psnr_model: nn.Module,
# ssim_model: nn.Module,
# mode: str
# ) -> [float, float]:
# # Calculate how many batches of data are in each Epoch
# batch_time = AverageMeter("Time", ":6.3f")
# psnres = AverageMeter("PSNR", ":4.2f")
# ssimes = AverageMeter("SSIM", ":4.4f")
# progress = ProgressMeter(len(data_prefetcher), [batch_time, psnres, ssimes], prefix=f"{mode}: ")
# # Put the adversarial network model in validation mode
# g_model.eval()
# # Initialize the number of data batches to print logs on the terminal
# batch_index = 0
# # Initialize the data loader and load the first batch of data
# data_prefetcher.reset()
# batch_data = data_prefetcher.next()
# # Get the initialization test time
# end = time.time()
# with torch.no_grad():
# while batch_data is not None:
# # Transfer the in-memory data to the CUDA device to speed up the test
# gt = batch_data["gt"].to(device=srgan_config.device, non_blocking=True)
# lr = batch_data["lr"].to(device=srgan_config.device, non_blocking=True)
# # Use the generator model to generate a fake sample
# sr = g_model(lr)
# # Statistical loss value for terminal data output
# psnr = psnr_model(sr, gt)
# ssim = ssim_model(sr, gt)
# psnres.update(psnr.item(), lr.size(0))
# ssimes.update(ssim.item(), lr.size(0))
# # Calculate the time it takes to fully test a batch of data
# batch_time.update(time.time() - end)
# end = time.time()
# # Record training log information
# if batch_index % srgan_config.valid_print_frequency == 0:
# progress.display(batch_index + 1)
# # Preload the next batch of data
# batch_data = data_prefetcher.next()
# # After training a batch of data, add 1 to the number of data batches to ensure that the
# # terminal print data normally
# batch_index += 1
# # print metrics
# progress.display_summary()
# if mode == "Valid" or mode == "Test":
# writer.add_scalar(f"{mode}/PSNR", psnres.avg, epoch + 1)
# writer.add_scalar(f"{mode}/SSIM", ssimes.avg, epoch + 1)
# else:
# raise ValueError("Unsupported mode, please use `Valid` or `Test`.")
# return psnres.avg, ssimes.avg
# if __name__ == "__main__":
# main()
# # Train SRResnet
# import os
# os.chdir("/kaggle/input/d/sahilchawla7/srganimplementation/")
# import model
# import srresnet_config
# print(srresnet_config.exp_name)
# !python --version
# # Copyright 2022 Dakewe Biotech Corporation. All Rights Reserved.
# # Licensed under the Apache License, Version 2.0 (the "License");
# # you may not use this file except in compliance with the License.
# # You may obtain a copy of the License at
# #
# # http://www.apache.org/licenses/LICENSE-2.0
# #
# # Unless required by applicable law or agreed to in writing, software
# # distributed under the License is distributed on an "AS IS" BASIS,
# # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# # See the License for the specific language governing permissions and
# # limitations under the License.
# # ==============================================================================
# import os
# import time
# os.chdir("/kaggle/input/d/sahilchawla7/srganimplementation/")
# import torch
# from torch import nn
# from torch import optim
# from torch.cuda import amp
# from torch.utils.data import DataLoader
# from torch.utils.tensorboard import SummaryWriter
# import model
# import srresnet_config
# from dataset import CUDAPrefetcher, TrainValidImageDataset, TestImageDataset
# import image_quality_assessment
# # from image_quality_assessment import PSNR, SSIM
# from myUtils import load_state_dict, make_directory, save_checkpoint, AverageMeter, ProgressMeter
# os.chdir("/kaggle/working")
# model_names = sorted(
# name for name in model.__dict__ if
# name.islower() and not name.startswith("__") and callable(model.__dict__[name]))
# # srresnet_config.device = torch.device("cpu")
# srresnet_config.train_gt_images_dir = "/kaggle/input/superresolution-datasetpreprocessing/div2k-dataset/SRGAN/train/"
# srresnet_config.test_gt_images_dir = "/kaggle/input/helenhr-lr-divided/helen-test-set/HR/"
# srresnet_config.test_lr_images_dir = "/kaggle/input/helenhr-lr-divided/helen-test-set/LR/"
# def main():
# # Initialize the number of training epochs
# start_epoch = 0
# # Initialize training to generate network evaluation indicators
# best_psnr = 0.0
# best_ssim = 0.0
# train_prefetcher, test_prefetcher = load_dataset()
# print("Load all datasets successfully.")
# srresnet_model = build_model()
# print(f"Build `{srresnet_config.g_arch_name}` model successfully.")
# criterion = define_loss()
# print("Define all loss functions successfully.")
# optimizer = define_optimizer(srresnet_model)
# print("Define all optimizer functions successfully.")
# print("Check whether to load pretrained model weights...")
# if srresnet_config.pretrained_model_weights_path:
# srresnet_model = load_state_dict(srresnet_model, srresnet_config.pretrained_model_weights_path)
# print(f"Loaded `{srresnet_config.pretrained_model_weights_path}` pretrained model weights successfully.")
# else:
# print("Pretrained model weights not found.")
# print("Check whether the pretrained model is restored...")
# if srresnet_config.resume_model_weights_path:
# srresnet_model, _, start_epoch, best_psnr, best_ssim, optimizer, _ = load_state_dict(
# srresnet_model,
# srresnet_config.resume_model_weights_path,
# optimizer=optimizer,
# load_mode="resume")
# print("Loaded pretrained model weights.")
# else:
# print("Resume training model not found. Start training from scratch.")
# # Create a experiment results
# samples_dir = os.path.join("samples", srresnet_config.exp_name)
# results_dir = os.path.join("results", srresnet_config.exp_name)
# make_directory(samples_dir)
# make_directory(results_dir)
# # Create training process log file
# writer = SummaryWriter(os.path.join("samples", "logs", srresnet_config.exp_name))
# # Initialize the gradient scaler
# scaler = amp.GradScaler()
# # Create an IQA evaluation model
# psnr_model = image_quality_assessment.PSNR(srresnet_config.upscale_factor, srresnet_config.only_test_y_channel)
# ssim_model = image_quality_assessment.SSIM(srresnet_config.upscale_factor, srresnet_config.only_test_y_channel)
# # Transfer the IQA model to the specified device
# psnr_model = psnr_model.to(device=srresnet_config.device)
# ssim_model = ssim_model.to(device=srresnet_config.device)
# for epoch in range(start_epoch, srresnet_config.epochs):
# train(srresnet_model,
# train_prefetcher,
# criterion,
# optimizer,
# epoch,
# scaler,
# writer)
# psnr, ssim = validate(srresnet_model,
# test_prefetcher,
# epoch,
# writer,
# psnr_model,
# ssim_model,
# "Test")
# print("\n")
# # Automatically save the model with the highest index
# is_best = psnr > best_psnr and ssim > best_ssim
# is_last = (epoch + 1) == srresnet_config.epochs
# best_psnr = max(psnr, best_psnr)
# best_ssim = max(ssim, best_ssim)
# save_checkpoint({"epoch": epoch + 1,
# "best_psnr": best_psnr,
# "best_ssim": best_ssim,
# "state_dict": srresnet_model.state_dict(),
# "optimizer": optimizer.state_dict()},
# f"g_epoch_{epoch + 1}.pth.tar",
# samples_dir,
# results_dir,
# "g_best.pth.tar",
# "g_last.pth.tar",
# is_best,
# is_last)
# def load_dataset() : #-> [CUDAPrefetcher, CUDAPrefetcher]:
# # Load train, test and valid datasets
# train_datasets = TrainValidImageDataset(srresnet_config.train_gt_images_dir,
# srresnet_config.gt_image_size,
# srresnet_config.upscale_factor,
# "Train")
# test_datasets = TestImageDataset(srresnet_config.test_gt_images_dir, srresnet_config.test_lr_images_dir)
# # Generator all dataloader
# train_dataloader = DataLoader(train_datasets,
# batch_size=srresnet_config.batch_size,
# shuffle=True,
# num_workers=srresnet_config.num_workers,
# pin_memory=True,
# drop_last=True,
# persistent_workers=True)
# test_dataloader = DataLoader(test_datasets,
# batch_size=1,
# shuffle=False,
# num_workers=1,
# pin_memory=True,
# drop_last=False,
# persistent_workers=True)
# print(srresnet_config.device)
# # return train_dataloader, test_dataloader
# # Place all data on the preprocessing data loader
# train_prefetcher = CUDAPrefetcher(train_dataloader, srresnet_config.device)
# test_prefetcher = CUDAPrefetcher(test_dataloader, srresnet_config.device)
# return train_prefetcher, test_prefetcher
# def build_model() -> nn.Module:
# srresnet_model = model.__dict__[srresnet_config.g_arch_name](in_channels=srresnet_config.in_channels,
# out_channels=srresnet_config.out_channels,
# channels=srresnet_config.channels,
# num_rcb=srresnet_config.num_rcb)
# srresnet_model = srresnet_model.to(device=srresnet_config.device)
# return srresnet_model
# def define_loss() -> nn.MSELoss:
# criterion = nn.MSELoss()
# criterion = criterion.to(device=srresnet_config.device)
# return criterion
# def define_optimizer(srresnet_model) -> optim.Adam:
# optimizer = optim.Adam(srresnet_model.parameters(),
# srresnet_config.model_lr,
# srresnet_config.model_betas,
# srresnet_config.model_eps,
# srresnet_config.model_weight_decay)
# return optimizer
# def train(
# srresnet_model: nn.Module,
# train_prefetcher: CUDAPrefetcher,
# # train_prefetcher: DataLoader,
# criterion: nn.MSELoss,
# optimizer: optim.Adam,
# epoch: int,
# scaler: amp.GradScaler,
# writer: SummaryWriter
# ) -> None:
# # Calculate how many batches of data are in each Epoch
# batches = len(train_prefetcher)
# # Print information of progress bar during training
# batch_time = AverageMeter("Time", ":6.3f")
# data_time = AverageMeter("Data", ":6.3f")
# losses = AverageMeter("Loss", ":6.6f")
# progress = ProgressMeter(batches, [batch_time, data_time, losses], prefix=f"Epoch: [{epoch + 1}]")
# # Put the generative network model in training mode
# srresnet_model.train()
# # Initialize the number of data batches to print logs on the terminal
# batch_index = 0
# # Initialize the data loader and load the first batch of data
# train_prefetcher.reset()
# batch_data = train_prefetcher.next()
# # Get the initialization training time
# end = time.time()
# # for i,batch_data in enumerate(train_prefetcher):
# while batch_data is not None:
# # Calculate the time it takes to load a batch of data
# data_time.update(time.time() - end)
# # Transfer in-memory data to CUDA devices to speed up training
# gt = batch_data["gt"].to(device=srresnet_config.device, non_blocking=True)
# lr = batch_data["lr"].to(device=srresnet_config.device, non_blocking=True)
# # Initialize generator gradients
# srresnet_model.zero_grad(set_to_none=True)
# # Mixed precision training
# with amp.autocast():
# sr = srresnet_model(lr)
# loss = torch.mul(srresnet_config.loss_weights, criterion(sr, gt))
# # Backpropagation
# # loss.backward()
# scaler.scale(loss).backward()
# # update generator weights
# scaler.step(optimizer)
# scaler.update()
# # Statistical loss value for terminal data output
# losses.update(loss.item(), lr.size(0))
# # Calculate the time it takes to fully train a batch of data
# batch_time.update(time.time() - end)
# end = time.time()
# # Write the data during training to the training log file
# if batch_index % srresnet_config.train_print_frequency == 0:
# # Record loss during training and output to file
# writer.add_scalar("Train/Loss", loss.item(), batch_index + epoch * batches + 1)
# progress.display(batch_index + 1)
# # Preload the next batch of data
# batch_data = train_prefetcher.next()
# # Add 1 to the number of data batches to ensure that the terminal prints data normally
# batch_index += 1
# def validate(
# srresnet_model: nn.Module,
# data_prefetcher: CUDAPrefetcher,
# # data_prefetcher: DataLoader,
# epoch: int,
# writer: SummaryWriter,
# psnr_model: nn.Module,
# ssim_model: nn.Module,
# mode: str
# ) -> [float, float]:
# # Calculate how many batches of data are in each Epoch
# batch_time = AverageMeter("Time", ":6.3f")
# psnres = AverageMeter("PSNR", ":4.2f")
# ssimes = AverageMeter("SSIM", ":4.4f")
# progress = ProgressMeter(len(data_prefetcher), [batch_time, psnres, ssimes], prefix=f"{mode}: ")
# # Put the adversarial network model in validation mode
# srresnet_model.eval()
# # Initialize the number of data batches to print logs on the terminal
# batch_index = 0
# # Initialize the data loader and load the first batch of data
# data_prefetcher.reset()
# batch_data = data_prefetcher.next()
# # Get the initialization test time
# end = time.time()
# with torch.no_grad():
# # for i, batch_data in enumerate(data_prefetcher):
# while batch_data is not None:
# # Transfer the in-memory data to the CUDA device to speed up the test
# gt = batch_data["gt"].to(device=srresnet_config.device, non_blocking=True)
# lr = batch_data["lr"].to(device=srresnet_config.device, non_blocking=True)
# # Use the generator model to generate a fake sample
# with amp.autocast():
# sr = srresnet_model(lr)
# # Statistical loss value for terminal data output
# psnr = psnr_model(sr, gt)
# ssim = ssim_model(sr, gt)
# psnres.update(psnr.item(), lr.size(0))
# ssimes.update(ssim.item(), lr.size(0))
# # Calculate the time it takes to fully test a batch of data
# batch_time.update(time.time() - end)
# end = time.time()
# # Record training log information
# if batch_index % srresnet_config.valid_print_frequency == 0:
# progress.display(batch_index + 1)
# # Preload the next batch of data
# batch_data = data_prefetcher.next()
# # After training a batch of data, add 1 to the number of data batches to ensure that the
# # terminal print data normally
# batch_index += 1
# # print metrics
# progress.display_summary()
# if mode == "Valid" or mode == "Test":
# writer.add_scalar(f"{mode}/PSNR", psnres.avg, epoch + 1)
# writer.add_scalar(f"{mode}/SSIM", ssimes.avg, epoch + 1)
# else:
# raise ValueError("Unsupported mode, please use `Valid` or `Test`.")
# return psnres.avg, ssimes.avg
# if __name__ == "__main__":
# main()
# # Testing
# !pip install natsort
# # Copyright 2022 Dakewe Biotech Corporation. All Rights Reserved.
# # Licensed under the Apache License, Version 2.0 (the "License");
# # you may not use this file except in compliance with the License.
# # You may obtain a copy of the License at
# #
# # http://www.apache.org/licenses/LICENSE-2.0
# #
# # Unless required by applicable law or agreed to in writing, software
# # distributed under the License is distributed on an "AS IS" BASIS,
# # WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied.
# # See the License for the specific language governing permissions and
# # limitations under the License.
# # ==============================================================================
# import os
# import cv2
# import torch
# from natsort import natsorted
# os.chdir("/kaggle/input/d/sahilchawla7/srganimplementation/")
# import imgproc
# import model
# import srgan_config
# import image_quality_assessment
# from myUtils import make_directory
# model_names = sorted(
# name for name in model.__dict__ if
# name.islower() and not name.startswith("__") and callable(model.__dict__[name]))
# os.chdir("/kaggle/working")
# srgan_config.mode = "test"
# # srgan_config.train_gt_images_dir = "/kaggle/input/superresolution-datasetpreprocessing/div2k-dataset/SRGAN/train/"
# # srgan_config.train_gt_images_dir = "/kaggle/input/div2ksmall/"
# # srgan_config.test_gt_images_dir = "/kaggle/input/set5dataset/Set5/GTmod12/"
# # CHANGE HERE FOR TESTING
# # srgan_config.lr_dir = f"/kaggle/input/urban100/Urban100/LRbicx{srgan_config.upscale_factor}/"
# # srgan_config.lr_dir = f"/kaggle/input/set5dataset/Set5/LRbicx{srgan_config.upscale_factor}/"
# # srgan_config.lr_dir = f"/kaggle/input/set14dataset/Set14/LRbicx{srgan_config.upscale_factor}/"
# srgan_config.lr_dir = f"/kaggle/input/randomtestimage/face_images_dataset/LRbicx{srgan_config.upscale_factor}/"
# # srgan_config.gt_dir = f"/kaggle/input/urban100/Urban100/GTmod12/"
# # srgan_config.gt_dir = f"/kaggle/input/set5dataset/Set5/GTmod12"
# # srgan_config.gt_dir = f"/kaggle/input/set14dataset/Set14/GTmod12"
# srgan_config.gt_dir = f"/kaggle/input/randomtestimage/face_images_dataset/GTmod12/"
# srgan_config.sr_dir = f"/kaggle/working/test/{srgan_config.exp_name}"
# srgan_config.g_model_weights_path = "/kaggle/input/srganmodels/g_best_gmsd1_mse1_adv001_epoch4.pth.tar"
# srgan_config.epochs = 4
# def main() -> None:
# # Initialize the super-resolution bsrgan_model
# g_model = model.__dict__[srgan_config.g_arch_name](in_channels=srgan_config.in_channels,
# out_channels=srgan_config.out_channels,
# channels=srgan_config.channels,
# num_rcb=srgan_config.num_rcb)
# g_model = g_model.to(device=srgan_config.device)
# print(f"Build `{srgan_config.g_arch_name}` model successfully.")
# # Load the super-resolution bsrgan_model weights
# checkpoint = torch.load(srgan_config.g_model_weights_path, map_location=lambda storage, loc: storage)
# g_model.load_state_dict(checkpoint["state_dict"])
# print(f"Load `{srgan_config.g_arch_name}` model weights "
# f"`{os.path.abspath(srgan_config.g_model_weights_path)}` successfully.")
# # Create a folder of super-resolution experiment results
# make_directory(srgan_config.sr_dir)
# # Start the verification mode of the bsrgan_model.
# g_model.eval()
# # Initialize the sharpness evaluation function
# psnr = image_quality_assessment.PSNR(srgan_config.upscale_factor, srgan_config.only_test_y_channel)
# ssim = image_quality_assessment.SSIM(srgan_config.upscale_factor, srgan_config.only_test_y_channel)
# # Set the sharpness evaluation function calculation device to the specified model
# psnr = psnr.to(device=srgan_config.device, non_blocking=True)
# ssim = ssim.to(device=srgan_config.device, non_blocking=True)
# # Initialize IQA metrics
# psnr_metrics = 0.0
# ssim_metrics = 0.0
# # ------------TEMPORARY TESTING--------------------
# lr_image_file = "/kaggle/input/celeba-dataset/img_align_celeba/img_align_celeba/000033.jpg"
# lr_tensor = imgproc.preprocess_one_image(lr_image_file, srgan_config.device)
# with torch.no_grad():
# sr_tensor = g_model(lr_tensor)
# sr_image = imgproc.tensor_to_image(sr_tensor, False, False)
# sr_image = cv2.cvtColor(sr_image, cv2.COLOR_RGB2BGR)
# sr_image_path = os.path.join(srgan_config.sr_dir, "000033.jpg")
# cv2.imwrite(sr_image_path, sr_image)
# # # ------------TESTING OVER-------------------------
# # # Get a list of test image file names.
# # file_names = natsorted(os.listdir(srgan_config.lr_dir))
# # # Get the number of test image files.
# # total_files = len(file_names)
# # for index in range(total_files):
# # lr_image_path = os.path.join(srgan_config.lr_dir, file_names[index])
# # sr_image_path = os.path.join(srgan_config.sr_dir, file_names[index])
# # gt_image_path = os.path.join(srgan_config.gt_dir, file_names[index])
# # print(f"Processing `{os.path.abspath(lr_image_path)}`...")
# # lr_tensor = imgproc.preprocess_one_image(lr_image_path, srgan_config.device)
# # gt_tensor = imgproc.preprocess_one_image(gt_image_path, srgan_config.device)
# # # Only reconstruct the Y channel image data.
# # with torch.no_grad():
# # sr_tensor = g_model(lr_tensor)
# # # Save image
# # sr_image = imgproc.tensor_to_image(sr_tensor, False, False)
# # sr_image = cv2.cvtColor(sr_image, cv2.COLOR_RGB2BGR)
# # cv2.imwrite(sr_image_path, sr_image)
# # # Cal IQA metrics
# # psnr_metrics += psnr(sr_tensor, gt_tensor).item()
# # ssim_metrics += ssim(sr_tensor, gt_tensor).item()
# # print("PSNR: " + str(psnr(sr_tensor, gt_tensor).item()))
# # print("SSIM: " + str(ssim(sr_tensor, gt_tensor).item()))
# # # Calculate the average value of the sharpness evaluation index,
# # # and all index range values are cut according to the following values
# # # PSNR range value is 0~100
# # # SSIM range value is 0~1
# # avg_psnr = 100 if psnr_metrics / total_files > 100 else psnr_metrics / total_files
# # avg_ssim = 1 if ssim_metrics / total_files > 1 else ssim_metrics / total_files
# # print(f"PSNR: {avg_psnr:4.2f} [dB]\n"
# # f"SSIM: {avg_ssim:4.4f} [u]")
# if __name__ == "__main__":
# main()
# # Dataset Preprocessing
# # Casia
# !pip install opencv-python-headless
import os
import cv2
import shutil
def casia_crop(fn):
return cv2.imread(fn)[29:-29, 29:-29, ::-1]
def resample(im, dsize=192):
ratio = im.shape[0] / dsize
s = 0.25 * ratio
filtered = cv2.GaussianBlur(im, ksize=(0, 0), sigmaX=s, sigmaY=s)
inter = cv2.INTER_CUBIC
lr = cv2.resize(filtered, dsize=(dsize, dsize), interpolation=inter)
return lr
img_dir = "/kaggle/input/casia-webface/casia-webface"
if not os.path.exists("/kaggle/working/casia-webface"):
os.mkdir("/kaggle/working/casia-webface")
if not os.path.exists("/kaggle/working/casia-webface/SRGAN"):
os.mkdir("/kaggle/working/casia-webface/SRGAN")
if not os.path.exists("/kaggle/working/casia-webface/SRGAN/train"):
os.mkdir("/kaggle/working/casia-webface/SRGAN/train")
counter = 0
for folder in os.listdir(img_dir):
for image in os.listdir(os.path.join(img_dir, folder)):
imagePath = os.path.join(img_dir, folder, image)
img = casia_crop(imagePath)
# print(os.path.join("/kaggle/working/casia-webface/SRGAN/train",image))
cv2.imwrite(
os.path.join("/kaggle/working/casia-webface/SRGAN/train", image), img
)
counter = counter + 1
if counter % 100 == 0:
print(counter)
# break
shutil.make_archive("casia-webface-zip", "zip", "/kaggle/working/casia-webface/")
shutil.rmtree("/kaggle/working/casia-webface/")
# # Helen
# import os
# import cv2
# # resampling function to derive the high-res and low-res reference images
# # of 192px and 24px from face crops of arbitrary resoulution
# def resample(im, dsize=192):
# ratio = im.shape[0] / dsize
# s = 0.25 * ratio
# filtered = cv2.GaussianBlur(im, ksize=(0, 0), sigmaX=s, sigmaY=s)
# inter = cv2.INTER_CUBIC
# lr = cv2.resize(filtered, dsize=(dsize, dsize), interpolation=inter)
# return lr
# img_dir = "/kaggle/input/helen-test-set/test"
# if not os.path.exists("/kaggle/working/helen-test-set"):
# os.mkdir("/kaggle/working/helen-test-set")
# if not os.path.exists("/kaggle/working/helen-test-set/LR"):
# os.mkdir("/kaggle/working/helen-test-set/LR")
# if not os.path.exists("/kaggle/working/helen-test-set/HR"):
# os.mkdir("/kaggle/working/helen-test-set/HR")
# counter = 0
# for image in os.listdir(img_dir):
# imagePath = os.path.join(img_dir,image)
# hr = resample(cv2.imread(imagePath), dsize=192)
# lr = resample(hr, dsize=48)
# cv2.imwrite(os.path.join("/kaggle/working/helen-test-set/LR",image), lr[:, :, ::-1])
# cv2.imwrite(os.path.join("/kaggle/working/helen-test-set/HR",image), hr[:, :, ::-1])
# counter = counter + 1
# if (counter%100==0):
# print(counter)
# # cv2.imwrite("celeba_hr.png", hr[:, :, ::-1])
# # cv2.imwrite("celeba_lr.png", lr[:, :, ::-1])
# # img_dir = "/opt/data/Helen/testset"
# # img_fns = [fn for fn in os.listdir(img_dir) if not fn.endswith(".pts")]
# # img_fn = os.path.join(img_dir, np.random.choice(img_fns))
# # img = helen_crop(img_fn)
# # hr = resample(img, dsize=192)
# # lr = resample(hr, dsize=24)
# # cv2.imwrite("helen_hr.png", hr[:, :, ::-1])
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
train = pd.read_csv("/kaggle/input/spaceship-titanic/train.csv")
test = pd.read_csv("/kaggle/input/spaceship-titanic/test.csv")
train.head()
test.head()
from pandas_profiling import ProfileReport
profile = ProfileReport(train)
profile.to_notebook_iframe()
train = train.drop(["Name", "Cabin", "FoodCourt", "ShoppingMall"], axis=1)
categorical = train[["PassengerId", "HomePlanet", "Destination"]]
numerical = train[["PassengerId", "Age", "RoomService", "Spa", "VRDeck"]]
boolean = train[["PassengerId", "CryoSleep", "VIP", "Transported"]]
# ## Categorical
categorical.head()
# variables HomePlanet et Destination
print(categorical["Destination"].isnull().sum())
print(categorical["HomePlanet"].isnull().sum())
# remplacement des lignes avec des catégories nulles
categorical[["Destination", "HomePlanet"]] = categorical[
["Destination", "HomePlanet"]
].fillna(categorical.mode().loc[0])
categorical["HomePlanet"].value_counts()
categorical["Destination"].value_counts()
from sklearn.pipeline import Pipeline
from sklearn.impute import SimpleImputer
from sklearn.preprocessing import OneHotEncoder
cat_pipeline = Pipeline([("one_hot", OneHotEncoder())])
cat_tr = cat_pipeline.fit_transform(categorical.drop("PassengerId", axis=1))
cat_feature_names = cat_pipeline.named_steps["one_hot"].get_feature_names_out(
["HomePlanet", "Destination"]
)
cat_tr_df = pd.DataFrame(cat_tr.toarray(), columns=cat_feature_names)
print(cat_tr_df.columns)
# ## BOOLEAN
# Remplacement des lignes avec des catégories nulles
boolean[["CryoSleep", "VIP"]] = boolean[["CryoSleep", "VIP"]].fillna(
boolean.mode().loc[0]
)
boolean["CryoSleep_True"] = boolean["CryoSleep"].astype(int)
boolean["CryoSleep_False"] = (1 - boolean["CryoSleep"]).astype(int)
boolean = boolean.drop("CryoSleep", axis=1)
boolean["VIP_True"] = boolean["VIP"].astype(int)
boolean["VIP_False"] = (1 - boolean["VIP"]).astype(int)
boolean = boolean.drop("VIP", axis=1)
# ## Numerical
from sklearn.preprocessing import StandardScaler
num_pipeline = Pipeline(
[
("inputer", SimpleImputer(strategy="median")),
("std_scaler", StandardScaler()),
]
)
numerical = pd.DataFrame(
num_pipeline.fit_transform(numerical), columns=numerical.columns
)
# ## Fusion des datas
df = pd.concat(
[
categorical["PassengerId"],
pd.DataFrame(cat_tr.toarray()),
numerical.iloc[:, 1:],
boolean.iloc[:, 1:],
],
axis=1,
)
df.head()
df = df.dropna(subset=["Transported"])
df = df.dropna(axis=0)
X = df.drop("Transported", axis=1)
y = df["Transported"]
X = X.drop("PassengerId", axis=1)
y.head()
y = y.astype(int)
y.head()
X.info()
X.rename(columns={0: "HomePlanet_Earth"}, inplace=True)
X.rename(columns={1: "HomePlanet_Europa"}, inplace=True)
X.rename(columns={2: "HomePlanet_Mars"}, inplace=True)
X.rename(columns={3: "Destination_55 Cancri e"}, inplace=True)
X.rename(columns={4: "Destination_PSO J318.5-22"}, inplace=True)
X.rename(columns={5: "Destination_TRAPPIST-1e"}, inplace=True)
print(X.isna().sum())
# ## Classifier
from sklearn.model_selection import cross_val_score
from sklearn.ensemble import RandomForestClassifier, GradientBoostingClassifier
from sklearn.tree import DecisionTreeClassifier
from xgboost import XGBClassifier
from sklearn.svm import SVC
from sklearn.metrics import accuracy_score
from sklearn.utils import shuffle
classifiers = [
("Random Forest", RandomForestClassifier(n_estimators=100, random_state=42)),
("Decision Tree", DecisionTreeClassifier(random_state=42)),
("Xgboost", XGBClassifier(use_label_encoder=False, random_state=42)),
("SVM", SVC(kernel="poly", random_state=42)),
]
for name, classifier in classifiers:
clf = classifier
cv_scores = cross_val_score(clf, X, y, cv=10, scoring="accuracy")
print(f"Average cross validation score for {name}:", np.mean(cv_scores))
print(f"Std cross_validation score for {name}", np.std(cv_scores))
print("-" * 20)
# ## ESAYONS UNE REDUCTION DE LA DIMENSION
from sklearn.decomposition import PCA
import matplotlib.pyplot as plt
pc = PCA(n_components=10)
pc.fit(X)
plt.figure(figsize=(10, 5))
plt.title("Principal Component Analysis")
plt.plot(pc.explained_variance_ratio_)
plt.legend("Explained Variance")
plt.xlabel("N components")
plt.ylabel("Explained Variance ratio")
plt.show()
X_new = pc.transform(X)
classifiers = [
("Random Forest", RandomForestClassifier(n_estimators=100, random_state=42)),
("Decision Tree", DecisionTreeClassifier(random_state=42)),
("Xgboost", XGBClassifier(use_label_encoder=False, random_state=42)),
("SVM", SVC(kernel="poly", random_state=42)),
]
for name, classifier in classifiers:
clf = classifier
cv_scores = cross_val_score(clf, X, y, cv=10, scoring="accuracy")
print(f"Average cross validation score for {name}:", np.mean(cv_scores))
print(f"Std cross_validation score for {name}", np.std(cv_scores))
print("-" * 20)
# ## MAKE PREDICTIONS
profile = ProfileReport(test)
profile.to_notebook_iframe()
test = test.drop(["Name", "Cabin", "FoodCourt", "ShoppingMall"], axis=1)
categorical_test = test[["PassengerId", "HomePlanet", "Destination"]]
numerical_test = test[["PassengerId", "Age", "RoomService", "Spa", "VRDeck"]]
boolean_test = test[["PassengerId", "CryoSleep", "VIP"]]
# categorical
categorical_test[["Destination", "HomePlanet"]] = categorical_test[
["Destination", "HomePlanet"]
].fillna(categorical.mode().loc[0])
cat_pipeline_test = Pipeline([("one_hot", OneHotEncoder())])
cat_test = cat_pipeline_test.fit_transform(categorical_test.drop("PassengerId", axis=1))
cat_feature_names_test = cat_pipeline_test.named_steps["one_hot"].get_feature_names_out(
["HomePlanet", "Destination"]
)
cat_test_df = pd.DataFrame(cat_test.toarray(), columns=cat_feature_names_test)
# Boolean
boolean_test[["CryoSleep", "VIP"]] = boolean_test[["CryoSleep", "VIP"]].fillna(
boolean_test.mode().loc[0]
)
boolean_test["CryoSleep_True"] = boolean_test["CryoSleep"].astype(int)
boolean_test["CryoSleep_False"] = (1 - boolean_test["CryoSleep"]).astype(int)
boolean_test = boolean_test.drop("CryoSleep", axis=1)
boolean_test["VIP_True"] = boolean_test["VIP"].astype(int)
boolean_test["VIP_False"] = (1 - boolean_test["VIP"]).astype(int)
boolean_test = boolean_test.drop("VIP", axis=1)
# numerical
num_pipeline_test = Pipeline(
[
("inputer", SimpleImputer(strategy="median")),
("std_scaler", StandardScaler()),
]
)
numerical_test = pd.DataFrame(
num_pipeline_test.fit_transform(numerical_test), columns=numerical_test.columns
)
# fusion
df_test = pd.concat(
[
categorical_test["PassengerId"],
pd.DataFrame(cat_test.toarray()),
numerical_test.iloc[:, 1:],
boolean_test.iloc[:, 1:],
],
axis=1,
)
df_test = df_test.dropna(axis=0)
df_test.rename(columns={0: "HomePlanet_Earth"}, inplace=True)
df_test.rename(columns={1: "HomePlanet_Europa"}, inplace=True)
df_test.rename(columns={2: "HomePlanet_Mars"}, inplace=True)
df_test.rename(columns={3: "Destination_55 Cancri e"}, inplace=True)
df_test.rename(columns={4: "Destination_PSO J318.5-22"}, inplace=True)
df_test.rename(columns={5: "Destination_TRAPPIST-1e"}, inplace=True)
pc_test = PCA(n_components=10)
pc.fit(df_test)
df_test = pc.transform(df_test)
# créer un modèle XGBoost
model = XGBClassifier()
model.fit(X_new, y)
# faire des prédictions sur les données de test
y_pred = model.predict(df_test)
# ## Submission
submission = pd.read_csv("/kaggle/input/spaceship-titanic/sample_submission.csv")
submission.head()
submission.shape
n_predictions = (y_pred > 0.5).astype(bool)
output = pd.DataFrame(
{"PassengerId": submission_id, "Transported": n_predictions.squeeze()}
)
output.head()
sample_submission_df = pd.read_csv(
"/kaggle/input/spaceship-titanic/sample_submission.csv"
)
sample_submission_df["Transported"] = n_predictions
sample_submission_df.to_csv("/kaggle/working/", index=False)
sample_submission_df.head()
|
# # PREDICTION OF SURGERY DURATION
# ## Introduction
# The goal of this competition is to predict the duration of a surgery. A regression model trained on labeled data will be extracted by using Borda Technology's IoT sensor and Borda Technology's database.
# ## Outline
# 1. Step 0 - Preparing data
# - 1.1 - Importing Libraries & Dataset
# - 1.2 - Assesing Data
# - 1.3 - Quality Issues
# - 1.4 - Tidiness Issues
# - 1.5 - Cleaning Data (issue-define-code-test)
# 2. Step 1 -
# 3. Step 2 -
# 4. Step 3 -
# 5. Step 4 -
# ## Step 0
# ### 1.1 Importing Libraries & Dataset
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import seaborn as sns
import matplotlib.pyplot as plt
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Read the csv file
# Note: Dataset do not have utf-8 encoding, so I encoded it as latin-1
df_train = pd.read_csv(
"/kaggle/input/prediction-of-surgery-duration/train.csv", encoding="latin-1"
)
df_train.head()
df_test = pd.read_csv(
"/kaggle/input/prediction-of-surgery-duration/test.csv", encoding="latin-1"
)
df_test.head()
# ### 1.2 Assesing Data
df_train.info()
# See null values
df_train.isna().sum()
# 221 null values in both of DiagnosticICD10Code and AnesthesiaType.
# I wondered whether these two exist null together in the dataset
# Let's check
df_train[df_train["DiagnosticICD10Code"].isna()]
# Yes, if a row contains null DiagnosticICD10Code then its AnesthesiaType is also null and vice versa
# To be sure, check again
df_train[df_train["DiagnosticICD10Code"].isna() & df_train["AnesthesiaType"].notnull()]
df_train[df_train["DiagnosticICD10Code"].notnull() & df_train["AnesthesiaType"].isna()]
# Check whether there exist empty sets {} under DiagnosticICD10Code column
df_train[df_train["DiagnosticICD10Code"] == "{}"]
# Check whether there exist empty sets {} under AnesthesiaType column
df_train[df_train["AnesthesiaType"] == "{}"]
# See how many types of Service exist
df_train["Service"].value_counts()
# See how many types of AnesthesiaType exist
df_train["AnesthesiaType"].value_counts()
# Also see the DiagnosticICD10Code frequencies
# meaning of DiagnosticICD10Code: https://www.icd10data.com/
df_train["DiagnosticICD10Code"].value_counts()
# Also see the SurgeryGroup frequencies
df_train["SurgeryGroup"].value_counts()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
oil_df = pd.read_csv("/kaggle/input/store-sales-time-series-forecasting/oil.csv")
sample_df = pd.read_csv(
"/kaggle/input/store-sales-time-series-forecasting/sample_submission.csv"
)
holidays_df = pd.read_csv(
"/kaggle/input/store-sales-time-series-forecasting/holidays_events.csv"
)
stores_df = pd.read_csv("/kaggle/input/store-sales-time-series-forecasting/stores.csv")
train_df = pd.read_csv("/kaggle/input/store-sales-time-series-forecasting/train.csv")
test_df = pd.read_csv("/kaggle/input/store-sales-time-series-forecasting/test.csv")
transactions_df = pd.read_csv(
"/kaggle/input/store-sales-time-series-forecasting/transactions.csv"
)
oil_df.info()
import matplotlib.pyplot as plt
plt.plot(oil_df.index, oil_df.dcoilwtico)
plt.show()
# Convert date column to datetime and set it as the index
oil_df["date"] = pd.to_datetime(oil_df["date"], format="%Y-%m-%d")
oil_df.set_index("date", inplace=True)
# Resample to daily frequency and interpolate missing values
oil_df = oil_df.resample("D").interpolate(method="time")
# Fill any remaining missing values with nearest data point
oil_df = oil_df.interpolate(method="nearest")
# Reset the index to a column
oil_df.reset_index(inplace=True)
holidays_df.info()
holidays_df = holidays_df[holidays_df.transferred == 0]
holidays_df.loc[holidays_df.type == "Transfer", "description"] = holidays_df.loc[
holidays_df.type == "Transfer", "description"
].str.replace("Traslado ", "")
type_map = {"Transfer": "Holiday"}
holidays_df["type"] = holidays_df["type"].map(type_map).fillna(holidays_df["type"])
stores_df.info()
transactions_df.info()
# import seaborn as sns
# g = sns.FacetGrid(transactions_df, col="store_nbr", col_wrap=4, height=2.5, aspect=1.5)
# g.map(plt.plot, "date", "transactions")
# plt.show()
train_df.info(show_counts=True)
test_df.info(show_counts=True)
train_missing_pct = train_df.isnull().mean() * 100
test_missing_pct = test_df.isnull().mean() * 100
print("Train DataFrame Missing Values (%)")
print(train_missing_pct[train_missing_pct > 0])
print("\nTest DataFrame Missing Values (%)")
print(test_missing_pct[test_missing_pct > 0])
data = pd.concat([train_df, test_df])
display(data)
data["date"] = pd.to_datetime(data["date"], format="%Y-%m-%d")
data.set_index("date", inplace=True)
data = data.join(stores_df.set_index("store_nbr"), on="store_nbr", how="left")
display(data)
data.columns
data["holiday"] = 0
for i in holidays_df.values:
if i[2] == "Holiday":
data.loc[data.index == i[0], "holiday"] = 1
from sklearn.preprocessing import OneHotEncoder
low_card_cols = ["type", "family", "city", "state"]
low_card_values = data.type.values
low_card_enc = OneHotEncoder(handle_unknown="ignore", sparse=False)
low_card_df = pd.DataFrame(low_card_enc.fit_transform(data[low_card_cols]))
low_card_df.index = data.index
data = pd.concat([data.drop(low_card_cols, axis=1), low_card_df], axis=1)
data = data.astype("float64")
display(data)
from sklearn.model_selection import train_test_split
from sklearn.tree import DecisionTreeRegressor
submission_df = data[data.sales.isna()]
train_test = data[data.sales.isna() == 0]
X = train_test.drop(["sales"], axis=1)
y = train_test[["sales"]]
X = X.dropna()
X.columns = X.columns.astype(str)
X.info(verbose=True, show_counts=True)
X_train, X_test, y_train, y_test = train_test_split(X, y)
model = DecisionTreeRegressor()
model.fit(X_train.drop("id", axis=1), y_train)
print(model.score(X_test.drop("id", axis=1), y_test))
submission_df.columns = submission_df.columns.astype(str)
result = model.predict(submission_df.drop(["id", "sales"], axis=1))
result_df = pd.DataFrame(result, index=submission_df.id, columns=["sales"])
result_df.index = result_df.index.astype(int)
result_df.to_csv("result.csv", index=True)
display(submission_df)
display(result_df)
|
import pandas as pd
import seaborn as sns
import plotly.express as pl
import matplotlib.pyplot as plt
import warnings
from sklearn.ensemble import ExtraTreesClassifier # Decison tree
from sklearn.feature_selection import SelectKBest, f_classif, chi2, f_regression
from sklearn.decomposition import PCA
from sklearn.preprocessing import MinMaxScaler
from sklearn.model_selection import train_test_split as tts
from sklearn.linear_model import LogisticRegression
warnings.filterwarnings("ignore")
df = pd.read_csv("/kaggle/input/credit-card-dataset/credit card.csv")
df.head()
df.shape
df.isnull().sum()
df.nunique()
df.info()
df.rename(columns={"default.payment.next.month": "payment"}, inplace=True)
df.head()
x = df.drop(["ID", "payment"], axis=1)
y = df[["payment"]]
y.value_counts()
23364 / (23364 + 6636)
# # Extra Trees Classifier
extra = ExtraTreesClassifier()
extra.fit(x, y)
extra.feature_importances_ # IG from decision tree
feature_importance = extra.feature_importances_
feature_importance # inpo gain
imp = pd.DataFrame(feature_importance, columns=["Gain_Score"])
imp.head()
x.columns
cols = pd.DataFrame(x.columns, columns=["Feature_Names"])
cols.head()
gains = pd.concat([cols, imp], axis=1)
gains.head()
gains.nlargest(15, "Gain_Score") # smallest()
gains.nsmallest(8, "Gain_Score")
newx = gains.nlargest(18, "Gain_Score")
newx.head()
newx.plot(kind="bar", color="red")
x.shape
features = pd.Series(extra.feature_importances_, index=x.columns)
plt.figure(figsize=(10, 6))
features.nlargest(20).plot(kind="bar", color="blue")
plt.savefig("score1.png")
features = pd.Series(extra.feature_importances_, index=x.columns)
plt.figure(figsize=(10, 6))
features.nlargest(20).plot(kind="barh", color="blue")
plt.savefig("score1.png")
# x = newx
x.corr()
x.describe()
x.columns
feature_names = x.columns
colname = x.corr().index
plt.figure(figsize=(25, 25))
sns.heatmap(df[colname].corr(), annot=True)
# # Select K Best
df = pd.read_csv("/kaggle/input/credit-card-dataset/credit card.csv")
df = df.rename(columns={"default.payment.next.month": "payment"})
x = df.drop(["ID", "payment"], axis=1)
y = df["payment"]
x.head()
model2 = SelectKBest(score_func=f_classif)
feature_score = model2.fit(x, y)
feature_score.scores_
cols = pd.DataFrame(feature_score.scores_, columns=["Feature_Scores"])
cols.head()
x.columns
col2 = pd.DataFrame(x.columns, columns=["Feature_Names"])
col2.head()
scores = pd.concat([col2, cols], axis=1)
scores
scores.nlargest(15, "Feature_Scores")
df3 = df.drop(
["LIMIT_BAL", "EDUCATION", "PAY_0"],
axis=1,
)
df3.to_csv("new.csv")
df
scores = pd.concat([col2, cols], axis=1)
scores
newx = scores.nlargest(15, "Feature_Scores")
newx
# # PCA
df = pd.read_csv("/kaggle/input/credit-card-dataset/credit card.csv")
df = df.rename(columns={"default.payment.next.month": "payment"})
x = df.drop(["ID", "payment"], axis=1)
y = df["payment"]
x
# # FEATURE SCALING
mx = MinMaxScaler() # feature_range=(2,5)
scaled_x = mx.fit_transform(x)
scaled_x.shape
scaled_x
pca = PCA(n_components=3)
x_pca = pca.fit_transform(scaled_x)
x_pca
features = pd.DataFrame(x_pca, columns=["pca1", "pca2", "pca3"])
features
pl.scatter_3d(features, x="pca1", y="pca2", z="pca3", color="pca1")
# # train_test_split
xtrain, xtest, ytrain, ytest = tts(features, y, test_size=0.30, random_state=1)
xtrain
# # Logistic Regression
lg = LogisticRegression()
lg.fit(xtrain, ytrain)
lg.score(xtest, ytest) # testiong
lg.score(xtrain, ytrain) # training
y.value_counts()
x.head()
# # Before
xtrain1, xtest2, ytrain1, ytest1 = tts(x, y, test_size=0.30, random_state=1)
xtrain1.head()
lg.fit(xtrain1, ytrain1)
lg.score(xtest2, ytest1)
lg.score(xtrain1, ytrain)
|
# # Motivation
# Movies have been an integral part of the modern day living. Over the years in past 4 decades, movie making has become $140 billion dollar industry worldwide.
# On the adjacent path, Internet technology advancements has enabled viewers to rate the movies in increasing numbers. Such ratings data collected over the years can reveal critical key factors that particular audience did or did not like , thus allowing broadcasters and movie makers to deploy such findings in restructuring their resources to aim better success rate of the movie they are working on.
# # Objective of this exploratory anlaysis
# Current analysis is aiming towards examining the available movies, ratings and tags data in integrated manner. Following are key objectives for this analysis.
# **Objective 1 : Trends analysis of the following sub-items**
# * Identify number of movies made for each genre
# * Number of movies released over the years
# * Change in movie quality (aggregate ratings) over the year
#
# **Objective 2 : Find the characteristics (keywords / tags) that are common among the most popular movies,**
# **Objective 3 : Identify the genres that attracted most users to rate the movies.**
# # Dataset used for analysis : movielens
# Input data files are available in the read-only "../input/" directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# **Data Source :**
#
# * Source Description: MovieLens web site (filename: ml-25m.zip)
# * Location: https://grouplens.org/datasets/movielens/
# **Available Data Entities :**
# * movies.csv
# * ratings.cs
# * tags.csv
# Preparing basic imports and reusable methods
# !pip install cufflinks
# !pip install plotly
# !pip install textblob
import pandas as pd
from matplotlib import pyplot as plt
import numpy as np
import seaborn as sns
import plotly as py
def screen_data_frame(df):
"""
funciton to print the common characteristics of the input data frame.
"""
print(f"No. of rows : {df.shape[0]} \nNo. of Columns : {df.shape[1]}")
print(f"Columns : {df.columns.to_list()}")
print("-" * 40, f"\nNull value count : \n{df.isnull().sum()}")
print("-" * 40, f"\nUnique value count : \n{df.nunique()}\n", "-" * 40)
movies = pd.read_csv("../input/movielens-dataset/movies.csv")
movies.sample(3)
screen_data_frame(movies)
# Movies data is clean, there are no missing values.
# Adding Year as new column in the data set
def strp_parenthesis(x):
"""
Function to clean up extra parenthesis at the beginning and end of the string.
"""
return x.str.strip("()")
movies["year"] = movies["title"].str.extract("(\(\d{4}\))").apply(strp_parenthesis)
# converting year string to datetime 'year' values
movies["year"] = pd.to_datetime(movies["year"], format="%Y")
movies.head()
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
# Load the dataset.
df = pd.read_csv("/kaggle/input/wholesale-customers-data/Wholesale customers data.csv")
X = df.iloc[:, 2:]
df.head()
df.describe()
df.info()
# Checking for null values.
df.isnull().sum()
# Removing any potential outliers.
from scipy import stats
z_scores = stats.zscore(df)
abs_z_scores = np.abs(z_scores)
filtered_entries = (abs_z_scores < 3).all(axis=1)
df = df[filtered_entries]
# Scale the features using standard scaler.
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
scaled_df = scaler.fit_transform(df)
scaled_df = pd.DataFrame(scaled_df, columns=df.columns)
# # Task A
# 1- Apply the K-Mean clustering method with Euclidean distance matrix, cluster count: 2
# 2- Display the properties of each cluster (# observations, homogeneity, heterogeneity, etc
# 3- Visualize the clusters
# Apply the K-Mean clustering method with Euclidean distance matrix, cluster count: 2
from sklearn.cluster import KMeans
kmeans = KMeans(n_clusters=2, random_state=0).fit(scaled_df)
cluster_labels = kmeans.labels_
silhouette_avg = silhouette_score(scaled_df, cluster_labels)
print(
"For n_clusters =",
n_clusters,
"and distance =",
dist,
", the average silhouette_score is :",
silhouette_avg,
)
# Display the properties of each cluster:
cluster_labels = kmeans.labels_
scaled_df["Cluster"] = cluster_labels
grouped = scaled_df.groupby("Cluster")
grouped.mean()
# Visualize the clusters:
plt.scatter(
scaled_df.iloc[:, 0], scaled_df.iloc[:, 1], c=kmeans.labels_, cmap="rainbow"
)
plt.scatter(kmeans.cluster_centers_[:, 0], kmeans.cluster_centers_[:, 1], color="black")
# calculate and display cluster properties
cluster_props = []
for i in range(2):
obs_count = len(np.where(kmeans.labels_ == i)[0])
homogeneity = np.mean(
data.iloc[kmeans.labels_ == i, 0].values
== data.iloc[kmeans.labels_ == i, 0].mean()
)
heterogeneity = np.mean(
data.iloc[kmeans.labels_ != i, 0].values
== data.iloc[kmeans.labels_ == i, 0].mean()
)
cluster_props.append(
{
"Cluster": i,
"Observation Count": obs_count,
"Homogeneity": homogeneity,
"Heterogeneity": heterogeneity,
}
)
cluster_props_df = pd.DataFrame(cluster_props)
print(cluster_props_df)
# # Task B
# Apply K-Means clustering with various distance matrices and cluster counts:
for dist in distances:
for n_clusters in num_clusters:
kmeans = KMeans(n_clusters=n_clusters, random_state=0).fit(scaled_df)
cluster_labels = kmeans.labels_
silhouette_avg = silhouette_score(scaled_df, cluster_labels)
print(
"For n_clusters =",
n_clusters,
"and distance =",
dist,
", the average silhouette_score is :",
silhouette_avg,
)
# This will apply K-Means clustering with various distance matrices and cluster counts, and compute the silhouette score for each combination. The silhouette score is a metric that measures how similar an object is to its own cluster compared to other clusters, and ranges from -1 to 1, with higher values indicating better cluster quality.
# Visualize the clusters:
kmeans = KMeans(n_clusters=2, random_state=0).fit(scaled_df)
plt.scatter(
scaled_df.iloc[:, 0], scaled_df.iloc[:, 1], c=kmeans.labels_, cmap="rainbow"
)
plt.scatter(kmeans.cluster_centers_[:, 0], kmeans.cluster_centers_[:, 1], color="black")
plt.title("K-Means Clustering (2 clusters)")
plt.show()
# Number of observations in each cluster
n_obs = []
for i in range(kmeans.n_clusters):
cluster_indices = np.where(kmeans.labels_ == i)[0]
n_obs.append(len(cluster_indices))
# Homogeneity of each cluster
homogeneity = []
for i in range(kmeans.n_clusters):
cluster_indices = np.where(kmeans.labels_ == i)[0]
cluster_data = scaled_df.iloc[cluster_indices, :]
homogeneity.append(
np.mean(np.sum((cluster_data - kmeans.cluster_centers_[i]) ** 2, axis=1))
)
# Heterogeneity of each cluster
heterogeneity = []
for i in range(kmeans.n_clusters):
other_cluster_indices = np.where(kmeans.labels_ != i)[0]
other_cluster_data = scaled_df.iloc[other_cluster_indices, :]
heterogeneity.append(
np.mean(np.sum((other_cluster_data - kmeans.cluster_centers_[i]) ** 2, axis=1))
)
# Display cluster properties
for i in range(kmeans.n_clusters):
print(
"Cluster {}: # Observations={}, Homogeneity={}, Heterogeneity={}".format(
i, n_obs[i], homogeneity[i], heterogeneity[i]
)
)
kmeans = KMeans(n_clusters=3, random_state=0).fit(scaled_df)
plt.scatter(
scaled_df.iloc[:, 0], scaled_df.iloc[:, 1], c=kmeans.labels_, cmap="rainbow"
)
plt.scatter(kmeans.cluster_centers_[:, 0], kmeans.cluster_centers_[:, 1], color="black")
plt.title("K-Means Clustering (3 clusters)")
plt.show()
# Number of observations in each cluster
n_obs = []
for i in range(kmeans.n_clusters):
cluster_indices = np.where(kmeans.labels_ == i)[0]
n_obs.append(len(cluster_indices))
# Homogeneity of each cluster
homogeneity = []
for i in range(kmeans.n_clusters):
cluster_indices = np.where(kmeans.labels_ == i)[0]
cluster_data = scaled_df.iloc[cluster_indices, :]
homogeneity.append(
np.mean(np.sum((cluster_data - kmeans.cluster_centers_[i]) ** 2, axis=1))
)
# Heterogeneity of each cluster
heterogeneity = []
for i in range(kmeans.n_clusters):
other_cluster_indices = np.where(kmeans.labels_ != i)[0]
other_cluster_data = scaled_df.iloc[other_cluster_indices, :]
heterogeneity.append(
np.mean(np.sum((other_cluster_data - kmeans.cluster_centers_[i]) ** 2, axis=1))
)
# Display cluster properties
for i in range(kmeans.n_clusters):
print(
"Cluster {}: # Observations={}, Homogeneity={}, Heterogeneity={}".format(
i, n_obs[i], homogeneity[i], heterogeneity[i]
)
)
kmeans = KMeans(n_clusters=4, random_state=0).fit(scaled_df)
plt.scatter(
scaled_df.iloc[:, 0], scaled_df.iloc[:, 1], c=kmeans.labels_, cmap="rainbow"
)
plt.scatter(kmeans.cluster_centers_[:, 0], kmeans.cluster_centers_[:, 1], color="black")
plt.title("K-Means Clustering (4 clusters)")
plt.show()
# Number of observations in each cluster
n_obs = []
for i in range(kmeans.n_clusters):
cluster_indices = np.where(kmeans.labels_ == i)[0]
n_obs.append(len(cluster_indices))
# Homogeneity of each cluster
homogeneity = []
for i in range(kmeans.n_clusters):
cluster_indices = np.where(kmeans.labels_ == i)[0]
cluster_data = scaled_df.iloc[cluster_indices, :]
homogeneity.append(
np.mean(np.sum((cluster_data - kmeans.cluster_centers_[i]) ** 2, axis=1))
)
# Heterogeneity of each cluster
heterogeneity = []
for i in range(kmeans.n_clusters):
other_cluster_indices = np.where(kmeans.labels_ != i)[0]
other_cluster_data = scaled_df.iloc[other_cluster_indices, :]
heterogeneity.append(
np.mean(np.sum((other_cluster_data - kmeans.cluster_centers_[i]) ** 2, axis=1))
)
# Display cluster properties
for i in range(kmeans.n_clusters):
print(
"Cluster {}: # Observations={}, Homogeneity={}, Heterogeneity={}".format(
i, n_obs[i], homogeneity[i], heterogeneity[i]
)
)
kmeans = KMeans(n_clusters=5, random_state=0).fit(scaled_df)
plt.scatter(
scaled_df.iloc[:, 0], scaled_df.iloc[:, 1], c=kmeans.labels_, cmap="rainbow"
)
plt.scatter(kmeans.cluster_centers_[:, 0], kmeans.cluster_centers_[:, 1], color="black")
plt.title("K-Means Clustering (5 clusters)")
plt.show()
# Number of observations in each cluster
n_obs = []
for i in range(kmeans.n_clusters):
cluster_indices = np.where(kmeans.labels_ == i)[0]
n_obs.append(len(cluster_indices))
# Homogeneity of each cluster
homogeneity = []
for i in range(kmeans.n_clusters):
cluster_indices = np.where(kmeans.labels_ == i)[0]
cluster_data = scaled_df.iloc[cluster_indices, :]
homogeneity.append(
np.mean(np.sum((cluster_data - kmeans.cluster_centers_[i]) ** 2, axis=1))
)
# Heterogeneity of each cluster
heterogeneity = []
for i in range(kmeans.n_clusters):
other_cluster_indices = np.where(kmeans.labels_ != i)[0]
other_cluster_data = scaled_df.iloc[other_cluster_indices, :]
heterogeneity.append(
np.mean(np.sum((other_cluster_data - kmeans.cluster_centers_[i]) ** 2, axis=1))
)
# Display cluster properties
for i in range(kmeans.n_clusters):
print(
"Cluster {}: # Observations={}, Homogeneity={}, Heterogeneity={}".format(
i, n_obs[i], homogeneity[i], heterogeneity[i]
)
)
# # Task C
# Apply the elbow method with 3-4 different distance matrices. Summarize the results in tabular format with
# graph visualization
# Drop the Channel and Region columns, since we're only interested in clustering based on the customer spending habits
data = df.drop(["Channel", "Region"], axis=1)
# Apply scaling to the data
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
scaled_data = scaler.fit_transform(data)
# Define a function to perform K-means clustering with a given number of clusters and distance metric
def kmeans_clustering(k, distance_metric):
model = KMeans(
n_clusters=k,
init="k-means++",
n_init=10,
max_iter=300,
random_state=0,
algorithm="auto",
)
model.fit(scaled_data)
silhouette_avg = silhouette_score(scaled_data, model.labels_)
return silhouette_avg
# Calculate the silhouette scores for different values of k and distance metrics
distance_metrics = ["euclidean", "manhattan", "chebyshev", "cosine"]
ks = range(2, 11)
silhouette_scores = {}
for metric in distance_metrics:
scores = []
for k in ks:
score = kmeans_clustering(k, metric)
scores.append(score)
silhouette_scores[metric] = scores
# Plot the silhouette scores for different values of k and distance metrics
plt.figure(figsize=(10, 8))
for metric, scores in silhouette_scores.items():
plt.plot(ks, scores, label=metric)
plt.title("Silhouette Scores for Different Distance Metrics")
plt.xlabel("Number of Clusters (k)")
plt.ylabel("Silhouette Score")
plt.legend()
plt.show()
# Display the silhouette scores in tabular format
silhouette_df = pd.DataFrame(silhouette_scores, index=ks)
print(silhouette_df)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import seaborn as sns
import matplotlib.pyplot as plt
import warnings
warnings.filterwarnings("ignore")
pd.set_option("display.max_columns", None)
df = pd.read_csv("/kaggle/input/zomato-eda/zomato.csv")
df.head()
# # **Deleting redundant columns & Repeating rows**
# * In Raw datasets there will be many unneccessary Columns and Repeating rows which we need to drop before procceding further.
# * In this dataset of zomato listed stores we need to delete the following columns
# 1. url
# 2. address
# 3. phone
# 4. dish_liked
# 5. reviews_list
# 6. menu_item
# 7. listed_in(city)
df1 = df.drop(
columns=[
"url",
"address",
"phone",
"dish_liked",
"reviews_list",
"menu_item",
"listed_in(city)",
]
)
df1.head()
# * lets drop Duplicate rows
df1["name"].drop_duplicates(inplace=True)
df1.shape
# * Renaming the columns
df1 = df1.rename(
columns={"approx_cost(for two people)": "cost2plate", "listed_in(type)": "type"}
)
df1
# # **Dealing with NaN values and Individual columns**
df1.isna().sum()
# # 1.Rate
# rate column is object right now containing strings.
# it contains,
# 1. '4.3/5'
# 2. 'NEW','-','nan'
# **goal**
# - deal with unwanted strings and Nan values -- convert 'new'and'-' into 'nan' and replace it with Mean values of rate for coresponding location and type
# - to convert data into float -- by spliting the values from '/'
df1["rate"].unique()
def handle_rate(value):
if value == "NEW" or value == "-":
return np.nan
else:
value = str(value).split("/")
value = value[0]
return float(value)
df1["rate"] = df1["rate"].apply(handle_rate)
df1["rate"].unique()
mask = df1[df1["rate"] != 0]
mask = mask[["rate", "location", "type"]]
grouped = mask.groupby(["type", "location"])["rate"].mean().round(1)
mask1 = df1["rate"] == 0
df1.loc[mask1, "rate"] = df1.loc[mask1].apply(
lambda row: grouped[row["type"], row["location"]], axis=1
)
df1[df1["rate"] == 0]
# df1['rate'].fillna(round(df1['rate'].mean(),1),inplace=True)
# df1['rate'].isna().sum()
# # 2. location
# * To deal with the NaN values of Location, we will retrieve the data from the preceviely droped column 'listed_in(city)' and will replace the na with that corresponding strings.
mask = df[df1["location"].isna()]
mask = mask[["location", "listed_in(city)", "name"]]
mask
mask.loc[mask["location"].isna(), "location"] = mask.loc[
mask["location"].isna(), "listed_in(city)"
]
mask
city_dict = mask.set_index("name")["listed_in(city)"].to_dict()
df1.loc[df1["location"].isna(), "location"] = (
df1.loc[df1["location"].isna(), "name"]
.map(city_dict)
.fillna(df1.loc[df1["location"].isna(), "location"])
)
df1["location"].isna().sum()
# * we have a large no of location available, with very less amount of stores at someof that places. To deal with it we can rename some locations as others.
mask = df["location"].value_counts()
def handle_loc(value):
if value in mask[mask < 300]:
return "Others"
else:
return value
df["location"] = df["location"].apply(handle_loc)
df["location"].value_counts()
# # 3. approx_cost(for two people)
mask = df1[df1["cost2plate"].isna()]
mask[["cost2plate", "location", "type"]]
df1["cost2plate"].unique()
def handle_c2p(value):
value = str(value)
if value == "nan":
return float(0)
elif "," in value:
value = value.replace(",", "")
return float(value)
else:
return float(value)
df1["cost2plate"] = df1["cost2plate"].apply(handle_c2p)
df1["cost2plate"].unique()
df1.loc[(df1["cost2plate"] != 0), "cost2plate"].describe()
mask = df1[df1["cost2plate"] != 0]
mask = mask[["cost2plate", "location", "type"]]
mask
df1["type"].isna().sum()
grouped = mask.groupby(["type", "location"])["cost2plate"].mean().round(0)
grouped
mask1 = df1["cost2plate"] == 0
df1.loc[mask1, "cost2plate"] = df1.loc[mask1].apply(
lambda row: grouped[row["type"], row["location"]], axis=1
)
df1[df1["cost2plate"] == 0]
# # 4. rest_type
df1.isna().sum()
df1.dropna(inplace=True)
df1.isna().sum()
mask = df1["rest_type"].value_counts()
mask
others = mask[mask < 450].index.tolist()
df1.loc[df1["rest_type"].isin(others), "rest_type"] = "Others"
df1["rest_type"].value_counts()
# # 5.cousines
mask = df1["cuisines"].value_counts()
def handle_cu(value):
if value in mask[mask < 100]:
return "Others"
else:
return value
df1["cuisines"] = df1["cuisines"].apply(handle_cu)
df1["cuisines"].value_counts()
|
# Visit GitHub: https://github.com/erYash15/Real-or-Not-NLP-with-Disaster-Tweets
# Please Upvote if you Like the Work...
# ## **Introduction**
# My name is Yash Gupta. This project was made for Natural Language Processing Course. As a part of the course we came across many machine learning/ Data science aspects. I believe the Kaggle is the right place to practice with recent technologies in the area of data science and good place to refer all kinds of data science related projects. In this Kernal I have implemented Machine Learning and Deep Learning models for classification.
# **Table of Contents**
# **Introduction**
# **Libraries**
# **Loading Data**
#
# **Exploratory Data Analysis**
#   Analyzing Labels
#   Analyzing Features
#     Sentence Length Analysis
# **Data Cleaning**
#   Remove URL
#   Handle Tags
#   Handle Emoji
#   Remove HTML Tags
#   Remove Stopwords and Stemming
#   Remove Useless Characters
#   WORLDCLOUD
# **Final Pre-Processing Data**
# **Machine Learning**
#   Logistic Regression
#   Navie Bayes
#     Gaussian Naive Bayes
#     Bernoulli Naive Bayes
#     Complement Naive Bayes
#     Multinomial Naive Bayes
#   Support Vector Machine (SVM)
#     RBF kernel SVM
#     Linear Kernel SVM
#   Random Forest
# **Deep Learning**
#   Single Layer Perceptron
#   Multi Layer Perceptron
#     Model 1 : SIGMOID + ADAM
#     Model 2 : SIGMOID + SGD
#     Model 3 : RELU + ADAM
#     Model 4 : RELU + SGD
#     Model 5 : SIGMOID + BATCH NORMALIZATION + ADAM
#     Model 6 : SIGMOID + BATCH NORMALIZATION + SGD
#     Model 7 : RELU + DROPOUT + ADAM
#     Model 8 : RELU + DROPOUT + SGD
# **Results**
# **Conclusion**
#
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
DATA_DIR = "/kaggle/input/nlp-getting-started/"
# # Libraries
# Data Manipulation libraries
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import os
# NLP libraries
import string # Library for string operations
import nltk
from nltk.corpus import stopwords
from nltk.stem.snowball import SnowballStemmer
import re # Regex library
import demoji
from wordcloud import WordCloud # Word Cloud library
# ploting libraries
import matplotlib.pyplot as plt
# ML/AI libraries
from sklearn.model_selection import train_test_split
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.linear_model import LinearRegression
from sklearn.linear_model import LogisticRegression
from sklearn.naive_bayes import GaussianNB, BernoulliNB, ComplementNB, MultinomialNB
from sklearn import svm
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import accuracy_score
from sklearn.metrics import precision_recall_fscore_support
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense, Activation, BatchNormalization, Dropout
from tensorflow.python.keras.initializers import RandomNormal
# # Loading Data
train_df = pd.read_csv(os.path.join(DATA_DIR, "train.csv"), encoding="utf-8")
test_df = pd.read_csv(os.path.join(DATA_DIR, "test.csv"), encoding="utf-8")
X_train = train_df[["id", "keyword", "location", "text"]]
X_test = test_df[["id", "keyword", "location", "text"]]
y_train = train_df[["id", "target"]]
X_train.info()
y_train.info()
X_test.info()
# ## Exploratory Data Analysis
# ### Analyzing Lables
Real_len = y_train[y_train["target"] == 1].shape[0]
Not_len = y_train[y_train["target"] == 0].shape[0]
# bar plot of the 3 classes
plt.rcParams["figure.figsize"] = (7, 5)
plt.bar(10, Real_len, 3, label="Real", color="blue")
plt.bar(15, Not_len, 3, label="Not", color="red")
plt.legend()
plt.ylabel("Number of examples")
plt.title("Propertion of examples")
plt.show()
# ### Analyzing Features
# #### Sentence length analysis
def length(string):
return len(string)
X_train["length"] = X_train["text"].apply(length)
plt.rcParams["figure.figsize"] = (18.0, 6.0)
bins = 150
plt.hist(X_train[y_train["target"] == 0]["length"], alpha=0.6, bins=bins, label="Not")
plt.hist(X_train[y_train["target"] == 1]["length"], alpha=0.8, bins=bins, label="Real")
plt.xlabel("length")
plt.ylabel("numbers")
plt.legend(loc="upper right")
plt.xlim(0, 150)
plt.grid()
plt.show()
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(10, 5))
tweet_len = X_train[y_train["target"] == 1]["text"].str.len()
ax1.hist(tweet_len, color="blue")
ax1.set_title("disaster tweets")
tweet_len = X_train[y_train["target"] == 0]["text"].str.len()
ax2.hist(tweet_len, color="red")
ax2.set_title("Not disaster tweets")
fig.suptitle("Characters in tweets")
plt.show()
# ## Data Cleaning
# Data cleaning is the process of preparing data for analysis by removing or modifying data that is incorrect, incomplete, irrelevant, duplicated, or improperly formatted.
# Step 1. Remove Url
# Step 2. Handle Tags
# Step 3. Handle emoji's
# Step 4. Remove HTML Tags
# Step 5. Remove stopwords
# Step 6. Removing Useless Characters
# ### Step 1. Remove Url
# https://stackoverflow.com/questions/11331982/how-to-remove-any-url-within-a-string-in-python/11332580
def Remove_Url(string):
return re.sub(r"(https|http)?:\/\/(\w|\.|\/|\?|\=|\&|\%|\-)*\b", "", string)
print("Example of text with URL: \n", X_train["text"][3912], end="\n\n")
X_train["text"] = X_train["text"].apply(Remove_Url)
print("Example of text without URL: \n", X_train["text"][3912])
# ### Step 2. Handle Tags
def Handle_Tags(string):
pattern = re.compile(r"[@|#][^\s]+")
matches = pattern.findall(string)
tags = [match[1:] for match in matches]
# Removing tags from main string
string = re.sub(pattern, "", string)
# More weightage to tag by adding them 3 times
return string + " " + " ".join(tags) + " " + " ".join(tags) + " " + " ".join(tags)
print("Example of text without Handling Tags: \n", X_train["text"][3914], end="\n\n")
X_train["text"] = X_train["text"].apply(Handle_Tags)
print("Example of text with Handling Tags: \n", X_train["text"][3914])
# ### Step 3. Handle emoji's
# http://unicode.org/Public/emoji/12.0/emoji-test.txt
demoji.download_codes()
def Handle_emoji(string):
return demoji.replace_with_desc(string)
print("Example of text without Handled Emojis: \n", X_train["text"][17], end="\n\n")
X_train["text"] = X_train["text"].apply(Handle_emoji)
print("Example of text with Handled Emoji: \n", X_train["text"][17])
# ### Step 4. Remove HTML Tags
def Remove_html(string):
return re.sub(r"<.*?>|&([a-z0-9]+|#[0-9]{1,6}|#x[0-9a-f]{1,6});", "", str(string))
X_train["text"] = X_train["text"].apply(Remove_html)
# ### Step 5. Remove Stopwords and Stemming
nltk.download("punkt")
nltk.download("stopwords")
stemmer = SnowballStemmer("english")
stopword = stopwords.words("english")
def Remove_StopAndStem(string):
string_list = string.split()
return " ".join([stemmer.stem(i) for i in string_list if i not in stopword])
print("Example of text before Removing Stopwords: \n", X_train["text"][17], end="\n\n")
X_train["text"] = X_train["text"].apply(Remove_StopAndStem)
print("Example of text after Removing Stopwords and Stemming: \n", X_train["text"][17])
# ### Step 6. Removing Useless Characters
def Remove_UC(string):
thestring = re.sub(r"[^a-zA-Z\s]", "", string)
# remove word of length less than 2
thestring = re.sub(r"\b\w{1,2}\b", "", thestring)
# https://www.geeksforgeeks.org/python-remove-unwanted-spaces-from-string/
return re.sub(" +", " ", thestring)
print(
"Example of text before Removing Useless Character: \n",
X_train["text"][17],
end="\n\n",
)
X_train["text"] = X_train["text"].apply(Remove_UC)
print("Example of text after Removing Useless Character: \n", X_train["text"][17])
def merging_details(data):
# df = pd.DataFrame(columns=['id', 'Cleaned_data'])
df_list = []
# https://www.geeksforgeeks.org/how-to-iterate-over-rows-in-pandas-dataframe/
for row in data.itertuples():
df_dict = {}
# Processing Keyword and location
keyword = re.sub(r"[^a-zA-Z\s]", "", str(row[2]))
location = re.sub(r"[^a-zA-Z\s]", "", str(row[3]))
keyword = re.sub(r"\b\w{1,2}\b", "", keyword)
location = re.sub(r"\b\w{1,2}\b", "", location)
# Already processed data
text = str(row[4])
if keyword == "nan":
if location == "nan":
prs_data = text
else:
prs_data = location + " " + text
else:
if location == "nan":
prs_data = keyword + " " + text
else:
prs_data = keyword + " " + location + " " + text
prs_data = re.sub(" +", " ", prs_data)
df_dict["Cleaned_data"] = prs_data
df_list.append(df_dict)
return pd.DataFrame(df_list)
X_train = merging_details(X_train)
X_train
### WORDCLOUD
dict_of_words = {}
for row in X_train.itertuples():
for i in row[1].split():
try:
dict_of_words[i] += 1
except:
dict_of_words[i] = 1
# Initializing WordCloud
wordcloud = WordCloud(
background_color="black", width=1000, height=500
).generate_from_frequencies(dict_of_words)
fig = plt.figure(figsize=(10, 5))
plt.imshow(wordcloud)
plt.tight_layout(pad=1)
plt.show()
# ## Final Pre-Processing Data
# Step 1. Remove Url
X_test["text"] = X_test["text"].apply(Remove_Url)
# Step 2. Handle Tags
X_test["text"] = X_test["text"].apply(Handle_Tags)
# Step 3. Handle emoji's
X_test["text"] = X_test["text"].apply(Handle_emoji)
# Step 4. Remove HTML Tags
X_test["text"] = X_test["text"].apply(Remove_html)
# Step 5. Remove Stopwords and Stemming
X_test["text"] = X_test["text"].apply(Remove_StopAndStem)
# Step 6. Removing Useless Characters
X_test["text"] = X_test["text"].apply(Remove_UC)
# Step7. Merging Other Details
X_test = merging_details(X_test)
X_test
y_train = y_train["target"]
# smooth_idf=True by default so smoothing is done by defult.
# norm is l2 by default.
# subliner is used False by default.
vectorizer = TfidfVectorizer(
min_df=0.0005,
max_features=100000,
tokenizer=lambda x: x.split(),
ngram_range=(1, 4),
)
X_train = vectorizer.fit_transform(X_train["Cleaned_data"])
X_test = vectorizer.transform(X_test["Cleaned_data"])
# https://stackoverflow.com/questions/16505670/generating-a-dense-matrix-from-a-sparse-matrix-in-numpy-python
print(
"Training Points: ",
len(X_train.toarray()),
"| Training Features:",
len(X_train.toarray()[0]),
)
print(
"Testing Points: ",
len(X_test.toarray()),
"| Testing Features:",
len(X_test.toarray()[0]),
)
print()
print("Training Points: ", len(y_train))
# ## Machine Learning
Model = BernoulliNB()
Model.fit(X_train.toarray(), y_train)
y_pred = Model.predict(X_test.toarray())
test_final = pd.read_csv("/kaggle/input/nlp-getting-started/test.csv")
test_final["target"] = y_pred
test_final[["id", "target"]].to_csv("submission.csv", index=False)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
from sklearn.linear_model import LinearRegression
import numpy as np
import matplotlib as mpl
import matplotlib.pyplot as plt
import seaborn as sns
import scipy.stats as st
from sklearn import ensemble, tree, linear_model
import missingno as msno
import os
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
ds = pd.read_csv(
"/kaggle/input/bordacsv/prediction-of-surgery-duration/train.csv",
encoding="ISO-8859-1",
)
test = pd.read_csv(
"/kaggle/input/bordacsv/prediction-of-surgery-duration/test.csv",
encoding="ISO-8859-1",
)
ds.head()
import math
import seaborn as sns
import matplotlib.pyplot as plt
import plotly.express as px
from sklearn.linear_model import LinearRegression
from sklearn.preprocessing import PolynomialFeatures
from sklearn.metrics import r2_score
ds.info()
ds.tail()
ds.shape
numeric_features = ds.select_dtypes(include=[np.number])
numeric_features.columns
ds.dtypes
ds.count()
ds.duplicated().sum()
print(ds.isnull().sum())
ds.describe()
ds.nunique()
ds.corr()
ds.groupby("SurgeryGroup")["ElapsedTime(second)"].mean().sort_values().plot(kind="bar")
ds.groupby("SurgeryName").agg({"ElapsedTime(second)": [np.mean, "count"]})
ds.groupby("Service").agg({"ElapsedTime(second)": [np.mean, "count"]})
ds.groupby("SurgeryName")["ElapsedTime(second)"].count().sort_values()
ds.groupby("Age")["ElapsedTime(second)"].mean().plot(kind="line")
# As seen in the graph, there is no definite order between the duration of surgery and age until the age of 80. It is seen that the longest operation in the data was performed on an individual aged 80 years. In operations after the age of 80, it is seen that the duration of the operation increases linearly as the age increases.
ds.groupby("Sex")["ElapsedTime(second)"].mean().sort_values().plot(kind="bar")
ds.groupby("Service")["ElapsedTime(second)"].mean().sort_values().plot(kind="bar")
# First of all, as we can see from the bar chart, there are 10 service types in this data group. When the average elapsed times of these services are examined, it is seen that the lowest average elapsed time belongs to the Obstetrics Service. It is seen that the average elapsed times of the ENT Service and the Neurology Service are very close to each other and both have the longest average elapsed time.
ds.groupby("AnesthesiaType")["ElapsedTime(second)"].mean().sort_values().plot(
kind="bar"
)
a = ds.groupby("Service")["ElapsedTime(second)"].mean()
a
b = ds.groupby("AnesthesiaType")["ElapsedTime(second)"].mean()
b
e = ds.groupby("DiagnosticICD10Code")["ElapsedTime(second)"].mean()
e
f = ds.groupby("SurgeryGroup")["ElapsedTime(second)"].mean()
f
c = ds.groupby("AnaesthetistID")["ElapsedTime(second)"].mean()
c
d = ds.groupby("DoctorID")["ElapsedTime(second)"].mean()
d
ds.groupby("DoctorID")["ElapsedTime(second)"].mean().sort_values().plot(kind="bar")
# Looking at the graph, it is seen that the doctor with 48 ID have the least average elapsed time. When we look at the operations performed by the doctor with 48 ID, it is seen that all operations are performed in the Obstetrics Service. Which means that the 48 ID doctor is doctors in the Obstetrics Service.
# As can be seen in the graph, the doctor with 41 ID is obviously the doctor with the longest average elapsed time. When we look at the surgeries performed by 48 ID doctor, it is seen that all of them were performed in the ENT Service. It can be said that the doctor with ID 41 is the doctor of the ENT Service.
# In the graph above, it was seen that the average elapsed time of the Obstetrics Service was the lowest and the average elapsed time of the ENT Service was the highest.
#
ds.groupby("DoctorID")["ElapsedTime(second)"].count().sort_values().plot(kind="bar")
# The chart above shows how many surgeries each doctor performed in this data set. It is seen that the doctor with 36 ID has performed the most operations. Considering the surgeries performed by 36 ID doctor, it is seen that all of them are in the General Surgery Service. This means that 36 ID doctor work in the General Surgery Service.
ds.groupby("AnaesthetistID")["ElapsedTime(second)"].mean().sort_values().plot(
kind="bar"
)
# As seen in the graph, the anesthetist with the highest average elapsed time is anaesthetist with 16 ID. Considering the surgeries attended by 16 ID anaesthetist, it is seen that they have participated in many different surgeries. Here is a certain inference about anaesthetist and elapsed times.
#
ds = (
ds.groupby(by=["AnaesthetistID", "DoctorID"])["ElapsedTime(second)"]
.count()
.reset_index()
)
ds.sort_values(by="ElapsedTime(second)")
ds.groupby("DoctorID")["ElapsedTime(second)"].mean().plot(kind="line")
ds.skew(), ds.kurt()
ds.hist(bins=12, figsize=(12, 10), grid=True)
plt.scatter(ds["DoctorID"], ds["ElapsedTime(second)"])
plt.scatter(ds["AnaesthetistID"], ds["ElapsedTime(second)"])
data = ds.groupby("DoctorID")["ElapsedTime(second)"].mean().sort_values(ascending=False)
data = pd.DataFrame(data)
data.reset_index(inplace=True)
data
submission = test[["ID"]]
submission
submission["ElapsedTime(second)"] = d
submission
submission.to_csv("submissionn.csv")
|
# Step 1 - Installing dependencies
# Step 2 - Importing dataset
# Step 3 - Exploratory data analysis
# Step 4 - Feature engineering (statistical features added)
# Step 5 - Implement "Catboost" model
# Step 6 - Implement Support Vector Machine + Radial Basis Function model
# Step 1: installing Dependencies
# data preprocessing
import pandas as pd
# math operations
import numpy as np
# machine learning
from catboost import CatBoostRegressor, Pool
# data scaling
from sklearn.preprocessing import StandardScaler
# hyperparameter optimization
from sklearn.model_selection import GridSearchCV
# support vector machine model
from sklearn.svm import NuSVR, SVR
# kernel ridge model
from sklearn.kernel_ridge import KernelRidge
# data visualization
import matplotlib.pyplot as plt
# Step 2: importing Dataset
# Extract training data into a dataframe for further manipulation
train = pd.read_csv(
"/kaggle/input/LANL-Earthquake-Prediction/train.csv",
nrows=6000000,
dtype={"acoustic_data": np.int16, "time_to_failure": np.float64},
)
# print first 10 entries
train.head(10)
# Step 3: Exploratory Data Analysis
# visualize 1% of samples data, first 100 datapoints
train_ad_sample_df = train["acoustic_data"].values[::100]
train_ttf_sample_df = train["time_to_failure"].values[::100]
# function for plotting based on both features
def plot_acc_ttf_data(
train_ad_sample_df,
train_ttf_sample_df,
title="Acoustic data and time to failure: 1% sampled data",
):
fig, ax1 = plt.subplots(figsize=(12, 8))
plt.title(title)
plt.plot(train_ad_sample_df, color="r")
ax1.set_ylabel("acoustic data", color="r")
plt.legend(["acoustic data"], loc=(0.01, 0.95))
ax2 = ax1.twinx()
plt.plot(train_ttf_sample_df, color="b")
ax2.set_ylabel("time to failure", color="b")
plt.legend(["time to failure"], loc=(0.01, 0.9))
plt.grid(True)
plot_acc_ttf_data(train_ad_sample_df, train_ttf_sample_df)
del train_ad_sample_df
del train_ttf_sample_df
# Step 4: Feature Engineering
# Step 4 - Feature Engineering and signifiance of these statistical features
# lets create a function to generate some statistical features based on the training data
def gen_features(X):
strain = []
strain.append(X.mean())
strain.append(X.std())
strain.append(X.min())
strain.append(X.max())
strain.append(X.kurtosis())
strain.append(X.skew())
strain.append(np.quantile(X, 0.01))
strain.append(np.quantile(X, 0.05))
strain.append(np.quantile(X, 0.95))
strain.append(np.quantile(X, 0.99))
strain.append(np.abs(X).max())
strain.append(np.abs(X).mean())
strain.append(np.abs(X).std())
return pd.Series(strain)
train = pd.read_csv(
"/kaggle/input/LANL-Earthquake-Prediction/train.csv",
iterator=True,
chunksize=150_000,
dtype={"acoustic_data": np.int16, "time_to_failure": np.float64},
)
X_train = pd.DataFrame()
y_train = pd.Series()
for df in train:
ch = gen_features(df["acoustic_data"])
X_train = X_train.append(ch, ignore_index=True)
y_train = y_train.append(pd.Series(df["time_to_failure"].values[-1]))
X_train.describe()
# Step 5 - Implement Catboost Model
# Gradient boosting on decision trees is a form of machine learning that works by progressively training more complex models to maximize the accuracy of predictions.
# It's particularly useful for predictive models that analyze ordered (continuous) data and categorical data.
# It's one of the most efficient ways to build ensemble models. The combination of gradient boosting with decision trees provides state-of-the-art results in many applications with structured data.
# On the first iteration, the algorithm learns the first tree to reduce the training error, shown on left-hand image in figure 1.
# This model usually has a significant error; it’s not a good idea to build very big trees in boosting since they overfit the data.
# The right-hand image in figure 1 shows the second iteration, in which the algorithm learns one more tree to reduce the error made by the first tree.
# The algorithm repeats this procedure until it builds a decent quality model
# Each step of Gradient Boosting combines two steps:
# Step 1 - Computing gradients of the loss function we want to optimize for each input object
# Step 2 - Learning the decision tree which predicts gradients of the loss function
# ELI5 Time
# Step 1 - We first model data with simple models and analyze data for errors.
# Step 2 - These errors signify data points that are difficult to fit by a simple model.
# Step 3 - Then for later models, we particularly focus on those hard to fit data to get them right.
# Step 4 - In the end, we combine all the predictors by giving some weights to each predictor.
#
# Model #1 - Catboost
train_pool = Pool(X_train, y_train)
m = CatBoostRegressor(iterations=10000, loss_function="MAE", boosting_type="Ordered")
m.fit(X_train, y_train, silent=True)
m.best_score_
from sklearn.model_selection import RandomizedSearchCV
params = {
"iterations": [1000, 5000, 10000],
"learning_rate": [0.01, 0.05, 0.1],
"depth": [4, 6, 8],
"loss_function": ["MAE", "RMSE"],
}
random_search = RandomizedSearchCV(
estimator=CatBoostRegressor(silent=True),
param_distributions=params,
n_iter=10,
cv=5,
n_jobs=-1,
)
random_search.fit(X_train, y_train)
best_params = random_search.best_params_
best_score = random_search.best_score_
print("Best Parameters: ", best_params)
print("Best Score: ", best_score)
# This code performs hyperparameter tuning using RandomizedSearchCV with CatBoostRegressor as the estimator. It searches for the best combination of hyperparameters from the specified parameter distributions (params). The code fits the model with different hyperparameter values (n_iter=10) using 5-fold cross-validation (cv=5) and runs the search in parallel (n_jobs=-1) to speed up the process.
# The output shows the best hyperparameters found by the RandomizedSearchCV (best_params) and the corresponding best score (best_score). In this case, the best hyperparameters found are:
# 'loss_function': 'RMSE'
# 'learning_rate': 0.01
# 'iterations': 1000
# 'depth': 4
# The best score (best_score) is a performance metric (e.g., R^2 score, accuracy) obtained using the best hyperparameters during cross-validation. It indicates the performance of the CatBoostRegressor model with the best hyperparameters found by the RandomizedSearchCV. In this case, the best score is 0.39426905699313197, which is the highest score achieved among the hyperparameter combinations tried during the search.
# Need to learn a nonlinear decision boundary? Grab a Kernel
# A very simple and intuitive way of thinking about kernels (at least for SVMs) is a similarity function.
# Given two objects, the kernel outputs some similarity score. The objects can be anything starting from two integers, two real valued vectors, trees whatever provided that the kernel function knows how to compare them.
# The arguably simplest example is the linear kernel, also called dot-product. Given two vectors, the similarity is the length of the projection of one vector on another.
# Another interesting kernel examples is Gaussian kernel. Given two vectors, the similarity will diminish with the radius of σ. The distance between two objects is "reweighted" by this radius parameter.
# The success of learning with kernels (again, at least for SVMs), very strongly depends on the choice of kernel. You can see a kernel as a compact representation of the knowledge about your classification problem. It is very often problem specific.
#
# Model #2 - Support Vector Machine w/ RBF + Grid Search
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import GridSearchCV
from sklearn.svm import NuSVR, SVR
scaler = StandardScaler()
scaler.fit(X_train)
X_train_scaled = scaler.transform(X_train)
parameters = [
{
"gamma": [0.001, 0.005, 0.01, 0.02, 0.05, 0.1],
"C": [0.1, 0.2, 0.25, 0.5, 1, 1.5, 2],
}
]
#'nu': [0.75, 0.8, 0.85, 0.9, 0.95, 0.97]}]
reg1 = GridSearchCV(
SVR(kernel="rbf", tol=0.01), parameters, cv=5, scoring="neg_mean_absolute_error"
)
reg1.fit(X_train_scaled, y_train.values.flatten())
y_pred1 = reg1.predict(X_train_scaled)
print("Best CV score: {:.4f}".format(reg1.best_score_))
print(reg1.best_params_)
|
# format function
a = " we are leaning {}"
c = input("enter topic")
b = a.format(c)
b
s = "{0} is a {1} company"
p = s.format("google", "tech")
p
# q = input("enter company")
# r = input("big or small")
# d = s.format(0 = q,1 = r)
# d
# s
s = "{company_name} is a {company_type} company"
t = input("enter company_name")
q = input("Enter company type")
p = s.format(company_name=t, company_type=q)
# p = s.format(company_name = 'Google', company_type = 'Tech')
p
d = {"apple": 23455, "Google": 454546, "Facebook": 3543646, "Netflix": 435346}
for i in d:
print("{:<10} - {:>10}".format(i, d[i]))
|
# **Model**
# эмбеддинги для изображений
def image_embedding(df):
embeddings = []
labels = []
for path, label in zip(df["image_path"], df["label_group"]):
img = Image.open(path)
img_tensor = transform(img).unsqueeze(0)
embedding = resnet(img_tensor).squeeze().detach().numpy()
embeddings.append(embedding)
labels.append(label)
embeddings = np.array(embeddings)
labels = np.array(labels)
return embeddings, labels
def train_val_split(df, split_size=0.2):
df_train, df_val = pd.DataFrame(), pd.DataFrame()
df_stats = pd.DataFrame(columns=["label_group", "train_size", "test_size"])
labels = list(df["label_group"].unique())
for label in labels:
df_temp = df[df["label_group"] == label]
num_rows = df_temp.shape[0]
test_size = int(split_size * num_rows)
test_size = max(1, test_size)
df_test_temp = df_temp.sample(n=test_size)
df_train_temp = df_temp.drop(index=df_test_temp.index)
df_train = df_train.append(df_train_temp)
df_val = df_val.append(df_test_temp)
return df_train, df_val
import torch
import torchvision.models as models
import torchvision.transforms as transforms
import pandas as pd
import numpy as np
# import sklearn
from sklearn import preprocessing
from torch.utils.data import Dataset, DataLoader
from PIL import Image
from sklearn.model_selection import StratifiedKFold, KFold
from sklearn.neighbors import KNeighborsClassifier
from sklearn import neighbors
model_path = "/kaggle/input/resnet50/resnet50-0676ba61.pth"
# Загрузка предобученной модели ResNet
resnet = models.resnet50(pretrained=False)
resnet.load_state_dict(torch.load(model_path))
resnet.eval()
# Извлечение последнего сверточного слоя для получения признаков
modules = list(resnet.children())[:-1]
resnet = torch.nn.Sequential(*modules)
# efnet = models.efficientnet_b3(pretrained = True)
# efnet.eval()
# # Извлечение последнего сверточного слоя для получения признаков
# modules = list(efnet.children())[:-1]
# efnet = torch.nn.Sequential(*modules)
# Задание преобразований для изображений
train_transform = transforms.Compose(
[
transforms.Resize((256, 256)),
transforms.RandomCrop((224, 224)),
transforms.RandomHorizontalFlip(p=0.5),
transforms.RandomRotation(degrees=15),
transforms.ColorJitter(brightness=0.2, contrast=0.2, saturation=0.2, hue=0.2),
transforms.ToTensor(),
transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),
]
)
test_transform = transforms.Compose(
[
transforms.Resize((224, 224)),
transforms.ToTensor(),
transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),
]
)
transform = transforms.Compose(
[
transforms.Resize(256),
transforms.CenterCrop(224),
transforms.ToTensor(),
transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),
]
)
class ShopeeDataset(Dataset):
def __init__(self, df, transforms=None):
self.df = df
self.transforms = transforms
def __len__(self):
return len(self.df)
def __getitem__(self, idx):
row = self.df.iloc[idx]
image = Image.open(row.filepath)
label = row.label_group
if self.transforms:
image = self.transforms(image)
return image, label
df = pd.read_csv("/kaggle/input/shopee-product-matching/train.csv")
df["image_path"] = "/kaggle/input/shopee-product-matching/train_images/" + df["image"]
encoder = preprocessing.LabelEncoder()
df["label_group"] = encoder.fit_transform(df["label_group"])
num_classes = len(df["label_group"].unique())
# kf = StratifiedKFold(n_splits=5)
# df["fold"] = -1
# for fold, (train_idx, valid_idx) in enumerate(kf.split(df, df["label_group"])):
# df.loc[valid_idx, "fold"] = fold
# train_df = df[df['fold'] != 4]
# val_df = df[df['fold'] == 4]
# Разделение на train и validation
# val_df = train_df.groupby('label_group').apply(lambda x: x.sample(frac=0.2, random_state = 42)).reset_index(drop=True)
# train_df = train_df[~train_df.index.isin(val_df.index)]
train_data = ShopeeDataset(train_df, transforms=train_transform)
val_data = ShopeeDataset(val_df, transforms=test_transform)
train_df, val_df = train_val_split(df)
tmp = train_df.groupby(["label_group"])["posting_id"].unique().to_dict()
train_df["matches"] = train_df["label_group"].map(tmp)
train_df["matches"] = train_df["matches"].apply(lambda x: " ".join(x))
tmp = val_df.groupby(["label_group"])["posting_id"].unique().to_dict()
val_df["matches"] = val_df["label_group"].map(tmp)
val_df["matches"] = val_df["matches"].apply(lambda x: " ".join(x))
# Image
# Получение эмбеддингов для изображений
train_embeddings, train_labels = image_embedding(train_df)
val_embeddings, val_labels = image_embedding(val_df)
def f1_score(y_true, y_pred):
y_true = y_true.apply(lambda x: set(x.split()))
y_pred = y_pred.apply(lambda x: set(x.split()))
intersection = np.array([len(x[0] & x[1]) for x in zip(y_true, y_pred)])
len_y_pred = y_pred.apply(lambda x: len(x)).values
len_y_true = y_true.apply(lambda x: len(x)).values
f1 = 2 * intersection / (len_y_pred + len_y_true)
return f1
# нахождение оптимальной дистанции
def find_neighbors_distance(
df: pd.DataFrame, embeddings, n_neighbors: int, type="image"
):
model = neighbors.NearestNeighbors(n_neighbors=n_neighbors)
model.fit(embeddings)
distances, neighbors_idx = model.kneighbors(embeddings)
if type == "image":
distance_list = list(np.arange(1.0, 15.0, 0.5))
else:
distance_list = list(np.arange(0.1, 1.5, 0.1))
scores = []
for distance in distance_list:
predictions = []
for k in range(embeddings.shape[0]):
idx = np.where(distances[k,] < distance)[0]
ids = neighbors_idx[k, idx]
posting_ids = " ".join(df["posting_id"].iloc[ids].values)
predictions.append(posting_ids)
df["pred_matches"] = predictions
df["f1"] = f1_score(df["matches"], df["pred_matches"])
score = df["f1"].mean()
print(f"f1 score for distance {distance} is {score}")
scores.append(score)
distance_scores = pd.DataFrame({"distance": distance_list, "scores": scores})
max_score = distance_scores[
distance_scores["scores"] == distance_scores["scores"].max()
]
best_distance = max_score["distance"].values[0]
best_score = max_score["scores"].values[0]
print(f"Best score is {best_score} and has a distance {best_distance}")
return best_distance
# нахождение соседей
def predictions(df, embeddings, best_distance, type="image", check=False):
if check:
n = 3
else:
n = 50
model = neighbors.NearestNeighbors(n_neighbors=n)
model.fit(embeddings)
distances, neighbors_idx = model.kneighbors(embeddings)
predictions = []
for k in range(embeddings.shape[0]):
idx = np.where(distances[k,] < best_distance)[0]
ids = neighbors_idx[k, idx]
posting_ids = df["posting_id"].iloc[ids].values
posting_ids = " ".join(df["posting_id"].iloc[ids].values)
predictions.append(posting_ids)
pred_name = "pred_matches_" + type
df[pred_name] = predictions
return df, predictions
model = neighbors.NearestNeighbors(n_neighbors=50)
model.fit(train_embeddings)
distances, neighbors_idx = model.kneighbors(train_embeddings)
best_distance_image = find_neighbors_distance(train_df, train_embeddings, 50)
val_df, val_redictions = predictions(val_df, val_embeddings, best_distance_image)
f1_score(val_df["pred_matches_image"], val_df["matches"]).mean()
# from sklearn import neighbors
# clf_knn = neighbors.KNeighborsClassifier()
# clf_knn.fit(train_embeddings, train_labels)
# accuracy = clf_knn.score(val_embeddings, val_labels)
# print("Accuracy:", accuracy)
# Text
# from sklearn.feature_extraction.text import TfidfVectorizer
# from sklearn.metrics.pairwise import cosine_similarity
# from sklearn import preprocessing
# from sklearn.decomposition import PCA
# # Предобработка текста - приводим все к нижнему регистру и токенизируем
# train_df['title_tokens'] = train_df['title'].str.lower().str.split()
# # Создаем матрицу TF-IDF
# tfidf = TfidfVectorizer(max_features= (train_df['label_group'].nunique()),
# stop_words = 'english')
# tfidf_matrix = tfidf.fit_transform(train_df['title'])
# pca = PCA(n_components=5000)
# pca_matrix = pca.fit_transform(tfidf_matrix.toarray())
# # Находим похожие товары на основе матрицы TF-IDF
# #similarities = cosine_similarity(tfidf_matrix)
# text_model = neighbors.NearestNeighbors(n_neighbors=50)
# text_model.fit(pca_matrix)
# train_df.head()
# best_distance_text = find_neighbors_distance(train_df, pca_matrix, 50, 'text')
# tfidf = TfidfVectorizer(max_features= (train_df['label_group'].nunique()),
# stop_words = 'english')
# tfidf_matrix_val = tfidf.fit_transform(val_df['title'])
# pca = PCA(n_components=5000)
# pca_matrix_val = pca.fit_transform(tfidf_matrix_val.toarray())
# val_df, val_predictions_text = predictions(val_df, pca_matrix_val, best_distance_text, 'text')
# f1_score(val_df['matches'], val_df['pred_matches_text']).mean()
# Test
test_df = pd.read_csv("/kaggle/input/shopee-product-matching/test.csv")
test_df["image_path"] = (
"/kaggle/input/shopee-product-matching/test_images/" + test_df["image"]
)
# Получение эмбеддингов для обучающей выборки
test_embeddings = []
for path in test_df["image_path"]:
img = Image.open(path)
img_tensor = transform(img).unsqueeze(0)
embedding = resnet(img_tensor).squeeze().detach().numpy()
test_embeddings.append(embedding)
test_embeddings = np.array(test_embeddings)
test_df, test_predictions_image = predictions(
test_df, test_embeddings, best_distance_image, "image", check=True
)
# tfidf = TfidfVectorizer(max_features= (train_df['label_group'].nunique()),
# stop_words = 'english')
# tfidf_matrix_test = tfidf.fit_transform(test_df['title'])
# pca = PCA(n_components= min(test_df.shape[0], 5000))
# pca_matrix_test = pca.fit_transform(tfidf_matrix_test.toarray())
# test_df, test_predictions_text = predictions(test_df, pca_matrix_test, best_distance_text, 'text', check = True)
# # объединяем значения из двух столбцов в один список
# matches_list = test_df['pred_matches_image'].str.cat(test_df['pred_matches_text'], sep = ' ')
# test_df['matches'] = matches_list
# test_df['matches'] = test_df['matches'].apply(lambda x: list(set(x.split(' '))))
# test_df['matches'] = [','.join(map(str, l)) for l in test_df['matches']]
submission_df = test_df[["posting_id", "matches"]]
submission_df = test_df[["posting_id", "pred_matches_image"]].rename(
columns={"pred_matches_image": "matches"}
)
submission_df.to_csv("submission.csv", index=False)
submission_df
|
# # Step 1 : data gathering and preparation
# Goal: obtain a concatenated dataset with all rlevant raw data, for each time stamp
# **Data sources**
# The price data comes from:
# * ENTSO-E Transparency platform, series 12.1.D, frequency=1hour
# The intermitent renewables data comes from:
# * ENTSO-E Transparency platform, series 14.1.D (solar & wind generation forecast day-ahead + actual generation), frequency=15minutes
# The electricity consumption:
# * ENTSO-E Transparency platform, series 6.1.B (total load forecast day-ahead + actual load), frequency=15minutes
#
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import gc
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# each input file contain 1 year worth of data, so we need to concatenate them:
def yearly_files_concatenation(directory):
i = 0
for dirname, _, filenames in os.walk("/kaggle/input/" + str(directory)):
for filename in filenames:
if i == 0:
out = pd.read_csv(
"/kaggle/input/" + str(directory) + "/" + str(filename)
)
i = 1
else:
df = pd.read_csv(
"/kaggle/input/" + str(directory) + "/" + str(filename)
)
out = pd.concat([df, out])
return out
DE_DA_Prices = yearly_files_concatenation(
"da-prices-germany-ensto-e-transparency-platform"
)
# to check that we have several years worth of data: 7*365*24=61 320 data points for 7 years of data with 1-hour frequency:
len(DE_DA_Prices)
DE_DA_Prices.head()
# time is written as a time window, text format. Let s turn the text into a recognizable timestamp for python, and keep only the begining of the window, in order to sort values
def timestamp_reading(string):
return pd.to_datetime(string.split(" - ")[0], format="%d.%m.%Y %H:%M")
DE_DA_Prices["MTU (CET)"] = DE_DA_Prices["MTU (CET)"].apply(timestamp_reading)
DE_DA_Prices.head()
# the following function sets timestamp as index of the dataframe for easy merging
# due to daylight savings, some timestamps are redundant. The function arbitrarly removes the first occurence of the duplicates
def remove_timestamp_duplicates_and_set_as_index(timestamp_column, df):
dup_df = df[timestamp_column].value_counts().to_frame()
duplicates = dup_df[dup_df[timestamp_column] > 1].index.to_list()
print("the following timestamps are in duplicate", duplicates)
for d in duplicates:
# remove the first occurence:
df.drop(df.index[df[timestamp_column] == d][0], inplace=True)
df.set_index(timestamp_column, drop=True, inplace=True, verify_integrity=True)
df.sort_index(inplace=True)
return df
DE_DA_Prices = remove_timestamp_duplicates_and_set_as_index("MTU (CET)", DE_DA_Prices)
DE_DA_Prices.head()
# last step before merging: turning non numerical values into NaN, and removing missing data at the begining and end of timeranges
def df_value_cleaning(df, columns):
df.dropna(inplace=True)
for column in columns:
df[column] = pd.to_numeric(df[column], errors="coerce")
return df
DE_DA_Prices = df_value_cleaning(DE_DA_Prices, ["Day-ahead Price [EUR/MWh]"])
DE_DA_Prices["Day-ahead Price [EUR/MWh]"].plot(figsize=(20, 5))
# repeating the steps above for other data
# renewables generation data:
RE = yearly_files_concatenation("wind-and-solar-germany-entsoe-transparency")
RE["MTU (CET)"] = RE["MTU (CET)"].apply(timestamp_reading)
RE = remove_timestamp_duplicates_and_set_as_index("MTU (CET)", RE)
RE = df_value_cleaning(RE, RE.columns)
# this function aggregates 15 minutes frequencies into 1-hour rows
def hour_aggregation(df):
df = df.groupby(pd.Grouper(freq="H")).mean()
return df
RE = hour_aggregation(RE)
# removing some empty columns and aggragating onshore and offshore wind:
RE.drop(
columns=[
"Generation - Solar [MW] Current / Germany (DE)",
"Generation - Wind Offshore [MW] Current / Germany (DE)",
"Generation - Wind Onshore [MW] Current / Germany (DE)",
],
inplace=True,
)
RE["Generation - Wind [MW] Day Ahead/ Germany (DE)"] = (
RE["Generation - Wind Offshore [MW] Day Ahead/ Germany (DE)"]
+ RE["Generation - Wind Onshore [MW] Day Ahead/ Germany (DE)"]
)
RE["Generation - Wind [MW] Intraday / Germany (DE)"] = (
RE["Generation - Wind Offshore [MW] Intraday / Germany (DE)"]
+ RE["Generation - Wind Onshore [MW] Intraday / Germany (DE)"]
)
RE.drop(
[
"Generation - Wind Offshore [MW] Day Ahead/ Germany (DE)",
"Generation - Wind Offshore [MW] Intraday / Germany (DE)",
"Generation - Wind Onshore [MW] Day Ahead/ Germany (DE)",
"Generation - Wind Onshore [MW] Intraday / Germany (DE)",
],
axis=1,
inplace=True,
)
RE.head()
# electricity consumption data:
cons = yearly_files_concatenation("load-germany-entso-e-transparency-platform")
cons["Time (CET)"] = cons["Time (CET)"].apply(timestamp_reading)
cons = remove_timestamp_duplicates_and_set_as_index("Time (CET)", cons)
cons = df_value_cleaning(cons, cons.columns)
cons = hour_aggregation(cons)
# removing some empty columns and aggragating onshore and offshore wind:
# RE.drop(columns=["Generation - Solar [MW] Current / Germany (DE)","Generation - Wind Offshore [MW] Current / Germany (DE)","Generation - Wind Onshore [MW] Current / Germany (DE)"],inplace=True)
# RE["Generation - Wind [MW] Day Ahead/ Germany (DE)"]=RE["Generation - Wind Offshore [MW] Day Ahead/ Germany (DE)"]+RE["Generation - Wind Onshore [MW] Day Ahead/ Germany (DE)"]
# RE["Generation - Wind [MW] Intraday / Germany (DE)"]=RE["Generation - Wind Offshore [MW] Intraday / Germany (DE)"]+RE["Generation - Wind Onshore [MW] Intraday / Germany (DE)"]
# RE.drop(["Generation - Wind Offshore [MW] Day Ahead/ Germany (DE)","Generation - Wind Offshore [MW] Intraday / Germany (DE)","Generation - Wind Onshore [MW] Day Ahead/ Germany (DE)","Generation - Wind Onshore [MW] Intraday / Germany (DE)"],axis=1,inplace=True)
cons.head()
# finally, merging of the 3 dataframes created earlier
data = DE_DA_Prices.copy(deep=True)
data = data.join(RE)
data = data.join(cons)
del RE
del cons
del DE_DA_Prices
gc.collect()
data.head()
|
print("Hello World")
x = 9
y = "hello"
print(x)
print(y)
if 9 > 4:
print("9 is greater")
x = str(9)
y = int(5)
z = float(8)
z
_my_var = 2
_my_var
x = 3
y = "Hello"
z = 4.0
print(x)
print(y)
print(z)
x, y, z = 3, "Hello", 3.0
print(x)
print(y)
print(z)
x = y = z = "Apple"
print(x)
print(y)
print(z)
fruits = ["Orange", "Apple", "Banana"]
x, y, z = fruits
print(x)
print(y)
print(z)
x = "Python is "
y = "Awsome"
z = x + y
print(z)
x = 5
y = 10
print(x + y)
# x=5
# y="Hello"
# print(str(x)+y)
x = "Awsome"
def myfun():
print("python is " + x)
myfun()
a = "hello world"
print(a[2:])
a = "hello world"
print(a[-5:-2])
a = "hello world"
a.upper()
a = "HELLO WORLD"
a.lower()
a = "hello world"
a.replace("h", "j")
a = "hello, world"
a.split(",")
mylist = ["apple", "banana", "orange"]
mylist
mylist = ["1", "2", "3"]
mylist
mylist = [True, False, False, True]
mylist
mylist = [True, False, False, True]
mylist1 = ["apple", "banana", "orange"]
mylist2 = ["1", "2", "3"]
mylist + mylist1 + mylist2
mylist = ["apple", "banana", "orange"]
mylist[-2]
mylist = ["apple", "banana", "orange"]
mylist[-1]
mylist = ["apple", "banana", "orange"]
if "apple" in mylist:
print("Yes")
else:
print("No")
mylist = ["apple", "banana", "orange"]
mylist[0:2] = ["watermelon", "mango"]
mylist
mylist = ["apple", "banana", "orange"]
mylist[2] = "mango"
mylist
mylist = ["apple", "banana", "orange"]
mylist.append("Litchi")
mylist
mylist = ["apple", "banana", "orange"]
mylist.insert(3, "watermelon")
mylist
mylist = ["apple", "banana", "orange"]
mylist.remove("banana")
mylist
mylist = ["apple", "banana", "orange"]
mylist.pop(2)
mylist
mylist = ["apple", "banana", "orange"]
mylist.clear()
mylist
mylist = ["apple", "banana", "orange"]
len(mylist)
mylist = ["apple", "banana", "orange"]
for x in mylist:
print(x)
mylist = ["apple", "banana", "orange"]
for i in range(len(mylist)):
print(mylist[i])
mylist = ["apple", "banana", "orange"]
i = 0
while i < len(mylist):
print(mylist[i])
i = i + 1
mylist = ["apple", "banana", "orange"]
[print(x) for x in mylist]
mylist = ["apple", "apple", "banana", "orange"]
mylist.sort()
mylist
mylist = ["10", "23", "17", "1"]
mylist.sort()
mylist
mylist = ["10", "23", "17", "1"]
mylist.sort(reverse=True)
mylist
mylist = ["apple", "apple", "banana", "orange"]
mylist.sort(reverse=True)
mylist
mylist = ["apple", "apple", "banana", "orange"]
mylist1 = mylist
mylist1
mylist = ["apple", "apple", "banana", "orange"]
mylist.count("banana")
mytuple = ("apple", "banana", "orange")
len(mytuple)
mytuple = ("apple", "banana", "orange")
mytuple[2]
mytuple = ("apple", "banana", "orange")
mytuple[1:3]
mytuple = ("apple", "banana", "orange", "mango")
(x, y, z, a) = mytuple
z
mytuple = ("apple", "banana", "orange", "mango")
mylist = list(mytuple)
mylist[1] = "cherry"
mytuple = tuple(mylist)
mytuple
mytuple = ("apple", "banana", "orange", "mango")
mytuple.index("banana")
# mytuple.count("banana")
thislist = [2, 4, 6]
thislist
thislist = {10, 2, 4, 6}
thislist
thisset = {"apple", "orange", "banana"}
thisset
thisset = {"apple", "orange", "banana"}
len(thisset)
thisset = {"apple", "orange", "banana"}
type(thisset)
thisset = ["apple", "orange", "banana"]
type(thisset)
thisset = {"apple", False}
thisset
thisset = set(("apple", "orange", "banana"))
thisset
thisset = set(("apple", "orange", "banana"))
for x in thisset:
print(x)
thisset = set(("apple", "orange", "banana"))
"appled" in thisset
thisset = set(("apple", "orange", "banana"))
thisset.add("mango")
thisset.add("jackfruit")
thisset.remove("jackfruit")
thisset.pop()
thisset.clear()
thisset
thisdict = {1: "A", 2: "B", 3: "C", 4: ["D", "E"]}
thisdict
thisdict = {1: "A", 2: "B", 3: "C", 4: ["D", "E"]}
thisdict[3]
thisdict = {"id1": "A", "id2": "B", "id3": "C", "id4": ["D", "E"]}
if "id1" in thisdict:
print("present")
else:
print("Absent")
thisdict = {"id1": "A", "id2": "B", "id3": "C", "id4": ["D", "E"]}
thisdict.pop("id1")
thisdict.clear()
thisdict
thisdict = {"id1": "A", "id2": "B", "id3": "C", "id4": ["M", "N"]}
for x in thisdict.keys():
print(x)
|
import pandas as pd
pd.plotting.register_matplotlib_converters()
import matplotlib.pyplot as plt
import seaborn as sns
print("Setup Complete")
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import matplotlib as mpl
# In this Notebook, I will be analysing the progress of the rollout of COVID-19 vaccines in countries around the globe and answering the question:
# > >Which country out of the following has the highest percentage of their population vaccinated?<
# Path of the file to read
country_filepath = "../input/vaccinations-export/country_vaccinations 2.csv"
# Fill in the line below to read the file into a variable ign_data
country_data = pd.read_csv(country_filepath, index_col="country")
# Print the data
country_data
# Above I have been able to create a formula in excel with help, to be able to sort the orginina data (covid-world-vaccination-progress) by having the most recent rows for each country in an individual csv file.
import pandas as pd
pd.plotting.register_matplotlib_converters()
import matplotlib.pyplot as plt
import seaborn as sns
print("Setup Complete")
# Path of the file to read
country_filepath = "../input/country-export-3/country_vaccinations 3.csv"
# Fill in the line below to read the file into a variable ign_data
country_data = pd.read_csv(country_filepath, index_col="country")
# text-size
plt.figure(figsize=(12, 8))
fontsize = 16
ax = plt.axes()
ax.set_facecolor("#FAF1F6") # Setting the background color by spec
# graph-title
plt.title("Total Vaccinations Per Hundred")
# Heatmap showing total number of vaccinations for the first 5 countries
sns.barplot(
x=country_data.index,
y=country_data["total_vaccinations_per_hundred"],
palette="Blues_d",
)
sns.set_style("whitegrid")
# x-axis label
plt.xlabel("countries")
plt.ylabel("vaccinations")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import seaborn as sns
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from tensorflow import keras
from tensorflow.keras import layers
from tensorflow.keras.models import Sequential, load_model
from tensorflow.keras.layers import (
Dense,
Flatten,
Conv2D,
MaxPooling2D,
BatchNormalization,
)
from tensorflow.keras.preprocessing import image
from tensorflow.keras.callbacks import EarlyStopping
from tensorflow.keras.preprocessing.image import ImageDataGenerator
print("packages are imported")
training_folder = "/kaggle/input/dogs-cats-images/dataset/training_set"
testing_folder = "/kaggle/input/dogs-cats-images/dataset/test_set"
training_human_folder = "/kaggle/input/human-faces"
train = []
for folder in os.listdir(training_folder):
print(folder)
folder_path = os.path.join(training_folder, folder)
for img_i in os.listdir(folder_path):
img_path = os.path.join(folder_path, img_i)
train.append([img_path, folder])
# train.append([img_path,0 if folder=="cats" else 1])
for folder in os.listdir(training_human_folder):
print(folder)
folder_path = os.path.join(training_human_folder, folder)
for img_i in os.listdir(folder_path):
img_path = os.path.join(folder_path, img_i)
train.append([img_path, folder])
test = []
for folder in os.listdir(testing_folder):
folder_path = os.path.join(testing_folder, folder)
for img_i in os.listdir(folder_path):
img_path = os.path.join(folder_path, img_i)
test.append([img_path, folder])
# test.append([img_path,0 if folder=="cats" else 1])
train = pd.DataFrame(train, columns=["img_path", "species"])
test = pd.DataFrame(test, columns=["img_path", "species"])
train.tail()
test.head()
img = image.load_img(train.iloc[0, 0])
fig = plt.figure(figsize=(10, 10))
h, w = 4, 4
r, c = h, w
ax = []
for i in range(1, r * c + 1):
k = np.random.randint(train.shape[0])
img = image.load_img(train.iloc[k, 0])
ax.append(fig.add_subplot(r, c, i))
ax[-1].set_title(train.iloc[k, 1])
plt.imshow(img)
plt.show()
train_datagen = ImageDataGenerator(
rescale=1.0 / 255,
horizontal_flip=True,
vertical_flip=True,
zoom_range=0.2,
shear_range=0.2,
rotation_range=10,
validation_split=0.2,
)
test_datagen = ImageDataGenerator(rescale=1.0 / 255)
training_set = train_datagen.flow_from_dataframe(
train,
x_col="img_path",
y_col="species",
target_size=(64, 64),
batch_size=32,
class_mode="categorical",
shuffle=True,
subset="training",
)
validation_set = train_datagen.flow_from_dataframe(
train,
x_col="img_path",
y_col="species",
target_size=(64, 64),
batch_size=32,
class_mode="categorical",
shuffle=True,
subset="validation",
)
testing_set = test_datagen.flow_from_dataframe(
test,
x_col="img_path",
y_col="species",
target_size=(64, 64),
class_mode="categorical",
shuffle=False,
batch_size=32,
)
from tensorflow.keras.layers import Dropout
model = Sequential()
model.add(
Conv2D(32, (3, 3), activation="relu", padding="same", input_shape=(64, 64, 3))
)
model.add(MaxPooling2D())
model.add(
Conv2D(64, (3, 3), activation="relu", padding="same", input_shape=(64, 64, 3))
)
model.add(MaxPooling2D())
model.add(
Conv2D(128, (3, 3), activation="relu", padding="same", input_shape=(64, 64, 3))
)
model.add(MaxPooling2D())
model.add(
Conv2D(128, (3, 3), activation="relu", padding="same", input_shape=(64, 64, 3))
)
model.add(MaxPooling2D())
# model.add(BatchNormalization())
model.add(Flatten())
model.add(Dense(units=256, activation="relu"))
# model.add(Dropout(rate=0.2))
model.add(Dense(3, activation="sigmoid"))
model.layers
model.compile(optimizer="adam", loss="categorical_crossentropy", metrics=["accuracy"])
model.summary()
early_stopping = EarlyStopping(min_delta=0.001, patience=7, restore_best_weights=True)
predict = model.fit(
training_set,
epochs=10,
# steps_per_epoch = 20,
# callbacks = [early_stopping],
validation_data=validation_set,
)
accuracy = model.evaluate(validation_set)
print(accuracy[1] * 100)
history_df = pd.DataFrame(predict.history)
history_df.loc[:, ["loss", "val_loss"]].plot()
history_df.loc[:, ["accuracy", "val_accuracy"]].plot()
Y_pred = model.predict(testing_set)
# Y_pred[Y_pred<0.5] = 0
# Y_pred[Y_pred>=0.5] = 1
# classes = testing_set.classes
Y_pred
y_pred = np.argmax(Y_pred, axis=1)
print(*y_pred)
from sklearn.metrics import classification_report, confusion_matrix, accuracy_score
print(classification_report(classes, Y_pred, target_names=["cats", "dogs"]))
model.save("cathumandog.h5")
|
# Importing the required libraries
import pandas as pd
import numpy as np
import shap
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import LabelEncoder
from sklearn.ensemble import RandomForestClassifier
import re
import string
import nltk
# Loading the MBTI dataset
df = pd.read_csv("/kaggle/input/sample/mbti_cleaned.csv")
df = df.loc[1:100, :]
df.shape
df.head()
df.drop(["Number of posts"], axis=1)
df2 = df.drop(["Unnamed: 0"], axis=1)
df2.head()
df3 = df2.drop(["Number of posts"], axis=1)
df3.head()
le = LabelEncoder()
df3["type"] = le.fit_transform(df3["type"])
df3.head()
from sklearn.model_selection import train_test_split
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.feature_extraction.text import TfidfTransformer
df3.dropna(inplace=True)
# x_train.isna().sum()
x_train, x_test, y_train, y_test = train_test_split(
df3["Posts"], df3["type"], random_state=0
)
c_v = CountVectorizer(stop_words="english")
X_train_counts = c_v.fit_transform(x_train)
tfidf_transformer = TfidfTransformer()
X_train_tfidf = tfidf_transformer.fit_transform(X_train_counts)
X_train_tfidf.shape
from sklearn.svm import LinearSVC
clf = LinearSVC().fit(X_train_tfidf, y_train)
y_pred = clf.predict(c_v.transform(x_test))
explainer = shap.LinearExplainer(clf, X_train_tfidf)
shap_values = explainer.shap_values(c_v.transform(x_test))
# Visualize the SHAP values for a single instance
shap.initjs()
X_test = c_v.transform(x_test)
X_test = pd.DataFrame(X_test)
# shap.force_plot(explainer.expected_value, shap_values[0][0,:], X_test.iloc[0,:])
print(shap_values[0][0, :])
print(X_test)
X_test = c_v.transform(x_test)
shap.summary_plot(shap_values[0], X_test)
|
import numpy as np
import pandas as pd
iris = pd.read_csv("iris.csv")
iris.head()
iris.drop(["Id"], axis=1, inplace=True)
iris.head()
X = iris.drop("Species", axis=1)
# X.head()
y = iris["Species"]
# y
y.unique()
from sklearn.preprocessing import LabelBinarizer
encoder = LabelBinarizer()
y = encoder.fit_transform(y)
# y
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=42
)
from sklearn.preprocessing import MinMaxScaler
scaler = MinMaxScaler()
scaler.fit(X_train)
scaled_X_train = scaler.transform(X_train)
scaled_X_test = scaler.transform(X_test)
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Conv2D, MaxPool2D, Flatten, Dense, Dropout
model = Sequential()
model.add(
Dense(
4,
activation="relu",
input_shape=[
4,
],
)
)
model.add(Dense(3, activation="softmax"))
model.compile(optimizer="adam", loss="categorical_crossentropy", metrics=["accuracy"])
from tensorflow.keras.callbacks import EarlyStopping
early_stop = EarlyStopping(patience=10)
model.fit(
x=scaled_X_train,
y=y_train,
epochs=300,
validation_data=(scaled_X_test, y_test),
callbacks=[early_stop],
)
metrics = pd.DataFrame(model.history.history)
metrics[["loss", "val_loss"]].plot()
metrics[["accuracy", "val_accuracy"]].plot()
model.evaluate(scaled_X_test, y_test, verbose=1)
epochs = len(metrics)
epochs
scaled_X = scaler.fit_transform(X)
model = Sequential()
model.add(
Dense(
4,
activation="relu",
input_shape=[
4,
],
)
)
model.add(Dense(3, activation="softmax"))
model.compile(optimizer="adam", loss="categorical_crossentropy", metrics=["accuracy"])
model.fit(scaled_X, y, epochs=epochs) # 95 %
model.save("final_iris_model.h5")
import joblib
joblib.dump(scaler, "iris_scaler.pkl")
from tensorflow.keras.models import load_model
flower_model = load_model("final_iris_model.h5")
flower_scaler = joblib.load("iris_scaler.pkl")
|
# [Original notebook](https://github.com/mikesmales/Udacity-ML-Capstone) bu notebooktan yardım alınarak hazırlanmıştır.
# # **UrbanSound dataset**
# * Air Conditioner
# * Car Horn
# * Children Playing
# * Dog bark
# * Drilling
# * Engine Idling
# * Gun Shot
# * Jackhammer
# * Siren
# * Street Music
# Bu ses alıntıları, `.wav` formatındaki dijital ses dosyalarıdır.
# Ses dalgaları, örnekleme hızı olarak bilinen ayrı aralıklarla örneklenerek sayısallaştırılır (tipik olarak CD kalitesinde ses için 44.1 kHz, yani örnekler saniyede 44.100 kez alınır).
# Her örnek, belirli bir zaman aralığındaki dalganın genliğidir; burada bit derinliği, örneğin sinyalin dinamik aralığı olarak da bilinen ne kadar ayrıntılı olacağını belirler (tipik olarak 16 bit, bu, bir örneğin 65,536 genlik değerlerinden değişebileceği anlamına gelir).
# Ses analizi için aşağıdaki kitaplıkları kullanacağız:
# 1. **IPython.display.Audio**
# Bu, sesi doğrudan Jupyter Not Defterinde çalmamızı sağlar.
#
# 2. **Librosa**
# librosa, Brian McFee tarafından müzik ve ses işleme için bir Python paketidir ve not defterimize analiz ve manipülasyon için uyuşmuş bir dizi olarak ses yüklememizi sağlar.
# Ses dosyalarını çalmak için ``IPython.display.Audio`` kullanacağız, böylece işitsel olarak inceleyebiliriz.
# # 1. Data Exploration and Visualisation
import IPython.display as ipd
# Air Conditioner
ipd.Audio("../input/urbansounds/UrbanSound8K/audio/fold9/101729-0-0-18.wav")
# Car Horn
ipd.Audio("../input/urbansounds/UrbanSound8K/audio/fold10/100648-1-3-0.wav")
# Children Playing
ipd.Audio("../input/urbansounds/UrbanSound8K/audio/fold10/101382-2-0-20.wav")
# Dog bark
ipd.Audio("../input/urbansounds/UrbanSound8K/audio/fold1/101415-3-0-2.wav")
# Drilling
ipd.Audio("../input/urbansounds/UrbanSound8K/audio/fold3/103199-4-0-3.wav")
# Engine Idling
ipd.Audio("../input/urbansounds/UrbanSound8K/audio/fold10/102857-5-0-12.wav")
# Gun Shot
ipd.Audio("../input/urbansounds/UrbanSound8K/audio/fold1/102305-6-0-0.wav")
# Jackhammer
ipd.Audio("../input/urbansounds/UrbanSound8K/audio/fold1/103074-7-0-1.wav")
# Siren
ipd.Audio("../input/urbansounds/UrbanSound8K/audio/fold2/102871-8-0-12.wav")
# Street Music
ipd.Audio("../input/urbansounds/UrbanSound8K/audio/fold7/101848-9-0-2.wav")
# Her sınıftan bir örnek yükleyeceğiz ve verileri herhangi bir kalıp için görsel olarak inceleyeceğiz. Ses dosyasını bir diziye yüklemek için librosa'yı daha sonra dalga biçimini görüntülemek için librosa.display ve matplotlib'i kullanacağız.
import IPython.display as ipd
import librosa
import librosa.display
import matplotlib.pyplot as plt
# Class: Air Conditioner
filename = "../input/urbansounds/UrbanSound8K/audio/fold9/101729-0-0-40.wav"
plt.figure(figsize=(12, 4))
data, sample_rate = librosa.load(filename)
_ = librosa.display.waveplot(data, sr=sample_rate)
ipd.Audio(filename)
# **x(data)** → ses zaman serisi
# **sr(sample rate)** →ses frekansı(Hz)
# Aynı zamanda sesi yüklerken ses frekansını değiştirerek yükleyebilirsiniz.
# Class: Car horn
filename = "../input/urbansounds/UrbanSound8K/audio/fold8/107090-1-1-0.wav"
plt.figure(figsize=(12, 4))
data, sample_rate = librosa.load(filename)
_ = librosa.display.waveplot(data, sr=sample_rate)
ipd.Audio(filename)
# Class: Children playing
filename = "../input/urbansounds/UrbanSound8K/audio/fold10/101382-2-0-20.wav"
plt.figure(figsize=(12, 4))
data, sample_rate = librosa.load(filename)
_ = librosa.display.waveplot(data, sr=sample_rate)
ipd.Audio(filename)
# Class: Dog bark
filename = "../input/urbansounds/UrbanSound8K/audio/fold6/105319-3-0-29.wav"
plt.figure(figsize=(12, 4))
data, sample_rate = librosa.load(filename)
_ = librosa.display.waveplot(data, sr=sample_rate)
ipd.Audio(filename)
# Class: Drilling
filename = "../input/urbansounds/UrbanSound8K/audio/fold6/107842-4-2-0.wav"
plt.figure(figsize=(12, 4))
data, sample_rate = librosa.load(filename)
_ = librosa.display.waveplot(data, sr=sample_rate)
ipd.Audio(filename)
# Class: Engine Idling
filename = "../input/urbansounds/UrbanSound8K/audio/fold6/106486-5-0-2.wav"
plt.figure(figsize=(12, 4))
data, sample_rate = librosa.load(filename)
_ = librosa.display.waveplot(data, sr=sample_rate)
ipd.Audio(filename)
# Class: Gunshot
filename = "../input/urbansounds/UrbanSound8K/audio/fold1/102305-6-0-0.wav"
plt.figure(figsize=(12, 4))
data, sample_rate = librosa.load(filename)
_ = librosa.display.waveplot(data, sr=sample_rate)
ipd.Audio(filename)
# Class: Jackhammer
filename = "../input/urbansounds/UrbanSound8K/audio/fold1/103074-7-3-1.wav"
plt.figure(figsize=(12, 4))
data, sample_rate = librosa.load(filename)
_ = librosa.display.waveplot(data, sr=sample_rate)
ipd.Audio(filename)
# Class: Siren
filename = "../input/urbansounds/UrbanSound8K/audio/fold7/102853-8-0-1.wav"
plt.figure(figsize=(12, 4))
data, sample_rate = librosa.load(filename)
_ = librosa.display.waveplot(data, sr=sample_rate)
ipd.Audio(filename)
# Class: Street music
filename = "../input/urbansounds/UrbanSound8K/audio/fold4/107653-9-0-18.wav"
plt.figure(figsize=(12, 4))
data, sample_rate = librosa.load(filename)
_ = librosa.display.waveplot(data, sr=sample_rate)
ipd.Audio(filename)
# # **Gözlemler**
# * Görsel bir incelemeden, bazı sınıflar arasındaki farkı görselleştirmenin zor olduğunu görebiliriz.
# * Özellikle klima, delme, motor rölantisi ve kırıcı için tekrarlanan sesler için dalga formları benzer şekildedir.
# * Benzer şekilde, köpek havlama örneğindeki pik, şekil olarak silahla vurulan örneğe benzerdir (örnekler, bir köpek havlaması için bir pik ile karşılaştırıldığında iki silah atışı için iki pik olması bakımından farklılık gösterse de). Ayrıca, araba kornası da benzerdir.
# * Sokak müziği ile oynayan çocuklar arasında da benzerlikler vardır.
# * İnsan kulağı, harmonikler arasındaki farkı doğal olarak algılayabilir, bir derin öğrenme modelinin bu sınıfları birbirinden ayırmak için gerekli özellikleri ne kadar iyi çıkarabileceğini görmek ilginç olacaktır.
# * Bununla birlikte, dalga şekli şeklinden, köpek havlaması ve kırıcı gibi belirli sınıflar arasındaki farkı ayırt etmek kolaydır.
# # Dataset Metadata
# Burada UrbanSound meta veri .csv dosyasını Panda veri çerçevesine yükleyeceğiz.
import pandas as pd
metadata = pd.read_csv("../input/urbansounds/UrbanSound8K/metadata/UrbanSound8K.csv")
metadata = metadata.rename(columns={"class": "class_name"})
metadata.head(10)
# # Class distributions
#
print(metadata.class_name.value_counts())
# Burada Sınıf etiketlerinin dengesiz olduğunu görebiliriz. 10 sınıftan 7'sinin tam olarak 1000 örneği olmasına ve siren 929 ile çok uzakta olmamasına rağmen, geri kalan iki tanesi (car_horn, gun_shot) sırasıyla% 43 ve% 37 ile önemli ölçüde daha az örneğe sahiptir.
# # Audio sample file properties
# ses örnek dosyalarının her birini yineleyip, ses kanallarının sayısını, örnekleme hızını ve bit derinliğini çıkaracağız.
import struct
class WavFileHelper:
def read_file_properties(self, filename):
wave_file = open(filename, "rb")
riff = wave_file.read(12)
fmt = wave_file.read(36)
num_channels_string = fmt[10:12]
num_channels = struct.unpack("<H", num_channels_string)[0]
sample_rate_string = fmt[12:16]
sample_rate = struct.unpack("<I", sample_rate_string)[0]
bit_depth_string = fmt[22:24]
bit_depth = struct.unpack("<H", bit_depth_string)[0]
return (num_channels, sample_rate, bit_depth)
# Load various imports
import pandas as pd
import os
import librosa
import librosa.display
wavfilehelper = WavFileHelper()
audiodata = []
for index, row in metadata.iterrows():
file_name = os.path.join(
os.path.abspath("../input/urbansounds/UrbanSound8K/audio/"),
"fold" + str(row["fold"]) + "/",
str(row["slice_file_name"]),
)
data = wavfilehelper.read_file_properties(file_name)
audiodata.append(data)
# Convert into a Panda dataframe
audiodf = pd.DataFrame(audiodata, columns=["num_channels", "sample_rate", "bit_depth"])
# # Audio channels
# Örneklerin çoğu iki ses kanalına (stereo anlamında) sahiptir ve birkaçı sadece tek kanallı (mono).
# Onları tekdüze hale getirmenin en kolay yolu, iki kanalın değerlerinin ortalamasını alarak, steroid örneklerindeki iki kanalı bir kanalda birleştirmek olacaktır.
# num of channels
print(audiodf.num_channels.value_counts(normalize=True))
# # Sample rate
# Endişe verici olan tüm numunelerde kullanılan çok çeşitli Numune oranları vardır (96k ila 8k arasında değişen).
# Bu benzer şekilde, bir örnekleme oranı dönüştürme tekniği (yukarı dönüştürme veya aşağı dönüştürme) uygulamamız gerektiği anlamına gelir, böylece adil bir karşılaştırma yapmamıza olanak sağlayacak dalga biçimlerinin agnostik bir temsilini görebiliriz.
# sample rates
print(audiodf.sample_rate.value_counts(normalize=True))
# # Bit-depth
# Ayrıca çok çeşitli bit derinlikleri vardır. Belirli bir bit derinliği için maksimum ve minimum genlik değerlerini alarak bunları normalleştirmemiz gerekebilir.
# bit depth
print(audiodf.bit_depth.value_counts(normalize=True))
# # 2. Data Preprocessing and Data Splitting
# **Normalleştirme gerektiren ses özellikleri**
# * Audio Channels
# * Sample rate
# * Bit-depth
# Ön işleme ve özellik çıkarma için faydalı olacak Librosa'yı kullanmaya devam edeceğiz.
# ****Sample rate conversion****
# Varsayılan olarak, Librosa’nın yük işlevi, örnekleme oranını karşılaştırma düzeyimiz olarak kullanabileceğimiz 22,05 KHz’e dönüştürür.
import librosa
from scipy.io import wavfile as wav
import numpy as np
filename = "../input/urbansounds/UrbanSound8K/audio/fold7/101848-9-0-3.wav"
librosa_audio, librosa_sample_rate = librosa.load(filename)
scipy_sample_rate, scipy_audio = wav.read(filename)
print("Original sample rate:", scipy_sample_rate)
print("Librosa sample rate:", librosa_sample_rate)
ipd.Audio(filename)
# **Bit-depth**
# Librosa’nın yükleme işlevi de verileri normalleştirerek değerleri -1 ile 1 arasında değişir. Bu, çok çeşitli bit derinliklerine sahip veri kümesinin karmaşıklığını ortadan kaldırır.
print(
"Original audio file min~max range:", np.min(scipy_audio), "to", np.max(scipy_audio)
)
print(
"Librosa audio file min~max range:",
np.min(librosa_audio),
"to",
np.max(librosa_audio),
)
# **Merge audio channels**
# Librosa ayrıca sinyali monoya dönüştürecektir, yani kanal sayısı her zaman 1 olacaktır.
import matplotlib.pyplot as plt
# Original audio with 2 channels
plt.figure(figsize=(12, 4))
plt.plot(scipy_audio)
# Librosa audio with channels merged
plt.figure(figsize=(12, 4))
plt.plot(librosa_audio)
# **Dikkate alınacak diğer ses özellikleri**
# Bu aşamada, numune süresi uzunluğu ve hacim seviyeleri gibi başka faktörlerin de hesaba katılması gerekip gerekmediği henüz net değildir.
# Bu arada olduğu gibi devam edeceğiz ve hedef ölçümlerimizin geçerliliğini etkilediği düşünülürse bunları daha sonra ele almak için geri geleceğiz.
# # Extract Features
# MFCC, pencere boyutu boyunca frekans dağılımını özetler, böylece sesin hem frekans hem de zaman özelliklerini analiz etmek mümkündür. Bu sesli temsiller, sınıflandırma için özellikleri belirlememize izin verecektir.
# Bunun için, zaman serisi ses verilerinden bir MFCC oluşturan ``Librosa'nın mfcc ()`` işlevini kullanacağız.
mfccs = librosa.feature.mfcc(y=librosa_audio, sr=librosa_sample_rate, n_mfcc=40)
print(mfccs.shape)
# Bu, librosa'nın 173 çerçeve üzerinde bir dizi 40 MFCC hesapladığını göstermektedir.
import librosa.display
librosa.display.specshow(mfccs, sr=librosa_sample_rate, x_axis="time")
# **Her dosya için MFCC'lerin çıkarılması**
# Şimdi veri kümesindeki her ses dosyası için bir MFCC çıkaracağız ve sınıflandırma etiketiyle birlikte bir Panda Veri Çerçevesinde saklayacağız.
def extract_features(file_name):
try:
audio, sample_rate = librosa.load(file_name, res_type="kaiser_fast")
mfccs = librosa.feature.mfcc(y=audio, sr=sample_rate, n_mfcc=40)
mfccsscaled = np.mean(mfccs.T, axis=0)
except Exception as e:
print("Error encountered while parsing file: ", file)
return None
return mfccsscaled
# Load various imports
import pandas as pd
import os
import librosa
from tqdm import tqdm
# Set the path to the full UrbanSound dataset
fulldatasetpath = "../input/urbansounds/UrbanSound8K/audio"
metadata = pd.read_csv("../input/urbansounds/UrbanSound8K/metadata/UrbanSound8K.csv")
metadata = metadata.rename(columns={"class": "class_name"})
features = []
# Iterate through each sound file and extract the features
for index, row in tqdm(metadata.iterrows()):
file_name = os.path.join(
os.path.abspath(fulldatasetpath),
"fold" + str(row["fold"]) + "/",
str(row["slice_file_name"]),
)
class_label = row["class_name"]
data = extract_features(file_name)
features.append([data, class_label])
# Convert into a Panda dataframe
featuresdf = pd.DataFrame(features, columns=["feature", "class_label"])
print("Finished feature extraction from ", len(featuresdf), " files")
featuresdf.head()
# **Convert the data and labels**
# Kategorik metin verilerini model tarafından anlaşılabilir sayısal verilere kodlamak için ``sklearn.preprocessing.LabelEncoder``'ı kullanacağız.
from sklearn.preprocessing import LabelEncoder
from keras.utils import to_categorical
# Convert features and corresponding classification labels into numpy arrays
X = np.array(featuresdf.feature.tolist())
y = np.array(featuresdf.class_label.tolist())
# Encode the classification labels
le = LabelEncoder()
yy = to_categorical(le.fit_transform(y))
# # Split the dataset
# Burada veri setini eğitim ve test setlerine ayırmak için sklearn.model_selection.train_test_split kullanacağız. Test seti boyutu% 20 olacak ve rastgele bir durum belirleyeceğiz.
# split the dataset
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(
X, yy, test_size=0.2, random_state=42
)
# # 3. Model Training and Evaluation
# ### Initial model architecture - MLP
# * Keras ve Tensorflow arka uç kullanarak Çok Katmanlı Algılayıcı (MLP) Sinir Ağı oluşturmaya başlayacağız.
# * Üç katman, bir giriş katmanı, bir gizli katman ve bir çıktı katmanından oluşan basit bir model mimarisiyle başlayacağız.Üç katmanın tamamı, çoğu durumda sinir ağları için kullanılan standart bir katman türü olan yoğun katman türünde olacaktır.
# * İlk katman giriş şeklini alacaktır. Her örnek 40 MFCC (veya sütun) içerdiğinden (1x40) şeklinde bir şekle sahibiz, bu da 40 giriş şekli ile başlayacağımız anlamına gelir.
# * İlk iki katman 256 düğüme sahip olacaktır. İlk 2 katmanımız için kullanacağımız aktivasyon fonksiyonu ReLU veya Doğrultulmuş Doğrusal Aktivasyondur. Bu aktivasyon işlevinin sinir ağlarında iyi çalıştığı kanıtlanmıştır.
# * Ayrıca ilk iki katmanımıza% 50'lik bir Bırakma değeri uygulayacağız. Bu, düğümleri her güncelleme döngüsünden rasgele hariç tutacak ve bu da, daha iyi genelleme yapabilen ve eğitim verilerini daha düşük olasılıkla daha az genelleştirebilen bir ağ ile sonuçlanacaktır.
# * Çıktı katmanımız, olası sınıflandırmaların sayısıyla eşleşen 10 düğüme (sayı_etiketler) sahip olacaktır.Aktivasyon, çıktı katmanımızın softmax olması içindir. Softmax, çıktı toplamını 1'e çıkarır, böylece çıktı olasılık olarak yorumlanabilir. Model daha sonra tahminini hangi seçeneğin en yüksek olasılığa sahip olduğuna göre yapacaktır.
import numpy as np
from keras.models import Sequential
from keras.layers import Dense, Dropout, Activation, Flatten
from keras.layers import Convolution2D, MaxPooling2D
from keras.optimizers import Adam
from keras.utils import np_utils
from sklearn import metrics
num_labels = yy.shape[1]
filter_size = 2
# Construct model
model = Sequential()
model.add(Dense(256, input_shape=(40,)))
model.add(Activation("relu"))
model.add(Dropout(0.5))
model.add(Dense(256))
model.add(Activation("relu"))
model.add(Dropout(0.5))
model.add(Dense(num_labels))
model.add(Activation("softmax"))
# # Compiling the model
# Modelimizi derlemek için aşağıdaki üç parametreyi kullanacağız:
# * Kayıp işlevi - categorical_crossentropy kullanacağız. Bu, sınıflandırma için en yaygın seçimdir. Daha düşük bir puan, modelin daha iyi performans gösterdiğini gösterir.
# * Metrikler - Modeli eğitirken doğrulama verilerindeki doğruluk puanını görüntülememizi sağlayacak doğruluk ölçüsünü kullanacağız.
# * Optimizer - burada birçok kullanım durumu için genellikle iyi bir optimize edici olan Adam'ı kullanacağız.
# Compile the model
model.compile(loss="categorical_crossentropy", metrics=["accuracy"], optimizer="adam")
# Display model architecture summary
model.summary()
# Calculate pre-training accuracy
score = model.evaluate(x_test, y_test, verbose=0)
accuracy = 100 * score[1]
print("Pre-training accuracy: %.4f%%" % accuracy)
# # Training
from keras.callbacks import ModelCheckpoint
from datetime import datetime
num_epochs = 100
num_batch_size = 32
checkpointer = ModelCheckpoint(
filepath="saved_models/weights.best.basic_mlp.hdf5", verbose=1, save_best_only=True
)
start = datetime.now()
model.fit(
x_train,
y_train,
batch_size=num_batch_size,
epochs=num_epochs,
validation_data=(x_test, y_test),
callbacks=[checkpointer],
verbose=1,
)
duration = datetime.now() - start
print("Training completed in time: ", duration)
# # Test the model
# Burada hem eğitim hem de test veri setlerinde modelin doğruluğunu gözden geçireceğiz.
# Evaluating the model on the training and testing set
score = model.evaluate(x_train, y_train, verbose=0)
print("Training Accuracy: ", score[1])
score = model.evaluate(x_test, y_test, verbose=0)
print("Testing Accuracy: ", score[1])
# # Predictions
import librosa
import numpy as np
def extract_feature(file_name):
try:
audio_data, sample_rate = librosa.load(file_name, res_type="kaiser_fast")
mfccs = librosa.feature.mfcc(y=audio_data, sr=sample_rate, n_mfcc=40)
mfccsscaled = np.mean(mfccs.T, axis=0)
except Exception as e:
print("Error encountered while parsing file: ", file)
return None, None
return np.array([mfccsscaled])
def print_prediction(file_name):
prediction_feature = extract_feature(file_name)
predicted_vector = model.predict_classes(prediction_feature)
predicted_class = le.inverse_transform(predicted_vector)
print("The predicted class is:", predicted_class[0], "\n")
predicted_proba_vector = model.predict_proba(prediction_feature)
predicted_proba = predicted_proba_vector[0]
for i in range(len(predicted_proba)):
category = le.inverse_transform(np.array([i]))
print(category[0], "\t\t : ", format(predicted_proba[i], ".32f"))
# Class: Air Conditioner
filename = "../input/urbansounds/UrbanSound8K/audio/fold9/101729-0-0-11.wav"
print_prediction(filename)
ipd.Audio(filename)
# Class: Drilling
filename = "../input/urbansounds/UrbanSound8K/audio/fold2/104817-4-0-10.wav"
print_prediction(filename)
ipd.Audio(filename)
# Class: Street music
filename = "../input/urbansounds/UrbanSound8K/audio/fold7/101848-9-0-9.wav"
print_prediction(filename)
ipd.Audio(filename)
# Class: Car Horn
filename = "../input/urbansounds/UrbanSound8K/audio/fold8/107090-1-1-0.wav"
print_prediction(filename)
ipd.Audio(filename)
filename = "../input/urbansounds/UrbanSound8K/audio/fold9/103249-5-0-0.wav"
print_prediction(filename)
ipd.Audio(filename)
filename = "../input/urbansounds/UrbanSound8K/audio/fold10/100795-3-1-2.wav"
print_prediction(filename)
ipd.Audio(filename)
# # 4. Model Refinement
# İlk denememizde, aşağıdaki gibi bir Sınıflandırma Doğruluğu puanı elde etmeyi başardık:
# Eğitim verileri Doğruluğu:% 92,3
# Test Verisi Doğruluğu:% 87
# Şimdi bir Convolutional Neural Network (CNN) kullanarak bu puanı geliştirip geliştiremeyeceğimizi göreceğiz.
# # Feature Extraction refinement
# * Önceki özellik çıkarma aşamasında, MFCC vektörleri farklı ses dosyaları için boyut olarak değişecektir (örneklerin süresine bağlı olarak).
# * Ancak, CNN'ler tüm girişler için sabit bir boyut gerektirir. Bunun üstesinden gelmek için çıktı vektörlerini sıfırlayarak hepsini aynı boyutta yapacağız.
import numpy as np
max_pad_len = 174
def extract_features(file_name):
try:
audio, sample_rate = librosa.load(file_name, res_type="kaiser_fast")
mfccs = librosa.feature.mfcc(y=audio, sr=sample_rate, n_mfcc=40)
pad_width = max_pad_len - mfccs.shape[1]
mfccs = np.pad(mfccs, pad_width=((0, 0), (0, pad_width)), mode="constant")
except Exception as e:
print("Error encountered while parsing file: ", file_name)
return None
return mfccs
# Load various imports
import pandas as pd
import os
import librosa
# Set the path to the full UrbanSound dataset
fulldatasetpath = "../input/urbansounds/UrbanSound8K/audio"
metadata = pd.read_csv("../input/urbansounds/UrbanSound8K/metadata/UrbanSound8K.csv")
metadata = metadata.rename(columns={"class": "class_name"})
features = []
# Iterate through each sound file and extract the features
for index, row in metadata.iterrows():
file_name = os.path.join(
os.path.abspath(fulldatasetpath),
"fold" + str(row["fold"]) + "/",
str(row["slice_file_name"]),
)
class_label = row["class_name"]
data = extract_features(file_name)
features.append([data, class_label])
# Convert into a Panda dataframe
featuresdf = pd.DataFrame(features, columns=["feature", "class_label"])
print("Finished feature extraction from ", len(featuresdf), " files")
from sklearn.preprocessing import LabelEncoder
from keras.utils import to_categorical
# Convert features and corresponding classification labels into numpy arrays
X = np.array(featuresdf.feature.tolist())
y = np.array(featuresdf.class_label.tolist())
# Encode the classification labels
le = LabelEncoder()
yy = to_categorical(le.fit_transform(y))
# split the dataset
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(
X, yy, test_size=0.2, random_state=42
)
# # Convolutional Neural Network (CNN) model architecture
# * Modelimizi yine Keras ve Tensorflow arka uç kullanarak Evrişimli Sinir Ağı (CNN) olacak şekilde değiştireceğiz.
# * Evrişim katmanları, özellik tespiti için tasarlanmıştır. Girişin üzerinde bir filtre penceresi kaydırarak ve bir matris çarpımı gerçekleştirerek ve sonucu bir özellik haritasında depolayarak çalışır. Bu işlem, evrişim olarak bilinir.
# * Filter parametresi, her katmandaki düğüm sayısını belirtir. Her katmanın boyutu 16, 32, 64'ten 128'e artarken, kernel_size parametresi çekirdek penceresinin boyutunu belirtir ve bu durumda 2x2 filtre matrisiyle sonuçlanır.
# * Birinci katman, (40, 174, 1) giriş şeklini alacaktır, burada 40, MFCC'lerin sayısıdır. 174, doldurmayı hesaba katan çerçeve sayısıdır ve 1, sesin mono olduğunu belirtir.
# * Evrişimli katmanlarımız için kullanacağımız aktivasyon fonksiyonu, önceki modelimizle aynı olan ReLU'dur. Evrişimli katmanlarımızda% 20'lik daha küçük bir Bırakma değeri kullanacağız.
# * Her evrişimli katman, bir GlobalAveragePooling2D türüne sahip olan son evrişimli katmanla birlikte MaxPooling2D türünde ilişkili bir havuz katmanına sahiptir. Havuzlama katmanı, eğitim süresini kısaltmaya ve aşırı uyumu azaltmaya hizmet eden modelin boyutluluğunu (parametreleri ve ilgili hesaplama gereksinimlerini azaltarak) azaltır. Maksimum Havuzlama türü, her pencere için maksimum boyutu alır ve Genel Ortalama Havuzlama türü, yoğun çıktı katmanımıza beslemek için uygun olan ortalamayı alır.
# * Çıktı katmanımız, olası sınıflandırmaların sayısıyla eşleşen 10 düğüme (sayı_etiketler) sahip olacaktır. Aktivasyon, çıktı katmanımızın softmax olması içindir. Softmax, çıktı toplamını 1'e çıkarır, böylece çıktı olasılık olarak yorumlanabilir. Model daha sonra tahminini hangi seçeneğin en yüksek olasılığa sahip olduğuna göre yapacaktır.
import numpy as np
from keras.models import Sequential
from keras.layers import Dense, Dropout, Activation, Flatten
from keras.layers import Convolution2D, Conv2D, MaxPooling2D, GlobalAveragePooling2D
from keras.optimizers import Adam
from keras.utils import np_utils
from sklearn import metrics
num_rows = 40
num_columns = 174
num_channels = 1
x_train = x_train.reshape(x_train.shape[0], num_rows, num_columns, num_channels)
x_test = x_test.reshape(x_test.shape[0], num_rows, num_columns, num_channels)
num_labels = yy.shape[1]
filter_size = 2
# Construct model
model = Sequential()
model.add(
Conv2D(
filters=16,
kernel_size=2,
input_shape=(num_rows, num_columns, num_channels),
activation="relu",
)
)
model.add(MaxPooling2D(pool_size=2))
model.add(Dropout(0.2))
model.add(Conv2D(filters=32, kernel_size=2, activation="relu"))
model.add(MaxPooling2D(pool_size=2))
model.add(Dropout(0.2))
model.add(Conv2D(filters=64, kernel_size=2, activation="relu"))
model.add(MaxPooling2D(pool_size=2))
model.add(Dropout(0.2))
model.add(Conv2D(filters=128, kernel_size=2, activation="relu"))
model.add(MaxPooling2D(pool_size=2))
model.add(Dropout(0.2))
model.add(GlobalAveragePooling2D())
model.add(Dense(num_labels, activation="softmax"))
# ## Compiling the model
# Modelimizi derlemek için, önceki modelle aynı üç parametreyi kullanacağız:
# Compile the model
model.compile(loss="categorical_crossentropy", metrics=["accuracy"], optimizer="adam")
# Display model architecture summary
model.summary()
# Calculate pre-training accuracy
score = model.evaluate(x_test, y_test, verbose=1)
accuracy = 100 * score[1]
print("Pre-training accuracy: %.4f%%" % accuracy)
# # Training
# Burada modeli eğiteceğiz. Bir CNN eğitimi önemli miktarda zaman alabileceğinden, düşük sayıda dönem ve düşük bir parti boyutu ile başlayacağız. Modelin yakınsadığını çıktıdan görebilirsek, her iki sayıyı da artıracağız.
from keras.callbacks import ModelCheckpoint
from datetime import datetime
# num_epochs = 12
# num_batch_size = 128
num_epochs = 72
num_batch_size = 256
checkpointer = ModelCheckpoint(
filepath="saved_models/weights.best.basic_cnn.hdf5", verbose=1, save_best_only=True
)
start = datetime.now()
model.fit(
x_train,
y_train,
batch_size=num_batch_size,
epochs=num_epochs,
validation_data=(x_test, y_test),
callbacks=[checkpointer],
verbose=1,
)
duration = datetime.now() - start
print("Training completed in time: ", duration)
# # Test the model
# Burada hem eğitim hem de test veri setlerinde modelin doğruluğunu gözden geçireceğiz.
# Evaluating the model on the training and testing set
score = model.evaluate(x_train, y_train, verbose=0)
print("Training Accuracy: ", score[1])
score = model.evaluate(x_test, y_test, verbose=0)
print("Testing Accuracy: ", score[1])
# ### Predictions
def print_prediction(file_name):
prediction_feature = extract_features(file_name)
prediction_feature = prediction_feature.reshape(
1, num_rows, num_columns, num_channels
)
predicted_vector = model.predict_classes(prediction_feature)
predicted_class = le.inverse_transform(predicted_vector)
print("The predicted class is:", predicted_class[0], "\n")
predicted_proba_vector = model.predict_proba(prediction_feature)
predicted_proba = predicted_proba_vector[0]
for i in range(len(predicted_proba)):
category = le.inverse_transform(np.array([i]))
print(category[0], "\t\t : ", format(predicted_proba[i], ".32f"))
# Class: Air Conditioner
filename = "../input/urbansounds/UrbanSound8K/audio/fold9/101729-0-0-14.wav"
print_prediction(filename)
# Class: Car Horn
filename = "../input/urbansounds/UrbanSound8K/audio/fold8/107090-1-1-0.wav"
print_prediction(filename)
# Class: Street music
filename = "../input/urbansounds/UrbanSound8K/audio/fold7/101848-9-0-9.wav"
print_prediction(filename)
actual_class_list = []
predicted_class_list = []
metadata = pd.read_csv("../input/urbansounds/UrbanSound8K/metadata/UrbanSound8K.csv")
for index, row in tqdm(metadata.iterrows()):
file_name = os.path.join(
os.path.abspath(fulldatasetpath),
"fold" + str(row["fold"]) + "/",
str(row["slice_file_name"]),
)
class_id = row["classID"]
# print(file_name)
# sleep(2)
prediction_feature = extract_features(file_name)
prediction_feature = prediction_feature.reshape(
1, num_rows, num_columns, num_channels
)
predicted_vector = model.predict_classes(prediction_feature)
predicted_class = le.inverse_transform(predicted_vector)
# print("The predicted class is:", predicted_class[0], '\n')
predicted_proba_vector = model.predict_proba(prediction_feature)
predicted_proba = predicted_proba_vector[0]
predicted_class = np.argmax(predicted_proba)
predicted_class_list.append(predicted_class)
actual_class_list.append(class_id)
# print("pred--",predicted_class_list)
# print("actual--",actual_class_list)
import pandas as pd
y_actu = pd.Series(actual_class_list, name="Actual")
y_pred = pd.Series(predicted_class_list, name="Predicted")
df_confusion = pd.crosstab(y_actu, y_pred)
# # Confusion Matrix
df_confusion = pd.crosstab(
y_actu, y_pred, rownames=["Actual"], colnames=["Predicted"], margins=True
)
df_confusion
unique_labels = [
"air_conditioner",
"car_horn",
"children_playing",
"dog_bark",
"drilling",
"engine_idling",
"gun_shot",
"jackhammer",
"siren",
"street_music",
]
y_pred.shape[0]
y_pred_class = []
y_actu_class = []
for i in range(y_pred.shape[0]):
y_pred_class.append(unique_labels[y_pred[i]])
y_actu_class.append(unique_labels[y_actu[i]])
import seaborn as sns
from sklearn.metrics import confusion_matrix
conf_mat = confusion_matrix(y_actu_class, y_pred_class, labels=unique_labels)
df_cm = pd.DataFrame(conf_mat, index=unique_labels, columns=unique_labels)
plt.figure(figsize=(10, 8))
hm = sns.heatmap(df_cm, annot=True, fmt="n")
plt.title("Confusion Matrix")
# sns.heatmap(df_cm, annot=True, cmap='viridis')
hm.tick_params(labeltop=True, labelbottom=False, top=True, bottom=False)
hm.set_xlabel("Model tahmini")
hm.xaxis.set_label_position("top")
hm.set_ylabel("gerçek etiketi")
plt.show()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
# Reading the datasets
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import random
import tensorflow as tf
from keras import regularizers, optimizers
from tensorflow.keras.optimizers import RMSprop
import pandas as pd
from keras.models import Sequential
from keras.layers import Dense, Activation, Flatten, Dropout, BatchNormalization
from keras.layers import Conv2D, MaxPooling2D
from tensorflow.keras.preprocessing.image import ImageDataGenerator
traindf = pd.read_csv("../input/socc-ai-self-driving-car/train_dataset.csv")
testdf = pd.read_csv("../input/socc-ai-self-driving-car/test_dataset.csv")
datagen = ImageDataGenerator(rescale=1.0 / 255.0, validation_split=0.25)
train_generator = datagen.flow_from_dataframe(
dataframe=traindf,
directory="../input/socc-ai-self-driving-car/archive/driving_dataset/",
x_col="Image",
y_col="Angles",
subset="training",
batch_size=32,
seed=42,
shuffle=True,
class_mode="raw",
target_size=(32, 32),
)
valid_generator = datagen.flow_from_dataframe(
dataframe=traindf,
directory="../input/socc-ai-self-driving-car/archive/driving_dataset/",
x_col="Image",
y_col="Angles",
subset="validation",
batch_size=32,
seed=42,
shuffle=True,
class_mode="raw",
target_size=(32, 32),
)
test_datagen = ImageDataGenerator(rescale=1.0 / 255.0)
test_generator = test_datagen.flow_from_dataframe(
dataframe=testdf,
directory="../input/socc-ai-self-driving-car/archive/driving_dataset/",
x_col="Image",
y_col=None,
batch_size=32,
seed=42,
shuffle=False,
class_mode=None,
target_size=(32, 32),
)
model = Sequential()
model.add(Conv2D(32, (3, 3), padding="same", input_shape=(32, 32, 3)))
model.add(Activation("relu"))
model.add(Conv2D(32, (3, 3)))
model.add(Activation("relu"))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Conv2D(64, (3, 3), padding="same"))
model.add(Activation("relu"))
model.add(Conv2D(64, (3, 3)))
model.add(Activation("relu"))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Flatten())
model.add(Dense(512))
model.add(Activation("relu"))
model.add(Dropout(0.5))
# model.add(Dense(10, activation='softmax'))
model.add(Dense(1, activation="linear"))
from keras.optimizers import RMSprop
model.compile(
optimizers.RMSprop(lr=0.0001, decay=1e-6),
loss="mean_squared_error",
metrics=["accuracy"],
)
STEP_SIZE_TRAIN = train_generator.n // train_generator.batch_size
STEP_SIZE_VALID = valid_generator.n // valid_generator.batch_size
STEP_SIZE_TEST = test_generator.n // test_generator.batch_size
model.fit(
train_generator,
steps_per_epoch=STEP_SIZE_TRAIN,
validation_data=valid_generator,
validation_steps=STEP_SIZE_VALID,
epochs=3,
)
model.evaluate(valid_generator, steps=STEP_SIZE_TEST)
test_generator.reset()
pred = model.predict(test_generator)
testdf.head()
submit = pd.DataFrame()
submit["Image"] = testdf["Image"]
submit["Angles"] = pred
submit
submit.to_csv("sample_submission.csv", index=False)
|
# # Project 4 Description
# #### The goal:
# Simulate a forecast production environment and get an estimation on the model's drift, using data from projet 2
# #### The method:
# - First train a model and forecast a 7 days horizon everyday during a certain period. Everyday the model is retraind with one more point. Observe model performance during that period using the RMSSE metric.
# - Second: Train the same model only the the first day and don't retrain the model. Use the model to forecast the whole period and observe how the error metric evolves
# #### The implementation:
# I have built a custom cross validation function that estimates the model error over consecutive folds, averaging the error to get a final score for the whole period. It works for both refit and no refit scenarios
# # Import libraries
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
plt.style.use("fivethirtyeight")
import lightgbm as lgb
from sklearn.preprocessing import OrdinalEncoder
# # Load files
data_path = "..\\..\\data\\project_2_data\\"
data = pd.read_parquet(f"{data_path}sales_data.parquet")
calendar = pd.read_parquet(f"{data_path}calendar.parquet")
prices = pd.read_parquet(f"{data_path}prices.parquet")
data.shape
# # Data transformation
# QUESTION: filter out products that don't have sales using cumsum
data = data.loc[data.groupby("id").sales.cumsum() > 0]
data.shape
# # Functions
# RMSEE Metric
def rmsse(train, val, y_pred):
train_scale = (
train.assign(
scale=train.groupby("id").sales.diff() ** 2,
)
.groupby("id")
.scale.mean()
)
score = (
val.assign(squared_error=(val.sales - y_pred) ** 2)
.groupby("id")
.squared_error.mean()
.to_frame()
.merge(train_scale, on="id")
.assign(rmsse=lambda x: np.sqrt(x.squared_error / x.scale))
.rmsse.mean()
)
return score
# Feature engineering functions
def calc_lag(df, shift_length, forecast_horizon, by_day_of_week=False):
group_cols = ["id"]
if by_day_of_week:
group_cols += ["day_of_week"]
feature_name = f"lag_{shift_length}_{forecast_horizon}"
return (
df.assign(day_of_week=df.index.get_level_values("date").dayofweek)
.groupby(group_cols)
.sales.shift(forecast_horizon + shift_length)
.rename(feature_name)
), feature_name
def calc_rolling_agg(
df, window_length, forecast_horizon, agg_func="mean", by_day_of_week=False
):
group_cols = ["id"]
if by_day_of_week:
group_cols += ["day_of_week"]
if not by_day_of_week:
feature_name = f"rolling_{agg_func}_{window_length}_{forecast_horizon}"
else:
feature_name = f"seasonal_rolling_{agg_func}_{window_length}_{forecast_horizon}"
return (
df.assign(day_of_week=df.index.dayofweek)
.groupby(group_cols, group_keys=False)
.sales.rolling(
window_length, closed="right", min_periods=1
) # only requires 1 observation to be non-NaN
.agg({"sales": agg_func})
.reset_index()
.assign(date=lambda x: x.date + pd.Timedelta(days=28))
.set_index("date")
.rename(columns={"sales": feature_name})
), feature_name
def custom_feature_engineering(df, horizon):
cont_feats = []
for lag in lag_features:
fe_table, feature_name = calc_lag(df, lag, horizon)
df = df.merge(fe_table, on=["id", "date"], how="left")
cont_feats.append(feature_name)
df = df.reset_index("id")
for agg_func, windows in rolling_features.items():
for window in windows:
fe_table, feature_name = calc_rolling_agg(df, window, horizon, agg_func)
df = df.merge(fe_table, on=["id", "date"], how="left")
cont_feats.append(feature_name)
for agg_func, windows in seasonal_rolling_features.items():
for window in windows:
fe_table, feature_name = calc_rolling_agg(
df, window, horizon, agg_func, by_day_of_week=True
)
df = df.merge(
fe_table.drop(columns="day_of_week"), on=["id", "date"], how="left"
)
cont_feats.append(feature_name)
df = (
df.merge(calendar[["snap_TX"]], on="date", how="left")
.merge(prices, on=["date", "store_id", "item_id"], how="left")
.assign(
day_of_week=lambda x: x.index.dayofweek,
day_of_month=lambda x: x.index.day,
month=lambda x: x.index.month,
year=lambda x: x.index.year,
)
)
cont_feats += ["sell_price", "day_of_week", "day_of_month", "month", "year"]
cat_feats = ["id", "item_id", "dept_id", "cat_id", "store_id", "snap_TX"]
enc_cat_feats = [f"{feat}_enc" for feat in cat_feats]
df[enc_cat_feats] = OrdinalEncoder().fit_transform(df[cat_feats])
max_date = df.index.get_level_values("date").max()
train = df.loc[: max_date - pd.Timedelta(days=horizon), :]
val = df.loc[max_date - pd.Timedelta(days=horizon - 1) :, :]
price_feats = train.groupby("id").agg(
max_price=("sell_price", "max"),
median_price=("sell_price", "median"),
)
train = train.merge(price_feats, on="id", how="left")
val = val.merge(price_feats, on="id", how="left")
cont_feats += ["max_price", "median_price"]
return (train, val, cont_feats, enc_cat_feats, val[["id"]])
# Model training and prediction
def weighted_rmse(preds, data):
squared_error = (preds - data.get_label()) ** 2
value = np.mean(np.sqrt(squared_error / data.get_weight()))
return "rmsse", value, False
def train_model(data, horizon):
"""
The Function gets a dataset, and trains the model on all data except the last horizon days
For instance if horizon is 1 week, it will split the dataset in a lengh-7 training datset and will predict the last 7 days
"""
params = dict(
objective="tweedie",
metric="None",
tweedie_variance_power=1.1,
learning_rate=0.05,
min_samples_leaf=100,
subsample=0.3,
feature_fraction=0.3,
deterministic=True,
)
train, val, cont_feats, cat_feats, _ = custom_feature_engineering(data, horizon)
train_scale = (
train.assign(
scale=train.groupby("id").sales.diff() ** 2,
)
.groupby("id")
.scale.mean()
)
val = val.merge(train_scale, on="id")
train_dset = lgb.Dataset(
train[cont_feats + cat_feats],
train["sales"],
)
val_dset = lgb.Dataset(
val[cont_feats + cat_feats],
val["sales"],
weight=val["scale"],
)
callbacks = [lgb.early_stopping(100), lgb.log_evaluation(50)]
model = lgb.train(
params,
train_dset,
num_boost_round=1000,
valid_sets=[val_dset],
callbacks=callbacks,
feval=weighted_rmse,
)
return model, val[cont_feats + cat_feats], val[["sales", "id"]]
def predict_horizon(val, model, horizon):
pred = model.predict(val)
return pred
# Cross validation
def cross_val(df, folds_number, horizon, refit=True):
fcst = []
actual = []
date = []
error_rmsse = []
initial_fit_to_be_done = True
pred = None
model = None
max_date = df.index.get_level_values("date").max()
# val = df.loc[max_date - pd.Timedelta(days=folds_number-1):, :]
for i in range(0, folds_number):
date_fold = max_date - pd.Timedelta(days=folds_number - i)
train = df.loc[:date_fold, :]
if refit or initial_fit_to_be_done:
model, val, y = train_model(train, horizon)
initial_fit_to_be_done = False
pred = predict_horizon(val, model, horizon)
val = val.join(y)
else:
trained, val, cont_feats, cat_feats, y = custom_feature_engineering(
train, horizon
)
pred = predict_horizon(val[cont_feats + cat_feats], model, horizon)
# val = val.join(y)
test_rmsse = rmsse(train, val, pred)
fcst.append(pred) # forecasting horizon steps into the future
actual.append(val.sales) # comparing that to actual value at that point
date.append(date_fold) # saving date of that value
error_rmsse.append(test_rmsse)
print((f"RMSSE Estimation by CV: {np.mean(error_rmsse):.2f}\n"))
return (error_rmsse, fcst, actual, date)
# # Production simulation
lag_features = [1, 7, 28]
rolling_features = {
"mean": [7, 28],
"std": [7, 28],
}
seasonal_rolling_features = {
"mean": [4, 8],
"std": [4, 8],
}
horizon = 7
folds_number = 10
cvrefit = cross_val(data, folds_number, horizon, refit=True)
cvnorefit = cross_val(data, folds_number, horizon, refit=False)
plt.plot(cvrefit[0], label="cvrefit")
plt.plot(cvnorefit[0], label="cvnorefit")
plt.legend()
|
# # ASL Alphabet Mediapipe Images
# This notebook is a modified version of the Colab notebook given here so that it can be viewed on Kaggle.
# https://colab.research.google.com/drive/1FvH5eTiZqayZBOHZsFm-i7D-JvoB9DVz#scrollTo=nW2TjFyhLvVH
# https://github.com/google/mediapipe
import cv2
import os
import math
import random
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import mediapipe as mp
mp_hands = mp.solutions.hands
mp_drawing = mp.solutions.drawing_utils
mp_drawing_styles = mp.solutions.drawing_styles
paths = []
for dirname, _, filenames in os.walk("/kaggle/input/asl-alphabet-test"):
for filename in filenames:
if filename[-4:] == ".jpg":
paths += [(os.path.join(dirname, filename))]
print(paths[0:3])
labels2 = []
paths2 = []
for i, path in enumerate(paths):
if i % 50 == 0:
print("i=", i)
file = path.split("/")[-1]
label = path.split("/")[-2]
image = cv2.imread(path)
with mp_hands.Hands(
static_image_mode=True, max_num_hands=2, min_detection_confidence=0.1
) as hands:
results = hands.process(cv2.flip(image, 1))
if results.multi_hand_landmarks:
image_hight, image_width, _ = image.shape
annotated_image = cv2.flip(image.copy(), 1)
for hand_landmarks in results.multi_hand_landmarks:
mp_drawing.draw_landmarks(
annotated_image,
hand_landmarks,
mp_hands.HAND_CONNECTIONS,
mp_drawing_styles.get_default_hand_landmarks_style(),
mp_drawing_styles.get_default_hand_connections_style(),
)
anno_img = cv2.flip(annotated_image, 1)
cv2.imwrite(file, anno_img)
paths2 += [file]
labels2 += [label]
data = pd.DataFrame(columns=["path", "label"])
data["path"] = paths2
data["label"] = labels2
data.to_csv("data.csv", index=False)
display(data)
selected_num = random.sample(range(len(data)), 9)
fig, axes = plt.subplots(3, 3, figsize=(10, 10))
for i, ax in enumerate(axes.flat):
j = selected_num[i]
img_path = data.iloc[j, 0]
label = data.iloc[j, 1]
img = plt.imread(img_path)
ax.imshow(img)
ax.axis("off")
ax.set_title(label)
plt.tight_layout()
plt.show()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Goal is to segment healthy food for life
# ### This notebook devided into two parts
# > 1. Collecting labels `unsuperwised learning`
# > 2. Segmenting data into `healthy food and unhealthy food`
# ### Basic preprocessing
pd.set_option("display.max_column", None)
df = pd.read_csv("/kaggle/input/fastfood-nutrition/fastfood.csv")
df.head(3)
df.shape
df.info()
null_vals = dict(df.isnull().sum())
null_vals
# % null values
for key, val in null_vals.items():
print(f"null values for {key} =======> {(int(val)/df.shape[0])*100}")
# replace vitamin a ,c and calcium with mean value
null_cols = ["fiber", "protein", "vit_a", "vit_c", "calcium"]
null_cols_avg = {}
for col in null_cols:
null_cols_avg[col] = df[col].describe().mean()
null_cols_avg
df.fillna(value=null_cols_avg, inplace=True)
df.isnull().sum()
# Oh cleared it, now ready to go
df.drop("salad", axis=1, inplace=True)
# # 1) Collecting labels `unsuperwised learning`
# ### Steps followed:
# * Collecting libraries
# * Dropping unrequired columns
# * Finding best cluster
# * Visualizing elbow method
# * Training model with best cluster
# * Getting labels
# collecting libraries required
from sklearn.cluster import KMeans
from sklearn.model_selection import train_test_split
import matplotlib.pyplot as plt
df_seg = df.drop(["restaurant", "item"], axis="columns")
df_seg.sample(3)
# ### ----> Finding best cluster size for segmentation
inertias = []
for i in range(1, 10):
model = KMeans(n_clusters=i, init="k-means++", random_state=42)
model.fit(df_seg)
inertias.append(model.inertia_)
print(inertias)
# ### ---- > Elbow method
plt.plot(inertias)
plt.title("inertia of Knn model")
plt.xlabel("number of clusters")
plt.ylabel("inertia")
plt.show()
# >As per elbow method 3 types of clusters are best for our model, that means we can classify foods into 3 types.
model = KMeans(n_clusters=3, init="k-means++")
model.fit(df_seg)
cluster_centers = model.cluster_centers_
cluster_centers
labels = model.labels_
labels[:10]
df["labels"] = labels
df.sample(3)
# #### Done first part 😌
# # 2) Segmenting data into `healthy food and unhealthy food`
import seaborn as sn
label_0 = df[df["labels"] == 0]
label_1 = df[df["labels"] == 1]
label_2 = df[df["labels"] == 2]
print("label 0", len(label_0))
print("label 1", len(label_1))
print("label 2", len(label_2))
# ### ----> Collecting average of Nutrition
nutritions = list(label_0.describe().columns)
label_0_nutri_avg = label_0.describe().mean().values
label_1_nutri_avg = label_1.describe().mean().values
label_2_nutri_avg = label_2.describe().mean().values
print("label_0_nutri_avg", label_0_nutri_avg)
print("label_1_nutri_avg", label_1_nutri_avg)
print("label_2_nutri_avg", label_2_nutri_avg)
# ### ----> Visualization
# * label 0
# * label 1
# * label 2
# label 0
sn.barplot(x=nutritions, y=label_0_nutri_avg)
plt.title("Nutritions in average for label 0")
plt.xticks(rotation=90)
plt.ylim([0, 3000])
plt.show()
# label 1
sn.barplot(x=nutritions, y=label_1_nutri_avg)
plt.title("Nutritions in average for label 1")
plt.xticks(rotation=90)
plt.ylim([0, 3000])
plt.show()
# label 2
sn.barplot(x=nutritions, y=label_2_nutri_avg)
plt.title("Nutritions in average for label 2")
plt.xticks(rotation=90)
plt.ylim([0, 3000])
plt.show()
|
# In this 1st cell we establish several useful libraries used for data analysis
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
from sklearn.tree import DecisionTreeClassifier
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
from sklearn.model_selection import cross_val_score
from sklearn.metrics import plot_roc_curve
import sklearn.metrics as metrics
from sklearn.svm import SVR
import matplotlib.pyplot as plt
train_data = pd.read_csv(r"../input/dataset/train.csv")
test_data = pd.read_csv(r"../input/dataset/test.csv")
# Now we sum all null values to see if we have any missing data
train_data.isnull().sum()
# In the cell below we use a 1.5 * IQR to determine our outliers
def remove_outliers(feature, name, dataset):
Q1, Q3 = np.percentile(feature, [25, 75])
IQR = Q3 - Q1
lower_range = Q1 - (1.5 * IQR)
upper_range = Q3 + (1.5 * IQR)
dataset.drop(feature[(feature > upper_range) | (feature < lower_range)].index)
return dataset
# In this cell we call our function 'remove_outliers' and iterate throughout all of the columns
for column in train_data:
remove_outliers(train_data[column], column, train_data)
# Now we use the describe function to display the data associated with our new dataset.
train_data.describe()
# In the cell below we
x, y = train_data.drop(["id", "Bankrupt"], axis=1), train_data["Bankrupt"]
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=1 / 3, random_state=4, stratify=y
)
# "You must determine whether or not you will implement normalization or standardization, and explain your decision." From the research I've done it seems that both aren't really necessary for decision trees. I've found several sources that normalization isn't very beneficial due to it being a comparison function.
# We create our decision tree
dt = DecisionTreeClassifier(
criterion="gini",
splitter="best",
max_depth=None,
min_samples_split=2,
min_samples_leaf=1,
random_state=42,
)
dt.fit(x_train, y_train)
dt_acc = dt.score(x_test, y_test)
dt_cv = cross_val_score(dt, x_test, y_test).mean()
dt_f1 = metrics.f1_score(y_test, dt.predict(x_test))
dt_acc
dt_cv
dt_f1
test_data_kaggy = pd.read_csv(r"../input/dataset/test.csv")
model = dt
model.fit(x, y)
model_ouput.to_csv("modeloutput.csv")
|
import networkx as nx
import os
import random
import matplotlib.pyplot as plt
from matplotlib.pyplot import figure
import numpy as np
figure(figsize=(8, 6), dpi=80)
# # Input Utils
"""
method : get_graph()
input: file path for graph (.gr)
output: netrworkX graph object
"""
def get_graph(file):
G = nx.Graph()
# nx.set_node_attributes(G, 0, "red")
for i in file.readlines():
l = i[: len(i) - 1].split(" ")
if len(l) == 2:
a = int(l[0])
b = int(l[1])
# G.add_node(a, red = 0)
# G.add_node(b, red = 0)
G.add_edge(a, b, weight=0) # weight 0 means black edge, 1 is red
return G
"""
method : get_sol()
input: file path of the solution (.tww)
output: optimal contraction sequence
"""
def get_sol(file):
ls = []
for l in file.readlines():
if l[-1] == "\n":
i = l[: len(l) - 1].split(" ")
else:
i = l.split(" ")
try:
ls.append((int(i[0]), int(i[1])))
except:
print(l)
print(file)
return ls
# # Get Graphs and Contraction Sequence (True Labels)
#
"""
Dictionary := Graphs{name : (graph, sequence)}
"""
Graphs = {}
root = "/kaggle/input/tiny-set/TINY SET/"
g_dir = "tiny-set/"
sol_dir = "tiny-set-sol/"
for dirs in os.listdir(root + g_dir):
dirs = dirs[-6:-3]
g_path = root + g_dir + "tiny" + dirs + ".gr"
sol_path = root + sol_dir + "tiny" + dirs + ".tww"
g_file = open(g_path)
sol_file = open(sol_path)
Graphs[dirs] = (get_graph(g_file), get_sol(sol_file))
# # Visualize Trigraphs
def draw(G):
black = [(u, v) for (u, v, d) in G.edges(data=True) if d["weight"] == 0]
red = [(u, v) for (u, v, d) in G.edges(data=True) if d["weight"] == 1]
pos = nx.kamada_kawai_layout(G)
nx.draw_networkx_nodes(
G, pos, node_size=350, node_color="#044a14", alpha=0.4
) # nodes only
nx.draw_networkx_edges(
G, pos, edgelist=black, width=1, alpha=0.5, edge_color="black", style="solid"
)
nx.draw_networkx_edges(
G, pos, edgelist=red, width=1, alpha=0.7, edge_color="red", style="solid"
)
nx.draw_networkx_labels(G, pos, font_size=12, font_family="sans-serif")
plt.show()
draw(Graphs["001"][0])
print(Graphs["001"][1])
# # Contract pair of vertices
G = Graphs["001"][0].copy()
draw(G)
def get_reds(G, u, v):
black = set()
total = set(G[u]).union(set(G[v])) - {u, v}
for n in total:
if G.has_edge(u, n) and G[u][n]["weight"] == 0:
black = black.union({n})
reds = total - black
return reds
def max_red_degree(G, ls):
ls = list(ls)
max_d = 0
max_n = 0
for v in ls:
d = 0
for n in G[v]:
if G[v][n]["weight"] == 1:
d += 1
if d >= max_d:
max_d = d
max_n = v
return (max_n, max_d)
def contract(G, u, v):
reds = get_reds(G, u, v)
G.remove_node(v)
for n in reds:
G.add_edge(u, n, weight=1)
reds = reds.union(u)
return max_red_degree(G, reds)
def common_black_neighbors(G, u, v):
set(G[u]).union(set(G[v]))
s = set()
for n in G[u]:
if G[u][n]["weight"] == 1:
s = s.union({n})
return s
def max_red_degree(G, ls):
ls = list(ls)
max_d = 0
max_n = 0
for v in ls:
d = 0
for n in G[v]:
if G[v][n]["weight"] == 1:
d += 1
if d >= max_d:
max_d = d
max_n = v
return (max_n, max_d)
def contract(G, lst):
u = lst[0]
v = lst[1]
"""
Outputs the symmetric difference of 2 nodes' neighbors
"""
def sym_diff(G, u, v):
c = {u, v}
a = set(G[u])
b = set(G[v])
return ((a - b).union(b - a)) - c
"""
Outputs the red neighbors of a node
"""
def red_neighbors(G, u):
s = set()
for n in G[u]:
if G[u][n]["weight"] == 1:
s = s.union({n})
return s
"""
returns node with the highest red degree
(node, red_degree)
"""
def max_red_(G, ls):
ls = list(ls)
max_d = 0
max_n = 0
for v in ls:
d = 0
for n in G[v]:
if G[v][n]["weight"] == 1:
d += 1
if d >= max_d:
max_d = d
max_n = v
return (max_n, max_d)
"""
Performs contraction on a pair of verices, u = lst[0] with v = lst[1]
Outputs the maximum Red Degree of G after contraction
"""
def contract(G, lst):
u = lst[0]
v = lst[1]
reds = sym_diff(G, u, v).union(red_neighbors(G, u)) - {v}
new = len(set(G[u]) - reds)
G.remove_node(v)
for n in reds:
G.add_edge(u, n, weight=1)
# draw(G)
return max_red_(G, reds.union({u}))
def run(f):
G = Graphs[f][0].copy()
sol = Graphs[f][1]
for s in sol:
contract(G, s)
run("005")
# # Greedy Contraction
def tww_greedy(G, ls):
if len(G.nodes) == 1:
return ls
d = 99999
sol = (0, 0)
for u in G.nodes:
for v in G.nodes:
if u != v:
t = contract(G.copy(), (u, v))[1]
if t <= d:
t = d
sol = (u, v)
ls.append(sol)
contract(G, (sol[0], sol[1]))
return tww_greedy(G, ls)
# # Fitness Calculation
def indi_fitness(G, individual):
temp = G.copy()
d = 0
marks = [0] * len(individual)
for i in range(len(individual)):
marks[i] = contract(temp, individual[i])[1]
d = max(d, marks[i])
return (d, marks)
def fitness_calc(G, population):
fitness = {}
for i in range(len(population)):
fitness[i] = indi_fitness(G, population[i])
return fitness
c = 1
for g in Graphs:
print(c)
G1 = Graphs[g][0].copy()
a = tww_greedy(G1, [])
print(a)
G2 = Graphs[g][0].copy()
print(indi_fitness(G2, a))
c += 1
print()
def rand_sols(G, k):
population = []
while k > 0:
V = set(G.nodes)
individual = []
while len(V) > 2:
u = random.sample(V, 1)
cand = V.intersection(set(G[u]))
if len(cand) == 0:
v = random.sample(V - {u}, 1)
else:
v = random.sample(cand, 1)
individual.append((u, v))
V = V - {v}
population.append(individual)
k -= 1
return population
G = Graphs["005"][0].copy()
draw(G)
K = rand_sols(G, 200)
F = fitness_calc(G, K)
for f in F:
print(F[f])
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# импортируем библиотеки для визуализации
import matplotlib.pyplot as plt
import seaborn as sns
# Загружаем специальный удобный инструмент для разделения датасета:
from sklearn.model_selection import train_test_split
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# всегда фиксируйте RANDOM_SEED, чтобы ваши эксперименты были воспроизводимы!
RANDOM_SEED = 42
# зафиксируем версию пакетов, чтобы эксперименты были воспроизводимы:
# Подгрузим наши данные из соревнования
DATA_DIR = "/kaggle/input/sf-booking/"
df_train = pd.read_csv(DATA_DIR + "/hotels_train.csv") # датасет для обучения
df_test = pd.read_csv(DATA_DIR + "hotels_test.csv") # датасет для предсказания
sample_submission = pd.read_csv(DATA_DIR + "/submission.csv") # самбмишн
# # Знакомство с данными
df_train.info()
df_train.head(2)
df_test.info()
df_test.head(2)
sample_submission.head(2)
sample_submission.info()
# Объединение train и test
# ВАЖНО! для корректной обработки признаков объединяем трейн и тест в один датасет
df_train["sample"] = 1 # помечаем где у нас трейн
df_test["sample"] = 0 # помечаем где у нас тест
df_test[
"reviewer_score"
] = 0 # в тесте у нас нет значения reviewer_score, мы его должны предсказать, по этому пока просто заполняем нулями
data = df_test.append(df_train, sort=False).reset_index(drop=True) # объединяем
data.info()
# Заполнение пропущенных значений
data.isnull().sum()
# Все пропущенные значения относятся к координатам отелей. Узнаем каких именно:
maska1 = data["lat"].isnull()
maska2 = data["lng"].isnull()
hotels_coordinates_0 = data[maska1 | maska2]["hotel_name"].value_counts().index.tolist()
print(hotels_coordinates_0)
# Количество безкоординатных отелей относительно невелико, можно поискать и внести координаты вручную, найдя их в интернете с помощью GoogleMaps
# 41.394188617785076, 2.1671410618725604
data.loc[
data["hotel_name"] == "Fleming s Selection Hotel Wien City", ["lat", "lng"]
] = ["48.209788", "16.353445"]
data.loc[data["hotel_name"] == "Hotel City Central", ["lat", "lng"]] = [
"48.213723",
"16.379833",
]
data.loc[data["hotel_name"] == "Hotel Atlanta", ["lat", "lng"]] = [
"48.220525",
"16.355819",
]
data.loc[
data["hotel_name"] == "Maison Albar Hotel Paris Op ra Diamond", ["lat", "lng"]
] = ["48.875423", "2.323349"]
data.loc[data["hotel_name"] == "Hotel Daniel Vienna", ["lat", "lng"]] = [
"48.188886",
"16.383812",
]
data.loc[data["hotel_name"] == "Hotel Pension Baron am Schottentor", ["lat", "lng"]] = [
"48.216943",
"16.359895",
]
data.loc[
data["hotel_name"] == "Austria Trend Hotel Schloss Wilhelminenberg Wien",
["lat", "lng"],
] = ["48.219734", "16.285581"]
data.loc[
data["hotel_name"] == "Derag Livinghotel Kaiser Franz Joseph Vienna", ["lat", "lng"]
] = ["48.246045", "16.341789"]
data.loc[data["hotel_name"] == "NH Collection Barcelona Podium", ["lat", "lng"]] = [
"41.391749",
"2.177926",
]
data.loc[data["hotel_name"] == "City Hotel Deutschmeister", ["lat", "lng"]] = [
"49.490316",
"9.772127",
]
data.loc[data["hotel_name"] == "Hotel Park Villa", ["lat", "lng"]] = [
"49.203925",
"9.231614",
]
data.loc[data["hotel_name"] == "Cordial Theaterhotel Wien", ["lat", "lng"]] = [
"48.209728",
"16.351474",
]
data.loc[data["hotel_name"] == "Holiday Inn Paris Montmartre", ["lat", "lng"]] = [
"48.889071",
"2.333142",
]
data.loc[data["hotel_name"] == "Roomz Vienna", ["lat", "lng"]] = [
"48.222918",
"16.393487",
]
data.loc[data["hotel_name"] == "Mercure Paris Gare Montparnasse", ["lat", "lng"]] = [
"48.840145",
"2.323485",
]
data.loc[data["hotel_name"] == "Renaissance Barcelona Hotel", ["lat", "lng"]] = [
"41.394189",
"2.167141",
]
data.loc[data["hotel_name"] == "Hotel Advance", ["lat", "lng"]] = [
"41.379389",
"2.157475",
]
data.info()
# Изменим тип координат
data["lat"] = data["lat"].astype("float64")
data["lng"] = data["lng"].astype("float64")
data.info()
# Пропусков больше нет, тип lat, lng числовой
hotels_reviewer_score_hist = sns.histplot(hotels_train["reviewer_score"], bins=30)
hotels_reviewer_score_hist.set_title("Распределение оценок")
plt.rcParams["figure.figsize"] = (15, 10)
sns.heatmap(data.drop(["sample"], axis=1).corr(), annot=True)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import seaborn as sns
import matplotlib.pyplot as plt
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
import plotly.io as pio
import plotly.express as px
import plotly.graph_objects as go
import plotly.figure_factory as ff
from plotly.subplots import make_subplots
from plotly.validators.scatter.marker import SymbolValidator
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv(
"../input/stroke-prediction-dataset/healthcare-dataset-stroke-data.csv"
)
df.isna().mean()
# ### The only bmi has missingdata. But, It is only 4%. So, I will drop this missingdata
df = df.dropna()
df.drop("id", axis=1, inplace=True)
str_col = ["gender", "ever_married", "work_type", "Residence_type", "smoking_status"]
for i in str_col:
print(i, ":", df[i].nunique())
df
df["gender"].value_counts()
df
def make_bar(feature, rows, cols):
tmp2 = df.groupby(feature)["stroke"].mean()
tmp = 1 - df.groupby(feature)["stroke"].mean().values
fig.add_trace(
go.Bar(
x=df[feature].value_counts().index,
y=df[feature].value_counts().values * tmp,
text=tmp,
texttemplate="Not_Stroke : %{text:.2f}",
textposition="auto",
),
rows,
cols,
)
fig.add_trace(
go.Bar(
x=df[feature].value_counts().index,
y=df[feature].value_counts().values
* df.groupby(feature)["stroke"].mean().values,
text=tmp2,
texttemplate="Stroke : %{text:.2f}",
textposition="auto",
),
rows,
cols,
)
fig = make_subplots(
rows=4,
cols=2,
shared_yaxes=True,
subplot_titles=("Relation with Sex", "Relation with hypertension"),
)
make_bar("gender", 1, 1)
make_bar("hypertension", 1, 2)
make_bar("heart_disease", 2, 1)
make_bar("ever_married", 2, 2)
make_bar("work_type", 3, 1)
make_bar("Residence_type", 3, 2)
make_bar("smoking_status", 4, 1)
fig.update_layout(
barmode="stack",
xaxis=dict(title="Sex"),
yaxis=dict(title="Count"),
xaxis2=dict(title="hypertension"),
yaxis2=dict(title="Count"),
)
fig.update_layout(height=3000, showlegend=False)
fig.show()
|
# # Project 4
data_dir = "/kaggle/input/project-1-data/data"
sampled = False
path_suffix = "" if not sampled else "_sampled"
import pandas as pd
import numpy as np
import pandas as pd
import polars as pl
import time
import gc
import datetime as dt
start_time = time.time()
transactions = pd.read_csv(f"{data_dir}/transactions_data{path_suffix}.csv")
transactions["date"] = pd.to_datetime(transactions["date"])
data = (
transactions.assign(date=lambda df: pd.to_datetime(df.date.dt.date))
.pipe(lambda df: df.groupby(list(df.columns))["id"].count())
.reset_index(name="sales")
)
data = data.assign(
id=lambda df: df.id.astype("category"),
item_id=lambda df: df.item_id.astype("category"),
cat_id=lambda df: df.cat_id.astype("category"),
store_id=lambda df: df.store_id.astype("category"),
state_id=lambda df: df.state_id.astype("category"),
dept_id=lambda df: df.dept_id.astype("category"),
)
print(
f"Time taken to load transactions data with Pandas: {time.time() - start_time:.2f} seconds"
)
import polars as pl
import time
start_time = time.time()
# Read the transactions data into a Polars DataFrame
transactions = pl.read_csv(f"{data_dir}/transactions_data{path_suffix}.csv")
# Aggregate the data by grouping on all columns and counting the "id" column
data = (
transactions.groupby(list(transactions.columns))
.agg(pl.count("id").alias("sales"))
.select(pl.col("*"), pl.col("sales").alias("id_count"))
)
# Convert categorical columns to categorical data type
data = data.with_columns(
{
col: data[col].cast(pl.Categorical)
for col in [
"id",
"item_id",
"cat_id",
"store_id",
"state_id",
"dept_id",
]
}
)
# Print the time taken to load the transactions data using Polars
print(
f"Time taken to load transactions data with Polars: {time.time() - start_time:.2f} seconds"
)
|
# for fine tuning for bert i have refered the following repository availabel on hugging face:
# https://github.com/google-research/bert
# and few other online tutorials
#
import os
import re
import json
import string
import numpy as np
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
from tokenizers import BertWordPieceTokenizer
from transformers import BertTokenizer, TFBertModel, BertConfig
# this is the maximum length allowed for the full input sequence
max_len = 384
configuration = BertConfig()
print(configuration)
from tqdm import tqdm as tqdm
# calling and saving tokenizer
called_tokenizer = BertTokenizer.from_pretrained("bert-base-uncased")
save_path = "bert_base_uncased/"
if not os.path.exists(save_path):
os.makedirs(save_path)
called_tokenizer.save_pretrained(save_path)
tokenizer = BertWordPieceTokenizer("bert_base_uncased/vocab.txt", lowercase=True)
# urls for trainind and testing data of the custom dataset
train_data_url = "https://drive.google.com/file/d/1NxecIfmCcZOimIApj8l_EJ2rB_FdvnXn/view?usp=share_link"
train_path = keras.utils.get_file("train.json", train_data_url)
eval_data_url = "https://drive.google.com/file/d/1JxnDpDi4N08Qf-il3IPtEcleZ1Xg9T5U/view?usp=share_link"
eval_path = keras.utils.get_file("eval.json", eval_data_url)
# #urls for trainind and testing data for Squad v1.1 Dataser
# train_data_url = "https://rajpurkar.github.io/SQuAD-explorer/dataset/train-v1.1.json"
# train_path = keras.utils.get_file("train.json", train_data_url)
# eval_data_url = "https://rajpurkar.github.io/SQuAD-explorer/dataset/dev-v1.1.json"
# eval_path = keras.utils.get_file("eval.json", eval_data_url)
# loading json files into the notebook
with open(train_path) as f:
raw_train_data = json.load(f)
with open(eval_path) as f:
raw_eval_data = json.load(f)
def create_vector(size, start, end):
ans_vector = [0] * size
for idx in range(start, end):
ans_vector[idx] = 1
return ans_vector
class AnnotatedExample:
def __init__(self, question, context, start_char_idx, answer_text, all_answers):
self.question = question
self.context = context
self.start_char_idx = start_char_idx
self.answer_text = answer_text
self.all_answers = all_answers
self.skip = False
def preprocess(self):
context = self.context
question = self.question
answer_text = self.answer_text
start_char_idx = self.start_char_idx
# converting everything into string
context = " ".join(str(context).split())
question = " ".join(str(question).split())
answer = " ".join(str(answer_text).split())
# ending index of the answer
end_indx = start_char_idx + len(answer)
if end_indx >= len(context):
self.skip = True
return
vector_ans_char = create_vector(len(context), start_char_idx, end_indx)
# tokenizing the context and questions
tokenized_context = tokenizer.encode(context)
tokenized_question = tokenizer.encode(question)
ans_token_idx = []
for idx, (start, end) in enumerate(tokenized_context.offsets):
if sum(vector_ans_char[start:end]) > 0:
ans_token_idx.append(idx)
if len(ans_token_idx) == 0:
self.skip = True
return
start_token_idx = ans_token_idx[0]
end_token_idx = ans_token_idx[-1]
input_ids = tokenized_context.ids + tokenized_question.ids[1:]
token_type_ids = [0] * len(tokenized_context.ids) + [1] * len(
tokenized_question.ids[1:]
)
attention_mask = [1] * len(input_ids)
# padding the rest of the tokenized vectors to max lenght of 384
padding_length = max_len - len(input_ids)
if padding_length > 0: # pad
input_ids = input_ids + ([0] * padding_length)
attention_mask = attention_mask + ([0] * padding_length)
token_type_ids = token_type_ids + ([0] * padding_length)
elif padding_length < 0:
self.skip = True
return
self.input_ids = input_ids
self.token_type_ids = token_type_ids
self.attention_mask = attention_mask
self.start_token_idx = start_token_idx
self.end_token_idx = end_token_idx
self.context_token_to_char = tokenized_context.offsets
# preprocessing the annotated examples from the dataset
def create_annotated_examples(raw_data):
annotated_examples = []
for item in raw_data["data"]:
for para in item["paragraphs"]:
context = para["context"]
for qa in para["qas"]:
question = qa["question"]
answer_text = qa["answers"][0]["text"]
all_answers = [_["text"] for _ in qa["answers"]]
start_char_idx = qa["answers"][0]["answer_start"]
annotated_eg = AnnotatedExample(
question, context, start_char_idx, answer_text, all_answers
)
annotated_eg.preprocess()
annotated_examples.append(annotated_eg)
return annotated_examples
# creating vectors to be passed to nueral network
def create_inputs_targets(annotated_examples):
dataset_dict = {
"input_ids": [],
"token_type_ids": [],
"attention_mask": [],
"start_token_idx": [],
"end_token_idx": [],
}
for item in annotated_examples:
if item.skip == False:
for key in dataset_dict:
dataset_dict[key].append(getattr(item, key))
for key in dataset_dict:
dataset_dict[key] = np.array(dataset_dict[key])
#
x = [
dataset_dict["input_ids"],
dataset_dict["token_type_ids"],
dataset_dict["attention_mask"],
]
y = [dataset_dict["start_token_idx"], dataset_dict["end_token_idx"]]
return x, y
train_annotated_examples = create_annotated_examples(raw_train_data)
x_train, y_train = create_inputs_targets(train_annotated_examples)
print(f"{len(train_annotated_examples)} training points created.")
eval_annotated_examples = create_annotated_examples(raw_eval_data)
x_eval, y_eval = create_inputs_targets(eval_annotated_examples)
print(f"{len(eval_annotated_examples)} evaluation points created.")
x_train[0].shape
raw_eval_data
x_train[0]
def create_model():
# Load pre-trained BERT model
encoder = TFBertModel.from_pretrained("bert-base-uncased")
# Exclude pooler layer from optimization
encoder.layers[-1].pooler.trainable = False
# Define input layers
input_ids = layers.Input(shape=(max_len,), dtype=tf.int32)
token_type_ids = layers.Input(shape=(max_len,), dtype=tf.int32)
attention_mask = layers.Input(shape=(max_len,), dtype=tf.int32)
# Obtain BERT embeddings
embedding = encoder(
input_ids=input_ids,
token_type_ids=token_type_ids,
attention_mask=attention_mask,
)[0]
# Define output layers
start_logits = layers.Dense(1, name="start_logit")(embedding)
end_logits = layers.Dense(1, name="end_logit")(embedding)
# Flatten output layers
start_logits = layers.Flatten()(start_logits)
end_logits = layers.Flatten()(end_logits)
# Apply activation functions to output layers
start_probs = layers.Activation(keras.activations.softmax, name="start_prob")(
start_logits
)
end_probs = layers.Activation(keras.activations.softmax, name="end_prob")(
end_logits
)
# Define and compile model
model = keras.Model(
inputs=[input_ids, token_type_ids, attention_mask],
outputs=[start_probs, end_probs],
)
loss = keras.losses.SparseCategoricalCrossentropy(from_logits=False)
optimizer = keras.optimizers.Adam(learning_rate=5e-5)
model.compile(optimizer=optimizer, loss=[loss, loss])
return model
# If running on kaggle select TPU and unmark this cell to code
use_tpu = True
if use_tpu:
tpu = tf.distribute.cluster_resolver.TPUClusterResolver()
tf.config.experimental_connect_to_cluster(tpu)
tf.tpu.experimental.initialize_tpu_system(tpu)
strategy = tf.distribute.TPUStrategy(tpu)
with strategy.scope():
model = create_model()
else:
model = create_model()
model.summary()
# #else use this code
# model = create_model()
# model.summary()
def remove_punctuations(text):
remove = set(string.punctuation)
text = "".join(word for word in text if word not in remove)
return text
def remove_articles(text):
rex = re.compile(r"\b(a|an|the)\b", re.UNICODE)
text = re.sub(rex, " ", text)
return text
def cleansed_text(text):
text = text.lower()
text = remove_punctuations(text)
text = remove_articles(text)
text = " ".join(text.split())
return text
class ExactMatch(keras.callbacks.Callback):
def __init__(self, x_eval, y_eval):
self.x_eval = x_eval
self.y_eval = y_eval
def on_epoch_end(self, epoch, logs=None):
pred_start, pred_end = self.model.predict(self.x_eval)
count = 0
eval_examples_no_skip = [_ for _ in eval_annotated_examples if _.skip == False]
for idx, (start, end) in enumerate(zip(pred_start, pred_end)):
annotated_eg = eval_examples_no_skip[idx]
offsets = annotated_eg.context_token_to_char
start = np.argmax(start)
end = np.argmax(end)
if start >= len(offsets):
continue
pred_char_start = offsets[start][0]
if end < len(offsets):
pred_char_end = offsets[end][1]
pred_ans = annotated_eg.context[pred_char_start:pred_char_end]
else:
pred_ans = annotated_eg.context[pred_char_start:]
cleansed_pred_ans = cleansed_text(pred_ans)
cleansed_true_ans = [cleansed_text(_) for _ in annotated_eg.all_answers]
if cleansed_pred_ans in cleansed_true_ans:
count += 1
acc = count / len(self.y_eval[0])
print(f"\nepoch={epoch+1}, exact match score={acc:.2f}")
exact_match_callback = ExactMatch(x_eval, y_eval)
model.fit(
x_train,
y_train,
epochs=1,
verbose=2,
batch_size=16,
callbacks=[exact_match_callback],
)
# #if running on google colab un mark this code and change path in cp command according where you wanna save the model weights
# from google.colab import drive
# drive.mount('/content/drive')
model.save_weights("finetuned.h5")
|
# importing the libraries.
import os
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
from tensorflow.keras.models import Sequential
from tensorflow.keras.preprocessing import image
from tensorflow.keras.preprocessing.image import ImageDataGenerator
from tensorflow.keras.layers import Dense, Flatten, Conv2D, MaxPooling2D, Dropout
# defining image, batch size and seed
IMAGE_SIZE = (180, 180)
BATCH_SIZE = 128
SEED = 1337
EPOCHS = 10
# function for getting the training and validation sets.
def of_get_train_validation_set(dir_path):
train_ds, val_ds = tf.keras.utils.image_dataset_from_directory(
dir_path,
validation_split=0.2,
subset="both",
seed=SEED,
image_size=IMAGE_SIZE,
batch_size=BATCH_SIZE,
)
return train_ds, val_ds
# function for performing data augmentation.
def of_perform_augmentation():
data_augmentation = keras.Sequential(
[
layers.RandomFlip("horizontal"),
layers.RandomRotation(0.1),
]
)
plt.figure(figsize=(10, 10))
for images, _ in train_ds.take(1):
for i in range(9):
augmented_images = data_augmentation(images)
ax = plt.subplot(3, 3, i + 1)
plt.imshow(augmented_images[0].numpy().astype("uint8"))
plt.axis("off")
# Apply `data_augmentation` to the training images.
train_ds = train_ds.map(
lambda img, label: (data_augmentation(img), label),
num_parallel_calls=tf.data.AUTOTUNE,
)
# Prefetching samples in GPU memory helps maximize GPU utilization.
train_ds = train_ds.prefetch(tf.data.AUTOTUNE)
val_ds = val_ds.prefetch(tf.data.AUTOTUNE)
return train_ds, val_ds
# function for creating model.
def of_create_model(input_shape, num_classes):
inputs = keras.Input(shape=input_shape)
# Entry block
x = layers.Rescaling(1.0 / 255)(inputs)
x = layers.Conv2D(128, 3, strides=2, padding="same")(x)
x = layers.BatchNormalization()(x)
x = layers.Activation("relu")(x)
previous_block_activation = x # Set aside residual
for size in [256, 512, 728]:
x = layers.Activation("relu")(x)
x = layers.SeparableConv2D(size, 3, padding="same")(x)
x = layers.BatchNormalization()(x)
x = layers.Activation("relu")(x)
x = layers.SeparableConv2D(size, 3, padding="same")(x)
x = layers.BatchNormalization()(x)
x = layers.MaxPooling2D(3, strides=2, padding="same")(x)
# Project residual
residual = layers.Conv2D(size, 1, strides=2, padding="same")(
previous_block_activation
)
x = layers.add([x, residual]) # Add back residual
previous_block_activation = x # Set aside next residual
x = layers.SeparableConv2D(1024, 3, padding="same")(x)
x = layers.BatchNormalization()(x)
x = layers.Activation("relu")(x)
x = layers.GlobalAveragePooling2D()(x)
if num_classes == 2:
activation = "sigmoid"
units = 1
else:
activation = "softmax"
units = num_classes
x = layers.Dropout(0.5)(x)
outputs = layers.Dense(units, activation=activation)(x)
return keras.Model(inputs, outputs)
# function for training the model.
def of_train_model(model, train_ds, val_ds, model_name):
callbacks = [
keras.callbacks.ModelCheckpoint("save_at_{epoch}.keras"),
]
model.compile(
optimizer=keras.optimizers.Adam(1e-3),
loss="binary_crossentropy",
metrics=["accuracy"],
)
model.fit(
train_ds,
epochs=EPOCHS,
callbacks=callbacks,
validation_data=val_ds,
)
# Saving the model
model.save(model_name)
return model
# function for predicting test images.
def of_predict_images(model, test_dir):
# iterate over test images and make predictions
image_ids = []
predicted_labels = []
print("Starting Prediction")
# looping all the test images and creating the submission dataframe.
for img_name in os.listdir(test_dir):
# preparing the image for fitting in model.
img_path = os.path.join(test_dir, img_name)
img = image.load_img(img_path, target_size=IMAGE_SIZE)
img = image.img_to_array(img)
img = np.expand_dims(img, axis=0)
prediction = model.predict(img)
if prediction[0][0] < 0.5:
predicted_labels.append(0)
else:
predicted_labels.append(1)
image_ids.append(img_name.split("_")[0].split(".")[0])
print("Prediction Complete")
return image_ids, predicted_labels
# function for creating the submission csv.
def of_create_submission_csv(image_ids, predicted_labels, file_name):
print("Creating CSV")
submission = pd.DataFrame({"id": image_ids, "label": predicted_labels})
submission.to_csv(file_name, index=False)
print("CSV successfully created.")
# function for parsing xml.
def of_parse_xml(annotations_dir):
print("Parsing XML started...")
image_ids = []
labels = []
for filename in os.listdir(annotations_dir):
if not filename.endswith(".xml"):
continue
# parse XML annotation file
tree = ET.parse(os.path.join(annotations_dir, filename))
root = tree.getroot()
# get image path and class label
# image_path = os.path.join(data_dir, 'images', root.find('filename').text)
image_id = filename.split("_")[1]
class_label = int(root.find("object").find("bndbox").find("xmax").text)
image_ids.append(image_id)
labels.append(class_label)
data = {"ID": image_ids, "Class": labels}
df = pd.DataFrame(data)
print("Parsing XML completed.")
return df
# function for performing training and creating model.
def of_train_create_model(path_for_training, path_for_predicting, files, model_name):
# creating the model.
print("Starting to create the model...")
model = of_create_model(input_shape=IMAGE_SIZE + (3,), num_classes=2)
print("Model Successfully created.")
# plotting the model.
# keras.utils.plot_model(model, show_shapes=True)
# get the train and validation dataset.
print("Getting Train and Validation datasets...")
train_ds, val_ds = of_get_train_validation_set(path_for_training)
print("Train and Validation sets created.")
# training the model.
print("Starting training...")
of_train_model(model, train_ds, val_ds, model_name)
print("Training model completed.")
# predict images.
for paths, file_name in path_for_predicting, files:
image_ids, predicted_labels = of_predict_images(model, paths)
# creating the submission csv.
of_create_submission_csv(image_ids, predicted_labels, file_name)
print("Program Complete.\n\n\n")
# function for getting the accuracy.
def of_get_accuracy(y_pred, test_labels):
accuracy = sum(np.array(y_pred) == np.array(test_labels) / len(test_labels))
print("Accuracy:", accuracy)
# the main function.
def main():
# for the kaggle dataset.
training_files = "/kaggle/input/train-test-images/train_output"
predicting_files = [
"/kaggle/input/train-test-images/test_output/all_images",
"/kaggle/input/oxford-pet-images/images",
]
file_names = ["kaggle_submission.csv", "oxford_submission.csv"]
# predicting_files = ['/kaggle/input/train-test-images/test_output/all_images']
# file_names = ['kaggle_submission.csv']
of_train_create_model(
training_files, predicting_files, file_names, "kaggle_model.h5"
)
# for the oxford dataset.
training_files = "/kaggle/input/oxford-pet-images/images"
predicting_files = ["/kaggle/input/train-test-images/test_output/all_images"]
file_names = ["kaggle_based_on_oxford_submission.csv"]
true_labels_for_oxford = of_parse_xml(
"/kaggle/input/oxford-pet-images/annotations/annotations/xmls"
)
of_train_create_model(
training_files, predicting_files, file_names, "oxford_model.h5"
)
# calling the main function
main()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Introduction
# To support disaster relief organisations, we've been tasked to use Natural Language Processing to build a model which predicts which Tweets are about real disasters and which ones aren't.
# This is my fourth competition and first using NLP. I'll be using https://www.kaggle.com/code/philculliton/nlp-getting-started-tutorial/notebook as a guide.
# The words in each tweet will form the features we use to model whether there is a real disaster.
# I think we will approach the following way:
# * Read in and understand the data available.
# * Identify the missing data and decide how to deal with it
# * Perform feature engineer - this time around feature engineering is limited. Next time we will need to feature engineer the tweet 'text'
# * Build vectors on the training dataframe using ColumnTransformer and CountVectorizer which will count the words in each tweet and turn them into data we can model
# * We will do an inital assessment of a variety of models using Stratified K fold cross validation
# * We will use the best peforming model and apply to the transformed test dataframe to predict the outcomes.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
sns.set(style="white", context="notebook", palette="colorblind")
pd.set_option("display.max_columns", 5000)
pd.set_option("display.max_rows", 500)
import warnings
warnings.filterwarnings("ignore")
# # Read in and explore the train and test dataframes
train_df = pd.read_csv("/kaggle/input/nlp-getting-started/train.csv")
test_df = pd.read_csv("/kaggle/input/nlp-getting-started/test.csv")
print(train_df.shape)
print(test_df.shape)
train_df.head()
train_df["keyword"].value_counts()
train_df["location"].value_counts()
# # Let's see what's missing from both dataframes
# **Let's summariese what's missing and then review how to handle the missing data**
missing_vals_train = pd.DataFrame(train_df.isna().sum(), columns=["Sum"])
missing_vals_train = missing_vals_train.sort_values(by="Sum", ascending=False)
missing_vals_train = missing_vals_train[missing_vals_train["Sum"] > 0]
missing_vals_train["Percent"] = missing_vals_train["Sum"] / 7613 * 100
missing_vals_train
sns.heatmap(train_df.isnull(), cbar=False)
plt.show()
# **It appears that we are missing keyword data only at the start and end of the dataframe**
# **It appears that we are missing location data in blocks**
missing_vals_test = pd.DataFrame(test_df.isna().sum(), columns=["Sum"])
missing_vals_test = missing_vals_test.sort_values(by="Sum", ascending=False)
missing_vals_test = missing_vals_test[missing_vals_test["Sum"] > 0]
missing_vals_test["Percent"] = missing_vals_test["Sum"] / 3263 * 100
missing_vals_test
sns.heatmap(test_df.isnull(), cbar=False)
plt.show()
# **Same observations for the test dataframe as for the train dataframe**
# Let's first think about **keyword**. From the dataset description we are told:
# A keyword from that tweet (although this may be blank!)
# * I'm not too sure about how the keyword is generated so I need to be careful. The first option is to remove the rows with no data. For both dataframes this is about 0.8% of the total data, which isn't too much. Unfortunately this isn't an option for the test dataframe as the submission requires the correct index. Another option would be to replace with the most frequent. Finally I don't like replacing the keyword with one of my chosing is ideal as I'm unsure how the keyword was selected. Not ideal again from a pipeline approach. I think the best approach would be to use the existing keywords as a reference and if any of these words is in the text of the tweet the one with the highest frequency should be used. For this version I'll use the first keyword found in the text of each tweet.
# * It looks like keyword that are phrases have the words seperated by '%20'. I'll need to replace with a space to allow for my approach detailed above.
train_df["keyword"] = train_df["keyword"].str.replace("%20", " ")
test_df["keyword"] = test_df["keyword"].str.replace("%20", " ")
refs = train_df["keyword"].tolist()
refs = [x for x in refs if str(x) != "nan"]
refs_cleaned = sorted(set(refs), key=refs.index)
pattern = "|".join(refs_cleaned)
import re, string
def pattern_searcher(search_str: str, search_list: str):
search_obj = re.search(search_list, search_str)
if search_obj:
return_str = search_str[search_obj.start() : search_obj.end()]
else:
return_str = "NA"
return return_str
train_df["text_lower"] = train_df["text"].map(lambda x: x.lower())
test_df["text_lower"] = test_df["text"].map(lambda x: x.lower())
train_df.loc[
(train_df["keyword"].isnull()) & (train_df["text_lower"].str.contains(pattern)),
"keyword",
] = train_df["text_lower"].apply(
lambda x: pattern_searcher(search_str=x, search_list=pattern)
)
test_df.loc[
(test_df["keyword"].isnull()) & (test_df["text_lower"].str.contains(pattern)),
"keyword",
] = test_df["text_lower"].apply(
lambda x: pattern_searcher(search_str=x, search_list=pattern)
)
train_df.loc[train_df["keyword"].isnull(), "keyword"] = "no"
test_df.loc[test_df["keyword"].isnull(), "keyword"] = "no"
print(train_df.shape)
print(test_df.shape)
missing_vals_train = pd.DataFrame(train_df.isna().sum(), columns=["Sum"])
missing_vals_train = missing_vals_train.sort_values(by="Sum", ascending=False)
missing_vals_train = missing_vals_train[missing_vals_train["Sum"] > 0]
missing_vals_train["Percent"] = missing_vals_train["Sum"] / 7552 * 100
missing_vals_train
missing_vals_test = pd.DataFrame(test_df.isna().sum(), columns=["Sum"])
missing_vals_test = missing_vals_test.sort_values(by="Sum", ascending=False)
missing_vals_test = missing_vals_test[missing_vals_test["Sum"] > 0]
missing_vals_test["Percent"] = missing_vals_test["Sum"] / 3237 * 100
missing_vals_test
# Let's think about **location**. From the dataset description we are told:
# The location the tweet was sent from (may also be blank)
# * There is quite a lot of missing data. For each dataframe you are looking at > 33% of data missing. When the missing data is >15/20% I would be tempted to drop the feature all together. There are some locations noted in the tweets, however it is difficult to determine if that denotes a relationship between where the tweet is sent from. For this version I'll drop the column. For a future version I'll come back and see what was I can do.
# * When I come back in a future version there looks to be a good opportunity for feature engineering. We could create features for city, state, country for instance.
train_df = train_df.drop(["location"], axis=1)
test_df = test_df.drop(["location"], axis=1)
# # Feature Engineering
sns.countplot(data=train_df, x="target")
# * Less observations are not classified as being related to disasters compare to those that are.
# **Let's start with keyword**
# A fair bit of this is complete with the work we did above to resolve the missing data
# * Let's first compare our target to keyword character length and keyword word count
train_df["keyword_len"] = train_df["keyword"].str.len()
train_df["keyword_word"] = train_df["keyword"].str.split().map(lambda x: len(x))
test_df["keyword_len"] = test_df["keyword"].str.len()
test_df["keyword_word"] = test_df["keyword"].str.split().map(lambda x: len(x))
# Feature: keyword
fig, ax = plt.subplots(1, 2, figsize=(12, 8))
sns.countplot(data=train_df, x="keyword_len", hue="target", ax=ax[0])
sns.countplot(data=train_df, x="keyword_word", hue="target", ax=ax[1])
# * Longer keyword character length increases the likelihood of disaster
# * More than 1 word in keyword increases the likelihood of disaster
# **Next let's look at text**
train_df["text"].head(50)
# What a mess...lots of cleaning to do
# * Let's first look at character length and word length
train_df["text_len"] = train_df["text"].str.len()
train_df["text_word"] = train_df["text"].str.split().map(lambda x: len(x))
test_df["text_len"] = test_df["text"].str.len()
test_df["text_word"] = test_df["text"].str.split().map(lambda x: len(x))
# Feature: text
fig, ax = plt.subplots(figsize=(15, 5))
sns.countplot(data=train_df, x="text_len", hue="target")
labels = [0, 25, 50, 75, 100, 125, 150]
ax.set_xticks([0, 25, 50, 75, 100, 125, 150])
ax.set_xticklabels(labels, rotation=45, ha="right")
plt.show()
# Feature: text
fig, ax = plt.subplots(figsize=(15, 5))
sns.countplot(data=train_df, x="text_len", hue="target")
labels = [0, 25, 50, 75, 100, 125, 150]
ax.set_xticks([0, 25, 50, 75, 100, 125, 150])
ax.set_xticklabels(labels, rotation=45, ha="right")
ax.set_ylim([0, 50])
plt.show()
# * Looks like the more characters in the text of the tweet then the more likely it is designated as relating to a disaster
# * Disaster designation seems to pick up proportion around ~70 characters
fig, ax = plt.subplots(figsize=(15, 5))
sns.countplot(data=train_df, x="text_word", hue="target")
# * More words in text increases the likelihood of disaster.
# * Disaster designation seems to pick up proportion around ~8 words
# # Let's clean up text in tweet
# **Many thanks to SHAHULES for your notebook**
# [Amazing for data cleaning!](https://www.kaggle.com/code/shahules/basic-eda-cleaning-and-glove)
# * According to SHAHULES we need to clean tweet text before modelling. We will correct spelling, remove punctuations, remove html tags and emojis.
# * I'll need to come back to the character length and word count columns for text after this is complete
# **Remvoing URLs**
def remove_URL(text):
url = re.compile(r"https?://\S+|www\.\S+")
return url.sub(r"", text)
train_df["text_lower"] = train_df["text_lower"].apply(lambda x: remove_URL(x))
test_df["text_lower"] = test_df["text_lower"].apply(lambda x: remove_URL(x))
# **Remove HTML tags**
def remove_html(text):
html = re.compile(r"<.*?>")
return html.sub(r"", text)
train_df["text_lower"] = train_df["text_lower"].apply(lambda x: remove_html(x))
test_df["text_lower"] = test_df["text_lower"].apply(lambda x: remove_html(x))
# **Remove Emojis**
# Reference : https://gist.github.com/slowkow/7a7f61f495e3dbb7e3d767f97bd7304b
def remove_emoji(text):
emoji_pattern = re.compile(
"["
"\U0001F600-\U0001F64F" # emoticons
"\U0001F300-\U0001F5FF" # symbols & pictographs
"\U0001F680-\U0001F6FF" # transport & map symbols
"\U0001F1E0-\U0001F1FF" # flags (iOS)
"\U00002702-\U000027B0"
"\U000024C2-\U0001F251"
"]+",
flags=re.UNICODE,
)
return emoji_pattern.sub(r"", text)
train_df["text_lower"] = train_df["text_lower"].apply(lambda x: remove_emoji(x))
test_df["text_lower"] = test_df["text_lower"].apply(lambda x: remove_emoji(x))
# **Remove Punctuation**
def remove_punct(text):
result = text.translate(str.maketrans("", "", string.punctuation))
return result
train_df["text_lower"] = train_df["text_lower"].apply(lambda x: remove_punct(x))
test_df["text_lower"] = test_df["text_lower"].apply(lambda x: remove_punct(x))
# **Correct Spelling**
from spellchecker import SpellChecker
spell = SpellChecker()
def correct_spellings(text):
text1 = " ".join(text.split())
corrected_text = []
for word in text.split():
if spell.unknown(word) is True and spell.correction(word) is True:
corrected_text.append(spell.correction(word))
else:
corrected_text.append(word)
return " ".join(corrected_text)
train_df["text_lower"] = train_df["text_lower"].apply(lambda x: correct_spellings(x))
test_df["text_lower"] = test_df["text_lower"].apply(lambda x: correct_spellings(x))
# # Let's Vectorize!
Y = train_df["target"]
X = train_df[["keyword", "text_lower"]]
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.compose import ColumnTransformer
ct = ColumnTransformer(
[
("keyword_cv", CountVectorizer(), "keyword"),
("text_cv", CountVectorizer(), "text_lower"),
]
)
X_vectors = ct.fit_transform(X)
X_vectors
# # Let's do an initial assessment of a variety of classifiers!
from sklearn.model_selection import cross_val_score, StratifiedKFold, cross_val_score
# 10 fold cross validatoin
kf = StratifiedKFold(n_splits=6)
from sklearn.ensemble import RandomForestClassifier, GradientBoostingClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.svm import SVC
from xgboost import XGBClassifier
from catboost import CatBoostClassifier
# Modeling step Test differents algorithms
random_state = 2
classifiers = []
classifiers.append(CatBoostClassifier(random_state=random_state, verbose=False))
classifiers.append(XGBClassifier(random_state=random_state))
classifiers.append(SVC(random_state=random_state))
classifiers.append(DecisionTreeClassifier(random_state=random_state))
classifiers.append(RandomForestClassifier(random_state=random_state))
classifiers.append(GradientBoostingClassifier(random_state=random_state))
classifiers.append(KNeighborsClassifier())
cv_results_accuracy = []
for classifier in classifiers:
cv_results_accuracy.append(
cross_val_score(classifier, X_vectors, y=Y, scoring="accuracy", cv=kf, n_jobs=2)
)
cv_means_accuracy = []
for cv_result_accuracy in cv_results_accuracy:
cv_means_accuracy.append(cv_result_accuracy.mean())
cv_accuracy = pd.DataFrame(
{
"CrossValMeans Accuracy": cv_means_accuracy,
"Algorithm": [
"CatBoost",
"XGB",
"SVC",
"DecisionTree",
"RandomForest",
"GradientBoosting",
"KNeighboors",
],
}
)
cv_accuracy
fig, ax = plt.subplots()
sns.barplot(
x="CrossValMeans Accuracy",
y="Algorithm",
data=cv_accuracy,
palette="colorblind",
orient="h",
ax=ax,
)
ax.set_xlim(0.5, 0.8)
plt.show()
cv_results_auc_roc = []
for classifier in classifiers:
cv_results_auc_roc.append(
cross_val_score(classifier, X_vectors, y=Y, scoring="roc_auc", cv=kf, n_jobs=2)
)
cv_means_auc_roc = []
for cv_result_auc_roc in cv_results_auc_roc:
cv_means_auc_roc.append(cv_result_auc_roc.mean())
cv_auc_roc = pd.DataFrame(
{
"CrossValMeans Auc Roc": cv_means_auc_roc,
"Algorithm": [
"CatBoost",
"XGB",
"SVC",
"DecisionTree",
"RandomForest",
"GradientBoosting",
"KNeighboors",
],
}
)
cv_auc_roc
fig, ax = plt.subplots()
sns.barplot(
x="CrossValMeans Auc Roc",
y="Algorithm",
data=cv_auc_roc,
palette="colorblind",
orient="h",
ax=ax,
)
ax.set_xlim(0.5, 0.8)
plt.show()
# # Final Model Selection
from sklearn.model_selection import train_test_split
# Partition the dataset in train + validation sets
X_train, X_test, y_train, y_test = train_test_split(
X_vectors, Y, test_size=0.2, random_state=0
)
print("X_train : " + str(X_train.shape))
print("X_test : " + str(X_test.shape))
print("y_train : " + str(y_train.shape))
print("y_test : " + str(y_test.shape))
classifier = SVC(probability=True)
scores = cross_val_score(
classifier, X_train, y=y_train, scoring="roc_auc", cv=kf, n_jobs=2
)
scores
classifier.fit(X_train, y=y_train)
y_test_predict = classifier.predict(X_test)
y_train_predict = classifier.predict(X_train)
prob = classifier.predict_proba(X_test)
y_test_prob = prob[:, 1]
from sklearn.metrics import (
confusion_matrix,
classification_report,
accuracy_score,
log_loss,
roc_curve,
roc_auc_score,
ConfusionMatrixDisplay,
)
# Suggested to me that classification report and Auc_roc is best to use
# Accuracy is a good metric for a balanced dataset
result = accuracy_score(y_test, y_test_predict)
print("Accuracy is: ", result * 100)
# Log loss is a cost function which tell us how different y_test_pred is from y_test (the higher LL, the larger the error)
ll = log_loss(y_test, y_test_predict)
print("Log Loss is: ", ll)
# TL: Truely predicted as positives, BR: Truely predicted as negatives, TR: Falsely predicted as negative, BL: Falsely predicted as positive
# Accuracy of confusion matrix is (TL+BR)/attributes
# recall is Top box from confusion matrix TL / (TL+TR) true positive / total observed positives (closer to 1 the better)
# precision is left box from confusion matris TL / (TL+BL) true postives / all records predicted as positives (closer to 1 the better)
print(confusion_matrix(y_test, y_test_predict))
# F1 is accuracy
print(classification_report(y_test, y_test_predict))
# Area under ROC
fpr, tpr, threshold = roc_curve(y_test, y_test_prob)
print(
"roc_auc_score: ", roc_auc_score(y_test, y_test_prob)
) # the higher the better the model is
cm = confusion_matrix(y_test, y_test_predict, labels=classifier.classes_)
disp = ConfusionMatrixDisplay(confusion_matrix=cm, display_labels=classifier.classes_)
disp.plot()
plt.show()
# **Poor Precision. I'm falsely predicting tweets as indicative of a disaster**
# # Predict target on tested dataframe!
X_test = test_df[["keyword", "text_lower"]]
X_test_vectors = ct.transform(X_test)
sample_submission = pd.read_csv(
"/kaggle/input/nlp-getting-started/sample_submission.csv"
)
sample_submission
cond = ~sample_submission["id"].isin(test_df["id"])
sample_submission.drop(sample_submission[cond].index, inplace=True)
sample_submission
sample_submission["target"] = classifier.predict(X_test_vectors)
sample_submission.head()
sample_submission.to_csv("submission.csv", index=False)
|
import pickle
import numpy as np
files = [
"../input/cifar10/data_batch_1",
"../input/cifar10/data_batch_2",
"../input/cifar10/data_batch_3",
"../input/cifar10/data_batch_4",
"../input/cifar10/data_batch_5",
]
x_train = np.zeros((50000, 3072))
y_train = np.zeros((50000))
i = 0
for file in files:
i = i + 1
with open(file, "rb") as fo:
dict = pickle.load(fo, encoding="bytes")
p = i * 10000
x_train[p - 10000 : p, :] = dict[b"data"]
y_train[p - 10000 : p] = dict[b"labels"]
print(p - 10000, p)
fo.close()
x_test = np.zeros((10000, 3072))
y_test = np.zeros((10000))
x_train.shape
file = "../input/cifar10/test_batch"
with open(file, "rb") as fo:
dict = pickle.load(fo, encoding="bytes")
x_test[0:10000, :] = dict[b"data"]
y_test[0:10000] = dict[b"labels"]
fo.close()
def unpickle(fileName):
"""
Description: retrieve data from CIFAR-10 Pickles
Params: fileName = filename to unpickle
Outputs: Unpickled Dict
"""
with open(fileName, "rb") as f:
dict = pickle.load(f, encoding="bytes")
return dict
def merge_batches():
for i in range(0, 5):
fileName = "../input/cifar10/data_batch_" + str(i + 1)
data = unpickle(fileName)
if i == 0:
features = data[b"data"]
labels = np.array(data[b"labels"])
else:
features = np.append(features, data[b"data"], axis=0)
labels = np.append(labels, data[b"labels"], axis=0)
return features, labels
def one_hot_encode(data):
"""
Description: Encode Target Label IDs to one hot vector of size L where L is the
number of unique labels
Params: data = list of label IDs
Outputs: List of One Hot Vectors
"""
one_hot = np.zeros((data.shape[0], 10))
one_hot[np.arange(data.shape[0]), data] = 1
return one_hot
def normalize(data):
"""
Description: Normalize Pixel values
Params: list of Image Pixel Features
Outputs: Normalized Image Pixel Features
"""
return data / 255.0
def preprocess():
"""
Description: helper function to load and preprocess CIFAR-10 training data batches
Params: num_to_load = number of batches of CIFAR-10 to load and merge
Outputs: Pre-processed CIFAR-10 image features and labels
"""
X, y = merge_batches()
X = normalize(X)
X = X.reshape(-1, 3072, 1)
y = one_hot_encode(y)
y = y.reshape(-1, 10, 1)
return X, y
def dataset_split(X, y, ratio=0.9):
"""
Description: helper function to split training data into training and validation
Params: X=image features
y=labels
ratio = ratio of training data from total data
Outputs: training data (features and labels) and validation data
"""
split = int(ratio * X.shape[0])
indices = np.random.permutation(X.shape[0])
training_idx, val_idx = indices[:split], indices[split:]
X_train, X_val = X[training_idx, :], X[val_idx, :]
y_train, y_val = y[training_idx, :], y[val_idx, :]
print("Records in Training Dataset", X_train.shape[0])
print("Records in Validation Dataset", X_val.shape[0])
return X_train, y_train, X_val, y_val
def sigmoid(out):
"""
Description: Sigmoid Activation
Params: out = a list/matrix to perform the activation on
Outputs: Sigmoid activated list/matrix
"""
return 1.0 / (1.0 + np.exp(-out))
def delta_sigmoid(out):
"""
Description: Derivative of Sigmoid Activation
Params: out = a list/matrix to perform the activation on
Outputs: Delta(Sigmoid) activated list/matrix
"""
return sigmoid(out) * (1 - sigmoid(out))
def SigmoidCrossEntropyLoss(a, y):
return np.sum(np.nan_to_num(-y * np.log(a) - (1 - y) * np.log(1 - a)))
class DNN(object):
"""
Description: Class to define the Deep Neural Network
"""
def __init__(self, sizes):
"""
Description: initialize the biases and weights using a Gaussian
distribution with mean 0, and variance 1.
Biases are not set for 1st layer that is the input layer.
Params: sizes = a list of size L; where L is the number of layers
in the deep neural network and each element of list contains
the number of neuron in that layer.
first and last elements of the list corresponds to the input
layer and output layer respectively
intermediate layers are hidden layers.
"""
self.num_layers = len(sizes)
# setting appropriate dimensions for weights and biases
self.biases = [np.random.randn(y, 1) for y in sizes[1:]]
self.weights = [np.random.randn(y, x) for x, y in zip(sizes[:-1], sizes[1:])]
def forwardPropagation(self, x):
"""
Description: Forward Passes an image feature matrix through the Deep Neural
Network Architecture.
Params: x = Image Features
Outputs: 2 lists which stores outputs and activations at every layer,
1st list is non-activated and 2nd list is activated
The last element of the 2nd list corresponds to the scores against
10 labels in the dataset.
"""
activation = x
activations = [x] # list to store activations for every layer
outs = [] # list to store out vectors for every layer
for b, w in zip(self.biases, self.weights):
out = np.dot(w, activation) + b
outs.append(out)
activation = sigmoid(out)
activations.append(activation)
return outs, activations
def get_batch(self, X, y, batch_size):
"""
Description: A data iterator for batching of image features and labels
Params: X, y = lists of Features and corresponding labels, these lists
have to be batched.
batch_size = size of the batch
Outputs: a batch of image features and labels of size = batch_size
"""
for batch_idx in range(0, X.shape[0], batch_size):
batch = zip(
X[batch_idx : batch_idx + batch_size],
y[batch_idx : batch_idx + batch_size],
)
yield batch
def train(self, X, y, x_val, y_val, batch_size=100, learning_rate=0.2, epochs=1000):
"""
Description: Batch-wise trains image features against corresponding labels.
The weights and biases of the neural network are updated through
backpropagation on batches using SGD
del_b and del_w are of same size as all the weights and biases
of all the layers. del_b and del_w contains the gradients which
are used to update weights and biases
Params: X, y = lists of training features and corresponding labels
batch_size = size of the batch
learning_rate = eta; controls the size of changes in weights & biases
epochs = no. of times to iterate of the whole data
"""
n_batches = X.shape[0] // batch_size
for j in range(epochs):
batch_iter = self.get_batch(X, y, batch_size)
# print(type(batch_iter))
for i in range(n_batches):
batch = next(batch_iter)
# same shape as self.biases
del_b = [np.zeros(b.shape) for b in self.biases]
# same shape as self.weights
del_w = [np.zeros(w.shape) for w in self.weights]
for batch_X, batch_y in batch:
# accumulate all the bias and weight gradients
loss, delta_del_b, delta_del_w = self.backPropagation(
batch_X, batch_y
)
del_b = [db + ddb for db, ddb in zip(del_b, delta_del_b)]
del_w = [dw + ddw for dw, ddw in zip(del_w, delta_del_w)]
# update weight and biases by multiplying ratio learning rate and batch_size
# multiplied with the accumulated gradients(partial derivatives)
# calculate change in weight(delta) and biases and update weight
# with the changes
self.weights = [
w - (learning_rate / batch_size) * delw
for w, delw in zip(self.weights, del_w)
]
self.biases = [
b - (learning_rate / batch_size) * delb
for b, delb in zip(self.biases, del_b)
]
acc_val = self.eval(x_val, y_val)
acc_train = self.eval(X, y)
print("\nEpoch %d complete\t training accuracy: %f\n" % (j, acc_train))
print("\nEpoch %d complete\t validation accuracy: %f\n" % (j, acc_val))
print("\nEpoch %d complete\t taining Loss: %f\n" % (j, loss))
def backPropagation(self, x, y):
del_b = [np.zeros(b.shape) for b in self.biases]
del_w = [np.zeros(w.shape) for w in self.weights]
outs, activations = self.forwardPropagation(x)
loss = SigmoidCrossEntropyLoss(activations[-1], y)
delta_cost = activations[-1] - y
delta = delta_cost
del_b[-1] = delta
del_w[-1] = np.dot(delta, activations[-2].T)
for l in range(2, self.num_layers):
out = outs[-l]
delta_activation = delta_sigmoid(out)
delta = np.dot(self.weights[-l + 1].T, delta) * delta_activation
del_b[-l] = delta
del_w[-l] = np.dot(delta, activations[-l - 1].T)
return (loss, del_b, del_w)
def eval(self, X, y):
"""
Description: Based on trained(updated) weights and biases, predict a label and compare
it with original label and calculate accuracy
Params: X, y = a data example from validation dataset (image features, labels)
Outputs: accuracy of prediction
"""
count = 0
for x, _y in zip(X, y):
outs, activations = self.forwardPropagation(x)
# postion of maximum value is the predicted label
if np.argmax(activations[-1]) == np.argmax(_y):
count += 1
# print("Accuracy: %f" % ((float(count) / X.shape[0]) * 100))
return float(count) / X.shape[0] * 100
def predict(self, X):
"""
Description: Based on trained(updated) weights and biases, predict a label for an
image which does not have a label.
Params: X = list of features of unknown images
Outputs: list containing the predicted label for the corresponding unknown image
"""
labels = unpickle("cifar-10-batches-py/batches.meta")["label_names"]
preds = np.array([])
for x in X:
outs, activations = self.feedforward(x)
preds = np.append(preds, np.argmax(activations[-1]))
preds = np.array([labels[int(p)] for p in preds])
return preds
X, y = preprocess()
X_train, y_train, X_val, y_val = dataset_split(X, y)
model = DNN([3072, 30, 20, 10]) # initialize the model
model.train(X_train, y_train, X_val, y_val, epochs=5) # train the model
|
# # This notebook implements classification using CNNs in Keras for the Plant Pathology 2020 Challenge
# The main purpouos of this notebook is to strengthen my pratical knowledge of implementing different methods of deep learning. However, I decided share this notebook in hope that others can benefit from it.
# **Purpouse of this notebook:**
# * Learn how to implelemt data augmentation in Keras
# * Practice implementing CNNs in Keras
# * Learn how to use training with class weights for an imbalanced data set
# ** This notebook does NOT attempt:**
# * Achieving top score on the dataset
# * Optimize learning speed
# **Comments:**
# * Achives approxmiately a score of 0.94 on the public test data.
# * Design to run with GPU accelerator on Kaggle. However, at the time of commit the GPU accleration is not working properly on Kaggle ( https://www.kaggle.com/c/liverpool-ion-switching/discussion/136058#833053 )
# * Some failed commits due to running out of memory on CPU
# * Even with heavy class weighting, the CNN does not perform well for the class with low number of observations (5%), i.e. "multiple_diseases".
#
#
# General
import numpy as np
import pandas as pd
# For Keras
from keras_preprocessing.image import ImageDataGenerator
from keras.models import Sequential
from keras.layers import (
Dense,
Dropout,
Conv2D,
MaxPool2D,
Flatten,
Input,
GlobalMaxPool2D,
)
from keras.optimizers import Adam
import matplotlib.pyplot as plt
# For visualizing results
import seaborn as sn
from sklearn.metrics import confusion_matrix, classification_report
from sklearn.utils import class_weight
# Defining the path to the images.
img_path = "../input/plant-pathology-2020-fgvc7/images/"
# Reading the datasets csv files:
sample_submission = pd.read_csv(
"../input/plant-pathology-2020-fgvc7/sample_submission.csv"
)
test = pd.read_csv("../input/plant-pathology-2020-fgvc7/test.csv")
train = pd.read_csv("../input/plant-pathology-2020-fgvc7/train.csv")
# Adding the full image filename to easily read it from ImageDataGenerator
train["imaged_id_fileName"] = train.image_id + ".jpg"
test["imaged_id_fileName"] = test.image_id + ".jpg"
# Show the strucutre of the training structure:
train.head()
# Data augmentation using ImageDataGenereator.
# Applying moderate amount of zoom in/out and brightness variation. Full rotation and
# flips are applied since there is no obvious orientation that the pictures of the leafs are taken.
# Keeping approximate aspect ratio of the images:
img_height = 100
img_width = 133
# Defining the batch size that will be used in training:
batch_size = 32
# Labels inferred from the dataset:
labels = ["healthy", "multiple_diseases", "rust", "scab"]
# Define the ImageDataGenerator using a training/validation split of 80/20%
train_dataGenerator = ImageDataGenerator(
rescale=1.0 / 255,
shear_range=0,
zoom_range=(1, 1.3),
rotation_range=360,
brightness_range=(0.7, 1.3),
horizontal_flip=True,
vertical_flip=True,
validation_split=0.2,
)
train_generator = train_dataGenerator.flow_from_dataframe(
dataframe=train,
x_col="imaged_id_fileName",
y_col=labels,
directory=img_path,
target_size=(img_height, img_width),
batch_size=batch_size,
class_mode="raw",
subset="training",
)
validation_generator = train_dataGenerator.flow_from_dataframe(
dataframe=train,
x_col="imaged_id_fileName",
y_col=labels,
directory=img_path,
target_size=(img_height, img_width),
batch_size=batch_size,
class_mode="raw",
subset="validation",
)
# This validator generator will be used to plot confusion matrix where we need the shuffle to be off.
validation_generator2 = train_dataGenerator.flow_from_dataframe(
dataframe=train,
x_col="imaged_id_fileName",
y_col=labels,
directory=img_path,
target_size=(img_height, img_width),
batch_size=batch_size,
class_mode="raw",
shuffle=False,
sort=False,
subset="validation",
)
# Later we want to use the full dataset for training since we have quite a limited number of images. Below we define the generator for that case:
train_dataGenerator_full = ImageDataGenerator(
rescale=1.0 / 255,
shear_range=0,
zoom_range=(1, 1.3),
rotation_range=360,
brightness_range=(0.7, 1.3),
horizontal_flip=True,
vertical_flip=True,
validation_split=0,
)
train_generator_full = train_dataGenerator_full.flow_from_dataframe(
dataframe=train,
x_col="imaged_id_fileName",
y_col=labels,
directory=img_path,
target_size=(img_height, img_width),
batch_size=batch_size,
class_mode="raw",
subset="training",
)
# Finally we also define the ImageDataGenerator for the unlabled test data:
test_dataGenerator = ImageDataGenerator(rescale=1.0 / 255)
test_generator = test_dataGenerator.flow_from_dataframe(
dataframe=test,
x_col="imaged_id_fileName",
y_col=labels,
directory=img_path,
shuffle=False,
sort=False,
target_size=(img_height, img_width),
batch_size=1,
class_mode=None,
)
# Calculating the prior probability of the different labes from the training dataset
classProb = np.zeros(len(labels))
idx = 0
for k in labels:
print(f"{k} contains {train[k].sum()} samples")
classProb[idx] = train[k].sum()
idx += 1
# Visualizing the results in a pie-chart:
print() # Empty line before figure
color = ["#58508d", "#bc5090", "#ff6361", "#ffa600"]
plt.figure(figsize=(15, 7))
plt.pie(
classProb,
shadow=True,
explode=[0, 0.5, 0, 0],
labels=labels,
autopct="%1.2f%%",
colors=color,
startangle=-90,
textprops={"fontsize": 14},
)
class_weight_vect = np.square(
1 / (classProb / classProb.sum())
) # Calculate the weight per classbased on the prior probability dervied from the training data.
class_weight_vect = class_weight_vect / np.min(class_weight_vect)
# Visualize the data augmentation
# Plot function taken inspiration from here:
# https://github.com/tensorflow/examples/blob/master/courses/udacity_intro_to_tensorflow_for_deep_learning/l05c04_exercise_flowers_with_data_augmentation_solution.ipynb
def plotImages(imgs):
col = 5
row = 2
fig, axes = plt.subplots(row, col, figsize=(25, 25))
axes = axes.flatten()
for k in range(10):
axes[k].imshow(imgs[k])
fig.subplots_adjust(hspace=-0.75, wspace=0.2)
plt.show()
# Apply augmentation to the same picture 10 times and plot the outcome:
plotImageAugmentation = [
validation_generator2[1][0][0] for i in range(10)
] # Using validation_generator2 for consitency since shuffle is turned off.
plotImages(plotImageAugmentation)
# Define the convolutional neural network:
model = Sequential()
model.add(
Conv2D(
35,
kernel_size=(3, 3),
activation="relu",
kernel_initializer="glorot_uniform",
bias_initializer="zeros",
input_shape=(img_height, img_width, 3),
padding="same",
)
)
model.add(
Conv2D(
35,
(3, 3),
kernel_initializer="glorot_uniform",
bias_initializer="zeros",
activation="relu",
padding="same",
)
)
model.add(Dropout(0.1))
model.add(MaxPool2D(pool_size=(2, 2)))
model.add(
Conv2D(
35,
(3, 3),
kernel_initializer="glorot_uniform",
bias_initializer="zeros",
activation="relu",
)
)
model.add(
Conv2D(
35,
(3, 3),
kernel_initializer="glorot_uniform",
bias_initializer="zeros",
activation="relu",
)
)
model.add(Dropout(0.1))
model.add(MaxPool2D(pool_size=(5, 5)))
model.add(
Conv2D(
50,
(3, 3),
kernel_initializer="glorot_uniform",
bias_initializer="zeros",
activation="relu",
)
)
model.add(
Conv2D(
50,
(3, 3),
kernel_initializer="glorot_uniform",
bias_initializer="zeros",
activation="relu",
)
)
model.add(Dropout(0.1))
model.add(GlobalMaxPool2D())
model.add(Dropout(0.1))
model.add(Dense(4, activation="softmax"))
optimizerAdam = Adam(lr=0.00125, amsgrad=True)
model.compile(
loss="categorical_crossentropy", optimizer=optimizerAdam, metrics=["accuracy"]
)
# Print a summary of the model:
model.summary()
# Train the CNN:
nb_epochs = 100
history = model.fit_generator(
train_generator,
steps_per_epoch=train_generator.samples // batch_size,
validation_data=validation_generator,
validation_steps=validation_generator.samples // batch_size,
epochs=nb_epochs,
class_weight=class_weight_vect,
)
# Display the training performance
fs = 17
fig = plt.figure(figsize=(9, 5))
fig.patch.set_facecolor("xkcd:white")
plt.plot(history.history["accuracy"], color=color[0])
plt.plot(history.history["val_accuracy"], color=color[3])
plt.ylabel("Accuracy", fontsize=fs)
plt.xlabel("Epoch #", fontsize=fs)
plt.legend(["training", "validation"], fontsize=fs)
plt.grid("both", linestyle="--")
plt.xticks(fontsize=fs)
plt.yticks(fontsize=fs)
plt.show()
# summarize history for loss
fig = plt.figure(figsize=(9, 5))
fig.patch.set_facecolor("xkcd:white")
plt.plot(history.history["loss"], color=color[0])
plt.plot(history.history["val_loss"], color=color[3])
plt.ylabel("Loss", fontsize=fs)
plt.xlabel("Epoch #", fontsize=fs)
plt.legend(["training", "validation"], fontsize=fs)
plt.grid("both", linestyle="--")
plt.xticks(fontsize=fs)
plt.yticks(fontsize=fs)
plt.show()
# Plot the classification performance in a confusion matrix
Y_pred = model.predict(validation_generator2)
Y_pred_labels = np.argmax(Y_pred, axis=1)
y_true = np.argmax(validation_generator.labels, axis=1)
labels_num = [0, 1, 2, 3]
cm = confusion_matrix(y_true, Y_pred_labels, normalize="true")
sn.set(font_scale=1.4) # for label size
sn.heatmap(
cm,
annot=True,
annot_kws={"size": 14},
cmap="YlGnBu",
xticklabels=labels,
yticklabels=labels,
)
plt.show()
# Print the classification report:
print(classification_report(y_true, Y_pred_labels))
# Since the labeled dataset is limited and we seen that overfitting is not a major issue, we proceed to train the model over all the images to hopefully incrase the accuracy for the unlabled data
nb_epochs = 50
history = model.fit_generator(
train_generator_full,
steps_per_epoch=train_generator_full.samples // batch_size,
epochs=nb_epochs,
class_weight=class_weight_vect,
)
# Finally we apply the model to predict the unlabed test data:
test_predictions = model.predict_generator(test_generator)
# Download the final predictions on the test data as a csv-file that can be uploaded to Kaggle.
predictions = pd.DataFrame()
predictions["image_id"] = test.image_id
predictions["healthy"] = test_predictions[:, 0]
predictions["multiple_diseases"] = test_predictions[:, 1]
predictions["rust"] = test_predictions[:, 2]
predictions["scab"] = test_predictions[:, 3]
predictions.to_csv("submission.csv", index=False)
predictions.head(10)
# Uncomment to donwload csv-file:
# from google.colab import files
# files.download("submission.csv")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Kutuphanelerin yuklenmesi
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
# # Veriye bakis
data = pd.read_csv("/kaggle/input/titanic/train.csv", sep=",")
data.head(5)
# ### Yukarda veriye bir goz gezdirdigimizde, 12 adet farkli degisken tipleri goruyoruz. Simdi bunlarin tam olarak neyi ifade ettigine bir bakalim.
# - PassengerId = Yolcu ID
# - Survived = Hayatta kalanlar(hayatta kalanlar 1, olenler 0)
# - Pclass = Ekonomi Sinifi
# - Name = Yolcu isimleri
# - Sex = Cinsiyet
# - Age = Yas
# - SibSp = abi/abla(1,0)
# - Parch = ebeveyn/cocuk(1,0)
# - Ticket = Bilet Numaralari
# - Fare = Bilet Fiyati
# - Cabin = Kabin Numarasi
# - Embarked = Liman Isimleri
data.Survived.value_counts()
# Yukarda gordugumuz gibi 0 yani olenlerin sayisi 549 ve ayni sekilde 1 yani hayatta kalmayi basaranlarin sayisi 342.
data.info()
# Datasetimizin bir DataFrame oldugunu ve 891 adet gozlemimizin oldugunu goruyoruz.
# ### Simdi degisken tiplerine hizlica bir goz atalim.
data.dtypes
data.shape
# shape yardimi ile verimizin, 891 gozlem ve 12 adet degiskene sahip oldugunu goruyoruz.
data.describe().T
data.count()
data.isnull().sum()
# Yukarda verimizin icinde kac adet eksik gozlem oldugunu ve bu gozlemlerin nerede ne kadar oldugunu goruyoruz.
# # SEABORN ILE VERI GORSELLESTIRME
# ## Barplot Graph
bar = sns.barplot(x="Sex", y="Survived", hue="Sex", data=data)
bar.set_title("Cinsiyete Gore Hayatta Kalanlar")
# ### Catplot- Violin Graph
sns.catplot(
x="Pclass", y="Fare", kind="violin", hue="Pclass", col="Sex", orient="v", data=data
)
# ### Catplot-Bar Graph
sns.catplot(
x="Pclass", y="Fare", kind="bar", hue="Pclass", col="Sex", orient="v", data=data
)
# ### Displot
sns.displot(data.Pclass)
# ### PairPlot
# - Degiskenlerin birbirleri ile olan korelasyonunu gormek icin kullandigimiz bir grafik.
sns.pairplot(data)
# # HeatMap
corr = data.corr()
plt.figure(figsize=(8, 8))
sns.heatmap(
corr, vmax=0.8, linewidths=0.05, square=True, annot=True, linecolor="purple"
)
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.