script
stringlengths 113
767k
|
|---|
import pandas as pd
import numpy as np
import warnings
import tensorflow as tf
import tensorflow.keras.backend as K
import keras_metrics
import spacy
from tensorflow import keras
from keras.models import Sequential
from keras.layers import Dense, SimpleRNN, Dropout, GRU, LSTM, Bidirectional
from sklearn.model_selection import train_test_split
from sklearn.metrics import (
accuracy_score,
f1_score,
precision_score,
recall_score,
classification_report,
confusion_matrix,
)
from sklearn.exceptions import DataConversionWarning, UndefinedMetricWarning
warnings.filterwarnings(action="ignore", category=DataConversionWarning)
warnings.filterwarnings(action="ignore", category=UndefinedMetricWarning)
tf.compat.v1.logging.set_verbosity(tf.compat.v1.logging.ERROR)
nlp = spacy.load("en_core_web_lg")
sarcasm_df = pd.read_csv("/kaggle/input/sarcasm-detection/Sarcasm Dataset.csv")
sarcasm_df = sarcasm_df.iloc[:, 1:]
sarcasm_df.head()
# LABELS
# sarcasm 0
# irony 1
# satire 2
# understatement 3
# overstatement 4
# rhetorical_question 5
tweets_lst = []
labels_lst = []
multiclass_df = sarcasm_df.loc[sarcasm_df["sarcastic"] == 1].drop(columns="sarcastic")
for itr in range(len(multiclass_df)):
if int(multiclass_df.iloc[itr].sarcasm) == 1:
tweets_lst.append(multiclass_df.iloc[itr].tweet), labels_lst.append(0)
if int(multiclass_df.iloc[itr].irony) == 1:
tweets_lst.append(multiclass_df.iloc[itr].tweet), labels_lst.append(1)
if int(multiclass_df.iloc[itr].satire) == 1:
tweets_lst.append(multiclass_df.iloc[itr].tweet), labels_lst.append(2)
if int(multiclass_df.iloc[itr].understatement) == 1:
tweets_lst.append(multiclass_df.iloc[itr].tweet), labels_lst.append(3)
if int(multiclass_df.iloc[itr].overstatement) == 1:
tweets_lst.append(multiclass_df.iloc[itr].tweet), labels_lst.append(4)
if int(multiclass_df.iloc[itr].overstatement) == 1:
tweets_lst.append(multiclass_df.iloc[itr].tweet), labels_lst.append(5)
dataset_df = pd.DataFrame({"tweet": tweets_lst, "label": labels_lst})
# #################### BINARY CLASSIFICATION ########################
doc_embeddings = []
for itr in dataset_df.tweet:
doc = nlp(str(itr))
doc_embeddings.append(doc.vector)
X_train, X_test, y_train, y_test = train_test_split(
np.asarray(doc_embeddings), np.asarray(dataset_df.label), test_size=0.25
)
X_train = np.expand_dims(X_train, -1)
X_test = np.expand_dims(X_test, -1)
y_train = tf.one_hot(y_train, depth=6)
################################# SIMPLE RNN DROUPOUT RATE 0.2 ########################################
model = keras.Sequential()
model.add(
SimpleRNN(
128, input_shape=(X_train.shape[1:]), activation="relu", return_sequences=True
)
)
model.add(Dropout(0.2))
model.add(SimpleRNN(128, activation="relu", return_sequences=False))
model.add(Dropout(0.2))
model.add(Dense(32, activation="relu"))
model.add(Dropout(0.2))
model.add(Dense(6, activation="softmax"))
# model.summary()
opt = tf.keras.optimizers.Adam(learning_rate=1e-3, decay=1e-5)
model.compile(loss="binary_crossentropy", optimizer=opt, metrics=["accuracy"])
print("Train...")
history = model.fit(X_train, y_train, validation_split=0.2, epochs=5)
predicted_y = np.argmax(model.predict(X_test), axis=1)
print("\nSIMPLE RNN WITH DROPOUT 0.2 WITH MICRO AVERAGE")
print("Accuray: ", accuracy_score(y_test, predicted_y))
print("F1 Score: ", f1_score(y_test, predicted_y, average="micro"))
print("Precision: ", precision_score(y_test, predicted_y, average="micro"))
print("Recall: ", recall_score(y_test, predicted_y, average="micro"))
print("\nSIMPLE RNN WITH DROPOUT 0.2 WITH MACRO AVERAGE")
print("Accuray: ", accuracy_score(y_test, predicted_y))
print("F1 Score: ", f1_score(y_test, predicted_y, average="macro"))
print("Precision: ", precision_score(y_test, predicted_y, average="macro"))
print("Recall: ", recall_score(y_test, predicted_y, average="macro"))
################################# SIMPLE RNN DROUPOUT RATE 0.4 ########################################
model = keras.Sequential()
model.add(
SimpleRNN(
128, input_shape=(X_train.shape[1:]), activation="relu", return_sequences=True
)
)
model.add(Dropout(0.4))
model.add(SimpleRNN(128, activation="relu", return_sequences=False))
model.add(Dropout(0.4))
model.add(Dense(32, activation="relu"))
model.add(Dropout(0.4))
model.add(Dense(6, activation="softmax"))
# model.summary()
opt = tf.keras.optimizers.Adam(learning_rate=1e-3, decay=1e-5)
model.compile(loss="binary_crossentropy", optimizer=opt, metrics=["accuracy"])
print("Train...")
history = model.fit(X_train, y_train, validation_split=0.2, epochs=5)
predicted_y = np.argmax(model.predict(X_test), axis=1)
print("\nSIMPLE RNN WITH DROPOUT 0.4 WITH MICRO AVERAGE")
print("Accuray: ", accuracy_score(y_test, predicted_y))
print("F1 Score: ", f1_score(y_test, predicted_y, average="micro"))
print("Precision: ", precision_score(y_test, predicted_y, average="micro"))
print("Recall: ", recall_score(y_test, predicted_y, average="micro"))
print("\nSIMPLE RNN WITH DROPOUT 0.4 WITH MACRO AVERAGE")
print("Accuray: ", accuracy_score(y_test, predicted_y))
print("F1 Score: ", f1_score(y_test, predicted_y, average="macro"))
print("Precision: ", precision_score(y_test, predicted_y, average="macro"))
print("Recall: ", recall_score(y_test, predicted_y, average="macro"))
################################# SIMPLE RNN DROUPOUT RATE 0.6 ########################################
model = keras.Sequential()
model.add(
SimpleRNN(
128, input_shape=(X_train.shape[1:]), activation="relu", return_sequences=True
)
)
model.add(Dropout(0.6))
model.add(SimpleRNN(128, activation="relu", return_sequences=False))
model.add(Dropout(0.6))
model.add(Dense(32, activation="relu"))
model.add(Dropout(0.6))
model.add(Dense(6, activation="softmax"))
# model.summary()
opt = tf.keras.optimizers.Adam(learning_rate=1e-3, decay=1e-5)
model.compile(loss="binary_crossentropy", optimizer=opt, metrics=["accuracy"])
print("Train...")
history = model.fit(X_train, y_train, validation_split=0.2, epochs=5)
predicted_y = np.argmax(model.predict(X_test), axis=1)
print("\nSIMPLE RNN WITH DROPOUT 0.6 WITH MICRO AVERAGE")
print("Accuray: ", accuracy_score(y_test, predicted_y))
print("F1 Score: ", f1_score(y_test, predicted_y, average="micro"))
print("Precision: ", precision_score(y_test, predicted_y, average="micro"))
print("Recall: ", recall_score(y_test, predicted_y, average="micro"))
print("\nSIMPLE RNN WITH DROPOUT 0.6 WITH MACRO AVERAGE")
print("Accuray: ", accuracy_score(y_test, predicted_y))
print("F1 Score: ", f1_score(y_test, predicted_y, average="macro"))
print("Precision: ", precision_score(y_test, predicted_y, average="macro"))
print("Recall: ", recall_score(y_test, predicted_y, average="macro"))
################################# SIMPLE RNN DROUPOUT RATE 0.8 ########################################
model = keras.Sequential()
model.add(
SimpleRNN(
128, input_shape=(X_train.shape[1:]), activation="relu", return_sequences=True
)
)
model.add(Dropout(0.8))
model.add(SimpleRNN(128, activation="relu", return_sequences=False))
model.add(Dropout(0.8))
model.add(Dense(32, activation="relu"))
model.add(Dropout(0.8))
model.add(Dense(6, activation="softmax"))
# model.summary()
opt = tf.keras.optimizers.Adam(learning_rate=1e-3, decay=1e-5)
model.compile(loss="binary_crossentropy", optimizer=opt, metrics=["accuracy"])
print("Train...")
history = model.fit(X_train, y_train, validation_split=0.2, epochs=5)
predicted_y = np.argmax(model.predict(X_test), axis=1)
print("\nSIMPLE RNN WITH DROPOUT 0.8 WITH MICRO AVERAGE")
print("Accuray: ", accuracy_score(y_test, predicted_y))
print("F1 Score: ", f1_score(y_test, predicted_y, average="micro"))
print("Precision: ", precision_score(y_test, predicted_y, average="micro"))
print("Recall: ", recall_score(y_test, predicted_y, average="micro"))
print("\nSIMPLE RNN WITH DROPOUT 0.8 WITH MACRO AVERAGE")
print("Accuray: ", accuracy_score(y_test, predicted_y))
print("F1 Score: ", f1_score(y_test, predicted_y, average="macro"))
print("Precision: ", precision_score(y_test, predicted_y, average="macro"))
print("Recall: ", recall_score(y_test, predicted_y, average="macro"))
# **#################### GRU ##############################**
################################# GRU DROUPOUT RATE 0.2 ########################################
model = keras.Sequential()
model.add(
GRU(128, input_shape=(X_train.shape[1:]), activation="relu", return_sequences=True)
)
model.add(Dropout(0.2))
model.add(GRU(128, activation="relu", return_sequences=False))
model.add(Dropout(0.2))
model.add(Dense(32, activation="relu"))
model.add(Dropout(0.2))
model.add(Dense(6, activation="softmax"))
# model.summary()
opt = tf.keras.optimizers.Adam(learning_rate=1e-3, decay=1e-5)
model.compile(loss="binary_crossentropy", optimizer=opt, metrics=["accuracy"])
print("Train...")
history = model.fit(X_train, y_train, validation_split=0.2, epochs=5)
predicted_y = np.argmax(model.predict(X_test), axis=1)
print("\nGRU WITH DROPOUT 0.2 WITH MICRO AVERAGE")
print("Accuray: ", accuracy_score(y_test, predicted_y))
print("F1 Score: ", f1_score(y_test, predicted_y, average="micro"))
print("Precision: ", precision_score(y_test, predicted_y, average="micro"))
print("Recall: ", recall_score(y_test, predicted_y, average="micro"))
print("\nGRU WITH DROPOUT 0.2 WITH MACRO AVERAGE")
print("Accuray: ", accuracy_score(y_test, predicted_y))
print("F1 Score: ", f1_score(y_test, predicted_y, average="macro"))
print("Precision: ", precision_score(y_test, predicted_y, average="macro"))
print("Recall: ", recall_score(y_test, predicted_y, average="macro"))
################################# GRU DROUPOUT RATE 0.4 ########################################
model = keras.Sequential()
model.add(
GRU(128, input_shape=(X_train.shape[1:]), activation="relu", return_sequences=True)
)
model.add(Dropout(0.4))
model.add(GRU(128, activation="relu", return_sequences=False))
model.add(Dropout(0.4))
model.add(Dense(32, activation="relu"))
model.add(Dropout(0.4))
model.add(Dense(6, activation="softmax"))
# model.summary()
opt = tf.keras.optimizers.Adam(learning_rate=1e-3, decay=1e-5)
model.compile(loss="binary_crossentropy", optimizer=opt, metrics=["accuracy"])
print("Train...")
history = model.fit(X_train, y_train, validation_split=0.2, epochs=5)
predicted_y = np.argmax(model.predict(X_test), axis=1)
print("\nGRU WITH DROPOUT 0.4 WITH MICRO AVERAGE")
print("Accuray: ", accuracy_score(y_test, predicted_y))
print("F1 Score: ", f1_score(y_test, predicted_y, average="micro"))
print("Precision: ", precision_score(y_test, predicted_y, average="micro"))
print("Recall: ", recall_score(y_test, predicted_y, average="micro"))
print("\nGRU WITH DROPOUT 0.4 WITH MACRO AVERAGE")
print("Accuray: ", accuracy_score(y_test, predicted_y))
print("F1 Score: ", f1_score(y_test, predicted_y, average="macro"))
print("Precision: ", precision_score(y_test, predicted_y, average="macro"))
print("Recall: ", recall_score(y_test, predicted_y, average="macro"))
################################# GRU DROUPOUT RATE 0.6 ########################################
model = keras.Sequential()
model.add(
GRU(128, input_shape=(X_train.shape[1:]), activation="relu", return_sequences=True)
)
model.add(Dropout(0.6))
model.add(GRU(128, activation="relu", return_sequences=False))
model.add(Dropout(0.6))
model.add(Dense(32, activation="relu"))
model.add(Dropout(0.6))
model.add(Dense(6, activation="softmax"))
# model.summary()
opt = tf.keras.optimizers.Adam(learning_rate=1e-3, decay=1e-5)
model.compile(loss="binary_crossentropy", optimizer=opt, metrics=["accuracy"])
print("Train...")
history = model.fit(X_train, y_train, validation_split=0.2, epochs=5)
predicted_y = np.argmax(model.predict(X_test), axis=1)
print("\nGRU WITH DROPOUT 0.6 WITH MICRO AVERAGE")
print("Accuray: ", accuracy_score(y_test, predicted_y))
print("F1 Score: ", f1_score(y_test, predicted_y, average="micro"))
print("Precision: ", precision_score(y_test, predicted_y, average="micro"))
print("Recall: ", recall_score(y_test, predicted_y, average="micro"))
print("\nGRU WITH DROPOUT 0.6 WITH MACRO AVERAGE")
print("Accuray: ", accuracy_score(y_test, predicted_y))
print("F1 Score: ", f1_score(y_test, predicted_y, average="macro"))
print("Precision: ", precision_score(y_test, predicted_y, average="macro"))
print("Recall: ", recall_score(y_test, predicted_y, average="macro"))
################################# GRU DROUPOUT RATE 0.8 ########################################
model = keras.Sequential()
model.add(
GRU(128, input_shape=(X_train.shape[1:]), activation="relu", return_sequences=True)
)
model.add(Dropout(0.8))
model.add(GRU(128, activation="relu", return_sequences=False))
model.add(Dropout(0.8))
model.add(Dense(32, activation="relu"))
model.add(Dropout(0.8))
model.add(Dense(6, activation="softmax"))
# model.summary()
opt = tf.keras.optimizers.Adam(learning_rate=1e-3, decay=1e-5)
model.compile(loss="binary_crossentropy", optimizer=opt, metrics=["accuracy"])
print("Train...")
history = model.fit(X_train, y_train, validation_split=0.2, epochs=5)
predicted_y = np.argmax(model.predict(X_test), axis=1)
print("\nGRU WITH DROPOUT 0.8 WITH MICRO AVERAGE")
print("Accuray: ", accuracy_score(y_test, predicted_y))
print("F1 Score: ", f1_score(y_test, predicted_y, average="micro"))
print("Precision: ", precision_score(y_test, predicted_y, average="micro"))
print("Recall: ", recall_score(y_test, predicted_y, average="micro"))
print("\nGRU WITH DROPOUT 0.8 WITH MACRO AVERAGE")
print("Accuray: ", accuracy_score(y_test, predicted_y))
print("F1 Score: ", f1_score(y_test, predicted_y, average="macro"))
print("Precision: ", precision_score(y_test, predicted_y, average="macro"))
print("Recall: ", recall_score(y_test, predicted_y, average="macro"))
# **######################LSTM########################**
################################# LSTM DROUPOUT RATE 0.2 ########################################
model = keras.Sequential()
model.add(
LSTM(128, input_shape=(X_train.shape[1:]), activation="relu", return_sequences=True)
)
model.add(Dropout(0.2))
model.add(LSTM(128, activation="relu", return_sequences=False))
model.add(Dropout(0.2))
model.add(Dense(32, activation="relu"))
model.add(Dropout(0.2))
model.add(Dense(6, activation="softmax"))
# model.summary()
opt = tf.keras.optimizers.Adam(learning_rate=1e-3, decay=1e-5)
model.compile(loss="binary_crossentropy", optimizer=opt, metrics=["accuracy"])
print("Train...")
history = model.fit(X_train, y_train, validation_split=0.2, epochs=5)
predicted_y = np.argmax(model.predict(X_test), axis=1)
print("\nLSTM WITH DROPOUT 0.2 WITH MICRO AVERAGE")
print("Accuray: ", accuracy_score(y_test, predicted_y))
print("F1 Score: ", f1_score(y_test, predicted_y, average="micro"))
print("Precision: ", precision_score(y_test, predicted_y, average="micro"))
print("Recall: ", recall_score(y_test, predicted_y, average="micro"))
print("\nLSTM WITH DROPOUT 0.2 WITH MACRO AVERAGE")
print("Accuray: ", accuracy_score(y_test, predicted_y))
print("F1 Score: ", f1_score(y_test, predicted_y, average="macro"))
print("Precision: ", precision_score(y_test, predicted_y, average="macro"))
print("Recall: ", recall_score(y_test, predicted_y, average="macro"))
################################# LSTM DROUPOUT RATE 0.4 ########################################
model = keras.Sequential()
model.add(
LSTM(128, input_shape=(X_train.shape[1:]), activation="relu", return_sequences=True)
)
model.add(Dropout(0.4))
model.add(LSTM(128, activation="relu", return_sequences=False))
model.add(Dropout(0.4))
model.add(Dense(32, activation="relu"))
model.add(Dropout(0.4))
model.add(Dense(6, activation="softmax"))
# model.summary()
opt = tf.keras.optimizers.Adam(learning_rate=1e-3, decay=1e-5)
model.compile(loss="binary_crossentropy", optimizer=opt, metrics=["accuracy"])
print("Train...")
history = model.fit(X_train, y_train, validation_split=0.2, epochs=5)
predicted_y = np.argmax(model.predict(X_test), axis=1)
print("\nLSTM WITH DROPOUT 0.4 WITH MICRO AVERAGE")
print("Accuray: ", accuracy_score(y_test, predicted_y))
print("F1 Score: ", f1_score(y_test, predicted_y, average="micro"))
print("Precision: ", precision_score(y_test, predicted_y, average="micro"))
print("Recall: ", recall_score(y_test, predicted_y, average="micro"))
print("\nLSTM WITH DROPOUT 0.4 WITH MACRO AVERAGE")
print("Accuray: ", accuracy_score(y_test, predicted_y))
print("F1 Score: ", f1_score(y_test, predicted_y, average="macro"))
print("Precision: ", precision_score(y_test, predicted_y, average="macro"))
print("Recall: ", recall_score(y_test, predicted_y, average="macro"))
################################# LSTM DROUPOUT RATE 0.6 ########################################
model = keras.Sequential()
model.add(
LSTM(128, input_shape=(X_train.shape[1:]), activation="relu", return_sequences=True)
)
model.add(Dropout(0.6))
model.add(LSTM(128, activation="relu", return_sequences=False))
model.add(Dropout(0.6))
model.add(Dense(32, activation="relu"))
model.add(Dropout(0.6))
model.add(Dense(6, activation="softmax"))
# model.summary()
opt = tf.keras.optimizers.Adam(learning_rate=1e-3, decay=1e-5)
model.compile(loss="binary_crossentropy", optimizer=opt, metrics=["accuracy"])
print("Train...")
history = model.fit(X_train, y_train, validation_split=0.2, epochs=5)
predicted_y = np.argmax(model.predict(X_test), axis=1)
print("\nLSTM WITH DROPOUT 0.6 WITH MICRO AVERAGE")
print("Accuray: ", accuracy_score(y_test, predicted_y))
print("F1 Score: ", f1_score(y_test, predicted_y, average="micro"))
print("Precision: ", precision_score(y_test, predicted_y, average="micro"))
print("Recall: ", recall_score(y_test, predicted_y, average="micro"))
print("\nLSTM WITH DROPOUT 0.6 WITH MACRO AVERAGE")
print("Accuray: ", accuracy_score(y_test, predicted_y))
print("F1 Score: ", f1_score(y_test, predicted_y, average="macro"))
print("Precision: ", precision_score(y_test, predicted_y, average="macro"))
print("Recall: ", recall_score(y_test, predicted_y, average="macro"))
################################# LSTM DROUPOUT RATE 0.8 ########################################
model = keras.Sequential()
model.add(
LSTM(128, input_shape=(X_train.shape[1:]), activation="relu", return_sequences=True)
)
model.add(Dropout(0.8))
model.add(LSTM(128, activation="relu", return_sequences=False))
model.add(Dropout(0.8))
model.add(Dense(32, activation="relu"))
model.add(Dropout(0.8))
model.add(Dense(6, activation="softmax"))
# model.summary()
opt = tf.keras.optimizers.Adam(learning_rate=1e-3, decay=1e-5)
model.compile(loss="binary_crossentropy", optimizer=opt, metrics=["accuracy"])
print("Train...")
history = model.fit(X_train, y_train, validation_split=0.2, epochs=5)
predicted_y = np.argmax(model.predict(X_test), axis=1)
print("\nLSTM WITH DROPOUT 0.8 WITH MICRO AVERAGE")
print("Accuray: ", accuracy_score(y_test, predicted_y))
print("F1 Score: ", f1_score(y_test, predicted_y, average="micro"))
print("Precision: ", precision_score(y_test, predicted_y, average="micro"))
print("Recall: ", recall_score(y_test, predicted_y, average="micro"))
print("\nLSTM WITH DROPOUT 0.8 WITH MACRO AVERAGE")
print("Accuray: ", accuracy_score(y_test, predicted_y))
print("F1 Score: ", f1_score(y_test, predicted_y, average="macro"))
print("Precision: ", precision_score(y_test, predicted_y, average="macro"))
print("Recall: ", recall_score(y_test, predicted_y, average="macro"))
# **################## BIDIRECTIONAL LSTM #######################**
################################# Bidirectional LSTM DROUPOUT RATE 0.2 ########################################
model = keras.Sequential()
model.add(
Bidirectional(
(
LSTM(
128,
input_shape=(X_train.shape[1:]),
activation="relu",
return_sequences=True,
)
)
)
)
model.add(Dropout(0.2))
model.add(Bidirectional((LSTM(128, activation="relu", return_sequences=False))))
model.add(Dropout(0.2))
model.add(Dense(32, activation="relu"))
model.add(Dropout(0.2))
model.add(Dense(6, activation="softmax"))
# model.summary()
opt = tf.keras.optimizers.Adam(learning_rate=1e-3, decay=1e-5)
model.compile(loss="binary_crossentropy", optimizer=opt, metrics=["accuracy"])
print("Train...")
history = model.fit(X_train, y_train, validation_split=0.2, epochs=5)
predicted_y = np.argmax(model.predict(X_test), axis=1)
print("\nBidirectional LSTM WITH DROPOUT 0.2 WITH MICRO AVERAGE")
print("Accuray: ", accuracy_score(y_test, predicted_y))
print("F1 Score: ", f1_score(y_test, predicted_y, average="micro"))
print("Precision: ", precision_score(y_test, predicted_y, average="micro"))
print("Recall: ", recall_score(y_test, predicted_y, average="micro"))
print("\nnBidirectional LSTM WITH DROPOUT 0.2 WITH MACRO AVERAGE")
print("Accuray: ", accuracy_score(y_test, predicted_y))
print("F1 Score: ", f1_score(y_test, predicted_y, average="macro"))
print("Precision: ", precision_score(y_test, predicted_y, average="macro"))
print("Recall: ", recall_score(y_test, predicted_y, average="macro"))
################################# Bidirectional LSTM DROUPOUT RATE 0.4 ########################################
model = keras.Sequential()
model.add(
Bidirectional(
(
LSTM(
128,
input_shape=(X_train.shape[1:]),
activation="relu",
return_sequences=True,
)
)
)
)
model.add(Dropout(0.4))
model.add(Bidirectional((LSTM(128, activation="relu", return_sequences=False))))
model.add(Dropout(0.4))
model.add(Dense(32, activation="relu"))
model.add(Dropout(0.4))
model.add(Dense(6, activation="softmax"))
# model.summary()
opt = tf.keras.optimizers.Adam(learning_rate=1e-3, decay=1e-5)
model.compile(loss="binary_crossentropy", optimizer=opt, metrics=["accuracy"])
print("Train...")
history = model.fit(X_train, y_train, validation_split=0.2, epochs=5)
predicted_y = np.argmax(model.predict(X_test), axis=1)
print("\nBidirectional LSTM WITH DROPOUT 0.4 WITH MICRO AVERAGE")
print("Accuray: ", accuracy_score(y_test, predicted_y))
print("F1 Score: ", f1_score(y_test, predicted_y, average="micro"))
print("Precision: ", precision_score(y_test, predicted_y, average="micro"))
print("Recall: ", recall_score(y_test, predicted_y, average="micro"))
print("\nnBidirectional LSTM WITH DROPOUT 0.4 WITH MACRO AVERAGE")
print("Accuray: ", accuracy_score(y_test, predicted_y))
print("F1 Score: ", f1_score(y_test, predicted_y, average="macro"))
print("Precision: ", precision_score(y_test, predicted_y, average="macro"))
print("Recall: ", recall_score(y_test, predicted_y, average="macro"))
################################# Bidirectional LSTM DROUPOUT RATE 0.6 ########################################
model = keras.Sequential()
model.add(
Bidirectional(
(
LSTM(
128,
input_shape=(X_train.shape[1:]),
activation="relu",
return_sequences=True,
)
)
)
)
model.add(Dropout(0.6))
model.add(Bidirectional((LSTM(128, activation="relu", return_sequences=False))))
model.add(Dropout(0.6))
model.add(Dense(32, activation="relu"))
model.add(Dropout(0.6))
model.add(Dense(6, activation="softmax"))
# model.summary()
opt = tf.keras.optimizers.Adam(learning_rate=1e-3, decay=1e-5)
model.compile(loss="binary_crossentropy", optimizer=opt, metrics=["accuracy"])
print("Train...")
history = model.fit(X_train, y_train, validation_split=0.2, epochs=5)
predicted_y = np.argmax(model.predict(X_test), axis=1)
print("\nBidirectional LSTM WITH DROPOUT 0.6 WITH MICRO AVERAGE")
print("Accuray: ", accuracy_score(y_test, predicted_y))
print("F1 Score: ", f1_score(y_test, predicted_y, average="micro"))
print("Precision: ", precision_score(y_test, predicted_y, average="micro"))
print("Recall: ", recall_score(y_test, predicted_y, average="micro"))
print("\nnBidirectional LSTM WITH DROPOUT 0.6 WITH MACRO AVERAGE")
print("Accuray: ", accuracy_score(y_test, predicted_y))
print("F1 Score: ", f1_score(y_test, predicted_y, average="macro"))
print("Precision: ", precision_score(y_test, predicted_y, average="macro"))
print("Recall: ", recall_score(y_test, predicted_y, average="macro"))
################################# Bidirectional LSTM DROUPOUT RATE 0.8 ########################################
model = keras.Sequential()
model.add(
Bidirectional(
(
LSTM(
128,
input_shape=(X_train.shape[1:]),
activation="relu",
return_sequences=True,
)
)
)
)
model.add(Dropout(0.8))
model.add(Bidirectional((LSTM(128, activation="relu", return_sequences=False))))
model.add(Dropout(0.8))
model.add(Dense(32, activation="relu"))
model.add(Dropout(0.8))
model.add(Dense(6, activation="softmax"))
# model.summary()
opt = tf.keras.optimizers.Adam(learning_rate=1e-3, decay=1e-5)
model.compile(loss="binary_crossentropy", optimizer=opt, metrics=["accuracy"])
print("Train...")
history = model.fit(X_train, y_train, validation_split=0.2, epochs=5)
predicted_y = np.argmax(model.predict(X_test), axis=1)
print("\nBidirectional LSTM WITH DROPOUT 0.8 WITH MICRO AVERAGE")
print("Accuray: ", accuracy_score(y_test, predicted_y))
print("F1 Score: ", f1_score(y_test, predicted_y, average="micro"))
print("Precision: ", precision_score(y_test, predicted_y, average="micro"))
print("Recall: ", recall_score(y_test, predicted_y, average="micro"))
print("\nnBidirectional LSTM WITH DROPOUT 0.8 WITH MACRO AVERAGE")
print("Accuray: ", accuracy_score(y_test, predicted_y))
print("F1 Score: ", f1_score(y_test, predicted_y, average="macro"))
print("Precision: ", precision_score(y_test, predicted_y, average="macro"))
print("Recall: ", recall_score(y_test, predicted_y, average="macro"))
|
# # PSS3, E12. Kidney Stone Prediction
# # Define the objetive
# ### Thanks [Kidney Stone Prediction based on Urine Analysis](https://www.kaggle.com/datasets/vuppalaadithyasairam/kidney-stone-prediction-based-on-urine-analysis) by [@vuppalaadithyasairam](https://www.kaggle.com/vuppalaadithyasairam)
# Determine if certain physical characteristics of the urine might be related to the formation of calcium oxalate crystals.
# The six physical characteristics of the urine are:
# (1) `gravity` specific gravity, the density of the urine relative to water.
# (2) `ph` pH, the negative logarithm of the hydrogen ion.
# (3) `osmo` osmolarity (mOsm), a unit used in biology and medicine but not in physical chemistry. Osmolarity is proportional to the concentration of molecules in solution.
# (4) `cond` conductivity (mMho milliMho). One Mho is one reciprocal Ohm. Conductivity is proportional to the concentration of charged ions in solution.
# (5) `urea` concentration in millimoles per litre.
# (6) `calc` calcium.
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
from pprint import pprint
class conf:
index = "id"
target = "target"
random = 2023
load_original = True
only_positive = False
np.random.seed(conf.random)
train_full = pd.read_csv(
"/kaggle/input/playground-series-s3e12/train.csv", index_col=conf.index
)
test_full = pd.read_csv(
"/kaggle/input/playground-series-s3e12/test.csv", index_col=conf.index
)
train = train_full.copy()
test = test_full.copy()
if conf.load_original:
print("Load external data...")
original_tr = pd.read_csv(
"/kaggle/input/kidney-stone-prediction-based-on-urine-analysis/kindey stone urine analysis.csv"
)
# original_te = pd.read_csv('/kaggle/input/media-campaign-cost-prediction/test_dataset.csv')
if conf.only_positive:
train = pd.concat(
[original_tr[original_tr[conf.target] == 1], train_full], ignore_index=True
)
# test = pd.concat([original_te[original_te[conf.target] == 1], test_full], ignore_index=True)
else:
train = pd.concat([original_tr, train_full], ignore_index=True)
# test = pd.concat([original_te, test_full], ignore_index=True)
features_ex = test.columns.tolist()
# # Explore the data
train_ex = train.copy()
test_ex = test.copy()
# ## No Missing Values
percent_missing = train_ex.isnull().sum() * 100 / len(train_ex)
missing_value_df = pd.DataFrame(
{"column_name": train_ex.columns, "percent_missing": percent_missing}
)
display(missing_value_df)
# ## No duplicate Values
train_ex.duplicated().value_counts()
train_ex[conf.target].value_counts()
# #### Descriptive statistics (min, max, mean, std, varciance, etc.) of each attirbutes
train_ex[features_ex].describe().T
# ## Visualize the data
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
ax = train_full[conf.target].plot.hist(alpha=0.5, figsize=(10, 6))
ax = original_tr[conf.target].plot.hist(alpha=0.5, figsize=(10, 6))
ax.legend(["syntetic", "original"])
plt.figure(figsize=(14, 8))
plt.suptitle("Frequency of attributes in train")
for i, col in enumerate(features_ex):
plt.subplot(3, 2, i + 1)
ax1 = train_full[col].plot.hist(alpha=0.5, density=True)
ax2 = original_tr[col].plot.hist(alpha=0.5, density=True)
ax1.set_title(col)
ax2.legend(["syntetic", "original"])
# sns.histplot(train_full[col], color='r', kde=True)
# sns.histplot(original_tr[col], color='g', kde=True)
plt.grid()
plt.tight_layout()
# ## Study the correlations
# #### Check feature importance using Mutual information
from sklearn.feature_selection import mutual_info_classif, mutual_info_regression
mi = mutual_info_classif(
train_ex[features_ex],
train_ex[[conf.target]][conf.target].values,
discrete_features="auto",
n_neighbors=3,
copy=True,
random_state=2,
)
ax = (
pd.DataFrame([mi], columns=features_ex)
.transpose()
.plot.bar(figsize=(8, 6), legend=False)
)
ax.grid()
ax.set_title("Calcium and Urea are the most important factors to Kidney Stone")
# #### For each low MI scored feature check interaction with other features
hi_estimated_mi = (
pd.DataFrame([mi], columns=features_ex)
.T.rename(columns={0: "mi_score"})
.sort_values(by="mi_score", ascending=False)
)
hi_estimated_mi = hi_estimated_mi[hi_estimated_mi.mi_score > 0.07].index.tolist()
low_mi_feat = features_ex.copy()
low_mi_feat.remove("calc")
low_mi = mutual_info_regression(
train_ex[low_mi_feat],
train_ex[["calc"]]["calc"].values,
discrete_features="auto",
n_neighbors=3,
copy=True,
random_state=2,
)
ax = (
pd.DataFrame([low_mi], columns=low_mi_feat)
.transpose()
.plot.bar(figsize=(8, 6), legend=False)
)
ax.grid()
ax.set_title("Gravity had an impact on Calcium")
from sklearn.feature_selection import f_regression
f_regression(train_ex[["gravity", "urea"]], train_ex[["calc"]]["calc"].values)
# #### Correlations between the variables
fig, ax = plt.subplots(figsize=(5, 4))
# https://www.kaggle.com/code/sergiosaharovskiy/ps-s3e11-2023-eda-and-submission?scriptVersionId=123851844&cellId=26
corr = train_ex[features_ex].corr()
mask = np.zeros_like(corr)
mask[np.triu_indices_from(mask)] = True
ax = sns.heatmap(
corr,
mask=mask,
vmin=-1,
vmax=1,
cmap=sns.diverging_palette(20, 220, as_cmap=True),
annot=True,
)
ax.set_title(
"All feature down the pH. and Urea are the most important factors to Kidney Stone"
)
plt.tight_layout()
plt.show()
# * `gravity` and `urea` have the same negative correlation over `pH`.
# * `gravity`, `osmo` and `urea` have the equal positive correlation over `calc`.
# * If `pH` arise the other ones features downs.
# # Prepare the data
# ## Data cleaning
# ### Fix or remove outliers
plt.figure(figsize=(7, 6))
plt.suptitle("Frequency of attributes in train")
for i, col in enumerate(features_ex):
plt.subplot(2, 3, i + 1)
ax1 = train_full[col].plot.box()
# ax2 = original_tr[col].plot.box()
# ax1.set_title(col)
# ax2.legend(['syntetic', 'original'])
# sns.histplot(train_full[col], color='r', kde=True)
# sns.histplot(original_tr[col], color='g', kde=True)
plt.grid()
plt.tight_layout()
# ## Feature engineering
# #### Combining the top features with other related features
# ##### Relationships among high importance features: `calc`, `urea`, `osmo`, `cond`
# F-test captures only linear dependency. On the other hand, mutual information can capture any kind of dependency between variables and it rates ([source](https://scikit-learn.org/stable/modules/feature_selection.html#univariate-feature-selection))
# We compute whether there is a statistically significant relationship between each feature and the target and the features that are related with the highest confidence
from sklearn.feature_selection import f_regression, mutual_info_regression
mi = mutual_info_classif(
train_ex[features_ex],
train_ex[[conf.target]][conf.target].values,
discrete_features="auto",
n_neighbors=3,
copy=True,
random_state=2,
)
pd.DataFrame([mi], columns=features_ex).T.rename(columns={0: "mi_score"}).sort_values(
by="mi_score", ascending=False
)
hi_estimated_mi = (
pd.DataFrame([mi], columns=features_ex)
.T.rename(columns={0: "mi_score"})
.sort_values(by="mi_score", ascending=False)
)
hi_estimated_mi = hi_estimated_mi[hi_estimated_mi.mi_score > 0.07].index.tolist()
# ### Mathematical transforms
# #### Are any relationships among numerical features expressed through mathematical formula?
# We consider 3 features and I try discover how each feature (target) depends on the other ones :
# target = f2(feature_1) + f2(feature_2),
# i.e
# `calc` = f1(`urea`)+f2(`osmo`)
# `urea` = f1(`calc`)+f2(`osmo`)
# `osmo`= f1(`calc`)+f2(`urea`)
# Acording to F-test only captures linear dependency over features `calc`, `urea` and `osmo`. Mutual Information can capture any kind of dependency between variables.
# So F-test and Mutual Information shows:
# - To `calc` = f1(`urea`)+f2(`osmo`) rates that both features have strong dependency. `osmo` have a little more dependence than `urea` .
#
# - To `urea` = f1(`calc`)+f2(`osmo`) rates that `osmo` have a strong dependence.
#
# - To `osmo`= f1(`calc`)+f2(`urea`) rates that `urea` have a strong dependence.
#
# [source](https://scikit-learn.org/stable/auto_examples/feature_selection/plot_f_test_vs_mi.html#sphx-glr-auto-examples-feature-selection-plot-f-test-vs-mi-py)
# https://scikit-learn.org/stable/auto_examples/feature_selection/plot_f_test_vs_mi.html#comparison-of-f-test-and-mutual-information
features_to_math_formula = hi_estimated_mi
for feat in features_to_math_formula:
other_feat = features_to_math_formula.copy()
other_feat.remove(feat)
X = train_ex[other_feat].to_numpy()
y = train_ex[[feat]][feat].values
f_test, _ = f_regression(X, y)
f_test /= np.max(f_test)
mi = mutual_info_regression(X, y)
mi /= np.max(mi)
n_feat = len(features_to_math_formula) - 1
plt.figure(figsize=(10, 3))
# plt.title('Top features with other related features with F-test and Mutual Information')
for i in range(n_feat):
plt.subplot(1, n_feat, i + 1)
plt.scatter(X[:, i], y, edgecolor="black", s=20)
plt.xlabel("{}".format(other_feat[i]), fontsize=8)
if i == 0:
plt.ylabel("{}".format(feat), fontsize=8)
plt.title("F-test={:.2f}, MI={:.2f}".format(f_test[i], mi[i]), fontsize=8)
plt.show()
# Need more work
# ##### Try find a mathematical equations of transforms through Ordinary least squares Linear Regression
from sklearn.linear_model import LinearRegression
lin_reg = LinearRegression()
X_math_trans_train = train_ex[["urea", "osmo"]]
y_math_trans_train = train_ex[["calc"]]["calc"].values
lin_reg.fit(X_math_trans_train, y_math_trans_train)
lin_reg.intercept_, lin_reg.coef_
X_new = np.array([[354], [698]])
lin_reg.predict(X_new.transpose())
# Need more work
# #### "Reshaping" features through powers or logarithms to create new features
math_trans_train = train_ex[["calc", "urea", "osmo", "target"]]
math_trans_train["calc_log10"] = train_ex["calc"].apply(lambda x: np.log10(x))
math_trans_train["urea_log10"] = train_ex["urea"].apply(lambda x: np.log10(x))
# math_trans_train['osmo'] = train_pr['osmo'].apply( lambda x: np.exp(x))
train_pr = math_trans_train[["calc_log10", "urea_log10", "target"]]
reshaping_feat = ["calc_log10", "urea_log10"]
# ### Creating features
# #### Ratio combinations
train_pr["calc_per_gravity"] = train_ex.calc / train_ex.gravity
train_pr["calc_per_urea"] = train_ex.calc / train_ex.urea
# ratio_train_pr['gravity_urea'] = ratio_train_pr.gravity/ratio_train_pr.urea
train_pr
# ### Group Transforms
# #### Aggregate information across multiple rows grouped by some category(feature).
from sklearn.preprocessing import KBinsDiscretizer
n_bins = 10
kbins_ordinal = KBinsDiscretizer(n_bins=n_bins, strategy="uniform", encode="ordinal")
train_kbins_ordinal = kbins_ordinal.fit_transform(train_ex[["calc", "urea"]])
df_train_kbins_ordinal = pd.DataFrame(
train_kbins_ordinal, columns=["calc_bin", "urea_bin"]
)
train_pr_kbins_ordinal = pd.concat(
[train_ex[["calc", "urea"]], df_train_kbins_ordinal], axis=1
)
calc_bins_means = (
train_pr_kbins_ordinal.groupby(["calc_bin"])["calc"]
.mean()
.to_frame()
.rename(columns={"calc": "calc_mean_per_bin"})
)
urea_bins_means = (
train_pr_kbins_ordinal.groupby(["urea_bin"])["urea"]
.mean()
.to_frame()
.rename(columns={"urea": "urea_mean_per_bin"})
)
means_per_bins = pd.merge(
df_train_kbins_ordinal, calc_bins_means, how="left", on="calc_bin"
)
means_per_bins = pd.merge(means_per_bins, urea_bins_means, how="left", on="urea_bin")
# #### Discretize continuous features into intervals with bins
bins_cols = []
for i in range(2 * n_bins):
bins_col = f"bins_{i}"
bins_cols.append(bins_col)
kbins = KBinsDiscretizer(n_bins=n_bins, strategy="uniform", encode="onehot-dense")
train_kbins = kbins.fit_transform(train_ex[["calc", "urea"]])
df_train_kbins = pd.DataFrame(train_kbins, columns=bins_cols)
train_pr = pd.concat([train_pr, df_train_kbins], axis=1)
# Get worse score if add `means_per_bins`
# train_pr = pd.concat([train_pr, means_per_bins[['calc_mean_per_bin', 'urea_mean_per_bin']]], axis=1)
# # Train many quick and dirty models from different categories
# ### From [Utility Script Competition](https://www.kaggle.com/general/10965) this [comment](https://www.kaggle.com/general/109651#633850) thanks to [chmaxx](https://www.kaggle.com/chmaxx)
import quick_classification
df_score_train_ex = quick_classification.score_models(
train_ex, conf.target, scoring_metric="roc_auc", verbose=False
)
df_scores = df_score_train_ex.rename(columns={"roc_auc": "train_ex"})
df_scores.index = df_scores.Classifier
df_scores = df_scores.drop(columns=["Classifier"])
hi_mi_and_target = hi_estimated_mi.copy()
hi_mi_and_target.append("target")
df_score_hi_estimated_mi = quick_classification.score_models(
train_ex[hi_mi_and_target], conf.target, scoring_metric="roc_auc", verbose=False
)
df_score_hi_estimated_mi = df_score_hi_estimated_mi.rename(
columns={"roc_auc": "hi_estimated_mi"}
)
df_score_hi_estimated_mi.index = df_score_hi_estimated_mi.Classifier
df_score_hi_estimated_mi = df_score_hi_estimated_mi.drop(columns=["Classifier"])
df_scores = pd.concat([df_score_hi_estimated_mi, df_scores], axis=1)
df_score_train_pr = quick_classification.score_models(
train_pr, conf.target, scoring_metric="roc_auc", verbose=False
)
df_score_train_pr = df_score_train_pr.rename(columns={"roc_auc": "train_pr"})
df_score_train_pr.index = df_score_train_pr.Classifier
df_score_train_pr = df_score_train_pr.drop(columns=["Classifier"])
df_scores = pd.concat([df_score_train_pr, df_scores], axis=1)
df_scores
df_scores.T.mean().sort_values(ascending=True).plot.barh(color="chocolate")
plt.title("Model comparison by mean (bigger bar is better)")
plt.xlim(0.4, 0.84)
# ---
# # Next sections are working in progress =)
# # Fine tune models
from sklearn.inspection import PartialDependenceDisplay
from sklearn.ensemble import HistGradientBoostingClassifier
from sklearn.metrics import roc_auc_score
from sklearn.model_selection import StratifiedShuffleSplit
from sklearn.model_selection import train_test_split
from sklearn.pipeline import make_pipeline
from sklearn.model_selection import GridSearchCV
from sklearn.preprocessing import StandardScaler
y = train_ex[conf.target]
X = train_ex.drop([conf.target], axis=1)
split = StratifiedShuffleSplit(n_splits=1, test_size=0.2, random_state=42)
for train_index, test_index in split.split(X, y):
strat_train_set = train_ex.loc[train_index]
strat_test_set = train_ex.loc[test_index]
X_train, X_test, y_train, y_test = (
strat_train_set[features_ex],
strat_test_set[features_ex],
strat_train_set[conf.target],
strat_test_set[conf.target],
)
# ##### Histogram-based Gradient Boosting Classification Tree
param_grid = {
"histgradientboostingclassifier__max_depth": [1, 2, 4, 10],
"histgradientboostingclassifier__random_state": [1, 100, 200],
"histgradientboostingclassifier__scoring": ["roc_auc"],
}
pipe = make_pipeline(StandardScaler(), HistGradientBoostingClassifier())
grid = GridSearchCV(pipe, param_grid, cv=5)
grid.fit(X_train, y_train)
y_pred = grid.predict_proba(X_test)[:, 1]
roc_auc_score(y_test, y_pred)
# ##### To plot the ROC curve,
from sklearn.metrics import RocCurveDisplay
RocCurveDisplay.from_predictions(y_test, y_pred)
grid.best_params_
# pprint(grid.cv_results_)
# ##### RandomForestClassifier
from sklearn.ensemble import RandomForestClassifier
for cv in np.arange(7, 13):
param_grid = {
"randomforestclassifier__n_estimators": [50, 100, 200],
"randomforestclassifier__random_state": [1, 2],
"randomforestclassifier__min_samples_split": [2, 5, 10],
# 'randomforestclassifier__min_samples_leaf': [1, 2, 3]
}
pipe = make_pipeline(StandardScaler(), RandomForestClassifier())
grid = GridSearchCV(pipe, param_grid, cv=cv)
grid.fit(X_train, y_train)
y_pred = grid.predict_proba(X_test)[:, 1]
print(
f"roc_auc_score:{roc_auc_score(y_test, y_pred)} | "
f"cv: {cv} | "
f"best_params:{grid.best_params_}"
)
# display(roc_auc_score(y_test, y_pred))
# ##### Linear classifiers (SVM, logistic regression, etc.) with SGD training
from sklearn.linear_model import LogisticRegression
param_grid = {
"logisticregression__penalty": ["l2", "l1"],
"logisticregression__random_state": [1, 2, 3],
"logisticregression__tol": [1e-4, 1e-3, 1e-2],
"logisticregression__solver": ["liblinear"],
}
pipe = make_pipeline(StandardScaler(), LogisticRegression())
grid = GridSearchCV(pipe, param_grid, cv=3)
grid.fit(X_train, y_train)
y_pred = grid.predict_proba(X_test)[:, 1]
roc_auc_score(y_test, y_pred)
grid.best_params_
# pprint(grid.cv_results_)
# ## Analyze the types of errors the models make
X_test["y_pred"] = y_pred
X_test["target"] = y_test
X_test["y_errors"] = (X_test.y_pred - X_test.target).abs().round(1)
X_test["y_errors"].value_counts()
X_test_errors = X_test[X_test.y_errors > 0.5]
X_test_errors_positive = X_test_errors[X_test_errors.target == 1]
# #### Probability calibration
# ##### Plot calibration of binary classifier
from sklearn.calibration import CalibrationDisplay
histGBC = HistGradientBoostingClassifier(max_depth=2, random_state=1).fit(
X_train, y_train
)
disp = CalibrationDisplay.from_estimator(histGBC, X_test, y_test)
plt.show()
from sklearn.calibration import CalibrationDisplay
y_prob = histGBC.predict_proba(X_test)[:, 1]
disp = CalibrationDisplay.from_predictions(y_test, y_prob)
plt.show()
fig = plt.figure()
ax1 = fig.add_subplot(2, 1, 1)
ax2 = fig.add_subplot(2, 1, 2)
ax1.hist(y_prob, bins=20, color="black", alpha=0.5)
ax1.set_title("Y Predicted")
ax2.hist(y_test, bins=20, color="blue", alpha=0.5)
ax2.set_title("Y True")
# ##### Calibration with isotonic regression or logistic regression
from sklearn.calibration import CalibratedClassifierCV
calibrated_histGBC = CalibratedClassifierCV(histGBC, cv=3)
calibrated_histGBC.fit(X_test, y_test)
y_calib = calibrated_histGBC.predict_proba(X)[:, 1]
# #### Utility Script: classification
#
import quick_classification
hi_mi_and_target = hi_estimated_mi.copy()
hi_mi_and_target.append("target")
quick_classification.score_models(
train_ex[hi_mi_and_target], conf.target, scoring_metric="roc_auc"
)
quick_classification.score_models(train_ex, conf.target, scoring_metric="roc_auc")
|
# # Read the data from the file Balance.csv
# # Use any instructions you need to check your data
import pandas as pd
df = pd.read_csv("/kaggle/input/teaching-balance-example/balance.csv")
df.head()
# # find the total balance in each customer account
df["Total"] = df["Balance"] + df["Deposite"]
df.head()
# # Find the total sum of the amounts
sum = df["Total"].mean()
sum
# # Find the owner of the maximum amount
mx = df["Total"].max()
x = df[df["Total"] == mx]
x
# # Add 10 percent to the total amount for all customers
df["Total"] = df["Total"] + 0.01 * df["Total"]
df.head()
|
# # **NETFLIX ANALYSIS**
import pandas as pd
import numpy as np
import matplotlib as mpl
import matplotlib.pyplot as plt
import seaborn as sb
import re
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
df = pd.read_csv("../input/netflix-shows/netflix_titles.csv")
df.head()
df.isnull().sum()
df["director"] = df["director"].fillna("Unknown")
df["cast"] = df["cast"].fillna("Unknown")
df["country"] = df["country"].fillna("Unknown")
df["rating"] = df["rating"].fillna("Unknown")
df["date_added"] = df["date_added"].fillna("Unknown")
# fill nulls with 'unknown'
# now check again
df.isnull().sum()
# check type
df.dtypes
# sliptting the year and month the movie was added to netflix
df["year_added"] = df["date_added"].apply(lambda x: x.split(" ")[-1])
df["month_added"] = df["date_added"].apply(lambda x: x.split(" ")[0])
df
data = pd.DataFrame(df.groupby("type")["title"].count())
data.reset_index(inplace=True)
data.columns = ["type", "value"]
data
|
#
# Mutual Information by Ryan Holbrook
# All script by Ryan Holbrook https://www.kaggle.com/mpwolke/exercise-mutual-information/edit
# Feature Engineering Kaggle Micro-course by Ryan Holbrook.
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# #Introduction
# In this exercise you'll identify an initial set of features in the Ames dataset to develop using mutual information scores and interaction plots.
# Setup feedback system
from learntools.core import binder
binder.bind(globals())
from learntools.feature_engineering_new.ex2 import *
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import seaborn as sns
from sklearn.feature_selection import mutual_info_regression
# Set Matplotlib defaults
plt.style.use("seaborn-whitegrid")
plt.rc("figure", autolayout=True)
plt.rc(
"axes",
labelweight="bold",
labelsize="large",
titleweight="bold",
titlesize=14,
titlepad=10,
)
# Load data
df = pd.read_csv("../input/fe-course-data/ames.csv")
# Utility functions from Tutorial
def make_mi_scores(X, y):
X = X.copy()
for colname in X.select_dtypes(["object", "category"]):
X[colname], _ = X[colname].factorize()
# All discrete features should now have integer dtypes
discrete_features = [pd.api.types.is_integer_dtype(t) for t in X.dtypes]
mi_scores = mutual_info_regression(
X, y, discrete_features=discrete_features, random_state=0
)
mi_scores = pd.Series(mi_scores, name="MI Scores", index=X.columns)
mi_scores = mi_scores.sort_values(ascending=False)
return mi_scores
def plot_mi_scores(scores):
scores = scores.sort_values(ascending=True)
width = np.arange(len(scores))
ticks = list(scores.index)
color = np.array(["C0"] * scores.shape[0])
# Color red for probes
idx = [i for i, col in enumerate(scores.index) if col.startswith("PROBE")]
color[idx] = "C3"
# Create plot
plt.barh(width, scores, color=color)
plt.yticks(width, ticks)
plt.title("Mutual Information Scores")
# #Review the meaning of mutual information by looking at a few features from the Ames dataset.
features = ["YearBuilt", "MoSold", "ScreenPorch"]
sns.relplot(
x="value",
y="SalePrice",
col="variable",
data=df.melt(id_vars="SalePrice", value_vars=features),
facet_kws=dict(sharex=False),
)
# #Understanding Mutual Information
# Based on the plots, which feature do you think would have the highest mutual information with SalePrice?
# Based on the plots, YearBuilt should have the highest MI score since knowing the year tends to constrain SalePrice to a smaller range of possible values. This is generally not the case for MoSold, however. Finally, since ScreenPorch is usually just one value, 0, on average it won't tell you much about SalePrice (though more than MoSold) .
# The Ames dataset has seventy-eight features -- a lot to work with all at once! Fortunately, you can identify the features with the most potential.
# Use the make_mi_scores function (introduced in the tutorial) to compute mutual information scores for the Ames features:
X = df.copy()
y = X.pop("SalePrice")
mi_scores = make_mi_scores(X, y)
# #Examine the scores using the functions in this cell. Look especially at top and bottom ranks.
print(mi_scores.head(20))
# print(mi_scores.tail(20)) # uncomment to see bottom 20
plt.figure(dpi=100, figsize=(8, 5))
plot_mi_scores(mi_scores.head(20))
# plot_mi_scores(mi_scores.tail(20)) # uncomment to see bottom 20
# #Examine MI Scores
# Do the scores seem reasonable? Do the high scoring features represent things you'd think most people would value in a home? Do you notice any themes in what they describe?
# Some common themes among most of these features are:
# Location: Neighborhood
# Size: all of the Area and SF features, and counts like FullBath and GarageCars
# Quality: all of the Qual features
# Year: YearBuilt and YearRemodAdd
# Types: descriptions of features and styles like Foundation and GarageType
# These are all the kinds of features you'll commonly see in real-estate listings (like on Zillow), It's good then that our mutual information metric scored them highly. On the other hand, the lowest ranked features seem to mostly represent things that are rare or exceptional in some way, and so wouldn't be relevant to the average home buyer.
# In this step you'll investigate possible interaction effects for the BldgType feature. This feature describes the broad structure of the dwelling in five categories:
# Bldg Type (Nominal): Type of dwelling
# 1Fam Single-family Detached
#
# 2FmCon Two-family Conversion; originally built as one-family dwelling
#
# Duplx Duplex
# TwnhsE Townhouse End Unit
# TwnhsI Townhouse Inside Unit
# The BldgType feature didn't get a very high MI score. A plot confirms that the categories in BldgType don't do a good job of distinguishing values in SalePrice (the distributions look fairly similar, in other words):
sns.catplot(x="BldgType", y="SalePrice", data=df, kind="boxen")
# Still, the type of a dwelling seems like it should be important information. Investigate whether BldgType produces a significant interaction with either of the following:
# GrLivArea - Above ground living area
# MoSold - Month sold
# YOUR CODE HERE:
feature = "GrLivArea"
sns.lmplot(
x=feature,
y="SalePrice",
hue="BldgType",
col="BldgType",
data=df,
scatter_kws={"edgecolor": "w"},
col_wrap=3,
height=4,
)
feature = "MoSold"
sns.lmplot(
x=feature,
y="SalePrice",
hue="BldgType",
col="BldgType",
data=df,
scatter_kws={"edgecolor": "w"},
col_wrap=3,
height=4,
)
# #The trend lines being significantly different from one category to the next indicates an interaction effect.
# #Discover Interactions
# From the plots, does BldgType seem to exhibit an interaction effect with either GrLivArea or MoSold?
# The trends lines within each category of BldgType are clearly very different, indicating an interaction between these features. Since knowing BldgType tells us more about how GrLivArea relates to SalePrice, we should consider including BldgType in our feature set.
# The trend lines for MoSold, however, are almost all the same. This feature hasn't become more informative for knowing BldgType.
# #Found the ten features with the highest MI scores.
mi_scores.head(10)
|
# # Générateur d'indices pour CodeNames
# ## Introduction
# CodeNames est un jeu de plateau se jouant en 2 équipes de 2 joueurs minimum.
# But : Dans chaque équipe, un espion doit faire deviner à son équipe une liste de mots (appelés Nom de Code) inscrit sur une « carte clé » commune aux deux espions.
# L’équipe qui trouve tous les noms de Code inscrits sur la carte Clé gagne la partie (l’équipe qui commence doit faire deviner 9 mots et l’autre équipe 8).
# 
# Déroulement :
# L'équipe avec le plus de cartes à deviner commence, ici ce sont les rouges.
# Les équipes sont composées d'agents et d'espions, dans le cas où il n'y a que 4 joueurs dans la partie, il y aura donc 1 agent et 1 espion dans chaque équipe.
# L'agent voit les cartes mais ne voit pas leur couleur.
# L'espion donne un indice suivi d'un chiffre qui va permettre à son agent de deviner les cartes clés associées. Le chiffre lui permet de savoir combien de cartes sont associées au mot donné.
# Par exemple l'espion Rouge pourrait proposer FILM 2 pour faire deviner les mots CINEMA et TITRE.
# Si l'agent découvre une carte alliée (Rouge dans ce cas), il peut continuer à découvrir d'autres cartes jusqu'à un total du nombre d'indice +1. Dans le cas de FILM 2 l'agent pourrait découvrir jusqu'à 3 cartes.
# Si l'agent découvre une carte adverse (Bleue dans ce cas) ou une carte blanche, il finit son tour, l'espion adverse peut proposer un indice.
# Si l'agent découvre la carte assassin (carte noire), la partie est perdu, les adversaires gagnent.
from gensim.models import KeyedVectors
import spacy
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# ## Création d'une grille
# On récupère dans un premier temps la liste de tous les mots contenus dans Codenames
file = open("/kaggle/input/mots-codenames/codenames.txt").readlines()
words = [s.replace("\n", "") for s in file]
print(words)
# Ensuite on va pouvoir générer notre liste de mots
import random
random.shuffle(words) # Mélanger la liste de manière aléatoire
words = words[:25]
words = list(map(str.lower, words))
# Maintenant on va assigner une couleur à chaque mot
red_words = words[:9]
blue_words = words[9:17]
white_words = words[17:24]
black_words = words[24:25]
print("les mots rouges sont : ", red_words)
print("les mots bleus sont : ", blue_words)
print("les mots blancs sont : ", white_words)
print("le mot noir est : ", black_words)
import numpy as np
import matplotlib.pyplot as plt
# Création de la grille
grid = np.random.randint(4, size=(5, 5))
# Création du graphique
fig, ax = plt.subplots()
random.shuffle(words)
n = 0
# Dessin des rectangles et ajout des labels
for i in range(len(grid)):
for j in range(len(grid[i])):
if words[n] in red_words:
color = "#f36722"
elif words[n] in blue_words:
color = "#01abc4"
elif words[n] in white_words:
color = "#f5d9b6"
else:
color = "#949494"
rect = plt.Rectangle((j, i), 1, 1, facecolor=color)
ax.add_patch(rect)
ax.text(j + 0.5, i + 0.5, words[n], ha="center", va="center")
n = n + 1
# Configuration du graphique
ax.set_xlim([0, 5])
ax.set_ylim([0, 5])
ax.set_aspect("equal")
ax.axis("off")
# Affichage de la grille
plt.show()
# Voilà, on peut à présent générer nos propres grilles de codenames, celà va nous être utile pour faire des test randomisés et mieux observer nos IA.
# ## Développement de l'IA
# Pour notre IA nous utiliserons gensim avec un modèle pré entrainé sur la langue francaise. Il est possible d'adapter le jeu dans n'importe quelle langue tant que nous avons une liste de mots disponible et un modèle pré entrainé disponible également.
# Le modèle utilisé est disponible ici : https://fauconnier.github.io/#data
# Chemin vers le modèle Word2Vec pré-entraîné
path = "/kaggle/input/word2vecfr/frWac_no_postag_no_phrase_500_cbow_cut100.bin"
# Charger le modèle
model = KeyedVectors.load_word2vec_format(path, binary=True)
# Trouver les 10 mots les plus similaires pour la liste de mots donnée
similar_words = model.most_similar(
positive=red_words, negative=blue_words + white_words + black_words, topn=10
)
# Afficher les résultats
for word, similarity in similar_words:
print(word, similarity)
# L'IA nous permet de récupérer les 10 premiers mots qui sont les plus proches avec tout ceux que l'on a à découvrir.
# Cette méthode n'est pas forcément la meilleure, les mots sont un peu trop hasardeux et on perd en précision.
# On ne peut pas demander à quelqu'un de trouver 8 ou 9 mots d'un coup en un seul indice.
# On va chercher une autre méthode
# ### Méthode n°1
# Ici on va essayer d'abord de trouver une corrélation forte entre nos propres mots
# Il est plus facile de faire deviner deux ou trois mots qui se ressemblent plutot que donner un indice trop général pour 9 mots qui sont pas vraiment liés
score = []
for i in range(len(red_words)):
for j in range(i, len(red_words)):
if i != j:
similarite = model.similarity(red_words[i], red_words[j])
print(red_words[i], "+", red_words[j], "=", similarite)
score.append([similarite, red_words[i], red_words[j]])
sorted_scores = sorted(score, key=lambda x: x[0], reverse=True)
print(sorted_scores)
# Maintenant nous avons les couples de mots les plus ressemblants avec leur scores respectifs.
# On va pouvoir filtrer un peu tout ca et retirer ceux qui ne vont pas du tout ensemble.
# On peut soit retirer les éléments en fonction d'un seuil, ici 0.12, soit les retirer en fonction du nombre de résultats que l'on veut garder (par exemple: les 5 premiers).
sorted_scores = [element for element in sorted_scores if element[0] >= 0.12]
print(sorted_scores)
score = []
for i in range(len(sorted_scores)):
for j in range(len(red_words)):
if red_words[j] not in sorted_scores[i][1:]:
similarite = model.n_similarity(sorted_scores[i][1:], red_words[j])
words = []
for word in sorted_scores[i][1:]:
words.append(word)
words.append(red_words[j])
score.append([similarite, words])
sorted_scores.append(sorted(score, key=lambda x: x[0], reverse=True))
print(sorted_scores)
# Maintenant nous avons agrandi nos possibilités à 3 mots à trouver pour l'indice donné. On remarque que plus le nombre de mots à trouver est élevé, plus le score a tendance à être faible, c'est normal.
# On pourrait essayer de rectifier ca en mettant un coefficient multiplicateur sur le score pour que l'IA prenne plus de risques et qu'elle essaie de faire deviner plus de mots d'un coup.
# Mais pour l'instant nous allons se concentrer sur la proposition d'indice.
#
model.most_similar(
positive=red_words, negative=blue_words + white_words + black_words, topn=10
)
# Au début du projet nous avions cette fonction qui nous permettait d'avoir une liste d'indice mais rien de très pertinent, essayons avec notre nouvelle méthode.
resultats = model.most_similar(
positive=sorted_scores[0][1:],
negative=blue_words + white_words + black_words,
topn=10,
)
resultats = [mot[0] for mot in resultats]
print(
"L'IA propose ",
resultats[0],
len(sorted_scores[0][1:]),
" pour trouver les mots :",
sorted_scores[0][1:],
)
|
# ---
# Analisis Penjualan Game di Seluruh Dunia
# ---
# ---
# image from Google
# Studi Kasus
# Pada tugas Exploratory Data Analysis ini, kami memilih untuk menganalisis data penjualan game. Data ini berisi 16600 judul game beserta data penjualannya. Melalui Exploratory Data Analysis, kami berusaha untuk mencari insight yang dapat diambil dari data tersebut.
# Import important library
# Sebelum masuk ke proses data preparation, kita perlu mengimport library-library penting yang akan digunakan dalam proses EDA, antara lain: numpy, pandas, matplotlib, dan sebagainya
import numpy as np
import pandas as pd
import scipy.stats as st
pd.set_option("display.max_columns", None)
import math
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_style("whitegrid")
import missingno as msno
from sklearn.preprocessing import StandardScaler
from scipy import stats
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Data Preparation
# Selanjutnya kita melakukan data preparation untuk menyiapkan data sehingga menjadi bentuk yang mudah untuk dianalisis
# Import data
# Kita import data menggunakan pandas, kemudian menggunakan perintah 'head' untuk menampilkan lima data teratas. Hal ini dilakukan agar kita bisa mengetahui gambaran data yang baru saja kita import
data = pd.read_csv("/kaggle/input/videogamesales/vgsales.csv")
data.head()
# Menghapus data tidak lengkap
# Data yang tidak lengkap perlu dihapus karena dapat menghambat analisis. Dalam data ini, data dari tahun 2016 tidak sepenuhnya lengkap, sehingga kita perlu menghapusnya
drop_row_index = data[data["Year"] > 2015].index
data = data.drop(drop_row_index)
# Mendeskripsi data
# Selanjutnya kita perlu mendapatkan informasi mengenai data secara lebih lengkap. Mulai dari jumlah data, bentuk, dan ada atau tidaknya data kosong yang perlu dihilangkan lagi
# Menggunakan perintah 'shape', kita dapat memperoleh informasi dimensi data
data.shape
# Menggunakan perintah 'info' kita dapat melihat atribut apa saja yang ada pada data
data.info()
# Berdasarkan informasi data yang didapat, kita dapat menyimpulkan bahwa atribut-atribut dalam data tersebut yaitu:
# * Peringkat - Peringkat penjualan secara keseluruhan (bilangan bulat)
#
# * Nama - Nama game
# * Platform - Platform rilis game (mis. PC, PS4, dll.)
# * Tahun - Tahun rilis game (desimal)
# * Genre - Genre permainan
# * Publisher - Penerbit game
# * NA_Sales - Penjualan di Amerika Utara (dalam jutaan) (desimal)
# * EU_Sales - Penjualan di Eropa (dalam jutaan) (desimal)
# * JP_Sales - Penjualan di Jepang (dalam jutaan) (desimal)
# * Other_Sales - Penjualan di seluruh dunia (dalam jutaan) (desimal)
# * Global_Sales - Total penjualan di seluruh dunia (desimal)
# data.describe()
# data.describe(include=['object', 'bool'])
# Selanjutnya, kita cari tahu apakah terdapat banyak data kosong
data.isnull().sum()
# Karena tidak terdapat banyak data kosong, kita dapat memulai melakukan analisis EDA
# # 1. Game bergenre apa yang paling banyak dibuat?
# Menggunakan fungsi 'value_count', kita dapat menghitung berdasarkan atribut tertentu (dalam hal ini yaitu genre)
data["Genre"].value_counts()
# Selanjutnya kita gunakan matplotlib untuk membuat chart dengan bentuk bar chart agar tampilan menjadi menarik dan mudah dimengerti
plt.figure(figsize=(15, 10))
sns.countplot(x="Genre", data=data, order=data["Genre"].value_counts().index)
plt.xticks(rotation=90)
# Jawaban
# Genre yang paling banyak gamenya adalah genre Action dengan jumlah 3196 game
# # 2. Tahun mana yang memiliki rilis game terbanyak?
# Menggunakan fungsi 'value_count', kita dapat menghitung berdasarkan atribut tertentu (dalam hal ini yaitu Year)
data["Year"].value_counts()
# Selanjutnya kita gunakan matplotlib untuk membuat chart dengan bentuk bar chart agar tampilan menjadi menarik dan mudah dimengerti
plt.figure(figsize=(15, 10))
sns.countplot(
x="Year",
data=data,
order=data.groupby(by=["Year"])["Name"].count().sort_values(ascending=False).index,
)
plt.xticks(rotation=90)
# Jawaban
# Tahun yang memiliki rilis game terbanyak yaitu tahun 2009 dengan jumlah game mencapai 1431
# # 3. Rilis game teratas dalam 5 tahun terakhir berdasarkan genre.
plt.figure(figsize=(30, 10))
sns.countplot(
x="Year", data=data, hue="Genre", order=data.Year.value_counts().iloc[:5].index
)
plt.xticks(size=16, rotation=90)
# Jawaban
# Dalam 5 tahun terakhir genre action selalu menjadi yang teratas dalam jumlah rilis game
# # 4. Tahun berapa yang memiliki penjualan tertinggi di seluruh dunia?
data_year = data.groupby(by=["Year"])["Global_Sales"].sum()
data_year = data_year.reset_index()
# data_year.sort_values(by=['Global_Sales'], ascending=False)
plt.figure(figsize=(15, 10))
sns.barplot(x="Year", y="Global_Sales", data=data_year)
plt.xticks(rotation=90)
# Jawaban
# 2008 menjadi tahun dengan jumlah penjualan tertinggi yang mencapai lebih dari 600 juta copy game
# # 5. Game bergenre apa yang paling banyak dirilis dalam satu tahun?
year_max_df = data.groupby(["Year", "Genre"]).size().reset_index(name="count")
year_max_idx = (
year_max_df.groupby(["Year"])["count"].transform(max) == year_max_df["count"]
)
year_max_genre = year_max_df[year_max_idx].reset_index(drop=True)
year_max_genre = year_max_genre.drop_duplicates(
subset=["Year", "count"], keep="last"
).reset_index(drop=True)
# year_max_genre
genre = year_max_genre["Genre"].values
# genre[0]
plt.figure(figsize=(30, 15))
g = sns.barplot(x="Year", y="count", data=year_max_genre)
index = 0
for value in year_max_genre["count"].values:
# print(asd)
g.text(
index,
value + 5,
str(genre[index] + "----" + str(value)),
color="#000",
size=14,
rotation=90,
ha="center",
)
index += 1
plt.xticks(rotation=90)
plt.show()
# Jawaban
# Genre yang paling banyak dirilis adalah game bergenre action dengan puncaknya pada tahun 2009 yang mencapai 272 game
# # 6. Game bergenre apa yang paling banyak terjual dalam satu tahun?
year_sale_dx = data.groupby(by=["Year", "Genre"])["Global_Sales"].sum().reset_index()
year_sale = (
year_sale_dx.groupby(by=["Year"])["Global_Sales"].transform(max)
== year_sale_dx["Global_Sales"]
)
year_sale_max = year_sale_dx[year_sale].reset_index(drop=True)
# year_sale_max
genre = year_sale_max["Genre"]
plt.figure(figsize=(30, 18))
g = sns.barplot(x="Year", y="Global_Sales", data=year_sale_max)
index = 0
for value in year_sale_max["Global_Sales"]:
g.text(
index,
value + 1,
str(genre[index] + "----" + str(round(value, 2))),
color="#000",
size=14,
rotation=90,
ha="center",
)
index += 1
plt.xticks(rotation=90)
plt.show()
# Jawaban
# Game action menjadi game dengan penjualan paling banyak dalam satu tahun. Puncaknya terjadi pada tahun 2009 yang mencapai 139.36 juta copy game terjual
# * Action and Sports are always in top. but in third whis is remarkable "Shooter", in count which was no 5
# # 7. Platformm mana yang memiliki penjualan tertinggi secara global
data_platform = data.groupby(by=["Platform"])["Global_Sales"].sum()
data_platform = data_platform.reset_index()
data_platform = data_platform.sort_values(by=["Global_Sales"], ascending=False)
# data_platform
plt.figure(figsize=(15, 10))
sns.barplot(x="Platform", y="Global_Sales", data=data_platform)
plt.xticks(rotation=90)
# Jawaban
# PS2 menjadi platform dengan jumlah penjualan game terbanyak yang mencapai lebih dari 1,2M copy
# # 9. Game apa yang memiliki penjualan terbanyak di seluruh dunia
top_game_sale = data.head(20)
top_game_sale = top_game_sale[["Name", "Year", "Genre", "Global_Sales"]]
top_game_sale = top_game_sale.sort_values(by=["Global_Sales"], ascending=False)
# top_game_sale
name = top_game_sale["Name"]
year = top_game_sale["Year"]
y = np.arange(0, 20)
plt.figure(figsize=(30, 18))
g = sns.barplot(x="Name", y="Global_Sales", data=top_game_sale)
index = 0
for value in top_game_sale["Global_Sales"]:
g.text(
index, value - 18, name[index], color="#000", size=14, rotation=90, ha="center"
)
index += 1
plt.xticks(y, top_game_sale["Year"], fontsize=14, rotation=90)
plt.xlabel("Release Year")
plt.show()
# Jawaban
# Game Wii Sports menduduki peringkat pertama sebagai game dengan penjualan terbanyak yang mana mencapai lebih dari 80 juta copy
# # 10. Seperti apa perbandingan penjualan game berdasarkan genre?
comp_genre = data[["Genre", "NA_Sales", "EU_Sales", "JP_Sales", "Other_Sales"]]
# comp_genre
comp_map = comp_genre.groupby(by=["Genre"]).sum()
# comp_map
plt.figure(figsize=(15, 10))
sns.set(font_scale=1)
sns.heatmap(comp_map, annot=True, fmt=".1f")
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.show()
# Jawaban
# Game dengan genre action memilki tingkat penjualan tertinggi dengan selisih cukup jauh dengan genre game lainya
# # 11. Relasi antar kolom
plt.figure(figsize=(13, 10))
sns.heatmap(data.corr(), cmap="Blues", annot=True, linewidth=3)
|
import os
import torch
import torchvision
import numpy as np
import pandas as pd
from PIL import Image
from torch import optim
import matplotlib.pyplot as plt
from torch.utils.data import DataLoader, Dataset
from sklearn.model_selection import train_test_split
import torchvision.models as models
import torch.nn as nn
import torch.optim as optim
import torch.nn.functional as F
from torch.autograd import Variable
from torch.utils.data.sampler import SubsetRandomSampler
import time
class FIDataset(Dataset):
"""Fashion Image Dataset"""
def __init__(self, dir, dataframe, transform, cat_lookup):
super(FIDataset, self).__init__()
self.dataframe = dataframe
self.dir = dir
self.transform = transform
self.cat_lookup = cat_lookup
def __getitem__(self, idx):
line = self.dataframe.iloc[idx]
cat = line.articleType
cat_id = self.cat_lookup[cat]
img_path = os.path.join(self.dir, str(line.id) + ".jpg")
img = Image.open(img_path).convert("RGB")
img_tensor = self.transform(img)
return img_tensor, cat_id
def __len__(self):
return len(self.dataframe)
def split_train_valid(train_data, valid_size):
# obtain training indices that will be used for validation
num_train = len(train_data)
indices = list(range(num_train))
np.random.shuffle(indices)
split = int(np.floor(valid_size * num_train))
train_idx, valid_idx = indices[split:], indices[:split]
# define samplers for obtaining training and validation batches
train_sampler = SubsetRandomSampler(train_idx)
valid_sampler = SubsetRandomSampler(valid_idx)
return train_sampler, valid_sampler
def imshow(img):
# helper function to un-normalize and display an image
img = img / 2 + 0.5 # unnormalize
plt.imshow(np.transpose(img, (1, 2, 0))) # convert from Tensor image
def plot_sample_data(dataloader, num, cat_lookup):
# obtain one batch of training images
dataiter = iter(dataloader)
images, labels = next(dataiter)
# convert to numpy for display
images = images.numpy()
labels = labels.numpy()
# plot the images in the batch, along with the corresponding labels
fig = plt.figure(figsize=(25, 4))
# display images
for idx in np.arange(num):
ax = fig.add_subplot(1, 5, idx + 1, xticks=[], yticks=[])
imshow(images[idx])
ax.set_title(cat_lookup[labels[idx]])
def plot_sample_data_model(dataloader, num, model, cat_lookup, use_cuda=True):
import torch
# obtain one batch of training images
dataiter = iter(dataloader)
images, labels = next(dataiter)
# convert to numpy for display
labels = labels.numpy()
# move model inputs to cuda, if GPU available
if use_cuda:
images = images.cuda()
else:
model = model.cpu()
# get sample outputs
output = model(images)
# convert output probabilities to predicted class
_, preds_tensor = torch.max(output, 1)
preds = (
np.squeeze(preds_tensor.numpy())
if not use_cuda
else np.squeeze(preds_tensor.cpu().numpy())
)
# plot the images in the batch, along with the corresponding labels
fig = plt.figure(figsize=(15, 6))
# display images
for idx in np.arange(num):
ax = fig.add_subplot(2, num / 2, idx + 1, xticks=[], yticks=[])
imshow(images[idx].cpu())
ax.set_title(
"{} ({})".format(cat_lookup[preds[idx]], cat_lookup[labels[idx]]),
color=("green" if preds[idx] == labels[idx].item() else "red"),
)
def plot_training_and_valid_loss(train_loss_history, valid_loss_history, n_epochs):
# Define a helper function for plotting training and valid loss
plt.title("Training and Validation Loss vs. Number of Training Epochs")
plt.xlabel("Training Epochs")
plt.ylabel("Loss")
plt.plot(range(1, n_epochs + 1), train_loss_history, label="Training Loss")
plt.plot(range(1, n_epochs + 1), valid_loss_history, label="Validation Loss")
plt.xticks(np.arange(1, n_epochs + 1, 1.0))
plt.legend()
plt.show()
def train(
n_epochs,
train_loader,
valid_loader,
model,
optimizer,
criterion,
use_cuda,
save_path,
scheduler=False,
):
"""returns trained model"""
since = time.time()
# initialize tracker for minimum validation loss
valid_loss_min = np.Inf
# initialize lists of values of train and valid losses over the training process
train_loss_history = []
valid_loss_history = []
for epoch in range(n_epochs):
# initialize variables to monitor training and validation loss
train_loss = 0.0
valid_loss = 0.0
###################
# train the model #
###################
# Call the learning rate scheduler if given
if scheduler:
scheduler.step()
model.train()
for batch_idx, (data, target) in enumerate(train_loader):
# move to GPU
if use_cuda:
data, target = data.cuda(), target.cuda()
optimizer.zero_grad()
# forward pass: compute predicted outputs by passing inputs to the model
output = model(data)
# calculate the batch loss
loss = criterion(output, target)
# backward pass: compute gradient of the loss with respect to model parameters
loss.backward()
# perform a single optimization step (parameter update)
optimizer.step()
# record the average training loss and add it to history
train_loss = train_loss + ((1 / (batch_idx + 1)) * (loss.data - train_loss))
train_loss_history.append(train_loss)
######################
# validate the model #
######################
model.eval()
for batch_idx, (data, target) in enumerate(valid_loader):
# move to GPU
if use_cuda:
data, target = data.cuda(), target.cuda()
# update the average validation loss and add it to history
output = model(data)
_, preds = torch.max(output, 1)
loss = criterion(output, target)
valid_loss = valid_loss + ((1 / (batch_idx + 1)) * (loss.data - valid_loss))
valid_loss_history.append(valid_loss)
# Print training/validation statistics
print(
"Epoch: {}/{} \tTraining Loss: {:.6f} \tValidation Loss: {:.6f}".format(
epoch, n_epochs, train_loss, valid_loss
)
)
# Save the model if validation loss has decreased
if valid_loss < valid_loss_min:
torch.save(model.state_dict(), save_path)
print(
"Validation loss decreased ({:.6f} --> {:.6f}). Saving model ...".format(
valid_loss_min, valid_loss
)
)
valid_loss_min = valid_loss
# Print training time
time_elapsed = time.time() - since
print(
"Training complete in {:.0f}m {:.0f}s".format(
time_elapsed // 60, time_elapsed % 60
)
)
# return trained model
return model, train_loss_history, valid_loss_history
def correct_top_k(output, target, topk=(1,)):
"""Returns a tensor with 1 if target in top-k best guesses and 0 otherwise"""
with torch.no_grad():
maxk = max(topk)
batch_size = target.size(0)
_, pred = output.topk(maxk, 1, True, True)
pred = pred.t()
correct = pred.eq(target.view(1, -1).expand_as(pred)).sum(0, keepdim=True)
return correct[0]
def test(test_loader, model, criterion, cat_lookup, use_cuda):
# initialize lists to monitor correct guesse and total number of data
test_loss = 0.0
class_correct = list(0.0 for i in range(len(cat_lookup)))
class_correct_top_5 = list(0.0 for i in range(len(cat_lookup)))
class_total = list(0.0 for i in range(len(cat_lookup)))
model.eval() # prep model for evaluation
for data, target in test_loader:
if use_cuda:
data, target, model = data.cuda(), target.cuda(), model.cuda()
# else:
# model = model.cpu()
# forward pass: compute predicted outputs by passing inputs to the model
output = model(data)
# calculate the loss
loss = criterion(output, target)
# update test loss
test_loss += loss.item() * data.size(0)
# get 1 if target in top-1 predictions
correct_top_1 = correct_top_k(output, target, topk=(1,))
# get 1 if target in top-5 predictions
correct_top_5 = correct_top_k(output, target, topk=(5,))
# calculate test accuracy for each object class
for i in range(len(target)):
label = target.data[i]
class_correct[label] += correct_top_1[i].item()
class_correct_top_5[label] += correct_top_5[i].item()
class_total[label] += 1
# calculate and print avg test loss
test_loss = test_loss / len(test_loader.sampler)
print("Test Loss: {:.6f}\n".format(test_loss))
class_accuracy_top_1 = {}
print("\nPrinting accuracy for each class")
for i in range(len(cat_lookup)):
if class_total[i] > 0:
accuracy_top_1 = 100 * class_correct[i] / class_total[i]
accuracy_top_5 = 100 * class_correct_top_5[i] / class_total[i]
class_accuracy_top_1[i] = accuracy_top_1
print(
"Test accuracy of %5s: \nTop-1 accuracy: %2d%% (%2d/%2d) \nTop-5 accuracy: %2d%% (%2d/%2d)".format()
% (
cat_lookup[i],
accuracy_top_1,
np.sum(class_correct[i]),
np.sum(class_total[i]),
accuracy_top_5,
np.sum(class_correct_top_5[i]),
np.sum(class_total[i]),
)
)
else:
print("Test Accuracy of %5s: N/A (no training examples)" % (cat_lookup[i]))
print("\nPrinting 5 classes with greatest top-1 accuracy")
sorted_class_accuracy = sorted(
class_accuracy_top_1.items(), key=lambda kv: kv[1], reverse=True
)
for i in range(5):
print(
"Test Accuracy of %5s: %2d%%"
% (
str(cat_lookup[sorted_class_accuracy[i][0]]),
sorted_class_accuracy[i][1],
)
)
print(
"\nTest Accuracy (Overall): \nTop-1 accuracy: %2d%% (%2d/%2d) \nTop-5 accuracy: %2d%% (%2d/%2d)"
% (
100.0 * np.sum(class_correct) / np.sum(class_total),
np.sum(class_correct),
np.sum(class_total),
100.0 * np.sum(class_correct_top_5) / np.sum(class_total),
np.sum(class_correct_top_5),
np.sum(class_total),
)
)
# create a df from styles.csv neglecting lines with error
DATASET_PATH = "/kaggle/input/fashion-product-images-dataset/fashion-dataset/"
styles = pd.read_csv(os.path.join(DATASET_PATH, "styles.csv"), error_bad_lines=False)
print(styles.head())
print(len(styles))
# Get the list of names of images that are available
imgs_available = os.listdir(DATASET_PATH + "/images")
print(len(imgs_available))
# Check if each entry in styles.csv has a corresponding images listing.
# If not, we remove it from the dataframe.
missing_img = []
for idx, line in styles.iterrows():
if not os.path.exists(os.path.join(DATASET_PATH, "images", str(line.id) + ".jpg")):
print(os.path.join(DATASET_PATH, "images", str(line.id) + ".jpg"))
missing_img.append(idx)
styles.drop(styles.index[missing_img], inplace=True)
print(len(styles))
# Check how many unique article types we have
uniquie_article_types = styles["articleType"].unique()
print(len(uniquie_article_types))
sorted_df = styles.groupby(["articleType"]).size().sort_values(ascending=False)
top_classes = sorted_df.nlargest(20)
display(top_classes)
# plot the distribution of top 20 classes
top_classes.plot.bar()
styles.dropna(inplace=True, subset=["year", "articleType"])
len(styles)
training_data = styles[styles["year"].astype("int") % 2 == 0]
testing_data = styles[styles["year"].astype("int") % 2 == 1]
top_classes_names = list(top_classes.index)
training_top20 = training_data[training_data.articleType.isin(top_classes_names)]
testing_top20 = testing_data[testing_data.articleType.isin(top_classes_names)]
# Find weights
total_number_of_samples = np.sum(top_classes.values)
weights = [total_number_of_samples / top_classes.values]
# Create a mapping between article types and their ids
cat_list = sorted_df.index
cat2num = {cat: i for i, cat in enumerate(cat_list)}
num2cat = {i: cat for i, cat in enumerate(cat_list)}
# Create data transform
transform = torchvision.transforms.Compose(
[
torchvision.transforms.Resize((224, 224)),
torchvision.transforms.ToTensor(),
torchvision.transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5)),
]
)
# number of subprocesses to use for data loading
num_workers = 8
# how many samples per batch to load
batch_size = 16
# percentage of training set to use as validation
valid_size = 0.2
# Construct datasets
top20_data = FIDataset(
os.path.join(DATASET_PATH, "images"), training_top20, transform, cat2num
)
# others_data = FIDataset(os.path.join(DATASET_PATH, 'images'), training_others, transform, cat2num)
test_top20 = FIDataset(
os.path.join(DATASET_PATH, "images"), testing_top20, transform, cat2num
)
# split the training data for 'top 20' classes into train and validation datasets
top20_train_sampler, top20_valid_sampler = split_train_valid(top20_data, valid_size)
# prepare data loaders
top20_train_loader = DataLoader(
top20_data,
batch_size=batch_size,
sampler=top20_train_sampler,
num_workers=num_workers,
)
top20_valid_loader = DataLoader(
top20_data,
batch_size=batch_size,
sampler=top20_valid_sampler,
num_workers=num_workers,
)
top20_test_loader = DataLoader(
test_top20, batch_size=batch_size, num_workers=num_workers
)
# Plot sample data from the training dataset of top 20 classes
plot_sample_data(top20_train_loader, 5, num2cat)
# Plot sample data from the training dataset of the remaining classes
plot_sample_data(top20_test_loader, 5, num2cat)
# Check if CUDA is available
use_cuda = torch.cuda.is_available()
# Specify model architecture
model_top20 = models.resnet50(pretrained=True)
# To reshape the network, we reinitialize the classifier’s linear layer
n_inp = model_top20.fc.in_features
model_top20.fc = nn.Linear(n_inp, len(top_classes_names))
if use_cuda:
model_top20 = model_top20.cuda()
print(model_top20.fc)
# Convert weights vector to a tensor
weights = torch.tensor(weights, dtype=torch.float)
if use_cuda:
weights = weights.cuda()
# Specify a loss function and optimizer
criterion = nn.CrossEntropyLoss(weight=weights)
optimizer = optim.SGD(model_top20.parameters(), lr=0.001)
n_epochs = 10
# train the model
model_top20, train_loss_history_top20, valid_loss_history_top20 = train(
n_epochs,
top20_train_loader,
top20_valid_loader,
model_top20,
optimizer,
criterion,
use_cuda,
"model_top20.pt",
)
plot_training_and_valid_loss(
train_loss_history_top20, valid_loss_history_top20, n_epochs
)
# Load the model that got the best validation accuracy
model_top20.load_state_dict(torch.load("model_top20.pt"))
plot_sample_data_model(top20_test_loader, 10, model_top20, num2cat, use_cuda=True)
top20_cat_lookup = {i: j for i, j in num2cat.items() if j in top_classes_names}
test(
top20_test_loader,
model_top20,
criterion,
cat_lookup=top20_cat_lookup,
use_cuda=True,
)
# Data augmentation
transform_augment = torchvision.transforms.Compose(
[
torchvision.transforms.Resize((224, 224)),
torchvision.transforms.RandomHorizontalFlip(),
torchvision.transforms.ToTensor(),
torchvision.transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5)),
]
)
# Construct datasets
test_data = FIDataset(
os.path.join(DATASET_PATH, "images"), testing_data, transform, cat2num
)
training_dataset = FIDataset(
os.path.join(DATASET_PATH, "images"), training_data, transform_augment, cat2num
)
valid_dataset = FIDataset(
os.path.join(DATASET_PATH, "images"), training_data, transform, cat2num
)
# split the training data into train and validation datasets
train_sampler, valid_sampler = split_train_valid(training_dataset, valid_size)
# prepare data loaders
train_loader = DataLoader(
training_dataset,
batch_size=batch_size,
sampler=train_sampler,
num_workers=num_workers,
)
valid_loader = DataLoader(
valid_dataset, batch_size=batch_size, sampler=valid_sampler, num_workers=num_workers
)
test_loader = DataLoader(test_data, batch_size=batch_size, num_workers=num_workers)
# Plot sample data from the training dataset of top 20 classes
plot_sample_data(train_loader, 5, num2cat)
# Plot sample data from the training dataset of top 20 classes
plot_sample_data(test_loader, 5, num2cat)
model_142 = model_top20
# Freeze the first 5 layers of the model
layers_to_freeze = 5
layer_count = 0
print("Layers to freeze: \n")
for child in model_142.children():
layer_count += 1
if layer_count <= layers_to_freeze:
print(child)
for param in child.parameters():
param.requires_grad = False
# To reshape the network, we reinitialize the classifier’s linear layer
n_inp = model_142.fc.in_features
model_142.fc = nn.Linear(n_inp, len(uniquie_article_types))
if use_cuda:
model_142 = model_142.cuda()
# Define the weights for all the 142 classes as before
total_number_of_samples = np.sum(sorted_df.values)
weights = [total_number_of_samples / sorted_df.values]
# Convert weights vector to a tensor
weights = torch.tensor(weights, dtype=torch.float)
if use_cuda:
weights = weights.cuda()
# Specify a loss function and optimizer
criterion = nn.CrossEntropyLoss(weight=weights)
optimizer = optim.SGD(model_142.parameters(), lr=0.001)
n_epochs = 20
# train the model
model_142, train_loss_history_142, valid_loss_history_142 = train(
n_epochs,
train_loader,
valid_loader,
model_142,
optimizer,
criterion,
use_cuda,
"model_142_augment2.pt",
)
plot_training_and_valid_loss(train_loss_history_142, valid_loss_history_142, n_epochs)
plot_sample_data_model(test_loader, 10, model_142, num2cat, use_cuda=True)
# Load the model that got the best validation accuracy
model_142.load_state_dict(torch.load("model_142_augment2.pt"))
test(test_loader, model_142, criterion, cat_lookup=num2cat, use_cuda=True)
|
# ## kandinsky
import torch
torch.set_grad_enabled(False)
import requests
from PIL import Image
from io import BytesIO
from kandinsky2 import get_kandinsky2
from PIL import Image
model = get_kandinsky2(
"cuda", task_type="text2img", model_version="2.1", use_flash_attention=False
)
response = requests.get(
"https://ih1.redbubble.net/image.2086350547.7750/flat,750x,075,f-pad,750x1000,f8f8f8.jpg"
)
# response = requests.get('https://nationaltoday.com/wp-content/uploads/2022/01/Ginger-Cat-640x514.jpg')
content_img = Image.open(BytesIO(response.content))
response = requests.get(
"https://hips.hearstapps.com/hmg-prod.s3.amazonaws.com/images/lord-of-the-rings-sean-bean-boromir-1584636601.jpg"
)
# response = requests.get('https://encrypted-tbn0.gstatic.com/images?q=tbn:ANd9GcRMtfKLzUTi-tBt_pzGzltUXO3muc7k3qjq0g&usqp=CAU')
style_img = Image.open(BytesIO(response.content))
images_texts = [style_img, content_img]
weights = [0.5, 0.5]
images = model.mix_images(
images_texts,
weights,
num_steps=50,
batch_size=1,
guidance_scale=5,
h=768,
w=768,
sampler="p_sampler",
prior_cf_scale=4,
prior_steps="5",
)
images[0]
|
import autosklearn.classification
import autosklearn.classification
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
pd.set_option("display.max_rows", None)
pd.set_option("display.max_columns", None)
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
train = pd.read_csv("/kaggle/input/digit-recognizer/train.csv")
test = pd.read_csv("/kaggle/input/digit-recognizer/test.csv")
train[0:1]
LABEL = "label"
y = train[LABEL]
train = train.drop(LABEL, axis=1) # Dropping label to normalize
train[0:1]
width = 28
def make_grid(row):
# print(row)
grid = np.zeros((width, width))
for x in range(width):
for y in range(width):
grid[y][x] = row[x * width + y]
# print(row[x*width + y],end=" ")
# print()
return grid
def make_grid_array(ar):
ar = np.array(ar)
num = len(ar)
grid = np.zeros((num, width, width))
for i in range(num):
# print(ar[i])
grid[i] = make_grid(ar[i])
return grid
val = 3
# print(y)
# print(y[val:val+1])
# print(make_grid(train[val:val+1]))
# train = make_grid_array(train)
# test = make_grid_array(test)
# print(train[0:3])
np.shape(train)
from autosklearn.metrics import (
accuracy,
f1,
roc_auc,
precision,
average_precision,
recall,
log_loss,
)
automl = autosklearn.classification.AutoSklearnClassifier(
time_left_for_this_task=600,
max_models_on_disc=5,
memory_limit=25000,
scoring_functions=[
roc_auc,
average_precision,
accuracy,
f1,
precision,
recall,
log_loss,
],
)
automl.fit(train, y)
y_hat = automl.predict(test)
submission = pd.read_csv("/kaggle/input/digit-recognizer/sample_submission.csv")
submission["Label"] = y_hat
y_hat
submission.to_csv("/kaggle/working/output.csv", index=False)
|
from fastai.vision.all import *
from pathlib import Path
import h5py
from matplotlib import cm
import matplotlib.pyplot as plt
import os
kaggle = os.environ.get("KAGGLE_KERNEL_RUN_TYPE", "")
path = "/kaggle/input/jet-images-train-val-test/jet-images_train.hdf5"
h5_file = h5py.File(path, "r")
signal_data = h5_file["signal"]
image_data = h5_file["image"]
signal_array = np.array(signal_data)
image_array = np.array(image_data)
len(image_array)
# Filter the image_array based on signal_array
filtered_images = image_array[signal_array == 1]
# Calculate the mean image
mean_image = np.mean(filtered_images, axis=0)
len(filtered_images)
plt.imshow(mean_image)
plt.show()
plt.imshow(filtered_images[7])
plt.show()
filtered_images_general = image_array[signal_array == 0]
mean_image_general = np.mean(filtered_images_general, axis=0)
plt.imshow(mean_image_general)
plt.show()
plt.imshow(filtered_images_general[22])
plt.show()
path_val = "/kaggle/input/jet-images-train-val-test/jet-images_micro.hdf5"
h5_file_val = h5py.File(path_val, "r")
signal_data_val = h5_file_val["signal"]
image_data_val = h5_file_val["image"]
signal_array = np.array(signal_data_val)
image_array = np.array(image_data_val)
print(len(signal_array))
print(len(image_array))
from sklearn.metrics.pairwise import cosine_similarity
from sklearn.metrics import accuracy_score
def mean_image_predict(threshold):
mean_image = np.mean(image_array[signal_array == 1], axis=0)
similarities = np.zeros(len(image_array))
for i, image_vec in enumerate(image_array):
similarities[i] = cosine_similarity(
image_vec.reshape(1, -1), mean_image.reshape(1, -1)
)
threshold = threshold
predicted_labels = np.where(similarities > threshold, 1, 0)
accuracy = accuracy_score(signal_array, predicted_labels)
return accuracy
test_num = [0.9, 0.81, 0.79, 0.6, 0.5, 0.4]
for i in test_num:
test = mean_image_predict(i)
print(f"Whit a treshold of {i} we have a accuracy: {test}")
preds = []
def predict(array):
general = np.abs(np.mean(mean_image_general) - np.mean(array))
boson = np.abs(np.mean(mean_image) - np.mean(array))
if boson < general:
preds.append(1.0)
else:
preds.append(0.0)
for image in image_data_val:
predict(image)
def accuracy(predictions, labels):
"""Calculate the accuracy of the predictions."""
num_correct = sum(
[1 for i in range(len(predictions)) if predictions[i] == labels[i]]
)
return num_correct / len(predictions)
accuracy(preds, signal_data)
|
# # Задача:
# На основе имеющихся в датасете данных о 515 000 отзывах на отели Европы обучить модель, которая должна предсказывать рейтинг отеля по данным сайта Booking.
# ## Проблема:
# Наличие отелей накручивающих себе рейтинг на Booking
# ## Цель проекта:
# Построить модель на основе алгоритмов машинного обучения, которая предсказывает рейтинг отеля
# ## 0. Подготовительный этап
# ЗАГРУЗКА НЕОБХОДИМЫХ БИБЛИОТЕК
# для работы с данными
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# для работы с признаками
# кодирования признаков
import category_encoders as ce
# нормализации признаков
from sklearn import preprocessing
# импортируем библиотеки для визуализации
import matplotlib.pyplot as plt
import seaborn as sns
# импортируем библиотеки для оценки тональности текста
import nltk
from nltk.sentiment.vader import SentimentIntensityAnalyzer
import time
# загружаем специальный удобный инструмент для разделения датасета:
from sklearn.model_selection import train_test_split
# библиотеки для работы с моделью
from sklearn.ensemble import (
RandomForestRegressor,
) # инструмент для создания и обучения модели
from sklearn import metrics # инструменты для оценки точности модели
# загружаем специальный удобный инструмент для работы с исходными данными:
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# фиксируем RANDOM_SEED, для воспроизводимости эксперимента
RANDOM_SEED = 42
# зафиксируем версию пакетов, чтобы эксперименты были воспроизводимы
# ## 1. Знакомимся с входными данными
# Подгрузим наши данные из соревнования
DATA_DIR = "/kaggle/input/sf-booking/"
df_train = pd.read_csv(DATA_DIR + "/hotels_train.csv") # датасет для обучения
df_test = pd.read_csv(DATA_DIR + "hotels_test.csv") # датасет для предсказания
sample_submission = pd.read_csv(DATA_DIR + "/submission.csv") # самбмишн
# Первоначальная версия датасета (df_train) содержит **17 полей** со следующей информацией:
# - hotel_address — адрес отеля;
# - review_date — дата, когда рецензент разместил соответствующий отзыв;
# - average_score — средний балл отеля, рассчитанный на основе последнего комментария за последний год;
# - hotel_name — название отеля;
# - reviewer_nationality — страна рецензента;
# - negative_review — отрицательный отзыв, который рецензент дал отелю;
# - review_total_negative_word_counts — общее количество слов в отрицательном отзыв;
# - positive_review — положительный отзыв, который рецензент дал отелю;
# - review_total_positive_word_counts — общее количество слов в положительном отзыве.
# - reviewer_score — оценка, которую рецензент поставил отелю на основе своего опыта;
# - total_number_of_reviews_reviewer_has_given — количество отзывов, которые рецензенты дали в прошлом;
# - total_number_of_reviews — общее количество действительных отзывов об отеле;
# - tags — теги, которые рецензент дал отелю;
# - days_since_review — количество дней между датой проверки и датой очистки;
# - additional_number_of_scoring — есть также некоторые гости, которые просто поставили оценку сервису, но не оставили отзыв. Это число указывает, сколько там действительных оценок без проверки.
# - lat — географическая широта отеля;
# - lng — географическая долгота отеля.
df_train.info()
# **Описание:**
# Признаки можно разделить на три группы:
# 1. Связанные с отелем признаки / Hotel Related Features (7)
# - hotel_name - full hotel name;
# - hotel_address - address: street, post code, city, country;
# - lat - hotel latitude coordinate;
# - lng - hotel longitude coordinate;
# - average_score - average hotel rating;
# - total_number_of_reviews - total number of hotel reviews;
# - additional_number_of_scoring - number of hotel scores without review.
# 2. Связанные с рецензентом признаки / Reviewer Related Features (3)
# - reviewer_nationality - reviewer nationality;
# - total_number_of_reviews_reviewer_has_given - total number of reviews reviewer has given;
# - tags - tags describing stay in the hotel.
# 3. Связанные с отзывом признаки / Review Related Features (6)
# - review_date date of review;
# - days_since_review difference in the number of days between review date and scrape date;
# - negative_review text of negative review;
# - review_total_negative_word_counts negative review words number;
# - positive_review text of positive review;
# - review_total_positive_word_counts positive review words number.
df_train.head(2)
# Тестовая версия датасета (df_test) содержит 16 полей (без 'reviewer_score' )
df_test.info()
df_test.head(2)
# Сабмишн содержит 128935 строки и два признака 'reviewer_score' и 'id'
sample_submission.head(2)
sample_submission.info()
# ## 2. Подготовка данных, которые будут использованы для обучения модели
# ВАЖНО! дkя корректной обработки признаков объединяем трейн и тест в один датасет
df_train["sample"] = 1 # помечаем где у нас трейн
df_test["sample"] = 0 # помечаем где у нас тест
df_test[
"reviewer_score"
] = 0 # в тесте у нас нет значения reviewer_score, мы его должны предсказать, по этому пока просто заполняем нулями
data = df_test.append(df_train, sort=False).reset_index(drop=True) # объединяем
data.info()
data.describe()
data.hist(figsize=(15, 15))
# Имеются следующие **"проблемы"** с представленными данными:
# - Отсутствуют некоторые значения в столбцах lat и lng.
# - Числовые значения в 9 столбцах: 'additional_number_of_scoring', 'average_score','review_total_negative_word_counts','total_number_of_reviews_reviewer_has_given', 'total_number_of_reviews', 'review_total_positive_word_counts', 'reviewer_scoretotal_number_of_reviews_reviewer_has_given', 'lat', 'lng'.
# - Значения типа object в 8 столбцах: 'hotel_address', review_date', 'hotel_name', 'reviewer_nationality', 'negative_review', 'positive_review', ' tags ', 'days_since_review'.
# ### **2.1 Работа с "нулевыми" значениями**
# проверим может быть пропущенные данные по местоположению отелей можно найти в других записях
no_place = set(data[(data["lat"] == 0) | (data["lng"] == 0)]["hotel_name"].values)
yes_place = set(data[(data["lat"] != 0) & (data["lng"] != 0)]["hotel_name"].values)
no_place.intersection(yes_place) # нет данных
# заполним пустые значения в координатах нулями
data[["lat", "lng"]] = data[["lat", "lng"]].fillna(0)
# ### **2.2 Преобразуем признаки**
# #### **2.2.1 Преобразование признака 'review_date' (дата написания признака)**
# изменим значение даты на тип данных datetime
data["review_date"] = pd.to_datetime(
data["review_date"], dayfirst=False, yearfirst=False
)
# определим номер дня года, когда написан отзыв и создадим новый признак
data["day_of_year"] = data["review_date"].apply(lambda x: x.timetuple().tm_yday)
# выделим год отзыва
data["rewiew_year"] = data["review_date"].dt.year
# преобразуем информацию о времени, прошедшем с момента
# написания отзыва, в числовое значение с помощью функции:
def time_review(arg):
word_list = arg.split() # преобразуем в список по пробелам
for word in word_list:
if word.isnumeric(): # выберем числовое значение
return int(word)
data["days_since_review"] = data["days_since_review"].apply(time_review)
# #### **2.2.2 Обработка тегов**
# из тегов выделим количество ночей проведенных клиентом в отеле
# создадим функцию, отыскивающую нужный тег и возвращающую количество дней
def days_number(arg):
tag_list = arg.split(",")
for tag in tag_list:
ind = tag.find("night")
if ind > 0:
word_list = tag.split()
for word in word_list:
if word.isnumeric():
return int(word)
data["accommodation_days"] = data["tags"].apply(days_number)
# создадим признак считающий количество слов в тегах
data["word_tegs_cnt"] = data["tags"].apply(lambda x: len(list(x.split(","))))
# удалим теги, отвечающий за количество ночей (мы уже выписали их)
# функция принимает строку, разбивает ее на теги, находит и удаляет те, в которых уrазано количество ночей возвращает список
def tags_no_nights(arg):
k = list(arg.split(","))
for tag in k:
ind = 0
ind = tag.find("night")
if ind > 0:
k.remove(tag)
arg = ",".join(k)
return arg
# очищаем теги от длительности пребывания, возможно они нам еще пригодятся
data["tags"] = data["tags"].apply(tags_no_nights)
# из отзывов выделим эмоциональную окраску текста в отзывах
nltk.downloader.download("vader_lexicon")
sent_analyzer = SentimentIntensityAnalyzer()
# в результате обработки выдается словарь с четырьмя парами ключ:значение
# по негативным отзывам
data["negative_review"] = data["negative_review"].apply(sent_analyzer.polarity_scores)
# по позитивным отзывам
data["positive_review"] = data["positive_review"].apply(sent_analyzer.polarity_scores)
# создадим новые признаки из негативнного отзыва
data["negative_neg"] = data["negative_review"].apply(lambda x: x["neg"])
data["negative_neu"] = data["negative_review"].apply(lambda x: x["neu"])
data["negative_pos"] = data["negative_review"].apply(lambda x: x["pos"])
data["negative_compound"] = data["negative_review"].apply(lambda x: x["compound"])
# создадим новые признаки из позитивного отзыва
data["positive_neg"] = data["positive_review"].apply(lambda x: x["neg"])
data["positive_neu"] = data["positive_review"].apply(lambda x: x["neu"])
data["positive_pos"] = data["positive_review"].apply(lambda x: x["pos"])
data["positive_compound"] = data["positive_review"].apply(lambda x: x["compound"])
data["negative_review"].head(10)
# в некоторых негативных отзывах указано "нет негатива", пометим их и создадим соответствующий признак
data["no_negative"] = 0
data[data["negative_review"] == "No Negative"] = 1
data["positive_review"].head(10)
# в некоторых позитивных отзывах указано "нет позитива", пометим их и создадим соответствующий признак
data["no_positive"] = 0
data[data["positive_review"] == "No Positive"] = 1
# удалим ненужные теперь признаки отзывов
data.drop(["negative_review", "positive_review"], axis=1, inplace=True)
# города местоположения отелей
data["city_name"] = data["hotel_address"].apply(
lambda x: x.split()[-2] if x.split()[-2] != "United" else x.split()[-5]
)
# ['London', 'Paris', 'Amsterdam', 'Milan', 'Vienna', 'Barcelona']
# можно использовать One-Hot-Coding
encoder = ce.OneHotEncoder(
cols=["city_name"], use_cat_names=True
) # указываем столбец для кодирования
type_bin = encoder.fit_transform(data["city_name"])
data = pd.concat([data, type_bin], axis=1)
# Зная город, можно использовать данные по его расположению для заполнения пропущенных координат
data[(data["city_name"] == "Amsterdam") & (data["lat"] == 0)].fillna(
{"lat": 52.362209, "lng": 4.885346}
)
data[(data["city_name"] == "Barcelona") & (data["lat"] == 0)].fillna(
{"lat": 41.389125, "lng": 2.169152}
)
data[(data["city_name"] == "London") & (data["lat"] == 0)].fillna(
{"lat": 51.510737, "lng": -0.139075}
)
data[(data["city_name"] == "Milan") & (data["lat"] == 0)].fillna(
{"lat": 45.479619, "lng": 9.191844}
)
data[(data["city_name"] == "Paris") & (data["lat"] == 0)].fillna(
{"lat": 48.863658, "lng": 2.326816}
)
data[(data["city_name"] == "Vienna") & (data["lat"] == 0)].fillna(
{"lat": 48.203368, "lng": 16.367176}
)
data["lat"].nunique
data.info()
# пустые значения пребывания заменим на медиану
data["accommodation_days"] = data["accommodation_days"].fillna(
data["accommodation_days"].median()
)
data.info()
plt.rcParams["figure.figsize"] = (25, 20)
sns.heatmap(data.drop(["sample"], axis=1).corr(), annot=True)
# Высокая корреляция между days_since_review - rewiew_year(-0.92), negative_neg - negative_neu (-0.94) и positive_pos - positive_neu(-0.92
data.nunique(dropna=False)
# Обработаем списки тегов
# введем признак страны для отелей
data["country"] = data["hotel_address"].apply(
lambda x: x.split()[-1] if x.split()[-1] != "Kingdom" else " ".join(x.split()[-2:])
)
# уберем пробелы в названии места жительства резидента
data["reviewer_nationality"] = data["reviewer_nationality"].apply(lambda x: x[1:-1])
# создадим признак,определяющий иностранец или "местный жительЭ писал отзыв"
data["local_reviewer"] = data.apply(
lambda x: 1 if x["reviewer_nationality"] == x["country"] else 0, axis=1
)
data["local_reviewer"].value_counts(normalize=True)
# используем бинарное кодирования для некоторых признаков
data["reviewer_nationality"].nunique(dropna=False)
bin_encoder = ce.BinaryEncoder(cols=["hotel_name"]) # указываем столбец для кодирования
type_bin = bin_encoder.fit_transform(data["hotel_name"])
data = pd.concat([data, type_bin], axis=1)
data.hist(figsize=(25, 25))
# создадим копию
data_copied = data.copy()
object_columns = [
"hotel_address",
"hotel_name",
"reviewer_nationality",
"negative_review",
"positive_review",
"tags",
"review_date",
"city_name",
"country",
]
data.info()
# убираем признаки которые еще не успели обработать,
# модель на признаках с dtypes "object" обучаться не будет, просто выберим их и удалим
object_columns = [s for s in data.columns if data[s].dtypes == "object"]
data.drop(object_columns, axis=1, inplace=True)
data.info()
data2 = data.copy()
# Провели нормализацию признаков by RobustScaler
# ```
# list_normal = ['average_score','additional_number_of_scoring','additional_number_of_scoring', 'review_total_positive_word_counts','total_number_of_reviews_reviewer_has_given','total_number_of_reviews']
# r_scaler = preprocessing.RobustScaler(copy=False)
# for col in list(data.columns):
# if col != 'reviewer_score' and col != 'sample':
# data[col] = r_scaler.fit_transform(data[[col]])
# ```
# По итогу нормализация данных признаков не привела к улучшению результата. Решено от нее отказаться.
# data['average_score', 'additional_number_of_scoring','additional_number_of_scoring' 'review_total_positive_word_counts','total_number_of_reviews_reviewer_has_given''total_number_of_reviews']
data.head()
data.describe()
# ## 3.
# Теперь выделим тестовую часть
train_data = data.query("sample == 1").drop(["sample"], axis=1).drop_duplicates()
test_data = data.query("sample == 0").drop(["sample"], axis=1)
y = train_data.reviewer_score.values # наш таргет
X = train_data.drop(["reviewer_score"], axis=1)
# Воспользуемся специальной функцие train_test_split для разбивки тестовых данных
# выделим 20% данных на валидацию (параметр test_size)
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=RANDOM_SEED
)
# проверяем
test_data.shape, train_data.shape, X.shape, X_train.shape, X_test.shape
# Создаём модель
model = RandomForestRegressor(
n_estimators=100, verbose=1, n_jobs=-1, random_state=RANDOM_SEED
)
# Обучаем модель на тестовом наборе данных
model.fit(X_train, y_train)
# Используем обученную модель для предсказания рейтинга ресторанов в тестовой выборке.
# Предсказанные значения записываем в переменную y_pred
y_pred = model.predict(X_test)
# Функция для рассчёта MAPE:
def mape(actual, pred):
actual, pred = np.array(actual), np.array(pred)
return np.mean(np.abs((actual - pred) / actual)) * 100
# Сравниваем предсказанные значения (y_pred) с реальными (y_test), и смотрим насколько они в среднем отличаются
# Метрика называется Mean Absolute Error (MAE) и показывает среднее отклонение предсказанных значений от фактических.
print("MAE:", metrics.mean_absolute_error(y_test, y_pred))
print("MAPE:", mape(y_test, y_pred))
# print('MAPE:', metrics.mean_absolute_percentage_error(y_test, y_pred))
plt.rcParams["figure.figsize"] = (10, 10)
feat_importances = pd.Series(model.feature_importances_, index=X.columns)
feat_importances.nlargest(25).plot(kind="barh")
test_data.sample(10)
test_data = test_data.drop(["reviewer_score"], axis=1)
sample_submission
predict_submission = model.predict(test_data)
predict_submission
list(sample_submission)
sample_submission["reviewer_score"] = predict_submission
sample_submission.to_csv("submission.csv", index=False)
sample_submission.head(10)
|
# # Imports
import os
import numpy as np
import matplotlib.pyplot as plt
from PIL import Image
from tqdm import tqdm
import torch
import torch.nn as nn
import torch.optim as optim
from torch.utils.data import Dataset, DataLoader
import torchvision.transforms as transforms
from torchsummary import summary
from torchvision.utils import save_image
# # Dataset
def load_image(root_dir: str, dir_name: str, file_name: str, extension: str) -> Image:
return Image.open(os.path.join(root_dir, dir_name, file_name + extension))
class CelebADataset(Dataset):
def __init__(self, dir, transform=None):
super().__init__()
self.root = dir
self.transform = transform
self.list_of_inputs = os.listdir(dir)
def __getitem__(self, idx):
to_tensor_transform = transforms.ToTensor()
input_image = load_image(self.root, r"", self.list_of_inputs[idx], "")
input_image = to_tensor_transform(input_image)
if self.transform:
input_image = self.transform(input_image)
return input_image.float()
def __len__(self):
return len(self.list_of_inputs)
transform = transforms.Compose(
[
transforms.Resize((64, 64)),
transforms.Normalize(mean=[0.5, 0.5, 0.5], std=[0.5, 0.5, 0.5]),
]
)
batch_size = 16
train_dataset = CelebADataset(
dir=r"/kaggle/input/celeba-dataset/img_align_celeba/img_align_celeba",
transform=transform,
)
train_dataloader = DataLoader(
train_dataset, batch_size=batch_size, shuffle=True, num_workers=8
)
# # Model
# I tested two generators: Upsample + Conv2d and ConvTranspose2d
class UpsampleGenerator(nn.Module):
def __init__(self):
super().__init__()
self.conv_0 = nn.Sequential(
nn.UpsamplingNearest2d(scale_factor=2),
nn.Conv2d(
in_channels=100, out_channels=512, kernel_size=3, stride=1, padding=1
),
nn.BatchNorm2d(512),
nn.ReLU(),
)
self.conv_1 = nn.Sequential(
nn.UpsamplingNearest2d(scale_factor=2),
nn.Conv2d(
in_channels=512, out_channels=256, kernel_size=3, stride=1, padding=1
),
nn.BatchNorm2d(256),
nn.ReLU(),
)
self.conv_2 = nn.Sequential(
nn.UpsamplingNearest2d(scale_factor=2),
nn.Conv2d(
in_channels=256, out_channels=128, kernel_size=3, stride=1, padding=1
),
nn.BatchNorm2d(128),
nn.ReLU(),
)
self.conv_3 = nn.Sequential(
nn.UpsamplingNearest2d(scale_factor=2),
nn.Conv2d(
in_channels=128, out_channels=64, kernel_size=3, stride=1, padding=1
),
nn.BatchNorm2d(64),
nn.ReLU(),
)
self.conv_4 = nn.Sequential(
nn.UpsamplingNearest2d(scale_factor=2),
nn.Conv2d(
in_channels=64, out_channels=3, kernel_size=3, stride=1, padding=1
),
nn.BatchNorm2d(3),
nn.ReLU(),
)
self.conv_5 = nn.Sequential(
nn.UpsamplingNearest2d(scale_factor=2),
nn.Conv2d(
in_channels=3, out_channels=3, kernel_size=3, stride=1, padding=1
),
nn.BatchNorm2d(3),
nn.ReLU(),
)
self.conv_6 = nn.Sequential(
nn.Conv2d(
in_channels=3, out_channels=3, kernel_size=3, stride=1, padding=1
),
nn.Tanh(),
)
# 3 x 64 x 64
def forward(self, x):
x = self.conv_0(x)
x = self.conv_1(x)
x = self.conv_2(x)
x = self.conv_3(x)
x = self.conv_4(x)
x = self.conv_5(x)
x = self.conv_6(x)
return x
class TransposeGenerator(nn.Module):
def __init__(self):
super().__init__()
self.conv_0 = nn.Sequential(
nn.ConvTranspose2d(
in_channels=100,
out_channels=512,
kernel_size=4,
stride=2,
padding=0,
bias=False,
),
nn.BatchNorm2d(512),
nn.ReLU(),
)
self.conv_1 = nn.Sequential(
nn.ConvTranspose2d(
in_channels=512,
out_channels=256,
kernel_size=4,
stride=2,
padding=1,
bias=False,
),
nn.BatchNorm2d(256),
nn.ReLU(),
)
self.conv_2 = nn.Sequential(
nn.ConvTranspose2d(
in_channels=256,
out_channels=128,
kernel_size=4,
stride=2,
padding=1,
bias=False,
),
nn.BatchNorm2d(128),
nn.ReLU(),
)
self.conv_3 = nn.Sequential(
nn.ConvTranspose2d(
in_channels=128,
out_channels=64,
kernel_size=4,
stride=2,
padding=1,
bias=False,
),
nn.BatchNorm2d(64),
nn.ReLU(),
)
self.conv_4 = nn.Sequential(
nn.ConvTranspose2d(
in_channels=64, out_channels=3, kernel_size=4, stride=2, padding=1
),
nn.Tanh(),
)
# 3 x 64 x 64
def forward(self, x):
x = self.conv_0(x)
x = self.conv_1(x)
x = self.conv_2(x)
x = self.conv_3(x)
x = self.conv_4(x)
return x
class Discriminator(nn.Module):
def __init__(self):
super().__init__()
self.conv_0 = nn.Sequential(
nn.Conv2d(
in_channels=3, out_channels=32, kernel_size=4, stride=2, padding=1
),
nn.LeakyReLU(0.2),
)
self.conv_1 = nn.Sequential(
nn.Conv2d(
in_channels=32, out_channels=64, kernel_size=4, stride=2, padding=1
),
nn.LeakyReLU(0.2),
)
self.conv_2 = nn.Sequential(
nn.Conv2d(
in_channels=64, out_channels=128, kernel_size=4, stride=2, padding=1
),
nn.LeakyReLU(0.2),
)
self.conv_3 = nn.Sequential(
nn.Conv2d(
in_channels=128, out_channels=256, kernel_size=4, stride=2, padding=1
),
nn.LeakyReLU(0.2),
)
self.conv_4 = nn.Sequential(
nn.Conv2d(
in_channels=256, out_channels=512, kernel_size=4, stride=2, padding=1
),
nn.LeakyReLU(0.2),
)
self.conv_5 = nn.Sequential(
nn.Conv2d(
in_channels=512, out_channels=256, kernel_size=4, stride=2, padding=1
),
nn.LeakyReLU(0.2),
)
# self.classifier = nn.Sequential(
# nn.Linear(in_features=256*2*2, out_features=1)
# )
self.conv_6 = nn.Sequential(
nn.Conv2d(
in_channels=256, out_channels=1, kernel_size=3, stride=1, padding=1
),
)
self.flatten = nn.Flatten()
def forward(self, x):
x = self.conv_0(x)
x = self.conv_1(x)
x = self.conv_2(x)
x = self.conv_3(x)
x = self.conv_4(x)
x = self.conv_5(x)
x = self.conv_6(x)
x = self.flatten(x)
# x = self.classifier(x)
return x
def calc_gp(discriminator, true, fake):
eps = torch.randn(true.shape[0], 1, 1, 1).to(device)
eps = eps.expand(true.size())
interpolation = eps * true + (1 - eps) * fake
interp_logits = discriminator(interpolation)
grad_outputs = torch.ones_like(interp_logits)
gradients = torch.autograd.grad(
outputs=interp_logits,
inputs=interpolation,
grad_outputs=grad_outputs,
create_graph=True,
retain_graph=True,
)[0]
gradients = gradients.view(true.shape[0], -1)
grad_norm = gradients.norm(2, 1)
gp = torch.mean((grad_norm - 1) ** 2)
return gp
# In paper "Improved Training of Wasserstein GANs" using .mean() on discriminator outputs is suggested for loss function to train both discriminator and generator. Discriminator also recieves gradient penalty.
def train_gan(
generator,
generator_loss,
generator_optimizer,
discriminator,
discriminator_loss,
discriminator_optimizer,
dataloader,
device="cpu",
epochs=1,
):
print(device)
device = torch.device(device)
generator.train()
discriminator.train()
generator_loss.to(device)
discriminator_loss.to(device)
generator.to(device)
discriminator.to(device)
losses_d = []
losses_g = []
real_scores = []
fake_scores = []
for epoch in range(epochs):
losses_d_running = []
losses_g_running = []
real_scores_running = []
fake_scores_running = []
for index, input_ in enumerate(tqdm(dataloader)):
input_ = input_.to(device)
# train critic
train_critic_loss = 0
real_score = 0
fake_score = 0
# training discriminator
discriminator_optimizer.zero_grad()
real_preds = discriminator(input_)
real_targets = torch.ones(input_.shape[0], 1, device=device)
real_loss = discriminator_loss(real_preds, real_targets)
real_score = real_loss.mean().item()
real_scores_running.append(real_score)
# generate fake images
noise = torch.randn(input_.shape[0], 100, 1, 1, device=device)
fake_img = generator(noise)
fake_preds = discriminator(fake_img)
fake_targets = -1 * torch.ones(input_.shape[0], 1, device=device)
fake_loss = discriminator_loss(fake_preds, fake_targets)
fake_score = fake_loss.mean().item()
fake_scores_running.append(fake_score)
# wgan-gp moment
gp = calc_gp(discriminator, input_, fake_img)
d_loss = real_loss + fake_loss + 10 * gp
losses_d_running.append(d_loss.item())
d_loss.backward()
discriminator_optimizer.step()
train_critic_loss += d_loss.item()
if index % 5 == 0:
# training generator
generator_optimizer.zero_grad()
noise = torch.randn(input_.shape[0], 100, 1, 1, device=device)
fake_img = generator(noise)
fake_preds = discriminator(fake_img)
targets = torch.ones(input_.shape[0], 1, device=device)
g_loss = generator_loss(fake_preds, targets)
g_loss.backward()
generator_optimizer.step()
losses_g_running.append(g_loss.item())
losses_g.append(np.mean(losses_g_running))
losses_d.append(np.mean(losses_d_running))
real_scores.append(np.mean(real_scores_running))
fake_scores.append(np.mean(fake_scores_running))
print(
"Epoch [{}/{}], loss_g: {:.4f}, loss_d: {:.4f}, real_score: {:.4f}, fake_score: {:.4f}".format(
epoch + 1,
epochs,
losses_g[-1],
losses_d[-1],
real_scores[-1],
fake_scores[-1],
)
)
return losses_g, losses_d, real_scores, fake_scores
class WassersteinLoss(nn.Module):
def __init__(self):
super().__init__()
def forward(self, preds, targets):
return -torch.mean(targets * preds)
loss = WassersteinLoss()
device = "cuda" if torch.cuda.is_available() else "cpu"
print(device)
# # Upsample training + results
up_d = Discriminator()
up_g = UpsampleGenerator()
up_d_optim = torch.optim.Adam(up_d.parameters(), lr=1 * 10e-5, betas=(0.5, 0.999))
up_g_optim = torch.optim.Adam(up_g.parameters(), lr=1 * 10e-5, betas=(0.5, 0.999))
# up_d_optim = torch.optim.RMSprop(up_d.parameters(), lr=1*10e-5)
# up_g_optim = torch.optim.RMSprop(up_g.parameters(), lr=1*10e-5)
up_d.to(device)
summary(up_d, (3, 64, 64))
g_loss, d_loss, real_score, fake_score = train_gan(
up_g, loss, up_g_optim, up_d, loss, up_d_optim, train_dataloader, device, epochs=40
)
plt.plot(g_loss, label="g")
plt.plot(d_loss, label="d")
plt.legend()
plt.show()
plt.plot(real_score, label="real score")
plt.plot(fake_score, label="fake score")
plt.legend()
plt.show()
noise = torch.randn(1, 100, 1, 1, device=device)
img = up_g(noise)
save_image(img[0], fp="output.png")
img = transforms.ToPILImage()(img[0] / 2 + 0.5)
img = np.array(img)
plt.imshow(img)
# # Convtranspose training + results
tr_g = TransposeGenerator()
tr_d = Discriminator()
tr_g_optim = torch.optim.Adam(tr_g.parameters(), lr=1 * 10e-4, betas=(0.0, 0.9))
tr_d_optim = torch.optim.Adam(tr_d.parameters(), lr=1 * 10e-4, betas=(0.0, 0.9))
g_loss, d_loss, real_score, fake_score = train_gan(
tr_g, loss, tr_g_optim, tr_d, loss, tr_d_optim, train_dataloader, device, epochs=20
)
plt.plot(g_loss, label="g")
plt.plot(d_loss, label="d")
plt.legend()
plt.show()
noise = torch.randn(1, 100, 1, 1, device=device)
img = tr_g(noise)
img = transforms.ToPILImage()(img[0] / 2 + 0.5)
img = np.array(img)
plt.imshow(img)
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import xgboost as xgb
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
import tensorflow as tf
from tensorflow import keras
from sklearn.model_selection import GridSearchCV
pd.set_option("display.max_rows", 200)
# ## Data Exploration
df_init = pd.read_csv("TrainingWiDS2021.csv", index_col=0)
# drop columns which have more than 30% Nan values
df = df_init.dropna(thresh=0.7 * len(df_init), axis="columns")
# columns removed
remove_cols = list(set(df_init.columns) - set(df.columns))
set(df.columns)
# df
print(df.dtypes.unique())
# readmission_status columns has all 0
for c in df.columns:
if df.dtypes[c] == "int64":
print(c, df[c].unique())
# Number of non-nan values in each column
df.count(axis=0)
# Exploring categorical columns
for c in df.columns:
if df.dtypes[c] == "object":
print("Categorical columns: ", c)
print("Null values: ", df[c].isna().sum())
print(df[c].value_counts())
print("--------")
# print(df[c].unique())
# one-hot encoding categorical columns
enc_df = pd.get_dummies(df)
# removing a few hospital_admit_source_ as they are not present in the training data
X = enc_df.drop(
[
"encounter_id",
"hospital_id",
"diabetes_mellitus",
"icu_id",
"readmission_status",
"hospital_admit_source_Acute Care/Floor",
"hospital_admit_source_ICU",
"hospital_admit_source_Observation",
"hospital_admit_source_Other",
"hospital_admit_source_PACU",
],
axis=1,
)
Y = enc_df["diabetes_mellitus"]
train_X, test_X, train_y, test_y = train_test_split(
X, Y, random_state=1, test_size=0.25
)
X.shape
# ## Classification using XGBoost
model = xgb.XGBClassifier(
n_estimators=300, max_depth=3, learning_rate=0.1, subsample=0.5
)
# Fit the model to the training set and predict on the test set
train_model = model.fit(train_X, train_y)
pred = train_model.predict(test_X)
model2 = xgb.XGBClassifier(
n_estimators=500, max_depth=3, learning_rate=0.1, subsample=0.5
)
# Fit the model to the training set and predict on the test set
train_model2 = model2.fit(train_X, train_y)
pred2 = train_model2.predict(test_X)
model3 = xgb.XGBClassifier(
n_estimators=500, max_depth=3, learning_rate=0.01, subsample=0.5
)
# Fit the model to the training set and predict on the test set
train_model3 = model3.fit(train_X, train_y)
pred3 = train_model3.predict(test_X)
print("Accuracy for model: %.2f" % (accuracy_score(test_y, pred) * 100))
print("Accuracy for model: %.2f" % (accuracy_score(test_y, pred2) * 100))
print("Accuracy for model: %.2f" % (accuracy_score(test_y, pred3) * 100))
# ## Predict probability using model 2
predprob2 = train_model2.predict_proba(test_X)
predprob2
pred2
# ## Grid search CV
# ## Feature Importance
# Plot feature importance
xgb.plot_importance(model2, max_num_features=100, height=0.2)
plt.rcParams["figure.figsize"] = [50, 20]
# plt.show()
plt.tight_layout()
# plt.savefig('xgboost_model1_featureimp.png',dpi=400)
# # Applying the model to the Unlabeled data
testdf = pd.read_csv("UnlabeledWiDS2021.csv", index_col=0)
testdf = testdf.drop(
[
"h1_arterial_ph_min",
"h1_glucose_min",
"urineoutput_apache",
"h1_diasbp_invasive_max",
"ph_apache",
"d1_bilirubin_max",
"h1_albumin_min",
"bilirubin_apache",
"d1_pao2fio2ratio_min",
"h1_lactate_max",
"h1_diasbp_invasive_min",
"d1_arterial_pco2_max",
"h1_calcium_min",
"paco2_for_ph_apache",
"d1_albumin_max",
"h1_hematocrit_max",
"h1_sysbp_invasive_max",
"h1_arterial_po2_min",
"h1_hemaglobin_max",
"h1_arterial_pco2_max",
"d1_mbp_invasive_max",
"d1_sysbp_invasive_min",
"h1_inr_max",
"d1_diasbp_invasive_max",
"h1_platelets_min",
"h1_potassium_max",
"h1_wbc_max",
"d1_pao2fio2ratio_max",
"h1_platelets_max",
"h1_hemaglobin_min",
"h1_pao2fio2ratio_max",
"h1_sodium_min",
"albumin_apache",
"h1_lactate_min",
"pao2_apache",
"d1_arterial_ph_min",
"d1_bilirubin_min",
"h1_hco3_max",
"h1_sysbp_invasive_min",
"fio2_apache",
"d1_diasbp_invasive_min",
"h1_mbp_invasive_max",
"h1_bilirubin_min",
"d1_arterial_ph_max",
"d1_albumin_min",
"h1_bun_min",
"h1_bun_max",
"d1_arterial_pco2_min",
"d1_inr_max",
"h1_arterial_po2_max",
"h1_inr_min",
"d1_arterial_po2_max",
"d1_lactate_max",
"d1_arterial_po2_min",
"h1_wbc_min",
"h1_arterial_ph_max",
"h1_mbp_invasive_min",
"h1_pao2fio2ratio_min",
"h1_sodium_max",
"paco2_apache",
"d1_sysbp_invasive_max",
"d1_inr_min",
"h1_hematocrit_min",
"h1_glucose_max",
"h1_bilirubin_max",
"h1_arterial_pco2_min",
"h1_calcium_max",
"h1_creatinine_max",
"h1_creatinine_min",
"h1_albumin_max",
"h1_potassium_min",
"d1_mbp_invasive_min",
"d1_lactate_min",
"h1_hco3_min",
],
axis=1,
)
testdf.shape
# writing the encounter_id into an array
enc_id = testdf["encounter_id"]
# one hot encoding
enc_testdf = pd.get_dummies(testdf)
testdf_X = enc_testdf.drop(
["encounter_id", "hospital_id", "icu_id", "readmission_status"], axis=1
)
print(testdf_X.columns)
# df['hospital_admit_source'].unique()
# set(X.columns) - set(testdf_X.columns)
# testdf['hospital_admit_source'].unique()
# Apply model to predict diabetes_mellitus
testdf_Y = model2.predict_proba(testdf_X)
# writing encounter_id and diabetes_mellitus prediction into data frame
finalx = enc_id.to_numpy()
finaly = testdf_Y[:, 1]
finaldict = {"encounter_id": finalx, "diabetes_mellitus": finaly}
finaldf = pd.DataFrame(data=finaldict)
finaldf
# writing dataframe into csv
finaldf.to_csv("Team_Noether_SubmissionWiDS2021.csv", index=False)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# - train.csv - Personal records for about two-thirds (~8700) of the passengers, to be used as training data.
# - PassengerId - A unique Id for each passenger. Each Id takes the form gggg_pp where gggg indicates a group the passenger is travelling with and pp is their number within the group. People in a group are often family members, but not always.
# - HomePlanet - The planet the passenger departed from, typically their planet of permanent residence.
# - CryoSleep - Indicates whether the passenger elected to be put into suspended animation for the duration of the voyage. Passengers in cryosleep are confined to their cabins.
# - Cabin - The cabin number where the passenger is staying. Takes the form deck/num/side, where side can be either P for Port or S for Starboard.
# - Destination - The planet the passenger will be debarking to.
# - Age - The age of the passenger.
# - VIP - Whether the passenger has paid for special VIP service during the voyage.
# - RoomService, FoodCourt, ShoppingMall, Spa, VRDeck - Amount the passenger has billed at each of the Spaceship Titanic's many luxury amenities.
# - Name - The first and last names of the passenger.
# - Transported - Whether the passenger was transported to another dimension. This is the target, the column you are trying to predict.
train = pd.read_csv("/kaggle/input/spaceship-titanic/train.csv")
test = pd.read_csv("/kaggle/input/spaceship-titanic/test.csv")
import numpy as np
import pandas as pd
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
# - ## Need to add steps note havent done that yet... will do later
# DATA EXPLORATION
# - passenger id is useful the groupnumber and passenger number can identify the missing HomePlanet and Destination using the data just above and below
#
train.head(5)
train_len = len(train)
print(len(train))
train.isnull().sum()
train.describe()
# Percentile meaning.....
# (Age).........................Same meaning for others
# 75% people are 38 or younger
# 50% people are 27 or younger
# cant use function for visualisn here as except Age all num hav very diff vals
# cay=tegorical also notworking
train.info()
# ### Age wrt Destination & Homeplanet
sns.histplot(data=train, x=train["Age"], hue=train["Destination"], kde=True)
# - Most have age 21-25 (Can fill Age wrt Destination)
sns.histplot(data=train, x=train["Age"], hue=train["HomePlanet"], bins=30, kde=True)
# - From Earth most are of age-22
# - From mars -25
# - From Europa 27
# ########################################################################################################
# #### -Transported is numerical ie it is in Boolean so need to convert it to Categorical ?? (No need actualy works fine normally)
# def impute_val(col):
# if col==True:
# return 'true'
# elif col== False:
# return 'false'
# train['Transported']= train['Transported'].apply(impute_val)
# train['Transported'].head()
# - function for categorical ddnt work
# ########################################################################################################
# ### CRYOSLEEP wrt Homeplanet, Destination
sns.countplot(data=train, x=train["CryoSleep"], hue=train["HomePlanet"])
sns.countplot(data=train, x=train["CryoSleep"], hue=train["Destination"])
# Cryosleep has no pattern wrt either HomePlanet or EndDestination (so use Mode for impute cryosleep)
# ### VIP wrt Homeplanet and Destination
sns.countplot(data=train, x=train["VIP"], hue="HomePlanet")
# - Earth has no VIPs , So if VIP is missing, HomePlanet is Earth VIP=FALSE (actually most are non VIP so VIp= False only)
# - If VIP is True HomePlanet missing most Vip from Europa so fill Europa
sns.countplot(data=train, x=train["VIP"], hue="Destination")
# - No VIP is going to PSO Destination
sns.countplot(data=train, x="Destination", hue="VIP") # hue=df['VIP'] no relation
# Most passengers going to Trappist (No pattern for VIP either
# sns.countplot(data=train,x=train['Cabin'])
# - Cabin has lot of unique values
test.info()
test.isnull().sum()
train.corr()
# - Merging both train and test for missing values operation
train_len = len(train)
# combine train and test to do all data processing in them together
df = pd.concat(
[train, test], axis=0
) # axis=0 => row wise concatenation 1=> column wise ie more columns added
df = df.reset_index(drop=True) # df is combination of both train and test
df.head()
df.tail()
# - transport is missing in the end b/c test doesn't have Transported column
df.isnull().sum()
# ### Histplot for all numerical values
df.hist(figsize=(20, 20), bins=20)
# - Highly Left skewed data so must use log transformation to get bell curve (~equal distribution)
# #### any transformation only after filling missing values
sns.kdeplot(df["Age"]) # sns.kdeplot(np.sqrt(df['Age']+1))
df["Age"].median()
df.isnull().sum()
# only Pid ,Transported we have full values
# # Testing sqrt transform
sns.histplot(data=df, x=np.sqrt(df["Spa"]), bins=30, hue=df["VIP"], color="dark")
sns.countplot(
data=train,
x=df["VIP"],
)
train["VIP"].value_counts()
train.pivot_table(
index="HomePlanet", columns="VIP", values="CryoSleep", aggfunc="count"
)
df["RoomService"].unique
# # Exploring Cabin
df["Cabin"]
cabininitial = train["Cabin"].str.split("/")
cabininitial.head(20)
# - How does this try except work look again appending.., Its correct each value of list goes to individual lists (since we have null values we are using try except
deck = []
decknum = []
side = []
for i in cabininitial:
try:
deck.append(i[0])
decknum.append(i[1])
side.append(i[2])
except: # when 14. [F, 2, P] 15 NaN
deck.append("NULL")
decknum.append("-1") # else -1 as indicator of nan value
side.append("NULL")
print(set(deck))
print("maximum decknum is:{}".format(max(decknum)), "minimum :{}".format(0), sep="\n")
print(set(side))
# - P=Port=Left
# - S=Starboard=Right
# - Find mean median mode and impute the nan
# - (as currently they are list not series so can't do value_counts )
# - can do mode for deck but cant do median/mean for deck no as it varies
import statistics
print(statistics.mode(deck))
print(statistics.mode(side))
Cabin2 = pd.DataFrame({"deck": deck, "decknum": decknum, "side": side})
# sns.histplot(x=Cabin2["decknum"],kde=True,bins=200)
# - decknum is maybe useless...
sns.countplot(x=Cabin2["deck"], hue=df["VIP"])
Cabin2["deck"].value_counts()
# VIPs are distributed in every Cabin ie- no special cabins for VIPs but in the same Cabins probably some deluxe rooms, T can be merged with F or G
Cabin2["deck"].value_counts()
Cabin2.loc("deck" == "T")
def impute_deck(cols):
deck = cols
if deck == "NULL":
return "F"
else:
return deck
Cabin2["deck"] = Cabin2["deck"].apply(impute_deck)
def impute_side(cols):
side = cols
if side == "NULL":
return "S"
else:
return side
Cabin2["side"] = Cabin2["side"].apply(impute_side)
Cabin2.head(16)
Cabin2 = Cabin2.drop(columns=["decknum"], axis=1)
Cabin2.head(10)
# ###################################################################################################
# ## Taking passenger group number-
Groupnum = df["PassengerId"].str.split("_")
Groupnum
Groupid = []
for i in Groupnum:
Groupid.append(i[0])
Groupid = pd.DataFrame({"Groupid": Groupid})
Groupid = Groupid.astype("int")
Groupid.head(10)
# ########################################################################################################
train["VIP"].value_counts()
df.columns
df["FoodCourt"].max() # 0 t0 30k
df.boxplot("Age")
df.boxplot(["RoomService", "FoodCourt", "ShoppingMall", "Spa", "VRDeck"])
# ## Imputing Missing Values
# ### Impute AGE
def age(cols): # cols has Age value if df[['Age'],['']] #
Age = cols
if pd.isnull(Age):
return 24
else:
return Age
df["Age"] = df["Age"].apply(age)
# ## Let's Try Binning the age
def age(cols): # cols has Age value if df[['Age'],['']] #
Age = cols
if Age <= 10:
return "Infant"
elif Age > 10 and Age <= 20:
return "Teen"
elif Age > 20 and Age <= 50:
return "Adult"
else:
return "elder"
df["Age"] = df["Age"].apply(age)
# # Impute HomePlanet
def Homeplanetimpute(cols):
HomePlanet = cols[0]
VIP = cols[1]
if pd.isnull(HomePlanet):
if VIP == "True":
return "Europa"
elif VIP == "False":
return "Earth"
else:
return "Earth"
else:
return HomePlanet
df["HomePlanet"] = df[["HomePlanet", "VIP"]].apply(Homeplanetimpute, axis=1)
# ## Imputing RoomService', 'FoodCourt', 'ShoppingMall', 'Spa', 'VRDeck'
df["RoomService"].median()
df["RoomService"].fillna(value=df["RoomService"].median(), inplace=True)
df["FoodCourt"].fillna(value=df["FoodCourt"].median(), inplace=True)
df["ShoppingMall"].fillna(value=df["ShoppingMall"].median(), inplace=True)
df["Spa"].fillna(value=df["Spa"].median(), inplace=True)
df["VRDeck"].fillna(value=df["VRDeck"].median(), inplace=True)
df.isnull().sum()
# ### Fill missing Destination and CryoSleep
print(df["Destination"].mode()[0])
df["Destination"].fillna(value=df["Destination"].mode()[0], inplace=True)
print(df["CryoSleep"].mode()[0])
df["CryoSleep"].fillna(value=df["CryoSleep"].median(), inplace=True)
print(df["VIP"].mode()[0])
df["VIP"].fillna(value=df["VIP"].mode()[0], inplace=True)
df.isnull().sum()
# ### Transforming The data (log transform used due to skewness)
# Checking the Skewness
df.skew() # VIP is categorical
df["RoomService"] = df["RoomService"].apply(lambda x: np.log(x + 1))
df["FoodCourt"] = df["FoodCourt"].apply(lambda x: np.log(x + 1))
df["ShoppingMall"] = df["ShoppingMall"].apply(lambda x: np.log(x + 1))
df["Spa"] = df["Spa"].apply(lambda x: np.log(x + 1))
df["VRDeck"] = df["VRDeck"].apply(lambda x: np.log(x + 1))
df.skew()
# - Skewness removed pretty much
# - VIP is Categorical data so its skewness doesnt matter
df.hist(bins=20, figsize=(20, 20))
# ## Feature Selection
# #### Dropping Name,Cabin,PassengerId
df.drop(["Name", "Cabin", "PassengerId"], inplace=True, axis=1)
df.columns
# ### Concatenating Cabin2 (Groupid didnt work well performance detoriated so not adding groupid)
df = pd.concat([df, Cabin2], axis=1)
df.head()
# ### Using LabelEncoder to convert categorical values to numbers
# - HomePLanet
# - Destination
# - VIP
# - Transported
# - deck
# - side
# - Age (as we binned it)
from sklearn import preprocessing
le = preprocessing.LabelEncoder()
cols = [
"HomePlanet",
"Destination",
"VIP",
"CryoSleep",
"Transported",
"deck",
"side",
"Age",
]
for col in cols:
df[col] = le.fit_transform(df[col])
# df['HomePlanet']= le.fit_transform(data['HomePlanet'])
df.head()
# ### Standardizing (Didnt work properly so didnt do standardization)
# - (Checking by both doing it and not Doing it choosing whichever gives better reults)
# - All our datas are in different units so Using StandardScaler to bring all the data into same scale
# (can be usefull sometimes)
"""from sklearn.preprocessing import StandardScaler
scaler=StandardScaler()
cols=df.columns
df=pd.DataFrame(scaler.fit_transform(df),columns=cols)"""
df
# # BLINK (just to stop execution mid way ie no purpose of blink)
# blink
# ### Seperating Train and Test
train = df.iloc[:train_len, :] # =>rows till 0 to train_len-1 , all columns
test = df.iloc[train_len:, :]
############################################ STOP ########################
# ### Seperating X and y ( Y= Transported)
# input split (What is this step) note wer using 'train' only
X = train.drop(columns=["Transported"], axis=1)
Y = train["Transported"]
# test run of standardization
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
cols = X.columns
X = pd.DataFrame(scaler.fit_transform(X), columns=cols)
# X.info()
from sklearn.model_selection import train_test_split, cross_val_score
# classify column
def classify(model):
x_train, x_test, y_train, y_test = train_test_split(
X, Y, test_size=0.25, random_state=42
)
model.fit(x_train, y_train)
print("Accuracy:", model.score(x_test, y_test))
score = cross_val_score(model, X, Y, cv=5) # will give 5 scores cv=5 => 5 folds
print("CV_score:", np.mean(score))
from sklearn.linear_model import LogisticRegression
model = LogisticRegression()
classify(model)
from sklearn.svm import SVC
model = SVC()
classify(model)
from sklearn.tree import DecisionTreeClassifier
model = DecisionTreeClassifier()
classify(model)
from sklearn.ensemble import RandomForestClassifier
model = RandomForestClassifier()
classify(model)
from sklearn.ensemble import ExtraTreesClassifier
model = ExtraTreesClassifier()
classify(model)
from xgboost import XGBClassifier
model = XGBClassifier()
classify(model)
from lightgbm import LGBMClassifier
model = LGBMClassifier()
classify(model)
from catboost import CatBoostClassifier
model = CatBoostClassifier(verbose=0) # verbose=0 else it will go deeplearning
classify(model)
# ## Complete Model Training with Full data
model = CatBoostClassifier(verbose=0)
model.fit(X, Y)
# input split for test data we dont need y as thats what we need to predict
X_test = test.drop(columns=["Transported"], axis=1)
pred = model.predict(X_test)
pred # our predicted Y_test (Transported) using the 'X'_test
# need to convert the values to Boolean type acc to submission.csv pattern ( if changed to categ)
transported = []
for i in pred:
if i == 1:
transported.append(True)
if i == 0:
transported.append(False)
transported[0:10]
sub = pd.read_csv("/kaggle/input/spaceship-titanic/sample_submission.csv")
sub.info()
# - this means we need pandas dataframe type object and PassengerId too
transported = list(transported)
PassengerId = pd.read_csv(
"/kaggle/input/spaceship-titanic/test.csv", usecols=["PassengerId"]
)
print(PassengerId[0:10])
print(type(PassengerId))
PassengerId = PassengerId["PassengerId"].tolist()
PassengerId[0:5]
transported2 = pd.DataFrame({"PassengerId": PassengerId, "transported": transported})
# pip show pandas
transported2.info()
# transported['transpored']=transported['transported'].astype('bool')
transported2
# Generate csv file
transported2.to_csv("submission.csv", index=False)
# Now click submit
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Define periods
PERIODS = [
(None, "2000-03-23"), # Period 1: before dot-bubble
("2000-03-24", "2002-10-04"), # Period 2 : dot-bubble
("2002-10-05", "2007-10-08"), # Period 3: between dot-bubble and 2008 crisis
("2007-10-09", "2009-03-09"), # Period 4: 2008 financial crisis
("2009-03-10", "2020-02-18"), # Period 5: between 2008 financial crisis and COVID
("2020-02-19", "2020-03-23"), # Period 6: COVID
("2020-02-19", None), # Period 7: after COVID
]
SPY_FILE = "/kaggle/input/daily-adjusted-spy-from-alpha-vantage/SPY.csv"
SPY_CONSTITUENTS = (
"/kaggle/input/daily-adjusted-spy-from-alpha-vantage/SPY_holdings_20-Mar-2023.csv"
)
PROGRESS = "/kaggle/input/alpha-vantage-daily-adjusted-prices/download_progress.csv"
DIR = "/kaggle/input/alpha-vantage-daily-adjusted-prices/"
def validate_price_data(price_df, symbol):
"""
validate Date and Adjusted Close Price of a symbol
Parameters:
- price_df (DataFrame) : DataFrame with date and prices
- symbol (string) : symbol of the equity you want to enquire e.g. 'IBM'
Return:
- True if price_df is valid
- False if price_df is invalid
"""
# check if any duplicated dates
if price_df["timestamp"].duplicated().any():
print("Duplicated timestamp for " + symbol)
return False
# check if any NULL price or prices <= 0
if (
price_df["adjusted_close"].isnull().any()
| (price_df["adjusted_close"] <= 0).any()
):
print("Invalid price on " + price_df["timestamp"] + " for " + symbol)
return False
if len(price_df) > 0:
return True
else:
return False
def transform_df(df):
"""
Transform DataFrame of a stock before processing:
1. 'timestamp' to datetime
2. Sort 'timestamp' in ascending order
3. Add a new column 'year' by converting 'timestamp'
4. Add a new column 'return_pct' to calculate the percentage change of 'adjusted_close'
5. Drop the first row of data as 'return_pct' is NA
Parameters:
- df (DataFrame) : DataFrame with date, prices of a stock / ETF
Return:
- DataFrame with 'timestamp', 'year', 'adjusted_close', 'return_pct'
"""
cols = ["timestamp", "year", "adjusted_close", "return_pct"]
df["timestamp"] = pd.to_datetime(df["timestamp"])
df = df.sort_values("timestamp")
df["year"] = df["timestamp"].dt.year + (df["timestamp"].dt.dayofyear - 1) / 365.25
df["return_pct"] = df["adjusted_close"].pct_change()
df = df.dropna()
return df.loc[:, cols]
def merge_df(df1, df2, suff1, suff2):
"""
Merge 2 DataFrames with inner join
Parameters:
- df1 (DataFrame) : DataFrame 1
- df2 (DataFrame) : DataFrame 2
- suff1 (string) : Suffix for columns of DataFrame 1
- suff2 (string) : Suffix for columns of DataFrame 2
Return:
- merged DataFrame
"""
return pd.merge(
df1, df2, on=["timestamp", "year"], how="inner", suffixes=(suff1, suff2)
)
import statsmodels.api as sm
def fit_linear_reg_model(x, y):
"""
Fit 2 variables to a linear regression model from statsmodels
Parameters:
- x (array_like) : independent variable
- y (array_like) : dependent variable
Return:
- fitted model
"""
x = sm.add_constant(x)
mdl = sm.OLS(y, x).fit()
return mdl
def calculate_stock_stats(symbol, df):
"""
Computes several statistical measures related to a particular stock or ETF
Parameters:
- symbol (string): The symbol for the stock/ETF of interest
- df (DataFrame): A DataFrame containing information about the investment and a benchmark
Return:
- DataFrame with the following values:
- symbol: symbol of the stock/ETF
- in_SPY: indicates whether the investment is part of the SPY index (TRUE) or not (FALSE)
- slope: the slope of a linear regression model that's fitted to the investment's adjusted price over time
- r2: the R-squared value of the linear regression model
- beta: the slope of a linear regression model that's fitted to the investment's adjusted price against the benchmark's adjusted price
- alpha: the intercept of the same linear regression model as Beta
- r2_with_benchmark: the R-squared value of the linear regression model that uses the investment's adjusted price and the SPY's adjusted price
- correlation: the correlation between the investment's adjusted price and the SPY's adjusted price.
"""
error = 0
if len(df) >= 2:
mdl_symbol = fit_linear_reg_model(df["year"], df["adjusted_close"])
mdl_against_index = fit_linear_reg_model(df["return_pct_spy"], df["return_pct"])
corr = df["return_pct"].corr(df["return_pct_spy"])
else:
error = 1
corr = 0
return pd.DataFrame(
{
"symbol": symbol,
"in_SPY": spy_holdings["Ticker"].isin([symbol]).any(),
"slope": mdl_symbol.params[1] if error != 1 else 0,
"r2": mdl_symbol.rsquared if error != 1 else 0,
"beta": mdl_against_index.params[1] if error != 1 else 0,
"alpha": mdl_against_index.params[0] if error != 1 else 0,
"r2_with_benchmark": mdl_against_index.rsquared if error != 1 else 0,
"correlation": corr,
},
index=[0],
)
from scipy.stats import ttest_ind
def calculate_periodic_stock_stats(symbol, df):
"""
Computes several statistical measures related to a particular stock or ETF in different periods
Parameters:
- symbol (string): The symbol for the stock/ETF of interest
- df (DataFrame): A DataFrame containing information about the investment and a benchmark
Return:
- DataFrame with the following values:
- symbol: symbol of the stock/ETF
- period: period number of the statistics
- t-statistic: t_statistic of one-tailed test between returns of the investment and a benchmark during that period
- p-value: p-value of the same one-tailed test during that period
- correlation: correlation between returns of the investment and a benchmark during that period
"""
results = []
# Perform one-tailed-test for each period
for i, period in enumerate(PERIODS):
start_date, end_date = period
if (start_date is not None) & (end_date is not None):
subset_df = df.loc[
(df["timestamp"] >= start_date) & (df["timestamp"] < end_date)
]
elif end_date is not None:
subset_df = df.loc[(df["timestamp"] <= end_date)]
else:
subset_df = df.loc[(df["timestamp"] >= start_date)]
t_stat, p_val = ttest_ind(
subset_df["return_pct"],
subset_df["return_pct_spy"],
equal_var=False,
alternative="greater",
)
p_val /= 2 # divide p-value by 2 for one-tailed test
corr = subset_df["return_pct"].corr(subset_df["return_pct_spy"])
results.append(
{
"symbol": symbol,
"period": f"Period {i+1}",
"t-statistic": t_stat,
"p-value": p_val,
"correlation": corr,
}
)
# print(f'Period {i+1} t-statistic: {t_stat:.3f}, p-value: {p_val:.4f}, Pearson Correlation: {corr:.4f}')
return pd.DataFrame(results)
SPY_df = pd.read_csv(SPY_FILE)
SPY_df = transform_df(SPY_df)
progress_df = pd.read_csv(PROGRESS)
downloaded = progress_df[progress_df["Status"] == "Downloaded"]["Symbol"]
spy_holdings = pd.read_csv(SPY_CONSTITUENTS)
results_df = pd.DataFrame({})
periodic_results_df = pd.DataFrame({})
for symbol in downloaded:
df = pd.read_csv(DIR + symbol + ".csv")
df = transform_df(df)
merged_df = merge_df(SPY_df, df, "_spy", "")
results_df = results_df.append(calculate_stock_stats(symbol, merged_df))
periodic_results_df = periodic_results_df.append(
calculate_periodic_stock_stats(symbol, merged_df)
)
results_df.reset_index()
results_df.to_csv("results.csv", index=False)
periodic_results_pivot = periodic_results_df.pivot(index="symbol", columns="period")
periodic_results_pivot.columns = [
f'{col[1]}-{col[0].replace(" ", "").lower()}'
for col in periodic_results_pivot.columns
]
# reset the index to have Symbol as a column again
periodic_results_pivot = periodic_results_pivot.reset_index()
periodic_results_pivot.to_csv("periodic_results.csv", index=False)
|
from fastai.vision.all import *
path = Path("/kaggle/input/25-indian-bird-species-with-226k-images")
path.ls()
files = get_image_files(path)
len(files)
dls = DataBlock(
blocks=(ImageBlock, CategoryBlock),
get_items=get_image_files,
splitter=RandomSplitter(valid_pct=0.2, seed=42),
get_y=parent_label,
item_tfms=[Resize(224, method="squish")],
).dataloaders(path, bs=32)
dls.show_batch(max_n=15)
learn = vision_learner(dls, resnet34, metrics=error_rate)
learn.fine_tune(2)
learn.show_results(max_n=12)
what_breed, _, prob = learn.predict(files[4500])
# files[4500]
print(f"The bird breed is: {what_breed}")
print(prob)
interp = Interpretation.from_learner(learn)
interp.plot_top_losses(9, figsize=(20, 10))
|
import numpy as np
import pandas as pd
import tensorflow as tf
from tensorflow import keras
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# # Custrom Normalization Layer
class LayerNormalization(keras.layers.Layer):
def __init__(self):
super().__init__()
def build(self, input_shape):
self.alpha = self.add_weight(
"alpha",
shape=input_shape[-1:],
dtype=tf.float32,
initializer=tf.constant_initializer(1),
)
self.beta = self.add_weight(
"beta",
shape=input_shape[-1:],
dtype=tf.float32,
initializer=tf.constant_initializer(0),
)
def call(self, inputs, epsilon=1e-3):
mean, variance = tf.nn.moments(inputs, axes=-1, keepdims=True)
std = tf.math.sqrt(variance)
return self.alpha * (inputs - mean) / (std + epsilon) + self.beta
def get_config(self):
base_config = super().get_config()
return {**base_config, "epsilon": self.epsilon}
tf.random.set_seed(42)
X = tf.random.uniform(
(50, 12),
)
my_layer = LayerNormalization()
std_layer = keras.layers.LayerNormalization()
print(tf.reduce_mean(my_layer(X)))
print(tf.reduce_mean(std_layer(X)))
random_alpha = np.random.rand(X.shape[-1])
random_beta = np.random.rand(X.shape[-1])
my_layer.set_weights([random_alpha, random_beta])
std_layer.set_weights([random_alpha, random_beta])
tf.reduce_mean(keras.losses.mean_absolute_error(std_layer(X), my_layer(X)))
|
# # Project 2
# We're going to continue from where we left off with Project 1. Project 1 left us with a daily time series for every product with no gaps -- exactly what we want for modeling!
data_path = "/kaggle/input/project-2-data/project_2_data"
data_path = "./project_2_data"
import pandas as pd
import numpy as np
data = pd.read_parquet(f"{data_path}/sales_data.parquet")
data.head()
data.shape
# We did some EDA in Project 1, but it was primarily focused on higher level patterns (i.e., at the department level). This time, spend some time doing EDA at the item level to see what kind of items you're dealing with.
# Some questions you may want to explore:
# 1. How do high-volume items compare to low-volume/itermittent items?
# 2. What sort of seasonal patterns are at play?
# 3. Do items from different departments show different patterns?
# 4. Does the same item show different behavior at different stores?
# These questions are just a starting point. Feel free to explore this any way you feel is necessary to make better models. The best EDA is done iteratively, so I encourage you to come back to this once you've started fitting models!
top10items = (
data.groupby("item_id")["sales"].sum().sort_values(ascending=False).index[:10]
)
btm10items = (
data.groupby("item_id")["sales"].sum().sort_values(ascending=False).index[-10:]
)
data_items = data.groupby(["item_id", "date"])["sales"].sum()
data_items.shape
import matplotlib.pyplot as plt
data_items.unstack(0)[top10items].rolling(7).sum().plot()
plt.show()
data_items.unstack(0)[btm10items].rolling(7).sum().plot()
plt.show()
pd.plotting.autocorrelation_plot(data_items.unstack(0)[top10items[0]])
pd.plotting.autocorrelation_plot(data_items["FOODS_2_320"])
# Before we get to modeling, let's create our evaluation setup. The models that we're going to create have a 28-day forecast horizon, and our goal is to best approximate "average" sales.
# The first step is to implement our evaluation metric. The original competition used a metric called RMSSE, or "Root Mean Squared Scaled Error." It's similar to the MASE metric that we discussed before, except that the metric optimizes better for "average" sales (as opposed to MASE, which optimizes for the median, since it's an absolute error metric). The competition actually used a weighted version of RMSSE which is techincally more robust, but we're going to stick to RMSSE. Here's what RMSSE looks like:
# $RMSSE = \sqrt{\frac{1}{h}\frac{\sum^{n+h}_{t=n+1} (Y_t - \hat{Y}_t)^2}{\frac{1}{n-1}\sum^n_{t=2} (Y_t - Y_{t-1})^2}}$
# where $Y_t$ is the actual future value of sales at date $t$, $\hat{Y}_t$ is your forecast for date $t$, $n$ is the number of dates in our training set, and $h$ is our forecast horizon (28 days, in our case).
# That looks intimidating! But, similarly to MASE, you can break it down into two parts:
# - The numerator: $\frac{1}{h}\sum^{n+h}_{t=n+1} (Y_t - \hat{Y}_t)^2$, which is just the MSE for every prediction in the validation set.
# - The denominator: $\frac{1}{n-1}\sum^n_{t=2} (Y_t - Y_{t-1})^2$, which is just the MSE over the entire training set if your forecast was a naive, one-day-ahead forecast. We refer to this as the "scale" since it's really just a benchmark -- errors less than this are better than the benchmark, and errors greater than this are worse.
# Of course, the "naive, one-day-ahead forecast" part only works if you calculate both the numerator and denominator separately for each `id`. So, the idea here is that you are effectively calculating an RMSSE value for each `id`, and then averaging those to get the final RMSSE.
# Last comment: there are products in the dataset that don't start showing sales for some time. For those products, the denominator is only supposed to be calculated after the first sale in the dataset. I'd recommend just dropping the records for those products until that first sales, which is straightforward to do using `.cumsum()` over `sales` while grouping by `id`.
# QUESTION: filter out products that don't have sales using cumsum
sales_cumul = data.groupby("item_id")["sales"].cumsum()
data = data[sales_cumul > 0]
# Here's how you should implement your RMSSE:
# 1. Create a function called `rmsse` that looks like this:
# `def rmsse(train, val, y_pred):`
# where:
# - `train` is the `pd.DataFrame` representing the training set
# - `val` is the `pd.DataFrame` representing the validation set
# - `y_pred` is either a `pd.Series` or `np.ndarray` that is the output of your model
# 2. Start by calculating the scale (i.e. denominator from above) for each `id` over the training set.
# 3. Then, calculate the MSE for each `id` over the validation set.
# 4. Merge the scale dataframe onto the dataframe that contains your validation MSE values.
# 5. Use the merged dataframe to calculate the RMSSE for each `id`, and finally return the average of all of those RMSSE values.
# Don't worry that you haven't split your data into training and validation sets yet. I gave you a test case below to see if your code is working before you move on. Also, don't be afraid to do this in a simple, looped fashion before refactoring it into more beautiful Pandas code. Take advantage of that test case!
# QUESTION: implement rmsse
def rmsse(train, val, y_pred):
# Calculate the denominator over the training set
df_scaled = (
train.groupby("id")["sales"]
.apply(lambda x: np.sum(np.square(np.diff(x))) / (len(x) - 1))
.reset_index()
)
df_scaled.columns = ["id", "scale"]
# Calculate MSE over the validation set
val["y_pred"] = y_pred
df_mse = (
val.groupby("id")
.apply(lambda x: np.mean(np.square(x["sales"] - x["y_pred"])))
.reset_index()
)
df_mse.columns = ["id", "mse"]
# Merge
df_merged = pd.merge(df_mse, df_scaled, on="id")
# Calculate the RMSSE
df_merged["rmsse"] = np.sqrt(df_merged["mse"] / df_merged["scale"])
# Return the average
return df_merged["rmsse"].mean()
num = lambda tr, val, pred, id="item_id": (val.sales - pred).pow(2).groupby(id).mean()
den = (
lambda tr, val, pred, id="item_id": tr.groupby(id)
.sales.diff()
.pow(2)
.groupby(id)
.mean()
)
rmsse = lambda tr, val, pred, id="item_id": np.sqrt(
num(tr, val, pred, id) / den(tr, val, pred, id)
).mean()
def test_rmsse():
"""
I've modified the dataframes below so they get the same indices, ie
like the arguments I will pass later
"""
test_train = pd.DataFrame(
{
"item_id": ["a", "a", "a", "b", "b", "b", "c", "c", "c"],
"sales": [3, 2, 5, 100, 150, 60, 10, 20, 30],
}
).set_index("item_id")
test_val = pd.DataFrame(
{
"item_id": ["a", "a", "a", "b", "b", "b", "c", "c", "c"],
"sales": [6, 1, 4, 200, 120, 270, 10, 20, 30],
}
).set_index("item_id")
test_y_pred = pd.Series(
[1, 2, 3, 180, 160, 240, 20, 30, 40],
index=["a", "a", "a", "b", "b", "b", "c", "c", "c"],
)
# test_y_pred = pd.Series([1, 2, 3, 180, 160, 240, 20, 30, 40])
rmss = rmsse(test_train, test_val, test_y_pred)
# print(rmss)
assert (
np.abs(rmss - 0.92290404515501) < 1e-6
), f"RMSSE should be 0.9229, we got {rmss}"
print("RMSSE implementation is correct!")
test_rmsse()
# # Fitting models
# From this point on, the project is a bit of a "choose your own adventure." There's a huge range of skill levels out there, and I want to provide you with a path that will meet you where you're at (but test you a little). At a minimum, though, you'll be fitting a LightGBM model.
# 1. If you're a beginner, use [`mlforecast`](https://nixtla.github.io/mlforecast/) (the sister package to `statsforecast`). It helps a lot with both feature engineering and model fitting, so you'll be able to try out a lot of options without getting bogged down in writing complex code. Focus your efforts on trying lots of different features/hyperparameters and seeing how they affect your model!
# If you want to go this route, here are the steps you should take:
# - Familiarize yourself with mlforecast [here](https://nixtla.github.io/mlforecast/)
# - Read the code in the below cell. This is your starting point!
# - Try adding other `date_features`, like the week of year and day of year.
# - Try adding `static_features=['item_id', 'dept_id', 'cat_id']` to `fcst.fit()`
# - Try out other rolling mean/std lengths and at different lags to see if they help. (You can import `rolling_std` from `window_ops.rolling`)
# - Try adding seasonal rolling means using the following code, which implements a 4 week seasonal rolling mean with a season length of 7 days:
# ```
# @njit
# def seasonal_rolling_mean(x):
# return seasonal_rolling_mean(x, season_length=7, window_size=4, min_samples=1)
# ```
# - Try out some difference and lag features.
# - Try adding variables from the other data files, such as price.
# 2. If you feel more comfortable, then I want you to not only try out different features/hyperparameters, but also compare modeling methods! Some things to try:
# - Features
# - Benefits from lag features vs. rolling window features
# - Which rolling window aggregations help
# - Comparing seasonal rolling features to non-seasonal
# - Features aggregated at the department/category level (but make sure to only calculate over the training set!)
# - Modeling
# - Simple 28-day forecast horizon LightGBM model
# - MLForecast's recursive strategy
# - The multi-horizon strategy (i.e. one model predicting 7 days out, a second model predicting 14 days out, etc.)
# - Deep learning models using [`neuralforecast`](https://nixtla.github.io/neuralforecast/) or `darts`
# 3. (Optional) no matter which group you fit into, try adding in calendar and price features from the other data files that I added!
prices = pd.read_parquet(f"{data_path}/prices.parquet")
prices.head()
data
data = pd.merge(
data.reset_index(), prices, on=["store_id", "item_id", "date"], how="left"
)
data
data["dayofyear"] = data["date"].dt.dayofyear
data["dayofweek"] = data["date"].dt.dayofweek
data["month"] = data["date"].dt.month
data = data.set_index(["date", "id"])
# Don't worry about any error outputs here, unless you get the same "Retrying" error as Project 1
from mlforecast import MLForecast
from statsforecast import StatsForecast
from sklearn.preprocessing import OrdinalEncoder
from numba import njit
from window_ops.rolling import rolling_mean, rolling_std
import lightgbm as lgb
# split into training and validation sets and conform the column names to what MLForecast expects
val = (
data.reset_index()
.groupby("id")
.tail(28)
.rename(
columns={
"date": "ds",
"id": "unique_id",
"sales": "y",
}
)
)
train = (
data.reset_index()
.drop(val.index)
.rename(
columns={
"date": "ds",
"id": "unique_id",
"sales": "y",
}
)
)
# label encode categorical features
cat_feats = ["unique_id", "item_id", "dept_id", "cat_id"]
enc_cat_feats = [f"{feat}_enc" for feat in cat_feats]
encoder = OrdinalEncoder()
train[enc_cat_feats] = encoder.fit_transform(train[cat_feats])
val[enc_cat_feats] = encoder.transform(val[cat_feats])
reference_cols = ["unique_id", "ds", "y", "sell_price"]
# add features to this list if you want to use them
features = reference_cols + enc_cat_feats
train = train[features]
val = val[features]
@njit
def rolling_mean_28(x):
return rolling_mean(x, window_size=28)
@njit
def rolling_mean_14(x):
return rolling_mean(x, window_size=14)
@njit
def rolling_mean_7(x):
return rolling_mean(x, window_size=7)
@njit
def rolling_std_7(x):
return rolling_std(x, window_size=7)
@njit
def rolling_std_14(x):
return rolling_std(x, window_size=14)
@njit
def rolling_std_28(x):
return rolling_std(x, window_size=28)
# feel free to tweak these parameters!
model_params = {
"verbose": -1,
"num_leaves": 256,
"n_estimators": 50,
"objective": "tweedie",
"tweedie_variance_power": 1.1,
}
models = [
lgb.LGBMRegressor(**model_params),
]
fcst = MLForecast(
models=models,
freq="D",
# dictionary reads like this:
# {number of days to lag the feature: [list of functions to apply to the lagged data]}
lag_transforms={
7: [rolling_mean_28, rolling_mean_14, rolling_mean_7],
7: [rolling_std_28, rolling_std_14, rolling_std_7],
},
date_features=["dayofweek", "dayofyear", "month"],
)
# don't worry about nul value warnings. LightGBM and XGBoost can handle it!
fcst.fit(
train,
id_col="unique_id",
time_col="ds",
target_col="y",
static_features=enc_cat_feats,
dropna=False,
)
predictions = fcst.predict(28, dynamic_dfs=[val[["ds", "unique_id", "sell_price"]]])
# plot the last 45 days of the training set, the validation set, and the predictions
plot_data = pd.concat(
[
train.groupby("unique_id").tail(45)[["unique_id", "ds", "y"]],
val[["unique_id", "ds", "y"]],
predictions,
]
)
# for some reason, MLForecast doesn't have this awesome plotting method!
StatsForecast.plot(plot_data)
# Original
StatsForecast.plot(plot_data)
rmsse(
train.rename({"y": "sales"}, axis=1).set_index(["unique_id", "ds"]),
val.rename({"y": "sales"}, axis=1).set_index(
["unique_id", "ds"]
), # we need those indices so subtraction matches rows using indices
predictions.set_index(["unique_id", "ds"])["LGBMRegressor"],
id="unique_id",
)
|
# ##### 데이터셋
# - CementComponent: 콘크리트에 혼합된 시멘트 양
# - BlastFurnaceSlag: 콘크리트에 혼합된 고로슬래그 양
# - FlyAshComponent: 콘크리트에 혼합된 플라이애쉬 양
# - WaterComponent: 콘크리트에 혼합된 물의 양
# - SuperplasticizerComponent: 콘크리트에 혼합된 고성능 플라스티서 양
# - CoarseAggregateComponent: 콘크리트에 혼합된 거친 골재 양
# - FineAggregateComponent: 콘크리트에 혼합된 미세 골재 양
# - AgeInDays: 콘크리트가 건조한 일 수
# - Strength: 콘크리트의 최종 강도(Target)
# #### 위 대회는 콘크리트 강도 예측 대회입니다.
# - 기존 대회였던 Concrete Strength Prediction 의 데이터셋을 딥러닝 모델을 거쳐 확장된 데이터셋이 제공됩니다. 기존 데이터셋 분포와는 큰 차이가 없으며 원한다면 자유롭게 기존 데이터셋과 병행해서 사용하는 것을 허락합니다.
import os
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from matplotlib.ticker import MaxNLocator
from sklearn.model_selection import KFold
from sklearn.pipeline import make_pipeline, Pipeline
from sklearn.compose import ColumnTransformer
from sklearn.preprocessing import StandardScaler
from sklearn.ensemble import RandomForestRegressor, VotingRegressor
from sklearn.linear_model import Ridge
import sklearn.gaussian_process.kernels as kernels
from sklearn.gaussian_process import GaussianProcessRegressor
from sklearn.ensemble import GradientBoostingRegressor
from sklearn.metrics import mean_squared_error
from sklearn.inspection import PartialDependenceDisplay
from sklearn.manifold import TSNE
from sklearn.decomposition import PCA
from fasteda import fast_eda
np.set_printoptions(linewidth=150, edgeitems=5)
result_list = [] # logging
# ##### Numpy set Printoptions Document
# - https://numpy.org/doc/stable/reference/generated/numpy.set_printoptions.html
for width, edge, config in zip([75, 150], [3, 5], ["default", "customized"]):
np.set_printoptions(linewidth=width, edgeitems=edge)
print(
f"[{config}] width: {width} edgeitem: {edge}\n {np.random.random(10000)}",
end="\n\n",
)
# - 중복된 데이터를 어떻게 다룰지에 대한 결론을 내리기 위해 분석을 수행해야 합니다(중복 데이터 삭제, 일부 샘플 가중치 설정, GroupKFold 사용 등). 그러나 현재는 중복 데이터 분석을 건너뛰고 "duplicate saga"를 참조하도록 되어 있습니다. 이는 중복 데이터가 있을 경우 발생할 수 있는 문제를 경험한 사례나 이야기를 의미할 수 있습니다.
original = pd.read_csv(
"/kaggle/input/predict-concrete-strength/ConcreteStrengthData.csv"
)
file_path = "/kaggle/input/playground-series-s3e9"
train = pd.read_csv(os.path.join(file_path, "train.csv"))
test = pd.read_csv(os.path.join(file_path, "test.csv"))
submission = pd.read_csv(os.path.join(file_path, "sample_submission.csv"))
# 경로 수정 필요
# train = pd.read_csv('C:/BNACode/Python Code/Regression with a Tabular Concrete Strength Dataset/train.csv')
# test = pd.read_csv('C:/BNACode/Python Code/Regression with a Tabular Concrete Strength Dataset/test.csv')
# submission = pd.read_csv('C:/BNACode/Python Code/Regression with a Tabular Concrete Strength Dataset/sample_submission.csv')
# original = pd.read_csv('C:/BNACode/Python Code/Regression with a Tabular Concrete Strength Dataset/ConcreteStrengthData.csv') # 원본 데이터
# ### EDA
train.head()
# Train Data 중복값
col_idxs = []
for col in train.columns:
col_list = []
col_list.append(col)
if col != "id":
col_list.append("id")
train_dup = train.drop(columns=col_list)
dup_idx = list(train_dup.loc[train_dup.duplicated(keep="first")].index)
print(f"# of duplicates: {len(dup_idx)} --> columns dropped: {col_list}")
if col == "Strength":
col_idxs.append(dup_idx)
ax = train.loc[sum(col_idxs, [])]["Strength"].hist(bins=100)
train.loc[sum(col_idxs, [])].sort_values("Strength").query(
"CementComponent == 540.0 and BlastFurnaceSlag == 0.0 and FlyAshComponent == 0.0 and AgeInDays == 90"
).T
ax = train["Strength"].hist(bins=100)
# Test Data 중복값
ool_idxs = []
for col in test.columns:
col_list = []
col_list.append(col)
if col != "id":
col_list.append("id")
test_dup = test.drop(columns=col_list)
dup_idx = list(test_dup.loc[test_dup.duplicated(keep="first")].index)
print(f"# of duplicates: {len(dup_idx)} --> columns dropped: {col_list}")
if col == "id":
ool_idxs.append(dup_idx)
test.loc[sum(ool_idxs, [])]
# Origin 데이터 중복값
ool_idxs = []
for col in original.columns:
col_list = []
col_list.append(col)
original_dup = original.drop(columns=col_list)
dup_idx = list(original_dup.loc[original_dup.duplicated(keep="first")].index)
print(f"# of duplicates: {len(dup_idx)} --> columns dropped: {col_list}")
if col == "Strength":
ool_idxs.append(dup_idx)
original.loc[sum(ool_idxs, [])]
train["is_train"] = 1
original["is_train"] = 1
test["is_train"] = 0
train_test = pd.concat([train, test], ignore_index=True) # train, test 데이터 합친거
train_test_orig = pd.concat(
[train.drop(columns="id"), test.drop(columns="id"), original], ignore_index=True
)
train_test_orig["CementComponent"] = train_test_orig["CementComponent"].fillna(
train_test_orig.iloc[:, 10]
)
train_test_orig = train_test_orig.iloc[:, :-1]
train_test
fast_eda(train_test, target="is_train")
# ### T-SNE
# - https://velog.io/@swan9405/%EB%A8%B8%EC%8B%A0%EB%9F%AC%EB%8B%9D-T-SNE-T-distributed-Stochastic-Neighbor-Embedding
# - https://amazelimi.tistory.com/13
train_orig = pd.concat([train.drop(columns="id"), original], ignore_index=True)
train_orig["cement"] = train_orig.iloc[:, 10] # CementComponent
train_orig["CementComponent"] = train_orig["CementComponent"].fillna(
train_orig["cement"]
) # CementComponent의 결측치 제거한 값
train_orig = train_orig.iloc[:, :-2]
ss = StandardScaler()
drop_cols = ["is_train", "Strength"]
X = train.drop(drop_cols, axis=1)
X_orig = train_orig.drop(drop_cols, axis=1)
X_train_ss = pd.DataFrame(ss.fit_transform(X))
X_orig_ss = pd.DataFrame(ss.fit_transform(X_orig))
y = train["Strength"].values
train_tsne = train[test.drop(columns="id").columns].values
test_tsne = test[test.drop(columns="id").columns].values
tsne = TSNE(n_components=2)
train_2D = tsne.fit_transform(train_tsne)
plt.style.use("dark_background")
plt.figure(figsize=(12, 8))
sns.scatterplot(
x=train_2D[:, 0],
y=train_2D[:, 1],
s=4,
hue=y,
linewidth=0,
palette=sns.color_palette("rainbow", as_cmap=True),
)
plt.grid(False)
plt.title("TSNE plot | train data")
plt.show()
# ### PCA n_components = 2 on train Data
pca = PCA(2)
X_train_ss2 = pca.fit_transform(X_train_ss)
pca2 = pd.DataFrame(
{
"PCA1": X_train_ss2[:, 0],
"PCA2": X_train_ss2[:, 1],
"Strength": train["Strength"],
}
)
plt.style.use("dark_background")
plt.figure(figsize=(12, 8))
sns.scatterplot(
data=pca2,
x="PCA1",
y="PCA2",
s=6,
hue="Strength",
linewidth=0,
palette=sns.color_palette("rainbow", as_cmap=True),
)
plt.grid(False)
plt.title("Data scaled with StandardScaler()")
plt.show()
# ### 분석
original = pd.read_csv(
"/kaggle/input/predict-concrete-strength/ConcreteStrengthData.csv"
)
file_path = "/kaggle/input/playground-series-s3e9"
train = pd.read_csv(os.path.join(file_path, "train.csv"))
test = pd.read_csv(os.path.join(file_path, "test.csv"))
submission = pd.read_csv(os.path.join(file_path, "sample_submission.csv"))
# train = pd.read_csv('C:/BNACode/Python Code/Regression with a Tabular Concrete Strength Dataset/train.csv')
# test = pd.read_csv('C:/BNACode/Python Code/Regression with a Tabular Concrete Strength Dataset/test.csv')
# submission = pd.read_csv('C:/BNACode/Python Code/Regression with a Tabular Concrete Strength Dataset/sample_submission.csv')
# original = pd.read_csv('C:/BNACode/Python Code/Regression with a Tabular Concrete Strength Dataset/ConcreteStrengthData.csv') # 원본 데이터
target = "Strength"
original_features = list(test.columns)
print(
f"train set 결측치: {train.isna().sum().sum()}, test set 결측치: {test.isna().sum().sum()}"
)
print(
f"train set 중복값: {train[test.columns].duplicated().sum()}, test set 중복값: {test.duplicated().sum()}"
)
# - 위에서 fasteda 라이브러리를 통해 확인함
# - 히스토그램에서 보면 모든 샘플은 시멘트, 물, 거친 골재 및 고급 골재를 포함한다.
# - 일부 샘플에서만 고로슬래그, 플라이애쉬 또는 슈퍼플라스틱 라이저가 포함되어있음
# - train, test 셋의 분포는 유사해 보임..
# - train set에서 blast furnance, fly ash, superplasticizer 에서 0값을 가짐
# - AgeInDays 컬럼에서 특이점이 보이는데, 건조 기간이 아래와 같이 특정 기간에 몰려있다는 점이다.
train[
(train.BlastFurnaceSlag == 0)
& (train.FlyAshComponent == 0)
& (train.SuperplasticizerComponent == 0)
].shape
print("AgeInDays 분포 확인")
(pd.concat([train, test], axis=0).AgeInDays.value_counts().sort_index()).plot(
kind="bar"
)
# - FlyAshComponent와 Superplasticizier와의 상관성이 높음
# - 하지만 물과의 상관성은 낮은것으로 보아 시멘트에 물은 적게 들어 가는것으로 보임
corr = train.corr()
plt.figure(figsize=(4, 4))
sns.heatmap(
corr,
linewidths=0.1,
fmt=".1f",
annot=True,
annot_kws={"size": 8},
cmap="PiYG",
center=0,
)
# plt.show()
corr2 = test.corr()
plt.figure(figsize=(4, 4))
sns.heatmap(
corr2,
linewidths=0.1,
fmt=".1f",
annot=True,
annot_kws={"size": 8},
cmap="PiYG",
center=0,
)
# ### 분석 모델 선택
# #### Linear Baseline
# - Ridge regression 모델을 baseline 모델로 사용하였다.
# - std scaler와 모델 순으로 파이프라인 적용
# - Pipeline 개념: https://zephyrus1111.tistory.com/254
#
def score_model(model, features_used, label=None):
"Cross-validate a model with feature selection"
score_list = []
oof = np.zeros_like(train[target])
kf = KFold()
for fold, (idx_tr, idx_va) in enumerate(kf.split(train)):
X_tr = train.iloc[idx_tr][features_used]
X_va = train.iloc[idx_va][features_used]
y_tr = train.iloc[idx_tr][target]
y_va = train.iloc[idx_va][target]
model.fit(X_tr, y_tr)
trmse = mean_squared_error(y_tr, model.predict(X_tr), squared=False)
y_va_pred = model.predict(X_va)
rmse = mean_squared_error(y_va, y_va_pred, squared=False)
if (
type(model) == Pipeline
and type(model.steps[-1][1]) == GaussianProcessRegressor
):
print("Kernel:", model.steps[-1][1].kernel_)
print(f"Fold {fold}: trmse = {trmse:.3f} rmse = {rmse:.3f}")
oof[idx_va] = y_va_pred
score_list.append(rmse)
rmse = sum(score_list) / len(score_list)
print(f"rmse 평균: {rmse:.3f}")
if label is not None:
global result_list
result_list.append((label, rmse, oof))
score_model(
model=make_pipeline(StandardScaler(), Ridge(70)), features_used=original_features
)
# Blast Furnace Slag, Fly Ash, 그리고 Superplasticizer 총 3가지 선택적 구성 요소의 존재를 나타내는 지표를 추가하면 RMSE가 조금 향상됨
for df in [train, test]:
df["hasBlastFurnaceSlag"] = df.BlastFurnaceSlag != 0
df["hasFlyAshComponent"] = df.FlyAshComponent != 0
df["hasSuperplasticizerComponent"] = df.SuperplasticizerComponent != 0
score_model(model=make_pipeline(StandardScaler(), Ridge(70)), features_used=df.columns)
# ### Random forests and partial dependency plots
# - 선형 Ridge모델 보다 랜포의 성능이 더 좋았고 과적합을 방지하기 위해 min_samples_leaf 값을 30으로 설정함
score_model(
model=RandomForestRegressor(n_estimators=300, min_samples_leaf=30, random_state=1),
features_used=original_features,
label="Random Forest",
)
# #### PDP는 비선형적인 종속관계에 대한 중요한 통찰력을 제공함
# - PDP plot에 대한 설명: https://westshine-data-analysis.tistory.com/m/134
# - 콘크리트는 최대 강도를 얻기 위해 1~2개월의 시간이 필요합니다. 더 오래 놔둬도 강도가 더 좋아지지 않습니다.
# - 물을 더 추가하면 강도가 감소하지만, 195 이상의 물은 강도에 영향을 미치지 않습니다.
# - 파란색 개별 조건부 예상치들은 평행하지 않습니다. 이는 중요한 feature 상호 작용의 존재를 나타냅니다.
#
model = RandomForestRegressor(n_estimators=300, min_samples_leaf=30, random_state=1)
model.fit(train[original_features], train[target])
features_for_pdp = original_features
fig, axs = plt.subplots(2, 5, figsize=(14, 10))
plt.suptitle("Partial Dependence", y=1.0)
PartialDependenceDisplay.from_estimator(
model,
train[original_features],
features_for_pdp,
pd_line_kw={"color": "red"},
ice_lines_kw={"color": "blue"},
kind="both",
ax=axs.ravel()[: len(features_for_pdp)],
)
plt.tight_layout(h_pad=0.3, w_pad=0.5)
plt.show()
# 콘크리트가 건조되어 있는 기간을 40일로, 물의 양을 195로 제한한 후에 linear model의 RMSE가 크게 개선된 된다.
for df in [train, test]:
df["clippedAge"] = df.AgeInDays.clip(None, 40)
score_model(
model=make_pipeline(StandardScaler(), Ridge(70)),
features_used=original_features
+ [
"hasBlastFurnaceSlag",
"hasFlyAshComponent",
"hasSuperplasticizerComponent",
"clippedAge",
],
)
for df in [train, test]:
df["clippedwater"] = df.WaterComponent.clip(195, None)
score_model(
model=make_pipeline(StandardScaler(), Ridge(30)), features_used=test.columns
)
# Flyash를 지우면 ridge 모델 성능이 올라감
ridge_features = [
"CementComponent",
"BlastFurnaceSlag",
"WaterComponent",
"SuperplasticizerComponent",
"CoarseAggregateComponent",
"FineAggregateComponent",
"AgeInDays",
"hasBlastFurnaceSlag",
"hasSuperplasticizerComponent",
"clippedAge",
"clippedwater",
]
score_model(
model=make_pipeline(StandardScaler(), Ridge(30)),
features_used=ridge_features,
label="Ridge",
)
# ### Gaussian process regression
# - 가우시안 회귀: https://sonsnotation.blogspot.com/2020/11/11-2-gaussian-progress-regression.html
# - 특정 변수들이 모델 예측에 중요하게 작용하는 것으로 나타났고 해당 변수들을 활용해 가우시안 회귀를 진행하면 ridge 모델보다 훨씬 좋은 성능을 나타냄
# - 이해하기 가장 어려운 부분 이므로 반드시 공부가 필요함
gp_features = ["CementComponent", "SuperplasticizerComponent", "AgeInDays"]
kernel = kernels.RBF(
length_scale=[1] * (len(gp_features) - 1) + [0.07], length_scale_bounds=(0.01, 10)
) + kernels.WhiteKernel(0.55)
score_model(
model=make_pipeline(
StandardScaler(),
GaussianProcessRegressor(kernel=kernel, normalize_y=True, random_state=1),
),
features_used=gp_features,
label="Gaussian Process",
)
# #### Gradient boosting
# - 그래디안 부스팅은 랜포 보다 더 나은 성능을 보여줌
gbr_params = {
"n_estimators": 550,
"max_depth": 4,
"learning_rate": 0.01,
"min_samples_leaf": 30,
"max_features": 3,
}
score_model(
model=GradientBoostingRegressor(**gbr_params, random_state=1),
features_used=original_features,
label="GradientBoostingRegressor",
)
# ColumnTransformer의 사용법:https://scribblinganything.tistory.com/661
ensemble_model = VotingRegressor(
[
(
"gb",
make_pipeline(
ColumnTransformer([("pt", "passthrough", original_features)]),
GradientBoostingRegressor(**gbr_params, random_state=1),
),
),
(
"rf",
make_pipeline(
ColumnTransformer([("pt", "passthrough", original_features)]),
RandomForestRegressor(
n_estimators=300, min_samples_leaf=30, random_state=1
),
),
),
(
"gp",
make_pipeline(
ColumnTransformer([("pt", "passthrough", gp_features)]),
StandardScaler(),
GaussianProcessRegressor(
kernel=kernel, normalize_y=True, random_state=1
),
),
),
(
"ridge",
make_pipeline(
ColumnTransformer([("pt", "passthrough", ridge_features)]),
StandardScaler(),
Ridge(30),
),
),
],
weights=[0.4, 0.3, 0.2, 0.1],
)
score_model(
model=ensemble_model,
features_used=test.columns,
label="GradientBoostingRegressor + RF + GP + Ridge",
)
result_df = pd.DataFrame(result_list, columns=["label", "rmse", "oof"])
result_df.drop_duplicates(subset="label", keep="last", inplace=True)
result_df.sort_values("rmse", inplace=True)
plt.figure(figsize=(6, len(result_df) * 0.3))
plt.title("Cross-validation scores")
plt.barh(np.arange(len(result_df)), result_df.rmse, color="lightgreen")
plt.gca().invert_yaxis()
plt.yticks(np.arange(len(result_df)), result_df.label)
plt.xticks(np.linspace(12, 12.3, 4))
plt.xlabel("RMSE")
plt.xlim(12, 12.3)
# plt.savefig('cv-scores.png', bbox_inches='tight')
plt.show()
|
# ## All Space Missions from 1957 | Data Analysis and Visualization
# Group Members:
# ➡️ Alex Tamboli (202011071)
# ➡️ Ashish Kumar Gupta (202011013)
# ➡️ Chinmay Bhalodia (202011016)
# ➡️ Nishesh Jain (202011050)
# ➡️ Prashant Kumar (202011058)
# ## Importing necessary libraries
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
plt.style.use("seaborn-dark")
# ## Analysis of dataset
# Let's know what the data is and how the data is.
# loading the dataset
df = pd.read_csv("/kaggle/input/all-space-missions-from-1957/Space_Corrected.csv")
df.head()
# `Unnamed: 0.1` and `Unnamed: 0` are of no use. We will drop them.
df = df.drop(["Unnamed: 0", "Unnamed: 0.1"], axis=1)
df.head()
# ## Visualization #1: Everything related to the Companies.
# ### 1) Number of Launches by Company
# First, we find the total number of unique companies.
number_of_companies = df["Company Name"].unique()
len(number_of_companies)
# Out of total 56 companies, we will see number of launches by top 10 companies.
company_count = df["Company Name"].value_counts()
top_companies = company_count[:10]
plt.figure(figsize=(20, 4))
plt.scatter(
x=top_companies.index,
y=[1] * len(top_companies),
s=top_companies.values * 10,
c="blue",
alpha=0.5,
)
for i, count in enumerate(top_companies.values):
plt.text(
top_companies.index[i],
1,
str(count),
ha="center",
va="center",
fontsize=12,
color="white",
)
plt.xticks(fontsize=12)
plt.xlim(-0.75)
plt.yticks([])
plt.title("Top 10 Companies by number of Launches.", fontsize=15)
plt.show()
# ### 2) Number of Launches by Company per Year
# We need to modify the dataframe to extract the year of launch from the date.
df["Datum"] = pd.to_datetime(df["Datum"], utc=True)
df["Year"] = df["Datum"].dt.year
df.head()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# 
# # Playground Series - Season 3, Episode 11
# ## Tabular Regression with a Media Campaign Cost Dataset
# ### Welcome to the 2023 edition of Kaggle's Playground Series!
# Thank you to everyone who participated in and contributed to Season 3 Playground Series so far!
# With the same goal to give the Kaggle community a variety of fairly light-weight challenges that can be used to learn and sharpen skills in different aspects of machine learning and data science, we will continue launching the Tabular Tuesday in March every Tuesday 00:00 UTC, with each competition running for 2 weeks. Again, these will be fairly light-weight datasets that are synthetically generated from real-world data, and will provide an opportunity to quickly iterate through various model and feature engineering ideas, create visualizations, etc..
# ### Synthetically-Generated Datasets
# Using synthetic data for Playground competitions allows us to strike a balance between having real-world data (with named features) and ensuring test labels are not publicly available. This allows us to host competitions with more interesting datasets than in the past. While there are still challenges with synthetic data generation, the state-of-the-art is much better now than when we started the Tabular Playground Series two years ago, and that goal is to produce datasets that have far fewer artifacts. Please feel free to give us feedback on the datasets for the different competitions so that we can continue to improve!
# #### Synthetically-Generated Datasets
# Root Mean Squared Log Error (RMLSE)
# Submissions are scored on the root mean squared log error (RMSLE) (the sklearn mean_squared_log_error with squared=False).
# #### Synthetically-Generated Datasets
# Submission File
# For each id in the test set, you must predict the value for the target cost. The file should contain a header and have the following format:
# ```Python
# id,cost
# 360336,99.615
# 360337,87.203
# 360338,101.111
# etc.
# ```
# **Start Date** - March 20, 2023
# **Entry Deadline** - Same as the Final Submission Deadline
# **Team Merger Deadline** - Same as the Final Submission Deadline
# **Final Submission Deadline** - April 3, 2023
# All deadlines are at 11:59 PM UTC on the corresponding day unless otherwise noted. The competition organizers reserve the right to update the contest timeline if they deem it necessary.
# 1 - import the necessary libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.linear_model import LinearRegression
from sklearn.preprocessing import StandardScaler
from sklearn.ensemble import RandomForestRegressor
from sklearn.metrics import (
mean_absolute_error,
mean_squared_error,
mean_squared_log_error,
r2_score,
)
from catboost import CatBoostRegressor
from xgboost import XGBRegressor
from lightgbm import LGBMRegressor
from mlxtend.regressor import StackingRegressor
# 2 - open training and test data
train = pd.read_csv("/kaggle/input/playground-series-s3e11/train.csv")
test = pd.read_csv("/kaggle/input/playground-series-s3e11/test.csv")
train.head(3)
# 3 - display descriptive statistics of training data
train.describe()
# 4 - look at the number of gaps in the training set
train.isna().sum() # as you can see the number of passes is 0
# 5 - look at the number of gaps in the test set
test.isna().sum() # as you can see the number of passes is 0
# 6 - look at the data type of the training set
train.info() # all data is of type float64
# 6a - Create boxplots of our features
sns.set(rc={"figure.figsize": (40, 6)})
sns.boxplot(train)
plt.ylim([0, 151])
# 7 - look at the correlation between features
correlation = train.corr()
correlation.style.background_gradient(cmap="PuRd")
# ### Features salad_bar and prepared_food have a very high correlation (about 1). To avoid multicollinearity, remove the prepared_food feature
# 8 - remove the feature 'prepared_food'
train.drop(columns="prepared_food", axis=1, inplace=True)
test.drop(columns="prepared_food", axis=1, inplace=True)
# 9 - checking the correlation after removing the feature 'prepared_food'
correlation = train.corr()
correlation.style.background_gradient(cmap="Blues")
# ### Features with a high correlation (more than 0.62) are no longer left
# 10 - look at the distribution of the dependent variable using kernel density estimation
sns.kdeplot(train.cost, fill=True, color="green")
# 11 - look at the distribution of the dependent variable using a histogram of the distribution
train["cost"].hist(bins=50, color="red")
# 12 - use the Shapiro-Wilk test to check the distribution of 'cost' for normality (ignore warnings)
import warnings
import scipy.stats as stats
warnings.filterwarnings("ignore")
result = stats.shapiro(train["cost"])
print(f"Shapiro-Wilk test statistic: {result[0]:.4f}")
print(f"p-value: {result[1]}")
# ### If p-value > 0.05, then we can accept the hypothesis of normal distribution. Otherwise, the hypothesis is rejected and the feature is not normally distributed.
# ### In our case, the distribution is not normal, because p-value is 0.
# 13 - To check if the distribution is normal, you can also use the Anderson-Darling test,
# which is more powerful than the Shapiro-Wilk test.
result = stats.anderson(train["cost"], dist="norm")
print(f"Anderson-Darling test statistic: {result[0]:.4f}")
print(f"critical values: {result[1]}")
print(f"Significance levels: {result[2]}")
# ### If the value of the test statistic is less than the critical value, then the hypothesis of normal distribution can be accepted. If the value of the statistic is greater, then the hypothesis is rejected, and the feature is not normally distributed.
# ### In our case, the distribution is not normal
# 14 - split the training dataset into X and y
X = train.drop(columns=["cost"], axis=1)
y = train.cost
# 15 - Standardizing data
sc = StandardScaler()
train_std = sc.fit_transform(X) # first we train
test_std = sc.transform(test)
# 16 - train a Linear regression model and look at the metrics
lin_reg = LinearRegression().fit(train_std, y)
predict = lin_reg.predict(train_std)
print(f"mean_absolute_error: {mean_absolute_error(y, predict):.4f}")
print(f"mean_squared_error: {mean_squared_error(y, predict):.4f}")
print(f"mean_squared_log_error: {mean_squared_log_error(y, predict):.4f}")
print(f"r2_score: {r2_score(y, predict):.4f}")
predict
# 17 - train a XGB regression model and look at the metrics
xgb_reg = XGBRegressor().fit(train_std, y)
predict = xgb_reg.predict(train_std)
print(f"mean_absolute_error: {mean_absolute_error(y, predict):.4f}")
print(f"mean_squared_error: {mean_squared_error(y, predict):.4f}")
print(f"mean_squared_log_error: {mean_squared_log_error(y, predict):.4f}")
print(f"r2_score: {r2_score(y, predict):.4f}")
predict
# 18 - initialize the models and train on the training set
model1 = LinearRegression()
model2 = CatBoostRegressor(random_state=42)
model3 = XGBRegressor(random_state=42)
model4 = LGBMRegressor(random_state=42)
model1.fit(train_std, y)
model2.fit(train_std, y)
model3.fit(train_std, y)
model4.fit(train_std, y)
# create an ensemble of models and predict on a test set
ensemble = StackingRegressor(
regressors=[model1, model2, model3, model4],
meta_regressor=XGBRegressor(n_estimators=100, random_state=42),
)
ensemble.fit(train_std, y)
# evaluate the accuracy of predictions on a test set
predict = ensemble.predict(train_std)
print(f"mean_absolute_error: {mean_absolute_error(y, predict):.4f}")
print(f"mean_squared_error: {mean_squared_error(y, predict):.4f}")
print(f"mean_squared_log_error: {mean_squared_log_error(y, predict):.4f}")
print(f"r2_score: {r2_score(y, predict):.4f}")
predict
# 18a - train random forest regression model and look at the metrics
reg_randfor = RandomForestRegressor(max_depth=5, random_state=42)
reg_randfor.fit(train_std, y)
predict = reg_randfor.predict(train_std)
print(f"mean_absolute_error: {mean_absolute_error(y, predict):.4f}")
print(f"mean_squared_error: {mean_squared_error(y, predict):.4f}")
print(f"mean_squared_log_error: {mean_squared_log_error(y, predict):.4f}")
print(f"r2_score: {r2_score(y, predict):.4f}")
predict
# 19 - train a Catboost regression model and look at the metrics
cat = CatBoostRegressor()
cat.fit(train_std, y)
predict = cat.predict(train_std)
print(f"mean_absolute_error: {mean_absolute_error(y, predict):.4f}")
print(f"mean_squared_error: {mean_squared_error(y, predict):.4f}")
print(f"mean_squared_log_error: {mean_squared_log_error(y, predict):.4f}")
print(f"r2_score: {r2_score(y, predict):.4f}")
predict
# 20 - the best submit result was shown by the catboost model. Let's train a test sample on it
cat = CatBoostRegressor()
cat.fit(train_std, y)
cat_predict = cat.predict(test_std)
cat_predict
# 21 - making submission's dataset
submission = pd.DataFrame()
submission["id"] = test.id
submission["cost"] = cat_predict
submission
# 22 - making submission's csv file
submission[["id", "cost"]].to_csv("CatReg submission PSs3e11.csv", index=False)
|
#
# * [1. IMPORTING LIBRARIES](#1)
#
# * [2. LOADING DATASET 📚](#2)
#
# * [3. Statistical information and general information about the data 🔍](#3)
# * [4. EDA 📊](#4)
# * [5. Data preprocessing 🔧](#5)
# # Importing libraries
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
plt.style.use(
"https://github.com/dhaitz/matplotlib-stylesheets/raw/master/pitayasmoothie-dark.mplstyle"
)
import warnings
warnings.filterwarnings("ignore")
import plotly.express as px
import plotly.graph_objects as go
from plotly.subplots import make_subplots
import seaborn as sns
#
# # Reading the data 📚
#
data = pd.read_csv(r"/kaggle/input/suicide-rates-overview-1985-to-2016/master.csv")
data.head() # show the first 5 rows
data.tail() # show the last 5 rows
#
# # Statistical information and general information about the data 🔍
#
data.shape
# **Observations 📋**
# 🔘 The data consists of **27820** and **12** columns (features)
data.describe().T # Statistical information for numirc data
data.describe(include="O").T # tatistical information for object data
data.info()
# **Observations 📋**
# 🔘 HDI for year has a lot of null values
data.columns
#
# # EDA 📊
#
data.nunique()
# 🔘 **Exploratory data analysis (EDA)** : is used by data scientists to analyze and investigate data sets and summarize their main characteristics, often employing data visualization methods.
#
sns.pairplot(data=data, hue="sex")
# **The ratio of males to females**
data.sex.value_counts()
sns.countplot(data=data, x="sex")
# **Observations 📋**
#
# 🔘 The ratio of the number of males and females in the data is equal
# 🔘 The column **Sex** is balanced
# **What are the most suicide age groups ?**
data.age.value_counts()
data.age.value_counts().plot.pie(
autopct="%1.1f%%", figsize=(10, 7), title="Distribution by age groups"
)
# **Observations 📋**
#
# 🔘 The number of suicide groups is almost equal in all age groups in the data
# **Distribution by gdp per capita**
px.violin(
data,
x="sex",
y="gdp_per_capita ($)",
color="sex",
box=True,
points="all",
color_discrete_sequence=["#71AEC2 ", "#D58989"],
title="Sex distribution by gdp per capita ",
template="plotly_dark",
)
# **Observations 📋**
#
# 🔘 The column **gdb per captia** is balanced
# **Distribution of country by suicides_no**
# **Distribution of country by population**
px.histogram(
data,
x="country",
y="population",
color="country",
height=700,
width=1500,
template="plotly_dark",
marginal="rug",
title="Distribution of country by population",
hover_data=["sex", "year"],
)
# **Distribution of Generation by gdp per capita**
data.generation.value_counts()
plt.figure(figsize=(20, 10))
sns.lineplot(data=data, x="generation", y="gdp_per_capita ($)")
plt.title("Distribution of Generation by gdp per capita")
# **Distribution of gdp per capita by suicides/100k pop**
px.scatter(
data,
x="gdp_per_capita ($)",
y="suicides/100k pop",
color="country",
template="plotly_dark",
title="Distribution of gdp per capita by suicides/100k pop",
hover_data=["year"],
)
# **Distribution of Countries by population**
px.choropleth(
data,
locations="country",
locationmode="country names",
color="population",
animation_frame="year",
template="plotly_dark",
title="Distribution of Countries by population",
)
px.treemap(
data,
path=["country"],
values="population",
title="Distribution of Countries by population",
template="plotly_dark",
)
px.choropleth(
data,
locations="country",
locationmode="country names",
color="suicides_no",
animation_frame="year",
template="plotly_dark",
title="Distribution of Countries by suicides_no",
)
px.treemap(
data,
path=["country"],
values="suicides_no",
title="Distribution of Countries by suicides_no",
template="plotly_dark",
)
px.choropleth(
data,
locations="country",
locationmode="country names",
color="gdp_per_capita ($)",
animation_frame="year",
template="plotly_dark",
title="Distribution of Countries by gdp per capita",
)
px.treemap(
data,
path=["country"],
values="gdp_per_capita ($)",
title="Distribution of Countries by gdp per capita",
template="plotly_dark",
)
#
# # Data preprocessing 🔧
# **Data cleaning**
data.isna().sum()
sns.heatmap(data.isna())
# **Observations 📋**
# 🔘 The column HDI for year has a lot of null values
# **Decisions 📋**
# 🔘 so it is better to be delted
data.drop("HDI for year", axis=1, inplace=True)
data.isna().sum()
data.head(2)
# **Observations 📋**
# 🔘 The column **country-year** is a combination of two coulmns ** country** and **year**
# **Decisions 📋**
# 🔘 so it is better to be delted
data.drop("country-year", axis=1, inplace=True)
data.head(2)
data[" gdp_for_year ($) "] = data[" gdp_for_year ($) "].apply(
lambda x: x.replace(",", "")
)
data[" gdp_for_year ($) "] = data[" gdp_for_year ($) "].astype(int)
px.choropleth(
data,
locations="country",
locationmode="country names",
color=" gdp_for_year ($) ",
animation_frame="year",
template="plotly_dark",
title="Distribution of Countries by gdp for year",
)
px.treemap(
data,
path=["country"],
values=" gdp_for_year ($) ",
title="Distribution of Countries by gdp for year",
template="plotly_dark",
)
data.head()
data["suicides_no"] = pd.cut(data["suicides_no"], 6, labels=[1, 2, 3, 4, 5, 6])
data["population"] = pd.cut(data["population"], 6, labels=[1, 2, 3, 4, 5, 6])
data["suicides/100k pop"] = pd.cut(
data["suicides/100k pop"], 6, labels=[1, 2, 3, 4, 5, 6]
)
data["gdp_per_capita ($)"] = pd.cut(
data["gdp_per_capita ($)"], 6, labels=[1, 2, 3, 4, 5, 6]
)
data[" gdp_for_year ($) "] = pd.cut(
data[" gdp_for_year ($) "], 6, labels=[1, 2, 3, 4, 5, 6]
)
# To clarify
data.population.unique()
# 🔘**let's encoding the data**
# 🔘 Models only work with numerical values. For this reason, it is necessary to convert the categorical values of the features into numerical ones, So the machine can learn from those data and gives the right model. This process of converting categorical data into numerical data is called Encoding.
# 🔘 **Label encoder** :Sklearn provides a very efficient tool for encoding the levels of categorical features into numeric values. LabelEncoder encode labels with a value between 0 and n_classes-1 where n is the number of distinct labels. If a label repeats it assigns the same value to as assigned earlier.
# **Note 📋**
# also we can use one hot encoder or pandas.get_dummies function with diffrent output but finally the same task
to_categorical = ["country", "year", "sex", "age", "generation"]
from sklearn.preprocessing import LabelEncoder
L_encoder = LabelEncoder()
for label in to_categorical:
data[label] = L_encoder.fit_transform(data[label])
data.head()
# **Data correlations**
plt.figure(figsize=(10, 7))
sns.heatmap(data.corr(), annot=True)
plt.title("Data Correlations")
|
# To download the Dataset with "wget", we use the following command.
# if the wget didin't work, you must install "WGET" in your OS
# ## 1) Importing Dataset and Libraries
# ### 1- 1) Importing Libraries
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
# data processing
from sklearn.preprocessing import Normalizer, StandardScaler
# data splitting
from sklearn.model_selection import (
train_test_split,
StratifiedKFold,
cross_validate,
KFold,
)
# data evaluation
from sklearn.metrics import r2_score, mean_squared_error
# data modeling
from sklearn.linear_model import LinearRegression
from sklearn.tree import DecisionTreeRegressor
from sklearn.ensemble import RandomForestRegressor
from xgboost import XGBRegressor
from lightgbm import LGBMRegressor
from catboost import CatBoostRegressor
# set SEED
SEED = 123
df = pd.read_csv(
"/kaggle/input/prediction-salesbased-on-advertising-costs/advertising.csv"
) # Loading dataset as DataFram with Pandas
df.head() # Looking top 5 row
df.info() # A summary of the dataset and columns
# Observaions:
# 1) All of features are numeric
# 2) The target variable is a numerical continuous type
# 3) So we must Use Regression Models
df.describe() # describtion of numeric features
# ### Visualization of Data
sns.scatterplot(x="Radio", y="Sales", data=df)
sns.scatterplot(x="TV", y="Sales", data=df)
sns.scatterplot(x="Newspaper", y="Sales", data=df)
plt.legend(labels=["Radio", "TV", "Newspaper"])
plt.xlabel("Advertising Costs")
plt.ylabel("Mount of Sales")
# ### Spliting data to features and target variable
X = df.drop("Sales", axis=1) # The Features
y = df["Sales"] # The Target
# ### Splitting Data and making KFOLD
X_train, X_val, y_train, y_val = train_test_split(
X, y, test_size=0.2, random_state=SEED, shuffle=True
)
# The shape of train and validation data
y_train.shape, y_val.shape
kf = KFold(n_splits=5, shuffle=True, random_state=SEED)
# ### Linear Regression
lin_reg = LinearRegression()
cv = cross_validate(
lin_reg,
X_train,
y_train,
cv=kf,
scoring=("r2", "neg_mean_squared_error"),
return_estimator=True,
)
# detect more metrics https://scikit-learn.org/stable/modules/model_evaluation.html#scoring-parameter
model_lin = cv["estimator"]
y_hats = [model.predict(X_val) for model in model_lin]
r2_lin = np.mean([r2_score(y_val, y_hat) for y_hat in y_hats])
print("R2 of linearregression: " f"{r2_lin:0.2%}")
# ### DecisionTree
dec = DecisionTreeRegressor()
cv = cross_validate(
dec,
X_train,
y_train,
cv=kf,
scoring=("r2", "neg_mean_squared_error"),
return_estimator=True,
)
model_dec = cv["estimator"]
y_hats = [model.predict(X_val) for model in model_dec]
r2_dec = np.mean([r2_score(y_val, y_hat) for y_hat in y_hats])
print("R2 of DecisionTree: " f"{r2_dec:0.2%}")
# ## RandomForest
rf = RandomForestRegressor()
cv = cross_validate(
rf,
X_train,
y_train,
cv=kf,
scoring=("r2", "neg_mean_squared_error"),
return_estimator=True,
)
model_rf = cv["estimator"]
y_hats = [model.predict(X_val) for model in model_rf]
r2_rf = np.mean([r2_score(y_val, y_hat) for y_hat in y_hats])
print("R2 of Randomforest: " f"{r2_rf:0.2%}")
# ### Light Gradiant Boosting Model
lgb = LGBMRegressor()
cv = cross_validate(
lgb,
X_train,
y_train,
cv=kf,
scoring=("r2", "neg_mean_squared_error"),
return_estimator=True,
)
model_lgb = cv["estimator"]
y_hats = [model.predict(X_val) for model in model_lgb]
r2_lgb = np.mean([r2_score(y_val, y_hat) for y_hat in y_hats])
print("R2 of LightGBM: " f"{r2_lgb:0.2%}")
# ### Extra Gradiant Boosting
xgb = XGBRegressor()
cv = cross_validate(
xgb,
X_train,
y_train,
cv=kf,
scoring=("r2", "neg_mean_squared_error"),
return_estimator=True,
)
model_xgb = cv["estimator"]
y_hats = [model.predict(X_val) for model in model_xgb]
r2_xgb = np.mean([r2_score(y_val, y_hat) for y_hat in y_hats])
print("R2 of XGB: " f"{r2_xgb:0.2%}")
# Table of Results
print("R2 of linearregression: " f"{r2_lin:0.2%}")
print("R2 of DecisionTree: " f"{r2_dec:0.2%}")
print("R2 of Randomforest: " f"{r2_rf:0.2%}")
print("R2 of LightGBM: " f"{r2_lgb:0.2%}")
print("R2 of XGB: " f"{r2_xgb:0.2%}")
|
# Aşağıda ihtiyacımız doğrultusunda kullanacağımız kütüphaneleri yükleyelim.
#
import numpy as np
import seaborn as sns
import pandas as pd
# Veri çerçevemizi bulunduğumuz dizinden yükleyelim ve bir veri çerçevesi haline getirerek df değişkenine atayalım. (pd.read_csv(...csv))
df = pd.read_csv("/kaggle/input/vizeodev/iris.csv")
# Veri çerçevesinin ilk 5 gözlemini görüntüleyelim.
df.head(5)
# Veri çerçevesinin kaç öznitelik ve kaç gözlemden oluştuğunu görüntüleyelim.
# Öznitelik sayısını (sütun sayısı) görüntüleme
oznitelik_sayisi = df.shape[1]
print("Öznitelik Sayısı: ", oznitelik_sayisi)
# Gözlem sayısını (satır sayısı) görüntüleme
gozlem_sayisi = df.shape[0]
print("Gözlem Sayısı: ", gozlem_sayisi)
# Veri çerçevesindeki değişkenlerin hangi tipte olduğunu ve bellek kullanımını görüntüleyelim.
# Değişkenlerin veri tiplerini görüntüleme
veri_tipleri = df.dtypes
print("Değişken Veri Tipleri:\n", veri_tipleri)
# Bellek kullanımını görüntüleme
bellek_kullanimi = df.memory_usage(deep=True).sum() / (
1024**2
) # MB cinsinden hesaplama
print("Toplam Bellek Kullanımı (MB):", bellek_kullanimi)
# Veri çerçevesindeki sayısal değişkenler için temel istatistik değerlerini görüntüleyelim.
# Standart sapma ve ortalama değerlerden çıkarımda bulunarak hangi değişkenlerin ne kadar varyansa sahip olduğu hakkında fikir yürütelim.
# Sayisal değişkenlerin temel istatistik değerlerini görüntüleme
sayisal_degiskenler = df.select_dtypes(include=np.number) # Sayisal değişkenleri seçme
temel_istatistikler = (
sayisal_degiskenler.describe()
) # Temel istatistik değerleri hesaplama
print("Temel İstatistik Değerler:\n", temel_istatistikler)
# Standart sapma ve ortalama değerlerden çikarimda bulunma
varyans = (
temel_istatistikler.loc["std"] ** 2
) # Standart sapmanin karesi varyansi temsil eder
ortalama = temel_istatistikler.loc["mean"]
cikarim = ortalama**2 - varyans
print("\nDeğişkenlerin Varyans ve Ortalama Değerlerden Çıkarımları:\n", cikarim)
# Veri çerçevesinde hangi öznitelikte kaç adet eksik değer olduğunu gözlemleyelim.
# Her bir özniteliğin eksik değer sayısını görüntüleme
eksik_deger_sayisi = df.isna().sum()
print("Eksik Değer Sayıları:\n", eksik_deger_sayisi)
# Sayısal değişkenler arasında korelasyon olup olmadığını göstermek için korelasyon matrisi çizdirelim. Korelasyon katsayıları hakkında fikir yürütelim.
# En güçlü pozitif ilişki hangi iki değişken arasındadır?
import matplotlib.pyplot as plt
# Korelasyon matrisini hesapla
corr = df.corr()
# Korelasyon matrisini ısı haritası olarak çizdir
sns.heatmap(corr, annot=True, cmap="coolwarm")
plt.title("Değişkenler Arasındaki Korelasyon Matrisi")
plt.show()
# Korelasyon katsayılarını daha iyi okuyabilmek için ısı haritası çizdirelim.
# Korelasyon matrisini hesaplayalım
corr_matrix = df.corr()
# Korelasyon matrisini büyük bir ısı haritası olarak çizdirme
plt.figure(figsize=(10, 8)) # Grafiğin boyutunu belirleme
sns.heatmap(
corr_matrix, annot=True, cmap="coolwarm", linewidths=0.5, annot_kws={"fontsize": 12}
)
plt.title("Korelasyon Matrisi")
plt.show()
# Veri çerçevemizin hedef değişkeninin "variety" benzersiz değerlerini görüntüleyelim.
# "variety" değişkeninin benzersiz değerlerini görüntüleme
variety_unique = df["variety"].unique()
print("variety Benzersiz Değerler: ", variety_unique)
# Veri çerçevemizin hedef değişkeninin "variety" benzersiz kaç adet değer içerdiğini görüntüleyelim.
# "variety" değişkeninin benzersiz değer sayısını görüntüleme
variety_unique_count = df["variety"].nunique()
print("variety Benzersiz Değer Sayısı: ", variety_unique_count)
# Veri çerçevesindeki sepal.width ve sepal.length değişkenlerinin sürekli olduğunu görüyoruz. Bu iki sürekli veriyi görselleştirmek için önce scatterplot kullanalım.
# Scatterplot çizimi
sns.scatterplot(x="sepal.width", y="sepal.length", data=df)
plt.xlabel("Sepal Width")
plt.ylabel("Sepal Length")
plt.title("Sepal Width vs Sepal Length Scatterplot")
plt.show()
# Aynı iki veriyi daha farklı bir açıdan frekanslarıyla incelemek için jointplot kullanarak görselleştirelim.
# Jointplot çizimi
sns.jointplot(x="sepal.width", y="sepal.length", data=df, kind="hex")
plt.xlabel("Sepal Width")
plt.ylabel("Sepal Length")
plt.title("Sepal Width vs Sepal Length Jointplot")
plt.show()
# Aynı iki veriyi scatterplot ile tekrardan görselleştirelim fakat bu sefer "variety" parametresi ile hedef değişkenine göre kırdıralım.
# 3 farklı renk arasında sepal değişkenleriyle bir kümeleme yapılabilir mi? Ne kadar ayırt edilebilir bunun üzerine düşünelim.
# Scatterplot çizimi
sns.scatterplot(
x="sepal.width", y="sepal.length", data=df, hue="variety", palette="Set1"
)
plt.xlabel("Sepal Width")
plt.ylabel("Sepal Length")
plt.title("Sepal Width vs Sepal Length Scatterplot (Colored by Variety)")
plt.show()
# value_counts() fonksiyonu ile veri çerçevemizin ne kadar dengeli dağıldığını sorgulayalım.
# "variety" değişkeninin sınıf frekanslarını hesapla
class_counts = df["variety"].value_counts()
# Sonuçları ekrana yazdır
print("Sınıf Frekansları:\n", class_counts)
# Keman grafiği çizdirerek sepal.width değişkeninin dağılımını inceleyin.
# Söz konusu dağılım bizim için ne ifade ediyor, normal bir dağılım olduğunu söyleyebilir miyiz?
# Sepal.width değişkeninin keman grafiğini çizdir
sns.violinplot(x="variety", y="sepal.width", data=df)
# Grafiği göster
plt.show()
# Daha iyi anlayabilmek için sepal.width üzerine bir distplot çizdirelim.
# Sepal.width değişkeni için distplot çizdir
sns.histplot(df["sepal.width"])
# Grafiği göster
plt.show()
# Üç çiçek türü için üç farklı keman grafiğini sepal.length değişkeninin dağılımı üzerine tek bir satır ile görselleştirelim.
# Üç çiçek türü için sepal.length değişkeninin keman grafiğini çizdir
sns.violinplot(x="variety", y="sepal.length", data=df)
# Grafiği göster
plt.show()
# Hangi çiçek türünden kaçar adet gözlem barındırıyor veri çerçevemiz?
# 50 x 3 olduğunu ve dengeli olduğunu value_counts ile zaten görmüştük, ancak bunu görsel olarak ifade etmek için sns.countplot() fonksiyonuna variety parametresini vereilm.
# Çiçek türlerinin görsel olarak ifade edildiği bir countplot çizdir
sns.countplot(data=df, x="variety")
plt.title("Çiçek Türlerine Göre Gözlem Sayısı")
plt.xlabel("Çiçek Türü")
plt.ylabel("Gözlem Sayısı")
plt.show()
# sepal.length ve sepal.width değişkenlerini sns.jointplot ile görselleştirelim, dağılımı ve dağılımın frekansı yüksek olduğu bölgelerini inceleyelim.
sns.jointplot(data=df, x="sepal.length", y="sepal.width", kind="hex", cmap="viridis")
plt.xlabel("Sepal Length")
plt.ylabel("Sepal Width")
plt.title("Sepal Length vs Sepal Width")
plt.show()
# Bir önceki hücrede yapmış olduğumuz görselleştirmeye kind = "kde" parametresini ekleyelim. Böylelikle dağılımın noktalı gösterimden çıkıp yoğunluk odaklı bir görselleştirmeye dönüştüğünü görmüş olacağız.
sns.jointplot(data=df, x="sepal.length", y="sepal.width", kind="kde", cmap="viridis")
plt.xlabel("Sepal Length")
plt.ylabel("Sepal Width")
plt.title("Sepal Length vs Sepal Width (KDE)")
plt.show()
# scatterplot ile petal.length ve petal.width değişkenlerinin dağılımlarını çizdirelim.
sns.scatterplot(
data=df, x="petal.length", y="petal.width", hue="variety", palette="viridis"
)
plt.xlabel("Petal Length")
plt.ylabel("Petal Width")
plt.title("Petal Length vs Petal Width")
plt.show()
# Aynı görselleştirmeye hue = "variety" parametresini ekleyerek 3. bir boyut verelim.
# Label Encoding ile variety değişkenini sayısal bir değişkene dönüştürme
df["variety_label"] = df["variety"].astype("category").cat.codes
# 3D scatterplot figürünü oluşturma
fig = plt.figure()
ax = fig.add_subplot(111, projection="3d")
# Veriyi 3D scatterplot ile çizme
ax.scatter(
df["petal.length"],
df["petal.width"],
df["variety_label"],
c=df["variety_label"],
cmap="viridis",
)
# Eksen etiketlerini ve başlığı ekleme
ax.set_xlabel("Petal Length")
ax.set_ylabel("Petal Width")
ax.set_zlabel("Variety")
plt.title("Petal Length vs Petal Width (with Variety)")
# Grafik gösterme
plt.show()
# sns.lmplot() görselleştirmesini petal.length ve petal.width değişkenleriyle implemente edelim. Petal length ile petal width arasında ne tür bir ilişki var ve bu ilişki güçlü müdür? sorusunu yanıtlayalım.
# petal.length ve petal.width değişkenleri arasındaki ilişkiyi görselleştirme
sns.lmplot(x="petal.length", y="petal.width", data=df)
# Başlığı ve eksen etiketlerini ekleme
plt.title("Petal Length vs Petal Width")
plt.xlabel("Petal Length")
plt.ylabel("Petal Width")
# Grafik gösterme
plt.show()
# Bu sorunun yanıtını pekiştirmek için iki değişken arasında korelasyon katsayısını yazdıralım.
import scipy.stats
# petal.length ve petal.width arasındaki korelasyon katsayısını hesapla
correlation, _ = scipy.stats.pearsonr(df["petal.length"], df["petal.width"])
# Korelasyon katsayısını yazdır
print(
"Petal Length ve Petal Width arasındaki Korelasyon Katsayısı: {:.2f}".format(
correlation
)
)
# Petal Length ile Sepal Length değerlerini toplayarak yeni bir total length özniteliği oluşturalım.
# Yeni özniteliği hesapla
df["total.length"] = df["petal.length"] + df["sepal.length"]
# Veri çerçevesini gözden geçir
print(df.head())
# total.length'in ortalama değerini yazdıralım.
# total.length sütununun ortalama değeri
total_length_mean = np.mean(df["total.length"])
# Sonucu ekrana yazdır
print("total.length ortalama değeri: ", total_length_mean)
# total.length'in standart sapma değerini yazdıralım.
# total.length sütununun standart sapma değeri
total_length_std = np.std(df["total.length"])
# Sonucu ekrana yazdır
print("total.length standart sapma değeri: ", total_length_std)
# sepal.length'in maksimum değerini yazdıralım.
# sepal.length sütununun maksimum değeri
sepal_length_max = df["sepal.length"].max()
# Sonucu ekrana yazdır
print("sepal.length maksimum değer: ", sepal_length_max)
# sepal.length'i 5.5'den büyük ve türü setosa olan gözlemleri yazdıralım.
# sepal.length'i 5.5'den büyük ve türü setosa olan gözlemleri filtrele
filtered_data = df[(df["sepal.length"] > 5.5) & (df["variety"] == "Setosa")]
# Filtrelenen veriyi yazdır
print(filtered_data)
# petal.length'i 5'den küçük ve türü virginica olan gözlemlerin sadece sepal.length ve sepal.width değişkenlerini ve değerlerini yazdıralım.
# petal.length'i 5'ten küçük ve türü virginica olan gözlemleri filtrele
filtered_data = df[(df["petal.length"] < 5) & (df["variety"] == "Virginica")]
# Filtrelenen verinin sadece sepal.length ve sepal.width sütunlarını ve değerlerini yazdır
filtered_data[["sepal.length", "sepal.width"]]
# Hedef değişkenimiz variety'e göre bir gruplama işlemi yapalım değişken değerlerimizin ortalamasını görüntüleyelim.
# "variety" değişkenine göre gruplama yaparak değişken değerlerinin ortalamasını hesapla
grouped_mean = df.groupby("variety").mean()
# Ortalama değerleri görüntüle
print(grouped_mean)
# Hedef değişkenimiz variety'e göre gruplama işlemi yaparak sadece petal.length değişkenimizin standart sapma değerlerini yazdıralım.
# "variety" değişkenine göre gruplama yaparak "petal.length" değişkeninin standart sapma değerlerini hesapla
grouped_std = df.groupby("variety")["petal.length"].std()
# Standart sapma değerlerini görüntüle
print(grouped_std)
|
#
# Rename Pandas Columns in DataFrame🔥💥
# Welcome! In this notebook
# Rename Columns is necessary for the data preprocessing stage. Everyone is using it for data analytics, machine learning, data engineering, and even software development. renaming columns in a Pandas dataframe is a simple and useful technique that can save time and make data analysis easier
# Thanks for visiting my notebook
# # All Method I cover this version
# - ✒️**Method 1**:
# ```bash
# Using Rename Function
# ```
# - ✒️**Method 2**:
# ```bash
# Replacing the Column String
# ```
# - ✒️**Method 3**:
# ```bash
# Assigning a List of Column Names
# ```
# - ✒️**Method 4**:
# ```bash
# Renaming Columns with a Dictionary
# ```
# - ✒️**Method 5**:
# ```bash
# Using axis-style parameters: upper and lower case
# ```
# - ✒️**Method 6**:
# ```bash
# Rename column names using DataFrame add_prefix() and add_suffix() functions
# ```
# # Make Data
import pandas as pd
info = {
"ID": [11, 12, 13],
"Name": ["Pritom Saha", "Himel Saha", "Pranajit Saha"],
"Age": [25, 27, 65],
}
# Dictionary into DataFrame
# Make some copy
dataset = pd.DataFrame(info)
dataset1 = pd.DataFrame(info)
dataset2 = pd.DataFrame(info)
dataset3 = pd.DataFrame(info)
dataset4 = pd.DataFrame(info)
dataset5 = pd.DataFrame(info)
dataset6 = pd.DataFrame(info)
dataset7 = pd.DataFrame(info)
dataset8 = pd.DataFrame(info)
print("\033[1m\033[31m{}\033[0m".format(dataset.head()))
# # Using Rename Function
# Syntax:
# ```bash
# dataset.rename(columns = {'OldName':'New Name '}, inplace = True)
# ```
# using columns
dataset1.rename(columns={"Name": "Full Name ", "Age": "Current Age"}, inplace=True)
print("\033[35m{}\033[0m".format(dataset1.head()))
# using axis = "columns"
dataset2.rename(
{"Name": "Full Name ", "Age": "Current_Age"}, axis="columns", inplace=True
)
print("\033[36m{}\033[0m".format(dataset2.head()))
# # Replacing the Column String
# Syntax:
# ```bash
# dataset.columns.str.replace('OldName', 'new_name')
# ```
dataset3.columns = dataset3.columns.str.replace("Name", "full_name")
print("\033[1m\033[31m{}\033[0m".format(dataset3.head()))
# # Assigning a List of Column Names
# Syntax:
# - Make all columns name list
# ```bash
# dataset.columns = [x for x in columnlist]
# ```
columnN = ["serial", "full_name", "Current_age"]
dataset4.columns = [x for x in columnN]
print("\033[1m\033[31m{}\033[0m".format(dataset4.head()))
# # Renaming Columns with a Dictionary
# Syntax:
# - First make a dictionary than use columns as this dictionary
# ```bash
# dataset.rename(columns=new_col_name, inplace=True)
# ```
new_col_name = {"ID": "Serial", "Name": "Full Name", "Age": "Years Old"}
dataset5.rename(columns=new_col_name, inplace=True)
print("\033[1m\033[31m{}\033[0m".format(dataset5.head()))
# # Using axis-style parameters:
# Syntax:
# ```bash
# dataset.rename(str.lower, axis='columns')
# dataset.rename(str.upper, axis='columns')
# ```
dataset6.rename(str.lower, axis="columns", inplace=True)
print("\033[1m\033[31m{}\033[0m".format(dataset6.head()))
dataset6.rename(str.upper, axis="columns", inplace=True)
print("\033[1m\033[31m{}\033[0m".format(dataset6.head()))
# # Rename column names using DataFrame add_prefix() and add_suffix() functions
# Syntax:
# ```bash
# # Prefix
# dataset.add_prefix('X')
# # Suffix
# dataset.add_suffix('X')
# ```
# Add prefix to the column names
dataset7 = dataset7.add_prefix("student_")
print("\033[1m\033[31m{}\033[0m".format(dataset7.head()))
dataset8 = dataset8.add_suffix("_student")
print("\033[33m{}\033[0m".format(dataset8.head()))
|
# # Data Dictionary - train.csv - dependent variable is survival
# * Variable Definition Key
# * survival Survival 0 = No, 1 = Yes
# * pclass Ticket class 1 = 1st, 2 = 2nd, 3 = 3rd
# * sex Sex
# * Age Age in years
# * sibsp # For children number of siblings. For parents number of spouses.
# * parch # For children number of parents. For parents number of children.
# * ticket Ticket number
# * fare Passenger fare
# * cabin Cabin number
# * embarked Port Embarkation C = Cherbourg, Q = Queenstown, S = Southampton
# Normally, all data is processed and then split with Sklearn test_train_split. Since both files are given, they are processed separately.
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Other imports besides Kaggle provided ones.
import seaborn as sn
# importing one hot encoder
from sklearn.preprocessing import OneHotEncoder
from sklearn.naive_bayes import GaussianNB
from sklearn.metrics import classification_report, confusion_matrix, accuracy_score
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
dash = "-" * 80
# Use this to allow all rows to be printed if you need to analyze the data.
# pd.set_option('display.max_rows', None)
# pd.set_option('display.max_columns',None)
# # Read in Competition Data
train_data = pd.read_csv("/kaggle/input/titanic/train.csv")
train_data.head()
test_data = pd.read_csv("/kaggle/input/titanic/test.csv")
test_data.head()
# ## Data Shape
print("Train Data: ", train_data.shape)
print("Test Data: ", test_data.shape)
# # Review Null Values
# Funtion to Iterate over column names to display how many null values for each column.
def Check_Dataframe_for_Null_Values(df_to_check):
for column in df_to_check:
print(
" Count of Null values for "
+ column
+ ": "
+ str(len(df_to_check[df_to_check[column].isna()]))
)
return
Check_Dataframe_for_Null_Values(train_data)
# **Training Data Nulls:**
# * Age: 177
# * Cabin: 687
# * Embarked: 2
Check_Dataframe_for_Null_Values(test_data)
# **Testing Data Nulls:**
# * Age: 86
# * Fare: 1
# * Cabin: 327
# # Feature Engineering and Populating Null Values
# ## Size of Family
# Adding Size of the Family to the training set in hopes that it will improve modeling.
# Add the siblings with the spouse and the passenger to get the size of the family aboart the Titanic.
train_data["family_size"] = train_data["SibSp"] + train_data["Parch"] + 1
test_data["family_size"] = train_data["SibSp"] + train_data["Parch"] + 1
# ## For Null Age, Fill in Age of Passenger by Ticket Class Assignment
# A boxplot to see approximate mean age by Ticket Class (Pclass)
sn.boxplot(x="Pclass", y="Age", data=train_data)
# Review distinct values of Pclass:
print(train_data["Pclass"].unique())
# Calculate Age based on Ticket Class
# To address null values for age we query for each class and then calcuate the mean.
df_class_1 = train_data.query("Pclass == 1", engine="python")
class_1_mean_age = int(df_class_1["Age"].mean())
df_class_2 = train_data.query("Pclass == 2", engine="python")
class_2_mean_age = int(df_class_2["Age"].mean())
df_class_3 = train_data.query("Pclass == 3", engine="python")
class_3_mean_age = int(df_class_3["Age"].mean())
print("Mean Age for Class 1", class_1_mean_age)
print("Mean Age for Class 2", class_2_mean_age)
print("Mean Age for Class 3", class_3_mean_age)
# ### Update Training and Testing data set with Mean Age
# if data is null and class 1 then update age with value, etc.
# train_data.query('Pclass == 1 and Age.isnull()' , engine='python')
# Update train and test separately.
train_data.loc[
train_data.query("Pclass == 1 and Age.isnull()", engine="python").index, "Age"
] = class_1_mean_age
test_data.loc[
test_data.query("Pclass == 1 and Age.isnull()", engine="python").index, "Age"
] = class_1_mean_age
train_data.loc[
train_data.query("Pclass == 2 and Age.isnull()", engine="python").index, "Age"
] = class_2_mean_age
test_data.loc[
test_data.query("Pclass == 2 and Age.isnull()", engine="python").index, "Age"
] = class_2_mean_age
train_data.loc[
train_data.query("Pclass == 3 and Age.isnull()", engine="python").index, "Age"
] = class_3_mean_age
test_data.loc[
test_data.query("Pclass == 3 and Age.isnull()", engine="python").index, "Age"
] = class_3_mean_age
# First Class - Age Set to 38 for Null
train_data.query(
"Pclass == 1 and Age == " + str(class_1_mean_age), engine="python"
).head(5)
# First Class - Age Set to 38 for Null
test_data.query(
"Pclass == 1 and Age == " + str(class_1_mean_age), engine="python"
).head(5)
# Second Class - Age Set to 29 for Null
train_data.query(
"Pclass == 2 and Age == " + str(class_2_mean_age), engine="python"
).head(5)
# Second Class - Age Set to 29 for Null
test_data.query(
"Pclass == 2 and Age == " + str(class_2_mean_age), engine="python"
).head(5)
# Third Class - Age Set to 25 for Null
train_data.query(
"Pclass == 3 and Age == " + str(class_3_mean_age), engine="python"
).head(5)
# Third Class - Age Set to 25 for Null
test_data.query(
"Pclass == 3 and Age == " + str(class_3_mean_age), engine="python"
).head(5)
# Review Null values again
Check_Dataframe_for_Null_Values(train_data)
# Review Null values again
Check_Dataframe_for_Null_Values(test_data)
# ## Title - Cateogorizing into 4 titles - Mr, Mrs, Miss and Master
# * This will replace with the following:
# * Mr = Adult male
# * Mrs = Adult female
# * Miss = Child female
# * Master = Child male
# https://triangleinequality.wordpress.com/2013/09/08/basic-feature-engineering-with-the-titanic-data/
# * An article on parsing out the titles of the passengers.
# This takes the full string, big_string and determines if the substring (ie. "Mr.") occurs. -1 means that the subtring was not found.
def substrings_in_string(big_string, substrings):
# print (big_string, substrings)
for substring in substrings:
if big_string.find(substring) != -1:
return substring
print(big_string)
return np.nan # np.nan will return a type of float but with no value
# creating a title column from name. These are the subtrings we will use to search the names
title_list = [
"Mrs",
"Mr",
"Master",
"Miss",
"Major",
"Rev",
"Dr",
"Ms",
"Mlle",
"Col",
"Capt",
"Mme",
"Countess",
"Don",
"Jonkheer",
]
# This will use map to go through train_data and test_data and then create a new column called Title with the title found in each name.
train_data["Title"] = train_data["Name"].map(
lambda x: substrings_in_string(x, title_list)
)
test_data["Title"] = test_data["Name"].map(
lambda x: substrings_in_string(x, title_list)
)
# This will replace each record with the following:
# Mr = Adult male
# Mrs = Adult female
# Miss = Child female
# Master = Child male
def replace_titles(x):
title = x["Title"]
if title in ["Don", "Major", "Capt", "Jonkheer", "Rev", "Col"]:
return "Mr"
elif title in ["Countess", "Mme"]:
return "Mrs"
elif title in ["Mlle", "Ms"]:
return "Miss"
elif title == "Dr":
if x["Sex"] == "Male":
return "Mr"
else:
return "Mrs"
else:
return title
# Categorizing into 4 groups - Mr, Mrs, Miss and Master
train_data["Title"] = train_data.apply(replace_titles, axis=1)
test_data["Title"] = test_data.apply(replace_titles, axis=1)
train_data.head(20)
test_data.head(20)
# big_string = "Braund, Mr. Owen Harris"
# substrings = "['Mrs', 'Mr', 'Master', 'Miss', 'Major', 'Rev', 'Dr', 'Ms', 'Mlle', 'Col', 'Capt', 'Mme', 'Countess', 'Don', 'Jonkheer']"
# print (big_string.find('substring'))
# ## Cabin column removed for this iteration
# https://www.kaggle.com/code/ccastleberry/titanic-cabin-features/notebook
# * Intersting article on how to process the Cabin Column with all of the missing data.
# * It's clear that many passengers who were not assigned a cabin were in the lower deck.
# * Meaning, these passengers were more likely not to survive.
# Removing the cabin column
train_data.drop(
[
"Cabin",
],
axis=1,
inplace=True,
)
test_data.drop(["Cabin"], axis=1, inplace=True)
## Remove Appropriate Columns Before Modeling
train_data.drop(
[
"Name",
],
axis=1,
inplace=True,
)
test_data.drop(["Name"], axis=1, inplace=True)
train_data.drop(
[
"PassengerId",
],
axis=1,
inplace=True,
)
# For the test_data, I'm not deleting passenger id because it's needed for submittal to competition.
train_data.drop(
[
"Ticket",
],
axis=1,
inplace=True,
)
test_data.drop(["Ticket"], axis=1, inplace=True)
Check_Dataframe_for_Null_Values(train_data)
Check_Dataframe_for_Null_Values(test_data)
# # One Hot Encoding
# creating one hot encoder object
OneHotEncoder = OneHotEncoder()
# ## Title
df_OneHotEncoded_train = pd.DataFrame(
OneHotEncoder.fit_transform(train_data[["Title"]]).toarray()
)
df_OneHotEncoded_test = pd.DataFrame(
OneHotEncoder.fit_transform(test_data[["Title"]]).toarray()
)
df_OneHotEncoded_train.rename(
columns={
0: "master-male-child",
1: "miss-female-child",
2: "mr-male-adult",
3: "mrs-female-adult",
},
inplace=True,
)
df_OneHotEncoded_test.rename(
columns={
0: "master-male-child",
1: "miss-female-child",
2: "mr-male-adult",
3: "mrs-female-adult",
},
inplace=True,
)
train_data = train_data.join(df_OneHotEncoded_train)
test_data = test_data.join(df_OneHotEncoded_test)
train_data
# ## Sex
df_OneHotEncoded_train = pd.DataFrame(
OneHotEncoder.fit_transform(train_data[["Sex"]]).toarray()
)
df_OneHotEncoded_test = pd.DataFrame(
OneHotEncoder.fit_transform(test_data[["Sex"]]).toarray()
)
df_OneHotEncoded_train.rename(columns={0: "sex-female", 1: "sex-male"}, inplace=True)
df_OneHotEncoded_test.rename(columns={0: "sex-female", 1: "sex-male"}, inplace=True)
train_data = train_data.join(df_OneHotEncoded_train)
test_data = test_data.join(df_OneHotEncoded_test)
train_data.head()
test_data.head()
# ## Embarked
# I can't include Embarked because test_data doesn't have any values for this. When I do one hot encoding
# it won't create the emb_4 column. I'm having to leave off Embarked.
# df_OneHotEncoded_train_emb=pd.DataFrame(OneHotEncoder.fit_transform(train_data[['Embarked']]).toarray())
# df_OneHotEncoded_test_emb=pd.DataFrame(OneHotEncoder.fit_transform(test_data[['Embarked']]).toarray())
# df_OneHotEncoded_train_emb.rename(columns={0: 'emb_0',1: 'emb_1',2: 'emb_2',3: 'emb_3'}, inplace=True)
# df_OneHotEncoded_test_emb.rename(columns={0: 'emb_0' ,1: 'emb_1',2: 'emb_2',3: 'emb_3'}, inplace=True)
# train_data=train_data.join(df_OneHotEncoded_train_emb)
# test_data=test_data.join(df_OneHotEncoded_test_emb)
train_data.head(5)
test_data.head(5)
# ## Remove Title, Sex and Embarked After One Hot Encoding
train_data.drop(
[
"Title",
],
axis=1,
inplace=True,
)
test_data.drop(["Title"], axis=1, inplace=True)
train_data.drop(
[
"Sex",
],
axis=1,
inplace=True,
)
test_data.drop(["Sex"], axis=1, inplace=True)
train_data.drop(
[
"Embarked",
],
axis=1,
inplace=True,
)
test_data.drop(["Embarked"], axis=1, inplace=True)
# # Final Data for Modeling
train_data.head(5)
test_data.head(5)
# # Correlations
sn.set(rc={"figure.figsize": (20, 8)})
# plotting correlation heatmap
dataplot = sn.heatmap(train_data.corr(), cmap="YlGnBu", annot=True)
# # Split Independent and Dependent Varibles for Training Data
x = train_data.iloc[:, 1:].values
y = train_data.iloc[:, 0].values
# # Scale the Data ( Independent Features)
print(type(test_data))
print(type(x))
# Only scale the independent variables.
Stand_Scale = StandardScaler()
# Fit Transform Training Data
x = Stand_Scale.fit_transform(x)
# Fit Transform Testing Data
test_data_scaled = Stand_Scale.fit_transform(test_data)
print("\nx after standarization:", x)
print(dash)
print("\ntest_data_scaled after standarization:", test_data_scaled)
# # Split Training Data into Training and Testing
# * We will run our final predict against test_data for submittal.
# * For now I'm splitting the training data to see how it will perform against a test set.
# split the training data to generate training and test data.
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.20, random_state=5
)
print("\nx_train", x_train.shape)
print("\ny_train", y_train.shape)
print(dash)
print("\nx_test", x_test.shape)
print("\ny_test", y_test.shape)
# # Gaussian Naive Bayes
# ### Fit Model
# -----------Gaussian generative model----------------
GNB_model = GaussianNB()
GNB_model.fit(x_train, y_train)
print(GNB_model)
# ### Predict Model
y_pred = GNB_model.predict(x_test)
print("Actual:", y_test)
print("Predicted:", y_pred)
# ### Show Accuracy Score and Confusion Matrix
print("Accuracy Score:", accuracy_score(y_test, y_pred))
# in your classification, Bayesian approaches like this can be a useful approach.
# -----performance..... Confusion Matrix (actual, predicted)
cm_GNB = confusion_matrix(y_test, y_pred)
print("Confusion matrix (y_test,y_pred\n", cm_GNB)
from sklearn import metrics
import matplotlib.pyplot as plt
# Receiver Operating Characteristic (ROC)
fpr, tpr, _ = metrics.roc_curve(y_test, y_pred)
auc = metrics.roc_auc_score(y_test, y_pred)
# create ROC curve
plt.plot(fpr, tpr, label="AUC=" + str(auc))
plt.legend(loc=4)
plt.title("Gaussian Naive Bayes - ROC( Receiver Operating Characteristic) curve")
plt.show()
print(auc)
print(fpr, "\n", tpr, "\n", _)
# # Making Final Predictions for Submittal
# ### test_data column names:
# * PassengerId
# * Pclass
# * Age
# * SibSp
# * Parch
# * Fare
# * family_size
# * master-male-child
# * miss-female-child
# * mr-male-adult
# * mrs-female-adult
# * sex-female
# * sex-male
test_data_scaled
# ### The test_data needs to be converted back into a data frame to review the null values.
# defining array column names and assigning to a dataframe.
test_data_scaled = pd.DataFrame(
test_data_scaled,
columns=[
"PassengerId",
"Pclass",
"Age",
"SibSp",
"Parch",
"Fare",
"family_size",
"master-male-child",
"miss-female-child",
"mr-male-adult",
"mrs-female-adult",
"sex-female",
"sex-male",
],
)
test_data_scaled.head(10)
# Checking for Null records before submitting
# Removing 1 null record in the test_data.
Check_Dataframe_for_Null_Values(test_data_scaled)
test_data_scaled["Fare"].describe().loc[["mean"]]
# Updated Fare to a Mean Value - It can't be deleted or the competition entry will fail.
# Find just the mean Fare value and put it in a variable.
f_mean = test_data_scaled.loc[:, "Fare"].mean()
f_mean
# Update test_data null values with mean Fare.
test_data_scaled["Fare"].fillna(f_mean, inplace=True)
# Verifying
Check_Dataframe_for_Null_Values(test_data_scaled)
# I need to drop passenger id since I'm not using it as an independent variable.
test_data_GNB = test_data_scaled.drop(
[
"PassengerId",
],
axis=1,
)
# Generate predictions for competition.
predictions = GNB_model.predict(test_data_GNB)
print("Predictions for Submittal:", predictions)
test_data.info()
# I need to include the Passenger ID that is not scaled so we are using the original test_data dataframe.
output = pd.DataFrame({"PassengerId": test_data.PassengerId, "Survived": predictions})
output.to_csv("submission.csv", index=False)
print("Your submission was successfully saved!")
|
import numpy as np
import pandas as pd
# from sklearn import *
from sklearn.naive_bayes import GaussianNB
from sklearn.metrics import accuracy_score
import glob
def get_id(file):
return file.split("/")[-1].split(".")[0]
def build_submission(files, model, features):
submission = []
for file in files:
df = pd.read_csv(file)
df["Id"] = get_id(file)
# df = df.fillna(0).reset_index(drop=True)
res = pd.DataFrame(
np.round(model.predict(df[features]), 3),
columns=["StartHesitation", "Turn", "Walking"],
)
df = pd.concat([df, res], axis=1)
# df['Id'] = df['Id'].astype(str) + '_' + df['Time'].astype(str)
submission.append(df[["Id", "StartHesitation", "Turn", "Walking"]])
count = 0
# def get_file_data(file):
def get_file_data(datasets):
data_frames = []
count = 0
for file in datasets:
if count > 30:
break
# print(file)
df = pd.read_csv(file)
df["Id"] = get_id(file)
df["Type"] = file.split("/")[-2]
# filter out rows where either Valid or Task is false
# try:
# df = df[(df['Valid'] == True) & (df['Task'] == True)]
# except:
# print("D")
data_frames.append(df)
count = count + 1
return pd.concat(data_frames)
train = glob.glob(
"/kaggle/input/tlvmc-parkinsons-freezing-gait-prediction/train/defog/**"
)
test = glob.glob("/kaggle/input/tlvmc-parkinsons-freezing-gait-prediction/test/**/**")
subjects = pd.read_csv(
"/kaggle/input/tlvmc-parkinsons-freezing-gait-prediction/subjects.csv"
)
tasks = pd.read_csv("/kaggle/input/tlvmc-parkinsons-freezing-gait-prediction/tasks.csv")
# train
train_data = get_file_data(train)
test_data = get_file_data(test)
train_data
test_data
print(train_data.columns)
# print(subjects.columns)
print(tasks.columns)
print(test_data.columns)
print("Merge taining")
train_data = pd.merge(train_data, tasks, on="Id")
train_data
print("merge testing")
test_data = pd.merge(test_data, tasks, on="Id")
test_data
print("training")
X = train_data[["AccV", "AccML", "AccAP"]]
X_test = test_data[["AccV", "AccML", "AccAP"]]
y = train_data["Valid"].values.ravel()
y_test = test_data[["Task"]]
# print(train_data.shape)
# print(test_data.shape)
# print(X.shape, y.shape)
print("Model area")
nb = GaussianNB()
nb.fit(X, y)
y_pred = nb.predict(X_test)
accuracy = accuracy_score(y_test, y_pred)
print("Naive Bayes accuracy:", accuracy)
path = "/kaggle/input/tlvmc-parkinsons-freezing-gait-prediction/"
sub = pd.read_csv(path + "sample_submission.csv")
test = glob.glob(path + "test/**/**")
count = 0
sub["t"] = 0
submission = []
for f in test:
print("Working test ", count)
if count > 30:
break
count = count + 1
df = pd.read_csv(f)
df["Id"] = f.split("/")[-1].split(".")[0]
df = df.fillna(0).reset_index(drop=True)
res = nb.predict(df[["AccV", "AccML", "AccAP"]])
n = len(res)
if n % 3 != 0:
res = np.concatenate([res, np.zeros(3 - n % 3)])
res = res.reshape(-1, 3).astype("float64")
res = pd.DataFrame(res, columns=["StartHesitation", "Turn", "Walking"])
df = pd.concat([df, res], axis=1)
df["Id"] = df["Id"].astype(str) + "_" + df["Time"].astype(str)
submission.append(df[["Id", "StartHesitation", "Turn", "Walking"]])
submission = pd.concat(submission)
submission = pd.merge(sub[["Id", "t"]], submission, how="left", on="Id").fillna(0.0)
submission[["Id", "StartHesitation", "Turn", "Walking"]].to_csv(
"submission.csv", index=False
)
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import missingno
from sklearn.preprocessing import LabelEncoder, OneHotEncoder
from sklearn.impute import KNNImputer
from sklearn.metrics import (
roc_auc_score,
roc_curve,
accuracy_score,
confusion_matrix,
log_loss,
plot_roc_curve,
auc,
precision_recall_curve,
)
from sklearn.cluster import KMeans
from sklearn.model_selection import StratifiedKFold
from sklearn.svm import SVC
from sklearn.linear_model import LogisticRegression
from sklearn.gaussian_process import GaussianProcessClassifier
from sklearn.ensemble import RandomForestClassifier
from xgboost import XGBClassifier
from functools import partial
from skopt import gp_minimize
from skopt import space
from skopt.plots import plot_convergence
sns.set_style("whitegrid")
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
import warnings
warnings.filterwarnings("ignore")
df_train = pd.read_csv(
"../input/hr-analytics-job-change-of-data-scientists/aug_train.csv"
)
df_test = pd.read_csv(
"../input/hr-analytics-job-change-of-data-scientists/aug_test.csv"
)
df_train.head()
df_test.head()
df_train.info()
df_test.info()
import matplotlib.pyplot as plt
import seaborn as sns
plt.figure(figsize=(10, 6))
ax = sns.countplot(x="company_type", data=df_train[df_train["target"] == 1])
plt.xticks(rotation=45, ha="right")
plt.xlabel("Company Type")
plt.ylabel("Count")
total = len(df_train[df_train["target"] == 1])
for p in ax.patches:
percentage = "{:.1f}%".format(100 * p.get_height() / total)
x = p.get_x() + p.get_width() / 2 - 0.1
y = p.get_height() + 10
ax.annotate(percentage, (x, y), size=12)
plt.show()
import matplotlib.pyplot as plt
import seaborn as sns
plt.figure(figsize=(10, 6))
ax = sns.countplot(x="company_size", data=df_train[df_train["target"] == 1])
plt.xlabel("Company Size")
plt.ylabel("Count")
total = len(df_train[df_train["target"] == 1])
for p in ax.patches:
percentage = "{:.1f}%".format(100 * p.get_height() / total)
x = p.get_x() + p.get_width() / 2 - 0.1
y = p.get_height() + 10
ax.annotate(percentage, (x, y), size=12)
plt.show()
plt.figure(figsize=(10, 6))
ax = sns.countplot(x="education_level", data=df_train[df_train["target"] == 1])
plt.xlabel("Education Level")
plt.ylabel("Count")
total = len(df_train[df_train["target"] == 1])
for p in ax.patches:
percentage = "{:.1f}%".format(100 * p.get_height() / total)
x = p.get_x() + p.get_width() / 2 - 0.1
y = p.get_height() + 10
ax.annotate(percentage, (x, y), size=12)
plt.show()
plt.figure(figsize=(10, 6))
ax = sns.countplot(x="relevent_experience", data=df_train[df_train["target"] == 1])
plt.xlabel("Relevant Experience")
plt.ylabel("Count")
total = len(df_train[df_train["target"] == 1])
for p in ax.patches:
percentage = "{:.1f}%".format(100 * p.get_height() / total)
x = p.get_x() + p.get_width() / 2 - 0.1
y = p.get_height() + 10
ax.annotate(percentage, (x, y), size=12)
plt.show()
plt.figure(figsize=(10, 6))
ax = sns.countplot(x="relevent_experience", data=df_train[df_train["target"] == 1])
plt.xlabel("Relevant Experience")
plt.ylabel("Count")
total = len(df_train[df_train["target"] == 1])
for p in ax.patches:
percentage = "{:.1f}%".format(100 * p.get_height() / total)
x = p.get_x() + p.get_width() / 2 - 0.1
y = p.get_height() + 10
ax.annotate(percentage, (x, y), size=12)
plt.show()
plt.figure(figsize=(10, 6))
ax = sns.countplot(x="enrolled_university", data=df_train[df_train["target"] == 1])
plt.xlabel("Enrolled University")
plt.ylabel("Count")
total = len(df_train[df_train["target"] == 1])
for p in ax.patches:
percentage = "{:.1f}%".format(100 * p.get_height() / total)
x = p.get_x() + p.get_width() / 2 - 0.1
y = p.get_height() + 10
ax.annotate(percentage, (x, y), size=12)
plt.show()
plt.figure(figsize=(10, 6))
ax = sns.countplot(x="major_discipline", data=df_train[df_train["target"] == 1])
plt.xlabel("Major Discipline")
plt.ylabel("Count")
total = len(df_train[df_train["target"] == 1])
for p in ax.patches:
percentage = "{:.1f}%".format(100 * p.get_height() / total)
x = p.get_x() + p.get_width() / 2 - 0.1
y = p.get_height() + 10
ax.annotate(percentage, (x, y), size=12)
plt.show()
df_train2 = df_train.copy()
experience_map = {
"<1": "0-5",
"1": "0-5",
"2": "0-5",
"3": "0-5",
"4": "0-5",
"5": "0-5",
"6": "6-10",
"7": "6-10",
"8": "6-10",
"9": "6-10",
"10": "6-10",
"11": "11-15",
"12": "11-15",
"13": "11-15",
"14": "11-15",
"15": "11-15",
"16": "16-20",
"17": "16-20",
"18": "16-20",
"19": "16-20",
"20": "16-20",
">20": ">20",
}
df_train2.loc[:, "experience"] = df_train2["experience"].map(experience_map)
category_order = ["0-5", "6-10", "11-15", "16-20", ">20"]
plt.figure(figsize=(10, 6))
ax = sns.countplot(
x="experience", data=df_train2[df_train2["target"] == 1], order=category_order
)
plt.xlabel("Experience")
plt.ylabel("Count")
total = len(df_train2[df_train2["target"] == 1])
for p in ax.patches:
percentage = "{:.1f}%".format(100 * p.get_height() / total)
x = p.get_x() + p.get_width() / 2 - 0.1
y = p.get_height() + 10
ax.annotate(percentage, (x, y), size=12)
plt.show()
plt.figure(figsize=(10, 6))
ax = sns.countplot(x="gender", data=df_train[df_train["target"] == 1])
plt.xlabel("Gender")
plt.ylabel("Percentage")
total = len(df_train[df_train["target"] == 1])
for p in ax.patches:
percentage = "{:.1f}%".format(100 * p.get_height() / total)
x = p.get_x() + p.get_width() / 2 - 0.1
y = p.get_height() + 10
ax.annotate(percentage, (x, y), size=12)
plt.show()
plt.figure(figsize=(10, 6))
ax = sns.countplot(x="last_new_job", data=df_train[df_train["target"] == 1])
plt.xlabel("Last New Job")
plt.ylabel("Percentage")
total = len(df_train[df_train["target"] == 1])
for p in ax.patches:
percentage = "{:.1f}%".format(100 * p.get_height() / total)
x = p.get_x() + p.get_width() / 2 - 0.1
y = p.get_height() + 10
ax.annotate(percentage, (x, y), size=12)
plt.show()
df_test["target"] = -1 # ทำเป้น -1
df_pre = pd.concat([df_train, df_test], axis=0).reset_index(drop=True)
df_test.head()
df_pre.info()
#
# **Label Encoding** refers to converting the labels into numeric form so as to convert it into the machine-readable form. Machine learning algorithms can then decide in a better way on how those labels must be operated. It is an important pre-processing step for the structured dataset in supervised learning.
# 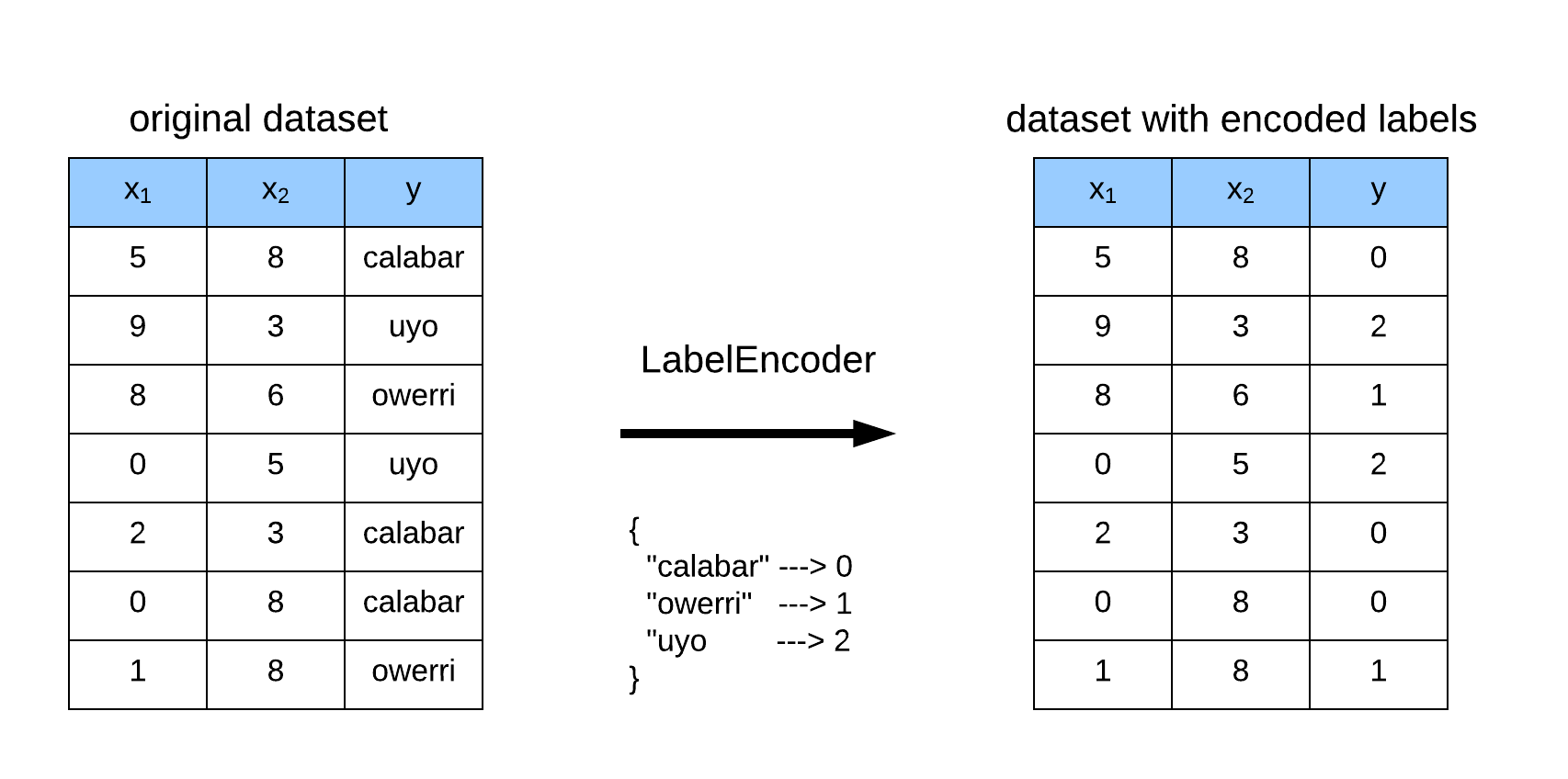
# We can do label Encoding From LabelEncoder of scikit-Learn but to do so first we have to impute missing values in data
from sklearn.preprocessing import LabelEncoder
df_lb = df_pre.copy()
df_lb["education_level"].value_counts()
# Fill nan values
df_lb.loc[:, "education_level"] = df_lb["education_level"].fillna("NONE")
# In above code cell i just create null values as new category "NONE"
# initialize LabelEncoder
lbl_enc = LabelEncoder()
# one hot encoder
df_lb.loc[:, "education_level"] = lbl_enc.fit_transform(df_lb["education_level"].values)
df_lb["education_level"].value_counts()
# Making Dictionaries of ordinal features
gender_map = {"Female": 2, "Male": 1, "Other": 0}
relevent_experience_map = {"Has relevent experience": 1, "No relevent experience": 0}
enrolled_university_map = {
"no_enrollment": 0,
"Full time course": 1,
"Part time course": 2,
}
education_level_map = {
"Primary School": 0,
"Graduate": 2,
"Masters": 3,
"High School": 1,
"Phd": 4,
}
major_map = {
"STEM": 0,
"Business Degree": 1,
"Arts": 2,
"Humanities": 3,
"No Major": 4,
"Other": 5,
}
experience_map = {
"<1": 0,
"1": 1,
"2": 2,
"3": 3,
"4": 4,
"5": 5,
"6": 6,
"7": 7,
"8": 8,
"9": 9,
"10": 10,
"11": 11,
"12": 12,
"13": 13,
"14": 14,
"15": 15,
"16": 16,
"17": 17,
"18": 18,
"19": 19,
"20": 20,
">20": 21,
}
company_type_map = {
"Pvt Ltd": 0,
"Funded Startup": 1,
"Early Stage Startup": 2,
"Other": 3,
"Public Sector": 4,
"NGO": 5,
}
company_size_map = {
"<10": 0,
"10/49": 1,
"100-500": 2,
"1000-4999": 3,
"10000+": 4,
"50-99": 5,
"500-999": 6,
"5000-9999": 7,
}
last_new_job_map = {"never": 0, "1": 1, "2": 2, "3": 3, "4": 4, ">4": 5}
# Transforming Categorical features into numarical features
df_pre.loc[:, "education_level"] = df_pre["education_level"].map(education_level_map)
df_pre.loc[:, "company_size"] = df_pre["company_size"].map(company_size_map)
df_pre.loc[:, "company_type"] = df_pre["company_type"].map(company_type_map)
df_pre.loc[:, "last_new_job"] = df_pre["last_new_job"].map(last_new_job_map)
df_pre.loc[:, "major_discipline"] = df_pre["major_discipline"].map(major_map)
df_pre.loc[:, "enrolled_university"] = df_pre["enrolled_university"].map(
enrolled_university_map
)
df_pre.loc[:, "relevent_experience"] = df_pre["relevent_experience"].map(
relevent_experience_map
)
df_pre.loc[:, "gender"] = df_pre["gender"].map(gender_map)
df_pre.loc[:, "experience"] = df_pre["experience"].map(experience_map)
# encoding city feature using label encoder
lb_en = LabelEncoder()
df_pre.loc[:, "city"] = lb_en.fit_transform(df_pre.loc[:, "city"])
df_pre.head()
df_pre.info()
colors = [
"#E1FEFE",
"#E3EBFD",
"#E7DCFC",
"#E0C7EE",
"#F8D5F8",
"#E0E0F3",
"#D4D2F2",
"#D4D2F2",
"#E5D0E2",
"#F5E8F7",
"#FFF5ED",
"#FAE0D8",
"#F0F4BF",
"#DFE1BE",
]
missingno.bar(df_pre, color=colors, sort="ascending", figsize=(10, 5), fontsize=12)
# Just to check number of null values of every column in data
for col in df_pre.columns:
null_val = df_pre[col].isnull().sum()
null_prec = (null_val * 100) / df_pre.shape[0]
print("> %s , Missing: %d (%.1f%%)" % (col, null_val, null_prec))
missingno.heatmap(df_pre, cmap="RdYlGn", figsize=(10, 5), fontsize=12)
df_pre1 = df_pre.copy()
knn_imputer = KNNImputer(n_neighbors=3)
X = np.round(knn_imputer.fit_transform(df_pre1))
df_pre1 = pd.DataFrame(X, columns=df_pre1.columns)
df_pre1.info()
df_pre1.head()
plt.figure(figsize=(15, 8))
plt.title("Before Imputation")
df_pre["city_development_index"].plot(kind="kde")
plt.show()
plt.figure(figsize=(15, 8))
plt.title("After Imputation")
df_pre1["city_development_index"].plot(kind="kde")
plt.show()
# So rather than using imputation on whole dataset just use it on those features having missing values.
# missing columns
missing_cols = df_pre.columns[df_pre.isna().any()].tolist()
missing_cols
# Above shown columns have missing values and all 8 are categorical features
# Now i would like make two different dataframes one having features with missing values and second having features without missing values. But there will be one common column enrollee_id so that later we can perform inner join on both dataframes
# dataframe having features with missing values
df_missing = df_pre[["enrollee_id"] + missing_cols]
# dataframe having features without missing values
df_non_missing = df_pre.drop(missing_cols, axis=1)
# k-Nearest Neighbour Imputation
knn_imputer = KNNImputer(n_neighbors=3)
X = np.round(knn_imputer.fit_transform(df_missing))
# Rounding them because these are categorical features
df_missing = pd.DataFrame(X, columns=df_missing.columns)
# now lets join both dataframes
df_pre2 = pd.merge(df_missing, df_non_missing, on="enrollee_id")
# If you remember i did concatenation between train and test data before preprocessing. Now after preprocessing of data we can seprate train and test data
train = df_pre2[df_pre2["target"] != -1].reset_index(drop=True)
test = df_pre2[df_pre2["target"] == -1].reset_index(drop=True)
X = train.drop(["enrollee_id", "target"], axis=1)
Y = train["target"]
# drop fake target feature from test data
test = test.drop("target", axis=1)
sns.countplot(train["target"], edgecolor="black")
# # Model
# In this notebook i would like to use [Extreme Gradient Boosting (XGBoost)](https://machinelearningmastery.com/extreme-gradient-boosting-ensemble-in-python/) Classifier
from sklearn.model_selection import train_test_split
X_train, X_val, y_train, y_val = train_test_split(X, Y, test_size=0.2, random_state=42)
clf = XGBClassifier()
clf.fit(X_train, y_train)
y_train_pred = clf.predict_proba(X_train)
y_train_pred_pos = y_train_pred[:, 1]
y_val_pred = clf.predict_proba(X_val)
y_val_pred_pos = y_val_pred[:, 1]
auc_train = roc_auc_score(y_train, y_train_pred_pos)
auc_test = roc_auc_score(y_val, y_val_pred_pos)
print(f"Train AUC Score {auc_train}")
print(f"Test AUC Score {auc_test}")
fpr, tpr, _ = roc_curve(y_val, y_val_pred_pos)
# Let's plot AUC Curve
def plot_auc_curve(fpr, tpr, auc):
plt.figure(figsize=(16, 6))
plt.plot(fpr, tpr, "b+", linestyle="-")
plt.fill_between(fpr, tpr, alpha=0.5)
plt.ylabel("True Postive Rate")
plt.xlabel("False Postive Rate")
plt.title(f"ROC Curve Having AUC = {auc}")
plot_auc_curve(fpr, tpr, auc_test)
# funtion to plot learning curves
def plot_learning_cuve(model, X, Y):
x_train, x_test, y_train, y_test = train_test_split(
X, Y, test_size=0.2, random_state=11
)
train_loss, test_loss = [], []
for m in range(200, len(x_train), 200):
model.fit(x_train.iloc[:m, :], y_train[:m])
y_train_prob_pred = model.predict_proba(x_train.iloc[:m, :])
train_loss.append(log_loss(y_train[:m], y_train_prob_pred))
y_test_prob_pred = model.predict_proba(x_test)
test_loss.append(log_loss(y_test, y_test_prob_pred))
plt.figure(figsize=(15, 8))
plt.plot(train_loss, "r-+", label="Training Loss")
plt.plot(test_loss, "b-", label="Test Loss")
plt.xlabel("Number Of Batches")
plt.ylabel("Log-Loss")
plt.legend(loc="best")
plt.show()
plot_learning_cuve(XGBClassifier(), X, Y)
# It's a high variance problem
sns.countplot(Y, edgecolor="black", palette=["#96deff", "#ffa3ed"])
# # Oversampling using SMOTE
# [SMOTE for Imbalanced Classification](https://machinelearningmastery.com/smote-oversampling-for-imbalanced-classification/)\
# [SMOTE implementation](https://imbalanced-learn.org/stable/generated/imblearn.over_sampling.SMOTE.html)
from imblearn.over_sampling import SMOTE
smote = SMOTE(random_state=402)
X_smote, Y_smote = smote.fit_resample(X, Y)
sns.countplot(Y_smote, edgecolor="black")
print(X_smote.shape)
X_train, X_val, y_train, y_val = train_test_split(
X_smote, Y_smote, test_size=0.2, random_state=42
)
clf = XGBClassifier()
clf.fit(X_train, y_train)
y_train_pred = clf.predict_proba(X_train)
y_train_pred_pos = y_train_pred[:, 1]
y_val_pred = clf.predict_proba(X_val)
y_val_pred_pos = y_val_pred[:, 1]
auc_train = roc_auc_score(y_train, y_train_pred_pos)
auc_test = roc_auc_score(y_val, y_val_pred_pos)
print(f"Train AUC Score {auc_train}")
print(f"Test AUC Score {auc_test}")
plot_learning_cuve(XGBClassifier(), X_smote, Y_smote)
# Let's try to increase more data to conquer overfitting using SMOTE
smote = SMOTE(random_state=446)
X_smote1, Y_smote1 = smote.fit_resample(X, Y)
X_final = pd.concat([X_smote, X_smote1], axis=0).reset_index(drop=True)
Y_final = pd.concat([Y_smote, Y_smote1], axis=0).reset_index(drop=True)
sns.countplot(Y_final, edgecolor="black")
print(X_final.shape)
X_train, X_val, y_train, y_val = train_test_split(
X_final, Y_final, test_size=0.2, random_state=42
)
clf = XGBClassifier()
clf.fit(X_train, y_train)
y_train_pred = clf.predict_proba(X_train)
y_train_pred_pos = y_train_pred[:, 1]
y_val_pred = clf.predict_proba(X_val)
y_val_pred_pos = y_val_pred[:, 1]
auc_train = roc_auc_score(y_train, y_train_pred_pos)
auc_test = roc_auc_score(y_val, y_val_pred_pos)
print(f"Train AUC Score {auc_train}")
print(f"Test AUC Score {auc_test}")
plot_learning_cuve(XGBClassifier(), X_final, Y_final)
#######################
from sklearn.metrics import f1_score
y_val_pred_bin = [1 if x >= 0.5 else 0 for x in y_val_pred_pos]
# calculate f1 score
f1 = f1_score(y_val, y_val_pred_bin)
print(f"F1 score: {f1}")
from sklearn.metrics import precision_recall_curve
y_val_pred_pos = clf.predict_proba(X_val)[:, 1]
precision, recall, thresholds = precision_recall_curve(y_val, y_val_pred_pos)
plt.figure(figsize=(8, 6))
plt.plot(recall, precision, color="blue", linewidth=2)
plt.xlabel("Recall", fontsize=14)
plt.ylabel("Precision", fontsize=14)
plt.title("Precision-Recall Curve", fontsize=16)
plt.show()
import matplotlib.pyplot as plt
from sklearn.metrics import plot_precision_recall_curve, plot_roc_curve
# Precision-Recall Curve
plot_precision_recall_curve(clf, X_val, y_val)
plt.title("Precision-Recall Curve")
# ROC Curve
plot_roc_curve(clf, X_val, y_val)
plt.title("Receiver Operating Characteristic (ROC) Curve")
from sklearn.metrics import confusion_matrix
import xgboost as xgb
model = xgb.XGBClassifier()
model.fit(X_train, y_train)
y_train_pred = model.predict(X_train)
y_val_pred = model.predict(X_val)
tn, fp, fn, tp = confusion_matrix(y_val, y_val_pred).ravel()
print("True Negative: ", tn)
print("False Positive: ", fp)
print("False Negative: ", fn)
print("True Positive: ", tp)
cols_to_plot = ["enrollee_id", "gender", "experience"]
import matplotlib.pyplot as plt
for col in cols_to_plot:
fig, ax = plt.subplots(figsize=(10, 5))
ax = sns.countplot(x="target", hue=col, data=df_pre2)
ax.set_title(f"Target vs {col}")
plt.show()
import matplotlib.pyplot as plt
import seaborn as sns
# Plot target vs last_new_job
plt.figure(figsize=(10, 6))
sns.countplot(x="target", hue="last_new_job", data=df_pre2)
# Plot target vs city
plt.figure(figsize=(10, 6))
sns.countplot(x="target", hue="city", data=df_pre2)
# Plot target vs gender
plt.figure(figsize=(10, 6))
sns.countplot(x="target", hue="gender", data=df_pre2)
# Plot target vs experience
plt.figure(figsize=(10, 6))
sns.boxplot(x="target", y="experience", data=df_pre2)
# Plot target vs city_development_index
plt.figure(figsize=(10, 6))
sns.boxplot(x="target", y="city_development_index", data=df_pre2)
# Plot target vs major_discipline
plt.figure(figsize=(10, 6))
sns.countplot(x="target", hue="major_discipline", data=df_pre2)
# Plot target vs enrolled_university
plt.figure(figsize=(10, 6))
sns.countplot(x="target", hue="enrolled_university", data=df_pre2)
import seaborn as sns
import matplotlib.pyplot as plt
sns.countplot(x="target", hue="relevent_experience", data=df_pre2)
plt.show()
# Plot target vs relevant_experience
plt.figure(figsize=(10, 6))
sns.countplot(x="target", hue="relevent_experience", data=df_pre2)
# Plot target vs education_level
plt.figure(figsize=(10, 6))
sns.countplot(x="target", hue="education_level", data=df_pre2)
# Plot target vs company_size
plt.figure(figsize=(10, 6))
sns.countplot(x="target", hue="company_size", data=df_pre2)
# Plot target vs enrollee_id
plt.figure(figsize=(10, 6))
sns.countplot(x="target", hue="enrollee_id", data=df_pre2)
# Plot target vs company_type
plt.figure(figsize=(10, 6))
sns.countplot(x="target", hue="company_type", data=df_pre2)
# Plot target vs training_hours
plt.figure(figsize=(10, 6))
sns.boxplot(x="target", y="training_hours", data=df_pre2)
|
# # Speech Emotion Recognition
# ### Algorithm used:
# * Gradient Boosting
# * Decision Tree
# * Gaussian Naive Bayes
# * MLP Classifier
# * Random Forest
# * AdaBoost
# * KNN
# * Logistic Regression
# * Support Vector Machine (RBF & Linear)
# * Stochastic Gradient Descent
# ### Databases used
# * Toronto emotional speech set (TESS)
# ### Reference
# * [Recognizing Speech Emotion Based on Acoustic
# Features Using Machine Learning](https://aiubedu60714-my.sharepoint.com/personal/manjurul_aiub_edu/Documents/Microsoft%20Teams%20Chat%20Files/Recognizing_Speech_Emotion_Based_on_Acoustic_Features_Using_Machine_Learning.pdf)
# ### Import Libraries
# Import necessary libraries
import glob
import os
import librosa
import time
import seaborn as sns
import numpy as np
import pandas as pd
from tqdm import tqdm
tess_emotions = ["angry", "disgust", "fear", "happy", "neutral", "ps", "sad"]
def extract_feature(file_name):
X, sample_rate = librosa.load(os.path.join(file_name), res_type="kaiser_fast")
result = np.array([])
stft = np.abs(librosa.stft(X))
chromas = np.mean(librosa.feature.chroma_stft(S=stft, sr=sample_rate).T, axis=0)
result = np.hstack((result, chromas))
mfccs = np.mean(librosa.feature.mfcc(y=X, sr=sample_rate, n_mfcc=40).T, axis=0)
result = np.hstack((result, mfccs))
mels = np.mean(
librosa.feature.melspectrogram(y=X, sr=sample_rate, n_mels=128).T, axis=0
)
result = np.hstack((result, mels))
return result
def load_data():
sound, emo = [], []
for file in glob.glob(
"/kaggle/input/toronto-emotional-speech-set-tess/tess toronto emotional speech set data/TESS Toronto emotional speech set data/*AF_*/*.wav"
):
file_name = os.path.basename(file)
emotion = file_name.split("_")[2][:-4] # split and remove .wav
sound.append(file)
emo.append(emotion)
return {"file": sound, "emotion": emo}
start_time = time.time()
Trial_dict = load_data()
print("--- Data loaded. Loading time: %s seconds ---" % (time.time() - start_time))
X = pd.DataFrame(Trial_dict["file"])
y = pd.DataFrame(Trial_dict["emotion"])
X.shape, y.shape
y.value_counts()
# X_features = X[0].swifter.progress_bar(enable=True).apply(lambda x: extract_feature(x))
X_features = []
for x in tqdm(X[0]):
# print(x)
X_features.append(extract_feature(x))
X_features = pd.DataFrame(X_features)
# renaming the label column to emotion
y = y.rename(columns={0: "emotion"})
# concatinating the attributes and label into a single dataframe
data = pd.concat([X_features, y], axis=1)
data.head()
# ## Shuffling data#reindexing to shuffle the data at random
# data = data.reindex(np.random.permutation(data.index))
# reindexing to shuffle the data at random
data = data.reindex(np.random.permutation(data.index))
# Storing shuffled ravdess and tess data to avoid loading again
data.to_csv("TESS_FEATURES.csv")
starting_time = time.time()
data = pd.read_csv("./TESS_FEATURES.csv")
print("data loaded in " + str(time.time() - starting_time) + "ms")
print(data.head())
data.shape
# printing all columns
data.columns
# dropping the column Unnamed: 0 to removed shuffled index
data = data.drop("Unnamed: 0", axis=1)
data.columns
# separating features and target outputs
X = data.drop("emotion", axis=1).values
y = data["emotion"].values
print(y)
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
X.shape, y.shape
np.unique(y)
# # Importing sklearn libraries
from sklearn.model_selection import train_test_split
from sklearn.metrics import classification_report, confusion_matrix, accuracy_score
from sklearn.preprocessing import StandardScaler
from sklearn.pipeline import Pipeline
# # Cross Validation Parameters
from sklearn.model_selection import RepeatedStratifiedKFold
from sklearn.model_selection import cross_val_score, cross_validate, cross_val_predict
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.50, random_state=0
)
cv = RepeatedStratifiedKFold(n_splits=5, n_repeats=10)
# # SVC - RBF
from sklearn.svm import SVC
svclassifier = SVC(kernel="rbf")
# ### Without Scaling
# Accuracy should be lower than scaled
import time
starting_time = time.time()
svclassifier.fit(X_train, y_train)
print("Trained model in %s ms " % str(time.time() - starting_time))
train_acc = float(svclassifier.score(X_train, y_train) * 100)
print("----train accuracy score %s ----" % train_acc)
test_acc = float(svclassifier.score(X_test, y_test) * 100)
print("----test accuracy score %s ----" % test_acc)
# ### With Scaling
# Setup the pipeline steps: steps
steps = [("scaler", StandardScaler()), ("SVM", SVC(kernel="rbf"))]
# Create the pipeline: pipeline
pipeline = Pipeline(steps)
# Fit the pipeline to the training set: svc_scaled
svc_scaled = pipeline.fit(X_train, y_train)
# Compute and print metrics
print("Accuracy with Scaling: {}".format(svc_scaled.score(X_test, y_test)))
# ### Cross Validation
cv_results2 = cross_val_score(pipeline, X, y, cv=cv, n_jobs=-1)
print(cv_results2)
print("Average:", np.average(cv_results2))
# ### Generalization check
train_acc = float(svc_scaled.score(X_train, y_train) * 100)
print("----train accuracy score %s ----" % train_acc)
test_acc = float(svc_scaled.score(X_test, y_test) * 100)
print("----test accuracy score %s ----" % test_acc)
scaled_predictions = svc_scaled.predict(X_test)
# ### Classification reports and Confusion Matrix
print(classification_report(y_test, scaled_predictions))
acc = float(accuracy_score(y_test, scaled_predictions)) * 100
print("----accuracy score %s ----" % acc)
cm = confusion_matrix(y_test, scaled_predictions)
ax = plt.subplot()
sns.heatmap(cm, annot=True, fmt="g", ax=ax)
# labels, title and ticks
ax.set_xlabel("Predicted labels")
ax.set_ylabel("True labels")
ax.set_title("Confusion Matrix (SVM - RBF)")
ax.xaxis.set_ticklabels(tess_emotions)
ax.yaxis.set_ticklabels(tess_emotions)
# # SVM - Linear
from sklearn.svm import LinearSVC
steps2 = [("scaler", StandardScaler()), ("LinearSVM", LinearSVC())]
svml = Pipeline(steps2)
svml_res = svml.fit(X_train, y_train)
print("Accuracy with Scaling: {}".format(svml.score(X_test, y_test)))
svml_train_acc = float(svml_res.score(X_train, y_train) * 100)
print("----train accuracy score %s ----" % svml_train_acc)
svml_test_acc = float(svml_res.score(X_test, y_test) * 100)
print("----test accuracy score %s ----" % svml_test_acc)
svml_score = cross_val_score(svml, X, y, cv=cv, n_jobs=-1)
print(svml_score)
print("Average:", np.average(svml_score))
svml_pred = svml_res.predict(X_test)
print(svml_pred)
print(classification_report(y_test, svml_pred))
acc_svml = float(accuracy_score(y_test, svml_pred)) * 100
print("----accuracy score %s ----" % acc_svml)
cm_svml = confusion_matrix(y_test, svml_pred)
# df_cm_svml = pd.DataFrame(cm_svml)
# sn.heatmap(df_cm_svml, annot=True, fmt='')
# plt.show()
ax = plt.subplot()
sns.heatmap(cm_svml, annot=True, fmt="g", ax=ax)
# labels, title and ticks
ax.set_xlabel("Predicted labels")
ax.set_ylabel("True labels")
ax.set_title("Confusion Matrix (SVM - Linear)")
ax.xaxis.set_ticklabels(tess_emotions)
ax.yaxis.set_ticklabels(tess_emotions)
# # Random Forest
from sklearn.ensemble import RandomForestClassifier
rfm = RandomForestClassifier()
rfm_score = cross_val_score(rfm, X, y, cv=cv, n_jobs=-1)
print(rfm_score)
print("Average:", np.average(rfm_score))
rfm_res = rfm.fit(X_train, y_train)
rfm_train_acc = float(rfm_res.score(X_train, y_train) * 100)
print("----train accuracy score %s ----" % rfm_train_acc)
rfm_test_acc = float(rfm_res.score(X_test, y_test) * 100)
print("----test accuracy score %s ----" % rfm_test_acc)
rfm_pred = rfm_res.predict(X_test)
print(classification_report(y_test, rfm_pred))
acc = float(accuracy_score(y_test, rfm_pred)) * 100
print("----accuracy score %s ----" % acc)
cm_rfm = confusion_matrix(y_test, rfm_pred)
ax = plt.subplot()
sns.heatmap(cm_rfm, annot=True, fmt="g", ax=ax)
# labels, title and ticks
ax.set_xlabel("Predicted labels")
ax.set_ylabel("True labels")
ax.set_title("Confusion Matrix (Random Forest)")
ax.xaxis.set_ticklabels(tess_emotions)
ax.yaxis.set_ticklabels(tess_emotions)
# # Naive Bayes
from sklearn.naive_bayes import GaussianNB
nbm = GaussianNB().fit(X_train, y_train)
nbm_train_acc = float(nbm.score(X_train, y_train) * 100)
print("----train accuracy score %s ----" % nbm_train_acc)
nbm_test_acc = float(nbm.score(X_test, y_test) * 100)
print("----test accuracy score %s ----" % nbm_train_acc)
nbm_score = cross_val_score(nbm, X, y, cv=cv, n_jobs=-1)
print(nbm_score)
print("Average:", np.average(nbm_score))
nbm_pred = nbm.predict(X_test)
print(nbm_pred)
print(classification_report(y_test, nbm_pred))
acc_nbm = float(accuracy_score(y_test, nbm_pred)) * 100
print("----accuracy score %s ----" % acc_nbm)
cm_nbm = confusion_matrix(y_test, nbm_pred)
ax = plt.subplot()
sns.heatmap(cm_nbm, annot=True, fmt="g", ax=ax)
# labels, title and ticks
ax.set_xlabel("Predicted labels")
ax.set_ylabel("True labels")
ax.set_title("Confusion Matrix (Gaussian Naive Bayes)")
ax.xaxis.set_ticklabels(tess_emotions)
ax.yaxis.set_ticklabels(tess_emotions)
# # Logistic Regression
# [Resource](https://www.datacamp.com/tutorial/understanding-logistic-regression-python)
from sklearn.linear_model import LogisticRegression
lrm = LogisticRegression(solver="liblinear").fit(X_train, y_train)
lrm_train_acc = float(lrm.score(X_train, y_train) * 100)
print("----train accuracy score %s ----" % lrm_train_acc)
lrm_test_acc = float(lrm.score(X_test, y_test) * 100)
print("----test accuracy score %s ----" % lrm_train_acc)
lrm_score = cross_val_score(lrm, X, y, cv=cv, n_jobs=-1)
print(lrm_score)
print("Average:", np.average(lrm_score))
lrm_pred = lrm.predict(X_test)
print(lrm_pred)
print(classification_report(y_test, lrm_pred))
acc_lrm = float(accuracy_score(y_test, lrm_pred)) * 100
print("----accuracy score %s ----" % acc_lrm)
cm_lrm = confusion_matrix(y_test, lrm_pred)
ax = plt.subplot()
sns.heatmap(cm_lrm, annot=True, fmt="g", ax=ax)
# labels, title and ticks
ax.set_xlabel("Predicted labels")
ax.set_ylabel("True labels")
ax.set_title("Confusion Matrix (Logistic Regression)")
ax.xaxis.set_ticklabels(tess_emotions)
ax.yaxis.set_ticklabels(tess_emotions)
# # Decision Tree
from sklearn.tree import DecisionTreeClassifier
dtc = DecisionTreeClassifier(max_depth=50)
dtc.fit(X_train, y_train)
dtc_train_acc = float(dtc.score(X_train, y_train) * 100)
print("----train accuracy score %s ----" % dtc_train_acc)
dtc_test_acc = float(dtc.score(X_test, y_test) * 100)
print("----test accuracy score %s ----" % dtc_train_acc)
dtc_score = cross_val_score(dtc, X, y, cv=cv, n_jobs=-1)
print(dtc_score)
print("Average:", np.average(dtc_score))
dtc_pred = dtc.predict(X_test)
print(dtc_pred)
print(classification_report(y_test, dtc_pred))
acc_dtc = float(accuracy_score(y_test, dtc_pred)) * 100
print("----accuracy score %s ----" % acc_dtc)
cm_dtc = confusion_matrix(y_test, dtc_pred)
ax = plt.subplot()
sns.heatmap(cm_dtc, annot=True, fmt="g", ax=ax)
# labels, title and ticks
ax.set_xlabel("Predicted labels")
ax.set_ylabel("True labels")
ax.set_title("Confusion Matrix (Decision Tree)")
ax.xaxis.set_ticklabels(tess_emotions)
ax.yaxis.set_ticklabels(tess_emotions)
# # AdaBoost
from sklearn.ensemble import AdaBoostClassifier
abc = AdaBoostClassifier().fit(X_train, y_train)
abc_train_acc = float(abc.score(X_train, y_train) * 100)
print("----train accuracy score %s ----" % abc_train_acc)
abc_test_acc = float(abc.score(X_test, y_test) * 100)
print("----test accuracy score %s ----" % abc_train_acc)
abc_score = cross_val_score(abc, X, y, cv=cv, n_jobs=-1)
print(abc_score)
print("Average:", np.average(abc_score))
abc_pred = abc.predict(X_test)
print(abc_pred)
# print(classification_report(y_test,abc_pred))
acc_abc = float(accuracy_score(y_test, abc_pred)) * 100
print("----accuracy score %s ----" % acc_abc)
cm_abc = confusion_matrix(y_test, abc_pred)
ax = plt.subplot()
sns.heatmap(cm_abc, annot=True, fmt="g", ax=ax)
# labels, title and ticks
ax.set_xlabel("Predicted labels")
ax.set_ylabel("True labels")
ax.set_title("Confusion Matrix (AdaBoost)")
ax.xaxis.set_ticklabels(tess_emotions)
ax.yaxis.set_ticklabels(tess_emotions)
# # SGD
from sklearn.linear_model import SGDClassifier
sgd = SGDClassifier().fit(X_train, y_train)
sgd_train_acc = float(sgd.score(X_train, y_train) * 100)
print("----train accuracy score %s ----" % sgd_train_acc)
sgd_test_acc = float(sgd.score(X_test, y_test) * 100)
print("----test accuracy score %s ----" % sgd_train_acc)
sgd_score = cross_val_score(sgd, X, y, cv=cv, n_jobs=-1)
print(sgd_score)
print("Average:", np.average(sgd_score))
sgd_pred = sgd.predict(X_test)
print(sgd_pred)
# print(classification_report(y_test,abc_pred))
acc_sgd = float(accuracy_score(y_test, sgd_pred)) * 100
print("----accuracy score %s ----" % acc_sgd)
cm_sgd = confusion_matrix(y_test, sgd_pred)
ax = plt.subplot()
sns.heatmap(cm_sgd, annot=True, fmt="g", ax=ax)
# labels, title and ticks
ax.set_xlabel("Predicted labels")
ax.set_ylabel("True labels")
ax.set_title("Confusion Matrix (Stochastic Gradient Descent)")
ax.xaxis.set_ticklabels(tess_emotions)
ax.yaxis.set_ticklabels(tess_emotions)
# # Gradient Boosting
from sklearn.ensemble import GradientBoostingClassifier
gbc = GradientBoostingClassifier().fit(X_train, y_train)
gbc_train_acc = float(gbc.score(X_train, y_train) * 100)
print("----train accuracy score %s ----" % gbc_train_acc)
gbc_test_acc = float(gbc.score(X_test, y_test) * 100)
print("----test accuracy score %s ----" % gbc_train_acc)
# Very time consuming
gbc_score = cross_val_score(gbc, X, y, cv=cv, n_jobs=-1)
print(gbc_score)
print("Average:", np.average(gbc_score))
gbc_pred = gbc.predict(X_test)
print(gbc_pred)
print(classification_report(y_test, gbc_pred))
acc_gbc = float(accuracy_score(y_test, gbc_pred)) * 100
print("----accuracy score %s ----" % acc_gbc)
cm_gbc = confusion_matrix(y_test, gbc_pred)
ax = plt.subplot()
sns.heatmap(cm_gbc, annot=True, fmt="g", ax=ax)
# labels, title and ticks
ax.set_xlabel("Predicted labels")
ax.set_ylabel("True labels")
ax.set_title("Confusion Matrix (Gradient Boosting)")
ax.xaxis.set_ticklabels(tess_emotions)
ax.yaxis.set_ticklabels(tess_emotions)
# # MLP Classifier
from sklearn.neural_network import MLPClassifier
steps3 = [("scaler", StandardScaler()), ("MLP", MLPClassifier())]
pipeline_mlp = Pipeline(steps3)
mlp = pipeline_mlp.fit(X_train, y_train)
print("Accuracy with Scaling: {}".format(mlp.score(X_test, y_test)))
mlp_train_acc = float(mlp.score(X_train, y_train) * 100)
print("----train accuracy score %s ----" % mlp_train_acc)
mlp_test_acc = float(mlp.score(X_test, y_test) * 100)
print("----test accuracy score %s ----" % mlp_train_acc)
mlp_res = cross_val_score(mlp, X, y, cv=cv, n_jobs=-1)
print(mlp_res)
print("Average:", np.average(mlp_res))
mlp_pred = mlp.predict(X_test)
print(mlp_pred)
print(classification_report(y_test, mlp_pred))
acc_mlp = float(accuracy_score(y_test, mlp_pred)) * 100
print("----accuracy score %s ----" % acc_mlp)
cm_mlp = confusion_matrix(y_test, mlp_pred)
ax = plt.subplot()
sns.heatmap(cm_mlp, annot=True, fmt="g", ax=ax)
# labels, title and ticks
ax.set_xlabel("Predicted labels")
ax.set_ylabel("True labels")
ax.set_title("Confusion Matrix (Multi Layer Perceptron)")
ax.xaxis.set_ticklabels(tess_emotions)
ax.yaxis.set_ticklabels(tess_emotions)
# # KNN
from sklearn.neighbors import KNeighborsClassifier
steps4 = [("scaler", StandardScaler()), ("KNN", KNeighborsClassifier())]
pipeline_knn = Pipeline(steps4)
knn = pipeline_mlp.fit(X_train, y_train)
print("Accuracy with Scaling: {}".format(knn.score(X_test, y_test)))
knn_train_acc = float(knn.score(X_train, y_train) * 100)
print("----train accuracy score %s ----" % knn_train_acc)
knn_test_acc = float(knn.score(X_test, y_test) * 100)
print("----test accuracy score %s ----" % knn_train_acc)
knn_res = cross_val_score(knn, X, y, cv=cv, n_jobs=-1)
print(knn_res)
print("Average:", np.average(knn_res))
knn_pred = knn.predict(X_test)
print(knn_pred)
print(classification_report(y_test, knn_pred))
acc_knn = float(accuracy_score(y_test, knn_pred)) * 100
print("----accuracy score %s ----" % acc_knn)
cm_knn = confusion_matrix(y_test, knn_pred)
ax = plt.subplot()
sns.heatmap(cm_knn, annot=True, fmt="g", ax=ax)
# labels, title and ticks
ax.set_xlabel("Predicted labels")
ax.set_ylabel("True labels")
ax.set_title("Confusion Matrix (K Nearest Neighbour)")
ax.xaxis.set_ticklabels(tess_emotions)
ax.yaxis.set_ticklabels(tess_emotions)
import pandas as pd
import numpy as np
import os
import seaborn as sns
import librosa
import librosa.display
from IPython.display import Audio
import tensorflow as tf
import keras
import warnings
warnings.filterwarnings("ignore")
import matplotlib.pyplot as plt
paths = []
labels = []
for dirname, _, filename in os.walk("/kaggle/input/toronto-emotional-speech-set-tess/"):
for filename in filename:
paths.append(os.path.join(dirname, filename))
label = filename.split("_")[-1]
label = label.split(".")[0]
labels.append(label)
df = pd.DataFrame()
df["speech_data"] = paths
df["label"] = labels
df.head()
# paste here
paths = []
labels = []
for dirname, _, filename in os.walk(
"/content/drive/MyDrive/TESS Toronto emotional speech set data"
):
for filename in filename:
paths.append(os.path.join(dirname, filename))
label = filename.split("_")[-1]
label = label.split(".")[0]
labels.append(label)
df = pd.DataFrame()
df["speech_data"] = paths
df["label"] = labels
df.head()
df["label"].hist(color="pink")
# defining the waveplot and spectogram
def waveplot(data, sampling_rate, emotion):
plt.figure(figsize=(8, 4))
plt.title(emotion, size=15)
librosa.display.waveshow(data, sr=sampling_rate)
plt.show()
def spectogram(data, sampling_rate, emotion):
frequency_domain = librosa.stft(data)
freq_db = librosa.amplitude_to_db((frequency_domain))
plt.figure(figsize=(8, 4))
plt.title(emotion, size=15)
librosa.display.specshow(freq_db, sr=sampling_rate, x_axis="time", y_axis="hz")
plt.colorbar()
# calling function waveplot and spectoram on all the emotions [angry ,disgust,fear,happy,neural,pleasent surprised ]
emotion = "angry"
path = np.array(df["speech_data"][df["label"] == emotion])[0]
data, sampling_rate = librosa.load(path)
waveplot(data, sampling_rate, emotion)
spectogram(data, sampling_rate, emotion)
Audio(path)
emotion = "disgust"
path = np.array(df["speech_data"][df["label"] == emotion])[0]
data, sampling_rate = librosa.load(path)
waveplot(data, sampling_rate, emotion)
spectogram(data, sampling_rate, emotion)
Audio(path)
emotion = "fear"
path = np.array(df["speech_data"][df["label"] == emotion])[0]
data, sampling_rate = librosa.load(path)
waveplot(data, sampling_rate, emotion)
spectogram(data, sampling_rate, emotion)
Audio(path)
emotion = "happy"
path = np.array(df["speech_data"][df["label"] == emotion])[0]
data, sampling_rate = librosa.load(path)
waveplot(data, sampling_rate, emotion)
spectogram(data, sampling_rate, emotion)
Audio(path)
emotion = "neutral"
path = np.array(df["speech_data"][df["label"] == emotion])[0]
data, sampling_rate = librosa.load(path)
waveplot(data, sampling_rate, emotion)
spectogram(data, sampling_rate, emotion)
Audio(path)
emotion = "ps"
path = np.array(df["speech_data"][df["label"] == emotion])[0]
data, sampling_rate = librosa.load(path)
waveplot(data, sampling_rate, emotion)
spectogram(data, sampling_rate, emotion)
Audio(path)
# data augmentation /characterisric tuning of data to make the model invariant to these changes
def noise(data):
noise_amp = 0.035 * np.random.uniform() * np.amax(data)
data = data + noise_amp * np.random.normal(size=data.shape[0])
return data
def stretch(data, rate):
return librosa.effects.time_stretch(data, rate=rate)
def shift(data):
shift_range = int(np.random.uniform(low=-5, high=5) * 1000)
return np.roll(data, shift_range)
def pitch(data, sampling_rate, pitch_factor):
return librosa.effects.pitch_shift(data, sr=sampling_rate, n_steps=pitch_factor)
path = np.array(df["speech_data"][df["label"] == "happy"])[2]
data, sample_rate = librosa.load(path)
# given audio
plt.figure(figsize=(8, 4))
waveplot(data, sampling_rate, "happy")
Audio(path)
# noise injection
x = noise(data)
plt.figure(figsize=(14, 4))
waveplot(x, sampling_rate, "happy")
Audio(x, rate=sampling_rate)
# streching
x = stretch(data, 0.8)
plt.figure(figsize=(14, 4))
waveplot(x, sampling_rate, "happy")
Audio(x, rate=sampling_rate)
# shifting
x = shift(data)
plt.figure(figsize=(14, 4))
waveplot(x, sampling_rate, "happy")
Audio(x, rate=sample_rate)
# pitch
x = pitch(data, sampling_rate, 0.7)
plt.figure(figsize=(8, 4))
waveplot(x, sampling_rate, "happy")
Audio(x, rate=sampling_rate)
# feature extraction
# noise injection
x = noise(data)
plt.figure(figsize=(14, 4))
waveplot(x, sampling_rate, "happy")
Audio(x, rate=sampling_rate)
# streching
x = stretch(data, 0.8)
plt.figure(figsize=(14, 4))
waveplot(x, sampling_rate, "happy")
Audio(x, rate=sampling_rate)
# shifting
x = shift(data)
plt.figure(figsize=(14, 4))
waveplot(x, sampling_rate, "happy")
Audio(x, rate=sample_rate)
# pitch
x = pitch(data, sampling_rate, 0.7)
plt.figure(figsize=(8, 4))
waveplot(x, sampling_rate, "happy")
Audio(x, rate=sampling_rate)
# feature extraction
# 1. MFCC
# 2. Zero Crossing Rate
# 3. RMS
# 4. Chroma_shift
# 5. MelSpectogram
def feature_extraction(data):
# ZCR
result = np.array([])
zcr = np.mean(librosa.feature.zero_crossing_rate(y=data).T, axis=0)
result = np.hstack((result, zcr))
# Chroma_stft
stft = np.abs(librosa.stft(data))
chroma_stft = np.mean(
librosa.feature.chroma_stft(S=stft, sr=sampling_rate).T, axis=0
)
result = np.hstack((result, chroma_stft))
# MFCC
mfcc = np.mean(librosa.feature.mfcc(y=data, sr=sampling_rate).T, axis=0)
result = np.hstack((result, mfcc))
# Root Mean Square Value
rms = np.mean(librosa.feature.rms(y=data).T, axis=0)
result = np.hstack((result, rms))
# MelSpectogram
mel = np.mean(librosa.feature.melspectrogram(y=data, sr=sampling_rate).T, axis=0)
result = np.hstack((result, mel))
return result
def get_features(path, rate_stretch, pitch_factor):
data, sampling_rate = librosa.load(path, duration=2.5, offset=0.6)
# original data
r1 = feature_extraction(data)
result = np.array(r1)
# noise induced audio
noise_data = noise(data)
r2 = feature_extraction(noise_data)
result = np.vstack((result, r2))
# sretched and pitched
new_data = stretch(data, rate_stretch)
data_stretch_pitch = pitch(new_data, sampling_rate, pitch_factor)
r3 = feature_extraction(data_stretch_pitch)
result = np.vstack((result, r3))
return result
X, Y = [], []
for path, emotion in zip(df.speech_data, df.label):
feature = get_features(path, 0.8, 0.7)
for ele in feature:
X.append(ele)
Y.append(emotion)
len(X), len(Y), df.speech_data.shape
Features = pd.DataFrame(X)
Features["labels"] = Y
Features.to_csv("features.csv", index=False)
Features.head()
# data preparation
X = Features.iloc[:, :-1].values
Y = Features["labels"].values
from sklearn.preprocessing import StandardScaler, OneHotEncoder
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import classification_report
from sklearn.metrics import accuracy_score
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(X, Y, random_state=0, shuffle=True)
x_train.shape, y_train.shape, x_test.shape, y_test.shape ## do till here
import keras
from keras.callbacks import ReduceLROnPlateau
from keras.models import Sequential
from keras.layers import (
Dense,
Conv1D,
MaxPooling1D,
Flatten,
Dropout,
BatchNormalization,
)
from keras.utils import np_utils, to_categorical
from keras.callbacks import ModelCheckpoint
encoder = OneHotEncoder()
Y = encoder.fit_transform(np.array(Y).reshape(-1, 1)).toarray()
x_train, x_test, y_train, y_test = train_test_split(X, Y, random_state=0, shuffle=True)
x_train.shape, y_train.shape, x_test.shape, y_test.shape
# data scaling
scaler = StandardScaler()
x_train = scaler.fit_transform(x_train)
x_test = scaler.transform(x_test)
x_train.shape, y_train.shape, x_test.shape, y_test.shape
# transformation of data for compatibility
x_train = np.expand_dims(x_train, axis=2)
x_test = np.expand_dims(x_test, axis=2)
x_train.shape, y_train.shape, x_test.shape, y_test.shape
# Applying CNN
model = Sequential()
model.add(
Conv1D(
256,
kernel_size=5,
strides=1,
padding="same",
activation="relu",
input_shape=(x_train.shape[1], 1),
)
)
model.add(MaxPooling1D(pool_size=5, strides=2, padding="same"))
model.add(Conv1D(256, kernel_size=5, strides=1, padding="same", activation="relu"))
model.add(MaxPooling1D(pool_size=5, strides=2, padding="same"))
model.add(Conv1D(128, kernel_size=5, strides=1, padding="same", activation="relu"))
model.add(MaxPooling1D(pool_size=5, strides=2, padding="same"))
model.add(Dropout(0.2))
model.add(Conv1D(64, kernel_size=5, strides=1, padding="same", activation="relu"))
model.add(MaxPooling1D(pool_size=5, strides=2, padding="same"))
model.add(Flatten())
model.add(Dense(units=32, activation="relu"))
model.add(Dropout(0.3))
model.add(Dense(units=7, activation="softmax"))
model.compile(optimizer="adam", loss="categorical_crossentropy", metrics=["accuracy"])
model.summary()
rlrp = ReduceLROnPlateau(
monitor="loss", factor=0.4, verbose=0, patience=2, min_lr=0.0000001
)
history = model.fit(
x_train,
y_train,
batch_size=56,
epochs=50,
validation_data=(x_test, y_test),
callbacks=[rlrp],
)
print(
"Accuracy of our model on test data : ",
model.evaluate(x_test, y_test)[1] * 100,
"%",
)
epochs = [i for i in range(50)]
fig, ax = plt.subplots(1, 2)
train_acc = history.history["accuracy"]
train_loss = history.history["loss"]
test_acc = history.history["val_accuracy"]
test_loss = history.history["val_loss"]
fig.set_size_inches(20, 6)
ax[0].plot(epochs, train_loss, label="Training Loss")
ax[0].plot(epochs, test_loss, label="Testing Loss")
ax[0].set_title("Training & Testing Loss")
ax[0].legend()
ax[0].set_xlabel("Epochs")
ax[1].plot(epochs, train_acc, label="Training Accuracy")
ax[1].plot(epochs, test_acc, label="Testing Accuracy")
ax[1].set_title("Training & Testing Accuracy")
ax[1].legend()
ax[1].set_xlabel("Epochs")
plt.show()
pred_test = model.predict(x_test)
y_pred = encoder.inverse_transform(pred_test)
y_test = encoder.inverse_transform(y_test)
df = pd.DataFrame(columns=["Predicted Labels", "Actual Labels"])
df["Predicted Labels"] = y_pred.flatten()
df["Actual Labels"] = y_test.flatten()
df.head(10)
def feature_extraction(data):
# ZCR
result = np.array([])
zcr = np.mean(librosa.feature.zero_crossing_rate(y=data).T, axis=0)
result = np.hstack((result, zcr))
# Chroma_stft
stft = np.abs(librosa.stft(data))
chroma_stft = np.mean(
librosa.feature.chroma_stft(S=stft, sr=sampling_rate).T, axis=0
)
result = np.hstack((result, chroma_stft))
# MFCC
mfcc = np.mean(librosa.feature.mfcc(y=data, sr=sampling_rate).T, axis=0)
result = np.hstack((result, mfcc))
# Root Mean Square Value
rms = np.mean(librosa.feature.rms(y=data).T, axis=0)
result = np.hstack((result, rms))
# MelSpectogram
mel = np.mean(librosa.feature.melspectrogram(y=data, sr=sampling_rate).T, axis=0)
result = np.hstack((result, mel))
return result
def get_features(path, rate_stretch, pitch_factor):
data, sampling_rate = librosa.load(path, duration=2.5, offset=0.6)
# original data
r1 = feature_extraction(data)
result = np.array(r1)
# noise induced audio
noise_data = noise(data)
r2 = feature_extraction(noise_data)
result = np.vstack((result, r2))
# sretched and pitched
new_data = stretch(data, rate_stretch)
data_stretch_pitch = pitch(new_data, sampling_rate, pitch_factor)
r3 = feature_extraction(data_stretch_pitch)
result = np.vstack((result, r3))
return result
X, Y = [], []
for path, emotion in zip(df.speech_data, df.label):
feature = get_features(path, 0.8, 0.7)
for ele in feature:
X.append(ele)
Y.append(emotion)
# # Comparison
paper_results = [
0.8986,
0.9886,
0.7911,
0.79,
0.9929,
0.9089,
0.2643,
0.9757,
0.9875,
0.9964,
0.9886,
] # From paper
results = [
np.average(cv_results2),
np.average(svml_score),
np.average(rfm_score),
np.average(nbm_score),
np.average(lrm_score),
np.average(dtc_score),
np.average(abc_score),
np.average(sgd_score),
np.average(gbc_score),
np.average(mlp_res),
np.average(knn_res),
]
algo_labels = [
"SVM (RBF)",
"SVM (Linear)",
"Random Forest",
"Naive Bayes",
"Logistic Regression",
"Decision Tree",
"AdaBoost",
"SGD",
"GradBoosting",
"MLP",
"KNN",
]
res_df = pd.DataFrame(results)
algo_df = pd.DataFrame(algo_labels)
pres_df = pd.DataFrame(paper_results)
algo_df = algo_df.rename(columns={0: "Classifier"})
res_df = res_df.rename(columns={0: "Acc"})
pres_df = pres_df.rename(columns={0: "Acc"})
res_df["Type"] = "Ours"
pres_df["Type"] = "Paper"
data_res1 = pd.concat([algo_df, res_df], axis=1)
data_res2 = pd.concat([algo_df, pres_df], axis=1)
data_res = pd.concat([data_res1, data_res2], axis=0)
data_res
sns.set_theme(style="whitegrid")
plt.figure(figsize=(20, 10))
ax = sns.barplot(
data=data_res,
x="Classifier",
y="Acc",
hue="Type",
)
ax.set_xlabel("Classifier Algorithms", fontsize=20)
ax.set_ylabel("Test Accuracy", fontsize=20)
ax.set_title(
"Comparison Between Different Algorithms Based on Test Accuracy (5-Fold Repeated CV Avg Test Accuracy (Ours) vs Paper) "
)
# plt.legend(bbox_to_anchor=(1.05, 1), loc=2, borderaxespad=0.)
ax.legend(loc="upper left", bbox_to_anchor=(1, 1), fontsize=15)
|
# ## Se cargan las librerias
import numpy as np
import pandas as pd
import time
import tensorflow as tf
from sklearn.metrics import mean_absolute_error, mean_squared_error
import matplotlib.pyplot as plt
import cartopy.crs as ccrs
import cartopy.feature as cfeatureÇ
param = ["t", "precip"]
zone = "NW"
path = (
"/kaggle/input/meteonet/NW_Ground_Stations/NW_Ground_Stations/NW_Ground_Stations_"
)
cols = ["number_sta", "lat", "lon", "date"] + param
df = pd.concat(
[
pd.read_csv(
path + "2016.csv",
usecols=cols,
parse_dates=["date"],
infer_datetime_format=True,
),
pd.read_csv(
path + "2017.csv",
usecols=cols,
parse_dates=["date"],
infer_datetime_format=True,
),
pd.read_csv(
path + "2018.csv",
usecols=cols,
parse_dates=["date"],
infer_datetime_format=True,
),
],
axis=0,
)
def create_time_steps(length):
return list(range(-length, 0))
# ## Mapa de la temperatura usando cartopy
date = "2016-01-01T06:00:00"
d_sub = df[df["date"] == date]
# Coordinates of studied area boundaries (in °N and °E)
lllat = 46.25 # lower left latitude
urlat = 51.896 # upper right latitude
lllon = -5.842 # lower left longitude
urlon = 2 # upper right longitude
# Assuming d_sub is a DataFrame containing 'lon', 'lat', and the parameter (e.g., 'temperature' and 'precipitation') columns
params = ["t", "precip"] # Replace these with the correct column names for your data
date = "2016-01-01T06:00:00" # Replace this with the correct date for your data
fig, axs = plt.subplots(
nrows=1, ncols=2, figsize=(18, 5), subplot_kw={"projection": ccrs.PlateCarree()}
)
for ax, param in zip(axs, params):
# Plot the data
scatter = ax.scatter(d_sub["lon"], d_sub["lat"], c=d_sub[param], cmap="jet")
fig.colorbar(
scatter, ax=ax, label=f"{param} (units)"
) # Replace 'units' with the appropriate unit for each parameter
ax.set_title(f"{date} - {param}")
ax.coastlines(resolution="50m", linewidth=1)
ax.add_feature(cfeatureÇ.BORDERS.with_scale("50m"))
# Adjust the plot to the area we defined
ax.set_xlim(lllon, urlon)
ax.set_ylim(lllat, urlat)
plt.show()
number_sta = 91200002
uni_data = df[(df["number_sta"] == number_sta)]["t"]
uni_data.index = df[(df["number_sta"] == number_sta)]["date"]
plt.figure(figsize=(10, 5))
plt.ylabel("Temperatura (ºK)")
plt.title("Evolución de la temperatura en la estación " + str(number_sta))
uni_data.plot(subplots=True)
plt.show()
number_sta = 91200002
uni_data_2 = df[(df["number_sta"] == number_sta)]["precip"]
uni_data_2.index = df[(df["number_sta"] == number_sta)]["date"]
plt.figure(figsize=(10, 5))
plt.ylabel("Precipitacion (km.m^2)")
plt.title("Evolución de la precipitacion en la estación " + str(number_sta))
uni_data_2.plot(subplots=True)
plt.show()
df
df = df[(df["number_sta"] == number_sta)][params + ["date"]]
df.index = df["date"]
df = df.drop(columns=["date"])
df
# Fill missing values with mean
for col in params:
df[col] = df[col].fillna(df[col].mean())
coeff_train = 0.7
TRAIN_SPLIT = round(df.shape[0] * coeff_train)
def multivariate_data(x_data, start_index, end_index, input_size, target_size):
data = []
labels = []
start_index = start_index + input_size
if end_index is None:
end_index = len(x_data) - target_size
for i in range(start_index, end_index):
data.append(
x_data[(i - input_size) : i, :]
) # Updated to handle multiple variables
labels.append(np.max(x_data[i : i + target_size, :], axis=0))
return np.array(data), np.array(labels)
univariate_past_history = 240
univariate_future_target = 240
x_train, y_train = multivariate_data(
df.to_numpy(), 0, TRAIN_SPLIT, univariate_past_history, univariate_future_target
)
x_val, y_val = multivariate_data(
df.to_numpy(), TRAIN_SPLIT, None, univariate_past_history, univariate_future_target
)
x_train_mean = np.mean(x_train, axis=(0, 1))
x_train_std = np.std(x_train, axis=(0, 1))
y_train_mean = np.mean(y_train, axis=0)
y_train_std = np.std(y_train, axis=0)
x_data_train = (x_train - x_train_mean) / x_train_std
x_data_val = (x_val - x_train_mean) / x_train_std
y_data_train = (y_train - y_train_mean) / y_train_std
y_data_val = (y_val - y_train_mean) / y_train_std
# Update the LSTM model to handle multiple outputs
simple_lstm_model = tf.keras.models.Sequential(
[
tf.keras.layers.LSTM(
64, input_shape=x_data_train.shape[-2:], return_sequences=True
),
tf.keras.layers.Dropout(0.3),
tf.keras.layers.LSTM(32, return_sequences=True),
tf.keras.layers.Dropout(0.3),
tf.keras.layers.LSTM(16, return_sequences=True),
tf.keras.layers.Dropout(0.3),
tf.keras.layers.LSTM(8),
tf.keras.layers.Dropout(0.3),
tf.keras.layers.Dense(2),
]
)
optimizer = tf.keras.optimizers.Adam(learning_rate=0.001)
simple_lstm_model.compile(optimizer=optimizer, loss="mse")
BATCH_SIZE = 64
BUFFER_SIZE = 1000
train_univariate = tf.data.Dataset.from_tensor_slices((x_data_train, y_data_train))
train_univariate = (
train_univariate.cache().shuffle(BUFFER_SIZE).batch(BATCH_SIZE).repeat()
)
val_univariate = tf.data.Dataset.from_tensor_slices((x_data_val, y_data_val))
val_univariate = val_univariate.batch(BATCH_SIZE).repeat()
EPOCHS = 5
STEPS_PER_EPOCH = (x_data_train.shape[0] / BATCH_SIZE // 1) + 1
VAL_STEPS = (x_data_val.shape[0] / BATCH_SIZE // 1) + 1
simple_lstm_model.fit(
train_univariate,
epochs=EPOCHS,
steps_per_epoch=STEPS_PER_EPOCH,
validation_data=val_univariate,
validation_steps=VAL_STEPS,
)
def baseline(history):
return np.mean(history, axis=0)
def show_result(index, plot_data, delta):
labels = ["History : x", "True Future : y", "Prediction", "Baseline : Max(History)"]
marker = [".-", "rx", "go", "go"]
colors = ["blue", "red", "green", "darkblue"]
time_steps = create_time_steps(plot_data[0].shape[0])
if delta:
future = delta
else:
future = 0
fig, (ax1, ax2) = plt.subplots(2, 1)
fig.suptitle("Sample example " + str(ind))
axes = [ax1, ax2]
ylabel = ["Standardized temperature", "Standardized precipitation"]
for j, ax in enumerate(axes):
for i, x in enumerate(plot_data):
if i:
ax.plot(
future,
plot_data[i][j],
marker[i],
markersize=10,
label=labels[i],
color=colors[i],
)
else:
ax.plot(
time_steps,
plot_data[i][:, j].flatten(),
marker[i],
label=labels[i],
color=colors[i],
)
ax.legend()
ax.set_xlim([time_steps[0], (future + 5) * 2])
ax.set_xlabel("Time-Step")
ax.set_ylabel(ylabel[j])
plt.show()
ind = 0
for x, y in val_univariate.take(3):
y_true = y[ind].numpy()
basel = baseline(x[ind].numpy())
print("mse baseline", mean_squared_error(y_true, basel))
print("mse LSTM", mean_squared_error(y_true, simple_lstm_model.predict(x)[ind]))
plot = show_result(
ind,
[
x[ind].numpy(),
y[ind].numpy(),
simple_lstm_model.predict(x)[ind],
baseline(x[ind].numpy()),
],
0,
)
|
# ## Intel Classification Problem:
# The dataset contains images of natural scenes around the world.
# This Data contains around 25k images of size 150x150 distributed under 6 categories.
# {'buildings' -> 0,
# 'forest' -> 1,
# 'glacier' -> 2,
# 'mountain' -> 3,
# 'sea' -> 4,
# 'street' -> 5 }
# The objectif of this project is to build a convolutional neural networks to classify these images into their respective category.
# We are first going to build a basic model, and then use transfer learning to improve performance.
# ## What are Convolutional Neural Networks?
# A Convolutional Neural Network (CNN) is a type of deep neural network that is designed to process data with a grid-like topology, such as images or videos. It is inspired by the structure and function of the visual cortex in the brain, which is responsible for processing visual information.
# CNNs consist of multiple layers of interconnected neurons that perform a series of mathematical operations on the input data. The core building blocks of a CNN are convolutional layers, pooling layers, and fully connected layers.
# Convolutional layers apply a set of filters to the input data, each filter detecting a specific pattern or feature in the input. The filters slide over the input data, performing element-wise multiplication and summing up the results to produce a feature map. The pooling layers then downsample the feature map, reducing its size while retaining the most important features. Finally, the fully connected layers process the output of the convolutional and pooling layers, using it to make a prediction.
# CNNs have proven to be very effective in image classification, object detection, and other computer vision tasks, and are widely used in various fields such as autonomous driving, medical image analysis, and natural language processing.
# ### Importing the required libraries:
import numpy as np
import matplotlib.pyplot as plt
from PIL import Image
import tensorflow as tf
import os
import cv2
from sklearn.model_selection import train_test_split
from sklearn.utils import shuffle
from keras.utils import np_utils
from tensorflow.keras.layers import Dense, Conv2D, MaxPool2D, Flatten, Dropout
from tensorflow.keras.models import Sequential, Model
from tensorflow.keras.optimizers import Adam
from keras.preprocessing.image import ImageDataGenerator
from tensorflow.keras.applications.vgg16 import VGG16
from tensorflow.keras.applications.vgg16 import preprocess_input
# ### Loading the images:
d = {}
X_train = []
train_path = "/kaggle/input/intel-image-classification/seg_train/seg_train"
for folder in os.listdir(train_path):
image_folder = train_path + "/" + folder
images = os.listdir(image_folder)
d[folder] = len(images)
for image in images:
image_path = image_folder + "/" + image
x = Image.open(image_path)
x = x.convert("RGB")
x = np.array(x)
img_resize = cv2.resize(x, (224, 224))
X_train.append(img_resize)
for key in d.keys():
print("Found {} images for {}!".format(d[key], key))
# ### Creating the train set:
X_train = np.array(X_train)
y_train = np.ones((14034,), dtype="int32")
y_train[:2512] = 3 # for moutain
y_train[2512:4894] = 5 # for street
y_train[4894:7085] = 0 # for buildings
y_train[7085:9359] = 4 # for sea
y_train[9359:11630] = 1 # for forest
y_train[11630:] = 2 # for glacier
X_train, y_train = shuffle(X_train, y_train, random_state=42)
def get_label(id):
label_dict = {
0: "buildings",
1: "forest",
2: "glacier",
3: "mountain",
4: "sea",
5: "street",
}
return label_dict[id]
j = np.random.randint(0, 10000)
plt.imshow(X_train[j])
plt.xlabel(get_label(y_train[j]))
y_train = np_utils.to_categorical(y_train, 6)
X_train, X_val, y_train, y_val = train_test_split(X_train, y_train, test_size=0.1)
# ### What is data augmentation :
# Data augmentation is a technique used in machine learning and computer vision to artificially increase the size of a training dataset by creating new data from existing data. The process involves applying a set of transformations to the original data, such as rotation, scaling, flipping, cropping, adding noise, or changing the brightness and contrast.
# The goal of data augmentation is to improve the performance of machine learning models by providing more training data to learn from, which can help prevent overfitting and improve generalization. By introducing variations in the training data, the model can learn to recognize the same object under different conditions, such as changes in lighting or viewpoint, and become more robust to these variations.
# ***Keras' ImageDataGenerator*** work as a python generator that allows us to generate new data on the fly, this allows you to train on an almost infinite stream of augmented image data without running out of memory.
aug = ImageDataGenerator(
preprocessing_function=preprocess_input,
rotation_range=25,
width_shift_range=0.1,
height_shift_range=0.1,
shear_range=0.2,
zoom_range=0.2,
horizontal_flip=True,
fill_mode="nearest",
)
val_aug = ImageDataGenerator(preprocessing_function=preprocess_input)
def create_model():
model = Sequential()
model.add(
Conv2D(
64,
(3, 3),
padding="same",
strides=(2, 2),
activation="relu",
input_shape=(224, 224, 3),
)
)
model.add(MaxPool2D(2, 2))
model.add(Conv2D(128, (3, 3), activation="relu"))
model.add(MaxPool2D(2, 2))
model.add(Conv2D(256, (3, 3), activation="relu"))
model.add(MaxPool2D(2, 2))
model.add(Conv2D(512, (3, 3), activation="relu"))
model.add(MaxPool2D(2, 2))
model.add(Conv2D(512, (3, 3), activation="relu"))
model.add(MaxPool2D(2, 2))
model.add(Flatten())
model.add(Dense(128, activation="relu"))
model.add(Dense(64, activation="relu"))
model.add(Dropout(0.5))
model.add(Dense(6, activation="softmax"))
return model
my_model = create_model()
my_model.summary()
my_model.compile(
optimizer=Adam(learning_rate=0.001),
loss="categorical_crossentropy",
metrics=["accuracy"],
)
# ### The problem with training with a large amount of images:
# Normally we would use X_train, y_train created previously to train our CNN, but since the amount of images is large a certain problem might arise which is:
# Memory constraints: It could happen that your Kaggle notebook does not have enough memory to handle large datasets. When working with a large dataset, it's important to ensure that you have enough memory to load the data and perform the necessary computations. When working with a dataset with over 14000 training images, the ressources available might not be able to handle it during training.
# To address these issues, we can try using a generator to load the images in batches rather than loading the entire dataset into memory at once.
# For that, we can use the method 'flow_from_directory' of ImageDataGenerator that will generate batches of data for training or evaluation from a directory structure.
train_data = aug.flow_from_directory(
"/kaggle/input/intel-image-classification/seg_train/seg_train",
target_size=(224, 224),
batch_size=128,
class_mode="categorical",
)
val_data = val_aug.flow_from_directory(
"/kaggle/input/intel-image-classification/seg_test/seg_test",
target_size=(224, 224),
batch_size=128,
shuffle=False,
class_mode="categorical",
)
# ### Model training:
my_model_history = my_model.fit(train_data, epochs=50, validation_data=val_data)
# ### Making predictions and visualizing errors::
predict = my_model.predict_generator(val_data, steps=3000)
y_pred_classes = np.argmax(predict, axis=1)
from sklearn.metrics import accuracy_score, confusion_matrix, ConfusionMatrixDisplay
print(accuracy_score(val_data.classes, y_pred_classes))
ConfusionMatrixDisplay(confusion_matrix(val_data.classes, y_pred_classes)).plot()
X_test = []
test_path = "/kaggle/input/intel-image-classification/seg_test/seg_test"
for folder in sorted(os.listdir(test_path)):
image_folder = test_path + "/" + folder
images = sorted(os.listdir(image_folder))
print(
"Loaded the images of -" + "{} having {} images. \n".format(folder, len(images))
)
for image in images:
image_path = image_folder + "/" + image
x = Image.open(image_path)
x = x.convert("RGB")
x = np.array(x)
img_resize = cv2.resize(x, (224, 224))
X_test.append(img_resize)
def plot_errors(true_label, predicted_label):
rows, cols = 1, 5
fig, ax = plt.subplots(rows, cols, figsize=(10, 3))
x = (y_pred_classes == predicted_label) & (val_data.classes == true_label)
y = np.where(x == True)[0]
z = np.random.randint(0, len(y), 5)
fig.suptitle(
"Predicted : {}, Real : {}".format(
get_label(predicted_label), get_label(true_label)
),
fontsize=11,
)
for col in range(cols):
ax[col].imshow(X_test[y[z[col]]])
ax[col].tick_params(
axis="both",
which="both",
bottom=False,
left=False,
top=False,
labelbottom=False,
labelleft=False,
)
plot_errors(0, 5)
plot_errors(4, 3)
plot_errors(3, 2)
plot_errors(2, 3)
# ## What is transfer learning?
# Transfer learning is a machine learning technique where a model trained on one task is re-purposed to solve a different but related task. Rather than starting the training process from scratch, the pre-trained model is used as a starting point and then fine-tuned or further trained on the new task.
# Transfer learning is based on the idea that features learned from one task can be useful for solving other related tasks. For example, a model trained to recognize objects in images can be fine-tuned to recognize specific types of objects in new images. By using transfer learning, the model can learn to perform the new task faster and with less training data than if it were trained from scratch.
# Transfer learning is widely used in computer vision. It can help to overcome the limitations of small datasets, reduce the amount of training time required, and improve the overall performance of the model.
# Here we are going to use VGG16 which is a known architecture for image classification tasks, and it has achieved state-of-the-art performance on several benchmarks.
base_model = VGG16(weights="imagenet", include_top=False, input_shape=(224, 224, 3))
base_model.trainable = False
inputs = tf.keras.Input(shape=(224, 224, 3))
x = Flatten()(base_model(inputs))
y = Dense(128, activation="relu")(x)
z = Dense(64, activation="relu")(y)
outputs = Dense(6, activation="softmax")(z)
model = Model(inputs, outputs)
model.summary()
model.compile(
optimizer=Adam(learning_rate=0.001),
loss="categorical_crossentropy",
metrics=["accuracy"],
)
history = model.fit(train_data, epochs=20, validation_data=val_data)
# ### Visualizing Loss and accuracy curves:
fig, ax = plt.subplots(1, 2, figsize=(10, 5))
ax[0].plot(history.epoch, history.history["loss"], label="Train Loss")
ax[0].plot(history.epoch, history.history["val_loss"], label="Test Loss")
ax[1].plot(history.epoch, history.history["accuracy"], label="Train Accuracy")
ax[1].plot(history.epoch, history.history["val_accuracy"], label="Test Accuracy")
ax[0].legend()
ax[1].legend()
plt.show()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
#
# # Project-3 .Booking reviews
# Прогнозирование рейтинга отеля на Booking.
# ## 1.1 Задание
# Представьте, что вы работаете дата-сайентистом в компании Booking. Одна из проблем компании — это нечестные отели, которые накручивают себе рейтинг. Одним из способов обнаружения таких отелей является построение модели, которая предсказывает рейтинг отеля. Если предсказания модели сильно отличаются от фактического результата, то, возможно, отель ведёт себя нечестно, и его стоит проверить. Вам поставлена задача создать такую модель.
# ### Описание данных
# |**Признак** | **Описание** |
# |------------------------------------------- |---------------------------------------------------------------------------|
# |*hotel_address* | адрес отеля
# |*review_date* | дата, когда рецензент разместил соответствующий отзыв
# |*average_score* |средний балл отеля, рассчитанный на основе последнего комментария за последний год
# |*hotel_name* |название отеля
# |*reviewer_nationality* |страна рецензента
# |*negative_review* |отрицательный отзыв, который рецензент дал отелю
# |*review_total_negative_word_counts* |общее количество слов в отрицательном отзыве
# |*positive_review* |положительный отзыв, который рецензент дал отелю
# |*review_total_positive_word_counts* |общее количество слов в положительном отзыве
# |*reviewer_score* |оценка, которую рецензент поставил отелю на основе своего опыта
# |*total_number_of_reviews_reviewer_has_given* |количество отзывов, которые рецензенты дали в прошлом
# |*total_number_of_reviews* |общее количество действительных отзывов об отеле
# |*tags* |теги, которые рецензент дал отелю
# |*days_since_review* |количество дней между датой проверки и датой создания датасета
# |*additional_number_of_scoring* |количество оценок без отзыва
# |*lat* |географическая широта отеля
# |*lng* |географическая долгота отеля
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import plotly.express as px
from plotly.subplots import make_subplots
import plotly.graph_objects as go
import seaborn as sns
import category_encoders as ce
from sklearn import metrics
from sklearn import preprocessing
from sklearn.ensemble import RandomForestRegressor
from sklearn.preprocessing import RobustScaler
from sklearn.model_selection import train_test_split
from textblob import TextBlob
from geopy.distance import geodesic as GD
pd.options.mode.chained_assignment = None
# ### 1.2 Загрузка данных
# всегда фиксируйте RANDOM_SEED, чтобы ваши эксперименты были воспроизводимы!
RANDOM_SEED = 42
# добавим пикселей для графиков, чтобы не так мыльно и уныло смотрелись
sns.set(rc={"figure.dpi": 300, "savefig.dpi": 300})
# форматируем вывод данных pandas, чтобы реже любоваться троеточием
pd.set_option("display.max_colwidth", None)
pd.set_option("display.float_format", "{:.3f}".format)
pd.set_option("display.max_rows", 50)
# Подгрузим наши данные из соревнования
DATA_DIR = "/kaggle/input/sf-booking/"
df_train = pd.read_csv(DATA_DIR + "/hotels_train.csv") # датасет для обучения
df_test = pd.read_csv(DATA_DIR + "hotels_test.csv") # датасет для предсказания
sample_submission = pd.read_csv(DATA_DIR + "/submission.csv") # самбмишн
# ВАЖНО! дря корректной обработки признаков объединяем трейн и тест в один датасет
df_train["sample"] = 1 # помечаем где у нас трейн
df_test["sample"] = 0 # помечаем где у нас тест
df_test[
"reviewer_score"
] = 0 # в тесте у нас нет значения reviewer_score, мы его должны предсказать, поэтому пока просто заполняем нулями
data = pd.concat([df_train, df_test], axis=0, ignore_index=True) # объединяем
data.info()
# Все признаки датасета можно условно разделить на три категории:
# 1. Признаки, описывающие отель
# 2. Признаки, описывающие рецензента
# 3. Признаки, описывающие отзыв
# ## 2. Анализ признаков, описывающих отель
# ### 2.1 hotel_address
data["hotel_address"].head()
# Значение признака имеет следующую структуру:
# * адрес
# * почтовый индекс
# * город
# * страна
# Используем эту информацию для создания новых признаков: 'hotel_city' и 'hotel_country'.
# из столбца с адресом отелей получаем новые признаки: страна и город отеля
data["hotel_country"] = data["hotel_address"].apply(
lambda x: x.split()[-1] if x.split()[-1] != "Kingdom" else " ".join(x.split()[-2:])
)
print(data["hotel_country"].value_counts())
print()
data["hotel_city"] = data["hotel_address"].apply(
lambda x: x.split()[-2] if x.split()[-1] != "Kingdom" else x.split()[-5]
)
print(data["hotel_city"].value_counts())
# Создаем функцию для визуализации категорийных признаков
def visual_cat(df, col, n=31):
"""A function for visualizing categorical features.
Builds a px.treemap for the number of reviews by category
and px.bar for the average score for these categories.
Args:
df (pandas.DataFrame): researched dataset
col (pandas.Series): researched future
n (int): number of categories
"""
tree_data = (
df.groupby(col)["sample"]
.count()
.sort_values(ascending=False)
.nlargest(n)
.reset_index(name="Counts")
)
bar_data = (
data.groupby(col)["reviewer_score"]
.mean()
.round(2)
.sort_values(ascending=False)
.nlargest(n)
.reset_index(name="Score")
)
fig_tree = px.treemap(
data_frame=tree_data,
path=[col],
values="Counts",
height=350,
width=600,
title="Количество отзывов по группам",
)
fig_tree.show()
fig_bar = px.bar(
data_frame=bar_data,
x=col,
y="Score",
color=col,
text="Score",
orientation="v",
height=350,
width=600,
title="Средний балл отелей по группам",
)
fig_bar.show()
visual_cat(df=data, col="hotel_country")
visual_cat(df=data, col="hotel_city", n=6)
# Признаки "hotel_city" и "hotel_country" полностью дублируют друг друга. Оставляем эти признаки для дальнейших исследований. Максимальный средний бал у отелей Испании, минимальный - у отелей Великобритании.
# ### 2.2 hotel_name
print(f"Количество уникальных названий отелей: {data['hotel_name'].nunique()}")
print(f"Количество уникальных адресов отелей: {data['hotel_address'].nunique()}")
# проверим есть ли отели с одинаковыми названиями, но разными адресами
grouped_hotels = (
data.groupby(["hotel_name", "hotel_address"])["sample"]
.count()
.sort_values(ascending=False)
)
display(
grouped_hotels.groupby("hotel_name").count().sort_values(ascending=False).head(5)
)
# получаем имена таких отелей и их количество
duplicated_hotel_name = (
grouped_hotels.groupby("hotel_name").count().sort_values(ascending=False).index[0]
)
n_of_duplicated = (
grouped_hotels.groupby("hotel_name").count().sort_values(ascending=False)[0]
)
print(
"\nВсего "
+ str(n_of_duplicated)
+ " отеля '"
+ duplicated_hotel_name
+ "' с различными адресами.\n"
)
display(
data[data["hotel_name"] == duplicated_hotel_name]["hotel_address"].value_counts()
)
# Переименуем отели 'Hotel Regina' добавляя название города
adress_to_rename_Barcelona = (
data[data["hotel_name"] == duplicated_hotel_name]["hotel_address"]
.value_counts()
.index[0]
)
adress_to_rename_Vienna = (
data[data["hotel_name"] == duplicated_hotel_name]["hotel_address"]
.value_counts()
.index[1]
)
adress_to_rename_Milan = (
data[data["hotel_name"] == duplicated_hotel_name]["hotel_address"]
.value_counts()
.index[2]
)
data.loc[
(data["hotel_address"] == adress_to_rename_Barcelona), "hotel_name"
] = "Hotel Regina Barcelona"
data.loc[
(data["hotel_address"] == adress_to_rename_Vienna), "hotel_name"
] = "Hotel Regina Vienna"
data.loc[
(data["hotel_address"] == adress_to_rename_Milan), "hotel_name"
] = "Hotel Regina Milan"
print("Всего " + str(data["hotel_name"].nunique()) + " уникальных отелей.")
# Из названия отеля мы можем узнать сеть, которой он принадлежит. Создаем новый признак 'hotel_chain'
largest_hotel_chains = [
"Hilton",
"Barriott",
"Hyatt",
"Sheraton",
"Holiday Inn",
"Best Western",
"Crowne Plaza",
"Radisson",
"Britannia",
"Mercure",
"NH",
"Novotel",
]
def get_hotel_chain(hotel_name):
"""A function to determine whether a hotel
belongs to one of the chains by its name.
Args:
hotel_name (str): hotel name
Returns:
chain (str): chain name
"""
for chain in largest_hotel_chains:
if chain in hotel_name:
return chain
return "Undefined chain"
data["hotel_chain"] = data["hotel_name"].apply(get_hotel_chain)
data["hotel_chain"].value_counts()
visual_cat(data, "hotel_chain", 12)
# Только четверть всех отелей принадлежит определенной сети. Самый высокий средний балл у отелей сети Crowne Plaza, самый низкий - у сети Britannia.
# ### 2.3 lat и lng
# Данные признаки имеют пропуски. Мы можем их заполнить, так как знаем название и адрес отеля.
# создаем списки индексов записей в которых есть пропуски координат отелей и проверяем их идентичность
hotel_index_without_lat = list(data[data["lat"].isna() == True].index)
hotel_index_without_lng = list(data[data["lng"].isna() == True].index)
print(hotel_index_without_lat == hotel_index_without_lng)
print(
f"Общее количество записей с пропущенными координатами {len(hotel_index_without_lat)}"
)
# создаем список имен отелей без координат
nul_coor_hotels = data[data["lat"].isna() == True]["hotel_name"].value_counts()
hotels_list = list(nul_coor_hotels.index)
hotels_list
# создаем словарь с координатами данных отелей
hotels_coor = {
"Fleming s Selection Hotel Wien City": [48.20953762534306, 16.353445198051627],
"Hotel City Central": [48.21373711264242, 16.379908198051773],
"Hotel Atlanta": [48.220589085614826, 16.355829326887893],
"Maison Albar Hotel Paris Op ra Diamond": [48.875416068138684, 2.3233490827330874],
"Hotel Daniel Vienna": [48.18897203791783, 16.38377969805083],
"Hotel Pension Baron am Schottentor": [48.21698150234599, 16.360153962634904],
"Austria Trend Hotel Schloss Wilhelminenberg Wien": [
48.22022075977996,
16.287605889524585,
],
"Derag Livinghotel Kaiser Franz Joseph Vienna": [
48.24986700746027,
16.35143806924949,
],
"NH Collection Barcelona Podium": [41.398032508061526, 2.178148084530765],
"City Hotel Deutschmeister": [48.22101628770906, 16.36661149805206],
"Hotel Park Villa": [48.233379, 16.345510],
"Cordial Theaterhotel Wien": [48.21117559983704, 16.35126149555466],
"Holiday Inn Paris Montmartre": [48.89504057540539, 2.3424128967362043],
"Roomz Vienna": [48.19326450823598, 16.44164878115289],
"Mercure Paris Gare Montparnasse": [48.84005228479027, 2.3235456428967862],
"Renaissance Barcelona Hotel": [41.40224270585638, 2.192538019639329],
"Hotel Advance": [41.38343167642923, 2.1629495978105107],
}
# создаем словарь с данными городов, в которых расположены эти отели: широта, долгота, площадь города
city_info = {
"London": [51.509792839719985, -0.11271152139656829, 1706],
"Barcelona": [41.38743213521596, 2.1878525754382365, 101],
"Paris": [48.86052268694293, 2.3378305054065533, 105],
"Amsterdam": [52.373503451385275, 4.8997592221035, 219],
"Vienna": [48.20959417455436, 16.369159260825704, 415],
"Milan": [45.46333908667008, 9.191085064501934, 183],
}
# создаем словарь с координатами аэропортов городов, в которых расположены эти отели
city_airport_coor = {
"London": [51.470970478463485, -0.4539386225916353],
"Barcelona": [41.29819139987498, 2.08407948816106],
"Paris": [49.00860015550327, 2.5529518146013754],
"Amsterdam": [52.311215140719774, 4.768994284266432],
"Vienna": [48.11370272268876, 16.575215723658697],
"Milan": [45.630608899633756, 8.72726519572734],
}
# Заполняем пропуски координат значениями словаря hotels_coor.
# После этого мы можем создать три новых признака:
# * удаленность от центра города
# * удаленность от аэропорта
# * отношение расстояния от отеля до центра города к площади этого города
# заполняем пропуски координат отелей в нашем датасете значениями из словаря
data["lat"] = data.apply(
lambda x: hotels_coor[x["hotel_name"]][0]
if x["hotel_name"] in hotels_list
else x["lat"],
axis=1,
)
data["lng"] = data.apply(
lambda x: hotels_coor[x["hotel_name"]][1]
if x["hotel_name"] in hotels_list
else x["lng"],
axis=1,
)
# создаем новые признаки: координаты центров городов, координаты аэропортов, площадь города
data["city_center_lat"] = data["hotel_city"].apply(lambda x: city_info[x][0])
data["city_center_lng"] = data["hotel_city"].apply(lambda x: city_info[x][1])
data["city_airport_lat"] = data["hotel_city"].apply(lambda x: city_airport_coor[x][0])
data["city_airport_lng"] = data["hotel_city"].apply(lambda x: city_airport_coor[x][1])
data["city_square"] = data["hotel_city"].apply(lambda x: city_info[x][2])
# Для удобства дальнейших преобразований объединим интересующие нас признаки в один датасет.
# создаем новый датасет
col_coor_list = [
"city_center_lat",
"city_center_lng",
"city_airport_lat",
"city_airport_lng",
"lat",
"lng",
"city_square",
]
data_coor = data[col_coor_list]
data_coor.info()
# Вычисляем расстояния от отеля до центра города и аэропорта, используя модуль geodesic библиотеки geopy.
# создаем новые признаки: растояние от отеля до центра города и растояние от отеля до аэропорта в метрах
data_coor["distance_to_center"] = data_coor.apply(
lambda x: round(
GD((x["lat"], x["lng"]), (x["city_center_lat"], x["city_center_lng"])).m
),
axis=1,
)
data_coor["distance_to_airport"] = data_coor.apply(
lambda x: round(
GD((x["lat"], x["lng"]), (x["city_airport_lat"], x["city_airport_lng"])).m
),
axis=1,
)
# посмотрим на статистику новых признаков
print(data_coor["distance_to_center"].describe())
print()
print(data_coor["distance_to_airport"].describe())
# Среди значений признаков distance_to_center и distance_to_airport нет выбросов. Все расстояния не выходят за пределы разумного для современной европейской столицы.
# Визуализируем распределение полученныч признаков.
# создаем функцию для визуализации распределения непрерывных признаков
def visual_num(df, col):
"""A function for visualizing numerical features.
Builds a px.histogram and px.box for the numerical feature.
Args:
df (pandas.DataFrame): researched dataset
col (pandas.Series): researched future
"""
future_hist = px.histogram(
df[col],
height=500,
width=700,
title="Распределение признака",
labels={"value": col},
marginal="box",
)
future_hist.show()
visual_num(data_coor, "distance_to_center")
visual_num(data_coor, "distance_to_airport")
# Также создадим признак,показывающий как условный радиус города относится к расстоянию от отеля до центра города.
data_coor["dist_square_ratio"] = round(
((data_coor["city_square"] * 1000000 / 3.14) ** 0.5)
/ data_coor["distance_to_center"],
2,
)
data_coor["dist_square_ratio"].describe()
# визуализируем полученный признак
visual_num(data_coor, "dist_square_ratio")
# Новые признаки распределены не нормально. Нормализуем их методом RobustScaler.
rb_scaler = preprocessing.RobustScaler()
data_coor["distance_to_center"] = rb_scaler.fit_transform(
data_coor[["distance_to_center"]]
)
data_coor["distance_to_airport"] = rb_scaler.fit_transform(
data_coor[["distance_to_airport"]]
)
data_coor["dist_square_ratio"] = rb_scaler.fit_transform(
data_coor[["dist_square_ratio"]]
)
# удаляем из data_coor признаки с координатами и объединяем с data
data_coor.drop(columns=col_coor_list, inplace=True)
data = pd.concat([data, data_coor], axis=1)
data.info()
# удаляем из нашего датасета признаки, которые нам больше не понадобятся
data.drop(
columns=[
"city_center_lat",
"city_center_lng",
"city_airport_lat",
"city_airport_lng",
"lat",
"lng",
],
inplace=True,
)
# ### 2.4 total_number_of_reviews
# Возможно это один из признаков надёжности отеля. Если отзывов много, то средний рейтинг можно считать объективным. Исследуем распределение этого признака.
visual_num(data, "total_number_of_reviews")
# Обратим внимание на отели, количество отзывов у которых около 17000. Узнаем их имена.
data[data["total_number_of_reviews"] > 15000]["hotel_name"].value_counts()
# Такое большое количество отзывов только у одого отеля. Выясняем его средний балл.
data[data["hotel_name"] == "Hotel Da Vinci"]["reviewer_score"].mean()
# Средний балл достаточно низкий. Такое большое количество отзывов вероятно обусловлено большим количеством недовольных клиентов. Создаем признак, указывающий на большое количество отзывов.
data["total_reviews"] = data["total_number_of_reviews"].apply(
lambda x: 1 if x > 15000 else 0
)
# ### 2.5 average_score
visual_num(data, "average_score")
# Распределение признака близко к нормальному. Отрицательных значения и значения больше десяти отсутствуют.
# ### 2.6 additional_number_of_scoring
visual_num(data, "additional_number_of_scoring")
# Создадим признак, показывающий отношение количества оценок без отзыва к общему количеству действительных отзывов об отеле.
data["review_ratio"] = (
data["additional_number_of_scoring"] / data["total_number_of_reviews"]
)
visual_num(data, "review_ratio")
# Доля оценок без отзыва не превышает сорок процентов. Выясним среднюю оценку отелей, у которых доля оценок бе отзыов близка к верхней границе полученного нами диапазона.
print(data[data["review_ratio"] > 0.39]["reviewer_score"].mean())
print(data[data["review_ratio"] < 0.39]["reviewer_score"].mean())
# Разница составляет более половины балла, что довольно существенно. Это может быть обусловлено психологией человека: недовольный клиент с большей вероятностью опишет то, что ему не понравилось. Удовлетворенный клиент скорее просто поставит высокую оценку, не вдаваясь в подробности. Создадим признак, указывающий на высокую долю оценок без отзывов.
data["top_review_ratio"] = data["review_ratio"].apply(lambda x: 1 if x > 0.39 else 0)
data["top_review_ratio"].value_counts()
# ## 3. Анализ признаков, описывающих рецензента
# ### 3.1 reviewer_nationality
# Создадим из признаков reviewer_nationality и hotel_country признак-флаг, указывающий на то, является ли человек, оставивший отзыв об отеле, гражданином страны, в которой этот отель расположен.
data["reviewer_nationality"] = data["reviewer_nationality"].apply(lambda s: s.strip())
print(
"Всего "
+ str(data["reviewer_nationality"].nunique())
+ " различных национальностей в датасете.\n"
)
data["reviewer_nationality"].value_counts()
# Возможно для некоторых национальностей имеет смысл создать отдельный признак-флаг. Сгруппируем наши данные по национальностям рецензентов и посмотрим на поставленные средние баллы первых двадцати по количеству отзывов
group_nationality = (
data.groupby("reviewer_nationality")["reviewer_score"]
.agg(["mean", "count"])
.round(2)
.sort_values(ascending=False, by="count")
.nlargest(20, columns="count")
)
group_nationality
# Максимальный средний балл у представителей США, минимальный - у представителей ОАЭ. Создаем два новых признака "national_is_usa" и "national_is_uae". Также введем признак для самой многочисленной группы национальностей рецензентов - "national_is_uk".
data["national_is_usa"] = data["reviewer_nationality"].apply(
lambda x: 1 if x == "United States of America" else 0
)
data["national_is_uae"] = data["reviewer_nationality"].apply(
lambda x: 1 if x == "United Arab Emirates" else 0
)
data["national_is_uk"] = data["reviewer_nationality"].apply(
lambda x: 1 if x == "United Kingdom" else 0
)
# Разделим рецензентов по следующему принципу: те, кто проживал в отеле своей страны и те, кто проживал в отеле заграницей.
# создаем признак-флаг и выводим соотношение резидентов и нерезидентов
data["local_reviewer"] = data.apply(
lambda x: 1 if x["reviewer_nationality"] == x["hotel_country"] else 0, axis=1
)
data["local_reviewer"].value_counts(normalize=True)
print(
f'Резидентов: {round(data["local_reviewer"].value_counts(normalize=True)[0]*100)} %\nНерезидентов:'
f'{round(data["local_reviewer"].value_counts(normalize=True)[1]*100)} %'
)
# визуализируем признак local_reviewer
visual_cat(data, "local_reviewer", 2)
# Средний балл отзыва, оставленного рецензентом-резидентом, выше среднего балла, оставленногого нерезидентом.
# Cреди национальностей, входящих в топ-20 по количеству рецензентов, есть 4 национальности, преобладающей религией которых является ислам.
# Создаем новый признак-флаг, определяющий то, что отзыв оставил представитель исламской страны.
a = ["United Arab Emirates", "Saudi Arabia", "Turkey", "Kuwait"]
data["reviewer_is_muslim"] = data["reviewer_nationality"].apply(
lambda x: 1 if x in a else 0
)
data["reviewer_is_muslim"].value_counts(normalize=True)
# визуализируем признак reviewer_is_muslim
visual_cat(data, "reviewer_is_muslim", 2)
# ### 3.2 total_number_of_reviews_reviewer_has_given
visual_num(data, "total_number_of_reviews_reviewer_has_given")
# Распределение признака похоже на логнормальное. Создадим из признака total_number_of_reviews_reviewer_has_given новый признак verified_reviewer. Будем считать надежным такого рецезента, общее количество отзывов у которого не менее 10.
data["verified_reviewer"] = data["total_number_of_reviews_reviewer_has_given"].apply(
lambda x: 1 if x >= 11 else 0
)
print(data["verified_reviewer"].value_counts())
visual_cat(data, "verified_reviewer", 2)
# Разница в средней оценке между группами минимальна.
# ### 3.3 review_date
# Создадим из признака review_date новые признаки: год, месяц, число месяца и день недели.
data["review_date"] = pd.to_datetime(data["review_date"])
data["year"] = data["review_date"].dt.year
data["day"] = data["review_date"].dt.day
data["month"] = data["review_date"].dt.month
data["weekday"] = data["review_date"].dt.weekday + 1 # Пн = 1, ..., Вс = 7
# Визуализируем новые признаки
fig, axes = plt.subplots(nrows=2, ncols=2, figsize=(12, 12))
plt.subplots_adjust(hspace=0.25)
histplot1 = sns.histplot(data=data, x="year", bins=3, ax=axes[0, 0])
histplot1.set_title("Распределение по году", fontsize=10)
histplot1.set_xlabel("Год")
histplot1.set_ylabel("Количество отзывов")
histplot1.set_xticklabels(histplot1.get_xticklabels(), rotation=30)
histplot2 = sns.histplot(data=data, x="month", bins=12, ax=axes[0, 1])
histplot2.set_title("Распределение по месяцам", fontsize=10)
histplot2.set_xlabel("Месяц")
histplot2.set_ylabel("Количество отзывов")
histplot3 = sns.histplot(data=data, x="day", bins=31, ax=axes[1, 0])
histplot3.set_title("Распределение по дням месяца", fontsize=10)
histplot3.set_xlabel("день")
histplot3.set_ylabel("Количество отзывов")
histplot4 = sns.histplot(data=data, x="weekday", bins=7, ax=axes[1, 1])
histplot4.set_title("Распределение по дням недели", fontsize=10)
histplot4.set_xlabel("День недели")
histplot4.set_ylabel("Количество отзывов")
# Из построенных графиков можно сделать следующие выводы:
# * все отзывы были сделаны с 2015 по 2017 годы
# * максимальное количество отзывов было сделано в 2016 году
# * чаще всего отзывы оставляют в июле-августе
# * на первые числа месяца приходится больше отзывов, чем на последние
# * самый популярный день недели для отзывов - вторник, наименее популярный - пятница
# ## 4. Признаки, описывающие отзыв
# ### 4.1 negative_review, positive_review
# Данные признаки будем анализировать с помощью библиотеки Textblob.
# создаем новый датасет
data_emotion = data[["negative_review", "positive_review"]]
# создаем функцию для получения новых признаков
def emotional_rate(x):
"""Text sentiment analysis function
Args:
x (str): parsed text
Returns:
tuple: tuple consisting of polarity and subjectivity values_
"""
tuple_emotion = TextBlob(x).polarity, TextBlob(x).subjectivity
return tuple_emotion
# применяем функцию к столбцам 'negative_review' и 'positive_review'
# получаем четыре новых признака: показатели "полярности" и "субъективности" для положительных и отрицательных отзывов
data_emotion["negative_polarity"] = data_emotion["negative_review"].apply(
lambda x: emotional_rate(x)[0]
)
data_emotion["negative_subjectivity"] = data_emotion["negative_review"].apply(
lambda x: emotional_rate(x)[1]
)
data_emotion["positive_polarity"] = data_emotion["positive_review"].apply(
lambda x: emotional_rate(x)[0]
)
data_emotion["positive_subjectivity"] = data_emotion["positive_review"].apply(
lambda x: emotional_rate(x)[1]
)
data_emotion.describe()
# Визуализируем распределение полученных признаков.
emotion_futures = [
"positive_polarity",
"negative_polarity",
"positive_subjectivity",
"negative_subjectivity",
]
for future in emotion_futures:
visual_num(data_emotion, future)
# Проверим, насколько точно данные признаки описывают содержание отзывов.
# удаляем признаки типа "object"
data_emotion.drop(columns=["negative_review", "positive_review"], axis=1, inplace=True)
# объединяем с исходным датасетом
data = pd.concat([data, data_emotion], axis=1)
# Посмотрим на разброс значений reviewer_score с положительным значением positive_polarity и отрицательным значением negative_polarity
data[(data["positive_polarity"] > 0) & (data["negative_polarity"] < 0)][
"reviewer_score"
].value_counts().sort_values(ascending=False)
# При данных условиях мы ожидаем увидеть высокие значения reviewer_score. Однако количество низких баллов(будем считать таковыми все, что ниже четырех баллов) значительно. Теперь посмотрим на самые часто встречающиеся отзывы с оценкой ниже пяти баллов при тех же условиях.
data[
(data["positive_polarity"] > 0)
& (data["negative_polarity"] < 0)
& (data["reviewer_score"] < 4)
& (data["reviewer_score"] != 0)
]["positive_review"].value_counts().sort_values(ascending=False).nlargest(20)
# Основная масса таких отзывов - это указание на удобное месторасположение отеля. Создадим новый признак 'good_location'.
def good_location(x):
a = " ".join(x.lower().split())
if a == "good location":
return 1
if a == "location was good":
return 1
if a == "location is good":
return 1
else:
return 0
data["good_location"] = data["positive_review"].apply(good_location)
data["good_location"].value_counts()
visual_cat(data, "good_location")
# Среднее значение признака reviewer_score в категории отзывов, где положительный отзыв ограничивается упоминанием удобного расположения отеля, значительно ниже.
# Теперь рассмотрим обратную ситуацию: нас интересует разброс значений reviewer_score с отрицательным значением positive_polarity и положительным значением negative_polarity.
data[(data["positive_polarity"] < 0) & (data["negative_polarity"] > 0)][
"reviewer_score"
].value_counts().sort_values(ascending=False)
# При данных условиях мы ожидаем увидеть низкие значения reviewer_score. Однако количество высоких баллов(будем считать таковыми все, что выше восьми баллов) значительно. Теперь посмотрим на самые часто встречающиеся положительные отзывы с оценкой выше восьми баллов при тех же условиях.
# Самое часто встречающееся значение положительного отзыва - 'No Positive'. Создадим новый признак, указывающий на равенство значения положительного отзыва 'No Positive'.
data["no_positive"] = data["positive_review"].apply(
lambda x: 1 if x == "No Positive" else 0
)
visual_cat(data, "no_positive")
# Среднее значение признака reviewer_score в категории отзывов, где положительный отзыв равен 'No Positive, значительно ниже.
# ### 4.2 tags
# Каждое значение признака tags является списком тегов в строковом формате. В тегах встречается много "мусора". Для начала избавимся от него.
def tags_cleaner(x):
"""The function removes non-numeric and non-alphabetic
characters from the elements of the spike.
Args:
x (list): tag list
Returns:
list: list of cleared tags
"""
drop_simbols = ["[", "]", "' ", " '"]
for simbol in drop_simbols:
x = x.replace(simbol, "")
return x.split(", ")
data["tags_cleaned"] = data["tags"].apply(tags_cleaner)
data["tags_cleaned"].head()
# Создаем новый признак, показывающий количество тэгов в отзыве
data["num_tegs"] = data["tags_cleaned"].apply(lambda x: len(x))
data["num_tegs"].value_counts()
# Максимальное количество тэгов в отзыве равно шести. Выводим на экран все отзывы с максимальным количеством тэгов
data[data["num_tegs"] == 6]["tags_cleaned"].value_counts()
# Из тэгов мы можем извлечь следующую информацию:
# 1. Тип поездки: командировка или отдых
# 2. Количество проживающих
# 3. Количество комнат в номере
# 4. Количество ночей в отеле
# 5. Где оставлен отзыв
# 6. Наличие окон в номере
# 7. Проживание с домашними животными или без
# создаем новый признак, указывающий на проживание с домашними животными
def pet_tag(x):
for tag in x:
if "pet" in tag:
return 1
return 0
data["with_pet"] = data["tags_cleaned"].apply(pet_tag)
print(data["with_pet"].value_counts())
visual_cat(data, "with_pet")
# Средний балл отзывов рецензентов с домашними животными ниже чем средний балл рецензентов без них.
# создаем новый признак, указывающий на то, кто проживал в отеле: один человек, два, семья или группа
def num_travelels(x):
for tag in x:
if "Solo" in tag:
return "one"
if "Couple" in tag:
return "two"
if "Family" in tag:
return "family"
if "Group" in tag:
return "group"
return "unknown"
data["num_travelers"] = data["tags_cleaned"].apply(num_travelels)
print(data["num_travelers"].value_counts())
visual_cat(data, "num_travelers")
# Больше всего отзывов в категории 'unknown'. В этой же категории самая высокая оценка. Самая низкая оценка в категории 'one'.
# создаем признак количества ночей проживания в отеле
def num_night(x):
for tag in x:
if "Stayed" in tag:
return int(tag.split()[1])
return 0
data["num_night"] = data["tags_cleaned"].apply(num_night)
print(data["num_night"].value_counts())
visual_num(data, "num_night")
# Распределение данного признака близко к логнормальному
# создаем признак, указывающий то, что отзыв оставлен в мобильном приложении
def mobile_review(x):
for tag in x:
if "Submitted" in tag:
return 1
return 0
data["mobile_review"] = data["tags_cleaned"].apply(mobile_review)
print(data["mobile_review"].value_counts())
visual_cat(data, "mobile_review")
# создаем признак, указывающий на то, с какой целью рецензент в отеле: командировка или отдых
def trip_tag(x):
for tag in x:
if "trip" in tag:
if "Business" in tag:
return "business"
elif "Leisure" in tag:
return "leisure"
return "unknown"
data["trip_tag"] = data["tags_cleaned"].apply(trip_tag)
print(data["trip_tag"].value_counts())
visual_cat(data, "trip_tag")
# Отдыхающиеся рецензенты в среднем ставят более высокую оценку, чем находящиеся в командировке.
def window(x):
for tag in x:
if "Window" in tag:
return 1
return 0
data["window"] = data["tags_cleaned"].apply(window)
print(data["window"].value_counts())
visual_cat(data, "window")
# ### 4.3 review_total_positive_word_counts, review_total_negative_word_counts
# Визуализируем распределение данных признаков
visual_num(data, "review_total_positive_word_counts")
visual_num(data, "review_total_negative_word_counts")
# Распределение признаков близко к логнормальному.
fig = px.scatter(
data,
x="review_total_negative_word_counts",
y="review_total_positive_word_counts",
title="График зависимости двух переменных",
width=700,
height=500,
)
fig.update_traces(marker={"size": 6})
fig.show()
# Зависимость переменных близка к обратной линейной функции.
# нормализуем данные признаки
rb_scaler = preprocessing.RobustScaler()
data["review_total_negative_word_counts"] = rb_scaler.fit_transform(
data[["review_total_negative_word_counts"]]
)
data["review_total_positive_word_counts"] = rb_scaler.fit_transform(
data[["review_total_positive_word_counts"]]
)
# ### 4.4 days_since_review
# Данный признак представлен в текстовом формате. Скорее всего влияние этого признака на обучение модели будет незначительным.
# приводим признак числовому типу данных
data["days_since_review"] = data["days_since_review"].apply(lambda x: int(x.split()[0]))
# строим графики распределения признака
visual_num(data, "days_since_review")
# ## 5. Отбор признаков
# На данный момент в нашем датасете один признак datetime и 13 признаков типа object. Закодируем созданные нами признаки и удалим все нечисловые признаки.
data_copy = data.copy()
# список признаков для кодирования
list_to_code = [
"hotel_country",
"hotel_city",
"hotel_chain",
"num_travelers",
"trip_tag",
"year",
"month",
"weekday",
]
for elem in list_to_code:
one_hot_encoder = ce.OneHotEncoder(cols=[elem], use_cat_names=True)
encoded = one_hot_encoder.fit_transform(data_copy[elem])
data_copy = pd.concat([data_copy, encoded], axis=1, join="inner")
data_copy.drop(columns="review_date", axis=1, inplace=True)
object_columns = [s for s in data_copy.columns if data_copy[s].dtypes == "object"]
data_copy.drop(object_columns, axis=1, inplace=True)
data_copy.info()
# Все признаки в нашем датасете числовые. Разделим его на два датасета: с числовыми непрерывными признаками и с числовыми категориальными признаками.
# список непрерывных признаков
num_future = [
"additional_number_of_scoring",
"average_score",
"review_total_negative_word_counts",
"total_number_of_reviews",
"review_total_positive_word_counts",
"total_number_of_reviews_reviewer_has_given",
"reviewer_score",
"distance_to_center",
"distance_to_airport",
"negative_polarity",
"positive_polarity",
"negative_subjectivity",
"positive_subjectivity",
"num_tegs",
"days_since_review",
"dist_square_ratio",
"city_square",
"review_ratio",
]
# список категориальных признаков
cat_future = []
for col in data_copy.columns:
if col not in num_future:
cat_future.append(col)
cat_data = data_copy[cat_future]
num_data = data_copy[num_future]
# Строим матрицы корелляций отдельно для непрерывных и отдельно для категориальных признаков
plt.figure(figsize=(10, 7))
corr_heatmap = sns.heatmap(num_data.corr(method="spearman"), annot=True, cmap="binary")
corr_heatmap.set_title("Матрица корреляции для непрерывных признаков ")
# Сильную корелляцию имеют три пары признаков:
# * total_number_of_reviews, additional_number_of_scoring
# * positive_subjectivity, positive_polarity
# * city_square, dist_square_ratio
# Однако удаление из датасета по одному признаку из каждой пары не привело к уменьшению MAPE
plt.figure(figsize=(23, 12))
corr_heatmap = sns.heatmap(cat_data.corr(method="spearman"), annot=True, cmap="binary")
corr_heatmap.set_title("Матрица корреляции для категориальных признаков ")
# Из-за большого количества признаков визуальное восприятие матрицы корелляции затруднено. Выводим на экран только признаки с корелляцией выше по модулю 0.7
# функция фильтрации матрицы корелляции
def correl(xtrain, thresh):
"""Function to filter correlation values
Args:
xtrain (pd.DataFrame): researched dataframe
thresh (float): filtration limit
Returns:
_pd.DataFrame: pairs of features with a higher correlation modulo a given limit
"""
cor = xtrain.corr(method="spearman")
c = cor.stack().sort_values(ascending=False).drop_duplicates()
all_cor = c[c.values != 1]
return all_cor[abs(all_cor) > thresh]
correl(xtrain=cat_data, thresh=0.7)
# Удаление признаков из каждой пары также не привело к уменьшению значения MAPE
# ## 6. Создание и проверка модели
train_data = data_copy.query("sample == 1").drop(["sample"], axis=1)
test_data = data_copy.query("sample == 0").drop(["sample"], axis=1)
y = train_data.reviewer_score.values # наш таргет
X = train_data.drop(["reviewer_score"], axis=1)
X = train_data.drop(columns="reviewer_score")
y = train_data["reviewer_score"]
# Воспользуемся специальной функцие train_test_split для разбивки тестовых данных
# выделим 20% данных на валидацию (параметр test_size)
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=RANDOM_SEED
)
# Импортируем необходимые библиотеки:
from sklearn.ensemble import (
RandomForestRegressor,
) # инструмент для создания и обучения модели
from sklearn import metrics # инструменты для оценки точности модели
# Создаём модель (НАСТРОЙКИ НЕ ТРОГАЕМ)
model = RandomForestRegressor(
n_estimators=100, verbose=1, n_jobs=-1, random_state=RANDOM_SEED
)
# Обучаем модель на тестовом наборе данных
model.fit(X_train, y_train)
# Используем обученную модель для предсказания рейтинга отелей в тестовой выборке.
# Предсказанные значения записываем в переменную y_pred
y_pred = model.predict(X_test)
# Сравниваем предсказанные значения (y_pred) с реальными (y_test), и смотрим насколько они в среднем отличаются
# Метрика называется mean absolute percentage error (MAPE) и показывает среднюю абсолютную процентную ошибку предсказанных значений от фактических.
print("MAPE:", metrics.mean_absolute_percentage_error(y_test, y_pred))
# в RandomForestRegressor есть возможность вывести самые важные признаки для модели
plt.rcParams["figure.figsize"] = (10, 10)
feat_importances = pd.Series(model.feature_importances_, index=X.columns)
feat_importances.nlargest(32).plot(kind="barh")
test_data = test_data.drop(["reviewer_score"], axis=1)
predict_submission = model.predict(test_data)
sample_submission["reviewer_score"] = predict_submission
sample_submission.to_csv("submission.csv", index=False)
sample_submission
|
# # CNN v2
# import models
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
from keras.models import Sequential
from keras.callbacks import EarlyStopping
from keras.layers.convolutional import Conv1D, MaxPooling1D
from keras.layers import Dense, LSTM, RepeatVector, Flatten, TimeDistributed, Dropout
from sklearn.preprocessing import MinMaxScaler
from keras.losses import (
mean_absolute_error,
mean_absolute_percentage_error,
mean_squared_error,
)
from sklearn.model_selection import train_test_split
import matplotlib.dates as mdates
color_pal = sns.color_palette()
plt.style.use("fivethirtyeight")
import os
# # import and Clean Data
# Read & fill empty Date
df = pd.read_csv("/kaggle/input/iraq2016/iraq2016.csv")
df.fillna(method="ffill", inplace=True)
df = df.set_index("datetime")
df.index = pd.to_datetime(df.index)
# df
# # split the data
train_data, test_data = train_test_split(
df, test_size=0.23, shuffle=False, random_state=42
)
# # Plotting
# create a list of the days of the month
# plot the train and test sets in different colors
plt.figure(figsize=(15, 5))
plt.plot(train_data.index, train_data["power"], label="Train", color="green")
plt.plot(test_data.index, test_data["power"], label="Test", color="orange")
plt.axvline(x=pd.Timestamp("2016-01-24 20:00:00"), color="black", linestyle="--")
plt.xlabel("Date")
plt.ylabel("Power")
plt.title("Power Consumption in 2016 (MW)")
plt.legend()
# format the x-axis as dates and show every 3 days
date_format = mdates.DateFormatter("%Y-%m-%d")
plt.gca().xaxis.set_major_formatter(date_format)
plt.gca().xaxis.set_major_locator(mdates.DayLocator(interval=7))
plt.gcf().autofmt_xdate()
plt.show()
data = df.loc["2016-01-25"]
# plot the data
plt.figure(figsize=(15, 5))
plt.plot(data.index, data["power"], color="blue")
plt.xlabel("Time")
plt.ylabel("Power")
plt.title("Power Consumption on January 25, 2016 (MW)")
plt.ylim(22000, 42000)
# # Feautre Creation
def create_features(df):
"""
Create time series features based on time series index.
"""
df = df.copy()
df["hour"] = df.index.hour
df["dayofweek"] = df.index.dayofweek
df["quarter"] = df.index.quarter
df["month"] = df.index.month
df["year"] = df.index.year
df["dayofyear"] = df.index.dayofyear
df["dayofmonth"] = df.index.day
df["weekofyear"] = df.index.isocalendar().week
return df
df = create_features(df)
df.head()
# # split data to train and test
features = ["dayofyear", "hour", "dayofweek", "quarter", "month", "year"]
labels = ["power"]
train_data, test_data = train_test_split(
df, test_size=0.23, shuffle=False, random_state=42
)
train_x = train_data[features]
train_y = train_data[labels]
test_x = test_data[features]
test_y = test_data[labels]
# # create windows
data = df["power"].values
scaler = MinMaxScaler()
data = scaler.fit_transform(df["power"].values.reshape(-1, 1))
# define the window size and stride
window_size = 24
stride = 1
# create windows and labels for the data
windows = []
labels = []
for i in range(0, len(data) - window_size, stride):
window = data[i : i + window_size]
label = data[i + window_size]
windows.append(window)
labels.append(label)
windows = np.array(windows)
labels = np.array(labels)
# split the data into a train set and a test set
train_ratio = 0.8
train_size = int(train_ratio * len(windows))
train_windows, train_labels = windows[:train_size], labels[:train_size]
test_windows, test_labels = windows[train_size:], labels[train_size:]
# split the data into a train set and a test set
train_ratio = 0.8
train_size = int(train_ratio * len(windows))
train_windows, train_labels = windows[:train_size], labels[:train_size]
test_windows, test_labels = windows[train_size:], labels[train_size:]
# reshape the data for the CNN layer
train_windows = train_windows.reshape(
(train_windows.shape[0], train_windows.shape[1], 1)
)
test_windows = test_windows.reshape((test_windows.shape[0], test_windows.shape[1], 1))
# # perform hybrid CNN-LSTM model
model = Sequential()
# define the model architecture
model = Sequential()
model.add(
Conv1D(filters=16, kernel_size=3, activation="relu", input_shape=(window_size, 1))
)
model.add(MaxPooling1D(pool_size=2))
model.add(
Conv1D(filters=32, kernel_size=3, activation="relu", input_shape=(window_size, 1))
)
model.add(MaxPooling1D(pool_size=2))
model.add(Dropout(0.2))
model.add(LSTM(units=64))
model.add(Dense(units=1))
# compile the model
model.compile(optimizer="adam", loss="mse", metrics=["mse", "mae", "mape"])
early_stopping_monitor = EarlyStopping(patience=3)
# train the model
history = model.fit(
train_windows,
train_labels,
epochs=7,
batch_size=12,
verbose=1,
callbacks=early_stopping_monitor,
)
# # Analyse Outputs
loss, mse, mae, mape = model.evaluate(test_windows, test_labels, verbose=0)
rmse = np.sqrt(mse)
# evaluate the model on the test set
predictions = model.predict(test_windows)
predictions = scaler.inverse_transform(predictions)
test_labels = scaler.inverse_transform(test_labels)
print(f"Mean Squared Error: {mse:.2f}")
print(f"Mean Absolute Error: {mae:.2f}")
print(f"Root Mean Squared Error: {rmse:.2f}")
print(f"Mean Absulote Precentage Error: {mape:.2f}%")
# # plot the result
# plot the predicted values and the actual values
plt.figure(figsize=(10, 6))
plt.plot(predictions, label="predictions")
plt.plot(test_labels, label="actual")
plt.legend()
plt.title("Predicted vs Actual Power Consumption")
plt.xlabel("Time Step")
plt.ylabel("Power (MW)")
plt.show()
labels = ["MSE", "MAE", "RMSE", "%(MAPE / 100) "]
values = [mse, mae, rmse, mape / 100]
position = range(len(values))
plt.barh(position, values, 0.4)
plt.yticks(position, labels)
plt.title("CNN-LSTM Evaluation Metrics")
plt.xlabel("Values")
plt.show()
# # save our prediction
# create a DataFrame with the predictions
predictions_df = pd.DataFrame(predictions, columns=["predictions"])
# read the original CSV file
df = pd.read_csv(
"/kaggle/input/iraq2016/iraq2016.csv", index_col="datetime", parse_dates=True
)
# merge the original DataFrame with the predictions DataFrame
df = pd.concat([df, predictions_df], axis=1)
# save the merged DataFrame to a CSV file
df.to_csv("iraq2016_prediction2.csv")
# # read our data
predict_df = pd.read_csv("/kaggle/working/iraq2016_prediction1.csv", parse_dates=True)
print(predict_df)
predict_df.isna().sum()
# plt.figure(figsize=(15, 5))
# # plt.plot(predict_df.index, predict_df['power'], label='Train', color='green')
# # plt.plot(predict_df.index, predict_df['predictions'], label='Train', color='green')
# plt.xlabel('Date')
# plt.ylabel('Power')
# plt.title('Predict in 2016 (MW)')
# plt.legend()
# # format the x-axis as dates and show every 3 days
# date_format = mdates.DateFormatter('%Y-%m-%d')
# plt.gca().xaxis.set_major_formatter(date_format)
# plt.gca().xaxis.set_major_locator(mdates.DayLocator(interval=7))
# plt.gcf().autofmt_xdate()
# plt.show()
|
#
# # Understanding The Dataset:
# Now from dataset description we have 2 datasets:
#
# 1. The Original Dataset
# 2. The Generated Dataset (train.csv, test.csv, sample.csv)
#
# ## Original Dataset Description:
#
# This dataset consists of **79 urine specimens** that were analyzed to determine if certain physical characteristics of the urine are related to the formation of calcium oxalate crystals, which are the most common type of kidney stones. The six physical characteristics of the urine that were analyzed are:
# 1. Specific gravity: The density of the urine relative to water.
# 2. pH: The negative logarithm of the hydrogen ion.
# 3. Osmolarity (mOsm): A unit used in biology and medicine but not in physical chemistry. Osmolarity is proportional to the concentration of molecules in solution.
# 4. Conductivity (mMho milliMho): One Mho is one reciprocal Ohm. Conductivity is proportional to the concentration of charged ions in solution.
# 5. Urea concentration in millimoles per litre.
# 6. Calcium concentration (CALC) in millimoles per litre.
# The objective of this dataset is to **predict the presence of kidney stones** based on the u**rine analysis results**.
#
# # EDA
#
# importing libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from matplotlib_inline.backend_inline import set_matplotlib_formats
import seaborn as sns
set_matplotlib_formats("svg")
sns.set_style("whitegrid")
original_df = pd.read_csv(
"/kaggle/input/kidney-stone-prediction-based-on-urine-analysis/kindey stone urine analysis.csv"
)
train_df = pd.read_csv("/kaggle/input/playground-series-s3e12/train.csv")
test_df = pd.read_csv("/kaggle/input/playground-series-s3e12/test.csv")
original_df.head()
train_df
test_df
print("The shape of Original Dataset:", original_df.shape)
print("The shape of Train Dataset:", train_df.shape)
print("The shape of Test Dataset:", test_df.shape)
# note that the shape of **Train Dataset** includes 8 columns because it includes id column
# let's see datatypes for each column:
print("The Original Dataset Info: ")
print()
print(original_df.info())
print("-" * 50)
print()
print("The Train Dataset Info: ")
print()
print(train_df.info())
print("-" * 50)
print()
print("The Test Dataset Info: ")
print()
print(test_df.info())
print("-" * 50)
# Great! from original, train, test datasets we see there's no null values and datatypes float64 and int64
# We can aslo check the null using other method such:
# > train.isnull().sum() #copy and run in a cell
# Let's now check duplicates
print("Duplicates for Original DataFrames:")
print(original_df[original_df.duplicated()])
print()
print("-" * 60)
print("Duplicates for Train DataFrames:")
print(train_df[train_df.duplicated()])
print()
print("-" * 60)
print("Duplicates for Test DataFrames")
print(test_df[test_df.duplicated()])
# let's see descriptive data using .describe()
original_df.describe()
train_df.describe()
train_df.nunique()
#
# ## Visualizations
# now time to visualization but before this, dropping id column
#
train_df_2 = train_df.drop("id", axis=1)
# ### Distributions OF Features
fig = plt.figure(figsize=(13, 11))
for i, col in enumerate(train_df_2.columns, 1):
plt.subplot(4, 2, i)
sns.histplot(
x=train_df_2[col],
kde=True,
color="#9080ff",
linewidth=0.8,
edgecolor="k",
alpha=0.7,
)
plt.title(f"Distribution of {col}")
plt.tight_layout()
fig.suptitle(
"Distributions OF Features Using Histograms",
fontsize=13.5,
fontweight="heavy",
y=1.02,
)
plt.show()
# From Histograms:
# 1. Gravity has a slightly positive skewness, meaning that the distribution is slightly skewed to the right.
# 2. Ph has a positive skewness, meaning that the distribution is skewed to the right, and the tail on the right side is longer or fatter than the left side.
# 3. Osmo has a slightly positive skewness, meaning that the distribution is slightly skewed to the right.
# 4. Cond has a negative skewness, meaning that the distribution is skewed to the left, and the tail on the left side is longer or fatter than the right side.
# 5. Urea has a slightly positive skewness, meaning that the distribution is slightly skewed to the right.
# 6. Calc has a positive skewness, meaning that the distribution is skewed to the right, and the tail on the right side is longer or fatter than the left side.
# 7. Target has a slightly positive skewness, meaning that the distribution is slightly skewed to the right.
_, axes = plt.subplots(1, 5, figsize=(15, 4))
for i, ax in enumerate(axes.flatten()):
sns.boxenplot(y=train_df_2[train_df_2.columns[i]], color="#ffb400", ax=axes[i])
_.suptitle("Boxen Plots For Features", fontsize=14, fontweight="heavy")
plt.tight_layout()
plt.show()
# ### Target vs Features
fig, axes = plt.subplots(1, 6, figsize=(15, 4))
for i, ax in enumerate(axes.flatten()):
sns.kdeplot(
data=train_df_2,
x=train_df_2.columns[i],
hue="target",
fill=True,
ax=ax,
palette=["#ffb400", "#9080ff"],
)
fig.suptitle("KDE Plots For Target vs. Features", fontsize=14, fontweight="heavy")
plt.tight_layout()
f = sns.pairplot(
train_df_2,
hue="target",
diag_kind="kde",
palette=["#ffb400", "#9080ff"],
height=4,
aspect=1.2,
)
f.fig.set_size_inches(15, 11)
f.fig.suptitle("Pair Plot & hue = target", fontsize=14, fontweight="heavy", y=1.05)
plt.legend()
plt.tight_layout()
plt.show()
f, ax = plt.subplots(1, 6, figsize=(15, 4))
for i in range(len(train_df_2.columns) - 1):
sns.boxplot(
x="target",
y=train_df_2.columns[i],
data=train_df_2,
ax=ax[i],
palette=["#ffb400", "#9080ff"],
)
ax[i].title.set_text("target vs {0}".format(train_df_2.columns[i]))
f.suptitle("Box Plots Output Vs. Numerical Features", fontsize=14, fontweight="heavy")
plt.tight_layout()
# ### Person Correlation Heatmap
plt.figure(figsize=(13, 7))
sns.heatmap(train_df_2.corr(), annot=True, cmap="Blues", linewidth=0.1)
plt.title("Person Correlation Heatmap", fontsize=14, fontweight="heavy")
plt.show()
#
# # Modeling Using Pycaret
#
#
test_df_2 = test_df.drop("id", axis=1)
train_df_3 = pd.concat([original_df, train_df_2])
train_df_3.shape
from pycaret.classification import *
s = setup(train_df_3, target="target", session_id=123)
best = compare_models()
plot_model(best, plot="confusion_matrix")
plot_model(best, plot="auc")
evaluate_model(best)
tuned_best = tune_model(best)
predictions = predict_model(tuned_best, data=test_df_2)
test_df
predictions
save_model(tuned_best, "my_pipeline")
#
# # Submission
#
#
my_sub = pd.DataFrame({"id": test_df["id"], "target": predictions["prediction_label"]})
my_sub
my_sub.to_csv("submission.csv")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# generating array of timestamps of comments:
baseline = 48640
commentsTimes = [48640]
for i in range(50):
newNumber = baseline + (i + 1) * 2560
commentsTimes.append(newNumber)
print(commentsTimes)
print(len(commentsTimes))
Set1comments = [
49,
43,
17,
15,
13,
45,
41,
1,
3,
25,
16,
42,
9,
24,
40,
32,
29,
2,
26,
46,
36,
18,
12,
34,
19,
4,
48,
50,
28,
23,
22,
14,
33,
6,
21,
7,
8,
5,
27,
47,
38,
37,
30,
11,
35,
39,
31,
20,
44,
10,
]
S1channels = pd.read_csv(
"/kaggle/input/EEGwithChannelsNOLabels/kaggle/working/S01channels.csv",
low_memory=False,
)
dfS1 = pd.DataFrame(S1channels)
dfS1 = dfS1.iloc[2:, :]
S1float = dfS1.astype(float) # NEEDED?
S1channelsOnly = S1float.iloc[:, 1:]
S1channelsOnly
subjIDs = [
"S01",
"S02",
"S03",
"S04",
"S05",
"S06",
"S07",
"S08",
"S10",
"S11",
"S12",
"S13",
"S14",
"S15",
"S16",
"S18",
"S19",
"S20",
"S21",
"S22",
"S23",
"S25",
"S26",
"S27",
"S28",
"S29",
"S31",
] # ,'S32','S33','S34']
def makeFileNames(headerArr):
nameArr = []
for i in range(len(headerArr)):
nameStr = headerArr[i] + "noHeadersNoTimes.csv"
nameArr.append(nameStr)
return nameArr
outputFiles = makeFileNames(subjIDs)
import csv
# remove headers from all files
input_files = sorted(
[
os.path.join(dirname, f)
for dirname, _, files in os.walk(
"/kaggle/input/EEGwithChannelsNOLabels/kaggle/working/"
)
for f in files
if f.startswith("S") and f.endswith(".csv")
]
)
# print(input_files)
# Create a list of output filenames based on the index of the input files
# new_csv_files = ['output{}.csv'.format(i) for i in range(len(input_files))]
output_dir = "/kaggle/working"
# Iterate over the input files and create new CSV files with the desired headers and data
for i, input_file in enumerate(input_files):
with open(input_file) as infile:
reader = csv.reader(infile)
with open(os.path.join(output_dir, outputFiles[i]), "w", newline="") as outfile:
writer = csv.writer(outfile)
# writer.writerow(headers)
for row in reader:
if row[2]:
writer.writerow(row[1:])
# looks like this one actually worked
print(len(os.listdir("/kaggle/working")))
# os.remove('%i')
# making list of segmented comments for boxplotting:
fiftyCommentsS1 = []
for i in range(len(commentsTimes) - 1): # -1 to allow for i+1 indexing
# print(commentsTimes[i],commentsTimes[i+1])
# print(S1channelsOnly[commentsTimes[i]:commentsTimes[i+1]])
comment = S1channelsOnly[commentsTimes[i] : commentsTimes[i + 1]]
fiftyCommentsS1.append(comment)
def makeArrays(headerArr):
nameArr = []
for i in range(len(headerArr)):
nameStr = headerArr[i] + "comments.csv"
nameArr.append(nameStr)
return nameArr
commentsFiles = makeArrayNames(subjIDs)
commentsFiles
# attempting to make plot of 30 subjs 50 comments
from ast import literal_eval
working_files = sorted(
[
os.path.join(dirname, f)
for dirname, _, files in os.walk("/kaggle/working/")
for f in files
if f.startswith("S") and f.endswith(".csv")
]
)
# print(working_files)
# commentsArrays = literal_eval(commentsFiles)
for i, file in enumerate(working_files):
for i in range(len(commentsTimes) - 1): # -1 to allow for i+1 indexing
# print(commentsTimes[i],commentsTimes[i+1])
# print(S1channelsOnly[commentsTimes[i]:commentsTimes[i+1]])
comment = file[commentsTimes[i] : commentsTimes[i + 1]]
commentsArr.append(comment)
commentsArray1
i = 1
for comment in fiftyCommentsS1:
plt.figure()
if Set1comments[i - 1] > 25:
plt.title("Comment #%i (CS)" % i)
elif Set1comments[i - 1] <= 25:
plt.title("Comment #%i (HS)" % i)
sns.violinplot(comment)
i += 1
i = 1
for comment in fiftyCommentsS1:
plt.figure()
if Set1comments[i - 1] > 25:
plt.title("Comment #%i (CS)" % i)
elif Set1comments[i - 1] <= 25:
plt.title("Comment #%i (HS)" % i)
sns.boxplot(comment)
i += 1
i = 1
for comment in fiftyCommentsS1:
plt.figure()
if Set1comments[i - 1] > 25:
plt.title("Comment #%i (CS)" % i)
elif Set1comments[i - 1] <= 25:
plt.title("Comment #%i (HS)" % i)
sns.scatterplot(comment)
i += 1
# rip from old notebook to try PCA on S1
# PCA Model
# probably only need n_components = 3 (explains roughly 80% of data [53/15/13{next 3 are 2/1/1}])
scaler = StandardScaler()
scaled_df = scaler.fit_transform(dataset)
print("scaled_df shape:", scaled_df.shape)
pca = PCA(n_components=7) # mle solves PCA so that n_components sum = 100 %
pca_vectors = pca.fit_transform(scaled_df)
print("PCA_vectors shape: ", pca_vectors.shape)
pca_var = 0
for index, var in enumerate(pca.explained_variance_ratio_):
pca_var += var
print(
"Explained Variance ratio by Principal Component ",
(index + 1),
" : ",
var * 100,
"%",
)
print("Total Variance Explained by PCA: ", pca_var * 100, "%")
print("components:", pca.components_)
print("singular values:", pca.singular_values_)
print("Number of features:", pca.n_features_in_)
plt.figure()
plt.plot(pca.explained_variance_ratio_)
plt.xticks(rotation="vertical")
plt.figure(figsize=(25, 8))
sns.scatterplot(x=pca_vectors[:, 0], y=pca_vectors[:, 1], hue=dataset["label"])
plt.title("Principal Components vs Class distribution", fontsize=16)
plt.ylabel("Principal Component 2", fontsize=16)
plt.xlabel("Principal Component 1", fontsize=16)
plt.xticks(rotation="vertical")
plt.clf()
# leftover from ModelBuildingTest
# boxplot attempt of S1
# plt.boxplot(S1labeled)
# works but has no scaling:
# S1float.boxplot()
# col-by-col try
# S1float['EEG.AF3'].plot(kind='box')
# works; split into baseline/comments
S1baseline = S1float[0:48640]
S1comment1 = S1float[48640:51200]
S1comment2 = S1float[commentsTimes[1] : commentsTimes[2]]
S1comment3 = S1float[commentsTimes[2] : commentsTimes[3]]
S1comment4 = S1float[commentsTimes[3] : commentsTimes[4]]
S1comments = [S1comment1, S1comment2, S1comment3, S1comment4]
print(S1baseline.shape, S1comment1.shape)
# S1baseline['EEG.AF3'].plot(kind='box', title='baselineAF3')
S1c1NoTime = S1comment1.iloc[:, 1:]
S1baseNoTime = S1baseline.iloc[:, 1:]
S1commentsNoTime = [
S1comment1.iloc[:, 1:15],
S1comment2.iloc[:, 1:15],
S1comment3.iloc[:, 1:15],
S1comment4.iloc[:, 1:15],
]
# Comment3 is first HS in S1 (2CS followed by 3HS)
|
# Ovo je druga obavezna bilježnica u okviru kolegija Dubinska analiza podataka. Cilj ove bilježnice je raditi sa značajkama train skuoa te stvoriti nove značajke koje će pridonjeti kvaliteti skupa podataka.
import numpy as np
import pandas as pd
# # 0. Priprema skupa podataka
train = pd.read_csv("/kaggle/input/dapprojekt23/train.csv")
del train["id"]
train = train.astype({"machine_name": "category"})
train = train.astype({"day": "uint16"})
train = train.astype({"broken": "uint16"})
train = train.astype({"total": "uint16"})
train = train.astype({"label": "uint8"})
train = train.loc[train["day"] > 365]
for machine in train.machine_name.unique():
anomaly_day = pom = train[
(train["machine_name"] == machine) & (train["label"] == 1)
].day.min()
pom = train[
(train["machine_name"] == machine)
& (train["label"] == 1)
& (train["day"] > anomaly_day)
].index
train.drop(pom, inplace=True)
strojevi = train.machine_name.unique()
data_to_be_appended = pd.DataFrame(
columns=["day", "broken", "total", "label", "machine_name"]
)
i = 1
for stroj in strojevi:
dani = range(366, train.loc[train["machine_name"] == stroj].day.max() + 1)
stroj_dani = train.loc[train["machine_name"] == stroj].day.unique()
dani_bez_podataka = np.setdiff1d(np.asarray(dani), np.asarray(stroj_dani))
machine_data = train.loc[(train["machine_name"] == stroj)]
i += 1
for dan in dani_bez_podataka:
total = abs(
int(
np.random.normal(
loc=machine_data.total.mean(), scale=machine_data.total.std()
)
)
)
broken = abs(
int(
np.random.normal(
loc=machine_data.broken.mean(), scale=machine_data.broken.std()
)
)
)
while broken >= total:
total = abs(
int(
np.random.normal(
loc=machine_data.total.mean(), scale=machine_data.total.std()
)
)
)
broken = abs(
int(
np.random.normal(
loc=machine_data.broken.mean(), scale=machine_data.broken.std()
)
)
)
day_before = machine_data.loc[(machine_data["day"] == dan - 1)]
day_after = machine_data.loc[(machine_data["day"] == dan + 1)]
if (
day_before.empty
or day_after.empty
or day_before.label.sum() != day_after.label.sum()
):
novi_red = {
"day": dan,
"broken": broken,
"total": total,
"label": 0,
"machine_name": stroj,
}
data_to_be_appended = data_to_be_appended.append(
novi_red, ignore_index=True
)
else:
broken = int((day_before.broken.sum() + day_after.broken.sum()) / 2)
total = int((day_before.total.sum() + day_after.total.sum()) / 2)
novi_red = {
"day": dan,
"broken": broken,
"total": total,
"label": day_before.label.sum(),
"machine_name": stroj,
}
data_to_be_appended = data_to_be_appended.append(
novi_red, ignore_index=True
)
train = train.append(data_to_be_appended, ignore_index=True)
train = train.astype({"machine_name": "category"})
train = train.astype({"day": "uint16"})
train = train.astype({"broken": "uint16"})
train = train.astype({"total": "uint16"})
train = train.astype({"label": "uint8"})
train.dtypes
# train.to_csv('train_fixed.csv', index=False)
# # 1. Cross-validation function 1
# Ovdje ću sada stvoriti prvu cross validation funkciju koja će koristiti GaussianNB, LogisticRegression, RIPPER, RandomForestClassifier, ExtraTreesClassifier i XGBClassifier modele.
import wittgenstein as lw
from xgboost import XGBClassifier
from sklearn.model_selection import KFold
from sklearn.naive_bayes import GaussianNB
from itertools import combinations
from sklearn.metrics import f1_score
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import RandomForestClassifier
from sklearn.ensemble import ExtraTreesClassifier
def gaussNB(train_x, train_y, test_x, test_y):
clf = GaussianNB()
clf.fit(train_x, train_y)
y_pred = clf.predict(test_x)
return f1_score(test_y, y_pred, average="macro")
def logReg(train_x, train_y, test_x, test_y):
clf = LogisticRegression(random_state=0).fit(train_x, train_y)
y_pred = clf.predict(test_x)
return f1_score(test_y, y_pred, average="macro")
def ripper(train_x, train_y, test_x, test_y):
ripper_clf = lw.RIPPER()
ripper_clf.fit(train_x, train_y)
y_pred = ripper_clf.predict(test_x)
return f1_score(test_y, y_pred, average="macro")
def RFC(train_x, train_y, test_x, test_y):
clf = RandomForestClassifier()
clf.fit(train_x, train_y)
y_pred = clf.predict(test_x)
return f1_score(test_y, y_pred, average="macro")
def ETC(train_x, train_y, test_x, test_y):
clf = ExtraTreesClassifier()
clf.fit(train_x, train_y)
y_pred = clf.predict(test_x)
return f1_score(test_y, y_pred, average="macro")
def XGB(train_x, train_y, test_x, test_y):
clf = XGBClassifier()
clf.fit(train_x, train_y)
y_pred = clf.predict(test_x)
return f1_score(test_y, y_pred, average="macro")
def cross_validation_1(data, num_folds):
data = data.drop(["machine_name"], axis=1)
pom = data
folds = []
for i in range(1, num_folds):
folds.append(pom.iloc[: int(pom.size / num_folds)])
pom = pom.iloc[int(pom.size / num_folds) :, :]
folds.append(pom)
gaussianNB_f1 = []
logReg_f1 = []
ripper_f1 = []
RFC_f1 = []
ETC_f1 = []
XGB_f1 = []
for fold in folds:
train = pd.concat([data, fold, fold]).drop_duplicates(keep=False)
train_x = train.drop(["label"], axis=1)
train_y = train["label"]
train_x = np.asarray(train_x)
train_y = np.asarray(train_y)
test_x = fold.drop(["label"], axis=1)
test_y = fold["label"]
test_x = np.asarray(test_x)
test_y = np.asarray(test_y)
gaussianNB_f1.append(gaussNB(train_x, train_y, test_x, test_y))
logReg_f1.append(logReg(train_x, train_y, test_x, test_y))
ripper_f1.append(ripper(train_x, train_y, test_x, test_y))
RFC_f1.append(RFC(train_x, train_y, test_x, test_y))
ETC_f1.append(ETC(train_x, train_y, test_x, test_y))
XGB_f1.append(XGB(train_x, train_y, test_x, test_y))
gaussianNB_f1.append(np.mean(gaussianNB_f1))
logReg_f1.append(np.mean(logReg_f1))
ripper_f1.append(np.mean(ripper_f1))
RFC_f1.append(np.mean(RFC_f1))
ETC_f1.append(np.mean(ETC_f1))
XGB_f1.append(np.mean(XGB_f1))
results = pd.DataFrame(
columns=[
"algorithm",
"Fold 1",
"Fold 2",
"Fold 3",
"Fold 4",
" Fold 5",
"average",
]
)
results.loc[len(results)] = np.append(["Gaussian_NB"], gaussianNB_f1)
results.loc[len(results)] = np.append(["LogisticRegresion"], logReg_f1)
results.loc[len(results)] = np.append(["RIPPER"], ripper_f1)
results.loc[len(results)] = np.append(["RandomForrestClassifier"], RFC_f1)
results.loc[len(results)] = np.append(["ExtraTreesClassifier"], ETC_f1)
results.loc[len(results)] = np.append(["XGBClassifier"], XGB_f1)
return results
# # 2. Run the cross-validation function 1
results = cross_validation_1(train, 5)
results.head(6)
# Najbolji model je logistička regresija.
X_test = pd.read_csv("/kaggle/input/dapprojekt23/test.csv")
"""del X_test['id']
del X_test['machine_name']
X=train.drop(["label", "machine_name"], axis=1)
y=list(train['label'])
X=np.asarray(X)
X_test=np.asarray(X_test)
clf = LogisticRegression(random_state=0).fit(X, y)
y_pred=clf.predict(X_test)
submission=pd.read_csv("/kaggle/input/dapprojekt23/test.csv")
submission['Predicted']=y_pred
del submission['day']
del submission['broken']
del submission['total']
del submission['machine_name']
submission.head()"""
# submission.to_csv('submission.csv',index=False)
# Rezultat nad test skupom je 0.49957.
# # 3. Cross-validation function 2
# Ovdje će podjela na grupe biti izvršena pazeći na imena mašina tako da su svi podatci za jednu mašinu u istoj grupi,a zatim će se provesti cross validacija.
def cross_validation_2(data, num_folds):
num_of_machines = len(data.machine_name.unique())
pom = data
folds = []
for i in range(1, num_folds):
fold = pom.loc[
pom["machine_name"].isin(
pom.machine_name.unique()[: int(num_of_machines / num_folds)]
)
]
fold = fold.drop(["machine_name"], axis=1)
folds.append(fold)
pom = pom.loc[
pom["machine_name"].isin(
pom.machine_name.unique()[int(num_of_machines / num_folds) :]
)
]
pom = pom.drop(["machine_name"], axis=1)
folds.append(pom)
gaussianNB_f1 = []
logReg_f1 = []
ripper_f1 = []
RFC_f1 = []
ETC_f1 = []
XGB_f1 = []
data = data.drop(["machine_name"], axis=1)
for fold in folds:
train = pd.concat([data, fold, fold]).drop_duplicates(keep=False)
train_x = train.drop(["label"], axis=1)
train_y = train["label"]
train_x = np.asarray(train_x)
train_y = np.asarray(train_y)
test_x = fold.drop(["label"], axis=1)
test_y = fold["label"]
test_x = np.asarray(test_x)
test_y = np.asarray(test_y)
gaussianNB_f1.append(gaussNB(train_x, train_y, test_x, test_y))
logReg_f1.append(logReg(train_x, train_y, test_x, test_y))
ripper_f1.append(ripper(train_x, train_y, test_x, test_y))
RFC_f1.append(RFC(train_x, train_y, test_x, test_y))
ETC_f1.append(ETC(train_x, train_y, test_x, test_y))
XGB_f1.append(XGB(train_x, train_y, test_x, test_y))
gaussianNB_f1.append(np.mean(gaussianNB_f1))
logReg_f1.append(np.mean(logReg_f1))
ripper_f1.append(np.mean(ripper_f1))
RFC_f1.append(np.mean(RFC_f1))
ETC_f1.append(np.mean(ETC_f1))
XGB_f1.append(np.mean(XGB_f1))
results = pd.DataFrame(
columns=[
"algorithm",
"Fold 1",
"Fold 2",
"Fold 3",
"Fold 4",
" Fold 5",
"average",
]
)
results.loc[len(results)] = np.append(["Gaussian_NB"], gaussianNB_f1)
results.loc[len(results)] = np.append(["LogisticRegresion"], logReg_f1)
results.loc[len(results)] = np.append(["RIPPER"], ripper_f1)
results.loc[len(results)] = np.append(["RandomForrestClassifier"], RFC_f1)
results.loc[len(results)] = np.append(["ExtraTreesClassifier"], ETC_f1)
results.loc[len(results)] = np.append(["XGBClassifier"], XGB_f1)
return results
# # 4. Run the cross-validation function 2
results = cross_validation_2(train, 5)
results.head(6)
# Vidimo da najbolje rezultate daje Gaussian_NB. Sada ću utrenirati GausianNB model nad cijelim train skupom i onda vidjeti kako radi and test skupom.
del X_test["id"]
del X_test["machine_name"]
X = train.drop(["label", "machine_name"], axis=1)
y = list(train["label"])
X = np.asarray(X)
X_test = np.asarray(X_test)
clf = GaussianNB()
clf.fit(X, y)
y_pred = clf.predict(X_test)
submission = pd.read_csv("/kaggle/input/dapprojekt23/test.csv")
submission["Predicted"] = y_pred
del submission["day"]
del submission["broken"]
del submission["total"]
del submission["machine_name"]
submission.head()
submission.to_csv("submission.csv", index=False)
|
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import numpy as np
import plotly.express as px
import warnings
warnings.filterwarnings("ignore")
# ## Accessing the data
df = pd.read_csv("/kaggle/input/virat-kohli-100s/Virat_Kohli_100s.csv")
df.columns
df.columns = [x.lower() for x in df.columns]
df.rename(
columns={
"strike rate": "strike_rate",
"host nation": "host_nation",
"team total": "team_total",
"wickets lost": "wickets_lost",
"not out": "not_out",
},
inplace=True,
)
df.info()
df.describe()
# ## Checking for null values
df.isnull().sum()
plt.figure(figsize=(18, 5), dpi=300)
sns.heatmap(df.isnull(), yticklabels=False, cbar=False, cmap="magma")
# #### No null values
## Checking Data types
df.dtypes
# Converting important string column to numeric values
df = pd.get_dummies(df, columns=["not_out", "motm", "win", "captain"])
df.drop(
["not_out_No", "motm_No", "win_Drawn", "win_No", "win_Tie", "captain_No"],
axis=1,
inplace=True,
)
df.rename(
columns={
"not_out_Yes": "not_out",
"motm_Yes": "motm",
"win_Yes": "win",
"captain_Yes": "captain",
},
inplace=True,
)
df.head(3) # 1-Yes 0-No
df.columns
plt.subplots(figsize=(10, 10))
sns.heatmap(df.corr(), cmap="OrRd", annot=True)
centuries = df.groupby("format").size()
fig, ax = plt.subplots(figsize=(10, 5))
# Horizontal Bar Plot
ax.barh(centuries.index, centuries.values)
# Add annotation to bars
for i in ax.patches:
plt.text(
i.get_width(),
i.get_y() + 0.4,
str(round((i.get_width()), 2)),
fontsize=10,
fontweight="bold",
color="grey",
)
# Show top values
ax.invert_yaxis()
# Add Plot Title
ax.set_title("Centuries", loc="left")
# Show Plot
plt.show()
a = df[["format", "strike_rate"]]
print("IPL", a.loc[a["format"] == "T20"].mean())
print("T20i", a.loc[df["format"] == "T20i"].mean())
print("ODI", a.loc[df["format"] == "ODI"].mean())
print("test", a.loc[df["format"] == "Test"].mean())
df.groupby(["format", "inning"]).count()[["number"]]
df.groupby(["format", "inning", "position"]).count()[["number"]]
df.groupby(["against"]).count()[["number"]]
centuries = df.groupby(["against"]).count()[["number"]].reset_index()
plt.figure(figsize=(25, 15), dpi=300)
sns.barplot(x="against", y="number", data=centuries)
plt.show()
# # Considering only countries
centuries.drop([4, 5, 6, 7, 9], inplace=True)
fig, ax = plt.subplots(figsize=(10, 5))
sns.barplot(x="against", y="number", data=centuries)
ax.set_title("Countries", loc="left")
plt.show()
format = []
against = []
avg_strike_rate = []
for i in df.loc[:, ["format", "against"]]["format"].unique():
for j in df.loc[:, ["format", "against"]]["against"].unique():
strike_rate_mean = df.loc[(df["format"] == i) & (df["against"] == j)][
"strike_rate"
].mean()
format.append(i)
against.append(j)
avg_strike_rate.append(strike_rate_mean)
country_cent = pd.DataFrame(
list(zip(format, against, avg_strike_rate)),
columns=["format", "against", "avg_strike_rate"],
)
country_cent.dropna(inplace=True)
country_cent.drop(country_cent[(country_cent["format"] == "T20")].index, inplace=True)
country_cent.drop(country_cent[(country_cent["format"] == "T20i")].index, inplace=True)
country_cent = country_cent.sort_values("against")
country_cent = country_cent.reset_index(drop=True)
country_cent
format_cent = df.groupby(["format", "against"]).count()[["number"]]
format_cent
new_format_cent = format_cent.drop(["T20", "T20i"])
new_format_cent
new_format_cent.reset_index(inplace=True)
new_format_cent.loc[len(new_format_cent.index)] = ["Test", "Zimbabwe", 0]
new_format_cent.loc[len(new_format_cent.index)] = ["Test", "Pakistan", 0]
new_format_cent = new_format_cent.sort_values("against")
new_format_cent = new_format_cent.reset_index(drop=True)
new_format_cent
fig = px.bar(new_format_cent, x="format", y="number", color="against")
fig.update_layout(
height=700,
width=700,
title_text="Comparison of centuries \
in different formats against nations",
)
fig.show()
cent_strike_rate = new_format_cent.sort_values("against")
cent_strike_rate["strike_rate"] = country_cent["avg_strike_rate"]
# Correcting errors and replacing missing values
cent_strike_rate.loc[17, "strike_rate"] = 106.481482
cent_strike_rate.loc[8, "strike_rate"] = np.nan
cent_strike_rate["strike_rate"] = cent_strike_rate["strike_rate"].fillna(0)
cent_strike_rate
print(
"Virat Kohli avg strike rate in ODI is",
cent_strike_rate[cent_strike_rate["format"] == "ODI"]["strike_rate"].mean(),
)
print(
"Virat Kohli avg strike rate in Test is",
cent_strike_rate[cent_strike_rate["format"] == "test"]["strike_rate"].mean(),
)
fig = px.bar(cent_strike_rate, x="format", y="strike_rate", color="against")
fig.update_layout(
height=700,
width=700,
title_text="Comparison of strike_rate \
in different formats against nations",
)
fig.show()
cent_nations = df.groupby(["format", "host_nation"]).count()[["number"]].reset_index()
fig = px.bar(cent_nations, x="format", y="number", color="host_nation")
fig.update_layout(height=700, width=700, title_text="Centuries in different nations")
fig.show()
cent_venue = (
df.groupby(["venue"]).count()[["number"]].reset_index().sort_values("venue")
)
cent_venue = cent_venue.sort_values(by="number", ascending=False)
fig = px.bar(cent_venue, x="venue", y="number", color="number")
fig.update_layout(title_text="Centuries in different venues across formats")
fig.show()
a = (
df.groupby(["format", "venue"])
.count()[["number"]]
.sort_values("venue")
.reset_index()
)
a[a["format"] == "ODI"]
df["freq_count"] = df.groupby("score")["score"].transform("count")
# ## Finding the score a at which Virat Kohli has been dismissed most times after scoring a century across formats
df.query("freq_count==5 and not_out==0")
df.query("freq_count==4 and not_out==0 and score!=107 and score!=103")
df.query("score>200")
print(
"Average team score in test when Virat scores a double century",
df.query("score>200")["team_total"].mean(),
)
print(
"Average team total when Virat scores a century in ODI",
df[df["format"] == "ODI"]["team_total"].mean(),
)
|
# # Website Ad-Clicks Prediction with Deployment
# *Author: Amitesh Tripathi & Sayali Lad*
# ## Table of Contents
# 1. Importing libraries
# 2. Importing and exploring the dataset
# 3. Data Pre-processing and Feature Engineering
# 4. Exploratory Data Analysis (EDA)
# 5. Feature Selection & Feature Scaling
# 6. Data Modeling
# 7. Model Evaluation
# 8. Conclusion
# 9. Reference
# ># **1- Importing necessary libraries**
# ---
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import time
import warnings
warnings.filterwarnings("ignore")
# ># **2- Importing and Exploring the dataset**
# ---
from google.colab import files
uploaded = files.upload()
df = pd.read_csv("Ad_click_prediction_train (1).csv")
df.head()
df.shape
tab_info = pd.DataFrame(df.dtypes).T.rename(index={0: "column type"})
tab_info = tab_info.append(
pd.DataFrame(df.isnull().sum()).T.rename(index={0: "null values (nb)"})
)
tab_info = tab_info.append(
pd.DataFrame(df.isnull().sum() / df.shape[0] * 100).T.rename(
index={0: "null values (%)"}
)
)
tab_info
# ># **3- Data Pre-processing and Feature Engineering**
# ---
df.drop(labels="product_category_2", axis=1, inplace=True)
sum(df["is_click"] == 1)
num_missing = df["city_development_index"].isnull().sum()
if num_missing > 0:
missing_indices = df["city_development_index"].isnull()
num_missing_values = num_missing
replace_values = np.random.choice(
[1, 2, 3, 4], size=num_missing_values, p=[0.25, 0.25, 0.25, 0.25]
)
df["city_development_index"][missing_indices] = replace_values
df.isna().sum()
df = df.dropna()
df.isna().sum()
sum(df["is_click"] == 1)
df.head()
from sklearn.preprocessing import LabelEncoder
LE = LabelEncoder()
df["gender"] = LE.fit_transform(df["gender"])
df["product"] = LE.fit_transform(df["product"])
df.info()
df
df["DateTime"] = pd.to_datetime(df["DateTime"])
df["DateTime"] = df["DateTime"].dt.hour
# df['day_of_week'] = df['DateTime'].dt.dayofweek
# df['hour_of_day'] = df['DateTime'].dt.hour
df.head()
df.info()
df.nunique()
print("not click:", df.loc[df["is_click"] == 0].shape[0])
print("clicked:", df.loc[df["is_click"] == 1].shape[0])
user_ctr = df.groupby("user_id")["is_click"].agg(["count", "sum"])
user_ctr["avg_ctr"] = user_ctr["sum"] / user_ctr["count"]
user_ctr = user_ctr.reset_index()
# Merge the average CTR back to the main dataset
df = df.merge(user_ctr[["user_id", "avg_ctr"]], on="user_id", how="left")
df.head()
import matplotlib.pyplot as plt
# Create a scatter plot of count vs. avg_ctr
plt.figure(figsize=(10, 6))
plt.scatter(user_ctr["count"], user_ctr["avg_ctr"], alpha=0.5)
# Customize the plot
plt.title("Click Counts vs. Average Click-Through Rate per User", fontsize=16)
plt.xlabel("Click Counts", fontsize=14)
plt.ylabel("Average Click-Through Rate", fontsize=14)
# Show the plot
plt.show()
from sklearn.ensemble import RandomForestClassifier
from sklearn.feature_selection import SelectFromModel
# Separate features and target variable
X = df.drop(columns=["is_click"])
y = df["is_click"]
# Create a random forest classifier
rf_clf = RandomForestClassifier(random_state=42)
# Train the classifier
rf_clf.fit(X, y)
# Identify the most important features
selector = SelectFromModel(rf_clf, prefit=True)
X_selected = selector.transform(X)
# Get the names of the selected features
selected_features = X.columns[selector.get_support()]
print("Selected Features:", selected_features)
# ># **4- Exploratory Data Analysis**
# ---
import matplotlib.pyplot as plt
import seaborn as sns
labels = ["Not Click", "Click"]
size = df["is_click"].value_counts()
colors = ["#66b3ff", "#ff9999"]
explode = [0, 0.1]
sns.set(style="whitegrid")
plt.figure(figsize=(8, 6))
plt.pie(
size,
colors=colors,
explode=explode,
labels=labels,
shadow=True,
autopct="%.2f%%",
startangle=90,
)
plt.axis("equal")
plt.title("Data Distribution", fontsize=16)
plt.legend(frameon=False, fontsize=12, loc="upper right")
sns.despine(left=True, bottom=True)
plt.tight_layout()
plt.show()
plt.figure(figsize=(12, 8))
sns.set(style="white")
# Generate a custom diverging colormap
cmap = sns.diverging_palette(230, 20, as_cmap=True)
# Draw the heatmap with the mask and correct aspect ratio
sns.heatmap(
df.corr(),
cmap=cmap,
annot=True,
fmt=".2f",
vmin=-1,
vmax=1,
center=0,
square=True,
linewidths=0.5,
cbar_kws={"shrink": 0.5},
)
plt.title("Correlation Heatmap", fontsize=16)
plt.xticks(rotation=45, ha="right")
plt.yticks(rotation=0)
plt.tight_layout()
plt.show()
import matplotlib.pyplot as plt
import seaborn as sns
# Map numeric gender values to string representations
df["gender_str"] = df["gender"].map({0: "Female", 1: "Male"})
plt.figure(figsize=(8, 6))
sns.set(style="whitegrid")
sns.countplot(x="is_click", hue="gender_str", data=df, palette="Set2")
# Customize the plot
plt.title("Click Counts by Gender", fontsize=16)
plt.xlabel("Click", fontsize=14)
plt.ylabel("Count", fontsize=14)
# Customize the legend
leg = plt.legend(title="Gender", title_fontsize=12, frameon=False)
for text in leg.get_lines():
text.set_linestyle("-")
text.set_linewidth(1.5)
sns.despine(left=True)
plt.tight_layout()
plt.show()
Male = df[df["gender"] == 1]
Male
Female = df[df["gender"] == 0]
# Calculate click rates
male_click_rate = round(100 * sum(Male["is_click"] == 1) / Male.shape[0], 2)
female_click_rate = round(100 * sum(Female["is_click"] == 1) / Female.shape[0], 2)
# Print click rates
print(f"Male click rate: {male_click_rate}%")
print(f"Female click rate: {female_click_rate}%")
# Create a DataFrame for visualization
click_rates = pd.DataFrame(
{"Gender": ["Male", "Female"], "Click Rate": [male_click_rate, female_click_rate]}
)
# Create a bar plot
plt.figure(figsize=(8, 6))
sns.set(style="whitegrid")
sns.barplot(x="Gender", y="Click Rate", data=click_rates, palette="Set2")
# Customize the plot
plt.title("Click Rate by Gender", fontsize=16)
plt.xlabel("Gender", fontsize=14)
plt.ylabel("Click Rate (%)", fontsize=14)
sns.despine(left=True)
plt.tight_layout()
plt.show()
Is_clicked = df[df["is_click"] == 1]
Is_clicked_hour = Is_clicked.groupby("DateTime").agg("sum").reset_index()
Is_clicked_hour["is_click"] = (
100 * Is_clicked_hour["is_click"] / sum(Is_clicked_hour["is_click"])
)
Is_clicked_hour
sns.lineplot(x="DateTime", y="is_click", data=Is_clicked_hour, palette="Set2")
plt.title("Click Counts by DateTime")
plt.xlabel("DateTime")
plt.ylabel("Count %")
plt.show()
plt.figure(figsize=(10, 6))
sns.set(style="whitegrid")
sns.countplot(
x="product",
data=df[df["is_click"] == 1],
palette="Set2",
order=df["product"].value_counts().index,
)
# Customize the plot
plt.title("Click Counts by Product", fontsize=16)
plt.xlabel("Product ID", fontsize=14)
plt.ylabel("Count", fontsize=14)
# Customize the x-axis labels
plt.xticks(rotation=45, ha="right", fontsize=12)
sns.despine(left=True)
plt.tight_layout()
plt.show()
import matplotlib.pyplot as plt
import seaborn as sns
plt.figure(figsize=(10, 6))
sns.set(style="whitegrid")
sns.countplot(
y="campaign_id",
data=df[df["is_click"] == 1],
palette="Set2",
order=df["campaign_id"].value_counts().index,
)
# Customize the plot
plt.title("Click Counts by Campaign ID", fontsize=16)
plt.xlabel("Counts", fontsize=14)
plt.ylabel("Campaign ID", fontsize=14)
# Customize the y-axis labels
plt.yticks(fontsize=12)
sns.despine(left=True)
plt.tight_layout()
plt.show()
# ># **5- Feature Selection & Feature Scaling**
# ---
from collections import Counter
from sklearn.datasets import make_classification
import matplotlib.pyplot as plt
import numpy as np
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
# Steps 1 and 2: Sampling and preparing the feature matrix and target vector
df_sampled = (
df.groupby("is_click")
.apply(lambda x: x.sample(n=7000 if x.name == 1 else 20000))
.reset_index(drop=True)
)
x = df_sampled.drop(["is_click", "session_id"], axis=1)
y = df_sampled["is_click"]
# Step 3: Splitting the data into train and test sets
x_train_1, x_test_1, y_train_1, y_test_1 = train_test_split(
x, y, test_size=0.3, shuffle=True
)
# Step 4: Standardizing the feature matrix using the training set and transforming both the training and test sets
scaler = StandardScaler()
x_train_1 = scaler.fit_transform(x_train_1)
x_test_1 = scaler.transform(x_test_1)
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from imblearn.over_sampling import SMOTE
# Steps 1 and 2: Sampling and preparing the feature matrix and target vector
df_2 = df.sample(n=30000)
x = df_2.drop(["is_click", "session_id"], axis=1)
y = df_2["is_click"]
# Step 3: Splitting the data into train and test sets
x_train_2, x_test_2, y_train_2, y_test_2 = train_test_split(
x, y, test_size=0.3, shuffle=True
)
# Step 4: Standardizing the feature matrix using the training set and transforming both the training and test sets
scaler = StandardScaler()
x_train_2 = scaler.fit_transform(x_train_2)
x_test_2 = scaler.transform(x_test_2)
# Step 5: Applying SMOTE to handle class imbalance
sm = SMOTE(random_state=2)
x_train_2, y_train_2 = sm.fit_resample(x_train_2, y_train_2)
pd.Series(y_train_2).value_counts()
pd.Series(y_test_2).value_counts()
x_train_2.shape
x_test_2.shape
# ># **6- Data Modelling**
# ---
from sklearn.linear_model import LogisticRegression
from sklearn.linear_model import SGDClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.svm import SVC
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import GridSearchCV
from sklearn.model_selection import StratifiedKFold
from collections import Counter
from sklearn.metrics import (
confusion_matrix,
precision_score,
recall_score,
f1_score,
roc_auc_score,
accuracy_score,
classification_report,
)
from sklearn.ensemble import BaggingClassifier, GradientBoostingClassifier
from xgboost import XGBClassifier
classifiers = {
"LogisticRegression": LogisticRegression(),
"KNN": KNeighborsClassifier(),
"DT": DecisionTreeClassifier(),
"RFC": RandomForestClassifier(),
"Bagging": BaggingClassifier(),
"SGD": SGDClassifier(),
"GBC": GradientBoostingClassifier(),
"xgb": XGBClassifier(),
}
def accuracy_score(X_train, y_train):
for key, classifier in classifiers.items():
classifier.fit(X_train, y_train)
training_score = cross_val_score(classifier, X_train, y_train, cv=5)
print(
"Classifier Name : ",
classifier.__class__.__name__,
" Training Score :",
round(training_score.mean(), 2) * 100,
"%",
)
accuracy_score(x_train_1, y_train_1)
accuracy_score(x_train_2, y_train_2)
def LR_gs(X_train, y_train):
# LR
LR_param = {"penalty": ["l1", "l2"], "C": [0.001, 0.01, 0.1, 1, 10]}
LR_gs = GridSearchCV(
LogisticRegression(), param_grid=LR_param, n_jobs=-1, scoring="accuracy"
)
LR_gs.fit(X_train, y_train)
LR_estimators = LR_gs.best_estimator_
return LR_estimators
def KNN_gs(X_train, y_train):
KNN_param = {
"n_neighbors": list(range(2, 5, 1)),
"algorithm": ["auto", "ball_tree", "kd_tree", "brute"],
}
KNN_gs = GridSearchCV(
KNeighborsClassifier(), param_grid=KNN_param, n_jobs=-1, scoring="accuracy"
)
KNN_gs.fit(X_train, y_train)
KNN_estimators = KNN_gs.best_estimator_
return KNN_estimators
def SVC_gs(X_train, y_train):
SVC_param = {"C": [0.5, 0.7, 0.9, 1], "kernel": ["rfb", "sigmod", "poly"]}
SVC_gs = GridSearchCV(SVC(), param_grid=SVC_param, n_jobs=-1, scoring="accuracy")
SVC_gs.fit(X_train, y_train)
SVC_estimators = SVC_gs.best_estimator_
return SVC_estimators
def DT_gs(X_train, y_train):
DT_param = {
"criterion": ["gini", "entropy"],
"max_depth": list(range(2, 5, 1)),
"min_samples_leaf": list(range(3, 7, 1)),
}
DT_gs = GridSearchCV(
DecisionTreeClassifier(), param_grid=DT_param, n_jobs=-1, scoring="accuracy"
)
DT_gs.fit(X_train, y_train)
DT_estimators = DT_gs.best_estimator_
return DT_estimators
def RFC_gs(X_train, y_train):
RFC_param = {
"n_estimators": [100, 150, 200],
"criterion": ["gini", "entropy"],
"max_depth": list(range(2, 5, 1)),
}
RFC_gs = GridSearchCV(
RandomForestClassifier(), param_grid=RFC_param, n_jobs=-1, scoring="accuracy"
)
RFC_gs.fit(X_train, y_train)
RFC_estimators = RFC_gs.best_estimator_
return RFC_estimators
def BAG_gs(X_train, y_train):
BAG_param = {"n_estimators": [10, 15, 20]}
BAG_gs = GridSearchCV(
BaggingClassifier(), param_grid=BAG_param, n_jobs=-1, scoring="accuracy"
)
BAG_gs.fit(X_train, y_train)
BAG_estimators = BAG_gs.best_estimator_
return BAG_estimators
def SGD_gs(X_train, y_train):
SGD_param = {"penalty": ["l2", "l1"], "max_iter": [1000, 1500, 2000]}
SGD_gs = GridSearchCV(
SGDClassifier(), param_grid=SGD_param, n_jobs=-1, scoring="accuracy"
)
SGD_gs.fit(X_train, y_train)
SGD_estimators = SGD_gs.best_estimator_
return SGD_estimators
def XGB_gs(X_train, y_train):
XGB_param = {"max_depth": [3, 4, 5, 6]}
XGB_gs = GridSearchCV(
XGBClassifier(), param_grid=XGB_param, n_jobs=-1, scoring="accuracy"
)
XGB_gs.fit(X_train, y_train)
XGB_estimators = XGB_gs.best_estimator_
return XGB_estimators
LR_best_estimator = LR_gs(x_train_1, y_train_1)
KNN_best_estimator = KNN_gs(x_train_1, y_train_1)
SVC_best_estimator = SVC_gs(x_train_1, y_train_1)
DT_best_estimator = DT_gs(x_train_1, y_train_1)
RFC_best_estimator = RFC_gs(x_train_1, y_train_1)
BAG_best_estimator = BAG_gs(x_train_1, y_train_1)
SGD_best_estimator = SGD_gs(x_train_1, y_train_1)
XGB_best_estimator = XGB_gs(x_train_1, y_train_1)
LR_best_estimator = LR_gs(x_train_2, y_train_2)
KNN_best_estimator = KNN_gs(x_train_2, y_train_2)
SVC_best_estimator = SVC_gs(x_train_2, y_train_2)
DT_best_estimator = DT_gs(x_train_2, y_train_2)
RFC_best_estimator = RFC_gs(x_train_2, y_train_2)
BAG_best_estimator = BAG_gs(x_train_2, y_train_2)
SGD_best_estimator = SGD_gs(x_train_2, y_train_2)
XGB_best_estimator = XGB_gs(x_train_2, y_train_2)
# ># **7- Model Evaluation**
# ---
from sklearn.metrics import accuracy_score
from sklearn.metrics import precision_recall_fscore_support
result_df1 = pd.DataFrame(
columns=["Accuracy", "F1-score", "Recall", "Precision", "AUC_ROC"],
index=["LR", "KNN", "DT", "RFC", "Bagging", "SGD", "XGB"],
)
def caculate(models, X_test, y_test):
accuracy_results = []
F1_score_results = []
Recall_results = []
Precision_results = []
AUC_ROC_results = []
for model in models:
y_pred = model.predict(X_test)
accuracy = accuracy_score(y_test, y_pred)
precision, recall, f1_score, _ = precision_recall_fscore_support(y_test, y_pred)
AUC_ROC = roc_auc_score(y_test, y_pred) # AUC
accuracy_results.append(accuracy)
F1_score_results.append(f1_score)
Recall_results.append(recall)
AUC_ROC_results.append(AUC_ROC)
Precision_results.append(precision)
return (
accuracy_results,
F1_score_results,
Recall_results,
AUC_ROC_results,
Precision_results,
)
best_models = [
LR_best_estimator,
KNN_best_estimator,
DT_best_estimator,
RFC_best_estimator,
BAG_best_estimator,
SGD_best_estimator,
XGB_best_estimator,
]
(
accuracy_results,
F1_score_results,
Recall_results,
AUC_ROC_results,
Precision_results,
) = caculate(best_models, x_test_1, y_test_1)
result_df1["Accuracy"] = accuracy_results
result_df1["F1-score"] = F1_score_results
result_df1["Recall"] = Recall_results
result_df1["Precision"] = Precision_results
result_df1["AUC_ROC"] = AUC_ROC_results
result_df1
best_models = [
LR_best_estimator,
KNN_best_estimator,
DT_best_estimator,
RFC_best_estimator,
BAG_best_estimator,
SGD_best_estimator,
XGB_best_estimator,
]
result_df2 = pd.DataFrame(
columns=["Accuracy", "F1-score", "Recall", "Precision", "AUC_ROC"],
index=["LR", "KNN", "DT", "RFC", "Bagging", "SGD", "XGB"],
)
(
accuracy_results,
F1_score_results,
Recall_results,
AUC_ROC_results,
Precision_results,
) = caculate(best_models, x_test_2, y_test_2)
result_df2["Accuracy"] = accuracy_results
result_df2["F1-score"] = F1_score_results
result_df2["Recall"] = Recall_results
result_df2["Precision"] = Precision_results
result_df2["AUC_ROC"] = AUC_ROC_results
result_df2
import seaborn as sns
import matplotlib.pyplot as plt
# Set the plot style and size
sns.set(style="whitegrid")
plt.figure(figsize=(10, 6))
# Create the barplot
ax = sns.barplot(data=result_df1, x=result_df1.index, y="AUC_ROC")
# Customize the plot
plt.title("AUC-ROC Scores for Different Models", fontsize=16)
plt.xlabel("Model", fontsize=14)
plt.ylabel("AUC-ROC Score", fontsize=14)
# Customize the y-axis ticks
ax.set_ylim([0, 1])
# Annotate the bars with the AUC-ROC scores
for p in ax.patches:
ax.annotate(
format(p.get_height(), ".2f"),
(p.get_x() + p.get_width() / 2.0, p.get_height()),
ha="center",
va="baseline",
fontsize=12,
color="black",
xytext=(0, 5),
textcoords="offset points",
)
# Remove the top and right spines
sns.despine(top=True, right=True)
# Show the plot
plt.show()
import seaborn as sns
import matplotlib.pyplot as plt
# Set the plot style and size
sns.set(style="whitegrid")
plt.figure(figsize=(10, 6))
# Create the barplot
ax = sns.barplot(data=result_df2, x=result_df2.index, y="AUC_ROC")
# Customize the plot
plt.title("AUC-ROC Scores for Different Models (Alternative Dataset)", fontsize=16)
plt.xlabel("Model", fontsize=14)
plt.ylabel("AUC-ROC Score", fontsize=14)
# Customize the y-axis ticks
ax.set_ylim([0, 1])
# Annotate the bars with the AUC-ROC scores
for p in ax.patches:
ax.annotate(
format(p.get_height(), ".2f"),
(p.get_x() + p.get_width() / 2.0, p.get_height()),
ha="center",
va="baseline",
fontsize=12,
color="black",
xytext=(0, 5),
textcoords="offset points",
)
# Remove the top and right spines
sns.despine(top=True, right=True)
# Show the plot
plt.show()
top_3_classifier_1 = result_df1.sort_values("AUC_ROC", ascending=False)[:3]
top_3_classifier_1
top_3_classifier_2 = result_df2.sort_values("AUC_ROC", ascending=False)[:3]
top_3_classifier_2
from sklearn.ensemble import VotingClassifier
voting_clf1 = VotingClassifier(
estimators=[
("XGB", XGB_best_estimator),
("DT", DT_best_estimator),
("RFC", RFC_best_estimator),
],
n_jobs=-1,
)
voting_clf1.fit(x_train_1, y_train_1)
voting_clf2 = VotingClassifier(
estimators=[
("RFC", RFC_best_estimator),
("SGD", SGD_best_estimator),
("DT", DT_best_estimator),
],
n_jobs=-1,
)
voting_clf2.fit(x_train_2, y_train_2)
from sklearn.metrics import classification_report, roc_auc_score
from sklearn.metrics import confusion_matrix, ConfusionMatrixDisplay
y_final_pred = voting_clf1.predict(x_test_1)
print(classification_report(y_test_1, y_final_pred))
# Compute confusion matrix
confusion_mat = confusion_matrix(y_test_1, y_final_pred, labels=[1, 0])
disp = ConfusionMatrixDisplay(
confusion_matrix=confusion_mat, display_labels=["Yes(1)", "No(0)"]
)
# Customize the plot style and size
sns.set(style="whitegrid")
plt.figure(figsize=(8, 6))
# Plot the confusion matrix
disp.plot(cmap="Blues")
# Customize the plot
plt.title("Confusion Matrix for Voting Classifier 1", fontsize=16)
plt.xlabel("Predicted label", fontsize=14)
plt.ylabel("True label", fontsize=14)
# Remove the top and right spines
sns.despine(top=True, right=True)
# Show the plot
plt.show()
# Calculate AUC-ROC score
AUC_ROC = roc_auc_score(y_test_1, y_final_pred)
print(AUC_ROC)
from sklearn.metrics import classification_report, roc_auc_score
from sklearn.metrics import confusion_matrix, ConfusionMatrixDisplay
y_final_pred = voting_clf2.predict(x_test_2)
print(classification_report(y_test_2, y_final_pred))
# Compute confusion matrix
confusion_mat = confusion_matrix(y_test_2, y_final_pred, labels=[1, 0])
disp = ConfusionMatrixDisplay(
confusion_matrix=confusion_mat, display_labels=["Yes(1)", "No(0)"]
)
# Customize the plot style and size
sns.set(style="whitegrid")
plt.figure(figsize=(8, 6))
# Plot the confusion matrix
disp.plot(cmap="Blues")
# Customize the plot
plt.title("Confusion Matrix for Voting Classifier 2", fontsize=16)
plt.xlabel("Predicted label", fontsize=14)
plt.ylabel("True label", fontsize=14)
# Remove the top and right spines
sns.despine(top=True, right=True)
# Show the plot
plt.show()
# Calculate AUC-ROC score
AUC_ROC = roc_auc_score(y_test_2, y_final_pred)
print(AUC_ROC)
|
import pandas as pd
import numpy as np
from sklearn.metrics.pairwise import cosine_similarity
import random
ratings = pd.read_csv(
"/kaggle/input/lab04-dataset/ccai422_lab03_part1_data.csv", index_col="Unnamed: 0"
)
ratings.head(5)
# The total number of data points
print("The number of data points in this dataset: " + str(len(ratings)))
# The number of items (i.e. movies) in the dataset
print(
"The number of items (i.e. movies) in the dataset: "
+ str(ratings["movie_id"].nunique())
)
# The number of users in the dataset
print("The number of users in the dataset: " + str(ratings["user_id"].nunique()))
# The average ratings per user
ratings_per_users = ratings.groupby("user_id").count()
print("The average ratings per user: " + str(round(ratings_per_users.mean()[0], 2)))
# The number of ratings/user
print("The below table shows the number of ratings per user\n")
print(ratings_per_users)
# Build the ratings matrix using pivot_table function
ratings = ratings.pivot_table(values="rating", index="user_id", columns="movie_id")
# Before starting computations, copy the original ratings into a dummy ratings matrix which will have all null values imputed to zeros
cos_matrix_dummy = ratings.copy()
# Rename the axis of the new matrix
cos_matrix_dummy = cos_matrix_dummy.rename_axis("user_id", axis=1).rename_axis(
None, axis=0
)
# Compute the mean rating per user
cos_matrix_dummy["mean"] = cos_matrix_dummy.mean(axis=1)
# Substract the mean from each item
cos_matrix_dummy.loc[:, cos_matrix_dummy.columns != "mean"] = (
cos_matrix_dummy.loc[:, cos_matrix_dummy.columns != "mean"]
- cos_matrix_dummy["mean"].values[:, None]
)
# Drop the newly added mean column from the data
cos_matrix_dummy.drop(columns="mean", inplace=True)
# Display the data centred around the mean
cos_matrix_dummy.head()
# Fill NaN values with zeros
cos_matrix_dummy.fillna(0, inplace=True)
# Compute the cosine similarity. Notice we take the transpose to compute the similarity between the items
cos_matrix_dummy_sim = cosine_similarity(cos_matrix_dummy.T)
# Build dataframe based on the similarity value. Note that the type of r_matrix_dummy_sim is numpy array
cos_matrix_dummy_sim = pd.DataFrame(
cos_matrix_dummy_sim,
columns=list(cos_matrix_dummy.T.index),
index=list(cos_matrix_dummy.T.index),
)
display(cos_matrix_dummy_sim)
# Optionally set a seed
random.seed(3)
# Select random user
random_user = random.randrange(len(ratings))
print("random user ID is {}".format(random_user))
# Retrieve ratings data for the randomly selected user
random_user_ratings = ratings[ratings.index == random_user]
# Retrieve unrated item for that the randomly selected user
unrated_items_for_random_user = random_user_ratings.columns[
random_user_ratings.isnull().all(0)
]
# Randomly select an unrated item whose rating will be predicted using item-based methods
random_unrated_item = random.choice(list(unrated_items_for_random_user))
print("Item {} is unrated by user{}".format(random_unrated_item, random_user))
# Retrieve all the items rated by the randomly selected user
rated_items_for_random_user = random_user_ratings.columns[
random_user_ratings.notnull().all(0)
]
# Show the similarity matrix between the randomly selected unrated item and rated items of the randomly selected user
filtered_col_sim = list(rated_items_for_random_user)
filtered_col_sim.append(random_unrated_item)
rated_unrated_sim_random_user = cos_matrix_dummy_sim.loc[
cos_matrix_dummy_sim.columns.isin(filtered_col_sim)
]
# Of those items find the top n neighbors to the randomely selected unrated item
topn = rated_unrated_sim_random_user.nlargest(3, random_unrated_item).index.tolist()[1:]
# Retrieve the ratings that the top neighbor items gave to the unrated item
neighbors_item_ratings_random_item = random_user_ratings.loc[:, topn]
# Get the similarity values for the top n similar items
neighbors_sim = rated_unrated_sim_random_user[[random_unrated_item]].nlargest(
3, random_unrated_item
)[1:]
print(
"The top neighbors for the item {} that is unrated by user {} are the items: {}".format(
random_unrated_item, random_user, topn
)
)
print(
"The ratings for the top {} neigbors items are: {}".format(
len(topn), *neighbors_item_ratings_random_item.values
)
)
print(
"The cosine similarities between item {} and items: {} are: {}".format(
random_unrated_item, topn, neighbors_sim.values
)
)
# Apply the formula
predicted_value_for_unrated_item = (
neighbors_sim.T.dot(neighbors_item_ratings_random_item.T).values[0]
/ neighbors_sim.sum()
)
predicted_value_for_unrated_item
|
# # What is stacking?
# Stacking is a popular machine learning technique that involves combining multiple models to improve predictive accuracy. It's a powerful tool that can help you squeeze out every last bit of performance from your data and models, and it's widely used in competitions and real-world applications.
# So, what exactly is stacking? Simply put, it involves training several different models on your data and then combining their predictions to make a final prediction. The idea is that by combining the strengths of different models, you can mitigate their weaknesses and produce more accurate results.
# The basic workflow for stacking is as follows:
# - Split your data into training and validation sets.
# - Train several different models on the training set.
# - Use the validation set to evaluate the performance of each model.
# - Combine the predictions of the models using a meta-model.
# - Use the meta-model to make final predictions on new data.
# There are several different ways to combine the predictions of the models, but one common approach is to use a simple linear regression or logistic regression model as the meta-model. The meta-model takes the predictions of the base models as input and learns how to combine them to produce a final prediction.
# One of the key benefits of stacking is that it can help you overcome overfitting, which is a common problem in machine learning. By combining the predictions of multiple models, you can reduce the risk of any one model memorizing the training data and performing poorly on new data.
# Another advantage of stacking is that it can be used with any type of machine learning model, including neural networks, decision trees, and support vector machines. This means that you can experiment with different models and use the ones that work best for your data.
# 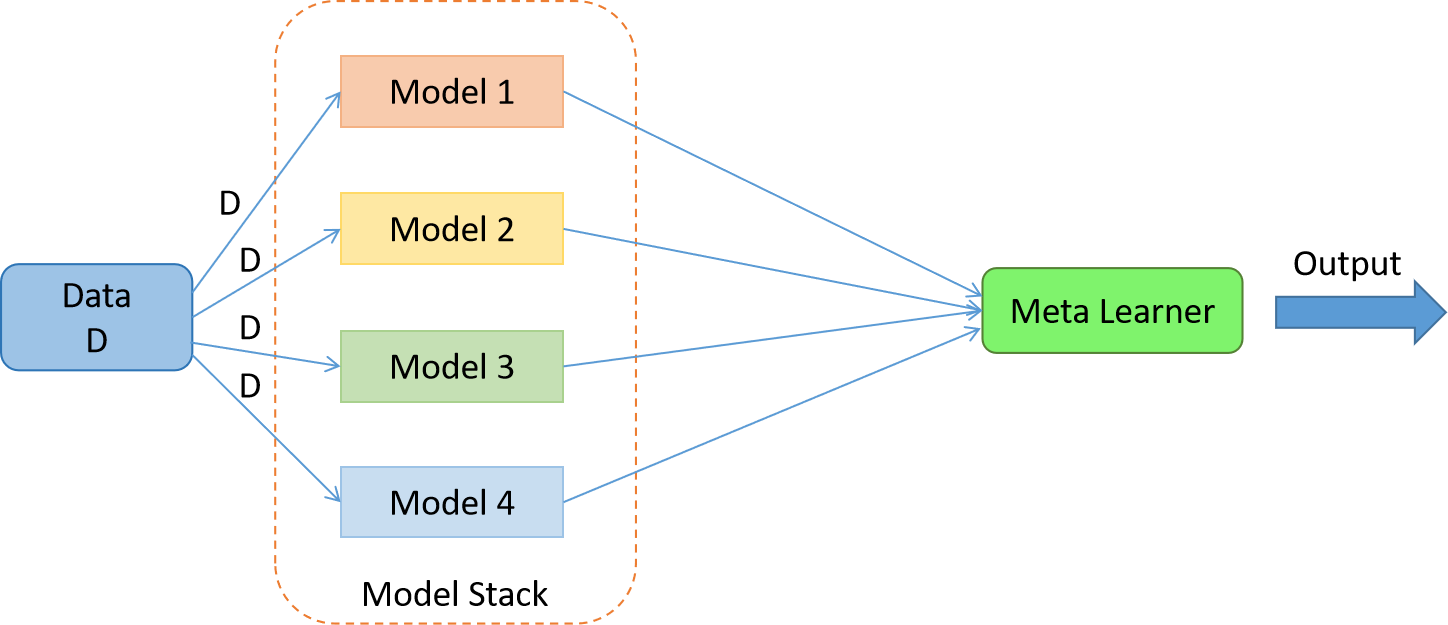
# # Import data and libraries
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import log_loss
from sklearn.svm import SVC
from sklearn.naive_bayes import GaussianNB
from xgboost import XGBClassifier
from vecstack import stacking
import pandas as pd
import numpy as np
# import data
X_train = pd.read_csv("/kaggle/input/playground-series-s3e12/train.csv")
y_train = X_train["target"]
X_test = pd.read_csv("/kaggle/input/playground-series-s3e12/test.csv")
test_id = X_test["id"]
X_train = X_train.drop(["id"], axis=1)
X_train = X_train.drop(["target"], axis=1)
X_test = X_test.drop(["id"], axis=1)
# # Initialize 1st level models
# Choose and set up a list of base models that will be used in the stacking process. In this notebook, we use logistic regression, random forest, Gaussian naive bayes, support vector machines, and XGBoost as our base models.
models = [
LogisticRegression(),
RandomForestClassifier(),
GaussianNB(),
SVC(probability=True),
XGBClassifier(),
]
# # Vecstack
# Vecstack is a Python library that provides a convenient and easy-to-use method for stacking machine learning models. Vecstack takes care of splitting the data, training the models, and generating the predictions for the next level (meta-model).
# The library is built on top of the scikit-learn library and is compatible with any scikit-learn model.
S_train_1, S_test_1 = stacking(
models, # list of models
X_train,
y_train,
X_test, # data
regression=False, # classification task (if you need regression - set to True)
mode="oof_pred", # mode: oof for train set, fit on full # train and predict test set once
needs_proba=True, # predict probabilities (if you need # class labels - set to False)
save_dir=".", # save result and log in current dir # (to disable saving - set to None)
metric=log_loss, # metric: callable
n_folds=10, # number of folds
stratified=True, # stratified split for folds
shuffle=True, # shuffle the data
random_state=0, # ensure reproducibility
verbose=2,
) # print all info
# # Level 2 Data
# Process the output from the base models to create the dataset for the meta-model. In this case, we extract the probability predictions for each class and keep only one probability column for each model, since the sum of probabilities for all classes is equal to 1.
S_train = S_train_1[:, 1::2]
S_test = S_test_1[:, 1::2]
print(f"S_train shape: {S_train.shape}")
print(f"S_test shape: {S_test.shape}")
# The new dataset is made of out-of-fold predictions of the 5 models, so it has 5 columns.
# # Level 2 Data
# Train a logistic regression model as the meta-model, which takes the predictions from the base models as input features and combines them for a final prediction. The idea is that the meta-model learns the optimal way to combine the base models' predictions, taking advantage of their strengths and compensating for their weaknesses.
# fit logreg
model = LogisticRegression()
model = model.fit(S_train, y_train)
# predict
y_pred = model.predict_proba(S_test)[:, 1]
# # Submit
# final submission
sub = pd.DataFrame()
sub["id"] = test_id
sub["target"] = y_pred
sub.to_csv("submission.csv", index=False)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import torch
import torch.nn.functional as F
from datasets import load_dataset
import torchvision
from denoising_diffusion_pytorch import Unet
from torch.optim import Adam
import tqdm
from torch.cuda.amp import GradScaler
scaler = GradScaler()
device = torch.device("cuda")
def linear_beta_schedule(timesteps):
beta_start = 0.0001
beta_end = 0.02
return torch.linspace(beta_start, beta_end, timesteps)
# ## Parameters of noise
timesteps = 500
betas = linear_beta_schedule(timesteps).to(device)
alphas = 1.0 - betas
alphas_cumprod = torch.cumprod(alphas, axis=0) # 累乘
sqrt_alphas_cumprod = torch.sqrt(alphas_cumprod)
sqrt_one_minus_alphas_cumprod = torch.sqrt(1.0 - alphas_cumprod)
sqrt_recip_alphas = torch.sqrt(1.0 / alphas)
alphas_cumprod_prev = F.pad(
alphas_cumprod[:-1], (1, 0), value=1.0
) # t时刻对应的alpht_{t-1}的累乘
posterior_variance = (
betas * (1.0 - alphas_cumprod_prev) / (1 - alphas_cumprod)
) # Xt时刻的分布方差
# ## Dataset
dataset = load_dataset("fashion_mnist")
image_size = 28
channels = 1
batch_size = 128
from torch.utils.data import DataLoader
transform = torchvision.transforms.Compose(
[
torchvision.transforms.RandomHorizontalFlip(),
torchvision.transforms.ToTensor(),
torchvision.transforms.Lambda(lambda t: (t * 2) - 1),
]
)
def transforms(examples):
examples["pixel_values"] = [
transform(image.convert("L")) for image in examples["image"]
]
del examples["image"]
return examples
transformed_dataset = dataset.with_transform(transforms).remove_columns("label")
dataloader = DataLoader(
transformed_dataset["train"], batch_size=batch_size, shuffle=True, num_workers=2
)
# ## Training stage: using $X_t$ to predict the noise $ Z_t$
def extract(a, t, x_shape):
bs = x_shape[0]
out = a.gather(-1, t)
return out.reshape(bs, *((1,) * (len(x_shape) - 1))).to(device) # (bs)->(bs,1,1,1)
epochs = 5
model = Unet(dim=18, channels=1, dim_mults=(1, 2, 4), resnet_block_groups=6).to(device)
optimizer = Adam(model.parameters(), lr=8e-5, betas=(0.9, 0.99))
lf = lambda x: (1 - x / (epochs - 1)) * (1.0 - 0.01) + 0.01 # linear
lr_scheduler = torch.optim.lr_scheduler.LambdaLR(optimizer, lr_lambda=lf)
model.train()
for epoch in range(epochs):
total_loss = 0.0
progress_bar = tqdm.tqdm(enumerate(dataloader), total=len(dataloader))
for step, batch in progress_bar:
batch = batch["pixel_values"].to(device)
bs = batch.size()[0]
t = torch.randint(0, timesteps, (bs,), device=device).long()
# generate noise
noise = torch.randn_like(batch).to(device)
# Adding Noise_T to X_0
sqrt_alphas_cumprod_t = extract(sqrt_alphas_cumprod, t, batch.shape)
sqrt_one_minus_alphas_cumprod_t = extract(
sqrt_one_minus_alphas_cumprod, t, batch.shape
)
x_t = sqrt_alphas_cumprod_t * batch + sqrt_one_minus_alphas_cumprod_t * noise
predicted_noise = model(x_t, t)
loss = F.smooth_l1_loss(noise, predicted_noise)
total_loss += loss.data
optimizer.zero_grad()
scaler.scale(loss).backward()
scaler.step(optimizer)
scaler.update()
mem = "%.3gG" % (
torch.cuda.memory_reserved() / 1e9 if torch.cuda.is_available() else 0
)
s = ("epoch: %10s" + "loss: %10.4g" + "Mem: %10s") % (
"%g/%g" % (epoch + 1, epochs),
total_loss / (step + 1),
mem,
)
progress_bar.set_description(s)
# ## 模型保存
import os
save = {"state_dict": model.state_dict()}
torch.save(save, os.path.join("/kaggle/working/", "weights.pth"))
# ## Sampling
# 
model = Unet(dim=18, channels=1, dim_mults=(1, 2, 4), resnet_block_groups=6).to(device)
model.load_state_dict(torch.load(args.model_path)["state_dict"])
img = torch.randn((1, 1, 28, 28), device=device) # X_t 随机高斯分布的噪声
model.eval()
imgs = []
for current_t in tqdm.tqdm(reversed(range(0, timesteps))):
bs = img.size()[0]
t = torch.full((bs,), current_t, device=device, dtype=torch.long)
# calculate the means and std of the distribution of X_{t-1}
betas_t = extract(betas, t, img.shape)
sqrt_one_minus_alphas_cumprod_t = extract(
sqrt_one_minus_alphas_cumprod, t, img.shape
)
sqrt_recip_alphas_t = extract(sqrt_recip_alphas, t, img.shape)
# get the prediction noise z_t
with torch.no_grad():
z_t = model(img, t)
mean = sqrt_recip_alphas_t * (img - betas_t / sqrt_one_minus_alphas_cumprod_t * z_t)
# 根据均值方差重参数化采样:
if current_t > 0:
posterior_variance_t = extract(posterior_variance, t, img.shape)
img = mean + torch.sqrt(posterior_variance_t) * torch.randn_like(img)
else:
img = mean
imgs.append(img.cpu().numpy())
def unnormalize(img):
return (img + 1) * 0.5
# ## Visualization
from matplotlib import pyplot as plt
plt.imshow(unnormalize(imgs[-1][0]).reshape(28, 28, 1), cmap="gray")
# ### Save as gif
import matplotlib.animation as animation
fig = plt.figure()
ims = []
for i in tqdm.tqdm(range(timesteps)):
im = plt.imshow(
unnormalize(imgs[i][0]).reshape(28, 28, 1), cmap="gray", animated=True
)
ims.append([im])
ims = ims[::10]
animate = animation.ArtistAnimation(
fig, ims, interval=50, blit=True, repeat_delay=10000
)
animate.save("/kaggle/working/diffusion_2.gif")
|
# 
# # Summary & Objective :
# ## Historical sales data for 45 Walmart stores located in different regions are available. There are certain events and holidays which impact sales on each day. The business is facing a challenge due to unforeseen demands and runs out of stock some times, due to inappropriate machine learning algorithm. Walmart would like to predict the sales and demand accurately. An ideal ML algorithm will predict demand accurately and ingest factors like economic conditions including CPI, Unemployment Index, etc. The objective is to determine the factors affecting the sales and to analyze the impact of markdowns around holidays on the sales.
# # Data Understanding :
# ## This is the historical data that covers sales from 2010-02-05 to 2012-11-01, in which you will find the following fields:
# ### Store - the store number
# ### Date - the week of sales
# ### Weekly_Sales - sales for the given store
# ### Holiday_Flag - whether the week is a special holiday week 1 – Holiday week 0 – Non-holiday week
# ### Temperature - Temperature on the day of sale
# ### Fuel_Price - Cost of fuel in the region
# ### CPI – Prevailing consumer price index
# ### Unemployment - Prevailing unemployment rate
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.preprocessing import StandardScaler, LabelEncoder, OneHotEncoder
import warnings
warnings.simplefilter("ignore")
df = pd.read_csv("/kaggle/input/wallmart-sale/Walmart.csv")
df.head()
df.shape
df.dtypes
df.info()
df.nunique()
df.isnull().sum()
labelencoder = LabelEncoder()
# Converting the categprical features to numerical features
for i in df.columns:
if df[i].dtype == "object":
df[i] = df[i].astype("category").cat.codes
df.info()
sns.displot(df.Weekly_Sales)
sns.pairplot(df)
df.corr()
# Linear Regression :
from sklearn.model_selection import train_test_split
from sklearn import metrics
from sklearn.linear_model import LinearRegression
X = df[["Store", "Fuel_Price", "CPI", "Unemployment", "Date"]]
Y = df["Weekly_Sales"]
X_train, X_test, Y_train, Y_test = train_test_split(X, Y, test_size=0.2)
print("Linear Regression:")
print()
reg = LinearRegression()
reg.fit(X_train, Y_train)
Y_pred = reg.predict(X_test)
print("Accuracy:", reg.score(X_train, Y_train) * 100)
print("Mean Absolute Error:", metrics.mean_absolute_error(Y_test, Y_pred))
print("Mean Squared Error:", metrics.mean_squared_error(Y_test, Y_pred))
print("Root Mean Squared Error:", np.sqrt(metrics.mean_squared_error(Y_test, Y_pred)))
# sns.scatterplot(Y_pred, Y_test)
import warnings
warnings.filterwarnings("ignore")
# Random Forest Regressor
from sklearn.ensemble import RandomForestRegressor
print("Random Forest Regressor:")
print()
rfr = RandomForestRegressor()
rfr.fit(X_train, Y_train)
Y_pred = rfr.predict(X_test)
print("Accuracy:", rfr.score(X_test, Y_test) * 100)
print("Mean Absolute Error:", metrics.mean_absolute_error(Y_test, Y_pred))
print("Mean Squared Error:", metrics.mean_squared_error(Y_test, Y_pred))
print("Root Mean Squared Error:", np.sqrt(metrics.mean_squared_error(Y_test, Y_pred)))
# sns.scatterplot(Y_pred, Y_test)
import warnings
warnings.filterwarnings("ignore")
# Here, Linear Regression is not an appropriate model to use which is clear from it's low accuracy. However, Random Forest Regression gives accuracy of over 95% , so, it is the best model to forecast demand
plt.figure(figsize=(20, 7))
barplot = sns.barplot(
x="Store",
y="Weekly_Sales",
data=df,
estimator=np.sum,
ci=None,
order=df.groupby("Store")
.agg("sum")
.reset_index()
.sort_values(by="Weekly_Sales", ascending=False)["Store"],
).set_title("Total Sales By Store")
plt.ylabel("Sales (millions)")
plt.show()
# Checking for outlier and NaN value
features_list = "Temperature, Fuel_Price, CPI, Unemployment, Date, Weekly_Sales".split(
", "
)
plt.figure(dpi=150)
count = 1
for feature in features_list:
plt.subplot(4, 2, count)
sns.boxplot(df[feature])
count += 1
plt.tight_layout()
plt.show()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
train = pd.read_csv("/kaggle/input/2023cnu-machinelearning/train.csv")
test = pd.read_csv("/kaggle/input/2023cnu-machinelearning/test.csv")
train
test
train.info()
test.info()
print("Train 크기:{}".format(train.shape))
print("Test 크기:{}".format(test.shape))
print("train 결측치 확인: ", train.isnull().sum())
print("----------------------------------------------------")
print("test 결측치 확인: ", test.isnull().sum())
df = train.corr()
df
sf = test.corr()
sf
import matplotlib.pyplot as plt
train.hist(bins=50, figsize=(20, 15))
plt.show()
# ## 데이터 전처리
# - 해당 데이터가 연속적인 시간의 값이기에 결측치를 앞 방향 값으로 채웠습니다.
# - 수치형 데이터가 문자로 표시되는d 값이 있을 수도 있기에 전부 수치형으로 변경하였습니다.
# 수치형 변수에 포함되는 데이터 타입 선정
numerics = ["int16", "int32", "int64", "float16", "float32", "float64"]
# 결측치는 위 데이터로 채우기
train[train.select_dtypes(include=numerics).columns] = train[
train.select_dtypes(include=numerics).columns
].fillna(method="ffill")
test[test.select_dtypes(include=numerics).columns] = test[
test.select_dtypes(include=numerics).columns
].fillna(method="ffill")
print("train_data 결측치 확인: ", train.isnull().sum())
print("----------------------------------------------------")
print("test_data 결측치 확인: ", test.isnull().sum())
test_not_index = test.drop(["index"], axis=1)
test_not_index
train_not_index = train.drop(["index"], axis=1)
train_not_index
# ## 검증 데이터로 분리
# 베이스 코드에는 모델의 성능을 비교해보기 위해 train 데이터에서 일부를 validation 데이터로 나눠서 진행
from sklearn.model_selection import train_test_split
train_data, val_data = train_test_split(
train_not_index, test_size=0.2, random_state=2023, shuffle=True
)
print(train_data.shape)
print(val_data.shape)
X_train = train_data.drop(["count"], axis=1)
X_val = val_data.drop(["count"], axis=1)
X_test = test_not_index
y_train = train_data["count"]
y_val = val_data["count"]
print("X_train의 shape: ", X_train.shape)
print("X_val의 shape: ", X_val.shape)
print("X_test의 shape: ", X_test.shape)
print("\t")
print("y_train의 shape: ", y_train.shape)
print("y_val의 shape: ", y_val.shape)
X_test
# ## 모델 학습
# 베이스라인에서는 간단하게 랜덤포레스트회귀를 사용하여 성능평가지표인 MAE 사용
from sklearn.ensemble import RandomForestRegressor
from sklearn.metrics import mean_absolute_error
rnd_clf = RandomForestRegressor(n_estimators=500, max_leaf_nodes=13, n_jobs=-1)
rnd_clf.fit(X_train, y_train)
y_rnd_pred = rnd_clf.predict(X_val)
print(rnd_clf.score(X_train, y_train))
print(rnd_clf.score(X_val, y_val))
print(mean_absolute_error(y_val, y_rnd_pred))
# # 결과 제출
# 예측한 결과값을 submission 파일로 생성
test_predicted = rnd_clf.predict(X_test)
X_result = pd.concat([test["index"], X_test], axis=1)
X_result
sample_sub = pd.DataFrame({"index": X_result["index"], "count": test_predicted})
sample_sub
sample_sub.to_csv("submission.csv", index=False)
|
# **Syntax Errors**
a = 90
print("The value of a is:", a)
if a > 80:
print("yes")
i = 10
while i > 0:
print(i)
i = i - 1
# **Runtime Error**
num = int(input("Enter the value of Num "))
den = int(input("Enter the value of Den"))
c = num / den
print(c) # division by 0 will create runtime error
# **Logical Error**
a = 90
b = 30
c = 60
f = (
a + b + c / 3
) # here logic of dividing is first put a+b+c in bracket then divide by 3 but by not doint so will create a logical error
print(f)
if a < 80: # by mistake wrote < other than > . This type of error is hard to find
print("yes")
else:
print("no")
|
# # Parte 1: Conceitos básicos
# ### Parte prática
# Neste material apresentamos ferramentas em Python para a manipulação de redes complexas. Vamos focar principalmente no uso do Networkx, biblioteca voltada para criação e manipulação de redes complexas.
# Os tópicos aqui apresentados consistem em conceitos básicos, iniciando com a criação de instâncias de redes complexas de diferentes tipos (com peso, sem peso, com direção e sem direção), manipulação dos vértices e arestas, geração de redes não direcionadas a partir de redes direcionadas.
# Essa biblioteca cobre as principais medidas e algoritmos conhecidos na Literatura de Redes Complexas. Quando estiver programando, em caso de dúvida recomendamos consultar a documentação disponível em https://networkx.org/documentation/stable/reference/index.html.
# Nosso curso focará no uso do Networkx, entretanto outras ferramentas estão disponíveis na web conforme listadas no [link](
# https://github.com/briatte/awesome-network-analysis).
"""
No google colab, o Networkx já está instalado.
"""
# !pip install networkx
import networkx as nx
# ## Criando redes sem direção e com direção
graph = nx.Graph() # aqui estamos criando um grafo vazio, sem vértices ou arestas
# sem direção ou atributos
label = "1"
graph.add_node(
label
) # podemos adicionar apenas um vértice com o label informado como argumento
graph.add_nodes_from([1, 2, 3]) # ou adicionar uma lista de vértices
print(graph)
graph.add_edge("1", 1) # usando os labels dos vértices, podemos adicionar arestas
graph.add_edges_from([("1", 2), (2, 3), (1, 3)]) # ou uma lista de vértices
print(graph)
# temos métodos análogos para o caso de redes dirigidas
dir_graph = nx.DiGraph() # rede vazia dirigida
label = 1
dir_graph.add_node(1) # adicionando vértices
dir_graph.add_nodes_from([2, 3, 4, 5])
dir_graph.add_edge(1, 2) # adicionando arestas direcionadas
dir_graph.add_edges_from([(2, 3), (3, 2), (3, 1), (5, 1), (3, 4)])
print(dir_graph)
# ## Visualizando as redes
# A visualização de redes complexas é uma área ampla por si só.
# Para redes pequenas, as ferramentas de visualização oferecidas pelo Networkx podem ser interessantes. Aqui vamos apresentar o básico que pode facilitar no entendimento dos métodos apresentados ao longo do material.
# vamos visualizar os dois grafos que criamos até o momento
import matplotlib.pyplot as plt
# Networkx usa o matplotlib para desenhar grafos
# por isso, podemos usar também as funções dessa outra biblioteca
pos = nx.circular_layout(
graph
) # aqui estamos definindo uma forma de calcula a posição dos vértices
nx.draw(
graph, pos=pos, node_color="orange"
) # além de passar a posição recém calculada,
# também podemos informar outros parâmetros como a cor dos vértices
nx.draw_networkx_labels(graph, pos=pos) # adicionamos os labels dos vértices
plt.title("graph") # função do matplotlib para adicionar título
plt.show() # depois de todas as informações para gerar a figura, vamos visualizá-la
# Agora o grafo direcionado que criamos anteriormente
pos = nx.circular_layout(dir_graph)
nx.draw(dir_graph, pos=pos, with_labels=True, node_size=500) # with_labels é uma forma
# mais simples de adicionar os labels do vértices
plt.title("dir_graph")
plt.show()
# ## Adicionando atributos
# Vértices e arestas podem possuir atributos.
# Em particular, o peso (weight) da conexão das arestas é um atributo muito estudado.
attr_graph = nx.Graph()
# adicionando atributos aos vértices
attr_graph.add_node(2, title="The Lord of the Rings", writer="Tolkien")
attr_graph.add_nodes_from(
[
(3, {"title": "Dune", "writer": "Frank Herbert"}),
(4, {"title": "Brave New World", "writer": "Huxley"}),
]
)
# adicionando atributos às arestas
attr_graph.add_edge(2, 3, weight=0.3)
attr_graph.add_edges_from([(2, 4, {"weight": 0.25})])
labels = nx.get_edge_attributes(attr_graph, "weight")
pos = nx.circular_layout(attr_graph)
nx.draw(attr_graph, pos=pos, with_labels=True, node_color="orange", node_size=600)
nx.draw_networkx_edge_labels(attr_graph, pos, edge_labels=labels)
plt.show()
# Mais informações sobre adição de atributos podem ser encontradas na documentação disponível em [link](https://networkx.org/documentation/networkx-1.7/tutorial/tutorial.html#adding-attributes-to-graphs-nodes-and-edges).
# Como consultar vértices e arestas:
print("Vértices do grafo attr_graph:")
# os vértices podem ser acessados da seguinte forma
for vtx in attr_graph.nodes():
print(vtx)
print("Forma alternativa de acessar os vértices do mesmo grafo:")
# dica: iterar sobre o objeto grafo também funciona
for vtx in attr_graph:
print(vtx)
print("Arestas do grafo:")
# de forma similar, para as arestas podemos fazer
for edge in attr_graph.edges():
print(edge)
# ## Matriz de adjacência
# O método adjacency_matrix retorna a matriz que representa as conexões do grafo.
# Para o caso de redes sem peso e sem direção, será uma matriz simétrica composta por 0s e 1s. No caso de redes sem peso e com direção, será uma matriz não simétrica de 0s e 1s. Finalmente, o valor 1 que normalmente indica a existência da conexão entre dois vértices será trocado pelo peso da conexão para as redes com peso.
# A representação que o Networkx usa é de uma matriz esparça. Para visualização intuitiva podemos transformá-la para versão densa. Nota: no caso de redes com muitos vértices, esse procedimento pode não ser viável, pois a matriz gerada pode ocupar muita memória.
# rede sem peso e sem direção
print("Sem peso, sem direção:")
print(nx.adjacency_matrix(graph).todense())
# rede com direção
print("Com direção:")
print(nx.adjacency_matrix(dir_graph).todense())
# rede sem direção
print("Com peso:")
print(nx.adjacency_matrix(attr_graph).todense())
# Nota: quando uma aresta sem peso informado é adicionada, o valor padrão do peso é 1
print("Lista de adjacência:")
for vtx, adj in attr_graph.adjacency():
print(vtx, adj)
# no caso de redes direcionadas, a lista de adjacência indica vértices vizinhos
# apontados pelo vértice em análise
# ## Rede de co-citação e acoplamento bibliográfico
# Uma forma de transformar uma rede direcionada em não direcionada é extraindo a rede de co-citação. A ideia geral consiste em conectar dois vértices A e B caso ambos sejam apontados por um terceiro vértice C. Podemos incluir peso nas conexões ao considerarmos o número de vértices C que apontam para A e B.
# O método apresentado a seguir é comumente utilizado para dados de citações de artigos. A ideia é análoga ao explicado anteriormente, artigos citados por um mesmo artigo devem ser conectados.
#
import numpy as np
from itertools import combinations
from collections import defaultdict
def co_citation(digraph):
di_nodes = digraph.nodes()
graph = nx.Graph()
graph.add_nodes_from(di_nodes)
edges = defaultdict(lambda: 0) # key: edge, value: edge weight
for node, adj in digraph.adjacency():
neig = list(adj.keys())
for edge in combinations(neig, 2):
edges[edge] += 1
graph.add_edges_from(list(edges.keys()))
nx.set_edge_attributes(graph, edges, "weight")
return graph
nx.draw(dir_graph, with_labels=True, node_color="orange")
plt.title("dir_graph")
plt.show()
co_cit_graph = co_citation(dir_graph)
nx.draw(co_cit_graph, with_labels=True, node_color="pink")
plt.title("co_cit_graph")
plt.show()
print("Informações do co_cit_graph:")
print(co_cit_graph.nodes())
print(co_cit_graph.edges())
print(nx.adjacency_matrix(co_cit_graph).todense())
# O acoplamento bibliográfico é outro tipo de rede também comum para dados de citações de artigos. Nesse caso, dois artigos são conectados quando citam outros artigos em comum. Uma forma de considerar peso nas conexões é o número de artigos C que A e B citam.
def get_bibliographic_coupling_graph(digraph):
di_nodes = digraph.nodes()
graph = nx.Graph()
graph.add_nodes_from(di_nodes)
edges = []
for node1, adj1 in digraph.adjacency():
for node2, adj2 in digraph.adjacency():
if node1 != node2:
co = set(adj1.keys()) & set(adj2.keys())
if len(co) > 0:
edges.append((node1, node2, len(co)))
graph.add_weighted_edges_from(edges)
return graph
nx.draw(dir_graph, with_labels=True, node_color="orange")
plt.title("dir_graph")
plt.show()
coupling_graph = get_bibliographic_coupling_graph(dir_graph)
nx.draw(coupling_graph, with_labels=True, node_color="pink")
plt.title("coupling_graph")
plt.show()
# ## Ciclos em redes direcionadas
# O Networkx tem métodos para a identificação dos ciclos em si, portanto, para confirmar se uma rede é ciclica ou acíclica podemos criar um métedo simples que confere a lista de saída do método já pronto.
# retorna um iterador sobre os ciclos do grafo
# Nota: esse método é específico para redes direcionadas
def has_cycle(graph):
return len(list(nx.simple_cycles(graph))) > 0
cycle_graph = nx.DiGraph()
cycle_graph.add_nodes_from([1, 2, 3, 4, 5])
cycle_graph.add_edges_from([(1, 2), (2, 3), (3, 1), (4, 5), (5, 2)])
nx.draw(cycle_graph, with_labels=True)
plt.show()
print(has_cycle(cycle_graph))
for cycle in nx.simple_cycles(cycle_graph):
print(cycle)
# ## Projeções de redes bipartidas
# Redes bipartidas são redes com vértides que podem ser divididos em grupos distintos, existindo apenas conexões entre vértices de grupos distintos. Para esse tipo de rede, podemos gerar um grafo de projeção selecionando um grupo de vértices que desejamos focar.
# A seguir, utilizamos um método do Networkx que gera a projeção dada uma rede bipartida. Dois vértices de um grupo são conectados caso estejam conectados a um terceiro vértice em comum do outro grupo.
bi_graph = nx.Graph()
bi_graph.add_edges_from([(1, "a"), (1, "b"), ("c", 2), ("d", 3), ("e", 2), (3, "b")])
pos = nx.bipartite_layout(bi_graph, nodes=[1, 2, 3])
nx.draw(bi_graph, pos=pos, with_labels=True, node_color="pink")
plt.title("rede bipartida")
plt.show()
projection_letters = nx.bipartite.projected_graph(bi_graph, ["a", "b", "c", "d", "e"])
nx.draw(projection_letters, with_labels=True, node_color="gray")
plt.title("projeção (vértices do tipo letras)")
plt.show()
projection_numbers = nx.bipartite.projected_graph(bi_graph, [1, 2, 3])
nx.draw(projection_numbers, with_labels=True, node_color="pink")
plt.title("projeção (vértices do tipo número)")
plt.show()
# ## Grau
# O grau de um vértice é o número de vizinhos que o vértice tem, isto é, vértices diretamente conectados. Utilizando os métodos do Networkx, é possível consultar o grau de um vértice específico ou obter uma lista com o grau de todos os vértices da rede. Para os casos de redes com peso e com direção, a medida pode ser adaptada, como mostrado a seguir.
print("grau dos vértices do grafo sem peso, sem direção: ")
for node in graph.degree(): # podemos iterar sobre todos os vértices
print(node) # tupla com o label do vértice e o grau correspondente
print(
"vértice '1', peso", graph.degree("1")
) # para saber o grau de um vértice em particular
print("vértice 3, peso", graph.degree(3))
print(
"grafo com peso:"
) # quando informamos o atributo que representa o peso das arestas
for node in attr_graph.degree(weight="weight"):
print(node)
print("grafo com direção:") # para o grafo com direção
print("- grau de entrada:")
for node in dir_graph.in_degree():
print(node)
print("- grau de saída:")
for node in dir_graph.out_degree():
print(node)
# utilizando o matplotlib podemos visualizar a distribuição do grau dos vértices de uma rede
import math
import matplotlib.pyplot as plt
degree_graph = nx.Graph()
degree_graph.add_nodes_from([1, 2, 3, 4, 5, 6])
degree_graph.add_edges_from([(1, 2), (3, 4), (5, 2), (5, 6), (2, 3), (4, 1), (5, 1)])
degrees = [val for (node, val) in degree_graph.degree()]
plt.hist(degrees)
plt.xlabel("grau")
plt.ylabel("ocorrência")
plt.show()
# # Parte 2: Medidas de redes
# Há métodos do Networkx para medidas apresentadas nas aulas teóricas, conforme apresentado a seguir.
# vamos utilizar o grafo a seguir para extrar as medidas de centralidade
metric_graph = nx.Graph()
metric_graph.add_edges_from([(1, 2), (1, 3), (1, 4), (1, 5), (5, 6), (5, 7)])
nx.draw(metric_graph, with_labels=True, node_color="pink")
plt.show()
# eigenvector centrality
print("eigenvector centrality:")
size_factor = []
for node, val in nx.eigenvector_centrality(metric_graph).items():
print("%d %.2f" % (node, val))
size_factor.append(val * 1000)
# vamos visualizar a rede com os vértices com tamanho proporcional
# à medida de centralidade
nx.draw(metric_graph, node_size=size_factor, with_labels=True)
plt.show()
# centralidade de Katz
print("centralidade de Katz:")
# os nomes dos parâmetros são diretamente adaptados do método apresentado na aula teórica
alpha = 0.01
beta = 1.0
size_factor = []
for node, val in nx.katz_centrality(metric_graph, alpha=alpha, beta=beta).items():
print("%d %.2f" % (node, val))
size_factor.append(val * 1000)
nx.draw(metric_graph, node_size=size_factor, with_labels=True)
plt.show()
# pagerank
print("pagerank:")
size_factor = []
for node, val in nx.pagerank(metric_graph).items():
print("%d %.2f" % (node, val))
size_factor.append(200 + val * 1200)
nx.draw(metric_graph, node_size=size_factor, with_labels=True)
plt.show()
# # Parte 3: Modelos de redes complexas
# Por vezes, pode ser útil gerar redes de maneira procedural. Alguns modelos de geração de redes são bem estabelecidos na Literatura por servirem de referência para comparar com redes de dados real, por exemplo.
# ## Rede aleatória
n = 100
e = 400
p = 0.15
er = nx.gnm_random_graph(
n, e
) # nesse caso são especificados número de vértices e arestas
gi = nx.gnp_random_graph(n, p) # aqui o número de vértices e a probabilidade de
# uma aresta ser criada entre dois vértices
nx.draw(er, node_size=7, edge_color="gray", alpha=0.5)
plt.title(
"gnm_random_graph (vertices=%d, arestas=%d)" % (len(er.nodes()), len(er.edges()))
)
plt.show()
nx.draw(gi, node_size=7, edge_color="gray", alpha=0.5)
plt.title(
"gnp_random_graph (vertices=%d, arestas=%d)" % (len(gi.nodes()), len(gi.edges()))
)
plt.show()
# ## Rede pequeno mundo (WS)
n = 150 # número de vértices
k = 5 # grau de conexão com vizinhos
p = 0.3 # probabilidade de realocar uma aresta
ws = nx.watts_strogatz_graph(n, k, p)
nx.draw(ws, node_size=7, edge_color="gray", alpha=0.5)
plt.title(
"watts_strogatz_graph (vertices=%d, arestas=%d)"
% (len(ws.nodes()), len(ws.edges()))
)
plt.show()
# ## Modelo de ligação preferencial (BA)
n = 100
m = 5
ba = nx.barabasi_albert_graph(n, m)
nx.draw(ba, node_size=7, edge_color="gray", alpha=0.5)
plt.title(
"barabasi_albert_graph (vertices=%d, arestas=%d)"
% (len(ba.nodes()), len(ba.edges()))
)
plt.show()
# Modelo de atratividade (disponível?)
# Modelo de fitness (disponível?)
# # Parte 5: Detecção de comunidades
# ## Método de Girvan Newman
# método Girvan Newman
# nesse caso, o método retorna um iterador com as comunidades encontradas a cada iteração do algoritmo
node_sets = None
for lvl, comms in enumerate(
nx.algorithms.community.centrality.girvan_newman(metric_graph)
):
print(lvl, comms)
if lvl == 1:
node_sets = comms
colors = dict()
for i, comm in enumerate(comms):
for vtx in comm:
colors[vtx] = i
# vamos colocar o grafo conforme as comunidades indentificadas no lvl = 1
node_color = []
for vtx in metric_graph.nodes():
node_color.append(colors[vtx])
nx.draw(metric_graph, node_color=node_color)
plt.show()
# ## Modularidade
# A medida de modularidade quantifica a qualidade de uma divisão de vértices em grupos seguindo a ideia de que grupos bem formados são aqueles com muitas conexões internas e poucas conexões com vértices de outros grupos.
# O método modularity do Networkx espera receber como parâmetros um grafo e a partição dos vértices do grafo.
# modularidade
# anteriormente salvamos na variável node_sets uma partição dos vértices gerada
# pelo algoritmo de Girvan Newman
print(node_sets)
nx.algorithms.community.quality.modularity(metric_graph, node_sets)
# vamos verificar a modularidade gerada para cada iteração do algoritmo
for lvl, comms in enumerate(
nx.algorithms.community.centrality.girvan_newman(metric_graph)
):
mod = nx.algorithms.community.quality.modularity(metric_graph, comms)
print(lvl, comms, mod)
# ## Greedy modularity optimization
# Neste caso, a ideia do algoritmo de Girvan Newman é utilizada, porém combinado com a medida de modularidade para indicar qual é a melhor partição encontrada.
# Ao contrário do método anterior que retorna um iterador, aqui já recebemos a partição ótima.
partition = nx.algorithms.community.modularity_max.greedy_modularity_communities(
metric_graph
)
def idx_partition(partition, node):
for i, p in enumerate(partition):
if node in p:
return i
return None
node_color = []
for node in metric_graph.nodes():
node_color.append(idx_partition(partition, node))
nx.draw(metric_graph, node_color=node_color)
plt.title("greedy_modularity_communities")
plt.show()
# ## Louvain
# A versão estável do Networkx ainda não possui uma implementação do método Louvain. Assim, utilizamos uma biblioteca alternativa chamada python-louvain
# que é compatível com Networkx.
import community.community_louvain as community_louvain # importando o método que vamos utilizar
partition_louvain = community_louvain.best_partition(metric_graph)
print(partition_louvain)
colors = list(partition_louvain.values())
nx.draw(metric_graph, with_labels=True, node_color=colors, cmap=plt.cm.Set2)
plt.title("community_louvain")
plt.show()
# ## Label propagation
# O método de label propagation implementado em asyn_lpa_communities retorna um iterador, onde cada item é uma comunidade identificada (conjunto de vértices identificados pelo label).
partition_iter = nx.community.label_propagation.asyn_lpa_communities(metric_graph)
partition = []
for comm in partition_iter:
partition.append(comm)
print(partition)
node_color = []
for node in metric_graph.nodes():
node_color.append(idx_partition(partition, node))
nx.draw(metric_graph, with_labels=True, node_color=node_color, cmap=plt.cm.Set3)
plt.title("asyn_lpa_communities")
plt.show()
# ## Normalized mutual Information (NMI)
# No contexto de detecção de comunidades, essa métrica pode ser utilizada para comparar uma partição identificada por algum algoritmo que estamos utilizando com a partição que sabemos previamente ser uma boa resposta (chamado também de ground truth).
# Essa medida não é necessariamente relacionada apenas com redes complexas, podendo ter um uso muito mais geral que não vamos entrar em detalhes nesse curso. Vamos utilizar a implementação disponível no sklearn.
# Normalized mutual Information
# !pip install sklearn
from sklearn.metrics.cluster import normalized_mutual_info_score as nmi
# vamos utilizar uma rede clássica da literatura
# https://en.wikipedia.org/wiki/Zachary%27s_karate_club
karate_club = nx.karate_club_graph()
# Cada vértice representa um aluno e a cor do vértice seria o grupo (professor)
# que o aluno pertence
ground_truth = [
"blue" if karate_club.nodes[v]["club"] == "Mr. Hi" else "red"
for v in karate_club.nodes()
]
nx.draw(karate_club, node_size=30, node_color=ground_truth)
plt.title("karate_club")
plt.show()
# vamos comparar a partição identificada pelo label propagation com o ground truth
partition_iter = nx.community.label_propagation.asyn_lpa_communities(karate_club)
partition = []
for comm in partition_iter:
partition.append(comm)
print(partition)
node_color = []
for node in karate_club.nodes():
node_color.append(idx_partition(partition, node))
karate_nmi = nmi(ground_truth, node_color)
print(karate_nmi)
nx.draw(karate_club, node_size=50, node_color=node_color, cmap=plt.cm.Set3)
plt.title("rede = karate club, método = asyn_lpa_communities, nmi = %.3f" % karate_nmi)
plt.show()
|
# ## Regressors
# There are many machine learning algorithms for Regression, but deciding best number of features and best model for getting best accuracy is typical. When we select a model for prediction, then we suffer problems related to underfitting and overfitting. In real scenario, there are more cases of overfitting, which we solve through regulariztion and hyperparameter tuning. Here, I implemented the following models to check best out of them for this cars dataset:
#
# 1. Linear Regression
# 2. Lasso Regression
# 3. Ridge Regression
# 4. Elastic Net Regression
# 5. Least Angle Regression
# 6. Stochastic Gradient Descent Regression
# 7. Support Vector Regression
# 8. K Neighbors Regression
# 9. Decision Tree Regression
# 10. Random Forest Regression
# 11. XG Boost Regression
from sklearn.linear_model import LinearRegression
from sklearn.linear_model import Lasso
from sklearn.linear_model import Ridge
from sklearn.linear_model import ElasticNet
from sklearn.linear_model import Lars
from sklearn.linear_model import SGDRegressor
from sklearn.svm import SVR
from sklearn.neighbors import KNeighborsRegressor
from sklearn.tree import DecisionTreeRegressor
from sklearn.ensemble import RandomForestRegressor
from xgboost import XGBRegressor
# ## Importing Libraries...
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import warnings
warnings.filterwarnings("ignore")
# ## Loading Data...
data = pd.read_csv("/kaggle/input/cars-dataset/mtcars2.csv")
data.head()
# ### Renaming column
data = data.rename(columns={"Unnamed: 0": "Model"})
data.head()
# ## EDA
data.shape
nulldata = data.isnull().sum()
nulldata[nulldata > 0]
data.describe()
data.info()
data = data.drop("Model", axis=1)
data.head()
data = data.rename(
columns={
"cyl": "cylinder",
"disp": "displacement",
"hp": "horsepower",
"wt": "weight",
}
)
data.head()
# ### Selecting main columns for analysis
data = data[["mpg", "cylinder", "displacement", "horsepower", "weight", "qsec"]]
data.head()
data.plot(kind="box", subplots=True, layout=(3, 3), figsize=(20, 12))
plt.show()
plt.subplots(figsize=(10, 8))
sns.heatmap(data.corr(), annot=True)
plt.show()
sns.pairplot(data, diag_kind="kde", kind="reg")
plt.show()
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from sklearn.metrics import r2_score
# ### Helper Functions
def build_model(
regression_fn, dataset, x_col, y_col, test_frac, preprocess_fn=None, show_plot=False
):
X = dataset[x_col]
Y = dataset[y_col]
if preprocess_fn is not None:
preprocess = preprocess_fn()
X = preprocess.fit_transform(X)
x_train, x_test, y_train, y_test = train_test_split(X, Y, test_size=test_frac)
model = regression_fn(x_train, y_train)
y_pred = model.predict(x_test)
print("Training Score : ", model.score(x_train, y_train))
print("Testing Score : ", r2_score(y_test, y_pred))
if show_plot is True:
plt.plot(y_pred, label="Predictions")
plt.plot(y_test.values, label="Actual")
plt.legend()
plt.show()
return {
"training score": model.score(x_train, y_train),
"testing score": r2_score(y_test, y_pred),
}
results_dict = {}
def compare_results():
for key in results_dict:
print("Regression : ", key)
print("Training Score : ", results_dict[key]["training score"])
print("Testing Score : ", results_dict[key]["testing score"])
print()
def overfitting():
overfitting_regression = []
print("These are the overfitting models : \n")
for key in results_dict:
if (
results_dict[key]["training score"]
> results_dict[key]["testing score"] + 0.02
):
print("Regression : ", key)
print("Training Score : ", results_dict[key]["training score"])
print("Testing Score : ", results_dict[key]["testing score"])
print()
overfitting_regression.append(key)
print("No. of Overfitting models : ", len(overfitting_regression))
for i in overfitting_regression:
print(i)
def underfitting():
underfitting_regression = []
print("These are the underfitting models : \n")
for key in results_dict:
if (
results_dict[key]["training score"]
< results_dict[key]["testing score"] - 0.02
):
print("Regression : ", key)
print("Training Score : ", results_dict[key]["training score"])
print("Testing Score : ", results_dict[key]["testing score"])
print()
underfitting_regression.append(key)
print("No. of Underfitting models : ", len(underfitting_regression))
for i in underfitting_regression:
print(i)
def balanced():
balanced_regression = []
print("Best models among all the models : \n")
for key in results_dict:
if (
results_dict[key]["training score"]
< results_dict[key]["testing score"] + 0.02
) and (
results_dict[key]["training score"]
> results_dict[key]["testing score"] - 0.02
):
print("Regression : ", key)
print("Training Score : ", results_dict[key]["training score"])
print("Testing Score : ", results_dict[key]["testing score"])
print()
balanced_regression.append(key)
print("No. of Balanced models : ", len(balanced_regression))
for i in balanced_regression:
print(i)
# ### Importing all Regressor Functions
from sklearn.linear_model import LinearRegression
from sklearn.linear_model import Lasso
from sklearn.linear_model import Ridge
from sklearn.linear_model import ElasticNet
from sklearn.linear_model import Lars
from sklearn.linear_model import SGDRegressor
from sklearn.svm import SVR
from sklearn.neighbors import KNeighborsRegressor
from sklearn.tree import DecisionTreeRegressor
from sklearn.ensemble import RandomForestRegressor
from xgboost import XGBRegressor
def linear_reg(x_train, y_train):
model = LinearRegression()
model.fit(x_train, y_train)
return model
def lasso_reg(x_train, y_train, alpha=0.5):
model = Lasso(alpha=alpha)
model.fit(x_train, y_train)
return model
def ridge_reg(x_train, y_train, alpha=0.5):
model = Ridge(alpha=alpha)
model.fit(x_train, y_train)
return model
def elastic_net_reg(
x_train, y_train, alpha=1, l1_ratio=0.5, warm_start=True, max_iter=10000
):
model = ElasticNet(
alpha=alpha, l1_ratio=l1_ratio, warm_start=warm_start, max_iter=max_iter
)
model.fit(x_train, y_train)
return model
def lars_reg(x_train, y_train, n_nonzero_coefs=4):
model = Lars(n_nonzero_coefs=n_nonzero_coefs)
model.fit(x_train, y_train)
return model
def sgd_reg(x_train, y_train, max_iter=10000, tol=1e-3):
model = SGDRegressor(max_iter=max_iter, tol=tol)
model.fit(x_train, y_train)
return model
def svr_reg(x_train, y_train, kernel="linear", epsilon=0.05, C=0.3):
model = SVR(kernel=kernel, epsilon=epsilon, C=C)
model.fit(x_train, y_train)
return model
def kneighbor_reg(x_train, y_train, n_neighbors=10):
model = KNeighborsRegressor(n_neighbors=n_neighbors)
model.fit(x_train, y_train)
return model
def decisiontree_reg(x_train, y_train, max_depth=2):
model = DecisionTreeRegressor(max_depth=max_depth)
model.fit(x_train, y_train)
return model
def randomforest_reg(x_train, y_train, max_depth=2):
model = RandomForestRegressor(max_depth=max_depth)
model.fit(x_train, y_train)
return model
def xgb_reg(x_train, y_train):
model = XGBRegressor()
model.fit(x_train, y_train)
return model
results_dict["Single Linear"] = build_model(
linear_reg, data, ["weight"], "mpg", 0.2, None, True
)
results_dict["Multi Linear all"] = build_model(
linear_reg,
data,
["cylinder", "displacement", "horsepower", "qsec", "weight"],
"mpg",
0.2,
StandardScaler,
True,
)
results_dict["Multi Linear 4 features"] = build_model(
linear_reg,
data,
["cylinder", "displacement", "horsepower", "weight"],
"mpg",
0.2,
StandardScaler,
True,
)
results_dict["Single Lasso"] = build_model(
lasso_reg, data, ["weight"], "mpg", 0.2, StandardScaler, True
)
results_dict["Multi Lasso all"] = build_model(
lasso_reg,
data,
["cylinder", "displacement", "horsepower", "qsec", "weight"],
"mpg",
0.2,
StandardScaler,
True,
)
results_dict["Multi Lasso 4 features"] = build_model(
lasso_reg,
data,
["cylinder", "displacement", "horsepower", "weight"],
"mpg",
0.2,
StandardScaler,
True,
)
results_dict["Single Ridge"] = build_model(
ridge_reg, data, ["weight"], "mpg", 0.2, StandardScaler, True
)
results_dict["Multi Ridge all"] = build_model(
ridge_reg,
data,
["cylinder", "displacement", "horsepower", "qsec", "weight"],
"mpg",
0.2,
StandardScaler,
True,
)
results_dict["Multi Ridge 4 features"] = build_model(
ridge_reg,
data,
["cylinder", "displacement", "horsepower", "weight"],
"mpg",
0.2,
None,
True,
)
results_dict["Single Elastic net"] = build_model(
elastic_net_reg, data, ["weight"], "mpg", 0.2, StandardScaler, True
)
results_dict["Multi Elastic net all"] = build_model(
elastic_net_reg,
data,
["cylinder", "displacement", "horsepower", "qsec", "weight"],
"mpg",
0.2,
StandardScaler,
True,
)
results_dict["Multi Elastic net 4 features"] = build_model(
elastic_net_reg,
data,
["cylinder", "displacement", "horsepower", "weight"],
"mpg",
0.2,
StandardScaler,
True,
)
results_dict["Single Lars"] = build_model(
lars_reg, data, ["weight"], "mpg", 0.2, StandardScaler, True
)
results_dict["Multi Lars all"] = build_model(
lars_reg,
data,
["cylinder", "displacement", "horsepower", "qsec", "weight"],
"mpg",
0.2,
StandardScaler,
True,
)
results_dict["Multi Lars 4 features"] = build_model(
lars_reg,
data,
["cylinder", "displacement", "horsepower", "weight"],
"mpg",
0.2,
None,
True,
)
results_dict["Single SVR"] = build_model(
svr_reg, data, ["weight"], "mpg", 0.2, StandardScaler, True
)
results_dict["Multi SVR all"] = build_model(
svr_reg,
data,
["cylinder", "displacement", "horsepower", "qsec", "weight"],
"mpg",
0.2,
StandardScaler,
True,
)
results_dict["Multi SVR 4 features"] = build_model(
svr_reg,
data,
["cylinder", "displacement", "horsepower", "weight"],
"mpg",
0.2,
StandardScaler,
True,
)
results_dict["Single SGD"] = build_model(
sgd_reg, data, ["weight"], "mpg", 0.2, StandardScaler, True
)
results_dict["Multi SGD all"] = build_model(
sgd_reg,
data,
["cylinder", "displacement", "horsepower", "qsec", "weight"],
"mpg",
0.2,
StandardScaler,
True,
)
results_dict["Multi SGD 4 features"] = build_model(
sgd_reg,
data,
["cylinder", "displacement", "horsepower", "weight"],
"mpg",
0.2,
StandardScaler,
True,
)
results_dict["Single Kneighbors"] = build_model(
kneighbor_reg, data, ["weight"], "mpg", 0.2, StandardScaler, True
)
results_dict["Multi Kneighbors all"] = build_model(
kneighbor_reg,
data,
["cylinder", "displacement", "horsepower", "qsec", "weight"],
"mpg",
0.2,
StandardScaler,
True,
)
results_dict["Multi Kneighbors 4 features"] = build_model(
kneighbor_reg,
data,
["cylinder", "displacement", "horsepower", "weight"],
"mpg",
0.2,
StandardScaler,
True,
)
results_dict["Single Decision tree"] = build_model(
decisiontree_reg, data, ["weight"], "mpg", 0.2, StandardScaler, True
)
results_dict["Multi Decision tree all"] = build_model(
decisiontree_reg,
data,
["cylinder", "displacement", "horsepower", "qsec", "weight"],
"mpg",
0.2,
StandardScaler,
True,
)
results_dict["Multi Decision tree 4 features"] = build_model(
decisiontree_reg,
data,
["cylinder", "displacement", "horsepower", "weight"],
"mpg",
0.2,
StandardScaler,
True,
)
results_dict["Single Random Forest"] = build_model(
randomforest_reg, data, ["weight"], "mpg", 0.2, StandardScaler, True
)
results_dict["Multi Random Forest all"] = build_model(
randomforest_reg,
data,
["cylinder", "displacement", "horsepower", "qsec", "weight"],
"mpg",
0.2,
StandardScaler,
True,
)
results_dict["Multi Random Forest 4 features"] = build_model(
randomforest_reg,
data,
["cylinder", "displacement", "horsepower", "weight"],
"mpg",
0.2,
StandardScaler,
True,
)
results_dict["Single xgb"] = build_model(
xgb_reg, data, ["weight"], "mpg", 0.2, StandardScaler, True
)
results_dict["Multi xgb all"] = build_model(
xgb_reg,
data,
["cylinder", "displacement", "horsepower", "qsec", "weight"],
"mpg",
0.2,
StandardScaler,
True,
)
results_dict["Multi xgb 4 features"] = build_model(
xgb_reg,
data,
["cylinder", "displacement", "horsepower", "weight"],
"mpg",
0.2,
StandardScaler,
True,
)
# ### Comparing all the results of applied Regressor functions with number of features
compare_results()
# ### Overfitting models
overfitting()
# ### Underfitting models
underfitting()
# ### Balanced Models
balanced()
# ### Finding best model with number of features with best training and testing score
results = pd.DataFrame(results_dict).transpose()
best = results[
(results["training score"] < results["testing score"] + 0.02)
& (results["training score"] > results["testing score"] - 0.02)
]
best_model = (
best.groupby(best.index)[["training score", "testing score"]]
.max()
.sort_values(by=["testing score"], ascending=False)
.head(1)
)
print(
f"Best Model with best training and best testing score for this dataset :\n\n{best_model}"
)
|
# # Exploring Ideas for ML
# ## Playground Series - S3, E12
# Install pycaret
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Import all the nesesary lebraries
from pycaret.classification import *
from imblearn.over_sampling import (
ADASYN,
BorderlineSMOTE,
KMeansSMOTE,
RandomOverSampler,
SMOTE,
SMOTENC,
SVMSMOTE,
)
from pathlib import Path # Import OS path libraries.
# I like to disable my Notebook Warnings.
import warnings
warnings.filterwarnings("ignore")
# Configure notebook display settings to only use 2 decimal places, tables look nicer.
pd.options.display.float_format = "{:,.2f}".format
pd.set_option("display.max_columns", 15)
pd.set_option("display.max_rows", 50)
# Define some of the notebook parameters for future experiment replication.
SEED = 42
def read_csv_to_dataframe(file_path, delimiter=",", encoding="utf-8", header="infer"):
"""
Read data from a CSV file and load it into a pandas DataFrame.
Parameters:
file_path (str): The file path to the CSV file.
delimiter (str): The delimiter used in the CSV file (default: ',').
encoding (str): The character encoding used in the CSV file (default: 'utf-8').
header (int or str): The row number to use as the header, or 'infer' to let pandas determine the header (default: 'infer').
Returns:
pandas.DataFrame: A DataFrame containing the data from the CSV file.
"""
return pd.read_csv(file_path, delimiter=delimiter, encoding=encoding, header=header)
# Example usage:
# Assuming 'file_path' is the path to your CSV file
# data = read_csv_to_dataframe(file_path)
TRN_PATH = "/kaggle/input/playground-series-s3e12/train.csv"
TST_PATH = "/kaggle/input/playground-series-s3e12/test.csv"
SUB_PATH = "/kaggle/input/playground-series-s3e12/sample_submission.csv"
ORG_PATH = "/kaggle/input/kidney-stone-prediction-based-on-urine-analysis/kindey stone urine analysis.csv"
trn_data = read_csv_to_dataframe(TRN_PATH)
tst_data = read_csv_to_dataframe(TST_PATH)
org_data = read_csv_to_dataframe(ORG_PATH)
org_data = org_data[org_data["target"] == 1]
sub_data = read_csv_to_dataframe(SUB_PATH)
import pandas as pd
def append_dataframes(df1, df2, reset_index=True):
"""
Append two pandas DataFrames along the rows.
Parameters:
df1 (pandas.DataFrame): The first DataFrame.
df2 (pandas.DataFrame): The second DataFrame.
reset_index (bool): Whether to reset the index of the resulting DataFrame (default: True).
Returns:
pandas.DataFrame: An appended DataFrame.
"""
appended_df = pd.concat([df1, df2], axis=0, ignore_index=reset_index)
return appended_df
trn_data = append_dataframes(trn_data, org_data)
def analyze_dataframe(df):
"""
Analyze a pandas DataFrame and provide a summary of its characteristics.
Parameters:
df (pandas.DataFrame): The input DataFrame to analyze.
Returns:
None
"""
print("DataFrame Information:")
print("----------------------")
display(df.info(verbose=True, show_counts=True))
print("\n")
print("DataFrame Description:")
print("----------------------")
display(df.describe(include="all"))
print("\n")
print("Number of Null Values:")
print("----------------------")
display(df.isnull().sum())
print("\n")
print("Number of Duplicated Rows:")
print("--------------------------")
display(df.duplicated().sum())
print("\n")
print("Number of Unique Values:")
print("------------------------")
display(df.nunique())
print("\n")
print("DataFrame Shape:")
print("----------------")
print(f"Rows: {df.shape[0]}, Columns: {df.shape[1]}")
# Example usage:
# Assuming 'data' is your DataFrame
# analyze_dataframe(data)
analyze_dataframe(trn_data)
TARGET = "target"
ignore = ["id", "target"]
numeric_feat = ["gravity", "ph", "osmo", "cond", "urea", "calc"]
categ_feat = [
feat for feat in trn_data.columns if feat not in numeric_feat and feat not in ignore
]
# features = categ_feat + numeric_feat
numeric_feat
categ_feat
numeric_feat = ["cond", "calc", "gravity"]
trn_data.columns
# Pycaret configuration.
clf = setup(
data=trn_data,
target=TARGET,
categorical_features=categ_feat,
numeric_features=numeric_feat,
normalize=True,
ignore_features=["id", "ph", "osmo", "urea"],
normalize_method="zscore",
fix_imbalance=True,
fix_imbalance_method=SMOTE(),
remove_outliers=True,
outliers_method="iforest",
fold_strategy="stratifiedkfold",
fold=20,
use_gpu=True,
session_id=123,
)
# Selecting what model should be trained.
best_model = compare_models(sort="auc", fold=10)
# Define the base models
ext = create_model("et", fold=20)
tuned_ext = tune_model(ext, fold=20)
plot_model(tuned_ext, plot="feature")
unseen_predictions_ext = predict_model(tuned_ext, data=tst_data, raw_score=True)
unseen_predictions_ext
# ...
sub_data["target"] = unseen_predictions_ext["prediction_score_1"]
sub_data.to_csv("pycaret_ext_submission.csv", index=False)
sub_data.head()
# ---
# Define the base models
lda = create_model("lda", fold=20)
tuned_lda = tune_model(lda, fold=20)
plot_model(tuned_lda, plot="feature")
unseen_predictions_lda = predict_model(tuned_lda, data=tst_data, raw_score=True)
unseen_predictions_lda
# ...
sub_data["target"] = unseen_predictions_lda["prediction_score_1"]
sub_data.to_csv("pycaret_lda_submission.csv", index=False)
sub_data.head()
|
# Assignment 7
# 1. Load your dataset from the “CO2 Emissions.csv” csv file.
# 2. Explore your dataset and list the name of the text columns.
# 3. Explore your dataset and check if there is any column with missing values.
# 4. Select your input variables (only select integer valued columns and columns without missing
# values) and ourput variable (hint: output colum is the one you want to predict).
# 5. Split your dataset as %80 training and %20 testing.
# 6. Scale your training data and testing data inputs.
# 7. Implement your neural network object using the ski-learn python library(you can design a
# network with one or 2 or 3 layers with any amount of the neurons).
# 8. Train (fit) your network.
# 9. Report the accuracy (R2 score) of the neural network for the test datasets.
# 10. Make a prediction for the given input values:
# Engine Size(L) = 5.6 Cylinders= 8 Fuel Consumption City (L/100 km) =17.5 Fuel Consumption Hwy
# (L/100 km) = 12 Fuel Consumption Comb (L/100 km) =15 and Fuel Consumption Comb (mpg) = 1
# **1. Loading data**
import pandas as pd
import warnings as w
w.filterwarnings("ignore")
co2_emissions = pd.read_csv("/kaggle/input/co2-emissions/CO2 Emissions.csv")
co2_emissions.head()
# **2. Explore the data**
co2_emissions.shape
co2_emissions.info()
co2_emissions.describe()
co2_emissions.columns
co2_emissions.nunique()
# **3. Divide the data into the training and test datasets**
X = co2_emissions[
[
"Engine Size(L)",
"Cylinders",
"Fuel Consumption City (L/100 km)",
"Fuel Consumption Hwy (L/100 km)",
"Fuel Consumption Comb (L/100 km)",
"Fuel Consumption Comb (mpg)",
]
]
y = co2_emissions["CO2 Emissions(g/km)"]
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
X_train.shape
X_test.shape
# **4. Scaling data**
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
X_train_scaled = scaler.fit_transform(X_train)
X_test_scaled = scaler.transform(X_test)
# **5. Implement the DL model**
from tensorflow.keras import Sequential
from tensorflow.keras.layers import Dense
from tensorflow.keras.layers import Flatten
from tensorflow.keras.optimizers import Adam
def build_model(hp):
model = Sequential()
model.add(Flatten(input_shape=(X_train_scaled.shape[1],)))
model.add(Dense(units=hp.Int("units", min_value=100, max_value=500, step=100)))
model.add(Dense(1, activation="relu"))
model.compile(
Adam(hp.Choice("learning_rate", [1e-2, 1e-3, 1e-4])), loss="mean_squared_error"
)
return model
# **6. Tune the hyper-parameters**
from kerastuner.tuners import RandomSearch
tuner = RandomSearch(build_model, objective="val_loss", max_trials=5)
tuner.search(X_train_scaled, y_train, epochs=5, validation_split=0.3)
best_model = tuner.get_best_models()[0]
tuner.results_summary()
# **7. Fitting the model to the training data**
best_model.fit(X_train_scaled, y_train)
# **8. Evaluate the quality of the trained model using test dataset**
best_model.evaluate(X_test_scaled, y_test)
# **9. Make prediction using the trained model**
best_model.predict([[5.6, 8, 17.5, 12, 15, 19]])
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import matplotlib.pyplot as plt
import torch
from torch.utils.data import TensorDataset, DataLoader, random_split
# In this notebook I will build my first Neural Net using Pytorch. For simplicity it will only consist of fully conncect layers.
# - Much of this notebook is based on chapter 4 of the Fastbook (https://course.fast.ai/Resources/book.html), which I can only recommend.
# - If you want to get familar with PyTorch quickly, check out this tutorial: https://pytorch.org/tutorials/beginner/deep_learning_60min_blitz.html
# ## 1. The Data
# The dataset consists out 28 by 28 pixel images.
# Load the dataframes
df_train = pd.read_csv("/kaggle/input/digit-recognizer/train.csv")
df_test = pd.read_csv("/kaggle/input/digit-recognizer/test.csv")
# Transform the data
y_train = df_train["label"]
X_train = np.array(df_train.drop("label", axis=1)).reshape(-1, 28, 28)
X_test = np.array(df_test).reshape(-1, 28, 28)
# We should transform our images to be between -1 and 1.
print(X_train.min(), X_train.max())
X_train = (2 / 255) * X_train - 1
print(X_train.min(), X_train.max())
# To prevent errors the data has to be converted to float32
X_train = X_train.astype("float32")
X_test = X_test.astype("float32")
# The target has to Integer for our loss function (https://stackoverflow.com/questions/60440292/runtimeerror-expected-scalar-type-long-but-found-float)
y_train = y_train.astype(int)
# Plot a sample
index = 2
plt.imshow(X_train[index])
plt.show()
print("The label of this picture is: " + str(y_train[index]))
# Before we can contiune with our model we have to convert our data into DataLoaders that PyTorch can use for training.
# Generate dataset
dataset = TensorDataset(torch.tensor(X_train), torch.tensor(y_train))
# split in train and validation set
g = torch.Generator().manual_seed(42)
train, val = random_split(dataset, [0.8, 0.2], generator=g)
train_loader = DataLoader(train)
val_loader = DataLoader(val)
# ## 2. The Model
# Now we define a simple fully connected Neutal Network. We have two hidden layers with 16 nodes each and add our nonlinearites by using the ReLU function. Our output layer has 10 units (one for each digit).
# The softmax layer was left out on purpose as it will be applied in the loss-function (https://stackoverflow.com/questions/55675345/should-i-use-softmax-as-output-when-using-cross-entropy-loss-in-pytorch).
import torch.nn as nn
import torch.nn.functional as F
class Net(nn.Module):
def __init__(self):
super().__init__()
self.fc1 = nn.Linear(28 * 28, 16)
self.fc2 = nn.Linear(16, 16)
self.fc3 = nn.Linear(16, 10)
def forward(self, x):
x = torch.flatten(
x, 1
) # flatten everything except the batch dimension (1 is the start_dims, so we don't flatten along the 0 dim which is the batch)
x = F.relu(self.fc1(x))
x = F.relu(self.fc2(x))
x = F.relu(self.fc3(x))
return x
net = Net()
# ## 3. Defining a Loss function and optimizer
# - As a loss function we will be using Classification Cross-Entropy Loss. A good explaination of this loss function can be found here: https://towardsdatascience.com/cross-entropy-loss-function-f38c4ec8643e
# - As a optimizer we will be using Stochastic Gradient Descent.
#
import torch.optim as optim
criterion = nn.CrossEntropyLoss()
optimizer = optim.SGD(
net.parameters(), lr=0.001, momentum=0.9
) # experiement with the learning rate, if train for longer it is often useful to decreae the learning rate after a few epochs
# data, labels = next(iter(train_loader))
# net(data)
# ## 4. Training the network
# We will now train our network. A detailed explained of all the steps can be found in fastbook chapter 4 (see above).
for epoch in range(5): # a epoch is one iteration over all the data
running_loss = 0.0
for i, data in enumerate(train_loader, 0):
# get the inputs (this is a batch, so lists of images & labels)
inputs, labels = data
# reset the gradient
optimizer.zero_grad()
# forward + backward + optimize
outputs = net(inputs) # forward
loss = criterion(outputs, labels) # loss
loss.backward() # backward
optimizer.step() # update
# print statistics
running_loss += loss.item()
if i % 2000 == 1999: # print every 2000 mini-batches
print(f"[{epoch + 1}, {i + 1:5d}] loss: {running_loss / 2000:.3f}")
running_loss = 0.0
# compute accuracy after each epoch
correct = 0
total = 0
with torch.no_grad():
for data in train_loader:
images, labels = data
output = net(images)
_, predicted = torch.max(outputs.data, 1)
total += labels.size(0)
correct += (predicted == labels).sum().item()
print(correct / total)
print("Training finished")
# ## 5. Testing on the validation set
# Now we compute the accuracy on the validation set.
train_iter = iter(train_loader)
image, label = next(train_iter)
net(image), label
correct = 0
total = 0
with torch.no_grad():
for data in train_loader:
images, labels = data
print(labels)
output = net(images)
_, predicted = torch.max(outputs.data, 1)
total += labels.size(0)
correct += (predicted == labels).sum().item()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
os.path.join(dirname, filename)
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# To Singapore With Love
# Deepest respect to Lee Kuan Yew 🙏🏽
#
# 
import pandas as pd
import numpy as np
import os
import plotly as plotly # Interactive Graphing Library for Python
import plotly.express as px
import plotly.graph_objects as go
from plotly.offline import init_notebook_mode, iplot, plot
init_notebook_mode(connected=True)
import matplotlib # 2D Plotting Library
import matplotlib.pyplot as plt
import seaborn as sns # Python Data Visualization Library based on matplotlib
import geopandas as gpd # Python Geospatial Data Library
plt.style.use("fivethirtyeight")
import folium
from folium.plugins import HeatMap
from folium.plugins import MarkerCluster
import warnings
warnings.simplefilter(action="ignore", category=FutureWarning)
from IPython.display import Image
# In Singapore 🇸🇬 : there are currently no licensing requirements for Airbnb properties here. BUT renting out your property on Airbnb might not be legal in the first place. Under the Planning Act, property rentals must be at least 3 months long (for private properties) or 6 months long (for HDB flats). HDB flats also cannot be rented out to tourists.
# In April, 2 men were fined $60,000 for renting out 4 condo apartments on Airbnb. The Singapore government is currently exploring options to allow short-term rentals in condos, but nothing confirmed yet. Until then, Airbnb hosts, you have been warned!
##
df = pd.read_csv("/kaggle/input/singapore-airbnb/listings.csv")
# Price Range
bins = [30, 50, 100, 150, 200, 300, 400, 500, 600, 700, 800, 900, 1000, 5000, 10000]
names = [
"<30",
"30-50",
"50-100",
"100-150",
"150-200",
"200-300",
"300-400",
"400-500",
"500-600",
"600-700",
"800-900",
"900-1000",
"1000-5000",
"5000-10000",
]
d = dict(enumerate(names, 1))
df["PriceRange"] = np.vectorize(d.get)(np.digitize(df["price"], bins))
### Missing values is replaced
df["reviews_per_month"] = df["reviews_per_month"].fillna(df["reviews_per_month"].mean())
df["last_review"] = df["last_review"].ffill()
# Singapore AirBnb Listed Properties
### Location point
plt.figure(figsize=(18, 12))
# plt.style.use("Solarize_Light2")
plt.style.use("fivethirtyeight")
map1 = sns.scatterplot(
df.longitude,
df.latitude,
hue=df.neighbourhood_group,
style=df.neighbourhood_group,
)
# map1.set_title('Singapore AirBnb Overview')
# Price Distribution Map
# plt.figure(figsize=(20,15))
# map2 = sns.scatterplot(x=df.longitude, y=df.latitude,
# hue=df.PriceRange, size=df.PriceRange,
# sizes=(20, 200), hue_norm=(0, 20),
# legend="full", data=df)
# map1.set_title('Singapore AirBnb Overview')
# Kaggle error not supporing above code
Image("/kaggle/input/singpore-geojson/price.png")
# Neighbourhood Price Distribution
# plt.figure(figsize=(20,15))
plt.style.use("fivethirtyeight")
map3 = sns.relplot(
data=df,
x="longitude",
y="latitude",
col="neighbourhood_group",
hue="PriceRange",
kind="scatter",
)
# Distribution of all listings in Singapore (Heatmap)
m = folium.Map([1.3521, 103.8198], zoom_start=10, tiles="stamenterrain")
HeatMap(
df[["latitude", "longitude"]].dropna(),
radius=8,
gradient={0.2: "blue", 0.4: "purple", 0.6: "orange", 1.0: "red"},
control_scale=True,
).add_to(m)
display(m)
# Distribution of all listings in Singapore (Count)
Singapore = folium.Map(
location=[1.3521, 103.8198], tiles="stamenterrain", zoom_start=10
)
marker_cluster = MarkerCluster().add_to(Singapore)
locations = df[["latitude", "longitude"]]
locationlist = locations.values.tolist()
for point in range(0, len(locationlist)):
folium.Marker(locationlist[point]).add_to(marker_cluster)
Singapore
# Using Singapore Community Data Map
# Source - Singapore Land Authority
import geopandas as gpd
import json
center = [1.3521, 103.8198]
m = folium.Map(location=center, zoom_start=10) # set map
# load district information
with open(
"/kaggle/input/singpore-geojson/community-use-sites-geojson.geojson",
mode="rt",
encoding="utf-8",
) as f:
geo = json.loads(f.read())
f.close()
# Add geojson to folium
folium.GeoJson(geo, name="Singapore").add_to(m)
MarkerCluster(
df[["latitude", "longitude"]].dropna(),
radius=10,
gradient={0.2: "blue", 0.4: "purple", 0.6: "orange", 1.0: "red"},
).add_to(m)
display(m)
plt.style.use("grayscale")
filter_df = df[
[
"neighbourhood_group",
"price",
"minimum_nights",
"calculated_host_listings_count",
"availability_365",
"room_type",
"PriceRange",
]
]
new_data = filter_df
y = new_data.columns
count = 0
plt.figure(figsize=(18, 18))
for i in range(2, 7):
plt.subplot(3, 3, count + 1)
sns.scatterplot(data=new_data, x="price", y=y[i], hue="room_type", palette="deep")
count = count + 1
sns.despine(offset=10, bottom=True, left=True)
## Will Update Soon ... !!
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import matplotlib.pyplot as plt
from sklearn import tree
from sklearn import metrics
from sklearn.tree import DecisionTreeClassifier
from sklearn.model_selection import train_test_split
import seaborn as sns
from sklearn.metrics import (
accuracy_score,
confusion_matrix,
classification_report,
precision_score,
recall_score,
f1_score,
)
# **READ DATA**
b = pd.read_csv("/kaggle/input/boston-house-prices/boston.csv")
b.head(b.info())
# **SCATTER PLOT AND HEATMAP TO SEE CORRELATION**
plt.scatter(x=b["AGE"], y=b["MEDV"], color="brown")
plt.xlabel = "Avg Number of rooms per dwelling"
plt.ylabel("Median value of home")
sns.heatmap(b.corr(), annot=False)
# **SPLIT DATA INTO FEATURE VARIABLE AND TARGET VARIABLE**
b.columns
feature_list = ["CRIM", "INDUS", "NOX", "RM", "AGE", "RAD", "TAX", "PTRATIO", "LSTAT"]
x = b[feature_list]
y = b["MEDV"]
x.head()
y.head()
# **SPLIT DATA INTO TRAIN AND TEST DATA**
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.2, random_state=5)
from sklearn.tree import DecisionTreeRegressor
regressor = DecisionTreeRegressor(
criterion="absolute_error", random_state=100, max_depth=4, min_samples_leaf=1
)
regressor.fit(x_train, y_train)
# **USE REGRESSOR TO PREDICT**
y_pred = regressor.predict(x_test)
y_test[1:10]
y_pred[1:10]
# **CHECK MODEL ACCURACY**
print("Accuracy:", mean_squared_error(y_test, y_pred))
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
input_folder = "/kaggle/input/tlvmc-parkinsons-freezing-gait-prediction"
import os
test_folders = [
os.path.join(input_folder, "test", "tdcsfog"),
os.path.join(input_folder, "test", "defog"),
]
submission_list = []
for folder in test_folders:
for fn in os.listdir(folder):
series_id = fn.split(".")[0]
series = pd.read_csv(os.path.join(folder, fn))
series["Id"] = series["Time"].apply(lambda time: f"{series_id}_{time}")
series["StartHesitation"] = 0
series["Turn"] = 0
series["Walking"] = 0
submission_list.append(series[["Id", "StartHesitation", "Turn", "Walking"]])
submission = pd.concat(submission_list).reset_index(drop=True)
submission.to_csv("submission.csv", index=False)
submission.head(n=100)
|
# <div style="color:white;
# display:fill;
# border-radius:5px;
# background-color:#c71585;
# font-size:200%;
# font-family:Verdana;
# letter-spacing:0.5px">
# <p style="padding: 30px;text-align:center;
# color:white;">
# 👩🏼💻 Recommendation Systems Tutorial 👩🏼💻
#
#
#
# <span style="padding: 30px;text-align:center;
# color:white;">
# 
#
# <div style="color:black;
# display:fill;
# border-radius:5px;
# background-color:#dda0dd;
# font-size:110%;
# font-family:Verdana;
# letter-spacing:0.5px">
# <p style="padding:2px;
# color:black;">
# ▶️Recommender System is a system that seeks to predict or filter preferences according to the user’s choices. Recommender systems are utilized in a variety of areas including movies, music, news, books, research articles, search queries, social tags, and products in general.
# Recommender systems produce a list of recommendations in any of the two ways –
# 💣Collaborative filtering: Collaborative filtering approaches build a model from the user’s past behavior (i.e. items purchased or searched by the user) as well as similar decisions made by other users. This model is then used to predict items (or ratings for items) that users may have an interest in.
#
# 💣Content-based filtering: Content-based filtering approaches uses a series of discrete characteristics of an item in order to recommend additional items with similar properties. Content-based filtering methods are totally based on a description of the item and a profile of the user’s preferences. It recommends items based on the user’s past preferences.
#
# ▶️They’re used by various large name companies like Google, Instagram, Spotify, Amazon, Reddit, Netflix etc. often to increase engagement with users and the platform. For example, Spotify would recommend songs similar to the ones you’ve repeatedly listened to or liked so that you can continue using their platform to listen to music. Amazon uses recommendations to suggest products to various users based on the data they have collected for that user.Recommender systems are often seen as a “black box”, the model created by these large companies are not very easily interpretable
#
#
#
# <div style="color:black;
# display:fill;
# border-radius:5px;
# background-color:#dda0dd;
# font-size:110%;
# font-family:Verdana;
# letter-spacing:0.5px">
# <p style="padding:2px;
# color:black;">
# ▶️Examples of Recommender Systems
#
# Some of the most popular examples of recommender systems include the ones used by Amazon, Netflix, and Spotify.
# ▶️Amazon’s recommender system is based on a combination of collaborative filtering and content-based algorithms. It uses past customer behavior to make recommendations for new products. Amazon’s recommender system is one of the most complex and sophisticated in the world.
#
# ▶️Netflix’s recommender system is also based on a combination of collaborative filtering and content-based algorithms. However, Netflix takes things a step further by also incorporating machine learning into its algorithm. This allows Netflix to make predictions about what a user might want to watch based on the behavior of other users.
#
# ▶️Spotify’s recommender system is based on collaborative filtering. It uses past user behavior to make recommendations for new songs to listen to.
#
#
#
#
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# import movie data set and rating data set look at columns and choosing columns
movie = pd.read_csv("/kaggle/input/movielens-20m-dataset/movie.csv")
movie.columns
movie = movie.loc[:, ["movieId", "title"]]
movie.head()
rating = pd.read_csv("/kaggle/input/movielens-20m-dataset/rating.csv")
rating.columns
rating = rating.loc[:, ["movieId", "userId", "rating"]]
rating.head()
# merge movie and rating data
data = pd.merge(movie, rating)
data.head()
data.shape
# <h2
# style="background-color:#db7093;color:white;font-family:newtimeroman;font-size:200%;border-radius: 5px 5px">limit too much data
data = data.iloc[:1000000, :]
# Calculate mean rating of all movies
data.groupby("title")["rating"].mean().sort_values(ascending=False).head(10)
# Calculate count rating of all movies
data.groupby("title")["rating"].count().sort_values(ascending=False).head(10)
# Creating dataframe with 'rating' count values
ratings = pd.DataFrame(data.groupby("title")["rating"].mean())
ratings["num of ratings"] = pd.DataFrame(data.groupby("title")["rating"].count())
ratings.head()
# Creating dataframe with 'rating' count values
ratings = pd.DataFrame(data.groupby("title")["rating"].mean())
ratings["num of ratings"] = pd.DataFrame(data.groupby("title")["rating"].count())
ratings.head()
# plot graph of 'num of ratings column'
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_style("white")
# plot graph of 'num of ratings column'
plt.figure(figsize=(10, 4))
ratings["num of ratings"].hist(bins=70)
# plot graph of 'ratings' column
plt.figure(figsize=(10, 4))
ratings["rating"].hist(bins=70)
# Sorting values according to the 'num of rating column'
moviemat = data.pivot_table(index="userId", columns="title", values="rating")
moviemat.head()
ratings.sort_values("num of ratings", ascending=False).head(10)
# make a pivot table in order to make rows are users and columns are movies. And values are rating
pivot_table = data.pivot_table(index=["userId"], columns=["title"], values="rating")
pivot_table.head(10)
# RESULT
movie_watched = pivot_table["Toy Story (1995)"]
similarity_with_other_movies = pivot_table.corrwith(
movie_watched
) # find correlation between "Bad Boys (1995)" and other movies
similarity_with_other_movies = similarity_with_other_movies.sort_values(ascending=False)
similarity_with_other_movies.head()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import seaborn as sns
import matplotlib.pyplot as plt
train_df = pd.read_csv("/kaggle/input/bike-sharing-demand/train.csv")
test_df = pd.read_csv("/kaggle/input/bike-sharing-demand/test.csv")
# # Display Data
train_df
train_df.isnull().sum()
# firt we will exctracte some useful features from datetime
train_df["datetime"] = pd.to_datetime(train_df["datetime"])
train_df["year"] = train_df.datetime.dt.year
train_df["month"] = train_df.datetime.dt.month
train_df["Day"] = train_df.datetime.dt.day
def hr_func(ts):
return ts.hour
train_df["hour"] = train_df["datetime"].apply(hr_func)
train_df.info()
train_df.hist(figsize=(15, 10))
train_df.var()
# # **Lets Start With Simple Visualization**
# fig, ax = plt.subplots(2, 2, figsize=(15, 10))
# sns.countplot(x='season', data=train_df, ax=ax[0,0]);
# sns.countplot(x='holiday', data=train_df, ax=ax[0,1]);
# sns.countplot(x='workingday', data=train_df, ax=ax[1,0]);
# sns.countplot(x='weather', data=train_df, ax=ax[1,1]);
train_df["registered"].sum()
train_df["casual"].sum()
train_df["count"].sum()
ig, ax = plt.subplots(figsize=(10, 5))
sns.barplot(data=train_df, x="season", y="count")
ax.set_title("Rented_bikes Vs season")
plt.show()
ig, ax = plt.subplots(figsize=(10, 5))
sns.barplot(data=train_df, x="Day", y="count")
ax.set_title("Rented_bikes Vs Day")
plt.show()
ig, ax = plt.subplots(figsize=(10, 5))
sns.barplot(data=train_df, x="month", y="count")
ax.set_title("Rented_bikes Vs month")
plt.show()
ig, ax = plt.subplots(figsize=(10, 5))
sns.barplot(data=train_df, x="year", y="count")
ax.set_title("Rented_bikes Vs year")
plt.show()
train_df[["season", "count"]].groupby(["season"], as_index=False).sum().sort_values(
by="count", ascending=False
)
# here we will extract weekday from days and see if weekend will be useful
import datetime as dt
import os
train_df["weekday"] = train_df[["datetime"]].apply(
lambda x: dt.datetime.strftime(x["datetime"], "%A"), axis=1
)
for i in range(len(train_df["weekday"])):
if train_df["weekday"][i] == "Sunday" or train_df["weekday"][i] == "Saturday":
train_df["weekday"][i] = 0
else:
train_df["weekday"][i] = 1
train_df["weekday"] = train_df[["weekday"]].astype(int)
# from histgram we can see number of rentals increase in the weekend
plt.hist(train_df.weekday)
# * we can see here number of rentals increase at 8-9 and also at 17-20 in working day
# * and when there is no working day we can see number of rentals increase at 10-18
workday_avg = pd.DataFrame(
train_df.groupby(["hour", "workingday"])["count"].mean()
).reset_index()
plt.figure(figsize=(10, 6))
sns.pointplot(data=workday_avg, x="hour", y="count", hue="workingday")
# draw a seaborn correlation heatmap to detect correlation between features for feature selection process
fig, ax = plt.subplots(figsize=(15, 15)) # Sample figsize in inches
sns.heatmap(train_df.corr(), annot=True, ax=ax)
# # Model Prediction
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.ensemble import RandomForestRegressor
X = train_df.drop(["datetime", "casual", "registered", "count", "Day"], axis=1)
y = train_df["count"]
X_train, X_val, y_train, y_val = train_test_split(X, y, test_size=0.3, random_state=42)
# Root Mean Squared Logarithmic Error
from math import sqrt
def RMSLE(y_pred, y_actual):
n = y_pred.size
RMSLE = sqrt(((np.log(y_pred + 1) - np.log(y_actual + 1)) ** 2).sum() / n)
return RMSLE
# # Linear Regression
lr = LinearRegression().fit(X_train, y_train)
train_pred = lr.predict(X_train)
val_pred = lr.predict(X_val)
train_score = RMSLE(train_pred, y_train)
val_score = RMSLE(val_pred, y_val)
print("Train score:", train_score)
print("validation score:", val_score)
# # Random Forest
# TRy to find the best hyperparameters for RandomForestRegressor
from sklearn.ensemble import RandomForestRegressor
from sklearn.model_selection import GridSearchCV
param_grid = {"n_estimators": [50, 60, 70, 80, 90, 100], "max_depth": [3, 7, 9, 10, 15]}
# kfold = (n_splits=3, shuffle=True, random_state=1)
BestModel = GridSearchCV(
estimator=RandomForestRegressor(),
param_grid=param_grid,
scoring="neg_root_mean_squared_error",
cv=5,
n_jobs=-1,
verbose=3,
)
BestModel.fit(X_train, y_train)
print(BestModel.best_score_, BestModel.best_estimator_, BestModel.best_params_)
rf = RandomForestRegressor(max_depth=9, n_estimators=200, random_state=42).fit(
X_train, y_train
)
tr_pred = rf.predict(X_train)
v_pred = rf.predict(X_val)
tr_score = RMSLE(tr_pred, y_train)
v_score = RMSLE(v_pred, y_val)
print("validation score:", v_score)
print("Train score:", tr_score)
# # Hist Gradient Boosting Regressor
from sklearn.experimental import enable_hist_gradient_boosting
from sklearn.ensemble import HistGradientBoostingRegressor
hist = HistGradientBoostingRegressor(max_depth=15, learning_rate=0.15).fit(
X_train, y_train
)
y_pred_hist_tr = hist.predict(X_train)
y_pred_hist_ts = hist.predict(X_val)
tr1_score = RMSLE(y_pred_hist_tr, y_train)
v1_score = RMSLE(y_pred_hist_ts, y_val)
print("validation score:", v1_score)
print("Train score:", tr1_score)
# # StackingRegressor
from sklearn.tree import DecisionTreeRegressor
from sklearn.linear_model import RidgeCV
from sklearn.svm import LinearSVR
from sklearn.ensemble import RandomForestRegressor
from sklearn.ensemble import StackingRegressor
from sklearn.linear_model import LinearRegression
from sklearn.linear_model import Ridge
from sklearn.neighbors import KNeighborsRegressor
from sklearn.ensemble import ExtraTreesRegressor
from sklearn.experimental import enable_hist_gradient_boosting
from sklearn.ensemble import HistGradientBoostingRegressor
estimators = [
("Hist", HistGradientBoostingRegressor(max_depth=9, learning_rate=0.15)),
("DT", DecisionTreeRegressor(max_depth=5, random_state=42)),
("Linear", Ridge(alpha=0.9)),
("Extra", ExtraTreesRegressor(max_depth=15, n_estimators=50, random_state=39)),
("ran", RandomForestRegressor(n_estimators=70, max_depth=15, random_state=39)),
]
reg = StackingRegressor(
estimators=estimators,
final_estimator=RandomForestRegressor(
n_estimators=60, max_depth=9, random_state=42
),
)
reg.fit(X_train, y_train)
#
y_pred_reg_tr = reg.predict(X_train)
y_pred_reg_ts = reg.predict(X_val)
R_reg = RMSLE(y_pred_reg_ts, y_val)
R1_reg = RMSLE(y_pred_reg_tr, y_train)
print("RMSLE of Random Forest Regressor :", R_reg)
print("RMSLE of what model learn :", R1_reg)
# # Test data
test_df["datetime"] = pd.to_datetime(test_df["datetime"])
test_df["year"] = test_df.datetime.dt.year
test_df["month"] = test_df.datetime.dt.month
test_df["Day"] = test_df.datetime.dt.day
def hr_func(ts):
return ts.hour
test_df["hour"] = test_df["datetime"].apply(hr_func)
test_df["weekday"] = test_df[["datetime"]].apply(
lambda x: dt.datetime.strftime(x["datetime"], "%A"), axis=1
)
for i in range(len(test_df["weekday"])):
if test_df["weekday"][i] == "Sunday" or test_df["weekday"][i] == "Saturday":
test_df["weekday"][i] = 0
else:
test_df["weekday"][i] = 1
test_df["weekday"] = test_df[["weekday"]].astype(int)
test_df = test_df.drop(["datetime", "Day"], axis=1)
test_df
# # Submission
y_test_predicted = reg.predict(test_df)
test = pd.read_csv("/kaggle/input/bike-sharing-demand/test.csv")
submission_df = pd.DataFrame({"datetime": test["datetime"], "count": y_test_predicted})
submission_df.to_csv("submission.csv", index=False)
submission_df
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Представим, что мы работаем дата-сайентистом в компании Booking. Одна из проблем компании — это нечестные отели, которые накручивают себе рейтинг. Одним из способов обнаружения таких отелей является построение модели, которая предсказывает рейтинг отеля. Если предсказания модели сильно отличаются от фактического результата, то, возможно, отель ведёт себя нечестно, и его стоит проверить.
# Немного о данных:
# 
# ## Загружаем данные
hotels_train = pd.read_csv("/kaggle/input/sf-booking/hotels_train.csv")
hotels_train.head()
hotels_test = pd.read_csv("/kaggle/input/sf-booking/hotels_test.csv")
hotels_test.head()
submission = pd.read_csv("/kaggle/input/sf-booking/submission.csv")
submission.head()
# Объединим тренировочный и тестовый наборы данных для того, чтобы было удобнее их обработать
hotels_train["sample"] = 1 # пометим тренировочную выборку
hotels_test["sample"] = 0 # пометим тестовую выборку
hotels_test["reviewer_score"] = 0 # добавим тестовому набору данных колонку с рейтингом
hotels = pd.concat([hotels_train, hotels_test], ignore_index=True)
hotels.head()
# # Заполнение пропущенных значений
# Посмотрим на наличие пропущенных значений в наборе данных
hotels.isnull().sum()
# Можно увидеть, что в некоторых отзывах у отелей нет координат. Посмотрим, у каких именно.
mask1 = hotels["lat"].isnull()
mask2 = hotels["lng"].isnull()
hotels_without_coordinates = (
hotels[mask1 | mask2]["hotel_name"].value_counts().index.tolist()
)
display(hotels_without_coordinates)
# Так как количество отелей, у которых нет координат, сравнительно небольшое, то самостоятельно поищем их координаты в интернете и заполним пропуски.
hotels.loc[
hotels["hotel_name"] == "Fleming s Selection Hotel Wien City", ["lat", "lng"]
] = ["48.206481", "16.363451"]
hotels.loc[hotels["hotel_name"] == "Hotel City Central", ["lat", "lng"]] = [
"48.213558",
"16.379923",
]
hotels.loc[hotels["hotel_name"] == "Hotel Atlanta", ["lat", "lng"]] = [
"48.220310",
"16.355881",
]
hotels.loc[
hotels["hotel_name"] == "Maison Albar Hotel Paris Op ra Diamond", ["lat", "lng"]
] = ["48.875223", "2.323385"]
hotels.loc[hotels["hotel_name"] == "Hotel Daniel Vienna", ["lat", "lng"]] = [
"48.188835",
"16.383810",
]
hotels.loc[
hotels["hotel_name"] == "Hotel Pension Baron am Schottentor", ["lat", "lng"]
] = ["48.216705", "16.359819"]
hotels.loc[
hotels["hotel_name"] == "Austria Trend Hotel Schloss Wilhelminenberg Wien",
["lat", "lng"],
] = ["48.219555", "16.285566"]
hotels.loc[
hotels["hotel_name"] == "Derag Livinghotel Kaiser Franz Joseph Vienna",
["lat", "lng"],
] = ["48.245914", "16.341188"]
hotels.loc[hotels["hotel_name"] == "NH Collection Barcelona Podium", ["lat", "lng"]] = [
"41.391430",
"2.177890",
]
hotels.loc[hotels["hotel_name"] == "City Hotel Deutschmeister", ["lat", "lng"]] = [
"48.220856",
"16.366642",
]
hotels.loc[hotels["hotel_name"] == "Hotel Park Villa", ["lat", "lng"]] = [
"48.233379",
"16.345510",
]
hotels.loc[hotels["hotel_name"] == "Cordial Theaterhotel Wien", ["lat", "lng"]] = [
"48.209530",
"16.351515",
]
hotels.loc[hotels["hotel_name"] == "Holiday Inn Paris Montmartre", ["lat", "lng"]] = [
"48.888860",
"2.333190",
]
hotels.loc[hotels["hotel_name"] == "Roomz Vienna", ["lat", "lng"]] = [
"48.222458",
"16.393538",
]
hotels.loc[
hotels["hotel_name"] == "Mercure Paris Gare Montparnasse", ["lat", "lng"]
] = ["48.839701", "2.323519"]
hotels.loc[hotels["hotel_name"] == "Renaissance Barcelona Hotel", ["lat", "lng"]] = [
"41.392429",
"2.167500",
]
hotels.loc[hotels["hotel_name"] == "Hotel Advance", ["lat", "lng"]] = [
"41.379389",
"2.157475",
]
# Удостоверимся, что пропусков больше нет
hotels.isnull().sum()
# # Исследование целевого признака
hotels_reviewer_score_hist = sns.histplot(hotels_train["reviewer_score"], bins=30)
hotels_reviewer_score_hist.set_title("Распределение оценок")
hotels_reviewer_score_box = sns.boxplot(hotels_train["reviewer_score"], orient="h")
hotels_reviewer_score_box.set_title("Распределение оценок")
# Можно заметить, что оценка отеля может лежать в диапазоне от 0 до 10. Коробчатая диаграмма показывает, что 50% оценок лежат между значениями примерно в 7.5 и 9.5.
# # Проектирование признаков
# Посмотрим на типы данных в нашем наборе
hotels.info()
# ## Количество дней с публикации отзыва (days_since_review)
# Преобразуем признак 'days_since_review' из object в int, оставив только число
hotels["days_since_review"] = hotels["days_since_review"].apply(
lambda x: int(x.split(" ")[0])
)
hotels["days_since_review"]
# ## Дата публикации отзыва (review_date)
# Преобразуем признак 'review_date' в datetime
hotels["review_date"] = pd.to_datetime(hotels["review_date"])
# Далее выделим в отдельные признаки месяц и сезон, в котором был написан отзыв
hotels["review_date_month"] = pd.to_datetime(hotels["review_date"]).dt.month
def get_season(x):
if x in [1, 2, 12]:
return 1
elif x in [3, 4, 5]:
return 2
elif x in [6, 7, 8]:
return 3
elif x in [9, 10, 11]:
return 4
hotels["review_date_season"] = hotels["review_date_month"].apply(get_season)
# Посмотрим, меняется ли средняя оценка и количество отзывов в зависимости от месяца
hotels_date_gb = (
hotels.groupby("review_date_month")["reviewer_score"].agg("mean").reset_index()
)
hotels_date_gb_line = sns.lineplot(
hotels_date_gb, x="review_date_month", y="reviewer_score"
)
hotels_date_gb_line.set_title("Зависимость оценки от месяца отзыва")
hotels_date_gb = (
hotels.groupby("review_date_month")["hotel_name"].agg("count").reset_index()
)
hotels_date_gb_line = sns.lineplot(
hotels_date_gb, x="review_date_month", y="hotel_name"
)
hotels_date_gb_line.set_title("Зависимость количества отзывов от месяца отзыва")
# Посмотрим, меняется ли средняя оценка и количество отзывов в зависимости от сезона
hotels_date_gb = (
hotels.groupby("review_date_season")["reviewer_score"].agg("mean").reset_index()
)
hotels_date_gb_line = sns.lineplot(
hotels_date_gb, x="review_date_season", y="reviewer_score"
)
hotels_date_gb_line.set_title(
"Зависимость средней оценки от сезона, в котором был написан отзыв"
)
hotels_date_gb = (
hotels.groupby("review_date_season")["hotel_name"].agg("count").reset_index()
)
hotels_date_gb_line = sns.lineplot(
hotels_date_gb, x="review_date_season", y="hotel_name"
)
hotels_date_gb_line.set_title(
"Зависимость количества отзывов от сезона, в котором был написан отзыв"
)
|
# **Import modules**
import pandas as pd
import numpy as np
import os
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
from tqdm.notebook import tqdm
warnings.filterwarnings("ignore")
import tensorflow as tf
from tensorflow.keras.utils import load_img
from keras.models import Sequential, Model
from keras.layers import Dense, Conv2D, Dropout, Flatten, MaxPooling2D, Input
# **Load the dataset**
BASE_DIR = "/content/imdb_crop"
def imShow(path):
import cv2
import matplotlib.pyplot as plt
image = cv2.imread(path)
height, width = image.shape[:2]
resized_image = cv2.resize(
image, (3 * width, 3 * height), interpolation=cv2.INTER_CUBIC
)
fig = plt.gcf()
fig.set_size_inches(18, 10)
plt.axis("off")
# plt.rcParams['figure.figsize'] = [10, 5]
plt.imshow(cv2.cvtColor(resized_image, cv2.COLOR_BGR2RGB))
def imShowCV(image):
import cv2
import matplotlib.pyplot as plt
height, width = image.shape[:2]
resized_image = cv2.resize(
image, (3 * width, 3 * height), interpolation=cv2.INTER_CUBIC
)
fig = plt.gcf()
fig.set_size_inches(18, 10)
plt.axis("off")
# plt.rcParams['figure.figsize'] = [10, 5]
plt.imshow(cv2.cvtColor(resized_image, cv2.COLOR_BGR2RGB))
# Printing the number of files that do not have '.mat' extension
import os
ages = os.scandir("/content/imdb_crop")
length = 0
for age in ages:
if ".mat" not in str(age.path):
length += len(os.listdir(age.path))
print(length)
import scipy.io
imdbMat = scipy.io.loadmat("imdb_crop/imdb.mat")
imdbPlace = imdbMat["imdb"][0][0]
print(imdbPlace)
print(len(imdbPlace))
for item in imdbPlace:
print(item)
print(len(item[0]))
# labels - age, gender, ethnicity
image_paths = []
age_labels = []
gender_labels = []
place = imdbPlace
where = "imdb_crop"
total = 0
for i in range(460723):
if i % 10000 == 0:
print(i)
bYear = int(place[0][0][i] / 365) # birth year
taken = place[1][0][i] # photo taken
path = place[2][0][i][0]
gender = place[3][0][i] # Female/Male
name = place[4][0][i] # Name
faceBox = place[5][0][i] # Face coords
faceScore = place[6][0][i] # Face score
secFaceScore = place[7][0][i] # Sec face score
# Calculating shit
age = taken - bYear
faceScore = str(faceScore)
secFaceScore = str(secFaceScore)
age_labels.append(age)
gender_labels.append(gender)
image_paths.append(path)
if (
"n" not in faceScore
): # n as in Inf; if true, implies that there isn't a face in the image
if "a" in secFaceScore: # a as in NaN; implies that no second face was found
if age >= 0:
try:
gender = int(gender)
total += 1
if i > 1500:
print("----------------------")
print(i)
print(bYear)
print(taken)
print("AGE", age)
print("NAME", name)
print("GENDER", gender)
print(faceBox)
print(faceScore)
print(secFaceScore)
print(age_labels)
print(gender_labels)
print(image_paths)
imShow(os.path.join(where, path))
break
except:
print("Failed with gender")
continue
print(total)
# Feature Extraction
def extract_features(images):
features = []
for image in tqdm(images):
img = load_img(image, grayscale=True)
img = img.resize((128, 128), Image.ANTIALIAS)
img = np.array(img)
features.append(img)
features = np.array(features)
# ignore this step if using RGB
features = features.reshape(len(features), 128, 128, 1)
return features
# convert to dataframe
df = pd.DataFrame()
df["image"], df["age"], df["gender"] = image_paths, age_labels, gender_labels
df.head()
import os
# iterate over all the files in the folder imdb_crop
for filename in os.listdir("imdb_crop"):
# create the full path to the file
# file_path = os.path.join(image_paths, filename)
temp = filename.split("/")
img_path_new = int(temp[1])
# apply the function to the file
X = extract_features(df["image"])
# create the full path to the output file
output_file_path = os.path.join("imdb_crop_output", filename)
X = extract_features(df["image"])
X.shape
# normalize the images
X = X / 255.0
y_gender = np.array(df["gender"])
y_age = np.array(df["age"])
input_shape = (128, 128, 1)
# Model Creation
# Model 1
inputs = Input((input_shape))
# convolutional layers
conv_1 = Conv2D(32, kernel_size=(3, 3), activation="relu")(inputs)
maxp_1 = MaxPooling2D(pool_size=(2, 2))(conv_1)
conv_2 = Conv2D(64, kernel_size=(3, 3), activation="relu")(maxp_1)
maxp_2 = MaxPooling2D(pool_size=(2, 2))(conv_2)
conv_3 = Conv2D(128, kernel_size=(3, 3), activation="relu")(maxp_2)
maxp_3 = MaxPooling2D(pool_size=(2, 2))(conv_3)
conv_4 = Conv2D(256, kernel_size=(3, 3), activation="relu")(maxp_3)
maxp_4 = MaxPooling2D(pool_size=(2, 2))(conv_4)
flatten = Flatten()(maxp_4)
# fully connected layers
dense_1 = Dense(256, activation="relu")(flatten)
dense_2 = Dense(256, activation="relu")(flatten)
dropout_1 = Dropout(0.3)(dense_1)
dropout_2 = Dropout(0.3)(dense_2)
output_1 = Dense(1, activation="sigmoid", name="gender_out")(dropout_1)
output_2 = Dense(1, activation="relu", name="age_out")(dropout_2)
model = Model(inputs=[inputs], outputs=[output_1, output_2])
model.compile(
loss=["binary_crossentropy", "mae"], optimizer="adam", metrics=["accuracy"]
)
# Train the model
# train model
history = model.fit(
x=X, y=[y_gender, y_age], batch_size=32, epochs=30, validation_split=0.2
)
# Save the model
model.save("model_IMDB-WIKI_30.h5")
|
# [birdclef-2023-pytorch-lightning-inference](https://www.kaggle.com/code/nischaydnk/birdclef-2023-pytorch-lightning-inference)
# [birdclef-2023-pytorch-lightning-training-w-cmap](https://www.kaggle.com/code/nischaydnk/birdclef-2023-pytorch-lightning-training-w-cmap)
# [audio-deep-learning-made-simple-sound-classification-step-by-step](https://towardsdatascience.com/audio-deep-learning-made-simple-sound-classification-step-by-step-cebc936bbe5)
# [signal_framing](https://superkogito.github.io/blog/2020/01/25/signal_framing.html)
import pandas as pd
import numpy as np
import torch
import torch.nn as nn
import torchaudio
from pathlib import Path
from sklearn import preprocessing
from torch.nn import init
from torch.utils.data import DataLoader, Dataset, random_split
download_path = "/kaggle/input/"
metadata_file = download_path + "/birdclef-2023/train_metadata.csv"
df = pd.read_csv(metadata_file)
df["relative_path"] = "train_audio/" + df["filename"].astype(str)
df = df[["relative_path", "primary_label"]]
birds = list(pd.get_dummies(df["primary_label"]).columns)
birds = np.transpose(birds)
le = preprocessing.LabelEncoder()
df[["classID"]] = df[["primary_label"]].apply(le.fit_transform)
del df["primary_label"]
# df.head()
# birds.shape
# df[['classID']].nunique(),df.classID.unique()
import math, random
import torch
import torchaudio
from torchaudio import transforms
from IPython.display import Audio
class AudioUtil:
# ----------------------------
# Load an audio file. Return the signal as a tensor and the sample rate
# ----------------------------
@staticmethod
def open(audio_file):
sig, sr = torchaudio.load(audio_file)
return (sig, sr)
# ----------------------------
# Convert the given audio to the desired number of channels
# ----------------------------
@staticmethod
def rechannel(aud, new_channel):
sig, sr = aud
if sig.shape[0] == new_channel:
# Nothing to do
return aud
if new_channel == 1:
# Convert from stereo to mono by selecting only the first channel
resig = sig[:1, :]
else:
# Convert from mono to stereo by duplicating the first channel
resig = torch.cat([sig, sig])
return (resig, sr)
# ----------------------------
# Since Resample applies to a single channel, we resample one channel at a time
# ----------------------------
@staticmethod
def resample(aud, newsr):
sig, sr = aud
if sr == newsr:
# Nothing to do
return aud
num_channels = sig.shape[0]
# Resample first channel
resig = torchaudio.transforms.Resample(sr, newsr)(sig[:1, :])
if num_channels > 1:
# Resample the second channel and merge both channels
retwo = torchaudio.transforms.Resample(sr, newsr)(sig[1:, :])
resig = torch.cat([resig, retwo])
return (resig, newsr)
# ----------------------------
# Pad (or truncate) the signal to a fixed length 'max_ms' in milliseconds
# ----------------------------
@staticmethod
def pad_trunc(aud, max_ms):
sig, sr = aud
num_rows, sig_len = sig.shape
max_len = sr // 1000 * max_ms
if sig_len > max_len:
# Truncate the signal to the given length
sig = sig[:, :max_len]
elif sig_len < max_len:
# Length of padding to add at the beginning and end of the signal
pad_begin_len = random.randint(0, max_len - sig_len)
pad_end_len = max_len - sig_len - pad_begin_len
# Pad with 0s
pad_begin = torch.zeros((num_rows, pad_begin_len))
pad_end = torch.zeros((num_rows, pad_end_len))
sig = torch.cat((pad_begin, sig, pad_end), 1)
return (sig, sr)
# ----------------------------
# Shifts the signal to the left or right by some percent. Values at the end
# are 'wrapped around' to the start of the transformed signal.
# ----------------------------
@staticmethod
def time_shift(aud, shift_limit):
sig, sr = aud
_, sig_len = sig.shape
shift_amt = int(random.random() * shift_limit * sig_len)
return (sig.roll(shift_amt), sr)
# ----------------------------
# Generate a Spectrogram
# ----------------------------
@staticmethod
def spectro_gram(aud, n_mels=64, n_fft=1024, hop_len=None):
sig, sr = aud
top_db = 80
# spec has shape [channel, n_mels, time], where channel is mono, stereo etc
spec = transforms.MelSpectrogram(
sr, n_fft=n_fft, hop_length=hop_len, n_mels=n_mels
)(sig)
# Convert to decibels
spec = transforms.AmplitudeToDB(top_db=top_db)(spec)
return spec
# ----------------------------
# Augment the Spectrogram by masking out some sections of it in both the frequency
# dimension (ie. horizontal bars) and the time dimension (vertical bars) to prevent
# overfitting and to help the model generalise better. The masked sections are
# replaced with the mean value.
# ----------------------------
@staticmethod
def spectro_augment(spec, max_mask_pct=0.1, n_freq_masks=1, n_time_masks=1):
_, n_mels, n_steps = spec.shape
mask_value = spec.mean()
aug_spec = spec
freq_mask_param = max_mask_pct * n_mels
for _ in range(n_freq_masks):
aug_spec = transforms.FrequencyMasking(freq_mask_param)(
aug_spec, mask_value
)
time_mask_param = max_mask_pct * n_steps
for _ in range(n_time_masks):
aug_spec = transforms.TimeMasking(time_mask_param)(aug_spec, mask_value)
return aug_spec
# Let’s walk through the steps as our data gets transformed, starting with an audio file:
# * The audio from the file gets loaded into a Numpy array of shape (num_channels, num_samples). Most of the audio is sampled at 44.1kHz and is about 4 seconds in duration, resulting in 44,100 * 4 = 176,400 samples. If the audio has 1 channel, the shape of the array will be (1, 176,400). Similarly, audio of 4 seconds duration with 2 channels and sampled at 48kHz will have 192,000 samples and a shape of (2, 192,000).
# * Since the channels and sampling rates of each audio are different, the next two transforms resample the audio to a standard 44.1kHz and to a standard 2 channels.
# * Since some audio clips might be more or less than 4 seconds, we also standardize the audio duration to a fixed length of 4 seconds. Now arrays for all items have the same shape of (2, 176,400)
# * The Time Shift data augmentation now randomly shifts each audio sample forward or backward. The shapes are unchanged.
# * The augmented audio is now converted into a Mel Spectrogram, resulting in a shape of (num_channels, Mel freq_bands, time_steps) = (2, 64, 344)
# * The SpecAugment data augmentation now randomly applies Time and Frequency Masks to the Mel Spectrograms. The shapes are unchanged.
# Thus, each batch will have two tensors, one for the X feature data containing the Mel Spectrograms and the other for the y target labels containing numeric Class IDs. The batches are picked randomly from the training data for each training epoch.
# * Each batch has a shape of (batch_sz, num_channels, Mel freq_bands, time_steps)
class SoundDS(Dataset):
def __init__(self, df, data_path):
self.df = df
self.data_path = str(data_path)
self.duration = 4000
self.sr = 44100
self.channel = 2
self.shift_pct = 0.4
def __len__(self):
return len(self.df)
# self.audio_length = self.duration*self.sr
def __getitem__(self, idx):
audio_file = self.data_path + self.df.loc[idx, "relative_path"]
class_id = self.df.loc[idx, "classID"]
aud = AudioUtil.open(audio_file)
reaud = AudioUtil.resample(aud, self.sr)
rechan = AudioUtil.rechannel(reaud, self.channel)
dur_aud = AudioUtil.pad_trunc(rechan, self.duration)
shift_aud = AudioUtil.time_shift(dur_aud, self.shift_pct)
sgram = AudioUtil.spectro_gram(shift_aud, n_mels=64, n_fft=1024, hop_len=None)
aug_sgram = AudioUtil.spectro_augment(
sgram, max_mask_pct=0.1, n_freq_masks=2, n_time_masks=2
)
return aug_sgram, class_id
data_path = download_path + "birdclef-2023/"
myds = SoundDS(df, data_path)
num_items = len(myds)
num_train = round(num_items * 0.8)
num_val = num_items - num_train
train_ds, val_ds = random_split(myds, [num_train, num_val])
train_dl = torch.utils.data.DataLoader(train_ds, batch_size=16, shuffle=True)
val_dl = torch.utils.data.DataLoader(val_ds, batch_size=16, shuffle=False)
import pandas as pd
import sklearn.metrics
def padded_cmap(solution, submission, padding_factor=5):
solution = solution.drop(["row_id"], axis=1, errors="ignore")
submission = submission.drop(["row_id"], axis=1, errors="ignore")
new_rows = []
for i in range(padding_factor):
new_rows.append([1 for i in range(len(solution.columns))])
new_rows = pd.DataFrame(new_rows)
new_rows.columns = solution.columns
padded_solution = pd.concat([solution, new_rows]).reset_index(drop=True).copy()
padded_submission = pd.concat([submission, new_rows]).reset_index(drop=True).copy()
score = sklearn.metrics.average_precision_score(
padded_solution.values,
padded_submission.values,
average="macro",
)
return score
class AudioClassifier(nn.Module):
# ----------------------------
# Build the model architecture
# ----------------------------
def __init__(self):
super().__init__()
conv_layers = []
# First Convolution Block with Relu and Batch Norm. Use Kaiming Initialization
self.conv1 = nn.Conv2d(2, 8, kernel_size=(5, 5), stride=(2, 2), padding=(2, 2))
self.relu1 = nn.ReLU()
self.bn1 = nn.BatchNorm2d(8)
init.kaiming_normal_(self.conv1.weight, a=0.1)
self.conv1.bias.data.zero_()
conv_layers += [self.conv1, self.relu1, self.bn1]
# Second Convolution Block
self.conv2 = nn.Conv2d(8, 16, kernel_size=(3, 3), stride=(2, 2), padding=(1, 1))
self.relu2 = nn.ReLU()
self.bn2 = nn.BatchNorm2d(16)
init.kaiming_normal_(self.conv2.weight, a=0.1)
self.conv2.bias.data.zero_()
conv_layers += [self.conv2, self.relu2, self.bn2]
# Second Convolution Block
self.conv3 = nn.Conv2d(
16, 32, kernel_size=(3, 3), stride=(2, 2), padding=(1, 1)
)
self.relu3 = nn.ReLU()
self.bn3 = nn.BatchNorm2d(32)
init.kaiming_normal_(self.conv3.weight, a=0.1)
self.conv3.bias.data.zero_()
conv_layers += [self.conv3, self.relu3, self.bn3]
# Second Convolution Block
self.conv4 = nn.Conv2d(
32, 64, kernel_size=(3, 3), stride=(2, 2), padding=(1, 1)
)
self.relu4 = nn.ReLU()
self.bn4 = nn.BatchNorm2d(64)
init.kaiming_normal_(self.conv4.weight, a=0.1)
self.conv4.bias.data.zero_()
conv_layers += [self.conv4, self.relu4, self.bn4]
# Linear Classifier
self.ap = nn.AdaptiveAvgPool2d(output_size=1)
self.lin = nn.Linear(in_features=64, out_features=264)
# Wrap the Convolutional Blocks
self.conv = nn.Sequential(*conv_layers)
# ----------------------------
# Forward pass computations
# ----------------------------
def forward(self, x):
# Run the convolutional blocks
x = self.conv(x)
# Adaptive pool and flatten for input to linear layer
x = self.ap(x)
x = x.view(x.shape[0], -1)
# Linear layer
x = self.lin(x)
# Final output
return x
myModel = AudioClassifier()
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
myModel = myModel.to(device)
next(myModel.parameters()).device
import gc
# ----------------------------
# Training Loop
# ----------------------------
def training(model, train_dl, num_epochs):
# Loss Function, Optimizer and Scheduler
criterion = nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(model.parameters(), lr=0.001)
scheduler = torch.optim.lr_scheduler.OneCycleLR(
optimizer,
max_lr=0.001,
steps_per_epoch=int(len(train_dl)),
epochs=num_epochs,
anneal_strategy="linear",
)
# Repeat for each epoch
for epoch in range(num_epochs):
running_loss = 0.0
correct_prediction = 0
total_prediction = 0
# Repeat for each batch in the training set
for i, data in enumerate(train_dl):
# Get the input features and target labels, and put them on the GPU
inputs, labels = data[0].to(device), data[1].to(device)
# Normalize the inputs
inputs_m, inputs_s = inputs.mean(), inputs.std()
inputs = (inputs - inputs_m) / inputs_s
# Zero the parameter gradients
optimizer.zero_grad()
# forward + backward + optimize
outputs = model(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
scheduler.step()
# Keep stats for Loss and Accuracy
running_loss += loss.item()
# Get the predicted class with the highest score
_, prediction = torch.max(outputs, 1)
# Count of predictions that matched the target label
correct_prediction += (prediction == labels).sum().item()
total_prediction += prediction.shape[0]
if i % 10 == 0: # print every 10 mini-batches
print("[%d, %5d] loss: %.3f" % (epoch + 1, i + 1, running_loss / 10))
# Print stats at the end of the epoch
num_batches = len(train_dl)
avg_loss = running_loss / num_batches
acc = correct_prediction / total_prediction
print(f"Epoch: {epoch}, Loss: {avg_loss:.2f}, Accuracy: {acc:.2f}")
print("Finished Training")
num_epochs = 2 # Just for demo, adjust this higher.
training(myModel, train_dl, num_epochs)
# 23/03
# Epoch: 0, Loss: 5.18, Accuracy: 0.04
# Epoch: 1, Loss: 4.56, Accuracy: 0.09
gc.collect()
torch.cuda.empty_cache()
torch.save(myModel.state_dict(), "r1001_birdclef2023.v.1.0.pth")
myModel.load_state_dict(torch.load("r1001_birdclef2023.v.1.0.pth"))
# evaluate on the validation set after each epoch
myModel.eval()
with torch.no_grad():
correct = 0
total = 0
# Repeat for each batch in the validation set
for i, data in enumerate(val_dl):
# Get the input features and target labels, and put them on the GPU
inputs, labels = data[0].to(device), data[1].to(device)
# Normalize the inputs
inputs_m, inputs_s = inputs.mean(), inputs.std()
inputs = (inputs - inputs_m) / inputs_s
outputs = myModel(inputs)
_, predicted = torch.max(outputs, 1)
total += predicted.shape[0]
correct += (predicted == labels).sum().item()
output_val = outputs.sigmoid().cpu().detach().numpy()
target_one_hot = torch.eye(264)[labels]
target_val = target_one_hot.numpy()
val_df = pd.DataFrame(target_val, columns=birds)
pred_df = pd.DataFrame(output_val, columns=birds)
avg_score = padded_cmap(val_df, pred_df, padding_factor=3)
print(f"cmAP score pad 3: {avg_score}")
accuracy = 100 * correct / total
print(f"Validation Accuracy: {accuracy:.2f}%")
myModel.train()
# 23/03 - Validation Accuracy: 8.53%
# (label_df == 1).any(axis=1).sum(), (predicted_df == 1).any(axis=1).sum()
# label_df.columns[label_df.eq(1).any()],predicted_df.columns[predicted_df.eq(1).any()]
# y_true, y_scores
# labels, birds
from torch.utils.data import DataLoader, Dataset
import numpy as np
class SoundTestDS(Dataset):
def __init__(self, df, data_path):
self.df = df
self.data_path = str(data_path)
self.duration = 4000
self.sr = 44100
self.channel = 2
self.shift_pct = 0.4
def __len__(self):
return len(self.df)
def audio_to_image(self, aud):
reaud = AudioUtil.resample(aud, 44100)
rechan = AudioUtil.rechannel(reaud, 2)
dur_aud = AudioUtil.pad_trunc(rechan, 4000)
shift_aud = AudioUtil.time_shift(dur_aud, 0.4)
sgram = AudioUtil.spectro_gram(shift_aud, n_mels=64, n_fft=1024, hop_len=None)
aug_sgram = AudioUtil.spectro_augment(
sgram, max_mask_pct=0.1, n_freq_masks=2, n_time_masks=2
)
return aug_sgram
def split_audio(self, aud, duration, step=None):
sig, sr = aud
audio_length = sr * 5
audios = []
step = None or audio_length
for i in range(audio_length, len(sig[0]) + step, step):
start = max(0, i - audio_length)
end = start + audio_length
audios.append(sig[0][start:end])
if len(audios[-1]) < audio_length:
audios = audios[:-1]
images = [self.audio_to_image((audio.unsqueeze(0), sr)) for audio in audios]
images = np.stack(images)
return images
def __getitem__(self, idx):
audio_file = self.data_path + self.df.loc[idx, "relative_path"]
aud = AudioUtil.open(audio_file)
return self.split_audio(aud, 5)
# import soundfile as sf
# import os
# # define the path to the audio file
# audio_file_path = "/kaggle/input/birdclef-2023/test_soundscapes/soundscape_29201.ogg"
# # define the duration of each segment in seconds
# segment_duration = 60
# # read the audio file
# audio_data, samplerate = sf.read(audio_file_path)
# # calculate the total number of segments
# total_segments = int(len(audio_data) / (segment_duration * samplerate))
# # create a directory to store the segments
# output_dir = "/kaggle/working/testaud"
# if not os.path.exists(output_dir):
# os.makedirs(output_dir)
# # split the audio file into segments and save each segment as a separate file
# for i in range(total_segments):
# segment_start = i * segment_duration * samplerate
# segment_end = segment_start + segment_duration * samplerate
# segment_data = audio_data[segment_start:segment_end]
# segment_file_name = os.path.join(output_dir, f"segment_{i}.ogg")
# sf.write(segment_file_name, segment_data, samplerate)
df_test = pd.DataFrame(
[
(path.stem, path.parent.absolute().stem + "/" + path.stem + ".ogg", -1)
for path in Path(download_path + "birdclef-2023/test_soundscapes/").glob(
"*.ogg"
)
],
columns=["filename", "relative_path", "classID"],
)
# df_test = pd.DataFrame(
# [(path.stem, path.parent.absolute().stem + '/' + path.stem + '.ogg',-1) for path in Path('/kaggle/working/testaud').glob("*.ogg")],
# columns = ["filename", "relative_path", "classID" ]
# )
# print(df_test.shape)
# df_test.head()
# test_dataset = SoundTestDS(df_test[['relative_path','classID']], '/kaggle/working/')#data_path)
test_dataset = SoundTestDS(df_test[["relative_path", "classID"]], data_path)
test_dataloader = DataLoader(test_dataset, batch_size=16, shuffle=False)
# dataset = train_ds
# # Iterate through the dataset
# for i in range(10):
# # Get the i-th sample from the dataset
# sample = dataset[i]
# # Extract the input (audio data) and target (label) from the sample
# input, target = sample
# # Do something with the input and target
# print(f'Sample {i}: Input shape: {input.shape}, Target: {target}')
# dataset = test_dataset
# # Iterate through the dataset
# for i in range(len(dataset)):
# # Get the i-th sample from the dataset
# sample = dataset[i]
# for j in range(len(sample)):
# aud = sample[j]
# # Extract the input (audio data) and target (label) from the sample
# input, _ = aud
# # Do something with the input and target
# print(f'Sample {i}: Audion {j}: Input shape: {input.shape}')
# Load your trained model
myModel.load_state_dict(torch.load("r1001_birdclef2023.v.1.0.pth"))
# Put the model in evaluation mode
myModel.eval()
# Iterate over the test set and make predictions
predictions = []
with torch.no_grad():
# Repeat for each batch in the test set
for i, data in enumerate(test_dataloader):
# Get the input features and target labels, and put them on the GPU
row = []
for j in range(len(data)):
inputs = data[j].to(device)
# Normalize the inputs
inputs_m, inputs_s = inputs.mean(), inputs.std()
inputs = (inputs - inputs_m) / inputs_s
# Make a prediction on the waveform tensor
outputs = myModel(inputs)
row.append(outputs.sigmoid().cpu().detach().numpy())
predictions.append(row)
myModel.train()
# len(filenames), len(predictions[0]),len(predictions[0][0]), predictions[0][0].shape,len(predictions[0][0][1])
# len(bird_cols)
filenames = df_test.filename.values.tolist()
bird_cols = list(birds)
sub_df = pd.DataFrame(columns=["row_id"] + bird_cols)
sub_df[bird_cols] = sub_df[bird_cols].astype(np.float32)
for i, file in enumerate(filenames):
pred = predictions[0][i]
num_rows = len(pred)
row_ids = [f"{file}_{(j+1)*5}" for j in range(num_rows)]
df = pd.DataFrame(columns=["row_id"] + bird_cols)
df["row_id"] = row_ids
df[bird_cols] = pred
sub_df = pd.concat([sub_df, df]).reset_index(drop=True)
# len(sub_df)
# sub_df[0:13]
sub_df.to_csv("submission.csv", index=False)
|
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
import seaborn as sns
import os
import sklearn
from sklearn.model_selection import train_test_split
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# ## Predicting Loan Default - When should credit be approved?
# **Problem statement**:
# The aim of this notebook is to create a simple program that can predict wether a credit should be approved or not, based on previous client data. The program should help the bank minimize risk with future clients.
# The model used will predict wether a client should have their loan approved or not, based on his or her history, using logistic regression for classification.
# **The data:**
# The data consists of a csv file containing records of clients from a private german bank. Includes the client profile (account balance, number of credits,...) and a variable **Creditability** (1 : credit-worthy 0 : not credit-worthy).
# A detailed description of the variables can be found [here](https://newonlinecourses.science.psu.edu/stat508/book/export/html/803).
# read and inspect dataset
data = pd.read_csv("../input/german-credit-risk/german_credit.csv")
data.head()
data.describe() # data summary
data.corr() # check for correlations with target variable
# new data will include the variables with highest correlation
# with dependent variable - creditability
x = data[
[
"Account Balance",
"Duration of Credit (month)",
"Payment Status of Previous Credit",
]
]
y = data["Creditability"]
# splitting data
# test = 80% of data
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.80, random_state=6
)
# using statsmodels for model and metrics
import statsmodels.api as sm
# building the model and fitting the training data
model = sm.Logit(y_train, x_train).fit()
# summary statistics of log. regression
model.summary()
pred_y = model.predict(x_test) # values predicted by model
pred_y.head()
# Our logistic regression function returns the likelihood of a credit being worthy, and so values can range from 0 to 1.
# We'll transform the likelyhood into a binary variable for classification.
# If a credit is more than 50% likely to be worthy, it will be labeled worthy.
# The function below can apply this process.
# function for turning likelihood into labels
def binary_classify(x): # takes int x returns output int label
x = round(x, 2)
if x >= 0.50:
return 1
return 0
pred = list(map(binary_classify, pred_y)) # apply function to all predictions
# ### Classification performance
# Now that we have used logistic regression to perform classification, we can check how accurate the predictions are:
# accuracy score
sklearn.metrics.accuracy_score(y_test, pred)
# The model makes 73.75% of predictions correct.
# ## Proposed solution
# In order to classify future clients as credit worthy or not, we can build a simple program that will take as inputs their account balance, credit duration and payment status of current credit. The program will then apply the previous model and classify the client, providing information to the bank of wether the credit should be approved or not.
# The formula used for classification was obtained previously with *statsmodels*.
# predictive function based on logistic model - returns likelyhood
def log_func(x):
balance, credit, pay_status = x
result = (0.6364 * balance) + (-0.05 * credit) + (0.2374 * pay_status)
return result
def binary_classify(x): # takes int x = likelihood returns output int label
x = round(x, 2)
if x >= 0.50:
return 1
return 0
def predict(x): # makes credit predictions
return binary_classify(log_func(x))
def print_result(x): # prints output to user
if predict(x) == 1:
return "Credit worthy"
return "Not credit worthy"
# Let's try to apply the program as an example.
# We can check the data of a specific client as a test:
test = data.iloc[97] # data from client
test = test[
[
"Creditability",
"Account Balance",
"Duration of Credit (month)",
"Payment Status of Previous Credit",
]
]
test
# Client data:
# * Creditability: 0 - was classified not credit worthy
# * Account Balance: 2
# * Duration of Credit (month): 36
# * Payment Status of Previous Credit: 3
# testing
input_test = (2, 36, 3) # client data
print("Result: " + print_result(input_test))
|
# Importing the relevant files
import os
import numpy as np
import cv2
from tqdm import tqdm
import random
import tensorflow as tf
from tensorflow.keras.optimizers import Adam
from tensorflow.keras.layers import Conv2D, MaxPool2D, Flatten, Dense
from tensorflow.keras.models import Sequential
from sklearn.metrics import confusion_matrix
from sklearn.utils import shuffle
# Setting the image size, epochs,classes and batch size
img = 150
epochs = 30
batch_size = 25
names = ["O", "R"]
encode_name = {name: i for i, name in enumerate(names)}
# Making a function to load the images and labels. I have used openCV for this purpose
def load_data():
datasets = [
"/kaggle/input/waste-classification-data/DATASET/TRAIN",
"/kaggle/input/waste-classification-data/DATASET/TEST",
]
output = []
for dataset in tqdm(datasets):
images = []
labels = []
for folder in os.listdir(dataset):
label = encode_name[folder]
if dataset == "/kaggle/input/waste-classification-data/DATASET/TRAIN":
img_set = random.sample(os.listdir(os.path.join(dataset, folder)), 7000)
else:
img_set = random.sample(os.listdir(os.path.join(dataset, folder)), 1000)
for file in img_set:
img_path = os.path.join(os.path.join(dataset, folder), file)
image = cv2.imread(img_path)
image = cv2.resize(image, (img, img))
images.append(image)
labels.append(label)
images = np.array(images, dtype=np.float32)
labels = np.array(labels, dtype=np.int32)
output.append((images, labels))
return output
# Loading the training and testing data
(train_images, train_labels), (test_images, test_labels) = load_data()
# Converting the training and testing images and labels to numpy arrays
train_images = np.array(train_images)
train_labels = np.array(train_labels)
test_images = np.array(test_images)
test_labels = np.array(test_labels)
# Printing the size of the images and labels
print(
f"Training images size:{train_images.shape}, Training labels size:{train_labels.shape}"
)
print(
f"Testing images size:{test_images.shape}, Testing labels size:{test_labels.shape}"
)
# Scaling the values of the images pixels to 0-1 to make the computation easier for our model
train_images, test_images = train_images / 255, test_images / 255
# Randomizing the training data
train_images, train_labels = shuffle(train_images, train_labels)
# Making the model architecture
model = Sequential(
[
Conv2D(
filters=32,
activation="relu",
input_shape=(img, img, 3),
padding="same",
kernel_size=(3, 3),
),
Conv2D(filters=32, activation="relu", padding="same", kernel_size=(3, 3)),
MaxPool2D(pool_size=(2, 2)),
Conv2D(filters=64, activation="relu", padding="same", kernel_size=(3, 3)),
Conv2D(filters=64, activation="relu", padding="same", kernel_size=(3, 3)),
MaxPool2D(pool_size=(2, 2)),
Conv2D(filters=128, activation="relu", padding="same", kernel_size=(3, 3)),
Conv2D(filters=128, activation="relu", padding="same", kernel_size=(3, 3)),
MaxPool2D(pool_size=(2, 2)),
Conv2D(filters=256, activation="relu", padding="same", kernel_size=(3, 3)),
Conv2D(filters=256, activation="relu", padding="same", kernel_size=(3, 3)),
MaxPool2D(pool_size=(2, 2)),
Conv2D(filters=256, activation="relu", padding="same", kernel_size=(3, 3)),
Conv2D(filters=256, activation="relu", padding="same", kernel_size=(3, 3)),
MaxPool2D(pool_size=(2, 2)),
Conv2D(filters=512, activation="relu", padding="same", kernel_size=(3, 3)),
Conv2D(filters=512, activation="relu", padding="same", kernel_size=(3, 3)),
MaxPool2D(pool_size=(2, 2)),
Conv2D(filters=512, activation="relu", padding="same", kernel_size=(3, 3)),
Conv2D(filters=512, activation="relu", padding="same", kernel_size=(3, 3)),
Flatten(),
Dense(units=4096, activation="relu"),
Dense(units=4096, activation="relu"),
Dense(units=1, activation="sigmoid"),
]
)
# Printing the model summary
model.summary()
# Saving the weights of the model
model.save("model.h5")
# Compiling the model. Specifying the optimizer to act on the data. The loss function(which could also have been sparse_categorical_crossentropy),since there are only two classes, I have used binary_crossentropy
model.compile(
optimizer=Adam(lr=0.0001), loss="binary_crossentropy", metrics=["accuracy"]
)
model.load_weights("model.h5")
# Training the model on the training data
model.fit(
x=train_images,
y=train_labels,
validation_split=0.3,
epochs=epochs,
batch_size=batch_size,
steps_per_epoch=100,
verbose=2,
)
# Using the model to make predictions on the testing data
p = model.predict(test_images)
# Using the confusion matrix to print the accuracy of those predictions
cm = confusion_matrix(y_true=test_labels, y_pred=np.round(p))
print(f"Accuracy:{(cm.trace()/cm.sum())*100}")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Importing Libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.model_selection import cross_val_score
from sklearn.naive_bayes import GaussianNB
from sklearn.linear_model import LogisticRegression
from sklearn import tree
from sklearn.neighbors import KNeighborsClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.svm import SVC
sns.set()
from sklearn.metrics import confusion_matrix, classification_report
from sklearn.neural_network import MLPClassifier
import warnings
warnings.filterwarnings("ignore")
# # Load DataSets
data = pd.read_csv("/kaggle/input/titanic/train.csv")
data_2 = pd.read_csv("/kaggle/input/titanic/test.csv")
data_3 = pd.read_csv("/kaggle/input/titanic/gender_submission.csv")
data
data_2
data_3
test = pd.merge(data_2, data_3, how="inner", on="PassengerId")
test
# # Data Manipuletion / Cleaning
# **As we can see that we wont able to find any informetion from passengerId or Name se we will drop these 2 columns.
# but there is chanse that we can find some informetion from ticket column. But before that we need to clean the data i.e. we need to process the null value
# if exists.**
sns.heatmap(data.isnull())
data.isnull().sum()
train = data.drop(["PassengerId", "Name", "Cabin"], axis=1)
sns.heatmap(test.isnull())
test.isnull().sum()
test = test.drop(["Cabin", "PassengerId", "Name"], axis=1)
# As we can see cabin contain huge number of null value so there is no point of using it and
# as we already discussed PassengerId and Name have no importaance in this modeling so we drop them. But we will fill the null value of age column with the mean of the age of pclass value
train.groupby("Pclass")["Age"].mean()
for i in range(len(train["Age"])):
if train["Age"].isnull()[i] == True:
if train["Pclass"][i] == 1:
train["Age"][i] = 38
elif train["Pclass"][i] == 2:
train["Age"][i] = 30
else:
train["Age"][i] = 25
else:
train["Age"][i] = train["Age"][i]
test.groupby("Pclass")["Age"].mean()
for i in range(len(test["Age"])):
if test["Age"].isnull()[i] == True:
if test["Pclass"][i] == 1:
test["Age"][i] = 40.918367
elif test["Pclass"][i] == 2:
test["Age"][i] = 28.7775
else:
test["Age"][i] = 24.027945
else:
test["Age"][i] = test["Age"][i]
train = train.dropna()
test = test.dropna()
# As we drop and fill the null values now we will process the ticket column and try to make them into different classes based on their initials and for those who contins numbers i consider the class low
ticket = train["Ticket"].tolist()
ticket[1].split(" ")[0][0]
catch = []
for i in range(len(ticket)):
catch.append(ticket[i].split(" ")[0][0])
for i in range(len(ticket)):
if catch[i] in ["1", "2", "3", "4", "5", "6", "7", "8", "9"]:
catch[i] = "low"
else:
catch[i] = catch[i]
train["Ticket"] = catch
data
ticket = test["Ticket"].tolist()
catch = []
for i in range(len(ticket)):
catch.append(ticket[i].split(" ")[0][0])
x = np.array(catch)
for i in range(len(ticket)):
if catch[i] in ["1", "2", "3", "4", "5", "6", "7", "8", "9"]:
catch[i] = "low"
else:
catch[i] = catch[i]
test["Ticket"] = catch
test
titanic = pd.concat([train, test], axis=0)
# # EDA
# As we have cleaned the data now we will try to find out some insight from the data. we will try to find out relation between different columns with Survived column.
sns.countplot(titanic, x=titanic["Survived"])
sns.countplot(titanic, x=titanic["Survived"], hue="Sex")
titanic.groupby("Sex")["Survived"].sum()
titanic.value_counts("Sex").reset_index()
male_survived = (
titanic.groupby("Sex")["Survived"].sum()[1]
/ titanic.value_counts("Sex").reset_index()[0][0]
) * 100
female_survived = (
titanic.groupby("Sex")["Survived"].sum()[0]
/ titanic.value_counts("Sex").reset_index()[0][1]
) * 100
print(round(male_survived, 2), "% of male passenger were survived the crash")
print(round(female_survived, 2), "% of female passenger were survived the crash")
sns.countplot(titanic, x=titanic["SibSp"], hue="Survived")
sns.countplot(titanic, x=titanic["SibSp"], hue="Sex")
sns.countplot(titanic, x=titanic["Survived"], hue="SibSp")
sns.countplot(titanic, x=titanic["Pclass"], hue="Sex")
sns.countplot(titanic, x=titanic["Pclass"], hue="Survived")
sns.countplot(titanic, x=titanic["Survived"], hue="Pclass")
titanic.groupby("Pclass")["Survived"].sum()
titanic.value_counts("Pclass").reset_index()
Pclass_1_survived = (
titanic.groupby("Pclass")["Survived"].sum()[0:1][1]
/ titanic.value_counts("Pclass").reset_index()[0][1]
) * 100
Pclass_2_survived = (
titanic.groupby("Pclass")["Survived"].sum()[1:2][2]
/ titanic.value_counts("Pclass").reset_index()[0][2]
) * 100
Pclass_3_survived = (
titanic.groupby("Pclass")["Survived"].sum()[2:3][3]
/ titanic.value_counts("Pclass").reset_index()[0][0]
) * 100
print(
round(Pclass_1_survived, 2),
"% of passenger were survived the crash who were travelling in class 1",
)
print(
round(Pclass_2_survived, 2),
"% of passenger were survived the crash who were travelling in class 2",
)
print(
round(Pclass_3_survived, 2),
"% of passenger were survived the crash who were travelling in class 3",
)
sns.pairplot(titanic, hue="Pclass")
sns.pairplot(titanic, hue="Survived")
sns.pairplot(titanic, hue="Sex")
# # Prepare The DataSet for ML Model
# We ahve some coluns like Sex, Ticket contains catagorical varible. to process them me need dummy
plt.rcParams["font.size"] = 11
corr = train.corr()
sns.heatmap(corr, annot=True, vmax=1, vmin=-1).set_title("Correlation Map")
plt.show()
Sex = pd.get_dummies(train["Sex"], drop_first=True)
Embarked = pd.get_dummies(train["Embarked"], drop_first=True)
train = pd.concat([train, Sex, Embarked], axis=1)
train["Embarked_Q"] = train["Q"]
train["Embarked_S"] = train["S"]
train = train.drop(["Q", "S"], axis=1)
Ticket = pd.get_dummies(train["Ticket"], drop_first=True)
train = pd.concat([train, Ticket], axis=1)
train = train.drop(["Sex", "Embarked", "Ticket"], axis=1)
plt.rcParams["font.size"] = 11
corr = test.corr()
sns.heatmap(corr, annot=True, vmax=1, vmin=-1).set_title("Correlation Map")
plt.show()
test = test.dropna()
Sex = pd.get_dummies(test["Sex"], drop_first=True)
Embarked = pd.get_dummies(test["Embarked"], drop_first=True)
test = pd.concat([test, Sex, Embarked], axis=1)
test["Embarked_Q"] = test["Q"]
test["Embarked_S"] = test["S"]
test = test.drop(["Q", "S"], axis=1)
Ticket = pd.get_dummies(test["Ticket"], drop_first=True)
test = pd.concat([test, Ticket], axis=1)
test = test.drop(["Sex", "Embarked", "Ticket"], axis=1)
x_test = test.drop("Survived", axis=1)
y_test = test["Survived"]
x_train = train.drop("Survived", axis=1)
y_train = train["Survived"]
# # Creating and Evaluating Different Models
# # Logistic Regression
from sklearn.linear_model import LogisticRegression
model = LogisticRegression()
model.fit(x_train, y_train)
model.score(x_train, y_train)
Logistic_score = model.score(x_test, y_test)
print("score of the Linear Regression model is:", model.score(x_test, y_test))
prediction = model.predict(x_test)
print(confusion_matrix(y_test, prediction))
sns.heatmap(confusion_matrix(y_test, prediction), annot=True)
print(classification_report(y_test, prediction))
# # Multinomial Naive_Bayes
#
from sklearn.naive_bayes import MultinomialNB
mnnb = MultinomialNB()
mnnb.fit(x_train, y_train)
print(mnnb.score(x_test, y_test))
prediction = mnnb.predict(x_test)
NB = mnnb.score(x_test, y_test)
print("score of the MultinomialNB model is:", mnnb.score(x_test, y_test))
sns.heatmap(confusion_matrix(y_test, prediction), annot=True)
print(classification_report(y_test, prediction))
# # MLP
MPL_model = MLPClassifier(
activation="logistic", max_iter=7000000000, verbose=1, learning_rate="adaptive"
)
MPL_model.fit(x_train, y_train)
plt.plot(MPL_model.loss_curve_)
plt.show()
mpl_score = MPL_model.score(x_test, y_test)
mpl_score
# # Random Forest
Rf = RandomForestClassifier()
Rf.fit(x_train, y_train)
print(Rf.score(x_test, y_test))
Rf_score = Rf.score(x_test, y_test)
predction = Rf.predict(x_test)
print(confusion_matrix(y_test, predction))
print(classification_report(y_test, predction))
sns.heatmap(confusion_matrix(y_test, predction), annot=True)
# # Decision Tree
Decion_tree_model = DecisionTreeClassifier()
Decion_tree_model.fit(x_train, y_train)
predction = Decion_tree_model.predict(x_test)
print(Decion_tree_model.score(x_test, y_test))
Decion_tree_model_score = Decion_tree_model.score(x_test, y_test)
print(confusion_matrix(y_test, predction))
print(classification_report(y_test, predction))
sns.heatmap(confusion_matrix(y_test, predction), annot=True)
# # KNN
knn = KNeighborsClassifier(n_neighbors=2)
knn.fit(x_train, y_train)
prediction = knn.predict(x_test)
print(knn.score(x_test, y_test))
knn_score = knn.score(x_test, y_test)
print(confusion_matrix(y_test, prediction))
print(classification_report(y_test, prediction))
sns.heatmap(confusion_matrix(y_test, prediction), annot=True)
print("Logistic_score:", Logistic_score)
print("NB:", NB)
print("mpl_score:", mpl_score)
print("Rf_score:", Rf_score)
print("Decion_tree_model_score:", Decion_tree_model_score)
print("knn_score:", knn_score)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
data_path = "/kaggle/input/bike-sharing-demand/"
train = pd.read_csv(data_path + "train.csv")
test = pd.read_csv(data_path + "test.csv")
submission = pd.read_csv(data_path + "sampleSubmission.csv")
# **데이터 확인**
train.head()
test.head()
# **casual, registered는 train data에만 있으므로 제거해도 되겠다**
train.info()
train.describe()
# **temp, atemp, humidity, windspeed는 max값이 다른 분위값에 비해 급격히 커지므로 이상치가 있을 것으로 예상됨**
train.isnull().sum()
# **많은 데이터가 묶인 datatime 데이터 쪼개기**
# **년, 월, 날짜, 시간대로 쪼개자**
train["year"] = train["datetime"].apply(lambda x: x.split()[0].split("-")[0])
train["month"] = train["datetime"].apply(lambda x: x.split()[0].split("-")[1])
train["day"] = train["datetime"].apply(lambda x: x.split()[0].split("-")[2])
train["hour"] = train["datetime"].apply(lambda x: x.split()[1].split(":")[0])
# **casual, registered 제거**
train.drop(columns=["casual", "registered"], inplace=True)
# **데이터 시각화**
import matplotlib.pyplot as plt
import seaborn as sns
train.head()
# **월별 대여량**
sns.pointplot(data=train, x="month", y="count", hue="year")
sns.pointplot(data=train, x="month", y="count")
# **계절별 대여량**
sns.pointplot(data=train, x="season", y="count", hue="year")
train["month"] = train["month"].astype(int)
train.loc[train["month"].isin([1, 2, 3]), "new_season"] = 1
train.loc[train["month"].isin([4, 5, 6]), "new_season"] = 2
train.loc[train["month"].isin([7, 8, 9]), "new_season"] = 3
train.loc[train["month"].isin([10, 11, 12]), "new_season"] = 4
train.head()
sns.pointplot(data=train, x="new_season", y="count")
# **1, 2, 3월이 봄(1), 4, 5, 6월이 여름(2), 7, 8, 9월이 가을(3), 10, 11, 12월이 겨울(4)인 것으로 보임**
# **월별 그래프와 계쩔별 그래프의 모양이 비슷하니 '계절'만 가져다가 사용하자**
# **날이 좋을 것으로 보이는 가을->여름->겨울-> 봄 순으로 자전거 사용량이 많음. 겨울이 더 사용량이 없을 것 같았지만 봄이 1, 2, 3월임을 감안하면 이때가 더 날이 안 좋거나 추울 수 있음**
# **시간의 흐름에 따라 숫자가 주어진 것이고 크고 작음의 관계는 없음. 원핫인코딩을 해야하지 않을까?**
train.head()
train.drop(columns=["month", "new_season"], inplace=True)
train.head()
# **시간별 대여량 확인**
sns.pointplot(data=train, x="hour", y="count", hue="year")
# **오전 7시, 8시의 대여량과 오후 17시, 18시의 대여량이 많음=> 출퇴근길(등하교길)에 자전거 대여량이 많을 것**
# **휴일이 영향을 주는지도 알아보자**
sns.pointplot(data=train, x="hour", y="count", hue="holiday")
# **holiday가 아니면 출퇴근 시간에 사용량이 많은 것 same**
# **holiday이면 오전 7시부터 오후 17시 정도까지 사용량이 꾸준히 늘어남.**
# **holiday에 대여량 이상치가 많음**
# **workingday-count**
sns.pointplot(data=train, x="hour", y="count", hue="workingday")
# **holiday와 마찬가지로 workingday가 아닌 날에는 오전 7시부터 오후 17시 정도까지 자전거 대여량이 꾸준히 늘어나서 유지됨**
# **결론: 보통은 출퇴근 길에 자전거 대여량 많음. 주말이나 휴일에는 오전 7시부터 시작하여 오후 5시 정도까지 대여량이 많음.**
# **working day와 holiday가 서로 상관관계가 높을까? 그렇다면 하나를 지워야 할 수도!**
sns.pointplot(data=train, x="day", y="count", hue="year")
# **날짜에서 특정한 패턴을 찾기는 어려움**
# **day feature는 버리자**
# **2012년에 2011년보다 전반적으로 자전거 대여량이 많았던 이유는 무엇일까? 휴일 수가 많았나? 날씨가 좋았나?**
train.groupby("year").mean()
# **holiday, workingday의 차이보다는 temp, atemp, humidity에서 좀 더 차이가 있지 않나 싶음.**
# **날씨의 영향을 분석해보자**
train.head()
sns.lineplot(data=train, x="weather", y="count")
# **4일 때 대여량이 많은 건 이상치 아닐까?. 신뢰구간도 없다.**
sns.lineplot(data=train, x="temp", y="count")
sns.lineplot(data=train, x="atemp", y="count")
sns.lineplot(data=train, x="humidity", y="count")
sns.lineplot(data=train, x="windspeed", y="count")
# **temp, atemp가 높을수록 대여량이 증가하는 경향, humidity는 높을수록 대여량이 감소하는 경향. 끈적끈적할 때 자전거 타고 싶지 않겠지**
# **weather가 4일 때 대여량이 높아지는 이상치가 있음. 무시해야 할 듯. 또한 windspeed가 강할 때 대여량이 높아지는 이상치 있음. 이 역시 무시해야 할 듯**
sns.regplot(data=train, x="weather", y="count", line_kws={"color": "red"})
sns.regplot(data=train, x="temp", y="count", line_kws={"color": "red"})
sns.regplot(data=train, x="atemp", y="count", line_kws={"color": "red"})
sns.regplot(data=train, x="humidity", y="count", line_kws={"color": "red"})
sns.regplot(data=train, x="windspeed", y="count", line_kws={"color": "red"})
# **windspeed에서 0과 10 사이가 크게 빈 것은 측정이 제대로 되지 못한 것이 아닐까?**
#
# **weather에서 4인데 대여량이 높아졌던 것은 이상치가 맞는 것으로 보임**
# **feature 사이의 상관관계 알아보기**
train.head()
# 수치형 데이터 = season, holiday, workingday, wearher, temp, atemp, humidity, windspeed, count 의 상관관계 계수 알아보기
train[
[
"season",
"holiday",
"workingday",
"weather",
"temp",
"atemp",
"humidity",
"windspeed",
"count",
"hour",
]
].corr()
corrMat = train[
[
"season",
"holiday",
"workingday",
"weather",
"temp",
"atemp",
"humidity",
"windspeed",
"count",
"hour",
]
].corr()
fig, ax = plt.subplots()
fig.set_size_inches(10, 10)
sns.heatmap(data=corrMat, annot=True) # annot을 넣으면 상관계수가 숫자로 표시됨
ax.set(title="Heatmap of Numerical Data")
# **feature 엔지니어링**
# 1. test data에는 없는 casual, registered 제거
# 2. datetime 쪼개기(연, 월, 일, 시간으로 나누기) / 출퇴근 시간에 대여량이 많은 것을 보면 '시간'이 대여량에 영향을 미칠 것. '월'은 season으로 대체할 것이기 때문에 제거. year, day는 count에 크게 영향을 미치지 않는 것으로 보이기 때문에 제거할 것.
# 3. season feature는 원핫인코딩으로 인코딩하기(숫자가 크고 작음을 의미하지 않으므로)
# 4. holiday와 workingday를 hue로 해서 시간별 대여량을 보면 그래프가 비슷하게 나타남. 두 feature이 서로 영향을 많이 미치면 하나를 제거하려고 했는데 heatmap을 보니 생각보다 수가 크지 않음. 우선은 두 가지 모두 두고 하자.
# 5. temp, atemp feature는 비슷한 개념인데 heatmap에서 서로 영향을 크게 주고받음을 알 수 있다. 하나는 빼버리자. (count와의 상관관계 점수도 같음). temp와 atemp는 높을수록 count가 높음
# 6. weather feature는 lineplot, rugplot을 보면 수가 커질수록(날씨가 나빠질수록) count가 줄어듦. 다만 weather가 4일때 200개 정도의 count가 있는 event는 오류로 보이므로 제거 예정
# 7. humidity feature는 커질수록 count가 감소함.
# 8. windspeed가 강할수록 count가 높아지는데(rugplot) lineplot을 보면 또 그게 아닌 것 같음. 그리고 rugplot 보면 사이에 크게 빈 곳이 있음. 결측치일까? windspeed feature 자체를 제거해야 하나?
data_path = "/kaggle/input/bike-sharing-demand/"
train = pd.read_csv(data_path + "train.csv")
test = pd.read_csv(data_path + "test.csv")
submission = pd.read_csv(data_path + "sampleSubmission.csv")
train.head()
test.head()
# 1. test data에는 없는 casual, registered 제거
train.drop(columns=["registered", "casual"], inplace=True)
train.head()
# 2. datetime 쪼개기(연, 월, 일, 시간으로 나누기) / 출퇴근 시간에 대여량이 많은 것을 보면 '시간'이 대여량에 영향을 미칠 것.
train["year"] = train["datetime"].apply(lambda x: int(x.split()[0].split("-")[0]))
train["month"] = train["datetime"].apply(lambda x: int(x.split()[0].split("-")[1]))
train["day"] = train["datetime"].apply(lambda x: int(x.split()[0].split("-")[2]))
train["hour"] = train["datetime"].apply(lambda x: int(x.split()[1].split(":")[0]))
test["year"] = test["datetime"].apply(lambda x: int(x.split()[0].split("-")[0]))
test["month"] = test["datetime"].apply(lambda x: int(x.split()[0].split("-")[1]))
test["day"] = test["datetime"].apply(lambda x: int(x.split()[0].split("-")[2]))
test["hour"] = test["datetime"].apply(lambda x: int(x.split()[1].split(":")[0]))
train.head()
test.head()
# 2. '월'은 season으로 대체할 것이기 때문에 제거. year, day는 count에 크게 영향을 미치지 않는 것으로 보이기 때문에 제거할 것.
train.drop(columns=["month", "year", "day", "datetime"], inplace=True)
test.drop(columns=["month", "year", "day", "datetime"], inplace=True)
train.head()
# 5. temp, atemp feature는 비슷한 개념인데 heatmap에서 서로 영향을 크게 주고받음을 알 수 있다. 하나는 빼버리자. (count와의 상관관계 점수도 같음). temp와 atemp는 높을수록 count가 높음
train.drop(columns="atemp", inplace=True)
test.drop(columns="atemp", inplace=True)
# 6. weather feature는 lineplot, rugplot을 보면 수가 커질수록(날씨가 나빠질수록) count가 줄어듦. 다만 weather가 4일때 200개 정도의 count가 있는 event는 오류로 보이므로 제거 예정
train = train[train["weather"] != 4]
# 8. windspeed가 강할수록 count가 높아지는데(rugplot) lineplot을 보면 또 그게 아닌 것 같음. 그리고 rugplot 보면 사이에 크게 빈 곳이 있음. 결측치일까? windspeed feature 자체를 제거해야 하나?
train.drop(columns="windspeed", inplace=True)
test.drop(columns="windspeed", inplace=True)
# 3. season feature는 원핫인코딩으로 인코딩하기(숫자가 크고 작음을 의미하지 않으므로)
# **원핫인코딩 결과 붙이기**
from sklearn.preprocessing import OneHotEncoder
encoder = OneHotEncoder()
train_encoded = encoder.fit_transform(train["season"].to_numpy().reshape(-1, 1))
test_encoded = encoder.fit_transform(test["season"].to_numpy().reshape(-1, 1))
train.drop(columns="season", inplace=True)
test.drop(columns="season", inplace=True)
from scipy import sparse
train_sprs = sparse.hstack((sparse.csr_matrix(train), train_encoded), format="csr")
test_sprs = sparse.hstack((sparse.csr_matrix(test), test_encoded), format="csr")
|
# # SALES ANALYTICS
# Hi! This is my Sales Analytics notebook for [Sales Product Data](https://www.kaggle.com/datasets/knightbearr/sales-product-data) from [Knightbearr](https://www.kaggle.com/knightbearr). The purpose is to provide analysis based on dataset provided by going through all the necessary step and methods in this notebook alongside its explanation. Since there is already task provided in the dataset description making it easier to explore different means to answer multiple questions and provide analysis. Also note that I'm still learning and there may be mistake and few things i might be overlooked. If that happen feel free comment and point them out.
# Thanks!
# # Table of Contents
# [SALES ANALYTICS](#SALES-ANALYTICS)
# - [LIBRARY IMPORT](#LIBRARY-IMPORT)
# - [DATA READING](#DATA-READING)
# - [DATA CLEANING](#DATA-CLEANING)
# - [DATA MANIPULATION](#DATA-MANIPULATION)
# - [QUESTIONS](#QUESTIONS)
# - [Annual Sales](#Annual-Sales)
# - [Monthly Sales](#Monthly-Sales)
# - [Sales by State & City](#Sales-by-State-and-City)
# - [Sales by State](#Sales-by-State)
# - [Sales by City](#Sales-by-City)
# - [Order Trend](#Order-Trend)
# - [Monthly Order Trend](#Monthly-Order-Trend)
# - [Daily Order Trend](#Daily-Order-Rrend)
# - [Hourly Order Trend](#Hourly-Order-Trend)
# - [Product Performance](#Product-Performance)
# - [Product Popularity](#Product-Popularity)
# - [Price List Comparision](#Price-List-Comparision)
# - [Product Sold by Order](#Product-Sold-by-Order)
# - [Probability](#Probability)
# - [Product Orders of Total Sales](#Product-Orders-of-Total-Sales)
# - [Product Associate Rules](#Product-Associate-Rules)
# - [SUMMARY](#TO-SUMMARIZE)
# - [REFERENCES](#REFERENCES)
# # LIBRARY IMPORT
# > Import all required library for files import/export, plotting, and association rules.
# import required library
import os
import squarify
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from matplotlib.dates import DateFormatter
# required library for one-hot matrix and association rules
from mlxtend.preprocessing import TransactionEncoder
from mlxtend.frequent_patterns import apriori, association_rules
# # DATA READING
# > Combine all related CSV data into a single dataframe(all_sales_df) then rename and lowercase all the column name.
# find file path directory
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# data path
dir_path = "../input/sales-product-data"
# create empty df for all sales
all_sales_df = pd.DataFrame()
# concat all csv in dir path
for filename in os.listdir(dir_path):
df = pd.read_csv(os.path.join(dir_path, filename))
all_sales_df = pd.concat([all_sales_df, df], ignore_index=True)
# rename column name
all_sales_df = all_sales_df.rename(mapper=str.strip, axis="columns")
all_sales_df = all_sales_df.rename(
columns={
"Order ID": "Order_id",
"Quantity Ordered": "Quantity",
"Price Each": "Price",
"Order Date": "Date",
"Purchase Address": "Address",
}
)
# lowercase column name
column_name = list(all_sales_df.columns)
column_name = [x.lower().strip() for x in column_name]
all_sales_df.columns = column_name
# show dataframe info
print(all_sales_df.info())
# show all_sales_df head
all_sales_df.head()
# # DATA CLEANING
# > First check for rows contain null and duplicates.
# Then check for non-numeric values in order_id, quantity, and price since the 3 column cannot be non-numeric.
# check for all rows contain null value
print("rows contain null:")
print(all_sales_df.isnull().sum())
# check duplicated rows
print("rows contain duplicates:", all_sales_df.duplicated().sum())
# check for non-numeric in order_id, quantity, and price
print(
"non-numeric in order_id:",
all_sales_df["order_id"]
.loc[pd.to_numeric(all_sales_df["order_id"], errors="coerce").isnull()]
.unique(),
)
print(
"non-numeric in quantity:",
all_sales_df["quantity"]
.loc[pd.to_numeric(all_sales_df["quantity"], errors="coerce").isnull()]
.unique(),
)
print(
"non-numeric in price:",
all_sales_df["price"]
.loc[pd.to_numeric(all_sales_df["price"], errors="coerce").isnull()]
.unique(),
)
# > There is a few consideration about dropping duplicated rows.
# Check duplicated rows and their duplicates.
# check order id in duplicated rows that are not null and is numeric
all_sales_df[
all_sales_df.duplicated(keep=False)
& all_sales_df["order_id"].notnull()
& all_sales_df["order_id"].str.isnumeric()
]["order_id"].head()
# check rows contain order id of 176585, 177795, 178158
all_sales_df[all_sales_df["order_id"].isin(["176585", "177795", "178158"])]
# >Now since order id 176585, 177795, and 178158 despite having quantity column it still have more than 1 of same rows each.
# Which doesn't make sense when the product is the same it should be added into 'quantity' instead of adding another duplicate row. Another thing to consider is that we can join the rows from duplicates into rows that are duplicated and add the value into quantity.
# But since it is still unclear, the next cells will continue to remove the duplicate rows.
# > If there is null and duplicated rows then it need to be dropped/removed.
# For non-numeric in mentioned columns, apply to_numeric to remove the rows dataframe.
# Then check the result by applying the previous methods for a new defined dataframe(clean_sales_df).
# drop null rows
clean_sales_df = all_sales_df.dropna(how="all")
# drop all duplicates rows
clean_sales_df = clean_sales_df[~clean_sales_df.duplicated()]
# drop non-numeric values in order_id, quantity, and price
clean_sales_df = clean_sales_df[
pd.to_numeric(clean_sales_df["order_id"], errors="coerce").notnull()
]
clean_sales_df = clean_sales_df[
pd.to_numeric(clean_sales_df["quantity"], errors="coerce").notnull()
]
clean_sales_df = clean_sales_df[
pd.to_numeric(clean_sales_df["price"], errors="coerce").notnull()
]
# print data cleaning result
# null result
print("rows contain null:")
print(clean_sales_df.isnull().sum())
# check duplicated rows
print("rows contain duplicates:", clean_sales_df.duplicated().sum())
# unique value result
print(
"non-numeric in order_id:",
clean_sales_df["order_id"]
.loc[pd.to_numeric(clean_sales_df["order_id"], errors="coerce").isnull()]
.unique(),
)
print(
"non-numeric in quantity:",
clean_sales_df["quantity"]
.loc[pd.to_numeric(clean_sales_df["quantity"], errors="coerce").isnull()]
.unique(),
)
print(
"non-numeric in price:",
clean_sales_df["price"]
.loc[pd.to_numeric(clean_sales_df["price"], errors="coerce").isnull()]
.unique(),
)
# # DATA MANIPULATION
# > Before analysis it's important to define data type for each column that will be called later on.
# Split city and state from 'address' into separate 'city' and 'state' column which will be required later for visualizations.
# Add 'total_sales' column from 'quantity'*'price'.
# change data type for column quantity, price, date, and address
clean_sales_df["quantity"] = clean_sales_df["quantity"].astype(int)
clean_sales_df["price"] = clean_sales_df["price"].astype(float)
clean_sales_df["date"] = pd.to_datetime(clean_sales_df["date"], format="%m/%d/%y %H:%M")
clean_sales_df["address"] = clean_sales_df["address"].astype(str)
# add city and state column
clean_sales_df["city"] = clean_sales_df["address"].apply(
lambda x: x.split(",")[1].strip()
)
clean_sales_df["state"] = clean_sales_df["address"].apply(
lambda x: x.split(",")[2].split(" ")[1].strip()
)
# add total sales column
clean_sales_df["total_sales"] = clean_sales_df["quantity"] * clean_sales_df["price"]
# > From here since data outside year 2019 is less significant.
# The following try to separate clean_sales_df(all data) and clean_sales_2019_df(2019 data).
# But we can check how many rows contain data outside 2019 by applying following method.
# find data outside 2019
print(
"number of rows outside 2019:",
len(clean_sales_df[clean_sales_df["date"].dt.year != 2019]),
)
# store 2019 data in separate dataframe
clean_sales_2019_df = pd.DataFrame(
clean_sales_df[clean_sales_df["date"].dt.year == 2019]
) # this will only include rows with year 2019
# print data manipulation result
# 2019 data
print("clean_sales_2019 data result:")
print(clean_sales_2019_df.info())
# data outside 2019
print(
"number of rows outside 2019 in clean_sales_2019_df:",
len(clean_sales_2019_df[clean_sales_2019_df["date"].dt.year != 2019]),
)
# > Reset the index for both dataframe and print data head to show dataframe result.
# reset data index for both dataframe
clean_sales_df = clean_sales_df.reset_index(drop=True)
clean_sales_2019_df = clean_sales_2019_df.reset_index(drop=True)
# check data head before analysis/visualization
clean_sales_df.head()
# # QUESTIONS
# ## Annual Sales
# > Task 1:
# > Q: What was the best Year for sales? How much was earned that Year?
# >
# > A: Create Annual Sales summary and find max/min values in Annual Sales
# > In this section clean_sales_df will be referred to find annual sales as annual_sales_df.
# Plot the annual_sales_df and find max/min values in annual_sales_df then print the result as conclusion.
# find annual sales summary and convert to dataframe
annual_sales_df = pd.DataFrame(
clean_sales_df.groupby(clean_sales_df["date"].dt.year)["total_sales"].sum()
)
annual_sales_df.reset_index(inplace=True)
# generate annual sales visual
with plt.style.context("ggplot"):
plt.figure(figsize=(4, 4))
sns.barplot(x="date", y="total_sales", data=annual_sales_df)
plt.ticklabel_format(style="plain", axis="y")
plt.title("Annual Sales", fontsize=20)
plt.xlabel("Year")
plt.ylabel("Sales")
# add data callouts
for x, y in enumerate(annual_sales_df["total_sales"]):
label = f"${y:,.2f}"
plt.annotate(
label,
(x, y),
textcoords="offset points",
xytext=(0, 1),
ha="center",
fontsize=12,
)
plt.show()
# find highest year and value in annual sales
highest_year = annual_sales_df["date"].iloc[annual_sales_df["total_sales"].idxmax()]
highest_value = annual_sales_df["total_sales"].max()
# find lowest year and value in annual sales
lowest_year = annual_sales_df["date"].iloc[annual_sales_df["total_sales"].idxmin()]
lowest_value = annual_sales_df["total_sales"].min()
# show result as visual
with plt.style.context("ggplot"):
plt.subplots(figsize=(8, 1))
plt.axis("off")
plt.text(
0.5,
0.5,
f"Highest Year: {highest_year}, Total Sales: ${highest_value:,.2f}\n"
f"Lowest Year: {lowest_year}, Total Sales: ${lowest_value:,.2f}",
fontsize=14,
ha="center",
va="center",
)
plt.title("Best & Worst Year", fontsize=20)
plt.show()
# print result as conclusion
print(
f"CONCLUSION\n"
f"Total sales generated by company: ${highest_value+lowest_value:,.2f}\n"
f"\n"
f"With the highest sales is in {highest_year}, generating: ${highest_value:,.2f}\n"
f"With the lowest sales is in {lowest_year}, generating: ${lowest_value:,.2f}"
)
# ## Monthly Sales
# > Task 2:
# > Q: What was the best month for sales? How much was earned that month?
# >
# > A: Create Monthly Sales summary and find largest/smallest values in Monthly Sales
# > In this section clean_sales_df will be referred to find monthly sales as monthly_sales_df.
# Plot the monthly_sales_df and find largest/smallest values in monthly_sales_df then print the result as conclusion.
# find monthly sales summary and convert to dataframe
monthly_sales_df = pd.DataFrame(
clean_sales_df.groupby(
[clean_sales_df["date"].dt.year, clean_sales_df["date"].dt.month]
)["total_sales"].sum()
)
monthly_sales_df.index = pd.to_datetime(
monthly_sales_df.index.map("{0[0]}-{0[1]}".format)
)
monthly_sales_df.index.name = "date"
monthly_sales_df = monthly_sales_df.reset_index()
# find monthly sales average
average_monthly_sales = monthly_sales_df["total_sales"].mean()
# generate monthly sales visual
with plt.style.context("ggplot"):
plt.figure(figsize=(16, 4))
sns.lineplot(x="date", y="total_sales", data=monthly_sales_df)
plt.gca().xaxis.set_major_formatter(DateFormatter("%y-%m"))
plt.ticklabel_format(style="plain", axis="y")
plt.title("Monthly Sales", fontsize=20)
plt.xlabel("Date")
plt.ylabel("Sales")
plt.xticks(monthly_sales_df["date"])
# add axh line
plt.axhline(y=average_monthly_sales, color="blue", linestyle="--", linewidth=1)
plt.text(
monthly_sales_df["date"].iloc[-12],
average_monthly_sales + 5,
f"Avg: ${average_monthly_sales:,.2f}",
color="blue",
fontsize=10,
ha="center",
va="bottom",
)
# add data callouts
for x, y in zip(monthly_sales_df["date"], monthly_sales_df["total_sales"]):
label = f"${y:,.2f}"
plt.annotate(
label, (x, y), textcoords="offset points", xytext=(2, 4), ha="center"
)
plt.show()
# find highest month and value in monthly sales
highest_months = monthly_sales_df.nlargest(3, "total_sales")
highest_months["date"] = pd.to_datetime(highest_months["date"]).dt.strftime("%B %Y")
highest_month_names = highest_months["date"].tolist()
highest_month_values = highest_months["total_sales"].tolist()
# find lowest month and value in monthly sales
lowest_months = monthly_sales_df.nsmallest(3, "total_sales")
lowest_months["date"] = pd.to_datetime(lowest_months["date"]).dt.strftime("%B %Y")
lowest_month_names = lowest_months["date"].tolist()
lowest_month_values = lowest_months["total_sales"].tolist()
# show result as visual
with plt.style.context("ggplot"):
plt.subplots(figsize=(8, 1))
plt.axis("off")
plt.text(
0.5,
0.5,
f"Highest Month: {highest_month_names[0]}, Total Sales: ${highest_month_values[0]:,.2f}\n"
f"Lowest Month: {lowest_month_names[0]}, Total Sales: ${lowest_month_values[0]:,.2f}",
fontsize=14,
ha="center",
va="center",
)
plt.title("Best & Worst Month", fontsize=20)
plt.show()
# print result as conclusion
print(
f"CONCLUSION\n"
f"Top 3 best month:\n{highest_months.to_string(index=False)}\n"
f"\n"
f"Bottom 3 worst month:\n{lowest_months.to_string(index=False)}"
)
# ## Sales by State and City
# > Task 3:
# > Q: What City had the highest number of sales?
# >
# > A: Create Sales by State/City and find largest/smallest values in Sales by State/City
# > This section separated by 2 for Sales by State and Sales by City from clean_sales_df.
# Purpose is to find both trend on state level and city level.
# Plot and find largest/smallest values in both dataframe then print the result as conclusion.
# ### Sales by State
# find sales by state and convert to dataframe
sales_by_state_df = pd.DataFrame(clean_sales_df.groupby("state")["total_sales"].sum())
sales_by_state_df = sales_by_state_df.sort_values(
by="total_sales", ascending=False
).reset_index()
# generate sales by state visual
with plt.style.context("fivethirtyeight"):
plt.figure(figsize=(16, 4))
sns.barplot(x="state", y="total_sales", data=sales_by_state_df, palette="husl")
plt.ticklabel_format(style="plain", axis="y")
plt.title("Sales by State", fontsize=20)
plt.xlabel("State")
plt.ylabel("Sales")
# add data callouts
for x, y in enumerate(sales_by_state_df["total_sales"]):
label = f"${y:,.2f}"
plt.annotate(
label,
(x, y),
textcoords="offset points",
xytext=(0, 2),
ha="center",
fontsize=10,
)
plt.show()
# > Following method is to create dictionary for state colors which is used by previous Sales by State plot.
# The resulted dictionary will be called in next Sales by City visual to refer state by its colors.
# create dictionary for state colors for use in sales by city visual
state_colors = {}
n_states = len(sales_by_state_df["state"])
palette = sns.color_palette(
"husl", n_colors=n_states
) # use same palette as previous state visual
for i, state in enumerate(sales_by_state_df["state"]):
state_colors[state] = palette[i]
# check the state colors
print(state_colors)
# find highest month and value in sales by state
highest_state = sales_by_state_df.nlargest(3, "total_sales")
highest_state_names = highest_state["state"].tolist()
highest_state_values = highest_state["total_sales"].tolist()
# find lowest month and value in sales by state
lowest_state = sales_by_state_df.nsmallest(3, "total_sales")
lowest_state_names = lowest_state["state"].tolist()
lowest_state_values = lowest_state["total_sales"].to_list()
# show result as visual
with plt.style.context("fivethirtyeight"):
plt.subplots(figsize=(8, 1))
plt.axis("off")
plt.text(
0.5,
0.5,
f"Highest State: {highest_state_names[0]}, Total Sales: ${highest_state_values[0]:,.2f}\n"
f"Lowest State: {lowest_state_names[0]}, Total Sales: ${lowest_state_values[0]:,.2f}",
fontsize=14,
ha="center",
va="center",
)
plt.title("Best & Worst State", fontsize=20)
plt.show()
# print result as conclusion
print(
f"CONCLUSION\n"
f"Top 3 best state sales:\n{highest_state.to_string(index=False)}\n"
f"\n"
f"Bottom 3 worst state sales:\n{lowest_state.to_string(index=False)}"
)
# ### Sales by City
# > There is a problem when referring y='city' in following cell for a same city name Portland, Maine and Portland Oregon which in return overwrite the y-axis.
# The solution is to rename the city name for 'Portland' by referring 'state' then return with new defined names.
# find sales by city and convert to dataframe
sales_by_city_df = pd.DataFrame(
clean_sales_df.groupby(["state", "city"])["total_sales"].sum()
)
sales_by_city_df = sales_by_city_df.sort_values(
by="total_sales", ascending=False
).reset_index()
# modify city named "Portland" before generating visual (to avoid overwrite)
sales_by_city_df.loc[
(sales_by_city_df["state"] == "ME") & (sales_by_city_df["city"] == "Portland"),
"city",
] = "Portland (Maine)"
sales_by_city_df.loc[
(sales_by_city_df["state"] == "OR") & (sales_by_city_df["city"] == "Portland"),
"city",
] = "Portland (Oregon)"
# generate visual for sales by city
with plt.style.context("fivethirtyeight"):
plt.figure(figsize=(16, 4))
sns.barplot(
x="total_sales",
y="city",
hue="state",
data=sales_by_city_df,
dodge=False,
palette=state_colors,
) # previous state_colors dict
plt.title("Sales by City", fontsize=20)
plt.xlabel("Sales")
plt.ylabel("City")
plt.legend(
title="State", bbox_to_anchor=(1.05, 0.5), loc="center left", borderaxespad=0.0
)
# add data callouts
for y, x in enumerate(sales_by_city_df["total_sales"]):
label = f"${x:,.2f}"
plt.annotate(
label,
(x, y),
textcoords="offset points",
xytext=(0, 0),
ha="left",
va="center",
fontsize=10,
)
plt.show()
# find highest month and value in sales by city
highest_city = sales_by_city_df.nlargest(3, "total_sales")
highest_city_names = highest_city["city"].tolist()
highest_city_values = highest_city["total_sales"].tolist()
# find lowest month and value in sales by city
lowest_city = sales_by_city_df.nsmallest(3, "total_sales")
lowest_city_names = lowest_city["city"].tolist()
lowest_city_values = lowest_city["total_sales"].tolist()
# show result as visual
with plt.style.context("fivethirtyeight"):
plt.subplots(figsize=(8, 1))
plt.axis("off")
plt.text(
0.5,
0.5,
f"Highest City: {highest_city_names[0]}, Total Sales: ${highest_city_values[0]:,.2f}\n"
f"Lowest City: {lowest_city_names[0]}, Total Sales: ${lowest_city_values[0]:,.2f}",
fontsize=14,
ha="center",
va="center",
)
plt.title("Highest & Lowest City Sales", fontsize=20)
plt.show()
# print result as conclusion
print(
f"CONCLUSION\n"
f"Top 3 best city sales:\n{highest_city.to_string(index=False)}\n"
f"\n"
f"Bottom 3 worst city sales:\n{lowest_city.to_string(index=False)}"
)
# ## Order Trend
# > Task 4:
# > Q: What time should we display adverstisement to maximize likelihood of customer's buying product?
# >
# > A: Find Monthly/Daily/Hourly Order Trend
# > This section will find the data related to orders by simply find count/unique number of order_id by month/day/hour.
# The data that will be referred here is clean_sales_2019_df(which also apply for the rest of the analysis).
# >
# > Despite order id cannot be duplicate in the dataset there is multiple rows with the same order id.
# The reason why it's not dropped is because each may contains different product.
# *not sure how to approach/may also need to change data cleaning section
# >
# > But using .count() it will return the duplicated order id as well.
# If we want to count the order id as unique number then we can call .nunique() which will be used in following sections.
# ### Monthly Order Trend
# find monthly order and convert to dataframe
monthly_order_df = pd.DataFrame(
clean_sales_2019_df.groupby(
[clean_sales_2019_df["date"].dt.year, clean_sales_2019_df["date"].dt.month]
)["order_id"].nunique()
)
monthly_order_df.index = pd.to_datetime(
monthly_order_df.index.map("{0[0]}-{0[1]}".format)
)
monthly_order_df.index.name = "date"
monthly_order_df = monthly_order_df.rename(
columns={"order_id": "total_orders"}
).reset_index()
# find average monthly order
average_monthly_order = monthly_order_df["total_orders"].mean()
# generate monthly order visual
with plt.style.context("bmh"):
plt.figure(figsize=(16, 4))
sns.lineplot(x="date", y="total_orders", data=monthly_order_df)
plt.gca().xaxis.set_major_formatter(DateFormatter("%Y-%m"))
plt.ticklabel_format(style="plain", axis="y")
plt.title("Monthly Order Trend", fontsize=20)
plt.xlabel("Date")
plt.ylabel("Order")
plt.xticks(monthly_order_df["date"])
# add axh line
plt.axhline(y=average_monthly_order, color="red", linestyle="--", linewidth=1)
plt.text(
monthly_order_df["date"].iloc[-12],
average_monthly_order + 5,
f"Avg: {average_monthly_order:,.2f}",
color="red",
fontsize=10,
ha="center",
va="bottom",
)
# add data callouts
for x, y in zip(monthly_order_df["date"], monthly_order_df["total_orders"]):
label = f"{(y):,}"
plt.annotate(
label, (x, y), textcoords="offset points", xytext=(2, 4), ha="center"
)
plt.show()
# find 3 highest value in monthly order
highest_monthly_orders = monthly_order_df.nlargest(3, "total_orders")
highest_monthly_orders["date"] = pd.to_datetime(
highest_monthly_orders["date"]
).dt.strftime("%B")
# find 3 lowest value in monthly order
lowest_monthly_orders = monthly_order_df.nsmallest(3, "total_orders")
lowest_monthly_orders["date"] = pd.to_datetime(
lowest_monthly_orders["date"]
).dt.strftime("%B")
# print result as conclusion
print(
f"CONCLUSION\n"
f"Months with highest total orders:\n{highest_monthly_orders.to_string(index=False)}\n"
f"\n"
f"Months with lowest total orders:\n{lowest_monthly_orders.to_string(index=False)}\n"
f"\n"
f"*use as references to maximize/prioritize advertisement"
)
# ### Daily Order Trend
# find daily order and convert to dataframe
daily_order_df = pd.DataFrame(
clean_sales_2019_df.groupby(clean_sales_2019_df["date"].dt.dayofweek)[
"order_id"
].nunique()
)
daily_order_df.columns = ["total_orders"]
daily_order_df.index = [
"Monday",
"Tuesday",
"Wednesday",
"Thursday",
"Friday",
"Saturday",
"Sunday",
]
daily_order_df.index.name = "day_of_week"
daily_order_df = daily_order_df.reset_index()
# find average daily order
average_daily_order = daily_order_df["total_orders"].mean()
# generate daily order visual
with plt.style.context("bmh"):
plt.figure(figsize=(16, 4))
sns.lineplot(x="day_of_week", y="total_orders", data=daily_order_df)
plt.ticklabel_format(style="plain", axis="y")
plt.title("Daily Order Trend", fontsize=20)
plt.xlabel("Day of Week")
plt.ylabel("Order")
# add axh line
plt.axhline(y=average_daily_order, color="red", linestyle="--", linewidth=1)
plt.text(
daily_order_df["day_of_week"].iloc[-7],
average_daily_order - 5,
f"Avg: {average_daily_order:,.2f}",
color="red",
fontsize=10,
ha="center",
va="top",
)
# add data callouts
for x, y in enumerate(daily_order_df["total_orders"]):
label = f"{(y):,}"
plt.annotate(
label, (x, y), textcoords="offset points", xytext=(2, 4), ha="center"
)
plt.show()
# find 3 highest value in daily orders
highest_daily_orders = daily_order_df.nlargest(3, "total_orders")
# find 3 lowest value in daily orders
lowest_daily_orders = daily_order_df.nsmallest(3, "total_orders")
# print result as conclusion
print(
f"CONCLUSION\n"
f"Days with highest total orders:\n{highest_daily_orders.to_string(index=False)}\n"
f"\n"
f"Days with lowest total orders:\n{lowest_daily_orders.to_string(index=False)}\n"
f"\n"
f"*use as references to maximize/prioritize advertisement"
)
# ### Hourly Order Trend
# find hourly order and convert to dataframe
hourly_order_df = pd.DataFrame(
clean_sales_2019_df.groupby(clean_sales_2019_df["date"].dt.hour)[
"order_id"
].nunique()
)
hourly_order_df.columns = ["total_orders"]
hourly_order_df.index = pd.to_datetime(hourly_order_df.index, format="%H").strftime(
"%H:%M"
)
hourly_order_df.index.name = "hour"
hourly_order_df = hourly_order_df.reset_index()
# find average hourly order
average_hourly_order = hourly_order_df["total_orders"].mean()
# generate hourly order visual
with plt.style.context("bmh"):
plt.figure(figsize=(16, 4))
sns.lineplot(x="hour", y="total_orders", data=hourly_order_df)
plt.ticklabel_format(style="plain", axis="y")
plt.title("Hourly Order Trend", fontsize=20)
plt.xlabel("Hour")
plt.ylabel("Order")
# add axh line
plt.axhline(y=average_hourly_order, color="red", linestyle="--", linewidth=2)
plt.text(
0.5,
average_hourly_order + 100,
f"Avg: {average_hourly_order:,.2f}",
color="red",
fontsize=10,
ha="center",
va="bottom",
)
# add data callouts
for x, y in enumerate(hourly_order_df["total_orders"]):
label = f"{y:,}"
plt.annotate(
label, (x, y), textcoords="offset points", xytext=(2, 4), ha="center"
)
plt.show()
# find 6 highest value in hourly orders
highest_hourly_orders = hourly_order_df.nlargest(6, "total_orders")
# find 6 lowest value in hourly orders
lowest_hourly_orders = hourly_order_df.nsmallest(6, "total_orders")
# print result as conclusion
print(
f"CONCLUSION\n"
f"Hours with highest total orders:\n{highest_hourly_orders.to_string(index=False)}\n"
f"\n"
f"Hours with lowest total orders:\n{lowest_hourly_orders.to_string(index=False)}\n"
f"\n"
f"*use as references to maximize/prioritize advertisement"
)
# ## Product Performance
# > Task 5:
# > Q: What product sold the most? Why do you think it sold the most?
# >
# > A: Find Products Popularity and Product Price List Comparision
# > Find data related to Product Popularity by calculate sum of quantity by each product.
# This return the total product being purchased.
# It also find price list by product which is used to create filled lines for comparision.
# ### Product Popularity
# create dataframe for product popularity
product_popularity_df = pd.DataFrame(
clean_sales_2019_df.groupby("product")["quantity"].sum()
)
product_popularity_df.index.name = "product"
product_popularity_df = product_popularity_df.sort_values(
by="quantity", ascending=False
)
product_popularity_df = product_popularity_df.reset_index()
# create dataframe for product price
product_price_df = pd.DataFrame(clean_sales_2019_df.groupby("product")["price"].first())
product_price_df = product_price_df.reset_index()
# merge the two dataframes on the product column
merged_popularity_price_df = pd.merge(
product_popularity_df, product_price_df, on="product"
)
# generate product sales visual
with plt.style.context("classic"):
fig, ax1 = plt.subplots(figsize=(16, 4))
sns.barplot(
x="product",
y="quantity",
data=merged_popularity_price_df,
palette="husl",
ax=ax1,
)
plt.ticklabel_format(style="plain", axis="y")
plt.title("Product Popularity", fontsize=20)
plt.xlabel("Product")
plt.ylabel("Quantity")
plt.xticks(rotation=80)
# add data callouts
for x, y in enumerate(merged_popularity_price_df["quantity"]):
label = f"{y:,}"
plt.annotate(
label,
(x, y),
textcoords="offset points",
xytext=(0, 4),
ha="center",
fontsize=12,
)
# plot product prices as filled line plot
ax2 = ax1.twinx()
ax2.plot(
merged_popularity_price_df["product"],
merged_popularity_price_df["price"],
color="red",
)
ax2.set_ylabel("Price")
ax2.fill_between(
merged_popularity_price_df["product"],
merged_popularity_price_df["price"],
alpha=0.2,
)
plt.show()
# > Similar to previous Sales by State/City method the following will store product colors as dictionary which will be used later on.
# create dictionary for product colors for use in product related visual
product_colors = {}
n_products = len(merged_popularity_price_df["product"])
palette = sns.color_palette(
"husl", n_colors=n_products
) # use same palette as previous product visual
for i, product in enumerate(merged_popularity_price_df["product"]):
product_colors[product] = palette[i]
# check the state colors
print(product_colors)
# find product sold most
product_sold_most = merged_popularity_price_df.nlargest(3, "quantity")
product_sold_most_names = product_sold_most["product"].tolist()
product_sold_most_quantities = product_sold_most["quantity"].tolist()
# find product sold least
product_sold_least = merged_popularity_price_df.nsmallest(3, "quantity")
product_sold_least_names = product_sold_least["product"].tolist()
product_sold_least_quantities = product_sold_least["quantity"].tolist()
# show result as visual
with plt.style.context("classic"):
plt.subplots(figsize=(8, 1))
plt.axis("off")
plt.text(
0.5,
0.5,
f"Most sold: {product_sold_most_names[0]}, Quantity: {product_sold_most_quantities[0]:,}\n"
f"Least sold: {product_sold_least_names[0]}, Quantity: {product_sold_least_quantities[0]:,}",
fontsize=14,
ha="center",
va="center",
)
plt.title("Most & Least Sold Product", fontsize=20)
plt.show()
# print result as conclusion
print(
f"CONCLUSION\n"
f"Top 3 most sold products:\n{product_sold_most.to_string(index=False)}\n"
f"\n"
f"Bottom 3 least sold products:\n{product_sold_least.to_string(index=False)}"
)
# ### Price List Comparision
# find price for each item list
product_list_df = pd.DataFrame(clean_sales_2019_df.groupby("product")["price"].first())
product_list_df = product_list_df.sort_values(by="price", ascending=False)
product_list_df = product_list_df.reset_index()
# find product price average
average_product_price = product_list_df["price"].mean()
# generate product list visual
with plt.style.context("classic"):
plt.figure(figsize=(16, 6))
sns.barplot(x="price", y="product", data=product_list_df, palette=product_colors)
plt.title("Product Price List", fontsize=20)
plt.xlabel("Price")
plt.ylabel("Product")
# add axv line
plt.axvline(x=average_product_price, color="red", linestyle="--", linewidth=1)
plt.text(
average_product_price + 5,
len(product_list_df) / 2,
f"Avg: ${average_product_price:.2f}",
color="red",
ha="left",
va="center",
)
plt.show()
# find expensive product
most_expensive_product = product_list_df.nlargest(3, "price")
# find cheap product
most_cheap_product = product_list_df.nsmallest(3, "price")
# print result as conclusion
print(
f"CONCLUSION\n"
f"Most expensive products:\n{most_expensive_product.to_string(index=False)}\n"
f"\n"
f"Most cheap products:\n{most_cheap_product.to_string(index=False)}"
)
# ### Product Sold by Order
# > Find average item each order and show how likely customers to buy more than 1 product in single order.
# find average quantity per order id
average_item_per_order = (
clean_sales_2019_df.groupby("order_id")["quantity"].mean().mean()
)
# show result as visual
with plt.style.context("classic"):
plt.subplots(figsize=(8, 1))
plt.axis("off")
plt.text(
0.5,
0.5,
f"{average_item_per_order:,.2f}",
fontsize=40,
ha="center",
va="center",
)
plt.title("Average Item Each Order", fontsize=20)
plt.show()
# ## Probability
# > Task 6:
# > Q: How much probability for next people will order certain products?
# >
# > A: Find Product Orders % of Total Sales
# >The following section is to find % product orders/sold compare to the total sales of all products.
# the result will give % of product which may define the likelihood of the product will be purchased next.
# This part is also the only section which used Squarify to plot the result as Treemap.
# ### Product Orders of Total Sales
# find total product sold
total_product_sold = product_popularity_df["quantity"].sum()
# find sizes and labels value in product popularity
sizes = product_popularity_df["quantity"]
labels = product_popularity_df["product"]
# calculate percentage for each product
percentages = [
f"{100*sizes[i]/sizes.sum():.2f}%\n{labels[i]}" for i in range(len(sizes))
]
# generate treemap visual for product sold percentage of total
with plt.style.context("seaborn-dark-palette"):
plt.figure(figsize=(16, 8))
squarify.plot(
sizes=sizes,
label=percentages,
alpha=0.7,
color=[product_colors[label] for label in labels],
) # get color from previous product color dict
plt.axis("off")
plt.title("Product % of Total", fontsize=20)
plt.show()
# print the result as conclusion
print(f"CONCLUSION\n" f"List of probability:")
# find percentages for product and print as conclusion
for p in percentages:
prob, label = p.split("\n")
print(f"{label} = {prob}")
# ## Product Associate Rules
# > Task 7:
# > Q: What products are most often sold together?
# >
# > A: Find Product Associate Rules using mlxtend library
# > First need to find the product sold by each order id and store them into series(product_by_order).
# Then transform the data into one-hot encoded matrix by applying TransactionEncoder().fit_transform().
# The result will be stored as dataframe(encoded_df).
# Encoded data then used to generate frequent item sets using apriori algorithm the minimum support are defined as following.
# Lastly sort the order by value in 'confidence'.
# group product by order id
product_by_order = clean_sales_2019_df.groupby("order_id")["product"].apply(list)
# convert data to one-hot encoded matrix
te = TransactionEncoder()
onehot = te.fit_transform(product_by_order)
encoded_df = pd.DataFrame(onehot, columns=te.columns_)
# identify frequent itemsets
frequent_itemsets = apriori(encoded_df, min_support=0.000015, use_colnames=True)
# generate association rules
rules = association_rules(frequent_itemsets, metric="confidence", min_threshold=0.5)
rules = rules.sort_values("confidence", ascending=False)
# print rules data head
rules.head()
# > Although the rules can be presented as tables.
# In this case the rules is generated as string following more descriptive explanation.
# There may be other method to visualize the rules waiting to be explored.
# But for now the following will do.
# create summary for each rule
# create list to store items
summaries = []
# iterrate throught available items in rules
for i, row in rules.iterrows():
antecedents = ", ".join(list(row["antecedents"]))
consequents = list(row["consequents"])[0]
support = row["support"]
confidence = row["confidence"]
lift = row["lift"]
leverage = row["leverage"]
conviction = row["conviction"]
# define ANSI escape codes for modifying the text's appearance
BLUE = "\033[94m"
GREEN = "\033[92m"
RED = "\033[91m"
END = "\033[0m"
# create summary in string
summary = (
f"Rule {i+1}:\n"
f"Customers who bought {BLUE}{antecedents}{END} are more likely to buy {RED}{consequents}{END}.\n"
f"This combination of products was purchased in {GREEN}{support*100:.3f}%{END} of all transactions.\n"
f"When customers buy {BLUE}{antecedents}{END}, the chance of them also buying {RED}{consequents}{END} is {GREEN}{confidence*100:.2f}%{END}.\n"
f"{RED}{consequents}{END} appears {GREEN}{lift:.2f}x{END} more likely when {BLUE}{antecedents}{END} appears compared to {RED}{consequents}{END} alone in general.\n"
f"The rule's significance, as measured by leverage, is {GREEN}{leverage:.5f}{END}.\n"
f"Dependence between {BLUE}{antecedents}{END} and {RED}{consequents}{END} is {GREEN}{conviction:.2f}{END}."
)
# append the summary
summaries.append(summary)
# join all summaries into a single string
summary_string = "\n\n".join(summaries)
# print summary
print(f"CONCLUSION\n\n{summary_string}")
|
# #### Now we are going to apply eigen cam on YOLO v5 model. There are certain things that we must consider before beginning with creation of the code for the interpretation of the model
# - **The reshape transform - This isnt necessary for this model since it directly will return 2D spatial tensor as it's output.**
# - **The target function - For eigen cam there won't be a a target function necessary for this, instead we will just apply PCA on the 2D activations**
# - **The target layer - We will be extracting the second last layer as the target layer since the final layer just deals with the detections.**
import warnings
warnings.filterwarnings("ignore")
warnings.simplefilter("ignore")
import torch
import cv2
import numpy as np
import requests
import torchvision.transforms as transforms
from pytorch_grad_cam import EigenCAM
from pytorch_grad_cam.utils.image import show_cam_on_image, scale_cam_image
from PIL import Image
model = torch.hub.load("ultralytics/yolov5", "yolov5s", pretrained=True)
# #### After playing with the model, we can find the second last layer in it
for modules in model.named_children():
print(modules)
print(type(model))
print(type(model.model))
print(type(model.model.model))
print(type(model.model.model.model))
print(model.model.model.model[-2])
import io
COLORS = np.random.uniform(0, 255, size=(80, 3))
def parse_detections(results):
detections = results.pandas().xyxy[0]
detections = detections.to_dict()
boxes, colors, names = [], [], []
for i in range(len(detections["xmin"])):
confidence = detections["confidence"][i]
if confidence < 0.2:
continue
xmin = int(detections["xmin"][i])
ymin = int(detections["ymin"][i])
xmax = int(detections["xmax"][i])
ymax = int(detections["ymax"][i])
name = detections["name"][i]
category = int(detections["class"][i])
color = COLORS[category]
boxes.append((xmin, ymin, xmax, ymax))
colors.append(color)
names.append(name)
return boxes, colors, names
def draw_detections(boxes, colors, names, img):
for box, color, name in zip(boxes, colors, names):
xmin, ymin, xmax, ymax = box
cv2.rectangle(img, (xmin, ymin), (xmax, ymax), color, 2)
cv2.putText(
img,
name,
(xmin, ymin - 5),
cv2.FONT_HERSHEY_SIMPLEX,
0.8,
color,
2,
lineType=cv2.LINE_AA,
)
return img
headers = {
"User-Agent": "Mozilla/5.0 (Windows NT 10.0; Win64; x64) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/112.0.0.0 Safari/537.36"
}
image_url = (
"https://upload.wikimedia.org/wikipedia/commons/f/f1/Puppies_%284984818141%29.jpg"
)
response = requests.get(image_url, stream=True, headers=headers)
img = np.array(Image.open(io.BytesIO(response.content)))
img = cv2.resize(img, (640, 640))
rgb_img = img.copy()
img = np.float32(img) / 255
transform = transforms.ToTensor()
tensor = transform(img).unsqueeze(0)
model = torch.hub.load("ultralytics/yolov5", "yolov5s", pretrained=True)
model.eval()
model.cpu()
target_layers = [model.model.model.model[-2]]
results = model([rgb_img])
boxes, colors, names = parse_detections(results)
detections = draw_detections(boxes, colors, names, rgb_img.copy())
Image.fromarray(detections)
# #### Creating the cam model and running it on the image.
cam = EigenCAM(model, target_layers, use_cuda=False)
grayscale_cam = cam(tensor)[0, :, :]
cam_image = show_cam_on_image(img, grayscale_cam, use_rgb=True)
Image.fromarray(cam_image)
# #### We can also remove the heatmap data outside the bounding boxes and scale them inside every bounding box
def renormalize_cam_in_bounding_boxes(
boxes, colors, names, image_float_np, grayscale_cam
):
"""Normalize the CAM to be in the range [0, 1]
inside every bounding boxes, and zero outside of the bounding boxes."""
renormalized_cam = np.zeros(grayscale_cam.shape, dtype=np.float32)
for x1, y1, x2, y2 in boxes:
renormalized_cam[y1:y2, x1:x2] = scale_cam_image(
grayscale_cam[y1:y2, x1:x2].copy()
)
renormalized_cam = scale_cam_image(renormalized_cam)
eigencam_image_renormalized = show_cam_on_image(
image_float_np, renormalized_cam, use_rgb=True
)
image_with_bounding_boxes = draw_detections(
boxes, colors, names, eigencam_image_renormalized
)
return image_with_bounding_boxes
renormalized_cam_image = renormalize_cam_in_bounding_boxes(
boxes, colors, names, img, grayscale_cam
)
Image.fromarray(renormalized_cam_image)
Image.fromarray(np.hstack((rgb_img, cam_image, renormalized_cam_image)))
|
# Diabetic Retinopathy Detection
# ## Table of Content
# 1 - Introduction and Create Workspace
#
# 1.1 Import Packages
# 1.2 Create Classes and Function
#
# 2 - Pre-Training on Diabetic Retinopathy 2015 data
#
# 2.1 Import Data
# 2.2 Explore Data
# 2.3 Preprocess and Augment Data
# 2.4 - Using EFfienctNetB7 on Diabetic Retinopathy 2015 data
#
# 2.4.1 Setup Model and Choose Hyperparameters
# 2.4.2 Train Model on Diabetic Retinopathy 2015 data
# 2.4.3 Evaluation
#
#
# 3 - Training on Diabetic Retinopathy 2019 data
#
# 3.1 Import Data
# 3.2 Explore Data
# 3.3 Preprocess and Augment Data
# 3.4 - Load Pre-Trained EFfienctNetB7 trained on 2015 data
#
# 3.4.1 Setup Model and Choose Hyperparameters
# 3.4.2 Train Model on Diabetic Retinopathy 2019 data
# 3.4.3 Evaluation
#
#
# # 1 - Introducation and Create Workspace
# Diabetic retinopathy (DR) is one of the leading causes of vision loss. According to a recent study from International Diabetes Federation, the global prevalence of DR among the individuals with diabetes for the period from 2015 to 2019 was at more than 25%. According to the World Health Organization, more than 300 million people worldwide have diabetes, and the disease
# prevalence has been rising rapidly in developing countries.
# Early detection and treatment are crucial steps towards preventing DR. Currently, detecting DR is a time-consuming process. The screening procedure requires a trained clinical expert to examine the fundus photographs of the patient’s retina. This creates delays in diagnosis and treatment of the disease. Automated evaluation of retina photographs can speed up the efficiency and coverage of the DR screening programs. This is especially relevant for developing countries, which often lack qualified medical stuff to perform the diagnosis.
# The project aims at developing a deep learning model for predicting the severity of DR disease based on the patient’s retina photograph. Previous research has explored the usage of deep learning for detecting DR and concluded that convolutional neural networks (CNNs) have high potential in this task . The Asia Pacific Tele-Ophthalmology Society (APTOS) has launched two Kaggle competitions with a goal of promoting the use of deep learning for DR detection and boosting the development of automated detection systems.
# GitHub Repository.
# The project leverages two data sets: 1. main data set used for modeling and evaluation. This data set is provided by APTOS. It has been employed in the APTOS 2019 Blindness Detection competition on Kaggle and is available for the download at the competition website: https://www.kaggle.com/c/aptos2019-blindness-detection/data. The data set includes 3,662 labeled retina images of clinical patients. The images are taken using a fundus photography technique.
# 2. supplementary data set for pre-training. This data set features 35,126 retina images labeled by a clinician using the same scale as the main data set. The data set has been used in the 2015 Diabetic Retinopathy Detection competition and is available for the download at the corresponding website: https://www.kaggle.com/c/diabetic-retinopathy-detection/data.
# ## 1.1 Import Packages
# importing libraries
import numpy as np
import pandas as pd
from PIL import Image, ImageFile
ImageFile.LOAD_TRUNCATED_IMAGES = True
import cv2
from tqdm import tqdm_notebook as tqdm
from functools import partial
import scipy as sp
import random
import time
import sys
import os
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn import metrics
from sklearn.metrics import confusion_matrix
import torch
import torchvision
import torch.nn as nn
import torch.optim as optim
from torch.optim import lr_scheduler
from torchvision import transforms, models, datasets
from torch.utils.data import Dataset
from torch.autograd import Variable
from efficientnet_pytorch import EfficientNet
import warnings
warnings.filterwarnings("ignore")
#
# ## 1.2 Create Classes and Function
# seed function
def seed_everything(seed=23):
# tests
assert isinstance(seed, int), "seed has to be an integer"
# randomness
os.environ["PYTHONHASHSEED"] = str(seed)
random.seed(seed)
np.random.seed(seed)
torch.manual_seed(seed)
torch.cuda.manual_seed(seed)
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = False
image_size = 256
# IMAGE PREPROCESSING
def prepare_image(path, sigmaX=10, do_random_crop=False):
"""
Preprocess image
"""
# import image
image = cv2.imread(path)
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
# perform smart crops
image = crop_black(image, tol=7)
if do_random_crop == True:
image = random_crop(image, size=(0.9, 1))
# resize and color
image = cv2.resize(image, (int(image_size), int(image_size)))
image = cv2.addWeighted(image, 4, cv2.GaussianBlur(image, (0, 0), sigmaX), -4, 128)
# circular crop
image = circle_crop(image, sigmaX=sigmaX)
# convert to tensor
image = torch.tensor(image)
image = image.permute(2, 1, 0)
return image
# CROP FUNCTIONS
def crop_black(img, tol=7):
"""
Perform automatic crop of black areas
"""
if img.ndim == 2:
mask = img > tol
return img[np.ix_(mask.any(1), mask.any(0))]
elif img.ndim == 3:
gray_img = cv2.cvtColor(img, cv2.COLOR_RGB2GRAY)
mask = gray_img > tol
check_shape = img[:, :, 0][np.ix_(mask.any(1), mask.any(0))].shape[0]
if check_shape == 0:
return img
else:
img1 = img[:, :, 0][np.ix_(mask.any(1), mask.any(0))]
img2 = img[:, :, 1][np.ix_(mask.any(1), mask.any(0))]
img3 = img[:, :, 2][np.ix_(mask.any(1), mask.any(0))]
img = np.stack([img1, img2, img3], axis=-1)
return img
def circle_crop(img, sigmaX=10):
"""
Perform circular crop around image center
"""
height, width, depth = img.shape
largest_side = np.max((height, width))
img = cv2.resize(img, (largest_side, largest_side))
height, width, depth = img.shape
x = int(width / 2)
y = int(height / 2)
r = np.amin((x, y))
circle_img = np.zeros((height, width), np.uint8)
cv2.circle(circle_img, (x, y), int(r), 1, thickness=-1)
img = cv2.bitwise_and(img, img, mask=circle_img)
return img
def random_crop(img, size=(0.9, 1)):
"""
Random crop
"""
height, width, depth = img.shape
cut = 1 - random.uniform(size[0], size[1])
i = random.randint(0, int(cut * height))
j = random.randint(0, int(cut * width))
h = i + int((1 - cut) * height)
w = j + int((1 - cut) * width)
img = img[i:h, j:w, :]
return img
class EyeData(Dataset):
# initialize
def __init__(
self, data, directory, transform=None, do_random_crop=True, itype=".png"
):
self.data = data
self.directory = directory
self.transform = transform
self.do_random_crop = do_random_crop
self.itype = itype
# length
def __len__(self):
return len(self.data)
# get items
def __getitem__(self, idx):
img_name = os.path.join(
self.directory, self.data.loc[idx, "id_code"] + self.itype
)
image = prepare_image(img_name, do_random_crop=self.do_random_crop)
image = self.transform(image)
label = torch.tensor(self.data.loc[idx, "diagnosis"])
return {"image": image, "label": label}
class Data(Dataset):
# initialize
def __init__(
self, data, directory, transform=None, do_random_crop=True, itype=".png"
):
self.data = data
self.directory = directory
self.transform = transform
self.do_random_crop = do_random_crop
self.itype = itype
# length
def __len__(self):
return len(self.data)
# get items
def __getitem__(self, idx):
img_name = os.path.join(
self.directory, self.data.loc[idx, "id_code"] + self.itype
)
image = cv2.imread(img_name)
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
image = crop_black(image, tol=7)
image = cv2.resize(image, (int(image_size), int(image_size)))
image = circle_crop(image, sigmaX=10)
image = torch.tensor(image)
image = image.permute(2, 1, 0)
image = self.transform(image)
label = torch.tensor(self.data.loc[idx, "diagnosis"])
return {"image": image, "label": label}
def init_model(train=True, trn_layers=2, model_name="enet_b7"):
"""
Initialize the model
"""
### training mode
if train == True:
# load pre-trained model
model = EfficientNet.from_pretrained("efficientnet-b7", num_classes=5)
model.load_state_dict(
torch.load(
"../input/diabetic-retinopathy-pre-training/models/model_{}.bin".format(
model_name, 1
)
)
)
# freeze first layers
for child in list(model.children())[:-trn_layers]:
for param in child.parameters():
param.requires_grad = False
# inference mode
if train == False:
# load pre-trained model
model = EfficientNet.from_pretrained("efficientnet-b7", num_classes=5)
model.load_state_dict(
torch.load(
"../input/diabetic-retinopathy-pre-training/models/model_{}.bin".format(
model_name, 1
)
)
)
# freeze all layers
for param in model.parameters():
param.requires_grad = False
### return model
return model
# GPU CHECK
train_on_gpu = torch.cuda.is_available()
if not train_on_gpu:
print("CUDA is not available. Training on CPU...")
device = torch.device("cpu")
else:
print("CUDA is available. Training on GPU...")
device = torch.device("cuda:0")
# RANDOMNESS
seed = 23
seed_everything(seed)
#
# # 2 - Pre-Training on Diabetic Retinopathy 2015 data
# ## 2.1 Import Data
# import data
train = pd.read_csv("../input/diabetic-retinopathy-resized/trainLabels.csv")
train.columns = ["id_code", "diagnosis"]
test = pd.read_csv("../input/aptos2019-blindness-detection/train.csv")
# check shape
print(train.shape, test.shape)
print("-" * 15)
print(train["diagnosis"].value_counts())
print("-" * 15)
print(test["diagnosis"].value_counts())
#
# ## 2.2 Explore Data
# CLASS DISTRIBUTION
# plot
fig = plt.figure(figsize=(15, 5))
plt.hist(train["diagnosis"])
plt.title("Class Distribution")
plt.ylabel("Number of examples")
plt.xlabel("Diagnosis")
# transformations
sample_trans = transforms.Compose(
[
transforms.ToPILImage(),
transforms.ToTensor(),
]
)
sample = Data(
data=train.iloc[0:10],
directory="../input/diabetic-retinopathy-resized/resized_train/resized_train",
transform=sample_trans,
itype=".jpeg",
)
# data loader
sample_loader = torch.utils.data.DataLoader(
dataset=sample, batch_size=10, shuffle=False, num_workers=4
)
# display images
for batch_i, data in enumerate(sample_loader):
# extract data
inputs = data["image"]
labels = data["label"].view(-1, 1)
# create plot
fig = plt.figure(figsize=(15, 7))
for i in range(len(labels)):
ax = fig.add_subplot(2, int(len(labels) / 2), i + 1, xticks=[], yticks=[])
plt.imshow(inputs[i].numpy().transpose(1, 2, 0))
ax.set_title(labels.numpy()[i])
break
# IMAGE SIZES
# placeholder
image_stats = []
# import loop
for index, observation in tqdm(train.iterrows(), total=len(train)):
# import image
img = cv2.imread(
"../input/diabetic-retinopathy-resized/resized_train/resized_train/{}.jpeg".format(
observation["id_code"]
)
)
# compute stats
height, width, channels = img.shape
ratio = width / height
# save
image_stats.append(
np.array((observation["diagnosis"], height, width, channels, ratio))
)
# construct DF
image_stats = pd.DataFrame(image_stats)
image_stats.columns = ["diagnosis", "height", "width", "channels", "ratio"]
# IMAGE SIZE DISTRIBUTION
fig = plt.figure(figsize=(15, 5))
# width
plt.subplot(1, 3, 1)
plt.hist(image_stats["width"])
plt.title("(a) Image Width")
plt.ylabel("Number of examples")
plt.xlabel("Width")
# height
plt.subplot(1, 3, 2)
plt.hist(image_stats["height"])
plt.title("(b) Image Height")
plt.ylabel("Number of examples")
plt.xlabel("Height")
# ratio
plt.subplot(1, 3, 3)
plt.hist(image_stats["ratio"])
plt.title("(c) Aspect Ratio")
plt.ylabel("Number of examples")
plt.xlabel("Ratio")
#
# ## 2.3 Preprocess and Augment Training Data
# TRANSFORMATIONS
# parameters
batch_size = 16
image_size = 256
# train transformations
train_trans = transforms.Compose(
[
transforms.ToPILImage(),
transforms.RandomRotation((-360, 360)),
transforms.RandomHorizontalFlip(),
transforms.RandomVerticalFlip(),
transforms.ToTensor(),
]
)
# validation transformations
valid_trans = transforms.Compose(
[
transforms.ToPILImage(),
transforms.ToTensor(),
]
)
# test transformations
test_trans = valid_trans
# EXAMINE FIRST BATCH (TRAIN)
# get dataset
sample = EyeData(
data=train.iloc[0:10],
directory="../input/diabetic-retinopathy-resized/resized_train/resized_train",
transform=train_trans,
itype=".jpeg",
)
# data loader
sample_loader = torch.utils.data.DataLoader(
dataset=sample, batch_size=batch_size, shuffle=True, num_workers=4
)
# display images
for batch_i, data in enumerate(sample_loader):
# extract data
inputs = data["image"]
labels = data["label"].view(-1, 1)
# create plot
fig = plt.figure(figsize=(20, 10))
for i in range(len(labels)):
ax = fig.add_subplot(2, int(len(labels) / 2), i + 1, xticks=[], yticks=[])
plt.imshow(inputs[i].numpy().transpose(1, 2, 0))
ax.set_title(labels.numpy()[i])
break
# EXAMINE FIRST BATCH (TEST)
# get dataset
sample = EyeData(
data=test.iloc[0:10],
directory="../input/aptos2019-blindness-detection/train_images",
transform=test_trans,
itype=".png",
do_random_crop=False,
)
# data loader
sample_loader = torch.utils.data.DataLoader(
dataset=sample, batch_size=batch_size, shuffle=False, num_workers=4
)
# display images
for batch_i, data in enumerate(sample_loader):
# extract data
inputs = data["image"]
# create plot
fig = plt.figure(figsize=(20, 10))
for i in range(10):
ax = fig.add_subplot(2, int(10 / 2), i + 1, xticks=[], yticks=[])
plt.imshow(inputs[i].numpy().transpose(1, 2, 0))
break
#
# ## 2.4 - Using EFfienctNet for Transfer Learning
# ### 2.4.1 - Setup Model and Choose Hyperparameters
# MODEL ARCHITECTURE
# model name
model_name = "enet_b7"
# initialization function
def init_pre_model(train=True):
"""
Initialize the model
"""
### training mode
if train == True:
# load pre-trained model
model = EfficientNet.from_pretrained("efficientnet-b7", num_classes=5)
### inference mode
if train == False:
# load pre-trained model
model = EfficientNet.from_name("efficientnet-b7")
model._fc = nn.Linear(model._fc.in_features, 5)
# freeze layers
for param in model.parameters():
param.requires_grad = False
### return model
return model
# check architecture
model = init_pre_model()
print(model)
# VALIDATION SETTINGS
# placeholders
oof_preds = np.zeros((len(test), 5))
# timer
cv_start = time.time()
# PARAMETERS
# loss function
criterion = nn.CrossEntropyLoss()
# epochs
max_epochs = 15
early_stop = 5
# learning rates
eta = 1e-3
# scheduler
step = 5
gamma = 0.5
#
# ## 2.4.2 - Train Model on Diabetic Retinopathy 2015 data
# DATA PREPARATION
# load splits
data_train = train
data_valid = test
# create datasets
train_dataset = EyeData(
data=data_train,
directory="../input/diabetic-retinopathy-resized/resized_train/resized_train",
transform=train_trans,
itype=".jpeg",
)
valid_dataset = EyeData(
data=data_valid,
directory="../input/aptos2019-blindness-detection/train_images",
transform=valid_trans,
itype=".png",
)
# create data loaders
train_loader = torch.utils.data.DataLoader(
train_dataset, batch_size=batch_size, shuffle=True, num_workers=4
)
valid_loader = torch.utils.data.DataLoader(
valid_dataset, batch_size=batch_size, shuffle=False, num_workers=4
)
#
# ## 2.4.3 - Evaluation
# RECHECK PERFORMANCE
# rounding
oof_preds_round = oof_preds.argmax(axis=1)
coef = [0.5, 1.5, 2.5, 3.5]
for i, pred in enumerate(oof_preds_round):
if pred < coef[0]:
oof_preds_round[i] = 0
elif pred >= coef[0] and pred < coef[1]:
oof_preds_round[i] = 1
elif pred >= coef[1] and pred < coef[2]:
oof_preds_round[i] = 2
elif pred >= coef[2] and pred < coef[3]:
oof_preds_round[i] = 3
else:
oof_preds_round[i] = 4
# compute kappa
oof_loss = criterion(
torch.tensor(oof_preds), torch.tensor(test["diagnosis"]).view(-1).type(torch.long)
)
oof_kappa = metrics.cohen_kappa_score(
test["diagnosis"], oof_preds_round.astype("int"), weights="quadratic"
)
print("OOF loss = {:.4f}".format(oof_loss))
print("OOF kappa = {:.4f}".format(oof_kappa))
# CONFUSION MATRIX
# construct confusion matrix
cm = confusion_matrix(test["diagnosis"], oof_preds_round)
cm = cm.astype("float") / cm.sum(axis=1)[:, np.newaxis]
annot = np.around(cm, 2)
# plot matrix
fig, ax = plt.subplots(figsize=(10, 10))
sns.heatmap(cm, cmap="Blues", annot=annot, lw=0.5)
ax.set_xlabel("Prediction")
ax.set_ylabel("Ground Truth")
ax.set_aspect("equal")
from sklearn.metrics import classification_report
# Classification Report Test
print(
"\n Classification Report in Test: \n",
classification_report(test["diagnosis"], oof_preds_round),
)
#
# # 3 - Training on Diabetic Retinopathy 2019 data
#
# ## 3.1 Import Data
# import data
train = pd.read_csv("../input/aptos2019-blindness-detection/train.csv")
test = pd.read_csv("../input/aptos2019-blindness-detection/sample_submission.csv")
# check shape
print(train.shape, test.shape)
print("-" * 15)
print(train["diagnosis"].value_counts(normalize=True))
# MODELING EPOCHS
# placeholders
val_kappas = []
val_losses = []
trn_losses = []
bad_epochs = 0
# initialize and send to GPU
model = init_pre_model()
model = model.to(device)
# optimizer
optimizer = optim.Adam(model.parameters(), lr=eta)
scheduler = lr_scheduler.StepLR(optimizer, step_size=step, gamma=gamma)
# training and validation loop
for epoch in range(max_epochs):
### PREPARATION
# timer
epoch_start = time.time()
# reset losses
trn_loss = 0.0
val_loss = 0.0
# placeholders
fold_preds = np.zeros((len(data_valid), 5))
# TRAINING
# switch regime
model.train()
# loop through batches
for batch_i, data in enumerate(train_loader):
# extract inputs and labels
inputs = data["image"]
labels = data["label"].view(-1)
inputs = inputs.to(device, dtype=torch.float)
labels = labels.to(device, dtype=torch.long)
optimizer.zero_grad()
# forward and backward pass
with torch.set_grad_enabled(True):
preds = model(inputs)
loss = criterion(preds, labels)
loss.backward()
optimizer.step()
# compute loss
trn_loss += loss.item() * inputs.size(0)
# INFERENCE
# switch regime
model.eval()
# loop through batches
for batch_i, data in enumerate(valid_loader):
# extract inputs and labels
inputs = data["image"]
labels = data["label"].view(-1)
inputs = inputs.to(device, dtype=torch.float)
labels = labels.to(device, dtype=torch.long)
# compute predictions
with torch.set_grad_enabled(False):
preds = model(inputs).detach()
fold_preds[
batch_i * batch_size : (batch_i + 1) * batch_size, :
] = preds.cpu().numpy()
# compute loss
loss = criterion(preds, labels)
val_loss += loss.item() * inputs.size(0)
# save predictions
oof_preds = fold_preds
# scheduler step
scheduler.step()
# EVALUATION
# evaluate performance
fold_preds_round = fold_preds.argmax(axis=1)
val_kappa = metrics.cohen_kappa_score(
data_valid["diagnosis"], fold_preds_round.astype("int"), weights="quadratic"
)
# save perfoirmance values
val_kappas.append(val_kappa)
val_losses.append(val_loss / len(data_valid))
trn_losses.append(trn_loss / len(data_train))
# EARLY STOPPING
# display info
print(
"- epoch {}/{} | lr = {} | trn_loss = {:.4f} | val_loss = {:.4f} | val_kappa = {:.4f} | {:.2f} min".format(
epoch + 1,
max_epochs,
scheduler.get_lr()[len(scheduler.get_lr()) - 1],
trn_loss / len(data_train),
val_loss / len(data_valid),
val_kappa,
(time.time() - epoch_start) / 60,
)
)
# check if there is any improvement
if epoch > 0:
if val_kappas[epoch] < val_kappas[epoch - bad_epochs - 1]:
bad_epochs += 1
else:
bad_epochs = 0
# save model weights if improvement
if bad_epochs == 0:
oof_preds_best = oof_preds.copy()
torch.save(model.state_dict(), "models/model_{}.bin".format(model_name))
# break if early stop
if bad_epochs == early_stop:
print(
"Early stopping. Best results: loss = {:.4f}, kappa = {:.4f} (epoch {})".format(
np.min(val_losses),
val_kappas[np.argmin(val_losses)],
np.argmin(val_losses) + 1,
)
)
print("")
break
# break if max epochs
if epoch == (max_epochs - 1):
print(
"Did not met early stopping. Best results: loss = {:.4f}, kappa = {:.4f} (epoch {})".format(
np.min(val_losses),
val_kappas[np.argmin(val_losses)],
np.argmin(val_losses) + 1,
)
)
print("")
break
# load best predictions
oof_preds = oof_preds_best
# print performance
print("")
print("Finished in {:.2f} minutes".format((time.time() - cv_start) / 60))
#
# ## 3.2 Explore Data
# CLASS DISTRIBUTION
# plot
fig = plt.figure(figsize=(15, 5))
plt.hist(train["diagnosis"])
plt.title("Class Distribution")
plt.ylabel("Number of examples")
plt.xlabel("Diagnosis")
# transformations
sample_trans = transforms.Compose(
[
transforms.ToPILImage(),
transforms.ToTensor(),
]
)
sample = Data(
data=train.iloc[0:10],
directory="../input/aptos2019-blindness-detection/train_images",
transform=sample_trans,
itype=".png",
)
# data loader
sample_loader = torch.utils.data.DataLoader(
dataset=sample, batch_size=10, shuffle=False, num_workers=4
)
# display images
for batch_i, data in enumerate(sample_loader):
# extract data
inputs = data["image"]
labels = data["label"].view(-1, 1)
# create plot
fig = plt.figure(figsize=(15, 7))
for i in range(len(labels)):
ax = fig.add_subplot(2, int(len(labels) / 2), i + 1, xticks=[], yticks=[])
plt.imshow(inputs[i].numpy().transpose(1, 2, 0))
ax.set_title(labels.numpy()[i])
break
# IMAGE SIZES
# placeholder
image_stats = []
# import loop
for index, observation in tqdm(train.iterrows(), total=len(train)):
# import image
img = cv2.imread(
"../input/aptos2019-blindness-detection/train_images/{}.png".format(
observation["id_code"]
)
)
# compute stats
height, width, channels = img.shape
ratio = width / height
# save
image_stats.append(
np.array((observation["diagnosis"], height, width, channels, ratio))
)
# construct DF
image_stats = pd.DataFrame(image_stats)
image_stats.columns = ["diagnosis", "height", "width", "channels", "ratio"]
# IMAGE SIZE DISTRIBUTION
fig = plt.figure(figsize=(15, 5))
# width
plt.subplot(1, 3, 1)
plt.hist(image_stats["width"])
plt.title("(a) Image Width")
plt.ylabel("Number of examples")
plt.xlabel("Width")
# height
plt.subplot(1, 3, 2)
plt.hist(image_stats["height"])
plt.title("(b) Image Height")
plt.ylabel("Number of examples")
plt.xlabel("Height")
# ratio
plt.subplot(1, 3, 3)
plt.hist(image_stats["ratio"])
plt.title("(c) Aspect Ratio")
plt.ylabel("Number of examples")
plt.xlabel("Ratio")
#
# ## 3.3 Preprocess and Augment Data
# TRANSFORMATIONS
# parameters
batch_size = 25
image_size = 256
# train transformations
train_trans = transforms.Compose(
[
transforms.ToPILImage(),
transforms.RandomRotation((-360, 360)),
transforms.RandomHorizontalFlip(),
transforms.RandomVerticalFlip(),
transforms.ToTensor(),
]
)
# valid transformations
valid_trans = transforms.Compose(
[
transforms.ToPILImage(),
transforms.ToTensor(),
]
)
# test transformations
test_trans = valid_trans
# EXAMINE FIRST BATCH (TRAIN)
# get dataset
sample = EyeData(
data=train.iloc[0:10],
directory="../input/aptos2019-blindness-detection/train_images",
transform=train_trans,
itype=".png",
)
# data loader
sample_loader = torch.utils.data.DataLoader(
dataset=sample, batch_size=batch_size, shuffle=True, num_workers=4
)
# display images
for batch_i, data in enumerate(sample_loader):
# extract data
inputs = data["image"]
labels = data["label"].view(-1, 1)
# create plot
fig = plt.figure(figsize=(20, 10))
for i in range(len(labels)):
ax = fig.add_subplot(2, int(len(labels) / 2), i + 1, xticks=[], yticks=[])
plt.imshow(inputs[i].numpy().transpose(1, 2, 0))
ax.set_title(labels.numpy()[i])
break
# EXAMINE FIRST BATCH (TEST)
# get dataset
sample = EyeData(
data=test.iloc[0:10],
directory="../input/aptos2019-blindness-detection/test_images",
transform=test_trans,
itype=".png",
do_random_crop=False,
)
# data loader
sample_loader = torch.utils.data.DataLoader(
dataset=sample, batch_size=batch_size, shuffle=False, num_workers=4
)
# display images
for batch_i, data in enumerate(sample_loader):
# extract data
inputs = data["image"]
# create plot
fig = plt.figure(figsize=(20, 10))
for i in range(10):
ax = fig.add_subplot(2, int(10 / 2), i + 1, xticks=[], yticks=[])
plt.imshow(inputs[i].numpy().transpose(1, 2, 0))
break
#
# ## 3.4 - Load Pre-Trained EFfienctNetB7 trained on 2015 data
# ### 3.4.1 Setup Model and Choose Hyperparameters
# MODEL ARCHITECTURE
# model name
model_name = "enet_b7"
# check architecture
model = init_model(model_name=model_name)
print(model)
# VALIDATION SETTINGS
from sklearn.model_selection import KFold, StratifiedKFold
# no. folds
num_folds = 4
# creating splits
skf = StratifiedKFold(n_splits=num_folds, shuffle=True, random_state=seed)
splits = list(skf.split(train["id_code"], train["diagnosis"]))
# placeholders
oof_preds = np.zeros((len(train), 1))
# timer
cv_start = time.time()
# PARAMETERS
# loss function
criterion = nn.CrossEntropyLoss()
# epochs
max_epochs = 15
early_stop = 5
# learning rates
eta = 1e-3
# scheduler
step = 5
gamma = 0.5
#
# ## 3.4.2 Train Model on Diabetic Retinopathy 2019 dat
# CROSS-VALIDATION LOOP
for fold in tqdm(range(num_folds)):
# DATA PREPARATION
# display information
print("-" * 30)
print("FOLD {}/{}".format(fold + 1, num_folds))
print("-" * 30)
# load splits
data_train = train.iloc[splits[fold][0]].reset_index(drop=True)
data_valid = train.iloc[splits[fold][1]].reset_index(drop=True)
# create datasets
train_dataset = EyeData(
data=data_train,
directory="../input/aptos2019-blindness-detection/train_images",
transform=train_trans,
itype=".png",
)
valid_dataset = EyeData(
data=data_valid,
directory="../input/aptos2019-blindness-detection/train_images",
transform=valid_trans,
itype=".png",
)
# create data loaders
train_loader = torch.utils.data.DataLoader(
train_dataset, batch_size=batch_size, shuffle=True, num_workers=4
)
valid_loader = torch.utils.data.DataLoader(
valid_dataset, batch_size=batch_size, shuffle=False, num_workers=4
)
# MODEL PREPARATION
# placeholders
val_kappas = []
val_losses = []
trn_losses = []
bad_epochs = 0
# load best OOF predictions
if fold > 0:
oof_preds = oof_preds_best.copy()
# initialize and send to GPU
model = init_model(train=True)
model = model.to(device)
# optimizer
optimizer = optim.Adam(model._fc.parameters(), lr=eta)
scheduler = lr_scheduler.StepLR(optimizer, step_size=step, gamma=gamma)
# TRAINING AND VALIDATION LOOP
for epoch in range(max_epochs):
## PREPARATION
# timer
epoch_start = time.time()
# reset losses
trn_loss = 0.0
val_loss = 0.0
# placeholders
fold_preds = np.zeros((len(data_valid), 1))
# TRAINING
# switch regime
model.train()
# loop through batches
for batch_i, data in enumerate(train_loader):
# extract inputs and labels
inputs = data["image"]
labels = data["label"].view(-1)
inputs = inputs.to(device, dtype=torch.float)
labels = labels.to(device, dtype=torch.long)
optimizer.zero_grad()
# forward and backward pass
with torch.set_grad_enabled(True):
preds = model(inputs)
loss = criterion(preds, labels)
loss.backward()
optimizer.step()
# compute loss
trn_loss += loss.item() * inputs.size(0)
# INFERENCE
# initialize
model.eval()
# loop through batches
for batch_i, data in enumerate(valid_loader):
# extract inputs and labels
inputs = data["image"]
labels = data["label"].view(-1)
inputs = inputs.to(device, dtype=torch.float)
labels = labels.to(device, dtype=torch.long)
# compute predictions
with torch.set_grad_enabled(False):
preds = model(inputs).detach()
_, class_preds = preds.topk(1)
fold_preds[
batch_i * batch_size : (batch_i + 1) * batch_size, :
] = class_preds.cpu().numpy()
# compute loss
loss = criterion(preds, labels)
val_loss += loss.item() * inputs.size(0)
# save predictions
oof_preds[splits[fold][1]] = fold_preds
# scheduler step
scheduler.step()
# EVALUATION
# evaluate performance
fold_preds_round = fold_preds
val_kappa = metrics.cohen_kappa_score(
data_valid["diagnosis"], fold_preds_round.astype("int"), weights="quadratic"
)
# save perfoirmance values
val_kappas.append(val_kappa)
val_losses.append(val_loss / len(data_valid))
trn_losses.append(trn_loss / len(data_train))
# EARLY STOPPING
# display info
print(
"- epoch {}/{} | lr = {} | trn_loss = {:.4f} | val_loss = {:.4f} | val_kappa = {:.4f} | {:.2f} min".format(
epoch + 1,
max_epochs,
scheduler.get_lr()[len(scheduler.get_lr()) - 1],
trn_loss / len(data_train),
val_loss / len(data_valid),
val_kappa,
(time.time() - epoch_start) / 60,
)
)
# check if there is any improvement
if epoch > 0:
if val_kappas[epoch] < val_kappas[epoch - bad_epochs - 1]:
bad_epochs += 1
else:
bad_epochs = 0
# save model weights if improvement
if bad_epochs == 0:
oof_preds_best = oof_preds.copy()
torch.save(
model.state_dict(),
"models/model_{}_fold{}.bin".format(model_name, fold + 1),
)
# break if early stop
if bad_epochs == early_stop:
print(
"Early stopping. Best results: loss = {:.4f}, kappa = {:.4f} (epoch {})".format(
np.min(val_losses),
val_kappas[np.argmin(val_losses)],
np.argmin(val_losses) + 1,
)
)
print("")
break
# break if max epochs
if epoch == (max_epochs - 1):
print(
"Did not meet early stopping. Best results: loss = {:.4f}, kappa = {:.4f} (epoch {})".format(
np.min(val_losses),
val_kappas[np.argmin(val_losses)],
np.argmin(val_losses) + 1,
)
)
print("")
break
# load best predictions
oof_preds = oof_preds_best
# print performance
print("")
print("Finished in {:.2f} minutes".format((time.time() - cv_start) / 60))
#
# ## 3.4.3 - Evaluation
#
# PLOT LOSS AND KAPPA DYNAMICS
sns.set()
# plot size
fig = plt.figure(figsize=(15, 5))
# plot loss dynamics
plt.subplot(1, 2, 1)
plt.plot(trn_losses, "red", label="Training")
plt.plot(val_losses, "green", label="Validation")
plt.xlabel("Epoch")
plt.ylabel("Loss")
plt.legend()
# plot kappa dynamics
plt.subplot(1, 2, 2)
plt.plot(val_kappas, "blue", label="Kappa")
plt.xlabel("Epoch")
plt.ylabel("Kappa")
plt.legend()
# RECHECK PERFORMANCE
# evaluate performance
oof_preds_round = oof_preds.copy()
oof_kappa = metrics.cohen_kappa_score(
train["diagnosis"], oof_preds_round.astype("int"), weights="quadratic"
)
print("OOF kappa = {:.4f}".format(oof_kappa))
# CONFUSION MATRIX
# construct confusion matrx
cm = confusion_matrix(train["diagnosis"], oof_preds_round)
cm = cm.astype("float") / cm.sum(axis=1)[:, np.newaxis]
annot = np.around(cm, 2)
# plot matrix
fig, ax = plt.subplots(figsize=(8, 6))
sns.heatmap(cm, cmap="Blues", annot=annot, lw=0.5)
ax.set_xlabel("Prediction")
ax.set_ylabel("Ground Truth")
ax.set_aspect("equal")
from sklearn.metrics import classification_report
# Classification Report Test
print(
"\n Classification Report in Test: \n",
classification_report(train["diagnosis"], oof_preds_round),
)
# PLOT LOSS AND KAPPA DYNAMICS
sns.set()
# plot size
fig = plt.figure(figsize=(15, 5))
# plot loss dynamics
plt.subplot(1, 2, 1)
plt.plot(trn_losses, "red", label="Training")
plt.plot(val_losses, "green", label="Validation")
plt.xlabel("Epoch")
plt.ylabel("Loss")
plt.legend()
# plot kappa dynamics
plt.subplot(1, 2, 2)
plt.plot(val_kappas, "blue", label="Kappa")
plt.xlabel("Epoch")
plt.ylabel("Kappa")
plt.legend()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
# # Homework 5
# First we make a list of a range of angles. Feel free to print 'theta' to see what it looks like
N = 121 # Want a lot of points. I made a point for every half angle between 30 and 90 degrees
theta = np.linspace(30, 90, N)
# Next we define our function. Where do we get it? Do you understand what it means?
def v(theta, x, g, h, H):
radians = theta * np.pi / 180 # np.cos/tan take radian values
return (x / np.cos(radians)) * np.sqrt(
(g / 2) / (x * np.tan(radians) + h - H)
) # maybe write this equation on scratch paper to see what it looks like
# Defining our constants
x = 15 # ft Distance to hoop
g = 32.174 # ft/s^2
h = 6 # ft Height of person
H = 10 # ft Height of hoop
rim_radius = 0.738 # ft
ball_radius = 0.398 # ft
import matplotlib.pyplot as plt
plt.plot(theta, v(theta, x, g, h, H), label="perfect")
plt.plot(theta, v(theta, x - rim_radius + ball_radius, g, h, H), label="min")
plt.plot(theta, v(theta, x + rim_radius - ball_radius, g, h, H), label="max")
plt.ylim(24.3, 27)
plt.xlim(38, 68)
# Label your own axes with units
plt.legend()
plt.grid() # to better see the plot
plt.show()
|
# # Predicting Airbnb Prices
# # Importing Data
import pandas as pd
import numpy as np
import matplotlib as mpl
import matplotlib.pyplot as plt
import seaborn as sns
import os
from sklearn import set_config
plt.style.use("ggplot")
pd.set_option("display.max_columns", 100)
# set_config(transform_output="pandas") #doesn't work here :(
os.chdir("/kaggle/input/airbnb-cleaned-europe-dataset")
df = pd.read_csv("Aemf1.csv")
df
df.info()
# # EDA
# ## Heat map
# Correlations between variables are visualized in the heat map below.
# - Cleaner rooms are rated higher.
# - Attractions and restraunts are found in the same locations.
# - City centers are further from the metro.
# Heat map
# optimizng range for color scale
min = df.corr().min().min()
max = df.corr()[df.corr() != 1].max().max()
# thresholding selected correlations
df_corr = df.corr()[np.absolute(df.corr()) > 0.3]
# Mask for selecting only bottom triangle
mask = np.triu(df_corr)
with plt.style.context("default"):
sns.heatmap(df_corr, vmin=min, vmax=max, mask=mask)
# ## Pair plots
# Distributions and relationships are plotted for selected parameters below:
# raw data
sns.pairplot(
df[
[
"City Center (km)",
"Metro Distance (km)",
"Attraction Index",
"Restraunt Index",
"Price",
]
][df["Price"] < 2000],
kind="hist",
corner=True,
)
# We see that these features are better represented in log space:
# rescaled data
df_trial = pd.DataFrame()
df_trial["City Center (km)"] = np.log(df["City Center (km)"])
df_trial["Metro Distance (km)"] = np.log(df["Metro Distance (km)"])
df_trial["Attraction Index"] = np.log(df["Attraction Index"])
df_trial["Restraunt Index"] = np.log(df["Restraunt Index"])
df_trial["Price"] = np.log(df["Price"])
sns.pairplot(df_trial[df_trial["Price"] < 2000], kind="hist", corner=True)
# ## RBF Definition
# We can replace features with an apparent radial price distribution with an radial basis function.
# Metro, city center and restraunt index appear to have radial price distributions
from sklearn.metrics.pairwise import rbf_kernel
rbf_metro = rbf_kernel(df_trial[["Metro Distance (km)"]], [[-0.5]], gamma=0.25)
rbf_city = rbf_kernel(df_trial[["City Center (km)"]], [[0.8]], gamma=2)
rbf_res = rbf_kernel(df_trial[["Restraunt Index"]], [[6]], gamma=3)
# visualizing metro rbf function
fig, ax1 = plt.subplots(1)
plt.bar(df_trial["Metro Distance (km)"], df["Price"])
plt.xlabel("Log Metro Distance (km)")
plt.ylabel("Price")
ax2 = ax1.twinx()
ax2.scatter(df_trial["Metro Distance (km)"], rbf_metro, color="k", s=0.5)
ax2.set_ylim([0, 3])
ax2.set_ylabel("Price rbf")
plt.show()
# visualizing city rbf function
fig, ax1 = plt.subplots(1)
plt.bar(df_trial["City Center (km)"], df["Price"])
ax2 = ax1.twinx()
ax2.scatter(df_trial["City Center (km)"], rbf_city, color="k", s=0.5)
ax2.set_ylim([0, 1])
plt.show()
# visualizing city rbf function
fig, ax1 = plt.subplots(1)
plt.bar(df_trial["Restraunt Index"], df["Price"])
ax2 = ax1.twinx()
ax2.scatter(df_trial["Restraunt Index"], rbf_res, color="k", s=0.5)
ax2.set_ylim([0, 1])
plt.show()
# updating df with rbf functions
df["rbf_metro"] = rbf_metro
df["rbf_city"] = rbf_city
df["rbf_res"] = rbf_res
# ## Visualizing categorical data
fig, ax = plt.subplots(1)
fig.set_size_inches(8, 4)
sns.boxplot(data=df, x="City", y="Price", showfliers=False)
ax.set_ylim([0, 1300])
plt.show()
sns.boxplot(data=df, x="Superhost", y="Price", showfliers=False)
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(8, 3))
plt.sca(ax1)
sns.boxplot(data=df, x="Room Type", y="Price", showfliers=False)
plt.sca(ax2)
sns.boxplot(data=df, x="Shared Room", y="Price", showfliers=False)
plt.tight_layout()
sns.boxplot(data=df, x="Day", y="Price", showfliers=False)
sns.boxplot(
data=df, x="Person Capacity", y="Price", showfliers=False
) # ,scatter_kws={'alpha':0.05},line_kws={"color": "black"})
plt.ylim([0, 1000])
plt.show()
# ## Visualizing consumer rankings
# sns.regplot(data = df[df["Price"]<2000], x='Cleanliness Rating', y='Price',scatter=True,scatter_kws={'alpha':0.05},line_kws={"color": "black"});
sns.jointplot(
x=df["Cleanliness Rating"],
y=np.log(df["Price"]),
kind="reg",
scatter_kws={"alpha": 0.05},
line_kws={"color": "black"},
)
plt.show()
# sns.regplot(data = df[df['Price']<2000], x='Guest Satisfaction', y='Price',scatter_kws={'alpha':0.05},line_kws={"color": "black"})
sns.jointplot(
x=df["Guest Satisfaction"],
y=np.log(df["Price"]),
kind="reg",
scatter_kws={"alpha": 0.05},
line_kws={"color": "black"},
)
plt.show()
# # Cleaning Data
# Three preprocessing transformations are defined for selecting different subsets of input features.
from sklearn.compose import ColumnTransformer
from sklearn.preprocessing import FunctionTransformer
from sklearn.preprocessing import OneHotEncoder
from sklearn.preprocessing import MinMaxScaler
from sklearn.base import BaseEstimator, TransformerMixin
from sklearn.pipeline import make_pipeline
from sklearn.model_selection import train_test_split
# defining functions for column transformer
cat_encoder = OneHotEncoder()
log_pipeline = make_pipeline(
FunctionTransformer(np.log), MinMaxScaler() # , inverse_func=np.exp),
)
def day_2_num(X):
return X == "Weekend"
def day_pipeline():
return make_pipeline(FunctionTransformer(day_2_num))
day_pipe = day_pipeline()
# defining standard column transformer
preprocessing = ColumnTransformer(
[
("day", day_pipe, ["Day"]),
(
"drop",
"drop",
[
"Normalised Attraction Index",
"Normalised Restraunt Index",
"rbf_metro",
"rbf_city",
"rbf_res",
],
),
(
"pass",
"passthrough",
["Private Room", "Shared Room", "Superhost", "Business", "Multiple Rooms"],
),
(
"maxscale",
MinMaxScaler(),
["Cleanliness Rating", "Bedrooms", "Guest Satisfaction"],
),
(
"log",
log_pipeline,
[
"Attraction Index",
"City Center (km)",
"Metro Distance (km)",
"Restraunt Index",
],
),
("cat", cat_encoder, ["Room Type", "City"]),
]
)
# column transformer with rbf functions instead of metro, city, and restraunts
preprocessing_rbf = ColumnTransformer(
[
("day", day_pipe, ["Day"]),
(
"drop",
"drop",
[
"Normalised Attraction Index",
"Normalised Restraunt Index",
"Metro Distance (km)",
"City Center (km)",
"Restraunt Index",
],
),
(
"pass",
"passthrough",
["Private Room", "Shared Room", "Superhost", "Business", "Multiple Rooms"],
),
(
"maxscale",
MinMaxScaler(),
["Cleanliness Rating", "Bedrooms", "Guest Satisfaction"],
),
("log", log_pipeline, ["Attraction Index"]),
("pass2", "passthrough", ["rbf_metro", "rbf_city", "rbf_res"]),
("cat", cat_encoder, ["Room Type", "City"]),
]
)
# column transformer with given normalized features
preprocessing_norm = ColumnTransformer(
[
("day", day_pipe, ["Day"]),
(
"drop",
"drop",
["Attraction Index", "Restraunt Index", "rbf_metro", "rbf_city", "rbf_res"],
),
(
"pass",
"passthrough",
["Private Room", "Shared Room", "Superhost", "Business", "Multiple Rooms"],
),
(
"maxscale",
MinMaxScaler(),
["Cleanliness Rating", "Bedrooms", "Guest Satisfaction"],
),
("pass2", "passthrough", ["Normalised Attraction Index"]),
("log", log_pipeline, ["City Center (km)", "Metro Distance (km)"]),
("pass3", "passthrough", ["Normalised Restraunt Index"]),
("cat", cat_encoder, ["Room Type", "City"]),
]
)
# naming output columns
names = pd.Series(
[
"Weekend",
"Private Room",
"Shared Room",
"Superhost",
"Business",
"Multiple Rooms",
"Cleanliness Rating",
"Bedrooms",
"Guest Satisfaction",
"Attraction Index",
"City Center",
"Metro Distance",
"Restraunt Index",
"Private room",
"Entire home/apt",
"Shared room",
"Amsterdam",
"Athens",
"Barcelona",
"Berlin",
"Budapest",
"Lisbon",
"Paris",
"Rome",
"Vienna",
]
)
# transforming data
df_processed = pd.DataFrame(preprocessing.fit_transform(df), columns=names)
# splitting data into test and train sets
y = df["Price"].copy()
X = df.drop(columns={"Price"})
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=10
)
# # Predicing Price
# importing packages
from sklearn.linear_model import LinearRegression, Ridge, ElasticNet
from sklearn.neighbors import KNeighborsRegressor
from sklearn.svm import SVR
from sklearn.ensemble import (
RandomForestRegressor,
ExtraTreesRegressor,
AdaBoostRegressor,
GradientBoostingRegressor,
)
from sklearn.metrics import mean_squared_error
import xgboost as xgb
from sklearn.decomposition import PCA
# ## Sampling regression models
# Common regression models were evaluated with six variations:
# 1. No rerescaling or feature engineering
# 2. Price target transformed to log-space
# 3. Dimensionality rediction via PCA
# 4. Feature engineering via replacement with RBF
# 5. Provided normalized resraunt and attraction indexes used
# 5. Combining variations 2-4
# unscaled y
np.random.seed(10)
models = [
Ridge(),
ElasticNet(),
SVR(),
KNeighborsRegressor(),
RandomForestRegressor(),
ExtraTreesRegressor(),
AdaBoostRegressor(),
GradientBoostingRegressor(),
xgb.XGBRegressor(),
]
model_names = [
"Ridge",
"ElasticNet",
"SVR",
"K-nearest neighbors",
"Random Forest",
"Extra Trees",
"Adaptive Boosting",
"Gradient Boosting",
"XGBoost",
]
error = []
for model in models:
sample_pipe = make_pipeline(preprocessing, model)
sample_pipe.fit(X_train, y_train)
y_pred = sample_pipe.predict(X_test)
error.append(mean_squared_error(y_test, y_pred))
# scaled y
scaled_error = []
for model in models:
scaled_pipe = make_pipeline(preprocessing, model)
scaled_pipe.fit(X_train, np.log(y_train))
y_pred = scaled_pipe.predict(X_test)
y_pred = np.exp(y_pred)
scaled_error.append(mean_squared_error(y_test, y_pred))
# with dimensionality reduction
pca = PCA(n_components=0.95)
pca_error = []
for model in models:
pca_pipe = make_pipeline(preprocessing, pca, model)
pca_pipe.fit(X_train, y_train)
y_pred = pca_pipe.predict(X_test)
pca_error.append(mean_squared_error(y_test, y_pred))
# with rbf
rbf_error = []
for model in models:
rbf_pipe = make_pipeline(preprocessing_rbf, model)
rbf_pipe.fit(X_train, y_train)
y_pred = rbf_pipe.predict(X_test)
rbf_error.append(mean_squared_error(y_test, y_pred))
# with normalized features
norm_error = []
for model in models:
norm_pipe = make_pipeline(preprocessing_norm, model)
norm_pipe.fit(X_train, y_train)
y_pred = norm_pipe.predict(X_test)
norm_error.append(mean_squared_error(y_test, y_pred))
# combination of all
combo_error = []
for model in models:
combo_pipe = make_pipeline(preprocessing_rbf, pca, model)
combo_pipe.fit(X_train, np.log(y_train))
y_pred = combo_pipe.predict(X_test)
y_pred = np.exp(y_pred)
combo_error.append(mean_squared_error(y_test, y_pred))
sample_results = pd.DataFrame(
[error, scaled_error, pca_error, rbf_error, norm_error, combo_error],
index=[
"Unscaled",
"Scaled",
"Reduced Dimensions",
"RBF Features",
"Normalized Features",
"Combination",
],
columns=model_names,
).T
sample_results.plot(kind="barh")
# plt.axvline((sum(error)+sum(scaled_error)+sum(pca_error))/(len(error)+len(scaled_error)+len(pca_error)), color='k', linestyle='--')
plt.title("Classifier RMSE")
plt.xlim([0, 250000])
plt.legend(loc=4, face_color="white")
plt.show()
# The top performing classifier was the random forest classifier with an unscaled price target.
# ## Optimizing the random forest regressor
from sklearn.model_selection import RandomizedSearchCV
from scipy.stats import loguniform, expon, uniform, randint
rf_pipeline = make_pipeline(
preprocessing, RandomForestRegressor(random_state=10, n_jobs=-1)
)
rf_params = {
"randomforestregressor__n_estimators": randint(70, 110),
"randomforestregressor__max_depth": randint(1, 10),
"randomforestregressor__min_weight_fraction_leaf": uniform(0, 0.1),
"randomforestregressor__max_features": ["auto", "sqrt", "log2"],
"randomforestregressor__max_leaf_nodes": randint(1, 10),
#'randomforestregressor__bootstrap':[True,False]
}
rf_search = RandomizedSearchCV(
rf_pipeline,
param_distributions=rf_params,
n_iter=10000,
cv=5,
scoring="neg_mean_squared_error",
random_state=15,
)
rf_search.fit(X_train, y_train)
score = rf_search.best_score_
params = rf_search.best_params_
score, params
baseline = error[5]
improvement = (baseline + rf_search.best_score_) / baseline * 100
baseline, improvement
# 23% improvement with bootstrap true and n_est = 85
from skopt import BayesSearchCV
from skopt.space import Real, Categorical, Integer
from skopt.plots import plot_objective
rf_pipeline = make_pipeline(
preprocessing, RandomForestRegressor(random_state=10, n_jobs=-1)
)
rf_search = {
"randomforestregressor__n_estimators": (1, 100, "randint"),
"randomforestregressor__max_depth": (1, 10, "randint"),
"randomforestregressor__min_weight_fraction_leaf": (0, 0.1, "uniform"),
#'randomforestregressor__max_features':['auto','sqrt','log2'],
"randomforestregressor__max_leaf_nodes": (1, 10, "randint"),
#'randomforestregressor__bootstrap':[True,False]
}
opt = BayesSearchCV(rf_pipeline, [(rf_search, 40)], cv=3)
opt.fit(X_train, y_train)
print("val. score: %s" % opt.best_score_)
print("test score: %s" % opt.score(X_test, y_test))
print("best params: %s" % str(opt.best_params_))
|
# * The images provided for this competition are not classified in folders, based on classes, provided as whole in train and test folder.
# * In this notebook we would be classifying and adding images to their respective class folders i.e either 'Benign' or 'Malignant' for the train images dataset.
# * This would help in the training of model as tensorflow requires image folder to train on image data.
# * We would need train.csv folder for classifying images.
import os
import shutil
import random
import re
import math
import time
import pandas as pd
import numpy as np
from os import listdir
from os.path import isfile, join
# Setting file paths for our notebook:
base_path = r"../input/siim-isic-melanoma-classification/jpeg"
train_dir = r"../input/siim-isic-melanoma-classification/jpeg/train"
test_dir = r"../input/siim-isic-melanoma-classification/jpeg/test"
img_stats_path = r"../input/siim-isic-melanoma-classification"
# Loading train and test data.
train = pd.read_csv(os.path.join(base_path, "train.csv"))
test = pd.read_csv(os.path.join(base_path, "test.csv"))
sample = pd.read_csv(os.path.join(base_path, "sample_submission.csv"))
# List containing all the names of images
train_images = [f for f in listdir(train_dir) if isfile(join(train_dir, f))]
test_images = [f for f in listdir(test_dir) if isfile(join(test_dir, f))]
# Labels for the image names
train_images_labels = []
for i in range(train.shape[0]):
train_images_labels.append(train["target"].iloc[i])
# Create folders with class names
src = r"../input/siim-isic-melanoma-classification"
# Create new train_class folder, in that benign n malignant folders
benign = r"../input/siim-isic-melanoma-classification\train_class\benign"
malignant = r"../input/siim-isic-melanoma-classification\train_class\malignant"
# Code for moving images from train folder to respective benign or malignant folder
for i in range(train.shape[0]):
if train["target"].iloc[i] == 0:
shutil.move(src + "\\" + train_images[i], benign + "\\" + train_images[i])
else:
shutil.move(src + "\\" + train_images[i], malignant + "\\" + train_images[i])
|
# # Imbalanced classification problem with anonimized data
# New Bulgarian University semester I project
# 1. Do an explarotary data analysis;
# 2. Reduce data dimensionality;
# 3. Acquire data from categorical features;
# 4. Scale the data;
# 5. Use logistic regression and SVM to find the best model available, while varying the hyperparameters via GridSearchCV or other methods. Use different metrics for the results;
# 6. *optional* Try solving the problem via XGBoost;
# 7. Test the best model on the testing dataset.
# # Exploratory Data Analysis
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.feature_selection import RFE
from sklearn.linear_model import LogisticRegression
from sklearn.preprocessing import (
StandardScaler,
MinMaxScaler,
MaxAbsScaler,
RobustScaler,
QuantileTransformer,
PowerTransformer,
)
from sklearn.metrics import (
accuracy_score,
precision_score,
recall_score,
f1_score,
roc_auc_score,
confusion_matrix,
)
from statsmodels.stats.outliers_influence import variance_inflation_factor
from sklearn.decomposition import PCA
from sklearn.model_selection import train_test_split
from sklearn.model_selection import (
KFold,
RepeatedKFold,
StratifiedKFold,
RandomizedSearchCV,
cross_val_score,
train_test_split,
TimeSeriesSplit,
GridSearchCV,
)
from numpy import mean
from numpy import std
from lightgbm import LGBMClassifier
import xgboost as xgb
from xgboost import XGBClassifier
from sklearn.svm import SVC
from imblearn.over_sampling import SMOTE
from sklearn.utils import resample
from scipy.stats import reciprocal, uniform
pd.options.display.max_rows = 999
pd.options.display.max_columns = 999
df_train = pd.read_csv("X_train.csv", encoding='"Windows-1251"')
df_test = pd.read_csv("X_test.csv", encoding='"Windows-1251"')
df_test.shape
df_train.shape
df_test["test_flag"] = 1
df_train["test_flag"] = 0
len(df_test[df_test["Bad_Flag"] == 1]) / len(df_test[df_test["Bad_Flag"] == 0])
len(df_test[df_test["Bad_Flag"] == 0])
len(df_train[df_train["Bad_Flag"] == 1]) / len(df_train[df_train["Bad_Flag"] == 0])
len(df_train[df_train["Bad_Flag"] == 0])
# The training and testing datasets will be concatenated only for the sake of data cleaning and feature engineering. The 'test_flag' was created in order to be able to separate the datasets after those steps. Care will be taken not to have any information leak from the testing dataset.
df_train = pd.concat([df_train, df_test])
df_train.shape
df_train.describe()
df_train.isnull().sum().sort_values(ascending=False)[:10]
df_train.x255[:10]
# It seems that x255 is a date type of feature. It will be transformed in the following section.
# # Data cleaning and feature engineering
df_train.x255 = pd.to_datetime(df_train.x255)
df_train.x255.mean()
df_train.x255 = df_train.x255.fillna(df_train.x255.mean())
df_train.x255.isnull().unique()
df_train.x255 = df_train["x255"].astype(str)
df_train["month"] = df_train.x255.str[5:7]
df_train["year"] = df_train.x255.str[:4]
df_train["day"] = df_train.x255.str[8:10]
df_train.day.unique()
df_train.month.unique()
df_train.year.unique()
df_train.year = df_train.year.astype(int)
df_train.year.unique()
df_train.month = df_train.month.astype(int)
df_train.month.unique()
df_train.day = df_train.day.astype(int)
df_train.day.unique()
df_train.shape
# Dropping columns with no more than 1 unique value, as they can not have explanatory power:
for col in df_train.columns:
if len(df_train[col].unique()) == 1:
df_train.drop(col, inplace=True, axis=1)
df_train.shape
df_train = df_train.drop(["x255"], axis=1)
df_train.x261.unique()
# Cleaning up features before One-hot encoding:
df_train.x273.unique()
features = ["x273", "x274", "x275", "x276"]
for feature in features:
df_train[feature] = df_train[feature].str.replace("Не", "0")
df_train[feature] = df_train[feature].str.replace("Да", "1")
df_train[feature] = df_train[feature].astype(bool)
df_train.x273
# Now those features are boolean and can be used in model training and prediction.
df_train.x2.value_counts()
df_train.x3.value_counts()
df_train.x4.value_counts()
features = []
X = df_train.drop(["Bad_Flag"], axis=1)
X = X.select_dtypes(exclude=["number", "bool_"])
X.head()
# Cleaning more features:
df_train.x195.value_counts()
df_train.x262.value_counts()
features = ["x195", "x262", "x253"]
for feature in features:
df_train[feature] = df_train[feature].str.replace("No", "0")
df_train[feature] = df_train[feature].str.replace("Yes", "1")
df_train[feature] = df_train[feature].astype(bool)
df_train.x262
df_train.x256.value_counts()
# Resetting index to enable concatenation later:
df_train = df_train.reset_index()
X = df_train.copy()
ind = X["index"]
X = X.select_dtypes(exclude=["number", "bool_"])
X.head()
# Proper encoding to be able to make sense of feature x193 could not be found.
# Transforming categorical data:
from sklearn.preprocessing import OneHotEncoder
enc = OneHotEncoder(drop="first")
enc_data = pd.DataFrame(
enc.fit_transform(X[X.columns.tolist()]).toarray(), columns=enc.get_feature_names()
)
# Drop_first was used to make sure that no multicollinearity occurs between features. This would be damaging especially to logistic regression and linear-based models in general.
enc_data
# Dropping original categorical variables:
df_train = df_train.drop(X.columns.tolist(), axis=1)
df_train.shape
df_train.head()
# Returning the index to enc_data to enable concatenation with df_train:
enc_data["index"] = ind
enc_data.head()
df_train = pd.concat([enc_data, df_train], axis=1)
df_train.shape
df_train.head()
df_train.x135.equals(df_train.x136)
# Even though pd.equals returns False, I suspect x135 and x136 are duplicate features.
df_train.x135.describe()
df_train.x136.describe()
# They are, therefore drop either one:
df_train = df_train.drop(["x135"], axis=1)
# Removing column unnamed:
df_train.drop(df_train.filter(regex="Unname"), axis=1, inplace=True)
df_train.head()
# Index not needed anymore:
df_train = df_train.drop(["index"], axis=1)
# Checking for duplicated columns as xgboost returns error for them:
duplicate_columns = df_train.columns[df_train.columns.duplicated()]
duplicate_columns
# No duplicates, proceed with next steps.
df_train.head()
# Preparing original df_train for anomaly removal.
df_train_og = df_train[df_train.test_flag == 0]
# # Anomaly removal
# Training with RFClassifier in order to find the most important features according to it (and remove anomalous observations from them) will be done on original training data to avoid information leakage from test data and thus preserve a 'true' testing dataset.
from sklearn.ensemble import RandomForestClassifier
X = df_train_og.drop(["Bad_Flag"], axis=1)
y = df_train_og.Bad_Flag
rf = RandomForestClassifier(n_estimators=100)
rf.fit(X, y)
# Get feature importances
importances = rf.feature_importances_
# Sort the feature importances in descending order
sorted_importances = np.argsort(importances)[::-1]
# Get the top k features
k = 10 # number of top features to consider
top_k_features = X.columns[sorted_importances[:k]]
# RFClassifier was used to find some important features and later use them for anomaly removal. It was important to fit the classifier on training data only (test_flag==0).
top_k_features
df_train_og.shape
# Outliers were removed using the interquartile range.
# Interquartile Range (IQR) is calculated via the difference between the 75th percentile and 25th percentile. The aim is to create a threshold beyond the 75th and 25th percentile. Beyond this threshold, instances will be detected as anomalies and deleted from the training dataset.
# The threshold is determined by multiplying a number (ex: 1.5) by the IQR. The higher this threshold is, the less outliers will be detected (multiplying by a higher number ex: 3), and the lower this threshold is the more outliers will be detected.
# The Tradeoff: The lower the threshold the more outliers will be removed. More "extreme outliers" should be removed, because there is a risk of information loss which will cause the models to perform worse.
# # -----> x1 Removing Outliers (Highest Negative Correlated with Labels)
x1_bad = df_train_og["x1"].loc[df_train_og["Bad_Flag"] == 1].values
q25, q75 = np.percentile(x1_bad, 25), np.percentile(x1_bad, 75)
print("Quartile 25: {} | Quartile 75: {}".format(q25, q75))
x1_iqr = q75 - q25
print("iqr: {}".format(x1_iqr))
x1_cut_off = x1_iqr * 1.5
x1_lower, x1_upper = q25 - x1_cut_off, q75 + x1_cut_off
print("Cut Off: {}".format(x1_cut_off))
print("x1 Lower: {}".format(x1_lower))
print("x1 Upper: {}".format(x1_upper))
outliers = [x for x in x1_bad if x < x1_lower or x > x1_upper]
print("Feature x1 Outliers for Bad Cases: {}".format(len(outliers)))
print("x1 outliers:{}".format(outliers))
df_train_og = df_train_og.drop(
df_train_og[(df_train_og["x1"] > x1_upper) | (df_train_og["x1"] < x1_lower)].index
)
print("----" * 44)
print("Size of new dataset", df_train_og.shape)
# # -----> x265 Removing Outliers (Highest Negative Correlated with Labels)
x265_bad = df_train_og["x265"].loc[df_train_og["Bad_Flag"] == 1].values
q25, q75 = np.percentile(x265_bad, 25), np.percentile(x265_bad, 75)
print("Quartile 25: {} | Quartile 75: {}".format(q25, q75))
x265_iqr = q75 - q25
print("iqr: {}".format(x265_iqr))
x265_cut_off = x265_iqr * 5
x265_lower, x265_upper = q25 - x265_cut_off, q75 + x265_cut_off
print("Cut Off: {}".format(x265_cut_off))
print("x265 Lower: {}".format(x265_lower))
print("x265 Upper: {}".format(x265_upper))
outliers = [x for x in x265_bad if x < x265_lower or x > x265_upper]
print("Feature x265 Outliers for Bad Cases: {}".format(len(outliers)))
print("x265 outliers:{}".format(outliers))
df_train_og = df_train_og.drop(
df_train_og[
(df_train_og["x265"] > x265_upper) | (df_train_og["x265"] < x265_lower)
].index
)
print("----" * 44)
print("Size of new dataset", df_train_og.shape)
top_k_features
# # -----> x268 Removing Outliers (Highest Negative Correlated with Labels)
x268_bad = df_train_og["x268"].loc[df_train_og["Bad_Flag"] == 1].values
q25, q75 = np.percentile(x268_bad, 25), np.percentile(x268_bad, 75)
print("Quartile 25: {} | Quartile 75: {}".format(q25, q75))
x268_iqr = q75 - q25
print("iqr: {}".format(x268_iqr))
x268_cut_off = x268_iqr * 5
x268_lower, x268_upper = q25 - x268_cut_off, q75 + x268_cut_off
print("Cut Off: {}".format(x268_cut_off))
print("x268 Lower: {}".format(x268_lower))
print("x268 Upper: {}".format(x268_upper))
outliers = [x for x in x268_bad if x < x268_lower or x > x268_upper]
print("Feature x268 Outliers for Bad Cases: {}".format(len(outliers)))
print("x268 outliers:{}".format(outliers))
df_train_og = df_train_og.drop(
df_train_og[
(df_train_og["x268"] > x268_upper) | (df_train_og["x268"] < x268_lower)
].index
)
print("----" * 44)
print("Size of new dataset", df_train_og.shape)
df_train_og.head()
# # Train-validation-test split
# The train-test(or validation) split should be done before any feature scaling and feature selection to avoid information leakage. This is because feature scaling and feature selection should only be performed on the training data and not on the test data. The test data is meant to be unseen data that provides a measure of how well the model generalizes to new data. By scaling and selecting features based on the test data, the test set is no longer truly unseen and can result in overestimating the performance of the model.
df_test = df_train[df_train.test_flag == 1]
df_train_og = df_train_og.drop(["test_flag"], axis=1)
df_train_split, df_val = train_test_split(
df_train_og, test_size=0.05, random_state=42, stratify=df_train_og.Bad_Flag
)
print(df_train_split.shape, df_val.shape)
df_test = df_test.drop(["test_flag"], axis=1)
df_test.shape
df_train_vif = df_train_split.copy()
df_val_vif = df_val.copy()
df_test_vif = df_test.copy()
df_train_vif_corr = df_train_split.copy()
df_val_vif_corr = df_val.copy()
df_test_vif_corr = df_test.copy()
df_train_corr = df_train_split.copy()
df_val_corr = df_val.copy()
df_test_corr = df_test.copy()
# # Variance Inflation Factor
# Variance Inflation Factor (VIF) is a statistical measure used to quantify the degree of multicollinearity among the independent variables in a multiple regression model. VIF is calculated as the ratio of the variance of a particular predictor in a multiple regression model to the variance of that predictor when it is considered alone. High VIF values indicate that a predictor is highly correlated with other predictors in the model, which can lead to instability in the regression coefficients and poor model performance.
def remove_high_vif_features(dataframe, threshold=5.0):
"""Remove features with high Variance Inflation Factor (VIF) from a DataFrame.
Args:
dataframe: pandas DataFrame
threshold: VIF threshold for removing features (default: 5.0)
Returns:
dataframe: pandas DataFrame with high VIF features removed
"""
# Convert dataframe to a matrix
X = dataframe.values
# Get the feature names
feature_names = dataframe.columns
# Compute the VIF for each feature
vif = [variance_inflation_factor(X, i) for i in range(X.shape[1])]
# Create a dictionary with the feature names and their corresponding VIF values
vif_dict = dict(zip(feature_names, vif))
# Create a list of features with VIF values greater than the threshold
high_vif_features = [
feature for feature, vif in vif_dict.items() if vif > threshold
]
# Remove the high VIF features from the DataFrame
dataframe = dataframe.drop(columns=high_vif_features)
return dataframe
X = df_train_vif.drop(["Bad_Flag"], axis=1)
y = df_train_vif.Bad_Flag
X = X.astype("float")
X = remove_high_vif_features(X, threshold=6.0)
# saving variables for direct later use
df_train_vif = X.join(y)
X_vif = X.copy()
y_vif = y.copy()
df_train_vif.shape
# A significant number of features were removed as a result of being multicolinear.
X_val_vif = df_val_vif.drop(["Bad_Flag"], axis=1)
X_val_vif = X_val_vif[X_vif.columns.tolist()]
X_val_vif.head()
df_val_vif = X_val_vif.join(df_val_vif.Bad_Flag)
X_test_vif = df_test_vif.drop(["Bad_Flag"], axis=1)
X_test_vif = X_test_vif[X.columns.tolist()]
X_test_vif.head()
df_test_vif = X_test_vif.join(df_test_vif.Bad_Flag)
# # Correlation
def remove_low_correlation_features(dataframe, target, threshold=0.1):
"""Remove features with low correlation to the target variable from a DataFrame.
Args:
dataframe: pandas DataFrame
target: target variable name
threshold: correlation threshold for removing features (default: 0.1)
Returns:
dataframe: pandas DataFrame with low correlation features removed
"""
# Compute the correlation matrix
corr_matrix = dataframe.corr()
# Create a list of features with correlation values less than the threshold
low_correlation_features = [
feature
for feature, corr in corr_matrix[target].iteritems()
if abs(corr) < threshold
]
# Remove the low correlation features from the DataFrame
dataframe = dataframe.drop(columns=low_correlation_features)
return dataframe
df_train_vif_corr = remove_low_correlation_features(
df_train_vif, "Bad_Flag", threshold=0.05
)
df_train_vif_corr.shape
X = df_train_vif_corr.drop(["Bad_Flag"], axis=1)
X = X.astype("float")
y = df_train_vif_corr.Bad_Flag
X_vif_corr = X.copy()
y_vif_corr = y.copy()
# y_vif_corr = pd.DataFrame(y_vif_corr)
# y_vif_corr.Bad_Flag.isna().unique()
X_test_vif_corr = X_test_vif[X_vif_corr.columns.tolist()]
X_test_vif_corr
df_test_vif_corr = X_test_vif_corr.join(df_test_vif_corr.Bad_Flag)
X_val_vif_corr = X_val_vif[X_vif_corr.columns.tolist()]
X_val_vif_corr
df_val_vif_corr = X_val_vif_corr.join(df_val_vif_corr.Bad_Flag)
df_train_corr = remove_low_correlation_features(
df_train_split, "Bad_Flag", threshold=0.05
)
X_corr = df_train_corr.drop(["Bad_Flag"], axis=1)
y_corr = df_train_corr.Bad_Flag
df_train_corr.shape
X_test_corr = df_test_corr.drop(["Bad_Flag"], axis=1)
X_test_corr = X_test_corr[X_corr.columns.tolist()]
X_test_corr
df_test_corr = X_test_corr.join(df_test_corr.Bad_Flag)
X_val_corr = df_val_corr.drop(["Bad_Flag"], axis=1)
X_val_corr = X_val_corr[X_corr.columns.tolist()]
X_val_corr
df_val_corr = X_val_corr.join(df_val_corr.Bad_Flag)
# # Downsampling
# Downsampling is a technique for handling imbalanced datasets in machine learning. The idea behind this technique is to balance the class distribution of the target variable by randomly removing instances from the majority class. The aim is to reduce the size of the majority class to match the size of the minority class, creating a more balanced dataset. This can help to improve the performance of some machine learning algorithms, as they may perform poorly on imbalanced datasets where the majority class dominates the training data.
df_majority = df_train_split[df_train_split.Bad_Flag == 0]
df_minority = df_train_split[df_train_split.Bad_Flag == 1]
# Downsample majority class
df_majority_downsampled = resample(
df_majority,
replace=False, # sample without replacement
n_samples=len(df_minority), # to match minority class
random_state=42,
) # reproducible results
# Combine minority class with downsampled majority class
df_downsampled = pd.concat([df_majority_downsampled, df_minority])
# Display new class counts
df_downsampled.Bad_Flag.value_counts()
df_majority_vif = df_train_vif[df_train_vif.Bad_Flag == 0]
df_minority_vif = df_train_vif[df_train_vif.Bad_Flag == 1]
# Downsample majority class
df_majority_downsampled_vif = resample(
df_majority_vif,
replace=False, # sample without replacement
n_samples=len(df_minority_vif), # to match minority class
random_state=42,
) # reproducible results
# Combine minority class with downsampled majority class
df_downsampled_vif = pd.concat([df_majority_downsampled_vif, df_minority_vif])
# Display new class counts
df_downsampled_vif.Bad_Flag.value_counts()
df_majority_vif_corr = df_train_vif_corr[df_train_vif_corr.Bad_Flag == 0]
df_minority_vif_corr = df_train_vif_corr[df_train_vif_corr.Bad_Flag == 1]
# Downsample majority class
df_majority_downsampled_vif_corr = resample(
df_majority_vif_corr,
replace=False, # sample without replacement
n_samples=len(df_minority_vif_corr), # to match minority class
random_state=42,
) # reproducible results
# Combine minority class with downsampled majority class
df_downsampled_vif_corr = pd.concat(
[df_majority_downsampled_vif_corr, df_minority_vif_corr]
)
# Display new class counts
df_downsampled_vif_corr.Bad_Flag.value_counts()
df_majority_corr = df_train_corr[df_train_corr.Bad_Flag == 0]
df_minority_corr = df_train_corr[df_train_corr.Bad_Flag == 1]
# Downsample majority class
df_majority_downsampled_corr = resample(
df_majority_corr,
replace=False, # sample without replacement
n_samples=len(df_minority_corr), # to match minority class
random_state=42,
) # reproducible results
# Combine minority class with downsampled majority class
df_downsampled_corr = pd.concat([df_majority_downsampled_corr, df_minority_corr])
# Display new class counts
df_downsampled_corr.Bad_Flag.value_counts()
# # Upsampling
# Upsampling is a technique used in imbalanced datasets to balance the class distribution. It involves increasing the number of instances in the minority class by duplicating existing instances. The goal is to make the number of instances in the minority class equal or close to the number of instances in the majority class, so that the class distribution becomes more balanced. This can help reduce the bias of the model towards the majority class and improve the model's performance in detecting the minority class. However, oversampling can lead to overfitting to the minority class and an increased computational burden,
df_majority = df_train_split[df_train_split.Bad_Flag == 0]
df_minority = df_train_split[df_train_split.Bad_Flag == 1]
# Upsample minority class
df_minority_upsampled = resample(
df_minority,
replace=True, # sample with replacement
n_samples=len(df_majority), # to match majority class
random_state=42,
) # reproducible results
# Combine majority class with upsampled minority class
df_upsampled = pd.concat([df_majority, df_minority_upsampled])
# Display new class counts
df_upsampled.Bad_Flag.value_counts()
df_majority_vif = df_train_vif[df_train_vif.Bad_Flag == 0]
df_minority_vif = df_train_vif[df_train_vif.Bad_Flag == 1]
# Downsample majority class
df_minority_upsampled_vif = resample(
df_minority_vif,
replace=True, # sample with replacement
n_samples=len(df_majority_vif), # to match minority class
random_state=42,
) # reproducible results
# Combine minority class with downsampled majority class
df_upsampled_vif = pd.concat([df_majority_vif, df_minority_upsampled_vif])
# Display new class counts
df_upsampled_vif.Bad_Flag.value_counts()
df_majority_vif_corr = df_train_vif_corr[df_train_vif_corr.Bad_Flag == 0]
df_minority_vif_corr = df_train_vif_corr[df_train_vif_corr.Bad_Flag == 1]
# Downsample majority class
df_minority_upsampled_vif_corr = resample(
df_minority_vif_corr,
replace=True, # sample with replacement
n_samples=len(df_majority_vif_corr), # to match minority class
random_state=42,
) # reproducible results
# Combine minority class with downsampled majority class
df_upsampled_vif_corr = pd.concat(
[df_majority_vif_corr, df_minority_upsampled_vif_corr]
)
# Display new class counts
df_upsampled_vif_corr.Bad_Flag.value_counts()
df_majority_corr = df_train_corr[df_train_corr.Bad_Flag == 0]
df_minority_corr = df_train_corr[df_train_corr.Bad_Flag == 1]
# Downsample majority class
df_minority_upsampled_corr = resample(
df_minority_corr,
replace=True, # sample with replacement
n_samples=len(df_majority_corr), # to match minority class
random_state=42,
) # reproducible results
# Combine minority class with downsampled majority class
df_upsampled_corr = pd.concat([df_majority_corr, df_minority_upsampled_corr])
# Display new class counts
df_upsampled_corr.Bad_Flag.value_counts()
# # Logistic Regression
# upsampled
X_up = df_upsampled.drop(["Bad_Flag"], axis=1)
y_up = df_upsampled.Bad_Flag
x_train = X_up.copy()
y_train = df_upsampled.Bad_Flag
x_val = df_val.drop(["Bad_Flag"], axis=1)
y_val = df_val.Bad_Flag
reg = LogisticRegression(max_iter=2000)
reg.fit(x_train, y_train)
def cmetrics(y_true, y_pred):
metrics = {}
metrics["accuracy"] = round(accuracy_score(y_true, y_pred), 2)
metrics["precision"] = round(precision_score(y_true, y_pred), 2)
metrics["recall"] = round(recall_score(y_true, y_pred), 2)
metrics["f1"] = round(f1_score(y_true, y_pred), 2)
metrics["AUC-ROC"] = round(roc_auc_score(y_true, y_pred), 2)
return metrics
from sklearn.metrics import confusion_matrix
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
def cmatrix(y_true, y_pred):
# Calculate the confusion matrix
cm = confusion_matrix(y_true, y_pred)
# Plot the confusion matrix
sns.heatmap(
cm,
annot=True,
xticklabels=["0", "1"],
yticklabels=["0", "1"],
cmap="Blues",
fmt="g",
)
plt.xlabel("Predicted Label")
plt.ylabel("True Label")
plt.title("Confusion Matrix")
plt.show()
y_hat_val = reg.predict(x_val)
print(cmetrics(y_val, y_hat_val))
cmatrix(y_val, y_hat_val)
from tabulate import tabulate
results = [["Log.Reg. default", round(f1_score(y_val, y_hat_val), 2)]]
print(tabulate(results, headers=["Algorithm", "f1"], numalign="left"))
cv_lr = StratifiedKFold(n_splits=5, random_state=42, shuffle=True)
params_lr = {
"C": np.logspace(-5, 5),
"penalty": ["l1", "l2", "elasticnet"],
"solver": ["lbfgs", "newton-cg"],
"fit_intercept": [True, False],
"l1_ratio": np.linspace(0, 1, 11),
}
reg_search = RandomizedSearchCV(
LogisticRegression(max_iter=2000),
params_lr,
scoring="f1",
cv=cv_lr,
random_state=42,
n_jobs=-1,
)
reg_search.fit(x_train, y_train)
reg_search.best_estimator_
reg_search.best_estimator_.solver
y_hat_val = reg_search.best_estimator_.predict(x_val)
print(cmetrics(y_val, y_hat_val))
cmatrix(y_val, y_hat_val)
results += [["Log.Reg. tuned", round(f1_score(y_val, y_hat_val), 2)]]
print(tabulate(results, headers=["Algorithm", "f1"], numalign="left"))
# # Logistic Regression with feature selection via VIF and low corr.
#
# upsampled
X_up = df_upsampled_vif_corr.drop(["Bad_Flag"], axis=1)
y_up = df_upsampled_vif_corr.Bad_Flag
x_train = X_up.copy()
y_train = y_up.copy()
x_val = df_val_vif_corr.drop(["Bad_Flag"], axis=1)
y_val = df_val_vif_corr.Bad_Flag
from sklearn.linear_model import LogisticRegression
reg = LogisticRegression(max_iter=2000)
reg.fit(x_train, y_train)
y_hat_val = reg.predict(x_val)
print(cmetrics(y_val, y_hat_val))
cmatrix(y_val, y_hat_val)
results += [["Log.Reg. VIF+corr default", round(f1_score(y_val, y_hat_val), 2)]]
print(tabulate(results, headers=["Algorithm", "f1"], numalign="left"))
params_lr = {
"C": np.logspace(-5, 5),
"penalty": ["l1", "l2", "elasticnet"],
"solver": ["newton-cg", "lbfgs", "liblinear", "saga"],
"fit_intercept": [True, False],
"l1_ratio": np.linspace(0, 1, 11),
}
reg_search = RandomizedSearchCV(
LogisticRegression(max_iter=2000),
params_lr,
scoring="f1",
cv=cv_lr,
random_state=42,
n_jobs=-1,
)
reg_search.fit(x_train, y_train)
reg_search.best_estimator_
y_hat_val = reg_search.best_estimator_.predict(x_val)
print(cmetrics(y_val, y_hat_val))
cmatrix(y_val, y_hat_val)
results += [["Log.Reg. VIF+corr Tuned", round(f1_score(y_val, y_hat_val), 2)]]
print(tabulate(results, headers=["Algorithm", "f1"], numalign="left"))
# Feature selection via combined VIF and removing lowly correlated to the target variable features does not offer improvement over using no feature selection techniques.
# # Logistic Regression with feature selection via VIF
# upsampled
X_up = df_upsampled_vif.drop(["Bad_Flag"], axis=1)
y_up = df_upsampled_vif.Bad_Flag
x_train = X_up.copy()
y_train = y_up.copy()
x_val = df_val_vif.drop(["Bad_Flag"], axis=1)
y_val = df_val_vif.Bad_Flag
from sklearn.linear_model import LogisticRegression
reg = LogisticRegression(max_iter=2000)
reg.fit(x_train, y_train)
y_hat_val = reg.predict(x_val)
print(cmetrics(y_val, y_hat_val))
cmatrix(y_val, y_hat_val)
results += [["Log.Reg. VIF default", round(f1_score(y_val, y_hat_val), 2)]]
print(tabulate(results, headers=["Algorithm", "f1"], numalign="left"))
reg_search = RandomizedSearchCV(
LogisticRegression(max_iter=2000),
params_lr,
scoring="f1",
cv=cv_lr,
random_state=42,
n_jobs=-1,
)
reg_search.fit(x_train, y_train)
reg_search.best_estimator_
y_hat_val = reg_search.best_estimator_.predict(x_val)
print(cmetrics(y_val, y_hat_val))
cmatrix(y_val, y_hat_val)
results += [["Log.Reg. VIF tuned", round(f1_score(y_val, y_hat_val), 2)]]
print(tabulate(results, headers=["Algorithm", "f1"], numalign="left"))
# Similar results to VIF+low corr.
# # Logistic Regression with feature selection via low corr.
# upsampled
X_up = df_upsampled_corr.drop(["Bad_Flag"], axis=1)
y_up = df_upsampled_corr.Bad_Flag
x_train = X_up.copy()
y_train = y_up.copy()
x_val = df_val_corr.drop(["Bad_Flag"], axis=1)
y_val = df_val_corr.Bad_Flag
from sklearn.linear_model import LogisticRegression
reg = LogisticRegression(max_iter=2000)
reg.fit(x_train, y_train)
y_hat_val = reg.predict(x_val)
print(cmetrics(y_val, y_hat_val))
cmatrix(y_val, y_hat_val)
results += [["Log.Reg. corr default", round(f1_score(y_val, y_hat_val), 2)]]
print(tabulate(results, headers=["Algorithm", "f1"], numalign="left"))
params_lr = {
"C": np.logspace(-5, 5),
"penalty": ["l1", "l2", "elasticnet"],
"solver": ["newton-cg", "lbfgs", "liblinear"],
"fit_intercept": [True, False],
"l1_ratio": np.linspace(0, 1, 11),
}
reg_search = RandomizedSearchCV(
LogisticRegression(max_iter=2000),
params_lr,
scoring="f1",
cv=cv_lr,
random_state=42,
n_jobs=-1,
)
reg_search.fit(x_train, y_train)
reg_search.best_estimator_
y_hat_val = reg_search.best_estimator_.predict(x_val)
print(cmetrics(y_val, y_hat_val))
cmatrix(y_val, y_hat_val)
results += [["Log.Reg. corr tuned", round(f1_score(y_val, y_hat_val), 2)]]
print(tabulate(results, headers=["Algorithm", "f1"], numalign="left"))
# Logistic regression does not provide better results when the lowly correlated features are removed.
# # SVM
# SVM takes long to compile - using downsampled (i.e. smaller) data and scaling helps to improve that.
# downsampled
X_dw = df_downsampled.drop(["Bad_Flag"], axis=1)
y_dw = df_downsampled.Bad_Flag
# scaling
scaler = MinMaxScaler()
scaler.fit(X_dw)
X_dw_scaled = scaler.transform(X_dw)
x_train = X_dw_scaled.copy()
y_train = y_dw.copy()
x_val = df_val.drop(["Bad_Flag"], axis=1)
x_val = scaler.transform(x_val)
y_val = df_val.Bad_Flag
from sklearn.svm import SVC
SVC = SVC()
SVC.fit(x_train, y_train)
y_hat_val = SVC.predict(x_val)
print(cmetrics(y_val, y_hat_val))
cmatrix(y_val, y_hat_val)
results += [["SVM default", round(f1_score(y_val, y_hat_val), 2)]]
print(tabulate(results, headers=["Algorithm", "f1"], numalign="left"))
params_svm = {
"C": np.logspace(-5, 5, 50),
"gamma": np.logspace(-5, 5, 50),
"kernel": ["linear", "poly", "rbf", "sigmoid"],
}
def spline(data_1, data_2):
kernel = np.ones((data_1.shape[0], data_2.shape[0]))
for d in range(data_1.shape[1]):
column_1 = data_1[:, d].reshape(-1, 1)
column_2 = data_2[:, d].reshape(-1, 1)
c_prod = column_1 * column_2.T
c_sum = column_1 + column_2.T
c_min = np.minimum(column_1, column_2.T)
kernel *= (
1.0
+ c_prod
+ c_prod * c_min
- c_sum / 2.0 * c_min**2.0
+ 1.0 / 3.0 * c_min**3.0
)
return kernel
# The spline kernel is not included in sk-learn library. Code taken from [here](https://github.com/gmum/pykernels/blob/master/pykernels/regular.py).
params_svm = {
"C": np.logspace(-5, 5, 50),
"gamma": np.logspace(-5, 5, 50),
"kernel": ["poly", "rbf", "sigmoid", spline],
}
# fewer parameters are included to reduce compilation time, otherwise script does not compile at all
cv_svm = StratifiedKFold(n_splits=5, random_state=42, shuffle=True)
svm_search_best = RandomizedSearchCV(
SVC, params_svm, scoring="f1", cv=cv_svm, random_state=42, n_jobs=-1
)
svm_search_best.fit(x_train, y_train)
svm_search_best.best_estimator_
svm_search_best.best_estimator_.kernel
y_pred = svm_search_best.best_estimator_.predict(x_val)
print(cmetrics(y_val.values, y_pred))
cmatrix(y_val, y_pred)
results += [["SVM tuned", round(f1_score(y_val.values, y_pred), 2)]]
print(tabulate(results, headers=["Algorithm", "f1"], numalign="left"))
# The results for SVM do not show improvement over logistic regression.
# # SVM with feature selection via VIF and low corr.
# downsampled
X_dw = df_downsampled_vif_corr.drop(["Bad_Flag"], axis=1)
y_dw = df_downsampled_vif_corr.Bad_Flag
# scaling
scaler = MinMaxScaler()
scaler.fit(X_dw)
X_dw_scaled = scaler.transform(X_dw)
x_train = X_dw_scaled.copy()
y_train = y_dw.copy()
x_val = df_val_vif_corr.drop(["Bad_Flag"], axis=1)
x_val = scaler.transform(x_val)
y_val = df_val_vif_corr.Bad_Flag
from sklearn.svm import SVC
SVC = SVC()
SVC.fit(x_train, y_train)
y_hat_val = SVC.predict(x_val)
print(cmetrics(y_val, y_hat_val))
cmatrix(y_val, y_hat_val)
results += [["SVM Vif+corr default", round(f1_score(y_val, y_hat_val), 2)]]
print(tabulate(results, headers=["Algorithm", "f1"], numalign="left"))
cv_svm = StratifiedKFold(n_splits=5, random_state=42, shuffle=True)
svm_search = RandomizedSearchCV(
SVC, params_svm, scoring="f1", cv=cv_svm, random_state=42, n_jobs=None
)
svm_search.fit(x_train, y_train)
svm_search.best_estimator_.kernel
y_pred = svm_search.best_estimator_.predict(x_val)
print(cmetrics(y_val, y_pred))
cmatrix(y_val, y_pred)
results += [["SVM Vif+corr tuned", round(f1_score(y_val, y_pred), 2)]]
print(tabulate(results, headers=["Algorithm", "f1"], numalign="left"))
# SVM performs very poorly with VIF + low correlation removed data. Interestingly, the spline kernel was also not chosen as the best estimator kernel.
# It was hence decided to experiment - perhaps removing the lowly correlated features removes some important features.
# # SVM with feature selection via VIF
# downsampled
X_dw = df_downsampled_vif.drop(["Bad_Flag"], axis=1)
y_dw = df_downsampled_vif.Bad_Flag
# scaling
scaler = MinMaxScaler()
scaler.fit(X_dw)
X_dw_scaled = scaler.transform(X_dw)
x_train = X_dw_scaled.copy()
y_train = y_dw.copy()
x_val = df_val_vif.drop(["Bad_Flag"], axis=1)
x_val = scaler.transform(x_val)
y_val = df_val_vif.Bad_Flag
from sklearn.svm import SVC
SVC = SVC()
SVC.fit(x_train, y_train)
y_hat_val = SVC.predict(x_val)
print(cmetrics(y_val, y_hat_val))
cmatrix(y_val, y_hat_val)
results += [["SVM VIF default", round(f1_score(y_val, y_hat_val), 2)]]
print(tabulate(results, headers=["Algorithm", "f1"], numalign="left"))
params_svm = {"kernel": ["poly", "rbf", "sigmoid", spline]}
# fewer parameters are included to reduce compilation time, otherwise script does not compile at all
cv_svm = StratifiedKFold(n_splits=5, random_state=42, shuffle=True)
svm_search = RandomizedSearchCV(
SVC, params_svm, scoring="f1", cv=cv_svm, random_state=42, n_jobs=-1
)
svm_search.fit(x_train, y_train)
svm_search.best_estimator_
y_pred = svm_search.best_estimator_.predict(x_val)
print(cmetrics(y_val.values, y_pred))
cmatrix(y_val, y_pred)
results += [["SVM VIF tuned", round(f1_score(y_val.values, y_pred), 2)]]
print(tabulate(results, headers=["Algorithm", "f1"], numalign="left"))
# # SVM with feature selection via low corr.
# downsampled
X_dw = df_downsampled_corr.drop(["Bad_Flag"], axis=1)
y_dw = df_downsampled_corr.Bad_Flag
# scaling
scaler = MinMaxScaler()
scaler.fit(X_dw)
X_dw_scaled = scaler.transform(X_dw)
x_train = X_dw_scaled.copy()
y_train = y_dw.copy()
x_val = df_val_corr.drop(["Bad_Flag"], axis=1)
x_val = scaler.transform(x_val)
y_val = df_val_corr.Bad_Flag
from sklearn.svm import SVC
SVC = SVC()
SVC.fit(x_train, y_train)
y_hat_val = SVC.predict(x_val)
print(cmetrics(y_val, y_hat_val))
cmatrix(y_val, y_hat_val)
results += [["SVM corr default", round(f1_score(y_val, y_hat_val), 2)]]
print(tabulate(results, headers=["Algorithm", "f1"], numalign="left"))
params_svm = {"kernel": ["poly", "rbf", "sigmoid", spline]}
# fewer parameters are included to reduce compilation time, otherwise script does not compile at all
cv_svm = StratifiedKFold(n_splits=5, random_state=42, shuffle=True)
svm_search = RandomizedSearchCV(
SVC, params_svm, scoring="f1", cv=cv_svm, random_state=42, n_jobs=-1
)
svm_search.fit(x_train, y_train)
svm_search.best_estimator_
y_pred = svm_search.best_estimator_.predict(x_val)
print(cmetrics(y_val.values, y_pred))
cmatrix(y_val, y_pred)
results += [["SVM corr tuned", round(f1_score(y_val.values, y_pred), 2)]]
print(tabulate(results, headers=["Algorithm", "f1"], numalign="left"))
# # XGBoost
# upsample
X_up = df_upsampled.drop(["Bad_Flag"], axis=1)
y_up = df_upsampled.Bad_Flag
x_train = X_up.copy()
y_train = df_upsampled.Bad_Flag
x_val = df_val.drop(["Bad_Flag"], axis=1)
y_val = df_val.Bad_Flag
eval_set = [(x_val, y_val)]
from xgboost import XGBClassifier
xgb = XGBClassifier()
xgb.fit(x_train.values, y_train.values)
y_hat_xgb_def = xgb.predict(x_val.values)
print(cmetrics(y_val, y_hat_xgb_def))
cmatrix(y_val, y_hat_xgb_def)
results += [["XGB default", round(f1_score(y_val, y_hat_xgb_def), 2)]]
print(tabulate(results, headers=["Algorithm", "f1"], numalign="left"))
params_xgb = {
"max_depth": [1, 2, 3, 4, 5, 6],
"min_child_weight": [1, 2, 3, 4],
} # parameters to be tried in the search
fix_params = {
"learning_rate": 0.2,
"n_estimators": 100,
"objective": "binary:logistic",
} # other parameters, fixed for the moment
cv_xgb = StratifiedKFold(n_splits=5, random_state=42, shuffle=True)
xgb = XGBClassifier(**fix_params)
xgb_search_best = RandomizedSearchCV(
xgb,
params_xgb,
n_iter=5,
scoring="f1",
cv=cv_xgb,
random_state=42,
refit=True,
n_jobs=-1,
)
xgb_search_best.fit(x_train, y_train)
xgb_search_best.best_params_
# The particular choice of min_child_weight does not have a significant impact on the F1-score. To be selected are the best results of this search, i.e., max_depth = 5, and min_child_weight = 1. Other parameters can also be searched for:
params_xgb = {
"subsample": [0.8, 0.9, 1],
"max_delta_step": [0, 1, 2, 4],
} # parameters to be tries in the search
fix_params = {
"learning_rate": 0.2,
"n_estimators": 100,
"objective": "binary:logistic",
"max_depth": 5,
"min_child_weight": 1,
} # other parameters, fixed for the moment
cv_xgb = StratifiedKFold(n_splits=5, random_state=42, shuffle=True)
xgb = XGBClassifier(**fix_params)
xgb_search_best = RandomizedSearchCV(
xgb,
params_xgb,
n_iter=5,
scoring="f1",
cv=cv_xgb,
random_state=42,
refit=True,
n_jobs=-1,
)
xgb_search_best.fit(x_train, y_train)
xgb_search_best.best_params_
# It is obtained that max_delta_step = 4 and subsample = 0.9 are (quasi)-optimal. Optimizing the learning rate:
#
params_xgb = {"learning_rate": [0.05, 0.1, 0.15, 0.2, 0.25, 0.3]}
fix_params = {
"n_estimators": 100,
"objective": "binary:logistic",
"max_depth": 5,
"min_child_weight": 1,
"max_delta_step": 4,
"subsample": 0.9,
} # other parameters, fixed for the moment
xgb = XGBClassifier(**fix_params)
xgb_search_best = RandomizedSearchCV(
xgb,
params_xgb,
n_iter=5,
scoring="f1",
cv=cv_xgb,
random_state=42,
refit=True,
n_jobs=-1,
)
xgb_search_best.fit(x_train, y_train)
xgb_search_best.best_params_
fix_params = {
"n_estimators": 100,
"objective": "binary:logistic",
"max_depth": 5,
"min_child_weight": 1,
"max_delta_step": 4,
"subsample": 0.9,
"learning_rate": 0.3,
}
xgb = XGBClassifier(**fix_params)
xgb_final = xgb.fit(x_train, y_train)
y_hat_xgb = xgb_final.predict(x_val)
print(cmetrics(y_val, y_hat_xgb))
cmatrix(y_val, y_hat_xgb)
results += [["XGB tuned", round(f1_score(y_val, y_hat_xgb), 2)]]
print(tabulate(results, headers=["Algorithm", "f1"], numalign="left"))
# # XGBoost with feature selection via VIF and low corr.
# Normally, XGboost is a boosting algorithm and should be able to select features (hence remove variables with low correlation with target variable) and not be affected by high VIF features as well. Despite that, it is interesting to compare cleaned and not cleaned data.
# upsampled
X_up = df_upsampled_vif_corr.drop(["Bad_Flag"], axis=1)
y_up = df_upsampled_vif_corr.Bad_Flag
x_train = X_up.copy()
y_train = y_up.copy()
x_val = df_val_vif_corr.drop(["Bad_Flag"], axis=1)
y_val = df_val_vif_corr.Bad_Flag
from xgboost import XGBClassifier
xgb = XGBClassifier()
xgb.fit(x_train, y_train)
y_hat_xgb_def = xgb.predict(x_val)
print(cmetrics(y_val, y_hat_xgb_def))
cmatrix(y_val, y_hat_xgb_def)
results += [["XGB VIF+corr def", round(f1_score(y_val, y_hat_xgb_def), 2)]]
print(tabulate(results, headers=["Algorithm", "f1"], numalign="left"))
xgb = XGBClassifier(**fix_params)
xgb_final_vif_corr = xgb.fit(x_train, y_train)
y_hat_xgb = xgb_final_vif_corr.predict(x_val)
print(cmetrics(y_val, y_hat_xgb))
cmatrix(y_val, y_hat_xgb)
results += [["XGB VIF+corr tuned", round(f1_score(y_val, y_hat_xgb), 2)]]
print(tabulate(results, headers=["Algorithm", "f1"], numalign="left"))
# # XGBoost with feature selection via VIF
# upsample
X_up = df_upsampled_vif.drop(["Bad_Flag"], axis=1)
y_up = df_upsampled_vif.Bad_Flag
x_train = X_up.copy()
y_train = y_up.copy()
x_val = df_val_vif.drop(["Bad_Flag"], axis=1)
y_val = df_val_vif.Bad_Flag
from xgboost import XGBClassifier
xgb = XGBClassifier()
xgb.fit(x_train, y_train)
y_hat_xgb_def = xgb.predict(x_val)
print(cmetrics(y_val, y_hat_xgb_def))
cmatrix(y_val, y_hat_xgb_def)
results += [["XGB VIF default", round(f1_score(y_val, y_hat_xgb_def), 2)]]
print(tabulate(results, headers=["Algorithm", "f1"], numalign="left"))
xgb = XGBClassifier(**fix_params)
xgb_final_vif = xgb.fit(x_train, y_train)
y_hat_xgb = xgb_final_vif.predict(x_val)
print(cmetrics(y_val, y_hat_xgb))
cmatrix(y_val, y_hat_xgb)
results += [["XGB VIF tuned", round(f1_score(y_val, y_hat_xgb), 2)]]
print(tabulate(results, headers=["Algorithm", "f1"], numalign="left"))
# # XGBoost with feature selection via low corr.
# upsample
X_up = df_upsampled_corr.drop(["Bad_Flag"], axis=1)
y_up = df_upsampled_corr.Bad_Flag
x_train = X_up.copy()
y_train = y_up.copy()
x_val = df_val_corr.drop(["Bad_Flag"], axis=1)
y_val = df_val_corr.Bad_Flag
eval_set = [(x_val, y_val)]
from xgboost import XGBClassifier
xgb = XGBClassifier()
xgb.fit(x_train, y_train)
y_hat_xgb_def = xgb.predict(x_val)
print(cmetrics(y_val, y_hat_xgb_def))
cmatrix(y_val, y_hat_xgb_def)
results += [["XGB corr default", round(f1_score(y_val, y_hat_xgb_def), 2)]]
print(tabulate(results, headers=["Algorithm", "f1"], numalign="left"))
xgb = XGBClassifier(**fix_params)
xgb_final_corr = xgb.fit(x_train, y_train)
y_hat_xgb = xgb_final_corr.predict(x_val)
print(cmetrics(y_val, y_hat_xgb))
cmatrix(y_val, y_hat_xgb)
results += [["XGB corr tuned", round(f1_score(y_val, y_hat_xgb), 2)]]
print(tabulate(results, headers=["Algorithm", "f1"], numalign="left"))
# # Testing
# DEFAULT XGB
# upsample
X_up = df_upsampled.drop(["Bad_Flag"], axis=1)
y_up = df_upsampled.Bad_Flag
x_train = X_up.copy()
y_train = df_upsampled.Bad_Flag
from xgboost import XGBClassifier
xgb = XGBClassifier()
xgb.fit(x_train, y_train)
X_test = df_test.drop(["Bad_Flag"], axis=1)
y_test = df_test.Bad_Flag
y_pred = xgb.predict(X_test)
print(cmetrics(y_test, y_pred))
print(confusion_matrix(y_test, y_pred))
# TUNED XGB
X_test = df_test.drop(["Bad_Flag"], axis=1)
y_test = df_test.Bad_Flag
X_up = df_upsampled.drop(["Bad_Flag"], axis=1)
y_up = df_upsampled.Bad_Flag
x_train = X_up.copy()
y_train = df_upsampled.Bad_Flag
y_pred = xgb_final.predict(X_test)
print(cmetrics(y_test, y_pred))
print(confusion_matrix(y_test, y_pred))
|
# Imports for project
import functools
import os
import tensorflow.compat.v2 as tf
import tensorflow_hub as hub
import matplotlib.pylab as plt
import numpy as np
# Loading model and setting the driectory of the content and style images
model = "/kaggle/input/arbitrary-image-stylization-v1/tensorflow1/256/2"
model = hub.load(model)
content_image = "/kaggle/input/100-bird-species/test/ABBOTTS BABBLER/1.jpg"
style_image = "/kaggle/input/abstract-art-gallery/Abstract_gallery/Abstract_gallery/Abstract_image_1002.jpg"
# Function for displaying the images
def show_image(images, titles=("",)):
img_num = len(images)
image_sizes = [image.shape[1] for image in images]
width = (image_sizes[0] * 6) // 320
plt.figure(figsize=(width * img_num, width))
for i in range(img_num):
plt.imshow(images[i][0])
plt.axis("off")
plt.title(titles[i] if len(titles) > i else "")
plt.show()
# Function to decode image directory and setting outputs
def decode_image(image):
img = tf.io.decode_image(tf.io.read_file(image), channels=3, dtype=tf.float32)[
tf.newaxis, ...
]
return img
outputs = model(
tf.constant(decode_image(content_image)), tf.constant(decode_image(style_image))
)
stylized_image = outputs[0]
# Displaying content, style and stylized images
show_image(
[decode_image(content_image), decode_image(style_image), stylized_image],
titles=["Content Image", "Style Image", "Stylized Image"],
)
|
# World happiness dataset has 5 years ranking of countries separately. The main object of this notebook is to work on my EDA skills. There is not a lot of data present, but this dataset might work best if you want to work on your EDA mainly, visualization skills.
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
# reading data from csv into pandas dataframe, as the files are differentiated based on years, i will be reading them seperately and in future i will be merging them based on requirements.
data15 = pd.read_csv("/kaggle/input/world-happiness/2015.csv")
data16 = pd.read_csv("/kaggle/input/world-happiness/2016.csv")
data17 = pd.read_csv("/kaggle/input/world-happiness/2017.csv")
data18 = pd.read_csv("/kaggle/input/world-happiness/2018.csv")
data19 = pd.read_csv("/kaggle/input/world-happiness/2019.csv")
data15.head()
# Lets see what are the features
data15.columns
data15.info()
data15.describe()
data15.describe(include="object")
# By above info and describe, there are 2 categorical features and 10 continuous features. non of them have any missing values. But if we focus a little into the continuous values, **Happiness ranking is not actually a continuous values where the values range from 1 to 158** where 1 is best and 158 is worst. we need to find a way to work with this data in future.
# # Understanding Data
# ### Not using Visualiation
data15["Region"].value_counts()
# In this 2015 dataset, Among 158 countries, 40 lie in Sub-Saharan Africa which is considerably 25%.
# ### Now Lets use Visualization
# **Continuous features**:
# * Happiness Score
# * Standard Error
# * Economy (GDP per Capita)
# * Family
# * Health (Life Expectancy)
# * Freedom
# * Trust (Government Corruption)
# * Generosity
# * Dystopia Residual
# **Categorical Features**:
# * Country
# * Region
# **Special Feature**:
# * Happiness Rank
sns.scatterplot(data=data15, x="Economy (GDP per Capita)", y="Happiness Rank")
|
# A language model is trained on large amounts of textual data to understand the patterns and structure of language. The primary goal of a language model is to predict the probability of the next word or sequence of words in a sentence given the previous words.
# Language models can be used for a variety of natural language processing (NLP) tasks, such as text classification, machine translation, text summarization, speech recognition, and sentiment analysis. There are many types of language models, ranging from simple n-gram models to more complex neural network-based models such as recurrent neural networks (RNNs) and transformers.
# The transformer architecture is currently mostly used for language models and can be divided into an encoder and/or decoder architecture depending on the specific task. In general, transformers are trained on a large quantity of unlabeled text using self-supervised learning. The training of a transformer model on a lot of data takes a lot of computational effort and the training of language models can get expensive very quickly. So, often the best way to have a task-specific transformer model is to use a pre-trained model from [Hugging Face](https://huggingface.co/) and fine-tune the model based on your data.
# Based on my work experience with invoices, fine-tuning a pre-existing model didn't work well. I received the best results for text classification after I finetuned a french base-model on german invoices. Nevertheless the overall F1-score wasn't worth the effort. I assume that the content and structure of an invoice differs too much from the training data (e.g. no continuous text and many numbers). Additional, the tokenizers of the pre-trained models are not optimied for invoices, so the context window of a transformer will contain less text, which makes the training less effective.
# I worked on text classification of invoices for multiple clients. I trained a base-model on a few million invoices (mostly german and english) and fine-tuned the base model for each client consists with around 2000 - 50000 invoices and 70 - 2000 labels. Initially I used the Longformer architecture ([Beltagy et al. 2020](https://arxiv.org/pdf/2004.05150.pdf)), but based on a [bug](https://github.com/pytorch/pytorch/issues/94810), I couldn't deploy the models. Besides its limitations, I used the BERT architecture [Devlin et al. 2019](https://arxiv.org/pdf/1810.04805.pdf).
# Obviously, I can't share anything from my work related data, so I use a german corpus and draft the training of a base-model. Another tutorial can be found [on Hugging Face](https://huggingface.co/blog/how-to-train).
import os
from pathlib import Path
from datasets import load_dataset, Dataset
import pandas as pd
from sklearn.metrics import f1_score, precision_score, recall_score, accuracy_score
import torch
from transformers.models.gpt2.tokenization_gpt2 import bytes_to_unicode
from transformers import BertTokenizerFast, DataCollatorForLanguageModeling
from transformers import BertConfig, BertForMaskedLM
from transformers import LineByLineTextDataset
from transformers import Trainer, TrainingArguments, EarlyStoppingCallback
from tokenizers import Tokenizer
from tokenizers import normalizers
from tokenizers.normalizers import NFD, Lowercase, StripAccents
from tokenizers import pre_tokenizers
from tokenizers.pre_tokenizers import Digits, Whitespace, ByteLevel, Punctuation
from tokenizers.processors import TemplateProcessing
from tokenizers import Tokenizer, models, trainers
from tqdm.auto import tqdm
os.environ["WANDB_DISABLED"] = str("true")
os.environ["TOKENIZERS_PARALLELISM"] = str("true")
# ## Split data
# Finding a good dataset is hard. I picked the [3 Million German Sentences](https://www.kaggle.com/datasets/rtatman/3-million-german-sentences) to showcase a short model training. Also, I don't expect any useful model here.
# Each sentence has an id and a timestamp. For training a model, this is not needed. Additional, I use around 3000 sentences for the evaluation for the model training. The datasets will fit in memory, but to able to scale, I will use a streaming approch.
def clean_text(examples):
return {"text": examples["text"].split("\t")[1].split(":")[-1].strip()}
dataset = load_dataset(
"text",
data_files="/kaggle/input/3-million-german-sentences/deu_news_2015_3M-sentences.txt",
split="train",
streaming=False,
)
dataset = dataset.map(clean_text)
dataset = dataset.train_test_split(test_size=0.01)
train_length = len(dataset["train"])
eval_length = len(dataset["test"])
# convenience beats best practice
df = pd.DataFrame({"text": dataset["train"]["text"]})
df.to_parquet("/kaggle/working/train_data.snap.parquet", compression="snappy")
df = pd.DataFrame({"text": dataset["test"]["text"]})
df.to_parquet("/kaggle/working/eval_data.snap.parquet", compression="snappy")
# del [dataset, df]
# Finally, let's "load" the data. Did I mention, that I really like the [datasets library](https://huggingface.co/docs/datasets/index).
# ## Tokenizer
# A tokenizer converts raw text into into smaller units, such as words or subwords, that can be used for training machine learning models. The tokenizer takes as input a string of text and outputs a sequence of tokens, each of which represents a distinct unit of meaning. The subword tokenizer breaks down words into smaller subword units. This is useful for handling out-of-vocabulary (OOV) words, which are words that are not in the training data.
# I use the [Byte-Pair Encoding tokenizer](https://huggingface.co/course/chapter6/5?fw=pt), which is a data compression algorithm, where the most common pair of consecutive bytes of data is replaced with a byte that does not occur in that data ([Gage 1994](https://www.derczynski.com/papers/archive/BPE_Gage.pdf), [Sennrich et al. 2016](https://arxiv.org/pdf/1508.07909.pdf)).
byte_to_unicode_map = bytes_to_unicode()
unicode_to_byte_map = dict((v, k) for k, v in byte_to_unicode_map.items())
base_vocab = list(unicode_to_byte_map.keys())
print(f"Size of our base vocabulary: {len(base_vocab)}")
print(f"First element: `{base_vocab[0]}`, last element: `{base_vocab[-1]}`")
# normalizer = normalizers.Sequence([NFD(),
# # Lowercase(),
# # StripAccents()
# ])
# pre_tokenizer = pre_tokenizers.Sequence([
# # ByteLevel(add_prefix_space=False),
# Whitespace()
# ])
# tokenizer.normalizer = normalizer
# tokenizer.pre_tokenizer = pre_tokenizer
from tokenizers import Tokenizer
from tokenizers.decoders import ByteLevel as ByteLevelDecoder
from tokenizers.models import BPE
from tokenizers.normalizers import Lowercase, NFKC, Sequence
from tokenizers.pre_tokenizers import ByteLevel
# First we create an empty Byte-Pair Encoding model (i.e. not trained model)
tokenizer = Tokenizer(BPE())
# Then we enable lower-casing and unicode-normalization
# The Sequence normalizer allows us to combine multiple Normalizer that will be
# executed in order.
tokenizer.normalizer = Sequence(
[
NFKC(),
# Lowercase()
]
)
# Our tokenizer also needs a pre-tokenizer responsible for converting the input to a ByteLevel representation.
tokenizer.pre_tokenizer = pre_tokenizers.Sequence(
[ByteLevel(add_prefix_space=False), Whitespace()]
)
# And finally, let's plug a decoder so we can recover from a tokenized input to the original one
# tokenizer.decoder = ByteLevelDecoder()
print(
tokenizer.normalizer.normalize_str("""Dies ist ein Beispielsatz in Deutsch Äoüß""")
)
# Here, we define the tokenization trainer. We define the size of the vocabular and the special tokens we use.
vocab_size = 32768
trainer = trainers.BpeTrainer(
vocab_size=vocab_size,
show_progress=True,
initial_alphabet=ByteLevel.alphabet(),
special_tokens=["[UNK]", "[CLS]", "[SEP]", "[PAD]", "[MASK]"],
)
# Normally, I avoid loading the dataset into memory to train a tokenizer. In this case, the tokenizer doesn't train with the batch_iterator. I don't know, what causes this problems, but for now, I
# iter_dataset = iter(train_dataset)
# def batch_iterator(batch_size=10):
# for _ in tqdm(range(0, round(train_length,-1), batch_size)):
# yield [next(iter_dataset)['text'] for _ in range(batch_size)]
train_dataset = load_dataset(
"parquet", data_files="/kaggle/working/train_data.snap.parquet", split="train"
)
dataset = load_dataset(
"text",
data_files="/kaggle/input/3-million-german-sentences/deu_news_2015_3M-sentences.txt",
split="train",
streaming=False,
)
# tokenizer.train_from_iterator(batch_iterator(), trainer=trainer)
tokenizer.train_from_iterator(dataset, trainer=trainer, length=len(train_dataset))
del [train_dataset]
print(
tokenizer.normalizer.normalize_str(
"wie geht es dir? Héllò hôw1 are äöü? 1234.12 1,2 12 euro po12"
)
)
print(
tokenizer.pre_tokenizer.pre_tokenize_str(
"wie geht es dir? Héllò hôw1 are äöü? 1234.12 1,2 12 euro po12"
)
)
output = tokenizer.encode(
"wie geht es dir Beispielsatz in Deutsch Äoüß? Héllò hôw1 are äöü? 1234.12 1,2 12 euro po12"
)
print(output.tokens)
tokenizer.get_vocab_size()
words = pd.read_csv(
"/kaggle/input/3-million-german-sentences/deu_news_2015_3M-words.csv", dtype=str
)
words.rename(columns={"!": "word", "53658": "count"}, inplace=True)
words = words[["word", "count"]]
words.head()
many_tokens = []
for word in tqdm(words["word"].tolist()):
if not isinstance(word, str):
continue
enc = tokenizer.encode(word)
if len(enc) > 2: # ignore cls and sep
many_tokens.append(word)
many_tokens[1]
len(many_tokens)
tokenizer.save(f"/kaggle/working/tokenizer.json")
# ## Model Training
train_dataset = load_dataset(
"parquet",
data_files="/kaggle/working/train_data.snap.parquet",
streaming=True,
split="train",
)
eval_dataset = load_dataset(
"parquet",
data_files="/kaggle/working/eval_data.snap.parquet",
streaming=True,
split="train",
)
model_path = "/kaggle/working/model"
use_mlm = True
mlm_probability = 0.2 # still keeping the 80 - 10 - 10 rule
batch_size = 16
seed, buffer_size = 42, 10_000
max_length = 512
block_size = 512
max_position_embeddings = 512
hidden_size = 768
num_hidden_layers = 2 # 12
num_attention_heads = 2 # 12
intermediate_size = 3072
drop_out = 0.1
config = BertConfig(
attention_window=[block_size] * num_attention_heads,
# mask_token_id = 4,
bos_token_id=1,
sep_token_id=2,
# pad_token_id = 3,
eos_token_id=2,
max_position_embeddings=max_position_embeddings,
hidden_size=hidden_size,
num_hidden_layers=num_hidden_layers,
num_attention_heads=num_attention_heads,
intermediate_size=intermediate_size,
hidden_act="gelu",
hidden_dropout_prob=drop_out,
attention_probs_dropout_prob=drop_out,
type_vocab_size=2,
initializer_range=0.02,
layer_norm_eps=1e-12,
vocab_size=vocab_size,
use_cache=True,
classifier_dropout=None,
onnx_export=False,
)
# train_dataset = load_dataset('parquet', data_files=f'../corpus/lufthansa.snap.parquet',
# streaming=True,
# split="train"
# )
# eval_dataset = load_dataset('parquet', data_files=f'../corpus/lufthansa.snap.parquet',
# streaming=True,
# split="train"
# )
def encode(examples, max_length, tokenizer):
return tokenizer.batch_encode_plus(
examples["text"],
padding=True,
truncation=True,
max_length=max_length,
# return_special_tokens_mask=True,
# return_tensors="pt"
)
tk_tokenizer = Tokenizer.from_file(f"/kaggle/working/tokenizer.json")
tokenizer = PreTrainedTokenizerFast(tokenizer_object=tk_tokenizer)
tokenizer.add_special_tokens(
{
"pad_token": "[PAD]",
"unk_token": "[UNK]",
"sep_token": "[SEP]",
"cls_token": "[CLS]",
"bos_token": "[CLS]",
"eos_token": "[SEP]",
"mask_token": "[MASK]",
}
)
train_dataset = train_dataset.map(
encode,
remove_columns=["text"],
batched=True,
batch_size=batch_size,
fn_kwargs={"max_length": max_length, "tokenizer": tokenizer},
)
eval_dataset = eval_dataset.map(
encode,
remove_columns=["text"],
batched=True,
batch_size=batch_size,
fn_kwargs={"max_length": max_length, "tokenizer": tokenizer},
)
train_dataset = train_dataset.with_format("torch")
eval_dataset = eval_dataset.with_format("torch")
data_collator = DataCollatorForLanguageModeling(
tokenizer=tokenizer, mlm=use_mlm, mlm_probability=mlm_probability
)
model = BertForMaskedLM(config=config)
print(f"n of parameters: {model.num_parameters()}")
# comp_model = torch.compile(model)
# print(f"n of parameters: {comp_model.num_parameters()}")
# ### Trainer config
early_stopping = EarlyStoppingCallback(
early_stopping_patience=3, early_stopping_threshold=0.02
)
callbacks = [early_stopping]
learning_rate = 1e-4 # bert
weight_decay = 1e-2 # bert
lr_scheduler_type = "linear"
num_train_epochs = 1 # 5 but training set is small
train_batch_size = 32
eval_batch_size = 32
gradient_accumulation_steps = 2
eval_accumulation_steps = 2
warmup_steps = 1_000
adam_beta1 = 0.9 # bert
adam_beta2 = 0.999 # bert
adam_epsilon = 1e-8 # bert
max_grad_norm = 1.0 # bert
max_steps = len(train_dataset) // train_batch_size # 1_000_000
print(max_steps)
training_args = TrainingArguments(
output_dir=model_path,
overwrite_output_dir=True,
learning_rate=learning_rate,
weight_decay=weight_decay,
lr_scheduler_type=lr_scheduler_type,
num_train_epochs=num_train_epochs,
adam_beta1=adam_beta1,
adam_beta2=adam_beta2,
adam_epsilon=adam_epsilon,
max_grad_norm=max_grad_norm,
evaluation_strategy="steps",
eval_steps=5_000,
max_steps=max_steps,
per_device_train_batch_size=train_batch_size, # depends on memory
per_device_eval_batch_size=eval_batch_size,
gradient_accumulation_steps=gradient_accumulation_steps,
# eval_accumulation_steps=eval_accumulation_steps,
save_strategy="steps",
save_steps=5_000,
save_total_limit=3,
prediction_loss_only=False,
report_to="tensorboard",
log_level="warning",
logging_strategy="steps",
# fp16 = True,
# fp16_full_eval=True,
load_best_model_at_end=True,
metric_for_best_model="loss",
greater_is_better=False,
push_to_hub=False,
dataloader_pin_memory=True,
)
def compute_metrics(eval_pred):
logits, labels = eval_pred
predictions = np.argmax(logits, axis=-1)
accuracy = accuracy_score(y_true=labels, y_pred=predictions)
recall = recall_score(y_true=labels, y_pred=predictions, average="weighted")
precision = precision_score(y_true=labels, y_pred=predictions, average="weighted")
f1 = f1_score(y_true=labels, y_pred=predictions, average="weighted")
return {"accuracy": accuracy, "precision": precision, "recall": recall, "f1": f1}
trainer = Trainer(
model=model,
args=training_args,
data_collator=data_collator,
train_dataset=train_dataset,
eval_dataset=eval_dataset,
# compute_metrics=compute_metrics,
tokenizer=tokenizer,
callbacks=callbacks,
)
trainer.train()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
print("Hello World!")
x = 5
y = "Boo Boo"
print(x)
y
x = 80
if x < 60:
print("x is smaller")
print("ABCD")
else:
print("x is greater")
print("EFGH")
x
x = 55
x
x = 5
y = str(5)
z = float(4)
x
y
z
x = 5
y = "Hello World"
z = 3.33
print(x)
print(y)
print(z)
x, y, z = 55, "Hello World 5", 3.335
print(x)
print(y)
print(z)
print(x, y, z)
print("The value of x is", x)
print("The value of y is " + y)
print("The value of y is", y)
print("The value of z is ", z)
x = "abcd "
y = "efgh"
z = x + y
z
|
# Roman numerals are written left to right with symbols of decreasing magnitude.
# They are broken down straightforwardly with the following components:
# Numerals=[I:1, IV:4, V:5, IX:9, X:10, XL:40, L:50, XC:90, C:100, CD:400, D:500, CM:900, M:1000]
# Task: construct any number 1-1000
# Make sure that the function only accepts numbers in the range 1-1000
# Anything else should return an error message
x = 5
type(x) is int
def check(x):
if type(x) is int and 1 <= x <= 1000:
return True
else:
return "No"
check(x)
a = 2000
b = "string"
c = 55.5
print(check(a))
print(check(b))
print(check(c))
# We will iterate over the symbols, appending them to a string while subtracting their value from x:
roman = ""
x = 100
x -= 10
roman += "X"
print(x)
print(roman)
# Constructing the generator, incorporating this check:
# Reminder of the symbols we need:
# [I:1, IV:4, V:5, IX:9, X:10, XL:40, L:50, XC:90, C:100, CD:400, D:500, CM:900, M:1000]
def RomanNumeral(x):
Numeral = "" # start with an empty string
if type(x) is int and 1 <= x <= 1000:
while x > 0:
if x == 1000:
x -= 1000
Numeral += "M"
elif 900 <= x < 1000:
x -= 900
Numeral += "CM"
elif 500 <= x < 900:
x -= 500
Numeral += "D"
elif 400 <= x < 500:
x -= 400
Numeral += "CD"
elif 100 <= x < 400:
x -= 100
Numeral += "C"
elif 90 <= x < 100:
x -= 90
Numeral += "XC"
elif 50 <= x < 90:
x -= 50
Numeral += "L"
elif 40 <= x < 50:
x -= 40
Numeral += "XL"
elif 10 <= x < 40:
x -= 10
Numeral += "X"
elif x == 9:
x -= 9
Numeral += "IX"
elif 5 <= x < 9:
x -= 5
Numeral += "V"
elif x == 4:
x -= 4
Numeral += "IV"
elif 1 <= x < 4:
x -= 1
Numeral += "I"
return Numeral
else:
return "Please input an integer between 1 and 1000"
print(RomanNumeral(145))
print(RomanNumeral(678))
print(RomanNumeral(999))
print(RomanNumeral(-100))
print(RomanNumeral(3000))
print(RomanNumeral("three hundred"))
print(RomanNumeral(300.00))
# Can the code be streamlined using a for loop:
Numerals = {
"I": 1,
"IV": 4,
"V": 5,
"IX": 9,
"X": 10,
"XL": 40,
"L": 50,
"XC": 90,
"C": 100,
"CD": 400,
"D": 500,
"CM": 900,
"M": 1000,
}
Numerals = dict(sorted(Numerals.items(), key=lambda x: x[1], reverse=True))
def RomanNumeral(x):
Numeral = "" # start with an empty string
if type(x) is int and 1 <= x <= 1000:
while x > 0:
for i in Numerals:
if x >= Numerals[i]:
x -= Numerals[i]
Numeral += i
break
return Numeral
else:
return "Please input an integer between 1 and 1000"
print(RomanNumeral(145))
print(RomanNumeral(678))
print(RomanNumeral(999))
print(RomanNumeral(-100))
print(RomanNumeral(3000))
print(RomanNumeral("three hundred"))
print(RomanNumeral(300.00))
|
# kutuphanelerin yuklenmesi
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# # OLCULERİN FORMULLERİ
# 
# 
# 
# # 1. Denklem
# ***b = 2.90, w1 = 0.04, w2 = 0.17, w3= 0.002***
#
# verinin yuklenmesi
df = pd.read_csv("../input/advertising-data/Advertising.csv")
# gereksiz degiskenlerin düsürulmes
df = df.drop("Unnamed: 0", axis=1)
# ilk 5 gozlem
df.head()
# tahminlerin hesaplanması
df[["yhat"]] = df.iloc[:, 0:3].apply(
lambda x: 2.90 + 0.04 * x[0] + 0.17 * x[1] + 0.002 * x[2], axis=1
)
# gercek ve tahmin arasındaki farkın hesaplanması
df[["difference"]] = df.apply(lambda x: x[3] - x[4], axis=1)
# farkların karelerinin alınması
df[["square"]] = df.apply(lambda x: x[5] ** 2, axis=1)
# mean squared error degerinin hesaplanması
mse1 = df.square.sum() / len(df)
# root mean square error degerinin hesaplanması
rmse1 = np.sqrt(mse1)
# mean absolute error degerinin hesaplanması
mae1 = np.abs(df.difference).sum() / len(df)
# mse , rmse ve mae'nin birlikte gosterimi
print(mse1, rmse1, mae1)
# # 2. Denklem
# ***b = 1.70, w1 = 0.09, w2 = 0.20, w3= 0.017***
# verinin yuklenmesi
df = pd.read_csv("../input/advertising-data/Advertising.csv")
# gereksiz degiskenlerin düsürulmes
df = df.drop("Unnamed: 0", axis=1)
# ilk 5 gozlem
df.head()
# tahminlerin hesaplanması
df[["yhat"]] = df.iloc[:, 0:3].apply(
lambda x: 1.70 + 0.09 * x[0] + 0.20 * x[1] + 0.017 * x[2], axis=1
)
# gercek ve tahmin arasındaki farkın hesaplanması
df[["difference"]] = df.apply(lambda x: x[3] - x[4], axis=1)
# farkların karelerinin alınması
df[["square"]] = df.apply(lambda x: x[5] ** 2, axis=1)
# mean squared error degerinin hesaplanması
mse2 = df.square.sum() / len(df)
# root mean square error degerinin hesaplanması
rmse2 = np.sqrt(mse2)
# mean absolute error degerinin hesaplanması
mae2 = np.abs(df.difference).sum() / len(df)
# mse , rmse ve mae'nin birlikte gosterimi
print(mse2, rmse2, mae2)
# Hesaplanan olculerin birlikte gosterilmesi
measure = pd.DataFrame(
[mse1, rmse1, mae1, mse2, rmse2, mae2],
index=["mse1", "rmse1", "mae1", "mse2", "rmse2", "mae2"],
)
# olculere isim verilmese
measure.columns = ["measures"]
# olculerin gosterimi
measure
|
# # Pandas revision - 101
# - This notebook contains Pandas revision
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
# ## Method Chaining
df = pd.read_csv(
"https://raw.githubusercontent.com/cajjster/data_files/main/vgsales.csv"
)
df.head()
df["Genre"].value_counts().sort_index()
df["Genre"].sort_index().value_counts().plot.bar()
# ## `loc` and `iloc`
type(df.iloc[4])
# - Interger location
df.iloc[4:10]
df.iloc[1:10:3] # start:stop:step
df.iloc[-1:]
df.iloc[-1:, 2:5] # r,c
# - Column name based
df.loc[:100, "NA_Sales"]
df.loc[:100, ["Platform", "NA_Sales"]]
df.loc[100, "NA_Sales"] = 4.55
df.loc[100, "NA_Sales"] # change or update particular value
# ## Filter with conditional logic
for i in df[["Name", "Platform", "Year", "Genre", "Publisher"]]:
print(i.upper())
print(df[i].unique())
print()
wii = df["Platform"] == "Wii"
wii.value_counts()
df.loc[wii]
df.loc[df["Platform"] == "PS2"]
# ## Video gammes those has higher number of sales in japan than EU
df.loc[df["JP_Sales"] > df["EU_Sales"], "Name"]
df.loc[(df["Platform"] == "Wii") & (df["Genre"] == "Role-Playing")].sort_values(
by="Global Sales", ascending=False
)
## Alternate wway for above technique
rpg = df["Genre"] == "Role-Playing"
df.loc[wii & rpg].reset_index(drop=True)
## Filter with OR
action = df["Genre"] == "Action"
shooter = df["Genre"] == "Shooter"
df.loc[action | shooter]
# filter with both or and condition
since_2010 = df["Year"] == 2010
df.loc[(since_2010) & (action | shooter)]
# not using () will give you different outputs
df.loc[since_2010 & action | shooter]
# #### isin()
top_3_pub = df["Publisher"].value_counts()[:3].index.tolist()
top_3_pub_isin = df.Publisher.isin(top_3_pub)
top_3_pub_df = df.loc[top_3_pub_isin]
top_3_pub_df
genres = df.Genre.isin(["Sports", "Action", "Shooter"])
sales_2M = df["Global Sales"] >= 2
df.loc[genres & sales_2M]
df.loc[genres & top_3_pub_isin & sales_2M]
## Between() allows us to specify a range'
between_2m_10_m = df["Global Sales"].between(2, 10)
y_90 = df["Year"].between(1990, 1999)
y_00 = df["Year"].between(2000, 2010)
y_10 = df["Year"].between(2010, 2019)
df.loc[between_2m_10_m & y_90, "Genre"].value_counts(
normalize=True
).sort_index().plot.bar()
df.loc[between_2m_10_m & y_00, "Genre"].value_counts(normalize=True)
df.loc[between_2m_10_m & y_10, "Genre"].value_counts(normalize=True).plot.bar()
## Leta find out which Racing games has the highest sales in EU compared to the NA.JP and Other Sales
# between 2000 & 2010
race = df["Genre"] == "Racing"
highest_EU = (
(df["EU_Sales"] > df["JP_Sales"])
& (df["EU_Sales"] > df["NA_Sales"])
& (df["EU_Sales"] > df["Other_Sales"])
)
y_00s = df["Year"].between(2000, 2019)
df.loc[race & highest_EU & y_00s, ["Name", "EU_Sales"]].sort_values(
by="EU_Sales", ascending=False
).head().set_index("Name")
# ## Most useful methods for data analysis
df.query("NA_Sales < JP_Sales")
df.query("Publisher == 'Ubisoft' and Year > 2014")
ps3 = df.query("Platform == 'PS3'")
# unique and nunique()
df.Platform.unique()
df.nunique()
df["Year"].nunique()
# for i in df.select_dtypes("object"):
# print(df[i].unique())
ps3.reset_index(drop=True, inplace=True)
ps3
ps3.set_index("Name", inplace=True)
ps3.rename({"EU_Sales": "Europe", "NA_Sales": "US"}, axis=1)
ps3.rename({"Call of Duty: Black Ops II ": "COD Black OPS 2"})
## nlargest and nsmallest
ps3.nlargest(n=10, columns="NA_Sales").loc[:, "NA_Sales"].plot.bar()
import matplotlib.pyplot as plt
import seaborn as sns
fig, ax = plt.subplots(2, 2, figsize=(20, 20))
ps3.nlargest(n=10, columns="NA_Sales").loc[:, "NA_Sales"].plot.bar(ax=ax[0][0])
ax[1][0].set_title("North America")
ps3.nlargest(n=10, columns="EU_Sales").loc[:, "EU_Sales"].plot.bar(ax=ax[0][1])
ax[0][1].set_title("Europe")
ps3.nlargest(n=10, columns="JP_Sales").loc[:, "JP_Sales"].plot.bar(ax=ax[1][0])
ax[1][0].set_title("Japan")
ps3.nlargest(n=10, columns="Other_Sales").loc[:, "Other_Sales"].plot.bar(ax=ax[1][1])
ax[1][1].set_title("Other")
fig.suptitle("Most sold PS3 games by Continent ", fontsize=20)
fig.tight_layout()
# fig, ax = plt.subplots(2,2,figsize=(20,20))
# ps3.nsmallest(n=5,columns='NA_Sales').loc[:,"NA_Sales"].plot.bar(ax=ax[0][0])
# ax[1][0].set_title("North America")
# ps3.nsmallest(n=5,columns='EU_Sales').loc[:,"EU_Sales"].plot.bar(ax=ax[0][1])
# ax[0][1].set_title("Europe")
# ps3.nsmallest(n=5,columns='JP_Sales').loc[:,"JP_Sales"].plot.bar(ax=ax[1][0])
# ax[1][0].set_title("Japan")
# ps3.nsmallest(n=5,columns='Other_Sales').loc[:,"Other_Sales"].plot.bar(ax=ax[1][1])
# ax[1][1].set_title("Other")
# fig.suptitle("Least sold PS3 games by Continent ", fontsize=20)
# fig.tight_layout()
ps3.nsmallest(n=5, columns="Global Sales")
top_100 = ps3.nlargest(n=100, columns="Global Sales")
top_100["Genre"].value_counts().plot.barh()
print(df.Year.min())
print(df.Year.max())
df["Global Sales"].quantile(0.99)
df.EU_Sales.agg({"min", "max", "mean", "median", "std", "sum", "count"})
xbox_sales = (
df.loc[df.Platform == "XB", ["NA_Sales", "Year"]].groupby("Year")["NA_Sales"].sum()
)
xbox_sales
xbox_sales.cumsum()
xbox_sales.cumsum().plot.area()
# cut and qcut
# cut gives you same bin size
pd.cut(x=df["Year"], bins=5).value_counts()
pd.cut(
x=df["Year"],
bins=[1980, 1989, 1999, 2009, 2019],
labels=["80s", "90s", "00s", "10s"],
).value_counts()
df["Decade"] = pd.cut(
x=df["Year"],
bins=[1980, 1989, 1999, 2009, 2019],
labels=["80s", "90s", "00s", "10s"],
include_lowest=True,
)
df
# qcut quantile cut
pd.qcut(x=df["Global Sales"], q=4, precision=50).value_counts()
sony = ["PS", "PS2", "PS3", "PS4", "PSP", "PSV"]
nintendo = ["N64", "GB", "GBA", "GC", "Wii", "Wiiu", "DS", "NES", "3DS", "SNES"]
microsoft = ["XB", "XB360", "XOne"]
pc = ["PC"]
atari = ["2600"]
panasonic = ["3DO"]
sega = ["DC", "GG", "SAT", "SCD"]
def categories(x):
if x in sony:
return "Sony"
elif x in nintendo:
return "Nintendo"
elif x in microsoft:
return "Microsoft"
elif x in pc:
return "PC"
elif x in sega:
return "Sega"
else:
return "Other"
df["Platform"].apply(categories).value_counts()
df["Platform_Company"] = df["Platform"].apply(categories)
df
import scipy.stats as stats
z_score = df.select_dtypes("float").apply(lambda x: stats.zscore(x))
z_score.mean()
z_score.std()
## correlation
corr_matrix = df.drop("Year", axis=1).corr()
corr_matrix
sns.heatmap(corr_matrix, annot=True)
import numpy as np
matrix = np.triu(corr_matrix)
sns.heatmap(corr_matrix, annot=True, mask=matrix)
# ## GroupBY
df.groupby("Genre")["NA_Sales"].mean()
df.Genre.unique()
print(df["NA_Sales"].mean())
df.loc[df["Genre"] == "Shooter", "NA_Sales"].mean()
sales_by_genre = df.groupby("Genre")[
["NA_Sales", "EU_Sales", "JP_Sales", "Other_Sales"]
].mean()
sales_by_genre
plt.figure(figsize=(12, 8))
sns.heatmap(sales_by_genre, annot=True, cmap="Greens")
df.groupby("Platform_Company")["JP_Sales"].agg("mean")
df.groupby("Platform_Company")["JP_Sales"].agg(["mean", "median", "count"])
df.groupby("Platform_Company")[["JP_Sales", "Global Sales"]].agg(
{"JP_Sales": ["mean", "median", "count"], "Global Sales": ["mean"]}
)
df.groupby(["Platform_Company", "Decade"])[["Global Sales"]].sum()
platform_sales_per_decade = (
df.groupby(["Decade", "Platform_Company"])[["Global Sales"]].sum().unstack()
)
df.groupby(["Decade", "Platform_Company"])[["Global Sales"]].sum().unstack(
-1
).plot.line()
platform_sales_per_decade.columns.get_level_values(1)
platform_sales_per_decade_columns = platform_sales_per_decade.columns.get_level_values(
1
)
platform_sales_per_decade_columns
platform_sales_per_decade.plot.line()
# - Crosstab: “Compute a simple cross-tabulation of two (or more) factors. By default computes a frequency table of the factors unless an array of values and an aggregation function are passed.”
# - Pivot Table: “Create a spreadsheet-style pivot table as a DataFrame. The levels in the pivot table will be stored in MultiIndex objects (Hierarchical indexes on the index and columns of the result DataFrame.
# ### Pivot Table
pd.pivot_table(df.drop("Year", axis=1), index="Platform_Company", aggfunc="mean")
## create multiple indexes and spcify its value and aggfunc
pd.pivot_table(df, index=["Platform_Company", "Decade"], aggfunc="mean")
# ### Crosstab
pd.crosstab(index=df["Platform_Company"], columns=df["Decade"])
# multiple indexes and a column
pd.crosstab(index=[df["Platform_Company"], df["Publisher"]], columns=df["Decade"])
df.columns
# single index and multple columns
pd.crosstab(
index=df["Platform_Company"], columns=[df["Genre"], df["Publisher"]], margins=True
)
# ### Pivot Table or CrossTab?
# Choose how you want to represent the data. At bare minimum, the pivot table compares the index and yields numerical values. Additionally, the crosstab compares the index and columns which yield the count.
# ## Join, Merge and Concat
# #### Split dataframe where we have Publisher is Activision
activision_df = df.loc[df["Publisher"] == "Activision"].copy()
ea_df = df.loc[df["Publisher"] == "Electronic Arts"].copy()
pd.merge(ea_df, activision_df, how="inner", on="Platform")
pd.concat([ea_df, activision_df], axis=0, ignore_index=True)
# ### Apply, Applymap and map
nyc = pd.read_csv(
"https://raw.githubusercontent.com/misraturp/Pandas-apply-vs.-map-vs.-applymap/main/NYC_Jobs.csv"
)
nyc.head()
nyc = nyc[
[
"Job ID",
"Civil Service Title",
"Agency",
"Posting Type",
"Job Category",
"Salary Range From",
"Salary Range To",
]
]
nyc.head()
## Capitalize the text
### Apply is good for applying a function to either axis or the
### whole dataframe to the function
capitalize = lambda x: x.upper()
df["Publisher"].apply(capitalize)
df
## difference between apply and applymap is that apply passes the dataframe to the function
## one axis at a time
## applymap passes one element at a time
def add_year(text):
return str(text) + "_2022"
nyc.applymap(add_year)
## map works on series and its main strength is to replace values
## so given a series like that
s = pd.Series(["cat", "dog", "NaN", "horse"])
s
# we can change the values using a dict only
s.map({"cat": "kitten", "dog": "puppy"})
# but you can also still pass a function to it
# this, however, can be done using apply too, so nothing special
# thus, map is mainly useful for when you want to completely map elements from
# one value to another
def change_word(title):
title = title.replace("DEPT", "kitten")
return title
nyc["Agency"].map(change_word)
nyc["Agency"].map("The position is created by {}".format)
# for both map and applymap, you can specify na_action to ignore,
# so that NA values will not even be passed to the function
# the result does not change
s.map({"cat": "kitten", "dog": "puppy"}, na_action="")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
df = pd.read_csv("/kaggle/input/student-marks-dataset/Student_Marks.csv")
df.sample(10)
# df[df["time_study"]==3.736]
df.info()
df.isnull().sum()
df.describe()
sns.heatmap(df.corr(), annot=True)
figure = plt.scatter(x=df["time_study"], y=df["Marks"])
plt.hist(x=df["number_courses"], bins=25, color="red")
figure = plt.scatter(x=df["number_courses"], y=df["Marks"])
x = df.drop(["Marks"], axis=1)
y = df["Marks"]
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.2, random_state=42
)
model = LinearRegression()
model.fit(x_train, y_train)
pred = model.predict(x_test)
from sklearn.metrics import r2_score, mean_squared_error, mean_absolute_error
r2_score(y_test, pred)
model.score(x_test, y_test)
df[df["time_study"] == 3.736]
features = [[3.736, 4]]
model.predict(features)
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sea
df = pd.read_csv("../input/titanic/train.csv")
df.shape
df.head(5)
# ## Analysing data
sea.countplot(x="Survived", data=df)
sea.countplot(x="Survived", hue="Sex", data=df)
sea.countplot(x="Survived", hue="Pclass", data=df)
df["Age"].plot.hist()
df["Fare"].plot.hist(bins=10, figsize=(10, 5))
df.info()
sea.countplot(x="SibSp", data=df)
# ## Data Wranglling
df.isnull()
df.isnull().sum()
sea.heatmap(df.isnull())
df.head(5)
df.drop("Cabin", axis=True, inplace=True)
df.head(2)
df.dropna(inplace=True)
sea.heatmap(df.isnull())
df.head(3)
# # **Normalization & Standardization**
df["Fare"] = (df["Fare"] - df["Fare"].min()) / (df["Fare"].max() - df["Fare"].min())
df["Age"] = (df["Age"] - df["Age"].mean()) / df["Age"].std()
df["Sex"] = df["Sex"].factorize()[0]
df["Sex"]
df["Embarked"] = df["Embarked"].factorize()[0]
df["Embarked"]
df.head(5)
df = df.drop(columns=["Ticket", "Name", "PassengerId"])
# # **TEST DATA**
df2 = pd.read_csv("../input/titanic/test.csv")
df2.isnull().sum()
df2
df2.head(3)
df2.isnull().sum()
df2.drop("Cabin", axis=1, inplace=True)
df2["Age"] = df2["Age"].fillna(df2["Age"].mean())
df2["Fare"] = df2["Fare"].fillna(df2["Fare"].mean())
df2.isnull().sum()
df2["Sex"] = df2["Sex"].factorize()[0]
df2["Embarked"] = df2["Embarked"].factorize()[0]
df2.head()
# # **Normalization & Standardization**
df2["Fare"] = (df2["Fare"] - df2["Fare"].min()) / (
df2["Fare"].max() - df2["Fare"].min()
)
df2["Age"] = (df2["Age"] - df2["Age"].mean()) / df2["Age"].std()
df2
df2.drop(["Name", "Ticket", "PassengerId"], axis=1, inplace=True)
df2.head(7)
# # **Training Data**
X_train = df.drop("Survived", axis=1)
Y_train = df["Survived"]
X_train.isnull().sum()
from sklearn.model_selection import train_test_split
train_x, val_x, train_y, val_y = train_test_split(X_train, Y_train)
from sklearn.linear_model import LogisticRegression
from sklearn.neighbors import KNeighborsClassifier
from sklearn.svm import SVC
from sklearn.ensemble import RandomForestClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.tree import DecisionTreeClassifier
from sklearn.metrics import accuracy_score
# Define the models
model_1 = LogisticRegression(random_state=1)
model_2 = RandomForestClassifier()
model_3 = KNeighborsClassifier()
model_4 = SVC()
model_5 = GaussianNB()
model_6 = DecisionTreeClassifier()
def score_model(models, X_train=train_x, X_val=val_x, y_train=train_y, y_val=val_y):
models.fit(X_train, y_train)
predictions = models.predict(X_val)
return accuracy_score(y_val, predictions)
models = [model_1, model_2, model_3, model_4, model_5, model_6]
for i in range(0, len(models)):
acc = score_model(models[i])
print("model {} accuracy {}".format(i + 1, acc))
model_1.fit(train_x, train_y)
pred = model_1.predict(df2)
sub = pd.read_csv("../input/titanic/gender_submission.csv")
sub.head(2)
submit5 = pd.DataFrame({"PassengerId": sub["PassengerId"], "Survived": pred})
submit5.to_csv("submit.csv", index=False)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
from sklearn.preprocessing import StandardScaler
from sklearn.metrics import mean_squared_error, r2_score, make_scorer
from sklearn.linear_model import LinearRegression, Lasso, Ridge
from sklearn.tree import DecisionTreeRegressor
from sklearn.ensemble import RandomForestRegressor, GradientBoostingRegressor
from sklearn.model_selection import GridSearchCV
import matplotlib.pyplot as plt
plt.style.use("fivethirtyeight")
# load data
df = pd.read_csv("/kaggle/input/insurance/insurance.csv")
df.sample(10)
# distribution of label
# Get the label column
label = df["charges"]
# Create a figure for 2 subplots (2 rows, 1 column)
fig, ax = plt.subplots(2, 1, figsize=(18, 10))
# Plot the histogram
ax[0].hist(label, bins=100)
ax[0].set_ylabel("Frequency")
# Add lines for the mean, median, and mode
ax[0].axvline(label.mean(), color="magenta", linestyle="dashed", linewidth=2)
ax[0].axvline(label.median(), color="cyan", linestyle="dashed", linewidth=2)
# Plot the boxplot
ax[1].boxplot(label, vert=False)
ax[1].set_xlabel("Charges")
# Add a title to the Figure
fig.suptitle("Charges Distribution")
# Show the figure
fig.show()
# box_plots
cat_features = ["age", "sex", "children", "smoker", "region"]
for col in cat_features:
df_outliers = df.charges.quantile(0.95)
df_outliers_removed = df[df.charges <= df_outliers]
df_outliers_removed.boxplot("charges", col, figsize=(20, 5))
plt.suptitle("")
plt.title("charges by " + col)
plt.show()
# scatter plot
df.plot.scatter("bmi", "charges", figsize=(8, 8))
correlation = df.corr()["charges"]["bmi"]
plt.title("bmi correlation with charges: " + str(correlation))
plt.show()
# one-hot encoding
df = pd.get_dummies(df)
# Separate features and labels
X, y = df.drop("charges", 1).values, df["charges"].values
print("Features:", X[:10], "\nLabels:", y[:10], sep="\n")
# scale features
scaler = StandardScaler()
X_scaled = scaler.fit_transform(X)
from sklearn.model_selection import train_test_split
# Split data 70%-30% into training set and test set
X_train, X_test, y_train, y_test = train_test_split(
X_scaled, y, test_size=0.30, random_state=0
)
print("Training Set: %d rows\nTest Set: %d rows" % (X_train.shape[0], X_test.shape[0]))
# Fit a linear regression model on the training set
model = LinearRegression().fit(X_train, y_train)
# Evaluate the model using the test data
predictions = model.predict(X_test)
mse = mean_squared_error(y_test, predictions)
print("MSE:", mse)
rmse = np.sqrt(mse)
print("RMSE:", rmse)
r2 = r2_score(y_test, predictions)
print("R2:", r2)
# Plot predicted vs actual
plt.scatter(y_test, predictions)
plt.xlabel("Actual Labels")
plt.ylabel("Predicted Labels")
plt.title("Insurance Cost Predictions")
# overlay the regression line
z = np.polyfit(y_test, predictions, 1)
p = np.poly1d(z)
plt.plot(y_test, p(y_test), color="magenta")
plt.show()
# Fit a lasso model on the training set
model = Lasso().fit(X_train, y_train)
# Evaluate the model using the test data
predictions = model.predict(X_test)
mse = mean_squared_error(y_test, predictions)
print("MSE:", mse)
rmse = np.sqrt(mse)
print("RMSE:", rmse)
r2 = r2_score(y_test, predictions)
print("R2:", r2)
# Plot predicted vs actual
plt.scatter(y_test, predictions)
plt.xlabel("Actual Labels")
plt.ylabel("Predicted Labels")
plt.title("Insurance Cost Predictions")
# overlay the regression line
z = np.polyfit(y_test, predictions, 1)
p = np.poly1d(z)
plt.plot(y_test, p(y_test), color="magenta")
plt.show()
# Fit a decision tree model on the training set
model = DecisionTreeRegressor().fit(X_train, y_train)
# Evaluate the model using the test data
predictions = model.predict(X_test)
mse = mean_squared_error(y_test, predictions)
print("MSE:", mse)
rmse = np.sqrt(mse)
print("RMSE:", rmse)
r2 = r2_score(y_test, predictions)
print("R2:", r2)
# Plot predicted vs actual
plt.scatter(y_test, predictions)
plt.xlabel("Actual Labels")
plt.ylabel("Predicted Labels")
plt.title("Insurance Cost Predictions")
# overlay the regression line
z = np.polyfit(y_test, predictions, 1)
p = np.poly1d(z)
plt.plot(y_test, p(y_test), color="magenta")
plt.show()
# Train the model
model = RandomForestRegressor().fit(X_train, y_train)
# Evaluate the model using the test data
predictions = model.predict(X_test)
mse = mean_squared_error(y_test, predictions)
print("MSE:", mse)
rmse = np.sqrt(mse)
print("RMSE:", rmse)
r2 = r2_score(y_test, predictions)
print("R2:", r2)
# Plot predicted vs actual
plt.scatter(y_test, predictions)
plt.xlabel("Actual Labels")
plt.ylabel("Predicted Labels")
plt.title("Insurance Cost Predictions")
# overlay the regression line
z = np.polyfit(y_test, predictions, 1)
p = np.poly1d(z)
plt.plot(y_test, p(y_test), color="magenta")
plt.show()
# Train the model
from sklearn.ensemble import GradientBoostingRegressor
# Fit a lasso model on the training set
model = GradientBoostingRegressor().fit(X_train, y_train)
# Evaluate the model using the test data
predictions = model.predict(X_test)
mse = mean_squared_error(y_test, predictions)
print("MSE:", mse)
rmse = np.sqrt(mse)
print("RMSE:", rmse)
r2 = r2_score(y_test, predictions)
print("R2:", r2)
# Plot predicted vs actual
plt.scatter(y_test, predictions)
plt.xlabel("Actual Labels")
plt.ylabel("Predicted Labels")
plt.title("Insurance Cost Predictions")
# overlay the regression line
z = np.polyfit(y_test, predictions, 1)
p = np.poly1d(z)
plt.plot(y_test, p(y_test), color="magenta")
plt.show()
# Use a Gradient Boosting algorithm
alg = GradientBoostingRegressor()
# Try these hyperparameter values
params = {
"learning_rate": [0.001, 0.01, 0.1, 0.5, 1.0],
"n_estimators": [20, 50, 100, 150],
}
# Find the best hyperparameter combination to optimize the R2 metric
score = make_scorer(r2_score)
gridsearch = GridSearchCV(alg, params, scoring=score, cv=5, return_train_score=True)
gridsearch.fit(X_train, y_train)
print("Best parameter combination:", gridsearch.best_params_, "\n")
# Get the best model
model = gridsearch.best_estimator_
print(model, "\n")
# Evaluate the model using the test data
predictions = model.predict(X_test)
mse = mean_squared_error(y_test, predictions)
print("MSE:", mse)
rmse = np.sqrt(mse)
print("RMSE:", rmse)
r2 = r2_score(y_test, predictions)
print("R2:", r2)
# Plot predicted vs actual
plt.scatter(y_test, predictions)
plt.xlabel("Actual Labels")
plt.ylabel("Predicted Labels")
plt.title("Insurance Cost Predictions")
# overlay the regression line
z = np.polyfit(y_test, predictions, 1)
p = np.poly1d(z)
plt.plot(y_test, p(y_test), color="magenta")
plt.show()
|
# Install dependencies
# -----------------------------------------------------------------------------
# Imports
# -----------------------------------------------------------------------------
import os, random, copy, yaml, pickle
from time import time, sleep
from tqdm import tqdm
from math import floor
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import pyarrow.parquet as pq
# torch imports
import torch
import torch.nn as nn
import torch.nn.init as init
import torch.nn.functional as F
from torch import Tensor
from torch.utils.data import random_split, IterableDataset
import torch_geometric
from torch_geometric.data import Data, Batch
from torch_geometric.utils import homophily
from torch_geometric.loader import DataLoader
from torch_geometric.nn import EdgeConv, knn_graph
from torch_scatter import scatter_add, scatter_mean, scatter_max, scatter_min
import pytorch_lightning as pl
# -----------------------------------------------------------------------------
# Helper function
# -----------------------------------------------------------------------------
import logging
import logging.config
import os, re, gc, psutil
def get_logger(name, msg):
"""
:param name: string
:param msg: DEBUG, INFO, WARNING, ERROR
:return: Logger() instance
"""
level = {
"DEBUG": logging.DEBUG,
"INFO": logging.INFO,
"WARNING": logging.WARNING,
"ERROR": logging.ERROR,
}
logging.basicConfig(
level=level[msg],
format="== %(name)s == %(asctime)s %(levelname)s:\t%(message)s",
datefmt="%H:%M:%S",
)
logger = logging.getLogger(name)
return logger
def walk_dir(dirname, batch_ids):
files = dict()
pattern = r"_(\d+)\.parquet"
if batch_ids is None:
batch_ids = list()
for base, _, names in os.walk(dirname):
for name in names:
match = re.findall(pattern, name)
batch_ids.append(int(match[0]))
files[int(match[0])] = os.path.join(base, name)
return files, batch_ids
for base, _, names in os.walk(dirname):
selected_files = dict()
for name in names:
match = re.findall(pattern, name)
if int(match[0]) in batch_ids:
selected_files[int(match[0])] = os.path.join(base, name)
files.update(selected_files)
return files, batch_ids
def memory_check(logger, msg=""):
gc.collect()
logger.debug(
f"memory usage {psutil.virtual_memory().used / 1024**3:.2f} "
f"of {psutil.virtual_memory().total / 1024**3:.2f} GB {msg}"
)
# -----------------------------------------------------------------------------
# Basic settings
# -----------------------------------------------------------------------------
# basic
BATCH_SIZE = 400
BATCHES_TEST = None #####################
EVENTS_PER_FILE = 200_000
# paths
BASE_PATH = "/kaggle/" ######################
PATH = os.path.join(BASE_PATH, "input", "icecube-neutrinos-in-deep-ice")
MODEL_PATH = os.path.join(BASE_PATH, "input", "ice-cube-model")
OUTPUT_PATH = os.path.join(BASE_PATH, "working")
TEST_PATH = os.path.join(PATH, "test")
META_PATH = os.path.join(OUTPUT_PATH, "test_meta")
# files
FILES_TEST, BATCHES_TEST = walk_dir(TEST_PATH, BATCHES_TEST)
FILE_META = os.path.join(PATH, "test_meta.parquet")
FILE_SENSOR_GEO = os.path.join(PATH, "sensor_geometry.csv")
FILE_GNNPre = os.path.join(MODEL_PATH, "official-pretrained.pth")
FILE_GNN = os.path.join(MODEL_PATH, "finetuned.ckpt")
FILE_BDT = os.path.join(MODEL_PATH, "BDT_clf.Baseline.0414.sklearn")
# -----------------------------------------------------------------------------
# Split meta file
# -----------------------------------------------------------------------------
if not os.path.exists(META_PATH):
os.mkdir(META_PATH)
meta_test = pd.read_parquet(FILE_META)
for i, df in meta_test.groupby("batch_id"):
print(f"processing {i} -> {df.shape}")
splitted = os.path.join(META_PATH, f"meta_{i}.parquet")
df.to_parquet(splitted)
# -----------------------------------------------------------------------------
# Some Logging
# -----------------------------------------------------------------------------
LOGGER = get_logger("IceCube", "DEBUG")
DEVICE = torch.device("cuda" if torch.cuda.is_available() else "cpu")
LOGGER.info(f"using {DEVICE}")
LOGGER.info(f"{len(FILES_TEST)} files for testing")
memory_check(LOGGER)
LOGGER.info(f"GNN model:{FILE_GNN}")
LOGGER.info(f"BDT model:{FILE_BDT}")
# -----------------------------------------------------------------------------
# Dataset
# -----------------------------------------------------------------------------
# sensor geometry
def prepare_sensors(scale=None):
sensors = pd.read_csv(FILE_SENSOR_GEO).astype(
{
"sensor_id": np.int16,
"x": np.float32,
"y": np.float32,
"z": np.float32,
}
)
if scale is not None and isinstance(scale, float):
sensors["x"] *= scale
sensors["y"] *= scale
sensors["z"] *= scale
return sensors
def angle_to_xyz(angles_b):
az, zen = angles_b.t()
x = torch.cos(az) * torch.sin(zen)
y = torch.sin(az) * torch.sin(zen)
z = torch.cos(zen)
return torch.stack([x, y, z], dim=1)
def xyz_to_angle(xyz_b):
x, y, z = xyz_b.t()
az = torch.arccos(x / torch.sqrt(x**2 + y**2)) * torch.sign(y)
zen = torch.arccos(z / torch.sqrt(x**2 + y**2 + z**2))
return torch.stack([az, zen], dim=1)
def angular_error(xyz_pred_b, xyz_true_b):
return torch.arccos(torch.clip_(torch.sum(xyz_pred_b * xyz_true_b, dim=1), -1, 1))
def angles2vector(df):
df["nx"] = np.sin(df.zenith) * np.cos(df.azimuth)
df["ny"] = np.sin(df.zenith) * np.sin(df.azimuth)
df["nz"] = np.cos(df.zenith)
return df
def vector2angles(n, eps=1e-8):
n = n / (np.linalg.norm(n, axis=1, keepdims=True) + eps)
azimuth = np.arctan2(n[:, 1], n[:, 0])
azimuth[azimuth < 0] += 2 * np.pi
zenith = np.arccos(n[:, 2].clip(-1, 1))
return np.concatenate([azimuth[:, np.newaxis], zenith[:, np.newaxis]], axis=1)
def series2tensor(series, set_device=None):
ret = torch.from_numpy(series.values).float()
if set_device is not None:
return ret.to(DEVICE)
return ret
def solve_linear(xw, yw, zw, xxw, yyw, xyw, yzw, zxw):
A = torch.tensor(
[
[xxw, xyw, xw],
[xyw, yyw, yw],
[xw, yw, 1],
]
)
b = torch.tensor([zxw, yzw, zw])
try:
coeff = torch.linalg.solve(A, b)
return coeff
except Exception:
LOGGER.debug("linear system not solvable")
return torch.zeros((3,))
def feature_extraction(df, fun=None, eps=1e-8): # list of variables
# sort by time
df.sort_values(["time"], inplace=True)
t = series2tensor(df.time)
c = series2tensor(df.charge)
x = series2tensor(df.x)
y = series2tensor(df.y)
z = series2tensor(df.z)
hits = t.numel() # hits
# weighted values
Sx = torch.sum(x)
Sxx = torch.sum(x * x)
Sxy = torch.sum(x * y)
Sy = torch.sum(y)
Syy = torch.sum(y * y)
Syz = torch.sum(y * z)
Sz = torch.sum(z)
Szx = torch.sum(z * x)
# error of plane estimate
coeff = solve_linear(Sx, Sy, Sz, Sxx, Syy, Sxy, Syz, Szx)
error = torch.sum((z - coeff[0] * x - coeff[1] * y - coeff[2]))
error = torch.square(error * 1e3) # error
# plane norm vector
norm_vec = torch.tensor([coeff[0], coeff[1], -1], dtype=torch.float)
norm_vec /= torch.sqrt(coeff[0] ** 2 + coeff[1] ** 2 + 1) # norm_vec -> (3, )
# delta t -> median time
dt = torch.quantile(
t, torch.tensor([0.15, 0.50, 0.85], dtype=torch.float)
) # dt -> (3, )
# charge centre (vector)
sumq = torch.sum(c) # sumq
meanq = sumq / hits # meanq
qv = torch.tensor(
[torch.sum(x * c), torch.sum(y * c), torch.sum(z * c)], dtype=torch.float
)
qv /= sumq # qv -> (3, )
# bright sensor ratio
bratio = c[c > 5 * meanq].numel() / hits # bratio
# grouping by time (remember to sort by time)
n_groups = 4 # xyzt -> (16, )
if hits > n_groups:
sec_len = floor(hits / n_groups)
remain_len = hits - (n_groups - 1) * sec_len
xyzt = series2tensor(df[["x", "y", "z", "time"]])
xyzt = torch.split(xyzt, [sec_len, sec_len, sec_len, remain_len])
xyzt = torch.concat([xx.mean(axis=0) for xx in xyzt])
else:
xyzt = torch.zeros(n_groups * 4)
_xxxx = list()
for i in range(hits):
_xxxx.append(x[i])
_xxxx.append(y[i])
_xxxx.append(z[i])
_xxxx.append(t[i])
xyzt[: hits * 4] = torch.tensor(_xxxx, dtype=torch.float)
# unique xyz
unique = torch.tensor(
[_x.unique().numel() for _x in [x, y, z]], dtype=torch.float
) # unique -> (3, )
# global features
glob_feat = torch.tensor(
[
hits,
error,
sumq,
meanq,
bratio,
],
dtype=torch.float,
)
return torch.concat([norm_vec, dt, qv, xyzt, unique, glob_feat]).unsqueeze(0)
def prepare_feature(df):
df = df.reset_index(drop=True)
# remove auxiliary
df = df[~df.auxiliary]
df.x *= 1e-3
df.y *= 1e-3
df.z *= 1e-3
df.time -= np.min(df.time)
return df[["time", "charge", "x", "y", "z"]]
# Dataset
class IceCube(IterableDataset):
def __init__(
self,
parquet_dir,
meta_dir,
chunk_ids,
batch_size=200,
max_pulses=200,
extra=False,
):
self.parquet_dir = parquet_dir
self.meta_dir = meta_dir
self.chunk_ids = chunk_ids
self.batch_size = batch_size
self.max_pulses = max_pulses
self.extra = extra
def __iter__(self):
# Handle num_workers > 1 and multi-gpu
is_dist = torch.distributed.is_initialized()
world_size = torch.distributed.get_world_size() if is_dist else 1
rank_id = torch.distributed.get_rank() if is_dist else 0
info = torch.utils.data.get_worker_info()
num_worker = info.num_workers if info else 1
worker_id = info.id if info else 0
num_replica = world_size * num_worker
offset = rank_id * num_worker + worker_id
chunk_ids = self.chunk_ids[offset::num_replica]
# Sensor data
sensor = prepare_sensors()
# Read each chunk and meta iteratively into memory and build mini-batch
for c, chunk_id in enumerate(chunk_ids):
data = pd.read_parquet(
os.path.join(self.parquet_dir, f"batch_{chunk_id}.parquet")
)
meta = pd.read_parquet(
os.path.join(self.meta_dir, f"meta_{chunk_id}.parquet")
)
eids = meta["event_id"].tolist()
eids_batches = [
eids[i : i + self.batch_size]
for i in range(0, len(eids), self.batch_size)
]
for batch_eids in eids_batches:
batch = []
# For each sample, extract features
for eid in batch_eids:
df = data.loc[eid]
df = pd.merge(df, sensor, on="sensor_id")
# sampling of pulses if number exceeds maximum
if len(df) > self.max_pulses:
df_pass = df[~df.auxiliary]
df_fail = df[df.auxiliary]
if len(df_pass) >= self.max_pulses:
df = df_pass.sample(self.max_pulses)
else:
df_fail = df_fail.sample(self.max_pulses - len(df_pass))
df = pd.concat([df_fail, df_pass])
df.sort_values(["time"], inplace=True)
t = series2tensor(df.time)
c = series2tensor(df.charge)
a = series2tensor(df.auxiliary)
x = series2tensor(df.x)
y = series2tensor(df.y)
z = series2tensor(df.z)
feat = torch.stack([x, y, z, t, c, a], dim=1)
batch_data = Data(
x=feat, n_pulses=len(feat), eid=torch.tensor([eid]).long()
)
if self.extra:
feats = feature_extraction(prepare_feature(df))
setattr(batch_data, "extra_feat", feats)
batch.append(batch_data)
yield Batch.from_data_list(batch)
del data
del meta
gc.collect()
# -----------------------------------------------------------------------------
# GraphNet GNN model
# -----------------------------------------------------------------------------
class MLP(nn.Sequential):
def __init__(self, feats):
layers = []
for i in range(1, len(feats)):
layers.append(nn.Linear(feats[i - 1], feats[i]))
layers.append(nn.LeakyReLU())
super().__init__(*layers)
class Model(pl.LightningModule):
def __init__(
self,
max_lr=1e-3,
num_warmup_step=1_000,
remaining_step=1_000,
):
super().__init__()
self.save_hyperparameters()
self.conv0 = EdgeConv(MLP([17 * 2, 128, 256]), aggr="add")
self.conv1 = EdgeConv(MLP([512, 336, 256]), aggr="add")
self.conv2 = EdgeConv(MLP([512, 336, 256]), aggr="add")
self.conv3 = EdgeConv(MLP([512, 336, 256]), aggr="add")
self.post = MLP([1024 + 17, 336, 256])
self.readout = MLP([768, 128])
self.pred = nn.Linear(128, 3)
def forward(self, data: Batch):
vert_feat = data.x
batch = data.batch
# x, y, z, t, c, a
# 0 1 2 3 4 5
vert_feat[:, 0] /= 500.0 # x
vert_feat[:, 1] /= 500.0 # y
vert_feat[:, 2] /= 500.0 # z
vert_feat[:, 3] = (vert_feat[:, 3] - 1.0e04) / 3.0e4 # time
vert_feat[:, 4] = torch.log10(vert_feat[:, 4]) / 3.0 # charge
edge_index = knn_graph(vert_feat[:, :3], 8, batch)
# Construct global features
hx = homophily(edge_index, vert_feat[:, 0], batch).reshape(-1, 1)
hy = homophily(edge_index, vert_feat[:, 1], batch).reshape(-1, 1)
hz = homophily(edge_index, vert_feat[:, 2], batch).reshape(-1, 1)
ht = homophily(edge_index, vert_feat[:, 3], batch).reshape(-1, 1)
means = scatter_mean(vert_feat, batch, dim=0)
n_p = torch.log10(data.n_pulses).reshape(-1, 1)
global_feats = torch.cat([means, hx, hy, hz, ht, n_p], dim=1) # [B, 11]
# Distribute global_feats to each vertex
_, cnts = torch.unique_consecutive(batch, return_counts=True)
global_feats = torch.repeat_interleave(global_feats, cnts, dim=0)
vert_feat = torch.cat((vert_feat, global_feats), dim=1)
# Convolutions
feats = [vert_feat]
# Conv 0
vert_feat = self.conv0(vert_feat, edge_index)
feats.append(vert_feat)
# Conv 1
edge_index = knn_graph(vert_feat[:, :3], k=8, batch=batch)
vert_feat = self.conv1(vert_feat, edge_index)
feats.append(vert_feat)
# Conv 2
edge_index = knn_graph(vert_feat[:, :3], k=8, batch=batch)
vert_feat = self.conv2(vert_feat, edge_index)
feats.append(vert_feat)
# Conv 3
edge_index = knn_graph(vert_feat[:, :3], k=8, batch=batch)
vert_feat = self.conv3(vert_feat, edge_index)
feats.append(vert_feat)
# Postprocessing
post_inp = torch.cat(feats, dim=1)
post_out = self.post(post_inp)
# Readout
readout_inp = torch.cat(
[
scatter_min(post_out, batch, dim=0)[0],
scatter_max(post_out, batch, dim=0)[0],
scatter_mean(post_out, batch, dim=0),
],
dim=1,
)
readout_out = self.readout(readout_inp)
# Predict
pred = self.pred(readout_out)
kappa = pred.norm(dim=1, p=2) + 1e-8
pred_x = pred[:, 0] / kappa
pred_y = pred[:, 1] / kappa
pred_z = pred[:, 2] / kappa
pred = torch.stack([pred_x, pred_y, pred_z, kappa], dim=1)
return pred
def train_or_valid_step(self, data, prefix):
pred_xyzk = self.forward(data) # [B, 4]
true_xyz = data.gt.view(-1, 3) # [B, 3]
loss = VonMisesFisher3DLoss()(pred_xyzk, true_xyz).mean()
error = angular_error(pred_xyzk[:, :3], true_xyz).mean()
self.log(
f"loss-{prefix}",
loss,
batch_size=len(true_xyz),
on_epoch=True,
prog_bar=True,
logger=True,
sync_dist=True,
)
self.log(
f"error-{prefix}",
error,
batch_size=len(true_xyz),
on_epoch=True,
prog_bar=True,
logger=True,
sync_dist=True,
)
return loss
def training_step(self, data, _):
return self.train_or_valid_step(data, "train")
def validation_step(self, data, _):
return self.train_or_valid_step(data, "valid")
def configure_optimizers(self):
optimizer = torch.optim.Adam(self.parameters(), lr=self.hparams.max_lr)
scheduler = torch.optim.lr_scheduler.SequentialLR(
optimizer,
schedulers=[
torch.optim.lr_scheduler.LinearLR(
optimizer, 1e-2, 1, self.hparams.num_warmup_step
),
torch.optim.lr_scheduler.LinearLR(
optimizer, 1, 1e-3, self.hparams.remaining_step
),
],
milestones=[self.hparams.num_warmup_step],
)
return {
"optimizer": optimizer,
"lr_scheduler": {
"scheduler": scheduler,
"interval": "step",
},
}
# -----------------------------------------------------------------------------
# GNN prediction
# -----------------------------------------------------------------------------
def predict_gnn(model):
test_set = IceCube(TEST_PATH, META_PATH, BATCHES_TEST, batch_size=400, extra=True)
test_loader = DataLoader(test_set, batch_size=1)
pred = None
eid = None
with torch.no_grad():
for i, data in tqdm(enumerate(test_loader)):
pred_xyzk = model(data.to(DEVICE))
angles = np.concatenate(
[
# +-------------+------------------------------------+----------------------+
# | x, y, z, kp | azimuth, zenith | extra features |
# +-------------+------------------------------------+----------------------+
pred_xyzk.cpu(),
xyz_to_angle(pred_xyzk[:, :3]).cpu(),
data.extra_feat.cpu()
# +-------------+------------------------------------+----------------------+
],
axis=1,
)
pred = angles if pred is None else np.concatenate([pred, angles])
eid = (
data.eid.cpu() if eid is None else np.concatenate([eid, data.eid.cpu()])
)
col_xyzk = ["x", "y", "z", "kappa"]
col_angles = ["azimuth", "zenith"]
col_norm_vec = ["ex", "ey", "ez"]
col_dt = ["dt_15", "dt_50", "dt_85"]
col_qv = ["qx", "qy", "qz"]
col_xyzt = [
"x0",
"y0",
"z0",
"t0",
"x1",
"y1",
"z1",
"t1",
"x2",
"y2",
"z2",
"t2",
"x3",
"y3",
"z3",
"t3",
]
col_unique = ["uniq_x", "uniq_y", "uniq_z"]
col_glob_feat = ["hits", "error", "sumq", "meanq", "bratio"]
col_extra = col_norm_vec + col_dt + col_qv + col_xyzt + col_unique + col_glob_feat
res = pd.DataFrame(pred, columns=col_xyzk + col_angles + col_extra)
res["azimuth"] = np.remainder(res["azimuth"], 2 * np.pi)
res["zenith"] = np.remainder(res["zenith"], 2 * np.pi)
res["event_id"] = eid
return res
model = Model.load_from_checkpoint(FILE_GNN)
LOGGER.info(f"loaded {FILE_GNN}")
model.eval()
model.freeze()
model.to(DEVICE)
reco_df = predict_gnn(model)
# -----------------------------------------------------------------------------
# GNN + Plane projection prediction
# -----------------------------------------------------------------------------
n_hat = reco_df[["x", "y", "z"]].to_numpy()
e = reco_df[["ex", "ey", "ez"]].to_numpy()
xe = np.sum(n_hat * e, axis=1, keepdims=True)
proj = n_hat - xe * e
proj /= np.linalg.norm(proj, axis=1, keepdims=True) + 1e-8
# -----------------------------------------------------------------------------
# GNN + Plane projection + BDT prediction
# -----------------------------------------------------------------------------
# reco_df inputs
reco_df["error"] = np.log10(reco_df["error"] + 1e-6)
reco_df["sumq"] = np.log10(reco_df["sumq"] + 1e-3)
reco_df["dt_50"] = np.log10(reco_df["dt_50"] + 1e-3)
reco_df["dt_85"] = np.log10(reco_df["dt_85"] + 1e-3)
reco_df["kappa"] = np.log10(reco_df["kappa"] + 1e-3)
columns = ["kappa", "zenith", "error", "sumq", "qz", "dt_50", "dt_85", "ez"]
reco = reco_df[columns].to_numpy()
xe = np.arccos(xe)
# trajectory display
col_xyzt = [
"x0",
"y0",
"z0",
"t0",
"x1",
"y1",
"z1",
"t1",
"x2",
"y2",
"z2",
"t2",
"x3",
"y3",
"z3",
"t3",
]
traj = reco_df[col_xyzt].values
traj = traj.reshape(-1, 4, 4)
v1 = (
1e3
* (traj[:, 1, :3] - traj[:, 0, :3])
/ (traj[:, 1, 3] - traj[:, 0, 3] + 1)[:, np.newaxis]
)
v2 = (
1e3
* (traj[:, 2, :3] - traj[:, 1, :3])
/ (traj[:, 2, 3] - traj[:, 1, 3] + 1)[:, np.newaxis]
)
v3 = (
1e3
* (traj[:, 3, :3] - traj[:, 2, :3])
/ (traj[:, 3, 3] - traj[:, 2, 3] + 1)[:, np.newaxis]
)
v1scale = np.linalg.norm(v1, axis=1, keepdims=True) + 1e-1
v2scale = np.linalg.norm(v2, axis=1, keepdims=True) + 1e-1
v3scale = np.linalg.norm(v3, axis=1, keepdims=True) + 1e-1
ev1 = np.sum(-v1 * e / v1scale, axis=1, keepdims=True)
ev2 = np.sum(-v2 * e / v2scale, axis=1, keepdims=True)
ev3 = np.sum(-v3 * e / v3scale, axis=1, keepdims=True)
ev1 = np.arccos(ev1)
ev2 = np.arccos(ev2)
ev3 = np.arccos(ev3)
vv12 = np.sum(v1 * v2 / v1scale / v2scale, axis=1, keepdims=True)
vv23 = np.sum(v2 * v3 / v2scale / v3scale, axis=1, keepdims=True)
vv31 = np.sum(v3 * v1 / v3scale / v1scale, axis=1, keepdims=True)
vavg = np.log10(np.mean((v1scale, v2scale, v3scale), axis=0))
evvv = np.mean((ev1, ev2, ev3), axis=0)
vvvv = np.mean((vv12, vv23, vv31), axis=0)
pos = np.mean(traj[:, :, :3], axis=1)
xyzq = reco_df[["qx", "qy", "qz"]].to_numpy()
distq = pos - xyzq
distq = np.linalg.norm(distq, axis=1, keepdims=True) + 1e-3
# load the model and predict
LOGGER.info("Loading BDT model...")
clf = pickle.load(open(FILE_BDT, "rb"))
LOGGER.info("Predicting...")
X = np.concatenate([reco, xe, ev1, ev2, ev3, vavg, evvv, vvvv, distq], axis=1)
X[np.isnan(X)] = 0
y_hat = clf.predict(X)[:, np.newaxis]
gnn = reco_df[["azimuth", "zenith"]].to_numpy()
fit = vector2angles(proj)
bdt = (gnn * ~y_hat) + fit * y_hat
LOGGER.info(f"GNN prediction\n{gnn}")
LOGGER.info(f"Plane prediction\n{fit}")
LOGGER.info(f"BDT prediction\n{bdt}")
submit_df = pd.DataFrame(bdt, columns=["azimuth", "zenith"])
submit_df["event_id"] = reco_df.event_id.values
submit_df = submit_df.set_index("event_id")
submit_df = submit_df.sort_values(["event_id"]) # sort by event_id
submit_df.to_csv(os.path.join(OUTPUT_PATH, "submission.csv"))
print(submit_df)
|
import numpy as np
import torch
from torch.utils.data import DataLoader, Dataset
import torch.nn as nn
from transformers import AutoTokenizer
# import os
# os.environ['PYTORCH_CUDA_ALLOC_CONF'] = 'max_split_size_mb:512'
dev = "cpu"
if torch.cuda.is_available():
dev = "cuda:0"
DEVICE = torch.device(dev)
MAX_LENGTH = 50
DEVICE
count = 0
english_lines = []
with open(
"../input/phomt-cs644/PhoMT/tokenization/train/train.en", encoding="utf-8"
) as f:
for line in f:
english_lines.append(" ".join(line.split()[:MAX_LENGTH]))
count = count + 1
if count > 500:
break
count = 0
vietnamese_lines = []
with open(
"../input/phomt-cs644/PhoMT/tokenization/train/train.vi", encoding="utf-8"
) as f:
for line in f:
vietnamese_lines.append(" ".join(line.split()[:MAX_LENGTH]))
count = count + 1
if count > 500:
break
print(english_lines[0])
print("--> len of english_lines is", len(english_lines))
print(vietnamese_lines[0])
print("--> len of vietnamese_lines is", len(vietnamese_lines))
# # 1.1 - Tokenize words with subword tokenizers
tokenizer_vi = AutoTokenizer.from_pretrained("vinai/phobert-base")
tokenizer_en = AutoTokenizer.from_pretrained("bert-base-uncased")
# ## 1.3 - Build encoder input, decoder input, decoder target
encoder_input_data = []
for line in vietnamese_lines:
encoder_input_data.append(tokenizer_vi.encode(line))
# print(encoder_input_data)
# break
decoder_input_data = []
decoder_target_data = []
for line in english_lines: # line is: \t xin chao bla bla \n
decoder_input_data.append(tokenizer_en.encode(line)[:-1])
# print(decoder_input_data)
decoder_target_data.append(tokenizer_en.encode(line)[1:])
# print(decoder_target_data)
# break
# shape of encoder_input_data, decoder_input_data, decoder_target_data are all (dataset_size, seq_len)
print(tokenizer_vi.decode(encoder_input_data[0]))
print(tokenizer_en.decode(decoder_input_data[0]))
print(tokenizer_en.decode(decoder_target_data[0]))
# ## 1.4 - Create Dataset and DataLoader
class PrepareDataset(Dataset):
def __init__(self, encoder_input_data, decoder_input_data, decoder_target_data):
self.encoder_input_indexes = encoder_input_data
self.decoder_input_indexes = decoder_input_data
self.decoder_target_indexes = decoder_target_data
def __len__(self):
return len(self.encoder_input_indexes)
def __getitem__(self, index):
encoder_input_indexes = self.encoder_input_indexes[index]
decoder_input_indexes = self.decoder_input_indexes[index]
decoder_target_indexes = self.decoder_target_indexes[index]
return (
torch.LongTensor(encoder_input_indexes),
torch.LongTensor(decoder_input_indexes),
torch.LongTensor(decoder_target_indexes),
)
def collate_fn(data): # batch, seq_len
encoder_input_indexes, decoder_input_indexes, decoder_target_indexes = zip(*data)
max_length = 0
for seq in encoder_input_indexes:
if len(seq) >= max_length:
max_length = len(seq)
for target in decoder_input_indexes:
if len(target) >= max_length:
max_length = len(target)
max_length = min([max_length, MAX_LENGTH])
length_enc_inp_indexes = []
for seq in encoder_input_indexes:
length_enc_inp_indexes.append(min([len(seq), max_length]))
# length_enc_inp_indexes = [min([len(seq), MAX_LENGTH]) for seq in encoder_input_indexes]
length_dec_inp_indexes = []
for label in decoder_input_indexes:
length_dec_inp_indexes.append(min([len(label), max_length]))
# length_dec_inp_indexes = [min([len(label), MAX_LENGTH]) for label in decoder_input_indexes]
length_dec_tar_indexes = []
for label in decoder_target_indexes:
length_dec_tar_indexes.append(min([len(label), max_length]))
# length_dec_tar_indexes = [min([len(label), MAX_LENGTH]) for label in decoder_target_indexes]
padded_enc_inp = torch.zeros(len(encoder_input_indexes), max_length).long()
padded_dec_inp = torch.zeros(len(encoder_input_indexes), max_length).long()
padded_dec_tar = torch.zeros(len(encoder_input_indexes), max_length).long()
# print(f"encoder_input_indexes shape is {padded_enc_inp.shape}")
# print(f"decoder_input_indexes shape is {padded_dec_inp.shape}")
# print(f"decoder_target_indexes shape is {padded_dec_tar.shape}\n")
# print(f"encoder_input_indexes shape is {encoder_input_indexes.shape}")
for i, seq in enumerate(encoder_input_indexes):
end = length_enc_inp_indexes[i]
# print(len(seq))
try:
padded_enc_inp[i, :end] = seq
except:
padded_enc_inp[i, :end] = seq[:max_length]
# print(f"decoder_input_indexes shape is {decoder_input_indexes.shape}")
for i, label in enumerate(decoder_input_indexes):
end = length_dec_inp_indexes[i]
# padded_dec_inp[i, :end-1] = label
# padded_dec_inp[i, end] = label
try:
padded_dec_inp[i, :end] = label
except:
padded_dec_inp[i, :end] = label[:max_length]
# print(f"decoder_target_indexes shape is {decoder_target_indexes.shape}")
for i, label in enumerate(decoder_target_indexes):
end = length_dec_tar_indexes[i]
try:
padded_dec_tar[i, :end] = label
except:
padded_dec_tar[i, :end] = label[:max_length]
return padded_enc_inp, padded_dec_inp, padded_dec_tar
batch_size = 64
train_dataset = PrepareDataset(
encoder_input_data, decoder_input_data, decoder_target_data
)
train_loader = DataLoader(train_dataset, batch_size=batch_size, collate_fn=collate_fn)
for enc_inp, dec_inp, dec_tar in train_loader:
print(enc_inp.shape)
print(dec_inp.shape)
print(dec_tar.shape)
break
# Encoder
class LSTMEncoder(
nn.Module
): # Here hidden_size is output size, which we can pass latent_size in
def __init__(
self, src_embed_size, src_vocab_size, hidden_size, num_layers, dropout
):
super(LSTMEncoder, self).__init__()
self.hidden_size = hidden_size
self.embedding_layer = nn.Embedding(
src_vocab_size, src_embed_size, padding_idx=0
)
self.lstm_layer = nn.LSTM(
input_size=src_embed_size,
hidden_size=hidden_size,
num_layers=num_layers,
batch_first=True,
)
self.dropout_layer = nn.Dropout(dropout)
def forward(self, input):
x = self.embedding_layer(input)
x = self.dropout_layer(x)
batch_size = x.size(0)
hidden, cell = self.init_hidden_cell_state(batch_size)
_, hidden_cell = self.lstm_layer(x, (hidden, cell))
return hidden_cell
def init_hidden_cell_state(self, batch_size):
hidden = torch.zeros(1, batch_size, self.hidden_size, device=DEVICE)
cell = torch.zeros(1, batch_size, self.hidden_size, device=DEVICE)
return hidden, cell
# Decoder
class LSTMDecoder(nn.Module):
def __init__(
self, tar_embed_size, tar_vocab_size, hidden_size, num_layers, dropout
):
super(LSTMDecoder, self).__init__()
self.hidden_size = hidden_size
self.embedding_layer = nn.Embedding(
tar_vocab_size, tar_embed_size, padding_idx=0
)
self.lstm_layer = nn.LSTM(
input_size=tar_embed_size,
hidden_size=hidden_size,
num_layers=num_layers,
batch_first=True,
)
self.dropout_layer = nn.Dropout(dropout)
# self.output_layer = nn.Linear(hidden_size, tar_vocab_size)
def forward(self, input, latent_state, latent_cell_state):
x = self.embedding_layer(input)
dec_out, hidden_cell = self.lstm_layer(x, (latent_state, latent_cell_state))
return dec_out, hidden_cell
class Seq2SeqModel(nn.Module):
def __init__(
self,
embed_size,
src_vocab_size,
tar_vocab_size,
hidden_size,
num_layers,
dropout,
):
super(Seq2SeqModel, self).__init__()
self.encoder = LSTMEncoder(
embed_size, src_vocab_size, hidden_size, num_layers, dropout
)
self.encoder = self.encoder.to(DEVICE)
self.decoder = LSTMDecoder(
embed_size, tar_vocab_size, hidden_size, num_layers, dropout
)
self.decoder = self.decoder.to(DEVICE)
self.output_layer = nn.Linear(hidden_size, tar_vocab_size)
def forward(self, inputs):
enc_inp, dec_inp = inputs
enc_hidd, enc_cell = self.encoder(enc_inp)
# print(f"enc_hidd is at {enc_hidd.device}")
# print(f"enc_cell is at {enc_cell.device}\n")
enc_hidd = enc_hidd.to(DEVICE)
enc_cell = enc_cell.to(DEVICE)
# print(f"enc_hidd is at {enc_hidd.device}")
# print(f"enc_cell is at {enc_cell.device}")
dec_out, hidden_cell = self.decoder(dec_inp, enc_hidd, enc_cell)
pred = self.output_layer(dec_out)
return pred
num_epochs = 2
src_vocab_size = tokenizer_vi.vocab_size
tar_vocab_size = tokenizer_en.vocab_size
# Hyperparameters
batch_size = 64
embed_size = 100
latent_size = 100
num_layers = 1
dropout = 0.33
lr = 2
mmt = 0.9
train_dataset = PrepareDataset(
encoder_input_data, decoder_input_data, decoder_target_data
)
train_loader = torch.utils.data.DataLoader(
train_dataset, batch_size=batch_size, collate_fn=collate_fn
)
seq2seq_model = Seq2SeqModel(
embed_size=embed_size,
src_vocab_size=src_vocab_size,
tar_vocab_size=tar_vocab_size,
hidden_size=latent_size,
num_layers=num_layers,
dropout=dropout,
)
seq2seq_model.to(DEVICE)
criterion = nn.CrossEntropyLoss(ignore_index=0)
optimizer = torch.optim.SGD(seq2seq_model.parameters(), lr=lr, momentum=mmt)
scheduler = torch.optim.lr_scheduler.ReduceLROnPlateau(
optimizer=optimizer, mode="min", verbose=True, patience=3, factor=0.2
)
training_loss = 0
for i in range(num_epochs):
training_loss = 0.0
seq2seq_model.train()
count = 1
for enc_inp, dec_inp, dec_tar in train_loader:
enc_inp = enc_inp.to(DEVICE)
dec_inp = dec_inp.to(DEVICE)
dec_tar = dec_tar.to(DEVICE)
optimizer.zero_grad()
# print(f"enc_inp shape is {enc_inp.shape}")
# print(f"dec_inp shape is {dec_inp.shape}\n")
preds = seq2seq_model((enc_inp, dec_inp))
preds = preds.contiguous().view(-1, tar_vocab_size)
dec_tar = dec_tar.view(-1)
# print(preds.shape)
# print(dec_tar.shape)
loss = criterion(preds, dec_tar)
loss.backward()
optimizer.step()
training_loss += loss.item() * enc_inp.size(0)
# print(f"Batch {count} \t Training loss: {training_loss / (count* enc_inp.size(0))}")
count += 1
scheduler.step(training_loss)
train_loss = training_loss / len(train_loader.dataset)
print(f"Epoch {i+1} \t Training loss: {train_loss}")
torch.save(seq2seq_model.state_dict(), "model_weights.pth")
# model = Seq2SeqModel(embed_size=embed_size,
# src_vocab_size=src_vocab_size,
# tar_vocab_size=tar_vocab_size,
# hidden_size=latent_size,
# num_layers=num_layers, dropout=dropout)
# model.to(DEVICE)
seq2seq_model.eval()
stcns = [
"Mọi đứa trẻ sinh ra trong khoảng nửa đêm đều sở hữu năng lực đặc biệt ",
"Bạn vưà bị loại ra khỏi tổng hợp gien trên quả đất .",
"Nên chúng tôi đã thực hiện một thí nghiệm , chúng tôi trồng cây những cây mẹ cùng với cây con",
"Tôi rất vui .",
"Đầu tiên , ta đều cần phải đi ra khỏi những khu rừng",
]
inputs = []
for line in stcns:
line_index = tokenizer_vi.encode(line)
# print(line_index)
# print(tokenizer_vi.decode(line_index))
inputs.append(line_index)
# print(input)
# print("".join([vietnamese_characters[i-1] for i in input[0]]))
sos = tokenizer_en.convert_tokens_to_ids(["[CLS]"])
for c, input in enumerate(inputs):
# print(input)
dec_inp = torch.LongTensor([sos])
dec_inp = dec_inp.to(DEVICE)
enc_inp = torch.LongTensor([input])
enc_inp = enc_inp.to(DEVICE)
for i in range(MAX_LENGTH):
# print(dec_inp.device)
pred = seq2seq_model(
(enc_inp, dec_inp)
) # batch_size x seq_len x tar_vocab_size
# print(pred.shape)
_, prediction_char = torch.max(pred[:, -1:, :].data, 2)
# print(f"prediction_char is {prediction_char}")
dec_inp = torch.concat([dec_inp, prediction_char], axis=1)
# print(f"dec_inp is {dec_inp}")
prediction_text = dec_inp.cpu().numpy()
# print(prediction_text)
print("\n", stcns[c])
print(tokenizer_en.decode(prediction_text[0]))
class Translator(nn.Module): # For inference
def __init__(self, seq2seq_model) -> None:
super(Translator, self).__init__()
self.seq2seq_model = seq2seq_model
# def forward(self, inputs):
# enc_inp, dec_inp = inputs
# enc_hidd, enc_cell = self.encoder(enc_inp)
# # print(f"enc_hidd is at {enc_hidd.device}")
# # print(f"enc_cell is at {enc_cell.device}\n")
# enc_hidd = enc_hidd.to(DEVICE)
# enc_cell = enc_cell.to(DEVICE)
# # print(f"enc_hidd is at {enc_hidd.device}")
# # print(f"enc_cell is at {enc_cell.device}")
# dec_out, hidden_cell = self.decoder(dec_inp, enc_hidd, enc_cell)
# pred = self.output_layer(dec_out)
# return pred
def forward(
self, input
): # encoder input, dec_inp is [\t] aka start token and will be iteratively fed to decoder
sos = english_characters["\t"]
dec_inp = torch.Tensor([[sos]], device=DEVICE)
for i in range(MAX_LENGTH):
pred = self.seq2seq_model(
(input, dec_inp)
) # batch_size x seq_len x tar_vocab_size
print(pred.shape)
|
# install mediapipe
import ultralytics
ultralytics.checks()
# import dependencies
import os
import subprocess
import IPython
from IPython.display import Video, display
import numpy as np
import pandas as pd
import cv2
import mediapipe as mp
def play_video(video_path: str):
frac = 0.65 # scaling factor for display
display(
Video(
data=video_path, embed=True, height=int(720 * frac), width=int(1280 * frac)
)
)
def detect_player_bbox(video_path: str) -> str:
"""
Annotates video with object detection bounding boxes using YOLOv8 model.
"""
video_name = video_path.split("/")[-1]
folder_name = "predict"
# YOLOv8 Object Detection
# you can edit the model if you want to use a different yolo8 model
os.system(f"yolo predict model=yolov8x.pt source={video_path} save_txt=True")
# path to latest output
max = 0
for dir_name in os.listdir("/kaggle/working/runs/detect"):
if dir_name != "predict":
if int(dir_name[-1]) > max:
max = int(dir_name[-1])
if max > 0:
folder_name += str(max)
output_path = f"/kaggle/working/runs/detect/{folder_name}/labels"
return output_path
def detect_player_bbox(video_path: str) -> str:
"""
Annotates video with object detection bounding boxes using YOLOv8 model.
"""
video_name = video_path.split("/")[-1]
folder_name = "predict"
# YOLOv8 Object Detection
# you can edit the model if you want to use a different yolo8 model
os.system(f"yolo predict model=yolov8x.pt source={video_path} save_txt=True")
# path to latest output
max = 0
for dir_name in os.listdir("/kaggle/working/runs/detect"):
if dir_name != "predict":
if int(dir_name[-1]) > max:
max = int(dir_name[-1])
if max > 0:
folder_name += str(max)
output_path = f"/kaggle/working/runs/detect/{folder_name}/labels"
return output_path
def bbox_labels_to_dataframe(bbox_labels_path: str) -> pd.DataFrame:
bbox_labels = {
"video_name": [],
"frame": [],
"class_id": [],
"center_x": [],
"center_y": [],
"width": [],
"height": [],
}
for filename in os.listdir(bbox_labels_path):
video_name = "_".join(filename.split("_")[0:3]) + ".mp4"
frame = filename.split("_")[-1]
frame = int(frame.split(".")[0])
with open(bbox_labels_path + "/" + filename, "r") as f:
for line in f:
line = line.split(" ")
class_id = int(line[0])
center_x = float(line[1])
center_y = float(line[2])
width = float(line[3])
height = float(line[4])
if class_id == 0: # if person
# append to dict
bbox_labels["video_name"].append(video_name)
bbox_labels["frame"].append(frame)
bbox_labels["class_id"].append(class_id)
bbox_labels["center_x"].append(center_x)
bbox_labels["center_y"].append(center_y)
bbox_labels["width"].append(width)
bbox_labels["height"].append(height)
return pd.DataFrame(bbox_labels)
def multi_pose_estimation(
video_path: str, bbox_labels: pd.DataFrame, verbose=True
) -> str:
"""
Performs multi-shot multi-pose estimation by obtianing person bbox from YOLOv8
and performing single pose estimation on the bbox crop.
"""
# intializing mediapipe utils
mp_drawing = mp.solutions.drawing_utils
mp_drawing_styles = mp.solutions.drawing_styles
mp_pose = mp.solutions.pose
# video name
video_name = video_path.split("/")[-1]
# VideoCapture Object
cap = cv2.VideoCapture(video_path)
# video variables
width = int(cap.get(cv2.CAP_PROP_FRAME_WIDTH))
height = int(cap.get(cv2.CAP_PROP_FRAME_HEIGHT))
fps = cap.get(cv2.CAP_PROP_FPS)
total_frames = bbox_labels["frame"].max()
# VideoWriter Object
output_path = "labeled_" + video_name
tmp_output_path = "tmp_" + output_path
out = cv2.VideoWriter(
tmp_output_path, cv2.VideoWriter_fourcc(*"MP4V"), fps, (width, height)
)
# check if camera opened successfully
if cap.isOpened() == False:
print("Error opening video file!")
# multipose estimation
with mp_pose.Pose(
min_detection_confidence=0.8, min_tracking_confidence=0.8
) as pose:
frame = 1
while cap.isOpened():
success, image = cap.read()
if success:
# selecting the frame
bbox_set = bbox_labels.query("frame==@frame")
# iterating through bboxs in the frame
for idx, annot in bbox_set.iterrows():
bbox_center_x = annot["center_x"] * width
bbox_center_y = annot["center_y"] * height
bbox_width = annot["width"] * width
bbox_height = annot["height"] * height
# finding top-left and bottom-right bbox cooridnates
bbox_top_left_x = int(bbox_center_x - (bbox_width / 2))
bbox_top_left_y = int(bbox_center_y - (bbox_height / 2))
bbox_bottom_right_x = int(bbox_center_x + (bbox_width / 2))
bbox_bottom_right_y = int(bbox_center_y + (bbox_height / 2))
# cropping image to bbox
image_crop = image[
bbox_top_left_y:bbox_bottom_right_y,
bbox_top_left_x:bbox_bottom_right_x,
]
# pose estimation
# set image as not writeable to improve perfromance
image_crop.flags.writeable = False
image_crop = cv2.cvtColor(image_crop, cv2.COLOR_BGR2RGB)
results = pose.process(image_crop)
# transposing results to be drawn on the original image
if results.pose_landmarks != None:
for landmark in results.pose_landmarks.landmark:
landmark.x = (
(abs(bbox_bottom_right_x - bbox_top_left_x) / width)
* landmark.x
) + (bbox_top_left_x / width)
landmark.y = (
(abs(bbox_bottom_right_y - bbox_top_left_y) / height)
* landmark.y
) + (bbox_top_left_y / height)
# draw the pose annotations on the image
# set image as writeable
image.flags.writeable = True
mp_drawing.draw_landmarks(
image,
results.pose_landmarks,
mp_pose.POSE_CONNECTIONS,
landmark_drawing_spec=mp_drawing_styles.get_default_pose_landmarks_style(),
)
# save video
out.write(image)
if verbose:
print(f"Frame: {frame}/{total_frames}")
frame += 1
else:
break
cap.release()
out.release()
# Not all browsers support the codec, we will re-load the file at tmp_output_path
# and convert to a codec that is more broadly readable using ffmpeg
if os.path.exists(output_path):
os.remove(output_path)
subprocess.run(
[
"ffmpeg",
"-i",
tmp_output_path,
"-crf",
"18",
"-preset",
"veryfast",
"-hide_banner",
"-loglevel",
"error",
"-vcodec",
"libx264",
output_path,
]
)
os.remove(tmp_output_path)
return output_path
video_path = "/kaggle/input/nfl-player-contact-detection/train/58168_003392_Endzone.mp4"
bbox_labels_path = detect_player_bbox(video_path)
bbox_labels = bbox_labels_to_dataframe(bbox_labels_path)
output_path = multi_pose_estimation(video_path, bbox_labels)
play_video(output_path)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# !pip install tensorflow
import sys
import numpy as np
import random as rn
import pandas as pd
import torch
from pytorch_pretrained_bert import BertModel
from torch import nn
# from torchnlp.datasets import imdb_dataset # --> We are using our own uploaded dataset.
from pytorch_pretrained_bert import BertTokenizer
from tensorflow.keras.preprocessing.sequence import pad_sequences
from torch.utils.data import TensorDataset, DataLoader, RandomSampler, SequentialSampler
from torch.optim import Adam
from torch.nn.utils import clip_grad_norm_
from IPython.display import clear_output
import matplotlib.pyplot as plt
rn.seed(321)
np.random.seed(321)
torch.manual_seed(321)
torch.cuda.manual_seed(321)
path = "../input/imdb-50k-movie-reviews-test-your-bert/"
train_data = pd.read_csv(path + "train.csv")
test_data = pd.read_csv(path + "test.csv")
# experimenting here with a sample of dataset, to avoid memory overflow.
train_data = train_data[:2000]
test_data = test_data[:500]
train_data = train_data.to_dict(orient="records")
test_data = test_data.to_dict(orient="records")
type(train_data)
train_texts, train_labels = list(
zip(*map(lambda d: (d["text"], d["sentiment"]), train_data))
)
test_texts, test_labels = list(
zip(*map(lambda d: (d["text"], d["sentiment"]), test_data))
)
len(train_texts), len(train_labels), len(test_texts), len(test_labels)
tokenizer = BertTokenizer.from_pretrained("bert-base-uncased", do_lower_case=True)
# tokenizer = torch.hub.load('huggingface/pytorch-transformers', 'tokenizer', 'bert-base-uncased')
train_tokens = list(
map(lambda t: ["[CLS]"] + tokenizer.tokenize(t)[:510] + ["[SEP]"], train_texts)
)
test_tokens = list(
map(lambda t: ["[CLS]"] + tokenizer.tokenize(t)[:510] + ["[SEP]"], test_texts)
)
len(train_tokens), len(test_tokens)
train_tokens_ids = pad_sequences(
list(map(tokenizer.convert_tokens_to_ids, train_tokens)),
maxlen=512,
truncating="post",
padding="post",
dtype="int",
)
test_tokens_ids = pad_sequences(
list(map(tokenizer.convert_tokens_to_ids, test_tokens)),
maxlen=512,
truncating="post",
padding="post",
dtype="int",
)
train_tokens_ids.shape, test_tokens_ids.shape
train_y = np.array(train_labels) == "pos"
test_y = np.array(test_labels) == "pos"
train_y.shape, test_y.shape, np.mean(train_y), np.mean(test_y)
train_masks = [[float(i > 0) for i in ii] for ii in train_tokens_ids]
test_masks = [[float(i > 0) for i in ii] for ii in test_tokens_ids]
class BertBinaryClassifier(nn.Module):
def __init__(self, dropout=0.1):
super(BertBinaryClassifier, self).__init__()
self.bert = BertModel.from_pretrained(
"bert-base-uncased", output_attentions=True
)
# self.bert = torch.hub.load('huggingface/pytorch-transformers', 'modelForSequenceClassification', 'bert-base-uncased', output_attention=True)
# self.bert = BertForSequenceClassification.from_pretrained('bert-base-uncased', output_attentions=True)
self.dropout = nn.Dropout(dropout)
self.linear = nn.Linear(768, 1)
self.sigmoid = nn.Sigmoid()
def forward(self, tokens, masks=None):
encoded_layers, pooled_output = self.bert(
tokens, attention_mask=masks, output_all_encoded_layers=True
)
dropout_output = self.dropout(pooled_output)
linear_output = self.linear(dropout_output)
proba = self.sigmoid(linear_output)
print(encoded_layers)
return proba
# ensuring that the model runs on GPU, not on CPU
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
device
str(torch.cuda.memory_allocated(device) / 1000000) + "M"
bert_clf = BertBinaryClassifier()
bert_clf = bert_clf.cuda() # running BERT on CUDA_GPU
str(torch.cuda.memory_allocated(device) / 1000000) + "M"
x = torch.tensor(train_tokens_ids[:3]).to(device)
y, pooled = bert_clf.bert(x, output_all_encoded_layers=False)
x.shape, y.shape, pooled.shape
y = bert_clf(x)
y.cpu().detach().numpy() # kinda Garbage Collector to free up used and cache space
# Cross- checking CUDA GPU Memory to ensure GPU memory is not overflowing.
str(torch.cuda.memory_allocated(device) / 1000000) + "M"
y, x, pooled = None, None, None
torch.cuda.empty_cache() # Clearing Cache space for fresh Model run
str(torch.cuda.memory_allocated(device) / 1000000) + "M"
# Setting hyper-parameters
BATCH_SIZE = 4
EPOCHS = 10
train_tokens_tensor = torch.tensor(train_tokens_ids)
train_y_tensor = torch.tensor(train_y.reshape(-1, 1)).float()
test_tokens_tensor = torch.tensor(test_tokens_ids)
test_y_tensor = torch.tensor(test_y.reshape(-1, 1)).float()
train_masks_tensor = torch.tensor(train_masks)
test_masks_tensor = torch.tensor(test_masks)
# str(torch.cuda.memory_allocated(device)/1000000 ) + 'M'
train_dataset = TensorDataset(train_tokens_tensor, train_masks_tensor, train_y_tensor)
train_sampler = RandomSampler(train_dataset)
train_dataloader = DataLoader(
train_dataset, sampler=train_sampler, batch_size=BATCH_SIZE
)
test_dataset = TensorDataset(test_tokens_tensor, test_masks_tensor, test_y_tensor)
test_sampler = SequentialSampler(test_dataset)
test_dataloader = DataLoader(test_dataset, sampler=test_sampler, batch_size=BATCH_SIZE)
param_optimizer = list(bert_clf.sigmoid.named_parameters())
optimizer_grouped_parameters = [{"params": [p for n, p in param_optimizer]}]
optimizer = Adam(bert_clf.parameters(), lr=3e-6)
# torch.cuda.empty_cache() # Clearing Cache space for a fresh Model run
# import time
# start = time.time()
# for epoch_num in range(EPOCHS):
# bert_clf.train()
# train_loss = 0
# for step_num, batch_data in enumerate(train_dataloader):
# token_ids, masks, labels = tuple(t.to(device) for t in batch_data)
# print(str(torch.cuda.memory_allocated(device)/1000000 ) + 'M')
# logits = bert_clf(token_ids, masks)
# loss_func = nn.BCELoss()
# batch_loss = loss_func(logits, labels)
# train_loss += batch_loss.item()
# bert_clf.zero_grad()
# batch_loss.backward()
# clip_grad_norm_(parameters=bert_clf.parameters(), max_norm=1.0)
# optimizer.step()
# clear_output(wait=True)
# print('Epoch: ', epoch_num + 1)
# print("\r" + "{0}/{1} loss: {2} ".format(step_num, len(train_data) / BATCH_SIZE, train_loss / (step_num + 1)))
# end = time.time()
# print("time elapsed: ",end - start)
bert_clf.eval()
bert_predicted = []
all_logits = []
with torch.no_grad():
for step_num, batch_data in enumerate(test_dataloader):
token_ids, masks, labels = tuple(t.to(device) for t in batch_data)
logits = bert_clf(token_ids, masks)
loss_func = nn.BCELoss()
loss = loss_func(logits, labels)
numpy_logits = logits.cpu().detach().numpy()
bert_predicted += list(numpy_logits[:, 0] > 0.5)
all_logits += list(numpy_logits[:, 0])
np.mean(bert_predicted)
print(classification_report(test_y, bert_predicted))
# print(bert_clf.out)
# print(bert_clf.bert.encoder)
# print(bert_clf.bert.encoder.layer[0].attention.self.query.weight)
# basic
import math
# PyTorch
import torch
import torch.nn.functional as F
# visualisation
import matplotlib.pyplot as plt
import seaborn as sns
# # random tensors function from https://rockt.github.io/2018/04/30/einsum
# def random_tensors(shape, num=1, requires_grad=False):
# tensors = [torch.randn(shape, requires_grad=requires_grad) for i in range(0,num)]
# return tensors[0] if num == 1 else tensors
# # dimensions = batch size x sequence length x embedding dimension
# Y = random_tensors([3, 5, 7])
# attention function based on http://nlp.seas.harvard.edu/2018/04/03/attention.html#attention
def attention_transf(query, key, value):
"Compute 'Scaled Dot Product Attention'"
# scaling factor for scores
d_k = query.size(-1)
# matrix multiplication to get every dot product for query and transposed key
scores = torch.matmul(query, key.transpose(-2, -1)) / math.sqrt(d_k)
# apply softmax of the scores on the last dimensions (= sequence length)
p_attn = F.softmax(scores, dim=-1)
# matrix multiplication of attention matrix and value matrix to get output based
out = torch.matmul(p_attn, value)
return out, p_attn
# run Y through attention function
# out, p_attn = attention_transf(Y, Y, Y)
out, p_attn = attention_transf(Y, Y, Y)
# print shapes
Y.shape, p_attn.shape, out.shape
# input dimmension = batch size x sequence length x embedding dimension
Y.shape
# transpose the last two dimension to enable matrix multiplication in the next step
Y.transpose(-2, -1).shape
# matrix multiplication for attention matrix
torch.matmul(Y, Y.transpose(-2, -1)).shape
# matrix multiplication for output
torch.matmul(torch.matmul(Y, Y.transpose(-2, -1)), Y).shape
# check attention matrix shape
p_attn.shape
# print attention matrix
p_attn
# print first attention vector = dimension of softmax
p_attn[0, 0]
# verify that softmax adds up to 1
p_attn[0, 0].sum().item()
# verify that softmax adds up to 1 for every row in the attention matrix
p_attn.sum(dim=2)
# input
sns.heatmap(
Y[0].numpy(), annot=True, cmap=sns.light_palette("blue", as_cmap=True), linewidths=1
).set_title("Input (= query, key, and value)")
# output
sns.heatmap(
out[0].numpy(),
annot=True,
cmap=sns.light_palette("green", as_cmap=True),
linewidths=1,
).set_title("Output")
p_attn[0, 0].view(1, -1)
Y[0, :, 0].view(-1, 1)
# attention matrix
sns.heatmap(
p_attn[0, 0].view(1, -1).numpy(),
annot=True,
cmap=sns.light_palette("orange", as_cmap=True),
linewidths=1,
)
# input
sns.heatmap(
Y[0, :, 0].view(-1, 1).numpy(),
annot=True,
cmap=sns.light_palette("blue", as_cmap=True),
linewidths=1,
)
# output
sns.heatmap(
out[0, 0, 0].view(1, 1).numpy(),
annot=True,
cmap=sns.light_palette("green", as_cmap=True),
linewidths=1,
)
|
# ## Binary Classification to predict 'Buggy'
# import libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.preprocessing import MinMaxScaler
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import accuracy_score, confusion_matrix
from sklearn.metrics import roc_curve, auc
from sklearn.metrics import precision_recall_curve
import xgboost as xgb
# read data from Excel file
df = pd.read_excel("/kaggle/input/completo/completo.xlsx")
df.head()
df.info()
print("Null Values: ", df.isnull().sum().sum())
df.describe()
df["Buggy"].value_counts
# Check for Duplicates
print("duplicated Values: ", df.duplicated().sum())
# remove duplicate values
df = df.drop_duplicates()
print("duplicated Values: ", df.duplicated().sum())
# drop unwanted columns
df = df.drop(["Project", "Hash", "LongName", "Number of Bugs"], axis=1)
df.isnull().sum().sum()
df.head()
df.shape
# handling outliers
# plot all columns in a boxplot
plt.figure(figsize=(29, 10))
df.boxplot()
plt.xticks(rotation=45)
plt.show()
# split the data into inputs and outputs
x = df.drop("Buggy", axis=1) # inputs/features
y = df["Buggy"] # output/target variable
x
y
scaler = StandardScaler()
# fit and transform the data
scaled_data = scaler.fit_transform(df)
# Split the data into training and testing sets
X_train, X_test, y_train, y_test = train_test_split(
x, y, test_size=0.2, random_state=42
)
# Before Excueting the random forest model we first need to make hyperparamter tuning
from xgboost import XGBRegressor
from sklearn.model_selection import GridSearchCV
xgb_model = XGBRegressor(random_state=2022)
search_space = {
"n_estimators": [100, 300],
"max_depth": [3, 7],
"min_samples_split": [25, 50],
"max_features": [2, 4],
}
GS = GridSearchCV(
estimator=xgb_model,
param_grid=search_space,
scoring=["r2", "neg_root_mean_squared_error"],
refit="r2",
cv=5,
verbose=4,
)
GS.fit(X_train, y_train)
print(GS.best_params_)
# ## Random Forest Classifier
# Create a random forest classifier with 300 trees
rf = RandomForestClassifier(
max_depth=35, max_features=20, min_samples_split=50, n_estimators=300
)
# Fit the model to the training data
rf.fit(X_train, y_train)
y_pred = rf.predict(X_test)
accuracy = accuracy_score(y_test, y_pred)
print("Accuracy: {:.2f}%".format(accuracy * 100))
# calculate confusion matrix
cm = confusion_matrix(y_test, y_pred)
# plot confusion matrix using heatmap
sns.heatmap(cm, annot=True, cmap="Blues")
# calculate ROC curve
fpr, tpr, thresholds = roc_curve(y_test, rf.predict_proba(X_test)[:, 1])
roc_auc = auc(fpr, tpr)
# plot ROC curve
plt.plot(fpr, tpr, label="ROC curve (AUC = {:.2f})".format(roc_auc))
plt.plot([0, 1], [0, 1], linestyle="--", label="Random guess")
plt.xlabel("False Positive Rate")
plt.ylabel("True Positive Rate")
plt.title("Receiver Operating Characteristic (ROC) Curve")
plt.legend()
# plot feature importances
fig, ax = plt.subplots(figsize=(12, 15))
ax.barh(range(len(rf.feature_importances_)), rf.feature_importances_)
ax.set_yticks(range(len(X_train.columns)))
ax.set_yticklabels(X_train.columns)
ax.set_xlabel("Importance")
ax.set_ylabel("Feature")
plt.show()
import shap
shap.initjs()
import joblib
explainer = shap.TreeExplainer(rf)
explainer = shap.TreeExplainer(rf)
shap_values = explainer.shap_values(np.array(X_test.iloc[0]))
Explainer = shap.force_plot(explainer.expected_value[1], shap_values[1], X_test.iloc[0])
Explainer
import lime
import lime.lime_tabular
explainer = lime.lime_tabular.LimeTabularExplainer(
X_test.values, feature_names=X_test, discretize_continuous=True
)
exp = explainer.explain_instance(
X_test.iloc[10], rf.predict_proba, num_features=4, top_labels=1
)
exp.show_in_notebook(show_table=True, show_all=False)
# # XGBoost
# Define model
model = xgb.XGBClassifier()
# Train model
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
accuracy = accuracy_score(y_test, y_pred)
print("Accuracy: {:.2f}%".format(accuracy * 100))
# calculate confusion matrix
cm = confusion_matrix(y_test, y_pred)
# plot confusion matrix using heatmap
sns.heatmap(cm, annot=True, cmap="Blues")
# # XGBoost with GPU usage
# Define model
xgb_gpu = xgb.XGBClassifier(tree_method="gpu_hist", gpu_id=0)
# Train model
xgb_gpu.fit(X_train, y_train)
y_pred = xgb_gpu.predict(X_test)
accuracy = accuracy_score(y_test, y_pred)
print("Accuracy: {:.2f}%".format(accuracy * 100))
# calculate confusion matrix
cm = confusion_matrix(y_test, y_pred)
# plot confusion matrix using heatmap
sns.heatmap(cm, annot=True, cmap="Blues")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
from tensorflow import keras
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
from tensorflow.keras import datasets, layers, models
import matplotlib.pyplot as plt
import tensorflow as tf
import numpy as np
import seaborn as sns
import pandas as pd
logdir = "log"
(train_images, train_labels), (test_images, test_labels) = datasets.mnist.load_data()
train_images = train_images.reshape((60000, 28, 28, 1))
test_images = test_images.reshape((10000, 28, 28, 1))
train_images, test_images = train_images / 255.0, test_images / 255.0
classes = [0, 1, 2, 3, 4, 5, 6, 7, 8, 9]
model = models.Sequential()
model.add(layers.Conv2D(32, (3, 3), activation="relu", input_shape=(28, 28, 1)))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(64, (3, 3), activation="relu"))
model.add(layers.MaxPooling2D((2, 2)))
model.add(layers.Conv2D(64, (3, 3), activation="relu"))
model.add(layers.Flatten())
model.add(layers.Dense(64, activation="relu"))
model.add(layers.Dense(10, activation="softmax"))
model.compile(
optimizer="adam", loss="sparse_categorical_crossentropy", metrics=["accuracy"]
)
model.fit(
x=train_images, y=train_labels, epochs=5, validation_data=(test_images, test_labels)
)
y_true = test_labels
y_pred = model.predict_classes(test_images)
|
from sklearn.model_selection import train_test_split
import pandas as pd
diamonds = pd.read_csv("../input/diamonds/diamonds.csv")
diamonds.head(10)
df = diamonds.copy()
X = df.drop(["price"], axis=1)
y = df.price
df.cut.value_counts()
df["cut"] = df["cut"].map(
{"Fair": 1, "Good": 2, "Very Good": 3, "Premium": 4, "Ideal": 5}
)
df.head()
df = pd.get_dummies(data=df, columns=["color", "clarity"])
df.head()
df.isnull().sum()
X = df.drop(["price"], axis=1)
y = df.price
from sklearn.tree import DecisionTreeRegressor
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, train_size=0.75, random_state=0
)
model = DecisionTreeRegressor(random_state=0)
model.fit(X_train, y_train)
from sklearn.metrics import mean_absolute_error
predictions = model.predict(X_test)
print(mean_absolute_error(y_test, predictions))
def get_mae(max_leaf_nodes, X_train, X_test, y_train, y_test):
model = DecisionTreeRegressor(max_leaf_nodes=max_leaf_nodes, random_state=0)
model.fit(X_train, y_train)
pred = model.predict(X_test)
mae = mean_absolute_error(y_test, pred)
return mae
for max_leaf_nodes in [5, 50, 500, 5000, 6000]:
my_mae = get_mae(max_leaf_nodes, X_train, X_test, y_train, y_test)
print("Max leaf nodes: %d \t\t Mean Absolute Error: %d" % (max_leaf_nodes, my_mae))
model = DecisionTreeRegressor(max_leaf_nodes=5000, random_state=0, criterion="mae")
model.fit(X_train, y_train)
predictions = model.predict(X_test)
print(mean_absolute_error(y_test, predictions))
model.score(X_train, y_train) # train_score
model.score(X_test, y_test) # test score
|
# # NCAA M Basketball Player / Team Stats Scraping
# #### This notebook can be used to scrape men's NCAA basketball player / team stats from https://www.sports-reference.com/
# ## Introduction
# As I got started on the March Machine Learning Mania 2021 - NCAAM contest, I thought it might be useful to bring in player stats to help make the winning predictions.
# As the compeition data didn't include player stats, I decided to create a notebook to get that data!
# ## Prep
## library imports
## linear algebra
import numpy as np
## file IO / processing
import pandas as pd
## system
import os
## requests
import requests
## results storage
from collections import defaultdict
## scraping
from bs4 import BeautifulSoup
## functions
def get_soup(url, parser="html.parser", verify=True):
"""
Gets the Beautifulsoup object which holds the data to be scrapped
Args:
url (str): URL of the page to be scrapped
parser (str): Type of parser to be used
Returns:
BeautifulSoup object
"""
request = get_request_response(url, verify=verify)
return BeautifulSoup(request.content, parser)
def get_request_response(url, verify=True):
"""
Requests the content to be scrapped
Args:
url (url): URL of the page to be scrapped
Returns:
request object
"""
return requests.get(url, verify=verify)
def find_all(soup, **kwargs):
"""
Returns all the PageElement tags which match a given criteria
See BeautifulSoup find_all documentation
Args:
soup (BeautifulSoup): BeautifulSoup object containing the web page content
Returns:
list of PageElements
"""
return soup.find_all(**kwargs)
def find(soup, **kwargs):
"""
Returns one PageElement tag which matches a given criteria
See BeautifulSoup find_all documentation
Args:
soup (BeautifulSoup): BeautifulSoup object containing the web page content
Returns:
PageElement instance
"""
return soup.find(**kwargs)
def find_per_game_table(soup):
"""
Returns per game stats table PageElement tag
See BeautifulSoup find_all documentation
Args:
soup (BeautifulSoup): BeautifulSoup object containing the web page content
Returns:
per game stats table PageElement tag instance
"""
return find(
soup, name="table", attrs={"class": "sortable stats_table", "id": "per_game"}
)
def find_stats_tags_from_soup(soup, name):
"""
Returns table PageElement tag of the chosen table
See BeautifulSoup find_all documentation
Args:
soup (BeautifulSoup): BeautifulSoup object containing the web page content
name (str): name of the table to be scraped
Returns:
table PageElement tag instance
"""
if name == "per game":
table_soup = find_per_game_table(soup)
else:
raise NotImplementedError("Invalid table name")
return find_all(soup=table_soup, name="td")
def get_stats(tags):
"""
Get stats from the table tags
Args:
tags (list(PageElement tag)): list of tag elements which contain player informations
returns:
dictionary of player stats
"""
game_stats = defaultdict(list)
for tag in tags:
game_stats[tag["data-stat"]].append(tag.string)
return game_stats
def get_stats_from_soup(soup, name):
"""
Get player stats from BeautifulSoup object
Args:
soup (BeautifulSoup): BeautifulSoup object containing the web page content
name (str): name of the table to be scraped
Returns:
dictionary of player stats
"""
tags = find_stats_tags_from_soup(soup, name=name)
return get_stats(tags)
def get_stats_frame_from_soup(soup, name):
"""
Get player stats DataFrame from BeautifulSoup object
Args:
soup (BeautifulSoup): BeautifulSoup object containing the web page content
name (str): name of the table to be scraped
Returns:
pandas DataFrame of player stats
"""
return pd.DataFrame(data=get_stats_from_soup(soup, name=name))
def scrape_per_game_stats_from_url(url, school=None, season=None):
"""
Get player stats from BeautifulSoup object with school and season
Args:
soup (BeautifulSoup): BeautifulSoup object containing the web page content
name (str): name of the table to be scraped
Returns:
pandas DataFrame of player stats
"""
soup = get_soup(url=url)
df = get_stats_frame_from_soup(soup=soup, name="per game")
df["School"] = school
df["Season"] = season
return df
# ## Example
## Gonzaga 2020-2021
url = "https://www.sports-reference.com/cbb/schools/gonzaga/2021.html#all_schools_per_game"
df = scrape_per_game_stats_from_url(url=url, school="Gonzaga", season="2020-2021")
df
## Duke 2018-2019
url = "https://www.sports-reference.com/cbb/schools/duke/2019.html#all_schools_per_game"
df = scrape_per_game_stats_from_url(url=url, school="Duke", season="2018-2019")
df
|
import os
print(os.getcwd())
os.listdir("/kaggle/input/monthly-data")
import pandas as pd
files = [file for file in os.listdir("/kaggle/input/monthly-data")]
files.sort()
files
all_month = pd.DataFrame()
for file in files:
df = pd.read_csv("/kaggle/input/monthly-data/" + file)
all_month = pd.concat([all_month, df])
all_month
all_month = all_month.dropna(how="all")
all_month
# all_month = all_month[all_month['Order_Date'].str[0:2] != 'Or']
# all_month
# all_month['Order_Date'] = pd.to_datetime(all_month['Order_Date'])
# all_month.head()
# all_month['month'] = pd.DatetimeIndex(all_month['Order_Date']).month
# all_month.head()
# all_month['Quantity_Ordered'] = pd.to_numeric(all_month['Quantity_Ordered'])
# all_month['Price_Each'] = pd.to_numeric(all_month['Price_Each'])
# all_month.info()
# all_month = all_month.drop(columns = 'Quantitiy_Ordered')
# all_month.info()
all_month["Order_ID"] = all_month["Order_ID"].astype("str")
all_month["Product"] = all_month["Product"].astype("str")
all_month["Purchase_Address"] = all_month["Purchase_Address"].astype("str")
all_month.info()
# all_month['ffp'] = all_month['Product'].str[0:4]
# all_month.head()
# all_month = all_month.drop(columns = 'ffp')
# all_month.head()
for_product = all_month.groupby("Product")
for_product.sum()
import matplotlib.pyplot as plt
product = [product for product, df in for_product]
quantity = for_product.sum()["Quantity_Ordered"]
plt.bar(product, quantity)
plt.xticks(product, rotation="vertical", size=6)
plt.show()
# all_month.loc[(all_month.Product == 'LG Dryer') | (all_month.Product == 'LG Washing Machine')]
all_month.info()
all_month["sales"] = all_month["Quantity_Ordered"] * all_month["Price_Each"]
# all_month.head()
all_month["Order_Date"] = pd.to_datetime(all_month["Order_Date"])
# all_month.info()
all_month["month"] = pd.DatetimeIndex(all_month["Order_Date"]).month
# all_month.head()
all_month.groupby("month").sum()
# import matplotlib.ticker as mtick
month_list = [month for month, df in all_month.groupby("month")]
sales_data = all_month.groupby("month").sum()["sales"]
plt.bar(month_list, sales_data)
plt.xticks(month_list)
# plt.gca().yaxis.set_major_formatter(mtick.FormatStrFormatter('%.1f'))
plt.ticklabel_format(style="plain")
plt.tight_layout()
plt.show()
import matplotlib.ticker as mtick
month_list = [month for month, df in all_month.groupby("month")]
sales_data = all_month.groupby("month").sum()["sales"]
plt.bar(month_list, sales_data)
plt.xticks(month_list)
# plt.gca().yaxis.set_major_formatter(mtick.FormatStrFormatter('%.f'))
plt.tight_layout()
plt.show()
all_month["city"] = all_month["Purchase_Address"].apply(
lambda x: x.split(",")[1] + " " + x.split(",")[2][0:3]
)
# all_month.head()
all_month.groupby("city").sum()
cities = [city for city, df in all_month.groupby("city")]
sales_data = all_month.groupby("city").sum()["sales"]
plt.bar(cities, sales_data)
plt.xticks(cities, rotation="vertical")
plt.ticklabel_format(style="plain", axis="y")
plt.show()
|
def fun(name, *args):
print(type(args))
print("first arg is ", name)
for i in args:
print(i)
fun("sai", "helllo", 1, 2, 3)
def myfun(name, **args):
print(type(args))
print("first arg is ", name)
for i, j in args.items():
print(i, j)
myfun("sai", age=20, city="hyderabad")
def myfun(name, city, *args, **kwargs):
print("first arg is ", name)
print(type(args))
for i in args:
print(i)
print(type(kwargs))
for i, j in kwargs.items():
print(i, " : ", j)
myfun("sai", "chennai", 20, 40, "comapany", Role="Ml Engineer", Date="20/30")
import os
os.path.join("sai", "man")
|
from pycocotools.coco import COCO
import requests
import os
# Set up data directories
dataDir = "check"
dataType = "train"
annFile = f"/kaggle/input/coco-dataset-annotation/instances_train2017.json"
imgDir = f"{dataDir}/{dataType}2017"
os.makedirs(imgDir, exist_ok=True)
# Initialize COCO API
coco = COCO(annFile)
# Get image ids for the 'chair' and 'bench' categories
catIds = coco.getCatIds(catNms=["chair", "bench"])
imgIds = coco.getImgIds(catIds=catIds)
# Download the images and annotations
for imgId in imgIds:
# Download image
img = coco.loadImgs(imgId)[0]
img_url = img["coco_url"]
img_name = img["file_name"]
with open(f"{imgDir}/{img_name}", "wb") as f:
f.write(requests.get(img_url).content)
# Download annotations
annIds = coco.getAnnIds(imgIds=imgId, catIds=catIds)
anns = coco.loadAnns(annIds)
annFile = f'{imgDir}/{img_name.split(".")[0]}.txt'
with open(annFile, "w") as f:
for ann in anns:
bbox = ann["bbox"]
x_center = (bbox[0] + bbox[2] / 2) / img["width"]
y_center = (bbox[1] + bbox[3] / 2) / img["height"]
width = bbox[2] / img["width"]
height = bbox[3] / img["height"]
catId = ann["category_id"]
f.write(f"{catId} {x_center} {y_center} {width} {height}\n")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Karakterleri 2. konumdan 5. konuma getirin (dahil değildir):
b = "Ciao!"
print(b[0:6])
# Not: İlk karakterin indeksi 0'dır.
b = "Buongiorno!"
print(b[:5])
b = "Merhaba Ben Yusuf yirmi dört yaşındayım."
print(b[14:19])
b = "Günaydın Trabzon!"
print(b[-5:-2])
a = "arkadaşlar Selam!"
print(a.upper())
a = "DÜNYANIN EN İYİ Asma Köprü Istanbul"
print(a.lower())
a = " Dünyanın en güzel şehri Istanbul! "
print(a.strip())
a = "Merhaba, Yusuf"
print(a.replace("değişecek_olan", "yerine_gelmesini_istediğimiz"))
a = "Merhaba, Ben, Yusuf"
print(a.split(",")) # returns ['Hello', ' World!']
b = a.split()
a = "YU"
b = "SUF"
c = a + b
print(c)
a = "Şampiyon"
b = "Trabzon"
c = a + " " + b # boşluk oluşturuma için kullanılıyor
print(c)
age = 28
txt = "My name is Yusuf, I am {}"
print(txt.format(age))
quantity = 10
itemno = 1500
price = 1000
myorder = "Zara da {} parça kıyafete {} yerine {} tl ödedim."
print(myorder.format(quantity, itemno, price))
txt = 'Benim memleketim karadenizde "Trabzon"un meşhur yemeği hamsi tavadır.'
txt
x = "Bize Her yer Trabzon"
print(len(x))
|
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
df = pd.read_csv(
"/kaggle/input/electricity-production-by-source-world/Electricity_Production_By_Source.csv"
)
df.head()
# loading ISO 3166 dataset to check full list of existing countries based on Alpha-3 codes.
iso_a3 = pd.read_csv(
"/kaggle/input/country-names-with-short-codes-a2-a3-iso/country code.csv",
encoding="ISO-8859-1",
)
iso_a3_list = list(iso_a3["Alpha-3 code"])
iso_a3.head()
# replace null values with 0
df = df.fillna(0)
# create a new column for total electricity production
df["Total"] = df.iloc[:, 3:].sum(axis=1)
# check if Code not matches with ISO Alpha-3 codes to detect Totals and Region Sub-totals in the dataset
SubTotal_codes = df[~df["Code"].isin(iso_a3_list)]
SubTotal_codes = (
SubTotal_codes.groupby(["Entity", "Code"])["Total"]
.sum()
.sort_values(ascending=False)
.reset_index()
)
# filter dataset only for rows which have a valid ISO code, so represent real country
df = df[df["Code"].isin(iso_a3_list)]
df_year = (
df.groupby("Year")[
[
"Electricity from coal (TWh)",
"Electricity from gas (TWh)",
"Electricity from hydro (TWh)",
"Electricity from other renewables (TWh)",
"Electricity from solar (TWh)",
"Electricity from oil (TWh)",
"Electricity from wind (TWh)",
"Electricity from nuclear (TWh)",
]
]
.sum()
.reset_index()
)
# Chart 1 - Electricity Production by Year and Source
sns.set_style("whitegrid")
# Define the label and color for each energy source
energy_sources = {
"Coal": "darkgray",
"Gas": "lightcoral",
"Hydro": "steelblue",
"Other Renewables": "yellowgreen",
"Solar": "gold",
"Oil": "darkred",
"Wind": "teal",
"Nuclear": "purple",
}
# Plot the bar chart
ax = sns.barplot(
x="Year",
y="Electricity from coal (TWh)",
data=df_year,
color=energy_sources["Coal"],
)
for source in list(energy_sources.keys())[1:]:
ax = sns.barplot(
x="Year",
y=f"Electricity from {source.lower()} (TWh)",
data=df_year,
color=energy_sources[source],
bottom=df_year[
[
f"Electricity from {prev.lower()} (TWh)"
for prev in list(energy_sources.keys())[
: list(energy_sources.keys()).index(source)
]
]
].sum(axis=1),
)
# Add the legend
handles = [
plt.Rectangle((0, 0), 1, 1, color=energy_sources[source])
for source in energy_sources
]
labels = list(energy_sources.keys())
ax.legend(handles, labels, loc="upper left", bbox_to_anchor=(1, 1))
ax.set_title("Electricity Production by Year and Source")
ax.set_ylabel("Electricity Production (TWh)")
plt.xticks(rotation=90)
plt.show()
# Select the data for the decade 2010-2019
df_decade = df[df["Year"].between(2010, 2019)]
# Group the data by Entity and Source of electricity
df_grouped = (
df_decade.groupby("Entity")[
[
"Electricity from coal (TWh)",
"Electricity from gas (TWh)",
"Electricity from hydro (TWh)",
"Electricity from other renewables (TWh)",
"Electricity from solar (TWh)",
"Electricity from oil (TWh)",
"Electricity from wind (TWh)",
"Electricity from nuclear (TWh)",
"Total",
]
]
.sum()
.reset_index()
)
# get top 10 by total production to limit the chart
top_10 = (
df_grouped.groupby("Entity")
.sum()
.sort_values(by="Total", ascending=False)
.reset_index()
.head(10)
)
# Create a stacked bar chart for production per source per country
fig, ax = plt.subplots(figsize=(10, 10))
sns.barplot(
x="Electricity from coal (TWh)",
y="Entity",
data=top_10,
label="Coal",
color="darkgray",
)
sns.barplot(
x="Electricity from gas (TWh)",
y="Entity",
data=top_10,
label="Gas",
color="lightcoral",
)
sns.barplot(
x="Electricity from hydro (TWh)",
y="Entity",
data=top_10,
label="Hydro",
color="steelblue",
)
sns.barplot(
x="Electricity from other renewables (TWh)",
y="Entity",
data=top_10,
label="Other Renewables",
color="yellowgreen",
)
sns.barplot(
x="Electricity from solar (TWh)",
y="Entity",
data=top_10,
label="Solar",
color="gold",
)
sns.barplot(
x="Electricity from oil (TWh)",
y="Entity",
data=top_10,
label="Oil",
color="darkred",
)
sns.barplot(
x="Electricity from wind (TWh)", y="Entity", data=top_10, label="Wind", color="teal"
)
sns.barplot(
x="Electricity from nuclear (TWh)",
y="Entity",
data=top_10,
label="Nuclear",
color="purple",
)
ax.set(
xlabel="Electricity Production (TWh)",
ylabel="Country",
title="Electricity Production by Source and Country (2010-2019)",
)
ax.legend(loc="lower right")
plt.show()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
from tensorflow.keras.datasets import fashion_mnist
from keras.preprocessing.image import ImageDataGenerator
from keras.layers import Conv2D, MaxPooling2D, Flatten, BatchNormalization
import random # for generating random numbers
from keras.models import Sequential # Model type to be used
from keras.layers.core import Dense, Dropout, Activation
from keras.utils import np_utils # NumPy related tools
from sklearn.model_selection import train_test_split
class_names = [
"T-shirt/top",
"Trouser",
"Pullover",
"Dress",
"Coat",
"Sandal",
"Shirt",
"Sneaker",
"Bag",
"Ankle boot",
]
(X_train, y_train), (X_test, y_test) = fashion_mnist.load_data()
print(f"X_train's shape is {X_train.shape}")
print(f"y_train's shape is {y_train.shape}")
print(f"y_train has {np.max(y_train) + 1} classes")
# We want to add channels to our X data, so that they
# are compatible with the Convolutional Layers
# The idea here is that we are working in greyscale
# So the image has 1 channel
# The image data is of shape (N, im_height, im_width, channels)
X_train = X_train.reshape(X_train.shape + (1,))
X_test = X_test.reshape(X_test.shape + (1,))
print(X_train.shape)
print(X_test.shape)
print(
f"The maximum value of X_train is {np.max(X_train)}. We don't want this, because it'll make training longer"
)
X_train = X_train / 255
X_test = X_test / 255
print(
f"Now the maximum value of X_train is {np.max(X_train)}. We have now scaled our training data."
)
# Let's take a look at some of these images
fig, ax = plt.subplots(2, 5)
for i in range(2):
for j in range(5):
ind = (i * 5) + j
ds_ex = np.where(y_train == ind)[0][0]
ax[i, j].imshow(X_train[ds_ex, ...])
ax[i, j].set_title(class_names[ind])
plt.show()
def to_one_hot(y):
"""
Input: y of shape (n_samples)
Output: y of shape (n_samples, n_classes)
"""
onehot = np.zeros((y.shape[0], len(class_names)))
onehot[np.arange(y.shape[0]), y] = 1
# for i in range(len(y)):
# onehot[i, y[i]] = 1
return onehot
y_train = to_one_hot(y_train)
y_test = to_one_hot(y_test)
# Over to you. Create a CNN to classify this
# Including validation set - reduces the test accuracy
# X_train, X_val, y_train, y_val = train_test_split(X_train, y_train, test_size=0.2, random_state=42)
# Now make the CNN model - use the same code as for MNIST digits dataset
model = Sequential() # Linear stacking of layers
# Convolution Layer 1
model.add(
Conv2D(32, (3, 3), input_shape=(28, 28, 1))
) # 32 different 3x3 kernels -- so 32 feature maps
model.add(BatchNormalization(axis=-1)) # normalize each feature map before activation
model.add(Activation("relu")) # activation
model.add(Dropout(0.2)) # 20% dropout of randomly selected nodes
# Convolution Layer 2
model.add(Conv2D(32, (3, 3))) # 32 different 3x3 kernels -- so 32 feature maps
model.add(BatchNormalization(axis=-1)) # normalize each feature map before activation
model.add(Activation("relu")) # activation
model.add(MaxPooling2D(pool_size=(2, 2))) # Pool the max values over a 2x2 kernel
model.add(Dropout(0.2)) # 20% dropout of randomly selected nodes
# Convolution Layer 3
model.add(Conv2D(64, (3, 3))) # 64 different 3x3 kernels -- so 64 feature maps
model.add(BatchNormalization(axis=-1)) # normalize each feature map before activation
model.add(Activation("relu")) # activation
model.add(Dropout(0.2)) # 20% dropout of randomly selected nodes
# Convolution Layer 4
model.add(Conv2D(64, (3, 3))) # 64 different 3x3 kernels -- so 64 feature maps
model.add(BatchNormalization(axis=-1)) # normalize each feature map before activation
model.add(Activation("relu")) # activation
model.add(MaxPooling2D(pool_size=(2, 2))) # Pool the max values over a 2x2 kernel
model.add(Flatten()) # Flatten final 4x4x64 output matrix into a 1024-length vector
model.add(Dropout(0.2)) # 20% dropout of randomly selected nodes
# Fully Connected Layer 5
model.add(Dense(512)) # 512 FCN nodes
model.add(BatchNormalization()) # normalization
model.add(Activation("relu")) # activation
model.add(Dropout(0.2)) # 20% dropout of randomly selected nodes
# Fully Connected Layer 6
model.add(Dense(10)) # final 10 FCN nodes
model.add(Activation("softmax")) # softmax activation
model.summary()
model.compile(loss="categorical_crossentropy", optimizer="adam", metrics=["accuracy"])
# Did worse with data augmentation
# gen = ImageDataGenerator(
# featurewise_center=False, # set input mean to 0 over the dataset
# samplewise_center=False, # set each sample mean to 0
# featurewise_std_normalization=False, # divide inputs by std of the dataset
# samplewise_std_normalization=False, # divide each input by its std
# zca_whitening=False, # apply ZCA whitening
# rotation_range=10, # randomly rotate images in the range (degrees, 0 to 180)
# zoom_range = 0.1, # Randomly zoom image
# width_shift_range=0.1, # randomly shift images horizontally (fraction of total width)
# height_shift_range=0.1, # randomly shift images vertically (fraction of total height)
# horizontal_flip=False, # randomly flip images
# vertical_flip=False) # randomly flip images
batch_sz = 32 # 32 seems to work best
num_epochs = 10
# model.fit(X_train, y_train, batch_size=batch_sz, epochs=num_epochs, validation_data=(X_val, y_val), verbose=1)
model.fit(X_train, y_train, batch_size=batch_sz, epochs=num_epochs, verbose=1)
# predict with the cnnmodel
score = model.evaluate(X_test, y_test)
print("Loss:", score[0])
print("Test accuracy:", score[1])
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import cudf
import cuml
from cuml.model_selection import train_test_split
from cuml.metrics import accuracy_score
import xgboost as xgb
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
time_slot_data_X = cudf.read_csv(
"/kaggle/input/tumai-x-aimunich-x-kkcompany/light_train_source_labels.csv"
)
time_slot_data_y = cudf.read_csv(
"/kaggle/input/tumai-x-aimunich-x-kkcompany/light_train_target_labels.csv"
)
time_slot_data_X.head()
time_slot_data_X.describe()
# Target Time Span
time_slot_data_y.head()
time_slot_data_y.describe()
# time_slot_cols = list(time_slot_data.columns)
# time_slot_cols.remove('user_id')
# time_slot_data[time_slot_cols].sum().plot.bar()
time_slot_cols_X = list(time_slot_data_X.columns)
time_slot_cols_X.remove("user_id")
time_slot_cols_y = list(time_slot_data_y.columns)
time_slot_cols_y.remove("user_id")
X = time_slot_data_X[time_slot_cols_X]
y = time_slot_data_y[time_slot_cols_y]
# split into training and testing sets
X_train, X_test, y_train_cudf, y_test_cudf = train_test_split(
X, y, test_size=0.2, random_state=42
)
cudf.DataFrame(X_train)
X_train = cudf.DataFrame(X_train)
X_test = cudf.DataFrame(X_test)
y_train = cudf.DataFrame(y_train_cudf)
y_test = cudf.DataFrame(y_test_cudf)
# create XGBoost classifier
clf = xgb.XGBClassifier(tree_method="gpu_hist", gpu_id=0)
# fit the classifier to the training data
clf.fit(X_train, y_train)
# make predictions on the testing data
y_pred = clf.predict(X_test)
# calculate accuracy of the predictions
accuracy = cuml.accuracy_score(y_test.values.ravel(), y_pred)
print("Accuracy:", accuracy)
# Create Submission
X_submission = cudf.read_csv(
"/kaggle/input/tumai-x-aimunich-x-kkcompany/light_test_source_labels.csv"
)
user_ids = X_submission["user_id"]
X_sub = X_submission[time_slot_cols_X]
y_sub = clf.predict(X_sub)
submission = pd.DataFrame(y_sub, columns=time_slot_cols_y)
submission["user_id"] = user_ids.to_numpy()
submission.to_csv("submission.csv")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
import pandas as pd
import numpy as np
from sklearn.ensemble import RandomForestRegressor
from sklearn.metrics import mean_absolute_percentage_error
train_df = pd.read_csv("/kaggle/input/gdz-elektrik-datathon-2023/train.csv")
submit_df = pd.read_csv(
"/kaggle/input/gdz-elektrik-datathon-2023/sample_submission.csv"
)
med_df = pd.read_csv("/kaggle/input/gdz-elektrik-datathon-2023/med.csv")
med_df["Yıl"] = pd.to_datetime(med_df["Tarih"]).dt.year
med_df["Ay"] = pd.to_datetime(med_df["Tarih"]).dt.month
med_df["Gün"] = pd.to_datetime(med_df["Tarih"]).dt.day
train_df["Yıl"] = pd.to_datetime(train_df["Tarih"]).dt.year
train_df["Ay"] = pd.to_datetime(train_df["Tarih"]).dt.month
train_df["Gün"] = pd.to_datetime(train_df["Tarih"]).dt.day
train_df["Saat"] = pd.to_datetime(train_df["Tarih"]).dt.hour
submit_df["Yıl"] = pd.to_datetime(submit_df["Tarih"]).dt.year
submit_df["Ay"] = pd.to_datetime(submit_df["Tarih"]).dt.month
submit_df["Gün"] = pd.to_datetime(submit_df["Tarih"]).dt.day
submit_df["Saat"] = pd.to_datetime(submit_df["Tarih"]).dt.hour
submit_df["Kesintili Günler"] = 0
train_df = train_df.drop("Tarih", axis=1)
submit_df = submit_df.drop("Tarih", axis=1)
med_df = med_df.drop("Tarih", axis=1)
med_df[["Yıl", "Ay", "Gün"]] = med_df[["Yıl", "Ay", "Gün"]].astype(int)
train_df["Kesintili Günler"] = train_df.apply(
lambda row: int(
(row[["Yıl", "Ay", "Gün"]] == med_df[["Yıl", "Ay", "Gün"]]).all(axis=1).any()
),
axis=1,
)
calendar = pd.read_csv(
"/kaggle/input/d/frtgnn/turkish-calendar/Turkish calendar.csv", sep=(";")
)
new_df = calendar.iloc[853:2557].copy()
new_df.loc[:, "Ay"] = pd.to_datetime(
new_df["CALENDAR_DATE"], format="%d.%m.%Y"
).dt.month
new_df.loc[:, "Yıl"] = pd.to_datetime(
new_df["CALENDAR_DATE"], format="%d.%m.%Y"
).dt.year
new_df = new_df.drop(["SEASON_SK", "SPECIAL_DAY_SK", "SPECIAL_DAY_SK2"], axis=1)
new_df["WEEKEND_FLAG"] = new_df["WEEKEND_FLAG"].replace(["N"], 0)
new_df["WEEKEND_FLAG"] = new_df["WEEKEND_FLAG"].replace(["Y"], 1)
new_df["RAMADAN_FLAG"] = new_df["RAMADAN_FLAG"].replace(["N"], 0)
new_df["RAMADAN_FLAG"] = new_df["RAMADAN_FLAG"].replace(["Y"], 1)
new_df["PUBLIC_HOLIDAY_FLAG"] = new_df["PUBLIC_HOLIDAY_FLAG"].replace(["N"], 0)
new_df["PUBLIC_HOLIDAY_FLAG"] = new_df["PUBLIC_HOLIDAY_FLAG"].replace(["Y"], 1)
new_df.rename(columns={"DAY_OF_MONTH": "Gün"}, inplace=True)
new_df.rename(columns={"DAY_OF_WEEK_SK": "Haftanın Günü"}, inplace=True)
new_df.rename(columns={"QUARTER_OF_YEAR": "Sezon"}, inplace=True)
new_df.rename(columns={"WEEKEND_FLAG": "Haftasonu - Haftaiçi"}, inplace=True)
new_df.rename(columns={"WEEK_OF_YEAR": "Yılın kaçıncı haftası"}, inplace=True)
new_df.rename(columns={"RAMADAN_FLAG": "Ramazan"}, inplace=True)
new_df.rename(columns={"RELIGIOUS_DAY_FLAG_SK": "Dini Gün"}, inplace=True)
new_df.rename(columns={"NATIONAL_DAY_FLAG_SK": "Ulusal Gün"}, inplace=True)
new_df.rename(columns={"PUBLIC_HOLIDAY_FLAG": "Resmi tatil"}, inplace=True)
new_df_submit = new_df.iloc[0:31]
new_df_train = new_df.iloc[31:1704]
merged_df = pd.merge(train_df, new_df_train, on=["Yıl", "Ay", "Gün"])
merged_df2 = pd.merge(submit_df, new_df_submit, on=["Yıl", "Ay", "Gün"])
merged_df
X_train = merged_df.drop("Dağıtılan Enerji (MWh)", axis=1)
X_train = X_train.drop("CALENDAR_DATE", axis=1)
y_train = merged_df["Dağıtılan Enerji (MWh)"]
X_test = merged_df2.drop("Dağıtılan Enerji (MWh)", axis=1)
X_test = X_test.drop("CALENDAR_DATE", axis=1)
rf_model = RandomForestRegressor(n_estimators=100, random_state=42)
rf_model.fit(X_train, y_train)
# Make predictions on the validation set
y_pred = rf_model.predict(X_test)
subm = pd.read_csv("/kaggle/input/gdz-elektrik-datathon-2023/sample_submission.csv")
subm["Dağıtılan Enerji (MWh)"] = y_pred
subm
subm.to_csv("submission6.csv", index=None)
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
train = pd.read_csv("/kaggle/input/aqueous-solubility-predictioin/train.csv")
test = pd.read_csv("/kaggle/input/aqueous-solubility-predictioin/test.csv")
train.info()
features_todrop = ["SD", "Ocurrences", "Group"]
label = train["Solubility"]
features = train.drop(columns=features_todrop)
features = features.drop(columns="Solubility")
num_features = features.select_dtypes(["float64", "int64"])
from sklearn.model_selection import train_test_split
features_train, features_valid, labels_train, labels_valid = train_test_split(
num_features, label, test_size=0.2, random_state=7
)
from xgboost import XGBRegressor
xgb_model = XGBRegressor(n_estimators=200)
xgb_model.fit(features_train, labels_train)
from sklearn.metrics import mean_squared_error
mean_squared_error(labels_valid, xgb_model.predict(features_valid))
test["Solubility"] = xgb_model.predict(test.select_dtypes(["float64", "int64"]))
test[["comp_id", "Solubility"]].to_csv("/kaggle/working/submission.csv", index=False)
|
import pandas as pd
import matplotlib as mpl
import matplotlib.pyplot as plt
from plotly.express import *
import plotly.express as px
df = pd.read_csv(
"/kaggle/input/top-50-us-tech-companies-2022-2023-dataset/Top 50 US Tech Companies 2022 - 2023.csv"
)
df.head(10)
# ### rename the columns so that they make sense. We can use rename() method by passing in a dictionary of old and new names
df.rename(
columns={
"Company Name": "Company",
"Founding Year": "Founding",
"Annual Revenue 2022-2023 (USD in Billions)": "Revenue_USD",
"Market Cap (USD in Trillions)": "MarketCap_with_Trillions",
"Employee Size": "Employee",
"Stock Name": "Stock_Name",
},
inplace=True,
)
df.columns
# # Data Wringling
df.head().info
df.describe()
df.shape
# ### extracting profits (Annual Revenue 2022-2023 (USD in Billions)) for this Tech Companies
df.head(10)
fig = px.bar(df, x="Revenue_USD", y="Company")
fig.show()
# ### Compare the trend of top 5 companies that contributed the most Revenue With US Dollars¶
inplace = True
df.sort_values(by=["Revenue_USD"], ascending=False, axis=0, inplace=True)
df.head()
# get the top 5 entries
df_top5 = df.head()
# transpose the dataframe
df_top5 = df_top5["Revenue_USD"].transpose()
print(df_top5)
import plotly.express as px
fig = px.bar(df_top5, y=["Revenue_USD"])
fig.show()
# ### Numbers of Employees for the first 10 Companies in DataFrame
df.head(10).index = df.head(10).index.map(
int
) # let's change the index values of df_CI to type integer for plotting
df.head(10).plot(kind="bar")
plt.title("distribution of Employee")
plt.ylabel("Founding")
plt.xlabel("Company")
plt.show()
# ### Another Compare the trend of top 10 companies that contributed the most MarketCap with Trillions¶
inplace = True
df.sort_values(by=["MarketCap_with_Trillions"], ascending=False, axis=0, inplace=True)
df.head(10)
df_top10 = df.head(10)
# transpose the dataframe
df_top10 = df_top10["MarketCap_with_Trillions"].transpose()
print(df_top10)
import plotly.express as px
fig = px.line(df_top10, y=["MarketCap_with_Trillions"])
fig.show()
|
# # Sentiment Analysis Jumia Reviews
# In this NLP project I'll be attempting to classify reviews on a particula product sold on the [Jumia](https://www.jumia.com.ng/) intopositive or negative type categories based off the text content and summary in the reviews.
# We will use the [Jumia dataset](https://www.kaggle.com/datasets/chibuzornwachukwu/product-ratings-oraimo-jumia-reviews) from Kaggle.
# This dataset covers the ratings on the [Oraimo](oraimo.com.ng) Freepods 3 over a year after its release.
# It is a decent product and the data pretty much buttresses this point.
# Check out and - if you can - classify the reviews of the customer base.
# - Customer Name - the name of the customer
# - Review Topic - the title and summary of each review
# - Review Body - the contents of the reviews
# - Rating(of 5) - product score by each customer (on a 1-5 scale)
# - Review date - date of each review
# ### Importing Dependencies
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
color = sns.color_palette()
import plotly.offline as py
py.init_notebook_mode(connected=True)
import plotly.graph_objs as go
import plotly.tools as tls
import plotly.express as px
# ### Wranging and Cleaning
jumia = pd.read_csv(
"/kaggle/input/product-ratings-oraimo-jumia-reviews/jumia_reviews_df_multi_page.csv"
)
jumia.head()
jumia.info()
jumia.rename({"Rating(of 5)": "rating"}, axis=1, inplace=True)
jumia.dropna(inplace=True)
# ### Exploring the reviews
fig = px.histogram(jumia, x="rating")
fig.update_traces(
marker_color="turquoise", marker_line_color="rgb(8,48,107)", marker_line_width=1.5
)
fig.update_layout(title_text="Product Ratings")
fig.show()
ratings = jumia["rating"].value_counts()
numbers = ratings.index
quantity = ratings.values
custom_colors = ["green", "yellowgreen", "orange", "tomato", "red"]
plt.figure(figsize=(10, 8))
plt.pie(quantity, labels=numbers, colors=custom_colors)
central_circle = plt.Circle((0, 0), 0.5, color="white")
fig = plt.gcf()
fig.gca().add_artist(central_circle)
plt.rc("font", size=12)
plt.title("Distribution of Jumia Product Ratings", fontsize=14)
plt.show()
# > More than half the customers that purchased the product loved it
import nltk
from nltk.corpus import stopwords
from wordcloud import WordCloud
# Creating Stopwords list
stopwords = set(stopwords.words("english"))
stopwords.update(["br", "href"])
textt = " ".join(review for review in jumia["Review Body"])
wordcloud = WordCloud(stopwords=stopwords).generate(textt)
plt.imshow(wordcloud, interpolation="bilinear")
plt.axis("off")
plt.show()
#### Removing the neutral reviews
jumia = jumia[jumia["rating"] != 3]
jumia["sentiment"] = jumia["rating"].apply(lambda rating: +1 if rating > 3 else -1)
# #### Positive vs Negative reviews
positive = jumia[jumia["sentiment"] == 1].dropna()
negative = jumia[jumia["sentiment"] == -1].dropna()
import nltk
from nltk.corpus import stopwords
stopwords = set(stopwords.words("english"))
stopwords.update(
[
"freepod",
"pod",
"Freepods",
"Freepod",
"stuff",
"href",
"good",
"great",
"sound",
"product",
"flavour",
"like",
"coffee",
"dog",
"flavor",
"buy",
"but",
]
)
## good and great removed because they were included in negative sentiment
pos = " ".join(review for review in positive["Review Body"])
wordcloud = WordCloud(stopwords=stopwords).generate(pos)
plt.imshow(wordcloud, interpolation="bilinear")
plt.axis("off")
plt.show()
import nltk
from nltk.corpus import stopwords
stopwords = set(stopwords.words("english"))
stopwords.update(
[
"freepod",
"pod",
"Freepods",
"Freepod",
"stuff",
"href",
"good",
"great",
"sound",
"product",
"flavour",
"like",
"coffee",
"dog",
"flavor",
"buy",
"but",
]
)
## good and great removed because they were included in negative sentiment
pos = " ".join(review for review in negative["Review Body"])
wordcloud = WordCloud(stopwords=stopwords).generate(pos)
plt.imshow(wordcloud, interpolation="bilinear")
plt.axis("off")
plt.show()
jumia["sentiment"] = jumia["sentiment"].replace({-1: "negative"})
jumia["sentiment"] = jumia["sentiment"].replace({1: "positive"})
fig = px.histogram(jumia, x="sentiment")
custom_colors = ["yellowgreen", "tomato"]
fig.update_traces(
marker_color=custom_colors, marker_line_color=custom_colors, marker_line_width=1.5
)
fig.update_layout(title_text="Product Sentiment")
fig.show()
# > The ratio of positive reviews is way higher than that of negative
# ### Even more cleaning
# > We "attack" the punctuation and unnecessary characters
def remove_punctuation(text):
final = "".join(u for u in text if u not in ("?", ".", ";", ":", "!", '"'))
return final
jumia["Review Body"] = jumia["Review Body"].apply(remove_punctuation)
jumia = jumia.dropna(subset=["Review Topic"])
jumia["Review Topic"] = jumia["Review Topic"].apply(remove_punctuation)
# #### Creating Bag of Words
# > Making use of the count vectorizer from sklearn which will transform the text in our data frame into a a sparse matrix of integers - a bag of words model that can be understood and used by the logistic regression model.
# random split train and test data
index = jumia.index
jumia["random_number"] = np.random.randn(len(index))
train = jumia[jumia["random_number"] <= 0.8]
test = jumia[jumia["random_number"] > 0.8]
# count vectorizer:
from sklearn.feature_extraction.text import CountVectorizer
vectorizer = CountVectorizer(token_pattern=r"\b\w+\b")
train_matrix = vectorizer.fit_transform(train["Review Topic"])
test_matrix = vectorizer.transform(test["Review Topic"])
# ### Split - Train - Test
# #### Spliting Dataframe using the following ratio
# - 80% for the training
# - 20% for the testing and evaluation
X_train = train_matrix
X_test = test_matrix
y_train = train["sentiment"]
y_test = test["sentiment"]
# Logistic Regression
from sklearn.linear_model import LogisticRegression
lr = LogisticRegression(solver="lbfgs", max_iter=6000)
# Feature Scaling
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler(with_mean=False)
X_train = scaler.fit_transform(X_train)
X_test = scaler.transform(X_test)
lr.fit(X_train, y_train)
predictions = lr.predict(X_test)
# #### Find accuracy, precision, recall:
from sklearn.metrics import confusion_matrix, classification_report
new = np.asarray(y_test)
confusion_matrix(predictions, y_test)
print(classification_report(predictions, y_test))
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
from matplotlib import pyplot as plt
import seaborn as sns
df_train = pd.read_csv("/kaggle/input/spaceship-titanic/train.csv")
print("Tamanho: ", df_train.shape)
df_train.head()
df_test = pd.read_csv("/kaggle/input/spaceship-titanic/test.csv")
# # Repassando pela análise de dados
# ### Ressaltando que para as variáveis categóricas é necessário transformar para numerico/booleano
df_train.describe()
df_train.corr()
sns.pairplot(
df_train,
hue="Transported",
vars=["Age", "RoomService", "FoodCourt", "Spa", "ShoppingMall", "VRDeck"],
)
# ### Visualizar os dados nulos
df_train.isna().sum()
df_train[df_train.isna().any(axis=1)]
df_train[df_train.isna().any(axis=1)].isna().sum()
#
# ### uma opção válida é excluir as linhas com dados nulos
# ## Age
sns.histplot(data=df_train, x="Age", kde=True)
sns.histplot(data=df_train, x="Age", kde=True, hue="Transported")
display(
df_train[["Transported", "HomePlanet", "Age"]]
.groupby(by=["Transported", "HomePlanet"])
.median()
.reset_index()
)
fig, axes = plt.subplots(nrows=3, ncols=1)
sns.histplot(
data=df_train,
x="Age",
kde=True,
hue="HomePlanet",
ax=axes[0],
hue_order=["Earth", "Europa", "Mars"],
).set_title("População")
sns.histplot(
data=df_train[df_train["Transported"] == True],
x="Age",
kde=True,
hue="HomePlanet",
ax=axes[1],
hue_order=["Earth", "Europa", "Mars"],
).set_title("Transportados")
sns.histplot(
data=df_train[df_train["Transported"] == False],
x="Age",
kde=True,
hue="HomePlanet",
ax=axes[2],
hue_order=["Earth", "Europa", "Mars"],
).set_title("Não transportados")
plt.subplots_adjust(top=1.7)
fig.set_figheight(7)
# Em transportados, há um aumento nas idades baixas
# #### Cryosleep
sns.countplot(data=df_train, x="CryoSleep", hue="Transported").set_title(
"Sono Criogênico"
)
# #### HomePlanet
df_train["earthling"] = df_train["HomePlanet"].map(
{"Earth": True, "Mars": False, "Europa": False}
)
df_test["earthling"] = df_train["HomePlanet"].map(
{"Earth": True, "Mars": False, "Europa": False}
)
df_train["HomePlanet_num"] = df_train["HomePlanet"].map(
{"Earth": 1, "Mars": 0, "Europa": -1}
)
df_test["HomePlanet_num"] = df_train["HomePlanet"].map(
{"Earth": 1, "Mars": 0, "Europa": -1}
)
fig, axes = plt.subplots(nrows=1, ncols=2)
sns.countplot(df_train, x="HomePlanet", hue="Transported", ax=axes[0]).set_title(
"HomePlanet"
)
sns.countplot(df_train, x="earthling", hue="Transported", ax=axes[1]).set_title(
"Earthling"
)
fig.set_figwidth(10)
# #### Destination
sns.countplot(df_train, x="Destination", hue="Transported")
df_train["Destination_num"] = df_train["Destination"].map(
{"TRAPPIST-1e": 1, "PSO J318.5-22": 0, "55 Cancri e": -1}
)
df_test["Destination_num"] = df_test["Destination"].map(
{"TRAPPIST-1e": 1, "PSO J318.5-22": 0, "55 Cancri e": -1}
)
# #### Cabin
df_train[["deck", "num", "side"]] = df_train["Cabin"].str.split("/", expand=True)
df_test[["deck", "num", "side"]] = df_test["Cabin"].str.split("/", expand=True)
sns.countplot(df_train, x="deck", hue="Transported")
df_train["deck_num"] = df_train["deck"].map(
{"F": 1, "E": 1, "B": -1, "C": -1, "A": 0, "G": 0, "D": 0}
)
df_test["deck_num"] = df_test["deck"].map(
{"F": 1, "E": 1, "B": -1, "C": -1, "A": 0, "G": 0, "D": 0}
)
sns.countplot(df_train, x="side", hue="Transported")
df_train["side_num"] = df_train["side"].map({"P": 1, "S": -1})
df_test["side_num"] = df_test["side"].map({"P": 1, "S": -1})
# # Seleção de variáveis importantes
# ['CryoSleep','Age',"VIP",'RoomService','FoodCourt','ShoppingMall','Spa','VRDeck','HomePlanet_num', 'Destination_num', 'deck_num','side_num']
colunas = [
"CryoSleep",
"Age",
"VIP",
"RoomService",
"FoodCourt",
"ShoppingMall",
"Spa",
"VRDeck",
"HomePlanet_num",
"Destination_num",
"deck_num",
"side_num",
]
# #### Decide-se da base de treino remover dados nulos e da base de teste preencher com a mediana
df_test[colunas] = df_test[colunas].fillna(df_test[colunas].median())
df_train = df_train.dropna()
Y_train = df_train["Transported"]
X_test_origin = df_test[colunas]
X_train = df_train[colunas]
# # Rodar o modelo
# #### Usando a biblioteca sklearn
# ### Separar dados de treino entre treino e validação
# 
from sklearn.model_selection import train_test_split
X = X_train
y = Y_train
# X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.2, shuffle=True)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, shuffle=True)
# ## Exemplificando com o algoritmo KNN (K-nearest neighbors)
# 
# ### Inicializar e rodar o modelo
from sklearn.neighbors import KNeighborsClassifier
# Inicializa o modelo
model = KNeighborsClassifier()
# O método 'fit' do sklearn treina o algoritmo com os dados de treino
model.fit(X_train, y_train)
# Para modelos de classificação, o método score do sklearn calcula a acurácia por padrão.
model.score(X_test, y_test)
# Resultados que o modelo está prevendo para a variável alvo.
y_pred = model.predict(X_test)
y_pred
from sklearn.metrics import accuracy_score
accuracy_score(y_test, y_pred)
# ## "Tunar" os parâmetros
# #### Modelo tem vários parâmetros, que você pode ajustar para o problema:
# https://scikit-learn.org/stable/modules/generated/sklearn.neighbors.KNeighborsClassifier.html
#
model = KNeighborsClassifier(n_neighbors=5)
model.fit(X_train, y_train)
model.score(X_test, y_test)
scores = {}
for n_neighbors in range(1, 51):
model = KNeighborsClassifier(n_neighbors=n_neighbors)
model.fit(X_train, y_train)
score = model.score(X_test, y_test)
scores[n_neighbors] = score
scores
plt.figure(figsize=(6, 4))
plt.plot(scores.keys(), scores.values(), "-bo")
plt.xlabel("k", fontsize=15)
plt.ylabel("Accuracy", fontsize=15)
plt.show(True)
best_k = max(scores, key=scores.get)
print("Melhor k:", best_k)
model = KNeighborsClassifier(n_neighbors=15)
model.fit(X_train, y_train)
model.score(X_test, y_test)
# ### GridSearchCV
from sklearn.model_selection import cross_val_score, GridSearchCV
param_grid = {
"n_neighbors": list(range(1, 51)),
}
grid = GridSearchCV(
KNeighborsClassifier(), param_grid, cv=5, scoring="accuracy", verbose=3
)
# grid_search_knn=grid.fit(X_train, y_train)
grid_search_knn = grid.fit(X, y)
print("\n Melhores parâmetros: ", grid_search_knn.best_params_)
print("\nScore: ", grid_search_knn.best_score_)
# ## Cross Validation
# 
from sklearn.model_selection import cross_validate
for n_neighbors in range(1, 51):
model = KNeighborsClassifier(n_neighbors=n_neighbors)
# realiza a validação cruzada. Atenção ao X e y usados
cv = cross_validate(model, X, y, cv=5)
print("k:", n_neighbors, "accurace:", cv["test_score"].mean())
score = cv["test_score"].mean()
scores[n_neighbors] = score
cv
best_k = max(scores, key=scores.get)
best_score = scores[best_k]
print(
f"Melhor k = {best_k} com score = {best_score}".format(
best_k=best_k, best_score=best_score
)
)
best_score == grid_search_knn.best_score_
# ## Random Forest
# > #### O objetivo de um SVM é encontrar o hiperplano de separação ideal o qual maximiza a margem da base de treinamento.
# ### Árvore de decisão
# 
# #### Problemas: overfitting e viés
# ### Floresta aleatória
# 
# https://www.ibm.com/topics/random-forest#:~:text=Random%20forest%20is%20a%20commonly,both%20classification%20and%20regression%20problems.
# #### Documentação do sklearn: https://scikit-learn.org/stable/modules/generated/sklearn.ensemble.RandomForestClassifier.html
from sklearn.ensemble import RandomForestClassifier
# random_state=42
model = RandomForestClassifier(random_state=42)
model.fit(X_train, y_train)
model.score(X_test, y_test)
param_grid = {
"n_estimators": [200, 500], # número de árvores na floresta
"max_features": [
"sqrt",
"log2",
], # número de features a se considerar para cada divisão
"max_depth": [7, 8, 9, 10], # profundidade máxima da árvore
}
grid = GridSearchCV(
estimator=RandomForestClassifier(random_state=42),
param_grid=param_grid,
cv=5,
verbose=1,
)
grid_search_RF = grid.fit(X_train, y_train)
print("\n Melhores parâmetros: ", grid_search_RF.best_params_)
print("\nScore: ", grid_search_RF.best_score_)
# ## SVM (Support Vector Machine)
# > #### O objetivo de um SVM é encontrar o hiperplano de separação ideal o qual maximiza a margem da base de treinamento.
# 
# 
# 
# 
# 
# http://www2.decom.ufop.br/imobilis/svm-entendendo-sua-matematica-parte-1-a-margem/#:~:text=O%20SVM%20%C3%A9%20uma%20t%C3%A9cnica%20de%20classifica%C3%A7%C3%A3o%20baseada%20em%20aprendizado,cada%20classe%20estejam%20previamente%20classificadas.
from sklearn.svm import SVC
model = SVC()
model.fit(X_train, y_train)
model.score(X_test, y_test)
# #### Documentação sklearn: https://scikit-learn.org/stable/modules/generated/sklearn.svm.SVC.html
import time
start = time.time()
param_grid = {
"C": [1, 10, 50], # paramêtro de regularização
"gamma": [1, 0.1, 0.005, 0.00001, 0.0000001], # coeficiente do kernel
"kernel": ["rbf"],
}
grid = GridSearchCV(SVC(), param_grid, cv=5, verbose=1)
grid_search_svm = grid.fit(X_train, y_train)
end = time.time()
print("\n Melhores parâmetros: ", grid_search_svm.best_params_)
print("\nScore: ", grid_search_svm.best_score_)
print("\nTotal time: ", end - start)
# # Modelo final
# #### Compara-se os resultados dos modelos acima e escolhe o que obteve o maior score
print("Knn: ", grid_search_knn.best_score_)
print("RandomForest: ", grid_search_RF.best_score_)
print("SVM: ", grid_search_svm.best_score_)
# #### RandomForest é escolhido por ter maior pontuação
# Com modelo já treinado, obtém-se a predição para a base de testes
# Obs: lembrar de fazer na base de testes as mesmas modificações de features (colunas) que foi feito na base de treino (transformar features, criar features novas, etc)
y_test_predict = grid_search_RF.predict(X_test_origin)
output = pd.DataFrame(
{"PassengerId": df_test["PassengerId"], "Transported": y_test_predict}
)
output
output.to_csv("submission.csv", index=False)
|
import numpy as np
import torch
import torch.nn as nn
import torch.optim as optim
import matplotlib.pyplot as plt
def get_data(a, seq_lenth):
x = np.cos(a)
y = np.sin(a)
x_data = []
y_data = []
sample_num = int(len(x) / seq_length) # sample_num的个数为x_data的个数
for i in range(sample_num):
x_data.append(x[i * seq_length : (i + 1) * seq_length])
y_data.append(y[i * seq_length : (i + 1) * seq_length])
x_data = torch.tensor(x_data, dtype=torch.float32).unsqueeze(2)
y_data = torch.tensor(y_data, dtype=torch.float32).unsqueeze(2)
return x_data, y_data
seq_length = 5 # 每个epoch的长度
pi = 3.14
a = np.linspace(0 * pi, 1.1 * pi, 11000)
b = np.linspace(1.1 * pi, 2 * pi, 9000)
x_train, y_train = get_data(a, seq_length)
x_test, y_test = get_data(b, seq_length)
# 训练集的范围是0-10pi
aa = np.linspace(-10 * pi, 10 * pi, 2000)
bb = np.linspace(10 * pi, 14 * pi, 400)
xx_train, yy_train = get_data(aa, seq_length)
xx_test, yy_test = get_data(bb, seq_length)
# RNN
class EncoderRNN(nn.Module):
def __init__(self, input_size, hidden_size):
super(EncoderRNN, self).__init__()
self.hidden_size = hidden_size
self.rnn = nn.RNN(input_size, hidden_size, batch_first=True)
def forward(self, x):
_, hidden = self.rnn(x)
return hidden
class DecoderRNN(nn.Module):
def __init__(self, hidden_size, output_size):
super(DecoderRNN, self).__init__()
self.hidden_size = hidden_size
self.rnn = nn.RNN(output_size, hidden_size, batch_first=True)
self.fc = nn.Linear(hidden_size, output_size)
def forward(self, x, hidden):
output, _ = self.rnn(x, hidden)
output = self.fc(output)
return output
# RNN
input_size = 1
hidden_size = 64
output_size = 1
encoder = EncoderRNN(input_size, hidden_size)
decoder = DecoderRNN(hidden_size, output_size)
criterion = nn.MSELoss()
encoder_optimizer = optim.Adam(encoder.parameters(), lr=0.001)
decoder_optimizer = optim.Adam(decoder.parameters(), lr=0.001)
# RNN,训练集的范围是0-1pi
import matplotlib.pyplot as plt
epochs = 3000
loss_val = []
for epoch in range(epochs):
encoder_optimizer.zero_grad()
decoder_optimizer.zero_grad()
hidden = encoder(x_train)
decoder_input = torch.zeros(x_train.size(0), seq_length, 1)
y_pred = decoder(decoder_input, hidden)
loss = criterion(y_pred, y_train)
loss_val.append(loss.item())
loss.backward()
encoder_optimizer.step()
decoder_optimizer.step()
if epoch % 100 == 0:
print(f"Epoch{epoch},Loss:{loss.item()}")
plt.plot(loss_val, label="loss")
plt.legend()
plt.show()
print("Finished Training")
# RNN,训练集的范围是0-1pi
encoder.eval()
decoder.eval()
with torch.no_grad():
hidden = encoder(x_train)
decoder_input = torch.zeros(x_train.size(0), seq_length, 1)
y_train_pred = decoder(decoder_input, hidden)
train_loss = criterion(y_train_pred, y_train)
print(f"Train Loss:{train_loss.item()}")
hidden = encoder(x_test)
decoder_input = torch.zeros(x_test.size(0), seq_length, 1)
y_test_pred = decoder(decoder_input, hidden)
test_loss = criterion(y_test_pred, y_test)
print(f"Test Loss:{test_loss.item()}")
# RNN,训练集的范围是0-1pi
import matplotlib.pyplot as plt
x1 = x_train.numpy().reshape(-1)
y1 = y_train.numpy().reshape(-1)
y1_ = y_train_pred.numpy().reshape(-1)
x2 = x_test.numpy().reshape(-1)
y2 = y_test.numpy().reshape(-1)
y2_ = y_test_pred.numpy().reshape(-1)
x1.shape
y1.shape
plt.plot(a, y1, label="train_ture")
plt.plot(a, y1_, label="train_pred")
plt.plot(b, y2, label="test_ture")
plt.plot(b, y2_, label="test_pred")
plt.legend()
plt.show()
# RNN,训练集的范围是-10到10pi
import matplotlib.pyplot as plt
epochs = 3000
loss_val = []
for epoch in range(epochs):
encoder_optimizer.zero_grad()
decoder_optimizer.zero_grad()
hidden = encoder(xx_train)
decoder_input = torch.zeros(xx_train.size(0), seq_length, 1)
yy_pred = decoder(decoder_input, hidden)
loss = criterion(yy_pred, yy_train)
loss_val.append(loss.item())
loss.backward()
encoder_optimizer.step()
decoder_optimizer.step()
if epoch % 100 == 0:
print(f"Epoch{epoch},Loss:{loss.item()}")
plt.plot(loss_val, label="loss")
plt.legend()
plt.show()
print("Finished Training")
# RNN,训练集的范围是-10到10pi
encoder.eval()
decoder.eval()
with torch.no_grad():
hidden = encoder(xx_train)
decoder_input = torch.zeros(xx_train.size(0), seq_length, 1)
yy_train_pred = decoder(decoder_input, hidden)
train_loss = criterion(yy_train_pred, yy_train)
print(f"Train Loss:{train_loss.item()}")
hidden = encoder(xx_test)
decoder_input = torch.zeros(xx_test.size(0), seq_length, 1)
yy_test_pred = decoder(decoder_input, hidden)
test_loss = criterion(yy_test_pred, yy_test)
print(f"Test Loss:{test_loss.item()}")
# RNN,训练集的范围是-10到10pi
import matplotlib.pyplot as plt
x1 = xx_train.numpy().reshape(-1)
y1 = yy_train.numpy().reshape(-1)
y1_ = yy_train_pred.numpy().reshape(-1)
x2 = xx_test.numpy().reshape(-1)
y2 = yy_test.numpy().reshape(-1)
y2_ = yy_test_pred.numpy().reshape(-1)
x1.shape
y1.shape
plt.plot(aa, y1, label="train_ture")
plt.plot(aa, y1_, label="train_pred")
plt.plot(bb, y2, label="test_ture")
plt.plot(bb, y2_, label="test_pred")
plt.legend()
plt.show()
# LSTM
class LstmRNN(nn.Module):
def __init__(self, input_size, hidden_size, output_size, num_layers):
super().__init__()
self.lstm = nn.LSTM(
input_size, hidden_size, num_layers
) # utilize the LSTM model in torch.nn
self.linear1 = nn.Linear(hidden_size, output_size) # 全连接层
def forward(self, _x):
x, _ = self.lstm(_x) # _x is input, size (seq_len, batch, input_size)
# s, b, h = x.shape # x is output, size (seq_len, batch, hidden_size)
# x = x.view(s * b, h)
x = self.linear1(x)
# x = x.view(s, b, -1)
return x
# LSTM
input_size = 1
hidden_size = 64
output_size = 1
num_layers = 1
lstm = LstmRNN(input_size, hidden_size, output_size, num_layers)
criterion = nn.MSELoss()
lstm_optimizer = optim.Adam(lstm.parameters(), lr=0.001)
# LSTM,训练集的范围是0-1pi
import matplotlib.pyplot as plt
epochs = 600
running_loss = 0.0
loss_val = []
for epoch in range(epochs):
lstm_optimizer.zero_grad()
y_pred = lstm(x_train)
loss = criterion(y_pred, y_train)
loss.backward()
lstm_optimizer.step()
loss_val.append(loss.item())
if epoch % 100 == 0:
print(f"Epoch{epoch},Loss:{loss.item()}")
plt.plot(loss_val, label="loss")
plt.legend()
plt.show()
print("Finished Training")
# LSTM,训练集的范围是0-1pi
lstm.eval()
with torch.no_grad():
y_train_pred = lstm(x_train)
train_loss = criterion(y_train_pred, y_train)
print(f"Train Loss:{train_loss.item()}")
y_test_pred = lstm(x_test)
test_loss = criterion(y_test_pred, y_test)
print(f"Test Loss:{test_loss.item()}")
# LSTM,训练集的范围是0-1pi
import matplotlib.pyplot as plt
x1 = x_train.numpy().reshape(-1)
y1 = y_train.numpy().reshape(-1)
y1_ = y_train_pred.numpy().reshape(-1)
x2 = x_test.numpy().reshape(-1)
y2 = y_test.numpy().reshape(-1)
y2_ = y_test_pred.numpy().reshape(-1)
x1.shape
y1.shape
plt.plot(a, y1, label="train_ture")
plt.plot(a, y1_, label="train_pred")
plt.plot(b, y2, label="test_ture")
plt.plot(b, y2_, label="test_pred")
plt.legend()
plt.show()
# LSTM,训练集的范围是-10到10pi
import matplotlib.pyplot as plt
epochs = 600
running_loss = 0.0
loss_val = []
for epoch in range(epochs):
lstm_optimizer.zero_grad()
yy_pred = lstm(xx_train)
loss = criterion(yy_pred, yy_train)
loss.backward()
lstm_optimizer.step()
loss_val.append(loss.item())
if epoch % 100 == 0:
print(f"Epoch{epoch},Loss:{loss.item()}")
plt.plot(loss_val, label="loss")
plt.legend()
plt.show()
print("Finished Training")
# LSTM,训练集的范围是-10到10pi
lstm.eval()
with torch.no_grad():
yy_train_pred = lstm(xx_train)
train_loss = criterion(yy_train_pred, yy_train)
print(f"Train Loss:{train_loss.item()}")
yy_test_pred = lstm(xx_test)
test_loss = criterion(yy_test_pred, yy_test)
print(f"Test Loss:{test_loss.item()}")
# LSTM,训练集的范围是-10到10pi
import matplotlib.pyplot as plt
x1 = xx_train.numpy().reshape(-1)
y1 = yy_train.numpy().reshape(-1)
y1_ = yy_train_pred.numpy().reshape(-1)
x2 = xx_test.numpy().reshape(-1)
y2 = yy_test.numpy().reshape(-1)
y2_ = yy_test_pred.numpy().reshape(-1)
x1.shape
y1.shape
plt.plot(aa, y1, label="train_ture")
plt.plot(aa, y1_, label="train_pred")
plt.plot(bb, y2, label="test_ture")
plt.plot(bb, y2_, label="test_pred")
plt.legend()
plt.show()
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.