script
stringlengths 113
767k
|
|---|
import torch
import requests
from PIL import Image
import matplotlib.pyplot as plt
from diffusers import StableDiffusionDepth2ImgPipeline
pipe = StableDiffusionDepth2ImgPipeline.from_pretrained(
"stabilityai/stable-diffusion-2-depth",
torch_dtype=torch.float16,
).to("cuda")
url = "http://images.cocodataset.org/val2017/000000039769.jpg"
init_image = Image.open(requests.get(url, stream=True).raw)
plt.imshow(init_image)
plt.show()
prompt = "two tigers"
n_propmt = "bad, deformed, ugly, bad anotomy"
image = pipe(
prompt=prompt, image=init_image, negative_prompt=n_propmt, strength=0.7
).images[0]
plt.imshow(image)
plt.show()
prompt = "two funny pink panthers"
n_propmt = "bad, deformed, ugly, bad anotomy"
another_image = pipe(
prompt=prompt, image=init_image, negative_prompt=n_propmt, strength=0.7
).images[0]
plt.imshow(another_image)
plt.show()
|
# # Assessing the Effect of Smoking On Individuals' Health Insurance Premiums
# This notebook illustrates the relationship between smoking and its effect on insurance premiums in the United States of America.
# ## Content
# Columns
# age: age of primary beneficiary
# sex: insurance contractor gender, female, male
# bmi: Body mass index, providing an understanding of body, weights that are relatively high or low relative to height,
# objective index of body weight (kg / m ^ 2) using the ratio of height to weight, ideally 18.5 to 24.9
# children: Number of children covered by health insurance / Number of dependents
# smoker: Smoking
# region: the beneficiary's residential area in the US, northeast, southeast, southwest, northwest.
# charges: Individual medical costs billed by health insurance
# ## A: Data acquisition and dataset preparation for analysis
import pandas as pd
import numpy as np
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
import matplotlib.pyplot as plt
from scipy import stats, integrate
import statsmodels.formula.api as smf
from sklearn.linear_model import LinearRegression
from sklearn import metrics
from sklearn.model_selection import train_test_split
# allow plots to appear directly in the notebook
import seaborn as sns
sns.set(color_codes=True)
insurance_df = pd.read_csv("../input/insurance/insurance.csv")
insurance_df.info()
insurance_df.head()
# Fortunately, there are no missing values in the dataset. There are three data types; integer, float, object, which only require two modifications. First changing sex column name to gender and charges to prices column;
# Second encoding data for sex and smoker columns as flowing:
# Gender column: male=0,female=1
# Smoker column: no=0, yes=1
insurance_df.rename(columns={"sex": "gender"}, inplace=True)
insurance_df.rename(columns={"charges": "prices"}, inplace=True)
insurance_df.head()
df1 = insurance_df[:]
print(df1["gender"].unique(), df1["smoker"].unique(), df1["region"].unique())
df1["gender"] = pd.get_dummies(df1["gender"])
df1["smoker"] = df1["smoker"].replace("yes", "1")
df1["smoker"] = df1["smoker"].replace("no", "0")
df1["smoker"] = df1["smoker"].astype(int)
df1.head()
# # B: Statictice information and Variables relationshipe
df1.describe()
# 18 years old is the minimum age of patients in the dataset and the maximum age is 64 years; this is a good point for the dataset, because the majority of smokers are in this scope. From min, max and quarters infer that gender is evenly distributed. Non-smokers outnumber smokers 4 to 1.
sns.heatmap(df1.corr(), cmap="Wistia", annot=True)
# Only smokers are highly correlated to charges and others are having low or no correlation. For better analysis at first, the age column categorized to Young Adult, Senior Adult, Elder and bmi column to obese and non-obese.
f, ax = plt.subplots(figsize=(8, 5))
sns.boxplot(x="region", y="prices", data=df1, dodge=False)
f, ax = plt.subplots(figsize=(8, 5))
sns.boxplot(x="gender", y="prices", data=df1, dodge=False)
f, ax = plt.subplots(figsize=(8, 5))
sns.boxplot(x="smoker", y="prices", data=df1, dodge=False)
df1["bmi30"] = np.nan
lst = [df1]
for col in lst:
col.loc[col["bmi"] < 30, "bmi30"] = "non_obese"
col.loc[col["bmi"] >= 30, "bmi30"] = "obese"
f, ax = plt.subplots(figsize=(8, 5))
sns.boxplot(x="bmi30", y="prices", data=df1, dodge=False)
df1_gb4 = df1.groupby(["bmi30"])["prices"].mean()
df1_gb4
df1["age_cat"] = np.nan
lst = [df1]
for col in lst:
col.loc[(col["age"] >= 18) & (col["age"] <= 35), "age_cat"] = "Young Adult"
col.loc[(col["age"] > 35) & (col["age"] <= 55), "age_cat"] = "Senior Adult"
col.loc[col["age"] > 55, "age_cat"] = "Elder"
f, ax = plt.subplots(figsize=(8, 5))
sns.boxplot(x="age_cat", y="prices", data=df1, dodge=False)
df1_gb1 = df1.groupby(["smoker", "gender"])["prices"].mean()
df1_gb1
sns.lmplot(x="smoker", y="prices", hue="gender", data=df1)
df1_gb2 = df1.groupby(["smoker", "age_cat"])["prices"].mean()
df1_gb2
sns.lmplot(x="smoker", y="prices", hue="age_cat", data=df1)
df1_gb3 = df1.groupby(["smoker", "bmi30"])["prices"].mean()
df1_gb3
sns.lmplot(x="smoker", y="prices", hue="bmi30", data=df1)
sns.lmplot(x="smoker", y="prices", hue="region", data=df1)
# RESULTS:
# * Prices are higher for older group ages and do not seem to be affected by gender.
# * Although obese and non-obese people have the same median prices, their average expenditure differ by almost 5000 U.S dollars.
# * We can disclose that region of origin doesn’t have much impact with the amount of prices.
# * The comorbidity between smoking and obesity has higher prices than smokers who are in shape.
# * Age and gender almost have the same effect for pricing of smokers.
# # Model Building
#
df1["region"] = df1["region"].replace("southwest", "1")
df1["region"] = df1["region"].replace("southeast", "2")
df1["region"] = df1["region"].replace("northwest", "3")
df1["region"] = df1["region"].replace("northeast", "4")
df1["region"] = df1["region"].astype(int)
sns.pairplot(
df1,
x_vars=["smoker", "bmi", "age", "region"],
y_vars="prices",
size=7,
aspect=0.7,
kind="reg",
)
# Based on former parts the model for prediction price could be multivariate linear regression:
# * Y = β_0+β_1 x_1+β_2 x_2+ β_3 x_3 +β_4 x_1 x_2+ β_5 x_1 x_3 + β_6 x_2 x_3
# ### Data normalization
# The values of several ranges are transformed into similar range by min-max method.
df1["smoker"] = (df1["smoker"] - df1["smoker"].min()) / (
df1["smoker"].max() - df1["smoker"].min()
)
df1["gender"] = (df1["gender"] - df1["gender"].min()) / (
df1["gender"].max() - df1["gender"].min()
)
df1["age"] = (df1["age"] - df1["age"].min()) / (df1["age"].max() - df1["age"].min())
df1["bmi"] = (df1["bmi"] - df1["bmi"].min()) / (df1["bmi"].max() - df1["bmi"].min())
df1["region"] = (df1["region"] - df1["region"].min()) / (
df1["region"].max() - df1["region"].min()
)
df1["prices"] = (df1["prices"] - df1["prices"].min()) / (
df1["prices"].max() - df1["prices"].min()
)
df1.head()
# ### Hypothesis Testing and p-values
lm1 = smf.ols(formula="prices ~ smoker", data=df1).fit()
lm1.params
lm1.pvalues
# create X and y
feature_cols = ["smoker"]
X = df1[feature_cols]
y = df1.prices
# instantiate and fit
lm2 = LinearRegression()
lm2.fit(X, y)
# print the coefficients
print(lm2.intercept_)
print(lm2.coef_)
lm1.pvalues
# ***
# The p-value for smoker is far less than 0.05, and so there is a relationship between smoker and prices.
# ### STATSMODELS
# ### Feature Selection
lm1 = smf.ols(formula="prices ~ smoker + bmi + age", data=df1).fit()
lm1.rsquared
lm1 = smf.ols(formula="prices ~ smoker + bmi + age + gender", data=df1).fit()
lm1.rsquared
lm1 = smf.ols(formula="prices ~ smoker + bmi + age + gender+ region", data=df1).fit()
lm1.rsquared
lm1.summary()
# Smoking, bmi and age have significant p-values,gender have insignificant p-values, p-values of region is acceptable.
# ### Model Evaluation Using Train/Test Split
# Train/test split with RMSE are used to see whether gender and region should be kept in the model. First we considered smokers, bmi and age; the MSE and R-square values then gender added and results show no significant value changes. eventually test done by region and we have a little changes that it can be ignored.
lm = LinearRegression()
x = df1[["smoker", "bmi", "age"]]
lm.fit(x, df1["prices"])
print(lm.intercept_)
print(lm.coef_)
X = df1[["smoker", "bmi", "age"]]
y = df1.prices
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=1)
lm2.fit(X_train, y_train)
y_pred = lm2.predict(X_test)
print(np.sqrt(metrics.mean_squared_error(y_test, y_pred)))
X = df1[["smoker", "bmi", "age", "gender"]]
y = df1.prices
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=1)
lm2.fit(X_train, y_train)
y_pred = lm2.predict(X_test)
print(np.sqrt(metrics.mean_squared_error(y_test, y_pred)))
X = df1[["smoker", "bmi", "age", "region"]]
y = df1.prices
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=1)
lm2.fit(X_train, y_train)
y_pred = lm2.predict(X_test)
print(np.sqrt(metrics.mean_squared_error(y_test, y_pred)))
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # DATA EDA
import matplotlib.pyplot as plt
import seaborn as sns
from tqdm import tqdm
plt.style.use("seaborn-colorblind")
# installing nb_black for autoformatting
BASE_DIR = "../input/asl-signs/"
train = pd.read_csv(f"{BASE_DIR}/train.csv")
# Train.csv has path, participant_id, sequence_id, sign
train.head()
# How many signs we have in the data set?
# 1. We have 250 Unique sign available.
# 2. Each sign have around 299 to 415 examples of each variable.
fig, ax = plt.subplots(figsize=(6, 6))
train["sign"].value_counts().sort_values(ascending=False).head(20).plot(
kind="barh", ax=ax, title="Top 50 signs in training dataset"
)
ax.set_xlabel("Number of Training Examples")
plt.show()
fig, ax = plt.subplots(figsize=(6, 6))
train["sign"].value_counts().sort_values(ascending=True).head(20).plot(
kind="barh", ax=ax, title="Bottom 50 signs in training dataset"
)
ax.set_xlabel("Number of Training Examples")
plt.show()
# # Parquet Landmark Data
train.query('sign =="listen"')
train.query('sign=="blow"').head()
# # Pull an example parquet file data--
# We have taken one value out of the all the files to check
example_fn = train.query("sign == 'listen'")["path"].values[0]
example_landmark = pd.read_parquet(f"{BASE_DIR}/{example_fn}")
example_landmark
unique_frames = example_landmark["frame"].nunique()
unique_types = example_landmark["type"].nunique()
types_in_video = example_landmark["type"].unique()
print(
f"This file has {unique_frames} unique frames and {unique_types} unique types:{types_in_video}"
)
# ## Lets compare bunch of parquet files what type of data we have.
# - We notice the number of frames is not consistent
# - Almost every file has 4 types of landmarks
listen_files = train.query('sign == "listen"')["path"]
for index, f in enumerate(listen_files):
example_landmark = pd.read_parquet(f"{BASE_DIR}/{f}")
unique_frames = example_landmark["frame"].nunique()
unique_types = example_landmark["type"].nunique()
types_in_video = example_landmark["type"].unique()
print(
f"This file has {unique_frames} unique frames and {unique_types} unique types:{types_in_video}"
)
if index == 20:
break
# ## Create Metadata for Training Dateset
N_PARQUETS_TO_READ = 1000
combined_meta = {}
for i, d in tqdm(train.iterrows(), total=len(train)):
file_path = d["path"]
example_landmark = pd.read_parquet(f"{BASE_DIR}/{file_path}")
meta = (
example_landmark.dropna(subset=["x", "y", "z"])["type"].value_counts().to_dict()
)
meta["frames"] = example_landmark["frame"].nunique()
xyz_meta = (
example_landmark.agg(
{
"x": ["min", "max", "mean"],
"y": ["min", "max", "mean"],
"z": ["min", "max", "mean"],
}
)
.unstack()
.to_dict()
)
for key in xyz_meta.keys():
new_key = key[0] + "_" + key[1]
meta[new_key] = xyz_meta[key]
combined_meta[file_path] = meta
if i == N_PARQUETS_TO_READ:
break
train_with_meta = train.merge(
pd.DataFrame(combined_meta).T.reset_index().rename(columns={"index": "path"}),
how="left",
)
# # What are the most frequent types of landmarks provided?
train_with_meta[["face", "pose", "left_hand", "right_hand"]].sum().sort_values().plot(
kind="barh"
)
(
train_with_meta.query("index < 1000").fillna(0)[
["face", "pose", "left_hand", "right_hand"]
]
> 0
).mean().plot(kind="barh")
# # Check one example?
example_fn = train.query("sign == 'shhh'")["path"].values[0]
example_landmark = pd.read_parquet(f"{BASE_DIR}/{example_fn}")
example_landmark.query("frame == 25")["type"].value_counts()
example_landmark.groupby("frame")["x"].isna()
example_landmark["no_xyz"] = example_landmark["x"].isna()
example_landmark.groupby("frame")["no_xyz"].sum().plot()
# # 3D plot of Landmarks from "shhh" example
import plotly.express as px
example_frame = example_landmark.query("frame == 16")
px.scatter_3d(example_frame, x="x", y="y", z="z", color="type")
# Evaluation
def load_relevant_data_subset(pq_path):
data_columns = ["x", "y", "z"]
data = pd.read_parquet(pq_path, columns=data_columns)
n_frames = int(len(data) / ROWS_PER_FRAME)
data = data.values.reshape(n_frames, ROWS_PER_FRAME, len(data_columns))
return data.astype(np.float32)
import tflite_runtime.interpreter as tflite
interpreter = tflite.Interpreter(model_path)
found_signatures = list(interpreter.get_signature_list().keys())
if REQUIRED_SIGNATURE not in found_signatures:
raise KernelEvalException("Required input signature not found.")
prediction_fn = interpreter.get_signature_runner("serving_default")
output = prediction_fn(inputs=frames)
sign = np.argmax(output["outputs"])
|
# импортируем необходимые библиотеки
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import category_encoders as ce # кодирование признаков
# импортируем библиотеки для визуализации
import matplotlib.pyplot as plt
import seaborn as sns
# импортируем библиотеки для парсинга данных
import requests
from bs4 import BeautifulSoup
from requests.adapters import HTTPAdapter
from urllib3.util.retry import Retry
# загружаем библиотеку для разделения датасета:
from sklearn.model_selection import train_test_split
# создаем директорию для хранения выходных данных
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# фиксируем RANDOM_SEED для воспроизводимости экспертиментов
RANDOM_SEED = 42
# также зафиксируем версию пакетов для вопроизводимости кода
# подгрузим данные из соревнования
DATA_DIR = "/kaggle/input/sf-booking/"
df_train = pd.read_csv(DATA_DIR + "/hotels_train.csv") # датасет для обучения
df_test = pd.read_csv(DATA_DIR + "hotels_test.csv") # датасет для предсказания
sample_submission = pd.read_csv(DATA_DIR + "/submission.csv") # самбмишн
# # **Знакомство с данными**
df_train.info()
# **Признаки**
# hotel_address - адрес отеля
# review_date - дата, когда рецензент разместил соответствующий отзыв
# average_score - средний балл отеля, рассчитанный на основе последнего комментария за последний год
# hotel_name - название отеля
# reviewer_nationality - национальность рецензента
# negative_review - отрицательный отзыв, который рецензент дал отелю
# review_total_negative_word_counts - общее количество слов в отрицательном отзыв
# positive_review - положительный отзыв, который рецензент дал отелю
# review_total_positive_word_counts - общее количество слов в положительном отзыве
# reviewer_score - оценка, которую рецензент поставил отелю на основе своего опыта
# total_number_of_reviews_reviewer_has_given - количество отзывов, которые рецензенты дали в прошлом
# total_number_of_reviews - общее количество действительных отзывов об отеле
# tags - теги, которые рецензент дал отелю
# days_since_review - продолжительность между датой проверки и датой очистки
# additional_number_of_scoring - есть также некоторые гости, которые просто поставили оценку сервису, а не оставили отзыв. Это число указывает, сколько там действительных оценок без проверки
# lat - широта отеля
# lng - долгота отеля
df_train.head(2)
df_test.info()
df_test.head(2)
sample_submission.head(2)
sample_submission.info()
# ВАЖНО! дря корректной обработки признаков объединяем трейн и тест в один датасет
df_train["sample"] = 1 # помечаем где у нас трейн
df_test["sample"] = 0 # помечаем где у нас тест
df_test[
"reviewer_score"
] = 0 # в тесте у нас нет значения reviewer_score, мы его должны предсказать, по этому пока просто заполняем нулями
data = df_test.append(df_train, sort=False).reset_index(drop=True) # объединяем
# # **Исследование данных**
data.info()
data.describe()
data.describe(include="object")
data.hist(figsize=(12, 8))
data.nunique(dropna=False)
plt.rcParams["figure.figsize"] = (15, 10)
sns.heatmap(data.drop(["sample"], axis=1).corr(), annot=True)
# Небольшое резюме по итогам знакомства с данными. Датасет содержит два признака, имеющих пропуски в данных - lat и lng. Кроме того, датасет содержит восемь признаков с данными типа object, которые будет необходимо преобразовать или удалить для дальнейшего обучения модели. Судя по графикам, числовые признаки распределены ненормально. Признаки содержат большое число уникальных значений. Признаки total_number_of_reviews и additional_number_of_scoring достаточно сильно скореллированы.
# # **Подготовка данных**
# Из признака с адресом отеля создадим новый признак, указывающий на то, находится ли отель в городе-миллионере.
# Составляем список европейских городов-миллионеров.
cities = [
"Istanbul",
"Moscow",
"London",
"Saint Petersburg",
"Berlin",
"Madrid",
"Kyiv",
"Rome",
"Baku",
"Bucharest",
"Paris",
"Minsk",
"Vienna",
"Warsaw",
"Hamburg",
"Budapest",
"Belgrade",
"Barcelona",
"Munich",
"Kharkiv",
"Milan",
"Sofia",
"Prague",
"Kazan",
"Nizhny Novgorod",
"Tbilisi",
"Birmingham",
"Samara",
"Ufa",
"Rostov-on-Don",
"Yerevan",
"Cologne",
"Voronezh",
"Perm",
"Odesa",
"Volgograd",
]
# Пишем функцию, которая проверяет наличие в адресе указания на соответствующий город.
def find_city(adress):
for c in cities:
if c.lower() in adress.lower():
return 1
return 0
data["large_city"] = data["hotel_address"].apply(find_city)
# Cоздаем признак, определяющий принадлежность отеля к сети отелей.
# Составляем список c наименованиями сетей отелей
large_hotel_chains = [
"Ibis",
"Best Western",
"Holiday Inn",
"Mercure",
"Premier Inn",
"Novotel",
"Hilton",
"NH",
"Travelodge",
"Four Seasons",
"Hyatt",
"InterContinental",
"Kempinski",
"Lotte",
"Marriott",
"Ritz",
"Millennium",
"Copthorne",
"Radisson",
"Wyndham",
"Accor",
"Whitbread",
"Melia",
"Louvre",
"TUI",
"Carlson",
"Motel One",
]
# Пишем функцию, которая проверяет наличие в наименовании отеля названия сети
def if_chain(hotel):
for h in large_hotel_chains:
if h.lower() in hotel.lower():
return 1
return 0
data["chain_hotel"] = data["hotel_name"].apply(if_chain)
# Создаем новый признак, который содержит информацию о месяце, в котором оставлен отзыв.
# создаем новый признак с месяцем отзыва
data["review_date"] = pd.to_datetime(data["review_date"])
data["review_month"] = data["review_date"].dt.month
# кодируем данный признак, чтобы он не был порядковым признаком
bin_encoder = ce.BinaryEncoder(cols=["review_month"])
month_bin = bin_encoder.fit_transform(data["review_month"])
data = pd.concat([data, month_bin], axis=1)
# удаляем первоначальные признаки
data = data.drop(["review_month", "review_date"], axis=1)
# Создаем новые самостоятельные признаки из признака с тегами.
# признак с информацией о типе поездки
trip_reg = "Leisure|Business"
data["trip_type"] = data["tags"].str.findall(trip_reg).str.get(0)
data["trip_type"] = data["trip_type"].fillna(data["trip_type"].mode()[0])
# признак с информацией о типе постояльца
traveler_reg = "Couple|Solo traveler|Group|Family with young children|Family with older children|Travelers with friends"
data["traveler_type"] = data["tags"].str.findall(traveler_reg).str.get(0)
# признак с информацией о типе комнаты
room_reg = "[a-zA-Z\s]+Room|\d\srooms|[a-zA-Z\s]+Suite|[a-zA-Z\s]+Appartment"
data["room_type"] = data["tags"].str.findall(room_reg).str.get(0)
data["room_type"] = data["room_type"].fillna("other")
# признак с информацией о количестве ночей
nights_reg = "Stayed\s([\d]*)"
data["nights_amount"] = data["tags"].str.findall(nights_reg).str.get(0)
data["nights_amount"] = data["nights_amount"].fillna(data["nights_amount"].mode()[0])
data["nights_amount"] = data["nights_amount"].astype("int64")
# признак с информацией о способе бронирования
data["mobile_submission"] = (
data["tags"].str.findall("Submitted from a mobile device").str.get(0)
)
# Закодируем признак с названиями отелей.
# кодируем столбец с названиями отелей при помощи двоичного кодировщика
bin_encoder = ce.BinaryEncoder(cols=["hotel_name"])
name_bin = bin_encoder.fit_transform(data["hotel_name"])
data = pd.concat([data, name_bin], axis=1)
# удаляем первоначальный признак
data = data.drop(["hotel_name"], axis=1)
# Закондируем признак с национальностью рецензента
# оставим только три самые популярные национальности остальные обозначим как 'other_nationalities'
popular_countries = data["reviewer_nationality"].value_counts().nlargest(3)
data["reviewer_nationality"] = data["reviewer_nationality"].apply(
lambda x: x if x in popular_countries else "other_nationalities"
)
# кодируем полученные данные
dummies_data = pd.get_dummies(data["reviewer_nationality"])
data = pd.concat([data, dummies_data], axis=1)
# удаляем первоначальный признак
data = data.drop(["reviewer_nationality"], axis=1)
# Закодируем признаки, содержащие информацию о негативном или позитивном отзыве.
# пишем функции, которые позволяют определить оставил ли рецензент негативные или позитивный отзыв отелю
def negative_reviews(review):
non_negative = ["no negative", " nothing", " none", " na", " n a", " "]
if review.lower() in non_negative:
return 0
return 1
def positive_reviews(review):
non_positive = ["no positive", " nothing", " none", " na", " n a", " "]
if review.lower() in non_positive:
return 0
return 1
data["negative_review"] = data["negative_review"].apply(negative_reviews)
data["positive_review"] = data["positive_review"].apply(positive_reviews)
# Закодируем признак, содержащий информацию о типе комнаты.
# создаем функцию, которая группирует популярные типы комнат
def get_room_type(room):
if "Superior" in room or "Deluxe" in room or "King" in room or "Queen" in room:
return "superior_or_deluxe"
if "Double" in room or "Twin" in room:
return "double_or_twin"
if "Single" in room or "Classic" in room:
return "single"
if "rooms" in room:
return "several_rooms"
return "else"
data["room_type"] = data["room_type"].apply(get_room_type)
# кодируем полученные данные
dummies_data = pd.get_dummies(data["room_type"])
data = pd.concat([data, dummies_data], axis=1)
# удаляем первоначальный признак
data = data.drop(["room_type"], axis=1)
# Кодируем признаки с типом поездки, постояльца и способа бронирования
data = pd.get_dummies(
data,
prefix=["trip", "traveler", "booking"],
columns=["trip_type", "traveler_type", "mobile_submission"],
)
# Преобразуем признак, показывающий разницу во времени между первым и последним отзывом
data["days_since_review"] = data["days_since_review"].apply(lambda x: x.split()[0])
data["days_since_review"] = data["days_since_review"].astype("int64")
# заполняем пропуски в признаках с данными о широте и долготе
data["lat"] = data["lat"].fillna(0, inplace=True)
data["lng"] = data["lng"].fillna(0, inplace=True)
# убираем признаки которые еще не успели обработать,
# модель на признаках с dtypes "object" обучаться не будет, просто выберим их и удалим
object_columns = [s for s in data.columns if data[s].dtypes == "object"]
data.drop(object_columns, axis=1, inplace=True)
data.info()
# Теперь выделим тестовую часть
train_data = data.query("sample == 1").drop(["sample"], axis=1)
test_data = data.query("sample == 0").drop(["sample"], axis=1)
y = train_data.reviewer_score.values # наш таргет
X = train_data.drop(["reviewer_score"], axis=1)
# Воспользуемся специальной функцие train_test_split для разбивки тестовых данных
# выделим 20% данных на валидацию (параметр test_size)
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=RANDOM_SEED
)
# проверяем
test_data.shape, train_data.shape, X.shape, X_train.shape, X_test.shape
# Импортируем необходимые библиотеки:
from sklearn.ensemble import (
RandomForestRegressor,
) # инструмент для создания и обучения модели
from sklearn import metrics # инструменты для оценки точности модели
# Создаём модель (НАСТРОЙКИ НЕ ТРОГАЕМ)
model = RandomForestRegressor(
n_estimators=100, verbose=1, n_jobs=-1, random_state=RANDOM_SEED
)
# Обучаем модель на тестовом наборе данных
model.fit(X_train, y_train)
# Используем обученную модель для предсказания рейтинга ресторанов в тестовой выборке.
# Предсказанные значения записываем в переменную y_pred
y_pred = model.predict(X_test)
# Сравниваем предсказанные значения (y_pred) с реальными (y_test), и смотрим насколько они в среднем отличаются
# Метрика называется Mean Absolute Error (MAE) и показывает среднее отклонение предсказанных значений от фактических.
print("MAPE:", metrics.mean_absolute_error(y_test, y_pred))
# в RandomForestRegressor есть возможность вывести самые важные признаки для модели
plt.rcParams["figure.figsize"] = (10, 10)
feat_importances = pd.Series(model.feature_importances_, index=X.columns)
feat_importances.nlargest(15).plot(kind="barh")
test_data.sample(10)
test_data = test_data.drop(["reviewer_score"], axis=1)
sample_submission
predict_submission = model.predict(test_data)
predict_submission
list(sample_submission)
sample_submission["reviewer_score"] = predict_submission
sample_submission.to_csv("submission.csv", index=False)
sample_submission.head(10)
|
# # confusion matrix
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.datasets import load_iris
from sklearn.model_selection import train_test_split
from sklearn.tree import DecisionTreeClassifier
from sklearn.metrics import confusion_matrix
iris = load_iris()
x_train, x_test, y_train, y_test = train_test_split(
iris.data, iris.target, test_size=0.2, random_state=42
)
clf = DecisionTreeClassifier()
clf.fit(x_train, y_train)
y_pred = clf.predict(x_test)
cm = confusion_matrix(y_test, y_pred)
labels = ["Setosa", "Versicolor", "Virginica"]
df_cm = pd.DataFrame(cm, index=labels, columns=labels)
plt.figure(figsize=(8, 6))
sns.heatmap(
df_cm, annot=True, cmap=plt.cm.Purples
) # annot=True matrisin kutu içindeki sayı değerlerini gösteriyor. 10,9,11 gibi
plt.xlabel("predicted")
plt.ylabel("actual")
plt.show()
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.tree import DecisionTreeClassifier
from sklearn.metrics import confusion_matrix
from sklearn.linear_model import LogisticRegression
from sklearn.datasets import make_classification
# veri oluşturma
x, y = make_classification(n_samples=1000, n_classes=2, random_state=42)
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.2, random_state=120
)
clf = LogisticRegression()
clf.fit(x_train, y_train)
y_pred = clf.predict(x_test)
cm = confusion_matrix(y_test, y_pred)
tn, fp, fn, tp = cm.ravel()
precision = tp / (tp + fp)
recall = tp / (tp + fn)
f1_score = 2 * (precision * recall) / (precision + recall)
accuracy = (tp + tn) / (tp + tn + fn + fp)
plt.figure(figsize=(8, 6))
plt.imshow(cm, cmap="Blues", interpolation="nearest")
plt.title("confusion matrix")
plt.colorbar()
tick_marks = np.arrange(2)
plt.xticks(tick_marks, ["Negative", "Positive"], rotation=45)
plt.yticks(tick_marks, ["Negative", "Positive"])
plt.xlabel("predicted")
plt.ylabel("actual")
plt.text(
0, 0, f"True Negative: {tn}", ha="center", va="center", color="white", fontsize=12
)
plt.text(
0, 1, f"false Negative: {fn}", ha="center", va="center", color="red", fontsize=12
)
plt.text(
1, 0, f"false positive: {fp}", ha="center", va="center", color="red", fontsize=12
)
plt.text(
1, 1, f"True positive: {tp}", ha="center", va="center", color="white", fontsize=12
)
plt.text(
2.5,
0,
f"Precision:{precision:.2f}",
ha="center",
va="center",
color="red",
fontsize=12,
)
plt.text(
2.5,
-0.2,
f"Recall:{recall:.2f}",
ha="center",
va="center",
color="red",
fontsize=12,
)
plt.text(
2.5,
-0.4,
f"F1 score:{f1_score:.2f}",
ha="center",
va="center",
color="red",
fontsize=12,
)
plt.text(
2.5,
-0.6,
f"Accuracy:{accuracy:.2f}",
ha="center",
va="center",
color="red",
fontsize=12,
)
plt.show()
|
# ***
# # CURSO DE ESTATÍSTICA - PARTE 1
# ***
# ## Trabalho de Análise Descritiva de um Conjunto de Dados
# Utilizando os conhecimentos adquiridos em nosso treinamento realize uma análise descritiva básica de um conjunto de dados retirados da Pesquisa Nacional por Amostra de Domicílios - 2015 do IBGE.
# Vamos construir histogramas, calcular e avaliar medidas de tendência central, medidas separatrizes e de dispersão dos dados.
# Siga o roteiro proposto e vá completando as células vazias. Procure pensar em mais informações interessantes que podem ser exploradas em nosso dataset.
# # DATASET DO PROJETO
# ***
# ### Pesquisa Nacional por Amostra de Domicílios - 2015
# A Pesquisa Nacional por Amostra de Domicílios - PNAD investiga anualmente, de forma permanente, características gerais da população, de educação, trabalho, rendimento e habitação e outras, com periodicidade variável, de acordo com as necessidades de informação para o país, como as características sobre migração, fecundidade, nupcialidade, saúde, segurança alimentar, entre outros temas. O levantamento dessas estatísticas constitui, ao longo dos 49 anos de realização da pesquisa, um importante instrumento para formulação, validação e avaliação de políticas orientadas para o desenvolvimento socioeconômico e a melhoria das condições de vida no Brasil.
# ### Fonte dos Dados
# https://ww2.ibge.gov.br/home/estatistica/populacao/trabalhoerendimento/pnad2015/microdados.shtm
# ### Variáveis utilizadas
# > ### Renda
# > ***
# Rendimento mensal do trabalho principal para pessoas de 10 anos ou mais de idade.
# > ### Idade
# > ***
# Idade do morador na data de referência em anos.
# > ### Altura (elaboração própria)
# > ***
# Altura do morador em metros.
# > ### UF
# > ***
# |Código|Descrição|
# |---|---|
# |11|Rondônia|
# |12|Acre|
# |13|Amazonas|
# |14|Roraima|
# |15|Pará|
# |16|Amapá|
# |17|Tocantins|
# |21|Maranhão|
# |22|Piauí|
# |23|Ceará|
# |24|Rio Grande do Norte|
# |25|Paraíba|
# |26|Pernambuco|
# |27|Alagoas|
# |28|Sergipe|
# |29|Bahia|
# |31|Minas Gerais|
# |32|Espírito Santo|
# |33|Rio de Janeiro|
# |35|São Paulo|
# |41|Paraná|
# |42|Santa Catarina|
# |43|Rio Grande do Sul|
# |50|Mato Grosso do Sul|
# |51|Mato Grosso|
# |52|Goiás|
# |53|Distrito Federal|
# > ### Sexo
# > ***
# |Código|Descrição|
# |---|---|
# |0|Masculino|
# |1|Feminino|
# > ### Anos de Estudo
# > ***
# |Código|Descrição|
# |---|---|
# |1|Sem instrução e menos de 1 ano|
# |2|1 ano|
# |3|2 anos|
# |4|3 anos|
# |5|4 anos|
# |6|5 anos|
# |7|6 anos|
# |8|7 anos|
# |9|8 anos|
# |10|9 anos|
# |11|10 anos|
# |12|11 anos|
# |13|12 anos|
# |14|13 anos|
# |15|14 anos|
# |16|15 anos ou mais|
# |17|Não determinados|
# ||Não aplicável|
# > ### Cor
# > ***
# |Código|Descrição|
# |---|---|
# |0|Indígena|
# |2|Branca|
# |4|Preta|
# |6|Amarela|
# |8|Parda|
# |9|Sem declaração|
# #### Observação
# ***
# > Os seguintes tratamentos foram realizados nos dados originais:
# > 1. Foram eliminados os registros onde a Renda era inválida (999 999 999 999);
# > 2. Foram eliminados os registros onde a Renda era missing;
# > 3. Foram considerados somente os registros das Pessoas de Referência de cada domicílio (responsável pelo domicílio).
# ***
# ***
# ### Utilize as células abaixo para importar as bibliotecas que precisar e para configurações gerais
# #### Sugestões: dplyr, ggplot2 etc.
# ### Importe o dataset e armazene o conteúdo em uma DataFrame
# ### Visualize o conteúdo do DataFrame
# ### Para avaliar o comportamento da variável RENDA vamos construir uma tabela de frequências considerando as seguintes classes em salários mínimos (SM)
# #### Descreva os pontos mais relevantes que você observa na tabela e no gráfico.
# Classes de renda:
# A ► Acima de 25 SM
# B ► De 15 a 25 SM
# C ► De 5 a 15 SM
# D ► De 2 a 5 SM
# E ► Até 2 SM
# Para construir as classes de renda considere que o salário mínimo na época da pesquisa era de R$ 788,00.
# #### Siga os passos abaixo:
# ### 1º Definir os intevalos das classes em reais (R$)
# ### 2º Definir os labels das classes
# ### 3º Construir a coluna de frequências
# ### 4º Construir a coluna de percentuais
# ### 5º Juntar as colunas de frequência e percentuais e ordenar as linhas de acordo com os labels das classes
# ### Construa um gráfico de barras para visualizar as informações da tabela de frequências acima
# #### Lembre-se de transformar a matriz de resultados em um data frame.
# > ### Conclusões
# Escreva suas conclusões aqui...
# ### Crie um histograma para as variáveis QUANTITATIVAS de nosso dataset
# #### Descreva os pontos mais relevantes que você observa nos gráficos (assimetrias e seus tipos, possíveis causas para determinados comportamentos etc.)
# > ### Conclusões
# Escreva suas conclusões aqui...
# ### Para a variável RENDA, construa um histograma somente com as informações das pessoas com rendimento até R$ 20.000,00
# ### Construa uma tabela de frequências e uma com os percentuais cruzando das variáveis SEXO e COR
# #### Avalie o resultado da tabela e escreva suas principais conclusões
# #### Utilize os vetores abaixo para renomear as linha e colunas das tabelas de frequências e dos gráficos em nosso projeto
sexo = c("Masculino", "Feminino")
cor = c("Indígena", "Branca", "Preta", "Amarela", "Parda")
anos_de_estudo = c(
"Sem instrução e menos de 1 ano",
"1 ano",
"2 anos",
"3 anos",
"4 anos",
"5 anos",
"6 anos",
"7 anos",
"8 anos",
"9 anos",
"10 anos",
"11 anos",
"12 anos",
"13 anos",
"14 anos",
"15 anos ou mais",
"Não determinados",
)
|
#
# # Vehicle insurance claim prediction using binary classification
# In this notebook, I'll be attempting to build a model that can predict whether or not a driver will make an insurance claim using a very complex dataset consisting of 27 features (columns) and 10,000+ instances (rows). Below is the following steps I'll follow.
# 1. **Exploring the data** - we'll look into the data to get more information on what it looks like
# 2. **Data preprocessing** - preparing the data for the model making sure there is no missing data, wrong formats, and imbalanced data.
# 3. **Choosing a model** - here I'll assess the performance of two models I have in mind, I'll use this to choose a better performing model
# 4. **Building the pipeline** - creating a pipeline to streamline the process
# 5. **Evaluating the model** - I'll be assessing the model's performance in depth and conducting cross validation
import numpy as np # linear algebra
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.preprocessing import StandardScaler, OneHotEncoder, LabelEncoder
from sklearn.metrics import (
roc_curve,
roc_auc_score,
confusion_matrix,
classification_report,
)
from sklearn.model_selection import (
train_test_split,
GridSearchCV,
cross_val_score,
KFold,
)
from sklearn.impute import SimpleImputer
from sklearn.linear_model import LogisticRegression
from sklearn.neighbors import KNeighborsClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.compose import ColumnTransformer
from sklearn.feature_selection import SelectKBest, chi2, mutual_info_classif
from sklearn.pipeline import make_pipeline
from sklearn.decomposition import PCA
from imblearn.under_sampling import RandomUnderSampler
import warnings
warnings.simplefilter(action="ignore", category=FutureWarning)
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(f"File path: {os.path.join(dirname, filename)}")
PATH = "/kaggle/input/car-insurance-claim-data/car_insurance_claim.csv"
df = pd.read_csv(PATH)
pd.set_option("display.max_columns", None)
# ### Below we'll look at some of the data stats
# Looking at the shape, we can see there's 27 features (columns) and 10,302 instances (rows or samples)
df.shape
# Data statistics such as the mean, min and max, and the standard deviation
df.describe()
df.describe(include="object")
# Data types of the features
print(df.dtypes)
print(df.columns)
# Percentage of missing data for each feature
print(df.isna().sum().sort_values(ascending=False) / len(df) * 100)
# First 7 rows and what the data looks like
df.head(7)
# ### Cleaning the data minimally to see the correlation and plot the relationships
# Removing all USD symbols ($) and changing the data types from objects to floats
cols = ["INCOME", "HOME_VAL", "OLDCLAIM", "CLM_AMT", "BLUEBOOK"]
for col in cols:
df[col] = df[col].str.replace(",", "").str.replace("$", "").astype("float")
# Also removing the 'z_' that are attatched to some of the data values to encode them later on
for col in df.columns:
if df[col].dtype == "object":
df[col] = df[col].str.replace("z_", "")
# If we take a look at the data now, we should see any currency value is now a float and there aren't any more 'z_' preceding any of the data entries
df.head(10)
# ### Below I'll take a look at the distribution of the target class and see how many have made a claim vs the amount that didn't so I could see if the data is imbalanced.
sns.countplot(x=df["CLAIM_FLAG"], data=df)
plt.show()
# We can see the number of customers who haven't made a claim is significantly higher than those who have. This could cause our machine learning model to become biased to the class that has more instances when our data is imbalanced.
# I have three options, to use the `RandomOversampler` tool which could lead to overfitting, or use the *undersampling* & *oversampling* technique but that's beyond the scope of this project so I'll leave it as is for now and come back to it later. I'll check to see if it could increase the accuracy of the model.
# ### Plotting data relationships
# Charts which give us more insight into the data
colz = ["EDUCATION", "OCCUPATION", "CAR_TYPE", "URBANICITY"]
for col in colz:
ax = sns.countplot(x=df[col], hue=df["CLAIM_FLAG"], data=df)
ax.set_xticklabels(ax.get_xticklabels(), rotation=90, ha="right")
plt.show()
# # Preprocessing the data to use in our model
scaler = StandardScaler()
encoder = LabelEncoder()
# Splitting the data before I encode and scale it to avoid data leakage
X = df.drop(["CLAIM_FLAG", "ID", "BIRTH", "CLM_AMT", "OLDCLAIM"], axis=1)
y = df["CLAIM_FLAG"]
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=12, shuffle=True
)
X_train, X_holdout, y_train, y_holdout = train_test_split(
X_train, y_train, test_size=0.1, random_state=42, shuffle=True
)
# There's a lot going on below, so I'll try to explain what each line does using comments
# 1. changing X_train and X_test to dataframes to find mean and modes
X_train = pd.DataFrame(X_train, columns=df.columns)
X_test = pd.DataFrame(X_test, columns=df.columns)
# 2. imputing the data - filling in missing values
X_train.fillna(X_train.mode().iloc[0], inplace=True)
X_test.fillna(X_train.mean(), inplace=True)
# 3. dropping target feature
X_train.drop(["ID", "CLAIM_FLAG", "BIRTH", "CLM_AMT", "OLDCLAIM"], axis=1, inplace=True)
X_test.drop(["ID", "CLAIM_FLAG", "BIRTH", "CLM_AMT", "OLDCLAIM"], axis=1, inplace=True)
# 4. Scaling the numerical features
numeric_features = []
for col in X.columns:
if X[col].dtype == "float" or X[col].dtype == "int":
numeric_features.append(col)
X_train[numeric_features] = scaler.fit_transform(X_train[numeric_features])
X_test[numeric_features] = scaler.fit_transform(X_test[numeric_features])
# 5. Encoding the categorical features
categorical_features = X.select_dtypes(include=["object"]).columns
for col in categorical_features:
X_train[col] = encoder.fit_transform(X_train[col])
X_test[col] = encoder.fit_transform(X_test[col])
# ### Undersampling the data
undersample = RandomUnderSampler(sampling_strategy="majority")
# undersampling
X_train_resampled, y_train_resampled = undersample.fit_resample(X_train, y_train)
X_test_resampled, y_test_resampled = undersample.fit_resample(X_test, y_test)
# Data target classes are now balanced
sns.countplot(x=y_train_resampled, hue=y_train_resampled)
plt.show()
# ### Choosing the 10 best features
selector = SelectKBest(mutual_info_classif, k=10)
X_new = selector.fit_transform(X_train_resampled, y_train_resampled)
selected_features_indices = selector.get_support(indices=True)
selected_features_names = X_train_resampled.columns[selected_features_indices]
X_train_resampled = X_train_resampled[selected_features_names]
X_test_resampled = X_test_resampled[selected_features_names]
# With all those steps done, below we can see our training data that's been encoded and scaled. It's now ready for our model
pca = PCA()
pca.fit(X_train)
# create range enumerating pca feats
features = range(pca.n_components_)
# create a bar plot of the variance
plt.bar(features, pca.explained_variance_)
plt.xticks(features)
plt.xlabel("pca feature")
plt.ylabel("variance")
plt.show()
X_train_resampled.head()
# # Choosing a model
# Now we can compare the performance of both models below. As you can see, although Logistic Regression and KNeighborsClassifier are both classification algorithms, Logistic Regression performs better.
# With the accuracy of Logisitic Regression at 98% and KNN at 87%, and the mean cross validation score as shown in the chart, I'll choose to work with the Logistic Regression model for this use case.
models = {
"Logistic Regression": LogisticRegression(),
"KNN": KNeighborsClassifier(),
"Random Forest": RandomForestClassifier(),
}
results = []
for model in models.values():
kf = KFold(n_splits=5, shuffle=True, random_state=12)
cv_score = cross_val_score(model, X_train_resampled, y_train_resampled, cv=kf)
results.append(cv_score)
plt.boxplot(results, labels=models.keys())
plt.show()
for name, model in models.items(): # names and values using .item()
model.fit(X_test, y_test)
test_score = model.score(X_test, y_test)
print("{} accuracy: {}".format(name, test_score))
# # Evaluating the model
logreg = LogisticRegression()
logreg.fit(X_train_resampled, y_train_resampled)
score = logreg.score(X_test_resampled, y_test_resampled)
y_pred = logreg.predict(X_test_resampled)
y_pred_proba = logreg.predict_proba(X_test_resampled)[:, 1]
importances = logreg.class_weight
print(f"Accuracy score: {score}")
print("\n")
print(classification_report(y_test_resampled, y_pred))
print("\n")
print("Confusion Matrix:")
print(confusion_matrix(y_test_resampled, y_pred))
# plotting confusion matrix
labels = ["True Neg", "False Pos", "False Neg", "True Pos"]
categories = ["Zero", "One"]
cm = confusion_matrix(y_test_resampled, y_pred)
cm = cm.astype("float") / cm.sum(axis=1)[:, np.newaxis]
sns.heatmap(
cm,
annot=True,
cmap="rocket_r",
xticklabels=categories,
yticklabels=categories,
fmt=".2f",
)
plt.xlabel("Predicted label")
plt.ylabel("True label")
plt.title("Confusion matrix")
plt.show()
# Above we can see the precision, recall, f1-score, and the confusion matrix. Accuracy is 68%.
# ### Plotting the ROC curve to validate performance
fpr, tpr, thresholds = roc_curve(y_test_resampled, y_pred_proba)
plt.plot([0, 1], [0, 1], "k--")
plt.plot(fpr, tpr)
plt.xlabel("False positive")
plt.ylabel("True positive")
plt.title("ROC Curve")
plt.show()
# ### Calculating AUC
print(f"AUC Score: {roc_auc_score(y_pred, y_pred_proba)}")
|
# # tensorflow "classes" not working on flow_from_dataframe #47281
# [GitHub Issue](https://github.com/tensorflow/tensorflow/issues/47281)
# Please correct me if I am wrong. Thanks!
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import glob
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# ## Load classmap
class_map = pd.read_csv("/kaggle/input/cnn-who-is-she/CNN_who_is_she/classmap.csv")
class_dict = dict(list(zip(class_map.classname, class_map.classnum)))
class_dict.keys()
# ## Load training image path
img_path = glob.glob(
"/kaggle/input/cnn-who-is-she/CNN_who_is_she/training_set/training_set/*/*.png"
)
class_y = []
for i in img_path:
y = i.split("/")[7]
class_y.append(y)
df = pd.DataFrame({"img_path": img_path, "class": class_y})
df
# ## ImageDataGenerator
from tensorflow.keras.preprocessing.image import ImageDataGenerator
train_datagen = ImageDataGenerator(
rescale=1.0 / 255,
rotation_range=10,
width_shift_range=0.05,
height_shift_range=0.05,
horizontal_flip=True,
vertical_flip=False,
fill_mode="wrap",
)
train_generator = train_datagen.flow_from_dataframe(
dataframe=df,
x_col="img_path",
y_col="class",
classes=["rika", "risa", "yui", "akane", "neru"],
target_size=(200, 200),
batch_size=32,
class_mode="categorical",
shuffle=False,
)
# ## Question
# - The classes parameter works on `flow_from_directory` but not `flow_from_dataframe` in TF 2.4.
# - The `train_generator.class_indices` output are not correspond my classes lists.
# class indices from `train_datagen.flow_from_dataframe`
train_generator.class_indices
# Here is the result I want
class_dict
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import plotly.express as px
import plotly.graph_objs as go
import folium
import pandas as pd
import requests
import json
import squarify
import seaborn as sns
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# An overview of the immigration data in the canadian_immegration_data.csv file
df = pd.read_csv("/kaggle/input/immigration-to-canada/canadian_immegration_data.csv")
# Display the first five rows of the dataset
print("First 5 Rows of Immigration Data:")
print(df.head())
# Display the shape of the dataset
print("Data Shape:")
print(df.shape)
# Display the data types of each column
print("Data Types:")
print(df.dtypes)
# Display summary statistics of the dataset
print("Summary Statistics:")
print(df.describe())
# Display the number of missing values in each column
print("Missing Values:")
print(df.isnull().sum())
# Get total immigration by year
total_by_year = df.set_index("Country").loc[:, "1980":"2013"].sum(axis=0)
# Calculate linear regression line
x = total_by_year.index.astype(int) # convert year strings to integers
y = total_by_year.values
slope, intercept = np.polyfit(x, y, 1)
trend_line = slope * x + intercept
# Create Plotly figure
fig = go.Figure()
# Add bar trace for total immigration
fig.add_trace(
go.Bar(
name="Total Immigration",
x=total_by_year.index,
y=total_by_year.values,
xperiodalignment="middle",
)
)
# Set the color of the bars
fig.update_traces(
marker_color="rgb(4, 194, 166)",
# marker_line_color='rgb(8,48,107)',
# marker_line_width=1.5, opacity=0.6
)
# Add line trace for total immigration
fig.add_trace(
go.Scatter(
name="Total Immigration",
mode="lines+markers",
x=total_by_year.index,
y=total_by_year.values,
marker=dict(symbol="star"),
)
)
# Add trend line trace to figure
fig.add_trace(
go.Scatter(
name="Trend Line",
mode="lines",
x=total_by_year.index,
y=trend_line,
line=dict(color="blue", dash="dot"),
)
)
# Set axis labels and title
fig.update_xaxes(title="Year", showgrid=True, ticklabelmode="period")
fig.update_yaxes(title="Total Number of Immigrants")
fig.update_layout(
title="Total Immigration to Canada from 1980 to 2013",
xaxis=dict(
title="Year", tickangle=50 # Set the angle of the tick labels to 50 degrees
),
)
# Show figure
fig.show()
# This chart shows the total immigration to Canada from 1980 to 2013. There are 2 ways to interact on the chart, you can click the 2 'Total Immigration' buttons in the top right corner to toggle if you want to view in bar or line format is fine.
# Overall, total immigration to Canada increased from 1980 with 99,137k to 2013 reaching 257,537k, with fluctuations in the middle affecting the overall shape, peaking in 2010 with 276,956k. It's interesting to note that the overall trend is upward, with total immigration more than doubling from 1980 to 2013.
fig = px.histogram(
df, x="Total", nbins=50, title="Histogram of Total Immigrants from 1980 to 2013"
)
fig.update_layout(xaxis_title="Number of Immigrants", yaxis_title="Number of Countries")
fig.show()
# This histogram shows the number of countries distributed by total immigrants to Canada from 1980 to 2013. Under 100k immigrants are the most with more than 140 countries. There are only 4 countries with over 500k immigrants.
top_5_countries = df[df["Total"].isin(df["Total"].nlargest(5))].sort_values(
"Total", ascending=False
)
# Create Plotly figure
fig = go.Figure()
# Loop through each country and add a trace to the figure
for country in top_5_countries["Country"]:
# Get the immigration data for the country
data = top_5_countries.loc[top_5_countries["Country"] == country, "1980":"2013"]
# Create a line trace for the data
trace = go.Scatter(x=data.columns, y=data.values[0], name=country)
# Add the trace to the figure
fig.add_trace(trace)
# Set the title and axis labels for the figure
fig.update_layout(
title="Immigration from Top 5 Countries (1980-2013)",
xaxis_title="Year",
yaxis_title="Number of Immigrants",
xaxis=dict(
title="Year", tickangle=50 # Set the angle of the tick labels to 50 degrees
),
)
# Show the figure
fig.show()
# This chart displays the migration trends of the top 5 countries from 1980 to 2013. The migration trend of the United Kingdom shows an initial upward trend, reaching a peak in 1994, followed by a significant decline, dropping below 10k in the years that followed.
# On the other hand, the migration trends of India, China, the Philippines, and Pakistan show more gradual growth until around 1993, with each country having its unique pattern. China's migration trend reached a peak of 42,584k in 2005, after which it declined. In contrast, the Philippines had its peak migration in 2010, reaching 38,617k.
# Overall, we observe an increasing trend in migration for these top 5 countries, but there is also a significant fluctuation in the migration trends over time, reflecting the various economic and political factors that drive migration patterns.
# Select the top 10 countries based on total immigration
top_10_countries = df[df["Total"].isin(df["Total"].nlargest(10))].sort_values(
"Total", ascending=True
)
# Create a bar trace for the total immigration of each country
trace = go.Bar(x=top_10_countries["Country"], y=top_10_countries["Total"])
# Create a figure and add the bar trace to it
fig = go.Figure(data=[trace])
# Set the title and axis labels for the figure
fig.update_layout(
title="Total Immigration from Top 10 Countries",
xaxis_title="Country",
yaxis_title="Number of Immigrants",
)
# Show the figure
fig.show()
# This chart shows the total immigration figures for the top 10 countries, with the top 4 countries having over 500k immigrants each. The Philippines had a total of 511,391k immigrants, while the United Kingdom had 551.5k immigrants. China had 659,962k immigrants, and India had the highest number of immigrants among the top 10 countries, with a total of 691,904k.
# Get the top 10 countries by Total column
top10_countries = df.sort_values(by="Total", ascending=False).head(10)
# Create a treemap
fig = px.treemap(
top10_countries,
path=["Country"],
values="Total",
color="Country",
color_discrete_sequence=px.colors.qualitative.Pastel,
)
# Update the layout
fig.update_layout(title="Top 10 Countries by Total Immigrants to Canada (1980-2013)")
# Increase the font size of the labels
fig.update_traces(textfont_size=18)
# Show the chart
fig.show()
# Another treemap chart shows the top countries by total immigrants to Canada (1980-2013)
# Download countries geojson file
URL = "https://cf-courses-data.s3.us.cloud-object-storage.appdomain.cloud/IBMDeveloperSkillsNetwork-DV0101EN-SkillsNetwork/Data%20Files/world_countries.json"
r = requests.get(URL)
world_geo = r.json()
# Dictionary mapping incorrect country names to correct names
country_map = {
"Bolivia (Plurinational State of)": "Bolivia",
"Democratic People's Republic of Korea": "North Korea",
"Guinea-Bissau": "Guinea Bissau",
"Iran (Islamic Republic of)": "Iran",
"Congo": "Republic of the Congo",
"Venezuela (Bolivarian Republic of)": "Venezuela",
"Côte d'Ivoire": "Ivory Coast",
"United Kingdom of Great Britain and Northern Ireland": "United Kingdom",
"Viet Nam": "Vietnam",
"Serbia": "Republic of Serbia",
"The former Yugoslav Republic of Macedonia": "Macedonia",
"Brunei Darussalam": "Brunei",
"Syrian Arab Republic": "Syria",
"Bahamas": "The Bahamas",
"Republic of Korea": "South Korea",
"Lao People's Democratic Republic": "Laos",
"Republic of Moldova": "Moldova",
}
# Use map() method to replace incorrect country names with correct names
df["Country"] = df["Country"].map(country_map).fillna(df["Country"])
# Create a map centered on Canada
map = folium.Map(location=[0, 0], zoom_start=2)
# Add the choropleth layer
folium.Choropleth(
geo_data=world_geo,
name="choropleth",
data=df,
columns=["Country", "Total"],
key_on="feature.properties.name",
fill_color="YlOrRd",
fill_opacity=0.7,
line_opacity=0.2,
legend_name="Total Immigrants",
).add_to(map)
# Add a layer control
folium.LayerControl().add_to(map)
# Display the map
map
# A choropleth map has been created using the Folium library in Python to display the total immigration figures for all countries in the world. The map uses color-coding to indicate the immigration figures, with the redder shade indicating higher immigration numbers.
# This type of visualization provides a geospatial perspective of where immigrants originate from, making it easier to identify regions or countries with the highest immigration rates.
region_totals = df.groupby(["Region"])["Total"].sum().reset_index()
region_totals.sort_values("Total", ascending=False, inplace=True)
# Create a bar chart of the regions
fig = px.bar(
region_totals,
x="Total",
y="Region",
color="Region",
orientation="h",
title="Immigration to Canada by Region from 1980 to 2013",
color_continuous_scale="matter",
)
fig.show()
# From the chart, we see a breakdown of immigrants to Canada by region from 1980 to 2013. The data shows that more than half a million immigrants came from four regions: South Asia, East Asia, Southeast Asia, and Northern Europe. In contrast, the number of immigrants from areas such as Australia and New Zealand, Melanesia, Central Asia, Polynesia, and Micronesia was lower, with under 40,000 immigrants from these regions during the same period.
# This pattern of immigration also reflects population density in different regions. For example, regions with higher population densities, such as Southern Asia, have contributed more immigrants to Canada than regions with lower population densities, such as Australia and New Zealand, Melanesia, Central Asia, Polynesia, and Micronesia.
# Group the data by development status and year, and sum the total immigrants
dev_df = (
df.groupby(["DevName"])
.sum(numeric_only=True)
.transpose()
.reset_index()
.rename(columns={"index": "Year"})
)
# drop the last row
dev_df = dev_df.drop(dev_df.index[-1])
# Create a line chart of the total immigrants by development status and year
fig = go.Figure()
fig.add_trace(
go.Scatter(
x=dev_df["Year"],
y=dev_df["Developing regions"],
mode="lines",
name="Developing regions",
)
)
fig.add_trace(
go.Scatter(
x=dev_df["Year"],
y=dev_df["Developed regions"],
mode="lines",
name="Developed regions",
)
)
fig.update_layout(
title="Total Immigrants by Development Status from 1980 to 2013",
xaxis_title="Year",
yaxis_title="Number of Immigrants",
xaxis=dict(
title="Year", tickangle=50 # Set the angle of the tick labels to 50 degrees
),
)
fig.show()
# This line graph shows the difference in the number of people immigrating to Canada from 1980 to 2013 between developed and developing countries. While developed countries hovered around 50k during this period, developing countries quadrupled the number of migrants, reaching more than 200k in migration in 2013.
fig = px.sunburst(df, path=["DevName", "Continent", "Region"], values="Total")
fig.update_layout(height=800, width=800)
fig.show()
# This sunburst chart is an extension of the previous line chart, giving us a more detailed look at the distribution of immigrants from developed and developing countries compared to total immigration to Canada. The donut circle next to the center are the continents, and next are the regions within these continents. Developing regions account for nearly three-quarters of that of developed regions. The countries of the island nation type, central Asia and East Asia account for a very small number compared to other countries.
df_continent = (
df.groupby(["Continent"])["Total"]
.sum()
.reset_index()
.sort_values("Total", ascending=False)
)
fig = px.bar(
df_continent,
x="Continent",
y="Total",
color="Continent",
title="Total Immigrants by Continent from 1980 to 2013",
)
fig.update_layout(xaxis_title="Continent", yaxis_title="Number of Immigrants")
fig.show()
fig = px.pie(values=df_continent["Total"], names=df_continent["Continent"])
fig.show()
# The bar graph and pie chart illustrate the total immigration to Canada from different continents between 1980 and 2013. The data shows that Asia accounted for the largest number of immigrants, with about 3.3 million individuals, representing over 51.8% of all immigrants to Canada during this period. Europe was the second largest contributor with around 1.4 million immigrants, which corresponds to roughly 22% of all immigrants.
# In contrast, the smallest numbers of immigrants came from Northern America and Oceania, with 241,142k and 55,174k respectively, accounting for only 3.76% and 0.861% of all immigrants to Canada. The pie chart highlights the differences in the distribution of immigrants across the continents, with Asia comprising more than half of all immigrants.
# Group the data by continent and sum the total immigrants
continent_totals = df.groupby("Continent").sum().sort_values("Total", ascending=False)
# Create a list of x-axis labels
x_labels = [str(year) for year in range(1980, 2014)]
# Create a line chart for each continent
data = []
for continent in continent_totals.index:
trace = go.Scatter(
x=x_labels, y=continent_totals.loc[continent], mode="lines", name=continent
)
data.append(trace)
# Create the layout for the chart
layout = go.Layout(
title="Total Immigration by Continent from 1980 to 2013",
xaxis=dict(
title="Year", tickangle=50 # Set the angle of the tick labels to 50 degrees
),
yaxis=dict(title="Number of Immigrants"),
)
# Create the figure and plot the chart
fig = go.Figure(data=data, layout=layout)
fig.show()
# The line graph depicts the number of immigrants who migrated to Canada from different continents between 1980 and 2013. The data shows that in 1980, Europe had the highest number of immigrants with nearly 40k, followed by Asia with approximately 31k. From 1980 to 1985, the number of immigrants from all continents decreased slightly before starting to increase again.
# Since 1985, the number of immigrants from Asia, Europe, and Latin America and the Caribbean has increased, with the most significant increases observed in Asia and Europe. Asia had a noticeable increase, reaching its highest peak in 2010 with nearly 164k immigrants. In contrast, Europe and Latin America and the Caribbean experienced a decreasing trend from 1992 onwards.
# Interestingly, the number of immigrants from Africa has continuously increased from 1980 to 2013. In contrast, Northern America and Oceania have maintained steady levels below 11k and 3k, respectively, throughout this period.
import plotly.express as px
def plot_continent_scatter(continent, color_continuous_scale=None):
continent_df = df[df["Continent"] == continent]
x_vals = continent_df.columns[4:-1]
y_vals = continent_df.iloc[:, 4:-1].sum(axis=0)
fig = px.scatter(
x=x_vals,
y=y_vals,
color=y_vals,
size=y_vals,
title=f"Total Immigrants from {continent} from 1980 to 2013",
labels={"x": "Year", "y": "Number of Immigrants"},
color_continuous_scale=color_continuous_scale,
trendline="ols",
)
fig.update_layout(xaxis={"tickangle": 50})
fig.show()
# Plot scatter charts for each continent
plot_continent_scatter("Asia", color_continuous_scale="matter")
plot_continent_scatter("Africa", color_continuous_scale="magenta")
plot_continent_scatter(
"Latin America and the Caribbean", color_continuous_scale="sunset"
)
plot_continent_scatter("Europe", color_continuous_scale="darkmint")
# These four scatter charts represent immigration to Canada from four different continents: Asia, Africa, Europe, and Latin America and the Caribbean. The charts share a common feature where each circle dot represents the number of immigrants for a given year, and the size and color of the circle dot correspond to the number of immigrants. Specifically, larger and darker circle dots indicate a higher number of immigrants.
# Moreover, each scatter chart has a trendline that shows the overall trend from 1980 to 2013. The trendline allows us to see whether the number of immigrants is increasing or decreasing over time. In general, we observe that Asia, Africa, and Latin America and the Caribbean have an upward trend, while Europe has a tendency to decrease despite some periods of rapid growth, such as from 1985 to 1990.
# Overall, these scatter charts provide a more detailed and nuanced view of immigration to Canada than the previous chart that displayed the total immigration numbers by continent. The scatter charts allow us to see the year-to-year fluctuations and the trends over time for each continent.
# Filter the dataset to include only the records from the Asia continent
asia_df = df[df["Continent"] == "Asia"]
# Group the records by country and sum the immigration values across all years
asia_grouped = asia_df.groupby("Country")["Total"].sum()
# Select the top 5 countries by total immigration
asia_top5 = asia_grouped.nlargest(5)
# Filter the data to include only the top 5 countries
top_5_df = asia_df[asia_df["Country"].isin(asia_top5.index)]
# Create a line plot for each of the top 5 countries to show the trend of immigration over the years
fig = go.Figure()
for country in top_5_df["Country"].unique():
df_country = top_5_df[top_5_df["Country"] == country]
fig.add_trace(
go.Scatter(
x=df_country.columns[4:-1],
y=df_country.iloc[:, 4:-1].sum(axis=0),
mode="lines",
name=country,
)
)
fig.update_layout(
title="Top 5 Immigration Countries from Asia from 1980 to 2013",
xaxis_title="Year",
yaxis_title="Number of Immigrants",
xaxis=dict(
title="Year", tickangle=50 # Set the angle of the tick labels to 50 degrees
),
)
fig.show()
# The chart provides insights into the immigration patterns of the top 5 Asian countries to Canada from 1980 to 2013. It is clear that China, India, and the Philippines are the top three countries with the most immigrants during this period. Despite some fluctuations, these three countries generally exhibit a consistent upward trend in the number of immigrants, with the Philippines experiencing a significant decline from 1993 to 1998. Iran and Pakistan also show an increasing trend, albeit with some fluctuations, with both countries having around 12,000 immigrants in 2013 compared to around 1,000 in 1980. These trends suggest that Canada has become an increasingly attractive destination for immigrants from Asia, particularly from China, India, and the Philippines.
# Filter the dataset to include only the records from the Africa continent
africa_df = df[df["Continent"] == "Africa"]
# Group the records by country and sum the immigration values across all years
africa_grouped = africa_df.groupby("Country")["Total"].sum()
# Select the top 5 countries by total immigration
africa_top5 = africa_grouped.nlargest(5)
# Filter the data to include only the top 5 countries
top_5_df = africa_df[africa_df["Country"].isin(africa_top5.index)]
# Create a line plot for each of the top 5 countries to show the trend of immigration over the years
fig = go.Figure()
for country in top_5_df["Country"].unique():
df_country = top_5_df[top_5_df["Country"] == country]
fig.add_trace(
go.Scatter(
x=df_country.columns[4:-1],
y=df_country.iloc[:, 4:-1].sum(axis=0),
mode="lines",
name=country,
)
)
fig.update_layout(
title="Top 5 Immigration Countries from Africa from 1980 to 2013",
xaxis_title="Year",
yaxis_title="Number of Immigrants",
xaxis=dict(
title="Year", tickangle=50 # Set the angle of the tick labels to 50 degrees
),
)
# Add text annotations
fig.add_annotation(
x=10.8,
y=4000,
xref="x",
yref="y",
text="Somalia Civil War and Famine",
font=dict(family="sans serif", size=16, color="purple"),
textangle=-80,
showarrow=False,
)
fig.show()
# The data in this chart presents the historical progression of emigration from the top 5 African countries from 1980 to 2013. The results indicate an overall rising trend of emigration from these countries during this period. However, the most significant increases in emigration were recorded in Algeria, Egypt, and Morocco.
# Somalia stands out as an exceptional case, with a substantial surge in emigration from 401 individuals in 1989 to 5,794 individuals in 1992. This period coincides with political turmoil and conflict that may have pushed individuals to seek refuge in Canada. Likewise, in the cases of Egypt and Morocco, the data suggests that economic and political challenges may have motivated some individuals to move to Canada, with a remarkable surge in emigration in 2010, with around 6,000 individuals compared to only a few hundred in 1980.
# On the other hand, South Africa experienced a moderate increase in emigration, with notable fluctuations from year to year. Overall, the findings indicate that the most significant increases in emigration from Africa occurred in Algeria, Egypt, and Morocco, which could be attributed to various factors such as political instability, economic hardships, or a desire for better opportunities.
# Filter the dataset to include only the records from the Latin America and the Caribbean continent
europe_df = df[df["Continent"] == "Latin America and the Caribbean"]
# Group the records by country and sum the immigration values across all years
europe_grouped = europe_df.groupby("Country").sum(numeric_only=True)
# Sort the resulting dataframe in descending order based on the total immigration values
europe_top5 = europe_grouped.sort_values("Total", ascending=False).head(5)
# Create a line plot for each of the top 5 countries to show the trend of immigration over the years
fig = go.Figure()
for country in europe_top5.index:
x_vals = europe_top5.columns[:-1]
y_vals = europe_top5.loc[country][:-1]
fig.add_trace(go.Scatter(x=x_vals, y=y_vals, mode="lines", name=country))
fig.update_layout(
title="Top 5 Immigration Countries from Latin America and the Caribbean from 1980 to 2013",
xaxis_title="Year",
yaxis_title="Number of Immigrants",
xaxis=dict(
title="Year", tickangle=50 # Set the angle of the tick labels to 50 degrees
),
)
# Add text annotations
fig.add_annotation(
x=29.5,
y=5200,
xref="x",
yref="y",
text="2010 Haiti Earthquake",
font=dict(family="sans serif", size=16, color="red"),
textangle=-76,
showarrow=False,
)
# Add text annotations
fig.add_annotation(
x=22.5,
y=4700,
xref="x",
yref="y",
text="Colombia Civil Wars 2000-2006",
font=dict(family="sans serif", size=16, color="purple"),
textangle=-63,
showarrow=False,
)
fig.show()
# This line chart portrays the immigration trends from five countries from Latin America and the Caribbean to Canada, highlighting that the most notable and pronounced increase in immigration occurred in Haiti, Colombia, and Mexico.
# Colombia experienced a period of internal conflict, drug trafficking, and violence from 1999 to 2006, which forced many Colombians to seek refuge in Canada. The conflict intensified during this period, leading to widespread violence, human rights abuses, and displacement of civilians.
# Haiti also saw a surge in immigration to Canada from 2009 to 2011, mainly due to a series of significant events, including a devastating earthquake in January 2010 that killed an estimated 200,000 people and displaced over a million others, a cholera outbreak that started in October 2010 and spread quickly throughout the country, and a presidential election that led to violence and dependency.
# In contrast, Jamaica and Guyana showed a relatively high number of immigrants to Canada in 1980, with around 3,000 people. However, the numbers fluctuated significantly, increasing sharply until 1995 and then fluctuating slightly and tending to decrease after 2000. Jamaica faced high inflation and debt, while Guyana was dealing with a declining economy and political unrest in 1987.
# Overall, the line chart indicates that the most significant increases in immigration to Canada came from Haiti, Colombia, and Mexico, which could be attributed to various factors such as political instability, economic hardship, and natural disasters.
# Filter the dataset to include only the records from the Europe continent
europe_df = df[df["Continent"] == "Europe"]
# Group the records by country and sum the immigration values across all years
europe_grouped = europe_df.groupby("Country").sum(numeric_only=True)
# Sort the resulting dataframe in descending order based on the total immigration values
europe_top5 = europe_grouped.sort_values("Total", ascending=False).head(5)
# Create a line plot for each of the top 5 countries to show the trend of immigration over the years
fig = go.Figure()
for country in europe_top5.index:
x_vals = europe_top5.columns[:-1]
y_vals = europe_top5.loc[country][:-1]
fig.add_trace(go.Scatter(x=x_vals, y=y_vals, mode="lines", name=country))
fig.update_layout(
title="Top 5 Immigration Countries from Europe from 1980 to 2013",
xaxis_title="Year",
yaxis_title="Number of Immigrants",
xaxis=dict(
title="Year", tickangle=50 # Set the angle of the tick labels to 50 degrees
),
)
fig.show()
# The line chart represents the immigration trends of the top 5 European countries to Canada from 1980 to 2013. The data shows that these countries experienced the most substantial volatility and increase in immigration during this period. Notably, the UK had the highest number of migrants compared to the other European countries, with fluctuations and increases peaking in 1994 at around 39,231k migrants.
# The increase in immigration can be attributed to several factors, including economic challenges faced by the UK during the 1980s and early 1990s, changes in Canada's immigration policies that made it easier for skilled workers and professionals to immigrate, and the strong cultural and historical connection between Canada and the UK, which may have made Canada a more appealing destination for UK immigrants who were looking to maintain cultural and familial ties.
# On the other hand, France and Romania experienced slow growth and remained at around 5,000 immigrants from 1980 to 2013. This could be due to several factors, including differences in culture and language, limited job opportunities, and strict immigration policies in both countries. Overall, the data suggests that the United Kingdom, Poland, France, Romania, and Portugal were the top European countries for immigration to Canada, with the UK being the most significant contributor to the trend.
country_totals = df[["Country", "Total"]]
top_15_countries = country_totals.sort_values("Total", ascending=False)[:15]
fig = go.Figure()
fig.add_trace(
go.Bar(x=top_15_countries["Total"], y=top_15_countries["Country"], orientation="h")
)
fig.update_layout(
title={
"text": "Top 15 Countries with Total Immigrants to Canada from 1980 to 2013",
"y": 0.9,
"x": 0.5,
"xanchor": "center",
"yanchor": "top",
},
xaxis_title="Number of Immigrants",
yaxis_title="Country",
title_font_size=16,
xaxis_tickfont_size=12,
)
# Set the color of the bars
fig.update_traces(
marker_color="rgb(4, 131, 194)",
marker_line_color="rgb(8,48,107)",
marker_line_width=1.5,
opacity=0.6,
)
fig.show()
# The horizontal bar chart shows the top 15 countries with the largest number of immigrants to Canada from 1980 to 2013. The Philippines, the United Kingdom, China, and India are the top four countries with the highest number of immigrants to Canada from 1980 to 2013, with more than 500,000 immigrants each. China and India had the highest number of immigrants among the top 15 countries, with 659,962 and 691,904 immigrants, respectively. On the other hand, Romania and Vietnam had fewer than 100,000 immigrants each during this period.
# Step 1: Get the data
iceland_df = df[df["Country"] == "Iceland"]
x_vals = iceland_df.columns[4:-1]
y_vals = iceland_df.iloc[:, 4:-1].sum(axis=0)
# Create the trace
trace = go.Bar(x=x_vals, y=y_vals)
# Create the layout
layout = go.Layout(
title="Icelandic Immigrants to Canada from 1980 to 2013",
xaxis=dict(
title="Year", tickangle=50 # Set the angle of the tick labels to 50 degrees
),
yaxis=dict(title="Number of Immigrants"),
)
# Create the figure
fig = go.Figure(data=[trace], layout=layout)
# Add the arrow and text annotations
fig.add_annotation(
x=32,
y=70,
xref="x",
yref="y",
ax=28,
ay=20,
axref="x",
ayref="y",
text="2008 - 2011 Financial Crisis",
font=dict(family="sans serif", size=16, color="red"),
textangle=-30,
showarrow=True,
arrowhead=5,
arrowsize=1,
arrowwidth=3,
arrowcolor="red",
)
# Set the color of the bars
fig.update_traces(
marker_color="rgb(194, 93, 4)",
marker_line_color="rgb(8,48,107)",
marker_line_width=1.5,
opacity=0.4,
)
# Show the figure
fig.show()
|
import numpy as np
import pandas as pd
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# # Using Kaggle Models for Inference
import tensorflow as tf
import tensorflow_hub as hub
import requests
from PIL import Image
from io import BytesIO
import matplotlib.pyplot as plt
import numpy as np
# @title Helper functions for loading image (hidden)
original_image_cache = {}
def preprocess_image(image):
image = np.array(image)
# reshape into shape [batch_size, height, width, num_channels]
img_reshaped = tf.reshape(
image, [1, image.shape[0], image.shape[1], image.shape[2]]
)
# Use `convert_image_dtype` to convert to floats in the [0,1] range.
image = tf.image.convert_image_dtype(img_reshaped, tf.float32)
return image
def load_image_from_url(img_url):
"""Returns an image with shape [1, height, width, num_channels]."""
user_agent = {"User-agent": "Colab Sample (https://tensorflow.org)"}
response = requests.get(img_url, headers=user_agent)
image = Image.open(BytesIO(response.content))
image = preprocess_image(image)
return image
def load_image(image_url, image_size=256, dynamic_size=False, max_dynamic_size=512):
"""Loads and preprocesses images."""
# Cache image file locally.
if image_url in original_image_cache:
img = original_image_cache[image_url]
elif image_url.startswith("https://"):
img = load_image_from_url(image_url)
else:
fd = tf.io.gfile.GFile(image_url, "rb")
img = preprocess_image(Image.open(fd))
original_image_cache[image_url] = img
# Load and convert to float32 numpy array, add batch dimension, and normalize to range [0, 1].
img_raw = img
if tf.reduce_max(img) > 1.0:
img = img / 255.0
if len(img.shape) == 3:
img = tf.stack([img, img, img], axis=-1)
if not dynamic_size:
img = tf.image.resize_with_pad(img, image_size, image_size)
elif img.shape[1] > max_dynamic_size or img.shape[2] > max_dynamic_size:
img = tf.image.resize_with_pad(img, max_dynamic_size, max_dynamic_size)
return img, img_raw
def show_image(image, title=""):
image_size = image.shape[1]
w = (image_size * 6) // 320
plt.figure(figsize=(w, w))
plt.imshow(image[0], aspect="equal")
plt.axis("off")
plt.title(title)
plt.show()
image_size = 224
dynamic_size = False
# Select an Image Classification model
# Select an Image Classification model
model_name = "vit-b8"
model_handle_map = {
"vit-b8": "/kaggle/input/vision-transformer/tensorflow2/vit-b8-classification/1",
# "evit-b8": "/kaggle/input/vision-transformer/tensorflow2/vit-b8-classification/1",
# "vit-b8": "/kaggle/input/efficientnet-v2/tensorflow2/imagenet1k-b0-classification/versions/1",
# "vit-b8": "https://kaggle.com/models/kaggle/vision-transformer/frameworks/TensorFlow2/variations/vit-b8-classification/",
# "vit-b8": "https://kaggle.com/models/kaggle/vision-transformer/frameworks/TensorFlow2/variations/vit-b8-classification/versions/1",
# "vit-b8": "https://tfhub.dev/sayakpaul/vit_b8_classification/1",
}
model_image_size_map = {
"vit-b8": 224,
}
model_handle = model_handle_map[model_name]
print(f"Selected model: {model_name} : {model_handle}")
max_dynamic_size = 512
if model_name in model_image_size_map:
image_size = model_image_size_map[model_name]
dynamic_size = False
print(f"Images will be converted to {image_size}x{image_size}")
else:
dynamic_size = True
print(
f"Images will be capped to a max size of {max_dynamic_size}x{max_dynamic_size}"
)
labels_file = (
"https://storage.googleapis.com/download.tensorflow.org/data/ImageNetLabels.txt"
)
# download labels and creates a maps
downloaded_file = tf.keras.utils.get_file("labels.txt", origin=labels_file)
classes = []
with open(downloaded_file) as f:
labels = f.readlines()
classes = [l.strip() for l in labels]
# Select an Input Image
# @title Select an Input Image
image_name = "turtle"
images_for_test_map = {
"turtle": "https://upload.wikimedia.org/wikipedia/commons/8/80/Turtle_golfina_escobilla_oaxaca_mexico_claudio_giovenzana_2010.jpg",
# by Claudio Giovenzana, CC BY-SA 3.0 <https://creativecommons.org/licenses/by-sa/3.0>, via Wikimedia Commons
}
img_url = images_for_test_map[image_name]
image, original_image = load_image(img_url, image_size, dynamic_size, max_dynamic_size)
show_image(image, "Scaled image")
classifier = hub.load(model_handle)
input_shape = image.shape
warmup_input = tf.random.uniform(input_shape, 0, 1.0)
# Everything is ready for inference. Here you can see the top 5 results from the model for the selected image.
# Run model on image
top_5 = tf.argsort(probabilities, axis=-1, direction="DESCENDING")[0][:5].numpy()
np_classes = np.array(classes)
# Some models include an additional 'background' class in the predictions, so
# we must account for this when reading the class labels.
includes_background_class = probabilities.shape[1] == 1001
for i, item in enumerate(top_5):
class_index = item if includes_background_class else item + 1
line = f"({i+1}) {class_index:4} - {classes[class_index]}: {probabilities[0][top_5][i]}"
print(line)
show_image(image, "")
# # Using Kaggle Models for Transfer Learning
# Select a model
model_name = "vit-b8"
model_handle_map = {
"vit-b8": "/kaggle/input/vision-transformer/tensorflow2/vit-b8-fe/1",
# "evit-b8": "/kaggle/input/vision-transformer/tensorflow2/vit-b8-fe/1",
# "vit-b8": "/kaggle/input/vision-transformer/tensorflow2/vit-b8-fe/versions/1",
# "vit-b8": "https://kaggle.com/models/kaggle/vision-transformer/frameworks/TensorFlow2/variations/vit-b8-fe/",
# "vit-b8": "https://kaggle.com/models/kaggle/vision-transformer/frameworks/TensorFlow2/variations/vit-b8-fe/versions/1",
# "vit-b8": "https://tfhub.dev/sayakpaul/vit_b8_fe/1",
}
model_image_size_map = {
"vit-b8": 224,
}
model_handle = model_handle_map.get(model_name)
pixels = model_image_size_map.get(model_name, 224)
print(f"Selected model: {model_name} : {model_handle}")
IMAGE_SIZE = (pixels, pixels)
print(f"Input size {IMAGE_SIZE}")
BATCH_SIZE = 16 # @param {type:"integer"}
# Select a dataset to fine-tune the model against
data_dir = tf.keras.utils.get_file(
"flower_photos",
"https://storage.googleapis.com/download.tensorflow.org/example_images/flower_photos.tgz",
untar=True,
)
def build_dataset(subset):
return tf.keras.preprocessing.image_dataset_from_directory(
data_dir,
validation_split=0.20,
subset=subset,
label_mode="categorical",
# Seed needs to provided when using validation_split and shuffle = True.
# A fixed seed is used so that the validation set is stable across runs.
seed=123,
image_size=IMAGE_SIZE,
batch_size=1,
)
train_ds = build_dataset("training")
class_names = tuple(train_ds.class_names)
train_size = train_ds.cardinality().numpy()
train_ds = train_ds.unbatch().batch(BATCH_SIZE)
train_ds = train_ds.repeat()
normalization_layer = tf.keras.layers.Rescaling(1.0 / 255)
preprocessing_model = tf.keras.Sequential([normalization_layer])
do_data_augmentation = False # @param {type:"boolean"}
if do_data_augmentation:
preprocessing_model.add(tf.keras.layers.RandomRotation(40))
preprocessing_model.add(tf.keras.layers.RandomTranslation(0, 0.2))
preprocessing_model.add(tf.keras.layers.RandomTranslation(0.2, 0))
# Like the old tf.keras.preprocessing.image.ImageDataGenerator(),
# image sizes are fixed when reading, and then a random zoom is applied.
# If all training inputs are larger than image_size, one could also use
# RandomCrop with a batch size of 1 and rebatch later.
preprocessing_model.add(tf.keras.layers.RandomZoom(0.2, 0.2))
preprocessing_model.add(tf.keras.layers.RandomFlip(mode="horizontal"))
train_ds = train_ds.map(lambda images, labels: (preprocessing_model(images), labels))
val_ds = build_dataset("validation")
valid_size = val_ds.cardinality().numpy()
val_ds = val_ds.unbatch().batch(BATCH_SIZE)
val_ds = val_ds.map(lambda images, labels: (normalization_layer(images), labels))
# Defining the model.
# All it takes is to put a linear classifier on top of the `feature_extractor_layer` with the Hub module.
# For speed, we start out with a non-trainable `feature_extractor_layer`, but you can also enable fine-tuning for greater accuracy.
do_fine_tuning = False
print("Building model with", model_handle)
model = tf.keras.Sequential(
[
# Explicitly define the input shape so the model can be properly
# loaded by the TFLiteConverter
tf.keras.layers.InputLayer(input_shape=IMAGE_SIZE + (3,)),
hub.KerasLayer(model_handle, trainable=do_fine_tuning),
tf.keras.layers.Dropout(rate=0.2),
tf.keras.layers.Dense(
len(class_names), kernel_regularizer=tf.keras.regularizers.l2(0.0001)
),
]
)
model.build((None,) + IMAGE_SIZE + (3,))
model.summary()
# Training the model
model.compile(
optimizer=tf.keras.optimizers.SGD(learning_rate=0.005, momentum=0.9),
loss=tf.keras.losses.CategoricalCrossentropy(from_logits=True, label_smoothing=0.1),
metrics=["accuracy"],
)
steps_per_epoch = train_size // BATCH_SIZE
validation_steps = valid_size // BATCH_SIZE
hist = model.fit(
train_ds,
epochs=5,
steps_per_epoch=steps_per_epoch,
validation_data=val_ds,
validation_steps=validation_steps,
).history
plt.figure()
plt.ylabel("Loss (training and validation)")
plt.xlabel("Training Steps")
plt.ylim([0, 2])
plt.plot(hist["loss"])
plt.plot(hist["val_loss"])
plt.figure()
plt.ylabel("Accuracy (training and validation)")
plt.xlabel("Training Steps")
plt.ylim([0, 1])
plt.plot(hist["accuracy"])
plt.plot(hist["val_accuracy"])
x, y = next(iter(val_ds))
image = x[0, :, :, :]
true_index = np.argmax(y[0])
plt.imshow(image)
plt.axis("off")
plt.show()
# Expand the validation image to (1, 224, 224, 3) before predicting the label
prediction_scores = model.predict(np.expand_dims(image, axis=0))
predicted_index = np.argmax(prediction_scores)
print("True label: " + class_names[true_index])
print("Predicted label: " + class_names[predicted_index])
|
from torch.utils.data import Dataset, DataLoader
import torch
import cv2
import glob
from torchvision.io import read_image
from clearml import Task, logger
from torchvision.transforms import ToTensor, Compose, Normalize
import numpy as np
from datetime import datetime
from torchvision import models
import time
import copy
import torch.nn as nn
import torch.optim as optim
from torch.optim import lr_scheduler
from torch.utils.tensorboard import SummaryWriter
from tqdm import tqdm
from sklearn.metrics import (
confusion_matrix,
classification_report,
roc_auc_score,
roc_curve,
)
import gc
from clearml import Task, Logger, Dataset
# Download dataset from ClearML
dataset_name = "Dataset_phones_splited_v1"
dataset_project = "Check in car"
dataset_path = Dataset.get(
dataset_name=dataset_name, dataset_project=dataset_project
).get_local_copy()
# Make task in ClearML
task = Task.init(
project_name="Check in car",
task_name="MobileNetV2_cls_v2",
tags=["classification", "MobileNetV2", "AdamW"],
)
logger = task.get_logger()
class Drivers_with_phone_Dataset(Dataset):
def __init__(self, img_dir, transform=None, target_transform=None):
# self.img_labels = annotations_file
self.img_dir = img_dir
file_list = glob.glob(self.img_dir + "*")
self.data = []
for class_path in file_list:
class_name = class_path.split("/")[-1]
for img_path in glob.glob(class_path + "/*.jpg"):
self.data.append([img_path, class_name])
self.class_map = {"cellphone": 1, "no_cellphone": 0}
self.transform = Compose(
[ToTensor(), Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])]
)
self.target_transform = target_transform
self.img_dim = (640, 480)
def __len__(self):
return len(self.data)
def __getitem__(self, idx):
img_path, class_name = self.data[idx]
img = cv2.imread(img_path)
# img = cv2.resize(img, self.img_dim)
class_id = self.class_map[class_name]
class_id = torch.tensor(class_id)
if self.transform:
img_tensor = self.transform(img)
return img_tensor, class_id
def train_one_epoch(epoch_index, tb_writer):
full_batch_loss = 0.0
total = 0
correct = 0
total_step = len(train_data_loader)
for i, data in enumerate(train_data_loader):
inputs, labels = data
inputs, labels = inputs.to(device), labels.to(device)
optimizer.zero_grad()
outputs = model(inputs)
loss = criterion(outputs, labels)
loss.backward()
# Adjust learning weights
optimizer.step()
# Gather data and report
full_batch_loss += float(loss.item())
# Acc check
_, pred = torch.max(outputs, dim=1)
correct += torch.sum(pred == labels).item()
total += labels.size(0)
torch.cuda.empty_cache()
# lr_scheduler.step()
train_acc_epoch = 100 * correct / total
avg_loss = full_batch_loss / total_step
del full_batch_loss
gc.collect()
torch.cuda.empty_cache()
return avg_loss, train_acc_epoch
# Const
n_epochs = 10
batch_size = 32
lr = 0.001
best_vloss = np.Inf
val_loss = []
val_acc = []
train_loss = []
train_acc = []
# Download data for testing and training
drivers_data_train = dataset_path + "/dataset_phones_splited/train/"
drivers_data_test = dataset_path + "/dataset_phones_splited/test/"
train_data = Drivers_with_phone_Dataset(drivers_data_train)
test_data = Drivers_with_phone_Dataset(drivers_data_test)
print(len(train_data), len(test_data))
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
train_data_loader = DataLoader(
train_data, batch_size=batch_size, shuffle=True, num_workers=0
)
test_data_loader = DataLoader(
test_data, batch_size=batch_size, shuffle=False, num_workers=0
)
# exp_lr_scheduler = lr_scheduler.StepLR(optimizer_ft, step_size=7, gamma=0.1)
# lr_scheduler = torch.optim.lr_scheduler.MultiStepLR(optimizer, milestones=[7,12,18], gamma=0.1)
for features, labels in train_data_loader:
print("Shape of batch of features: ", features.shape)
print("Shape of the corresponding labels: ", labels.shape)
break
# Model, optomizer, loss function
model = models.mobilenet_v2(pretrained=False)
n = model.classifier[1].in_features
model.classifier = nn.Linear(n, 2)
model = model.to(device)
criterion = nn.CrossEntropyLoss()
optimizer = optim.AdamW(model.parameters(), lr=lr)
timestamp = datetime.now().strftime("%Y%m%d_%H%M%S")
writer = SummaryWriter("runs/MobileNet_v2_{}".format(timestamp))
epoch_number = 0
for epoch in range(n_epochs):
print("EPOCH {}:".format(epoch_number + 1))
torch.cuda.empty_cache()
# On gradients
model.train(True)
avg_loss, train_acc_epoch = train_one_epoch(epoch_number, writer)
# Off gradients
model.train(False)
torch.cuda.empty_cache()
with torch.no_grad():
print("Validation phase")
running_vloss = 0.0
total_v = 0
correct_v = 0
for i, vdata in enumerate(test_data_loader):
vinputs, vlabels = vdata
vinputs, vlabels = vinputs.to(device), vlabels.to(device)
voutputs = model(vinputs)
vloss = criterion(voutputs, vlabels)
running_vloss += float(vloss.item())
_, pred_v = torch.max(voutputs, dim=1)
correct_v += torch.sum(pred_v == vlabels).item()
total_v += vlabels.size(0)
torch.cuda.empty_cache()
avg_vloss = running_vloss / len(test_data_loader)
val_acc_epoch = 100 * correct_v / total_v
print("LOSS train {} valid {}".format(avg_loss, avg_vloss))
print("ACC train {} valid {}".format(train_acc_epoch, val_acc_epoch))
del running_vloss
gc.collect()
# Metrics
train_loss.append(avg_loss)
val_loss.append(float(avg_vloss))
train_acc.append(train_acc_epoch)
val_acc.append(val_acc_epoch)
# When Kaggle is down
# json.dump(train_loss, open("train_loss.json", "w"), indent=4)
# json.dump(val_loss, open("val_loss.json", "w"), indent=4)
# json.dump(train_acc, open("train_acc.json", "w"), indent=4)
# json.dump(val_acc, open("val_acc.json", "w"), indent=4)
# Log loss and accuracy to Clear ML
logger.report_scalar(
"Training vs. Validation Loss",
"Training",
iteration=epoch_number + 1,
value=avg_loss,
)
logger.report_scalar(
"Training vs. Validation Loss",
"Validation",
iteration=epoch_number + 1,
value=avg_vloss,
)
logger.report_scalar(
"Training vs. Validation Accuracy",
"Training",
iteration=epoch_number + 1,
value=train_acc_epoch,
)
logger.report_scalar(
"Training vs. Validation Accuracy",
"Validation",
iteration=epoch_number + 1,
value=val_acc_epoch,
)
# Track best performance, and save the model's state
if avg_vloss < best_vloss:
best_vloss = avg_vloss
model_path = "model_{}_{}".format(timestamp, "best")
torch.save(model.state_dict(), model_path)
epoch_number += 1
# # Test, Confusion Matrix, ROC-AUC
# # Code To Test Pretrained Model
# drivers_data_test= "/kaggle/input/dataset-phones-splited/dataset_phones_splited/test/"
# test_data = Drivers_with_phone_Dataset(drivers_data_test)
# test_data_loader = DataLoader(test_data, batch_size=32,
# shuffle=False, num_workers=0)
# device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
# model = models.mobilenet_v2()
# n = model.classifier[1].in_features
# model.classifier = nn.Linear(n, 2)
# model = model.to(device)
# model.load_state_dict(torch.load("/kaggle/input/model-resnet/model_20230328_082109_best"))
# Test Model
y_pred_list = []
y_test = []
y_pred_proba = []
with torch.no_grad():
model.eval()
for X_batch, y_batch in test_data_loader:
X_batch = X_batch.to(device)
y_test_pred = model(X_batch)
_, y_pred_tags = torch.max(y_test_pred, dim=1)
y_pred_list.append(y_pred_tags.cpu().numpy())
y_test.append(y_batch.cpu().numpy())
y_pred_proba.append(y_test_pred)
# for conf matr and roc
y_pred_list = [a.squeeze().tolist() for a in y_pred_list]
y_test = [a.squeeze().tolist() for a in y_test]
# one big list
y_pred_list_flat = [item for sublist in y_pred_list for item in sublist]
y_test_flat = [item for sublist in y_test for item in sublist]
# Calc Confusion matrix and log to Clear ML
confusion_matrix_1 = confusion_matrix(y_test_flat, y_pred_list_flat)
logger.report_matrix(
"Confusion_matrix_",
"ignored",
matrix=confusion_matrix_1,
xaxis="Predicted lable",
yaxis="True label",
xlabels=["no phones", "phone"],
ylabels=["no phones", "phone"],
yaxis_reversed=True,
)
# Calc Roc-Auc
fpr, tpr, thresholds = roc_curve(y_test_flat, y_pred_list_flat)
auc = roc_auc_score(y_test_flat, y_pred_list_flat)
logger.report_scatter2d(
"ROC AUC Curve",
"ROC",
scatter=zip(fpr, tpr),
yaxis="tpr",
xaxis="fpr",
mode="lines+markers",
)
logger.flush()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import sklearn
from sklearn.model_selection import train_test_split
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn import tree
from sklearn import preprocessing
from sklearn.metrics import roc_auc_score
from sklearn.metrics import f1_score
from sklearn.metrics import accuracy_score
from sklearn.model_selection import GridSearchCV
from sklearn.preprocessing import StandardScaler
import seaborn as sns
import matplotlib.pyplot as plt
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Here I input the training & testing data. I split the training data into its respective features and target.
# Then I split the testing data features, ignoring the 'id' column.
train_data = pd.read_csv("/kaggle/input/cap-4611-spring-21-assignment-1/train.csv")
features = train_data.columns[2:]
X = train_data.loc[:, features]
y = train_data.loc[:, "Bankrupt"]
train_data.head()
test_data = pd.read_csv("/kaggle/input/cap-4611-spring-21-assignment-1/test.csv")
x_features = test_data.columns[1:]
x_test = test_data.loc[:, x_features]
test_data.head()
# Below, I check for any null/missing values in the dataset. These checks have never found any missing values. Because of this, I decided not to implement any handling of missing values. If the datasets were to change, and missing values were found, then I would manually inspect the data and figure out a solution. This could be dropping a row, dropping a feature, or replacing the missing value with a metric.
# Check and handle missing values.
if train_data.isnull().any().any():
# Handle missing values.
print("Missing values.")
if test_data.isnull().any().any():
# Handle missing values.
print("Missing values.")
# Below is where I utilize a boxplot to showcase how to check for outliers. You can see clearly there exist outliers in the data, but I decided not to handle them since they are valid datapoints & removing them does not affect my models' performance.
# Check for outliers here using boxplots.
import seaborn as sns
sns.set_theme(style="whitegrid")
f, axs = plt.subplots(1, 3, figsize=(10, 3))
sns.boxplot(x=X["No-credit interval"], ax=axs[0])
sns.boxplot(x=X["liability to equity"], ax=axs[1])
sns.boxplot(x=X["liability to equity"], ax=axs[2])
f.tight_layout()
# Below, I have coded an example of standardization, which I would rather use instead of normalization. But since I am not using regression, gradient descent, or building neural networks, there is no need to scale the data. Trees are virtually indifferent to scaling since the split does not depend on multiple features.
# For this dataset, I would rather have used standardization, because I feel this dataset would be largely affected by smushing everything into a range between 0-1.
# Very simple example showcasing standardization.
"""scaler = StandardScaler()
scaler.fit(X)
scaler.transform(X)"""
# In the cell below, I begin by splitting the training data into pieces, so that I can effectively fit and train both models. I then build the models, and fit them to the training portion of the split. Following that, I use predict_proba [:, 1], so that my predictions vector can represent the probability of being bankrupt. The predict() function returns the binary representation instead, which is then used to calculate the various metrics.
# **Explanation for model parameters:**
# criterion is "entropy" because it represents information gain
# max_depth is usually a smaller number because a tall tree can result in overfitting
# n_estimators is 1000 because I wanted a large amount of trees working together
# The rest of the parameters were found by utilizing GridSearchCV
# Split the training data.
X_train, X_test, y_train, y_test = train_test_split(X, y)
# Build a decision tree, and fit it.
clf = tree.DecisionTreeClassifier(
criterion="entropy", max_depth=4, splitter="best", max_leaf_nodes=7
)
clf.fit(X_train, y_train)
# Build a random forest, and fit it.
rf = RandomForestClassifier(
criterion="entropy",
max_depth=5,
max_leaf_nodes=25,
n_estimators=1000,
min_samples_split=2,
)
rf.fit(X_train, y_train)
# Here is an example of a GridSearch I used to better my parameters. Commented it out because it took me a few minutes to complete.
"""rf_CV = RandomForestClassifier(criterion = "entropy")
param_grid = {'max_leaf_nodes': np.array([25, 35, 40]), 'n_estimators': np.array([1000])}
rf_cv = GridSearchCV(rf_CV, param_grid, cv = 5, verbose = 1)
rf_cv.fit(X, y)
print(rf_cv.best_params_)"""
# Below are the metrics for both models.
y_predi_true = clf.predict_proba(X_test)[:, 1]
y_predict_clf = clf.predict(X_test)
print("Decision tree ROC AUC score:", roc_auc_score(y_test, y_predi_true))
print("Decision tree accuracy score:", accuracy_score(y_test, y_predict_clf))
print("Decision tree F1 score:", f1_score(y_test, y_predict_clf))
print()
y_predi_true_rf = rf.predict_proba(X_test)[:, 1]
y_predict_rf = rf.predict(X_test)
print("Random forest ROC AUC score:", roc_auc_score(y_test, y_predi_true_rf))
print("Random forest accuracy score:", accuracy_score(y_test, y_predict_rf))
print("Random forest F1 score:", f1_score(y_test, y_predict_rf))
# I decided to go with a random forest model. After a multitude of runs, the random forest consistently had a higher ROC AUC score and accuracy score.
# Here, build the final model, using the parameters from the training models.
model = RandomForestClassifier(
criterion="entropy",
max_depth=5,
max_leaf_nodes=25,
n_estimators=1000,
min_samples_split=2,
)
model.fit(X, y)
y_pred_true = model.predict_proba(x_test)[:, 1]
output = pd.DataFrame({"id": test_data.id, "Bankrupt": y_pred_true})
output.to_csv("my_submission.csv", index=False)
print("Your submission was successfully saved!")
|
# # EDA of Bigquery Google Analytics Data
# **Objective :**
# Analyzing Google Analytics data from Bigquery using SQL.
# **Data Source :**
# The sample dataset contains Google Analytics 360 data from the Google Merchandise Store, a real ecommerce store. The Google Merchandise Store sells Google branded merchandise.
# Traffic source data: information about where website visitors originate. This includes data about organic traffic, paid search traffic, display traffic, etc.
# Content data: information about the behavior of users on the site. This includes the URLs of pages that visitors look at, how they interact with content, etc.
# Transactional data: information about the transactions that occur on the Google Merchandise Store website.
# **Work Flow :**
# 1. Extract data and confirm structure/contents
# 2. Explore and analyze data
# 3. Visualize insights and interpret results
# Import necessary libraries
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
sns.set()
from plotly.offline import init_notebook_mode, iplot
init_notebook_mode(connected=True)
import plotly.graph_objs as go
import plotly.express as px
# # 1. Data Extraction
from google.cloud import bigquery
# Create client object
client = bigquery.Client()
# Create dataset reference
dataset_ref = client.dataset("google_analytics_sample", project="bigquery-public-data")
# Retrieve dataset from reference
dataset = client.get_dataset(dataset_ref)
# View tables in dataset
[x.table_id for x in client.list_tables(dataset)][:5]
# Create table reference
table_ref_20160801 = dataset_ref.table("ga_sessions_20160801")
# Retrieve table from reference
table_20160801 = client.get_table(table_ref_20160801)
# View columns
client.list_rows(table_20160801, max_results=5).to_dataframe()
# **Schema Interaction**
# Display schemas
print("SCHEMA field for the 'totals' column:\n")
print(table_20160801.schema[5])
print("\nSCHEMA field for the 'trafficSource' column:\n")
print(table_20160801.schema[6])
print("\nSCHEMA field for the 'device' column:\n")
print(table_20160801.schema[7])
print("\nSCHEMA field for the 'geoNetwork' column:\n")
print(table_20160801.schema[8])
print("\nSCHEMA field for the 'customDimensions' column:\n")
print(table_20160801.schema[9])
print("\nSCHEMA field for the 'hits' column:\n")
print(table_20160801.schema[10])
# # 2. Explore and Analyze Data
# Analyzing the most frequently visited landing pages and their respective bounce rates.
# A high bounce rate can indicate either:
# 1. Users are not seeing what they expect when they enter the site, and Search Engine Optimization (SEO) needs to be done
# 2. There is a functionality or display issue with the landing page
# 「hitNumber=1」Indicates the first hit of a session
query = """
SELECT
hits.page.pagePath AS landing_page,
COUNT(*) AS views,
SUM(totals.bounces)/COUNT(*) AS bounce_rate
FROM
`bigquery-public-data.google_analytics_sample.ga_sessions_*`,
UNNEST(hits) AS hits
WHERE
_TABLE_SUFFIX BETWEEN '20160801' AND '20170801'
AND
hits.type='PAGE'
AND
hits.hitNumber=1
GROUP BY landing_page
ORDER BY views DESC
LIMIT 10
"""
result = client.query(query).result().to_dataframe()
result.head(10)
# Looking at the bounce rates, it looks like Men's Apparel and Bags are comparatively lower than the other pages.
# We can also see that the YouTube and Drinkware pages have a lot of views, but have a high bounce rate, indicating either a problem with the page's functionality, or users not seeing what they were expecting to when they entered the site.
# Analyzing Exit rate
# A high exit rate can indicate either:
# 1. Users are not seeing what they expect when they navigate to a page
# 2. There is a functionality or display issue with the page
query = """
SELECT
hits.page.pagePath AS page,
COUNT(*) AS views,
SUM(totals.bounces)/COUNT(*) AS exit_rate
FROM
`bigquery-public-data.google_analytics_sample.ga_sessions_*`,
UNNEST(hits) AS hits
WHERE
_TABLE_SUFFIX BETWEEN '20160801' AND '20170801'
AND
hits.type='PAGE'
GROUP BY page
ORDER BY views DESC
"""
result = client.query(query).result().to_dataframe()
result.head(20)
# Compared with other pages, exit rates for pages related to Men's Apparel are slightly high, and the exit rate for YouTube is very high.
# Checking sessions by browser and device to see compatibility issues if.
query = """
SELECT
device.Browser AS browser,
COUNT(*) AS sessions,
SUM(totals.bounces)/COUNT(*) AS exit_rate
FROM
`bigquery-public-data.google_analytics_sample.ga_sessions_*`,
UNNEST(hits) AS hits
WHERE
_TABLE_SUFFIX BETWEEN '20160801' AND '20170801'
GROUP BY browser
ORDER BY sessions DESC
LIMIT 10
"""
result = client.query(query).result().to_dataframe()
result.head(10)
# Among the top 5 browsers, the exit rate for Chrome is comparatively low at 9%, while the other browsers are roughly 7% - 8% higher with the exception of Internet Explorer, having an exit rate of 19%.
query = """
SELECT
device.deviceCategory AS device,
COUNT(*) AS sessions,
SUM(totals.bounces)/COUNT(*) AS exit_rate
FROM
`bigquery-public-data.google_analytics_sample.ga_sessions_*`,
UNNEST(hits) AS hits
WHERE
_TABLE_SUFFIX BETWEEN '20160801' AND '20170801'
GROUP BY device
ORDER BY sessions DESC
"""
result = client.query(query).result().to_dataframe()
result.head(10)
# Looking at the exit rates, we can see that Mobile is about 2.5% higher than other devices. It's possible that the site is not optimized for mobile viewing.
# Checking session and transaction data for each source of traffic to site.
query = """
SELECT
trafficSource.medium AS medium,
COUNT(*) AS sessions,
SUM(totals.bounces)/COUNT(*) AS exit_rate,
SUM(totals.transactions) AS transactions,
SUM(totals.totalTransactionRevenue)/1000000 AS total_revenue,
SUM(totals.transactions)/COUNT(*) AS conversion_rate
FROM
`bigquery-public-data.google_analytics_sample.ga_sessions_*`,
UNNEST(hits) AS hits
WHERE
_TABLE_SUFFIX BETWEEN '20160801' AND '20170801'
GROUP BY medium
ORDER BY sessions DESC
LIMIT 10
"""
result = client.query(query).result().to_dataframe()
result.head(10)
# Most of the traffic is either Organic or Referral.
# Also the conversion rate for CPC and CPM is quite high compared to other sources.
# Identifing the customer's conversion path through the site, as well as areas in the conversion process where users are leaving the site.
# Aggregate hits by action type
query = """
SELECT
CASE WHEN hits.eCommerceAction.action_type = '1' THEN 'Click through of product lists'
WHEN hits.eCommerceAction.action_type = '2' THEN 'Product detail views'
WHEN hits.eCommerceAction.action_type = '5' THEN 'Check out'
WHEN hits.eCommerceAction.action_type = '6' THEN 'Completed purchase'
END AS action,
COUNT(fullVisitorID) AS users,
FROM
`bigquery-public-data.google_analytics_sample.ga_sessions_*`,
UNNEST(hits) AS hits,
UNNEST(hits.product) AS product
WHERE
_TABLE_SUFFIX BETWEEN '20160801' AND '20170801'
AND
(
hits.eCommerceAction.action_type != '0'
AND
hits.eCommerceAction.action_type != '3'
AND
hits.eCommerceAction.action_type != '4'
)
GROUP BY action
ORDER BY users DESC
"""
result = client.query(query).result().to_dataframe()
result.head(10)
# Create funnel graph
fig = go.Figure(
go.Funnel(
y=result["action"],
x=result["users"],
textposition="inside",
textinfo="value+percent initial",
)
)
fig.update_layout(title_text="Google Merchandise Store Conversion Path")
fig.show()
# 69% of users who viewed the product details proceeded to the check out page.
# Of those users, only 30% actually completed a purchase.
# Identify Best-Selling Product Categories and Forecast Demand
# The best-selling categories are related to Apparel, with Men's-T-Shirts being near the top. Also the Office, Electronics, and Water Bottles and Tumblers are selling well.
# 7-day moving average for transactions for Mens t-shirt.
query = """
SELECT
product.v2ProductCategory AS category,
SUM(totals.transactions) AS transactions,
SUM(totals.totalTransactionRevenue)/1000000 AS total_revenue
FROM
`bigquery-public-data.google_analytics_sample.ga_sessions_*`,
UNNEST(hits) AS hits,
UNNEST(hits.product) AS product
WHERE
_TABLE_SUFFIX BETWEEN '20160801' AND '20170801'
GROUP BY category
ORDER BY total_revenue DESC
LIMIT 10
"""
cat_result = client.query(query).result().to_dataframe()
cat_result.head(10)
query = """
WITH daily_mens_tshirt_transactions AS
(
SELECT
date,
SUM(totals.transactions) AS transactions
FROM
`bigquery-public-data.google_analytics_sample.ga_sessions_*`,
UNNEST(hits) AS hits,
UNNEST(hits.product) AS product
WHERE
_TABLE_SUFFIX BETWEEN '20160801' AND '20170801'
AND
product.v2ProductCategory = "Home/Apparel/Men's/Men's-T-Shirts/"
GROUP BY date
ORDER BY date
)
SELECT
date,
AVG(transactions)
OVER (
ORDER BY date
ROWS BETWEEN CURRENT ROW AND 7 FOLLOWING
) AS avg_transactions
FROM
daily_mens_tshirt_transactions
"""
result = client.query(query).result().to_dataframe()
result["date"] = pd.to_datetime(result["date"])
result.plot(
y="avg_transactions",
x="date",
kind="line",
title="Men's T-Shirts Weekly Moving Average",
)
query = """
WITH daily_drinkware_transactions AS
(
SELECT
date,
SUM(totals.transactions) AS transactions
FROM
`bigquery-public-data.google_analytics_sample.ga_sessions_*`,
UNNEST(hits) AS hits,
UNNEST(hits.product) AS product
WHERE
_TABLE_SUFFIX BETWEEN '20160801' AND '20170801'
AND
product.v2ProductCategory = "Home/Drinkware/Water Bottles and Tumblers/"
GROUP BY date
ORDER BY date
)
SELECT
date,
AVG(transactions)
OVER (
ORDER BY date
ROWS BETWEEN CURRENT ROW AND 7 FOLLOWING
) AS avg_transactions
FROM
daily_drinkware_transactions
"""
result = client.query(query).result().to_dataframe()
result["date"] = pd.to_datetime(result["date"])
result.plot(
y="avg_transactions", x="date", kind="line", title="Drinkware Weekly Moving Average"
)
query = """
WITH daily_electronics_transactions AS
(
SELECT
date,
SUM(totals.transactions) AS transactions
FROM
`bigquery-public-data.google_analytics_sample.ga_sessions_*`,
UNNEST(hits) AS hits,
UNNEST(hits.product) AS product
WHERE
_TABLE_SUFFIX BETWEEN '20160801' AND '20170801'
AND
product.v2ProductCategory = "Home/Electronics/"
GROUP BY date
ORDER BY date
)
SELECT
date,
AVG(transactions)
OVER (
ORDER BY date
ROWS BETWEEN CURRENT ROW AND 7 FOLLOWING
) AS avg_transactions
FROM
daily_electronics_transactions
"""
result = client.query(query).result().to_dataframe()
result["date"] = pd.to_datetime(result["date"])
result.plot(
y="avg_transactions",
x="date",
kind="line",
title="Electronics Weekly Moving Average",
)
query = """
WITH daily_office_transactions AS
(
SELECT
date,
SUM(totals.transactions) AS transactions
FROM
`bigquery-public-data.google_analytics_sample.ga_sessions_*`,
UNNEST(hits) AS hits,
UNNEST(hits.product) AS product
WHERE
_TABLE_SUFFIX BETWEEN '20160801' AND '20170801'
AND
product.v2ProductCategory = "Home/Office/"
GROUP BY date
ORDER BY date
)
SELECT
date,
AVG(transactions)
OVER (
ORDER BY date
ROWS BETWEEN CURRENT ROW AND 7 FOLLOWING
) AS avg_transactions
FROM
daily_office_transactions
"""
result = client.query(query).result().to_dataframe()
result["date"] = pd.to_datetime(result["date"])
result.plot(
y="avg_transactions", x="date", kind="line", title="Office Weekly Moving Average"
)
# Office and Electronics products seem to have steady demand year-round, Drinkware seems to see a spike in demand in December and March, and demand for Men's T-Shirts seem to increase in September, March, and August.
# # 3. Visualize Insights
# Most visited landing pages and their respective bounce rates:
query = """
SELECT
hits.page.pagePath AS landing_page,
COUNT(*) AS views,
SUM(totals.bounces)/COUNT(*) AS bounce_rate
FROM
`bigquery-public-data.google_analytics_sample.ga_sessions_*`,
UNNEST(hits) AS hits
WHERE
_TABLE_SUFFIX BETWEEN '20160801' AND '20170801'
AND
hits.type='PAGE'
AND
hits.hitNumber=1
GROUP BY landing_page
ORDER BY views DESC
LIMIT 10
"""
result = client.query(query).result().to_dataframe()
fig, ax = plt.subplots(figsize=(10, 6))
result.plot(
y=["bounce_rate"],
x="landing_page",
kind="barh",
legend=False,
title="Bounce Rates for Top 10 Landing Pages",
ax=ax,
)
ax.set_ylabel("")
plt.show()
# Browser and Device
query = """
SELECT
device.Browser AS browser,
COUNT(*) AS sessions,
SUM(totals.bounces)/COUNT(*) AS exit_rate
FROM
`bigquery-public-data.google_analytics_sample.ga_sessions_*`,
UNNEST(hits) AS hits
WHERE
_TABLE_SUFFIX BETWEEN '20160801' AND '20170801'
GROUP BY browser
ORDER BY sessions DESC
LIMIT 7
"""
result = client.query(query).result().to_dataframe()
fig, ax = plt.subplots(figsize=(12, 7))
result.plot(
y=["sessions", "exit_rate"],
x="browser",
kind="bar",
secondary_y="exit_rate",
ax=ax,
mark_right=False,
title="Sessions and Exit Rates by Browser",
)
# sns.barplot(data=result, x='browser', y='sessions')
# sns.barplot(data=result, x='browser', y='exit_rate')
ax.set_xticklabels(labels=result["browser"], rotation=45)
ax.set_xlabel("")
ax.legend(loc=(1.1, 0.55))
plt.legend(loc=(1.1, 0.5))
plt.show()
query = """
SELECT
device.deviceCategory AS device,
COUNT(*) AS sessions,
SUM(totals.bounces)/COUNT(*) AS exit_rate
FROM
`bigquery-public-data.google_analytics_sample.ga_sessions_*`,
UNNEST(hits) AS hits
WHERE
_TABLE_SUFFIX BETWEEN '20160801' AND '20170801'
GROUP BY device
ORDER BY sessions DESC
"""
result = client.query(query).result().to_dataframe()
fig, ax = plt.subplots(figsize=(10, 6))
result.plot(
y=["sessions", "exit_rate"],
x="device",
kind="bar",
title="Exit Rate by Device",
secondary_y="exit_rate",
ax=ax,
)
ax.set_xlabel("")
ax.set_xticklabels(labels=result["device"], rotation=45)
plt.show()
query = """
SELECT
trafficSource.medium AS medium,
COUNT(*) AS sessions,
SUM(totals.bounces)/COUNT(*) AS exit_rate,
SUM(totals.transactions) AS transactions,
SUM(totals.totalTransactionRevenue)/1000000 AS total_revenue,
SUM(totals.transactions)/COUNT(*) AS conversion_rate
FROM
`bigquery-public-data.google_analytics_sample.ga_sessions_*`,
UNNEST(hits) AS hits
WHERE
_TABLE_SUFFIX BETWEEN '20160801' AND '20170801'
GROUP BY medium
ORDER BY sessions DESC
LIMIT 10
"""
result = client.query(query).result().to_dataframe()
fig, ax = plt.subplots(figsize=(10, 7))
result.plot(
y=["total_revenue", "conversion_rate"],
x="medium",
kind="bar",
secondary_y="conversion_rate",
ax=ax,
)
ax.set_xticklabels(labels=result["medium"], rotation=45)
plt.show()
fig.show()
# Identify Best-Selling Product Categories and Forecast Demand
fig, ax = plt.subplots(figsize=(10, 6))
cat_result.plot(
y="total_revenue", x="category", kind="barh", title="Revenue by Category", ax=ax
)
ax.set_ylabel("")
plt.show()
fig, axes = plt.subplots(nrows=2, ncols=2, figsize=(20, 14))
# Men's T-shirts
query1 = """
WITH daily_mens_tshirt_transactions AS
(
SELECT
date,
SUM(totals.transactions) AS transactions
FROM
`bigquery-public-data.google_analytics_sample.ga_sessions_*`,
UNNEST(hits) AS hits,
UNNEST(hits.product) AS product
WHERE
_TABLE_SUFFIX BETWEEN '20160801' AND '20170801'
AND
product.v2ProductCategory = "Home/Apparel/Men's/Men's-T-Shirts/"
GROUP BY date
ORDER BY date
)
SELECT
date,
AVG(transactions)
OVER (
ORDER BY date
ROWS BETWEEN CURRENT ROW AND 7 FOLLOWING
) AS avg_transactions
FROM
daily_mens_tshirt_transactions
"""
result1 = client.query(query1).result().to_dataframe()
result1["date"] = pd.to_datetime(result1["date"])
ax1 = result1.plot(
y="avg_transactions",
x="date",
kind="line",
title="Men's T-Shirts Weekly Moving Average",
ax=axes[0, 0],
)
# Drinkware
query2 = """
WITH daily_drinkware_transactions AS
(
SELECT
date,
SUM(totals.transactions) AS transactions
FROM
`bigquery-public-data.google_analytics_sample.ga_sessions_*`,
UNNEST(hits) AS hits,
UNNEST(hits.product) AS product
WHERE
_TABLE_SUFFIX BETWEEN '20160801' AND '20170801'
AND
product.v2ProductCategory = "Home/Drinkware/Water Bottles and Tumblers/"
GROUP BY date
ORDER BY date
)
SELECT
date,
AVG(transactions)
OVER (
ORDER BY date
ROWS BETWEEN CURRENT ROW AND 7 FOLLOWING
) AS avg_transactions
FROM
daily_drinkware_transactions
"""
result2 = client.query(query2).result().to_dataframe()
result2["date"] = pd.to_datetime(result2["date"])
result2.plot(
y="avg_transactions",
x="date",
kind="line",
title="Drinkware Weekly Moving Average",
ax=axes[0, 1],
)
# Office Supplies
query3 = """
WITH daily_office_transactions AS
(
SELECT
date,
SUM(totals.transactions) AS transactions
FROM
`bigquery-public-data.google_analytics_sample.ga_sessions_*`,
UNNEST(hits) AS hits,
UNNEST(hits.product) AS product
WHERE
_TABLE_SUFFIX BETWEEN '20160801' AND '20170801'
AND
product.v2ProductCategory = "Home/Office/"
GROUP BY date
ORDER BY date
)
SELECT
date,
AVG(transactions)
OVER (
ORDER BY date
ROWS BETWEEN CURRENT ROW AND 7 FOLLOWING
) AS avg_transactions
FROM
daily_office_transactions
"""
result3 = client.query(query3).result().to_dataframe()
result3["date"] = pd.to_datetime(result3["date"])
result3.plot(
y="avg_transactions",
x="date",
kind="line",
title="Office Weekly Moving Average",
ax=axes[1, 0],
)
# Electronics
query4 = """
WITH daily_electronics_transactions AS
(
SELECT
date,
SUM(totals.transactions) AS transactions
FROM
`bigquery-public-data.google_analytics_sample.ga_sessions_*`,
UNNEST(hits) AS hits,
UNNEST(hits.product) AS product
WHERE
_TABLE_SUFFIX BETWEEN '20160801' AND '20170801'
AND
product.v2ProductCategory = "Home/Electronics/"
GROUP BY date
ORDER BY date
)
SELECT
date,
AVG(transactions)
OVER (
ORDER BY date
ROWS BETWEEN CURRENT ROW AND 7 FOLLOWING
) AS avg_transactions
FROM
daily_electronics_transactions
"""
result4 = client.query(query4).result().to_dataframe()
result4["date"] = pd.to_datetime(result4["date"])
result4.plot(
y="avg_transactions",
x="date",
kind="line",
title="Electronics Weekly Moving Average",
ax=axes[1, 1],
)
|
# ## İş Problemi
# Özellikleri belirtildiğinde kişilerin diyabet hastası olup olmadıklarını tahmin edebilecek bir makine öğrenmesi modeli geliştirilmesi istenmektedir. Modeli geliştirmeden önce gerekli olan veri analizi ve özellik mühendisliği adımlarını gerçekleştirmeniz beklenmektedir.
# ## Veri Seti Hikayesi
# Veri seti ABD'deki Ulusal Diyabet-Sindirim-Böbrek Hastalıkları Enstitüleri'nde tutulan büyük veri setinin parçasıdır. ABD'deki Arizona Eyaleti'nin en büyük 5. şehri olan Phoenix şehrinde yaşayan 21 yaş ve üzerinde olan Pima Indian kadınları üzerinde yapılan diyabet araştırması için kullanılan verilerdir. Hedef değişken "outcome" olarak belirtilmiş olup; 1 diyabet test sonucunun pozitif oluşunu, 0 ise negatif oluşunu belirtmektedir.
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.preprocessing import (
MinMaxScaler,
RobustScaler,
StandardScaler,
LabelEncoder,
)
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split
from sklearn.metrics import (
accuracy_score,
precision_score,
recall_score,
f1_score,
roc_auc_score,
)
pd.set_option("display.max_column", None)
pd.set_option("display.width", 500)
pd.set_option("display.float_format", lambda x: "%.2f" % x)
# ## Keşifçi Veri Analizi
#
df_ = pd.read_csv("/kaggle/input/diabetescsv/diabetes.csv")
df = df_.copy()
def check_df(dataframe, head=5):
print("###########Shape############")
print(dataframe.shape)
print("###########Dtypes###########")
print(dataframe.dtypes)
print("###########NA################")
print(dataframe.isnull().sum())
print("##########Describe###########")
print(dataframe.describe().T)
print("##########Head###############")
print(dataframe.head(head))
check_df(df)
# ## Numerik ve Kategorik Değişkenler
def grab_col_names(dataframe, cat_th=10, car_th=20):
cat_cols = [col for col in dataframe.columns if dataframe[col].dtype == "O"]
num_but_cat = [
col
for col in dataframe.columns
if dataframe[col].dtype != "O" and dataframe[col].nunique() < cat_th
]
cat_but_car = [
col
for col in dataframe.columns
if dataframe[col].dtype == "O" and dataframe[col].nunique() > car_th
]
cat_cols = cat_cols + num_but_cat
cat_cols = [col for col in cat_cols if col not in cat_but_car]
num_cols = [col for col in dataframe.columns if dataframe[col].dtype != "O"]
num_cols = [col for col in num_cols if col not in num_but_cat]
print(f"Observations: {dataframe.shape[0]}")
print(f"Variables: {dataframe.shape[1]}")
print(f"cat_cols: {len(cat_cols)}")
print(f"num_cols: {len(num_cols)}")
print(f"cat_but_car: {len(cat_but_car)}")
print(f"num_but_cat: {len(num_but_cat)}")
return cat_cols, num_cols, cat_but_car
cat_cols, num_cols, cat_but_car = grab_col_names(df)
def cat_summary(dataframe, col_name, plot=False):
print(
pd.DataFrame(
{
col_name: dataframe[col_name].value_counts(),
"Ratio": 100 * dataframe[col_name].value_counts() / len(dataframe),
}
)
)
if plot:
sns.countplot(x=dataframe[col_name], data=dataframe)
plt.show()
cat_summary(df, "Outcome")
def num_summary(dataframe, col_name, plot=False):
quantiles = [0.05, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, 0.95, 0.99]
print(dataframe[col_name].describe(quantiles).T)
if plot:
dataframe[col_name].hist(bins=20)
plt.xlabel(col_name)
plt.title(col_name)
plt.show()
for col in num_cols:
num_summary(df, col)
# ## Hedef Değişken Analizi
def target_summary_with_num(dataframe, target, col_name):
print(dataframe.groupby(target).agg({col_name: "mean"}), end="\n\n\n")
for col in num_cols:
target_summary_with_num(df, "Outcome", col)
# ## Aykırı Gözlem Analizi
def outlier_tresholds(dataframe, col, q1=0.25, q3=0.75):
qauntile1 = dataframe[col].quantile(q1)
quantile3 = dataframe[col].quantile(q3)
iqr = quantile3 - qauntile1
low_limit = qauntile1 - 1.5 * iqr
up_limit = quantile3 + 1.5 * iqr
return low_limit, up_limit
def check_outlier(dataframe, col_name):
low_limit, up_limit = outlier_tresholds(dataframe, col_name)
if dataframe[
(dataframe[col_name] < low_limit) | (dataframe[col_name] > up_limit)
].any(axis=None):
return True
else:
return False
for col in num_cols:
print(check_outlier(df, col))
# ## Eksik Değer Analizi
def missing_values_table(dataframe, na_name=False):
na_cols = [col for col in dataframe.columns if dataframe[col].isnull().sum() > 0]
n_miss = dataframe[na_cols].isnull().sum().sort_values(ascending=False)
ratio = (dataframe[na_cols].isnull().sum() / dataframe.shape[0] * 100).sort_values(
ascending=False
)
missing_df = pd.concat(
[n_miss, np.round(ratio, 2)], axis=1, keys=["n_miss", "ratio"]
)
print(missing_df, end="\n")
if na_name:
return na_cols
missing_values_table(df)
# ## Korelasyon Analizi
df.corr()
f, ax = plt.subplots(figsize=[18, 13])
sns.heatmap(df.corr(), annot=True, fmt=".2f", ax=ax)
ax.set_title("Correlation Matrix")
plt.show()
# ## Feature Engineering
# #### Eksik ve Aykırı Gözlemler
zero_cols = [
col
for col in df.columns
if df[col].min() == 0 and col not in ["Pregnancies", "Outcome"]
]
for col in zero_cols:
df[col] = np.where(df[col] == 0, np.nan, df[col])
na_columns = missing_values_table(df, na_name=True)
def missing_vs_target(dataframe, target, na_columns):
temp_df = dataframe.copy()
for col in na_columns:
temp_df[col + "_NA_FLAG"] = np.where(temp_df[col].isnull(), 1, 0)
na_flags = temp_df.loc[:, temp_df.columns.str.contains("_NA_")].columns
for col in na_flags:
print(
pd.DataFrame(
{
"TARGET_MEAN": temp_df.groupby(col)[target].mean(),
"Count": temp_df.groupby(col)[target].count(),
}
),
end="\n\n\n",
)
missing_vs_target(df, "Outcome", na_columns)
for col in zero_cols:
df.loc[df[col].isnull(), col] = df[col].median()
missing_values_table(df)
# ## Yeni Değişkenler Oluşturulması
df.loc[(df["Age"] >= 21) & (df["Age"] < 50), "NEW_AGE_CAT"] = "mature"
df.loc[(df["Age"] >= 50), "NEW_AGE_CAT"] = "senior"
df["NEW_BMI"] = pd.cut(
x=df["BMI"],
bins=[0, 18.5, 24.9, 29.9, 100],
labels=["Underweight", "Healthy", "Overweight", "Obese"],
)
df["NEW_GLUCOSE"] = pd.cut(
x=df["Glucose"],
bins=[0, 140, 200, 300],
labels=["Normal", "Prediabetes", "Diabetes"],
)
df.loc[
(df["BMI"] < 18.5) & ((df["Age"] >= 21) & (df["Age"] < 50)), "NEW_AGE_BMI_NOM"
] = "underweightmature"
df.loc[(df["BMI"] < 18.5) & (df["Age"] >= 50), "NEW_AGE_BMI_NOM"] = "underweightsenior"
df.loc[
((df["BMI"] >= 18.5) & (df["BMI"] < 25)) & ((df["Age"] >= 21) & (df["Age"] < 50)),
"NEW_AGE_BMI_NOM",
] = "healthymature"
df.loc[
((df["BMI"] >= 18.5) & (df["BMI"] < 25)) & (df["Age"] >= 50), "NEW_AGE_BMI_NOM"
] = "healthysenior"
df.loc[
((df["BMI"] >= 25) & (df["BMI"] < 30)) & ((df["Age"] >= 21) & (df["Age"] < 50)),
"NEW_AGE_BMI_NOM",
] = "overweightmature"
df.loc[
((df["BMI"] >= 25) & (df["BMI"] < 30)) & (df["Age"] >= 50), "NEW_AGE_BMI_NOM"
] = "overweightsenior"
df.loc[
(df["BMI"] > 18.5) & ((df["Age"] >= 21) & (df["Age"] < 50)), "NEW_AGE_BMI_NOM"
] = "obesemature"
df.loc[(df["BMI"] > 18.5) & (df["Age"] >= 50), "NEW_AGE_BMI_NOM"] = "obesesenior"
df.loc[
(df["Glucose"] < 70) & ((df["Age"] >= 21) & (df["Age"] < 50)), "NEW_AGE_GLUCOSE_NOM"
] = "lowmature"
df.loc[(df["Glucose"] < 70) & (df["Age"] >= 50), "NEW_AGE_GLUCOSE_NOM"] = "lowsenior"
df.loc[
((df["Glucose"] >= 70) & (df["Glucose"] < 100))
& ((df["Age"] >= 21) & (df["Age"] < 50)),
"NEW_AGE_GLUCOSE_NOM",
] = "normalmature"
df.loc[
((df["Glucose"] >= 70) & (df["Glucose"] < 100)) & (df["Age"] >= 50),
"NEW_AGE_GLUCOSE_NOM",
] = "normalsenior"
df.loc[
((df["Glucose"] >= 100) & (df["Glucose"] <= 125))
& ((df["Age"] >= 21) & (df["Age"] < 50)),
"NEW_AGE_GLUCOSE_NOM",
] = "hiddenmature"
df.loc[
((df["Glucose"] >= 100) & (df["Glucose"] <= 125)) & (df["Age"] >= 50),
"NEW_AGE_GLUCOSE_NOM",
] = "hiddensenior"
df.loc[
(df["Glucose"] > 125) & ((df["Age"] >= 21) & (df["Age"] < 50)),
"NEW_AGE_GLUCOSE_NOM",
] = "highmature"
df.loc[(df["Glucose"] > 125) & (df["Age"] >= 50), "NEW_AGE_GLUCOSE_NOM"] = "highsenior"
def set_insulin(dataframe, col_name="Insulin"):
if 16 <= dataframe[col_name] <= 166:
return "Normal"
else:
return "Abnormal"
df["NEW_INSULIN_SCORE"] = df.apply(set_insulin, axis=1)
df["NEW_GLUCOSE*INSULIN"] = df["Glucose"] * df["Insulin"]
df["NEW_GLUCOSE*PREGNANCIES"] = df["Glucose"] * df["Pregnancies"]
df.columns = [col.upper() for col in df.columns]
check_df(df)
# ## Encoding
cat_cols, num_cols, cat_but_car = grab_col_names(df)
def label_encoder(dataframe, binary_cols):
le = LabelEncoder()
dataframe[binary_cols] = le.fit_transform(dataframe[binary_cols])
return dataframe
binary_cols = [
col for col in df.columns if df[col].dtype == "O" and df[col].nunique() == 2
]
for col in binary_cols:
df = label_encoder(df, col)
def one_hot_encoder(dataframe, cat_cols, drop_first=True):
dataframe = pd.get_dummies(dataframe, columns=cat_cols, drop_first=drop_first)
return dataframe
cat_cols = [
col for col in cat_cols if col not in binary_cols and col not in ["OUTCOME"]
]
df = one_hot_encoder(df, cat_cols)
df.shape
# ## Standartlaştırma
ss = StandardScaler()
df[num_cols] = ss.fit_transform(df[num_cols])
df.head()
# ## Model Kurma
y = df["OUTCOME"]
X = df.drop("OUTCOME", axis=1)
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.30, random_state=17
)
rfc = RandomForestClassifier(random_state=46)
rfc.fit(X_train, y_train)
y_pred = rfc.predict(X_test)
print(f"Accuracy: {round(accuracy_score(y_pred, y_test), 2)}")
print(f"Recall: {round(recall_score(y_pred,y_test),3)}")
print(f"Precision: {round(precision_score(y_pred,y_test), 2)}")
print(f"F1: {round(f1_score(y_pred,y_test), 2)}")
print(f"Auc: {round(roc_auc_score(y_pred,y_test), 2)}")
# ## Özellik Önemi
def plot_importance(model, features, num=len(X), save=False):
feature_imp = pd.DataFrame(
{"Value": model.feature_importances_, "Feature": features.columns}
)
print(feature_imp.sort_values("Value", ascending=False))
plt.figure(figsize=(10, 20))
sns.set(font_scale=1)
sns.barplot(
x="Value",
y="Feature",
data=feature_imp.sort_values("Value", ascending=False)[0:num],
)
plt.title("Features")
plt.tight_layout()
plt.show()
if save:
plt.savefig("importances.png")
plot_importance(rfc, X)
|
import numpy as np
import pandas as pd
TRAIN_PATH = "/kaggle/input/titanic/train.csv"
ID = "PassengerId"
TARGET = "Survived"
train = pd.read_csv(TRAIN_PATH)
train.head()
# # get mean survival percent by [Sex,Pclass]
train.pivot_table(index=["Sex", "Pclass"], values="Survived").unstack()
train.groupby(["Sex", "Pclass"])["Survived"].mean().unstack()
train.groupby(["Sex", "Pclass"])["Survived"].aggregate("mean").unstack()
# # get total survival count by [Sex,Pclass]
train.groupby(["Sex", "Pclass"])["Survived"].sum().unstack()
train.groupby(["Sex", "Pclass"])["Survived"].aggregate("sum").unstack()
|
# # But what is semantic search?
# Semantic search is a technique for searching text data using meaning or context to match the search query with relevant results. Unlike traditional search algorithms that rely solely on keywords, semantic search uses Natural Language Processing (NLP) and Machine Learning (ML) to understand the intent behind the search query and the context of the text data. This results in more accurate and relevant search results compared to keyword-based search.
# In this notebook, we will explore the basics of semantic. If you want to experiment with semantic search hit the copy and edit button.
# *Upvote the notebook if you found it usefull ❤️!*
# # Importing Packages
import numpy as np
import pandas as pd
# # Installing datasets, evalute, transformers and faiss (Facebook AI Similarity Search).
# ## Loading IMDB Movies Dataset
df = pd.read_csv(
"/kaggle/input/imdb-dataset-of-top-1000-movies-and-tv-shows/imdb_top_1000.csv"
)
df.columns
# We only need Series_Title, Genre, Overview and Director for search purpose. Because in most of the cases we search movies by these features.
df = df[["Series_Title", "Genre", "Overview", "Director"]]
df.head()
# ## Converting pandas dataframe to Huggingface dataset.
# Because it is easy to use and we can use Huggingface tokenizers and models directly on huggingface dataset objects.
from datasets import Dataset
movie_dataset = Dataset.from_pandas(df)
movie_dataset
# Concatenating all the text field so that we can make a single embedding vector for all the relevant data.
#
def concatenate_text(data):
return {
"text": data["Series_Title"]
+ "\n"
+ data["Genre"]
+ "\n"
+ data["Overview"]
+ "\n"
+ data["Director"]
}
movie_dataset = movie_dataset.map(concatenate_text)
movie_dataset
# ### Result of concatenation
movie_dataset["text"][0]
# ## Importing Model and Tokenizer from HuggingFace
from transformers import AutoTokenizer, TFAutoModel
model_ckpt = "sentence-transformers/multi-qa-mpnet-base-dot-v1"
tokenizer = AutoTokenizer.from_pretrained(model_ckpt)
model = TFAutoModel.from_pretrained(model_ckpt, from_pt=True)
# > We need a single vector for our data so we need to average our token embeddings.One popular approach is to perform CLS pooling on our model’s outputs, where we simply collect the last hidden state for the special [CLS] token. The following function does the trick for us:
def cls_pooling(model_output):
return model_output.last_hidden_state[:, 0]
def get_embeddings(text_list):
encoded_input = tokenizer(
text_list, padding=True, truncation=True, return_tensors="tf"
)
encoded_input = {k: v for k, v in encoded_input.items()}
model_output = model(**encoded_input)
return cls_pooling(model_output)
# ## Debugging the Output
# embedding = get_embeddings(movie_dataset['text'][0])
# embedding
# # Now let's apply the function to the whole dataset.
# ### This will take some time so be patient 🙃.
embeddings_dataset = movie_dataset.map(
lambda x: {"embeddings": get_embeddings(x["text"]).numpy()[0]}
)
# Debugging
embeddings_dataset
# Uncomment to debug the output
# embeddings_dataset['embeddings'][0]
# # Using FAISS for efficient similarity search
embeddings_dataset.add_faiss_index(column="embeddings")
# # Testing
question = "what is Batman?"
question_embedding = get_embeddings([question]).numpy()
question_embedding.shape
scores, samples = embeddings_dataset.get_nearest_examples(
"embeddings", question_embedding, k=5
)
samples_df = pd.DataFrame.from_dict(samples)
samples_df["scores"] = scores
samples_df.sort_values("scores", ascending=False, inplace=True)
samples_df
# # Results
for _, row in samples_df.iterrows():
print(f"Series Title: {row.Series_Title}")
print(f"Overview: {row.Overview}")
print(f"Genre: {row.Genre}")
print(f"Scores: {row.scores}")
print("=" * 50)
print()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
x = 10
print(type(x))
x = 6 # int
y = 9.9 # float
z = 9j # complex
print(type(x))
print(type(y))
print(type(z))
x = 88
y = 8394477568343433224342234243234
z = -54343424532245324
print(type(x))
print(type(y))
print(type(z))
x = 324.32
y = 9878.443
z = -232332.098789
print(type(x))
print(type(y))
print(type(z))
x = 213123e432
y = 123123e4
z = -2312.7e100
print(type(x))
print(type(y))
print(type(z))
x = 75 + 23j
y = 7j
z = -8j
print(type(x))
print(type(y))
x = 88
y = 123.32
z = 98j
a = float(x)
b = int(y)
c = complex(x)
print(a)
print(b)
print(c)
print(type(a))
print(type(b))
print(type(c))
import random
print(random.randrange(4000, 20000))
x = int(67) # x will be 1
y = int(4.5) # y will be 2
z = int("7") # z will be 3
print(x)
print(y)
print(z)
x = float(6)
y = float(6.1)
z = float("8")
w = float("8.8")
print(x)
print(y)
print(z)
print(w)
x = str("dodo1") # x will be 's1'
y = str(1) # y will be '2'
z = str(6.1) # z will be '3.0'
print(x)
print(y)
print(z)
print("Ciao")
print("Ciao")
a = "Ciao"
print(a)
a = """Sabah kalktım, kahvaltı ettim,sporuma gittim ve kendimi çok iyi hissediyorum."""
print(a)
a = "Bu gün günlerden Trabzon,en büyük çimbom başka büyük yok,Konsantrasyon ." ""
print(a)
a = "Ciao!"
print(a[1])
for x in "Trabzon":
print(x)
a = "Ciao!"
print(len(a))
txt = "We are tedted to show our strengths not our weaknesses!"
print("weaknesses" in txt)
txt = "We are tedted to show our strengths not our weaknesses!"
if "weaknesses" in txt:
print("Yes, 'weaknesses' is present.")
txt = "We are tedted to show our strengths not our weaknesses!"
print("beatiful" not in txt)
txt = "We are tedted to show our strengths not our weaknesses!"
if "beatiful" not in txt:
print("No, 'beatiful' is NOT present.")
|
from fastai.vision.all import *
from pathlib import Path
import h5py
from matplotlib import cm
import matplotlib.pyplot as plt
import os
kaggle = os.environ.get("KAGGLE_KERNEL_RUN_TYPE", "")
path = "/kaggle/input/jet-images-train-val-test/jet-images_train.hdf5"
classes = ["general", "W-boson"]
h5_file = h5py.File(path, "r")
signal_data = h5_file["signal"]
image_data = h5_file["image"]
signal_array = np.array(signal_data)
image_array = np.array(image_data)
len(image_array)
# Filter the image_array based on signal_array
filtered_images = image_array[signal_array == 1]
# Calculate the mean image
mean_image = np.mean(filtered_images, axis=0)
len(filtered_images)
plt.imshow(mean_image)
plt.show()
plt.imshow(filtered_images[7])
plt.show()
filtered_images_general = image_array[signal_array == 0]
mean_image_general = np.mean(filtered_images_general, axis=0)
plt.imshow(mean_image_general)
plt.show()
plt.imshow(filtered_images_general[22])
plt.show()
mean_vector = np.mean(image_array[signal_array == 1], axis=0)
predicted_labels = np.zeros(len(image_array))
for i in range(len(image_array)):
distance = np.linalg.norm(image_array[i] - mean_vector)
if distance < 5:
predicted_labels[i] = 1
accuracy = np.mean(predicted_labels == signal_array)
print(f"Accuracy: {accuracy}")
predicted_labels
from sklearn.metrics.pairwise import cosine_similarity
from sklearn.metrics import accuracy_score
def mean_image_predict(threshold):
mean_image = np.mean(image_array[signal_array == 1], axis=0)
similarities = np.zeros(len(image_array))
for i, image_vec in enumerate(image_array):
similarities[i] = cosine_similarity(
image_vec.reshape(1, -1), mean_image.reshape(1, -1)
)
threshold = threshold
predicted_labels = np.where(similarities > threshold, 1, 0)
accuracy = accuracy_score(signal_array, predicted_labels)
return accuracy
from sklearn.metrics import accuracy_score
# Compute the accuracy score
accuracy = accuracy_score(signal_array, predicted_labels)
print(f"Accuracy: {accuracy}")
test_num = [1.0, 0.9, 0.8, 0.7, 0.6, 0.5, 0.4, 0, 0.3, 0.2, 0.1]
for i in test_num:
test = mean_image_predict(i)
print(f"Whit a treshold of {i} we have a accuracy: {test[0]}")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
pass
# print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pickle
import random
import warnings
from pathlib import Path
import numpy as np
from skimage import io
from tqdm import tqdm, tqdm_notebook
from PIL import Image
import torch
import torchvision
from torchvision import transforms
from multiprocessing.pool import ThreadPool
from sklearn.preprocessing import LabelEncoder
from torch.utils.data import Dataset, DataLoader
import torch.nn as nn
from matplotlib import colors, pyplot as plt
# в sklearn не все гладко, чтобы в colab удобно выводить картинки
# мы будем игнорировать warnings
warnings.filterwarnings(action="ignore", category=DeprecationWarning)
SEED = 1937
random.seed(SEED)
np.random.seed(SEED)
torch.manual_seed(SEED)
torch.cuda.manual_seed(SEED)
torch.backends.cudnn.deterministic = True
# работаем на видеокарте
DEVICE = torch.device("cuda")
# пути до тестовой и обучающей выборок
TRAIN_DIR = Path("/kaggle/input/journey-springfield/train/simpsons_dataset")
TEST_DIR = Path("/kaggle/input/journey-springfield/testset/testset")
# масштаб изображений
RESCALE_SIZE = 224
# Датасет для работы с данными
class SimpsonsDataset(Dataset):
"""
Датасет с картинками, который паралельно подгружает их из папок
производит скалирование и превращение в торчевые тензоры
"""
def __init__(self, files, mode, transforms, rescale_size=224):
super().__init__()
# список файлов для загрузки
self.files = sorted(files)
# режим работы
self.mode = mode
self.rescale_size = rescale_size
self.transforms = transforms
available_modes = ["train", "val", "test"]
if self.mode not in available_modes:
print(f"{self.mode} is not correct; correct modes: {available_modes}")
raise NameError
self.len_ = len(self.files)
self.label_encoder = LabelEncoder()
if self.mode != "test":
self.labels = [path.parent.name for path in self.files]
self.label_encoder.fit(self.labels)
with open("label_encoder.pkl", "wb") as le_dump_file:
pickle.dump(self.label_encoder, le_dump_file)
def __len__(self):
return self.len_
def load_sample(self, file):
image = Image.open(file)
image.load()
return image
def __getitem__(self, index):
x = self.load_sample(self.files[index])
x = self.transforms(x)
if self.mode == "test":
return x
else:
label = self.labels[index]
label_id = self.label_encoder.transform([label])
y = label_id.item()
return x, y
def _prepare_sample(self, image):
image = image.resize((self.rescale_size, self.rescale_size))
return np.array(image)
# Создаем датасеты
from sklearn.model_selection import train_test_split
train_val_files = sorted(list(TRAIN_DIR.rglob("*.jpg")))
test_files = sorted(list(TEST_DIR.rglob("*.jpg")))
train_val_labels = [path.parent.name for path in train_val_files]
train_files, val_files = train_test_split(
train_val_files, test_size=0.25, stratify=train_val_labels
)
# Примеры картинок и лэйблов
def imshow(inp, title=None, plt_ax=plt, default=False):
"""Imshow для тензоров"""
inp = inp.numpy().transpose((1, 2, 0))
mean = np.array([0.485, 0.456, 0.406])
std = np.array([0.229, 0.224, 0.225])
inp = std * inp + mean
inp = np.clip(inp, 0, 1)
plt_ax.imshow(inp)
if title is not None:
plt_ax.set_title(title)
plt_ax.grid(False)
fig, ax = plt.subplots(nrows=3, ncols=3, figsize=(8, 8), sharey=True, sharex=True)
for fig_x in ax.flatten():
random_characters = int(np.random.uniform(0, 1000))
im_val, label = val_dataset[random_characters]
img_label = " ".join(
map(
lambda x: x.capitalize(),
val_dataset.label_encoder.inverse_transform([label])[0].split("_"),
)
)
imshow(im_val.data.cpu(), title=img_label, plt_ax=fig_x)
# Создание модели
# За основу возьмем предобученную ResNet34
from torchvision.models import resnet34, ResNet34_Weights
weights = ResNet34_Weights.DEFAULT
model = resnet34(weights=weights)
print(model)
# Изменим выходной слой для получения нужного числа классов
n_classes = len(np.unique(train_val_labels))
model.fc = torch.nn.Linear(512, n_classes)
# Заморозим все слои, кроме 3-х последних
for parameter in model.parameters():
parameter.requires_grad = False
layers_to_unfreeze = ["fc", "avgpool", "layer4"]
for layer_to_unfreeze in layers_to_unfreeze:
layer = getattr(model, layer_to_unfreeze)
for parameter in layer.parameters():
parameter.requires_grad = True
# Обучение
from sklearn.metrics import f1_score
def calculate_f1_score(labels: torch.Tensor, preds: torch.Tensor) -> float:
labels = labels.cpu().numpy()
preds = preds.cpu().numpy()
return f1_score(labels, preds, average="micro")
def fit_epoch(model, train_loader, criterion, optimizer):
model.train()
running_loss = 0.0
running_f1_score = 0
batch_count = 0
pbar = tqdm(train_loader)
for inputs, labels in pbar:
inputs = inputs.to(DEVICE)
labels = labels.to(DEVICE)
optimizer.zero_grad()
outputs = model(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
preds = torch.argmax(outputs, 1)
running_loss += loss.item()
running_f1_score += calculate_f1_score(preds, labels)
batch_count += 1
train_loss = running_loss / batch_count
train_f1_score = running_f1_score / batch_count
return train_loss, train_f1_score
def eval_epoch(model, val_loader, criterion):
model.eval()
running_loss = 0.0
running_f1_score = 0
batch_count = 0
pbar = tqdm(val_loader)
for inputs, labels in pbar:
inputs = inputs.to(DEVICE)
labels = labels.to(DEVICE)
with torch.set_grad_enabled(False):
outputs = model(inputs)
loss = criterion(outputs, labels)
preds = torch.argmax(outputs, 1)
running_loss += loss.item()
running_f1_score += calculate_f1_score(preds, labels)
batch_count += 1
val_loss = running_loss / batch_count
vall_f1_score = running_f1_score / batch_count
return val_loss, vall_f1_score
def save_model(model):
torch.save(model.state_dict(), "best.pth")
epochs = 20
model = model.to(DEVICE)
optimizer = torch.optim.Adam(model.parameters(), lr=1e-4)
loss_function = torch.nn.CrossEntropyLoss()
scheduler = torch.optim.lr_scheduler.StepLR(optimizer, 10, gamma=0.1)
preprocess = weights.transforms()
train_dataset = SimpsonsDataset(train_files, mode="train", transforms=preprocess)
val_dataset = SimpsonsDataset(val_files, mode="val", transforms=preprocess)
train_dataloader = DataLoader(train_dataset, batch_size=128, shuffle=True)
val_dataloader = DataLoader(val_dataset, batch_size=128)
best_f1_score = 0
train_history = []
val_history = []
for epoch in range(1, epochs + 1):
print("Epoch {}:".format(epoch))
train_loss, train_f1 = fit_epoch(model, train_dataloader, loss_function, optimizer)
train_history.append((train_loss, train_f1))
print("Train. Loss: {}, F1-score: {}".format(train_loss, train_f1))
with torch.no_grad():
val_loss, val_f1 = eval_epoch(model, val_dataloader, loss_function)
print("Validation. Loss: {}, F1-score: {}".format(val_loss, val_f1))
val_history.append((val_loss, val_f1))
if val_f1 > best_f1_score:
print("Is best result. Save model.")
save_model(model)
scheduler.step()
# График функции потерь
train_loss = [_[0] for _ in train_history]
val_loss = [_[0] for _ in val_history]
plt.plot(train_loss, label="Train loss")
plt.plot(val_loss, label="Validation loss")
plt.legend()
plt.xlabel("epochs")
plt.ylabel("loss")
plt.show()
# График F1-score
train_f1 = [_[1] for _ in train_history]
val_f1 = [_[1] for _ in val_history]
plt.plot(train_f1, label="Train F1-score")
plt.plot(val_f1, label="Validation F1-score")
plt.legend()
plt.xlabel("epochs")
plt.ylabel("F1-score")
plt.show()
# Сабмит
import pandas as pd
def load_best_weights(model):
model.load_state_dict(torch.load("best.pth"))
def predict(model, test_loader):
with torch.no_grad():
logits = []
for inputs in test_loader:
inputs = inputs.to(DEVICE)
model.eval()
outputs = model(inputs).cpu()
logits.append(outputs)
probs = nn.functional.softmax(torch.cat(logits), dim=-1).numpy()
return probs
def create_submission(preds, files):
submit = pd.DataFrame(columns=["Id"])
submit["Id"] = files
submit["Expected"] = preds
submit.to_csv("submission.csv", index=False)
load_best_weights(model)
model.eval()
label_encoder = pickle.load(open("label_encoder.pkl", "rb"))
test_dataset = SimpsonsDataset(test_files, mode="test", transforms=preprocess)
test_loader = DataLoader(test_dataset, shuffle=False, batch_size=64)
probs = predict(model, test_loader)
preds = label_encoder.inverse_transform(np.argmax(probs, axis=1))
test_filenames = [path.name for path in test_dataset.files]
create_submission(preds, test_filenames)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# how are you
# how you are
# are you how ?
from fuzzywuzzy import fuzz
from fuzzywuzzy import process
choices = [
"This is data science lecture",
"how far is mumbai to delhi",
"distance of mumbai to delhi",
"data science",
]
process.extract("data science lecture This is", choices, limit=2)
process.extractOne("new york jets", choices)
import spacy
nlp = spacy.load("en_core_web_sm")
sentence = "i want to eat apple"
doc = nlp(sentence)
for ent in doc.ents:
print(ent.text, ent.start_char, ent.end_char, ent.label_)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import locale
def getpreferredencoding(do_setlocale=True):
return "UTF-8"
locale.getpreferredencoding = getpreferredencoding
import pandas as pd
import re
import emoji
import numpy as np
from emoji_translate.emoji_translate import Translator
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.neighbors import KNeighborsClassifier
from sklearn.model_selection import train_test_split
from string import punctuation
from sklearn.metrics import classification_report
import pickle5
import torch.nn as nn
import torch
import torch.optim as optim
import pytorch_lightning as pl
emo = Translator(exact_match_only=False, randomize=True)
stop_ward = ward.split(",")
word_to_index = {"<PAD>": 0, "<UNK>": 1}
SEQ_LEN = 25
def pre_process(line):
# for j in range(len(listx)):
# line = re.sub(r"\p{S}+", "", line)
delimiter = " "
tokens = line.split()
filtered_tokens = [token for token in tokens if token not in stop_ward]
# Join the tokens back into a string
processed_text = " ".join(filtered_tokens)
processed_text = "".join([x for x in processed_text if x not in punctuation])
processed_text = "".join(
(" " + c + " ") if c in emoji.UNICODE_EMOJI["en"] else c for c in processed_text
)
# processed_text = emoji.emojize(processed_text,delimiters=(delimiter,delimiter))
# listx[j] = processed_text
processed_text = emo.demojify(processed_text)
# processed_text.split(delimiter)
return processed_text
df = pd.read_csv("/kaggle/input/design-lab-train/hindi_train_val.csv")
df["text"] = df["text"].apply(pre_process)
y = df["label"]
X = df["text"]
vectorizer = TfidfVectorizer()
# x_train, x_val, y_train, y_val = train_test_split(X,y, test_size=0.1, random_state=42)
x_train = vectorizer.fit_transform(
X.values
) # you can perform the buil_preprocessor() to perform the preprocessing
# x_val = vectorizer.transform(x_val.values)
neigh = KNeighborsClassifier(n_neighbors=19)
neigh.fit(x_train, y)
# print(f" the best score : {best_score} is with the k : {best_k}")
pickle5.dump(neigh, open("knn.pkl", "wb"))
# # TESTING
df_test = pd.read_csv("/kaggle/input/design-lab-test/hindi_test.csv")
df_train = pd.read_csv("/kaggle/input/design-lab-train/hindi_train_val.csv")
df_test.merge(df_train, on="text")
df_test["text"] = df_test["text"].apply(pre_process)
y_test = df_test["label"]
X_test = df_test["text"]
vectorizer = TfidfVectorizer()
df_train["text"] = df_train["text"].apply(pre_process)
y_train = df_train["label"]
X_train = df_train["text"]
Fit = vectorizer.fit(X_train.values)
X_test = Fit.transform(X_test.values)
knn = pickle5.load(open("/kaggle/working/knn.pkl", "rb"))
y_pred = knn.predict(X_test)
from sklearn.metrics import f1_score, accuracy_score
print(
f"the macro f1_score : {f1_score(y_test,y_pred)} and acc_score : {accuracy_score(y_test,y_pred)} "
)
df_test = pd.read_csv("/kaggle/input/design-lab-test/hindi_test.csv")
df_train = pd.read_csv("/kaggle/input/design-lab-train/hindi_train_val.csv")
df_test.merge(df_train, on="text")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
if filename.endswith("jpg"):
continue
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
BASE_PATH = (
"/kaggle/input/multi-label-classification-competition-2023/COMP5329S1A2Dataset"
)
import re
import re
def read_csv(path, n_columns=2):
data = []
with open(path, "r") as f:
for line in f.readlines():
if not re.match("^\d+\.jpg", line):
continue
ImageID = line.split(",")[0]
if n_columns == 2:
Labels = line.split(",")[1]
data.append({"ImageID": ImageID, "Labels": Labels})
else:
data.append({"ImageID": ImageID})
return pd.DataFrame(data)
train_df = read_csv(f"{BASE_PATH}/train.csv")
train_df
test_df = read_csv(f"{BASE_PATH}/test.csv", 1)
test_df.head(4)
import random
def rand_label():
num = random.randrange(1, 20)
while num == 12:
num = random.randrange(1, 20)
return num
import random
random.seed(10)
data = []
for index, row in enumerate(test_df.itertuples()):
n_labels = random.randrange(1, 3)
labels = []
for ix in range(n_labels):
labels.append(rand_label())
labels.sort()
data.append(
{
"ImageID": f"{index}.jpg",
"Labels": "_".join([str(label) for label in labels]),
}
)
pd.DataFrame(data).to_csv("submission.csv", index=False)
with open("submission.csv", "w") as f:
f.write("\n".join(data))
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import sklearn
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
titanic_df = pd.read_csv("/kaggle/input/titanic-survival-master/titanic.csv")
titanic_df.head()
titanic_df.shape
titanic_df.drop(["PassengerId", "Name", "Ticket", "Cabin"], "columns", inplace=True)
titanic_df.head()
titanic_df.isnull().any()
titanic_df[titanic_df.isnull().any(axis=1)].count()
titanic_df = titanic_df.dropna()
titanic_df[titanic_df.isnull().any(axis=1)].count()
titanic_df.describe()
fig, ax = plt.subplots(figsize=(12, 8))
plt.scatter(titanic_df["Age"], titanic_df["Survived"])
plt.xlabel("Age")
plt.ylabel("Survived")
fig, ax = plt.subplots(figsize=(12, 8))
plt.scatter(titanic_df["Fare"], titanic_df["Survived"])
plt.xlabel("Fare")
plt.ylabel("Survived")
pd.crosstab(titanic_df["Sex"], titanic_df["Survived"])
pd.crosstab(titanic_df["Pclass"], titanic_df["Survived"])
titanic_data_corr = titanic_df.corr()
titanic_data_corr
fig, ax = plt.subplots(figsize=(12, 10))
sns.heatmap(titanic_data_corr, annot=True)
# ## If a categorical (non-number) variable/feature has some orders like Male-Female, t-shirt sizes like S,M,L,XL than we can use labelEncoder
from sklearn import preprocessing
label_encoding = preprocessing.LabelEncoder()
titanic_df["Sex"] = label_encoding.fit_transform(titanic_df["Sex"].astype(str))
titanic_df.head()
# ## In classes_ values that which come first is 0, the next is 1 and so on. Hence Female is 0 and male is 1
label_encoding.classes_
# ## Since Emabrked is string datatpe and and has no order since this contains city names so we should use pd.get_dummies function to make them 0's and 1's
titanic_df = pd.get_dummies(titanic_df, columns=["Embarked"])
titanic_df.head()
# ## We will shuffle the data with df.sample(frac=1) function. Here "frac=1" means it will consider all the data in the dataframe. After that we will reset the index so that original index will appear as column. Then we will drop the original index column by passing paramieter drop=True in reset_index method.
# ## Shuffle is done so as to remove any inherent pattern in original data.
titanic_df = titanic_df.sample(frac=1).reset_index(drop=True)
titanic_df.head()
titanic_df.shape
from sklearn.model_selection import train_test_split
X = titanic_df.drop("Survived", axis=1)
Y = titanic_df["Survived"]
x_train, x_test, y_train, y_test = train_test_split(X, Y, test_size=0.2)
x_train.shape, y_train.shape
# ## "penalty" parameter is used to regularize the model if it becomes overly complex. "C" value determines the 'Inverse of Regularization strength'. Smaller value indicates stronger regularization.Lastly "solver" value determines the optimization algorithm. 'libliner' works better with smaller data set as with here.So it is used here.
from sklearn.linear_model import LogisticRegression
logistic_model = LogisticRegression(penalty="l2", C=1.0, solver="liblinear").fit(
x_train, y_train
)
y_pred = logistic_model.predict(x_test)
pred_results = pd.DataFrame({"y_test": y_test, "y_pred": y_pred})
pred_results
titanic_crosstab = pd.crosstab(pred_results.y_pred, pred_results.y_test)
titanic_crosstab
from sklearn.metrics import accuracy_score
from sklearn.metrics import precision_score
from sklearn.metrics import recall_score
# ## Accuracy shows: how many of the predicted value did the model get right ?
# ## Precision shows: How many of the passengers our model thought survived actually did survive ?
# ## Recall shows : How many of the actual survivors did the model correctly predict?
acc = accuracy_score(y_test, y_pred)
prec = precision_score(y_test, y_pred)
recall = recall_score(y_test, y_pred)
print("Accuracy score:", acc)
print("Precision score:", prec) # Very less False-Positive exists ,so it's good
print("Recall value:", recall) # A little high False-Negative
TP = titanic_crosstab[1][1]
TN = titanic_crosstab[0][0]
FP = titanic_crosstab[0][1]
FN = titanic_crosstab[1][0]
TP, TN, FP, FN
titanic_df.head()
titanic_df.to_csv("/kaggle/working/titanic_processed.csv")
|
# # **This is the practice - Power BI: Displaying Live System Performance using Power BI, SQL and Python**
# **Previously, create the Performance table (this is an empty table).
# this table has fields like Time, cpu_usage, memory_usage, cpu_interrupts, cpu_calls, memory_used, memory_free, bytes_sent, bytes_received, and disk_usage. By the following Python code, I try to input data into this table, Performance.**
# **First, the code imports some Python libraries:**
import psutil
import time
import pyodbc
# * psutil is a library for retrieving system information, such as CPU usage, memory usage, and disk usage.
# * time is a library for working with time and timing functions.
# * pyodbc is a library for connecting to Microsoft SQL Server databases using Python.
# **Next, the code establishes a connection to a SQL Server database:**
con = pyodbc.connect(
"Driver={SQL Server};"
"Server=DESKTOP-3Q7QOV2\SQLEXPRESS;"
"Database=System_Information;"
"Trusted_Connection=yes;"
)
# * This code creates a connection object (con) that connects to a SQL Server database.
# * The Driver={SQL Server} parameter specifies that the connection should use the SQL Server driver.
# * The Server parameter specifies the name of the SQL Server instance to connect to. In this case, it's 'DESKTOP-3Q7QOV2\SQLEXPRESS'.
# * The Database parameter specifies the name of the database to connect to. In this case, it's 'System_Information'.
# * The Trusted_Connection=yes parameter specifies that the connection should use Windows authentication.
# **After connecting to the database, the code enters an infinite loop using a while loop:**
while 1 == 1:
# This code creates a loop that will run indefinitely.
# **Within the loop, the code retrieves system information using the psutil library:**
cpu_usage = psutil.cpu_percent()
memory_usage = psutil.virtual_memory()[2]
cpu_interrupts = psutil.cpu_stats()[1]
cpu_calls = psutil.cpu_stats()[3]
memory_used = psutil.virtual_memory()[3]
memory_free = psutil.virtual_memory()[4]
bytes_sent = psutil.net_io_counters()[0]
bytes_received = psutil.net_io_counters()[1]
disk_usage = psutil.disk_usage("/")[3]
# These lines of code retrieve various system information, such as CPU usage, memory usage, network traffic, and disk usage.
# **Next, the code executes an SQL query to insert the system information into a database table:**
cursor.execute(
"insert into Performance values (GETDATE(),"
+ str(cpu_usage)
+ ","
+ str(memory_usage)
+ ","
+ str(cpu_interrupts)
+ ","
+ str(cpu_calls)
+ ","
+ str(memory_used)
+ ","
+ str(memory_free)
+ ","
+ str(bytes_sent)
+ ","
+ str(bytes_received)
+ ","
+ str(disk_usage)
+ ")"
)
# * This code uses the cursor object to execute an SQL query that inserts the system information into a database table named "Performance".
# * The GETDATE() function retrieves the current date and time.
# * The various system information variables are inserted into the table using string concatenation.
# **Finally, the code commits the transaction and sleeps for one second before looping again:**
con.commit()
print(memory_usage)
time.sleep(1)
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import xgboost as xgb
from sklearn.model_selection import cross_val_score, TimeSeriesSplit, KFold
from sklearn.metrics import mean_squared_error
from sklearn.model_selection import KFold
import lightgbm as lgb
import catboost as cb
# # Load Data
train_raw = pd.read_csv(
"/kaggle/input/yasmines-meteorological-mystery-dsc-psut-comp/train_data.csv"
)
test_raw = pd.read_csv(
"/kaggle/input/yasmines-meteorological-mystery-dsc-psut-comp/test_data.csv"
)
train_raw.isnull().sum()
test_raw.isnull().sum()
target_min = "min_feels_like"
target_max = "max_feels_like"
targets = [target_min, target_max]
# # Cross Validation
xgb_params = {
"seed": 2,
"objective": "reg:squarederror",
"max_depth": 6,
"eta": 0.09,
"subsample": 0.65,
"colsample_bytree": 1,
}
def plot_importances(model):
importances = model.feature_importances_
sorted_indices = importances.argsort()[::-1]
feature_names = X_train.columns[sorted_indices]
plt.figure(figsize=(10, 5))
plt.barh(range(X_train.shape[1]), importances[sorted_indices])
plt.yticks(range(X_train.shape[1]), feature_names)
plt.ylabel("Feature")
plt.xlabel("Importance")
plt.title("Feature Importances")
plt.show()
# ## min_feels_like
def create_features_min(df):
df["date"] = pd.to_datetime(df["date"])
df["humidity_dew_mul"] = df["humidity"] * df["dew"]
df["humidity_pressure_mul"] = df["humidity"] * df["pressure"]
df["humidity_dew_diff"] = df["humidity"] - df["dew"]
df["humidity_dew_div"] = df["dew"] / df["humidity"]
df["month"] = df["date"].dt.month
df["season"] = (df["month"] % 12 + 3) // 3
df["season_group_mean_humidity"] = df.groupby("season")["humidity"].transform(
"mean"
)
df["year"] = df["date"].dt.year
df["year_group_mean_humidity"] = df.groupby("year")["humidity"].transform("mean")
df["year_group_mean_dew"] = df.groupby("year")["dew"].transform("mean")
df["year_group_mean_pressure"] = df.groupby("year")["pressure"].transform("mean")
df["pressure_cloudcover_diff"] = df["pressure"] - df["cloudcover"] # min
# df['col3'] = df['humidity'] - df['humidity'].shift(7) #min
# df['col3'] = df['humidity'] - df['humidity'].shift(28) #min
# df['col'] = df['humidity'] - df['season_group_mean_humidity'] #min
return df
def handle_missing_min(df):
df = df.dropna().reset_index(drop=True)
return df
def drop_features_min(df):
cols_drop = ["sunrise", "sunset", "date"]
df = df.drop(cols_drop, axis=1)
return df
def feature_engineering_min(train_raw, test_raw):
train = handle_missing_min(train_raw)
test = handle_missing_min(test_raw)
train = create_features_min(train)
test = create_features_min(test)
train = drop_features_min(train)
test = drop_features_min(test)
X = train.drop(targets, axis=1)
y_min = train[target_min]
y_max = train[target_max]
return X, y_min, y_max, test
X_min, y_min_min, y_max_min, test_min = feature_engineering_min(
train_raw.copy(), test_raw.copy()
)
n_splits = 5
# tscv = TimeSeriesSplit(n_splits=n_splits)
kf = KFold(n_splits=n_splits, shuffle=True, random_state=42)
min_scores = []
for fold, (train_index, test_index) in enumerate(kf.split(X_min)):
X_train, X_val = X_min.iloc[train_index], X_min.iloc[test_index]
y_train_min, y_val_min = y_min_min[train_index], y_min_min[test_index]
y_train_max, y_val_max = y_max_min[train_index], y_max_min[test_index]
xgb_reg_min = xgb.XGBRegressor(**xgb_params)
xgb_reg_min.fit(X_train, y_train_min)
y_pred_min = xgb_reg_min.predict(X_val)
min_mse = mean_squared_error(y_val_min, y_pred_min)
min_scores.append(min_mse)
print(f"Fold {fold+1} | min MSE: {min_mse:.4f}")
print(f"{n_splits} Folds | Average min MSE: {np.average(min_scores)}")
# ### Predict test
xgb_reg_min = xgb.XGBRegressor(**xgb_params)
xgb_reg_min.fit(X_min, y_min_min)
xgb_y_test_min = xgb_reg_min.predict(test_min)
plot_importances(xgb_reg_min)
# ## max_feels_like
def create_features_max(df):
df["date"] = pd.to_datetime(df["date"])
df["humidity_dew_mul"] = df["humidity"] * df["dew"]
df["humidity_pressure_mul"] = df["humidity"] * df["pressure"]
df["humidity_dew_diff"] = df["humidity"] - df["dew"]
df["humidity_dew_div"] = df["dew"] / df["humidity"]
df["month"] = df["date"].dt.month
df["season"] = (df["month"] % 12 + 3) // 3
df["season_group_mean_humidity"] = df.groupby("season")["humidity"].transform(
"mean"
)
df["year"] = df["date"].dt.year
df["year_group_mean_humidity"] = df.groupby("year")["humidity"].transform("mean")
df["year_group_mean_dew"] = df.groupby("year")["dew"].transform("mean")
df["year_group_mean_pressure"] = df.groupby("year")["pressure"].transform("mean")
df["col"] = (df["precipcover"] + df["cloudcover"]) / 2 # max
# df['col2'] = df['pressure'] - df['cloudcover'] #min
# df['col2'] = df['pressure'] - df['visibility'] #max
return df
def handle_missing_max(df):
df = df.dropna().reset_index(drop=True)
return df
def drop_features_max(df):
cols_drop = ["sunrise", "sunset", "date"]
df = df.drop(cols_drop, axis=1)
return df
def feature_engineering_max(train_raw, test_raw):
train = handle_missing_max(train_raw)
test = handle_missing_max(test_raw)
train = create_features_max(train)
test = create_features_max(test)
train = drop_features_max(train)
test = drop_features_max(test)
X = train.drop(targets, axis=1)
y_min = train[target_min]
y_max = train[target_max]
return X, y_min, y_max, test
X_max, y_min_max, y_max_max, test_max = feature_engineering_max(
train_raw.copy(), test_raw.copy()
)
n_splits = 5
# tscv = TimeSeriesSplit(n_splits=n_splits)
kf = KFold(n_splits=n_splits, shuffle=True, random_state=42)
max_scores = []
for fold, (train_index, test_index) in enumerate(kf.split(X_max)):
X_train, X_val = X_max.iloc[train_index], X_max.iloc[test_index]
y_train_min, y_val_min = y_min_max[train_index], y_min_max[test_index]
y_train_max, y_val_max = y_max_max[train_index], y_max_max[test_index]
xgb_reg_max = xgb.XGBRegressor(**xgb_params)
xgb_reg_max.fit(X_train, y_train_max)
y_pred_max = xgb_reg_max.predict(X_val)
max_mse = mean_squared_error(y_val_max, y_pred_max)
max_scores.append(max_mse)
print(f"Fold {fold+1} | min MSE: {min_mse:.4f} ")
print(f"{n_splits} Folds | Average max MSE: {np.average(max_scores)}")
# ### Predict test
xgb_reg_max = xgb.XGBRegressor(**xgb_params)
xgb_reg_max.fit(X_max, y_max_max)
xgb_y_test_max = xgb_reg_max.predict(test_max)
plot_importances(xgb_reg_max)
# # Make Submission
submission = pd.read_csv(
"/kaggle/input/yasmines-meteorological-mystery-dsc-psut-comp/sample_sub.csv"
)
submission[target_min] = xgb_y_test_min
submission[target_max] = xgb_y_test_max
submission.to_csv("xgb_sep_models.csv", index=False)
|
# 
# # Welcome to HPA. Part 1.
#
# Here we will get acquainted with the data and the topic of the competition,so that everything becomes Clearly Explained!!!
# > Добро пожаловать в HPA. Часть 1.
# > Здесь мы познакомимся с данными и темой конкурса, чтобы все стало предельно ясно!!!
# The purpose of the competition: to classify different human cells by images.
# Cells are the basic structural units of all living organisms. The bustling activity of the human cell is maintained as proteins perform specific tasks in designated compartments, called organelles.
# I want to give you quality study material where everything is well structured and explained. To do this, I need to receive feedback from you in the form of comments. And also, if you like it, support the notebook - press the upvote button! =) Thanks!
# > Цель соревнования: классифицировать различные клетки человека по изображениям.
# > Клетки - это основные структурные единицы всех живых организмов. Активная активность человеческой клетки поддерживается, поскольку белки выполняют определенные задачи в определенных отсеках, называемых органеллами.
# > Я хочу давать вам качественный материал для изучения, где всё хорошо структруированно и объяснено. Для этого мне необходимо получать от вас фидбек в виде комментариев. А также, если это вам нравится - поддержите ноутбук и жмите кнопку "за" !=) Спасибо!
# # 1. Data
# Let us have a look at competiton data directory.
# > Давайте посмотрим на каталог данных соревнования.
import os
import pandas as pd
from glob import glob
from tqdm.notebook import tqdm
# directory
print("Competition Data/Files")
ROOT = "../input/hpa-single-cell-image-classification/"
os.listdir(ROOT)
# # Train
# Мы имеем 2 тренировочных набора ('train' и 'train.csv'):
# >****train**** - здесь находится набор изображений с клетками человека. Для каждой клетки существует 4 изображения. Это связано с распознованием внутренностей клетки, ведь каждая органелла(компонент клетки) хорошо видна лишь в своём спектре.
# 
# >****train.csv**** - содержит набор меток органелл к каждой клетке. Эти файлы выглядят вот так:
train_df = pd.read_csv(os.path.join(ROOT, "train.csv"))
train_df.head()
#
# What do these labels represent?
# | Label | Name | Description |
# |-|-|-|
# | 0. | Nucleoplasm | The nucleus is found in the center of cell and can be identified with the help of the signal in the blue nucleus channel. A staining of the nucleoplasm may include the whole nucleus or of the nucleus without the regions known as nucleoli (Class 2). |
# | 1. | Nuclear membrane | The nuclear membrane appears as a thin circle around the nucleus. It is not perfectly smooth and sometimes it is also possible to see the folds of the membrane as small circles or dots inside the nucleus. |
# | 2. | Nucleoli | Nucleoli can be seen as slightly elongated circular areas in the nucleoplasm, which usually display a much weaker staining in the blue DAPI channel. The number and size of nucleoli varies between cell types. |
# | 3. | Nucleoli fibrillar center | Nucleoli fibrillary center can appear as a spotty cluster or as a single bigger spot in the nucleolus, depending on the cell type. |
# | 4. | Nuclear speckles | Nuclear speckles can be seen as irregular and mottled spots inside the nucleoplasm. |
# | 5. | Nuclear bodies | Nuclear bodies are visible as distinct spots in the nucleoplasm. They vary in shape, size and numbers depending on the type of bodies as well as cell type, but are usually more rounded compared to nuclear speckles. |
# | 6. | Endoplasmic reticulum | The endoplasmic reticulum (ER) is recognized by a network-like staining in the cytosol, which is usually stronger close to the nucleus and weaker close to the edges of the cell. The ER can be identified with the help of the staining in the yellow ER channel. |
# | 7. | Golgi apparatus | The Golgi apparatus is a rather large organelle that is located next to the nucleus, close to the centrosome, from which the microtubules in the red channel originate. It has a folded ribbon-like appearance, but the shape and size can vary between cell types, and in response to cellular various processes. |
# | 8. | Intermediate filaments | Intermediate filaments often exhibit a slightly tangled structure with strands crossing every so often. They can appear similar to microtubules, but do not match well with the staining in the red microtubule channel. Intermediate filaments may extend through the whole cytosol, or be concentrated in an area close to the nucleus. |
# | 9. | Actin filaments | Actin filaments can be seen as long and rather straight bundles of filaments or as branched networks of thinner filaments. They are usually located close to the edges of the cells. |
# | 10. | Microtubules | Microtubules are seen as thin strands that stretch throughout the whole cell. It is almost always possible to detect the center from which they all originate (the centrosome). And yes, as you might have guessed, this overlaps the staining in the red channel. |
# | 11. | Mitotic spindle | The mitotic spindle can be seen as an intricate structure of microtubules radiating from each of the centrosomes at opposite ends of a dividing cell (mitosis). At this stage, the chromatin of the cell is condensed, as visible by intense DAPI staining. The size and exact shape of the mitotic spindle changes during mitotic progression, clearly reflecting the different stages of mitosis. |
# | 12. | Centrosome | This class includes centrosomes and centriolar satellites. They can be seen as a more or less distinct staining of a small area at the origin of the microtubules, close to the nucleus. When a cell is dividing, the two centrosomes move to opposite ends of the cell and form the poles of the mitotic spindle. |
# | 13. | Plasma membrane | This class includes plasma membrane and cell junctions. Both are at the outer edge of the cell. Plasma membrane sometimes appears as a more or less distinct edge around the cell, occasionally with characteristic protrusions or ruffles. In some cell lines, the staining can be uniform across the entire cell. Cell junctions can be observed at contact sites between neighboring cells. |
# | 14. | Mitochondria | Mitochondria are small rod-like units in the cytosol, which are often distributed in a thread-like pattern along microtubules. |
# | 15. | Aggresome | An aggresome can be seen as a dense cytoplasmic inclusion, which is usually found close to the nucleus, in a region where the microtubule network is disrupted. |
# | 16. | Cytosol | The cytosol extends from the plasma membrane to the nuclear membrane. It can appear smooth or granular, and the staining is often stronger close to the nucleus. |
# | 17. | Vesicles and punctate cytosolic patterns | This class includes small circular compartments in the cytosol: Vesicles, Peroxisomes (lipid metabolism), Endosomes (sorting compartments), Lysosomes (degradation of molecules or eating up dead molecules), Lipid droplets (fat storage), Cytoplasmic bodies (distinct granules in the cytosol). They are highly dynamic, varying in numbers and size in response to environmental and cellular cues. They can be round or more elongated. |
# | 18. | Negative | This class include negative stainings and unspecific patterns. This means that the cells have no green staining (negative), or have staining but no pattern can be deciphered from the staining (unspecific). |
# >
# >
# >Что означают эти ярлыки?
# >
# > | Этикетка | Имя | Описание |
# > |-|-|-|
# > |0.| Нуклеоплазма | Ядро находится в центре клетки и может быть идентифицировано с помощью сигнала в синем канале ядра. Окрашивание нуклеоплазмы может охватывать все ядро или ядро без участков, известных как ядрышки (класс 2). |
# > |1.| Ядерная мембрана | Ядерная мембрана выглядит как тонкий круг вокруг ядра. Он не идеально гладкий, и иногда можно также увидеть складки мембраны в виде маленьких кружков или точек внутри ядра. |
# > | 2. | Ядрышки | Ядрышки можно рассматривать как слегка удлиненные круглые области в нуклеоплазме, которые обычно имеют гораздо более слабое окрашивание в синем канале DAPI. Количество и размер ядрышек варьируется в зависимости от типа клеток. |
# > | 3. | Фибриллярный центр ядрышек | Фибриллярный центр ядрышек может выглядеть как пятнистый кластер или как единое более крупное пятно в ядрышке, в зависимости от типа клетки. |
# > | 4. | Ядерные спеклы | Ядерные спеклы можно рассматривать как неправильные и пестрые пятна внутри нуклеоплазмы. |
# > | 5. | Ядерные тела | Ядерные тела видны как отдельные пятна в нуклеоплазме. Они различаются по форме, размеру и количеству в зависимости от типа тел, а также от типа клеток, но обычно имеют более округлую форму по сравнению с ядерными крапинками. |
# > | 6. | Эндоплазматический ретикулум | Эндоплазматический ретикулум (ЭР) распознается по сетчатому окрашиванию в цитозоле, которое обычно сильнее вблизи ядра и слабее вблизи краев клетки. ER можно идентифицировать с помощью окрашивания желтого канала ER. |
# > | 7. | Аппарат Гольджи | Аппарат Гольджи представляет собой довольно крупную органеллу, которая находится рядом с ядром, недалеко от центросомы, от которой берут начало микротрубочки в красном канале. Он имеет вид свернутой ленты, но форма и размер могут варьироваться в зависимости от типа клеток и в ответ на различные клеточные процессы. |
# > | 8. | Промежуточные волокна | Промежуточные волокна часто демонстрируют слегка запутанная структура с частыми перекрещивающимися нитями. Они могут казаться похожими на микротрубочки, но не соответствуют окрашиванию красного канала микротрубочек. Промежуточные филаменты могут проходить через весь цитозоль или концентрироваться в области, близкой к ядру. |
# > | 9. | Актиновые нити | Актиновые филаменты можно увидеть как длинные и довольно прямые пучки филаментов или как разветвленные сети из более тонких филаментов. Обычно они располагаются близко к краям ячеек. |
# > | 10. | Микротрубочки | Микротрубочки представляют собой тонкие нити, тянущиеся по всей клетке. Почти всегда можно обнаружить центр, из которого все они происходят (центросома). И да, как вы могли догадаться, это перекрывает окрашивание в красном канале. |
# > | 11. | Митотическое веретено | Митотическое веретено можно рассматривать как сложную структуру микротрубочек, исходящих от каждой из центросом на противоположных концах делящейся клетки (митоз). На этом этапе хроматин клетки конденсируется, что видно по интенсивному окрашиванию DAPI. Размер и точная форма митотического веретена меняются во время митотической прогрессии, четко отражая различные стадии митоза. |
# > | 12. | Центросома | Этот класс включает центросомы и центриолярные сателлиты. Их можно увидеть как более или менее отчетливое окрашивание небольшого участка в начале микротрубочек, рядом с ядром. Когда клетка делится, две центросомы перемещаются к противоположным концам клетки и образуют полюса митотического веретена. |
# > | 13. | Плазменная мембрана | Этот класс включает плазматические мембраны и соединения клеток. Оба находятся на внешнем краю ячейки. Плазменная мембрана иногда выглядит как более или менее отчетливый край вокруг клетки, иногда с характерными выступами или складками. В некоторых клеточных линиях окрашивание может быть равномерным по всей клетке. Соединения клеток можно наблюдать в местах контакта между соседними клетками. |
# > | 14. | Митохондрии | Митохондрии маленькие палочковидные единиц цитозоля, которые часто распределены в виде нитей вдоль микротрубочек. |
# > | 15. | Агрессивный | Агресома может рассматриваться как плотное цитоплазматическое включение, которое обычно находится рядом с ядром, в области, где нарушена сеть микротрубочек. |
# > | 16. | Цитозоль | Цитозоль простирается от плазматической мембраны до ядерной мембраны. Он может казаться гладким или зернистым, а окраска часто более сильна вблизи ядра. |
# > | 17. | Пузырьки и точечные узоры цитозола | Этот класс включает небольшие кольцевые компартменты в цитозоле: везикулы, пероксисомы (липидный обмен), эндосомы (отсеки сортировки), лизосомы (разрушение молекул или поедание мертвых молекул), липидные капли (накопление жира), цитоплазматические тела (отдельные гранулы в цитозоль). Они очень динамичны, различаются по количеству и размеру в зависимости от внешних и клеточных сигналов. Они могут быть круглыми или более удлиненными. |
# > | 18. | Отрицательный | К этому классу относятся отрицательные окрашивания и неспецифические шаблоны. Это означает, что клетки не окрашены в зеленый цвет (отрицательно) или имеют окрашивание, но по окрашиванию нельзя определить рисунок (неспецифический). |
# * It looks like this.
# >На изображении это выглядит примерно так.
# 
# # **Submission**
#
sample_sub = pd.read_csv(os.path.join(ROOT, "sample_submission.csv"))
sample_sub.head()
# The file that we will have to send for verification: sample_submission.csv - contains ID, sizes and a string containing predictions(The PredicitionString column contains a string of label, confidence score for that classified label/organelle and mask information ecoded in RLE encoding and we are doing this for every cell present in the image. It may be possible that some of the cells in the image may not have the protein of interest.)
# > Файл, который мы должны будем отправить на проверку:
# > sample_submission.csv - содержит ID, размеры и строку, в которой записаны предсказания(Столбец PredicitionString содержит строку метки, оценку достоверности для этой классифицированной метки / органеллы и информацию о маске, в кодировке RLE, и мы делаем это для каждой клетки, присутствующей на изображении. Возможно, некоторые из клеток на изображении могут не содержать интересующий белок.)
#
print(
f"We have {sample_sub.shape[0]} rows and {sample_sub.shape[1]} columns in our sample_sub.csv."
)
# At the end, I suggest you relax and watch this video.
# > В конце предлогаю вам расслабиться и посмотреть это видео.
#
from IPython.display import HTML
HTML(
'<center><iframe width="950" height="450" src="https://www.youtube.com/embed/P4gz6DrZOOI" frameborder="0" allow="accelerometer; autoplay; clipboard-write; encrypted-media; gyroscope; picture-in-picture" allowfullscreen></iframe></center>'
)
|
# # Rakam Tanima CNN
# ---
#
import keras
import tensorflow
from keras.datasets import mnist
from keras.models import Sequential
from keras.layers import Dense, Dropout, Flatten
from keras.layers import Conv2D, MaxPooling2D
from tensorflow.keras.utils import to_categorical
from tensorflow.keras import optimizers
import matplotlib.pyplot as plt
import numpy as np
(x_train, y_train), (x_test, y_test) = mnist.load_data()
print(x_test)
# ### Veriden örnekleri görselleştirme
plt.figure(figsize=(14, 14))
x, y = 10, 4
for i in range(40):
plt.subplot(y, x, i + 1)
plt.imshow(x_train[i], cmap="gray")
plt.show()
print(x_train.shape)
print(x_test.shape)
img_rows, img_cols = x_train.shape[1], x_train.shape[2]
x_train = x_train.reshape(x_train.shape[0], img_rows, img_cols, 1)
x_test = x_test.reshape(x_test.shape[0], img_rows, img_cols, 1)
input_shape = (img_rows, img_cols, 1)
print(x_train.shape)
print(x_test.shape)
num_classes = 10
y_train = tensorflow.keras.utils.to_categorical(y_train, num_classes)
y_test = tensorflow.keras.utils.to_categorical(y_test, num_classes)
# ## MODEL OLUŞTURMA
model = Sequential()
epochs = 6
batch_size = 128
# **Katmanların oluşturulması**
model.add(Conv2D(32, kernel_size=(3, 3), activation="relu", input_shape=input_shape))
model.add(Conv2D(64, (3, 3), activation="relu"))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Flatten())
model.add(Dense(128, activation="relu"))
model.add(Dropout(0.5))
model.add(Dense(num_classes, activation="softmax"))
# Modell Görselleştirme
#
model.summary()
model.compile(
loss=keras.losses.categorical_crossentropy,
optimizer=tensorflow.keras.optimizers.Adadelta(),
metrics=["accuracy"],
)
# ### Eğitim İşlemleri
model_history = model.fit(
x_train,
y_train,
batch_size=batch_size,
epochs=epochs,
verbose=1,
validation_data=(x_test, y_test),
)
plt.plot(model_history.history["accuracy"], label="Accuracy")
plt.plot(model_history.history["val_accuracy"], label="Validation Accuracy")
plt.legend()
plt.show
plt.plot(model_history.history["loss"], label="Loss")
plt.plot(model_history.history["val_loss"], label="Validation Loss")
plt.legend()
plt.show()
model.save("saved_mnist_model")
score = model.evaluate(x_test, y_test, verbose=1)
print("Test Loss:", score[0])
print("Test Accuracy:", score[1])
# **Rastgele değer için test işlemi**
np.random.seed(1)
random = np.random.randint(0, len(x_test), 1)
test_image = x_test[random]
print("Secilen Liste Görünümü: ", y_test[random])
holder = y_test[random]
result = np.where(holder == 1.0)
print("Secilen Sayi: ", result[1][0])
plt.imshow(test_image.reshape(28, 28), cmap="gray", vmin=2, vmax=255)
# # Kayıtlı Modeli Yükle ve Kullan
# ---
import keras
import numpy as np
from keras.datasets import mnist
from keras.models import load_model
import tensorflow
import matplotlib.pyplot as plt
model = keras.models.load_model("/kaggle/input/mnist-saved-model/saved_mnist_model")
(x_train, y_train), (x_test, y_test) = mnist.load_data()
num_classes = 10
img_rows, img_cols = x_train.shape[1], x_train.shape[2]
x_train = x_train.reshape(x_train.shape[0], img_rows, img_cols, 1)
x_test = x_test.reshape(x_test.shape[0], img_rows, img_cols, 1)
input_shape = (img_rows, img_cols, 1)
y_train = tensorflow.keras.utils.to_categorical(y_train, num_classes)
y_test = tensorflow.keras.utils.to_categorical(y_test, num_classes)
score = model.evaluate(x_test, y_test, verbose=0)
print("Test Loss:", score[0])
print("Test Accuracy:", score[1])
np.random.seed(1)
random = np.random.randint(0, len(x_test), 1)
test_image = x_test[random]
print("Secilen Liste Görünümü: ", y_test[random])
holder = y_test[random]
result = np.where(holder == 1.0)
print("Secilen Sayi: ", result[1][0])
plt.imshow(test_image.reshape(28, 28), cmap="gray", vmin=2, vmax=255)
|
# ## Import
# Directive pour afficher les graphiques dans Jupyter
# Pandas : librairie de manipulation de données
# NumPy : librairie de calcul scientifique
# MatPlotLib : librairie de visualisation et graphiques
import pandas as pd
import numpy as np
from matplotlib import pyplot as plt
import seaborn as sns
import time
from sklearn import model_selection
from sklearn.metrics import (
classification_report,
confusion_matrix,
roc_curve,
roc_auc_score,
auc,
accuracy_score,
)
from sklearn.preprocessing import StandardScaler, MinMaxScaler
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import RandomForestClassifier, ExtraTreesClassifier
from xgboost import XGBClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.metrics import *
from sklearn.model_selection import *
from sklearn import datasets
# ## Tensorflow
import tensorflow as tf
from tensorflow.keras.models import Sequential, load_model
from tensorflow.keras.layers import InputLayer, Dense, Dropout, Flatten
from tensorflow.keras.optimizers import Adam
from tensorflow.keras.activations import relu, swish, softmax
from tensorflow.keras.utils import to_categorical
from tensorflow.keras.losses import categorical_crossentropy
# ## Data Logging
df = pd.read_csv("/kaggle/input/microbes-dataset/microbes.csv")
# ## Information of the Data
df.head()
df.shape
df.columns
df.info()
df.count()
df.describe()
df["Solidity"]
df["microorganisms"]
df = df.drop(["Unnamed: 0"], axis=1)
# ### the id is meaningless for the process of machine learning
df.shape
sns.boxplot(data=df, x="microorganisms", y="Solidity")
sns.scatterplot(data=df, x="Solidity", y="Eccentricity", hue="microorganisms")
sns.kdeplot(data=df, x="Centroid1", y="Centroid2", hue="microorganisms", fill=True)
# ### Adjusting one by one and looking at the situation is too primitive, so the for loop is used.
def affichage_data(df, column, label):
print("count of NaN:", df[column].isna().sum())
# isnull is not suitable for sum,but it is the same as isnull
plt.figure(figsize=(15, 5))
plt.boxplot(df[column].value_counts(), labels=[label], vert=False)
plt.show()
def afffichage_class(df, column, label):
print("count of NaN: ", df[column].isna().sum())
plt.figure(figsize=(15, 5))
_ = df[column].value_counts().plot.barh()
plt.xlabel("Occurrences")
plt.show()
def affichage_relation(df, x_column, y_column):
sns.set(rc={"figure.figsize": (20, 10)})
sns.boxenplot(data=df, x=x_column, y=y_column)
plt.show()
# #### Because there are two types of data, float and object, which require different ways of drawing, different functions are predefined for different situations.
for x in df.columns:
print(x)
if df[x].dtype == "float64":
affichage_data(df, x, x)
else:
afffichage_class(df, x, x)
# #### show relationship of the class and the values
for x in df.columns:
print(x)
if df[x].dtype == "float":
affichage_relation(df, x, "microorganisms")
# ### After trying various variables, I found that the ten classification objectives of microbial species had a high degree of coincidence for a single variable
# ### there are 20+ variables ,if we try these combinations of variales one by one, it will cost a lot of time.
# ### I'm going to try to see what happens when you categorize things directly through a simple machine learning method
y = df["microorganisms"]
X = df.drop(["microorganisms"], axis=1)
# #### Fitting all the data causes kaggle to crash
df1 = df.sample(1000)
y = df1["microorganisms"]
X = df1.drop(["microorganisms"], axis=1)
start_time = time.time()
model1 = LogisticRegression(solver="liblinear")
cross_val_score(model1, X, y, cv=20).mean()
end_time = time.time()
print(end_time - start_time)
cross_val_score(model1, X, y, cv=20).mean()
model2 = DecisionTreeClassifier()
cross_val_score(model2, X, y, cv=20).mean()
model3 = RandomForestClassifier()
cross_val_score(model3, X, y, cv=20).mean()
model4 = ExtraTreesClassifier()
cross_val_score(model4, X, y, cv=20).mean()
# #### in xgbclassifier,label of y is exceped as [0,1,2...],so we need to relabel it
model5 = XGBClassifier()
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
y = le.fit_transform(y)
cross_val_score(model5, X, y, cv=20, error_score="raise").mean()
# #### The method of linear regression will occupy a lot of time and has a lower accuracy, on the one hand, because of the large amount of data, on the other hand, because of the high collinearity of data, which is difficult to fit.
# #### Then let me see the relationship between those variables
df.corr()
sns.set(rc={"figure.figsize": (30, 30)})
_ = sns.heatmap(df.corr(), annot=True, vmin=-1, vmax=1, center=0).set(
title="Correlations between features"
)
sns.clustermap(abs(df.corr()), vmin=-1, vmax=1, center=0)
sns.clustermap(df.corr(), vmin=-1, vmax=1, center=0)
# ### As you can see, many variables are almost linearly correlated.If you use Bayesian or decision tree methods, this will not cause much impact, but if you use regression method, it will cause great harm
# #### in ordre to visualize la number of the target of classfication ,i will make a one-hot encoding
# ### one-hot encoding
def exclude_onehot_columns(columns, onehot_columns):
return [
c
for c in columns
if len([ohc for ohc in onehot_columns if c.startswith(ohc)]) == 0
]
def get_onehot_columns(columns, onehot_columns):
return [
c
for c in columns
if len([ohc for ohc in onehot_columns if c.startswith(ohc)]) >= 1
]
one_hot_columns = ["microorganisms"]
linear_columns = exclude_onehot_columns(df.columns, one_hot_columns)
one_hot_df = pd.get_dummies(df[one_hot_columns])
df = df.drop(one_hot_columns, axis=1)
df = df.join(one_hot_df)
scaler = StandardScaler()
scaler.fit(df[linear_columns])
df[linear_columns] = scaler.transform(df[linear_columns])
df.sample(10)
# ### data split
y_columns = get_onehot_columns(df.columns, one_hot_columns)
y = df[y_columns]
X = df.drop(y_columns, axis=1)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
# ### RandomForest
from sklearn.metrics import roc_auc_score, precision_recall_fscore_support
crit = ["gini", "entropy"]
for cr in crit:
clf = RandomForestClassifier(criterion=cr)
clf.fit(X_train, y_train)
y_pred = clf.predict(X_test)
auc = roc_auc_score(y_test, y_pred)
pr, re, fs, su = precision_recall_fscore_support(y_test, y_pred, average="macro")
print("-" * 40)
print(cr)
print(f"auc:{auc}")
print(f"precision:{pr}")
print(f"recall:{re}")
print(f"f1_score:{fs}")
# ### DecisionTree
for cr in crit:
clf = DecisionTreeClassifier(criterion=cr)
clf.fit(X_train, y_train)
y_pred = clf.predict(X_test)
auc = roc_auc_score(y_test, y_pred)
pr, re, fs, su = precision_recall_fscore_support(y_test, y_pred, average="macro")
print("-" * 40)
print(cr)
print(f"auc:{auc}")
print(f"precision:{pr}")
print(f"recall:{re}")
print(f"f1_score:{fs}")
# ### ExtraTree
for cr in crit:
clf = ExtraTreesClassifier(criterion=cr)
clf.fit(X_train, y_train)
y_pred = clf.predict(X_test)
auc = roc_auc_score(y_test, y_pred)
pr, re, fs, su = precision_recall_fscore_support(y_test, y_pred, average="macro")
print("-" * 40)
print(cr)
print(f"auc:{auc}")
print(f"precision:{pr}")
print(f"recall:{re}")
print(f"f1_score:{fs}")
# ### XGBClassifier
import warnings
warnings.filterwarnings("ignore")
boos = ["gbtree", "gblinear"]
for bs in boos:
bs = XGBClassifier(booster=bs)
bs.fit(X_train, y_train)
y_pred = bs.predict(X_test)
auc = roc_auc_score(y_test, y_pred)
pr, re, fs, su = precision_recall_fscore_support(y_test, y_pred, average="macro")
print("-" * 40)
print(cr)
print(f"auc:{auc}")
print(f"precision:{pr}")
print(f"recall:{re}")
print(f"f1_score:{fs}")
# #### model with classification linear is not suitable in case without data collinear processing
# ## model dpl
model = Sequential()
model.add(Dense(256, input_shape=[X_train.shape[1]], activation="sigmoid"))
model.add(Dense(256, activation="sigmoid"))
model.add(Dense(256, activation="sigmoid"))
model.add(Dense(256, activation="sigmoid"))
model.add(Dense(y_train.shape[1], activation=softmax))
model.compile(
optimizer=Adam(learning_rate=0.001),
loss=categorical_crossentropy,
metrics=["accuracy"],
)
model.summary()
history = model.fit(X_train, y_train, epochs=150, validation_split=0.2)
plt.plot(history.history["accuracy"])
plt.plot(history.history["val_accuracy"])
plt.title("model accuracy")
plt.ylabel("accuracy")
plt.xlabel("epoch")
plt.legend(["train", "test"], loc="upper left")
plt.show()
plt.plot(history.history["loss"])
plt.plot(history.history["val_loss"])
plt.title("model loss")
plt.ylabel("loss")
plt.xlabel("epoch")
plt.legend(["train", "test"], loc="upper left")
plt.show()
model.predict(X_test)
y_pred = to_categorical(np.argmax(model.predict(X_test), axis=1), y_test.shape[1])
auc = roc_auc_score(y_test, y_pred)
pr, re, fs, su = precision_recall_fscore_support(y_test, y_pred, average="macro")
print(f"auc:{auc}")
print(f"precision:{pr}")
print(f"recall:{re}")
print(f"f1_score:{fs}")
|
# # Imports
# The task is to predict library by question title on StackOverflow.
from lets_plot import *
import numpy as np
import pandas as pd
import os
import torch
np.random.seed(42)
LetsPlot.setup_html()
# # SetUp
if torch.cuda.is_available():
device = torch.device("cuda")
print(f"Using GPU : {torch.cuda.get_device_name(0)}")
else:
device = torch.device("cpu")
print(f"Using CPU")
# # Data
# ## Exploration
path_to_train_data = "/kaggle/input/stackoverlow-data/train.csv"
path_to_test_data = "/kaggle/input/stackoverlow-data/test.csv"
train_data = pd.read_csv(path_to_train_data)
test_data = pd.read_csv(path_to_test_data)
print(train_data.shape)
train_data.head()
# As we can see there are no null values in our train dataset. Also we can notice that some questions have the same title and at all we got 24 libraries (classes).
print(f"Null values:\n{train_data.isna().sum()}")
print(f"Unique:\n{train_data.nunique()}")
print(test_data.shape)
test_data.head()
label_to_id = {k: v for v, k in enumerate(set(train_data.lib))}
id_to_label = {v: k for k, v in label_to_id.items()}
label_to_id
frequency = pd.DataFrame(
{
"Library": list(label_to_id.keys()),
"Count": [
len(train_data[train_data.lib == lib]) for lib in list(label_to_id.keys())
],
}
)
(
ggplot(
frequency,
aes(x=frequency.Library, weight=frequency.Count, fill=frequency.Library),
)
+ geom_bar()
+ labs(x="Library", y="Count")
)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# **Importing all the required libraries**
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler, MinMaxScaler
from sklearn.metrics import mean_absolute_error, mean_squared_error
import numpy as np
import pandas as pd
from pandas import DataFrame
import missingno as msno
from datetime import datetime
from numpy.random import multivariate_normal as mvnrnd
from scipy.stats import wishart
from scipy.stats import invwishart
from numpy.linalg import inv as inv
import scipy.io
import time
from sklearn.decomposition import PCA
import matplotlib.pyplot as plt
from minepy import MINE
import torch
from torch import nn
from torch.autograd import Variable
from sklearn.linear_model import RidgeCV
from math import sqrt
from statsmodels.tsa.seasonal import seasonal_decompose
from statsmodels.tsa.stattools import adfuller
from statsmodels.graphics.tsaplots import plot_acf
from statsmodels.graphics.tsaplots import plot_pacf
from pykalman import KalmanFilter
from sklearn.ensemble import RandomForestRegressor
import random
# # Data import
# We use the data of Aquifer_Auser as an example to demonstrate our model.
df = pd.read_csv("/kaggle/input/acea-water-prediction/Aquifer_Auser.csv")
df["date"] = df["Date"].apply(lambda x: datetime.strptime(x, "%d/%m/%Y"))
target_variable = [
"Depth_to_Groundwater_SAL",
"Depth_to_Groundwater_CoS",
"Depth_to_Groundwater_LT2",
]
columns_name = df.columns.values.tolist()
rain_list = [a for a in columns_name if a.startswith("Rain")]
Temp_list = [a for a in columns_name if a.startswith("Temperature")]
Depth_list = [a for a in columns_name if a.startswith("Depth")]
n_row = df.shape[0]
msno.matrix(df)
rain_list + Temp_list
# from the figure above, we can see that there are a huge number of missing values, and the density of these values is quite large. For the variables that will be used as predicted values, many sparse missing values are scattered in them. We decided to remove the rows with large density of missing values first, and then fill in the sparse missing values.
# We found that there are some continuous zero values in the temperature variable and depth variable, which is quite abnormal. To prevent these zero values from being caused by measurement errors, we turn them into missing values.
for i in range(len(Depth_list)):
df[df[[Depth_list[i]]] == 0] = np.nan
for i in range(len(Temp_list)):
df[df[[Temp_list[i]]] == 0] = np.nan
# We created some graphs to observe the missing value distribution.
# missing value in rainfall variables
for i in range(len(rain_list)):
nullnum = df[["Date", rain_list[i]]].isnull().sum(axis=1).to_numpy()
plt.plot(np.array(list(range(n_row))), nullnum, label=rain_list[i])
plt.legend()
plt.show()
# missing value in Depth variables
for i in range(len(Depth_list)):
nullnum = df[["Date", Depth_list[i]]].isnull().sum(axis=1).to_numpy()
plt.plot(np.array(list(range(n_row))), nullnum, label=Depth_list[i])
plt.legend()
plt.show()
# missing value in temperature variables
for i in range(len(Temp_list)):
nullnum = df[["Date", Temp_list[i]]].isnull().sum(axis=1).to_numpy()
plt.plot(np.array(list(range(n_row))), nullnum, label=Temp_list[i])
plt.legend()
plt.show()
# according to the four graphs above, we select all the data between the 4685th row and the 7000th row.
dfsecond = df[4685:7000]
msno.matrix(dfsecond)
# # Feature Engineering
# Rainfall variables and temperature variables are important independent variables in this dataset,
# but their influence on the Depth_to_Groundwater variable may lag behind. So we need to find how many days will it take for their impact on the target variables to be reflected in the value.
# Take 'Depth_to_Groundwater_CoS' as excample, shift the data in this variable 31 times and create a variable after each shift.
dfTestLag = dfsecond[rain_list + Temp_list]
for i in range(0, 31):
dfTestLag["CoS" + str(i)] = dfsecond["Depth_to_Groundwater_CoS"].shift(-1 * i)
targetlistCoS = [
"CoS0",
"CoS1",
"CoS2",
"CoS3",
"CoS4",
"CoS5",
"CoS6",
"CoS7",
"CoS8",
"CoS9",
"CoS10",
"CoS11",
"CoS12",
"CoS13",
"CoS14",
"CoS15",
"CoS16",
"CoS17",
"CoS18",
"CoS19",
"CoS20",
"CoS21",
"CoS22",
"CoS23",
"CoS24",
"CoS25",
"CoS26",
"CoS27",
"CoS28",
"CoS29",
"CoS30",
]
# We use ridgeCV regression to fit the independent variables to 31 new variables to create 31 models,
# then we test the rmse of each model, draw a plot, and choose the point with the smallest rmse value
# to determine the lag value.
dfTestLag = dfTestLag.ffill().bfill()
CoSList = []
for i in range(len(targetlistCoS)):
YCoS = dfTestLag[targetlistCoS[i]]
X = dfTestLag[rain_list + Temp_list]
X_train1, X_test1, y_train1, y_test1 = train_test_split(
X, YCoS, test_size=0.2, random_state=i
)
ridgecv = RidgeCV(alphas=[0.01, 0.1, 0.5, 1, 5, 7, 10, 30, 100, 200])
model = ridgecv.fit(X_train1, y_train1)
y_pred1 = model.predict(X_test1)
rms1 = sqrt(mean_squared_error(y_test1, y_pred1))
CoSList.append(rms1)
dfLT2 = pd.DataFrame(CoSList, columns=["rms"])
plt.title("CoS lag")
plt.ylabel("RMS")
plt.xlabel("lag")
plt.plot(dfLT2["rms"])
# We used the same method to test the lag value of other target variables, then we chose '28' as the lag value of target variables.
# Bayesian Temporal Matrix Factorization (BTMF)
# We chose BTMF method to fill in the missing values of Depth_to_Groundwater variables. The BTMF method has better performance in filling the missing values of long-term time series data sets.
dfmeasure = dfsecond[
[
"Depth_to_Groundwater_LT2",
"Depth_to_Groundwater_SAL",
"Depth_to_Groundwater_PAG",
"Depth_to_Groundwater_CoS",
"Depth_to_Groundwater_DIEC",
]
]
dfdens = dfmeasure[
[
"Depth_to_Groundwater_LT2",
"Depth_to_Groundwater_SAL",
"Depth_to_Groundwater_PAG",
"Depth_to_Groundwater_CoS",
"Depth_to_Groundwater_DIEC",
]
]
dfdens["Depth_to_Groundwater_LT2"] = dfdens["Depth_to_Groundwater_LT2"].interpolate()
dfdens["Depth_to_Groundwater_SAL"] = dfdens["Depth_to_Groundwater_SAL"].interpolate()
dfdens["Depth_to_Groundwater_PAG"] = dfdens["Depth_to_Groundwater_PAG"].interpolate()
dfdens["Depth_to_Groundwater_CoS"] = dfdens["Depth_to_Groundwater_CoS"].interpolate()
dfdens["Depth_to_Groundwater_DIEC"] = dfdens["Depth_to_Groundwater_DIEC"].interpolate()
dfdens = dfdens.ffill().bfill()
dfdens = np.delete(
dfdens.to_numpy().T, range(len(dfsecond) - (len(dfsecond) // 28) * 28), axis=1
)
dfdealMis = np.delete(
dfmeasure.fillna(0).to_numpy().T,
range(len(dfsecond) - (len(dfsecond) // 28) * 28),
axis=1,
)
def kr_prod(a, b):
return np.einsum("ir, jr -> ijr", a, b).reshape(a.shape[0] * b.shape[0], -1)
def cov_mat(mat):
dim1, dim2 = mat.shape
new_mat = np.zeros((dim2, dim2))
mat_bar = np.mean(mat, axis=0)
for i in range(dim1):
new_mat += np.einsum("i, j -> ij", mat[i, :] - mat_bar, mat[i, :] - mat_bar)
return new_mat
def ten2mat(tensor, mode):
return np.reshape(np.moveaxis(tensor, mode, 0), (tensor.shape[mode], -1), order="F")
def mat2ten(mat, tensor_size, mode):
index = list()
index.append(mode)
for i in range(tensor_size.shape[0]):
if i != mode:
index.append(i)
return np.moveaxis(np.reshape(mat, list(tensor_size[index]), order="F"), 0, mode)
def mnrnd(M, U, V):
"""
Generate matrix normal distributed random matrix.
M is a m-by-n matrix, U is a m-by-m matrix, and V is a n-by-n matrix.
"""
dim1, dim2 = M.shape
X0 = np.random.rand(dim1, dim2)
P = np.linalg.cholesky(U)
Q = np.linalg.cholesky(V)
return M + np.matmul(np.matmul(P, X0), Q.T)
def BTMF(dense_mat, sparse_mat, init, rank, time_lags, maxiter1, maxiter2):
"""Bayesian Temporal Matrix Factorization, BTMF."""
W = init["W"]
X = init["X"]
d = time_lags.shape[0]
dim1, dim2 = sparse_mat.shape
pos = np.where((dense_mat != 0) & (sparse_mat == 0))
position = np.where(sparse_mat != 0)
binary_mat = np.zeros((dim1, dim2))
binary_mat[position] = 1
beta0 = 1
nu0 = rank
mu0 = np.zeros((rank))
W0 = np.eye(rank)
tau = 1
alpha = 1e-6
beta = 1e-6
S0 = np.eye(rank)
Psi0 = np.eye(rank * d)
M0 = np.zeros((rank * d, rank))
W_plus = np.zeros((dim1, rank))
X_plus = np.zeros((dim2, rank))
X_new_plus = np.zeros((dim2 + 1, rank))
A_plus = np.zeros((rank, rank, d))
mat_hat_plus = np.zeros((dim1, dim2 + 1))
for iters in range(maxiter1):
W_bar = np.mean(W, axis=0)
var_mu_hyper = (dim1 * W_bar) / (dim1 + beta0)
var_W_hyper = inv(
inv(W0)
+ cov_mat(W)
+ dim1 * beta0 / (dim1 + beta0) * np.outer(W_bar, W_bar)
)
var_Lambda_hyper = wishart(df=dim1 + nu0, scale=var_W_hyper, seed=None).rvs()
var_mu_hyper = mvnrnd(var_mu_hyper, inv((dim1 + beta0) * var_Lambda_hyper))
var1 = X.T
var2 = kr_prod(var1, var1)
var3 = tau * np.matmul(var2, binary_mat.T).reshape(
[rank, rank, dim1]
) + np.dstack([var_Lambda_hyper] * dim1)
var4 = (
tau * np.matmul(var1, sparse_mat.T)
+ np.dstack([np.matmul(var_Lambda_hyper, var_mu_hyper)] * dim1)[0, :, :]
)
for i in range(dim1):
inv_var_Lambda = inv(var3[:, :, i])
W[i, :] = mvnrnd(np.matmul(inv_var_Lambda, var4[:, i]), inv_var_Lambda)
if iters + 1 > maxiter1 - maxiter2:
W_plus += W
Z_mat = X[np.max(time_lags) : dim2, :]
Q_mat = np.zeros((dim2 - np.max(time_lags), rank * d))
for t in range(np.max(time_lags), dim2):
Q_mat[t - np.max(time_lags), :] = X[t - time_lags, :].reshape([rank * d])
var_Psi = inv(inv(Psi0) + np.matmul(Q_mat.T, Q_mat))
var_M = np.matmul(var_Psi, np.matmul(inv(Psi0), M0) + np.matmul(Q_mat.T, Z_mat))
var_S = (
S0
+ np.matmul(Z_mat.T, Z_mat)
+ np.matmul(np.matmul(M0.T, inv(Psi0)), M0)
- np.matmul(np.matmul(var_M.T, inv(var_Psi)), var_M)
)
Sigma = invwishart(
df=nu0 + dim2 - np.max(time_lags), scale=var_S, seed=None
).rvs()
A = mat2ten(mnrnd(var_M, var_Psi, Sigma).T, np.array([rank, rank, d]), 0)
if iters + 1 > maxiter1 - maxiter2:
A_plus += A
Lambda_x = inv(Sigma)
var1 = W.T
var2 = kr_prod(var1, var1)
var3 = tau * np.matmul(var2, binary_mat).reshape(
[rank, rank, dim2]
) + np.dstack([Lambda_x] * dim2)
var4 = tau * np.matmul(var1, sparse_mat)
for t in range(dim2):
Mt = np.zeros((rank, rank))
Nt = np.zeros(rank)
if t < np.max(time_lags):
Qt = np.zeros(rank)
else:
Qt = np.matmul(
Lambda_x,
np.matmul(ten2mat(A, 0), X[t - time_lags, :].reshape([rank * d])),
)
if t < dim2 - np.min(time_lags):
if t >= np.max(time_lags) and t < dim2 - np.max(time_lags):
index = list(range(0, d))
else:
index = list(
np.where(
(t + time_lags >= np.max(time_lags))
& (t + time_lags < dim2)
)
)[0]
for k in index:
Ak = A[:, :, k]
Mt += np.matmul(np.matmul(Ak.T, Lambda_x), Ak)
A0 = A.copy()
A0[:, :, k] = 0
var5 = X[t + time_lags[k], :] - np.matmul(
ten2mat(A0, 0),
X[t + time_lags[k] - time_lags, :].reshape([rank * d]),
)
Nt += np.matmul(np.matmul(Ak.T, Lambda_x), var5)
var_mu = var4[:, t] + Nt + Qt
if t < np.max(time_lags):
inv_var_Lambda = inv(var3[:, :, t] + Mt - Lambda_x + np.eye(rank))
else:
inv_var_Lambda = inv(var3[:, :, t] + Mt)
X[t, :] = mvnrnd(np.matmul(inv_var_Lambda, var_mu), inv_var_Lambda)
mat_hat = np.matmul(W, X.T)
X_new = np.zeros((dim2 + 1, rank))
if iters + 1 > maxiter1 - maxiter2:
X_new[0:dim2, :] = X.copy()
X_new[dim2, :] = np.matmul(
ten2mat(A, 0), X_new[dim2 - time_lags, :].reshape([rank * d])
)
X_new_plus += X_new
mat_hat_plus += np.matmul(W, X_new.T)
tau = np.random.gamma(
alpha + 0.5 * sparse_mat[position].shape[0],
1 / (beta + 0.5 * np.sum((sparse_mat - mat_hat)[position] ** 2)),
)
rmse = np.sqrt(
np.sum((dense_mat[pos] - mat_hat[pos]) ** 2) / dense_mat[pos].shape[0]
)
if (iters + 1) % 200 == 0 and iters < maxiter1 - maxiter2:
print("Iter: {}".format(iters + 1))
print("RMSE: {:.6}".format(rmse))
print()
W = W_plus / maxiter2
X_new = X_new_plus / maxiter2
A = A_plus / maxiter2
mat_hat = mat_hat_plus / maxiter2
if maxiter1 >= 100:
final_mape = (
np.sum(np.abs(dense_mat[pos] - mat_hat[pos]) / dense_mat[pos])
/ dense_mat[pos].shape[0]
)
final_rmse = np.sqrt(
np.sum((dense_mat[pos] - mat_hat[pos]) ** 2) / dense_mat[pos].shape[0]
)
print("Imputation MAPE: {:.6}".format(final_mape))
print("Imputation RMSE: {:.6}".format(final_rmse))
print()
return mat_hat, W, X_new, A
sparse_mat = dfdealMis
dense_mat = dfdens
import time
start = time.time()
dim1, dim2 = sparse_mat.shape
rank = 10
time_lags = np.array([1, 2, (len(dfsecond) // 28)])
init = {"W": 0.1 * np.random.rand(dim1, rank), "X": 0.1 * np.random.rand(dim2, rank)}
maxiter1 = 1100
maxiter2 = 100
a, b, c, d = BTMF(dense_mat, sparse_mat, init, rank, time_lags, maxiter1, maxiter2)
end = time.time()
print("Running time: %d seconds" % (end - start))
# After imputation we got 'a', which is the array contains all 'Depth_to_Groundwater' variable values
# after filling in the missing values.
# We use this array to replace the original data of 'Depth_to_Groundwater' variable in the dataset.
# 1.
a = np.delete(a, -1, axis=1)
dfRainfall = dfsecond[rain_list].to_numpy()
dfRainfall = np.delete(
dfRainfall, range(len(dfsecond) - (len(dfsecond) // 28) * 28), axis=0
)
dfTemp = dfsecond[Temp_list].to_numpy()
dfTemp = np.delete(dfTemp, range(len(dfsecond) - (len(dfsecond) // 28) * 28), axis=0)
pdate = pd.DataFrame(dfsecond["date"].values.astype("float32"), columns=["Date"])
dfDate = pdate["Date"].to_numpy()
dfDate = np.delete(dfDate, range(len(dfsecond) - (len(dfsecond) // 28) * 28), axis=0)
dfDate = dfDate.reshape(-1, 1)
a = a.T
wholedata = np.hstack((a, dfRainfall, dfTemp, dfDate))
wholelist = Depth_list + rain_list + Temp_list + ["date"]
newFrame = DataFrame(wholedata, index=None, columns=wholelist)
# PCA
# We found there are too many rainfall variables, so we decided to use PCA to reduce the number of these variables.
test = newFrame[rain_list]
test = test.ffill().bfill()
fig = plt.figure()
ax = fig.add_axes([0, 0, 1, 1])
pca = PCA(n_components=10)
pca.fit(test)
v = pca.explained_variance_ratio_.round(2)
ax.bar(range(1, 11), v)
plt.xlabel("PCA")
plt.title("variance")
plt.show()
# We chose the first two PCA to replace the rainfall variables.
Xpca = PCA(n_components=2).fit_transform(test)
pf = pd.DataFrame(Xpca, columns=["PCA1", "PCA2"])
newFrame["PCA1"] = pf["PCA1"]
newFrame["PCA2"] = pf["PCA2"]
for i in range(len(rain_list)):
newFrame = newFrame.drop(rain_list[i], axis=1)
newFrame = newFrame.ffill().bfill()
# We use ".shift(-28)" to creat there new variables, which are the target variables for the prediction model of this dataset.
lag_target = []
for i in range(len(target_variable)):
newFrame[target_variable[i] + "28"] = newFrame[target_variable[i]].shift(-28)
lag_target.append(target_variable[i] + "28")
newFrame
# Correlation Matrix of the new dataset
# We checked MIC values between all the variables, and delete those variables had higher MIC values with some other variables.
def MIC_matirx(dataframe, mine):
data_array = np.array(dataframe)
n = len(data_array[0, :])
output = np.zeros([n, n])
for i in range(n):
for j in range(n):
mine.compute_score(data_array[:, i], data_array[:, j])
output[i, j] = mine.mic()
output[j, i] = mine.mic()
mic_value = pd.DataFrame(output)
return mic_value
mine = MINE(alpha=0.6, c=15)
Matrix_mic_value = MIC_matirx(newFrame, mine)
def HeatMap(DataFrame):
colormap = plt.cm.RdBu
plt.figure(figsize=(14, 12))
plt.title("MIC", y=1.05, size=15)
sns.heatmap(
DataFrame.astype(float),
linewidths=0.1,
vmax=1.0,
square=True,
cmap=colormap,
linecolor="white",
annot=True,
)
plt.show()
HeatMap(Matrix_mic_value)
newFrame.columns.values.tolist()
# According to the MIC matrix above, we can delete columns named 'Temperature_Ponte_a_Moriano' and 'Temperature_Lucca_Orto_Botanico'.
newFrame = newFrame.drop(
["Temperature_Ponte_a_Moriano", "Temperature_Lucca_Orto_Botanico"], axis=1
)
# # Seasonal and Trend
dfseason = pd.Series(
newFrame["Depth_to_Groundwater_LT2"].tolist(), index=newFrame["date"].tolist()
)
decomposition = seasonal_decompose(
dfseason, model="additive", period=365, two_sided=False
)
decomposition.plot()
plt.show()
# We use the ADF to check the Stationarity of the target variable.
adfuller(dfseason)
dfseasonshift = dfseason.shift(-1)
dfseasondff = dfseason - dfseasonshift
dfseasondff = dfseasondff.dropna(inplace=False)
adfuller(dfseasondff)
plot_acf(dfseasondff)
plot_pacf(dfseasondff)
columns_name = newFrame.columns.values.tolist()
Depth_list = [a for a in columns_name if a.startswith("Depth")]
Temp_list = [a for a in columns_name if a.startswith("Temperature")]
# # Kalman filter
# Errors such as measurement errors will add some noise to the data. Which will affect the accuracy of prediction.
# We used Kalman filter to remove this noise in Depth_to_Groundwater variables and temperature variables.
def Kalman1D(data, damping=1):
observation_covariance = damping
first_value = data[0]
transition_matrix = 1
transition_covariance = 0.1
first_value
kf = KalmanFilter(
initial_state_mean=first_value,
initial_state_covariance=observation_covariance,
observation_covariance=observation_covariance,
transition_covariance=transition_covariance,
transition_matrices=transition_matrix,
)
pred_state, state_cov = kf.smooth(data)
return pred_state
dffull = newFrame[: len(newFrame) - 28]
orenArray = dffull["Temperature_Orentano"].to_numpy()
orenkal = Kalman1D(orenArray, 0.1)
plt.plot(np.array(list(range(len(newFrame) - 28))), orenArray, label="measured")
plt.plot(np.array(list(range(len(newFrame) - 28))), orenkal, label="kal")
plt.legend()
plt.show()
# The orange line show the temperature value after the noise is eliminated.
# We can see that it has become smoother than the blue line which contains the original value.
dffullkal = dffull.drop(Depth_list + Temp_list, axis=1)
for i in range(len(Depth_list)):
DepthArray = dffull[Depth_list[i]].to_numpy()
Depthkal = Kalman1D(DepthArray, 0.1)
kallist = map(lambda x: x[0], Depthkal)
Depthkalseries = pd.Series(kallist)
dffullkal[Depth_list[i]] = Depthkalseries
for i in range(len(Temp_list)):
TempArray = dffull[Temp_list[i]].to_numpy()
Tempkal = Kalman1D(TempArray, 0.1)
kallist = map(lambda x: x[0], Tempkal)
Tempkalseries = pd.Series(kallist)
dffullkal[Temp_list[i]] = Tempkalseries
dffullkal
# # LSTM
# Recurrent Neural Network (RNN) is a neural network used to process sequence data. Compared with the general neural network, it can process the data of the sequence change.
# Long short-term memory (Long short-term memory, LSTM) is a special RNN, mainly to solve the problem of gradient disappearance and gradient explosion in the training process of long sequences. Simply put, LSTM can perform better in longer sequences than ordinary RNNs.
class lstm_reg(nn.Module):
def __init__(self, input_size, hidden_size, output_size=1, num_layers=2):
super(lstm_reg, self).__init__()
self.rnn = nn.LSTM(input_size, hidden_size, num_layers, dropout=0.3)
self.reg = nn.Linear(hidden_size, output_size)
def forward(self, x):
x, _ = self.rnn(x)
s, b, h = x.shape
x = x.view(s * b, h)
x = self.reg(x)
x = x.view(s, b, -1)
return x
dffullkal.columns.values.tolist()
# Take 'Depth_to_Groundwater_SAL28' as the example.
# if we want to predict the value of 'Depth_to_Groundwater_SAL28', the features we need is 'date','PCA1','PCA2','Depth_to_Groundwater_SAL', 'Temperature_Orentano','Temperature_Monte_Serra'.
n_test = int(((len(dffullkal) - 28) / 28) // 5 * 28)
n_train = len(dffullkal) - 28 - n_test
dftrain = dffullkal[:n_train]
dftest = dffullkal[n_train : len(dffullkal) - 28]
dftrainX = dftrain[
[
"date",
"PCA1",
"PCA2",
"Depth_to_Groundwater_SAL",
"Temperature_Orentano",
"Temperature_Monte_Serra",
]
]
n_feature = len(dftrainX.columns.values.tolist())
dflistX = np.reshape(dftrainX.values.tolist(), (28, -1, n_feature))
dftrainY = dftrain["Depth_to_Groundwater_SAL28"]
dflistY = np.reshape(dftrainY.values.tolist(), (28, -1, 1))
dflistX = dflistX.astype("float32")
dflistY = dflistY.astype("float32")
tensorx = torch.from_numpy(dflistX)
tensory = torch.from_numpy(dflistY)
net = lstm_reg(n_feature, 100)
criterion = nn.MSELoss()
optimizer = torch.optim.Adam(net.parameters(), lr=1e-2)
for e in range(100):
var_x = Variable(tensorx)
var_y = Variable(tensory)
out = net(var_x)
loss = criterion(out, var_y)
optimizer.zero_grad()
loss.backward()
torch.nn.utils.clip_grad_norm_(net.parameters(), 1.1)
optimizer.step()
if (e + 1) % 10 == 0:
print("Epoch: {}, Loss: {:.5f}".format(e + 1, loss.data))
dftestX = dftest[
[
"date",
"PCA1",
"PCA2",
"Depth_to_Groundwater_SAL",
"Temperature_Orentano",
"Temperature_Monte_Serra",
]
]
dftestlistX = np.reshape(dftestX.values.tolist(), (28, -1, n_feature))
dftestY = dftest["Depth_to_Groundwater_SAL28"]
dftestlistY = np.reshape(dftestY.values.tolist(), (28, -1, 1))
dftestlistX = dftestlistX.astype("float32")
dftestlistY = dftestlistY.astype("float32")
tensortestx = torch.from_numpy(dftestlistX)
tensortesty = torch.from_numpy(dftestlistY)
testvar_x = Variable(tensortestx)
testvar_y = Variable(tensortesty)
nettest = net.eval()
pred_teste = nettest(testvar_x)
loss = criterion(pred_teste, testvar_y)
print("Epoch: {}, Loss: {:.5f}".format("mse", loss.data))
a = nn.L1Loss()
maeloss = a(pred_teste, testvar_y)
print("Epoch: {}, Loss: {:.5f}".format("mae", maeloss.data))
# # Create Method
# referance table
# We used the missingno library to visualize the missing values of each table, and make a line chart to observe the distribution of missing values, and finally determine the range of data used to build the prediction model in each table.
# We refer to the introduction of each table in the 'datasets_description.xlsx' to determine the output of each table and the variables(except for the outputs themselves) that may be used to predict these outputs.
# We make this information into a table so that it can be used when needed.
referdata = {
"table": [
"Aquifer_Auser",
"Aquifer_Doganella",
"Aquifer_Luco",
"Aquifer_Petrignano",
"Lake_Bilancino",
"River_Arno",
"Water_Spring_Amiata",
"Water_Spring_Lupa",
"Water_Spring_Madonna_di_Canneto",
],
"start": [
4685,
3075,
6540,
1000,
1000,
2250,
5600,
600,
1600,
], # Get data begin from which row
"end": [
7000,
3950,
6950,
5223,
6000,
3450,
7487,
4199,
2500,
], # Stop getting data after reaching which row
"feature": [
["Rain", "Temperature", "date"],
["Rain", "Temperature", "date"],
["Rain", "Temperature", "date"],
["Rain", "Temperature", "date"],
["Rain", "Temperature", "date"],
["Rain", "Temperature", "date"],
["Rain", "Temperature", "date"],
["Rain", "date"],
["Rain", "Temperature", "date"],
],
}
referdf = DataFrame(referdata)
referdf
# create a class to collect all the methods of processing data
class data_cook:
def __init__(self, dataframe, start, end, target_variable):
"""
target_variable contains the name of all the variables which cound be used as target variable in this table.
"""
self.dfsecond = dataframe[
start:end
] # the start and end number could be check in the table 'referdf'.
self.columns_name = dataframe.columns.values.tolist()
self.Rain_list = [a for a in self.columns_name if a.startswith("Rain")]
self.Depth_list = [a for a in self.columns_name if a.startswith("Depth")]
self.Temp_list = [a for a in self.columns_name if a.startswith("Temperature")]
self.Flow_list = [a for a in self.columns_name if a.startswith("Flow")]
self.n_row = dataframe.shape[0]
self.target_list = target_variable
def BasicInformation(self):
msno.matrix(self.dfsecond)
print(self.columns_name)
# BTMF
def FillNullBTMF(self, nullValue_list):
"""
the nullValue_list contain the columns' name which we want to fill the null value.
"""
for i in range(len(self.Depth_list)):
self.dfsecond[self.dfsecond[[self.Depth_list[i]]] == 0] = np.nan
for i in range(len(self.Temp_list)):
self.dfsecond[self.dfsecond[[self.Temp_list[i]]] == 0] = np.nan
for i in range(len(self.Flow_list)):
self.dfsecond[self.dfsecond[[self.Flow_list[i]]] == 0] = np.nan
dfmeasure = self.dfsecond[nullValue_list]
dfdens = dfmeasure[nullValue_list]
for i in range(len(nullValue_list)):
dfdens[nullValue_list[i]] = dfdens[nullValue_list[i]].interpolate()
dfdens = dfdens.ffill().bfill()
dfdens = np.delete(
dfdens.to_numpy().T,
range(len(self.dfsecond) - (len(self.dfsecond) // 28) * 28),
axis=1,
)
dfdealMis = np.delete(
dfmeasure.fillna(0).to_numpy().T,
range(len(self.dfsecond) - (len(self.dfsecond) // 28) * 28),
axis=1,
)
def kr_prod(a, b):
return np.einsum("ir, jr -> ijr", a, b).reshape(a.shape[0] * b.shape[0], -1)
def cov_mat(mat):
dim1, dim2 = mat.shape
new_mat = np.zeros((dim2, dim2))
mat_bar = np.mean(mat, axis=0)
for i in range(dim1):
new_mat += np.einsum(
"i, j -> ij", mat[i, :] - mat_bar, mat[i, :] - mat_bar
)
return new_mat
def ten2mat(tensor, mode):
return np.reshape(
np.moveaxis(tensor, mode, 0), (tensor.shape[mode], -1), order="F"
)
def mat2ten(mat, tensor_size, mode):
index = list()
index.append(mode)
for i in range(tensor_size.shape[0]):
if i != mode:
index.append(i)
return np.moveaxis(
np.reshape(mat, list(tensor_size[index]), order="F"), 0, mode
)
def mnrnd(M, U, V):
"""
Generate matrix normal distributed random matrix.
M is a m-by-n matrix, U is a m-by-m matrix, and V is a n-by-n matrix.
"""
dim1, dim2 = M.shape
X0 = np.random.rand(dim1, dim2)
P = np.linalg.cholesky(U)
Q = np.linalg.cholesky(V)
return M + np.matmul(np.matmul(P, X0), Q.T)
def BTMF(dense_mat, sparse_mat, init, rank, time_lags, maxiter1, maxiter2):
"""Bayesian Temporal Matrix Factorization, BTMF."""
W = init["W"]
X = init["X"]
d = time_lags.shape[0]
dim1, dim2 = sparse_mat.shape
pos = np.where((dense_mat != 0) & (sparse_mat == 0))
position = np.where(sparse_mat != 0)
binary_mat = np.zeros((dim1, dim2))
binary_mat[position] = 1
beta0 = 1
nu0 = rank
mu0 = np.zeros((rank))
W0 = np.eye(rank)
tau = 1
alpha = 1e-6
beta = 1e-6
S0 = np.eye(rank)
Psi0 = np.eye(rank * d)
M0 = np.zeros((rank * d, rank))
W_plus = np.zeros((dim1, rank))
X_plus = np.zeros((dim2, rank))
X_new_plus = np.zeros((dim2 + 1, rank))
A_plus = np.zeros((rank, rank, d))
mat_hat_plus = np.zeros((dim1, dim2 + 1))
for iters in range(maxiter1):
W_bar = np.mean(W, axis=0)
var_mu_hyper = (dim1 * W_bar) / (dim1 + beta0)
var_W_hyper = inv(
inv(W0)
+ cov_mat(W)
+ dim1 * beta0 / (dim1 + beta0) * np.outer(W_bar, W_bar)
)
var_Lambda_hyper = wishart(
df=dim1 + nu0, scale=var_W_hyper, seed=None
).rvs()
var_mu_hyper = mvnrnd(
var_mu_hyper, inv((dim1 + beta0) * var_Lambda_hyper)
)
var1 = X.T
var2 = kr_prod(var1, var1)
var3 = tau * np.matmul(var2, binary_mat.T).reshape(
[rank, rank, dim1]
) + np.dstack([var_Lambda_hyper] * dim1)
var4 = (
tau * np.matmul(var1, sparse_mat.T)
+ np.dstack([np.matmul(var_Lambda_hyper, var_mu_hyper)] * dim1)[
0, :, :
]
)
for i in range(dim1):
inv_var_Lambda = inv(var3[:, :, i])
W[i, :] = mvnrnd(
np.matmul(inv_var_Lambda, var4[:, i]), inv_var_Lambda
)
if iters + 1 > maxiter1 - maxiter2:
W_plus += W
Z_mat = X[np.max(time_lags) : dim2, :]
Q_mat = np.zeros((dim2 - np.max(time_lags), rank * d))
for t in range(np.max(time_lags), dim2):
Q_mat[t - np.max(time_lags), :] = X[t - time_lags, :].reshape(
[rank * d]
)
var_Psi = inv(inv(Psi0) + np.matmul(Q_mat.T, Q_mat))
var_M = np.matmul(
var_Psi, np.matmul(inv(Psi0), M0) + np.matmul(Q_mat.T, Z_mat)
)
var_S = (
S0
+ np.matmul(Z_mat.T, Z_mat)
+ np.matmul(np.matmul(M0.T, inv(Psi0)), M0)
- np.matmul(np.matmul(var_M.T, inv(var_Psi)), var_M)
)
Sigma = invwishart(
df=nu0 + dim2 - np.max(time_lags), scale=var_S, seed=None
).rvs()
A = mat2ten(
mnrnd(var_M, var_Psi, Sigma).T, np.array([rank, rank, d]), 0
)
if iters + 1 > maxiter1 - maxiter2:
A_plus += A
Lambda_x = inv(Sigma)
var1 = W.T
var2 = kr_prod(var1, var1)
var3 = tau * np.matmul(var2, binary_mat).reshape(
[rank, rank, dim2]
) + np.dstack([Lambda_x] * dim2)
var4 = tau * np.matmul(var1, sparse_mat)
for t in range(dim2):
Mt = np.zeros((rank, rank))
Nt = np.zeros(rank)
if t < np.max(time_lags):
Qt = np.zeros(rank)
else:
Qt = np.matmul(
Lambda_x,
np.matmul(
ten2mat(A, 0), X[t - time_lags, :].reshape([rank * d])
),
)
if t < dim2 - np.min(time_lags):
if t >= np.max(time_lags) and t < dim2 - np.max(time_lags):
index = list(range(0, d))
else:
index = list(
np.where(
(t + time_lags >= np.max(time_lags))
& (t + time_lags < dim2)
)
)[0]
for k in index:
Ak = A[:, :, k]
Mt += np.matmul(np.matmul(Ak.T, Lambda_x), Ak)
A0 = A.copy()
A0[:, :, k] = 0
var5 = X[t + time_lags[k], :] - np.matmul(
ten2mat(A0, 0),
X[t + time_lags[k] - time_lags, :].reshape([rank * d]),
)
Nt += np.matmul(np.matmul(Ak.T, Lambda_x), var5)
var_mu = var4[:, t] + Nt + Qt
if t < np.max(time_lags):
inv_var_Lambda = inv(
var3[:, :, t] + Mt - Lambda_x + np.eye(rank)
)
else:
inv_var_Lambda = inv(var3[:, :, t] + Mt)
X[t, :] = mvnrnd(np.matmul(inv_var_Lambda, var_mu), inv_var_Lambda)
mat_hat = np.matmul(W, X.T)
X_new = np.zeros((dim2 + 1, rank))
if iters + 1 > maxiter1 - maxiter2:
X_new[0:dim2, :] = X.copy()
X_new[dim2, :] = np.matmul(
ten2mat(A, 0), X_new[dim2 - time_lags, :].reshape([rank * d])
)
X_new_plus += X_new
mat_hat_plus += np.matmul(W, X_new.T)
tau = np.random.gamma(
alpha + 0.5 * sparse_mat[position].shape[0],
1 / (beta + 0.5 * np.sum((sparse_mat - mat_hat)[position] ** 2)),
)
rmse = np.sqrt(
np.sum((dense_mat[pos] - mat_hat[pos]) ** 2)
/ dense_mat[pos].shape[0]
)
if (iters + 1) % 200 == 0 and iters < maxiter1 - maxiter2:
print("Iter: {}".format(iters + 1))
print("RMSE: {:.6}".format(rmse))
print()
W = W_plus / maxiter2
X_new = X_new_plus / maxiter2
A = A_plus / maxiter2
mat_hat = mat_hat_plus / maxiter2
if maxiter1 >= 100:
final_mape = (
np.sum(np.abs(dense_mat[pos] - mat_hat[pos]) / dense_mat[pos])
/ dense_mat[pos].shape[0]
)
final_rmse = np.sqrt(
np.sum((dense_mat[pos] - mat_hat[pos]) ** 2)
/ dense_mat[pos].shape[0]
)
print("Imputation MAPE: {:.6}".format(final_mape))
print("Imputation RMSE: {:.6}".format(final_rmse))
print()
return mat_hat, W, X_new, A
sparse_mat = dfdealMis
dense_mat = dfdens
if np.isnan(sparse_mat).any() == False:
self.dfsecond = self.dfsecond.reset_index(drop=True)
pdate = pd.DataFrame(
self.dfsecond["date"].values.astype("float32"), columns=["Datefloat"]
)
self.dfsecond["Datefloat"] = pdate["Datefloat"]
return self.dfsecond
start = time.time()
dim1, dim2 = sparse_mat.shape
rank = 10
time_lags = np.array([1, 2, (len(self.dfsecond) // 28)])
init = {
"W": 0.1 * np.random.rand(dim1, rank),
"X": 0.1 * np.random.rand(dim2, rank),
}
maxiter1 = 1100
maxiter2 = 100
a, b, c, d = BTMF(
dense_mat, sparse_mat, init, rank, time_lags, maxiter1, maxiter2
)
end = time.time()
print("Running time: %d seconds" % (end - start))
a = np.delete(a, -1, axis=1)
dfRainfall = self.dfsecond[self.Rain_list].to_numpy()
dfRainfall = np.delete(
dfRainfall,
range(len(self.dfsecond) - (len(self.dfsecond) // 28) * 28),
axis=0,
)
dfFlow = self.dfsecond[self.Flow_list].to_numpy()
dfFlow = np.delete(
dfFlow, range(len(self.dfsecond) - (len(self.dfsecond) // 28) * 28), axis=0
)
dfTemp = self.dfsecond[self.Flow_list].to_numpy()
dfTemp = np.delete(
dfTemp, range(len(self.dfsecond) - (len(self.dfsecond) // 28) * 28), axis=0
)
pdate = pd.DataFrame(
self.dfsecond["date"].values.astype("float32"), columns=["Datefloat"]
)
dfDate = pdate["Datefloat"].to_numpy()
dfDate = np.delete(
dfDate, range(len(self.dfsecond) - (len(self.dfsecond) // 28) * 28), axis=0
)
dfDate = dfDate.reshape(-1, 1)
a = a.T
wholedata = np.hstack((a, dfRainfall, dfTemp, dfDate))
wholelist = self.Depth_list + self.Rain_list + self.Temp_list + ["Datefloat"]
newFrame = DataFrame(wholedata, index=None, columns=wholelist)
self.dfsecond = newFrame
self.dfsecond = self.dfsecond.reset_index(drop=True)
return self.dfsecond
def PCA_trans(self, feature_list):
if len(feature_list) <= 2:
return self.dfsecond
test = self.dfsecond[feature_list].ffill().bfill()
Xpca = PCA(n_components=2).fit_transform(test)
pf = pd.DataFrame(Xpca, columns=["PCA1", "PCA2"])
self.dfsecond["PCA1"] = pf["PCA1"]
self.dfsecond["PCA2"] = pf["PCA2"]
for i in range(len(feature_list)):
self.dfsecond = self.dfsecond.drop(feature_list[i], axis=1)
self.dfsecond = self.dfsecond.ffill().bfill()
self.PCA_list = ["PCA1", "PCA2"]
return self.dfsecond
# use.shift(-28) made target variable
def target_made(self, potential_list):
self.lag_target = []
"""
potential_list contains the name of all the variables which cound be seen as output in this table.
"""
for i in range(len(potential_list)):
name = potential_list[i] + "28"
self.dfsecond[name] = self.dfsecond[potential_list[i]].shift(-28)
self.lag_target.append(name)
return self.dfsecond
def MICMethod(self):
"""
use MICMethod to delete those variables which have higher MIC values with some other variables.
"""
mine = MINE(alpha=0.6, c=15)
deldep_feature = []
deltem_feature = []
delflow_feature = []
if (len(self.Depth_list) != 0) & (self.target_list[0] not in self.Depth_list):
dataDepth = self.dfsecond[self.Depth_list]
data_array = np.array(dataDepth)
n = len(data_array[0, :])
for i in range(n):
for j in range(n):
mine.compute_score(data_array[:, i], data_array[:, j])
if (mine.mic() >= 0.9) & (i != j):
if self.Depth_list[j] not in deldep_feature:
deldep_feature.append(self.Depth_list[i])
break
if len(self.Temp_list) != 0:
dataTem = self.dfsecond[self.Temp_list]
data_array = np.array(dataTem)
n = len(data_array[0, :])
for i in range(n):
for j in range(n):
mine.compute_score(data_array[:, i], data_array[:, j])
if (mine.mic() >= 0.9) & (i != j):
if self.Temp_list[j] not in deltem_feature:
deltem_feature.append(self.Temp_list[i])
break
if (len(self.Flow_list) != 0) & (self.target_list[0] not in self.Flow_list):
dataflow = self.dfsecond[self.Flow_list]
data_array = np.array(dataflow)
n = len(data_array[0, :])
for i in range(n):
for j in range(n):
mine.compute_score(data_array[:, i], data_array[:, j])
if (mine.mic() >= 0.9) & (i != j):
if self.Flow_list[j] not in delflow_feature:
delflow_feature.append(self.Flow_list[i])
break
if len(self.PCA_list):
datapca = self.dfsecond[["PCA1", "PCA2"]]
data_array = np.array(datapca)
mine.compute_score(data_array[:, 0], data_array[:, 1])
if mine.mic() >= 0.9:
delpca_feature = ["PCA2"]
self.dfsecond = self.dfsecond.drop(
deldep_feature + deltem_feature + delpca_feature + delflow_feature, axis=1
)
for i in range(len(deldep_feature)):
self.Depth_list.remove(deldep_feature[i])
for i in range(len(deltem_feature)):
self.Temp_list.remove(deltem_feature[i])
for i in range(len(delflow_feature)):
self.Flow_list.remove(delflow_feature[i])
self.PCA_list = ["PCA1"]
return self.dfsecond
def KalmanCook(self):
"""
used Kalman filter to remove noise in Depth_to_Groundwater,flow, and temperature variables.
"""
def Kalman1D(data, damping=1):
observation_covariance = damping
first_value = data[0]
transition_matrix = 1
transition_covariance = 0.1
first_value
kf = KalmanFilter(
initial_state_mean=first_value,
initial_state_covariance=observation_covariance,
observation_covariance=observation_covariance,
transition_covariance=transition_covariance,
transition_matrices=transition_matrix,
)
pred_state, state_cov = kf.smooth(data)
return pred_state
dfreborn = self.dfsecond.drop(
self.Depth_list + self.Temp_list + self.Flow_list, axis=1
)
for i in range(len(self.Depth_list)):
tryArray = self.dfsecond[self.Depth_list[i]].to_numpy()
trykal = Kalman1D(tryArray, 0.1)
kallist = map(lambda x: x[0], trykal)
trykalseries = pd.Series(kallist)
dfreborn[self.Depth_list[i]] = trykalseries
for i in range(len(self.Temp_list)):
tryArray = self.dfsecond[self.Temp_list[i]].to_numpy()
trykal = Kalman1D(tryArray, 0.1)
kallist = map(lambda x: x[0], trykal)
trykalseries = pd.Series(kallist)
dfreborn[self.Temp_list[i]] = trykalseries
for i in range(len(self.Flow_list)):
tryArray = self.dfsecond[self.Flow_list[i]].to_numpy()
trykal = Kalman1D(tryArray, 0.1)
kallist = map(lambda x: x[0], trykal)
trykalseries = pd.Series(kallist)
dfreborn[self.Flow_list[i]] = trykalseries
self.dfsecond = dfreborn
return self.dfsecond, self.lag_target
def LSTMGo(self, target_variable):
"""
Target_variable is the name of the variable which would be used as dependent variable in the LSTM model.
This method will print out the results of the training phase and the test phase, and return the forcast
results of the last 28 days.
"""
self.target_variable = target_variable
self.n_test = int(((len(self.dfsecond) - 28) / 28) // 5 * 28)
self.n_train = int(((len(self.dfsecond) - 28) / 28) // 5 * 4 * 28)
self.dftrain = self.dfsecond[: self.n_train]
self.dftest = self.dfsecond[self.n_train : self.n_test + self.n_train]
if target_variable.startswith("Depth"):
fake_target = [a for a in self.Depth_list if target_variable.startswith(a)][
0
]
else:
fake_target = [a for a in self.Flow_list if target_variable.startswith(a)][
0
]
self.feature_name = (
["Datefloat"] + self.PCA_list + self.Temp_list + [fake_target]
)
dftrainX = self.dftrain[self.feature_name]
n_feature = len(dftrainX.columns.values.tolist())
dflistX = np.reshape(dftrainX.values.tolist(), (28, -1, n_feature))
dftrainY = self.dftrain[target_variable]
dflistY = np.reshape(dftrainY.values.tolist(), (28, -1, 1))
dflistX = dflistX.astype("float32")
dflistY = dflistY.astype("float32")
tensorx = torch.from_numpy(dflistX)
tensory = torch.from_numpy(dflistY)
net = lstm_reg(n_feature, 100)
criterion = nn.MSELoss()
optimizer = torch.optim.Adam(net.parameters(), lr=1e-2)
for e in range(100):
var_x = Variable(tensorx)
var_y = Variable(tensory)
out = net(var_x)
loss = criterion(out, var_y)
optimizer.zero_grad()
loss.backward()
torch.nn.utils.clip_grad_norm_(
net.parameters(), 1.1
) # gradient clipping, used to avoid Exploding Gradients
optimizer.step()
if (e + 1) % 10 == 0:
print("Epoch: {}, Loss: {:.5f}".format(e + 1, loss.data))
dftestX = self.dftest[self.feature_name]
n_feature = len(dftestX.columns.values.tolist())
dftestlistX = np.reshape(dftestX.values.tolist(), (28, -1, n_feature))
dftestY = self.dftest[self.target_variable]
dftestlistY = np.reshape(dftestY.values.tolist(), (28, -1, 1))
dftestlistX = dftestlistX.astype("float32")
dftestlistY = dftestlistY.astype("float32")
tensortestx = torch.from_numpy(dftestlistX)
tensortesty = torch.from_numpy(dftestlistY)
testvar_x = Variable(tensortestx)
testvar_y = Variable(tensortesty)
nettest = net.eval()
pred_teste = nettest(testvar_x)
loss = criterion(pred_teste, testvar_y)
print("Epoch: {}, Loss: {:.5f}".format("mse", loss.data))
a = nn.L1Loss()
maeloss = a(pred_teste, testvar_y)
print("Epoch: {}, Loss: {:.5f}".format("mae", maeloss.data))
dfpre = self.dfsecond.tail(28)
dfpreX = dfpre[self.feature_name]
n_feature = len(dfpreX.columns.values.tolist())
dfprelistX = np.reshape(dfpreX.values.tolist(), (28, -1, n_feature))
dfprelistX = dfprelistX.astype("float32")
tensorprex = torch.from_numpy(dfprelistX)
prevar_x = Variable(tensorprex)
preY = net(prevar_x)
return preY
class lstm_reg(nn.Module):
def __init__(self, input_size, hidden_size, output_size=1, num_layers=2):
super(lstm_reg, self).__init__()
self.rnn = nn.LSTM(input_size, hidden_size, num_layers, dropout=0.3)
self.reg = nn.Linear(hidden_size, output_size)
def forward(self, x):
x, _ = self.rnn(x)
s, b, h = x.shape
x = x.view(s * b, h)
x = self.reg(x)
x = x.view(s, b, -1)
return x
def output_y_hc(self, x, hc):
y, hc = self.rnn(x, hc) # y, (h, c) = self.rnn(x)
s, b, h = y.size()
y = y.view(s * b, h)
y = self.reg(y)
y = y.view(s, b, -1)
return y, hc
# **2- Random Forest**
# **2.1- Predicting the Depth_to_Groundwater_SAL 28**
# dftrain containing the training dataset while dftest containing the testing dataset.
n_train = len(dffullkal) - 28 - n_test
dftrain = dffullkal[:n_train]
dftest = dffullkal[n_train : len(dffullkal) - 28]
dftrainX = dftrain[
[
"date",
"PCA1",
"PCA2",
"Depth_to_Groundwater_SAL",
"Temperature_Orentano",
"Temperature_Monte_Serra",
]
]
n_feature = len(dftrainX.columns.values.tolist())
dflistX = np.reshape(dftrainX.values.tolist(), (28, -1, n_feature))
dftrainY = dftrain["Depth_to_Groundwater_SAL28"]
dflistY = np.reshape(dftrainY.values.tolist(), (28, -1, 1))
dflistX = dflistX.astype("float32")
dflistY = dflistY.astype("float32")
tensorx = torch.from_numpy(dflistX)
tensory = torch.from_numpy(dflistY)
dftestX = dftest[
[
"date",
"PCA1",
"PCA2",
"Depth_to_Groundwater_SAL",
"Temperature_Orentano",
"Temperature_Monte_Serra",
]
]
dftestlistX = np.reshape(dftestX.values.tolist(), (28, -1, n_feature))
dftestY = dftest["Depth_to_Groundwater_SAL28"]
dftestlistY = np.reshape(dftestY.values.tolist(), (28, -1, 1))
dftestlistX = dftestlistX.astype("float32")
dftestlistY = dftestlistY.astype("float32")
tensortestx = torch.from_numpy(dftestlistX)
tensortesty = torch.from_numpy(dftestlistY)
testvar_x = Variable(tensortestx)
testvar_y = Variable(tensortesty)
# Instantiate model with 1000 decision trees
rf = RandomForestRegressor(n_estimators=1000, random_state=42)
# Train the model on training data
rf.fit(dftrainX, dftrainY)
predictions = rf.predict(dftestX)
# RMSE
print("RMSE for predicting the Depth_to_Groundwater_SAL28 using the Random Forest")
RMSE = mean_squared_error(dftestY, predictions, squared=False)
print(RMSE)
# MAE
print("MAE for predicting the Depth_to_Groundwater_SAL28 using the Random Forest")
MAE = mean_absolute_error(dftestY, predictions)
print(MAE)
plt.figure()
plt.plot(dftestY, predictions, "bo")
plt.grid()
plt.xlabel("True")
plt.ylabel("Predicted")
plt.title("Predicting the Depth_to_Groundwater_SAL 28")
plt.show()
# **3- The Steepest Descent algorithm**
# **3.1- Predicting the Depth_to_Groundwater_SAL28**
w_hat_old = np.ones(len(dffullkal.columns))
rd = [random.randint(1, 100) for i in range(1, 9)]
w_hat = np.array(rd) # generating the same random value
y_hat_trainSAL = np.dot(dftrainX, w_hat_old)
# calculating the gradient
X_train_transposeSAL = dftrainX.T
gradient_w_hat = -2 * np.dot(dftrainX.T, dftrainY) + 2 * np.dot(
dftrainX.T, np.dot(dftrainX, w_hat)
)
# hessian matrix at point w_hat
Hessian = 4 * np.dot(X_train_transposeSAL, dftrainX)
iterations = 0
max_iterations = 1e4
old_error = []
# %%
# using the Steepest Descent algorithm
w_hat_old = np.ones(len(dftrainX.columns))
rd = [random.randint(1, 100) for i in range(1, 9)]
w_hat = np.array(rd) # generating the same random value
y_hat_trainSAL = np.dot(dftrainX, w_hat_old)
# calculating the gradient
X_train_transposeSAL = dftrainX.T
gradient_w_hat = -2 * np.dot(dftrainX.T, dftrainY) + 2 * np.dot(
dftrainX.T, np.dot(dftrainX, w_hat)
)
# hessian matrix at point w_hat
Hessian = 4 * np.dot(X_train_transposeSAL, dftrainX)
iterations = 0
max_iterations = 1e4
old_error = []
while np.linalg.norm(w_hat - w_hat_old) > 1e-8 and iterations < max_iterations:
iterations += 1
# old_error += [train_error]
w_hat_old = w_hat
gamma = np.linalg.norm(gradient_w_hat) ** 2 / np.dot(
np.dot(gradient_w_hat.T, Hessian), gradient_w_hat
)
w_hat = w_hat - gamma * gradient_w_hat # update the guess
# train_error=np.linalg.norm(np.dot(X_train,w_hat)- y_train)**2
# hyp=np.dot(X_train, w_hat)
# v = -y_train+hyp
gradient_w_hat = -2 * np.dot(dftrainX.T, dftrainY) + 2 * np.dot(
dftrainX.T, np.dot(dftrainX, w_hat)
)
y_hat_trainSAL = np.dot(dftrainX, w_hat)
y_hat_testSAL = np.dot(dftestX, w_hat)
# MAE
print(
"MAE for predicting the Depth_to_Groundwater_SAL28 using the Steepest Descent in testing phase"
)
MAE = mean_absolute_error(dftestY, y_hat_testSAL)
print(MAE)
# RMSE
print(
"RMSE for predicting the Depth_to_Groundwater_SAL28 using the Steepest Descent in testing phase"
)
RMSE = mean_squared_error(dftestY, y_hat_testSAL, squared=False)
print(RMSE)
plt.figure()
plt.plot(dftestY, y_hat_testSAL, "bo")
plt.grid()
plt.xlabel("True")
plt.ylabel("Predicted")
plt.title("Depth_to_Groundwater_SAL28 prediction with Steepest Descent Algorithm ")
plt.show()
|
#
# From Game Play
# Predict Student Performance
# AUTHOR: Mojahid Ahmad
#
#
# Table of Contents
# 1. Introduction
#
# Problem Statement
# Data Description
#
#
# 2. Import Libraries
# 3. Basic Exploration
#
# Read Dataset
# Some Information
# Data Visualization
#
#
# 4. Machine Learning Model
# 5. Conclusion
# 6. Author Message
# Introduction
# Problem Statement
# Learning is meant to be fun, which is where game-based learning comes in. This educational approach allows students to engage with educational content inside a game framework, making it enjoyable and dynamic. Although game-based learning is being used in a growing number of educational settings, there are still a limited number of open datasets available to apply data science and learning analytic principles to improve game-based learning.
# Most game-based learning platforms do not sufficiently make use of knowledge tracing to support individual students. Knowledge tracing methods have been developed and studied in the context of online learning environments and intelligent tutoring systems. But there has been less focus on knowledge tracing in educational games.
# Competition host Field Day Lab is a publicly-funded research lab at the Wisconsin Center for Educational Research. They design games for many subjects and age groups that bring contemporary research to the public, making use of the game data to understand how people learn. Field Day Lab's commitment to accessibility ensures all of its games are free and available to anyone. The lab also partners with nonprofits like The Learning Agency Lab, which is focused on developing science of learning-based tools and programs for the social good.
# If successful, you'll enable game developers to improve educational games and further support the educators who use these games with dashboards and analytic tools. In turn, we might see broader support for game-based learning platforms.
# Data description
# This competition uses the Kaggle's time series API. Test data will be delivered in groupings that do not allow access to future data. The objective of this competition is to use time series data generated by an online educational game to determine whether players will answer questions correctly. There are three question checkpoints (level 4, level 12, and level 22), each with a number of questions. At each checkpoint, you will have access to all previous test data for that section.
# |No | Columns name | Meaning |
# |:---| :--- |:--- |
# | 1 | session_id | the ID of the session the event took place in |
# | 2 | index | the index of the event for the session |
# | 3 | elapsed_time | how much time has passed (in milliseconds) between the start of the session and when the event was recorded |
# | 4 | event_name | the name of the event type |
# | 5 | name | the event name (e.g. identifies whether a notebook_click is is opening or closing the notebook) |
# | 6 | level | what level of the game the event occurred in (0 to 22) |
# | 7 | page | the page number of the event (only for notebook-related events) |
# | 8 | room_coor_x | the coordinates of the click in reference to the in-game room (only for click events) |
# | 9 | room_coor_y | the coordinates of the click in reference to the in-game room (only for click events) |
# | 10 | screen_coor_x | the coordinates of the click in reference to the player’s screen (only for click events) |
# | 11 | screen_coor_y | the coordinates of the click in reference to the player’s screen (only for click events) |
# | 12 | hover_duration | how long (in milliseconds) the hover happened for (only for hover events) |
# | 13 | text | the text the player sees during this event |
# | 14 | fqid | the fully qualified ID of the event |
# | 15 | room_fqid | the fully qualified ID of the room the event took place in |
# | 16 | text_fqid | the fully qualified ID of the |
# | 17 | fullscreen | whether the player is in fullscreen mode |
# | 18 | hq | whether the game is in high-quality |
# | 19 | music | whether the game music is on or off |
# | 20 | level_group | which group of levels - and group of questions - this row belongs to (0-4, 5-12, 13-22) |
# Import libraries
import plotly.express as px
import matplotlib.pyplot as plt
import seaborn as sns
import missingno as msno
import pandas as pd
import numpy as np
import matplotlib
from plotly.subplots import make_subplots
import plotly.graph_objects as go
# theme_colors = ['#901272', '#94A832', '#6C7CD3', '#F16262', '#4C5760']
theme_colors = [
"#901272",
"#94A832",
"#6C7CD3",
"#F16262",
"#4C5760",
"#DB8F00",
"#008B8B",
"#8B008B",
"#9ACD32",
"#4682B4",
]
import warnings
warnings.filterwarnings("ignore")
#
# Data Exploration
# Read Dataset
def read_dataset():
train = pd.read_csv(
"/kaggle/input/predict-student-performance-from-game-play/train.csv"
)
print("Train data imported successfully!")
train_labels = pd.read_csv(
"/kaggle/input/predict-student-performance-from-game-play/train_labels.csv"
)
print("Train labels data imported successfully!")
test = pd.read_csv(
"/kaggle/input/predict-student-performance-from-game-play/test.csv"
)
print("Test data imported sucessfully!")
return train, train_labels, test
raw_train_df, raw_train_labels_df, raw_test_df = read_dataset()
|
# # Entrance
# ***Variable Types***
# * **Numerical variables**: Age variable can be decimal or discrete, such as the Square Meter variable.
# * **Categorical Variables (Nominal, Ordinal)**: Female, Male; They are variables that express a class, such as Survive, Unable to Survive. If there is no difference between the classes, for example, Female, Male Nominal Categorical variable. Educational status, such as middle school, high school, undergraduate, graduate, is an ordinal variable.
# * **Dependent Variable (target, dependent, output, response)**: The target variable in the problem we are interested in is called. For example, being sick or not is the target variable in a disease prediction. In house or car price estimation, the prices of houses or cars are the variable to be reached.
# * **Independent Variable (feature, independent, input, column, predictor, explanatory)** : Other variables that we assume affect the target.
# ***Learning Types***
# * **Reinforcement Learning**: An example can be given as an example of learning by trial and error, by hitting and punishing while trying to get out of a closed section. A driverless car is an example.
# * **Supervised Learning**: It is also called supervised learning. If there is a dependent variable in the data set, it is called supervised learning.
# * **Unsupervised Learning**: Also called unsupervised learning. If there is no dependent variable in the data set, it is called unsupervised learning. Here, a kind of clustering is done and the result is tried to be reached.
# # Types of Problems
# * **In regression problems**, the dependent variable is numerical.
# * **Sınıflandırma problemlerinde** bağımlı değişken kategoriktir.
# # Model Validation
# * **Holdout Method (Test Set Method)**
# In this method, the original data set is divided into training set and test set. The model is modeled with the training set and then the model is tested with the unseen test set.
# * **K-Fold Cross Validation**
# If the data set is rich, the holdout method is used if there is enough data. I'm talking about 100000 data.
# K-Fold Cross Validation can be used in two ways. Let's examine them now.
# 1. For example, the original data set is divided into 5 (the number of divisions is an example, it can be 5-6). It is trained with 4 pieces and tested with 1 piece. It is decided according to the success evaluation averages of the 5 possible combinations.
# 2. The original data set is first divided into training set and test set. Cross validation is done by dividing the separated training set into 5 parts, for example. In the last case
# The success evaluation is made by inserting the test set into the allocated test set.
#
# # Bias — Variance Tradeoff
# **Building a Model**: It is to extract the relationship between the dependent variable and the independent variable, the essence and direction of the relationship.
# **Overfitting**: The model memorizes the data. High variance means high variability.
# **Underfitting**: The model cannot learn the data. High Bias i.e. it is closer to some observations.
# What we are looking for is to build the right model with low bias and low variance. It is aimed to learn the self-state of the data set.
# **Model Complexity**:
# Although it varies according to the model, for example, adding exponential expressions in linear models, that is, refining the model, with the number of branches in **tree methods**, with the number of iterations in tree methods based on optimization methods, for example, with the number of layers, cell number, epoch in **artificial neural networks**. As the parameters are increased, the model complexity will increase.
# # Linear Regression
# Purpose here, dependent and independent variable
# or to model the relationship between variables linearly.
#
# y(hat)=b + w1x1+ w2x2 + ...
#
# What is learned is actually constants and weights. These values are the core of the data, expressing the pattern.
# **b is beta**, constant, bias, intercept; **The expression w** can appear as weight, weight, coefficient.
# ***Finding the weights***
# The weights are found by finding the b and w values that can minimize the sum or average of the squares of the differences between the actual values and the predicted values.
# It is studied on the question of where to put the right optimum way close to the points where these data are located.
# **Achievement Evaluation (MSE, RMSE, MAE)**
# MSE, RMSE, MAE calculations are used while evaluating success in the regression model.
# ***MSE*** : In order to eliminate the measurement problem in the Mean Squared Error indicator, squaring is performed. It is acceptable for the mean of these values to be close to the dependent variable. In this case, since the square is taken, the effect of the errors increases a lot. THE SMALLER THE VALUE, THE BETTER.
# ***RMSE***: It aims to reduce the effect of squaring each error more accurately in the Root Mean Squared Error indicator. It is the square root of the MSE expression.
# ***In the MAE*** : Mean Absolute Error indicator, the absolute values of each error are taken and averaged.
# There is no question of which of these indicators is better. Each of these are separate metrics. One is preferred and referenced. Whichever is lower, I can't take it and use it.
# # Applying Linear Regression
#
import numpy as np
import pandas as pd
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
import matplotlib.pyplot as plt
import seaborn as sns
pd.set_option("display.float_format", lambda x: "%.2f" % x)
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_squared_error, mean_absolute_error
from sklearn.model_selection import train_test_split, cross_val_score
df = pd.read_csv("/kaggle/input/advertising-data/advertising.csv")
df.shape
df.head()
df.describe().T
X = df[["TV"]]
y = df[["sales"]]
reg_model = LinearRegression().fit(X, y)
reg_model.intercept_[0]
reg_model.coef_[0][0]
reg_model.intercept_[0] + reg_model.coef_[0][0] * 200
reg_model.intercept_[0] + reg_model.coef_[0][0] * 300
# MSE
y_pred = reg_model.predict(X)
mean_squared_error(y, y_pred)
y.mean()
y.std()
# RMSE
np.sqrt(mean_squared_error(y, y_pred))
# MAE
mean_absolute_error(y, y_pred)
# R-KARE
reg_model.score(X, y)
|
from duckduckgo_search import ddg_images
from fastcore.all import *
from fastdownload import download_url
from fastai.vision.all import *
from urllib.error import HTTPError
def search_images(search_term, max_images=30):
print(f"\nSearching for '{search_term}'")
return L(ddg_images(search_term, max_results=max_images)).itemgot("image")
def predict_search(search_term):
search_urls = search_images(search_term, max_images=5)
filename = "searched_image.jpg"
for i in range(5):
try:
download_url(search_urls[i], filename, show_progress=False)
is_car, _, probs = learner.predict(filename)
print(f"This is a: {is_car}.")
print(f"Probability it is a car: {probs[0]:.4f}")
break
except HTTPError:
if i == 4:
printf("Cannot find any images for this search term")
else:
print(f"Image {search_urls[i]} not found, trying next...")
search_terms = "motorbike", "car"
path = Path("car_or_not")
for search_term in search_terms:
dest = path / search_term
dest.mkdir(exist_ok=True, parents=True)
download_images(dest, urls=search_images(f"regular {search_term}"))
download_images(dest, urls=search_images(f"sport {search_term}"))
download_images(dest, urls=search_images(f"classic {search_term}"))
resize_images(path / search_term, max_size=400, dest=path / search_term)
failed = verify_images(get_image_files(path))
failed.map(Path.unlink)
len(failed)
dataloaders = DataBlock(
blocks=(ImageBlock, CategoryBlock),
get_items=get_image_files,
splitter=RandomSplitter(valid_pct=0.2, seed=43),
get_y=parent_label,
item_tfms=[Resize(192, method="squish")],
).dataloaders(path, bs=32)
dataloaders.show_batch(max_n=8)
learner = vision_learner(dataloaders, resnet18, metrics=error_rate)
learner.fine_tune(3)
predict_search("volvo xc40")
predict_search("seat ibiza")
predict_search("honda cb650f")
predict_search("honda africa twin")
predict_search("citroen xara picasso")
|
# # Project 4
# My goal is to try some polars operations including filtering, indexing, updating string and numeric columns, updating a column based on a window function; and compare polars, Dask to pandas on creating some feature enginering
data_path = "/kaggle/input/dataSales/project_2_data"
import pandas as pd
import numpy as np
import polars as pl
import dask.dataframe as dd
import time
from datetime import timedelta
# Pandas
# Load sales data
data_pd = pd.read_parquet(f"{data_path}/sales_data.parquet")
# Load Prices data
prices_pd = pd.read_parquet(f"{data_path}/prices.parquet")
prices_pd = prices_pd.reset_index(drop=False)
# Merge prices data
data_with_prices_pd = pd.merge(
data_pd.reset_index(), prices_pd, how="left", on=["date", "store_id", "item_id"]
).set_index(["date", "id"])
# Dask
# Load sales data
data_dd = dd.read_parquet(f"{data_path}/sales_data.parquet", npartitions=4)
# Load Prices data
prices_dd = dd.read_parquet(f"{data_path}/prices.parquet", npartitions=4)
# Merge prices data
data_with_prices_dd = dd.merge(
data_dd, prices_dd, on=["date", "store_id", "item_id"], how="left"
)
print(data_dd.npartitions)
# Polars
# Load sales data
data_pl = pl.read_parquet(f"{data_path}/sales_data.parquet")
# Load Prices data
prices_pl = pl.read_parquet(f"{data_path}/prices.parquet")
# Merge prices data
data_with_prices_pl = data_pl.join(
prices_pl, on=["date", "store_id", "item_id"], how="left"
)
# ## Some test on basic polars operations (filter, select ...)
data_dd.head()
data_pl.head()
print(data_pl.head())
data_pl.head().to_pandas()
data_pl[0, :]
data_pl[:, ["dept_id", "sales"]].head()
data_pl.select(["dept_id", "sales"]).head()
data_pl.filter(pl.col("sales") > 600).head()
data_pl.filter(pl.col("dept_id").is_in(["HOUSEHOLD_2", "HOUSEHOLD_1"])).head()
data_pl.select([pl.col("dept_id").n_unique()])
data_pl.select(
[
pl.col("sales").min().alias("min"),
pl.col("sales").mean().alias("mean"),
pl.col("sales").median().alias("median"),
pl.col("sales").max().alias("max"),
pl.col("sales").std().alias("std_dev"),
]
)
data_pl.select([pl.col("sales")]).describe()
price = data_pl.select([pl.col("sales")])
price.to_pandas().hist(bins=40)
probs = [0, 0.25, 0.5, 0.75, 1]
percentiles = [price.quantile(prob)[0, 0] for prob in probs]
pd.DataFrame(dict(probs=probs, percentiles=percentiles))
data_pl.head()
# Update a string column
data_pl.with_columns(
(pl.col("item_id") + "_" + pl.col("store_id")).alias("unique_id2")
).head()
# Update a numeric column
data_pl.with_columns(
[
(pl.col("item_id") + "_" + pl.col("store_id")).alias("unique_id2"),
(pl.col("sales") + 50).alias("price_50"),
]
).tail()
# Create a column with a constant (based on the mean)
data_pl.select([pl.all(), pl.col("sales").mean().alias("price_avg")]).head()
# Update values based on window function
data_pl.with_columns(
[pl.col("sales").mean().over("dept_id").alias("price_by_dept_id_code")]
).tail()
# Computing the difference between the sales price and the dept_id average.
df_dept_window = data_pl.select(
[
pl.col("dept_id"),
pl.col("sales"),
pl.col("sales").mean().over("dept_id").alias("price_dept_id"),
(pl.col("sales") / pl.col("sales").mean().over("dept_id") - 1).alias(
"price_div_dept_average"
),
]
)
df_dept_window.head()
df_dept_window.sample(1000000).to_pandas().price_div_dept_average.hist(
bins=30, range=[0, 5]
)
data_pl.head()
# ## Feature enginer comparison
data_pd.head()
# Pandas
data_pd = data_pd.reset_index()
data_pd["rolling_7_mean"] = (
data_pd.groupby("id", as_index=False)["sales"].rolling(7).mean()["sales"]
)
data_pd["rolling_14_mean"] = (
data_pd.groupby("id", as_index=False)["sales"].rolling(14).mean()["sales"]
)
data_pd["rolling_28_mean"] = (
data_pd.groupby("id", as_index=False)["sales"].rolling(28).mean()["sales"]
)
data_pd["rolling_7_std"] = (
data_pd.groupby("id", as_index=False)["sales"].rolling(7).std()["sales"]
)
data_pd["rolling_14_std"] = (
data_pd.groupby("id", as_index=False)["sales"].rolling(14).std()["sales"]
)
data_pd["rolling_28_std"] = (
data_pd.groupby("id", as_index=False)["sales"].rolling(28).std()["sales"]
)
data_pd.tail()
data_dd.head()
data_dd.dtypes
# Dask dataframe
data_dd = data_dd.reset_index(drop=True)
data_dd = data_dd.assign(
lag_7=data_dd["sales"].shift(7),
lag_21=data_dd["sales"].shift(21),
lag_28=data_dd["sales"].shift(28),
sales=data_dd["sales"] - data_dd["sales"].shift(7),
rolling_7_mean=data_dd["sales"].rolling(window=7).mean(),
rolling_21_mean=data_dd["sales"].rolling(window=21).mean(),
rolling_28_mean=data_dd["sales"].rolling(window=28).mean(),
rolling_7_std=data_dd["sales"].rolling(window=7).std(),
rolling_21_std=data_dd["sales"].rolling(window=21).std(),
rolling_28_std=data_dd["sales"].rolling(window=28).std(),
)
data_dd = data_dd.compute() # Compute the Dask DataFrame to get back a pandas DataFrame
data_dd.tail()
# Polars
data_pl = data_pl.with_columns(
[
(pl.col("sales").shift(7).over("id").alias("lag_7")),
(pl.col("sales").shift(21).over("id").alias("lag_21")),
(pl.col("sales").shift(28).over("id").alias("lag_28")),
(pl.col("sales") - pl.col("sales").shift(7).over("id")).alias("sales"),
(
pl.col("sales")
.rolling_mean(window_size=7)
.over("id")
.alias("rolling_7_mean")
),
(
pl.col("sales")
.rolling_mean(window_size=21)
.over("id")
.alias("rolling_21_mean")
),
(
pl.col("sales")
.rolling_mean(window_size=28)
.over("id")
.alias("rolling_28_mean")
),
(pl.col("sales").rolling_std(window_size=7).over("id").alias("rolling_7_std")),
(
pl.col("sales")
.rolling_std(window_size=21)
.over("id")
.alias("rolling_21_std")
),
(
pl.col("sales")
.rolling_std(window_size=28)
.over("id")
.alias("rolling_28_std")
),
]
)
print(data_pl.tail())
|
# # Deployment on AWS
# In this phase we will deploy our models and data pipeline.
# We are going to use Amazon Web Services (AWS), where there are various ways to deploy our project:
# - Run an EC2 instance with a web server
# - Build a container image and deploy it with Lambda + API Gateway (to manage prediction requests) and ECS + Fargate (for our ETL process)
# - Use AWS SageMaker (in this case we would have to rewrite a lot of code)
# We are going with Lambda and ECS, because it's faster and cheaper.
# Hence we will see how to setup:
# - AWS S3, to store trained models.
# - AWS Lambda (+ API Gateway), to expose HTTP endpoints to request predictions
# - AWS ECS + Fargate, to run the ETL process + training of new models
# For starters, we focus on creating a docker image with all the functionalities we need.
# Then we are going to setup all configurations for predicting results with Lambda.
# To conclude we are going to setup all configurations for ETL and training.
# ## Simple Storage Service (S3)
# AWS S3 (Simple Storage Service) is a cloud-based object storage service provided by Amazon Web Services (AWS). It allows users to store and retrieve data from anywhere on the internet, using a simple web interface or API calls.
# S3 is designed to provide high durability, scalability, and availability for storing and retrieving large amounts of data, ranging from a few kilobytes to multiple terabytes. It offers multiple storage classes to meet different data access needs, including Standard, Infrequent Access (IA), and Glacier.
# S3 also provides features such as versioning, encryption, and access control, making it a secure and reliable solution for storing and managing data. It can be used for a variety of use cases, including website hosting, data backup and archiving, content distribution, and big data analytics.
# S3 is integrated with other AWS services, such as Amazon EC2, AWS Lambda, AWS Glue, Amazon CloudFront, and Amazon SNS, making it a flexible and powerful solution for building a wide range of cloud-based applications.
# We simply create a new **bucket** within AWS S3 dashboard. We will store there our trained models.
# https://aws.amazon.com/s3/
# ## Docker
# We will use Docker as our container engine.
# A container is portable software that runs in a virtualized environment independent from:
# - Platform (cloud, desktop, on premises, ...)
# - Operating System (Windows, Linux, MacOS, ...)
# - Programming Language (Python, Java, NodeJS, ...)
# A Dockerfile is used to build a container image, which is stored in a registry.
# The images in our registry can then be used to run containers (an instance of that image).
# Let's create a Dockerfile, following AWS documentation.
# https://docs.aws.amazon.com/lambda/latest/dg/images-create.html
# The base image *public.ecr.aws/lambda/python:3.7* has a predefined entrypoint that wrap our code an allows it to run inside a lambda handler. This is required when using a container in a Lambda function.
# We copy requirements.txt and run pip install to install our dependencies.
# **LAMBDA_TASK_ROOT** is an environment variable defined in the AWS image that refers to /var/task, using --target "${LAMBDA_TASK_ROOT}" will install dependencies under /var/task.
# We also copy all our project in ${LAMBDA_TASK_ROOT}.
# CMD specify the command to be executed by the container. The entrypoint specified by the AWS Lambda base image requires as CMD a function that handles the Lambda function.
def handler(event, context):
# do stuff
return {"statusCode": 200, "body": "data"}
# ### Upload image to Elastic Container Registry (ECR)
# ECR is the AWS Container Registry, where we are going to upload our images.
# https://aws.amazon.com/ecr/
# Before building the image we create a new repository in AWS ECR.
# This will give us a URI similar to [account_id].dkr.ecr.eu-south-1.amazonaws.com/[repository_name].
# We can use this to push a container image with the Docker CLI.
# First we have to login with AWS CLI credentials. For convenience we created a *portfolio* profile with the AWS CLI to authenticate with our AWS resources.
# Then we create the image with docker build command, tagging the image with [account_id].dkr.ecr.eu-south-1.amazonaws.com/[repository_name]:[tag].
# Let's push the image to ECR.
# Every time we push a new image, AWS creates automatically a sha identifier associated with the container for versioning purposes.
# Now we can use the image saved under [account_id].dkr.ecr.eu-south-1.amazonaws.com/porfolio:version_1 with AWS resources like Lambda and ECS.
# ## AWS Lambda
# AWS Lambda is a serverless compute service provided by Amazon Web Services (AWS). It allows us to run our code in the cloud without managing servers, operating systems, or infrastructure.
# https://aws.amazon.com/lambda/
# ### Setup Lambda
# To setup a Lambda function to act as a serverless predictor we a Lambda using our container image.
# Selecting *Create function* from AWS Lambda dashboard we can choose *Container image* as configuration option.
# Here we can choose the *function name*, we choose **predict** and specify a *Container image URI*.
# With the new Lambda function created we need to add an **API Gateway** as trigger of the function. This allows us to send request to a specified endpoint that will call the Lambda handler method as specify in the CMD line of the Dockerfile of our image.
# Once set the API Gateway we will have an endpoint similar to: https://[api_gateway_id].execute-api.eu-south-1.amazonaws.com/default/predict.
# We can use this URL in a simple HTTP request to get the predicted values.
# ### Predict with Lambda
# To predict with lambda we need to define the app.handler method, inside the app.py file.
# This handler method will take *event* and *context* as input parameters and then call a predict method.
# app.py
def handler(event, context):
"""
:param event: event parameters passed by Lambda handler
:param context: context parameters passed by Lambda handler
:return: json that Lambda will wrap as body of an HTTP response
Note that request will have this body:
event['body'] : {
"model":["HYBRID" | "LINEAR" | "ARIMA" ], the model we want to use for prediction
"target": targetXXX, # the target we want to predict
"start_date": YYYY-MM-DD, the start date of the predicition
"end_date": YYYY-MM-DD, the end date of the prediction
}
"""
try:
d = json.loads(event["body"])
if d is not None:
r = predict(
d["model"],
d["target"],
start_date=pd.Timestamp(d["start_date"]),
end_date=pd.Timestamp(d["end_date"]),
)
return {"statusCode": 200, "body": json.dumps(r)}
except Exception as e:
error = e
return {"statusCode": 400, "body": e}
# The handler reads the event parameter and call the predict method passing the chosen model, target and dates.
# For this example, as we said in Modeling we use the hybrid model with exogenous features.
# This predict method takes the input parameters,
# load the trained model stored on S3 and then predict values on specified dates.
from portfolio_optimization_helper import get_df_from_table
import pandas as pd
from sklearn.preprocessing import StandardScaler
from sklearn.decomposition import PCA
from statsmodels.tsa.deterministic import DeterministicProcess
import pickle
from portfolio_optimization_helper import get_object_from_s3
from kaggle_secrets import UserSecretsClient
user_secrets = UserSecretsClient()
BUCKET_ID = user_secrets.get_secret("BUCKET_ID")
def standardize_features(df):
scaled_features = StandardScaler().fit_transform(df.values)
return pd.DataFrame(scaled_features, index=df.index, columns=df.columns)
def apply_pca(df, pca_components):
idx = df.index
cols = [f"featurePCA{i + 1}" for i in range(pca_components)]
df = PCA(n_components=pca_components).fit_transform(df)
df = pd.DataFrame(df, columns=cols)
df.index = idx
return df
def get_X(df, cols):
X = df[cols].dropna()
X = standardize_features(X)
pca_components = int(len(cols) / 2)
X = apply_pca(X, pca_components)
return X
def make_lags(df, lags):
res = df.copy()
old_cols = df.columns
for i in range(1, lags + 1):
for col in old_cols:
res[f"{col}_lag_{i}"] = res[col].shift(i)
return res.drop(old_cols, axis=1)
def predict(model_name, target, start_date, end_date):
"""
:param model_name: model name
:param target: target name
:param start_date: start date of the prediction
:param end_date: end date of the prediction
:return:
"""
df = get_df_from_table("pivot")
df["date"] = pd.to_datetime(df["date"])
df = df.sort_values(by="date").set_index("date").apply(pd.to_numeric).asfreq("MS")
feature_columns = [col for col in df.columns if "feature" in col]
if model_name == "HYBRID":
y = df[[target]].dropna()
X = get_X(df, feature_columns)
y, X = y.align(X, join="inner", axis=0)
dp = DeterministicProcess(
index=y.index,
constant=True,
order=2,
drop=True,
seasonal=False,
)
X_dp = dp.in_sample()
lags = 1
X_lags = make_lags(y, lags=lags)
X_lags = pd.concat([X_lags, make_lags(X, lags=lags)], axis=1)
X_lags = X_lags.fillna(0.0)
X_dp = X_dp[(X_dp.index >= start_date) & (X_dp.index <= end_date)]
X_lags = X_lags[(X_lags.index >= start_date) & (X_lags.index <= end_date)]
model_trend_season = pickle.loads(
get_object_from_s3(f"hybrid_trend_estimator-{target}", BUCKET_ID)
)
y_hat = pd.DataFrame(
model_trend_season.predict(X_dp), index=X_dp.index, columns=[target]
)
model_cycle = pickle.loads(
get_object_from_s3(f"hybrid_cycle_estimator-{target}", BUCKET_ID)
)
y_hat += pd.DataFrame(
model_cycle.predict(X_lags), index=X_lags.index, columns=[target]
)
return y_hat
predict("HYBRID", "target266", "2020-01-01", "2020-08-01")
# Now that are Lambda is ready, we need to deploy our ETL + Training to AWS in order to train our models, so that we can use them with Lambda to serve predictions.
# ## Elastic Container Service (ECS)
# AWS Elastic Container Service (ECS) is a fully-managed container orchestration service provided by Amazon Web Services. It allows users to easily run and scale containerized applications in the cloud using Docker containers.
# With ECS, users can create and manage clusters of EC2 instances, which can then be used to deploy and run containerized applications. Users can define and configure their containerized applications using task definitions, which specify the Docker image, container port mappings, and other settings required to run the application.
# ECS also includes a service scheduler that allows users to easily deploy, manage, and scale their applications across multiple instances. The service scheduler monitors the health of instances and containers and automatically replaces failed instances or containers to maintain availability and reliability.
# https://aws.amazon.com/ecs/
# ### AWS Fargate
# We will use AWS Fargate to deploy our container.
# AWS Fargate is a serverless compute engine provided by Amazon Web Services (AWS) for running containers. It allows users to run Docker containers without the need to manage the underlying infrastructure, including servers, operating systems, or clusters.
# This solution is ideal to because it allows execution of long running task (like data collection, ETL, model training, ...).
# https://aws.amazon.com/fargate/
# ### ECS setup
# First we need to create an **ECS cluster**, *ecs_portolio_cluster*.
# An Amazon ECS cluster groups together tasks, and services, and allows for shared capacity and common configurations. All of your tasks, services, and capacity must belong to a cluster.
# Then we create a new **task definition**, *portfolio_etl_training*.
# A task definition is required to run Docker containers in Amazon ECS. When creating a task definition we can specify the URI of our container, we are going to use the same image we are using for Lambda: [account_id].dkr.ecr.eu-south-1.amazonaws.com/porfolio:version_1.
# Then we can specify a new *ENTRYPOINT* to execute a different command on the container. The new entry point is: *python,aws_fargate.py*, that call the aws_fargate.py file containing the code to execute for ETL to update our data and then call the training method to train our models and store them on the S3 bucket.
def training():
# Get data from pivot table and transform them to be ready for modeling.
feature_columns, target_columns, df = get_df(stationary=True, mode="diff")
# For each target, train a new model and store its pickle serialization to S3
for t_col in target_columns:
X = get_X(df, feature_columns)
y = df[[target]].dropna()
y, X = y.align(X, join="inner", axis=0)
dp = DeterministicProcess(
index=y.index,
constant=True,
order=2,
drop=True,
seasonal=False,
)
X_dp = dp.in_sample()
X_lags = make_lags(y, lags=lags)
X_lags = pd.concat([X_lags, make_lags(X, lags=lags)], axis=1)
X_lags = X_lags.fillna(0.0)
model = LinearRegression()
model.fit(X_dp, y)
# model.pushs3
put_object_to_s3(
pickle.dumps(model), BUCKET_ID, f"hybrid_trend_estimator-{target}"
)
y_hat = pd.DataFrame(model.predict(X_dp), index=y.index, columns=y.columns)
y -= y_hat
model = GradientBoostingRegressor()
model.fit(X_lags, y.values.ravel())
# model.pushs3
put_object_to_s3(
pickle.dumps(model), BUCKET_ID, f"hybrid_cycle_estimator-{target}"
)
def aws_fargate():
# donwload data from sources: FRED, Yahoo Finance, Investing.com and OECD
extract()
# Clean and trasform data then load them into pivot table on PostgresSQL
transform_and_load()
# train our models with the new training data and save them into S3
training()
# This task can be manually executed from ECS dashboard or can be scheduled to repeat periodically.
# Now that we have our data pipeline and prediction endpoint in place let's use it to do some prediction.
# requests.get => prediction
import requests
import pandas as pd
from configparser import ConfigParser
from portfolio_optimization_helper import get_df_from_table
from kaggle_secrets import UserSecretsClient
user_secrets = UserSecretsClient()
url = user_secrets.get_secret("lambda_api_gateway_url")
target = "target266"
r = requests.post(
url,
json={
"model": "HYBRID",
"target": target,
"start_date": "2020-01-01",
"end_date": "2020-08-01",
},
)
df = pd.DataFrame(r.json())
df.columns = ["date", target]
df["date"] = pd.to_datetime(df["date"])
df[target] = pd.to_numeric(df[target])
df = df.set_index("date")
df
# Let's plot the prediction against actual values.
import matplotlib.pyplot as plt
# plot actual + predictions
df_actual = get_df_from_table("pivot")
df_actual["date"] = pd.to_datetime(df_actual["date"])
df_actual = (
df_actual.sort_values(by="date")
.set_index("date")
.apply(pd.to_numeric)
.asfreq("MS")[[target]]
)
df_actual = df_actual.rename({target: f"{target}_actual"}, axis=1)
df_actual = df_actual[
(df_actual.index >= pd.Timestamp("2017-01-01"))
& (df_actual.index <= pd.Timestamp("2020-08-01"))
]
df_join = pd.merge(df, df_actual, how="right", left_index=True, right_index=True)
plt.plot(df_join.index, df_join[target], color="red", label="prediction")
plt.plot(df_join.index, df_join[f"{target}_actual"], color="blue", label="actual")
plt.legend()
plt.show()
# ## Project conclusion
# We started our journey with this question in mind.
# **Problem**: Is it possible to use macroeconomic data to "predict" an optimal asset allocation for a portfolio to achieve better risk-adjusted returns?
# To answer this we have tried different modeling techniques, but we have seen that the prediction accuracy is not extraordinary.
# We had assumed, at the beginning, that the "main" feature predictors would have been the exogenous features, but we found out that the target past values (aka endogenous features) also help the model accuracy by far.
# What kind of returns can we expect by using our model to perform portfolio asset allocation?
# Let's find out for an example period (2010-2020).
# We use these targets both as target for our model and benchmark for our portfolio.
# - Equity: SP500, target259
# - Real Estate: HousingPrices OECD, target267
# - Commodity: Gold, target266
# - Bonds: 10Y Treasury, target256
# - Cash: 3Mo bill, target71
def position_by_returns(data):
"""
For this example we want only long position.
data contains expected returns for each of our targets.
to compute portfolio position (in percent terms) we divide each
target expected return by the sum of the POSITIVE expected returns.
We ignore negative expected returns as it would involve short positions.
example
data = {"t1": 0.3, "t2": 0.2, "t3": -0.1}
sum of positive returns = 0.5
result = {"t1": 0.6, "t2": 0.4, "t3": 0}
"""
result = {}
sum_returns = 0
for k, v in data.items():
if v > 0:
sum_returns += v
for k, v in data.items():
if v > 0:
result[k] = v / sum_returns
else:
result[k] = 0
return result
def get_current_price(df, column, year):
"""
get the df[column] price on the last date of the specified year.
"""
return df.loc[pd.Timestamp(f"{year}-12-01"), column]
def get_pivot_df():
"""
get df from postgreSQL table "pivot"
"""
df = get_df_from_table("pivot")
df["date"] = pd.to_datetime(df["date"])
df = df.sort_values(by="date").set_index("date").apply(pd.to_numeric).asfreq("MS")
return df
def get_prediction_df(url, json):
r = requests.post(url, json=json)
df = pd.DataFrame(r.json())
df.columns = ["date", json["target"]]
df["date"] = pd.to_datetime(df["date"])
df[json["target"]] = pd.to_numeric(df[json["target"]])
df = df.set_index("date")
return df
def compute_portfolio_value(portfolio, returns, year):
"""
compute portfolio mark-to-market value by multiplying the assets
contained in the portfolio for their respective current market prices
cash price is = 1 + short-term interest rate
bond price is computed from the long-term interest rate
assuming 10y maturity, zero-coupon and face value = 1000
"""
portfolio_value = 0
for k, v in portfolio.items():
# for cash
if k == "target71":
price = 1 + returns[k]
else:
price = get_current_price(df, k, year)
# for bond
if k == "target256":
price = 1000 / pow(1 + price / 100, 10)
portfolio_value += v * price
return portfolio_value
def compute_return(pred_df, target, initial_value):
"""
compute the returns as a simple ROI calculation based on
final value and initial value.
cash return is the average yield of short-term 3Mo bills.
bond price is computed from the long-term interest rate
assuming 10y maturity, zero-coupon and face value = 1000
"""
final_value = pred_df.iloc[-1, 0]
# for cash
if target == "target71":
return (final_value + initial_value) / 2
else:
# for bond
if target == "target256":
initial_value = 1000 / pow(1 + initial_value / 100, 10)
final_value = 1000 / pow(1 + final_value / 100, 10)
return final_value / initial_value - 1
def allocate_portfolio_current(allocation, df, year, portfolio_value):
"""
calculate asset quantity to allocate in the portfolio based on
their current price.
We have only long positions, so the computation is fairly easy.
"""
portfolio_current_value = 0
for k, v in allocation.items():
# keep only long positions
if v > 0:
# for cash
if k == "target71":
price = 1
else:
price = get_current_price(df, k, year)
# for bond
if k == "target256":
price = 1000 / pow(1 + price / 100, 10)
portfolio_current[k] = portfolio_value * v / price
portfolio_current_value += portfolio_value * v
# print(k, v, price, portfolio_value * v / price)
else:
portfolio_current[k] = 0
return portfolio_current
target_list = ["target259", "target267", "target266", "target256", "target71"]
model = "HYBRID"
# Beginning year for simulation
start_year = 2010
simulation_years = 10
all_predictions_df = None
# get all predictions
for t in target_list:
if all_predictions_df is None:
all_predictions_df = get_prediction_df(
url,
{
"model": model,
"target": t,
"start_date": f"{start_year}-01-01",
"end_date": f"{start_year+simulation_years}-12-01",
},
)
else:
all_predictions_df = pd.merge(
all_predictions_df,
get_prediction_df(
url,
{
"model": model,
"target": t,
"start_date": f"{start_year}-01-01",
"end_date": f"{start_year+simulation_years}-12-01",
},
),
how="inner",
left_index=True,
right_index=True,
)
all_predictions_df.tail()
import datetime
portfolio_start_value = 100
portfolio_value = 0
portfolio_history = []
portfolio_current = {}
# get pivot from postgreSQL
df = get_pivot_df()[target_list]
for i in range(simulation_years):
# print(f"STEP {i+1}")
start_date = f"{start_year + i}-01-01"
end_date = f"{start_year + i}-12-01"
if i == 0:
portfolio_value = portfolio_start_value
else:
# get current portfolio value
portfolio_value = compute_portfolio_value(
portfolio_current, returns, start_year + i - 1
)
portfolio_history.append(
{"date": datetime.date(start_year + i, 1, 1), "Portfolio": portfolio_value}
)
returns = {}
for t in target_list:
# get prediction for the target in the current period
pred_df = all_predictions_df.copy()[[t]]
pred_df = pred_df[
(pred_df.index >= pd.Timestamp(start_date))
& (pred_df.index <= pd.Timestamp(end_date))
]
initial_value = df.loc[pd.Timestamp(start_date), t]
# calculate asset return
returns[t] = compute_return(pred_df, t, initial_value)
# calculate asset % allocation based on their returns
allocation = position_by_returns(returns)
# print("ALLOCATION", allocation)
# calculate asset quantity allocation based on their price
portfolio_current = allocate_portfolio_current(
allocation, df, start_year + i - 1, portfolio_value
)
# print("PORTFOLIO", portfolio_current)
# Compute final portfolio value
portfolio_value = compute_portfolio_value(portfolio_current, returns, start_year + i)
portfolio_history.append(
{"date": datetime.date(start_year + i + 1, 1, 1), "Portfolio": portfolio_value}
)
portfolio_df = pd.DataFrame(portfolio_history)
portfolio_df["date"] = pd.to_datetime(portfolio_df["date"])
# join to have our portfolio and benchmark values in the same df
portfolio_df = pd.merge(df, portfolio_df, how="inner", left_index=True, right_on="date")
portfolio_df = portfolio_df.set_index("date")
# Transform bond yield to bond price
portfolio_df["target256"] = portfolio_df["target256"].apply(
lambda x: 1000 / pow(1 + x / 100, 10)
)
# Cash
portfolio_df["target71"] += 1
legend_names = {
"target259": "Equities",
"target267": "Real Estate",
"target266": "Gold",
"target256": "Bonds",
"target71": "Cash",
}
for t in target_list:
# Cash, the cumulative return is the cumulate product of the returns
if t == "target71":
portfolio_df[t] = portfolio_df[t].cumprod() * portfolio_start_value
else:
portfolio_df[t] = (
portfolio_df[t] / portfolio_df[t].iloc[0] * portfolio_start_value
)
portfolio_df = portfolio_df.rename(legend_names, axis=1)
portfolio_df.plot()
plt.legend()
plt.show()
|
# run this command then restart the session to get the correct version for opencv
# dont forget to comment before restarting
import os
import cv2
import numpy as np
import matplotlib.pyplot as plt
src_folder = "/kaggle/input/images-dataset/data/"
def load_data(path):
images = os.listdir(path)
data = []
for image in images:
i = cv2.imread(os.path.join(path, image))
i = cv2.resize(i, (128, 128)) # resize the image to a 128x128
i = cv2.cvtColor(i, cv2.COLOR_BGR2GRAY)
# i = i/255.0 #cant do this cv2.detectAndCompute requires 8bit vals
data.append(i)
return np.array(data)
cats = load_data(src_folder + "cats")
dogs = load_data(src_folder + "dogs")
horses = load_data(src_folder + "horses")
# 0->cat
# 1->dog
# 2->horse
num_cats = cats.shape[0]
num_dogs = dogs.shape[0]
num_horses = horses.shape[0]
dataset = np.concatenate((cats, dogs, horses))
samples = dataset.shape[0]
labels = np.zeros((samples))
labels[num_cats : num_cats + num_dogs] = 1
labels[num_cats + num_dogs :] = 2
from sklearn.model_selection import train_test_split
from tensorflow.keras.utils import to_categorical
X_train, X_test, y_train, y_test = train_test_split(dataset, labels, test_size=0.3)
y_train = to_categorical(y_train, 3)
y_test = to_categorical(y_test, 3)
def applySIFT(data):
sift = cv2.xfeatures2d.SIFT_create()
if len(data.shape) < 3: # to take care of a single image instance
data = data[np.newaxis]
sift_key_points = []
sift_descriptors = []
for image in data:
sift_kp, sift_des = sift.detectAndCompute(image, None)
sift_key_points.append(sift_kp)
sift_descriptors.append(sift_des)
return (sift_key_points, sift_descriptors)
def applySURF(data):
surf = cv2.xfeatures2d.SURF_create()
if len(data.shape) < 3:
data = data[np.newaxis]
surf_key_points = []
surf_descriptors = []
for image in data:
surf_kp, surf_des = surf.detectAndCompute(image, None)
surf_key_points.append(surf_kp)
surf_descriptors.append(surf_des)
return (surf_key_points, surf_descriptors)
def applyHOG(data):
pass
(kp_sift, des_sift) = applySIFT(X_train)
(kp_surf, des_surf) = applySURF(X_train)
img_kp_sift = cv2.drawKeypoints(X_train[5], kp_sift[0], None, (255, 0, 0), 4)
img_kp_surf = cv2.drawKeypoints(X_train[5], kp_surf[0], None, (255, 0, 0), 4)
# Display the image with keypoints using Matplotlib
plt.imshow(img_kp_sift)
plt.imshow(img_kp_surf)
from sklearn.cluster import KMeans
# no of clusters=no of visual words
km_sift = KMeans(n_clusters=250)
km_surf = KMeans(n_clusters=250)
visual_words_codebook_sift = np.vstack(des_sift).astype(float)
visual_words_codebook_surf = np.vstack(des_surf).astype(float)
km_est_sift = km_sift.fit(visual_words_codebook_sift)
km_est_sift = km_sift.fit(visual_words_codebook_sift)
def create_freq_matrix(descriptors, km_estimator):
freq_matrix = []
n_clusters = km_estimator.n_clusters
for des in descriptors:
clusters = km_estimator.predict(
des.astype(float)
) # clusters are numbered from 0 to n_clusters-1
# now i know which clusters is this descriptor closest to,
# i can now convert it to a freq representation
freq_this = np.zeros((1, n_clusters))
cluster_num, freq = np.unique(clusters, return_counts=True)
freq_this[0][
cluster_num
] = freq # place the occr freq of that cluster at the particular index
freq_matrix.append(freq_this)
return np.vstack(freq_matrix)
freq_sift = create_freq_matrix(des_sift, km_est_sift)
freq_surf = create_freq_matrix(des_surf, km_est_surf)
from sklearn.feature_extraction.text import TfidfTransformer
tfidf_sift = TfidfTransformer()
tfidf_sift.fit(freq_sift)
tfidf_X_train_sift = tfidf_sift.transform(freq_sift)
tfidf_surf = TfidfTransformer().fit(freq_surf)
tfidf_X_train_surf = tfidf_surf.transform(freq_surf)
from sklearn.ensemble import RandomForestClassifier
RF_sift = RandomForestClassifier()
RF_sift.fit(tfidf_X_train_sift, y_train)
RF_surf = RandomForestClassifier()
RF_surf.fit(tfidf_X_train_surf, y_train)
from tensorflow.keras import Sequential
from tensorflow.keras.layers import Dense
from tensorflow.keras.regularizers import L2
from tensorflow import random
from tensorflow import sparse
random.set_seed(19)
ANN_sift = Sequential(
[
Dense(250, activation=None),
Dense(32, activation="relu", kernel_regularizer=L2(0.01)),
Dense(32, activation="relu", kernel_regularizer=L2(0.01)),
Dense(3, activation="softmax"),
]
)
ANN_surf = Sequential(
[
Dense(250, activation=None),
Dense(32, activation="sigmoid"),
Dense(16, activation="sigmoid"),
Dense(3, activation="softmax"),
]
)
ANN_sift.compile(
loss="categorical_crossentropy", optimizer="adam", metrics=["accuracy"]
)
ANN_surf.compile(
loss="categorical_crossentropy", optimizer="adam", metrics=["accuracy"]
)
ANN_sift.fit(tfidf_X_train_sift.todense(), y_train, epochs=100)
ANN_surf.fit(tfidf_X_train_surf.todense(), y_train, epochs=100)
(_, des_sift_test) = applySIFT(X_test)
freq_sift_test = create_freq_matrix(des_sift_test, km_est_sift)
tfidf_X_test_sift = tfidf_sift.transform(freq_sift_test)
(_, des_surf_test) = applySURF(X_test)
freq_surf_test = create_freq_matrix(des_surf_test, km_est_surf)
tfidf_X_test_surf = tfidf_surf.transform(freq_surf_test)
y_predicted_RF_SIFT = RF_sift.predict(tfidf_X_test_sift)
y_predicted_ANN_SIFT = ANN_sift.predict(freq_sift_test)
y_predicted_RF_SURF = RF_surf.predict(tfidf_X_test_surf)
y_predicted_ANN_SURF = ANN_surf.predict(tfidf_X_test_surf.todense())
from sklearn.metrics import (
accuracy_score,
confusion_matrix,
log_loss,
f1_score,
roc_curve,
)
y_perform_RF_SIFT = {
"accuracy_score": accuracy_score(
y_test.argmax(axis=1), y_predicted_RF_SIFT.argmax(axis=1)
),
"confusion_matrix": confusion_matrix(
y_test.argmax(axis=1), y_predicted_RF_SIFT.argmax(axis=1)
),
"log_loss": log_loss(y_test, y_predicted_RF_SIFT),
"f1_score": f1_score(
y_test.argmax(axis=1), y_predicted_RF_SIFT.argmax(axis=1), average="micro"
),
}
y_perform_RF_SURF = {
"accuracy_score": accuracy_score(
y_test.argmax(axis=1), y_predicted_RF_SURF.argmax(axis=1)
),
"confusion_matrix": confusion_matrix(
y_test.argmax(axis=1), y_predicted_RF_SURF.argmax(axis=1)
),
"log_loss": log_loss(y_test, y_predicted_RF_SURF),
"f1_score": f1_score(
y_test.argmax(axis=1), y_predicted_RF_SURF.argmax(axis=1), average="micro"
),
}
y_perform_ANN_SURF = {
"accuracy_score": accuracy_score(
y_test.argmax(axis=1), y_predicted_ANN_SURF.argmax(axis=1)
),
"confusion_matrix": confusion_matrix(
y_test.argmax(axis=1), y_predicted_ANN_SURF.argmax(axis=1)
),
"log_loss": log_loss(y_test, y_predicted_ANN_SURF),
"f1_score": f1_score(
y_test.argmax(axis=1), y_predicted_ANN_SURF.argmax(axis=1), average="micro"
),
}
y_perform_ANN_SIFT = {
"accuracy_score": accuracy_score(
y_test.argmax(axis=1), y_predicted_ANN_SIFT.argmax(axis=1)
),
"confusion_matrix": confusion_matrix(
y_test.argmax(axis=1), y_predicted_ANN_SIFT.argmax(axis=1)
),
"log_loss": log_loss(y_test, y_predicted_ANN_SIFT),
"f1_score": f1_score(
y_test.argmax(axis=1), y_predicted_ANN_SIFT.argmax(axis=1), average="micro"
),
}
print("RF_SURF")
for key, value in y_perform_RF_SURF.items():
print(key, ":\n", value)
print("RF_SIFT")
for key, value in y_perform_RF_SIFT.items():
print(key, ":\n", value)
print("ANN_SIFT")
for key, value in y_perform_ANN_SIFT.items():
print(key, ":\n", value)
print("ANN_SURF")
for key, value in y_perform_ANN_SURF.items():
print(key, ":\n", value)
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
dataset_raw = pd.read_csv(
"/kaggle/input/higher-education-predictors-of-student-retention/dataset.csv"
)
print(dataset_raw.shape)
dataset_raw.head().T
dataset_raw.info()
dataset_raw.describe().T
dataset = dataset_raw.copy()
dataset.shape
dataset.Target.value_counts()
target = dataset["Target"]
features = dataset.drop(["Target"], axis=1)
target.shape, features.shape
from sklearn.model_selection import train_test_split, KFold
kf = KFold(n_splits=20, shuffle=True, random_state=2304)
for tr_idx, te_idx in kf.split(features):
X_train, X_test = features.iloc[tr_idx], features.iloc[te_idx]
y_train, y_test = target.iloc[tr_idx], target.iloc[te_idx]
X_train.shape, X_test.shape, y_train.shape, y_test.shape
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
y_train = le.fit_transform(y_train)
print(y_train)
y_test = le.fit_transform(y_test)
y_test
from xgboost import XGBClassifier
model = XGBClassifier(
n_estimators=40, random_state=2304, eval_metric="mlogloss"
) # use_label_encoder=False
model.fit(X_train, y_train)
print(model.score(X_train, y_train))
print(model.score(X_test, y_test))
pred_proba = model.predict_proba(X_test)[:, 1]
pred_proba[:10]
pred_label = model.predict(X_test)
pred_label[:100]
y_test[:100]
classes = np.unique(y_train)
classes
from yellowbrick.classifier import confusion_matrix
plt.figure(figsize=(3, 3))
confusion_matrix(model, X_train, y_train, X_test, y_test, classes=classes)
plt.show()
import seaborn as sns
XGBClassifier_importances_values = model.feature_importances_
XGBClassifier_importances = pd.Series(
XGBClassifier_importances_values, index=X_train.columns
)
XGBClassifier_top34 = XGBClassifier_importances.sort_values(ascending=False)[:34]
plt.figure(figsize=(8, 6))
plt.title("Feature importances Top 34")
sns.barplot(x=XGBClassifier_top34, y=XGBClassifier_top34.index)
plt.show()
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import re
ratings_path = "/kaggle/input/anime-recommendation-database-2020/animelist.csv"
data_path = "/kaggle/input/anime-recommendation-database-2020/anime.csv"
# The dataset that we are dealing with is divided into two parts, the first part which we are going to store as `anime_ratings` consists of the individual user ratings for different animes
# The second part, `anime_data` consists of overall information about the anime such as genre, ratings, name, etc.
# # Preprocessing
anime_ratings = pd.read_csv(ratings_path)
anime_data = pd.read_csv(data_path)
anime_data.info()
# 1) `anime_data` contains:
# * MAL_ID: MyAnimelist ID of the anime. (e.g. 1)
# * Name: full name of the anime. (e.g. Cowboy Bebop)
# * Score: average score of the anime given from all users in MyAnimelist database. (e.g. 8.78)
# * Genres: comma separated list of genres for this anime. (e.g. Action, Adventure, Comedy, Drama, Sci-Fi, Space)
# * English name: full name in english of the anime. (e.g. Cowboy Bebop)
# * Japanese name: full name in japanses of the anime. (e.g. カウボーイビバップ)
# * Type: TV, movie, OVA, etc. (e.g. TV)
# * Episodes': number of chapters. (e.g. 26)
# * Aired: broadcast date. (e.g. Apr 3, 1998 to Apr 24, 1999)
# * Premiered: season premiere. (e.g. Spring 1998)
# * Producers: comma separated list of produducers (e.g. Bandai Visual)
# * Licensors: comma separated list of licensors (e.g. Funimation, Bandai Entertainment)
# * Studios: comma separated list of studios (e.g. Sunrise)
# * Source: Manga, Light novel, Book, etc. (e.g Original)
# * Duration: duration of the anime per episode (e.g 24 min. per ep.)
# * Rating: age rate (e.g. R - 17+ (violence & profanity))
# * Ranked: position based in the score. (e.g 28)
# * Popularity: position based in the the number of users who have added the anime to their list. (e.g 39)
# * Members: number of community members that are in this anime's "group". (e.g. 1251960)
# * Favorites: number of users who have the anime as "favorites". (e.g. 61,971)
# * Watching: number of users who are watching the anime. (e.g. 105808)
# * Completed: number of users who have complete the anime. (e.g. 718161)
# * On-Hold: number of users who have the anime on Hold. (e.g. 71513)
# * Dropped: number of users who have dropped the anime. (e.g. 26678)
# * Plan to Watch': number of users who plan to watch the anime. (e.g. 329800)
# * Score-10': number of users who scored 10. (e.g. 229170)
# * Score-9': number of users who scored 9. (e.g. 182126)
# * Score-8': number of users who scored 8. (e.g. 131625)
# * Score-7': number of users who scored 7. (e.g. 62330)
# * Score-6': number of users who scored 6. (e.g. 20688)
# * Score-5': number of users who scored 5. (e.g. 8904)
# * Score-4': number of users who scored 4. (e.g. 3184)
# * Score-3': number of users who scored 3. (e.g. 1357)
# * Score-2': number of users who scored 2. (e.g. 741)
# * Score-1': number of users who scored 1. (e.g. 1580)
anime_data.head()
# As we are going to build a simplistic model, we drop most of our data to allow for easier computation:
anime_data = anime_data[
["MAL_ID", "Name", "Score", "Genres", "Type", "Episodes", "Members"]
]
anime_data.rename(columns={"MAL_ID": "anime_id"}, inplace=True)
anime_data.info()
anime_ratings.info()
anime_ratings.drop(anime_ratings.iloc[:, 3:], axis=1, inplace=True)
anime_ratings.info()
anime_ratings.anime_id.nunique()
# anime_complete = pd.merge(anime_data,anime_ratings,on='anime_id')
# anime_complete=anime_complete.rename(columns={'rating':'user_rating','Score':'total_rating'})
# anime_complete.info()
# The problem being faced above is that the resultant df is too big to work with (8.1+ GB!)
# To fix this, we downsample the anime_ratings df and then try creating the `anime_complete` df
anime_ratings = anime_ratings.sample(frac=0.2)
anime_ratings.info()
anime_ratings.anime_id.nunique()
# We see that some animes are complete lost, we proceed anyway as it's not a significant number
# We combine the two seperate dataframes into a single dataframe:
anime_complete = pd.merge(anime_data, anime_ratings, on="anime_id")
anime_complete = anime_complete.rename(
columns={"rating": "user_rating", "Score": "total_rating", "Name": "anime_title"}
)
anime_complete.info()
anime_complete.isna().sum()
import matplotlib.pyplot as plt
# Count the number of occurrences of each anime name
top_10_anime = anime_complete["anime_title"].value_counts().nlargest(10)
palette = sns.color_palette("rocket", len(top_10_anime))
# Create the bar chart
plt.bar(top_10_anime.index, top_10_anime.values, color=palette)
# Set the title and labels
plt.title("Top 10 Anime by User Rating Count")
plt.xlabel("Anime Name")
plt.ylabel("User Rating Count")
# Rotate the x-axis labels for better readability
plt.xticks(rotation=40, ha="right")
# Show the plot
plt.show()
top_10_anime = (
anime_complete.sort_values(by="Members", ascending=False)
.drop_duplicates(subset="anime_title")
.head(10)
)
palette = sns.color_palette("rocket", len(top_10_anime))
# Create a bar chart with the anime titles and the number of members
plt.bar(top_10_anime["anime_title"], top_10_anime["Members"], color=palette)
# Set the title and labels
plt.title("Top 10 Anime by Number of Members")
plt.xlabel("Anime Title")
plt.ylabel("Number of Members")
# Rotate the x-axis labels for better readability
plt.xticks(rotation=40, ha="right")
# Show the plot
plt.show()
anime_features = anime_complete.copy()
anime_features.head()
anime_features.isnull().sum()
user_id_counts = anime_features["user_id"].value_counts()
user_id_counts
user_id_counts.describe()
# In order to consider only the reviewers of "trusted" members, the ones we are considering trustworthy are those who have reviewed a certain number (100 in our case) of animes, the rest of the reviewers may be dropped.
# Keep only the rows for which the user_id appears at least 200 times
anime_features = anime_features[
anime_features["user_id"].isin(user_id_counts[user_id_counts >= 100].index)
]
anime_features.user_id.nunique()
# Since the title text was not found to be clean we a function to clean the title names using regex:
def text_cleaning(text):
text = re.sub(r""", "", text)
text = re.sub(r".hack//", "", text)
text = re.sub(r"'", "", text)
text = re.sub(r"A's", "", text)
text = re.sub(r"I'", "I'", text)
text = re.sub(r"&", "and", text)
return text
anime_features["anime_title"] = anime_features["anime_title"].apply(text_cleaning)
anime_pivot = anime_features.pivot_table(
index="anime_title", columns="user_id", values="user_rating"
).fillna(0)
anime_pivot.head()
# # Collaborative Filtering
# Collaborative filtering is a type of recommendation algorithm that predicts a user's preference for an item by finding patterns in the preferences of similar users. It works by analyzing a large dataset of user-item interactions, such as ratings or purchase histories, and then identifies users with similar patterns of interactions. The algorithm then uses these similarities to recommend items that the target user has not interacted with, but that similar users have rated highly. Collaborative filtering is widely used in recommendation systems for online retailers, streaming services, and social media platforms, among others.
# ## Cosine Similarity using KNN
# Cosine similarity is a measure of similarity between two non-zero vectors of an inner product space. In the context of recommendation systems, cosine similarity is often used to determine how similar two items or users are based on their feature vectors.
anime_data["Name"] = anime_data["Name"].apply(text_cleaning)
# Sparse matrix was created to optimize the memory usage and computational efficiency of the model. The user-anime matrix can be very large and the majority of the entries are likely to be zeros, which means that they don't contribute to the similarity computations. By converting the matrix to a sparse format, we can represent it using less memory, and perform computations only on the non-zero entries. This can significantly speed up the model training and recommendation generation processes.
#
# Import the necessary libraries
from scipy.sparse import csr_matrix
from sklearn.neighbors import NearestNeighbors
# Convert the pivot table to a sparse matrix format
anime_matrix = csr_matrix(anime_pivot.values)
# Create a NearestNeighbors model using cosine similarity and brute-force algorithm
model_knn = NearestNeighbors(metric="cosine", algorithm="brute")
# Fit the model to the data (i.e., the sparse matrix)
model_knn.fit(anime_matrix)
# import pickle
# with open('model.pkl', 'wb') as f:
# pickle.dump(model_knn, f)
# with open('anime_pivot.pkl', 'wb') as f:
# pickle.dump(anime_pivot, f)
# Select a random anime title, or input your own title here
anime_title = np.random.choice(anime_pivot.index)
# Print the selected anime title
print(f"Randomly selected anime title: {anime_title} \n")
# Find the row index of the selected anime title
query_index = anime_pivot.index.get_loc(anime_title)
# Use the fitted KNN model to find the 6 nearest neighbors to the selected row
distances, indices = model_knn.kneighbors(
anime_pivot.iloc[query_index, :].values.reshape(1, -1), n_neighbors=6
)
# Print the recommendations for the selected row
print(f"Recommendations for {anime_pivot.index[query_index]}:\n")
# Iterate over the nearest neighbors and print their names and distances
for i, (distance, index) in enumerate(
zip(distances.flatten()[1:], indices.flatten()[1:])
):
print(f"{i+1}: {anime_pivot.index[index]}, with distance of {distance}")
# Code above is doing the following:
# * It selects a random row index from the pivot table using np.random.choice() and assigns it to query_index.
# * It uses the fitted KNN model to find the 6 nearest neighbors to the selected row by calling the kneighbors() method and passing the values of the selected row as a reshaped array. The distances and indices of the nearest neighbors are assigned to distances and indices, respectively.
# * It prints the recommendations for the selected row by using an f-string to format the row index.
# * It iterates over the nearest neighbors (excluding the selected row itself) and prints their names and distances using an f-string and the zip() function to iterate over the distances and indices arrays simultaneously. The enumerate() function is used to add a counter to the loop starting from 1.
def give_rec_knn(
anime_title=np.random.choice(anime_pivot.index), anime_pivot=anime_pivot
):
# Print the selected anime title
print(f"Randomly selected anime title: {anime_title} \n")
# Find the row index of the selected anime title
query_index = anime_pivot.index.get_loc(anime_title)
# Use the fitted KNN model to find the 6 nearest neighbors to the selected row
distances, indices = model_knn.kneighbors(
anime_pivot.iloc[query_index, :].values.reshape(1, -1), n_neighbors=6
)
# Print the recommendations for the selected row
print(f"Recommendations for {anime_pivot.index[query_index]}:\n")
# Iterate over the nearest neighbors and print their names and distances
for i, (distance, index) in enumerate(
zip(distances.flatten()[1:], indices.flatten()[1:])
):
print(f"{i+1}: {anime_pivot.index[index]}, with distance of {distance}")
give_rec_knn("Steins;Gate")
give_rec_knn()
# # Content Based Filtering
# Content-based filtering is a recommendation system technique that recommends items based on their intrinsic features or attributes. It identifies items similar to the ones the user has shown interest in and recommends them. For example, a movie recommendation system might recommend other movies with similar genres, actors, directors, or plot themes to those previously watched by the user. Content-based filtering does not rely on the preferences of other users and can work well for new or niche items with little user data. However, it may suffer from limited diversity in recommendations and inability to capture serendipitous recommendations.
from sklearn.feature_extraction.text import TfidfVectorizer
# Initialize the TfidfVectorizer with various parameters
tfv = TfidfVectorizer(
min_df=3,
max_features=None,
strip_accents="unicode",
analyzer="word",
token_pattern=r"\w{1,}",
ngram_range=(1, 3),
stop_words="english",
)
# Fill NaN values in the 'Genres' column with an empty string
anime_data["Genres"] = anime_data["Genres"].fillna("")
# Split the 'Genres' column by comma and convert to string format
genres_str = anime_data["Genres"].str.split(",").astype(str)
# Use the TfidfVectorizer to transform the genres_str into a sparse matrix
tfv_matrix = tfv.fit_transform(genres_str)
# Print the shape of the sparse matrix
print(tfv_matrix.shape)
# TF-IDF stands for Term Frequency-Inverse Document Frequency.
# TF-IDF vectorizer is a specific implementation of the TF-IDF technique. It is a commonly used technique in natural language processing to convert a collection of text documents into numerical feature vectors, which can be used for machine learning tasks like text classification or clustering.
# * It first counts the number of occurrences of each word (term) in each document (text) in the collection, and then * calculates a weight for each term based on how frequently it appears across all documents.
# * The weight is higher for terms that appear frequently in a particular document, but not so much in other documents.
# * The weight is also higher for terms that appear less frequently across all documents.
# This helps to give more importance to words that are relevant to a specific document and less importance to common words that are not specific to any document.
#
from sklearn.metrics.pairwise import sigmoid_kernel
# Compute the sigmoid kernel
sig = sigmoid_kernel(tfv_matrix, tfv_matrix)
# The sigmoid kernel is a type of kernel function used in machine learning for non-linear classification and regression. It maps the data into a higher-dimensional space and computes the dot product between two data points in that space.
# * The sigmoid_kernel is a similarity function that computes the sigmoid kernel between two input feature vectors.
# * It is commonly used in machine learning for non-linear classification and regression tasks.
# * The sigmoid kernel function takes two feature vectors as input and computes a value between 0 and 1, where 1 indicates a high degree of similarity between the two vectors and 0 indicates no similarity.
# * The sigmoid kernel function applies the sigmoid function to the dot product of the two feature vectors, which transforms the dot product into a value between 0 and 1.
# * The `sigmoid_kernel` is being used to compute the similarity between anime genres based on their TF-IDF feature vectors, which can be used for content-based recommendation systems.
# Create a Pandas Series object where the index is the anime names and the values are the indices in anime_data
indices = pd.Series(anime_data.index, index=anime_data["Name"])
# Remove duplicates in the index (i.e., duplicate anime names)
indices = indices.drop_duplicates()
def give_rec_cbf(title, sig=sig):
# Get the index corresponding to anime title
idx = indices[title]
# Get the pairwsie similarity scores
sig_scores = list(enumerate(sig[idx]))
# Sort the anime based on similarity scores
sig_scores = sorted(sig_scores, key=lambda x: x[1], reverse=True)
# Get the indices of top 10 most similar anime excluding the input anime
anime_indices = [i[0] for i in sig_scores[1:11]]
# Create dataframe of top 10 recommended anime
top_anime = pd.DataFrame(
{
"Anime name": anime_data["Name"].iloc[anime_indices].values,
"Rating": anime_data["Score"].iloc[anime_indices].values,
}
)
return top_anime
give_rec_cbf("One Piece")
|
# # **Shop Customer | Data Cleaning, K-Means Clustering**
# ## **1. Data Exploration**
# ### **1.1. Importing the modules**
import pandas as pd
import seaborn as sns
import plotly.express as px
import plotly.offline as py
import matplotlib.pyplot as plt
import numpy as np
# from sklearn.preprocessing import OneHotEncoder
from sklearn import decomposition
import category_encoders as ce
from sklearn.cluster import KMeans
# from sklearn.metrics import silhouette_score
from scipy.stats import zscore
from yellowbrick.cluster import KElbowVisualizer
import plotly.graph_objs as go
py.init_notebook_mode(connected=True)
# ### **1.2. Importing dataset**
df = pd.read_csv("/kaggle/input/customers-dataset/Customers.csv")
df.head()
# ### **1.3. Data Cleaning and Preprocessing**
# **Number of rows** and **number of columns** of dataframe:
num_rows, num_cols = df.shape
print("Num rows: ", num_rows)
print("Num cols: ", num_cols)
# **The meaning of each row**: Represents the shopping information of a customer at the store. There is no row that carries any other meaning.
df.head(1)
# Check for any duplicate rows:
df.duplicated().sum()
# **The meaning of each column**:
# * `Customer ID`: Customer ID
# * `Gender`: Gender
# * `Age`: Age
# * `Annual Income`: Annual income (unit: $)
# * `Spending Score`: Score assigned by the store based on customer behavior and spending nature. Ranges from 0-100.
# * `Profession`: Occupation
# * `Work Experience`: Work experience (unit: years)
# * `Family Size`: Number of family members
# Perform columns name standardization for convenience in future usage:
df = df.rename(
columns={
"Annual Income ($)": "Annual_Income",
"Spending Score (1-100)": "Spending_Score",
"Work Experience": "Work_Experience",
"Family Size": "Family_Size",
}
)
df.columns
# **The data types of each column are as follows:**
df.dtypes
# **Check for missing values in each column:**
df.isna().sum()
# There are 35 missing values in column `Profession`.
# **Distribution of values in numerical columns:**
# Based on the data type check in the previous step, there are 6 columns that seem to belong to the numeric group: `CustomerID`, `Age`, `Annual_Income`, `Spending_Score`, `Work_Experience`, `Family_Size`. However, the `CustomerID` column is actually of categorical type (as the ID is coded as numbers).
# Below is a basic statistical table summarizing the distribution of values in the numeric attributes:
df[
["Age", "Annual_Income", "Spending_Score", "Work_Experience", "Family_Size"]
].describe()
# Overall, the youngest customers have an age of 0. Let's try querying:
df[df["Age"] == 0]
# It appears that the data collector has filled in the value of 0 for customers with missing age information.
# **Distribution of values in categorical columns:**
# There are 3 columns belonging to the categorical group, which are: `CustomerID`, `Gender`, `Profession`
# Check for duplicate values in the `CustomerID` column:
df["CustomerID"].duplicated().sum()
# Each customer ID is unique, there is no duplication.
# Excluding the `CustomerID` attribute, I have a basic statistical summary of the value distributions of the remaining categorical attributes:
df[["Gender", "Profession"]].describe()
# **To inspect the values in each column of the dataset and perform preprocessing steps as necessary.**
# **Column `CustomerID`**: Based on the results of the above checks, all values in the CustomerID column are unique, so there is no need for any preprocessing.
# **Column `Gender`**:
fig = go.Figure(
data=[
go.Pie(
labels=["Male", "Female"],
values=[
df[df["Gender"] == "Male"]["Gender"].count(),
df[df["Gender"] == "Female"]["Gender"].count(),
],
pull=[0, 0.05],
)
]
)
fig.update_layout(autosize=False, width=450, height=450, title="Gender Distribution")
fig.show()
df.loc[df["Age"] == 0, "Age"] = int(df[df["Age"] > 0]["Age"].median())
# **Column `Profession`**:
plt.figure(figsize=(11, 4))
ax = sns.countplot(
x="Profession",
data=df,
palette="colorblind",
order=df["Profession"].value_counts().index,
)
ax.bar_label(ax.containers[0])
# Missing values in the `Profession` column will be imputed with a new value of "Other". Imputing with a new value instead of using the existing values will help preserve the distribution of the original values. Additionally, using the value "Other" will avoid the need to retrain the machine learning model if new values for `Profession` appear in the future.
df["Profession"] = df["Profession"].fillna("Other")
df["Profession"].isna().sum()
plt.figure(figsize=(11, 4))
data = df["Profession"].astype(str).value_counts()
x = data.index.tolist()
y = data.values
bar = sns.barplot(y=x, x=y, palette="colorblind")
for n, i in enumerate(y):
porc = (
f'{round((i/sum(df["Profession"].astype(str).value_counts().values))*100, 2)}%'
)
bar.annotate(
f" {i} ({porc})",
xy=(i, n),
verticalalignment="center",
horizontalalignment="left",
fontsize=8,
)
bar.spines["top"].set_visible(False)
bar.spines["right"].set_visible(False)
plt.title("Customer's Profession")
bar.set_ylabel("Profession", horizontalalignment="center")
bar.set_xlabel("Counts", horizontalalignment="right")
plt.show()
# **Column `Age`**:
fig = px.histogram(df, x="Age")
fig.show()
# The distribution appears to be uniform from 0 to 99 years old, but this uniformity seems illogical because the minimum working age is at least 18 years old. Let's attempt a query:
df.query("Age<18")
# Replace customers who are under 18 years old with the mean age by Profession and Gender:
mean_age = df.groupby(["Gender", "Profession"])["Age"].mean()
df["Age"] = df.apply(
lambda row: int(mean_age[(row["Gender"], row["Profession"])])
if row["Age"] < 18
else row["Age"],
axis=1,
)
# **Column `Annual_Income`**:
fig = px.violin(df, x="Annual_Income", width=800, height=300, box=True)
fig.show()
fig = px.box(df, y="Annual_Income", x="Age")
fig.show()
# Overall, there are some outliers present which may represent unique cases. Therefore, I will not process them.
# **Column `Spending_Score`**:
fig = px.violin(df, x="Spending_Score", width=800, height=300, box=True)
fig.show()
# **Columns `Work_Experience`**:
sns.countplot(data=df, x="Work_Experience")
# Based on the graph, we can see that `Work_Experience` is skewed to the right and there is an outlier value of 17. Let's query these rows:
df[df["Work_Experience"] == 17]
# Although there was a few rows with a slightly abnormal `Age` value compared to `Work_Experience`, the outliers overall seem reasonable (since the `Profession` is "Artist"), so I decided to keep these rows.
fig = px.box(df, y="Work_Experience", x="Age")
fig.show()
# There are quite a few outliers when visualizing `Work_Experience` in combination with the `Age` column. Upon closer inspection of these outliers, we can observe several issues with the data, such as an 8-year-old having 14 years of work experience, a 19-year-old having 17 years of work experience, etc.
df[df["Work_Experience"] - df["Age"] >= 0]
# Remove the rows where `Work_Experience` is greater than or equal to `Age`:
df = df.drop(df[df["Work_Experience"] - df["Age"] >= 0].index)
# **Column `Family_Size`:**
fig = px.violin(df, x="Family_Size", width=800, height=300, box=True)
fig.show()
# Dataframe after preprocessing:
df
# ## **2. Building a machine learning mode**
# Apply K-Means clustering algorithm to segment customers into distinct clusters.
# ### **2.1. Prepare dataset:**
# For the `numerical` columns: Check the distribution skewness of each data column.
df.drop(columns="CustomerID").skew(numeric_only=True)
# `Work_Experience` is highly skewed to the right. I will perform skewness reduction using **Square root** transformation.
df["Work_Experience"] = np.sqrt(df["Work_Experience"])
plt.hist(np.sqrt(df["Work_Experience"]))
# The relationship between numerical variables:
display(df.drop(columns="CustomerID").corr())
sns.pairplot(
df[["Age", "Annual_Income", "Spending_Score", "Work_Experience", "Family_Size"]],
height=5,
)
# For `categorical` columns: Apply One-hot Encoding technique to convert all of them to `numerical`.
encoder = ce.OneHotEncoder(
cols=["Gender", "Profession"], return_df=True, use_cat_names=True
)
df_encoded = encoder.fit_transform(df)
# Remove`CustomerID`
df_encoded = df_encoded.drop(columns="CustomerID")
# Data types of the columns:
df_encoded.dtypes
# Z-Score Normalization:
df_encoded = df_encoded.apply(zscore)
df_encoded = df_encoded.values
df_encoded
# ### **2. Training the Model**
# Number of clusters
max_clusters = 20
df_encoded.shape
visualizer = KElbowVisualizer(KMeans(), k=(2, max_clusters))
visualizer.fit(df_encoded)
plt.xticks([i for i in range(2, max_clusters + 1)])
visualizer.show()
visualizer = KElbowVisualizer(KMeans(), k=(2, max_clusters), metric="silhouette")
visualizer.fit(df_encoded)
plt.xticks([i for i in range(2, max_clusters + 1)])
visualizer.show()
# It seems unreasonable to use 17 variables to build a K-means model. Both **the Elbow Method** and **the Silhouette Score** suggest a larger number of clusters than the original goal of grouping customers in a shop. To reduce the number of variables used in the model, I will attempt to apply PCA to perform dimensionality reduction.
pca = decomposition.PCA()
X_train_pca = pca.fit_transform(df_encoded)
# Determine explained variance using explained_variance_ration_ attribute
exp_var_pca = pca.explained_variance_ratio_
# Cumulative sum of eigenvalues; This will be used to create step plot for visualizing the variance explained by each principal component.
cum_sum_eigenvalues = np.cumsum(exp_var_pca)
# Create the visualization plot
plt.bar(
range(0, len(exp_var_pca)),
exp_var_pca,
alpha=0.5,
align="center",
label="Individual explained variance",
)
plt.step(
range(0, len(cum_sum_eigenvalues)),
cum_sum_eigenvalues,
where="mid",
label="Cumulative explained variance",
)
plt.ylabel("Explained variance ratio")
plt.xlabel("Principal component index")
plt.xticks([i for i in range(0, df_encoded.shape[1])])
plt.legend(loc="best")
plt.tight_layout()
plt.show()
pca = decomposition.PCA(n_components=3)
df_pca = pca.fit_transform(df_encoded)
visualizer = KElbowVisualizer(KMeans(), k=(2, 20))
visualizer.fit(df_pca)
plt.xticks([i for i in range(2, max_clusters + 1)])
visualizer.show()
visualizer = KElbowVisualizer(KMeans(), k=(2, 20), metric="silhouette")
visualizer.fit(df_pca)
plt.xticks([i for i in range(2, max_clusters + 1)])
visualizer.show()
kmeans = KMeans(n_clusters=4)
y_kmeans = kmeans.fit_predict(df_pca)
y_kmeans
df_results = pd.DataFrame(df_pca).rename(
columns={0: "X_axis", 1: "Y_axis", 2: "Z_axis"}
)
df_results["y_kmeans"] = y_kmeans
df_results["ID"] = df["CustomerID"]
df_results
# ### **3. Model Visualization:**
fig = px.scatter_3d(
df_results, x="X_axis", y="Y_axis", z="Z_axis", color="y_kmeans", hover_name="ID"
)
fig.update_traces(marker_size=2)
fig.show()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# ## Import Dataset
df = pd.read_csv(
"/kaggle/input/bank-note-authentication-uci-data/BankNote_Authentication.csv"
)
df.head()
list(df)
# ## Select Features and Output variable
X = df.drop(["class"], axis=1)
Y = df.drop(["variance", "skewness", "curtosis", "entropy"], axis=1)
X.head()
# ## Data Split
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(
X, Y, train_size=0.3, random_state=1
)
# ## Model Creation and fitting
from sklearn.ensemble import RandomForestClassifier
clf = RandomForestClassifier()
clf.fit(x_train, y_train)
# ## Model Prediction
y_pred = clf.predict(x_test)
# ## Accuracy
from sklearn.metrics import accuracy_score
accuracy_score(y_test, y_pred)
|
# ## Guiding notes:
# Current status: We have prepared alldf and tdf with all protein and peptide features and all 16 updrs possible outputs in vdf. The code builds one model for each Validation output regression, and then tests prediction on training the features with MI scores of 0.02 on each of those regression targets. We also include additional features for months 6, 12, and 24 added only to extra rows for those features (this seemed to make a big difference in the SMAPE score).
# Best results and what was tried in summary form:
# 1. Using RandomForestRegressor (tried DNN, LinearRegressor (LR did better than DDN, and LR is poor)).
# 2. The MI summary scores of 0.02 or higher are now in use.
# 3. As of 4/9/2021, RFR worked better (smape 0.78) with Protein data. Just adding Peptide data does not seem to help.
# 4. We may be underfitting, the internal test suggests a SMAPE of .61, but validation date is doing better at .57
# Three ideas for improvement:
# 1. Find clues in the Protein and Peptide data for some feature engineering,
# 2. Add in Peptide data, the run on 4/10/2023 is using only Protein data,
# 3. Consider alternatives where estimates of later months can also use prior months to improve accuracy, and
# 4. We are not yet using the matric on UPDRS estimation when 2 and 3 have medicine affecting them.
# Reminder: Not every clinical data update (visits with UPDRS updates) includes protein and peptide lab work
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
proteinsdf = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/train_proteins.csv"
)
peptitesdf = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/train_peptides.csv"
)
clinicaldf = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/train_clinical_data.csv"
)
supplemedf = pd.read_csv(
"/kaggle/input/amp-parkinsons-disease-progression-prediction/supplemental_clinical_data.csv"
)
generateNewtdfAndvdf = True # expensive data engineering
# generateNewtdfAndvdf = False # save time by loading prior engineered data
print("Proteinsdf")
print(proteinsdf.describe())
print(proteinsdf.info())
print(proteinsdf.nunique())
print(proteinsdf.head())
print("Peptidesdf")
print(peptitesdf.describe())
print(peptitesdf.info())
print(peptitesdf.nunique())
print(peptitesdf.head())
print("Clinical Data")
print(clinicaldf.describe())
print(clinicaldf.info())
print(clinicaldf.nunique())
print(clinicaldf.head(10))
print("Supplemental Data")
print(supplemedf.describe())
print(supplemedf.info())
print(supplemedf.nunique())
print(supplemedf.head())
# ## We are going to convert Peptides and Protienes into enumerates
# We will keep them numerically close to each other by starting the base at 100,000 and incrementing proteine names and peptide names to numbers from there. We will give the neural net enough width in its dense layers to adjust weights accordingly. What follows is a structure to preserve the enumeration mappings between training and test submission generation.
class CatVarStorageLabels:
_instance = None
catvarmaps = {}
def __new__(cls):
if cls._instance is None:
cls._instance = super().__new__(cls)
return cls._instance
def getCatVarLabelMapping(self, catVarName, catVarLabel):
catDetail = catVarName + "_" + str(catVarLabel)
retval = -1
try:
retval = self.catvarmaps[catDetail]
except:
pass
return retval
def setCatVarLabelMapping(self, catVarName, catVarLabel, catVarMapped):
catDetail = catVarName + "_" + str(catVarLabel)
self.catvarmaps[catDetail] = catVarMapped
return
def reset(self):
self.catvarmaps = {}
return
def print(self):
print(self.catvarmaps)
return
catvars = CatVarStorageLabels()
catvars.setCatVarLabelMapping("Cabin", 1, 0.333)
catvars.print()
print(catvars.getCatVarLabelMapping("Cabin", 1))
print(catvars.getCatVarLabelMapping("Cabin", 2))
catvars.reset()
catvars.print()
# ## Prepare a single data frame to use for training
clinProtein = pd.merge(clinicaldf, proteinsdf, on=["visit_id"], how="outer")
alldf = pd.merge(clinProtein, peptitesdf, on=["visit_id", "UniProt"], how="outer")
print("All Data")
print(alldf.describe())
print(alldf.info())
print(alldf.nunique())
print(alldf.head())
# ## Data Cleanup and enumerating the categorical data
# 1. Create a feature set for the abundance of each unique proteine and peptide included in the training set.
# 2. For each patient visit that has at least a updrs set of ratings 6, 12 and 24 months in the future, create a row for training to predict the 16 total updrs values (for each per visit).
# 3. Replace NaN with 0.0
# After this we will prepare a validation set and then begin training. Quick note, the validation set needs to follow the 0, 6, 12 and 24 month spacing. The plan is to sample out training also in those sets to feed to a Neural Network (e.g. 12, 18, 24, 36 months are also a case of 0, 6, 12, and 24 months).
# I'm going to try writing a DF with 227 features one for each UniProt, and 968 features, one for each Peptide, times 4 (once for each 0, 6, 12 and 24 month spaced sample) and train them on the expected 4 updrs outputs. The value in each feature will be the NPX or abundance value. Right now, we are using only the Protein data.
#
# first, build the feature index for each UniProt and Peptide
featureid = 0
for i in range(len(alldf)):
if catvars.getCatVarLabelMapping("FeatureID", alldf.loc[i, "UniProt"]) == -1:
catvars.setCatVarLabelMapping("FeatureID", alldf.loc[i, "UniProt"], featureid)
featureid = featureid + 1
if catvars.getCatVarLabelMapping("FeatureID6m", alldf.loc[i, "UniProt"]) == -1:
catvars.setCatVarLabelMapping("FeatureID6m", alldf.loc[i, "UniProt"], featureid)
featureid = featureid + 1
if catvars.getCatVarLabelMapping("FeatureID12m", alldf.loc[i, "UniProt"]) == -1:
catvars.setCatVarLabelMapping(
"FeatureID12m", alldf.loc[i, "UniProt"], featureid
)
featureid = featureid + 1
if catvars.getCatVarLabelMapping("FeatureID24m", alldf.loc[i, "UniProt"]) == -1:
catvars.setCatVarLabelMapping(
"FeatureID24m", alldf.loc[i, "UniProt"], featureid
)
featureid = featureid + 1
# if catvars.getCatVarLabelMapping('FeatureID', alldf.loc[i, 'Peptide']) == -1:
# catvars.setCatVarLabelMapping('FeatureID', alldf.loc[i, 'Peptide'], featureid)
# featureid = featureid + 1
print("The inputs will be", featureid, "features.")
# Create a training dataframe (tdf) with all features create above.
# build a blank data frame (tdf)
# what we need to do is predict the next 3 updrs sets (there are 12 outputs)
tdf = pd.DataFrame({})
allfeatures = []
for i in range(featureid):
featurename = "F" + str(i)
tdf[featurename] = 0.0 # consider making ndf and tdf.concat([tdf, ndf], axis=1)
allfeatures.insert(len(allfeatures), featurename)
# these are the features we have at month 0 for a test patient with
# which to predict the next 6, 12, and 24 month visits updrs 1,2,3,4 ratings
print(tdf.head())
print(allfeatures)
# The code below fills out tdf, with a parallel vdf (validate data frame). The vdf has the 16 updrs values to be predicated, based upon the protein (for now no peptide data is included). The tdf has one row for each clinical month that is evenly divisible by 6 and has at least 4 updrs examples available. The tdf creates a second row for months known to be month 6, 12, and 24 with additional features for those months.
# Now we need to scan alldf once for each row to add to tdf
# To do this we will select 4 months per patient space 0, 6, 12 and 24
# and do this for all patients. Each such combination makes one row in tdf
# Taking advantage that visit_id is patient_id _ month
import math
vdf = pd.DataFrame({})
def loadTdfVdf(tdf, vdf, alldf, clinicaldf, production):
month0found = 0
curpat = alldf.loc[0, "patient_id_x"]
kstart = 0
runlength = 0 # how many in a row - used to detect earlier month with no lab work
parallel_entries = 0
for i in range(len(clinicaldf)):
if production and i % 3 > 0:
continue # in production mode only process the every fourth line for its patient_x and month0found
month0found = clinicaldf.loc[i, "visit_month"]
if month0found % 6 != 0:
continue # be literal, keep with 0, 6, 12, and 24 months
if clinicaldf.loc[i, "patient_id"] != curpat:
continue # move to the next patient as last patient does not have enough forecasting data to use in training
# gather the 16 updrs values to train on - if we no longer have matches, then skip to the next patient
candidateOutput = {
"month": 0,
"updrs_1_0": 0.0,
"updrs_2_0": 0.0,
"updrs_3_0": 0.0,
"updrs_4_0": 0.0,
"updrs_1_6": 0.0,
"updrs_2_6": 0.0,
"updrs_3_6": 0.0,
"updrs_4_6": 0.0,
"updrs_1_12": 0.0,
"updrs_2_12": 0.0,
"updrs_3_12": 0.0,
"updrs_4_12": 0.0,
"updrs_1_24": 0.0,
"updrs_2_24": 0.0,
"updrs_3_24": 0.0,
"updrs_4_24": 0.0,
}
findpat = curpat
j = i
rowsleft = 3
## Must test for and impute NaN values - they show up in y_train...
candidateOutput["month"] = month0found
if not production:
if not math.isnan(clinicaldf.loc[j, "updrs_1"]):
candidateOutput["updrs_1_0"] = clinicaldf.loc[j, "updrs_1"]
if not math.isnan(clinicaldf.loc[j, "updrs_2"]):
candidateOutput["updrs_2_0"] = clinicaldf.loc[j, "updrs_2"]
if not math.isnan(clinicaldf.loc[j, "updrs_3"]):
candidateOutput["updrs_3_0"] = clinicaldf.loc[j, "updrs_3"]
if not math.isnan(clinicaldf.loc[j, "updrs_4"]):
candidateOutput["updrs_4_0"] = clinicaldf.loc[j, "updrs_4"]
while (
(j < len(clinicaldf))
and (clinicaldf.loc[j, "patient_id"] == findpat)
and (rowsleft > 0)
):
if clinicaldf.loc[j, "visit_month"] == month0found + 6:
if not math.isnan(clinicaldf.loc[j, "updrs_1"]):
candidateOutput["updrs_1_6"] = clinicaldf.loc[j, "updrs_1"]
if not math.isnan(clinicaldf.loc[j, "updrs_2"]):
candidateOutput["updrs_2_6"] = clinicaldf.loc[j, "updrs_2"]
if not math.isnan(clinicaldf.loc[j, "updrs_3"]):
candidateOutput["updrs_3_6"] = clinicaldf.loc[j, "updrs_3"]
if not math.isnan(clinicaldf.loc[j, "updrs_4"]):
candidateOutput["updrs_4_6"] = clinicaldf.loc[j, "updrs_4"]
rowsleft = rowsleft - 1
if clinicaldf.loc[j, "visit_month"] == month0found + 12:
if not math.isnan(clinicaldf.loc[j, "updrs_1"]):
candidateOutput["updrs_1_12"] = clinicaldf.loc[j, "updrs_1"]
if not math.isnan(clinicaldf.loc[j, "updrs_2"]):
candidateOutput["updrs_2_12"] = clinicaldf.loc[j, "updrs_2"]
if not math.isnan(clinicaldf.loc[j, "updrs_3"]):
candidateOutput["updrs_3_12"] = clinicaldf.loc[j, "updrs_3"]
if not math.isnan(clinicaldf.loc[j, "updrs_4"]):
candidateOutput["updrs_4_12"] = clinicaldf.loc[j, "updrs_4"]
rowsleft = rowsleft - 1
if clinicaldf.loc[j, "visit_month"] == month0found + 24:
if not math.isnan(clinicaldf.loc[j, "updrs_1"]):
candidateOutput["updrs_1_24"] = clinicaldf.loc[j, "updrs_1"]
if not math.isnan(clinicaldf.loc[j, "updrs_2"]):
candidateOutput["updrs_2_24"] = clinicaldf.loc[j, "updrs_2"]
if not math.isnan(clinicaldf.loc[j, "updrs_3"]):
candidateOutput["updrs_3_24"] = clinicaldf.loc[j, "updrs_3"]
if not math.isnan(clinicaldf.loc[j, "updrs_4"]):
candidateOutput["updrs_4_24"] = clinicaldf.loc[j, "updrs_4"]
rowsleft = rowsleft - 1
j = j + 1
else:
rowsleft = 0
j = j + 3
if rowsleft > 0: # not enough left to train forcasting
if j >= len(clinicaldf):
break # we processed the last patient; avoid out of bounds index
curpat = clinicaldf.loc[j, "patient_id"]
continue
else:
print(
"Data collection for patient id", curpat, "with base month", month0found
)
# if we have a training line, then build the features for it - advance i one past the end of this patient and month
# since we did not abort with a continue, we should be safe to find the
# patient_id and months ahead, just us them to fill out a new row for the training set
candidateInput = {}
for k in range(featureid):
featurename = "F" + str(k)
candidateInput[featurename] = 0.0
startedProcessing = False
for k in range(kstart, len(alldf)):
if (
alldf.loc[k, "patient_id_x"] == curpat
): # looking at the right patient
if alldf.loc[k, "visit_month_x"] == month0found:
protid = catvars.getCatVarLabelMapping(
"FeatureID", alldf.loc[k, "UniProt"]
)
if protid == -1:
stophere = 0
stophere = 1 / stophere
# peptid = catvars.getCatVarLabelMapping('FeatureID', alldf.loc[k, 'Peptide'])
candidateInput["F" + str(protid)] = alldf.loc[k, "NPX"]
# candidateInput["F"+str(peptid)] = alldf.loc[k, "PeptideAbundance"]
startedProcessing = True
else:
if startedProcessing:
kstart = k # we can start at the new next patient's row
break # we are on to the next patient, we can stop
else:
if startedProcessing:
kstart = k # we can start at the new next patient's row
runlength = 0
break # we are on to the next patient, we can stop
if startedProcessing:
zerocount = (
0 # yes, some clinical updates on updrs does not have lab results
)
for inp in candidateInput:
if math.isnan(candidateInput[inp]) or candidateInput[inp] == 0:
zerocount = zerocount + 1
if zerocount < len(candidateInput): # we have lab results
vdf = vdf.append(candidateOutput, ignore_index=True)
tdf = tdf.append(candidateInput, ignore_index=True)
runlength = runlength + 1
if (
month0found == 6 and runlength > 0
): # add an extra row using FeatureId6m (odd F#)
vdf = vdf.append(candidateOutput, ignore_index=True)
tdf = tdf.append(candidateInput, ignore_index=True)
therow = len(tdf) - 1
for kk in range(featureid):
if kk % 4 == 0:
tdf.iloc[therow, kk + 1] = tdf.iloc[therow, kk]
tdf.iloc[therow, kk] = tdf.iloc[therow - 1, kk]
if (
month0found == 12 and runlength > 1
): # add an extra row using FeatureId6m (odd F#)
vdf = vdf.append(candidateOutput, ignore_index=True)
tdf = tdf.append(candidateInput, ignore_index=True)
therow = len(tdf) - 1
for kk in range(featureid):
if kk % 4 == 0:
tdf.iloc[therow, kk + 2] = tdf.iloc[therow, kk]
tdf.iloc[therow, kk + 1] = tdf.iloc[therow - 1, kk]
tdf.iloc[therow, kk] = tdf.iloc[therow - 1, kk]
if (
month0found == 24 and runlength > 2
): # add an extra row using FeatureId6m (odd F#)
vdf = vdf.append(candidateOutput, ignore_index=True)
tdf = tdf.append(candidateInput, ignore_index=True)
therow = len(tdf) - 1
for kk in range(featureid):
if kk % 4 == 0:
tdf.iloc[therow, kk + 3] = tdf.iloc[therow, kk]
tdf.iloc[therow, kk + 2] = tdf.iloc[therow - 1, kk]
tdf.iloc[therow, kk + 1] = tdf.iloc[therow - 1, kk]
tdf.iloc[therow, kk] = tdf.iloc[therow - 1, kk]
else:
runlength = 0
print(
"No lab results detected - will not be able to train on this."
)
parallel_entries = parallel_entries + 1
return tdf, vdf
tdf, vdf = loadTdfVdf(tdf, vdf, alldf, clinicaldf, False)
vdf_months = vdf["month"] # keep a parallel copy of which month each row represents
vdf.drop("month", axis=1, inplace=True) # but remove it until it is needed
def drop_nan_cols(df):
dropped_cols = []
for col in df.columns:
if df[col].isnull().any():
df.drop(col, axis=1, inplace=True)
dropped_cols.append(col)
return dropped_cols
dropsmade = drop_nan_cols(tdf)
print("Columns dropped are ", dropsmade)
for i in range(len(dropsmade)):
try:
allfeatures.remove(dropsmade[i])
except:
pass
# Use Mutual Information Scores to pick features for each of the 16 UPDRS values to be predicted for sample protein data.
from sklearn.feature_selection import mutual_info_regression
import pandas as pd
def rank_features_by_mutual_info(tdf: pd.DataFrame, y: pd.Series):
mi_scores = mutual_info_regression(tdf, y)
mi_scores = pd.Series(mi_scores, name="MI Scores", index=tdf.columns)
mi_scores = mi_scores.sort_values(ascending=False)
return list(mi_scores.items())
def top_features(feature_scores):
top_features_list = []
for feature in feature_scores:
if feature[1] > 0.02 or len(top_features_list) < 8: # SMAPE/all .90 using 0.05.
top_features_list.append(feature[0])
else:
if len(top_features_list) == 0:
top_features_list.append(feature[0])
break
return top_features_list
vfeatures_num = [
"updrs_1_0",
"updrs_2_0",
"updrs_3_0",
"updrs_4_0",
"updrs_1_6",
"updrs_2_6",
"updrs_3_6",
"updrs_4_6",
"updrs_1_12",
"updrs_2_12",
"updrs_3_12",
"updrs_4_12",
"updrs_1_24",
"updrs_2_24",
"updrs_3_24",
"updrs_4_24",
]
# goal, get all 16, and combine the top features off each to see what
# the list looks like and how well the algorithm works to regress on them...
tdf_subset = []
tdf_feat_subset = []
for vfeat in vfeatures_num:
miscores = rank_features_by_mutual_info(tdf, vdf[vfeat])
usefeats = top_features(miscores)
print("Total features in", vfeat, "=", len(usefeats))
tdfsub = tdf[usefeats].copy()
tdf_subset.append(tdfsub)
tdf_feat_subset.append(usefeats)
# Build a pipeline to transform the tdf and vdf data to make it safe for models to use.
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler, MinMaxScaler, OneHotEncoder
from sklearn.impute import SimpleImputer
from sklearn.pipeline import make_pipeline
from sklearn.decomposition import PCA
from sklearn.compose import make_column_transformer
vfeatinx = 0
shape_to_use = []
X_train_list = []
X_valid_list = []
y_train_list = []
y_valid_list = []
y_train_month_list = []
y_valid_month_list = []
def prepare_features_for_models(
productionflag,
vfeatinx,
shape_to_use,
X_train_list,
X_valid_list,
y_train_list,
y_valid_list,
vdf_months,
y_train_month_list,
y_valid_month_list,
):
for vfeat in vfeatures_num:
features_num = tdf_feat_subset[vfeatinx]
features_cat = []
vfeatures_cat = []
# Before and instead of using the built in split, we need to have separate training sets
# for each of the month numbers 0, 6, 12, 18, 24, 30, and 36 (7 sets in all).
# Set aside one in five of each for validation, and the rest for training.
# NN training does not separate them, and proteins along the way change causing random
# results, the aim here is to elliminate the randomness.
# The Month0 only data
tdf0 = tdf_subset[vfeatinx].copy()
vdf0 = vdf.copy()
transformer_num = make_pipeline(
SimpleImputer(
strategy="constant", fill_value=0.0
), # there are a few missing values
# PCA(n_components=16), # this destroyed the validation results SMAPE 1.92 (either not used correctly or not appropriate)
# MinMaxScaler(), # for some reason, this results in poor predictions
# StandardScaler(),
)
transformer_cat = make_pipeline(
SimpleImputer(strategy="constant", fill_value=0),
OneHotEncoder(handle_unknown="ignore"),
)
vtransformer_num = make_pipeline(
SimpleImputer(
strategy="constant", fill_value=0.0
), # there are a few missing values
# StandardScaler(),
)
vtransformer_cat = make_pipeline(
SimpleImputer(strategy="constant", fill_value=0),
OneHotEncoder(handle_unknown="ignore"),
)
preprocessor = make_column_transformer(
(transformer_num, features_num),
(transformer_cat, features_cat),
)
vpreprocessor = make_column_transformer(
(transformer_num, vfeatures_num),
(transformer_cat, vfeatures_cat),
)
if productionflag:
X_train = preprocessor.fit_transform(tdf0)
y_train = vpreprocessor.fit_transform(vdf0)
y_train_month_list.append(vdf_months)
X_train_list.append(X_train)
y_train_list.append(y_train)
input_shape_num = [X_train.shape[1]]
vfeatinx = vfeatinx + 1
print("The input shape is ", input_shape_num)
shape_to_use.append(X_train.shape[1])
else:
vdf0["month"] = vdf_months
X_train, X_valid, y_train, y_valid = train_test_split(
tdf0,
vdf0,
# stratify=vdf,
train_size=0.8,
random_state=283,
) # was 103 showing SMAPE of 63.3 and 57.3
vdf0.drop("month", axis=1, inplace=True) # but remove it until it is needed
y_train_month = y_train["month"]
y_train.drop(
"month", axis=1, inplace=True
) # but remove it until it is needed
y_valid_month = y_valid["month"]
y_valid.drop(
"month", axis=1, inplace=True
) # but remove it until it is needed
X_train = preprocessor.fit_transform(X_train)
X_valid = preprocessor.fit_transform(X_valid)
y_train = vpreprocessor.fit_transform(y_train)
y_valid = vpreprocessor.fit_transform(y_valid)
X_train_list.append(X_train)
X_valid_list.append(X_valid)
y_train_list.append(y_train)
y_valid_list.append(y_valid)
y_train_month_list.append([y_train_month])
# print("UPDRS ", vfeatinx, "value", y_train_month)
# print("y_train_month_list", y_train_month_list)
y_valid_month_list.append([y_valid_month])
input_shape_num = [X_train.shape[1]]
vfeatinx = vfeatinx + 1
# print("The input shape is ", input_shape_num)
shape_to_use.append(X_train.shape[1])
prepare_features_for_models(
False,
vfeatinx,
shape_to_use,
X_train_list,
X_valid_list,
y_train_list,
y_valid_list,
vdf_months,
y_train_month_list,
y_valid_month_list,
)
# ## Build one model to train for each updrs to predict (16 total).
from sklearn.linear_model import LinearRegression
from sklearn.ensemble import RandomForestRegressor
from sklearn.ensemble import GradientBoostingRegressor
from sklearn.linear_model import SGDRegressor
from sklearn.linear_model import BayesianRidge
from xgboost.sklearn import XGBRegressor
# maybe revisit DDM, but note the DDM was at 1.47,
# LinearRegression pulled off a score of 1.17, and
# RandomForestRegressor made it to a SMAPE of 0.84 (the best by far)
# all without hyper tunning.
# We really do need to clean up the input features to optimize the
# regression results.
# Adding in all peptides made RandomForestRegressor drop to 0.93
# (first we will optimize with just the protein abundance info)
modelList = []
def create_models(modelList):
for i in range(16):
# model = LinearRegression() # aweful scores - scatter plot is all over the place
# model = RandomForestRegressor(n_estimators = 1000, max_depth=7, min_samples_split=10, min_samples_leaf=2, max_features=None, random_state = 0)
model = RandomForestRegressor(n_estimators=500, random_state=0) # best results
# model = GradientBoostingRegressor() # seems to underperform relative to RFR
# model = SGDRegressor() # aweful - the results are SMAPE > 1.0
# model = BayesianRidge() # better than SGCRegressor, but close to SMAPE of 1.0
# model = XGBRegressor() # seems to overfit, but is also a clear diagnol and horizontal line with total misses on prediction
modelList.append(model)
create_models(modelList)
# ## Fit each of the 16 models and then verify the prediction SMAPE score we anticipate from it
import matplotlib.pyplot as plt
import numpy as np
from scipy import stats
# early_stopping = keras.callbacks.EarlyStopping(
# patience=5,
# min_delta=0.001,
# restore_best_weights=True,
# )
def get_elements_at_index(lists, index):
result = []
for inner_list in lists:
result.append([inner_list[index]])
return result
i = 0
sumpart = 0
totalruns = 0
sumpart2 = 0
totalruns2 = 0
for model in modelList:
y_train_inner = np.array(get_elements_at_index(y_train_list[i], i)).ravel()
y_valid_inner = np.array(get_elements_at_index(y_valid_list[i], i)).ravel()
history = model.fit(X_train_list[i], y_train_inner)
tt_pred = model.predict(
X_train_list[i]
) # let's find out if we are over or under running consistently
sumdiff = 0.0
raisedneg = False
raisedover = False
for jj in range(len(tt_pred)):
sumdiff = sumdiff + (y_train_inner[jj] - tt_pred[jj])
if tt_pred[jj] < 0 and not raisedneg:
print("Some predictions are negative")
tt_pred[jj] = 0
raisedneg = True
if tt_pred[jj] > 100 and not raisedover:
print("Some predictions are greater than 100")
tt_pred[jj] = 100
raisedover = True
nomin2 = abs(y_train_inner[jj] - tt_pred[jj])
denomin2 = abs(y_train_inner[jj]) + abs(tt_pred[jj])
if denomin2 > 0:
sumpart2 = sumpart2 + (nomin2 / (denomin2 / 2.0))
totalruns2 = totalruns2 + 1
print("The average", vfeatures_num[i], "is off by", sumdiff / len(tt_pred))
print("Training performance...")
plt.scatter(y_train_inner, tt_pred)
plt.title("Scatter plot of training to trained predictions")
plt.xlabel("Training value")
plt.ylabel("Predicted value")
plt.show()
print("Median of training values ", np.median(tt_pred))
print("Mode of training values ", stats.mode(tt_pred), "of", len(tt_pred))
sumdiff = 0
y_pred = model.predict(X_valid_list[i])
for j in range(len(y_pred)):
y_pred[j] = int(y_pred[j])
sumdiff = sumdiff + (y_valid_inner[j] - y_pred[j])
if y_pred[j] < 0:
y_pred[j] = 0
if y_pred[j] > 100:
y_pred[j] = 100
nomin = abs(y_valid_inner[j] - y_pred[j])
denomin = abs(y_valid_inner[j]) + abs(y_pred[j])
if denomin > 0:
sumpart = sumpart + (nomin / (denomin / 2.0))
totalruns = totalruns + 1
print(
"The average validation prediction",
vfeatures_num[i],
"is off by",
sumdiff / len(y_pred),
)
print("Validation performance...")
plt.scatter(y_valid_inner, y_pred)
plt.title("Scatter plot of training to validatation predictions")
plt.xlabel("Validation value")
plt.ylabel("Predicted value")
plt.show()
print("Median of predicted values ", np.median(y_pred))
print("Mode of predicted values ", stats.mode(y_pred), "of", len(y_pred))
i = i + 1
smape = (1 / totalruns2) * sumpart2
print("SMAPE train = ", smape)
smape = (1 / totalruns) * sumpart
print("SMAPE valid = ", smape)
#
# We need to rework this data, a set of results show an indication where Random Forest Regressor is
# just returning a value near the average or median value which suggests the protein data is not
# correct to predict the value. These may belong to the next 6 months set (but were still month 0 when recorded.)
# ## Reset the environment and train models using the feature sets already established with all of the training data.
# Code to rebuild the models (all 16 of them) trained with all of the available training data
vfeatinx = 0
shape_to_use = []
X_train_list = []
X_valid_list = []
y_train_list = []
y_valid_list = []
y_train_month_list = []
y_valid_month_list = []
prepare_features_for_models(
True,
vfeatinx,
shape_to_use,
X_train_list,
X_valid_list,
y_train_list,
y_valid_list,
vdf_months,
y_train_month_list,
y_valid_month_list,
)
modelList = []
create_models(modelList)
# ## initialize for a new set of outputs
vfeatinxL2 = 0
shape_to_useL2 = []
X_train_listL2 = []
X_valid_listL2 = []
y_train_listL2 = []
y_valid_listL2 = []
y_train_month_listL2 = []
y_valid_month_listL2 = []
tdfL2 = pd.DataFrame(columns=tdf.columns)
vdfL2 = pd.DataFrame(columns=vdf.columns)
newLocList = []
vdfL2.info()
tdfL2.info()
# ## Fit all the data to each of the 16 models and confirm that the training data predictions looks similar to those when we split (a sanit test of the model).
i = 0
sumpart2 = 0
totalruns2 = 0
for model in modelList:
y_train_inner = np.array(get_elements_at_index(y_train_list[i], i)).ravel()
history = model.fit(X_train_list[i], y_train_inner)
tt_pred = model.predict(
X_train_list[i]
) # let's find out if we are over or under running consistently
sumdiff = 0.0
raisedneg = False
raisedover = False
for jj in range(len(tt_pred)):
sumdiff = sumdiff + (y_train_inner[jj] - tt_pred[jj])
if tt_pred[jj] < 0 and not raisedneg:
print("Some predictions are negative")
tt_pred[jj] = 0
raisedneg = True
if tt_pred[jj] > 60 and not raisedover:
print("Some predictions are greater than 60")
tt_pred[jj] = 60
raisedover = True
# estimate the SMAPE's summation
nomin2 = abs(y_train_inner[jj] - tt_pred[jj])
denomin2 = abs(y_train_inner[jj]) + abs(tt_pred[jj])
if denomin2 > 0:
sumpart2 = sumpart2 + (nomin2 / (denomin2 / 2.0))
totalruns2 = totalruns2 + 1
sumdiff = sumdiff / len(tt_pred)
print("The average", vfeatures_num[i], "is off by", sumdiff)
# if totalruns == 0:
plt.scatter(y_train_inner, tt_pred)
plt.title("Scatter plot of training to trained predictions")
plt.xlabel("Training value")
plt.ylabel("Predicted value")
plt.show()
print("Median of training values ", np.median(tt_pred))
print("Mode of training values ", stats.mode(tt_pred), "of", len(tt_pred))
i = i + 1
# history_df = pd.DataFrame(history.history)
# history_df.loc[:, ['loss', 'val_loss']].plot()
smape = (1 / totalruns2) * sumpart2
print("SMAPE train = ", smape)
# print("A look at tdf")
# print(tdf.info())
# print(tdf.describe())
# print(tdf.nunique())
# print(tdf.head(30))
# print(tdf.tail(30))
# print("A look at vdf")
# print(vdf.info())
# print(vdf.describe())
# print(vdf.nunique())
# print(vdf.head(30))
# print(vdf.tail(30))
vfeatinx = 0
tt_pred_list = []
import amp_pd_peptide
env = amp_pd_peptide.make_env() # initialize the environment
iter_test = env.iter_test() # an iterator which loops over the test files
for test, test_peptides, test_proteins, sample_submission in iter_test:
myclinProtein = pd.merge(test, test_proteins, on=["visit_id"], how="outer")
myalldf = pd.merge(
myclinProtein, test_peptides, on=["visit_id", "UniProt"], how="outer"
)
tdf, vdf = loadTdfVdf(tdf, vdf, myalldf, test, True)
print("tdf", tdf.head(32))
print("test", test.head(20))
for model in modelList:
tdf0 = tdf[tdf_feat_subset[vfeatinx]].copy()
# fitting transform
transformer_num = make_pipeline(
SimpleImputer(strategy="constant", fill_value=0.0),
)
transformer_cat = make_pipeline(
SimpleImputer(strategy="constant", fill_value=0),
OneHotEncoder(handle_unknown="ignore"),
)
vtransformer_num = make_pipeline(
SimpleImputer(
strategy="constant", fill_value=0.0
), # there are a few missing values
# StandardScaler(),
)
vtransformer_cat = make_pipeline(
SimpleImputer(strategy="constant", fill_value=0),
OneHotEncoder(handle_unknown="ignore"),
)
preprocessor = make_column_transformer(
(transformer_num, features_num),
(transformer_cat, features_cat),
)
vpreprocessor = make_column_transformer(
(transformer_num, vfeatures_num),
(transformer_cat, vfeatures_cat),
)
tdf0 = preprocessor.fit_transform(tdf0)
# end fitting transform
vfeatinx = vfeatinx + 1
tt_pred = model.predict(
tdf0
) # let's find out if we are over or under running consistently
for jj in range(len(tt_pred)):
if tt_pred[jj] < 0 and not raisedneg:
tt_pred[jj] = 0
if tt_pred[jj] > 60 and not raisedover:
tt_pred[jj] = 60
tt_pred_list.append(tt_pred)
rateloc = 1 # hard coded
for i in range(len(tt_pred)):
if i % 16 == 0:
sample_submission.iloc[i, rateloc] = tt_pred_list[0][i]
sample_submission.iloc[i + 1, rateloc] = tt_pred_list[4][i]
sample_submission.iloc[i + 2, rateloc] = tt_pred_list[8][i]
sample_submission.iloc[i + 3, rateloc] = tt_pred_list[12][i]
sample_submission.iloc[i + 4, rateloc] = tt_pred_list[1][i]
sample_submission.iloc[i + 5, rateloc] = tt_pred_list[5][i]
sample_submission.iloc[i + 6, rateloc] = tt_pred_list[9][i]
sample_submission.iloc[i + 7, rateloc] = tt_pred_list[13][i]
sample_submission.iloc[i + 8, rateloc] = tt_pred_list[2][i]
sample_submission.iloc[i + 9, rateloc] = tt_pred_list[6][i]
sample_submission.iloc[i + 10, rateloc] = tt_pred_list[10][i]
sample_submission.iloc[i + 11, rateloc] = tt_pred_list[14][i]
sample_submission.iloc[i + 12, rateloc] = tt_pred_list[3][i]
sample_submission.iloc[i + 13, rateloc] = tt_pred_list[7][i]
sample_submission.iloc[i + 14, rateloc] = tt_pred_list[11][i]
sample_submission.iloc[i + 15, rateloc] = tt_pred_list[15][i]
sample_prediction_df["rating"] = np.arange(
len(sample_prediction)
) # make your predictions her
env.predict(sample_submission) # register your predictions
pd.read_csv("/kaggle/working/submission.csv")
|
#
# Introduction
# Welcome to my notebook!
# Ensemble is a technique of combining multiple models to improve performance and reduce noises of models.
# Ensemble is a powerful technique to improve your score in Kaggle Competitions.
# In this notebook, I compare some ensemble methods, Majority Voting, Mean Method, Brending, Stacking.
# Import Library
#
import warnings
warnings.filterwarnings("ignore") # Ignore all warnings
from IPython.display import clear_output
from collections import Counter
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# for plot result
import matplotlib.pyplot as plt
import seaborn as sns
# preprocess, split data, model, and metrics
from sklearn.preprocessing import LabelEncoder, StandardScaler
from sklearn.model_selection import StratifiedKFold, cross_validate, train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.neighbors import KNeighborsClassifier
from sklearn.svm import SVC
from sklearn.ensemble import (
RandomForestClassifier,
GradientBoostingClassifier,
HistGradientBoostingClassifier,
StackingClassifier,
)
from sklearn.metrics import accuracy_score, roc_auc_score
# scipy
import scipy
# catboost
from catboost import CatBoostClassifier
# XGBoost
from xgboost import XGBClassifier
# lightgbm
from lightgbm import LGBMClassifier
# tensorflow
import tensorflow as tf
# pytorch
import torch
# Check the path of data
import os
for dirname, _, filenames in os.walk("/kaggle/input/titanic"):
for filename in filenames:
print(os.path.join(dirname, filename))
TEST_SIZE = 0.2
SHUFFLE = True
SEED = 42 # for Reproducibililty
#
# Read Data
#
train_df = pd.read_csv("/kaggle/input/titanic/train.csv")
#
# Feature Engineering
#
def titanic_preprocessing(input_df):
df = input_df.copy()
# missin values
# Age is int(or float) so use mean or median
# Embarked is caterogy(label) so use mode
df["Age"] = df["Age"].fillna(df["Age"].median())
df["Embarked"] = df["Embarked"].fillna(df["Embarked"].mode()[0])
df["Fare"] = df["Fare"].fillna(df["Fare"].median()) # Fare is missing in test data
# extract title
df["Title"] = df["Name"].str.extract(" ([A-Za-z]+)\.", expand=False)
df["Title"] = df["Title"].replace(
[
"Lady",
"Countess",
"Capt",
"Col",
"Don",
"Dr",
"Major",
"Rev",
"Sir",
"Jonkheer",
"Dona",
],
"Rare",
)
df["Title"] = df["Title"].replace("Mlle", "Miss")
df["Title"] = df["Title"].replace("Ms", "Miss")
df["Title"] = df["Title"].replace("Mme", "Mrs")
# Label Encoding
# let the string category feature to integer
le = LabelEncoder()
df["Sex"] = le.fit_transform(df["Sex"])
df["Embarked"] = le.fit_transform(df["Embarked"])
df["Title"] = le.fit_transform(df["Title"])
# Standardize
# Scaling int or float feature, it improve predict_probaion for especially Neural Network Model.
sc = StandardScaler()
df[["Age", "Fare"]] = sc.fit_transform(df[["Age", "Fare"]])
# Get the number of passenger in each group
df["PassengersInGroup"] = (
df["SibSp"] + df["Parch"] + 1
) # Siblings/Spouses + Parent/Children + him/herself
df["IsAlone"] = df["PassengersInGroup"].apply(lambda x: 1 if x == 1 else 0)
# drop sibsp and parch because these have high correlation for PassengersInGroup
df = df.drop(columns=["PassengerId", "Name", "Ticket", "Cabin", "SibSp", "Parch"])
return df
train_data = titanic_preprocessing(train_df)
train_data.head()
# target column
target_col = "Survived"
# columns, using for predict_probaion
feature_cols = train_data.columns.tolist()
feature_cols.remove(target_col)
X, y = np.array(train_data[feature_cols]), np.array(train_data[target_col])
train_X, val_X, train_y, val_y = train_test_split(
X, y, test_size=TEST_SIZE, shuffle=SHUFFLE, random_state=SEED
)
#
# Define Models
# Define models we use in ensemble.
#
lr_model = LogisticRegression(random_state=SEED)
kn_model = KNeighborsClassifier()
svc_model = SVC(probability=True, random_state=SEED)
rf_model = RandomForestClassifier(random_state=SEED)
gb_model = GradientBoostingClassifier(random_state=SEED)
hgb_model = HistGradientBoostingClassifier(random_state=SEED)
cb_model = CatBoostClassifier(random_seed=SEED)
xgb_model = XGBClassifier(random_state=SEED)
lgb_model = LGBMClassifier(random_state=SEED)
#
# Before trying Ensemble, I will check each model's performances.
#
lr_model.fit(train_X, train_y)
kn_model.fit(train_X, train_y)
svc_model.fit(train_X, train_y)
rf_model.fit(train_X, train_y)
gb_model.fit(train_X, train_y)
hgb_model.fit(train_X, train_y)
cb_model.fit(train_X, train_y)
xgb_model.fit(train_X, train_y)
lgb_model.fit(train_X, train_y)
clear_output()
lr_pred = lr_model.predict(val_X)
kn_pred = kn_model.predict(val_X)
svc_pred = svc_model.predict(val_X)
rf_pred = rf_model.predict(val_X)
gb_pred = gb_model.predict(val_X)
hgb_pred = hgb_model.predict(val_X)
cb_pred = cb_model.predict(val_X)
xgb_pred = xgb_model.predict(val_X)
lgb_pred = lgb_model.predict(val_X)
lr_acc = accuracy_score(val_y, lr_pred)
kn_acc = accuracy_score(val_y, kn_pred)
svc_acc = accuracy_score(val_y, svc_pred)
rf_acc = accuracy_score(val_y, rf_pred)
gb_acc = accuracy_score(val_y, gb_pred)
hgb_acc = accuracy_score(val_y, hgb_pred)
cb_acc = accuracy_score(val_y, cb_pred)
xgb_acc = accuracy_score(val_y, xgb_pred)
lgb_acc = accuracy_score(val_y, lgb_pred)
print(f"Logistic Regression: {lr_acc:.4}")
print(f"K-Neibors: {kn_acc:.4}")
print(f"SupportVector Machine: {svc_acc:.4}")
print(f"Random Forest: {rf_acc:.4}")
print(f"Gradient Boosting: {gb_acc:.4}")
print(f"Hist Gradient Boosting: {hgb_acc:.4}")
print(f"CatBoost: {cb_acc:.4}")
print(f"XGBoost: {xgb_acc:.4}")
print(f"LightGBM: {lgb_acc:.4}")
#
# LightGBM: 0.8547 is the maximum aucuracy.
# Also check roc_auc_score.
#
lr_proba = lr_model.predict_proba(val_X)[:, 1]
kn_proba = kn_model.predict_proba(val_X)[:, 1]
svc_proba = svc_model.predict_proba(val_X)[:, 1]
rf_proba = rf_model.predict_proba(val_X)[:, 1]
gb_proba = gb_model.predict_proba(val_X)[:, 1]
hgb_proba = hgb_model.predict_proba(val_X)[:, 1]
cb_proba = cb_model.predict_proba(val_X)[:, 1]
xgb_proba = xgb_model.predict_proba(val_X)[:, 1]
lgb_proba = lgb_model.predict_proba(val_X)[:, 1]
lr_auc = roc_auc_score(val_y, lr_proba)
kn_auc = roc_auc_score(val_y, kn_proba)
svc_auc = roc_auc_score(val_y, svc_proba)
rf_auc = roc_auc_score(val_y, rf_proba)
gb_auc = roc_auc_score(val_y, gb_proba)
hgb_auc = roc_auc_score(val_y, hgb_proba)
cb_auc = roc_auc_score(val_y, cb_proba)
xgb_auc = roc_auc_score(val_y, xgb_proba)
lgb_auc = roc_auc_score(val_y, lgb_proba)
print(f"Logistic Regression: {lr_auc:.4}")
print(f"K-Neibors: {kn_auc:.4}")
print(f"SupportVector Machine: {svc_auc:.4}")
print(f"Random Forest: {rf_auc:.4}")
print(f"Gradient Boosting: {gb_auc:.4}")
print(f"Hist Gradient Boosting: {hgb_auc:.4}")
print(f"CatBoost: {cb_auc:.4}")
print(f"XGBoost: {xgb_auc:.4}")
print(f"LightGBM: {lgb_auc:.4}")
#
# Gradient Boosting: 0.9122 is the maximum roc_auc_score.
# Ensemble
# Now let's explore ensemble. I try 1 methods, majority voting,
# Majority Voting
# Simple method for classification is majority voting.
# It get mode value between models and decide for predict_probaion values. It reduces each model noise.
#
y_pred = np.stack(
[
lr_pred,
kn_pred,
svc_pred,
rf_pred,
gb_pred,
hgb_pred,
cb_pred,
xgb_pred,
lgb_pred,
]
).T
max_voting = np.apply_along_axis(scipy.stats.mode, 1, y_pred)[:, 0]
max_voting_auc = roc_auc_score(val_y, max_voting)
print(f"Majority Voting: {max_voting_auc:.4}")
#
# Valiadtion score decreases but Public Score increases. This mean that majority voting reduces noise.
# Mean
# getting mean of some models also reduces noises. There are some mean methods.
#
y_proba = np.stack(
[
lr_proba,
kn_proba,
svc_proba,
rf_proba,
gb_proba,
hgb_proba,
cb_proba,
xgb_proba,
lgb_proba,
]
).T
# arithmetric mean
arithmetic = y_proba.mean(axis=1)
arithmetic_auc = roc_auc_score(val_y, arithmetic)
print(f"Arithmetic Mean: {arithmetic_auc:.4f}")
# geometric mean
geometric = y_proba.prod(axis=1) ** (1 / y_proba.shape[1])
geometric_auc = roc_auc_score(val_y, geometric)
print(f"Geometric Mean: {geometric_auc:.4f}")
# harmonic mean
harmonic = 1.0 / np.mean(1.0 / (y_proba + 1e-5), axis=1)
harmonic_auc = roc_auc_score(val_y, harmonic)
print(f"Harmonic Mean: {harmonic_auc:.4f}")
# mean of powers
n = 3
mean_of_powers = np.mean(y_proba**n, axis=1) ** (1 / n)
mean_of_powers_auc = roc_auc_score(val_y, mean_of_powers)
print(f"Mean of Powers: {mean_of_powers_auc:.4f}")
# logarithmic mean
logarithmic = np.expm1(np.mean(np.log1p(y_proba), axis=1))
logarithmic_auc = roc_auc_score(val_y, logarithmic)
print(f"Logarithmic Mean: {logarithmic_auc:.4f}")
#
# Weighted Mean
# Next method is weighted mean. It takes correlation of model predictions and set weights based on the correlation. It effectively reduce variance of models.
#
# get correlation matrix.
# set the diagonal values to 0
# take average
# get the sum of inverse value
# normalize sum
#
#
y_proba = np.stack(
[
lr_proba,
kn_proba,
svc_proba,
rf_proba,
gb_proba,
hgb_proba,
cb_proba,
xgb_proba,
lgb_proba,
]
).T
cormat = np.corrcoef(y_proba.T)
np.fill_diagonal(cormat, 0.0)
W = 1 / np.mean(cormat, axis=1)
W = W / sum(W)
weighted = y_proba.dot(W)
weighted_auc = roc_auc_score(val_y, weighted)
print(f"Weighted Mean: {weighted_auc:.4f}")
#
# Brending
# Next method is brending. It is one of weighted mean. It decide weight by holdout and meta-estimator.
#
# sample holdout dataset from train data.
# train models by train data.
# predict from holdout data.
# train meta-estimator by prediction of holdout data.
# get weight by meta-estimator and prediction of validation data.
#
# Brending with Linear Meta Estimator
# We can use linear or non-linear estimator for meta-estimator, so I check the difference of these results. At first, liear meta-estimator.
# 1. sample blend and holdout data from trian data.
#
X_blend, X_holdout, y_blend, y_holdout = train_test_split(
train_X, train_y, test_size=0.1, shuffle=SHUFFLE, random_state=SEED
)
#
# 2. train models by blend data.
#
lr_model = LogisticRegression(random_state=SEED)
kn_model = KNeighborsClassifier()
svc_model = SVC(probability=True, random_state=SEED)
rf_model = RandomForestClassifier(random_state=SEED)
gb_model = GradientBoostingClassifier(random_state=SEED)
hgb_model = HistGradientBoostingClassifier(random_state=SEED)
cb_model = CatBoostClassifier(random_seed=SEED)
xgb_model = XGBClassifier(random_state=SEED)
lgb_model = LGBMClassifier(random_state=SEED)
lr_model.fit(X_blend, y_blend)
kn_model.fit(X_blend, y_blend)
svc_model.fit(X_blend, y_blend)
rf_model.fit(X_blend, y_blend)
gb_model.fit(X_blend, y_blend)
hgb_model.fit(X_blend, y_blend)
cb_model.fit(X_blend, y_blend)
xgb_model.fit(X_blend, y_blend)
lgb_model.fit(X_blend, y_blend)
clear_output()
#
# 3. predict holdout data.
#
y_proba = np.stack(
[
lr_model.predict_proba(X_holdout)[:, 1],
kn_model.predict_proba(X_holdout)[:, 1],
svc_model.predict_proba(X_holdout)[:, 1],
rf_model.predict_proba(X_holdout)[:, 1],
gb_model.predict_proba(X_holdout)[:, 1],
hgb_model.predict_proba(X_holdout)[:, 1],
cb_model.predict_proba(X_holdout)[:, 1],
xgb_model.predict_proba(X_holdout)[:, 1],
lgb_model.predict_proba(X_holdout)[:, 1],
]
).T
#
# 4. train meta-estimator by prediction.
#
sc = StandardScaler()
X_meta = sc.fit_transform(y_proba)
meta_estimator = LogisticRegression(solver="liblinear", random_state=0)
meta_estimator.fit(X_meta, y_holdout)
print(meta_estimator.coef_)
#
# 5. predict validation data by meta-estimator.
#
y_proba = np.stack(
[
lr_model.predict_proba(val_X)[:, 1],
kn_model.predict_proba(val_X)[:, 1],
svc_model.predict_proba(val_X)[:, 1],
rf_model.predict_proba(val_X)[:, 1],
gb_model.predict_proba(val_X)[:, 1],
hgb_model.predict_proba(val_X)[:, 1],
cb_model.predict_proba(val_X)[:, 1],
xgb_model.predict_proba(val_X)[:, 1],
lgb_model.predict_proba(val_X)[:, 1],
]
).T
blending = meta_estimator.predict_proba(y_proba)[:, 1]
blending_auc = roc_auc_score(val_y, blending)
print(f"Blending with Linear: {blending_auc:.4f}")
#
# Brending with Non-Linear Meta Estimator
# We can use non-linear meta-estimator.
#
X_blend, X_holdout, y_blend, y_holdout = train_test_split(
train_X, train_y, test_size=0.1, shuffle=SHUFFLE, random_state=SEED
)
lr_model = LogisticRegression(random_state=SEED)
kn_model = KNeighborsClassifier()
svc_model = SVC(probability=True, random_state=SEED)
rf_model = RandomForestClassifier(random_state=SEED)
gb_model = GradientBoostingClassifier(random_state=SEED)
hgb_model = HistGradientBoostingClassifier(random_state=SEED)
cb_model = CatBoostClassifier(random_seed=SEED)
xgb_model = XGBClassifier(random_state=SEED)
lgb_model = LGBMClassifier(random_state=SEED)
lr_model.fit(X_blend, y_blend)
kn_model.fit(X_blend, y_blend)
svc_model.fit(X_blend, y_blend)
rf_model.fit(X_blend, y_blend)
gb_model.fit(X_blend, y_blend)
hgb_model.fit(X_blend, y_blend)
cb_model.fit(X_blend, y_blend)
xgb_model.fit(X_blend, y_blend)
lgb_model.fit(X_blend, y_blend)
clear_output()
y_proba = np.stack(
[
lr_model.predict_proba(X_holdout)[:, 1],
kn_model.predict_proba(X_holdout)[:, 1],
svc_model.predict_proba(X_holdout)[:, 1],
rf_model.predict_proba(X_holdout)[:, 1],
gb_model.predict_proba(X_holdout)[:, 1],
hgb_model.predict_proba(X_holdout)[:, 1],
cb_model.predict_proba(X_holdout)[:, 1],
xgb_model.predict_proba(X_holdout)[:, 1],
lgb_model.predict_proba(X_holdout)[:, 1],
]
).T
sc = StandardScaler()
X_meta = sc.fit_transform(y_proba)
meta_estimator = RandomForestClassifier(random_state=0)
meta_estimator.fit(X_meta, y_holdout)
y_proba = np.stack(
[
lr_model.predict_proba(val_X)[:, 1],
kn_model.predict_proba(val_X)[:, 1],
svc_model.predict_proba(val_X)[:, 1],
rf_model.predict_proba(val_X)[:, 1],
gb_model.predict_proba(val_X)[:, 1],
hgb_model.predict_proba(val_X)[:, 1],
cb_model.predict_proba(val_X)[:, 1],
xgb_model.predict_proba(val_X)[:, 1],
lgb_model.predict_proba(val_X)[:, 1],
]
).T
blending = meta_estimator.predict_proba(y_proba)[:, 1]
blending_auc = roc_auc_score(val_y, blending)
print(f"Blending with Non-Linear: {blending_auc:.4f}")
#
# Ensemble Selection
# To reduce the risk of over-fitting, Ensemble Selection is effective.
#
# start with trained models and holdout dataset.
# test each models by holdout dataset and select effective models.
#
# If score does not improve stop selection and use the models.
#
#
X_blend, X_holdout, y_blend, y_holdout = train_test_split(
train_X, train_y, test_size=0.5, shuffle=SHUFFLE, random_state=SEED
)
lr_model = LogisticRegression(random_state=SEED)
kn_model = KNeighborsClassifier()
svc_model = SVC(probability=True, random_state=SEED)
rf_model = RandomForestClassifier(random_state=SEED)
gb_model = GradientBoostingClassifier(random_state=SEED)
hgb_model = HistGradientBoostingClassifier(random_state=SEED)
cb_model = CatBoostClassifier(random_seed=SEED)
xgb_model = XGBClassifier(random_state=SEED)
lgb_model = LGBMClassifier(random_state=SEED)
lr_model.fit(X_blend, y_blend)
kn_model.fit(X_blend, y_blend)
svc_model.fit(X_blend, y_blend)
rf_model.fit(X_blend, y_blend)
gb_model.fit(X_blend, y_blend)
hgb_model.fit(X_blend, y_blend)
cb_model.fit(X_blend, y_blend)
xgb_model.fit(X_blend, y_blend)
lgb_model.fit(X_blend, y_blend)
clear_output()
y_proba = np.stack(
[
lr_model.predict_proba(X_holdout)[:, 1],
kn_model.predict_proba(X_holdout)[:, 1],
svc_model.predict_proba(X_holdout)[:, 1],
rf_model.predict_proba(X_holdout)[:, 1],
gb_model.predict_proba(X_holdout)[:, 1],
hgb_model.predict_proba(X_holdout)[:, 1],
cb_model.predict_proba(X_holdout)[:, 1],
xgb_model.predict_proba(X_holdout)[:, 1],
lgb_model.predict_proba(X_holdout)[:, 1],
]
).T
model_names = [
"LogisticRegresssion",
"KNeiborsClassifier",
"SVC",
"RandomForestClassifier",
"GradientBoostingClassifier",
"HistGradientBoostingClassifier",
"CatBoostClassifier",
"XGBClassifier",
"LGBMClassifier",
]
iterations = 100
baseline = 0.5
print(f"Start Baseline = {baseline:.4f}")
models = []
for i in range(iterations):
challengers = list()
for j in range(y_proba.shape[1]):
new_proba = np.stack(y_proba[:, models + [j]])
score = roc_auc_score(y_holdout, np.mean(new_proba, axis=1))
challengers.append([score, j])
challengers = sorted(challengers, key=lambda x: x[0], reverse=True)
best_score, best_model = challengers[0]
if best_score > baseline:
print(f"Adding {model_names[best_model]} to Ensemble: ", end="")
print(f"ROC-AUC Score = {best_score:.4f}")
models.append(best_model)
baseline = best_score
else:
print(f"Cannot improve scores anymore - Stop Iteration")
break
freqs = Counter(models)
weights = {key: freq / len(models) for key, freq in freqs.items()}
weights = np.array([weights.get(i, 0.0) for i in range(y_proba.shape[1])])
print(weights)
lr_model.fit(train_X, train_y)
kn_model.fit(train_X, train_y)
svc_model.fit(train_X, train_y)
rf_model.fit(train_X, train_y)
gb_model.fit(train_X, train_y)
hgb_model.fit(train_X, train_y)
cb_model.fit(train_X, train_y)
xgb_model.fit(train_X, train_y)
lgb_model.fit(train_X, train_y)
clear_output()
y_proba = np.stack(
[
lr_model.predict_proba(val_X)[:, 1],
kn_model.predict_proba(val_X)[:, 1],
svc_model.predict_proba(val_X)[:, 1],
rf_model.predict_proba(val_X)[:, 1],
gb_model.predict_proba(val_X)[:, 1],
hgb_model.predict_proba(val_X)[:, 1],
cb_model.predict_proba(val_X)[:, 1],
xgb_model.predict_proba(val_X)[:, 1],
lgb_model.predict_proba(val_X)[:, 1],
]
).T
y_proba = y_proba.dot(weights)
ensemble_selection_auc = roc_auc_score(val_y, y_proba)
print(f"Ensemble Selection ROC-AUC: {ensemble_selection_auc:.4f}")
#
# Stacking
# Next method is Stacking. This method is to reduce bias, not variance. This point is different from other previous methods.
# Another different point is that Stacking don't require the model with equivalent level of predictive powers.
# 1. k-fold cross validation and predict each validation dataset.
#
models = [
lr_model,
kn_model,
svc_model,
rf_model,
gb_model,
hgb_model,
cb_model,
xgb_model,
lgb_model,
]
skf = StratifiedKFold(n_splits=10, shuffle=SHUFFLE, random_state=SEED)
first_lvl_oof = np.zeros((len(train_X), len(models)))
first_lvl_preds = np.zeros((len(val_X), len(models)))
for k, (train_idx, val_idx) in enumerate(skf.split(train_X, train_y)):
X_kf_train, X_kf_val, y_kf_train, y_kf_val = (
train_X[train_idx, :],
train_X[val_idx, :],
train_y[train_idx],
train_y[val_idx],
)
for i in range(len(models)):
models[i].fit(X_kf_train, y_kf_train)
first_lvl_oof[val_idx, i] = models[i].predict_proba(X_kf_val)[:, 1]
clear_output()
#
# 2. train whole train dataset.
#
for i in range(len(models)):
models[i].fit(train_X, train_y)
first_lvl_preds[:, i] = models[i].predict_proba(val_X)[:, 1]
clear_output()
#
# 3. use same models and add stacking prediction values to train data.
#
second_lvl_oof = np.zeros((len(train_X), len(models)))
second_lvl_preds = np.zeros((len(val_X), len(models)))
for k, (train_idx, val_idx) in enumerate(skf.split(train_X, train_y)):
# add cross validation prediction to train data
skip_X_train = np.hstack([train_X, first_lvl_oof])
X_kf_train, X_kf_val, y_kf_train, y_kf_val = (
skip_X_train[train_idx, :],
skip_X_train[val_idx, :],
train_y[train_idx],
train_y[val_idx],
)
for i in range(len(models)):
models[i].fit(X_kf_train, y_kf_train)
second_lvl_oof[val_idx, i] = models[i].predict_proba(X_kf_val)[:, 1]
clear_output()
#
# 4. train by whole train dataset again.
#
skip_X_train = np.hstack([train_X, first_lvl_oof])
skip_X_val = np.hstack([val_X, first_lvl_preds])
for i in range(len(models)):
models[i].fit(skip_X_train, train_y)
second_lvl_preds[:, i] = models[i].predict_proba(skip_X_val)[:, 1]
clear_output()
#
# 5. mean second stacking prediction.
#
arithmetic = second_lvl_preds.mean(axis=1)
print(f"Stacking ROC-AUC: {roc_auc_score(val_y, arithmetic):.4f}")
#
# Define whole steps to the function.
#
def stacking_ensemble(models, train_X, train_y, test_X):
skf = StratifiedKFold(n_splits=10, shuffle=SHUFFLE, random_state=SEED)
# first train
first_lvl_oof = np.zeros((len(train_X), len(models)))
first_lvl_preds = np.zeros((len(test_X), len(models)))
for k, (train_idx, val_idx) in enumerate(skf.split(train_X, train_y)):
X_kf_train, X_kf_val, y_kf_train, y_kf_val = (
train_X[train_idx, :],
train_X[val_idx, :],
train_y[train_idx],
train_y[val_idx],
)
for i in range(len(models)):
models[i].fit(X_kf_train, y_kf_train)
first_lvl_oof[val_idx, i] = models[i].predict_proba(X_kf_val)[:, 1]
# first prediction
for i in range(len(models)):
models[i].fit(train_X, train_y)
first_lvl_preds[:, i] = models[i].predict_proba(test_X)[:, 1]
# second train
second_lvl_oof = np.zeros((len(train_X), len(models)))
second_lvl_preds = np.zeros((len(test_X), len(models)))
for k, (train_idx, val_idx) in enumerate(skf.split(train_X, train_y)):
# add cross validation prediction to train data
skip_X_train = np.hstack([train_X, first_lvl_oof])
X_kf_train, X_kf_val, y_kf_train, y_kf_val = (
skip_X_train[train_idx, :],
skip_X_train[val_idx, :],
train_y[train_idx],
train_y[val_idx],
)
for i in range(len(models)):
models[i].fit(X_kf_train, y_kf_train)
second_lvl_oof[val_idx, i] = models[i].predict_proba(X_kf_val)[:, 1]
# second prediction
skip_X_train = np.hstack([train_X, first_lvl_oof])
skip_X_test = np.hstack([test_X, first_lvl_preds])
for i in range(len(models)):
models[i].fit(skip_X_train, train_y)
second_lvl_preds[:, i] = models[i].predict_proba(skip_X_test)[:, 1]
# prediction
arithmetic = second_lvl_preds.mean(axis=1)
return arithmetic
#
# Submit Your Results
# submit what you want.
#
test_df = pd.read_csv("/kaggle/input/titanic/test.csv")
test_data = titanic_preprocessing(test_df)
test_X = np.array(test_data[feature_cols])
submission_df = pd.read_csv("/kaggle/input/titanic/gender_submission.csv")
lr_model = LogisticRegression(random_state=SEED)
kn_model = KNeighborsClassifier()
svc_model = SVC(probability=True, random_state=SEED)
rf_model = RandomForestClassifier(random_state=SEED)
gb_model = GradientBoostingClassifier(random_state=SEED)
hgb_model = HistGradientBoostingClassifier(random_state=SEED)
cb_model = CatBoostClassifier(random_seed=SEED)
xgb_model = XGBClassifier(random_state=SEED)
lgb_model = LGBMClassifier(random_state=SEED)
models = [
lr_model,
kn_model,
svc_model,
rf_model,
gb_model,
hgb_model,
cb_model,
xgb_model,
lgb_model,
]
y_proba = stacking_ensemble(models, X, y, test_X)
clear_output()
y_pred = [1 if proba > 0.5 else 0 for proba in y_proba]
submission_df[target_col] = y_pred
submission_df.to_csv("submission.csv", index=False)
|
# ### Trying to run a GPU Notebook and exiting/killing it after a crash or a good execution to stop spending GPU quota
# (So far I cannot achieve to make the Session really stop, liberating GPU resources and stopping the GPU quota timer.)
import torch
import os
import sys
from time import sleep
from tqdm.notebook import tqdm
print(torch.cuda.device_count())
torch.__version__
# Some mock GPU compute
def my_random_gpu_computation():
for i in tqdm(range(10)):
x = torch.randn(10000, 10000, device="cuda")
y = torch.randn(10000, 10000, device="cuda")
print(f"{i}: {torch.mean(x * y)}")
sleep(0.5)
# Making sure that always ends up exiting
try:
my_random_gpu_computation()
except Exception as e:
print(f"An error occurred: {e}")
finally:
print("Exiting the notebook trying to save GPU quota.")
sleep(0.5)
os._exit(0)
# sys.exit()
# !exit
|
# ## 1. Reading the file:
# #### Firstly, I collected all the data and saved it in the excel file using the csv format. After that, I uploaded the file in a website called kaggle.
# #### In the beginning, we have to write a code that reads the excel file, so I wrote this code to read the excel file that i have loaded into the program.
import pandas as pd
df = pd.read_csv("/kaggle/input/car-prices-in-oman/car-prices-in-oman.csv")
df
# ### 1.1 Converting data into a clear table:
# #### During the collection of data, I wrote all the information in one column. When I implemented it in the program it printed all the information in a row, which is the same way that I wrote in the excel file. I currently wrote this code to convert this data and information into a clear and an understandable table.
city = []
state = []
car_make = []
Model = []
year = []
Regional_Specs = []
SpecsTransmission = []
fuel_type = []
color = []
Condition = []
Kilometers = []
Paint = []
Body_Condition = []
Car_License = []
Insurance = []
Payment_Method = []
price = []
for _, row in df.iterrows():
str = row["Ad"].split(":")
if str[1].find("Neighborhood"):
b = str[1].replace("Neighborhood", "")
city.append(b.strip())
if str[2].find("Car Make:"):
b = str[2].replace("Car Make", "")
state.append(b.strip())
if str[3].find("Model"):
b = str[3].replace("Model", "")
car_make.append(b.strip())
if str[4].find("Year"):
b = str[4].replace("Year", "")
Model.append(b.strip())
if str[5].find("Regional Specs"):
b = str[5].replace("Regional Specs", "")
year.append(int(b))
if str[6].find("SpecsTransmission"):
b = str[6].replace("SpecsTransmission", "")
Regional_Specs.append(b.strip())
if str[7].find("Fuel"):
b = str[7].replace("Fuel", "")
SpecsTransmission.append(b.strip())
if str[8].find("Color"):
b = str[8].replace("Color", "")
fuel_type.append(b.strip())
if str[9].find("Condition"):
b = str[9].replace("Condition", "")
color.append(b.strip())
if str[10].find("Kilometers"):
b = str[10].replace("Kilometers", "")
Condition.append(b.strip())
if str[11].find("Paint"):
b = str[11].replace("Paint", "")
Kilometers.append(b.strip())
if str[12].find("Body Condition"):
b = str[12].replace("Body Condition", "")
Paint.append(b.strip())
if str[13].find("Car License"):
b = str[13].replace("Car License", "")
Body_Condition.append(b.strip())
if str[14].find("Insurance"):
b = str[14].replace("Insurance", "")
Car_License.append(b.strip())
if str[15].find("Payment Method"):
b = str[15].replace("Payment Method", "")
Insurance.append(b.strip())
if str[16].find("Price"):
b = str[16].replace("Price", "")
Payment_Method.append(b.strip())
try:
if str[17].find("OMR"):
b = str[17].replace("OMR", "")
b = b.replace(" ", "")
b = b.replace(",", "")
price.append(float(b))
except IndexError or AttributeError:
price.append(None)
df.drop("Ad", axis=1, inplace=True)
df["city"] = city
df["state"] = state
df["Company"] = car_make
df["Model"] = Model
df["year"] = year
df["Regional Specs"] = Regional_Specs
df["fuel type"] = fuel_type
df["color"] = color
df["Condition"] = Condition
df["Kilometers"] = Kilometers
df["Paint"] = Paint
df["Body Condition"] = Body_Condition
df["Car License"] = Car_License
df["Insurance"] = Insurance
df["Payment Method"] = Payment_Method
df["Price"] = price
df
# ## **2.Data Cleansing and Improvement:**
# ### Find if there are some duplicated data:
# #### When working with data, we need to make sure that it does not contain any repeated data. As a result, I wrote this code that prints the repeated data.
df.loc[df.duplicated()]
df.duplicated().sum()
# ## **3.Process Missing data:**
# ### 3.1 Find out the missing data:
# #### Before starting to work on any data, the data must be fully written, there shouldn’t be anything missing. Therefore, I wrote this code to verify if the data is missing or not.
df.isna().sum()
# #### It is clear from the previous code that the missing data is only in the prince values. Therefore, it is true that there were 9 missing values in the field “price”.
# #### There are several ways to find solutions for the missing data, such as, deleting this missing information from the data. However, this isn’t a really good idea, because the number of data is really small ( 250 only). Therefore, it is better to find another solution. On the other hand, the more the data the better it is, and the results are more accurate.
x = df.isna().sum()
cnt = 0
for temp in x.values:
if temp > 0:
print(x.index[cnt], x.values[cnt])
cnt += 1
# ### 3.2 Print Cars with no price:
# #### There is another way to compensate for the price of the unknown cars, such as finding the mean, medium, or mode.
# #### However, firstly, we need to know the car model that contains the missing price. I wrote this code to print all the cars that contains the missing price.
temp = df[df["Price"].isna()]
temp
# ### 3.3 Print the most frequent car models with no price:
# #### Based on the previous code, we must know the most repeated car model with the missing price. I wrote this code and I added [0] to print the most repeated car model. However, if we did not add the [0], it will print all the car models that contains the missing price.
t1 = temp["Model"].mode()[0]
t1
# ### 3.4 Print all cars contain the model "yaris":
# #### It is clear that the most repeated car model that contains the missing price is “yaris”. As a result, I wrote this code to print the yaris car model.
allyaris = df.copy()
allyaris1 = allyaris[allyaris["Model"] == t1]
allyaris1
# ### 3.5 Find out the missing data:
# #### After printing all the “Yaris” car model. I checked the missing data once again. It appeared that the price=9, it is clear that all the cars with the missing price are of the “Yaris” type.
allyaris1.isna().sum()
# ### 3.6 Delete cars with a lost price:
# #### We deleted “Yaris” cars that contains missing prices from the table allyaris to calculate the mean, median, and mode of the “Yaris” cars
new = allyaris1.dropna()
new
# ### 3.7 Find out the missing data:
# #### Before calculating the mean, mode, and median. We must make sure that there is not missing data in the table.
new.isna().sum()
# ### 3.8 Calculating mean, median, mode for Yaris models:
# #### After making sure that there isn’t any missing data. I wrote this code in 2 ways to calculate the mean, mode, and median of the “Yaris” car model.
mean = new["Price"].mean()
median = new["Price"].median()
mode = new["Price"].mode()[0]
print(mean, median, mode)
mean = new["Price"].mean()
median = new["Price"].median()
mode = new["Price"].mode()[0]
print(mean)
print(median)
print(mode)
# ### 3.9 Print mean in cars with a missing price:
# #### After displaying the results of the mean, median, and mode, we will enter the mean value in the empty cells in the price column, i.e. put the mean value in the missing price of the cars field. I wrote this code that prints the table and puts in the missing price value the middle value.
# #### The 'inplace=True' to commit our change in the same dataset
df["Price"].fillna(mean, inplace=True)
df
# ### 3.10 Check for any missing data:
# #### After printing the table and inserting the mean value in the missing price of the cars field, you should check again whether there is missing data
new.isna().sum()
# ### 3.11 Delete date column from table:
# #### It became clear from the previous code that there is no missing data. Currently, we can work on the data, make comparisons, and convert the data into numbers, but initially in the data there is a column named date, which is the date of publication of the advertisement, and this column is not important, so I wrote this code to delete the date column from the data table.
dff = df.copy()
dff = dff.drop(["Date"], axis=1)
dff.head()
# ## **4.Data Visualisation**
import matplotlib.pyplot as plt
# ### 4.1 Print all car models in the data without repeating:
# #### I will make a comparison between the model of car with the number of cars, so I started writing this code to print for all model of cars in the data without repeating
df["Model"].unique()
# ### 4.2 Comparison of car models and their number:
# #### I wrote the code here to make a comparison between the model and the number of cars of each model in the data
element = []
counter = []
Model1 = set(df["Model"])
for e in Model1:
element.append(e)
counter.append(Model.count(e))
x = element
y = counter
plt.figure(figsize=(10, 6))
plt.pie(y, labels=x)
plt.show()
# ### 4.3 Converting the drawing into a table between car models and their number:
# #### The number of cars was not indicated in the previous code, so I wrote this code that prints a table containing model and number of car
combinelist = list(zip(x, y))
mytable = pd.DataFrame(combinelist, columns=["Model", "Number of car"])
mytable
# ### 4.4 Print all cars with Camry model:
# #### I will make a comparison between Camry and Yaris through the mean value. The mean value has been calculated for Yaris cars. Now I will calculate the mean for Camry cars.
# #### Initially, I will print all Camry models.
allCamry = df.copy()
allCamry1 = allCamry[allCamry["Model"] == "Camry"]
allCamry1
# ### 4.5 Mean calculation for Camry models:
# #### After printing all Camry models, I wrote this code to calculate the mean
mean = allCamry1["Price"].mean()
mean
# ### 4.6 Comparison between the Yaris model and the Camry model through mean:
# #### After calculating the mean for the Yaris and Camry cars, I wrote this code to draw a comparison between the Camry and Yaris through the mean
x = ["Camry", "Yaris"]
y = [5594.736842105263, 2821.6129032258063]
plt.bar(x, y)
plt.show()
df.head()
# ## **5.Converting data into numbers:**
df1 = dff.copy()
def change_to_numeric(x, df1):
temp = pd.get_dummies(df1[x])
df3 = pd.concat([df1, temp], axis=1)
df3.drop([x], axis=1, inplace=True)
return df3
df2 = change_to_numeric("city", df1)
df3 = change_to_numeric("state", df2)
df4 = change_to_numeric("Company", df3)
df5 = change_to_numeric("Model", df4)
df6 = change_to_numeric("Regional Specs", df5)
df7 = change_to_numeric("color", df6)
df8 = change_to_numeric("Payment Method", df7)
df9 = change_to_numeric("fuel type", df8)
df10 = change_to_numeric("Paint", df9)
df10["Condition"].unique()
df10["Condition"] = df10["Condition"].replace(["Used"], 0)
df10["Condition"] = df10["Condition"].replace(["New"], 1)
df10
df10["Condition"].unique()
df10["Insurance"].unique()
df10["Insurance"] = df10["Insurance"].replace(["Comprehensive Insurance"], 2)
df10["Insurance"] = df10["Insurance"].replace(["Compulsory Insurance"], 1)
df10["Insurance"] = df10["Insurance"].replace(["Not Insured"], 0)
df10
df10["Insurance"].unique()
df10["Car License"].unique()
df10["Car License"] = df10["Car License"].replace(["Licensed"], 1)
df10["Car License"] = df10["Car License"].replace(["Not Licensed"], 0)
df10["Car License"].unique()
df10
df10["Car License"].unique()
df10["Body Condition"].unique()
df10["Body Condition"] = df10["Body Condition"].replace(
["Excellent with no defects"], 5
)
df10["Body Condition"] = df10["Body Condition"].replace(
["Good (body only has minor blemishes)"], 4
)
df10["Body Condition"] = df10["Body Condition"].replace(["Fair (body needs work)"], 3)
df10["Body Condition"] = df10["Body Condition"].replace(
["Poor (severe body damages)"], 2
)
df10["Body Condition"] = df10["Body Condition"].replace(["Other"], 1)
df10
newkilometers = []
for _, row in df10.iterrows():
try:
x = row["Kilometers"].split("-")
d1 = x[0]
d2 = x[1]
d1 = d1.replace(",", "")
d1 = d1.replace("+", "")
d2 = d2.replace(",", "")
d2 = d2.replace("+", "")
d1 = int(d1)
d2 = int(d2)
val = d1 + 1 + (d2 - d1) // 2
newkilometers.append(val)
except IndexError:
x = row["Kilometers"]
d1 = x[0]
d1 = d1.replace(",", "")
d1 = d1.replace("+", "")
if d1 == "":
val = 0
newkilometers.append(val)
else:
val = int(d1)
newkilometers.append(val)
df10.drop(["Kilometers"], axis=1, inplace=True)
df10["Kilometers"] = newkilometers
df10
df10.head()
df11 = df10[["year", "Body Condition"]]
df11.head()
df11 = df10[
["year", "Condition", "Body Condition", "Car License", "Insurance", "Price"]
]
df11.head()
import seaborn as sns
df11 = df10[
["year", "Condition", "Body Condition", "Car License", "Insurance", "Price"]
]
df11.corr()["Price"]
_, ax = plt.subplots(figsize=(14, 12))
colormap = sns.diverging_palette(220, 10, as_cmap=True)
_ = sns.heatmap(
df11.corr(),
cmap=colormap,
square=True,
cbar_kws={"shrink": 0.9},
ax=ax,
annot=True,
linewidths=0.1,
vmax=1.0,
linecolor="white",
annot_kws={"fontsize": 12},
)
plt.title("Pearson Correlation of Features", y=1.05, size=15)
import pandas as pd
from sklearn.tree import DecisionTreeClassifier
from sklearn.model_selection import train_test_split
data = df10.copy()
data
from sklearn.model_selection import train_test_split
x = df10.drop(columns=["Price"])
y = df10["Price"]
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.2)
from sklearn.metrics import accuracy_score
from sklearn import linear_model
model = linear_model.LinearRegression()
model.fit(x_train, y_train)
p = model.predict(x_test)
# score = accuracy_score(y_test, p)
# score
# print ( y_test, p)
print(p)
x = df10.drop(columns=["Price"])
y = df10["Price"]
w = x.columns.values.tolist()
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.2)
from sklearn.metrics import accuracy_score
from sklearn import linear_model
model = linear_model.LinearRegression()
model.fit(x_train, y_train)
p = model.predict(x_test)
# score = accuracy_score(y_test, p)
# score
# print ( y_test, p)
print(p)
x_test.shape
x_train.shape
xd = df1["Price"]
yd = df1["Model"]
plt.scatter(xd, yd)
plt.show()
df10
pd.set_option("display.max_columns", None)
pd.set_option("display.width", 100)
display(df.loc[[0]])
display(df10.loc[[0]])
xt = [
2011,
0,
5,
1,
2,
0,
0,
0,
0,
0,
0,
0,
1,
0,
0,
0,
0,
0,
1,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
1,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
1,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
1,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
1,
1,
0,
0,
0,
1,
0,
1,
0,
0,
0,
175000,
]
listmyoptions = zip(w, x)
zipped_listmyoptions = list(listmyoptions)
print(zipped_listmyoptions)
plspredict = model.predict([xt])
print("the price for car is : ", plspredict)
pd.set_option("display.max_columns", None)
pd.set_option("display.width", 100)
display(df.loc[[2]])
display(df10.loc[[2]])
xt1 = [
2019,
0,
1,
1,
2,
0,
0,
0,
0,
0,
0,
0,
1,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
1,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
1,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
1,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
1,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
1,
1,
0,
0,
0,
1,
0,
0,
0,
1,
0,
35000,
]
listmyoptions = zip(w, xt1)
zipped_listmyoptions = list(listmyoptions)
print(zipped_listmyoptions)
plspredict = model.predict([xt1])
print("the price for car is : ", plspredict)
pd.set_option("display.max_columns", None)
pd.set_option("display.width", 100)
display(df.loc[[20]])
display(df10.loc[[20]])
xt2 = [
2011,
0,
5,
1,
1,
0,
0,
0,
0,
0,
0,
0,
1,
0,
0,
0,
0,
0,
0,
0,
0,
0,
1,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
1,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
1,
0,
0,
1,
0,
0,
0,
0,
1,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
1,
0,
0,
0,
1,
0,
1,
0,
0,
0,
15000,
]
listmyoptions = zip(w, xt2)
zipped_listmyoptions = list(listmyoptions)
print(zipped_listmyoptions)
plspredict = model.predict([xt2])
print("the price for car is : ", plspredict)
dx = pd.read_csv("/kaggle/input/car-price-testing1/car_1.csv")
dx
dx.loc[df.duplicated()]
dx.duplicated().sum()
dx.isna().sum()
dx1 = dx.copy()
def change_to_numeric(x, dx1):
temp = pd.get_dummies(dx1[x])
dx3 = pd.concat([dx1, temp], axis=1)
dx3.drop([x], axis=1, inplace=True)
return dx3
dx2 = change_to_numeric("city", dx1)
dx3 = change_to_numeric("state", dx2)
dx4 = change_to_numeric("Company", dx3)
dx5 = change_to_numeric("Model", dx4)
dx6 = change_to_numeric("Regional Specs", dx5)
dx7 = change_to_numeric("color", dx6)
dx8 = change_to_numeric("Payment Method", dx7)
dx9 = change_to_numeric("fuel type", dx8)
dx10 = change_to_numeric("Paint", dx9)
dx10["Condition"].unique()
dx10["Condition"] = dx10["Condition"].replace(["Used"], 0)
dx10["Condition"] = dx10["Condition"].replace(["New"], 1)
dx10
dx10["Condition"].unique()
dx10["Insurance"].unique()
dx10["Insurance"] = dx10["Insurance"].replace(["Comprehensive Insurance"], 2)
dx10["Insurance"] = dx10["Insurance"].replace(["Compulsory Insurance"], 1)
dx10["Insurance"] = dx10["Insurance"].replace(["Not Insured"], 0)
dx10
dx10["Insurance"].unique()
dx10["Car License"].unique()
dx10["Car License"] = dx10["Car License"].replace(["Licensed"], 1)
dx10["Car License"] = dx10["Car License"].replace(["Not Licensed"], 0)
dx10["Car License"].unique()
dx10
dx10["Car License"].unique()
dx10["Body Condition"].unique()
dx10["Body Condition"] = dx10["Body Condition"].replace(
["Excellent with no defects"], 5
)
dx10["Body Condition"] = dx10["Body Condition"].replace(
["Good (body only has minor blemishes)"], 4
)
dx10["Body Condition"] = dx10["Body Condition"].replace(["Fair (body needs work)"], 3)
dx10["Body Condition"] = dx10["Body Condition"].replace(
["Poor (severe body damages)"], 2
)
dx10["Body Condition"] = dx10["Body Condition"].replace(["Other"], 1)
dx10
newkilometers = []
for _, row in dx10.iterrows():
try:
x = row["Kilometers"].split("-")
d1 = x[0]
d2 = x[1]
d1 = d1.replace(",", "")
d1 = d1.replace("+", "")
d2 = d2.replace(",", "")
d2 = d2.replace("+", "")
d1 = int(d1)
d2 = int(d2)
val = d1 + 1 + (d2 - d1) // 2
newkilometers.append(val)
except IndexError:
x = row["Kilometers"]
d1 = x[0]
d1 = d1.replace(",", "")
d1 = d1.replace("+", "")
if d1 == "":
val = 0
newkilometers.append(val)
else:
val = int(d1)
newkilometers.append(val)
dx10.drop(["Kilometers"], axis=1, inplace=True)
dx10["Kilometers"] = newkilometers
dx10
pd.set_option("display.max_columns", None)
pd.set_option("display.width", 100)
display(dx.loc[[2]])
display(dx10.loc[[2]])
xt3 = [2014, 0, 5, 1, 1, 1, 1, 0, 0, 1, 0, 0, 1, 0, 0, 1, 0, 0, 1, 1, 0, 1, 1, 0, 2]
listmyoptions = zip(w, xt3)
zipped_listmyoptions = list(listmyoptions)
print(zipped_listmyoptions)
plspredict = model.predict([xt3])
print("the price for car is : ", plspredict)
|
# # ABOUT DATASET 📁
# In response to the COVID-19 pandemic, the White House and a coalition of leading research groups have prepared the COVID-19 Open Research Dataset (CORD-19).CORD-19 is a resource of over 400,000 scholarly articles, including over 150,000 with full text, about COVID-19, SARS-CoV-2, and related coronaviruses. This freely available dataset is provided to the global research community to apply recent advances in natural language processing and other AI techniques to generate new insights in support of the ongoing fight against this infectious disease.
# # NOTEBOOK GOAL 🎯
# In this notebook , we will find related articles by using Topic modelling. Here I am using Latent Dirichlet Allocation(LDA).
# LDA is an example of topic model and is used to classify text in a document to a particular topic. It builds a topic per document model and words per topic model, modeled as Dirichlet distributions.
#
# import libraries
import os
import re
import numpy as np
import pandas as pd
import json
from pprint import pprint
import random
import string
from nltk import word_tokenize
from nltk.corpus import stopwords
from gensim.corpora import Dictionary
pd.set_option("display.max_rows", 100)
pd.set_option("display.max_columns", 100)
pd.set_option("display.max_colwidth", 100)
documents_dir = "../input/CORD-19-research-challenge/document_parses/pdf_json/"
filenames = os.listdir(documents_dir)
print("Number of documents :", len(filenames))
random.shuffle(filenames)
file = json.load(
open(
"../input/CORD-19-research-challenge/document_parses/pdf_json/0000028b5cc154f68b8a269f6578f21e31f62977.json",
"rb",
)
)
pprint(file["metadata"]["title"])
# # Step 1 : Data Cleaning
def clean(text):
text = str(text).lower()
text = re.sub(r"\[.*?\]", "", text)
text = re.sub(r"\(.*?\)", "", text)
text = re.sub(r"\s+", " ", text)
text = re.sub(r"\w*\d\w*", "", text)
text = re.sub(r"\w+…|…", "", text) # Remove ellipsis (and last word)
text = re.sub(f"[{re.escape(string.punctuation)}]", "", text)
return text
def remove_stopwords_and_tokenize(text):
my_stopwords = set(stopwords.words("english"))
tokens = word_tokenize(text) # tokenize
tokens = [t for t in tokens if not t in my_stopwords] # Remove stopwords
tokens = [t for t in tokens if len(t) > 1] # Remove short tokens
return tokens
def parse_body_text(body_text):
body = ""
for item in body_text:
body += item["section"]
body += "\n\n"
body += item["text"]
body += "\n\n"
body = clean(body)
tokens = remove_stopwords_and_tokenize(body)
return body, tokens
all_text = []
all_tokens = []
all_titles = []
for i, filename in enumerate(filenames[:1000]):
filepath = documents_dir + filename
file = json.load(open(filepath, "rb"))
text, tokens = parse_body_text(file["body_text"])
all_text.append(text)
all_tokens.append(tokens)
all_titles.append(file["metadata"]["title"])
data = pd.DataFrame()
data["text"] = all_text
data["tokens"] = all_tokens
data["doc_id"] = filenames[:1000]
data["title"] = all_titles
del all_text, all_tokens, all_titles
data.head(2)
# # Step 2 : Apply LDA model
# Create a dictionary representation of the documents.
dictionary = Dictionary(data["tokens"])
# Filter out words that occur less than 20 documents, or more than 50% of the documents.
dictionary.filter_extremes(no_below=20, no_above=0.5)
# Bag-of-words representation of the documents.
corpus = [dictionary.doc2bow(doc) for doc in data["tokens"]]
from gensim.models import LdaModel
# Build LDA model
lda_model = LdaModel(
corpus=corpus,
id2word=dictionary,
num_topics=20,
random_state=100,
chunksize=200,
passes=100,
)
lda_model.print_topics()[:5]
lda_model[corpus][0]
# # Step 4: Results
# ## Document - Topic Table
def get_document_topic_table(lda_model, corpus, texts=data):
# Init output
document_topic_df = pd.DataFrame()
# Get main topic in each document
for i, row_list in enumerate(lda_model[corpus]):
row = sorted(row_list, key=lambda x: (x[1]), reverse=True)
topic_num = row[0][0]
prop_topic = row[0][1]
wp = lda_model.show_topic(topic_num)
topic_keywords = ", ".join([word for word, prop in wp])
document_topic_df.at[i, "best_topic"] = topic_num
document_topic_df.at[i, "prop_topic"] = prop_topic
document_topic_df.at[i, "topic_keywords"] = topic_keywords
document_topic_df.at[i, "document_num"] = i
return document_topic_df
document_topic_df = get_document_topic_table(
lda_model=lda_model, corpus=corpus, texts=data["tokens"]
)
document_topic_df.head(2)
# ## Recommend k topics
def get_topic_id(doc_id):
for i, row in data.iterrows():
if row["doc_id"] == doc_id:
# print(document_topic_df["best_topic"][i])
return document_topic_df["best_topic"][i]
return -1
def get_matching_topics_docs(topic_id):
matched_topics = []
for i, row in document_topic_df.iterrows():
if row["best_topic"] == topic_id:
topic_prop_doc = (topic_id, row["prop_topic"], i)
matched_topics.append(topic_prop_doc)
return matched_topics
def get_top_k_topics(matched_topics, k):
top_k = sorted(matched_topics, key=lambda x: [x[1]], reverse=True)
print(top_k[:k])
k_topics_df = pd.DataFrame(columns=["doc_id", "topic_id", "topic_prop", "title"])
i = 0
for topic_id, topic_prop, doc_num in top_k[:k]:
k_topics_df.at[i, "doc_id"] = data["doc_id"][doc_num]
k_topics_df.at[i, "topic_id"] = topic_id
k_topics_df.at[i, "topic_prop"] = topic_prop
k_topics_df.at[i, "title"] = data["title"][doc_num]
i += 1
return k_topics_df
def recommend_k_topics(doc_id, k):
topic_id = get_topic_id(doc_id)
if topic_id != -1:
matched_topics = get_matching_topics_docs(topic_id)
return get_top_k_topics(matched_topics, k)
k_topics_df = recommend_k_topics("328401206bf2e3657e352ad5c5a2e566cc09736d.json", 5)
k_topics_df
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("../input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import shutil
src_beng_train_data = "../input/bengalidata/Bengali.traineddata"
dest_beng_train_data = "/usr/share/tesseract-ocr/4.00/tessdata/"
shutil.copy(src_beng_train_data, dest_beng_train_data)
filenames = os.listdir("/usr/share/tesseract-ocr/4.00/tessdata/")
print(filenames)
# img = cv2.imread('../input/bonglekha/bong.jpeg')
# gray = cv2.cvtColor(img, cv2.COLOR_BGR2GRAY)
# plt.imshow(gray,cmap='gray', vmin=0, vmax=255)
# ret,thrshld = cv2.threshold(img,127,255,cv2.THRESH_BINARY) # binary thresholding
beng_text = "আমরা বাঙালিরা মিষ্টি খেতে ভালোবাসি। আমরা খুব বই ও পড়ি। আমি ছোট থাকতে টেনিদা, ফেলুদা, আর সমরেশ বাবুর বই খুব পড়তাম। এখন আর বেশি বই পড়তে পারি না । রসগোল্লা ৩ আর পাই না"
# beng_text = pytesseract.image_to_string(img, lang='Bengali' )
print(beng_text)
eng_text = "We Bengalis like to eat sweets. We read a lot of books. When I was small, I used to read Tenida, Feluda and Babur Bhoy's books a lot. Now I can't read as many books. I can't eat Rasgulla either."
# beng_text = pytesseract.image_to_string(img, lang='Bengali' )
print(eng_text)
def get_open_ai_key():
from kaggle_secrets import UserSecretsClient
user_secrets = UserSecretsClient()
secret_value = user_secrets.get_secret("api_key")
return secret_value
# Function to translate bengali to English
def ask_a_question(question, context, my_language):
import openai
openai.api_key = get_open_ai_key()
prompt = "Answer the question based on the context below in {my_language} \n\n Context: {context} \n\n question:{question} \n\n Answer:".format(
question=question, context=context, my_language=my_language
)
res = openai.Completion.create(
engine="text-davinci-003",
prompt=prompt,
temperature=0,
max_tokens=400,
top_p=1,
frequency_penalty=0,
presence_penalty=0,
stop=None,
)
return res["choices"][0]["text"].strip()
# Function to translate bengali to English
def translate_through_gpt(prompt):
import openai
openai.api_key = get_open_ai_key()
prompt = "Translate the text into english \n\n Text:{text} \n\n Translated:".format(
text=prompt
)
res = openai.Completion.create(
engine="text-davinci-003",
prompt=prompt,
temperature=0,
max_tokens=400,
top_p=1,
frequency_penalty=0,
presence_penalty=0,
stop=None,
)
return res["choices"][0]["text"].strip()
# Function to summarize bengali content
def summarize_through_gpt(prompt, my_language="bengali"):
import openai
openai.api_key = get_open_ai_key()
prompt = (
"Summarize the text in {my_language} \n\n Text:{text} \n\n Summary:".format(
text=prompt, my_language=my_language
)
)
res = openai.Completion.create(
engine="text-davinci-003",
prompt=prompt,
temperature=0,
max_tokens=400,
top_p=1,
frequency_penalty=0,
presence_penalty=0,
stop=None,
)
return res["choices"][0]["text"].strip()
print(summarize_through_gpt(beng_text))
print(translate_through_gpt(beng_text))
# print(ask_a_question("What does bengalis like to eat?"))
# print(ask_a_question("What does bengalis like to read?",beng_text,"bengali"))
print(ask_a_question("What does bengalis like to read?", beng_text, "Spanish"))
print(ask_a_question("What does bengalis like to read?", beng_text, "English"))
print(summarize_through_gpt(eng_text, "english"))
|
# ***BUSINESS GOAL :***
# In this notebook, we will explore a dataset of credit card customers and use clustering techniques to group customers based on their characteristics and behavior, with a particular focus on those who have churned (or have a high likelihood of doing so). By leveraging the power of clustering algorithms, we aim to uncover insights that can help credit card companies better understand their customers and develop effective strategies to retain them. So let's dive in!
# **STEP 1: READING AND UNDERSTANDING DATA**
# import all libraries and dependencies for dataframe and visualization
import pandas as pd
import numpy as np
from numpy import unique
from numpy import where
from numpy import mean
import matplotlib.pyplot as plt
import seaborn as sns
from yellowbrick.cluster import SilhouetteVisualizer
import warnings
warnings.filterwarnings("ignore")
from datetime import datetime, timedelta
# import all libraries and dependencies for machine learning
from sklearn.model_selection import train_test_split
from sklearn import preprocessing
from sklearn.preprocessing import MinMaxScaler
from sklearn.linear_model import LogisticRegressionCV
from sklearn.feature_selection import mutual_info_classif
from sklearn.metrics import confusion_matrix, classification_report, accuracy_score
from sklearn.cluster import KMeans
from sklearn.metrics import silhouette_score
from statsmodels.stats.outliers_influence import variance_inflation_factor
from statsmodels.tools.tools import add_constant
from sklearn.datasets import make_classification
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import RepeatedStratifiedKFold
from imblearn.ensemble import BalancedBaggingClassifier
from sklearn.cluster import Birch
from sklearn.cluster import AgglomerativeClustering
df = pd.read_csv("BankChurners.csv")
df.shape
df.head()
df.info()
# **STEP 2 :DATA CLEANING**
# droping costumer id beacause it's insignificant
df = df.drop("CLIENTNUM", axis=1)
print(df.isnull().sum())
# no missing data
# cheking for duplicated rows
print(df.loc[df.duplicated()])
# no dplicated rows
# CHEKING OUTLIERS :
data_1 = df["Dependent_count"] # small
data_2 = df["Months_on_book"] # small
data_3 = df["Total_Relationship_Count"] # small
data_4 = df["Months_Inactive_12_mon"] # smallx
data_5 = df["Credit_Limit"] # large
data_6 = df["Total_Revolving_Bal"] # large
data_7 = df["Avg_Open_To_Buy"] # large
data_8 = df["Total_Amt_Chng_Q4_Q1"] # smallx
data_9 = df["Total_Trans_Ct"] # small
data_10 = df["Total_Ct_Chng_Q4_Q1"] # smallx
data_11 = df["Avg_Utilization_Ratio"] # smallx
data_12 = df["Total_Trans_Amt"] # large
data_s = [data_1, data_2, data_3, data_9]
data_l = [data_5, data_6, data_7, data_12]
data_sx = [data_4, data_8, data_10, data_11]
general = [data_s, data_l, data_sx]
for gen in general:
fig = plt.figure(figsize=(10, 7))
# Creating axes instance
ax = fig.add_axes([0, 0, 1, 1])
# Creating plot
bp = ax.boxplot(gen)
# show plot
plt.show()
# as we can see we can't plot all features in the same figure due to large diffrence in values so i divided them in 3 categories large small and smallx
# as we expected there's a large diffrence between variables distribution Total_Amt_Chng_Q4_Q1 ,Total_Ct_Chng_Q4_Q1,
# Months_on_book (+/-) ,Credit_Limit,Avg_Open_To_Buy,Total_Trans_Amt (the last 3 are : all oultiers are above the 3rd quartline )
# **STEP 3 : DATA VIZUALIZATION**
sns.countplot(x="Attrition_Flag", data=df)
plt.show()
sns.countplot(x="Gender", data=df)
plt.show()
plt.figure(figsize=(8, 10))
sns.countplot(x="Income_Category", data=df)
plt.show()
plt.figure(figsize=(8, 10))
sns.countplot(x="Income_Category", data=df, hue="Attrition_Flag")
plt.show()
sns.histplot(x="Marital_Status", hue="Attrition_Flag", data=df)
plt.show()
# histogram of Customer_Age
sns.histplot(x="Customer_Age", data=df, kde=True)
plt.title("age distribution ")
# Adding the legends
plt.show()
sns.histplot(x="Customer_Age", data=df, kde=True, hue="Attrition_Flag")
plt.show()
sns.histplot(x="Customer_Age", data=df, kde=True, hue="Income_Category")
plt.show()
sns.histplot(x="Credit_Limit", data=df, kde=True, hue="Attrition_Flag")
plt.show()
sns.barplot(
x="Credit_Limit", y="Income_Category", hue="Attrition_Flag", data=df, palette="Set1"
)
plt.show()
# WE TAKE THIS
sns.set_theme(style="ticks")
# Plot the orbital period with horizontal boxes
sns.boxplot(
x="Credit_Limit",
y="Income_Category",
data=df,
whis=[0, 100],
width=0.6,
palette="Set1",
)
# Add in points to show each observation
sns.stripplot(
x="Credit_Limit",
y="Income_Category",
data=df,
size=4,
linewidth=0,
hue="Attrition_Flag",
)
# Tweak the visual presentation
ax.set(ylabel="")
sns.despine(trim=True, left=True)
plt.show()
sns.lineplot(x="Avg_Utilization_Ratio", y="Credit_Limit", data=df)
plt.show()
sns.histplot(x="Total_Revolving_Bal", data=df, kde=True, hue="Attrition_Flag")
plt.show()
sns.set_theme(style="ticks")
# Plot the orbital period with horizontal boxes
sns.boxplot(
x="Avg_Utilization_Ratio",
y="Income_Category",
data=df,
whis=[0, 100],
width=0.6,
palette="Set1",
)
# Add in points to show each observation
sns.stripplot(
x="Avg_Utilization_Ratio",
y="Income_Category",
data=df,
size=4,
linewidth=0,
hue="Attrition_Flag",
)
# Tweak the visual presentation
ax.set(ylabel="")
sns.despine(trim=True, left=True)
plt.show()
sns.distplot(df["Total_Revolving_Bal"], kde=True)
plt.show()
sns.boxplot(x="Income_Category", y="Total_Revolving_Bal", hue="Attrition_Flag", data=df)
plt.show()
sns.boxplot(x="Attrition_Flag", y="Total_Revolving_Bal", data=df)
plt.show()
sns.violinplot(data=df, x="Income_Category", y="Months_on_book", palette="Set1")
plt.show()
sns.countplot(data=df, x="Months_Inactive_12_mon", hue="Attrition_Flag")
plt.show()
sns.set_theme(style="ticks")
# Plot the orbital period with horizontal boxes
sns.boxplot(
x="Avg_Open_To_Buy",
y="Income_Category",
data=df,
whis=[0, 100],
width=0.6,
palette="Set1",
)
# Add in points to show each observation
sns.stripplot(
x="Avg_Open_To_Buy",
y="Income_Category",
data=df,
size=4,
linewidth=0,
hue="Attrition_Flag",
)
# Tweak the visual presentation
ax.set(ylabel="")
sns.despine(trim=True, left=True)
plt.show()
sns.histplot(x="Avg_Open_To_Buy", data=df, kde=True, hue="Attrition_Flag")
plt.show()
sns.boxplot(x="Income_Category", y="Avg_Open_To_Buy", hue="Attrition_Flag", data=df)
plt.show()
sns.barplot(x="Income_Category", y="Avg_Open_To_Buy", hue="Attrition_Flag", data=df)
plt.show()
sns.countplot(x="Card_Category", data=df, hue="Attrition_Flag")
plt.show()
sns.scatterplot(x="Avg_Open_To_Buy", y="Credit_Limit", data=df)
plt.show()
sns.histplot(x="Avg_Open_To_Buy", kde=True, data=df)
sns.histplot(x="Credit_Limit", kde=True, data=df)
plt.show()
sns.boxplot(
y="Avg_Utilization_Ratio",
x="Income_Category",
hue="Attrition_Flag",
data=df[df.Avg_Utilization_Ratio > 0.3],
)
plt.show()
|
# # ⚕️ Binary Classification with a Kidney Stone Prediction Dataset ⚕️
# ## 📋 About The Dataset
# This dataset can be used to predict the presence of kidney stones based on urine analysis.
# The 79 urine specimens, were analyzed in an effort to determine if certain physical characteristics of the urine might be related to the formation of calcium oxalate crystals.
# The six physical characteristics of the urine are:
# * **specific gravity** (gravity) the density of the urine relative to water
# * **pH** (ph) - the negative logarithm of the hydrogen ion
# * **osmolarity** (osmo) - a unit used in biology and medicine but not inphysical chemistry. Osmolarity is proportional to the concentration of molecules in solution
# * **conductivity** (condo) - Conductivity is proportional to the concentration of charged ions in solution
# * **urea concentration** (urea) - in millimoles per litre
# * **calcium concentration** (calc) - in millimoles per litre
# The dataset for this competition (both train and test) was generated from a deep learning model trained on the [Kidney Stone Prediction based on Urine Analysis dataset](https://www.kaggle.com/datasets/vuppalaadithyasairam/kidney-stone-prediction-based-on-urine-analysis).
# # Imports
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import pathlib
import random
import math
import sys
import os
import gc
from copy import deepcopy
from functools import partial
from itertools import combinations
# Model selection, cross validation, and performance evaluation
from category_encoders import (
OneHotEncoder,
OrdinalEncoder,
CountEncoder,
CatBoostEncoder,
)
from sklearn.model_selection import RepeatedStratifiedKFold, RepeatedKFold
from sklearn.base import BaseEstimator, TransformerMixin
from imblearn.under_sampling import RandomUnderSampler
from sklearn.model_selection import train_test_split
from sklearn.model_selection import cross_val_score
from sklearn.preprocessing import StandardScaler
from sklearn.feature_selection import RFECV
from sklearn.decomposition import PCA
from sklearn.metrics import log_loss, auc
from sklearn.cluster import KMeans
import sklearn.metrics as metrics
# from sklearn.metrics import auc
# from sklearn.metrics import RocCurveDisplay
from sklearn.ensemble import StackingClassifier, VotingClassifier
import sklearn
# import umap
# # Hypertuning
# import optuna
# Classifiers
from sklearn.dummy import DummyClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.svm import SVC
from sklearn.neural_network import MLPClassifier
from sklearn.gaussian_process import GaussianProcessClassifier
from sklearn.gaussian_process.kernels import RBF
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier, AdaBoostClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.discriminant_analysis import QuadraticDiscriminantAnalysis
from sklearn.linear_model import LogisticRegression
from lightgbm import LGBMClassifier
from xgboost import XGBClassifier
from catboost import CatBoostClassifier
import catboost
from pygam import GAM, LogisticGAM, s, f, te
from tqdm.auto import tqdm
import warnings
warnings.filterwarnings("ignore", category=UserWarning)
sns.set_theme(font="Roboto")
sns.set_style("white")
# List available files in the project's environment
for file in pathlib.Path("/kaggle/input").rglob("*"):
if file.is_file():
print(file)
print(100 * "-")
print(f"Python \tv{sys.version}")
print(f"Pandas \tv{pd.__version__}")
print(f"Numpy \tv{np.__version__}")
print(f"Scikit-Learn\tv{sklearn.__version__}")
# random seed/state for reproducability
RANDOM = 12345
np.random.seed(RANDOM)
# CSV file paths
ORIGINAL_SET = "/kaggle/input/kidney-stone-prediction-based-on-urine-analysis/kindey stone urine analysis.csv"
ROOT_PATH = pathlib.Path("/kaggle/input/playground-series-s3e12")
TRAINING_SET = pathlib.Path(ROOT_PATH / "train.csv")
TESTING_SET = pathlib.Path(ROOT_PATH / "test.csv")
SAMPLE_SUBMISSION = pathlib.Path(ROOT_PATH / "sample_submission.csv")
# Pandas DataFrame Object for training data
df_train = pd.read_csv(TRAINING_SET, index_col="id")
# Pandas DataFrame Object for testing (hold-out) data
df_test = pd.read_csv(TESTING_SET, index_col="id")
# Pandas DataFrame Object for original datset
df_original = pd.read_csv(ORIGINAL_SET)
print("- ORIGINAL DATASET -\n")
display(df_original.head(5))
print("- TRAIN DATASET -\n")
display(df_train.head(5))
print("\n- TEST DATASET -\n")
display(df_test.head(5))
# # Data Understanding
print(f"Data shape in original data: \t{df_original.shape}")
print(f"Data shape in test data: \t{df_test.shape}")
print(f"Data shape in train data: \t{df_train.shape}")
print(f"\nMissing values in original dataset: \t{df_original.isnull().sum().sum()}")
print(f"Missing values in test dataset: \t{df_test.isnull().sum().sum()}")
print(f"Missing values in train dataset: \t{df_train.isnull().sum().sum()}")
print(
f"\nNumber of duplicates in original dataset: \t{df_original.duplicated().sum()}"
)
print(f"Number of duplicates in test dataset: \t{df_test.duplicated().sum()}")
print(f"Number of duplicates in train dataset: \t{df_train.duplicated().sum()}")
print(f"\nOriginal dataset datatypes:")
print(f"{pd.value_counts(df_original.dtypes)}")
print(f"\nTest dataset datatypes:")
print(f"{pd.value_counts(df_test.dtypes)}")
print(f"\nTrain dataset datatypes:")
print(f"{pd.value_counts(df_train.dtypes)}")
print("\n- Information for Original Dataset -\n")
display(df_original.info())
print("\n- Information for Testing Dataset -\n")
display(df_test.info())
print("\n- Information for Training Dataset -\n")
display(df_train.info())
# ## Statistical Overview
print("\n- Description for Original Dataset -\n")
display(df_original.describe().T)
print("\n- Description for Testing Dataset -\n")
display(df_test.describe().T)
print("\n- Description for Training Dataset -\n")
display(df_train.describe().T)
# ## Original vs. Training Data Duplicates(?)
df_train_original = pd.concat([df_original, df_train]).reset_index(drop=True)
print(
f"Number of duplicates between train and original dataset: {df_train_original.duplicated().sum()}"
)
# No duplicates found between train and original dataset so I will test if concatenating them improves model performance.
# # Feature Understanding
# ## Target Balance
df_original["data"] = "original"
df_train["data"] = "train"
df_train_original = pd.concat([df_original, df_train]).reset_index(drop=True)
sns.countplot(
df_train_original, x="target", hue="data", palette=("dodgerblue", "orange")
)
plt.suptitle(f"Target Balance (Original vs Train)")
plt.tight_layout()
plt.show()
original_balance = (
df_original[df_original["target"] == 1].count()[0]
/ df_original[df_original["target"] == 0].count()[0]
)
train_balance = (
df_train[df_train["target"] == 1].count()[0]
/ df_train[df_train["target"] == 0].count()[0]
)
print(f"Target ratio in Original Dataset: {original_balance.round(2)}")
print(f"Target ratio in Training Dataset: {train_balance}")
df_test["data"] = "test"
df_all = pd.concat([df_original, df_train, df_test]).reset_index(drop=True)
# ## Univariate Analysis
features = []
print("Feature Unique Values:")
for feature in df_all.columns:
if (feature != "target") and (feature != "data"):
print(f"{feature} unique values: {len(np.unique(df_all[feature]))}")
features.append(feature)
no_num = 0
no_cat = 0
n_cols = 3
n_rows = (len(features) - 1) // n_cols + 1
fig, axes = plt.subplots(nrows=n_rows, ncols=n_cols, figsize=(20, 10))
for i, feature in enumerate(features):
row = i // n_cols
col = i % n_cols
ax = axes[row, col]
no_num += 1
sns.histplot(
df_all,
x=feature,
hue="data",
kde=True,
bins=50,
ax=ax,
palette=("dodgerblue", "orange", "seagreen"),
multiple="layer",
)
# {“layer”, “dodge”, “stack”, “fill”}
ax.set_title(f"{feature} kernel density estimate (KDE)")
ax.set_xlabel("")
ax.set_ylabel("")
plt.legend()
plt.tight_layout()
plt.show()
print(f"No. of Numerical Features: {no_num}")
print(f"No. of Categorical Features: {no_cat}\n")
# Training and test data have similar distributions (that stand to be standardized)- original data is too small to see comparison.
# ## Boxplots for Numerical Features
fig, axes = plt.subplots(nrows=n_rows, ncols=n_cols, figsize=(20, 10))
for i, feature in enumerate(features):
row = i // n_cols
col = i % n_cols
ax = axes[row, col]
no_cat += 1
sns.boxplot(
data=df_all,
y=feature,
x="data",
ax=ax,
palette=("dodgerblue", "orange", "seagreen"),
orient="v",
)
ax.set_title(f"{feature} Box Plot")
i += 1
plt.legend()
plt.tight_layout()
plt.show()
# - Training data has few outliers in *gravity* and *ph*.
# - Test dat has few outliers in *gravity*, *ph*, and *calc*.
# # Feature Relationships - Bivariate Analysis
sns.pairplot(
df_all, hue="data", corner=True, palette=("dodgerblue", "orange", "seagreen")
)
mask0 = np.triu(np.ones_like(df_original.corr(), dtype=bool))
plt.subplots(figsize=(15, 8))
h2 = sns.heatmap(df_original.corr(), mask=mask0, annot=True, fmt=".2f", cmap="coolwarm")
h2.set_title(f"Original Dataset Correlation Matrix", fontsize=14)
mask1 = np.triu(np.ones_like(df_test.corr(), dtype=bool))
plt.subplots(figsize=(15, 8))
h2 = sns.heatmap(df_test.corr(), mask=mask1, annot=True, fmt=".2f", cmap="coolwarm")
h2.set_title(f"Test Dataset Correlation Matrix", fontsize=14)
mask2 = np.triu(np.ones_like(df_train.corr(), dtype=bool))
plt.subplots(figsize=(15, 8))
h1 = sns.heatmap(df_train.corr(), mask=mask2, annot=True, fmt=".2f", cmap="coolwarm")
h1.set_title(f"Train Dataset Correlation Matrix", fontsize=14)
# Target has fair correlation with *calc* and *gravity* in training data.
# Strong correlation between *gravity*, *urea* and *osmo*.
# # Model Training & Analysis
# ## Data Splitting
X = df_train[features]
y = df_train["target"]
X_ = df_train_original[features]
y_ = df_train_original["target"]
X_train, X_val, y_train, y_val = train_test_split(
X, y, test_size=0.2, random_state=RANDOM
)
X_train_, X_val_, y_train_, y_val_ = train_test_split(
X_, y_, test_size=0.2, random_state=RANDOM
)
print(
f"Training set has {X_train.shape[0]} examples and validation set has {X_val.shape[0]}"
)
print(
f"Training set with original data has {X_train_.shape[0]} examples and validation set has {X_val_.shape[0]}"
)
names = [
"Nearest_Neighbors",
"Linear_SVM",
"SVM",
"Gaussian_Process",
"Decision_Tree",
"Random_Forest",
"Multi-layer_Perceptron",
"Ada_Boost",
"Naive_Bayes",
"Quadratic_Discriminant_Analysis",
"Logistic_Regressor",
"LightGBM",
"XGBoost",
"CatBoost",
]
classifiers = [
KNeighborsClassifier(),
SVC(kernel="linear", probability=True, random_state=RANDOM),
SVC(probability=True, random_state=RANDOM),
GaussianProcessClassifier(random_state=RANDOM),
DecisionTreeClassifier(random_state=RANDOM),
RandomForestClassifier(random_state=RANDOM),
MLPClassifier(random_state=RANDOM),
AdaBoostClassifier(random_state=RANDOM),
GaussianNB(),
QuadraticDiscriminantAnalysis(),
LogisticRegression(random_state=RANDOM),
LGBMClassifier(objective="binary", n_jobs=-1, random_state=RANDOM),
XGBClassifier(eval_metric="auc", random_state=RANDOM, verbosity=0),
CatBoostClassifier(eval_metric="AUC", random_state=RANDOM, verbose=0),
]
# ## Feature Engineering - ***(to do)***
#### from https://www.kaggle.com/competitions/playground-series-s3e12/discussion/399441
# # Ratio of calcium concentration to urea concentration:
# df['calc_urea_ratio'] = df['calc'] / df['urea']
# # Ratio of specific gravity to osmolarity:
# df['gravity_osm_ratio'] = df['gravity'] / df['osmo']
# # Product of calcium concentration and osmolarity:
# df['calc_osm_product'] = df['calc'] * df['osmo']
# # Product of specific gravity and conductivity:
# df['gravity_cond_product'] = df['gravity'] * df['cond']
# # Ratio of calcium concentration to specific gravity:
# df['calc_gravity_ratio'] = df['calc'] / df['gravity']
# # Ratio of urea concentration to specific gravity:
# df['urea_gravity_ratio'] = df['urea'] / df['gravity']
# # Product of osmolarity and conductivity:
# df['osm_cond_product'] = df['osmo'] * df['cond']
# # Ratio of calcium concentration to osmolarity:
# df['calc_osm_ratio'] = df['calc'] / df['osmo']
# # Ratio of urea concentration to osmolarity:
# df['urea_osm_ratio'] = df['urea'] / df['osmo']
# # Product of specific gravity and urea concentration:
# df['gravity_urea_product'] = df['gravity'] * df['urea']
# ## Feature Scaling
# Testing baseline model with and without feature standardization for comparison.
scaler = StandardScaler()
X_train_scaled = scaler.fit_transform(X_train)
X_val_scaled = scaler.transform(X_val)
X_train_scaled_ = scaler.fit_transform(X_train_)
X_val_scaled_ = scaler.transform(X_val_)
print(
f"Scaled training set has {X_train_scaled.shape[0]} examples and scaled validation set has {X_val_scaled.shape[0]}"
)
print(
f"Scaled training set with original data has {X_train_scaled_.shape[0]} examples and scaled validation set has {X_val_scaled_.shape[0]}"
)
eval_sets = ["training", "scaled_training", "with_original", "with_original_scaled"]
results = {
"training": [],
"scaled_training": [],
"with_original": [],
"with_original_scaled": [],
}
def evaluate_model(model, X, y):
# prepare the cross-validation procedure
cv = RepeatedKFold(n_splits=10, n_repeats=10, random_state=RANDOM)
# evaluate model
scores = cross_val_score(model, X, y, scoring="roc_auc", cv=cv)
return scores
def compare_eval_sets(model):
for i in range(0, 4):
if i == 0:
model.fit(X_train, y_train)
features = X_val
target = y_val
elif i == 1:
model.fit(X_train_scaled, y_train)
features = X_val_scaled
target = y_val
elif i == 2:
model.fit(X_train_, y_train_)
features = X_val_
target = y_val_
elif i == 3:
model.fit(X_train_scaled_, y_train_)
features = X_val_scaled_
target = y_val_
scores = evaluate_model(model, features, target)
results[eval_sets[i]] = [*results[eval_sets[i]], *scores]
for i in tqdm(range(len(classifiers))):
compare_eval_sets(classifiers[i])
df_eval = pd.DataFrame(results)
fig, ax = plt.subplots(figsize=(7, 5))
v = sns.boxplot(data=df_eval)
v.set_title("Comparison of All Models' ROC-AUC with Various Preprocessing")
v.set(xlabel="Preprocess Dataset", ylabel="ROC-AUC Score")
plt.show()
display(df_eval.mean().sort_values(ascending=False).round(4).reset_index())
# **Models perform best with original data included and standardized.**
# ## Model Performance vs. Outliers
print("Outliers in training data")
X_train_.query("ph > 7.5").shape
outlier_ids = X_train_.query("ph > 7.5").index.values
df_out = df_train_original.drop(outlier_ids).reset_index(drop=True)
X_out = df_out[features]
y_out = df_out["target"]
X_train_out, X_val_out, y_train_out, y_val_out = train_test_split(
X_out, y_out, test_size=0.2, random_state=RANDOM
)
scaler = StandardScaler()
X_train_out = scaler.fit_transform(X_train_out)
X_val_out = scaler.transform(X_val_out)
outlier_labels = ["full data", "data without outliers"]
scores = []
def compare_outliers(model):
for i in range(0, 2):
if i == 0:
model.fit(X_train_out, y_train_out)
features = X_val_out
target = y_val_out
elif i == 1:
model.fit(X_train_scaled_, y_train_)
features = X_val_scaled_
target = y_val_
score = evaluate_model(model, features, target)
for s in score:
scores.append({"data_set": outlier_labels[i], "roc_auc": s})
for i in tqdm(range(len(classifiers))):
compare_outliers(classifiers[i])
df_eval = pd.DataFrame(scores)
fig, ax = plt.subplots(figsize=(7, 5))
v = sns.boxplot(data=df_eval, y="data_set", x="roc_auc")
v.set_title("Model Performance With and Without Outliers")
v.set(xlabel="ROC-AUC Score", ylabel="Dataset")
display(df_eval.groupby("data_set")["roc_auc"].aggregate(np.median).reset_index())
# **Improved mean model performance after 'ph' outlier removal from training data.**
# # Model Cross-Validated Baselines
scores = []
for i in tqdm(range(len(classifiers))):
classifiers[i].fit(X_train_out, y_train_out)
score = evaluate_model(classifiers[i], X_val_out, y_val_out)
for s in score:
scores.append({"model": names[i], "roc_auc": s})
df_eval = pd.DataFrame(scores)
fig, ax = plt.subplots(figsize=(7, 5))
order = (
df_eval.groupby(["model"])["roc_auc"]
.aggregate(np.median)
.reset_index()
.sort_values("roc_auc")
)
v = sns.boxplot(
data=df_eval, y="model", x="roc_auc", whis=[0, 100], order=order["model"]
)
v.set_title("Comparison of All Models' baseline ROC-AUC")
v.set(xlabel="ROC-AUC Score", ylabel="Model Algorithm")
plt.show()
# # Baseline Ensemble
# Train baseline model with the top 4 performing models.
X_out_ = scaler.fit_transform(X_out)
def plot_roc_auc(model, features, target):
pred_proba_ = model.predict_proba(features)[:, 1]
pred_target = model.predict(features)
# ROC/Accuracy
fpr, tpr, roc_thresholds = metrics.roc_curve(target, pred_proba_)
roc_auc = metrics.roc_auc_score(target, pred_proba_)
plt.plot(fpr, tpr, lw=2, alpha=0.4)
return fpr, tpr, roc_auc
def eval_roc_auc_(model):
cv = RepeatedKFold(n_splits=10, n_repeats=10, random_state=RANDOM)
tprs = []
aucs = []
mean_fpr = np.linspace(0, 1, 100)
fig, ax = plt.subplots(figsize=(6, 6))
for fold, (train, test) in enumerate(cv.split(X_out_, y_out)):
model.fit(X_out_[train], y_out[train])
fpr, tpr, roc_auc = plot_roc_auc(model, X_out_[test], y_out[test])
interp_tpr = np.interp(mean_fpr, fpr, tpr)
interp_tpr[0] = 0.0
tprs.append(interp_tpr)
aucs.append(roc_auc)
ax.plot([0, 1], [0, 1], "k--", label="chance level (AUC = 0.5)")
mean_tpr = np.mean(tprs, axis=0)
mean_tpr[-1] = 1.0
mean_auc = auc(mean_fpr, mean_tpr)
std_auc = np.std(aucs)
ax.plot(
mean_fpr,
mean_tpr,
color="b",
label=r"Mean ROC (AUC = %0.4f $\pm$ %0.4f)" % (mean_auc, std_auc),
lw=4,
alpha=0.8,
)
std_tpr = np.std(tprs, axis=0)
tprs_upper = np.minimum(mean_tpr + std_tpr, 1)
tprs_lower = np.maximum(mean_tpr - std_tpr, 0)
ax.fill_between(
mean_fpr,
tprs_lower,
tprs_upper,
color="grey",
alpha=0.4,
label=r"$\pm$ 1 std. dev.",
)
ax.set(
xlabel="False Positive Rate",
ylabel="True Positive Rate",
title=f"Mean ROC curve",
)
plt.axis("square")
plt.legend(loc="lower right")
plt.show()
estimators = [
("catboost", classifiers[13]),
("SVM", classifiers[2]),
("lightGBM", classifiers[11]),
("random_forest", classifiers[5]),
]
# ## Voting Classifier
vc = VotingClassifier(estimators=estimators, voting="soft")
eval_roc_auc_(vc)
# ## Stacked Generalization
sc = StackingClassifier(estimators=estimators)
eval_roc_auc_(sc)
# #### Voting classifier has better average ROC-AUC score.
# # Feature Selection
# Recursive feature elimination with cross-validation
cv = RepeatedKFold(n_splits=10, n_repeats=10, random_state=RANDOM)
selector = RFECV(estimator=classifiers[11], cv=cv, scoring="roc_auc") # lightgbm
selector = selector.fit(X_out, y_out)
print("Optimal number of features: ", selector.n_features_)
print("Best features : ", X.columns[selector.support_].values)
# #### Recursive feature elimination with cross-validation shows that models in the ensemble perform (on average) best with all the features.
# ## Train Final Model
# # best score was with original data included, without outliers, and scaled
X_out = scaler.fit_transform(X_out)
vc.fit(X_out, y_out)
# # Submission
# # prepare test data for prediction
df_test = df_test.drop("data", axis=1)
df_test = scaler.transform(df_test)
predictions = vc.predict(df_test)
submission_ids = pd.read_csv(TESTING_SET)["id"]
submission = pd.DataFrame(
{
"id": submission_ids,
"target": predictions,
}
)
submission.to_csv("submission.csv", index=False)
# Double check if the CSV file before submitting.
print("Target Balance")
fig, axes = plt.subplots(nrows=1, ncols=2, figsize=(10, 4))
sns.countplot(submission, ax=axes[0], x="target").set_title("Submission Target Balance")
sns.countplot(df_out, ax=axes[1], x="target").set_title("Training Data Target Balance")
plt.legend()
plt.tight_layout()
plt.show()
|
import pandas as pd
a = pd.read_csv("/kaggle/input/videogamesales/vgsales.csv")
a.head(3)
a = pd.read_csv("/kaggle/input/videogamesales/vgsales.csv", index_col=["Rank"])
a.head(3)
a["Global_Sales"]
a["Global_Sales"].mean()
b = a[["Name", "Global_Sales"]]
b.head()
# **concepts of axis**
d = {"student": ["Bala", "Raja", "Aalini"], "marks": [40, 25, 30], "age": [12, 30, 10]}
d
l = ["Bala", "Raja", "Aalini"]
df = pd.DataFrame(d, index=l)
df
df.mean()
df.mean(axis=1) # calculate row-wise
# **mathematical operations**
a = pd.read_csv("/kaggle/input/videogamesales/vgsales.csv", index_col=["Rank"])
a
a.mean()
a.mean(axis=1)
a.median()
a["Global_Sales"].sum()
a["Global_Sales"].mean()
a["Global_Sales"].max()
a["Global_Sales"].min()
a["Global_Sales"].prod()
a["Global_Sales"].std()
a["Global_Sales"].median()
a["Global_Sales"] = a["Global_Sales"].add(10)
a["Global_Sales"] # permanent change
a["Global_Sales"].mul(2)
a["Global_Sales"].div(2)
a["Global_Sales"].div(2, fill_value=10)
|
# # Data Dictionary - train.csv - dependent variable is survival
# * Variable Definition Key
# * survival Survival 0 = No, 1 = Yes
# * pclass Ticket class 1 = 1st, 2 = 2nd, 3 = 3rd
# * sex Sex
# * Age Age in years
# * sibsp # For children number of siblings. For parents number of spouses.
# * parch # For children number of parents. For parents number of children.
# * ticket Ticket number
# * fare Passenger fare
# * cabin Cabin number
# * embarked Port Embarkation C = Cherbourg, Q = Queenstown, S = Southampton
# Normally, all data is processed and then split with Sklearn test_train_split. Since both files are given, they are processed separately.
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Other imports besides Kaggle provided ones.
import seaborn as sn
# importing one hot encoder
from sklearn.preprocessing import OneHotEncoder
from sklearn.metrics import classification_report, confusion_matrix, accuracy_score
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression, LogisticRegressionCV
import matplotlib.pyplot as plt
dash = "-" * 80
# Use this to allow all rows to be printed if you need to analyze the data.
# pd.set_option('display.max_rows', None)
# pd.set_option('display.max_columns',None)
# # Read in Competition Data
train_data = pd.read_csv("/kaggle/input/titanic/train.csv")
train_data.head(100)
test_data = pd.read_csv("/kaggle/input/titanic/test.csv")
test_data.head()
# ## Data Shape
print("Train Data: ", train_data.shape)
print("Test Data: ", test_data.shape)
# # Review Null Values
# Funtion to Iterate over column names to display how many null values for each column.
def Check_Dataframe_for_Null_Values(df_to_check):
for column in df_to_check:
print(
" Count of Null values for "
+ column
+ ": "
+ str(len(df_to_check[df_to_check[column].isna()]))
)
return
Check_Dataframe_for_Null_Values(train_data)
# **Training Data Nulls:**
# * Age: 177
# * Cabin: 687
# * Embarked: 2
Check_Dataframe_for_Null_Values(test_data)
# **Testing Data Nulls:**
# * Age: 86
# * Fare: 1
# * Cabin: 327
# # Feature Engineering and Populating Null Values
# ## Size of Family
# Adding Size of the Family to the training set in hopes that it will improve modeling.
# Add the siblings with the spouse and the passenger to get the size of the family aboart the Titanic.
train_data["family_size"] = train_data["SibSp"] + train_data["Parch"] + 1
test_data["family_size"] = train_data["SibSp"] + train_data["Parch"] + 1
# ## For Null Age, Fill in Age of Passenger by Average Age by Ticket Class Assignment
# A boxplot to see approximate mean age by Ticket Class (Pclass)
sn.boxplot(x="Pclass", y="Age", data=train_data)
# Review distinct values of Pclass:
print(train_data["Pclass"].unique())
# Calculate Age based on Ticket Class
# To address null values for age we query for each class and then calcuate the mean.
df_class_1 = train_data.query("Pclass == 1", engine="python")
class_1_mean_age = int(df_class_1["Age"].mean())
df_class_2 = train_data.query("Pclass == 2", engine="python")
class_2_mean_age = int(df_class_2["Age"].mean())
df_class_3 = train_data.query("Pclass == 3", engine="python")
class_3_mean_age = int(df_class_3["Age"].mean())
print("Mean Age for Class 1", class_1_mean_age)
print("Mean Age for Class 2", class_2_mean_age)
print("Mean Age for Class 3", class_3_mean_age)
# ### Update Training and Testing data set with Mean Age
# if data is null and class 1 then update age with value, etc.
# train_data.query('Pclass == 1 and Age.isnull()' , engine='python')
# Update train and test separately.
train_data.loc[
train_data.query("Pclass == 1 and Age.isnull()", engine="python").index, "Age"
] = class_1_mean_age
test_data.loc[
test_data.query("Pclass == 1 and Age.isnull()", engine="python").index, "Age"
] = class_1_mean_age
train_data.loc[
train_data.query("Pclass == 2 and Age.isnull()", engine="python").index, "Age"
] = class_2_mean_age
test_data.loc[
test_data.query("Pclass == 2 and Age.isnull()", engine="python").index, "Age"
] = class_2_mean_age
train_data.loc[
train_data.query("Pclass == 3 and Age.isnull()", engine="python").index, "Age"
] = class_3_mean_age
test_data.loc[
test_data.query("Pclass == 3 and Age.isnull()", engine="python").index, "Age"
] = class_3_mean_age
# First Class - Age Set to 38 for Null
train_data.query(
"Pclass == 1 and Age == " + str(class_1_mean_age), engine="python"
).head(5)
# First Class - Age Set to 38 for Null
test_data.query(
"Pclass == 1 and Age == " + str(class_1_mean_age), engine="python"
).head(5)
# Second Class - Age Set to 29 for Null
train_data.query(
"Pclass == 2 and Age == " + str(class_2_mean_age), engine="python"
).head(5)
# Second Class - Age Set to 29 for Null
test_data.query(
"Pclass == 2 and Age == " + str(class_2_mean_age), engine="python"
).head(5)
# Third Class - Age Set to 25 for Null
train_data.query(
"Pclass == 3 and Age == " + str(class_3_mean_age), engine="python"
).head(5)
# Third Class - Age Set to 25 for Null
test_data.query(
"Pclass == 3 and Age == " + str(class_3_mean_age), engine="python"
).head(5)
# Review Null values again
Check_Dataframe_for_Null_Values(train_data)
# Review Null values again
Check_Dataframe_for_Null_Values(test_data)
# ## Title - Categorizing into 4 titles - Mr, Mrs, Miss and Master
# * This will replace with the following:
# * Mr = Adult male
# * Mrs = Adult female
# * Miss = Child female or unmarried female
# * Master = Child male
# https://triangleinequality.wordpress.com/2013/09/08/basic-feature-engineering-with-the-titanic-data/
# * An article on parsing out the titles of the passengers.
# This takes the full string, big_string and determines if the substring (ie. "Mr.") occurs. -1 means that the subtring was not found.
def substrings_in_string(big_string, substrings):
# print (big_string, substrings)
for substring in substrings:
if big_string.find(substring) != -1:
return substring
print(big_string)
return np.nan # np.nan will return a type of float but with no value
# creating a title column from name. These are the subtrings we will use to search the names
title_list = [
"Mrs",
"Mr",
"Master",
"Miss",
"Major",
"Rev",
"Dr",
"Ms",
"Mlle",
"Col",
"Capt",
"Mme",
"Countess",
"Don",
"Jonkheer",
]
# This will use map to go through train_data and test_data and then create a new column called Title with the title found in each name.
train_data["Title"] = train_data["Name"].map(
lambda x: substrings_in_string(x, title_list)
)
test_data["Title"] = test_data["Name"].map(
lambda x: substrings_in_string(x, title_list)
)
# This will replace each record with the following:
# Mr = Adult male
# Mrs = Adult female
# Miss = Child female or unmarried female
# Master = Child male
def replace_titles(x):
title = x["Title"]
if title in ["Don", "Major", "Capt", "Jonkheer", "Rev", "Col"]:
return "Mr"
elif title in ["Countess", "Mme"]:
return "Mrs"
elif title in ["Mlle", "Ms"]:
return "Miss"
elif title == "Dr":
if x["Sex"] == "Male":
return "Mr"
else:
return "Mrs"
else:
return title
# Categorizing into 4 groups - Mr, Mrs, Miss and Master
train_data["Title"] = train_data.apply(replace_titles, axis=1)
test_data["Title"] = test_data.apply(replace_titles, axis=1)
train_data.head(10)
test_data.head(10)
# ## Cabin
# * With so many missing values were going to do a simple has_cabin feature. Reviewing the data, it seems third class passengers had no cabin assignment.
# First Class List of Passengers with no cabin assignment
df_class_1 = train_data.query("Pclass == 1", engine="python")
df_class_1_with_null_cabin = df_class_1.query("Cabin.isnull()", engine="python")
df_class_1_with_null_cabin.info()
# Second Class List of Passengers with no cabin assignment
df_class_2 = train_data.query("Pclass == 2", engine="python")
df_class_2_with_null_cabin = df_class_2.query("Cabin.isnull()", engine="python")
df_class_2_with_null_cabin.info()
# Third Class List of Passengers with no cabin assignment
df_class_3 = train_data.query("Pclass == 3", engine="python")
df_class_3_with_null_cabin = df_class_3.query("Cabin.isnull()", engine="python")
df_class_3_with_null_cabin.info()
# Return yes for cabin and no if cabin is null or "nan"
# nan is a float value representing null.
def check_for_cabin_assignment(cabin_name):
if str(cabin_name) == "nan":
return "no"
return "yes"
# add new column, has_cabin with yes or no.
train_data["has_cabin"] = train_data["Cabin"].map(
lambda x: check_for_cabin_assignment(x)
)
test_data["has_cabin"] = test_data["Cabin"].map(lambda x: check_for_cabin_assignment(x))
Check_Dataframe_for_Null_Values(train_data)
Check_Dataframe_for_Null_Values(test_data)
# # Replace Fare with Mean Value for Competition Test Data
# Find mean value of the Fare.
test_data["Fare"].describe().loc[["mean"]]
# Updated Fare to a Mean Value - It can't be deleted or the competition entry will fail.
# Find just the mean Fare value and put it in a variable.
f_mean = test_data.loc[:, "Fare"].mean()
f_mean
# Update test_data null values with mean Fare.
test_data["Fare"].fillna(f_mean, inplace=True)
Check_Dataframe_for_Null_Values(test_data)
# # One Hot Encoding
# creating one hot encoder object
OneHotEncoder = OneHotEncoder()
# ## Pclass
# * I'm going to encode this and treat it as categorical since it has only 3 choices. I question which is better, to leave it as is or to one-hot-encode it.
# * 1 - 1st class passenger
# * 2 - 2nd class passenger
# * 3 - 3rd class passenger
df_OneHotEncoded_train = pd.DataFrame(
OneHotEncoder.fit_transform(train_data[["Pclass"]]).toarray()
)
df_OneHotEncoded_test = pd.DataFrame(
OneHotEncoder.fit_transform(test_data[["Pclass"]]).toarray()
)
df_OneHotEncoded_train.rename(
columns={0: "first_class", 1: "second_class", 2: "third_class"}, inplace=True
)
df_OneHotEncoded_test.rename(
columns={0: "first_class", 1: "second_class", 2: "third_class"}, inplace=True
)
train_data = train_data.join(df_OneHotEncoded_train)
test_data = test_data.join(df_OneHotEncoded_test)
# ## Title
df_OneHotEncoded_train = pd.DataFrame(
OneHotEncoder.fit_transform(train_data[["Title"]]).toarray()
)
df_OneHotEncoded_test = pd.DataFrame(
OneHotEncoder.fit_transform(test_data[["Title"]]).toarray()
)
df_OneHotEncoded_train.rename(
columns={
0: "master-male-child",
1: "miss-female-child_or_unmarried",
2: "mr-male-adult",
3: "mrs-female-adult",
},
inplace=True,
)
df_OneHotEncoded_test.rename(
columns={
0: "master-male-child",
1: "miss-female-child_or_unmarried",
2: "mr-male-adult",
3: "mrs-female-adult",
},
inplace=True,
)
train_data = train_data.join(df_OneHotEncoded_train)
test_data = test_data.join(df_OneHotEncoded_test)
# ## Sex
df_OneHotEncoded_train = pd.DataFrame(
OneHotEncoder.fit_transform(train_data[["Sex"]]).toarray()
)
df_OneHotEncoded_test = pd.DataFrame(
OneHotEncoder.fit_transform(test_data[["Sex"]]).toarray()
)
df_OneHotEncoded_train.rename(columns={0: "sex-female", 1: "sex-male"}, inplace=True)
df_OneHotEncoded_test.rename(columns={0: "sex-female", 1: "sex-male"}, inplace=True)
train_data = train_data.join(df_OneHotEncoded_train)
test_data = test_data.join(df_OneHotEncoded_test)
train_data.head()
test_data.head()
# ## Cabin
df_OneHotEncoded_train = pd.DataFrame(
OneHotEncoder.fit_transform(train_data[["has_cabin"]]).toarray()
)
df_OneHotEncoded_test = pd.DataFrame(
OneHotEncoder.fit_transform(test_data[["has_cabin"]]).toarray()
)
df_OneHotEncoded_train.rename(columns={0: "cabin_no", 1: "cabin_yes"}, inplace=True)
df_OneHotEncoded_test.rename(columns={0: "cabin_no", 1: "cabin_yes"}, inplace=True)
train_data = train_data.join(df_OneHotEncoded_train)
test_data = test_data.join(df_OneHotEncoded_test)
# ## Embarked
# I can't include Embarked because test_data doesn't have any values for this. When I do one hot encoding
# it won't create the emb_4 column. I'm having to leave off Embarked.
# df_OneHotEncoded_train_emb=pd.DataFrame(OneHotEncoder.fit_transform(train_data[['Embarked']]).toarray())
# df_OneHotEncoded_test_emb=pd.DataFrame(OneHotEncoder.fit_transform(test_data[['Embarked']]).toarray())
# df_OneHotEncoded_train_emb.rename(columns={0: 'emb_0',1: 'emb_1',2: 'emb_2',3: 'emb_3'}, inplace=True)
# df_OneHotEncoded_test_emb.rename(columns={0: 'emb_0' ,1: 'emb_1',2: 'emb_2',3: 'emb_3'}, inplace=True)
# train_data=train_data.join(df_OneHotEncoded_train_emb)
# test_data=test_data.join(df_OneHotEncoded_test_emb)
# ## Remove Pclass, Title, Sex, Cabin and Embarked After One Hot Encoding. Also, other irrelevant columns.
train_data.drop(
[
"Pclass",
],
axis=1,
inplace=True,
)
test_data.drop(["Pclass"], axis=1, inplace=True)
train_data.drop(
[
"Title",
],
axis=1,
inplace=True,
)
test_data.drop(["Title"], axis=1, inplace=True)
train_data.drop(
[
"Sex",
],
axis=1,
inplace=True,
)
test_data.drop(["Sex"], axis=1, inplace=True)
train_data.drop(
[
"Cabin",
],
axis=1,
inplace=True,
)
test_data.drop(["Cabin"], axis=1, inplace=True)
train_data.drop(
[
"has_cabin",
],
axis=1,
inplace=True,
)
test_data.drop(["has_cabin"], axis=1, inplace=True)
train_data.drop(
[
"Embarked",
],
axis=1,
inplace=True,
)
test_data.drop(["Embarked"], axis=1, inplace=True)
train_data.drop(
[
"Name",
],
axis=1,
inplace=True,
)
test_data.drop(["Name"], axis=1, inplace=True)
train_data.drop(
[
"PassengerId",
],
axis=1,
inplace=True,
)
# For the test_data, I'm not deleting passenger id because it's needed for submittal to competition.
train_data.drop(
[
"Ticket",
],
axis=1,
inplace=True,
)
test_data.drop(["Ticket"], axis=1, inplace=True)
# # Final Data for Modeling
train_data.head(5)
test_data.head(5)
# # Correlations
sn.set(rc={"figure.figsize": (20, 8)})
# plotting correlation heatmap
dataplot = sn.heatmap(train_data.corr(), cmap="YlGnBu", annot=True)
# # Separating Independent and Dependent Variables for Training Data
x = train_data.iloc[:, 1:].values
y = train_data.iloc[:, 0].values
train_data
# # Scale or Standardize the Data ( Independent Features)
# Only scale the independent variables.
Stand_Scale = StandardScaler()
# Fit Transform Training Data
x = Stand_Scale.fit_transform(x)
# removing passenger id for test_data_scaled.
test_data_temp = (
test_data.copy()
) # leaving passenger id with test_data. It's needed for competition submittal.
test_data_temp.drop(
[
"PassengerId",
],
axis=1,
inplace=True,
)
# Fit Transform Testing Data ( Will be submitted to competition at the end.)
test_data_scaled = Stand_Scale.fit_transform(test_data_temp)
print("\nx after standarization:", x)
print(dash)
print("\ntest_data_scaled after standarization:", test_data_scaled)
# # Split Training Data into Training and Testing
# * We will run our final predict against test_data for submittal to competition.
# * For now I'm splitting the training data to see how it will perform against a test set.
# split the training data to generate training and test data.
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.20, random_state=5
)
print("\nx_train", x_train.shape)
print("\ny_train", y_train.shape)
print(dash)
print("\nx_test", x_test.shape)
print("\ny_test", y_test.shape)
# ###
# # Regularization
# * Regularization is way to prevent a model from "over-learning" or "over-weighting" a feature
# * Strong regularization can effectively eliminate features from the model entirely.
# * There are two common types of regularization: "LASSO" or "L1", and "Ridge" or "L2". The main thing to know, for now, is that "L1" will push coefficients to 0 effectively removing features completely. "L2" will not reduce coefficients all the way to zero.
# * To apply regularization to our model, all we need to do is change the C value.
# * The smaller the value of C, the stronger the regularization.
reg_params = [10, 5, 1, 0.1, 0.05, 0.01] # Selecting a few values c values to try.
train_scores = []
coefs = []
for c in reg_params:
# Create and fit a model with the value of c
logReg = LogisticRegression(
C=c, max_iter=1000, penalty="l1", solver="liblinear"
) # Penalty is the L1 or Lasso.
logReg.fit(x_train, y_train)
coefs.append(logReg.coef_.reshape(-1))
# Find the accuracy on the training data
train_data_accuracy = accuracy_score(logReg.predict(x_train), y_train)
train_scores.append(train_data_accuracy)
# ## Plotting Accuracy on Training Data With Respect To Regularization
fig, ax = plt.subplots(1, 1, figsize=(14, 7))
ax.plot(reg_params, train_scores, label="train scores", marker="o")
ax.set_xscale("log")
ax.legend()
ax.set_title("Accuracy on Training Data With Respect To Regularization")
ax.set_xlabel("C Value")
ax.set_ylabel("Accuracy")
# 10^-2 = .01
# 5*10^-2 = .05
# 10^-1 = .1
# 10^0 = 1
# 10^1 = 10
fig, ax = plt.subplots(1, 1, figsize=(14, 7))
ax.plot(reg_params, coefs, marker="o")
ax.set_xscale("log")
ax.set_title("Coefficient Values With Respect to Regularization")
ax.set_ylabel("Coefficient Value")
ax.set_xlabel("C Value")
# ## Refining C values to see the best result
# Trying a range of c values by stepping .01.
reg_params = np.arange(0.01, 1, 0.01) # arange = start, stop, step
train_scores = []
test_scores = []
coefs = []
for c in reg_params:
# Create and fit a model with the value of c
# logReg = LogisticRegression(C=c, penalty='l1') # Penalty is the L1 or Lasso.
logReg = LogisticRegression(
C=c, max_iter=1000, penalty="l1", solver="liblinear"
) # Penalty is the L1 or Lasso.
logReg.fit(x_train, y_train)
coefs.append(logReg.coef_.reshape(-1))
# Find the accuracy on the training data
train_data_accuracy = accuracy_score(logReg.predict(x_train), y_train)
train_scores.append(train_data_accuracy)
reg_params
fig, ax = plt.subplots(1, 1, figsize=(14, 7))
ax.plot(reg_params, train_scores, label="train scores", marker="o")
# ax.set_xscale('log')
ax.legend()
ax.set_title("Accuracy on Training Data With Respect To Regularization")
ax.set_xlabel("C Value")
ax.set_ylabel("Accuracy")
# # Applying Cross Validated Regularization Optimization with Scikit-Learn
# * Utilizing LogisticRegressionCV
# * http://scikit-learn.org/stable/modules/generated/sklearn.linear_model.LogisticRegressionCV.html
# Trying a range of c values by stepping .01.
# reg_params are the c-values to try.
reg_params = np.arange(0.01, 1, 0.01) # arange = start, stop, step
# cv=5 is how many slices of the test data to iterate through for cross validation.
logRegCV = LogisticRegressionCV(Cs=reg_params, penalty="l1", cv=5, solver="liblinear")
logRegCV.fit(x_train, y_train)
print(logRegCV.scores_)
print("***********")
print(logRegCV.scores_[1].mean(axis=0))
print("***********")
logRegCV.C_
# ### Above our optimal C value is .25
fig, axs = plt.subplots(1, 1, figsize=(14, 7))
axs.plot(logRegCV.Cs_, logRegCV.scores_[1].mean(axis=0), marker="o")
# axs.set_xscale('log')
axs.set_title("Accuracy on Training with Cross Validation and Regularization.")
axs.set_xlabel("C Value")
axs.set_ylabel("Accuracy")
axs.grid()
logRegCV.scores_[1].mean(axis=0).max()
# ### Reviewing which coefficients have been pushed to zero
# Printing list of all column names in training data except for dependent variable. (Survived)
print(train_data.iloc[:, 1:].columns)
# print the coefficients!
print(logRegCV.coef_)
# now print out the columns that correspond to the non-zero coefficients
for coef, col in zip(logRegCV.coef_[0], train_data.iloc[:, 1:].columns):
if coef != 0:
print("{:.3f} * {}".format(coef, col))
# How many features have we retained in the model?
print(
"We have reduced from {} features to {}".format(
x_train.shape[1], sum(logRegCV.coef_[0] != 0)
)
)
# * For his iteration I added if a passenger has a cabin or not (yes or no) which was then one hot encoded.
# * Newly removed are:
# * Fare
#
# * Removed Again:
# * Parch, second_class, miss_female_child_or_unmarried
# * Added back after being removed from last iteration:
# * mr_male_adult
# Regularization can be so strong that it will "push" coefficients all the way to 0 - effectively removing the feature from the model
# # Fit the Logistic Regression Model
logRegCV.fit(x_train, y_train)
# # Accuracy of Logistic Regression Model on the test data.
logRegSDAccuracy = accuracy_score(logRegCV.predict(x_test), y_test)
print(
"logRegCV has an accuracy {:.2f}% on the test set.(12 features)".format(
logRegSDAccuracy * 100
)
)
# ### Comparing List of 'Predictions' to 'Actual' on Test Data.
y_pred = logRegCV.predict(x_test)
print("Act:", y_test) # actual
print("Pre:", y_pred) # predicted
# ### Show Accuracy Score and Confusion Matrix
print("Accuracy Score:", accuracy_score(y_test, y_pred))
# -----performance..... Confusion Matrix (actual, predicted)
cm_LR = confusion_matrix(y_test, y_pred)
print("Confusion matrix (y_test,y_pred\n", cm_LR)
from sklearn import metrics
import matplotlib.pyplot as plt
# Receiver Operating Characteristic (ROC)
fpr, tpr, _ = metrics.roc_curve(y_test, y_pred)
auc = metrics.roc_auc_score(y_test, y_pred)
# create ROC curve
plt.plot(fpr, tpr, label="AUC=" + str(auc))
plt.legend(loc=4)
plt.title(
"Logistic Regression with Regularization, Cross Validation and Standardization - ROC( Receiver Operating Characteristic) curve"
)
plt.show()
print(auc)
print(fpr, "\n", tpr, "\n", _)
# # Making Final Predictions for Submittal
# ### test_data column names:
#
test_data.columns
test_data_scaled
# defining array column names and assigning to a dataframe. xxxxx
# test_data_scaled = pd.DataFrame(test_data_scaled, columns = ['PassengerId','Pclass','Age','SibSp','Parch','Fare','family_size','master-male-child','miss-female-child','mr-male-adult','mrs-female-adult','sex-female','sex-male'])
# Generate predictions for competition.
predictions = logRegCV.predict(test_data_scaled)
print("Predictions for Submittal:", predictions)
test_data.info()
# I need to include the Passenger ID that is not scaled so we are using the original test_data dataframe.
output = pd.DataFrame({"PassengerId": test_data.PassengerId, "Survived": predictions})
output.to_csv("submission.csv", index=False)
print("Your submission was successfully saved!")
|
# # Simple howto notebook
# Lets say we have build a module we want to use in our notebook. First we upload the module as a data set, this is then uploaded in the folder `/kaggle/input/`. Since our working directory is `/kaggle/working/` we need to copy our module explicitly there in order to use it.
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
from shutil import copyfile
import matplotlib.pyplot as plt
import cv2 as cv
# We load explicitly md so we can print markdown from code
from IPython.display import Markdown as md
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
# import os
# for dirname, _, filenames in os.walk('/kaggle/input'):
# for filename in filenames:
# print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# We use `copyfile` to copy the module.
# copy our file into the working directory
copyfile(src="/kaggle/input/hpamodule1/module_1.py", dst="/kaggle/working/module_1.py")
import module_1 as hpm
# Lets see how the module works.
# Set data folders and files
hpa_data = hpm.HPA(
data_folder="/kaggle/input/hpa-single-cell-image-classification", img_folder="test"
)
print("Lets see what is the first row from the submission file:")
print(hpa_data.sample_sub_pd.loc[0, :])
# Load image
r_img, g_img, b_img, _ = hpa_data.get_rgby_images(hpa_data.sample_sub_pd.loc[0, "ID"])
print(f"We load an image channel as a np array of shape {r_img.shape}")
# Show the image
b_blur_img = cv.GaussianBlur(b_img, (15, 15), 0)
thres, b_blur_thres_img = cv.threshold(
b_blur_img, 0, 255, cv.THRESH_BINARY + cv.THRESH_OTSU
)
images = [b_img, b_blur_img, b_blur_thres_img]
fig = plt.figure(figsize=(23, 23))
for i in range(3):
ax = fig.add_subplot(131 + i)
ax.imshow(images[i])
plt.show()
|
# # 🔬 Overview
# 👀 Object detection is an important task in computer vision that involves identifying and localizing objects of interest in images or videos. 💻 YOLOv5 is a state-of-the-art object detection algorithm that has gained popularity in recent years due to its speed, accuracy, and simplicity. In this 🤩 project, we aim to use YOLOv5 to detect two classes of objects - faces with 👓 and without 👓.
# # Data Collection: Gathering Images 🖼️📷
# For the data collection step, I used the Unsplash API to gather a large set of face images with and without glasses. 📷 I obtained permission from Unsplash to use their API for this purpose. ✔️ The API allowed me to easily search for and download high-quality images that met my criteria. 🔎 I then curated the dataset by manually filtering out any irrelevant images.
# The final dataset consists of a set of face images with and without glasses, and is now available on Kaggle for anyone to use.
# ## Data Labeling Process 🏷️📝
# For the data labeling process, I used a website called makesense.ai. 🖥️ This website provides a free platform for labeling images for object detection in YOLO format. With this tool, I was able to upload my set of face images and manually annotate each image with the presence or absence of glasses. 👓 The tool generated a .txt file for each image with the corresponding annotations. 📄
# 
# # Training The Model Using Ultralytics YOLOv5 🚀
# To train my glasses detection model, I followed the steps provided in the YOLOv5 GitHub Wiki. 📚 These steps included setting up the environment, preparing the dataset and annotations, and configuring the training parameters. I used the YOLOv5 implementation by Ultralytics, which provided me with a well-documented and easy-to-use framework for training custom object detection models. With YOLOv5, I was able to efficiently train my model on the annotated face images with and without glasses. For more information, you can find the steps I followed in the YOLOv5 GitHub Wiki here: https://github.com/ultralytics/yolov5/wiki/Train-Custom-Data
# Feel free to edit the notebook as needed so that you can test the implementation for yourself.
import os
import shutil
# Define the path to your dataset
dataset_path = "/kaggle/input/glasses-detection-yolo-format/data"
# Create a directory for the images and labels in the working directory
output_path = "/kaggle/working"
image_path = os.path.join(output_path, "images")
os.makedirs(image_path, exist_ok=True)
label_path = os.path.join(output_path, "labels")
os.makedirs(label_path, exist_ok=True)
for filename in os.listdir(dataset_path):
if filename.endswith(".jpg") or filename.endswith(".jpeg"):
# If the file is an image, copy it to the images directory
src_path = os.path.join(dataset_path, filename)
dst_path = os.path.join(image_path, filename)
shutil.copy(src_path, dst_path)
elif filename.endswith(".txt"):
# If the file is a label, copy it to the labels directory
src_path = os.path.join(dataset_path, filename)
dst_path = os.path.join(label_path, filename)
shutil.copy(src_path, dst_path)
import yaml
# Define the path to the YAML file
yaml_path = "/kaggle/working/yolov5/data/my_dataset.yaml"
# Define the contents of the YAML file
data = dict(
train="../yolov5/images",
val="../yolov5/images",
nc=2,
names=["no_glasses", "with_glasses"],
)
# Write the YAML file
with open(yaml_path, "w") as f:
yaml.dump(data, f)
yolov5_path = "/kaggle/working/yolov5"
shutil.move("/kaggle/working/images", yolov5_path)
shutil.move("/kaggle/working/labels", yolov5_path)
# ### Predicting On Test Images 🔮
# 👀 In this section, I'll be putting our pre-trained model to the test by detecting glasses on both individual portraits and group photos! 💪
# 💾 By sharing the pre-trained model weights, my aim is to help you save significant time and GPU resources since you can use the model for predictions without the need to train it from scratch. 🚀
# 👉 You can access the pre-trained model through this link: https://www.kaggle.com/datasets/mohamedchahed/yolov5-model-for-glasses-detection/settings?datasetId=3105852 🌟
#
import PIL
import matplotlib.pyplot as plt
import random
detections_dir = "runs/detect/yolo_group/"
detection_images = [os.path.join(detections_dir, x) for x in os.listdir(detections_dir)]
random_detection_image = PIL.Image.open(random.choice(detection_images))
plt.figure(figsize=(30, 30))
plt.imshow(random_detection_image)
plt.xticks([])
plt.yticks([])
import random
detections_dir = "runs/detect/yolo_indivs3/"
detection_images = [os.path.join(detections_dir, x) for x in os.listdir(detections_dir)]
for i in range(3):
random_detection_image = PIL.Image.open(random.choice(detection_images))
plt.figure(figsize=(20, 20))
plt.imshow(random_detection_image)
plt.xticks([])
plt.yticks([])
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
from fastai.vision.all import *
from ipywidgets import widgets
import cv2
# import fastai
# import torch
# path
path = Path("../input/pnevmoniya")
path.ls()
# get_image_files(path)
# verify_images(path)
# datablock
diagnosis = DataBlock(
blocks=(ImageBlock, CategoryBlock),
get_items=get_image_files,
splitter=RandomSplitter(valid_pct=0.2, seed=33),
get_y=parent_label,
item_tfms=Resize(224),
)
# dataloader
dls = diagnosis.dataloaders(path / "train")
dls.train.show_batch(max_n=24, nrows=4)
# learn
learn = cnn_learner(dls, resnet34, metrics=accuracy)
learn.fine_tune(4)
# tekshirish
interp = ClassificationInterpretation.from_learner(learn)
interp.plot_confusion_matrix()
# show top loses
interp.plot_top_losses(9, figsize=(15, 10))
test_data = "../input/pnevmoniya/test/"
df = pd.read_csv("../input/pnevmoniya/sample_solution.csv")
df
for i in range(0, 624):
label = df.iloc[i, 0]
img = cv2.imread(test_data + label)
pred, pred_id, prob = learn.predict(img)
df.iloc[i, 1] = pred
df
mapping = {"PNEUMONIA": 1, "NORMAL": 0}
df = df.replace({"labels": mapping})
df
df.to_csv("diagnosis_submission.csv", index=False)
|
# # Introduction to Kmeans Clustering
# Machine learning algorithms can be broadly classified into two categories - supervised and unsupervised learning. There are other categories also like semi-supervised learning and reinforcement learning. But, most of the algorithms are classified as supervised or unsupervised learning. The difference between them happens because of presence of target variable. In unsupervised learning, there is no target variable. The dataset only has input variables which describe the data. This is called unsupervised learning.
# K-Means clustering is the most popular unsupervised learning algorithm. It is used when we have unlabelled data which is data without defined categories or groups. The algorithm follows an easy or simple way to classify a given data set through a certain number of clusters, fixed apriori. K-Means algorithm works iteratively to assign each data point to one of K groups based on the features that are provided. Data points are clustered based on feature similarity.
#
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
data = pd.read_csv("/kaggle/input/mallcustomersdataset/Mall_Customers.csv")
data.head()
data.info()
# # Drop Column
data = data.drop(["CustomerID"], axis=1)
data.info()
data.head()
# # EDA
import matplotlib.pyplot as plt
import seaborn as sns
sns.kdeplot(data=data)
from sklearn import preprocessing
le = preprocessing.LabelEncoder()
data["Genre"] = le.fit_transform(data["Genre"])
data.head()
from sklearn.cluster import KMeans
km = KMeans(n_clusters=4)
km.fit(data)
y_pred = km.fit_predict(data)
y_pred
wcss = []
for i in range(1, 11):
clustering = KMeans(n_clusters=i, init="k-means++", random_state=42)
clustering.fit(data)
wcss.append(clustering.inertia_)
ks = [1, 2, 3, 4, 5, 6, 7, 8, 9, 10]
sns.lineplot(x=ks, y=wcss)
fig, axes = plt.subplots(nrows=1, ncols=2, figsize=(15, 5))
sns.scatterplot(ax=axes[0], data=data, x="Age", y="Annual Income (k$)").set_title(
"Without clustering"
)
sns.scatterplot(
ax=axes[1], data=data, x="Age", y="Annual Income (k$)", hue=clustering.labels_
).set_title("Using the elbow method")
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
data.isnull().sum()
# # Heatmap
# Heat map Heat map is used to find out the correlation between different features in the dataset.High positive or negative value shows that the features have high correlation.This helps us to select the parmeters for machine learning
import seaborn as sns
plt.figure(figsize=(7, 4))
sns.heatmap(data.corr(), annot=True)
plt.show()
|
# Credit Card Fraud Detection Using Machine Learning
#!pip install tpot
#!pip install scikit-learn-intelex
#!pip install mlxtend
#!pip install tpot
#!pip install xgboost
#!pip install hyperopt
#!pip install imblearn
#!pip install xlwt
#!pip install seaborn
#!pip install xgboost==1.7
#!pip install matplotlib
from sklearnex import (
patch_sklearn,
) # This is a library to accelerate existing scikit-learn code
patch_sklearn() # For more info: https://github.com/intel/scikit-learn-intelex
from collections import Counter
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
import seaborn as sns
import xgboost as xgb
from mlxtend.classifier import StackingCVClassifier
from scipy.stats import uniform, randint
from sklearn.metrics import (
classification_report,
confusion_matrix,
roc_curve,
roc_auc_score,
ConfusionMatrixDisplay,
f1_score,
precision_score,
recall_score,
accuracy_score,
average_precision_score,
)
from sklearn.neighbors import KNeighborsClassifier
from sklearn.linear_model import LogisticRegression, SGDClassifier
from sklearn.model_selection import (
RandomizedSearchCV,
cross_val_score,
KFold,
StratifiedKFold,
train_test_split,
)
from sklearn.preprocessing import RobustScaler, power_transform
from sklearn.tree import DecisionTreeClassifier
from sklearn.svm import SVC
from sklearn.ensemble import (
VotingClassifier,
GradientBoostingClassifier,
RandomForestClassifier,
ExtraTreesClassifier,
)
from sklearn.naive_bayes import GaussianNB
from sklearn.neural_network import MLPClassifier
from sklearn.manifold import TSNE
from sklearn.decomposition import PCA, TruncatedSVD
from imblearn.pipeline import make_pipeline
from imblearn.under_sampling import RandomUnderSampler, NearMiss
from imblearn.over_sampling import ADASYN
import hyperopt
from hyperopt.pyll import scope
from hyperopt import fmin, hp, tpe, Trials, space_eval, STATUS_OK
from warnings import filterwarnings
filterwarnings("ignore")
np.random.seed(0)
# General information about the data:
credit_card = pd.read_csv("creditcard.csv")
pd.set_option("display.float_format", "{:.2f}".format)
credit_card.head()
credit_card.describe()
credit_card.info()
# Class needed to be categorical.
credit_card["Class"] = credit_card["Class"].astype("category")
# Let' see if there are any null. (There isn't.)
print("Total None values:", credit_card.isna().any().sum())
# Let's see how many transactions are fraudulent.
credit_card["Class"].value_counts()
# It's are extremly imbalanced
def get_axes_list(length, column_number=2):
fig, axes_list = get_fig_and_axes_list(length, column_number=column_number)
return axes_list
def get_fig_and_axes_list(plot_count, column_number=2):
"""
This function takes in the number of subplots to be plotted and the desired number of columns for the subplot grid.
It then calculates the number of rows required and generates a matplotlib figure with the given number of subplots
in a grid with the desired number of columns.
Args:
- plot_count: int, the number of subplots to be plotted
- column_number: int, the number of columns in the subplot grid. Default value is 2.
Returns:
- fig: matplotlib Figure object, the generated figure
- axes_list: list of matplotlib Axes objects, the axes of the subplots in the figure
"""
reminder_num = plot_count % column_number
row_num = (plot_count // column_number) + (reminder_num > 0)
axes_list = []
row_number_alignment = np.ones((row_num, column_number), dtype="int")
if reminder_num != 0:
row_number_alignment[-1, -(column_number - reminder_num) :] = 0
coefficient = 3.5 if plot_count == 1 else 4.2
col_size = coefficient * column_number
row_size = coefficient * row_num
fig = plt.figure(figsize=(col_size, row_size), layout="constrained")
spec = fig.add_gridspec(row_num, column_number)
for i in range(row_num):
for j in range(column_number):
if row_number_alignment[i, j] == 1:
ax = fig.add_subplot(spec[i, j])
axes_list.append(ax)
return fig, axes_list
ax = get_axes_list(1)[0]
sns.set_palette("PuRd_r")
bar = sns.countplot(x=credit_card["Class"], ax=ax)
ax.set_xticklabels(["No Fraud", "Fraud"])
ax.set_xlabel("Class")
ax.set_ylabel("Count")
bar.set_title("Distribution of credit card frauds")
plt.show()
# Distribution by time:
axes = get_axes_list(2, 2)
trans_plots = []
trans_plots.append(
sns.histplot(credit_card["Time"][credit_card.Class == 1], bins=50, ax=axes[0])
)
trans_plots[0].set_title("Fraudulent Transactions")
trans_plots[0].tick_params(labelrotation=75)
trans_plots.append(
sns.histplot(credit_card["Time"][credit_card.Class == 0], bins=50, ax=axes[1])
)
trans_plots[1].set_title("Non-Fraudulent Transactions")
trans_plots[1].tick_params(labelrotation=75)
plt.show()
# #### Let's apply a necessary transform for Amount. It's highly skewed. We apply power transformation(basically you can think that taking the logarithm for this example).
data_transformed = power_transform(credit_card[["Amount"]])
axes = get_axes_list(2, 2)
rel_plots = []
rel_plots.append(sns.kdeplot(x=credit_card["Amount"], fill=True, alpha=1, ax=axes[0]))
rel_plots[0].set_title("Before Transforming Amount Data")
rel_plots.append(
sns.kdeplot(x=data_transformed.ravel(), fill=True, alpha=1, ax=axes[1])
)
rel_plots[1].set_title("After Transforming Amount Data")
sns.despine()
plt.show()
credit_card["Amount"] = data_transformed # Apply the transformation.
# Split the dataset
X = credit_card.drop("Class", axis=1)
y = credit_card["Class"].values
rand_state = 42
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=rand_state, stratify=y
)
# Scale the data
scaler = RobustScaler()
X_train_scaled = scaler.fit_transform(X_train)
X_test_scaled = scaler.transform(X_test)
# Our results DataFrame
results = pd.DataFrame(
columns=[
"Function Name",
"Accuracy",
"Precision",
"Recall",
"F1",
"TN",
"FP",
"FN",
"TP",
"ROC-AUC Score",
"Precision-Recall Score",
"Train CV Score",
]
)
kf = StratifiedKFold(
n_splits=5, shuffle=True, random_state=rand_state
) # We use stratified becuse of imbalanced dataset.
kf_for_test = StratifiedKFold(
n_splits=6, shuffle=True, random_state=0
) # We use another one for testing different cross validations
# Some functions to tune algorithms :)
def tune_log_reg(X=X_train_scaled, y=y_train, scoring="average_precision", cv=kf):
log_reg_params = {
"solver": ["liblinear"],
# "penalty": ['l1', "l2"],
"C": [0.001, 0.01, 0.1, 1, 10, 100],
"class_weight": [{0: 1, 1: w} for w in list(range(1, 100, 40))],
}
grid_log_reg = RandomizedSearchCV(
LogisticRegression(max_iter=100000, n_jobs=-1),
log_reg_params,
cv=kf,
n_iter=2,
n_jobs=-1,
scoring=scoring,
verbose=1,
)
grid_log_reg.fit(
X, y
) # If you have time, don't hesitate to increase number of iterations
return grid_log_reg.best_estimator_
def tune_dec_tree(X=X_train_scaled, y=y_train, scoring="average_precision", cv=kf):
tree_params = {
"criterion": ["gini", "entropy"],
"max_depth": list(range(2, 5, 1)),
"min_samples_leaf": list(range(5, 7, 1)),
"class_weight": [{0: 1, 1: w} for w in list(range(1, 100, 20))],
}
grid_tree = RandomizedSearchCV(
DecisionTreeClassifier(),
tree_params,
cv=kf,
n_iter=7,
n_jobs=-1,
scoring=scoring,
verbose=1,
)
grid_tree.fit(X, y)
return grid_tree.best_estimator_
def tune_knn(X=X_train_scaled, y=y_train, scoring="average_precision", cv=kf):
knears_params = {
"n_neighbors": list(range(2, 5, 1)),
"metric": ["minkowski", "euclidean", "manhattan"],
"weights": ["uniform", "distance"],
}
grid_knears = RandomizedSearchCV(
KNeighborsClassifier(n_jobs=-1),
knears_params,
cv=kf,
n_iter=20,
scoring=scoring,
verbose=1,
)
grid_knears.fit(X, y)
return grid_knears.best_estimator_
def tune_xgb(X=X_train_scaled, y=y_train, scoring="average_precision", cv=kf):
params = {
"colsample_bytree": uniform(0.7, 0.1),
"gamma": uniform(0, 0.5),
"learning_rate": uniform(0.03, 0.5),
"max_depth": randint(2, 25),
"n_estimators": randint(100, 250),
"subsample": uniform(0.7, 0.3),
"max_delta_step": randint(1, 10),
}
grid_xgb = RandomizedSearchCV(
xgb.XGBClassifier(
objective="binary:logistic",
eval_metric=average_precision_score,
n_jobs=-1,
tree_method="hist",
random_state=rand_state,
),
params,
cv=kf,
n_iter=20,
scoring=scoring,
)
grid_xgb.fit(X, y)
return grid_xgb.best_estimator_
def tune_sgd_bayes(X=X_train_scaled, y=y_train, scoring="average_precision", cv=kf):
def objective_for_minimization(search_space, X=X, y=y, scoring_objective=scoring):
model = SGDClassifier(
**search_space, max_iter=int(1e9), n_jobs=-1, random_state=rand_state
)
best_score = cross_val_score(
model, X, y, scoring=scoring_objective, cv=kf, n_jobs=-1
).mean()
loss = 1 - best_score
return {"loss": loss, "status": STATUS_OK}
search_space = {
"penalty": hp.choice("penalty", ["l1", "l2", "elasticnet"]),
"alpha": hp.uniform("alpha", 0.00001, 3),
"learning_rate": hp.choice(
"learning_rate", ["constant", "optimal", "invscaling", "adaptive"]
),
"eta0": hp.uniform("n_estimators", 0, 3),
"power_t": hp.uniform("power_t", -0.1, 1),
"validation_fraction": hp.uniform("validation_fraction", 0.1, 0.3),
"loss": hp.choice("loss", ["modified_huber"]),
"class_weight": {0: 1, 1: scope.int(hp.uniform("class_weight", 1, 100))},
}
# implement Hyperopt
algorithm = tpe.suggest
best_params = fmin(
fn=objective_for_minimization, space=search_space, algo=algorithm, max_evals=40
)
fun_params_dict = space_eval(search_space, best_params)
return SGDClassifier(**fun_params_dict, n_jobs=-1)
# If we want to cross-validate our data, wee need to do it during cross-validation. The reason is not to contaminate our training data with sampled data! This function resolves the problem.
def cross_val_score_for_sampling(
model,
X_train,
y_train,
cv=kf_for_test,
results=results,
undersample=False,
oversample=False,
stack=False,
other=False,
):
scoring_metrics = [
"accuracy",
"precision",
"recall",
"f1",
"roc_auc",
"average_precision",
]
scores_dict = {metric: [] for metric in scoring_metrics}
sampling_methods = {
"undersample": NearMiss(sampling_strategy="majority", n_jobs=-1, version=3),
"oversample": ADASYN(random_state=rand_state),
}
sample_pipeline = make_pipeline(model)
if undersample:
sample_pipeline.steps.insert(
0, ("undersample", sampling_methods["undersample"])
)
if oversample:
sample_pipeline.steps.insert(0, ("oversample", sampling_methods["oversample"]))
for train_idx, test_idx in cv.split(X_train, y_train):
sample_model = sample_pipeline.fit(X_train[train_idx], y_train[train_idx])
sample_prediction_proba = sample_model.predict_proba(X_train[test_idx])[:, 1]
sample_prediction = np.round(sample_prediction_proba)
scores_dict["accuracy"].append(
accuracy_score(y_train[test_idx], sample_prediction)
)
scores_dict["precision"].append(
precision_score(y_train[test_idx], sample_prediction)
)
scores_dict["recall"].append(recall_score(y_train[test_idx], sample_prediction))
scores_dict["f1"].append(f1_score(y_train[test_idx], sample_prediction))
scores_dict["roc_auc"].append(
roc_auc_score(y_train[test_idx], sample_prediction_proba)
)
scores_dict["average_precision"].append(
average_precision_score(y_train[test_idx], sample_prediction_proba)
)
return scores_dict
def get_scoring_name(scoring):
equal = {
"accuracy": "Accuracy",
"precisione": "Precision",
"recall": "Recall",
"f1": "F1",
"roc_auc": "ROC AUC",
"average_precision": "AP AUC",
}
return equal[scoring]
def get_training_cross_validated_score(
model,
X_train,
y_train,
undersample=False,
oversample=False,
stack=False,
other=False,
scoring="Accuracy",
):
try:
if undersample or oversample or stack:
training_cross_validated_score = np.mean(
cross_val_score_for_sampling(
model,
X_train,
y_train,
undersample=undersample,
oversample=oversample,
stack=stack,
other=other,
)[scoring]
)
elif other:
return None
else:
training_cross_validated_score = cross_val_score(
model, X_train, y=y_train, cv=kf, scoring=scoring, n_jobs=-1
).mean()
return training_cross_validated_score
except Exception as e:
return None
def print_scores(
models,
X_train=X_train_scaled,
X_test=X_test_scaled,
y_train=y_train,
y_test=y_test,
result_prefix="",
results=results,
undersample=False,
oversample=False,
stack=False,
other=False,
scoring="average_precision",
):
"""
Plot confusion matrices and evaluation metrics for a dictionary of models.
Parameters:
-----------
models : dict
A dictionary of scikit-learn models to evaluate.
X_train : array-like, shape (n_samples, n_features), default=X_train_scaled
The training input samples.
X_test : array-like, shape (n_samples, n_features), default=X_test_scaled
The testing input samples.
y_train : array-like, shape (n_samples,), default=y_train
The target values of the training input samples.
y_test : array-like, shape (n_samples,), default=y_test
The target values of the testing input samples.
result_prefix : str, default=""
A prefix to add to the function name in the results DataFrame.
results : pandas.DataFrame, default=results
A DataFrame to store the results.
undersample : bool, default=False
Whether to perform undersampling during cross-validation on training set.
oversample : bool, default=False
Whether to perform oversampling during cross-validation on training set.
stack : bool, default=False
Whether Stacking Classifier will be used during cross-validation on training set.
other : bool, default=False
Whether cross-validation will be used on training set.
scoring : str or callable, default="average_precision"
The scoring metric to use for cross validation.
Returns:
--------
None
"""
ax_list = get_axes_list(len(models))
ax_counter = 0
for i, (name, model) in enumerate(models.items()):
model_name = model.__class__.__name__
ax_of_model = ax_list[i]
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
y_pred = np.round(y_pred)
accuracy = accuracy_score(y_test, y_pred)
precision = precision_score(y_test, y_pred)
recall = recall_score(y_test, y_pred)
f1 = f1_score(y_test, y_pred)
tn, fp, fn, tp = confusion_matrix(y_test, y_pred).ravel()
if hasattr(model, "predict_proba"):
predicted_prob = model.predict_proba(X_test)[:, 1]
roc_auc = roc_auc_score(y_test, predicted_prob)
average_precision = average_precision_score(y_test, predicted_prob)
else:
predicted_prob = None
roc_auc = None
average_precision = None
training_cross_validated_score = get_training_cross_validated_score(
model,
X_train,
y_train,
undersample=undersample,
oversample=oversample,
stack=stack,
other=other,
scoring=scoring,
)
scoring_name = get_scoring_name(scoring)
scoring_variable = locals()[scoring]
if training_cross_validated_score and scoring_variable:
ax_of_model.set_title(
f"{model_name} \n ({scoring_name}: {scoring_variable:3.2f} || Train {scoring_name}: {scoring_variable:3.2f})",
fontdict={"fontsize": 10},
)
elif scoring_variable:
ax_of_model.set_title(
f"{model_name} \n ({scoring_name} score: {scoring_variable:3.2f})"
)
else:
ax_of_model.set_title(f"{model_name}")
function_name = f"{model_name}{result_prefix}"
function_location = results[results["Function Name"] == function_name].index
index_to_insert = (
len(results.index) if function_location.empty else function_location[0]
)
results.loc[index_to_insert] = [
function_name,
accuracy,
precision,
recall,
f1,
tn,
fp,
fn,
tp,
roc_auc,
average_precision,
training_cross_validated_score,
]
labels_ = ["No Fraud", "Fraud"]
ConfusionMatrixDisplay.from_predictions(
y_test,
y_pred,
display_labels=labels_,
ax=ax_of_model,
cmap="Blues",
xticks_rotation="vertical",
)
# print(classification_report(y_test, y_pred)) #If you want to get classification report.
plt.show()
# # Modeling
# First try with default parameters.
models = {
"Random Forest": RandomForestClassifier(n_jobs=-1, random_state=rand_state),
"Extra": ExtraTreesClassifier(n_jobs=-1),
"XGB Classifier": xgb.XGBClassifier(
eval_metric=average_precision_score,
n_jobs=-1,
tree_method="hist",
random_state=rand_state,
),
"XGB Random Forest Classifier": xgb.XGBRFClassifier(
objective="binary:logistic",
eval_metric=average_precision_score,
tree_method="hist",
random_state=rand_state,
),
"Logistic Regression": LogisticRegression(n_jobs=-1),
"KNN": KNeighborsClassifier(n_jobs=-1),
"SGD": SGDClassifier(n_jobs=-1, loss="log"),
}
print_scores(models)
# #### And try Voting Classifier.
# A voting classifier is a machine learning technique that combines the predictions of multiple models to make a final prediction. This ensemble approach can improve the accuracy and stability of predictions, especially when individual models are prone to error or have biases.
clf_meta = xgb.XGBClassifier(
eval_metric=average_precision_score,
n_jobs=-1,
tree_method="hist",
random_state=rand_state,
)
classifiers_stack = [
ExtraTreesClassifier(n_jobs=-1),
RandomForestClassifier(n_jobs=-1),
xgb.XGBClassifier(
eval_metric=average_precision_score,
n_jobs=-1,
tree_method="hist",
random_state=rand_state,
),
xgb.XGBRFClassifier(
objective="binary:logistic",
eval_metric=average_precision_score,
n_jobs=-1,
tree_method="hist",
random_state=rand_state,
),
LogisticRegression(
solver="liblinear", n_jobs=-1, max_iter=int(1e9), class_weight="balanced"
),
KNeighborsClassifier(n_jobs=-1),
SGDClassifier(
loss="modified_huber", n_jobs=-1, max_iter=int(1e9), class_weight="balanced"
),
]
clf_stack = StackingCVClassifier(
classifiers=classifiers_stack,
meta_classifier=clf_meta,
cv=kf,
verbose=False,
use_probas=True,
use_features_in_secondary=True,
)
vc = {"Stack": clf_stack}
print_scores(vc, stack=True, result_prefix="_voting_stack")
# #### Tuning
# Now let's try tuning according to AP score.
tree_clf_ap = tune_dec_tree()
# log_reg_ap = tune_log_reg()
knears_neighbors_ap = tune_knn()
xgb_est_ap = tune_xgb()
sgd_est_ap = tune_sgd_bayes()
models_with_params = {
"Decision Tree": tree_clf_ap,
"KNN": knears_neighbors_ap,
# "Logistic Regression": log_reg_ap,
"XGB": xgb_est_ap,
"SGD": sgd_est_ap,
}
print_scores(models_with_params, result_prefix="_tuned_ap")
classifiers_tuned_ap = [
tree_clf_ap,
knears_neighbors_ap,
xgb_est_ap,
sgd_est_ap,
] # You can include log_reg_ap
clf_stack_tuned_ap = StackingCVClassifier(
classifiers=classifiers_tuned_ap,
meta_classifier=clf_meta,
cv=kf,
verbose=False,
use_probas=True,
use_features_in_secondary=True,
)
vc = {"Stack": clf_stack_tuned_ap}
print_scores(vc, stack=True, result_prefix="_voting_stack_tuned_ap")
# # Oversampling
# If we apply ADASYN(It's a oversampling algorithm technique for imbalanced datasets.):
method = ADASYN(random_state=rand_state)
X_train_resampled, y_train_resampled = method.fit_resample(X_train_scaled, y_train)
models = {
"Random Forest": RandomForestClassifier(n_jobs=-1, random_state=rand_state),
"Extra Trees": ExtraTreesClassifier(n_jobs=-1),
"XGB Classifier": xgb.XGBClassifier(
eval_metric=average_precision_score,
n_jobs=-1,
tree_method="hist",
random_state=rand_state,
),
"XGB Random Forest Classifier": xgb.XGBRFClassifier(
objective="binary:logistic",
eval_metric=average_precision_score,
tree_method="hist",
random_state=rand_state,
),
"Logistic Regression": LogisticRegression(n_jobs=-1),
"KNN": KNeighborsClassifier(n_jobs=-1),
"SGD": SGDClassifier(n_jobs=-1, loss="log"),
}
print_scores(
models,
X_train=X_train_resampled,
y_train=y_train_resampled,
oversample=True,
result_prefix="_adasyn",
)
classifiers = [
("Extra Trees", ExtraTreesClassifier(n_jobs=-1)),
("Random Forest", RandomForestClassifier(n_jobs=-1)),
(
"XGB Classifier",
xgb.XGBClassifier(
eval_metric=average_precision_score, n_jobs=-1, tree_method="hist"
),
),
(
"XGB Random Forest Classifier",
xgb.XGBRFClassifier(
objective="binary:logistic",
n_jobs=-1,
tree_method="hist",
eval_metric=average_precision_score,
),
),
("Logistic Regression", LogisticRegression(max_iter=100000, n_jobs=-1)),
("KNN", KNeighborsClassifier(n_jobs=-1)),
("Decision Tree", DecisionTreeClassifier()),
("SGD", SGDClassifier(loss="log", n_jobs=-1)),
]
vc = {"Vote": VotingClassifier(estimators=classifiers, voting="soft", n_jobs=-1)}
print_scores(
vc,
X_train=X_train_resampled,
y_train=y_train_resampled,
oversample=True,
result_prefix="_voting_adasyn",
)
vc = {"Stack": clf_stack}
print_scores(
vc,
X_train=X_train_resampled,
y_train=y_train_resampled,
oversample=True,
result_prefix="_voting_stack_adasyn",
)
# #### Tuning
# Let's see if we can get any better by tuning.
# log_reg_adasyn_ap = tune_log_reg(X=X_train_resampled, y=y_train_resampled)
tree_clf_adasyn_ap = tune_dec_tree(X=X_train_resampled, y=y_train_resampled)
knears_neighbors_adasyn_ap = tune_knn(X=X_train_resampled, y=y_train_resampled)
xgb_est_adasyn_ap = tune_xgb(X=X_train_resampled, y=y_train_resampled)
models_with_params = {
"Decision Tree": tree_clf_adasyn_ap,
"KNN": knears_neighbors_adasyn_ap,
# "Logistic Regression": log_reg_adasyn_ap, #takes too long
"XGB": xgb_est_adasyn_ap,
"SGD": sgd_est_ap,
}
print_scores(
models_with_params,
X_train=X_train_resampled,
y_train=y_train_resampled,
oversample=True,
result_prefix="_adasyn_tuned_ap",
)
classifiers = [
("XGB Classifier", xgb_est_adasyn_ap),
# ("Logistic Regression", log_reg_adasyn_ap),
("KNN", knears_neighbors_adasyn_ap),
("Decision Tree", tree_clf_adasyn_ap),
]
vc = {"Vote": VotingClassifier(estimators=classifiers, voting="soft", n_jobs=-1)}
print_scores(
vc,
X_train=X_train_resampled,
y_train=y_train_resampled,
oversample=True,
result_prefix="_voting_adasyn_tuned_ap",
)
# # Undersampling
# First we'll do random undersampling for dimensionality reduction and correlation matrix then use Near Miss algorithm for actual undersampling modeling.
X_scaled_for_rus = credit_card.drop(columns="Class")
y_scaled_for_rus = credit_card.Class
X_scaled_for_rus = scaler.fit_transform(X_scaled_for_rus)
rus = RandomUnderSampler(random_state=rand_state)
X_rus, y_rus = rus.fit_resample(X_scaled_for_rus, y_scaled_for_rus)
Counter(y_rus)
# #### If we apply Dimensionality Reduction techniques:
# T-SNE Implementation
X_reduced_tsne = TSNE(n_components=2, random_state=rand_state, n_jobs=-1).fit_transform(
X_rus
)
# PCA Implementation
X_reduced_pca = PCA(n_components=2, random_state=rand_state).fit_transform(X_rus)
# TruncatedSVD
X_reduced_svd = TruncatedSVD(n_components=2, random_state=rand_state).fit_transform(
X_rus
)
f, (ax1, ax2, ax3) = plt.subplots(1, 3, figsize=(24, 6.5))
f.suptitle("Clusters by Dimensionality Reduction", fontsize=14)
# t-SNE scatter plot
ax1.scatter(
X_reduced_tsne[:, 0],
X_reduced_tsne[:, 1],
c=(y_rus == 0),
label="No Fraud",
linewidths=2,
)
ax1.scatter(
X_reduced_tsne[:, 0],
X_reduced_tsne[:, 1],
c=(y_rus == 1),
label="Fraud",
linewidths=2,
)
ax1.set_title("t-SNE", fontsize=14)
ax1.grid(True)
ax1.legend()
# PCA scatter plot
ax2.scatter(
X_reduced_pca[:, 0],
X_reduced_pca[:, 1],
c=(y_rus == 0),
label="No Fraud",
linewidths=2,
)
ax2.scatter(
X_reduced_pca[:, 0],
X_reduced_pca[:, 1],
c=(y_rus == 1),
label="Fraud",
linewidths=2,
)
ax2.set_title("PCA", fontsize=14)
ax2.grid(True)
ax2.legend()
# TruncatedSVD scatter plot
ax3.scatter(
X_reduced_svd[:, 0],
X_reduced_svd[:, 1],
c=(y_rus == 0),
label="No Fraud",
linewidths=2,
)
ax3.scatter(
X_reduced_svd[:, 0],
X_reduced_svd[:, 1],
c=(y_rus == 1),
label="Fraud",
linewidths=2,
)
ax3.set_title("Truncated SVD", fontsize=14)
ax3.grid(True)
ax3.legend()
plt.show()
# #### Let's apply NearMiss(It's a undersampling algorithm technique for imbalanced datasets.):
nm = NearMiss(version=3)
X_train_nm, y_train_nm = nm.fit_resample(X_train_scaled, y_train)
Counter(y_train_nm)
new_df = pd.DataFrame(X_train_nm, columns=credit_card.drop("Class", axis=1).columns)
new_df["Class"] = y_train_nm
# See correlation matrix. (We did this right after undersampling to see correlations clearly.)
plt.figure(figsize=(20, 20))
corr = new_df.corr()
mask = np.triu(np.ones_like(corr, dtype=bool)) # Generate a mask for the upper triangle
ax = sns.heatmap(
corr, cmap="PuOr", mask=mask, center=0, linewidths=1, annot=True, fmt=".2f"
)
plt.show()
# Now try algorithms and Voting Classifier:
models = {
"Random Forest": RandomForestClassifier(n_jobs=-1, random_state=rand_state),
"Extra": ExtraTreesClassifier(n_jobs=-1),
"XGB Classifier": xgb.XGBClassifier(
eval_metric=average_precision_score,
n_jobs=-1,
tree_method="hist",
random_state=rand_state,
),
"XGB Random Forest Classifier": xgb.XGBRFClassifier(
objective="binary:logistic",
n_jobs=-1,
tree_method="hist",
eval_metric=average_precision_score,
random_state=rand_state,
),
"Logistic Regression": LogisticRegression(n_jobs=-1),
"KNN": KNeighborsClassifier(n_jobs=-1),
"Decision Tree": DecisionTreeClassifier(),
"SGD": SGDClassifier(loss="log", n_jobs=-1),
"Support Vector Classifier": SVC(probability=True),
}
print_scores(
models,
X_train=X_train_nm,
y_train=y_train_nm,
undersample=True,
result_prefix="_us",
)
vc = {"Stack": clf_stack}
print_scores(
vc,
X_train=X_train_nm,
y_train=y_train_nm,
undersample=True,
result_prefix="_voting_stack_us",
)
# # Bonus: TPOT
# Last but not least. Let's try TPOT. It's is a Python Automated Machine Learning tool that optimizes machine learning pipelines using genetic programming. Let's try it.
try:
from tpot import TPOTClassifier
# If you have time, then don't hesitate increasing generations ,offspring_size and population_size
TPOT_model_us = TPOTClassifier(
generations=30,
offspring_size=20,
scoring="average_precision",
cv=kf,
population_size=20,
verbosity=0,
n_jobs=-1,
random_state=rand_state,
)
models = {"TPOT": TPOT_model_us}
print_scores(
models,
X_train=X_train_nm,
y_train=y_train_nm,
other=True,
result_prefix="_us_TPOT",
)
except Exception as e:
print(e, "Packages have conficlict.")
TPOT_model_us.fitted_pipeline_
# Drop accuracy because we don't need because of imbalanced dataset
results.drop("Accuracy", axis=1).sort_values(
by="Precision-Recall Score", ascending=False
).style.format(precision=3).highlight_max(
subset=["Precision-Recall Score", "ROC-AUC Score", "Train CV Score"],
color="lightgreen",
)
# Best results highlited. I will be waiting for your feedback :)
results.to_excel("results_7.1.xls", index=False)
|
# #### Import required libraries
import numpy as np
import pandas as pd
from sklearn.model_selection import train_test_split
from sklearn.utils.class_weight import compute_class_weight
from tensorflow.keras import Model
from tensorflow.keras.layers import (
LayerNormalization,
MultiHeadAttention,
Dropout,
LayerNormalization,
Conv1D,
Dense,
GlobalAveragePooling1D,
Input,
Embedding,
)
from tensorflow.keras.optimizers import Adam
from tensorflow.keras.callbacks import EarlyStopping
# #### Define variables
window_size = 256
# #### Read data
sequence_data = pd.read_csv(
"/kaggle/input/introduction-to-sequence-data-analytics-vidhya/sequence_data.csv"
)
sequence_labels = pd.read_csv(
"/kaggle/input/introduction-to-sequence-data-analytics-vidhya/sequence_labels.csv"
)
values = pd.read_csv(
"/kaggle/input/introduction-to-sequence-data-analytics-vidhya/possible_values.csv"
)
sequence_data
sequence_labels
values
# #### Preprocessing
values = (
values
# Ensure that index is numbered 0, 1, etc
.reset_index(drop=True)
# Use index as column (label encoding)
.reset_index()
# Rename column 'index' to 'label'
.rename(columns={"index": "value_label"})
)
# Labels should start with 1.
# 0 is reserved for missing values which will be masked.
values["value_label"] += 1
values
sequence_data = pd.merge(left=sequence_data, right=values, on="value", how="left")
sequence_data
sequence_data = sequence_data.drop(columns=["value"])
sequence_data
for i in range(1, window_size):
sequence_data[f"step_{i}"] = (
sequence_data["value_label"].shift(i).fillna(0).astype(int)
)
sequence_data
sequence_data = sequence_data.rename(columns={"value_label": "step_0"})
sequence_data
sequence_data = sequence_data.groupby(by=["ID"]).tail(1).reset_index(drop=True)
sequence_data
sequence_data = pd.merge(
left=sequence_data, right=sequence_labels, on=["ID"], how="left"
)
sequence_data
sequence_data = sequence_data.drop(columns=["timestamp"])
sequence_data
input_cols = [f"step_{i}" for i in range(window_size)]
input_cols.reverse()
input_cols[:10]
sequence_data = sequence_data[["ID"] + input_cols + ["outcome"]]
sequence_data
X_train, X_valid, y_train, y_valid = train_test_split(
sequence_data.drop(columns=["outcome"]),
sequence_data["outcome"],
test_size=300,
random_state=0,
)
X_train
# #### Build and train model
def transformer_encoder(inputs, head_size, num_heads, ff_dim, dropout=0):
# Normalization and Attention
x = LayerNormalization(epsilon=1e-6)(inputs)
x = MultiHeadAttention(key_dim=head_size, num_heads=num_heads, dropout=dropout)(
x, x
)
x = Dropout(dropout)(x)
res = x + inputs
# Feed Forward Part
x = LayerNormalization(epsilon=1e-6)(res)
x = Conv1D(filters=ff_dim, kernel_size=1, activation="relu")(x)
x = Dropout(dropout)(x)
x = Conv1D(filters=inputs.shape[-1], kernel_size=1)(x)
return x + res
def build_model(
input_shape,
head_size,
num_heads,
ff_dim,
num_transformer_blocks,
mlp_units,
dropout=0,
mlp_dropout=0,
):
inputs = Input(shape=input_shape)
embeddings = Embedding(
input_dim=values.shape[0] + 1, # Number of codes + 1
output_dim=64, # Desired embedding size
trainable=True,
mask_zero=True, # Mask 0s (padding)
)(inputs)
x = embeddings
for _ in range(num_transformer_blocks):
x = transformer_encoder(x, head_size, num_heads, ff_dim, dropout)
x = GlobalAveragePooling1D(data_format="channels_first")(x)
for dim in mlp_units:
x = Dense(dim, activation="relu")(x)
x = Dropout(mlp_dropout)(x)
outputs = Dense(1, activation="sigmoid")(x)
return Model(inputs, outputs)
model = build_model(
input_shape=(window_size,),
head_size=256,
num_heads=4,
ff_dim=4,
num_transformer_blocks=4,
mlp_units=[128],
mlp_dropout=0.4,
dropout=0.25,
)
model.compile(
loss="binary_crossentropy",
optimizer=Adam(learning_rate=1e-4),
metrics=["binary_accuracy"],
)
model.summary()
class_weights = compute_class_weight(class_weight="balanced", classes=[0, 1], y=y_train)
class_weights = dict(zip([0, 1], class_weights))
callbacks = [EarlyStopping(patience=10, restore_best_weights=True)]
history = model.fit(
X_train[input_cols].values,
y_train.values,
epochs=5,
batch_size=64,
class_weight=class_weights,
callbacks=callbacks,
)
model.evaluate(X_valid[input_cols].values, y_valid.values, verbose=1)
# #### Evaluate model
############# USE PRECISION AS METRIC
############# SHOW CONFUSION MATRIX AND OTHER STUFF
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import numpy as np
import pandas as pd
import matplotlib
import matplotlib.pyplot as plt
import seaborn as sns
sns.set(rc={"figure.figsize": (10, 8)})
vodafoneTrain = pd.read_csv("../input/vodafone-age/train_age.csv", sep=",")
vodafoneTrain.head(10)
vodafoneTrain.groupby("target")[
"fb_count",
"intagram_count",
"viber_count",
"youtube_count",
"skype_count",
"steam_count",
"twitch_count",
"badoo_count",
"netflix_count",
"telegram_count",
].mean().plot(kind="bar", figsize=(30, 10))
|
# **Web scraping latest pokedex**
# The project is done to practice myself with web scraping and numpy/pandas with a familliar data.
# Following is a notebook to webscrape pokemon data from https://pokemondb.net/ . The step-by-step process is recorded
# Import packages
import requests
from bs4 import BeautifulSoup
import numpy as np
import pandas as pd
# Saving the webpage as url and 'scrape' from it
url = "https://pokemondb.net/pokedex/all"
response = requests.get(url)
soup = BeautifulSoup(response.content, "html.parser")
# Testing
# obtain webpage header
header = soup.find("h1")
print(header)
# obtain webpage intro
intro = soup.find("div", attrs={"class": "panel panel-intro"})
print(intro)
# Select pokedex (data source)
pokedex = soup.find("table", id="pokedex").find_all("tr")
# Selecting the first pokemon, Bulbasaur
pokedex[1]
# Test to get the respective values
# pokemon no.
pokedex[1].find_all("td")[0].get_text()
# pokemon name
pokedex[1].find_all("td")[1].get_text()
# pokemon types
pokedex[1].find_all("td")[2].get_text()
# pokemon hp
pokedex[1].find_all("td")[3].get_text()
# pokemon attack
pokedex[1].find_all("td")[4].get_text()
# Create function to collect data into list
dex = []
def pokemon_stats(pokedex, number):
pokemon = pokedex[number].find_all("td")
pokemon_id = pokemon[0].get_text()
name = pokemon[1].get_text()
types = pokemon[2].get_text()
hp = pokemon[3].get_text()
attack = pokemon[4].get_text()
defence = pokemon[5].get_text()
sp_attack = pokemon[6].get_text()
sp_defence = pokemon[7].get_text()
speed = pokemon[8].get_text()
stats = [pokemon_id, name, types, hp, attack, defence, sp_attack, sp_defence, speed]
dex.append(stats)
return dex
# Collect all pokemon data
def create_pokedex(data_source):
# get the total number of pokemons
total_pokemons = len(pokedex)
# loop the number of total number of pokemons
for number in range(1, total_pokemons):
pokemon_stats(pokedex, number)
create_pokedex(pokedex)
# Checking
dex[1193]
# Change into dataframe
completedex = pd.DataFrame(
columns=[
"pokemon_id",
"names",
"types",
"hp",
"attack",
"defence",
"sp_attack",
"sp_defence",
"speed",
],
data=dex,
)
completedex
# Checking data types
checkdatatype = completedex["speed"]
datatypelist = []
for i in checkdatatype:
datatypelist.append(type(i))
set(datatypelist)
# Change data types
completedex[
["pokemon_id", "hp", "attack", "defence", "sp_attack", "sp_defence", "speed"]
] = completedex[
["pokemon_id", "hp", "attack", "defence", "sp_attack", "sp_defence", "speed"]
].astype(
int
)
completedex.loc[1]
# Saving the dataframe into csv
completedex.to_csv("pokedex.csv", index=False)
# **Pokemon abilities**
# Below is to scrape data from bulbapedia on pokemon abilities
ability_url = "https://bulbapedia.bulbagarden.net/wiki/List_of_Pok%C3%A9mon_by_Ability"
ability_response = requests.get(ability_url)
ability_soup = BeautifulSoup(ability_response.content, "html.parser")
# Testing
# obtain webpage header
header = ability_soup.find("h1")
print(header)
ability_table = ability_soup.select("tbody")
ability_table[1]
# The above code gives Kanto (First generation pokemons), as there is another 'tbody' before each table
# Hence, here, we will have to '+1' to obtain table for each generation
kanto = ability_table[1]
johto = ability_table[3]
hoenn = ability_table[5]
sinnoh = ability_table[7]
unova = ability_table[9]
kalos = ability_table[11]
alola = ability_table[13]
galar = ability_table[15]
paldea = ability_table[17]
# Testing
kanto.select("tr")[1]
kanto.select("tr")[1].select("td")[5]
# From the above code, we have obtained tables according to generation.
# The order starting with 0, will be the header, then the first pokemon.
# To get all the pokemons, it starts with 1.
# By selecting the 0~5, we are able to obtain each column data of the pokemon. Next we gather all data as shown below
pokemon_id = kanto.select("tr")[1].select("td")[0].get_text()
image = kanto.select("tr")[1].select("td")[1].select("a")[0].select("img")[0].get("src")
name = kanto.select("tr")[1].select("td")[2].get_text()
ability1 = kanto.select("tr")[1].select("td")[3].get_text()
ability2 = kanto.select("tr")[1].select("td")[4].get_text()
hidden_ability = kanto.select("tr")[1].select("td")[5].get_text()
example = [pokemon_id, image, name, ability1, ability2, hidden_ability]
example
# Now, to create a function that compiles all into a data frame
ability_database = []
def ability(generation, number, gen_name):
ability_data = generation.select("tr")[number].select("td")
pokemon_id = ability_data[0].get_text()
image = ability_data[1].select("a")[0].select("img")[0].get("src")
name = ability_data[2].get_text()
ability1 = ability_data[3].get_text()
ability2 = ability_data[4].get_text()
hidden_ability = ability_data[5].get_text()
abilities = [pokemon_id, image, name, ability1, ability2, hidden_ability, gen_name]
ability_database.append(abilities)
return ability_database
# Testing
ability(kanto, 1, "kanto")
ability(kanto, 2, "Kanto")
ability_database
ability_database = []
# The function works, now to make a function that loops through all generations, and also to add a column of their respective generation
# get the total number of pokemons, subtract 1 to remove the header row
len(kanto.select("tr")) - 1
ability(kanto, len(kanto.select("tr")) - 1, "kanto")
ability_database
ability_database = []
# loop the number of total number of pokemons
def gen_loop(generation, gen_name):
total_pokemons = len(generation.select("tr")) - 1
for number in range(1, total_pokemons):
ability(generation, number, gen_name)
# Testing
gen_loop(kanto, "Kanto")
ability_database
ability_database = []
gen_loop(kanto, "Kanto")
gen_loop(johto, "Johto")
gen_loop(hoenn, "Hoenn")
gen_loop(sinnoh, "Sinnoh")
gen_loop(unova, "Unova")
gen_loop(kalos, "Kalos")
gen_loop(alola, "Alola")
gen_loop(galar, "Galar")
gen_loop(paldea, "Paldea")
complete_abilitydex = pd.DataFrame(
columns=[
"pokemon_id",
"image",
"names",
"ability1",
"ability2",
"hidden_ability",
"gen",
],
data=ability_database,
)
complete_abilitydex
# Combine all gen, and add generation column
complete_abilitydex.replace("\n", "", regex=True)
complete_abilitydex["pokemon_id"].replace("#", "", regex=True, inplace=True)
complete_abilitydex.to_csv("ability.csv", index=False)
# The following is code to scrape data from pokemon.com/us about the height and weight information for standard form pokemons
pokemon_list_url = "https://www.pokemon.com/us/pokedex"
pokemonlistsoup = BeautifulSoup(requests.get(pokemon_list_url).content, "html.parser")
pokemonlistsoup.find_all("h1")
# Getting the html part with all pokemon names
pokemon_list_raw = pokemonlistsoup.find_all("noscript")[1].select("ul")[0]
pokemon_list_raw.select("li")[0].find("a").get("href")
pokemon_list = []
for pokemon in range(0, (len(pokemon_list_raw.select("li")) - 1)):
pokemon_list.append(pokemon_list_raw.select("li")[pokemon].find("a").get("href"))
pokemon_list
# Getting the webpage for each pokemon
pokemon_size_url = []
for pokemons in pokemon_list:
pokemon_size_url.append("https://www.pokemon.com" + pokemons)
pokemon_size_url
testurl = "https://www.pokemon.com/us/pokedex/Bulbasaur"
testsoup = BeautifulSoup(requests.get(testurl).content, "html.parser")
testdata = testsoup.find_all(
"div", attrs={"class": "pokemon-ability-info color-bg color-lightblue match active"}
)[0].select("li")
testdata[0].select("span")[1].get_text()
testdata[1].select("span")[1].get_text()
testdata[3].select("span")[1].get_text()
testsoup.find_all("title")[0].get_text().split(" | ")[0]
morphtable = []
def pokemon_morphology(url):
testurl = url
testsoup = BeautifulSoup(requests.get(testurl).content, "html.parser")
testdata = testsoup.find_all(
"div",
attrs={"class": "pokemon-ability-info color-bg color-lightblue match active"},
)[0].select("li")
names = testsoup.find_all("title")[0].get_text().split(" | ")[0]
height = testdata[0].select("span")[1].get_text()
weight = testdata[1].select("span")[1].get_text()
category = testdata[3].select("span")[1].get_text()
morph = [names, height, weight, category]
morphtable.append(morph)
# Test
pokemon_morphology("https://www.pokemon.com/us/pokedex/Bulbasaur")
pokemon_morphology("https://www.pokemon.com/us/pokedex/Ivysaur")
morphtable
# Loop through the list of pokemon websites created previously, in pokemon_size_url
morphtable = []
for pokemonurl in pokemon_size_url:
pokemon_morphology(pokemonurl)
morphtable[1000]
morphology = pd.DataFrame(
columns=["names", "height", "weight", "category"], data=morphtable
)
# adding one to the index number, so bulbasaur starts with 1
morphology.index = np.arange(1, len(morphology) + 1)
morphology
morphology.to_csv("morphology.csv")
# Next, we can also try to obtain the movest data from bulbapedia, which has a complete list of moves and easy to scrape
moves_url = "https://bulbapedia.bulbagarden.net/wiki/List_of_moves"
movesoup = BeautifulSoup(requests.get(moves_url).content, "html.parser")
movepool = movesoup.select("table", {"class": "sortable roundy jquery-tablesorter"})[
0
].select("tbody tr tr")
movepool[0]
len(movepool)
movepool[1].find_all("td")
# try to get the link as well
movepool[1].find_all("td")[1].find("a").get("href")
moveset = []
def movesets(number):
all_moves = movepool[number].find_all("td")
link = all_moves[1].find("a").get("href")
move = all_moves[1].get_text().replace("\n", "")
types = all_moves[2].get_text().replace("\n", "")
category = all_moves[3].get_text().replace("\n", "")
pp = all_moves[4].get_text().replace("\n", "")
power = all_moves[5].get_text().replace("\n", "")
accuracy = all_moves[6].get_text().replace("\n", "")
the_move = [link, move, types, category, pp, power, accuracy]
moveset.append(the_move)
movesets(3)
moveset
moveset = []
for total_moves in range(1, len(movepool)):
movesets(total_moves)
# test
moveset[5]
all_movesets = pd.DataFrame(
columns=["link", "move", "types", "category", "pp", "power", "accuracy"],
data=moveset,
)
all_movesets.to_csv("moveset.csv")
# After getting the full list of movesets, time to get the pokemons that learn these moves. The final codes are not executed as there are too many webpages to scrape
moveset_url = []
for link in all_movesets["link"]:
moveset_url.append("https://bulbapedia.bulbagarden.net/" + link)
moveset_url[0]
testurl = "https://bulbapedia.bulbagarden.net/wiki/Thunder_Wave_(move)"
testsoup = BeautifulSoup(requests.get(testurl).content, "html.parser")
testsoup.find_all("table", {"class": "roundy"})[0].find_all(
"tr", {"style": "background:#fff"}
)[0]
all_moveset = []
def allmoves(url, moveset_no, total_pokemons):
testsoup = BeautifulSoup(requests.get(url).content, "html.parser")
move_info = (
testsoup.find_all("table", {"class": "roundy"})[moveset_no]
.find_all("tr", {"style": "background:#fff"})[total_pokemons]
.find_all
)
move_name = testsoup.select("title")[0].get_text().split(" (move)")[0]
pokemon_name = move_info("td")[2].get_text().replace("\n", "")
egg_group1 = move_info("td")[4].get_text().replace("\n", "")
egg_group2 = move_info("td")[5].get_text().replace("\n", "")
if move_info("th")[-1].get_text() == "\n":
lvl = move_info("th")[-2].get_text()
else:
lvl = move_info("th")[-1].get_text()
moves = [move_name, pokemon_name, egg_group1, egg_group2, lvl]
all_moveset.append(moves)
allmoves("https://bulbapedia.bulbagarden.net/wiki/Thunder_Wave_(move)", 1, 291)
all_moveset
all_moveset = []
def loopmoves(url, moveset_no):
testsoup = BeautifulSoup(requests.get(url).content, "html.parser")
for i in range(
0,
len(
testsoup.find_all("table", {"class": "roundy"})[moveset_no].find_all(
"tr", {"style": "background:#fff"}
)
),
):
allmoves(url, moveset_no, i)
loopmoves("https://bulbapedia.bulbagarden.net/wiki/Thunder_Wave_(move)", 2)
all_moveset[0]
for moveset_no in range(0, 3):
try:
loopmoves(moveset_no)
except:
pass
def one_move(url):
for moveset_no in range(0, 4):
try:
loopmoves(url, moveset_no)
except:
pass
# The following codes are most likely to be correct, but it takes too much time to run.
all_moveset = []
for all_the_moves in moveset_url:
one_move(all_the_moves)
moves_data = pd.DataFrame(
columns=["move", "pokemon", "egg_group1", "egg_group2", "lvl"], data=all_moveset
)
moves_data.to_csv("moves.csv")
|
# # SURVIVAL AFTER TITANIC ACCIDENT
# 
import pandas as pd
import numpy as np
from math import *
import seaborn as sns
from tensorflow import keras
from keras.callbacks import Callback
from tensorflow.keras.optimizers import Adam
from keras.layers import Dense
from sklearn.preprocessing import OrdinalEncoder, LabelEncoder
from tensorflow.keras.models import Sequential
from tensorflow.keras.utils import to_categorical
df = pd.read_csv("/kaggle/input/titanic/train.csv")
df_test = pd.read_csv("/kaggle/input/titanic/test.csv")
df
df.info()
df.isna().sum()
df["Embarked"] = df["Embarked"].fillna("S")
df["Age"] = df["Age"].fillna(float(df["Age"].mean()))
df
df.isna().sum()
df_new = df.drop(["PassengerId", "Name", "Ticket", "Cabin"], axis=1)
df_new
encode = OrdinalEncoder()
df_new[["Sex", "Embarked"]] = encode.fit_transform(df_new[["Sex", "Embarked"]])
df_new["Sex"] = to_categorical(df_new["Sex"])
df_new.info()
df_new.Embarked.value_counts()
x_train = df_new.drop("Survived", axis=1).values
y_train = df_new["Survived"].values
x_train.shape, y_train.shape
model = Sequential(
[
Dense(64, input_shape=[7], activation="relu"),
Dense(64, activation="relu"),
Dense(1, name="OUTPUT", activation="softmax"),
]
)
num_epoch = 1000
model.compile(optimizer=Adam(), loss="bce", metrics=["mae"])
hist = model.fit(x_train, y_train, epochs=num_epoch, batch_size=16)
df_test.isna().sum()
df_test = df_test.drop(["Name", "Ticket", "Cabin"], axis=1)
df_test = df_test.interpolate()
df_test.isna().sum()
onehot = LabelEncoder()
df_test["Sex"] = onehot.fit_transform(df_test["Sex"])
df_test["Sex"] = to_categorical(df_test["Sex"])
df_test["Embarked"] = onehot.fit_transform(df_test["Embarked"])
df_test
df_test.Embarked.value_counts()
df_test.Embarked = to_categorical(df_test.Embarked)
x_test = df_test.drop("PassengerId", axis=1).values
ID = df_test["PassengerId"].values
predictions = kms.predict(x_test)
predictions
submit = pd.DataFrame({"PassengerId": ID, "Survived": predictions})
submit
submit.to_csv("submission.csv", index=False)
|
# ## Forest Cover Type Prediction
# _[Kaggle competition](https://www.kaggle.com/c/forest-cover-type-prediction/overview)_
# Author: Piotr Cichacki
# Date: 18.02.2021
# 1) Goal: to predict the forest cover type from strictly cartographic variables.
# 2) Data description
# The training set (15120 observations) contains both features information and the cover type. Each observation is a 30m x 30m patch.
# There are 4 binary columns for wilderness area and 40 binary columns for soil type in which 0 = absence and 1 = presence.
# Seven cover types (our target variable): spruce/fir, lodgepole pine, ponderosa pine, cottonwood/willow, aspen, douglas-fir, krummholz.
#
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_style("whitegrid")
import warnings
warnings.filterwarnings("ignore")
train = pd.read_csv("../input/forest-cover-type-prediction/train.csv")
train.drop("Id", axis=1, inplace=True)
test = pd.read_csv("../input/forest-cover-type-prediction/test.csv")
# ### Quick overview of our data
print("Training data shpae: ", train.shape)
print("Test data shpae: ", test.shape)
train.head(10)
train.info()
train.describe()
print(
"There are missing values in the training dataset: ",
train.isnull().sum().values.sum() > 0,
)
print(
"There are missing values in the test dataset: ",
test.isnull().sum().values.sum() > 0,
)
# #### Conclusions:
# We have 56 columns in our train dataset but only 12 attributes (without ID and our target variable) because there are 4 columns dedicated to wilderness area and 40 columns dedicated to soil type.
# All attributes are of type int (wilderness area and soil type columns are binary which means that they can have only value 0 or 1).
# We do not have to deal with missing values in our datasets.
# ### Our target variable: cover type
plt.title("Distribution of cover type")
sns.barplot(
train["Cover_Type"].value_counts().index, train["Cover_Type"].value_counts().values
)
plt.show()
# We can see that we have the same number of occurences for each type of cover.
# ### Exploratory data analysis
# At the beginning I will convert our train dataset to have Soil Type and Wilderness Area in single columns. Then I will present data in contingency table to check whether there is significant difference in proportions between groups.
soil_type = train.loc[:, "Soil_Type1":"Soil_Type40"].stack()
soil_type = pd.Series(soil_type[soil_type != 0].index.get_level_values(1))
for i in range(soil_type.size):
soil_type.values[i] = int((soil_type.values[i])[9:])
wilderness_area = train.loc[:, "Wilderness_Area1":"Wilderness_Area4"].stack()
wilderness_area = pd.Series(
wilderness_area[wilderness_area != 0].index.get_level_values(1)
)
for i in range(wilderness_area.size):
wilderness_area.values[i] = int((wilderness_area.values[i])[15:])
data = pd.concat(
[train.iloc[:, 0:10], wilderness_area, soil_type, train["Cover_Type"]], axis=1
)
data = data.rename(columns={0: "Wilderness_Area", 1: "Soil_Type"})
data.head()
pd.crosstab(data["Wilderness_Area"], data["Cover_Type"])
sns.catplot(data=data, kind="count", x="Cover_Type", hue="Wilderness_Area")
plt.title("Distribution of cover type between wilderness areas")
plt.show()
pd.crosstab(data["Cover_Type"], data["Soil_Type"])
# We can see that wilderness area and soil type have significant influence on cover type because there are a lot of zeros in our tables which means that certain types of cover occur only in certain conditions.
# Now let's focus on remaining attributes.
data.groupby(["Cover_Type"]).mean()
columns = data.columns[:-3]
for column in columns:
sns.displot(
data, x=data[column], hue="Cover_Type", kind="kde", fill=True, palette="Paired"
)
plt.title(column + " distribution between cover types")
plt.show()
for column in columns:
sns.boxplot(x="Cover_Type", y=column, data=data, palette="Paired")
plt.title(column + " distribution between cover types")
plt.show()
from sklearn.preprocessing import StandardScaler
from sklearn.preprocessing import MinMaxScaler
scaler = MinMaxScaler()
train["Slope"] = scaler.fit_transform(np.array(train["Slope"]).reshape(-1, 1))
columns = [
"Elevation",
"Aspect",
"Horizontal_Distance_To_Hydrology",
"Vertical_Distance_To_Hydrology",
"Horizontal_Distance_To_Roadways",
"Hillshade_9am",
"Hillshade_Noon",
"Hillshade_3pm",
]
scaler = MinMaxScaler()
for column in columns:
train[column] = scaler.fit_transform(np.array(train[column]).reshape(-1, 1))
plt.figure(figsize=(10, 10))
sns.heatmap(data.corr().round(2), annot=True)
plt.show()
# ### Building model
#
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
train.drop(["Cover_Type"], axis=1), train["Cover_Type"], random_state=42
)
from sklearn.linear_model import LogisticRegression
lr = LogisticRegression(C=1)
lr.fit(X_train, y_train)
print("Accuracy on training set: ", lr.score(X_train, y_train))
print("Accuracy on test set: ", lr.score(X_test, y_test))
from sklearn.ensemble import RandomForestClassifier
rfc = RandomForestClassifier(n_estimators=1000)
rfc.fit(X_train, y_train)
print("Accuracy on training set: ", rfc.score(X_train, y_train))
print("Accuracy on test set: ", rfc.score(X_test, y_test))
# Let's now evaluate our model using K-fold cross-validation.
from sklearn.model_selection import cross_val_score
cross_val_score(rfc, X_train, y_train, cv=3, scoring="accuracy")
# The next step is to build confusion matrix to see how our model make predictions.
from sklearn.model_selection import cross_val_predict
y_train_pred = cross_val_predict(rfc, X_train, y_train, cv=5)
from sklearn.metrics import confusion_matrix
conf_matrix = confusion_matrix(y_train, y_train_pred)
conf_matrix
plt.rcParams["figure.figsize"] = (8, 8)
plt.matshow(conf_matrix, interpolation="nearest", cmap="plasma")
plt.title("Confusion matrix", fontdict={"fontsize": 12})
plt.colorbar()
plt.show()
row_sums = conf_matrix.sum(axis=1, keepdims=True)
norm_conf_matrix = conf_matrix / row_sums
np.fill_diagonal(norm_conf_matrix, 0)
plt.rcParams["figure.figsize"] = (8, 8)
plt.matshow(norm_conf_matrix, interpolation="nearest", cmap="plasma")
plt.title(
"Plot of the errors\n (divided by number of observations in the corresponding class)",
fontdict={"fontsize": 12},
)
plt.colorbar()
plt.show()
# From above plot we can see that many Lodgepole Pine (1 type) are classified as Spruce/Fir (0 type) and another way around.
# ### Making predictions
submission = pd.DataFrame()
submission["Id"] = test["Id"]
submission["Cover_Type"] = rfc.predict(test.drop("Id", axis=1))
submission.set_index("Id", inplace=True)
submission.to_csv("submission.csv")
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import plotly.graph_objects as go
import plotly.express as px
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import (
confusion_matrix,
accuracy_score,
recall_score,
precision_score,
f1_score,
)
from sklearn.metrics import classification_report
from sklearn.preprocessing import LabelEncoder
import statsmodels.api as sm
from scipy import stats
from sklearn.model_selection import cross_val_score
plt.rcParams["font.sans-serif"] = ["SimHei"]
plt.rcParams["axes.unicode_minus"] = False
# 繪製sankey plot function
def get_sankey_df(df, cat_cols=[], value_cols=""):
colorNumList = []
labelList = []
for catCol in cat_cols:
labelListTemp = list(set(df[catCol].values))
colorNumList.append(len(labelListTemp))
labelList = labelList + labelListTemp
labelList = list(dict.fromkeys(labelList))
for i in range(len(cat_cols) - 1):
if i == 0:
sourceTargetDf = df[[cat_cols[i], cat_cols[i + 1], value_cols]]
sourceTargetDf.columns = ["source", "target", "count"]
else:
tempDf = df[[cat_cols[i], cat_cols[i + 1], value_cols]]
tempDf.columns = ["source", "target", "count"]
sourceTargetDf = pd.concat([sourceTargetDf, tempDf])
sourceTargetDf = (
sourceTargetDf.groupby(["source", "target"])
.agg({"count": "sum"})
.reset_index()
)
# add index for source-target pair
sourceTargetDf["sourceID"] = sourceTargetDf["source"].apply(
lambda x: labelList.index(x)
)
sourceTargetDf["targetID"] = sourceTargetDf["target"].apply(
lambda x: labelList.index(x)
)
return {"label_list": labelList, "df": sourceTargetDf}
# # About
# 本次根據 IBM Watson Marketing Customer Value Data 做分析,主要有兩個目的,
# 1. 用戶畫像描繪
# - 用戶畫像能夠幫助利益相關者了解自身產品的使用者特性與狀況。
# - 精準行銷,不同產品類型可能有不同受眾。
# - 透過洞察,看出客戶的潛藏需求,提供更貼心、更好的服務。
# 2. 再續訂預測
# - 以機器學習模型建立分類模型,幫助判斷當前客戶是否會再續訂目前的服務或方案。
# - 提早看出客戶再續約意願,可以過濾出無續約意願的客戶,把成本投入其他高價值客戶身上。
# - 針對續約意願高的客戶,推出更客製化服務,增加黏著度。
# ## 變數說明
# - 客戶編號, customer id
# - 州, 居住的州
# - Customer Lifetime Value, 客戶的生命週期價值
# - Response, 是否願意續約(是,否)
# - Coverage, 產品覆蓋範圍(基本、進階、高級)
# - Education, 教育程度(高中或以下, 大學, 碩士, 博士)
# - Effective To Date, 生效日期(保單)
# - EmploymentStatus, 就業程度(就業中, 待業, 病假, 殘疾人士, 退休)
# - Gender, 性別
# - Income, 年收入
# - Location Code, 位置代碼(郊區, 鄉村, 城市)
# - Mariage Status, 婚姻狀況(單身, 已婚, 離婚)
# - Monthly Premium Auto, 每月平均繳納金額
# - Months Since Last Claim, 自上次索賠以來的月數
# - Months Since Policy Inception, 自產品生效以來的月數
# - Number of Open Complaints, 未解決的索賠數量
# - Number of Policies, 保單數量
# - Policy Type, 產品類型(個人, 公司, 特殊)
# - Policy 各產品類型的級別(L1, L2, L3)
# - Renew Offer Type 續訂邀約類型(offer 1,offer 2,offer 3,offer 4)
# - Sales Channel 銷售渠道(代理商、分行, 客服中心, 網路)
# - Total Claim Amount 累積索賠金額
# - Vehicle Class 車輛類型(四門, 雙門, SUV, 跑車, 豪華SUV, 豪車)
# - Vehicle Size 車輛類型(大, 中, 小)
# # 載入套件、數據
df = pd.read_csv(
"../input/ibm-watson-marketing-customer-value-data/WA_Fn-UseC_-Marketing-Customer-Value-Analysis.csv"
)
df.head()
df.shape
pd.set_option("display.max_columns", None)
df.head()
# # Part 1. 用戶畫像分析 Persona
# ## 資料清洗與檢驗
# 此數據無重複值或重複客戶,也沒發現遺失值。
# chech if there is any duplicated customer
df["Customer"].duplicated().any()
# percentage of missing value
df.apply(lambda x: sum(x.isnull()) / len(x), axis=0)
# ### 離群值檢驗
# 以盒鬚圖方法檢驗離群值,設定資料值小於 Q1 - 3xIQR 或大於 Q3 + 3xIQR 為離群值
# 有部分變數存在離群值,後續依情形排除。
def outlier_dectect(df, column, cutoff_rate):
global lower, upper
q1, q3 = np.quantile(df[column], 0.25), np.quantile(df[column], 0.75)
IQR = q3 - q1
cut_off = IQR * cutoff_rate
lower, upper = q1 - cut_off, q3 + cut_off
print("The lower bound value is", lower)
print("The upper bound value is", upper)
df1 = df[df[column] > upper]
df2 = df[df[column] < lower]
return print("Total number of outliers are", df1.shape[0] + df2.shape[0])
df_remove_outlier = df.copy()
df_remove_outlier.loc[
df_remove_outlier["Customer Lifetime Value"] >= 16414.04, "Customer Lifetime Value"
] = 16414
df_remove_outlier.loc[
df_remove_outlier["Monthly Premium Auto"] >= 170.5, "Monthly Premium Auto"
] = 170.5
df_remove_outlier.loc[
df_remove_outlier["Monthly Premium Auto"] <= 6.5, "Monthly Premium Auto"
] = 6.5
df_remove_outlier.loc[
df_remove_outlier["Total Claim Amount"] >= 960.3997, "Total Claim Amount"
] = 960.3997
out_index = df[
(df["Customer Lifetime Value"] >= 16414.04)
| (df["Monthly Premium Auto"] >= 170.5)
| (df["Monthly Premium Auto"] <= 6.5)
| (df["Total Claim Amount"] >= 960.3997)
].index
df_remove_outlier = df.drop(axis=0, labels=out_index)
for col in df.columns[(df.dtypes == "int64") | (df.dtypes == "float64")]:
print(f"Outlier detection for `{col}`:")
outlier_dectect(df, col, 1.5)
print("\n")
# ## Quick EDA
# ### 客戶性別
# 兩性別比例無明顯差異
gender_df = df.groupby("Gender")["Customer"].count()
gender_df = gender_df.to_frame()
gender_df.plot(kind="pie", subplots=True, autopct="%1.1f%%")
plt.title("Customer gender distribution")
plt.show()
# ### 客戶居住地區
# 多數客戶選擇居住在郊區,又以California最多人居住。
loc_df = df.groupby(["Location Code", "State"])["Customer"].count()
loc_df = loc_df.to_frame()
loc_df = loc_df.reset_index()
loc_data_for_sankey = get_sankey_df(loc_df, ["Location Code", "State"], "Customer")
colors = [
"rgb(249, 226, 175)",
"rgb(0, 159, 189)",
"rgb(33, 0, 98)",
"rgb(119, 3, 123)",
"rgb(250, 112, 112)",
"rgb(251, 242, 207)",
"rgb(198, 235, 197)",
"rgb(178, 164, 255)",
]
fig = go.Figure(
data=[
go.Sankey(
node=dict(
pad=15,
thickness=20,
line=dict(color="black", width=0.5),
label=loc_data_for_sankey["label_list"],
color=colors,
),
link=dict(
source=loc_data_for_sankey["df"]["sourceID"],
target=loc_data_for_sankey["df"]["targetID"],
value=loc_data_for_sankey["df"]["count"],
),
)
]
)
fig.update_layout(title_text="Sankey diagram - 客戶居住地區分佈", font_size=15, width=600)
fig.show()
# ### 客戶教育程度
# 有碩士、博士學歷的是少數,大多客戶為學士或以下的學歷。
edu_df = df.groupby("Education")["Customer"].count()
edu_df = edu_df.to_frame().reset_index()
edu_df
fig = px.bar(edu_df, x="Education", y="Customer")
fig.update_layout(
title_text="客戶教育程度分佈",
barmode="stack",
xaxis={"categoryorder": "total descending"},
width=600,
)
fig.show()
# ### 持有汽車種類
# 4門、雙門、SUV占多數的中型車占多數。
#
vehicel_df = df.groupby(["Vehicle Size", "Vehicle Class"])["Customer"].count()
vehicel_df = vehicel_df.to_frame()
vehicel_df = vehicel_df.reset_index()
vehicel_df
fig = px.histogram(
vehicel_df, x="Vehicle Class", y="Customer", color="Vehicle Size", barmode="group"
)
fig.update_layout(
title_text="客戶持有汽車類型分佈",
barmode="stack",
xaxis={"categoryorder": "total descending"},
width=600,
)
fig.show()
# ### 收入
# 收入的分配極度右偏,有2.5成客戶收入不到 2000美元
fig = px.histogram(df, x="Income", width=600)
fig.show()
# ### 婚姻狀態
marriage_df = df.groupby("Marital Status")["Customer"].count()
marriage_df = marriage_df.to_frame().reset_index()
fig = px.bar(marriage_df, x="Marital Status", y="Customer")
fig.update_layout(
title_text="客戶婚姻狀態分佈",
barmode="stack",
xaxis={"categoryorder": "total descending"},
width=600,
)
fig.show()
# ### 職業類型
employstatus_df = df.groupby("EmploymentStatus")["Customer"].count()
employstatus_df = employstatus_df.to_frame().reset_index()
fig = px.bar(employstatus_df, x="EmploymentStatus", y="Customer")
fig.update_layout(
title_text="客戶職業類型分佈",
barmode="stack",
xaxis={"categoryorder": "total descending"},
width=600,
)
fig.show()
# 因為這些類別變數是有序性的,也有部分連續型變數嚴重右偏,故以pd.factorize給予所有變數label encoding,再以Spearman觀察所有變數的相關性
df_corr = df.drop(["Response", "Customer"], axis=1).apply(lambda x: pd.factorize(x)[0])
df_corr = df_corr.corr("spearman").round(3)
# 從相關性可發現一些有趣的事實:
# - 顧客終身價值 與**收入**、**累積索賠金額**有低至中度相關
# - 累積索賠金額 與**客戶地區**有中度相關
# - 每月平均繳納金額 與**產品涵蓋類型**、**持有汽車種類**有中度相關
plt.figure(figsize=(15, 12))
ax = sns.heatmap(df_corr, annot=True, cmap="Blues")
plt.title("Spearman correlation between variables")
plt.show()
# 顧客終身價值(CLV)能夠幫助我們決定多少成本應該投入在這些客戶身上,結合產品生效以來月數(Months Since Policy Inception)可以看客戶的忠誠度,接下來以兩變數做市場區分
df_cus_seg = df.copy()
df_cus_seg["CLV seg"] = df_cus_seg["Customer Lifetime Value"].apply(
lambda x: "high" if x > df_cus_seg["Customer Lifetime Value"].median() else "low"
)
df_cus_seg["policy duration seg"] = df_cus_seg["Months Since Policy Inception"].apply(
lambda x: "high"
if x > df_cus_seg["Months Since Policy Inception"].median()
else "low"
)
fig = df_cus_seg.loc[
(df_cus_seg["CLV seg"] == "high") & (df_cus_seg["policy duration seg"] == "high")
].plot.scatter(
x="Months Since Policy Inception", y="Customer Lifetime Value", c="red", logy=True
)
df_cus_seg.loc[
(df_cus_seg["CLV seg"] == "low") & (df_cus_seg["policy duration seg"] == "high")
].plot.scatter(
ax=fig,
x="Months Since Policy Inception",
y="Customer Lifetime Value",
c="blue",
logy=True,
)
df_cus_seg.loc[
(df_cus_seg["CLV seg"] == "high") & (df_cus_seg["policy duration seg"] == "low")
].plot.scatter(
ax=fig,
x="Months Since Policy Inception",
y="Customer Lifetime Value",
c="yellow",
logy=True,
)
df_cus_seg.loc[
(df_cus_seg["CLV seg"] == "low") & (df_cus_seg["policy duration seg"] == "low")
].plot.scatter(
ax=fig,
x="Months Since Policy Inception",
y="Customer Lifetime Value",
c="grey",
logy=True,
)
fig.set_ylabel("log(CLV)")
fig.set_xlabel("Months Since Policy Inception")
fig.set_title("Customer Segmentation - Based on CLV and Months Since Policy Inception")
plt.show()
response_rate_by_cus_seg = (
df_cus_seg.loc[df_cus_seg["Response"] == 1]
.groupby(["CLV seg", "policy duration seg"])
.count()["Customer"]
/ df_cus_seg.groupby(["CLV seg", "policy duration seg"]).count()["Customer"]
)
response_rate_by_cus_seg = (
response_rate_by_cus_seg.to_frame()
.reset_index()
.rename(columns={"Customer": "Response (%)"})
)
response_rate_by_cus_seg["Response (%)"] = (
response_rate_by_cus_seg["Response (%)"] * 100
)
# 將客戶分成4群後,依各組分別觀察其回應率,可以看到保單生效月數長,其回應率較高,也就是說長期投保的客戶有較高的回應率。
# 另外,低終身價值但是保單生效月數長的客戶有最高的回應率。
fig = px.bar(
response_rate_by_cus_seg,
x="CLV seg",
y="Response (%)",
color="policy duration seg",
width=600,
barmode="group",
title="分群後 - 客戶回應率",
)
fig.show()
# # Part 2. 用戶續訂預測
# ## Quick EDA
# ## Target: Response rate
response_df = df.groupby("Response")["Customer"].count()
response_df = response_df.to_frame().reset_index()
response_df = response_df.assign(percentage=lambda x: x.Customer / len(df) * 100)
response_df["percentage"] = response_df["percentage"].apply(
lambda x: "{0:1.2f}%".format(x)
)
fig = px.bar(response_df, x="Response", y="Customer", text="percentage")
fig.update_layout(
title_text="客戶回應狀況分佈",
barmode="stack",
xaxis={"categoryorder": "total descending"},
width=600,
)
fig.show()
# ### Response Rate V.S. Education
fig = px.histogram(
df,
x="Response",
y="Customer",
color="Education",
barmode="group",
histfunc="count",
width=600,
)
fig.update_layout(title_text="客戶回應狀況 V.S. 教育程度")
fig.show()
# ### Response Rate V.S. Sales Channel
fig = px.histogram(
df,
x="Response",
y="Customer",
color="Sales Channel",
barmode="group",
histfunc="count",
width=600,
)
fig.update_layout(title_text="客戶回應狀況 V.S. 銷售渠道")
fig.show()
# ### Response Rate V.S. Total Claim Amount
outlier_dectect(df, "Total Claim Amount", 1.5)
total_claim_remove_out_df = df[df["Total Claim Amount"] < 960.3997]
fig = px.box(total_claim_remove_out_df, x="Response", y="Total Claim Amount", width=600)
fig.update_layout(title_text="客戶回應狀況 V.S. 累積索賠金額")
fig.show()
# ### Response Rate V.S. Income
fig = px.box(total_claim_remove_out_df, x="Response", y="Income", width=600)
fig.update_layout(title_text="客戶回應狀況 V.S. 收入")
fig.show()
# ### Response Rate V.S. EmploymentStatus
fig = px.histogram(
df,
x="Response",
y="Customer",
color="EmploymentStatus",
barmode="group",
histfunc="count",
width=600,
)
fig.update_layout(title_text="客戶回應狀況 V.S. 就業狀況")
fig.show()
# ### Response rate V.S. Policy
fig = px.histogram(
df,
x="Response",
y="Customer",
color="Policy",
barmode="group",
histfunc="count",
width=600,
)
fig.update_layout(title_text="客戶回應狀況 V.S. 產品類型的級別")
fig.show()
df.Response = df.Response.apply(lambda X: 0 if X == "No" else 1)
plt.figure(figsize=(10, 6))
sns.heatmap(numeric_df.corr(), annot=True, cmap="Blues")
plt.show()
lb = LabelEncoder()
for col in cat_df.columns:
cat_df[col] = lb.fit_transform(cat_df[col])
all_df = pd.concat([cat_df, numeric_df], axis=1)
X = all_df.drop(axis=1, labels="Response")
y = all_df["Response"]
lr_model = sm.Logit(y, X)
lr_model_fit = lr_model.fit()
lr_model_fit.summary()
lr_model_fit.pvalues[lr_model_fit.pvalues < 0.05]
# ## Prediction
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=42
)
# #### Logistic Regression
from sklearn.preprocessing import MinMaxScaler
scaler = MinMaxScaler()
X_train_scaled = scaler.fit_transform(X_train)
X_test_scaled = scaler.transform(X_test)
lr = LogisticRegression()
lr.fit(X_train_scaled, y_train)
lr_pred = lr.predict(X_test_scaled)
print(confusion_matrix(lr_pred, y_test))
print("Accuracy score:", accuracy_score(lr_pred, y_test))
print(classification_report(lr_pred, y_test))
scores = cross_val_score(lr, X, y, cv=5)
print("\n")
print(
"%0.2f accuracy with a standard deviation of %0.2f" % (scores.mean(), scores.std())
)
# #### Random Forest
from sklearn.ensemble import RandomForestClassifier
rfc = RandomForestClassifier()
rfc.fit(X_train, y_train)
rfc_pred = rfc.predict(X_test)
print(confusion_matrix(rfc_pred, y_test))
print("Accuracy score:", accuracy_score(rfc_pred, y_test))
print(classification_report(rfc_pred, y_test))
scores = cross_val_score(rfc, X, y, cv=5)
print("\n")
print(
"%0.2f accuracy with a standard deviation of %0.2f" % (scores.mean(), scores.std())
)
feature_imp = rfc.feature_importances_.round(3)
feature_imp_ = pd.Series(feature_imp, index=X.columns).sort_values(ascending=False)
plt.figure(figsize=(8, 8))
sns.barplot(x=feature_imp_.values, y=feature_imp_.index)
plt.title("Random forest feature importance")
plt.xlabel("Gini importance")
plt.show()
# #### Xgboost
import xgboost as xgb
xgb = xgb.XGBClassifier(objective="binary:logistic", random_state=42)
xgb.fit(X_train, y_train)
xgb_pred = xgb.predict(X_test)
print(confusion_matrix(xgb_pred, y_test))
print("Accuracy score:", accuracy_score(xgb_pred, y_test))
print(classification_report(xgb_pred, y_test))
feature_imp = xgb.feature_importances_.round(3)
feature_imp_ = pd.Series(feature_imp, index=X.columns).sort_values(ascending=False)
plt.figure(figsize=(8, 8))
sns.barplot(x=feature_imp_.values, y=feature_imp_.index)
plt.title("Xgboost feature importance")
plt.xlabel("Average gain")
plt.show()
from sklearn.svm import SVC
svm = SVC(gamma="scale", kernel="rbf", class_weight="balanced")
svm.fit(X_train_scaled, y_train)
svm_pred = svm.predict(X_test_scaled)
print(confusion_matrix(svm_pred, y_test))
print("Accuracy score:", accuracy_score(svm_pred, y_test))
print(classification_report(svm_pred, y_test))
def get_chi_square_res(df, x="", y=""):
res = pd.crosstab(df[x], df[y], margins=False)
chi2, p, dof, ex = stats.chi2_contingency(res, correction=False)
return p
lst = []
for i in cat_df.columns:
p = get_chi_square_res(df, i, "Response")
lst.append(p)
chi_square_test_res = pd.Series(lst, index=cat_df.columns).to_frame(
name="Chi-square test's p-value"
)
chi_imp_cat_col = chi_square_test_res[
chi_square_test_res["Chi-square test's p-value"] < 0.05
].index
# #### build model with chi-square test important features
X_train, X_test, y_train, y_test = train_test_split(
X[chi_imp_cat_col.append(numeric_df.drop(axis=1, labels="Response").columns)],
y,
test_size=0.2,
random_state=42,
)
rfc = RandomForestClassifier()
rfc.fit(X_train, y_train)
rfc_pred = rfc.predict(X_test)
print(confusion_matrix(rfc_pred, y_test))
print("Accuracy score:", accuracy_score(rfc_pred, y_test))
print("\n")
print(classification_report(rfc_pred, y_test))
scores = cross_val_score(rfc, X, y, cv=5)
print("\n")
print(
"%0.2f accuracy with a standard deviation of %0.2f" % (scores.mean(), scores.std())
)
def get_point_biserial(df, x="", y=""):
point_biserial = stats.pointbiserialr(df[x], df[y])
return point_biserial.correlation
lst = []
for i in numeric_df.columns:
res = get_point_biserial(df, i, "Response")
lst.append(res)
pd.Series(lst, index=numeric_df.columns).to_frame("point_biserial_corr")
State = pd.get_dummies(df["State"], drop_first=True)
Coverage = pd.get_dummies(df["Coverage"], drop_first=True)
Education = pd.get_dummies(df["Education"], drop_first=True)
EmploymentStatus = pd.get_dummies(df["EmploymentStatus"], drop_first=True)
Gender = pd.get_dummies(df["Gender"], drop_first=True)
Marital_Status = pd.get_dummies(df["Marital Status"], drop_first=True)
Policy_Type = pd.get_dummies(df["Policy Type"], drop_first=True)
Policy = pd.get_dummies(df["Policy"], drop_first=True)
Renew_Offer_Type = pd.get_dummies(df["Renew Offer Type"], drop_first=True)
Sales_Channel = pd.get_dummies(df["Sales Channel"], drop_first=True)
Vehicle_Class = pd.get_dummies(df["Vehicle Class"], drop_first=True)
Vehicle_Size = pd.get_dummies(df["Vehicle Size"], drop_first=True)
cat_encoded_df = pd.concat(
[
State,
Coverage,
Education,
EmploymentStatus,
Gender,
Marital_Status,
Policy_Type,
Policy,
Renew_Offer_Type,
Sales_Channel,
Vehicle_Class,
Vehicle_Size,
],
axis=1,
)
cat_encoded_df
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn import model_selection
from sklearn import svm
# **Read Data**
df = pd.read_csv(
"/kaggle/input/admission-based-on-exam-scores/admission_basedon_exam_scores.csv"
)
# **PRINT SHAPE AND DETAILS OF DATA**
print(df.shape)
df.head(df.info())
# **Capture and store data of admitted students**
a = df[df["Admission status"] == 1]
print("Training examples with admission status 1:", a.shape[0])
a.head(3)
# **Capture and store data of non admitted students**
na = df[df["Admission status"] == 0]
print("Training examples with admission status 1:", na.shape[0])
na.head(3)
# **Visualization**
def plot_title(title):
plt.figure(figsize=(10, 6))
plt.scatter(
a["Exam 1 marks"], a["Exam 2 marks"], color="green", label="Admitted Students"
)
plt.scatter(
na["Exam 1 marks"],
na["Exam 2 marks"],
color="red",
label="Non Admitted Students",
)
plt.xlabel("Exam 1 marks")
plt.ylabel("Exam 2 marks")
plt.title(title)
plt.legend()
plot_title("Admitted vs Non Admitted Students")
|
import numpy as np
from sklearn.linear_model import LinearRegression
import matplotlib.pyplot as plt
import pandas as pd
# price prediction based on the features
df = pd.read_csv("../input/car-data/Car_Data.csv")
print(type(df))
df.head()
"""
preprocessing we have to do on data
1. drop the columns with id or other features that leads to memorize the data > id, name, ...
* company name is ok but the unique name is not
* linear reg is a weak algo > doesn't memorize but NN does it
2. convert all of the non numeric columns into numeric mapping
"""
df2 = df
# no inplace
# df2 = df2.drop('car_ID', axis=1)
# df2 = df2.drop('CarName', axis=1)
df2 = df2.drop(["car_ID", "CarName"], axis=1)
df2.head()
df = df.drop(["car_ID", "CarName"], axis=1)
df.head(6)
df.tail()
df.columns # iterables
# df['fueltype']
s = df.fueltype
print(type(s))
print(s)
# mathematic describe for numeric columns
# counts the nonNaN data > NaN data > missing value > we have to fill them or drop them
df.describe()
# x=index, y=value > plt.plot > can accepts the series > numeric
plt.plot(df.price, "bo")
plt.hist(df.price)
# both data numeric and string
plt.hist(df.fueltype)
# visualize two numeric values based on each other
plt.plot(df.price, df.carlength, "bo")
plt.xlabel("price")
plt.ylabel("carheight")
# degree 2, log, ... are good for fitting the data
plt.plot(df.price, df.wheelbase, "bo")
plt.xlabel("price")
plt.ylabel("carheight")
plt.grid()
# line is good for fittng data
df.fueltype.value_counts()
df.fueltype.unique()
def pow2(a):
return a**2
b = [1, 2, 3, 4, 5]
list(map(pow2, b))
def t(x):
if x == "gas":
return 0
else:
return 1
# 1. write map function > series == array == list
# df.fueltype = df.fueltype.map(t)
df.fueltype = df.fueltype.map(lambda x: 0 if x == "gas" else 1)
df.head()
df.fueltype.value_counts()
# 2. filtering
a = np.array([1, 2, 3, 4, 5, 6, 7])
print(a < 4)
# t/f as an index
print(a[a < 4])
df.fueltype = df.fueltype.map(lambda x: "gas" if x == 0 else "diesel")
df.fueltype[df.fueltype == "gas"] = 0
df.fueltype[df.fueltype == "diesel"] = 1
df.head()
df.fueltype.value_counts()
df.fueltype.unique()
df.fueltype
# it can be gas or 0
plt.plot(df.price[df.fueltype == 0], df.highwaympg[df.fueltype == 0], "bo")
plt.plot(df.price[df.fueltype == 1], df.highwaympg[df.fueltype == 1], "ro")
plt.plot(df.price[df.drivewheel == "rwd"], df.highwaympg[df.drivewheel == "rwd"], "bo")
plt.plot(df.price[df.drivewheel == "fwd"], df.highwaympg[df.drivewheel == "fwd"], "ro")
plt.plot(df.price[df.drivewheel == "4wd"], df.highwaympg[df.drivewheel == "4wd"], "go")
# 3. get dummies > the previous approaches good for 2 values not more than that
# more than one value > one hot encoding > 4 values = 4 columns
df.fueltype = df.fueltype.map(lambda x: "gas" if x == 0 else "diesel")
df.head()
# it doesn't change df
pd.get_dummies(df, columns=["fueltype"]) # remove fuel > fueltype_diesel fueltype_gas
pd.get_dummies(
df, columns=["fueltype"], drop_first=True
) # rather than 2 cols > one col > remove fuel > fueltype_gas
pd.get_dummies(df, prefix=["new"], columns=["fueltype"], drop_first=True)
pd.get_dummies(
df, columns=["fueltype"], drop_first=True, dummy_na=True
) # add another column > nan or not the value
# we have two main dtype for series > object + numeric
for c in df.columns:
if df[c].dtype == "object":
print("-" * 10)
print(c)
print(df[c].unique())
# for al of the columns which is binary > one col is enough
# for more than two value > we need al columns > dropfirst = false
binary_columns = ["enginelocation", "doornumber", "aspiration", "fueltype"]
df = pd.get_dummies(df, prefix=binary_columns, columns=binary_columns, drop_first=True)
df.head()
for c in df.columns:
if df[c].dtype == "object":
print("-" * 10)
print(c)
print(len(df[c].value_counts()))
binary_columns = []
categorical_columns = []
for c in df.columns:
if df[c].dtype == "object":
if len(df[c].value_counts()) == 2:
binary_columns.append(c)
elif len(df[c].value_counts()) > 2:
categorical_columns.append(c)
print(binary_columns)
print(categorical_columns)
df = pd.get_dummies(df, prefix=binary_columns, columns=binary_columns, drop_first=True)
df = pd.get_dummies(df, prefix=categorical_columns, columns=categorical_columns)
df.head()
df.columns
# # Training a model
# prepare data
df = pd.read_csv("../input/car-data/Car_Data.csv")
df = df.drop(["car_ID", "CarName"], axis=1)
binary_columns = []
categorical_columns = []
for c in df.columns:
if df[c].dtype == "object":
if len(df[c].value_counts()) == 2:
binary_columns.append(c)
elif len(df[c].value_counts()) > 2:
categorical_columns.append(c)
df = pd.get_dummies(df, prefix=binary_columns, columns=binary_columns, drop_first=True)
df = pd.get_dummies(df, prefix=categorical_columns, columns=categorical_columns)
print(df.shape)
# series is array > not matrix > we convert both x and y to matrix
y = df.price.to_numpy().reshape(-1, 1)
df = df.drop("price", axis=1)
# convert df to numpy
X = df.to_numpy()
print(X.shape)
print(y.shape)
from sklearn import preprocessing
# normalize all of the samples of one feature > 0
X = preprocessing.normalize(X, axis=0)
X.min(), X.max(), X.shape
lr_model = LinearRegression()
lr_model.fit(X, y)
print(len(lr_model.coef_[0]), lr_model.intercept_)
h = lr_model.predict(X)
MSE = np.mean((y - h) ** 2) / 2
print(MSE**0.5)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Install Packages
# Load Libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import torch
from transformers import BertTokenizer, BertForSequenceClassification
from torch.utils.data import DataLoader, RandomSampler, SequentialSampler
from keras.utils import pad_sequences
from sklearn.model_selection import train_test_split
import bert
import tensorflow as tf
import tensorflow_hub as hub
from tensorflow.keras.utils import to_categorical
from tensorflow.keras.models import Model
from tensorflow.keras.layers import Input, Dense, Dropout
from tensorflow.keras.optimizers import Adam
from tensorflow.keras.callbacks import ModelCheckpoint, TensorBoard
from tqdm import tqdm
import joblib
print("TensorFlow Version:", tf.__version__)
print("Hub version: ", hub.__version__)
pd.set_option("display.max_colwidth", 1000)
# Import train and test dataset
d2_train_data = pd.read_csv("/kaggle/input/tweets/Tweets.csv")
d1_test_data = pd.read_csv("/kaggle/input/nlp-getting-started/test.csv")
# Create train dataset copy
d2_train_df = d2_train_data
# Print Train data info
print("First five rows in train datset:\n\n", d2_train_data.head())
print("\n")
print("Total rows and columns in train dataset:\n\n", d2_train_data.shape)
print("\n")
print(d2_train_data.info())
print("\n")
print("Summary of train dataset:\n\n", d2_train_data.describe())
print("\n")
print("Null values in train dataset:\n\n", d2_train_data.isnull().sum())
print("\n")
# Print Test data info
print(d1_test_data.head())
print("\n")
print(d1_test_data.shape)
print("\n")
print(d1_test_data.info())
print("\n")
# Check the target class balance
print(d2_train_data["sentiment"].value_counts()) # It's a balanced dataset
print("\n")
sns.countplot(x=d2_train_data.sentiment, width=0.5)
plt.title("sentiment Distribution")
# Check Unique 'Text' values in both train and test dataset
print("Count of Unique Texts in Train Data:", d2_train_data["text"].nunique())
print(d2_train_data.shape)
print("Count of Unique Texts in Test Data:", d1_test_data["text"].nunique())
print(d1_test_data.shape)
## Both train and test dataset have duplicate texts
# Check the first text in train data
d2_train_data["textID"][0]
# Set "Id" column as index in train data
d2_train_data = d2_train_data.set_index("textID")
d2_train_data.head(2)
# Perform drop duplicate on train data to get unique data
# train_data = train_data.drop_duplicates()
# print("Updated Train Data:", train_data.shape)
# print("Original Train Data:", train_df.shape)
# Create new "sentiment" column. 0 = Negative and 1 = positive
def label_change(x):
if x == "neutral":
return 0
if x == "negative":
return 1
if x == "positive":
return 2
d2_train_data["label"] = d2_train_data["sentiment"].apply(lambda x: label_change(x))
print(d2_train_data.head())
print("\n")
print(d2_train_data["sentiment"].value_counts())
print(d2_train_data["label"].value_counts())
# Create copy of original train dataset
d2_training_data = d2_train_data
d2_training_data.head(2)
d2_training_data = d2_training_data.dropna()
# ## Pre-Trained Bert Model
# Split the train dataset into train and val sets
d2_train_text, d2_val_text, d2_train_labels, d2_val_labels = train_test_split(
d2_training_data["text"].tolist(), d2_training_data["label"].tolist(), test_size=0.2
)
# Load the BERT tokenizer and model
tokenizer = BertTokenizer.from_pretrained("bert-base-uncased")
bert_model = BertForSequenceClassification.from_pretrained(
"bert-base-uncased", num_labels=3
)
# Define the maximum sequence length and batch size
MAX_LEN = 500
BATCH_SIZE = 32
# Tokenize the text and add special tokens
train_tokens = tokenizer.batch_encode_plus(
d2_train_text,
add_special_tokens=True,
max_length=MAX_LEN,
truncation=True,
padding=True,
return_attention_mask=True,
return_tensors="pt",
)
val_tokens = tokenizer.batch_encode_plus(
d2_val_text,
add_special_tokens=True,
max_length=MAX_LEN,
truncation=True,
padding=True,
return_attention_mask=True,
return_tensors="pt",
)
# Create PyTorch datasets
train_dataset = torch.utils.data.TensorDataset(
train_tokens["input_ids"],
train_tokens["attention_mask"],
torch.tensor(d2_train_labels),
)
val_dataset = torch.utils.data.TensorDataset(
val_tokens["input_ids"], val_tokens["attention_mask"], torch.tensor(d2_val_labels)
)
# Create PyTorch dataloaders
train_dataloader = DataLoader(
train_dataset, sampler=RandomSampler(train_dataset), batch_size=BATCH_SIZE
)
val_dataloader = DataLoader(
val_dataset, sampler=SequentialSampler(val_dataset), batch_size=BATCH_SIZE
)
train_tokens = tokenizer.batch_encode_plus(
d2_train_text,
add_special_tokens=True,
max_length=MAX_LEN,
truncation=True,
padding=True,
return_attention_mask=True,
return_tensors="pt",
)
val_tokens = tokenizer.batch_encode_plus(
d2_val_text,
add_special_tokens=True,
max_length=MAX_LEN,
truncation=True,
padding=True,
return_attention_mask=True,
return_tensors="pt",
)
train_dataset = torch.utils.data.TensorDataset(
train_tokens["input_ids"],
train_tokens["attention_mask"],
torch.tensor(d2_train_labels),
)
val_dataset = torch.utils.data.TensorDataset(
val_tokens["input_ids"], val_tokens["attention_mask"], torch.tensor(d2_val_labels)
)
train_dataloader = DataLoader(
train_dataset, sampler=RandomSampler(train_dataset), batch_size=BATCH_SIZE
)
val_dataloader = DataLoader(
val_dataset, sampler=SequentialSampler(val_dataset), batch_size=BATCH_SIZE
)
# Set the device to run on (GPU if available)
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
bert_model.to(device)
# Define the optimizer and loss function
optimizer = torch.optim.AdamW(bert_model.parameters(), lr=2e-5, eps=1e-8)
loss_fn = torch.nn.CrossEntropyLoss()
# Train the model
num_epochs = 5
for epoch in range(num_epochs):
bert_model.train()
train_loss = 0.0
for batch in train_dataloader:
input_ids = batch[0].to(device)
attention_mask = batch[1].to(device)
labels = batch[2].to(device)
optimizer.zero_grad()
outputs = bert_model(input_ids, attention_mask=attention_mask, labels=labels)
loss = outputs.loss
logits = outputs.logits
loss.backward()
optimizer.step()
train_loss += loss.item()
print(f"Epoch {epoch+1} Training Loss: {train_loss/len(train_dataloader)}")
# Assume "model" is a trained Scikit-learn model
joblib.dump(bert_model, "bert_model.pkl")
# Evaluate the model on the val set
bert_model.eval()
with torch.no_grad():
val_loss = 0.0
val_correct = 0
predictions = []
for batch in val_dataloader:
input_ids = batch[0].to(device)
attention_mask = batch[1].to(device)
labels = batch[2].to(device)
outputs = bert_model(input_ids, attention_mask=attention_mask, labels=labels)
loss = outputs.loss
logits = outputs.logits
probs = torch.nn.functional.softmax(logits, dim=-1)
preds = torch.argmax(probs, dim=-1)
predictions.extend(preds.cpu().numpy())
from sklearn.metrics import classification_report
# Generate the classification report
report = classification_report(d2_val_labels, predictions)
print(report)
# Model prediction on test dataset
d1_test_data["sentiment"] = 0
# Tokenize the text and add special tokens
test_tokens = tokenizer.batch_encode_plus(
d1_test_data["text"].tolist(),
add_special_tokens=True,
max_length=MAX_LEN,
truncation=True,
padding=True,
return_attention_mask=True,
return_tensors="pt",
)
# Pad the input sequences to have equal lengths
test_input_ids = pad_sequences(
test_tokens["input_ids"],
maxlen=MAX_LEN,
dtype="long",
value=0,
truncating="post",
padding="post",
)
test_attention_masks = pad_sequences(
test_tokens["attention_mask"],
maxlen=MAX_LEN,
dtype="long",
value=0,
truncating="post",
padding="post",
)
# Create PyTorch dataset
test_dataset = torch.utils.data.TensorDataset(
torch.tensor(test_input_ids), torch.tensor(test_attention_masks)
)
# Create PyTorch dataloader
test_dataloader = DataLoader(
test_dataset, sampler=SequentialSampler(test_dataset), batch_size=BATCH_SIZE
)
# Set the model to evaluation mode
bert_model.eval()
# Lists to store predictions
predictions = []
# Deactivate gradient computations
with torch.no_grad():
for batch in test_dataloader:
input_ids = batch[0].to(device)
attention_mask = batch[1].to(device)
# Forward pass
outputs = bert_model(input_ids, attention_mask=attention_mask)
# Get the predicted class
_, preds = torch.max(outputs[0], dim=1)
# Append predictions to the list
predictions.extend(preds.tolist())
# Add the predicted labels to the test dataset
d1_test_data["sentiment"] = np.array(predictions)
d1_test_data.head()
# Save the results to a new CSV file
bert_result = d1_test_data[["id", "sentiment"]]
bert_result.to_csv("Submission_BertModel.csv", index=False)
|
#
# # Project Connect: Bridging the Education Gap for Marginalized Students in Trinidad and Tobago
# ## ✨Team Pandalytics✨
# 🐼 Giatri Lalla
# 🐼 Kirstin Sylvester
# 🐼 Arianne Baliramsingh
# 🐼 Kira Mohammed
# ## Table of contents 📖
# * Introduction
# * Methodology
# * Results
#
# ## Introduction
# This project helps migrant/disenfranchised children of school age, access formal education through the publishing of a carefully curated public Dashboard with information for the parents, members in a Government Office and the public to help with this task. The focus group used is the Venezuelan migrant population in Trinidad and Tobago.
#
# ## Methodology
# This project used a combination of methods such as feature correlation to determine the factors influencing the settlement locations of Venezuelan migrants in Trinidad. This was then used to determine the variance of the population in these areas.
#
# ### 1. Communities in Trinidad were ranked on attractiveness factors represented by a numerical score.
#
import pandas as pd
# Load the Excel file
data_file = pd.ExcelFile("/kaggle/input/datatest/data.xlsx")
# Read data from the Excel file into a Pandas DataFrame
df = data_file.parse("Sheet1", index_col="Community")
# Calculate the average score for each community
unweighted_scores = df.mean(axis=1)
# Rank communities
ranked_communities = unweighted_scores.sort_values(ascending=False)
# Print a rank list of all communities
print("Community rank list:")
for rank, (community, score) in enumerate(ranked_communities.iteritems()):
print(f"{rank+1}. {community}: {score:.2f}")
import pandas as pd
import matplotlib.pyplot as plt
# Load the Excel file
data_file = pd.ExcelFile("/kaggle/input/datatest/data.xlsx")
# Read data from the Excel file into a Pandas DataFrame
df = data_file.parse("Sheet1", index_col="Community")
# Calculate the average score for each community
unweighted_scores = df.mean(axis=1)
# Rank communities
ranked_communities = unweighted_scores.sort_values(ascending=False)
# Create a bar chart of community rankings
plt.barh(ranked_communities.index, ranked_communities.values)
plt.xlabel("Attractiveness Rating")
plt.ylabel("Community")
plt.title("Community Attractiveness Rankings")
plt.show()
# ### 2. Community ranks were compared against each other.
import pandas as pd
import matplotlib.pyplot as plt
# Load the Excel file
data_file = pd.ExcelFile("/kaggle/input/datatest/data.xlsx")
# Read data from the Excel file into a Pandas DataFrame
df = data_file.parse("Sheet1", index_col="Community")
# Calculate the average score for each community
unweighted_scores = df.mean(axis=1)
# Rank communities
ranked_communities = unweighted_scores.rank(ascending=False, method="dense")
df["Rank"] = ranked_communities.astype(int)
# Reverse the order of the y-axis so that the communities with the highest rank are listed at the top of the chart
ranked_communities = ranked_communities.iloc[::-1]
# Create a horizontal bar chart of community rankings
plt.barh(ranked_communities.index, ranked_communities.values)
plt.xlabel("Rank", fontsize=12)
plt.ylabel("Community", fontsize=12)
plt.title("Community Rankings", fontsize=14)
plt.grid(axis="x", linestyle="--")
plt.gca().invert_yaxis()
plt.show()
# ### 3. Feature Importance Analysis
import pandas as pd
import matplotlib.pyplot as plt
# Load the Excel file
data_file = pd.ExcelFile("/kaggle/input/datatest/data.xlsx")
# Read data from the Excel file into a Pandas DataFrame
df = data_file.parse("Sheet1", index_col="Community")
# Calculate the average score for each community
unweighted_scores = df.mean(axis=1)
# Rank communities
ranked_communities = unweighted_scores.rank(ascending=False, method="dense")
df["Rank"] = ranked_communities.astype(int)
# Calculate the correlation between each feature and the ranking
correlations = df.corrwith(ranked_communities)
# Select top 10 variables with highest absolute correlation values
top_10 = correlations.abs().nlargest(10)
# Create a bar chart of feature importance for top 10 variables
ax = top_10.plot(kind="barh", figsize=(8, 6), color="blue", alpha=0.8)
# Set x-axis label
ax.set_xlabel("Correlation with Rank", fontsize=12)
# Set y-axis label
ax.set_ylabel("Feature", fontsize=12)
# Set chart title
ax.set_title("Feature Importance Analysis", fontsize=14)
# Set ticks font size
ax.tick_params(axis="both", which="major", labelsize=10)
# Remove top and right spines
ax.spines["top"].set_visible(False)
ax.spines["right"].set_visible(False)
# Add horizontal grid lines
ax.grid(axis="x")
plt.show()
# ### 4. Correlating features with Attractiveness Rating
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
# Load the Excel file
data_file = pd.ExcelFile("/kaggle/input/datatest/data.xlsx")
# Read data from the Excel file into a Pandas DataFrame
df = data_file.parse("Sheet1", index_col="Community")
# Calculate the average score for each community
unweighted_scores = df.mean(axis=1)
# Rank communities
ranked_communities = unweighted_scores.rank(ascending=False, method="dense")
df["Rank"] = ranked_communities.astype(int)
# Calculate the correlation between each feature and the AR
correlations = df.corrwith(unweighted_scores)
# Get the top 11 features with the highest correlation with the AR
top_features = correlations.abs().sort_values(ascending=False)[:11]
# Print the top 11 features with their correlation with the AR
print("Top 11 features affecting the attractiveness rating (AR):\n")
for feature in top_features.index:
print(f"{feature}: {correlations[feature]:.2f}")
# Create a heatmap showing the AR against the top 11 features
fig, ax = plt.subplots(figsize=(8, 6))
heatmap_data = df[top_features.index.tolist() + ["Rank"]].corr()
sns.heatmap(heatmap_data, cmap="coolwarm", annot=True, ax=ax)
# Set the title and axis labels
ax.set_title("Attractiveness Rating vs. Top 11 Features")
ax.set_xlabel("Features")
ax.set_ylabel("Features")
# Save the heatmap as a PNG image
plt.savefig("heatmap.png", dpi=300)
# ## Results
# ### 1. Population change analysis (2000-2010)
import pandas as pd
# Read the CSV file into a Pandas DataFrame, specifying the data type for '2000 Population' and '2010 Population' as integers
data = pd.read_csv(
"/kaggle/input/dataset3/data.csv",
dtype={"2000 Population": int, "2010 Population": int},
sep=",",
thousands=" ",
)
# Calculate the population change percentage for each community from 2000 to 2010
data["Population Change"] = (
(data["2010 Population"] - data["2000 Population"]) / data["2000 Population"]
) * 100
# Print the top 10 communities with the highest population change percentage
print(data.sort_values("Population Change", ascending=False).head(10))
# ### 2. Population change analysis (2023)
# Read the CSV file into a Pandas DataFrame, specifying the data type for '2000 Population' and '2010 Population' as integers
data = pd.read_csv(
"/kaggle/input/dataset3/data.csv",
dtype={"2000 Population": int, "2010 Population": int},
sep=",",
thousands=" ",
)
# Calculate the population change percentage for each community from 2000 to 2010
data["Population Change"] = (
(data["2010 Population"] - data["2000 Population"]) / data["2000 Population"]
) * 100
# Calculate the estimated population for 2023 based on the population change percentage
data["2023 Population"] = data["2010 Population"] * (
(data["Population Change"] / 100) + 1
)
# Print the top 10 communities with the highest estimated population for 2023
print(data.sort_values("2023 Population", ascending=False).head(10))
|
# ## Multi-Class Classification:
# In machine learning, multi-class classification refers to the problem of classifying instances into one of three or more classes. For example, a multi-class classifier might be trained to classify an image as one of several different types of animals, such as a dog, cat, or bird. This is in contrast to binary classification that we saw in our supervised learning workshop, which is the problem of classifying instances into one of two classes.
# In this tutorial we are going to use the MNIST dataset.
# MNIST is a dataset of handwritten digits, comprising a training set of 60,000 examples and a test set of 10,000 examples . It is widely used as a benchmark for image classification and machine learning algorithms, as well as a simple dataset for testing and experimenting with machine learning techniques.
# Each example in the MNIST dataset consists of a 28x28 grayscale image of a handwritten digit (0-9) and a corresponding label indicating the correct digit.
# The MNIST dataset is a popular choice for testing machine learning algorithms because it is relatively small, easy to work with, and well-studied. It is also widely available, with many libraries and frameworks including built-in support for loading and working with the MNIST dataset.
# ### Loading the data:
#
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.tree import DecisionTreeClassifier
from sklearn.metrics import accuracy_score, confusion_matrix, classification_report
import warnings
warnings.filterwarnings("ignore") # this is to ignore warnings generated by code
train = "/kaggle/input/digit-recognizer/train.csv"
mnist = pd.read_csv(train)
mnist.head()
# ### Getting the Features and the Target:
# Since this is a classification problem, it is crucial to identify the target; meaning the variable we are trying the predict, and the features we are going to use for our predictions, Scikit-learn makes it easy for us by dividing the MNIST dataset into "data" and "target", the features we are using in our prediction is in the values of the dictionnary key "data" and our target is in the values of the dictionnary key "target".
X, y = mnist.drop(["label"], axis=1), mnist["label"]
# #### A bit of exploration :
#
y.unique()
# We can see that our target has 10 categories, which corresponds to the digits, this is our target; what we are trying to predict, remember that our objectif is to see what handwritten digit corresponds to!
first_image = X.iloc[0]
first_image = first_image.to_numpy().reshape(28, 28)
plt.imshow(first_image, cmap="binary")
plt.axis("off")
plt.show
# That allows us to see that every observation in the dataset corresponds to a hand written digit, in this case it corresponds to a badly written 1.
# Now that we've understood a bit more what our objectif entails and what our data is, we can start dealing with our problem.
# ### Splitting the data into a train and test set:
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=42
)
# ## Understanding how multiclass classification works:
# Remember that in binary classification, we have two classes (example A and B), with one being the positive class and the other being the negative class. A binary classification can be a classifier that distinguishes between A and NOT A, for example in the MNIST dataset we can build a classifier that distinguishes between 5 and not 5.
# In Multiclass classification there are more than two classes so there has to be a way to work around it.
# There are several approaches to performing multiclass classification, including one-vs-all and one-vs-one.
# In the one-versus-all (OVA) approach, a separate binary classifier is trained for each class, with the negative class being the combination of all classes other than the one being considered. For example, if you have a dataset with three classes (A, B, and C), the OVA approach would train three binary classifiers:
# * One classifier to distinguish between class A and the combination of classes B and C.
# * One classifier to distinguish between class B and the combination of classes A and C.
# * One classifier to distinguish between class C and the combination of classes A and B.
# At test time, all of the classifiers are applied to the test data and the class that results in the highest score is chosen as the prediction.
# In the one-versus-one (OVO) approach, a separate binary classifier is trained for each pair of classes. For example, if you have a dataset with three classes (A, B, and C), the OVO approach would train three binary classifiers:
# * One classifier to distinguish between classes A and B.
# * One classifier to distinguish between classes A and C.
# * One classifier to distinguish between classes B and C.
# At test time, all of the classifiers are applied to the test data and the class that wins the most pairwise comparisons is chosen as the prediction.
# The OVA approach is generally faster to train and predict than the OVO approach, but the OVO approach is generally more accurate. The choice between the two approaches depends on the specific problem you are trying to solve and the trade-off you are willing to make between speed and accuracy.
# ### Training a Logistic regression model:
# You must've came across Logistic Regression, which is a widely known classifier, we saw its application on a binary classification problem, here we are going to apply it to a multiclass classification problem.
# Logistic regression can be used for multiclass classification. By default, logistic regression uses a one-versus-all (OVA) approach to multiclass classification, where a separate binary classifier is trained for each class, with the negative class being the combination of all classes other than the one being considered.
#
lrc = LogisticRegression(
multi_class="multinomial"
) # we add the multi_class argument to do multiclass classification
lrc.fit(X_train, y_train)
some_digit = X_test.iloc[[0]]
some_digit_pred = lrc.predict(some_digit)
some_digit_pred
y_test.iloc[[0]]
# Looks like our model predicted correctly in this one case! Now let's see our model's performance a bit more closely!
y_pred = lrc.predict(X_test)
# #### Accuracy score and confusion matrix:
print(accuracy_score(y_test, y_pred))
confusion = confusion_matrix(y_test, y_pred)
sns.heatmap(confusion, annot=True, fmt="d")
plt.xlabel("Predicted")
plt.ylabel("True")
plt.show()
# #### Classification report:
print(classification_report(y_test, y_pred))
# Looks like our model does pretty well, it manages to have a good accuracy score, and also great precision, recall and f1 score on every class! The confusion matrix also shows us that the class are predicted well most of the time. We can notice few problems however:
# * The model is confused more often when it comes to 8 and 5, for example a lot of 5's get wronly classified as 8 or 3, that might because of the similarities of the two. This could be a problem with the way that the handwritten digit was drawn.
# If you want to force ScikitLearn to use one-versus-one or one-versus-all, you can use the OneVsOneClassifier or OneVsRestClassifier classes.
from sklearn.multiclass import OneVsOneClassifier
ovo_lrc = OneVsOneClassifier(LogisticRegression(random_state=42))
ovo_lrc.fit(X_train, y_train)
ovo_lrc.predict(some_digit)
# ### Trying out other models:
# It is generally good practice to try out other models before settling on one model in particular, because a model can be performing well, but be slightly worse than another model, and that slight improvement can make the difference.
# #### Decision Tree Classifier:
# Decision trees works by constructing a tree-like model of decisions based on features of an instance. Each internal node in the tree represents a "test" on an attribute, each branch represents the outcome of the test, and each leaf node represents a class label. The tree is constructed by starting at the root node and then adding internal nodes and leaves as needed to split the instances into their appropriate classes.
# To classify a new instance, the algorithm starts at the root of the tree and traverses the tree until it reaches a leaf node. The class label associated with that leaf node is then returned as the prediction for the instance.
# Decision trees are popular because they are easy to understand and interpret, and they can handle both numerical and categorical data. They are also relatively fast to train, and they can scale well to large datasets.
# Example :
# 
dtc = DecisionTreeClassifier(random_state=42)
dtc.fit(X_train, y_train)
dtc_pred = dtc.predict(X_test)
# #### Accuracy score and Confusion Matrix:
print(accuracy_score(y_test, dtc_pred))
confusion = confusion_matrix(dtc_pred, y_pred)
sns.heatmap(confusion, annot=True, fmt="d")
plt.xlabel("Predicted")
plt.ylabel("True")
plt.show()
# #### Classification report:
#
from sklearn.metrics import classification_report
print(classification_report(y_test, dtc_pred))
|
import pandas as pd
import csv
import os
print(os.listdir())
# DATAFRAME
#
pd.DataFrame({"Yes": [50, 21], "No": [131, 2]})
# DataFrame with noninteger values
#
pd.DataFrame(
{
"Bob": ["I liked it.", "It was awful."],
"Ashley": ["Pretty good.", "Bland."],
"Travis": ["It was okay.", "I did not like it."],
}
)
# Data Frame with an row name
pd.DataFrame(
{
"Bob": ["I liked it.", "It was awful."],
"Ashley": ["Pretty good.", "Bland."],
"Travis": ["It was okay.", "I did not like it."],
},
index=[
"Clydes Brew",
"Clydes Coffee",
],
)
# A series, kind of like a dataframe but less
#
pd.Series([1, 2, 3, 4, 5])
# A series when you only need one column of data and a row name
#
pd.Series(
[90, 95, 40], index=["Python grade", "Linux Grade", "Assembly Grade"], name="Grades"
)
# A dataframe is just a bunch of series glued together.
# read from a csv file
pandemic_stats = pd.read_csv(
"/kaggle/input/pandemic-details-of-world/pandemic_details_of_world.csv"
)
pandemic_stats.shape
# we probably want to preview our material but not look at all of it
# we can do that with the head command
# head() will default to 5 rows. or we can add an int to decide how many rows we want to preview
pandemic_stats.head(2)
# the first column here is unnammed because it just represents the row number. we should establish that's what that is so we don't double document
pandemic_stats = pd.read_csv("pandemic_stats.csv", index_col=0)
pandemic_stats.head(2)
# our dataframe works similar to python objects. this allows us to access parts of our table the same way
pandemic_stats.head(3).Disease
pandemic_stats["Death toll"][1]
# let's select data based on a row number
pandemic_stats.iloc[1]
pandemic_stats.loc[0][1]
# iloc is for index location and works similar but different than the loc command
pandemic_stats.loc[0:1, 0]
pandemic_stats.iloc[0:1, 0]
pandemic_stats.iloc[:3, 2]
pandemic_stats.head(1)
# the first digit is the range of rows. : alone means everything or if inbetween ints it can be a range
pandemic_stats.iloc[:3, 2:5]
pandemic_stats.loc[0, "Location"]
pandemic_stats.loc[:4, ["Disease", "Date"]]
# For information about choosing between loc and iloc, then check out the powerpoint
# Manipulating the index
pandemic_stats.set_index("Epidemics/pandemics").head(3)
# Conditional Selection
pandemic_stats.Location == "Worldwide"
# More useful combine location and conditional selection
pandemic_stats.loc[pandemic_stats.Location == "Worldwide"].head()
# combining conditions, notice it's only '|' pipe or '&'
pandemic_stats.loc[
(pandemic_stats.Location == "Worldwide") & (pandemic_stats.Disease == "COVID-19")
]
pandemic_stats.loc[
(pandemic_stats.Location == "Worldwide") | (pandemic_stats.Disease == "COVID-19")
]
# you can basically do the same thing but use the built in pandas .isin
pandemic_stats.loc[pandemic_stats.Location.isin(["Asia", "North America"])]
# there is a .isnull and a .notnull as well
pandemic_stats.loc[pandemic_stats["Death toll"].notnull()]
# assigning data
pandemic_stats["Date"] = "today"
pandemic_stats.head()
# reset
pandemic_stats = pd.read_csv(
"/kaggle/input/pandemic-details-of-world/pandemic_details_of_world.csv"
)
pandemic_stats.head()
|
# # Prompt 范式Pattern-Exploiting Training
# 1. [训练数据介绍](#1)
# 2. [BertForMaskedLM使用](#2)
# 3. [超参数](#3)
# 4. [数据预处理](#4)
# 4.1[数据集类](#4.1)
# 4.2[dataloader](#4.2)
# 5. [训练](#5)
#
# # 1. 训练数据 [↑](#top)
# 训练数据如下:
# ```
# 电池时间短,无线网卡上不去,模板是10寸的,不是8.9的,没有想象中小。 0
# 小巧,美观 1
# ```
# 前面用`很[MASK],`做拼接:
# ```
# 很[MASK],电池时间短,无线网卡上不去,模板是10寸的,不是8.9的,没有想象中小。
# 很[MASK],小巧,美观
# ```
# 丢到BertForMaskedLM
# 训练时让分别让两个MASK经过linear 一个越接近好越好,一个越接近差越好。
# 推理时比较映射到两个词的概率,选概率大的
# # 2. BertForMaskedLM [↑](#top)
# BertForMaskedLM是Hugging Face的Transformers库中的一个类,它是基于谷歌公司发布的BERT(Bidirectional Encoder Representations from Transformers)模型的预训练模型,用于填充被遮盖(masked)的词汇。
# 在自然语言处理(NLP)中,填空题通常是一种常见的语言理解任务。在这种任务中,模型需要根据上下文来预测被遮盖的词汇,以此来理解文本中的含义。BertForMaskedLM库为这种任务提供了一种预训练模型。
# BertForMaskedLM库中的模型通过输入一段文本和一些被遮盖的词汇来生成文本中每个被遮盖的词汇的可能预测值。它通过一个双向Transformer编码器来建模上下文,并使用一个全连接层来预测每个被遮盖的词汇的可能预测值。这使得模型能够在填空任务中表现出色,并在多种NLP任务中取得了最先进的性能。
# 总之,BertForMaskedLM库提供了一个基于BERT预训练模型的方法来填补被遮盖的词汇,并在多个NLP任务中取得了很好的效果。
#
# BertForMaskedLM可以理解成bert 模型后面接了一个linear把bert 的 768维向量转成单词表长度维数
# [文档链接](https://huggingface.co/docs/transformers/v4.27.2/en/model_doc/bert#transformers.BertForMaskedLM)
import pdb
from transformers import AutoTokenizer, BertForMaskedLM
import torch
tokenizer = AutoTokenizer.from_pretrained(
"/kaggle/input/huggingface-bert/bert-base-uncased"
)
model = BertForMaskedLM.from_pretrained(
"/kaggle/input/huggingface-bert/bert-base-uncased"
)
print("vocab_size", tokenizer.vocab_size)
print("输出向量的维度", model.cls.predictions.decoder.out_features)
inputs = tokenizer("The capital of France is [MASK].", return_tensors="pt")
with torch.no_grad():
logits = model(**inputs).logits
# pdb.set_trace()
# 找到mask 再句子中分词后的索引
mask_token_index = (inputs.input_ids == tokenizer.mask_token_id)[0].nonzero(
as_tuple=True
)[0]
predicted_token_id = logits[0, mask_token_index].argmax(axis=-1)
tokenizer.decode(predicted_token_id)
"paris"
labels = tokenizer("The capital of France is Paris.", return_tensors="pt")["input_ids"]
# mask labels of non-[MASK] tokens
labels = torch.where(inputs.input_ids == tokenizer.mask_token_id, labels, -100)
outputs = model(**inputs, labels=labels)
round(outputs.loss.item(), 2)
model
model = BertForMaskedLM.from_pretrained(
"/kaggle/input/huggingface-bert/bert-base-chinese"
)
model
tokenizer = tokenizer = AutoTokenizer.from_pretrained(
"/kaggle/input/huggingface-bert/bert-base-chinese"
)
tokenizer
# # 3. 超参数[↑](#top)
import os
def fix_seed(seed):
import numpy as np
import random
random.seed(seed)
os.environ["PYTHONHASHSEED"] = str(seed)
np.random.seed(seed)
torch.manual_seed(seed)
torch.cuda.manual_seed(seed)
torch.backends.cudnn.deterministic = True
fix_seed(199)
max_length = 200
batch_size = 16
lr = 1e-4
lr = 1e-5
model_path = "/kaggle/input/huggingface-bert/bert-base-chinese"
device = "cuda"
train_path = "/kaggle/input/sentiment/sentiment.train.data"
valid_path = "/kaggle/input/sentiment/sentiment.valid.data"
test_path = "/kaggle/input/sentiment/sentiment.test.data"
train_num = 20
valid_num = None # 取全部
prefix = "很好,"
mask_index = 2 # CLS 很 好 好的索引是2
epochs = 100
# # 4. 数据预处理 [↑](#top)
# ## 4.1 数据集类 [↑](#top)
import torch
import torch.nn.utils.rnn as rnn_utils
class Dataset(torch.utils.data.Dataset):
def __init__(self, input_ids, attention_masks, target_ids, data_length):
self.input_ids = input_ids
self.attention_masks = attention_masks
self.target_ids = target_ids
self.data_length = data_length
def __getitem__(self, idx):
if self.target_ids is None:
return self.input_ids[idx], self.attention_masks[idx]
else:
return self.input_ids[idx], self.attention_masks[idx], self.target_ids[idx]
def __len__(self):
return self.data_length
# ## 4.2 创建dataloader [↑](#top)
from transformers.models.bert import BertTokenizerFast
tokenizer = BertTokenizerFast.from_pretrained(model_path)
def pattern_data(data, label):
all_input_ids = []
all_attention_mask = []
all_target_ids = []
for d in zip(data, label):
# [CLS] ..... [SEP]
# 在句子前面加 ‘很好,’
text = prefix + d[0]
encoding = tokenizer(text, return_tensors="np")
# 输入值的下标
input_ids = encoding["input_ids"][0]
# 输出值的下标
target_ids = [-100] * len(input_ids)
attention_mask = encoding["attention_mask"][0]
# positive
if d[1] == 1:
input_ids[mask_index] = tokenizer.mask_token_id
target_ids[mask_index] = tokenizer.convert_tokens_to_ids("好")
# negative
elif d[1] == 0:
input_ids[mask_index] = tokenizer.mask_token_id
target_ids[mask_index] = tokenizer.convert_tokens_to_ids("差")
all_input_ids.append(torch.tensor(input_ids[:max_length]))
all_attention_mask.append(torch.tensor(attention_mask[:max_length]))
all_target_ids.append(torch.tensor(target_ids[:max_length]))
return all_input_ids, all_attention_mask, all_target_ids
# 创建dataloader
def get_data_loader(data_path, data_num=None):
train_text = []
train_label = []
if not data_num:
data_num = float("inf")
with open(data_path, encoding="utf-8") as file:
for index, line in enumerate(file.readlines()):
t, l = line.strip().split("\t")
train_text.append(t)
train_label.append(int(l))
if index == data_num:
break
train_dataset = Dataset(*pattern_data(train_text, train_label), len(train_text))
return torch.utils.data.DataLoader(
train_dataset, batch_size=batch_size, collate_fn=collate_fn
)
def collate_fn(batch):
input_ids = rnn_utils.pad_sequence(
list(zip(*batch))[0], batch_first=True, padding_value=tokenizer.pad_token_id
)
attention_masks = rnn_utils.pad_sequence(
list(zip(*batch))[1], batch_first=True, padding_value=0
)
target_ids = rnn_utils.pad_sequence(
list(zip(*batch))[2], batch_first=True, padding_value=-100
)
if len(list(zip(*batch))) == 3:
return input_ids, attention_masks, target_ids
else:
return input_ids, attention_masks
train_loader = get_data_loader(train_path, data_num=train_num)
valid_loader = get_data_loader(valid_path)
# # 5 训练
from transformers import AdamW
from tqdm import tqdm
model = BertForMaskedLM.from_pretrained(
"/kaggle/input/huggingface-bert/bert-base-uncased"
)
model = model.to(device)
optimizer = AdamW(model.parameters(), lr=lr)
def train_step(data, mode):
input_ids = data[0].to(device).long()
attention_mask = data[1].to(device).long()
target_ids = data[2].to(device).long()
outputs = model(input_ids, attention_mask=attention_mask, labels=target_ids)
if mode == "dev":
output = outputs.logits
# 得到标签
labels = target_ids[:, mask_index].cpu().numpy()
# 标签映射
label_dict = {
tokenizer.convert_tokens_to_ids("好"): 1,
tokenizer.convert_tokens_to_ids("差"): 0,
}
labels = [label_dict[l] for l in labels]
# 取label对应的logits
pos_logits = output[
:, mask_index, tokenizer.convert_tokens_to_ids("好")
].unsqueeze(0)
neg_logits = output[
:, mask_index, tokenizer.convert_tokens_to_ids("差")
].unsqueeze(0)
# 这里需要注意位置,neg才是0
logits = torch.cat([neg_logits, pos_logits], dim=0)
# 取最大的下标也就是label
y_pred = torch.argmax(logits, dim=0).cpu().numpy()
return outputs.loss, y_pred, labels
return outputs.loss
from sklearn.metrics import (
accuracy_score,
f1_score,
recall_score,
precision_score,
classification_report,
)
def calculate_metrics(y_true, y_pred):
acc = 0
recall = 0
precision = 0
f1 = 0
acc = accuracy_score(y_true, y_pred)
recall = recall_score(y_true, y_pred, average="macro")
precision = precision_score(y_true, y_pred, average="macro")
f1 = f1_score(y_true, y_pred, average="macro")
return {"acc": acc, "recall": recall, "precision": precision, "f1": f1}
def train_func(loader):
pbar = tqdm(loader)
for batch in pbar:
optimizer.zero_grad()
loss = train_step(batch, mode="train")
loss.backward()
optimizer.step()
pbar.update()
pbar.set_description(f"loss:{loss.item():.4f}")
def dev_func(loader):
dev_loss = 0
all_label = []
all_pred = []
metrics = {}
for batch in tqdm(loader):
with torch.no_grad():
loss, output, label = train_step(batch, "dev")
dev_loss += loss.item()
if output is not None:
all_label.extend(label)
all_pred.extend(output)
# 打印评价指标
if all_pred is not None:
metrics = calculate_metrics(all_label, all_pred)
print(metrics)
return metrics["acc"]
def main():
dev_metric = 0
for epoch in range(epochs):
print(f"***********epoch: {epoch + 1}***********")
train_func(train_loader)
metric = dev_func(valid_loader)
if dev_metric < metric:
dev_metric = metric
print(f"best acc:{dev_metric}")
main()
|
from keras.preprocessing import sequence
from keras.models import Sequential
from keras.layers import Dense, Embedding
from keras.layers import LSTM
from keras.datasets import imdb
(x_train, y_train), (x_test, y_test) = imdb.load_data(num_words=2000)
from keras.utils import pad_sequences
x_train = pad_sequences(x_train, maxlen=15)
x_test = pad_sequences(x_test, maxlen=15)
model = Sequential()
model.add(Embedding(2000, 128))
model.add(LSTM(128, dropout=0.2, recurrent_dropout=0.2))
model.add(Dense(1, activation="sigmoid"))
model.compile(loss="binary_crossentropy", optimizer="adam", metrics=["accuracy"])
model.fit(x_train, y_train, batch_size=32, epochs=7, validation_data=(x_test, y_test))
score, acc = model.evaluate(x_test, y_test, batch_size=32)
print("Test score:", score)
print("Test accuracy:", acc)
|
import pandas as pd
from tqdm import tqdm
import nltk
import spacy
from sklearn.model_selection import train_test_split
from sklearn.metrics import (
f1_score,
accuracy_score,
classification_report,
confusion_matrix,
)
from sklearn.feature_extraction.text import CountVectorizer, TfidfVectorizer
from sklearn.ensemble import GradientBoostingClassifier, RandomForestClassifier
import seaborn as sns
from matplotlib import pyplot as plt
import numpy as np
from wordcloud import WordCloud
pd.options.plotting.backend = "matplotlib"
df_train = pd.read_csv("/kaggle/input/nlp-getting-started/train.csv")
df_test = pd.read_csv("/kaggle/input/nlp-getting-started/test.csv")
df_train.tail(20)
df_train.info()
# df_train = df_train.dropna()
df_train.describe()
df_train.target.value_counts().plot(kind="bar")
nltk.download("stopwords")
snlp = spacy.load("en_core_web_sm")
def lemmatizer(text):
result = [word.lemma_ for word in snlp(text)]
return " ".join(result)
def preprocess(df):
df["text_lemma"] = df.text.apply(lemmatizer)
return df
stopwords = list(
set(snlp.Defaults.stop_words).union(set(nltk.corpus.stopwords.words("english")))
)
# # Count Vectorizer
vec_model = CountVectorizer(stop_words=stopwords, ngram_range=(1, 2))
df_train = preprocess(df_train)
negative_cases = "".join(df_train[df_train["target"] == 0]["text_lemma"].values)
positive_cases = "".join(df_train[df_train["target"] == 1]["text_lemma"].values)
fig, axs = plt.subplots(1, 2, figsize=(15, 10))
wc1 = WordCloud(background_color="white").generate(negative_cases)
wc2 = WordCloud(background_color="white").generate(positive_cases)
axs[0].imshow(wc1, interpolation="bilinear")
axs[0].set_title("Negative")
axs[1].imshow(wc2, interpolation="bilinear")
axs[1].set_title("Positive")
plt.show()
vectors = vec_model.fit_transform(df_train.text_lemma)
X = vectors.toarray()
y = df_train["target"].values
# vec_model.vocabulary_
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=42
)
X_train.shape, X_test.shape, y_train.shape, y_test.shape
# # Random Forest
random_forest = RandomForestClassifier(random_state=42)
random_forest.fit(X_train, y_train)
y_pred = random_forest.predict(X_test)
print(classification_report(y_test, y_pred))
cnf_matrix = confusion_matrix(y_test, y_pred)
cmap = sns.color_palette("Blues", as_cmap=True)
sns.heatmap(
cnf_matrix,
annot=True,
fmt="g",
cmap=cmap,
xticklabels=["Negativo", "Positivo"],
yticklabels=["Negativo", "Positivo"],
)
plt.title("Matriz de Confusão")
plt.xlabel("Valor Previsto")
plt.ylabel("Valor Real")
# - 848 amostras foram classificadas corretamente como pertencentes à classe negativa (ou verdadeiros negativos).
# - 36 amostras foram classificadas erroneamente como pertencentes à classe positiva, mas na verdade pertencem à classe negativa (ou falsos positivos).
# - 304 amostras foram classificadas erroneamente como pertencentes à classe negativa, mas na verdade pertencem à classe positiva (ou falsos negativos).
# - 336 amostras foram classificadas corretamente como pertencentes à classe positiva (ou verdadeiros positivos).
# # Submit dataset
df_new = pd.DataFrame()
df_new["id"] = df_test["id"]
vectors_test = vec_model.transform(df_test["text"])
df_new["target"] = random_forest.predict(vectors_test)
df_new
df_new.target.value_counts().plot(kind="bar")
df_new.shape
df_test2 = df_test.merge(df_new)
df_test2.head()
negative_cases = "".join(df_test2[df_test2["target"] == 0]["text"].values)
positive_cases = "".join(df_test2[df_test2["target"] == 1]["text"].values)
fig, axs = plt.subplots(1, 2, figsize=(15, 10))
wc1 = WordCloud(background_color="white").generate(negative_cases)
wc2 = WordCloud(background_color="white").generate(positive_cases)
axs[0].imshow(wc1, interpolation="bilinear")
axs[0].set_title("Negative")
axs[1].imshow(wc2, interpolation="bilinear")
axs[1].set_title("Positive")
plt.show()
df_new.to_csv("/kaggle/working/submission.csv", index=False)
|
# ### Analiza statystyk piłkarskich z portalu Transfermarkt - Wybór najlepszych zawodników do kadry narodowej
# W tym projekcie wykorzystam dane z bazy SQL, a następnie przeprowadzę analizę danych, korzystając z indeksów i rankingów. Celem projektu jest wybranie najlepszych piłkarzy dla każdej pozycji, którzy mogliby reprezentować drużynę narodową.
#
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import seaborn as sns
import matplotlib.pyplot as plt
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Read CSVs into dataframes
players = pd.read_csv("/kaggle/input/player-scores/players.csv")
clubs = pd.read_csv("/kaggle/input/player-scores/clubs.csv")
competitions = pd.read_csv("/kaggle/input/player-scores/competitions.csv")
appearances = pd.read_csv("/kaggle/input/player-scores/appearances.csv")
games = pd.read_csv("/kaggle/input/player-scores/games.csv")
# import sqlalchemy and create a sqlite engine
from sqlalchemy import create_engine
engine = create_engine("sqlite://", echo=False)
players.to_sql("players", con=engine)
clubs.to_sql("clubs", con=engine)
competitions.to_sql("competitions", con=engine)
appearances.to_sql("appearances", con=engine)
games.to_sql("games", con=engine)
pd.options.display.float_format = "{:.2f}".format
# ## Zapytanie SQL pozyskujące dane zawodników z bazy Transfermarkt
query = engine.execute(
"""SELECT
t1.player_name,
t1.country_of_citizenship,
t1.position,
t1.sub_position,
t1.sum_minutes_played,
t1.sum_goals,
t1.sum_assists,
t1.market_value_in_eur,
t1.GoalsPerMinute,
t1.AssistsPerMinute,
t1.LoseGoalsPerMinute,
PERCENT_RANK() OVER (PARTITION BY t1.sub_position ORDER BY t1.GoalsPerMinute) AS GoalsPercentile,
PERCENT_RANK() OVER (PARTITION BY t1.sub_position ORDER BY t1.AssistsPerMinute) AS AssistsPercentile,
PERCENT_RANK() OVER (PARTITION BY t1.sub_position ORDER BY t1.LoseGoalsPerMinute) AS LoseGoalsPercentile
FROM (
SELECT
player_name,
country_of_citizenship,
position,
sub_position,
SUM(minutes_played) AS sum_minutes_played,
SUM(goals) AS sum_goals,
SUM(assists) AS sum_assists,
SUM(games.away_club_goals),
market_value_in_eur,
COALESCE(SUM(minutes_played)/NULLIF(SUM(goals),0), 9999) AS GoalsPerMinute,
COALESCE(SUM(minutes_played)/NULLIF(SUM(assists),0), 9999) AS AssistsPerMinute,
COALESCE(SUM(minutes_played)/NULLIF(SUM(games.away_club_goals),0), 9999) AS LoseGoalsPerMinute
FROM players
INNER JOIN games ON appearances.game_id = games.game_id
INNER JOIN appearances ON appearances.player_id = players.player_id
WHERE games.season = '2021'
GROUP BY player_name, sub_position
HAVING SUM(minutes_played) >= 1500
) AS t1
ORDER BY t1.GoalsPerMinute"""
)
df = pd.DataFrame(query.fetchall())
df.columns = query.keys()
df
# ## Wykresy
# ### Pokazujące co jaki czas piłkarze na konkretnych pozycjach strzelaja,asystuja czy traca bramki
import matplotlib.pyplot as plt
# Filter out rows where sum_goals == 0 to avoid division by zero errors
df_filteredAtk = df[df["position"] == "Attack"]
df_filteredAtk = df_filteredAtk[df_filteredAtk["sum_goals"] > 1]
# Calculate minutes per goal
minutes_per_goal = df_filteredAtk["sum_minutes_played"] / df_filteredAtk["sum_goals"]
# Plot histogram
plt.hist(minutes_per_goal, bins=50)
plt.title("Minutes per Goal Histogram")
plt.xlabel("Minutes per Goal")
plt.ylabel("Frequency")
plt.show()
df_filteredDF = df[df["position"].isin(["Defender", "Goalkeeper"])]
df_filteredDF.loc[df["position"] == "Goalkeeper", "sub_position"] = "Goalkeeper"
sns.boxplot(x="sub_position", y="LoseGoalsPerMinute", data=df_filteredDF)
plt.xticks(rotation=90)
plt.show()
# df_filteredAtk = df[df['position'] == 'Attack']
df2 = df[df["sum_goals"] > 1]
# df_filteredAtk = df_filteredAtk[df_filteredAtk['sum_goals'] > 1]
sns.boxplot(x="sub_position", y="GoalsPerMinute", data=df2)
plt.xticks(rotation=90)
plt.show()
# df_filteredM = df[df['position'] == 'Midfield']
# df_filteredM = df_filteredM[df_filteredM['sum_assists'] > 1]
df2 = df[df["sum_assists"] > 1]
sns.boxplot(x="sub_position", y="AssistsPerMinute", data=df2)
plt.xticks(rotation=90)
plt.show()
df2 = df[df["sum_assists"] > 1]
sns.boxplot(x="sub_position", y="AssistsPerMinute", data=df2)
plt.xticks(rotation=90)
plt.show()
# ## Stworzenie Indeksów
# Aby ułatwić wybór odpowiednich piłkarzy, będziemy tworzyć indeksy dla każdej z pozycji na boisku: bramkarzy, obrońców, pomocników i napastników. Indeksy te będą uwzględniały statystyki dotyczące gry w danej pozycji, takie jak liczba straconych/oddanych bramek, liczba asyst, liczba rozegranych minut, itp.
# Dzięki tym indeksom, będziemy mogli wybrać piłkarzy, którzy osiągają najlepsze wyniki na swojej pozycji i zasługują na powołanie do kadry. Indeksy dla każdej pozycji będą opierać się na różnych kryteriach i będą wykorzystywać różne wagi dla poszczególnych statystyk, aby lepiej odzwierciedlać rolę i wymagania danej pozycji na boisku.
#
# Tworzenie kolumny 'GoalkeeperIndex' na podstawie wag
df["AttackIndex"] = df.apply(
lambda row: (1 - row["GoalsPercentile"]) * 0.8
+ (1 - row["AssistsPercentile"]) * 0.2
if row["position"] == "Attack"
else 0,
axis=1,
)
# Sortowanie wg wartości GoalkeeperIndex
df_Attackers = df[df["position"] == "Attack"].sort_values(
by="AttackIndex", ascending=False
)
# Tworzenie kolumny 'MidfielderIndex' na podstawie wag
df["MidfielderIndex"] = df.apply(
lambda row: (1 - row["LoseGoalsPercentile"]) * 0.5
+ (1 - row["AssistsPercentile"]) * 0.25
+ (1 - row["GoalsPercentile"]) * 0.25
if row["position"] == "Midfield"
else 0,
axis=1,
)
# Sortowanie wg wartości MidfielderIndex
df_midfielders = df[df["position"] == "Midfield"].sort_values(
by="MidfielderIndex", ascending=False
)
# Tworzenie kolumny 'DefenderIndex' na podstawie wag
df["DefenderIndex"] = df.apply(
lambda row: (1 - row["LoseGoalsPercentile"]) * 0.85
+ (1 - row["AssistsPercentile"]) * 0.05
+ (1 - row["GoalsPercentile"]) * 0.1
if row["position"] == "Defender"
else 0,
axis=1,
)
# Sortowanie wg wartości DefenderIndex
df_defenders = df[df["position"] == "Defender"].sort_values(
by="DefenderIndex", ascending=False
)
# Tworzenie kolumny 'GoalkeeperIndex' na podstawie wag
df["GoalkeeperIndex"] = df.apply(
lambda row: (1 - row["LoseGoalsPercentile"])
if row["position"] == "Goalkeeper"
else 0,
axis=1,
)
# Sortowanie wg wartości GoalkeeperIndex
df_goalkeepers = df[df["position"] == "Goalkeeper"].sort_values(
by="GoalkeeperIndex", ascending=False
)
df_goalkeepers = df_goalkeepers.loc[
df["country_of_citizenship"] == "England",
[
"player_name",
"country_of_citizenship",
"position",
"sub_position",
"sum_minutes_played",
"LoseGoalsPerMinute",
"market_value_in_eur",
"GoalkeeperIndex",
],
]
df_defenders = df_defenders.loc[
df["country_of_citizenship"] == "England",
[
"player_name",
"country_of_citizenship",
"position",
"sub_position",
"sum_minutes_played",
"sum_goals",
"sum_assists",
"LoseGoalsPerMinute",
"market_value_in_eur",
"DefenderIndex",
],
]
df_midfielders = df_midfielders.loc[
df["country_of_citizenship"] == "England",
[
"player_name",
"country_of_citizenship",
"position",
"sub_position",
"sum_minutes_played",
"sum_goals",
"sum_assists",
"market_value_in_eur",
"MidfielderIndex",
],
]
df_Attackers = df_Attackers.loc[
df["country_of_citizenship"] == "England",
[
"player_name",
"country_of_citizenship",
"position",
"sub_position",
"sum_minutes_played",
"sum_goals",
"sum_assists",
"market_value_in_eur",
"AttackIndex",
],
]
df_goalkeepers.sort_values(
by=["position", "GoalkeeperIndex"], ascending=[True, False], inplace=True
)
df_defenders.sort_values(
by=["sub_position", "DefenderIndex"], ascending=[True, False], inplace=True
)
df_midfielders.sort_values(
by=["sub_position", "MidfielderIndex"], ascending=[True, False], inplace=True
)
df_Attackers.sort_values(
by=["sub_position", "AttackIndex"], ascending=[True, False], inplace=True
)
# ## Prezentacja wyników
# Po stworzeniu indeksów dla każdej pozycji, wybierzemy piłkarzy z najlepszymi wynikami w każdym indeksie i przedstawimy je trenerowi jako propozycje do powołania do kadry.
top_goalkeepers = df_goalkeepers.groupby("position").head(5)
top_goalkeepers
top_defenders = df_defenders.groupby("sub_position").head(5)
top_defenders
top_midfielders = df_midfielders.groupby("sub_position").head(5)
top_midfielders
top_attackers = df_Attackers.groupby("sub_position").head(5)
top_attackers
|
import numpy as np # linear algebra
import pandas as pd
import seaborn as sns
from sklearn.model_selection import cross_val_score, train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_squared_error
from sklearn.neighbors import KNeighborsRegressor
import xgboost
import matplotlib.pyplot as plt
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv("/kaggle/input/housesalesprediction/kc_house_data.csv")
df1 = pd.read_csv("/kaggle/input/housesalesprediction/kc_house_data.csv")
# Let's use the head method to get a brief overview
df.head()
# We have 21 features to work with, but the columns of 'id, 'date' won't be of much use, let's discard them
df.drop(columns=["id", "date"], inplace=True)
# The dataset is of a particular county located in Seattle, there are 3 features that give us info on the location,namely the latitude, longitude and zipcode.Since all the houses are from the same area, including location information will hardly give us differentiating information, so let's drop them.
df.drop(columns=["lat", "long", "zipcode"], inplace=True)
df.columns
# We have 16 features left, all of which appear important so let's do some quick EDA to determine what to prioritize
sns.scatterplot(x=df["sqft_living"], y=df["price"])
fig, (sp1, sp2, sp3, sp4, sp5, sp6) = plt.subplots(1, 6, figsize=(10, 6))
sp1.scatter(df["sqft_living"], df["price"])
sp2.scatter(df["sqft_lot"], df["price"])
sp3.scatter(df["sqft_above"], df["price"])
sp4.scatter(df["sqft_basement"], df["price"])
sp5.scatter(df["sqft_living15"], df["price"])
sp6.scatter(df["sqft_lot15"], df["price"])
sp1.set_xlabel("sqft_living", fontsize=8)
sp1.set_ylabel("price", fontsize=8)
sp2.set_xlabel("sqft_lot", fontsize=8)
sp2.set_ylabel("price", fontsize=8)
sp3.set_xlabel("sqft_above", fontsize=8)
sp3.set_ylabel("price", fontsize=8)
sp4.set_xlabel("sqft_basement", fontsize=8)
sp4.set_ylabel("price", fontsize=8)
sp5.set_xlabel("sqft_living15", fontsize=8)
sp5.set_ylabel("price", fontsize=8)
sp6.set_xlabel("sqft_lot15", fontsize=8)
sp6.set_ylabel("price", fontsize=8)
sp3.set_title("Correlation of house area based features", fontsize=15, pad=40)
plt.show()
# Let's examine the no_of_bedrooms and bathrooms, to see how they are correlated with the target
sns.scatterplot(x=df["bedrooms"], y=df["price"])
sns.scatterplot(x=df["bathrooms"], y=df["price"])
# Grade parameter is very important as it directly gives us information on the overall quality of the house, which means it will play an important role in our predictor, so let's check it out.
sns.countplot(x=df["grade"])
sns.scatterplot(x=df["grade"], y=df["price"])
sns.countplot(x=df["view"])
sns.countplot(x=df["waterfront"])
# Only very few houses have a "view" and a "waterfront", so they can be safely ignored and dropped from the dataset.
df.drop(columns=["waterfront", "view"], inplace=True)
# Let's plot a correlation table and a heatmap to get a general idea on how the features are correlated
df_corr = df.corr()
df.corr().T
# write about the correlation table
sns.heatmap(df_corr)
# Let's begin building the model now, first we sperate the target and the dataset into training and test sets.
df_target = df["price"]
df.drop(columns=["price"], inplace=True)
X_train, X_test, y_train, y_test = train_test_split(
df1[features], df_target, test_size=0.3
)
regressors = [
[LinearRegression(), "Linear Regression"],
[KNeighborsRegressor(), "KNeighborsRegressor"],
[xgboost.XGBRegressor(), "XGB Regressor"],
]
for regressor in regressors:
model = regressor[0]
model.fit(X_train, y_train)
model_name = regressor[1]
score_list = []
pred = model.predict(X_test)
score = model.score(X_test, y_test)
score_list.append(score)
print(model_name, "model score: " + str(round(score * 100, 2)) + "%")
# Now let's try different models and check their performance metrics, starting from a simple Linear Regression and trying out more complex models
lin_reg = LinearRegression()
lin_reg.fit(X_train, y_train)
baseline_pred = lin_reg.predict(X_test)
lin_mse = mean_squared_error(y_test, baseline_pred)
print(lin_mse)
lin_reg.score(X_test, y_test)
knn = KNeighborsRegressor(n_neighbors=2)
knn.fit(X_train, y_train)
baseline_pred_knn = knn.predict(X_test)
print(baseline_pred)
knn.score(X_test, y_test)
xgb = xgboost.XGBRegressor(
n_estimators=100,
learning_rate=0.08,
gamma=0,
subsample=0.75,
colsample_bytree=1,
max_depth=7,
)
xgb.fit(X_train, y_train)
baseline_pred_xgb = xgb.predict(X_test)
print(baseline_pred_xgb)
score_xgb = xgb.score(X_test, y_test)
print(score_xgb)
|
import tensorflow as tf
from tensorflow.keras import models, layers
import matplotlib.pyplot as plt
import numpy as np
import pathlib
import os
os.environ["TF_CPP_MIN_LOG_LEVEL"] = "2" # to disable all debugging logs
# Global initialization of some imp variables
IMAGE_SIZE = 256
BATCH_SIZE = 32
CHANNELS = 3
EPOCHS = 50
train = tf.keras.preprocessing.image_dataset_from_directory(
"/kaggle/input/tomato/Tomato_images/train",
shuffle=True,
image_size=(IMAGE_SIZE, IMAGE_SIZE),
batch_size=BATCH_SIZE,
)
val = tf.keras.preprocessing.image_dataset_from_directory(
"/kaggle/input/tomato/Tomato_images/val",
shuffle=True,
image_size=(IMAGE_SIZE, IMAGE_SIZE),
batch_size=BATCH_SIZE,
)
test = tf.keras.preprocessing.image_dataset_from_directory(
"/kaggle/input/tomato/Tomato_images/test",
shuffle=True,
image_size=(IMAGE_SIZE, IMAGE_SIZE),
batch_size=BATCH_SIZE,
)
class_names = train.class_names
class_names
# Number of Batches = (total number of files belonging to all classes / Batch_Size)
len(train)
# prints Elements in dataset: here 1st element is image and 2nd index of that image.
print(train)
for image_batch, labels_batch in train.take(1):
print(image_batch.shape)
print(labels_batch.numpy())
plt.figure(figsize=(20, 10))
for image_batch, labels_batch in train.take(1):
for i in range(15):
ax = plt.subplot(3, 5, i + 1)
plt.imshow(image_batch[i].numpy().astype("uint8"))
plt.title(class_names[labels_batch[i]])
plt.axis("off")
# print("Size of Data is: {0} \nBatch size of Training Data is: {1}\nBatch size of Validation Data is: {2} \nBatch size of Testing Data is: {3} " .format(len(dataset), len(train), len(val), len(test)))
train.take(41)
# print("Size of Data is: {0} \nBatch size of Training Data is: {1}\nBatch size of Validation Data is: {2} \nBatch size of Testing Data is: {3} " .format(len(dataset), len(train_ds), len(val_ds), len(test_ds)))
train = train.cache().shuffle(1000).prefetch(buffer_size=tf.data.AUTOTUNE)
val = val.cache().shuffle(1000).prefetch(buffer_size=tf.data.AUTOTUNE)
test = test.cache().shuffle(1000).prefetch(buffer_size=tf.data.AUTOTUNE)
resize_and_rescale = tf.keras.Sequential(
[
layers.experimental.preprocessing.Resizing(IMAGE_SIZE, IMAGE_SIZE),
layers.experimental.preprocessing.Rescaling(1.0 / 255),
]
)
data_augmentation = tf.keras.Sequential(
[
layers.experimental.preprocessing.RandomFlip("horizontal_and_vertical"),
layers.experimental.preprocessing.RandomRotation(0.2),
]
)
input_shape = (BATCH_SIZE, IMAGE_SIZE, IMAGE_SIZE, CHANNELS)
n_classes = 3
model = models.Sequential(
[
resize_and_rescale,
data_augmentation,
layers.Conv2D(
32, kernel_size=(3, 3), activation="relu", input_shape=input_shape
),
layers.MaxPooling2D((2, 2)),
layers.Conv2D(64, kernel_size=(3, 3), activation="relu"),
layers.MaxPooling2D((2, 2)),
layers.Conv2D(64, kernel_size=(3, 3), activation="relu"),
layers.MaxPooling2D((2, 2)),
layers.Conv2D(64, (3, 3), activation="relu"),
layers.MaxPooling2D((2, 2)),
layers.Conv2D(64, (3, 3), activation="relu"),
layers.MaxPooling2D((2, 2)),
layers.Conv2D(32, (3, 3), activation="relu"),
layers.MaxPooling2D((2, 2)),
layers.Flatten(),
layers.Dense(32, activation="relu"),
layers.Dense(n_classes, activation="softmax"),
]
)
model.build(input_shape=input_shape)
model.summary()
model.compile(
optimizer="adam",
loss=tf.keras.losses.SparseCategoricalCrossentropy(from_logits=False),
metrics=["accuracy"],
)
history = model.fit(
train, batch_size=BATCH_SIZE, validation_data=val, verbose=1, epochs=50
)
# # VGG16
from tensorflow.keras.applications.vgg16 import VGG16
input_shape = (IMAGE_SIZE, IMAGE_SIZE, CHANNELS)
VGG_16 = VGG16(input_shape=input_shape, weights="imagenet", include_top=False)
VGG_16 = models.Sequential(
[
resize_and_rescale,
data_augmentation,
vgg1,
layers.Flatten(),
# layers.Dense(64, activation='relu'),
layers.Dense(n_classes, activation="softmax"),
]
)
VGG_16.compile(
optimizer="adam",
loss=tf.keras.losses.SparseCategoricalCrossentropy(from_logits=False),
metrics=["accuracy"],
)
history = VGG_16.fit(
train, batch_size=BATCH_SIZE, validation_data=val, verbose=1, epochs=30
)
scores = VGG_16.evaluate(test)
scores
# # VGG19
from tensorflow.keras.applications.vgg19 import VGG19
input_shape = (IMAGE_SIZE, IMAGE_SIZE, CHANNELS)
VGG_19 = VGG19(input_shape=input_shape, weights="imagenet", include_top=False)
VGG_19 = models.Sequential(
[
resize_and_rescale,
data_augmentation,
vgg1,
layers.Flatten(),
# layers.Dense(64, activation='relu'),
layers.Dense(n_classes, activation="softmax"),
]
)
VGG_19.compile(
optimizer="adam",
loss=tf.keras.losses.SparseCategoricalCrossentropy(from_logits=False),
metrics=["accuracy"],
)
history = VGG_19.fit(
train, batch_size=BATCH_SIZE, validation_data=val, verbose=1, epochs=30
)
scores = VGG_19.evaluate(test)
scores
# # VGG21
VGG_21 = models.Sequential(
[
resize_and_rescale,
data_augmentation,
vgg1,
layers.Conv2D(64, kernel_size=(3, 3), activation="relu"),
layers.MaxPooling2D((2, 2)),
layers.Flatten(),
layers.Dense(64, activation="relu"),
layers.Dense(n_classes, activation="softmax"),
]
)
VGG_21.compile(
optimizer="adam",
loss=tf.keras.losses.SparseCategoricalCrossentropy(from_logits=False),
metrics=["accuracy"],
)
history = VGG_21.fit(
train, batch_size=BATCH_SIZE, validation_data=val, verbose=1, epochs=10
)
VGG_21.summary()
scores = VGG_21.evaluate(test)
scores
train_loss = history.history["loss"]
train_acc = history.history["accuracy"]
val_loss = history.history["val_loss"]
val_acc = history.history["val_accuracy"]
# graphs for accuracy and loss of training and validation data
plt.figure(figsize=(15, 15))
plt.subplot(2, 3, 1)
plt.plot(range(10), train_acc, label="Training Accuracy")
plt.plot(range(10), val_acc, label="Validation Accuracy")
plt.legend(loc="lower right")
plt.title("Training and Validation Accuracy")
plt.subplot(2, 3, 2)
plt.plot(range(10), train_loss, label="Training Loss")
plt.plot(range(10), val_loss, label="Validation Loss")
plt.legend(loc="upper right")
plt.title("Training and Validation Loss")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
test = pd.read_csv(
r"/kaggle/input/test-competition-2783456756923/airline_tweets_train.csv"
)
test = test.rename(columns={"airline_sentiment": "Category"})
test["Category"][:1000] = "neutral"
test["Category"][1000:2000] = "positive"
test["Category"][2000:] = "negative"
test.to_csv(r"test_submission.csv")
|
# ### 1. 安装依赖库
# ### 2. 导入依赖库
import jieba
import pandas as pd
import numpy as np
import tensorflow as tf
from tensorflow.keras.preprocessing.sequence import pad_sequences
from sklearn.metrics import classification_report
# ### 3. 数据加载
import shutil
import os
if os.path.exists("data") != True:
shutil.copytree("/kaggle/input/nlp-ea-data/", "./data")
train_data = pd.read_csv("data/data/train.tsv", sep="\t")
valid_data = pd.read_csv("data/data/dev.tsv", sep="\t")
test_data = pd.read_csv("data/data/test.tsv", sep="\t")
x_train, y_train = train_data.text_a.values, train_data.label.values # 训练集
x_valid, y_valid = valid_data.text_a.values, valid_data.label.values # 验证集
x_test, y_test = test_data.text_a.values, test_data.label.values # 测试集
train_data
x_train, y_train
# ### 4. 构建词汇表
vocab = set()
cut_docs = train_data.text_a.apply(lambda x: jieba.cut(x)).values
for doc in cut_docs:
for word in doc:
if word.strip():
vocab.add(word.strip())
# 将词表写入本地vocab.txt文件
with open("data/data/vocab.txt", "w") as file:
for word in vocab:
file.write(word)
file.write("\n")
len(vocab)
list(vocab)[:10]
# ### 5. 定义配置参数
class Config:
embedding_dim = 300 # 词向量维度
max_seq_len = 200 # 文章最大词数
vocab_file = "data/data/vocab.txt" # 词汇表文件路径
config = Config()
# ### 6. 定义预处理类
class Preprocessor:
def __init__(self, config):
self.config = config
# 初始化词和id的映射词典,预留0给padding字符,1给词表中未见过的词
token2idx = {"[PAD]": 0, "[UNK]": 1} # {word:id}
with open(config.vocab_file, "r") as reader:
for index, line in enumerate(reader):
token = line.strip()
token2idx[token] = index + 2
self.token2idx = token2idx
def transform(self, text_list):
# 文本分词,并将词转换成相应的id, 最后不同长度的文本padding长统一长度,后面补0
idx_list = [
[
self.token2idx.get(word.strip(), self.token2idx["[UNK]"])
for word in jieba.cut(text)
]
for text in text_list
]
idx_padding = pad_sequences(idx_list, self.config.max_seq_len, padding="post")
return idx_padding
preprocessor = Preprocessor(config)
preprocessor.transform(["性价比不错,交通方便。", "房间太小。其他的都一般。"])
# ### 7. 定义模型类
class TextCNN(object):
def __init__(self, config):
self.config = config
self.preprocessor = Preprocessor(config)
self.class_name = {0: "负面", 1: "正面"}
def build_model(self):
# 模型架构搭建
idx_input = tf.keras.layers.Input((self.config.max_seq_len,))
input_embedding = tf.keras.layers.Embedding(
len(self.preprocessor.token2idx),
self.config.embedding_dim,
input_length=self.config.max_seq_len,
mask_zero=True,
)(idx_input)
convs = []
for kernel_size in [3, 4, 5]:
c = tf.keras.layers.Conv1D(128, kernel_size, activation="relu")(
input_embedding
)
c = tf.keras.layers.GlobalMaxPooling1D()(c)
convs.append(c)
fea_cnn = tf.keras.layers.Concatenate()(convs)
fea_cnn_dropout = tf.keras.layers.Dropout(rate=0.4)(fea_cnn)
fea_dense = tf.keras.layers.Dense(128, activation="relu")(fea_cnn_dropout)
output = tf.keras.layers.Dense(2, activation="softmax")(fea_dense)
model = tf.keras.Model(inputs=idx_input, outputs=output)
model.compile(
loss="sparse_categorical_crossentropy",
optimizer="adam",
metrics=["accuracy"],
)
model.summary()
self.model = model
def fit(
self,
x_train,
y_train,
x_valid=None,
y_valid=None,
epochs=5,
batch_size=128,
callbacks=None,
**kwargs
):
# 训练
self.build_model()
x_train = self.preprocessor.transform(x_train)
valid_data = None
if x_valid is not None and y_valid is not None:
x_valid = self.preprocessor.transform(x_valid)
valid_data = (x_valid, y_valid)
self.model.fit(
x=x_train,
y=y_train,
validation_data=valid_data,
batch_size=batch_size,
epochs=epochs,
callbacks=callbacks,
**kwargs
)
def evaluate(self, x_test, y_test):
# 评估
x_test = self.preprocessor.transform(x_test)
y_pred_probs = self.model.predict(x_test)
y_pred = np.argmax(y_pred_probs, axis=-1)
result = classification_report(y_test, y_pred, target_names=["负面", "正面"])
print(result)
def single_predict(self, text):
# 预测
input_idx = self.preprocessor.transform([text])
predict_prob = self.model.predict(input_idx)[0]
predict_label_id = np.argmax(predict_prob)
predict_label_name = self.class_name[predict_label_id]
predict_label_prob = predict_prob[predict_label_id]
return predict_label_name, predict_label_prob
def load_model(self, ckpt_file):
self.build_model()
self.model.load_weights(ckpt_file)
# ### 8. 启动训练
# 定义early stop早停回调函数
patience = 6
early_stop = tf.keras.callbacks.EarlyStopping(monitor="val_loss", patience=patience)
# 定义checkpoint回调函数
checkpoint_prefix = "./checkpoints/textcnn_imdb_ckpt"
checkpoint_callback = tf.keras.callbacks.ModelCheckpoint(
filepath=checkpoint_prefix, save_weights_only=True, save_best_only=True
)
# 初始化模型类,启动训练
textcnn = TextCNN(config)
textcnn.fit(
x_train,
y_train,
x_valid,
y_valid,
epochs=50,
callbacks=[early_stop, checkpoint_callback],
) # 训练
# ### 9. 测试评估
textcnn.evaluate(x_test, y_test) # 测试集评估
# ### 10. 离线加载预测
textcnn = TextCNN(config)
textcnn.load_model(checkpoint_prefix)
textcnn.single_predict("外观很漂亮,出人意料地漂亮,做工非常好") # 单句预测
textcnn.single_predict("书的内容没什么好说的,主要是纸张、印刷太差,所用的纸非常粗糙比一般的盗版书还要差,裁的也不好。") # 单句预测
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import seaborn as sns
import matplotlib.pyplot as plt
sns.set()
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
train = pd.read_csv("/kaggle/input/jdc-2023-dataset/train.csv")
train.head()
train.info()
train.dtypes
train.isna().sum()
train.shape
train.describe(include=["int64", "float64"])
train.describe(include=["object"])
train.nunique(dropna=True)
def convert_plinth_area(x):
if x == "More than 1000 ft^2":
return 1000
elif type(x) == str:
if "ft" in x:
return int(x.split(" ")[0])
elif type(x) == int:
return x
else:
return np.nan
def convert_floors(x):
if x in [
"1st Story",
"floor one",
"Has 1 floor",
"one story",
"1",
"Floor 1",
"floor 1st",
"one",
"Just 1 floor",
]:
return 1
elif x in [
"floor two",
"Two Floor",
"two",
" just 2 floor",
"There is 2 Floor/Story",
"Floor two",
"Floor 2",
"2",
"two story",
"floor second",
"2 floor",
]:
return 2
elif x in [
"Floor 3",
"Three floor",
"floor third",
"3.00",
" has 3 Floor",
"three Story",
"Floor-three",
"Three",
]:
return 3
elif x in [
"four Floor",
" has Four fl",
"Fl four",
"4",
"Floor 4",
"floor four",
"Four Story",
"Four",
]:
return 4
elif x in [
"fifth",
"Fl Five",
"5",
"Floor Fifth",
"five Floor",
" Has Five fl",
"Has Five fl",
]:
return 5
elif x in ["6"]:
return 6
elif x in ["7"]:
return 7
elif x in ["8"]:
return 8
elif x in ["9"]:
return 9
else:
return x
def convert_foundation(x):
value_map = {
"Bamboo or Timber": "Bamboo/Timber",
"Bamboo/TImber": "Bamboo/Timber",
"Clay Sand Mixed mortar-Stone/Brick": "Clay mortar-Stone/Brick",
"Cement-Stone or Cement-Brick": "Cement-Stone/Brick",
"RC": "Reinforced Concrete",
"Other": "Others",
}
if x in value_map:
return value_map[x]
else:
return x
def convert_roof(x):
# Create a dictionary to map similar values to their group
value_map = {
"Bamboo/Timber Light roof": "Bamboo/Timber Light roof",
"Wood Light Roof or Bamboo Heavy Roof": "Wood/Bamboo Light/Heavy roof",
"Wood Light Roof or Bamboo Light Roof": "Wood/Bamboo Light/Heavy roof",
"Bamboo or Timber Light roof": "Bamboo/Timber Light roof",
"Bamboo/TImber-Light Roof": "Bamboo/Timber Light roof",
"Reinforced Brick Slab/rcc/rbc": "Reinforced Brick Slab/rcc/rbc",
"Bamboo/Timber Heavy roof": "Bamboo/Timber Heavy roof",
"reinforced cement concrete/rb/rbc": "Reinforced cement concrete/rb/rbc",
"Bamboo or Timber Heavy roof": "Bamboo/Timber Heavy roof",
"Bamboo/TImber-Heavy Roof": "Bamboo/Timber Heavy roof",
"rcc/rb/rbc": "Reinforced cement concrete/rb/rbc",
"Reinforced brick concrete/rcc/rbc": "Reinforced cement concrete/rb/rbc",
}
# Replace similar values with their group
if x in value_map:
return value_map[x]
else:
return x
def convert_gfloor(value):
# Create a dictionary to map similar values to their group
value_map = {
"Clay": "Clay",
"Mud": "Mud",
"Brick or Stone": "Brick/Stone",
"Reinforced Concrete": "Reinforced Concrete",
"mud": "Mud",
"reinforced concrete": "Reinforced Concrete",
"RC": "Reinforced Concrete",
"soil, water, loam Mixed": "Soil/Loam/Water Mixed",
"brick/stone": "Brick/Stone",
"Other": "Other",
"Lumber": "Lumber",
"Brick/Stone": "Brick/Stone",
"TImber": "Timber",
"Timber": "Timber",
"Wood": "Lumber", # Grouping Wood and Lumber together
}
# Replace similar values with their group
if value in value_map:
return value_map[value]
else:
return value
def convert_ofloor(value):
# Create a dictionary to map similar values to their group
value_map = {
"TImber/Bamboo-Mud": "Timber/Bamboo-Mud",
"Wood-Mud or Bamboo Mud": "Wood/Bamboo-Mud",
"Timber Mud or Bamboo-Mud": "Timber/Bamboo-Mud",
"Not applicable": "Other",
"Wood or Bamboo Mud": "Wood/Bamboo-Mud",
"rcc/rb/rbc": "RCC/RB/RBC",
"Wood-Plank": "Wood-Plank",
"Timber-Planck": "Timber-Plank",
"Lumber-plank": "Wood-Plank",
"Reinforced brick concrete/rcc/rbc": "RCC/RB/RBC",
"reinforced cement concrete/rb/rbc": "RCC/RB/RBC",
}
# Replace similar values with their group
if value in value_map:
return value_map[value]
else:
return value
def convert_status(x):
# Create a dictionary to map similar values to their group
value_map = {
"Private Use": "Private",
"Private": "Private",
"Prvt": "Private",
"Privste": "Private",
"Public Space": "Public",
"Public": "Public",
"Unknown": "Unknown",
"Public Use": "Public",
"Institutionals": "Institutional",
"Unspecified": "Unknown",
"Institutional Use": "Institutional",
"Other": "Other",
"Institutional": "Institutional",
}
# Replace similar values with their group
if x in value_map:
return value_map[x]
else:
return x
def convert_residing(x):
if x == "None":
return 0
elif type(x) == str:
return int(float(x))
else:
return x
train_clean = train.copy()
train_clean["floors_before_eq (total)"] = train["floors_before_eq (total)"].apply(
convert_floors
)
train_clean["plinth_area (ft^2)"] = train["plinth_area (ft^2)"].apply(
convert_plinth_area
)
train_clean["type_of_foundation"] = train["type_of_foundation"].apply(
convert_foundation
)
train_clean["type_of_roof"] = train["type_of_roof"].apply(convert_roof)
train_clean["type_of_ground_floor"] = train["type_of_ground_floor"].apply(
convert_gfloor
)
train_clean["type_of_other_floor"] = train["type_of_other_floor"].apply(convert_ofloor)
train_clean["legal_ownership_status"] = train["legal_ownership_status"].apply(
convert_status
)
train_clean["no_family_residing"] = train["no_family_residing"].apply(convert_residing)
train_clean.head()
numerik = train_clean.select_dtypes(include=["int64", "float64"])
kategorik = train_clean.select_dtypes(include=["object"])
numerik_columns = numerik.columns
print(numerik.columns)
print(len(numerik.columns))
kategorik_columns = kategorik.columns
print(kategorik.columns)
print(len(kategorik.columns))
fig, axes = plt.subplots(4, 3, figsize=(12, 14))
fig.suptitle("Numerik Data Distribution\n\n")
sns.boxplot(train_clean, x=numerik_columns[1], ax=axes[0, 0], color="r")
axes[0, 0].set_title(numerik_columns[1])
sns.boxplot(train_clean, x=numerik_columns[2], ax=axes[0, 1], color="y")
axes[0, 1].set_title(numerik_columns[2])
sns.boxplot(train_clean, x=numerik_columns[3], ax=axes[0, 2], color="g")
axes[0, 2].set_title(numerik_columns[3])
sns.boxplot(train_clean, x=numerik_columns[4], ax=axes[1, 0], color="b")
axes[1, 0].set_title(numerik_columns[4])
sns.boxplot(train_clean, x=numerik_columns[5], ax=axes[1, 1], color="k")
axes[1, 1].set_title(numerik_columns[5])
sns.boxplot(train_clean, x=numerik_columns[6], ax=axes[1, 2], color="c")
axes[1, 2].set_title(numerik_columns[6])
sns.boxplot(train_clean, x=numerik_columns[7], ax=axes[2, 0], color="m")
axes[2, 1].set_title(numerik_columns[7])
sns.boxplot(train_clean, x=numerik_columns[8], ax=axes[2, 1], color="m")
axes[2, 1].set_title(numerik_columns[8])
sns.boxplot(train_clean, x=numerik_columns[9], ax=axes[2, 2], color="m")
axes[2, 2].set_title(numerik_columns[9])
sns.boxplot(train_clean, x=numerik_columns[10], ax=axes[3, 1], color="m")
axes[3, 1].set_title(numerik_columns[10])
fig.delaxes(axes[3, 0])
fig.delaxes(axes[3, 2])
plt.tight_layout()
plt.show()
fig, axes = plt.subplots(4, 3, figsize=(12, 14))
fig.suptitle("Numerik Data Distribution\n\n")
sns.histplot(train_clean, x=numerik_columns[1], kde=True, ax=axes[0, 0], color="r")
axes[0, 0].set_title(numerik_columns[1])
sns.histplot(train_clean, x=numerik_columns[2], kde=True, ax=axes[0, 1], color="y")
axes[0, 1].set_title(numerik_columns[2])
sns.histplot(train_clean, x=numerik_columns[3], kde=True, ax=axes[0, 2], color="g")
axes[0, 2].set_title(numerik_columns[3])
sns.histplot(train_clean, x=numerik_columns[4], kde=True, ax=axes[1, 0], color="b")
axes[1, 0].set_title(numerik_columns[4])
sns.histplot(train_clean, x=numerik_columns[5], kde=True, ax=axes[1, 1], color="k")
axes[1, 1].set_title(numerik_columns[5])
sns.histplot(train_clean, x=numerik_columns[6], kde=True, ax=axes[1, 2], color="c")
axes[1, 2].set_title(numerik_columns[6])
sns.histplot(train_clean, x=numerik_columns[7], kde=True, ax=axes[2, 0], color="m")
axes[2, 1].set_title(numerik_columns[7])
sns.histplot(train_clean, x=numerik_columns[8], kde=True, ax=axes[2, 1], color="m")
axes[2, 1].set_title(numerik_columns[8])
sns.histplot(train_clean, x=numerik_columns[9], kde=True, ax=axes[2, 2], color="m")
axes[2, 2].set_title(numerik_columns[9])
sns.histplot(train_clean, x=numerik_columns[10], kde=True, ax=axes[3, 1], color="m")
axes[3, 1].set_title(numerik_columns[10])
fig.delaxes(axes[3, 0])
fig.delaxes(axes[3, 2])
plt.tight_layout()
plt.show()
train_clean[
["type_of_reinforcement_concrete", "wall_binding", "wall_material", "damage_grade"]
]
sns.heatmap(numerik.corr(), annot=True, fmt=".2f", annot_kws={"fontsize": 10})
plt.title("\nKorelasi Data Numerik\n")
# sns.set(font_scale=3)
# plt.subplots_adjust(top=0.92, bottom=0.08, left=0.12, right=0.95)
# # Target Analysis
train_clean["damage_grade"].value_counts()
sns.countplot(train_clean, x="damage_grade")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import seaborn as sns
import matplotlib.pyplot as plt
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
df = pd.read_csv("/kaggle/input/smart-watch-prices/Smart watch prices.csv")
print("Shape", df.shape)
print("Columns", df.columns)
df.head()
# Datase info
# Data types
df.info()
df.describe()
# null values
print(df.isnull().any())
print(df.isnull().sum())
# Unique values in all columns
for columns in df.columns:
print(columns, ":", df[columns].nunique())
# Brands
# Top 10 selling brands
brand_counts = df["Brand"].value_counts()
brand_counts.head(10)
# Less selling brands
brand_counts.tail(10)
brand_counts.plot.bar()
# Set the axis labels and title
plt.xlabel("Brand")
plt.ylabel("Count")
plt.title("Value Counts of Brands")
plt.show()
# Models
Garmin_models = df[df["Brand"] == "Garmin"]
print(Garmin_models.shape)
Garmin_models.head()
# Most sell Models from Garmin Brand
print("Number of Unique values:", Garmin_models["Model"].nunique())
print("Unique values:", Garmin_models["Model"].unique())
print("Unique values:", Garmin_models["Model"].value_counts())
# Top selling Model
Model = df["Model"].value_counts()
Model.head(10)
# Most sell model Brand
df["Brand"][df["Model"] == "Sense"].value_counts()
# Most sell model Brand
df["Brand"][df["Model"] == "Falster 3"].value_counts()
Model.head(10).plot.bar()
# Set the axis labels and title
plt.xlabel("Model")
plt.ylabel("Count")
plt.title("Value Counts of Model")
plt.show()
# Less selling Model
Model.tail(10)
# Operating System
# Most used os
os = df["Operating System"].value_counts()
os.head(10)
os.head(10).plot.bar()
# Set the axis labels and title
plt.xlabel("os")
plt.ylabel("Count")
plt.title("Value Counts of os")
plt.show()
#
os_brand = df[["Brand", "Model"]][df["Operating System"] == "Wear OS"]
os_brand.nunique()
# Connectivity
print("Number of unique values:", df["Connectivity"].nunique())
print("Unique values:", df["Connectivity"].unique())
print(df["Connectivity"].value_counts())
df["Connectivity"].value_counts().plot.bar()
# Set the axis labels and title
plt.xlabel("os")
plt.ylabel("Count")
plt.title("Value Counts of Brands")
plt.show()
# Display Type
# Display Type
print("Number of unique values:", df["Display Type"].nunique())
print("Top 10 most used Display:\n", df["Display Type"].value_counts().head(10))
print("Less used Display:\n", df["Display Type"].value_counts().tail(10))
df.head()
# Numerical Values
num_list = [
"Display Size (inches)",
"Resolution",
"Water Resistance (meters)",
"Battery Life (days)",
]
df[
[
"Display Size (inches)",
"Resolution",
"Water Resistance (meters)",
"Battery Life (days)",
]
].head()
df[num_list].info()
for columns in num_list:
print(columns, ":", df[columns].nunique())
# Display Size
print("Maximum Display Size:", df["Display Size (inches)"].max(), "inches")
print("Minimum Display Size:", df["Display Size (inches)"].min(), "inches")
df["Resolution"].value_counts().plot.bar()
# Set the axis labels and title
plt.xlabel("os")
plt.ylabel("Count")
plt.title("Value Counts of Brands")
plt.show()
df["Water Resistance (meters)"].value_counts()
water_r = df[df["Water Resistance (meters)"] == "200"]
water_r.nunique()
sns.histplot(x="Water Resistance (meters)", data=df, bins=10)
# all watches with 50 meters Water Resistance
print("shape:", water_r.shape)
water_r.head()
df["Battery Life (days)"].value_counts()
df["Battery Life (days)"].value_counts().tail()
# all watches with 14 day battery life
battery = df[df["Battery Life (days)"] == "60"]
print("Shape:", battery.shape)
battery.tail()
# Category
df_yn = df[["Heart Rate Monitor", "GPS", "NFC"]]
df_yn.head()
for col in df_yn:
print(df[col].value_counts(), "\n")
df["Price (USD)"].nunique()
|
# [论文链接](https://arxiv.org/pdf/1909.03227.pdf)
# [代码链接](https://github.com/weizhepei/CasRel)
# [本篇参考链接](https://github.com/Onion12138/CasRelPyTorch)
# # 论文简介
# ### 这个模型要解决的问题:
# 
# 以往的模型是第一种。
# 但是一个人既是一个电影的导演优势一个电影的演员,就不太好处理。
# 一个人同时出生于中国和北京也不好处理
# ### 模型架构
# 
# Jackie R Brown was born in washington, the capital city if united states of Amarica .
# 一共计算四个损失
# 前两个简单的损失:
# - label 的构建:
# 
# - predct 计算:
# 整个句子经过bert编码后变为`[seq len, bert dim]` 经过两个线性层然后sigmoid后变为 两个`[seq len, 1]` 这两个predict 跟label越接近越好
# 后两个损失
# 假如现在关系只有三种:出生地,工作地,首都
# 现在我要寻找Jackie R Brown的所有关系
# 可以取出Jackie和 Brown的编码取平均`[1,bert_dim]`然后跟整个句子相加`[seq len,bert_dim]`
# 然后经过一个线性层后燃sigmiod 变为`[seq len,3*2]`
# 
## 缺陷
不能搞定嵌套命名实体识别
# # 数据集格式
# ```json
# {
# "text": "笔 名:木斧原 名:杨莆曾 用 名:穆新文、牧羊、寒白、洋漾出生日期:1931—职 业:作家、诗人性 别: 男民 族: 回族政治面貌:中共党员 祖 籍:固原县出 生 地:成都",
# "spo_list":
# [{"predicate": "民族", "object_type": "文本", "subject_type": "人物", "object": "回族", "subject": "木斧"},
# {"predicate": "出生日期", "object_type": "日期", "subject_type": "人物", "object": "1931", "subject": "木斧"},
# {"predicate": "出生地", "object_type": "地点", "subject_type": "人物", "object": "成都", "subject": "木斧"}]
# }
# ```
import torch
from fastNLP import Vocabulary
from transformers import BertTokenizer, AdamW
from collections import defaultdict
from random import choice
import json
class Config:
"""
句子最长长度是294 这里就不设参数限制长度了,每个batch 自适应长度
"""
def __init__(self):
self.device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
self.bert_path = "../input/huggingface-bert/bert-base-chinese"
self.num_rel = 18 # 关系的种类数
self.train_data_path = "../input/baidurelationshipextraction/train.json"
self.dev_data_path = "../input/baidurelationshipextraction/dev.json"
self.test_data_path = "../input/baidurelationshipextraction/test.json"
self.batch_size = 16 # 批次的个数
self.rel_dict_path = "../input/baidurelationshipextraction/rel.json"
id2rel = json.load(
open(self.rel_dict_path, encoding="utf8")
) # 从json文件中读取数据,返回为dict(字典)对象
self.rel_vocab = Vocabulary(unknown=None, padding=None)
self.rel_vocab.add_word_lst(
list(id2rel.values())
) # 关系到id的映射,Vocabulary用于将文本转化为index
self.tokenizer = BertTokenizer.from_pretrained(
self.bert_path
) # 初始化 BertTokenizer
self.device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
self.learning_rate = 1e-5 # 设定学习率
self.bert_dim = 768 # ????维度
self.epochs = 10 # 所有训练数据训练的次数
# # 数据预处理
from torch.utils.data import Dataset, DataLoader
import json
def collate_fn(batch):
# batch是一个列表,其中是一个一个的元组,每个元组是dataset中_getitem__的结果
batch = list(zip(*batch)) # 与 zip 相反,可理解为解压
text = batch[0]
triple = batch[1]
del batch # 删除变量而不删除数据本身
return text, triple
class MyDataset(Dataset): # 创建一个Dataset的子类MyDataset 其中的 getitem和len方法必须重写
def __init__(self, path):
super().__init__() # 调用父类的init方法,同样可以使用super()去调用父类的其他方法,__init__()是构造方法
self.dataset = [] # 给实例中增加dataset变量
with open(path, encoding="utf8") as F:
for line in F:
line = json.loads(line) # 字符串转化为字典
self.dataset.append(line)
def __getitem__(self, item):
content = self.dataset[item]
text = content["text"]
spo_list = content["spo_list"]
return text, spo_list
def __len__(self):
return len(self.dataset)
def create_data_iter(config):
train_data = MyDataset(config.train_data_path) # ??????
dev_data = MyDataset(config.dev_data_path)
test_data = MyDataset(config.test_data_path)
train_iter = DataLoader(
train_data, batch_size=config.batch_size, shuffle=True, collate_fn=collate_fn
)
dev_iter = DataLoader(
dev_data, batch_size=config.batch_size, shuffle=True, collate_fn=collate_fn
)
test_iter = DataLoader(
test_data, batch_size=config.batch_size, shuffle=False, collate_fn=collate_fn
)
return train_iter, dev_iter, test_iter
class Batch:
def __init__(self, config):
self.tokenizer = config.tokenizer
self.num_relations = config.num_rel
self.rel_vocab = config.rel_vocab
self.device = config.device
def __call__(self, text, triple):
text = self.tokenizer(text, padding=True).data
batch_size = len(text["input_ids"])
seq_len = len(text["input_ids"][0])
sub_head = []
sub_tail = []
sub_heads = []
sub_tails = []
obj_heads = []
obj_tails = []
sub_len = []
sub_head2tail = []
for batch_index in range(batch_size):
inner_input_ids = text["input_ids"][batch_index] # 单个句子变成索引后
inner_triples = triple[batch_index]
(
inner_sub_heads,
inner_sub_tails,
inner_sub_head,
inner_sub_tail,
inner_sub_head2tail,
inner_sub_len,
inner_obj_heads,
inner_obj_tails,
) = self.create_label(inner_triples, inner_input_ids, seq_len)
sub_head.append(inner_sub_head)
sub_tail.append(inner_sub_tail)
sub_len.append(inner_sub_len)
sub_head2tail.append(inner_sub_head2tail)
sub_heads.append(inner_sub_heads)
sub_tails.append(inner_sub_tails)
obj_heads.append(inner_obj_heads)
obj_tails.append(inner_obj_tails)
input_ids = torch.tensor(text["input_ids"]).to(self.device)
mask = torch.tensor(text["attention_mask"]).to(self.device)
sub_head = torch.stack(sub_head).to(self.device)
sub_tail = torch.stack(sub_tail).to(self.device)
sub_heads = torch.stack(sub_heads).to(self.device)
sub_tails = torch.stack(sub_tails).to(self.device)
sub_len = torch.stack(sub_len).to(self.device)
sub_head2tail = torch.stack(sub_head2tail).to(self.device)
obj_heads = torch.stack(obj_heads).to(self.device)
obj_tails = torch.stack(obj_tails).to(self.device)
return {
"input_ids": input_ids,
"mask": mask,
"sub_head2tail": sub_head2tail,
"sub_len": sub_len,
}, {
"sub_heads": sub_heads,
"sub_tails": sub_tails,
"obj_heads": obj_heads,
"obj_tails": obj_tails,
}
def create_label(self, inner_triples, inner_input_ids, seq_len):
inner_sub_heads, inner_sub_tails = torch.zeros(seq_len), torch.zeros(seq_len)
inner_sub_head, inner_sub_tail = torch.zeros(seq_len), torch.zeros(seq_len)
inner_obj_heads = torch.zeros((seq_len, self.num_relations))
inner_obj_tails = torch.zeros((seq_len, self.num_relations))
inner_sub_head2tail = torch.zeros(seq_len) # 随机抽取一个实体,从开头一个词到末尾词的索引
# 因为数据预处理代码还待优化,会有不存在关系三元组的情况,
# 初始化一个主词的长度为1,即没有主词默认主词长度为1,
# 防止零除报错,初始化任何非零数字都可以,没有主词分子是全零矩阵
inner_sub_len = torch.tensor([1], dtype=torch.float)
# 主词到谓词的映射
s2ro_map = defaultdict(list)
for inner_triple in inner_triples:
inner_triple = (
self.tokenizer(inner_triple["subject"], add_special_tokens=False)[
"input_ids"
],
self.rel_vocab.to_index(inner_triple["predicate"]),
self.tokenizer(inner_triple["object"], add_special_tokens=False)[
"input_ids"
],
)
sub_head_idx = self.find_head_idx(inner_input_ids, inner_triple[0])
obj_head_idx = self.find_head_idx(inner_input_ids, inner_triple[2])
if sub_head_idx != -1 and obj_head_idx != -1:
sub = (sub_head_idx, sub_head_idx + len(inner_triple[0]) - 1)
# s2ro_map保存主语到谓语的映射
s2ro_map[sub].append(
(
obj_head_idx,
obj_head_idx + len(inner_triple[2]) - 1,
inner_triple[1],
)
) # {(3,5):[(7,8,0)]} 0是关系
if s2ro_map:
for s in s2ro_map:
inner_sub_heads[s[0]] = 1
inner_sub_tails[s[1]] = 1
sub_head_idx, sub_tail_idx = choice(list(s2ro_map.keys()))
inner_sub_head[sub_head_idx] = 1
inner_sub_tail[sub_tail_idx] = 1
inner_sub_head2tail[sub_head_idx : sub_tail_idx + 1] = 1
inner_sub_len = torch.tensor(
[sub_tail_idx + 1 - sub_head_idx], dtype=torch.float
)
for ro in s2ro_map.get((sub_head_idx, sub_tail_idx), []):
inner_obj_heads[ro[0]][ro[2]] = 1
inner_obj_tails[ro[1]][ro[2]] = 1
return (
inner_sub_heads,
inner_sub_tails,
inner_sub_head,
inner_sub_tail,
inner_sub_head2tail,
inner_sub_len,
inner_obj_heads,
inner_obj_tails,
)
@staticmethod
def find_head_idx(source, target):
target_len = len(target)
for i in range(len(source)):
if source[i : i + target_len] == target:
return i
return -1
# # model
import torch.nn as nn
import torch
from transformers import BertModel
class CasRel(nn.Module):
def __init__(self, config):
super(CasRel, self).__init__()
self.config = config
self.bert = BertModel.from_pretrained(self.config.bert_path)
self.sub_heads_linear = nn.Linear(self.config.bert_dim, 1)
self.sub_tails_linear = nn.Linear(self.config.bert_dim, 1)
self.obj_heads_linear = nn.Linear(self.config.bert_dim, self.config.num_rel)
self.obj_tails_linear = nn.Linear(self.config.bert_dim, self.config.num_rel)
self.alpha = 0.25
self.gamma = 2
def get_encoded_text(self, token_ids, mask):
encoded_text = self.bert(token_ids, attention_mask=mask)[0]
return encoded_text
def get_subs(self, encoded_text):
pred_sub_heads = torch.sigmoid(self.sub_heads_linear(encoded_text))
pred_sub_tails = torch.sigmoid(self.sub_tails_linear(encoded_text))
return pred_sub_heads, pred_sub_tails
def get_objs_for_specific_sub(self, sub_head2tail, sub_len, encoded_text):
# sub_head_mapping [batch, 1, seq] * encoded_text [batch, seq, dim]
sub = torch.matmul(sub_head2tail, encoded_text) # batch size,1,dim
sub_len = sub_len.unsqueeze(1)
sub = sub / sub_len # batch size, 1,dim
encoded_text = encoded_text + sub
# [batch size, seq len,bert_dim] -->[batch size, seq len,relathion counts]
pred_obj_heads = torch.sigmoid(self.obj_heads_linear(encoded_text))
pred_obj_tails = torch.sigmoid(self.obj_tails_linear(encoded_text))
return pred_obj_heads, pred_obj_tails
def forward(self, input_ids, mask, sub_head2tail, sub_len):
"""
:param token_ids:[batch size, seq len]
:param mask:[batch size, seq len]
:param sub_head:[batch size, seq len]
:param sub_tail:[batch size, seq len]
:return:
"""
encoded_text = self.get_encoded_text(input_ids, mask)
pred_sub_heads, pred_sub_tails = self.get_subs(encoded_text)
sub_head2tail = sub_head2tail.unsqueeze(1) # [[batch size,1, seq len]]
pred_obj_heads, pre_obj_tails = self.get_objs_for_specific_sub(
sub_head2tail, sub_len, encoded_text
)
return {
"pred_sub_heads": pred_sub_heads,
"pred_sub_tails": pred_sub_tails,
"pred_obj_heads": pred_obj_heads,
"pred_obj_tails": pre_obj_tails,
"mask": mask,
}
def compute_loss(
self,
pred_sub_heads,
pred_sub_tails,
pred_obj_heads,
pred_obj_tails,
mask,
sub_heads,
sub_tails,
obj_heads,
obj_tails,
):
rel_count = obj_heads.shape[-1]
rel_mask = mask.unsqueeze(-1).repeat(1, 1, rel_count)
loss_1 = self.loss_fun(pred_sub_heads, sub_heads, mask)
loss_2 = self.loss_fun(pred_sub_tails, sub_tails, mask)
loss_3 = self.loss_fun(pred_obj_heads, obj_heads, rel_mask)
loss_4 = self.loss_fun(pred_obj_tails, obj_tails, rel_mask)
return loss_1 + loss_2 + loss_3 + loss_4
def loss_fun(self, logist, label, mask):
count = torch.sum(mask)
logist = logist.view(-1)
label = label.view(-1)
mask = mask.view(-1)
alpha_factor = torch.where(torch.eq(label, 1), 1 - self.alpha, self.alpha)
focal_weight = torch.where(torch.eq(label, 1), 1 - logist, logist)
loss = -(torch.log(logist) * label + torch.log(1 - logist) * (1 - label)) * mask
return torch.sum(focal_weight * loss) / count
def load_model(config):
device = config.device
model = CasRel(config)
model.to(device)
# prepare optimzier
param_optimizer = list(model.named_parameters())
no_decay = ["bias", "LayerNorm.bias", "LayerNorm.weight"]
optimizer_grouped_parameters = [
{
"params": [
p for n, p in param_optimizer if not any(nd in n for nd in no_decay)
],
"weight_decay": 0.01,
},
{
"params": [
p for n, p in param_optimizer if any(nd in n for nd in no_decay)
],
"weight_decay": 0.0,
},
]
optimizer = AdamW(optimizer_grouped_parameters, lr=config.learning_rate, eps=10e-8)
sheduler = None
return model, optimizer, sheduler, device
# # train
import pandas as pd
from tqdm import tqdm
def train_epoch(model, train_iter, dev_iter, optimizer, batch, best_triple_f1, epoch):
for step, (text, triple) in enumerate(train_iter):
model.train()
inputs, labels = batch(text, triple)
logist = model(**inputs)
loss = model.compute_loss(**logist, **labels)
model.zero_grad()
loss.backward()
optimizer.step()
if step % 500 == 1:
(
sub_precision,
sub_recall,
sub_f1,
triple_precision,
triple_recall,
triple_f1,
df,
) = test(model, dev_iter, batch)
if triple_f1 > best_triple_f1:
best_triple_f1 = triple_f1
torch.save(model.state_dict(), "best_f1.pth")
print(
"epoch:{},step:{},sub_precision:{:.4f}, sub_recall:{:.4f}, sub_f1:{:.4f}, triple_precision:{:.4f}, triple_recall:{:.4f}, triple_f1:{:.4f},train loss:{:.4f}".format(
epoch,
step,
sub_precision,
sub_recall,
sub_f1,
triple_precision,
triple_recall,
triple_f1,
loss.item(),
)
)
print(df)
return best_triple_f1
def train(model, train_iter, dev_iter, optimizer, config):
epochs = config.epochs
best_triple_f1 = 0
for epoch in range(epochs):
best_triple_f1 = train_epoch(
model, train_iter, dev_iter, optimizer, batch, best_triple_f1, epoch
)
def test(model, dev_iter, batch):
model.eval()
df = pd.DataFrame(
columns=["TP", "PRED", "REAL", "p", "r", "f1"], index=["sub", "triple"]
)
df.fillna(0, inplace=True)
for text, triple in tqdm(dev_iter):
inputs, labels = batch(text, triple)
logist = model(**inputs)
pred_sub_heads = convert_score_to_zero_one(logist["pred_sub_heads"])
pred_sub_tails = convert_score_to_zero_one(logist["pred_sub_tails"])
sub_heads = convert_score_to_zero_one(labels["sub_heads"])
sub_tails = convert_score_to_zero_one(labels["sub_tails"])
batch_size = inputs["input_ids"].shape[0]
obj_heads = convert_score_to_zero_one(labels["obj_heads"])
obj_tails = convert_score_to_zero_one(labels["obj_tails"])
pred_obj_heads = convert_score_to_zero_one(logist["pred_obj_heads"])
pred_obj_tails = convert_score_to_zero_one(logist["pred_obj_tails"])
for batch_index in range(batch_size):
pred_subs = extract_sub(
pred_sub_heads[batch_index].squeeze(),
pred_sub_tails[batch_index].squeeze(),
)
true_subs = extract_sub(
sub_heads[batch_index].squeeze(), sub_tails[batch_index].squeeze()
)
pred_ojbs = extract_obj_and_rel(
pred_obj_heads[batch_index], pred_obj_tails[batch_index]
)
true_objs = extract_obj_and_rel(
obj_heads[batch_index], obj_tails[batch_index]
)
df["PRED"]["sub"] += len(pred_subs)
df["REAL"]["sub"] += len(true_subs)
for true_sub in true_subs:
if true_sub in pred_subs:
df["TP"]["sub"] += 1
df["PRED"]["triple"] += len(pred_ojbs)
df["REAL"]["triple"] += len(true_objs)
for true_obj in true_objs:
if true_obj in pred_ojbs:
df["TP"]["triple"] += 1
df.loc["sub", "p"] = df["TP"]["sub"] / (df["PRED"]["sub"] + 1e-9)
df.loc["sub", "r"] = df["TP"]["sub"] / (df["REAL"]["sub"] + 1e-9)
df.loc["sub", "f1"] = (
2 * df["p"]["sub"] * df["r"]["sub"] / (df["p"]["sub"] + df["r"]["sub"] + 1e-9)
)
sub_precision = df["TP"]["sub"] / (df["PRED"]["sub"] + 1e-9)
sub_recall = df["TP"]["sub"] / (df["REAL"]["sub"] + 1e-9)
sub_f1 = 2 * sub_precision * sub_recall / (sub_precision + sub_recall + 1e-9)
df.loc["triple", "p"] = df["TP"]["triple"] / (df["PRED"]["triple"] + 1e-9)
df.loc["triple", "r"] = df["TP"]["triple"] / (df["REAL"]["triple"] + 1e-9)
df.loc["triple", "f1"] = (
2
* df["p"]["triple"]
* df["r"]["triple"]
/ (df["p"]["triple"] + df["r"]["triple"] + 1e-9)
)
triple_precision = df["TP"]["triple"] / (df["PRED"]["triple"] + 1e-9)
triple_recall = df["TP"]["triple"] / (df["REAL"]["triple"] + 1e-9)
triple_f1 = (
2 * triple_precision * triple_recall / (triple_precision + triple_recall + 1e-9)
)
return (
sub_precision,
sub_recall,
sub_f1,
triple_precision,
triple_recall,
triple_f1,
df,
)
def extract_sub(pred_sub_heads, pred_sub_tails):
subs = []
heads = torch.arange(0, len(pred_sub_heads))[pred_sub_heads == 1]
tails = torch.arange(0, len(pred_sub_tails))[pred_sub_tails == 1]
for head, tail in zip(heads, tails):
if tail >= head:
subs.append((head.item(), tail.item()))
return subs
def extract_obj_and_rel(obj_heads, obj_tails):
obj_heads = obj_heads.T
obj_tails = obj_tails.T
rel_count = obj_heads.shape[0]
obj_and_rels = (
[]
) # [(rel_index,strart_index,end_index),(rel_index,strart_index,end_index)]
for rel_index in range(rel_count):
obj_head = obj_heads[rel_index]
obj_tail = obj_tails[rel_index]
objs = extract_sub(obj_head, obj_tail)
if objs:
for obj in objs:
start_index, end_index = obj
obj_and_rels.append((rel_index, start_index, end_index))
return obj_and_rels
def convert_score_to_zero_one(tensor):
tensor[tensor >= 0.5] = 1
tensor[tensor < 0.5] = 0
return tensor
# # main
if __name__ == "__main__":
config = Config()
train_data = MyDataset(config.train_data_path)
model, optimizer, sheduler, device = load_model(config)
train_iter, dev_iter, test_iter = create_data_iter(config)
batch = Batch(config)
train(model, train_iter, dev_iter, optimizer, config)
# len(train_iter) 3498
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import matplotlib.pyplot as plt
from sklearn import tree
from sklearn import metrics
from sklearn.tree import DecisionTreeClassifier
from sklearn.model_selection import train_test_split
from sklearn.metrics import (
accuracy_score,
confusion_matrix,
classification_report,
precision_score,
recall_score,
f1_score,
)
df = pd.read_csv("/kaggle/input/review/diabetes.csv")
df.head(df.info())
# **Split dataset into feature and target variables**
df.columns
feature_cols = [
"Pregnancies",
"Insulin",
"BMI",
"Age",
"Glucose",
"BloodPressure",
"DiabetesPedigreeFunction",
]
x = df[feature_cols]
y = df.Outcome
x
y
# **Splitting dataset into training set and test set**
# 70% Training set and 30% test set
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.3, random_state=1)
x_test.head(x_test.info())
# **Create decision tree classifier object**
clf = DecisionTreeClassifier()
# **Train Decision Tree Classifier**
clf = clf.fit(x_train, y_train)
# **Predict response of the dataset**
y_pred = clf.predict(x_test)
# **Check model accuracy (Accuracy score,confusion matric,precision score,recall score,f-1 score,classification report)**
print("Accuracy:", metrics.accuracy_score(y_test, y_pred))
confusion_matrix = metrics.confusion_matrix(y_test, y_pred)
print(confusion_matrix)
print("Precison Score:", format(precision_score(y_test, y_pred)))
print("Recall Score:", format(recall_score(y_test, y_pred)))
print("F1-Score:", format(f1_score(y_test, y_pred)))
print(classification_report(y_test, y_pred))
# **Plot Decision Tree**
fig = plt.figure(figsize=(45, 12))
tree.plot_tree(clf, feature_names=feature_cols, filled=True, fontsize=10)
plt.show()
|
# Import all required Libraries
from sklearn.metrics import roc_curve, roc_auc_score, ConfusionMatrixDisplay
import shutil
import warnings
import re
import math
import os
from sklearn.model_selection import KFold
from plotly.subplots import make_subplots
import plotly.io as pio
import plotly.graph_objects as go
import plotly.express as px
import seaborn as sns
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from matplotlib.gridspec import GridSpec
import cv2
from sklearn.metrics import roc_curve, confusion_matrix
import glob
from tqdm.notebook import tqdm
import seaborn as sns
import tensorflow as tf
from tensorflow import keras
from PIL import Image
from tensorflow.keras.preprocessing.image import ImageDataGenerator
import random
from random import seed
import math
plt.style.use("fivethirtyeight")
pio.templates.default = "plotly_dark"
import tensorflow as tf
from kaggle_datasets import KaggleDatasets
import tensorflow.keras.backend as K
import efficientnet.tfkeras as efn
warnings.filterwarnings("ignore")
# Global Variables
global_vars = {
"DEVICE": "tpu",
"NFOLDS": 5,
"BATCH_SIZE": 32,
"IMAGE_SIZE": 512,
"EXTERNAL_DATA": True,
"AUTO": tf.data.AUTOTUNE,
"LR_MIN": 0.00001,
"LR_MAX": 0.00005,
"STEP_SIZE": 5,
"LR_METHOD": "triangular",
"EPOCHS": 12,
}
# Set up TPU
# Locate tpu
tpu = tf.distribute.cluster_resolver.TPUClusterResolver()
# connect
tf.config.experimental_connect_to_cluster(tpu)
# initialize
tf.tpu.experimental.initialize_tpu_system(tpu)
# TPU Strategy
strategy = tf.distribute.TPUStrategy()
replicas = strategy.num_replicas_in_sync
print("Num of Replicas: ", replicas)
isic2019 = pd.read_csv("../input/isic2019-512x512/train.csv")
melanoma = pd.read_csv("../input/melanoma-512x512/train.csv")
train = pd.concat((melanoma.drop("patient_code", axis=1), isic2019)).reset_index(
drop=True
)
test = pd.read_csv("../input/siim-isic-melanoma-classification/test.csv")
cols = test.columns
df = pd.concat([train[cols], test[cols]], ignore_index=True).reset_index(drop=True)
df.head()
GCS_PATH = KaggleDatasets().get_gcs_path("melanoma-512x512")
train_files = np.sort(tf.io.gfile.glob(GCS_PATH + "/train*.tfrec"))
test_files = np.sort(tf.io.gfile.glob(GCS_PATH + "/test*.tfrec"))
if global_vars["EXTERNAL_DATA"]:
GCS_PATH2 = KaggleDatasets().get_gcs_path("isic2019-512x512")
GCS_TRAIN = KaggleDatasets().get_gcs_path("siim-isic-melanoma-classification")
GCS_TRAIN = GCS_TRAIN + "/jpeg/train"
GCS_HAIRS = KaggleDatasets().get_gcs_path("melanoma-hairs")
hair_images = os.listdir("../input/melanoma-hairs")
hair_images = [GCS_HAIRS + "/" + image_name for image_name in hair_images]
hair_images = tf.convert_to_tensor(hair_images)
df.info()
# Create Figure
fig = plt.figure(constrained_layout=True, figsize=(20, 10))
color = "#8B0000"
# Create grid:
grid = GridSpec(ncols=4, nrows=2, figure=fig)
# Plot Gender Distribution on first grid
ax1 = fig.add_subplot(grid[0, :2])
ax1.set_title("Gender Distribution")
sns.countplot(
df.sex.sort_values(ignore_index=True),
alpha=0.9,
ax=ax1,
color=color,
)
# Plot Anatom Site General Challenge Distribution on second grid.
ax2 = fig.add_subplot(grid[0, 2:])
sns.countplot(
df["anatom_site_general_challenge"],
alpha=0.9,
ax=ax2,
color=color,
order=df["anatom_site_general_challenge"].value_counts().index,
)
ax2.set_title("Anatom Site Challenge Distribution")
plt.xticks(rotation=20)
# Plot Distribution of Age on third grid.
ax3 = fig.add_subplot(grid[1, :])
sns.distplot(df["age_approx"], color=color, ax=ax3)
ax3.set_title("Age Distribution")
plt.show()
# **Observations:-**
# 1. Men and Female have almost same counts.
# 2. In Anatom Site Challenge there are different types of torso.
# 3. Age follows Normal Distribution.
# Donut Chart of Target
fig = go.Figure()
fig.add_trace(
go.Pie(
labels=train["benign_malignant"].value_counts().index,
values=train["benign_malignant"].value_counts().values,
hole=0.4,
marker_colors=["#008B8B", "#FF7F50"],
)
)
fig.update_layout(
title_text="Donut Chart of Target",
title_font_size=30,
annotations=[dict(x=0.49, y=0.5, text="Target", font_size=20, showarrow=False)],
)
fig.show()
# **There is Class Imbalance**
print("No of rows where age is zero =", len(df[df["age_approx"] == 0]))
# Age distribution by target and sex
color1 = "#008B8B"
color2 = "#DC143C"
grid = GridSpec(nrows=1, ncols=4)
figure = plt.figure(figsize=(15, 9), constrained_layout=True)
ax1 = figure.add_subplot(grid[0, :2])
b, m = (
train.loc[train["target"] == 0, "age_approx"],
train.loc[train["target"] == 1, "age_approx"],
)
sns.distplot(b, ax=ax1, color=color1, label="Benign")
sns.distplot(m, ax=ax1, color=color2, label="Malignant")
ax1.set_title("Age Distribution by Target")
ax1.legend()
ax2 = figure.add_subplot(grid[0, 2:])
m, f = (
df.loc[df["sex"] == "male", "age_approx"],
df.loc[df["sex"] == "female", "age_approx"],
)
sns.distplot(f, ax=ax2, color=color1, label="Female")
sns.distplot(m, ax=ax2, color=color2, label="Male")
ax2.set_title("Age Distribution by sex")
ax2.legend()
plt.show()
# Sunburst Chart
colors = ["#a2ef44", "#31aa75", "#fcd47d", "#b23256"]
fig = px.sunburst(
data_frame=train.dropna(),
path=["target", "sex", "anatom_site_general_challenge"],
color="sex",
color_discrete_sequence=colors,
maxdepth=-1,
title="Sunburst Chart Benign/Malignant > Sex > Location",
)
fig.update_traces(textinfo="label+percent parent")
fig.show()
# Pivot Tables
print(
"Pivot Table where index=anatom site challenge, columns=sex, values=target, aggfunc=count\n"
)
print(
pd.pivot_table(
data=train,
index="anatom_site_general_challenge",
columns="sex",
values="target",
aggfunc="count",
fill_value=0,
)
)
print("-" * 80)
print(
"Pivot Table where index=anatom site challenge, columns=[sex,target], values=age_approx, aggfunc=mean\n"
)
print(
pd.pivot_table(
data=train,
index="anatom_site_general_challenge",
columns=["sex", "target"],
values="age_approx",
fill_value=0,
)
)
a = set(test["patient_id"].unique()).intersection(set(train["patient_id"].unique()))
if len(a) == 0:
print("There is no patient in test set that was present in train set.")
else:
print("There are some patients in test which were present in train set.")
# Plot some benign and Malignant Images
df = pd.read_csv("../input/siim-isic-melanoma-classification/train.csv")
benign_images = df.loc[df.target == 0, "image_name"]
malignant_images = df.loc[df.target == 1, "image_name"]
random_benign_images = [np.random.choice(benign_images) + ".jpg" for i in range(9)]
random_malignant_images = [
np.random.choice(malignant_images) + ".jpg" for i in range(9)
]
figure = plt.figure(figsize=(20, 10), tight_layout=True)
folder_path = "../input/siim-isic-melanoma-classification/jpeg/train"
print("Benign Images")
for i in range(9):
figure.add_subplot(3, 3, i + 1)
image = plt.imread(os.path.join(folder_path, random_benign_images[i]))
plt.imshow(image)
plt.axis("off")
plt.show()
figure = plt.figure(figsize=(20, 10), tight_layout=True)
print("Malignant Images")
for i in range(9):
figure.add_subplot(3, 3, i + 1)
image = plt.imread(os.path.join(folder_path, random_malignant_images[i]))
plt.imshow(image)
plt.axis("off")
plt.show()
fig = plt.figure(figsize=(16, 8))
fig.add_subplot(1, 2, 1)
benign = train.loc[train.target == 0]
sample_img = benign["image_name"][0] + ".jpg"
folder_path = "../input/siim-isic-melanoma-classification/jpeg/train"
raw_image = plt.imread(os.path.join(folder_path, sample_img))
plt.imshow(raw_image, cmap="gray")
plt.colorbar()
plt.title("Benign Image")
print(f"Image dimensions: {raw_image.shape[0],raw_image.shape[1]}")
print(
f"Maximum pixel value : {raw_image.max():.1f} ; Minimum pixel value:{raw_image.min():.1f}"
)
print(
f"Mean value of the pixels : {raw_image.mean():.1f} ; Standard deviation : {raw_image.std():.1f}"
)
fig.add_subplot(1, 2, 2)
# _ = plt.hist(raw_image.ravel(),bins = 256, color = 'orange')
_ = plt.hist(raw_image[:, :, 0].ravel(), bins=256, color="red", alpha=0.5)
_ = plt.hist(raw_image[:, :, 1].ravel(), bins=256, color="Green", alpha=0.5)
_ = plt.hist(raw_image[:, :, 2].ravel(), bins=256, color="Blue", alpha=0.5)
_ = plt.xlabel("Intensity Value")
_ = plt.ylabel("Count")
_ = plt.legend(["Red_Channel", "Green_Channel", "Blue_Channel"])
plt.show()
f = plt.figure(figsize=(16, 8))
f.add_subplot(1, 2, 1)
malignant = train.loc[train.target == 1].reset_index(drop=True)
sample_img = malignant["image_name"][0] + ".jpg"
raw_image = plt.imread(os.path.join(folder_path, sample_img))
plt.imshow(raw_image, cmap="gray")
plt.colorbar()
plt.title("Malignant Image")
print(f"Image dimensions: {raw_image.shape[0],raw_image.shape[1]}")
print(
f"Maximum pixel value : {raw_image.max():.1f} ; Minimum pixel value:{raw_image.min():.1f}"
)
print(
f"Mean value of the pixels : {raw_image.mean():.1f} ; Standard deviation : {raw_image.std():.1f}"
)
f.add_subplot(1, 2, 2)
# _ = plt.hist(raw_image.ravel(),bins = 256, color = 'orange',)
_ = plt.hist(raw_image[:, :, 0].ravel(), bins=256, color="red", alpha=0.5)
_ = plt.hist(raw_image[:, :, 1].ravel(), bins=256, color="Green", alpha=0.5)
_ = plt.hist(raw_image[:, :, 2].ravel(), bins=256, color="Blue", alpha=0.5)
_ = plt.xlabel("Intensity Value")
_ = plt.ylabel("Count")
_ = plt.legend(["Red_Channel", "Green_Channel", "Blue_Channel"])
plt.show()
def get_rotation_matrix(rotation):
# Convert degrees to radians
rotation = math.pi * rotation / 180.0
# Sine,Cosine,one,zero
sin = tf.math.sin(rotation)
cos = tf.math.cos(rotation)
one = tf.constant(1, dtype=tf.float32)
zero = tf.constant(0, dtype=tf.float32)
# Rotation Matrix
rotation_matrix = tf.reshape(
tf.concat([[cos, -sin, zero, sin, cos, zero, zero, zero, one]], axis=0), (3, 3)
)
return rotation_matrix
def Transform(image, rotation):
DIM = image.shape[1]
XDIM = DIM % 2
# List Destination Pixel indices
x = tf.repeat(tf.range(DIM // 2, -DIM // 2, -1), DIM)
y = tf.tile(tf.range(-DIM // 2, DIM // 2), [DIM])
z = tf.ones([DIM * DIM], dtype=tf.int32)
idx = tf.stack([x, y, z])
# Rotate destination pixels
m = get_rotation_matrix(rotation)
m = K.dot(m, tf.cast(idx, tf.float32))
m = tf.cast(m, tf.int32)
m = K.clip(m, -DIM // 2 + XDIM + 1, DIM // 2)
# Find Original Pixels
idx3 = tf.stack([DIM // 2 - m[0,], DIM // 2 - 1 + m[1,]])
image = tf.gather_nd(image, tf.transpose(idx3))
return tf.reshape(image, (DIM, DIM, 3))
a = "../input/siim-isic-melanoma-classification/jpeg/train/ISIC_0015719.jpg"
b = "../input/siim-isic-melanoma-classification/jpeg/train/ISIC_0052212.jpg"
a = cv2.imread(a)
a = cv2.resize(a, (512, 512))
b = cv2.imread(b)
b = cv2.resize(b, (512, 512))
img = Transform(a, 50)
plt.figure(figsize=(10, 6))
ax = plt.subplot(1, 2, 1)
ax.imshow(a)
ax.set_title("Original Image")
ax1 = plt.subplot(1, 2, 2)
ax1.imshow(img)
ax1.set_title("Rotated Image")
image = plt.imread(
"../input/siim-isic-melanoma-classification/jpeg/train/ISIC_0075663.jpg"
)
hue_image = tf.image.adjust_hue(image, delta=0.1)
plt.figure(figsize=(15, 9))
ax = plt.subplot(1, 2, 1)
ax.set_title("Original Image")
ax.imshow(image)
ax1 = plt.subplot(1, 2, 2)
ax1.set_title("Image with random Hue")
ax1.imshow(hue_image)
brightness_image = tf.image.adjust_brightness(image, delta=0.3)
plt.figure(figsize=(15, 9))
ax = plt.subplot(1, 2, 1)
ax.set_title("Original Image")
ax.imshow(image)
ax1 = plt.subplot(1, 2, 2)
ax1.set_title("Image with random Brightness")
ax1.imshow(brightness_image)
saturation_image = tf.image.adjust_saturation(image, saturation_factor=1.5)
plt.figure(figsize=(15, 9))
ax = plt.subplot(1, 2, 1)
ax.set_title("Original Image")
ax.imshow(image)
ax1 = plt.subplot(1, 2, 2)
ax1.set_title("Image with random Saturation")
ax1.imshow(saturation_image)
contrast_image = tf.image.adjust_contrast(image, contrast_factor=1.0)
plt.figure(figsize=(15, 9))
ax = plt.subplot(1, 2, 1)
ax.set_title("Original Image")
ax.imshow(image)
ax1 = plt.subplot(1, 2, 2)
ax1.set_title("Image with random Constrast")
ax1.imshow(contrast_image)
def cutout(
input_img,
p=0.5,
s_l=0.02,
s_h=0.4,
r_1=0.3,
r_2=1 / 0.3,
v_l=0,
v_h=255,
pixel_level=False,
):
img_h, img_w, img_c = input_img.shape
while True:
s = np.random.uniform(s_l, s_h) * img_h * img_w
r = np.random.uniform(r_1, r_2)
w = int(np.sqrt(s / r))
h = int(np.sqrt(s * r))
left = np.random.randint(0, img_w)
top = np.random.randint(0, img_h)
if left + w <= img_w and top + h <= img_h:
break
c = np.random.uniform(v_l, v_h)
input_img[top : top + h, left : left + w, :] = c
return input_img
a = "../input/siim-isic-melanoma-classification/jpeg/train/ISIC_0015719.jpg"
b = "../input/siim-isic-melanoma-classification/jpeg/train/ISIC_0052212.jpg"
a = cv2.imread(a)
a = cv2.resize(a, (512, 512))
b = cv2.imread(b)
b = cv2.resize(b, (512, 512))
plt.figure(figsize=(10, 6))
ax = plt.subplot(1, 2, 1)
ax.imshow(a)
ax.set_title("Original Image")
img = cutout(a)
ax1 = plt.subplot(1, 2, 2)
ax1.imshow(img)
ax1.set_title("Cutout Image")
a = "../input/siim-isic-melanoma-classification/jpeg/train/ISIC_0015719.jpg"
b = "../input/siim-isic-melanoma-classification/jpeg/train/ISIC_0052212.jpg"
a = cv2.imread(a)
a = cv2.resize(a, (512, 512))
b = cv2.imread(b)
b = cv2.resize(b, (512, 512))
DIM = 512
c = tf.random.uniform([], 0, 1)
img = tf.cast(a * c + (1 - c) * b, tf.int32)
plt.figure(figsize=(10, 6))
ax = plt.subplot(1, 3, 1)
ax.imshow(a)
ax.set_title("Original Image A")
ax1 = plt.subplot(1, 3, 2)
ax1.imshow(b)
ax1.set_title("Original Image B")
ax2 = plt.subplot(1, 3, 3)
ax2.imshow(img)
ax2.set_title("Image A Mixup with B")
a = "../input/siim-isic-melanoma-classification/jpeg/train/ISIC_0015719.jpg"
b = "../input/siim-isic-melanoma-classification/jpeg/train/ISIC_0052212.jpg"
a = cv2.imread(a)
a = cv2.resize(a, (512, 512))
b = cv2.imread(b)
b = cv2.resize(b, (512, 512))
DIM = 512
x = tf.cast(tf.random.uniform([], 0, DIM), tf.int32)
y = tf.cast(tf.random.uniform([], 0, DIM), tf.int32)
c = tf.random.uniform([], 0, 1)
WIDTH = tf.cast(DIM * tf.math.sqrt(c), tf.int32)
ya = tf.math.maximum(0, y - WIDTH // 2)
yb = tf.math.minimum(DIM, y + WIDTH // 2)
xa = tf.math.maximum(0, x - WIDTH // 2)
xb = tf.math.minimum(DIM, x + WIDTH // 2)
one = a[ya:yb, 0:xa, :]
two = b[ya:yb, xa:xb, :]
three = a[ya:yb, xb:, :]
img = tf.concat((one, two, three), axis=1)
img = tf.concat((a[0:ya, :, :], img, a[yb:DIM, :, :]), axis=0)
plt.figure(figsize=(10, 6))
ax = plt.subplot(1, 3, 1)
ax.imshow(a)
ax.set_title("Original Image A")
ax1 = plt.subplot(1, 3, 2)
ax1.imshow(b)
ax1.set_title("Original Image B")
ax2 = plt.subplot(1, 3, 3)
ax2.imshow(img)
ax2.set_title("Image A CutMix with B")
def hair_aug(input_image, label):
# Unnormalize: Returning the image from 0-1 to 0-255:
image = tf.identity(input_image)
image = tf.multiply(image, 255)
# Select Random Number of Hairs to Augment
random_number = tf.random.uniform([], 0, maxval=21, dtype=tf.int32)
scale = tf.cast(500 / 256, tf.int32)
for i in range(random_number):
# Randomly select a image to augment
hair_image_name = hair_images[
tf.random.uniform([], 0, maxval=tf.shape(hair_images)[0], dtype=tf.int32)
]
hair = tf.io.read_file(hair_image_name)
hair = tf.image.decode_jpeg(hair)
# Resize hair to new_width,new_scale
new_height = scale * tf.shape(hair)[0]
new_width = scale * tf.shape(hair)[1]
hair = tf.image.resize(hair, [new_height, new_width])
# Perform Augmentation on hair
hair = tf.image.random_flip_left_right(hair)
hair = tf.image.random_flip_up_down(hair)
n_rot = tf.random.uniform(shape=[], maxval=4, dtype=tf.int32)
hair = tf.image.rot90(hair, k=n_rot)
new_height, new_width = tf.shape(hair)[0], tf.shape(hair)[1]
# Top Left Coordinates
left_h = tf.random.uniform([], 0, 512 - new_height + 1, dtype=tf.int32)
left_w = tf.random.uniform([], 0, 512 - new_width + 1, dtype=tf.int32)
# Select Region of Interest
roi = image[left_h : left_h + new_height, left_w : left_w + new_width]
# Convert the hair image to grayscale (slice to remove the trainsparency channel)
hair2gray = tf.image.rgb_to_grayscale(hair[:, :, :3])
# Threshold:
mask = hair2gray > 10
# Get Image Background and just hair of hair image and add both.
image_bg = tf.multiply(
roi, tf.cast(tf.image.grayscale_to_rgb(~mask), dtype=tf.float32)
)
hair_fg = tf.multiply(
tf.cast(hair[:, :, :3], dtype=tf.int32),
tf.cast(tf.image.grayscale_to_rgb(mask), dtype=tf.int32),
)
dst = tf.add(image_bg, tf.cast(hair_fg, tf.float32))
# Generate paddings
paddings = tf.stack(
[
[left_h, 512 - (left_h + new_height)],
[left_w, 512 - (left_w + new_width)],
[0, 0],
]
)
dst_padded = tf.pad(dst, paddings)
# Create a boolean mask with zeros at the pixels of the augmentation segment and ones everywhere else
mask_img = tf.pad(tf.ones_like(dst), paddings)
mask_img = ~tf.cast(mask_img, dtype=tf.bool)
# Make a hole in the original image at the location of the augmentation segment
hole = tf.multiply(image, tf.cast(mask_img, dtype=tf.float32))
# Add hole and dst_padded
image = tf.add(hole, dst_padded)
# Normalize image
img = tf.multiply(image, 1 / 255)
return img, label
image_path = tf.io.gfile.glob(GCS_TRAIN + "/*.jpg")[0]
image = tf.io.read_file(image_path)
image = tf.image.decode_jpeg(image)
image = tf.image.resize(image, (512, 512))
img, _ = hair_aug(image, 0)
fig = plt.figure(figsize=(10, 6))
ax1 = fig.add_subplot(1, 2, 1)
ax1.imshow(tf.cast(image, tf.int32))
ax2 = fig.add_subplot(1, 2, 2)
ax2.imshow(tf.cast(img, tf.int32))
df = pd.read_csv("../input/siim-isic-melanoma-classification/train.csv")
image_names = list(df["image_name"])
image_names = [
os.path.join(GCS_TRAIN, image_name + ".jpg") for image_name in image_names
]
image_names = tf.convert_to_tensor(image_names)
targets = list(df["target"])
targets = tf.convert_to_tensor(targets)
def get_random_sample():
idx = tf.random.uniform([], 0, len(df), dtype=tf.int32)
image_name = image_names[idx]
target = targets[idx]
image = tf.io.read_file(image_name)
image = tf.image.decode_jpeg(image)
image = tf.image.resize(
image, (global_vars["IMAGE_SIZE"], global_vars["IMAGE_SIZE"])
)
image = tf.cast(image / 255.0, dtype=tf.float32)
target = tf.cast(target, dtype=tf.int64)
return image, target
# Augmentation functions
def get_transformation_matrix(
rotation, shear, height_zoom, width_zoom, height_shift, width_shift
):
# Convert degree to radians
rotation = math.pi * rotation / 180.0
shear = math.pi * shear / 180.0
# ROTATION MATRIX
c1 = tf.math.cos(rotation)
s1 = tf.math.sin(rotation)
one = tf.constant([1], dtype="float32")
zero = tf.constant([0], dtype="float32")
rotation_matrix = tf.reshape(
tf.concat([c1, s1, zero, -s1, c1, zero, zero, zero, one], axis=0), [3, 3]
)
# SHEAR MATRIX
c2 = tf.math.cos(shear)
s2 = tf.math.sin(shear)
shear_matrix = tf.reshape(
tf.concat([one, s2, zero, zero, c2, zero, zero, zero, one], axis=0), [3, 3]
)
# ZOOM MATRIX
zoom_matrix = tf.reshape(
tf.concat(
[
one / height_zoom,
zero,
zero,
zero,
one / width_zoom,
zero,
zero,
zero,
one,
],
axis=0,
),
[3, 3],
)
# SHIFT MATRIX
shift_matrix = tf.reshape(
tf.concat(
[one, zero, height_shift, zero, one, width_shift, zero, zero, one], axis=0
),
[3, 3],
)
return K.dot(K.dot(rotation_matrix, shear_matrix), K.dot(zoom_matrix, shift_matrix))
def Spatial_Transformation(image, label, DIM=global_vars["IMAGE_SIZE"]):
XDIM = DIM % 2
rotation = 180.0 * tf.random.normal([1], dtype="float32")
shear = 2.0 * tf.random.normal([1], dtype="float32")
height_zoom = 1.0 + tf.random.normal([1], dtype="float32") / 8.0
width_zoom = 1.0 + tf.random.normal([1], dtype="float32") / 8.0
height_shift = 8.0 * tf.random.normal([1], dtype="float32")
width_shift = 8.0 * tf.random.normal([1], dtype="float32")
m = get_transformation_matrix(
rotation, shear, height_zoom, width_zoom, height_shift, width_shift
)
# List Destination Pixels
x = tf.repeat(tf.range(DIM // 2, -DIM // 2, -1), DIM)
y = tf.tile(tf.range(-DIM // 2, DIM // 2), [DIM])
z = tf.ones([DIM * DIM], dtype="int32")
idx = tf.stack([x, y, z])
# ROTATE DESTINATION PIXELS ONTO ORIGIN PIXELS
idx2 = K.dot(m, tf.cast(idx, dtype="float32"))
idx2 = K.cast(idx2, dtype="int32")
idx2 = K.clip(idx2, -DIM // 2 + XDIM + 1, DIM // 2)
# FIND ORIGIN PIXEL VALUES
idx3 = tf.stack([DIM // 2 - idx2[0,], DIM // 2 - 1 + idx2[1,]])
d = tf.gather_nd(image, tf.transpose(idx3))
return tf.reshape(d, [DIM, DIM, 3]), label
def Color_Distortion(image, label):
image = tf.image.random_flip_left_right(image)
image = tf.image.random_hue(image, 0.01)
image = tf.image.random_saturation(image, 0.7, 1.3)
image = tf.image.random_contrast(image, 0.8, 1.2)
image = tf.image.random_brightness(image, 0.1)
return image, label
def Mixup_Augmentation(image, label):
# image=>(dim,dim,3) label=>(1,1)
image2, label2 = get_random_sample()
d = tf.random.uniform([], 0, 1, seed=42)
d1 = tf.cast(d, tf.float32)
d2 = tf.cast(d, tf.int64)
image = image * d1 + (1 - d1) * image2
label = label * d2 + (1 - d2) * label2
label = tf.cast(label, tf.int64)
return image, label
def Cutout_Augmentation(image, label):
p = 0.5
s_l = 0.02
s_h = 0.4
r_1 = 0.3
r_2 = 1 / 0.3
v_l = 0
v_h = 255
img_h, img_w = global_vars["IMAGE_SIZE"], global_vars["IMAGE_SIZE"]
while True:
s = np.random.uniform(s_l, s_h) * img_h * img_w
r = np.random.uniform(r_1, r_2)
w = int(np.sqrt(s / r))
h = int(np.sqrt(s * r))
left = np.random.randint(0, img_w)
top = np.random.randint(0, img_h)
if left + w <= img_w and top + h <= img_h:
break
c = np.random.uniform(v_l, v_h)
img = tf.reshape(tf.repeat(c, h * w * 3), (h, w, 3))
one = image[top : top + h, 0:left]
two = img
three = image[top : top + h, left + w : global_vars["IMAGE_SIZE"]]
img = tf.concat([one, two, three], axis=1)
return tf.concat((image[0:top, :, :], img, image[top + h :, :, :]), axis=0), label
def CutMix_Augmentation(image, label):
# image=>(dim,dim,3)
# label=>(1,1)
sample_image, sample_label = get_random_sample()
x, y = tf.cast(
tf.random.uniform([], 0, global_vars["IMAGE_SIZE"], seed=42), tf.int32
), tf.cast(tf.random.uniform([], 0, global_vars["IMAGE_SIZE"], seed=42), tf.int32)
b = tf.random.uniform([], 0, 1)
WIDTH = tf.cast(global_vars["IMAGE_SIZE"] * tf.math.sqrt(1 - b), tf.int32)
xmin, ymin = tf.math.maximum(0, x - WIDTH // 2), tf.math.maximum(0, y - WIDTH // 2)
xmax, ymax = tf.math.minimum(
global_vars["IMAGE_SIZE"], x + WIDTH // 2
), tf.math.minimum(global_vars["IMAGE_SIZE"], y + WIDTH // 2)
a = image[ymin:ymax, 0:xmin, :]
b = sample_image[ymin:ymax, xmin:xmax, :]
c = image[ymin:ymax, xmax:, :]
img = tf.concat((a, b, c), axis=1)
img = tf.concat(
[image[0:ymin, :, :], img, image[ymax : global_vars["IMAGE_SIZE"], :, :]],
axis=0,
)
a = tf.cast(
WIDTH * WIDTH / global_vars["IMAGE_SIZE"] / global_vars["IMAGE_SIZE"], tf.int64
)
label = label * a + (1 - a) * sample_label
label = tf.cast(label, tf.int64)
return img, label
def InformationDelection_Augmentation(image, label):
random_number = tf.cast(tf.random.uniform([], 1, 6, seed=42), tf.int64)
if random_number == 1:
return CutMix_Augmentation(image, label)
elif random_number == 2:
return Cutout_Augmentation(image, label)
elif random_number == 3:
return Mixup_Augmentation(image, label)
elif random_number == 4:
return hair_aug(image, label)
else:
return image, label
def InformationDelection_Augmentation(image, label):
random_number = tf.cast(tf.random.uniform([], 1, 4, seed=42), tf.int64)
if random_number == 1:
return Mixup_Augmentation(image, label)
elif random_number == 2:
return hair_aug(image, label)
else:
return image, label
# Data Pipeline
def read_labeled_tfrecord(example):
"""
This Function decodes a labeled tfrecord example.
returns image,target
"""
tfrecord_format = {
"image": tf.io.FixedLenFeature([], tf.string),
"image_name": tf.io.FixedLenFeature([], tf.string),
"patient_id": tf.io.FixedLenFeature([], tf.int64),
"sex": tf.io.FixedLenFeature([], tf.int64),
"age_approx": tf.io.FixedLenFeature([], tf.int64),
"anatom_site_general_challenge": tf.io.FixedLenFeature([], tf.int64),
"diagnosis": tf.io.FixedLenFeature([], tf.int64),
"target": tf.io.FixedLenFeature([], tf.int64),
}
example = tf.io.parse_single_example(example, tfrecord_format)
return example["image"], example["target"]
def read_unlabeled_tfrecord(example, image_name):
"""
This Function decodes a unlabeled tfrecord example.
returns image,image_name
"""
tfrecord_format = {
"image": tf.io.FixedLenFeature([], tf.string),
"image_name": tf.io.FixedLenFeature([], tf.string),
}
example = tf.io.parse_single_example(example, tfrecord_format)
return example["image"], example["image_name"] if image_name else 0
def prepare_image(image, label, augment=True, dim=256):
image = tf.image.decode_jpeg(image, channels=3)
image = tf.cast(image, dtype=tf.float32) / 255.0
if augment:
image, label = Spatial_Transformation(image, label, DIM=dim)
image, label = Color_Distortion(image, label)
image, label = InformationDelection_Augmentation(image, label)
image = tf.reshape(image, [dim, dim, 3])
return image, label
def get_dataset(
files,
labeled=True,
shuffle=True,
repeat=False,
augment=False,
batch_size=32,
dim=256,
return_image_name=True,
):
dataset = tf.data.TFRecordDataset(files, num_parallel_reads=global_vars["AUTO"])
dataset = dataset.cache()
if repeat:
dataset = dataset.repeat()
if shuffle:
dataset = dataset.shuffle(1024 * 8)
options = tf.data.Options()
options.experimental_deterministic = False
dataset = dataset.with_options(options)
if labeled:
dataset = dataset.map(
read_labeled_tfrecord, num_parallel_calls=global_vars["AUTO"]
)
else:
dataset = dataset.map(
lambda example: read_unlabeled_tfrecord(example, return_image_name),
num_parallel_calls=global_vars["AUTO"],
)
dataset = dataset.map(
lambda x, y: prepare_image(x, y, augment, dim),
num_parallel_calls=global_vars["AUTO"],
)
dataset = dataset.batch(batch_size * replicas)
dataset = dataset.prefetch(global_vars["AUTO"])
return dataset
def count_data_items(filenames):
return np.sum([int(file.split(".tfrec")[0].split("-")[2]) for file in filenames])
train_dataset = get_dataset(
train_files,
repeat=True,
augment=True,
batch_size=global_vars["BATCH_SIZE"],
dim=global_vars["IMAGE_SIZE"],
)
# Cylic Learning Rate Callback
def cylic(epoch, a, init_lr=0.0001, max_lr=0.001, step_size=3):
cycle = np.floor(1 + epoch / (2 * step_size))
x = np.abs(epoch / step_size - 2 * cycle + 1)
lr = init_lr + (max_lr - init_lr) * np.maximum(0, (1 - x)) * a(cycle)
return lr
# Learning-Rate Schedule
class LearningRateSchedule(tf.keras.callbacks.Callback):
def __init__(self, step_size, init_lr, max_lr, method, gamma=1):
super().__init__()
self.step_size = step_size
self.init_lr = init_lr
self.max_lr = max_lr
self.method = method
self.gamma = gamma
if self.method == "triangular":
self.a = lambda x: 1
self.schedule = cylic
elif self.method == "triangular2":
self.a = lambda x: 1 / (2 ** (x - 1))
self.schedule = cylic
elif self.method == "exp_range":
self.a = lambda x: gamma ** (x)
self.schedule = cylic
def on_train_begin(self, logs={}):
K.set_value(self.model.optimizer.learning_rate, self.init_lr)
def on_epoch_end(self, epoch, logs={}):
lr = self.schedule(epoch, self.a, self.init_lr, self.max_lr, self.step_size)
K.set_value(self.model.optimizer.learning_rate, lr)
# Plot Charts Function
def plot_charts(y_true, y_prob, history, fold):
losses = history.history["loss"]
auc = history.history["auc"]
y_pred = np.array([0 if i <= 0.5 else 1 for i in y_prob])
cm = confusion_matrix(y_true, y_pred)
grid = GridSpec(1, 8)
Figure = plt.figure(figsize=(15, 5), constrained_layout=True)
# Plot Line Chart of Loss and ROC AUC
plt.suptitle(f"{fold}", fontsize=20)
ax1 = Figure.add_subplot(grid[0, :2])
ax1.plot(np.arange(len(losses)), losses, "-o", label="Loss", color="#DC143C")
ax1.legend(loc=(0.6, 0.5))
ax1.set_title("Line Chart of Loss and ROC AUC")
ax12 = Figure.gca().twinx()
ax12.plot(np.arange(len(auc)), auc, "-o", label="AUC", color="#1E90FF")
ax12.legend(loc=(0.6, 0.43))
ax12.grid(False)
# Plot Confusion Matrix
ax2 = Figure.add_subplot(grid[0, 3:5])
sns.heatmap(cm, cbar=False, annot=True, ax=ax2)
ax2.set_xticklabels([])
ax2.set_yticklabels([])
ax2.set_title("Confusion Matrix")
# Plot ROC AUC Curve
fpr, tpr, _ = roc_curve(y_true, y_prob)
ax3 = Figure.add_subplot(grid[0, 6:8])
ax3.plot(fpr, tpr, "-o", color="#B22222")
ax3.fill_between(fpr, tpr, alpha=0.5, color="#F08080")
ax3.set_title("ROC AUC Curve")
plt.show()
return Figure
def get_model(dim=512):
inp = tf.keras.layers.Input(shape=(dim, dim, 3))
base = efn.EfficientNetB7(
input_shape=(dim, dim, 3), weights="imagenet", include_top=False
)
x = base(inp)
x = tf.keras.layers.GlobalAveragePooling2D()(x)
x = tf.keras.layers.Dense(1, activation="sigmoid")(x)
model = tf.keras.Model(inputs=inp, outputs=x)
opt = tf.keras.optimizers.Adam(learning_rate=0.001)
loss = tf.keras.losses.BinaryCrossentropy(label_smoothing=0.05)
model.compile(optimizer=opt, loss=loss, metrics=["AUC"])
return model
def count_data_items(filenames):
n = [
int(re.compile(r"-([0-9]*)\.").search(filename).group(1))
for filename in filenames
]
return np.sum(n)
LR_START = 0.00001
LR_MAX = 0.00005
LR_MIN = 0.00001
LR_RAMPUP_EPOCHS = 5
LR_SUSTAIN_EPOCHS = 0
LR_DECAY = 0.8
def lr_schedule(epoch):
if epoch < LR_RAMPUP_EPOCHS:
lr = (LR_MAX - LR_START) / LR_RAMPUP_EPOCHS * epoch + LR_START
elif epoch < LR_RAMPUP_EPOCHS + LR_SUSTAIN_EPOCHS:
lr = LR_MAX
else:
lr = (LR_MAX - LR_MIN) * LR_DECAY ** (
epoch - LR_RAMPUP_EPOCHS - LR_SUSTAIN_EPOCHS
) + LR_MIN
return lr
# Train Loop
def TrainModel():
kfold = KFold(n_splits=5, shuffle=True)
y_prediction = np.zeros((count_data_items(test_files), 1))
y_validation = np.zeros((count_data_items(train_files), 1))
losses = []
test_dataset = get_dataset(
test_files,
repeat=False,
shuffle=False,
dim=global_vars["IMAGE_SIZE"],
batch_size=global_vars["BATCH_SIZE"],
labeled=False,
return_image_name=False,
)
roc_auc_scores = []
if os.path.exists("SavedModels"):
shutil.rmtree("SavedModels")
os.mkdir("SavedModels")
for i, (train_index, val_index) in enumerate(kfold.split(np.arange(15))):
# Get all Files
Train_Files = tf.io.gfile.glob(
[GCS_PATH + "/train%.2i*.tfrec" % x for x in train_index]
)
Validation_Files = tf.io.gfile.glob(
[GCS_PATH + "/train%.2i*.tfrec" % x for x in train_index]
)
if global_vars["EXTERNAL_DATA"]:
Train_Files += tf.io.gfile.glob(
[GCS_PATH2 + "/train%.2i*.tfrec" % x for x in train_index * 2 + 1]
)
Train_Files += tf.io.gfile.glob(
[GCS_PATH2 + "/train%.2i*.tfrec" % x for x in train_index * 2]
)
np.random.shuffle(Train_Files)
# Get Dataset
train_dataset = get_dataset(
Train_Files,
repeat=True,
augment=True,
batch_size=global_vars["BATCH_SIZE"],
dim=global_vars["IMAGE_SIZE"],
)
val_dataset = get_dataset(
Validation_Files,
repeat=False,
augment=False,
shuffle=False,
batch_size=global_vars["BATCH_SIZE"],
dim=global_vars["IMAGE_SIZE"],
)
# Get Model
K.clear_session()
with strategy.scope():
model = get_model(dim=global_vars["IMAGE_SIZE"])
print("Model Sucessfully Loaded")
# Callbacks
ckpt = tf.keras.callbacks.ModelCheckpoint(
f"SavedModels/EfficientNet7{i+1}.h5",
save_best_only=True,
mode="min",
save_weights_only=True,
)
es = tf.keras.callbacks.EarlyStopping(
mode="min", patience=3, restore_best_weights=True
)
lr_scheduler = tf.keras.callbacks.LearningRateScheduler(lr_schedule)
callbacks = [ckpt, es, lr_scheduler]
# Calculate steps_per_epoch
steps_per_epoch = int(
count_data_items(Train_Files) / (global_vars["BATCH_SIZE"] * replicas)
)
# Fit Model
model.fit(
train_dataset,
validation_data=val_dataset,
epochs=15,
steps_per_epoch=steps_per_epoch,
callbacks=callbacks,
)
y_prediction[:, 0] += model.predict(test_dataset)[:, 0]
y_prediction[:, 0] = y_prediction[:, 0] / 5
return y_prediction
y_prediction = TrainModel()
from livelossplot import PlotLossesKeras
# Creating a config class to store all the configurations
class config:
# Image and Tabular data paths
DIRECTORY_PATH = "/kaggle/input/siim-isic-melanoma-classification/"
TRAINING_SAMPLES_FOLDER = DIRECTORY_PATH + "jpeg/train/"
TESTING_SAMPLES_FOLDER = DIRECTORY_PATH + "jpeg/test/"
TRAIN_FULL_DATA = DIRECTORY_PATH + "train.csv"
TEST_FULL_DATA = DIRECTORY_PATH + "test.csv"
# New directory path for image data
WORK_DIRECTORY = "dataset/"
TRAIN_IMAGES_FOLDER = WORK_DIRECTORY + "training_set/"
TEST_IMAGES_FOLDER = WORK_DIRECTORY + "test_set/"
VALIDATION_IMAGES_FOLDER = WORK_DIRECTORY + "validation_set/"
# Input parameters for data preprocessing
TARGET_NAME = "target"
TRAIN_SIZE = 0.80
VALIDATION_SIZE = 0.10
TEST_SIZE = 0.10
SEED = 42
# Tensorflow settings for model training
IMAGE_HEIGHT = 256
IMAGE_WIDTH = 256
NO_CHANNELS = 3
BATCH_SIZE = 256
EPOCHS = 20
DROPOUT = 0.5
LEARNING_RATE = 0.01
PATIENCE = 5
# Creating folders for training and validation data
dataset_home = "./dataset/"
subdirs = ["training_set/", "test_set/", "validation_set/"]
for subdir in subdirs:
labeldirs = ["benign", "malignant"]
for labeldir in labeldirs:
newdir = dataset_home + subdir + labeldir
os.makedirs(newdir, exist_ok=True)
# Splitting the dataset into train, test and validation set
test_examples = train_examples = validation_examples = 0
seed(config.SEED)
for record in open(config.TRAIN_FULL_DATA).readlines()[1:]:
split_record = record.split(",")
image_name = split_record[0]
target = split_record[7]
random_num = random.random()
if random_num < config.TRAIN_SIZE:
destination = config.TRAIN_IMAGES_FOLDER
train_examples += 1
# print('inif 1')
elif random_num < 0.9:
destination = config.VALIDATION_IMAGES_FOLDER
validation_examples += 1
else:
destination = config.TEST_IMAGES_FOLDER
test_examples += 1
if int(target) == 0:
shutil.copy(
config.TRAINING_SAMPLES_FOLDER + image_name + ".jpg",
destination + "benign/" + image_name + ".jpg",
)
elif int(target) == 1:
shutil.copy(
config.TRAINING_SAMPLES_FOLDER + image_name + ".jpg",
destination + "malignant/" + image_name + ".jpg",
)
# Preparing the data and performing Data Augmentation
train_datagen = ImageDataGenerator(
rescale=1.0 / 255,
shear_range=0.2,
zoom_range=(0.95, 0.95),
rotation_range=15,
horizontal_flip=True,
vertical_flip=True,
data_format="channels_last",
dtype=tf.float32,
)
validation_datagen = ImageDataGenerator(rescale=1.0 / 255, dtype=tf.float32)
train_generator = train_datagen.flow_from_directory(
directory=config.TRAIN_IMAGES_FOLDER,
target_size=(config.IMAGE_HEIGHT, config.IMAGE_WIDTH),
color_mode="rgb",
batch_size=config.BATCH_SIZE,
class_mode="binary",
shuffle=True,
)
validation_generator = validation_datagen.flow_from_directory(
directory=config.VALIDATION_IMAGES_FOLDER,
target_size=(config.IMAGE_HEIGHT, config.IMAGE_WIDTH),
color_mode="rgb",
batch_size=config.BATCH_SIZE,
class_mode="binary",
shuffle=True,
)
# Metrics to use for compiling the model
METRICS = [keras.metrics.AUC(name="auc")]
# Building a transfer learning model using Keras
base_model = tf.keras.applications.inception_resnet_v2.InceptionResNetV2(
include_top=False, classes=2
)
# 2. Freeze the base model(so the underlying pre-trained patterns aren't updated during training)
base_model.trainable = False
# 3. Create Inputs into our model
inputs = tf.keras.layers.Input(
shape=(config.IMAGE_HEIGHT, config.IMAGE_WIDTH, config.NO_CHANNELS),
name="input_layer",
)
# 4. If using ResNet50V2, add this to speed up convergence, remove for EfficientNet
x = tf.keras.layers.experimental.preprocessing.Rescaling(1.0 / 255)(inputs)
# 5. Pass the inputs to the base_model
x = base_model(inputs)
print(f"shape after passing inputs throught base modelL{x.shape}")
# 6. Average pool the outputs of the base model(aggreate all the most important information, reduce number of computation)
x = tf.keras.layers.GlobalAveragePooling2D(name="global_average_pooling")(x)
print(f"Shape after GlobalAveragingPooling2D: {x.shape}")
# 7. Create output activation layer
outputs = tf.keras.layers.Dense(1, activation="sigmoid", name="output_layer")(x)
# 8. Combine the inputs and outputs into a model
model_0 = tf.keras.Model(inputs, outputs)
# 9. Compile the model
model_0.compile(
optimizer="adam", loss=keras.losses.BinaryCrossentropy(), metrics=METRICS
)
# Compiling the transfer learning model
model_0.compile(
optimizer=keras.optimizers.Adam(),
loss=keras.losses.BinaryCrossentropy(),
metrics=METRICS,
)
# Training the model
history = model_0.fit(
train_generator,
epochs=config.EPOCHS,
validation_data=validation_generator,
validation_freq=1,
)
# Building a transfer learning model using Keras
base_model = tf.keras.applications.DenseNet121(include_top=False, classes=2)
base_model.trainable = False
inputs = tf.keras.layers.Input(
shape=(config.IMAGE_HEIGHT, config.IMAGE_WIDTH, config.NO_CHANNELS),
name="input_layer",
)
x = tf.keras.layers.experimental.preprocessing.Rescaling(1.0 / 255)(inputs)
x = base_model(inputs)
print(f"shape after passing inputs throught base modelL{x.shape}")
x = tf.keras.layers.GlobalAveragePooling2D(name="global_average_pooling")(x)
print(f"Shape after GlobalAveragingPooling2D: {x.shape}")
outputs = tf.keras.layers.Dense(1, activation="sigmoid", name="output_layer")(x)
model_1 = tf.keras.Model(inputs, outputs)
model_1.compile(
optimizer="adam", loss=keras.losses.BinaryCrossentropy(), metrics=METRICS
)
# Training the model
history = model_1.fit(
train_generator,
epochs=config.EPOCHS,
validation_data=validation_generator,
validation_freq=1,
)
# Building a transfer learning model using Keras
base_model = tf.keras.applications.VGG16(include_top=False, classes=2)
base_model.trainable = False
inputs = tf.keras.layers.Input(
shape=(config.IMAGE_HEIGHT, config.IMAGE_WIDTH, config.NO_CHANNELS),
name="input_layer",
)
x = tf.keras.layers.experimental.preprocessing.Rescaling(1.0 / 255)(inputs)
x = base_model(inputs)
print(f"shape after passing inputs throught base modelL{x.shape}")
x = tf.keras.layers.GlobalAveragePooling2D(name="global_average_pooling")(x)
print(f"Shape after GlobalAveragingPooling2D: {x.shape}")
outputs = tf.keras.layers.Dense(1, activation="sigmoid", name="output_layer")(x)
model_2 = tf.keras.Model(inputs, outputs)
model_2.compile(
optimizer="adam", loss=keras.losses.BinaryCrossentropy(), metrics=METRICS
)
# Training the model
history = model_2.fit(
train_generator,
epochs=config.EPOCHS,
validation_data=validation_generator,
validation_freq=1,
)
y_pred = model.predict(test_imgs)
cm = confusion_matrix(test_labels, y_pred)
cm_display = ConfusionMatrixDisplay(
confusion_matrix=cm, display_labels=["Melanoma", "Non-Melanoma"]
)
cm_display.plot()
plt.show()
total1 = sum(sum(cm))
#####from confusion matrix calculate accuracy
accuracy1 = (cm[0, 0] + cm[1, 1]) / total1
print("Accuracy : ", accuracy1)
sensitivity1 = cm[0, 0] / (cm[0, 0] + cm[0, 1])
print("Sensitivity : ", sensitivity1)
specificity1 = cm[1, 1] / (cm[1, 0] + cm[1, 1])
print("Specificity : ", specificity1)
y_pred = model_0.predict(test_imgs)
cm = confusion_matrix(test_labels, y_pred)
cm_display = ConfusionMatrixDisplay(
confusion_matrix=cm, display_labels=["Melanoma", "Non-Melanoma"]
)
cm_display.plot()
plt.show()
total1 = sum(sum(cm))
#####from confusion matrix calculate accuracy
accuracy1 = (cm[0, 0] + cm[1, 1]) / total1
print("Accuracy : ", accuracy1)
sensitivity1 = cm[0, 0] / (cm[0, 0] + cm[0, 1])
print("Sensitivity : ", sensitivity1)
specificity1 = cm[1, 1] / (cm[1, 0] + cm[1, 1])
print("Specificity : ", specificity1)
y_pred = model_1.predict(test_imgs)
cm = confusion_matrix(test_labels, y_pred)
cm_display = ConfusionMatrixDisplay(
confusion_matrix=cm, display_labels=["Melanoma", "Non-Melanoma"]
)
cm_display.plot()
plt.show()
total1 = sum(sum(cm))
#####from confusion matrix calculate accuracy
accuracy1 = (cm[0, 0] + cm[1, 1]) / total1
print("Accuracy : ", accuracy1)
sensitivity1 = cm[0, 0] / (cm[0, 0] + cm[0, 1])
print("Sensitivity : ", sensitivity1)
specificity1 = cm[1, 1] / (cm[1, 0] + cm[1, 1])
print("Specificity : ", specificity1)
y_pred = model_2.predict(test_imgs)
cm = confusion_matrix(test_labels, y_pred)
cm_display = ConfusionMatrixDisplay(
confusion_matrix=cm, display_labels=["Melanoma", "Non-Melanoma"]
)
cm_display.plot()
plt.show()
total1 = sum(sum(cm))
#####from confusion matrix calculate accuracy
accuracy1 = (cm[0, 0] + cm[1, 1]) / total1
print("Accuracy : ", accuracy1)
sensitivity1 = cm[0, 0] / (cm[0, 0] + cm[0, 1])
print("Sensitivity : ", sensitivity1)
specificity1 = cm[1, 1] / (cm[1, 0] + cm[1, 1])
print("Specificity : ", specificity1)
|
# # Predictive Maintenance
# ## Import Libraries
# Data Analysis
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
# Machine Learning tools
from sklearn.preprocessing import MinMaxScaler
from sklearn.metrics import (
confusion_matrix,
classification_report,
mean_absolute_error,
r2_score,
)
from sklearn.model_selection import GridSearchCV
# Machine Learning algos
from sklearn.linear_model import LinearRegression
from sklearn.svm import SVR
from sklearn.ensemble import RandomForestRegressor
from sklearn.neighbors import KNeighborsRegressor
# Keras for RNN
from keras.models import load_model
from keras.callbacks import EarlyStopping
from keras.models import Sequential
from keras.layers import Dense, Dropout, ConvLSTM2D, LSTM, Activation
# ## Data Import
# Load the data in kaggle
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Save as dataframe
train_df = pd.read_csv(
"/kaggle/input/preventive-to-predicitve-maintenance/Train_Data_CSV.csv"
)
test_df = pd.read_csv(
"/kaggle/input/preventive-to-predicitve-maintenance/Test_Data_CSV.csv"
)
# ## Data Exploration
train_df.head()
test_df.head()
train_df.describe()
# We got 50 diffrent time series with a total of 39420 time sets in our train data.
# Lets check for missing values with df.info:
train_df.info()
test_df.info()
# ### Plot The Data
# We can plot the data for each individual time series in one plot
plt.figure(figsize=(20, 10))
for data_no, df in train_df.groupby("Data_No"):
plt.plot(df["Time"], df["Differential_pressure"])
plt.xlabel("Time", fontsize=20)
plt.ylabel("Differential Pressure", fontsize=20)
plt.tick_params(axis="both", labelsize=16)
plt.title("Differential Pressure over Time", fontsize=30)
# We can see some time series reaches 600. This are the run to fail data in the train set. On these curves we want to predict, when it reaches 600 = failure.
plt.figure(figsize=(20, 10))
for data_no, df in train_df.groupby("Data_No"):
plt.plot(df["Time"], df["Flow_rate"])
# There are two diffrent flow rates but it does not change over time. The flow rate is constant.
train_df["Dust_feed"].value_counts(ascending=True).plot(kind="barh")
# Dust feed is also constant, but can have 8 different values. Some are underrepresented.
# ### Correlation
# Let's check the correlation of the data. We take a look at the test data because we are interested in the RUL. Also time and Data No does not belong here.
# Create correlation matrix
corr_matrix = test_df[["Differential_pressure", "Flow_rate", "Dust_feed", "RUL"]].corr()
# Farbliche Abgrenzung hinzufügen
corr_matrix.style.background_gradient(cmap="coolwarm")
# ## Data Preparation
# # Reshape the data to sequences
# Since we are dealing with time series data and also have 50 different time series, we want to prepare the data as followed:
# For reshaping the data credits to Michael Holting: https://www.kaggle.com/code/michaelhotaling2/calculating-remaining-useful-life-using-an-rnn
# This function splits the input data into sequences and returns them as a generator
def x_reshape(df, columns, sequence_length):
data = df[columns].values
num_elements = data.shape[0]
for start, stop in zip(
range(0, num_elements - sequence_length - 20),
range(sequence_length, num_elements - 20),
):
yield (data[start:stop, :])
# This function creates sequences of input data
def get_x_slices(df, feature_columns):
# Reshape the data to (samples, time steps, features)
feature_list = [
list(x_reshape(df[df["Data_No"] == i], feature_columns, 20))
for i in range(1, df["Data_No"].nunique() + 1)
if len(df[df["Data_No"] == i]) > 20
]
feature_array = np.concatenate(list(feature_list), axis=0).astype(np.float64)
length = len(feature_array) // 128
return feature_array[: length * 128]
# This function creates labels for the output data
def y_reshape(df, sequence_length, columns=["Differential_pressure"]):
data = df[columns].values
num_elements = data.shape[0]
return data[sequence_length + 20 : num_elements, :]
# This function creates sequences of output data
def get_y_slices(df):
label_list = [
y_reshape(df[df["Data_No"] == i], 20)
for i in range(1, df["Data_No"].nunique() + 1)
]
label_array = np.concatenate(label_list).astype(np.float64)
length = len(label_array) // 128
return label_array[: length * 128]
scaler = MinMaxScaler()
X_train = get_x_slices(train_df, ["Differential_pressure"])
# X_train = scaler.fit_transform(X_train.reshape(-1, X_train.shape[-1])).reshape(X_train.shape)
X_test = get_x_slices(test_df, ["Differential_pressure"])
# X_test = scaler.transform(X_test.reshape(-1, X_test.shape[-1])).reshape(X_test.shape)
y_train = get_y_slices(train_df)
y_test = get_y_slices(test_df)
print(X_train.shape)
print(X_test.shape)
print(y_train.shape)
print(y_test.shape)
# Drop the last axis
X_train = np.squeeze(X_train, axis=2)
y_train = np.squeeze(y_train, axis=1)
X_test = np.squeeze(X_test, axis=2)
y_test = np.squeeze(y_test, axis=1)
print(X_train.shape)
print(X_test.shape)
print(y_train.shape)
print(y_test.shape)
# We want to have a validation set so we just split of the last 10% of the train data for that. (Normally you want to shuffle it or use random sets)
# split train data in 90% train, 10% validation
train_size = int(len(X_train) * 0.9)
X_train, X_val = X_train[:train_size], X_train[train_size:]
y_train, y_val = y_train[:train_size], y_train[train_size:]
print("X Shape:", X_train.shape)
print("X Shape:", X_val.shape)
# Scale the data to a value between 0 and 1
scaler = MinMaxScaler()
X_train = scaler.fit_transform(X_train)
X_val = scaler.transform(X_val)
X_test = scaler.transform(X_test)
# ## Use ML
# #### Define Evaluation
# R2 score & MAE
def evaluate(y_true, y_pred, label="test"):
mae = mean_absolute_error(y_true, y_pred)
r2 = r2_score(y_true, y_pred)
print("{} set MAE:{}, R2:{}".format(label, mae, r2))
# ### Linear Regression
lin_reg = LinearRegression()
lin_reg.fit(X_train, y_train)
y_lr_train = lin_reg.predict(X_train) # Prediction on train data
evaluate(y_train, y_lr_train, label="train")
y_lr_test = lin_reg.predict(X_test) # Prediction on test data
evaluate(y_test, y_lr_test, label="test")
# ### Support Vector
sv = SVR(kernel="rbf")
sv.fit(X_train, y_train)
y_sv_train = sv.predict(X_train) # Prediction on train data
evaluate(y_train, y_sv_train, label="train")
y_sv_test = sv.predict(X_test) # Prediction on test data
evaluate(y_test, y_sv_test, label="test")
param_grid = {
"C": [
0.1,
1,
10,
],
"gamma": [
0.1,
0.01,
0.001,
],
"kernel": ["rbf", "linear"],
}
grid_search = GridSearchCV(
sv, param_grid, cv=5, scoring="neg_mean_squared_error", return_train_score=True
)
grid_search.fit(X_train, y_train)
grid_search.best_params_
final_model = grid_search.best_estimator_
cvres = grid_search.cv_results_
for mean_score, params in zip(cvres["mean_test_score"], cvres["params"]):
print(np.sqrt(-mean_score), params)
sv = SVR(kernel="rbf")
sv.fit(X_train, y_train)
y_sv_train = sv.predict(X_train) # Prediction on train data
evaluate(y_train, y_sv_train, label="train")
y_sv_test = sv.predict(X_test) # Prediction on test data
evaluate(y_test, y_sv_test, label="test")
# ### Random Forrest
rf = RandomForestRegressor(max_features="sqrt", n_estimators=30, random_state=42)
rf.fit(X_train, y_train)
y_rf_train = rf.predict(X_train) # Prediction on train data
evaluate(y_train, y_rf_train, label="train")
y_rf_test = rf.predict(X_test) # Prediction on test data
evaluate(y_test, y_rf_test, label="test")
param_grid = [
{"n_estimators": [3, 10, 30], "max_features": [2, 4, 6, 8]},
{"bootstrap": [False], "n_estimators": [3, 10], "max_features": [2, 3, 4]},
]
grid_search = GridSearchCV(
rf, param_grid, cv=5, scoring="neg_mean_squared_error", return_train_score=True
)
grid_search.fit(X_train, y_train)
grid_search.best_params_
final_model = grid_search.best_estimator_
cvres = grid_search.cv_results_
for mean_score, params in zip(cvres["mean_test_score"], cvres["params"]):
print(np.sqrt(-mean_score), params)
# ### KNN
knn = KNeighborsRegressor()
knn.fit(X_train, y_train)
y_knn_train = knn.predict(X_train) # Prediction on train data
evaluate(y_train, y_knn_train, label="train")
y_knn_test = knn.predict(X_test) # Prediction on test data
evaluate(y_test, y_knn_test, label="test")
# ### Evaluation
pred_failures = y_lr_test >= 400
real_failures = y_test >= 550
# Visualize the confusion matrix
fig, ax = plt.subplots(figsize=(8, 6))
ax = sns.heatmap(
confusion_matrix(real_failures, pred_failures),
annot=True,
annot_kws={"size": 14, "weight": "bold"},
fmt="d",
cbar=False,
cmap="Blues",
)
ax.set_xticklabels(["OK", "Near Failure"])
ax.set_yticklabels(["OK", "Near Failure"], va="center")
plt.tick_params(axis="both", labelsize=14, length=0)
plt.ylabel("Actual", size=14, weight="bold")
plt.xlabel("Predicted", size=14, weight="bold")
plt.show()
# Plot true and predicted values
plt.figure(figsize=(30, 15))
plt.plot(y_lr_test, label="Pred", color="red")
plt.plot(y_test, label="True", color="blue")
plt.legend()
plt.show()
# nur einen teil plotten:
y_lr_test_part = y_lr_test[1200:1600]
y_test_part = y_test[1200:1600]
# Plot true and predicted values
plt.figure(figsize=(30, 15))
plt.plot(y_lr_test_part, label="Pred", color="red")
plt.plot(y_test_part, label="True", color="blue")
plt.legend()
plt.show()
# ## Reccurent Neural Network
# Building the RNN
# An RNN is a neural network.The predicted value will be the remaining useful life after those n observations. We will need to reshape our data for the RNN by using the functions below.
# ### Reshape the data for input in a LSTM RNN
# An LSTM layer needs a special input shape for its data. It has to be a 3D array: (number of samples, time steps, number of features)
scaler = MinMaxScaler()
X_train = get_x_slices(train_df, ["Differential_pressure", "Flow_rate"])
X_train = scaler.fit_transform(X_train.reshape(-1, X_train.shape[-1])).reshape(
X_train.shape
)
X_test = get_x_slices(test_df, ["Differential_pressure", "Flow_rate"])
X_test = scaler.transform(X_test.reshape(-1, X_test.shape[-1])).reshape(X_test.shape)
y_train = get_y_slices(train_df)
y_test = get_y_slices(test_df)
# split train data in 90% train, 10% validation
train_size = int(len(X_train) * 0.9)
X_train, X_val = X_train[:train_size], X_train[train_size:]
y_train, y_val = y_train[:train_size], y_train[train_size:]
print("X Shape:", X_train.shape)
print("X Shape:", X_val.shape)
LSTM_model = Sequential()
LSTM_model.add(
LSTM(
input_shape=(X_train.shape[1], X_train.shape[2]),
units=16,
return_sequences=False,
)
)
LSTM_model.add(Dropout(0.2))
# LSTM_model.add(LSTM(units=64,
# return_sequences=True))
# LSTM_model.add(Dropout(0.2))
# LSTM_model.add(LSTM(units=32,
# return_sequences=False))
# LSTM_model.add(Dropout(0.2))
LSTM_model.add(Dense(units=1))
LSTM_model.add(Dropout(0.2))
LSTM_model.add(Activation("linear"))
LSTM_model.compile(loss="mean_squared_error", optimizer="adam", metrics=["mse", "mae"])
LSTM_model.summary()
es = EarlyStopping(monitor="val_mse", patience=2, verbose=True)
history = LSTM_model.fit(
X_train, y_train, epochs=999, validation_data=(X_val, y_val), callbacks=[es]
)
fig, [ax1, ax2] = plt.subplots(1, 2, figsize=(16, 4))
ax1.plot(
np.arange(1, len(history.history["loss"]) + 1),
history.history["loss"],
color="blue",
label="Mean Squared Error",
)
ax1.plot(
np.arange(1, len(history.history["val_loss"]) + 1),
history.history["val_loss"],
color="black",
label="Val Mean Squared Error",
)
ax1.legend()
ax2.plot(
np.arange(1, len(history.history["mae"]) + 1),
history.history["mae"],
color="blue",
label="Mean Absolute Error",
)
ax2.plot(
np.arange(1, len(history.history["val_mae"]) + 1),
history.history["val_mae"],
color="black",
label="Val Mean Absolute Error",
)
ax2.legend()
pred = LSTM_model.predict(X_test)
print(pred)
y_pred = pred.reshape(1, -1)[0]
print(y_pred.shape)
print(y_test)
pred_failures = pred >= 400
real_failures = y_test >= 400
print(real_failures.shape)
print(pred_failures.shape)
# Visualize the confusion matrix
fig, ax = plt.subplots(figsize=(4, 3))
ax = sns.heatmap(
confusion_matrix(real_failures, pred_failures),
annot=True,
annot_kws={"size": 14, "weight": "bold"},
fmt="d",
cbar=False,
cmap="Blues",
)
ax.set_xticklabels(["OK", "Defective"])
ax.set_yticklabels(["OK", "Defective"], va="center")
plt.tick_params(axis="both", labelsize=14, length=0)
plt.ylabel("Actual", size=14, weight="bold")
plt.xlabel("Predicted", size=14, weight="bold")
plt.show()
plt.figure(figsize=(16, 9))
sns.scatterplot(
x=pred.reshape(1, -1)[0],
y=y_test.reshape(1, -1)[0],
hue=y_test.reshape(1, -1)[0] - pred.reshape(1, -1)[0],
palette="inferno",
s=10,
)
plt.xlim(0, 140)
plt.ylim(0, 140)
plt.plot([0, 140], [0, 140], linestyle="--", color="red")
# Plot true and predicted values
plt.figure(figsize=(16, 9))
plt.plot(pred, label="Pred", color="red")
plt.plot(y_test, label="True", color="blue")
plt.legend()
plt.show()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.preprocessing import StandardScaler
import warnings
warnings.filterwarnings("ignore")
import cufflinks as cf
cf.go_offline()
cf.set_config_file(offline=False, world_readable=True)
from tensorflow.keras.layers import BatchNormalization
from keras.backend import dropout
from keras.models import Model, load_model, Sequential
from tensorflow.keras.models import Model
from keras.layers import Input, Dense, Dropout
import warnings
warnings.filterwarnings("ignore")
import cufflinks as cf
cf.go_offline()
cf.set_config_file(
offline=False, world_readable=True
) # link pandas to plotly and add the iplot method
all_data = pd.read_csv("/kaggle/input/lung-cancer-detection/survey lung cancer.csv")
all_data
all_data.info()
# # Processing data
all_data["LUNG_CANCER"] = all_data.LUNG_CANCER.map({"YES": 1, "NO": 0})
all_data.columns
columns = [
"GENDER",
"AGE",
"SMOKING",
"YELLOW_FINGERS",
"ANXIETY",
"PEER_PRESSURE",
"CHRONIC DISEASE",
"FATIGUE ",
"ALLERGY ",
"WHEEZING",
"ALCOHOL CONSUMING",
"COUGHING",
"SHORTNESS OF BREATH",
"SWALLOWING DIFFICULTY",
"CHEST PAIN",
"LUNG_CANCER",
]
df = pd.get_dummies(all_data[columns])
df
sc = StandardScaler()
df_NoScaled = df[["AGE"]]
df.drop(["AGE"], axis=1, inplace=True)
df_NoScaled = df_NoScaled.values
df_NoScaled = sc.fit_transform(df_NoScaled)
df_NoScaled = pd.DataFrame(df_NoScaled, columns=["AGE"])
df = pd.concat([df_NoScaled, df], axis=1)
df.head(2)
df
df_lung_cancer = df["LUNG_CANCER"]
model_df = df.drop(["LUNG_CANCER"], axis=1)
df_lung_cancer
# # Data Visualization
plt.figure(figsize=(16, 8))
sns.set_style("whitegrid")
plt.title(
"Grouping People by YELLOW FINGERS ",
fontsize=30,
fontweight="bold",
y=1.05,
)
plt.xlabel("YELLOW_FINGERS", fontsize=25)
plt.ylabel("Count", fontsize=25)
sns.countplot(x="YELLOW_FINGERS", data=all_data, palette="hls")
plt.show()
gender = all_data.groupby(all_data["GENDER"])["LUNG_CANCER"].sum()
df_gender = pd.DataFrame({"labels": gender.index, "values": gender.values})
colors = ["pink", "blue"]
df_gender.iplot(
kind="pie",
labels="labels",
values="values",
title="LUNG CANCER among Gender",
colors=colors,
)
cp = all_data.groupby(all_data["CHEST PAIN"])["LUNG_CANCER"].sum()
df_cp = pd.DataFrame({"labels": cp.index, "values": cp.values})
colors = ["yellow", "red"]
df_cp.iplot(
kind="pie",
labels="labels",
values="values",
title="LUNG CANCER among CHEST PAIN",
colors=colors,
)
wz = all_data.groupby(all_data["WHEEZING"])["LUNG_CANCER"].sum()
df_wz = pd.DataFrame({"labels": wz.index, "values": wz.values})
colors = ["yellow", "red"]
df_cp.iplot(
kind="pie",
labels="labels",
values="values",
title="LUNG CANCER among WHEEZING",
colors=colors,
)
# # Model building
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(
model_df, df_lung_cancer, test_size=0.20, shuffle=True
)
print("x_train shape: ", x_train.shape)
print("x_test shape: ", x_test.shape)
print("y_train shape: ", y_train.shape)
print("y_test shape: ", y_test.shape)
print("Number of classes ", len(np.unique(y_train)))
from sklearn.ensemble import RandomForestClassifier
def rf(x_train, y_train, n_estimators=300):
rndforest = RandomForestClassifier(n_estimators=n_estimators, n_jobs=-1)
rndforest.fit(x_train, y_train)
return rndforest
random_forest_en = rf(x_train, y_train, n_estimators=300)
print("Training accuracy:", random_forest_en.score(x_train, y_train))
print("Validation accuracy", random_forest_en.score(x_test, y_test))
import itertools
def plot_confusion_matrix(
model,
X,
y,
class_names,
file_name,
normalize=False,
title="CONFUSION MATRIX",
cmap=plt.cm.Greys,
):
y_pred = model.predict(X)
# Compute confusion matrix
cnf_matrix = confusion_matrix(y, y_pred)
np.set_printoptions(precision=1)
plt.figure(figsize=(18, 16))
if normalize:
cnf_matrix = cnf_matrix.astype("float") / cnf_matrix.sum(axis=1)[:, np.newaxis]
print("CONFUSION MATRIX")
else:
print("CONFUSION MATRIX")
plt.imshow(cnf_matrix, interpolation="nearest", cmap=cmap)
plt.title("CONFUSION MATRIX")
plt.colorbar()
tick_marks = np.arange(len(class_names))
plt.xticks(tick_marks, class_names, rotation=90)
plt.yticks(tick_marks, class_names)
fmt = ".2f" if normalize else "d"
thresh = cnf_matrix.max() / 2.0
for i, j in itertools.product(
range(cnf_matrix.shape[0]), range(cnf_matrix.shape[1])
):
plt.text(
j,
i,
format(cnf_matrix[i, j], fmt),
horizontalalignment="center",
color="white" if cnf_matrix[i, j] > thresh else "black",
)
plt.ylabel("CLASS")
plt.xlabel("PREDICTED CLASS")
plt.tight_layout()
plt.savefig(file_name + ".png")
plt.show()
from sklearn.metrics import confusion_matrix
LABELS = ["YES LUNG CANACER", "NO LUNG CANCER"]
plot_confusion_matrix(
random_forest_en,
x_test,
y_test,
class_names=LABELS,
file_name="ConfussionMatrix",
normalize=True,
)
# # NOW....WITH NEURAL NETWORKS
print(x_train.shape[0], x_train.shape[1])
model = Sequential()
model.add(Dense(128, activation="relu", input_shape=(16,)))
model.add(Dropout(0.2))
model.add(Dense(256, activation="relu"))
model.add(Dropout(0.2))
model.add(Dense(256, activation="relu"))
model.add(Dense(1, activation="sigmoid"))
model.summary()
from keras.callbacks import ModelCheckpoint, EarlyStopping
early_stop = EarlyStopping(monitor="val_loss", mode="min", verbose=1, patience=20)
model.compile(optimizer="adam", loss="binary_crossentropy", metrics=["accuracy"])
history = model.fit(
x_train,
y_train,
batch_size=512,
epochs=100,
validation_data=(x_test, y_test),
callbacks=[early_stop],
)
acc = history.history["accuracy"]
val_acc = history.history["val_accuracy"]
loss = history.history["loss"]
val_loss = history.history["val_loss"]
plt.plot(loss, label="Training Loss")
plt.plot(val_loss, label="Validation Loss")
plt.title("Training And Validation Loss")
plt.ylabel("LOSS")
plt.xlabel("EPOCH")
plt.legend(["TRAIN Loss", "VALIDATION Loss"], loc="upper left")
plt.rcParams["figure.figsize"] = [16, 9]
plt.legend()
plt.show()
plt.plot(acc, label="Training Acc")
plt.plot(val_acc, label="Validation Acc")
plt.title("Training And Validation Accuracy")
plt.ylabel("ACCURACY")
plt.xlabel("EPOCH")
plt.legend(["TRAIN Accuracy", "VALIDATION Accuracy"], loc="upper left")
plt.rcParams["figure.figsize"] = [16, 9]
plt.legend()
plt.show()
|
# # Intro
# *Disclaimer: I am not an expert of any sort in astrophysics. These are just findings from the papers that were recommended to read and my personal observations. I apologies for any errors / mistakes in this notebook.*
# In this notebook we do a quick overview of classification problem, literature overview and suggest some solutions for classification.
# **What are the different events?**
# Per year, [IceCube reads out roughly](https://storage.googleapis.com/kaggle-forum-message-attachments/1958559/18618/kaggle_webinar_small.pdf):
# 1. 10^10 events caused by atmospheric muons
# 2. 10^9 events caused by noise
# 3. 100 000 events from atmospheric neutrinos
# 4. A handful of very high energy events likely to be of astrophysical origin
#
#
#
# The above is the split of events in the real life, however, in our Kaggle competition we have synthetic data and from what we know from the [presentation](https://storage.googleapis.com/kaggle-forum-message-attachments/1958559/18618/kaggle_webinar_small.pdf): **every event has a neutrino in them (either atmospheric or of astrophysical origin)**, some events have noise + atmospheric muons and thus are hard to reconstruct.
# ## We can classify the events as following:
# 1. Events that are easy to reconstruct (no dominating noise, atmospheric muons)
# 2. Events that are hard to reconstruct (a lot of noise and/or atmospheric muons)
# ## What do these events look like? Showers/Cascades and Tracks
# Neutrino interactions in IceCube have two primary topologies:
# * **Showers (or Cascades)**: When a high-energy neutrino collides with a nucleus, it produces a cascade of particles, including charged particles such as electrons and muons, as well as gamma rays and other types of radiation. These particles then continue to collide with other nuclei in the atmosphere, producing more particles and radiation in a cascading effect.
# * **Secondary muon tracks** are created when neutrinos crash into other particles. This impact causes a big explosion of even smaller particles called hadrons. In this explosion, some of the particles are muons that leave behind a special track that we can detect.
# Approximately 80% of the observed events would appear as [showers](https://arxiv.org/pdf/1311.5238.pdf).
# ## Example of neutrino-induced particles shower
# Thanks to [edguy99](https://www.kaggle.com/edguy99) for the construction of these animations. ([forum discussion](https://www.kaggle.com/competitions/icecube-neutrinos-in-deep-ice/discussion/388858))
# 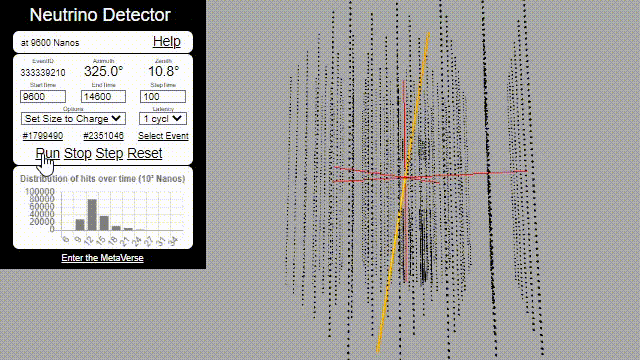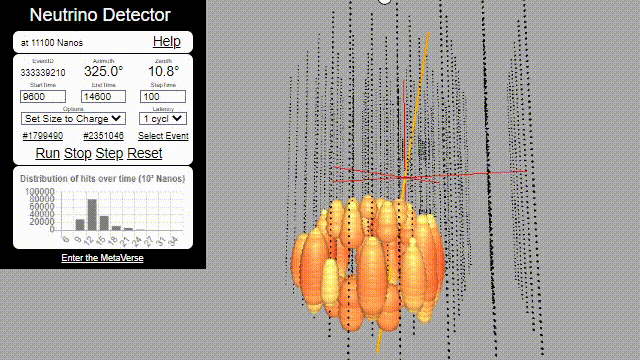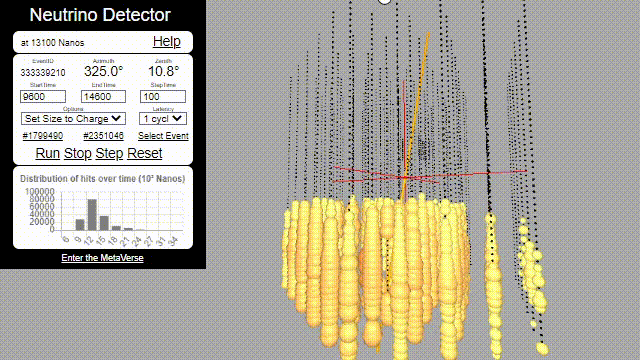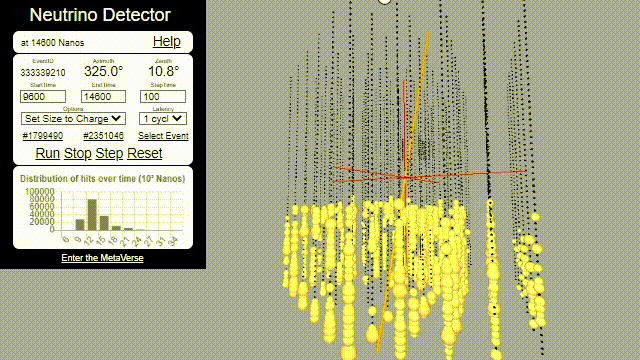
# In here we see an example of 'cascade-like' behavior - the neutrino 'explodes' inside the detector and deposits its energy in there.
# Here is an illustration from a [paper](https://arxiv.org/pdf/1311.4767.pdf):
# 
# ## Example of neutrino-induced muon-track
# 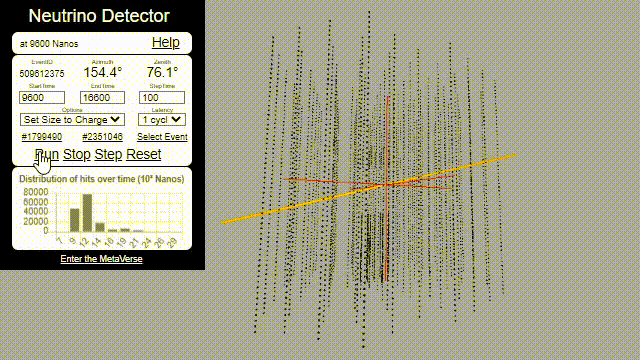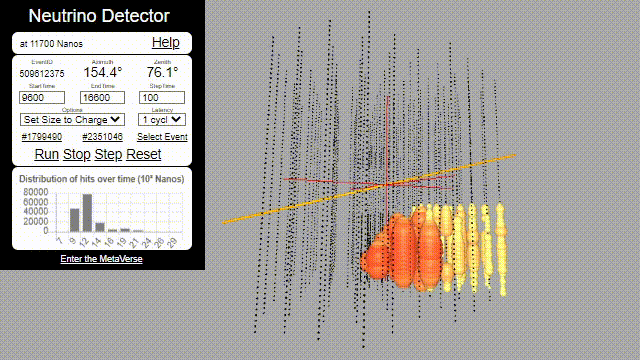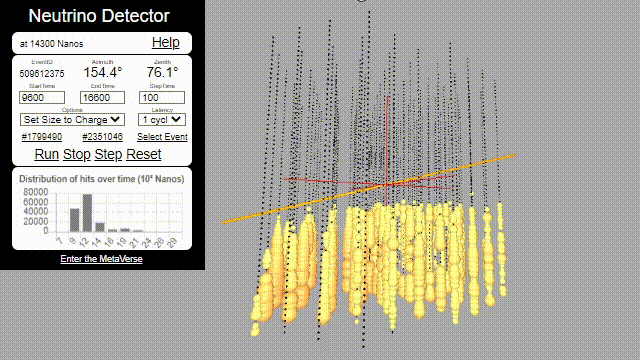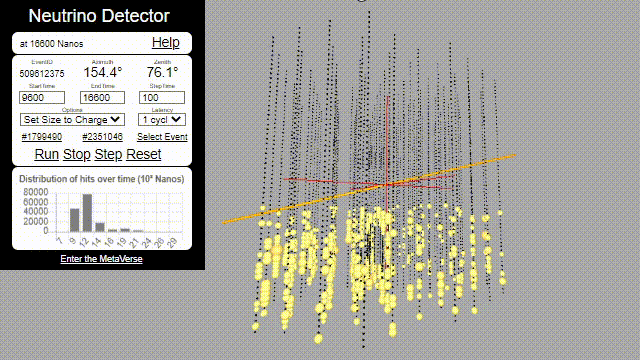
# Here is an illustration from [paper](https://arxiv.org/pdf/1311.4767.pdf) with captions that reads that muon has started in the detector and escaped through one of the facets:
# 
# # What are hard to reconstruct events?
# We know that each event has a neutrino in it, however, in some cases these neutrinos are hard to detect. We want to concentrate specifically on classifying the atmospheric muons.
# Animation of hard to detect neutrinos - you can see the line of neutrinos which is kind of hard to make sense given the lightened up detectors.
# 
# **What do we know about atmospheric muons?**:
# * The primary mechanism for separating the cosmic ray muons from the neu156 trino muons is reconstructing the muon track and determining whether the muon was traveling downwards into the Earth or upwards out of the Earth. Because neutrinos can penetrate through the Earth but cosmic ray muons cannot, it follows that a muon traveling out of the Earth must have been generated by a neutrino. Thus, by selecting only the muons that are reconstructed as up-going,the cosmic ray muons can, in principle, be removed from the data. Because the number of cosmic ray muons overwhelms the number of neutrino muons, high accuracy is critical for preventing erroneous reconstruction of cosmic ray muons as neutrino-induced
# * Atmospheric muons can in principle be excluded by simple geometrical considerations. To exclude the contribution from downward-going atmospheric muons, it is sufficient to identify upward-going events, which can only be produced by neutrinos.
# * This distinction requires a reliable determination of the elevation angle, because downward-going atmospheric muons outnumber upward-going atmospheric neutrinos by 5 to 6 orders of magnitude for typical neutrino telescope installation depths. [Source](https://arxiv.org/abs/1105.4116)
# # Literature overview: finding atmospheric muons
# ## Evidence for High-Energy Extraterrestrial Neutrinos at the IceCube Detector
# url: https://arxiv.org/pdf/1311.5238.pdf
# **Short summary**:
# * The paper studied a number of high energy events and concluded that they were inconsistent with those expected from the atmospheric muon and concluded its extraterrestrial origin.
# * The main metric was the total charge (the larger charge is more likely to be extraterrestrial)
# * Introducing the veto-layers (gray-out) that are used as a "filter"
# 
# * The shaded region in the middle contains ice of high dust concentration . Because of the high degree of light absorption in this region, near horizontal events could have entered here without being tagged at the sides of the detector without a dedicated tagging region.
# **Takeaways**:
# * Like in the given work we can filter out atmospheric muons by restricting the region of origin of the event. Given that the muons come from above or from the sides (muons don't penetrate the earth crest)
# # An algorithm for the reconstruction of neutrino-induced showers in the ANTARES neutrino telescope
# url: https://arxiv.org/abs/1708.03649
# **Short summary**:
# * The rate at which electron neutrinos are produced in the atmosphere at the energy of interest of neutrino telescopes (1 TeV to 1000 TeV) is more than a factor of 10 less compared to atmospheric muon neutrinos.
# * High-energy muons can travel straight for several kilometres through the rock and water surrounding the detector. Showers, on the other hand, deposit all their energy within a few metres from their interaction vertex.
# * In this paper, an algorithm optimised for accurate reconstruction of energy and direction of shower events in the ANTARES detector is presented.
# * **Several criterion chosed to classify correctly the events of 'showers', the list is below in the table:**
# 
# * **Interesting criterion that we can use in our study**:
# **Containment + M-Estimator**:
# * Reconstructing atmospheric muons with a shower algorithm often produces “shower positions” that lie far away from the detector boundary and have a large MEst value. A rough selection on position and reconstruction quality reduces the amount of background by 70 % already before the direction fit.
# **GridFit Ratio**
# * The GridFit algorithm was developed for another [paper](https://www.nikhef.nl/pub/services/biblio/theses_pdf/thesis_EL_Visser.pdf). It is used here to suppress down-going muon events. In a first step, it segments the full solid angle in 500 directions. For each direction, the number of hits compatible with a muon track from this direction is determined.
# * A lower value, therefore, means a higher likelihood of this event to be a down-going muon. A selection criterion combining the GridFit ratio and the number of selected shower hits was devised to further suppress the atmospheric muon background.
# **Takeaways**:
# * We can see that some kind of geometrical containment is used in this paper as well (restricting the region where the event comes from / occurs)
# * Another point is that they use angular restrictions to account for up-going movements of neutrinos both through angle of the shower and GridFit ratio
# # Literature Overview Summary
# ## Classification:
# * In our view it is worth to classify at least two kind of events:
# 1. Events that are easy to reconstruct (no dominating noise, atmospheric muons)
# 2. Events that are hard to reconstruct (a lot of noise and/or atmospheric muons)
# Given the paper overview we would suggest to do geometrical containment classification - it both should be time efficient and easy to implement, thus:
# 1. **Events that are easy to reconstruct**:
# 1. **Geometry**: Originated within the IceCube (cut off the boarders as per veto regions in the picture above)
# 2. **Direction** Account for up-going movement (GridFit or some kind of rough approximation of angular movement could be used)
# 2. **Events that are hard to reconstruct**:
# 1. Anything else
#
#
#
# It is worth noting that energy criterion could be used to identify higher energy events that occur from neutrino coming from the space
# ## References:
# 1. [Evidence for High-Energy Extraterrestrial Neutrinos at the IceCube Detector](https://arxiv.org/pdf/1311.5238.pdf)
# 2. [An algorithm for the reconstruction of neutrino-induced showers in the ANTARES neutrino telescope](https://arxiv.org/abs/1708.03649)
# 3. [Neutrinos from the Milky Way](https://www.nikhef.nl/pub/services/biblio/theses_pdf/thesis_EL_Visser.pdf)
# # Code: Classification
import numpy as np
import pandas as pd
import polars as pl
import seaborn as sns
import math
from pathlib import Path
from matplotlib import pyplot as plt
from tqdm import tqdm
PATH_INPUT = Path("/kaggle/input/icecube-neutrinos-in-deep-ice")
train_batch_id = 1
print("Training batch", train_batch_id)
batch_path = "train/batch_" + str(train_batch_id) + ".parquet"
train_batch = pl.scan_parquet(PATH_INPUT / batch_path).lazy()
df_train_meta = pl.scan_parquet(PATH_INPUT / "train_meta.parquet").lazy()
df_sensor_geometry = (
pl.scan_csv(PATH_INPUT / "sensor_geometry.csv")
.with_columns(pl.col("sensor_id").cast(pl.Int16))
.lazy()
)
# # Geometrical containment
# * **In order to classify by geometrical containment we do the following identificaiton of the sensors**:
# 1. The edge sensors from the sides (`sides`)
# 2. The bottom sensors (`bottom`)
# 3. The top 5 sensors (`top`)
DISTANCE = 470
sides = (
df_sensor_geometry.with_columns(
[(((pl.col("x") ** 2 + pl.col("y") ** 2) ** 0.5)).alias("xy_distance")]
)
.filter(pl.col("xy_distance") > DISTANCE)
.collect()
)
# need to delete these points
delete_points = sides.filter(
(pl.col("x") > 400) & (pl.col("x") < 500) & (pl.col("y") < 200) & (pl.col("y") > 0)
)
# need to add these points
add_points = df_sensor_geometry.filter(
(pl.col("x") > 100)
& (pl.col("x") < 180)
& (pl.col("y") > 350)
& (pl.col("y") < 420)
).collect()
plt.scatter(
df_sensor_geometry.select([pl.col("x")]).collect(),
df_sensor_geometry.select([pl.col("y")]).collect(),
)
plt.scatter(sides.select([pl.col("x")]), sides.select([pl.col("y")]))
plt.scatter(
delete_points.select(pl.col("x")), delete_points.select(pl.col("y")), c="black"
)
plt.scatter(add_points.select(pl.col("x")), add_points.select(pl.col("y")), c="gray")
plt.title("Identifying the edges")
plt.show()
delete_set = set(delete_points.select(pl.col("sensor_id")).to_pandas()["sensor_id"])
add_set = set(add_points.select(pl.col("sensor_id")).to_pandas()["sensor_id"])
sides_set = set(sides.select(pl.col("sensor_id")).to_pandas()["sensor_id"])
sides_set = (sides_set - delete_set) | add_set
plt.scatter(
df_sensor_geometry.select(pl.col("x")).collect(),
df_sensor_geometry.select([pl.col("y")]).collect(),
)
plt.scatter(
df_sensor_geometry.filter(pl.col("sensor_id").is_in(list(sides_set)))
.select(pl.col("x"))
.collect(),
df_sensor_geometry.filter(pl.col("sensor_id").is_in(list(sides_set)))
.select(pl.col("y"))
.collect(),
)
plt.title("Getting the edges out")
plt.show()
Z_BOTTOM = -500
Z_TOP = 450
top_sensors = df_sensor_geometry.filter(pl.col("z") > Z_TOP).collect()
bottom_sensors = df_sensor_geometry.filter(pl.col("z") < Z_BOTTOM).collect()
top_set = set(top_sensors.select(pl.col("sensor_id")).to_pandas()["sensor_id"])
bottom_set = set(bottom_sensors.select(pl.col("sensor_id")).to_pandas()["sensor_id"])
plt.scatter(
df_sensor_geometry.select(pl.col("x")).collect(),
df_sensor_geometry.select(pl.col("z")).collect(),
marker="o",
s=3,
)
plt.scatter(
bottom_sensors.select(pl.col("x")),
bottom_sensors.select(pl.col("z")),
marker="o",
s=3,
)
plt.scatter(
top_sensors.select(pl.col("x")), top_sensors.select(pl.col("z")), marker="o", s=3
)
plt.title("Identifying top and bottom")
plt.show()
# ## Analysing data using the geo containment criterion
# * Let's see how many events originated within the cube
def add_sides(dataf, account_for_aux):
if account_for_aux == False:
return (
dataf.groupby(["event_id"])
.agg([pl.col("sensor_id").first()])
.with_columns(
[
pl.col("sensor_id").is_in(list(sides_set)).alias("side"),
pl.col("sensor_id").is_in(list(top_set)).alias("top"),
pl.col("sensor_id").is_in(list(bottom_set)).alias("bottom"),
]
)
)
if account_for_aux == True:
return (
dataf.filter(pl.col("auxiliary") == False)
.groupby(["event_id"])
.agg([pl.col("sensor_id").first()])
.with_columns(
[
pl.col("sensor_id").is_in(list(sides_set)).alias("side"),
pl.col("sensor_id").is_in(list(top_set)).alias("top"),
pl.col("sensor_id").is_in(list(bottom_set)).alias("bottom"),
]
)
)
def for_plot(dataf):
return (
dataf.with_columns(
[
(pl.col("top").count() / pl.col("top").count()).alias("total"),
(
(
pl.col("side").sum()
+ pl.col("top").sum()
+ pl.col("bottom").sum()
)
/ pl.col("top").count()
).alias("outside_events"),
(pl.col("side").sum() / pl.col("side").count()).alias("side_ratio"),
(pl.col("top").sum() / pl.col("top").count()).alias("top_ratio"),
(pl.col("bottom").sum() / pl.col("top").count()).alias("bottom_ratio"),
]
)
.head(1)
.select(["total", "outside_events", "side_ratio", "top_ratio", "bottom_ratio"])
)
account_for_aux = False
temp1 = (
train_batch.pipe(add_sides, account_for_aux).pipe(for_plot).collect().to_pandas()
)
ax = sns.barplot(data=temp1)
ax.bar_label(ax.containers[0])
plt.title("Events by geometrical classification (aux=True)")
plt.show()
# * We see that overall we have about 39% of events generated on the edges on the IceCube (not accounting for auxiliary sensors)
account_for_aux = True
temp1 = (
train_batch.pipe(add_sides, account_for_aux).pipe(for_plot).collect().to_pandas()
)
ax = sns.barplot(data=temp1)
ax.bar_label(ax.containers[0])
plt.title("Events by geometrical classification (aux=False)")
plt.show()
# * If we account for auxiliary sensors we get to the about 70% 'hard to reconstruct' events
# # Direction of movement
# * We want to distinguish between 'up-going' and 'down-going' events
# * The easiest way to do it is to compare the first two sensors within an event on 'z' axis.
def join_tables(dataf, data_geometry):
return dataf.join(data_geometry, on="sensor_id")
def time_rank(dataf, account_for_aux):
if account_for_aux == True:
return (
dataf.filter(pl.col("auxiliary") == False)
.with_columns([pl.col("time").rank().over("event_id").alias("time_rank")])
.filter(pl.col("time_rank").is_in([1, 1.5, 2]))
)
else:
return dataf.with_columns(
[pl.col("time").rank().over("event_id").alias("time_rank")]
).filter(pl.col("time_rank").is_in([1, 1.5, 2]))
def add_direction(dataf):
return (
dataf.groupby("event_id")
.agg([pl.col("z").head(1).alias("first"), pl.col("z").tail(1).alias("second")])
.with_columns(
[
(pl.col("second").arr.explode() - pl.col("first").arr.explode()).alias(
"direction"
)
]
)
.with_columns(
[
(pl.col("direction") > 0).alias("upgoing"),
(pl.col("direction") == 0).alias("horizontal"),
(pl.col("direction") < 0).alias("downgoing"),
]
)
.select(pl.col("*").sort_by("event_id"))
)
def for_plot_direction(dataf):
return (
dataf.with_columns(
[
(pl.col("direction").count() / pl.col("direction").count()).alias(
"total"
),
(pl.col("upgoing").sum() / pl.col("upgoing").count()).alias("up_ratio"),
(pl.col("downgoing").sum() / pl.col("downgoing").count()).alias(
"down_ratio"
),
(pl.col("horizontal").sum() / pl.col("horizontal").count()).alias(
"horizontal_ratio"
),
]
)
.head(1)
.select(["total", "down_ratio", "horizontal_ratio", "up_ratio"])
)
account_for_aux = False
temp_2 = (
train_batch.pipe(join_tables, df_sensor_geometry)
.pipe(time_rank, account_for_aux)
.pipe(add_direction)
.pipe(for_plot_direction)
.head(1)
.collect()
.to_pandas()
)
ax = sns.barplot(data=temp_2)
ax.bar_label(ax.containers[0])
plt.title("Events by geometrical classification (aux=False)")
plt.show()
# * If we don't account for auxiliary sensors we have about equal portions of up-going and down-going events
account_for_aux = True
temp_2 = (
train_batch.pipe(join_tables, df_sensor_geometry)
.pipe(time_rank, account_for_aux)
.pipe(add_direction)
.pipe(for_plot_direction)
.head(1)
.collect()
.to_pandas()
)
ax = sns.barplot(data=temp_2)
ax.bar_label(ax.containers[0])
plt.title("Events by geometrical classification (aux=False)")
plt.show()
# * However, if we get rid of auxiliary sensors we see that about 40% of events are down-going, 31% horizontal and 28% up-going
# # Constructing features for classification
# * We want to come up with a feature that shows how good one event is for a candidate for 'easy to reconstruct' or 'hard for reconstruct'
# * Idially it is a continuous variable and lets say beween 1 and 0 where 1 - 'hard to reconstruct' event and 0 - 'easy to reconstruct' event.
# * We add the following weights to constuct this feature:
# * **Geo-containment**:
# * 1.0 - event originated on the top
# * 0.75 - event originated on the sides
# * 0.25 - event originated on the bottom
# * **Direction**:
# * 0.5 - down-going
# * 0.25 - horizontal
# * 0.0 - up-going
# * We normalize to [0,1] and do two separate features with accounting for auxiliary sensors and not
#
def join_two_features(dataf, df_train_batch, account_for_aux):
return dataf.join(df_train_batch.pipe(add_sides, account_for_aux), on="event_id")
def classification_feature(dataf, account_for_aux):
if account_for_aux == True:
return dataf.with_columns(
[
(
pl.col("horizontal") * 0.25
+ pl.col("downgoing") * 0.5
+ pl.col("side") * 0.75
+ pl.col("top")
+ pl.col("bottom") * 0.25
).alias("hard_to_reconstruct_aux_on")
]
).select(
[
pl.col("event_id"),
pl.col("hard_to_reconstruct_aux_on")
/ pl.col("hard_to_reconstruct_aux_on").max(),
]
)
if account_for_aux == False:
return dataf.with_columns(
[
(
pl.col("horizontal") * 0.25
+ pl.col("downgoing") * 0.5
+ pl.col("side") * 0.75
+ pl.col("top")
+ pl.col("bottom") * 0.25
).alias("hard_to_reconstruct_aux_off")
]
).select(
[
pl.col("event_id"),
pl.col("hard_to_reconstruct_aux_off")
/ pl.col("hard_to_reconstruct_aux_off").max(),
]
)
account_for_aux = False
temp_2 = (
train_batch.pipe(join_tables, df_sensor_geometry)
.pipe(time_rank, account_for_aux)
.pipe(add_direction)
.pipe(join_two_features, train_batch, account_for_aux)
.pipe(classification_feature, account_for_aux)
)
account_for_aux = True
temp_3 = (
train_batch.pipe(join_tables, df_sensor_geometry)
.pipe(time_rank, account_for_aux)
.pipe(add_direction)
.pipe(join_two_features, train_batch, account_for_aux)
.pipe(classification_feature, account_for_aux)
)
df_classification = temp_2.join(temp_3, on="event_id", how="left").collect().to_pandas()
df_classification.to_csv("classification_features.csv")
sns.histplot(data=df_classification["hard_to_reconstruct_aux_off"], bins=10, alpha=0.7)
sns.histplot(data=df_classification["hard_to_reconstruct_aux_on"], bins=10, alpha=0.5)
plt.title("Events classification histogram")
plt.show()
# # Code for copy-paste
# * For convinience here is copy-paste friendly code to generate these features to add to your notebook
import numpy as np
import pandas as pd
import polars as pl
import seaborn as sns
import math
from pathlib import Path
PATH_INPUT = Path("/kaggle/input/icecube-neutrinos-in-deep-ice")
train_batch_id = 1
print("Training batch", train_batch_id)
batch_path = "train/batch_" + str(train_batch_id) + ".parquet"
train_batch = pl.scan_parquet(PATH_INPUT / batch_path).lazy()
df_train_meta = pl.scan_parquet(PATH_INPUT / "train_meta.parquet").lazy()
df_sensor_geometry = (
pl.scan_csv(PATH_INPUT / "sensor_geometry.csv")
.with_columns(pl.col("sensor_id").cast(pl.Int16))
.lazy()
)
DISTANCE = 470
sides = (
df_sensor_geometry.with_columns(
[(((pl.col("x") ** 2 + pl.col("y") ** 2) ** 0.5)).alias("xy_distance")]
)
.filter(pl.col("xy_distance") > DISTANCE)
.collect()
)
# need to delete these points
delete_points = sides.filter(
(pl.col("x") > 400) & (pl.col("x") < 500) & (pl.col("y") < 200) & (pl.col("y") > 0)
)
# need to add these points
add_points = df_sensor_geometry.filter(
(pl.col("x") > 100)
& (pl.col("x") < 180)
& (pl.col("y") > 350)
& (pl.col("y") < 420)
).collect()
delete_set = set(delete_points.select(pl.col("sensor_id")).to_pandas()["sensor_id"])
add_set = set(add_points.select(pl.col("sensor_id")).to_pandas()["sensor_id"])
sides_set = set(sides.select(pl.col("sensor_id")).to_pandas()["sensor_id"])
sides_set = (sides_set - delete_set) | add_set
Z_BOTTOM = -500
Z_TOP = 450
top_sensors = df_sensor_geometry.filter(pl.col("z") > Z_TOP).collect()
bottom_sensors = df_sensor_geometry.filter(pl.col("z") < Z_BOTTOM).collect()
top_set = set(top_sensors.select(pl.col("sensor_id")).to_pandas()["sensor_id"])
bottom_set = set(bottom_sensors.select(pl.col("sensor_id")).to_pandas()["sensor_id"])
def add_sides(dataf, account_for_aux):
if account_for_aux == False:
return (
dataf.groupby(["event_id"])
.agg([pl.col("sensor_id").first()])
.with_columns(
[
pl.col("sensor_id").is_in(list(sides_set)).alias("side"),
pl.col("sensor_id").is_in(list(top_set)).alias("top"),
pl.col("sensor_id").is_in(list(bottom_set)).alias("bottom"),
]
)
)
if account_for_aux == True:
return (
dataf.filter(pl.col("auxiliary") == False)
.groupby(["event_id"])
.agg([pl.col("sensor_id").first()])
.with_columns(
[
pl.col("sensor_id").is_in(list(sides_set)).alias("side"),
pl.col("sensor_id").is_in(list(top_set)).alias("top"),
pl.col("sensor_id").is_in(list(bottom_set)).alias("bottom"),
]
)
)
def join_tables(dataf, data_geometry):
return dataf.join(data_geometry, on="sensor_id")
def time_rank(dataf, account_for_aux):
if account_for_aux == True:
return (
dataf.filter(pl.col("auxiliary") == False)
.with_columns(
[pl.col("time").rank("ordinal").over("event_id").alias("time_rank")]
)
.filter(pl.col("time_rank").is_in([1, 2]))
)
else:
return dataf.with_columns(
[pl.col("time").rank("ordinal").over("event_id").alias("time_rank")]
).filter(pl.col("time_rank").is_in([1, 2]))
def add_direction(dataf):
return (
dataf.groupby("event_id")
.agg([pl.col("z").head(1).alias("first"), pl.col("z").tail(1).alias("second")])
.with_columns(
[
(pl.col("second").arr.explode() - pl.col("first").arr.explode()).alias(
"direction"
)
]
)
.with_columns(
[
(pl.col("direction") > 0).alias("upgoing"),
(pl.col("direction") == 0).alias("horizontal"),
(pl.col("direction") < 0).alias("downgoing"),
]
)
.select(pl.col("*").sort_by("event_id"))
)
def join_two_features(dataf, df_train_batch, account_for_aux):
return dataf.join(df_train_batch.pipe(add_sides, account_for_aux), on="event_id")
def classification_feature(dataf, account_for_aux):
if account_for_aux == True:
return dataf.with_columns(
[
(
pl.col("horizontal") * 0.25
+ pl.col("downgoing") * 0.5
+ pl.col("side") * 0.75
+ pl.col("top")
+ pl.col("bottom") * 0.25
).alias("hard_to_reconstruct_aux_on")
]
).select(
[
pl.col("event_id"),
pl.col("hard_to_reconstruct_aux_on")
/ pl.col("hard_to_reconstruct_aux_on").max(),
]
)
if account_for_aux == False:
return dataf.with_columns(
[
(
pl.col("horizontal") * 0.25
+ pl.col("downgoing") * 0.5
+ pl.col("side") * 0.75
+ pl.col("top")
+ pl.col("bottom") * 0.25
).alias("hard_to_reconstruct_aux_off")
]
).select(
[
pl.col("event_id"),
pl.col("hard_to_reconstruct_aux_off")
/ pl.col("hard_to_reconstruct_aux_off").max(),
]
)
account_for_aux = False
temp_2 = (
train_batch.pipe(join_tables, df_sensor_geometry)
.pipe(time_rank, account_for_aux)
.pipe(add_direction)
.pipe(join_two_features, train_batch, account_for_aux)
.pipe(classification_feature, account_for_aux)
)
account_for_aux = True
temp_3 = (
train_batch.pipe(join_tables, df_sensor_geometry)
.pipe(time_rank, account_for_aux)
.pipe(add_direction)
.pipe(join_two_features, train_batch, account_for_aux)
.pipe(classification_feature, account_for_aux)
)
df_classification = temp_2.join(temp_3, on="event_id", how="left").collect().to_pandas()
df_classification.to_csv("classification_features.csv")
df_classification
|
# # Análise de clientes
# Vamos explorar os dados de crédito dos clientes de uma instituição financeira. O intuito é explicar a coluna 'default', que aponta se um cliente é adimplente (default = 0) ou inadimplente (default = 1). Buscamos com essa análise entender o porquê de um cliente deixar de pagar suas dívidas com base no comportamento dos seus atributos, como salário, escolaridade e movimentação financeira.
# Temos como variável resposta o atributo de interesse 'default' e os demais atributos temos como variáveis preditoras.
# Abaixo nós temos a descrição completa dos atributos.
# | Coluna | Descrição |
# | ------- | --------- |
# | id | Número da conta |
# | default | Indica se o cliente é adimplente (0) ou inadimplente (1) |
# | idade | --- |
# | sexo | --- |
# | depedentes | --- |
# | escolaridade | --- |
# | estado_civil | --- |
# | salario_anual | Faixa do salario mensal multiplicado por 12 |
# | tipo_cartao | Categoria do cartao: blue, silver, gold e platinium |
# | meses_de_relacionamento | Quantidade de meses desde a abertura da conta |
# | qtd_produtos | Quantidade de produtos contratados |
# | iteracoes_12m | Quantidade de iteracoes com o cliente no último ano |
# | meses_inatico_12m | Quantidade de meses que o cliente ficou inativo no último ano |
# | limite_credito | Valor do limite do cartão de crédito |
# | valor_transacoes_12m | Soma total do valor das transações no cartão de crédito no último ano |
# | qtd_transacoes_12m | Quantidade total de transações no cartão de crédito no último ano |
# ## Importando blibliotecas
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
# ## Carregando a base de dados com pandas
df = pd.read_csv(
"/kaggle/input/base-de-dados/Python_M10_support material.csv", na_values="na"
)
df.head(n=10)
# ## Exploração dos dados
# Entendendo a estrutura do conjunto de dados.
df.shape
df[df["default"] == 0].shape
df[df["default"] == 1].shape
# Descobrindo a proporção de clientes adimplentes e inadimplentes
qtd_total, _ = df.shape
qtd_adimplentes, _ = df[df["default"] == 0].shape
qtd_inadimplentes, _ = df[df["default"] == 1].shape
print(
f"A proporção de clientes adimplentes: {round(100 * qtd_adimplentes/qtd_total, 2)}%"
)
print(
f"A proporção de clientes inadimplentes: {round(100 * qtd_inadimplentes/qtd_total, 2)}%"
)
# Colunas e respectivos tipos de dados
df.dtypes
# Atributos categóricos
df.select_dtypes("object").describe().transpose()
# Atributos numéricos
df.drop("id", axis=1).select_dtypes("number").describe().transpose()
# ## Transformação e limpeza dos dados
# Correção dos tipos de dados das colunas 'limite_credito' e 'valor_transacoes_12m', passando de 'object' para 'float'.
df[["limite_credito", "valor_transacoes_12m"]].dtypes
df[["limite_credito", "valor_transacoes_12m"]].head(n=5)
# Transformando em 'float' e substituindo os '.' e ','
fn = lambda valor: float(valor.replace(".", "").replace(",", "."))
# Verificando
valores_originais = ["12.691,51", "8.256,96", "3.418,56", "3.313,03", "4.716,22"]
valores_limpos = list(map(fn, valores_originais))
print(valores_originais)
print(valores_limpos)
# Aplicando nas colunas
df["valor_transacoes_12m"] = df["valor_transacoes_12m"].apply(fn)
df["limite_credito"] = df["limite_credito"].apply(fn)
df.dtypes
# Atributos categóricos
df.select_dtypes("object").describe().transpose()
# Atributos numéricos
df.drop("id", axis=1).select_dtypes("number").describe().transpose()
# ## Remoção dos Dados Faltantes
df.dropna(inplace=True)
df.shape
df[df["default"] == 0].shape
df[df["default"] == 1].shape
# Nova proporção de clientes adimplentes e inadimplentes
qtd_total_novo, _ = df.shape
qtd_adimplentes_novo, _ = df[df["default"] == 0].shape
qtd_inadimplentes_novo, _ = df[df["default"] == 1].shape
print(
f"A proporcão adimplentes ativos é de {round(100 * qtd_adimplentes / qtd_total, 2)}%"
)
print(
f"A nova proporcão de clientes adimplentes é de {round(100 * qtd_adimplentes_novo / qtd_total_novo, 2)}%"
)
print("")
print(
f"A proporcão clientes inadimplentes é de {round(100 * qtd_inadimplentes / qtd_total, 2)}%"
)
print(
f"A nova proporcão de clientes inadimplentes é de {round(100 * qtd_inadimplentes_novo / qtd_total_novo, 2)}%"
)
# ## Visualização de Dados
sns.set_style("whitegrid")
df_adimplente = df[df["default"] == 0]
df_inadimplente = df[df["default"] == 1]
# ### Visualização dos atributos categóricos
# Visualização dos atributos categóricos
df.select_dtypes("object").head(n=5)
# Visualizando a relação da variável resposta 'default' com os atributos categóricos
# Começando pelo atributo 'escolaridade'
coluna = "escolaridade"
titulos = [
"Escolaridade dos Clientes",
"Escolaridade dos Clientes Adimplentes",
"Escolaridade dos Clientes Inadimplentes",
]
eixo = 0
max_y = 0
max = df.select_dtypes("object").describe()[coluna]["freq"] * 1.1
figura, eixos = plt.subplots(1, 3, figsize=(20, 5), sharex=True)
for dataframe in [df, df_adimplente, df_inadimplente]:
df_to_plot = dataframe[coluna].value_counts().to_frame()
df_to_plot.rename(columns={coluna: "frequencia_absoluta"}, inplace=True)
df_to_plot[coluna] = df_to_plot.index
df_to_plot.sort_values(by=[coluna], inplace=True)
df_to_plot.sort_values(by=[coluna])
f = sns.barplot(
x=df_to_plot[coluna], y=df_to_plot["frequencia_absoluta"], ax=eixos[eixo]
)
f.set(title=titulos[eixo], xlabel=coluna.capitalize(), ylabel="Frequência Absoluta")
f.set_xticklabels(labels=f.get_xticklabels(), rotation=90)
_, max_y_f = f.get_ylim()
max_y = max_y_f if max_y_f > max_y else max_y
f.set(ylim=(0, max_y))
eixo += 1
figura.show()
# Agora com o atributo 'salario_anual'
coluna = "salario_anual"
titulos = [
"Salário Anual dos Clientes",
"Salário Anual dos Clientes Adimplentes",
"Salário Anual dos Clientes Inadimplentes",
]
eixo = 0
max_y = 0
max = df.select_dtypes("object").describe()[coluna]["freq"] * 1.1
figura, eixos = plt.subplots(1, 3, figsize=(20, 5), sharex=True)
for dataframe in [df, df_adimplente, df_inadimplente]:
df_to_plot = dataframe[coluna].value_counts().to_frame()
df_to_plot.rename(columns={coluna: "frequencia_absoluta"}, inplace=True)
df_to_plot[coluna] = df_to_plot.index
df_to_plot.sort_values(by=[coluna], inplace=True)
df_to_plot.sort_values(by=[coluna])
f = sns.barplot(
x=df_to_plot[coluna], y=df_to_plot["frequencia_absoluta"], ax=eixos[eixo]
)
f.set(title=titulos[eixo], xlabel=coluna.capitalize(), ylabel="Frequência Absoluta")
f.set_xticklabels(labels=f.get_xticklabels(), rotation=90)
_, max_y_f = f.get_ylim()
max_y = max_y_f if max_y_f > max_y else max_y
f.set(ylim=(0, max_y))
eixo += 1
figura.show()
# ### Visualização dos Atributos Numéricos
# Aqui nós já iremos analisar a relação entre a variável resposta 'default' com os atributos numéricos.
df.drop(["id", "default"], axis=1).select_dtypes("number").head(n=5)
# Começando pelo atributo 'qtd_transacoes_12m'
coluna = "qtd_transacoes_12m"
titulos = [
"Qtd. de Transações no Último Ano",
"Qtd. de Transações no Último Ano de Adimplentes",
"Qtd. de Transações no Último Ano de Inadimplentes",
]
eixo = 0
max_y = 0
figura, eixos = plt.subplots(1, 3, figsize=(20, 5), sharex=True)
for dataframe in [df, df_adimplente, df_inadimplente]:
f = sns.histplot(x=coluna, data=dataframe, stat="count", ax=eixos[eixo])
f.set(title=titulos[eixo], xlabel=coluna.capitalize(), ylabel="Frequência Absoluta")
_, max_y_f = f.get_ylim()
max_y = max_y_f if max_y_f > max_y else max_y
f.set(ylim=(0, max_y))
eixo += 1
figura.show()
# Agora com o atributo 'valor_transacoes_12m'
coluna = "valor_transacoes_12m"
titulos = [
"Valor das Transações no Último Ano",
"Valor das Transações no Último Ano de Adimplentes",
"Valor das Transações no Último Ano de Inadimplentes",
]
eixo = 0
max_y = 0
figura, eixos = plt.subplots(1, 3, figsize=(20, 5), sharex=True)
for dataframe in [df, df_adimplente, df_inadimplente]:
f = sns.histplot(x=coluna, data=dataframe, stat="count", ax=eixos[eixo])
f.set(title=titulos[eixo], xlabel=coluna.capitalize(), ylabel="Frequência Absoluta")
_, max_y_f = f.get_ylim()
max_y = max_y_f if max_y_f > max_y else max_y
f.set(ylim=(0, max_y))
eixo += 1
figura.show()
# Valor de Transações nos Últimos 12 Meses x Quantidade de Transações nos Últimos 12 Meses
f = sns.relplot(
x="valor_transacoes_12m", y="qtd_transacoes_12m", data=df, hue="default"
)
_ = f.set(
title="Relação entre Valor e Quantidade de Transações no Último Ano",
xlabel="Valor das Transações no Último Ano",
ylabel="Quantidade das Transações no Último Ano",
)
|
# for linear algebra and scientific calculation
import numpy as np
# for data processing and manipulation of data structure
import pandas as pd
# for Box-Cox Transformation
from scipy import stats
# for min_max scaling
from mlxtend.preprocessing import minmax_scaling
# plotting modules
import seaborn as sns
import matplotlib.pyplot as plt
# set seed for reproducibility
np.random.seed(0)
# split a dataset into train and test sets
from sklearn.model_selection import train_test_split
# sklearn is machine learning library for python
from sklearn.model_selection import KFold
from sklearn.preprocessing import MinMaxScaler
from sklearn.svm import SVR
# Linear Regression
from sklearn.linear_model import LinearRegression
from sklearn.ensemble import ExtraTreesRegressor
# importing the kaggle input to read the data set.
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# # ** Reading first data csv file and showing the output. **
car_data_first = pd.read_csv("../input/vehicle-dataset-from-cardekho/car data.csv")
car_data_first.head()
# ** Check for Null/Missing Values/Duplicate Values(drop if any) **
car_data_first.isnull()
car_data_first.isnull().sum()
# ** By running above code, we can see there is no missing/null values in our first dataset/csv. **
# # ** Reading Second data csv file and showing the output. **
car_data_second = pd.read_csv(
"../input/vehicle-dataset-from-cardekho/CAR DETAILS FROM CAR DEKHO.csv"
)
car_data_second.head()
# ** Check for Null/Missing Values/Duplicate Values(drop if any) **
car_data_second.isnull()
car_data_second.isnull().sum()
# ** By running above code, we can see there is no missing/null values in our second dataset/csv. **
# # ** Reading Third data csv file and showing the output. **#
car_data_third = pd.read_csv(
"../input/vehicle-dataset-from-cardekho/Car details v3.csv"
)
car_data_third.head()
# ** Check for Null/Missing Values/Duplicate Values(drop if any) **
car_data_second.isnull()
car_data_second.isnull().sum()
# #By running above code, we can see there is no missing/null values in our third dataset/csv.
# # ** Rescale the features. **
# select the Present_Price or Selling_Price column
_Present_Price = car_data_first.Present_Price
# scale the goals from 0 to 1
scaled_data = minmax_scaling(_Present_Price, columns=[0])
# plot the original & scaled data together to compare
fig, ax = plt.subplots(1, 2)
sns.histplot(car_data_first.Present_Price, ax=ax[0])
ax[0].set_title("Original Data")
sns.histplot(scaled_data, ax=ax[1])
ax[1].set_title("Scaled data")
# #Notice that the shape of the data doesn't change, but that instead of ranging from 0 to 75+, it now ranges from 0 to 1
# # **** Extract X as all columns except the last column and Y as the last column. ****
_X_ = car_data_first.loc[:, car_data_first.columns != "Owner"]
_X_.head()
_Y_ = car_data_first.loc[:, car_data_first.columns == "Owner"]
_Y_.head()
# Note: Since the last column has all values as zero, so out is not reflecting anything.
# # ** Split the data into a training set and testing set. **
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.2, random_state=2)
print(x_train.shape, x_test.shape, y_train.shape, y_test.shape)
# # ** Perform 10-fold cross-validation **
X = car_data_first.iloc[:, [1, 2, 3, 4]]
Y = car_data_first.iloc[:, 3]
sc = MinMaxScaler(feature_range=(0, 1))
X = sc.fit_transform(X)
scores = []
best_svr = SVR(kernel="rbf")
cv = KFold(n_splits=10, random_state=42, shuffle=True)
for train_index, test_index in cv.split(X):
print("Train Index: ", train_index, "\n")
print("Test Index: ", test_index)
X_train, X_test, Y_train, Y_test = (
X[train_index],
X[test_index],
Y[train_index],
Y[test_index],
)
best_svr.fit(X_train, Y_train)
scores.append(best_svr.score(X_test, Y_test))
best_svr.fit(X_train, Y_train)
scores.append(best_svr.score(X_test, Y_test))
print(np.mean(scores))
# # ** Train a Linear regression model for the dataset. **
lr = LinearRegression()
lr.fit(x_train, y_train)
y_pred = lr.predict(x_test)
print(
np.concatenate((y_pred.reshape(len(y_pred), 1), y_test.reshape(len(y_test), 1)), 1)
)
# # ** Visualize training and test results. **
plt.figure(figsize=[15, 10])
plt.plot(y_pred, label="Predicted")
plt.plot(y_test, label="Actual_test")
plt.legend()
plt.title("Linear Regression Model")
# # ** Compute the accuracy **
from sklearn.metrics import r2_score
lr_r2 = r2_score(y_test, y_pred)
print(lr_r2)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import torch
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
def mingradient(x):
# x = torch.tensor([4.], requires_grad=True) # Начальные значения x
lr = 0.05 # скорость оптимизации
for iteration in range(100):
with torch.no_grad(): # pytorch запрещает изменять тензоры, для которых подсчитывается градиент,
# torch.no_grad() позволяет исполнить код, не отслеживая градиент, в т.ч. изменить значения в тензоре
if x.grad is not None: # На первой итерации x.grad == None
x.grad.zero_() # устанавливаем градиент в нуль (в противном случае результат следующего вычисления градиента прибавится к предыдущему)
f = (2 * x + 1) * (x - 1**2) / x**2 # Вычисляем минимизируемую функцию
# Выводим (x.data - тензор, разделяющий память с x, но без отслеживания градиента, используем его, чтобы выводились только данные_
# Если в тензоре только один элемент, item позволяет получить его как число
# print(x.data, f.item())
# Вычисляем градиент
f.backward()
with torch.no_grad():
# Делаем шаг (сдвигаем параметры в направлении, противоположном градиенту, на величину, пропорциональную частным производным и скорости обучения)
x -= lr * x.grad
print(x.data, f.item())
def maxgradient(x):
# x = torch.tensor([4.], requires_grad=True) # Начальные значения x
lr = 0.05 # скорость оптимизации
for iteration in range(100):
with torch.no_grad(): # pytorch запрещает изменять тензоры, для которых подсчитывается градиент,
# torch.no_grad() позволяет исполнить код, не отслеживая градиент, в т.ч. изменить значения в тензоре
if x.grad is not None: # На первой итерации x.grad == None
x.grad.zero_() # устанавливаем градиент в нуль (в противном случае результат следующего вычисления градиента прибавится к предыдущему)
f = (2 * x + 1) * (x - 1**2) / x**2 # Вычисляем минимизируемую функцию
# Выводим (x.data - тензор, разделяющий память с x, но без отслеживания градиента, используем его, чтобы выводились только данные_
# Если в тензоре только один элемент, item позволяет получить его как число
# print(x.data, f.item())
# Вычисляем градиент
f.backward()
with torch.no_grad():
# Делаем шаг (сдвигаем параметры в направлении, противоположном градиенту, на величину, пропорциональную частным производным и скорости обучения)
x += lr * x.grad
print(x.data, f.item())
x = torch.tensor([3.0], requires_grad=True) # Начальные значения x
mingradient(x)
x = torch.tensor([3.0], requires_grad=True) # Начальные значения x
maxgradient(x)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv("/kaggle/input/cancer-data/Cancer_Data.csv")
# # First look at our data (no vizualization cus im too lazy)
df.sample(20)
# # Let's see if there are null values in our data
df.info()
# # Let's clean data by simply dropping useless columns
df = df.drop(["id", "Unnamed: 32"], axis=1)
# # Feed all data to our model ignoring high correlation between some of our features
from sklearn.model_selection import train_test_split, cross_val_score
from sklearn.compose import ColumnTransformer, make_column_selector as selector
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import OneHotEncoder, StandardScaler
from sklearn.ensemble import RandomForestClassifier
# # Split our data to features (X) and target (Y)
X = df.drop("diagnosis", axis=1)
Y = df["diagnosis"]
# # Defining transformations for our categorical and numerical data
num_transformer = Pipeline(steps=[("scale", StandardScaler())])
cat_transformer = Pipeline(steps=[("code", OneHotEncoder())])
processor = ColumnTransformer(
transformers=[
("num_transf", num_transformer, selector(dtype_exclude="category")),
("cat_transf", cat_transformer, selector(dtype_include="category")),
]
)
# # Creating model in a pipeline class
clf = Pipeline(
steps=[("transform_data", processor), ("model", RandomForestClassifier())]
)
# # Splitting data into training and test sets
X_train, X_test, Y_train, Y_test = train_test_split(X, Y, test_size=0.2)
# # Fitting and evaluating our model
clf.fit(X_train, Y_train)
print("model score: %.3f" % clf.score(X_test, Y_test))
# # Evaluating using cross_val_score
print(cross_val_score(RandomForestClassifier(), X, Y, cv=5))
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
from numpy import random as rn
from datetime import datetime, timedelta
event_values = [
[
"",
"",
"",
"",
"",
"",
"",
"",
"",
]
]
df = pd.DataFrame(
event_values,
columns=[
"person_id",
"vech_no",
"vech_name",
"State",
"Date of Event",
"Accident",
"Killed",
"Injured",
"Reg_vech",
],
index=range(150000),
)
len(df)
for i in range(0, len(df)):
df["person_id"].values[i] = i + 1
df.head()
Vechiles_list = [
"Alto",
"Swift",
"Vento",
"Rapid",
"I20",
"Jeep Compass",
"City",
"Civic",
"Verna",
"Nexon",
"Activa",
"Scooty peep",
"Splendor",
"Pulsar",
"R15",
"KTM",
"Dominar",
"Dio",
"RE",
"Yamaha RX",
"Tata Marcopolo Motors",
"JBM Motor Limited",
"Solaris Bus & Coach SA",
"Olectra",
"Greyhound Lines",
"SRS Bus",
"VRL",
"KPN",
"Jabbar Travels",
"MTC Bus",
"Mahindra Bolero Camper",
"Tata Yodha Pickup",
"Mahindra Bolero Pikup 4×4",
"Isuzu S-CAB",
"Isuzu S-CAB",
"Isuzu S-CAB",
"Toyota Tundra",
"Ram 1500",
"Ford F-150",
"Tata Ace gold",
"Auto 1",
"Auto 2",
"Auto 3",
"Auto 4",
"Auto 5",
"Auto 6",
"Auto 7",
"Auto 8",
"Auto 9",
"Auto 10",
]
## 1.5l total rows in our sample size
## 50 car vechiles are there in the list that need to be filled
rn.seed(0)
first = rn.choice(Vechiles_list, size=len(Vechiles_list))
first
from numpy import random
rn.seed(0)
df["vech_name"] = rn.choice(Vechiles_list, size=len(df))
# random.seed(0)
# second = random.choice(Vechiles_list,size=len(Vechiles_list))
## let's take TN for example and fill the vechile_id
## 88
## let's fill the slot with 6 digt number, where 1st 2 digits are between 0 and 88, last 4 are between 0000 and 9999
rn.seed(0)
## RTO code
rto = []
for i in range(0, len(df)):
n = random.randint(1, 88)
rto.append(n)
## Vechile Number
vechile_digits = []
for i in range(0, len(df)):
n = random.randint(1000, 9999)
vechile_digits.append(n)
# print(randomlist)
for i in range(0, len(df)):
df["vech_no"].loc[i] = "TN" + str(rto[i]) + "-" + str(vechile_digits[i])
## state we will hardcode it as TN for now
df["State"] = "TN"
start = datetime.now()
end = start + timedelta(days=1)
random_date = start + (end - start) * random.random()
for i in range(0, len(df)):
rn.seed(0)
start = datetime.now()
end = start + timedelta(days=365)
random_date = start + (end - start) * random.random()
df["Date of Event"].values[i] = pd.to_datetime(random_date)
df.applyhead()
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import xgboost as xgb
import statsmodels.api as sm
from sklearn.model_selection import RepeatedStratifiedKFold
from sklearn.metrics import roc_auc_score
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import StackingClassifier, ExtraTreesClassifier
from xgboost import XGBClassifier
sns.set_theme(style="white", palette="viridis")
pal = sns.color_palette("viridis")
train = pd.read_csv(r"../input/playground-series-s3e12/train.csv")
test_1 = pd.read_csv(r"../input/playground-series-s3e12/test.csv")
orig_train = pd.read_csv(
r"../input/kidney-stone-prediction-based-on-urine-analysis/kindey stone urine analysis.csv"
)
train.drop("id", axis=1, inplace=True)
test = test_1.drop("id", axis=1)
# # Knowing Your Data
# ## Descriptive Statistics
train.head()
test.head()
orig_train.head()
desc = train.describe().T
desc["nunique"] = train.nunique()
desc["%unique"] = desc["nunique"] / len(train) * 100
desc["null"] = train.isna().sum()
desc
desc = orig_train.describe().T
desc["nunique"] = orig_train.nunique()
desc["%unique"] = desc["nunique"] / len(orig_train) * 100
desc["null"] = train.isna().sum()
desc
# # Duplicates
print(
f"There are {train.duplicated(subset = list(train)[0:-1]).value_counts()[0]} non-duplicate values out of {train.count()[0]} rows in train dataset"
)
print(
f"There are {test.duplicated().value_counts()[0]} non-duplicate values out of {test.count()[0]} rows in test dataset"
)
print(
f"There are {orig_train.duplicated(subset = list(train)[0:-1]).value_counts()[0]} non-duplicate values out of {orig_train.count()[0]} rows in original train dataset"
)
# ## Distribution
fig, ax = plt.subplots(3, 2, figsize=(10, 10), dpi=300)
ax = ax.flatten()
for i, column in enumerate(test.columns):
sns.kdeplot(train[column], ax=ax[i], color=pal[0])
sns.kdeplot(test[column], ax=ax[i], color=pal[2])
sns.kdeplot(orig_train[column], ax=ax[i], color=pal[1])
ax[i].set_title(f"{column} Distribution")
ax[i].set_xlabel(None)
fig.suptitle("Distribution of Feature\nper Dataset\n", fontsize=24, fontweight="bold")
fig.legend(["Train", "Test", "Original Train"])
plt.tight_layout()
# **Key points:**
# 1. Train and test datasets have similar distribution so we can trust the CV.
# 2. Train and original train datasets also have relatively similar distribution, but we need to confirm this with adversarial validation.
fig, ax = plt.subplots(3, 2, figsize=(10, 10), dpi=300)
ax = ax.flatten()
for i, column in enumerate(test.columns):
sns.kdeplot(
data=train,
x=column,
ax=ax[i],
color=pal[0],
fill=True,
legend=False,
hue="target",
)
ax[i].set_title(f"{column} Distribution")
ax[i].set_xlabel(None)
ax[i].set_ylabel(None)
fig.suptitle("Distribution of Features per Class\n", fontsize=24, fontweight="bold")
fig.legend(["Crystal (1)", "No Crystal (0)"])
plt.tight_layout()
fig, ax = plt.subplots(3, 2, figsize=(10, 10), dpi=300)
ax = ax.flatten()
for i, column in enumerate(test.columns):
sns.kdeplot(
data=orig_train,
x=column,
ax=ax[i],
color=pal[0],
fill=True,
legend=False,
hue="target",
)
ax[i].set_title(f"{column} Distribution")
ax[i].set_xlabel(None)
ax[i].set_ylabel(None)
fig.suptitle("Distribution of Features per Class\n", fontsize=24, fontweight="bold")
fig.legend(["Crystal (1)", "No Crystal (0)"])
plt.tight_layout()
# **Key points:** It looks like `gravity`, `urea`, and `calc` have positive impact on `target`. We can confirm this causal relationship later.
fig, ax = plt.subplots(1, 2, figsize=(16, 5))
ax = ax.flatten()
ax[0].pie(
train["target"].value_counts(),
shadow=True,
explode=[0, 0.1],
autopct="%1.f%%",
textprops={"size": 20, "color": "white"},
)
sns.countplot(data=train, y="target", ax=ax[1])
ax[1].yaxis.label.set_size(20)
plt.yticks(fontsize=12)
ax[1].set_xlabel("Count", fontsize=20)
plt.xticks(fontsize=12)
fig.suptitle("Target Feature in Train Dataset", fontsize=25, fontweight="bold")
plt.tight_layout()
fig, ax = plt.subplots(1, 2, figsize=(16, 5))
ax = ax.flatten()
ax[0].pie(
orig_train["target"].value_counts(),
shadow=True,
explode=[0, 0.1],
autopct="%1.f%%",
textprops={"size": 20, "color": "white"},
)
sns.countplot(data=orig_train, y="target", ax=ax[1])
ax[1].yaxis.label.set_size(20)
plt.yticks(fontsize=12)
ax[1].set_xlabel("Count", fontsize=20)
plt.xticks(fontsize=12)
fig.suptitle("Target Feature in Original Train Dataset", fontsize=25, fontweight="bold")
plt.tight_layout()
# **Key points:** `target` has relatively balanced distribution
# ## Correlation
def heatmap(dataset, label=None):
corr = dataset.corr()
plt.figure(figsize=(14, 10), dpi=300)
mask = np.zeros_like(corr)
mask[np.triu_indices_from(mask)] = True
sns.heatmap(corr, mask=mask, annot=True, annot_kws={"size": 14}, cmap="viridis")
plt.yticks(fontsize=14)
plt.xticks(fontsize=14)
plt.title(f"{label} Dataset Correlation Matrix\n", fontsize=25, weight="bold")
plt.show()
heatmap(train, "Train")
heatmap(test, "Test")
heatmap(orig_train, "Original Train")
# **Key points:**
# 1. All features except `ph` are correlated with each others.
# 2. `target` is moderately correlated with `calc` and `gravity`
# # Causal Relationship
sm.Logit(train["target"], sm.add_constant(train.drop("target", axis=1))).fit().summary()
# **Key points:** Only `calc` has significant impact on `target`.
# **Additional note:** There might be a better way to determine a feature importance. However, due to how small our data is, I assume a simple method like this will work well. Don't do this for more complex dataset though.
# # Feature Engineering
# Let's combine both original and train dataset and see how many duplicates are there.
train = pd.concat([train, orig_train])
print(
f"There are {train.duplicated(subset = list(train)[0:-1]).value_counts()[0]} non-duplicate values out of {train.count()[0]} rows in train dataset"
)
# Now we should delete the duplicates.
train.drop_duplicates(subset=list(train)[0:-1], inplace=True, keep="first")
X = train.copy()
y = X.pop("target")
seed = 42
splits = 10
repeats = 10
k = RepeatedStratifiedKFold(n_splits=splits, random_state=seed, n_repeats=repeats)
np.random.seed(seed)
# Let's try to make our model as simple as possible by only using significant features.
X = X[["calc"]]
test = test[["calc"]]
# # Model
# I'll try using `StackingClassifier` for our model. Because the competition uses AUROC to evaluate our submission, we should use `roc_auc_score` as the parameter value for our metric in XGBoost. Because of the small dataset size, I'll use Repeated Stratified KFold for cross-validation.
xgb_params = {
"seed": seed,
"objective": "binary:logistic",
"eval_metric": "auc",
"tree_method": "exact",
"n_jobs": -1,
"max_depth": 2,
"eta": 0.01,
"n_estimators": 100,
}
predictions = np.zeros(len(test))
train_scores, val_scores = [], []
k = RepeatedStratifiedKFold(n_splits=splits, random_state=seed, n_repeats=repeats)
for fold, (train_idx, val_idx) in enumerate(k.split(X, y)):
stack = StackingClassifier(
[
("xgb", XGBClassifier(**xgb_params)),
("lr", LogisticRegression(penalty="l2", solver="saga", max_iter=10000)),
("ext", ExtraTreesClassifier(random_state=seed, min_samples_leaf=10)),
]
)
stack.fit(X.iloc[train_idx], y.iloc[train_idx])
train_preds = stack.predict_proba(X.iloc[train_idx])[:, 1]
val_preds = stack.predict_proba(X.iloc[val_idx])[:, 1]
train_score = roc_auc_score(y.iloc[train_idx], train_preds)
val_score = roc_auc_score(y.iloc[val_idx], val_preds)
train_scores.append(train_score)
val_scores.append(val_score)
predictions += stack.predict_proba(test)[:, 1] / splits / repeats
# print(f'Fold {fold // repeats} Iteration {fold % repeats}: val ROC = {val_score:.5f} | train ROC = {train_score:.5f}')
print(
f"Average val ROC = {np.mean(val_scores):.5f} | train ROC {np.mean(train_scores):.5f}"
)
stack_preds = predictions.copy()
# # Submission
test_1.drop(list(test_1.drop("id", axis=1)), axis=1, inplace=True)
test_1["target"] = stack_preds
test_1.to_csv("submission.csv", index=False)
|
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from matplotlib.ticker import FuncFormatter
df = pd.read_csv("/kaggle/input/hiv-cases-in-philippines/HIV-New-Cases-Philippines.csv")
df.dropna(inplace=True)
# Adding population as of 2020 from Philippine Statistics Office
df["Population as of 2020"] = [
179660,
13484462,
5301139,
3685744,
12422172,
16195042,
3228558,
6082165,
7954723,
8081988,
4547150,
3875576,
5022768,
5243536,
4901486,
2804788,
4404288,
]
df
df.columns
df_combined = df.drop(columns=["Region", "Population as of 2020"])
df_combined = df_combined.stack().reset_index(drop=True)
fig = plt.figure(figsize=(15, 7.5))
fig.patch.set_facecolor("ghostwhite")
sns.set_theme(style="darkgrid", palette="dark")
ax = sns.histplot(
data=df_combined,
kde=True,
)
plt.xticks(np.arange(-100, 600, 25))
plt.yticks(np.arange(0, 100, 5))
ax.set_xlabel("HIV cases per month")
ax.set(xlim=(0, 600), ylim=(0, 100))
plt.show()
# Histogram shows the distribution of cases per month. It is left skewed and shows that most HIV cases per month are below 25 mark.
sns.pairplot(
vars=[
"Jan-22",
"Feb-22",
"Mar-22",
"Apr-22",
"May-22",
"Jun-22",
"Jul-22",
"Aug-22",
"Sep-22",
"Oct-22",
"Nov-22",
"Dec-22",
"Jan-23",
],
hue="Region",
data=df,
kind="scatter",
)
plt.show()
# Pairplots are mostly consistent in distribution for monthly basis
new_df = pd.DataFrame(
{
"Region": df["Region"],
"Total Cases": df.drop(columns=["Region", "Population as of 2020"])
._get_numeric_data()
.sum(axis=1),
"Population as of 2020": df["Population as of 2020"],
}
)
new_df
new_df.describe()
fig = plt.figure(figsize=(15, 7.5))
fig.patch.set_facecolor("ghostwhite")
sns.set_theme(style="darkgrid")
ax = sns.barplot(
y="Region", x="Total Cases", data=new_df.sort_values("Total Cases", ascending=False)
)
ax.get_xaxis().set_major_formatter(FuncFormatter(lambda x, p: format(int(x), ",")))
plt.xticks(np.arange(0, 3_700, 200), rotation=45)
plt.title("HIV Cases per Region as of 2022")
plt.show()
# NCR has the highest total population in the country. But I wanted to see how much the population is affecting these results.
fig = plt.figure(figsize=(15, 7.5))
fig.patch.set_facecolor("ghostwhite")
sns.set_theme(style="darkgrid")
ax = sns.barplot(
y="Region",
x="Population as of 2020",
data=new_df.sort_values("Population as of 2020", ascending=False),
)
ax.get_xaxis().set_major_formatter(FuncFormatter(lambda x, p: format(int(x), ",")))
# plt.ticklabel_format(style = 'plain', axis ='x')
plt.xticks(np.arange(0, 17_000_000, 1_000_000), rotation=45)
plt.title("Population per Region as of 2020")
plt.show()
# Surprisingly, Region 4A has a higher population than NCR back in 2020
# We wanted to see the correlation between the population and the HIV cases. To determine how much of a factor it is to the number of cases
#
sns.heatmap(
new_df.corr().round(2), annot=True, vmin=-1, vmax=1, center=0, cmap="YlGnBu"
)
plt.show()
# The correlation between the total number of cases and population is very strong with an R of 0.89 and R-square of 0.80. With this, we might want to take population into consideration and determine the prevalence rate for each region.
fig = plt.figure(figsize=(15, 7.5))
fig.patch.set_facecolor("ghostwhite")
ax = sns.scatterplot(
data=new_df, x="Total Cases", y="Population as of 2020", hue="Region"
)
sns.move_legend(ax, "upper left", bbox_to_anchor=(1, 1))
ax.get_yaxis().set_major_formatter(FuncFormatter(lambda x, p: format(int(x), ",")))
plt.xticks(np.arange(0, 4000, 250))
plt.title("Scatterplot for Population as of 2020 and Total Cases")
plt.show()
from scipy import stats
import math
fig = plt.figure(figsize=(15, 7.5))
fig.patch.set_facecolor("ghostwhite")
x_data = new_df["Population as of 2020"]
y_data = new_df["Total Cases"]
slope, intercept, r_value, p_value, std_err = stats.linregress(y_data, x_data)
ax = sns.regplot(
data=new_df,
x="Total Cases",
y="Population as of 2020",
color="b",
line_kws={
"label": """
y = {0:.1f}x + {1:.1f}
R = {2:.2f}
R-square = {3:.2f}
""".format(
slope, intercept, r_value, math.pow(r_value, 2)
)
},
)
ax.legend()
sns.move_legend(ax, "upper left", bbox_to_anchor=(0, 1))
ax.get_yaxis().set_major_formatter(FuncFormatter(lambda x, p: format(int(x), ",")))
plt.xticks(np.arange(0, 4000, 250))
plt.show()
# Let's consider the population size and get the pravelance rate per region
# Incident Rate
df1 = pd.DataFrame(
{
"Region": new_df["Region"],
"Prevalence Rate": (new_df["Total Cases"] / new_df["Population as of 2020"])
* 100,
}
)
df1.sort_values("Prevalence Rate", ascending=False)
bg_color = "whitesmoke"
fig = plt.figure(figsize=(20, 10))
fig.patch.set_facecolor(bg_color)
_, _, autotexts = plt.pie(
new_df["Total Cases"],
labels=new_df["Region"],
autopct="%1.0f%%",
pctdistance=0.94,
labeldistance=1.03,
)
for text in autotexts:
text.set_color("white")
text.set_fontsize(10)
centre_circle = plt.Circle((0, 0), 0.72, fc=bg_color)
fig = plt.gcf()
fig.gca().add_artist(centre_circle)
plt.title("Total Cases per Region")
plt.show()
# NCR has the highest total cases and BARMM has the lowest based on the given dataset. However, this is not accounting for the population in each region.
fig = plt.figure(figsize=(20, 10))
fig.patch.set_facecolor(bg_color)
plt.pie(
df1["Prevalence Rate"],
labels=new_df["Region"],
autopct="%1.0f%%",
pctdistance=0.67,
labeldistance=1.03,
)
centre_circle = plt.Circle((0, 0), 0.72, fc=bg_color)
fig = plt.gcf()
fig.gca().add_artist(centre_circle)
plt.title("Prevalence Rate per Region Distribution")
plt.show()
fig = plt.figure(figsize=(15, 7.5))
fig.patch.set_facecolor("ghostwhite")
sns.set_theme(style="darkgrid", palette="deep")
sns.barplot(
y="Region",
x="Prevalence Rate",
data=df1.sort_values("Prevalence Rate", ascending=False),
)
plt.title("Prevalence Rate Chart")
plt.show()
# It seems CAR has the highest prevalence rate when population is taken to consideration while BARMM still has the lowest prevalence rate for HIV.
df_per_month = pd.DataFrame(
{
"Month": df.drop(columns="Region").columns,
"Total Cases": df.drop(columns="Region").sum(axis=0),
}
)
df_per_month.reset_index(drop=True, inplace=True)
df_per_month.drop(index=13, inplace=True)
# Adding Growth Rate to the DataFrame
df_per_month["Growth Rate"] = df_per_month["Total Cases"].pct_change()
df_per_month.fillna(0, inplace=True)
df_per_month
fig = plt.figure(figsize=(15, 7.5))
fig.patch.set_facecolor("ghostwhite")
sns.set_theme(style="darkgrid", palette="deep")
ax = sns.lineplot(x="Month", y=df_per_month["Total Cases"], data=df_per_month)
x = df_per_month.index
y = df_per_month["Total Cases"]
ax.grid(axis="x")
z = np.polyfit(x, y, 1)
p = np.poly1d(z)
ax = plt.gca()
ax.text(
0.6,
0.15,
f"""Average Growth Rate: {df_per_month['Growth Rate'].mean().round(2) * 100}%""",
transform=ax.transAxes,
color="green",
)
ax.get_yaxis().set_major_formatter(FuncFormatter(lambda x, p: format(int(x), ",")))
plt.xticks(rotation=45)
plt.plot(x, p(x), "r--")
plt.title("Total Cases of HIV per Month as of 2022")
plt.show()
|
# Load Data
# WordCloud
# Sentiment Analysis with DL model, predict +ve/-ve
# WordCloud with +ve, -ve respectively --> extract important words
# user network--> perform sentiment analysis
# user social network--> by text on same business and friends
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import json
from glob import glob
import warnings
warnings.filterwarnings("ignore")
# Libraries for Sentiment Analysis
import nltk
from nltk.corpus import stopwords
import re, string
from nltk.stem import WordNetLemmatizer
import tensorflow as tf
from tensorflow.keras.preprocessing.text import Tokenizer
from sklearn.metrics import (
accuracy_score,
precision_score,
recall_score,
confusion_matrix,
roc_curve,
auc,
classification_report,
)
from sklearn.svm import LinearSVC
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.naive_bayes import MultinomialNB
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import train_test_split
business = pd.read_json(
"../input/yelp-dataset/yelp_academic_dataset_business.json", lines=True
) # read business dataset
business.sample(5) # draw sample, take a look of the dataset
# The review data is too large to read in normally, so we will read it in by chunks of 1000 at a time
# Empty list to store each chunk
chunks = []
# Specify dtypes
r_dtypes = {
"stars": np.float16,
"useful": np.int32,
"funny": np.int32,
"cool": np.int32,
}
# Store the json chunks
reader = pd.read_json(
"../input/yelp-dataset/yelp_academic_dataset_review.json",
lines=True,
chunksize=1000,
dtype=r_dtypes,
orient="records",
)
# For loop to append each chunk into the chunks list
for chunk in reader:
chunks.append(chunk)
# Concatenate each chunk into a single dataframe
reviews = pd.concat(chunks, ignore_index=True)
reviews = reviews[reviews.stars != 3] # drop stars ==3
len(reviews)
# Recode 1,2 stars as 0 (negative sentiment)
# Recode 4,5 stars as 1 (positive sentiment)
reviews["sentiment"] = reviews["stars"].replace({1: 0, 2: 0, 4: 1, 5: 1})
reviews["sentiment"] = reviews["sentiment"].astype(int)
# Importing necessary libraries
import nltk
import pandas as pd
from textblob import Word
from nltk.corpus import stopwords
from sklearn.preprocessing import LabelEncoder
from sklearn.metrics import classification_report, confusion_matrix, accuracy_score
from keras.models import Sequential
from keras.preprocessing.text import Tokenizer
from tensorflow.keras.preprocessing.sequence import pad_sequences
from keras.layers import Dense, Embedding, LSTM, SpatialDropout1D
from sklearn.model_selection import train_test_split
# Pre-Processing the text
def cleaning(df, stop_words):
# Converting all text to lower case
df["text"] = df["text"].apply(lambda x: " ".join(x.lower() for x in x.split()))
# Replacing the digits/numbers
df["text"] = df["text"].str.replace("d", "")
# Removing stop words
df["text"] = df["text"].apply(
lambda x: " ".join(x for x in x.split() if x not in stop_words)
)
return df
reviews_sample = reviews.sample(n=200000) # take sample of 200,000
reviews_sample
stop_words = stopwords.words(
"english"
) # stopwords are defined in from nltk.corpus import stopwords
data_cleaned = cleaning(reviews_sample, stop_words) # apply the function in the dataset
X = data_cleaned["text"] # text as features
y = data_cleaned["sentiment"] # sentiment as labels
# Split to training and testing set
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=11
)
# Set the maximum number of words to consider in the vocabulary
max_vocab = 20000
# Create a tokenizer object with the specified maximum vocabulary size
tokenizer = Tokenizer(num_words=max_vocab)
# Fit the tokenizer on the training data to create a vocabulary index based on word frequency
tokenizer.fit_on_texts(X_train)
# Convert the text data in the training set into sequences of integers using the fitted vocabulary
X_train = tokenizer.texts_to_sequences(X_train)
# Convert the text data in the testing set into sequences of integers using the fitted vocabulary
X_test = tokenizer.texts_to_sequences(X_test)
# Pad sequences in X_train to a fixed length of 300 using zeros as padding
X_train = tf.keras.preprocessing.sequence.pad_sequences(
X_train, padding="post", maxlen=300
)
# Pad sequences in X_test to a fixed length of 300 using zeros as padding
X_test = tf.keras.preprocessing.sequence.pad_sequences(
X_test, padding="post", maxlen=300
)
import tensorflow as tf
# Define a sequential model architecture for a convolutional neural network
CNN_LSTM = tf.keras.Sequential(
[
# Add an embedding layer with a specified vocabulary size, embedding dimension, and input length
tf.keras.layers.Embedding(max_vocab, 128, input_length=300),
# Add a dropout layer to prevent overfitting and improve generalization
tf.keras.layers.Dropout(0.5),
# Add a 1D convolutional layer with a specified number of filters, kernel size, and activation function
tf.keras.layers.Conv1D(64, 5, activation="relu"),
# Add a max pooling layer to reduce the spatial dimensions of the output of the convolutional layer
tf.keras.layers.MaxPooling1D(pool_size=4),
# Add a long short-term memory (LSTM) layer with a specified number of units
tf.keras.layers.LSTM(128),
# Add a dense layer with a single output unit and a sigmoid activation function for binary classification
tf.keras.layers.Dense(1, activation="sigmoid"),
]
)
# Print a summary of the model architecture
CNN_LSTM.summary()
# Define an early stopping callback to stop training when validation loss stops improving, we wait 3 epoch
early_stop = tf.keras.callbacks.EarlyStopping(
monitor="val_loss", patience=3, restore_best_weights=True
)
# Compile the CNN-LSTM model with a binary cross-entropy loss function, Adam optimizer, and accuracy metric
CNN_LSTM.compile(loss="binary_crossentropy", optimizer="adam", metrics=["accuracy"])
# Train the CNN-LSTM model on the training data for a specified number of epochs, with a validation split for monitoring performance
model = CNN_LSTM.fit(
X_train,
y_train,
epochs=10,
validation_split=0.3,
batch_size=128,
shuffle=True,
callbacks=[early_stop],
)
# Get the training history of the model
history_model = model.history
# Extract the training and validation accuracy and loss values for plotting
acc = history_model["accuracy"]
val_acc = history_model["val_accuracy"]
loss = history_model["loss"]
val_loss = history_model["val_loss"]
epochs = model.epoch
# Plot the training and validation loss over the epochs
plt.figure(figsize=(10, 10)) # figure size
plt.plot(epochs, loss, "r", label="Training loss") # plot training loss
plt.plot(epochs, val_loss, "b", label="Validation loss") # plot validation loss
plt.title("Training and Validation Loss", size=20) # Title of the plot
plt.xlabel("Epochs", size=20) # label of x-axis
plt.ylabel("Loss", size=20) # label of y-axis
plt.legend(prop={"size": 20}) # plot the legend
plt.show()
plt.figure(figsize=(10, 10)) # figure size
plt.plot(epochs, acc, "g", label="Training acc") # plot training loss
plt.plot(epochs, val_acc, "b", label="Validation acc") # plot validation loss
plt.title("Training and Validation accuracy", size=20) # Title of the plot
plt.xlabel("Epochs", size=20) # label of x-axis
plt.ylabel("Accuracy", size=20) # label of y-axis
plt.legend(prop={"size": 20}) # plot the legend
plt.ylim((0.5, 1)) # set the value of y to be at least 0.5
plt.show()
# evaluates the performance of the CNN-LSTM model on the test data
CNN_LSTM.evaluate(X_test, y_test)
# Generate predictions on the test data using the trained CNN-LSTM model
pred_CNN_LSTM = CNN_LSTM.predict(X_test)
# Convert the continuous probability predictions to binary predictions using a threshold of 0.5
predictions = []
for i in pred_CNN_LSTM:
if i >= 0.5:
predictions.append(1)
else:
predictions.append(0)
# Evaluate the performance of the model on the test data using accuracy, precision, and recall
print("Accuracy on testing set:", accuracy_score(predictions, y_test))
print("Precision on testing set:", precision_score(predictions, y_test))
print("Recall on testing set:", recall_score(predictions, y_test))
# Generate the confusion matrix for the model predictions on the test data
matrix = confusion_matrix(predictions, y_test, normalize="all")
# Plot the confusion matrix using a heatmap
plt.figure(figsize=(5, 5))
ax = plt.subplot()
sns.set(font_scale=1)
sns.heatmap(matrix, annot=True, ax=ax)
# Set the axis labels, title, and tick labels for the plot
ax.set_xlabel("Predicted Sentiment", size=10)
ax.set_ylabel("True Sentiment", size=10)
ax.set_title("Confusion Matrix", size=15)
ax.xaxis.set_ticklabels(["negative", "positive"], size=15)
ax.yaxis.set_ticklabels(["negative", "positive"], size=15)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
from sklearn.metrics import r2_score, mean_squared_error, mean_absolute_error
from sklearn.model_selection import train_test_split
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.naive_bayes import MultinomialNB
df = pd.read_csv("/kaggle/input/language-detection/Language Detection.csv")
df.sample(30)
df.info()
df.describe()
df.corr()
df.shape
df["Language"].value_counts()
df.isnull().sum()
df.columns
x = np.array(df["Text"])
y = np.array(df["Language"])
cv = CountVectorizer()
x = cv.fit_transform(x)
df.shape
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.2, random_state=42
)
model = MultinomialNB()
model.fit(x_train, y_train)
model.predict(x_test)
model.score(x_test, y_test)
user = input("Enter a Text: ")
data = cv.transform([user]).toarray()
output = model.predict(data)
print(output)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
data = pd.read_csv("/kaggle/input/recidivism-forecasting/train.csv")
data = data.dropna()
data = data[
[
"ID",
"Gang_Affiliated",
"Supervision_Risk_Score_First",
"Dependents",
"Prison_Offense",
"Prior_Arrest_Episodes_Felony",
"Prior_Arrest_Episodes_Violent",
"Recidivism_Arrest",
]
]
data["Gang_Affiliated"] = data["Gang_Affiliated"].fillna(False)
data["Gang_Affiliated"] = data["Gang_Affiliated"].astype(int) # Over a thousand are GA
data["Supervision_Risk_Score_First"] = data["Supervision_Risk_Score_First"].fillna(0)
data["Supervision_Risk_Score_First"] = data["Supervision_Risk_Score_First"].astype(int)
data["Dependents"] = data["Dependents"].fillna(0)
data["Dependents"] = data["Dependents"].replace("3 or more", 3)
data["Dependents"] = data["Dependents"].astype(int)
# data["Prison_Offense"] = data["Prison_Offense"].fillna("Other")
data["Violent_Offense"] = data["Prison_Offense"].apply(
lambda x: 1 if x == "Violent/Non-Sex" or x == "Violent/Sex" else 0
)
data = data.drop(["Prison_Offense"], axis=1)
data["Prior_Arrest_Episodes_Felony"] = data["Prior_Arrest_Episodes_Felony"].fillna(0)
data["Prior_Arrest_Episodes_Felony"] = data["Prior_Arrest_Episodes_Felony"].replace(
"10 or more", 10
)
data["Prior_Arrest_Episodes_Felony"] = data["Prior_Arrest_Episodes_Felony"].astype(int)
data["Prior_Arrest_Episodes_Violent"] = data["Prior_Arrest_Episodes_Violent"].fillna(0)
data["Prior_Arrest_Episodes_Violent"] = data["Prior_Arrest_Episodes_Violent"].replace(
"3 or more", 10
)
data["Prior_Arrest_Episodes_Violent"] = data["Prior_Arrest_Episodes_Violent"].astype(
int
)
data
ytrain = data["Recidivism_Arrest"]
Xtrain = data.drop(["Recidivism_Arrest"], axis=1)
from sklearn.linear_model import LogisticRegression
estimator = LogisticRegression(solver="liblinear")
estimator.fit(Xtrain, ytrain)
data["prediction"] = estimator.predict(Xtrain)
data["prediction_round"] = data["prediction"].apply(lambda x: round(x))
data.groupby("Supervision_Risk_Score_First")["prediction_round"].mean()
submission = data.filter(["ID", "Recidivism_Arrest"], axis=1)
submission
# submission.to_csv('submission.csv',index=False)
|
import csv
from Bio import SeqIO
import pickle
import numpy as np
import pandas as pd
from xgboost import XGBClassifier
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
import xgboost
from sklearn import model_selection
from sklearn.metrics import accuracy_score
from sklearn.preprocessing import LabelEncoder
from sklearn.metrics import (
plot_confusion_matrix,
classification_report,
plot_precision_recall_curve,
plot_roc_curve,
)
# # Converting Amino acid sequence to DNA sequence for 3 organisms
def Reading_Fasata_file(path, sample):
Sequence = []
# Open the FASTA file
fasta_file = path
with open(fasta_file, "r") as handle:
# Parse the sequences using SeqIO
records = SeqIO.parse(handle, "fasta")
# Loop over each record and print its ID and sequence
for record in records:
if len(record.seq) <= 303:
Sequence.append(str(record.seq))
df = pd.DataFrame(data=Sequence, columns=["Amino acid"])
# take 300 dna sequence to test
df = df[:sample]
print(df.shape)
df.head()
return df
from Bio.Seq import Seq
def convert_AA_to_dna(df_Ecoli):
# Define a dictionary to map amino acids to codons
codon_table = {
"A": ["GCT", "GCC", "GCA", "GCG"],
"R": ["CGT", "CGC", "CGA", "CGG", "AGA", "AGG"],
"N": ["AAT", "AAC"],
"D": ["GAT", "GAC"],
"C": ["TGT", "TGC"],
"Q": ["CAA", "CAG"],
"E": ["GAA", "GAG"],
"G": ["GGT", "GGC", "GGA", "GGG"],
"H": ["CAT", "CAC"],
"I": ["ATT", "ATC", "ATA"],
"L": ["TTA", "TTG", "CTT", "CTC", "CTA", "CTG"],
"K": ["AAA", "AAG"],
"M": ["ATG"],
"F": ["TTT", "TTC"],
"P": ["CCT", "CCC", "CCA", "CCG"],
"S": ["TCT", "TCC", "TCA", "TCG", "AGT", "AGC"],
"T": ["ACT", "ACC", "ACA", "ACG"],
"W": ["TGG"],
"Y": ["TAT", "TAC"],
"V": ["GTT", "GTC", "GTA", "GTG"],
"*": ["TAA", "TAG", "TGA"],
}
dnas = []
# Loop over each amino acid sequence and translate to DNA
for seq in df_Ecoli["Amino acid"]:
# Remove the trailing asterisk
seq = seq[:-1]
# Translate the amino acid sequence to a DNA sequence
dna_seq = "".join(codon_table[aa][0] for aa in seq)
# Print the resulting DNA sequence
dnas.append(dna_seq)
return dnas
# Ecoli
df_Ecoli = Reading_Fasata_file("/kaggle/input/ecoli-fasta/SmProt2_Ecoli_Ribo.fa", 422)
df_Ecoli["DNA_sequence"] = convert_AA_to_dna(df_Ecoli)
# Zebrafish
df_Zebrafish = Reading_Fasata_file(
"/kaggle/input/zebrafish-fasta/SmProt2_zebrafish_Ribo.fa", 2481
)
df_Zebrafish["DNA_sequence"] = convert_AA_to_dna(df_Zebrafish)
# mouse
df_mouse = Reading_Fasata_file("/kaggle/input/mouse-fasta/SmProt2_mouse_Ribo.fa", 6451)
df_mouse["DNA_sequence"] = convert_AA_to_dna(df_mouse)
def Reading_ncFasata_file(path, sample):
Sequence = []
# Open the FASTA file
fasta_file = path
with open(fasta_file, "r") as handle:
# Parse the sequences using SeqIO
records = SeqIO.parse(handle, "fasta")
# Loop over each record and print its ID and sequence
for record in records:
if len(record.seq) <= 303:
Sequence.append(str(record.seq))
df = pd.DataFrame(data=Sequence, columns=["dnaseq"])
# take 300 dna sequence to test
df = df[:sample]
print(df.shape)
df.head()
return df
ncEcoli = Reading_ncFasata_file(
"/kaggle/input/noncoding-ecoli/ecoli_AND_so_rna_type_namencRNA_AND_so_rna_type_nameSncRNA_AND_entry_typeSequence.fasta",
10,
)
ncMouse = Reading_ncFasata_file(
"/kaggle/input/noncoding-mouse/Mus_musculus.GRCm39.ncrna.fa", 10
)
ncZebrafish = Reading_ncFasata_file(
"/kaggle/input/noncode-zebrafish/NONCODEv5_zebrafish.fa", 10
)
ncEcoli
# # Feature Engineering
class kmer_featurization:
def __init__(self, k):
"""
k: the "k" in k-mer
"""
self.k = k
self.letters = ["A", "T", "C", "G"]
self.multiplyBy = 4 ** np.arange(
k - 1, -1, -1
) # the multiplying number for each digit position in the k-number system
self.n = 4**k # number of possible k-mers
def obtain_kmer_feature_for_a_list_of_sequences(
self, seqs, write_number_of_occurrences=False
):
"""
Given a list of m DNA sequences, return a 2-d array with shape (m, 4**k) for the 1-hot representation of the kmer features.
Args:
write_number_of_occurrences:
a boolean. If False, then in the 1-hot representation, the percentage of the occurrence of a kmer will be recorded; otherwise the number of occurrences will be recorded. Default False.
"""
kmer_features = []
for seq in seqs:
this_kmer_feature = self.obtain_kmer_feature_for_one_sequence(
seq.upper(), write_number_of_occurrences=write_number_of_occurrences
)
kmer_features.append(this_kmer_feature)
kmer_features = np.array(kmer_features)
return kmer_features
def obtain_kmer_feature_for_one_sequence(
self, seq, write_number_of_occurrences=False
):
"""
Given a DNA sequence, return the 1-hot representation of its kmer feature.
Args:
seq:
a string, a DNA sequence
write_number_of_occurrences:
a boolean. If False, then in the 1-hot representation, the percentage of the occurrence of a kmer will be recorded; otherwise the number of occurrences will be recorded. Default False.
"""
number_of_kmers = len(seq) - self.k + 1
kmer_feature = np.zeros(self.n)
for i in range(number_of_kmers):
this_kmer = seq[i : (i + self.k)]
this_numbering = self.kmer_numbering_for_one_kmer(this_kmer)
kmer_feature[this_numbering] += 1
if not write_number_of_occurrences:
kmer_feature = kmer_feature / number_of_kmers
return kmer_feature
def kmer_numbering_for_one_kmer(self, kmer):
"""
Given a k-mer, return its numbering (the 0-based position in 1-hot representation)
"""
digits = []
for letter in kmer:
digits.append(self.letters.index(letter))
digits = np.array(digits)
numbering = (digits * self.multiplyBy).sum()
return numbering
def make_dna(df):
feats = []
for i in df["DNA_sequence"]:
featurizer = kmer_featurization(
4
) # create an instance of the kmer_featurization class, with k being the desired length of the k-mer
kmer_feature = featurizer.obtain_kmer_feature_for_one_sequence(i)
feats.append(kmer_feature)
# obtain the k-mer feature for the sequence seq
df_modified = pd.DataFrame(feats)
return df_modified
df_Ecoli_train = make_dna(df_Ecoli)
df_mouse_train = make_dna(df_mouse)
df_zebrafish_train = make_dna(df_Zebrafish)
df_Ecoli_train["label"] = 1
df_mouse_train["label"] = 1
df_zebrafish_train["label"] = 1
df_zebrafish_train.head()
# read train file
train = pd.read_csv("/kaggle/input/train/train_feature.csv")
train.head()
train.drop(columns=["orfID"])
Ecoli = df_Ecoli_train.iloc[0:50, :]
mouse = df_mouse_train.iloc[0:50, :]
zebrafish = df_zebrafish_train.iloc[0:50, :]
Ecoli = Ecoli.rename(columns={i: str(i) for i in range(256)})
mouse = mouse.rename(columns={i: str(i) for i in range(256)})
zebrafish = zebrafish.rename(columns={i: str(i) for i in range(256)})
new_train = pd.concat([train, Ecoli, mouse, zebrafish], ignore_index=True)
new_train.head()
# read test file
test = pd.read_csv("/kaggle/input/test-feature/test_feature.csv")
test.head()
import numpy as np
from sklearn.metrics import roc_auc_score, roc_curve
import matplotlib.pyplot as plt
def plot_roc_curve(true_y, y_prob):
"""
plots the roc curve based of the probabilities
"""
fpr, tpr, thresholds = roc_curve(true_y, y_prob)
plt.plot(fpr, tpr)
plt.xlabel("False Positive Rate")
plt.ylabel("True Positive Rate")
# # Training Model with Human Organism
target = "label"
# split features and target from each other
y_train = new_train[target]
X_train = new_train.drop(columns=["label", "orfID"])
y_test = test[target]
X_test = test.drop(columns=["label", "orfID"])
def report(model, X_test, y_test):
preds = model.predict_proba(X_test)
# Convert predicted probabilities to predicted class labels
y_pred = (preds[:, 1] > 0.5).astype(int)
# Calculate accuracy score
accuracy = accuracy_score(y_test, y_pred)
print("Accuracy:", accuracy)
print(classification_report(y_test, y_pred))
plot_roc_curve(y_test, y_pred)
try:
print(f"model 1 AUC score: {roc_auc_score(y_test, y_pred)}")
except ValueError:
pass
# fit model no training data
model = xgboost.XGBClassifier(
learning_rate=0.1, n_estimators=100, max_depth=5, probability=True
)
model.fit(X_train, y_train)
print(model)
# make predictions for test data
report(model, X_test, y_test)
# # Test with mammalians species
def Tested(df):
# df=pd.read_csv(path,index_col=0 )
# df['label']=1
y_test = df[target]
X_test = df.drop(columns=["label"])
return X_test, y_test
X_test_Zebrafish, y_test_Zebrafish = Tested(df_zebrafish_train)
report(model, X_test_Zebrafish, y_test_Zebrafish)
X_test_mouse, y_test_mouse = Tested(df_mouse_train)
report(model, X_test_mouse, y_test_mouse)
X_test_Ecoli, y_test_Ecoli = Tested(df_Ecoli_train)
report(model, X_test_Ecoli, y_test_Ecoli)
# import pickle
# #save model
# .dump(model, open("/kaggle/working/XGboost", 'wb') )
plot_roc_curve(y, y_proba)
print(f"model 1 AUC score: {roc_auc_score(y, y_proba)}")
|
# ```python
# # Last updated: Wed, February 24, 2021 - 13:07
# ############################################################
# # Reference:
# # https://www.kaggle.com/dansbecker/model-validation
# # https://www.youtube.com/watch?v=ZiKrbm-haoA
# ############################################################
# Validating the model
# =====
# # Does the model accurately predicts what we are trying to predict?
# """
# In the previous lesson, L3 - Your First Machine Learning Model
# We are using the testing data to retrieve the output, that is why the predicted value
# is 100% same as the training set output
# WHY SAME?
# Because the model already seen the exact input and outputs
# HOW TO FIX THIS?
# We can split some of the data from the testing set into testing set and validation data
# Split the prediction target and the features into 4 subsets
# - prediction target training data (Used to train the model)
# - feature training data (Used to train the model)
# - prediction target validation data (Used to validate the model)
# - feature validation data (Used to validate the model)
# """
# # Randomly splitted by 'from sklearn.model_selection import train_test_split'
# train_X, val_X, train_y, val_y = train_test_split(X, y, random_state = 0)
# """
# The validation data is the data that our model will not learn during its first training
# After the first training with the splitted training set, we use the splitted validation data
# to validate it with our trained_model and check with the output to see how accurate it is
# """
# How to validate the quality of our model?
# =====
# # Using Mean Absolute Error (MAE)
# MAE = See how much our model is off by this amount of value
# """
# # Formula as below
# error = actual_predicted
# absolute_error = |actual_predicted|
# mean_absolute_error = sum (|actual_predicted|) / total number of observations
# """
# ``` Lesson 4 Tutorial
# # Step 1: Splitting the data
# # ==========================
# # Import pandas
# import pandas as pd
# # Import the train_test_split function and uncomment
# from sklearn.model_selection import train_test_split
# # fill in and uncomment
# train_X, val_X, train_y, val_y = train_test_split(X, y, random_state=1)
# # Step 2: Specify and Fit the Model
# # =================================
# # Import DecisionTreeRegressor
# from sklearn.tree import DecisionTreeRegressor
# # Specify the model
# iowa_model = DecisionTreeRegressor(random_state=1)
# # Fit iowa_model with the training data.
# iowa_model.fit(train_X, train_y)
# # Step 3: Make Predictions with Validation data
# # =============================================
# val_prediction = iowa_model.predict(val_X)
# # print the top few validation predictions
# print(val_predictions[:5])
# # print the top few actual prices from validation data
# print(val_y.head())
# # Step 4: Calculate the Mean Absolute Error in Validation Data
# # ============================================================
# from sklearn.metrics import mean_absolute_error
# val_mae = mean_absolute_error(val_y, val_predictions)
# print(val_mae)
# # The lower the MAE the better but becareful of data overfitting
# # Normally the without splitting the training data for validation, we will get lower MAE
# # Overfitting will occurs
# ```
# RECAP
# =====
# 1. There are many metrics for summarizing model quality
# 2. mean_absolute_error() is used to calculate MAE, it is a function from scikit-leanr.metrics module
# 3. Validation data - Withold data from our model and use it to test our model accuracy.
# ```
# You've built a model. But how good is it?
# In this lesson, you will learn to use model validation to measure the quality of your model. Measuring model quality is the key to iteratively improving your models.
# # What is Model Validation
# You'll want to evaluate almost every model you ever build. In most (though not all) applications, the relevant measure of model quality is predictive accuracy. In other words, will the model's predictions be close to what actually happens.
# Many people make a huge mistake when measuring predictive accuracy. They make predictions with their *training data* and compare those predictions to the target values in the *training data*. You'll see the problem with this approach and how to solve it in a moment, but let's think about how we'd do this first.
# You'd first need to summarize the model quality into an understandable way. If you compare predicted and actual home values for 10,000 houses, you'll likely find mix of good and bad predictions. Looking through a list of 10,000 predicted and actual values would be pointless. We need to summarize this into a single metric.
# There are many metrics for summarizing model quality, but we'll start with one called **Mean Absolute Error** (also called **MAE**). Let's break down this metric starting with the last word, error.
# The prediction error for each house is:
# ```
# error=actual−predicted
# ```
#
# So, if a house cost \$150,000 and you predicted it would cost \$100,000 the error is \$50,000.
# With the MAE metric, we take the absolute value of each error. This converts each error to a positive number. We then take the average of those absolute errors. This is our measure of model quality. In plain English, it can be said as
# > On average, our predictions are off by about X.
# To calculate MAE, we first need a model. That is built in a hidden cell below, which you can review by clicking the `code` button.
# Data Loading Code Hidden Here
import pandas as pd
# Load data
melbourne_file_path = "../input/melbourne-housing-snapshot/melb_data.csv"
melbourne_data = pd.read_csv(melbourne_file_path)
# Filter rows with missing price values
filtered_melbourne_data = melbourne_data.dropna(axis=0)
# Choose target and features
y = filtered_melbourne_data.Price
melbourne_features = [
"Rooms",
"Bathroom",
"Landsize",
"BuildingArea",
"YearBuilt",
"Lattitude",
"Longtitude",
]
X = filtered_melbourne_data[melbourne_features]
from sklearn.tree import DecisionTreeRegressor
# Define model
melbourne_model = DecisionTreeRegressor()
# Fit model
melbourne_model.fit(X, y)
# Once we have a model, here is how we calculate the mean absolute error:
from sklearn.metrics import mean_absolute_error
predicted_home_prices = melbourne_model.predict(X)
mean_absolute_error(y, predicted_home_prices)
# # The Problem with "In-Sample" Scores
# The measure we just computed can be called an "in-sample" score. We used a single "sample" of houses for both building the model and evaluating it. Here's why this is bad.
# Imagine that, in the large real estate market, door color is unrelated to home price.
# However, in the sample of data you used to build the model, all homes with green doors were very expensive. The model's job is to find patterns that predict home prices, so it will see this pattern, and it will always predict high prices for homes with green doors.
# Since this pattern was derived from the training data, the model will appear accurate in the training data.
# But if this pattern doesn't hold when the model sees new data, the model would be very inaccurate when used in practice.
# Since models' practical value come from making predictions on new data, we measure performance on data that wasn't used to build the model. The most straightforward way to do this is to exclude some data from the model-building process, and then use those to test the model's accuracy on data it hasn't seen before. This data is called **validation data**.
# # Coding It
# The scikit-learn library has a function `train_test_split` to break up the data into two pieces. We'll use some of that data as training data to fit the model, and we'll use the other data as validation data to calculate `mean_absolute_error`.
# Here is the code:
from sklearn.model_selection import train_test_split
# split data into training and validation data, for both features and target
# The split is based on a random number generator. Supplying a numeric value to
# the random_state argument guarantees we get the same split every time we
# run this script.
train_X, val_X, train_y, val_y = train_test_split(X, y, random_state=0)
# Define model
melbourne_model = DecisionTreeRegressor()
# Fit model
melbourne_model.fit(train_X, train_y)
# get predicted prices on validation data
val_predictions = melbourne_model.predict(val_X)
print(mean_absolute_error(val_y, val_predictions))
|
import pandas as pd
import numpy as np
import sklearn as sn
import matplotlib.pyplot as plt
from sklearn import preprocessing
from sklearn.tree import DecisionTreeClassifier
import sklearn.tree as tree
# ### To read the dataset we should use Pandas library
df = pd.read_csv("/kaggle/input/drugs-a-b-c-x-y-for-decision-trees/drug200.csv")
df.head()
df.shape
df.describe()
df.info()
df.isna().sum()
df["Drug"].value_counts()
# ## Feature set
df.columns
# To use scikit-learn library, we have to convert the Pandas data frame to a Numpy array (Notice: we have read our dataset with the help of pandas library):
# X as the Feature Matrix (data of df)
# y as the response vector (Drug)
X = df[["Age", "Sex", "BP", "Cholesterol", "Na_to_K"]].values
X[0:5]
Y = df["Drug"].values
Y[0:5]
# ## Normalization
# As you may figure out, some features in this dataset are categorical, such as Sex or BP. Unfortunately, Sklearn Decision Trees does not handle categorical variables. We can still convert these features to numerical values using LabelEncoder to convert the categorical variable into numerical variables.
df["Sex"].value_counts()
df["BP"].value_counts()
df["Cholesterol"].value_counts()
from sklearn import preprocessing
le_sex = preprocessing.LabelEncoder()
le_sex.fit(["F", "M"])
X[:, 1] = le_sex.transform(X[:, 1])
le_BP = preprocessing.LabelEncoder()
le_BP.fit(["LOW", "NORMAL", "HIGH"])
X[:, 2] = le_BP.transform(X[:, 2])
le_Chol = preprocessing.LabelEncoder()
le_Chol.fit(["NORMAL", "HIGH"])
X[:, 3] = le_Chol.transform(X[:, 3])
X[0:5]
# ## Setting up the Decision Tree
# We will be using train/test split on our decision tree. Let's import train_test_split from sklearn.cross_validation.
from sklearn.model_selection import train_test_split
# Notice: The X and y are the arrays required before the split, the test_size represents the ratio of the testing dataset, and the random_state ensures that we obtain the same splits.
X_trainset, X_testset, Y_trainset, Y_testset = train_test_split(
X, Y, test_size=0.3, random_state=3
)
# ## Modeling
# We will first create an instance of the DecisionTreeClassifier called drugTree.
# Inside of the classifier, specify criterion="entropy" so we can see the information gain of each node.
drugTree = DecisionTreeClassifier(criterion="entropy", max_depth=4)
drugTree.fit(X_trainset, Y_trainset)
# ## Prediction
# Let's make some predictions on the testing dataset and store it into a variable called predTre
predTree = drugTree.predict(X_testset)
print(predTree[0:5])
print(Y_testset[0:5])
# ## Evaluation
# Next, let's import metrics from sklearn and check the accuracy of our model.
from sklearn import metrics
print("DecisionTrees's Accuracy: ", metrics.accuracy_score(Y_testset, predTree))
# ## Visualization
# Let's visualize the tree
tree.plot_tree(drugTree)
plt.show()
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
# About Dataset:
# 1. Age: in years
# 2. Sex: 1 = male; 0 = female
# 3. Chest pain type:
# Value 1: typical angina
# Value 2: atypical angina
# Value 3: non-anginal pain
# Value 4: asymptomatic
# 4. BP: resting blood pressure (in mm Hg on admission to the hospital)
# 5. Cholesterol : serum cholestoral in mg/dl
# 6. FBS: fasting blood sugar > 120 mg/dl (1 = true; 0 = false)
# 7. EKG results: resting electrocardiographic results
# Value 0: normal
# Value 1: aving ST-T wave abnormality (T wave inversions and/or ST elevation or depression of > 0.05 mV)
# Value 2: showing probable or definite left ventricular hypertrophy by Estes' criteria
# 8. MAx HR: maximum heart rate achieved
# 9. Excercise angina: exercise induced angina (1 = yes; 0 = no)
# 10. Slope of ST: oldpeak = ST depression induced by exercise relative to rest
# 11. Number of vessels fluro: number of major vessels (0-3) colored by flourosopy
# 12. Thallium: 3 = normal; 6 = fixed defect; 7 = reversable defect
df = pd.read_csv("../input/heart-disease-prediction/Heart_Disease_Prediction.csv")
df.head()
df.describe().T
df.info()
df.columns.values
df.nunique()
# > # EDA - Esploratory Data Analysis
# Converting object targets to boolean
target = df["Heart Disease"].map({"Presence": 1, "Absence": 0})
inputs = df.drop(["Heart Disease"], axis=1)
# Correlation matrix - The 'Slope of ST' is highly linearly correlated with 'ST depression'
plt.suptitle("Correlation Map/Pearson correlation coefficient")
sns.heatmap(df.iloc[:, 1:-1].corr())
plt.show()
# Heart disease based on age - Older people have more chance to have heart disease
plt.suptitle("Age")
sns.scatterplot(data=df, x="Age", y=np.zeros(len(df["Age"])), hue=target)
plt.show()
# Heart disease based on Gender - It can be observe that males have more opportunities to have heart disease than female
ax = sns.countplot(x="Heart Disease", hue="Sex", data=df)
legend_labels, _ = ax.get_legend_handles_labels()
ax.legend(legend_labels, ["Female", "Male"], bbox_to_anchor=(1, 1))
plt.show()
# Heart disease based on Chest pain type - 4th type of chest pain dominate in heart disease
plt.suptitle("Chest pain type vs Heart Disease")
sns.countplot(data=df, x="Heart Disease", hue="Chest pain type")
plt.show()
# Heart Disease based on BP - Persons with high BP have more chance to get heart disease
plt.figure(figsize=(10, 5))
plt.subplot(1, 2, 1)
plt.suptitle("BP")
sns.scatterplot(data=df, x="BP", y=np.zeros(len(df["BP"])), hue=target)
plt.subplot(1, 2, 2)
sns.scatterplot(data=df, x="BP", y="Age", hue=target)
plt.show()
# Cholesterol - Higer Cholesterol does not influence on heart disease
plt.suptitle("Cholesterol")
sns.scatterplot(
data=df, x="Cholesterol", y=np.zeros(len(df["Cholesterol"])), hue=target
)
plt.show()
# FBS over 120 - Also increased FBS over 120 does not imply on heart disease prediction
ax = sns.countplot(x="Heart Disease", hue="FBS over 120", data=df)
plt.show()
# EKG results - The 2nd value of EKG could influence on heart disease prediction
plt.suptitle("EKG results vs Heart Disease")
sns.countplot(data=df, x="Heart Disease", hue="EKG results")
plt.show()
# Max HR - From the first graph we can observe that people with lower HR max have a higher likelihood of heart disease than those with higher HR max. Furthermore, we can observe explicit cut of/threshold where below 120 HR max objects have a higher probability to have problem with heart.
plt.figure(figsize=(10, 5))
plt.subplot(1, 2, 1)
plt.suptitle("Max HR")
sns.scatterplot(data=df, x="Max HR", y=np.zeros(len(df["Max HR"])), hue=target)
plt.subplot(1, 2, 2)
sns.scatterplot(data=df, x="Max HR", y="Age", hue=target)
plt.show()
# Excercise angina - Chest pain after a high excercise intensity or stress can casues a presence of heart failure
plt.suptitle("Excercise angina vs Heart Disease")
sns.countplot(data=df, x="Heart Disease", hue="Exercise angina")
plt.show()
# ST depression - Increased ST deprsesion increase heart disease. Nevetheless it can be observe on the bottom figure that males have higher probability of having depression.
plt.suptitle("ST Depression")
sns.scatterplot(
data=df, x="ST depression", y=np.zeros(len(df["ST depression"])), hue=target
)
ax = sns.catplot(x="Sex", y="ST depression", kind="box", data=df)
plt.show()
# Slope of ST
plt.suptitle("Slope of ST vs Heart Disease")
sns.countplot(data=df, x="Heart Disease", hue="Slope of ST")
plt.show()
# **Balancing the dataset with standarization**
one_target = int(np.sum(target))
zero_counter = 0
indices_to_remove = []
for i in range(target.shape[0]):
if target[i] == 0:
zero_counter += 1
if zero_counter > one_target:
indices_to_remove.append(i)
print("Indices before balancing data:", target.shape[0])
print("Idices to delete:", len(indices_to_remove))
balanced_inputs = inputs.drop(indices_to_remove, axis=0)
balanced_targets = target.drop(indices_to_remove, axis=0)
# reset indices
reset_inputs = balanced_inputs.reset_index(drop=True)
reset_targets = balanced_targets.reset_index(drop=True)
print("Inputs after balancing data:", reset_inputs.shape[0])
print("Targets after balancing data:", reset_targets.shape[0])
balanced_inputs.head()
# > # Applying different models
# Standarization
from sklearn.preprocessing import MinMaxScaler
scaled_inputs = MinMaxScaler().fit_transform(balanced_inputs)
# Spliting the data set
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
scaled_inputs, balanced_targets, test_size=0.2
)
# **Linear Regression**
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_squared_error, r2_score
lr = LinearRegression()
lr.fit(X_train, y_train)
predicted = lr.predict(X_test)
RMSE = np.sqrt(mean_squared_error(y_test, predicted))
r2 = r2_score(y_test, predicted)
print("Root mean squared error: ", RMSE)
print("r2: ", r2)
# **Logistic Regression**
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import accuracy_score
from sklearn import metrics
from sklearn.metrics import roc_curve
logit = LogisticRegression()
logit.fit(X_train, y_train)
predicted_logit = logit.predict(X_test)
LogisticRegressionScore = accuracy_score(predicted_logit, y_test)
plt.figure()
metrics.plot_roc_curve(logit, X_test, y_test)
plt.title("Receiver Operating Characteristic (ROC)")
plt.show()
print("Logistic Regression score: ", LogisticRegressionScore)
# **Gaussian Naive Bayes**
from sklearn.naive_bayes import GaussianNB
gauss = GaussianNB()
gauss.fit(X_train, y_train)
gauss_pred = gauss.predict(X_test)
gauss_score = accuracy_score(gauss_pred, y_test)
plt.figure()
metrics.plot_roc_curve(gauss, X_test, y_test)
plt.title("Receiver Operating Characteristic (ROC)")
plt.show()
print("Gaussian Naive Bayes score: ", gauss_score)
# **KNeighborsClassifier**
from sklearn.neighbors import KNeighborsClassifier
KNC = KNeighborsClassifier(n_neighbors=2)
KNC.fit(X_train, y_train)
KNC_pred = KNC.predict(X_test)
KNC_accuracy = metrics.accuracy_score(y_test, KNC_pred)
print("KNeighbourClassifier score: ", KNC_accuracy)
# **Random Forest Classifier**
from sklearn.ensemble import RandomForestClassifier
rnd_clf = RandomForestClassifier(n_estimators=500, max_leaf_nodes=16, n_jobs=-1)
rnd_clf.fit(X_train, y_train)
rnd_clf_pred = rnd_clf.predict(X_test)
rnd_clf_accuracy = metrics.accuracy_score(y_test, rnd_clf_pred)
print("RandomForest score: ", rnd_clf_accuracy)
# **Bagging Decision Tree**
from sklearn.ensemble import BaggingClassifier
from sklearn.tree import DecisionTreeClassifier
bag_clf = BaggingClassifier(
DecisionTreeClassifier(),
n_estimators=500,
oob_score=True,
max_samples=100,
bootstrap=True,
n_jobs=-1,
)
bag_clf.fit(X_train, y_train)
bag_clf_oob = bag_clf.oob_score_
bag_clf_pred = bag_clf.predict(X_test)
bag_clf_accuracy = metrics.accuracy_score(y_test, bag_clf_pred)
print(
"Bagging Decision Tree score: ", bag_clf_accuracy, "Out of the bag: ", bag_clf_oob
)
# > # Model's accuracy
data = {
"Estimators": [
"Linear Regression",
"Logistic Regression",
"Gaussian Naive Bayes",
"K-Nearest Neighbor",
"Random Forest",
"Bagging Decision Tree",
],
"Accuracy": [
r2,
LogisticRegressionScore,
gauss_score,
KNC_accuracy,
rnd_clf_accuracy,
bag_clf_accuracy,
],
}
data = pd.DataFrame(data)
data.sort_values("Accuracy", ascending="False")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import warnings
warnings.filterwarnings("ignore")
# 
# > In 2020, the COVID-19 pandemic has come to dominate the media, both domestically and abroad. Alongside increased attention on the pandemic, has come the viral spread of COVID-19 fake news online.
# > In this notebook, I propose to build a Covid-19 news classification model using different tools:
# > - Word2Vec
# > - CountVectorizer
# > - Tf-idf
# > - Fasttext
# > - MultinomialNB
# download dataset
import pandas as pd
df = pd.read_csv("Constraint_Train.csv")
df.head()
# We have a dataset that contains three columns: id, tweet and class tag.
# # PYCARET
# > ## Let's use pycaret to select the best classifier
from pycaret.classification import *
setup(data=df, target="label")
compare_models(sort="F1", n_select=5)
# > This function trains all models in the model library and compares the score against each other using K-block cross-validation (default 10 blocks). Evaluation indicators are used as follows:
# >
# > For classification: Accuracy, AUC, Recall, Precision, F1, Kappa, TT (Sec).
# ### Logistic Regression, Naive Bayes, Dummy Classifier show best F-measure.
# > # Text preprocessing with NLTK
import numpy as np
from nltk.tokenize import word_tokenize
from tqdm import tqdm
import nltk
nltk.download("punkt")
import nltk
nltk.download("stopwords")
from nltk.corpus import stopwords
import re
import pymorphy2
# create function for preparing tweets
mystopwords = stopwords.words("english")
m = pymorphy2.MorphAnalyzer()
ru_words = re.compile("[A-Za-z]+")
def words_only(text):
return " ".join(ru_words.findall(text))
def lemmatize(text):
try:
return " ".join([m.parse(w)[0].normal_form for w in text.lower().split()])
except:
return " "
def remove_stopwords(text, mystopwords=mystopwords):
try:
return " ".join([token for token in text.split() if not token in mystopwords])
except:
return ""
def preprocess(text):
return remove_stopwords(lemmatize(words_only(text.lower())))
from tqdm.auto import tqdm
tqdm.pandas()
df["new_tweet"] = df.tweet.astype("str").progress_apply(preprocess)
# let's see what we got
df.head()
df.new_tweet[0]
sentences = [word_tokenize(text.lower()) for text in tqdm(df.new_tweet)]
from wordcloud import WordCloud
big_string = ""
for i in range(len(df.new_tweet)):
big_string += df.new_tweet[i] + " "
import matplotlib.pyplot as plt
from wordcloud import WordCloud, STOPWORDS
wordCloud = WordCloud(
width=1000, height=1000, random_state=1, background_color="black"
).generate(big_string)
plt.figure(figsize=(5, 5))
plt.imshow(wordCloud)
# Word Cloud is a data visualization technique used for representing text data in which the size of each word indicates its frequency or importance.
# > # WORD2VEC
from gensim.models.word2vec import Word2Vec
# ## Let's ask the model for 10 nearest neighbors for the word "vacine" and the coefficient of cosine proximity:
# check the model for semantic similarity
model_tweets.wv.most_similar(positive=["vaccine"], topn=10)
# normalize vectors (.init_sims() method in new version)
model_tweets.wv.get_normed_vectors()
def get_text_embedding(text):
result = []
for word in word_tokenize(text.lower()):
if word in model_tweets.wv:
result.append(model_tweets.wv[word])
if len(result):
result = np.average(result, axis=0)
else:
result = np.zeros(500)
return result
features = [get_text_embedding(text) for text in tqdm(df.new_tweet)]
from sklearn.linear_model import LogisticRegression
from sklearn.linear_model import RidgeClassifier
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
features, df.label, test_size=0.3, random_state=0
)
model = RidgeClassifier(alpha=0.001)
model.fit(X_train, y_train)
from sklearn.metrics import classification_report
predicted = model.predict(X_test)
print(classification_report(y_test, predicted))
# > # COUNTVECTORIZER METHOD
from sklearn.feature_extraction.text import CountVectorizer
vec = CountVectorizer()
bow = vec.fit_transform(df.new_tweet)
X_train, X_test, y_train, y_test = train_test_split(bow, df.label, test_size=0.3)
model = LogisticRegression()
model.fit(X_train, y_train)
predicted = model.predict(X_test)
print(classification_report(y_test, predicted))
# > # TF-IDF METHOD
from sklearn.pipeline import Pipeline
from sklearn.linear_model import LogisticRegression
from sklearn.feature_extraction.text import TfidfTransformer
clf = Pipeline(
[
("vect", CountVectorizer()),
("tfidf", TfidfTransformer()),
("clf", RidgeClassifier()),
]
)
X_train, X_test, y_train, y_test = train_test_split(
df.new_tweet, df.label, test_size=0.3
)
clf.fit(X_train, y_train)
pred = clf.predict(X_test)
print(classification_report(pred, y_test))
# > # FASTTEXT METHOD
import fasttext
X = df.new_tweet.tolist()
y = df.label.tolist()
X, y = np.array(X), np.array(y)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
print("total train examples %s" % len(y_train))
print("total test examples %s" % len(y_test))
with open("data.train.txt", "w+") as outfile:
for i in range(len(X_train)):
outfile.write("__label__" + y_train[i] + " " + X_train[i] + "\n")
with open("test.txt", "w+") as outfile:
for i in range(len(X_test)):
outfile.write("__label__" + y_test[i] + " " + X_test[i] + "\n")
classifier = fasttext.train_supervised("data.train.txt")
result = classifier.test("test.txt")
print("P@1:", result[1])
print("R@1:", result[2])
print("Number of examples:", result[0])
# > # MultinomialNB
from sklearn.naive_bayes import MultinomialNB
from sklearn.metrics import accuracy_score
from sklearn.decomposition import TruncatedSVD
X_train, X_test, y_train, y_test = train_test_split(
df.new_tweet, df.label, test_size=0.25, random_state=1
)
nb_classifier = Pipeline([("vect", CountVectorizer()), ("clf", MultinomialNB())])
# Fit the classifier to the training data
nb_classifier.fit(X_train, y_train)
# Create the predicted tags: pred
pred = nb_classifier.predict(X_test)
# Calculate the accuracy score: score
score = accuracy_score(y_test, pred)
print(score)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
files = []
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
files.append(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
nfdata = pd.read_csv(files[1])
data = pd.read_csv(files[1])
nfdata.info()
nfdata.head()
for index, data in nfdata.items():
if index == "genres":
print(data)
import json
# for data in nfdata['genres']:
# # print(data,type(data))
# # print("{ 'data' : "+ data + "}")
# # d = json.loads("{ 'data' : "+ data + "}")
# break
print(nfdata["genres"].apply(lambda x: x.split(",")))
def split_genre(item):
return item.replace("[", "").replace("]", "").split(",")
genre = nfdata["genres"].apply(split_genre)
# print(genre) #aa no use nai
actgen = list()
for item in genre:
for d in item:
actgen.append(d.replace("'", "").strip())
# print(actgen) #aa no use nai
actgen = set(actgen)
# print(actgen) #aa no use nai
actgen = list(actgen)
actgen.remove("")
print(actgen)
print(len(actgen))
# newdata = pd.get_dummies(actgen)
# print(newdata)
# pd.get_dummies(nfdata, prefix=actgen[:8])
rows, col = nfdata.shape
for d in actgen:
nfdata[d] = np.zeros(rows)
nfdata.head()
nfdata.shape
for index in nfdata.index:
for d in actgen:
if d in nfdata.loc[index, "genres"]:
nfdata.loc[index, d] = 1
actgen.extend(["genres"])
# nfdata[actgen].head()
nfdata.corr
nfdata.corr()
# nfdata.hist()
actdata = list()
def split_genre(item):
return item.replace("[", "").replace("]", "").split(",")
new_data = nfdata["production_countries"].apply(split_genre)
# print(new_data)
for data in new_data:
for d in data:
actdata.append(d.replace("'", "").strip())
# print(actdata)
actdata = set(actdata)
# print(actdata)
actdata = list(actdata)
actdata.remove("")
print(actdata)
print(len(actdata))
# for data in nfdata['production_countries']:
# # print(data)
# actdata.append(data.replace("]","").replace("[","").replace("'",""))
# actdata=set(actdata)
# print(actdata)
row, col = nfdata.shape
for d in actdata:
nfdata[d] = np.zeros(rows)
nfdata.head()
for index in nfdata.index:
for d in actdata:
if d in nfdata.loc[index, "production_countries"]:
nfdata.loc[index, d] = 1
actdata.extend(["production_countries"])
nfdata.corr
nfdata[actdata].describe()
actdata
count = 1
year = list()
for i in nfdata["release_year"]:
year.append(i)
count = count + 1
# print(year)
year = set(year)
year = list(year)
print(year)
# print(len(year))
row, col = nfdata.shape
# print(row)
# print(col)
for d in year:
nfdata[str(d)] = np.zeros(row)
for index in nfdata.index:
for d in year:
if str(d) == nfdata.loc[index, "release_year"]:
nfdata.loc[index, str(d)] = 1
year.extend(["release_year"])
nfdata.corr
rate = list()
for i in nfdata.age_certification:
rate.append(i)
rate = set(rate)
print(rate)
print(len(rate))
data = list()
new_data = list()
for i in nfdata.genres:
data.append(i.replace("]", "").replace("[", "").replace("'", "").split(","))
for i in data:
for j in i:
new_data.append(j.strip())
new_data = set(new_data)
new_data = list(new_data)
new_data.remove("")
print(new_data)
print(len(new_data))
rows, col = nfdata.shape
print(rows)
print(col)
nfdata.columns
for d in new_data:
# print(d)
# # break
nfdata[d] = np.zeros(rows)
nfdata[new_data].head()
data = list()
new_data = list()
for i in nfdata.production_countries:
data.append(i.replace("]", "").replace("[", "").replace("'", "").split(","))
for i in data:
for j in i:
new_data.append(j.strip())
new_data = set(new_data)
new_data.remove("")
new_data = list(new_data)
rows, cols = nfdata.shape
for d in new_data:
nfdata[d] = np.zeros(rows)
nfdata.head()
nfdata.shape
for index in nfdata.index:
for d in new_data:
if d in nfdata.loc[index, "production_countries"]:
nfdata.loc[index, d] = 1
new_data.extend(["production_countries"])
nfdata.corr
# nfdata.shape
# print(new_data)
# print(len(new_data))
# new_data=list()
# new_data.info(new_data)
nfdata[nfdata["UK"] == 1]
nfdata[new_data].head()
# nfdata[nfdata['US'] == '1.0']['release_year'].value_counts().to_frame()
USA_count = nfdata[nfdata["US"] == 1][["release_year"]].value_counts().to_frame()
USA_count.reset_index(level=[0], inplace=True)
INDIA_count = nfdata[nfdata["IN"] == 1][["release_year"]].value_counts().to_frame()
INDIA_count.reset_index(level=[0], inplace=True)
NEWZLAND_count = nfdata[nfdata["NZ"] == 1][["release_year"]].value_counts().to_frame()
NEWZLAND_count.reset_index(level=[0], inplace=True)
JAPAN_count = nfdata[nfdata["JP"] == 1][["release_year"]].value_counts().to_frame()
JAPAN_count.reset_index(level=[0], inplace=True)
LEBNON_count = (
nfdata[nfdata["Lebanon"] == 1][["release_year"]].value_counts().to_frame()
)
LEBNON_count.reset_index(level=[0], inplace=True)
USA_count = USA_count.rename(columns={0: "count"})
INDIA_count = INDIA_count.rename(columns={0: "count"})
NEWZLAND_count = NEWZLAND_count.rename(columns={0: "count"})
JAPAN_count = JAPAN_count.rename(columns={0: "count"})
LEBNON_count = LEBNON_count.rename(columns={0: "count"})
# INDIA_count=nfdata[nfdata['IN']==1]['release_year'].value_counts().to_frame()
# NEWZLAND_count=nfdata[nfdata['NZ']==1]['release_year'].value_counts().to_frame()
# JAPAN_count=nfdata[nfdata['JP']==1]['release_year'].value_counts().to_frame()
# LEBNON_count=nfdata[nfdata['Lebanon']==1]['release_year'].value_counts().to_frame()
# print(INDIA_count)
USA_count = USA_count.sort_values(by=["release_year"], ascending=False)
INDIA_count = INDIA_count.sort_values(by=["release_year"], ascending=False)
NEWZLAND_count = NEWZLAND_count.sort_values(by=["release_year"], ascending=False)
JAPAN_count = JAPAN_count.sort_values(by=["release_year"], ascending=False)
LEBNON_count = LEBNON_count.sort_values(by=["release_year"], ascending=False)
# data = data.sort_values(by=['release_year'],ascending=False)
# data = data.sort_values(by=['release_year'],ascending=False)
# data = data.sort_values(by=['release_year'],ascending=False)
# data = data.sort_values(by=['release_year'],ascending=False)
# print(USA_count[:10]['release_year'])
fig, ax = plt.subplots()
ax.plot(USA_count[:10]["release_year"], USA_count[:10]["count"], label="us")
ax.plot(INDIA_count[:10]["release_year"], INDIA_count[:10]["count"], label="india")
ax.plot(
NEWZLAND_count[:10]["release_year"], NEWZLAND_count[:10]["count"], label="newzland"
)
ax.plot(JAPAN_count[:10]["release_year"], JAPAN_count[:10]["count"], label="japan")
ax.plot(LEBNON_count[:10]["release_year"], LEBNON_count[:10]["count"], label="lebenon")
# ax.plot(xtv['release_year'], xtv['count'], label="TV Show")
ax.legend()
plt.show()
# data=list()
# for i in nfdata.tmdb_popularity:
# # print(i)
# data.append(i)
# # print(data)
# data=set(data)
# print(data)
data = list()
data1 = list()
# print(data)
for i in nfdata.genres:
data.append(i.replace("[", "").replace("]", "").replace("'", "").split(","))
# print(data)
for i in data:
for j in i:
# print(j.strip())
data1.append(j.strip())
# break
data1 = set(data1)
#
# print(data1)
data1.remove("")
# print(data1)
data1 = list(data1)
rows, cols = nfdata.shape
for i in data1:
nfdata[d] = np.zeros(rows)
nfdata.head()
nfdata.shape
for index in nfdata.index:
for d in data1:
if d in nfdata.loc[index, "genres"]:
nfdata.loc[index, d] = 1
data1.extend(["genres"])
nfdata.corr
ratingcols = ["bad", "fair", "good", "excellent"]
rows, cols = nfdata.shape
# print(rows)
# print(cols)
for d in ratingcols:
nfdata[d] = np.zeros(rows)
# for index in newnf.index:
# if
nfdata.head()
newnf = list()
newnf = nfdata.drop(columns=["genres"])
newnf.head()
newnf[["tmdb_popularity", "tmdb_score"]].describe()
# Bad - 0-2
# Fair - 3-5
# Good - 6-8
# Exellent - 9-10
ratingcols = ["Bad", "Fair", "Good", "Excellent"]
rows, col = newnf.shape
for d in ratingcols:
newnf[d] = np.zeros(rows)
for index in newnf.index:
if 0 <= newnf.loc[index, "tmdb_score"] < 3:
newnf.loc[index, "Bad"] = 1.0
elif 3 <= newnf.loc[index, "tmdb_score"] < 6:
newnf.loc[index, "Fair"] = 1.0
elif 6 <= newnf.loc[index, "tmdb_score"] < 9:
newnf.loc[index, "Good"] = 1.0
elif 9 <= newnf.loc[index, "tmdb_score"] <= 10:
newnf.loc[index, "Excellent"] = 1.0
newnf[ratingcols].describe()
print(newnf[newnf["Good"] == 1].agg(np.size))
print()
print(newnf.agg(np.size).head())
# newnf[newnf['Fair'] ==1].describe()
newnf.agg(np.size).describe()
# actgen.remove('genres')
print(actgen)
print(ratingcols)
# newnf[ratingcols + actgen].corr()
year = []
all = []
# nfdata[nfdata['release_year']].dropna
# rel_year=nfdata[nfdata['release_year']]
# print(rel_year)
for i in nfdata["release_year"]:
year.append(i)
year = list(set(year))
# print(year)
data = []
for i in year:
movie = nfdata[nfdata["type"] == "MOVIE"][nfdata["release_year"] == i].agg(np.size)[
"id"
]
show = nfdata[nfdata["type"] == "SHOW"][nfdata["release_year"] == i].agg(np.size)[
"id"
]
data.append({"release_year": i, "movie": movie, "show": show})
newdata = pd.DataFrame(data[-10:])
newdata
# print(show_yr,movie,show)
# break
# if(nfdata['release_year']==i):
# for i in year:
# if(nfdata[nfdata['release_year'] == i].groupby('i').agg(np.size)):
# print(nfdata[nfdata['release_year'] == i].groupby('i').agg(np.size))
# movie=nfdata[nfdata['type']=='MOVIE'].groupby('release_year').agg(np.size)
# show=nfdata[nfdata['type']=='SHOW'].groupby('release_year').agg(np.size)
# print(show['id'])
# print(movie['id'])
# print(show['id'])
for i in nfdata["release_year"]:
year.append(i)
year = list(set(year))
# print(year)
country = []
for i in nfdata[actdata]:
country.append(i)
print(country)
# data = []
# for i in year:
# movie=nfdata[nfdata['type']=='MOVIE'][nfdata['release_year'] == i].agg(np.size)['id']
# show=nfdata[nfdata['type']=='SHOW'][nfdata['release_year'] == i].agg(np.size)['id']
# data.append({'release_year' : i,'movie': movie,'show' : show})
# newdata = pd.DataFrame(data[-10:])
# newdata['tot']=newdata['movie']+newdata['show']
# newdata
nfdata
country = list()
new_data = list()
for i in nfdata.production_countries:
country.append(i.replace("]", "").replace("[", "").replace("'", "").split(","))
for i in country:
for j in i:
new_data.append(j.strip())
new_data = set(new_data)
new_data = list(new_data)
new_data.remove("")
print(new_data)
print(len(new_data))
nfdata
# data = nfdata[['type', 'release_year']]
# data = data.value_counts().to_frame()
# # data.reset_index(level=[0,1], inplace=True)
# data
# for i in new_data:
# print(i)
spdata = nfdata[["production_countries", "release_year"]]
spdata = spdata.value_counts().to_frame()
# data.reset_index(level=[0,2], inplace=True)
row, col = spdata.shape
# print(row,col)
spdata["tot"] = np.zeros(row)
xmv = nfdata[nfdata["type"] == "MOVIE"]
xtv = nfdata[nfdata["type"] == "SHOW"]
# tot=xmv+xtv
# spdata['tot'] = xmv+xtv
# spdata['tot']
spdata = nfdata[["production_countries", "release_year"]]
spdata = spdata.value_counts().to_frame()
spdata
data = nfdata[["type", "release_year"]]
data = data.value_counts().to_frame()
data.reset_index(level=[0, 1], inplace=True)
data = data.rename(columns={0: "count"})
data = data.sort_values(by=["release_year"], ascending=False)
xmv = data[data["type"] == "MOVIE"][:10]
xtv = data[data["type"] == "SHOW"][:10]
print(xmv)
print(xtv)
fig, ax = plt.subplots()
ax.plot(xmv["release_year"], xmv["count"], label="Movie")
ax.plot(xtv["release_year"], xtv["count"], label="TV Show")
ax.legend()
plt.show()
xmv = data[data["type"] == "MOVIE"].count()
xtv = data[data["type"] == "SHOW"].count()
print(xmv)
print(xtv)
print(tot)
for i in year:
nfdata[nfdata["release_year"] == i]
# print(nfdata[nfdata['release_year']==i])
# if(nfdata['release_year']==2021):
show = nfdata[nfdata["type"] == "SHOW"].count()
movie = nfdata[nfdata["type"] == "MOVIE"].count()
plt.style.use("_mpl-gallery-nogrid")
x = [show["id"], movie["id"]]
y = ["show", "movie"]
fig, ax = plt.subplots()
plt.style.use("classic")
plt.title("show content Vs movies content")
ax.pie(x, labels=y, autopct="%1.2f%%")
|
# ## Spotify EDA & Feature Engineering
# ### Table of Contents
# * [Datasets merge](#section-one)
# * [Exploratory Data Analysis](#section-two)
# - [Data cleaning](#subsection-one)
# - [Numerical features](#subsection-two)
# - [Categorical features](#subsection-three)
# * [Feature Engineering](#section-three)
# - [Missing data](#subsection-three-one)
# - [Date features](#subsection-three-two)
# - [Encoding](#subsection-three-three)
# * [Conclusion](#section-four)
# ### Description
# Spotify is the world's largest music streaming service provider with over 489 million active users. As a result, the amount of data
# generated by Spotify is literally tremendous. Every day new and new tracks, podcasts and albums come on service board. More than 100 million tracks
# of any type are available on demand. That's why it gives a great opportunity to start our journey with manipulating and analyzing data. This is my first Kaggle notebook where I try to dig into data and gain some insights about music industry. And this is what I aim at by performing this Exploratory Data Analysis.
# ### Main goal
# As said earlier, the main goal is to analyze Spotify dataset at different angles. In fact, this analysis is comprised of 2 datasets
# merged together, namely: tracks and artists. In many analyses, there's only one dataset with all available data.
# Here, I join two various datasets and when it's done I proceed with appropriate analysis. This project is divided into 3 parts:
# - Datasets merge
# - EDA
# - Feature Engineering
# With regard to Feature Engineering I just touched the topic performing some general engineering. Depending on prospective
# ML model to evaluate FE should be adjusted accordingly.
# Wrapping up, in this notebook, I will be carrying out EDA with elements of FE which should be practical and easy to understand.
# This analysis has a potential to grow and expand, especially in machine learning direction.
# ### 1. Datasets merge
# We start off with merging two datasets available publicly: tracks.csv and artists.csv.
# Detailed columns description is available on Spotify website:
# - Audio tracks - https://developer.spotify.com/documentation/web-api/reference/get-audio-features
# - Artists - https://developer.spotify.com/documentation/web-api/reference/get-an-artist
# Let's catch a glimpse at both datasets separately:
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
df_tracks = pd.read_csv("/kaggle/input/spotify-datasets/tracks.csv")
df_artists = pd.read_csv("/kaggle/input/spotify-datasets/artists.csv")
df_tracks.info()
# We have 20 features in dataset. Most of them are numeric ones.
df_tracks.head()
df_artists.info()
df_artists.head()
df_tracks.shape
df_artists.shape
# Artists dataset definitely outnumbers tracks dataset in terms of number of rows.
# Before we join the tables we should check if there are any duplicates:
dupl_tracks = df_tracks.duplicated().value_counts()
dupl_artists = df_artists.duplicated().value_counts()
check_dupl = f"duplicated tracks: {dupl_tracks} \n duplicated artists: {dupl_artists}"
print(check_dupl)
# There are no duplicates, we can join datasets. We want to merge tables by id and id_artists columns. We need one id_artists
# value in a row, unfortunately there are multiple ids in an array. First, we need to clean id_artists column by removing
# square brackets:
df_tracks["id_artists"] = (
df_tracks["id_artists"]
.str.replace("\['", "")
.str.replace("']", "")
.str.replace("'", "")
)
# Let's find all records with more than one artist id. We return all rows where comma's index is not equal -1: if comma appears in text then its index will be different than -1:
df_tracks[df_tracks["id_artists"].str.find(",") != -1]
# Before merge let's analyze genres column. This column will be added to the final, merged dataset:
unique_genres = df_artists["genres"].nunique()
print(f"Number of unique genres: {unique_genres}")
# Since the number of unique values is really big let's focus on values which account for the majority of the column:
df_artists["genres"].value_counts().sort_values(ascending=False)
# As we can see most of the column values are empty arrays []. Let's check the ratio:
num = df_artists["genres"].value_counts()["[]"]
perc = df_artists["genres"].value_counts(normalize=True)["[]"]
print(
f"Total number of empty entries is {num}, which accounts for {perc:.2%} of the column."
)
# We don't want empty arrays in the column and we don't want to show them in the final, merged dataset either.
# Replace them with NaN values. At the same time extract first element from each array in a row - it's gonna be our top genre.
# Genres column is a string of lists so first we need to convert them to list object.
# After that we will be able to extract first element of an array to be our final genre. We use ast.literal_eval to perform this
# operation:
import ast
def get_top_element(x):
if x == "[]":
return np.nan
return ast.literal_eval(x)[0]
df_artists["top_genre"] = df_artists["genres"].apply(get_top_element)
# Now display our top genres without NaN values:
df_artists[df_artists["top_genre"].notna()]
# It's quite obvious we are going to merge artists dataset by id column. We should make sure that there are no missings,
# duplicates or other unexpected entries:
miss = df_artists["id"].isnull().sum()
dupl = df_artists["id"].duplicated().sum()
print(f"There are {miss} missing values and {dupl} duplicated values.")
# Usually, so that to check if there are any unexpected values we coerce non-numerical values to NaN, then count all nulls to check
# if there're any. In this case we just count values and order in descending order to check if there are no 'strange' entries
# (like single signs e.g. !, @, $ , numbers only, short strings etc.):
df_artists["id"].value_counts().sort_values(ascending=False)
# Since id column is crucial in merging operation we want to double check if length of each id is the same throughout the whole
# column:
# id length = 22
df_artists["id"].str.len().sort_values(ascending=False)
# Make sure there is no id less that 22 signs.
df_artists[
df_artists["id"].str.len().sort_values(ascending=False).apply(lambda x: x < 22)
]
# Our id column is ready to join with df_tracks dataset, all ids are of the same length. Now, let's keep going with cleaning
# our df_tracks dataset. We want to extract first 'id' occurence in id_artists column - that's because we need to join both
# tables by one, unique id, not 2 or more id-s. At the same time we want to extract first occurance in artists column - top
# artist. We need to keep both column - artists and top_artist and keep them in final dataset:
df_tracks["id_artists"] = df_tracks["id_artists"].str.split(",").str[0]
df_tracks["top_artist"] = df_tracks["artists"].apply(get_top_element)
# Before we merge both tables let's check for missing values or anomalies in key column used to merge datsets - id_artists:
nulls_artists = df_tracks["id_artists"].isnull().sum()
dupl_artists = df_tracks["id_artists"].duplicated().sum()
# Duplicated artists are ok, we accept them. In fact we need to check if there are no duplicates in id column:
dupl_id = df_tracks["id"].duplicated().sum()
null_dupl = f"There are {nulls_artists} null values and {dupl_artists} duplicated values in id_artists. Also {dupl_id} duplicated id."
print(null_dupl)
# All rows are unique. Our dataset is ready for merge.
# It's time to join both tables. Now check if merge keys are unique in a dataset on a right side by adding validate parameter:
cols_artists = ["id", "followers", "popularity", "top_genre"]
df_merged = df_tracks.merge(
df_artists[cols_artists],
how="left",
left_on="id_artists",
right_on="id",
validate="many_to_one",
)
df_merged
# Let's compare the length of both datasets - merged and df_tracks. We used left join so we kept all rows from df_tracks. That's
# why we compare if their length is the same:
merged_check = df_merged.shape[0]
tracks_check = df_tracks.shape[0]
merged_txt = f"df_merged length: {merged_check}, df_tracks length: {tracks_check}"
print(merged_txt)
# Either dataset has the same length. As a final check let's compare 'joined' columns - id_artists and id_y.
# We are particularly interested if there are any not-joined artists from df_artists:
nulls_id_y = df_merged["id_y"].isnull().sum()
print(f'There are {nulls_id_y} in "id_y" column.')
# As we can see there are 12415 NaN values - there are no such artists in df_artists. Let's find these rows:
df_merged[df_merged["id_y"].isnull()]
# We notice that even though an artist exists in df_tracks (top_artists column) there is no such artists id in df_artists.
# For now, we have properly joined datasets. We can perform EDA.
# ### 2. Exploratory Data Analysis
# #### 2.1 Data cleaning
# Here we conduct some preliminary analysis based on new, merged dataset. First, let's take a look on a first few rows:
df_merged.sample(5).T
# Based on the sample above we can assume that cleaning and data transforming is needed on the whole dataset.
# This is what we are going to do now:
df_merged.shape
# There are 25 columns. We need to drop some of them later.
df_merged.info()
# It's most likely that some columns would require data type conversion. Summary below shows very little missing values
# which we will inspect later in more details.
# Remove unnecessary columns. Here we have some duplicated columns resulting from merging of 2 datasets. Let's create a new
# dataset (we will keep artists column since it comprises a list of all artists performing a song. Perhaps we will need it later):
cols_to_drop = ["id_x", "id_artists", "id_y"]
df = df_merged.drop(cols_to_drop, axis=1)
df.head()
# Rename columns to make them more clear:
dict_col_names = {
"name": "track_name",
"popularity_x": "track_popularity",
"popularity_y": "artist_popularity",
"followers": "artist_followers",
}
df.rename(columns=dict_col_names, inplace=True)
df.head()
# We want to rearrange columns as to make dataframe more concise and logic. We also keep 'artists' column for future reference:
cols = df.columns.tolist()
new_cols = (
cols[:1]
+ cols[18:19]
+ cols[4:5]
+ cols[21:22]
+ cols[5:6]
+ cols[1:2]
+ cols[20:21]
+ cols[19:20]
+ cols[2:3]
+ cols[3:4]
+ cols[6:18]
)
df = df[new_cols]
df.head()
# Check if there are any duplicates. Let's see how it looks on the whole dataset. We notice that duplicated rows have
# same track_name and top_artist but they vary in some of the remaining columns (e.g various realese_date, duration,
# followers etc.). As an example we can provide 'King - Acoustic' and their song 'Years & Years' - we have 7 same rows
# which differ by release_date and track_popularity.
# We are not interested in dropping all such rows because there will still be duplicated track_name together with top_artist.
# We need to drop rows based on those 2 columns solely:
dupl_rows = df.duplicated().sum()
dupl_rows_proper = df.duplicated(subset=["track_name", "top_artist"]).value_counts()
print(
f"There are {dupl_rows} duplicated rows in the whole dataset. \n There are {dupl_rows_proper} duplicated rows in 2 cols."
)
df[(df.track_name == "King - Acoustic") & (df.top_artist == "Years & Years")]
# Some of the duplicated rows differ by followers or other columns. I think we should take the row where release_date is the
# newest, most up-to-date. Next, return rest of the columns along with it. We then keep this duplicated value ('first' argument). So now we need to sort the datframe by track_name, top_artist and release_date where the last one will be sorted in descending order (by date, from newest to oldest). Then, if we keep first duplicate we will keep the newest one and drop oldest ones. Before that we need to convert release_date column to date type:
df["release_date"] = pd.to_datetime(df["release_date"])
df.sort_values(
["track_name", "top_artist", "release_date"],
ascending=[True, True, False],
inplace=True,
)
df.drop_duplicates(subset=["track_name", "top_artist"], inplace=True)
# Now let's check for duplicates in crucial columns - track_name & top_artist. See we have only False values which is what we
# expected:
df.duplicated(subset=["track_name", "top_artist"]).value_counts()
# Just for making sure let's check how it all looks based on the example provided before. As we can see there is only one row
# with the latest release_date. We can assume that all remaining columns consist of the latest data as well:
df[(df.track_name == "King - Acoustic") & (df.top_artist == "Years & Years")]
# Now let's focus on column types and think if we should cast any of them:
df.dtypes
df.head()
# We have changed release_date to datetime64 so it's ok already. All column types seem allright but we could check if
# float64 could be converted to int as we don't need precision on artist_popularity or artist_followers columns.
# Let's check first if there is any decimal precision different than zero in those columns. That would mean that precision
# exists and might be relevant (e.g. artist_popularity 53.0 but could be 53.8. That's why we check for precision != 0):
def check_for_zero_precision(x):
if x % 1 == 0:
return 0
else:
return 1
cols = ["artist_popularity", "artist_followers"]
arr = []
for col in cols:
decims = df[col].apply(check_for_zero_precision)
count_non_zero = decims[decims != 0].count()
print(f"{col}: {count_non_zero}")
# As we can see there is a decimal precision other than zero in both columns so we won't convert it to int.
# We could also change numeric types to lighter ones (like int32) but we don't and won't have performance issues so keep it as it is.
# As a final step we need to change duration from ms to seconds by dividing the column by 1000. Column type will change
# automatically to float64. We also want to change column name to duration_sec:
df["duration_ms"] = df["duration_ms"] / 1000
df.rename(columns={"duration_ms": "duration_sec"}, inplace=True)
# Let's check if we have any missing data:
count_all_nulls = df.isnull().sum().sort_values(ascending=False).loc[lambda x: x > 0]
missings_ratio = count_all_nulls / df.shape[0] * 100
missings_ratio
# We notice that most of the missing values are concentrated in top_genre column. Loc helps us filter non-zero values
# (we keep only these where any missings are detected).
# Now, let's take a deeper look into missing values in given columns. We will use missingno library for that. Let's start with
# matrix method which shows how missing values are distributed throughout the whole dataframe. We have 3 columns
# with missing data:
import missingno as msn
msn.matrix(df)
# Nullity by column:
msn.bar(df)
# Heatmap measuring nullity correlation comes in handy when we need to assess relationship between features in terms of nullity.
# It's clear that artist_popularity and artists_followers are correlated 100% positively - data presence and absence are exactly
# the same in both columns. Top_genre affects artist_popularity and artists_followers at the same level - 40% of data entries
# are present in these both columns. However, from logical point of view lack of genre has no real impact on lack of artist
# or track data:
msn.heatmap(df)
# We will make decision on what to do with missing values later on, in feature engineering part of this analysis.
# #### 2.2 EDA - numerical features
# #### Univariate
# Quick glimpse on dataframe data types:
df.info()
# Let's explore every single column:
df.describe()
# Let's visualize features using histograms with density curves. Axes.flat simplify indexing. Zip joins lists and creates
# one list. We need to use histplot instead of displot here beacuse only histplot is axes-level function, displot is not
# (figure-level). We need to draw a random sample from our datset since it's too large to generate so many plots.
# Distributions vary a lot for each feature:
import statsmodels.api as sm
import scipy.stats as stats
sns.set_style("darkgrid")
num_cols = df.select_dtypes(include=np.number)
cols = num_cols.columns.tolist()
df_sample = df.sample(n=100000)
fig, axes = plt.subplots(4, 5, figsize=(15, 12), layout="constrained")
for col, axs in zip(cols, axes.flat):
sns.histplot(data=df_sample[col], kde=True, stat="density", ax=axs, alpha=0.4)
plt.show()
# Let's take a look at kurtosis and skewness.
# Kurtosis - the greater the value (> 0) the more slim the distribution (and more outliers) and more values are similar
# to each other. It also means that a lot of values revolve around mean value. The lower the value the flatter the
# distribution is - it also means that values don't revolve around mean and are more spread (sparse).
# The highest kurtosis (tallest distribution): duration_sec, artist_followers, explicit, energy or speechiness.
# Values are similar here. For the first glance normal distibution (between 1 and -1) is in tempo, artist_popularity and
# danceability columns:
df.agg("kurtosis").transpose().sort_values()
# Skewness - the greater the value (> 0) the more right-skewed the distribution and more data is concentrated in lower values
# Right-skewed: duration_sec, artist_followers, explicit etc. Quite symmetric (close to normal distribution, skew ~ 0):
# energy, danceability, valence, artist_popularity, tempo:
df.agg("skew").transpose().sort_values()
# Let's now analyze numeric columns by specific data type: int
cols = df.select_dtypes(np.int64)
cols_names = cols.columns.tolist()
fig, axes = plt.subplots(3, 2, figsize=(15, 12), layout="constrained")
for col, axs in zip(cols_names, axes.flat):
sns.countplot(x=df[col], order=df[col].value_counts().index, ax=axs)
plt.show()
# Excluding 'track_popularity' rest of int columns may be treated as categorical features since they have a few unique values.
# We can convert those features into categorical ones. Since the list of possible values is pretty small and because there are
# a lot of repetitions, we could make this faster by using a category data type (except for track_popularity). Let's get back to
# our original dataframe and data types. There are some object columns as potential candidates for categoricals. However, there
# are a bunch of possible values for each of object features (e.g. artists) so we won't make it categorical:
cat_cols = ["explicit", "key", "mode", "time_signature"]
for col in cat_cols:
df[col] = pd.Categorical(df[col])
# Let's take a look at data types after casting:
df.info()
# #### Bivariate
# *Numerical-Numerical*
# We use pairplot to visualize relationship between numerical features. It helps in assessing where the correlation occurs.
# This plot also shows us which features are good candidates for categoricals (vertical/horizontal dot-like lines on the plot).
# We need to add alpha parameter to show areas where data is focused on the plot - it also allows us to assess correlation.
# The more data the lower alpha level. We can see correlation between some features but in this case it would be better to
# show real numbers:
df_sample = df.sample(n=100000)
sns.pairplot(df_sample, corner=True, plot_kws={"alpha": 0.02})
plt.show()
# Create correlation matrix to express relationship between numerical features in numbers:
pd.set_option("display.max_rows", 500)
corr_matrix = df.corr()
corr_matrix.unstack().sort_values()
# We notice that the highest negative correlation is between:
# - energy and acousticness (-0.72)
# - loudness and acousticness (-0.52)
# Medium negative correlation between:
# - instrumentalness and loudness (-0.33)
# - acousticness and track_popularity (-0.37)
# The highest positive correlation is between:
# - energy and loudness (0.76)
# - artist_popularity and track_popularity (0.53)
# - danceability and valence (0.53)
# Medium positive correlation between:
# - artist_followers and artist_popularity (0.42)
# - energy and valence (0.37)
# - loudness and track_popularity (0.33)
# - energy and track_popularity (0.3)
# We can focus on those particular (and strong) relations and move analysis forward in this direction. Those features have the
# biggest impact on our analysis.
# Let's use heatmap to display correlation matrix:
plt.figure(figsize=(12, 8))
sns.heatmap(corr_matrix, annot=True, linewidths=0.8)
plt.show()
# We calculated correlations between features above and noticed that some of them are more correlated than the others.
# We can consider them as most significant in terms of further analysis and prospective ML modelling. Let's focus on some of
# those key fetaures and try to fit distribution to them:
from fitter import Fitter
from scipy import stats
distr = [
"cauchy",
"chi2",
"expon",
"exponpow",
"gamma",
"beta",
"lognorm",
"logistic",
"norm",
"powerlaw",
"rayleigh",
"uniform",
]
# Energy - based on SSE beta distribution is the best one:
dist_fitter = Fitter(df["energy"], distributions=distr)
dist_fitter.fit()
dist_fitter.summary()
# Loudness - based on SSE logistic distribution is the best one:
dist_fitter = Fitter(df["loudness"], distributions=distr)
dist_fitter.fit()
dist_fitter.summary()
# Danceability - based on SSE beta distribution is the best one:
dist_fitter = Fitter(df["danceability"], distributions=distr)
dist_fitter.fit()
dist_fitter.summary()
# *Numerical-Categorical*
# We have a few categorical features which can be displayed on boxplots. Explicit refers to explicit content (0/1 - no explicit/
# explicit content). We can measure it in terms of other numeric columns:
num_cols = df.select_dtypes(np.number)
cols_names = num_cols.columns.tolist()
fig, axes = plt.subplots(4, 4, figsize=(12, 10), layout="constrained")
for col, axs in zip(cols_names, axes.flat):
sns.boxplot(x=df[col], y=df["explicit"], ax=axs)
plt.show()
# As we can see above the greatest difference between groups is for features: track_popularity (seems the more
# explicit content the more popular track, same for artist_popularity), danceability, energy (kind of). It's also quite obvious
# that for accousticness the explicit content is rather marginal and that for speechiness there is greater range of explicit box since
# we have words in a song.
# Now we can do a preliminary feature engineering and extract year from date column. That will be our new feature
# for which we create box plots. Let's make it categorical as well:
df["song_year"] = df["release_date"].dt.year
df["song_year"] = pd.Categorical(df["song_year"])
df.info()
# Let's visualize how most significant features have changed over the more than a century:
cols_names = ["duration_sec", "acousticness", "loudness", "energy"]
fig, axes = plt.subplots(4, 1, figsize=(15, 10), layout="constrained")
for col, axs in zip(cols_names, axes.flat):
sns.lineplot(x=df["song_year"], y=df[col], ax=axs)
plt.show()
# We can clearly see that song duration has increased greatly, same for energy and loudness. The last two had been on increase
# since 60's Accousticness has been on decrease since 50's. This is most likely the result of a new wave of rock'n roll music,
# louder and definitely more energetic. Now let's go into more details and analyze all numerical features using boxplots:
num_cols = df.select_dtypes(np.number)
cols_names = num_cols.columns.tolist()
fig, axes = plt.subplots(13, 1, figsize=(20, 30), layout="constrained")
for col, axs in zip(cols_names, axes.flat):
sns.boxplot(x=df["song_year"], y=df[col], ax=axs)
axs.tick_params(axis="x", rotation=90)
plt.show()
# These plots are very interesting. We can clearly see how particular features have changed over years. Track popularity has increased
# which is quite obvious from today's perspective. Energy is also on increase, it has changed remarkably in '60s. Speechiness is also really high in the
# beginning of a century, then sharply decreases. Accousticness - in the beginning of a century most songs were acoustic and it
# has changed a lot over the years. Up until '60s most songs were instrumental, then it changed abruptly. It has a lot to do
# with rock 'n roll revolution.
# Boxplots also show how values range for particular years. Take acousticness and see that in the
# beginning of XX century almost all songs were very acoustic. It had changed with rock'n roll revolution - tracks started varying
# a lot. It worked the other way round regarding instrumentalness - the same period of time and extremely low feature level
# comparing to 1900-1960. And of course there are a lot of outliers for each feature.
# #### 2.3 EDA - categorical features
# #### Univariate
# Now choose and analyze categorical data:
df_cat = df.select_dtypes(exclude=np.number)
df_cat.describe()
# Now check our table with most frequent category in each year. We can see that we have an array in year 1993. Why? Because
# we have 2 categories with the same frequency (126). That's why they are bind together into one array. We can replace one of
# those values to another one, then we would have one more top_genre (127, not 126) in given category. After that operation we will have one category per year:
most_freq_cat = (
df.groupby("song_year")["top_genre"].agg(pd.Series.mode).to_frame().reset_index()
)
most_freq_cat.head(15)
# For clarity - these 2 categories are most frequent:
df[df["song_year"] == 1993]["top_genre"].value_counts().sort_values(ascending=False)
# Choose random row with 'hoerspiel' as top_genre and replace this field with 'classic bollywood'. Then check if it's replaced correctly:
df.loc[432390, ["top_genre"]] = ["classic bollywood"]
df.loc[432390]
# Let's make sure if 'classic bollywood' outnumbers 'hoerspiel' genre. As we can see now we have 127 'classic bollywood' rows vs. 124 rows of 'hoerspiel' and this is what we wanted to achieve:
df[df["song_year"] == 1993]["top_genre"].value_counts().sort_values(ascending=False)
# Let's analyze top_genre. We see that adult_standards, album_rock, hoerspiel and classical genres were the most popular
# over the years. We see that these 4 were popular for more than 55% of all years (more than 60 years):
most_freq_cat = (
df.groupby("song_year")["top_genre"].agg(pd.Series.mode).to_frame().reset_index()
)
plt.figure(figsize=(15, 7), layout="constrained")
plt.xticks(rotation=45)
sns.countplot(
data=most_freq_cat,
x="top_genre",
order=most_freq_cat["top_genre"].value_counts().index,
)
most_freq_cat.groupby("top_genre")["top_genre"].value_counts().sort_values(
ascending=False
) / len(most_freq_cat)
# #### Multivariate
# We have previosuly detected relationships between some numeric variables and shown how they are distributed throughout the years
# on boxplots. Energy and accousticness are interesting cases, we see how they correlate with other features and how the have
# changed over the years. Let's see how the features' mean values are distributed in reference to two categorical features - song_year and top_genre:
aggregates = df.groupby("song_year").agg(
genres=("top_genre", pd.Series.mode),
energy=("energy", np.mean),
acousticness=("acousticness", np.mean),
)
pivot_energy = pd.pivot_table(
data=aggregates, index="song_year", columns="genres", values="energy"
)
plt.figure(figsize=(20, 20))
sns.heatmap(pivot_energy, annot=True, cmap="coolwarm")
plt.title("Song year and genre by energy")
plt.show()
# This heatmap shows us many interesting things. First of all it shows us graphically how top genres were distributed over the
# years - in which years they were popular and when they stopped to be popular. Secondly, how the core numeric feature (energy)
# has changed in years of given genre popularity. Thirdly, heatmap shows us general tendency and patterns in music industry
# over the years.
# For energy we can see that adult standards, classical, tango and rock music genres were popular for many following years.
# For example, tango was popular 9 years in a row. Also pop music was very popular at the time (blank years are filled with
# very similar k-pop and c-pop). Looking at mentioned genres we can also see how energy changed during genre's popularity
# period. For example adult standards increased from 0.29 to 0.45, rock from 0.45 to 0.54. Overall impression is that energy has
# increased dramatically from the beginning of a century. We also see the huge popularity of rock music in late 60's and 70's
# which is easily explainable - this is the period in time when rock'n roll started becoming very popular type of music.
# Now let's do the same for accousticness:
pivot_acc = pd.pivot_table(
data=aggregates, index="song_year", columns="genres", values="acousticness"
)
plt.figure(figsize=(20, 20))
sns.heatmap(pivot_acc, annot=True, cmap="coolwarm")
plt.title("Song year and genre by acousticness")
plt.show()
# We can notice reverse tendency comparing to energy. Accousticness gets lower and lower as the time passes. And even within
# genres those values decresed significantly. For example rock music is much less acoustic in 1979 than it was in 1969.
# ### 3. Feature Engineering
# #### 3.1 Missing data
# *Categorical features*
# Let's check again if we have any missing data in cat vars. We have a lot of top_genre values missing. As we calculated before
# this number accounts for 11% of all column's data:
cat_cols = df.select_dtypes(include=["object", "category"])
cat_cols.isnull().sum().sort_values().loc[lambda x: x > 0]
df[df["top_genre"].isnull()]
# Here we create generic function which fills in all missings. We group data by year and, depending if it's numeric or object
# column, we fill in NaN with mean or most frequent value:
def mapNullsWithValues(data, col_key, col_vals):
to_df = pd.DataFrame()
if (data[col_vals].dtype == object) | (data[col_vals].dtype == "category"):
to_df = (
df.groupby(col_key)[col_vals].agg(pd.Series.mode).to_frame().reset_index()
)
elif (data[col_vals].dtype == "float64") | (data[col_vals].dtype == "int64"):
to_df = df.groupby(col_key)[col_vals].median().to_frame().reset_index()
col_keys = to_df.iloc[:, 0].to_numpy()
col_values = to_df.iloc[:, 1].to_numpy()
dictionary = dict(zip(col_keys, col_values))
data[col_vals].fillna(data[col_key].map(dictionary), inplace=True)
# Now we use that function to fill in missing data with the most popular genre in each year. Genres differ a lot given a decade or even a year:
mapNullsWithValues(df, "song_year", "top_genre")
# Now check if all nulls are filled:
df["top_genre"].isnull().sum()
# It's ok, we have 0 missing values.
# *Numerical features*
# Let's check again if we have any missing data in num vars. We have 2 features which we can fill in with data. It's 2% of
# the whole column. Let's check mean and median for those columns:
cat_cols = df.select_dtypes(include=np.number)
cat_cols.isnull().sum().sort_values().loc[lambda x: x > 0] / len(df)
# Now, use our function again to fill in all missing values with median for each abovementioned column grouped by year.
# Let's check if we have no missing values after that:
mapNullsWithValues(df, "song_year", "artist_popularity")
mapNullsWithValues(df, "song_year", "artist_followers")
nan_pop = df["artist_popularity"].isnull().sum()
nan_foll = df["artist_followers"].isnull().sum()
print(
f"There are {nan_pop} missing values in 'artist_popularity' column and {nan_foll} in 'artist_followers' column."
)
# As we can see we have filled all NaNs correctly.
# #### 3.2 Date features
# We have extracted a year from release_date, now we can add additional column - a decade. Having this we can also analyze
# how numerical features have changed over decades:
year_int = df["song_year"].astype(int)
mod_year = year_int % 10
df["decade"] = year_int - mod_year
df.head(10)
# Having this extra column we can analyze features from another perspective. As an example - let's analyze valence and explicit.
# Valence describes the positiveness of a track. The higher valence the sound is more positive, the lower valence the track is
# more negative. Explicit is obvious - 1 means explict, 0 - non explicit:
plt.figure(figsize=(9, 6))
sns.barplot(data=df, x="decade", y="valence")
plt.show()
# As we noticed the most positive tracks come from the first decades of XX century and also from years 1960-2000.
# Lower valence can be spotted in 40's and 50's which is most likely related to II World War and depression/sadness
# that came along with it.
# #### 3.3 Encoding
# Depending on what's the purpose of ML model we can create different categorical features based on existing features. W don't
# have ordinal features. On the other hand we have lots of unique values in top_genre column and also in song_year column
# and we can use OrdinalEncoder here. Using this library let's make label encoding. We won't make hot-encoding, we have too
# many categories in columns (e.g. top_genre) so we would genrate too many new columns:
import category_encoders as ce
encoder = ce.OrdinalEncoder()
df[["top_genre", "song_year"]] = encoder.fit_transform(df[["top_genre", "song_year"]])
df.head(10).transpose()
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.