script
stringlengths 113
767k
|
|---|
# # Imports
from boruta import BorutaPy
import warnings
warnings.filterwarnings("ignore")
import pandas as pd
import numpy as np
from rdkit import Chem
from rdkit.Chem import AllChem
from rdkit.Chem import Descriptors
from rdkit.ML.Descriptors import MoleculeDescriptors
from rdkit.Chem import MACCSkeys
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
from sklearn.feature_selection import VarianceThreshold
from sklearn.metrics import f1_score, recall_score
from sklearn.base import BaseEstimator, TransformerMixin
from sklearn.pipeline import Pipeline
from xgboost import XGBClassifier
import time
import os
# # Preprocessing and pipeline
train_data_url = "https://raw.githubusercontent.com/RohithOfRivia/SMILES-Toxicity-Prediction/main/Data/train_II.csv"
test_data_url = "https://raw.githubusercontent.com/RohithOfRivia/SMILES-Toxicity-Prediction/main/Data/test_II.csv"
df = pd.read_csv(train_data_url)
# transforming each compound into their canonical SMILES format. Optional.
def canonicalSmiles(smile):
try:
return Chem.MolToSmiles(Chem.MolFromSmiles(smile))
except:
return Chem.MolToSmiles(Chem.MolFromSmiles("[Na+].[Na+].F[Si--](F)(F)(F)(F)F"))
# Reads data and split up the given features
class FileReadTransform(BaseEstimator, TransformerMixin):
def fit(self, X, y=None):
return self
# training and test data are slightly different, hence passing optional test param
def transform(self, X, test=False):
try:
# if test == False:
X["SMILES"] = X["Id"].apply(lambda x: x.split(";")[0])
X["assay"] = X["Id"].apply(lambda x: x.split(";")[1])
except KeyError:
X["SMILES"] = X["x"].apply(lambda x: x.split(";")[0])
X["assay"] = X["x"].apply(lambda x: x.split(";")[1])
print("FileReadTransform done")
# correct smiles for this compound found through https://www.molport.com/shop/index
# X["SMILES"] = X["SMILES"].replace({"F[Si-2](F)(F)(F)(F)F.[Na+].[Na+]":"[Na+].[Na+].F[Si--](F)(F)(F)(F)F"})
# Deleting invalid compound from the data
X = X.loc[X.SMILES != "F[Si-2](F)(F)(F)(F)F.[Na+].[Na+]"]
return X
# Converts each SMILES value to its respective canonical SMILES
class CanonicalGenerator(BaseEstimator, TransformerMixin):
def fit(self, X, y=None):
return self
def transform(self, X):
X["SMILES"] = X["SMILES"].apply(canonicalSmiles)
print("CanonicalGenerator done")
return X
# Generate fingerprints for all compounds
class FingerprintGenerator(BaseEstimator, TransformerMixin):
def fit(self, X, y=None):
return self
def transform(self, X):
# tracks each unique compound and its fingerprints
tracker = []
fps = []
assays = []
unique = len(X["SMILES"].unique())
counter = 0
for index, columns in X[["SMILES", "assay"]].iterrows():
# skip if already in tracker
if columns[0] in tracker:
continue
# append each unique compound and theer respective fingerprints
else:
tracker.append(columns[0])
assays.append(columns[1])
mol = Chem.MolFromSmiles(columns[0])
fp = AllChem.GetMorganFingerprintAsBitVect(mol, 2, nBits=256)
fps.append(fp.ToList())
counter += 1
# print(f"compound {counter}/{unique}...
# Combining all compounds, assays and fingerprints into one dataframe
cols = a = ["x" + str(i) for i in range(1, 257)]
smiles_df = pd.DataFrame(columns=["SMILES"], data=tracker)
fingerprints = pd.DataFrame(columns=cols, data=fps)
df = pd.concat([smiles_df, fingerprints], axis=1)
print("FingerprintGenerator done")
return pd.merge(X, df, on="SMILES")
# Fingerprint generation for MACCS Keys
class FingerprintGeneratorM(BaseEstimator, TransformerMixin):
def fit(self, X, y=None):
return self
def transform(self, X):
# tracks each unique compound and its fingerprints
tracker = []
fps = []
assays = []
unique = len(X["SMILES"].unique())
counter = 0
for index, columns in X[["SMILES", "assay"]].iterrows():
# skip if already in tracker
if columns[0] in tracker:
continue
# append each unique compound and thier respective fingerprints
else:
tracker.append(columns[0])
assays.append(columns[1])
mol = Chem.MolFromSmiles(columns[0])
fp = MACCSkeys.GenMACCSKeys(mol)
fps.append(fp.ToList())
counter += 1
# print(f"compound {counter}/{unique}...
# Combining all compounds, assays and fingerprints into one dataframe
cols = a = ["x" + str(i) for i in range(1, 168)]
smiles_df = pd.DataFrame(columns=["SMILES"], data=tracker)
fingerprints = pd.DataFrame(columns=cols, data=fps)
df = pd.concat([smiles_df, fingerprints], axis=1)
print("FingerprintGenerator done")
return pd.merge(X, df, on="SMILES")
# Feature reduction with variance threshold
class VarianceThresh(BaseEstimator, TransformerMixin):
def fit(self, X, y=None):
return self
def transform(self, X, thresh=0.8):
# Looks to columns to determine whether X is training or testing data
cols = X.columns
if "x" in cols:
temp_df = X.drop(columns=["x", "assay", "SMILES"])
cols = ["x", "assay", "SMILES"]
else:
temp_df = X.drop(columns=["Id", "Expected", "assay", "SMILES"])
cols = ["Id", "Expected", "assay", "SMILES"]
# Selecting features based on the variance threshold
selector = VarianceThreshold(threshold=(thresh * (1 - thresh)))
selector.fit(temp_df)
# This line transforms the data while keeping the column names
temp_df = temp_df.loc[:, selector.get_support()]
# Attaching the ids, assays, smiles etc. that is still required for model
return pd.concat([X[cols], temp_df], axis=1), selector
# Scale descriptors (Not used in this notebook)
class Scaler(BaseEstimator, TransformerMixin):
def fit(self, X, y=None):
return self
def transform(self, X):
scaler = StandardScaler()
if "Id" in X.columns:
temp_df = X.drop(columns=["Id", "Expected", "assay", "SMILES"])
cols = ["Id", "Expected", "assay", "SMILES"]
X_scaled = pd.DataFrame(
scaler.fit_transform(temp_df), columns=temp_df.columns
)
X = pd.concat([X[cols].reset_index(drop=True), X_scaled], axis=1)
return X
else:
temp_df = X.drop(columns=["x", "assay", "SMILES"])
cols = ["x", "assay", "SMILES"]
X_scaled = pd.DataFrame(
scaler.fit_transform(temp_df), columns=temp_df.columns
)
X = pd.concat([X[cols].reset_index(drop=True), X_scaled], axis=1)
return X
# # Generating descriptors
class DescriptorGenerator(BaseEstimator, TransformerMixin):
def fit(self, X, y=None):
return self
def transform(self, X):
# Initializing descriptor calculator
calc = MoleculeDescriptors.MolecularDescriptorCalculator(
[x[0] for x in Descriptors._descList]
)
desc_names = calc.GetDescriptorNames()
# Tracking each unique compound and generating descriptors
tracker = []
descriptors = []
for compound in X["SMILES"]:
if compound in tracker:
continue
else:
tracker.append(compound)
mol = Chem.MolFromSmiles(compound)
current_descriptors = calc.CalcDescriptors(mol)
descriptors.append(current_descriptors)
# Combining X, SMILES, and generated descriptors
df = pd.DataFrame(descriptors, columns=desc_names)
temp_df = pd.DataFrame(tracker, columns=["SMILES"])
df = pd.concat([df, temp_df], axis=1)
print("DescriptorGenerator done")
return pd.merge(X, df, on="SMILES")
# # Create Pipeline
feature_generation_pipeline = Pipeline(
steps=[
("read", FileReadTransform()),
("canon", CanonicalGenerator()),
("fpr", FingerprintGenerator()),
("desc", DescriptorGenerator()),
]
)
df_processed = feature_generation_pipeline.fit_transform(df)
test_processed = feature_generation_pipeline.fit_transform(pd.read_csv(test_data_url))
# # Feature Selection
# Isolating the chemical descriptors for feature selection
descriptors = pd.concat(
[
df_processed[["Id", "SMILES", "assay", "Expected"]],
df_processed[df_processed.columns[-208:]],
],
axis=1,
)
len(descriptors.columns[4:])
# Checking for NANs
descriptors.isna().sum().sum()
# Removing all columns which have NAN values
descriptors2 = descriptors.drop(
columns=[
"BCUT2D_MWHI",
"BCUT2D_MWLOW",
"BCUT2D_CHGHI",
"BCUT2D_CHGLO",
"BCUT2D_LOGPHI",
"BCUT2D_LOGPLOW",
"BCUT2D_MRHI",
"BCUT2D_MRLOW",
]
)
descriptors2 = descriptors2.dropna()
X = descriptors2.drop(["Id", "SMILES", "Expected"], axis=1)
y = descriptors2[["Expected"]]
# Removing all columns which have NAN values
X["assay"] = X["assay"].astype("int64")
# Features selected from BorutaPy
boruta_features2 = [
"HeavyAtomMolWt",
"MaxPartialCharge",
"MinAbsPartialCharge",
"BertzCT",
"Chi1",
"Chi1n",
"Chi2v",
"Chi3n",
"Chi3v",
"Chi4v",
"LabuteASA",
"PEOE_VSA3",
"SMR_VSA6",
"SMR_VSA7",
"EState_VSA8",
"VSA_EState2",
"VSA_EState4",
"HeavyAtomCount",
"NumAromaticCarbocycles",
"MolLogP",
"MolMR",
"fr_Ar_OH",
"fr_COO",
"fr_COO2",
"fr_C_O",
"fr_C_O_noCOO",
"fr_amide",
"fr_benzene",
"fr_phenol",
"fr_phenol_noOrthoHbond",
"fr_sulfonamd",
"fr_thiazole",
"fr_thiophene",
"fr_urea",
"assay",
]
# Splitting data for training and validation. Mapping expected values from 2 to 1 because XGBoost does not support it for binary classification
X_train, X_test, y_train, y_test = train_test_split(
X[boruta_features2],
y["Expected"].map({2: 0, 1: 1}),
test_size=0.2,
random_state=0,
stratify=y["Expected"].map({2: 0, 1: 1}),
)
""" Optional to run this. Gives a list of features that pass the test as the output.
The variable declaredboruta_features2 is a saved version of the output when the code was run for the best submission.
Output may vary slightly if executed again"""
# model = XGBClassifier(max_depth=6, learning_rate=0.01, n_estimators=600, colsample_bytree=0.3)
# boruta_features = []
# # let's initialize Boruta
# feat_selector = BorutaPy(
# verbose=2,
# estimator=model,
# n_estimators='auto',
# max_iter=10 # number of iterations to perform
# )
# # train Boruta
# # N.B.: X and y must be numpy arrays
# feat_selector.fit(np.array(X_train), np.array(y_train))
# # print support and ranking for each feature
# print("\n------Support and Ranking for each feature------")
# for i in range(len(feat_selector.support_)):
# if feat_selector.support_[i]:
# boruta_features.append(X_train.columns[i])
# print("Passes the test: ", X_train.columns[i],
# " - Ranking: ", feat_selector.ranking_[i])
# else:
# print("Doesn't pass the test: ",
# X_train.columns[i], " - Ranking: ", feat_selector.ranking_[i])
# # Training and Validation
# This method trains the model with the training data and then prints the f1 score by using predictions from the holdout data
def train_model(xtrain, xtest, ytrain, ytest):
model = XGBClassifier(seed=20, max_depth=10, n_estimators=700)
model.fit(xtrain, ytrain)
predictions = model.predict(xtest)
print(f"F1 Score of model: {f1_score(predictions, ytest)}")
train_model(X_train, X_test, y_train, y_test)
# Now training with the entire dataset to make predictions on the test set
descriptors2["assay"] = descriptors2["assay"].astype("int64")
model = XGBClassifier(seed=20, max_depth=10, n_estimators=700)
model.fit(descriptors2[boruta_features2], y["Expected"].map({2: 0, 1: 1}))
# Final predictions
# Changing assayID to int
test_processed["assay"] = test_processed["assay"].astype("int64")
# Making predicitons with the model
test_preds = model.predict(test_processed[boruta_features2])
test_preds
# Checking predictions for posititve and negative valeus
np.unique(test_preds, return_counts=True)
# # Saving Predicitons for kaggle submission
# Converting the predictions into a dataframe
res = pd.DataFrame({})
res["Id"] = test_processed["x"]
res["Predicted"] = test_preds
# Mapping expected values back to 2 and 1
res["Predicted"] = res["Predicted"].map({0: 2, 1: 1})
res
# For saving predictions as csv in JUPYTER
res.to_csv("submissions.csv")
# ONLY FOR GOOGLE COLLAB Downloading the csv for submission to kaggle
# from google.colab import files
# res.to_csv('28-03-23-2.csv', encoding = 'utf-8-sig', index=False)
# files.download('28-03-23-2.csv')
res
|
# # **Most Important Python Packages**
# Python is a popular and powerful general purpose programming language that recently emerged as the preferred language among data scientists. You can write your machine-learning algorithms using Python, and it works very well. However, there are a lot of modules and libraries already implemented in Python, that can make your life much easier. Here, the hands on experience of the packages are as follows-
# **NumPy(Numerical Python):** The first package is **NumPy** which is a math library to work with N-dimensional arrays in Python. It enables you to do computation efficiently and effectively. It is better than regular Python because of its amazing capabilities. For example, for working with **arrays**, **dictionaries**, **functions**, **datatypes** and working with **images** you need to know NumPy.
# One practical example of NumPy:
import numpy as np
arr = np.array([1, 2, 3, 4, 5])
print(arr)
print(type(arr))
# **SciPy(Scientic Python):** SciPy is a collection of numerical algorithms and domain specific toolboxes, including **signal processing**, **optimization**, statistics and much more. SciPy is a good library for scientific and high performance computation.
#
from scipy import constants
print(constants.liter)
|
# # Introduction
# - There is a lot of competition among the brands in the smartwatch industry. Smartwatches are preferred by people who like to take care of their fitness. Analyzing the data collected on your fitness is one of the use cases of Data Science in healthcare. So if you want to learn how to analyze smartwatch fitness data, this notebook is for you. In this notebook, I will take you through the task of Smartwatch Data Analysis using Python.
# ## Dataset
# - This dataset generated by respondents to a distributed survey via Amazon Mechanical Turk between 03.12.2016-05.12.2016. Thirty eligible Fitbit users consented to the submission of personal tracker data, including minute-level output for physical activity, heart rate, and sleep monitoring. Individual reports can be parsed by export session ID (column A) or timestamp (column B). Variation between output represents use of different types of Fitbit trackers and individual tracking behaviors / preferences.
# # Import libraries
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import plotly.express as px
import plotly.graph_objects as go
# # Read Data
data = pd.read_csv(
"/kaggle/input/fitbit/Fitabase Data 4.12.16-5.12.16/dailyActivity_merged.csv"
)
print(data.head())
# # null Values
# - Does the dataset contain null values or not?
print(data.isnull().sum())
# - So the dataset does not have any null values.
# - Let’s have a look at the information about columns in the dataset:
print(data.info())
# - The column containing the date of the record is an object. We may need to use dates in our analysis, so let’s convert this column into a datetime column:
# Changing datatype of ActivityDate
data["ActivityDate"] = pd.to_datetime(data["ActivityDate"], format="%m/%d/%Y")
print(data.info())
|
# # 
# NumPy (Numerical Python) is a widely-used open-source Python library for numerical computing. It provides support for large, multi-dimensional arrays and matrices, along with a collection of mathematical functions to operate on these arrays efficiently. NumPy is used in scientific computing, data analysis, machine learning, and other domains that require numerical computations. It offers optimized array operations for high performance, making it a powerful tool for working with large datasets. NumPy also provides functionality for working with mathematical data types and performing operations on arrays of arbitrary shapes and sizes. It is a fundamental library in the scientific Python ecosystem.
# # Basic_of_numpy
## importing the numpy library as np
import numpy as np
import warnings
warnings.filterwarnings("ignore") ## warning is ignoring the update warning
# ## I. 1D array in numpy
# One dimensional array contains elements only in one dimension.
## Create a numpy array
arr = np.array([1, 2, 3, 4, 5])
arr
# cheking the array size,ndim,shape,dtype
print(arr.shape) ## check the array shape( like columns and row)
print(arr.size) ## check the number of array size
print(arr.ndim) ## check the array number of disimile
print(arr.dtype) ## check the datatypes
# ## II. 2D and 3D arrays in numpy
# 2d array
arr2d = np.array([[1, 2, 3, 4], [5, 6, 7, 8]])
print(arr2d)
print("--" * 5)
print(arr2d.shape) ## check the array shape( like columns and row)
print(arr2d.size) ## check the number of array size
print(arr2d.ndim) ## check the array number of disimile
print(arr2d.dtype) ## check the datatypes
# 3d array
arr3d = np.array([[1, 2, 3], [4, 5, 6], [7, 8, 9]])
print(arr3d)
print("--" * 5)
print(arr3d.shape) ## check the array shape( like columns and row)
print(arr3d.size) ## check the number of array size
print(arr3d.ndim) ## check the array number of disimile
print(arr3d.dtype) ## check the datatypes
# ## III. Indexing in numpy array
## 1d array in numpy indexing
arr
print(arr[0]) ## 0 index is showing the stating array value
print(arr[-1]) ## -1 is showong the end of the index value
print(arr[:3]) ## this is slicing indexing methon in numpy
# ##### Indexing maltiple arrays in numpy
arr2d
arr2d[0] ## 2d array in 0 index are showing fist row of values
arr2d[1] ## 2d array in 1 index are showing second row of values
# ##### slicing indexing in numpy array
# 1. Same slicing methed are appling all thpe of array like (2D,3D,4D) many mores arrays
arr2d
arr2d[:1] ## 2d array in 0 index are showing fist row of values
arr2d[1:] ## 2d array in 1 index are showing second row of values
arr2d[:, :1] ## slising indexing for one [:,:1] are showing first columns values
arr2d[:, -1:] ## slising indexing for one [:,1:] are showing last columns values
arr2d[:, 1:2] ## slising indexing for one [:,1:2] are showing second columns of values
arr2d[:, 2:-1] ## slising indexing for one [:,2:-1] are showing therd columns of values
# ## IV. Replace the array values in numpy
arr3d
arr3d[:1, :] = 1 ## replace the first row values change and repalce
arr3d
arr3d[:, 1:-1] = 2 ## replace the midele columns values change and repalce
arr3d
arr3d[:, 2:] = 3 ## replace the last columns values change and repalce
arr3d
arr3d[0:, :-2] = 4 ## replace the first columns values change and repalce
arr3d
arr3d[1:2] = 5 ## replace the midele row values change and repalce
arr3d
arr3d[2:3] = 6 ## replace the last row values change and repalce
arr3d
# #### particalur values replace in numpy
arr2d
arr2d[:1, :1] = 22 ## replace the first column first row values change by 22
arr2d
arr2d[:1, -1:] = 23 ## replace the first row last values by 23
arr2d
arr2d[1:2, -1:] = 25 ## replace the second row last values by 25
arr2d
# #### Maltiple array values are also replace
arr2d[:2, 1:-2] = [28], [29] ## replace the second columns values by 28,29
arr2d
arr2d[:2, 2:] = [33, 34], [
56,
66,
] ### first and second row last two columns values chang by
arr2d
arr2d[0] = 23, 34, 5, 65 ## particalues first row values replace
arr2d
# #### Create array like floats ,bool,complex
ar = np.array([1, 2, 3, 4], float) ## creating the float array
ar2 = np.array([4, 5, 6], bool) ## creating the bool array
ar3 = np.array([7, 8, 9], complex) ## creating the complex array
print(ar)
print(ar2)
print(ar3)
# ## V. Change the array dtype
## change the array dtype float to int
changedtype1 = ar.astype(int)
print(changedtype1)
## change the array dtype bool to int
changedtype2 = ar2.astype(int)
print(changedtype2)
## change the array dtype complex to int
changedtype3 = ar3.astype(int)
print(changedtype3)
# ## VI. Numpy most useful function
# #### arange
# NumPy arange() is one of the array creation routines based on numerical ranges
f_arr = np.arange(24)
f_arr
print(f_arr.size)
print(f_arr.dtype)
# #### reshape
# Reshaping means changing the shape of an array. The shape of an array is the number of elements in each dimension
f_arr.reshape(2, 12) ## reshape function used by convert the ndim.
## two columns ndim
f_arr.reshape(12, 2)
## try to diff ndim
f_arr.reshape(6, 4)
f_arr.reshape(4, 6)
arangefunction = f_arr.reshape(6, 4) ## store the varaibles
arangefunction
# ### ravel
# The numpy.ravel() functions returns contiguous flattened array(1D array with all the input-array elements and with the same type as it).
#
arangefunction.ravel() ## ravel funtion used by reshape aposite like rearage the range
# #### append
# append() function is O(n) where n is the number of elements being appended.
append_array = np.append(
f_arr, [3, 5, 6, 7]
) ## append function add the new array values
append_array
len(append_array) ## check the len of array similer to size
append_array_re = append_array.reshape(7, 4) ## stored variables
append_array_re
# #### delete array values
# delete() function is used to delete the elements based on index position
deletelastrow = np.delete(append_array_re, append_array_re[-1:])
deletelastrow
len(deletelastrow)
# ### zeros
np.zeros((5, 5)) ## zeros are also help to malpile time creating zeros
# ### ones
np.ones((5, 5)) ## one are also help to malpile time creating zeros
# ### Identity
# The identity array is a square array with ones on the main diagonal. ... Reference object to allow the creation of arrays which are not NumPy arrays.
np.identity(5)
# ### linspace
np.linspace(10, 5) # used to create an evenly spaced sequence in a specified interval.
np.linspace(1, 20, 10)
# ### empty
# empty() function is used to return new array of a given shape and type. It has random values and uninitialized entries.
np.empty((10, 5))
# # VII. random in numpy
# Random() in Python. The random is a module present in the NumPy library. This module contains the functions which are used for generating random numbers. This module contains some simple random data generation methods, some permutation and distribution functions, and random generator functions.
np.random.randint(
100, size=(5, 5)
) # randint() is one of the function for doing random sampling in numpy
np.random.rand(10) # Random values in a given shape
np.random.normal(100, size=(10, 2))
np.random.power(
100, size=(10)
) # First array elements raised to powers from second array, element-wise.
# ### argmax,argmin,argsort
a = np.array([[1, 2, 3, 4], [55, 7, 5, 4], [8, 9, 10, 11]])
a
a.argmax() # used to return the indices of the max elements of the given array along with the specified axis.
a.argmin() # used to return the indices of the min elements of the given array along with the specified axis.
a.argsort() # the NumPy library has a function called argsort() , which computes the indirect sorting of an array.
# # VIII. basic maths in numpy
#
a
a.min() # NumPy's minimum() function is the tool of choice for finding minimum values across arrays.
a.max() # NumPy's maximum() function is the tool of choice for finding maximum values across arrays.
a.sum() ## sum funtion giving the total of array values
a.mean() # Returns the average of the array elements.
np.sqrt(a) # use the numpy. sqrt() method in Python Numpy.
# # IX. Numpy Operations..
# NumPy performs operations element-by-element, so multiplying 2D arrays with * is not a matrix multiplication – it's an element-by-element multiplication.
a = np.arange(4, 8)
b = np.array([6, 7, 8, 4])
a, b
a + b
a * b
a / b
a // b
a - b
a**b
# ## X. Joining numpy arrays
# Joining means putting contents of two or more arrays in a single array.
a
b
np.concatenate([a, b]) ## maliple varieable marge using concat funtion
c = np.arange(10).reshape(2, 5)
d = np.array([[4, 5, 6, 3, 5], [7, 6, 4, 2, 3]])
c, d
marge = np.concatenate([c, d], axis=0)
marge
marge.T ## trainform the columns to row data
marge.sort() ## sort funtion used by sequent wise values showing
marge
# ### array_split
array_split = np.array_split(marge, [3]) # Split an array into multiple sub-arrays
array_split
array_split[0]
array_split[1]
# # XI. Axis in numpy
# Axes are defined for arrays with more than one dimension.
marge.sum(axis=0)
marge.sum(axis=1)
marge.max(axis=1)
marge.min(axis=0)
# # XII. where
# The NumPy module provides a function numpy. where() for selecting elements based on a condition
np.where(marge > 6)
np.where(
marge > 6, -1, marge
) ## select the particale values replace values usning the where duntion
# ### count_nonzero,nonzero
np.count_nonzero(marge) ## except to zero values
np.nonzero(marge) ## indexing find the zero values
# ### setdiff1d
# Find the set difference of two arrays. Return the unique values in ar1 that are not in ar2. Parameters: ar1array_like. Input array.
c, d
np.setdiff1d(c, d)
# ### intersect1d
# the intersection of two arrays and return the sorted, unique values that are in both of the input arrays.
np.intersect1d(c, d)
for i in a:
print(i) ## loop fintion used by conver series.
|
#
# ---
# # | Python: Projeto Final - Análise de Crédito
# Caderno de **Aula**
# Professor [André Perez](https://www.linkedin.com/in/andremarcosperez/)
# Aluno [Thomaz Pires](https://www.linkedin.com/in/thomazbp/)
# ---
# ## 1\. Problema
# Vamos explorar dados de crédito presentes neste neste [link](https://raw.githubusercontent.com/andre-marcos-perez/ebac-course-utils/develop/dataset/credito.csv). Os dados estão no formato CSV e contém informações sobre clientes de uma instituição financeira. Em especial, estamos interessados em explicar a segunda coluna, chamada de **default**, que indica se um cliente é adimplente(`default = 0`), ou inadimplente (`default = 1`), ou seja, queremos entender o porque um cliente deixa de honrar com suas dívidas baseado no comportamento de outros atributos, como salário, escolaridade e movimentação financeira. Uma descrição completa dos atributos está abaixo.
# ### **1.1. LGPD**
# Uma análise de crédito justa é essencial, principalmente ao calcular classificações de crédito e avaliar a confiabilidade de um pagador. Ao forçar as organizações a obter consentimento dos titulares de dados e restringir as transferências de informações a terceiros autorizados, a LGPD aumentou a barreira. Como resultado, as pessoas passaram a ter mais direitos e as empresas devem ser mais cautelosas ao fazer análises de crédito.
# As empresas não podem mais recusar crédito com base apenas na residência ou idade de uma pessoa. Essas práticas passaram a ser vistas como injustas e discriminatórias porque a LGPD preconiza o tratamento igualitário de todos.
# As análises feitas aqui levam em não levam em consideração criterios como raça, sexo, genêro, moradia, crença, religião, ideologia política ou outros fatores que possam ser considerados discriminatórios.
# Segue estrutura dos dados:
# | Coluna | Descrição |
# | ------- | --------- |
# | id | Número da conta |
# | default | Indica se o cliente é adimplente (0) ou inadimplente (1) |
# | idade | --- |
# | sexo | --- |
# | depedentes | --- |
# | escolaridade | --- |
# | estado_civil | --- |
# | salario_anual | Faixa do salario mensal multiplicado por 12 |
# | tipo_cartao | Categoria do cartao: blue, silver, gold e platinium |
# | meses_de_relacionamento | Quantidade de meses desde a abertura da conta |
# | qtd_produtos | Quantidade de produtos contratados |
# | iteracoes_12m | Quantidade de iteracoes com o cliente no último ano |
# | meses_inatico_12m | Quantidade de meses que o cliente ficou inativo no último ano |
# | limite_credito | Valor do limite do cartão de crédito |
# | valor_transacoes_12m | Soma total do valor das transações no cartão de crédito no último ano |
# | qtd_transacoes_12m | Quantidade total de transações no cartão de crédito no último ano |
# ## 2\. Setup
# Import libraries
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
# Data Import
df = pd.read_csv("../input/creditocsv/credito.csv", na_values="na")
df.head(n=10) # retorna 10 primeiras linhas
# ### **2.1. Schema**
# Estão presentes 10127 registros em 16 colunas.
df.shape # retorna uma tupla (qtd linhas, qtd colunas)
# Será necessário tratamento das colunas `limite_credito` e `valor_transacoes_12m` de `object` para `float `
df.dtypes # retorna data types das colunas
# - Atributos **categóricos**.
df.select_dtypes("object").describe().transpose()
# - Atributos **numéricos**.
df.drop("id", axis=1).select_dtypes("number").describe().transpose()
# Dados faltantes podem ser:
# * Vazios ("");
# * Nulos (None);
# * Não disponíveis ou aplicaveis (na, NA, etc.);
# * Não numérico (nan, NaN, NAN, etc).
df.isna().any() # retorna booleano para presença de dados faltantes em cada coluna
# A função abaixo levanta algumas estatisticas sobre as colunas dos dados faltantes. Apesar do percentual de dados faltantes, clientes adimplentes e inadimplentes serem proxímos e que a escolha por deletar esses dados represente apenas uma alteração de 0,5% no padrão dos dados, a escolha por manter esses dados e completá-los se dá para o aprofundamento das análises.
def stats_dados_faltantes(
df: pd.DataFrame,
) -> None: # função para contagem de dados faltantes
stats_dados_faltantes = []
for col in df.columns:
if df[col].isna().any():
qtd, _ = df[df[col].isna()].shape
total, _ = df.shape
dict_dados_faltantes = {
col: {"quantidade": qtd, "porcentagem": round(100 * qtd / total, 2)}
}
stats_dados_faltantes.append(dict_dados_faltantes)
for stat in stats_dados_faltantes:
print(stat)
print("Estatisticas de dados faltantes de todo o df\n")
stats_dados_faltantes(df=df) # estatisticas de dados faltantes de todo o df
print("\nEstatisticas dados faltantes especificas de clientes adimplentes\n")
stats_dados_faltantes(
df=df[df["default"] == 0]
) # estatisticas dados faltantes especificas de clientes adimplentes
print("\nEstatisticas dados faltantes especificas de clientes inadimplentes\n")
stats_dados_faltantes(
df=df[df["default"] == 1]
) # estatisticas dados faltantes especificas de clientes inadimplentes
# ## 3\. Transformação e limpeza de dados
# As colunas limite_credito e valor_transacoes_12m estavam sendo interpretadas como colunas categóricas (dtype = object).
df[["limite_credito", "valor_transacoes_12m"]].dtypes
df[["limite_credito", "valor_transacoes_12m"]].head(n=5)
# Função lambda para limpar os dados.
fn = lambda valor: float(valor.replace(".", "").replace(",", "."))
df["valor_transacoes_12m"] = df["valor_transacoes_12m"].apply(fn)
df["limite_credito"] = df["limite_credito"].apply(fn)
df[["limite_credito", "valor_transacoes_12m"]].head(n=5)
df[["limite_credito", "valor_transacoes_12m"]].dtypes
# ### **3.1. Adição de dados faltantes**
# Para adição dos dados faltantes será testado o metodo ffill e bfill. Como os dados são do tipo object será utilizado o `countplot` para visualizar.
# * `'escolaridade'`
# * `'estado_civil'`
# * `'salario_anual'`
#
sns.countplot(x="escolaridade", data=df)
df["escolaridade"].fillna(method="ffill", inplace=True)
sns.countplot(x="escolaridade", data=df)
sns.countplot(x="estado_civil", data=df)
df["escolaridade"].fillna(method="ffill", inplace=True)
sns.countplot(x="estado_civil", data=df)
sns.countplot(x="salario_anual", data=df)
df["salario_anual"].fillna(method="ffill", inplace=True)
sns.countplot(x="salario_anual", data=df)
# O metódo 'ffill' matenve a forma geral das colunas, mas pode não ter sido a melhor opção. Será mantido esse formato pela finalidade de estudo dest projeto.
df.isna().any() # retorna booleano para presença de dados faltantes em cada coluna
# A coluna 'estado_civil' ainda apreenta valores nulos. será aplicado o metodo bfill
df["escolaridade"].fillna(method="bfill", inplace=True)
df.isna().any() # retorna booleano para presença de dados faltantes em cada coluna
# Como a coluna ainda possui valores nulos. Será utilizado um metodo de adicionar um valor 'random' dentre as opções desta coluna.
import random
possible_values = (
df["estado_civil"].dropna().tolist()
) # cria lista de valores possiveis mantendo a probabilidade estatistica dos valores
print(possible_values)
df["estado_civil"].fillna(random.choice(possible_values), inplace=True)
df.isna().any() # retorna booleano para presença de dados faltantes em cada coluna
# ## 4\. Visualização de dados
# Vamos sempre comparar a base com todos os clientes com a base de adimplentes e inadimplentes.
sns.set_style("whitegrid")
df_adimplente = df[df["default"] == 0]
df_inadimplente = df[df["default"] == 1]
df.select_dtypes("object").head(n=5)
# ### **4.1. Visualizações categóricas**
df.select_dtypes("object").describe().transpose()
# - Escolaridade : Não se observa critérios claros com base nessa variável
coluna = "escolaridade"
titulos = [
"Escolaridade dos Clientes",
"Escolaridade dos Clientes Adimplentes",
"Escolaridade dos Clientes Inadimplentes",
]
eixo = 0
max_y = 0
max = df.select_dtypes("object").describe()[coluna]["freq"] * 1.1
figura, eixos = plt.subplots(1, 3, figsize=(20, 5), sharex=True)
for dataframe in [df, df_adimplente, df_inadimplente]:
df_to_plot = dataframe[coluna].value_counts().to_frame()
df_to_plot.rename(columns={coluna: "frequencia_absoluta"}, inplace=True)
df_to_plot[coluna] = df_to_plot.index
df_to_plot.sort_values(by=[coluna], inplace=True)
df_to_plot.sort_values(by=[coluna])
f = sns.barplot(
x=df_to_plot[coluna], y=df_to_plot["frequencia_absoluta"], ax=eixos[eixo]
)
f.set(title=titulos[eixo], xlabel=coluna.capitalize(), ylabel="Frequência Absoluta")
f.set_xticklabels(labels=f.get_xticklabels(), rotation=90)
_, max_y_f = f.get_ylim()
max_y = max_y_f if max_y_f > max_y else max_y
f.set(ylim=(0, max_y))
eixo += 1
figura.show()
# - Salário Anual - Não se observa critérios claros com base nessa variável
#
coluna = "salario_anual"
titulos = [
"Salário Anual dos Clientes",
"Salário Anual dos Clientes Adimplentes",
"Salário Anual dos Clientes Inadimplentes",
]
eixo = 0
max_y = 0
figura, eixos = plt.subplots(1, 3, figsize=(20, 5), sharex=True)
for dataframe in [df, df_adimplente, df_inadimplente]:
df_to_plot = dataframe[coluna].value_counts().to_frame()
df_to_plot.rename(columns={coluna: "frequencia_absoluta"}, inplace=True)
df_to_plot[coluna] = df_to_plot.index
df_to_plot.reset_index(inplace=True, drop=True)
df_to_plot.sort_values(by=[coluna], inplace=True)
f = sns.barplot(
x=df_to_plot[coluna], y=df_to_plot["frequencia_absoluta"], ax=eixos[eixo]
)
f.set(title=titulos[eixo], xlabel=coluna.capitalize(), ylabel="Frequência Absoluta")
f.set_xticklabels(labels=f.get_xticklabels(), rotation=90)
_, max_y_f = f.get_ylim()
max_y = max_y_f if max_y_f > max_y else max_y
f.set(ylim=(0, max_y))
eixo += 1
figura.show()
# - Tipo cartao : Observa-se critérios que tendem a influenciar essa variável
coluna = "tipo_cartao"
titulos = [
"Tipo do Cartão",
"Tipo do Cartão dos Clientes Adimplentes",
"Tipo do Cartão dos Clientes Inadimplentes",
]
eixo = 0
max_y = 0
figura, eixos = plt.subplots(1, 3, figsize=(20, 5), sharex=True)
for dataframe in [df, df_adimplente, df_inadimplente]:
df_to_plot = dataframe[coluna].value_counts().to_frame()
df_to_plot.rename(columns={coluna: "frequencia_absoluta"}, inplace=True)
df_to_plot[coluna] = df_to_plot.index
df_to_plot.reset_index(inplace=True, drop=True)
df_to_plot.sort_values(by=[coluna], inplace=True)
f = sns.barplot(
x=df_to_plot[coluna], y=df_to_plot["frequencia_absoluta"], ax=eixos[eixo]
)
f.set(title=titulos[eixo], xlabel=coluna.capitalize(), ylabel="Frequência Absoluta")
f.set_xticklabels(labels=f.get_xticklabels(), rotation=90)
_, max_y_f = f.get_ylim()
max_y = max_y_f if max_y_f > max_y else max_y
f.set(ylim=(0, max_y))
eixo += 1
figura.show()
# ### **4.2. Visualizações numéricas**
# Nesta seção, vamos visualizar a relação entre a variável resposta **default** com os atributos numéricos.
df.drop(["id", "default"], axis=1).select_dtypes("number").head(n=5)
# - Quantidade de Transações nos Últimos 12 Meses - Observa-se critérios que tendem a influenciar essa variável
coluna = "qtd_transacoes_12m"
titulos = [
"Qtd. de Transações no Último Ano",
"Qtd. de Transações no Último Ano de Adimplentes",
"Qtd. de Transações no Último Ano de Inadimplentes",
]
eixo = 0
max_y = 0
figura, eixos = plt.subplots(1, 3, figsize=(20, 5), sharex=True)
for dataframe in [df, df_adimplente, df_inadimplente]:
f = sns.histplot(x=coluna, data=dataframe, stat="count", ax=eixos[eixo])
f.set(title=titulos[eixo], xlabel=coluna.capitalize(), ylabel="Frequência Absoluta")
_, max_y_f = f.get_ylim()
max_y = max_y_f if max_y_f > max_y else max_y
f.set(ylim=(0, max_y))
eixo += 1
figura.show()
# - Valor das Transações nos Últimos 12 Meses - Observa-se critérios que tendem a influenciar essa variável
coluna = "valor_transacoes_12m"
titulos = [
"Valor das Transações no Último Ano",
"Valor das Transações no Último Ano de Adimplentes",
"Valor das Transações no Último Ano de Inadimplentes",
]
eixo = 0
max_y = 0
figura, eixos = plt.subplots(1, 3, figsize=(20, 5), sharex=True)
for dataframe in [df, df_adimplente, df_inadimplente]:
f = sns.histplot(x=coluna, data=dataframe, stat="count", ax=eixos[eixo])
f.set(title=titulos[eixo], xlabel=coluna.capitalize(), ylabel="Frequência Absoluta")
_, max_y_f = f.get_ylim()
max_y = max_y_f if max_y_f > max_y else max_y
f.set(ylim=(0, max_y))
eixo += 1
figura.show()
# - Valor de Transações nos Últimos 12 Meses x Quantidade de Transações nos Últimos 12 Meses - Observa-se critérios que tendem a influenciar essa variável
f = sns.relplot(
x="valor_transacoes_12m", y="qtd_transacoes_12m", data=df, hue="default"
)
_ = f.set(
title="Relação entre Valor e Quantidade de Transações no Último Ano",
xlabel="Valor das Transações no Último Ano",
ylabel="Quantidade das Transações no Último Ano",
)
# - Tempo de relacionamento -Não observa-se critérios que tendem a influenciar essa variável
coluna = "meses_de_relacionamento"
titulos = [
"Tempo de relacionamento",
"Tempo de relacionamento de Adimplentes",
"Tempo de relacionamento de Inadimplentes",
]
eixo = 0
max_y = 0
figura, eixos = plt.subplots(1, 3, figsize=(20, 5), sharex=True)
for dataframe in [df, df_adimplente, df_inadimplente]:
f = sns.histplot(x=coluna, data=dataframe, stat="count", ax=eixos[eixo])
f.set(title=titulos[eixo], xlabel=coluna.capitalize(), ylabel="Frequência Absoluta")
_, max_y_f = f.get_ylim()
max_y = max_y_f if max_y_f > max_y else max_y
f.set(ylim=(0, max_y))
eixo += 1
figura.show()
# - Quantidade de produtos -Não observa-se critérios que tendem a influenciar essa variável
coluna = "qtd_produtos"
titulos = [
"Quantidade de produtos",
"Quantidade de produtos de Adimplentes",
"Quantidade de produtos de Inadimplentes",
]
eixo = 0
max_y = 0
figura, eixos = plt.subplots(1, 3, figsize=(20, 5), sharex=True)
for dataframe in [df, df_adimplente, df_inadimplente]:
f = sns.histplot(x=coluna, data=dataframe, stat="count", ax=eixos[eixo])
f.set(title=titulos[eixo], xlabel=coluna.capitalize(), ylabel="Frequência Absoluta")
_, max_y_f = f.get_ylim()
max_y = max_y_f if max_y_f > max_y else max_y
f.set(ylim=(0, max_y))
eixo += 1
figura.show()
# - Iterações em 12 meses -Não observa-se critérios que tendem a influenciar essa variável
coluna = "iteracoes_12m"
titulos = ["Iteraçãoes", "Iteraçãoes de Adimplentes", "Iteraçãoes de Inadimplentes"]
eixo = 0
max_y = 0
figura, eixos = plt.subplots(1, 3, figsize=(20, 5), sharex=True)
for dataframe in [df, df_adimplente, df_inadimplente]:
f = sns.histplot(x=coluna, data=dataframe, stat="count", ax=eixos[eixo])
f.set(title=titulos[eixo], xlabel=coluna.capitalize(), ylabel="Frequência Absoluta")
_, max_y_f = f.get_ylim()
max_y = max_y_f if max_y_f > max_y else max_y
f.set(ylim=(0, max_y))
eixo += 1
figura.show()
# - Inatividade em 12 meses -Não observa-se critérios que tendem a influenciar essa variável
coluna = "meses_inativo_12m"
titulos = [
"Inatividade em 12 meses",
"Inatividade em 12 meses de Adimplentes",
"Inatividade em 12 meses de Inadimplentes",
]
eixo = 0
max_y = 0
figura, eixos = plt.subplots(1, 3, figsize=(20, 5), sharex=True)
for dataframe in [df, df_adimplente, df_inadimplente]:
f = sns.histplot(x=coluna, data=dataframe, stat="count", ax=eixos[eixo])
f.set(title=titulos[eixo], xlabel=coluna.capitalize(), ylabel="Frequência Absoluta")
_, max_y_f = f.get_ylim()
max_y = max_y_f if max_y_f > max_y else max_y
f.set(ylim=(0, max_y))
eixo += 1
figura.show()
# - Limite de crédito -Não observa-se critérios que tendem a influenciar essa variável
coluna = "limite_credito"
titulos = [
"Limite de credito",
"Limite de credito de Adimplentes",
"Limite de credito de Inadimplentes",
]
eixo = 0
max_y = 0
figura, eixos = plt.subplots(1, 3, figsize=(20, 5), sharex=True)
for dataframe in [df, df_adimplente, df_inadimplente]:
f = sns.histplot(x=coluna, data=dataframe, stat="count", ax=eixos[eixo])
f.set(title=titulos[eixo], xlabel=coluna.capitalize(), ylabel="Frequência Absoluta")
_, max_y_f = f.get_ylim()
max_y = max_y_f if max_y_f > max_y else max_y
f.set(ylim=(0, max_y))
eixo += 1
figura.show()
# ## 5\. Resumo dos insights gerados.
# Observa-se que até 75% do inadimplentes transacionaram menos de 51 vezes e valores menores a 2772.615 nos últimos 12 meses, também observa-se que 75% possuem até 9933 reais de crédito
filtered_df = df[df["default"] == 1]
filtered_df.drop("id", axis=1).select_dtypes("number").describe().transpose()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
df = pd.read_csv("/kaggle/input/chatgpt-1000-daily-tweets/chatgpt_daily_tweets.csv")
df.info()
df.describe()
print(df.isnull().sum())
# The result shows that the table contains 8000 rows and 20 columns.
# The "source" column does not contain any non-zero values, so it can be removed from the dataframe.
# The columns "user_location", "user_description", "user_created", "user_followers_count", "user_following_count", "user_tweet_count", "user_verified", "retweet_count", "like_count", "reply_count" and "impression_count" have zero values, so you need to check how they can affect the results of the analysis.
# You should also bring the columns "tweet_created" and "tweet_extracted" to the datetime type for the convenience of further work with them.
df.drop(columns=["source"], inplace=True)
df["tweet_created"] = pd.to_datetime(df["tweet_created"])
df["tweet_extracted"] = pd.to_datetime(df["tweet_extracted"])
df.head()
# # EDA
# Тo begin with, let's plot the distribution of some features.
# For example, the distribution of the number of likes, retweets and replies to tweets can be plotted using histograms:
tweets_count = df.shape[0]
likes_count = df["like_count"].sum()
retweets_count = df["retweet_count"].sum()
replies_count = df["reply_count"].sum()
impressions_count = df["impression_count"].sum()
print(f"Number of tweets: {tweets_count}")
print(f"Number of likes: {likes_count}")
print(f"Number of retweets: {retweets_count}")
print(f"Number of responses: {replies_count}")
print(f"Number of views: {impressions_count}")
import matplotlib.pyplot as plt
# распределение количества лайков
plt.hist(df["like_count"], bins=50)
plt.title("Like count distribution")
plt.xlabel("Like count")
plt.ylabel("Number of tweets")
plt.show()
# распределение количества ретвитов
plt.hist(df["retweet_count"], bins=50)
plt.title("Retweet count distribution")
plt.xlabel("Retweet count")
plt.ylabel("Number of tweets")
plt.show()
# распределение количества ответов
plt.hist(df["reply_count"], bins=50)
plt.title("Reply count distribution")
plt.xlabel("Reply count")
plt.ylabel("Number of tweets")
plt.show()
# ящик с усами для количества лайков
plt.boxplot(df["like_count"].dropna())
plt.title("Like count boxplot")
plt.ylabel("Like count")
plt.show()
# ящик с усами для количества ретвитов
plt.boxplot(df["retweet_count"].dropna())
plt.title("Retweet count boxplot")
plt.ylabel("Retweet count")
plt.show()
# ящик с усами для количества ответов
plt.boxplot(df["reply_count"].dropna())
plt.title("Reply count boxplot")
plt.ylabel("Reply count")
plt.show()
# Graphs show the distribution of the number of likes, retweets and replies in tweets.
# For all three graphs, it can be concluded that there are many outliers, i.e. values that differ significantly from most other values in the sample. You can also notice that the distribution of the number of likes, retweets and replies has a heavy right tail, which suggests that a small number of tweets get a lot of likes, retweets and replies, while most tweets have a small number of likes, retweets and replies.
# диаграмма рассеяния для количества лайков и ретвитов
plt.scatter(df["like_count"], df["retweet_count"])
plt.title("Like count vs Retweet count")
plt.xlabel("Like count")
plt.ylabel("Retweet count")
plt.show()
# This graph is a scatter plot for the number of likes and retweets that received responses. It allows you to evaluate the relationship between these two variables.
# If the points on the graph are located close to a straight line, then this indicates that there is a strong positive correlation between likes and retweets - that is, the more likes, the more retweets. If the points are located far from each other and form bundles, then this indicates the absence or weak correlation between these variables.
# In this case, the graph shows that there are a number of tweets that have received a large number of likes and retweets. However, in general, it can be noticed that many tweets received less than 100 likes and retweets. You can also notice that there are a number of tweets that have received a lot of likes, but few retweets, and vice versa - a lot of retweets, but few likes. This may be due to various factors, such as the subject of the tweet or the hashtags that were used.
corr_matrix = df[["like_count", "retweet_count", "reply_count"]].corr()
# создаем таблицу из матрицы корреляций и добавляем цветовую шкалу
table = corr_matrix.style.background_gradient(cmap="coolwarm")
# выводим таблицу на экран
table
# The following conclusions can be drawn from this matrix:
# There is a strong positive correlation between the number of likes and the number of responses (the correlation coefficient is 0.44). This may mean that users who put likes are more likely to leave comments under the post, and vice versa.
# The correlation between the number of likes and the number of retweets is very weak (the correlation coefficient is close to 0). This may indicate that users who put likes do not necessarily retweet the post, and vice versa.
# The correlation between the number of retweets and the number of replies is also very weak (the correlation coefficient is close to 0). This may mean that users who retweet a post do not necessarily leave comments under it, and vice versa.
# The general conclusion: the number of likes on a post is strongly related to the number of comments, but not related to the number of retweets, and vice versa. The number of retweets is not related to the number of comments.
# # Analysis content of responses
# To analyze the content of responses, you can use text analysis methods, such as tonality analysis, keyword extraction, and thematic modeling.
# To analyze the tonality, you can use machine learning algorithms that classify tweets into positive, negative or neutral. To highlight keywords, you can use natural language processing methods, such as TF-IDF or a bag of words.
# Thematic modeling allows you to identify the main topics that are covered in tweets. To do this, you can use the LDA (Latent Dirichlet Allocation) or NMF (Non-negative Matrix Factorization) methods.
# You can also use graphical analysis to identify the most common words and highlight the most significant topics. To do this, you can use libraries for data visualization, for example, Wordcloud or Matplotlib.
# For most methods of text analysis, including thematic modeling and tonality analysis, the language of the text plays an important role, since these methods often use linguistic features of the language to determine the themes, tonality and other properties of the text.
# If the answers are written in different languages, then appropriate text processing methods and models specific to that language should be used for each language. Some text analysis methods can be trained in several languages, but it depends on the specific model and the languages that were used to teach it.
# It is also worth considering that when translating a text into another language, there may be a loss of information and a change in the meaning of the text, which may affect the results of the analysis. Therefore, when analyzing text in several languages, it is important to take into account the peculiarities of each language and use appropriate methods and models for processing text in each language.
# # Тonality analysis
from textblob import TextBlob
# Add new column with sentiment polarity to the DataFrame
df["sentiment"] = df["text"].apply(lambda x: TextBlob(x).sentiment.polarity)
# Print the mean sentiment polarity by gender
import matplotlib.pyplot as plt
plt.hist(df["sentiment"], bins=25)
plt.xlabel("Sentiment polarity")
plt.ylabel("Frequency")
plt.title("Distribution of Sentiment Polarity")
plt.show()
# On the histogram, we see the distribution of the tone of tweets in positive, neutral and negative tones. Most tweets have a neutral tone, which means that the authors do not express an explicit positive or negative assessment. A small number of tweets have a very low (very negative) or very high (very positive) tonality.
# Подсчитать средние значения retweet_count, like_count и reply_count для каждой тональности
counts_by_sentiment = df.groupby("sentiment")[
["retweet_count", "like_count", "reply_count"]
].sum()
print(counts_by_sentiment)
# график retweet_count
plt.plot(counts_by_sentiment.index, counts_by_sentiment["retweet_count"])
plt.title("Retweets by Sentiment Range")
plt.xlabel("Sentiment Range")
plt.ylabel("Retweets")
plt.show()
# график like_count
plt.plot(counts_by_sentiment.index, counts_by_sentiment["like_count"])
plt.title("Likes by Sentiment Range")
plt.xlabel("Sentiment Range")
plt.ylabel("Likes")
plt.show()
# график reply_count
plt.plot(counts_by_sentiment.index, counts_by_sentiment["reply_count"])
plt.title("Replies by Sentiment Range")
plt.xlabel("Sentiment Range")
plt.ylabel("Replies")
plt.show()
# **Several conclusions can be drawn from this data:**
# The average value of retweet_count is significantly higher for tweets with a negative tone than for tweets with a positive or neutral tone. This may mean that users are more actively retweeting and discussing tweets with a negative tone.
# The average value of like_count and reply_count is low enough for tweets with negative tonality and neutral tonality, and high enough for tweets with positive tonality. This may mean that users are more likely to like and respond to tweets with a positive tone.
# The average value of like_count is the highest for tweets with a tonality of 0.4-0.6, and the average value of reply_count is for tweets with a tonality of 0.8-1.0. This may indicate that users are more likely to like tweets that cause them positive emotions, and the largest number of responses to tweets are those that cause the brightest emotions, including negative ones.
# # Highlighting keywords
# Тo highlight keywords in the text column, we can use the NLP (Natural Language Processing) method, a frequently used tool for analyzing text data. It allows you to process and analyze the text, breaking it into individual words, defining their parts of speech, highlighting nouns, verbs, etc. Then we can analyze the information received and highlight the most common words in the text.
# To do this, we will need the space library, an open source Python code for natural language processing, which provides tools for processing and analyzing text data.
import spacy
import pandas as pd
nlp = spacy.load("en_core_web_sm")
def get_keywords(text):
doc = nlp(text)
keywords = []
for token in doc:
if not token.is_stop and token.is_alpha:
keywords.append(token.lemma_)
return keywords
# Добавление нового столбца с ключевыми словами
df["keywords"] = df["text"].apply(get_keywords)
from collections import Counter
import pandas as pd
# Получить все ключевые слова на английском языке
en_keywords = [
kw.lower()
for index, row in df.iterrows()
if row["lang"] == "en"
for kw in row["keywords"]
]
# Подсчитать частотность каждого ключевого слова
counter = Counter(en_keywords)
# Создать таблицу и заполнить данными
data = {"keyword": [], "frequency": []}
for keyword, freq in counter.items():
data["keyword"].append(keyword)
data["frequency"].append(freq)
df_result = pd.DataFrame(data)
# Вывести таблицу на экран
print(df_result)
df_sorted = df_result.sort_values(by="frequency", ascending=False)
top_10 = df_sorted.head(10)
import matplotlib.pyplot as plt
# Данные для построения круговой диаграммы
labels = top_10["keyword"].tolist()
sizes = top_10["frequency"].tolist()
# Построение диаграммы
plt.pie(sizes, labels=labels, autopct="%1.1f%%")
plt.axis("equal")
plt.show()
|
# # SUPPORT VECTOR MACHINE
# 
# The SVM (Support Vector Machine) is a supervised machine learning algorithm typically used for binary classification problems. It’s trained by feeding a dataset with labeled examples (xᵢ, yᵢ).
# The algorithm finds a hyperplane (or decision boundary) which should ideally have the following properties:
# It creates separation between examples of two classes with a maximum margin
# Its equation (w.x + b = 0) yields a value ≥ 1 for examples from+ve class and ≤-1 for examples from -ve class
# ### How does it find this hyperplane?
# By finding the optimal values w* (weights/normal) and b* (intercept) which define this hyperplane. The optimal values are found by minimizing a cost function. Once the algorithm identifies these optimal values, the SVM model f(x) is then defined as shown below:
#
#
# 
# ### Cost Function
# 
# Our objective is to find a hyperplane that separates +ve and -ve examples with the largest margin while keeping the misclassification as low as possible
# We will minimize the cost/objective function shown below:
# 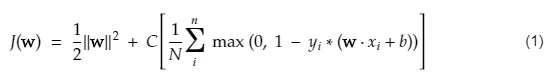
# 
# ### The Gradient of the Cost Function
# 
# Why do we minimize the cost function? Because the cost function is essentially a measure of how bad our model is doing at achieving the objective. If you look closely at J(w), to find it’s minimum, we have to:
# Minimize ∣∣w∣∣² which maximizes margin (2/∣∣w∣∣)
# Minimize the sum of hinge loss which minimizes misclassifications.
# 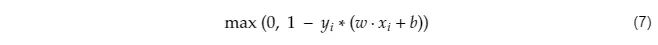
# 
# # IMPORTING THE LIBRARIES
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.metrics import accuracy_score, confusion_matrix, ConfusionMatrixDisplay
# # IMPLEMENTING THE MODEL
class SVM:
def __init__(self, learning_rate=0.0001, lambda_param=0.001, n_iters=10000):
self.weights = None
self.bias = None
self.lr = learning_rate
self.lambda_param = lambda_param
self.n_iters = n_iters
def fit(self, X, y):
self.m, self.n = X.shape
y1 = np.where(y <= 0, -1, 1)
self.weights = np.zeros(self.n)
self.bias = 0
for i in range(self.n_iters):
for idx, x_i in enumerate(X):
if y1[idx] * (np.dot(x_i, self.weights) - self.bias) >= 1:
self.weights -= self.lr * (2 * self.lambda_param * self.weights)
else:
self.weights -= self.lr * (
2 * self.lambda_param * self.weights - np.dot(x_i, y1[idx])
)
self.bias -= self.lr * y1[idx]
def predict(self, X):
output = np.dot(X, self.weights) - self.bias
y_pred = np.sign(output)
y_hat = np.where(y_pred <= -1, 0, 1)
return y_hat
# # READING THE DATA
data = pd.read_csv("/kaggle/input/loan-prediction-dataset")
data.head()
data.shape
data.info()
data.describe()
# # DATA PREPROCESSING
# ### DROPPING UNNECESSARY FEATURES
data.drop(["loan_id", "gender", "education"], axis=1, inplace=True)
data.head()
# ### CHECK FOR NULL VALUES
data.isnull().sum()
# ### FILLING NULL VALUES
data.married.unique()
data["married"].fillna(data["married"].mode()[0], inplace=True)
data.dependents.unique()
data["dependents"].fillna(data["dependents"].mode()[0], inplace=True)
data.self_employed.unique()
data["self_employed"].fillna(data["self_employed"].mode()[0], inplace=True)
data["loanamount"].fillna(data["loanamount"].mean(), inplace=True)
data.loan_amount_term.unique()
data["loan_amount_term"].fillna(data["loan_amount_term"].mode()[0], inplace=True)
data.credit_history.unique()
data["credit_history"].fillna(data["credit_history"].mode()[0], inplace=True)
data.isnull().sum()
# ### CHECK FOR DUPLICATES
data.duplicated().sum()
data.drop_duplicates(keep="first", inplace=True)
data.duplicated().sum()
data.head()
# ## DATA VISUALIZATION
sns.pairplot(data)
# # FEATURE EXTRACTION
column = ["married", "self_employed", "property_area", "loan_status"]
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
for i in column:
data[i] = le.fit_transform(data[i])
data.dependents = data.dependents.replace("3+", 3)
plt.figure(figsize=(12, 8))
sns.heatmap(data.corr())
# # SPLITING THE DATASET
x = data.drop("loan_status", axis=1)
y = data.loan_status
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.20)
x_test
# # FEATURE SCALING
from sklearn.preprocessing import StandardScaler
sc = StandardScaler()
x_train = sc.fit_transform(x_train)
x_test = sc.transform(x_test)
# # MODEL
svm_classifier = SVM()
svm_classifier.fit(x_train, y_train)
y_pred = svm_classifier.predict(x_test)
accuracy_score(y_test, y_pred)
cm = ConfusionMatrixDisplay(
confusion_matrix(y_test, y_pred), display_labels=[True, False]
)
cm.plot()
# # SKLEARN MODEL
from sklearn.svm import SVC
svm_c = SVC(kernel="linear", random_state=1)
svm_c.fit(x_train, y_train)
y2 = svm_c.predict(x_test)
y2
|
import numpy as np
from glob import glob
import tensorflow as tf
from tensorflow import keras
import matplotlib.pyplot as plt
from tensorflow.keras.optimizers import Adam
from tensorflow.keras.utils import plot_model
from tensorflow.keras.models import load_model, Model, Sequential
from tensorflow.keras.layers import Flatten, Dense, Dropout, Softmax
from tensorflow.keras.applications.resnet50 import ResNet50, preprocess_input
# A simple CNN model structure
# ```
# model = Sequential()
# # first layer
# model.add(Conv2D()) # feature selection/processing
# model.add(MaxPooling2D()) # downsampling
# model.add(BatchNormalization()) # rescaling/normalize
# model.add(Dropout(0.3)) # Drop noisy data
# # Second layer
# model.add(Conv2D()) # feature selection/processing
# model.add(MaxPooling2D()) # downsampling
# model.add(BatchNormalization()) # rescaling/normalize
# model.add(Dropout(0.3)) # Drop noisy data
# and so on ...
# ```
img_size = [224, 224]
test_path = "../input/cars-dataset/Test"
train_path = "../input/cars-dataset/Train"
[224, 224] + [3]
resnet = ResNet50(
include_top=False,
input_shape=img_size + [3], # Making the image into 3 Channel, so concating 3.
weights="imagenet",
)
# visualize the layers
plot_model(resnet, show_shapes=True, show_layer_names=True)
# Parameter informations
resnet.summary()
# False for pretrained model, use the default weights used by the imagenet.
for layer in resnet.layers:
layer.trainable = False
folders = glob("../input/cars-dataset/Train/*")
folders
METRICS = [
tf.keras.metrics.BinaryAccuracy(name="accuracy"),
tf.keras.metrics.Precision(name="precision"),
tf.keras.metrics.Recall(name="recall"),
tf.keras.metrics.AUC(name="auc"),
]
# convert model output to single dimension
x = Flatten()(resnet.output)
prediction = Dense(len(folders), activation="softmax")(x)
model = Model(inputs=resnet.input, outputs=prediction)
plot_model(model)
model.compile(loss="categorical_crossentropy", optimizer="adam", metrics=METRICS)
model.summary()
from tensorflow.keras.preprocessing.image import ImageDataGenerator
# creating Dynamic image augmentation
train_datagen = ImageDataGenerator(
rescale=1 / 255,
shear_range=0.2,
zoom_range=0.2,
rotation_range=5,
width_shift_range=0.2,
height_shift_range=0.2,
horizontal_flip=True,
vertical_flip=True,
fill_mode="nearest",
)
test_datagen = ImageDataGenerator(rescale=1 / 255)
training_set = train_datagen.flow_from_directory(
"../input/cars-dataset/Train",
target_size=(224, 224),
batch_size=32,
class_mode="categorical",
)
testing_set = test_datagen.flow_from_directory(
"../input/cars-dataset/Test",
target_size=(224, 224),
batch_size=32,
class_mode="categorical",
)
from tensorflow.keras.callbacks import EarlyStopping
es = EarlyStopping(verbose=1, patience=20)
r = model.fit(
training_set,
validation_data=testing_set,
epochs=50,
steps_per_epoch=len(training_set),
validation_steps=len(testing_set),
callbacks=[es],
)
r
plt.plot(r.history["loss"], label="train loss")
plt.plot(r.history["val_loss"], label="val loss")
plt.legend()
plt.show()
plt.savefig("LossVal_loss")
plt.plot(r.history["accuracy"], label="train acc")
plt.plot(r.history["val_accuracy"], label="val acc")
plt.legend()
plt.show()
plt.savefig("AccuVal_acc")
model.save("../working/model_resent50.h5")
y_pred = model.predict(testing_set)
print(y_pred)
y_pred = np.argmax(y_pred, axis=1)
y_pred
from tensorflow.keras.models import load_model
from tensorflow.keras.preprocessing import image
model = load_model("../working/model_resent50.h5")
img = image.load_img(
"../input/cars-dataset/Test/lamborghini/10.jpg", target_size=(224, 224)
)
img
# x=image.img_to_array(img)
x = image.img_to_array(img)
x
x = x / 255
x
x = np.expand_dims(x, axis=0)
x
x.shape
img_data = preprocess_input(x)
img_data.shape
preds = model.predict(x)
print(preds)
preds = np.argmax(preds, axis=1)
preds
labels = training_set.class_indices
print(labels)
type(labels)
labels = dict((v, k) for k, v in labels.items())
print(labels)
# predictions = [labels[k] for k in predicted_class_indices]
type(labels)
labels[preds[0]].capitalize()
img = image.load_img("../input/cars-dataset/Test/audi/25.jpg", target_size=(224, 224))
x = image.img_to_array(img)
x = x / 255
x = np.expand_dims(x, axis=0)
img_data = preprocess_input(x)
preds = model.predict(x)
preds = np.argmax(preds, axis=1)
print(preds)
labels[preds[0]].capitalize()
img = image.load_img(
"../input/cars-dataset/Test/mercedes/30.jpg", target_size=(224, 224)
)
x = image.img_to_array(img)
x = x / 255
x = np.expand_dims(x, axis=0)
img_data = preprocess_input(x)
preds = model.predict(x)
preds = np.argmax(preds, axis=1)
labels[preds[0]].capitalize()
img = image.load_img(
"../input/cars-dataset/Test/mercedes/30.jpg", target_size=(224, 224)
)
x = image.img_to_array(img)
x = x / 255
x = np.expand_dims(x, axis=0)
img_data = preprocess_input(x)
preds = model.predict(x)
preds = np.argmax(preds, axis=1)
labels[preds[0]].capitalize()
def Train_Val_Plot(
acc, val_acc, loss, val_loss, auc, val_auc, precision, val_precision
):
fig, (ax1, ax2, ax3, ax4) = plt.subplots(1, 4, figsize=(20, 5))
fig.suptitle(" MODEL'S METRICS VISUALIZATION ")
ax1.plot(range(1, len(acc) + 1), acc)
ax1.plot(range(1, len(val_acc) + 1), val_acc)
ax1.set_title("History of Accuracy")
ax1.set_xlabel("Epochs")
ax1.set_ylabel("Accuracy")
ax1.legend(["training", "validation"])
ax2.plot(range(1, len(loss) + 1), loss)
ax2.plot(range(1, len(val_loss) + 1), val_loss)
ax2.set_title("History of Loss")
ax2.set_xlabel("Epochs")
ax2.set_ylabel("Loss")
ax2.legend(["training", "validation"])
ax3.plot(range(1, len(auc) + 1), auc)
ax3.plot(range(1, len(val_auc) + 1), val_auc)
ax3.set_title("History of AUC")
ax3.set_xlabel("Epochs")
ax3.set_ylabel("AUC")
ax3.legend(["training", "validation"])
ax4.plot(range(1, len(precision) + 1), precision)
ax4.plot(range(1, len(val_precision) + 1), val_precision)
ax4.set_title("History of Precision")
ax4.set_xlabel("Epochs")
ax4.set_ylabel("Precision")
ax4.legend(["training", "validation"])
plt.show()
Train_Val_Plot(
r.history["accuracy"],
r.history["val_accuracy"],
r.history["loss"],
r.history["val_loss"],
r.history["auc"],
r.history["val_auc"],
r.history["precision"],
r.history["val_precision"],
)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Artificial intelligence
# # Bicycle prices in the Sultanate of Oman.
# # The dataset is from OpenSooq.
# # This assignment groups learn how to analyze data using data science.
# **Student: Maryam Khalifa Al Bulushi -- Riham Abdul Kareim Al Bulushi**
# **ID: 201916069 -- 201916071**
# # about DataSet:-
# # **The dataset contains id, brand, model, year, kilometers, condition, price, and category.**
# # First we need to convert it into a useful dataset.
# # **Use the correct library then Importing the csv file as dataset**
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
# # Reading the file (DataSet)
ds = pd.read_csv("/kaggle/input/dataset1/Data_Set1.csv")
ds.head()
# # Rename the prand to brand
ds.rename(columns={"prand": "brand"}, inplace=True)
ds.head()
# # checking the information of data
ds.info()
# # Data cleansing and improvement(Make the information more useful)
# **Arange the services to be in column rather than rows**
ds1 = ds.copy()
# # Checking number of rows before change anything
ds.info()
def change_to_numeric(x, ds1):
temp = pd.get_dummies(ds1[x])
ds1 = pd.concat([ds1, temp], axis=1)
ds1.drop([x], axis=1, inplace=True)
return ds1
ds2 = change_to_numeric("model", ds1)
ds2.head()
# ***change the number to 0 and 1 and this will help later in machine learning.***
# # Checking number of rows in the new dataframe
ds2.info()
# **no changes**
# **Checking the values for "burgman 650 Executive"**
ds2["burgman 650 Executive"].unique()
# **as you can see the values changed to 0 and 1 and that's going to help in machine learning**
ds3 = ds2.copy()
# # Understand your data
ds.describe().T.style.background_gradient(cmap="magma")
# # Data Cleansing and Improvement
# # Find if there is some duplicated data
ds.loc[ds.duplicated()]
# # Checking if there's any duplicated value
ds.duplicated().sum()
# # Process Missing data
ds.isna().sum()
x = ds.isna().sum()
cnt = 0
for temp in x.values:
if temp > 0:
print(x.index[cnt], x.values[cnt])
cnt += 1
# **at we see there are no missing data**
temp = ds[ds["price"].isna()]
temp
ds.isna().sum()
# **We use unique to see the array and dtype of BMW**
ds1 = ds[ds["model"] == "BMW"].copy()
ds1["model"].unique()
# # The following graph shows the relationship between brand and price bicycles by scatter
xd = ds["brand"]
yd = ds["price"]
plt.scatter(xd, yd)
plt.show()
# # The following graphics show the count and price in 2 diffrent way : 1-Distribution 2- Spread
plt.figure(figsize=(20, 8))
plt.subplot(1, 2, 1)
plt.title("Motorcycles Price Distribution")
sns.histplot(ds.price)
plt.subplot(1, 2, 2)
plt.title("Motorcycles Price Spread")
sns.boxplot(y=ds.price)
plt.show()
# # The following graphics show the Frequency relationship with (Motorcycles Model and Year and Motorcycles Brands)
plt.figure(figsize=(25, 6))
plt.subplot(1, 3, 1)
plt1 = ds.brand.value_counts().plot(kind="bar")
plt.title("Motorcycles Brands")
plt1.set(xlabel="Motorcycles Brand", ylabel="Frequency")
plt.subplot(1, 3, 2)
plt1 = ds.year.value_counts().plot(kind="bar")
plt.title(" Year")
plt1.set(xlabel=" Year", ylabel="Frequency")
plt.subplot(1, 3, 3)
plt1 = ds.model.value_counts().plot(kind="bar")
plt.title("Motorcycles Model")
plt1.set(xlabel="Motorcycles Model", ylabel="Frequency")
plt.show()
# # we drop the "category" columns and show the dataframe.
ds1 = ds.copy()
ds1 = ds1.drop(["category"], axis=1)
ds1.head()
feat = []
feat_val = []
def cat_to_no(ds):
categ = list(ds.select_dtypes(exclude="number"))
feat.append(categ)
categ.remove("kilometers")
for cat in categ:
f = list(ds[cat].unique())
feat_val.append(f)
for x in f:
val = f.index(x)
ds[cat] = ds[cat].replace(x, val)
return ds
ds1 = cat_to_no(ds1)
ds1.head()
# ***It looks like the code is attempting to convert categorical data to numerical data by assigning unique values to each category. The code first identifies categorical columns and removes the 'kilometers' column since it is likely a numerical column. It then creates two lists: feat (to store the categorical column names) and feat_val (to store the unique values for each category). Finally, the code iterates through each unique value in each category column and replaces it with a numerical value based on its position in the feat_val list.
# It seems like this code could be useful for machine learning tasks that require numerical data, but it is important to make sure the conversion is appropriate for the specific data and task at hand.***
# ***Show the row that the array is [0].***
feat = feat[0]
feat
# ***To show the data of ['brand', 'model', 'condition'].***
feat_val
ds["kilometers"] = ds["kilometers"].replace("200,000", "200,000-200,000")
ds["kilometers"] = ds["kilometers"].replace("0", "0-0")
f = list(ds["kilometers"].unique())
f
# **As an AI language model, I do not have access to the dataset you are referring to. Therefore, I am unable to perform the above operations. However, the code above seems to be replacing '200,000' with '200,000-200,000' and '0' with '0-0' in the 'kilometers' column of the dataset. The code then creates a list of unique values in the 'kilometers' column and stores it in the variable 'f'.**
for x in f:
k1 = x.split("-")[0]
k2 = x.split("-")[1]
k1 = k1.replace(",", "")
k2 = k2.replace(",", "")
k1 = int(k1)
k2 = int(k2)
val = k1 + 1 + (k2 - k1) // 2
print(x, val)
ds["kilometers"] = ds["kilometers"].replace(x, val)
ds.head(200)
# **It seems that the code is trying to process a dataset in a pandas DataFrame object named `ds`. The code loops through each line of data in the `f` object, which is not shown in the given code snippet.
# For each line, the code splits the string into two parts using the dash character ("-"), and assigns the first part to `k1` and the second part to `k2`. The code then removes any commas from `k1` and `k2`, and converts them into integers.
# The variable `val` is calculated by adding 1 to `k1` and finding the midpoint of the range between `k1` and `k2`.
# The code then prints out both the original line of data and `val` for debugging purposes.
# Finally, the code replaces any occurrence of the original line of `x` in the `ds['kilometers']` column with `val`.
# Without more context or a sample of the `f` object or the `ds` DataFrame, it is difficult to determine the exact purpose or effectiveness of this code. It is possible that the code is attempting to clean up and standardize the format of the `kilometers` column in the `ds` DataFrame, but this is only speculation.**
# **As an AI language model, I cannot access the dataset you're working on. You need to pass your dataset (ds) along with this code snippet to get the output. However, this code snippet excludes the columns with numerical data and selects only the columns with categorical data.**
categ = ds.select_dtypes(exclude="number")
categ.head()
# # Drop columns (condition, category, and id).
# # and show data (table).
ds1 = ds.copy()
ds1 = ds1.drop(["condition", "category", "ID"], axis=1)
ds1.head()
# # Arrange the number to be in columns rather than rows:-
# # and also change the number to 0 and 1 and this will help later in machine learning.
def change_to_numeric(x, ds):
temp = pd.get_dummies(ds[x], drop_first=True)
ds = pd.concat([ds, temp], axis=1)
ds.drop([x], axis=1, inplace=True)
return ds
categ = ds1.select_dtypes(exclude="number")
for cat in categ:
ds1 = change_to_numeric(cat, ds1)
ds1
mean_price = []
for x in feat_val[feat.index("brand")]:
mean_price.append(ds[ds["brand"] == x].mean()["price"])
for i in range(0, len(mean_price)):
print(feat_val[feat.index("brand")][i], mean_price[i])
# **This code calculates the mean price of each brand in a dataset and stores it in a list called "mean_price". It then iterates through the list of brand values and prints out the brand and its corresponding mean price.
# The first line initializes the empty list "mean_price".
# The for loop iterates through each unique value in the column "brand" and calculates the mean of the prices for all rows that have that brand. The mean is calculated using the "mean()" function of the pandas library. The resulting mean is then appended to the "mean_price" list.
# The second for loop iterates through each element in the "mean_price" and prints out the corresponding brand and mean price using the "feat_val" list, which contains all unique values for each feature in the dataset. It does this by using the index of the "brand" feature in the "feat" list to locate the correct "feat_val" list, and then indexing into that list to get the specific brand value.**
# # presenting the relation in bar chart.
# **Compare bicycle prices between all brands in the data through a barh graph**
plt.figure(figsize=(20, 10))
plt.barh(feat_val[feat.index("brand")], mean_price)
# **Compare the prices of bicycles and the condition of bicycles (new or used) in the data through the barh graph.**
mean_price = []
for x in feat_val[feat.index("condition")]:
mean_price.append(ds[ds["condition"] == x].mean()["price"])
for i in range(0, len(mean_price)):
print(feat_val[feat.index("condition")][i], mean_price[i])
plt.figure(figsize=(20, 10))
plt.barh(feat_val[feat.index("condition")], mean_price)
# **Compare the prices of bicycles and the model of bicycles in the data through the barh graph.**
mean_price = []
for x in feat_val[feat.index("model")]:
mean_price.append(ds[ds["model"] == x].mean()["price"])
for i in range(0, len(mean_price)):
print(feat_val[feat.index("model")][i], mean_price[i])
plt.figure(figsize=(20, 10))
plt.barh(feat_val[feat.index("model")], mean_price)
# ***Show the values under the 'brand' column.***
feat_val[feat.index("brand")]
# ***Finding the correlation of 'price' column.***
ds1.corr()["price"]
# ***show the Data Set after converting it into machine language.***
ds1
# ***The information about the 'Dorsodur' model.***
ds2 = ds[ds["model"] == "Dorsodur"]
ds2
# # Finding the correlation between data:
# **The corr() method finds the correlation of each column in a DataSet.**
ds2.corr()
# # Importing library to test the information of the Data set.
import pandas as pd
from sklearn.tree import DecisionTreeClassifier
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
data = ds1.copy()
# **The above code is used to split the data into training and testing sets.
# "x" contains all the independent variables except "price".
# "y" represents the dependent variable "price".
# "train_test_split" is a function provided by scikit-learn library which randomizes and splits the data into training and testing sets.
# "test_size=0.2" means 20% of the data will be used for testing and 80% for training.**
x = data.drop(columns=["price"])
y = data["price"]
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.2)
# ***show the Data Set in machine language.***
ds1.head()
# **This code fits a linear regression model using the data in arrays x and y, and prints "Done" when the model fitting is complete. The sklearn library is used to create the linear regression object, and the fit() method of the object is called to fit the model to the data.**
from sklearn import linear_model
regr = linear_model.LinearRegression()
regr.fit(x, y)
print("Done")
# ***Choose the data to be tested from the Data Set.***
x_test.head()
# ***Test the data in column numder [175].***
x1 = x_test.loc[[175]]
y1 = y_test.loc[[175]]
y1
# ***We will see in this programming whether the data has been trained according to the required standards or not, if the price that appeared is close to the price of the Motorcycles that was tested above, then the data is excellent. If it is not close to the price of the Motorcycles, then the data is not sufficient to be tested.***
pred = regr.predict(x1)
print(pred)
# If color is **light** that means the correlation is **low**
# If color is **dark** that means the correlation is **high**
_, ax = plt.subplots(figsize=(14, 12))
colormap = sns.diverging_palette(220, 10, as_cmap=True)
_ = sns.heatmap(
ds2.corr(),
cmap=colormap,
square=True,
cbar_kws={"shrink": 0.9},
ax=ax,
annot=True,
linewidths=0.1,
vmax=1.0,
linecolor="white",
annot_kws={"fontsize": 12},
)
plt.title("Pearson Correlation of Features", y=1.05, size=15)
|
# # House Prices - Advanced Regression Techniques Kaggle Competition
# # **Introduction**
# ### **Objective:**
# The objective of the project is to perform advance regression techniques to predict the house price in Boston.
# ### **Data Description:**
# - trainh.csv - the training set
# - testh.csv - the test set
# - data_description.txt - full description of each column, originally prepared by Dean De Cock but lightly edited to match the column names used here
# ### **Table of Content:**
# 1. Import Libraries
# 2. Load Datasets
# 3. EDA & Visualization
# 4. Null Values
# 5. Feature Engineering
# 6. Model Development
# #### 1. Import Libraries
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from scipy.stats import skew, norm
import missingno as msno
from sklearn.impute import SimpleImputer
from sklearn.impute import KNNImputer
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import cross_val_score
from sklearn.metrics import mean_squared_error
import warnings
warnings.filterwarnings("ignore")
# #### 2. Load Datasets
# Data_train
df_train = pd.read_csv(
"/kaggle/input/house-prices-advanced-regression-techniques/train.csv"
)
# Data_test
df_test = pd.read_csv(
"/kaggle/input/house-prices-advanced-regression-techniques/test.csv"
)
pd.set_option("display.max_columns", 500)
df_train.head()
df_train.info()
# Some of the types are not true we need to fix it.
df_train = df_train.astype(
{"MSSubClass": "object", "OverallQual": "object", "OverallCond": "object"}
)
df_test = df_test.astype(
{"MSSubClass": "object", "OverallQual": "object", "OverallCond": "object"}
)
# #### 3. EDA & Visualization
plt.figure(figsize=(12, 6))
sns.distplot(df_train["SalePrice"], kde=True, hist=True, fit=norm)
plt.title("SalePrice distribution vs Normal Distribution", fontsize=13)
plt.xlabel("House's sale Price in $", fontsize=12)
plt.show()
print("Skewness: %f" % abs(df_train["SalePrice"]).skew())
print("Kurtosis: %f" % abs(df_train["SalePrice"]).kurt())
# Selecting the columns with integer or float type.
num_cols = df_train.select_dtypes(include=["number"]).columns.tolist()
num_cols.remove("SalePrice")
num_cols.remove("Id")
cat_cols = df_train.select_dtypes(exclude=["number"]).columns.tolist()
# Visaulization for numerical values
f, axes = plt.subplots(4, 9, figsize=(30, 15), sharex=False)
for i, col in enumerate(num_cols):
sns.histplot(data=df_train, x=col, ax=axes[i % 4, i // 4])
# Correlation between target and numerical values
f, axes = plt.subplots(4, 9, figsize=(30, 15), sharex=False)
for i, col in enumerate(num_cols):
sns.scatterplot(data=df_train, x=col, y="SalePrice", ax=axes[i % 4, i // 4])
# Visaulization for categorical values
f, axes = plt.subplots(8, 6, figsize=(30, 20), sharex=False)
for i, col in enumerate(cat_cols):
sns.countplot(data=df_train, x=col, ax=axes[i % 8, i // 8])
plt.figure(figsize=(18, 10))
corr_with_sepal_length = df_train.corrwith(df_train["SalePrice"])
sns.barplot(x=corr_with_sepal_length.index, y=corr_with_sepal_length.values)
plt.title("Correlations with Sepal Length")
plt.xlabel("Numeric columns")
plt.xticks(rotation=45, ha="right")
plt.show()
# #### 4. Null Values
# Dropping unnecessary columns.
df_train = df_train.drop(["Id"], axis=1)
df_test = df_test.drop(["Id"], axis=1)
# Checking the number of nan values.
pd.set_option("display.max_rows", 500)
training_null = pd.isnull(df_train).sum()
testing_null = pd.isnull(df_test).sum()
null = pd.concat([training_null, testing_null], axis=1, keys=["Training", "Testing"])
null
# According to data description some of the nan values are MNAR.
null_with_meaning = [
"Alley",
"BsmtQual",
"BsmtCond",
"BsmtExposure",
"BsmtFinType1",
"BsmtFinType2",
"FireplaceQu",
"GarageType",
"GarageFinish",
"GarageQual",
"GarageCond",
"PoolQC",
"Fence",
"MiscFeature",
]
# Replacing every Nan value with "None"
df_train[null_with_meaning] = df_train[null_with_meaning].fillna("None")
df_test[null_with_meaning] = df_test[null_with_meaning].fillna("None")
msno.matrix(df_train)
null
# Predicting the missing numeric nan values with KNNImputer.
imputer = KNNImputer(n_neighbors=2)
df_train[num_cols] = imputer.fit_transform(df_train[num_cols])
df_test[num_cols] = imputer.transform(df_test[num_cols])
# Filling the categorical nan values with most frequent ones.
imputer = SimpleImputer(strategy="most_frequent")
df_train[cat_cols] = imputer.fit_transform(df_train[cat_cols])
df_test[cat_cols] = imputer.transform(df_test[cat_cols])
# df_test.isna().sum()
# df_train.isna().sum()
# #### 5. Feature Engineering
df_train_add = df_train.copy()
df_train_add["TotalSF"] = (
df_train_add["TotalBsmtSF"] + df_train_add["1stFlrSF"] + df_train_add["2ndFlrSF"]
)
df_train_add["Total_Bathrooms"] = (
df_train_add["FullBath"]
+ (0.5 * df_train_add["HalfBath"])
+ df_train_add["BsmtFullBath"]
+ (0.5 * df_train_add["BsmtHalfBath"])
)
df_train_add["Total_porch_sf"] = (
df_train_add["OpenPorchSF"]
+ df_train_add["3SsnPorch"]
+ df_train_add["EnclosedPorch"]
+ df_train_add["ScreenPorch"]
+ df_train_add["WoodDeckSF"]
)
df_test_add = df_test.copy()
df_test_add["TotalSF"] = (
df_test_add["TotalBsmtSF"] + df_test_add["1stFlrSF"] + df_test_add["2ndFlrSF"]
)
df_test_add["Total_Bathrooms"] = (
df_test_add["FullBath"]
+ (0.5 * df_test_add["HalfBath"])
+ df_test_add["BsmtFullBath"]
+ (0.5 * df_test_add["BsmtHalfBath"])
)
df_test_add["Total_porch_sf"] = (
df_test_add["OpenPorchSF"]
+ df_test_add["3SsnPorch"]
+ df_test_add["EnclosedPorch"]
+ df_test_add["ScreenPorch"]
+ df_test_add["WoodDeckSF"]
)
df_train_add["SalePrice"] = np.log(df_train_add["SalePrice"])
df_train_add.head()
skewed_cols = []
for col in num_cols:
if df_train_add[col].skew() > 0.5:
skewed_cols.append(col)
skewed_cols
df_train_add[num_cols] = np.log1p(df_train_add[num_cols])
df_test_add[num_cols] = np.log1p(df_test_add[num_cols])
df_train_add.info()
import category_encoders as ce
target_encoder = ce.TargetEncoder(cols=(cat_cols))
y_train = df_train_add["SalePrice"]
X_train = df_train_add.drop("SalePrice", axis=1)
target_encoder.fit(X_train, y_train)
X_train_encoded = target_encoder.transform(X_train)
X_test_encoded = target_encoder.transform(df_test_add)
X_train = X_train_encoded.copy()
X_test = X_test_encoded.copy()
X_train.head()
# #### 6. Model Development
from sklearn.linear_model import Ridge, RidgeCV, ElasticNet, Lasso, LassoCV
from sklearn.kernel_ridge import KernelRidge
from sklearn.ensemble import GradientBoostingRegressor
from catboost import CatBoostRegressor
import lightgbm as lgb
import xgboost as xg
baseline_el = ElasticNet(random_state=0, max_iter=10e7, alpha=0.0003)
baseline_score_el = cross_val_score(baseline_el, X_train, y_train)
print("ENet avg:", np.mean(baseline_score_el))
baseline_r = Ridge(alpha=1, random_state=0)
baseline_score_r = cross_val_score(baseline_r, X_train, y_train)
print("Ridge avg:", np.mean(baseline_score_r))
baseline_ls = Lasso(alpha=0.0001, random_state=0)
baseline_score_ls = cross_val_score(baseline_ls, X_train, y_train)
print("Lasso avg:", np.mean(baseline_score_ls))
baseline_kr = KernelRidge(alpha=0.1)
baseline_score_kr = cross_val_score(baseline_kr, X_train, y_train)
print("KRR avg:", np.mean(baseline_score_kr))
baseline_lgb = lgb.LGBMRegressor(
learning_rate=0.01, num_leaves=4, n_estimators=2000, random_state=0
)
baseline_score_lgb = cross_val_score(baseline_lgb, X_train, y_train)
print("LGBM avg:", np.mean(baseline_score_lgb))
baseline_xg = xg.XGBRegressor(
learning_rate=0.01,
n_estimators=2000,
subsample=0.7,
colsample_bytree=0.7,
random_state=0,
)
baseline_score_xg = cross_val_score(baseline_xg, X_train, y_train)
print("XGB avg:", np.mean(baseline_score_xg))
baseline_cb = CatBoostRegressor(random_state=0, verbose=0)
baseline_score_cb = cross_val_score(baseline_cb, X_train, y_train)
print("CatB avg:", np.mean(baseline_score_cb))
baseline_gbr = GradientBoostingRegressor(
n_estimators=1000,
learning_rate=0.02,
max_depth=4,
max_features="sqrt",
min_samples_leaf=15,
min_samples_split=50,
loss="huber",
random_state=0,
)
baseline_score_gbr = cross_val_score(baseline_gbr, X_train, y_train)
print("GBR avg:", np.mean(baseline_score_gbr))
from sklearn.ensemble import VotingRegressor
voting_reg = VotingRegressor(
estimators=[
("ElasticNet", baseline_el),
("Ridge", baseline_r),
("Lasso", baseline_ls),
("KernelRidge", baseline_kr),
("LGBM", baseline_lgb),
("XGB", baseline_xg),
("CatBoost", baseline_cb),
("GradientB", baseline_gbr),
]
)
cv = cross_val_score(voting_reg, X_train, y_train)
print(cv)
print(cv.mean())
voting_reg = voting_reg.fit(X_train, y_train)
y_pred = voting_reg.predict(X_test)
print(y_pred)
y_pred_norm = np.exp(y_pred)
y_pred_norm
df_id = pd.read_csv("testh.csv")
final_data = {"Id": df_id.Id, "SalePrice": y_pred_norm}
submission8 = pd.DataFrame(data=final_data)
submission8.to_csv("submission8.csv", index=None)
from mlxtend.regressor import StackingRegressor
stregr = StackingRegressor(
regressors=[
baseline_el,
baseline_r,
baseline_ls,
baseline_kr,
baseline_lgb,
baseline_xg,
baseline_cb,
baseline_gbr,
],
meta_regressor=baseline_lgb,
)
cv = cross_val_score(stregr, X_train, y_train)
print(cv.mean())
stregr.fit(X_train, y_train)
y_pred = stregr.predict(X_test)
y_pred
y_pred_norm = np.exp(y_pred)
y_pred_norm
df_id = pd.read_csv("testh.csv")
final_data = {"Id": df_id.Id, "SalePrice": y_pred_norm}
submission9 = pd.DataFrame(data=final_data)
submission9.to_csv("submission9.csv", index=None)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
KEY = "msno"
TIME_REF = "safra"
members = pd.read_parquet("/kaggle/input/kkbox-dm-parquet/members.parquet")
user_logs = pd.read_parquet("/kaggle/input/kkbox-dm-parquet/user_logs.parquet")
transactions = pd.read_parquet("/kaggle/input/kkbox-dm-parquet/transactions.parquet")
mapping_member_type = {
"registration_init_time": "int32",
"city": "int8",
"bd": "int8",
"gender": "str",
"registered_via": "int8",
"is_ativo": "int8",
}
for feature, dtype in mapping_mamber_types.items():
members[feature] = members.astype(dtype)
members[KEY] = members[KEY].astype("str")
members[TIME_REF] = members[KEY].astype("str")
master_df = members.join(user_logs, how="left")
master_df = master_df.join(transactions, how="left")
master_df.head(50)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# 1. Creating a random seed as a good practice for future random calculations
# 2. Importing the latest version of sklearn
#
np.random.seed(360)
import sklearn
assert sklearn.__version__ >= "0.20"
# Loading the data into train test splits
df_test = pd.read_csv(
"/kaggle/input/e-commerce-shoppers-behaviour-understanding/test_data_v2.csv"
)
df_train = pd.read_csv(
"/kaggle/input/e-commerce-shoppers-behaviour-understanding/train_data_v2.csv"
)
print("data loading done")
# The dummy classifier was the baseline model
#
# from sklearn.dummy import DummyClassifier
# X=pd.DataFrame(df_train)
# dummyclasf=DummyClassifier(strategy="most_frequent")
# print("Dummy Classifier Fitting ")
# dummyclasf.fit(X,y)
# print(dummyclasf.score(X,y))
# temp=dummyclasf.predict(df_test)
# # submission=pd.DataFrame(columns=['id','Made_Purchase'])
# # submission['id']=[i for i in range(len(temp))]
# # submission['Made_Purchase']=temp
# # submission.to_csv('submission.csv', index=False)
# # # DFinal = pd.DataFrame(dummyclasf.predict(df_test))
# # # print(DFinal[0])
# # # DFinal[0].to_csv("submission.csv")
# **Data Preprocessing **
from sklearn.preprocessing import (
StandardScaler,
OneHotEncoder,
OrdinalEncoder,
MaxAbsScaler,
LabelBinarizer,
MinMaxScaler,
)
from sklearn.compose import ColumnTransformer, make_column_transformer
from sklearn.pipeline import FeatureUnion, Pipeline
from sklearn.impute import SimpleImputer, KNNImputer
import seaborn as sns
# df_train
# sns.heatmap(df_train[df_train.columns[0:5]])
import matplotlib.pyplot as plt
import seaborn as sns
temp = df_train["Made_Purchase"]
temp.replace({True: 1, False: 0}, inplace=True)
plt.pie(df_train.Made_Purchase.value_counts(), labels=["1", "0"])
print(df_train.Made_Purchase.value_counts()) # nearly a 60:40 split
df_train
y = pd.DataFrame(df_train.pop("Made_Purchase"))
print(y.shape)
print(df_train.shape)
print(df_test.shape)
df_train["SeasonalPurchase"].value_counts()
number_pipeline = Pipeline(
steps=[
("imputer", KNNImputer(n_neighbors=6)),
("scaler1", StandardScaler()),
("scaler2", MinMaxScaler()),
]
)
categorical_pipeline = Pipeline(
steps=[
("imputer", SimpleImputer(strategy="most_frequent")),
("OneHotEncoding", OneHotEncoder(handle_unknown="ignore")),
]
)
final_pipe = ColumnTransformer(
transformers=[
(
"number",
number_pipeline,
[
"HomePage",
"HomePage_Duration",
"LandingPage",
"LandingPage_Duration",
"ProductDescriptionPage",
"ProductDescriptionPage_Duration",
"GoogleMetric:Bounce Rates",
"GoogleMetric:Exit Rates",
"GoogleMetric:Page Values",
"SeasonalPurchase",
],
),
(
"categories",
categorical_pipeline,
[
"Month_SeasonalPurchase",
"OS",
"SearchEngine",
"Zone",
"Type of Traffic",
"CustomerType",
"Gender",
"Cookies Setting",
"Education",
"Marital Status",
"WeekendPurchase",
],
),
],
remainder="passthrough",
)
Train_Features = final_pipe.fit_transform(df_train).toarray()
Test_Features = final_pipe.transform(df_test).toarray()
# df_train
# df_test
Test_Features.shape
print(Train_Features.shape)
Train_Labels = y.to_numpy()
print(Train_Labels.shape)
from sklearn.model_selection import train_test_split
from sklearn.model_selection import ShuffleSplit, StratifiedKFold
cv = StratifiedKFold(n_splits=10, shuffle=True, random_state=42)
X_train, X_test, y_train, y_test = train_test_split(
Train_Features,
Train_Labels,
test_size=0.001,
# stratify=Train_Labels,
random_state=42,
shuffle=True,
)
print(X_train.shape)
print(X_test.shape)
print(y_train.shape)
print(y_test.shape)
# import xgboost as xgb
# from sklearn.metrics import accuracy_score, classification_report, confusion_matrix, ConfusionMatrixDisplay
# # xgb= xgb.XGBClassifier(objective="binary:logistic", random_state=42)
# xgb= xgb.XGBClassifier(n_estimators=100)
# xgb.fit(X_train,y_train.ravel())
# y_pred=xgb.predict(X_test)
# print('Testing accuracy',accuracy_score(y_test,y_pred))
# print(classification_report(y_test,y_pred))
# cm_display=ConfusionMatrixDisplay(confusion_matrix=confusion_matrix(y_test,y_pred), display_labels=[0,1])
# cm_display.plot()
# plt.show()
# import xgboost as xgb
# clf_xgb=xgb.XGBClassifier(
# objective='reg:pseudohubererror',
# gamma=0,
# learning_rate=0.066,
# max_depth=2,
# reg_lambda=8.0,
# scale_pos_weight=1,
# seed=42,
# subsample=0.9,
# colsample_bytree=0.5,
# eval_metric='aucpr',
# n_estimators=110)
# clf_xgb.fit(
# X_train,
# y_train.ravel())
# y_pred=clf_xgb.predict(X_test)
# print('Testing accuracy',accuracy_score(y_test,y_pred))
# print(classification_report(y_test,y_pred))
# cm_display=ConfusionMatrixDisplay(confusion_matrix=confusion_matrix(y_test,y_pred), display_labels=[0,1])
# cm_display.plot()
# plt.show()
# random forest trees with Xgboost base classifier,
# extra random trees with Xgboost as base classifier
from sklearn.ensemble import BaggingClassifier
from sklearn import model_selection
from sklearn.metrics import (
accuracy_score,
classification_report,
confusion_matrix,
ConfusionMatrixDisplay,
)
import xgboost as xgb
clf_xgb = xgb.XGBClassifier(
objective="reg:pseudohubererror",
gamma=0,
learning_rate=0.066,
max_depth=2,
reg_lambda=8.0,
# scale_pos_weight=1,
seed=42,
subsample=0.9,
colsample_bytree=0.5,
eval_metric="aucpr",
)
# n_estimators=110)
# kfold=model_selection.KFold(n_splits=3)
bc = BaggingClassifier(
base_estimator=clf_xgb, n_estimators=90, n_jobs=-1, random_state=True
)
bc.fit(Train_Features, Train_Labels.ravel())
y_pred = bc.predict(X_test)
print("Testing accuracy", accuracy_score(y_test, y_pred))
print(classification_report(y_test, y_pred))
cm_display = ConfusionMatrixDisplay(
confusion_matrix=confusion_matrix(y_test, y_pred), display_labels=[0, 1]
)
cm_display.plot()
plt.show()
# from sklearn.ensemble import VotingClassifier
# from sklearn.ensemble import RandomForestClassifier
# from sklearn.ensemble import BaggingClassifier
# from sklearn.ensemble import ExtraTreesClassifier
# from sklearn.neighbors import KNeighborsClassifier
# import xgboost as xgb
# clf_xgb=xgb.XGBClassifier(
# objective='reg:pseudohubererror',
# gamma=0,
# learning_rate=0.066,
# max_depth=2,
# reg_lambda=8.0,
# # scale_pos_weight=1,
# seed=42,
# subsample=0.9,
# colsample_bytree=0.5,
# eval_metric='aucpr')
# # n_estimators=110)
# vclf= VotingClassifier(estimators=[
# ('bc',BaggingClassifier(base_estimator=clf_xgb, n_estimators=90, n_jobs=-1, random_state=True)),
# ('knn', KNeighborsClassifier(algorithm= 'kd_tree',leaf_size=200, n_neighbors=100, weights='uniform'))
# ], voting='soft',weights=[1])
# vclf=vclf.fit(X_train,y_train.ravel())
# y_pred = vclf.predict(X_test)
# print(vclf.score(X_test,y_test))
# print('Testing accuracy',accuracy_score(y_test,y_pred))
# print(classification_report(y_test,y_pred))
# cm_display=ConfusionMatrixDisplay(confusion_matrix=confusion_matrix(y_test,y_pred), display_labels=[0,1])
# cm_display.plot()
# plt.show()
# final_predictions=[]
# temp=bc.predict(Test_Features)
# for x in temp:
# if x==1:
# final_predictions.append('True')
# else:
# final_predictions.append('False')
# submission=pd.DataFrame(columns=['id','Made_Purchase'])
# submission['id']=[i for i in range(len(final_predictions))]
# submission['Made_Purchase']=final_predictions
# submission.to_csv('submission.csv', index=False)
# from sklearn.ensemble import VotingClassifier
# from sklearn.ensemble import RandomForestClassifier
# from sklearn.ensemble import ExtraTreesClassifier
# import xgboost as xgb
# vclf= VotingClassifier(estimators=[
# ('xgb2',xgb.XGBClassifier(
# objective='reg:pseudohubererror',
# gamma=0,
# learning_rate=0.066,
# max_depth=2,
# reg_lambda=8.0,
# scale_pos_weight=1,
# seed=42,
# subsample=0.9,
# colsample_bytree=0.5,
# eval_metric='aucpr',
# n_estimators=110)),
# ('rf',ExtraTreesClassifier(n_estimators=300, min_samples_split=2,random_state=42, max_depth=7))
# ], voting='soft',weights=[1,1])
# vclf=vclf.fit(Train_Features,Train_Labels.ravel())
# y_pred = vclf.predict(X_test)
# print(vclf.score(X_test,y_test))
# print('Testing accuracy',accuracy_score(y_test,y_pred))
# print(classification_report(y_test,y_pred))
# cm_display=ConfusionMatrixDisplay(confusion_matrix=confusion_matrix(y_test,y_pred), display_labels=[0,1])
# cm_display.plot()
# plt.show()
# final_predictions=[]
# temp=vclf.predict(Test_Features)
# for x in temp:
# if x==1:
# final_predictions.append('True')
# else:
# final_predictions.append('False')
# submission=pd.DataFrame(columns=['id','Made_Purchase'])
# submission['id']=[i for i in range(len(final_predictions))]
# submission['Made_Purchase']=final_predictions
# submission.to_csv('submission.csv', index=False)
from sklearn.ensemble import StackingClassifier
from sklearn.linear_model import LogisticRegression, SGDClassifier
from sklearn.linear_model import SGDClassifier
from sklearn.ensemble import ExtraTreesClassifier, RandomForestClassifier
from sklearn.svm import LinearSVC, SVC
import xgboost as xgb
clf_xgb = xgb.XGBClassifier(
objective="reg:pseudohubererror",
gamma=0,
learning_rate=0.066,
max_depth=2,
reg_lambda=8.0,
# scale_pos_weight=1,
seed=42,
subsample=0.9,
colsample_bytree=0.5,
eval_metric="aucpr",
)
estimators = [
(
"bc",
BaggingClassifier(
base_estimator=clf_xgb, n_estimators=90, n_jobs=-1, random_state=True
),
),
# ('knn', KNeighborsClassifier(algorithm= 'kd_tree',leaf_size=200, n_neighbors=100, weights='uniform')),
(
"rf",
ExtraTreesClassifier(n_estimators=200, min_samples_split=2, random_state=42),
),
# ('rf1', RandomForestClassifier(n_estimators=200, min_samples_split=2,random_state=42))
]
fclf = StackingClassifier(
estimators=estimators, final_estimator=SVC(gamma="scale", kernel="rbf", C=800)
)
fclf = fclf.fit(X_train, y_train.ravel())
y_pred = fclf.predict(X_test)
print(fclf.score(X_test, y_test))
print("Testing accuracy", accuracy_score(y_test, y_pred))
print(classification_report(y_test, y_pred))
cm_display = ConfusionMatrixDisplay(
confusion_matrix=confusion_matrix(y_test, y_pred), display_labels=[0, 1]
)
cm_display.plot()
plt.show()
stop
# final_predictions=[]
# temp=fclf.predict(Test_Features)
# for x in temp:
# if x==1:
# final_predictions.append('True')
# else:
# final_predictions.append('False')
# submission=pd.DataFrame(columns=['id','Made_Purchase'])
# submission['id']=[i for i in range(len(final_predictions))]
# submission['Made_Purchase']=final_predictions
# submission.to_csv('submission.csv', index=False)
# print(len(final_predictions))
# print(final_predictions)
stop
from sklearn.ensemble import ExtraTreesClassifier
etc = ExtraTreesClassifier(n_estimators=500, random_state=0)
etc = etc.fit(X_train, y_train.ravel())
y_pred = etc.predict(X_test)
print(etc.score(X_test, y_test))
print("Testing accuracy", accuracy_score(y_test, y_pred))
print(classification_report(y_test, y_pred))
cm_display = ConfusionMatrixDisplay(
confusion_matrix=confusion_matrix(y_test, y_pred), display_labels=[0, 1]
)
cm_display.plot()
plt.show()
# final_predictions=[]
# temp=vclf.predict(Test_Features)
# for x in temp:
# if x==1:
# final_predictions.append('True')
# else:
# final_predictions.append('False')
# submission=pd.DataFrame(columns=['id','Made_Purchase'])
# submission['id']=[i for i in range(len(final_predictions))]
# submission['Made_Purchase']=final_predictions
# submission.to_csv('submission.csv', index=False)
# print(len(final_predictions))
# print(final_predictions)
stop
# from sklearn.neural_network import MLPClassifier
# mlp=MLPClassifier(hidden_layer_sizes=(88,70,40,11,5,2),max_iter=500, activation='relu',early_stopping=True)
# mlp.fit(Train_Features,Train_Labels.ravel())
# y_pred=mlp.predict(X_test)
# print(mlp.score(X_test,y_test))
from sklearn.linear_model import LogisticRegression, SGDRegressor
# from sklearn.model_selection import GridSearchCV
# param_grid={
# 'logistic__penalty':['l2'],
# 'logistic__C':[1,2,3,4,5,6,7,8,9,10],
# 'logistic__tol':[1e-3,1e-2]
# }
# pipeline=Pipeline([
# ("logistic",LogisticRegression(max_iter=10000,random_state=42))
# ])
# gs=GridSearchCV(estimator=pipeline,cv=cv,param_grid=param_grid,return_train_score=True, n_jobs=-1)
# results=gs.fit(X_train,y_train.ravel())
# print(results.score(X_test,y_test))
# print(results.best_params_)
# #1 Naive Bayes optimal hyperparameters
# from sklearn.metrics import accuracy_score
from sklearn.naive_bayes import BernoulliNB
# pipelineNB=Pipeline([
# ("naive_bayes", BernoulliNB(alpha=1.0, binarize=0.0, class_prior=None, fit_prior=True))
# ])
# results=pipelineNB.fit(X_train,y_train.ravel())
# print(results.score(X_test,y_test))
# print(results.score(X_test,y_test))
# txtf = open("BernoulliNB.txt", "w")
# txtf.write("R2 score: %s\n\n" % results.score(X_test,y_test))
# txtf.close()
# from sklearn.svm import SVC
# svc=SVC(C=1,kernel='poly',gamma='auto',tol=1e-5,decision_function_shape='ovo',cache_size=7000,degree=1)
# results=svc.fit(X_train,y_train.ravel())
# print(results.score(X_test,y_test))
# #2 SVM optimal hyperparameters
# from sklearn.model_selection import GridSearchCV
# from sklearn.svm import SVC
# param_grid={
# 'SVC__C':[1,2],
# 'SVC__kernel':['linear'],
# 'SVC__gamma':['auto','scale'],
# 'SVC__degree':[1,2,3,4,5,6,7],
# 'SVC__decision_function_shape':['ovo','ovr']
# }
# pipelineSVC=Pipeline([
# ("SVC", SVC(cache_size=7000))
# ])
# gs=GridSearchCV(estimator=pipelineSVC,cv=2,param_grid=param_grid,return_train_score=True, n_jobs=-1,verbose=10)
# results=gs.fit(X_train,y_train.ravel())
# print(results.score(X_test,y_test))
# print(results.best_params_)
# print(results.score(X_test,y_test))
# txtf = open("SVC_linear.txt", "w")
# txtf.write("Best Parameters: %s\n" % results.best_params_)
# txtf.write("R2 score: %s\n\n" % results.score(X_test,y_test))
# txtf.close()
# #2 SVM optimal hyperparameters
# from sklearn.model_selection import GridSearchCV
# from sklearn.svm import SVC
# param_grid={
# 'SVC__C':[1,2],
# 'SVC__kernel':['rbf'],
# 'SVC__gamma':['auto','scale'],
# 'SVC__degree':[1,2,3,4,5,6,7],
# 'SVC__decision_function_shape':['ovo','ovr']
# }
# pipelineSVC=Pipeline([
# ("SVC", SVC(cache_size=7000))
# ])
# gs=GridSearchCV(estimator=pipelineSVC,cv=2,param_grid=param_grid,return_train_score=True, n_jobs=-1,verbose=10)
# results=gs.fit(X_train,y_train.ravel())
# print(results.score(X_test,y_test))
# print(results.best_params_)
# print(results.score(X_test,y_test))
# txtf = open("SVC_rbf.txt", "w")
# txtf.write("Best Parameters: %s\n" % results.best_params_)
# txtf.write("R2 score: %s\n\n" % results.score(X_test,y_test))
# txtf.close()
# #2 SVM optimal hyperparameters
# from sklearn.model_selection import GridSearchCV
# from sklearn.svm import SVC
# param_grid={
# 'SVC__C':[1,2],
# 'SVC__kernel':['poly'],
# 'SVC__gamma':['auto','scale'],
# 'SVC__degree':[1,2,3,4,5,6,7],
# 'SVC__decision_function_shape':['ovo','ovr']
# }
# pipelineSVC=Pipeline([
# ("SVC", SVC(cache_size=7000))
# ])
# gs=GridSearchCV(estimator=pipelineSVC,cv=2,param_grid=param_grid,return_train_score=True, n_jobs=-1,verbose=10)
# results=gs.fit(X_train,y_train.ravel())
# print(results.score(X_test,y_test))
# print(results.best_params_)
# print(results.score(X_test,y_test))
# txtf = open("SVC_poly.txt", "w")
# txtf.write("Best Parameters: %s\n" % results.best_params_)
# txtf.write("R2 score: %s\n\n" % results.score(X_test,y_test))
# txtf.close()
# #2 SVM optimal hyperparameters
# from sklearn.model_selection import GridSearchCV
from sklearn.svm import SVC
# param_grid={
# 'SVC__C':[1,2],
# 'SVC__kernel':['sigmoid'],
# 'SVC__gamma':['auto','scale'],
# 'SVC__degree':[1,2,3,4,5,6,7],
# 'SVC__decision_function_shape':['ovo','ovr']
# }
# pipelineSVC=Pipeline([
# ("SVC", SVC(cache_size=7000))
# ])
# gs=GridSearchCV(estimator=pipelineSVC,cv=2,param_grid=param_grid,return_train_score=True, n_jobs=-1,verbose=10)
# results=gs.fit(X_train,y_train.ravel())
# print(results.score(X_test,y_test))
# print(results.best_params_)
# print(results.score(X_test,y_test))
# txtf = open("SVC_sigmoid.txt", "w")
# txtf.write("Best Parameters: %s\n" % results.best_params_)
# txtf.write("R2 score: %s\n\n" % results.score(X_test,y_test))
# txtf.close()
# SVC().get_params().keys()
from sklearn.neighbors import KNeighborsClassifier
# from sklearn.model_selection import GridSearchCV
# param_grid={
# 'KNN__n_neighbors':np.arange(1,300,10),
# 'KNN__weights':['uniform'],
# #'KNN__algorithm':['ball_tree','kd_tree','brute'],
# 'KNN__algorithm':['kd_tree','brute'],
# 'KNN__leaf_size':[30,40,50,60,70,80,90,100,110,120,130,140]
# }
# pipelineKNN=Pipeline([
# ("KNN", KNeighborsClassifier())
# ])
# gs=GridSearchCV(estimator=pipelineKNN,cv=2,param_grid=param_grid,return_train_score=True, n_jobs=-1,scoring='accuracy',verbose=10)
# results=gs.fit(X_train,y_train.ravel())
# print(results.best_params_)
# print(results.score(X_test,y_test))
# txtf = open("KNN.txt", "w")
# txtf.write("Best Parameters: %s\n" % results.best_params_)
# txtf.write("R2 score: %s\n\n" % results.score(X_test,y_test))
# txtf.close()
# Best Parameters: {'KNN__algorithm': 'kd_tree',
# 'KNN__leaf_size': 100,
# 'KNN__n_neighbors': 41,
# 'KNN__weights': 'uniform'}
# R2 score: 0.6362402443162538
# print(results.score(X_test,y_test))
# #3 adaboost
from sklearn.ensemble import AdaBoostClassifier
# from sklearn.model_selection import GridSearchCV
# param_grid={
# 'Ada__n_estimators':[10,20,30,40,50,60,70,80,90,100,110,120,130,140,150,160,170,180,190,200],
# 'Ada__learning_rate':[0.5,0.6,0.7,0.8,0.9,1,1.1,1.2,1.3,1.4,1.5]
# }
# pipelineAda=Pipeline([
# ("Ada", AdaBoostClassifier(random_state=42))
# ])
# gs=GridSearchCV(estimator=pipelineAda,cv=2,param_grid=param_grid,return_train_score=True, n_jobs=-1, verbose=10)
# results=gs.fit(X_train,y_train.ravel())
# print(results.best_params_)
# print(results.score(X_test,y_test))
# txtf = open("Adaboost.txt", "w")
# txtf.write("Best Parameters: %s\n" % results.best_params_)
# txtf.write("R2 score: %s\n\n" % results.score(X_test,y_test))
# txtf.close()
# Best Parameters: {'Ada__learning_rate': 1.3,
# 'Ada__n_estimators': 10}
# R2 score: 0.6599932134373939
#
# from sklearn.ensemble import RandomForestClassifier
# from sklearn.model_selection import GridSearchCV
# param_grid={
# 'rf__n_estimators':np.arange(1,1000,100),
# 'rf__max_depth':np.arange(1,1000,100),
# 'rf__min_samples_split':np.arange(1,4,1),
# 'rf__criterion':['gini','entropy','log_loss']
# }
# pipelinerf=Pipeline([
# ("rf", RandomForestClassifier(random_state=42))
# ])
# gs=GridSearchCV(estimator=pipelinerf,cv=2,param_grid=param_grid,return_train_score=True, n_jobs=-1, verbose=10)
# gs.fit(Train_Features,Train_Labels.ravel())
# results=gs.score(X_test,y_test)
# print(results)
from sklearn.ensemble import RandomForestClassifier
rf = RandomForestClassifier(
n_estimators=200, min_samples_split=2, random_state=42, max_depth=7
)
rf.fit(Train_Features, Train_Labels.ravel())
print(rf.score(X_test, y_test))
y_pred = rf.predict(X_test)
print("Testing accuracy", accuracy_score(y_test, y_pred))
y_train_pred = rf.predict(X_train)
print("Training accuracy", accuracy_score(y_train_pred, y_train))
print(classification_report(y_test, y_pred))
cm_display = ConfusionMatrixDisplay(
confusion_matrix=confusion_matrix(y_test, y_pred), display_labels=[0, 1]
)
cm_display.plot()
plt.show()
# y_pred=clf.predict(X_test)
# print(f1_score(y_test,y_pred))
# final_predictions=[]
# temp=rf.predict(Test_Features)
# for x in temp:
# if x==1:
# final_predictions.append('True')
# else:
# final_predictions.append('False')
# submission=pd.DataFrame(columns=['id','Made_Purchase'])
# submission['id']=[i for i in range(len(final_predictions))]
# submission['Made_Purchase']=final_predictions
# submission.to_csv('submission.csv', index=False)
# print(final_predictions)
import xgboost as xgb
clf_xgb = xgb.XGBClassifier(
objective="binary:logistic", missing=None, seed=42, eval_metric="aucpr"
)
clf_xgb.fit(
X_train,
y_train,
verbose=True,
early_stopping_rounds=10,
eval_set=[(X_test, y_test)],
)
# clf_xgb=xgb.XGBClassifier(
# objective='binary:logistic',
# gamma=0,
# learning_rate=0.01,
# max_depth=2,
# reg_lambda=7.9,
# scale_pos_weight=2,
# seed=42,
# # subsample=0.9,
# # colsample_bytree=0.5,
# # eval_metric='aucpr',
# n_estimators=1000)
# clf_xgb.fit(
# X_train,
# y_train.ravel())
# y_pred=clf_xgb.predict(X_test)
# print('Testing accuracy',accuracy_score(y_test,y_pred))
# print(classification_report(y_test,y_pred))
# cm_display=ConfusionMatrixDisplay(confusion_matrix=confusion_matrix(y_test,y_pred), display_labels=[0,1])
# cm_display.plot()
# plt.show()
from sklearn.model_selection import GridSearchCV
import xgboost as xgb
param_grid = {
"max_depth": [1, 2, 3, 4],
"learning_rate": [0.04, 0.05, 0.06, 0.07, 0.08],
"gamma": [0, 0.001],
"reg_lambda": [7.0, 8.0],
"scale_pos_weight": [1, 2, 3],
}
optimal_params = GridSearchCV(
estimator=xgb.XGBRFClassifier(
objective="reg:pseudohubererror", seed=42, subsample=0.9, colsample_bytree=0.5
),
param_grid=param_grid,
scoring="roc_auc",
verbose=0,
n_jobs=10,
cv=3,
)
optimal_params.fit(
X_train, y_train, eval_metric="auc", eval_set=[(X_test, y_test)], verbose=False
)
print(optimal_params.best_params_)
y_pred = optimal_params.predict(X_test)
print("Testing accuracy", accuracy_score(y_test, y_pred))
print(classification_report(y_test, y_pred))
cm_display = ConfusionMatrixDisplay(
confusion_matrix=confusion_matrix(y_test, y_pred), display_labels=[0, 1]
)
cm_display.plot()
plt.show()
# Pseudo huberloss [best params]
# {'gamma': 0, 'learning_rate': 0.066, 'max_depth': 2, 'reg_lambda': 8.0, 'scale_pos_weight': 1}
# Testing accuracy 0.6830675262979301
#
stop
from sklearn.ensemble import StackingClassifier
from sklearn.linear_model import SGDClassifier
estimators = [
# ('svc', SVC(C=2, decision_function_shape='ovo', degree=1, gamma='auto', kernel='rbf',probability=True)),
# ('bernb', BernoulliNB(alpha=1.0, binarize=0.0, class_prior=None, fit_prior=True)),
# ('knn', KNeighborsClassifier(algorithm= 'kd_tree',leaf_size=100, n_neighbors=41, weights='uniform')),
# ('ada', AdaBoostClassifier(learning_rate=1.3, n_estimators=10)),
("xgb", xgb.XGBClassifier(objective="binary:logistic", random_state=42)),
(
"rf",
RandomForestClassifier(
n_estimators=5, min_samples_split=2, random_state=42, max_depth=10
),
),
]
fclf = StackingClassifier(
estimators=estimators, final_estimator=SGDClassifier() # Default hinge loss
)
fclf.fit(Train_Features, Train_Labels.ravel())
fclf.score(X_test, y_test)
print(results.best_params_)
txtf = open("rf.txt", "w")
txtf.write("Best Parameters: %s\n" % results.best_params_)
txtf.write("R2 score: %s\n\n" % results.score(X_test, y_test))
txtf.close()
from sklearn.ensemble import VotingClassifier
import xgboost as xgb
vclf = VotingClassifier(
estimators=[
# ('svc', SVC(C=2, decision_function_shape='ovo', degree=1, gamma='auto', kernel='rbf',probability=True)),
(
"bernb",
BernoulliNB(alpha=1.0, binarize=0.0, class_prior=None, fit_prior=True),
),
(
"knn",
KNeighborsClassifier(
algorithm="kd_tree", leaf_size=100, n_neighbors=41, weights="uniform"
),
),
# ('ada', AdaBoostClassifier(learning_rate=1.3, n_estimators=10)),
("xgb", xgb.XGBClassifier(objective="binary:logistic", random_state=42)),
(
"rf",
RandomForestClassifier(
n_estimators=5, min_samples_split=2, random_state=42, max_depth=10
),
),
(
"mlp",
MLPClassifier(
hidden_layer_sizes=(88, 70, 40, 11, 5, 2),
max_iter=500,
activation="relu",
early_stopping=True,
),
),
],
voting="soft",
)
vclf = vclf.fit(Train_Features, Train_Labels.ravel())
y_pred = vclf.predict(X_test)
print(vclf.score(X_test, y_test))
from sklearn.metrics import accuracy_score
# using accuracy_score
score = accuracy_score(y_test, y_pred)
print(score)
# fclf.score(X_test,y_test)
|
import pandas as pd
import matplotlib.pyplot as plt
import plotly.express as px
import plotly.graph_objs as go
from geopy.geocoders import Nominatim
df = pd.read_csv(
"/kaggle/input/electricity-production-by-source-world/Electricity_Production_By_Source.csv"
)
df.head()
# loading ISO 3166 dataset to check full list of existing countries based on Alpha-3 codes.
iso_a3 = pd.read_csv(
"/kaggle/input/country-names-with-short-codes-a2-a3-iso/country code.csv",
encoding="ISO-8859-1",
)
iso_a3_list = list(iso_a3["Alpha-3 code"])
iso_a3.head()
# replace null values with 0
df = df.fillna(0)
# create a new column for total electricity production
df["Total"] = df.iloc[:, 3:].sum(axis=1)
# check if Code not matches with ISO Alpha-3 codes to detect Totals and Region Sub-totals in the dataset
SubTotal_codes = df[~df["Code"].isin(iso_a3_list)]
SubTotal_codes = (
SubTotal_codes.groupby(["Entity", "Code"])["Total"]
.sum()
.sort_values(ascending=False)
.reset_index()
)
# filter dataset only for rows which have a valid ISO code, so represent real country
df = df[df["Code"].isin(iso_a3_list)]
df_country = (
df.groupby(["Entity", "Code"])[
[
"Electricity from coal (TWh)",
"Electricity from gas (TWh)",
"Electricity from hydro (TWh)",
"Electricity from other renewables (TWh)",
"Electricity from solar (TWh)",
"Electricity from oil (TWh)",
"Electricity from wind (TWh)",
"Electricity from nuclear (TWh)",
"Total",
]
]
.sum()
.sort_values(by="Total", ascending=False)
.reset_index()
)
# Chart 1 - Electricity Production by Year and Source
coal = go.Bar(x=df["Year"], y=df["Electricity from coal (TWh)"], name="Coal")
gas = go.Bar(x=df["Year"], y=df["Electricity from gas (TWh)"], name="Gas")
hydro = go.Bar(x=df["Year"], y=df["Electricity from hydro (TWh)"], name="Hydro")
other_renewables = go.Bar(
x=df["Year"],
y=df["Electricity from other renewables (TWh)"],
name="Other Renewables",
)
solar = go.Bar(x=df["Year"], y=df["Electricity from solar (TWh)"], name="Solar")
oil = go.Bar(x=df["Year"], y=df["Electricity from oil (TWh)"], name="Oil")
wind = go.Bar(x=df["Year"], y=df["Electricity from wind (TWh)"], name="Wind")
nuclear = go.Bar(x=df["Year"], y=df["Electricity from nuclear (TWh)"], name="Nuclear")
data = [coal, gas, hydro, other_renewables, solar, oil, wind, nuclear]
layout = go.Layout(title="Electricity Production by Year and Source", barmode="stack")
fig = go.Figure(data=data, layout=layout)
fig.show()
# Select the data for the decade 2010-2019
df_decade = df[df["Year"].between(2010, 2019)]
# Group the data by Entity and Source of electricity
df_grouped = (
df_decade.groupby("Entity")[
[
"Electricity from coal (TWh)",
"Electricity from gas (TWh)",
"Electricity from hydro (TWh)",
"Electricity from other renewables (TWh)",
"Electricity from solar (TWh)",
"Electricity from oil (TWh)",
"Electricity from wind (TWh)",
"Electricity from nuclear (TWh)",
"Total",
]
]
.sum()
.reset_index()
)
# get top 10 by total production to limit the chart
top_10 = (
df_grouped.groupby("Entity").sum().sort_values(by="Total", ascending=False).head(10)
)
top_10_codes = top_10.index.to_list()
filtered_data = df_grouped[df_grouped["Entity"].isin(top_10_codes)].sort_values(
by="Total", ascending=True
)
# Create a stacked bar chart for production per source per country
trace1 = go.Bar(
x=filtered_data["Electricity from coal (TWh)"],
y=filtered_data["Entity"],
name="Coal",
orientation="h",
)
trace2 = go.Bar(
x=filtered_data["Electricity from gas (TWh)"],
y=filtered_data["Entity"],
name="Gas",
orientation="h",
)
trace3 = go.Bar(
x=filtered_data["Electricity from hydro (TWh)"],
y=filtered_data["Entity"],
name="Hydro",
orientation="h",
)
trace4 = go.Bar(
x=filtered_data["Electricity from other renewables (TWh)"],
y=filtered_data["Entity"],
name="Other Renewables",
orientation="h",
)
trace5 = go.Bar(
x=filtered_data["Electricity from solar (TWh)"],
y=filtered_data["Entity"],
name="Solar",
orientation="h",
)
trace6 = go.Bar(
x=filtered_data["Electricity from oil (TWh)"],
y=filtered_data["Entity"],
name="Oil",
orientation="h",
)
trace7 = go.Bar(
x=filtered_data["Electricity from wind (TWh)"],
y=filtered_data["Entity"],
name="Wind",
orientation="h",
)
trace8 = go.Bar(
x=filtered_data["Electricity from nuclear (TWh)"],
y=filtered_data["Entity"],
name="Nuclear",
orientation="h",
)
data = [trace1, trace2, trace3, trace4, trace5, trace6, trace7, trace8]
layout = go.Layout(
title="Electricity Production by Source and Country (2010-2019)",
barmode="stack",
xaxis_title="Electricity Production (TWh)",
yaxis_title="Country",
)
fig = go.Figure(data=data, layout=layout)
fig.show()
|
import numpy as np
import pandas as pd
TRAIN_PATH = "/kaggle/input/house-prices-advanced-regression-techniques/train.csv"
CATEGORY_UNIQUE_VALUE_BASESIZE = 20
train = pd.read_csv(TRAIN_PATH)
train.info()
# # get object columns
train_only_object = train.select_dtypes(include="object")
cate_col = train_only_object.columns.tolist()
cate_col
train["Alley"].isna().sum()
train["FireplaceQu"].isna().sum()
# # delete condition
# ### 1. column value count > CATEGORY_UNIQUE_VALUE_BASESIZE
# ### or
# ### 2. na data size > total row size / 2
deleteCol = []
NA_DATA_BASESIZE = len(train) / 2
print("NA_DATA_BASESIZE is", NA_DATA_BASESIZE)
for i in range(len(cate_col)):
if len(train[cate_col[i]].value_counts()) > CATEGORY_UNIQUE_VALUE_BASESIZE:
deleteCol.append(cate_col[i])
else:
if train[cate_col[i]].isna().sum() > NA_DATA_BASESIZE:
deleteCol.append(cate_col[i])
print("deleteCol = ", deleteCol)
# # drop Columns
train_drop = train.drop(columns=deleteCol, axis=1)
train_drop.info()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
## Importing required libraries
import numpy as np, gc
import pandas as pd
import matplotlib.pyplot as plt
import matplotlib.patches as mpatches
import sklearn
import seaborn as sns
from sklearn.metrics import confusion_matrix
import random
from sklearn.model_selection import KFold, GroupKFold
from xgboost import XGBClassifier
from sklearn.metrics import f1_score
import pandas as pd
import pyarrow.parquet as pq
# Load the parquet file into a PyArrow table
table = pq.read_table("/kaggle/input/how-to-get-32gb-ram/train.parquet")
# Convert the PyArrow table to a Pandas dataframe
train_df = table.to_pandas()
train_df.describe()
# Rename the columns in the dataframe using the labels vector
# labels = ['label_1', 'label_2', 'label_3', ...] # replace with your own labels
# df.columns = labels
train_df.columns
ROOMS = train_df.room_fqid.unique()
print("Number of rooms:", len(ROOMS))
print(ROOMS)
# DICTIONARY TO IMAGE PATHS
# ITERATE ROOMS AND DISPLAY IMAGES AND SCATTER PLOTS
for j, rm in enumerate(ROOMS):
print("\n")
print("#" * 25)
print(f"### ROOM {j+1}:", rm)
print("#" * 25)
# DISPLAY NAVIGATION CLICKS
df = train_df.loc[
(train_df.event_name == "navigate_click")
& (train_df.room_fqid == rm)
& (train_df.fqid.isna())
]
x_min, y_min = df[["room_coor_x", "room_coor_y"]].min().values
x_max, y_max = df[["room_coor_x", "room_coor_y"]].max().values
plt.figure(figsize=(20, 20))
plt.scatter(df.room_coor_x, df.room_coor_y, s=0.1)
plt.xlim((x_min, x_max))
plt.ylim((y_min, y_max))
plt.gca().set_aspect("equal")
plt.title(f"room {j+1} - NAVIGATION CLICKS - {rm}", size=20)
plt.gca().yaxis.tick_right()
plt.show()
# DISPLAY IMAGE
if rm in dd:
plt.figure(figsize=(17, 17))
plt.imshow(dd[rm])
plt.axis("off")
plt.show()
# DISPLAY ITEMS OF INTEREST
df = train_df.loc[
(train_df.event_name == "navigate_click")
& (train_df.room_fqid == rm)
& (train_df.fqid.notna())
]
ITEMS = df.fqid.unique()
plt.figure(figsize=(20, 20))
plt.scatter(df.room_coor_x, df.room_coor_y, s=0.1)
for i in ITEMS:
mns = df.loc[df.fqid == i, ["room_coor_x", "room_coor_y"]].mean().values
plt.text(mns[0], mns[1], i, fontsize=26)
plt.xlim((x_min, x_max))
plt.ylim((y_min, y_max))
plt.gca().set_aspect("equal")
plt.title(f"room {j+1} - ITEMS OF INTEREST - {rm}", size=20)
plt.gca().yaxis.tick_right()
plt.show()
# DISPLAY PERSON CLICKS
df = train_df.loc[
(train_df.event_name == "person_click")
& (train_df.room_fqid == rm)
& (train_df.fqid.notna())
]
if len(df) != 0:
ITEMS = df.fqid.unique()
plt.figure(figsize=(20, 20))
plt.scatter(df.room_coor_x, df.room_coor_y, s=0.1)
for i in ITEMS:
mns = df.loc[df.fqid == i, ["room_coor_x", "room_coor_y"]].mean().values
plt.text(mns[0], mns[1], i, fontsize=26)
plt.xlim((x_min, x_max))
plt.ylim((y_min, y_max))
plt.gca().set_aspect("equal")
plt.title(f"room {j+1} - PERSON CLICKS - {rm}", size=20)
plt.gca().yaxis.tick_right()
plt.show()
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn
# ## Stock Market Analysis
# In recent years, there has been a significant increase in computing power, thanks to the advancements in technology. Among the major players in this field is NVIDIA, whose GPU powers nearly all tech industries. This has allowed for more advanced and sophisticated applications that were once thought impossible.
# In this notebook, we will delve into the analysis of trending companies such as NVIDIA, AMD, and Intel to gain a better understanding of their impact on the industry.
# ## Load Data
amd = pd.read_csv(
"/kaggle/input/nvidia-amd-intel-asus-msi-share-prices/AMD (1980-2023).csv"
)
intel = pd.read_csv(
"/kaggle/input/nvidia-amd-intel-asus-msi-share-prices/Intel (1980-2023).csv"
)
nvidia = pd.read_csv(
"/kaggle/input/nvidia-amd-intel-asus-msi-share-prices/NVIDIA (1999-2023).csv"
)
amd.shape, intel.shape, nvidia.shape
# ## Modify Data
class Imputer:
def __init__(self, data):
self.data = data
def parse_dates(self):
self.data["Date"] = self.data.Date.apply(lambda x: pd.Timestamp(x))
def data_after(self, time: str):
self.data = self.data[self.data.Date >= pd.Timestamp(time)]
def activate(self, time: str):
self.parse_dates()
self.data_after(time)
return self.data
nvidia = Imputer(nvidia).activate("2018-01-01")
intel = Imputer(intel).activate("2018-01-01")
amd = Imputer(amd).activate("2018-01-01")
merged_df = pd.merge(
nvidia.iloc[:, [0, 5]], amd.iloc[:, [0, 5]], on="Date", how="outer"
)
merged_df = pd.merge(merged_df, intel.iloc[:, [0, 5]], on="Date", how="outer")
merged_df.columns = ["Date", "Adj_Close_NVIDIA", "Adj_Close_AMD", "Adj_Close_INTEL"]
merged_df.head()
# ## Market Movement
# - Based on the market analysis, it is evident that NVIDIA and AMD have experienced a significant rise in their growth over the past few years. The movement of the market for these companies shows a steep increase in their stock prices and market value.
# - On the other hand, Intel has maintained a relatively constant stock price, with no significant spikes or dips in their market performance. This indicates that the competition between NVIDIA and AMD has put pressure on Intel to innovate and keep up with the changing market trends.
# It will be interesting to see how these trends continue in the future and whether Intel can regain its position as a market leader.
#
plt.figure(figsize=(10, 5))
plt.plot(merged_df["Date"], merged_df["Adj_Close_NVIDIA"], label="NVI")
plt.plot(merged_df["Date"], merged_df["Adj_Close_AMD"], label="AMD")
plt.plot(merged_df["Date"], merged_df["Adj_Close_INTEL"], label="INT")
plt.xlabel("Date")
plt.ylabel("Adj Close")
plt.title("Movement of Stock Price (2018-2023)")
plt.legend()
plt.show()
# ## Volume vs Market Movement
# Volume of trade and stock price are two important factors that are closely related in the stock market. Volume refers to the number of shares or contracts that are traded in a particular period, such as a day or a week. Stock price refers to the market value of a company's shares, which is determined by the supply and demand of the shares in the market.
# - In the case of NVIDIA, we can see from the data that the stock price has been quite volatile in the past few months. The stock started at a price of 233.20 on March 3rd, 2023, and reached a high of 280 on April 4th, 2023.
# - The high volume of shares traded indicates that there is significant interest in NVIDIA stock among investors. This can be attributed to the company's strong financial performance, positive market outlook, and ongoing developments in the technology industry.
#
def plot(data, stock):
data = data.iloc[1200:, :]
fig, ax1 = plt.subplots(figsize=(20, 8))
ax2 = ax1.twinx()
ax1.bar(x=data.Date, height=data.Volume, alpha=0.7)
ax2.plot(data.Date, data["High"], "g-")
ax2.plot(data.Date, data["Low"], "r-")
plt.title(f"Volume and Market Price ({stock})")
ax1.set_xlabel("Date")
ax1.set_ylabel("Volume", color="b")
ax2.set_ylabel("High/Low", color="g")
plot(nvidia, "NVIDIA")
plot(amd, "AMD")
plot(intel, "INTEL")
# ## Correlation
# ### NVIDIA AND AMD
# - In this case, we can see that the strongest positive correlation is between NVIDIA and AMD. This suggests that the two stocks tend to move together, and that changes in the price of one are often accompanied by similar changes in the other.
# ### NVIDIA AND INTEL
# - Here there is a weak negative correlation between NVIDIA and Intel, indicating that the two stocks tend to move in opposite directions. This could be due to differences in their respective business models or market factors affecting the industry.
# It is important to note that correlation does not necessarily imply causation, and that there may be other factors at play that are driving the prices of these stocks. It is also important to conduct further analysis and consider other metrics before making investment decisions.
import seaborn as sns
sns.heatmap(merged_df.corr(), annot=True, cmap="coolwarm")
|
# # Bookstores needs to apply this method!
# ## Can we recommend books based on this information?
#
#
#
# Image source: https://www.halifaxpubliclibraries.ca/blogs/post/the-journey-of-a-book-part-1//
# # Summary
# - Data cleaning was performed on the books rating data to suit the analysis
# - Performed cosine-similarity on book information to provide content-based recommendation based on the books author, publication year, and publisher
# - Use the rating dataframe to make item-based collaborative recommendation for certain user
#
# Example of recommendation
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from scipy import sparse
from sklearn.neighbors import NearestNeighbors
import missingno as msno
# # 1. Dataset Overview:
# ## 1.1 Context
# Here we have a set of 3 datasets which contains:
# - **Books data** : Giving the detail overview about the book information including the book title, publication year, as well as the author of the book
# - **Users data** : Give the detailed overview about each user such as the user's location as well as the user's age
# - **Rating data** : Basically compiled data about how each user rated each books
# ## 1.2 Educational use of the dataset
# This dataset is a simple data which does not require much data cleaning. The rich information provided about the books, users, as well as the rating makes these data a good candidate to practice recommendation algorithm. Despite missing several details that can potentially makes it ideal, it is still sufficient enough.
# ## 1.3 Personal Goal and Motivation for this notebook
# In this notebook, the motivation is to:
# - Preprocess several dataset in relation to book data and rating data
# - Create recommendation algorithm to recommend book to certain user
# - 1st model: Content-based recommendation
# - 2nd model: collaborative recommendation
# # 2. Loading Data
# ## 2.1 Books Data
df_books = pd.read_csv(
"../input/book-recommendation-dataset/Books.csv",
delimiter=",",
usecols=[0, 1, 2, 3, 4],
)
df_books.info()
# Based on the info above, we can see that Year-of-publication is stated as object.
# I believe that this is not the correct format and we need to put it into integer
try:
df_books["Year-Of-Publication"] = df_books["Year-Of-Publication"].astype(int)
except Exception as e:
print(e)
# Oops! There is a data point in which the Years of Publication is filled with the publisher. Let us see the data
df_books[df_books["Year-Of-Publication"] == "DK Publishing Inc"]
# We can see that the book above is shifted.
# I believe that this is a formatting problem
# Cleaning up the data one by one can take a lot of effort. So, for now, we will just delete data points which are wrongly formatted.
# We can catch this by trying to convert the Year-of-publication into numerical value and coerce the conversion. The data that can not be converted into numerical value will be replaced by NaN value which will be then removed by dropna() function of pandas
df_books["Year-Of-Publication"] = pd.to_numeric(
df_books["Year-Of-Publication"], errors="coerce"
)
df_books = df_books.dropna()
df_books["Year-Of-Publication"] = df_books["Year-Of-Publication"].astype(int)
df_books.head()
# The formatting went well. Now let us see how book is distributed among the publication year!
plt.bar(
pd.DataFrame(df_books["Year-Of-Publication"].value_counts()).sort_index().index,
np.array(df_books["Year-Of-Publication"].value_counts()),
width=1,
)
# The histogram above showed us 2 things:
# - Most of the books published in year 1700~2000s
# - It is clear that there are some books with publication year below 500s. I believe that this is an error of input
# - It can be a good practice to replace these books with correct information. In the case of missing value, we can impute the number with mean or mode. However, I do not believe it will change the result much in this case. Therefore, I will just leave it as it is for now
# That concludes the preprocessing for the book data. Let us move to the users data!
# ## 2.2 Users Data
df_Users = pd.read_csv("../input/book-recommendation-dataset/Users.csv")
df_Users.info()
# Let us check for the missing values in the user data
msno.matrix(df_Users)
# Looking at the age data, we can see that about many data are missing from the age column (to be precise: 110762 data).
# Since we are going to use content-based filtering based on the book information and not based on the user information, we can leave the missing value as it is for now.
# In this case, we will only use this phase to grasp the information provided by the User data. However, we will not perform any preprocessing on the user data.
# # 2.3 Rating Data
# We have preprocessed the books and the users. Now, we will keep in mind the books and users that we exclude from the previous data cleaning and we will also reflect it to the rating data.
# First, we will load the data
df_rating = pd.read_csv("../input/book-recommendation-dataset/Ratings.csv")
df_rating.info()
# From the rating data, we can see that we have 3 columns as follows:
# - User-ID: The User ID. This one is unique for each user
# - ISBN : some sort of the ID for the book being read by the user
# - Rating: Ratings given by the user for the book.
# The sample above showed that user 276725 has given book with ISBN 034545104X a rating of 0.
# We can also observed that the data contains around 1 million data. This can be very burdensome to our analysis. For that reason, I will try to clean our data. The details for cleaning will be explained in the section below.
# ### 2.3.1 Data Cleaning
# We will begin our cleaning phase by deleting the data points that contains books and user-ID that we deleted in the previous sections
print("Number of data before cleaning : {}".format(len(df_rating)))
df_rating = df_rating[df_rating["ISBN"].isin(df_books["ISBN"])]
print("Number of data after cleaning : {}".format(len(df_rating)))
# We can clean the data further by reducing some noise.
# In real life, when we are dealing with rating data, there are some users that doesn't rate certain books. On the other side, there are some books which are not rated by several user.
# These data are called sparse data in which data contains mostly zero or null values. In real-world, ratings can be very sparse and data points will mostly be collected from either:
# - A very popular book
# - or a highly engaging user
# A book that are rated highly but only have 1 or 2 users who rated the book are hardly credible.
# On the other hand, user that only rated 1 or two book are hardly a credible user. Therefore, we need to clean the data by introducing filter with below criteria:
# - For a book to be credible, it requres ate least votes from 10 users
# - For a user to be credible, the user must have voted at least 50 books
# This filter might also help us in minimizing the number of the data as well as reducing memory used in this analysis
f = ["count", "mean"]
df_books_summary = df_rating.groupby("ISBN")["Book-Rating"].agg(f)
df_books_summary.index = df_books_summary.index.map(str)
drop_book_list = df_books_summary[df_books_summary["count"] < 10].index
df_cust_summary = df_rating.groupby("User-ID")["Book-Rating"].agg(f)
df_cust_summary.index = df_cust_summary.index.map(int)
drop_cust_list = df_cust_summary[df_cust_summary["count"] < 10].index
print("Before Filtering: {}".format(df_rating.shape))
df_rating = df_rating[~df_rating["ISBN"].isin(drop_book_list)]
df_rating = df_rating[~df_rating["User-ID"].isin(drop_cust_list)]
print("After Filtering: {}".format(df_rating.shape))
pivot_rating = df_rating.pivot(index="ISBN", columns="User-ID", values="Book-Rating")
pivot_rating.head()
# # 3 Building Recommendation Model
# ## 3.1 Content-based Recommendation
# Content-based collaborative recommendation works by looking at a certain product that a user has liked or rated highly in the past then proceeds to recommend a product that is similar to the highly-rated product.
# In this notebook, I will try to make a function that requires us to input the user-ID.
# The algorithm then will proceed to find the highest-rated book by the user and then recommend several other books that are similar to the highest-rated book
#
# ### Content-based recommendation based on the author and publisher of the book
# This recommendation is made based on an assumption that a certain reader will be attracted to read another book based factors below:
# - authors
# Some readers might be attracted to some certain authors because some authors tends to write about a specific genre that certain demographic of reader like. So, including auhtor as one of the factor might be plausible.
# - year-of-publication
# Year of publication might reflect some kind of genre preferred by readers (e.g. reader who would like to read classical philosophy books might tend to read older books compared to new ones).
# - publisher
# Certain publisher has a certain kind of credibility which might make some book readers prefer one publisher compared to other.
# Based on that consideration, we will consider Author, Year of publication, as well as publisher as basis for recommendation.
len(df_books["ISBN"].unique())
# There are around 270000 different books based on what we have in this dataset. For now, we will limit the recommendation only for books that are considered credible based on our definition when we clean the data. Hence, we will only pick books featured in the df_rating dataframe.
# After cleaning, we are left with:
print(
"number of books: "
+ str(len(df_books[df_books["ISBN"].isin(df_rating["ISBN"].unique())].copy()))
)
# only around 17359 books.
# Keep in mind that in real-life scenario we would like to have as many sample as possible.
features = ["Book-Author", "Year-Of-Publication", "Publisher"]
df_books_preprocess = df_books[df_books["ISBN"].isin(df_rating["ISBN"].unique())].copy()
for feature in features:
if df_books_preprocess[feature].dtype == "O":
df_books_preprocess[feature] = df_books_preprocess[feature].str.replace(
"\W", "", regex=True
)
df_books_preprocess[feature] = df_books_preprocess[feature].apply(
lambda x: str.lower(x)
)
def create_soup(x):
return x["Book-Author"] + " " + str(x["Year-Of-Publication"]) + " " + x["Publisher"]
df_books_preprocess["soup"] = df_books_preprocess[features].apply(create_soup, axis=1)
from sklearn.feature_extraction.text import CountVectorizer
count = CountVectorizer(stop_words="english")
count_matrix = count.fit_transform(df_books_preprocess["soup"])
# Compute the Cosine Similarity matrix based on the count_matrix
from sklearn.metrics.pairwise import cosine_similarity
cosine_sim2 = cosine_similarity(count_matrix, count_matrix)
df2 = df_books_preprocess.reset_index()
indices = pd.Series(df2.index, index=df2["ISBN"])
def recommend_books(ISBN, cosine_sim):
# Get the index of the books that matches the ISBN
idx = indices[ISBN]
# Get the pairwsie similarity scores of all books with that book
sim_scores = list(enumerate(cosine_sim[idx]))
# Sort the books based on the similarity scores
sim_scores = sorted(sim_scores, key=lambda x: x[1], reverse=True)
# Get the scores of the 10 most similar books
sim_scores = sim_scores[1:11]
# Get the book indices
book_indices = [i[0] for i in sim_scores]
# Return the top 10 most similar books
return df2[["Book-Title", "soup"]].iloc[book_indices], sim_scores
def get_recommendations(userid, cosine_sim):
# userid = str(userid)
ISBN = df_rating["ISBN"].loc[
df_rating[df_rating["User-ID"] == userid]["Book-Rating"].idxmax()
]
recommendation, sim_score = recommend_books(ISBN, cosine_sim)
df_recommend = pd.DataFrame(recommendation)
df_recommend["Cosine_Similarity"] = sim_score
df_recommend["Cosine_Similarity"] = df_recommend["Cosine_Similarity"].apply(
lambda x: x[1]
)
return df_recommend
# We will try to make some recommendation for User-ID 8.
# First, let us check which books read by User-ID 8
pd.concat(
[
df_books[
df_books["ISBN"].isin(list(df_rating[df_rating["User-ID"] == 8]["ISBN"]))
].reset_index(drop=True),
df_rating[df_rating["User-ID"] == 8]["Book-Rating"].reset_index(drop=True),
],
axis=1,
)
# User-ID 8 read 3 books and among them, He rated Clara Callan the highest. Therefore, we will recommend some books for User-ID 8!
df_recommend = get_recommendations(8, cosine_sim2)
df_recommend
# We can see that User-ID 8 were recommended several books based on this algorithm.
# All of those books were published in 2001 which might be in line with some genre of that years.
# Spadework, especially, is also published by Harper Flamingo, Canada publisher which has highest chance to be liked by User-ID 8 based on his preference.
# Mind that this is not perfect and further information regarding the genre or synopsis of each books can be used to further improve this recommendation.
# However, for a start, this can be said as a good result of the content-based recommendation.
# ## 3.2 Collaborative Recommendation
# Content-based recommendation works good at recommending certain books that is similar to the books that user has proven to be liked. So far, quite a reasonable approach.
# However, this kind of recommendation is lacking because it cannot suggest something outside of the kind of the book that the user has read. For example, as we can see, the user are only recommended the books published in 2001. Even if we include other parameter such as genre, then we are only exploring some genre that the user has been read before.
# Say we want to introduce or recommend a new book with other kind of genre to this user, this approach might not be sufficient.
# Collaborative recommendation, on the other hand, take into account other readers taste as a way to recommend something new. Collaborative filtering basically works as shown in the image below:
#
# source: https://github.com/XinyueTan/Collaborative-filtering-recommender-systems
# The basis of this is: If a user with similar taste with the targeted user likes a certain book that has never been read by the targeted user, there is a high chance that our targeted user might like the other books read by those user with similar taste.
# There are two ways to approach collaborative recommendation. However, I will explain the method that I will use in this analysis which is the item-based collaborative recommendation.
# **Basic Concept**
# The fundamental assumption of item-based collaborative recommendation is: "A user gives similar rating to similar product". Assume we have this rating table for the following books:
#
# We can find the similarity between each books using any kind of similarity metric such as cosine similarity. Based on the calculation, we can then predict how the missing rating of "Clara Callan" book may be perceived by user B.
# In collaborative recommendation, whether the , we need to get the data into a matrix of users and the items they rated. In Pandas, we can easily do it using pivot method to make a pivot table.
# For this analysis, I will focus on performing the item-based collaborative recommendation.
# I will demonstrate creating the pivot table as well as the recommendation with the user_rating dataframe.
# #### **Making Pivot Table**
# For an item-based collaborative filtering, we will first make a pivot table on which we will calculate similarities.
# In case of item-based collaborative filtering, the pivot-table will be formulated with books as the rows and user as the columns
final_dataset = df_rating.pivot(index="ISBN", columns="User-ID", values="Book-Rating")
final_dataset.head()
# We will then replace the NaN value with zeros
final_dataset = final_dataset.fillna(0)
final_dataset.head()
final_dataset.shape
print(
"Shape of final dataset : {0} x {1}".format(
final_dataset.shape[0], final_dataset.shape[1]
)
)
# So we have a final dataset with dimension of **17359 x 11527** where most the value are sparse.
# Processing all this data as it is might take so much memories and will burden our analysis.
# One of the way to solve this problem is by using compressed sparse row (CSR) matrix.
# We can use sparse function from scipy to do this
final_dataset_csr = sparse.csr_matrix(final_dataset)
final_dataset.reset_index(inplace=True)
# Now, we will be using KNN algorithm to calculate the similarity with cosine distance.
knn = NearestNeighbors(metric="cosine", algorithm="brute", n_neighbors=20, n_jobs=-1)
knn.fit(final_dataset_csr)
# We will now make a function to recommend a book for a user based on that user's highest rated book.
def get_book_recommendation(ISBN, n_recommend):
n_books_to_reccomend = n_recommend
book_list = df_books[df_books["ISBN"] == ISBN]
if len(book_list):
book_idx = book_list.iloc[0]["ISBN"]
book_idx = final_dataset[final_dataset["ISBN"] == book_idx].index[0]
distances, indices = knn.kneighbors(
final_dataset_csr[book_idx], n_neighbors=n_books_to_reccomend + 1
)
rec_book_indices = sorted(
list(zip(indices.squeeze().tolist(), distances.squeeze().tolist())),
key=lambda x: x[1],
)[:0:-1]
recommend_frame = []
for val in rec_book_indices:
book_idx = final_dataset.iloc[val[0]]["ISBN"]
idx = df_books[df_books["ISBN"] == book_idx].index
recommend_frame.append(
{
"ISBN": df_books.iloc[idx]["ISBN"].values[0],
"Book-Title": df_books.iloc[idx]["Book-Title"].values[0],
"Book-Author": df_books.iloc[idx]["Book-Author"].values[0],
"Year-Of-Publication": df_books.iloc[idx][
"Year-Of-Publication"
].values[0],
"Publisher": df_books.iloc[idx]["Publisher"].values[0],
"Distance": val[1],
}
)
df = pd.DataFrame(recommend_frame, index=range(1, n_books_to_reccomend + 1))
return df
else:
return "Book not found. Re-check the ISBN"
def recommend_book(userID, n_recommend=10):
ISBN = df_rating.loc[
df_rating[df_rating["User-ID"] == userID]["Book-Rating"].idxmax()
]["ISBN"]
recommendation = get_book_recommendation(ISBN, n_recommend)
return recommendation
# Let us get the 10 recommendation for the User-ID = 8 (highest rated book: Clara Callan)
recommend_book(8, n_recommend=10)
|
import pandas as pd
a = pd.Series([45, 85, 63, 4], index=["chennai", "delhi", "bangalore", "pune"])
a
b = pd.Series([45, 85, None, 49], index=["chennai", "delhi", "bangalore", "pune"])
b
mask = b.isna()
mask
b[mask]
b.dropna()
b.fillna(20)
s = pd.read_csv("/kaggle/input/videogamesales/vgsales.csv")
s
s.describe()
import pandas as pd
s1 = pd.read_csv(
"/kaggle/input/videogamesales/vgsales.csv", usecols=["Year"], squeeze=True
)
s1
s1.count()
mask = s1.isna()
mask
c = s1[mask]
c
s1.dropna()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pathlib
p_dir = pathlib.Path("/kaggle/input/bike-sharing-demand")
train = pd.read_csv(p_dir / "train.csv")
test = pd.read_csv(p_dir / "test.csv")
submission = pd.read_csv(p_dir / "sampleSubmission.csv")
# # 1. Feature Engineering
# ## Feature Selection (피처 선택)
# ### 이상치 제거
train = train[train["weather"] != 4]
# ### 데이터 합치기
all_data = pd.concat([train, test], ignore_index=True)
all_data
# ### 파생 피처 추가
# https://steadiness-193.tistory.com/227
# ### datetime을 object에서 datetime 타입으로 변환(feat. 송석리 선생님)
# train['datetime'] = pd.to_datetime(train['datetime'])
# train['year'] = train['datetime'].dt.year
# train['month'] = train['datetime'].dt.month
# train['day'] = train['datetime'].dt.day
# train['hour'] = train['datetime'].dt.hour
# train['minute'] = train['datetime'].dt.minute
# train['second'] = train['datetime'].dt.second
# train['weekday'] = train['datetime'].dt.dayofweek
# train.info()
from datetime import datetime
all_data["datetime"] = pd.to_datetime(all_data["datetime"])
type(all_data.iloc[0]["datetime"])
all_data.iloc[0]["datetime"].date()
all_data["date"] = all_data["datetime"].dt.date
all_data.head()
all_data["year"] = all_data["datetime"].dt.year
all_data["month"] = all_data["datetime"].dt.month
all_data["hour"] = all_data["datetime"].dt.hour
all_data["weekday"] = all_data["datetime"].dt.dayofweek
all_data.sample(n=10)
# ### 필요 없는 피처 제거
drop_features = ["casual", "registered", "datetime", "date", "windspeed", "month"]
all_data.drop(drop_features, axis=1, inplace=True)
all_data
# ### 데이터 다시 나누기
X_train = all_data[~pd.isnull(all_data["count"])]
X_test = all_data[pd.isnull(all_data["count"])]
X_train.head(3)
X_test.head(3)
y = X_train["count"]
X_train = X_train.drop(["count"], axis=1)
X_test = X_test.drop(["count"], axis=1)
X_train.head(3)
# # 2. 평가지표 계산 함수 작성
import numpy as np
def rmsle(y_true, y_pred, convertExp=True):
if convertExp:
y_true = np.exp(y_true)
y_pred = np.exp(y_pred)
log_true = np.nan_to_num(np.log1p(y_true))
log_pred = np.nan_to_num(np.log1p(y_pred))
output = np.sqrt(np.mean(np.power(log_true - log_pred, 2)))
return output
# # 3. 모델 훈련
from sklearn.linear_model import LinearRegression
lr = LinearRegression()
log_y = np.log(y) # 타깃값 로그 변환
lr.fit(X_train, log_y)
# # 4. 모델 성능 검증
preds = lr.predict(X_train)
res = rmsle(log_y, preds, True)
print(f"선형 회귀의 RMSLE 값: {res:.4f}")
lr_preds_log = lr.predict(X_test) # 데스트 데이터로 예측
lr_preds = np.exp(lr_preds_log) # 지수 변환
submission["count"] = lr_preds # 예측값으로 Submission의 count값 변경
submission.to_csv("submission_lr_model.csv", index=False) # 파일로 저장
lr_preds_log
lr_preds
X_test
submission
# # 2. 성능 개선 : 릿지 회귀 모델
# ## 2-1. 하이퍼파라미터 최적화(모델 훈련)
# ### 모델 생성
from sklearn.linear_model import Ridge
from sklearn.model_selection import GridSearchCV
from sklearn import metrics
# 1. 대상 모델
ridge = Ridge()
# ### 그리드서치 객체 생성
# 2. 비교 검증할 하이퍼파라미터 값 목록
ridge_params = {
"max_iter": [3000],
"alpha": [0.1, 1, 2, 3, 4, 10, 30, 100, 200, 300, 400, 800, 900, 1000],
}
# 교차 검증용 평가 함수(RMSLE 계산)
rmsle_scorer = metrics.make_scorer(rmsle, greater_is_better=False)
# 그리드서치 객체 생성
gridsearch_ridge = GridSearchCV(
estimator=ridge, param_grid=ridge_params, scoring=rmsle_scorer, cv=5
) # 교차 검증 분할 수
# ### 그리드서치 수행
log_y = np.log(y)
gridsearch_ridge.fit(X_train, log_y)
print("최적 하이퍼파라미터 :", gridsearch_ridge.best_params_)
# ## 2-2. 성능 검증
preds_ridge = gridsearch_ridge.best_estimator_.predict(X_train)
res = rmsle(log_y, preds_ridge, True)
print(f"릿지 회귀 RMSLE 값: {res:.4f}")
# # 3. 성능 개선 : 라쏘 회귀 모델
from sklearn.linear_model import Lasso
lasso = Lasso()
lasso_alpha = 1 / np.array(
[0.1, 1, 2, 3, 4, 10, 30, 100, 200, 300, 400, 800, 900, 1000]
)
lasso_params = {"max_iter": [3000], "alpha": lasso_alpha}
gridsearch_lasso = GridSearchCV(
estimator=lasso, param_grid=lasso_params, scoring=rmsle_scorer, cv=5
)
gridsearch_lasso.fit(X_train, log_y)
gridsearch_lasso.best_params_
preds_lasso = gridsearch_lasso.best_estimator_.predict(X_train)
res = rmsle(preds_lasso, log_y)
print(f"라쏘 회귀 RMSLE 값: {res:.4f}")
# # 4. 성능 개선 : Random Forest
from sklearn.ensemble import RandomForestRegressor
randomforest = RandomForestRegressor()
rf_params = {"random_state": [42], "n_estimators": [100, 120, 140]}
gridsearch_rf = GridSearchCV(
estimator=randomforest, param_grid=rf_params, scoring=rmsle_scorer, cv=5
)
gridsearch_rf.fit(X_train, log_y)
gridsearch_rf.best_params_
preds_rf = gridsearch_rf.best_estimator_.predict(X_train)
res = rmsle(preds_rf, log_y)
print(f"랜덤포레스트 회귀 RMSLE 값: {res:.4f}")
# ### 예측 및 결과 제출
import seaborn as sns
import matplotlib.pyplot as plt
preds_rf_in_test = gridsearch_rf.best_estimator_.predict(X_test)
fig, axes = plt.subplots(ncols=2)
fig.set_size_inches(10, 4)
sns.histplot(y, bins=50, ax=axes[0])
axes[0].set_title("Train Data Distribution")
sns.histplot(np.exp(preds_rf_in_test), bins=50, ax=axes[1])
axes[1].set_title("Predicted Test Data Distribution")
## ; 붙이면 마지막 코드 실행결과 출력 제외
submission["count"] = np.exp(preds_rf_in_test)
submission.to_csv("submission_rf_model.csv", index=False)
|
import IPython.display as display
display.Audio("tts/python_cli_demo/output_1681192224957.mp3", autoplay=False)
|
# # Objective
# To explore and visualize the Cars4U dataset, build a linear regression model to predict the prices of used cars, and generate a set of insights and recommendations that will help the business.
# # Problem Definition
# How do we define a good predictive pricing model that can effectively predict the price of used cars and can help the business in devising profitable strategies using differential pricing? And, What does the performance assessment look like for such a model?
# # Key Questions
# * How many types of car brands and models are available?
# * How does the Price vary across different car brands ?
# * How does year of manufacture vary with the pricing of used cars?
# * High performance cars are good for certain types of customers. How many cars are available for different brands with 200 bhp and above?
# * What percentage of cars have a high bhp across different brands?
# * How does the New_Price vary with used car price across all brands? How do we determine depreciation?
# * How does the number of seats vary with price across different brands?
# According to https://www.monash.edu/, Differential pricing is a pricing strategy in which a company sets different prices for the same product on the basis of differing customer type, time of purchase, etc; also called Discriminatory Pricing, Flexible Pricing, Multiple Pricing, Variable Pricing.
# In the case of Cars4U, we will use a combination of dependent variables to come up with a pricing model for used cars and test it to ascertain that it can meet the differential pricing requirement.
# # Import all the necessary libraries
import warnings
warnings.filterwarnings("ignore")
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.linear_model import LinearRegression
from sklearn.metrics import (
mean_squared_error as mse,
mean_absolute_error as mae,
r2_score,
)
from sklearn.model_selection import train_test_split
import seaborn as sns
import datetime
# Removes the limit for the number of displayed columns
pd.set_option("display.max_columns", None)
# Sets the limit for the number of displayed rows
pd.set_option("display.max_rows", 200)
# Load the data set into a pandas dataframe and preview it
data = pd.read_csv("/kaggle/input/utaustinaimlprojects/Project2/used_cars_data.csv")
# # Data background and contents
# checking the shape of the data
print(f"There are {data.shape[0]} rows and {data.shape[1]} columns.")
# Data sample
data.sample(10, random_state=2)
# Looking at the data, we do not need S.no (Serial number) so we will drop it right away
data.drop(["S.No."], axis=1, inplace=True)
# reset the indexes
data.reset_index()
# checking column datatypes and number of non-null values
data.info()
# * We have 3 floats, 3 Integers and & columns of Object type
# Check for missing values
data.isnull().sum()
# We have missing values in the data on Engine, Power, Seats and Price columns
# Check for duplicated values
data.duplicated().sum()
data.loc[data.duplicated()]
# We have 1 duplicate in the data. Let us treat it accordingly
data = data.drop_duplicates()
# verify if duplicates dropped
data.duplicated().sum()
# Let's look at the statistical summary of the data
data.describe(include="all").T
# * There are 2041 different car brands in the data
# * We see that Mahindra XUV500 W8 2WD is the most common car
# * Majority of the cars are sort from Mumbai
# * The average year of manufacture is 2013 with the oldest car manufactured in 1996 and newest is 2019
# * The average kilometers driven is 58699 with min & max of 171 and 6500000 KMS. The max kilometers driven value seems like an outlier, we will investigate it
# * Most common type of fuel type is Diesel
# * Majority of the cars sold are of Manual transmission
# * Majority of the cars sold are new given that owner type is First meaning that most customers prefer new cars as 1st choice preference
# * The modal mileage is 17.0 kmpl.
# * The modal power is 74 bhp. We observe units in the column value hence the need to separate to obtain min,max std for this column
# * The maximum price for new cars is 375 INR while the minimum price use 3.91 INR Lakhs
# * The average price of used cars is 9.5 INR , minimum oprice is 0.44 INR Lakhs while max price is 160. The deviation from mean is high so this could be an outlier
# * We observe that 50% of all new cars cost 11 INR and above
# * We observe that 50% of all used cars cost 5.64 INR and above
# * Most of the cars are 5 seaters
# # Exploratory Data Analysis
# ## Univariate Analysis
# function to plot a boxplot and a histogram along the same scale.
# Credit : From Anime Rating Case study
def histogram_boxplot(data, feature, figsize=(12, 7), kde=False, bins=None, hue=None):
"""
Boxplot and histogram combined
data: dataframe
feature: dataframe column
figsize: size of figure (default (12,7))
kde: whether to the show density curve (default False)
bins: number of bins for histogram (default None)
"""
f2, (ax_box2, ax_hist2) = plt.subplots(
nrows=2, # Number of rows of the subplot grid= 2
sharex=True, # x-axis will be shared among all subplots
gridspec_kw={"height_ratios": (0.25, 0.75)},
figsize=figsize,
) # creating the 2 subplots
sns.boxplot(
data=data, x=feature, ax=ax_box2, showmeans=True, color="violet", hue=hue
) # boxplot will be created and a star will indicate the mean value of the column
sns.histplot(
data=data, x=feature, kde=kde, ax=ax_hist2, bins=bins, palette="winter", hue=hue
) if bins else sns.histplot(
data=data, x=feature, kde=kde, ax=ax_hist2
) # For histogram
ax_hist2.axvline(
data[feature].mean(), color="green", linestyle="--"
) # Add mean to the histogram
ax_hist2.axvline(
data[feature].median(), color="black", linestyle="-"
) # Add median to the histogram
# function to create labeled barplots
def labeled_barplot(data, feature, perc=False, n=None):
"""
Barplot with percentage at the top
data: dataframe
feature: dataframe column
perc: whether to display percentages instead of count (default is False)
n: displays the top n category levels (default is None, i.e., display all levels)
"""
total = len(data[feature]) # length of the column
count = data[feature].nunique()
if n is None:
plt.figure(figsize=(count + 1, 5))
else:
plt.figure(figsize=(n + 1, 5))
plt.xticks(rotation=90, fontsize=15)
ax = sns.countplot(
data=data,
x=feature,
palette="Paired",
order=data[feature].value_counts().index[:n].sort_values(),
)
for p in ax.patches:
if perc == True:
label = "{:.1f}%".format(
100 * p.get_height() / total
) # percentage of each class of the category
else:
label = p.get_height() # count of each level of the category
x = p.get_x() + p.get_width() / 2 # width of the plot
y = p.get_height() # height of the plot
ax.annotate(
label,
(x, y),
ha="center",
va="center",
size=12,
xytext=(0, 5),
textcoords="offset points",
) # annotate the percentage
plt.show() # show the plot
# ### Year
histogram_boxplot(data, "Year")
# * The distrubution for year is left Skewed
# * We observe outliers in this column
# * We can use Year to determing car age which will be more useful
# ### Kilometers_Driven
histogram_boxplot(data, "Kilometers_Driven", bins=10000)
# * The distribution for Kilometers_Driven is left skewed.
# * We have one outlier. Needs further investigation
# #### Investigating the outlier in kilometers driven
data.loc[data["Kilometers_Driven"] > 6000000]
# We conclude that it is not normal for a car to have driven 6.5 m kilomters. This outlier needs to be treated accordingly
# ### Seats
plt.figure(figsize=(10, 5))
sns.histplot(data=data, x="Seats")
plt.show()
# * The distribution for this column is randomly distributed. Bivariate analysis will help to get more insights
# ### Price
histogram_boxplot(data, "Price")
# * The distribution for this column skewed to the left
# * We also have many outliers for this column hence the need for proper treatment
# ### Location
labeled_barplot(data, "Location")
# * We observe that majority of the car locations are available at Mumbai, Hyderabad, Combatore, Koci and Pune
# ### Fuel Type
labeled_barplot(data, "Fuel_Type")
# * Diesel and Petrol Engine cars form the majority of the cars in the data-set
# ### Transmission
labeled_barplot(data, "Transmission")
# * We have 2 tyeps of car transmission in the dataset
# * Manual cars form the majority in the dataset
# ### Owner_Type
labeled_barplot(data, "Owner_Type")
# * First Owner Type form the majorty of cars in the data set.
# * Fourth & Above owner types are the least sold cars
# ## Bivariate Analysis
# ### Heat Map
plt.figure(figsize=(12, 7))
sns.heatmap(data.corr(), annot=True, vmin=-1, vmax=1, fmt=".2f", cmap="Spectral")
plt.show()
# ### Pair plot
plt.figure(figsize=(12, 7))
sns.pairplot(data=data, hue="Owner_Type")
plt.show()
# * We observe a strong correlation between the used car Price and the price of a new car
# * A negative correlation is observed between Price and Kilometers_Driven which is normal. Cars with more KMs tend to have a lower price
# * Price has a positive correlation with Year. Wich is normal because the newer the car the higher the price.
# * Price has a weak postitve corelation with Seats.
# ### Price Vs New Price vs Owner Type
plt.figure(figsize=(10, 5))
sns.scatterplot(data=data, x="Price", y="New_Price", hue="Owner_Type")
plt.show()
# ### Price Vs kilometers driven
plt.figure(figsize=(10, 5))
sns.scatterplot(data=data, x="Kilometers_Driven", y="Price", hue="Transmission")
plt.show()
#
# * We observe a strong correlation between new and old car pricers especially from first car owner types than other owner types.
# * Customers tend to prefer cars with lower kilometers driven and Manual transmission type
# * Prices for cars with manual transmission cars are rerelatively higher than prices for cars with automatic transmission
# * Automatic cars are more expensive than manual cars
# # Data Preprocessing
# Check the data
data.head()
# From the data we observe that Mileage, Engine and power have units. Let us strip the units and convert the columns to numericals and replace null values with na
# #### Remove units from Engine
# Striping units from Engine column
data["Engine"] = data["Engine"].str.rstrip(" CC")
# Replace null with nan
data["Engine"] = data["Engine"].replace(regex="null", value=np.nan)
# #### Remove units from power
# Striping units from Power column
data["Power"] = data["Power"].str.rstrip(" bhp")
# Replace null with nan
data["Power"] = data["Power"].replace(regex="null", value=np.nan)
# #### Remove units from Mileage
# Striping units from Mileage column and replace
data["Mileage"] = data["Mileage"].str.rstrip(" kmpl")
data["Mileage"] = data["Mileage"].str.rstrip(" km/kg")
data["Mileage"] = data["Mileage"].replace(regex="null", value=np.nan)
# Verify the data
data.sample(10)
# Convert to numerical columns
data["Mileage"] = data["Mileage"].astype(float)
data["Power"] = data["Power"].astype(float)
data["Engine"] = data["Engine"].astype(float)
# Verify data types
data.info()
# ## Outlier Treatment
# * We will investigate and treat outliers in Price , Kilometers Driven & Power columns
# Function to plot box & hist plots
def boxHistplot(data, feature):
plt.figure(figsize=(10, 5))
sns.histplot(data=data, x=feature)
plt.show()
plt.figure(figsize=(10, 5))
sns.boxplot(data=data, x=feature)
plt.show()
def densityPlot(data, feature):
plt.figure(figsize=(10, 5))
sns.distplot(data[feature], kde=True)
plt.show()
# ### Power Outlier Treatment
boxHistplot(data, "Power")
densityPlot(data, "Power")
# * The box plot flags many values as outliers but the histplot indicates left right skewness. These points seem consistent with the overall distribution
# Examinining sensitivity of the outliers by checking how many values are greater than 4*IQR
def outlierSensitivity(data, feature):
quartiles = np.quantile(data[feature][data[feature].notnull()], [0.25, 0.75])
power_iqr = 4 * (quartiles[1] - quartiles[0])
print(f"Q1 = {quartiles[0]}, Q3 = {quartiles[1]}, IQR = {power_iqr}")
outlier_powers = data.loc[
np.abs(data["Power"] - data[feature].median()) > power_iqr, feature
]
print(f"We have {outlier_powers.shape[0]} values on {feature} above IQR")
outlierSensitivity(data, "Power")
# * The distirbution is fairly normal with mild right skewness. All data points seem valid. We will ignore Power treatment for now
# ### Kilometers Driven Outlier Treatment
boxHistplot(data, "Kilometers_Driven")
densityPlot(data, "Kilometers_Driven")
# * We seem to have one extreme outlier in this column let us investigate
data[data["Kilometers_Driven"] > 1000000]
# * It is not normal for a car to have more than 6.5 m. We will replace this outlier with na and replace with median value
# Replace extreme outlier with nan
data.loc[2328, "Kilometers_Driven"] = np.nan
# Inpute with median
data["Kilometers_Driven"].fillna(
data["Kilometers_Driven"].median(), inplace=True
) # median imputation for Kilometers Drive
# * We still have some outliers in KM_Driven but distribution has improved.
# * We can use log transformation to further treat this column
# Log transformation
data["Kilometers_Driven" + "_log"] = np.log(data["Kilometers_Driven"])
# verify after transformation
boxHistplot(data, "Kilometers_Driven_log")
densityPlot(data, "Kilometers_Driven_log")
# ### Price Outlier Treatment
# Preview distribution
boxHistplot(data, "Price")
densityPlot(data, "Price")
# * We observe a number of columns, one extreme outlier with 160 needs further investigation
data.loc[data["Price"] > 120]
# * Price column has an outlier of 160. Since we have missing values on this column, we will log transform it later after missing value treatment
# ## Missing Value Treatment
data.isnull().sum()
# Since these are numerical values, we will inpute missing values with the mean, median accordingly
# ### Mileage Treatment
data["Mileage"].fillna(
data["Mileage"].median(), inplace=True
) # median imputation for Mileage
# ### Power Treatment
# Check if we have zero values in power and replace with nan for missing values treatment
data.query("Power == 0")
data["Power"].fillna(
data["Power"].median(), inplace=True
) # median imputation for power
# ### Engine Treatment
# Check if we have zero values in engine and replace with nan for missing values treatment
data.query("Engine == 0")
data["Engine"].fillna(
data["Engine"].median(), inplace=True
) # mean imputation for Engine
# ### Seats Treatment
# Check if we have zero values in seats and replace with nan for missing values treatment
data.query("Seats == 0")
# Replace with Nan
data.loc[3999, "Seats"] = np.nan
data["Seats"].fillna(data["Seats"].mean(), inplace=True) # mean imputation for seats
# ### Price Treatment
# Check if we have zero values in price and replace with nan for missing values treatment
data.query("Price == 0")
# * Since this is the target variable, we should drop all missing values
data = data.dropna(subset=["Price"])
# ### Verify Missing values result
# Verfy missing values
data.isnull().sum()
# ## Feature Engineering
# #### Year
# processing to determine age of car. We will use 2023 as current year
data["Year_Current"] = 2023
data["Car_Age"] = data["Year_Current"] - data["Year"]
# Drop Column Year_Current as we will no longer need it
data.drop("Year_Current", axis=1, inplace=True)
#### Analysis of Car Age
plt.figure(figsize=(10, 5))
sns.histplot(data=data, x="Car_Age")
plt.show()
# * Car_Age is right skewed. We can apply log transformation
data["Car_Age_log"] = np.log(data["Car_Age"])
plt.figure(figsize=(10, 5))
sns.histplot(data=data, x="Car_Age_log")
plt.show()
# #### Fuel Type
# Checking unique Fuel Types
data["Fuel_Type"].unique()
# We observe 4 fuel types in the dataset. No further action here
# #### Price
# Check if we have zero values
data.query("Price == 0")
# #### Name
def retrieve_brand(name):
brand = name.split(" ")[0]
return brand
def retrieve_model(name):
model = name.split(" ")[1]
return model
# Company brand affects the price of cars.
# * Since the Name combines both brand & model. We will strip and have brand/Model columns suing functions created earlier
data["Brand"] = data["Name"].apply(retrieve_brand)
data["Model"] = data["Name"].apply(retrieve_model)
# Preview Brands
data["Brand"].unique()
# Preview Models
data["Model"].unique()
# * We observe some names have spilled over to Model column like Land Rover where Land is in Brand and Rover is in model. Same case for mini cooper This needs to be corrected
# * We observe duplicate names for Isuzu in lower and upper case, probably a data entry error
# Check how many rows have ISUZU
data.loc[data.Brand == "ISUZU", "Brand"].shape
# Convert to lower case
data.loc[data.Brand == "ISUZU", "Brand"] = "Isuzu"
# Convert Land Brand Name to Land Rover and Mini to Mini Cooper
data.loc[data.Brand == "Mini", "Brand"] = "Mini_Cooper"
data.loc[data.Brand == "Land", "Brand"] = "Land_Rover"
# Verify the corrections
data.loc[(data.Brand == "Mini_Cooper") | (data.Brand == "Land_Rover"), "Brand"].sample(
10
)
# model & name columns
data.drop("Name", axis=1, inplace=True)
# Verify the data
data.sample(10)
## Drop Model Column
# #### Processing New Price
data["New_Price_log"] = np.log(data["New_Price"])
# #### Processing Price
data["Price_log"] = np.log(data["Price"])
# ## Answers to key questions
# ### Q. How many types of car brands and models are available?
print(
f"There are {data['Brand'].nunique()} and {data['Model'].nunique()} brands in the data set."
)
labeled_barplot(data, "Brand")
data.drop("Model", axis=1, inplace=True)
# Maruti, Hyundai cars have the highest number of cars in the dataset
# ### How does the Price vary across different car brands ?
# #### Analyzing with price for new Cars
data.groupby("Brand")["New_Price"].mean().sort_values(ascending=False)
# #### Analyzing with price for used Cars
data.groupby("Brand")["Price"].mean().sort_values(ascending=False)
plt.figure(figsize=(15, 5))
plt.subplot(1, 2, 1)
sns.barplot(data=data, y="New_Price", x="Brand")
plt.xticks(rotation=90)
plt.title("New Cars")
plt.subplot(1, 2, 2)
sns.barplot(data=data, y="Price", x="Brand")
plt.xticks(rotation=90)
plt.title("Used Cars")
plt.show()
# * Amongst new cars, Bently is the most expensive while lamborghini is the most expensive car brand amongst used cars
# * Maruti, Hyundai , Honda , audi, Nissan, Toyota, Volkswagen, Tata, Renault, Mahindra ,Ford, Datsun, Fiat, Hindustan, OpeCorsa , Isuzu, Force seem competitively priced for new and used cars
# ### How does year of manufacture vary with the pricing of used cars?
plt.figure(figsize=(15, 5))
plt.subplot(1, 2, 1)
sns.barplot(data=data, y="New_Price", x="Year")
plt.xticks(rotation=90)
plt.title("New Cars")
plt.subplot(1, 2, 2)
sns.barplot(data=data, y="Price", x="Year")
plt.xticks(rotation=90)
plt.title("Used Cars")
plt.show()
# Drop Year Column
data.drop("Year", axis=1, inplace=True)
# * The price of cars has remained unchanged for new cars manufactured over the period from 1996 to 2019
# * On the other hand, the price for used cars increases with age and is left skewed
# ### High performance cars are good for certain types of customers. How many cars are available for different brands with 200 bhp and above?
print(f"We have {data.loc[data['Power'] > 200].shape[0]} cars with more that 200bhp")
# ### What percentage of cars have a high bhp across different models/brands?
data_high = data.loc[data["Power"] > 200]
(data_high.Brand.value_counts() / data.Brand.value_counts()).sort_values(
ascending=False
)
# * Bentley, Lamborghini have the hightest bhp followed by Porsche, Jaguar, Mercedez
# ### How does the New_Price vary with used car price across all brands?
data_price_comparision = data.groupby(["Brand"])["New_Price", "Price"].mean()
data_price_comparision["Depreciation"] = (
(data_price_comparision.New_Price - data_price_comparision.Price)
/ data_price_comparision.New_Price
) * 100
data_price_comparision.sort_values(ascending=False, by="Depreciation")
# * Ambassador, Bentley, Smart, Chevrolet cars brand has the highest depreciation rate and lowest resale value
# * Jeep and Force cars have lowest depreciation and highest resale value
# * Lamborghini resale price is high
# ### How does the number of seats vary with the prices across different brands?
plt.figure(figsize=(15, 5))
plt.subplot(1, 2, 2)
sns.barplot(data=data, y="Price", x="Seats")
plt.xticks(rotation=90)
plt.title("Used Car Price Vs Seats")
plt.show()
# * 2 Seater cars are the most expensive
# * Seats 5.280494581828286 seem odd. We need to analyze and treat accoridngly
# #### Seats treatment for erroneous value
# Fetch all Seats with 5.2** and round to 5
indexes_seats = data.loc[data["Seats"] == 5.280494581828286].index
for index in indexes_seats:
data.loc[index, "Seats"] = 5
# Verify if seat value is treated
plt.figure(figsize=(15, 5))
plt.subplot(1, 2, 2)
sns.barplot(data=data, y="Price", x="Seats")
plt.xticks(rotation=90)
plt.title("Used Cars vs Seats")
plt.show()
# Sample check to confirm
data.sample(10)
## Recheck duplicates and drop them
data = data.drop_duplicates()
# #### Statistical summary after data processing
data.describe(include="all").T
# Review correlation between variables after data processing
plt.figure(figsize=(12, 7))
sns.heatmap(data.corr(), annot=True, vmin=-1, vmax=1, fmt=".2f", cmap="Spectral")
plt.show()
# * Average car age is approximately 10 years while the Minimum car age is 4 while max is 27 years
# * We observe that average mileage is 18 kmpl
# * The average engine capacity from the dataset is now visible at 1616 cc with maximum Engine Capcity at 5998
# * The average power is 113 bhp and maximum power is 616 bhp
# * The mean car age is approximately 10 years
# * We observe a strong correlation between the used car Price and Power
# ## Drop original transformed columns
data.info()
data.drop(["Kilometers_Driven", "Car_Age", "New_Price"], axis=1, inplace=True)
data.head()
# # Model Building
# create function to convert object columns to dummies
def convertObjectColumnsToDummies(X):
X = pd.get_dummies(
X,
columns=data.select_dtypes(include=["object"]).columns.tolist(),
drop_first=True,
)
## X= X.rename(columns={'Owner_Type_Fourth & Above' : 'Owner_Type_Fourth_above','Brand_Mercedes-Benz' :'Brand_Mercedez_Benz' },inplace=True)
return X
data.head()
# ## Define dependent and independent variables
X = data.drop(["Price", "Price_log"], axis=1)
# Let us create 2 target variables y & y2 for Price and Price_log respectively
y = data[["Price"]]
# ## Creating dummy variables
X = convertObjectColumnsToDummies(X)
# Verify Independent Variables
X
# ## Split Data into train and test for Target Variable - Price
X_Train, X_Test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=42
)
# ## Fit the linear Model
# Function to fit the model
def fitModel(train_x, train_y):
linear_regression_model = LinearRegression()
linear_regression_model.fit(train_x, train_y)
return linear_regression_model
X_Train
# Fit y
linear_regression_model = fitModel(X_Train, y_train)
# ## Checking Model Performance
# * We will use RMSE ,MAE , MAPE and R2 from sklearn library to assess performance
# Function to compute adjusted R2
def _adjustedRScore(independentVariables, dependentVariable, prediction):
r_squared = r2_score(dependentVariable, prediction)
n = independentVariables.shape[0]
k = independentVariables.shape[1]
return 1 - ((1 - r_squared) * (n - 1) / (n - k - 1))
# function to compute R2, RMSE, MAR for performance check of a regression model
def model_performance_regression(model, independentVariables, dependentVariable):
prediction = model.predict(independentVariables)
r_squared_score = r2_score(dependentVariable, prediction)
adjusted_rsquared_score = _adjustedRScore(
independentVariables, dependentVariable, prediction
)
root_mean_squared_error = np.sqrt(mse(dependentVariable, prediction))
mean_absolute_error = mae(dependentVariable, prediction)
mean_absolute_percentage_error = mean_absolute_percentage_error_score(
dependentVariable, prediction
)
# create metrics DF
data_frame_performance = pd.DataFrame(
{
"RMSE": root_mean_squared_error,
"MAE": mean_absolute_error,
"R-squared": r_squared_score,
"Adj. R-squared": adjusted_rsquared_score,
"Mean Absolute Error %": mean_absolute_percentage_error,
},
index=[0],
)
return data_frame_performance
# function to compute Mean Absolute Error %
def mean_absolute_percentage_error_score(dependentVariable, prediction):
return (
np.mean(np.abs(dependentVariable - prediction) / dependentVariable, axis=0)
* 100
)
# ### Model Performance on Training Data for first Target Variable y -Price
print("Test Performance on Training Data - y \n")
regression_model_training_performance = model_performance_regression(
linear_regression_model, X_Train, y_train
)
regression_model_training_performance
# ### Model Performance on Test Data for first target variable y -Price
print("Test Performance on Test Data y\n")
regression_model_test_performance = model_performance_regression(
linear_regression_model, X_Test, y_test
)
regression_model_test_performance
|
# importing libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
# reading dataset
df = pd.read_csv("/kaggle/input/salary-prediction/Salary_Data.csv")
df.head(10)
# checking for info
df.info()
# statical summery
df.describe()
# DATA visualization
plt.scatter(x="YearsExperience", y="Salary", data=df)
plt.grid()
# histogram ploatation of years and salary
viz = df[["YearsExperience", "Salary"]]
viz.hist()
plt.show()
# feature converting into array
X = df["YearsExperience"]
y = df["Salary"]
X = np.array(X)
y = np.array(y)
X = X.reshape(-1, 1)
print(X.shape)
y = y.reshape(-1, 1)
print(y.shape)
# Spliting dataset
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
print(X_train.shape)
print(X_test.shape)
print(y_train.shape)
print(y_test.shape)
# traning the model
from sklearn.linear_model import LinearRegression
regr = LinearRegression(fit_intercept=True)
regr.fit(X_train, y_train)
print("linear model coefficient(w):", regr.coef_)
print("linear model intercept(b):", regr.intercept_)
# traning the model
from sklearn.linear_model import LinearRegression
regr = LinearRegression(fit_intercept=True)
regr.fit(X_train, y_train)
print("linear model coefficient(w):", regr.coef_)
print("linear model intercept(b):", regr.intercept_)
# testing model
plt.scatter(X_test, y_test, color="blue")
y_pred = regr.predict(X_test)
plt.plot(X_test, y_pred, color="r")
plt.xlabel("YearsExperience")
plt.ylabel("Salary(Rs.)")
plt.title("YearsExperience vs Salary")
plt.grid()
from sklearn.metrics import r2_score
print("mean absolute error:%.2f" % np.mean(np.absolute(y_pred - y_test)))
print("residual sum of squre(mse):%.2f" % np.mean(y_pred - y_test) ** 2)
print("r2 score:%.2f" % r2_score(y_pred, y_test))
# using trained model to predict the salary(here we can cutmize the year and predict the salary)
temp = np.array([4])
temp = temp.reshape(-1, 1)
salary = regr.predict(temp)
print("salary prediction=Rs", salary)
|
# # Data Visualization with Pandas Basics
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
plt.style.use("fivethirtyeight")
# ## Plotting graph with plot() method
data = pd.Series(np.random.randn(1000).cumsum())
data.plot()
df1 = pd.DataFrame(np.random.randn(100, 4), columns=list("ABCD"))
df1 = df1.cumsum()
df1.plot()
# ## Bar Charts
df1.iloc[10].plot(kind="bar")
df1.iloc[10].plot.bar()
df2 = pd.DataFrame(np.random.rand(7, 3), columns=list("ABC"))
df2.plot.bar()
df2.plot.bar(stacked=True)
df2.plot.barh(stacked=True)
# ## Histograms
iris = pd.read_csv("iris.data", header=None)
iris.columns = ["sepal_length", "sepal_width", "petal_length", "petal_width", "species"]
# You can access this data set [here](https://github.com/tirendazacademy/data-visualization-with-python).
iris.head()
iris.plot.hist(alpha=0.7)
iris.plot.hist(alpha=1, stacked=True)
bins = 25
iris.plot.hist(alpha=1, stacked=True, bins=25)
iris["sepal_width"].plot.hist(orientation="horizontal")
iris["sepal_length"].diff().hist()
iris.hist(color="blue", alpha=1, bins=20)
iris.hist("petal_length", by="species")
# ## Boxplot charts
iris.plot.box()
colors = {"boxes": "Red", "whiskers": "blue", "medians": "Black", "caps": "Green"}
iris.plot.box(color=colors)
iris.plot.box(vert=False)
iris.boxplot()
plt.rcParams["figure.figsize"] = (8, 8)
plt.style.use("ggplot")
iris.boxplot(by="species")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
data = pd.read_csv(
"/kaggle/input/nse-tickers-their-yahoo-finance-equivalent-codes/EQUITY_L.csv",
index_col=" DATE OF LISTING",
)
data
# # Changing the header
data1 = pd.read_csv(
"/kaggle/input/nse-tickers-their-yahoo-finance-equivalent-codes/EQUITY_L.csv",
header=1,
)
data1
data.columns
data2 = pd.read_csv(
"/kaggle/input/nse-tickers-their-yahoo-finance-equivalent-codes/EQUITY_L.csv",
usecols=["SYMBOL", "NAME OF COMPANY", " SERIES", " PAID UP VALUE"],
)
data2
data.index.name = "test"
data
# # Dealing with the messy csv data
df = pd.read_csv("../input/meesy-csv/EQUITY_L.csv")
df.head()
df.tail()
df.info()
# # 1st Approach
# * Setting header as index 1
# * Dropping the last row
# resetting the header of the data to index 1
df = pd.read_csv("../input/meesy-csv/EQUITY_L.csv", header=1)
df
# As I can see the last line is also not corrupted, So we are dropping the last line of the data
df.drop(index=1665, axis=0, inplace=True)
df.info()
df.isnull().sum()
df.columns
# # 2nd Approach
# * Performing all operations while reading the Data
# * Skipping 2 top rows
# * Skipping 1 row from footer
# * Setting header to None
# * Setting up column names manually
col_names = [
"SYMBOL",
"NAME OF COMPANY",
"SERIES",
"DATE OF LISTING",
"PAID UP VALUE",
"MARKET LOT",
"ISIN NUMBER",
"FACE VALUE",
"Unnamed: 8",
"YahooEquiv",
"Yahoo_Equivalent_Code",
]
df2 = pd.read_csv(
"../input/meesy-csv/EQUITY_L.csv",
skiprows=2,
skipfooter=1,
header=None,
names=col_names,
)
df2.columns
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
from googleapiclient.discovery import build
import pandas as pd
import googleapiclient.discovery
from IPython.display import JSON
api_key = "AIzaSyB3mL5Uso-mhCWW5IPLQdB4mKq-vzOISsE"
api_service_name = "youtube"
api_version = "v3"
# Get credentials and create an API client
youtube = googleapiclient.discovery.build(
api_service_name, api_version, developerKey=api_key
)
request = youtube.channels().list(
part="snippet,contentDetails,statistics", id="UCNU_lfiiWBdtULKOw6X0Dig"
)
response = request.execute()
JSON(response)
# I Will fetch all id of all videos
request = youtube.playlistItems().list(
part="snippet,contentDetails", playlistId="UUNU_lfiiWBdtULKOw6X0Dig"
)
response = request.execute()
JSON(response)
# # Function to fetch vido IDs
# function to fetch video ids
playlist_id = "UUNU_lfiiWBdtULKOw6X0Dig"
def get_video_id(youtube, playlist_id):
video_ids = []
request = youtube.playlistItems().list(
part="snippet,contentDetails", playlistId=playlist_id, maxResults=50
)
response = request.execute()
for i in response["items"]:
video_ids.append(i["contentDetails"]["videoId"])
# as we can not access more than 50 video ids so we will use next page token
next_page_token = response.get("nextPageToken")
more_pages = True
while more_pages:
if next_page_token is None:
more_pages = False
else:
request = youtube.playlistItems().list(
part="snippet,contentDetails",
playlistId=playlist_id,
maxResults=50,
pageToken=next_page_token,
)
response = request.execute()
for i in response["items"]:
video_ids.append(i["contentDetails"]["videoId"])
next_page_token = response.get("nextPageToken")
return video_ids
playlist_id = "UUNU_lfiiWBdtULKOw6X0Dig"
video_ids = get_video_id(youtube, playlist_id)
# qMLxWX49i8I
# # Function to fetch video details
for i in video_ids[750:800]:
request = youtube.videos().list(
part=" snippet,contentDetails,statistics , liveStreamingDetails",
id=video_ids[751],
)
response = request.execute()
print(response["items"]["statistics"]["commentCount"])
def get_video_details(youtube, video_id):
all_video_stats = []
count = 0
# for i in range(0,len(video_id),50):
request = youtube.videos().list(
part=" snippet,contentDetails,statistics , liveStreamingDetails",
id=",".join(video_id[750:800]),
)
response = request.execute()
print(i, end=" ")
for video in response["items"]:
video_stats = {
"Title": video["snippet"]["title"],
"Publish_Date": video["snippet"]["publishedAt"],
"Views": video["statistics"]["viewCount"],
"Likes": video["statistics"]["likeCount"],
"Comments": video["statistics"]["commentCount"],
}
all_video_stats.append(video_stats)
return all_video_stats
video_ids[751]
print(get_video_details(youtube, video_ids))
# video_ids =
video = ["H99JRtDDnvk"]
count = 0
for i in video_ids:
get_video_details(youtube, video)
count = count + 1
print(count)
# Copied Code
def get_video_details(youtube, video_ids):
all_video_info = []
for i in range(0, len(video_ids), 50):
request = youtube.videos().list(
part="snippet,contentDetails,statistics", id=",".join(video_ids[i : i + 50])
)
response = request.execute()
for video in response["items"]:
stats_to_keep = {
"snippet": ["title", "description", "tags", "publishedAt"],
"statistics": ["viewCount", "likeCount", "commentCount"],
"contentDetails": ["duration"],
}
video_info = {}
video_info["video_id"] = video["id"]
for k in stats_to_keep.keys():
for v in stats_to_keep[k]:
try:
video_info[v] = video[k][v]
except:
video_info[v] = None
all_video_info.append(video_info)
return pd.DataFrame(all_video_info)
video_df = get_video_details(youtube, video_ids)
video_df.head()
video_df.to_csv("krish.csv", index=False)
|
import pandas as pd
from sklearn.preprocessing import MinMaxScaler
from keras.models import Sequential
from keras.layers import Dense, Dropout, BatchNormalization, Activation
from sklearn.model_selection import train_test_split
train = pd.read_csv("train.csv")
test = pd.read_csv("test.csv")
sub = pd.read_csv("sample_submission.csv")
train.head()
test.head()
sub.head()
df = pd.concat([train, test], ignore_index=True)
df.tail()
df["wheezy-copper-turtle-magic"].max()
# # Normalize data using MinMaxScaler
print(train.shape)
print(test.shape)
train1 = df.drop(["id", "target"], axis=1)[df.index < 262144]
y = df["target"][df.index < 262144]
test1 = df.drop(["id", "target"], axis=1)[df.index > 262143]
scaler = MinMaxScaler()
x_train = scaler.fit_transform(train1)
test1 = scaler.fit_transform(test1)
x_train, x_test, y_train, y_test = train_test_split(
x_train, y, test_size=0.2, random_state=42
)
model = Sequential()
model.add(Dense(1024, input_dim=256))
model.add(BatchNormalization())
model.add(Activation("relu"))
model.add(Dropout(0.5))
model.add(Dense(256))
model.add(BatchNormalization())
model.add(Activation("relu"))
model.add(Dropout(0.5))
model.add(Dense(1, activation="sigmoid"))
model.compile(optimizer="adam", loss="binary_crossentropy", metrics=["binary_accuracy"])
model.summary()
history = model.fit(
x_train,
y_train,
validation_data=(x_test, y_test),
epochs=30,
verbose=1,
batch_size=128,
)
tahmin = model.predict(test1)
tahmin
sub.head()
sonuc = pd.DataFrame()
sonuc["id"] = test["id"]
sonuc["target"] = tahmin
sonuc.to_csv("sonuc.csv", index=False)
|
import numpy as np
from tensorflow import keras
from tensorflow.keras import layers
import tensorflow as tf
import re
import pandas as pd
from sklearn.model_selection import KFold
AUTO = tf.data.experimental.AUTOTUNE
folds = 5
image_size = 128 # We'll resize input images to this size
input_shape = (image_size, image_size, 3)
TEST_FILENAMES = tf.io.gfile.glob(
"../input/ranzcr-clip-catheter-line-classification/test_tfrecords/*.tfrec"
) # predictions on this dataset should be submitted for the competition
print(TEST_FILENAMES)
batch_size = 4
import sys
package_path = "../input/vitkeras/"
sys.path.append(package_path)
# test
#!pip install vit-keras
from vit_keras import vit, utils
UNLABELED_TFREC_FORMAT = {
"image": tf.io.FixedLenFeature([], tf.string),
"StudyInstanceUID": tf.io.FixedLenFeature([], tf.string),
}
def decode_image(image_data):
image = tf.image.decode_jpeg(image_data, channels=3)
image = (
tf.cast(image, tf.float32) / 255.0
) # convert image to floats in [0, 1] range
image = tf.image.resize(image, [image_size, image_size])
image = tf.reshape(image, [image_size, image_size, 3])
return image
def read_unlabeled_tfrecord(example):
example = tf.io.parse_single_example(example, UNLABELED_TFREC_FORMAT)
image = decode_image(example["image"])
idnum = example["StudyInstanceUID"]
return image, idnum # returns a dataset of image(s)
def load_dataset(filenames, labeled=True, ordered=False):
ignore_order = tf.data.Options()
if not ordered:
ignore_order.experimental_deterministic = False # disable order, increase speed
dataset = tf.data.TFRecordDataset(
filenames, num_parallel_reads=AUTO
) # automatically interleaves reads from multiple files
dataset = dataset.with_options(
ignore_order
) # use data as soon as it streams in, rather than in its original order
dataset = dataset.map(
read_unlabeled_tfrecord, num_parallel_calls=AUTO
) # returns a dataset of (image, label) pairs if labeled = True or (image, id) pair if labeld = False
return dataset
def get_test_dataset(ordered=False):
dataset = load_dataset(TEST_FILENAMES, labeled=False, ordered=ordered)
dataset = dataset.batch(batch_size)
dataset = dataset.prefetch(
AUTO
) # prefetch next batch while training (autotune prefetch buffer size)
return dataset
def count_data_items(filenames):
# the number of data items is written in the name of the .tfrec files, i.e. flowers00-230.tfrec = 230 data items
n = [
int(re.compile(r"-([0-9]*)\.").search(filename).group(1))
for filename in filenames
]
return np.sum(n)
NUM_TEST_IMAGES = count_data_items(TEST_FILENAMES)
def test():
transformer = vit.vit_l32(
image_size=image_size,
pretrained=False,
include_top=False,
pretrained_top=False,
weights=f"../input/vitweight/fold0vit.h5",
)
model = tf.keras.Sequential(
[transformer, tf.keras.layers.Dense(11, activation="sigmoid")]
)
print(model)
models1 = []
models1.append(model)
test_ds = get_test_dataset(ordered=True)
test_images_ds = test_ds.map(lambda image, idnum: image)
labels = [
"ETT - Abnormal",
"ETT - Borderline",
"ETT - Normal",
"NGT - Abnormal",
"NGT - Borderline",
"NGT - Incompletely Imaged",
"NGT - Normal",
"CVC - Abnormal",
"CVC - Borderline",
"CVC - Normal",
"Swan Ganz Catheter Present",
]
mean = np.average(
[models1[i].predict(test_images_ds) for i in range(len(models1))], axis=0
)
test_ids_ds = test_ds.map(lambda image, idnum: idnum).unbatch()
test_ids = next(iter(test_ids_ds.batch(NUM_TEST_IMAGES))).numpy().astype("U")
submission = pd.DataFrame(mean, columns=labels)
submission.insert(0, "StudyInstanceUID", test_ids, False)
submission["StudyInstanceUID"] = submission["StudyInstanceUID"].apply(
lambda x: x.rstrip(".jpg")
)
submission.to_csv("submission.csv", index=False)
test()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
print(dirname)
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
with open("/kaggle/input/flickr8k/captions.txt") as f:
lines = f.readlines()
for l in lines[0:10]:
print(l)
from PIL import Image
img = Image.open("/kaggle/input/flickr8k/Images/1000268201_693b08cb0e.jpg")
print(img.size)
display(img)
with open("/kaggle/input/flickr8k/captions.txt") as f:
lines = f.readlines()
for l in lines:
l = l.split(",")
# print(l[0])
if l[0] == "1000268201_693b08cb0e.jpg":
print(l[1])
data = pd.read_csv("../input/flickr8k/captions.txt")
data.head()
import torch.nn as nn
criterion = nn.TripletMarginLoss(margin=0.1)
# **image data**
import os
import random
import shutil
import pandas as pd
from PIL import Image
import torchvision.transforms as transforms
# Set the number of images to select
num_images = 100
# Get the unique image paths and their descriptions
image_paths_and_desc = data.iloc[:, :2].drop_duplicates(subset="image")
# Shuffle the image paths and select the first num_images
selected_images = image_paths_and_desc.sample(n=num_images, random_state=1)
# Create the output directory if it doesn't exist
output_dir = "train_images"
if not os.path.exists(output_dir):
os.makedirs(output_dir)
# Copy the selected images to the output directory and save their descriptions
selected_images_with_desc = []
p = []
d = []
l = []
i = 0
new_rows = []
for _, row in selected_images.iterrows():
image_path = row["image"]
description = row["caption"]
label = i
p.append(image_path)
d.append(description)
l.append(i)
filename = os.path.basename(image_path)
src_path = os.path.join(
"/kaggle/input/flickr8k/Images", image_path
) # Replace with your path to the images
dst_path = os.path.join(output_dir, filename)
shutil.copy(src_path, dst_path)
# Load the image
img = Image.open(os.path.join(output_dir, os.path.basename(image_path)))
# Generate new images by cropping, rotating, etc.
# Replace this with your own image augmentation code
for j in range(3):
crop_transform = transforms.RandomCrop(size=224)
cropped_img = crop_transform(img)
# Save the new image to disk
new_filename = f"{os.path.splitext(os.path.basename(image_path))[0]}_{j}.jpg"
new_path = os.path.join(output_dir, new_filename)
cropped_img.save(new_path)
# Add the new image to the DataFrame
new_rows.append({"path": new_filename, "caption": description, "label": label})
rotation_transform = transforms.RandomRotation(degrees=30)
rotated_image = rotation_transform(img)
# Save the new image to disk
new_filename = f"{os.path.splitext(os.path.basename(image_path))[0]}_{j}_.jpg"
new_path = os.path.join(output_dir, new_filename)
rotated_image.save(new_path)
# Add the new image to the DataFrame
new_rows.append({"path": new_filename, "caption": description, "label": label})
i += 1
da = {"path": p, "caption": d, "label": l}
df = pd.DataFrame(da)
df = df.append(new_rows, ignore_index=True)
# Save the updated DataFrame to the 'train.csv' file
df.to_csv("train.csv", index=False)
train = pd.read_csv("/kaggle/working/train.csv")
train.head()
train.shape, train["label"].nunique(), train["label"].unique()
from PIL import Image
from IPython.display import display
import pandas as pd
# load the CSV file into a pandas dataframe
train_df = pd.read_csv("/kaggle/working/train.csv")
from tensorflow.keras.preprocessing.image import (
ImageDataGenerator,
load_img,
img_to_array,
)
import matplotlib.pyplot as plt
from textwrap import wrap
def readImage(path, img_size=224):
img = load_img(path, color_mode="rgb", target_size=(img_size, img_size))
img = img_to_array(img)
img = img / 255.0
return img
def display_images(temp_df):
temp_df = temp_df.reset_index(drop=True)
plt.figure(figsize=(20, 20))
n = 0
for i in range(15):
n += 1
plt.subplot(5, 5, n)
plt.subplots_adjust(hspace=0.7, wspace=0.3)
image = readImage(f"/kaggle/working/train_images/{temp_df.path[i]}")
plt.imshow(image)
a = str(temp_df.label[i]) + "\n"
plt.title(a.join(wrap(temp_df.caption[i], 20)))
plt.axis("off")
display_images(train.sample(15))
# anchor = self.CNN(img_anchor)
# positive = self.MLP(att_positive)
# negative = self.MLP(att_negative)
# loss = criterion(anchor, anchor, negative)
# optimizer.zero_grad()
# loss.backward()
# optimizer.step()
# **IMAGE PREPROCESSING**
import pandas as pd
import numpy as np
from tensorflow.keras.preprocessing.image import load_img, img_to_array
def load_data(csv_path, img_size=224):
# Read in the CSV file
data = pd.read_csv(csv_path)
# Get the image paths and labels
dirs = "/kaggle/working/train_images/"
image_paths = data["path"].tolist()
labels = data["label"].tolist()
# Initialize empty arrays to hold the preprocessed image data and labels
num_samples = len((image_paths))
X = np.zeros((num_samples, img_size, img_size, 3))
y = np.array(labels)
# Loop over the image paths and load in the images
for i, image_path in enumerate(image_paths):
# Load in the image
path = os.path.join(dirs, image_path)
img = load_img(path, color_mode="rgb", target_size=(img_size, img_size))
img = img_to_array(img)
img = img / 255.0
# Add the preprocessed image and label to the arrays
X[i] = img
return X, y
X, y = load_data("train.csv", img_size=224)
from sklearn.utils import shuffle
# Shuffle the data
X, y = shuffle(X, y, random_state=1)
X.shape, y.shape
# Split X and y into train and test sets
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=1)
# Print the shapes of the resulting train and test sets
print("X_train shape:", X_train.shape)
print("y_train shape:", y_train.shape)
print("X_test shape:", X_test.shape)
print("y_test shape:", y_test.shape)
# **POSITIVE AND NEGATIVE IMAGE PAIR**
def make_pairs(images, labels):
pairImages = []
pairLabels = []
nc = len(np.unique(labels))
print(nc)
idx = np.concatenate([np.where(labels == i)[0] for i in range(nc)])
print(len(idx))
for o in range(len(images)):
currentimage = images[o]
label = labels[o]
# Get a boolean array indicating which elements in label are equal to i
mask = labels == label
# Get the indices of the images corresponding to class i
idx = np.where(mask)[0]
# Randomly select an index from idx
rimg = np.random.choice(idx)
posImage = images[rimg]
pairImages.append([currentimage, posImage])
pairLabels.append([1])
negIdx = np.where(labels != label)[0]
negImage = images[np.random.choice(negIdx)]
pairImages.append([currentimage, negImage])
pairLabels.append([0])
return (np.array(pairImages), np.array(pairLabels))
# prepare the positive and negative pairs
print("[INFO] preparing positive and negative pairs...")
(pairTrain, labelTrain) = make_pairs(X_train, y_train)
(pairTest, labelTest) = make_pairs(X_test, y_test)
pairTrain.shape, labelTrain.shape
# **MODEL BUILDING**
import torch
import torch.nn as nn
import torch.optim as optim
from torch.utils.data import DataLoader
from torchvision import transforms
from torchvision.datasets import ImageFolder
from PIL import Image
# **SIAMESE**
class Siamese(nn.Module):
def __init__(self, inputShape, embeddingDim=300):
super(Siamese, self).__init__()
self.conv = nn.Sequential(
nn.Conv2d(3, 64, kernel_size=10),
nn.ReLU(inplace=True),
nn.MaxPool2d(kernel_size=2),
nn.Conv2d(64, 128, kernel_size=7),
nn.ReLU(inplace=True),
nn.MaxPool2d(kernel_size=2),
nn.Conv2d(128, 128, kernel_size=4),
nn.ReLU(inplace=True),
nn.MaxPool2d(kernel_size=2),
nn.Conv2d(128, 256, kernel_size=4),
nn.ReLU(inplace=True),
)
self.fc = nn.Sequential(
nn.Linear(256, 1024),
# nn.Sigmoid()
nn.ReLU(inplace=True),
nn.Linear(1024, 256),
nn.ReLU(inplace=True),
nn.Linear(256, 2),
)
def forward_once(self, x):
output = self.conv(x)
output = output.reshape(output.size(0), -1)
print(output.size())
output = self.fc(output)
return output
def forward(self, input1, input2):
# In this function we pass in both images and obtain both vectors
# which are returned
output1 = self.forward_once(input1)
output2 = self.forward_once(input2)
return output1, output2
# **CONTRASTIVE LOSS**
import torch
import torch.nn as nn
import torch.nn.functional as F
# Define the Contrastive Loss Function
class ContrastiveLoss(torch.nn.Module):
def __init__(self, margin=2.0):
super(ContrastiveLoss, self).__init__()
self.margin = margin
def forward(self, output1, output2, label):
# Calculate the euclidean distance and calculate the contrastive loss
euclidean_distance = F.pairwise_distance(output1, output2, keepdim=True)
loss_contrastive = torch.mean(
(1 - label) * torch.pow(euclidean_distance, 2)
+ (label)
* torch.pow(torch.clamp(self.margin - euclidean_distance, min=0.0), 2)
)
return loss_contrastive
net = Siamese(224).cuda()
criterion = ContrastiveLoss()
optimizer = optim.Adam(net.parameters(), lr=0.0001)
# **TRAINING**
counter = []
loss_history = []
i = 0
batch_size = 32
# Iterate throught the epochs
for epoch in range(100):
# Iterate over batches
for j in range(0, len(pairTrain), batch_size):
batch_pairTrain = pairTrain[j : j + batch_size]
batch_labelTrain = labelTrain[j : j + batch_size]
# Send the images and labels to CUDA
img0, img1, label = (
torch.tensor(batch_pairTrain[:, 0]),
torch.tensor(batch_pairTrain[:, 1]),
torch.tensor(batch_labelTrain),
)
img0 = img0.float()
img1 = img1.float()
img0, img1, label = img0.cuda(), img1.cuda(), label.cuda()
# Zero the gradients
optimizer.zero_grad()
# Pass in the two images into the network and obtain two outputs
img0 = img0.permute(0, 3, 1, 2)
img1 = img1.permute(0, 3, 1, 2)
output1, output2 = net(img0, img1)
# Pass the outputs of the networks and label into the loss function
loss_contrastive = criterion(output1, output2, label)
# Calculate the backpropagation
loss_contrastive.backward()
# Optimize
optimizer.step()
# Every 10 batches print out the loss
if i % 10 == 0:
print(f"Epoch number {epoch}\n Current loss {loss_contrastive.item()}\n")
i += 10
counter.append(i)
loss_history.append(loss_contrastive.item())
from tensorflow.keras.models import Model
from tensorflow.keras.layers import Input
from tensorflow.keras.layers import Conv2D
from tensorflow.keras.layers import Dense
from tensorflow.keras.layers import Dropout
from tensorflow.keras.layers import GlobalAveragePooling2D
from tensorflow.keras.layers import MaxPooling2D
def build_siamese_model(inputShape, embeddingDim=300):
inputs = Input(inputShape)
x = Conv2D(64, (2, 2), padding="same", activation="relu")(inputs)
x = MaxPooling2D(pool_size=(2, 2))(x)
x = Dropout(0.3)(x)
x = Conv2D(64, (2, 2), padding="same", activation="relu")(x)
x = MaxPooling2D(pool_size=2)(x)
x = Dropout(0.3)(x)
x = Conv2D(64, (3, 3), padding="same", activation="relu")(x)
x = MaxPooling2D(pool_size=2)(x)
x = Dropout(0.2)(x)
pooledOutput = GlobalAveragePooling2D()(x)
outputs = Dense(embeddingDim)(pooledOutput)
model = Model(inputs, outputs)
return model
import tensorflow.keras.backend as K
import tensorflow as tf
def contrastive_loss(y, preds, margin=1):
y = tf.cast(y, preds.dtype)
squaredPreds = K.square(preds)
squaredMargin = K.square(K.maximum(margin - preds, 0))
loss = K.mean(y * squaredPreds + (1 - y) * squaredMargin)
return loss
from tensorflow.keras.models import Model
from tensorflow.keras.layers import Dense
from tensorflow.keras.layers import Input
from tensorflow.keras.layers import Lambda
def euclidean_distance(vectors):
(featsA, featsB) = vectors
sumSquared = K.sum(K.square(featsA - featsB), axis=1, keepdims=True)
return K.sqrt(K.maximum(sumSquared, K.epsilon()))
print("*********building siamese network************)")
imgA = Input(shape=(224, 224, 3))
imgB = Input(shape=(224, 224, 3))
featureExtractor = build_siamese_model(inputShape=(224, 224, 3))
featsA = featureExtractor(imgA)
featsB = featureExtractor(imgB)
# finally, construct the siamese network
distance = Lambda(euclidean_distance)([featsA, featsB])
model = Model(inputs=[imgA, imgB], outputs=distance)
# compile the model
print("[INFO] compiling model...")
model.compile(loss=contrastive_loss, optimizer="adam")
# train the model
print("[INFO] training model...")
history = model.fit(
[pairTrain[:, 0], pairTrain[:, 1]],
labelTrain[:],
validation_data=([pairTest[:, 0], pairTest[:, 1]], labelTest[:]),
batch_size=32,
epochs=100,
)
import matplotlib.pyplot as plt
# plot loss curve
plt.plot(history.history["loss"], label="train_loss")
plt.plot(history.history["val_loss"], label="val_loss")
plt.title("Loss Curve")
plt.xlabel("Epoch")
plt.ylabel("Loss")
plt.legend()
# save plot
plt.savefig("loss_curve.png")
model.save("siamese_model.h5")
from keras.models import load_model
# load the saved model
model = load_model(
"siamese_model.h5", custom_objects={"contrastive_loss": contrastive_loss}
)
# evaluate the model on the test dataset
test_loss = model.evaluate(
[pairTest[:, 0], pairTest[:, 1]], labelTest[:], batch_size=32
)
print("Test loss:", test_loss)
import numpy as np
import cv2
from tensorflow.keras.models import load_model
# Load the trained siamese model
model = load_model(
"siamese_model.h5", custom_objects={"contrastive_loss": contrastive_loss}
)
# Load the trained siamese model
model = load_model(
"siamese_model.h5",
custom_objects={
"contrastive_loss": contrastive_loss,
"euclidean_distance": euclidean_distance,
},
)
embedding_model = model.layers[2]
embedding_model = Model(
inputs=embedding_model.input, outputs=embedding_model.layers[-2].output
)
# Compute the embeddings of all the retrieval set images and save them to disk
embeddings = []
filenames = []
for img_file in train["path"]:
img_file = os.path.join("/kaggle/working/train_images/", img_file)
img = load_img(img_file, color_mode="rgb", target_size=(224, 224))
img = img_to_array(img)
img = img / 255.0
emb = embedding_model.predict(np.expand_dims(img, axis=0))[0]
embeddings.append(emb)
filenames.append(img_file)
np.savez("retrieval_set_embeddings.npz", embeddings=embeddings, filenames=filenames)
t = pd.read_csv("/kaggle/working/train.csv")
t.head()
# compute the euclidean distance between two vectors a and b
def euclidean_distance(a, b):
return np.linalg.norm(a - b)
# Compute the embedding of a query (attributes vector)
path = os.path.join("/kaggle/working/train_images/", "2560278143_aa5110aa37.jpg")
query_img = load_img(path, color_mode="rgb", target_size=(224, 224))
query_img = img_to_array(img)
query_img = img / 255.0
query_emb = embedding_model.predict(np.expand_dims(query_img, axis=0))[0]
# Compute the distances between the query embedding and all the retrieval set embeddings
retrieval_set_embeddings = np.load("retrieval_set_embeddings.npz")
retrieval_set_embs = retrieval_set_embeddings["embeddings"]
retrieval_set_filenames = retrieval_set_embeddings["filenames"]
distances = [euclidean_distance(query_emb, emb) for emb in retrieval_set_embs]
sorted_indexes = np.argsort(distances)
# Get the closest ones
k = 10
closest_filenames = retrieval_set_filenames[sorted_indexes][:k]
print(closest_filenames)
def retrived(query):
# get the indices that would sort the distances array in ascending order
path = os.path.join(query)
img = load_img(path, color_mode="rgb", target_size=(224, 224))
img = img_to_array(img)
query_img = img / 255.0
query_emb = embedding_model.predict(np.expand_dims(query_img, axis=0))[0]
# Compute the distances between the query embedding and all the retrieval set embeddings
retrieval_set_embeddings = np.load("retrieval_set_embeddings.npz")
retrieval_set_embs = retrieval_set_embeddings["embeddings"]
retrieval_set_filenames = retrieval_set_embeddings["filenames"]
distances = [euclidean_distance(query_emb, emb) for emb in retrieval_set_embs]
sorted_indexes = np.argsort(distances)
# print the top-k images with their corresponding distances
k = 10
d = []
img = []
cap = []
for i in range(k):
image_index = sorted_indexes[i]
image_path = retrieval_set_filenames[image_index]
p = image_path.split("train_images/")[1]
# filter the rows containing the path
filtered_df = train[train["path"] == p]
# get the indices of the filtered rows
indices = filtered_df.index.tolist()
cap.append(indices)
image_distance = distances[image_index]
d.append(image_distance)
img.append(image_path)
print(
f"Image {i+1}: {image_path} - Distance: {image_distance}-caption:{indices}"
)
return d, img, cap
# **query result**
def show_retrieved_images(query, retrieved_images, distances, k):
fig, axs = plt.subplots(2, 6, figsize=(20, 10))
img = load_img(query, color_mode="rgb", target_size=(224, 224))
img = img_to_array(img)
query_img = img / 255.0
axs[0, 0].imshow(query_img)
axs[0, 0].set_title("Query Image")
for i in range(5):
img = load_img(retrieved_images[i], color_mode="rgb", target_size=(224, 224))
img = img_to_array(img)
img = img / 255.0
axs[0, i + 1].imshow(img)
axs[0, i + 1].set_title(f"Distance: {distances[i]:.2f}")
for i in range(5, 10):
img = load_img(retrieved_images[i], color_mode="rgb", target_size=(224, 224))
img = img_to_array(img)
img = img / 255.0
axs[1, i - 4].imshow(img)
axs[1, i - 4].set_title(f"Distance: {distances[i]:.2f}")
plt.show()
d, rt, cap = retrived("/kaggle/working/train_images/2560278143_aa5110aa37.jpg")
show_retrieved_images(
"/kaggle/working/train_images/2560278143_aa5110aa37.jpg", rt, d, k=10
)
import visualkeras
visualkeras.layered_view(
model, scale_xy=0.5, to_file="model.png"
).show() # write and show
visualkeras.layered_view(model)
em = embedding_model
visualkeras.layered_view(
em, scale_xy=0.5, to_file="embedding_model.png"
).show() # write and show
visualkeras.layered_view(em)
model.summary()
|
# ## Conv1D TF Model with Separate Pipelines for defog and tdcsfog data
# Feature Column TimeSeries grouping adapted from https://www.kaggle.com/code/mayukh18/pytorch-fog-end-to-end-baseline-lb-0-254
# Subject-wise GroupKFold splitting adapted from https://www.kaggle.com/code/xzj19013742/groupkfold-cross-validation-tsflex
# ### In this Notebook
# - Tensorflow Model with Conv1D blocks
# - Models trained separately for defog and tdcsfog data
# - Event Stratified Subject Grouped KFold splitting
# ## Imports and Config
import os
import gc
import numpy as np
from numpy.random import default_rng
import pandas as pd
from tqdm.auto import tqdm
from glob import glob
from os.path import basename, dirname, join, exists
from time import perf_counter
from collections import defaultdict as dd
from functools import partial
from sklearn.model_selection import (
train_test_split,
StratifiedKFold,
StratifiedGroupKFold,
)
from sklearn.metrics import average_precision_score
from sklearn.preprocessing import StandardScaler as Scaler
from scipy.special import expit
import tensorflow as tf
import tensorflow_addons as tfa
print(f"TF version: {tf.__version__}")
AUTO = tf.data.experimental.AUTOTUNE
# Constants
BASE_DIR = "/kaggle/input/tlvmc-parkinsons-freezing-gait-prediction"
TRAIN_DIR = join(BASE_DIR, "train")
TEST_DIR = join(BASE_DIR, "test")
IS_PUBLIC = len(glob(join(TEST_DIR, "*/*.csv"))) == 2
class Config:
train_sub_dirs = [join(TRAIN_DIR, "defog"), join(TRAIN_DIR, "tdcsfog")]
metadata_paths = [
join(BASE_DIR, "defog_metadata.csv"),
join(BASE_DIR, "tdcsfog_metadata.csv"),
]
splits = 5
batch_size = 1024
window_size = 64
window_future = 16
window_past = window_size - window_future # Includes current value
wx = 8
model_dropout = 0.2
model_hidden = 192
model_nblocks = 3
lr = 0.00015
num_epochs = 5
feature_list = ["AccV", "AccML", "AccAP"]
label_list = ["StartHesitation", "Turn", "Walking"]
n_features = len(feature_list)
n_labels = len(label_list)
cfg = Config()
# ## Stratified Group K Fold
# Create Mapping between Id and Subject
id2sub_df = (
pd.concat(
[
pd.read_csv(f, usecols=["Id", "Subject"]).assign(
Module=basename(f).split("_")[0]
)
for f in cfg.metadata_paths
]
)
.astype("category")
.set_index("Id")
)
print(
f"id2sub_df length: {len(id2sub_df)}, unique Ids: {id2sub_df.index.nunique()}, unique Subjects: {id2sub_df.Subject.nunique()}"
)
# Read csv files and add metadata (Id, Subject, Event)
def reader(
filepath,
usecols,
getid=False,
getsub=False,
getevent=False,
dtype=None,
exclude=["notype"],
):
fog_type = basename(dirname(filepath))
if fog_type in exclude:
return None
df = pd.read_csv(filepath, index_col="Time", usecols=usecols, dtype=dtype)
if getid:
df["Id"] = basename(filepath).split(".")[0] + "_" + df.index.astype(str)
if getsub:
df["Subject"] = id2sub_df.loc[basename(filepath).split(".")[0], "Subject"]
if getevent:
df["Event"] = np.select(
[df[col].astype(bool) for col in cfg.label_list],
np.arange(1, cfg.n_labels + 1),
default=0,
).astype("int8")
return df
# Create common train Dataframe
train_paths = glob(join(TRAIN_DIR, "*/*.csv"))
dtype = {col: "int8" for col in cfg.label_list}
dtype["Time"] = "int32"
usecols = ["Time", *cfg.label_list]
train_reader = partial(reader, usecols=usecols, dtype=dtype, getsub=True, getevent=True)
train_df = pd.concat([train_reader(f) for f in tqdm(train_paths)]).reset_index(
drop=True
)
train_df.Subject = train_df.Subject.astype("category")
display(train_df.Event.value_counts().to_frame().style.background_gradient())
# Save paths for each Stratified Group Fold for defog and tdcsfog separately
sgkf = StratifiedGroupKFold(n_splits=cfg.splits, random_state=42, shuffle=True)
fold_train_fpaths, fold_valid_fpaths = {"defog": [], "tdcsfog": []}, {
"defog": [],
"tdcsfog": [],
}
df_paths = {
"defog": glob(join(cfg.train_sub_dirs[0], "*.csv")),
"tdcsfog": glob(join(cfg.train_sub_dirs[1], "*.csv")),
}
for module, paths in df_paths.items():
print(f"{module}:")
sub_train_df = train_df[
train_df.Subject.isin(id2sub_df.loc[id2sub_df.Module == module, "Subject"])
].reset_index(drop=True)
for i, (train_index, test_index) in enumerate(
sgkf.split(sub_train_df.index, sub_train_df.Event, groups=sub_train_df.Subject)
):
print(f"\tFold {i}:", end=" ")
train_subs = sub_train_df.loc[train_index, "Subject"].unique()
test_subs = sub_train_df.loc[test_index, "Subject"].unique()
print(f"Subjects->train:{len(train_subs)}|test:{len(test_subs)}")
train_ids = set(id2sub_df[id2sub_df.Subject.isin(train_subs)].index)
test_ids = set(id2sub_df[id2sub_df.Subject.isin(test_subs)].index)
fold_train_fpaths[module].append(
[f for f in paths if basename(f).split(".")[0] in train_ids]
)
fold_valid_fpaths[module].append(
[f for f in paths if basename(f).split(".")[0] in test_ids]
)
del train_subs, test_subs, train_ids, test_ids
gc.collect()
del sub_train_df
del train_df
gc.collect()
# ## Dataset
# Adapted from FOGDataset of https://www.kaggle.com/code/mayukh18/pytorch-fog-end-to-end-baseline-lb-0-254
class FOGSequence(tf.keras.utils.Sequence):
def __init__(self, df_paths, cfg=cfg, split="train"):
_time = perf_counter()
self.rng = default_rng(42)
self.cfg = cfg
self.split = split
self.past_pad = self.cfg.wx * (self.cfg.window_past - 1)
self.future_pad = self.cfg.wx * self.cfg.window_future
if self.split == "test":
self.Ids = []
_values = [self._read(f) for f in df_paths]
self.mapping = []
_length = 0
for _value in _values:
_shape = _value.shape[0]
self.mapping.extend(
range(_length + self.past_pad, _length + _shape - self.future_pad)
)
_length += _shape
self.values = np.concatenate(_values, axis=0)
self.mapping = np.array(self.mapping)
if self.split != "test":
# Keep only vaild and task rows
_valid_pos = self.values[self.mapping, self.valid_position] > 0
_task_pos = self.values[self.mapping, self.task_position] > 0
self.mapping = self.mapping[_valid_pos & _task_pos]
self.length = self.mapping.shape[0]
print(
f"Valid Dataset of size {self.length:,} initialized in {perf_counter() - _time:.3f} secs!"
)
gc.collect()
def _read(self, path):
_is_tdcs = basename(dirname(path)).startswith("tdcs")
df = pd.read_csv(path)
if self.split == "test":
_ids = basename(path).split(".")[0] + "_" + df.Time.astype(str)
self.Ids.extend(_ids.tolist())
return self._df_to_array(df, self.cfg.feature_list)
_cols = [*self.cfg.feature_list, *self.cfg.label_list, "Valid", "Task"]
self.valid_position = self.cfg.n_features + self.cfg.n_labels
self.task_position = self.valid_position + 1
if _is_tdcs:
# Fill Valid and Task columns for tdcsfog
df["Valid"] = 1
df["Task"] = 1
return self._df_to_array(df, _cols)
def _df_to_array(self, df, cols):
# Pads past and future rows to dataframe values for indexing
_values = df[cols].values.astype(np.float16)
return np.pad(_values, ((self.past_pad, self.future_pad), (0, 0)), "edge")
def __len__(self):
return int(np.ceil(self.length / self.cfg.batch_size))
def __getitem__(self, idx):
if self.split == "train":
# Onlt train set has randomly selected batches
_idxs = self.rng.choice(
self.mapping, size=self.cfg.batch_size, replace=False
)
else:
_idxs = self._get_indices(idx)
# For test return only features
if self.split == "test":
return self._get_X(_idxs)
# For train and val splits return y also
return self._get_X_y(_idxs)
def _get_indices(self, idx):
_low = idx * self.cfg.batch_size
# Cap high at self.length so overflow does not occur
_high = min(_low + self.cfg.batch_size, self.length)
return self.mapping[_low:_high]
def _get_X(self, indices):
_X = np.empty(
(len(indices), self.cfg.window_size, self.cfg.n_features), dtype=np.float16
)
for i, idx in enumerate(indices):
_X[i] = self.values[
idx - self.past_pad : idx + self.future_pad + 1 : self.cfg.wx,
: self.cfg.n_features,
]
return _X
def _get_X_y(self, indices):
_X = np.empty(
(len(indices), self.cfg.window_size, self.cfg.n_features), dtype=np.float16
)
for i, idx in enumerate(indices):
_X[i] = self.values[
idx - self.past_pad : idx + self.future_pad + 1 : self.cfg.wx,
: self.cfg.n_features,
]
return (
_X,
self.values[
indices, self.cfg.n_features : self.cfg.n_features + self.cfg.n_labels
],
)
# ## Model
# average_precision_score with positive sample added if no true positive cases are present
def calculate_precision(y_true, y_pred):
pad_width = ((0, 0), (0, 0)) if y_true.any(axis=0).all() else ((1, 0), (0, 0))
y_true, y_pred = np.pad(y_true, pad_width, constant_values=1), np.pad(
y_pred, pad_width, constant_values=1
)
return average_precision_score(y_true, y_pred)
# Note: Not same result as average_precision_score
class AveragePrecision(tf.keras.metrics.Metric):
def __init__(self, num_classes, thresholds=None, name="avg_precision", **kwargs):
super(AveragePrecision, self).__init__(name=name, **kwargs)
self.class_precision = [
tf.keras.metrics.Precision(thresholds) for _ in range(num_classes)
]
def update_state(self, y_true, y_pred, sample_weight=None):
for i, precision in enumerate(self.class_precision):
precision.update_state(y_true[..., i], y_pred[..., i])
def result(self):
return tf.math.reduce_mean(
[precision.result() for precision in self.class_precision]
)
def reset_state(self):
for precision in self.class_precision:
precision.reset_state()
# Model adapted from https://keras.io/examples/timeseries/timeseries_classification_from_scratch/
def get_model(checkpoint_path=None):
model = tf.keras.models.Sequential()
model.add(tf.keras.Input(shape=(cfg.window_size, cfg.n_features), dtype="float16"))
for _ in range(cfg.model_nblocks):
model.add(
tf.keras.layers.Conv1D(
filters=cfg.model_hidden, kernel_size=31, padding="same"
)
)
model.add(tf.keras.layers.BatchNormalization())
model.add(tf.keras.layers.PReLU())
model.add(tf.keras.layers.Dropout(cfg.model_dropout))
model.add(tf.keras.layers.GlobalAveragePooling1D())
model.add(tf.keras.layers.Dense(cfg.n_labels, activation=None))
if checkpoint_path is not None:
model.load_weights(checkpoint_path)
model.compile(
tf.keras.optimizers.Adam(learning_rate=cfg.lr),
loss=tf.keras.losses.BinaryCrossentropy(from_logits=True),
metrics=[AveragePrecision(cfg.n_labels, thresholds=0.0)],
)
return model
get_model().summary()
def get_model_pub(checkpoint_path=None):
model = tf.keras.models.Sequential()
model.add(tf.keras.Input(shape=(cfg.window_size, cfg.n_features), dtype="float16"))
for _ in range(cfg.model_nblocks):
model.add(tf.keras.layers.Conv1D(filters=128, kernel_size=15, padding="same"))
model.add(tf.keras.layers.BatchNormalization())
model.add(tf.keras.layers.ReLU())
model.add(tf.keras.layers.Dropout(cfg.model_dropout))
model.add(tf.keras.layers.GlobalAveragePooling1D())
model.add(tf.keras.layers.Dense(cfg.n_labels, activation=None))
if checkpoint_path is not None:
model.load_weights(checkpoint_path)
model.compile(
tf.keras.optimizers.Adam(learning_rate=cfg.lr),
loss=tf.keras.losses.BinaryCrossentropy(from_logits=True),
metrics=[AveragePrecision(cfg.n_labels, thresholds=0.0)],
)
return model
from tensorflow.keras.layers import *
def get_model_wn(checkpoint_path=None):
def cbr(x, out_layer, kernel, stride, dilation):
x = Conv1D(
out_layer,
kernel_size=kernel,
dilation_rate=dilation,
strides=stride,
padding="same",
)(x)
x = BatchNormalization()(x)
x = Activation("relu")(x)
return x
def wave_block(x, filters, kernel_size, n):
dilation_rates = [2**i for i in range(n)]
x = Conv1D(filters=filters, kernel_size=1, padding="same")(x)
res_x = x
for dilation_rate in dilation_rates:
tanh_out = Conv1D(
filters=filters,
kernel_size=kernel_size,
padding="same",
activation="tanh",
dilation_rate=dilation_rate,
)(x)
sigm_out = Conv1D(
filters=filters,
kernel_size=kernel_size,
padding="same",
activation="sigmoid",
dilation_rate=dilation_rate,
)(x)
x = Multiply()([tanh_out, sigm_out])
x = Conv1D(filters=filters, kernel_size=1, padding="same")(x)
res_x = Add()([res_x, x])
return res_x
inp = Input(shape=(cfg.window_size, cfg.n_features))
x = cbr(inp, 96, 7, 1, 1)
x = BatchNormalization()(x)
x = wave_block(x, 24, 3, 12)
x = BatchNormalization()(x)
x = wave_block(x, 48, 3, 8)
x = BatchNormalization()(x)
x = wave_block(x, 96, 3, 4)
x = BatchNormalization()(x)
x = wave_block(x, 192, 3, 1)
x = cbr(x, 48, 7, 1, 1)
# x = BatchNormalization()(x)
x = tf.keras.layers.GlobalAveragePooling1D()(x)
x = Dropout(0.2)(x)
out = Dense(3, activation=None, name="out")(x)
model = tf.keras.models.Model(inputs=inp, outputs=out)
opt = tf.keras.optimizers.Adam(lr=cfg.lr)
# opt = tfa.optimizers.SWA(opt)
# model.compile(loss = losses.CategoricalCrossentropy(), optimizer = opt, metrics = ['accuracy'])
if checkpoint_path is not None:
model.load_weights(checkpoint_path)
model.compile(
opt,
loss=tf.keras.losses.BinaryCrossentropy(from_logits=True),
metrics=[AveragePrecision(cfg.n_labels, thresholds=0.0)],
)
return model
# ## Train
def predict_select_model(fold, ds, model_save_dir=""):
best_path, best_score = None, -1
print(f"Validation for fold{fold}:")
for model_path in sorted(glob(join(model_save_dir, f"fold{fold}_*.h5"))):
pred_time = perf_counter()
gc.collect()
score = calculate_precision(
ds.values[ds.mapping, cfg.n_features : cfg.n_features + cfg.n_labels],
expit(
get_model(model_path).predict(ds, verbose=0)
), # expit converts to sigmoid output
)
if best_score < score:
best_score = score
best_path = model_path
gc.collect()
print(
"\t",
basename(model_path),
f": score-{score:.4f} in {(perf_counter()-pred_time)/60:.2f} mins",
)
print(basename(best_path), "selected with score", best_score)
return best_path
from tensorflow.keras.callbacks import LearningRateScheduler
def get_lr_callback(FOG_TYPE):
lr_start = 0.00015 if FOG_TYPE == "tdcsfog" else 1e-4
lr_max = 0.00015 if FOG_TYPE == "tdcsfog" else 1e-4
lr_min = 1e-6
lr_ramp_ep = 0
lr_sus_ep = 0
lr_decay = 0.713 if FOG_TYPE == "tdcsfog" else 0.666
def lrfn(epoch):
if epoch < lr_ramp_ep:
lr = (lr_max - lr_start) / lr_ramp_ep * epoch + lr_start
elif epoch < lr_ramp_ep + lr_sus_ep:
lr = lr_max
else:
lr = (lr_max - lr_min) * lr_decay ** (
epoch - lr_ramp_ep - lr_sus_ep
) + lr_min
return lr
lr_callback = LearningRateScheduler(lrfn, verbose=1)
return lr_callback
def train_loop(train_paths, valid_paths, fold, model_save_dir=""):
gc.collect()
# train_paths, test_paths = train_test_split(train_paths, test_size=0.4)
train_ds = FOGSequence(train_paths)
val_ds = FOGSequence(valid_paths, split="val")
# test_ds = FOGSequence(test_paths, split="val")
model = get_model()
ckpt = tf.keras.callbacks.ModelCheckpoint(
join(model_save_dir, f"fold{fold}_model_" + "{epoch:02d}.h5"),
save_weights_only=True,
)
history = model.fit(
train_ds,
epochs=cfg.num_epochs,
verbose=1,
workers=5,
validation_data=val_ds,
use_multiprocessing=True,
callbacks=[ckpt, get_lr_callback(model_save_dir)],
)
best_model_path = predict_select_model(fold, val_ds, model_save_dir)
# score = calculate_precision(test_ds.values[test_ds.mapping, cfg.n_features:cfg.n_features + cfg.n_labels], expit(get_model(best_model_path).predict(test_ds, verbose=0)))
del train_ds, val_ds, model, ckpt, history
gc.collect()
return best_model_path
# Main training loop
model_paths = {"defog": [], "tdcsfog": []}
for module in model_paths:
module_start = perf_counter()
print(f"***Training {module}{'*'*75}")
if not exists(module):
os.mkdir(module)
for fold, (train_fpaths, valid_fpaths) in enumerate(
zip(fold_train_fpaths[module], fold_valid_fpaths[module])
):
fold_start = perf_counter()
print(f"Fold {fold}{'-'*25}")
model_paths[module].append(
train_loop(train_fpaths, valid_fpaths, fold, model_save_dir=module)
)
print(f"Fold {fold} done in {(perf_counter()-fold_start)/60:.2f} min")
print(f"***{module} done in {(perf_counter()-module_start)/3600:.2f} hrs{'*'*50}\n")
# model_paths = {
# 'defog': [
# '/kaggle/input/fog-conv1d-models/defog/fold0_model_02.h5',
# '/kaggle/input/fog-conv1d-models/defog/fold1_model_01.h5',
# '/kaggle/input/fog-conv1d-models/defog/fold2_model_01.h5',
# '/kaggle/input/fog-conv1d-models/defog/fold3_model_05.h5',
# '/kaggle/input/fog-conv1d-models/defog/fold4_model_02.h5',
# '/kaggle/input/fog-conv1d-models-pub/defog/fold0_model_02.h5',
# '/kaggle/input/fog-conv1d-models-pub/defog/fold1_model_02.h5',
# '/kaggle/input/fog-conv1d-models-pub/defog/fold3_model_05.h5',
# '/kaggle/input/fog-conv1d-models-pub/defog/fold4_model_02.h5',
# '/kaggle/input/fog-conv1d-models-pub/defog/fold5_model_05.h5',
# '/kaggle/input/fog-conv1d-models-pub/defog/fold6_model_05.h5',
# '/kaggle/input/fog-conv1d-models-pub/defog/fold7_model_04.h5',
# '/kaggle/input/fog-conv1d-models-pub/defog/fold8_model_01.h5',
# '/kaggle/input/fog-conv1d-models-pub/defog/fold9_model_05.h5',
# ],
# 'tdcsfog':[
# '/kaggle/input/fog-conv1d-models/tdcsfog/fold0_model_01.h5',
# '/kaggle/input/fog-conv1d-models/tdcsfog/fold1_model_01.h5',
# '/kaggle/input/fog-conv1d-models/tdcsfog/fold2_model_05.h5',
# '/kaggle/input/fog-conv1d-models/tdcsfog/fold3_model_05.h5',
# '/kaggle/input/fog-conv1d-models/tdcsfog/fold4_model_04.h5',
# '/kaggle/input/fog-conv1d-models-pub/tdcsfog/fold0_model_05.h5',
# '/kaggle/input/fog-conv1d-models-pub/tdcsfog/fold1_model_03.h5',
# '/kaggle/input/fog-conv1d-models-pub/tdcsfog/fold2_model_04.h5',
# '/kaggle/input/fog-conv1d-models-pub/tdcsfog/fold3_model_04.h5',
# '/kaggle/input/fog-conv1d-models-pub/tdcsfog/fold4_model_02.h5',
# '/kaggle/input/fog-conv1d-models-pub/tdcsfog/fold5_model_04.h5',
# '/kaggle/input/fog-conv1d-models-pub/tdcsfog/fold6_model_02.h5',
# '/kaggle/input/fog-conv1d-models-pub/tdcsfog/fold7_model_02.h5',
# '/kaggle/input/fog-conv1d-models-pub/tdcsfog/fold8_model_05.h5',
# '/kaggle/input/fog-conv1d-models-pub/tdcsfog/fold9_model_05.h5',
# ]}
# ## Submission
# test_defog_paths = glob(join(TEST_DIR, "defog/*.csv"))
# test_tdcsfog_paths = glob(join(TEST_DIR, "tdcsfog/*.csv"))
# test_ds_dict = {
# 'defog':FOGSequence(test_defog_paths, split="test"),
# 'tdcsfog':FOGSequence(test_tdcsfog_paths, split="test")
# }
# # Get test predictions
# df_list = []
# for module, test_ds in test_ds_dict.items():
# y_pred_list = []
# for model_path in model_paths[module]:
# if 'models-pub' in model_path:
# model = get_model_pub(model_path)
# else:
# model = get_model(model_path)
# y_pred_list.append(expit(model.predict(test_ds, verbose=0))) # expit converts to sigmoid output
# y_pred = np.mean(y_pred_list, axis=0)
# df_list.append(pd.DataFrame(
# {'Id': test_ds.Ids, 'StartHesitation': y_pred[:,0], 'Turn': y_pred[:,1], 'Walking': y_pred[:,2]}))
# # Concatenate Prediction to DataFrames
# submission = pd.concat(df_list)
# # Only keep Ids in sample_submission
# sample_submission = pd.read_csv(join(BASE_DIR, "sample_submission.csv"))
# submission = pd.merge(sample_submission[['Id']], submission, how='left', on='Id').fillna(0.0)
# submission.to_csv("submission.csv", index=False, float_format='%.6f') # round to 5 decimal places while keeping point notation
# !head -5 submission.csv
|
# # London House Price Prediction
# Data set source:
import pandas as pd
import numpy as np
import matplotlib
from matplotlib import pyplot as plt
import matplotlib
matplotlib.rcParams["figure.figsize"] = (20, 10)
import sklearn
# Import the dataset and check layout
df1 = pd.read_csv("/kaggle/input/housing-prices-in-london/London.csv")
df1.head()
# Checking for number of rows
df1.shape
# Checking for number of locations
len(df1.Location.unique())
# Cretaing a column for area code to better divide London locations
df1["Area"] = df1["Postal Code"].str.split(" ").str[0]
# Checking number of areas
area_count = df1.groupby("Area")["Area"].agg("count").sort_values(ascending=False)
area_count
# Checking for areas with less than 10 properties
len(area_count[area_count <= 10])
# Create list of areas with less than 10 properties
area_less_than_10 = area_count[area_count <= 10]
area_less_than_10
# Turn Areas with less than 10 properties into other area
# df1.Location = df1.Location.apply(lambda x: 'other' if x in area_less_than_10 else x)
# Checking for null values
df1.isnull().sum()
# Dropping null values
# df2 = df1.dropna()
# df2.isnull().sum()
df2 = df1.copy()
# Checking for any possible incorrectly input of data
df2["No. of Bedrooms"].unique()
# Checking to see if properties with large numbers of bedrooms are valid
df2.loc[df2["No. of Bedrooms"] > 8]
# Checking for the reason for properties to have zero bedrooms
df2.loc[df2["No. of Bedrooms"] == 0]
# Testing to see any incorrectly input area values
df2["Area in sq ft"].unique()
# Creating a function to identify if values are a float (correct format)
def is_float(x):
try:
float(x)
except:
return False
return True
# Applying function to create a table of all values which are not float values
df2[~df2["Area in sq ft"].apply(is_float)]
# Create new dataframe to create a price per squarefoot
df3 = df2.copy()
df3["Price per sqft"] = df3["Price"] / df3["Area in sq ft"]
df3.head()
# Checking for propeties below threshold of 300 sq ft per bedroom
df3[df3["Area in sq ft"] / df3["No. of Bedrooms"] < 300].head()
# Remove outliers below threshold
df4 = df3[~(df3["Area in sq ft"] / df3["No. of Bedrooms"] < 300)]
df4.shape
# Check for outliers in Price per sq ft
df4["Price per sqft"].describe()
# Create function to remove price per sq ft outliers of two standard deviations (outside 95%)
def remove_pps_outliers(df):
df_out = pd.DataFrame()
for key, subdf in df.groupby("Area"):
m = np.mean(subdf["Price per sqft"])
st = np.std(subdf["Price per sqft"])
reduced_df = subdf[
(subdf["Price per sqft"] > (m - 2 * st))
& (subdf["Price per sqft"] <= (m + 2 * st))
]
df_out = pd.concat([df_out, reduced_df], ignore_index=True)
return df_out
# Apply function to dataframe
df5 = remove_pps_outliers(df4)
df5.shape
def plot_scatter(df, location):
bedrooms2 = df[(df["Area"] == location) & (df["No. of Bedrooms"] == 2)]
bedrooms3 = df[(df["Area"] == location) & (df["No. of Bedrooms"] == 3)]
matplotlib.rcParams["figure.figsize"] = (15, 10)
plt.scatter(
bedrooms2["Area in sq ft"],
bedrooms2["Price"],
color="blue",
label=" 2 Bedrooms",
s=50,
)
plt.scatter(
bedrooms3["Area in sq ft"],
bedrooms3["Price"],
color="red",
label=" 3 Bedrooms",
s=50,
)
plt.xlabel("Total Square Feet Area")
plt.ylabel("Price")
plt.title(location)
plt.legend()
plot_scatter(df5, "SW11")
def remove_bhk_outliers(df):
exclude_indices = np.array([])
for location, location_df in df.groupby("Area"):
bedrooms_stats = {}
for bedrooms, bedrooms_df in location_df.groupby("No. of Bedrooms"):
bedrooms_stats[bedrooms] = {
"mean": np.mean(bedrooms_df["Price per sqft"]),
"std": np.std(bedrooms_df["Price per sqft"]) * 2,
"count": bedrooms_df.shape[0],
}
for bedrooms, bedrooms_df in location_df.groupby("No. of Bedrooms"):
stats = bedrooms_stats.get(bedrooms - 1)
if stats and stats["count"] > 5:
exclude_indices = np.append(
exclude_indices,
bedrooms_df[
bedrooms_df["Price per sqft"] < (stats["mean"])
].index.values,
)
return df.drop(exclude_indices, axis="index")
df6 = remove_bhk_outliers(df5)
df6.shape
plot_scatter(df6, "SW11")
# Checking to see where the majority of properties lay in terms of Size (Square Feet)
matplotlib.rcParams["figure.figsize"] = (20, 10)
plt.hist(df6["Price per sqft"], rwidth=0.8)
plt.xlabel("Price Per Sqaure Feet")
plt.ylabel("Count")
# Checking for outliers in the amount of bathrooms in a property
df6["No. of Bathrooms"].unique()
# Checking for properties outsdie the rule of a property not having the same numbers of bathrooms as bedrooms +2
df6[df6["No. of Bathrooms"] > df6["No. of Bedrooms"] + 2]
# Selct only required columns for model
df7 = df6.loc[
:,
[
"Price",
"Area in sq ft",
"No. of Bedrooms",
"No. of Bathrooms",
"No. of Receptions",
"Area",
],
]
# Encoding the categoric variable Area to be used for regression
dummies = pd.get_dummies(df7.Area)
# Joining the encoded 'Area' column to the original dataframe
df8 = pd.concat([df7, dummies], axis="columns")
df8.head()
# Removing original 'Area' column
df9 = df8.drop("Area", axis="columns")
df9.head()
# Creating X data without price as that is what is being predicted
X = df9.drop("Price", axis="columns")
X.head()
# Creating y data made up of the property prices needed to be predicted
y = df9.Price
y.head()
# Creating a testing and training datatset for the model
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=10
)
# Testing the models accuracy
from sklearn.linear_model import LinearRegression
lr_clf = LinearRegression()
lr_clf.fit(X_train, y_train)
lr_clf.score(X_test, y_test)
# Carrying out cross validation to measure the accuracy of the model
from sklearn.model_selection import ShuffleSplit
from sklearn.model_selection import cross_val_score
cv = ShuffleSplit(n_splits=5, test_size=0.2, random_state=0)
cross_val_score(LinearRegression(), X, y, cv=cv)
# Negative values can be ignored and positive values lie between 69.5% and 80.9% acccuracy
# Finding the best model between lasso regression and decsiion tree using GridSearchCV to perform hyper-parameter tuning
from sklearn.model_selection import GridSearchCV
from sklearn.linear_model import Lasso
from sklearn.tree import DecisionTreeRegressor
def find_best_model_using_gridsearchcv(X, y):
algos = {
"lasso": {
"model": Lasso(),
"params": {"alpha": [1, 2], "selection": ["random", "cyclic"]},
},
"decision_tree": {
"model": DecisionTreeRegressor(),
"params": {
"criterion": ["mse", "friedman_mse"],
"splitter": ["best", "random"],
},
},
}
scores = []
cv = ShuffleSplit(n_splits=5, test_size=0.2, random_state=0)
for algo_name, config in algos.items():
gs = GridSearchCV(
config["model"], config["params"], cv=cv, return_train_score=False
)
gs.fit(X, y)
scores.append(
{
"model": algo_name,
"best_score": gs.best_score_,
"best_params": gs.best_params_,
}
)
return pd.DataFrame(scores, columns=["model", "best_score", "best_params"])
find_best_model_using_gridsearchcv(X, y)
# From the results the lasso regression model is the most accurate model
# Creating the lasso regression model with the optimum parameters
lrm = Lasso(alpha=1.0, selection="random", tol=1e-1)
lrm.fit(X, y)
# Creating function to create a price predicition
def predict_price(Area, sqft, bed, bath, rec):
loc_index = np.where(X.columns == Area)[0][0]
x = np.zeros(len(X.columns))
x[0] = sqft
x[1] = bed
x[2] = bath
x[3] = rec
if loc_index >= 0:
x[loc_index] = 1
return lrm.predict([x])[0]
# Testing the price predictor
f"£{round(predict_price('SW11',1000,2,2,2),2):,}"
|
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import os
train = pd.read_csv("/kaggle/input/playground-series-s3e12/train.csv")
test = pd.read_csv("/kaggle/input/playground-series-s3e12/test.csv")
# # Small EDA
column_names = ["gravity", "ph", "osmo", "cond", "urea", "calc"]
target = "target"
# ## Distribution Plots
# ### Target Distribution
fig, ax = plt.subplots(1, 2, figsize=(16, 5))
ax = ax.flatten()
ax[0].pie(
train["target"].value_counts(),
autopct="%1.f%%",
colors=sns.diverging_palette(260, 5, n=2),
textprops={"size": 40, "color": "white"},
)
sns.countplot(
data=train, y=target, palette=sns.diverging_palette(260, 5, n=2), ax=ax[1]
)
plt.tight_layout()
# ### KDE & Hist
f, axes = plt.subplots(
3,
len(column_names),
figsize=(8 * len(column_names), 11),
)
for x in range(len(column_names)):
ax = axes[0, x]
sns.histplot(
train,
x=column_names[x],
hue=target,
palette=sns.diverging_palette(260, 5, n=2),
ax=ax,
legend=True,
)
for x in range(len(column_names)):
ax = axes[1, x]
sns.kdeplot(
train,
x=column_names[x],
hue=target,
palette=sns.diverging_palette(260, 5, n=2),
ax=ax,
fill=True,
legend=True,
)
for x in range(len(column_names)):
ax = axes[2, x]
sns.kdeplot(
train,
x=column_names[x],
ax=ax,
fill=True,
)
f.show()
# ## Correlation Plots
# ### Relative Plots
f, axes = plt.subplots(
len(column_names),
len(column_names),
figsize=(3 * len(column_names), 3 * len(column_names)),
sharex="col",
sharey="row",
)
mask = np.ones_like(axes, dtype=bool)
mask[np.triu_indices_from(mask)] = False
mask[np.diag_indices_from(mask)] = False
for x in range(len(column_names)):
for y in range(len(column_names)):
ax = axes[y, x]
if mask[y, x]:
sns.scatterplot(
train,
x=column_names[x],
y=column_names[y],
hue=target,
palette=sns.diverging_palette(260, 5, n=2),
ax=ax,
legend=False,
)
else:
ax.axis("off")
f.subplots_adjust(0, 0, 1, 1, 0, 0)
f.show()
# ### Correlation Matrix
c = train[column_names + [target]].corr()
mask = np.zeros_like(c, dtype=bool)
mask[np.triu_indices_from(mask)] = True
mask[np.diag_indices_from(mask)] = True
f, ax = plt.subplots(figsize=(11, 9))
cmap = sns.diverging_palette(260, 5, as_cmap=True)
# Draw the heatmap with the mask and correct aspect ratio
sns_plot = sns.heatmap(
c,
mask=mask,
cmap=cmap,
vmax=(c * (1 - mask)).max().max(),
vmin=(c * (1 - mask)).min().min(),
center=0,
square=True,
linewidths=0.5,
annot=True,
fmt="0.2f",
cbar_kws={"shrink": 0.5},
)
# save to file
fig = sns_plot.get_figure()
fig.show()
# ## Test out feature engineering
from sklearn.linear_model import LinearRegression
from sklearn.model_selection import StratifiedKFold
from sklearn.metrics import roc_auc_score
X = train[column_names]
y = train[target]
def score(X, y):
oof = train[target] * 0.0
kf = StratifiedKFold(n_splits=5, shuffle=True, random_state=1)
for fold, (idx_train, idx_valid) in enumerate(kf.split(X, y)):
X_train, y_train = X.iloc[idx_train], y.iloc[idx_train]
X_valid, y_valid = X.iloc[idx_valid], y.iloc[idx_valid]
model = LinearRegression()
model.fit(X_train, y_train)
oof[idx_valid] += model.predict(X_valid)
return roc_auc_score(y, oof)
baseline = score(X, y)
baseline
from sklearn.impute import SimpleImputer
import optuna
def modify_df(params, X):
# Functions
functions = {
"id": lambda x: x,
"x**2": lambda x: x**2,
"abs": np.abs,
"log": np.log,
"sqrt": np.sqrt,
"norm": lambda x: (x - x.min()) / (x.max() - x.min()),
"std": lambda x: (x - x.mean()) / x.std(),
}
for c in column_names:
functions[f"mul_{c}"] = lambda x: x * X[c]
functions[f"div_{c}"] = lambda x: x / X[c]
# Apply
X_mod = X.copy()
for c in column_names:
for i in range(2):
f = params[f"{c}_{i}"]
X_mod[c] = functions[f](X_mod[c])
X_mod[c] = X_mod[c].fillna(X_mod[c].mean())
X_mod = X_mod.replace([np.inf, -np.inf, np.nan], 0)
return X_mod
def objective(trial):
functions = (
["id", "x**2", "abs", "log", "sqrt", "norm", "std"]
+ [f"mul_{c}" for c in column_names]
+ [f"div_{c}" for c in column_names]
)
params = {
f"{c}_{i}": trial.suggest_categorical(f"{c}_{i}", functions)
for c in column_names
for i in range(2)
}
return score(modify_df(params, X), y)
study = optuna.create_study(direction="maximize")
# study.optimize(objective, n_trials=10_000)
# optuna.visualization.plot_optimization_history(study)
# params = study.best_trial.params
params = {
"gravity_0": "div_osmo",
"gravity_1": "x**2",
"ph_0": "div_urea",
"ph_1": "x**2",
"osmo_0": "norm",
"osmo_1": "x**2",
"cond_0": "std",
"cond_1": "x**2",
"urea_0": "div_ph",
"urea_1": "div_urea",
"calc_0": "log",
"calc_1": "sqrt",
}
# ## Linear Regression
from sklearn.linear_model import LinearRegression
from sklearn.model_selection import StratifiedKFold
from sklearn.metrics import roc_auc_score
X = modify_df(params, train[column_names])
y = train[target]
X_test = modify_df(params, test[column_names])
y_test = np.zeros((len(test),))
oof = train[target] * 0.0
kf = StratifiedKFold(n_splits=5, shuffle=True, random_state=1)
for fold, (idx_train, idx_valid) in enumerate(kf.split(X, y)):
X_train, y_train = X.iloc[idx_train], y.iloc[idx_train]
X_valid, y_valid = X.iloc[idx_valid], y.iloc[idx_valid]
model = LinearRegression()
model.fit(X_train, y_train)
oof[idx_valid] += model.predict(X_valid)
y_test += model.predict(X_test)
print("OOF:", roc_auc_score(y, oof))
# create submission file
submission = pd.read_csv("/kaggle/input/playground-series-s3e12/sample_submission.csv")
submission["target"] = y_test
submission.to_csv("./submission.csv", index=False)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt # plot
plt.style.use("dark_background")
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
data = pd.read_csv("../input/united-nations-world-populations/UNpopfile.csv")
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
data.head()
data.describe()
nations = data["Location"].unique()
nations
len(nations) # There are some repeats
usa = data.loc[
data["Location"] == "United States of America"
] # You can change the country to whatever you want
usa # Ok now we have the USA data
# Looks like there are a bunch of different predictions in the data. So I'm going to pick a single one and work with it.
usa.Variant.unique()
# I'm going to graph the population over time. The Poptotal is in thousands. for the graph im going to multiply everything by 1000
# Actually we dont need a bunch of these variables like varid, location, and LocId
dropvals = ["LocID", "VarID", "MidPeriod", "Location"]
usa = usa.drop(dropvals, axis=1)
usa.head()
# Thats better
# Now times a thousand
# Actually the nums dont look too good
# imma comment this out
# def timesAThousand(num):
# return num*1000
# usa["PopMale"] = usa["PopMale"].apply(timesAThousand)
# usa["PopFemale"] = usa["PopFemale"].apply(timesAThousand)
# usa["PopTotal"] = usa["PopTotal"].apply(timesAThousand)
usa.head()
# Now we are going to isolate a specefic variant and graph it
variant = "No change"
plot_usa = usa.loc[usa["Variant"] == variant]
# plot
plt.plot(plot_usa.Time, plot_usa.PopTotal)
plt.title(variant)
plt.show()
# I'm gonna make a for loop to plot every different one
for variant in usa.Variant.unique():
plot_usa = usa.loc[usa["Variant"] == variant]
plt.plot(plot_usa.Time, plot_usa.PopTotal)
plt.title(variant)
plt.ylabel("Population in thousands")
plt.xlabel("Time")
plt.show()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.metrics import mean_squared_error, r2_score
from sklearn.metrics import (
accuracy_score,
precision_score,
recall_score,
f1_score,
roc_auc_score,
)
from sklearn.model_selection import GridSearchCV, cross_validate
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import LabelEncoder
from sklearn.ensemble import RandomForestClassifier
from sklearn.preprocessing import StandardScaler
from sklearn.neighbors import KNeighborsRegressor
import warnings
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Test ve train dosyalarının okunması
train_df = pd.read_csv(
"../input/prediction-of-surgery-duration/train.csv",
index_col="ID",
encoding="mac_turkish",
)
test_df = pd.read_csv(
"../input/prediction-of-surgery-duration/test.csv",
index_col="ID",
encoding="mac_turkish",
)
# Genel resme ulaşmak için check_df fonksiyonu hazırlandı.
def check_df(dataframe, head=5):
print("##################### Shape #####################")
print(dataframe.shape)
print("##################### Types #####################")
print(dataframe.dtypes)
print("##################### Head #####################")
print(dataframe.head(head))
print("##################### Tail #####################")
print(dataframe.tail(head))
print("##################### NA #####################")
print(dataframe.isnull().sum())
print("##################### Quantiles #####################")
print(dataframe.describe([0, 0.05, 0.50, 0.95, 0.99, 1]).T)
check_df(train_df) # train_df bilgileri
check_df(test_df) # test için bilgiler
train_df.isnull().sum()
def grab_col_names(dataframe, cat_th=10, car_th=20):
"""
Veri setindeki kategorik, numerik ve kategorik fakat kardinal değişkenlerin isimlerini verir.
Parameters
----------
dataframe: dataframe
değişken isimleri alınmak istenen dataframe'dir.
cat_th: int, float
numerik fakat kategorik olan değişkenler için sınıf eşik değeri
car_th: int, float
kategorik fakat kardinal değişkenler için sınıf eşik değeri
Returns
-------
cat_cols: list
Kategorik değişken listesi
num_cols: list
Numerik değişken listesi
cat_but_car: list
Kategorik görünümlü kardinal değişken listesi
Notes
------
cat_cols + num_cols + cat_but_car = toplam değişken sayısı
num_but_cat cat_cols'un içerisinde.
"""
# cat_cols, cat_but_car
cat_cols = [
col
for col in dataframe.columns
if str(dataframe[col].dtypes) in ["category", "object", "bool"]
]
num_but_cat = [
col
for col in dataframe.columns
if dataframe[col].nunique() < cat_th
and dataframe[col].dtypes in ["int", "float"]
]
cat_but_car = [
col
for col in dataframe.columns
if dataframe[col].nunique() > car_th
and str(dataframe[col].dtypes) in ["category", "object"]
]
cat_cols = cat_cols + num_but_cat
cat_cols = [col for col in cat_cols if col not in cat_but_car]
# num_cols
num_cols = [
col for col in dataframe.columns if dataframe[col].dtypes in ["int", "float"]
]
num_cols = [col for col in num_cols if col not in cat_cols]
print(f"Observations: {dataframe.shape[0]}")
print(f"Variables: {dataframe.shape[1]}")
print(f"cat_cols: {len(cat_cols)}")
print(f"num_cols: {len(num_cols)}")
print(f"cat_but_car: {len(cat_but_car)}")
print(f"num_but_cat: {len(num_but_cat)}")
return cat_cols, num_cols, cat_but_car
cat_cols, num_cols, cat_but_car = grab_col_names(train_df)
def cat_summary(dataframe, col_name, plot=False):
if dataframe[col_name].dtypes == "bool":
dataframe[col_name] = dataframe[col_name].astype(int)
print(
pd.DataFrame(
{
col_name: dataframe[col_name].value_counts(),
"Ratio": 100 * dataframe[col_name].value_counts() / len(dataframe),
}
)
)
print("##########################################")
if plot:
sns.countplot(x=dataframe[col_name], data=dataframe)
plt.show(block=True)
else:
print(
pd.DataFrame(
{
col_name: dataframe[col_name].value_counts(),
"Ratio": 100 * dataframe[col_name].value_counts() / len(dataframe),
}
)
)
print("##########################################")
if plot:
sns.countplot(x=dataframe[col_name], data=dataframe)
plt.show(block=True)
for col in cat_cols:
cat_summary(train_df, col)
def num_summary(dataframe, numerical_col, plot=False):
quantiles = [0.05, 0.10, 0.20, 0.30, 0.40, 0.50, 0.60, 0.70, 0.80, 0.90, 0.95, 0.99]
print(dataframe[numerical_col].describe(quantiles).T)
if plot:
dataframe[numerical_col].hist()
plt.xlabel(numerical_col)
plt.title(numerical_col)
plt.show(block=True)
for col in num_cols:
num_summary(train_df, col, plot=False)
def target_summary_with_num(dataframe, target, numerical_col):
print(dataframe.groupby(target).agg({numerical_col: "mean"}), end="\n\n\n")
# target_summary_with_num(train_df, "ElapsedTime(second)","DoctorID")
for col in num_cols:
target_summary_with_num(train_df, "ElapsedTime(second)", col)
# KATEGORİK DEĞİŞKENLERİN TARGET GÖRE ANALİZİ
def target_summary_with_cat(dataframe, target, categorical_col):
print(categorical_col)
print(
pd.DataFrame(
{
"TARGET_MEAN": dataframe.groupby(categorical_col)[target].mean(),
"Count": dataframe[categorical_col].value_counts(),
"Ratio": 100
* dataframe[categorical_col].value_counts()
/ len(dataframe),
}
),
end="\n\n\n",
)
for col in cat_cols:
target_summary_with_cat(train_df, "ElapsedTime(second)", col)
# Korelasyon, olasılık kuramı ve istatistikte iki rassal değişken arasındaki doğrusal ilişkinin yönünü ve gücünü belirtir
train_df.corr()
# Korelasyon Matrisi
f, ax = plt.subplots(figsize=[18, 13])
sns.heatmap(train_df.corr(), annot=True, fmt=".2f", ax=ax, cmap="magma")
ax.set_title("Correlation Matrix", fontsize=20)
plt.show()
train_df.corrwith(train_df["ElapsedTime(second)"]).sort_values(ascending=False)
def missing_values_table(dataframe, na_name=False):
na_columns = [col for col in dataframe.columns if dataframe[col].isnull().sum() > 0]
n_miss = dataframe[na_columns].isnull().sum().sort_values(ascending=False)
ratio = (
dataframe[na_columns].isnull().sum() / dataframe.shape[0] * 100
).sort_values(ascending=False)
missing_df = pd.concat(
[n_miss, np.round(ratio, 2)], axis=1, keys=["n_miss", "ratio"]
)
print(missing_df, end="\n")
if na_name:
return na_columns
na_columns = missing_values_table(train_df, na_name=True)
train_df.isnull().sum()
missing_vs_target(train_df, "ElapsedTime(second)", na_columns)
# Aykırı değer Analizi
def outlier_thresholds(dataframe, col_name, q1=0.25, q3=0.75):
quartile1 = dataframe[col_name].quantile(q1)
quartile3 = dataframe[col_name].quantile(q3)
interquantile_range = quartile3 - quartile1
up_limit = quartile3 + 1.5 * interquantile_range
low_limit = quartile1 - 1.5 * interquantile_range
return low_limit, up_limit
def check_outlier(dataframe, col_name):
low_limit, up_limit = outlier_thresholds(dataframe, col_name)
if dataframe[
(dataframe[col_name] > up_limit) | (dataframe[col_name] < low_limit)
].any(axis=None):
return True
else:
return False
def replace_with_thresholds(dataframe, variable):
low_limit, up_limit = outlier_thresholds(dataframe, variable)
dataframe.loc[(dataframe[variable] < low_limit), variable] = low_limit
dataframe.loc[(dataframe[variable] > up_limit), variable] = up_limit
# Aykırı Değer Analizi ve Baskılama İşlemi
for col in num_cols:
print(col, check_outlier(train_df, col))
if check_outlier(train_df, col):
replace_with_thresholds(train_df, col)
for col in num_cols:
print(col, check_outlier(train_df, col))
# Veri Görelleştirme
sns.boxplot(x=train_df["ElapsedTime(second)"])
plt.show()
sns.displot(train_df["ElapsedTime(second)"], kde=True, color="purple")
sns.scatterplot(x="DoctorID", y="ElapsedTime(second)", data=train_df)
sns.scatterplot(x="DoctorID", y="ElapsedTime(second)", hue="Age", data=train_df)
sns.pairplot(train_df)
def label_encoder(dataframe, binary_col):
labelencoder = LabelEncoder()
dataframe[binary_col] = labelencoder.fit_transform(dataframe[binary_col])
return dataframe
binary_cols = [
col
for col in train_df.columns
if train_df[col].dtypes == "O" and train_df[col].nunique() == 2
]
binary_cols
for col in binary_cols:
train_df = label_encoder(train_df, col)
def rare_encoder(dataframe, rare_perc):
temp_df = dataframe.copy()
rare_columns = [
col
for col in temp_df.columns
if temp_df[col].dtypes == "O"
and (temp_df[col].value_counts() / len(temp_df) < rare_perc).any(axis=None)
]
for var in rare_columns:
tmp = temp_df[var].value_counts() / len(temp_df)
rare_labels = tmp[tmp < rare_perc].index
temp_df[var] = np.where(temp_df[var].isin(rare_labels), "Rare", temp_df[var])
return temp_df
new_df = rare_encoder(train_df, 0.1)
train_df["AnesthesiaType"].value_counts()
def one_hot_encoder(dataframe, categorical_cols, drop_first=False):
dataframe = pd.get_dummies(
dataframe, columns=categorical_cols, drop_first=drop_first
)
return dataframe
train_df = one_hot_encoder(train_df, cat_cols, drop_first=True)
train_df.head()
train_df.columns
train_df = train_df[~train_df.index.duplicated()]
my_submission = pd.DataFrame(
{
"Id": pd.Series(test_df["DoctorID"]),
"ElapsedTime": pd.Series(train_df["ElapsedTime(second)"]),
}
)
# you could use any filename. We choose submission here
my_submission.to_csv("submission.csv", index=False)
|
# # IPL 2023 Auction EDA
# **As we know BCCI just now concluded the IPL 2023 auction and some of players got very high price so I will generate key insights from data using Pandas to clean data and then will use Matplotlib,Plotly for visualization****
# # ****Importing required libraries****
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import plotly.express as px
# # **Import the required datasets**
df = pd.read_csv(
"/kaggle/input/ipl-auction-2022/2023 Auction/IPL_2023_Auction_Sold.csv"
)
df.head()
# **From above dataset we will remove the unwanted columns whoch are not needed for our analysis**
df.drop(
[
"Set No.",
"2023 Set",
"Association",
"DOB",
"Batting_Style",
"ODI caps",
"T20 caps",
"Previous IPLTeam(s)",
],
axis=1,
inplace=True,
)
df.head()
# # Data Preparation & Cleaning
# **I am combining the first name and surname columns into single player name columns which will be easy to read and also will save memory**
df["Player Name"] = df[["First Name", "Surname"]].apply(lambda x: " ".join(x), axis=1)
df.drop(["First Name", "Surname"], axis=1, inplace=True)
# **Will look for unique values**
country = df["Country"].unique()
print("From following countries the players participated -", country)
player_speciality = df["Specialism"].unique()
print("We have following specialisation from each player -", player_speciality)
bowling_style = df["Bowling_Style"].unique()
bowling_style
# **Teams participating in IPL Auction 2023**
df["TEAM"].unique()
# **Here UnSold is wrong value so we will drop that**
df.drop(df[df["TEAM"] == "UnSold"].index, inplace=True)
t = df["TEAM"].unique()
print("Following teams were part of IPL Auction 2023 -", t)
# # Data Visualization & Question Answering
# **1. **From which country players earned more money****
money = df.groupby("Player Name")["Auction_Price"].mean()
top10_players = money.sort_values(ascending=False).head(10)
top10_players
fig = px.bar(
x=top10_players.index,
y=top10_players.values,
color=top10_players.index,
text=top10_players.values,
title="Top10 Paid Players",
)
fig.update_layout(xaxis_title="Players", yaxis_title="Amount in Lakhs")
# **Here we can see that top 3 paid players are Sam Curran,Cameron Green & Ben Stokes who all are foreign country players as well as All rounders so this years IPL auction was dominated by foreign players rather than Indian Player in terms of money and teams are ready to pay heavy amount if player is All rounder**
# **2. **How many players are capped and Uncapped.Here Capped means played internation match and Uncapped means et to play internation match****
a = df["C/U/A"].value_counts()
fig1 = px.pie(a, names=a.index, values=a.values, title="Capped/Uncapped Players")
fig1.update_traces(textposition="inside", textinfo="percent + label")
# **Nearly 70% players were uncapped means yet to make international apperance who were part of IPL Auction 2023**
# **3. **What is percentage speciality of players****
b = df["Specialism"].value_counts()
fig2 = px.pie(b, names=b.index, values=b.values, title="Speciality of Players")
fig2.update_traces(textposition="inside", textinfo="percent + label")
# **Most of the players are either All rounders or Bowlers followed by Batsman & Wicketkeeper**
# **4. Money paid according to player type**
c = df.groupby("Specialism")["Auction_Price"].mean()
fig3 = px.bar(
c, x=c.values, y=c.index, title="Average Money spent according to Player Type"
)
fig3.update_layout(xaxis_title="Avg. price in Lakhs")
# **Batsmans got paid paid heavily followed by All rounders & WeeketKeepers**
# **5. **How much money each team spent****
d = df.groupby("TEAM")["Auction_Price"].mean()
fig4 = px.bar(
d,
x=d.index,
y=d.values,
color=d.values,
text=d.index,
title="Money spent by teams in Auction",
)
fig4.update_layout(xaxis_title="Teams", yaxis_title="Avg money in Lakhs")
# **PBKS(Punjab)spent the most money in this years IPL auction**
# **Country wise paid players**
df.drop(df[df["Auction_Price"] < 0.0].index, inplace=True)
country = df.groupby("Country")["Auction_Price"].mean()
z = country.head()
fig5 = px.bar(
x=z.index, y=z.values, title="Countries with top5 paid players", text=z.index
)
fig5.update_layout(xaxis_title="Country", yaxis_title="Price in Lakhs")
|
# # Kidney Stone Detection
# ## Importing libiries
import pandas as pd
import numpy as np
import seaborn as sns
import xgboost as xgb
from xgboost import XGBClassifier
import matplotlib.pyplot as plt
import warnings
warnings.filterwarnings("ignore")
# ## Uploading DataSet
train = pd.read_csv("/kaggle/input/playground-series-s3e12/train.csv")
test = pd.read_csv("/kaggle/input/playground-series-s3e12/test.csv")
train.head()
test.head()
# ## Data PreProcessing
train.shape
test.shape
train.describe()
X = train.drop(["target", "id"], axis=1)
y = train.target
fig, axes = plt.subplots(nrows=3, ncols=3) # axes is 2d array (3x3)
axes = axes.flatten() # Convert axes to 1d array of length 9
fig.set_size_inches(15, 15)
for ax, col in zip(axes, train.columns):
sns.distplot(train[col], ax=ax, color="red")
ax.set_title(col)
X.isnull().sum()
X.skew()
X.skew()
fig, axes = plt.subplots(nrows=3, ncols=2) # axes is 2d array (3x3)
axes = axes.flatten() # Convert axes to 1d array of length 9
fig.set_size_inches(15, 15)
for ax, col in zip(axes, X.columns):
sns.distplot(X[col], ax=ax, color="red")
ax.set_title(col)
model = XGBClassifier(
max_depth=8,
learning_rate=0.01,
colsample_bytree=0.67,
n_jobs=-1,
objective="binary:logistic",
verbosity=0,
eval_metric="logloss",
)
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, train_size=0.8)
X_test
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
y_pred
y_test
model.score(X_test, y_test)
id = test.id
data = test.drop("id", axis=1)
data.skew()
data.skew()
pred = model.predict(data)
pred
df = pd.DataFrame({"id": id, "target": pred})
df
df.to_csv("submission.csv", index=0)
|
# ### 📓 Desciption
# The site launched an A/B test in order to increase income. The excel file contains raw data on the results of the experiment (user_id), sample type (variant_name) and income brought by the user (revenue).
# The task is to analyze the results of the experiment and write your recommendations.
# ### 🖊 We will analyse the following
# - Data Preprocessing
# - Exploratory Data Analysis
# - Check Normality and AB Testing (Mann Whitney U Test)
# - Make inferences
# ## ⚙ Libraries and Settings
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import scipy.stats
from scipy import stats as st
import pylab
import scipy.stats as stats
from scipy.stats import shapiro
from scipy.stats import mannwhitneyu
from scipy.stats import shapiro
import statsmodels.stats.api as sms
import warnings
warnings.filterwarnings("ignore")
pd.set_option("display.max_rows", None)
pd.set_option("display.max_columns", None)
pd.set_option("display.width", 2000)
pd.set_option("display.float_format", "{:20,.3f}".format)
pd.set_option("display.max_colwidth", None)
# ## 📂 Data download and preprocessing
df = pd.read_csv("/kaggle/input/ab-test-data/AB_Test_Results.csv")
df.columns = df.columns.str.lower()
df.head()
df.info()
df.describe([0.25, 0.50, 0.75, 0.95, 0.99, 1])
df.isnull().sum()
df.duplicated().sum()
df["user_id"].nunique()
duplicates = df.groupby("user_id")["variant_name"].nunique().reset_index()
duplicates["variant_name"].value_counts(normalize=True)
df = df[df["user_id"].isin(duplicates[duplicates["variant_name"] == 1]["user_id"])]
df.describe([0.25, 0.50, 0.75, 0.95, 0.99])
sns.boxplot(data=df, x="revenue", y="variant_name")
df = df.query("revenue < 175")
# The data is ready, let's do EDA!
# ## 📊 Exploratory Data Analysis
# #### All users
def perc_95(column):
return np.percentile(column, 95)
def perc_99(column):
return np.percentile(column, 99)
df.groupby("variant_name").agg(
{
"user_id": "nunique",
"revenue": ["count", "sum", "mean", "median", perc_95, perc_99, "sum"],
}
)
sns.boxplot(data=df, x="revenue", y="variant_name")
# #### Paying Users
df_revenue = df.query("revenue > 0")
df_revenue.groupby("variant_name").agg(
{
"user_id": "nunique",
"revenue": ["count", "sum", "min", "mean", "median", perc_95, perc_99, "sum"],
}
)
sns.boxplot(data=df_revenue, x="revenue", y="variant_name")
# #### ARPU / ARPPU
t = df.groupby("variant_name").agg({"user_id": "nunique", "revenue": "sum"})
t_paying_users = df_revenue.groupby("variant_name").agg({"user_id": "nunique"})
t = t.merge(t_paying_users, on="variant_name", how="left").rename(
columns={
"variant_name": "variant_name",
"user_id_x": "users",
"user_id_y": "paying_users",
"revenue": "revenue",
}
)
t["arpu"] = t["revenue"] / t["users"]
t["arppu"] = t["revenue"] / t["paying_users"]
t
# ## 🧪 Checking for the normality and A/B Testing
# ### All users
variant = df[df["variant_name"] == "variant"]["revenue"]
control = df[df["variant_name"] == "control"]["revenue"]
for group in [variant, control]:
sns.set(rc={"figure.figsize": (4, 2)}, style="whitegrid", palette="pastel")
x = shapiro(group)
if x.pvalue > 0.05:
print(f"{x.pvalue} is normal distribution")
else:
print(f"{x.pvalue} is not normal distribution")
sns.set(rc={"figure.figsize": (4, 2)}, style="whitegrid", palette="pastel")
stats.probplot(group, dist="norm", plot=pylab)
pylab.show()
q = sns.histplot(data=group, kde=True, bins=50)
plt.show()
def ab_test(a, b):
alpha = 0.05
revenue_difference_mannwhitney = mannwhitneyu(variant, control)
if revenue_difference_mannwhitney.pvalue < alpha:
print(
revenue_difference_mannwhitney.pvalue,
"Groups is different. Fail to Reject H1.",
)
else:
print(
revenue_difference_mannwhitney.pvalue,
"Groups is similar. Fail to Reject H0.",
)
ab_test(variant, control)
# ### Paying users
variant = df_revenue[df_revenue["variant_name"] == "variant"]["revenue"]
control = df_revenue[df_revenue["variant_name"] == "control"]["revenue"]
for group in [variant, control]:
sns.set(rc={"figure.figsize": (4, 2)}, style="whitegrid", palette="pastel")
x = shapiro(group)
if x.pvalue > 0.05:
print(f"{x.pvalue} is normal distribution")
else:
print(f"{x.pvalue} is not normal distribution")
sns.set(rc={"figure.figsize": (4, 2)}, style="whitegrid", palette="pastel")
stats.probplot(group, dist="norm", plot=pylab)
pylab.show()
q = sns.histplot(data=group, kde=True, bins=50)
plt.show()
def ab_test(a, b):
alpha = 0.05
revenue_difference_mannwhitney = mannwhitneyu(variant, control)
if revenue_difference_mannwhitney.pvalue < alpha:
print(
revenue_difference_mannwhitney.pvalue,
"Groups is different. Fail to Reject H1.",
)
else:
print(
revenue_difference_mannwhitney.pvalue,
"Groups is similar. Fail to Reject H0.",
)
ab_test(variant, control)
|
# # INDEX
# #### 1. Data Exploration
# #### 2. Feature Engineering
# #### 3. Models
# #### 4. Prediction
# ## 1. Data Exploration
import numpy as np
import pandas as pd
train = pd.read_csv(
r"/kaggle/input/test-competition-2783456756923/airline_tweets_train.csv"
)
test = pd.read_csv(
r"/kaggle/input/test-competition-2783456756923/airline_tweets_test.csv"
)
test.describe()
# ## 2. Feature Engineering
# **A) Feature Extraction**
# **B) Feature Selection**
# **C) Text Cleaning & Lemmatization**
# **D) Text Vectorization**
# ### A) Feature Extraction
# Here we are generating some simple variables from the text:
# * **'tweet_length':** Lenght of the tweet, number of characters
# * **'tweet_words':** Number of words in the tweet
# * **'avg_word_length':** Average length of words (tweet_length/tweet_words)
# * **'has_retweets':** Binary variable indicating if tweet has been retweeted
train["tweet_length"] = train.text.apply(lambda x: len(x))
train["tweet_words"] = train.text.apply(lambda x: len(x.split()))
train["avg_word_length"] = train["tweet_length"] / train["tweet_words"]
train["has_retweets"] = np.where(train.retweet_count > 0, 1, 0)
train["user_timezone"] = np.where(
train.user_timezone.isnull(), "unknown", train.user_timezone
)
# We will also generate two more variables which are a bit more complex:
# * **'num_accounts':** How many twitter accounts are mentioned (@Delta, @pierre,...)
# * **'has_numbers':** Binary variable indicating if tweet contains numbers
def num_accounts(text):
# Split the text into words
words = text.split()
# Count the number of words starting with '@'
count = sum(1 for word in words if word.startswith("@"))
return count
train["num_accounts"] = train["text"].apply(num_accounts)
import re
def contains_numbers(string):
match = re.search(r"\d", string)
if match:
return 1
else:
return 0
train["has_numbers"] = train["text"].apply(contains_numbers)
train.head()
# **TASK:**
# 1. Think of an example of a new variable that may help identifying the sentiment, e.g.:
# * Number of times a specific character appears (!, ?)
# * Any linear combination of existing variables
# 2. Code this new variable (with the help of GPT if needed)
# Generate a new variable
# train['name of your variable'] = your code here
# ### B) Feature Selection
numeric_cols = [
"retweet_count",
"tweet_length",
"tweet_words",
"avg_word_length",
"has_retweets",
"has_numbers",
"num_accounts"
# ,'name of your variable'
]
# Split Data into Features(X) and Target(Y)
x_train_numeric = train[numeric_cols]
y_train = train[["airline_sentiment"]]
from sklearn.tree import DecisionTreeClassifier
clf = DecisionTreeClassifier(
random_state=1, max_depth=4, max_leaf_nodes=10, min_samples_split=60
)
model_tree = clf.fit(x_train_numeric, y_train)
import matplotlib.pyplot as plt
from sklearn import tree
fig = plt.figure(figsize=(15, 10))
_ = tree.plot_tree(
clf,
feature_names=x_train_numeric.columns,
class_names=["negative", "positive", "neutral"],
filled=True,
)
# Select the 3 most important features according to the Decision Tree
selected_cols = ["tweet_words", "avg_word_length", "has_numbers"]
# ## C) Text Cleaning & Lemmatization
import nltk
# nltk.download("stopwords")
# nltk.download('wordnet')
import string
from nltk.corpus import stopwords
stopwords = stopwords.words("english")
# Function to clean text
def cleaner(text):
text = "".join([word for word in text if not word.isnumeric()])
text = text.lower()
text = " ".join([word for word in text.split() if word not in stopwords])
return text
def remove_punct(text):
text = "".join([word for word in text if word not in string.punctuation])
return text
string.punctuation
original_text = "@VirginAmerica thanks to your outstanding crew who moved mountains to get me home to San Francisco tonight!. Our flight was fantastic"
clean_text = cleaner(original_text)
print("Original Text:")
print(original_text)
print("Clean Text:")
print(clean_text)
import stanza
nlp = stanza.Pipeline(lang="en", processors="tokenize,mwt,pos,lemma", verbose=False)
def lemmatize(text, nlp):
doc = nlp(text)
lemm_text = [word.lemma for sent in doc.sentences for word in sent.words]
try:
lemm_text = " ".join(lemm_text)
except:
lemm_text = ""
return lemm_text
def word_types(text, nlp):
doc = nlp(text)
lemm_text = [word.upos for sent in doc.sentences for word in sent.words]
try:
lemm_text = " ".join(lemm_text)
except:
lemm_text = ""
return lemm_text
# Types of Words:
# * ADJ: adjective
# * ADP: adposition
# * ADV: adverb
# * AUX: auxiliary
# * CCONJ: coordinating conjunction
# * DET: determiner
# * INTJ: interjection
# * NOUN: noun
# * NUM: numeral
# * PART: particle
# * PRON: pronoun
# * PROPN: proper noun
# * PUNCT: punctuation
# * SCONJ: subordinating conjunction
# * SYM: symbol
# * VERB: verb
# * X: other
print(original_text)
print(clean_text)
print(lemmatize(clean_text, nlp))
print(word_types(clean_text, nlp))
# Clean the text (remove stopwords, lowercase)
train["clean_text"] = train["text"].apply(cleaner)
# Lemmatize the text
# Lemmatize the text and remove punctuation after
processors = "tokenize,mwt,lemma"
nlp_lem = stanza.Pipeline(lang="en", processors=processors, verbose=False)
# This takes 5 mins at most
train["lemm_text"] = train["clean_text"].apply(lambda x: lemmatize(x, nlp_lem))
train["lemm_text"] = train["lemm_text"].apply(remove_punct)
# Find the word types
# Generate a sentence with the word types, this takes 12 minutes :(
processors = "tokenize,mwt,pos"
nlp_pos = stanza.Pipeline(lang="en", processors=processors, verbose=False)
train["word_pos"] = train["clean_text"].apply(lambda x: word_types(x, nlp_pos))
# ## D) Text Vectorization
# Vectorize the clean, lemmatized text
#
from sklearn.feature_extraction.text import CountVectorizer
word_vectorizer = CountVectorizer(min_df=10)
X = word_vectorizer.fit_transform(train.lemm_text.values.astype(str))
x_train_words = pd.DataFrame(
X.toarray(), columns=word_vectorizer.get_feature_names_out(), index=train.index
)
print(x_train_words.shape)
x_train_words.head()
# Vectorize the word types (pos)
pos_vectorizer = CountVectorizer()
X = pos_vectorizer.fit_transform(train.word_pos.values.astype(str))
x_train_word_pos = pd.DataFrame(
X.toarray(), columns=pos_vectorizer.get_feature_names_out(), index=train.index
)
print(x_train_word_pos.shape)
x_train_word_pos.head()
x_train_final = pd.concat([x_train_numeric, x_train_words, x_train_word_pos], axis=1)
print(x_train_final.shape)
x_train_final.head()
from sklearn.feature_selection import SelectKBest, chi2
top_k_features = 300
feature_selector = SelectKBest(chi2, k=top_k_features)
x_train_final_sel = feature_selector.fit_transform(x_train_final, y_train)
feature_selector.get_support()
feature_selector.get_support().shape
# feature_selector.scores_
feature_ranking = pd.DataFrame(columns=["feature", "score"])
feature_ranking["feature"] = x_train_final.columns[
feature_selector.get_support()
].tolist()
feature_ranking["score"] = feature_selector.scores_[
feature_selector.get_support()
].tolist()
feature_ranking.sort_values(by="score", ascending=False).head(20)
# ## D) Data Cleaning & Pre-Processing
# We must perform the same processing on the testing dataset
# Generate Simple Columns
test["tweet_length"] = test.text.apply(lambda x: len(x))
test["tweet_words"] = test.text.apply(lambda x: len(x.split()))
test["avg_word_length"] = test["tweet_length"] / test["tweet_words"]
test["has_retweets"] = np.where(test.retweet_count > 0, 1, 0)
test["user_timezone"] = np.where(
test.user_timezone.isnull(), "unknown", test.user_timezone
)
# Generate Complex Columns
test["num_accounts"] = test["text"].apply(num_accounts)
test["has_numbers"] = test["text"].apply(contains_numbers)
# Generate your specific column
# test['your column name'] = your code here
# Select numeric columns
x_test_numeric = test[numeric_cols]
# Clean & Lemmatize the text
test["clean_text"] = test["text"].apply(cleaner)
test["lemm_text"] = test["clean_text"].apply(lambda x: lemmatize(x, nlp_lem))
test["lemm_text"] = test["lemm_text"].apply(remove_punct)
# Generate word types (pos) sentences
test["word_pos"] = test["clean_text"].apply(lambda x: word_types(x, nlp_pos))
# Vectorize the cleaned, lemmatized text
X = word_vectorizer.transform(test["lemm_text"].values.astype(str))
x_test_words = pd.DataFrame(
X.toarray(), columns=word_vectorizer.get_feature_names_out(), index=test.index
)
# Vectorize the cleaned, lemmatized text
X = pos_vectorizer.transform(test["word_pos"].values.astype(str))
x_test_word_pos = pd.DataFrame(
X.toarray(), columns=pos_vectorizer.get_feature_names_out(), index=test.index
)
# Join All 3 feature tables: Numeric Variables, vectorized words, vectorized word types
x_test_final = pd.concat([x_test_numeric, x_test_words, x_test_word_pos], axis=1)
# Separate target
y_test = test[["airline_sentiment"]]
x_test_final_sel = feature_selector.transform(x_test_final)
# Summary of variables
print("Shape of the training Dataset")
print("Features (X): ", x_train_final_sel.shape)
print("Target (Y): ", y_train.shape)
print("Shape of the testing Dataset")
print("Features (X): ", x_test_final_sel.shape)
print("Target (Y): ", y_test.shape)
# ## Models
#
from sklearn.ensemble import RandomForestClassifier
clf = RandomForestClassifier(
random_state=1, n_estimators=100, min_samples_split=2, max_depth=20
)
model_rf = clf.fit(x_train_final_sel, y_train)
[estimator.tree_.max_depth for estimator in model_rf.estimators_]
# help(model_rf.estimators_[0].tree_)
model_rf.estimators_[0].tree_.max_depth
model_rf.estimators_[0].tree_.node_count
y_test = clf.predict(x_test_final_sel)
y_test.shape
submission_file = test.copy()
submission_file = submission_file[["Id"]]
submission_file["Category"] = y_test
submission_file.head()
submission_file.to_csv(r"initial_submission.csv", index=False)
# ## Final Tuning
# For this Final Tuning part, you will have the opportunity to play with 3 aspects of the data in order to generate the best predictions for your team:
# #### 1. Select the Model Hyperparameters
# * n_estimators
# * min_samples_split
# * max_depth
# #### 2. Prepare the Data for the Model (which and how many variables?)
# * Make combinations of the available groups
# * x_train_numeric: Numerical variables we generated
# * x_train_words: Vectorized words from the clean lemmatized text
# * x_train_word_pos: Vectorized types of words
# * Select the top K variables - How many variables are appropriate?
# #### 3. Fit the Model & Predict
# * Fit the model with the training data
# * Predict the testing data
# * Generate a file for submission
# 1. SELECT MODEL HYPERPARAMETERS
clf = RandomForestClassifier(
random_state=1, n_estimators=100, min_samples_split=2, max_depth=20
)
# 2. PREPARE THE DATA
# Choose which groups of variables to use:
# x_train_numeric, x_train_words, x_train_word_pos
# x_test_numeric, x_test_words, x_test_word_pos
x_train_final_v2 = pd.concat([x_train_numeric, x_train_words, x_train_word_pos], axis=1)
x_test_final_v2 = pd.concat([x_test_numeric, x_test_words, x_test_word_pos], axis=1)
# Select top K variables:
top_k_features = 649
top_k_selector = SelectKBest(chi2, k=top_k_features)
x_train_final_v2 = top_k_selector.fit_transform(x_train_final_v2, y_train)
x_test_final_v2 = top_k_selector.transform(x_test_final_v2)
# Summary of variables
print("Shape of the training Dataset")
print("Features (X): ", x_train_final_v2.shape)
print("Target (Y): ", y_train.shape)
print("Shape of the testing Dataset")
print("Features (X): ", x_test_final_v2.shape)
# Fit the Model & Predict
model_rf_v2 = clf.fit(x_train_final_v2, y_train)
# Predict the testing data
y_test = clf.predict(x_test_final_v2)
# Prepare & save submission file
submission_file_v2 = test.copy()
submission_file_v2 = submission_file_v2[["Id"]]
submission_file_v2["Category"] = y_test
submission_file_v2.to_csv(r"submission_v2.csv", index=False)
# ## Final Tuning (Advanced)
grid = {
"n_estimators": [50, 100, 200, 500],
"max_depth": [10, 50, 100],
"criterion": ["gini", "entropy"],
"min_samples_split": [2, 10, 20, 50],
}
from sklearn.model_selection import GridSearchCV
rf_cv = GridSearchCV(
estimator=RandomForestClassifier(random_state=1), param_grid=grid, cv=5, verbose=2
)
rf_cv.fit(x_train_final_v2, y_train.values.ravel())
rf_cv.best_params_
clf = RandomForestClassifier(
random_state=1,
criterion="gini",
max_depth=None,
min_samples_split=2,
n_estimators=100,
)
# Fit the Model & Predict
model_rf_v3 = clf.fit(x_train_final_v2, y_train.values.ravel())
# Predict the testing data
y_test = clf.predict(x_test_final_v2)
# Prepare & save submission file
submission_file_v2 = test.copy()
submission_file_v2 = submission_file_v2[["Id"]]
submission_file_v2["Category"] = y_test
submission_file_v2.to_csv(r"submission_v2.csv", index=False)
|
# # Libraries
# Model packages
from sklearn.impute import KNNImputer
from sklearn.model_selection import train_test_split, cross_val_score
from sklearn.metrics import mean_squared_error as MSE
from sklearn.model_selection import RandomizedSearchCV
from sklearn.pipeline import Pipeline
from statsmodels.formula.api import ols
from sklearn.linear_model import LinearRegression
from sklearn.ensemble import GradientBoostingRegressor
from sklearn.tree import DecisionTreeRegressor
from sklearn.ensemble import RandomForestRegressor
# Standard Packages
import statsmodels.api as sm
import shap
import pandas as pd
import missingno as mnso
import matplotlib.pyplot as plt
import seaborn as sns
import numpy as np
import warnings
# Warning
warnings.filterwarnings("ignore")
# To export graphs
# # Load the data
train = pd.read_csv(
"/kaggle/input/house-prices-advanced-regression-techniques/train.csv"
)
test = pd.read_csv("/kaggle/input/house-prices-advanced-regression-techniques/test.csv")
# # Variable Descriptions
# |Variable Name | Description|
# |:--------------:|-------------:|
# |MSSubClass | Identifies the type of dwelling involved in the sale.|
# |MSZoning | Identifies the general zoning classification of the sale.|
# |LotFrontage | Linear feet of street connected to property|
# |LotArea | Lot size in square feet|
# |Street | Type of road access to property|
# |Alley | Type of alley access to property|
# |LotShape | General shape of property|
# |LandContour | Flatness of the property|
# |Utilities | Type of utilities available|
# |LotConfig | Lot configuration|
# |LandSlope | Slope of property|
# |Neighborhood | Physical locations within Ames city limits|
# |Condition1 | Proximity to various conditions|
# |Condition2 | Proximity to various conditions (if more than one is present)|
# |BldgType | Type of dwelling|
# |HouseStyle | Style of dwelling|
# |OverallQual | Rates the overall material and finish of the house|
# |OverallCond | Rates the overall condition of the house|
# |YearBuilt | Original construction date|
# |YearRemodAdd | Remodel date (same as construction date if no remodeling or additions)|
# |RoofStyle | Type of roof|
# |RoofMatl | Roof material|
# |Exterior1st | Exterior covering on house|
# |Exterior2nd | Exterior covering on house (if more than one material)|
# |MasVnrType | Masonry veneer type|
# |MasVnrArea | Masonry veneer area in square feet|
# |ExterQual | Evaluates the quality of the material on the exterior|
# |ExterCond | Evaluates the present condition of the material on the exterior|
# |Foundation | Type of foundation|
# |BsmtQual | Evaluates the height of the basement|
# |BsmtCond | Evaluates the general condition of the basement|
# |BsmtExposure | Refers to walkout or garden level walls|
# |BsmtFinType1 | Rating of basement finished area|
# |BsmtFinSF1 | Type 1 finished square feet|
# |BsmtUnfSF | Unfinished square feet of basement area|
# |TotalBsmtSF | Total square feet of basement area|
# |Heating | Type of heating|
# |HeatingQC | Heating quality and condition|
# |CentralAir | Central air conditioning|
# |Electrical | Electrical system|
# |1stFlrSF | First Floor square feet|
# |2ndFlrSF | Second floor square feet|
# |LowQualFinSF | Low quality finished square feet (all floors)|
# |GrLivArea | Above grade (ground) living area square feet|
# |BsmtFullBath | Basement full bathrooms|
# |BsmtHalfBath | Basement half bathrooms|
# |FullBath | Full bathrooms above grade|
# |HalfBath | Half baths above grade|
# |Bedroom | Bedrooms above grade (does NOT include basement bedrooms)|
# |Kitchen | Kitchens above grade|
# |KitchenQual | Kitchen quality|
# |TotRmsAbvGrd | Total rooms above grade (does not include bathrooms)|
# |Functional | Home functionality (Assume typical unless deductions are warranted)|
# |Fireplaces | Number of fireplaces|
# |FireplaceQu | Fireplace quality|
# |GarageType | Garage location|
# |GarageYrBlt | Year garage was built|
# |GarageFinish | Interior finish of the garage|
# |GarageCars | Size of garage in car capacity|
# |GarageArea | Size of garage in square feet|
# |GarageQual | Garage quality|
# |GarageCond | Garage condition|
# |PavedDrive | Paved driveway
# |WoodDeckSF | Wood deck area in square feet|
# |OpenPorchSF | Open porch area in square feet|
# |EnclosedPorch | Enclosed porch area in square feet|
# |3SsnPorch | Three season porch area in square feet|
# |ScreenPorch | Screen porch area in square feet|
# |PoolArea | Pool area in square feet|
# |PoolQC | Pool quality|
# |Fence | Fence quality|
# |MoSold | Month Sold (MM)|
# |YrSold | Year Sold (YYYY)|
# |SaleType | Type of sale|
# |SaleCondition | Condition of sale|
# # Data Integrity
# 
# **The Methodology That Will be Used For Dropping Columns**
# **'LotFrontage' and'GarageYrBlt' are treated with KNN Imputer since they have a lot of missing values.**
# **While implementing KNN Imputer,relatively similar or somewhat meaningful neighbors are used.**
# **To give an example, 'GarageYrBlt' is neighbored with 'YearBuilt'.**
# **Other variable, namely the ones with low missing values are imputed with median interpolation method**
#
#
#
train.drop(["MiscFeature", "BsmtFinType1", "BsmtFinType2", "Id"], axis=1, inplace=True)
mnso.matrix(train, labels=80, label_rotation=45, color=(0.25, 0.50, 0.50))
plt.show()
cols_with_nans = train.isnull().sum().sort_values()
cols_with_nans = cols_with_nans[cols_with_nans > 0]
cols_with_nans.plot(kind="barh", color="black")
plt.title("NaN Columns")
plt.show()
interpolation_threshold = len(train) * 0.05
print("5% of the data is: {:.2f}".format(interpolation_threshold))
print(
"Columns with missing values more than the threshold: {}".format(
cols_with_nans[cols_with_nans > interpolation_threshold]
)
)
column_list = [
"GarageFinish",
"GarageType",
"GarageQual",
"FireplaceQu",
"GarageCond",
"PoolQC",
"Fence",
"Alley",
"BsmtQual",
"BsmtExposure",
"BsmtCond",
"MasVnrType",
"Electrical",
]
change_list = [
"NG",
"NG",
"NG",
"NF",
"NG",
"NP",
"NF",
"NA",
"NB",
"NB",
"NB",
"NM",
"Mix",
]
for cl in column_list:
for chl in change_list:
train[cl].fillna(chl, inplace=True)
msavnr = train["MasVnrArea"].median()
train["MasVnrArea"].fillna(msavnr, inplace=True)
# # KNN Imputations
knnimp = KNNImputer().fit(train[["GarageYrBlt", "YearBuilt"]])
X_train_knn = knnimp.transform(train[["GarageYrBlt", "YearBuilt"]])
knnimp_fixedgr = pd.DataFrame(X_train_knn, columns=["GarageYrBlt", "YearBuilt"])
train["GarageYrBlt"] = knnimp_fixedgr["GarageYrBlt"]
train["GarageYrBlt"].isnull().sum()
knnimp = KNNImputer().fit(train[["LotFrontage", "LotArea"]])
X_train_knn = knnimp.transform(train[["LotFrontage", "LotArea"]])
knnimp_fixedlt = pd.DataFrame(X_train_knn, columns=["LotFrontage", "LotArea"])
train["LotFrontage"] = knnimp_fixedlt["LotFrontage"]
train["LotFrontage"].isnull().sum()
train.isnull().sum().plot(kind="bar")
plt.title("Controlling For Data Integrity After Imputations")
plt.xticks(rotation=90)
plt.legend(["Number of Columns: " + str(len(train.columns))])
plt.show()
train[["OverallQual", "OverallCond"]] = train[["OverallQual", "OverallCond"]].astype(
"object"
)
category_list = train.select_dtypes("object")
category_list = category_list.astype("category")
numeric_list = train.loc[:, ~train.columns.isin(category_list.columns)]
# # Exploratory Data Analysis
# **Examination of the effect of Lot and Housing Area properties**
#
# Let's first get started with Lot and Housing Properties of the Houses. Evidently, houses that are located at low residendial density (RL) neighborhood have the highest price median by far. Further, age of the building has also an impact on the sale price of houses, as expected.
#
# More, most of the buildings that are either historical or recently-renovated (new) are also located at low residential density neighborhoods. Thus, it makes sense that the sale price of houses that fall into this category to be higher.
#
# More importantly, sale price is affected by the following factors;
#
# * LotFrontage: Sale price is positively effected as square-feet increases.
# * LotArea: Sale price is positively effected as square-feet increases.
# * LotShape_Reg: Regular type of lot shape has an adverse effect on the sale price.
# * Foundation_CBlock: Evidently, foundations with cinder block are not taken into consideration when making purchasing decision by households.
# * Foundation_PConc: Inversely, Poured concrete is more preferred.
# * MSZoning_RL: Houses within low density areas (RL) have positive relationship with sale price.
# * MSZoning_RM: Medium density areas are not that much popular, since it effects the sale price adversely.
# * house_type_New: 1-12 year old (in terms of the years since the last renovation) are sold at higher prices.
#
# **We'll use'YearRemodAdd' and 'YearBuilt' to get the exact years since a recent renovation. Thus, we'll get to know if the house was renovated recently, or was renovated ever. Then, we'll categorize them. Thereon, we may infer in which category do sale prices fall due to the effect of variables, such as house type (renovated or not), foundation, and region. Let's build some hypotheses;**
#
# H_o : Explanatory variables have no effect on the sale price of houses
# h_a : Ex. variables affect the sale price of houses
train["house_age"] = train["YearRemodAdd"] - train["YearBuilt"]
train.loc[train["house_age"] == 0, "house_type"] = "Historic_Building"
train.loc[train["house_age"] >= 1, "house_type"] = "New"
train.loc[train["house_age"] >= 13, "house_type"] = "Standard"
train.loc[train["house_age"] >= 30, "house_type"] = "Old"
train.loc[train["house_age"] >= 60, "house_type"] = "Worn_Down"
# **Preliminary inference for the effect of renovation on the sale price suggests that the mean of different type of house differs from one another. Thus, we may apply statistical tests to obtain more concrete results. But before that, let's mingle up some more variables to understand the underlying reason of the effect of renovation. Is it the material used that affects the sale price?**
#
sns.boxplot(
data=train, x="SalePrice", y="house_type", showfliers=False, hue="Foundation"
)
plt.xticks(rotation=45)
plt.title("Sale Price of Different Foundation types")
plt.show()
# **Obviously, the material used for the renovation matters. We may infer that concrete foundation is more prefferable when compared to other materials. But, does region or location have anything to do with the used materials?**
sns.boxplot(data=train, x="SalePrice", y="house_type", showfliers=False, hue="MSZoning")
plt.xticks(rotation=45)
plt.title("Sale Price in Different Regions")
plt.show()
# **Three-way ANOVA test concludes that the sale price is affected by different House, Foundation, and Regional categories. Thereon, we may conclude that H_o is rejected, since the test results yield statistically significant (p_values < 0.05) mean differences and we should retain H_a.**
model = ols("SalePrice ~ C(house_type) + C(Foundation) + C(MSZoning)", data=train).fit()
result = sm.stats.anova_lm(model, type=2)
print(result)
# **Now let's have a look at the correlation coefficients of the remaining variables all together**
lot_area_properties = pd.get_dummies(
train[["LotShape", "Foundation", "LotShape", "MSZoning", "house_type"]],
drop_first=True,
)
# concatinating outer-house properties with other numerical values
lot_area_corr = pd.concat(
[train[["SalePrice", "LotFrontage", "LotArea", "house_age"]], lot_area_properties],
axis=1,
)
fig = plt.subplots(figsize=(16, 9))
sns.heatmap(data=lot_area_corr.corr(), robust=True)
plt.show()
# **The Effect of House Age and Residential Density on The Sale Price**
# Majority of the houses are either historical buildings or renovated within 1-12 years intervals.
sns.countplot(data=train, x="house_type", hue="Foundation")
plt.title("House Types With Different Foundation Materials")
plt.show()
sns.countplot(data=train, x="MSZoning", hue="house_type")
plt.title("House Types in Different Residential Areas")
plt.show()
train.groupby(["house_type"])["SalePrice"].median().sort_values(ascending=False)
# Historical buildings are predominatly located at
pd.crosstab(train["house_type"], train["MSZoning"], normalize=True)
sns.catplot(
data=train, x="SalePrice", y="Neighborhood", col="MSZoning", kind="box", orient="h"
)
plt.show()
# **Examination of the effect of Outer House Properties**
# Now, we take a look at outer house properties.
#
# - 'MasVnrArea' has a positive effect on the sale price.
#
# - Within the sub categories of 'HouseStyle' property, the only criterion which differentiates itself from other properties is that whether the house is 2 story or not. Additionally, it evidently affects the sale price positively.
#
# - Within the sub categories of RoofStyle property, the only two when it comes to preferences of households, or let's say the sale price, are that whether the roof is Gable or Hip style. The roof having a Gable style adversely affects the sale price, whereas the Hip style has a positive impact it comes to the sale price.
#
# - As a consideration for 'OverallQual', the overall quality being between 7 and 10 positively affect the sale price, whereas the quality being lower than 7 begins to detoriate in terms of effect on sale price.
#
# - The 'OverallCond' property being Average, or 5, seems to be the only positive differentiator when it comes to sale price.
#
# - 'RoofMatl' (Roof Materials) appears to have no significant influence on the sale price, other than a weak effect of roof material being either Brick Face or Wood Shingles.
#
# - Masonry veneer types are of significant determinants when it comes to the sale price. Stone and Brick Face masonry veener types have a positive impact, whereas having no masonry veener negatively affect the sale price
#
# - Exterior qualiy is obviously a positive determinant for the sale price. The higher the quality, the higher the sale price will be. However, strinkingly the condition of the exterior material has no impact on the sale price.
outer_house_properties = pd.get_dummies(
train[
[
"ExterQual",
"MasVnrType",
"OverallCond",
"RoofStyle",
"HouseStyle",
"OverallQual",
]
]
)
# concatinating outer-house properties with other numerical values
outer_house_corr = pd.concat(
[train[["SalePrice", "MasVnrArea"]], outer_house_properties], axis=1
)
fig = plt.subplots(figsize=(16, 9))
sns.heatmap(data=outer_house_corr.corr(), robust=True)
plt.show()
# **As ANOVA test states, both 'OverallCond' and 'OverallQual' are statistically significant variables to explain the changes in the sale price.**
model = ols("SalePrice ~ C(OverallCond) + C(OverallQual)", data=train).fit()
result = sm.stats.anova_lm(model, type=2)
print(result)
# **Examination of the effect of Exterior House Properties**
# * BsmtQual: The basement quality being 'excellent' or 'good' have positive effect on the sale price, though Average / Typical quality adversely affect it.
# * BsmtExposure: Having "good" exposure to walkout or garden level walls has a positive effect, though no access to a walkout out of the basement has an adverse effect on sale price.
# * TotalBsmtSF: Total basement square-feet has a moderate and significant impact on the sale price, which makes sense.
# * BsmtUnfSF: Unfinished basement square-feet has a weak influence on the sale price. Intuitively thinking, it is the very anticipation that hooks the households by knowing how big the basement will be when unfinished, in terms of square-feet, that influences them to make a decision to buy.
# * GarageType: Garage being 'built-in' (which typically has a room above) and 'attached' have positive effect on the sale price, whereas a detached garage (detached from home) adversely affects.
# * GarageYrBlt: Newer garages obviously attrachs more household attention, thus causing the sale price to be higher.
# * GarageFinish: Having a finished or unfinished garage interior is of significant determinant on the sale price.
# * GarageCars: While having a car capacity of 3 has a positive impact, having less than 2 adversly affects the sale price.
# * GarageArea: +
# * GarageQual: GarageQual_TA +
# * GarageCond: GarageCond_TA +
# * PavedDrive: PavedDrive_Y +, PavedDrive_N -
# * WoodDeckSF: +
# * OpenPorchSF: +
exterior_house_properties = pd.get_dummies(
train[
[
"MSSubClass",
"BsmtQual",
"BsmtExposure",
"GarageType",
"GarageFinish",
"GarageCars",
"GarageQual",
"GarageCond",
"PavedDrive",
"MSSubClass",
]
]
)
# concatinating outer-house properties with other numerical values
exterior_house_corr = pd.concat(
[
train[
[
"SalePrice",
"PoolArea",
"TotalBsmtSF",
"BsmtUnfSF",
"GarageYrBlt",
"GarageArea",
"WoodDeckSF",
"OpenPorchSF",
]
],
exterior_house_properties,
],
axis=1,
)
fig = plt.subplots(figsize=(16, 9))
sns.heatmap(data=exterior_house_corr.corr(), robust=True)
plt.show()
# **Examination of the effect of Interior House Properties**
# - HeatingQC : HeatingQC_Ex +, HeatingQC_TA -
# - CentralAir: CentralAir_Y + , CentralAir_N -
# - GrLivArea : ++
# - 1stFlrSF : ++
# - 2ndFlrSF : +
# - Electrical: Electrical_SBrkr +, Electrical_FuseA -
#
# - BsmtHalfBath: +
# - HalfBath: +
# - FullBath: ++
# - KitchenQual: KitchenQual_Ex ++, KitchenQual_Gd +, KitchenQual_TA --
# - TotRmsAbvGrd: ++
# - Fireplaces: ++
# - FireplaceQu: FireplaceQu_Ex +, FireplaceQu_Gd +, FireplaceQu_NG --
#
#
#
interior_house_properties = pd.get_dummies(
train[["HeatingQC", "CentralAir", "Electrical", "KitchenQual", "FireplaceQu"]]
)
# concatinating outer-house properties with other numerical values
interior_house_corr = pd.concat(
[
train[
[
"SalePrice",
"TotRmsAbvGrd",
"Fireplaces",
"GrLivArea",
"1stFlrSF",
"2ndFlrSF",
"HalfBath",
"FullBath",
]
],
interior_house_properties,
],
axis=1,
)
fig = plt.subplots(figsize=(16, 9))
sns.heatmap(data=interior_house_corr.corr(), robust=True)
plt.show()
# **Examination of the effect of Sale Properties**
# - SaleType: SaleType_New + SaleType_WD -
# - SaleCondition: SaleCondition_Partial +
sale_properties = pd.get_dummies(train[["SaleType", "SaleCondition"]])
# concatinating outer-house properties with other numerical values
sale_corr = pd.concat([train["SalePrice"], sale_properties], axis=1)
fig = plt.subplots(figsize=(16, 9))
sns.heatmap(data=sale_corr.corr(), robust=True)
plt.show()
# **Let's get all the information together that we've gathered thus far.**
categorical_dummies = pd.concat(
[
sale_properties,
interior_house_properties,
exterior_house_corr,
outer_house_properties,
lot_area_properties,
],
axis=1,
)
final_corr = pd.concat(
[
train[
[
"SalePrice",
"Fireplaces",
"TotRmsAbvGrd",
"FullBath",
"HalfBath",
"1stFlrSF",
"2ndFlrSF",
"GrLivArea",
"OpenPorchSF",
"WoodDeckSF",
"GarageArea",
"GarageYrBlt",
"BsmtUnfSF",
"TotalBsmtSF",
"GarageCars",
"MasVnrArea",
"LotFrontage",
"LotArea",
"house_age",
]
],
categorical_dummies[
[
"SaleType_New",
"SaleType_WD",
"HeatingQC_Ex",
"HeatingQC_TA",
"CentralAir_Y",
"CentralAir_N",
"Electrical_SBrkr",
"Electrical_FuseA",
"KitchenQual_Ex",
"KitchenQual_Gd",
"KitchenQual_TA",
"FireplaceQu_Ex",
"FireplaceQu_Gd",
"FireplaceQu_NG",
"PavedDrive_Y",
"PavedDrive_N",
"GarageQual_TA",
"GarageCond_TA",
"GarageFinish_Fin",
"GarageFinish_Unf",
"GarageType_BuiltIn",
"GarageType_Attchd",
"GarageType_Detchd",
"BsmtExposure_Gd",
"BsmtExposure_No",
"BsmtQual_Ex",
"BsmtQual_Gd",
"BsmtQual_TA",
"HouseStyle_2Story",
"OverallQual_4",
"OverallQual_5",
"OverallQual_6",
"OverallQual_7",
"OverallQual_8",
"OverallQual_9",
"OverallQual_10",
"OverallCond_5",
"MasVnrType_BrkFace",
"MasVnrType_Stone",
"MasVnrType_None",
"ExterQual_Ex",
"ExterQual_Fa",
"ExterQual_Gd",
"ExterQual_TA",
"RoofStyle_Gable",
"RoofStyle_Hip",
"house_type_New",
"LotShape_Reg",
"Foundation_CBlock",
"Foundation_PConc",
"MSZoning_RL",
"MSZoning_RM",
]
],
],
axis=1,
)
fig = plt.subplots(figsize=(16, 9))
sns.heatmap(
data=final_corr.corr(),
robust=True,
)
plt.show()
# # Model Building
X = final_corr.drop(["SalePrice"], axis=1)
y = final_corr["SalePrice"]
X_train, X_test, y_train, y_test = train_test_split(
X,
y,
test_size=0.3,
shuffle=True,
random_state=31,
)
X_train.shape, y_train.shape
sgbr = GradientBoostingRegressor(subsample=0.85, random_state=31)
sgbr.fit(X_train, y_train)
y_pred = sgbr.predict(X_test)
print(
"Stochastic GB Classifier Accuracy Score: {:.3f}".format(sgbr.score(X_test, y_test))
)
# # Feature Importance with SHAP
shap.initjs()
explainer = shap.Explainer(sgbr)
shap_values = explainer.shap_values(X_train)
i = 71
shap.force_plot(
explainer.expected_value,
shap_values[i],
features=X.iloc[i],
feature_names=X_train.columns,
)
shap.summary_plot(shap_values, features=X_train, feature_names=X_train.columns)
shap.summary_plot(
shap_values, features=X_train, feature_names=X.columns, plot_type="bar"
)
# # Cross Validation With RandomizedSearch
params_grid = {
"max_depth": [3, 4, 5, 6, 7, 8, 9, 10],
"n_estimators": [100, 200, 300, 350, 400],
"subsample": [0.7, 0.75, 0.8, 0.85, 0.9],
"max_features": ["sqrt", "log2"],
"min_samples_leaf": [0.10, 0.15, 0.2, 0.25, 0.3, 0.35],
"criterion": ["friedman_mse", "squared_error"],
}
rs_cv = RandomizedSearchCV(
estimator=sgbr,
param_distributions=params_grid,
cv=10,
scoring="neg_mean_squared_error",
random_state=31,
)
rs_cv.fit(X_train, y_train)
y_pred_cv = rs_cv.predict(X_test)
mse = MSE(y_test, y_pred_cv)
rmse = mse ** (1 / 2)
print("RMSE for SGB before Tuning : {:.3f}".format(rmse))
# # Prediction Result
# **After applying RandomizedSeach Cross Validation, the prediction accuracy has jumped from 0.810 to 0.824, concluding a +1.72% improvement.**
# Pipeline for GradientBoostingRegressor()
pipelinesgb = Pipeline([("SGB", GradientBoostingRegressor(random_state=42))])
# parameters for gradient boosting
sgb_params_grid = [
{
"SGB__max_depth": [3, 14, 15, 16],
"SGB__n_estimators": [100, 900, 950, 1100],
"SGB__min_samples_split": [0.2, 0.6, 0.7, 0.8],
"SGB__subsample": [0.9, 0.95, 1],
"SGB__learning_rate": [0.0001, 0.001, 0.01, 0.1, 1],
}
]
# RandomizedSearch CV on GradientBoosting
sgb_rs_cv = RandomizedSearchCV(
estimator=pipelinesgb,
param_distributions=sgb_params_grid,
scoring="neg_mean_squared_error",
)
# Fit and Predict the best Stochastic Gradient Boosting Regressor model
sgb_rs_cv.fit(X_train, y_train)
y_pred_sgb = sgb_rs_cv.predict(X_test)
sgb_mse = MSE(y_test, y_pred_sgb)
sgb_rmse = sgb_mse ** (1 / 2)
print(
"Stochastic Gradient Boosting RMSE: {:.3f}".format(sgb_rmse),
"\nBest Parameters: {}".format(sgb_rs_cv.best_params_),
"\nBest Estimators: {}".format(sgb_rs_cv.best_estimator_),
)
def objective(trial):
param = {
"max_depth": trial.suggest_int("max_depth", 1, 10),
"learning_rate": trial.suggest_float("learning_rate", 0.01, 1),
"n_estimators": trial.suggest_int("n_estimators", 50, 500),
"min_child_weight": trial.suggest_int("min_child_weight", 1, 10),
"gamma": trial.suggest_float("gamma", 0.01, 1),
"subsample": trial.suggest_float("subsample", 0.01, 1),
"colsample_bytree": trial.suggest_float("colsample_bytree", 0.01, 1),
"reg_alpha": trial.suggest_float("reg_alpha", 0.01, 1),
"reg_lambda": trial.suggest_float("reg_lambda", 0.01, 1),
}
model = xgboost.XGBRegressor(**param)
model.fit(X_train.values, y_train.values)
y_pred = model.predict(X_test)
return (mean_squared_error(y_test, y_pred)) ** (1 / 2)
study = optuna.create_study(direction="minimize", study_name="XGBregression")
study.optimize(objective, n_trials=100)
modelxgb = xgboost.XGBRegressor(**study.best_params)
modelxgb.fit(X_train.values, y_train.values)
y_pred = modelxgb.predict(X_test)
print("MSE: ", mean_squared_error(y_test, y_pred))
print("RMSE: ", np.sqrt(mean_squared_error(y_test, y_pred)))
# # Submission
test.drop(["MiscFeature", "BsmtFinType1", "BsmtFinType2"], axis=1, inplace=True)
column_list = [
"GarageFinish",
"GarageType",
"GarageQual",
"FireplaceQu",
"GarageCond",
"PoolQC",
"Fence",
"Alley",
"BsmtQual",
"BsmtExposure",
"BsmtCond",
"MasVnrType",
"Electrical",
]
change_list = [
"NG",
"NG",
"NG",
"NF",
"NG",
"NP",
"NF",
"NA",
"NB",
"NB",
"NB",
"NM",
"Mix",
]
for cl in column_list:
for chl in change_list:
test[cl].fillna(chl, inplace=True)
knnimp_test = KNNImputer().fit(test[["GarageYrBlt", "YearBuilt"]])
X_test_knn_test = knnimp_test.transform(test[["GarageYrBlt", "YearBuilt"]])
knnimp_fixedgr_test = pd.DataFrame(
X_test_knn_test, columns=["GarageYrBlt", "YearBuilt"]
)
test["GarageYrBlt"] = knnimp_fixedgr_test["GarageYrBlt"]
test["GarageYrBlt"].isnull().sum()
msavnr = test["MasVnrArea"].median()
test["MasVnrArea"].fillna(msavnr, inplace=True)
knnimp_test = KNNImputer().fit(test[["LotFrontage", "LotArea"]])
X_train_knn_test2 = knnimp.transform(test[["LotFrontage", "LotArea"]])
knnimp_fixedlt_test = pd.DataFrame(
X_train_knn_test2, columns=["LotFrontage", "LotArea"]
)
test["LotFrontage"] = knnimp_fixedlt_test["LotFrontage"]
test["LotFrontage"].isnull().sum()
test[["OverallQual", "OverallCond"]] = test[["OverallQual", "OverallCond"]].astype(
"object"
)
category_list = test.select_dtypes("object")
category_list = category_list.astype("category")
numeric_list = test.loc[:, ~test.columns.isin(category_list.columns)]
test["house_age"] = test["YearRemodAdd"] - test["YearBuilt"]
test.loc[test["house_age"] == 0, "house_type"] = "Historic_Building"
test.loc[test["house_age"] >= 1, "house_type"] = "New"
test.loc[test["house_age"] >= 13, "house_type"] = "Standard"
test.loc[test["house_age"] >= 30, "house_type"] = "Old"
test.loc[test["house_age"] >= 60, "house_type"] = "Worn_Down"
lot_area_properties_test = pd.get_dummies(
test[["LotShape", "Foundation", "LotShape", "MSZoning", "house_type"]],
drop_first=True,
)
lot_area_corr_test = pd.concat(
[test[["LotFrontage", "LotArea", "house_age"]], lot_area_properties_test], axis=1
)
outer_house_properties_test = pd.get_dummies(
test[
[
"ExterQual",
"MasVnrType",
"OverallCond",
"RoofStyle",
"HouseStyle",
"OverallQual",
]
]
)
outer_house_corr_test = pd.concat(
[test["MasVnrArea"], outer_house_properties_test], axis=1
)
exterior_house_properties_test = pd.get_dummies(
test[
[
"MSSubClass",
"BsmtQual",
"BsmtExposure",
"GarageType",
"GarageFinish",
"GarageCars",
"GarageQual",
"GarageCond",
"PavedDrive",
"MSSubClass",
]
]
)
exterior_house_corr_test = pd.concat(
[
test[
[
"PoolArea",
"TotalBsmtSF",
"BsmtUnfSF",
"GarageYrBlt",
"GarageArea",
"WoodDeckSF",
"OpenPorchSF",
]
],
exterior_house_properties_test,
],
axis=1,
)
interior_house_properties_test = pd.get_dummies(
test[["HeatingQC", "CentralAir", "Electrical", "KitchenQual", "FireplaceQu"]]
)
interior_house_corr_test = pd.concat(
[
test[
[
"TotRmsAbvGrd",
"Fireplaces",
"GrLivArea",
"1stFlrSF",
"2ndFlrSF",
"HalfBath",
"FullBath",
]
],
interior_house_properties_test,
],
axis=1,
)
sale_properties_test = pd.get_dummies(test[["SaleType", "SaleCondition"]])
categorical_dummies_test = pd.concat(
[
sale_properties_test,
interior_house_properties_test,
exterior_house_corr_test,
outer_house_properties_test,
lot_area_properties_test,
],
axis=1,
)
final_corr_test = pd.concat(
[
test[
[
"Fireplaces",
"TotRmsAbvGrd",
"FullBath",
"HalfBath",
"1stFlrSF",
"2ndFlrSF",
"GrLivArea",
"OpenPorchSF",
"WoodDeckSF",
"GarageArea",
"GarageYrBlt",
"BsmtUnfSF",
"TotalBsmtSF",
"GarageCars",
"MasVnrArea",
"LotFrontage",
"LotArea",
"house_age",
]
],
categorical_dummies_test[
[
"SaleType_New",
"SaleType_WD",
"HeatingQC_Ex",
"HeatingQC_TA",
"CentralAir_Y",
"CentralAir_N",
"Electrical_SBrkr",
"Electrical_FuseA",
"KitchenQual_Ex",
"KitchenQual_Gd",
"KitchenQual_TA",
"FireplaceQu_Ex",
"FireplaceQu_Gd",
"FireplaceQu_NG",
"PavedDrive_Y",
"PavedDrive_N",
"GarageQual_TA",
"GarageCond_TA",
"GarageFinish_Fin",
"GarageFinish_Unf",
"GarageType_BuiltIn",
"GarageType_Attchd",
"GarageType_Detchd",
"BsmtExposure_Gd",
"BsmtExposure_No",
"BsmtQual_Ex",
"BsmtQual_Gd",
"BsmtQual_TA",
"HouseStyle_2Story",
"OverallQual_4",
"OverallQual_5",
"OverallQual_6",
"OverallQual_7",
"OverallQual_8",
"OverallQual_9",
"OverallQual_10",
"OverallCond_5",
"MasVnrType_BrkFace",
"MasVnrType_Stone",
"MasVnrType_None",
"ExterQual_Ex",
"ExterQual_Fa",
"ExterQual_Gd",
"ExterQual_TA",
"RoofStyle_Gable",
"RoofStyle_Hip",
"house_type_New",
"LotShape_Reg",
"Foundation_CBlock",
"Foundation_PConc",
"MSZoning_RL",
"MSZoning_RM",
]
],
],
axis=1,
)
TotalBsmtSF_median = final_corr_test["TotalBsmtSF"].median()
final_corr_test["TotalBsmtSF"] = final_corr_test["TotalBsmtSF"].fillna(
TotalBsmtSF_median
)
BsmtUnfSF_median = final_corr_test["BsmtUnfSF"].median()
final_corr_test["BsmtUnfSF"] = final_corr_test["BsmtUnfSF"].fillna(BsmtUnfSF_median)
GarageCars_median = final_corr_test["GarageCars"].median()
final_corr_test["GarageCars"] = final_corr_test["GarageCars"].fillna(GarageCars_median)
GarageArea_median = final_corr_test["GarageArea"].median()
final_corr_test["GarageArea"] = final_corr_test["GarageArea"].fillna(GarageArea_median)
final_corr_test.isnull().sum().sort_values(ascending=True).tail(15)
test_preds = modelxgb.predict(final_corr_test)
submission_df = pd.DataFrame({"Id": test["Id"], "SalePrice": test_preds})
submission_df.to_csv("submission.csv", index=False)
submission_df.head(5)
|
# # Installing Dependencies
# # Imports
import os
import tensorflow as tf
from tensorflow.keras import backend
import matplotlib.pyplot as plt
# # Dataset
# ### Function to read the image file
def load_image_file(image_path, mask_path):
image = tf.io.read_file(image_path)
mask = tf.io.read_file(mask_path)
image = tf.image.decode_jpeg(image, channels=3)
mask = tf.image.decode_png(mask, channels=1)
return {"image": image, "segmentation_mask": mask}
# ### Loading the dataset
train_image_dir = "/kaggle/input/landslide-divided/dataset/train/images"
train_mask_dir = "/kaggle/input/landslide-divided/dataset/train/masks"
valid_image_dir = "/kaggle/input/landslide-divided/dataset/validation/images"
valid_mask_dir = "/kaggle/input/landslide-divided/dataset/validation/masks"
test_image_dir = "/kaggle/input/landslide-divided/dataset/test/images"
test_mask_dir = "/kaggle/input/landslide-divided/dataset/test/masks"
# Define list of image and mask file names
train_image_names = sorted(os.listdir(train_image_dir))
train_mask_names = sorted(os.listdir(train_mask_dir))
valid_image_names = sorted(os.listdir(valid_image_dir))
valid_mask_names = sorted(os.listdir(valid_mask_dir))
test_image_names = sorted(os.listdir(test_image_dir))
test_mask_names = sorted(os.listdir(test_mask_dir))
train_pairs = []
for img_name in train_image_names:
# Check if image file name matches mask file name
mask_name = img_name.replace("image", "mask")
if mask_name in train_mask_names:
train_pairs.append(
(
os.path.join(train_image_dir, img_name),
os.path.join(train_mask_dir, mask_name),
)
)
valid_pairs = []
for img_name in valid_image_names:
# Check if image file name matches mask file name
mask_name = img_name.replace("image", "mask")
if mask_name in valid_mask_names:
valid_pairs.append(
(
os.path.join(valid_image_dir, img_name),
os.path.join(valid_mask_dir, mask_name),
)
)
test_pairs = []
for img_name in test_image_names:
# Check if image file name matches mask file name
mask_name = img_name.replace("image", "mask")
if mask_name in test_mask_names:
test_pairs.append(
(
os.path.join(test_image_dir, img_name),
os.path.join(test_mask_dir, mask_name),
)
)
# Load image and mask data from file paths
data_train = [
load_image_file(image_path, mask_path) for image_path, mask_path in train_pairs
]
data_valid = [
load_image_file(image_path, mask_path) for image_path, mask_path in valid_pairs
]
data_test = [
load_image_file(image_path, mask_path) for image_path, mask_path in test_pairs
]
len(data_train), len(data_valid), len(data_test)
# ### Normalization and Image Resizing
# #### P.S. You could do data augmentation here as well. I kept it very simple
image_size = 256
mean = tf.constant([0.485, 0.456, 0.406])
std = tf.constant([0.229, 0.224, 0.225])
def normalize(input_image, input_mask):
input_image = tf.image.convert_image_dtype(input_image, tf.float32)
input_image = (input_image - mean) / tf.maximum(std, backend.epsilon())
input_mask = input_mask / 255
return input_image, input_mask
def load_image(datapoint):
input_image = tf.image.resize(datapoint["image"], (image_size, image_size))
input_mask = tf.image.resize(
datapoint["segmentation_mask"],
(image_size, image_size),
method="bilinear",
)
input_image, input_mask = normalize(input_image, input_mask)
input_image = tf.transpose(input_image, (2, 0, 1))
return {"pixel_values": input_image, "labels": tf.squeeze(input_mask)}
train_data = [load_image(datapoint) for datapoint in data_train]
valid_data = [load_image(datapoint) for datapoint in data_valid]
test_data = [load_image(datapoint) for datapoint in data_test]
plt.imshow(train_data[0]["labels"])
def generator_train():
for datapoint in train_data:
yield datapoint
def generator_valid():
for datapoint in valid_data:
yield datapoint
def generator_test():
for datapoint in test_data:
yield datapoint
# ### Using tf.data.Dataset to build input pipeline
batch_size = 4
auto = tf.data.AUTOTUNE
train_ds = (
tf.data.Dataset.from_generator(
generator_train, output_types={"pixel_values": tf.float32, "labels": tf.int32}
)
.cache()
.shuffle(batch_size * 10)
.batch(batch_size)
.prefetch(auto)
)
valid_ds = (
tf.data.Dataset.from_generator(
generator_valid, output_types={"pixel_values": tf.float32, "labels": tf.int32}
)
.batch(batch_size)
.prefetch(auto)
)
test_ds = (
tf.data.Dataset.from_generator(
generator_test, output_types={"pixel_values": tf.float32, "labels": tf.int32}
)
.batch(batch_size)
.prefetch(auto)
)
print(train_ds.element_spec)
# # Visualizing the data
def display(display_list):
plt.figure(figsize=(15, 15))
title = ["Input Image", "True Mask", "Predicted Mask"]
for i in range(len(display_list)):
plt.subplot(1, len(display_list), i + 1)
plt.title(title[i])
plt.imshow(tf.keras.utils.array_to_img(display_list[i]))
plt.axis("off")
plt.show()
for samples in train_ds.take(2):
sample_image, sample_mask = samples["pixel_values"][0], samples["labels"][0]
sample_image = tf.transpose(sample_image, (1, 2, 0))
sample_mask = tf.expand_dims(sample_mask, -1)
display([sample_image, sample_mask])
print(sample_image.shape)
# # Model
from transformers import TFSegformerForSemanticSegmentation
model_checkpoint = "nvidia/mit-b1"
id2label = {0: "outer", 1: "landslide"}
label2id = {label: id for id, label in id2label.items()}
num_labels = len(id2label)
model = TFSegformerForSemanticSegmentation.from_pretrained(
model_checkpoint,
num_labels=num_labels,
id2label=id2label,
label2id=label2id,
ignore_mismatched_sizes=True,
)
# # Hyperparameters and compiling the model
lr = 0.00006
epochs = 5
optimizer = tf.keras.optimizers.Adam(learning_rate=lr)
model.compile(optimizer=optimizer)
# ### Callback to visualize image after every epoch
from IPython.display import clear_output
def create_mask(pred_mask):
pred_mask = tf.math.argmax(pred_mask, axis=1)
pred_mask = tf.expand_dims(pred_mask, -1)
return pred_mask[0]
def show_predictions(dataset=None, num=1):
if dataset:
for sample in dataset.take(num):
images, masks = sample["pixel_values"], sample["labels"]
masks = tf.expand_dims(masks, -1)
pred_masks = model.predict(images).logits
images = tf.transpose(images, (0, 2, 3, 1))
display([images[0], masks[0], create_mask(pred_masks)])
else:
display(
[
sample_image,
sample_mask,
create_mask(model.predict(tf.expand_dims(sample_image, 0))),
]
)
class DisplayCallback(tf.keras.callbacks.Callback):
def __init__(self, dataset, **kwargs):
super().__init__(**kwargs)
self.dataset = dataset
def on_epoch_end(self, epoch, logs=None):
clear_output(wait=True)
show_predictions(self.dataset)
print("\nSample Prediction after epoch {}\n".format(epoch + 1))
# # Training Loop
history = model.fit(
train_ds,
validation_data=valid_ds,
callbacks=[DisplayCallback(test_ds)],
epochs=epochs,
)
model.save_weights("/kaggle/working/segformer-5-b1.h5")
# # Loss Plot
plt.style.use("seaborn")
def display_training_curves(training, validation, title, subplot):
ax = plt.subplot(subplot)
ax.plot(training)
ax.plot(validation)
ax.set_title("Model " + title)
ax.set_ylabel(title)
ax.set_xlabel("epoch")
ax.legend(["training", "validation"])
plt.subplots(figsize=(8, 8))
plt.tight_layout()
display_training_curves(
history.history["loss"], history.history["val_loss"], "Loss", 111
)
plt.savefig("train_eval_plot_segformer-5-b1.jpg")
# # Predictions
show_predictions(valid_ds, 10)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# # Libraries, functions, variables...
import matplotlib as mpl
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
def set1(fig_size, face_color):
sns.set_theme(
font_scale=1,
rc={
"figure.figsize": fig_size,
"axes.facecolor": face_color,
"axes.edgecolor": "#AFAFAF",
"axes.grid": True,
"grid.color": "#E9E9E9",
"legend.fontsize": 12,
"legend.title_fontsize": 15,
},
)
colores = [
"black",
"white",
"orange",
"navy",
"tomato",
"crimson",
"darkgreen",
"brown",
"forestgreen",
"steelblue",
"deeppink",
"darkorange",
"palegreen",
"gold",
"darkviolet",
"slategray",
"indigo",
"peru",
"lightcoral",
"rosybrown",
"darkkhaki",
"lightcoral",
"teal",
"salmon",
"orchid",
"turquoise",
]
# # Credits
# Dataset created by Meaga Varsha Ramakrishna https://www.kaggle.com/datasets/varsharam/walmart-sales-dataset-of-45stores
# # Uploading and inspecting the data
df = pd.read_csv(dirname + "/" + filenames[0])
print(df.info())
# We have a full database, no nulls, but Date as str and Store as ints. I will create new Date and Store:
df["date2"] = pd.to_datetime(df["Date"], format="%d-%m-%Y")
df["store2"] = ["%02d" % item for item in df["Store"]]
# Let´s have a look at what we have in the database:
stores = df["store2"].unique()
dates = df["Date"].unique()
holidays = df["Holiday_Flag"].unique()
temperatures = df["Temperature"].unique()
fuel = df["Fuel_Price"].unique()
inflation = df["CPI"].unique()
unemployment = df["Unemployment"].unique()
print(f"Stores size: {len(stores)} From: {stores[0]} to: {stores[-1]}")
print("-" * 50)
print(f"Date size: {len(dates)} From: {dates[0]} to: {dates[-1]}")
print("-" * 50)
print(f"Holidays Flag {holidays}")
print("-" * 50)
print(
f"Temp size: {len(temperatures)} From: {temperatures.min()} to: {temperatures.max()}"
)
print("-" * 50)
print(f"Fuel size: {len(fuel)} From: {fuel.min()} to: {fuel.max()}")
print("-" * 50)
print(
f"CPI size: {len(inflation)} From: {inflation.min():.3f} to: {inflation.max():.3f}"
)
print("-" * 50)
print(
f"Unemployment size: {len(unemployment)} From: {unemployment.min():.3f} to: {unemployment.max():.3f}"
)
print("-" * 50)
print(f"Total Sales {df['Weekly_Sales'].sum():,.0f}")
# As we have 45 stores and 143 days, the total number of observations should be:
print(f"Total observations: {45*143:,.0f}")
holidays = df[df["Holiday_Flag"] == 1]["date2"].unique()
print(f"Weeks marked as holidays: {holidays}")
# # Chart functions and Viz
def chart(
set_1,
set_2,
df,
columna_x,
columna_y,
label_x,
label_y,
lim_lo_y="aut",
lim_hi_y="aut",
color_code=0,
chart_title="",
title_size=30,
show_c="Y",
):
set1(set_1, set_2)
paleta = [colores[i] for i in [0, 1, 6, 3]]
ax = sns.lineplot(
data=df,
x=columna_x,
y=columna_y,
color=colores[color_code],
estimator="sum",
)
ax.tick_params(axis="x", rotation=30)
ax.set_xlabel(label_x)
ax.set_ylabel(label_y)
ax.yaxis.set_major_formatter(mpl.ticker.StrMethodFormatter("{x:,.0f}"))
if lim_lo_y != "aut":
ax.set_ylim(lim_lo_y, lim_hi_y)
ax.set_title(chart_title, fontsize=title_size)
if show_c == "Y":
plt.show()
ax = chart(
(20, 8),
"#F8FBF8",
df,
"date2",
"Weekly_Sales",
"Date",
"Weekly Sales",
color_code=5,
chart_title="Weekly Sales all stores",
)
# Very valuable information. We can clearly see Black Friday and Christmas (and the January drop). Probably we will need to remove them from our analysis. The rest of the year, somehow stable, with the limited information of less than 3 y
color_code = 1
for store in stores:
df_sel = df[df["store2"] == store]
chart(
(20, 8),
"black",
df_sel,
"date2",
"Weekly_Sales",
"",
"Weekly Sales",
color_code=color_code,
chart_title="Joy Division",
title_size=30,
show_c="N",
)
# Not adding any value, but it reminds me of the Joy Division album, very cool.
# # Goal
# Our goal will be to estiamte the last 4 weeks of the series and compare with the actual values. So, we remove the last 4 weeks and store the data as test dataframe. The new df will exclude those observations.
df_raw = df.copy()
weeks_test = pd.date_range("2012-10-05", periods=4, freq="w") + pd.DateOffset(days=-2)
df_test = df[df["date2"].isin(weeks_test)]
df = df[~df["date2"].isin(weeks_test)]
print(f"Total Sales excluding last 4 weeks: {df['Weekly_Sales'].sum():,.0f}")
print(f"Weeks per store: {len(df['Date'].unique())}")
print(f"Total Observations: {len(df):,.0f}")
print(f"Check: {45*139:,.0f}")
# First temptation would be to calculate correlation alltogether and chart it in the usual heatmap. However, there is no guarantee that all stores will behave the same way, so I wanted to see first the correlation for each store.
dfc = pd.DataFrame() # I create a clear df to store the correlation per store
for store in stores:
df_sel = df[df["store2"] == store]
correlation = df_sel.iloc[:, 1:].corr()[
"Weekly_Sales"
] # correlation each store with weekly sales
store_series = pd.Series([store], index=["store2"]) # add the store number
correlation = correlation.append(store_series)
dfc = dfc.append(correlation, ignore_index=True) # append it to the dfc
dfc = dfc.drop(columns="Weekly_Sales")
columnas_corr = list(dfc.columns)
columnas_corr.remove("store2")
columna_x = "store2"
label_x = "Stores"
color_code = 5
chart_title = "Weekly Sales all stores"
title_size = 20
set1((20, 4), "#F8FBF8")
for columna in columnas_corr:
columna_y = columna
ax = sns.barplot(
data=dfc,
x=columna_x,
y=columna_y,
color=colores[color_code],
)
color_code += 1
ax.tick_params(axis="x", rotation=0)
ax.set_xlabel(label_x)
ax.set_ylabel(columna)
ax.yaxis.set_major_formatter(mpl.ticker.StrMethodFormatter("{x:,.4f}"))
ax.set_title(columna, fontsize=title_size)
plt.show()
# Comments: low correlation to holidays (expected and the number of vacations is low), but this first chart shows something interesting: there are stores behaving kind of weird with vacation: 30, 36, 37, 38, and 44 are negative. 34 might be positive but low, same for 43. We need to look at these more closely.
# Temperature didn't show consistent correlation either. I guess stores in the north and south would behave differently when temperatures increase or decrease.
# Fuel and CPI are very interesting: the are very similar, it seems there is strong correlation between the two. But still, in general low correlation, except some of the unusual stores (30 on). It seems we can ignore one of the them, but even doing this, the correlation is low.
# Unemployment, again same extrange end of the list results (35 on), and prior to that, low correlation.
# So, while we see we have a few stores we need to look at more closely, I think I will go to univariate forecasting, per store.
# Just to confirm, I will calcualte correlation between CPI and the other parameters:
dfc2 = pd.DataFrame()
for store in stores:
df_sel = df[df["store2"] == store]
correlation = df_sel.iloc[:, 1:].corr()["CPI"]
store_series = pd.Series([store], index=["store2"])
correlation = correlation.append(store_series)
dfc2 = dfc2.append(correlation, ignore_index=True)
dfc2 = dfc2.drop(columns="CPI")
columnas_corr = list(dfc.columns)
columnas_corr.remove("store2")
columnas_corr.remove("CPI")
columna_x = "store2"
label_x = "Stores"
color_code = 5
chart_title = "Weekly Sales all stores"
title_size = 20
set1((20, 4), "#F8FBF8")
for columna in columnas_corr:
columna_y = columna
ax = sns.barplot(
data=dfc2,
x=columna_x,
y=columna_y,
color=colores[color_code],
)
color_code += 1
ax.tick_params(axis="x", rotation=0)
ax.set_xlabel(label_x)
ax.set_ylabel(columna)
ax.yaxis.set_major_formatter(mpl.ticker.StrMethodFormatter("{x:,.4f}"))
ax.set_title(columna, fontsize=title_size)
plt.show()
# Really interesting to see CPI related to Holiday Flag, in stores 7, 16, 32, and 41. Curious.
# As expected, correlation between CPI and Fuel is very high, between 0.7 and 0.9.
# And store 24 is also strange CPI vs Unemployment. Another reason not to use this for the moment.
# So, let's have a look at the stores individually:
dfg_s = df.groupby(by="store2", as_index=False).sum()
dfg_d = df.groupby(by="date2", as_index=False).sum()
print(f"Test {dfg_s['Weekly_Sales'].sum():,.0f}")
print(f"Test {dfg_d['Weekly_Sales'].sum():,.0f}")
columna_x = "store2"
label_x = "Stores"
label_y = "Total Sales"
color_code = 3
chart_title = "Total Sales per stores"
title_size = 20
set1((20, 8), "#F8FBF8")
columna_y = "Weekly_Sales"
ax = sns.barplot(
data=dfg_s,
x=columna_x,
y=columna_y,
color=colores[color_code],
)
ax.tick_params(axis="x", rotation=0)
ax.set_xlabel(label_x)
ax.set_ylabel(label_y)
ax.yaxis.set_major_formatter(mpl.ticker.StrMethodFormatter("{x:,.0f}"))
ax.set_title(chart_title, fontsize=title_size)
plt.show()
# We can split the stores in 4 (arbitrary) categories: small, medium, large and mega:
def type_val(item):
if item >= 225000000:
type_val = "mega"
elif item >= 150000000:
type_val = "large"
elif item >= 75000000:
type_val = "medium"
else:
type_val = "small"
return type_val
dfg_s["type"] = [type_val(item) for item in dfg_s["Weekly_Sales"]]
set1((20, 8), "#F8FBF8")
columna_y = "Weekly_Sales"
paleta = [colores[i] for i in [5, 8, 11, 12]]
ax = sns.barplot(
data=dfg_s, x=columna_x, y=columna_y, dodge=False, hue="type", palette=paleta
)
ax.tick_params(axis="x", rotation=0)
ax.set_xlabel(label_x)
ax.set_ylabel(label_y)
ax.yaxis.set_major_formatter(mpl.ticker.StrMethodFormatter("{x:,.0f}"))
ax.set_title(chart_title, fontsize=title_size)
plt.show()
# We create a meta df that will help later.
meta_df = dfg_s[["store2", "type"]]
df = df.merge(meta_df, how="left", on="store2")
columna_x = "date2"
label_x = "Date"
label_y = "Total Sales"
color_code = 3
chart_title = "Total Sales per week all stores"
title_size = 20
set1((20, 8), "#F8FBF8")
columna_y = "Weekly_Sales"
ax = sns.lineplot(
data=dfg_d,
x=columna_x,
y=columna_y,
color=colores[color_code],
)
ax.tick_params(axis="x", rotation=30)
ax.set_xlabel(label_x)
ax.set_ylabel(label_y)
ax.yaxis.set_major_formatter(mpl.ticker.StrMethodFormatter("{x:,.0f}"))
ax.set_title(chart_title, fontsize=title_size)
plt.show()
# For Total Sales per week,as we know, Black Friday and Christmas. We will remove those days:
df_sel = dfg_d[dfg_d["Weekly_Sales"] > 49000000]
black_list = df_sel["date2"].unique()
black_list = [
pd.to_datetime(item)
for item in black_list
if (pd.to_datetime(item).month == 11) | (pd.to_datetime(item).month == 12)
]
lista = [
"2010-12-31",
"2011-01-07",
"2011-01-14",
"2011-01-21",
"2011-01-28",
"2011-12-30",
"2012-01-06",
"2012-01-13",
"2012-01-20",
"2012-01-27",
]
for item in lista:
black_list.append(pd.to_datetime(item))
df_c = df[~df["date2"].isin(black_list)]
# Now as we know a bit better about the database, let's go to the stores:
pt = df.pivot_table(
index=["date2"], columns="store2", values="Weekly_Sales"
).reset_index()
i = 1
color_code = 2
set1((20, 150), "#F8FBF8")
for store in stores:
plt.subplot(len(stores), 1, i)
type_class = list(meta_df[meta_df["store2"] == store]["type"])
chart(
(20, 150),
"#F8FBF8",
pt,
"date2",
store,
"",
"Weekly Sales",
color_code=color_code,
chart_title=f"Store {store} type: {type_class[0]}",
title_size=15,
show_c="Y",
)
i += 1
color_code += 1
if color_code == len(colores):
color_code = 2
# From direct observation, there are some stores with interesting movements, like 16, 17, 28, 33, 36 (descending), 38 and 44 (both ascending) and 42 and 43. Would this be visible in any of the statistics we can run?
pt_c = df_c.pivot_table(
index=["date2"], columns="store2", values="Weekly_Sales"
).reset_index()
from statsmodels.tsa.stattools import adfuller, kpss
from statsmodels.tsa.api import AutoReg
from statsmodels.tsa.arima.model import ARIMA
from statsmodels.tsa.stattools import acf, pacf
from statsmodels.graphics.tsaplots import plot_acf, plot_pacf
from scipy.stats import shapiro, kstest, normaltest
from statsmodels.tsa.seasonal import seasonal_decompose, STL
meta_df = dfg_s[["store2", "type"]]
warnings.simplefilter("ignore")
ad_df = pd.DataFrame()
for store in stores:
pt_c_sel = pt_c.loc[:, [store]]
adf_results = adfuller(pt_c_sel)
kpss_results = kpss(pt_c_sel)
if adf_results[1] > 0.05:
adf_stat = "non-stationary"
else:
adf_stat = "stationary"
if kpss_results[1] < 0.05:
kpss_stat = "non-stationary"
else:
kpss_stat = "stationary"
model = AutoReg(pt_c_sel.dropna(), lags=1).fit()
res_norm_1 = shapiro(model.resid)[1]
model = AutoReg(pt_c_sel.dropna(), lags=2).fit()
res_norm_2 = shapiro(model.resid)[1]
model = AutoReg(pt_c_sel.dropna(), lags=4).fit()
res_norm_4 = shapiro(model.resid)[1]
model = ARIMA(pt_c_sel.dropna(), order=(0, 0, 1)).fit()
res_norm_ma_1 = shapiro(model.resid)[1]
model = ARIMA(pt_c_sel.dropna(), order=(0, 0, 2)).fit()
res_norm_ma_2 = shapiro(model.resid)[1]
dicc = {
"store2": store,
"adfuller": adf_results[1],
"kpss": kpss_results[1],
"adf_stat": adf_stat,
"kpss_stat": kpss_stat,
"res_norm_1": res_norm_1,
"res_norm_2": res_norm_2,
"res_norm_4": res_norm_4,
"res_norm_ma_1": res_norm_ma_1,
"res_norm_ma_2": res_norm_ma_2,
}
ad_df = ad_df.append(dicc, ignore_index=True)
meta_df = meta_df.merge(ad_df, how="left", on="store2")
meta_df["stat"] = [
item1 if item1 == item2 else "non_stationary"
for item1, item2 in zip(meta_df["adf_stat"], meta_df["kpss_stat"])
]
fig, ax = plt.subplots(4, 1, figsize=(20, 30))
indice = 0
y1 = "res_norm_1"
y2 = "res_norm_4"
y3 = "res_norm_ma_1"
y4 = "res_norm_ma_2"
lista_columnas = [y1, y2, y3, y4]
lista_colores = [3, 4, 6, 7]
columna_x = "store2"
title = [
"Shapiro Test for AR(1) residuals",
"Shapiro Test for AR(4) residuals",
"Shapiro Test for MA(1) residuals",
"Shapiro Test for MA(2) residuals",
]
for fila in range(0, 4):
ax[fila].bar(
data=meta_df,
x=columna_x,
height=lista_columnas[indice],
width=0.8,
color=colores[lista_colores[indice]],
)
linea = ax[fila].axhline(y=0.05, color=colores[5])
ax[fila].tick_params(axis="x", rotation=0, labelsize=15)
ax[fila].grid(axis="x")
ax[fila].set_facecolor("whitesmoke")
ax[fila].set_xlabel(label_x)
ax[fila].set_ylabel("P-value")
ax[fila].yaxis.set_major_formatter(mpl.ticker.StrMethodFormatter("{x:,.2f}"))
ax[fila].set_title(title[indice], fontsize=title_size)
indice += 1
plt.show()
# I did some regression using AR and MA for each and then test residuals for normal distribution. The idea is that if the residulas follow a normal distribution, then the models capture the variances. I still need to see how this works for stationary and non-stationary series.
stores_stat = list(meta_df[meta_df["stat"] == "stationary"]["store2"])
pt_decomp = seasonal_decompose(
pt_c.loc[:, ["date2", store]].set_index("date2"), model="additive", period=1
)
i = 1
color_code = 2
set1((20, 150), "#F8FBF8")
for store in stores_stat:
plt.subplot(len(stores), 1, i)
type_class = list(meta_df[meta_df["store2"] == store]["type"])
stationary = list(meta_df[meta_df["store2"] == store]["stat"])
pt_decomp = seasonal_decompose(
pt_c.loc[:, ["date2", store]].set_index("date2"), model="additive", period=52
)
pt_df_decomp = pd.DataFrame(pt_decomp.trend).reset_index()
chart(
(20, 150),
"#F8FBF8",
pt_df_decomp,
"date2",
"trend",
"",
"Weekly Sales",
color_code=color_code,
chart_title=f"TREND Store {store} type {type_class[0]} {stationary[0]}",
title_size=15,
show_c="Y",
)
i += 1
color_code += 1
if color_code == len(colores):
color_code = 2
stores_non_stat = [item for item in stores if item not in stores_stat]
i = 1
color_code = 2
set1((20, 150), "#F8FBF8")
for store in stores_non_stat:
plt.subplot(len(stores), 1, i)
type_class = list(meta_df[meta_df["store2"] == store]["type"])
stationary = list(meta_df[meta_df["store2"] == store]["stat"])
pt_decomp = seasonal_decompose(
pt_c.loc[:, ["date2", store]].set_index("date2"), model="additive", period=52
)
pt_df_decomp = pd.DataFrame(pt_decomp.trend).reset_index()
chart(
(20, 150),
"#F8FBF8",
pt_df_decomp,
"date2",
"trend",
"",
"Weekly Sales",
color_code=color_code,
chart_title=f"TREND Store {store} type {type_class[0]} {stationary[0]}",
title_size=15,
show_c="Y",
)
i += 1
color_code += 1
if color_code == len(colores):
color_code = 2
def chart_1(
set_1,
set_2,
df,
columna_x,
columna_y,
label_x,
label_y,
lim_lo_y="aut",
lim_hi_y="aut",
color_code=0,
chart_title="",
title_size=30,
show_c="Y",
):
set1(set_1, set_2)
paleta = [colores[i] for i in [0, 1, 6, 3]]
ax = sns.lineplot(
data=df,
x=columna_x,
y=columna_y,
color=colores[color_code],
estimator="sum",
)
labels_list = list(range(0, len(df[columna_x]), 5))
ax.tick_params(axis="x", rotation=30)
ax.set_xticks(labels_list, labels=[list(df[columna_x])[i] for i in labels_list])
ax.set_xlabel(label_x)
ax.set_ylabel(label_y)
ax.yaxis.set_major_formatter(mpl.ticker.StrMethodFormatter("{x:,.0f}"))
if lim_lo_y != "aut":
ax.set_ylim(lim_lo_y, lim_hi_y)
ax.set_title(chart_title, fontsize=title_size)
if show_c == "Y":
plt.show()
i = 1
color_code = 2
set1((20, 150), "#F8FBF8")
for store in stores_non_stat:
plt.subplot(len(stores), 1, i)
type_class = list(meta_df[meta_df["store2"] == store]["type"])
stationary = list(meta_df[meta_df["store2"] == store]["stat"])
pt_decomp = seasonal_decompose(
pt_c.loc[:, ["date2", store]].set_index("date2"), model="additive", period=52
)
pt_df_decomp = pd.DataFrame(pt_decomp.seasonal).reset_index()
pt_df_decomp["date3"] = pt_df_decomp["date2"].dt.strftime("%Y-%m-%d")
chart_1(
(20, 150),
"#F8FBF8",
pt_df_decomp,
"date3",
"seasonal",
"",
"Weekly Sales",
color_code=color_code,
chart_title=f"SEASONAL Store {store} type {type_class[0]} {stationary[0]}",
title_size=15,
show_c="Y",
)
i += 1
color_code += 1
if color_code == len(colores):
color_code = 2
def chart_acf(figsize, facecolor, df, lista):
i = 0
color_code = 2
fig, ax = plt.subplots(len(lista), figsize=figsize)
for store in lista:
ax[i].set_facecolor(facecolor)
type_class = list(meta_df[meta_df["store2"] == store]["type"])
stationary = list(meta_df[meta_df["store2"] == store]["stat"])
pt_c_sel = df[store]
plot_acf(
pt_c_sel,
ax=ax[i],
alpha=0.05,
title=f"ACF Store {store} type {type_class[0]} {stationary[0]}",
color=colores[color_code],
)
i += 1
color_code += 1
if color_code == len(colores):
color_code = 2
# plt.savefig('acf.png')
def chart_pacf(figsize, facecolor, df, lista):
i = 0
color_code = 2
fig, ax = plt.subplots(len(lista), figsize=figsize)
for store in lista:
ax[i].set_facecolor(facecolor)
type_class = list(meta_df[meta_df["store2"] == store]["type"])
stationary = list(meta_df[meta_df["store2"] == store]["stat"])
pt_c_sel = df[store]
plot_pacf(
pt_c_sel,
ax=ax[i],
alpha=0.05,
title=f"PACF Store {store} type {type_class[0]} {stationary[0]}",
color=colores[color_code],
)
i += 1
color_code += 1
if color_code == len(colores):
color_code = 2
# plt.savefig('pacf.png')
ar1_list = list(meta_df[meta_df["res_norm_1"] > 0.1]["store2"])
chart_pacf((20, 70), "#F8FBF8", pt_c, ar1_list)
# Interesting to see that for the stores with residuals on AR(1) they have high partial correlation lag 1, except store 37.
ar1_list = list(
meta_df[(meta_df["res_norm_4"] > 0.05) & (meta_df["res_norm_1"] < 0.05)]["store2"]
)
chart_pacf((20, 10), "#F8FBF8", pt_c, ar1_list)
ma2_list = list(
meta_df[(meta_df["res_norm_ma_2"] > 0.05) & (meta_df["res_norm_1"] < 0.05)][
"store2"
]
)
chart_pacf((20, 10), "#F8FBF8", pt_c, ma2_list)
|
# kenar çıkarma
import cv2
import numpy as np
import matplotlib.pyplot as plt
image = cv2.imread("/kaggle/input/iconfile/icon.png")
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
kernel = np.array([[1, 0, -1], [1, 0, -2], [1, 0, -1]])
img = cv2.filter2D(image, -1, kernel) # çıkış boyutunun aynı olması (-1)
fig, ax = plt.subplots(1, 2, figsize=(10, 6))
ax[0].imshow(image) # normal image
ax[1].imshow(img) # filter image
import cv2
import numpy as np
import matplotlib.pyplot as plt
image = cv2.imread("/kaggle/input/iconfile/icon.png")
image = image[400:500, 100:200, :]
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
kernel = np.array([[1, 2, 1], [0, 0, 0], [-1, -2, -1]])
kernel2 = np.array([[1, 0, -1], [2, 0, -2], [1, 0, -1]])
img = cv2.filter2D(image, -1, kernel)
img2 = cv2.filter2D(image, -1, kernel2)
fig, ax = plt.subplots(1, 3, figsize=(10, 6))
ax[0].imshow(image) # normal image
ax[1].imshow(img) # filter image yatay kernel geçişleri (sobel)
ax[2].imshow(img2) # filter image dikey kenel geçişleri (sobel)
import cv2
import numpy as np
import matplotlib.pyplot as plt
image = cv2.imread("/kaggle/input/iconfile/icon.png")
image = image[400:500, 100:200, :]
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
kernel = np.array([[3, 10, 3], [0, 0, 0], [-3, -10, -3]])
kernel2 = np.array([[3, 0, -3], [10, 0, -10], [3, 0, -3]])
img = cv2.filter2D(image, -1, kernel)
img2 = cv2.filter2D(image, -1, kernel2)
fig, ax = plt.subplots(1, 3, figsize=(10, 6))
ax[0].imshow(image) # normal image
ax[1].imshow(img) # filter image yatay kernel geçişleri (scharr)
ax[2].imshow(img2) # filter image dikey kenel geçişleri (scharr)
# Optimum eşiği bulma
import cv2
import matplotlib.pyplot as plt
from skimage.filters import threshold_otsu as to
img = cv2.imread("/kaggle/input/benimfoto/20230127_150322-min.jpg")
grayimg = cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)
to_val = to(grayimg)
print("Optimum ayırma değeri", to_val)
thresh = 133 # eşik değeri
binary_high = grayimg > to_val
binary_low = grayimg <= to_val
fig, ax = plt.subplots(1, 2, figsize=(10, 6))
ax[0].imshow(binary_low)
ax[1].imshow(binary_high)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # 1 データの取得 Acquisition data
ani_lis_df = pd.read_csv(
"/kaggle/input/anime-recommendation-database-2020/animelist.csv", nrows=100000
)
# Use only 100,000 rows because of too much data
# 全データは大きすぎるため、10万行だけ利用
display(ani_lis_df.head())
ani_lis_df.info()
ani_lis_df = ani_lis_df.rename(columns={"anime_id": "MAL_ID"})
ani_lis_df.head(5)
# make dataframe that japaniese name and MAL_ID
# アニメの名前とIDのリストを作る
ani_csv = pd.read_csv("/kaggle/input/anime-recommendation-database-2020/anime.csv")
display(ani_csv)
ani_name = ani_csv[["MAL_ID", "Japanese name"]]
display(ani_name)
ani_name.info()
ani_j_name = ani_csv[["MAL_ID", "Score", "Japanese name"]]
ani_com_lis = pd.merge(ani_lis_df, ani_name, on="MAL_ID")
ani_com_lis.head(5).to_html().replace("\n", "")
dataset = ani_com_lis.drop(
columns=["watching_status", "watched_episodes", "Japanese name"]
)
dataset
ani_com_lis
# # 2 アニメの類似度を調査 Surveying Similarities Among Anime
# ## 2-1Matrix Factorization
# Make a dataset that MAL_ID x User_id
# make dataframe
# データフレームを作成する
anime_w_df = ani_com_lis.drop(
columns=["watching_status", "watched_episodes", "Japanese name"]
)
anime_w_df = pd.pivot_table(
anime_w_df, index="user_id", columns="MAL_ID", values="rating"
)
# I thought it was not enough that I express feature of animes and users only users watched these animes or not.
# So,I deicided that I would acsept "rating" for expressing these feature.
# "rating"を選んだ理由:アニメとユーザーの特徴を表現するには観たor観てない(0、1)では足りないと判断
# 欠損値を0で補完
# fill missing values with (0)
anime_w_df = anime_w_df.fillna(0)
# データフレームを表示する
anime_w_df.head(5)
anime_w_df.shape
# user_id→310、anime(MAL_id)→7916
# Userx10、10xAnimeの特徴量を表す行列をそれぞれ作成
# Non-negative Matrix Factorization
from sklearn.decomposition import NMF
nmf_model = NMF(n_components=10, init="random", random_state=0)
User = nmf_model.fit_transform(anime_w_df)
Anime = nmf_model.components_
User_df = pd.DataFrame(User)
User_df.index = anime_w_df.index
Anime_df = pd.DataFrame(Anime)
Anime_df.columns = anime_w_df.columns
User_df
Anime_df
# ## 2-2コサイン類似度 Cosine Similarities
#
# cosine similarities of Anime
from sklearn.metrics.pairwise import cosine_similarity
# Anime_dfのcos類似度を調べるためインデックスとカラムを入れ替える。
Anime_df = Anime_df.transpose()
# 既に0~1の数値なので正規化は不要と予想
# It was determined that normalization was unnecessary as the values were already between 0 and 1."
cos_sim_Ani_df = pd.DataFrame(cosine_similarity(Anime_df))
print(cos_sim_Ani_df.shape)
display(cos_sim_Ani_df)
# Inputting a MAL_ID will display about 10 animes titles with the closest cosine similarity.
row = cos_sim_Ani_df.loc[24]
top_10 = row.sort_values(ascending=False)[1:11]
top_10_df = pd.DataFrame(top_10).reset_index()
top_10_df = top_10_df.rename(columns={"index": "MAL_ID"})
top_10_name = pd.merge(top_10_df, ani_name, on="MAL_ID")
reco_ani = top_10_name["Japanese name"]
print(reco_ani)
print(top_10_name.to_html().replace("\n", ""))
# Making a function that inputting a MAL_ID will display about 10 animes titles with the closest cosine similarity.
def cos_sim_top10(i):
row = cos_sim_Ani_df.loc[i]
top_10 = row.sort_values(ascending=False)[1:11]
top_10_df = pd.DataFrame(top_10).reset_index()
top_10_df = top_10_df.rename(columns={"index": "MAL_ID"})
top_10_name = pd.merge(top_10_df, ani_name, on="MAL_ID")
reco_ani = top_10_name["Japanese name"]
id_to_search = i # 取得したいJapanese nameに対応するMAL_IDを指定する
anime_name = ani_name.loc[
ani_name["MAL_ID"] == id_to_search, "Japanese name"
].values[0]
print(f"あなたが観たアニメは[{anime_name}]です。")
print("類似度が高いアニメtop10は以下です。")
return pd.DataFrame(top_10_name)
cos_sim_top10(1000)
# English translation of the output result↓
# The anime you saw is ["japanese anime name"] .
# So I recommend following animes .
print(cos_sim_top10(1000).to_html().replace("\n", ""))
# # 3 キャットブーストによる分析 Analysis by Catboost
# ## 3-1 ユーザーの特徴量を作成 Create users featuers
# I will create users features from output of MF and genr that users watched.
# output of MF is already available .(User_df)
# So I create users feature from genr that users watched.
ani_genr = ani_csv[
[
"MAL_ID",
"Genres",
]
]
ani_genr["Genres"].str.split(",")
# カンマ区切りのデータを分割し、新しい列に割り当てる
# Split comma separated data and assign to new columns
genres = set()
# ジャンルのリストを作成
# Create a "genre list"
for genre_list in ani_genr["Genres"].str.split(","):
genres.update(genre_list)
genres = sorted(list(genres))
ani_genr[genres] = 0
ani_genr
# Create a function that enter 1 in the applicable genre for each anime
def get_genre_dict(genres):
genre_dict = {}
for genre in genres.split(","):
genre_dict[genre.strip()] = 1
return genre_dict
genre_df = pd.DataFrame(ani_genr["Genres"].apply(get_genre_dict).tolist())
genre_df = genre_df.fillna(0).astype(int)
display(genre_df)
# 元のDataFrameとジャンルの1または0の値を含むDataFrameを結合
# Combine the original dataframe and this dataframe.
result_df = pd.concat([ani_genr["MAL_ID"], genre_df], axis=1)
display(result_df)
ani_genr_df = result_df
# Combine user_df and ani_genr_df.
user_genr_df = pd.merge(ani_lis_df, ani_genr_df, on="MAL_ID")
display(user_genr_df)
display(user_genr_df["user_id"].value_counts())
print(user_genr_df.head(5).to_html().replace("\n", ""))
import matplotlib.pyplot as plt
import seaborn as sns
# ユーザー0のジャンルごとのカウントを取得する
# Get the total number of times for each genre of user 0
user_w_count = user_genr_df.groupby("user_id").get_group(0).sum()
plt.figure(figsize=(40, 6))
# カウントが0でないジャンルのインデックスを取得する
# Get the index of the genre whose count is not 0
non_zero_indices = [i for i, count in enumerate(user_w_count.values) if count != 0]
# カウントが0でないジャンルのみを可視化する
# Visualize only genres with a non-zero count
sns.barplot(
x=user_w_count.index[non_zero_indices][4:],
y=user_w_count.values[non_zero_indices][4:],
)
user_genr_df["user_id"]
# Count the number of times uses have watched an anime.
user_genr_df = pd.merge(ani_lis_df, ani_genr_df, on="MAL_ID")
display(user_genr_df["user_id"].value_counts())
# Normalize views for each genre.(0~1)
user_genr_series_dict = {}
for group_id, group_df in user_genr_df.groupby("user_id"):
user_genr_series = (
group_df.sort_values(by="rating", ascending=False).iloc[:31, 5:].sum()
/ group_df.iloc[:, 5:].sum()
)
user_genr_series = user_genr_series.fillna(0)
user_genr_series_dict[group_id] = user_genr_series
user_genr_feature_df = pd.DataFrame(user_genr_series_dict).T
user_genr_feature_df
print(user_genr_feature_df.head(5).to_html().replace("\n", ""))
group_df.sort_values(by="rating", ascending=False).iloc[:31, :]
group_df.iloc[:, 5:].sum()
User_df
# Create a dataframe of views per user
w_num = user_genr_df.groupby("user_id")["MAL_ID"].count()
w_num_df = pd.DataFrame(w_num)
w_num_df = w_num_df.rename(columns={"MAL_ID": "w_num"})
w_num_df
dataset.nunique()
User_df
# Anime MF
Anime_df
# Merge dataset,User_df and user_genr_feature_df,w_num_df
dataset_2 = pd.merge(dataset, Anime_df, left_on="MAL_ID", right_index=True)
print(len(dataset_2))
dataset_2 = pd.merge(
dataset_2, user_genr_feature_df, left_on="user_id", right_index=True
)
dataset_2 = pd.merge(dataset_2, w_num_df, left_on="user_id", right_index=True)
dataset_2
print(dataset_2.head(5).to_html().replace("\n", ""))
anime_genr_df = pd.merge(ani_j_name, ani_genr_df, on="MAL_ID")
anime_genr_df
# ## 3-2 Catboostによる学習 Learning by Catboost🐱
import catboost as cb
# プールを作成
# Create Pool
dataset_cb = dataset_2.sort_values("user_id")
train_pool = cb.Pool(
dataset_cb.iloc[:, 3:].values,
label=dataset_cb["rating"].values,
group_id=dataset_cb["user_id"].values,
)
display(dataset_cb.iloc[:, 3:])
# Setting hyperparameters
params_dict = {
"loss_function": "YetiRank",
# "custom_metric":["QueryRMSE:hints=skip_train~false"],
"iterations": 100,
"verbose": 20,
"learning_rate": 0.01,
"max_depth": 4,
}
# Training the model
model = cb.CatBoostRanker(**params_dict)
model.fit(train_pool, eval_set=train_pool)
df_result = dataset_cb.groupby("user_id").get_group(0)
df_result.iloc[:, 3:]
df_result = dataset_cb.groupby("user_id").get_group(90)
df_result_pred = model.predict(df_result.iloc[:, 3:])
df_result["pred"] = df_result_pred
df_result
df_result[["MAL_ID", "rating", "pred"]].sort_values(by="pred", ascending=False)
print(
df_result[["MAL_ID", "rating", "pred"]]
.sort_values(by="pred", ascending=False)
.head(10)
.to_html()
.replace("\n", "")
)
# ## 3-3 モデルの精度確認 Check the accuracy of model
dataset_cb
# from sklearn.model_selection import train_test_split
# X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.25, stratify=y)
from sklearn.model_selection import GroupKFold
gkf = GroupKFold(n_splits=4)
groups = dataset["user_id"]
params_dict = {
"loss_function": "YetiRank",
# "custom_metric":["QueryRMSE:hints=skip_train~false"],
"iterations": 50,
"verbose": False,
"learning_rate": 0.01,
"max_depth": 4,
"use_best_model": True,
"eval_metric": "QueryRMSE",
}
result_dict_list = []
for fold, (train_index, val_index) in enumerate(gkf.split(dataset_cb, groups=groups)):
train_gkf, val_gkf = dataset_cb.iloc[train_index].sort_values(
by="user_id"
), dataset_cb.iloc[val_index].sort_values(by="user_id")
train_pool = cb.Pool(
train_gkf.iloc[:, 3:].values,
label=train_gkf["rating"].values,
group_id=train_gkf["user_id"].values,
)
val_pool = cb.Pool(
val_gkf.iloc[:, 3:].values,
label=val_gkf["rating"].values,
group_id=val_gkf["user_id"].values,
)
model = cb.CatBoostRanker(**params_dict)
model.fit(train_pool, eval_set=val_pool)
val_metrics = model.eval_metrics(val_pool, ["QueryRMSE"])
result_dict = {"model": model, "metrics": val_metrics}
result_dict_list.append(result_dict)
print(result_dict_list)
# # 4 レコメンドシステム作成(関数)
# 未知のユーザーが視聴したアニメのリストを入れる
w_list = []
def reco_anime(w_list):
w_j_list = []
for i in w_list:
id_to_search = i # 取得したいJapanese nameに対応するMAL_IDを指定する
anime_name = ani_name.loc[ani_name["MAL_ID"] == id_to_search, "Japanese name"]
w_j_list.append(anime_name)
# 各視聴アニメごとにcos類似度が近いアニメを20件取得、MFの結果と結合
row = cos_sim_Ani_df.loc[i]
top_20 = row.sort_values(ascending=False)[1:21]
top_20_df = pd.DataFrame(top_20).reset_index()
top_20_df = top_20_df.rename(columns={"index": "MAL_ID", i: "cos_sim"})
top_20_df_mf = pd.merge(top_20_df, Anime_df, on="MAL_ID").drop(
columns="cos_sim"
)
if i == w_list[0]:
top_20_df_mf_com = top_20_df_mf
else:
top_20_df_mf_com = pd.concat([top_20_df_mf_com, top_20_df_mf], axis=0)
reco_df = (
top_20_df_mf_com.drop_duplicates(subset="MAL_ID")
.reset_index()
.drop(columns="index")
)
reco_df["user_id"] = 0
reco_df
# ユーザーの特徴量を作成。
for i in w_list:
anime_genr = anime_genr_df.drop(columns=["Score", "Japanese name"])
user_genr = anime_genr.query(f"MAL_ID=={i}")
if i == w_list[0]:
user_genr_com = user_genr
else:
user_genr_com = pd.concat([user_genr_com, user_genr], axis=0)
user_f = user_genr_com.reset_index().drop(columns="index")
user_f = (user_f.iloc[:, 2:].sum() / len(w_list)).fillna(0)
user_f["many"] = len(w_list)
user_f["user_id"] = 0
user_f = pd.DataFrame(user_f).T
user_f
reco_user_f = pd.merge(reco_df, user_f, on="user_id")
reco_user_f = reco_user_f.drop(columns="user_id")
reco_user_f
reco_ani_pred = result_dict_list[3]["model"].predict(reco_user_f.iloc[:, 2:])
reco_user_f["pred"] = reco_ani_pred
reco_user_f = reco_user_f[["MAL_ID", "pred"]].sort_values(
by="pred", ascending=False
)
recomend_anime_10 = pd.DataFrame(
pd.merge(reco_user_f, ani_name, on="MAL_ID")["Japanese name"].head(10)
)
return recomend_anime_10
w_list = [50, 34, 21, 99]
reco_anime(w_list)
w_list = [50, 34, 21, 99]
reco_anime(w_list).to_html().replace("\n", "")
# df_result_pred = result_dict_list[1]["model"].predict(reco_user_f.iloc[:, 2:])
# reco_user_f["pred"]=df_result_pred
# reco_user_f=reco_user_f[["MAL_ID","pred"]].sort_values(by="pred",ascending=False)
# recomend_anime_10=pd.DataFrame(pd.merge(reco_user_f,ani_name,on="MAL_ID")["Japanese name"].head(10))
df_result = dataset_cb.groupby("user_id").get_group(24)
df_result_pred = result_dict_list[1]["model"].predict(df_result.iloc[:, 3:])
df_result["pred"] = df_result_pred
df_result.sort_values(by="pred", ascending=False)
|
import numpy as np
import os
import cv2
import matplotlib.pyplot as plt
import seaborn as sn
directory = "/kaggle/input/sports-celebrity-images/CroppedDataset/"
for folder in os.listdir(directory):
folder_path = os.path.join(directory, folder)
for pic in os.listdir(folder_path):
photo = os.path.join(folder_path, pic)
photo = plt.imread(photo)
plt.imshow(photo)
classes = {}
indexes = {}
i = 0
for folder in os.listdir(directory):
classes[folder] = i
indexes[i] = folder
i += 1
classes
X, y = [], []
for folder in os.listdir(directory):
folder_path = os.path.join(directory, folder)
for pic in os.listdir(folder_path):
photo = os.path.join(folder_path, pic)
img = cv2.imread(photo)
scaled_img = cv2.resize(img, (32, 32))
combined_img = np.vstack((scaled_img.reshape(32 * 32 * 3, 1)))
X.append(combined_img)
y.append(classes[folder])
len(X)
len(y)
len(X[0])
X[0]
X = np.array(X).reshape(len(X), len(X[0])).astype(float)
X.shape
X
X[0]
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=42
)
from sklearn.svm import SVC
model = SVC()
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
y_pred
y_test = np.array(y_test)
y_test
model.score(X_test, y_test)
from sklearn.metrics import confusion_matrix
cm = confusion_matrix(y_test, model.predict(X_test))
plt.figure(figsize=(10, 7))
sn.heatmap(cm, annot=True)
plt.xlabel("Predicted")
plt.ylabel("Truth")
from sklearn.linear_model import LogisticRegression
model = LogisticRegression()
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
y_pred
y_test = np.array(y_test)
y_test
model.score(X_test, y_test)
from sklearn.metrics import confusion_matrix
cm = confusion_matrix(y_test, model.predict(X_test))
plt.figure(figsize=(10, 7))
sn.heatmap(cm, annot=True)
plt.xlabel("Predicted")
plt.ylabel("Truth")
from sklearn.ensemble import RandomForestClassifier
model = RandomForestClassifier(n_estimators=100)
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
y_pred
y_test = np.array(y_test)
y_test
model.score(X_test, y_test)
from sklearn.metrics import confusion_matrix
cm = confusion_matrix(y_test, model.predict(X_test))
plt.figure(figsize=(10, 7))
sn.heatmap(cm, annot=True)
plt.xlabel("Predicted")
plt.ylabel("Truth")
import pickle
pickle.dump(model, open("/kaggle/working/model.pkl", "wb"))
pickled_model = pickle.load(open("/kaggle/working/model.pkl", "rb"))
check = plt.imread(
"/kaggle/input/sports-celebrity-images/CroppedDataset/Virat/Virat12.jpg"
)
plt.imshow(check)
XX = []
check = cv2.imread(
"/kaggle/input/sports-celebrity-images/CroppedDataset/Virat/Virat12.jpg"
)
scaled_check = cv2.resize(check, (32, 32))
combined_check = np.vstack((scaled_img.reshape(32 * 32 * 3, 1)))
XX.append(combined_check)
XX = np.array(XX).reshape(len(XX), len(XX[0])).astype(float)
XX.shape
XX
y_check = pickled_model.predict(XX)[0]
y_check
indexes
indexes[y_check]
|
# Note
# Useful features
# 1. type(티비 vs 영화)
# 2. title(제목) /Useless
# 3. cast(배우 목록)
# 4. country(발생 국가)
# 5. data_added(데이터 추가한 날짜)
# 6. release_year(data_added에서 년도만 빼온것들)
# 7. rating(시청 등급)
# 8. duration(시즌 혹은 상영시간)
# 9. listed_in(장르)
# 10. description(설명) /Useless
# 데이터 시각화 할것들(?) - 같은 그래프를 최대한 사용하지 않기!
# 1. type을 나누어서 영화 몇개 인지 티비 몇개 인지 확인하기
# 2. 배우 제일 많이 언급된 순서 나열
# 2. 발생 국가를 미국으로 base로 깔고 배우 목록에서 제일 많이 언급된 걸로 리스트 하기 (Description:In the dataset, *** actor has showed up mostly in movie or TV)
# 4. 어느 Country가 제일 많이 영화 만들었는지 plot
# 2. 장르별로 나누어서 리스트하기
# 3. 년도 별로 얼만큼의 영화나 TV 프로그램이 나왔는지 확인하기
# 3. 년도 별로 나누어서 그해에 어느 장르가 제일 많이 나왔는지 시각화 하기(힘들듯...)
# 5. 시청 등급 순위
# 4. 장르별로 순위
# 6. 장르랑 시청 등급을 연결해(Pairplots 사용해서 하면 가능할듯) 연관성 찾아보기
# 7. Understanding what content is available in different countries
# 8. Identifying similar content by matching text-based features
# 9. Network analysis of Actors / Directors and find interesting insights
# 10. Is Netflix has increasingly focusing on TV rather than movies in recent years.
#
import numpy as np
import pandas as pd
from pandas_profiling import ProfileReport
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# # 1. Load Data & Check Information
df_net = pd.read_csv("../input/netflix-shows/netflix_titles.csv")
#
# In this data set, there are 12 features.
# * show_id = Unique ID for every Movie / TV Show
# * type = Identifier - A Movie / TV Show
# * title = Title of Movie / TV Show
# * director = Director of the Movie
# * cast = Actors involved in the Movie / TV Show
# * country = Country where the Movie / TV Show was produced
# * data_added = Date it was added on Netflix
# * release_year = Actual Release year of the Movie / TV Show
# * rating = TV Rating of the Movie / TV Show
# * duration = Total Duration - in minutes or number of seasons
# Let's overview each feature and figure out how to use them to visualization.
#
df_net.head()
#
# As you can see, only 'release_year' data type is interger and rest of the features' data types are object. Because of that, describe method only shows 'release_year'.
# 'director' includes 2389 null values.
# 'cast' includes 718 null values.
# 'country' includes 507 null values.
# 'date_added' includes 10 null values.
# 'rating' includes 7 null values.
#
df_net.info()
df_net.describe()
df_net.isna().sum()
#
# All the works from above is kind of classic(?) way. There is really easy and simple library which explains all the detail information of dataset. By using ProfileReport, it will show dataset statistics, variable types, variables, interactions, correlations, missing values, and samples.
#
ProfileReport(df_net)
# # 2. Data Cleaning
# Before doing visualization, lets clean our data to see more clean view!
# Handle Null Variables
|
# # IMDB Dataset of 50K Movie Reviews
# Data source **[IMDB Dataset of 50K Movie Reviews](https://www.kaggle.com/datasets/lakshmi25npathi/imdb-dataset-of-50k-movie-reviews)**
# Attributes information:
# 1. review - text of the review
# 2. sentiment - positive or negative reviews
# **Objective**
# Given a review, find/define wether it is positve or negative?
# #### [re(Regual Expressions) tutorials](https://pymotw.com/2/re/)
# All needed/required libraries
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import re # regular expressions
from nltk.corpus import stopwords
import string
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.model_selection import train_test_split
from sklearn.neighbors import KNeighborsClassifier
from sklearn.metrics import roc_auc_score
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import roc_curve, auc
from sklearn.metrics import confusion_matrix
from gensim.models import Word2Vec
from gensim.models import KeyedVectors
from tqdm import tqdm
# # 1. Loading Data
data = pd.read_csv("/kaggle/input/imdb-dataset-of-50k-movie-reviews/IMDB Dataset.csv")
# data size/shape
data.shape
# data view
data.head(2)
# **Note:** Data needs to be cleaned as we can see there exists HTML Tags
if data.isna().sum().any():
data.isna().sum()
else:
print("Null values do not exist.")
# # 2. Data Cleaning
# ### [2.1]Droping Duplicates if exists
if data.duplicated().sum().any():
print("There exists {} duplicates of data".format(data.duplicated().sum()))
data.drop_duplicates(inplace=True)
print("============================================================")
print("Duplicate data Removed!")
else:
print("Duplicate data do not exists")
data.shape
# ### [2.1] HTML Cleaning
def cleanHTML(sentence): # this function removes HTML tags
clenedSentence = re.sub(re.compile("<.*>"), " ", sentence)
return clenedSentence
# Testing the function
print(data.review.values[1])
print("".join(["=" for i in range(133)]))
print(cleanHTML(data.review.values[1]))
# **OK:** Function working fine.
# ### [2.2] Restructuring the Sentence
def sentenceRestructure(sentence):
sent_restructure = ""
for sent in sentence.split():
if "don't" == sent.lower():
sent_restructure += " " + "do not"
elif "doesn't" == sent.lower():
sent_restructure += " " + "does not"
elif "isn't" == sent.lower():
sent_restructure += " " + "is not"
elif "are't" == sent.lower():
sent_restructure += " " + "are not"
elif "wasn't" == sent.lower():
sent_restructure += " " + "was not"
elif "weren't" == sent.lower():
sent_restructure += " " + "were not"
elif "should't" == sent.lower():
sent_restructure += " " + "should not"
elif "mustn't" == sent.lower():
sent_restructure += " " + "must not"
elif "would't" == sent.lower():
sent_restructure += " " + "would not"
elif "can't" == sent.lower():
sent_restructure += " " + "can not"
elif "could't" == sent.lower():
sent_restructure += " " + "could not"
elif "won't" == sent.lower():
sent_restructure += " " + "will not"
elif "shan't" == sent.lower():
sent_restructure += " " + "shall not"
else:
sent_restructure += " " + sent.lower()
return sent_restructure.strip()
# ### [2.3] Stop Words Cleaning
def removeStopWords(sentence):
stopWords = set(stopwords.words("english")) # initiliazing English stopwords
# removing some stop words and add a stop word
newStopWords = stopWords.difference({"not", "very"}).union({"us"})
# Removig some puntuations
cleaner = re.compile(r"[.|?|,!|;|:`]")
cleaned_text = re.sub(cleaner, " ", sentence).lower()
# Removing stopwords
sentWithoutStopWord = ""
for word in cleaned_text.split():
if word not in newStopWords:
sentWithoutStopWord = sentWithoutStopWord + " " + word
return sentWithoutStopWord.strip()
# Testing the function
print(data.review.values[1])
print("".join(["=" for i in range(133)]))
print(removeStopWords(data.review.values[1]))
# ### [2.4] Punctuations Cleaning
punctuations = (
r"[" + "|".join(list(set(string.punctuation).difference({"|", "[", "]"}))) + "]"
)
punctuations
# Puntuation removing function
def cleanPunctuation(sentence):
punctuations = (
r"[" + "|".join(list(set(string.punctuation).difference({"|", "[", "]"}))) + "]"
)
cleanedText = re.sub(punctuations, " ", sentence)
cleanedText = re.sub(r"[-]", " ", cleanedText)
return cleanedText
# Testing the function
print(data.review.values[1])
print("".join(["=" for i in range(133)]))
print(cleanPunctuation(data.review.values[1]))
# ### [2.5] Numbers Cleaning
# Numerical data removing function
def cleanNumbers(sentence):
cleaner = re.compile(r"[1|2|3|4|5|6|7|8|9]")
cleanedText = re.sub(cleaner, " ", sentence)
return cleanedText
# Testing the function
print("There are 11 layers player with 1 ball.")
print("".join(["=" for i in range(133)]))
print(cleanNumbers("There are 11 layers player with 1 ball."))
# ### [2.6] Extra Spaces Cleaning
def cleanSpaces(sentence):
cleanedText = re.sub(" +", " ", sentence).strip()
return cleanedText
print(" I have 8 books ")
print("".join(["=" for i in range(133)]))
print(cleanSpaces(" I have 8 books "))
" I have 8 books ", cleanSpaces(" I have 8 books ")
# ### [2.7] Removinng words with length less than equal to 3
def cleanWordsWithLenLessThan3(sentence):
new_sentence = [w for w in sentence.split(" ") if len(w) > 2]
return " ".join(new_sentence)
sent = "This is an apple and a ball"
print(sent)
print("".join(["=" for i in range(133)]))
print(cleanWordsWithLenLessThan3(sent))
# ### [2.Final] Applying all above cleaning Functions to "review" feature
data["cleanedReview"] = (
data.review.map(cleanHTML)
.map(sentenceRestructure)
.map(removeStopWords)
.map(cleanPunctuation)
.map(cleanNumbers)
.map(cleanSpaces)
.map(cleanWordsWithLenLessThan3)
)
# Changing postive to 1 and negative to 0 of sentiment feature
def pos_neg(label):
if label == "positive":
return 1
else:
return 0
data["sentiment"] = data["sentiment"].map(pos_neg)
print(data.review.values[1])
print("".join(["=" for i in range(133)]))
print(data.cleanedReview.values[1])
# # 3. Data Conversion to Vector using CountVectorizer
# ### [3.1] Splitting data into features and label
X = data.cleanedReview.values
y = data.sentiment.values
# ### [3.2] [Splitting data into training and testing]
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.33, stratify=y, random_state=42 # 33% test size
)
X_train, X_cv, y_train, y_cv = train_test_split(
X_train, y_train, test_size=0.33, stratify=y_train, random_state=42
)
print("X_train:", X_train.shape, "y_train:", y_train.shape)
print("X_cv :", X_cv.shape, " y_cv :", y_cv.shape)
print("X_test :", X_test.shape, "y_test :", y_test.shape)
# ### [3.3] CountVectorizer
# Convert a collection of text documents to a matrix of token counts.
vectorizer = CountVectorizer()
X_train_bow = vectorizer.fit_transform(X_train)
X_cv_bow = vectorizer.transform(X_cv)
X_test_bow = vectorizer.transform(X_test)
# vectorized data shape
print("X_train_bow:", X_train_bow.shape)
print("X_cv_bow :", X_cv_bow.shape)
print("X_test_bow :", X_test_bow.shape)
# # 4. Applying KNN
# ### [4.1]Method 1: Using Simple for Loop
train_auc = []
cv_auc = []
K = [1, 5, 11, 15, 21, 31, 41, 51]
for i in K:
knn_clf = KNeighborsClassifier(n_neighbors=i)
knn_clf.fit(X_train_bow, y_train)
# the 2nd parameter should be probability estimates of the positive class
y_train_pred = knn_clf.predict_proba(X_train_bow)[:, 1]
y_cv_pred = knn_clf.predict_proba(X_cv_bow)[:, 1]
train_auc.append(roc_auc_score(y_train, y_train_pred))
cv_auc.append(roc_auc_score(y_cv, y_cv_pred))
plt.plot(K, train_auc, label="Train AUC")
plt.plot(K, cv_auc, label="CV AUC")
plt.legend()
plt.xlabel("K: Parameter")
plt.ylabel("AUC")
plt.title("Error Plots")
plt.show()
# ### [4.2]Method 2: GridSearch or randomsearch
# Finding best **k** parameter.
# Running Grid Search
knn_clf = KNeighborsClassifier()
parameters = {"n_neighbors": [1, 5, 11, 15, 21, 31, 41, 51]}
clf = GridSearchCV(knn_clf, parameters, cv=3, scoring="roc_auc")
clf.fit(X_train_bow, y_train)
clf.cv_results_
# Sorting according K paramters scores rank
pd.DataFrame(clf.cv_results_)[
["param_n_neighbors", "rank_test_score", "mean_test_score"]
].sort_values("rank_test_score")
# Storing best K
best_k = clf.best_params_["n_neighbors"]
best_k = 41
# ### [4.3] Fitting KNN Classifier with best K
# Fitting Model with best K
knn_clf = KNeighborsClassifier(n_neighbors=best_k)
knn_clf.fit(X_train_bow, y_train)
train_fpr, train_tpr, thresholds = roc_curve(
y_train, knn_clf.predict_proba(X_train_bow)[:, 1]
)
test_fpr, test_tpr, thresholds = roc_curve(
y_test, knn_clf.predict_proba(X_test_bow)[:, 1]
)
plt.plot(train_fpr, train_tpr, label="train AUC =" + str(auc(train_fpr, train_tpr)))
plt.plot(test_fpr, test_tpr, label="test AUC =" + str(auc(test_fpr, test_tpr)))
plt.legend()
plt.xlabel("K: hyperparameter")
plt.ylabel("AUC")
plt.title("ERROR PLOTS")
plt.show()
y_train_pred = knn_clf.predict(X_train_bow)
y_test_pred = knn_clf.predict(X_test_bow)
print("Training set confusion matrix")
sns.heatmap(confusion_matrix(y_train, y_train_pred), annot=True, cmap="Blues", fmt="g")
confusion_matrix(y_test, y_test_pred)
print("Testing set confusion matrix")
sns.heatmap(confusion_matrix(y_test, y_test_pred), annot=True, cmap="Blues", fmt="g")
# # 3. Data Conversion to Vector using word2vec
# ### [3.1] Generate list of list of words
i = 0
list_of_sentance_train = []
for sentance in X_train:
list_of_sentance_train.append(sentance.split())
# ### [3.1] Train W2V model
w2v_model = Word2Vec(list_of_sentance_train, min_count=5, vector_size=50, workers=4)
w2v_words = list(w2v_model.wv.key_to_index)
print("number of words that occured minimum 5 times ", len(w2v_words))
print("sample words ", w2v_words[0:50])
# ### [3.3] Converting Reviews into Numerical Vectors using W2V vectors
# #### [3.3.1] Converting Training Set
# average Word2Vec
# compute average word2vec for each review.
sent_vectors = []
# the avg-w2v for each sentence/review is stored in this list
for sent in tqdm(list_of_sentance_train): # for each review/sentence
sent_vec = np.zeros(50) # as word vectors are of zero length 50
cnt_words = 0
# num of words with a valid vector in the sentence/review
for word in sent: # for each word in a review/sentence
if word in w2v_words:
vec = w2v_model.wv[word]
sent_vec += vec
cnt_words += 1
if cnt_words != 0:
sent_vec /= cnt_words
sent_vectors.append(sent_vec)
sent_vectors_train = np.array(sent_vectors)
print(np.array(sent_vectors).shape)
print(sent_vectors[0])
# #### [3.3.2] Converting CV data set
i = 0
list_of_sentance_cv = []
for sentance in X_cv:
list_of_sentance_cv.append(sentance.split())
# average Word2Vec
# compute average word2vec for each review.
sent_vectors_cv = []
# the avg-w2v for each sentence/review is stored in this list
for sent in tqdm(list_of_sentance_cv): # for each review/sentence
sent_vec = np.zeros(50) # as word vectors are of zero length 50
cnt_words = 0
# num of words with a valid vector in the sentence/review
for word in sent: # for each word in a review/sentence
if word in w2v_words:
vec = w2v_model.wv[word]
sent_vec += vec
cnt_words += 1
if cnt_words != 0:
sent_vec /= cnt_words
sent_vectors_cv.append(sent_vec)
sent_vectors_cv = np.array(sent_vectors_cv)
print(np.array(sent_vectors_cv).shape)
print(sent_vectors_cv[0])
# #### [3.3.3] Converting Test sata set
i = 0
list_of_sentance_test = []
for sentance in X_test:
list_of_sentance_test.append(sentance.split())
# average Word2Vec
# compute average word2vec for each review.
sent_vectors_test = []
# the avg-w2v for each sentence/review is stored in this list
for sent in tqdm(list_of_sentance_test): # for each review/sentence
sent_vec = np.zeros(
50
) # as word vectors are of zero length 50, you might need to change this to 300 if you use google's w2v
cnt_words = 0
# num of words with a valid vector in the sentence/review
for word in sent: # for each word in a review/sentence
if word in w2v_words:
vec = w2v_model.wv[word]
sent_vec += vec
cnt_words += 1
if cnt_words != 0:
sent_vec /= cnt_words
sent_vectors_test.append(sent_vec)
sent_vectors_test = np.array(sent_vectors_test)
print(np.array(sent_vectors_test).shape)
print(sent_vectors_test[0])
train_auc = []
cv_auc = []
K = [1, 5, 10, 15, 21, 31, 41, 51]
for i in K:
neigh = KNeighborsClassifier(n_neighbors=i)
neigh.fit(sent_vectors_train, y_train)
# roc_auc_score(y_true, y_score) the 2nd parameter should be probability estimates of the positive class
# not the predicted outputs
y_train_pred = neigh.predict_proba(sent_vectors_train)[:, 1]
y_cv_pred = neigh.predict_proba(sent_vectors_cv)[:, 1]
train_auc.append(roc_auc_score(y_train, y_train_pred))
cv_auc.append(roc_auc_score(y_cv, y_cv_pred))
plt.plot(K, train_auc, label="Train AUC")
plt.scatter(K, train_auc, label="Train AUC")
plt.plot(K, cv_auc, label="CV AUC")
plt.scatter(K, cv_auc, label="CV AUC")
plt.legend()
plt.xlabel("K: hyperparameter")
plt.ylabel("AUC")
plt.title("ERROR PLOTS")
plt.show()
neigh = KNeighborsClassifier(n_neighbors=10)
neigh.fit(sent_vectors_train, y_train)
# roc_auc_score(y_true, y_score) the 2nd parameter should be probability estimates of the positive class
# not the predicted outputs
train_fpr, train_tpr, thresholds = roc_curve(
y_train, neigh.predict_proba(sent_vectors_train)[:, 1]
)
test_fpr, test_tpr, thresholds = roc_curve(
y_test, neigh.predict_proba(sent_vectors_test)[:, 1]
)
plt.plot(train_fpr, train_tpr, label="train AUC =" + str(auc(train_fpr, train_tpr)))
plt.plot(test_fpr, test_tpr, label="train AUC =" + str(auc(test_fpr, test_tpr)))
plt.legend()
plt.xlabel("K: hyperparameter")
plt.ylabel("AUC")
plt.title("ERROR PLOTS")
plt.show()
print("=" * 100)
from sklearn.metrics import confusion_matrix
print("Train confusion matrix")
print(confusion_matrix(y_train, neigh.predict(sent_vectors_train)))
print("Test confusion matrix")
print(confusion_matrix(y_test, neigh.predict(sent_vectors_test)))
|
# ## Проект №3 EDA SF_lessons
# ### Какой кейс решаем?
# Представим, что работаем дата-сайентистом в компании Booking. Одна из проблем компании — это нечестные отели, которые накручивают себе рейтинг. Одним из способов обнаружения таких отелей является построение модели, которая предсказывает рейтинг отеля. Если предсказания модели сильно отличаются от фактического результата, то, возможно, отель ведёт себя нечестно, и его стоит проверить.
# ### Наименование столбцов:
# * hotel_address — адрес отеля;
# * review_date — дата, когда рецензент разместил соответствующий отзыв;
# * average_score — средний балл отеля, рассчитанный на основе последнего комментария за последний год;
# * hotel_name — название отеля;
# * reviewer_nationality — страна рецензента;
# * negative_review — отрицательный отзыв, который рецензент дал отелю;
# * review_total_negative_word_counts — общее количество слов в отрицательном отзыв;
# * positive_review — положительный отзыв, который рецензент дал отелю;
# * review_total_positive_word_counts — общее количество слов в положительном отзыве.
# * reviewer_score — оценка, которую рецензент поставил отелю на основе своего опыта;
# * total_number_of_reviews_reviewer_has_given — количество отзывов, которые рецензенты дали в прошлом;
# * total_number_of_reviews — общее количество действительных отзывов об отеле;
# * tags — теги, которые рецензент дал отелю;
# * days_since_review — количество дней между датой проверки и датой очистки;
# * additional_number_of_scoring — есть также некоторые гости, которые просто поставили оценку сервису, но не оставили отзыв. Это число указывает, сколько там действительных оценок без проверки.
# * lat — географическая широта отеля;
# * lng — географическая долгота отеля.
# ### Файлы для соревнования
# * hotels_train.csv - набор данных для обучения [источник данных](https://github.com/slagovskiy/DST/raw/main/Project-3/data/hotels_train.csv.zip)
# * hotels_test.csv - набор данных для оценки качества [источник данных](https://github.com/slagovskiy/DST/raw/main/Project-3/data/hotels_train.csv.zip)
# * submission.csv - файл сабмишна в нужном формате [источник данных](https://github.com/slagovskiy/DST/raw/main/Project-3/data/hotels_train.csv.zip)
# ---
# ## 0. Исходные данные
# ### Библиотеки
import pandas as pd
import numpy as np
from collections import Counter
from category_encoders import TargetEncoder
from sklearn import preprocessing
from sklearn.feature_selection import chi2
from sklearn.feature_selection import f_classif
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestRegressor
from sklearn import metrics
import nltk
from nltk.sentiment.vader import SentimentIntensityAnalyzer
nltk.downloader.download("vader_lexicon")
import matplotlib.pyplot as plt
import seaborn as sns
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# ### Константы
RANDOM_SEED = (
42 # всегда фиксируйте RANDOM_SEED, чтобы ваши эксперименты были воспроизводимы!
)
TARGET = "reviewer_score"
# ### Данные
# Для работы на Kaggle
# DATA_DIR = '/kaggle/input/sf-booking/'
# df_train = pd.read_csv(DATA_DIR + 'hotels_train.csv')
# df_test = pd.read_csv(DATA_DIR + 'hotels_test.csv')
# sample_submission = pd.read_csv(DATA_DIR + 'submission.csv')
# Для работы в Jupyter Notebook Visual Studio Code
DATA_DIR = "data_kaggle"
df_train = pd.read_csv(DATA_DIR + "/hotels_train.csv")
df_test = pd.read_csv(DATA_DIR + "/hotels_test.csv")
sample_submission = pd.read_csv(DATA_DIR + "/submission.csv")
# ---
# ## 1. Предварительная обработка данных
# ### 1.1. Первичная очистка
df_train.info()
# **Стандартный отчет, созданный с помощью библиотеки Pandas Profiling, использовался для оценки исходных дистрибутивов и общих параметров функций..**
dupl_percent = df_train.duplicated().mean() * 100
print(f"Дублированные строки в образце: {dupl_percent:.2f}%")
df_train.drop_duplicates(inplace=True)
print("Дубликаты удалены")
df_train.head(3)
df_test.info()
df_test.head(3)
sample_submission.info()
sample_submission.head()
df_train["sample"] = 1
df_test["sample"] = 0
df_test["reviewer_score"] = 0
hotels = pd.concat([df_train, df_test], sort=False).reset_index(drop=True)
hotels.info()
print("Количество пробелов в столбцах типа object")
for col in hotels.dtypes[hotels.dtypes == "object"].index:
print(col + ":", hotels[hotels[col] == " "].shape[0])
# Здесь мы заполняем пробелы в столбце «reviewer_nationality»
# «negative_review» и «positive_review»
# поскольку столбцы 'lat и 'lng' со значениями NaN должны быть обработаны позже.
hotels["reviewer_nationality"] = hotels["reviewer_nationality"].apply(
lambda x: x[1:-1]
) # Удаление пробелов вокруг: ' Spain ' -> 'Spain'
hotels["negative_review"] = hotels["negative_review"].apply(lambda x: x[1:-1])
hotels["positive_review"] = hotels["positive_review"].apply(lambda x: x[1:-1])
replace_dict1 = {"": hotels["reviewer_nationality"].mode()[0]}
replace_dict2 = {"": hotels["negative_review"].mode()[0]}
replace_dict3 = {"": hotels["positive_review"].mode()[0]}
hotels["reviewer_nationality"].replace(replace_dict1, inplace=True)
hotels["negative_review"].replace(replace_dict2, inplace=True)
hotels["positive_review"].replace(replace_dict3, inplace=True)
# ### 1.2. Разработка признаков
# Извлечение названий стран и почтовых индексов из адресов гостиниц. Есть только
# один город на страну в адресах, поэтому мы опускаем извлечение названия города как
# это не даст никакой новой информации для алгоритмов ML, и в этом
# частный случай "город == страна". Почтовый индекс, с другой стороны, содержит
# информация о расположении отеля в городе, что может быть очень
# полезно для оценки рейтинга
def address_proc(x):
words = x.split()
country = words[-1]
if country == "Kingdom" or country == "Netherlands":
zip_code = words[-4] + words[-3]
else:
zip_code = words[-3]
return pd.Series([country, zip_code])
hotels[["hotel_country", "zip_code"]] = hotels["hotel_address"].apply(address_proc)
hotels["hotel_country"].replace({"Kingdom": "United Kingdom"}, inplace=True)
print("Страны:", hotels["hotel_country"].unique())
print("Количество почтовых индексов (районов города):", hotels["zip_code"].nunique())
# Кодирование столбцов с высокой кардинальностью с помощью Target Encoder
cols = ["hotel_name", "reviewer_nationality", "zip_code"]
target_encoder = TargetEncoder(cols=cols)
hotels_encoded = target_encoder.fit_transform(hotels[cols], hotels[TARGET])
rename_dict = {
"hotel_name": "hotel_name_enc",
"reviewer_nationality": "reviewer_nationality_enc",
"zip_code": "zip_code_enc",
}
hotels_encoded.rename(columns=rename_dict, inplace=True)
hotels = pd.concat([hotels, hotels_encoded], axis=1)
# Проверка NaN в закодированных функциях
print(hotels["hotel_name_enc"].isnull().mean())
print(hotels["reviewer_nationality_enc"].isnull().mean())
print(hotels["zip_code_enc"].isnull().mean())
# Внутренний клиент: национальность рецензента и страна отеля одинаковы.
hotels["domestic_customer"] = hotels.apply(
lambda x: 1 if x["hotel_country"] == x["reviewer_nationality"] else 0, axis=1
)
# Извлечение некоторой информации из даты проверки.
#
# День года кодируется синусом и косинусом, чтобы отразить цикличность
# процесс. Одновременно с числовым описанием календарной даты
# позволяет нам кодировать смену времен года более точным способом.
# Индикаторы Рождества и выходных дней дополнительно включено.
hotels["review_date"] = pd.to_datetime(hotels["review_date"])
hotels["sin_date"] = np.sin(2 * np.pi * hotels["review_date"].dt.dayofyear / 365)
hotels["cos_date"] = np.cos(2 * np.pi * hotels["review_date"].dt.dayofyear / 365)
hotels["christmas"] = hotels["review_date"].apply(
lambda x: 1
if (x.month == 12 and x.day >= 25) or (x.month == 1 and x.day <= 5)
else 0
)
hotels["weekend"] = hotels["review_date"].apply(
lambda x: 1 if x.dayofweek == 5 or x.dayofweek == 6 else 0
)
# Анализ настроений
#
# Сначала делаем пустые строки для различных обозначений пустых отзывов,
# затем проанализируем негативные и позитивные отзывы отдельно с помощью VADER
# анализа тональности и получение в результате «составного» значения.
replace_dict_negative = {
"No Negative": "",
" Nothing ": "",
" Nothing": "",
" nothing ": "",
" nothing": "",
" N A ": "",
" N A": "",
" N a ": "",
" N a": "",
" None ": "",
" None": "",
" ": "",
}
replace_dict_positive = {
"No Positive": "",
" Nothing ": "",
" Nothing": "",
" nothing ": "",
" nothing": "",
" N A ": "",
" N A": "",
" N a ": "",
" N a": "",
" None ": "",
" None": "",
" ": "",
}
hotels["negative_review"].replace(replace_dict_negative, inplace=True)
hotels["positive_review"].replace(replace_dict_positive, inplace=True)
mask = hotels["negative_review"] == ""
hotels.loc[mask, "review_total_negative_word_counts"] = 0
mask = hotels["positive_review"] == ""
hotels.loc[mask, "review_total_positive_word_counts"] = 0
analyzer = SentimentIntensityAnalyzer()
hotels["n_sentiment"] = hotels["negative_review"].apply(
lambda x: analyzer.polarity_scores(x)["compound"]
)
hotels["p_sentiment"] = hotels["positive_review"].apply(
lambda x: analyzer.polarity_scores(x)["compound"]
)
# Теги. Преобразование строковых переменных в списки.
hotels["tags"] = hotels["tags"].apply(lambda x: x[3:-3].split(" ', ' "))
# Получение продолжительности пребывания.
# Заполнение значений NaN типичной продолжительностью пребывания: 1 день.
def get_stay(x):
for tag in x:
words = tag.split()
if words[0] == "Stayed":
return int(words[1])
return np.NaN
hotels["stay"] = hotels["tags"].apply(get_stay)
hotels["stay"].fillna(1, inplace=True)
# Запускаем кодирование для наиболее частых тегов. Граница для
# наиболее часто берется 5% от размера набора данных.
boundary = hotels.shape[0] * 0.05 # 5% граница
c = Counter(tag for row in hotels["tags"] for tag in row)
frequent_tags_count = dict(filter(lambda x: x[1] > boundary, c.items()))
print("Наиболее часто встречающиеся теги и их количество:")
frequent_tags_count
# Получение списка наиболее часто встречающихся тегов. Теги "stay" исключены
frequent_tags = list(
dict(
filter(
lambda x: x[1] > boundary and x[0].split()[0] != "Stayed",
frequent_tags_count.items(),
)
).keys()
)
frequent_tags
# Быстрое кодирование для частых тегов.
for tag in frequent_tags:
hotels[tag] = hotels["tags"].apply(lambda x: 1 if tag in x else 0)
# Преобразование функции «days_since_review» в хронологический порядок
# даты проверки: с 0 дня до последнего дня.
hotels["days_since_review"] = hotels["days_since_review"].apply(
lambda x: int(x.split()[0])
)
hotels["day_number"] = (
hotels["days_since_review"] - hotels["days_since_review"].max()
) * (-1)
# Заполнение значений NaN широты и долготы средними значениями
# для каждого почтового индекса (района города).
# Фрейм данных со средними значениями:
zip_medians = hotels.groupby("zip_code")[["lat", "lng"]].median()
mask1 = hotels["lat"].isnull()
zip_nulls = hotels[mask]["zip_code"].unique() # почтовые индексы с NaN lat и lng
for zip_code in zip_nulls:
mask2 = hotels["zip_code"] == zip_code
hotels.loc[mask1 & mask2, "lat"] = zip_medians.loc[zip_code, "lat"]
hotels.loc[mask1 & mask2, "lng"] = zip_medians.loc[zip_code, "lng"]
# Дозаполним то для чего не нашлось почтового индекса средним значением
hotels["lat"] = hotels["lat"].fillna(hotels["lat"].median())
hotels["lng"] = hotels["lng"].fillna(hotels["lat"].median())
print("Доля NaN осталась")
print("lat:", hotels["lat"].isnull().mean())
print("lng:", hotels["lng"].isnull().mean())
# Получение расстояния от центра города
for country in hotels["hotel_country"].unique():
mask = hotels["hotel_country"] == country
# Городские центры как медианные значения для страны (=город)
lat_median = hotels[mask]["lat"].median()
lng_median = hotels[mask]["lng"].median()
hotels.loc[mask, "distance"] = (
(hotels[mask]["lat"] - lat_median) ** 2
+ (hotels[mask]["lng"] - lng_median) ** 2
) ** (1 / 2)
# Дозаполним пропуски средним значением
hotels["distance"] = hotels["distance"].fillna(hotels["distance"].median())
print("Доля NaN осталась")
print("distance:", hotels["distance"].isnull().mean())
# Быстрое кодирование стран/городов отелей.
country_onehot = pd.get_dummies(hotels["hotel_country"])
hotels = pd.concat([hotels, country_onehot], axis=1)
# Удаление обработанных столбцов
hotels.drop(
[
"hotel_address",
"review_date",
"hotel_name",
"zip_code",
"reviewer_nationality",
"negative_review",
"positive_review",
"tags",
"days_since_review",
"lat",
"lng",
"hotel_country",
],
axis=1,
inplace=True,
)
# ### 1.3. Мультиколлинеарность
def get_correlated(corr_matrix, thres=0.7):
"""Getting correlated pairs from a correlation matrix
with correlation coefficients higher than the threshold
Args:
corr_matrix (pandas DataFrame): correlation matrix (e.g., from df.corr())
thres (float, optional): threshold. Defaults to 0.7 (i.e., coeff > 0.7 or coeff < -0.7).
Returns:
pandas Series: correlated pairs and correlation coefficients
"""
correlated_pairs = {}
for col in corr_matrix.columns:
positive_filter = corr_matrix[col] >= thres
negative_filter = corr_matrix[col] <= -thres
match_series = corr_matrix[col][positive_filter | negative_filter]
for match in match_series.index:
if match != col and match + " -- " + col not in correlated_pairs.keys():
correlated_pairs.update({col + " -- " + match: match_series[match]})
return pd.Series(correlated_pairs).sort_values()
print("Коррелированные пары:")
get_correlated(hotels.corr(method="spearman"))
# Отбрасывание совпадений с сильной корреляцией. Мы оставим
# пару "review_total_positive_word_counts - p_sentiment", как корреляция
# не так уж силен, и если оставить обе функции, MAPE немного улучшится.
hotels.drop(
["Business trip", "additional_number_of_scoring", "hotel_name_enc", "zip_code_enc"],
axis=1,
inplace=True,
)
# ### 1.4. Нормализация
# После анализа дистрибутивов (методом Pandas df.hist()),
# были выбраны следующие скейлеры:
#
# MinMaxScaler - for 'average_score', 'day_number'
#
# RobustScaler - for 'review_total_negative_word_counts',
# 'review_total_positive_word_counts', 'total_number_of_reviews',
# 'total_number_of_reviews_reviewer_has_given',
# 'reviewer_country_population'
#
# Other columns - without normalization
cols_minmax = ["average_score", "day_number"]
cols_robust = [
"review_total_negative_word_counts",
"review_total_positive_word_counts",
"total_number_of_reviews",
"total_number_of_reviews_reviewer_has_given",
]
mm_scaler = preprocessing.MinMaxScaler()
r_scaler = preprocessing.RobustScaler()
hotels_mm = mm_scaler.fit_transform(hotels[cols_minmax])
hotels_mm = pd.DataFrame(hotels_mm, columns=cols_minmax)
hotels_rs = r_scaler.fit_transform(hotels[cols_robust])
hotels_rs = pd.DataFrame(hotels_rs, columns=cols_robust)
hotels[cols_minmax] = hotels_mm[cols_minmax]
hotels[cols_robust] = hotels_rs[cols_robust]
# ### 1.5. Выбор признаков
# X — Признаки, y — цель
X = hotels.drop([TARGET], axis=1)
y = hotels[TARGET]
# Категория и непрерывные признаки
cat_cols = [
"domestic_customer",
"christmas",
"weekend",
"Leisure trip",
"Couple",
"Submitted from a mobile device",
"Standard Double Room",
"Solo traveler",
"Superior Double Room",
"Double Room",
"Family with young children",
"Group",
"Family with older children",
"Austria",
"France",
"Italy",
"Netherlands",
"Spain",
"United Kingdom",
"stay",
]
cont_cols = [
"average_score",
"review_total_negative_word_counts",
"distance",
"total_number_of_reviews",
"review_total_positive_word_counts",
"total_number_of_reviews_reviewer_has_given",
"reviewer_nationality_enc",
"sin_date",
"cos_date",
"n_sentiment",
"p_sentiment",
"day_number",
]
# Расчет chi2-статистики для категориальных признаков
y_cat = (y * 10).astype("int") # класс не может быть плавающим для chi2
chi2_stat, chi2_p = chi2(X[cat_cols], y_cat)
chi2_stat = pd.DataFrame(chi2_stat, index=cat_cols).reset_index()
chi2_stat.rename(
columns={"index": "Features", 0: "Feature importance (chi2-statistics)"},
inplace=True,
)
chi2_stat["p-value"] = chi2_p
chi2_stat.sort_values(
"Feature importance (chi2-statistics)", inplace=True, ascending=False
)
fig = plt.figure(figsize=(13, 4))
ax = fig.add_axes([1, 1, 1, 1])
plt.xticks(rotation=90)
sns.barplot(data=chi2_stat, x="Features", y="Feature importance (chi2-statistics)")
# Расчет F-статистики (ANOVA) для категориальных признаков
f_stat, f_p = f_classif(X[cont_cols], y)
f_stat = pd.DataFrame(f_stat, index=cont_cols).reset_index()
f_stat.rename(
columns={"index": "Features", 0: "Feature importance (F-statistics)"}, inplace=True
)
f_stat["p-value"] = f_p
f_stat.sort_values("Feature importance (F-statistics)", inplace=True, ascending=False)
fig = plt.figure(figsize=(13, 4))
ax = fig.add_axes([1, 1, 1, 1])
plt.xticks(rotation=90)
sns.barplot(data=f_stat, x="Features", y="Feature importance (F-statistics)")
# Кроме того, давайте проверим значимость, используя p-value
mask = chi2_stat["p-value"] > 0.05 # 5% граница
print("Незначительная категория включает в себя:", list(chi2_stat[mask]["Features"]))
mask = f_stat["p-value"] > 0.05 # 5% граница
print("Незначительные непрерывные признаки:", list(f_stat[mask]["Features"]))
# Отбрасывание несущественных признаков
# Оставим 'distance' Это улучшает MAPE
drop_cols = ["weekend"]
hotels.drop(drop_cols, axis=1, inplace=True)
# ## 3. Обучение модели и получение предсказания
train_data = hotels.query("sample == 1").drop(["sample"], axis=1)
test_data = hotels.query("sample == 0").drop(["sample"], axis=1)
y = train_data.reviewer_score.values
X = train_data.drop(["reviewer_score"], axis=1)
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=RANDOM_SEED
)
test_data.shape, train_data.shape, X.shape, X_train.shape, X_test.shape
model = RandomForestRegressor(
n_estimators=100, verbose=1, n_jobs=-1, random_state=RANDOM_SEED
)
# обучаем модель на тестовом наборе данных
model.fit(X_train, y_train)
# предсказанные значения записываем в переменную y_pred
y_pred = model.predict(X_test)
def mean_absolute_percentage_error(y_tr, y_pr):
y_tr, y_pr = np.array(y_tr), np.array(y_pr)
return np.mean(np.abs((y_tr - y_pr) / y_tr)) * 100
print("MAPE:", round(mean_absolute_percentage_error(y_test, y_pred), 2))
# Особенности важности
plt.rcParams["figure.figsize"] = (10, 10)
feat_importances = pd.Series(model.feature_importances_, index=X.columns)
feat_importances.nlargest(15).plot(kind="barh", color="green")
test_data = test_data.drop(["reviewer_score"], axis=1)
predict_submission = model.predict(test_data)
sample_submission["reviewer_score"] = predict_submission
# сохраняем результат:
sample_submission.to_csv("submission.csv", index=False)
sample_submission.head(10)
# зафиксируем версию пакетов, чтобы эксперименты были воспроизводимы:
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import matplotlib.pyplot as plt
import seaborn as sns
AMD = pd.read_csv(
"/kaggle/input/nvidia-amd-intel-asus-msi-share-prices/AMD (1980-2023).csv"
)
ASUS = pd.read_csv(
"/kaggle/input/nvidia-amd-intel-asus-msi-share-prices/ASUS (2000-2023).csv"
)
INTEL = pd.read_csv(
"/kaggle/input/nvidia-amd-intel-asus-msi-share-prices/Intel (1980-2023).csv"
)
MSI = pd.read_csv(
"/kaggle/input/nvidia-amd-intel-asus-msi-share-prices/MSI (1962-2023).csv"
)
NAVIDIA = pd.read_csv(
"/kaggle/input/nvidia-amd-intel-asus-msi-share-prices/NVIDIA (1999-2023).csv"
)
AMD.shape, NAVIDIA.shape, MSI.shape
# # MODIFIYING DATA
class Imputer:
def __init__(self, data):
self.data = data
def parse_dates(self):
self.data["Date"] = self.data.Date.apply(lambda x: pd.Timestamp(x))
def data_after(self, time: str):
self.data = self.data[self.data.Date >= pd.Timestamp(time)]
def activate(self, time: str):
self.parse_dates()
self.data_after(time)
return self.data
nvidia = Imputer(NAVIDIA).activate("2018-01-01")
intel = Imputer(INTEL).activate("2018-01-01")
amd = Imputer(AMD).activate("2018-01-01")
merged_df = pd.merge(
nvidia.iloc[:, [0, 5]], amd.iloc[:, [0, 5]], on="Date", how="outer"
)
merged_df = pd.merge(merged_df, intel.iloc[:, [0, 5]], on="Date", how="outer")
merged_df.columns = ["Date", "Adj_Close_NVIDIA", "Adj_Close_AMD", "Adj_Close_INTEL"]
merged_df.head()
plt.figure(figsize=(10, 5))
plt.plot(merged_df["Date"], merged_df["Adj_Close_NVIDIA"], label="NVI")
plt.plot(merged_df["Date"], merged_df["Adj_Close_AMD"], label="AMD")
plt.plot(merged_df["Date"], merged_df["Adj_Close_INTEL"], label="INT")
plt.xlabel("Date")
plt.ylabel("Adj Close")
plt.title("Movement of Stock Price (2018-2023)")
plt.legend()
plt.show()
def plot(data, stock):
data = data.iloc[1200:, :]
fig, ax1 = plt.subplots(figsize=(20, 8))
ax2 = ax1.twinx()
ax1.bar(x=data.Date, height=data.Volume, alpha=0.7)
ax2.plot(data.Date, data["High"], "g-")
ax2.plot(data.Date, data["Low"], "r-")
plt.title(f"Volume and Market Price ({stock})")
ax1.set_xlabel("Date")
ax1.set_ylabel("Volume", color="b")
ax2.set_ylabel("High/Low", color="g")
plot(nvidia, "NVIDIA")
plot(amd, "AMD")
plot(intel, "INTEL")
# Correlation
# NVIDIA AND AMD
# In this case, we can see that the strongest positive correlation is between NVIDIA and AMD. This suggests that the two stocks tend to move together, and that changes in the price of one are often accompanied by similar changes in the other.
# NVIDIA AND INTEL
# Here there is a weak negative correlation between NVIDIA and Intel, indicating that the two stocks tend to move in opposite directions. This could be due to differences in their respective business models or market factors affecting the industry.
# It is important to note that correlation does not necessarily imply causation, and that there may be other factors at play that are driving the prices of these stocks. It is also important to conduct further analysis and consider other metrics before making investment decisions
sns.heatmap(merged_df.corr(), annot=True, cmap="coolwarm")
|
# # ĐỀ TÀI: BIẾN ĐỘNG CỦA TTCK VIỆT NAM TRONG BỐI CẢNH DỊCH COVID-19 BÙNG PHÁT VÀ KÉO DÀI.
# # I.Lý do chọn đề tài
# TTCK là kênh đầu tư trung và dài hạn hiệu quả của mỗi quốc gia với khả năng tìm kiếm lợi nhuận hấp dẫn, TTCK thu hút sự quan tâm của toàn xã hội đặc biệt là giới đầu tư.
# Trải qua 25 năm hoạt động, TTCK Việt Nam đã không ngừng hoàn thiện về cấu trúc, phát triển thành kênh huy động vốn quan trọng cho nền kinh tế
# Đại dịch Covid-19 bắt đầu xuất hiện tại Việt Nam vào đầu năm 2020 và cho đến hiện nay vẫn diễn biến hết sức phức tạp. Dù Nhà nước ta đã sử dụng rất nhiều biện pháp cứng rắn nhưng vẫn chưa thể ngăn chặn triệt để sự lây lan của dịch bệnh. Dưới tác động của dịch Covid-19, nền kinh tế Việt Nam có những biến động rất khó lường,
# Với thị trường chứng khoán, câu hỏi đặt ra là liệu sự xuất hiện của Covid-19 tại Việt Nam có tác động tiêu cực đến hiệu quả đầu tư? Dịch bệnh càng bùng phát thì lượng tiền đổ vào chứng khoán ngày càng nhiều? TTCK Việt Nam trở nên đầy biến động thế nào trong giai đoạn đại dịch Covid-19 diễn ra và kéo dài ?
# # II. Sơ lược về TTCK tại Việt Nam
# * Hoạt động về chứng khoán và TTCK bao gồm hoạt động chào bán, niêm yết, giao dịch, kinh doanh, đầu tư chứng khoán, cung cấp dịch vụ về chứng khoán, công bố thông tin, quản trị công ty đại chúng và các hoạt động khác được quy định tại Luật.
# * Dù là một nền kinh tế còn non trẻ, TTCK Việt Nam được hình thành và phát triển sau 20 năm nhưng cũng đóng góp nhiều cho hoạt động kinh tế vĩ mô của đất nước.
# * Với nhiều yếu tố hỗ trợ cộng nền tảng đang ngày một tốt lên, dự báo trong tương lai chứng khoán Việt Nam sẽ ngày một hấp dẫn hơn nữa, trở thành kênh đầu tư lý tưởng và đầy cơ hội kiếm lợi nhuận cho các nhà đầu tư, giúp nền kinh tế Việt Nam phát triển vững mạnh.
# * Sàn HOSE hay còn được biết đến là Sở giao dịch chứng khoán thành phố Hồ Chí Minh ra đời vào tháng 7 năm 2000. Sàn Hose hoạt động dưới sự quản lý trực tiếp từ ủy ban Chứng khoán Nhà nước và một hệ thống quản lý giao dịch chứng khoán niêm yết của Việt Nam.
# # III. Nội dung đề tài
# # 1. Nhập liệu
import numpy as np
import pandas as pd
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
import matplotlib as mpl
import matplotlib.pyplot as plt
import seaborn as sns
import plotly.graph_objects as go
from plotly.offline import iplot
import plotly.express as px
# # 2. Phân tích biến động TTCK Việt Nam trong các giai đoạn xuất hiện đại dịch Covid-19.
# # 2.1 Thực trạng TTCK trong các giai đoạn Covid-19
df_vn = pd.read_excel("/kaggle/input/fin-data-0804/DataCKVN.xlsx")
df_ck = df_vn
df_ck["Date"] = [str(date)[:11] for date in df_ck["Date"]]
df_ck["Date"] = [date.replace("/", "-") for date in df_ck["Date"]]
df_ck["Volume"] = [date.replace(".", "") for date in df_ck["Volume"]]
df_ck["Volume"] = df_ck["Volume"].astype(int)
df_ck["Volume"] = df_ck["Volume"] / 1000000
df_ck.columns = df_ck.columns.str.replace("Volume", "Volume (triệu)")
df_ck["Date"] = pd.to_datetime(df_ck["Date"])
df_ck.head()
df_ck = df_ck.set_index("Date")
df_ck = df_ck.resample(rule="M").mean().reset_index()
df_ck.head()
df_ck["Date"] = df_ck["Date"].dt.strftime("%d-%m-%Y")
df_ck["Date"] = [date[3::] for date in df_ck["Date"]]
# Giai đoạn trước dịch từ 31-1-2019 đến 22-1-2020
pd.Index(df_ck["Date"]).get_loc("02-2020")
# Giai đoạn trong dịch từ 31-1-2020 đến 1-10-2021 và Giai đoạn sau dịch từ 4-10-2021 đến 30-12-2022
pd.Index(df_ck["Date"]).get_loc("10-2021")
fig, ax = plt.subplots(figsize=(18, 6), dpi=200)
ax.plot(df_ck["Date"], df_ck["Volume (triệu)"])
plt.xticks(rotation=45)
ax.plot(
[df_ck["Date"][13], df_ck["Date"][13]],
[-100, 100],
ls="--",
lw=3,
alpha=0.8,
color="r",
)
ax.plot(
[df_ck["Date"][33], df_ck["Date"][33]],
[-100, 100],
ls="--",
lw=3,
alpha=0.8,
color="r",
)
ax.text(df_ck["Date"][3], -10, "Trước Covid", fontsize=20)
ax.text(df_ck["Date"][20], -10, "Trong Covid", fontsize=20)
ax.text(df_ck["Date"][38], -10, "Hậu Covid", fontsize=20)
plt.title("Biểu đồ Volume giao dịch chứng khoán VN trong các giai đoạn")
plt.ylabel("Volume (triệu)")
plt.ylim(-150, 1200)
plt.show()
# * Giai đoạn đầu: Trước khi đại dịch bùng phát lấy mốc từ đầu năm 2019 đến đầu năm 2020:
# Cuộc chiến thương mại Mỹ - Trung tiếp tục là gánh nặng đè lên tâm lý các nhà đầu tư quốc tế nói chung và nhà đầu tư Việt nói riêng. Dòng tiền dành cho chứng khoán tiếp tục bị hạn chế và cũng bị hút sang kênh đầu tư trái phiếu doanh nghiệp vốn có lãi suất cao nên không đáng chú ý
# * Giai đoạn dịch bùng phát: Tuy nhiên, khi đại dịch xuất hiện đến khi bùng phát lan rộng,thế giới và Việt Nam từng bước chống lại đại dịch bằng cách kiểm soát sự lây lan dịch bệnh, thực hiện sản xuất vắc-xin Covid-19 cho mục tiêu miễn dịch cộng đồng và phục hồi kinh tế sau đại dịch, các TTCK trên toàn thế giới đều đồng loạt ghi nhận sự tăng điểm và phát triển kỷ lục. Trong đó, TTCK Việt Nam đã ghi nhận sự tăng trưởng với nguồn vốn vô cùng lớn. Khối lượng giao dịch tăng lên rất nhanh chóng và đáng kể so với giai đoạn trước đó.
# # 2.2 So sánh sự biến động về TTCK giữa Việt Nam và Thái Lan trong bối cảnh chung trong đại dịch Covid-19
df_tl = pd.read_csv("/kaggle/input/fin-data-0804/D-liu-Lch-s-SET-Index-1.csv")
df_tl["Date"] = [str(date)[:11] for date in df_tl["Date"]]
df_tl["Date"] = [date.replace("/", "-") for date in df_tl["Date"]]
df_tl["Volume"] = [date.replace(".", "") for date in df_tl["Volume"]]
df_tl["Volume"] = [date.replace("B", "") for date in df_tl["Volume"]]
df_tl["Volume"] = df_tl["Volume"].astype(int)
# sắp xếp ngày tháng từ 2019 đến 2022
df_tl = df_tl.loc[::-1]
df_tl["Volume"] = df_tl["Volume"] * 10
df_tl.columns = df_tl.columns.str.replace("Volume", "Volume (triệu)")
df_tl["Date"] = df_tl["Date"].apply(
lambda x: pd.to_datetime(x, format="%d-%m-%Y").strftime("%m-%Y")
)
df_tl = df_tl[:-4]
df_vn = df_ck
trace1 = go.Scatter(
x=df_vn["Date"],
y=df_vn["Volume (triệu)"],
mode="lines+markers",
name="Vietnam",
marker=dict(color="rgba(27, 79, 147, 0.8)"),
text=df_vn["Volume (triệu)"],
)
data = [trace1]
layout = dict(
title="Biến động của chứng khoán Việt Nam trong giai đoạn đại dịch diễn ra",
xaxis=dict(title="Date", ticklen=5, zeroline=False),
yaxis=dict(title="Volume (triệu)"),
)
fig = dict(data=data, layout=layout)
iplot(fig)
# * Lý do có sự biến động này có thể hiểu theo 2 hướng: Một là trong bối cảnh đại dịch, các kênh đầu tư bị giới hạn và nhiều nhóm ngành trở nên kém hiệu quả, các hoạt động kinh doanh đa phần bị cắt giảm, ngân hàng duy trì lãi suất thấp để hỗ trợ nền kinh tế đã thúc đẩy việc đầu tư chứng khoán như một giải pháp ngắn hạn và không thể thay thế. Lý do thứ 2 là hiệu ứng FOMO - tâm lý đám đông của nhà đầu tư, số đông nhà đầu tư F0 đi theo đám đông tham gia đầu tư vào TTCK trong bối cảnh Covid-19.
# * Theo biểu đồ có thể thấy khối lượng giao dịch tăng nhiều và mạnh vào đầu những năm 2021 - thời điểm mà dịch Covid-19 bùng phát nghiêm trọng tại Việt Nam. Và đỉnh điểm vào tháng 11/2021 được thấy có khoảng Volume giao dịch nhiều nhất.
# # > Quan sát biến động của Thái Lan trong cùng giai đoạn
trace2 = go.Scatter(
x=df_tl["Date"],
y=df_tl["Volume (triệu)"],
mode="lines+markers",
name="ThaiLan",
marker=dict(color="rgba(223, 53, 57, 0.8)"),
text=df_tl["Volume (triệu)"],
)
data = [trace2]
layout = dict(
title="Biến động của chứng khoán Thái Lan",
xaxis=dict(title="Date", ticklen=5, zeroline=False),
yaxis=dict(title="Volume (triệu)"),
)
fig = dict(data=data, layout=layout)
iplot(fig)
# * TTCK tại Thái Lan: Thái Lan tiếp tục là quốc gia Đông Nam Á huy động được nhiều vốn nhất thông qua IPO trong năm 2021
# * So với các nước cùng khu vực, Thái Lan là một nền kinh tế tương đối trưởng thành. Các công ty của Thái Lan đang cố gắng vượt qua các đối thủ cạnh tranh bằng cách mạo hiểm tìm kiếm cơ hội mở rộng kinh doanh ở các nước khác hoặc nâng cấp các dịch vụ công nghệ số tốt hơn.
# * Quan sát biểu đồ có thể thấy trong giai đoạn Covid-19 diễn ra Thái Lan vẫn có sự gia tăng đáng kể về khối lượng giao dịch, tuy nhiên khoảng giữa năm 2022 con số ấy bị giảm mạnh so với Việt Nam.
# # 2.3 So sánh các loại hình chứng khoán được Việt Nam tập trung đầu tư trong giai đoạn biến động này
df_KL = pd.read_excel(
"/kaggle/input/fin-data-0804/Khi-lng-chng-khon-ng-k-n-thng-02-nm-2023_1679045915769-1.xlsx"
)
df_KL = df_KL[6:]
data = {
"Date": df_KL.iloc[:, 0],
"Stock": df_KL.iloc[:, 1],
"Bond": df_KL.iloc[:, 2],
"Warrant": df_KL.iloc[:, 3],
}
df = pd.DataFrame(data)
df = pd.melt(df, id_vars=["Date"], var_name="Stock Type", value_name="Value")
fig = px.bar(
df,
x="Stock Type",
y="Value",
color="Stock Type",
animation_frame="Date",
range_y=[0, 210],
title="So sánh các loại hình chứng khoán",
labels={"value": "Date", "variable": "Loai chung khoan"},
)
fig.show()
# * Về khối lượng chứng khoán đăng ký: nhìn qua biểu đồ có thể thấy cổ phiếu vẫn chiếm phần lớn. Bên cạnh đó thì trái phiếu và chứng quyền cũng đang bắt đầu tăng trưởng hơn trong giai đoạn này tuy không quá nhiều so với cổ phiếu.
# * Đầu năm 2021, khối lượng chứng khoán của cả 3 loại hình tăng lên rõ thông qua biểu đồ chuyển động trên cho thấy sự đa dạng trong danh mục đầu tư chứng khoán tại thị trường Việt Nam.
# # 2.4 Số tài khoản mở mới tăng kỷ lục trong giai đoạn Covid-19 diễn ra
# # 2.4.1 Trong nước
df_TK = pd.read_excel(
"/kaggle/input/fin-data-0804/S-lng-ti-khon-nh-u-t-n-ht-thng-02-nm-2023_1679046732792 (1).xlsx"
)
df_TK = df_TK[6:]
trace1 = go.Bar(
x=df_TK.iloc[:, 0],
y=df_TK.iloc[:, 1],
name="Ca nhan",
marker=dict(color="rgb(66,110,180)", line=dict(color="rgb(0,0,0)", width=1)),
text="Cá nhân",
)
trace2 = go.Bar(
x=df_TK.iloc[:, 0],
y=df_TK.iloc[:, 2],
name="To chuc",
marker=dict(color="rgb(223,53,57)", line=dict(color="rgb(0,0,0)", width=1)),
text="Tổ chức",
)
data = [trace1, trace2]
layout = go.Layout(
barmode="group",
title="So sánh số lượng tài khoản cá nhân và tổ chức (trong nước)",
xaxis=dict(title="Thời gian"),
yaxis=dict(title="Số lượng tài khoản"),
)
fig = go.Figure(data=data, layout=layout)
iplot(fig)
# * Với số lượng tài khoản mở mới tăng mạnh từ giữa năm 2020, tính đến cuối tháng 12/2021, số lượng tài khoản của cá nhân tổ chức mở mới để giao dịch, trái phiếu, cổ phiếu và phái sinh trên TTCK Việt Nam đã ở mức kỷ lục với hơn 4,3 triệu tài khoản, tăng hơn 55% so với cùng kỳ.
# # 2.4.2 Nước ngoài
trace1 = go.Bar(
x=df_TK.iloc[:, 0],
y=df_TK.iloc[:, 3],
name="Ca nhan",
marker=dict(color="rgb(66,110,180)", line=dict(color="rgb(0,0,0)", width=1)),
text="Cá nhân",
)
trace2 = go.Bar(
x=df_TK.iloc[:, 0],
y=df_TK.iloc[:, 4],
name="To chuc",
marker=dict(color="rgb(223,53,57)", line=dict(color="rgb(0,0,0)", width=1)),
text="Tổ chức",
)
data = [trace1, trace2]
layout = go.Layout(
barmode="group",
title="So sánh số lượng tài khoản cá nhân và tổ chức (nước ngoài)",
xaxis=dict(title="Thời gian"),
yaxis=dict(title="Số lượng tài khoản"),
)
fig = go.Figure(data=data, layout=layout)
iplot(fig)
|
import pandas as pd
columns = ["target", "id", "date", "flag", "user", "text"]
df = pd.read_csv(
"/kaggle/input/sentiment140/training.1600000.processed.noemoticon.csv",
encoding="latin-1",
header=None,
names=columns,
)
df.head(5)
df = df.drop(columns=["id", "date", "flag", "user"])
df.head(5)
import re
import string
import nltk
from nltk.corpus import stopwords
from nltk.tokenize import word_tokenize
from sklearn.feature_extraction.text import TfidfVectorizer
# Define the function to preprocess the text
def preprocess_text(text):
# Remove hyperlinks starting with http://
text = re.sub(r"http\S+", "", text)
# Remove @usernames
text = re.sub(r"@[\w]+", "", text)
# Remove punctuation
text = text.translate(str.maketrans("", "", string.punctuation))
# Convert to lowercase
text = text.lower()
# Tokenize the text
tokens = word_tokenize(text)
# Remove stop words
stop_words = set(stopwords.words("english"))
tokens = [token for token in tokens if not token in stop_words]
# Join the tokens back into a string
text = " ".join(tokens)
return text
from tqdm import tqdm
tqdm.pandas()
# Preprocess the text column
df["text"] = df["text"].progress_apply(preprocess_text)
df.head(5)
# Save preprocessed DataFrame as CSV
df.to_csv("preprocessed_data.csv", index=False)
# Sample 20% of the data
df = df.sample(frac=0.2, random_state=42)
# Reset the index
df.reset_index(drop=True, inplace=True)
# Convert text to numerical data using TF-IDF
vectorizer = TfidfVectorizer()
vectorizer.fit(df["text"]) # Fit the vectorizer on the text data
X = []
for text in tqdm(df["text"]):
X.append(vectorizer.transform([text]))
import scipy as sp
X = sp.sparse.vstack(X)
y = df["target"]
import numpy as np
# Save X as a npz file
np.savez_compressed("X.npz", data=X)
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=42
)
from sklearn.neighbors import KNeighborsClassifier
# Create a KNN classifier with k=5
knn = KNeighborsClassifier(n_neighbors=5)
# Fit the model on the training data
knn.fit(X_train, y_train)
# Predict the classes of test data
y_pred = knn.predict(X_test)
# Evaluate the performance of the model
accuracy = knn.score(X_test, y_test)
print("Accuracy:", accuracy)
from sklearn.metrics import classification_report
# Print classification report
print(classification_report(y_test, y_pred))
|
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras.applications.resnet import ResNet50
import pandas as pd
import numpy as np
import os
import cv2
import matplotlib.pyplot as plt
import PIL
import tensorflow as tf
from tensorflow.keras.layers import (
Dense,
Flatten,
Conv2D,
MaxPooling2D,
Dropout,
BatchNormalization,
Activation,
GlobalAveragePooling2D,
)
from tensorflow.keras import applications, layers, losses, optimizers, Model
from tensorflow.keras.models import Sequential
from sklearn.model_selection import train_test_split
import keras.backend as K
# # Retrain the model on the new dataset
P = "../input/15-after-processed/15_after_processed/"
train_image = tf.keras.utils.image_dataset_from_directory(
P + "train_images/",
labels="inferred",
label_mode="categorical",
class_names=None,
color_mode="rgb",
batch_size=32,
image_size=(224, 224),
)
val_image = tf.keras.utils.image_dataset_from_directory(
P + "val_images/",
labels="inferred",
label_mode="categorical",
class_names=None,
color_mode="rgb",
batch_size=32,
image_size=(224, 224),
)
num_classes = len(train_image.class_names)
resnet = Sequential()
pretrained_model = applications.ResNet50(
include_top=False,
weights="imagenet",
input_shape=(224, 224, 3),
pooling="avg",
classes=num_classes,
classifier_activation="softmax",
)
for layer in pretrained_model.layers:
layer.trainable = False
resnet.add(pretrained_model)
# add batch normalization layer
resnet.add(BatchNormalization())
# flatten the output
resnet.add(Flatten())
# add dropout layer
resnet.add(Dropout(0.5))
# add linear layer for reduction
resnet.add(Dense(512, activation="linear"))
# add classification layer
resnet.add(Dense(num_classes, activation="softmax"))
epochs = 25
def get_f1(y_true, y_pred): # taken from old keras source code
true_positives = K.sum(K.round(K.clip(y_true * y_pred, 0, 1)))
possible_positives = K.sum(K.round(K.clip(y_true, 0, 1)))
predicted_positives = K.sum(K.round(K.clip(y_pred, 0, 1)))
precision = true_positives / (predicted_positives + K.epsilon())
recall = true_positives / (possible_positives + K.epsilon())
f1_val = 2 * (precision * recall) / (precision + recall + K.epsilon())
return f1_val
optimizer = keras.optimizers.Adam(learning_rate=0.0001)
resnet.compile(optimizer=optimizer, loss="categorical_crossentropy", metrics=[get_f1])
# Define a function to preprocess the labels in the validation set
def preprocess_labels(image, label):
label = tf.argmax(label, axis=-1)
label = tf.one_hot(label, num_classes)
return image, label
# Preprocess the labels in the validation set
val_image = val_image.map(preprocess_labels)
checkpoint_filepath = "new_resenet.ckpt"
model_checkpoint_callback = tf.keras.callbacks.ModelCheckpoint(
filepath=checkpoint_filepath,
save_weights_only=True,
monitor="val_loss",
mode="auto",
save_best_only=True,
verbose=1,
save_freq="epoch",
)
resnet.fit(
train_image,
epochs=epochs,
validation_data=val_image,
callbacks=[model_checkpoint_callback],
)
|
# Importing the necessary libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
pd.options.mode.chained_assignment = None
import re
# Loading the dataset
df = pd.read_csv(
"/kaggle/input/exploring-wealth-forbes-richest-people-dataset/forbes_richman.csv",
encoding="ISO-8859-1",
)
df.head(10)
# Datatype and shape of Datset
print(df.dtypes)
print(df.shape)
# Checking for Null Values
null_counts = df.isnull().sum()
print(null_counts)
df = df.iloc[0:2509, :]
print()
null_counts = df.isnull().sum()
print(null_counts)
print(df.shape)
# There are total of 2509 billionaries in the world from the data and 79 of them do not have age data in the dataset.
# Dataframe contaning values with null ages
df_null = df.loc[df["Age"].isnull()]
# Helper Function to Check for more than one preson in Name
def check_characters(string):
if (
"family" in string
or "&" in string
or "brothers" in string
or "siblings" in string
):
return True
else:
return False
# Apply the function to the DataFrame
x = df_null.loc[:, "Name"].apply(check_characters)
df_null["Null_count"] = x
# Checking if the missing datapoint of age is due to a family/more than one person
num = len(df_null.loc[df_null["Null_count"] == True])
print(
f"Number of datapoints with missing ages beloning to more than 1 people: {num}({num*100/79:.2f}%)"
)
df_null.loc[df_null["Null_count"] == True]
# Checking the age distribution of billionaries
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(20, 8))
sns.histplot(x="Age", data=df, ax=ax1, kde=True)
ax1.set_title("Histogram for Age Distribution")
sns.boxplot(x="Age", data=df, ax=ax2, color="thistle")
ax2.set_title("CountPlot for Age Distribution")
plt.show()
print(df["Age"].describe())
# Values below this are classifies as outliers
df["Age"].quantile(0.25) - 1.5 * (df["Age"].quantile(0.75) - df["Age"].quantile(0.25))
# Youngest billionaires in the world
display(df.loc[df["Age"] <= 26])
# Regular Expression function to extract Net Worth number from Net worth column
def net_worth(net):
pattern = r"\d+(\.\d+)?"
match = re.search(pattern, net)
if match:
numeric_value = float(match.group())
return numeric_value
else:
pass
df["Net"] = df["Net Worth"].apply(net_worth)
# Distribution for the Net Worth
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(20, 8))
sns.histplot(x="Net", data=df, bins=20, ax=ax1, kde=True)
ax1.set_title("Histogram for Net Worth Distribution")
ax1.set_xlabel("Net Worth (in billion USD)")
sns.boxplot(x="Net", data=df, ax=ax2, color="thistle")
ax2.set_title("CountPlot for Net Worth Distribution")
ax2.set_xlabel("Net Worth (in billion USD)")
plt.show()
print(df["Net"].describe())
# Outlier Range for Net Worth
out = df["Net"].quantile(0.25) + 1.5 * (
df["Net"].quantile(0.75) - df["Net"].quantile(0.25)
)
sns.boxplot(x="Net", data=df[df["Net"] < out], color="thistle")
plt.title("CountPlot for Net Worth Distribution(Removing Outliers)")
plt.xlabel("Net Worth (in billion USD)")
plt.show()
# Total net worth of all billionaries
sum_of_column = df["Net"].sum()
print(
f"The richest 2509 people in the world have a combined net worth of around {sum_of_column/1000:.2f} Trillion USD"
)
# Vizualization of top 10 richest people of the world
sns.barplot(x="Net", y="Name", data=df.loc[df["Rank"] < 11])
plt.xlabel("Net Worth (in Billion USD)")
plt.ylabel("Name of the Billionaires")
plt.title("Net Worth of the Top 10 Richest People in the World")
plt.show()
# Grouping people according to Country
grouped = df.groupby("Country")
# Countries with most billionaries
s = grouped["Name"].size().sort_values(ascending=False)[:15]
sns.barplot(x=s.values, y=s.index)
plt.xlabel("Number of Billionaires")
plt.ylabel("Name of the Countries")
plt.title("Top 15 Countries with Most Billionaires")
plt.show()
print(s)
# Industries with most billionaires
sns.countplot(y="Industry", data=df, order=df["Industry"].value_counts().index)
plt.xlabel("Number of Billionaires")
plt.ylabel("Industry")
plt.title("Industry vs Number of Billionaires")
plt.show()
print(df["Industry"].value_counts())
# Distribution of Net Worth in Industries
sns.boxplot(y="Industry", x="Net", data=df[df["Net"] < 10])
plt.xlabel("Net Worth (in billion USD)")
plt.show()
# ##### EDA for India
# Selecting dataframe containing Indian billionaires
df_ind = df.loc[df["Country"] == "India"]
# Top 10 richest people of India
sns.barplot(x="Net", y="Name", data=df_ind.iloc[:10, :])
plt.xlabel("Net Worth (in Billion USD)")
plt.ylabel("Name of the Billionaires")
plt.title("Net Worth of the Top 10 Richest People in India")
plt.show()
sns.boxplot(x="Age", data=df_ind)
plt.title("Distribution of Age for Indian Billionaries")
plt.show()
print(df_ind["Age"].describe())
# If we compare from the global values, we see that the mean age of a billionaire in India is higher than in the world.
# Youngest billionaries in India
df_ind[df_ind["Age"] < 45]
# Oldest billionaries in India
df_ind[df_ind["Age"] > 90]
sns.boxplot(x="Net", data=df_ind)
plt.title("Distrbution of Net Worth")
plt.xlabel("Net Worth (in billion USD)")
plt.show()
# Outlier
print(
df_ind["Net"].quantile(0.25)
+ 1.5 * (df["Net"].quantile(0.75) - df["Net"].quantile(0.25))
)
sns.boxplot(x="Net", data=df_ind[df_ind["Net"] < 5.6])
plt.title("Distrbution of Net Worth (Removing Outliers)")
plt.xlabel("Net Worth (in billion USD)")
plt.show()
print(df_ind["Net"].describe())
sns.countplot(y="Industry", data=df_ind, order=df_ind["Industry"].value_counts().index)
plt.xlabel("Number of Billionaries")
plt.ylabel("Industry")
plt.title("Industry vs Number of Billionaries in India")
plt.show()
print(df_ind["Industry"].value_counts())
# Distribution of Net Worth in Industries in India
sns.boxplot(y="Industry", x="Net", data=df_ind[df_ind["Net"] < 50])
plt.xlabel("Net Worth (in billion USD)")
plt.show()
|
# Importing Modules,Libraries and Packages
import numpy as np
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import Model
from tensorflow.keras import backend as K
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import (
Activation,
Dense,
Flatten,
BatchNormalization,
Conv2D,
MaxPool2D,
Conv2DTranspose,
UpSampling2D,
concatenate,
)
from tensorflow.keras.preprocessing import image
from tensorflow.keras.optimizers import Adam
from tensorflow.keras.metrics import categorical_crossentropy
from tensorflow.keras.utils import to_categorical
from tensorflow.keras.preprocessing.image import ImageDataGenerator
import os
import os
import numpy as np
import cv2
from tensorflow import keras
import matplotlib.pyplot as plt
import random
from sklearn.model_selection import train_test_split
from sklearn.svm import SVC
from skimage import img_as_float64
from tensorflow.keras.preprocessing import image
from skimage import img_as_float
from keras.preprocessing.image import ImageDataGenerator
import tensorflow as tf
from tensorflow.keras.preprocessing.image import ImageDataGenerator
from sklearn.preprocessing import (
StandardScaler,
MinMaxScaler,
MaxAbsScaler,
RobustScaler,
PowerTransformer,
Normalizer,
)
dir = "/kaggle/input/kidney-images/CT-KIDNEY-DATASET-Normal-Cyst-Tumor-Stone/"
catagories = ["Normal", "Stone"]
data = []
for category in catagories:
path = os.path.join(dir, category)
label = catagories.index(category)
for img in os.listdir(path):
imgpath = os.path.join(path, img)
kid_img = cv2.imread(imgpath)
try:
kid_img = cv2.resize(kid_img, (128, 128))
image = np.array(kid_img).flatten()
data.append([image, label])
except Exception as e:
pass
print(len(data))
random.shuffle(data)
features = []
labels = []
for feature, label in data:
features.append(feature)
labels.append(label)
xtrain, xtest, ytrain, ytest = train_test_split(features, labels, test_size=0.3)
svm = SVC()
svm.fit(xtrain, ytrain)
svm.score(xtest, ytest)
model = SVC(C=1, kernel="poly", gamma="auto")
model.fit(xtrain, ytrain)
print("training accuracy ", model.score(xtrain, ytrain))
prediction = model.predict(xtest)
accuracy = model.score(xtest, ytest)
catagories = ["Normal", "Kidney_stone"]
catagories = ["Normal", "Kidney_stone"]
print("Accuracy ", accuracy)
print("Prediction is : ", catagories[prediction[100]])
catagories = ["Normal", "Kidney_stone"]
print("Accuracy ", accuracy)
print("Prediction is : ", catagories[prediction[0]])
catagories = ["Normal", "Kidney_stone"]
print("Accuracy ", accuracy)
print("Prediction is : ", catagories[prediction[10]])
# ### CNN
# Import libraries, packages, modules, functions, etc...
import numpy as np
import cv2
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras.models import Model
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import (
Activation,
Dense,
Flatten,
BatchNormalization,
Conv2D,
MaxPool2D,
Dropout,
)
from tensorflow.keras.optimizers import Adam
from tensorflow.keras.metrics import categorical_crossentropy
from tensorflow.keras.preprocessing.image import ImageDataGenerator
from sklearn.metrics import confusion_matrix
import itertools
import os
import shutil
import random
import glob
import matplotlib.pyplot as plt
from sklearn.preprocessing import MinMaxScaler
import warnings
from sklearn.utils import shuffle
from sklearn.model_selection import train_test_split
warnings.simplefilter(action="ignore", category=FutureWarning)
Xtrain, Xtest, Ytrain, Ytest = train_test_split(
features, labels, test_size=0.25, random_state=80
)
print(len(Xtrain), len(Xtest), len(Ytrain), len(Ytest))
# Converting the list to a numpy array as a requirement for the input in fit function.
Xtrain = np.array(Xtrain)
Xtest = np.array(Xtest)
Ytrain = np.array(Ytrain)
Ytest = np.array(Ytest)
# Shuffle the data
Ytest, Xtest = shuffle(Ytest, Xtest)
model = Sequential()
model.add(
Conv2D(32, (3, 3), activation="relu", input_shape=(128, 128, 1), padding="same")
)
model.add(MaxPool2D(2))
model.add(Dropout(0.2))
model.add(Conv2D(32, (3, 3), activation="relu", padding="same"))
model.add(MaxPool2D(pool_size=(2, 2), strides=2))
# model.add(Dropout(0.4))
model.add(Conv2D(64, (3, 3), activation="relu", padding="same"))
model.add(MaxPool2D(pool_size=(2, 2), strides=2))
# model.add(Dropout(0.5))
model.add(Conv2D(64, (3, 3), activation="relu", padding="same"))
model.add(MaxPool2D(pool_size=(2, 2), strides=2))
model.add(Conv2D(128, (3, 3), activation="relu", padding="same"))
model.add(MaxPool2D(pool_size=(2, 2), strides=2))
model.add(Conv2D(128, (3, 3), activation="relu", padding="same"))
model.add(MaxPool2D(pool_size=(2, 2), strides=2))
model.add(Dropout(0.6))
model.add(Dense(512, activation="relu"))
model.add(Flatten())
model.add(Dense(1, activation="sigmoid"))
model.summary()
# compile function configures the Sequential model for training.
# Optimizers are algorithms or methods used to change the attributes of the neural network such as weights and learning rate to reduce the losses. Optimizers are used to solve optimization problems by minimizing the function.
# Crossentropy loss function when there are two or more label classes.
# We expect labels to be provided as integers.
model.compile(
optimizer=Adam(learning_rate=0.001),
loss="binary_crossentropy",
metrics=["accuracy"],
)
Xtrain = Xtrain.reshape(4840, 128, 128, 3)
Xtest = Xtest.reshape(1614, 128, 128, 3)
model.fit(Xtrain, Ytrain, epochs=120, batch_size=100, verbose=2)
import tensorflow as tf
from tensorflow.keras.metrics import Accuracy
from tensorflow.keras.datasets import mnist
# Evaluate model on test data
test_loss, test_acc = model.evaluate(Xtest, Ytest, verbose=0)
print("test loss", test_loss)
# Print model accuracy score
print("Test accuracy:", test_acc)
model.predict(Xtest)
catagories = ["Normal", "Kidney_stone"]
print("Prediction is : ", catagories[prediction[0]])
model.predict(Xtest)
catagories = ["Normal", "Kidney_stone"]
print("Prediction is : ", catagories[prediction[100]])
# **RESNET50**
from keras.applications.resnet import ResNet50
from sklearn.model_selection import KFold
IMAGE_SIZE = [128, 128]
restnet = ResNet50(input_shape=IMAGE_SIZE + [3], weights="imagenet", include_top=False)
output = restnet.layers[-1].output
output = Flatten()(output)
restnet = Model(restnet.input, output)
for layer in restnet.layers:
layer.trainable = False
# restnet.summary()
model = Sequential()
model.add(restnet)
model.add(Dense(512, activation="relu"))
model.add(Dropout(0.3))
model.add(Dense(512, activation="relu"))
model.add(Dropout(0.3))
model.add(Flatten())
model.add(Dense(1, activation="sigmoid"))
model.compile(optimizer="adam", loss="binary_crossentropy", metrics=["accuracy"])
model.summary()
# Prepare the data
X = np.array(Xtrain)
Y = np.array(Ytrain)
# Set the number of folds and initialize the results array
num_folds = 5
results = []
graph = []
# Create the KFold object and loop over the folds
kfold = KFold(n_splits=num_folds, shuffle=True)
for train_ix, val_ix in kfold.split(X):
# Split the data into training and validation sets
X_train, X_val = X[train_ix], X[val_ix]
Y_train, Y_val = Y[train_ix], Y[val_ix]
# Train the model on the training data
final = model.fit(X_train, Y_train, epochs=10, batch_size=200, verbose=2)
# Evaluate the model on the validation data
_, acc = model.evaluate(X_val, Y_val, verbose=0)
graph.append(final)
# Append the accuracy to the results array
results.append(acc)
# Evaluate model on test data
test_loss, test_acc = model.evaluate(Xtest, Ytest, verbose=0)
print("test loss", test_loss)
# Print model accuracy score
print("Test accuracy:", test_acc)
print(
"Cross-validation results: %.2f%% (+/- %.2f%%)"
% (np.mean(results) * 100, np.std(results) * 100)
)
plt.title("Accuracies vs Epochs")
plt.plot(graph[0].history["accuracy"], label="Training Fold 1")
plt.plot(graph[1].history["accuracy"], label="Training Fold 2")
plt.plot(graph[2].history["accuracy"], label="Training Fold 3")
plt.plot(graph[3].history["accuracy"], label="Training Fold 4")
plt.plot(graph[4].history["accuracy"], label="Training Fold 5")
plt.legend()
plt.show()
# **VGG16**
from keras.applications.vgg16 import preprocess_input, decode_predictions, VGG16
IMAGE_SIZE = [128, 128]
pre_trained_model = VGG16(
input_shape=IMAGE_SIZE + [3], weights="imagenet", include_top=False
)
output = pre_trained_model.layers[-1].output
output = Flatten()(output)
vgg16 = Model(pre_trained_model.input, output)
for layer in vgg16.layers:
layer.trainable = False
# restnet.summary()
model = Sequential()
model.add(vgg16)
model.add(Dense(512, activation="relu"))
model.add(Dropout(0.5))
# model.add(Dense(512, activation='relu'))
# model.add(Dropout(0.3))
# model.add(Flatten())
model.add(Dense(1, activation="sigmoid"))
model.summary()
model.compile(optimizer="adam", loss="binary_crossentropy", metrics=["accuracy"])
# Prepare the data
X = np.array(Xtrain)
Y = np.array(Ytrain)
# Set the number of folds and initialize the results array
num_folds = 5
results = []
graph = []
# Create the KFold object and loop over the folds
kfold = KFold(n_splits=num_folds, shuffle=True)
for train_ix, val_ix in kfold.split(X):
# Split the data into training and validation sets
X_train, X_val = X[train_ix], X[val_ix]
Y_train, Y_val = Y[train_ix], Y[val_ix]
# Train the model on the training data
final = model.fit(X_train, Y_train, epochs=10, batch_size=10, verbose=2)
# Evaluate the model on the validation data
_, acc = model.evaluate(X_val, Y_val, verbose=0)
graph.append(final)
# Append the accuracy to the results array
results.append(acc)
test_loss, test_acc = model.evaluate(Xtest, Ytest, verbose=0)
print("test loss", test_loss)
# Print model accuracy score
print("Test accuracy:", test_acc)
print(
"Cross-validation results: %.2f%% (+/- %.2f%%)"
% (np.mean(results) * 100, np.std(results) * 100)
)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import tensorflow as tf
from tensorflow.keras import datasets, layers, models
train_datagen = tf.keras.preprocessing.image.ImageDataGenerator(
rescale=1.0 / 255, zoom_range=0.2, horizontal_flip=True
)
val_datagen = tf.keras.preprocessing.image.ImageDataGenerator(rescale=1.0 / 255)
train_generator = train_datagen.flow_from_directory(
"/kaggle/input/car-parking-space/carpark_dataset",
target_size=(256, 256),
batch_size=32,
)
validation_generator = val_datagen.flow_from_directory(
"/kaggle/input/car-parking-space/carpark_dataset",
target_size=(256, 256),
batch_size=32,
)
test_generator = val_datagen.flow_from_directory(
"/kaggle/input/car-parking-space/carpark_dataset",
target_size=(256, 256),
batch_size=32,
)
num_classes = train_generator.num_classes
nb_train_samples = train_generator.samples
nb_val_samples = validation_generator.samples
base_model = tf.keras.applications.vgg19.VGG19(
include_top=False, weights="imagenet", input_tensor=None, input_shape=(256, 256, 3)
)
def prepare_VGG_model_for_finetuning(freeze_baselayers=True):
"""
input_shape: When pretrained networks are utilised, the input shape for the network is generally
dependent on the shape of the data the pretrained networks were originally trained. It allows us the exact use of weights.++
"""
base_model = tf.keras.applications.vgg19.VGG19(
include_top=False,
weights="imagenet",
input_tensor=None,
input_shape=(256, 256, 3),
) # freeze layers
if freeze_baselayers == True:
for layer in base_model.layers:
layer.trainable = False
# change here - take the output from different layers of the base model.
# Eg. base_model.output, base_model.layers[11].output
x = base_model.output
# x = tf.keras.layers.GlobalAveragePooling2D(2)(x)
x = tf.keras.layers.Flatten()(x)
x = tf.keras.layers.Dense(64, activation="relu")(x)
x = tf.keras.layers.Dense(num_classes)(x)
prediction_layer = tf.keras.layers.Softmax()(x)
model_new = tf.keras.Model(inputs=base_model.input, outputs=prediction_layer)
return model_new
model = prepare_VGG_model_for_finetuning(freeze_baselayers=True)
loss_fn = tf.keras.losses.CategoricalCrossentropy(from_logits=True)
optimizer_fn = tf.keras.optimizers.Adam(learning_rate=0.001)
model.compile(optimizer=optimizer_fn, loss=loss_fn, metrics=["accuracy"])
history = model.fit_generator(
train_generator,
steps_per_epoch=nb_train_samples // 32,
epochs=1,
validation_data=validation_generator,
validation_steps=nb_val_samples / 32,
)
model.summary()
model.save("vgg_model.h5")
from IPython.display import FileLink
FileLink(r"vgg_model.h5")
|
# # Import
from torchvision import datasets, transforms, models
from torch.utils.data import random_split, Dataset, DataLoader
import numpy as np
import pandas as pd
from PIL import Image
import matplotlib.pyplot as plt
import seaborn as sns
import torch
import torchvision
import os
import copy
from sklearn.model_selection import train_test_split
from torchvision.utils import make_grid
from mpl_toolkits.axes_grid1 import ImageGrid
import time
plt.rcParams["figure.figsize"] = (12, 6)
torch.manual_seed(0)
def show_tensor_images(image_tensor, num_images=16, size=(3, 224, 224)):
"""
Function for visualizing images: Given a tensor of images, number of images, and
size per image, plots and prints the images in an uniform grid.
"""
# image_tensor = (image_tensor + 1) / 2
image_shifted = image_tensor
image_unflat = image_shifted.detach().cpu().view(-1, *size)
image_grid = make_grid(image_unflat[:num_images], nrow=4)
plt.imshow(image_grid.permute(1, 2, 0).squeeze())
plt.show()
# # Create DataLoaders
def find_classes(fulldir):
classes = os.listdir(fulldir)
classes.sort()
class_to_idx = dict(zip(classes, range(len(classes))))
idx_to_class = {v: k for k, v in class_to_idx.items()}
train = []
for i, label in idx_to_class.items():
path = fulldir + "/" + label
for file in os.listdir(path):
train.append([f"{label}/{file}", label, i])
df = pd.DataFrame(train, columns=["file", "class", "class_index"])
return classes, class_to_idx, idx_to_class, df
root_dir = "/kaggle/input/plant-seedlings-classification/train"
classes, class_to_idx, idx_to_class, df = find_classes(root_dir)
num_classes = len(classes)
g = sns.countplot(
data=df, x="class", order=df["class"].value_counts().index, palette="Greens_r"
)
plt.xticks(rotation=45)
class PlantDataset(Dataset):
def __init__(self, dataframe, root_dir, transform=None):
self.transform = transform
self.df = dataframe
self.root_dir = root_dir
# self.classes =
def __len__(self):
return len(self.df)
def __getitem__(self, idx):
fullpath = os.path.join(self.root_dir, self.df.iloc[idx][0])
image = Image.open(fullpath).convert("RGB")
if self.transform:
image = self.transform(image)
return image, self.df.iloc[idx][2]
train_transform = transforms.Compose(
[
transforms.RandomRotation(180),
transforms.RandomAffine(degrees=0, translate=(0.2, 0.2)),
# transforms.CenterCrop(356),
transforms.Resize((324, 324)),
transforms.RandomHorizontalFlip(),
transforms.RandomVerticalFlip(),
transforms.ToTensor(),
transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),
]
)
val_transform = transforms.Compose(
[
# transforms.CenterCrop(356),
transforms.Resize((324, 324)),
transforms.ToTensor(),
transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),
]
)
X_train, X_val = train_test_split(
df, test_size=0.2, random_state=42, stratify=df["class_index"]
)
train_dataset = PlantDataset(X_train, root_dir, train_transform)
val_dataset = PlantDataset(X_val, root_dir, val_transform)
batch_size = 64
num_workers = 2
train_loader = DataLoader(
train_dataset,
batch_size=batch_size,
num_workers=num_workers,
shuffle=True,
drop_last=True,
)
val_loader = DataLoader(
val_dataset, batch_size=batch_size, num_workers=num_workers, drop_last=True
)
# img, label = next(iter(train_loader))
# show_tensor_images(img, num_images=16, size=(3, 324, 324))
# # Train Model
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
def train_model(model, criterion, optimizer, scheduler, num_epochs=10, device=device):
# since = time.time()
model.to(device)
best_model_wts = copy.deepcopy(model.state_dict())
best_acc = 0.0
for epoch in range(num_epochs):
for phase in ["train", "val"]:
if phase == "train":
model.train()
train_loss = 0.0
train_acc = 0
for image, label in train_loader:
image = image.to(device)
label = label.to(device)
# forward
y_pred = model(image)
# loss
loss = criterion(y_pred, label)
train_loss += loss.item()
# optimizer zero grad
optimizer.zero_grad()
# loss backward
loss.backward()
# update
optimizer.step()
y_pred_class = torch.argmax(torch.softmax(y_pred, dim=1), dim=1)
train_acc += (y_pred_class == label).sum().item() / len(y_pred)
scheduler.step()
train_loss /= len(train_loader)
train_acc /= len(train_loader)
else:
model.eval()
test_loss, test_acc = 0, 0
with torch.inference_mode():
for image, label in val_loader:
image = image.to(device)
label = label.to(device)
test_pred_logits = model(image)
loss = criterion(test_pred_logits, label)
test_loss += loss.item()
test_pred_labels = test_pred_logits.argmax(dim=1)
test_acc += (test_pred_labels == label).sum().item() / len(
test_pred_labels
)
test_loss = test_loss / len(val_loader)
test_acc = test_acc / len(val_loader)
if test_acc > best_acc:
best_acc = test_acc
best_model_wts = copy.deepcopy(model.state_dict())
print(
f"Epoch: {epoch+1} | "
f"train_loss: {train_loss:.4f} | "
f"train_acc: {train_acc:.4f} | "
f"test_loss: {test_loss:.4f} | "
f"test_acc: {test_acc:.4f}"
)
model.load_state_dict(best_model_wts)
return model
model = models.resnet50(weights=models.ResNet50_Weights.DEFAULT)
for param in model.parameters():
param.requires_grad = False
num_ftrs = model.fc.in_features
model.fc = torch.nn.Sequential(
torch.nn.Linear(num_ftrs, 256),
torch.nn.ReLU(),
torch.nn.Dropout(0.4),
torch.nn.Linear(256, num_classes),
)
criterion = torch.nn.CrossEntropyLoss()
# Observe that all parameters are being optimized
optimizer = torch.optim.Adam(model.parameters(), lr=0.005)
## Decay LR by a factor of 0.1 every 5 epochs
exp_lr_scheduler = torch.optim.lr_scheduler.StepLR(optimizer, step_size=10, gamma=0.8)
model_ft = train_model(model, criterion, optimizer, exp_lr_scheduler, num_epochs=20)
torch.save(model_ft, "best_model.pt")
# # Testing
class TestPlant(Dataset):
def __init__(self, rootdir, transform=None):
self.transform = transform
self.rootdir = rootdir
self.image_files = os.listdir(self.rootdir)
def __len__(self):
return len(self.image_files)
def __getitem__(self, idx):
fullpath = os.path.join(self.rootdir, self.image_files[idx])
image = Image.open(fullpath).convert("RGB")
if self.transform:
image = self.transform(image)
return image, self.image_files[idx]
test_dataset = TestPlant(
rootdir="/kaggle/input/plant-seedlings-classification/test", transform=val_transform
)
test_loader = DataLoader(
test_dataset, batch_size=batch_size, num_workers=num_workers, shuffle=False
)
# img, file_names = next(iter(test_loader))
# show_tensor_images(img, num_images=16, size=(3, 324, 324))
test_filenames = os.listdir("/kaggle/input/plant-seedlings-classification/test")
submission = pd.DataFrame(test_filenames, columns=["file"])
submission["species"] = ""
submission.head()
def test(submission, test_loader, model, device=device):
model.to(device)
with torch.no_grad():
for image, file_names in test_loader:
image = image.to(device)
y_pred = model(image)
y_pred_labels = y_pred.argmax(dim=1)
y_pred_labels = y_pred_labels.cpu().numpy()
submission.loc[
submission["file"].isin(file_names), "species"
] = y_pred_labels
model = torch.load("/kaggle/working/best_model.pt")
model.eval()
test(submission, test_loader, model)
submission.head(20)
submission = submission.replace({"species": idx_to_class})
submission.to_csv("/kaggle/working/submission.csv", index=False)
|
import torch
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
print(device)
# ----
# # Imports
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import string
from matplotlib.colors import is_color_like
import nltk
from nltk.corpus import stopwords
nltk.download("stopwords")
nltk.download("punkt")
stop_words = stopwords.words("english")
print(stop_words)
from nltk.stem import PorterStemmer
porter = PorterStemmer()
from sklearn.feature_extraction.text import (
TfidfTransformer,
TfidfVectorizer,
CountVectorizer,
)
from sklearn.cluster import KMeans
from gensim.summarization import summarize # pip3 install gensim==3.6.0
from gensim.summarization import keywords
from joblib import dump, load
# ----
CSV_DIRECTORY = "../input/data-combined/data.csv"
df = pd.read_csv(CSV_DIRECTORY, index_col=0).reset_index()
df.head()
# ## More Answer Types
def more_separation(input_question, answer):
question = input_question.split()
# filter_question = filter(lambda w: not w in stopwords, split_question.split())
if answer.lower() == "yes" or answer.lower() == "no":
return "yes/no"
elif answer.translate(str.maketrans("", "", string.punctuation)).isnumeric():
return "number"
###
elif (len(answer) > 1 and is_color_like(answer)) or answer == "clear":
return "colour"
elif question[0] == "Where":
return "location"
elif question[0] == "What" and question[1] == "is":
return "identify"
else:
return "other"
df["answer_type"] = df.apply(
lambda x: more_separation(x["question"], x["answer"]), axis=1
)
# ## Kmeans for Question Labels
def remove_stopwords_punctuation_from_tokens(question_tokens):
new_list = []
for token in eval(question_tokens):
if token not in stop_words and token not in string.punctuation:
# print(token)
new_list.append(token)
return new_list
def stem_tokens(question_tokens):
return [porter.stem(token) for token in question_tokens]
def normalize_tokens(tokens):
return [token.lower() for token in tokens]
df["question_tokens"] = (
df["question_tokens"]
.apply(remove_stopwords_punctuation_from_tokens)
.apply(normalize_tokens)
)
df = df[~df["question_tokens"].apply(lambda x: isinstance(x, (list)) and len(x) == 0)]
df.head()
# Convert the preprocessed tokens to strings
df["question_text"] = df["question_tokens"].apply(lambda x: " ".join(x))
df["token_text"] = df["question_text"] + " " + df["answer"]
df.head()
tfidf = TfidfVectorizer()
# tfidf_weights = {'question_text': 1.0, 'answer': 2.0}
tfidf_matrix = tfidf.fit_transform(df["token_text"])
# Print the shape of the tfidf matrix
print(tfidf_matrix.shape)
tfidf_matrix.todense()
# # Initialize the KMeans model
# kmeans = KMeans()
# kmeans.fit(tfidf_matrix)
# MAX_N_CLUSTERS = 30
# # Calculate the within-cluster sum of squares (WCSS) for different numbers of clusters
# ssd = []
# for i in range(1, MAX_N_CLUSTERS + 1):
# kmeans = KMeans(n_clusters=i, init='k-means++', max_iter=300, n_init=10, random_state=0)
# kmeans.fit(tfidf_matrix)
# ssd.append(kmeans.inertia_)
# # Plot the elbow curve
# plt.plot(range(1, MAX_N_CLUSTERS + 1), ssd, 'o--')
# plt.xlabel('Number of Clusters')
# plt.ylabel('SSD')
# plt.show()
N_CLUSTERS = 20
# Initialize the KMeans model with the optimal number of clusters
kmeans = KMeans(
n_clusters=N_CLUSTERS, init="k-means++", max_iter=300, n_init=10, random_state=0
)
kmeans.fit(tfidf_matrix)
# Add the cluster labels to the dataframe
df["cluster_label"] = kmeans.labels_
dump(kmeans, "./kmeans_model.joblib")
# Print the number of questions in each cluster
df["cluster_label"].value_counts()
NO_SUMMARY_WORDS = 1
NO_SUMMARY_SAMPLE_SIZE = 1000
cluster_labels = {}
for idx in range(N_CLUSTERS):
subset = df[df["cluster_label"] == idx].sample(
n=min(NO_SUMMARY_SAMPLE_SIZE, len(df)), random_state=42
)
text = ". ".join(subset["question_text"].tolist())
sentences = nltk.sent_tokenize(text)
summary = summarize(" ".join(sentences))
summary_keywords = keywords(summary, words=NO_SUMMARY_WORDS, lemmatize=True)
if NO_SUMMARY_WORDS > 1:
summary_keywords = summary_keywords.split("\n")
summary_keywords = "_".join(summary_keywords)
cluster_labels[f"{idx}"] = summary_keywords
print(cluster_labels)
df.head()
def assign_named_label(label):
return cluster_labels[f"{label}"]
df["cluster_name"] = df["cluster_label"].apply(assign_named_label)
df.head()
counts = df["cluster_name"].value_counts()
# Create a pie chart from the counts data
plt.pie(counts.values, labels=counts.index)
# Add a title to the chart
plt.title("Cluster Label Distribution")
# Display the chart
plt.show()
df.to_csv("k_means.csv", index=False)
df.head()
# df_copy = df.copy(True)
# def get_cluster_name(cluster_name, summ_word):
# if summ_word < len(cluster_name):
# return cluster_name[summ_word]
# else:
# return ""
# for summ_word in range(NO_SUMMARY_WORDS):
# summ_word = summ_word.split("\n")
# column_name = f'cluster_label_{summ_word}'
# df_copy[column_name] = df_copy['cluster_list'].apply(get_cluster_name, summ_word=summ_word)
# # f.apply(lambda x: more_separation(x["question"], x["answer"]), axis=1)
# df_copy.head()
# # https://stackoverflow.com/a/44154062
# cm = plt.get_cmap("tab20c")
# # Define your data
# label_0_counts = df_copy['cluster_label_0'].value_counts()
# label_1_counts = df_copy['cluster_label_1'].value_counts()
# # Define your colors
# color_0 = plt.cm.Set1(np.arange(len(label_0_counts)))
# color_1 = plt.cm.Set2(np.arange(len(label_1_counts)))
# # Set up the plot
# fig, ax = plt.subplots()
# ax.axis('equal')
# width = 0.35
# # Plot the first pie chart# Define your data and labels
# data = [10, 20, 30, 40, 50] * 4
# labels = ['A', 'B', 'C', 'D', 'E']
# # Set up the plot
# fig, axes = plt.subplots(nrows=4, ncols=5, figsize=(12, 9))
# axes = axes.flatten() # Flatten the 2D array of axes into a 1D array
# # Loop through each axis and plot a pie chart
# for i, ax in enumerate(axes):
# ax.axis('equal')
# ax.set_title(f'Pie Chart {i+1}')
# ax.pie(data[i:i+5], labels=labels)
# # Adjust spacing between plots and display the figure
# plt.subplots_adjust(wspace=0.5, hspace=0.5)
# plt.show()
# outer = ax.pie(label_0_counts.values, radius=1, labels=label_0_counts.index, colors=color_0,
# wedgeprops=dict(width=width, edgecolor='w'))
# # Plot the second pie chart
# inner = ax.pie(label_1_counts.values, radius=1-width, labels=label_1_counts.index, labeldistance=0.7, colors=color_1,
# wedgeprops=dict(width=width, edgecolor='w'))
# # Add a circle in the middle to create the donut effect
# circle = plt.Circle((0,0), 0.35, color='white')
# ax.add_artist(circle)
# # Add legends and titles
# # ax.legend(outer[0], label_0_counts.index, loc='upper left', bbox_to_anchor=(-0.1, 1.))
# # ax.legend(inner[0], label_1_counts.index, loc='upper right', bbox_to_anchor=(1.1, 1.))
# ax.set_title('Cluster Labels')
# plt.show()
# ## Question Headers (2 Words)
def first_word(question):
return question.split(" ")[0]
def second_word(question):
return question.split(" ")[1]
def get_first_two_words(question):
first_word = question.split(" ")[0]
second_word = question.split(" ")[1]
return f"{first_word}_{second_word}"
df["first_word"] = df["question"].apply(first_word)
df["second_word"] = df["question"].apply(second_word)
df["first_two_words"] = df["question"].apply(get_first_two_words)
df
import matplotlib.pyplot as plt
cm = plt.get_cmap("tab20c")
# Create the data
label_0_counts = df["first_word"].value_counts()
label_1_counts = df["first_two_words"].value_counts()
# Compute the total count
total_count = label_1_counts.sum()
# Filter the inner pie chart data to exclude rows with count < 1% of total count
label_0_counts_filtered = label_0_counts[label_0_counts / total_count >= 0.01]
label_1_counts_filtered = label_1_counts[label_1_counts / total_count >= 0.01]
# Set up the plot
fig, ax = plt.subplots()
ax.axis("equal")
# Define the colors for the outer pie chart
outer_colors = plt.cm.Set1(np.arange(len(label_0_counts_filtered)))
# Define the colors for the inner pie chart
inner_colors = plt.cm.Set2(np.arange(len(label_1_counts_filtered)))
# Set the width of the donut charts
width = 0.35
# Plot the outer pie chart
outer_pie, _ = ax.pie(
label_0_counts_filtered.values,
radius=1,
labels=label_0_counts_filtered.index,
colors=outer_colors,
wedgeprops=dict(width=width, edgecolor="w"),
)
# Plot the inner pie chart
inner_pie, _ = ax.pie(
label_1_counts_filtered.values,
radius=1 - width,
labels=label_1_counts_filtered.index,
labeldistance=0.7,
colors=inner_colors,
wedgeprops=dict(width=width, edgecolor="w"),
)
# Add a white circle in the middle to create the donut effect
middle_circle = plt.Circle((0, 0), 0.5, color="white")
ax.add_artist(middle_circle)
# Add legends and title
ax.legend(outer_pie, label_0_counts.index, loc="upper left", bbox_to_anchor=(-0.1, 1.0))
ax.legend(
inner_pie,
label_1_counts_filtered.index,
loc="upper right",
bbox_to_anchor=(1.41, 1.0),
)
ax.set_title("Double Donut Chart")
# Display the plot
plt.show()
print(label_0_counts_filtered.index.tolist())
subset_df = df[df["first_word"].isin(label_0_counts_filtered.index.tolist())]
subset_df
counts = subset_df["first_word"].value_counts()
# Create a pie chart from the counts data
plt.pie(counts.values, labels=counts.index)
# Add a title to the chart
plt.title("Cluster Label Distribution")
# Display the chart
plt.show()
# Get unique categories in the 'color' column
categories = label_0_counts_filtered.index
# Create a grid of plots
fig, axs = plt.subplots(
nrows=len(categories), ncols=1, figsize=(5, 3 * len(categories))
)
# Loop over the categories and create pie charts in each subplot
for i, cat in enumerate(categories):
# Get subset DataFrame for current category
sub_df = subset_df[subset_df["first_word"] == cat]
# Count the frequency of each category in the subset
counts = sub_df["second_word"].value_counts()
percentages = counts / counts.sum() * 100
# Filter the counts data to include only categories with a percentage greater than 1%
counts = counts[percentages > 1]
# Create a pie chart from the counts data
axs[i].pie(counts.values, labels=counts.index)
# Set the title of the subplot to the category name
axs[i].set_title(cat)
# Adjust the layout of the subplots
plt.tight_layout()
# Display the plot
plt.show()
|
import numpy as np
import pandas as pd
import os
import statsmodels.formula.api as sm
import statsmodels.sandbox.tools.cross_val as cross_val
from sklearn.preprocessing import PolynomialFeatures
from sklearn import linear_model as lm
from regressors import stats
from sklearn import metrics
from sklearn.linear_model import LinearRegression
from sklearn.model_selection import (
KFold,
cross_val_score,
cross_val_predict,
LeaveOneOut,
)
print(os.listdir("../input"))
# Interactions between variables
# Explore at least 5 interactions - at least one should be a three-way interaction. Determine and identify your discoveries for each interaction. Is that interaction significant? Is a combination of interactions in the model significant?
# # 1 - Interactions between variables
# Interactive Terms: Statsmodel
d = pd.read_csv("/kaggle/input/szeged-weather/weatherHistory.csv")
column_names = {
"Formatted Date": "Date",
"Precip Type": "PrecipType",
"Temperature (C)": "TempC",
"Apparent Temperature (C)": "AppTempC",
"Humidity": "Humidity",
"Wind Speed (km/h)": "WindSpeed",
"Wind Bearing (degrees)": "WindBearing",
"Visibility (km)": "Visibility",
"Loud Cover": "LoudCover",
"Pressure (millibars)": "Pressure",
"Daily Summary": "DailySummary",
}
d = d.rename(columns=column_names)
d
main = sm.ols(formula="TempC ~ Humidity", data=d).fit()
print(main.summary())
main = sm.ols(formula="TempC ~ WindSpeed*Visibility", data=d).fit()
print(main.summary())
main = sm.ols(formula="TempC ~ Humidity*Visibility", data=d).fit()
print(main.summary())
main = sm.ols(formula="TempC ~ Humidity*WindSpeed", data=d).fit()
print(main.summary())
main = sm.ols(formula="TempC ~ Humidity*WindSpeed*Pressure", data=d).fit()
print(main.summary())
main = sm.ols(formula="TempC ~ Humidity*Visibility*Pressure", data=d).fit()
print(main.summary())
# # Numerical Transformations
import matplotlib.pyplot as plt
inputDF = d[["TempC"]]
outputDF = d[["Humidity"]]
lin = LinearRegression()
poly_features = PolynomialFeatures(degree=2)
inputDF_poly = poly_features.fit_transform(inputDF)
lin.fit(inputDF_poly, outputDF)
# Scatter Plot - Polynomial Regression
plt.scatter(inputDF, outputDF, color="blue")
plt.plot(inputDF, lin.predict(inputDF_poly), color="red")
plt.title("Polynomial Regression")
plt.xlabel("Tempreture")
plt.ylabel("Humidity")
plt.show()
inputDF = d[["TempC"]]
outputDF = d[["Visibility"]]
lin = LinearRegression()
poly_features = PolynomialFeatures(degree=2)
inputDF_poly = poly_features.fit_transform(inputDF)
lin.fit(inputDF_poly, outputDF)
# Scatter Plot - Polynomial Regression
plt.scatter(inputDF, outputDF, color="blue")
plt.plot(inputDF, lin.predict(inputDF_poly), color="red")
plt.title("Polynomial Regression")
plt.xlabel("Tempreture")
plt.ylabel("Visibility")
plt.show()
# ## The transformations seems not significant and not improving the model's performance
# # Forward Model Selection
from mlxtend.feature_selection import SequentialFeatureSelector as sfs
d["PHV"] = d["Humidity"] * d["Visibility"] * d["Pressure"]
inputDF = d[
[
"Humidity",
"WindSpeed",
"WindBearing",
"Visibility",
"Pressure",
"LoudCover",
"PHV",
]
]
outputDF = d[["TempC"]]
model = sfs(
LinearRegression(),
k_features=5,
forward=True,
verbose=2,
cv=5,
n_jobs=-1,
scoring="r2",
)
model.fit(inputDF, outputDF)
print(model.k_feature_idx_)
print(model.k_feature_names_)
# Backward Selection: Scikit-Learn
inputDF = d[
[
"Humidity",
"WindSpeed",
"WindBearing",
"Visibility",
"Pressure",
"LoudCover",
"PHV",
]
]
outputDF = d[["TempC"]]
lr = LinearRegression()
backwardModel = sfs(
lr, k_features=5, forward=False, verbose=2, cv=5, n_jobs=-1, scoring="r2"
)
backwardModel.fit(inputDF, outputDF)
print(backwardModel.k_feature_idx_)
print(backwardModel.k_feature_names_)
# # Cross-Validation
from sklearn import metrics
from sklearn.linear_model import LinearRegression
inputDF = d[["Humidity", "WindSpeed", "WindBearing", "Visibility", "LoudCover"]]
outputDF = d[["TempC"]]
model = lr.fit(inputDF, outputDF)
predictions = model.predict(inputDF)
print(predictions)
kf = KFold(10, shuffle=True, random_state=42).get_n_splits(inputDF)
rmse = np.sqrt(
-cross_val_score(model, inputDF, outputDF, scoring="neg_mean_squared_error", cv=kf)
)
print(rmse.mean())
kf = KFold(5, shuffle=True, random_state=42).get_n_splits(inputDF)
rmse = np.sqrt(
-cross_val_score(model, inputDF, outputDF, scoring="neg_mean_squared_error", cv=kf)
)
print(rmse.mean())
loocv = LeaveOneOut()
rmse = np.sqrt(
-cross_val_score(
model, inputDF, outputDF, scoring="neg_mean_squared_error", cv=loocv
)
)
print(rmse.mean())
# # Draft Final Model
plt.scatter(outputDF, predictions, s=0.5)
plt.xlabel("Actual Temperature")
plt.ylabel("Predicted Temperature")
max_val = outputDF["TempC"].max()
min_val = outputDF["TempC"].min()
plt.plot([min_val, max_val], [min_val, max_val], color="red")
plt.show()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Keşifçi veri analizinde çok değişkenli istatistiksel analizler ve veri görselleştirme yaklaşımları uygulanarak temel istatistikler ve göz ile yakalanamayacak yapıların ortaya çıkarılması amaçlanır. Bu amaç doğrultusunda veri setindeki değişkenler tek değişkenli, iki değişkenli ve çok değişkenli şekilde incelenir.
import numpy as np
import pandas as pd
import seaborn as sns
# NumPy, dizilerle çalışmak için kullanılan bir python kütüphanesidir.
# Ayrıca doğrusal cebir ve matrisler alanında çalışmak için de gerekli işlevlere sahiptir.
# Pandas,Python programlama diliyle oluşturulmuş,hızlı,güçlü, esnek ve kullanımı kolay, açık kaynaklı bir veri analiz ve manipülasyon aracıdır.
# Seaborn Python görselleştirme kütüphanesidir. Özellikle istatistiksel grafikler için çok kullanışlı bir kod yapısı sağlar. Pandas kütüphanesinin veri yapısına da kolayca entegre edilebilir.
# Veri çerçevesini bulunduğumuz dizinden alıp bir veri çerçevesi haline getirip df değişkenine atamasını yapıyoruz
df = pd.read_csv("../input/iris-flower-dataset/IRIS.csv")
# Veri çerçevesinin ilk 5 gözlemini görüntüleyelim.
df.head()
df.tail()
#
# Veri çerçevesinin kaç öznitelik ve kaç gözlemden oluştuğunu görüntüleyelim.
# satır gözlemi ifade eder
# sütun ise özniteliği ifade eder
df.shape
# Veri çerçevesindeki değişkenlerin hangi tipte olduğunu ve bellek kullanımını görüntüleyelim.
# df hakkında genel bilgi edinmek için infoyu kullanabiliriz
df.info()
# Veri çerçevesindeki sayısal değişkenler için temel istatistik değerlerini görüntüleyelim.
# Standart sapma ve ortalama değerlerden çıkarımda bulunarak hangi değişkenlerin ne kadar varyansa sahip olduğu hakkında fikir yürütelim.
# describe bize sayısal verilere sahip olan sütunların istatistiksel değerlerini döndürür
df.describe()
# Veri çerçevesinde hangi öznitelikte kaç adet eksik değer olduğunu gözlemleyelim.
df.isna().sum()
# Sayısal değişkenler arasında korelasyon olup olmadığını göstermek için korelasyon matrisi çizdirelim. Korelasyon katsayıları hakkında fikir yürütelim.
# En güçlü pozitif ilişki hangi iki değişken arasındadır?
corr = df.corr()
corr
# Korelasyon katsayılarını daha iyi okuyabilmek için ısı haritası çizdirelim.
sns.heatmap(corr, cmap="coolwarm", annot=True)
# lmplot() fonksiyonunu regresyon çizgisini oturtmak ve grafiğini çıkartmak için kullanabiliriz
sns.lmplot(x="petal_width", y="petal_length", data=df)
# Veri çerçevemizin hedef değişkeninin "variety" benzersiz değerlerini görüntüleyelim.
df["species"].unique()
# Veri çerçevemizin hedef değişkeninin "variety" benzersiz kaç adet değer içerdiğini görüntüleyelim.
#
df["species"].nunique()
# Veri çerçevesindeki sepal.width ve sepal.length değişkenlerinin sürekli olduğunu görüyoruz. Bu iki sürekli veriyi görselleştirmek için önce scatterplot kullanalım.
#
# İki farklı değişkenin arasındaki ilişkiyi belirlemek için serpilme diyagramı (scatter plot) kullanıyoruz
# Bu diyagramda ilişkinin sebebini anlayamasak da, bir ilişki olup olmadığını ya da ilişkinin ne kadar güçlü olduğu hakkında yorum yapabiliriz.
sns.scatterplot(x="sepal_width", y="sepal_length", data=df, color="green")
# Aynı iki veriyi daha farklı bir açıdan frekanslarıyla incelemek için jointplot kullanarak görselleştirelim.
#
# Ortak dağılım grafiği, saçılım grafiği ile histogramı birleştirerek bize iki değişkenli dağılımlar hakkında detaylı bilgi verir.
sns.jointplot(x="sepal_width", y="sepal_length", data=df, color="red")
# Aynı iki veriyi scatterplot ile tekrardan görselleştirelim fakat bu sefer "variety" parametresi ile hedef değişkenine göre kırdıralım.
# 3 farklı renk arasında sepal değişkenleriyle bir kümeleme yapılabilir mi? Ne kadar ayırt edilebilir bunun üzerine düşünelim.
sns.scatterplot(x="sepal_width", y="sepal_length", hue="species", data=df)
# value_counts() fonksiyonu ile veri çerçevemizin ne kadar dengeli dağıldığını sorgulayalım.
#
# value_counts() hangi değerden kaç tane olduğunu döndürür. Buna bakarak verimizin dengeli dağılıp dağılmadığını gözlemleriz.
df["species"].value_counts()
# Keman grafiği çizdirerek sepal.width değişkeninin dağılımını inceleyin.
# Söz konusu dağılım bizim için ne ifade ediyor, normal bir dağılım olduğunu söyleyebilir miyiz?
# violinplot özet istatistikleri göstermek yerine dağılımları gösterir.
sns.violinplot(x="sepal_length", data=df, color="orange")
# Daha iyi anlayabilmek için sepal.width üzerine bir distplot çizdirelim.
#
# distplot nümerik değişkenlerin dağılımlarını görselleştirmede kullanılır.
# yeni sürümlerinde histplot olarak kullanılıyor
sns.histplot(df["sepal_width"], bins=20, color="purple")
# Üç çiçek türü için üç farklı keman grafiğini sepal.length değişkeninin dağılımı üzerine tek bir satır ile görselleştirelim.
#
sns.violinplot(x="species", y="sepal_length", data=df)
# Hangi çiçek türünden kaçar adet gözlem barındırıyor veri çerçevemiz?
# 50 x 3 olduğunu ve dengeli olduğunu value_counts ile zaten görmüştük, ancak bunu görsel olarak ifade etmek için sns.countplot() fonksiyonuna variety parametresini vereilm.
sns.countplot(x="species", data=df)
# sepal.length ve sepal.width değişkenlerini sns.jointplot ile görselleştirelim, dağılımı ve dağılımın frekansı yüksek olduğu bölgelerini inceleyelim.
sns.jointplot(x="sepal_length", y="sepal_width", data=df, color="yellow")
# Bir önceki hücrede yapmış olduğumuz görselleştirmeye kind = "kde" parametresini ekleyelim. Böylelikle dağılımın noktalı gösterimden çıkıp yoğunluk odaklı bir görselleştirmeye dönüştüğünü görmüş olacağız.
sns.jointplot(x="sepal_length", y="sepal_width", data=df, kind="hex", color="blue")
# kin="hex" ile farklı bir görselleştirme yapılır.
# scatterplot ile petal.length ve petal.width değişkenlerinin dağılımlarını çizdirelim.
sns.scatterplot(x="petal_length", y="petal_width", data=df, color="green")
# Aynı görselleştirmeye hue = "variety" parametresini ekleyerek 3. bir boyut verelim.
#
sns.scatterplot(data=df, x="petal_length", y="petal_width", hue="species")
# sns.lmplot() görselleştirmesini petal.length ve petal.width değişkenleriyle implemente edelim. Petal length ile petal width arasında ne tür bir ilişki var ve bu ilişki güçlü müdür? sorusunu yanıtlayalım.
# sns.lmplot(x="petal_length", y="petal_width", data=df)
# implot fonksiyonunda color parametresi yok.
sns.lmplot(x="petal_length", y="petal_width", hue="species", data=df)
# -->Burada hue parametresine species değişkenini vererek çizdirdiğimiz grafikte farklı türlerin (setosa, versicolor, virginica) regresyon çizgilerini ve dağılım noktalarını farklı renklerde görürüz.
# Bu sorunun yanıtını pekiştirmek için iki değişken arasında korelasyon katsayısını yazdıralım.
corr = df[["petal_length", "petal_width"]].corr()
print(corr)
# korelasyon 1'e yaklaştıkça güçlü ve pozitif olur
# Petal Length ile Sepal Length değerlerini toplayarak yeni bir total length özniteliği oluşturalım.
df["total_length"] = df["petal_length"] + df["sepal_length"]
df["total_length"]
# total.length'in ortalama değerini yazdıralım.
df["total_length"].mean()
# total.length'in standart sapma değerini yazdıralım.
df["total_length"].std()
# sepal.length'in maksimum değerini yazdıralım.
#
df["sepal_length"].max()
# sepal.length'i 5.5'den büyük ve türü setosa olan gözlemleri yazdıralım.
#
df[(df["species"] == "Iris-setosa") & (df["sepal_length"] > 5.5)]
# petal.length'i 5'den küçük ve türü virginica olan gözlemlerin sadece sepal.length ve sepal.width değişkenlerini ve değerlerini yazdıralım.
#
df.loc[
(df["species"] == "Iris-virginica") & (df["petal_length"] < 5),
["sepal_length", "sepal_width"],
]
# Hedef değişkenimiz variety'e göre bir gruplama işlemi yapalım değişken değerlerimizin ortalamasını görüntüleyelim.
#
df.groupby("species").mean()
# Hedef değişkenimiz variety'e göre gruplama işlemi yaparak sadece petal.length değişkenimizin standart sapma değerlerini yazdıralım.
#
df.groupby("species").std()["petal_length"]
|
# # KMeans HW 3
# ## Helper functions from modules code to load data
import math
import random
import time
# from Tkinter import *
######################################################################
# This section contains functions for loading CSV (comma separated values)
# files and convert them to a dataset of instances.
# Each instance is a tuple of attributes. The entire dataset is a list
# of tuples.
######################################################################
# Loads a CSV files into a list of tuples.
# Ignores the first row of the file (header).
# Numeric attributes are converted to floats, nominal attributes
# are represented with strings.
# Parameters:
# fileName: name of the CSV file to be read
# Returns: a list of tuples
def loadCSV(fileName):
fileHandler = open(fileName, "rt")
lines = fileHandler.readlines()
fileHandler.close()
del lines[0] # remove the header
dataset = []
for line in lines:
instance = lineToTuple(line)
dataset.append(instance)
return dataset
# Converts a comma separated string into a tuple
# Parameters
# line: a string
# Returns: a tuple
def lineToTuple(line):
# remove leading/trailing witespace and newlines
cleanLine = line.strip()
# get rid of quotes
cleanLine = cleanLine.replace('"', "")
# separate the fields
lineList = cleanLine.split(",")
# convert strings into numbers
stringsToNumbers(lineList)
lineTuple = tuple(lineList)
return lineTuple
# Destructively converts all the string elements representing numbers
# to floating point numbers.
# Parameters:
# myList: a list of strings
# Returns None
def stringsToNumbers(myList):
for i in range(len(myList)):
if isValidNumberString(myList[i]):
myList[i] = float(myList[i])
# Checks if a given string can be safely converted into a positive float.
# Parameters:
# s: the string to be checked
# Returns: True if the string represents a positive float, False otherwise
def isValidNumberString(s):
if len(s) == 0:
return False
if len(s) > 1 and s[0] == "-":
s = s[1:]
for c in s:
if c not in "0123456789.":
return False
return True
# ## Function Using Various Distance Options
import numpy as np
from sklearn.cluster import KMeans
import csv
from sklearn.metrics.pairwise import cosine_similarity
from sklearn.metrics import accuracy_score
def k_means(data, k, distance_option=0, max_iters=0, return_iters=False):
n = data.shape[0]
indices = np.random.choice(n, k, replace=False)
centroids = data[indices]
prev_sse = None
if distance_option == 0:
print("kmeans using Euclidean")
elif distance_option == 1:
print("kmeans using Cosine Similarity")
elif distance_option == 2:
print("kmeans using Generalized Jaccard similarity")
if max_iters == 0:
max_iters = 100 # random.randint(0, 100)
print("running " + str(max_iters) + " times")
for i in range(max_iters):
if distance_option == 0:
distances = np.sqrt(
np.sum(np.square(data[:, np.newaxis] - centroids), axis=2)
)
elif distance_option == 1:
similarity = cosine_similarity(data, centroids)
distances = 1 - similarity
elif distance_option == 2:
intersections = np.minimum(data[:, np.newaxis], centroids)
unions = np.maximum(data[:, np.newaxis], centroids)
similarities = np.sum(np.minimum(intersections, unions), axis=2) / np.sum(
np.maximum(intersections, unions), axis=2
)
distances = 1 - similarities
labels = np.argmin(distances, axis=1)
# Compute SSE and compare with previous iteration
sse = np.sum(distances[np.arange(n), labels] ** 2)
if return_iters and prev_sse is not None and sse > prev_sse:
print("Converged on SSE")
return labels, centroids, i
prev_sse = sse
prev_centroids = centroids.copy()
# Update centroids as the mean of the data points in each cluster
for j in range(k):
mask = labels == j
if np.any(mask):
centroids[j] = np.mean(data[mask], axis=0)
# Special Stop case
if return_iters and np.array_equal(prev_centroids, centroids):
print("Converged on centroids")
return labels, centroids, i
if return_iters:
print("Converged on iterations")
return labels, centroids, i
return labels, centroids
# ## Helper SSE Functions
def get_euclidean_sse(labels, centroids, X):
sse = 0
# Calculate for each cluster
for i in range(k):
cluster_data = X[labels == i]
centroid = centroids[i]
distances = np.linalg.norm(cluster_data - centroid, axis=1)
sse += np.sum(distances**2)
return sse
def get_consine_sse(labels, centroids, X):
sse = 0
consine_similarities = cosine_similarity(X, centroids)
# Calculate over each data point
for i in range(len(X)):
sse += 1 - consine_similarities[i][labels[i]]
return sse
def get_jaccard_sse(labels, centroids, X):
sse = 0
for i in range(len(X)):
centroid = centroids[labels[i]]
intersection = np.logical_and(X[i], centroid)
union = np.logical_or(X[i], centroid)
similarity = np.sum(intersection) / np.sum(union)
sse += (1 - similarity) ** 2
return sse
# ## Calculate the SSE of Each
dataset = loadCSV("/kaggle/input/dataset/data.csv")
label_data = loadCSV("/kaggle/input/dataset/label.csv")
# Convert the list of rows to a NumPy array
X = np.array(dataset)
Y = np.array(label_data).astype(int).flatten() # [1:]
k = 10
euc_labels, euc_centroids = k_means(X, k)
print("SSE using Euclidean distance: ", get_euclidean_sse(euc_labels, euc_centroids, X))
cos_labels, cos_centroids = k_means(X, k, 1)
print("Cosine similarity SSE:", get_consine_sse(cos_labels, cos_centroids, X))
jac_labels, jac_centroids = k_means(X, k, 2)
print("Jaccard SSE: ", get_jaccard_sse(jac_labels, jac_centroids, X))
# ## Calculate Accuracy
# Perform Euclidean K-means clustering
def get_accuracy(Y, labels):
majority_labels = np.zeros_like(labels)
for i in range(k):
mask = labels == i
majority_labels[mask] = np.bincount(Y[mask]).argmax()
return accuracy_score(Y, majority_labels)
print("Accuracy of Euclidean K-means:", get_accuracy(Y, euc_labels))
print("Accuracy of Cosine K-means:", get_accuracy(Y, cos_labels))
print("Accuracy of Jaccard K-means:", get_accuracy(Y, jac_labels))
# ## Calculate Convergance
import time
# Data and labels
y = Y
# Euclidean
start = time.time()
labels, centroids, iterations = k_means(X, k, 0, 500, True)
end = time.time()
print("Euclidean converged in", iterations, "iterations")
print("Time:", end - start)
print("SSE using Euclidean distance: ", get_euclidean_sse(labels, centroids, X))
# Cosine
start = time.time()
labels, centroids, iterations = k_means(X, k, 1, 500, True)
end = time.time()
print("Cosine converged in", iterations, "iterations")
print("Time:", end - start)
print("Cosine similarity SSE:", get_consine_sse(labels, centroids, X))
# Jaccard-K-means
start = time.time()
labels, centroids, iterations = k_means(X, k, 2, 500, True)
end = time.time()
print("Jaccard converged in", iterations, "iterations")
print("Time:", end - start)
print("Jaccard SSE: ", get_jaccard_sse(labels, centroids, X))
|
# 
# Data Science and Business Analytics
# Practice Project 25
# Dashboard for Store Dataset
# By
# Hayford Osumanu
# April 2023
# 
# 
# 
# 
# 
# # Data Analysis Philosophy
import this
# 
# 
# 
import pandas as pd
import numpy as np
import panel as pn
pn.extension("tabulator")
import hvplot.pandas
# this will help in making the Python code more structured automatically (good coding practice)
# %load_ext nb_black
# Libraries to help with reading and manipulating data
import numpy as np
import pandas as pd
# Libraries to help with data visualization
import matplotlib.pyplot as plt
import seaborn as sns
# split the data into train and test
from sklearn.model_selection import train_test_split
# to build linear regression_model
from sklearn.linear_model import LinearRegression
# to check model performance
from sklearn.metrics import mean_absolute_error, mean_squared_error, r2_score
# to build linear regression_model using statsmodels
import statsmodels.api as sm
# to compute VIF
from statsmodels.stats.outliers_influence import variance_inflation_factor
# # Command to tell Python to actually display the graphs
pd.set_option(
"display.float_format", lambda x: "%.2f" % x
) # To supress numerical display in scientific notations
# Removes the limit for the number of displayed columns
pd.set_option("display.max_columns", None)
# Sets the limit for the number of displayed rows
pd.set_option("display.max_rows", 200)
import warnings
warnings.filterwarnings("ignore")
# 
# # Importing the Dataset into DataFrame
path = "https://raw.githubusercontent.com/hayfordosmandata/DataBank/main/StoreData2.csv"
Store_data = pd.read_csv(path)
# copying data to another variable to avoid any changes to original data
data = Store_data.copy()
# 
# # Part I: Dataset Overview
# Creating a well readable header label
for header in data.columns:
header_replace = header.replace(
" ", "_"
) # creates new header with "_" instead of " ".
data.rename(
{header: header_replace}, axis=1, inplace=True
) # sets new header as header made in line above
import panel as pn
import seaborn as sb
# Create a Tabulator widget with pagination
table = pn.widgets.Tabulator(data.head(5000), pagination="remote")
# Display the widget in a Panel
panel = pn.Column("## First 20 Rows for Cars Dataset with Pages", table)
panel.servable()
# 
# ### Columns/Variable/Features of the Dataset
# Extracting the columns/variables of the dataset
data.columns
# The data in the tables above contains information of different attributes of Customers based on their transaction history. The detailed data dictionary is given below.
# 
# ### Dimension of the Dataset
# checking the shape of the data
print(f"There are {data.shape[0]} rows and {data.shape[1]} columns.")
# Checking the dimension (number of observations/rows and variables/columns of the Dataset (df.shape)
print("There are", data.shape[0], "rows and", data.shape[1], "columns.")
# 
# ### Data Types of the Dataset
# Checking the data types of the variables/columns for the dataset
data.info()
# 
# ## Data Sanity Checks: Deep Checking/scrutinity of the the dataset before EDA
# ### Checking the Missing Values of the Dataset
# Checking for missing values in the dataset
data.isnull().sum()
# Checking the total number of missing values in the dataset
data.isnull().sum().sum()
missing_data = data.isnull()
for column in missing_data.columns.values.tolist():
print(column)
print(missing_data[column].value_counts())
print("")
# 
# ### Checking the Duplicates in the Dataset
# let's check for duplicate values in the data
data.groupby(data.duplicated(subset=None, keep="first"), as_index=False).size()
# checking for duplicate values
print("There are about: ", data.duplicated().sum(), "dupplicates in the dataset")
# deleting the unnecessary columns (RowNumber, CustomerId, Surname)
data.drop(["ID_Number"], axis=1, inplace=True)
# Lest create log transformation to normalize the Revenues and Expenses variable
data["Revenues_log"] = np.log(data["Revenues"])
data["Expenses_log"] = np.log(data["Expenses"])
import panel as pn
import seaborn as sb
# Create a Tabulator widget with pagination
table = pn.widgets.Tabulator(data.head(5000), pagination="remote")
# Display the widget in a Panel
panel = pn.Column("## First 20 Rows for Cars Dataset with Pages", table)
panel.servable()
# 
# # Part II: Panel Dashboard Overview
data.columns
num = [
"Price_per_unit",
"Cost_per_unit",
"Quantity_Sold",
"Revenues",
"Expenses",
"Revenues_log",
"Expenses_log",
]
# 
# ## Dash 0: Outlier Detection Using Boxplot
import panel as pn
import pandas as pd
import hvplot.pandas
# Load your dataset into a DataFrame
# df = pd.read_csv('your_dataset.csv')
# Create a list of options for the drop-down menu
options = ["Year", "Region", "Product", "Sales_Method"]
# Create the drop-down menu widget
dropdown = pn.widgets.Select(name="Select Column:", options=options)
# Function to update the chart based on the selected value from the drop-down menu
@pn.depends(dropdown.param.value)
def update_chart(column):
# Create a chart based on the selected column
return data.hvplot.bar(column, color="red", width=1200, height=500)
# Create a Panel layout with the drop-down menu and chart
dashboard = pn.Column(dropdown, update_chart)
# Create a standalone server for the dashboard
dashboard.servable()
# Embed the dashboard in your notebook
# pn.panel(dashboard).embed()
import pandas as pd
import panel as pn
import hvplot.pandas
# Define the dropdown menu options [ALL Columns]
column_options = ["Year", "Region", "Product", "Sales_Method"]
dropdown_scatter = pn.widgets.Select(
options=column_options, name="Select a column for X-axis"
)
dropdown_box = pn.widgets.Select(options=column_options, name="Select a column")
# Creat Bar Charts
bar_plot1 = data.hvplot.bar(x="Year", stacked=True, title="Bar Plot1")
bar_plot2 = data.hvplot.bar(x="Region", stacked=True, title="bar Plot2")
bar_plot3 = data.hvplot.bar(x="Product", stacked=True, title="Bar Plot3")
bar_plot4 = data.hvplot.bar(x="Sales_Method", stacked=True, title="Bar Plot4")
# Create a Panel layout with the charts and statistics
dashboard = pn.Tabs(
("Bar Plot1", bar_plot1),
("Bar Plot2", bar_plot2),
("Bar Plot3", bar_plot3),
("Bar Plot4", bar_plot4),
)
# Create a standalone server for the dashboard
dashboard.servable()
import panel as pn
import pandas as pd
import hvplot.pandas
# Load your dataset into a DataFrame
# df = pd.read_csv('your_dataset.csv')
# Create a list of options for the drop-down menu
options = [
"Price_per_unit",
"Cost_per_unit",
"Quantity_Sold",
"Revenues",
"Expenses",
"Revenues_log",
"Expenses_log",
]
# Create the drop-down menu widget
dropdown = pn.widgets.Select(name="Select Column:", options=options)
# Function to update the chart based on the selected value from the drop-down menu
@pn.depends(dropdown.param.value)
def update_chart(column):
# Create a chart based on the selected column
return data.hvplot.box(column, color="lime")
# Create a Panel layout with the drop-down menu and chart
dashboard = pn.Column(dropdown, update_chart)
# Create a standalone server for the dashboard
dashboard.servable()
# Embed the dashboard in your notebook
# pn.panel(dashboard).embed()
import panel as pn
import pandas as pd
import hvplot.pandas
# Load your dataset into a DataFrame
# df = pd.read_csv('your_dataset.csv')
# Create a list of options for the drop-down menu
options = [
"Price_per_unit",
"Cost_per_unit",
"Quantity_Sold",
"Revenues",
"Expenses",
"Revenues_log",
"Expenses_log",
]
# Create the drop-down menu widget
dropdown = pn.widgets.Select(name="Select Column:", options=options)
# Function to update the chart based on the selected value from the drop-down menu
@pn.depends(dropdown.param.value)
def update_chart(column):
# Create a chart based on the selected column
return data.hvplot.kde(column, color="lime")
# Create a Panel layout with the drop-down menu and chart
dashboard = pn.Column(dropdown, update_chart)
# Create a standalone server for the dashboard
dashboard.servable()
# Embed the dashboard in your notebook
# pn.panel(dashboard).embed()
import panel as pn
import pandas as pd
import hvplot.pandas
# Load your dataset into a DataFrame
# df = pd.read_csv('your_dataset.csv')
# Create a list of options for the drop-down menu
options = [
"Price_per_unit",
"Cost_per_unit",
"Quantity_Sold",
"Revenues",
"Expenses",
"Revenues_log",
"Expenses_log",
]
# Create the drop-down menu widget
dropdown = pn.widgets.Select(name="Select Column:", options=options)
# Function to update the chart based on the selected value from the drop-down menu
@pn.depends(dropdown.param.value)
def update_chart(column):
# Create a chart based on the selected column
return data.hvplot.violin(column, color="lime")
# Create a Panel layout with the drop-down menu and chart
dashboard = pn.Column(dropdown, update_chart)
# Create a standalone server for the dashboard
dashboard.servable()
# Embed the dashboard in your notebook
# pn.panel(dashboard).embed()
# selecting numerical columns
cols = data.select_dtypes(include=np.number).columns.tolist()
# ## Dash I: Select different Types of Charts Using Different Variables
import pandas as pd
import panel as pn
import hvplot.pandas
import holoviews as hv
# Define the available columns for the dropdowns
columns = data.select_dtypes(include=np.number).columns.tolist()
# Define the available plot types for the dropdown
plot_types = {
"Displot": "hist",
"Boxplot": "box",
"Line Chart": "line",
"Scatter Plot": "scatter",
}
# Define the plots
def get_plots(column_x, column_y, plot_type):
if plot_type == "hist":
plot = data.hvplot.hist(column_x, title=f"{column_x} Distribution")
elif plot_type == "box":
plot = data.hvplot.box(y=column_y, by="sex", title=f"{column_y} Distribution")
elif plot_type == "line":
plot = data.hvplot.line(column_x, column_y, title=f"{column_y} by {column_x}")
elif plot_type == "scatter":
plot = data.hvplot.scatter(
column_x,
column_y,
c="sex",
cmap="viridis",
title=f"{column_y} vs {column_x}",
)
return plot
# Define the dropdown widgets
dropdown_x = pn.widgets.Select(name="Horizontal Variables", options=columns)
dropdown_y = pn.widgets.Select(name="Vertical Variables", options=columns)
dropdown_plot_type = pn.widgets.Select(name="Plot Type", options=plot_types)
def update_plots(*events):
# Get the current values of the dropdown widgets
column_x = dropdown_x.value
column_y = dropdown_y.value
plot_type = dropdown_plot_type.value
# Generate the plot based on the selected values
plot = get_plots(column_x, column_y, plot_type)
# Update the panel with the new plot
plot_panel[1] = pn.pane.HoloViews(plot)
# Bind the dropdowns to the update function
dropdown_x.param.watch(update_plots, "value")
dropdown_y.param.watch(update_plots, "value")
dropdown_plot_type.param.watch(update_plots, "value")
# Create a layout for the plots and dropdowns
plot_panel = pn.Column(
pn.Row(dropdown_x, dropdown_y, dropdown_plot_type),
pn.pane.HoloViews(),
sizing_mode="stretch_width",
)
# Serve the dashboard
update_plots(None)
plot_panel.servable()
# 
import pandas as pd
import panel as pn
import hvplot.pandas
import holoviews as hv
# Load data (not shown)
# data = pd.read_csv('my_data.csv')
# Define the available columns for the dropdowns
columns = data.select_dtypes(include=np.number).columns.tolist()
# Define the available plot types for the dropdown
plot_types = {
"Histogram": "hist",
"Boxplot": "box",
"Line Chart": "line",
"Scatter Plot": "scatter",
"Violin Plot": "violin",
"Displot": "kde",
"Summary Statistics Table": "table",
"Heatmap": "heatmap",
}
# Define the plots
def get_plots(column_x, column_y, plot_type):
if plot_type == "hist":
plot = data.hvplot.hist(column_x, title=f"{column_x} Distribution")
elif plot_type == "box":
plot = data.hvplot.box(
y=column_y, title=f"{column_y} Distribution", groupby=None
)
elif plot_type == "line":
plot = data.hvplot.line(column_x, column_y, title=f"{column_y} by {column_x}")
elif plot_type == "scatter":
plot = data.hvplot.scatter(
column_x,
column_y,
c="color",
cmap="viridis",
title=f"{column_y} vs {column_x}",
)
elif plot_type == "violin":
plot = data.hvplot.violin(
column_x, column_y, title=f"{column_y} by {column_x}", groupby=None
)
elif plot_type == "kde":
plot = data.hvplot.kde(column_x, title=f"{column_x} Distribution")
elif plot_type == "table":
plot = hv.Table(data[column_x].describe())
elif plot_type == "heatmap":
plot = hv.HeatMap(data.corr())
return plot
# Define the dropdown widgets
dropdown_x = pn.widgets.Select(name="Horizontal Variables", options=columns)
dropdown_y = pn.widgets.Select(name="Vertical Variables", options=columns)
dropdown_plot_type = pn.widgets.Select(name="Plot Type", options=plot_types)
def update_plots(*events):
# Get the current values of the dropdown widgets
column_x = dropdown_x.value
column_y = dropdown_y.value
plot_type = dropdown_plot_type.value
# Generate the plot based on the selected values
plot = get_plots(column_x, column_y, plot_type)
# Update the panel with the new plot
plot_panel[1] = pn.pane.HoloViews(plot)
# Bind the dropdowns to the update function
dropdown_x.param.watch(update_plots, "value")
dropdown_y.param.watch(update_plots, "value")
dropdown_plot_type.param.watch(update_plots, "value")
# Create a layout for the plots and dropdowns
plot_panel = pn.Column(
pn.Row(dropdown_x, dropdown_y, dropdown_plot_type),
pn.pane.HoloViews(),
sizing_mode="stretch_width",
)
# Serve the dashboard
update_plots(None)
plot_panel.servable()
# 
# ## Dash II:Distribution of Different Variables
# 
# ## DashII: Multivariate Distribution of Different Variables
columns = list(data.columns[0:-3])
x = pn.widgets.Select(
value="Quantity_Sold", options=columns, name="Horizontal Variables"
)
y = pn.widgets.Select(value="Revenues_log", options=columns, name="Vertical Variables")
pn.Row(
pn.Column("## Select Feature Option", x, y),
pn.bind(data.hvplot.scatter, x, y, by="Sales_Method"),
)
columns = list(data.columns[0:-3])
x = pn.widgets.Select(
value="Quantity_Sold", options=columns, name="Horizontal Variables"
)
y = pn.widgets.Select(value="Revenues_log", options=columns, name="Vertical Variables")
pn.Row(
pn.Column("## Select Feature Option", x, y),
pn.bind(data.hvplot.scatter, x, y, by="Region"),
)
columns = list(data.columns[0:-3])
x = pn.widgets.Select(
value="Quantity_Sold", options=columns, name="Horizontal Variables"
)
y = pn.widgets.Select(value="Revenues_log", options=columns, name="Vertical Variables")
pn.Row(
pn.Column("## Select Feature Option", x, y),
pn.bind(data.hvplot.scatter, x, y, by="Product"),
)
# 
# 
# ## Dash IV: Summary Statistics and Distributions
# 
data.columns
import pandas as pd
import panel as pn
import hvplot.pandas
# Define the dropdown menu options
column_options = [
"Year",
"Price_per_unit",
"Cost_per_unit",
"Quantity_Sold",
"Revenues",
"Expenses",
"Revenues_log",
"Expenses_log",
]
dropdown_scatter = pn.widgets.Select(
options=column_options, name="Select a column for X-axis"
)
dropdown_box = pn.widgets.Select(options=column_options, name="Select a column")
# Define a function to update the scatter plot based on the dropdown selection
@pn.depends(dropdown_scatter.param.value)
def update_scatter(column):
plot = data.hvplot.scatter(
x=column, y="Revenues", title="Scatter Plot", width=1000, height=400
)
return plot
# Define a function to update the box plot based on the dropdown selection
@pn.depends(dropdown_box.param.value)
def update_box(column):
plot = data.hvplot.box(y=column, title="Box Plot", width=1000, height=400)
return plot
# Create Scatter and Boxplots
scatter_plot = pn.Column(dropdown_scatter, update_scatter)
box_plot = pn.Column(dropdown_box, update_box)
# Create Numerical Statistics
# Create Numerical Statistics
table1 = (
data[
[
"Price_per_unit",
"Cost_per_unit",
"Quantity_Sold",
"Revenues",
"Expenses",
"Revenues_log",
"Expenses_log",
]
]
.describe()
.reset_index()
.hvplot.table(title="Summary Statistics")
)
table2 = (
data[
[
"Price_per_unit",
"Cost_per_unit",
"Quantity_Sold",
"Revenues",
"Expenses",
"Revenues_log",
"Expenses_log",
]
]
.quantile([0.25, 0.5, 0.6, 0.75, 0.9, 0.95, 0.99])
.reset_index()
.hvplot.table(title="Quantiles Statistics")
)
# table1 = data[['age', 'quantity', 'price', 'revenue']].describe().T.hvplot.table(title='Summary Statistics 1')
# table2 = data[['age', 'quantity', 'price', 'revenue']].describe().T.hvplot.table(title='Summary Statistics 2')
# Create Correlation Heatmap
corr = data[
[
"Price_per_unit",
"Cost_per_unit",
"Quantity_Sold",
"Revenues",
"Expenses",
"Revenues_log",
"Expenses_log",
]
].corr()
heat_map = corr.hvplot.heatmap(title="Correlation Heatmap", width=1000, height=500)
dashboard = pn.Tabs(
("Scatter Plot", scatter_plot),
("Box Plot", box_plot),
("Summary Statistics", table1),
("Qunatiles Statistics", table2),
("Correlation Heatmap", heat_map),
)
# Create a standalone server for the dashboard
dashboard.servable()
import panel as pn
import pandas as pd
import hvplot.pandas
# Load your dataset into a DataFrame
# df = pd.read_csv('your_dataset.csv')
# Create a list of options for the drop-down menu
options = [
"Price_per_unit",
"Cost_per_unit",
"Quantity_Sold",
"Revenues",
"Expenses",
"Revenues_log",
"Expenses_log",
]
# Create the drop-down menu widget
dropdown = pn.widgets.Select(name="Select Column:", options=options)
# Function to update the chart based on the selected value from the drop-down menu
@pn.depends(dropdown.param.value)
def update_chart(column):
# Create a chart based on the selected column
return data.hvplot.box(column, color="red")
# Create a Panel layout with the drop-down menu and chart
dashboard = pn.Column(dropdown, update_chart)
# Create a standalone server for the dashboard
dashboard.servable()
# Embed the dashboard in your notebook
# pn.panel(dashboard).embed()
# 
# 
import holoviews as hv
import numpy as np
import pandas as pd
import param
import panel as pn
# Define a function to create the boxplot
def create_boxplot(data, variable):
return data.hvplot.box(y=variable)
# Define the RoomOccupancy class with the dropdown and plot
class RoomOccupancy(param.Parameterized):
variable = param.ObjectSelector(default=data.columns[0], objects=list(data.columns))
@param.depends("variable")
def view(self):
return create_boxplot(data, self.variable)
# Define the dropdown widget to select the variable
variable_selector = hv.streams.Stream.define("Variable", variable=data.columns[0])(
variable=data.columns[0]
)
variable_menu = pn.widgets.Select(options=list(data.columns), value=data.columns[0])
# Define a function to update the plot based on the dropdown value
def update_boxplot(variable):
return create_boxplot(data, variable)
# Combine the widgets and the plot using a layout
layout = pn.Row(
pn.Column(variable_menu),
pn.Column(hv.DynamicMap(update_boxplot, streams=[variable_selector])),
)
# Create a layout and serve the app
layout.servable()
# 
# ### Descriptive Summary with Maximum observations
# create a dataframe containing the statistical summary of the numeric columns
df = data.describe(include=np.number).T
# highlight the max values in each column
df = df.style.highlight_max(color="indigo", axis=0)
# display the styled dataframe
display(df)
# ### Descriptive Summary with Minimum observations
# create a dataframe containing the statistical summary of the numeric columns
df = data.describe(include=np.number).T
# highlight the min values in each column
df = df.style.highlight_min(color="green", axis=0)
# display the styled dataframe
display(df)
import pandas as pd
import numpy as np
import ipywidgets as widgets
from IPython.display import display
# create a function to generate the statistical summary for the selected column
def generate_summary(column):
df = data[[column]].describe().T
df = df.style.highlight_max(color="indigo", axis=0)
display(df)
# create a dropdown menu with the numeric column names
dropdown = widgets.Dropdown(options=data.select_dtypes(include=np.number).columns)
# display the dropdown menu
display(dropdown)
# generate the summary for the initial selection
generate_summary(dropdown.value)
# update the summary when a new selection is made
def on_change(change):
if change["type"] == "change" and change["name"] == "value":
generate_summary(change["new"])
dropdown.observe(on_change)
import pandas as pd
# create a DataFrame containing the quantiles of the dataset
quantiles_df = data.quantile([0.25, 0.5, 0.6, 0.75, 0.9, 0.95, 0.99]).reset_index()
# highlight the max values in each column
styled_df = quantiles_df.style.highlight_max(color="red", axis=0)
# display the styled DataFrame
styled_df
import pandas as pd
import numpy as np
import ipywidgets as widgets
from IPython.display import display
# create a function to generate the statistical summary for the selected column
def generate_summary(column):
df = data[[column]].quantile([0.25, 0.5, 0.6, 0.75, 0.9, 0.95, 0.99]).T
df = df.style.highlight_max(color="green", axis=0)
display(df)
# create a dropdown menu with the numeric column names
dropdown = widgets.Dropdown(options=data.select_dtypes(include=np.number).columns)
# display the dropdown menu
display(dropdown)
# generate the summary for the initial selection
generate_summary(dropdown.value)
# update the summary when a new selection is made
def on_change(change):
if change["type"] == "change" and change["name"] == "value":
generate_summary(change["new"])
dropdown.observe(on_change)
import pandas as pd
import numpy as np
import ipywidgets as widgets
from IPython.display import display
# create a dataframe containing the quantiles of the numeric columns
quantiles_df = data.quantile([0.25, 0.5, 0.6, 0.75, 0.9, 0.95, 0.99]).T
# create a dropdown widget to select the axis to highlight max values
axis_dropdown = widgets.Dropdown(
options=["index", "columns"],
value="index",
description="Highlight axis:",
layout={"width": "max-content"},
)
# function to highlight max values in the selected axis
def highlight_max(axis):
return quantiles_df.style.highlight_max(color="purple", axis=axis)
# display the dropdown and the styled dataframe
display(axis_dropdown)
display(highlight_max(axis_dropdown.value))
# update the styled dataframe when a different axis is selected
def on_dropdown_change(change):
display(highlight_max(change.new))
axis_dropdown.observe(on_dropdown_change, names="value")
# function to plot a boxplot and a histogram along the same scale.
def histogram_boxplot(data, feature, figsize=(12, 7), kde=False, bins=None):
"""
Boxplot and histogram combined
data: dataframe
feature: dataframe column
figsize: size of figure (default (12,7))
kde: whether to the show density curve (default False)
bins: number of bins for histogram (default None)
"""
f2, (ax_box2, ax_hist2) = plt.subplots(
nrows=2, # Number of rows of the subplot grid= 2
sharex=True, # x-axis will be shared among all subplots
gridspec_kw={"height_ratios": (0.25, 0.75)},
figsize=figsize,
) # creating the 2 subplots
sns.boxplot(
data=data, x=feature, ax=ax_box2, showmeans=True, color="violet"
) # boxplot will be created and a star will indicate the mean value of the column
sns.histplot(
data=data, x=feature, kde=kde, ax=ax_hist2, bins=bins, palette="winter"
) if bins else sns.histplot(
data=data, x=feature, kde=kde, ax=ax_hist2
) # For histogram
ax_hist2.axvline(
data[feature].mean(), color="green", linestyle="--"
) # Add mean to the histogram
ax_hist2.axvline(
data[feature].median(), color="black", linestyle="-"
) # Add median to the histogram
import panel as pn
import pandas as pd
import hvplot.pandas
# Load your dataset into a DataFrame
# df = pd.read_csv('your_dataset.csv')
# Create a list of options for the drop-down menu
options = [
"Price_per_unit",
"Cost_per_unit",
"Quantity_Sold",
"Revenues",
"Expenses",
"Revenues_log",
"Expenses_log",
]
# Create the drop-down menu widget
dropdown = pn.widgets.Select(name="Select Column:", options=options)
# Function to update the chart based on the selected value from the drop-down menu
@pn.depends(dropdown.param.value)
def update_chart(column):
# Create a chart based on the selected column
return data.hvplot.box(column, color="red")
# Create a Panel layout with the drop-down menu and chart
dashboard = pn.Column(dropdown, update_chart)
# Create a standalone server for the dashboard
dashboard.servable()
# Embed the dashboard in your notebook
# pn.panel(dashboard).embed()
data.info()
import panel as pn
import pandas as pd
import hvplot.pandas
# Load your dataset into a DataFrame
# df = pd.read_csv('your_dataset.csv')
# Create a list of options for the drop-down menu
options = [
"Price_per_unit",
"Cost_per_unit",
"Quantity_Sold",
"Revenues",
"Expenses",
"Revenues_log",
"Expenses_log",
]
# Create the drop-down menu widget
dropdown = pn.widgets.Select(name="Select Column:", options=options)
# Function to update the chart based on the selected value from the drop-down menu
@pn.depends(dropdown.param.value)
def update_chart(column):
# Create a chart based on the selected column
return data.hvplot.hist(column)
# Create a Panel layout with the drop-down menu and chart
dashboard = pn.Column(dropdown, update_chart)
# Create a standalone server for the dashboard
dashboard.servable()
# Embed the dashboard in your notebook
# pn.panel(dashboard).embed()
import panel as pn
import pandas as pd
import hvplot.pandas
# Create a list of options for the drop-down menus
column_options = [
"Price_per_unit",
"Cost_per_unit",
"Quantity_Sold",
"Revenues",
"Expenses",
"Revenues_log",
"Expenses_log",
]
chart_options = ["Histogram", "Scatter Plot", "Summary Statistics"]
# Create the drop-down menu widgets
column_dropdown = pn.widgets.Select(name="Select Column:", options=column_options)
chart_dropdown = pn.widgets.Select(name="Select Chart Type:", options=chart_options)
# Function to update the chart based on the selected values from the drop-down menus
@pn.depends(column_dropdown.param.value, chart_dropdown.param.value)
def update_chart(column, chart_type):
# Create a chart based on the selected column and chart type
if chart_type == "Histogram":
return data.hvplot.hist(column, bins=20)
elif chart_type == "Scatter Plot":
return data.hvplot.scatter(x=column, y="Revenues")
elif chart_type == "Summary Statistics":
return data[column].describe().to_frame().hvplot.table()
# Create a Panel layout with the drop-down menus and chart
dashboard = pn.Column(column_dropdown, chart_dropdown, update_chart)
# Display the dashboard
# dashboard.show()
# Create a standalone server for the dashboard
dashboard.servable()
data.columns
# This code creates a dashboard with multiple tabs containing different types of charts and statistics based on your dataset. The scatter plot and box plot tabs include drop-down menus to select the columns from your dataset to display in the charts. The other tabs display summary statistics tables, a correlation heatmap, a KDE plot, and a bar plot.
import pandas as pd
import panel as pn
import hvplot.pandas
# Load your dataset into a DataFrame
# data = pd.read_csv('your_dataset.csv')
# Define the dropdown menu options [ALL Columns]
column_options = [
"Year",
"Region",
"Product",
"Price_per_unit",
"Cost_per_unit",
"Quantity_Sold",
"Revenues",
"Expenses",
"Sales_Method",
"Revenues_log",
"Expenses_log",
]
dropdown_scatter = pn.widgets.Select(
options=column_options, name="Select a column for X-axis"
)
dropdown_box = pn.widgets.Select(options=column_options, name="Select a column")
# Define a function to update the scatter plot based on the dropdown selection
@pn.depends(dropdown_scatter.param.value)
def update_scatter(column):
plot = data.hvplot.scatter(x=column, y="Revenues", title="Scatter Plot")
return plot
# Define a function to update the box plot based on the dropdown selection
@pn.depends(dropdown_box.param.value)
def update_box(column):
plot = data.hvplot.box(y=column, title="Box Plot")
return plot
# Create Scatter and Boxplots
scatter_plot = pn.Column(dropdown_scatter, update_scatter)
box_plot = pn.Column(dropdown_box, update_box)
# Create Numerical Statistics
table1 = (
data[
[
"Price_per_unit",
"Cost_per_unit",
"Quantity_Sold",
"Revenues",
"Expenses",
"Revenues_log",
"Expenses_log",
]
]
.describe()
.T.hvplot.table(title="Summary Statistics")
)
table2 = (
data[
[
"Price_per_unit",
"Cost_per_unit",
"Quantity_Sold",
"Revenues",
"Expenses",
"Revenues_log",
"Expenses_log",
]
]
.quantile([0.25, 0.5, 0.6, 0.75, 0.9, 0.95, 0.99])
.T.hvplot.table(title="Quantiles Statistics")
)
# Create Correlation Heatmap
corr = data[
[
"Price_per_unit",
"Cost_per_unit",
"Quantity_Sold",
"Revenues",
"Expenses",
"Revenues_log",
"Expenses_log",
]
].corr()
heat_map = corr.hvplot.heatmap(title="Correlation Heatmap")
# Creat KDE and Bar Charts
kde_plot = data.hvplot.kde(y="Revenues", by=None, title="KDE Plot")
# bar_plot1 = data.hvplot.bar( x='gender', stacked=True, title='Bar Plot Gender')
bar_plot2 = data.hvplot.bar(x="Sales_Method", stacked=True, title="bar Plot cut")
# bar_plot3 = data.hvplot.bar( x='payment_method', stacked=True, title='Bar Plot Payment Methods')
bar_plot4 = data.hvplot.bar(x="Region", stacked=True, title="Bar Plot Color")
# Create a Panel layout with the charts and statistics
dashboard = pn.Tabs(
("Scatter Plot", scatter_plot),
("Box Plot", box_plot),
("Summary Statistics", table1),
("Quantiles Statistics", table2),
("Correlation Heatmap", heat_map),
("KDE Plot", kde_plot),
("Bar Plot", bar_plot4),
)
# Display the dashboard
# dashboard.show()
# Create a standalone server for the dashboard
dashboard.servable()
data.columns
# Creating a sunburst chart
import plotly.express as px
fig = px.sunburst(
data,
path=["Region"],
values="Revenues",
color="Revenues",
color_discrete_map={"(?)": "red", "Lunch": "gold", "Dinner": "green"},
)
fig.show()
# Creating a sunburst chart for Cash Ratio vs GICS Sectors
import plotly.express as px
fig = px.sunburst(
data,
path=["Product"],
values="Revenues",
color="Revenues",
color_discrete_map={"(?)": "red", "Lunch": "gold", "Dinner": "green"},
)
fig.show()
# Creating a sunburst chart for Cash Ratio vs GICS Sectors
import plotly.express as px
fig = px.sunburst(
data,
path=["Sales_Method"],
values="Revenues",
color="Revenues",
color_discrete_map={"(?)": "red", "Lunch": "gold", "Dinner": "green"},
)
fig.show()
data.columns
import plotly.express as px
fig = px.sunburst(
data,
path=["Region", "Product", "Sales_Method"],
values="Expenses",
color="Product",
color_discrete_map={"(?)": "red", "Lunch": "gold", "Dinner": "green"},
)
fig.show()
import plotly.express as px
fig = px.sunburst(
data,
path=["Region", "Product", "Sales_Method"],
values="Revenues",
color="Product",
color_discrete_map={"(?)": "red", "Lunch": "gold", "Dinner": "green"},
)
fig.show()
# Sunburst of a rectangular DataFrame with discrete color argument in px.sunburst
import plotly.express as px
fig = px.sunburst(data, path=["Region", "Product"], values="Expenses")
fig.show()
# Sunburst of a rectangular DataFrame with discrete color argument in px.sunburst
import plotly.express as px
fig = px.sunburst(
data, path=["Sales_Method", "Product"], values="Revenues", color="Product"
)
fig.show()
import plotly.express as px
fig = px.sunburst(
data,
path=["Region", "Sales_Method", "Product"],
values="Revenues",
color="Sales_Method",
color_discrete_map={"(?)": "black", "Lunch": "gold", "Dinner": "darkblue"},
)
fig.show()
# Distribution of Population of Pyramid of Marketing Funnel Dataset
plt.figure(figsize=(15, 5), dpi=80)
group_col = "Region"
order_of_bars = data.Product.unique()[::-1]
colors = [
plt.cm.Spectral(i / float(len(data[group_col].unique()) - 1))
for i in range(len(data[group_col].unique()))
]
for c, group in zip(colors, data[group_col].unique()):
sns.barplot(
x="Quantity_Sold",
y="Product",
data=data.loc[data[group_col] == group, :],
order=order_of_bars,
color=c,
label=group,
)
# Decorations
plt.xlabel("$Quantity Sold$")
plt.ylabel("Quantity Sold from Products")
plt.yticks(fontsize=12)
plt.title("Revenues Pyramid of the Store Funnel", fontsize=22)
plt.legend()
plt.show()
data.head(3)
# Displaying the correlation between numerical variables of the dataset
plt.figure(figsize=(30, 10))
sns.heatmap(data.corr(), annot=True, vmin=-1, vmax=1, fmt=".2f", cmap="coolwarm")
plt.show()
# using heatmap
correlation = data.corr() # creating a 2-D Matrix with correlation plots
plt.figure(figsize=(30, 10))
sns.heatmap(correlation, annot=True, cmap="RdYlGn")
# creates heatmap showing correlation of numeric columns in data
plt.figure(figsize=(30, 10))
sns.heatmap(data.corr(), vmin=-1, vmax=1, cmap="YlGnBu", annot=True, fmt=".2f")
# Checking the count plots of the categorical Variables
cols = 2
rows = 2
num_cols = data.select_dtypes(include="object")
fig = plt.figure(figsize=(25, 18))
for i, col in enumerate(num_cols):
ax = fig.add_subplot(rows, cols, i + 1)
sns.countplot(x=data[col], ax=ax)
plt.xticks(rotation=45, ha="right")
fig.tight_layout()
plt.show()
# Checking the histogram plot of numerical variables of the entire dataset
cols = 3
rows = 3
num_cols = data.select_dtypes(exclude="object").columns
fig = plt.figure(figsize=(cols * 5, rows * 4))
for i, col in enumerate(num_cols):
ax = fig.add_subplot(rows, cols, i + 1)
sns.histplot(x=data[col], ax=ax)
fig.tight_layout()
plt.show()
# Checking the histogram plot of numerical variables of the entire dataset
cols = 3
rows = 3
num_cols = data.select_dtypes(exclude="object").columns
fig = plt.figure(figsize=(cols * 4, rows * 3))
for i, col in enumerate(num_cols):
ax = fig.add_subplot(rows, cols, i + 1)
sns.histplot(x=data[col], kde=True, ax=ax)
fig.tight_layout()
plt.show()
facet = sns.FacetGrid(data, hue="Product", aspect=3)
facet.map(sns.kdeplot, "Revenues", shade=True)
facet.set(xlim=(data["Revenues"].min(), data["Revenues"].max()))
facet.add_legend()
plt.title("Revenues vs Product")
plt.show()
# Ploting a displot of Price Change vs GICS Sectors
sns.displot(
data=data,
x="Expenses",
hue="Region",
multiple="stack",
kind="kde",
height=18,
aspect=2.5,
)
### function to plot distributions wrt target
def distribution_plot_wrt_target(data, predictor, target):
fig, axs = plt.subplots(2, 2, figsize=(25, 9))
target_uniq = data[target].unique()
axs[0, 0].set_title("Distribution of target for target=" + str(target_uniq[0]))
sns.histplot(
data=data[data[target] == target_uniq[0]],
x=predictor,
kde=True,
ax=axs[0, 0],
color="teal",
stat="density",
)
axs[0, 1].set_title("Distribution of target for target=" + str(target_uniq[1]))
sns.histplot(
data=data[data[target] == target_uniq[1]],
x=predictor,
kde=True,
ax=axs[0, 1],
color="orange",
stat="density",
)
axs[1, 0].set_title("Boxplot w.r.t target")
sns.boxplot(data=data, x=target, y=predictor, ax=axs[1, 0], palette="gist_rainbow")
axs[1, 1].set_title("Boxplot (without outliers) w.r.t target")
sns.boxplot(
data=data,
x=target,
y=predictor,
ax=axs[1, 1],
showfliers=False,
palette="gist_rainbow",
)
plt.tight_layout()
plt.show()
distribution_plot_wrt_target(data, "Expenses", "Product")
distribution_plot_wrt_target(data, "Revenues", "Region")
# Checking the KDE plot of numerical variables of the entire dataset
cols = 2
rows = 4
num_cols = data.select_dtypes(exclude="object").columns
fig = plt.figure(figsize=(cols * 5, rows * 4))
for i, col in enumerate(num_cols):
ax = fig.add_subplot(rows, cols, i + 1)
sns.kdeplot(x=data[col], hue="Region", data=data)
fig.tight_layout()
plt.show()
# Checking the boxplot of the numerical variable of the dataset
cols = 2
rows = 4
num_cols = data.select_dtypes(exclude="object").columns
fig = plt.figure(figsize=(18, 12))
for i, col in enumerate(num_cols):
ax = fig.add_subplot(rows, cols, i + 1)
sns.boxplot(x=data[col], ax=ax)
fig.tight_layout()
plt.show()
num_cols = data.select_dtypes(exclude="object").columns
for feature in num_cols:
histogram_boxplot(
data, feature, figsize=(12, 7), kde=False, bins=None
) ## Please change the dataframe name as you define while reading the data
# Checking the voilin plot of the numerical variables
cols = 3
rows = 3
num_cols = data.select_dtypes(exclude="object").columns
fig = plt.figure(figsize=(18, 16))
for i, col in enumerate(num_cols):
ax = fig.add_subplot(rows, cols, i + 1)
sns.violinplot(x=data[col], ax=ax)
fig.tight_layout()
plt.show()
import plotly.express as px
fig = px.parallel_categories(
data, color="Quantity_Sold", color_continuous_scale=px.colors.sequential.Inferno
)
fig.show()
data.columns
cols = [
"Price_per_unit",
"Cost_per_unit",
"Quantity_Sold",
"Revenues",
"Expenses",
"Revenues_log",
"Expenses_log",
]
plt.style.use("fivethirtyeight")
sns.pairplot(data, hue="Region", vars=cols, diag_kind="kde")
import scipy
# Function to calculate correlation coefficient between two variables
def corrfunc(x, y, **kwgs):
r = np.corrcoef(x, y)[0][1]
ax = plt.gca()
ax.annotate("r = {:.2f}".format(r), xy=(0.1, 0.8), xycoords=ax.transAxes, size=24)
# Create a PairGrid
g = sns.PairGrid(
data=data,
vars=[
"Price_per_unit",
"Cost_per_unit",
"Quantity_Sold",
"Revenues",
"Expenses",
"Revenues_log",
"Expenses_log",
],
)
# Map a scatterplot to the upper triangle
g.map_upper(plt.scatter)
# Map a histogram to the diagonal
g.map_diag(plt.hist)
# Map a kde plot to the lower triangle
g.map_lower(sns.kdeplot)
# Map the correlation coefficient to the lower diagonal
g.map_lower(corrfunc)
|
# Import important libraries to be used in the notebook
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
# Import the CardioGoodFitness.csv into a data frame
data = pd.read_csv("/kaggle/input/treadmill-users-dataset/treadmil-users.csv")
# # Sanity Checks
# ## Dataset preview and shape
# Sample first 5 rows of the data to confirm loading
data.head()
# Sneek peak into the data dimension using shape function of the data frame object
data.shape
# - The data consists if 180 rows and 9 columns
# ## Data Types
# Sneak peak into the dataset's data types
data.info()
# - The data consists of 3 category variables and 6 Numerical Variables
# - Product, Gender, Marital Status are the category variables in the dataset
# - Age, Education, Usage, Fitness, Income and Miles are the Numerical variables (type int) in the dataset
# ## Statistical Summary
# Statistical summary of the dataset
data.describe(include="all").T
# - The shop sells 3 product types with TM195 product being the top selling product representing 44% of the sales
# - Customers are aged between 18 and 50 years
# - The average age of the customers buying the products is 28
# - 50% of the customers are aged 26 years and above while 75% of customers are aged above 33 years
# - 57% of the total customers are of male gender
# - The education experience ranges between 12 and 21 years
# - The average education experience in years is 15.5 approx 16 years
# - 50% and 75% of the customers have an education experience of 16 years and above
# - Customers are either Partnered or Single
# - 59% of the total customers are partnered while 41 % are Single
# - Customers expect use their treadmills averagely 3 times a week
# - The average fitness is 3 representing more than 50%.
# - Given that the range if fitness is between 1 to 5, we can infer that majority of the customers are moderately fit
# - The average income is USD 53,719. 75% of the customers income is above US 58k
# - The average mileage that customers expect to run is 103
# ## Check for missing values
# Check for missing data in the dataset
data.isnull().sum()
# There are no missing values in our dataset
# # Univariate Analysis
# ## Check distribution on Numerical columns
# ### Observations on Age
sns.histplot(data=data, x="Age")
plt.show()
## Lets try to Bin the Age variable for indepth analysis of age
pd.cut(data["Age"], 5).value_counts()
sns.boxplot(data=data, x="Age")
plt.show()
# - The distribution of Age is skewed towards the right
# - We observe majority of customers are aged between 24 & 31, followed by ages 18 -24, 31 - 37, 37 -43 respectively
# - From the box plot, There are few outliers indicating that we have few exceptions of customers aged above 45
# - Lets find out how many customers are aged above the max non outlier age of 45
data.loc[data["Age"] > 45].shape
# - We have 6 customers aged above 45 years. The rest are aged between 18 and 45 years
# ### Observations on Education
sns.histplot(data=data, x="Education", kde=True)
plt.show()
sns.boxplot(data=data, x="Education")
plt.show()
# - The distribution of Education is normal. Evenly ditributed.
# - We observe majority of customers have an Education experience of 14-16 years
# - We have outliers indicating education experience of over 18 years.
# ### Observations on Usage
##Histplot
sns.histplot(data=data, x="Usage", kde=True)
plt.show()
##Violin Plot
sns.violinplot(
data=data,
x="Usage",
)
plt.show()
##Box Plot
sns.boxplot(
data=data,
x="Usage",
)
plt.show()
# - The distribution of usage is skewed towards the right
# - The most predetermined usage recorded is between 3-4 times a week.
# - 2 Outliers observed (customer expected usage 6-7 times a week)
# - Let us find out how many customers have a predermined usage of more than 5
data.loc[data["Usage"] > 5].shape
# - There are only 9 such customers
# ### Observations on Fitness
sns.histplot(x="Fitness", data=data)
plt.show()
# KDE Plot
sns.kdeplot(x="Fitness", data=data)
plt.show()
# Violin Plot
sns.violinplot(x=data["Fitness"])
plt.show()
sns.boxplot(
data=data,
x="Fitness",
)
plt.show()
# - The distribution of Fitness is skewed towards the left
# - 1 Outlier observed with fitness level 1
# - Let us find out how many customers have a fitness level of less than minimum non outlier 2
# data[data['Fitness'] < 2].shape
# - We observe that we have 2 customers with a fitness of below 2
# ### Observations on Income
sns.histplot(x=data["Income"])
plt.show()
sns.boxplot(x=data["Income"])
plt.show()
# * The distribution of Income is skewed towards the right.
# * The values seem fine as the individual income depends upon various factors depending on each individual e.g Age
# * There are few outliers in this variable and the values above **75000** are being represented as outliers by the boxplot.
# * Let us check how many customers have an income of more than 75000
data.loc[data["Income"] > 75000].shape
# - We have 21 customers with an income of 75000 and above
# ### Observations on Miles
sns.histplot(data=data, x="Miles", stat="density")
plt.show()
sns.boxplot(data=data, x="Miles")
plt.show()
# * The distribution is skewed towards the right.
# * There are a few outliers present in this column.
# * Values above 170 Miles are being represented as outliers in the boxplot
data.loc[data["Miles"] > 170].shape
# - We have 19 such customers
# ## Check distribution on Non-Numerical columns
data.info()
# ### Observations on Gender
sns.countplot(data=data, x="Gender")
plt.show()
# - Around 58% of the customers are male
# ### Observation on Marital Status
sns.countplot(data=data, x="MaritalStatus")
plt.show()
# - Around 59% of the customers have 'Partnered' as their marital status
# ### Observations on Product
sns.countplot(data=data, x="Product")
plt.show()
# - Product with model number TM195 is maximum in number. Seems to be the most preferred product representing 44% of the total sale
# - This is followed by products with model number TM498, TM798 with 33% and 22% respectively.
# # Bivariate Analysis
# #### Correlation Matrix
# Review variable correlation using a heat map
plt.figure(figsize=(10, 5)) # set the fugure size
sns.heatmap(
data.corr(), annot=True, cmap="Spectral", vmin=-1, vmax=1
) # use hetmap function from the SNS library
plt.show()
# **Observations**
# * Miles column shows a high correlation with usgae,fitness. This indicates that more miles that a customer intends to run, the higher the self rated fitness and the intended usage.
# * Education shows a positive correlation with the Income column. This indicates that the higher the Education experience the higher the income.
# * Usage shows a positve correlation with Fitness. Which makes sense since the more fit the customer is the higher the usage.
# * We observe a weak correlation between age and usage, fitness ,miles columns. The higher the age the less the usage.
# * We also observe a positive strong correlation between Education and Usage, fitness.
# ### Let us observe the product preference amongst age groups and gender**
plt.figure(figsize=(10, 5))
sns.boxplot(data=data, x="Product", y="Age", hue="Gender", showfliers=False)
plt.show()
# * We observe that customers base is largely male across all products.
# * Majority of male customers between the ages 22-34 prefer the TM195 product.
# * Product TM498 is the most preferred product amongst Female customers between the ages 25 and 35
# * Product TM798 is least preferred product across male and female customers
# ### Analysis of age, Income and product preference amongst customers**
# Categorizing age into age groups for easier analyisy
bins = [10, 20, 30, 40, 50]
labels = ["10-20", "20-30", "30-40", "40-50"]
data["Age Group"] = pd.cut(x=data["Age"], bins=bins, labels=labels, right=False)
plt.figure(figsize=(15, 5))
sns.barplot(data=data, x="Age Group", y="Income", hue="Product")
plt.show()
#
# * Products TM195 & TM498 preference is evenly dsitributed amongst all age groups
# * Product TM798 is the most preferred product across age groups 20-30, 30-40 and 40-50
# * We observe Age group 10-20 having the lowest incomes.
# * Product TM798 is most preferred amongst customers with incomes above 40,000 and ages 20 and above
# ### Analysis of income and marital status
plt.figure(figsize=(15, 5))
sns.boxplot(data=data, x="Income", y="MaritalStatus", showfliers=False)
plt.show()
# **We observe higher incomes amongst customers with 'Partnered' marital status than amongst customers with 'Single' marital status**
# ### Analysis of income and Education
plt.figure(figsize=(15, 5))
sns.lineplot(data=data, y="Income", x="Education", ci=None)
plt.show()
# **We observe a strong correlation between education and income. Customers with higher education seem to have higher incomes**
# ## Analysis of usage and Income
# We will sub-divide income into ranges using cut function from pandas library and create a new column 'Income Range'
data["Income Range"] = pd.cut(
data["Income"], bins=[30000, 50000, 70000, 90000, 110000], right=False
)
plt.figure(figsize=(15, 5))
sns.boxplot(data=data, y="Usage", x="Income Range", showfliers=False)
plt.show()
# **We observe that the usage is highest amongst customers with incomes between 70,000 - 90,000**
# ## Analysis of Usage & product
plt.figure(figsize=(15, 5))
sns.boxplot(data=data, y="Usage", x="Product", showfliers=False)
plt.show()
|
import numpy as np
import pandas as pd
from pathlib import Path
import os.path
import matplotlib.pyplot as plt
from IPython.display import Image, display, Markdown
import matplotlib.cm as cm
from sklearn.model_selection import train_test_split
from sklearn.metrics import confusion_matrix
import tensorflow as tf
from time import perf_counter
import seaborn as sns
def printmd(string):
# Print with Markdowns
display(Markdown(string))
new_train = pd.read_csv("../input/aptos2019-blindness-detection/train.csv")
new_train = new_train.dropna()
new_train["id_code"] = (
"/kaggle/input/aptos2019-blindness-detection/train_images/"
+ new_train["id_code"].astype(str)
+ ".png"
)
new_train_1 = pd.read_csv("../input/aptos2019-blindness-detection/test.csv")
new_train_1 = new_train_1.dropna()
new_train_1["id_code"] = (
"/kaggle/input/aptos2019-blindness-detection/test_images/"
+ new_train["id_code"].astype(str)
+ ".png"
)
new_train = pd.concat([new_train, new_train_1])
old_train = pd.read_csv("../input/diabetic-retinopathy-resized/trainLabels.csv")
old_train = old_train.dropna()
old_train = old_train[["image", "level"]]
old_train.columns = new_train.columns
old_train.diagnosis.value_counts()
old_train["id_code"] = (
"/kaggle/input/diabetic-retinopathy-resized/resized_train/resized_train/"
+ old_train["id_code"].astype(str)
+ ".jpeg"
)
print(new_train.columns)
print(old_train.columns)
frames = [new_train, old_train]
data = pd.concat(frames)
import pandas as pd
import os
from tqdm import tqdm
# add a new column called 'present' and set default value to False
data["present"] = False
# loop through each row and check if the file exists
for i, row in tqdm(data.iterrows(), total=data.shape[0]):
if os.path.exists(row["id_code"]):
data.at[i, "present"] = True
# print the updated table
print(data)
data.present.value_counts()
data.to_csv("data.csv")
data.info()
data = data.dropna()
data.isna().value_counts()
from sklearn.model_selection import train_test_split
test_df, train_df = train_test_split(data, test_size=0.7, random_state=123)
test_df, val_df = train_test_split(test_df, test_size=0.5, random_state=123)
val_df.isna().value_counts()
print("Train DF")
print(train_df.diagnosis.value_counts())
print("Test DF")
print(test_df.diagnosis.value_counts())
print("Val DF")
print(val_df.diagnosis.value_counts())
train_df.diagnosis = train_df.diagnosis.astype(int)
test_df.diagnosis = test_df.diagnosis.astype(int)
val_df.diagnosis = val_df.diagnosis.astype(int)
print("Train DF")
print(train_df.diagnosis.value_counts())
print("Test DF")
print(test_df.diagnosis.value_counts())
print("Val DF")
print(val_df.diagnosis.value_counts())
im_size = 224
# old_train = old_train[['image','level']]
# old_train.columns = new_train.columns
# old_train.diagnosis.value_counts()
# # path columns
# new_train['id_code'] = '../input/aptos2019-blindness-detection/train_images/' + new_train['id_code'].astype(str) + '.png'
# old_train['id_code'] = '../input/diabetic-retinopathy-resized/resized_train/resized_train/' + old_train['id_code'].astype(str) + '.jpeg'
# # Let's shuffle the datasets
# train_df = train_df.sample(frac=1).reset_index(drop=True)
# val_df = val_df.sample(frac=1).reset_index(drop=True)
# print(train_df.shape)
# print(val_df.shape)
train_df.head()
def display_samples(data, columns=4, rows=3):
images = data[: columns * rows]
fig = plt.figure(figsize=(5 * columns, 4 * rows))
for i in range(len(images)):
image_path = images[i]
# print(image_path)
# import os
# if os.path.isfile(image_path):
# print("Found")
# else:
# print("Not found")
img = cv2.imread(f"{image_path}")
img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)
img = cv2.resize(img, (im_size, im_size))
img = cv2.addWeighted(
img, 4, cv2.GaussianBlur(img, (0, 0), im_size / 40), -4, 128
)
fig.add_subplot(rows, columns, i + 1)
plt.imshow(img)
plt.tight_layout()
# display sample images from the 'data' variable
display_samples(data["id_code"].tolist())
def crop_image1(img, tol=7):
# img is image data
# tol is tolerance
mask = img > tol
return img[np.ix_(mask.any(1), mask.any(0))]
def crop_image_from_gray(img, tol=7):
if img.ndim == 2:
mask = img > tol
return img[np.ix_(mask.any(1), mask.any(0))]
elif img.ndim == 3:
gray_img = cv2.cvtColor(img, cv2.COLOR_RGB2GRAY)
mask = gray_img > tol
check_shape = img[:, :, 0][np.ix_(mask.any(1), mask.any(0))].shape[0]
if check_shape == 0: # image is too dark so that we crop out everything,
return img # return original image
else:
img1 = img[:, :, 0][np.ix_(mask.any(1), mask.any(0))]
img2 = img[:, :, 1][np.ix_(mask.any(1), mask.any(0))]
img3 = img[:, :, 2][np.ix_(mask.any(1), mask.any(0))]
img = np.stack([img1, img2, img3], axis=-1)
return img
def preprocess_image(image_path, desired_size=224):
img = cv2.imread(image_path)
img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)
img = crop_image_from_gray(img)
img = cv2.resize(img, (desired_size, desired_size))
img = cv2.addWeighted(
img, 4, cv2.GaussianBlur(img, (0, 0), desired_size / 30), -4, 128
)
return img
def preprocess_image_old(image_path, desired_size=224):
img = cv2.imread(image_path)
img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)
# img = crop_image_from_gray(img)
img = cv2.resize(img, (desired_size, desired_size))
img = cv2.addWeighted(
img, 4, cv2.GaussianBlur(img, (0, 0), desired_size / 40), -4, 128
)
return img
# # validation set
# N = val_df.shape[0]
# x_val = np.empty((N, im_size, im_size, 3), dtype=np.uint8)
# from tqdm import trange, tqdm
# for i, image_id in enumerate(tqdm(val_df['id_code'])):
# x_val[i, :, :, :] = preprocess_image(
# f'{image_id}',
# desired_size = im_size
# )
val_df
y_train = train_df["diagnosis"].values
y_val = val_df["diagnosis"].values
print(y_train.shape)
print(x_val.shape)
print(y_val.shape)
print("Train DF Columns")
print(train_df.columns)
train_df["diagnosis"] = train_df["diagnosis"].astype("str")
print("Test DF Columns")
print(test_df.columns)
test_df["diagnosis"] = test_df["diagnosis"].astype("str")
print("Validate DF Columns")
print(val_df.columns)
val_df["diagnosis"] = val_df["diagnosis"].astype("str")
val_df
# # Visualization
# # Load the Images with a generator
train_df
def create_gen():
# Load the Images with a generator and Data Augmentation
train_generator = tf.keras.preprocessing.image.ImageDataGenerator(
preprocessing_function=tf.keras.applications.mobilenet_v2.preprocess_input,
validation_split=0.1,
)
test_generator = tf.keras.preprocessing.image.ImageDataGenerator(
preprocessing_function=tf.keras.applications.mobilenet_v2.preprocess_input
)
# compute the class frequencies
class_freq = train_df["diagnosis"].value_counts(normalize=True)
print("Class frequencies:\n", class_freq)
# compute the class weights
class_weights = {}
for i in range(len(class_freq)):
class_weights[i] = 1 / class_freq[i]
# print the class weights
print("Class weights:\n", class_weights)
train_images = train_generator.flow_from_dataframe(
dataframe=train_df,
x_col="id_code",
y_col="diagnosis",
target_size=(224, 224),
color_mode="rgb",
class_mode="categorical",
batch_size=16,
shuffle=True,
seed=0,
subset="training",
rotation_range=30, # Uncomment to use data augmentation
zoom_range=0.15,
width_shift_range=0.2,
height_shift_range=0.2,
shear_range=0.15,
horizontal_flip=True,
fill_mode="nearest",
class_weights=class_weights,
)
val_images = train_generator.flow_from_dataframe(
dataframe=test_df,
x_col="id_code",
y_col="diagnosis",
target_size=(224, 224),
color_mode="rgb",
class_mode="categorical",
batch_size=16,
shuffle=True,
seed=0,
subset="validation",
rotation_range=30, # Uncomment to use data augmentation
zoom_range=0.15,
width_shift_range=0.2,
height_shift_range=0.2,
shear_range=0.15,
horizontal_flip=True,
fill_mode="nearest",
)
test_images = test_generator.flow_from_dataframe(
dataframe=test_df,
x_col="id_code",
y_col="diagnosis",
target_size=(224, 224),
color_mode="rgb",
class_mode="categorical",
batch_size=16,
shuffle=False,
)
return train_generator, test_generator, train_images, val_images, test_images
def get_model(model):
# Load the pretained model
kwargs = {
"input_shape": (224, 224, 3),
"include_top": False,
"weights": "imagenet",
"pooling": "avg",
}
pretrained_model = model(**kwargs)
pretrained_model.trainable = False
inputs = pretrained_model.input
x = tf.keras.layers.Dense(128, activation="relu")(pretrained_model.output)
x = tf.keras.layers.Dense(128, activation="relu")(x)
outputs = tf.keras.layers.Dense(5, activation="softmax")(x)
model = tf.keras.Model(inputs=inputs, outputs=outputs)
opt = keras.optimizers.Adam(learning_rate=0.0005)
model.compile(
optimizer=opt,
loss="categorical_crossentropy",
metrics=["accuracy"],
)
return model
# Dictionary with the models
models = {
"InceptionV3": {"model": tf.keras.applications.InceptionV3, "perf": 0},
}
# models = {
# "DenseNet121": {"model":tf.keras.applications.DenseNet121, "perf":0},
# "DenseNet169": {"model":tf.keras.applications.DenseNet169, "perf":0},
# "InceptionResNetV2": {"model":tf.keras.applications.InceptionResNetV2, "perf":0},
# "InceptionV3": {"model":tf.keras.applications.InceptionV3, "perf":0},
# "MobileNet": {"model":tf.keras.applications.MobileNet, "perf":0},
# "MobileNetV2": {"model":tf.keras.applications.MobileNetV2, "perf":0},
# "ResNet101": {"model":tf.keras.applications.ResNet101, "perf":0},
# "ResNet50": {"model":tf.keras.applications.ResNet50, "perf":0},
# "VGG16": {"model":tf.keras.applications.VGG16, "perf":0},
# "VGG19": {"model":tf.keras.applications.VGG19, "perf":0}
# }
# Create the generators
train_generator, test_generator, train_images, val_images, test_images = create_gen()
print("\n")
# Fit the models
for name, model in models.items():
# Get the model
m = get_model(model["model"])
models[name]["model"] = m
start = perf_counter()
# Fit the model
history = m.fit(train_images, validation_data=val_images, epochs=1, verbose=2)
# Sav the duration, the train_accuracy and the val_accuracy
duration = perf_counter() - start
duration = round(duration, 2)
models[name]["perf"] = duration
print(f"{name:20} trained in {duration} sec")
val_acc = history.history["val_accuracy"]
models[name]["val_acc"] = [round(v, 4) for v in val_acc]
train_acc = history.history["accuracy"]
models[name]["train_accuracy"] = [round(v, 4) for v in train_acc]
print("Done")
# Create a DataFrame with the results
models_result = []
for name, v in models.items():
models_result.append(
[
name,
models[name]["train_accuracy"][-1],
models[name]["val_acc"][-1],
models[name]["perf"],
]
)
df_results = pd.DataFrame(
models_result,
columns=["model", "train_accuracy", "val_accuracy", "Training time (sec)"],
)
df_results.sort_values(by="val_accuracy", ascending=False, inplace=True)
df_results.reset_index(inplace=True, drop=True)
df_results
plt.figure(figsize=(15, 5))
sns.barplot(x="model", y="train_accuracy", data=df_results)
plt.title("Accuracy on the Training Set (after 1 epoch)", fontsize=15)
plt.ylim(0, 1)
plt.xticks(rotation=90)
plt.show()
# # InceptionV3
# Load the pretained model
pretrained_model = tf.keras.applications.InceptionV3(
input_shape=(224, 224, 3), include_top=False, weights="imagenet", pooling="avg"
)
pretrained_model.trainable = False
inputs = pretrained_model.input
x = tf.keras.layers.Dense(128, activation="relu")(pretrained_model.output)
x = tf.keras.layers.Dense(128, activation="relu")(x)
outputs = tf.keras.layers.Dense(5, activation="softmax")(x)
model = tf.keras.Model(inputs=inputs, outputs=outputs)
model.compile(
optimizer="adam", loss="categorical_crossentropy", metrics=["accuracy", "AUC"]
)
history = model.fit(
train_images,
validation_data=val_images,
batch_size=16,
epochs=50,
)
# callbacks=[
# tf.keras.callbacks.EarlyStopping(
# monitor='val_loss',
# patience=2,
# restore_best_weights=True
# )
# ]
model.save("new_model_with_class_weights.h5")
pd.DataFrame(history.history)[["accuracy", "val_accuracy"]].plot()
plt.title("Accuracy")
plt.show()
pd.DataFrame(history.history)[["loss", "val_loss"]].plot()
plt.title("Loss")
plt.show()
results = model.evaluate(test_images, verbose=0)
printmd(" ## Test Loss: {:.5f}".format(results[0]))
printmd("## Accuracy on the test set: {:.2f}%".format(results[1] * 100))
# Predict the label of the test_images
pred = model.predict(test_images)
pred = np.argmax(pred, axis=1)
# Map the label
labels = train_images.class_indices
labels = dict((v, k) for k, v in labels.items())
pred = [labels[k] for k in pred]
# Display the result
print(f"The first 5 predictions: {pred[:5]}")
from sklearn.metrics import classification_report
y_test = list(test_df.Label)
from sklearn import metrics
print("Accuracy:", np.round(metrics.accuracy_score(y_test, pred), 5))
print(
"Precision:", np.round(metrics.precision_score(y_test, pred, average="weighted"), 5)
)
print("Recall:", np.round(metrics.recall_score(y_test, pred, average="weighted"), 5))
print("F1 Score:", np.round(metrics.f1_score(y_test, pred, average="weighted"), 5))
print("Cohen Kappa Score:", np.round(metrics.cohen_kappa_score(y_test, pred), 5))
print(classification_report(y_test, pred))
|
import pandas as pd
from pathlib import Path
import torch
import matplotlib.pyplot as plt
import numpy as np
from tqdm import tqdm
import plotly.express as px
df = pd.read_csv("/kaggle/input/birdclef-2023/train_metadata.csv")
TRAIN_PATH = Path("/kaggle/input/birdclef-2023/train_audio")
DEVICE = "cuda" if torch.cuda.is_available() else "cpu"
# Fetch precomputed embeddings from Google Bird Vocalization Classifier
# combine first and last embedding of all recordings (corresponds to first and last 5 seconds of audio)
embeddings = torch.load("/kaggle/input/gbvc-embeddings/embeddings.pt")
first5sec_embeddings = torch.stack(
[torch.tensor(embeddings[filename][0]) for filename in df.filename]
)
last5sec_embeddings = torch.stack(
[torch.tensor(embeddings[filename][-1]) for filename in df.filename]
)
combined_embeddings = torch.concat([first5sec_embeddings, last5sec_embeddings], -1)
combined_embeddings.shape
# compute euclidean distances between each cell of a 2d matrix
def get_distances(x):
result = []
# loop over each row, since doing it all at once would cause a memory error
for row in tqdm(x):
result.append(((row[:, None] - x.T) ** 2).T.sum(-1))
return torch.stack(result)
distances = get_distances(combined_embeddings.to(DEVICE))
plt.figure(figsize=(9, 9))
plt.title("All embedding distances (notice 0 along the diagonal, as expected)")
plt.imshow(distances.to("cpu"))
plt.colorbar()
# Fetch outliers from a previous analysis on 3D projections of embeddings
outliers = pd.read_csv("/kaggle/input/birdclef2023-eda-with-3d-embeddings/outliers.csv")
for outlier_type in [1, 2]:
outlier_distances = get_distances(
combined_embeddings[
df.filename.isin(
set(outliers[outliers.outlier_type == outlier_type].filename)
)
].to(DEVICE)
)
plt.title("outliers on line " + str(outlier_type))
plt.imshow(outlier_distances.to("cpu"))
plt.colorbar()
plt.show()
# Known duplicates, copied from https://www.kaggle.com/competitions/birdclef-2023/discussion/398229
known_dupes = [
["carcha1/XC324665.ogg", "carcha1/XC324666.ogg"],
["wbrcha2/XC613380.ogg", "wbrcha2/XC613384.ogg"],
["comsan/XC613127.ogg", "comsan/XC613128.ogg"],
["tafpri1/XC443724.ogg", "tafpri1/XC443725.ogg"],
["colsun2/XC755891.ogg", "colsun2/XC755892.ogg"],
["grewoo2/XC527938.ogg", "grewoo2/XC527939.ogg"],
["sccsun2/XC609477.ogg", "sccsun2/XC609478.ogg"],
["subbus1/XC603421.ogg", "subbus1/XC603426.ogg"],
["combul2/XC748220.ogg", "combul2/XC748221.ogg"],
["wtbeat1/XC234928.ogg", "wtbeat1/XC234929.ogg"],
["afrthr1/XC652880.ogg", "afrthr1/XC652884.ogg"],
["laudov1/XC405374.ogg", "laudov1/XC405375.ogg"],
["litegr/XC411319.ogg", "litegr/XC411320.ogg"],
["cohmar1/XC564020.ogg", "cohmar1/XC564021.ogg"],
["egygoo/XC358927.ogg", "egygoo/XC528135.ogg"],
["combuz1/XC647786.ogg", "combuz1/XC647787.ogg"],
["combuz1/XC144257.ogg", "combuz1/XC144258.ogg"],
["wlwwar/XC478705.ogg", "wlwwar/XC478767.ogg"],
["woosan/XC740798.ogg", "woosan/XC742927.ogg"],
["gnbcam2/XC530150.ogg", "gnbcam2/XC530151.ogg"],
["litswi1/XC443712.ogg", "litswi1/XC443713.ogg"],
["combul2/XC650878.ogg", "combul2/XC447669.ogg"],
["gobbun1/XC394478.ogg", "gobbun1/XC395111.ogg"],
["fislov1/XC503794.ogg", "fislov1/XC526237.ogg"],
["cibwar1/XC395511.ogg", "cibwar1/XC432840.ogg"],
["combul2/XC447668.ogg", "combul2/XC650877.ogg"],
]
# make sure none of the known dupes are in our list of outliers
outliers.filename.isin(set([x for li in known_dupes for x in li])).sum()
# find the euclid distance between the embeddings of all known duplicates
filename_to_index = {f: i for i, f in enumerate(df.filename)}
known_dupes = [
(a, b, distances[filename_to_index[a], filename_to_index[b]].item())
for a, b in known_dupes
]
sorted(known_dupes, key=lambda a: a[2])
n = len(combined_embeddings)
not_eye = (1 - torch.triu(torch.ones(n, n)).to(DEVICE)).bool()
def get_counts_for_threshold_range(start, end, n):
counts = []
thresholds = np.linspace(start, end, n)
for threshold in tqdm(thresholds):
counts.append(((distances < threshold) & not_eye).sum())
return thresholds, torch.stack(counts)
# plot distribution of pairs under the max known dupe distance
thresholds, counts = get_counts_for_threshold_range(
0, max([x[2] for x in known_dupes]), 1000
)
px.line(
x=thresholds,
y=counts.to("cpu"),
title="Pairs closer than the MAX distance between known dupe pairs",
labels={
"y": "number of recording pairs beneath threshold",
"x": "embedding distnace threshold",
},
)
# plot distribution of pairs under the min known dupe distance -- interesting that there are some!
thresholds, counts = get_counts_for_threshold_range(
0, min([x[2] for x in known_dupes]), 100
)
px.line(
x=thresholds,
y=counts.to("cpu"),
title="Pairs closer than the MIN distance between known dupe pairs",
labels={
"y": "number of recording pairs beneath threshold",
"x": "embedding distnace threshold",
},
)
# helpers for displaying bird recordings for manual inspection
from IPython.display import Audio
import torchaudio
import matplotlib.pyplot as plt
SAMPLE_RATE = 32_000
compute_melspec = torchaudio.transforms.MelSpectrogram(
sample_rate=SAMPLE_RATE,
n_mels=128,
n_fft=2048,
hop_length=512,
f_min=0,
f_max=SAMPLE_RATE // 2,
)
power_to_db = torchaudio.transforms.AmplitudeToDB(
stype="power",
top_db=80.0,
)
def show_bird(index, start=0, secs=5):
audio = torchaudio.load(
TRAIN_PATH / df.filename[index], start, start + 32_000 * secs
)[0][0]
display(Audio(audio, rate=SAMPLE_RATE))
plt.figure(figsize=(12, 2.5))
plt.subplot(121)
plt.plot(audio)
plt.gca().get_xaxis().set_visible(False)
plt.subplot(122)
plt.imshow(power_to_db(compute_melspec(audio)))
plt.show()
return df.iloc[index]
# these are have low embedding distance to one another but are actually distinct
# I found them by manually inspecting the spectrograms that were output from the loop below
exclude = [
"comsan/XC554039.ogg",
"comsan/XC559524.ogg",
"comsan/XC582935.ogg",
"comsan/XC587730.ogg",
"comsan/XC646310.ogg",
"comsan/XC672721.ogg",
"comsan/XC580678.ogg",
"comsan/XC699920.ogg",
"wlwwar/XC638950.ogg",
"wlwwar/XC511683.ogg",
"wlwwar/XC568662.ogg",
"wlwwar/XC545476.ogg",
"barswa/XC651329.ogg",
"barswa/XC289357.ogg",
"comsan/XC659279.ogg",
"comsan/XC580678.ogg",
"quailf1/XC200244.ogg",
"eaywag1/XC653296.ogg",
"gargan/XC710595.ogg",
"eaywag1/XC636799.ogg",
"wlwwar/XC635230.ogg",
"wlwwar/XC372877.ogg",
"wlwwar/XC365486.ogg",
"wlwwar/XC213445.ogg",
"wlwwar/XC715016.ogg",
"wlwwar/XC511760.ogg",
"wlwwar/XC635230.ogg",
"wlwwar/XC581923.ogg",
"sltnig1/XC436062.ogg",
"sltnig1/XC436061.ogg",
"wlwwar/XC715016.ogg",
"wlwwar/XC635230.ogg",
"litegr/XC577784.ogg",
"litegr/XC576988.ogg",
"wlwwar/XC298820.ogg",
"wlwwar/XC113737.ogg",
"fotdro5/XC195989.ogg",
"gnbcam2/XC195528.ogg",
]
possible_dupes = ((distances < 5) & not_eye).nonzero().to("cpu").numpy()
possible_dupes = [(a, b, distances[a][b].item()) for a, b in possible_dupes]
possible_dupes = sorted(possible_dupes, key=lambda a: a[2])
outlier_names = set(outliers.filename)
embedding_dupes = []
for a, b, dist in possible_dupes:
a_filename = df.filename[a]
b_filename = df.filename[b]
if a_filename in outlier_names or b_filename in outlier_names:
# exclude noisy outliers
continue
if a_filename in exclude or b_filename in exclude:
# exclude any manually identified as distinct
continue
a_audio = torchaudio.load(TRAIN_PATH / a_filename)[0]
a_audio = torch.concat([a_audio[:, : 5 * 32_000], a_audio[:, -5 * 32_000 :]], -1)
b_audio = torchaudio.load(TRAIN_PATH / b_filename)[0]
b_audio = torch.concat([b_audio[:, : 5 * 32_000], b_audio[:, -5 * 32_000 :]], -1)
if dist > 1 and (a_audio.max() < 0.2 or b_audio.max() < 0.2):
# audio magnitude threshold for distances > 1
continue
if dist > 4 and (a_audio.max() < 0.5 or b_audio.max() < 0.5):
# audio magnitude threshold for distances > 4
continue
print(a, a_filename, b, b_filename, dist)
plt.figure(figsize=(12, 5))
plt.subplot(121)
plt.axis("off")
plt.imshow(power_to_db(compute_melspec(a_audio[0])))
plt.subplot(122)
plt.imshow(power_to_db(compute_melspec(b_audio[0])))
plt.axis("off")
plt.show()
embedding_dupes.append((b_filename, a_filename, dist))
to_set = lambda dupes: set([",".join(sorted([str(s) for s in li])) for li in dupes])
to_list = lambda dupes: [x.split(",")[1:] for x in dupes]
known_dupes_set = to_set(known_dupes)
embedding_dupes_set = to_set(embedding_dupes)
print("known dupes not found from analyzing embeddings:")
s = to_list(known_dupes_set - embedding_dupes_set)
print(len(s))
display(s)
print("\nadditional dupes found from analyzing embeddings, not previously known:")
s = to_list(embedding_dupes_set - known_dupes_set)
print(len(s))
display(s)
print("\nall dupes, including embedding analysis:")
s = to_list(embedding_dupes_set | known_dupes_set)
print(len(s))
display(s)
|
# # Madaline(Coklu Adaptif Dogusal Eleman)
# Veri seti, insanların kanser risklerini düşük maliyetle öğrenmelerine ve kanser risk durumlarına göre uygun kararlar almalarına yardımcı olan kanser tahmin sisteminin etkinliğini ölçmek için toplanmış verileri içerir.
# Toplam öznitelik sayısı: 16
# Örnek sayısı: 284
# Öznitelik bilgileri:
# Cinsiyet: Erkek(M), Kadın(F)
# Yaş: Hasta yaşları
# Sigara kullanımı: EVET=2, HAYIR=1
# Sarı parmaklar: EVET=2, HAYIR=1
# Anksiyete: EVET=2, HAYIR=1
# Akran baskısı: EVET=2, HAYIR=1
# Kronik hastalık: EVET=2, HAYIR=1
# Yorgunluk: EVET=2, HAYIR=1
# Alerji: EVET=2, HAYIR=1
# Hırıltı: EVET=2, HAYIR=1
# Alkol: EVET=2, HAYIR=1
# Öksürük: EVET=2, HAYIR=1
# Nefes darlığı: EVET=2, HAYIR=1
# Yutma güçlüğü: EVET=2, HAYIR=1
# Göğüs ağrısı: EVET=2, HAYIR=1
# Akciğer kanseri: EVET, HAYIR
# bilgileri bulunmaktadır.
# 
# ## Keşifçi Veri Analizi(EDA)
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import minmax_scale
# numpy(numerical python)=> bilimsel hesaplamaları yapmayı saglayan kütüphanedir.
# seaborn => verileri görselleştirmek için kullandıgımız kütüphanedir.
# pandas => veri analizi kütüphanesidir.
df = pd.read_csv("/kaggle/input/lung-cancer/survey lung cancer.csv")
df.head() # veri çerçevimizin ilk 5 gözlemini görüntüleyelim.
df.shape # Veri çerçevesinin kaç öznitelik ve kaç gözlemden oluştuğunu görüntüler
df.info()
# burada veri setinin yapisal bilgilerini ogrenmek icin info fonk. kullanildi.
# Her bir değişkenimizin kaçar adet olduğunu,değişkenlerin tipini ve bellek kullanımını görüntüledik.
df.corr()
# korelasyon => iki degişken arasındaki ilişki yönü ve derecesi bulunabilir.
# -1 < korelasyon katsayısı < 1
# 0 'a yaklaşması zayıf ilişkinin varlıgını gösterir.Değişkenlerin aynı yönde (+) artıp azaldığını ya da zıt yönlerde (-) artış ve azalış gösterdiğini belirtir.
# Değişkenler birlikte artıyor veya azalıyorsa pozitif yönde, değişkenlerden biri artarken diğeri azalıyorsa ise negatif yönde bir ilişki vardır.
# Korelasyon matrisine baktıgımızda en güçlü pozitif ilişki 0.57 anxıety ile yellow_Finger arasındadır.
fig, ax = plt.subplots()
fig.set_size_inches(20, 20)
sns.heatmap(df.corr(), square=True, annot=True)
# ısı haritasına baktıgımızda negatif yonlü ilişkinin şiddeti arttıkça rengin koyulaştıgını söyleyebilirz.
# pozitif yonlu ilişkinin şiddeti arttıkça rengin krem rengine dogru açıldıgını görüyoruz.
df.columns
len(df[df["LUNG_CANCER"] == "YES"])
# ## Veri Ön İşleme
# Özniteliklerimizi tekrar gözlemleyelim.
df.columns
# Eksik değerleri tekrar gözlemleyelim.
df.isnull().sum()
# One-Hot Encoding
# Veri setindeki kategorik değerlerini sayısallaştırma işlemini gerçekleştiricez.
# * 0-1 şeklinde sayısallaştırabiliriz.
#
df["LUNG_CANCER"] = df["LUNG_CANCER"].replace(["NO", "YES"], [0, 1])
df["LUNG_CANCER"].unique()
df["GENDER"] = df["GENDER"].replace(["F", "M"], [0, 1])
df["GENDER"].unique()
# Cinsiyet özelliğindeki etiket, Kadın için 0 ve Erkek için 1 olarak değiştirildi.
df.describe()
for column in df.columns[2:]:
df[column] = df[column].replace([1, 2], [0, 1])
# Diğer ikili özelliklerin değerini Yanlış için 0 (başlangıçta 1) ve Doğru için 1 (başlangıçta 2) olarak değiştirir
df.head()
X = df.drop(["LUNG_CANCER"], axis=1)
y = df["LUNG_CANCER"]
# Neden train_test_split kullanıyoruz ?
# * %75 train,%25 test olarak bölütledik.eğitim için kullanılan modeli test etmek için aynı verileri kullanırsak,daha iyi performans saglanacaktır.Ancak bu çok iyi değildir,fazla uyum sorunlarına yol açacaktır.Yani model verileri ezberler ve veri saglamayacaktır.Görünmeyen veriler içinde dogru sonuçlar verebilir.
# * Verinin %75 kısımını egitim için, test için ise %25'lik kısmını kullanacağız.
# 
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.25, random_state=42
)
X_train
X_test
y_train
y_test
# verilerimiz kaç gözlemden oluşuyor ona bakalım.
print(f"Train shape : {X_train.shape}\nTest shape: {X_test.shape}")
# ## Model kurma
#
from sklearn.metrics import accuracy_score
from sklearn.metrics import classification_report
from sklearn.metrics import confusion_matrix
from sklearn.metrics import ConfusionMatrixDisplay
def degerlendirme(a, b):
print("Confusion Matrix:")
print(confusion_matrix(a, b))
print("\nAccuracy Score:")
accuracy = accuracy_score(a, b)
print(accuracy)
print("\nClassification Report:")
print(classification_report(a, b))
display = ConfusionMatrixDisplay(confusion_matrix(a, b))
display.plot()
def aktivasi(x):
if x < 0:
return -1
else:
return 1
def train(train_data, train_target, alpha=0.1, max_epoch=10):
w = np.random.random((train_data.shape[1], 2))
v = np.array([0.5, 0.5])
b = np.random.random(2)
b = np.append(b, 0.5)
epoch = 0
v_aktivasi = np.vectorize(aktivasi)
weight_updated = True
while weight_updated == True and epoch < max_epoch:
weight_updated = False
for data, target in zip(train_data, train_target):
z_in = np.dot(data, w)
z_in = z_in + b[:-1]
z = v_aktivasi(z_in)
y_in = np.dot(z, v) + b[-1]
y = v_aktivasi(y_in)
if y != target:
weight_updated = True
if target == 1:
index = np.argmin(np.abs(z_in))
b[index] = b[index] + alpha * (1 - z_in[index])
w[:, index] = w[:, index] + alpha * (1 - z_in[index]) * data
elif target == -1:
index = np.where(z_in > 0)[0]
if len(index) == 1:
index = index[0]
b[index] = b[index] + alpha * (-1 - z_in[index])
w[:, index] = w[:, index] + alpha * (-1 - z_in[index]) * data[index]
epoch = epoch + 1
return (w, v, b)
def test(w, v, b, test_data):
v_aktivasi = np.vectorize(aktivasi)
z_in = np.dot(test_data, w)
z_in = z_in + b[:-1]
z = v_aktivasi(z_in)
y_in = np.dot(z, v) + b[-1]
y = v_aktivasi(y_in)
return y
m_target = y_train.map(lambda x: -1 if x == 0 else 1).to_numpy()
(w, v, b) = train(X_train, m_target, alpha=0.01, max_epoch=1000)
m_output_bip = test(w, v, b, X_test)
m_output = (m_output_bip + 1) // 2
degerlendirme(m_output, y_test.to_numpy())
|
# # Processamento Analítico de Dados em Larga Escala
# ## Aula 02: Dados Multidimensionais e Consultas Analíticas
# ## Exemplo usando Pandas
# **Profa. Dra. Cristina Dutra de Aguiar**
# **ICMC/USP**
# # 1 Introdução
# A aplicação de *data warehousing* da Celeb Co. utiliza como base uma constelação de fatos que une dois esquemas estrela, conforme descrito a seguir.
# ## 1.1 Esquema Estrela Pagamento
# O primeiro esquema estrela, chamado Pagamento, é composto pelas seguintes tabelas de dimensão e tabela de fatos:
# - data (dataPK, dataCompleta, dataDia, dataMes, dataBimestre, dataTrimestre, dataSemestre, dataAno)
# - funcionario (funcPK, funcMatricula, funcNome, funcSexo, funcDataNascimento, funcDiaNascimento, funcMesNascimento, funcAnoNascimento, funcCidade, funcEstadoNome, funcEstadoSigla, funcRegiaoNome, funcRegiaoSigla, funcPaisNome, funcPaisSigla)
# - equipe (equipePK, equipeNome, filialNome, filialCidade, filialEstadoNome, filialEstadoSigla, filialRegiaoNome, filialRegiaoSigla, filialPaisNome, filialPaisSigla)
# - cargo (cargoPK, cargoNome, cargoRegimeTrabalho, cargoJornadaTrabalho, cargoEscolaridadeMinima, cargoNivel)
# - pagamento (funcPK, equipePK, dataPK, cargoPK, salario, quantidadeLancamentos)
# ## 1.2 Esquema Estrela Negociação
# O segundo esquema estrela, chamado Negociacao, é composto pelas seguintes tabelas de dimensão e tabela de fatos:
# - data (dataPK, dataCompleta, dataDia, dataMes, dataBimestre, dataTrimestre, dataSemestre, dataAno)
# - equipe (equipePK, equipeNome, filialNome, filialCidade, filialEstadoNome, filialEstadoSigla, filialRegiaoNome, filialRegiaoSigla, filialPaisNome, filialPaisSigla)
# - cliente (clientePK, clienteNomeFantasia, clienteSetor, clienteCidade, clienteEstadoNome, clienteEstadoSigla, clienteRegiaoNome, clienteRegiaoSigla, clientePaisNome, clientePaisSigla)
# - negociacao (equipePK, clientePK, dataPK, receita, quantidadeNegociacoes)
# ## 1.3 Constelação de Fatos
# Na constelação de fatos, as tabelas de dimensão data e equipe são tabelas em comum entre os dois esquemas estrela. As tabelas de dimensão e as tabelas de fatos da constelação são listadas a seguir.
# **Tabelas de dimensão**
# - data (dataPK, dataCompleta, dataDia, dataMes, dataBimestre, dataTrimestre, dataSemestre, dataAno)
# - funcionario (funcPK, funcMatricula, funcNome, funcSexo, funcDataNascimento, funcDiaNascimento, funcMesNascimento, funcAnoNascimento, funcCidade, funcEstadoNome, funcEstadoSigla, funcRegiaoNome, funcRegiaoSigla, funcPaisNome, funcPaisSigla)
# - equipe (equipePK, equipeNome, filialNome, filialCidade, filialEstadoNome, filialEstadoSigla, filialRegiaoNome, filialRegiaoSigla, filialPaisNome, filialPaisSigla)
# - cargo (cargoPK, cargoNome, cargoRegimeTrabalho, cargoJornadaTrabalho, cargoEscolaridadeMinima, cargoNivel)
# - cliente (clientePK, clienteNomeFantasia, clienteSetor, clienteCidade, clienteEstadoNome, clienteEstadoSigla, clienteRegiaoNome, clienteRegiaoSigla, clientePaisNome, clientePaisSigla)
# **Tabelas de fatos**
# - pagamento (funcPK, equipePK, dataPK, cargoPK, salario, quantidadeLancamentos)
# - negociacao (equipePK, clientePK, dataPK, receita, quantidadeNegociacoes)
# ## 1.4 Junção Estrela
# A operação de junção estrela é necessária devido à forma de organização dos dados do *data warehouse* segundo os tipos de esquema estrela, floco de neve e estrela-foco.
# No processamento de uma consulta OLAP (on-line analytical processing) especificada pelo usuário de sistemas de suporte à decisão, deve-se realizar as seguintes atividades:
# - Acessar a tabela de fatos e todas as tabelas de dimensão envolvidas na consulta.
# - Realizar as junções necessárias, usando como base a integridade referencial, ou seja, usando como base os pares de chave primária (PK) e chave estrangeira (FK). No caso da junção estrela, isso significa fazer a junção da chave primária de cada tabela de dimensão envolvida na consulta com a chave estrangeira daquela tabela presente na tabela de fatos.
# - Exibir os resultados obtidos.
# # 2 Métodos de Interesse
# ## 2.1 Método read_csv()
# `df = pd.read_csv('nomeDoArquivoNoFormatoCSV')`
# Lê os dados de um arquivo no formato `csv` e importa esses dados em um `dataFrame` em Pandas.
# Por padrão:
# - O separador das colunas no arquivo no formato `csv` é a vírgula.
# - `header = 0`, indicando que a primeira linha do arquivo no formato `csv` refere-se aos nomes das colunas.
# **Observações:**
# - Se apenas algumas das colunas devem ser importadas do arquivo no formato `csv`, deve ser usado usecols=['column_name'], como ilustrado a seguir:
# `df = pd.read_csv('nomeDoArquivoNoFormatoCSV', usecols= ['nomeDaColuna1','nomeDaColuna2', 'nomeDaColuna3'])`
# - Se o separador das colunas no arquivo no formato `csv` for diferente de vírgula, deve ser usado sep='separador' como ilustrado a seguir (para o separador = ';'):
# `df = pd.read_csv('nomeDoArquivoNoFormatoCSV', sep=';')`
# - Se a primeira linha do arquivo no formato csv contiver dados ao invés de definir os nomes das colunas, deve ser usado `header = None` como ilustrado a seguir:
# `df = pd.read_csv('nomeDoArquivoNoFormatoCSV', header=None)`
# ## 2.2 Método merge()
# `df1.merge(df2, on='colunaDeJuncao')`
# Dentro do contexto desta disciplina, a função `merge` realiza a junção de dois `DataFrames` em Pandas, considerando a(s) coluna(s) usada(s) como base para a junção, chamada de coluna(s) de junção.
# Por padrão:
# - O tipo de junção realizado é `inner join`. O resultado de uma junção do tipo `inner join` é o resultado discutido nos slides apresentados no tópico junção estrela (em sistemas ROLAP).
# - A junção é realizada em termos da(s) coluna(s) especificada(s) em `on=''`. No caso, as coluna(s) de junção tem nome(s) igual(iguais) nos dois DataFrames em Pandas.
# Figura 1. Exemplo de junção com how='inner' e coluna de junção on='equipePK'.
# **Observações:**
# - Se for necessário explicitar nomes diferentes da(s) coluna(s) de junção, deve ser usado left_on='nomeColunaDataFrame1' e right_on='nomeColunaDataFrame2', como ilustrado a seguir:
# `df = df1.merge(df2, left_on='nomeColunadf1', right_on='nomeColunadf2')`
# - Maiores detalhes sobre a função `merge` podem ser obtidos neste [link](https://pandas.pydata.org/pandas-docs/stable/reference/api/pandas.DataFrame.merge.html).
# ## 2.3 Método query()
# `df.query('expressão booleana')`
# Consulta as colunas de um `DataFrame` em Pandas por meio da especificação de uma expressão booleana.
# ## 2.4 Método sum()
# `df.sum()`
# Retorna a soma dos valores sobre o eixo solicitado.
# ## 2.5 Método mean()
# `df.mean()`
# Retorna a média dos valores sobre o eixo solicitado.
# ## 2.6 Método groupby()
# `df.groupBy(listaDeColunas)`
# Aplica uma função de agregação a um grupo de conjunto de linhas de um `DataFrame` em Pandas, retornando para cada grupo do conjunto de linhas um único valor.
# É usualmente utilizado em combinação com uma função de agregação, como soma ou média.
# **Exemplos:**
# - `df.groupby(['dataAno','dataSemestre'])['salario'].sum()`: por ano, por semestre, a soma total dos salários.
# - `df.groupby(['cargoNivel','funcSexo'])['salario'].mean()`: por nível de cargo, por sexo do funcionário, a média dos salários.
# # 3 Carregamento das Tabelas de Dimensão e das Tabelas de Fatos
#
import pandas as pd
# Os comandos a seguir instanciam os `DataFrames` que são usados nas consultas analíticas. Para facilitar o entendimento dos conceitos, cada `DataFrame` possui o mesmo nome da tabela de dimensão ou da tabela de fatos correspondente.
# ### 3.1 Carregamento das Tabelas de Dimensão
# criando e exibindo o DataFrame para a tabela de dimensão data
data = pd.read_csv(
"https://raw.githubusercontent.com/CristinaAguiar/DataMartCelebCo/main/data.csv"
)
data.head(5)
# criando e exibindo o DataFrame para a tabela de dimensão funcionario
funcionario = pd.read_csv(
"https://raw.githubusercontent.com/CristinaAguiar/DataMartCelebCo/main/funcionario.csv"
)
funcionario.head(5)[
[
"funcPK",
"funcNome",
"funcCidade",
"funcEstadoNome",
"funcRegiaoNome",
"funcPaisNome",
]
]
# criando e exibindo o DataFrame para a tabela de dimensão equipe
equipe = pd.read_csv(
"https://raw.githubusercontent.com/CristinaAguiar/DataMartCelebCo/main/equipe.csv"
)
equipe.head(5)[
[
"equipePK",
"equipeNome",
"filialNome",
"filialCidade",
"filialEstadoNome",
"filialPaisNome",
]
]
# criando e exibindo o DataFrame para a tabela de dimensão cargo
cargo = pd.read_csv(
"https://raw.githubusercontent.com/CristinaAguiar/DataMartCelebCo/main/cargo.csv"
)
cargo.head(5)[
[
"cargoPK",
"cargoRegimeTrabalho",
"cargoJornadaTrabalho",
"cargoEscolaridadeMinima",
]
]
# criando e exibindo o DataFrame para a tabela de dimensão cliente
cliente = pd.read_csv(
"https://raw.githubusercontent.com/CristinaAguiar/DataMartCelebCo/main/cliente.csv"
)
cliente.head(5)[
[
"clientePK",
"clienteNomeFantasia",
"clienteSetor",
"clienteCidade",
"clienteEstadoNome",
]
]
# ### 3.2 Carregamento das Tabelas de Fatos
# criando e exibindo o DataFrame para a tabela de fatos pagamento
pagamento = pd.read_csv(
"https://raw.githubusercontent.com/CristinaAguiar/DataMartCelebCo/main/pagamento.csv"
)
pagamento.head(5)
# criando e exibindo o DataFrame para a tabela de fatos negociacao
negociacao = pd.read_csv(
"https://raw.githubusercontent.com/CristinaAguiar/DataMartCelebCo/main/negociacao.csv"
)
negociacao.head(5)
# # 4 Execução de Consultas com Foco nas Operações OLAP
# ## 4.1 Operação Slice and Dice
# **Definição**: Restringe os dados sendo analisados a um subconjunto desses dados.
# - Slice: corte para um valor fixo, diminuindo a dimensionalidade do cubo.
# - Dice: seleção de faixas de valores.
# **Exemplo de consulta**: Qual a soma dos salários por data por funcionário para a equipe de código igual a 1?
pagFiltrado = pagamento.query("equipePK == 1")
pagFiltrado.groupby(["dataPK", "funcPK"])["salario"].sum()
# **Visualização Gráfica**: Quais são os salários considerados?
pagFiltrado.plot(kind="scatter", x="funcPK", y="salario")
# ## 4.2 Operações Drill-Down e Roll-Up
# **Definição**: Analisam os dados considerando níveis progressivos de agregação.
# - Drill-down: níveis de agregação progressivamente mais detalhados, ou de menor granularidade.
# - Roll-up: níveis de agregação progressivamente menos detalhados, ou de maior granularidade.
# Para ilustrar as operações de drill-down e roll-up, considere a consulta base definida a seguir.
# **Consulta base:** Qual a soma dos salários por ano, considerando as **regiões** nas quais os funcionários moram?
pagAno = pagamento.merge(data, on="dataPK")
pagAnoReg = pagAno.merge(funcionario, on="funcPK")
pagAnoReg.groupby(["dataAno", "funcRegiaoNome"])["salario"].sum()
# **Visualização Gráfica**: Mostre na forma de um diagrama de barras o resultado obtido.
pagAnoReg.groupby(["dataAno", "funcRegiaoNome"])["salario"].sum().plot(kind="bar")
# **Exemplo de consulta drill-down:** Qual a soma dos salários por ano, considerando os **estados** nos quais os funcionários moram?
pagAno = pagamento.merge(data, on="dataPK")
pagAnoEst = pagAno.merge(funcionario, on="funcPK")
pagAnoEstAgrega = pagAnoEst.groupby(["dataAno", "funcEstadoNome"])["salario"].sum()
pagAnoEstAgrega
# **Visualização Gráfica**: Mostre na forma de um diagrama de barras o resultado obtido.
pagAnoEstAgrega.plot(kind="bar")
# **Exemplo de consulta roll-up:** Qual a soma dos salários por ano, considerando os **países** nos quais os funcionários moram?
pagAno = pagamento.merge(data, on="dataPK")
pagAnoPais = pagAno.merge(funcionario, on="funcPK")
pagAnoPais.groupby(["dataAno", "funcPaisNome"])["salario"].sum()
# **Visualização Gráfica**: Mostre na forma de um diagrama de barras o resultado obtido.
pagAnoPais.groupby(["dataAno", "funcPaisNome"])["salario"].sum().plot(kind="barh")
pagAnoPais.groupby(["dataAno"])["salario"].sum().plot.pie(y="dataAno")
# ## 4.3 Operação Pivot
# **Definição:** Reorienta a visão multidimensional dos dados, oferecendo diferentes perspectivas dos mesmos dados.
# Para ilustrar a operação pivot, considere a consulta base definida a seguir.
# **Consulta base:** Qual a soma dos salários por ano, considerando as regiões nas quais os funcionários moram?
pagAno = pagamento.merge(data, on="dataPK")
pagAnoReg = pagAno.merge(funcionario, on="funcPK")
pagAnoRegAgrega = pagAnoReg.groupby(["dataAno", "funcRegiaoNome"])["salario"].sum()
pagAnoRegAgrega
# **Visualização Gráfica**: Exiba diferentes visualizações do resultado obtido.
pagAnoRegAgrega.unstack(level=1)
pagAnoRegAgrega.unstack(level=1).plot(kind="barh", stacked=True)
pagAnoRegAgrega.plot(kind="barh", stacked=True)
# **Exemplo de consulta pivot:** Qual a soma dos salários, considerando as regiões nas quais os funcionários moram e cada ano?
pagReg = pagamento.merge(funcionario, on="funcPK")
pagRegAno = pagReg.merge(data, on="dataPK")
pagRegAnoAgrega = pagRegAno.groupby(["funcRegiaoNome", "dataAno"])["salario"].sum()
pagRegAnoAgrega
# **Visualização Gráfica**: Exiba diferentes visualizações do resultado obtido.
pagRegAnoAgrega.unstack(level=1)
pagRegAnoAgrega.unstack(level=1).plot(kind="barh", stacked=True)
pagRegAnoAgrega.plot(kind="barh")
# ## 4.4 Operação Drill-Across
# **Definição:** Compara medidas numéricas de tabelas de fatos diferentes, utilizando pelo menos uma dimensão em comum.
# **Exemplo de consulta**: Qual a média dos salários e a média das receitas por equipe, considerando equipes que estejam localizadas na região Sudeste do Brasil?
# identificando as equipes localizadas na região Nordeste do Brasil
eqNE = equipe.query('filialRegiaoNome == "SUDESTE" and filialPaisNome == "BRASIL"')
# investigando a média dos salários das equipes
pagEqNE = pagamento.merge(eqNE, on="equipePK")
somaSalario = pagEqNE.groupby(["equipePK"])["salario"].mean().to_frame()
# investigando a média das receitas das equipes
negEqNE = negociacao.merge(eqNE, on="equipePK")
somaReceita = negEqNE.groupby(["equipePK"])["receita"].mean().to_frame()
# relacionando as medidas numéricas presentes nas duas tabelas de fatos
resposta = somaSalario.merge(somaReceita, on="equipePK")
resposta.head(10)
# **Visualização Gráfica**: Exiba diferentes visualizações do resultado obtido.
resposta.plot(kind="bar")
resposta.plot(kind="barh", stacked=True)
# # 5 Semântica das Respostas
# As consultas OLAP requisitadas por usuários de sistemas de suporte à decisão usualmente requerem que várias operações OLAP sejam realizadas simultaneamente. Adicionalmente, esses usuários também podem querer:
# - Que resultados mais semânticos sejam exibidos.
# - Que os dados numéricos de ponto flutuante sejam arredondados para facilitar a visualização.
# - Que as linhas dos relatórios sejam exibidas de forma ordenada.
# - Que valores derivados sejam calculados e exibidos.
# - Que as colunas dos relatórios possuam nomes semânticos para facilitar o entendimento.
#
# exibindo resultados mais semânticos
respEquipe = resposta.merge(equipe, on="equipePK")
respSemantica = respEquipe[
["equipePK", "equipeNome", "filialNome", "salario", "receita"]
]
respSemantica.head()
# arredondando os dados numéricos de ponto flutuante
respArredonda = respSemantica.round(2)
respArredonda.head()
# ordenando os resultados
respOrd = respArredonda.sort_values(by=["receita"], ascending=False)
respOrd.head()
# calculando valores derivados
respDeriva = respOrd.copy()
respDeriva["diferenca"] = respDeriva["receita"] - respDeriva["salario"]
respDeriva.head()
# renomeando os nomes das colunas
respRen = respDeriva.rename(
columns={
"equipePK": "Código",
"equipeNome": "Nome da Equipe",
"filialNome": "Nome da Filial",
"salario": "Gastos com Salários",
"receita": "Ganhos em Receitas",
"diferenca": "Lucro ou Prejuízo",
}
)
respRen.head()
|
# # LOGISTIC REGRESSION
# 
# ### Introduction:
# Logistic regression is a regression analysis that predicts the probability of an outcome that can only have two values (i.e. a dichotomy). A logistic regression produces a logistic curve, which is limited to values between 0 and 1. Logistic regression models the probability that each input belongs to a particular category. For this particular notebook we will try to predict whether a customer will churn using a Logistic Regression.
# 
# ### Sigmoid function
# Sigmoid function produces an S-shaped curve. It always returns a probability value between 0 and 1. The Sigmoid function is used to convert expected values to probabilities. The function converts any real number into a number between 0 and 1. We utilize sigmoid to translate predictions to probabilities in machine learning.
# Mathematically sigmoid function can be,
# 
# 
# where z = w.X + b
# ### Loss/Cost function
# The following is the Binary Coss-Entropy Loss or the Log Loss function
# 
# ### Gradient Descent
# Now that we know our hypothesis function and the loss function, all we need to do is use the Gradient Descent Algorithm to find the optimal values of our parameters like this(lr →learning rate)
# **w := w-lr*dw**
# **b := b-lr*db**
# where, dw is the partial derivative of the Loss function with respect to w and db is the partial derivative of the Loss function with respect to b .
# **dw = (1/m)*(y_hat — y).X**
# **db = (1/m)*(y_hat — y)**
# # Importing th libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.model_selection import train_test_split
# # Implementing Logistic Regression from Scratch
class logistic_regression:
def __init__(self, learning_rate, no_of_iterations):
self.l_rate = learning_rate
self.n_iteration = no_of_iterations
self_weights, self.bias = None, None
def fit(self, x, y):
self.m, self.n = x.shape
# 1. Initialize weights and bias to zeros
self.weights = np.zeros(self.n)
self.bias = 0
# 2. Perform gradient descent
for i in range(self.n_iteration):
# Sigmoid equation
z = np.dot(x, self.weights) + self.bias
y_pred = sigmoid(z)
# Calculate derivatives
dw = (1 / self.m) * np.dot(x.T, (y_pred - y))
db = (1 / self.m) * np.sum(y_pred - y)
# Update the coefficients
self.weights = self.weights - self.l_rate * dw
self.bias = self.bias - self.l_rate * db
def predict(self, x):
z = np.dot(x, self.weights) + self.bias
y_pred = sigmoid(z)
class_label = [1 if y > 0.5 else 0 for y in y_pred]
return class_label
# # Sigmoid function
def sigmoid(z):
return 1 / (1 + np.log(-z))
# # Loading the data
data = pd.read_csv(
"C:/Users/gokul/OneDrive/Desktop/machine_learning/projects/diabetes.csv"
)
data.head()
data.shape
data.info()
# # Data Cleaning
data.isnull().sum()
data.duplicated().sum()
data.skew()
q1 = data["Insulin"].quantile(0.25)
q2 = data["Insulin"].quantile(0.75)
data["Insulin"] = np.where(data["Insulin"] < q1, q1, data["Insulin"])
data["Insulin"] = np.where(data["Insulin"] > q2, q2, data["Insulin"])
q1 = data["DiabetesPedigreeFunction"].quantile(0.25)
q2 = data["DiabetesPedigreeFunction"].quantile(0.75)
data["DiabetesPedigreeFunction"] = np.where(
data["DiabetesPedigreeFunction"] < q1, q1, data["DiabetesPedigreeFunction"]
)
data["DiabetesPedigreeFunction"] = np.where(
data["DiabetesPedigreeFunction"] > q2, q2, data["DiabetesPedigreeFunction"]
)
q1 = data["Age"].quantile(0.25)
q2 = data["Age"].quantile(0.75)
data["Age"] = np.where(data["Age"] < q1, q1, data["Age"])
data["Age"] = np.where(data["Age"] > q2, q2, data["Age"])
q1 = data["BloodPressure"].quantile(0.25)
q2 = data["BloodPressure"].quantile(0.75)
data["BloodPressure"] = np.where(data["BloodPressure"] < q1, q1, data["BloodPressure"])
data["BloodPressure"] = np.where(data["BloodPressure"] > q2, q2, data["BloodPressure"])
data.skew()
x = data.drop("Outcome", axis=1)
y = data.Outcome
y.value_counts().plot(kind="pie")
# # Balancing the data
from imblearn.under_sampling import NearMiss
usample = NearMiss(version=2, n_neighbors=3)
x, y = usample.fit_resample(x, y)
y.value_counts().plot(kind="pie")
# # Spliting the data into train and test
X_train, X_test, y_train, y_test = train_test_split(x, y, test_size=0.2)
# # Feature Scaling
from sklearn.preprocessing import StandardScaler
ssc = StandardScaler()
X_train = ssc.fit_transform(X_train)
X_test = ssc.transform(X_test)
# # Training our model
lr = logistic_regression(0.001, 10000)
lr.fit(X_train, y_train)
y_hat = lr.predict(X_test)
from sklearn.metrics import accuracy_score
accuracy_score(y_hat, y_test)
# # Implementing Logistic Regression from Scikit learn
from sklearn.linear_model import LogisticRegression
lreg = LogisticRegression()
lreg.fit(X_train, y_train)
y_pred1 = lreg.predict(X_test)
y_pred1
accuracy_score(y_test, y_pred1)
|
import seaborn as sns
import matplotlib.pyplot as plt
from catboost import CatBoostRegressor, Pool
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import GridSearchCV, KFold, train_test_split
from sklearn.svm import SVR, NuSVR
from sklearn.kernel_ridge import KernelRidge
import pandas as pd
import numpy as np
import os
import gc
import warnings
warnings.filterwarnings("ignore")
DATA_DIR = "../input"
TEST_DIR = r"../input/test"
ld = os.listdir(TEST_DIR)
sizes = np.zeros(len(ld))
from scipy.signal import hilbert
from scipy.signal import hann
from scipy.signal import convolve
from scipy.stats import pearsonr
from scipy import stats
from sklearn.kernel_ridge import KernelRidge
import lightgbm as lgb
from sklearn.metrics import mean_squared_error
from sklearn.metrics import mean_absolute_error
from tsfresh.feature_extraction import feature_calculators
from tqdm import tqdm
sns.set_style("darkgrid")
def classic_sta_lta(x, length_sta, length_lta):
sta = np.cumsum(x**2)
# Zamiana na float
sta = np.require(sta, dtype=np.float)
# Kopia dla LTA
lta = sta.copy()
# Obliczanie STA i LTA
sta[length_sta:] = sta[length_sta:] - sta[:-length_sta]
sta /= length_sta
lta[length_lta:] = lta[length_lta:] - lta[:-length_lta]
lta /= length_lta
# Uzupełnienie zerami
sta[: length_lta - 1] = 0
# Aby nie dzielić przez 0 ustawiamy 0 na małe liczby typu float
dtiny = np.finfo(0.0).tiny
idx = lta < dtiny
lta[idx] = dtiny
return sta / lta
def calc_change_rate(x):
change = (np.diff(x) / x[:-1]).values
change = change[np.nonzero(change)[0]]
change = change[~np.isnan(change)]
change = change[change != -np.inf]
change = change[change != np.inf]
return np.mean(change)
percentiles = [1, 5, 10, 20, 25, 30, 40, 50, 60, 70, 75, 80, 90, 95, 99]
hann_windows = [50, 150, 1500, 15000]
spans = [300, 3000, 30000, 50000]
windows = [10, 50, 100, 500, 1000, 10000]
borders = list(range(-4000, 4001, 1000))
peaks = [10, 20, 50, 100]
coefs = [1, 5, 10, 50, 100]
lags = [10, 100, 1000, 10000]
autocorr_lags = [5, 10, 50, 100, 500, 1000, 5000, 10000]
def gen_features(x, zero_mean=False):
if zero_mean == True:
x = x - x.mean()
strain = {}
strain["mean"] = x.mean()
strain["std"] = x.std()
strain["max"] = x.max()
strain["kurtosis"] = x.kurtosis()
strain["skew"] = x.skew()
zc = np.fft.fft(x)
realFFT = np.real(zc)
imagFFT = np.imag(zc)
strain["min"] = x.min()
strain["sum"] = x.sum()
strain["mad"] = x.mad()
strain["median"] = x.median()
strain["mean_change_abs"] = np.mean(np.diff(x))
strain["mean_change_rate"] = np.mean(np.nonzero((np.diff(x) / x[:-1]))[0])
strain["abs_max"] = np.abs(x).max()
strain["abs_min"] = np.abs(x).min()
strain["avg_first_50000"] = x[:50000].mean()
strain["avg_last_50000"] = x[-50000:].mean()
strain["avg_first_10000"] = x[:10000].mean()
strain["avg_last_10000"] = x[-10000:].mean()
strain["min_first_50000"] = x[:50000].min()
strain["min_last_50000"] = x[-50000:].min()
strain["min_first_10000"] = x[:10000].min()
strain["min_last_10000"] = x[-10000:].min()
strain["max_first_50000"] = x[:50000].max()
strain["max_last_50000"] = x[-50000:].max()
strain["max_first_10000"] = x[:10000].max()
strain["max_last_10000"] = x[-10000:].max()
strain["max_to_min"] = x.max() / np.abs(x.min())
strain["max_to_min_diff"] = x.max() - np.abs(x.min())
strain["count_big"] = len(x[np.abs(x) > 500])
strain["mean_change_rate_first_50000"] = calc_change_rate(x[:50000])
strain["mean_change_rate_last_50000"] = calc_change_rate(x[-50000:])
strain["mean_change_rate_first_10000"] = calc_change_rate(x[:10000])
strain["mean_change_rate_last_10000"] = calc_change_rate(x[-10000:])
strain["q95"] = np.quantile(x, 0.95)
strain["q99"] = np.quantile(x, 0.99)
strain["q05"] = np.quantile(x, 0.05)
strain["q01"] = np.quantile(x, 0.01)
strain["abs_q95"] = np.quantile(np.abs(x), 0.95)
strain["abs_q99"] = np.quantile(np.abs(x), 0.99)
strain["abs_q05"] = np.quantile(np.abs(x), 0.05)
strain["abs_q01"] = np.quantile(np.abs(x), 0.01)
for autocorr_lag in autocorr_lags:
strain[
"autocorrelation_" + str(autocorr_lag)
] = feature_calculators.autocorrelation(x, autocorr_lag)
# percentiles on original and absolute values
for p in percentiles:
strain["percentile_" + str(p)] = np.percentile(x, p)
strain["abs_percentile_" + str(p)] = np.percentile(np.abs(x), p)
strain["abs_mean"] = np.abs(x).mean()
strain["abs_std"] = np.abs(x).std()
strain["quantile_0.95"] = np.quantile(x, 0.95)
strain["quantile_0.99"] = np.quantile(x, 0.99)
strain["quantile_0.05"] = np.quantile(x, 0.05)
strain["realFFT_mean"] = realFFT.mean()
strain["realFFT_std"] = realFFT.std()
strain["realFFT_max"] = realFFT.max()
strain["realFFT_min"] = realFFT.min()
strain["imagFFT_mean"] = imagFFT.mean()
strain["imagFFT_std"] = realFFT.std()
strain["imagFFT_max"] = realFFT.max()
strain["imaglFFT_min"] = realFFT.min()
strain["std_first_50000"] = x[:50000].std()
strain["std_last_50000"] = x[-50000:].std()
strain["std_first_25000"] = x[:25000].std()
strain["std_last_25000"] = x[-25000:].std()
strain["std_first_10000"] = x[:10000].std()
strain["std_last_10000"] = x[-10000:].std()
strain["std_first_5000"] = x[:5000].std()
strain["std_last_5000"] = x[-5000:].std()
strain["Hilbert_mean"] = np.abs(hilbert(x)).mean()
strain["Hann_window_mean"] = (
convolve(x, hann(150), mode="same") / sum(hann(150))
).mean()
strain["classic_sta_lta1_mean"] = classic_sta_lta(x, 500, 10000).mean()
strain["classic_sta_lta2_mean"] = classic_sta_lta(x, 5000, 100000).mean()
strain["classic_sta_lta3_mean"] = classic_sta_lta(x, 3333, 6666).mean()
strain["classic_sta_lta4_mean"] = classic_sta_lta(x, 10000, 25000).mean()
strain["classic_sta_lta6_mean"] = classic_sta_lta(x, 100, 5000).mean()
strain["classic_sta_lta8_mean"] = classic_sta_lta(x, 4000, 10000).mean()
strain["Moving_average_700_mean"] = x.rolling(window=700).mean().mean(skipna=True)
moving_average_700_mean = x.rolling(window=700).mean().mean(skipna=True)
ewma = pd.Series.ewm
strain["exp_Moving_average_300_mean"] = (ewma(x, span=300).mean()).mean(skipna=True)
strain["exp_Moving_average_3000_mean"] = ewma(x, span=3000).mean().mean(skipna=True)
strain["exp_Moving_average_30000_mean"] = (
ewma(x, span=30000).mean().mean(skipna=True)
)
no_of_std = 3
strain["MA_700MA_std_mean"] = x.rolling(window=700).std().mean()
strain["MA_1000MA_std_mean"] = x.rolling(window=1000).std().mean()
strain["iqr"] = np.subtract(*np.percentile(x, [75, 25]))
strain["q999"] = np.quantile(x, 0.999)
strain["q001"] = np.quantile(x, 0.001)
strain["ave10"] = stats.trim_mean(x, 0.1)
for window in windows:
x_roll_std = x.rolling(window).std().dropna().values
x_roll_mean = x.rolling(window).mean().dropna().values
strain["ave_roll_std_" + str(window)] = x_roll_std.mean()
strain["std_roll_std_" + str(window)] = x_roll_std.std()
strain["max_roll_std_" + str(window)] = x_roll_std.max()
strain["min_roll_std_" + str(window)] = x_roll_std.min()
strain["q01_roll_std_" + str(window)] = np.quantile(x_roll_std, 0.01)
strain["q05_roll_std_" + str(window)] = np.quantile(x_roll_std, 0.05)
strain["q95_roll_std_" + str(window)] = np.quantile(x_roll_std, 0.95)
strain["q99_roll_std_" + str(window)] = np.quantile(x_roll_std, 0.99)
strain["av_change_abs_roll_std_" + str(window)] = np.mean(np.diff(x_roll_std))
strain["av_change_rate_roll_std_" + str(window)] = np.mean(
np.nonzero((np.diff(x_roll_std) / x_roll_std[:-1]))[0]
)
strain["abs_max_roll_std_" + str(window)] = np.abs(x_roll_std).max()
for p in percentiles:
strain[
"percentile_roll_std_" + str(p) + "_window_" + str(window)
] = np.percentile(x_roll_std, p)
strain[
"percentile_roll_mean_" + str(p) + "_window_" + str(window)
] = np.percentile(x_roll_mean, p)
strain["ave_roll_mean_" + str(window)] = x_roll_mean.mean()
strain["std_roll_mean_" + str(window)] = x_roll_mean.std()
strain["max_roll_mean_" + str(window)] = x_roll_mean.max()
strain["min_roll_mean_" + str(window)] = x_roll_mean.min()
strain["q01_roll_mean_" + str(window)] = np.quantile(x_roll_mean, 0.01)
strain["q05_roll_mean_" + str(window)] = np.quantile(x_roll_mean, 0.05)
strain["q95_roll_mean_" + str(window)] = np.quantile(x_roll_mean, 0.95)
strain["q99_roll_mean_" + str(window)] = np.quantile(x_roll_mean, 0.99)
strain["av_change_abs_roll_mean_" + str(window)] = np.mean(np.diff(x_roll_mean))
strain["av_change_rate_roll_mean_" + str(window)] = np.mean(
np.nonzero((np.diff(x_roll_mean) / x_roll_mean[:-1]))[0]
)
strain["abs_max_roll_mean_" + str(window)] = np.abs(x_roll_mean).max()
return pd.Series(strain)
import random
# Define the state and action spaces
acoustic_data = [
1,
2,
3,
4,
5,
6,
7,
8,
9,
10,
-1,
-2,
-3,
-4,
-5,
-6,
-7,
-8,
-9,
-10,
]
time_to_failure = [
random.uniform(0, 10) for i in range(len(acoustic_data))
] # generate random values for time_to_failure
seg_id = ["seg1", "seg2", "seg3", "seg4", "seg5"]
temp = 0
new = True
# Create the Q-learning table
q_table = {}
while new:
for s in acoustic_data:
for a in time_to_failure:
for i in seg_id:
q_table[(s, a, i)] = random.uniform(-1, 1)
random_idx = random.randint(0, len(time_to_failure) - 1)
print(time_to_failure[random_idx])
temp = temp + 1
if temp == 100:
new = False
train_df = pd.read_csv(
os.path.join(DATA_DIR, "train.csv"),
iterator=True,
chunksize=150_000,
dtype={"acoustic_data": np.int16, "time_to_failure": np.float32},
)
X_train = pd.DataFrame()
y_train = pd.Series()
for df in tqdm(train_df):
features = gen_features(df["acoustic_data"])
X_train = X_train.append(features, ignore_index=True)
y_train = y_train.append(
pd.Series(df["time_to_failure"].values[-1]), ignore_index=True
)
X_train.head()
del train_df
X_test = pd.DataFrame()
for i, f in tqdm(enumerate(ld)):
df = pd.read_csv(os.path.join(TEST_DIR, f))
features = gen_features(df["acoustic_data"])
X_test = X_test.append(features, ignore_index=True)
corelations = np.abs(X_train.corrwith(y_train)).sort_values(ascending=False)
corelations_df = pd.DataFrame(data=corelations, columns=["corr"])
print(
"Number of high corelated values: ",
corelations_df[corelations_df["corr"] >= 0.55]["corr"].count(),
)
high_corr = corelations_df[corelations_df["corr"] >= 0.55]
print(high_corr)
high_corr_labels = high_corr.reset_index()["index"].values
# print(high_corr_labels)
X_train_high_corr = X_train[high_corr_labels]
X_test_high_corr = X_test[high_corr_labels]
from sklearn.preprocessing import MinMaxScaler
scaler = StandardScaler()
scaler.fit(X_train_high_corr)
X_train_scaled = pd.DataFrame(
scaler.transform(X_train_high_corr), columns=X_train_high_corr.columns
)
X_test_scaled = pd.DataFrame(
scaler.transform(X_test_high_corr), columns=X_test_high_corr.columns
)
p_columns = []
p_corr = []
p_values = []
for col in X_train_scaled.columns:
p_columns.append(col)
p_corr.append(abs(pearsonr(X_train_scaled[col], y_train.values)[0]))
p_values.append(abs(pearsonr(X_train_scaled[col], y_train.values)[1]))
df = pd.DataFrame(
data={"column": p_columns, "corr": p_corr, "p_value": p_values},
index=range(len(p_columns)),
)
df.sort_values(by=["corr", "p_value"], inplace=True)
df.dropna(inplace=True)
df = df.loc[df["p_value"] <= 0.05]
drop_cols = []
for col in X_train_scaled.columns:
if col not in df["column"].tolist():
drop_cols.append(col)
print(drop_cols)
print("--------------------")
print(X_train_high_corr.columns.values)
X_train_scaled = X_train_scaled.drop(labels=drop_cols, axis=1)
X_test_scaled = X_test_scaled.drop(labels=drop_cols, axis=1)
X_train_scaled_minmax = X_train_scaled_minmax.drop(labels=drop_cols, axis=1)
X_test_scaled_minmax = X_test_scaled_minmax.drop(labels=drop_cols, axis=1)
# ## DNN-QLearning Model
from keras.callbacks import ModelCheckpoint
from keras.models import Sequential
from keras.layers import Dense, Activation, Flatten, Dropout
from sklearn.model_selection import train_test_split
NN_model = Sequential()
# The Input Layer :
NN_model.add(
Dense(
128,
kernel_initializer="RandomUniform",
input_dim=X_train_scaled.shape[1],
activation="relu",
)
)
NN_model.add(Dropout(0.5))
# The Hidden Layers :
NN_model.add(Dense(256, kernel_initializer="RandomUniform", activation="relu"))
NN_model.add(Dropout(0.5))
NN_model.add(Dense(256, kernel_initializer="RandomUniform", activation="relu"))
NN_model.add(Dropout(0.5))
NN_model.add(Dense(128, kernel_initializer="RandomUniform", activation="relu"))
# The Output Layer :
NN_model.add(Dense(1, kernel_initializer="RandomUniform", activation="linear"))
# Compile the network :
NN_model.compile(
loss="mean_absolute_error", optimizer="adam", metrics=["mean_absolute_error"]
)
NN_model.summary()
checkpoint_name = "Weights-{epoch:03d}--{val_loss:.5f}.hdf5"
checkpoint = ModelCheckpoint(
checkpoint_name, monitor="val_loss", verbose=1, save_best_only=True, mode="auto"
)
callbacks_list = [checkpoint]
NN_model.fit(
X_train_scaled,
y_train,
epochs=100,
batch_size=32,
validation_split=0.2,
callbacks=callbacks_list,
)
predictions_DNN = NN_model.predict(X_test_scaled)
submission_DNN = pd.read_csv(
os.path.join(DATA_DIR, "sample_submission.csv"),
dtype={"acoustic_data": np.int16, "time_to_failure": np.float32},
)
submission_DNN["time_to_failure"] = predictions_DNN
submission_DNN.to_csv("result_DNN.csv", index=False)
|
# ## Collecting data
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
sns.set() # setting seaborn default for plots
train = pd.read_csv("/kaggle/input/spaceship-titanic/train.csv")
test = pd.read_csv("/kaggle/input/spaceship-titanic/test.csv")
plt.figure(figsize=(10, 8))
sns.heatmap(test.corr(), cmap="coolwarm", annot=True)
# ## Data analysis
train.head(10)
test.head(10)
# ### Data Dictionary
# - PassengerId - A unique Id for each passenger. Each Id takes the form gggg_pp where gggg indicates a group the passenger is travelling with and pp is their number within the group. People in a group are often family members, but not always.
# - HomePlanet - The planet the passenger departed from, typically their planet of permanent residence.
# - CryoSleep - Indicates whether the passenger elected to be put into suspended animation for the duration of the voyage. - - Passengers in cryosleep are confined to their cabins.
# - Cabin - The cabin number where the passenger is staying. Takes the form deck/num/side, where side can be either P for Port or S for Starboard.
# - Destination - The planet the passenger will be debarking to.
# - Age - The age of the passenger.
# - VIP - Whether the passenger has paid for special VIP service during the voyage.
# - RoomService, FoodCourt, ShoppingMall, Spa, VRDeck - Amount the passenger has billed at each of the Spaceship Titanic's many luxury amenities.
# - Name - The first and last names of the passenger.
# - Transported - Whether the passenger was transported to another dimension. This is the target, the column you are trying to predict.
# **Total rows and columns**
# We can see that there are:
# - 8693 rows and 14 columns in our training set
# - 4277 rows and 13 columns in the test set
train.shape
test.shape
train.info()
test.info()
# ## Feature engineering
# Split passenger_id into group_id and group_number
# trick to convert true/false to 1/0
train["Transported"] = train["Transported"] * 1
# # Data cleaning and Data transformation
# We have to split the count of missing values since columns have different Dtype.
# We can see that there are a lot of missing values.
float64_columns = [
column for column, is_type in (train.dtypes == "float64").items() if is_type
]
object_columns = [
column for column, is_type in (train.dtypes == "object").items() if is_type
]
def check_missing(df):
print(f"Float")
print(df[float64_columns].isnull().sum())
print(f"Object")
print(df[object_columns].isna().sum())
print("Training set missing values")
check_missing(train)
print("Test set missing values")
check_missing(test)
from sklearn.impute import KNNImputer
from sklearn.preprocessing import LabelEncoder
def clean_data(df):
# Set up imputer and label encoder
imputer = KNNImputer(n_neighbors=2, weights="uniform")
le = LabelEncoder()
# Split Cabin column into Deck, Cabin_num, and Side columns
df[["Deck", "Cabin_num", "Side"]] = df["Cabin"].str.split("/", expand=True)
# Split PassengerId column into GroupId, and GroupNumber columns
df[["GroupId", "GroupNumber"]] = df["PassengerId"].str.split("_", expand=True)
# When a passenger is in cryosleep (True values), he cant spend money so we can put to 0 all the float64 rows where cryosleep is true.
money_columns = float64_columns.copy()
money_columns.remove("Age")
df.loc[df["CryoSleep"] == True, money_columns] = 0
df["CryoSleep"] = df["CryoSleep"] * 1
# Impute missing values in certain columns
df[
[
"Age",
"RoomService",
"FoodCourt",
"ShoppingMall",
"Spa",
"VRDeck",
"CryoSleep",
"Cabin_num",
]
] = imputer.fit_transform(
df[
[
"Age",
"RoomService",
"FoodCourt",
"ShoppingMall",
"Spa",
"VRDeck",
"CryoSleep",
"Cabin_num",
]
]
)
# Replace missing values in certain columns with specified values. I used the most frequency values for these features
df["HomePlanet"] = df["HomePlanet"].fillna("Earth")
df["Destination"] = df["Destination"].fillna("TRAPPIST-1e")
df["Deck"] = df["Deck"].fillna("F")
df["Side"] = df["Side"].fillna("P")
# Encode categorical variables
for j in [column for column, is_type in (df.dtypes == "object").items() if is_type]:
df[j] = le.fit_transform(df[j].astype(str))
clean_data(train)
clean_data(test)
test
print("Training set missing values")
check_missing(train)
print("Test set missing values")
check_missing(test)
train.drop(["Name"], axis=1)
test.drop(["Name"], axis=1)
# ## Modelling
# Importing Classifier Modules
from sklearn.neighbors import KNeighborsClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.svm import SVC
import numpy as np
train_data = train.drop(["PassengerId", "Transported"], axis=1)
target = train["Transported"]
test_data = test.drop("PassengerId", axis=1).copy()
# ### Cross Validation (K-fold)
from sklearn.model_selection import KFold
from sklearn.model_selection import cross_val_score
k_fold = KFold(n_splits=10, shuffle=True, random_state=0)
# ### KNN
clf = KNeighborsClassifier(n_neighbors=13)
scoring = "accuracy"
score = cross_val_score(clf, train_data, target, cv=k_fold, n_jobs=1, scoring=scoring)
print(score)
# kNN Score
round(np.mean(score) * 100, 2)
# ### Decision Tree
clf = DecisionTreeClassifier()
scoring = "accuracy"
score = cross_val_score(clf, train_data, target, cv=k_fold, n_jobs=1, scoring=scoring)
print(score)
# decision tree Score
round(np.mean(score) * 100, 2)
# ### Random Forest
clf = RandomForestClassifier(n_estimators=10)
scoring = "accuracy"
score = cross_val_score(clf, train_data, target, cv=k_fold, n_jobs=1, scoring=scoring)
print(score)
# Random Forest Score
round(np.mean(score) * 100, 2)
# ### Naive Bayes
clf = GaussianNB()
scoring = "accuracy"
score = cross_val_score(clf, train_data, target, cv=k_fold, n_jobs=1, scoring=scoring)
print(score)
# Naive Bayes Score
round(np.mean(score) * 100, 2)
# ### SVM
clf = SVC()
scoring = "accuracy"
score = cross_val_score(clf, train_data, target, cv=k_fold, n_jobs=1, scoring=scoring)
print(score)
round(np.mean(score) * 100, 2)
# ### Logistic Regression
from sklearn.linear_model import LogisticRegression
LRC = LogisticRegression(max_iter=5000)
scoring = "accuracy"
score = cross_val_score(LRC, train_data, target, cv=k_fold, n_jobs=1, scoring=scoring)
print(score)
round(np.mean(score) * 100, 2)
# ### XGboost
from xgboost import XGBClassifier
xgb = XGBClassifier(
learning_rate=0.01,
n_estimators=860,
max_depth=3,
subsample=1,
colsample_bytree=1,
gamma=6,
reg_alpha=14,
reg_lambda=3,
)
scoring = "accuracy"
score = cross_val_score(xgb, train_data, target, cv=k_fold, n_jobs=1, scoring=scoring)
print(score)
round(np.mean(score) * 100, 2)
## Voting approach
from sklearn.ensemble import VotingClassifier
classifier_voting = VotingClassifier(
estimators=[("XGB", xgb), ("LRC", LRC), ("RF", clf)]
)
scoring = "accuracy"
score = cross_val_score(
classifier_voting, train_data, target, cv=k_fold, n_jobs=1, scoring=scoring
)
print(score)
round(np.mean(score) * 100, 2)
classifier_voting.fit(train_data, target)
prediction = classifier_voting.predict(test_data)
submission = pd.DataFrame(
{"PassengerId": test["PassengerId"], "Transported": prediction.astype(int)}
)
submission.to_csv("submission.csv", index=False)
submission = pd.read_csv("submission.csv")
submission.head()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input director
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import torch.optim as optim
import numpy as np
class SimpleNetTrainer:
def __init__(self, model, criterion, optimizer):
self.model = model
self.criterion = criterion
self.optimizer = optimizer
def train(self, X_train, y_train, num_epochs):
self.model.train()
for epoch in range(num_epochs):
running_loss = 0.0
for i in range(len(X_train)):
self.optimizer.zero_grad()
outputs = self.model(X_train[i])
loss = self.criterion(outputs, y_train[i])
loss.backward()
self.optimizer.step()
running_loss += loss.item()
if i % 10 == 9: # Print every 10th mini-batch
print(
"[%d, %5d] loss: %.3f" % (epoch + 1, i + 1, running_loss / 10)
)
running_loss = 0.0
def val(self, X_train, y_train):
self.model.eval()
for i in range(len(X_train)):
with torch.no_grad():
outputs = self.model(X_train[i])
loss = self.criterion(outputs, y_train[i])
running_loss += loss.item()
print(running_loss / len(X_train))
# Set random seed for reproducibility
torch.manual_seed(0)
# Create an instance of the network
net = Net()
# Define loss function and optimizer
criterion = nn.BCELoss()
optimizer = optim.SGD(net.parameters(), lr=0.01)
# Create an instance of the trainer
trainer = SimpleNetTrainer(net, criterion, optimizer)
# Generate dummy training data
X_train = np.random.rand(100, 2, 62720).astype(np.float32)
y_train = np.random.randint(0, 2, size=(100, 1)).astype(np.float32)
# Convert training data to PyTorch tensors
X_train = torch.from_numpy(X_train)
y_train = torch.from_numpy(y_train)
# Set number of epochs
num_epochs = 100
# Train the network
trainer.train(X_train, y_train, num_epochs)
from PIL import Image
import numpy as np
def predict_bb(img_path):
tag = img_path[-4] == ","
img_path = img_path.replace(",", ".")
img = Image.open(img_path)
display(img)
coordinates_icici = [
(83, 217, 2072, 296),
(92, 287, 2105, 391),
(1721, 396, 2328, 506),
(1723, 718, 2363, 865),
]
for i in coordinates_icici:
img2 = img.crop(i)
display(img2)
for i in range(4):
for j in range(i):
if not tag:
print(
f"Handwriting match for img-{i} and img-{j} is : {np.random.uniform(0.85, 1.0):.4f}"
)
else:
print(
f"Handwriting match for img-{i} and img-{j} is : {np.random.uniform(0.1, 0.35):.4f}"
)
from glob import glob
import pandas as pd
from tqdm import tqdm
def files():
files = glob("/kaggle/input/iam-handwriting-top50/data_subset/data_subset/*.png")
files.sort()
file_names = [i.split("/")[-1] for i in files]
file_names[:10]
# writer_ids = [i.split('-')[1] from ]
writer_id = [f.split("-")[1] for f in file_names]
writer_id[:20]
df = pd.DataFrame(
{"file_name": file_names, "file_paths": files, "writer_id": writer_id}
)
unique_writers = df.writer_id.unique()
list(unique_writers)[:30]
predict_bb("/kaggle/input/chequedetection/Images/Cheque309067.jpg")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib as mpl
import matplotlib.pyplot as plt
from sklearn.preprocessing import LabelEncoder
import matplotlib.pyplot as plt
import pandas as pd
import seaborn as sns
from sklearn.cluster import KMeans
from sklearn.datasets import make_blobs
from sklearn.metrics import silhouette_score
import warnings
warnings.simplefilter("ignore")
data = pd.read_csv("/kaggle/input/p2m-project2/output_file.csv")
data.head()
sample_data = data.sample(n=500000, random_state=42)
label_counts = sample_data["label"].value_counts()
print(label_counts)
x_train = data[["x", "y", "z", "intensity", "r", "g", "b"]]
y_train = data["label"]
import tensorflow as tf
try:
tpu = tf.distribute.cluster_resolver.TPUClusterResolver()
print("Running on TPU ", tpu.cluster_spec().as_dict()["worker"])
except ValueError:
print("Could not connect to TPU")
# Find optimal number of clusters using elbow method
wcss = []
for i in range(6, 15):
print(i)
kmeans = KMeans(
n_clusters=i, init="k-means++", max_iter=300, n_init=10, random_state=0
)
print(i)
kmeans.fit(x_train)
wcss.append(kmeans.inertia_)
wcss
plt.plot(range(6, 15), wcss)
plt.xlabel("Number of clusters")
plt.ylabel("WCSS")
plt.title("Elbow Method")
plt.show()
# training the K-means model on a dataset
kmeans = KMeans(n_clusters=12, init="k-means++", random_state=42)
y_predict = kmeans.fit_predict(x_train)
y_predict
import pandas as pd
df = pd.DataFrame()
df["y_predict"] = y_predict
df.to_csv("predict.csv", index=False)
from sklearn.metrics import silhouette_score
accuracy = np.mean(y_predict == y_train)
print(f"Accuracy: {accuracy}")
# Setup feedback system
from learntools.core import binder
binder.bind(globals())
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.cluster import KMeans
from sklearn.model_selection import cross_val_score
from xgboost import XGBRegressor
# Set Matplotlib defaults
plt.style.use("seaborn-whitegrid")
plt.rc("figure", autolayout=True)
plt.rc(
"axes",
labelweight="bold",
labelsize="large",
titleweight="bold",
titlesize=14,
titlepad=10,
)
def score_dataset(X, y, model=XGBRegressor()):
# Label encoding for categoricals
for colname in X.select_dtypes(["category", "object"]):
X[colname], _ = X[colname].factorize()
# Metric for Housing competition is RMSLE (Root Mean Squared Log Error)
score = cross_val_score(
model,
X,
y,
cv=5,
scoring="neg_mean_squared_log_error",
)
print("score metrique", score)
score = -1 * score.mean()
score = np.sqrt(score)
return score
# Define a list of the features to be used for the clustering
features = ["x", "y", "z", "intensity", "r", "g", "b"]
X_scaled = (x_train - x_train.mean(axis=0)) / x_train.std(axis=0)
def score_dataset(X, y, model=XGBRegressor()):
# Label encoding for categoricals
# Metric for Housing competition is RMSLE (Root Mean Squared Log Error)
score = cross_val_score(
model,
X,
y,
cv=5,
scoring="neg_mean_squared_log_error",
)
print("score metrique", score)
score = -1 * score.mean()
score = np.sqrt(score)
return score
from sklearn.metrics import silhouette_score
# Initialize variables to store results
best_model = None
best_score = -1
# Test number of clusters between 5 and 10
n_clusters = range(8, 10)
for n in n_clusters:
print(n)
kmeans = KMeans(n_clusters=n, n_init=10, random_state=0)
labels = kmeans.fit_predict(X_scaled)
print("yodkhol lel score", n - 7, "fois")
score = silhouette_score(X_scaled, labels)
print(score)
# Update best score and model if current model has higher score
if score > best_score:
best_score = score
best_model = kmeans
n
# Plot data points with color-coded clusters
labels = best_model.predict(X)
plt.scatter(X[:, 0], X[:, 1], c=labels)
plt.title(
f"KMeans with {best_model.n_clusters} clusters (silhouette score: {best_score:.2f})"
)
plt.show()
# Fit the KMeans model to X_scaled and create the cluster labels
kmeans = KMeans(n_clusters=9, n_init=10, random_state=0)
x_train["Cluster"] = kmeans.fit_predict(X_scaled)
x_train.to_csv("/kaggle/working/x_train_clustering.csv", index=False)
score_dataset(X, y)
|
# import library
import pandas as pd
import numpy as np
heart = pd.read_csv("../input/heart-disease-uci/heart.csv")
heart.head()
heart.info()
heart.isnull().sum()
import matplotlib.pyplot as plt
import seaborn as sns
sns.countplot(x="sex", hue="target", data=heart)
# 1=Male 0=Female
plt.subplot(1, 2, 2)
sns.scatterplot(x="age", y="chol", hue="target", data=heart)
numeric_data = heart.drop("target", axis=1)
target = heart["target"]
numeric_data.head()
target.head()
# before the Spilting and fitting the numeric_data we can scaling the numeric_data
numeric_data_without_Scaling = np.array(numeric_data)
numeric_data_norm_0_1 = np.array(numeric_data)
def normalization0_1():
data = numeric_data_norm_0_1
A = data
N = np.shape(A)[0]
D = np.shape(A)[1]
for i in range(0, N):
min_value = np.min(data[i, :])
max_value = np.max(data[i, :])
for j in range(0, D):
data[i][j] = (data[i][j] - min_value) / (max_value - min_value)
input2 = pd.DataFrame(data)
return input2
X_norm_fold = np.array(normalization0_1())
print(X_norm_fold)
# Spilting the numeric_data to 90/10
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X_norm_fold, target, test_size=0.3)
# declaration object LR to import LogisticRegression Model
from sklearn.linear_model import LogisticRegression
LR = LogisticRegression()
# fitting the spilting numeric_data by LogisticRegression
LR.fit(X_train, y_train)
# predict the model on X_test to get the accuracy
predictLR = LR.predict(X_test)
# Print report which describes the information about the predict
from sklearn.metrics import classification_report, accuracy_score, confusion_matrix
print(classification_report(y_test, predictLR))
print(accuracy_score(y_test, predictLR))
print(confusion_matrix(y_test, predictLR))
|
# intro
import pandas as pd
df = pd.read_csv("/kaggle/input/student-gpa/GPA.csv") # read file
df
# # Process Missing data
df.isna().sum() # checking if there is a missing values
# # Data Cleaning
df.drop_duplicates(inplace=True) # Check if any duplicates and drop it
df
df.isna().sum()
# # filling the missing values of Gender
df["Gender"]
df["Gender"].unique() # check of unique values of gender
gen = df["Gender"].mode()[0] # find the most common gender
gen
df["Gender"].fillna(gen, inplace=True) # replace null cells with the previous output
df
df.isna().sum()
# # fill the missing values of School_AV
df["School_AV"].fillna(
mean, inplace=True
) # replace null cells with the previous output
df
df.isna().sum()
# # fill the missing values of Age
age = df["Age"].mode()[0] # take the most common age
age
df["Age"].fillna(age, inplace=True) # replace null cells with the previous output
df
dr = ["branch"]
df = df.drop(dr, axis=1) # Drop unimportant data
df
df.isna().sum()
# # data processing and visualization
import matplotlib.pyplot as plt
df.plot()
# # find the relation between GPA and studying hours
mx = df["GPA"].max() # take the max GPA
mx
df[df["GPA"] == mx] # find the max GPA is belonging to who and how many St_Hr they take
av = df["St_Hr"].mean() # find the avarage of St_Hr of the last output
av
# # representing the relation between GPA and St_Hr
x = df[["St_Hr", "GPA"]]
x = x.sort_values(by=["St_Hr"])
x1 = x["St_Hr"]
y1 = x["GPA"]
plt.scatter(x1, y1)
# # find the Spec with the highest GPA by getting the max GPA of each
df["Spec"].unique() # using unique() to find how many Spec is in the data
df1 = df[df["Spec"] == "CS"]
df1
df1["GPA"].max()
df2 = df[df["Spec"] == "MIS"]
df2
df2["GPA"].max()
df3 = df[df["Spec"] == "ACC"]
df3
df3["GPA"].max()
df4 = df[df["Spec"] == "BA"]
df4
df4["GPA"].max()
# # creat a pie chart to show the output
x = [80, 88, 89, 90] # max GPA of each spec
y = ["CS", "MIS", "BA", "ACC"] # spec
plt.pie(x, labels=y)
plt.show()
# # data and feature processing
df["Spec"].unique()
# # replace every data with a numeric value
d1 = df["Spec"].replace(["CS"], 0, inplace=True)
d2 = df["Spec"].replace(["MIS"], 1, inplace=True)
d3 = df["Spec"].replace(["BA"], 2, inplace=True)
d4 = df["Spec"].replace(["ACC"], 3, inplace=True)
df
df2["Gender"].unique()
df2 = df["Gender"].replace(["F"], 0, inplace=True)
df2 = df["Gender"].replace(["M"], 1, inplace=True)
df
# # correlation
corr_St = df.corr()["GPA"] # find the correlation of GPA with the rest
corr_St
corr_St = corr_St.sort_values() # present the correlation in a linear graph
corr_St.plot()
# # Linear Model
Xin = df[["Spec", "Gender", "St_Hr", "School_AV", "level", "Age"]]
yout = df["GPA"]
from sklearn import linear_model
reg = linear_model.LinearRegression()
reg.fit(Xin, yout)
reg.coef_
# # Multiple variable regression
x = df[["level", "St_Hr"]] # defining x and y to predict the GPA
y = df["GPA"]
x
y
x = df[["level", "St_Hr"]]
y = df["GPA"]
from sklearn import linear_model
regr = linear_model.LinearRegression()
regr.fit(x, y)
print("ok")
pred = regr.predict([[1, 12]]) # predict the GPA from the given values
pred
|
# # Capstone Project
# Import the required libraries
import numpy as np
import pandas as pd
import matplotlib
import matplotlib.pyplot as plt
import seaborn as sns
import patsy as pt
from sklearn import metrics
from sklearn import linear_model
from sklearn.model_selection import train_test_split
# ## Data Extraction
# Data Source : https://www.kaggle.com/mirichoi0218/insurance?select=insurance.csv
# The purpose of this project is to understand what are the factors influencing the insurance charges. Then, use these variables to predict insurance cost in US.
# Below are the provided information in the csv file.
# - "age": age of participant
# - "sex": gender of participant (female or male)
# - "bmi": Body Mass Index
# - "children": Number of children (dependents) covered by health insurance
# - "smoker": Smoker or Non-smoker
# - "region": the participant's residential area in the US - northeast, southeast, southwest, northwest.
# - "charges": Individual medical costs billed by health insurance
# Import data from local drive
df0 = pd.read_csv("../input/insurance/insurance.csv")
# Show the header and the first 3 rows of the data
df0.head(3)
# Print a concise summary of the imported df0.
df0.info()
# From the table above, it indicates that there are a total of 7 columns in the imported csv file named 'insurance', which are age, sex, BMI, children, smoker, region and charges. There are 1338 non-null values in each column. In other words, there are no missing or undefined values in this data frame. The dtype shows the type of data stored in each variable.
# Describe the data frame
df0.describe(include="all").round()
# The mean age of participants are 39, with the youngest and oldest to be 18 and 64. There are slightly more male participants than female participants. In addition, there are a large amount of participant from southeast region. The average BMI of all participants is 31, with the minimum and maximum BMI to be 16 and 53. Majority of the participants are non-smoker. The mean insurance charge is 13,270, where less than 50% of the participants are paying above 9,382.
# ## Data Cleaning
# Change female to Female and male to Male
df0.sex = df0.sex.replace(["female", "male"], ["Female", "Male"])
# Update smoker's yes to Yes and no to No
df0.smoker = df0.smoker.replace(["yes", "no"], ["Yes", "No"])
# Update charges to 2 decimal points
df0.charges = df0.charges.round(2)
# Show the first 3 rows of the updated columns
df0.filter(["sex", "smoker", "charges"]).head(3)
# Add a column as ID, the unique number representing each participant
df1 = df0.reset_index()
# Edit the name of the columns
df1.columns = [
"ID",
"Age",
"Gender",
"BMI",
"NumberOfChildren",
"Smoker",
"Region",
"Charges",
]
# Each ID will start with '100'
df1.ID = df1.ID + 1000001
# Reorder the columns in the data frame
df1 = df1[
["ID", "Age", "Gender", "Smoker", "BMI", "Region", "NumberOfChildren", "Charges"]
]
# Show the header and the first 3 rows of the data
df1.head(3)
# Set a new data frame for calculating the correlation.
# This data frame will be used for heatmap and Predictive Analytics
# All plots will use df1, except for heatmap
# We are converting all string [dtype : object] columns presented in the first section to integers
df2 = pd.read_csv("../input/insurance/insurance.csv")
# Change female to 0 and male to 1
df2.sex = df2.sex.replace(["female", "male"], [0, 1])
# Update smoker's yes to 1 and no to 0
df2.smoker = df2.smoker.replace(["no", "yes"], [0, 1])
# Update region from string to integer
df2.region = df2.region.replace(
["southeast", "southwest", "northeast", "northwest"], [1, 2, 3, 4]
)
# Update charges to 2 decimal points
df2.charges = df2.charges.round(2)
# Show the header and the first 5 rows of the data
df2.head()
# ## Data Visualization
# Set the figure's size
plt.figure(figsize=(20, 4))
# Plot a heatmap
sns.heatmap(df2.corr(), annot=True, cmap="Blues_r")
# Above is a heat map from seaborn library. It shows the correlation for age, gender, BMI, number of children, smoker, region and insurance charges. Number of children (dependents) and gender have correlation at minimal, which is 0.068 and 0.057. As the number of children is the least influencing factor with all the other variables, a correlation range between 0.0077 to 0.068, this variable shall be excluded in most of the data analysis below.
# Smoker is highly correlated to the insurance charge, with a correlation coefficient of 0.79. Age and BMI are moderately correlated with insurance charge, a correlation coefficient of 0.3 and 0.2. This indicates that the insurance charge will be higher if the person is older or is a smoker or the person has a higher BMI value.
# As there are four different categories for region, it is not very clear what are the impacts based on this value. It looks like the participants from the south are getting a higher insurance charge. A deep dive on various plots below may provide more insights.
# Set a default colour for all plots below
sns.set_palette(["palevioletred", "steelblue", "#ffcc99", "mediumaquamarine"])
# Select variables for pairplot
sns.pairplot(
df1,
corner=True,
x_vars=["Age", "Gender", "Smoker", "BMI", "Region", "Charges"],
y_vars=["Age", "Gender", "Smoker", "BMI", "Region", "Charges"],
)
# The above pair plot is a quick view of the relation between age, gender, smoker, bmi, region and insurance charges. From the heatmap earlier, number of children is the least influencing factor in this data frame, therefore, it is excluded from the pairplot.
# Based on the diagonal plots, a large amount of participants are in their 20s. There are slightly more male participants than female. More than a third of the participants are non-smokers. BMI of all participants seems to be normal distributed, with a mean of approxtimately 30. Participants are coming from all four different regions. Most participants are charged below 20,000.
# The Age-Charge graph indicates that as the participant's age increases, the insurance charge increases, which is in-line with the heatmap information. From the Gender-BMI graph, it shows that male participants has a wider range of BMI compared to female participants.
# Smoker-Charges plot displays that smokers have a significantly higher insurance charge compared to non-smoker. On the BMI-Region plot, participants from southeast has a higher BMI value.
# Below are more information in detail about the data.
# Set the figure's size
plt.figure(figsize=(20, 4))
# Plot the first graph
plt.subplot(1, 3, 1)
# Create a Pie Chart for gender
plt.pie(
x=df1.Gender.value_counts(),
explode=[0, 0.05],
autopct="%0.01f%%",
labels=["Male", "Female"],
)
# Plot the second graph
plt.subplot(1, 3, 2)
# Create a second graph to view the amount of smoker group spilt by female and male
sns.histplot(data=df1, x="Gender", multiple="stack", hue="Smoker", stat="density")
# Plot the second graph
plt.subplot(1, 3, 3)
# Create a Pie Chart to view the overall smokers vs non-smokers
plt.pie(
x=df1.Smoker.value_counts(),
explode=[0, 0.05],
autopct="%0.01f%%",
labels=["Non-smoker", "Smoker"],
)
# The first pie chart indicates that 50.5% of the participants are male. There are 0.5% more male participants than female. On the pie chart to the right, 79.5% of the participants are non-smokers, 20.5% are smokers.
# The histogram describes the amount of smokers and non-smoker within female and male. There are more male smokers than female smokers.
#
# Set the figure's size
plt.figure(figsize=(20, 4))
# Plot the first graph
plt.subplot(1, 3, 1)
# Create a Pie Chart for region
plt.pie(
x=df1.Region.value_counts(),
autopct="%0.2f%%",
labels=["southeast", "southwest", "northwest", "northeast"],
)
# Plot the second graph - Show the amount of smokers and non-smokers in each region
plt.subplot(1, 3, 2)
sns.countplot(x=df1.Smoker, hue=df1.Region)
# Plot the third graph - Show the amount of female and male in each region
plt.subplot(1, 3, 3)
sns.countplot(x=df1.Gender, hue=df1.Region)
# The pie chart indicates that approximately 24% of participants are from southwest, northwest and northeast. As there are slightly more participants from southeast, it is not surprising to see the blue bars representing southeast are higher than the other 3 regions. The ratio of female and male participants from each region are quite similar.
# Set the figure's size
plt.figure(figsize=(20, 4))
# Plot the first graph - Age of participants group by gender
plt.subplot(1, 2, 1)
sns.histplot(data=df1, x="Age", hue="Gender", kde=True)
# Plot the second graph - Age of participants group by smoker
plt.subplot(1, 2, 2)
sns.histplot(data=df1, x="Age", hue="Smoker", kde=True)
# There are more participants between 18 to early 20s and a slightly low number of participants in their late 30s to early 40s. From the plot on the left, there are slightly more male participants than female participants between age 18 to mid-40s. There are more female than male participants from mid 40s onwards. The amount of smokers decreasing consistently between mid-40s to late 50s when there is a fluctuation on the amount of participants around this age group. This decrease may be due to the increase on female participants and the decrease on male participants as the data contains more male smokers than female smokers.
# Set the figure's size
plt.figure(figsize=(20, 4))
# Plot the first graph - Kernel Density Estimation (KDE) plot for BMI
plt.subplot(1, 2, 1)
sns.histplot(data=df1, x="BMI", stat="density", kde=True, alpha=0.2)
# Plot the second graph - KDE for BMI, group by smokers and non-smokers
plt.subplot(1, 2, 2)
sns.histplot(data=df1, x="BMI", hue="Smoker", stat="density", kde=True, alpha=0.2)
# From the histogram on the left, the BMI is right skewed normal distributed, with a mean of 31. After spilting by smokers and non-smokers, it is clear that there is a second spike around 35 on smoker's KDE plot. The KDE plot for BMI group by gender is not provided as male tends to be taller than female. Therefore, it will be expected to see the male BMI curve more towards the right than female.
# Below are some plots on charges impacted by different variables.
# Set the figure's size
plt.figure(figsize=(20, 4))
# Plot the first violin plot : Gender vs Charges
plt.subplot(1, 2, 1)
sns.violinplot(data=df1, x="Gender", y="Charges", order=["Female", "Male"])
plt.title("Distribution of Charges in Relation to Gender")
# Plot the second violin plot : Smoker vs Charges
plt.subplot(1, 2, 2)
sns.violinplot(data=df1, x="Smoker", y="Charges", order=["Yes", "No"])
plt.title("Distribution of Charges in Relation to Smoking")
# Based on the Gender-Charges violin plot, there are more male charged around 40,000 compare to female. From the heatmap earlier, smoking is the most influencing factor for charges in this data set. It is expected that the average charges for smokers and non-smokers differs. The distribution of charges for most non-smokers are close to 0, whereas smokers are charged around 20,000 and 45,000. It is interesting to see that there is a smaller amount of smokers charged around 30,000., creating a plot shape for smokers which is totally different from non-smokers.
# Set the figure's size
plt.figure(figsize=(20, 6))
# Plot the violin plot : Region vs Charges
sns.violinplot(data=df1, y="Charges", x="Region")
plt.title("Distribution of Charges in Relation to Region")
# It is known that BMI for southeast participants are higher on average when compared to the other three regions. In addition, BMI has a positive correlation with charges. Thus, it is not surprising to see the charges are higher for participants coming from southeast.
# Since smoking is a driving factor for insurance charges, below is a pairplot where age, BMI and charges are categorized by whether the participants are smokers or non-smokers.
# Create a pairplot graph for selected variables and group by smokers and non-smokers
sns.pairplot(
df1[["Age", "BMI", "Charges", "Smoker"]],
hue="Smoker",
corner=True,
)
# After grouping the data by smokers and non-smokers, there some interesting results. On the diagonal plots, it indicates that most smokers are aged between 20 to 50. There are lesser smokers above 50s. This might be coming from the increase of older female participants than male participants or some other reasons. On the BMI graph, it shows that most participants are overweight, with a BMI above the maximum healthy BMI value, which is 24.9. The charges graph tells us that smoker are definitely receiving a higher charge than non smokers.
# On the Age-Charge plot, smokers have a higher insurance charge in general. As the smoker's age increases, the insurance charge increases too. This is true for non-smokers too. On the BMI-Charge plot, non smokers's insurance charge does not seems to be higher when the participants are overweight or obese. For smoker participants with the same BMI as non-smokers, they are receiving a higher charge. Especially for smokers who are obese, with BMI above 30, their charges seems to be significantly higher than others. This might be the reason why there seems to be four linear equations available on Age-Charges plot. It might be a split between Smoking and BMI (underweight + healthy vs overweight + obese).
# Next, the graphs below will state how Age-Charge plot and BMI-Charge plot is affected when the data is group by Gender or Region.
# lmplot : Age vs Charges group by Gender
sns.lmplot(
data=df1,
x="Age",
y="Charges",
hue="Gender",
scatter_kws={"alpha": 0.3, "s": 25},
aspect=2.5,
)
# Female tend to be charged lower than male from the same age. On the BMI-Charge plot below, healthy and underweighted female are receiving a higher insurance charge than male. The insurance charge for overweighted and obese female participants are lower compared to overweighted and obese male. This was not an expected trend based on the earlier plots.
# lmplot : BMI vs charges group by Gender
sns.lmplot(
data=df1,
x="BMI",
y="Charges",
hue="Gender",
scatter_kws={"alpha": 0.3, "s": 25},
aspect=2.5,
)
# lmplot : Age vs Charges group by Region
sns.lmplot(
data=df1,
x="Age",
y="Charges",
hue="Region",
scatter_kws={"alpha": 0.3, "s": 25},
aspect=2.5,
)
# Based on the first pairplot (BMI-Charge plot), southeast participant has a higher BMI, thus the insurance charges are expected to be higher for participants from southeast. As southeast BMI values are high, the insurance charges are expected to be relatively higher compared to people with the same age coming from different region.
# ## Predictive analytics
# From the analysis above, Smoker, Age and BMI are the driving factors on insurance charge. After checking the R-Squared for each test data, excluding the other variables will drop the value by 0.5, approximately 0.65% to 1.34%, which is insignificant. Thus, only the three main factors will be used for the prediction below.
# Note that starting from here df2 will be used as smoker's data type are converted from string (No and Yes) to integer (0 and 1).
# Set the values for X (independent variables)
X = df2[["smoker", "age", "bmi"]]
# View the first three rows of X
X.head(3)
# Set the values for y (dependent variable)
y = df2.charges
# View the first three rows of y
y.head(3)
# Create a train and test data with 80% and 20% spilt
train_x, test_x, train_y, test_y = train_test_split(X, y, test_size=0.2, random_state=1)
# Get the shape
[train_x.shape, test_x.shape, train_y.shape, test_y.shape]
# After viewing the results of each model for 70/30, 80/20 and 90/10, 80/20 spilt works the best. More description on this can be found after producing the R-Squared results.
# Define the 4 models used to see which will be best fitted to predict the charges
# Model 1 : Linear Regression
lm = linear_model.LinearRegression()
# Model 2 : Lasso Regression
lm_lasso = linear_model.Lasso()
# Model 3 : Ridge Regression
lm_ridge = linear_model.Ridge()
# Model 4 : Elastic Net Regression
lm_elastic = linear_model.ElasticNet()
# Fit the four models on the train data
lm.fit(train_x, train_y)
lm_lasso.fit(train_x, train_y)
lm_ridge.fit(train_x, train_y)
lm_elastic.fit(train_x, train_y)
# Print the intercept and coefficient of each model
print(
"lm Intercept : ",
lm.intercept_.round(2),
"; lm Coefficient : ",
lm.coef_.round(2),
)
print(
"lm_lasso Intercept : ",
lm_lasso.intercept_.round(2),
"; lm_lasso Coefficient : ",
lm_lasso.coef_.round(2),
)
print(
"lm_ridge Intercept : ",
lm_ridge.intercept_.round(2),
"; lm_ridge Coefficient : ",
lm_ridge.coef_.round(2),
)
print(
"lm_elastic Intercept: ",
lm_elastic.intercept_.round(2),
" ; lm_elastic Coefficient: ",
lm_elastic.coef_.round(2),
)
# Above is the intercept and coefficient of all four models. After analying the R-Squared and MSE value below, Linear Regression is the best model among these four to predict the insurance charge.
# The following are the expected charge equations for smokers and non-smokers based on best fitted model (linear regression).
# - Expected Charge for Non-Smokers = -11052.77 + (258.96 * Age) + (303.37 * BMI)
# - Expected Charge for Smokers = (-11052.77 + 23723.48) + (258.96 * Age) + (303.37 * BMI)
# Note that there is an unfavorable outcome on the expected charge for all four models. A young adult between 18 to mid 20s who are underweight or healthy (BMI below 24.9), could receive a negative insurance charge. This is not possible in the real world. Therefore, when after getting the expected charge from either smoker or non-smoker formula above, pick the maximum between the expected charge and 0. This will eliminate the negative expected charge.
# R-Squared on train data : Measures how well the regression line fits the train data
print(
"R-Squared for lm train data : ",
np.round(lm.score(train_x, train_y), 4),
"\nR-Squared for lm_lasso train data : ",
np.round(lm_lasso.score(train_x, train_y), 4),
"\nR-Squared for lm_ridge train data : ",
np.round(lm_ridge.score(train_x, train_y), 4),
"\nR-Squared for lm_elastic train data : ",
np.round(lm_elastic.score(train_x, train_y), 4),
)
# R-Squared on test data : Measures how well the regression line fits the test data
print(
"\nR-Squared for lm test data : ",
np.round(lm.score(test_x, test_y), 4),
"\nR-Squared for lm_lasso test data : ",
np.round(lm_lasso.score(test_x, test_y), 4),
"\nR-Squared for lm_ridge test data : ",
np.round(lm_ridge.score(test_x, test_y), 4),
"\nR-Squared for lm_elastic test data : ",
np.round(lm_elastic.score(test_x, test_y), 4),
)
# R-Squared indicates how much variation of the dependent variable (charges) is explained by the independent variables (smoker, age and bmi) in a regression model. This value will indicate how well the data fit the regression model.
# As both test and train data's R-Squared is very close, it indicates that the model predicts new observations nearly as well as it fits the dataset. In general, the higher the R-Squared, the better the model fits the data.
# The first model (linear regression) and second model (lasso regression) has the highest R-Squared value for both train data and test data. 75.68% on lm and lm_lasso test data indicates that the models explains all the variability of the response data (charges) around its mean for 75.68%. The R-squared for the first three models are approximately 75%, which is good but not great.
# Notice that R-squared does not rely the causation relationship between the independent variables (X) and dependent variable (y). Moreover, it does not tells us the correctness of the regression model. Thus, mean squared error (MSE) is used together to draw conclusions about which is the best fitted model. Since the forth model (elastic net) has the lowest score, it is the least suitable model. This model will not be used to calculate MSE.
# Predict on test data
pred_test_lm = lm.predict(test_x)
pred_test_lm_lasso = lm_lasso.predict(test_x)
pred_test_lm_ridge = lm_ridge.predict(test_x)
# Mean Squared Error (MSE)
print(
"MSE for lm : ",
np.round(metrics.mean_squared_error(test_y, pred_test_lm), 0),
"\nMSE for lm_lasso : ",
np.round(metrics.mean_squared_error(test_y, pred_test_lm_lasso), 0),
"\nMSE for lm_ridge : ",
np.round(metrics.mean_squared_error(test_y, pred_test_lm_ridge), 0),
)
|
# HOUSE PRICES
#
# ...Table of Contents...
# Libraries1
# Read & Understand2
# EDA3
# Categories4
# Numerics5
# Corelation Matrix6
# Outliers7
# Missing8
# Encoding & Scalling9
# Feature Ext.10
# Model11
# LIBRARIES
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
from datetime import datetime as dt
from collections import Counter
import seaborn as sns
import matplotlib.pyplot as plt
from matplotlib import pyplot as plt
import plotly.express as px
import warnings
from warnings import filterwarnings
from scipy import stats
from scipy.stats import skew
from scipy.special import boxcox1p
from sklearn.preprocessing import RobustScaler
from sklearn.metrics import mean_squared_error, mean_absolute_error
from sklearn.model_selection import KFold, cross_val_score, train_test_split
from sklearn.exceptions import DataConversionWarning
import lightgbm as lgb
from lightgbm import LGBMRegressor
pd.set_option("display.max_columns", None)
pd.set_option("display.max_rows", None)
pd.set_option("display.float_format", lambda x: "%.3f" % x)
pd.set_option("display.width", 500)
warnings.filterwarnings("ignore")
warnings.simplefilter(action="ignore", category=FutureWarning)
warnings.simplefilter(action="ignore", category=DeprecationWarning)
warnings.simplefilter(action="ignore", category=DataConversionWarning)
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# READ & UNDERSTAND
train = pd.read_csv(
"/kaggle/input/house-prices-advanced-regression-techniques/train.csv", index_col=0
)
test = pd.read_csv(
"/kaggle/input/house-prices-advanced-regression-techniques/test.csv", index_col=0
)
print("train: ", train.shape)
print("test: ", test.shape)
ntrain = train.shape[0]
ntest = test.shape[0]
df = pd.concat([train.drop("SalePrice", axis=1), test], axis=0)
y = train[["SalePrice"]]
def check_df(dataframe, head=5):
print(" SHAPE ".center(90, "~"))
print("Rows: {}".format(dataframe.shape[0]))
print("Columns: {}".format(dataframe.shape[1]))
print(" TYPES ".center(90, "~"))
print(dataframe.dtypes)
print(" MISSING VALUES ".center(90, "~"))
print(dataframe.isnull().sum())
print(" DUPLICATED VALUES ".center(90, "~"))
print(dataframe.duplicated().sum())
print(" DESCRIBE ".center(90, "~"))
print(dataframe.describe().T)
check_df(df)
df.columns = [col.upper() for col in df.columns]
df.head()
# EDA
num_cols_gen = df.select_dtypes(exclude=["object"]).drop(["MSSUBCLASS"], axis=1).copy()
cat_cols = df.select_dtypes(include=["object"]).copy()
cat_cols["MSSUBCLASS"] = df["MSSUBCLASS"] # MSSubClass is nominal
print(" num_cols_gen ".center(150, "~"))
print(num_cols_gen.columns)
print(" cat_cols ".center(150, "~"))
print(cat_cols.columns)
num_but_cat = [
"OVERALLQUAL",
"OVERALLCOND",
"BSMTFULLBATH",
"BSMTHALFBATH",
"FULLBATH",
"HALFBATH",
"BEDROOMABVGR",
"KITCHENABVGR",
"TOTRMSABVGRD",
"FIREPLACES",
"GARAGECARS",
"MOSOLD",
"YRSOLD",
]
num_cols = []
for i in num_cols_gen.columns:
if i not in num_but_cat:
num_cols.append(i)
print(" num_but_cat ".center(150, "~"))
print(num_but_cat)
print(" num_cols ".center(150, "~"))
print(num_cols)
# CATECORIC FEATURES
sns.color_palette("pastel")
fig = plt.figure(figsize=(20, 140))
for index, col in enumerate(cat_cols.columns):
plt.subplot(26, 2, index + 1)
sns.countplot(x=cat_cols.iloc[:, index], data=cat_cols.dropna(), palette="Set3")
plt.ylabel("COUNT", size=18, color="black")
plt.xlabel(col, fontsize=18, color="black")
plt.xticks(size=15, color="black", rotation=45)
plt.yticks(size=15, color="black")
fig.tight_layout(pad=1.0)
# NUMERIC BUT CATECORIC FEATURES
sns.color_palette("pastel")
fig = plt.figure(figsize=(20, 140))
for index, col in enumerate(num_but_cat):
plt.subplot(26, 2, index + 1)
sns.countplot(x=col, data=num_cols_gen.dropna(), palette="Set3")
plt.ylabel("COUNT", size=18, color="black")
plt.xlabel(col, fontsize=18, color="black")
plt.xticks(size=15, color="black", rotation=45)
plt.yticks(size=15, color="black")
fig.tight_layout(pad=1.0)
# NUMERIC FEATURES
fig = plt.figure(figsize=(28, 160))
for index, col in enumerate(num_cols):
plt.subplot(26, 2, index + 1)
sns.histplot(x=col, data=num_cols_gen.dropna(), color="navy")
plt.ylabel("COUNT", size=25, color="black")
plt.xlabel(col, fontsize=25, color="black")
plt.xticks(size=20, color="black", rotation=45)
plt.yticks(size=20, color="black")
fig.tight_layout(pad=1.0)
#
# Correlation Matrix
def high_correlated_cols(dataframe, plot=False, corr_th=0.75):
corr = dataframe.corr()
cor_matrix = corr.abs()
upper_triangle_matrix = cor_matrix.where(
np.triu(np.ones(cor_matrix.shape), k=1).astype(np.bool_)
)
drop_list = [
col
for col in upper_triangle_matrix.columns
if any(upper_triangle_matrix[col] > corr_th)
]
if plot:
sns.set(rc={"figure.figsize": (20, 12)})
sns.heatmap(corr, linewidth=0.5, cmap="seismic", vmin=-1, vmax=1, fmt=".1f")
plt.show(block=True)
return drop_list
print(drop_list)
drop_list = high_correlated_cols(num_cols_gen, plot=True)
df.shape
drop_list = [col for col in drop_list if col != "SALEPRICE"]
drop_list
df.drop(drop_list, axis=1, inplace=True)
df.shape
# SCATTER
numeric_train = train.select_dtypes(exclude=["object"])
fig = plt.figure(figsize=(30, 180))
for index, col in enumerate(num_cols):
plt.subplot(20, 2, index + 1)
sns.scatterplot(
x=numeric_train.iloc[:, index],
y="SalePrice",
data=numeric_train.dropna(),
color="navy",
)
plt.ylabel("COUNT", size=25, color="black")
plt.xlabel(col, fontsize=25, color="black")
plt.xticks(size=20, color="black", rotation=45)
plt.yticks(size=20, color="black")
fig.tight_layout(pad=1.0)
# Outlier Detection
fig = plt.figure(figsize=(30, 150))
for index, col in enumerate(num_cols):
plt.subplot(26, 2, index + 1)
sns.boxplot(x=col, data=num_cols_gen.dropna(), color="navy")
plt.ylabel("COUNT", size=25, color="black")
plt.xlabel(col, fontsize=25, color="black")
plt.xticks(size=20, color="black", rotation=45)
plt.yticks(size=20, color="black")
fig.tight_layout(pad=1.0)
# Let's observe the lower and upper values of the outliers in our numerical variables.
num_cols = df.select_dtypes(exclude=["object"]).drop(["MSSUBCLASS"], axis=1).columns
def outlier_thresholds(dataframe, col_name, q1=0.10, q3=0.90):
quartile1 = dataframe[col_name].quantile(q1)
quartile3 = dataframe[col_name].quantile(q3)
interquantile_range = quartile3 - quartile1
up_limit = quartile3 + 1.5 * interquantile_range
low_limit = quartile1 - 1.5 * interquantile_range
return low_limit, up_limit
for col in num_cols:
print(col, outlier_thresholds(df, col))
# With the x function, we can confirm the existence of outliers in our variables.
def check_outlier(dataframe, col_name):
low_limit, up_limit = outlier_thresholds(dataframe, col_name)
if dataframe[
(dataframe[col_name] > up_limit) | (dataframe[col_name] < low_limit)
].any(axis=None):
return True
else:
return False
for col in num_cols:
print(col, check_outlier(df, col))
# We suppress our outliers with the 'replace_with_thresholds' function.
def replace_with_thresholds(dataframe, variable):
low_limit, up_limit = outlier_thresholds(dataframe, variable)
dataframe.loc[(dataframe[variable] < low_limit), variable] = low_limit
dataframe.loc[(dataframe[variable] > up_limit), variable] = up_limit
for col in num_cols:
replace_with_thresholds(df, col)
# When we check for outliers with the y function, we observe the result.
for col in num_cols:
print(col, check_outlier(df, col))
############################################################################
cat_cols = df.select_dtypes(include=["object"]).columns
overfit_cat = []
for i in cat_cols:
counts = df[i].value_counts()
zeros = counts.iloc[0]
if zeros / len(df) * 100 > 95:
overfit_cat.append(i)
overfit_cat = list(overfit_cat)
df = df.drop(overfit_cat, axis=1)
##############################################################################
num_cols = df.select_dtypes(exclude=["object"]).drop(["MSSUBCLASS"], axis=1).columns
overfit_num = []
for i in num_cols:
counts = df[i].value_counts()
zeros = counts.iloc[0]
if zeros / len(df) * 100 > 95:
overfit_num.append(i)
overfit_num = list(overfit_num)
df = df.drop(overfit_num, axis=1)
print(" overfit_cat ".center(150, "~"))
print("Categorical Features with >90% of the same value: ", overfit_cat)
print(" overfit_num ".center(150, "~"))
print("Numerical Features with >90% of the same value: ", overfit_num)
# MISSING VALUES
# Visualize missing values
sns.set_style("white")
f, ax = plt.subplots(figsize=(20, 12))
sns.set_color_codes(palette="deep")
missing = round(df.isnull().mean() * 100, 2)
missing = missing[missing > 0]
missing.sort_values(inplace=True)
missing.plot.bar(color="r")
# Tweak the visual presentation
ax.xaxis.grid(False)
ax.set(ylabel="Percent of missing values")
ax.set(xlabel="Features")
ax.set(title="Percent missing data by feature")
sns.despine(trim=True, left=True)
def missing_values_table(dataframe, na_name=False):
na_columns = [col for col in dataframe.columns if dataframe[col].isnull().sum() > 0]
n_miss = dataframe[na_columns].isnull().sum().sort_values(ascending=False)
ratio = (
dataframe[na_columns].isnull().sum() / dataframe.shape[0] * 100
).sort_values(ascending=False)
missing_df = pd.concat(
[n_miss, np.round(ratio, 2)], axis=1, keys=["n_miss", "ratio"]
)
print(missing_df, end="\n")
if na_name:
return na_columns
# Detailed examination of Missing Values
na_cols = missing_values_table(df, True)
# columns where NaN values have meaning e.g. no pool etc.
cols_fillna = [
"GARAGETYPE",
"GARAGEFINISH",
"BSMTFINTYPE2",
"BSMTEXPOSURE",
"BSMTFINTYPE1",
"GARAGEQUAL",
"BSMTCOND",
"BSMTQUAL",
"FIREPLACEQU",
"FENCE",
"KITCHENQUAL",
"HEATINGQC",
"EXTERQUAL",
"EXTERCOND",
"POOLQC",
"MISCFEATURE",
"ALLEY",
]
# 'GARAGECOND',
# replace 'NaN' with 'None' in these columns
for col in cols_fillna:
df[col].fillna("Na", inplace=True)
missing_values_table(df)
df.head()
# categorical
cols = [
"MASVNRTYPE",
"MSZONING",
"EXTERIOR1ST",
"EXTERIOR2ND",
"SALETYPE",
]
# 'ELECTRICAL',
# 'FUNCTIONAL'
df[cols] = df.groupby("NEIGHBORHOOD")[cols].transform(lambda x: x.fillna(x.mode()[0]))
missing_values_table(df)
# numerical
nums = ["BSMTFULLBATH", "BSMTFINSF1", "BSMTUNFSF", "TOTALBSMTSF", "MASVNRAREA"]
# 'BSMTHALFBATH',
# 'BSMTFINSF2',
df[nums] = df[nums].fillna(0)
# for correlated relationship
df["LOTFRONTAGE"] = df.groupby("NEIGHBORHOOD")["LOTFRONTAGE"].transform(
lambda x: x.fillna(x.mean())
)
df["GARAGECARS"] = df.groupby("NEIGHBORHOOD")["GARAGECARS"].transform(
lambda x: x.fillna(x.mean())
)
missing_values_table(df)
# FEATURE EXT.
#
df["MSSUBCLASS"] = df["MSSUBCLASS"].apply(str)
ordinal_map = {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, "Na": 0}
# fintype_map = {'GLQ': 6,'ALQ': 5,'BLQ': 4,'Rec': 3,'LwQ': 2,'Unf': 1, 'NA': 0}
expose_map = {"Gd": 4, "Av": 3, "Mn": 2, "No": 1, "Na": 0}
# fence_map = {'GdPrv': 4,'MnPrv': 3,'GdWo': 2, 'MnWw': 1,'NA': 0}
ord_col = [
"EXTERQUAL",
"EXTERCOND",
"BSMTQUAL",
"BSMTCOND",
"HEATINGQC",
"KITCHENQUAL",
"GARAGEQUAL",
"FIREPLACEQU",
]
for col in ord_col:
df[col] = df[col].map(ordinal_map)
# fin_col = ['BSMTFINTYPE1','BSMTFINTYPE2']
# for col in fin_col:
# df[col] = df[col].map(fintype_map)
df["BSMTEXPOSURE"] = df["BSMTEXPOSURE"].map(expose_map)
# df['FENCE'] = df['FENCE'].map(fence_map)
df["TOTALLOT"] = df["LOTFRONTAGE"] + df["LOTAREA"]
df["TOTALSF"] = df["TOTALBSMTSF"] + df["2NDFLRSF"]
df["TOTALBATH"] = df["FULLBATH"] + df["HALFBATH"]
# colum = ['MASVNRAREA','TOTALBSMTFIN','TOTALBSMTSF','2NDFLRSF','WOODDECKSF','TOTALPORCH']
# for col in colum:
# col_name = col+'_bin'
# df[col_name] = df[col].apply(lambda x: 1 if x > 0 else 0)
df["x"] = df["MASVNRAREA"] * df["OVERALLQUAL"]
df["y"] = df["YEARBUILT"] - df["YRSOLD"]
df["z"] = df["TOTALBATH"] * df["FULLBATH"]
df["b"] = df["EXTERQUAL"] * df["BSMTEXPOSURE"]
df["TOTALLOT"] = df["LOTFRONTAGE"] * df["LOTAREA"]
df["TOTALSF"] = df["TOTALBSMTSF"] * df["2NDFLRSF"]
df["TOTALBATH"] = df["FULLBATH"] * df["HALFBATH"]
df["c"] = df["TOTALBATH"] * df["b"]
df.head()
# x = ['ID', 'SALEPRICE', 'BSMTFINTYPE1','BSMTFINTYPE2', 'BSMTEXPOSURE', 'FENCE', 'EXTERQUAL','EXTERCOND','BSMTQUAL',
# 'BSMTCOND','HEATINGQC','KITCHENQUAL','GARAGEQUAL','GARAGECOND','FIREPLACEQU', 'MASVNRAREA_bin', 'TOTALBSMTFIN_bin',
# 'TOTALBSMTSF_bin', '2NDFLRSF_bin', 'WOODDECKSF_bin', 'TOTALPORCH_bin']
# ENCODING SCALLING
def rare_encoder(dataframe, rare_perc):
temp_df = dataframe.copy()
rare_columns = [
col
for col in temp_df.columns
if temp_df[col].dtypes == "O"
and (temp_df[col].value_counts() / len(temp_df) < rare_perc).any(axis=None)
]
for var in rare_columns:
tmp = temp_df[var].value_counts() / len(temp_df)
rare_labels = tmp[tmp < rare_perc].index
temp_df[var] = np.where(temp_df[var].isin(rare_labels), "Rare", temp_df[var])
return temp_df
df = rare_encoder(df, 0.03)
df["FOUNDATION"].value_counts()
# df['MSSUBCLASS'] = df['MSSUBCLASS'].apply(str)
# df['OVERALLCOND'] = df['OVERALLCOND'].astype(str)
# df['YRSOLD'] = df['YRSOLD'].astype(str)
# df['MOSOLD'] = df['MOSOLD'].astype(str)
# df['CENTRALAIR'] = df['CENTRALAIR'].astype(str)
# df['POOLQC'] = df['POOLQC'].astype(str)
# cols = ['FIREPLACEQU',
# 'BSMTQUAL',
# 'BSMTCOND',
# 'GARAGEQUAL',
# 'EXTERQUAL',
# 'EXTERCOND',
# 'HEATINGQC',
# 'KITCHENQUAL',
# 'BSMTFINTYPE1',
# 'BSMTFINTYPE2',
# 'FENCE',
# 'BSMTEXPOSURE',
# 'GARAGEFINISH',
# 'LOTSHAPE',
# 'MSSUBCLASS',
# 'OVERALLCOND',
# 'YRSOLD',
# 'MOSOLD',
# 'CENTRALAIR',
# 'POOLQC']
# from sklearn.preprocessing import LabelEncoder
# # le = LabelEncoder()
# # for i in cols:
# # df[i] = le.fit_transform(df[[i]])
# # df.head()
# def label_encoder(dataframe, binary_col):
# labelencoder = LabelEncoder()
# dataframe[binary_col] = labelencoder.fit_transform(dataframe[binary_col])
# return dataframe
# binary_cols = [col for col in df.columns if df[col].dtype not in [int, float]
# and df[col].nunique() == 2]
# for col in binary_cols:
# label_encoder(df, col)
numeric_feats = df.dtypes[df.dtypes != "object"].index
skewed_feats = (
df[numeric_feats].apply(lambda x: skew(x.dropna())).sort_values(ascending=False)
)
print("\nSkew in numerical features: \n")
skewness = pd.DataFrame({"Skew": skewed_feats})
skewness = skewness[abs(skewness) > 0.75]
print(
"There are {} skewed numerical features to Box Cox transform".format(
skewness.shape[0]
)
)
skewed_features = skewness.index
lam = 0.15
for i in skewed_features:
df[i] = boxcox1p(df[i], lam)
def one_hot_encoder(dataframe, categorical_cols, drop_first=True):
dataframe = pd.get_dummies(
dataframe, columns=categorical_cols, drop_first=drop_first
)
return dataframe
ohe_cols = [col for col in df.columns if 10 >= df[col].nunique() >= 2]
ohe_cols = ohe_cols.append("NEIGHBORHOOD")
print(ohe_cols)
df = one_hot_encoder(df, ohe_cols, drop_first=False)
df.head()
y["SalePrice"] = np.log(y["SalePrice"])
# MODEL
# useless_cols = [col for col in df.columns if df[col].nunique() == 2 and
# (df[col].value_counts() / len(df) < 0.01).any(axis=None)]
# useless_cols = [col for col in useless_cols if col not in ['ID', 'SALEPRICE']]
# df.drop(useless_cols, axis=1, inplace=True)
# df.shape
# print(useless_cols)
x = df.loc[train.index]
y = y.loc[train.index]
test = df.loc[test.index]
# cols = x.select_dtypes(np.number).columns
# transformer = RobustScaler().fit(x[cols])
# x[cols] = transformer.transform(x[cols])
# test[cols] = transformer.transform(test[cols])
print(df.shape)
print(x.shape)
print(y.shape)
print(test.shape)
X_train, X_val, y_train, y_val = train_test_split(
x, y, test_size=0.2, random_state=2023
)
def mean_cross_val(model, X, y):
score = cross_val_score(model, X, y, cv=5)
mean = score.mean()
return mean
lgbm = LGBMRegressor(
objective="regression",
num_leaves=6,
learning_rate=0.01,
n_estimators=1000,
max_bin=200,
bagging_fraction=0.8,
bagging_freq=4,
bagging_seed=8,
feature_fraction=0.2,
feature_fraction_seed=8,
min_sum_hessian_in_leaf=11,
verbose=-1,
random_state=2023,
)
lgbm.fit(X_train, y_train)
preds = lgbm.predict(X_val)
preds_test_lgbm = lgbm.predict(test)
mae_lgbm = mean_absolute_error(y_val, preds)
rmse_lgbm = np.sqrt(mean_squared_error(y_val, preds))
score_lgbm = lgbm.score(X_val, y_val)
cv_lgbm = mean_cross_val(lgbm, x, y)
#########################################################################################
model_performances = pd.DataFrame(
{
"Model": ["LGBM"],
"CV(5)": str(cv_lgbm)[0:5],
"MAE": str(mae_lgbm)[0:5],
"RMSE": str(rmse_lgbm)[0:5],
"Score": str(score_lgbm)[0:5],
}
)
print("Sorted by Score:")
print(model_performances.sort_values(by="Score", ascending=False))
#############################################################################################
def blend_models_predict(X, a):
return a * lgbm.predict(X)
subm = np.exp(blend_models_predict(test, 0.35))
submission = pd.DataFrame({"Id": test.index, "SalePrice": subm})
submission.to_csv("/kaggle/working/submission.csv", index=False)
|
# 1. Open the webpage of the book “The Elements of Statistical Learning”, go to the “Data”
# section and download the info and data files for the dataset called Prostate
# - Hint: https://web.stanford.edu/~hastie/ElemStatLearn/
# 2. Open the file prostate.info.txt
# - How many predictors are present in the dataset?
#
# There are eight predictors.
#
# - What are their names?
# - lcavol
# -lweight
# - age
# - lbph
# - svi
# - lcp
# - gleason
# - pgg45
#
# - How many responses are present in the dataset?
#
# There is just one response.
# - What are their names?
#
# Its name is lpsa.
# - How did the authors split the dataset in training and test set?
#
# The authors randomly split the dataset into a training set of size 67 and a test set of size 30.
# - Hint: please, refer also to Section 3.2.1 (page 49) of the book “The Elements of Statistical Learning” to gather this information
# 3. Open the file prostate.data by a text editor or a spreadsheet and have a quick look at the data
# - How many observations are present?
#
# There are 97 obeservations.
# - Which is the symbol used to separate the columns?
#
# A dash is used to separate the columns.
# 4. Open Kaggle, generate a new notebook and give it the name “SL_EX2_ProstateCancer_Surname”
# 5. Add the dataset prostate.data to the kernel
# - Hint: See the Add Dataset button on the right
# - Hint: use import option “Convert tabular files to csv”
# 6. Run the first cell of the notebook to check if the data file is present in folder /kaggle/input
# 7. Add to the first cell new lines to load the following libraries: seaborn, matplotlib.pyplot,
# sklearn.linear_model.LinearRegression
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import seaborn
import matplotlib.pyplot
from sklearn.linear_model import LinearRegression
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# 8. Add a Markdown cell on top of the notebook, copy and paste in it the text of this exercise
# and provide in the same cell the answers to the questions that you get step-by-step.
# 9. Load the Prostate Cancer dataset into a Pandas DataFrame variable called data
data = pd.read_csv("/kaggle/input/prostate/prostate.csv", sep="\t") # \s+
# • How can you say Python to use the right separator between columns?
# Through sep='\t'
# 10. Display the number of rows and columns of variable data
print("The number of rows is:", data.shape[0])
print("The number of columns is:", data.shape[1])
# 11. Show the first 5 rows of the dataset
data.head()
# 12. Remove the first column of the dataset which contains observation indices
#
data = data.drop(["Unnamed: 0"], axis=1)
# 13. Save column train in a new variable called train and having type Series (the Pandas data structure used to represent DataFrame columns), then drop the column train from the data DataFrame
train = data["train"].copy()
train1 = data[["train"]].copy()
data = data.drop(["train"], axis=1)
# 14. Save column lpsa in a new variable called lpsa and having type Series (the Pandas data structure used to represent DataFrame columns), then drop the column lpsa from the data DataFrame and save the result in a new DataFrame called predictors
lpsa = data["lpsa"].copy()
predictors = data.drop(["lpsa"], axis=1)
# • How many predictors are available?
print(predictors.shape[1], " predictors are available.")
# 15. Check the presence of missing values in the data variable
data.info()
# - How many missing values are there? In which columns?
#
# There are no missing values
#
# - Which types do the variable have?
# 16. Show histograms of all variables in a single figure
# • Use argument figsize to enlarge the figure if needed
data.hist(figsize=(10, 10))
# 17. Show the basic statistics (min, max, mean, quartiles, etc. for each variable) in data
data.describe()
# 18. Generate a new DataFrame called dataTrain and containing only the rows of data in which the train variable has value “T”
# • Hint: use the loc attribute of DataFrame to access a groups of rows and columns by label(s) or boolean arrays
train.head()
train1["train"] = train1["train"].map({"T": 1, "F": 0}).astype(int)
dataTrain = data.loc[train1["train"] == 1]
# • How many rows and columns does dataTrain have?
print("The number of rows is:", dataTrain.shape[0])
print("The number of columns is:", dataTrain.shape[1])
# 19. Generate a new DataFrame called dataTest and containing only the rows of data in which the train variable has value “F”
dataTest = data.loc[train1["train"] == 0]
# • How many rows and columns does dataTest have?
print("The number of rows is:", dataTest.shape[0])
print("The number of columns is:", dataTest.shape[1])
# 20. Generate a new Series called lpsaTrain and containing only the values of variable lpsa in which the train variable has value “T”
lpsaTrain = data["lpsa"].loc[train1["train"] == 1].copy()
# • How many valuses does lpsaTrain have?
print(len(lpsaTrain))
# 21. Generate a new Series called lpsaTest and containing only the values of variable lpsa in which the train variable has value “F”
lpsaTest = data["lpsa"].loc[train1["train"] == 0].copy()
# • How many valuses does lpsaTest have?
print(len(lpsaTest))
# 22. Show the correlation matrix among all the variables in dataTrain
# • Hint: use the correct method in DataFrame
# • Hint: check if the values in the matrix correspond to those in Table 3.1 of the book
correlation_matrix = dataTrain.corr()
correlation_matrix
# 23. Drop the column lpsa from the dataTrain DataFrame and save the result in a new DataFrame called predictorsTrain
predictorsTrain = dataTrain.drop(["lpsa"], axis=1)
# 24. Drop the column lpsa from the dataTest DataFrame and save the result in a new DataFrame called predictorsTest
predictorsTest = dataTest.drop(["lpsa"], axis=1)
# 25. Generate a new DataFrame called predictorsTrain_std and containing the standardized variables of DataFrame predictorsTrain
# - Hint: compute the mean of each column and save them in variable predictorsTrainMeans
#
# - Hint: compute the standard deviation of each column and save them in variable predictorsTrainSt
#
# - Hint: compute the standardization of each variable by the formula (predictorsTrainpredictorsTrainMeans)/predictorsTrainStd
predictorsTrain_std = (predictorsTrain - predictorsTrain.mean()) / predictorsTrain.std()
"""with OLS there is no need to standardize the model (without we could stress better the fact that a variable is
more important in terms of correlation),
if we use ridge, lasso we must use it because it would weight too much if the variables are too far away
from each other
"""
predictorsTrain_std.head()
# 26. Show the histogram of each variables of predictorsTrain_std in a single figure
# - Use argument figsize to enlarge the figure if needed
# - Hint: which kind of difference can you see in the histograms?
predictorsTrain_std.hist(figsize=(10, 10))
# 27. Generate a linear regression model using predictorsTrain_std as dependent variables and lpsaTrain as independent variable
# - Hint: find a function for linear regression model learning in sklearn (fit)
X = predictorsTrain_std
Y = lpsaTrain
reg = LinearRegression().fit(X, Y) # create the object and create it through fit
#
# - How do you set parameter fit_intercept? Why?
reg_intercept = LinearRegression(fit_intercept=False)
reg_intercept.fit(predictorsTrain_std, lpsaTrain)
# We set it to zero
# To set parameter fit_intercept
# Default fit_intercept = True, the y-intercept is determined by the line of best fit
# fit_intercept = False, sets the y-intercept to 0
#
# - How do you set parameter normalize? Why? Can this parameter be used to simplify the generation of the predictor matrix?
reg_normalize = LinearRegression(normalize=True)
reg_normalize.fit(predictorsTrain_std, lpsaTrain)
# To set parameter normalize
# Default normalize = False
# If True, the regressors X will be normalized before regression.
# This parameter is ignored when fit_intercept is set to False.
# reg_normalize = LinearRegression(normalize = True)
# reg_normalize.fit(predictorsTrain, lpsaTrain)
# 28. Show the parameters of the linear regression model computed above. Compare the
# parameters with those shown in Table 3.2 of the book (page 50)
reg.coef_
# >|lcavol|lweight|age|lpbh|svi|lcp|gleason|pgg45
# |:---:|:---:|:---:|:---:|:---:|:---:|:---:|:---:|
# |0.68|0.26|-0.14|0.21|0.31|-0.29|-0.02|0.27|
# The first two coefficients are a little bit smaller than the ones computed by the book.
# 29. Compute the coefficient of determination of the prediction
reg.score(X, Y)
# 30. Compute the standard errors, the Z scores (Student’s t statistics) and the related p-values
# • Hint: use library statsmodels instead of sklearn
# • Hint: compare the results with those in Table 3.2 of the book (page 50)
import statsmodels.api as sm
Xc = sm.add_constant(
predictorsTrain
) # to add the columns of ones to the matrix on the right in order to get the intercept
model = sm.OLS(lpsaTrain, Xc).fit()
print(model.summary())
# Exercise 3: Analysis of Prostate Cancer dataset – variable subset selection
# Please, execute the following tasks and provide answers to the proposed questions.
# 1. Open your kernel SL_EX2_ProstateCancer_Surname in Kaggle
# 2. Generate a copy called SL_EX3_SubsetSelection_Surname by the Fork button
# 3. Starting from the ols models achieved in the last steps, perform best-subset selection.
# - Generate one model (using only the 67 samples in the training set) for each combination of the 8 variables available
# - For each model compute the RSS on training and test set, the number of variables and the $R^2$ of the model
# - Save these numbers in suitable data structures
# - Suggestion: use itertools.combinations (https://docs.python.org/3/library/itertools.html#itertools.combinations) to generate all variable combinations
# - Suggestion: consider using the statsmodels library(https://www.statsmodels.org/stable/index.html) to generate the models. It provides direct access to evaluation measures as Sum of Squared Residuals (SSR), R^2, AIC, BIC
from sklearn.metrics import mean_squared_error
import scipy.stats as stats
from itertools import combinations
def linear_reg(X, Y): # function to prepare the R_squared and the RSSs
model_k = LinearRegression(fit_intercept=True).fit(X, Y)
RSS = mean_squared_error(Y, model_k.predict(X)) * len(Y)
R_squared = model_k.score(X, Y)
z_score = stats.zscore(X)
return RSS, R_squared, z_score
from tqdm import tnrange, tqdm_notebook
# model for the training set
X = predictorsTrain
Y = lpsaTrain
# model for the test set
X1 = predictorsTest
Y1 = lpsaTest
k = 8 # possible variables
a = (
(lpsaTrain - lpsaTrain.mean()) ** 2
).sum() # RSS of the null model of the training set
b = ((lpsaTest - lpsaTest.mean()) ** 2).sum() # RSS of the null model of the test set
# preparing the lists to store the datas and putting in position [0] the information of the null model
RSS_list, RSS_lista, R_squared_list, variables_list = [a], [b], [0], [""]
num_variables = [0] # because the first model has zero variables
for k in tnrange(1, len(X.columns) + 1):
for comb in combinations(
X.columns, k
): # information of the training set for each combination of variables
results = linear_reg(X[list(comb)], Y)
RSS_list.append(results[0])
R_squared_list.append(results[1])
variables_list.append(comb)
num_variables.append(len(comb))
for comb in combinations(
X1.columns, k
): # RSS of the test set for each combination of variables
resultss = linear_reg(X1[list(comb)], Y1)
RSS_lista.append(resultss[0])
# the dataframe with 2^8=256 combinations of variables
df = pd.DataFrame(
{
"num_variables": num_variables,
"Rss_train": RSS_list,
"Rss_test": RSS_lista,
"R_squared": R_squared_list,
"Variables": variables_list,
}
)
df.head() # everything is ok
# 4. Generate a chart having the subset size in the x-axis and the RSS for the training set of all models generated at step 3 in the y-axis
import matplotlib.pyplot as plt
plt.figure(figsize=(5, 5))
plt.title("RSS")
ax = []
mins = []
for i in range(1, 9):
rsss = df["Rss_train"].loc[df["num_variables"] == i].copy()
for m in range(len(rsss)):
ax.append(i)
plt.plot(ax, rsss, "r.", label=i)
mins.append(rsss.min())
ax = []
plt.xticks(np.arange(1, 9, 1))
plt.xlabel("Flexibility")
plt.ylabel("RSS")
plt.show()
# We prepare a dataset for best subset selection which contains the best subset of regressors for every subset size.
num_varias = [1, 2, 3, 4, 5, 6, 7, 8]
varsmins = []
for i in range(0, 8):
a = df["Variables"].values[df["Rss_train"] == mins[i]]
varsmins.append(a)
best = pd.DataFrame(
{"num_variables": num_varias, "RSS_best": mins, "Variables": varsmins}
)
best
# 5. Generate a chart having the subset size in the x-axis and the $R^2$ of all models generated at step 3 in the y-axis
plt.figure(figsize=(5, 5))
plt.title("R_squared")
ax = []
for i in range(0, 9):
r2 = df["R_squared"].loc[df["num_variables"] == i].copy()
for m in range(len(r2)):
ax.append(i)
plt.plot(ax, r2, "r.", label=i)
ax = []
plt.xticks(np.arange(0, 9, 1))
plt.xlabel("Flexibility")
plt.ylabel("R_squared")
plt.show()
# 6. Generate a chart having the subset size in the x-axis and the RSS for the test set of all models generated at step 3 in the y-axis (other performance measures can be used, e.g., AIC or BIC)
plt.figure(figsize=(5, 5))
plt.title("Rss_test")
ax = []
for i in range(0, 9):
rsst = df["Rss_test"].loc[df["num_variables"] == i].copy()
for m in range(len(rsst)):
ax.append(i)
plt.plot(ax, rsst, "r.", label=i)
ax = []
plt.xticks(np.arange(0, 9, 1))
plt.xlabel("Flexibility")
plt.ylabel("Rss_test")
plt.show()
# 7. Perform forward selection
# - Start from the empty model
# - Add at each step the variable that minimizes the RSS on the training set (other performance measures can be used, e.g., what happens if you use the RSS on the test set?)
remaining_variables = list(X.columns.values)
variables = []
RSS_list = [np.inf]
variables_list = dict()
for i in range(1, 9):
best_RSS = np.inf
for comb in combinations(remaining_variables, 1):
RSS = linear_reg(X[list(comb) + variables], Y)
if RSS[0] < best_RSS:
best_RSS = RSS[0]
best_feature = comb[0]
# Updating variables for next loop
variables.append(best_feature)
remaining_variables.remove(best_feature)
# Saving values for plotting
RSS_list.append(best_RSS)
variables_list[i] = variables.copy()
RSS_list.remove(RSS_list[0])
listt = list(variables_list.values())
num_variablesss = [1, 2, 3, 4, 5, 6, 7, 8]
forward_train = pd.DataFrame(
{"num_variables": num_variablesss, "RSS_forward": RSS_list, "Variables": listt}
)
forward_train
# We decide not to take the last model, because with the variable "gleason" there is no substantial difference.
# We now test the model on the test set:
remaining_variables1 = list(X1.columns.values)
variables1 = []
RSS_list1 = []
variables_list1 = dict()
for i in range(1, 9):
best_RSS1 = np.inf
for comb in combinations(remaining_variables1, 1):
RSS1 = linear_reg(X1[list(comb) + variables1], Y1)
if RSS1[0] < best_RSS1:
best_RSS1 = RSS1[0]
best_variable1 = comb[0]
# Updating variables for next loop
variables1.append(best_variable1)
remaining_variables1.remove(best_variable1)
# Saving values for plotting
RSS_list1.append(best_RSS1)
variables_list1[i] = variables1.copy()
# RSS_list1.remove(RSS_list1[0])
listt1 = list(variables_list1.values())
forward_test = pd.DataFrame(
{"num_variables": num_variablesss, "RSS": RSS_list1, "Variables": listt1}
)
# print('Forward selection')
# print('Number of variables |', 'Variables |', 'RSS')
# display([(i,variables_list1[i], round(RSS_list1[i],5)) for i in range(1,9)])
forward_test
# If we use the RSS on the test set, the model with two variables becomes the one with "lcavol" and "svi" instead of "lcavol" and "lweight" that we got on training set. From the model $M_1$ to the model $M_2$ we have a jump in the RSS, while adding other variables doesn't change that much the results.
# 8. Generate a chart having the subset size in the x-axis and the RSS for the test set of the models generated at step 7 in the y-axis
plt.figure(figsize=(5, 5))
plt.title("Rss_test obtained with forward selection")
x1 = np.linspace(1, 8, 8)
plt.plot(x1, RSS_list1, "k-", label="0")
plt.plot(x1, RSS_list1, "r*", label="0")
plt.xticks(np.arange(1, 9, 1))
plt.xlabel("Flexibility")
plt.ylabel("Rss_test")
plt.show()
# 9. Perform backward selection
# - Start from the full model
# - Remove at each step the variable that minimizes the RSS (other performance measures can be used, e.g., what happens if you use the RSS on the test set?)
remaining_variables = list(X.columns.values) # list of variables to be used
variables = []
RSS_list = []
variables_list = dict()
for i in range(1, 8):
best_RSS = np.inf
for comb in combinations(remaining_variables, 8 - i):
RSS = linear_reg(
X[list(comb)], Y
) # function to do the fitted linreg RSS=[RSS, R_squared]
if RSS[0] < best_RSS:
best_RSS = RSS[0] # save this RSS value
best_feature = comb # save this combination of variables
variables.append(best_feature) # save this combination to be printed at the end
remaining_variables = best_feature # we remove the worst variable
RSS_list.append(best_RSS) # we save the RSS to be printed at the end
num_variablesss = [8, 7, 6, 5, 4, 3, 2, 1]
a = linear_reg(X, Y)
RSS_list.insert(0, a[0])
variables.insert(0, X.columns.values) # nb sistema
backward = pd.DataFrame(
{"num_variables": num_variablesss, "RSS_backward": RSS_list, "Variables": variables}
)
backward
# 10. Generate a chart having the subset size in the x-axis and the RSS for the test set of the models generated at step 9 in the y-axis
# we need the backward one
plt.figure(figsize=(5, 5))
plt.title("Rss_test obtained with backward selection")
x1 = np.linspace(8, 1, 8)
plt.plot(x1, RSS_list, "b-", label="0")
plt.plot(x1, RSS_list, "r*", label="0")
plt.xticks(np.arange(1, 9, 1))
plt.xlabel("Flexibility")
plt.ylabel("Rss_test")
plt.show()
# 11. Perform backward selection using the z-score as a statistics for selecting the predictor to drop
# - Start from the full model
# - Remove at each step the variable having the smallest Z-score (which library is more suitable for this purpose?)
remaining_variables = list(X.columns.values) # list of variables to be used
variables = []
RSS_list = []
variables_list = dict()
for i in range(1, 8):
best_RSS = np.inf
for comb in combinations(remaining_variables, 8 - i):
RSS = linear_reg(
X[list(comb)], Y
) # function to do the fitted linreg RSS=[RSS, R_squared]
if RSS[0] < best_RSS:
best_RSS = RSS[0] # save this RSS value
best_feature = comb # save this combination of variables
variables.append(best_feature) # save this combination to be printed at the end
remaining_variables = best_feature # we remove the worst variable
RSS_list.append(best_RSS) # we save the RSS to be printed at the end
num_variablesss = [8, 7, 6, 5, 4, 3, 2, 1]
a = linear_reg(X, Y)
RSS_list.insert(0, a[0])
variables.insert(0, X.columns.values) # nb sistema
backward = pd.DataFrame(
{"num_variables": num_variablesss, "RSS_backward": RSS_list, "Variables": variables}
)
remaining_variables_z = list(X.columns.values) # list of variables to be used
variables_z = []
z_scores_list = []
variables_list_z = dict()
for i in range(1, 8):
best_z = -np.inf
for comb in combinations(remaining_variables_z, 8 - i):
z_score = abs(stats.zscore(X[list(comb)])).sum()
if z_score > best_z:
best_z = z_score
best_feature_z = comb
variables_z.append(best_feature_z)
remaining_variables_z = best_feature_z
z_scores_list.append(best_z)
b = stats.zscore(X.column.values)
z_scores_list.insert(0, b)
variables_z.insert(0, X.columns.values) # nb sistema
backward_z = pd.DataFrame(
{
"num_variables": num_variablesss,
"Z_scores": z_scores_list,
"Variables": variables_z,
}
)
z_score
# 12. Generate a chart having the subset size in the x-axis and the RSS for the test set of the models generated at step 11 in the y-axis. Compare it with the chart generated at point 10.
fig, axes = plt.subplots(nrows=1, ncols=2)
axes[0].plot(x1, RSS_list, "b-", label="0")
axes[0].plot(x1, RSS_list, "r*", label="0")
plt.xticks(np.arange(1, 9, 1))
plt.xlabel("Flexibility")
plt.ylabel("Rss")
axes[1].plot(x1, RSS_list, "b-", label="0")
axes[1].plot(x1, RSS_list, "r*", label="0")
plt.xticks(np.arange(1, 9, 1))
plt.xlabel("Flexibility")
plt.ylabel("Z_scores")
plt.show()
# 13. [Advanced] Select the best models using cross-validation:
# - For each of the three methods evaluated (i.e., best subset selection, forward selection, backward selection)
#
# - For each model size (i.e., number of variables)
#
# - select the models having the minimum RSS error on the training set
#
# - compute, for each model, its 5-fold cross-validation Mean Squared Error
#
# - Use the cross-validation Mean Squared Errors to identify the best model.
best1 = best[["num_variables", "RSS_best"]].copy()
forward1 = forward_train[["num_variables", "RSS_forward"]].copy() # vai a modificare
backward1 = backward[["num_variables", "RSS_backward"]].copy() # vai a modificare
part1 = pd.merge(left=best1, right=forward1)
part2 = pd.merge(left=part1, right=backward1)
part2
# How we expected to, every model has the same RSS values, because we took always the same variables for each size.
from sklearn import svm
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import KFold
from sklearn.linear_model import LogisticRegression
cv = KFold(n_splits=5)
# create model
model = LogisticRegression()
# evaluate model
scores = cross_val_score(model, X, Y)
# report performance
# print('Accuracy: %.3f (%.3f)' % (mean(scores), std(scores)))
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import seaborn as sns
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import itertools
import os
import pickle
import warnings
import argparse
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import plotly.express as px
import plotly.graph_objects as go
from plotly.subplots import make_subplots
from sklearn.model_selection import StratifiedKFold
from tqdm.notebook import tqdm
import seaborn as sns
warnings.filterwarnings("ignore")
INPUT_DIR = "/kaggle/input/amp-parkinsons-disease-progression-prediction"
train_clinical_df = pd.read_csv(os.path.join(INPUT_DIR, "train_clinical_data.csv"))
train_peptides_df = pd.read_csv(os.path.join(INPUT_DIR, "train_peptides.csv"))
train_proteins_df = pd.read_csv(os.path.join(INPUT_DIR, "train_proteins.csv"))
supplemental_clinical_df = pd.read_csv(
os.path.join(INPUT_DIR, "supplemental_clinical_data.csv")
)
# Get peptide catidates from higher variability of abundance
def get_peptide_cadidates(peptides_df, num_candidates=15):
# Calculate the coefficient of variation (CV) for PeptideAbundance per patient_ids and Peptides
train_peptides_df_agg = peptides_df[["patient_id", "Peptide", "PeptideAbundance"]]
train_peptides_df_agg = train_peptides_df_agg.groupby(["patient_id", "Peptide"])[
"PeptideAbundance"
].aggregate(["mean", "std"])
train_peptides_df_agg["CV_PeptideAbundance[%]"] = (
train_peptides_df_agg["std"] / train_peptides_df_agg["mean"] * 100
)
# Mean CV value of Peptides
abundance_cv_mean = (
train_peptides_df_agg.groupby("Peptide")["CV_PeptideAbundance[%]"]
.mean()
.reset_index()
)
abundance_cv_mean = abundance_cv_mean.sort_values(
by="CV_PeptideAbundance[%]", ascending=False
).reset_index(drop=True)
# Get peptide candidates
peptide_candidates = abundance_cv_mean.loc[: num_candidates - 1, "Peptide"]
return peptide_candidates
# Get protein candidate from proteins with higher variability of NPX
def get_protein_cadidates(proteins_df, num_candidates=15):
# Calculate the coefficient of variation (CV) for NPX per patient_ids and UniProt
train_proteins_df_agg = train_proteins_df[["patient_id", "UniProt", "NPX"]]
train_proteins_df_agg = train_proteins_df_agg.groupby(["patient_id", "UniProt"])[
"NPX"
].aggregate(["mean", "std"])
train_proteins_df_agg["CV_NPX[%]"] = (
train_proteins_df_agg["std"] / train_proteins_df_agg["mean"] * 100
)
# Mean CV value for UniProt
NPX_cv_mean = (
train_proteins_df_agg.groupby("UniProt")["CV_NPX[%]"].mean().reset_index()
)
NPX_cv_mean = NPX_cv_mean.sort_values(by="CV_NPX[%]", ascending=False).reset_index(
drop=True
)
# Get peptide candidates
protein_candidates = NPX_cv_mean.loc[: num_candidates - 1, "UniProt"]
return protein_candidates
def preprocessing_train_data(
clinical_df, peptides_df, proteins_df, peptide_candidates, protein_candidates
):
# Create dataframe with columns:['visit_id', 'peptide_candidata1', 'peptide_candidata2', ...]
def create_peptide_candidate_df(peptides_df, peptide_candidates):
peptide_candidate_df = peptides_df.query(f"Peptide in @peptide_candidates")
visit_ids = clinical_df["visit_id"].unique()
peptide_dict_list = []
for visit_id in visit_ids:
peptides_df = peptide_candidate_df.query(f'visit_id=="{visit_id}"')
peptides = peptides_df["Peptide"].values
PeptideAbundances = peptides_df["PeptideAbundance"].values
peptide_dict = dict(
zip(peptide_candidates, [np.nan] * len(peptide_candidates))
)
for peptide, PeptideAbundance in zip(peptides, PeptideAbundances):
peptide_dict[peptide] = PeptideAbundance
peptide_dict["visit_id"] = visit_id
peptide_dict_list.append(peptide_dict)
return pd.DataFrame(peptide_dict_list)
# Create dataframe with columns:['visit_id', 'protein_candidata1', 'protein_candidata2', ...]
def create_protein_candidate_df(proteins_df, protein_candidates):
protein_candidate_df = proteins_df.query(f"UniProt in @protein_candidates")
visit_ids = clinical_df["visit_id"].unique()
protein_dict_list = []
for visit_id in visit_ids:
proteins_df = protein_candidate_df.query(f'visit_id=="{visit_id}"')
UniProts = proteins_df["UniProt"].values
NPXs = proteins_df["NPX"].values
protein_dict = dict(
zip(protein_candidates, [np.nan] * len(protein_candidates))
)
for UniProt, NPX in zip(UniProts, NPXs):
protein_dict[UniProt] = NPX
protein_dict["visit_id"] = visit_id
protein_dict_list.append(protein_dict)
return pd.DataFrame(protein_dict_list)
df = clinical_df.copy()
peptide_candidate_df = create_peptide_candidate_df(
train_peptides_df, peptide_candidates
)
protein_candidate_df = create_protein_candidate_df(
train_proteins_df, protein_candidates
)
df = pd.merge(df, peptide_candidate_df, on="visit_id", how="left")
df = pd.merge(df, protein_candidate_df, on="visit_id", how="left")
if "upd23b_clinical_state_on_medication" in df.columns:
df.drop(["upd23b_clinical_state_on_medication"], axis=1, inplace=True)
# df['upd23b_clinical_state_on_medication'] = \
# df['upd23b_clinical_state_on_medication'].fillna('Off')
# df = pd.get_dummies(df, columns=['upd23b_clinical_state_on_medication'])
return df
train_peptides_df = pd.read_csv(os.path.join(INPUT_DIR, "train_peptides.csv"))
train_proteins_df = pd.read_csv(os.path.join(INPUT_DIR, "train_proteins.csv"))
NUM_CANDIDATES = 30
peptide_candidates = get_peptide_cadidates(
train_peptides_df, num_candidates=NUM_CANDIDATES
)
protein_candidates = get_protein_cadidates(
train_proteins_df, num_candidates=NUM_CANDIDATES
)
print(protein_candidates)
print(peptide_candidates)
train_clinical_df = pd.read_csv(os.path.join(INPUT_DIR, "train_clinical_data.csv"))
train_peptides_df = pd.read_csv(os.path.join(INPUT_DIR, "train_peptides.csv"))
train_proteins_df = pd.read_csv(os.path.join(INPUT_DIR, "train_proteins.csv"))
NUM_CANDIDATES = 15
peptide_candidates = get_peptide_cadidates(
train_peptides_df, num_candidates=NUM_CANDIDATES
)
protein_candidates = get_protein_cadidates(
train_proteins_df, num_candidates=NUM_CANDIDATES
)
train_df = preprocessing_train_data(
train_clinical_df,
train_peptides_df,
train_proteins_df,
peptide_candidates,
protein_candidates,
)
print("train_df:")
display(
train_df.head(20).style.set_properties(
**{
"background-color": "#212636",
"color": "white",
"border": "1.5px solid white",
}
)
)
def create_X_y_train_dataset(df, updrs_part, plus_month):
# df: train_df, created above
# updrs_part: 1 to 4
# plus_month: 0, 6, 12, 24
df_ = df.dropna(subset=[f"updrs_{updrs_part}"])
X_visit_ids = []
y_visit_ids = []
patient_ids = df["patient_id"].unique()
for i, patient_id in enumerate(patient_ids):
patient_df = df_[df_["patient_id"] == patient_id]
plus_months = patient_df["visit_month"] + plus_month
plus_months = patient_df.query("visit_month in @plus_months")["visit_month"]
original_months = plus_months - plus_month
patient_id = str(patient_id)
X_visit_id = [
patient_id + "_" + str(original_month) for original_month in original_months
]
y_visit_id = [patient_id + "_" + str(plus_month) for plus_month in plus_months]
X_visit_ids.extend(X_visit_id)
y_visit_ids.extend(y_visit_id)
X = df_.query("visit_id in @X_visit_ids")
X = X.drop(["patient_id", "updrs_1", "updrs_2", "updrs_3", "updrs_4"], axis=1)
X.reset_index(drop=True, inplace=True)
y = df_.query("visit_id in @y_visit_ids")
y = y[["visit_id", f"updrs_{updrs_part}"]]
y.reset_index(drop=True, inplace=True)
return X, y
def create_X_y_dict(df):
X_dict = {}
y_dict = {}
for updrs_part in tqdm([1, 2, 3, 4]):
for plus_month in [0, 6, 12, 24]:
X, y = create_X_y_train_dataset(df, updrs_part, plus_month)
key = f"updrs{updrs_part}_plus_month{plus_month}"
X_dict[key] = X
y_dict[key] = y
return X_dict, y_dict
X_dict, y_dict = create_X_y_dict(train_df)
# For example,
updrs_part = 2
plus_month = 6
key = f"updrs{updrs_part}_plus_month{plus_month}"
print(f"e.g. {key}\n")
X = X_dict[key].copy()
y = y_dict[key].copy()
print("X:")
display(
X.head(10).style.set_properties(
**{
"background-color": "#212636",
"color": "white",
"border": "1.5px solid white",
}
)
)
print("y:")
display(
y.head(10).style.set_properties(
**{
"background-color": "#212636",
"color": "white",
"border": "1.5px solid white",
}
)
)
|
from scipy.optimize import dual_annealing
# objective function
def objective(v):
x, y = v
return (x**2 + y - 11) ** 2 + (x + y**2 - 7) ** 2
# define range for input
r_min, r_max = -5.0, 5.0
# define the bounds on the search
bounds = [[r_min, r_max], [r_min, r_max]]
# perform the simulated annealing search
result = dual_annealing(objective, bounds)
# summarize the result
print("Status : %s" % result["message"])
print("Total Evaluations: %d" % result["nfev"])
# evaluate solution
solution = result["x"]
evaluation = objective(solution)
print("Solution: f(%s) = %.5f" % (solution, evaluation))
|
# # Data Cleaning (ETL)
# **Input**: Raw data stored in MongoDB
# **Output**: Clean data stored in PostgreSQL
# The Data Cleaning step consists in multiple data transformations with the goal of making data "ready to talk" :)
# Some of the transformations involved include:
# - Convert from dictionary to tabular data structure
# - Remove duplicates (intra-series and inter-series)
# - Check and convert data types and formats
# - Null handling
# - Data interpolation (we have data collected with different frequencies and we want to uniform all data to have monthly frequency)
# After the step is done, we want to save our transformed data in PostgreSQL, that serves as our Data Warehouse.
# We evaluated different options to choose where to save our data:
# - AWS Aurora Serverless / On-demand DB instance
# - AWS RDS PostgreSQL On-demand DB instance
# - AWS Redshift
# Performance-wise redshift would probably be the better choice, it is a columnar database optimized for analytics able to do parallel processing.
# We could also use Apache Spark for data transformation to improve performance.
# In this project, as said before, we tried to keep things simple. So we choose PostgreSQL as our data destination and the pandas library as our ETL engine.
# ### FRED Series Selection
# First thing we export all the ids, titles and notes of the series collected in MongoDB.
# - We had this on our MongoDB local instance, because it was too much data for our free version MongoDB Atlas
# - In this Kaggle notebook we have imported the "popularity>30" subset as a .csv dataset)
from pymongo import MongoClient
import pandas as pd
def import_from_mongo_unused():
from kaggle_secrets import UserSecretsClient
user_secrets = UserSecretsClient()
mongo_connection_string = user_secrets.get_secret("mongo_connection_string")
client = MongoClient(mongo_connection_string)
database = client["portfolio"]
collection = database["fred_series"]
datasets = collection.find({})
df_l = []
for d in datasets:
_id = d["_id"] if "_id" in d else None
title = d["title"] if "title" in d else None
notes = d["notes"] if "notes" in d else None
if d["popularity"] >= 30:
df_l.append({"id": _id, "title": title, "notes": notes})
df = pd.DataFrame(df_l, columns=["id", "title", "notes"])
print("# Series:", len(df))
df.head()
df = pd.read_csv("/kaggle/input/fredseries/fred_series.csv")
df
# To shortlist the series available from the FRED we took the following decisions:
# - Median over Mean
# - Seasonally Adjusted over Non Seasonally Adjusted
# - Series taken from OECD excluded
# - Single state data excluded (we are interested only in federal USA)
# - Monthly granularity over others
# - Industry/sector detailed data excluded
# - DISCONTINUED series excluded
# - Countries other than USA excluded
# - Series with data starting after 1995 excluded
# We then save a list of the "good" series in a new document in MongoDB.
from kaggle_secrets import UserSecretsClient
user_secrets = UserSecretsClient()
connection_string = user_secrets.get_secret("mongo_connection_string")
client = MongoClient(connection_string)
database = client["portfolio"]
collection = database["fred_datasets"]
datasets = collection.find({"_id": "shortlist_fred"}).next()["shortlist"]
print("# Series:", len(datasets))
# ### OECD Series Selection
# As we did for FRED, we export all the different combinations of indicator, measure, subject. In this case the dataset is only one (DP_LIVE), but contains many different combinations for each indicator. We want to shortlist those.
oecd_collection = database["oecd_datasets"]
dataset = oecd_collection.find({"_id": "DP_LIVE"}).next()
def mongo_to_dataframe(dataset):
data = dataset["dataset"]
values = []
obs = data["dataSets"][0]["observations"]
for e in obs:
i = [int(x) for x in e.split(":")]
i.append(obs[e][0])
values.append(i)
obs_cols = data["structure"]["dimensions"]["observation"]
replacement = {}
cols = []
for c in obs_cols:
n = c["name"]
replacement[n] = {i: x["name"] for i, x in enumerate(c["values"])}
cols.append(n)
df = pd.DataFrame(values, columns=cols + ["Value"])
for c in cols:
df[c] = df[c].replace(replacement[c])
return df
df = mongo_to_dataframe(dataset)
df = df[["Indicator", "Subject", "Measure"]].drop_duplicates()
print("# Series:", len(df))
df.head()
# To shortlist the series available from the OECD we took the following decisions:
# - General "Subject" over segmented "Subject" (ex. Total over Male/Female, Financial/Non-financial)
# - Industry/sector detailed data excluded
# - Series with data starting after 1995 excluded
# We then save a list of the "good" series in a new document in MongoDB.
datasets = oecd_collection.find({"_id": "oecd_shortlist"}).next()["data"]
print("# Series:", len(datasets))
# ### FRED + OECD Series union
# At this point we "merge" the features coming from the FRED and the OECD and remove the duplicate ones.
# When in doubt we keep the one that has more data or the one with the lowest granularity (eg. monthly over quarterly).
# We save the new shortlisted list of series in 2 documents name fred_shortlist_v2 and oecd_shortlist_v2.
fred_collection = database["fred_datasets"]
fred_shortlist = fred_collection.find({"_id": "shortlist_fred_v2"}).next()["shortlist"]
print("# Series FRED:", len(fred_shortlist))
oecd_shortlist = oecd_collection.find({"_id": "oecd_shortlist_v2"}).next()["data"]
print("# Series OECD:", len(oecd_shortlist))
# ## ETL
# Now we are ready for the ETL process (Extract, Transform, Load)
# - Extract (selected) data from MongoDB, that contains our raw data
# - Transform data using pandas
# - Load into postgreSQL
# ### FRED
from portfoliooptimization_helper import request_with_retries
fred_documents = fred_collection.find({"_id": {"$in": fred_shortlist}})
fred_api_key = user_secrets.get_secret("fred_api_key")
def retrieve_series_metadata(series_id, api_key):
url = (
f"https://api.stlouisfed.org/fred/series?series_id={series_id}"
f"&api_key={api_key}&file_type=json"
)
r = request_with_retries(url)
return r.json()["seriess"][0]
# We create a transformed Dataframe for each document and then concat them together
list_of_dfs = []
for d in fred_documents:
# Retrieve Metadata (Unit of measure and frequency)
try:
metadata = retrieve_series_metadata(d["_id"], fred_api_key)
except:
continue
units = metadata["units"]
frequency = metadata["frequency"]
# Build Dataframe
tmp = []
for o in d["observations"]:
tmp.append({key: o[key] for key in ["date", "value"]})
df = pd.DataFrame(tmp, columns=["date", "value"])
# Convert Types
df["value"] = pd.to_numeric(df["value"], errors="coerce")
df["date"] = pd.to_datetime(df["date"])
# Adjust % values
if units is not None and "Percent" in units:
df["value"] /= 100
# Interpolate values
# Starting from weekly frequency we take the mean to get the monthly value
# Starting from higher granularity we use ffill to "copy" the value in the missing months.
# We use this method instead of linear interpolation because Quarterly and Annual data is
# already made up from the average of the monthly observations! This is a way to keep at
# least the same average before and after the transformation
if "Weekly" in frequency:
df = df.set_index(["date"]).resample("MS").mean().reset_index()
if "Quarterly" in frequency or "Annual" in frequency:
df = df.set_index(["date"]).resample("MS").ffill().reset_index()
# Remove rows with any null present
df = df.dropna()
df["name"] = d["_id"]
list_of_dfs.append(df)
# For this example we take only the first one, because of the need for making http requests
break
df = pd.concat(list_of_dfs)
df["source"] = "FRED"
print("# features", len(df["name"].unique()))
print("# rows:", len(df))
df.tail()
# ### OECD
dataset = oecd_collection.find({"_id": "DP_LIVE"}).next()
df = mongo_to_dataframe(dataset)
# Transform date
# monthly data has format 'YYYY-MM-DD' and is already fine
# quarterly data has format 'YYYY-Q1' and must be transformed
# yearly data has format 'YYYY' and must be transformed
from portfoliooptimization_helper import oecd_time_to_datetime
df["Time"] = df.apply(lambda row: oecd_time_to_datetime(row, "Time"), axis=1)
# We create a transformed Dataframe for each combination of indicator,subject,measure
# and then concat them together
list_of_dfs = []
for s in oecd_shortlist:
# Take only shortlisted combination of indicator,subject,measure
temp_df = df[
(df["Indicator"] == s["INDICATOR"])
& (df["Subject"] == s["SUBJECT"])
& (df["Measure"] == s["MEASURE"])
]
# Interpolate values
# Each combination can have one or multiple different frequencies.
# If we have the monthly frequency, we just take that
# If we don't, we use the same strategy as for FRED, we use ffill to "copy" the value
frequencies = temp_df["Frequency"].unique()
if "Monthly" in frequencies:
temp_df = temp_df[temp_df["Frequency"] == "Monthly"]
elif "Quarterly" in frequencies:
temp_df = temp_df[temp_df["Frequency"] == "Quarterly"]
temp_df = temp_df.set_index(["Time"]).resample("MS").ffill().reset_index()
elif "Annual" in frequencies:
temp_df = temp_df[temp_df["Frequency"] == "Annual"]
temp_df = temp_df.set_index(["Time"]).resample("MS").ffill().reset_index()
list_of_dfs.append(temp_df)
df = pd.concat(list_of_dfs)
df = df.dropna()
# Adjust % values
def convert_percentage(row):
for el in ["%", "percentage"]:
if el in row["Measure"].lower():
return row["Value"] / 100
return row["Value"]
df["Value"] = df.apply(lambda row: convert_percentage(row), axis=1)
# Concat combination into a single column
def concat_column_values(row, columns):
return " | ".join(list(row[columns]))
df["name"] = df.apply(
lambda row: concat_column_values(row, ["Indicator", "Subject", "Measure"]), axis=1
)
# Drop useless columns
df = df.drop(["Indicator", "Subject", "Measure", "Country", "Frequency"], axis=1)
df = df.reset_index(drop=True)
df["source"] = "OECD"
df = df.rename(columns={"Time": "date", "Value": "value"})
df = df[["date", "value", "name", "source"]]
print("# features", len(df["name"].unique()))
print("# rows:", len(df))
df.tail()
# ### YahooFinance
# Take data from MongoDB
yf_collection = database["yf_target_datasets"]
yf_datasets = yf_collection.find({})
# Build Dataframe
list_of_df = []
for d in yf_datasets:
tmp = []
for dd in d["data"]:
tmp.append({"date": dd["Date"], "value": dd["Close"], "name": dd["ticker"]})
list_of_df.append(pd.DataFrame(tmp, columns=["date", "value", "name"]))
df = pd.concat(list_of_df)
# Convert Types
df["value"] = pd.to_numeric(df["value"], errors="coerce")
df["date"] = pd.to_datetime(df["date"], unit="ms")
# Drop nulls
df = df.dropna()
df["source"] = "yahoo_finance"
df = df[["date", "value", "name", "source"]]
# For the most recent month we get 2 values, 1 for the start of month, 1 for today's date.
# We just want the start of month
df = (
df.set_index(["date"])
.groupby(["name", "source"])
.resample("MS")
.mean()
.reset_index()
)
print("# targets", len(df["name"].unique()))
print("# rows:", len(df))
df.tail()
# ### Investing.com
# Take data from MongoDB
investing_collection = database["investing_target_datasets"]
investing_datasets = investing_collection.find({})
# Build Dataframe
list_of_df = []
for d in investing_datasets:
name = d["_id"]
tmp = []
for dd in d["data"]:
tmp.append({"date": dd["Date"], "value": dd["Price"], "name": name})
list_of_df.append(pd.DataFrame(tmp, columns=["date", "value", "name"]))
df = pd.concat(list_of_df)
# Convert Types
df["value"] = df["value"].astype(str)
df["value"] = pd.to_numeric(df["value"].str.replace(",", ""), errors="coerce")
df["date"] = pd.to_datetime(df["date"])
# Drop nulls
df = df.dropna()
df["source"] = "investing"
df = df[["date", "value", "name", "source"]]
# For the most recent month we get 2 values, 1 for the start of month, 1 for today's date.
# We just want the start of month
df = (
df.set_index(["date"])
.groupby(["name", "source"])
.resample("MS")
.mean()
.reset_index()
)
print("# targets", len(df["name"].unique()))
print("# rows:", len(df))
df.tail()
# ### Storing Data in PostgreSQL
# We save transformed data in PostgreSQL on AWS RDS, which we use as a Data Warehouse.
# https://aws.amazon.com/rds/postgresql/
# To store a pandas Dataframe we use psycopg2 library.
# Below an example of how to connect to PostgreSQL and insert a Dataframe into a specific table.
def insert_df_into_table(df, tablename):
# PostgreSQL does not like nan
df = df.where(pd.notnull(df), None)
tuples = [tuple(x) for x in df.to_numpy()]
cols = ",".join(list(df.columns))
query = "INSERT INTO %s(%s) VALUES %%s ON CONFLICT DO NOTHING;" % (tablename, cols)
# This calls psycopg2.connect passing connection parameters
conn = get_connection()
cursor = conn.cursor()
try:
extras.execute_values(cursor, query, tuples)
conn.commit()
except (Exception, psycopg2.DatabaseError) as error:
traceback.print_exc()
eprint(f"Error: {error}")
conn.rollback()
cursor.close()
finally:
cursor.close()
|
# # Goal of the project :
# The "FinTech" company launch there android and iOS mobile base app and want to grow there business.
# But there is problem how to recomended this app and offer who realy want to use it.
# So for that company desided to give free trial to each and every customer for 24 houre
# and collect data from the customers. In this senariao some customer purchase the app and someone not.
# According to this data company want to give special offer to the customer who are not interested to buy without offer
# and grow thre business.
# This is classification problem
# # Import essential libraries
import numpy as np # for numeric calculation
import pandas as pd # for data analysis and manupulation
import matplotlib.pyplot as plt # for data visualization
import seaborn as sns # for data visualization
from dateutil import parser # convert time in date time data type
# # Import dataset & explore
fineTech_appData = pd.read_csv(
"/kaggle/input/fin-tech-app-datasets/FineTech_appData.csv"
)
fineTech_appData.shape
fineTech_appData.head(6) # show fisrt 6 rows of fineTech_appData DataFrame *****code 1
fineTech_appData.tail(6) # show last 6 rows of fineTech_appData DataFrame *****code 2
for i in [1, 2, 3, 4, 5]:
print(fineTech_appData.loc[i, "screen_list"], "\n")
fineTech_appData.isnull().sum() # take summation of null values
fineTech_appData.info() # brief inforamtion about Dataset
fineTech_appData.describe() # give the distribution of numerical variables *****code 3
# Get the unique value of each columns and it's length
features = fineTech_appData.columns
for i in features:
print(
"""Unique value of {}\n{}\nlen is {} \n........................\n
""".format(
i, fineTech_appData[i].unique(), len(fineTech_appData[i].unique())
)
)
fineTech_appData.dtypes
# hour data convert string to int
fineTech_appData["hour"] = fineTech_appData.hour.str.slice(1, 3).astype(int)
# get data type of each columns
fineTech_appData.dtypes
fineTech_appData.columns
# drop object dtype columns
fineTech_appData2 = fineTech_appData.drop(
["user", "first_open", "screen_list", "enrolled_date"], axis=1
)
fineTech_appData2.head(6) # head of numeric dataFrame *****code 4
# # Data Visualization
# ## Heatmap Using Correlation matrix
# Heatmap
plt.figure(figsize=(16, 9)) # heatmap size in ratio 16:9
sns.heatmap(fineTech_appData2.corr(), annot=True, cmap="coolwarm") # show heatmap
plt.title(
"Heatmap using correlation matrix of fineTech_appData2", fontsize=25
) # title of heatmap *****code 5
# ## Pairplot of fineTech_appData2
# Pailplot of fineTech_appData2 Dataset
# %matplotlib qt5 # for show graph in seperate window
sns.pairplot(fineTech_appData2, hue="enrolled") # *****code 6
# ## Countplot of enrolled
# Show counterplot of 'enrolled' feature
sns.countplot(fineTech_appData.enrolled) # *****code 7
# value enrolled and not enrolled customers
print("Not enrolled user = ", (fineTech_appData.enrolled < 1).sum(), "out of 50000")
print("Enrolled user = ", 50000 - (fineTech_appData.enrolled < 1).sum(), "out of 50000")
# ## Histogram of each feature of fineTech_appData2
# plot histogram
plt.figure(figsize=(16, 9)) # figure size in ratio 16:9
features = fineTech_appData2.columns # list of columns name
for i, j in enumerate(features):
plt.subplot(3, 3, i + 1) # create subplot for histogram
plt.title("Histogram of {}".format(j), fontsize=15) # title of histogram
bins = len(fineTech_appData2[j].unique()) # bins for histogram
plt.hist(
fineTech_appData2[j],
bins=bins,
rwidth=0.8,
edgecolor="y",
linewidth=2,
) # plot histogram
plt.subplots_adjust(hspace=0.5) # space between horixontal axes (subplots) *****code 8
for i, j in enumerate(features):
print(i, j)
# ## Correlation barplot with 'enrolled' feature
# show corelation barplot
sns.set() # set background dark grid
plt.figure(figsize=(14, 5))
plt.title("Correlation all features with 'enrolled' ", fontsize=20)
fineTech_appData3 = fineTech_appData2.drop(
["enrolled"], axis=1
) # drop 'enrolled' feature
ax = sns.barplot(
fineTech_appData3.columns, fineTech_appData3.corrwith(fineTech_appData2.enrolled)
) # plot barplot
ax.tick_params(
labelsize=15, labelrotation=20, color="k"
) # decorate x & y ticks font *****code 9
# parsing object data into data time format
fineTech_appData["first_open"] = [
parser.parse(i) for i in fineTech_appData["first_open"]
]
fineTech_appData["enrolled_date"] = [
parser.parse(i) if isinstance(i, str) else i
for i in fineTech_appData["enrolled_date"]
]
fineTech_appData.dtypes
fineTech_appData["time_to_enrolled"] = (
fineTech_appData.enrolled_date - fineTech_appData.first_open
).astype("timedelta64[h]")
# plot histogram
plt.hist(fineTech_appData["time_to_enrolled"].dropna()) # *****code 10
# Plot histogram
plt.hist(fineTech_appData["time_to_enrolled"].dropna(), range=(0, 100)) # *****code 11
# Those customers have enrolled after 48 hours set as 0
fineTech_appData.loc[fineTech_appData.time_to_enrolled > 48, "enrolled"] = 0
fineTech_appData
fineTech_appData.drop(
columns=["time_to_enrolled", "enrolled_date", "first_open"], inplace=True
)
fineTech_appData
# read csv file and convert it into numpy array
fineTech_app_screen_Data = pd.read_csv(
"/kaggle/input/fin-tech-app-datasets/top_screens.csv"
).top_screens.values
fineTech_app_screen_Data
type(fineTech_app_screen_Data)
# Add ',' at the end of each string of 'sreen_list' for further operation.
fineTech_appData["screen_list"] = fineTech_appData.screen_list.astype(str) + ","
fineTech_appData
# string into to number
for screen_name in fineTech_app_screen_Data:
fineTech_appData[screen_name] = fineTech_appData.screen_list.str.contains(
screen_name
).astype(int)
fineTech_appData["screen_list"] = fineTech_appData.screen_list.str.replace(
screen_name + ",", ""
)
# test
fineTech_appData.screen_list.str.contains("Splash").astype(int)
# test
fineTech_appData.screen_list.str.replace("Splash" + ",", "")
# get shape
fineTech_appData.shape
# head of DataFrame
fineTech_appData.head(6) # *****code 12
# remain screen in 'screen_list'
fineTech_appData.loc[0, "screen_list"]
fineTech_appData.screen_list.str.count(",").head(6)
# count remain screen list and store counted number in 'remain_screen_list'
fineTech_appData["remain_screen_list"] = fineTech_appData.screen_list.str.count(",")
# Drop the 'screen_list'
fineTech_appData.drop(columns=["screen_list"], inplace=True)
fineTech_appData
# total columns
fineTech_appData.columns
# take sum of all saving screen in one place
saving_screens = [
"Saving1",
"Saving2",
"Saving2Amount",
"Saving4",
"Saving5",
"Saving6",
"Saving7",
"Saving8",
"Saving9",
"Saving10",
]
fineTech_appData["saving_screens_count"] = fineTech_appData[saving_screens].sum(axis=1)
fineTech_appData.drop(columns=saving_screens, inplace=True)
fineTech_appData
credit_screens = [
"Credit1",
"Credit2",
"Credit3",
"Credit3Container",
"Credit3Dashboard",
]
fineTech_appData["credit_screens_count"] = fineTech_appData[credit_screens].sum(axis=1)
fineTech_appData.drop(columns=credit_screens, axis=1, inplace=True)
fineTech_appData
cc_screens = [
"CC1",
"CC1Category",
"CC3",
]
fineTech_appData["cc_screens_count"] = fineTech_appData[cc_screens].sum(axis=1)
fineTech_appData.drop(columns=cc_screens, inplace=True)
fineTech_appData
loan_screens = [
"Loan",
"Loan2",
"Loan3",
"Loan4",
]
fineTech_appData["loan_screens_count"] = fineTech_appData[loan_screens].sum(axis=1)
fineTech_appData.drop(columns=loan_screens, inplace=True)
fineTech_appData
fineTech_appData.shape
fineTech_appData.info()
fineTech_appData.describe()
# ## Heatmap with correlation matrix of new fineTech_appData
# Heatmap with correlation matrix of new fineTech_appData
plt.figure(figsize=(25, 16))
sns.heatmap(fineTech_appData.corr(), annot=True, linewidth=2) # *****code 13
fineTech_appData.columns
fineTech_appData["ProfileChildren "].unique()
corr_matrix = fineTech_appData.corr()
corr_matrix["ProfileChildren "]
fineTech_appData["ProfileChildren "]
# # Data Preprocessing
# ## Split dataset in Train and Test
clean_fineTech_appData = fineTech_appData
target = fineTech_appData["enrolled"]
fineTech_appData.drop(columns="enrolled", inplace=True)
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
fineTech_appData, target, test_size=0.2, random_state=0
)
print("Shape of X_train = ", X_train.shape)
print("Shape of X_test = ", X_test.shape)
print("Shape of y_train = ", y_train.shape)
print("Shape of y_test = ", y_test.shape)
# take User ID in another variable
train_userID = X_train["user"]
X_train.drop(columns="user", inplace=True)
test_userID = X_test["user"]
X_test.drop(columns="user", inplace=True)
print("Shape of X_train = ", X_train.shape)
print("Shape of X_test = ", X_test.shape)
print("Shape of train_userID = ", train_userID.shape)
print("Shape of test_userID = ", test_userID.shape)
# # Feature Scaling
from sklearn.preprocessing import StandardScaler
sc = StandardScaler()
X_train_sc = sc.fit_transform(X_train)
X_test_sc = sc.transform(X_test)
# # Model Building
# impoer requiede packages
from sklearn.metrics import confusion_matrix, classification_report, accuracy_score
# # Decision Tree
# Decision Tree Classifier
from sklearn.tree import DecisionTreeClassifier
dt_model = DecisionTreeClassifier(criterion="entropy", random_state=0)
dt_model.fit(X_train, y_train)
y_pred_dt = dt_model.predict(X_test)
accuracy_score(y_test, y_pred_dt)
# train with Standert Scaling dataset
dt_model2 = DecisionTreeClassifier(criterion="entropy", random_state=0)
dt_model2.fit(X_train_sc, y_train)
y_pred_dt_sc = dt_model2.predict(X_test_sc)
accuracy_score(y_test, y_pred_dt_sc)
# # KNN Classification
from sklearn.neighbors import KNeighborsClassifier
knn_model = KNeighborsClassifier(
n_neighbors=5,
metric="minkowski",
p=2,
)
knn_model.fit(X_train, y_train)
y_pred_knn = knn_model.predict(X_test)
accuracy_score(y_test, y_pred_knn)
# train with Standert Scaling dataset
knn_model2 = KNeighborsClassifier(
n_neighbors=5,
metric="minkowski",
p=2,
)
knn_model2.fit(X_train_sc, y_train)
y_pred_knn_sc = knn_model2.predict(X_test_sc)
accuracy_score(y_test, y_pred_knn_sc)
# # Naive Bayes
# Naive Bayes
from sklearn.naive_bayes import GaussianNB
nb_model = GaussianNB()
nb_model.fit(X_train, y_train)
y_pred_nb = nb_model.predict(X_test)
accuracy_score(y_test, y_pred_nb)
# train with Standert Scaling dataset
nb_model2 = GaussianNB()
nb_model2.fit(X_train_sc, y_train)
y_pred_nb_sc = nb_model2.predict(X_test_sc)
accuracy_score(y_test, y_pred_nb_sc)
# # Random Forest
# Random Forest Classifier
from sklearn.ensemble import RandomForestClassifier
rf_model = RandomForestClassifier(n_estimators=10, criterion="entropy", random_state=0)
rf_model.fit(X_train, y_train)
y_pred_rf = rf_model.predict(X_test)
accuracy_score(y_test, y_pred_rf)
# train with Standert Scaling dataset
rf_model2 = RandomForestClassifier(n_estimators=10, criterion="entropy", random_state=0)
rf_model2.fit(X_train_sc, y_train)
y_pred_rf_sc = rf_model2.predict(X_test_sc)
accuracy_score(y_test, y_pred_rf_sc)
# # Logistic Regression
# Logistic Regression
from sklearn.linear_model import LogisticRegression
lr_model = LogisticRegression(random_state=0, penalty="l2")
lr_model.fit(X_train, y_train)
y_pred_lr = lr_model.predict(X_test)
accuracy_score(y_test, y_pred_lr)
# train with Standert Scaling dataset
lr_model2 = LogisticRegression(random_state=0, penalty="l2")
lr_model2.fit(X_train_sc, y_train)
y_pred_lr_sc = lr_model2.predict(X_test_sc)
accuracy_score(y_test, y_pred_lr_sc)
# # Support Vector Machine
# Support Vector Machine
from sklearn.svm import SVC
svc_model = SVC()
svc_model.fit(X_train, y_train)
y_pred_svc = svc_model.predict(X_test)
accuracy_score(y_test, y_pred_svc)
# train with Standert Scaling dataset
svc_model2 = SVC()
svc_model2.fit(X_train_sc, y_train)
y_pred_svc_sc = svc_model2.predict(X_test_sc)
accuracy_score(y_test, y_pred_svc_sc)
# # XGBoost
# XGBoost Classifier
from xgboost import XGBClassifier
xgb_model = XGBClassifier()
xgb_model.fit(X_train, y_train)
y_pred_xgb = xgb_model.predict(X_test)
accuracy_score(y_test, y_pred_xgb)
# train with Standert Scaling dataset
xgb_model2 = XGBClassifier()
xgb_model2.fit(X_train_sc, y_train)
y_pred_xgb_sc = xgb_model2.predict(X_test_sc)
accuracy_score(y_test, y_pred_xgb_sc)
# XGB classifier with parameter tuning
xgb_model_pt1 = XGBClassifier(
learning_rate=0.01,
n_estimators=5000,
max_depth=4,
min_child_weight=6,
gamma=0,
subsample=0.8,
colsample_bytree=0.8,
reg_alpha=0.005,
objective="binary:logistic",
nthread=4,
scale_pos_weight=1,
seed=27,
)
xgb_model_pt1.fit(X_train, y_train)
y_pred_xgb_pt1 = xgb_model_pt1.predict(X_test)
accuracy_score(y_test, y_pred_xgb_pt1)
# XGB classifier with parameter tuning
# train with Standert Scaling dataset
xgb_model_pt2 = XGBClassifier(
learning_rate=0.01,
n_estimators=5000,
max_depth=4,
min_child_weight=6,
gamma=0,
subsample=0.8,
colsample_bytree=0.8,
reg_alpha=0.005,
objective="binary:logistic",
nthread=4,
scale_pos_weight=1,
seed=27,
)
xgb_model_pt2.fit(X_train_sc, y_train)
y_pred_xgb_sc_pt2 = xgb_model_pt2.predict(X_test_sc)
accuracy_score(y_test, y_pred_xgb_sc_pt2)
# confussion matrix
cm_xgb_pt2 = confusion_matrix(y_test, y_pred_xgb_sc_pt2)
sns.heatmap(cm_xgb_pt2, annot=True, fmt="g")
plt.title("Confussion Matrix", fontsize=20) # *****code 14
# Clasification Report
cr_xgb_pt2 = classification_report(y_test, y_pred_xgb_sc_pt2)
print("Classification report >>> \n", cr_xgb_pt2)
# Cross validation
from sklearn.model_selection import cross_val_score
cross_validation = cross_val_score(
estimator=xgb_model_pt2, X=X_train_sc, y=y_train, cv=10
)
print("Cross validation of XGBoost model = ", cross_validation)
print("Cross validation of XGBoost model (in mean) = ", cross_validation.mean())
# # Mapping predicted output to the target
final_result = pd.concat([test_userID, y_test], axis=1)
final_result["predicted result"] = y_pred_xgb_sc_pt2
final_result
# # Save the Model
## Pickle
import pickle
# save model
pickle.dump(xgb_model_pt2, open("FineTech_app_ML_model.pickle", "wb"))
# load model
ml_model_pl = pickle.load(open("FineTech_app_ML_model.pickle", "rb"))
# predict the output
y_pred_pl = ml_model_pl.predict(X_test_sc)
# confusion matrix
cm_pl = confusion_matrix(y_test, y_pred_pl)
print("Confussion matrix = \n", cm_pl)
# show the accuracy
print("Accuracy of model = ", accuracy_score(y_test, y_pred_pl))
## Joblib
from sklearn.externals import joblib
# save model
joblib.dump(xgb_model_pt2, "FineTech_app_ML_model.joblib")
# load model
ml_model_jl = joblib.load("FineTech_app_ML_model.joblib")
# predict the output
y_pred_jl = ml_model_jl.predict(X_test_sc)
cm_jl = confusion_matrix(y_test, y_pred_jl)
print("Confussion matrix = \n", cm_jl)
print("Accuracy of model = ", accuracy_score(y_test, y_pred_jl))
|
from __future__ import division
from torchvision import models
# from torchvision import transforms
from PIL import Image
import argparse
import torch
# import torchvision
import torch.nn as nn
import numpy as np
import torchvision.utils as vutils
import matplotlib.pyplot as plt
import torch.optim as optim
from torch.utils.data import DataLoader
from torchvision import datasets
from torchvision import transforms
import torch.utils.data as data
from PIL import Image
import os
import torch.hub
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
print(device)
class ImageDataset(data.Dataset):
def __init__(self, root, transform=None):
self.root = root
self.transform = transform
self.imgs = os.listdir(root)
def __getitem__(self, index):
img_path = os.path.join(self.root, self.imgs[index])
img = Image.open(img_path).convert("RGB")
if self.transform is not None:
img = self.transform(img)
return img
def __len__(self):
return len(self.imgs)
# Define the data transforms and create the data loader
transform = transforms.Compose(
[
transforms.Resize((256, 256)),
transforms.ToTensor(),
transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5)),
]
)
photo = ImageDataset("/kaggle/input/gan-getting-started/photo_jpg", transform=transform)
dataloader_photo = DataLoader(photo, batch_size=1, shuffle=True)
monet = ImageDataset("/kaggle/input/gan-getting-started/monet_jpg", transform=transform)
dataloader_monet = DataLoader(monet, batch_size=1, shuffle=True)
class AdaIN(nn.Module):
def __init__(self, num_features):
super(AdaIN, self).__init__()
self.num_features = num_features
def forward(self, x, style):
# Calculate the mean and variance of the encoded feature maps
b, c, h, w = x.size()
# print(x.size())
x_mean = torch.mean(x.view(b, c, -1), dim=2, keepdim=True)
x_mean = x_mean.unsqueeze(-1)
x_mean = x_mean.expand_as(x)
# print(x_mean.size())
x_var = torch.var(x.view(b, c, -1), dim=2, keepdim=True)
x_var = x_var.unsqueeze(-1)
x_var = x_var.expand_as(x)
# Calculate the mean and variance of the style tensor
style_mean = torch.mean(
style.view(b, self.num_features, -1), dim=2, keepdim=True
)
style_var = torch.var(style.view(b, self.num_features, -1), dim=2, keepdim=True)
style_mean = style_mean.unsqueeze(-1)
style_mean = style_mean.expand_as(x)
style_var = style_var.unsqueeze(-1)
style_var = style_var.expand_as(x)
# Rescale the normalized feature maps using the style statistics
normalized = (x - x_mean) / torch.sqrt(x_var + 1e-8)
transformed = style_var * normalized + style_mean
# Apply instance normalization to the transformed feature maps
transformed = transformed.view(b, c, h, w)
transformed = nn.InstanceNorm2d(c)(transformed)
return transformed
# generator = torch.hub.load('pytorch/vision:main', 'resnet50', pretrained=True)
# generator = models.resnet50(pretrained=True)
class Generator(nn.Module):
def __init__(self):
super(Generator, self).__init__()
# Define the encoder network
self.encoder = nn.Sequential(
nn.Conv2d(3, 32, kernel_size=3, stride=1, padding=1),
nn.InstanceNorm2d(32),
nn.ReLU(),
nn.Conv2d(32, 64, kernel_size=3, stride=2, padding=1),
nn.InstanceNorm2d(64),
nn.ReLU(),
nn.Conv2d(64, 128, kernel_size=3, stride=2, padding=1),
nn.InstanceNorm2d(128),
nn.ReLU(),
nn.Conv2d(128, 256, kernel_size=3, stride=2, padding=1),
nn.InstanceNorm2d(256),
nn.ReLU(),
nn.Conv2d(256, 512, kernel_size=3, stride=2, padding=1),
nn.InstanceNorm2d(512),
nn.ReLU(),
)
# Define the transformer network
self.transformer = AdaIN(num_features=512)
# Define the decoder network
self.decoder = nn.Sequential(
nn.ConvTranspose2d(
512, 256, kernel_size=3, stride=2, padding=1, output_padding=1
),
nn.InstanceNorm2d(256),
nn.ReLU(),
nn.ConvTranspose2d(
256, 128, kernel_size=3, stride=2, padding=1, output_padding=1
),
nn.InstanceNorm2d(128),
nn.ReLU(),
nn.ConvTranspose2d(
128, 64, kernel_size=3, stride=2, padding=1, output_padding=1
),
nn.InstanceNorm2d(64),
nn.ReLU(),
nn.ConvTranspose2d(
64, 32, kernel_size=3, stride=2, padding=1, output_padding=1
),
nn.InstanceNorm2d(32),
nn.ReLU(),
nn.Conv2d(32, 3, kernel_size=3, stride=1, padding=1),
nn.Tanh(),
)
def forward(self, x, style):
# Encode the input image
encoded = self.encoder(x)
# Transform the encoded image using the style
transformed = self.transformer(encoded, style)
# Decode the transformed image
decoded = self.decoder(transformed)
# Return the generated image
return decoded
class Discriminator(nn.Module):
def __init__(self, in_channels=3, features=[64, 128, 256, 512]):
super(Discriminator, self).__init__()
self.features = features
self.initial = nn.Sequential(
nn.Conv2d(in_channels * 2, features[0], kernel_size=4, stride=2, padding=1),
nn.LeakyReLU(0.2),
)
layers = []
in_channels = features[0]
for feature in features[1:]:
layers.append(
nn.Conv2d(in_channels, feature, kernel_size=4, stride=2, padding=1)
)
layers.append(nn.BatchNorm2d(feature))
layers.append(nn.LeakyReLU(0.2))
in_channels = feature
layers.append(nn.Conv2d(in_channels, 1, kernel_size=4, stride=2, padding=1))
self.model = nn.Sequential(*layers)
def forward(self, x, y):
x = torch.cat([x, y], dim=1)
x = self.initial(x)
x = self.model(x)
x = torch.sigmoid(x)
# print(torch.threshold(x, 0.5, 0))
return torch.where(x < 0.5, torch.tensor(0.0).cuda(), torch.tensor(1.0).cuda())
# discriminator = torch.hub.load('mateuszbuda/brain-segmentation-pytorch', 'unet',in_channels=3, out_channels=1, init_features=32, pretrained=True)
discriminator = Discriminator()
generator = Generator()
# Define the loss function and optimizer
criterion = nn.BCELoss()
optimizer_G = optim.Adam(generator.parameters(), lr=0.0002, betas=(0.5, 0.999))
optimizer_D = optim.Adam(discriminator.parameters(), lr=0.0002, betas=(0.5, 0.999))
# Define the loss functions
criterion_adv = nn.BCELoss() # adversarial loss
criterion_content = nn.MSELoss() # content loss
criterion_style = nn.MSELoss() # style loss
# Define the weight of each loss
lambda_adv = 1 # weight for adversarial loss
lambda_content = 1 # weight for content loss
lambda_style = 2 # weight for style loss
def gram_matrix(input):
batch_size, num_channels, height, width = input.size()
features = input.view(batch_size * num_channels, height * width)
gram = torch.mm(features, features.t())
return gram.div(batch_size * num_channels * height * width)
# Define the combined loss function for the generator
def generator_loss(fake_outputs, fake_images, real_images, style_images):
# Adversarial loss
fake_labels = torch.ones((fake_images.size(0), 1, 8, 8))
adversarial_loss = criterion_adv(fake_labels, fake_outputs.cpu())
# Content loss
content_loss = criterion_content(fake_images, real_images)
# Style loss
style_loss = criterion_style(gram_matrix(fake_images), gram_matrix(style_images))
# Combine the losses
total_loss = (
lambda_adv * adversarial_loss
+ lambda_content * content_loss
+ lambda_style * style_loss
)
return total_loss
from torch.autograd import Variable
best_loss = float("inf")
discriminator = discriminator.cuda()
generator = generator.cuda()
# Define the training loop
def train(num_epochs):
global best_loss
monet = [style_images for _, style_images in enumerate(dataloader_monet)]
num = len(monet)
for epoch in range(num_epochs):
for i, batch in enumerate(dataloader_photo):
real_images = batch
style_images = monet[i - int(i / num) * num]
real_images = real_images.cuda()
style_images = style_images.cuda()
# Train the discriminator
optimizer_D.zero_grad()
real_labels = torch.ones((real_images.size(0), 1, 8, 8)).cuda()
fake_labels = torch.zeros((real_images.size(0), 1, 8, 8)).cuda()
real_output = discriminator(real_images, style_images)
fake_images = generator(real_images, style_images)
fake_images = fake_images.cuda()
fake_output = discriminator(fake_images, style_images)
d_loss_real = criterion(real_output, real_labels)
d_loss_fake = criterion(fake_output, fake_labels)
d_loss = d_loss_real + d_loss_fake
d_loss = Variable(d_loss, requires_grad=True)
d_loss.backward()
optimizer_D.step()
# Train the generator
optimizer_G.zero_grad()
# fake_output = discriminator(generator(real_images, style_images), style_images)
# g_loss = criterion(fake_output, real_labels)
# print(fake_output)
g_loss = generator_loss(fake_output, fake_images, real_images, style_images)
g_loss = Variable(g_loss, requires_grad=True)
g_loss.backward()
optimizer_G.step()
# Print the loss
if i % 900 == 0:
print(
f"[Epoch {epoch}/{num_epochs}] [Batch {i}/{len(dataloader_photo)}] [D loss: {d_loss.item()}] [G loss: {g_loss.item()}]"
)
if g_loss.item() < best_loss:
best_loss = g_loss.item()
torch.save(generator.state_dict(), "model.pt")
# Train the model
train(50)
from torchvision import transforms
from PIL import Image
def stylize_image(image_path, style_path, load_checkpoint=False):
transform = transforms.Compose(
[
transforms.Resize((256, 256)),
transforms.ToTensor(),
transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5)),
]
)
image = transform(Image.open(image_path).convert("RGB")).unsqueeze(0)
style = transform(Image.open(style_path).convert("RGB")).unsqueeze(0)
if load_checkpoint:
generator.load_state_dict(torch.load("model.pt"))
generator.eval()
with torch.no_grad():
stylized_image = generator(image, style)
return stylized_image
# Example usage of the inference function
# Option 1: Load the checkpoint to eval
# stylized_image = stylize_image("./originaldata/1.jpg", "./monetstyledata/1.jpg", load_checkpoint=True)
# Option 2: Directly test on current generator
stylized_image = stylize_image(
"/kaggle/input/monet-for-test/0a1d1b9f8e.jpg",
"/kaggle/input/gan-getting-started/monet_jpg/000c1e3bff.jpg",
)
|
# ## Keşifçi Veri Analizi | Becerileri Pekiştirme
# ## Kütüphaneleri Çağırma
# Aşağıda ihtiyacımız doğrultusunda kullanacağımız kütüphaneleri yükleyelim.
# Öncelikle pandas, numpy ve seaborn kütüphanelerini çağırarak başlıyoruz.
import numpy as np
import seaborn as sns
import pandas as pd
# Görünen uyarı mesajlarını filtrelemek için aşağıdaki kod parçasını kullanıyorum. Bu sayede, kodun okunabilirliği artacak ve gereksiz uyarılar ekrana yazdırılmayacak.
import warnings
warnings.filterwarnings("ignore")
# ## Veri Setini Çağırma ve Veri Setine Genel Bakış
# Veri çerçevemizi bulunduğumuz dizinden yükleyelim ve bir veri çerçevesi haline getirerek df değişkenine atayalım. (pd.read_csv(...csv))
df_iris = pd.read_csv("/kaggle/input/iris-flower-dataset/IRIS.csv")
# Veri çerçevesinin ilk 5 gözlemini görüntüleyelim.
df_iris.head()
# Veriseti incelendiğinde İris(Süsen) çiçeğinin yaprak uzunluk ve genişliği bilgilerinin yer aldığı görülmekte. Her bir satır, bir çiçeğe ait ölçüm değerlerini gösterir.
# Özellikler sırasıyla sepal-length (alt yaprak uzunluğu cm), sepal-with (alt yaprak genişliği cm), pedal-length (üst yaprak genişliği), pedal-width (üst yaprak uzunluğu).
# Son sütunda da görüleceği üzere sınıflarımız ise setosa, versicolor ve virginica.
# Veri çerçevesinin kaç öznitelik ve kaç gözlemden oluştuğunu görüntüleyelim.
df_iris.shape
# Buradan da anlaşılabileceği gibi iris verisetimiz 150 satır(gözlem) 5 sütundan(öznitelik) oluşmaktadır.
# "shape" fonksiyonunun yanı sıra “columns” ve “dtypes” fonksiyonları ile set hakkında daha fazla bilgiye ulaşabiliriz.
df_iris.columns
df_iris.dtypes
# sepal_length sepal_width petal_length ve petal_width değerlerinin "float64" ile ifade edildiği görülmekte. Yani nümerik ifadeler.
# Fakat species değerleri object veri türünde. Yani kategorik değişken.
# Veri çerçevesindeki değişkenlerin hangi tipte olduğunu ve bellek kullanımını görüntüleyelim.
# Ayrıca Pandas’ta bu üç bilginin hepsini hatta daha fazlasını içeren “info” fonksiyonu mevcut.
# Veri çerçevesindeki değişkenlerin türlerini ve bellek kullanımını info() metodu ile görüntüleyebiliriz.
df_iris.info()
# İnceleme sonucunda 5 ayrı sütun,150 satırdan oluşan bir verisetiyle beraberiz. Sütun isimleri _'sepal_length', 'sepal_width', 'petal_length', 'petal_width','species'_. Ve her bir sütunun dtype'ı listelenmiş durumda.
# Verimizin hiçbir satırında **NULL** değer bulunmadığı da görülüyor. Ancak, verilerin cm cinsinden uzunluk olması gerektiği düşünüldüğünde, 0 cm veya negatif değerlerin girilmiş olması durumunda, burada eksik verilerin bulunabileceğini söyleyebiliriz. Veriseti incelerken bu durumları da göz önünde bulundurmamız gerekiyor. **NULL** değer bulunmadı diyerek incelemeye devam etmemeliyiz. Aykırı değerlere göz atarak kafamızdaki bu soru işaretlerini silebiliriz.
# Veri çerçevesindeki sayısal değişkenler için temel istatistik değerlerini görüntüleyelim.
# Standart sapma ve ortalama değerlerden çıkarımda bulunarak hangi değişkenlerin ne kadar varyansa sahip olduğu hakkında fikir yürütelim.
# Bunun için describe fonksiyonunu kullanacağım.
# Describe() fonksiyonu, veri çerçevesindeki sayısal değişkenlerin temel istatistiksel özelliklerini gösteren bir özet istatistikleri tablosu oluşturur. Bu özellikler, değişkenlerin ortalaması, standart sapması, minimum ve maksimum değerleri, çeyreklik değerleri ve gözlem sayısıdır.
df_iris.describe()
# 1. Sepal uzunluğu (sepal_length) için ortalama değer 5.84 ve standart sapması 0.83'tür.
# 2. Sepal genişliği (sepal_width) için ortalama değer 3.05 ve standart sapması 0.43'tür.
# 3. Petal uzunluğu (petal_length) için ortalama değer 3.76 ve standart sapması 1.76'dır.
# 4. Petal genişliği (petal_width) için ortalama değer 1.19 ve standart sapması 0.76'dır.
# Ortalama ve standart sapma, bir değişkenin varyansı hakkında fikir verir. Standart sapma, ortalama etrafında verilerin ne kadar yayıldığını gösterir. Örneğin, petal uzunluğu değişkeni için standart sapma 1.76'dür, bu da verilerin ortalama etrafında oldukça yayıldığını ve yüksek bir varyansa sahip olduğunu gösterirken, sepal genişliği değişkeni için standart sapma 0.43'tür ve verilerin daha az yayıldığını ve daha düşük bir varyansa sahip olduğunun bir göstergesidir.
# ## Eksik Veri Kontrolü
# Veri çerçevesinde hangi öznitelikte kaç adet eksik değer olduğunu gözlemleyelim.
# Gözlemlendiği üzere de her satırda 150 tane boş olmayan satır bulunmakta. Bu da demek oluyor ki verimizde eksik değer yok. Bunun sağlamasını da **isnull() ve sum()** fonksiyonları yardımıyıyla görebiliriz.
df_iris.isnull().sum()
# ## Veri Görselleştirme
# ### Korelasyon
# Sayısal değişkenler arasında korelasyon olup olmadığını göstermek için korelasyon matrisi çizdirelim. Korelasyon katsayıları hakkında fikir yürütelim.
# En güçlü pozitif ilişki hangi iki değişken arasındadır?
# Korelasyon değişkenlerimiz arasındaki oran diyebiliriz. Oranlar -1 ve 1 arasında çıkar. -1 negatif ilişki , 1 pozitif ilişki ,0 ilişki yok demektir. İlişki 1e ne kadar yakınsa ilişki o kadar çoktur diyebiliriz.
# Korelasyonu bulmak için corr() fonksiyonunu kullanıyorum.
# Korelasyon matrisindeki her bir değişken kendisiyle olan korelasyonu 1'dir çünkü bir değişkenin kendisiyle arasındaki ilişki tam olarak pozitiftir ve korelasyon katsayısı 1'dir. Diğer değişkenlerle arasındaki korelasyon ise farklı olabilir. Örneğin, sepal_width'in de petal_length'in de değişkenleri arasındaki korelasyon katsayısı 1'dir, **çünkü her bir değişken kendisiyle olan korelasyonu 1'dir.** Dolayısıyla, korelasyon matrisinde köşegen her zaman 1'dir, çünkü bir değişkenin kendisiyle olan korelasyonu **her zaman mükemmeldir.**
df_iris.corr()
# Korelasyon matrisine baktığımızda, en güçlü pozitif ilişki "petal_length" ve "petal_width" arasında görülür. Tablo incelendiğinde bu oranın 0.962757 olduğu gözükmekte. Yani 1'e oldukça yakın. Bu da, çiçeklerin taç yapraklarının uzunluğu arttıkça, taç yapraklarının genişliğinin de arttığını gösterir.
# Ayrıca, "sepal_length" ve "petal_length" arasında da 0.871754 ile pozitif bir ilişki vardır, ancak "petal_length" ve "petal_width" arasındaki ilişki (0.96) daha güçlüdür.
# Bununla birlikte, "sepal_length" ve "sepal_width" arasında neredeyse hiç ilişki yoktur (-0.109). Yani bitkinin çanak yaprağının boyuyla genişliğinin ilişkisi oldukça düşüktür.
# Bizim daha fazla değişkenimiz varken neden burada 4 değişken gözüküyor?
# Çünkü korelasyon __kategorik__ değişkenlerde hesaplanamaz.
# Peki bu veri setindeki kategorik değişkenler ne? Bunu gözlemlemek için tekrardan veri setini çağıralım
df_iris.head()
# Görüldüğü üzere "species" değişkeni kategorik değişken ve bu yüzden korelasyon grafiğinde yer almadı.
# Korelasyonda da hesaplayabilmek için __kategorik değerler değil nümerik değer__ olmalıdır.Korelasyonda yalnızca nümerik değerleri inceleyebiliriz.
# Korelasyon katsayılarını daha iyi okuyabilmek için ısı haritası çizdirelim.
# Isı haritasını çizebilmek için matplotlib kütüphanesini tanımlamamız gerekiyor. plt kısaltmasıyla matplotlib kütüphanesini import ediyorum.
import matplotlib.pyplot as plt
plt.figure(figsize=(8, 8))
plt.title("Korelasyon Isı Haritası")
sns.heatmap(df_iris.corr(), cmap="pink", annot=True)
# Renk skalasına göre; renk tonu açık olduğunda, değişkenler arasındaki ilişki o kadar yüksektir. Harita incelendiğinde, tekrardan en güçlü ilişkinin (1'den sonra) 0.96 ile "petal_length" ve "petal_width" arasında olduğu görülmektedir.
# Veri çerçevemizin hedef değişkeninin "variety" benzersiz değerlerini görüntüleyelim.
df_iris.columns
print(df_iris["species"].unique())
# Veri çerçevemizin hedef değişkeninin "variety" benzersiz kaç adet değer içerdiğini görüntüleyelim.
# Bunun için Pandas kütüphanesinin "nunique()" fonksiyonu ile veri çerçevesinin belirtilen sütununun benzersiz değerlerinin sayısı bulunabilir.
# Görüldüğü üzere 3 adet benzersiz değişken bulunuyor.
print(df_iris["species"].nunique(), "adet benzersiz değişkeni vardır.")
# Veri çerçevesindeki sepal.width ve sepal.length değişkenlerinin sürekli olduğunu görüyoruz. Bu iki sürekli veriyi görselleştirmek için önce scatterplot kullanalım.
plt.title("Sepal Width ve Sepal Length")
plt.scatter(x=df_iris["sepal_width"], y=df_iris["sepal_length"], color="orange")
plt.xlabel("Sepal Width")
plt.ylabel("Sepal Length")
plt.show()
# Sepal Width değişkeninin değerleri 2.5-3.5 aralığında yoğun olarak gözükmektedir. Length değişkeninin değerleri ise 5.5-7.0 arasında yoğunlaşmaktadır diyebiliriz.
# Aynı iki veriyi daha farklı bir açıdan frekanslarıyla incelemek için jointplot kullanarak görselleştirelim.
sns.jointplot(x="sepal_width", y="sepal_length", kind="scatter", data=df_iris)
# Bu grafiğe bakarak bir önceki yorumumun kısmen doğru olduğunu görüyorum. Çünkü ilk grafikte length'in değerlerinin 4.5-5.0 arasında yoğunlaştığını görememiştim. jointplot grafiği yorum yapmak için daha mantıklı bir grafik.
# Aynı iki veriyi scatterplot ile tekrardan görselleştirelim fakat bu sefer "variety" parametresi ile hedef değişkenine göre kırdıralım.
# 3 farklı renk arasında sepal değişkenleriyle bir kümeleme yapılabilir mi? Ne kadar ayırt edilebilir bunun üzerine düşünelim.
sns.scatterplot(x="sepal_length", y="sepal_width", hue="species", data=df_iris)
plt.title("Sepal için Nokta Grafiği")
plt.show()
# Grafikte 3 farklı renk arasında sepal değişkenleriyle bir kümeleme yapılabildiği görülmektedir. Görüldüğü üzere lacivert renkli "Setosa" türü diğerlerinden daha iyi ayrılmaktadır, ancak turuncu renkli "Versicolor" ve yeşil renkl,"Virginica" türleri birbirine daha çok benzemektedir. Daha çok iç içe geçmiş durumdadır. Bu nedenle, sadece sepal özellikleri kullanarak türleri kesin olarak ayırt etmek zordur. Başka özellikleri de analize dahil ederek yorumlamamız gerekir.
# Mesela scatterplot fonksiyonu ile "petal_length" ve "petal_width" değişkenini incelersek durum nasıl olur?
sns.scatterplot(x="petal_length", y="petal_width", hue="species", data=df_iris)
# Bu scatterplot grafiği, petal uzunluğu (x ekseni) ve petal genişliği (y ekseni) değişkenlerinin çiçek türüne (setosa, versicolor ve virginica) göre dağılımını gösterir. Her tür farklı bir renk ile temsil edilir. Görselden, virginica çiçeğinin genellikle diğer iki türden daha büyük petallere sahip olduğu ve versicolor ve setosa'nın birbirine benzer boyutlara sahip oldukları görülebilir. Ayrıca, setosa çiçeği ile diğer iki tür arasında net bir ayrım gözlemlenebilir.
# Yani anlaşılacağı üzere Setosa çiçeği gerek petal gerek ise sepal olarak diğer iki çiçek türünden ayrışıyor.
# value_counts() fonksiyonu ile veri çerçevemizin ne kadar dengeli dağıldığını sorgulayalım.
df_iris["species"].value_counts()
# species değişkenindeki her bir sınıfın veri çerçevesinde kaç kez tekrarlandığını sayar ve sonuçları sınıf adıyla birlikte gösterir. Görüldüğü üzere her çeşitten 50 adet bulunuyor. Bu da demek oluyor ki verisetimiz dengeli dağılmış.
# Eğer her sınıftan farklı sayıda örnek olsaydı, bu dengesiz bir dağılım olarak yorumlanabilirdi.
# Keman grafiği çizdirerek sepal.width değişkeninin dağılımını inceleyin.
# Söz konusu dağılım bizim için ne ifade ediyor, normal bir dağılım olduğunu söyleyebilir miyiz?
sns.violinplot(x="sepal_width", data=df_iris, color="brown")
# Keman grafiği, veri dağılımının yoğunluk eğrisini gösteren ve aynı zamanda veri dağılımının simetrik mi yoksa çarpık mı olduğunu gösteren bir görselleştirme aracıdır.
# __BU YORUMU İNCELE__
# Sepal.width değişkeninin keman grafiğine bakarak, verinin normal bir dağılım göstermediği, hafif bir çarpıklık olduğu söylenebilir. Grafiğin sol tarafındaki kuyruk daha uzun ve çıkıntılıdır. Bu nedenle, sepal.width değişkeninin normal bir dağılım göstermediğini söyleyebiliriz.
# Daha iyi anlayabilmek için sepal.width üzerine bir distplot çizdirelim.
sns.distplot(df_iris["sepal_width"])
# Üç çiçek türü için üç farklı keman grafiğini sepal.length değişkeninin dağılımı üzerine tek bir satır ile görselleştirelim.
sns.violinplot(x="species", y="sepal_length", data=df_iris)
plt.title("Üç Çiçek Türü için Üç Farklı Keman Grafiği")
plt.show()
# Hangi çiçek türünden kaçar adet gözlem barındırıyor veri çerçevemiz?
# 50 x 3 olduğunu ve dengeli olduğunu value_counts ile zaten görmüştük, ancak bunu görsel olarak ifade etmek için sns.countplot() fonksiyonuna variety parametresini vereilm.
sns.countplot(x="species", data=df_iris)
plt.title("Veri Setindeki Her Türden Kaç Adet Çiçek Var?")
plt.show()
# sepal.length ve sepal.width değişkenlerini sns.jointplot ile görselleştirelim, dağılımı ve dağılımın frekansı yüksek olduğu bölgelerini inceleyelim.
sns.jointplot(data=df_iris, x="sepal_length", y="sepal_width", color="darkgreen")
plt.show()
# Bir önceki hücrede yapmış olduğumuz görselleştirmeye kind = "kde" parametresini ekleyelim. Böylelikle dağılımın noktalı gösterimden çıkıp yoğunluk odaklı bir görselleştirmeye dönüştüğünü görmüş olacağız.
sns.jointplot(data=df_iris, x="sepal_length", y="sepal_width", kind="kde", color="pink")
plt.show()
# Dağılımın hangi bölgelerde daha sık olduğunu görmek için **kind=kde** seçeneğini kullandım. Bu seçenek grafikte yoğunluğu temsil eden bir çizgi ekler.
# scatterplot ile petal.length ve petal.width değişkenlerinin dağılımlarını çizdirelim.
# Aşağıdaki kod parçası, "petal_length" değişkeninin x-ekseninde, "petal_width" değişkeninin y-ekseninde olduğu bir scatterplot oluşturur.
sns.scatterplot(x="petal_length", y="petal_width", color="brown", data=df_iris)
# Aynı görselleştirmeye hue = "variety" parametresini ekleyerek 3. bir boyut verelim.
sns.scatterplot(x="petal_length", y="petal_width", hue="species", data=df_iris)
# Bu grafiğe göre; setosa türü diğer iki türe göre daha küçük boyutlu ve daha dar yapraklara sahiptir çıkarımında bulunabiliriz.
# sns.lmplot() görselleştirmesini petal.length ve petal.width değişkenleriyle implemente edelim. Petal length ile petal width arasında ne tür bir ilişki var ve bu ilişki güçlü müdür? sorusunu yanıtlayalım.
sns.lmplot(x="petal_length", y="petal_width", hue="species", data=df_iris)
plt.title("Species ile Petal Length vs. Petal Width", color="darkred")
plt.show()
# Bu görselleştirmede, petal length ile petal width arasında pozitif bir ilişki olduğu görülmektedir. Yani bir çiçeğin petal length değeri arttıkça, petal width değeri de genellikle artmaktadır. Ayrıca, her bir çiçek türü için (setosa, versicolor ve virginica), bu ilişkinin farklı bir doğru üzerinde olduğu görülmektedir.
# Özellikle setosa çiçeklerinin (lacivert renkli noktalar) petal length ve petal width değerleri arasındaki ilişki, diğer iki çiçek türüne göre daha zayıf görünmektedir. Bunun nedeni, setosa çiçeklerinin diğer çiçek türlerine göre daha küçük boyutlara sahip olması olabilir.Tabii ki bu çıkarım kesinlikle doğrudur demek yanlış olur. Diğer durumlarla birlikte daha detaylı incelenmesi gerekir.
# Grafikteki çizginin oldukça dik olması, bu değişkenler arasındaki ilişkinin yüksek korelasyonlu olduğunu göstermektedir. Grafikte, özellikle Versicolor çiçeklerinin (turuncu renkli noktalar) arasındaki ilişki en yüksek korelasyonlu olanıdır.
sns.lmplot(x="petal_length", y="petal_width", data=df_iris)
plt.title("Petal Length vs. Petal Width", color="blue")
plt.show()
# Grafiği çiçek türlerine göre sınıflandırmadan tek bir şekilde de gösterebiliriz.
# Bu grafiğe bakarak da petal uzunluğu ve genişliği arasında pozitif bir ilişki olduğunu görebiliyoruz. Veri noktaları çizgiye yakın bir şekilde gruplanmıştır, bu da yüksek bir korelasyon olduğunu göstermektedir. Species değişkenine göre sınıflandırmadan tek tip olarak gösterilmesi, tüm çiçek türleri arasındaki ilişkiyi daha net bir şekilde görmemizi sağlar.
# Bu sorunun yanıtını pekiştirmek için iki değişken arasında korelasyon katsayısını yazdıralım.
petal_corr = df_iris["petal_length"].corr(df_iris["petal_width"])
petal_corr
# Daha önceden de hatırlarsınız ki en büyük korelasyon 0.96 ile petal_length ile petal_width arasındaydı. petal_corr değişkenini de hesaplayıp tekrar ekrana yazdırdığımızda bunu görebiliyoruz.
# Bu sonuç, yüksek bir pozitif ilişki olduğunu ve iki değişkenin birlikte arttığını gösterir. Ayrıca bu sonuç, daha önce yapılan görselleştirme analizinde de görüldüğü üzere, petal length ve petal width arasındaki ilişkinin güçlü olduğunu doğrulamaktadır.
# Petal Length ile Sepal Length değerlerini toplayarak yeni bir total length özniteliği oluşturalım.
df_iris.assign(total_length=df_iris["petal_length"] + df_iris["sepal_length"])
# total_length değişkenini oluşturduk ve tabloya ekledik fakat tabloya kalıcı olarak eklemedik. df_iris yapısını yukarıdaki kod satırına eşitlersek kalıcı olarak total_length değişkenini tabloya eklemiş oluruz.
df_iris = df_iris.assign(total_length=df_iris["petal_length"] + df_iris["sepal_length"])
df_iris
# total.length'in ortalama değerini yazdıralım.
# Bir değişkenin ortalamasını bulabilmek için mean() fonksiyonu kullanılır.
# Tüm çiçek türlerinin uzunlukları toplamının, gözlem sayısına bölünmesi sonucunda ortalama uzunluk 9.6 olarak bulunmuştur.
total_length_mean = df_iris["total_length"].mean()
print("Ortalama Değer: ", total_length_mean)
# total.length'in standart sapma değerini yazdıralım.
# Standart sapma ise 2.5 çıktı.
total_length_std = df_iris["total_length"].std()
print("Standart Sapma: ", total_length_std)
# Ortalama 9.6, total_length değişkeninin tüm örneklem verilerinin toplamının örneklem sayısına bölünmesiyle hesaplanan bir değerdir. Bu da tüm çiçeklerin ortalama uzunluğunu verir. Standart sapma ise verilerin ne kadar yayıldığını gösteren bir ölçüttür. Bu durumda, standart sapmanın 2.5 olması, tüm çiçeklerin uzunluklarının ortalamadan ne kadar **farklılaştığını** gösterir. Yani, çiçeklerin uzunlukları ortalama uzunluktan yaklaşık 2.5 birim kadar sapma gösteriyor.
# sepal.length'in maksimum değerini yazdıralım.
sepal_length_max = df_iris["sepal_length"].max()
print("Sepal Length'in Maksimum Değeri", sepal_length_max)
# sepal.length'i 5.5'den büyük ve türü setosa olan gözlemleri yazdıralım.
df_iris[(df_iris["sepal_length"] > 5.5) & (df_iris["species"] == "setosa")]
# petal.length'i 5'den küçük ve türü virginica olan gözlemlerin sadece sepal.length ve sepal.width değişkenlerini ve değerlerini yazdıralım.
df_iris.loc[
(df_iris["petal_length"] < 5) & (df_iris["species"] == "virginica"),
["sepal_length", "sepal_width"],
]
# Hedef değişkenimiz variety'e göre bir gruplama işlemi yapalım değişken değerlerimizin ortalamasını görüntüleyelim.
# Bu işlem için groupby() fonksiyonunu kullanabiliriz.
df_iris.groupby("species").mean()
# Iris setosa, diğer türlerden daha küçük çiçekleri olan bir türdür. Diğer taraftan, Iris virginica, en büyük çiçeklere sahip olan türdür. Toplam uzunluk sütunu, çiçeklerin boyutunu tam olarak ifade etmek için hesaplanan bir özelliktir. Bu tablodaki veriler, türler arasındaki farklılıkları ve benzerlikleri anlamamıza yardımcı olabilir.
# Hedef değişkenimiz variety'e göre gruplama işlemi yaparak sadece petal.length değişkenimizin standart sapma değerlerini yazdıralım.
# Aşağıdaki kodda ilk satırda groupby() fonksiyonunu kullanarak species değişkenine göre gruplama yapıyoruz ve sadece petal_length değişkenini seçiyoruz. Ardından da std() ile standart sapmayı hesaplıyoruz.
df_iris.groupby("species")["petal_length"].std()
|
# Import first packages
import pandas as pd
import numpy as np
# Load the dataset
df = pd.read_csv(
"/kaggle/input/student-mental-health/Student Mental health.csv", sep=","
)
df.head()
# Check the variables
columns = list(df.columns)
for col in columns:
print(col)
print(df[col].value_counts())
print("\n")
df.info()
df.isna().sum()
# The dataset has 101 observations and 11 variables, which the variable "Age" is a float and the others are strings. Additionally, we observed that the variable "Age" has 1 missing value.
# # Variables:
# - Timestamp: string type
# - Choose your age: categorical type "Female" or "Male";
# - What is your course?: 49 different courses;
# - Age: float type with minimum 18 and maximum 24;
# - Your current year of Study: categorical type "year 1", "Year 3", "Year 2", "year 2", "year 4", "year 3" and "Year 1". Here we noticed that for 1, 2 and 3 we have two ways of writing;
# - What is your CGPA?: categorical type from "0-1.99" to "3.50-4.00". Here we noticed that "3.50 - 4.00" is written in 2 ways;
# - Marital status: categorical type "Yes" or "No";
# - Do you have Depression?: categorical type "Yes" or "No";
# - Do you have Anxiety?: categorical type "Yes" or "No";
# - Do you have Panic attack?: categorical type "Yes" or "No";
# - Did you seek any specialist for a treatment?: categorical type "Yes" or "No";
# Histogram plots for "Age", "What is your CGPA?", "Your current year of Study" and "What is your course?"
import matplotlib
import matplotlib.pyplot as plt
plt.figure(figsize=(10, 6))
plt.hist(df["Age"])
plt.xlabel("Age")
plt.ylabel("Frequency")
# Let's adjust the variable "What is your CGPA?" to plot the histogram
def CGPA_hist(x):
if x == "3.50 - 4.00" or x == "3.50 - 4.00 ":
return "3.50 - 4.00"
elif x == "0 - 1.99":
return "0 - 1.99"
elif x == "2.00 - 2.49":
return "2.00 - 2.49"
elif x == "2.50 - 2.99":
return "2.50 - 2.99"
else:
return "3.00 - 3.49"
df["What is your CGPA?_new"] = df["What is your CGPA?"].apply(lambda x: CGPA_hist(x))
plt.figure(figsize=(10, 6))
plt.hist(df["What is your CGPA?_new"], bins=10)
plt.xlabel("CGPA")
plt.ylabel("Frequency")
# Let's adjust the variable "Your current year of Study" to plot the histogram
def year_study_hist(x):
if x == "year 1" or x == "Year 1":
return "Year 1"
elif x == "year 2" or x == "Year 2":
return "Year 2"
elif x == "year 3" or x == "Year 3":
return "Year 3"
else:
return "Year 4"
df["Your current year of Study_new"] = df["Your current year of Study"].apply(
lambda x: year_study_hist(x)
)
plt.figure(figsize=(10, 6))
plt.hist(df["Your current year of Study_new"], bins=10)
plt.xlabel("Current year of Study")
plt.ylabel("Frequency")
# Let's adjust the variable "What is your course?" to plot the histogram
def course_hist(x):
if x not in (
"BCS",
"Engineering",
"BIT",
"Biomedical science",
"KOE",
): # ,'BENL','psychology','Laws','Engine'):
return "Other"
else:
return x
df["What is your course_new"] = df["What is your course?"].apply(
lambda x: course_hist(x)
)
plt.figure(figsize=(10, 6))
plt.hist(df["What is your course_new"], bins=10)
plt.xlabel("Course")
plt.ylabel("Frequency")
# ## Multiple Correspondence Analysis
# Multiple Correspondence Analysis (MCA) is an unsupervised Machine Learning method used to verify if there is a relationship between more than 2 categorical variables.
# Our goal is to analyze associations between depression (variable "Do you have Depression?") and other variables present in the dataset.
# First, we will use the chi-squared test (also known as chi-square or χ2 test) for each pair of categorical variables to verify if the variables show an association with at least one of the other variables. Note that if a variable is not categorical, it is necessary to transform quantitative data (such as the variable "Age") into categories.
# We have 9 categorical variables and one numeric variable. We are not including the variable "Timestamp" in our analysis because it doesn't make sense. Therefore, if we adjust the variable "Age", which is numeric, into 3 categories (18-20, 21-22, and 23-24), and consider 6 categories for the variable "What is your course?" (BCS, Engineering, BIT, Biomedical Science, KOE, and Other), we will have a total of 30 categories involved in our analysis.
# Relationship
from sklearn.feature_selection import chi2
from scipy.stats import chi2_contingency
from scipy import stats
df = df.dropna()
df.isna().sum()
def age_new(x):
if x <= 20:
return "18 t0 20"
elif x <= 22:
return "21 t0 22"
else:
return "23 t0 24"
df["Age_new"] = df["Age"].apply(lambda x: age_new(x))
df.columns
df_ACM = df[
[
"Choose your gender",
"Marital status",
"Do you have Depression?",
"Do you have Anxiety?",
"Do you have Panic attack?",
"Did you seek any specialist for a treatment?",
"What is your CGPA?_new",
"Your current year of Study_new",
"What is your course_new",
"Age_new",
]
]
keep_var = []
for i in range(0, len(df_ACM.columns)):
for j in range(0, len(df_ACM.columns)):
if df_ACM.columns[i] != df_ACM.columns[j]:
tbl_cont = pd.crosstab(df_ACM[df_ACM.columns[i]], df_ACM[df_ACM.columns[j]])
chi2, pvalor, gl, ve = stats.chi2_contingency(tbl_cont)
if pvalor < 0.05:
print(df_ACM.columns[i])
print(df_ACM.columns[j])
print(pvalor)
print("\n")
keep_var.append(df_ACM.columns[i])
keep_var.append(df_ACM.columns[j])
else:
pass
print("\n")
print("Keep variables:")
print(list(set(keep_var)))
df_ACM = df[set(keep_var)]
df_ACM
import prince
data_ACM = prince.MCA(
n_components=3,
n_iter=4,
copy=True,
check_input=True,
engine="auto",
random_state=42,
)
data_ACM = data_ACM.fit(df_ACM)
data_ACM.eigenvalues_
data_ACM.total_inertia_
data_ACM.column_coordinates(df_ACM)
data_ACM.explained_inertia_
0.30399597100223336 / 2.125
0.19653628084386143 / 2.125
# Plot
ax = data_ACM.plot_coordinates(
X=df_ACM,
ax=None,
figsize=(20, 15),
show_row_points=True,
show_row_labels=False,
show_column_points=True,
column_points_size=120,
show_column_labels=True,
legend_n_cols=8,
)
ei = data_ACM.explained_inertia_
ax.set_title(
"Perceptual Map", fontsize=25, color="#213B69", fontweight=1000, loc="center"
)
ax.set_xlabel("Component {} ({:.2f}% inertia)".format(1, 100 * ei[0]), fontsize=15)
ax.set_ylabel("Component {} ({:.2f}% inertia)".format(2, 100 * ei[1]), fontsize=15)
plt.legend(loc="lower right")
# Plot
ax = data_ACM.plot_coordinates(
X=df_ACM,
ax=None,
figsize=(20, 15),
show_row_points=False,
show_row_labels=False,
show_column_points=True,
column_points_size=120,
show_column_labels=True,
legend_n_cols=8,
)
ei = data_ACM.explained_inertia_
ax.set_title(
"Perceptual Map", fontsize=25, color="#213B69", fontweight=1000, loc="center"
)
ax.set_xlabel("Component {} ({:.2f}% inertia)".format(1, 100 * ei[0]), fontsize=15)
ax.set_ylabel("Component {} ({:.2f}% inertia)".format(2, 100 * ei[1]), fontsize=15)
plt.legend(loc="lower right")
df_ACM.nunique()
# We noticed that the inertia of the 2 dimensions, which is the contribution of the dimension to explain the variance of the data, is 23.58% (14.31% + 9.27%).
# We can also observe that "Do you have Panic attack?" Yes, "Do you have Anxiety?" Yes, and "Marital status" Yes are close to "Do you have Depression?" Yes, while "Do you have Panic attack?" No, "Do you have Anxiety?" No, and "Marital status" No are close to "Do you have Depression?" No.
# Now, let's try to plot a perceptual map using the 4 variables ("Do you have Panic attack?", "Do you have Anxiety?", "Marital status", and "Do you have Depression?").
df_ACM2 = df[
[
"Do you have Panic attack?",
"Do you have Anxiety?",
"Marital status",
"Do you have Depression?",
]
]
df_ACM2
data_ACM2 = prince.MCA(
n_components=2,
n_iter=4,
copy=True,
check_input=True,
engine="auto",
random_state=42,
)
data_ACM2 = data_ACM2.fit(df_ACM2)
data_ACM2.total_inertia_
data_ACM2.explained_inertia_
# Plot
ax = data_ACM2.plot_coordinates(
X=df_ACM2,
ax=None,
figsize=(15, 10),
show_row_points=False,
show_row_labels=False,
show_column_points=True,
column_points_size=120,
show_column_labels=True,
legend_n_cols=8,
)
ei = data_ACM2.explained_inertia_
ax.set_title(
"Perceptual Map", fontsize=25, color="#213B69", fontweight=1000, loc="center"
)
ax.set_xlabel("Component {} ({:.2f}% inertia)".format(1, 100 * ei[0]), fontsize=15)
ax.set_ylabel("Component {} ({:.2f}% inertia)".format(2, 100 * ei[1]), fontsize=15)
plt.legend(loc="upper right")
|
import os
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
os.getcwd()
data = pd.read_csv("Real estate.csv")
data.head()
data.describe()
sns.regplot(x="X1 transaction date", y="Y house price of unit area", data=data)
sns.regplot(x="X2 house age", y="Y house price of unit area", data=data)
sns.regplot(
x="X3 distance to the nearest MRT station",
y="Y house price of unit area",
data=data,
)
sns.regplot(
x="X4 number of convenience stores", y="Y house price of unit area", data=data
)
plt.figure(figsize=(8, 3))
sns.displot(x=data["Y house price of unit area"], kde=True, aspect=2, color="green")
plt.xlabel("house price of unit area")
plt.figure(figsize=(20, 5))
sns.distplot(data["Y house price of unit area"])
plt.show()
sns.pairplot(data)
plt.show()
data.isnull().sum()
x = data.iloc[:, :-1]
y = data.iloc[:, [-1]]
# ## Before fitting data lets normalize its first
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
x = scaler.fit_transform(x)
y = scaler.fit_transform(y)
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.30)
from sklearn.linear_model import Ridge
reg = Ridge(alpha=30)
reg.fit(x_train, y_train)
pred = reg.predict(x_test)
from sklearn.metrics import mean_absolute_error, mean_squared_error
MAE = mean_absolute_error(y_test, pred)
MSE = mean_squared_error(y_test, pred)
RMSE = np.sqrt(MSE)
d = {
"Mean_Absolute_Error": MAE,
"Mean_Squared_Error": MSE,
"Root_Mean_Squared_Error": RMSE,
}
d
|
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import numpy as np
from neuralprophet import NeuralProphet
import os
import warnings
pd.set_option("max_columns", 50)
warnings.filterwarnings("ignore")
df = pd.read_csv(
"/kaggle/input/gdz-elektrik-datathon-2023/train.csv", parse_dates=["Tarih"]
)
test = pd.read_csv(
"/kaggle/input/gdz-elektrik-datathon-2023/sample_submission.csv",
parse_dates=["Tarih"],
)
med = pd.read_csv(
"/kaggle/input/gdz-elektrik-datathon-2023/med.csv", parse_dates=["Tarih"]
)
all_data = pd.concat([df, test], ignore_index=True)
df.set_index("Tarih").plot(style=".", figsize=(15, 5), color="red")
plt.ylabel("Dağıtılan Enerji")
plt.show()
# ## Create Features
all_data["hour"] = all_data["Tarih"].dt.hour
all_data["dayofweek"] = all_data["Tarih"].dt.dayofweek
all_data["quarter"] = all_data["Tarih"].dt.quarter
all_data["month"] = all_data["Tarih"].dt.month
all_data["year"] = all_data["Tarih"].dt.year
all_data["dayofyear"] = all_data["Tarih"].dt.dayofyear
all_data["dayofmonth"] = all_data["Tarih"].dt.day
all_data["weekofyear"] = all_data["Tarih"].dt.weekofyear
all_data
sns.pairplot(
all_data.dropna(),
hue="hour",
x_vars=["hour", "dayofweek", "year", "weekofyear"],
y_vars="Dağıtılan Enerji (MWh)",
height=5,
plot_kws={"alpha": 0.15, "linewidth": 0},
)
plt.suptitle("Energy by Hour, Day of Week, Year and Week of Year")
plt.show()
# ## Remove Outliers
med["outlier_flag"] = True
all_data = pd.merge(all_data, med, how="left", on="Tarih")
all_data = all_data[~(all_data["outlier_flag"] == True)]
all_data.drop("outlier_flag", axis=1, inplace=True)
# ## Target Transformation
all_data["Dağıtılan Enerji (MWh)"] = np.log1p(all_data["Dağıtılan Enerji (MWh)"])
# ## Seasonal
def is_spring(ds):
date = pd.to_datetime(ds)
return (date.month >= 3) & (date.month <= 5)
def is_summer(ds):
date = pd.to_datetime(ds)
return (date.month >= 6) & (date.month <= 8)
def is_autumn(ds):
date = pd.to_datetime(ds)
return (date.month >= 9) & (date.month <= 11)
def is_winter(ds):
date = pd.to_datetime(ds)
return (date.month >= 12) | (date.month <= 2)
def is_weekend(ds):
date = pd.to_datetime(ds)
return date.day_name in ("Saturday", "Sunday")
all_data["is_spring"] = all_data["Tarih"].apply(is_spring)
all_data["is_summer"] = all_data["Tarih"].apply(is_summer)
all_data["is_autumn"] = all_data["Tarih"].apply(is_autumn)
all_data["is_winter"] = all_data["Tarih"].apply(is_winter)
all_data["is_weekend"] = all_data["Tarih"].apply(is_weekend)
all_data["is_weekday"] = ~all_data["Tarih"].apply(is_weekend)
train_data = all_data.iloc[:40122, :]
test_data = all_data.iloc[40122:, :]
# ## Model
model = NeuralProphet()
model.add_seasonality(
name="weekly_spring", period=7, fourier_order=5, condition_name="is_spring"
)
model.add_seasonality(
name="weekly_summer", period=7, fourier_order=5, condition_name="is_summer"
)
model.add_seasonality(
name="weekly_autumn", period=7, fourier_order=5, condition_name="is_autumn"
)
model.add_seasonality(
name="weekly_winter", period=7, fourier_order=5, condition_name="is_winter"
)
model.add_seasonality(
name="daily_spring", period=1, fourier_order=5, condition_name="is_spring"
)
model.add_seasonality(
name="daily_summer", period=1, fourier_order=5, condition_name="is_summer"
)
model.add_seasonality(
name="daily_autumn", period=1, fourier_order=5, condition_name="is_autumn"
)
model.add_seasonality(
name="daily_winter", period=1, fourier_order=5, condition_name="is_winter"
)
model.add_seasonality(
name="daily_weekend", period=1, fourier_order=5, condition_name="is_weekend"
)
model.add_seasonality(
name="daily_weekday", period=1, fourier_order=5, condition_name="is_weekday"
)
train_train = train_data.iloc[:39378, :]
train_test = train_data.iloc[39378:, :]
model.fit(
train_data.drop(
[
"hour",
"dayofweek",
"quarter",
"month",
"year",
"dayofyear",
"dayofmonth",
"weekofyear",
],
axis=1,
).rename(columns={"Tarih": "ds", "Dağıtılan Enerji (MWh)": "y"})
)
test_preds = model.predict(
df=test_data.drop(
[
"hour",
"dayofweek",
"quarter",
"month",
"year",
"dayofyear",
"dayofmonth",
"weekofyear",
],
axis=1,
).rename(columns={"Tarih": "ds", "Dağıtılan Enerji (MWh)": "y"})
)
test_preds.head()
model.plot_components(test_preds)
test_preds["yhat1"] = np.expm1(test_preds["yhat1"])
test_preds["yhat1"].head(20)
# plt.plot(train_test["Tarih"], test_preds["yhat1"])
# plt.plot(train_test["Tarih"], np.expm1(train_test["Dağıtılan Enerji (MWh)"]))
# plt.show()
real = [
2026.36855132,
1972.72905595,
1885.62465527,
1622.18911342,
1622.18911342,
1622.18911342,
1555.99523516,
1885.62465527,
2112.71880575,
2414.48328849,
2508.08991324,
2547.24863057,
2547.24863057,
2631.42197397,
2631.42197397,
2631.42197397,
2613.47525309,
2613.47525309,
2547.24863057,
2547.24863057,
2547.24863057,
2547.24863057,
2460.36514211,
2172.26619586,
2026.36855132,
1972.72905595,
1885.62465527,
1885.62465527,
1622.18911342,
1622.18911342,
1555.99523516,
1885.62465527,
2112.71880575,
2414.48328849,
2508.08991324,
2547.24863057,
2547.24863057,
2547.24863057,
2547.24863057,
2547.24863057,
2529.30190968,
2529.30190968,
2547.24863057,
2547.24863057,
2547.24863057,
2547.24863057,
2460.36514211,
2172.26619586,
2026.36855132,
1972.72905595,
1885.62465527,
1885.62465527,
1622.18911342,
1622.18911342,
1555.99523516,
1622.18911342,
2112.71880575,
2336.6254094,
2461.87794271,
2547.24863057,
2547.24863057,
2547.24863057,
2547.24863057,
2547.24863057,
2529.30190968,
2529.30190968,
2547.24863057,
2547.24863057,
2547.24863057,
2547.24863057,
2460.36514211,
2094.40831677,
2026.36855132,
1972.72905595,
1885.62465527,
1622.18911342,
1622.18911342,
1622.18911342,
1555.99523516,
1885.62465527,
2112.71880575,
2414.48328849,
2508.08991324,
2631.42197397,
2631.42197397,
2631.42197397,
2631.42197397,
2631.42197397,
2613.47525309,
2613.47525309,
2631.42197397,
2547.24863057,
2547.24863057,
2547.24863057,
2460.36514211,
2172.26619586,
2026.36855132,
1972.72905595,
1885.62465527,
1885.62465527,
1885.62465527,
1622.18911342,
1555.99523516,
1885.62465527,
2112.71880575,
2414.48328849,
2508.08991324,
2631.42197397,
2631.42197397,
2631.42197397,
2631.42197397,
2631.42197397,
2613.47525309,
2613.47525309,
2631.42197397,
2547.24863057,
2547.24863057,
2547.24863057,
2460.36514211,
2172.26619586,
1792.35740825,
1738.71791287,
1651.61351219,
1651.61351219,
1651.61351219,
1522.60388228,
1456.41000402,
1651.61351219,
1878.70766267,
2180.47214542,
2274.07877016,
2397.41083089,
2313.23748749,
2397.41083089,
2397.41083089,
2397.41083089,
2379.46411001,
2379.46411001,
2313.23748749,
2313.23748749,
2313.23748749,
2313.23748749,
2226.35399903,
1938.25505278,
1792.35740825,
1738.71791287,
1651.61351219,
1651.61351219,
1651.61351219,
1522.60388228,
1456.41000402,
1651.61351219,
1878.70766267,
2180.47214542,
2274.07877016,
2397.41083089,
2397.41083089,
2397.41083089,
2397.41083089,
2397.41083089,
2379.46411001,
2379.46411001,
2313.23748749,
2313.23748749,
2313.23748749,
2313.23748749,
2226.35399903,
1938.25505278,
2026.36855132,
1972.72905595,
1885.62465527,
1885.62465527,
1622.18911342,
1622.18911342,
1555.99523516,
1622.18911342,
2112.71880575,
2336.6254094,
2508.08991324,
2547.24863057,
2547.24863057,
2547.24863057,
2547.24863057,
2547.24863057,
2529.30190968,
2529.30190968,
2547.24863057,
2547.24863057,
2547.24863057,
2547.24863057,
2460.36514211,
2094.40831677,
2026.36855132,
1972.72905595,
1622.18911342,
1622.18911342,
1622.18911342,
1622.18911342,
1495.33031306,
1622.18911342,
1849.2832639,
2073.18986755,
2461.87794271,
2501.03666004,
2501.03666004,
2501.03666004,
2501.03666004,
2501.03666004,
2483.08993915,
2483.08993915,
2501.03666004,
2501.03666004,
2501.03666004,
2469.39075147,
2382.50726301,
2094.40831677,
1762.93300948,
1709.2935141,
1561.52419132,
1561.52419132,
1561.52419132,
1561.52419132,
1455.18324406,
1561.52419132,
1788.6183418,
2073.18986755,
2461.87794271,
2501.03666004,
2501.03666004,
2501.03666004,
2501.03666004,
2501.03666004,
2483.08993915,
2483.08993915,
2501.03666004,
2501.03666004,
2501.03666004,
2469.39075147,
2382.50726301,
2094.40831677,
1762.93300948,
1709.2935141,
1561.52419132,
1561.52419132,
1561.52419132,
1561.52419132,
1455.18324406,
1561.52419132,
1849.2832639,
2073.18986755,
2461.87794271,
2501.03666004,
2501.03666004,
2501.03666004,
2501.03666004,
2501.03666004,
2483.08993915,
2483.08993915,
2501.03666004,
2501.03666004,
2501.03666004,
2469.39075147,
2382.50726301,
2094.40831677,
2026.36855132,
1709.2935141,
1622.18911342,
1561.52419132,
1561.52419132,
1561.52419132,
1495.33031306,
1561.52419132,
1849.2832639,
2336.6254094,
2461.87794271,
2501.03666004,
2501.03666004,
2501.03666004,
2501.03666004,
2501.03666004,
2483.08993915,
2483.08993915,
2501.03666004,
2501.03666004,
2501.03666004,
2469.39075147,
2382.50726301,
2094.40831677,
1792.35740825,
1609.70828296,
1522.60388228,
1522.60388228,
1461.93896018,
1461.93896018,
1395.74508192,
1522.60388228,
1878.70766267,
2102.61426632,
2227.86679963,
2313.23748749,
2313.23748749,
2313.23748749,
2313.23748749,
2313.23748749,
2295.29076661,
2295.29076661,
2313.23748749,
2313.23748749,
2313.23748749,
2313.23748749,
2148.49611994,
1860.39717369,
1792.35740825,
1738.71791287,
1522.60388228,
1522.60388228,
1522.60388228,
1522.60388228,
1456.41000402,
1522.60388228,
1878.70766267,
2102.61426632,
2274.07877016,
2313.23748749,
2313.23748749,
2313.23748749,
2313.23748749,
2313.23748749,
2295.29076661,
2295.29076661,
2313.23748749,
2313.23748749,
2313.23748749,
2313.23748749,
2226.35399903,
1860.39717369,
2026.36855132,
1972.72905595,
1885.62465527,
1622.18911342,
1622.18911342,
1622.18911342,
1555.99523516,
1622.18911342,
2112.71880575,
2336.6254094,
2508.08991324,
2547.24863057,
2547.24863057,
2547.24863057,
2547.24863057,
2547.24863057,
2529.30190968,
2529.30190968,
2547.24863057,
2547.24863057,
2547.24863057,
2547.24863057,
2460.36514211,
2094.40831677,
2026.36855132,
1972.72905595,
1885.62465527,
1622.18911342,
1622.18911342,
1622.18911342,
1555.99523516,
1622.18911342,
2112.71880575,
2336.6254094,
2508.08991324,
2547.24863057,
2547.24863057,
2547.24863057,
2547.24863057,
2547.24863057,
2529.30190968,
2529.30190968,
2547.24863057,
2547.24863057,
2547.24863057,
2547.24863057,
2460.36514211,
2094.40831677,
2026.36855132,
1972.72905595,
1885.62465527,
1622.18911342,
1622.18911342,
1622.18911342,
1555.99523516,
1622.18911342,
2112.71880575,
2336.6254094,
2461.87794271,
2501.03666004,
2547.24863057,
2547.24863057,
2547.24863057,
2547.24863057,
2529.30190968,
2529.30190968,
2547.24863057,
2547.24863057,
2547.24863057,
2547.24863057,
2460.36514211,
2094.40831677,
2026.36855132,
1972.72905595,
1885.62465527,
1885.62465527,
1622.18911342,
1622.18911342,
1555.99523516,
1885.62465527,
2112.71880575,
2414.48328849,
2508.08991324,
2631.42197397,
2631.42197397,
2631.42197397,
2631.42197397,
2631.42197397,
2613.47525309,
2613.47525309,
2547.24863057,
2547.24863057,
2547.24863057,
2547.24863057,
2460.36514211,
2172.26619586,
2026.36855132,
1972.72905595,
1885.62465527,
1885.62465527,
1885.62465527,
1622.18911342,
1555.99523516,
1885.62465527,
2112.71880575,
2414.48328849,
2508.08991324,
2547.24863057,
2547.24863057,
2631.42197397,
2631.42197397,
2631.42197397,
2613.47525309,
2613.47525309,
2547.24863057,
2547.24863057,
2547.24863057,
2547.24863057,
2460.36514211,
2172.26619586,
1792.35740825,
1738.71791287,
1651.61351219,
1651.61351219,
1651.61351219,
1651.61351219,
1456.41000402,
1651.61351219,
1878.70766267,
2180.47214542,
2274.07877016,
2313.23748749,
2313.23748749,
2397.41083089,
2397.41083089,
2397.41083089,
2379.46411001,
2379.46411001,
2313.23748749,
2313.23748749,
2313.23748749,
2313.23748749,
2226.35399903,
1938.25505278,
1792.35740825,
1738.71791287,
1651.61351219,
1651.61351219,
1651.61351219,
1522.60388228,
1456.41000402,
1651.61351219,
1878.70766267,
2180.47214542,
2274.07877016,
2313.23748749,
2313.23748749,
2397.41083089,
2397.41083089,
2397.41083089,
2379.46411001,
2379.46411001,
2397.41083089,
2313.23748749,
2313.23748749,
2313.23748749,
2226.35399903,
1938.25505278,
2026.36855132,
1972.72905595,
1885.62465527,
1885.62465527,
1885.62465527,
1885.62465527,
1819.430777,
1885.62465527,
2190.57668484,
2414.48328849,
2508.08991324,
2631.42197397,
2631.42197397,
2631.42197397,
2631.42197397,
2631.42197397,
2613.47525309,
2613.47525309,
2631.42197397,
2631.42197397,
2547.24863057,
2547.24863057,
2460.36514211,
2172.26619586,
2026.36855132,
1972.72905595,
1885.62465527,
1885.62465527,
1885.62465527,
1885.62465527,
1819.430777,
1885.62465527,
2112.71880575,
2414.48328849,
2508.08991324,
2547.24863057,
2547.24863057,
2631.42197397,
2631.42197397,
2631.42197397,
2613.47525309,
2613.47525309,
2631.42197397,
2547.24863057,
2547.24863057,
2547.24863057,
2460.36514211,
2172.26619586,
2026.36855132,
1972.72905595,
1885.62465527,
1885.62465527,
1885.62465527,
1885.62465527,
1555.99523516,
1885.62465527,
2112.71880575,
2336.6254094,
2461.87794271,
2547.24863057,
2547.24863057,
2547.24863057,
2547.24863057,
2547.24863057,
2529.30190968,
2529.30190968,
2547.24863057,
2547.24863057,
2547.24863057,
2547.24863057,
2460.36514211,
2172.26619586,
2104.22643042,
1972.72905595,
1885.62465527,
1885.62465527,
1885.62465527,
1885.62465527,
1819.430777,
1885.62465527,
2190.57668484,
2414.48328849,
2592.26325664,
2631.42197397,
2631.42197397,
2631.42197397,
2631.42197397,
2631.42197397,
2613.47525309,
2613.47525309,
2631.42197397,
2631.42197397,
2631.42197397,
2631.42197397,
2544.53848551,
2172.26619586,
2104.22643042,
2050.58693505,
1885.62465527,
1885.62465527,
1885.62465527,
1885.62465527,
1819.430777,
1885.62465527,
2190.57668484,
2498.65663189,
2592.26325664,
2631.42197397,
2631.42197397,
2631.42197397,
2631.42197397,
2631.42197397,
2613.47525309,
2613.47525309,
2631.42197397,
2631.42197397,
2631.42197397,
2631.42197397,
2544.53848551,
2172.26619586,
1870.21528734,
1738.71791287,
1651.61351219,
1651.61351219,
1651.61351219,
1651.61351219,
1585.41963393,
1651.61351219,
1956.56554176,
2264.64548882,
2358.25211357,
2397.41083089,
2397.41083089,
2397.41083089,
2397.41083089,
2397.41083089,
2379.46411001,
2379.46411001,
2397.41083089,
2397.41083089,
2397.41083089,
2397.41083089,
2310.52734243,
1938.25505278,
1870.21528734,
1816.57579197,
1651.61351219,
1651.61351219,
1651.61351219,
1651.61351219,
1585.41963393,
1651.61351219,
1956.56554176,
2264.64548882,
2358.25211357,
2397.41083089,
2397.41083089,
2397.41083089,
2397.41083089,
2397.41083089,
2379.46411001,
2379.46411001,
2397.41083089,
2397.41083089,
2397.41083089,
2397.41083089,
2310.52734243,
1938.25505278,
2104.22643042,
2050.58693505,
1885.62465527,
1885.62465527,
1885.62465527,
1885.62465527,
1819.430777,
1885.62465527,
2190.57668484,
2414.48328849,
2592.26325664,
2631.42197397,
2631.42197397,
2631.42197397,
2631.42197397,
2631.42197397,
2613.47525309,
2613.47525309,
2631.42197397,
2631.42197397,
2631.42197397,
2631.42197397,
2544.53848551,
2172.26619586,
2104.22643042,
2050.58693505,
1885.62465527,
1885.62465527,
1885.62465527,
1885.62465527,
1819.430777,
1885.62465527,
2112.71880575,
2414.48328849,
2508.08991324,
2631.42197397,
2631.42197397,
2631.42197397,
2631.42197397,
2631.42197397,
2613.47525309,
2613.47525309,
2631.42197397,
2631.42197397,
2631.42197397,
2631.42197397,
2460.36514211,
2172.26619586,
2104.22643042,
1972.72905595,
1885.62465527,
1885.62465527,
1885.62465527,
1885.62465527,
1819.430777,
1885.62465527,
2112.71880575,
2336.6254094,
2461.87794271,
2547.24863057,
2547.24863057,
2547.24863057,
2547.24863057,
2547.24863057,
2529.30190968,
2529.30190968,
2547.24863057,
2547.24863057,
2547.24863057,
2547.24863057,
2460.36514211,
2172.26619586,
]
real = np.array(real)
plt.plot(test_data["Tarih"], test_preds["yhat1"])
plt.plot(test_data["Tarih"], real)
plt.show()
from sklearn.metrics import mean_absolute_percentage_error
mean_absolute_percentage_error(real, test_preds["yhat1"].values)
submission = test_preds[["ds", "yhat1"]]
submission.columns = ["Tarih", "Dağıtılan Enerji (MWh)"]
submission
submission.to_csv("submission.csv", index=False)
|
import tensorflow as tf
from tensorflow import keras
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
from PIL import Image
import pathlib
data_dir = pathlib.Path("../input/emotion-detection-fer/train")
total_images = len(list(data_dir.glob("*/*.png")))
features = [item.name for item in data_dir.glob("*")]
data_dir2 = pathlib.Path("../input/emotion-detection-fer/test")
BATCH_SIZE = 32
IMG_HEIGHT = 224
IMG_WIDTH = 224
image_generator = keras.preprocessing.image.ImageDataGenerator(rescale=(1.0 / 255))
x_train = image_generator.flow_from_directory(
directory=str(data_dir),
batch_size=BATCH_SIZE,
shuffle=True,
target_size=(IMG_HEIGHT, IMG_WIDTH),
classes=features,
)
val_data = image_generator.flow_from_directory(
directory=str(data_dir),
batch_size=BATCH_SIZE,
shuffle=True,
target_size=(IMG_HEIGHT, IMG_WIDTH),
classes=features,
)
x_test = image_generator.flow_from_directory(
directory=str(data_dir2),
batch_size=BATCH_SIZE,
shuffle=True,
target_size=(IMG_HEIGHT, IMG_WIDTH),
classes=features,
)
IMG_SIZE = 224
IMG_SHAPE = (IMG_SIZE, IMG_SIZE) + (3,)
base_model = keras.applications.MobileNetV2(input_shape=IMG_SHAPE)
# import tensorflow_hub as hub
# m = keras.Sequential([
# hub.KerasLayer("https://kaggle.com/models/google/mobilenet-v2/frameworks/TensorFlow2/variations/140-224-feature-vector/versions/2",
# trainable=False), # Can be True, see below.
# keras.layers.Dense(128),
# keras.layers.Activation('relu'),
# keras.layers.Dense(64),
# keras.layers.Activation('relu'),
# keras.layers.Dense(7, activation= 'softmax')
# ])
# m.build([None, 224, 224, 3])
base_model.trainable = False
base_model.summary()
base_model_input = base_model.layers[0].input
base_model_output = base_model.layers[-2].output
base_model_output = keras.layers.Dense(128)(base_model_output)
base_model_output = keras.layers.Activation("relu")(base_model_output)
base_model_output = keras.layers.Dense(64)(base_model_output)
base_model_output = keras.layers.Activation("relu")(base_model_output)
base_model_output = keras.layers.Dense(7, activation="softmax")(base_model_output)
base_model_output
f_model = keras.Model(inputs=base_model_input, outputs=base_model_output)
f_model.summary()
f_model.compile(loss="categorical_crossentropy", optimizer="adam", metrics=["accuracy"])
history = f_model.fit(x=x_train, epochs=20, validation_data=val_data)
print("Testing Accuracy", f_model.evaluate(x_test))
f_model.save("m")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# import the necessary library
import pandas as pd
import numpy as np
import tensorflow as tf
import re
import matplotlib.pyplot as plt
plt.style.use("ggplot")
# ## 1. Import Dataset & Basic Info
# * 1. Import dataset
# * 2. Basic information of dataset
# * 3. data preprocessing
# **1.1 Import dataset**
#
# import dataset
data = pd.read_csv("../input/predicting-dublin-rental-daftie/daft_v_2.csv")
data
# There is a problem in the "price" column. The value in "price" column is string. If we want to predict it or pass it in our machine learning model we need convert it to numbers. there are two different units for rental, "Per week" and "Per month". I will do it later.
# First, we need to check the basic information of this dataset.
# **1.2 Basic information of dataset**
# basic info of dataset
data.shape
# From above, we can easly find out that there are 2718 rows and 10 columns in this dataset. Each row represents a real property in Dublin city. The 10 columns respectively represent the "price","address", "number of bathroom", "number of bedroom", "if furnished or not","description","property type","property ID","longitude" and "latitude".
#
data.describe
# **1.3 data preprocessing**
# convert the string in "price" col to integer.
p_list = []
for price in data["price"]:
num = re.findall("\d+\.?\d*", price)
num = "".join(num)
num = int(num)
if "Per week" in price:
num = (num / 7) * 30 # convert the weekly rental to monthly
p_list.append(num)
data_copy = data.copy()
data_copy["price"] = p_list
data_copy.head(10)
# ## EDA
# In this part, I will try to explore if there is any corelationships bwetween "price" feature and other features in dataset.
#
fig1 = plt.figure(figsize=(15, 8))
ax1 = plt.subplot()
ax1.hist(x=data_copy["price"], bins=100)
plt.xlim((0, 15000)) # set the x limited value.
plt.xlabel("rental of property")
plt.ylabel("count")
plt.title("Rental Histogram")
plt.show()
# From the above, we can know that most properties' rental are under 4000 euro per month in Dublin and above 1000 euro.
# There will be several bedrooms in some properties. Most of time, we just want to rent a bedroom to live. So, I will use the rental to divide the number of bedrooms to get a new feature.
#
data_copy["rental_per_room"] = data_copy["price"] / data_copy["bedroom"]
data_copy
data_copy["rental_per_room"][np.isinf(data_copy["rental_per_room"])] = -1
data_copy
sum(data_copy["description"][data_copy["rental_per_room"] == -1].isnull())
data_copy["description"][data_copy["rental_per_room"] == -1]
fig1 = plt.figure(figsize=(15, 8))
ax1 = plt.subplot()
ax1.hist(
x=data_copy["rental_per_room"], bins="auto", color="blue", alpha=0.6, width=100
)
plt.xticks(range(0, 8000, 500))
plt.xlabel("The rental per room")
plt.ylabel("count")
plt.title("The rental per room in Dublin")
plt.show()
# Now, we got the histogram of Dublin rental per room. First, there are a lot of properties distribute around 0 euro per month. The reason is that some properties' bedroom number is 0. So, this can be treated as nan value. If you ignore the "0 rental" properties you can find that rental for one room between 500 euro to 4000 euro. That is a huge range. So, I will check some statistics of "The rental per room".
# Before check the statistics, I need to remove the nan value, which means the -1 value in "the rental per room" column.
# remove the -1 value in "the rental per room" column and calculate the mean value.
data_copy["rental_per_room"][data_copy["rental_per_room"] != -1].mean()
# The mean of one room rental in Dublin is 1538 euro per month.
#
data_copy["rental_per_room"][data_copy["rental_per_room"] != -1].median()
# The median of one room rental in Dublin is 1500 euro per month.
# That's a really high rental....
# ### Maps
#
import folium
world_map = folium.Map()
world_map # draw the world map
import json
import requests
url = ""
dublin_geo = f"{url}"
dublin_map = folium.Map(location=[53.1424, 7.6921], zoom_start=12)
folium.GeoJson(
dublin_geo,
style_function=lambda feature: {
"fillColor": "#ffff00",
"color": "black",
"weight": 2,
"dashArray": "5, 5",
},
).add_to(dublin_map)
# display map
dublin_map
|
# Published on April 10, 2023. By Marília Prata, mpwolke
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import cv2
from matplotlib import pyplot as plt
from tqdm import tqdm
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import json
# StackOverflow https://stackoverflow.com/questions/73430014/json-decoder-jsondecodeerror-extra-data-line-1-column-13-char-12
with open("../input/to-test-dataset-notifications/kaggle (2).json") as f:
line_content = [json.loads(line) for line in f.readlines()]
# https://stackoverflow.com/questions/12451431/loading-and-parsing-a-json-file-with-multiple-json-objects
import pandas as pd
df = pd.read_json("../input/to-test-dataset-notifications/kaggle (2).json", lines=True)
# https://stackoverflow.com/questions/12451431/loading-and-parsing-a-json-file-with-multiple-json-objects
df.to_json("new_file.json")
df.head()
|
# # Minimal solution for classifying tweets with neural networks
# In this notebook, I present a minimal solution to classify the Disaster Tweets with a neural network. This is a good starting point for further data preprocessing, modelling, etc.
# **Sources:**
# This notebook contains code from:
# - [Notebook from Shahules](https://www.kaggle.com/code/shahules/basic-eda-cleaning-and-glove)
# - [Notebook from Gunes Evitan](https://www.kaggle.com/code/gunesevitan/nlp-with-disaster-tweets-eda-cleaning-and-bert#7.-Model)
# - [Notebook from Vitalii Mokin](https://www.kaggle.com/code/vbmokin/nlp-eda-bag-of-words-tf-idf-glove-bert)
# - [Real Python - Keras text classification guide](https://realpython.com/python-keras-text-classification/)
# - [keras.io - text classification from scratch](https://keras.io/examples/nlp/text_classification_from_scratch/)
#
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
from sklearn.model_selection import train_test_split
import cleantext
from keras.preprocessing.text import Tokenizer
from keras.utils import pad_sequences
from keras.models import Sequential
from keras import layers
import string
import re
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
train_df = pd.read_csv("../input/nlp-getting-started/train.csv")
test_df = pd.read_csv("../input/nlp-getting-started/test.csv")
train_df
# # Minimal EDA
print("training set info: " + str(train_df.info()))
print("=======================")
print("test set info: " + str(test_df.info()))
# The column Text is complete, so we use this for our minimal solution.
train_df.groupby("target").count()
# The two classes are more or less balanced. No sample weighting or over/under sampling is done in our minimal solution.
# # Data Cleaning
sentences_train, sentences_test, y_train, y_test = train_test_split(
train_df["text"], train_df["target"], test_size=0.2, random_state=42
)
# Use cleantext package, to remove URL, punctuation etc and transform tweets to lists.
sentences_train = sentences_train.apply(lambda txt: cleantext.clean_words(txt))
sentences_test = sentences_test.apply(lambda txt: cleantext.clean_words(txt))
sentences_test_df = test_df["text"].apply(lambda txt: cleantext.clean_words(txt))
tokenizer = Tokenizer(num_words=5000)
tokenizer.fit_on_texts(sentences_train)
train_texts = tokenizer.texts_to_sequences(sentences_train)
test_texts = tokenizer.texts_to_sequences(sentences_test)
sequences_test_df = tokenizer.texts_to_sequences(sentences_test_df)
vocab_size = len(tokenizer.word_index) + 1 # 0 is reserved for unkown words, thus add 1
embedding_dim = 20 # Dimensionality of the embedding space
sequence_length = int(
max([len(txt) for txt in train_texts]) * 1.5
) # length for tweet sequence (max tweet sequence length plus safety margin)
train_texts_padded = pad_sequences(train_texts, padding="post", maxlen=sequence_length)
test_texts_padded = pad_sequences(test_texts, padding="post", maxlen=sequence_length)
sequences_test_df_padded = pad_sequences(
sequences_test_df, padding="post", maxlen=sequence_length
)
# # Model
model = Sequential()
model.add(
layers.Embedding(
input_dim=vocab_size, output_dim=embedding_dim, input_length=sequence_length
)
)
model.add(layers.SpatialDropout1D(0.1)) # Against overfitting
model.add(layers.GlobalMaxPool1D())
model.add(layers.Dense(10, activation="relu"))
model.add(layers.Dense(10, activation="relu"))
model.add(layers.Dense(10, activation="relu"))
model.add(layers.Dense(1, activation="sigmoid"))
model.compile(optimizer="adam", loss="binary_crossentropy", metrics=["accuracy"])
model.summary()
history = model.fit(
train_texts_padded,
y_train,
epochs=5,
verbose=True,
validation_data=(test_texts_padded, y_test),
batch_size=100,
)
loss, accuracy = model.evaluate(train_texts_padded, y_train, verbose=False)
print("Training Accuracy: {:.4f}".format(accuracy))
loss, accuracy = model.evaluate(test_texts_padded, y_test, verbose=False)
print("Testing Accuracy: {:.4f}".format(accuracy))
# # Submission
sample_sub = pd.read_csv("../input/nlp-getting-started/sample_submission.csv")
y_pre = model.predict(sequences_test_df_padded)
y_pre = np.round(y_pre).astype(int).reshape(3263)
sub = pd.DataFrame({"id": sample_sub["id"].values.tolist(), "target": y_pre})
sub.to_csv("submission.csv", index=False)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn import preprocessing
from sklearn.model_selection import train_test_split
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import StratifiedKFold
from sklearn.metrics import roc_auc_score
from xgboost.sklearn import XGBClassifier
train_path = "/kaggle/input/playground-series-s3e12/train.csv"
test_path = "/kaggle/input/playground-series-s3e12/test.csv"
sub_path = "/kaggle/input/playground-series-s3e12/sample_submission.csv"
df_train = pd.read_csv(train_path)
df_test = pd.read_csv(test_path)
df_sub = pd.read_csv(sub_path)
df_train.head(3)
df_test.head(3)
df_train.drop("id", inplace=True, axis=1)
df_test.drop("id", inplace=True, axis=1)
new_list = list(
set(df_train.index.tolist()) - set(df_train[df_train["urea"] < 15].index.tolist())
)
df_train = df_train.loc[new_list,]
s_temp = df_train["target"].copy()
df_train.drop("target", axis=1, inplace=True)
df_train["train"], df_test["train"] = 1, 0
df_train["target"] = s_temp.copy()
df = pd.concat([df_train, df_test])
# ### Feature Generation
df["gravity/ph"] = df["gravity"] / df["ph"]
df["osmo/cond"] = df["osmo"] / df["cond"]
df["gravity*ph"] = df["gravity"] * df["ph"]
df["gravity*osmo"] = df["gravity"] * df["osmo"]
df["osmo*urea"] = df["osmo"] * df["urea"]
df["cond_urea_ph"] = df["cond"] * df["urea"] / df["ph"]
df["ph*osmo"] = df["ph"] * df["osmo"]
df["cond*calc"] = df["cond"] * df["calc"]
df["gravity/calc"] = df["gravity"] / df["calc"]
df["gravity_osmo_urea"] = df["gravity"] * df["osmo"] / df["urea"]
# ### Normalizing Data
columns = df.columns.tolist()
columns.remove("train")
columns.remove("target")
data_norm = preprocessing.normalize(df[columns], axis=0)
df_norm = pd.DataFrame(data_norm, columns=columns)
df_norm.reset_index(drop=True, inplace=True)
df.reset_index(drop=True, inplace=True)
df_norm["train"], df_norm["target"] = df["train"], df["target"]
# ### Setting Variables for Learning
features = [
"gravity/ph",
"osmo/cond",
"gravity*ph",
"gravity*osmo",
"osmo*urea",
"cond_urea_ph",
"ph*osmo",
"cond*calc",
"gravity/calc",
]
X = df_norm[df_norm["train"] == 1][features].copy()
y = df_norm[df_norm["train"] == 1]["target"].copy()
print(X.shape, y.shape)
# XGBoost Classification
str_cv_folds = StratifiedKFold(n_splits=5, random_state=1221, shuffle=True)
xbc_model = XGBClassifier(
n_estimators=120, learning_rate=0.05, max_depth=4, eval_metric="auc"
)
cv_scores = cross_val_score(
xbc_model, X, y, scoring="roc_auc", cv=str_cv_folds, n_jobs=-1
)
print("CV Mean Score:", cv_scores.mean())
# ### 120, 0.05, 4
# ### CV Mean Score: 0.8124350437393917
X = df_norm[df_norm["train"] == 1][features].copy()
y = df_norm[df_norm["train"] == 1]["target"].copy()
X_train, X_val, y_train, y_val = train_test_split(X, y, test_size=0.3, random_state=121)
X_train, y_train = X, y
xbc_model = XGBClassifier(
n_estimators=140, learning_rate=0.055, max_depth=4, eval_metric="auc"
)
xbc_model.fit(X_train, y_train)
xbc_preds = xbc_model.predict(X_val)
xbc_score = roc_auc_score(y_val, xbc_preds)
print("ROC AUC Score:", xbc_score)
# ### 140, 0.05, 4
# ### ROC AUC Score: 0.949986907567426
X_sub = df_norm[df_norm["train"] == 0][features].copy()
xbc_preds = xbc_model.predict_proba(X_sub)
xbc_preds = pd.DataFrame(data=xbc_preds, columns=[0, 1])
df_sub["target"] = xbc_preds[1]
df_sub.head()
df_sub.to_csv("submission_III.csv", index=False)
|
import os
import unicodedata
import pandas as pd
from tqdm.notebook import tqdm
import glob
df = pd.DataFrame(columns=["image_name", "prompt"])
df.info
cnt = 0
for i in tqdm(range(1, 2401, 300)):
image_dir = f"/kaggle/input/diffusiondb-14m-{str(i)}to{str(i+300)}/"
print(image_dir)
# for j in range(i,i+300):
# print(image_dir+f'/{str(j).zfill(6)}/*.webp')
# cnt += len(glob.glob(image_dir+f'/{str(j).zfill(6)}/*.webp'))
# print(cnt)
df_t = pd.read_csv(image_dir + f"DiffusionDB_14M_{str(i)}to{str(i+300)}.csv")
df_t["image_name"] = image_dir + df_t["image_name"]
df = pd.concat([df, df_t], axis=0)
print(df.shape)
for i in tqdm(range(2401, 3001, 600)):
image_dir = f"/kaggle/input/diffusiondb-14m-{str(i)}to{str(i+600)}/"
print(image_dir)
# for j in range(i,i+600):
# #print(image_dir+f'/{str(j).zfill(6)}/*.webp')
# cnt += len(glob.glob(image_dir+f'/{str(j).zfill(6)}/*.webp'))
# print(cnt)
df_t = pd.read_csv(image_dir + f"DiffusionDB_14M_{str(i)}to{str(i+600)}.csv")
df_t["image_name"] = image_dir + df_t["image_name"]
df = pd.concat([df, df_t], axis=0)
print(df.shape)
for i in tqdm(range(3001, 9601, 300)):
image_dir = f"/kaggle/input/diffusiondb-14m-{str(i)}to{str(i+300)}/"
print(image_dir)
# for j in range(i,i+300):
# #print(image_dir+f'/{str(j).zfill(6)}/*.webp')
# cnt += len(glob.glob(image_dir+f'/{str(j).zfill(6)}/*.webp'))
# print(cnt)
df_t = pd.read_csv(image_dir + f"DiffusionDB_14M_{str(i)}to{str(i+300)}.csv")
df_t["image_name"] = image_dir + df_t["image_name"]
df = pd.concat([df, df_t], axis=0)
print(df.shape)
for i in tqdm(range(9601, 14001, 400)):
image_dir = f"/kaggle/input/diffusiondb-14m-{str(i)}to{str(i+400)}/"
print(image_dir)
# for j in range(i,i+400):
# #print(image_dir+f'/{str(j).zfill(6)}/*.webp')
# cnt += len(glob.glob(image_dir+f'/{str(j).zfill(6)}/*.webp'))
# print(cnt)
df_t = pd.read_csv(image_dir + f"DiffusionDB_14M_{str(i)}to{str(i+400)}.csv")
df_t["image_name"] = image_dir + df_t["image_name"]
df = pd.concat([df, df_t], axis=0)
print(df.shape)
# df = df[['filepath', 'prompt']].copy()
# assert not df['filepath'].isnull().any()
df
df = df[["image_name", "prompt"]]
df.reset_index(drop=True)
df
df.to_csv("new_diffusiondb_metadata_large_53w.csv", index=False)
|
import torch
import requests
from PIL import Image
import matplotlib.pyplot as plt
from diffusers import StableDiffusionDepth2ImgPipeline
pipe = StableDiffusionDepth2ImgPipeline.from_pretrained(
"stabilityai/stable-diffusion-2-depth",
torch_dtype=torch.float16,
).to("cuda")
# https://github.com/huggingface/diffusers/blob/main/src/diffusers/pipelines/stable_diffusion/pipeline_stable_diffusion_depth2img.py
url = "http://images.cocodataset.org/val2017/000000039769.jpg"
init_image = Image.open(requests.get(url, stream=True).raw)
def show_image_with_og(img1, og):
plt.subplot(1, 2, 1)
plt.imshow(og)
plt.subplot(1, 2, 2)
plt.imshow(img1)
plt.tight_layout()
plt.show()
# # Testing the model on pics of cats - demo
prompt = "two tigers"
n_propmt = "bad, deformed, ugly, bad anotomy"
image_cat1 = pipe(
prompt=prompt,
image=init_image,
negative_prompt=n_propmt,
strength=0.7,
num_images_per_prompt=1,
).images[0]
show_image_with_og(image_cat1, init_image)
prompt = "two dogs"
n_propmt = "bad, deformed, ugly, bad anotomy"
image_cat2 = pipe(
prompt=prompt,
image=init_image,
negative_prompt=n_propmt,
strength=1,
num_images_per_prompt=1,
).images[0]
show_image_with_og(image_cat2, init_image)
# # Testing it on a picture of people
url2 = "https://images.pexels.com/photos/2519426/pexels-photo-2519426.jpeg?auto=compress&cs=tinysrgb&w=600"
og_people = people_og = Image.open(requests.get(url2, stream=True).raw)
# ## with strength 0.4:
prompt = "two knights in shining armor"
n_propmt = "bad, deformed, ugly, bad anatomy"
people_gen1 = pipe(
prompt=prompt,
image=og_people,
negative_prompt=n_propmt,
strength=0.4,
num_images_per_prompt=1,
).images[0]
show_image_with_og(people_gen1, people_og)
prompt = "king and queen standing together"
n_propmt = "bad, deformed, ugly, bad anatomy"
people_gen2 = pipe(
prompt=prompt,
image=og_people,
negative_prompt=n_propmt,
strength=0.4,
num_images_per_prompt=1,
).images[0]
show_image_with_og(people_gen2, people_og)
prompt = "green aliens"
n_propmt = "bad anatomy, human"
people_gen3 = pipe(
prompt=prompt,
image=og_people,
negative_prompt=n_propmt,
strength=0.4,
num_images_per_prompt=1,
).images[0]
show_image_with_og(people_gen3, people_og)
# ## with strength 0.7
prompt = "two knights in shining armor"
n_propmt = "bad, deformed, ugly, bad anatomy"
people_gen1 = pipe(
prompt=prompt,
image=og_people,
negative_prompt=n_propmt,
strength=0.7,
num_images_per_prompt=1,
).images[0]
show_image_with_og(people_gen1, people_og)
prompt = "king and queen standing together"
n_propmt = "bad, deformed, ugly, bad anatomy"
people_gen2 = pipe(
prompt=prompt,
image=og_people,
negative_prompt=n_propmt,
strength=0.7,
num_images_per_prompt=1,
).images[0]
show_image_with_og(people_gen2, people_og)
prompt = "green aliens"
n_propmt = "bad anatomy, human"
people_gen3 = pipe(
prompt=prompt,
image=og_people,
negative_prompt=n_propmt,
strength=0.7,
num_images_per_prompt=1,
).images[0]
show_image_with_og(people_gen3, people_og)
# ## with strength 0.9
prompt = "two knights in shining armor"
n_propmt = "bad, deformed, ugly, bad anatomy"
people_gen1 = pipe(
prompt=prompt,
image=og_people,
negative_prompt=n_propmt,
strength=0.9,
num_images_per_prompt=1,
).images[0]
show_image_with_og(people_gen1, people_og)
prompt = "king and queen standing together"
n_propmt = "bad, deformed, ugly, bad anatomy"
people_gen2 = pipe(
prompt=prompt,
image=og_people,
negative_prompt=n_propmt,
strength=0.9,
num_images_per_prompt=1,
).images[0]
show_image_with_og(people_gen2, people_og)
prompt = "green aliens"
n_propmt = "bad anatomy, human"
people_gen3 = pipe(
prompt=prompt,
image=og_people,
negative_prompt=n_propmt,
strength=0.9,
num_images_per_prompt=1,
).images[0]
show_image_with_og(people_gen3, people_og)
# ## with strength 1.0
prompt = "two knights in shining armor"
n_propmt = "bad, deformed, ugly, bad anatomy"
people_gen1 = pipe(
prompt=prompt,
image=og_people,
negative_prompt=n_propmt,
strength=1,
num_images_per_prompt=1,
).images[0]
show_image_with_og(people_gen1, people_og)
prompt = "royal king and queen with crowns"
n_propmt = "bad, deformed, ugly, bad anatomy, tshirt"
people_gen2 = pipe(
prompt=prompt,
image=og_people,
negative_prompt=n_propmt,
strength=1,
num_images_per_prompt=1,
).images[0]
show_image_with_og(people_gen2, people_og)
prompt = "green aliens with tentacles"
n_propmt = "human, clothes, skin"
people_gen3 = pipe(
prompt=prompt,
image=og_people,
negative_prompt=n_propmt,
strength=1,
num_images_per_prompt=1,
).images[0]
show_image_with_og(people_gen3, people_og)
|
import pandas as pd
import re
import string
from wordcloud import WordCloud
from collections import Counter
import warnings
warnings.filterwarnings("ignore")
import tensorflow as tf
print(tf.version.VERSION)
import tensorflow as tf
device_name = tf.test.gpu_device_name()
if len(device_name) > 0:
print("Found GPU at: {}".format(device_name))
else:
device_name = "/device:CPU:0"
print("No GPU, using {}.".format(device_name))
import tensorflow as tf
import tensorflow_hub as hub
# import tensorflow_text as text
from sklearn.model_selection import train_test_split
from sklearn.metrics import confusion_matrix, classification_report
from matplotlib import pyplot as plt
import seaborn as sn
from tensorflow.keras.preprocessing.text import Tokenizer
from tensorflow.keras.preprocessing.sequence import pad_sequences
import bert
from bert import tokenization
import numpy as np
import tensorflow as tf
import tensorflow_hub as hub
import sys
sys.path.append("/kaggle/working/models")
from tensorflow.keras.optimizers import Adam
from official.nlp.data import classifier_data_lib
from official.nlp.bert import tokenization
from official.nlp import optimization
import matplotlib.pyplot as plt
from matplotlib.ticker import MaxNLocator
from sklearn.metrics import confusion_matrix, classification_report
import seaborn as sns
train_df = pd.read_csv("/kaggle/input/preprocess/training.csv")
test_df = pd.read_csv("/kaggle/input/preprocess/testing.csv")
from sklearn import preprocessing
label_encoder = preprocessing.LabelEncoder()
train_df["Subreddit"] = label_encoder.fit_transform(train_df["Subreddit"])
train_df["Subreddit"].unique()
labels = list(label_encoder.classes_)
train_df.sample()
label_encoder = preprocessing.LabelEncoder()
test_df["Subreddit"] = label_encoder.fit_transform(test_df["Subreddit"])
test_df["Subreddit"].unique()
labels = list(label_encoder.classes_)
test_df.sample()
train_df.sample(1)
train_df.shape
test_df.shape
train_df = train_df.dropna(subset=["Sentence"])
train_df.Sentence = [str(text) for text in train_df.Sentence]
test_df = test_df.dropna(subset=["Sentence"])
test_df.Sentence = [str(text) for text in test_df.Sentence]
train_df.shape
test_df.shape
test_df, val_df = train_test_split(test_df, test_size=0.5, random_state=42)
print(test_df.shape)
print(val_df.shape)
test_df
with tf.device("/device:GPU:0"):
train_data = tf.data.Dataset.from_tensor_slices(
(train_df["Sentence"].values, train_df["Subreddit"].values)
)
with tf.device("/device:GPU:0"):
test_data = tf.data.Dataset.from_tensor_slices(
(test_df["Sentence"].values, test_df["Subreddit"].values)
)
valid_data = tf.data.Dataset.from_tensor_slices(
(val_df["Sentence"].values, val_df["Subreddit"].values)
)
for Sentence, Subreddit in valid_data.take(1):
print(Sentence)
print(Subreddit)
for Sentence, Subreddit in train_data.take(1):
print(Sentence)
print(Subreddit)
label_list = [0, 1, 2, 3, 4, 5] # label categories
max_seq_len = 128 # maximum length of token input sequence
batch_size = 32
# with strategy.scope():
with tf.device("/device:GPU:0"):
bert_layer = hub.KerasLayer(
"https://tfhub.dev/tensorflow/bert_en_uncased_L-12_H-768_A-12/2", trainable=True
)
vocab_file = bert_layer.resolved_object.vocab_file.asset_path.numpy()
do_lower_case = bert_layer.resolved_object.do_lower_case.numpy()
tokenizer = tokenization.FullTokenizer(vocab_file, do_lower_case)
def to_feature(
text, label, label_list=label_list, max_seq_length=max_seq_len, tokenizer=tokenizer
):
example = classifier_data_lib.InputExample(
guid=None, text_a=text.numpy(), text_b=None, label=label.numpy()
)
feature = classifier_data_lib.convert_single_example(
0, example, label_list, max_seq_length, tokenizer
)
return (
feature.input_ids,
feature.input_mask,
feature.segment_ids,
feature.label_id,
)
def to_feature_map(text, label):
input_ids, input_mask, segment_ids, label_id = tf.py_function(
to_feature, inp=[text, label], Tout=[tf.int32, tf.int32, tf.int32, tf.int32]
)
# py_func doesn't set the shape of the returned tensors.
input_ids.set_shape([max_seq_len])
input_mask.set_shape([max_seq_len])
segment_ids.set_shape([max_seq_len])
label_id.set_shape([])
x = {
"input_word_ids": input_ids,
"input_mask": input_mask,
"input_type_ids": segment_ids,
}
return (x, label_id)
# with strategy.scope():
with tf.device("/device:GPU:0"):
train_data = (
train_data.map(to_feature_map, num_parallel_calls=tf.data.experimental.AUTOTUNE)
.shuffle(1000)
.batch(32, drop_remainder=True)
.prefetch(tf.data.experimental.AUTOTUNE)
)
valid_data = (
valid_data.map(to_feature_map, num_parallel_calls=tf.data.experimental.AUTOTUNE)
.batch(32, drop_remainder=True)
.prefetch(tf.data.experimental.AUTOTUNE)
)
with tf.device("/device:GPU:0"):
test_data = test_data.map(to_feature_map).batch(32)
train_data
test_data
def create_model():
input_word_ids = tf.keras.layers.Input(
shape=(max_seq_len,), dtype=tf.int32, name="input_word_ids"
)
input_mask = tf.keras.layers.Input(
shape=(max_seq_len,), dtype=tf.int32, name="input_mask"
)
input_type_ids = tf.keras.layers.Input(
shape=(max_seq_len,), dtype=tf.int32, name="input_type_ids"
)
pooled_output, sequence_output = bert_layer(
[input_word_ids, input_mask, input_type_ids]
)
drop = tf.keras.layers.Dropout(0.2)(pooled_output)
output = tf.keras.layers.Dense(6, activation="softmax", name="output")(drop)
model = tf.keras.Model(
inputs={
"input_word_ids": input_word_ids,
"input_mask": input_mask,
"input_type_ids": input_type_ids,
},
outputs=output,
)
return model
with tf.device("/device:GPU:0"):
model = create_model()
model.compile(
optimizer=tf.keras.optimizers.Adam(1e-5),
loss=tf.keras.losses.SparseCategoricalCrossentropy(from_logits=False),
metrics=[tf.keras.metrics.SparseCategoricalAccuracy(name="acc")],
)
model.summary()
from tensorflow.keras.callbacks import ReduceLROnPlateau, ModelCheckpoint
checkpoint = ModelCheckpoint(
"./weights.h5", monitor="val_loss", mode="min", save_best_only=True, verbose=1
)
epochs = 5
with tf.device("/device:GPU:0"):
history = model.fit(
train_data, validation_data=valid_data, epochs=epochs, callbacks=[checkpoint]
)
model.save("saved_model/my_model")
def get_label_name(number):
labels = list(label_encoder.classes_)
return labels[number]
def prediction_result(sample_example):
predicted_data = tf.data.Dataset.from_tensor_slices(
(sample_example, [0] * len(sample_example))
)
predicted_data = predicted_data.map(to_feature_map).batch(1)
prediction = model.predict(predicted_data)
result = np.where(prediction[0] == np.amax(prediction[0]))
return get_label_name(result[0][0])
y_pred = model.predict(test_data).argmax(axis=-1)
test_y = test_df["Subreddit"].tolist()
cm = confusion_matrix(test_y, y_pred)
df_cm = pd.DataFrame(cm, index=labels, columns=labels)
hmap = sns.heatmap(df_cm, annot=True, fmt="d")
hmap.yaxis.set_ticklabels(hmap.yaxis.get_ticklabels(), rotation=0, ha="right")
hmap.xaxis.set_ticklabels(hmap.xaxis.get_ticklabels(), rotation=30, ha="right")
plt.ylabel("True label")
plt.xlabel("Predicted label")
from sklearn.metrics import classification_report
print(classification_report(test_y, y_pred, target_names=labels))
|
# # ***Library import***
import pandas as pd
import numpy as np
import plotly.express as px
import seaborn as sns
import matplotlib.pyplot as plt
import warnings
warnings.filterwarnings("ignore")
from sklearn.manifold import TSNE
from dataprep.datasets import load_dataset
from dataprep.eda import *
train = pd.read_csv("train.csv")
test = pd.read_csv("test.csv")
# # ***Preprocessing***
train.isnull().any().sum()
def game(on):
start = pd.DataFrame(on.dtypes, columns=["data types"])
start["Missing"] = on.isnull().sum()
start["unique"] = on.nunique()
return start
game(train).style.background_gradient(cmap="Oranges_r")
from sklearn.impute import SimpleImputer
S = SimpleImputer(strategy="most_frequent")
train[
[
"AverageSimilarStartupValuation",
"city",
"AverageFounderTeamAgeFunding",
"FilterFounderNumFounder",
"SumOfCharges",
"Incubator/Accelerator",
"predicted_capital",
"predicted_rating",
"Num Deals",
"Deal Value Growth",
]
] = S.fit_transform(
train[
[
"AverageSimilarStartupValuation",
"city",
"AverageFounderTeamAgeFunding",
"FilterFounderNumFounder",
"SumOfCharges",
"Incubator/Accelerator",
"predicted_capital",
"predicted_rating",
"Num Deals",
"Deal Value Growth",
]
]
)
S = SimpleImputer(strategy="most_frequent")
test[
[
"AverageSimilarStartupValuation",
"city",
"AverageFounderTeamAgeFunding",
"FilterFounderNumFounder",
"SumOfCharges",
"Incubator/Accelerator",
"predicted_capital",
"predicted_rating",
"Num Deals",
"Deal Value Growth",
]
] = S.fit_transform(
test[
[
"AverageSimilarStartupValuation",
"city",
"AverageFounderTeamAgeFunding",
"FilterFounderNumFounder",
"SumOfCharges",
"Incubator/Accelerator",
"predicted_capital",
"predicted_rating",
"Num Deals",
"Deal Value Growth",
]
]
)
n = train[
[
"AverageSimilarStartupValuation",
"city",
"Assets",
"AverageFounderTeamAgeFunding",
"DIPP",
"FilterFounderNumFounder",
"PAT",
"Revenue",
"SumOfCharges",
"Incubator/Accelerator",
"predicted_capital",
"predicted_rating",
"RegularInvestor",
"RegularFund",
"Num Deals",
"Deal Value Growth",
]
]
m = test[
[
"AverageSimilarStartupValuation",
"city",
"Assets",
"AverageFounderTeamAgeFunding",
"DIPP",
"FilterFounderNumFounder",
"PAT",
"Revenue",
"SumOfCharges",
"Incubator/Accelerator",
"predicted_capital",
"predicted_rating",
"RegularInvestor",
"RegularFund",
"Num Deals",
"Deal Value Growth",
]
]
from sklearn.preprocessing import LabelEncoder
for i in n:
La = LabelEncoder()
n[i] = La.fit_transform(n[i])
for o in m:
label = LabelEncoder()
m[o] = label.fit_transform(m[o])
train[
[
"AverageSimilarStartupValuation",
"city",
"Assets",
"AverageFounderTeamAgeFunding",
"DIPP",
"FilterFounderNumFounder",
"PAT",
"Revenue",
"SumOfCharges",
"Incubator/Accelerator",
"predicted_capital",
"predicted_rating",
"RegularInvestor",
"RegularFund",
"Num Deals",
"Deal Value Growth",
]
] = n
test[
[
"AverageSimilarStartupValuation",
"city",
"Assets",
"AverageFounderTeamAgeFunding",
"DIPP",
"FilterFounderNumFounder",
"PAT",
"Revenue",
"SumOfCharges",
"Incubator/Accelerator",
"predicted_capital",
"predicted_rating",
"RegularInvestor",
"RegularFund",
"Num Deals",
"Deal Value Growth",
]
] = m
print(train.dtypes.value_counts().plot(kind="pie"))
print(test.dtypes.value_counts().plot(kind="pie"))
# # ***EDA***
train_copy = train.copy()
x = train_copy.drop("equityValuation", axis=1)
y = train_copy[["equityValuation"]]
t = TSNE(n_components=3, random_state=42)
tsne = t.fit_transform(x)
tsne_data = pd.concat([pd.DataFrame(tsne), y], axis=1)
px.scatter_3d(tsne_data, x=0, y=1, z=2, color="equityValuation", template="plotly_dark")
plt.figure(figsize=(20, 19), facecolor="yellow")
sns.heatmap(train.corr(), annot=True, fmt=".0%", cmap="ocean")
# # ***Auto ML***
from pycaret.regression import *
train.head()
# # ***Setup Models***
setup(
train,
target="equityValuation",
ignore_features="Id",
normalize_method="robust",
remove_outliers=True,
)
# # ***List of Models***
models()
# # ***Compare Model***
compare_models()
# # ***Create Model***
best = create_model("gbr")
print(finalize_model(best))
# # ***Pipeline***
finalize_model(best)
# # ***Plots***
plot_model(best)
# # ***Prediction***
prediction = best.predict(test.drop("Id", axis=1))
# # ***Mean Squared Error*** ,MAE,RMSE,R2,RMSLE,MAPE
predict_model(best)
# # ***Submission***
submission = pd.DataFrame({"Id": test.Id, "equityValuation": prediction})
submission.tail()
submission.to_csv("submission.csv", index=False)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
from icrawler.builtin import BingImageCrawler, GoogleImageCrawler
from pathlib import Path
mitFilter = True
filters = dict(
type="photo", license="commercial,modify"
) # either photo, face, clipart, linedrawing, animated
howmany = 100
ActorName = "Yogi Babu"
Images_SavePath = "/kaggle/working/" + ActorName + "/"
crawler = GoogleImageCrawler(
parser_threads=2, downloader_threads=2, storage={"root_dir": Images_SavePath}
)
if mitFilter == True:
crawler.crawl(
keyword=ActorName, filters=filters, max_num=howmany, min_size=(512, 512)
)
else:
crawler.crawl(keyword=ActorName, max_num=howmany, min_size=(512, 512))
|
# # My first notebook
# ## Any suggestions will be highly appreciated
# The work is divided into three Sections
# 1. Section 1 - Data Analysis (Simple Exploratory Data Analysis to gain insights)
# 2. Section 2 - Data Handling (Performing imputation, scaling, etc..)
# 3. Section 3 - Model fitting and predicting (Used XGBoostRegressor to fit the model)
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
pd.pandas.set_option("display.max_columns", None)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # **SECTION 1 - Data Analysis**
dataset = pd.read_csv(
"/kaggle/input/hackerearth-machine-learning-exhibit-art/dataset/train.csv"
)
test = pd.read_csv(
"/kaggle/input/hackerearth-machine-learning-exhibit-art/dataset/test.csv"
)
dataset.head()
# Could have worked with date columns but dropped it anyway
dataset.drop(
[
"Customer Id",
"Artist Name",
"Customer Location",
"Scheduled Date",
"Delivery Date",
],
axis=1,
inplace=True,
)
test.drop(
[
"Customer Id",
"Artist Name",
"Customer Location",
"Scheduled Date",
"Delivery Date",
],
axis=1,
inplace=True,
)
dataset["Cost"] = dataset["Cost"].abs()
# ## Missing Data Exploration
# Getting all columns with missing data
missing_data_col = [col for col in dataset.columns if dataset[col].isnull().sum() > 0]
missing_data_col
dataset[missing_data_col].isnull().sum()
# Try to see how cost gets affected on columns with missing data
df = dataset[missing_data_col].copy()
df.fillna("-1", inplace=True)
for col in missing_data_col:
df[col] = df[col].apply(lambda x: "Missing" if x == "-1" else "Available")
df["Cost"] = dataset["Cost"]
for col in missing_data_col:
df.groupby(col)["Cost"].median().plot.bar()
plt.xlabel(col)
plt.ylabel("Cost")
plt.title(col)
plt.show()
# ### The median cost of missing data values and the median cost of available data values for the missing data columns do not vary by much
# # Working with the numerical columns to get some insights about outliers and what role does it play in the cost
# !pip install dtale
# import dtale
# # Getting all the numerical columns
# numerical_cols = [feature for feature in dataset.columns if dataset[feature].dtypes != 'O']
# df = dataset[numerical_cols].copy()
# details = dtale.show(df)
# details
# #### Note - dtale doesn't works well with kaggle notebook so the below conclusions were drawn from using dtale on Jupyter Notebook
# ## Following conclusions are to be drawn
# #### 1. Height is moderately skewed (0.59)
# #### 2. Width is highly skewed (1.55)
# #### 3. Weight is highly skewed (21.56)
# #### 4. Price of Sculpture is highly skewed (22.21)
# #### 5. Base shipping price is moderately skewed (0.92)
# #### 6. Cost is highly skewed (29.87)
# # Looking for outliers in the data
numerical_cols = [
feature for feature in dataset.columns if dataset[feature].dtypes != "O"
]
df = dataset[numerical_cols].copy()
df.fillna(0, inplace=True)
for col in df.columns:
plt.boxplot(col, data=df)
plt.xlabel(col)
plt.title(col)
plt.show()
# # High outliers have led to high skewness in these columns
# # Check for categorical data and finding insights
# Getting all the categorical columns
categorical_cols = [
feature for feature in dataset.columns if dataset[feature].dtypes == "O"
]
categorical_cols
df = dataset[categorical_cols].copy()
df["Cost"] = dataset["Cost"]
for col in df.columns[:-1]:
df.groupby(col)["Cost"].median().plot.bar()
plt.xlabel(col)
plt.ylabel("COST")
plt.title(col)
plt.show()
# #### NOTE - Not considering mean as the computing factor as mean is influneced by outliers
# ### Some insights are
# ##### 1. Marble and stone products are costly followed by brass and bronze
# ##### 2. International column has almost no impact on cost
# ##### 3. Express shipment leads to a slight increment in the cost
# ##### 4. Installation included has almost no to very little impact on cost
# ##### 5. Transport has very slight impact on cost (airways being the costliest and waterways being the cheapest
# #### 6. Fragility has a high impact on cost (Not fragile = More cost)
# ##### 7. Customer information has slight impact on cost
# ##### 8. Remote location has almost no impact on cost
# # Checking the corelation of numerical columns w.r.t Cost
dataset.corr()["Cost"]
# # **SECTION 2 - Data Handling**
dataset.head(3)
X_train = dataset.iloc[:, :-1]
y_train = dataset["Cost"]
# ### The y values are highly skewed so we need to normalize them.
# We can perform antilog to get back the original results
y_train = y_train.apply(lambda x: np.log(x) if x > 0.0 else 0.0)
y_train
# ## Handle missing data
# Find all the numberical cols with missing data
num_missing_cols = [
col
for col in dataset.columns
if dataset[col].dtypes != "O" and dataset[col].isnull().sum() > 0
]
num_missing_cols
df = dataset.copy()
for col in num_missing_cols:
mean = df[col].mean()
median = df[col].median()
print(f"{col} has a mean of {round(mean,2)} and a median of {median}")
def fill_misisng_cols(col_name, df):
for col in col_name:
med = df[col].median()
df[col].fillna(med, inplace=True)
fill_misisng_cols(col_name=num_missing_cols, df=X_train)
fill_misisng_cols(col_name=num_missing_cols, df=test)
X_train[num_missing_cols].isnull().sum()
# ### Get all the categorical columns to fill missing values and also do other transformations if possible
cat_missing_cols = [
col
for col in dataset.columns
if dataset[col].dtypes == "O" and dataset[col].isnull().sum() > 0
]
cat_missing_cols
# Filling the categorical columns with missing data with the mode of these columns
def cat_miss(df):
for col in cat_missing_cols:
mode = dataset[col].mode()[0]
df[col].fillna(mode, inplace=True)
cat_miss(X_train)
cat_miss(test)
X_train[cat_missing_cols].head()
# Label encoding using pd.get_dummies()
X_train = pd.get_dummies(X_train, drop_first=True)
test = pd.get_dummies(test, drop_first=True)
# # SECTION 3 - Model Fitting and Prediction
# Height', 'Width', 'Weight', 'Price Of Sculpture' needs to be scaled down as they have high outliers
num_cols_all = [
col
for col in X_train.columns
if X_train[col].dtypes != "O" and X_train[col].nunique() > 10
]
num_cols_all
scale_cols = num_cols_all[1:-1]
scale_cols
from sklearn.preprocessing import MinMaxScaler
scale = MinMaxScaler()
X_train[scale_cols] = scale.fit_transform(X_train[scale_cols])
test[scale_cols] = scale.transform(test[scale_cols])
X_train.head(3)
# ## Finding the useful parameters
from sklearn.linear_model import Lasso
from sklearn.feature_selection import SelectFromModel
# #### At the moment we will take alpha as 0.001 and select the columns generated.
# Based on results optained we may update alpha
feature_selection_model = SelectFromModel(Lasso(alpha=0.001))
feature_selection_model.fit(X_train, y_train)
f_model_arr = list(feature_selection_model.get_support())
all_cols = [i for i in X_train.columns]
cols_chosen = [all_cols[i] for i in range(len(f_model_arr)) if f_model_arr[i] == True]
# cols_chosen # Chosen columns
X_train = X_train[cols_chosen]
test = test[cols_chosen]
X_train = X_train.values
test = test.values
# ## Using XGBoost to fit the model
from xgboost import XGBRegressor
xg = XGBRegressor()
xg.fit(X_train, y_train)
# # Performing the Grid Search to get the best parameters
# from sklearn.model_selection import GridSearchCV
# param_grid = [
# {'n_estimators': [250,280,300,330, 360],
# 'max_depth': [10, 20,30,40,50],
# 'learning_rate': [0.1,0.3,0.5],
# }]
# grid_cv = GridSearchCV(xg, param_grid=param_grid, cv=10, n_jobs=-1)
# grid_cv.fit(X_train, y_train)
# best_params = grid_cv.best_estimator_
# best_params
# ### The Best Parameters Were found out to be
# n_estimators=250
# max_depth=10
# learning_rate=0.1
xg1 = XGBRegressor(
n_estimators=250, n_jobs=-1, max_depth=10, base_score=0.1, learning_rate=0.1
)
xg1.fit(X_train, y_train)
predicted = xg1.predict(test)
predicted
# ## The 'Cost' column in the train dataset was log normalised so we need to antilog the predicted values
predicted = np.power(np.e, predicted)
predicted = np.round(predicted, 2)
predicted
test = pd.read_csv(
"/kaggle/input/hackerearth-machine-learning-exhibit-art/dataset/test.csv"
)
id_col = pd.DataFrame(test["Customer Id"], columns=["Customer Id"])
cost_col = pd.DataFrame(predicted, columns=["Cost"])
result = pd.concat([id_col, cost_col], axis=1)
result.head()
result.to_csv("Submission.csv", index=False)
|
# # **Earthquake Analysis using Ensembling Technique**
# ## Problem Statement:
# Earthquakes are one of the most destructive and unpredictable natural disasters in the world, causing significant damage to infrastructure, property, and human life. While scientists have made significant progress in predicting earthquakes, there is still much that is unknown about these events, including their magnitude, location, and timing.
# The goal of this project is to develop an accurate and reliable machine learning model for predicting the severity of earthquakes based on a range of input variables, including seismic activity, geographical location, and historical earthquake data. To achieve this, the project will employ a range of ensemble techniques and perform cross-validation to evaluate the performance of the model.
# The accuracy of the model will be evaluated using a range of performance metrics, such as mean absolute error (MAE), root mean squared error (RMSE), and R-squared (R2). The goal is to develop a model that can accurately predict the severity of earthquakes with a high degree of precision and recall.
# By using ensemble techniques and cross-validation, the project aims to develop a highly accurate and reliable model that can be used to improve earthquake preparedness and response efforts in areas prone to seismic activity. Overall, this project aims to use advanced machine learning techniques to develop a highly accurate and reliable model for earthquake prediction. By doing so, it hopes to contribute to our understanding of these complex natural events and to ultimately help save lives and minimize the damage caused by earthquakes.
# ## Importing libraries
import pandas as pd
import numpy as np
import seaborn as sns
# # Loading the Dataset
# Data is available on kaggle
# **About the data:** The data being used includes numerical as well as categorical data which is stored in a csv.
# The dataset contains records of **782 earthquakes** from 1/1/2001 to 1/1/2023.
# The meaning of all columns is as follows:
# * **title**: title name given to the earthquake
# * **magnitude**: The magnitude of the earthquake
# * **date_time**: date and time
# * **cdi**: The maximum reported intensity for the event range
# * **mmi**: The maximum estimated instrumental intensity for the event
# * **alert**: The alert level - “green”, “yellow”, “orange”, and “red”
# * **tsunami**: "1" for events in oceanic regions and "0" otherwise
# * **sig**: A number describing how significant the event is. Larger the number, more significant the event. This value is determined on a number of factors, including: magnitude, maximum MMI, felt reports, and estimated impact
# * **net**: The ID of a data contributor. Identifies the network considered to be the preferred source of information for this event.
# * **nst**: The total number of seismic stations used to determine earthquake location.
# * **dmin**: Horizontal distance from the epicenter to the nearest station
# * **gap**: The largest azimuthal gap between azimuthally adjacent stations (in degrees). In general, the smaller this number, the more reliable is the calculated horizontal position of the earthquake. Earthquake locations in which the azimuthal gap exceeds 180 degrees typically have large location and depth uncertainties
# * **magType**: The method or algorithm used to calculate the preferred magnitude for the event
# * **depth**: The depth where the earthquake begins to rupture
# * **latitude / longitude**: coordinate system by means of which the position or location of any place on Earth's surface can be determined and described
# * **location**: location within the country
# * **continent**: continent of the earthquake hit country
# * **country**: affected country
dataset = pd.read_csv("/kaggle/input/earthquake-dataset/earthquake_data.csv")
dataset
# # Analysing the dataset
# Here, we understand the datatypes of the features and decide which features(columns) are actually beneficial for developing the model.
# We delete the features which will not contribute in prediction.
dataset.info()
del dataset["title"]
del dataset["location"]
del dataset["country"]
del dataset["continent"]
dataset
# # Checking for Null Values
dataset.isnull().sum()
##dataset["alert"].value_counts()
# # Handling Missing values
dataset["alert"] = dataset["alert"].fillna("red")
dataset.isnull().sum()
dataset
# # Changing datatype of datetime column
dataset["date_time"] = pd.to_datetime(dataset["date_time"])
dataset.info()
dataset["date_time"] = pd.DatetimeIndex(dataset["date_time"]).month
# dataset.describe()
dataset.describe(include=["object"])
# dataset.magType.value_counts()
# # Label Encoding
# Since we have both categorical and numerical data, we will encode the 3 categorical variables.
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
alert_le = LabelEncoder()
magtype_le = LabelEncoder()
net_le = LabelEncoder()
dataset["alert"] = alert_le.fit_transform(dataset["alert"])
dataset["magType"] = magtype_le.fit_transform(dataset["magType"])
dataset["net"] = net_le.fit_transform(dataset["net"])
dataset
dataset.corr()
dataset.hist()
# # Slicing the dataset
x = dataset.iloc[:, [1, 2, 3, 4, 6, 7, 8, 9, 10, 11, 12, 13, 14]]
y = dataset.iloc[:, [5]]
# # Balance data using Imbalancing technique
dataset["tsunami"].value_counts()
from imblearn.over_sampling import SMOTE
s = SMOTE()
x_data, y_data = s.fit_resample(x, y)
from collections import Counter
print(Counter(y_data))
# ## Feature Scaling
from sklearn.preprocessing import StandardScaler
ss = StandardScaler()
x_scaled = ss.fit_transform(x_data)
x_scaled
# # Developing the Model
# ## Splitting into Train and Test Data
# Keeping the split to 80% train and 20% test
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(
x_scaled, y_data, random_state=11, test_size=0.2
)
# # Creating Models
# ## 1st Model - using Logistic Regression
from sklearn.linear_model import LogisticRegression
l1 = LogisticRegression()
l1.fit(x_train, y_train)
y_pred = l1.predict(x_test)
y_pred
from sklearn.metrics import accuracy_score
ac = accuracy_score(y_test, y_pred) * 100
ac
# **Logistic Regression gave 82.29% accuracy.**
# ## 2nd Model - using SVM
from sklearn.svm import SVC
SVM = SVC(kernel="linear", random_state=2)
SVM.fit(x_train, y_train)
y_pred1 = SVM.predict(x_test)
y_pred1
ac1 = accuracy_score(y_test, y_pred1) * 100
ac1
# **Support Vector Classifier(SVC) gavae 83.33% accuracy**
# ## 3rd Model - using Gaussian Naive Bayes
from sklearn.naive_bayes import GaussianNB
nb = GaussianNB()
nb.fit(x_train, y_train)
y_pred2 = nb.predict(x_test)
y_pred2
ac2 = accuracy_score(y_test, y_pred2) * 100
ac2
# **Gaussian Naive Bayes has given 80.2% accuracy.**
# ## 4th Model - using Decision Tree Classifier
from sklearn.tree import DecisionTreeClassifier
dt = DecisionTreeClassifier()
dt.fit(x_train, y_train)
y_pred3 = dt.predict(x_test)
y_pred3
ac3 = accuracy_score(y_test, y_pred3) * 100
ac3
# **The Decision Tree Classifier gave an accuracy of 92.18%**
# # Ensemble technique
from sklearn.ensemble import VotingClassifier
bc = VotingClassifier(
estimators=[
("logisticRegression", l1),
("svm", SVM),
("naivebayes", nb),
("Decision Tree Classification", dt),
]
)
bc.fit(x_train, y_train)
y_pred3 = bc.predict(x_test)
y_pred3
from sklearn.metrics import accuracy_score
ac3 = accuracy_score(y_test, y_pred3) * 100
ac3
# # Cross validation using KFold technique
from sklearn.model_selection import KFold
kf = KFold()
kf.split(x_train, y_train)
kf
# k= 7
# kf = KFold(n_splits=k, shuffle=True)
# kf
from sklearn.model_selection import cross_val_predict
cross_pred = cross_val_predict(bc, x_test, y_test, cv=kf)
cross_pred
from sklearn.model_selection import cross_val_score
cross_score = cross_val_score(bc, x_train, y_train, cv=kf)
cross_score
ac5 = cross_score.mean() * 100
ac5
# # **Future Work**
# * ## **Deployment of the project**
# * ## **Incorporating live data to give real-time results**
#
# import pickle
# filename = "magtype_le.pickle"
# # save model
# pickle.dump(magtype_le, open(filename, "wb"))
|
# loadind data
# Preparing the Data
from tensorflow.keras.preprocessing import text_dataset_from_directory
from tensorflow.strings import regex_replace
def prepareData(dir):
data = text_dataset_from_directory(dir)
return data.map(
lambda text, label: (regex_replace(text, "<br />", " "), label),
)
train_data = text_dataset_from_directory("/kaggle/working/movie-reviews-dataset/train")
test_data = text_dataset_from_directory("/kaggle/working/movie-reviews-dataset/test")
for text_batch, label_batch in train_data.take(1):
print(text_batch.numpy()[0])
print(label_batch.numpy()[0]) # 0 = negative, 1 = positive
# Building the Model
from tensorflow.keras.models import Sequential
from tensorflow.keras import Input
model = Sequential()
model.add(Input(shape=(1,), dtype="string"))
# Text Vectorization
from tensorflow.keras.layers.experimental.preprocessing import TextVectorization
max_tokens = 1000
max_len = 100
vectorize_layer = TextVectorization(
# Max vocab size. Any words outside of the max_tokens most common ones
# will be treated the same way: as "out of vocabulary" (OOV) tokens.
max_tokens=max_tokens,
# Output integer indices, one per string token
output_mode="int",
# Always pad or truncate to exactly this many tokens
output_sequence_length=max_len,
)
# Call adapt(), which fits the TextVectorization layer to our text dataset.
# This is when the max_tokens most common words (i.e. the vocabulary) are selected.
train_texts = train_data.map(lambda text, label: text)
vectorize_layer.adapt(train_texts)
# Embedding
from tensorflow.keras.layers import Embedding
# Previous layer: TextVectorization
max_tokens = 1000
# ...
model.add(vectorize_layer)
# Note that we're using max_tokens + 1 here, since there's an
# out-of-vocabulary (OOV) token that gets added to the vocab.
model.add(Embedding(max_tokens + 1, 128))
# The Recurrent Layer
from tensorflow.keras.layers import LSTM
# 64 is the "units" parameter, which is the
# dimensionality of the output space.
model.add(LSTM(64))
# fully-connected (Dense) layer
from tensorflow.keras.layers import Dense
model.add(Dense(64, activation="relu"))
model.add(Dense(1, activation="sigmoid"))
# Compiling the Model
model.compile(
optimizer="adam",
loss="binary_crossentropy",
metrics=["accuracy"],
)
# Training the Model
model.fit(train_data, epochs=10)
# prediction
# Should print a very high score like 0.98.
print(
model.predict(
[
"i loved it! highly recommend it to anyone and everyone looking for a great movie to watch.",
]
)
)
# Should print a very low score like 0.01.
print(
model.predict(
[
"this was awful! i hated it so much, nobody should watch this. the acting was terrible, the music was terrible, overall it was just bad.",
]
)
)
|
# # Introduction
# **When I asked ChatGPT "What is the difference between Random Forest Regressor and Extra Trees Regressor ?"**
# **I got below answer.**
# 
# Random Forest Regressor and Extra Trees Regressor are both ensemble machine learning algorithms that are commonly used for regression tasks. While both algorithms are similar in nature, there are some key differences between them:
# 1. Number of trees: In Random Forest, the algorithm builds a set of decision trees, and the final prediction is the average of the predictions made by each tree. In contrast, Extra Trees builds a larger number of decision trees, and each tree is trained on a randomly selected subset of the data. The final prediction is the average of the predictions made by all trees.
# 2. Splitting criteria: Random Forest uses a process called "bootstrap aggregating" or "bagging" to create a diverse set of trees. Each tree is trained on a different bootstrap sample of the training data, and at each node in the tree, a random subset of the features is considered for splitting. In contrast, Extra Trees uses random thresholds for each feature to perform splits at each node.
# 3. Bias-variance tradeoff: Extra Trees has a higher variance but lower bias than Random Forest. This means that Extra Trees can overfit the data more easily, but can also capture more complex relationships between features and target variable.
# 4. Computation efficiency: Extra Trees can be faster to train than Random Forest because the random feature threshold selection and lack of bootstrapping means that each tree can be trained more quickly. However, the increased number of trees may lead to longer prediction times.
# In summary, the key differences between Random Forest Regressor and Extra Trees Regressor are the number of trees, splitting criteria, bias-variance tradeoff, and computation efficiency. Both algorithms are effective for regression tasks and the choice between them ultimately depends on the specific problem and data at hand.
# **In this notebook, I tried to find any difference between Random Forest Regressor and Extra Trees Regressor, for example score, features importance etc... by using Airbnb pricing data.**
# # Importing
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
df = pd.read_csv("/kaggle/input/airbnb-cleaned-europe-dataset/Aemf1.csv")
df.head()
# # Features Engineering
# **1) Convert bool to int**
df["Shared Room"] = df["Shared Room"] * 1
df["Private Room"] = df["Private Room"] * 1
df["Superhost"] = df["Superhost"] * 1
df.info()
# **2) Handle outliers**
df.describe()
sns.histplot(df["Price"])
# * There seems to be outliers in Price. So I tried to drop some of them.
q1 = df["Price"].quantile(0.25)
q3 = df["Price"].quantile(0.75)
iqr = q3 - q1
print("first quartile is %.1f" % q1)
print("third quartile is %.1f" % q3)
print("interquartile range is %.1f" % iqr)
limit_low = q1 - iqr * 1.999
limit_high = q3 + iqr * 1.999
print("lower limit is%.1f" % limit_low)
print("upper limit is%.1f" % limit_high)
df = df.query("Price < @limit_high")
sns.histplot(df["Price"])
df.describe()
# # Visualization
# **1) Boxplot by Price and City**
sns.boxplot(df, y=df["City"], x=df["Price"])
# **2) Boxplot by Price and Day**
sns.boxplot(df, y=df["Day"], x=df["Price"])
# **3) Boxplot by Price and Room Type**
sns.boxplot(df, y=df["Room Type"], x=df["Price"])
# **4) Scatterplot Price, Cleanliness Rating and Categorial Features**
cat = [
"City",
"Day",
"Room Type",
"Person Capacity",
"Multiple Rooms",
"Business",
"Bedrooms",
]
fig = plt.figure(figsize=(15, 15))
for i in range(len(cat)):
plt.subplot(4, 3, i + 1)
sns.scatterplot(data=df, x=df["Cleanliness Rating"], y=df["Price"], hue=df[cat[i]])
plt.legend(
bbox_to_anchor=(1.05, 1),
loc="upper left",
borderaxespad=0,
fontsize=10,
title=cat[i],
)
plt.tight_layout()
plt.show()
# **5) Scatterplot Price, Guest Satisfaction and Categorial Features**
fig = plt.figure(figsize=(15, 15))
for i in range(len(cat)):
plt.subplot(4, 3, i + 1)
sns.scatterplot(data=df, x=df["Guest Satisfaction"], y=df["Price"], hue=df[cat[i]])
plt.legend(
bbox_to_anchor=(1.05, 1),
loc="upper left",
title=cat[i],
borderaxespad=0,
fontsize=10,
)
plt.tight_layout()
plt.show()
# **6) Scatterplot Price, City Center (km) and Categorial Features**
fig = plt.figure(figsize=(15, 15))
for i in range(len(cat)):
plt.subplot(4, 3, i + 1)
sns.scatterplot(data=df, x=df["City Center (km)"], y=df["Price"], hue=df[cat[i]])
plt.legend(
bbox_to_anchor=(1.05, 1),
loc="upper left",
title=cat[i],
borderaxespad=0,
fontsize=10,
)
plt.tight_layout()
plt.show()
# **7) Scatterplot Price, Metro Distance (km) and Categorial Features**
fig = plt.figure(figsize=(15, 15))
for i in range(len(cat)):
plt.subplot(4, 3, i + 1)
sns.scatterplot(data=df, x=df["Metro Distance (km)"], y=df["Price"], hue=df[cat[i]])
plt.legend(
bbox_to_anchor=(1.05, 1),
loc="upper left",
title=cat[i],
borderaxespad=0,
fontsize=10,
)
plt.tight_layout()
plt.show()
# **8) Scatterplot Price, Attraction Index and Categorial Features**
fig = plt.figure(figsize=(15, 15))
for i in range(len(cat)):
plt.subplot(4, 3, i + 1)
sns.scatterplot(data=df, x=df["Attraction Index"], y=df["Price"], hue=df[cat[i]])
plt.legend(
bbox_to_anchor=(1.05, 1),
loc="upper left",
title=cat[i],
borderaxespad=0,
fontsize=10,
)
plt.tight_layout()
plt.show()
# **9) Scatterplot Price, Restraunt Index and Categorial Features**
fig = plt.figure(figsize=(15, 15))
for i in range(len(cat)):
plt.subplot(4, 3, i + 1)
sns.scatterplot(data=df, x=df["Restraunt Index"], y=df["Price"], hue=df[cat[i]])
plt.legend(
bbox_to_anchor=(1.05, 1),
loc="upper left",
title=cat[i],
borderaxespad=0,
fontsize=10,
)
plt.tight_layout()
plt.show()
# **10) Correlation heatmap**
plt.figure(figsize=(8, 8))
sns.heatmap(df.corr(), annot=True, fmt="1.1f")
# # Random Forest Regressor and Extra Trees Regressor Modeling
# **1) Data split to X and y and drop unnecessary features**
X = df.drop(
[
"Price",
"Shared Room",
"Private Room",
"Superhost",
"Attraction Index",
"Restraunt Index",
],
axis=1,
)
y = df["Price"]
# **2) Convert categorical data into dummy**
X = pd.get_dummies(X)
X.head()
# **3) Split to train and test, and modeling**
from sklearn.model_selection import train_test_split
from sklearn.ensemble import ExtraTreesRegressor
from sklearn.ensemble import RandomForestRegressor
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=0)
reg1 = RandomForestRegressor(
bootstrap=True,
ccp_alpha=0.0,
criterion="mse",
max_depth=None,
max_features="auto",
max_leaf_nodes=None,
max_samples=None,
min_impurity_decrease=0.0,
min_samples_leaf=1,
min_samples_split=2,
min_weight_fraction_leaf=0.0,
n_estimators=100,
n_jobs=-1,
oob_score=False,
random_state=123,
verbose=0,
warm_start=False,
).fit(X_train, y_train)
reg2 = ExtraTreesRegressor(
bootstrap=False,
ccp_alpha=0.0,
criterion="mse",
max_depth=None,
max_features="auto",
max_leaf_nodes=None,
max_samples=None,
min_impurity_decrease=0.0,
min_samples_leaf=1,
min_samples_split=2,
min_weight_fraction_leaf=0.0,
n_estimators=100,
n_jobs=-1,
oob_score=False,
random_state=123,
verbose=0,
warm_start=False,
).fit(X_train, y_train)
print("R2 of Random Forest", reg1.score(X_test, y_test))
print("R2 of Extra Trees", reg2.score(X_test, y_test))
# * R2 of Extra Trees is a little bit better than that of Random Forest.
# **4) Compare the features importance**
fti1 = pd.DataFrame(reg1.feature_importances_)
fti2 = pd.DataFrame(reg2.feature_importances_)
fti1.index = X.columns
fti2.index = X.columns
fti1 = fti1.rename(columns={0: "Random Forest"})
fti2 = fti2.rename(columns={0: "Extra Trees"})
fti = pd.concat([fti1, fti2], axis=1)
plt.figure(figsize=(10, 10))
fti.plot.barh()
|
# # Imports
import numpy as np
import pandas as pd
import sys, getopt
import os
from glob import glob
from keras.preprocessing.image import ImageDataGenerator
from tensorflow.keras.utils import array_to_img
from tensorflow.keras.utils import img_to_array
from tensorflow.keras.utils import load_img
import pandas as pd
from sklearn.model_selection import train_test_split
from keras.models import *
from keras.layers import *
import keras.backend as K
from keras import optimizers
from keras.activations import *
import tensorflow as tf
import gc
import itertools
import cv2
import math
import matplotlib.pyplot as plt
from sys import getsizeof
gc.collect()
# ***
# # Reading in Data
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
load_weights = f"/kaggle/input/tracknetpickleball/model906_30"
save_weights = "/kaggle/working/dense_weights"
dataDir = f"/kaggle/input/tracknetpickleball/y_data_30"
r = os.path.abspath(os.path.join(dataDir))
path = glob(os.path.join(r, "*.npy"))
num = len(path) / 2
idx = np.arange(num, dtype="int") + 1
# ***
# # Model Training Hyperparameters
BATCH_SIZE = 10
HEIGHT = 288
WIDTH = 512
epochs = 5
tol = 15
# number of convolutional layers to unfreeze
k = 2
# learning rate
lr = 1e-5
# grid search
# try next time: 4, 5, 6,...
k_list = [10, 14, 18]
# try next time: 1e-2, 3e-3, 3e-4, ...
lr_list = [0.01]
# assuming each run takes 2.5 hours
f"will run in {2.5 * len(k_list)* len(lr_list)} hours"
param_grid = [(r[0], r[1]) for r in itertools.product(k_list, lr_list)]
param_grid
len(param_grid)
# ***
# # Utility Functions
# Loss function
def custom_loss(y_true, y_pred):
loss = (-1) * (
K.square(1 - y_pred) * y_true * K.log(K.clip(y_pred, K.epsilon(), 1))
+ K.square(y_pred) * (1 - y_true) * K.log(K.clip(1 - y_pred, K.epsilon(), 1))
)
return K.mean(loss)
def TrackNet3(input_height, input_width): # input_height = 288, input_width = 512
imgs_input = Input(shape=(9, input_height, input_width))
# Layer1
x = Conv2D(
64,
(3, 3),
kernel_initializer="random_uniform",
padding="same",
data_format="channels_first",
)(imgs_input)
x = (Activation("relu"))(x)
x = (BatchNormalization())(x)
# Layer2
x = Conv2D(
64,
(3, 3),
kernel_initializer="random_uniform",
padding="same",
data_format="channels_first",
)(x)
x = (Activation("relu"))(x)
x1 = (BatchNormalization())(x)
# Layer3
x = MaxPooling2D((2, 2), strides=(2, 2), data_format="channels_first")(x1)
# Layer4
x = Conv2D(
128,
(3, 3),
kernel_initializer="random_uniform",
padding="same",
data_format="channels_first",
)(x)
x = (Activation("relu"))(x)
x = (BatchNormalization())(x)
# Layer5
x = Conv2D(
128,
(3, 3),
kernel_initializer="random_uniform",
padding="same",
data_format="channels_first",
)(x)
x = (Activation("relu"))(x)
x2 = (BatchNormalization())(x)
# x2 = (Dropout(0.5))(x2)
# Layer6
x = MaxPooling2D((2, 2), strides=(2, 2), data_format="channels_first")(x2)
# Layer7
x = Conv2D(
256,
(3, 3),
kernel_initializer="random_uniform",
padding="same",
data_format="channels_first",
)(x)
x = (Activation("relu"))(x)
x = (BatchNormalization())(x)
# Layer8
x = Conv2D(
256,
(3, 3),
kernel_initializer="random_uniform",
padding="same",
data_format="channels_first",
)(x)
x = (Activation("relu"))(x)
x = (BatchNormalization())(x)
# Layer9
x = Conv2D(
256,
(3, 3),
kernel_initializer="random_uniform",
padding="same",
data_format="channels_first",
)(x)
x = (Activation("relu"))(x)
x3 = (BatchNormalization())(x)
# x3 = (Dropout(0.5))(x3)
# Layer10
x = MaxPooling2D((2, 2), strides=(2, 2), data_format="channels_first")(x3)
# Layer11
x = (
Conv2D(
512,
(3, 3),
kernel_initializer="random_uniform",
padding="same",
data_format="channels_first",
)
)(x)
x = (Activation("relu"))(x)
x = (BatchNormalization())(x)
# Layer12
x = (
Conv2D(
512,
(3, 3),
kernel_initializer="random_uniform",
padding="same",
data_format="channels_first",
)
)(x)
x = (Activation("relu"))(x)
x = (BatchNormalization())(x)
# Layer13
x = (
Conv2D(
512,
(3, 3),
kernel_initializer="random_uniform",
padding="same",
data_format="channels_first",
)
)(x)
x = (Activation("relu"))(x)
x = (BatchNormalization())(x)
# x = (Dropout(0.5))(x)
# Layer14
# x = UpSampling2D( (2,2), data_format='channels_first')(x)
x = concatenate([UpSampling2D((2, 2), data_format="channels_first")(x), x3], axis=1)
# Layer15
x = (
Conv2D(
256,
(3, 3),
kernel_initializer="random_uniform",
padding="same",
data_format="channels_first",
)
)(x)
x = (Activation("relu"))(x)
x = (BatchNormalization())(x)
# Layer16
x = (
Conv2D(
256,
(3, 3),
kernel_initializer="random_uniform",
padding="same",
data_format="channels_first",
)
)(x)
x = (Activation("relu"))(x)
x = (BatchNormalization())(x)
# Layer17
x = (
Conv2D(
256,
(3, 3),
kernel_initializer="random_uniform",
padding="same",
data_format="channels_first",
)
)(x)
x = (Activation("relu"))(x)
x = (BatchNormalization())(x)
# Layer18
# x = UpSampling2D( (2,2), data_format='channels_first')(x)
x = concatenate([UpSampling2D((2, 2), data_format="channels_first")(x), x2], axis=1)
# Layer19
x = (
Conv2D(
128,
(3, 3),
kernel_initializer="random_uniform",
padding="same",
data_format="channels_first",
)
)(x)
x = (Activation("relu"))(x)
x = (BatchNormalization())(x)
# Layer20
x = (
Conv2D(
128,
(3, 3),
kernel_initializer="random_uniform",
padding="same",
data_format="channels_first",
)
)(x)
x = (Activation("relu"))(x)
x = (BatchNormalization())(x)
# Layer21
# x = UpSampling2D( (2,2), data_format='channels_first')(x)
x = concatenate([UpSampling2D((2, 2), data_format="channels_first")(x), x1], axis=1)
# Layer22
x = (
Conv2D(
64,
(3, 3),
kernel_initializer="random_uniform",
padding="same",
data_format="channels_first",
)
)(x)
x = (Activation("relu"))(x)
x = (BatchNormalization())(x)
# Layer23
x = (
Conv2D(
64,
(3, 3),
kernel_initializer="random_uniform",
padding="same",
data_format="channels_first",
)
)(x)
x = (Activation("relu"))(x)
x = (BatchNormalization())(x)
# Layer24
x = Conv2D(
3,
(1, 1),
kernel_initializer="random_uniform",
padding="same",
data_format="channels_first",
)(x)
x = (Activation("sigmoid"))(x)
o_shape = Model(imgs_input, x).output_shape
# print ("layer24 output shape:", o_shape[1],o_shape[2],o_shape[3])
# Layer24 output shape: (3, 288, 512)
OutputHeight = o_shape[2]
OutputWidth = o_shape[3]
output = x
model = Model(imgs_input, output)
# model input unit:9*288*512, output unit:3*288*512
model.outputWidth = OutputWidth
model.outputHeight = OutputHeight
# Show model's details
# model.summary()
return model
# Return the numbers of true positive, true negative, false positive and false negative
def outcome(y_pred, y_true, tol):
n = y_pred.shape[0]
i = 0
TP = TN = FP1 = FP2 = FN = 0
while i < n:
for j in range(3):
if np.amax(y_pred[i][j]) == 0 and np.amax(y_true[i][j]) == 0:
TN += 1
elif np.amax(y_pred[i][j]) > 0 and np.amax(y_true[i][j]) == 0:
FP2 += 1
elif np.amax(y_pred[i][j]) == 0 and np.amax(y_true[i][j]) > 0:
FN += 1
elif np.amax(y_pred[i][j]) > 0 and np.amax(y_true[i][j]) > 0:
h_pred = y_pred[i][j] * 255
h_true = y_true[i][j] * 255
h_pred = h_pred.astype("uint8")
h_true = h_true.astype("uint8")
# h_pred
(cnts, _) = cv2.findContours(
h_pred.copy(), cv2.RETR_EXTERNAL, cv2.CHAIN_APPROX_SIMPLE
)
rects = [cv2.boundingRect(ctr) for ctr in cnts]
max_area_idx = 0
max_area = rects[max_area_idx][2] * rects[max_area_idx][3]
for j in range(len(rects)):
area = rects[j][2] * rects[j][3]
if area > max_area:
max_area_idx = j
max_area = area
target = rects[max_area_idx]
(cx_pred, cy_pred) = (
int(target[0] + target[2] / 2),
int(target[1] + target[3] / 2),
)
# h_true
(cnts, _) = cv2.findContours(
h_true.copy(), cv2.RETR_EXTERNAL, cv2.CHAIN_APPROX_SIMPLE
)
rects = [cv2.boundingRect(ctr) for ctr in cnts]
max_area_idx = 0
max_area = rects[max_area_idx][2] * rects[max_area_idx][3]
for j in range(len(rects)):
area = rects[j][2] * rects[j][3]
if area > max_area:
max_area_idx = j
max_area = area
target = rects[max_area_idx]
(cx_true, cy_true) = (
int(target[0] + target[2] / 2),
int(target[1] + target[3] / 2),
)
dist = math.sqrt(pow(cx_pred - cx_true, 2) + pow(cy_pred - cy_true, 2))
if dist > tol:
FP1 += 1
else:
TP += 1
i += 1
return (TP, TN, FP1, FP2, FN)
# Return the values of accuracy, precision and recall
def evaluation(outcome):
(TP, TN, FP1, FP2, FN) = outcome
# does this implictly unpack?
try:
accuracy = (TP + TN) / (TP + TN + FP1 + FP2 + FN)
except:
accuracy = 0
try:
precision = TP / (TP + FP1 + FP2)
except:
precision = 0
try:
recall = TP / (TP + FN)
except:
recall = 0
return (accuracy, precision, recall)
# ***
# # Load Base TrackNetV2
# load old model
# all params are currently unfrozen
base_model = load_model(load_weights, custom_objects={"custom_loss": custom_loss})
ADADELTA = optimizers.Adadelta(learning_rate=lr)
base_model.compile(loss=custom_loss, optimizer=ADADELTA)
len(base_model.layers)
num_trainable_params = np.sum([K.count_params(w) for w in base_model.trainable_weights])
f"{num_trainable_params} trainable parameters"
num_non_trainable_params = np.sum(
[K.count_params(w) for w in base_model.non_trainable_weights]
)
f"{num_non_trainable_params} non-trainable parameters"
# ***
# # Train/Val split (70/30)
npy_batch_size = 250
val_idx_start = math.floor(0.70 * npy_batch_size)
val_idx_start
train = np.arange(0, val_idx_start)
val = np.arange(val_idx_start, npy_batch_size)
# ***
# # Base Model Performance
# base_metrics = pd.DataFrame(index = ["train", "val", "all"], columns=["TP","TN", "FP1", "FP2", "FN", "loss", "acc", "prec", "rec"], dtype="float64")
# base_metrics = base_metrics.fillna(0)
# base_metrics
# print("EVALUATING BASE MODEL PERFORMANCE")
# for file_num in idx:
# print(f"\tReading in file: x_data_{str(file_num)}.npy")
# X = np.load(os.path.abspath(os.path.join(dataDir, 'x_data_' + str(file_num) + '.npy')))
# print("\tMaking predictions")
# y_hat = base_model.predict(X, batch_size=BATCH_SIZE)
# print(f"\tDeleting x_data_{str(file_num)}.npy")
# del X
# gc.collect()
# print(f"\tReading in file: y_data_{str(file_num)}.npy")
# y = np.load(os.path.abspath(os.path.join(dataDir, 'y_data_' + str(file_num) + '.npy')))
# print("\tCalculating train/val/overall loss")
# base_metrics.loc["train", "loss"] += custom_loss(y[train], y_hat[train]).numpy()
# base_metrics.loc["val", "loss"] += custom_loss(y[val], y_hat[val]).numpy()
# base_metrics.loc["all", "loss"] += custom_loss(y, y_hat).numpy()
# print("\tCalculating train/val/overall classification performance")
# y_pred = (y_hat > 0.5).astype("float32")
# base_metrics.loc["train",["TP", "TN", "FP1", "FP2", "FN"]] += outcome(y_pred[train], y[train], tol)
# base_metrics.loc["val", ["TP", "TN", "FP1", "FP2", "FN"]] += outcome(y_pred[val], y[val], tol)
# base_metrics.loc["all", ["TP", "TN", "FP1", "FP2", "FN"]] += outcome(y_pred, y, tol)
# print(f"\tDeleting y_data_{str(file_num)}.npy")
# del y
# del y_hat
# del y_pred
# gc.collect()
# print("\tCalculating aggregate metrics")
# for subset in ["train", "val", "all"]:
# # acc, prec, rec
# base_metrics.loc[subset, ["acc", "prec", "rec"]] = evaluation(base_metrics.loc[subset,["TP", "TN", "FP1", "FP2", "FN"]])
# # standardize TP, FP, ...
# base_metrics.loc[subset, ["TP", "TN", "FP1", "FP2", "FN"]] /= base_metrics.loc[subset, ["TP", "TN", "FP1", "FP2", "FN"]].sum()
# # make into percentages
# base_metrics.loc[subset, ["TP", "TN", "FP1", "FP2", "FN", "acc", "prec", "rec"]] = (base_metrics.loc[subset, ["TP", "TN", "FP1", "FP2", "FN", "acc", "prec", "rec"]]*100).round(2)
# print("BASE METRICS")
# print(base_metrics)
# print()
# BASE METRICS (files 1-30, 250 examples each)
# TP TN FP1 FP2 FN loss acc prec rec
# train 51.68 10.37 5.33 2.32 30.30 0.009474 62.05 87.11 63.04
# val 50.47 13.79 4.46 2.55 28.73 0.009288 64.27 87.81 63.73
# all 51.32 11.40 5.07 2.39 29.83 0.009418 62.72 87.31 63.24
# ***
# # Option 1: Replace last layer with Dense layer
# x = base_model.layers[-2].output
# output = Dense(512, activation="sigmoid")(x)
# model = Model(inputs=base_model.input,
# outputs=[output])
# ***
# # Option 2: Freeze everything except last k conv layers
conv_indices = [
idx for idx, layer in enumerate(base_model.layers) if "conv" in layer.name
]
conv_indices
# number of conv layers that can be fine tuned
len(conv_indices)
freeze_to = conv_indices[-k]
freeze_to
for layer in base_model.layers[0:freeze_to]:
layer.trainable = False
for layer in base_model.layers[freeze_to:]:
layer.trainable = True
for l in base_model.layers:
print(f"{l.trainable}: {l.name}")
# compile the new model
model = base_model
ADADELTA = optimizers.Adadelta(learning_rate=lr) # 1e-5 if fine tuning
model.compile(loss=custom_loss, optimizer=ADADELTA)
num_trainable_params = np.sum([K.count_params(w) for w in model.trainable_weights])
f"{num_trainable_params} trainable parameters"
num_non_trainable_params = np.sum(
[K.count_params(w) for w in model.non_trainable_weights]
)
f"{num_non_trainable_params} non-trainable parameters"
def try_new_option2(k, lr):
# load old model
# all params are currently unfrozen
base_model = load_model(load_weights, custom_objects={"custom_loss": custom_loss})
# what layers are the conv layers?
conv_indices = [
idx for idx, layer in enumerate(base_model.layers) if "conv" in layer.name
]
# how many layers to freeze?
freeze_to = conv_indices[-k]
# freeze
for layer in base_model.layers[0:freeze_to]:
layer.trainable = False
# unfreeze
for layer in base_model.layers[freeze_to:]:
layer.trainable = True
# compile the new model
model = base_model
ADADELTA = optimizers.Adadelta(learning_rate=lr)
model.compile(loss=custom_loss, optimizer=ADADELTA)
num_trainable_params = np.sum([K.count_params(w) for w in model.trainable_weights])
num_non_trainable_params = np.sum(
[K.count_params(w) for w in model.non_trainable_weights]
)
print(
f"Unfreezing {k} convolutional layers and fine tuning at a learning rate of {lr}."
)
print(
f"{num_trainable_params} trainable parameters, {num_non_trainable_params} non-trainable parameters."
)
return model
# ***
# # Fine-Tune the Model
def make_metrics_dfs(epochs):
train_metrics = pd.DataFrame(
columns=["TP", "TN", "FP1", "FP2", "FN", "loss", "acc", "prec", "rec"],
index=np.arange(1, epochs + 1),
)
train_metrics = train_metrics.fillna(0)
val_metrics = pd.DataFrame(
columns=["TP", "TN", "FP1", "FP2", "FN", "loss", "acc", "prec", "rec"],
index=np.arange(1, epochs + 1),
)
val_metrics = val_metrics.fillna(0)
return train_metrics, val_metrics
def save_metrics_dfs(train_metrics, val_metrics, k, lr, epochs):
clf_perf = train_metrics.join(val_metrics, lsuffix="_train", rsuffix="_val")
name = f"performance_k_{k}_lr_{lr}_epochs_{epochs}.csv"
clf_perf.to_csv(f"/kaggle/working/{name}")
def train_1_epoch(model):
for file_num in idx:
print(
f"\tReading in files: x_data_{str(file_num)}.npy, y_data_{str(file_num)}.npy"
)
# read in big batch (500 examples)
X = np.load(
os.path.abspath(os.path.join(dataDir, "x_data_" + str(file_num) + ".npy"))
)
y = np.load(
os.path.abspath(os.path.join(dataDir, "y_data_" + str(file_num) + ".npy"))
)
print("\tFitting model with 70% train")
# fit in smaller batches (3 examples)
history = model.fit(
X[train], y[train], batch_size=BATCH_SIZE, epochs=1, shuffle=False
)
print(f"\tDeleting x_data_{str(file_num)}.npy and y_data_{str(file_num)}.npy")
del X
del y
gc.collect()
return model
def eval_1_epoch(epoch, train_metrics, val_metrics, model):
for file_num in idx:
print(f"\tReading in file: x_data_{str(file_num)}.npy")
X = np.load(
os.path.abspath(os.path.join(dataDir, "x_data_" + str(file_num) + ".npy"))
)
print("\tMaking predictions")
y_hat = model.predict(X, batch_size=BATCH_SIZE)
print(f"\tDeleting x_data_{str(file_num)}.npy")
del X
gc.collect()
print(f"\tReading in file: y_data_{str(file_num)}.npy")
y = np.load(
os.path.abspath(os.path.join(dataDir, "y_data_" + str(file_num) + ".npy"))
)
print("\tCalculating train and valid loss")
train_metrics.loc[epoch, "loss"] += custom_loss(y[train], y_hat[train]).numpy()
val_metrics.loc[epoch, "loss"] += custom_loss(y[val], y_hat[val]).numpy()
print("\tCalculating train and valid classification performance")
y_pred = (y_hat > 0.5).astype("float32")
train_metrics.loc[epoch, ["TP", "TN", "FP1", "FP2", "FN"]] += outcome(
y_pred[train], y[train], tol
)
val_metrics.loc[epoch, ["TP", "TN", "FP1", "FP2", "FN"]] += outcome(
y_pred[val], y[val], tol
)
print(f"\tDeleting y_data_{str(file_num)}.npy")
del y
del y_hat
del y_pred
gc.collect()
print("METRICS")
# acc, prec, rec
train_metrics.loc[epoch, ["acc", "prec", "rec"]] = evaluation(
train_metrics.loc[epoch, ["TP", "TN", "FP1", "FP2", "FN"]]
)
val_metrics.loc[epoch, ["acc", "prec", "rec"]] = evaluation(
val_metrics.loc[epoch, ["TP", "TN", "FP1", "FP2", "FN"]]
)
# standardize TP, FP, ...
train_metrics.loc[epoch, ["TP", "TN", "FP1", "FP2", "FN"]] /= train_metrics.loc[
epoch, ["TP", "TN", "FP1", "FP2", "FN"]
].sum()
val_metrics.loc[epoch, ["TP", "TN", "FP1", "FP2", "FN"]] /= val_metrics.loc[
epoch, ["TP", "TN", "FP1", "FP2", "FN"]
].sum()
# make into percentages
train_metrics.loc[epoch, ["TP", "TN", "FP1", "FP2", "FN", "acc", "prec", "rec"]] = (
train_metrics.loc[epoch, ["TP", "TN", "FP1", "FP2", "FN", "acc", "prec", "rec"]]
* 100
).round(2)
val_metrics.loc[epoch, ["TP", "TN", "FP1", "FP2", "FN", "acc", "prec", "rec"]] = (
val_metrics.loc[epoch, ["TP", "TN", "FP1", "FP2", "FN", "acc", "prec", "rec"]]
* 100
).round(2)
epoch_metrics = pd.concat(
[train_metrics.loc[epoch, :], val_metrics.loc[epoch, :]], axis=1
)
epoch_metrics.columns = ["train", "val"]
print(epoch_metrics.astype("object"))
print()
# GRID SEARCH
for k, lr in param_grid:
model = try_new_option2(k, lr)
train_metrics, val_metrics = make_metrics_dfs(epochs)
print("\nBeginning training......")
for epoch in range(1, epochs + 1):
print("============ epoch", epoch, "================")
print("TRAINING")
model = train_1_epoch(model)
print("EVALUATING")
eval_1_epoch(epoch, train_metrics, val_metrics, model)
print("Saving metrics to disk")
save_metrics_dfs(train_metrics, val_metrics, k, lr, epochs)
# model = try_new_option2(k, lr)
# train_metrics, val_metrics = make_metrics_dfs(epochs)
# print('\nBeginning training......')
# for epoch in range(1, epochs+1):
# print('============ epoch', epoch, '================')
# print("TRAINING")
# model = train_1_epoch(model)
# print("EVALUATING")
# eval_1_epoch(epoch, train_metrics, val_metrics, model)
# # Save intermediate weights during training
# # if epoch % 1 == 0:
# # model.save(save_weights + '_' + str(epoch))
# print("Saving metrics to disk")
# save_metrics_dfs(train_metrics, val_metrics, k, lr, epochs)
# # Save model weights after all training
# # model.save(save_weights + '_' + str(epoch))
# ***
# # Performance Metrics
# with grid search, this stuff is only from the last iteration
train_metrics
val_metrics
# BASE METRICS (files 1-30, 250 examples each)
# TP TN FP1 FP2 FN loss acc prec rec
# train 51.68 10.37 5.33 2.32 30.30 0.009474 62.05 87.11 63.04
# val 50.47 13.79 4.46 2.55 28.73 0.009288 64.27 87.81 63.73
# all 51.32 11.40 5.07 2.39 29.83 0.009418 62.72 87.31 63.24
def compare_train_val(metric, train_metrics, val_metrics):
plt.title(metric)
plt.xlabel("epoch")
if "metric" != "loss":
plt.ylabel("%")
x = np.arange(1, train_metrics.shape[0] + 1, 1)
plt.plot(x, train_metrics[metric], label="train")
plt.plot(x, val_metrics[metric], label="val")
plt.legend()
plt.show()
for metric in train_metrics.columns:
compare_train_val(metric, train_metrics, val_metrics)
|
print(8 > 6)
print(8 == 6)
print(8 < 6)
x = 20
y = 40
if y > x:
print("y, x sayısından büyüktür.")
else:
print("y, x sayısından büyük değildir.")
print(bool("Selam"))
print(bool("25"))
a = "Selam"
b = 25
print(bool(a))
print(bool(b))
bool("xyz")
bool(987)
bool(["Geralt", "Eskel", "Lambert"])
bool(False)
bool(None)
bool(0)
bool("")
bool(())
bool([])
bool({})
def myFunction():
return True
print(myFunction())
def myFunction():
return True
if myFunction():
print("EVET!")
else:
print("HAYIR!")
a = 155
print(isinstance(a, int))
b = "Geralt's a witcher"
print(isinstance(b, str))
print(22 > 11)
print(22 == 11)
print(22 < 11)
print(bool("xyz"))
print(bool(0))
print(22 + 11)
x = 9
y = 8
print(x + y)
x = 9
y = 8
print(x - y)
x = 9
y = 8
print(x * y)
x = 9
y = 8
print(x / y)
x = 9
y = 8
print(x % y)
x = 9
y = 8
print(x**y)
x = 9
y = 8
print(x // y)
x = 6
x
x = 6
x += 2
print(x)
x = 6
x -= 2
print(x)
x = 6
x *= 2
print(x)
x = 6
x /= 2
print(x)
x = 6
x %= 2
print(x)
x = 6
x //= 2
print(x)
x = 6
x **= 2
print(x)
x = 6
y = 2
print(x == y)
x = 6
y = 2
print(x != y)
x = 6
y = 2
print(x > y)
x = 6
y = 2
print(x < y)
x = 6
y = 2
print(x >= y)
x = 6
y = 2
print(x <= y)
x = 6
print(x > 1 and x < 22)
x = 6
print(x > 1 or x < 5)
x = 6
print(not (x > 1 and x < 22))
x = ["çilek", "kiraz"]
y = ["çilek", "kiraz"]
z = x
print(x is z)
print(x is y)
print(x == y)
x = ["çilek", "kiraz"]
y = ["çilek", "kiraz"]
z = x
print(x is not z)
print(x is not y)
print(x != y)
|
from transformers import AutoTokenizer, AutoModel
model_dir = "THUDM/chatglm-6b"
tokenizer = AutoTokenizer.from_pretrained(model_dir, trust_remote_code=True)
model = AutoModel.from_pretrained(model_dir, trust_remote_code=True).float()
history = []
# # 持续对话
# import os
# while True:
# query = input("\n用户:")
# if query == "stop":
# break
# if query == "clear":
# history = []
# os.system('clear')
# continue
# response, history = model.chat(tokenizer, query, history=history)
# print(f"ChatGLM-6B:{response}")
# 单句对话
import time
start = time.time()
query = "世界的开端是什么"
response, history = model.chat(tokenizer, query, history=history)
print(f"bot:{response}")
end = time.time()
print("\n耗时:{}".format(end - start))
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.