script
stringlengths 113
767k
|
|---|
# for basic mathematics operation
import numpy as np
import pandas as pd
from pandas import plotting
# for visualizations
import matplotlib.pyplot as plt
import seaborn as sns
plt.style.use("fivethirtyeight")
# for interactive visualizations
import plotly.offline as py
from plotly.offline import init_notebook_mode, iplot
import plotly.graph_objs as go
from plotly import tools
init_notebook_mode(connected=True)
import plotly.figure_factory as ff
# for path
import os
for dirname, _, filenames in os.walk("\kaggle\input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# print(os.listdir('kaggle\input'))
# Connecting to the house sales data present in the kaggle input directory.
os.chdir(r"/kaggle/input/telco-customer-churn")
data = pd.read_csv("WA_Fn-UseC_-Telco-Customer-Churn.csv")
data_1 = ff.create_table(data.head())
data.head(10)
desc = ff.create_table(data.describe())
py.iplot(desc)
data.isnull().any().any()
plt.rcParams["figure.figsize"] = (15, 10)
plotting.andrews_curves(data.drop("tenure", axis=1), "gender")
plt.title("Andrew Curves for Gender", fontsize=20)
plt.show()
import warnings
warnings.filterwarnings("ignore")
plt.rcParams["figure.figsize"] = (18, 8)
plt.subplot(1, 2, 1)
sns.set(style="whitegrid")
sns.distplot(data["Annual Income (k$)"])
plt.title("Distribution of Annual Income", fontsize=20)
plt.xlabel("Range of Annual Income")
plt.ylabel("Count")
plt.subplot(1, 2, 2)
sns.set(style="whitegrid")
sns.distplot(data["Age"], color="red")
plt.title("Distribution of Age", fontsize=20)
plt.xlabel("Range of Age")
plt.ylabel("Count")
plt.show()
labels = ["Female", "Male"]
size = data["gender"].value_counts()
colors = ["lightgreen", "orange"]
explode = [0, 0.1]
plt.rcParams["figure.figsize"] = (9, 9)
plt.pie(
size, colors=colors, explode=explode, labels=labels, shadow=True, autopct="%.2f%%"
)
plt.title("gender", fontsize=20)
plt.axis("off")
plt.legend()
plt.show()
plt.rcParams["figure.figsize"] = (15, 8)
sns.countplot(data["tenure"], palette="hsv")
plt.title("Distribution of Age", fontsize=20)
plt.show()
plt.rcParams["figure.figsize"] = (20, 8)
sns.countplot(data["MonthlyCharges"], palette="rainbow")
plt.title("Distribution of Annual Income", fontsize=20)
plt.show()
plt.rcParams["figure.figsize"] = (20, 8)
sns.countplot(data["TotalCharges"], palette="copper")
plt.title("Distribution of Spending Score", fontsize=20)
plt.show()
plt.rcParams["figure.figsize"] = (15, 8)
sns.heatmap(data.corr(), annot=True)
plt.title("Heatmap for the Data", fontsize=20)
plt.show()
# Gender vs Spendscore
plt.rcParams["figure.figsize"] = (18, 7)
sns.boxenplot(data["gender"], data["TotalCharges"], palette="Blues")
plt.title("Gender vs TotalCharges", fontsize=20)
plt.show()
plt.rcParams["figure.figsize"] = (18, 7)
sns.violinplot(data["gender"], data["MonthlyCharges"], palette="rainbow")
plt.title("Gender vs Spending Score", fontsize=20)
plt.show()
plt.rcParams["figure.figsize"] = (18, 7)
sns.stripplot(data["gender"], data["tenure"], palette="Purples", size=10)
plt.title("Gender vs Spending Score", fontsize=20)
plt.show()
x = data["MonthlyCharges"]
y = data["tenure"]
z = data["TotalCharges"]
sns.lineplot(x, y, color="blue")
sns.lineplot(x, z, color="pink")
plt.title("Annual Income vs Age and Spending Score", fontsize=20)
plt.show()
x = data.iloc[:, [3, 4]].values
# let's check the shape of x
print(x.shape)
from sklearn.cluster import KMeans
wcss = []
for i in range(1, 11):
km = KMeans(n_clusters=i, init="k-means++", max_iter=300, n_init=10, random_state=0)
km.fit(x)
wcss.append(km.inertia_)
plt.plot(range(1, 11), wcss)
plt.title("The Elbow Method", fontsize=20)
plt.xlabel("No. of Clusters")
plt.ylabel("wcss")
plt.show()
km = KMeans(n_clusters=5, init="k-means++", max_iter=300, n_init=10, random_state=0)
y_means = km.fit_predict(x)
plt.scatter(x[y_means == 0, 0], x[y_means == 0, 1], s=100, c="pink", label="miser")
plt.scatter(x[y_means == 1, 0], x[y_means == 1, 1], s=100, c="yellow", label="general")
plt.scatter(x[y_means == 2, 0], x[y_means == 2, 1], s=100, c="cyan", label="target")
plt.scatter(
x[y_means == 3, 0], x[y_means == 3, 1], s=100, c="magenta", label="spendthrift"
)
plt.scatter(x[y_means == 4, 0], x[y_means == 4, 1], s=100, c="orange", label="careful")
plt.scatter(
km.cluster_centers_[:, 0],
km.cluster_centers_[:, 1],
s=200,
c="blue",
label="centeroid",
)
plt.style.use("fivethirtyeight")
plt.title("K Means Clustering", fontsize=20)
plt.xlabel("TotalCharges")
plt.ylabel("tenure")
plt.legend()
plt.grid()
plt.show()
import scipy.cluster.hierarchy as sch
dendrogram = sch.dendrogram(sch.linkage(x, method="ward"))
plt.title("Dendrogam", fontsize=20)
plt.xlabel("Customers")
plt.ylabel("Ecuclidean Distance")
plt.show()
from sklearn.cluster import AgglomerativeClustering
hc = AgglomerativeClustering(n_clusters=5, affinity="euclidean", linkage="ward")
y_hc = hc.fit_predict(x)
plt.scatter(x[y_hc == 0, 0], x[y_hc == 0, 1], s=100, c="pink", label="miser")
plt.scatter(x[y_hc == 1, 0], x[y_hc == 1, 1], s=100, c="yellow", label="general")
plt.scatter(x[y_hc == 2, 0], x[y_hc == 2, 1], s=100, c="cyan", label="target")
plt.scatter(x[y_hc == 3, 0], x[y_hc == 3, 1], s=100, c="magenta", label="spendthrift")
plt.scatter(x[y_hc == 4, 0], x[y_hc == 4, 1], s=100, c="orange", label="careful")
plt.scatter(
km.cluster_centers_[:, 0],
km.cluster_centers_[:, 1],
s=50,
c="blue",
label="centeroid",
)
plt.style.use("fivethirtyeight")
plt.title("Hierarchial Clustering", fontsize=20)
plt.xlabel("TotalCharges")
plt.ylabel("tenure")
plt.legend()
plt.grid()
plt.show()
x = data.iloc[:, [2, 4]].values
x.shape
from sklearn.cluster import KMeans
wcss = []
for i in range(1, 11):
kmeans = KMeans(
n_clusters=i, init="k-means++", max_iter=300, n_init=10, random_state=0
)
kmeans.fit(x)
wcss.append(kmeans.inertia_)
plt.rcParams["figure.figsize"] = (15, 5)
plt.plot(range(1, 11), wcss)
plt.title("K-Means Clustering(The Elbow Method)", fontsize=20)
plt.xlabel("TotalCharges")
plt.ylabel("tenure")
plt.grid()
plt.show()
kmeans = KMeans(n_clusters=4, init="k-means++", max_iter=300, n_init=10, random_state=0)
ymeans = kmeans.fit_predict(x)
plt.rcParams["figure.figsize"] = (10, 10)
plt.title("Cluster of Ages", fontsize=30)
plt.scatter(
x[ymeans == 0, 0], x[ymeans == 0, 1], s=100, c="pink", label="Usual Customers"
)
plt.scatter(
x[ymeans == 1, 0], x[ymeans == 1, 1], s=100, c="orange", label="Priority Customers"
)
plt.scatter(
x[ymeans == 2, 0],
x[ymeans == 2, 1],
s=100,
c="lightgreen",
label="Target Customers(Young)",
)
plt.scatter(
x[ymeans == 3, 0], x[ymeans == 3, 1], s=100, c="red", label="Target Customers(Old)"
)
plt.scatter(
kmeans.cluster_centers_[:, 0], kmeans.cluster_centers_[:, 1], s=50, c="black"
)
plt.style.use("fivethirtyeight")
plt.xlabel("tenure")
plt.ylabel("TotalCharges")
plt.legend()
plt.grid()
plt.show()
x = data[["tenure", "TotalCharges", "MonthlyCharges"]].values
km = KMeans(n_clusters=5, init="k-means++", max_iter=300, n_init=10, random_state=0)
km.fit(x)
labels = km.labels_
centroids = km.cluster_centers_
data["labels"] = labels
trace1 = go.Scatter3d(
x=data["Age"],
y=data["Spending Score (1-100)"],
z=data["Annual Income (k$)"],
mode="markers",
marker=dict(
color=data["labels"],
size=10,
line=dict(color=data["labels"], width=12),
opacity=0.8,
),
)
df = [trace1]
layout = go.Layout(
title="Character vs Gender vs Alive or not",
margin=dict(l=0, r=0, b=0, t=0),
scene=dict(
xaxis=dict(title="tenure"),
yaxis=dict(title="MonthlyCharges"),
zaxis=dict(title="TotalCharges"),
),
)
fig = go.Figure(data=df, layout=layout)
py.iplot(fig)
|
import numpy as np # linear algebra
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from ast import literal_eval
import json
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Cleaning
# Loading csv file into a Pandas DataFrame
movies = pd.read_csv("../input/the-movies-dataset/movies_metadata.csv")
creadits = pd.read_csv("../input/the-movies-dataset/credits.csv")
keywords = pd.read_csv("../input/the-movies-dataset/keywords.csv")
movies.head()
movies.info()
# Through using the info() function, Dataframe Movies has 45466 rows and 24 columns corresponding to 24 features
# ### Check the number of null values present in each feature
movies.isnull().sum()
# Because in columns like belongs_to_collection,homepage,tagline there are too many null values, so we will proceed to delete them
# ## Handling missing value
movies = movies.drop(
[
"belongs_to_collection",
"homepage",
"tagline",
],
axis=1,
)
movies[movies["title"].isnull()]
# So there are 6 lines in the title column containing null values,We will remove them.
movies.dropna(subset=["title"], inplace=True)
# Similar to column 'production_companies' and 'spoken_language'
movies.dropna(subset=["production_companies"], axis="rows", inplace=True)
movies.dropna(subset=["spoken_languages"], axis="rows", inplace=True)
# ### converts json list to list of inputs (from the label specified with 'wanted' parameter)
# We can define this pattern with a regex, compile it and use it to find all words with that pattern in each column by applying a lambda function. If this was confusing, you can jump to the final result to see the cleaned data and compare it to the output above to see what we are trying to accomplish.
import re
regex = re.compile(r": '(.*?)'")
movies["genres"] = movies["genres"].apply(lambda x: str(x))
movies["genres"] = movies["genres"].apply(lambda x: ", ".join(regex.findall(x)))
# *Extracting relevant substring from production_companies, production_countries and spoken_languages as done with genres above.*
movies["production_companies"] = movies["production_companies"].apply(lambda x: str(x))
movies["production_countries"] = movies["production_countries"].apply(lambda x: str(x))
movies["spoken_languages"] = movies["spoken_languages"].apply(lambda x: str(x))
movies["production_companies"] = movies["production_companies"].apply(
lambda x: ", ".join(regex.findall(x))
)
movies["production_countries"] = movies["production_countries"].apply(
lambda x: ", ".join(regex.findall(x))
)
movies["spoken_languages"] = movies["spoken_languages"].apply(
lambda x: ", ".join(regex.findall(x))
)
# Remove wrong value data in column id
id_errors = []
for index, row in movies.iterrows():
row["id"] = row["id"].split("-")
if len(row["id"]) > 1:
id_errors.append(index)
movies = movies.drop(id_errors)
movies = movies.reset_index(drop=True)
# ### Converting the date to datetime and using year to create a new column
movies["release_date"] = pd.to_datetime(movies["release_date"], errors="coerce")
movies["year"] = movies["release_date"].dt.year
movies.dropna(inplace=True)
|
import tensorflow as tf
import tensorflow_datasets as tfds
from keras.preprocessing.text import Tokenizer
from tensorflow.keras import layers
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
import scipy
from sklearn.compose import make_column_transformer
from sklearn.preprocessing import MinMaxScaler, OneHotEncoder, LabelEncoder
from sklearn.model_selection import train_test_split
import random
import os
import io
import zipfile
from tensorflow.keras.preprocessing.image import ImageDataGenerator
import urllib.request
import cv2
# Setup the train and test directories
train_dir = "/kaggle/input/3classdataset/train"
test_dir = "/kaggle/input/3classdataset/test"
# Use ImageDataGenerator to create Train and Test Datsets with data augmentation built in
train_gen = tf.keras.preprocessing.image.ImageDataGenerator(
rescale=1.0 / 255,
shear_range=0.2, # shear the image
zoom_range=0.2, # zoom into the image
width_shift_range=0.2, # shift the image width ways
height_shift_range=0.2, # shift the image height ways
horizontal_flip=True,
) # flip the image on the horizontal axis
test_gen = tf.keras.preprocessing.image.ImageDataGenerator(rescale=1.0 / 255)
training_data = train_gen.flow_from_directory(
train_dir,
batch_size=32, # number of images to process at a time
target_size=(224, 224), # convert all images to be 224 x 224
class_mode="categorical", # type of problem we're working
shuffle=True,
seed=42,
)
testing_data = test_gen.flow_from_directory(
test_dir,
batch_size=32, # number of images to process at a time
target_size=(224, 224), # convert all images to be 224 x 224
class_mode="categorical", # type of problem we're working
shuffle=True,
seed=42,
)
# Create callbacks
early_stopping = tf.keras.callbacks.EarlyStopping(
monitor="val_loss", patience=3, restore_best_weights=True
)
lr_callback = tf.keras.callbacks.ReduceLROnPlateau(
monitor="val_loss", factor=0.5, patience=2, verbose=0
)
# 1. Create base model with tf.keras.applications
base_model = tf.keras.applications.efficientnet_v2.EfficientNetV2M(
include_top=False, include_preprocessing=False
)
# 2. Freeze the base model (so the undelying pre-trained patterns aren't updated during training )
base_model.trainable = True
# 3. Create inputs into our model
inputs = tf.keras.layers.Input(shape=(224, 224, 3), name="input_layer", dtype="float32")
# 4. If using a model like ResNet50V2, add this to speed up convergence, remove for EfficientNet
# x = tf.keras.layers.experimental.preprocessing.Rescaling(1./255)(inputs)
# 5. Pass the inputs to the base_model
x = base_model(inputs)
x = tf.keras.layers.GlobalAveragePooling2D(name="global_average_pooling_layer")(x)
# 7. Create the output activation layer
outputs = tf.keras.layers.Dense(3, activation="softmax", name="output_layer")(x)
model = tf.keras.Model(inputs=inputs, outputs=outputs)
model.summary()
model.compile(
loss="categorical_crossentropy",
optimizer=tf.keras.optimizers.Adam(),
metrics=["accuracy"],
)
model_history = model.fit(
training_data,
validation_data=(testing_data),
epochs=10,
callbacks=[early_stopping, lr_callback],
)
model.evaluate(training_data)
model.save_weights("weights.h5")
model.evaluate(training_data)
# 1. Create base model with tf.keras.applications
base_model = tf.keras.applications.efficientnet_v2.EfficientNetV2M(
include_top=False, include_preprocessing=False
)
# 2. Freeze the base model (so the undelying pre-trained patterns aren't updated during training )
base_model.trainable = True
# 3. Create inputs into our model
inputs = tf.keras.layers.Input(shape=(224, 224, 3), name="input_layer", dtype="float32")
# 4. If using a model like ResNet50V2, add this to speed up convergence, remove for EfficientNet
# x = tf.keras.layers.experimental.preprocessing.Rescaling(1./255)(inputs)
# 5. Pass the inputs to the base_model
x = base_model(inputs)
x = tf.keras.layers.GlobalAveragePooling2D(name="global_average_pooling_layer")(x)
# 7. Create the output activation layer
outputs = tf.keras.layers.Dense(1, activation="sigmoid", name="output_layer")(x)
model2 = tf.keras.Model(inputs=inputs, outputs=outputs)
model2.compile(
loss="binary_crossentropy",
optimizer=tf.keras.optimizers.Adam(),
metrics=["accuracy"],
)
model2.load_weights("/kaggle/working/weights.h5")
model2.evaluate(testing_data)
IMAGE_SIZE = 224
BATCH_SIZE = 64
datagen = tf.keras.preprocessing.image.ImageDataGenerator(
rescale=1.0 / 255, validation_split=0.2
)
train_generator = datagen.flow_from_directory(
train_dir,
target_size=(IMAGE_SIZE, IMAGE_SIZE),
batch_size=BATCH_SIZE,
subset="training",
)
val_generator = datagen.flow_from_directory(
test_dir,
target_size=(IMAGE_SIZE, IMAGE_SIZE),
batch_size=BATCH_SIZE,
subset="validation",
)
batch_images, batch_labels = next(val_generator)
logits = model(batch_images)
truth = np.argmax(batch_labels, axis=1)
prediction = np.argmax(logits, axis=1)
keras_accuracy = tf.keras.metrics.Accuracy()
keras_accuracy(prediction, truth)
print("Raw model accuracy: {:.3%}".format(keras_accuracy.result()))
prediction
model.save("model_3_classes.h5")
|
# # Project 4
# We're going to use Polars instead of Pandas to perform the transformations we did on projects 1 & 2. We are then going to compare how long they take to perform similar tasks.
import pandas as pd
import polars as pl
import numpy as np
import time
from datetime import timedelta
pl.Config.set_fmt_str_lengths(200)
data1_path = "/kaggle/input/project-1-data/data"
sampled = False
path_suffix = "" if not sampled else "_sampled"
data2_path = "/kaggle/input/project-2-data/project_2_data"
# # Loading data
# Load the transactions data from csv using each library.
# ## Polars
start_time = time.monotonic()
polar_transactions = pl.read_csv(
f"{data1_path}/transactions_data{path_suffix}.csv"
).with_columns(
pl.col("date").str.strptime(pl.Date, fmt="%Y-%m-%d %H:%M:%S", strict=False)
)
end_time = time.monotonic()
polars_loading_time = timedelta(seconds=end_time - start_time)
print(polars_loading_time)
# ## Pandas
start_time = time.monotonic()
pandas_transactions = pd.read_csv(f"{data1_path}/transactions_data{path_suffix}.csv")
pandas_transactions["date"] = pd.to_datetime(pandas_transactions["date"])
end_time = time.monotonic()
pandas_loading_time = timedelta(seconds=end_time - start_time)
print(pandas_loading_time)
# # Processing data
# Process the transactions data such that we have sales information even when no sales were performed, for all combination of dates and ids. We then compare with a prepared sales file to check if the processing was done correct on both occasions.
# ## Polars
start_time = time.monotonic()
polar_data = (
polar_transactions.with_columns(pl.lit(1).alias("sales"))
.groupby(list(polar_transactions.columns))
.sum()
)
max_date = polar_data.with_columns(pl.col("date")).max()["date"][0]
min_date = polar_data.with_columns(pl.col("date")).min()["date"][0]
date_range = pl.date_range(min_date, max_date, "1d")
polars_MultiIndex = pl.DataFrame({"date": date_range}).join(
polar_data.select(pl.col("id").unique()), how="cross"
)
polar_data = (
polar_data.join(polars_MultiIndex, on=["date", "id"], how="outer")
.with_columns(pl.col("sales").fill_null(0))
.sort("id", "date")
)
filled_data = (
polar_data.lazy()
.groupby("id")
.agg(
pl.col("date"),
pl.col("item_id").forward_fill(),
pl.col("dept_id").forward_fill(),
pl.col("cat_id").forward_fill(),
pl.col("store_id").forward_fill(),
pl.col("state_id").forward_fill(),
)
.explode(["date", "item_id", "dept_id", "cat_id", "store_id", "state_id"])
.sort(["id", "date"])
)
polar_data = (
polar_data.lazy()
.join(filled_data, on=["date", "id"], how="outer")
.with_columns(
pl.col("item_id").fill_null(pl.col("item_id_right")),
pl.col("dept_id").fill_null(pl.col("dept_id_right")),
pl.col("cat_id").fill_null(pl.col("cat_id_right")),
pl.col("store_id").fill_null(pl.col("store_id_right")),
pl.col("state_id").fill_null(pl.col("state_id_right")),
)
.select(polar_data.columns)
# We can remove the initial data without sales we would not need the cumsum trick.
.drop_nulls()
.collect()
)
end_time = time.monotonic()
polars_processing_time = timedelta(seconds=end_time - start_time)
print(polars_processing_time)
# ## Pandas
start_time = time.monotonic()
pandas_data = (
pandas_transactions.assign(sales=1, date=lambda df: df.date.dt.floor("d"))
.groupby(list(pandas_transactions.columns))
.sum()
.reset_index()
.set_index(["date", "id"])
.sort_index()
)
min_date = pandas_data.index.get_level_values("date").min()
max_date = pandas_data.index.get_level_values("date").max()
dates_to_select = pd.date_range(min_date, max_date, freq="1D")
ids = pandas_data.index.get_level_values("id").unique()
index_to_select = pd.MultiIndex.from_product(
[dates_to_select, ids], names=["date", "id"]
)
pandas_data = pandas_data.reindex(index_to_select)
pandas_data = pandas_data.loc[pandas_data.groupby("id").sales.cumsum() > 0]
data_ids = pandas_data.index.get_level_values("id").str.split("_")
data_id = pd.DataFrame(data_ids.tolist())
data_id.index = pandas_data.index
pandas_data.sales = pandas_data.sales.astype(np.int64)
item_id = data_id[0] + "_" + data_id[1] + "_" + data_id[2]
pandas_data["item_id"].update(item_id)
dept_id = data_id[0] + "_" + data_id[1]
pandas_data["dept_id"].update(dept_id)
cat_id = data_id[0]
pandas_data["cat_id"].update(cat_id)
store_id = data_id[3] + "_" + data_id[4]
pandas_data["store_id"].update(store_id)
state_id = data_id[3]
pandas_data["state_id"].update(state_id)
end_time = time.monotonic()
pandas_processing_time = timedelta(seconds=end_time - start_time)
print(pandas_processing_time)
# ## Comparison
# We compare each dataframe with the expected results. They should all match.
#
def test_sales_eq(data):
assert (
pd.read_csv(
f"{data1_path}/sales_data{path_suffix}.csv", usecols=["date", "id", "sales"]
)
# pd.read_parquet(f"{data2_path}/sales_data.parquet")[['sales']].reset_index()
.assign(date=lambda df: pd.to_datetime(df.date))
.merge(data, on=["date", "id"], how="left", suffixes=("_actual", "_predicted"))
.fillna({"sales_actual": 0, "sales_predicted": 0})
.assign(sales_error=lambda df: (df.sales_actual - df.sales_predicted).abs())
.sales_error.sum()
< 1e-6
), "Your version of sales does not match the original sales data."
print("Comparing POLARS")
test_sales_eq(polar_data.to_pandas())
print(" - matched.")
print("Comparing PANDAS")
test_sales_eq(pandas_data)
print(" - matched.")
# # Feature Engineering
# We now have to add date features, calendar and price features to the dataframes.
# ## Polars
start_time = time.monotonic()
pl_calendar = (
pl.read_parquet(f"{data2_path}/calendar.parquet")
.with_columns(pl.col("date").cast(pl.Date))
.lazy()
)
pl_prices = (
pl.read_parquet(f"{data2_path}/prices.parquet")
.with_columns(pl.col("date").cast(pl.Date))
.lazy()
)
pl_data = (
polar_data.lazy()
.drop(["state_id"])
.join(pl_prices, on=["date", "store_id", "item_id"], how="left")
.join(pl_calendar, on=["date"], how="left")
.with_columns(pl.col("date").dt.weekday().alias("day_of_week"))
.with_columns(pl.col("date").dt.day().alias("day_of_month"))
.with_columns(pl.col("date").dt.week().alias("week"))
.with_columns(pl.col("date").dt.month().alias("month"))
.with_columns(pl.col("date").dt.quarter().alias("quarter"))
.with_columns(pl.col("date").dt.year().alias("year"))
.with_columns(
pl.col("item_id", "dept_id", "cat_id", "store_id").cast(pl.Categorical)
)
.rename({"date": "ds", "id": "unique_id", "sales": "y"})
.collect()
)
event_cols = ["event_name_1", "event_name_2", "event_type_1", "event_type_2"]
cat_feats = ["unique_id", "item_id", "dept_id", "cat_id"]
cat_feats.extend(event_cols)
enc_cat_feats = [f"{feat}_enc" for feat in cat_feats]
encoding_feats = [
pl.UInt32 if col in ["unique_id", "item_id", "dept_id", "store_id"] else pl.UInt8
for col in cat_feats
]
# encoding since OrdinalEncoder does not seem to work with polars
for col, enc_col, encoding in zip(cat_feats, enc_cat_feats, encoding_feats):
pl_data = pl_data.with_columns(
pl.col(col).cast(pl.Categorical).to_physical().cast(encoding).alias(enc_col)
)
numeric_features = ["sell_price"]
reference_cols = ["unique_id", "ds", "y"]
# add features to this list if you want to use them
features = reference_cols + enc_cat_feats + numeric_features
pl_data = pl_data[features]
end_time = time.monotonic()
polars_feature_time = timedelta(seconds=end_time - start_time)
print(polars_feature_time)
# ## Pandas
from sklearn.preprocessing import OrdinalEncoder
start_time = time.monotonic()
pd_calendar = pd.read_parquet(f"{data2_path}/calendar.parquet")
pd_prices = pd.read_parquet(f"{data2_path}/prices.parquet")
pd_data = (
pandas_data.reset_index()
.drop(["state_id"], axis=1)
.merge(pd_prices, on=["date", "store_id", "item_id"], how="left")
.merge(pd_calendar, on=["date"], how="left")
.set_index("date")
.assign(
day_of_week=lambda d: d.index.dayofweek,
day_of_month=lambda d: d.index.day,
week=lambda d: d.index.isocalendar().week,
month=lambda d: d.index.month,
year=lambda d: d.index.year,
quarter=lambda d: d.index.quarter,
item_id=lambda d: d.item_id.astype("category"),
dept_id=lambda d: d.dept_id.astype("category"),
cat_id=lambda d: d.cat_id.astype("category"),
store_id=lambda d: d.store_id.astype("category"),
event_name_1=lambda d: d.event_name_1.astype("category"),
event_name_2=lambda d: d.event_name_2.astype("category"),
event_type_1=lambda d: d.event_type_1.astype("category"),
event_type_2=lambda d: d.event_type_2.astype("category"),
)
.reset_index()
.rename(columns={"id": "unique_id", "date": "ds", "sales": "y"})
)
# label encode categorical features
event_cols = ["event_name_1", "event_name_2", "event_type_1", "event_type_2"]
cat_feats = ["unique_id", "item_id", "dept_id", "cat_id"]
cat_feats.extend(event_cols)
enc_cat_feats = [f"{feat}_enc" for feat in cat_feats]
encoder = OrdinalEncoder()
pd_data[enc_cat_feats] = encoder.fit_transform(pd_data[cat_feats])
numeric_feature = ["sell_price"]
reference_cols = ["unique_id", "ds", "y"]
# add features to this list if you want to use them
features = reference_cols + enc_cat_feats + numeric_features
pd_data = pd_data[features]
end_time = time.monotonic()
pandas_feature_time = timedelta(seconds=end_time - start_time)
print(pandas_feature_time)
# # Results
# We now check the times for each of the parts to see results.
metrics = pd.DataFrame(
{
"loading": [
polars_loading_time.total_seconds(),
pandas_loading_time.total_seconds(),
],
"processing": [
polars_processing_time.total_seconds(),
pandas_processing_time.total_seconds(),
],
"feature_eng": [
polars_feature_time.total_seconds(),
pandas_feature_time.total_seconds(),
],
},
index=["polars", "pandas"],
)
metrics["total"] = metrics["loading"] + metrics["processing"] + metrics["feature_eng"]
# metrics["total"].apply(lambda x: x.total_seconds()) / sum( metrics["total"].apply(lambda x: x.total_seconds()))
metrics.loc["ratio", "loading"] = (
metrics.loc["pandas", "loading"] / metrics.loc["polars", "loading"]
)
metrics.loc["ratio", "processing"] = (
metrics.loc["pandas", "processing"] / metrics.loc["polars", "processing"]
)
metrics.loc["ratio", "feature_eng"] = (
metrics.loc["pandas", "feature_eng"] / metrics.loc["polars", "feature_eng"]
)
metrics.loc["ratio", "total"] = (
metrics.loc["pandas", "total"] / metrics.loc["polars", "total"]
)
metrics
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv("/kaggle/input/bitcoin-and-fear-and-greed/dataset.csv")
df
# # **Bitcoin Fear and greed days split overall**
# Define the colors for each bar
colors = ["red", "blue", "green", "purple", "orange"]
bar_chart = df["Value_Classification"].hist()
bar_chart.set_title("Bitcoin fear and greed index 1 Feb to 31 Mar 2023")
bar_chart.set_ylabel("Number of days")
print(df["Value_Classification"].value_counts())
# # **Bitcoin Fear and greed per month**
df["Date"] = pd.to_datetime(df["Date"])
# Extract the short name of the month from the date column
df["month"] = df["date"].dt.strftime("%b")
df
df.pivot_table(
index=[["month", "Value_Classification"]],
values="Value_Classification",
aggfunc="count",
)
|
# ### Build your own Neural Network from scratch using only Numpy
# Deep Learning has become a popular topic in recent times. It involves emulating the neural structure of the human brain through a network of nodes called a **Neural Network**. While our brain's neurons have physical components like nucleus, dendrites, and synapses, Neural Network neurons are interconnected and have weights and biases assigned to them.
# A neural network typically consists of an input layer, an output layer, and one or more hidden layers. In conventional neural networks, all nodes in these layers are interconnected to form a dense network. However, there are cases where certain nodes in the network are not connected to others, which are referred to as **Sparse Neural Networks**. InceptionNet models for image classification use Sparse Neural Networks. The following figure illustrates the structure of a neural network.
# 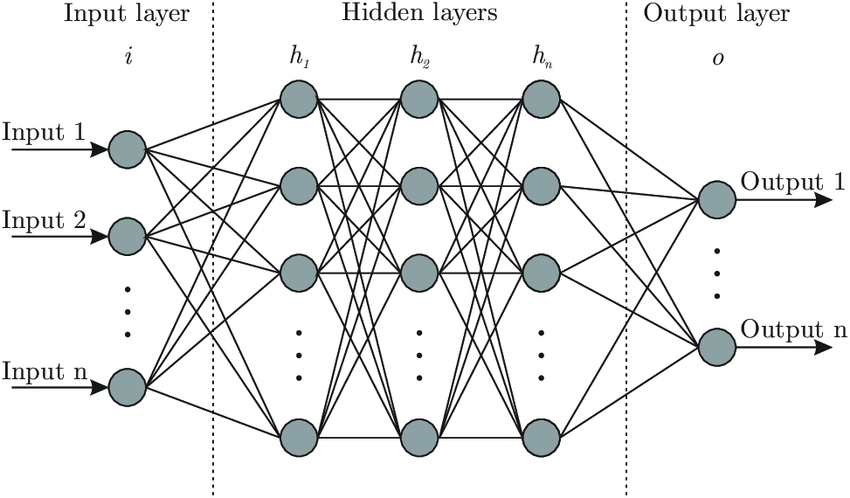
# The neurons will be activated or fired, which is the input will be passed through the neuron to the next layer. Each neuron inside the neural network will have a linear function using weights and biases like the following equation. The input will be transformed using the function below and produce a new output to the next layer.
# 
# You can see at the end there will be something called as the ***Activation function***. An activation function is a function which decides wheather the neuron needs to be activated or not. Some people say, a neuron without an activation function is just a linear regression model. There are several activation functions called Sigmoid, Softmax, Tanh, ReLu and many more. We'll see about activation functions in detail later.
# let's start to build a neural network from scratch. We'll get into code without much further ado. Let's start by importing numpy library for linear algebra functions.
#
import numpy as np # linear algebra
# Numpy is a python library which can be used to implement linear algebra functions. So for creating a neural network we need the base for building a nueral network, neurons. We'll create a class neuron to implement the weights and biases.
class Neuron:
def __init__(self, weights, bias):
self.weights = weights
self.bias = bias
def feedforward(self, x):
return np.dot(weights, x) + bias
weights = np.array([0, 1])
bias = 1
n = Neuron(weights, bias)
n.feedforward([1, 1])
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
import tensorflow as tf
from keras.preprocessing.image import ImageDataGenerator
from keras.utils import to_categorical
from keras.models import Sequential
from keras.layers import Conv2D, MaxPooling2D, BatchNormalization, Dropout
from keras.layers import Dense, Flatten
import cv2
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
import os
print(os.listdir("../input"))
category = ["cat", "dog"]
EPOCHS = 50
IMGSIZE = 128
BATCH_SIZE = 32
STOPPING_PATIENCE = 15
VERBOSE = 1
MODEL_NAME = "cnn_50epochs_imgsize128"
OPTIMIZER = "adam"
TRAINING_DIR = "/kaggle/working/train"
TEST_DIR = "/kaggle/working/test"
for img in os.listdir(TRAINING_DIR)[7890:]:
img_path = os.path.join(TRAINING_DIR, img)
img_arr = cv2.imread(img_path, cv2.IMREAD_GRAYSCALE)
img_arr = cv2.resize(img_arr, (IMGSIZE, IMGSIZE))
plt.imshow(img_arr, cmap="gray")
plt.title(img.split(".")[0])
break
def create_train_data(path):
X = []
y = []
for img in os.listdir(path):
if img == os.listdir(path)[7889]:
continue
img_path = os.path.join(path, img)
img_arr = cv2.imread(img_path, cv2.IMREAD_GRAYSCALE)
img_arr = cv2.resize(img_arr, (IMGSIZE, IMGSIZE))
img_arr = img_arr / 255.0
cat = np.where(img.split(".")[0] == "dog", 1, 0)
X.append(img_arr)
y.append(cat)
X = np.array(X).reshape(-1, IMGSIZE, IMGSIZE, 1)
y = np.array(y)
return X, y
X, y = create_train_data(TRAINING_DIR)
print(f"features shape {X.shape}.\nlabel shape {y.shape}.")
y = to_categorical(y, 2)
print(f"features shape {X.shape}.\nlabel shape {y.shape}.")
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=1 / 3)
X_train.shape
y_train.shape
model = Sequential()
model.add(Conv2D(32, kernel_size=(3, 3), activation="relu", input_shape=(28, 28, 1)))
model.add(Conv2D(64, (3, 3), activation="relu"))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Flatten())
model.add(Dense(128, activation="relu"))
model.add(Dropout(0.5))
model.add(Dense(10, activation="softmax"))
model.compile(loss="categorical_crossentropy", optimizer="Adam", metrics=["accuracy"])
model.fit(X_train, y_train, epochs=10, validation_data=(X_test, y_test))
|
from keras.layers import Input, Conv2D, Lambda, merge, Dense, Flatten, MaxPooling2D
from keras.models import Model, Sequential
from keras.regularizers import l2
from keras import backend as K
from keras.optimizers import SGD, Adam
from keras.losses import binary_crossentropy
import sklearn.metrics as sm
import numpy.random as rng
import numpy as np
import pandas as pd
import os
import pickle
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.utils import shuffle
import os
from keras.datasets import mnist
def loop(p, q, low, high):
m = []
cat = {}
n = 0
for j in range(low, high):
r = np.where(q == j)
cat[str(j)] = [n, None]
t = np.random.choice(r[0], 20)
c = []
for i in t:
c.append(np.resize(p[i], (35, 35)))
n += 1
cat[str(j)][1] = n - 1
m.append(np.stack(c))
return m, cat
def createdataset(low, high, t):
(a, b), (f, g) = mnist.load_data()
m = []
cat = {}
if t == 0:
m, cat = loop(a, b, low, high)
else:
m, cat = loop(f, g, low, high)
m = np.stack(m)
return m, cat
train, cat_train = createdataset(0, 6, 0)
test, cat_test = createdataset(6, 10, 0)
print(train.shape)
print(test.shape)
print(cat_train.keys())
print(cat_test.keys())
def W_init(shape, name=None, dtype=None):
"""Initialize weights as in paper"""
values = rng.normal(loc=0, scale=1e-2, size=shape)
return K.variable(values, name=name)
# //TODO: figure out how to initialize layer biases in keras.
def b_init(shape, name=None, dtype=None):
"""Initialize bias as in paper"""
values = rng.normal(loc=0.5, scale=1e-2, size=shape)
return K.variable(values, name=name)
nclass, nexample, row, col = train.shape
input_shape = (row, col, 1)
left_input = Input(input_shape)
right_input = Input(input_shape)
# build convnet to use in each siamese 'leg'
convnet = Sequential()
convnet.add(
Conv2D(
64,
(10, 10),
activation="relu",
input_shape=input_shape,
kernel_initializer=W_init,
kernel_regularizer=l2(2e-4),
)
)
convnet.add(MaxPooling2D())
# convnet.add(Conv2D(128,(7,7),activation='relu',kernel_regularizer=l2(2e-4),kernel_initializer=W_init, bias_initializer=b_init))
# convnet.add(MaxPooling2D())
convnet.add(
Conv2D(
128,
(4, 4),
activation="relu",
kernel_initializer=W_init,
kernel_regularizer=l2(2e-4),
bias_initializer=b_init,
)
)
# convnet.add(MaxPooling2D())
# convnet.add(Conv2D(256,(4,4),activation='relu',kernel_initializer=W_init,kernel_regularizer=l2(2e-4),bias_initializer=b_init))
convnet.add(Flatten())
convnet.add(
Dense(
4096,
activation="sigmoid",
kernel_regularizer=l2(1e-3),
kernel_initializer=W_init,
bias_initializer=b_init,
)
)
# call the convnet Sequential model on each of the input tensors so params will be shared
encoded_l = convnet(left_input)
encoded_r = convnet(right_input)
# layer to merge two encoded inputs with the l1 distance between them
L1_layer = Lambda(lambda tensors: K.abs(tensors[0] - tensors[1]))
# call this layer on list of two input tensors.
L1_distance = L1_layer([encoded_l, encoded_r])
prediction = Dense(1, activation="sigmoid")(L1_distance)
siamese_net = Model(inputs=[left_input, right_input], outputs=prediction)
optimizer = Adam(0.00006)
# //TODO: get layerwise learning rates and momentum annealing scheme described in paperworking
siamese_net.compile(loss="binary_crossentropy", optimizer=optimizer)
siamese_net.count_params()
# nclass,nexample,row,col = train.shape
# input_shape = (row,col, 1)
# left_input = Input(input_shape)
# right_input = Input(input_shape)
# #build convnet to use in each siamese 'leg'
# convnet = Sequential()
# convnet.add(Conv2D(64,(10,10),activation='relu',input_shape=input_shape,kernel_regularizer=l2(2e-4)))
# convnet.add(MaxPooling2D())
# # convnet.add(Conv2D(128,(7,7),activation='relu',kernel_regularizer=l2(2e-4),kernel_initializer=W_init, bias_initializer=b_init))
# # convnet.add(MaxPooling2D())
# convnet.add(Conv2D(128,(4,4),activation='relu',kernel_regularizer=l2(2e-4)))
# # convnet.add(MaxPooling2D())
# # convnet.add(Conv2D(256,(4,4),activation='relu',kernel_initializer=W_init,kernel_regularizer=l2(2e-4),bias_initializer=b_init))
# convnet.add(Flatten())
# convnet.add(Dense(4096,activation="sigmoid",kernel_regularizer=l2(1e-3)))
# #call the convnet Sequential model on each of the input tensors so params will be shared
# encoded_l = convnet(left_input)
# encoded_r = convnet(right_input)
# #layer to merge two encoded inputs with the l1 distance between them
# L1_layer = Lambda(lambda tensors:K.abs(tensors[0] - tensors[1]))
# #call this layer on list of two input tensors.
# L1_distance = L1_layer([encoded_l, encoded_r])
# prediction = Dense(1,activation='sigmoid')(L1_distance)
# siamese_net = Model(inputs=[left_input,right_input],outputs=prediction)
# siamese_net.load_weights("/kaggle/input/mnistweights/weights")
# optimizer = Adam(0.00006)
# #//TODO: get layerwise learning rates and momentum annealing scheme described in paperworking
# siamese_net.compile(loss="binary_crossentropy",optimizer=optimizer)
# siamese_net.count_params()
class Siamese_Loader:
"""For loading batches and testing tasks to a siamese net"""
def __init__(self):
self.data = {"train": train, "val": test}
self.categories = {"train": cat_train, "val": cat_test}
def get_batch(self, batch_size, s="train"):
"""Create batch of n pairs, half same class, half different class"""
X = self.data[s]
n_classes, n_examples, w, h = X.shape
# randomly sample several classes to use in the batch
categories = rng.choice(n_classes, size=(batch_size,), replace=False)
# initialize 2 empty arrays for the input image batch
pairs = [np.zeros((batch_size, h, w, 1)) for i in range(2)]
# initialize vector for the targets, and make one half of it '1's, so 2nd half of batch has same class
targets = np.zeros((batch_size,))
targets[batch_size // 2 :] = 1
for i in range(batch_size):
category = categories[i]
idx_1 = rng.randint(0, n_examples)
pairs[0][i, :, :, :] = X[category, idx_1].reshape(w, h, 1)
idx_2 = rng.randint(0, n_examples)
# pick images of same class for 1st half, different for 2nd
if i >= batch_size // 2:
category_2 = category
else:
# add a random number to the category modulo n classes to ensure 2nd image has
# ..different category
category_2 = (category + rng.randint(1, n_classes)) % n_classes
pairs[1][i, :, :, :] = X[category_2, idx_2].reshape(w, h, 1)
return pairs, targets
def make_oneshot_task(self, N, s="val", language=None):
"""Create pairs of test image, support set for testing N way one-shot learning."""
X = self.data[s]
n_classes, n_examples, w, h = X.shape
indices = rng.randint(0, n_examples, size=(N,))
# if language is not None:
# low, high = self.categories[s][language]
# if N > high - low:
# raise ValueError("This language ({}) has less than {} letters".format(language, N))
# categories = rng.choice(range(low,high),size=(N,),replace=False)
# else:#if no language specified just pick a bunch of random letters
# categories = rng.choice(range(n_classes),size=(N,),replace=False)
categories = rng.choice(range(n_classes), size=(N,), replace=True)
true_category = categories[0]
ex1, ex2 = rng.choice(n_examples, replace=False, size=(2,))
test_image = np.asarray([X[true_category, ex1, :, :]] * N).reshape(N, w, h, 1)
support_set = X[categories, indices, :, :]
support_set[0, :, :] = X[true_category, ex2]
support_set = support_set.reshape(N, w, h, 1)
targets = np.zeros((N,))
targets[np.where(categories == true_category)] = 1
targets, test_image, support_set = shuffle(targets, test_image, support_set)
pairs = [test_image, support_set]
return pairs, targets
def test_oneshot(self, model, N, k, i, s="val", verbose=0):
"""Test average N way oneshot learning accuracy of a siamese neural net over k one-shot tasks"""
n_correct = 0
if verbose:
print("iteration no.{}".format(i))
print(
"Evaluating model on {} random {} way one-shot learning tasks ...".format(
k, N
)
)
sum = 0.0
sum1 = 0.0
for i in range(k):
inputs, targets = self.make_oneshot_task(N, s)
probs = model.predict(inputs)
probability = []
for i in probs:
probability.append(round(i[0]))
probability = np.array(probability)
a = sm.confusion_matrix(targets, probability)
if len(a) > 1:
sum += sm.accuracy_score(targets, probability)
sum1 += sm.f1_score(targets, probability)
else:
sum += 1.0
sum1 += 1.0
percent = (sum / k) * 100.0
F1_score = sum1 / k
if verbose:
print(
"Got an average of {}% Accuracy in {} way one-shot learning accuracy".format(
round(percent, 2), N
)
)
print(
"Got an average of {} F1-Score in {} way one-shot learning accuracy".format(
round(F1_score, 2), N
)
)
return percent, F1_score
# Instantiate the class
loader = Siamese_Loader()
def concat_images(X):
"""Concatenates a bunch of images into a big matrix for plotting purposes."""
a, b, c, d = X.shape
X = np.resize(X, (a, 28, 28, d))
nc, h, w, _ = X.shape
X = X.reshape(nc, h, w)
n = np.ceil(np.sqrt(nc)).astype("int8")
img = np.zeros((n * w, n * h))
x = 0
y = 0
for example in range(nc):
img[x * w : (x + 1) * w, y * h : (y + 1) * h] = X[example]
y += 1
if y >= n:
y = 0
x += 1
return img
def plot_oneshot_task(pairs):
"""Takes a one-shot task given to a siamese net and"""
fig, (ax1, ax2) = plt.subplots(2)
ax1.matshow(np.resize(pairs[0][0], (28, 28)), cmap="gray")
img = concat_images(pairs[1])
ax1.get_yaxis().set_visible(False)
ax1.get_xaxis().set_visible(False)
ax2.matshow(img, cmap="gray")
plt.xticks([])
plt.yticks([])
plt.show()
# example of a one-shot learning task
pairs, targets = loader.make_oneshot_task(10, "train", "0")
plot_oneshot_task(pairs)
# Training loop
os.chdir(r"/kaggle/working/")
print("!")
evaluate_every = 1 # interval for evaluating on one-shot tasks
loss_every = 50 # interval for printing loss (iterations)
batch_size = 4
n_iter = 10000
N_way = 10 # how many classes for testing one-shot tasks>
n_val = 250 # how many one-shot tasks to validate on?
best = -1
s = -1
print("training")
for i in range(1, n_iter):
(inputs, targets) = loader.get_batch(batch_size)
loss = siamese_net.train_on_batch(inputs, targets)
print("Loss is = {}".format(round(loss, 2)))
if i % evaluate_every == 0:
print("evaluating")
val_acc, score = loader.test_oneshot(siamese_net, N_way, n_val, i, verbose=True)
if val_acc >= best or s >= score:
print("saving")
siamese_net.save(r"weights") # weights_path = os.path.join(PATH, "weights")
best = val_acc
if i % loss_every == 0:
print("iteration {}, training loss: {:.2f},".format(i, loss))
ways = np.arange(1, 30, 2)
resume = False
val_accs, train_accs, valscore, trainscore = [], [], [], []
trials = 400
i = 0
for N in ways:
train, trains = loader.test_oneshot(
siamese_net, N, trials, i, "train", verbose=True
)
val, vals = loader.test_oneshot(siamese_net, N, trials, i, "val", verbose=True)
val_accs.append(val)
train_accs.append(train)
valscore.append(vals)
trainscore.append(trains)
i += 1
from statistics import mean
print("The Average testing Accuracy is {}%".format(round(mean(val_accs), 2)))
print("The Average testing F1-Score is {}".format(round(mean(valscore), 2)))
plt.figure(1)
plt.plot(ways, train_accs, "b", label="Siamese(train set)")
plt.plot(ways, val_accs, "r", label="Siamese(val set)")
plt.xlabel("Number of possible classes in one-shot tasks")
plt.ylabel("% Accuracy")
plt.title("MNIST One-Shot Learning performace of a Siamese Network")
# box = plt.get_position()
# plt.set_position([box.x0, box.y0, box.width * 0.8, box.height])
plt.legend(loc="center left", bbox_to_anchor=(1, 0.5))
plt.show()
# inputs,targets = loader.make_oneshot_task(10,"val")
plt.figure(2)
plt.plot(ways, trainscore, "g", label="Siamese(train set)")
plt.plot(ways, valscore, "r", label="Siamese(val set)")
plt.xlabel("Number of possible classes in one-shot tasks")
plt.ylabel("F1-Score")
plt.title("MNIST One-Shot Learning F1 Score of a Siamese Network")
# box = plt.get_position()
# plt.set_position([box.x0, box.y0, box.width * 0.8, box.height])
plt.legend(loc="center left", bbox_to_anchor=(1, 0.5))
plt.show()
|
import numpy as np
import pandas as pd
import seaborn as sns
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split, GridSearchCV
from sklearn.linear_model import LogisticRegression
from sklearn.neighbors import KNeighborsClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import cross_val_score, cross_val_predict
from sklearn.svm import SVC
from sklearn.metrics import classification_report, confusion_matrix
df = pd.read_csv("../input/diabetes.csv")
df.head()
X = df.drop("Outcome", axis=1)
X = StandardScaler().fit_transform(X)
y = df["Outcome"]
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.25, random_state=0
)
model = SVC()
parameters = [
{"kernel": ["rbf"], "gamma": [1e-3, 1e-4], "C": [1, 10, 100, 1000]},
{"kernel": ["linear"], "C": [1, 10, 100, 1000]},
]
grid = GridSearchCV(estimator=model, param_grid=parameters, cv=5)
grid.fit(X_train, y_train)
roc_auc = np.around(
np.mean(cross_val_score(grid, X_test, y_test, cv=5, scoring="roc_auc")), decimals=4
)
print("Score: {}".format(roc_auc))
model1 = RandomForestClassifier(n_estimators=1000)
model1.fit(X_train, y_train)
predictions = cross_val_predict(model1, X_test, y_test, cv=5)
print(classification_report(y_test, predictions))
print(confusion_matrix(y_test, predictions))
score1 = np.mean(cross_val_score(model, X_test, y_test, cv=5, scoring="roc_auc"))
np.around(score1, decimals=4)
model2 = KNeighborsClassifier()
model2.fit(X_train, y_train)
predictions = cross_val_predict(model2, X_test, y_test, cv=5)
print(classification_report(y_test, predictions))
print(confusion_matrix(y_test, predictions))
score2 = np.around(
np.mean(cross_val_score(model2, X_test, y_test, cv=5, scoring="roc_auc")),
decimals=4,
)
print("Score : {}".format(score2))
model3 = LogisticRegression()
parameters = {"C": [0.001, 0.01, 0.1, 1, 10, 100]}
grid = GridSearchCV(estimator=model3, param_grid=parameters, cv=5)
grid.fit(X_train, y_train)
score3 = np.around(
np.mean(cross_val_score(model3, X_test, y_test, cv=5, scoring="roc_auc")),
decimals=4,
)
print("Score : {}".format(score3))
names = []
scores = []
names.extend(["SVC", "RF", "KNN", "LR"])
scores.extend([roc_auc, score1, score2, score3])
algorithms = pd.DataFrame({"Name": names, "Score": scores})
algorithms
print("Most accurate:\n{}".format(algorithms.max()))
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
import pandas as pd
import numpy as np
import re
from tqdm import tqdm_notebook
from sklearn.preprocessing import StandardScaler, OneHotEncoder, LabelEncoder
from sklearn.model_selection import train_test_split, GridSearchCV, RandomizedSearchCV
import pickle
from sklearn.impute import SimpleImputer
from sklearn.metrics import roc_auc_score
from sklearn.model_selection import KFold
from xgboost import XGBClassifier
from sklearn.metrics import roc_auc_score
from sklearn.model_selection import train_test_split
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
df = pd.read_csv("/kaggle/input/widsdatathon2020/training_v2.csv")
test = pd.read_csv("/kaggle/input/widsdatathon2020/unlabeled.csv")
print("Shape of the data is {}".format(df.shape))
print("Shape of the test data is {}".format(test.shape))
# * I dropped the columns which had more than 20 % missing values
# * After analysing the data I divided columns into following sections
target_column = "hospital_death"
doubtful_columns = [
"cirrhosis",
"diabetes_mellitus",
"immunosuppression",
"hepatic_failure",
"leukemia",
"lymphoma",
"solid_tumor_with_metastasis",
"gcs_unable_apache",
]
cols_with_around_70_percent_zeros = [
"intubated_apache",
"ventilated_apache",
]
cols_with_diff_dist_in_test = ["hospital_id", "icu_id"]
selected_columns = [
"d1_spo2_max",
"d1_diasbp_max",
"d1_temp_min",
"h1_sysbp_max",
"gender",
"heart_rate_apache",
"weight",
"icu_stay_type",
"d1_mbp_max",
"h1_resprate_max",
"d1_heartrate_min",
"apache_post_operative",
"apache_4a_hospital_death_prob",
"d1_mbp_min",
"apache_4a_icu_death_prob",
"d1_sysbp_max",
"icu_type",
"apache_3j_bodysystem",
"h1_sysbp_min",
"h1_resprate_min",
"d1_resprate_max",
"h1_mbp_min",
"ethnicity",
"arf_apache",
"resprate_apache",
"map_apache",
"temp_apache",
"icu_admit_source",
"h1_spo2_min",
"d1_spo2_min",
"d1_resprate_min",
"h1_mbp_max",
"height",
"age",
"h1_diasbp_max",
"d1_sysbp_min",
"pre_icu_los_days",
"d1_heartrate_max",
"d1_diasbp_min",
"apache_2_bodysystem",
"gcs_eyes_apache",
"apache_2_diagnosis",
"gcs_motor_apache",
"d1_temp_max",
"h1_spo2_max",
"h1_heartrate_max",
"bmi",
"d1_glucose_min",
"h1_heartrate_min",
"gcs_verbal_apache",
"apache_3j_diagnosis",
"d1_glucose_max",
"h1_diasbp_min",
]
print(f"Total number of diff. dist. columns are {len(cols_with_diff_dist_in_test)}")
print(
f"Total number of columns with 70% 0s are {len(cols_with_around_70_percent_zeros)}"
)
print(f"Total number of doubtful columns are {len(doubtful_columns)}")
print(f"Total number of selected columns are {len(selected_columns)}")
(
len(selected_columns)
+ len(cols_with_around_70_percent_zeros)
+ len(cols_with_diff_dist_in_test)
+ len(doubtful_columns)
)
# Dividing Columns into Categories
continuous_columns = [
"d1_spo2_max",
"d1_diasbp_max",
"d1_temp_min",
"h1_sysbp_max",
"heart_rate_apache",
"weight",
"d1_mbp_max",
"h1_resprate_max",
"d1_heartrate_min",
"apache_4a_hospital_death_prob",
"d1_mbp_min",
"apache_4a_icu_death_prob",
"d1_sysbp_max",
"h1_sysbp_min",
"h1_resprate_min",
"d1_resprate_max",
"h1_mbp_min",
"resprate_apache",
"map_apache",
"temp_apache",
"h1_spo2_min",
"d1_spo2_min",
"d1_resprate_min",
"h1_mbp_max",
"height",
"age",
"h1_diasbp_max",
"d1_sysbp_min",
"pre_icu_los_days",
"d1_heartrate_max",
"d1_diasbp_min",
"gcs_eyes_apache",
"gcs_motor_apache",
"d1_temp_max",
"h1_spo2_max",
"h1_heartrate_max",
"bmi",
"d1_glucose_min",
"h1_heartrate_min",
"gcs_verbal_apache",
"d1_glucose_max",
"h1_diasbp_min",
]
binary_columns = [
"apache_post_operative",
"arf_apache",
"cirrhosis",
"diabetes_mellitus",
"immunosuppression",
"hepatic_failure",
"leukemia",
"lymphoma",
"solid_tumor_with_metastasis",
"gcs_unable_apache",
"intubated_apache",
"ventilated_apache",
]
categorical_columns = [
"icu_stay_type",
"icu_type",
"apache_3j_bodysystem",
"ethnicity",
"gender",
"icu_admit_source",
"apache_2_bodysystem",
"apache_2_diagnosis",
"apache_3j_diagnosis",
]
high_cardinality_columns = ["hospital_id", "icu_id"]
print(
len(continuous_columns)
+ len(binary_columns)
+ len(categorical_columns)
+ len(high_cardinality_columns)
)
columns_to_be_used = list(
set(
doubtful_columns
+ cols_with_around_70_percent_zeros
+ cols_with_diff_dist_in_test
+ selected_columns
)
)
print(f"Total columns to be used initially are {len(columns_to_be_used)}")
categorical_columns = list(set(categorical_columns))
continuous_columns = list(set(continuous_columns))
binary_columns = list(set(binary_columns))
high_cardinality_columns = list(set(high_cardinality_columns))
print(f"Total categorical columns to be used initially are {len(categorical_columns)}")
print(f"Total continuous columns to be used initially are {len(continuous_columns)}")
print(f"Total binary_columns to be used initially are {len(binary_columns)}")
print(
f"Total high_cardinality_columns to be used initially are {len(high_cardinality_columns)}"
)
# Taking subset of the database with the above selected columns
df_train, Y_tr = df[columns_to_be_used], df[target_column]
df_test = test[columns_to_be_used]
print(df_train.shape, Y_tr.shape, df_test.shape)
# Using Label Encoder to encode text into integer classes
# for categorical label encoding
cat_labenc_mapping = {col: LabelEncoder() for col in categorical_columns}
for col in tqdm_notebook(categorical_columns):
df_train[col] = df_train[col].astype("str")
cat_labenc_mapping[col] = cat_labenc_mapping[col].fit(
np.unique(df_train[col].unique().tolist() + df_test[col].unique().tolist())
)
df_train[col] = cat_labenc_mapping[col].transform(df_train[col])
for col in tqdm_notebook(categorical_columns):
print()
df_test[col] = df_test[col].astype("str")
df_test[col] = cat_labenc_mapping[col].transform(df_test[col])
# ## Imputation
# 1. Imputing missing values in continuous columns by Median
# * I am considering high cardinality columns as continuous columns only
# 2. Imputing missing values in categorical columns by Mode
# imputing
# for categorical
cat_col2imputer_mapping = {
col: SimpleImputer(strategy="most_frequent") for col in categorical_columns
}
# for continuous
cont_col2imputer_mapping = {
col: SimpleImputer(strategy="median") for col in continuous_columns
}
# for binary
bin_col2imputer_mapping = {
col: SimpleImputer(strategy="most_frequent") for col in binary_columns
}
# for high cardinality
hicard_col2imputer_mapping = {
col: SimpleImputer(strategy="median") for col in high_cardinality_columns
}
all_imp_dicts = [
cat_col2imputer_mapping,
cont_col2imputer_mapping,
bin_col2imputer_mapping,
hicard_col2imputer_mapping,
]
# fitting imputers
for imp_mapping_obj in tqdm_notebook(all_imp_dicts):
for col, imp_object in imp_mapping_obj.items():
data = df_train[col].values.reshape(-1, 1)
imp_object.fit(data)
# transofrming imputed columns
# fitting imputers
for imp_mapping_obj in tqdm_notebook(all_imp_dicts):
for col, imp_object in imp_mapping_obj.items():
data = df_train[col].values.reshape(-1, 1)
data = imp_object.transform(data)
df_train[col] = list(
data.reshape(
-1,
)
)
# inputing on test
for imp_mapping_obj in tqdm_notebook(all_imp_dicts):
for col, imp_object in imp_mapping_obj.items():
data = df_test[col].values.reshape(-1, 1)
data = imp_object.transform(data)
df_test[col] = list(
data.reshape(
-1,
)
)
# Using sklearn's train test split to create validation set
# train_test split
X_train, X_eval, Y_train, Y_eval = train_test_split(
df_train, Y_tr, test_size=0.15, stratify=Y_tr
)
X_train.shape, X_eval.shape, Y_train.shape, Y_eval.shape
# ## Hyper Parameter Tuning
# ### Step 1. Finding n_estimators after fixing other parameters
# - max_depth = 5 : This should be between 3-10. I’ve started with 5 but you can choose a different number as well. 4-6 can be good starting points.
# - min_child_weight = 1 : A smaller value is chosen because it is a highly imbalanced class problem and leaf nodes can have smaller size groups.
# - gamma = 0 : A smaller value like 0.1-0.2 can also be chosen for starting. This will anyways be tuned later.
# > - subsample, colsample_bytree = 0.8 : This is a commonly used used start value. Typical values range between 0.5-0.9.
# - scale_pos_weight = 1: Because of high class imbalance.
#
# tuning tree specific features
gkf = KFold(n_splits=3, shuffle=True, random_state=42).split(X=X_train, y=Y_train)
fit_params_of_xgb = {
"early_stopping_rounds": 100,
"eval_metric": "auc",
"eval_set": [(X_eval, Y_eval)],
# 'callbacks': [lgb.reset_parameter(learning_rate=learning_rate_010_decay_power_099)],
"verbose": 100,
}
# A parameter grid for XGBoost
params = {
"booster": ["gbtree"],
"learning_rate": [0.1],
"n_estimators": range(100, 500, 100),
"min_child_weight": [1],
"gamma": [0],
"subsample": [0.8],
"colsample_bytree": [0.8],
"max_depth": [5],
"scale_pos_weight": [1],
}
xgb_estimator = XGBClassifier(
objective="binary:logistic",
# silent=True,
)
gsearch = GridSearchCV(
estimator=xgb_estimator,
param_grid=params,
scoring="roc_auc",
n_jobs=-1,
cv=gkf,
verbose=3,
)
# gsearch = RandomizedSearchCV(
# estimator=xgb_estimator,
# param_distributions=params,
# scoring='roc_auc',
# n_jobs=-1,
# cv=gkf, verbose=3
# )
xgb_model = gsearch.fit(X=X_train, y=Y_train, **fit_params_of_xgb)
gsearch.best_params_, gsearch.best_score_
# - Now we will fix this n_estimator=200 and learning_rate=0.1 value and find out others.
# ## Step 2. Finding min_child_weight and max_depth
gkf = KFold(n_splits=3, shuffle=True, random_state=42).split(X=X_train, y=Y_train)
fit_params_of_xgb = {
"early_stopping_rounds": 100,
"eval_metric": "auc",
"eval_set": [(X_eval, Y_eval)],
# 'callbacks': [lgb.reset_parameter(learning_rate=learning_rate_010_decay_power_099)],
"verbose": 100,
}
# A parameter grid for XGBoost
params = {
"booster": ["gbtree"],
"learning_rate": [0.1],
"n_estimators": [300],
"gamma": [0],
"subsample": [0.8],
"colsample_bytree": [0.8],
"scale_pos_weight": [1],
"max_depth": range(2, 7, 2),
"min_child_weight": range(2, 8, 2),
}
xgb_estimator = XGBClassifier(
objective="binary:logistic",
silent=True,
)
gsearch = GridSearchCV(
estimator=xgb_estimator,
param_grid=params,
scoring="roc_auc",
n_jobs=-1,
cv=gkf,
verbose=3,
)
xgb_model = gsearch.fit(X=X_train, y=Y_train, **fit_params_of_xgb)
gsearch.best_params_, gsearch.best_score_
# ## Tuning Gamma
gkf = KFold(n_splits=3, shuffle=True, random_state=42).split(X=X_train, y=Y_train)
fit_params_of_xgb = {
"early_stopping_rounds": 100,
"eval_metric": "auc",
"eval_set": [(X_eval, Y_eval)],
# 'callbacks': [lgb.reset_parameter(learning_rate=learning_rate_010_decay_power_099)],
"verbose": 100,
}
# A parameter grid for XGBoost
params = {
"booster": ["gbtree"],
"learning_rate": [0.1],
"n_estimators": [300],
"subsample": [0.8],
"colsample_bytree": [0.8],
"scale_pos_weight": [1],
"max_depth": [4],
"min_child_weight": [6],
"gamma": [0, 0.01, 0.01],
}
xgb_estimator = XGBClassifier(
objective="binary:logistic",
silent=True,
)
gsearch = GridSearchCV(
estimator=xgb_estimator,
param_grid=params,
scoring="roc_auc",
n_jobs=-1,
cv=gkf,
verbose=3,
)
# gsearch = RandomizedSearchCV(
# estimator=xgb_estimator,
# param_distributions=params,
# scoring='roc_auc',
# n_jobs=-1,
# cv=gkf, verbose=3
# )
xgb_model = gsearch.fit(X=X_train, y=Y_train, **fit_params_of_xgb)
gsearch.best_params_, gsearch.best_score_
# ## Tuning subsample and colsample_bytree
gkf = KFold(n_splits=3, shuffle=True, random_state=42).split(X=X_train, y=Y_train)
fit_params_of_xgb = {
"early_stopping_rounds": 100,
"eval_metric": "auc",
"eval_set": [(X_eval, Y_eval)],
# 'callbacks': [lgb.reset_parameter(learning_rate=learning_rate_010_decay_power_099)],
"verbose": 100,
}
# A parameter grid for XGBoost
params = {
"booster": ["gbtree"],
"learning_rate": [0.1],
"n_estimators": [300],
"scale_pos_weight": [1],
"max_depth": [4],
"min_child_weight": [6],
"gamma": [0],
"subsample": [i / 10.0 for i in range(2, 5)],
"colsample_bytree": [i / 10.0 for i in range(8, 10)],
}
xgb_estimator = XGBClassifier(
objective="binary:logistic",
silent=True,
)
gsearch = GridSearchCV(
estimator=xgb_estimator,
param_grid=params,
scoring="roc_auc",
n_jobs=-1,
cv=gkf,
verbose=3,
)
# gsearch = RandomizedSearchCV(
# estimator=xgb_estimator,
# param_distributions=params,
# scoring='roc_auc',
# n_jobs=-1,
# cv=gkf, verbose=3
# )
xgb_model = gsearch.fit(X=X_train, y=Y_train, **fit_params_of_xgb)
gsearch.best_params_, gsearch.best_score_
# ## Tuning reg_alpha
gkf = KFold(n_splits=3, shuffle=True, random_state=42).split(X=X_train, y=Y_train)
fit_params_of_xgb = {
"early_stopping_rounds": 100,
"eval_metric": "auc",
"eval_set": [(X_eval, Y_eval)],
# 'callbacks': [lgb.reset_parameter(learning_rate=learning_rate_010_decay_power_099)],
"verbose": 100,
}
# A parameter grid for XGBoost
params = {
"booster": ["gbtree"],
"learning_rate": [0.1],
"n_estimators": [300],
"scale_pos_weight": [1],
"max_depth": [4],
"min_child_weight": [6],
"gamma": [0],
"subsample": [0.4],
"colsample_bytree": [0.8],
"reg_alpha": [1, 0.5, 0.1, 0.08],
}
xgb_estimator = XGBClassifier(
objective="binary:logistic",
silent=True,
)
gsearch = GridSearchCV(
estimator=xgb_estimator,
param_grid=params,
scoring="roc_auc",
n_jobs=-1,
cv=gkf,
verbose=3,
)
# gsearch = RandomizedSearchCV(
# estimator=xgb_estimator,
# param_distributions=params,
# scoring='roc_auc',
# n_jobs=-1,
# cv=gkf, verbose=3
# )
xgb_model = gsearch.fit(X=X_train, y=Y_train, **fit_params_of_xgb)
gsearch.best_params_, gsearch.best_score_
# ## Reducing learning Rate and adding more Trees
gkf = KFold(n_splits=3, shuffle=True, random_state=42).split(X=X_train, y=Y_train)
fit_params_of_xgb = {
"early_stopping_rounds": 100,
"eval_metric": "auc",
"eval_set": [(X_eval, Y_eval)],
# 'callbacks': [lgb.reset_parameter(learning_rate=learning_rate_010_decay_power_099)],
"verbose": 100,
}
# A parameter grid for XGBoost
params = {
"booster": ["gbtree"],
"learning_rate": [0.01],
"n_estimators": range(1000, 6000, 1000),
"scale_pos_weight": [1],
"max_depth": [4],
"min_child_weight": [6],
"gamma": [0],
"subsample": [0.4],
"colsample_bytree": [0.8],
"reg_alpha": [0.08],
}
xgb_estimator = XGBClassifier(
objective="binary:logistic",
silent=True,
)
gsearch = GridSearchCV(
estimator=xgb_estimator,
param_grid=params,
scoring="roc_auc",
n_jobs=-1,
cv=gkf,
verbose=3,
)
xgb_model = gsearch.fit(X=X_train, y=Y_train, **fit_params_of_xgb)
gsearch.best_params_, gsearch.best_score_
|
# Data processing
import numpy as np
import pandas as pd
from keras.utils.np_utils import to_categorical
from sklearn.model_selection import train_test_split
from keras.preprocessing.image import ImageDataGenerator
# Dealing with warnings
import warnings
warnings.filterwarnings("ignore")
# Plotting the data
import matplotlib.pyplot as plt
import seaborn as sns
sns.set(style="whitegrid")
# Layers of NN
from keras.models import Sequential
from keras.layers.convolutional import Conv2D, MaxPooling2D
from keras.layers.core import Dense, Dropout, Activation, Flatten
# Optimizers
from keras.optimizers import SGD
# Metrics
from sklearn.metrics import log_loss, confusion_matrix
import itertools
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Load train and test data
train = pd.read_csv("../input/digit-recognizer/train.csv")
test = pd.read_csv("../input/digit-recognizer/test.csv")
train.head()
# Shape of training data
train.shape
# 42k images, each with 784 features (pixels) of each images
# Shape of training data
test.shape
# 28k images, each with 784 features (pixels) of each images
# Checking for any null values in training data
train[train.isna().any(1)]
# Checking for any null values in testing data
test[test.isna().any(1)]
# Count of each label
sns.countplot(train["label"])
# Showing image
import random
sample_index = random.choice(range(0, 42000))
sample_image = train.iloc[sample_index, 1:].values.reshape(28, 28)
plt.imshow(sample_image)
plt.grid("off")
plt.show()
# Showing with gray map
plt.imshow(sample_image, cmap=plt.get_cmap("gray"))
plt.grid("off")
plt.show()
# Preparing the data
y = train[["label"]]
X = train.drop(labels=["label"], axis=1)
# Standardizing the values
X = X / 255.0
# Making class labels as one hot encoding
y_ohe = to_categorical(y)
# Splitting the data
X_train, X_test, y_train, y_test = train_test_split(
X, y_ohe, stratify=y_ohe, test_size=0.3
)
# Verifying shape of datasets
X_train.shape, X_test.shape, y_train.shape, y_test.shape
# ### Normal ANN without using convolution
# Sequential model is linear stackk of layers.
# Since the model needs to know the shape of input it is receiving, the first layer of model will have the
# input_shape parameter.
# Dense is our normal fully connected layer.
# Dropout layer is applying dropout to a fraction of neurons in particular layer, that means making their weight as 0.
# activation is relu for hidden layer and softmax for output layer (class probabilities).
# Making a basic vanilla neural network to do classification
model = Sequential()
model.add(Dense(units=256, input_shape=(784,), activation="relu"))
model.add(Dropout(rate=0.2))
model.add(Dense(units=256, activation="relu"))
model.add(Dropout(rate=0.2))
model.add(Dense(units=128, activation="relu"))
model.add(Dropout(rate=0.2))
model.add(Dense(units=64, activation="relu"))
model.add(Dense(units=10, activation="softmax"))
# Compile the model
# Before compiling the model we can actually set some parameters of whichever optimizer we choose
model.compile(loss="categorical_crossentropy", optimizer="adam", metrics=["accuracy"])
# Summary of the model
model.summary()
history = model.fit(
x=X_train,
y=y_train,
batch_size=32,
epochs=30,
verbose=1,
validation_data=(X_test, y_test),
)
# Plotting the train and validation set accuracy and error
plt.figure(figsize=(20, 6))
plt.subplot(1, 2, 1)
plt.plot(history.history["accuracy"])
plt.plot(history.history["val_accuracy"])
plt.title("model accuracy")
plt.ylabel("accuracy")
plt.xlabel("epoch")
plt.legend(["train", "validation"], loc="center")
plt.subplot(1, 2, 2)
plt.plot(history.history["loss"])
plt.plot(history.history["val_loss"])
plt.title("model loss")
plt.ylabel("loss")
plt.xlabel("epoch")
plt.legend(["train", "validation"], loc="center")
plt.show()
# Predicting the output probabilities
y_pred = model.predict(x=X_test, batch_size=32, verbose=1)
# Calculating log loss
log_loss(y_test, y_pred)
# The log loss is very good
# Converting the probabilitiies as such that highest value will get 1 else 0
y_pred = np.round(y_pred, 2)
b = np.zeros_like(y_pred)
b[np.arange(len(y_pred)), y_pred.argmax(1)] = 1
# Now calcualting the log loss
log_loss(y_test, b)
# It has increased now
# ### Using normal CNN without using Data Augmentation
# CNN stands for convolutional NN majorly due to the convolution operator we have. It works with 2D input array
# (actually 3D as channel value is also expected).
# It has majorly two parts convolution and pooling (normally max-pooling, but we have other poolings also).
# In convolution layer, we convolve inout image with something called kernels, which help us identify features of
# images like edges, corners, round shapes etc. The output of this is called feature map.
# Convolution happens through strides, we can have values for strides and we also have padding as kernels will have
# situation where they will not have data fitting the image properly.
# We have two types of padding, VALID and SAME.
# VALID padding means no padding, so in that case there will be loss of information. Valid actually means that it will
# only take valid data for the operatioon and will not add any padding.
# SAME padding means zero padding. Here in this case, we actually add 0 padding to match our kernel size with our data
# we will not lose any information in this case.
# This applies to conv layers and pool layers both.
# In a typical CNN, we can have multiple conv layers followed by pool layers to get the feature maps. In hidden layers
# it is typical to have more kernels to get the complex feature maps of the data.
# CNN lacks one thing which Hinton tried to solve in CapsuleNet and that is relative location of elements of images.
# CNN is not good with identifying relative location of image components
# Let's talk about channels before we moeve ahead. Channels are basically a dimension which lets CNN identify colorful
# images. For a colored RGB image, there are 3 channels. Any RGB image can be divided into 3 different images of
# different channels and then they are stacked upon each other. We can also see channels as some aspect of information
# in 3rd dimension. For RGB we have 3 channels and for gray map we have 1 channel.
# Reshaping the data for CNN
X_train = X_train.values.reshape(-1, 28, 28, 1)
X_test = X_test.values.reshape(-1, 28, 28, 1)
# We will implement classic LeNet from Lecun's paper.
# It has two layers having conv, followed by pooling.
model = Sequential()
# First Conv and Pooling layer
model.add(
Conv2D(filters=20, kernel_size=(5, 5), padding="same", input_shape=(28, 28, 1))
)
model.add(Activation("relu"))
model.add(MaxPooling2D(pool_size=(2, 2), strides=(2, 2), padding="same"))
# Second conv and pooling layer
model.add(Conv2D(filters=50, kernel_size=(5, 5), padding="same"))
model.add(Activation("relu"))
model.add(MaxPooling2D(pool_size=(2, 2), strides=(2, 2), padding="same"))
# First fully connected layer
model.add(Flatten())
model.add(Dense(units=500))
model.add(Activation("relu"))
# Second fully connected layer
model.add(Dense(units=10))
model.add(Activation("softmax"))
from keras.optimizers import SGD
opt = SGD(lr=0.01)
model.compile(loss="categorical_crossentropy", optimizer=opt, metrics=["accuracy"])
model.summary()
history = model.fit(
x=X_train,
y=y_train,
batch_size=128,
verbose=1,
epochs=30,
validation_data=(X_test, y_test),
)
plt.figure(figsize=(20, 6))
plt.subplot(1, 2, 1)
plt.plot(history.history["accuracy"])
plt.plot(history.history["val_accuracy"])
plt.title("model accuracy")
plt.ylabel("accuracy")
plt.xlabel("epoch")
plt.legend(["train", "validation"], loc="center")
plt.subplot(1, 2, 2)
plt.plot(history.history["loss"])
plt.plot(history.history["val_loss"])
plt.title("model loss")
plt.ylabel("loss")
plt.xlabel("epoch")
plt.legend(["train", "validation"], loc="center")
plt.show()
y_pred = model.predict(x=X_test, batch_size=32, verbose=1)
# Calculating log loss
log_loss(y_test, y_pred)
# ### LeNet CNN with augmented images generated by ImageDataGenerator
from keras.preprocessing.image import ImageDataGenerator
# Imagedatagenerator DOES NOT USE ORIGINAL IMAGES, instead it uses all the augmented images which replace original
# images
augdata = ImageDataGenerator(
rotation_range=30, # rotating images
zoom_range=0.2, # zoom range
width_shift_range=0.3, # width shift range
height_shift_range=0.3, # height shift
shear_range=0.20,
horizontal_flip=True,
vertical_flip=True,
fill_mode="nearest",
)
# steps_per_epoch means number of batch iterations before a training epoch can be marked complete.
# Normally it should be data_size/batch_size. But when we have huge amount of augmented data, we might want to have
# less number of iterations as it would be time consuming and data is anyway augmented randomly so it should not be
# a problem of information loss.
# We will create a new model for that
history = model.fit_generator(
generator=augdata.flow(x=X_train, y=y_train, batch_size=32),
validation_data=(X_test, y_test),
steps_per_epoch=np.ceil(X_train.shape[0] / 32),
epochs=30,
)
plt.figure(figsize=(20, 6))
plt.subplot(1, 2, 1)
plt.plot(history.history["accuracy"])
plt.plot(history.history["val_accuracy"])
plt.title("model accuracy")
plt.ylabel("accuracy")
plt.xlabel("epoch")
plt.legend(["train", "validation"], loc="best", shadow=True)
# plt.show()
plt.subplot(1, 2, 2)
plt.plot(history.history["loss"])
plt.plot(history.history["val_loss"])
plt.title("model loss")
plt.ylabel("loss")
plt.xlabel("epoch")
plt.legend(["train", "validation"], loc="best", shadow=True)
plt.show()
y_pred = model.predict(x=X_test, batch_size=32, verbose=1)
# Calculating log loss
from sklearn.metrics import log_loss
log_loss(y_test, y_pred)
# ### A little complex model than LeNet
#
model = Sequential()
# First Conv and Pooling layer
model.add(
Conv2D(
filters=30,
kernel_size=(2, 2),
padding="same",
input_shape=(28, 28, 1),
activation="relu",
)
)
model.add(Conv2D(filters=30, kernel_size=(2, 2), padding="same", activation="relu"))
model.add(MaxPooling2D(pool_size=(2, 2), strides=(2, 2), padding="same"))
# Second conv and pooling layer
model.add(Conv2D(filters=50, kernel_size=(4, 4), padding="same", activation="relu"))
model.add(Conv2D(filters=50, kernel_size=(4, 4), padding="same", activation="relu"))
model.add(MaxPooling2D(pool_size=(2, 2), strides=(2, 2), padding="same"))
# First fully connected layer
model.add(Flatten())
model.add(Dense(units=500))
model.add(Activation("relu"))
# Second fully connected layer
model.add(Dense(units=10))
model.add(Activation("softmax"))
model.compile(loss="categorical_crossentropy", optimizer=opt, metrics=["accuracy"])
history = model.fit_generator(
generator=augdata.flow(x=X_train, y=y_train, batch_size=32),
validation_data=(X_test, y_test),
steps_per_epoch=np.ceil(X_train.shape[0] / 32),
epochs=30,
)
plt.figure(figsize=(20, 6))
plt.subplot(1, 2, 1)
plt.plot(history.history["accuracy"])
plt.plot(history.history["val_accuracy"])
plt.title("model accuracy")
plt.ylabel("accuracy")
plt.xlabel("epoch")
plt.legend(["train", "validation"], loc="best", shadow=True)
# plt.show()
plt.subplot(1, 2, 2)
plt.plot(history.history["loss"])
plt.plot(history.history["val_loss"])
plt.title("model loss")
plt.ylabel("loss")
plt.xlabel("epoch")
plt.legend(["train", "validation"], loc="best", shadow=True)
plt.show()
# We can see that our model is able to generalize well, though more accuracy can be achieved if we train for higher
# epoch
y_pred = model.predict(x=X_test, batch_size=32, verbose=1)
# Calculating log loss
from sklearn.metrics import log_loss
log_loss(y_test, y_pred)
def plot_confusion_matrix(
cm, classes, normalize=False, title="Confusion matrix", cmap=plt.cm.Blues
):
"""
This function prints and plots the confusion matrix.
Normalization can be applied by setting `normalize=True`.
"""
plt.figure(figsize=(6, 6))
plt.imshow(cm, interpolation="nearest", cmap=cmap)
plt.title(title)
plt.colorbar()
plt.grid("off")
tick_marks = np.arange(len(classes))
plt.xticks(tick_marks, classes, rotation=45)
plt.yticks(tick_marks, classes)
if normalize:
cm = cm.astype("float") / cm.sum(axis=1)[:, np.newaxis]
thresh = cm.max() / 2.0
for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):
plt.text(
j,
i,
cm[i, j],
horizontalalignment="center",
color="white" if cm[i, j] > thresh else "black",
)
plt.tight_layout()
plt.ylabel("True label")
plt.xlabel("Predicted label")
# Predict the values from the validation dataset
y_pred = model.predict(X_test)
# Convert predictions classes to one hot vectors
y_pred_classes = np.argmax(y_pred, axis=1)
# Convert validation observations to one hot vectors
y_true = np.argmax(y_test, axis=1)
# compute the confusion matrix
confusion_mtx = confusion_matrix(y_true, y_pred_classes)
# plot the confusion matrix
plot_confusion_matrix(confusion_mtx, classes=range(10))
# We can see that 2 is mistaken as 5 a lot and vice versa. Similar is the case with 6 and 9. So augmenting the data
# in this case, is it good or we should not do things like horizontal or vertical flip etc.
sns.set(style="white")
errors = y_pred_classes - y_true != 0
y_pred_classes_errors = y_pred_classes[errors]
y_pred_errors = y_pred[errors]
y_true_errors = y_true[errors]
X_val_errors = X_test[errors]
def display_errors(errors_index, img_errors, pred_errors, obs_errors):
"""This function shows 6 images with their predicted and real labels"""
n = 0
nrows = 2
ncols = 3
fig, ax = plt.subplots(nrows, ncols, sharex=True, sharey=True)
for row in range(nrows):
for col in range(ncols):
error = errors_index[n]
ax[row, col].imshow(
(img_errors[error]).reshape((28, 28)), cmap=plt.get_cmap("gray")
)
ax[row, col].set_title(
"Predicted label :{}\nTrue label :{}".format(
pred_errors[error], obs_errors[error]
)
)
n += 1
# Probabilities of the wrong predicted numbers
y_pred_errors_prob = np.max(y_pred_errors, axis=1)
# Predicted probabilities of the true values in the error set
true_prob_errors = np.diagonal(np.take(y_pred_errors, y_true_errors, axis=1))
# Difference between the probability of the predicted label and the true label
delta_pred_true_errors = y_pred_errors_prob - true_prob_errors
# Sorted list of the delta prob errors
sorted_dela_errors = np.argsort(delta_pred_true_errors)
# Top 6 errors
most_important_errors = sorted_dela_errors[-6:]
# Show the top 6 errors
display_errors(
most_important_errors, X_val_errors, y_pred_classes_errors, y_true_errors
)
|
from sympy import *
from math import *
import numpy as np
import sympy as sp
import matplotlib.pyplot as plt
from matplotlib.patches import Wedge
def eq_solve(
differential_equation, function, independent_variable, ics, display_=False
):
ans = dsolve(
differential_equation,
function,
ics={function.subs(independent_variable, ics[0]): ics[1]},
)
if display_:
display(ans)
return str(ans).split(",")[1].strip()[:-1]
def eq_solve_ideal(
differential_equation,
function,
independent_variable,
ics,
range_,
t_step,
display_=False,
):
ans = eq_solve(differential_equation, function, independent_variable, ics, display_)
t = range_[0]
true_val = []
while t <= range_[1]:
t += t_step
true_val.append(round(eval(ans), 8))
return np.array(true_val)
def _euler(
differential_equation, dependent_variable, independent_variable, ics, range_, step
):
f = lambdify(dependent_variable, differential_equation)
d_f = lambdify(dependent_variable, differential_equation.diff(dependent_variable))
d2_f = lambdify(
dependent_variable, differential_equation.diff(dependent_variable, 2)
)
start = range_[0]
end = range_[1]
x1 = ics
x3 = ics
x2 = ics
t = start
x1_arr = []
x2_arr = []
x3_arr = []
t_arr = []
while t <= end:
x1 = x1 + f(x1) * step
x2 = x2 + f(x2) * step + f(x2) * d_f(x2) * (step**2) / 2
x3 = (
x3
+ f(x3) * step
+ f(x3) * d_f(x3) * (step**2) / 2
+ ((f(x3) * (d_f(x3) ** 2)) + ((f(x3) ** 2) * d2_f(x3))) * (step**3) / 6
)
x1_arr.append(x1)
x2_arr.append(x2)
x3_arr.append(x3)
t += step
t_arr.append(t)
return np.array(x1_arr), np.array(x2_arr), np.array(x3_arr), np.array(t_arr)
def _euler_na(
differential_equation, dependent_variable, independent_variable, ics, range_, step
):
f = lambdify((dependent_variable, independent_variable), differential_equation)
g = diff(
differential_equation, independent_variable
) + differential_equation * diff(differential_equation, dependent_variable)
g_ = lambdify((dependent_variable, independent_variable), g)
h = g.diff(independent_variable) + differential_equation * g.diff(
dependent_variable
)
h_ = lambdify((dependent_variable, independent_variable), h)
print(g_, g, h_, h, sep="\n")
start = range_[0]
end = range_[1]
x1 = ics
x3 = ics
x2 = ics
t = start
x1_arr = []
x2_arr = []
x3_arr = []
t_arr = []
while t <= end:
x1 = x1 + f(x1, t) * step
x2 = x2 + f(x2, t) * step + g_(x2, t) * (step**2) / 2
x3 = (
x3
+ f(x3, t) * step
+ g_(x3, t) * (step**2) / 2
+ h_(x3, t) * (step**3) / 6
)
x1_arr.append(x1)
x2_arr.append(x2)
x3_arr.append(x3)
t += step
t_arr.append(t)
return np.array(x1_arr), np.array(x2_arr), np.array(x3_arr), np.array(t_arr)
def print_error(array, text):
if text:
print("\nError (", text, "):")
else:
print("Error analysis:")
min_err = np.min(array)
max_err = np.max(array)
if min_err >= 0:
pass
elif max_err <= 0:
min_err, max_err = max_err, min_err
else:
min_abs_err = np.min(np.abs(array))
max_abs_err = np.max(np.abs(array))
if min_abs_err in array:
min_err = min_abs_err
else:
min_err = -min_abs_err
if max_abs_err in array:
max_err = max_abs_err
else:
max_err = -max_abs_err
print("Local error: ", np.sum(np.abs(array)))
print("Global error:", np.linalg.norm(array) / sqrt(len(array)))
print("Mean of error: ", np.mean(array))
print("Standard deviation: ", np.std(array))
print("Minimum error: ", min_err)
print("Maximum error: ", max_err, "\n")
def print_steps(ans, steps):
print(
ans[0][:steps],
ans[1][:steps],
ans[2][:steps],
ans[3][:steps],
ans[4][:steps],
ans[5][:steps],
ans[6][:steps],
ans[7][:steps],
sep="\n",
)
def test(
function,
dependent_variable,
independent_variable,
ics,
range_,
step,
analysis=True,
show_steps=0,
diff_eq=None,
function_=None,
inde_var=None,
solve_symbolically=False,
true_function=None,
true_array=None,
plot_val=False,
display=False,
save_fig=None,
base_solver=_euler,
**fig_kwargs
):
"""
if true_function == None and solve_symbolically == False and true_array == None:
raise("Enter atleast one param out of true_function, solve_symbolically, true_array", ValueError)
if not ((true_function != None and solve_symbolically == False and true_array == None) or \
(true_function == None and solve_symbolically != False and true_array == None) or \
(true_function == None and solve_symbolically == False and true_array != None)):
raise("Enter only one param out of true_function, solve_symbolically, true_array", ValueError)"""
a, b, c, t = base_solver(
function, dependent_variable, independent_variable, ics, range_, step
)
ideal = true_array
if true_function != None:
ideal = [true_function(i) for i in t]
elif solve_symbolically:
ideal = eq_solve_ideal(
diff_eq,
function_,
inde_var,
ics=[range_[0], ics],
display_=display,
range_=range_,
t_step=step,
)
error1 = ideal - a
error2 = ideal - b
error3 = ideal - c
if plot_val == True:
f, ax = plt.subplots(1, 2, **fig_kwargs)
# f, ax = plt.subplots(1, 2, figsize=(30, 10))
ax[0].plot(t, a, "r")
ax[0].plot(t, b, "b")
ax[0].plot(t, c, "g")
ax[0].plot(t, ideal, "y")
ax[0].legend(["Euler", "Taylor (degree: 2)", "Taylor (degree: 3)", "Ideal"])
ax[1].plot(t, error1, "r")
ax[1].plot(t, error2, "b")
ax[1].plot(t, error3, "g")
ax[1].legend(
["Error Euler", "Error Taylor (degree: 2)", "Error Taylor (degree: 3)"]
)
ax[0].set_xlabel("t", fontsize=18)
ax[0].set_ylabel("x(t)", fontsize=18)
ax[1].set_xlabel("t", fontsize=18)
ax[1].set_ylabel("Error", fontsize=18)
ax[0].grid()
ax[1].grid()
for ax1 in ax:
for tick in ax1.xaxis.get_major_ticks():
tick.label.set_fontsize(14)
for tick in ax1.yaxis.get_major_ticks():
tick.label.set_fontsize(14)
plt.show()
if save_fig:
f.savefig(save_fig, dpi=300)
elif plot_val == "error":
f, ax = plt.subplots(1, 3, **fig_kwargs)
# f, ax = plt.subplots(1, 3, figsize=(40, 10))
ax[0].plot(t, error1, "r")
ax[1].plot(t, error2, "b")
ax[2].plot(t, error3, "g")
ax[0].set_xlabel("t", fontsize=18)
ax[0].set_ylabel("Error: Euler", fontsize=18)
ax[1].set_xlabel("t", fontsize=18)
ax[1].set_ylabel("Error Taylor (degree: 2)", fontsize=18)
ax[2].set_xlabel("t", fontsize=18)
ax[2].set_ylabel("Error Taylor (degree: 3)", fontsize=18)
ax[0].grid()
ax[1].grid()
ax[2].grid()
for ax1 in ax:
for tick in ax1.xaxis.get_major_ticks():
tick.label.set_fontsize(14)
for tick in ax1.yaxis.get_major_ticks():
tick.label.set_fontsize(14)
plt.show()
if save_fig:
f.savefig(save_fig, dpi=100)
if analysis:
print_error(error1, "Euler")
print_error(error2, "Taylor (degree: 2)")
print_error(error3, "Taylor (degree: 3)")
if show_steps:
print_steps(ans, show_steps)
return t, a, b, c, ideal, error1, error2, error3
def give_points(arr, x, tol=1e-12):
stable = []
unstable = []
ND = []
tot_points_id = []
for i in range(len(arr)):
if abs(arr[i]) < tol:
if arr[i - 1] > 0 and arr[i + 1] < 0:
stable.append(round(x[i], 3))
elif arr[i - 1] < 0 and arr[i + 1] > 0:
unstable.append(round(x[i], 3))
else:
ND.append(round(x[i], 3))
tot_points_id.append(i)
return stable, unstable, ND, tot_points_id
def _euler_over_flow_controlled(
differential_equation,
dependent_variable,
independent_variable,
ics,
range_,
step,
penalty=5,
):
f = lambdify(dependent_variable, differential_equation)
d_f = lambdify(dependent_variable, differential_equation.diff(dependent_variable))
d2_f = lambdify(
dependent_variable, differential_equation.diff(dependent_variable, 2)
)
start = range_[0]
end = range_[1]
x1 = ics
x3 = ics
x2 = ics
t = start
x1_arr = []
x2_arr = []
x3_arr = []
t_arr = []
while t <= end:
try:
x1 = x1 + f(x1) * step
x2 = x2 + f(x2) * step + f(x2) * d_f(x2) * (step**2) / 2
x3 = (
x3
+ f(x3) * step
+ f(x3) * d_f(x3) * (step**2) / 2
+ ((f(x3) * (d_f(x3) ** 2)) + ((f(x3) ** 2) * d2_f(x3)))
* (step**3)
/ 6
)
except:
if penalty != 0:
return (
np.array(x1_arr[:-penalty]),
np.array(x2_arr[:-penalty]),
np.array(x3_arr[:-penalty]),
np.array(t_arr[:-penalty]),
)
else:
break
x1_arr.append(x1)
x2_arr.append(x2)
x3_arr.append(x3)
t += step
t_arr.append(t)
return np.array(x1_arr), np.array(x2_arr), np.array(x3_arr), np.array(t_arr)
def test_phase(
function,
dependent_variable,
independent_variable,
ics_arr,
range_,
step,
phase_range,
phase_step,
plot_val="val",
save_fig=None,
save_phase_fig=None,
tol=1e-12,
dpi=100,
dpi_phase=100,
calculate_points=False,
stable=None,
unstable=None,
ND=None,
tick_points=None,
markersize=160,
bbox_to_anchor=None,
ND_arr=None,
penalty=5,
leg_stable=None,
leg_unstable=None,
leg_ND=None,
ics_arr_sym=None,
legend_fontsize=None,
**fig_kwargs
):
eu_arr = []
ty2_arr = []
ty3_arr = []
t_arr = []
d_f = lambdify(dependent_variable, function)
for ics in ics_arr:
a, b, c, t = _euler_over_flow_controlled(
function,
dependent_variable,
independent_variable,
ics,
range_,
step,
penalty=5,
)
eu_arr.append(a)
ty2_arr.append(b)
ty3_arr.append(c)
t_arr.append(t)
if plot_val == "phase":
x = list(np.arange(-1.0001, -0.999, 0.0001))
ideal_d_f = [d_f(i) for i in x]
ideal_d_f_eu = [d_f(i) for i in a]
ideal_d_f_ty1 = [d_f(i) for i in b]
ideal_d_f_ty2 = [d_f(i) for i in c]
f, ax = plt.subplots(1, 1, **fig_kwargs)
ax.plot(x, ideal_d_f)
ax.scatter(a, ideal_d_f_eu)
ax.scatter(b, ideal_d_f_ty1)
ax.scatter(c, ideal_d_f_ty2)
ax.legend(["Ideal", "Euler", "Taylor (degree: 2)", "Taylor (degree: 3)"])
ax.set_ylabel("dx/dt", fontsize=18)
ax.set_xlabel("x", fontsize=18)
plt.grid()
print(ideal_d_f_eu[-1], ideal_d_f_ty1[-1], ideal_d_f_ty2[-1])
elif plot_val == "val":
f, ax = plt.subplots(1, 1, **fig_kwargs)
x = np.arange(phase_range[0], phase_range[1], phase_step)
d_x = [d_f(i) for i in x]
if calculate_points:
stable, unstable, ND, tot_points_id = give_points(d_x, x, tol=tol)
if not leg_stable and stable != []:
print("leg_stable depricated")
leg_stable = ",".join(map(lambda x: str(round(x, 2)), stable))
if not leg_unstable and unstable != []:
print("leg_unstable depricated")
leg_unstable = ",".join(map(lambda x: str(round(x, 2)), unstable))
if not leg_ND and ND != []:
print("leg_ND depricated")
leg_ND = ",".join(map(lambda x: str(round(x, 2)), ND))
print(stable, unstable, ND)
if stable != []:
ax.scatter(
stable, np.zeros(len(stable)), s=markersize, c="black", label=leg_stable
)
if unstable != []:
ax.scatter(
unstable,
np.zeros(len(unstable)),
s=markersize,
marker="o",
facecolors="none",
edgecolors="black",
label=leg_unstable,
)
if ND != []:
marker_style = dict(
color="black",
linestyle=":",
marker="o",
markersize=markersize // 10,
markerfacecoloralt="white",
)
if not ND_arr:
ax.plot(
ND,
np.zeros(len(ND)),
fillstyle="right",
**marker_style,
label=leg_ND
)
else:
for i in range(len(ND)):
ax.plot(
[ND[i]], [0], label=leg_ND, fillstyle=ND_arr[i], **marker_style
)
ax.legend(prop={"size": legend_fontsize})
if calculate_points:
tot_points_id.insert(0, 0)
tot_points_id.append(len(x) - 1)
for idx in range(len(tot_points_id) - 1):
idx = (tot_points_id[idx] + tot_points_id[idx + 1]) // 2
if d_x[idx] > 0:
ax.scatter([x[idx]], [0], marker="$>$", c="k", s=markersize)
elif d_x[idx] < 0:
ax.scatter([x[idx]], [0], marker="$<$", c="k", s=markersize)
else:
for point in tick_points:
if d_f(point) > 0:
ax.scatter([point], [0], marker="$>$", c="k", s=markersize)
else:
ax.scatter([point], [0], marker="$<$", c="k", s=markersize)
ax.plot(x, d_x)
plt.grid()
plt.axhline(y=0, color="k")
plt.axvline(x=0, color="k")
ax.set_ylabel(r"$\frac{dx}{dt}$", rotation=0, fontsize=18)
ax.yaxis.set_label_coords(-0.06, 0.5)
ax.set_xlabel(r"$x$", fontsize=18)
plt.title("Phase plot")
if save_phase_fig:
plt.savefig(save_phase_fig + ".png", dpi=dpi_phase)
if not ics_arr_sym:
ics_arr_sym = list(map(lambda x: round(x, 2), ics_arr))
plt.figure(**fig_kwargs)
for i in range(len(eu_arr)):
plt.plot(t_arr[i], eu_arr[i])
plt.grid()
plt.legend(ics_arr_sym, bbox_to_anchor=bbox_to_anchor)
plt.title("Euler analysis plot")
plt.ylabel(r"$x$", rotation=0, fontsize=18)
plt.xlabel(r"$t$", fontsize=18)
if save_fig:
plt.savefig(save_fig + "eu.png", dpi=dpi)
plt.figure(**fig_kwargs)
for i in range(len(ty2_arr)):
plt.plot(t_arr[i], ty2_arr[i])
plt.grid()
plt.legend(ics_arr_sym, bbox_to_anchor=bbox_to_anchor)
plt.title("Taylor (D:2) analysis plot")
plt.ylabel(r"$x$", rotation=0, fontsize=18)
plt.xlabel(r"$t$", fontsize=18)
if save_fig:
plt.savefig(save_fig + "ty2.png", dpi=dpi)
plt.figure(**fig_kwargs)
for i in range(len(ty3_arr)):
plt.plot(t_arr[i], ty3_arr[i])
plt.grid()
plt.legend(ics_arr_sym, bbox_to_anchor=bbox_to_anchor)
plt.title("Taylor (D:3) analysis plot")
plt.ylabel(r"$x$", rotation=0, fontsize=18)
plt.xlabel(r"$t$", fontsize=18)
if save_fig:
plt.savefig(save_fig + "ty3.png", dpi=dpi)
# **Part 4:**
t = Symbol("t")
x = Function("x")(t)
f = Function("f")(x)
f = x**2 - 1
ics_arr = [1.000001, 0.9999, 0, -0.9999, -1.001, -3]
range_ = [0, 8]
time_step = 0.1
range_phase = [-2, 2]
phase_step = 0.001
test_phase(
f,
x,
t,
ics_arr,
range_,
time_step,
range_phase,
phase_step,
figsize=(10, 10),
save_fig="p1_",
save_phase_fig="p1_phase",
calculate_points=True,
)
t = Symbol("t")
x = Function("x")(t)
f = Function("f")(x)
f = -(x**3)
ics_arr = [0.001, -0.015, 1, -1]
range_ = [0, 2000]
time_step = 0.1
range_phase = [-5, 5]
phase_step = 0.001
l_temp = [-3, 3]
test_phase(
f,
x,
t,
ics_arr,
range_,
time_step,
range_phase,
phase_step,
figsize=(12, 12),
save_fig="p2_",
save_phase_fig="p2_phase",
stable=[0],
unstable=[],
ND=[],
tick_points=l_temp,
)
t = Symbol("t")
x = Function("x")(t)
f = Function("f")(x)
f = x**3
ics_arr = [0.105, -0.105, 0.11, -0.11]
range_ = [0, 40]
time_step = 0.01
range_phase = [-5, 5]
phase_step = 0.001
stable = []
unstable = [0]
ND = []
# ND_arr = ['left']
ticks = [-3, 3]
test_phase(
f,
x,
t,
ics_arr,
range_,
time_step,
range_phase,
phase_step,
figsize=(12, 12),
save_fig="p3_",
save_phase_fig="p3_phase",
stable=stable,
unstable=unstable,
ND=ND,
tick_points=ticks,
) # , ND_arr=ND_arr)
t = Symbol("t")
x = Function("x")(t)
f = Function("f")(x)
f = x**2
ics_arr = [0.007, -0.012, 0.011, -0.017]
range_ = [0, 57]
time_step = 0.001
range_phase = [-2, 2]
phase_step = 0.001
stable = []
unstable = []
ND = [0]
ND_arr = ["left"]
ticks = [-1, 1]
test_phase(
f,
x,
t,
ics_arr,
range_,
time_step,
range_phase,
phase_step,
figsize=(10, 10),
save_fig="p4_",
save_phase_fig="p4_phase",
stable=stable,
unstable=unstable,
ND=ND,
tick_points=ticks,
ND_arr=ND_arr,
)
t = Symbol("t")
x = Function("x")(t)
f = Function("f")(x)
f = 4 * x**2 - 16
ics_arr = [-3, -2.001, 0, -1, 1, 2.001, 2.0011, 1.9]
range_ = [0, 0.5]
time_step = 0.001
range_phase = [-4, 4]
phase_step = 0.001
stable = [-2]
unstable = [2]
ND = []
ND_arr = []
ticks = [0, 3, -3]
test_phase(
f,
x,
t,
ics_arr,
range_,
time_step,
range_phase,
phase_step,
figsize=(10, 10),
save_fig="p5_",
save_phase_fig="p5_phase",
stable=stable,
unstable=unstable,
ND=ND,
tick_points=ticks,
ND_arr=ND_arr,
)
t = Symbol("t")
x = Function("x")(t)
f = Function("f")(x)
f = 1 - x**14
ics_arr = [-0.999, 0, 1.2, 0.8, -1.00001, -1.000001]
range_ = [0, 2.5]
time_step = 0.0001
range_phase = [-1.1, 1.1]
phase_step = 0.001
stable = [1]
unstable = [-1]
ND = []
ND_arr = []
ticks = [0, -1.2, 1.2]
test_phase(
f,
x,
t,
ics_arr,
range_,
time_step,
range_phase,
phase_step,
figsize=(10, 10),
save_fig="p6_",
save_phase_fig="p6_phase",
stable=stable,
unstable=unstable,
ND=ND,
tick_points=ticks,
ND_arr=ND_arr,
penalty=0,
)
t = Symbol("t")
x = Function("x")(t)
f = Function("f")(x)
f = x - x**3
ics_arr = [-4, 4, -1.5, 0.5, 0.01, -0.01, 0.1, -0.1, 0.5, 1.5]
range_ = [0, 7]
time_step = 0.01
range_phase = [-1.8, 1.8]
phase_step = 0.001
stable = [1, -1]
unstable = [0]
ND = []
ND_arr = []
ticks = [0.5, -0.5, 1.5, -1.5]
test_phase(
f,
x,
t,
ics_arr,
range_,
time_step,
range_phase,
phase_step,
figsize=(10, 10),
save_fig="p7_",
save_phase_fig="p7_phase",
stable=stable,
unstable=unstable,
ND=ND,
tick_points=ticks,
ND_arr=ND_arr,
penalty=0,
)
t = Symbol("t")
x = Function("x")(t)
f = Function("f")(x)
f = sp.exp(x) * sp.sin(x)
ics_arr = [0.001, -0.9, -2, -pi - 1, -2 * pi + 0.001]
ics_arr_sym = ["0.001", "-0.9", "-2", "$-\pi - 1$", "$-2\pi + 0.001$"]
range_ = [0, 150]
time_step = 0.05
range_phase = [-7, 0.2]
phase_step = 0.001
leg_stable = r"$-\pi\hspace{2}(2k+1)\pi$"
leg_unstable = r"$-2\pi,0\hspace{2}2k\pi$"
test_phase(
f,
x,
t,
ics_arr,
range_,
time_step,
range_phase,
phase_step,
figsize=(10, 10),
save_fig="p8_",
save_phase_fig="p8_phase",
stable=[-pi],
unstable=[0, -2 * pi],
ND=[],
tick_points=[0.4, -pi / 2, -3 * pi / 2, -2 * pi + -0.4],
bbox_to_anchor=(0.9, 0.9),
leg_stable=leg_stable,
leg_unstable=leg_unstable,
ics_arr_sym=ics_arr_sym,
)
t = Symbol("t")
x = Function("x")(t)
f = Function("f")(x)
f = 1 + 0.5 * sp.sin(x)
ics_arr = list(range(-4, 5))
range_ = [0, 20]
time_step = 0.01
range_phase = [-5, 5]
phase_step = 0.001
test_phase(
f,
x,
t,
ics_arr,
range_,
time_step,
range_phase,
phase_step,
figsize=(12, 12),
save_fig="p9_",
save_phase_fig="p9_phase",
stable=[],
unstable=[],
ND=[],
tick_points=[-2.5, 2.5],
)
t = Symbol("t")
x = Function("x")(t)
f = Function("f")(x)
f = 1 - 2 * sp.cos(x)
temp = pi / 3
temp2 = 2 * pi
ics_arr = [
temp + 0.01,
temp - 0.01,
temp + temp2 + 0.01,
temp + temp2 - 0.01,
temp - temp2 + 0.01,
temp - temp2 - 0.01,
]
ics_arr_sym = [
r"$ \frac{\pi}{3} + 0.01$",
r"$ \frac{\pi}{3} - 0.01$",
r"$ \frac{7\pi}{3} + 0.01$",
r"$ \frac{7\pi}{3} - 0.01$",
r"$ \frac{-5\pi}{3} + 0.01$",
r"$ \frac{-5\pi}{3} - 0.01$",
]
range_ = [0, 7]
time_step = 0.01
range_phase = [-10, 10]
phase_step = 0.001
stable = [-pi / 3, -pi / 3 - 2 * pi, -pi / 3 + 2 * pi]
leg_stable = r"$\frac{-7\pi}{3},\frac{-\pi}{3},\frac{5\pi}{3}$"
unstable = [pi / 3, pi / 3 + 2 * pi, pi / 3 - 2 * pi]
leg_unstable = r"$\frac{-5\pi}{3},\frac{\pi}{3},\frac{7\pi}{3}$"
ND = []
ND_arr = []
ticks = [0, pi, -pi, -2 * pi, 2 * pi, 3 * pi, -3 * pi]
test_phase(
f,
x,
t,
ics_arr,
range_,
time_step,
range_phase,
phase_step,
figsize=(10, 10),
save_fig="p10_",
save_phase_fig="p10_phase",
stable=stable,
unstable=unstable,
ND=ND,
tick_points=ticks,
ND_arr=ND_arr,
penalty=0,
ics_arr_sym=ics_arr_sym,
leg_stable=leg_stable,
leg_unstable=leg_unstable,
legend_fontsize=15,
)
# **Part 5:**
t = Symbol("t")
x = Function("x")(t)
f = Function("f")(x)
m = 110 # kg
g = 9.8 # ms^-2
k = 0.18
A = g
B = k / m
f = A - B * x * x
t_f = lambda t: sqrt(A / B) * tanh(sqrt(A * B) * t)
ans = test(
f,
x,
t,
0,
[0, 50],
0.001,
true_function=t_f,
plot_val=True,
display=True,
analysis=False,
save_fig="S51.png",
figsize=(20, 10),
)
ans = test(
f,
x,
t,
0,
[0, 50],
0.001,
true_function=t_f,
plot_val="error",
display=True,
analysis=True,
save_fig="S51ee.png",
figsize=(40, 10),
)
t = Symbol("t")
x = Function("x")(t)
f = Function("f")(x)
v0 = 4
P = 400 # ms^-2
m = 70
b = 0.5
A = 0.33
rho = 1.25
B = k / m
f = P / (m * x)
t_f = lambda t: sqrt(v0**2 + (2 * P * t / m))
ans = test(
f,
x,
t,
4,
[0, 5],
0.001,
true_function=t_f,
plot_val=True,
display=True,
analysis=False,
save_fig="S3C.png",
figsize=(20, 10),
)
ans = test(
f,
x,
t,
4,
[0, 5],
0.001,
true_function=t_f,
plot_val="error",
display=True,
analysis=True,
save_fig="S3Ce.png",
figsize=(40, 10),
)
# set 5 problem 2b
t = Symbol("t")
x = Function("x")(t)
f = Function("f")(x)
v0 = 4
P = 400 # ms^-2
m = 70
b = 0.5
A = 0.33
rho = 1.25
B = k / m
f = P / (m * x) - ((b * rho * A) / m) * x**2
t_f = lambda t: 0
ans = test(
f,
x,
t,
4,
[0, 50],
0.001,
true_function=t_f,
plot_val=True,
display=True,
analysis=False,
save_fig="S3C.png",
figsize=(20, 10),
)
ans = test(
f,
x,
t,
4,
[0, 50],
0.001,
true_function=t_f,
plot_val="error",
display=True,
analysis=True,
save_fig="S3Ce.png",
figsize=(40, 10),
)
# **Examples Templates:**
t = Symbol("t")
x = Symbol("x")
f = Function("f")(x, t)
f = t**2 - x
y = Function("y")(t)
# dsolve(y.diff(t) + y - t**2, y, ics={y.subs(t, 0):1})
diff_eq = y.diff(t) + y - t**2
ans = test(
f,
x,
t,
1,
[0, 2],
0.01,
display=True,
base_solver=_euler_na,
plot_val=True, # save_fig="Output.png",
figsize=(20, 10),
solve_symbolically=True,
function_=y,
inde_var=t,
diff_eq=diff_eq,
analysis=False,
)
ans = test(
f,
x,
t,
1,
[0, 2],
0.01,
display=True,
base_solver=_euler_na,
plot_val=True, # save_fig="Output.png",
figsize=(20, 10),
solve_symbolically=True,
function_=y,
inde_var=t,
diff_eq=diff_eq,
)
x = Symbol("x")
f = Function("f")(x)
f = -1 * x * x
a, b, c, t_ = _euler(f, x, None, 1, [1, 2], 0.01)
ideal = 1 / t_
ans = test(
f,
x,
None,
1,
[1, 2],
0.01,
true_array=ideal,
plot_val=True,
display=True, # save_fig="Output.png",
figsize=(20, 10),
)
t = Symbol("t")
x = Function("x")(t)
f = Function("f")(x)
f = -1 * x * x
diff_eq = x.diff(t) + x * x # = 0 explicitly
ans = test(
f,
x,
None,
1,
[1, 2],
0.01,
solve_symbolically=True,
function_=x,
inde_var=t,
diff_eq=x.diff(t) + x * x,
plot_val=True,
display=True, # save_fig="Output.png",
figsize=(20, 10),
)
t = Symbol("t")
x = Function("x")(t)
f = Function("f")(x)
f = -1 * x * x
true_function = lambda t: 1 / t
ans = test(
f,
x,
None,
1,
[1, 2],
0.01,
true_function=true_function,
plot_val=True,
display=True, # save_fig="Output.png",
figsize=(20, 10),
)
x = Symbol("x")
t = Symbol("t")
f = Function("f")(x, t)
f = -x + sp.exp(-t)
a, b, c, t_ = _euler_na(f, x, t, 1, [0, 2], 0.01)
ideal = (1 + t_) * np.exp(-t_)
ans = test(
f,
x,
t,
1,
[0, 2],
0.01,
display=True,
true_array=ideal,
base_solver=_euler_na,
plot_val=True, # save_fig="Output.png",
figsize=(20, 10),
)
t = Symbol("t")
x = Symbol("x")
f = Function("f")(x, t)
f = -x + sp.exp(-t)
y = Function("y")(t) # depricated x as y
ans = test(
f,
x,
t,
1,
[0, 2],
0.01,
display=True,
base_solver=_euler_na,
plot_val=True, # save_fig="Output.png",
figsize=(20, 10),
solve_symbolically=True,
function_=y,
inde_var=t,
diff_eq=y.diff(t) + y - sp.exp(-t),
)
t = Symbol("t")
x = Symbol("x")
f = Function("f")(x, t)
f = -x + sp.exp(-t)
ans = test(
f,
x,
t,
1,
[0, 2],
0.01,
display=True,
true_function=lambda t: (t + 1) * exp(-t),
base_solver=_euler_na,
plot_val=True, # save_fig="Output.png",
figsize=(20, 10),
)
|
import pandas as pd
train = pd.read_csv("../input/otto-group-product-classification-challenge/train.csv")
test = pd.read_csv("../input/otto-group-product-classification-challenge/test.csv")
train.shape, test.shape
from pandas_profiling import ProfileReport
train_profile = ProfileReport(
train, title="Pandas Profiling Report", html={"style": {"full_width": True}}
)
train_profile.to_notebook_iframe()
|
# 
# ## Context
# ## Real estate appraisal, property valuation or land valuation is the process of developing an opinion of value for real property (usually market value). Real estate transactions often require appraisals because they occur infrequently and every property is unique (especially their condition, a key factor in valuation), unlike corporate stocks, which are traded daily and are identical (thus a centralized Walrasian auction like a stock exchange is unrealistic). The location also plays a key role in valuation. However, since property cannot change location, it is often the upgrades or improvements to the home that can change its value. Appraisal reports form the basis for mortgage loans, settling estates and divorces, taxation, and so on. Sometimes an appraisal report is used to establish a sale price for a property.
# ## Content
# ## Ask a home buyer to describe their dream house, and they probably won't begin with the height of the basement ceiling or the proximity to an east-west railroad. But this playground competition's dataset proves that much more influences price negotiations than the number of bedrooms or a white-picket fence. With 79 explanatory variables describing (almost) every aspect of residential homes in Ames, Iowa, this competition challenges you to predict the final price of each home.
# ## Data Dictionary
# <div style = "color: White; display: fill;
# border-radius: 5px;
# background-color: #009FBD;
# font-size: 100%;
# font-family: Verdana">
#
# 📌 SalePrice: the property's sale price in dollars. This is the target variable
# 📌 MSSubClass: The building class
# 📌 MSZoning: The general zoning classification
# 📌 LotFrontage: Linear feet of street connected to property
# 📌 LotArea: Lot size in square feet
# 📌 Street: Type of road access
# 📌 Alley: Type of alley access
# 📌 LotShape: General shape of property
# 📌 LandContour: Flatness of the property
# 📌 Utilities: Type of utilities available
# 📌 LotConfig: Lot configuration
# 📌 LandSlope: Slope of property
# 📌 Neighborhood: Physical locations within Ames city limits
# 📌 Condition1: Proximity to main road or railroad
# 📌 Condition2: Proximity to main road or railroad (if a second is present)
# 📌 BldgType: Type of dwelling
# 📌 HouseStyle: Style of dwelling
# 📌 OverallQual: Overall material and finish quality
# 📌 OverallCond: Overall condition rating
# 📌 YearBuilt: Original construction date
# 📌 YearRemodAdd: Remodel date
# 📌 RoofStyle: Type of roof
# 📌 RoofMatl: Roof material
# 📌 Exterior1st: Exterior covering on house
# 📌 Exterior2nd: Exterior covering on house (if more than one material)
# 📌 MasVnrType: Masonry veneer type
# 📌 MasVnrArea: Masonry veneer area in square feet
# 📌 ExterQual: Exterior material quality
# 📌 ExterCond: Present condition of the material on the exterior
# 📌 Foundation: Type of foundation
# 📌 BsmtQual: Height of the basement
# 📌 BsmtCond: General condition of the basement
# 📌 BsmtExposure: Walkout or garden level basement walls
# 📌 BsmtFinType1: Quality of basement finished area
# 📌 BsmtFinSF1: Type 1 finished square feet
# 📌 BsmtFinType2: Quality of second finished area (if present)
# 📌 BsmtFinSF2: Type 2 finished square feet
# 📌 BsmtUnfSF: Unfinished square feet of basement area
# 📌 TotalBsmtSF: Total square feet of basement area
# 📌 Heating: Type of heating
# 📌 HeatingQC: Heating quality and condition
# 📌 CentralAir: Central air conditioning
# 📌 Electrical: Electrical system
# 📌 1stFlrSF: First Floor square feet
# 📌 2ndFlrSF: Second floor square feet
# 📌 LowQualFinSF: Low quality finished square feet (all floors)
# 📌 GrLivArea: Above grade (ground) living area square feet
# 📌 BsmtFullBath: Basement full bathrooms
# 📌 BsmtHalfBath: Basement half bathrooms
# 📌 FullBath: Full bathrooms above grade
# 📌 HalfBath: Half baths above grade
# 📌 Bedroom: Number of bedrooms above basement level
# 📌 Kitchen: Number of kitchens
# 📌 KitchenQual: Kitchen quality
# 📌 TotRmsAbvGrd: Total rooms above grade (does not include bathrooms)
# 📌 Functional: Home functionality rating
# 📌 Fireplaces: Number of fireplaces
# 📌 FireplaceQu: Fireplace quality
# 📌 GarageType: Garage location
# 📌 GarageYrBlt: Year garage was built
# 📌 GarageFinish: Interior finish of the garage
# 📌 GarageCars: Size of garage in car capacity
# 📌 GarageArea: Size of garage in square feet
# 📌 GarageQual: Garage quality
# 📌 GarageCond: Garage condition
# 📌 PavedDrive: Paved driveway
# 📌 WoodDeckSF: Wood deck area in square feet
# 📌 OpenPorchSF: Open porch area in square feet
# 📌 EnclosedPorch: Enclosed porch area in square feet
# 📌 3SsnPorch: Three season porch area in square feet
# 📌 ScreenPorch: Screen porch area in square feet
# 📌 PoolArea: Pool area in square feet
# 📌 PoolQC: Pool quality
# 📌 Fence: Fence quality
# 📌 MiscFeature: Miscellaneous feature not covered in other categories
# 📌 MiscVal: Dollar value of miscellaneous feature
# 📌 MoSold: Month Sold
# 📌 YrSold: Year Sold
# 📌 SaleType: Type of sale
# 📌 SaleCondition: Condition of sale
#
# ## Importing Libraries
import pandas as pd
pd.pandas.set_option("display.max_columns", None)
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
colors = [
"#FF5F00",
"#AD7BE9",
"#03C988",
"#0081B4",
"#F48484",
"#03C988",
"#CC3636",
"#2DCDDF",
"#FFB200",
"#D5CEA3",
"#54B435",
"#EA047E",
"#FFF80A",
"#1D1CE5",
"#FF0032",
]
# ## Loading and Checking the Dataset
df_train = pd.read_csv(
"/kaggle/input/house-prices-advanced-regression-techniques/train.csv"
)
df_train.sample(10)
# ## Shape of the Dataset
df_train.shape
# ## Checking the Columns/Features
df_train.columns
# ## Information about the Dataset
df_train.info()
# ## Looking at the data, we can observe that there are 35 features with integer values, 3 features with floating-point values, and 43 features with object/string values. Therefore, it would be useful to examine how many features in this dataset are numerical and how many are categorical.
# ## Checking if there is any Duplicate values present in this Dataset
df_train.duplicated().sum()
# ## Checking if there is any missing values present in the Dataset
df_train.isnull().sum()
plt.figure(figsize=(20, 6))
sns.heatmap(df_train.isnull())
# ## Based on the heatmap displayed above, it is evident that some columns have a significant number of missing values. Therefore, I intend to investigate which columns contain what percentage of null values.
features_containing_null_values = []
for features in df_train.columns:
if df_train[features].isnull().sum() > 1:
features_containing_null_values.append(features)
null_percentage = (
df_train[features_containing_null_values].isnull().sum() / df_train.shape[0] * 100
).sort_values(ascending=False)
null_percentage
numerical_features = df_train.select_dtypes(include=["int", "float"]).columns.tolist()
print("Number of Numerical features:", len(numerical_features))
print(numerical_features)
categorical_features = df_train.select_dtypes(include=["object"]).columns.tolist()
print("Number of Categorical features:", len(categorical_features))
print(categorical_features)
|
# # **Profitable App Profiles for the App Store and Google Play Markets**
# This project aims to analyze mobile apps data from the Apple App Store and Android Google Play markets to help developers identify the types of apps that are most likely to attract the most users.
# The data from the Apple App Store data set was collected in July 2017 and can be found [here](https://dq-content.s3.amazonaws.com/350/AppleStore.csv).
# The data from the Android Google Play dataset was collected in August 2018 and can be found [here](https://dq-content.s3.amazonaws.com/350/googleplaystore.csv).
# Apple Store data set #
opened_file = open("/kaggle/input/app-store-apple-data-set-10k-apps/AppleStore.csv")
from csv import reader
read_file = reader(opened_file)
apple_apps = list(read_file)
apple_header = apple_apps[0]
apple_data = apple_apps[1:]
# Google Play Store data sets #
opened_file = open("/kaggle/input/google-play-store-apps/googleplaystore.csv")
read_file = reader(opened_file)
android_apps = list(read_file)
android_header = android_apps[0]
android_data = android_apps[1:]
# Function to display a certain range of rows + number of rows and columns from data
def explore_data(dataset, start, end, rows_and_columns=False):
dataset_slice = dataset[start:end]
for row in dataset_slice:
print(row)
print("\n") # Adds a new (empty) line after each row
if rows_and_columns:
print("Number of rows:", len(dataset))
print("Number of columns:", len(dataset[0]))
# |Apple Column|Description
# |-|-
# |id|App ID
# |track_name|App Name
# |size_bytes|App Size (in Bytes)
# |currency|Currency Type
# |price|Price Amount
# |rating_count_tot|User Rating Counts (all versions)
# |rating_count_ver|User Rating Counts (current version)
# |user_rating|Average User Rating value (all versions)
# |user_rating_ver|Average User Rating value (current version)
# |ver|Lateset Version Code
# |cont_rating|Content Rating
# |prime_genre|Primary Genre
# |sup_devices.num|Number of supporting devices
# |ipadSc_urls.num|Number of screenshots showed for display
# |lang.num|Number of supported languages
# |vpp_lic|Vpp Device Based Licensing Enabled
print(apple_header)
print("\n")
explore_data(apple_data, 0, 5, True)
# There are 7,197 apps on the App Store data set, each with 16 columns. The columns that are most pertinent to our analysis include `price`, `rating_count_tot`, `user_rating`, `cont_rating`, `prime_genre`, and `lang_num`.
# Next, we will take a look at the Android Google Play data set.
print(android_header)
print("\n")
explore_data(android_data, 0, 5, True)
# There are 10,841 apps on the Google Play data set, each with 13 columns. The columns that are most pertinent to our analysis include `Category`, `Rating`, `Reviews`, `Installs`, `Price`, `Content Rating`, and `Genres`.
# ### Data Cleaning
# Upon inspecting the dedicated [discussion section](https://www.kaggle.com/lava18/google-play-store-apps/discussion) for the Google Play data set, we can see that there is an error in the data set in row 10,472.
print(android_data[10472])
print(len(android_data))
del android_data[10472] # don't run more than once
print(len(android_data))
# After identifying and removing the error, we will inspect our data set for any duplicate values.
duplicate_android_apps = []
unique_android_apps = []
for app in android_data:
name = app[0]
if name in unique_android_apps:
duplicate_android_apps.append(name)
else:
unique_android_apps.append(name)
print("Number of duplicate apps:", len(duplicate_android_apps))
print("\n")
print("Examples of duplicate apps:", duplicate_android_apps[:10])
# The Google Play data set contains 1,181 duplicates. We will utilize the Reviews column (`app[3]`) as a criterion for isolating the unique apps. The higher the number of reviews, the more recent the data should be. Therefore, we can systematically isolate the unique apps by retaining the rows with the highest number of reviews and removing the other entries for any given app.
reviews_max = {}
for app in android_data:
name = app[0]
n_reviews = float(app[3])
if name in reviews_max and reviews_max[name] < n_reviews:
reviews_max[name] = n_reviews
elif name not in reviews_max:
reviews_max[name] = n_reviews
print(len(reviews_max))
# The number of unique apps with the highest number of reviews for any given app in the Google Play data set is 9,659. Let's check if this aligns with our expected value by subtracting the number of rows (`len(android_data)`) with the number of duplicates (1,181).
print("Expected Length:", len(android_data) - 1181)
# Our expected length matches the length of the `reviews_max` dictionary. Let's now remove the duplicate rows. In the code cell below:
# - We start by intializing two empty lists, `android_clean` and `already_added`.
# - We loop through the `android_data` data set, and for each iteration:
# - We extract the name of the app and the number of reviews.
# - We add the current row (`app`) to `android_clean`, and the app name (`name`) to `already_added` if:
# - The number of reviews in the current row equals the number of reviews in reviews_max dictionary for the corresponding app; and if:
# - The name of the app in the current row is not in the `already_added` list. This supplementary condition is needed to account for cases in which two or more of the same app has the same number of reviews.
android_clean = []
already_added = []
for app in android_data:
name = app[0]
n_reviews = float(app[3])
if (n_reviews == reviews_max[name]) and (name not in already_added):
android_clean.append(app)
already_added.append(name)
print(len(android_clean))
# ### Removing Non-English Apps
# We are only interested in isolating free, English apps, which means the names of the apps should only include letters from the English alphabet, digits from 0 - 9, punctuation marks (!, ?, ., etc), and other symbols (*, +, etc).
# All these characters that are specific to the English language are encoded using the ASCII standard. Each ASCII character has a corresponding number between 0 and 127 associated with it, and we can leverage that to build a function that checks an app name and tells us whether it contains non-ASCII characters.
# We built a function `english_check` and used the built-in function `ord()` to check for the corresponding ASCII encoding number of each character and determine if the string in our input satisfies our criteria for an English name.
# To account for app names that contain emojis or other symbols that fall outside our ASCII range, we'll only omit an app if its name contains more than three non-ASCII characters.
def english_check(string):
ascii = 0
for character in string:
if ord(character) > 127:
ascii += 1
if ascii > 3:
return False
else:
return True
print(english_check("Instachat 😜"))
print(english_check("Docs To Go™ Free Office Suite"))
print(english_check("爱奇艺PPS -《欢乐颂2》电视剧热播"))
# In the code cell below: After accounting for and removing non-English apps with more than three non-ASCII characters in the app name, we are left with 9,614 Android apps and 6,183 iOS apps.
android_english = []
apple_english = []
for app in android_clean:
name = app[0]
if english_check(name):
android_english.append(app)
for app in apple_data:
name = app[1]
if english_check(name):
apple_english.append(app)
explore_data(android_english, 0, 3, True)
print("\n")
explore_data(apple_english, 0, 3, True)
# ### Isolating Free Apps
# Our data sets contain both free and non-free apps, and we'll need to isolate only the free apps for our analysis. Below, we isolate the free apps for both our data sets.
android_final = []
apple_final = []
for app in android_english:
price = str(app[7])
if price == "0":
android_final.append(app)
for app in apple_english:
price = str(app[4])
if price == "0.0":
apple_final.append(app)
print(len(android_final))
print(len(apple_final))
# After isolating for free apps, we are left with 8,864 Android apps and 3,222 iOS apps that are ready for analysis.
# So far, we have accomplished the following data cleaning tasks:
# - Remove inaccurate data
# - Remove duplicate app entries
# - Remove non-English apps
# - Remove paid apps
# As mentioned in the introduction, our goal is to identify the types of apps that are most likely to attract the most users. Because our main source of revenue consists of in-app ads, attracting the most users in apps directly affects our revenue. To minimize risk and overhead, our validation strategy for an app idea consists of three steps:
# 1. Build a minimal version of the app for Android, and add it to Google Play.
# 2. If the app is well received by users, we develop it further.
# 3. If the app is profitable from in-app ads after six months, we build an iOS version of the app and add it to the App Store.
# ### Data Analysis: Most Common Apps by Genre
# We will begin our analysis of the data by determining the most common app genres by market. To do this, we will build two functions to analyze frequency tables:
# - One function to generate frequency tables that show percentages
# - One function to display the percentages in descending order
def freq_table(dataset, index):
table = (
{}
) # dictionary with count of unique values in a column that is specified by the index value.
total = 0
for app in dataset:
column = app[index]
total += 1
if column in table:
table[column] += 1
else:
table[column] = 1
percentage = {}
for key in table:
percent = (table[key] / total) * 100
percentage[key] = percent
return percentage
def display_table(dataset, index):
table = freq_table(dataset, index)
table_display = []
for key in table:
key_val_as_tuple = (table[key], key)
table_display.append(key_val_as_tuple)
table_sorted = sorted(table_display, reverse=True)
for entry in table_sorted:
print(entry[1], ":", entry[0])
# ### Most Common Apps by Genre in the App Store
# The most common genre of free English apps in the App Store is **Games** (58.16%). The next most common is **Entertainment** (7.88%). Among those that are in English and free, the App Store appears to have a significantly greater number of apps designed for fun (Games, Entertainment, Social Networking) than apps designed for practical uses (Education, Shopping, Utilities, Productivity, Lifestyle, etc). However, a large number of apps designed for fun does not imply that there is also a large customer base for those apps.
display_table(apple_final, -5) # prime_genre
# ### Most Common Apps by Genre in Google Play
# The most common genre of free English apps in Google Play is **Tools** (8.45%), followed by **Entertainment** (6.07%), and **Education** (5.35%). Compared to the App Store market, Google Play offers a more extensive variety of apps designed for practical use.
display_table(android_final, -4) # Genres
display_table(android_final, 1) # Category
# ### Most Popular Apps by Genre on Google Play
# Now, let's determine which kind of apps has the most users. For the Google Play data set, we will use the **Installs** column. This information is missing for the App Store data set, so we will use the total number of user ratings `rating_count_tot` as a proxy for the number of users.
display_table(android_final, 5) # Installs column
# A problem with the `Installs` column is that it does not precisely reflect the number of installs by users and is instead rounded down from open-ended numbers (100+, 1,000+, 5,000+, etc). However, for our purposes we don't need precision but to get an idea of the type of apps that attract the most users. For our analysis, we will consider an app with 100,000+ installs as 100,000, 1,000,000+ installs as 1,000,000, and so on. Additionally, in order to perform calculations, the values in the `Installs` column will be converted into `float`, which entails removing commas and plus sign characters.
android_categories = freq_table(android_final, 1)
for category in android_categories:
total = 0
len_category = 0
for app in android_final:
category_app = app[1]
if category_app == category:
n_installs = app[5]
n_installs = n_installs.replace(",", "")
n_installs = float(n_installs.replace("+", ""))
total += n_installs
len_category += 1
avg_n_installs = total / len_category
print(category, ":", avg_n_installs)
# In Google Play, the app category with the greatest number of installs is **Communication** (38,456,119 installs). The app category with the second largest number of installs is **Video Players** (24,727,872 installs).
for app in android_final:
if app[1] == "VIDEO_PLAYERS":
print(app[0], ":", app[5])
# ### Most Popular Apps by Genre on the App Store
apple_genres = freq_table(apple_final, -5)
for genre in apple_genres:
total = 0
len_genre = 0
for app in apple_final:
genre_app = app[-5]
if genre_app == genre:
n_ratings = float(app[5])
total += n_ratings
len_genre += 1
avg_n_ratings = total / len_genre
print(genre, ":", avg_n_ratings)
# The **Navigation** genre has the highest average number of user ratings (86,090) in the App Store. However, this figure is heavily influenced by Waze and Google Maps which make up about half a million user ratings. The genre with the second highest average number of user ratings is **Reference** (74,942), with nearly a million user ratings belonging to the Bible app.
for app in apple_final:
if app[-5] == "Navigation":
print(app[1], ":", app[5])
for app in apple_final:
if app[-5] == "Reference":
print(app[1], ":", app[5])
|
# This dataset Cotains the data of 1000 big Corporations in the world published annualy by Fortune Magazine.This is a kernel in Process and I will be updating the kernel in coming days.
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
import matplotlib.pyplot as plt
# **Importing data with Pandas**
df = pd.read_csv("../input/fortune1000/fortune1000.csv", index_col="Rank")
df.head()
# We have used rank column as our index
# **Groupby**
sectors = df.groupby("Sector")
sectors
# **Type**
type(sectors)
sectors
# **Number of Groupings**
len(sectors)
# There are 21 industry sectors in the dataset
# **Other method to count the number of Sectors**
df.nunique()
df["Sector"].nunique()
# **Sizing the Grouping**
sectors.size().sort_values(ascending=False).plot(kind="bar")
# So here we have go the information of number of companies in each sector.For companies are 139 are there in financial sector and least 15 nos being in apparel sector.
# **Extrating first row of every Sector**
sectors.first()
# **Getting the information of Last row of all sectors**
sectors.last()
# Now we have information on the last row of all the sectors
# **Groups on Groupby**
sectors.groups
df.loc[24]
# Here we have got the index number of all the rows in each group.We can see that the index 2 falls in the Aerospace and Defense.
# **Get Group**
sectors.get_group("Energy")
# Using the get_group command we are able to get the rows values in a particular sectors.
# **Max**
sectors.max()
# We can see that we have got the company arranged alphabetically.W is max alphebatically so we can see company like Woodward getting reported.
# **Min**
sectors.min()
# We can see that we have got the company arranged alphabetically.A is min alphebatically so we can see company like B/E Aerospace getting reported.
# **Sum**
sectors.sum()
# So for numerical columns we have got the sum of the Revenue,Profits and Employees for each sector
# **Mean**
sectors.mean()
# So for numerical columns we have got the mean of the Revenue,Profits and Employees for each sector
sectors["Employees"].sum()
sectors[["Profits", "Employees"]].sum()
# **Grouping by Multiple Columns**
sector = df.groupby(["Sector", "Industry"])
sector.size()
# Here in addition to sector our data is grouped by industry too.
# **Sum of Multi Groupby**
sector.sum()
sector["Revenue"].sum()
sector["Employees"].mean()
# We can do the sum,max,min,mean etc operation on a list of columns as shown above.
# **Add Method**
sectors.agg({"Revenue": "sum", "Profits": "sum", "Employees": "mean"})
# By using a dictionary on groupy by we can have prefered operation on each column usiing agg method
# **Multiple operations to Multiple Columns**
sectors.agg(["size", "sum", "mean"])
|
import pandas as pd
train = pd.read_csv("/kaggle/input/dog-breed-identification/labels.csv")
train.head()
train["id"] = train["id"] + ".jpg"
train.head()
train["id"] = "/kaggle/input/dog-breed-identification/train/" + train["id"]
train.head()
# sort label
labels = train.sort_values("breed")
labels.head()
# take unique labels
class_names = list(labels.breed.unique())
class_names[:5]
import shutil, os
newpath = r"./train"
if not os.path.exists(newpath):
os.makedirs(newpath)
# creating subfolders
for c in class_names:
dest = r"./train/" + str(c)
os.makedirs(dest)
for i in list(labels[labels["breed"] == c]["id"]): # Image Id
get_image = os.path.join(
"/kaggle/input/dog-breed-identification/train", i
) # Path to Images
move_image_to_cat = shutil.copy(get_image, dest)
newpath = r"./test"
if not os.path.exists(newpath):
os.makedirs(newpath)
import glob
src_dir = "/kaggle/input/dog-breed-identification/test"
dst_dir = "./test"
for jpgfile in glob.iglob(os.path.join(src_dir, "*.jpg")):
shutil.copy(jpgfile, dst_dir)
test = pd.read_csv("/kaggle/input/dog-breed-identification/sample_submission.csv")
test = test[["id"]]
test.head()
test["id"] = test["id"] + ".jpg"
test.head()
sol = pd.read_csv("/kaggle/input/dog-breed-identification/sample_submission.csv")
sol.head()
sol["id"] = sol["id"] + ".jpg"
sol.head()
train.to_csv("./train.csv", index=False)
test.to_csv("./test.csv", index=False)
sol.to_csv("./sample_submission.csv", index=False)
|
# **Revolut Analytical Challenge**
# Congratulations on making it to the Challenge stage of the application process! The goal of this
# home task is to assess your ability to generate great ideas and solve problems that our team
# has faced in the past!
# There are two independent tasks in this challenge:
# * Improving KYC (main task)
# * Catching fraud (bonus task)
# Your submission should contain:
# 1. Improving KYC report in pdf / doc / html (up to 1 page or 5 slides, optional appendix)
# 1. Optional: Catching fraud report in pdf / doc / html (up to 2 pages or 3 slides)
# 1. Optional: Supporting materials (calculation files, additional analysis, etc)
# If the files are too big please upload to external storage (gdrive, dropbox, etc.). Make sure to
# allow public access and send the coordinator the link.
# Deadline: 7 Days from the date you received the email
# ### Task definition
# **Task 1: Improving KYC**
# As a financial institution regulated by the FCA, Revolut has the obligation to verify the identity of all customers who want to open a Revolut account. Each prospective customer has to go through a Know Your Customer (KYC) process by submitting a government-issued photo ID and a facial picture of themself to our partner, Veritas. Veritas then would perform 2 checks:
# * Document check: To verify that the photo ID is valid and authentic;
# * Facial Similarity check: To verify that the face in the picture is the same with that on the submitted ID.
# The customer will ‘pass’ the KYC process and get onboarded if the results of both Document and Facial Similarity checks are ‘clear’. If the result of any check is not ‘clear’, the customer has to submit all the photos again.
# The “pass rate” is defined as the number of customers who pass both the KYC process divided by the number of customers who attempt the process. Each customer has up to 2 attempts.The pass rate has decreased substantially in the recent period. Please write a report that outlines the root causes and solutions.
# Relevant files:
# * facial_similarity_reports.csv - Reports of all Facial Similarity checks
# * doc_reports.csv - Reports of all Document checks
# * veritas.html - The API documentation of Veritas explaining some terms used in the reports (you might need to download it to your computer and open it with a browser)
# The candidate is free to use Excel or any scripting language to parse and analyse the data.
# Please show all your work (including your code if applicable) and assumptions.
# # Importing the libraries needed
from mpl_toolkits.mplot3d import Axes3D
import numpy as np # linear algebra
import os # accessing directory structure
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
from sklearn.preprocessing import StandardScaler
import matplotlib.pyplot as plt # plotting
import seaborn as sns # visualization tool
import json # for parse "properties" parameter
import warnings
warnings.filterwarnings("ignore")
# ### We add functions for mapping values and preparing our dataframes
# Function for maping datasets.
# We will change all 'consider' value to 1, and 'clear' = 0 for **map_encode_docs** list
def result_encode(features, dataset):
for feature in features:
dataset[feature] = dataset[feature].map(result_map)
# function for clean and prepare dataframe
def prepare_data(dataframe):
dataframe.created_at = pd.to_datetime(
dataframe.created_at, errors="coerce", format="%Y-%m-%d %H:%M:%S"
)
dataframe.fillna(np.nan)
map_encode_all = [
"result_doc",
"visual_authenticity_result_doc",
"image_integrity_result",
"police_record_result",
"compromised_document_result",
"face_detection_result",
"image_quality_result",
"supported_document_result",
"conclusive_document_quality_result",
"colour_picture_result",
"data_validation_result",
"data_consistency_result",
"data_comparison_result",
"face_comparison_result",
"facial_image_integrity_result",
"visual_authenticity_result_face",
"result_face",
]
result_map = {"clear": 0, "unidentified": 1, "consider": 1}
# ## Reading **doc_reports.csv** (from veritas documentation)
# The document report is composed of data integrity, visual authenticity and police record checks. It checks the internal and external consistency of the most recent identity document provided by the applicant to identify potential discrepancies.
# In addition, any data extracted from the document through OCR is returned in the properties attribute.
# The document report combines software and an expert team to maximise fraud detection. The majority of documents will be processed instantly. However, when document analysis falls back to expert review, the report status will be delivered asynchronously via webhook notifications.
# **Expert review is required when we encounter images that use sophisticated counterfeiting techniques, or the image is of poor quality (blurred, low resolution, obscured, cropped, or held at an unreadable angle).**
# **BREAKDOWN**
# * **visual_authenticity** - Asserts whether visual, non-textual, elements are correct given the type of document
# * **image_integrity** - Asserts whether the document was of sufficient quality to verify
# * **data_validation** - Asserts whether algorithmically-validatable elements are correct e.g. MRZ lines and document numbers
# * **data_consistency** - Asserts whether data represented in multiple places on the document is consistent e.g. between MRZ lines and OCR extracted text on passports
# * **data_comparison** - Asserts whether data on the document is consistent with data provided by an applicant (either through Veritas’s applicant form or when creating an applicant through the API)
# * **police_record** - Asserts whether the document has been identified as lost, stolen or otherwise compromised
# * **compromised_document** - Asserts whether the image of the document has been found in our internal database of compromised documents
# A result of clear in the conclusive_document_quality breakdown of image_integrity will assert if the document was of enough quality to be able to perform a fraud inspection. A result of consider will mean that even if sub breakdowns of visual_authenticity fail, we cannot positively say the document is fraudulent or not (in cases such as parts of the document are not visible).
#
document = pd.read_csv(
"../input/kyc-challenge/doc_reports.csv", delimiter=",", index_col=0
)
document.dataframeName = "doc_reports.csv"
prepare_data(document)
document.head(2)
# ## Reading **facial_similarity_reports.csv** (from veritas documentation)
# The facial similarity check will compare the most recent live photo or live video provided by the applicant to the photo in the most recent identity document provided.
# **BREAKDOWN**
# * **face_comparison** - Asserts whether the face in the document matches the face in the live photo or live video
# * **image_integrity** - Asserts whether the quality of the uploaded files and the content contained within them were sufficient to perform a face comparison
# * **visual_authenticity** - Asserts whether the live photo or live video is not a spoof (such as photos of printed photos or photos of digital screens)
# **PROPERTIES**
# The score property is a number between 0 and 1 that expresses how similar the two faces are, where 1 is a perfect match. If the report is completed manually or image integrity fails then the score property will not be present. The score only measures how similar the faces are, and does not make an assessment of the nature of the photo or video. If tampering (such as photos of printed photos or photos of digital screens) is detected the applicant will be rejected independently of the facial similarity score.
# **STANDARD**
# The standard variant uses live photos. The photo needs to be a live photo taken at the time of check submission, so that it can assess whether the holder of the identity document is the same person as the one on the document.
faces = pd.read_csv(
"../input/kyc-challenge/facial_similarity_reports.csv", delimiter=",", index_col=0
)
faces.dataframeName = "facial_similarity_reports.csv"
prepare_data(faces)
faces.head(2)
# ## Cleaning dataframes and dropping columns
mb = pd.merge(
document,
faces,
on="attempt_id",
how="left",
suffixes=("_doc", "_face"),
validate="one_to_one",
)
mb = mb.drop(
[
"user_id_face",
"created_at_face",
"sub_result",
"properties_face",
],
axis=1,
)
mb.head()
# ### Our goal is to find out which of the verification stages most often leads to the 'consider' for the **result_doc** column
# ### Calculate the proportion of **clear/consider** users for all time
mb.result_doc.value_counts(normalize=True).plot.bar()
# ## Mapping string values
# We will change all 'consider' value to 1, and 'clear' = 0 for **map_encode_all** list
result_encode(map_encode_all, mb)
mb.fillna(0, inplace=True)
mb.head()
# # Now dataframe 'mb' is ready to explore
# ## How many people register into the system per time?
plt.rcParams["figure.figsize"] = (18, 7)
mb.groupby(pd.Grouper(key="created_at_doc", freq="D"))["user_id_doc"].count().plot()
# ### Getting correlation dataframe for values
data_corr = mb.corr(method="pearson")
data_corr = data_corr.apply(lambda x: [y if y >= 0.3 else np.nan for y in x])
# correlation map plotting
f, ax = plt.subplots(figsize=(13, 13))
sns.heatmap(
data_corr, annot=True, linewidths=5, fmt=".3f", ax=ax, cmap="Reds", center=0.8
)
plt.show()
print(data_corr.apply(lambda x: [y if y >= 0.3 else np.nan for y in x]))
data_corr
# ### For **'result_doc'** column very high correlation coefficient **(0,935)** have next stage of KYC:
# * **image_integrity_result** - Asserts whether the document was of sufficient quality to verify.
# ### For **'result_face'** column very high correlation coefficient **(0,951)** have next stage of KYC:
# * **facial_image_integrity_result** - Asserts whether the quality of the uploaded files and the content contained within them were sufficient to perform a face comparison
# ### **These parameters have a great influence on the final KYC result.**
# # The failure rate for the individual stages of verification, grouped by day.
# ### As we can see, in mid-October, the failure rate for "image_integrity_result" increased to 1. This combined with other indicators suggests a system wide failure of some sort. Further investigation in that direction required
mb.groupby(pd.Grouper(key="created_at_doc", freq="D"))[map_encode_all].mean().plot()
suspected_params = [
"image_integrity_result",
"facial_image_integrity_result",
"image_quality_result",
]
mb.groupby(pd.Grouper(key="created_at_doc", freq="D"))[suspected_params].mean().plot()
# Now we need to parse '**properties**' column. All '**None**' results we'll replacing to '**NaN**', delete all bad symbols and replace single quotes to double - as JSON library required.
mb.properties_doc = mb.properties_doc.apply(lambda row: row.replace("None", '"NaN"'))
mb["properties_doc"] = mb.properties_doc.apply(
lambda x: x.strip("'<>()").replace("'", '"')
)
# cleaning
mb.properties_doc = mb.properties_doc.apply(lambda row: row.replace("None", '"NaN"'))
mb["properties_doc"] = mb.properties_doc.apply(
lambda x: x.strip("'<>()").replace("'", '"')
)
# loading
mb["properties_doc"] = mb["properties_doc"].apply(json.loads, strict=False)
# parsing
mb = mb.drop("properties_doc", 1).assign(
**pd.DataFrame(mb.properties_doc.values.tolist())
)
# and get dates from new columns
mb.date_of_expiry = pd.to_datetime(
mb.date_of_expiry, errors="coerce", format="%Y-%m-%d"
)
mb.issuing_date = pd.to_datetime(mb.issuing_date, errors="coerce", format="%Y-%m")
# Sort Dataframe by date
mb = mb.set_index(keys="created_at_doc")
mb.head()
# ## 10 Most often **image_integrity_result** failure grouped by **issuing country**
mb[(mb["result_doc"] == 1) & (mb["image_integrity_result"] == 1)][
"issuing_country"
].value_counts()[:10].plot(kind="bar")
# ### Most often **image_integrity_result** failure grouped by **nationality**
mb[(mb["result_doc"] == 1) & (mb["image_integrity_result"] == 1)][
"nationality"
].value_counts()[:10].plot(kind="bar")
# ### Most often **image_integrity_result** failure grouped by **document_type**
mb[(mb["result_doc"] == 1) & (mb["image_integrity_result"] == 1)][
"document_type"
].value_counts()[:10].plot(kind="bar")
# ### We can see that users make more than 2 attempts at the verification which is not allowed
attempts = mb.groupby(["user_id_doc"])["attempt_id"].count().reset_index(name="count")
attempts.groupby(["count"])["user_id_doc"].count()[:10].plot(kind="bar")
# # Let's take a closer look at the problem period.
suspect_data = mb["2017-10-10":"2017-10-25"]
# ## Most often **image_integrity_result** failure grouped by **issuing country**
suspect_data[suspect_data["image_integrity_result"] == 1][
"issuing_country"
].value_counts()[:10].plot(kind="bar")
# ## Most often **image_integrity_result** failure grouped by **nationality**
suspect_data[suspect_data["image_integrity_result"] == 1]["nationality"].value_counts()[
:10
].plot(kind="bar")
# ## Most often **image_integrity_result** failure grouped by **document_type**
suspect_data[suspect_data["image_integrity_result"] == 1][
"document_type"
].value_counts()[:10].plot(kind="bar")
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import datetime as dt, time
from operator import itemgetter
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression, LogisticRegression, BayesianRidge
from sklearn.preprocessing import (
PolynomialFeatures,
StandardScaler,
OneHotEncoder,
LabelEncoder,
)
from sklearn.pipeline import make_pipeline
from sklearn.neighbors import KNeighborsClassifier
from sklearn import metrics
input_data = pd.read_csv("../input/avocado-prices/avocado.csv")
print(input_data.head())
regiongroups_data = {}
region_names = []
date_max_limit = dt.datetime.strptime("2020-01-01", "%Y-%m-%d")
date_max_limit = dt.datetime.timetuple(date_max_limit)
date_max_limit = time.mktime(date_max_limit) * 0.001
# This time we convert the data of interest from pandas frame into list of tuples.
# Dates are covnerted into seconds.
for i in range(len(input_data)):
new_line = input_data.iloc[i]
region = new_line["region"]
# if region == "WestTexNewMexico":
# continue
date_seconds = dt.datetime.strptime(new_line["Date"], "%Y-%m-%d")
date_seconds = dt.datetime.timetuple(date_seconds)
date_seconds = time.mktime(date_seconds)
ave_price = new_line["AveragePrice"]
total_avocados = new_line["Total Volume"]
avo_type = new_line["type"]
if region not in regiongroups_data:
regiongroups_data[region] = []
region_names.append(region)
t = float(date_seconds) # /date_max_limit
regiongroups_data[region].append((t, ave_price, total_avocados, avo_type))
# Now we will generate the plots for each city showing average price behavior for conventional and organic avocados over total volumes vs. observation time (consder it arbitrary, repsented in seconds).
# * We will also atempt to use X_in, y_in are used for attempt to predict the city as:
# X_in.append([price_weighted, total_volume]) where 'price_weighted', 'total_volume' are the average weigthed price and total volume correspondingly.
# y_in.append(label) where labels identify the city.
#
# * 'np_data_by_city' list is used for estimating correlation between the cities on the behavior of 'price_weighted'
fig, a = plt.subplots(6, 9, figsize=(40, 55))
n = 0
X_in = []
y_in = []
np_data_by_city = np.empty(
[len(region_names), int(len(regiongroups_data["Albany"]) / 2)]
) # len town, #len of prices
for i in range(6):
for j in range(9):
idx = 9 * n + j
label = region_names[idx]
a[i][j].set_title(label)
date_conv_idx = [
regiongroups_data[label][k][0]
for k in range(len(regiongroups_data[label]))
if regiongroups_data[label][k][3] == "conventional"
]
date_orga_idx = [
regiongroups_data[label][k][0]
for k in range(len(regiongroups_data[label]))
if regiongroups_data[label][k][3] == "organic"
]
n_orga = len(date_orga_idx)
n_conv = len(date_conv_idx)
indices, date_idx_sorted = zip(
*sorted(enumerate(date_conv_idx), key=itemgetter(1))
)
indices_sh, date_sh_idx_sorted = zip(
*sorted(enumerate(date_orga_idx), key=itemgetter(1))
)
ave_price_conv_idx_sorted = []
ave_price_orga_idx_sorted = []
vol_conv_idx_sorted = []
vol_orga_idx_sorted = []
shift = len(indices)
l = 0 # conv index
p = 0 # org index
for ix in range(shift):
vol_orga = 0
vol_conv = 0
if date_idx_sorted[l] != date_sh_idx_sorted[p]:
if n_orga > n_conv:
l -= 1
if n_conv > n_orga:
p -= 1
ave_price_conv_idx_sorted.append(regiongroups_data[label][indices[l]][1])
vol_conv = regiongroups_data[label][indices[l]][2]
ave_price_orga_idx_sorted.append(
regiongroups_data[label][indices_sh[p] + shift][1]
)
vol_orga = regiongroups_data[label][indices_sh[p] + shift][2]
p += 1
l += 1
total_volume = float(vol_orga + vol_conv)
price_weighted = (
float(
(
ave_price_conv_idx_sorted[-1] * vol_conv
+ ave_price_orga_idx_sorted[-1] * vol_orga
)
)
/ total_volume
)
# X_in, y_in are used in attempts to predict the city
X_in.append([price_weighted, total_volume])
y_in.append(label)
# np_data_by_city is used for estimating correlation between the cities
np_data_by_city[idx, ix] = price_weighted
a[i][j].plot(date_idx_sorted, ave_price_conv_idx_sorted)
a[i][j].plot(date_idx_sorted, ave_price_orga_idx_sorted)
n += 1
# We can easily see that spikes affect price behavior most of the cities in a very similar manner. Though some of the are less affected. Let us try to isolate those cities.
# First we shall check if the average weighted prices vs. observation time have similar behavior.
fig, a = plt.subplots(6, 9, figsize=(40, 55))
n = 0
for i in range(6):
for j in range(9):
idx = 9 * n + j
label = region_names[idx]
a[i][j].set_title(label)
a[i][j].plot(range(len(np_data_by_city[idx, :])), np_data_by_city[idx, :])
n += 1
plt.show()
# We can see that 'price_weighted' preserves overall behavior seen with separate organic and conventional prices. Let us try perform a simple prediction using KNN and linear regression ti see if it could bring any result and worth continuing with it.
y_in = np.array(y_in)
scaler = StandardScaler(copy=True, with_mean=True, with_std=True)
X_in_s = scaler.fit_transform(X_in)
# withinout encoder
X_train, X_test, y_train, y_test = train_test_split(
X_in_s, y_in, test_size=0.2, shuffle=True
)
for nn in range(2, 50, 2):
model_knn = KNeighborsClassifier(n_neighbors=nn)
model_knn.fit(X_train, y_train)
y_pred = model_knn.predict(X_test)
print(
"KNN accuracy for nn= {} is {};".format(
nn, metrics.accuracy_score(y_test, y_pred)
)
)
# It has been seen that similar score has been reached with other tts. Not satisfying result is rather expected due do a very similar behavior of weighted prices in most of the cities.
yenc = LabelEncoder()
y_in_1d = np.reshape(y_in, (y_in.size, 1))
y_in_enc = yenc.fit_transform(y_in_1d)
print(y_in_enc)
scaler = StandardScaler(copy=True, with_mean=True, with_std=True)
X_in_s = scaler.fit_transform(X_in)
X_in_train, X_test, y_train, y_test = train_test_split(
X_in_s, y_in_enc, test_size=0.1, shuffle=True
)
print(X_in_train[0:1])
lr_model = LogisticRegression(
multi_class="multinomial", solver="lbfgs", penalty="l2", max_iter=500, tol=1e-6
)
lr_model.fit(X_in_train, y_train)
y_pred = lr_model.predict(X_test)
accuracy = np.sum(y_pred == y_test) / y_test.shape[0]
print("accuracy ", accuracy)
# The same is the case for the linear regression below, not a surprise..
# Now we will estimate correlation coefficients between the cities consedring weighted average prices
corrcoef = np.corrcoef(np_data_by_city)
plt.subplots(figsize=(20, 15))
heatm = sns.heatmap(
corrcoef,
cbar=True,
annot=True,
fmt=".2f", # annot_kws={'size':15},
yticklabels=region_names,
xticklabels=region_names,
)
plt.show()
# There are few locations less affected by common price trends. Those are BuffaloRochester, Pittsburgh and Syracuse..
# Check the same with clsutered heatmap.
clustm = sns.clustermap(corrcoef)
# clustm.ax_heatmap.set_xlabel(region_names)
# clustm.ax_heatmap.set_ylabel(region_names)
# plt.setp(clustm.ax_heatmap.get_ylabel(), rotation=0)
# clustm.ax_heatmap.set_ylabel(region_names, rotation=0)
plt.show()
print("Coeff corr all: ", corrcoef)
|
# Analysis of Pokémon Database
# Creator: alopez247
# Notebook Author: João Paulo Ribeiro dos Santos (joaopauloribsantos)
# This notebook is intended to apply some concepts and methods that I am learning. For this reason, that it will be updated frequently, until I can answer two questions that have always puzzled me about pokemons:
#
# What stats really define a legendary pokemon?
# Which attributes stands out most in my favorite pokemon?
#
#
# Libraries Import
# The following code expresses the main libraries that we will use on this notebook
import numpy as np
import pandas as pd
import sklearn as sk
import matplotlib.pyplot as plt
import seaborn as sns
from matplotlib.colors import ListedColormap
from IPython.display import Image
from sklearn import preprocessing
# Notebook Configurations
pd.set_option("display.max_columns", None)
# Database Import
# In the code below we are creating a dataframe from the pokemon database
df_pokemon = pd.read_csv("../input/pokemon/pokemon_alopez247.csv")
# Dataframe Operations/ Analysis
# Dataframe dimension
df_pokemon.shape
# 721 Rows and 23 Columns
# Columns of dataframe
df_pokemon.columns
# Columns Types
df_pokemon.dtypes
# Viewing the 3 first rows
df_pokemon.head(3)
# Viewing the 3 last rows
df_pokemon.tail(3)
# The column 'Number' on the dataframe is like the pokemon ID, so we can consider it the index of the dataframe
df_pokemon.set_index(["Number"], inplace=True)
# Handling Null/ Nan Values
# Verifying if exists NAN values on dataset
null_columns = df_pokemon.columns[df_pokemon.isnull().any()]
df_pokemon[null_columns].isnull().sum()
# Percentage of Null/ Nan Values per column in dataframe
df_pokemon[null_columns].isnull().sum() * 100 / len(df_pokemon)
# Creating a HeatMap plot to show the null of all values in the entire dataframe
plt.figure(figsize=(20, 10))
pl = sns.heatmap(df_pokemon.isnull(), cmap="Greens", cbar=False)
pl.set_xticklabels(pl.get_xticklabels(), rotation=30)
plt.show()
# Veifying types of data in column Type_2
df_pokemon["Type_2"].value_counts(dropna=False)
# Veifying types of data in column Pr_Male
df_pokemon["Pr_Male"].value_counts(dropna=False)
# Veifying types of data in column Egg_Group_2
df_pokemon["Egg_Group_2"].value_counts(dropna=False)
# According to website bulbapedia (https://bulbapedia.bulbagarden.net/wiki/Egg_Group):
## Egg Groups are categories which determine which Pokémon are able to interbreed.
## The concept was introduced in Generation II, along with breeding. Similar to types,
## a Pokémon may belong to either one or two Egg Groups
# Replacing null values
df_pokemon["Egg_Group_2"].fillna("Undiscovered", inplace=True)
# According to website bulbapedia (https://bulbapedia.bulbagarden.net/wiki/%3F%3F%3F_(type):
## The ??? type is a type that exists only in Generations II, III, and IV.
## It was removed in the Generation V games and has not returned.
# Replacing null values
df_pokemon["Type_2"].fillna("???", inplace=True)
# Checking more about the column 'Pr_Male'
# Pr_Male = Probability of a pokemon being male
df_pokemon[df_pokemon["Pr_Male"].isnull()].loc[
:, ["Name", "Type_1", "Type_2", "isLegendary", "hasGender"]
]
# The pokemons that hasn't gender are the same pokemons who doesn't have probability of being male
df_pokemon[df_pokemon["hasGender"] == False & df_pokemon["Pr_Male"].isnull()].loc[
:, ["Name", "Type_1", "Type_2", "isLegendary", "hasGender"]
]
df_pokemon.columns
# As the variable 'Pr_Male' is totally dependent on the variable 'Has_Gender',
# and until that moment there is no reason to consider it in the analysis,
# that the variable will be disregarded in a new dataframe.
# Creating the new Dataframe
df_pokemon_an_01 = df_pokemon.drop(["Pr_Male"], axis=1)
df_pokemon_an_01.head(3)
# Exploratory Data Analysis (EDA)
df_pokemon_eda = df_pokemon_an_01.drop(["Name"], axis=1)
# Show the main dataframe statiscs
df_pokemon_eda.describe()
# The variables 'Name' and describes only categorical pokemon characteristics,
# as well as the number of football player's shirts
# The previous code showed the boxplot of all variables / columns.
plt.figure(figsize=(20, 15))
ax = sns.boxplot(data=df_pokemon_eda)
ax.set_xticklabels(pl.get_xticklabels(), rotation=30)
plt.show()
# Generating a table with the correlation of all variables
df_pokemon_eda[
[
"Total",
"HP",
"Attack",
"Defense",
"Sp_Atk",
"Sp_Def",
"Speed",
"Generation",
"Height_m",
"Weight_kg",
"Catch_Rate",
]
].corr()
# As we saw earlier, the correlation between some variables is significantly weak, however,
# there are some columns with a relatively high correlation, such as 'Total' and 'Attack.'
ax = sns.lmplot(x="Attack", y="Total", data=df_pokemon_eda)
plt.show()
# Detecting the outliers
# In this section, we will create a function that returns the main data related to outliers
def fn_validate_catching_outliers_values(p_df_dataframe, p_column):
"""
Description:
Validates information related to the
dataframe and its column, before proceeding
with the function 'fn_catching_outliers'.
Keyword arguments:
p_df_dataframe -- the dataframe
p_column -- the dataframe column
Return:
None
Exception:
Validates that the dataframe is empty;
Validates whether the column exists on the dataframe;
Validates whether the column is a numeric type
"""
if p_df_dataframe.empty:
raise Exception("The dataframe is empty")
if p_column not in p_df_dataframe.columns:
raise Exception("The column does not exist in the dataframe")
if not np.issubdtype(p_df_dataframe[p_column].dtype, np.number):
raise Exception("The informed column doesn't have the numeric type.")
def fn_catching_outliers(p_df_dataframe, p_column):
"""
Description:
Function that locates outliers in an informed dataframe.
Keyword arguments:
p_df_dataframe -- the dataframe
p_column -- the dataframe column
Return:
df_with_outliers -- Dataframe with the outliers located
df_without_outliers -- Dataframe without the outilers
Exception:
None
"""
# Check if the information passed is valid.
fn_validate_catching_outliers_values(p_df_dataframe, p_column)
# Calculate the first and the third qurtile of the dataframe
quartile_1, quartile_3 = np.percentile(p_df_dataframe[p_column], [25, 75])
# Calculate the interquartile value
iqr = quartile_1 - quartile_3
# Generating the fence hig and low values
fence_high = quartile_3 + (1.5 * iqr)
fence_low = quartile_1 - (1.5 * iqr)
# And Finally we are generating two dataframes, onde with the outliers values and the second with the values within values
df_without_outliers = p_df_dataframe[
(p_df_dataframe[p_column] < fence_low) & (p_df_dataframe[p_column] > fence_high)
]
df_with_outliers = p_df_dataframe[
~p_df_dataframe.isin(df_without_outliers)
].dropna()
if df_with_outliers.empty:
print("No outliers were detected.")
return df_with_outliers, df_without_outliers
df_pokemon_out, _ = fn_catching_outliers(df_pokemon_eda, "Attack")
df_pokemon_out.head(3)
# To provide greater accuracy to the model, it will be necessary to apply some statistical methods to
# the categorical variables, such as 'dummies', 'label enconding', etc ...
# Identify the amount of unique data per non-numeric column.
df_pokemon_eda[df_pokemon_eda.select_dtypes(exclude=np.number).columns].nunique()
# Given that categorical variables / columns have more than 10 different types of values,
# it will be necessary to apply the scikit-leran method / function, label encondig.
encoder = preprocessing.LabelEncoder()
categorical_columns = [
"Type_1",
"Type_2",
"Color",
"Egg_Group_1",
"Egg_Group_2",
"Body_Style",
]
for col in categorical_columns:
df_pokemon_lb_encoding = encoder.fit_transform(df_pokemon_eda[col])
df_pokemon_eda["encoder_" + col] = pd.DataFrame(
df_pokemon_lb_encoding, columns=["encoder_" + col]
)
df_pokemon_eda.head(3)
df_pokemon_eda.dtypes
# Generating a table with the correlation of all variables
df_pokemon_eda[df_pokemon_eda.select_dtypes(exclude=["object"]).columns].corr()
|
# ## Cifar 10を AlexNetで実装
# > ImagenetをDLするのは実際に大変なので、実際に使われているモデルをcifar10用に一部修正して実験する。
#
# Imagenetのコードを変更する
import argparse
import os
import random
import shutil
import time
import warnings
import torch
import torch.nn as nn
import torch.nn.parallel
import torch.backends.cudnn as cudnn
import torch.distributed as dist
import torch.optim
import torch.multiprocessing as mp
import torch.utils.data
import torch.utils.data.distributed
import torchvision.transforms as transforms
import torchvision.datasets as datasets
import torchvision.models as models
import torchvision
model_names = sorted(
name
for name in models.__dict__
if name.islower() and not name.startswith("__") and callable(models.__dict__[name])
)
parser = argparse.ArgumentParser(description="PyTorch ImageNet Training")
# parser.add_argument('data', metavar='DIR',
# help='path to dataset')
parser.add_argument(
"-a",
"--arch",
metavar="ARCH",
default="resnet18",
choices=model_names,
help="model architecture: " + " | ".join(model_names) + " (default: resnet18)",
)
# Google colab のデフォルトで引数が存在しているようで、これを指定しないとエラーになる。
# スクリプトとして実行する場合は不要
parser.add_argument("-f", "--fdumy", default=None, help="dummy for google colab")
parser.add_argument(
"-j",
"--workers",
default=4,
type=int,
metavar="N",
help="number of data loading workers (default: 4)",
)
parser.add_argument(
"--epochs", default=90, type=int, metavar="N", help="number of total epochs to run"
)
parser.add_argument(
"--start-epoch",
default=0,
type=int,
metavar="N",
help="manual epoch number (useful on restarts)",
)
parser.add_argument(
"-b",
"--batch-size",
default=256,
type=int,
metavar="N",
help="mini-batch size (default: 256), this is the total "
"batch size of all GPUs on the current node when "
"using Data Parallel or Distributed Data Parallel",
)
parser.add_argument(
"--lr",
"--learning-rate",
default=0.1,
type=float,
metavar="LR",
help="initial learning rate",
dest="lr",
)
parser.add_argument("--momentum", default=0.9, type=float, metavar="M", help="momentum")
parser.add_argument(
"--wd",
"--weight-decay",
default=1e-4,
type=float,
metavar="W",
help="weight decay (default: 1e-4)",
dest="weight_decay",
)
parser.add_argument(
"-p",
"--print-freq",
default=10,
type=int,
metavar="N",
help="print frequency (default: 10)",
)
parser.add_argument(
"--resume",
default="",
type=str,
metavar="PATH",
help="path to latest checkpoint (default: none)",
)
parser.add_argument(
"-e",
"--evaluate",
dest="evaluate",
action="store_true",
help="evaluate model on validation set",
)
parser.add_argument(
"--pretrained", dest="pretrained", action="store_true", help="use pre-trained model"
)
parser.add_argument(
"--world-size",
default=-1,
type=int,
help="number of nodes for distributed training",
)
parser.add_argument(
"--rank", default=-1, type=int, help="node rank for distributed training"
)
parser.add_argument(
"--dist-url",
default="tcp://224.66.41.62:23456",
type=str,
help="url used to set up distributed training",
)
parser.add_argument(
"--dist-backend", default="nccl", type=str, help="distributed backend"
)
parser.add_argument(
"--seed", default=None, type=int, help="seed for initializing training. "
)
parser.add_argument("--gpu", default=None, type=int, help="GPU id to use.")
parser.add_argument(
"--multiprocessing-distributed",
action="store_true",
help="Use multi-processing distributed training to launch "
"N processes per node, which has N GPUs. This is the "
"fastest way to use PyTorch for either single node or "
"multi node data parallel training",
)
print("oooooooooKKKKKKKKKKKKKK")
best_acc1 = 0
def main():
args = parser.parse_args()
if args.seed is not None:
random.seed(args.seed)
torch.manual_seed(args.seed)
cudnn.deterministic = True
warnings.warn(
"You have chosen to seed training. "
"This will turn on the CUDNN deterministic setting, "
"which can slow down your training considerably! "
"You may see unexpected behavior when restarting "
"from checkpoints."
)
if args.gpu is not None:
warnings.warn(
"You have chosen a specific GPU. This will completely "
"disable data parallelism."
)
if args.dist_url == "env://" and args.world_size == -1:
args.world_size = int(os.environ["WORLD_SIZE"])
args.distributed = args.world_size > 1 or args.multiprocessing_distributed
ngpus_per_node = torch.cuda.device_count()
if args.multiprocessing_distributed:
# Since we have ngpus_per_node processes per node, the total world_size
# needs to be adjusted accordingly
args.world_size = ngpus_per_node * args.world_size
# Use torch.multiprocessing.spawn to launch distributed processes: the
# main_worker process function
mp.spawn(main_worker, nprocs=ngpus_per_node, args=(ngpus_per_node, args))
else:
# Simply call main_worker function
main_worker(args.gpu, ngpus_per_node, args)
def main_worker(gpu, ngpus_per_node, args):
global best_acc1
args.gpu = gpu
if args.gpu is not None:
print("Use GPU: {} for training".format(args.gpu))
if args.distributed:
if args.dist_url == "env://" and args.rank == -1:
args.rank = int(os.environ["RANK"])
if args.multiprocessing_distributed:
# For multiprocessing distributed training, rank needs to be the
# global rank among all the processes
args.rank = args.rank * ngpus_per_node + gpu
dist.init_process_group(
backend=args.dist_backend,
init_method=args.dist_url,
world_size=args.world_size,
rank=args.rank,
)
# create model
if args.pretrained:
print("=> using pre-trained model '{}'".format(args.arch))
model = models.__dict__[args.arch](pretrained=True)
else:
print("=> creating model '{}'".format(args.arch))
model = models.__dict__[args.arch]()
if args.distributed:
# For multiprocessing distributed, DistributedDataParallel constructor
# should always set the single device scope, otherwise,
# DistributedDataParallel will use all available devices.
if args.gpu is not None:
torch.cuda.set_device(args.gpu)
model.cuda(args.gpu)
# When using a single GPU per process and per
# DistributedDataParallel, we need to divide the batch size
# ourselves based on the total number of GPUs we have
args.batch_size = int(args.batch_size / ngpus_per_node)
args.workers = int((args.workers + ngpus_per_node - 1) / ngpus_per_node)
model = torch.nn.parallel.DistributedDataParallel(
model, device_ids=[args.gpu]
)
else:
model.cuda()
# DistributedDataParallel will divide and allocate batch_size to all
# available GPUs if device_ids are not set
model = torch.nn.parallel.DistributedDataParallel(model)
elif args.gpu is not None:
torch.cuda.set_device(args.gpu)
model = model.cuda(args.gpu)
else:
# DataParallel will divide and allocate batch_size to all available GPUs
if args.arch.startswith("alexnet") or args.arch.startswith("vgg"):
model.features = torch.nn.DataParallel(model.features)
model.cuda()
else:
model = torch.nn.DataParallel(model).cuda()
# define loss function (criterion) and optimizer
criterion = nn.CrossEntropyLoss().cuda(args.gpu)
optimizer = torch.optim.SGD(
model.parameters(),
args.lr,
momentum=args.momentum,
weight_decay=args.weight_decay,
)
# optionally resume from a checkpoint
if args.resume:
if os.path.isfile(args.resume):
print("=> loading checkpoint '{}'".format(args.resume))
if args.gpu is None:
checkpoint = torch.load(args.resume)
else:
# Map model to be loaded to specified single gpu.
loc = "cuda:{}".format(args.gpu)
checkpoint = torch.load(args.resume, map_location=loc)
args.start_epoch = checkpoint["epoch"]
best_acc1 = checkpoint["best_acc1"]
if args.gpu is not None:
# best_acc1 may be from a checkpoint from a different GPU
best_acc1 = best_acc1.to(args.gpu)
model.load_state_dict(checkpoint["state_dict"])
optimizer.load_state_dict(checkpoint["optimizer"])
print(
"=> loaded checkpoint '{}' (epoch {})".format(
args.resume, checkpoint["epoch"]
)
)
else:
print("=> no checkpoint found at '{}'".format(args.resume))
cudnn.benchmark = True
# Data loading code
# traindir = os.path.join(args.data, 'train')
# valdir = os.path.join(args.data, 'val')
normalize = transforms.Normalize(
mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]
)
# Datasetのみ cifar10に変更
transform = transforms.Compose(
[transforms.ToTensor(), transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5))]
)
train_dataset = torchvision.datasets.CIFAR10(
root="./data", train=True, download=True, transform=transform
)
val_dataset = torchvision.datasets.CIFAR10(
root="./data", train=False, download=True, transform=transform
)
# train_dataset = datasets.ImageFolder(
# traindir,
# transforms.Compose([
# transforms.RandomResizedCrop(224),
# transforms.RandomHorizontalFlip(),
# transforms.ToTensor(),
# normalize,
# ]))
if args.distributed:
train_sampler = torch.utils.data.distributed.DistributedSampler(train_dataset)
else:
train_sampler = None
train_loader = torch.utils.data.DataLoader(
train_dataset,
batch_size=args.batch_size,
shuffle=(train_sampler is None),
num_workers=args.workers,
pin_memory=True,
sampler=train_sampler,
)
val_loader = torch.utils.data.DataLoader(
val_dataset,
batch_size=args.batch_size,
shuffle=False,
num_workers=args.workers,
pin_memory=True,
)
if args.evaluate:
validate(val_loader, model, criterion, args)
return
for epoch in range(args.start_epoch, args.epochs):
if args.distributed:
train_sampler.set_epoch(epoch)
adjust_learning_rate(optimizer, epoch, args)
# train for one epoch
train(train_loader, model, criterion, optimizer, epoch, args)
# evaluate on validation set
acc1 = validate(val_loader, model, criterion, args)
# remember best acc@1 and save checkpoint
is_best = acc1 > best_acc1
best_acc1 = max(acc1, best_acc1)
if not args.multiprocessing_distributed or (
args.multiprocessing_distributed and args.rank % ngpus_per_node == 0
):
save_checkpoint(
{
"epoch": epoch + 1,
"arch": args.arch,
"state_dict": model.state_dict(),
"best_acc1": best_acc1,
"optimizer": optimizer.state_dict(),
},
is_best,
)
def train(train_loader, model, criterion, optimizer, epoch, args):
batch_time = AverageMeter("Time", ":6.3f")
data_time = AverageMeter("Data", ":6.3f")
losses = AverageMeter("Loss", ":.4e")
top1 = AverageMeter("Acc@1", ":6.2f")
top5 = AverageMeter("Acc@5", ":6.2f")
progress = ProgressMeter(
len(train_loader),
[batch_time, data_time, losses, top1, top5],
prefix="Epoch: [{}]".format(epoch),
)
# switch to train mode
model.train()
end = time.time()
for i, (images, target) in enumerate(train_loader):
# measure data loading time
data_time.update(time.time() - end)
if args.gpu is not None:
images = images.cuda(args.gpu, non_blocking=True)
target = target.cuda(args.gpu, non_blocking=True)
# compute output
output = model(images)
loss = criterion(output, target)
# measure accuracy and record loss
acc1, acc5 = accuracy(output, target, topk=(1, 5))
losses.update(loss.item(), images.size(0))
top1.update(acc1[0], images.size(0))
top5.update(acc5[0], images.size(0))
# compute gradient and do SGD step
optimizer.zero_grad()
loss.backward()
optimizer.step()
# measure elapsed time
batch_time.update(time.time() - end)
end = time.time()
if i % args.print_freq == 0:
progress.display(i)
def validate(val_loader, model, criterion, args):
batch_time = AverageMeter("Time", ":6.3f")
losses = AverageMeter("Loss", ":.4e")
top1 = AverageMeter("Acc@1", ":6.2f")
top5 = AverageMeter("Acc@5", ":6.2f")
progress = ProgressMeter(
len(val_loader), [batch_time, losses, top1, top5], prefix="Test: "
)
# switch to evaluate mode
model.eval()
with torch.no_grad():
end = time.time()
for i, (images, target) in enumerate(val_loader):
if args.gpu is not None:
images = images.cuda(args.gpu, non_blocking=True)
target = target.cuda(args.gpu, non_blocking=True)
# compute output
output = model(images)
loss = criterion(output, target)
# measure accuracy and record loss
acc1, acc5 = accuracy(output, target, topk=(1, 5))
losses.update(loss.item(), images.size(0))
top1.update(acc1[0], images.size(0))
top5.update(acc5[0], images.size(0))
# measure elapsed time
batch_time.update(time.time() - end)
end = time.time()
if i % args.print_freq == 0:
progress.display(i)
# TODO: this should also be done with the ProgressMeter
print(
" * Acc@1 {top1.avg:.3f} Acc@5 {top5.avg:.3f}".format(top1=top1, top5=top5)
)
return top1.avg
def save_checkpoint(state, is_best, filename="checkpoint.pth.tar"):
torch.save(state, filename)
if is_best:
shutil.copyfile(filename, "model_best.pth.tar")
class AverageMeter(object):
"""Computes and stores the average and current value"""
def __init__(self, name, fmt=":f"):
self.name = name
self.fmt = fmt
self.reset()
def reset(self):
self.val = 0
self.avg = 0
self.sum = 0
self.count = 0
def update(self, val, n=1):
self.val = val
self.sum += val * n
self.count += n
self.avg = self.sum / self.count
def __str__(self):
fmtstr = "{name} {val" + self.fmt + "} ({avg" + self.fmt + "})"
return fmtstr.format(**self.__dict__)
class ProgressMeter(object):
def __init__(self, num_batches, meters, prefix=""):
self.batch_fmtstr = self._get_batch_fmtstr(num_batches)
self.meters = meters
self.prefix = prefix
def display(self, batch):
entries = [self.prefix + self.batch_fmtstr.format(batch)]
entries += [str(meter) for meter in self.meters]
print("\t".join(entries))
def _get_batch_fmtstr(self, num_batches):
num_digits = len(str(num_batches // 1))
fmt = "{:" + str(num_digits) + "d}"
return "[" + fmt + "/" + fmt.format(num_batches) + "]"
def adjust_learning_rate(optimizer, epoch, args):
"""Sets the learning rate to the initial LR decayed by 10 every 30 epochs"""
lr = args.lr * (0.1 ** (epoch // 30))
for param_group in optimizer.param_groups:
param_group["lr"] = lr
def accuracy(output, target, topk=(1,)):
"""Computes the accuracy over the k top predictions for the specified values of k"""
with torch.no_grad():
maxk = max(topk)
batch_size = target.size(0)
_, pred = output.topk(maxk, 1, True, True)
pred = pred.t()
correct = pred.eq(target.view(1, -1).expand_as(pred))
res = []
for k in topk:
correct_k = correct[:k].view(-1).float().sum(0, keepdim=True)
res.append(correct_k.mul_(100.0 / batch_size))
return res
if __name__ == "__main__":
main()
|
# ### Entrance Hall
# The Research Data analysis kernel is focused on exploring the relationship between various factors and surgery time. Surgery is a crucial and intricate medical procedure, and understanding the factors that affect its duration is important for improving patient outcomes and optimizing healthcare resources. This kernel utilizes various data analysis techniques, including data cleaning and preprocessing, exploratory data analysis, and statistical modeling, to gain insights into the key factors that affect surgery time. By examining these factors, we can identify areas for improvement in surgical procedures and ultimately enhance patient care.
# #### Import Series
# - Pandas:
# Pandas is an open-source Python library used for data manipulation and analysis. It provides easy-to-use data structures such as data frames and series, which allow users to work with and manipulate large datasets.
# - NumPy:
# NumPy is a Python library that provides support for large, multi-dimensional arrays and matrices, along with a range of mathematical functions to operate on them. NumPy is widely used in scientific computing and data analysis applications.
# - Plotly:
# Plotly is a Python library used to create interactive visualizations and data analysis tools. It provides a wide range of visualizations, including scatter plots, line graphs, heatmaps, and 3D graphs. Plotly is highly customizable and can be used for creating dashboards, reports, and other visualizations.
# Basic data library for python
import pandas as pd
import numpy as np
# Plotly : One of the popular visulation libarary on python
from plotly.offline import init_notebook_mode, iplot
import chart_studio.plotly.plotly as py
init_notebook_mode(connected=True)
import plotly.graph_objects as go
import plotly.express as px
from plotly.subplots import make_subplots
# Sklearn : Machine Learning Library
from sklearn.experimental import enable_iterative_imputer
from sklearn.impute import IterativeImputer
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# In the end we import the our data set
# Train data
data = pd.read_csv(
"/kaggle/input/borda-academy-transformed-train/Transformed_Train_Data.csv"
)
# I want to add some columns in here to inspect in the future
data["LenSurgeryCode"] = data[data.columns[11:18]].sum(axis=1)
data.LenSurgeryCode.replace(0, np.nan, inplace=True)
dict_transform = dict(
zip(
pd.Series(data.SurgeryName.unique()).dropna(),
range(len(data.SurgeryName.unique())),
)
)
data["SurgeryCode"] = data.SurgeryName.map(dict_transform)
# ### Data Inspectation Stage
# Data visualization and inspection play a crucial role in the data science workflow. By visualizing the data, we can quickly identify patterns,
# outliers, and trends that may not be immediately apparent in raw data. This helps us to make informed decisions about how to preprocess the data and
# select appropriate machine learning models. Moreover, data visualization can also be used to effectively communicate insights to non-technical stakeholders.
# By creating clear and visually appealing charts and graphs, we can help others understand complex data sets and make data-driven decisions.
# Therefore, data visualization and inspection are essential tools for exploratory data analysis and model interpretation.
# #### Missing Data Imputation
# Let's define the our str columns to drop it
str_col = ["ID", "DiagnosticICD10Code", "SurgeryGroup", "SurgeryName"]
# Now we can get rid of them
num_data = data.drop(str_col, axis=1)
# Little check
num_data.head(2)
# let's define our imputer
imputer_model = IterativeImputer()
# Wuick check unique value on our most nasty column
print(num_data.A1.unique())
# As you can see it has only one value. That causes normalization state errors because you can't decide something into zero.
num_data.drop("A1", axis=1, inplace=True)
# Now we can throw our data into our KNN imputer model
imputed_data = imputer_model.fit_transform(num_data)
# Quick check
imputed_data
# Let's create our Data Fream again
num_data = pd.DataFrame(imputed_data, columns=num_data.columns)
# Quick check
num_data.head()
# #### Numerical Data Analysis
# ###### Age vs Surgery Time
graph = go.Figure()
for val in data["LenSurgeryCode"].unique():
graph.add_trace(
go.Scatter(
x=data.loc[data.LenSurgeryCode == val].AnesthesiaType,
y=data.loc[data.LenSurgeryCode == val]["ElapsedTime(second)"],
mode="markers",
name=val,
)
)
graph.update_layout(title="Age & Elapsed Time")
graph.show()
# ###### Surgery Group Fourier vs Surgery Time
graph = go.Figure()
graph.add_trace(
go.Scatter(
x=data.SurgeryGroup_Fourier, y=data["ElapsedTime(second)"], mode="markers"
)
)
graph.update_layout(title="SurgeryGroup_Fourier & Elapsed Time")
graph.show()
# ###### Lenght of Surgery Group Code vs Surgery Time
fig = px.violin(
data,
y="ElapsedTime(second)",
x="LenSurgeryCode",
color="LenSurgeryCode",
box=True,
points="all",
hover_data=data.columns,
)
fig.show()
# ###### Anesthesia Type vs Surgery Time
graph = go.Figure()
graph.add_trace(
go.Scatter(
x=data.AnesthesiaType,
y=data["ElapsedTime(second)"],
mode="markers",
marker_color=data["ElapsedTime(second)"],
)
)
graph.update_layout(title="Anesthesia Type Code & Elapsed Time")
graph.show()
# ###### Service Code vs Surgery Time
graph = go.Figure()
graph.add_trace(
go.Violin(x=data["Service"], y=data["ElapsedTime(second)"], line_color="seagreen")
)
graph.update_traces(box_visible=False)
graph.update_layout(title="Service Code & Elapsed Time", violinmode="group")
graph.show()
# ###### Doctor ID vs Surgery Time
graph = go.Figure()
graph.add_trace(
go.Scatter(
x=data.DoctorID,
y=data["ElapsedTime(second)"],
mode="markers",
marker_color=data["ElapsedTime(second)"],
)
)
graph.update_layout(title="Doctor ID & Elapsed Time")
graph.show()
# ###### Surgery Code vs Surgery Time
graph = go.Figure()
graph.add_trace(
go.Scatter(
x=data.SurgeryCode,
y=data["ElapsedTime(second)"],
mode="markers",
marker_color=data["ElapsedTime(second)"],
)
)
graph.update_layout(title="SurgeryCode & Elapsed Time")
graph.show()
# #### Hand made PCA application
# Feature engineering is a crucial step in the machine learning process that involves transforming raw data into meaningful features
# that can improve the performance of predictive models. This process can include creating new features based on existing ones,
# encoding categorical variables, scaling numerical data, and more. Feature engineering can help to reduce noise,
# improve model accuracy, and increase interpretability.
# PCA (Principal Component Analysis) is a technique used for dimensionality reduction in which the original features of a dataset
# are transformed into a new set of features that capture the maximum amount of variation in the data. PCA is particularly useful
# when dealing with high-dimensional data, where it can be difficult to identify relevant features or avoid overfitting.
# By reducing the dimensionality of the data, PCA can simplify the analysis and visualization of complex datasets,
# as well as improve the performance of predictive models by reducing the risk of overfitting.
num_data.head()
# Let's change the our columns names quickly
num_data = num_data.rename(
columns={
"ElapsedTime(second)": "SurgeryTime",
"SurgeryGroup_Fourier": "SGroup_F",
"Lenght_ICD10_Code": "Lgn_ICD10",
"ICD10_Fourier": "ICD10_F",
"LenSurgeryCode": "L_SrgyCode",
}
)
features = num_data.loc[:, num_data.columns[: len(num_data.columns) - 1]]
fig = go.Figure(
data=go.Splom(
dimensions=[dict(label=col, values=num_data[col]) for col in features],
showupperhalf=False,
text=num_data.SurgeryTime,
marker=dict(
color=num_data.SurgeryTime,
line_color="white",
line_width=0.5,
opacity=0.6,
colorscale="Greys",
),
)
)
fig.update_layout(
title="Surgery Time PCA",
width=3000,
height=2000,
)
fig.show()
# #### Big Boss PCA application
# Just define the our PCA columns
list_PCA_col = ["Age", "Service", "DoctorID", "Lgn_ICD10", "L_SrgyCode", "SurgeryCode"]
# Let's create the our pca data
data_PCA = num_data[list_PCA_col]
# And now we can give quick chak of it
data_PCA.head()
from sklearn.decomposition import PCA
# data_PCA.drop('SurgeryTime', axis=1, inplace=True)
# Create our pca model
pca_model = PCA(n_components=2, whiten=True)
# Throw the our pca data into our model
pca_model.fit(data_PCA)
# Now we can get back the pca resualt
pca_res = pca_model.transform(data_PCA)
# Quick variance ratio on our new data
print(f"Protected variation : {sum(pca_model.explained_variance_ratio_)} ")
print(f"Ratio between the columns : {pca_model.explained_variance_ratio_}")
# Let's create our Data Fream again
pca_res_data = pd.DataFrame(pca_res, columns=["Pca_col_One", "Pca_col_Two"])
# Add the or Surgery time to make graph more understanable
pca_res_data["SurgeryTime"] = num_data.SurgeryTime
# Quick check
pca_res_data.head()
# Create the our little graph to check our resualt
graph = go.Figure(
data=go.Scatter(
x=pca_res_data.Pca_col_One,
y=pca_res_data.Pca_col_Two,
mode="markers",
marker=dict(color=pca_res_data.SurgeryTime, colorscale="ylorrd"),
)
)
# Wuba laba lab lab
graph.show()
|
import plotly.express as px
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
df1 = pd.read_csv("/kaggle/input/superbowl-history-1967-2020/superbowl.csv")
df1.head()
df1.isna().sum()
df1["Diff"] = df1["Winner Pts"] - df1["Loser Pts"]
fig = px.scatter(
df1,
x="Loser",
y="Winner",
size="Diff",
hover_name="Stadium",
color="Date",
title=" a 1967-2020 games resume",
)
fig.show()
rb2 = df1.State.value_counts()
rbg2 = rb2.reset_index()
rbg2.rename(columns={"index": "State", "State": "num of played match"}, inplace=True)
rbg2
import plotly.graph_objects as go
rbg2.set_index("State", inplace=True)
r30 = rbg2.rename(
index={
"Florida": "FL",
"California": "CA",
"Louisiana": "LA",
"Texas": "TX",
"Arizona": "AZ",
"Georgia": "GA",
"Michigan": "MI",
"Minnesota": "MN",
"Indiana": "IN",
"New Jersey": "NZ",
}
)
final = r30.reset_index()
fig = go.Figure(
data=go.Choropleth(
locations=final["State"], # Spatial coordinates
z=final["num of played match"].astype(float),
locationmode="USA-states",
colorscale="Blues",
)
)
fig.update_layout(
title_text="1967-2020 number of played games per State (USA)",
geo_scope="usa",
)
fig.show()
a = (
pd.DataFrame(df1.MVP.value_counts())
.reset_index()
.rename(columns={"index": "Player"})
.max()
)
print("Most Valuable Player 1967-2020 is", a[0], " with a total of: ", a[1])
rb1 = df1.Winner.value_counts()
rbg1 = rb1.reset_index()
rbg1.rename(columns={"index": "Team", "Winner": "num of won games"}, inplace=True)
import plotly.express as px
fig = px.line(
rbg1, x="Team", y="num of won games", title=" number of won games by team 1967-2020"
)
fig.show()
rb = df1.Loser.value_counts()
rbg = rb.reset_index()
rbg.rename(columns={"index": "Team", "Loser": "num of lost games"}, inplace=True)
rr = rbg.head(5)
a = rr.sort_index(axis=0, ascending=False)
fig = px.bar(
a,
y="Team",
x="num of lost games",
orientation="h",
title=" TOP FIVE LOSING TEAMS 1967-2020",
)
fig.show()
nn = pd.merge(rbg, rbg1, on="Team", how="inner")
nn["total finals"] = nn.sum(axis=1, skipna=True)
nn = nn.sort_values(by="total finals", ascending=False).head()
fig = px.pie(
nn,
names="Team",
values="total finals",
hole=0.6,
title=" TOP FIVE TEAMS THAT MADE IT TO FINALS 1967-2020",
)
fig.show()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import seaborn as sns
import matplotlib.pyplot as plt
df = pd.read_csv("/kaggle/input/drug-classification/drug200.csv")
df.head()
df.info()
# # EDA
df.isnull().sum()
df["Sex"].nunique()
sns.countplot(data=df, x="Sex")
df["Sex"].value_counts()
df.groupby("Sex").size().plot(kind="pie", autopct="%.2f")
df["BP"].nunique()
df["BP"].value_counts()
df["BP"].value_counts().plot(kind="bar")
sns.countplot(data=df, x="BP")
df["BP"].value_counts().plot(kind="pie", autopct="%0.0f%%")
df["Cholesterol"].value_counts()
df["Cholesterol"].value_counts().plot(kind="pie", autopct="%.2f")
df.head()
df["Drug"].value_counts()
df["Drug"].value_counts().plot(kind="pie", autopct="%.2f")
df.groupby(by=["Drug"]).size().plot(kind="bar")
# df['Age'].plot(kind='bar')
df.groupby(by=["Age"]).count()["Drug"].head(5)
df.groupby(by=["Age"]).count()["Drug"].plot(kind="line")
# # Feature Engineering
df.head(2)
gender = pd.get_dummies(df["Sex"])
gender.head(2)
bp = pd.get_dummies(df["BP"])
bp.head()
chol = pd.get_dummies(df["Cholesterol"])
chol.head(2)
# drug = pd.get_dummies(df['Drug'])
# drug.head()
df = pd.concat([df, gender, bp, chol], axis=1)
df.head()
df.drop(["Sex", "BP", "Cholesterol"], axis=1, inplace=True)
df.head()
# # Training & Testing
from sklearn.model_selection import train_test_split
X = df.drop(["Drug"], axis=1)
y = df["Drug"]
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=101
)
from sklearn.linear_model import LogisticRegression
lomodel = LogisticRegression()
lomodel.fit(X_train, y_train)
predictions = lomodel.predict(X_test)
predictions
y_test.head()
# # Model Evaluation
from sklearn.metrics import classification_report
print(classification_report(y_test, predictions))
from sklearn.metrics import confusion_matrix
print(confusion_matrix(y_test, predictions))
X.head()
y.value_counts()
# # My Predictions
lomodel.score(X_test, y_test)
lomodel.predict([[3, 3, 0, 1, 0, 1, 0, 1, 0]])
|
# The Gini coefficient is a measure of income distribution dispersion in a country. Values closer to 100% mean that there is more inequality, while values close to 0 mean that income in a country is evenly spread. In this notebook we want to study relationship between the Gini index (World Bank estimate) and different economic indicators such as:
# * Inflation, consumer prices (annual %)
# * Tax revenue (% of GDP)
# * Time required to start a business (days)
# * Unemployment, total (% of total labor force)
# The outcome is to understand if a change in fiscal, monetary or other policies can impact the degree of income inequality in a country, as measured by the Gini coeffcient.
# #### Ideas
# * Don't use year as indep var, use as group
# * Region varying intercept same slope
# * Region varying intercept varying slope
# * Time dimension?
# * Use BigQuer data
import numpy as np
import pandas as pd
import seaborn as sns
import statsmodels.api as sm
from sklearn.metrics import mean_squared_error
import statsmodels.formula.api as smf
import sqlite3
# ## Load and explore data
# Load WDI data from BigQuery
import bq_helper
from bq_helper import BigQueryHelper
# https://www.kaggle.com/sohier/introduction-to-the-bq-helper-package
wdi = bq_helper.BigQueryHelper(
active_project="patents-public-data", dataset_name="worldbank_wdi"
)
bq_assistant = BigQueryHelper("patents-public-data", "worldbank_wdi")
bq_assistant.list_tables()
query_str = """SELECT country_code, year, indicator_code, indicator_value from `patents-public-data.worldbank_wdi.wdi_2016`
WHERE year BETWEEN 1960 AND 2015 AND
indicator_code IN ('SL.UEM.TOTL.NE.ZS','FP.CPI.TOTL.ZG',
'IC.REG.DURS','GC.TAX.TOTL.GD.ZS','SI.POV.GINI')
AND indicator_value<>0
"""
# Estimate query size
bq_assistant.estimate_query_size(query_str)
wdi_df = wdi.query_to_pandas_safe(query_str)
# Count how many values we have for each indicator (across countries and years)
wdi_df.groupby("indicator_code").count()
# See top rows and size of DataFrame
wdi_df.head()
wdi_df.shape
# Pivot dataframe so the indicators are on columns
wdi_df_piv = wdi_df.pivot_table(
index=["country_code", "year"],
columns=["indicator_code"],
values=["indicator_value"],
fill_value=np.nan,
).reset_index()
wdi_df_piv.shape
wdi_df_piv.head()
wdi_df_piv.columns = ["country_code", "year"] + list(wdi_df_piv.columns.droplevel())[2:]
wdi_df_piv.head()
wdi_df_piv.columns
# Rearrange and rename columns
wdi_df_mod = wdi_df_piv[
[
"country_code",
"SI.POV.GINI",
"year",
"FP.CPI.TOTL.ZG",
"GC.TAX.TOTL.GD.ZS",
"IC.REG.DURS",
"SL.UEM.TOTL.NE.ZS",
]
]
wdi_df_mod.columns = [
"CountryCode",
"Gini",
"Year",
"Inflat",
"TaxRev",
"BusDay",
"Unempl",
]
wdi_df_mod.head()
# Describe data
wdi_df_mod.describe()
# Count missing data
wdi_df_mod.groupby(["Year"]).count()
# Correlation
sns.set(rc={"figure.figsize": (11.7, 8.27)})
wdi_corr = wdi_df_mod.iloc[:, 1:].corr()
mask = np.zeros(wdi_corr.shape, dtype=bool)
mask[np.tril_indices(len(mask))] = True
sns.heatmap(wdi_corr, annot=True, mask=mask)
sns.set(font_scale=1)
# Pair plot
sns.pairplot(wdi_df_mod)
# Distribution of Gini
wdi_df_mod["Gini"].median()
sns.distplot(wdi_df_mod["Gini"].dropna())
sns.distplot(np.log(wdi_df_mod["Gini"].dropna()))
# ## Models
# ### Pooled OLS model
wdi_df_clean = wdi_df_mod.dropna()
from sklearn.preprocessing import RobustScaler
rob_sc = RobustScaler()
# Apply log transform on Gini and scale independent variables
Gini_log = np.log(wdi_df_clean["Gini"])
X_sc = rob_sc.fit_transform(wdi_df_clean.iloc[:, 2:])
wdi_sc = pd.concat(
[
wdi_df_clean.iloc[:, 0],
wdi_df_clean.iloc[:, 2],
Gini_log,
pd.DataFrame(
X_sc,
index=wdi_df_clean.index,
columns=[x + "_sc" for x in wdi_df_clean.iloc[:, 2:].columns],
),
],
axis=1,
)
wdi_sc.shape
wdi_sc.head()
# Correlation and pair plot with transformed data
wdi_corr_sc = wdi_sc.iloc[:, 2:].corr()
mask = np.zeros(wdi_corr_sc.shape, dtype=bool)
mask[np.tril_indices(len(mask))] = True
sns.heatmap(wdi_corr_sc, annot=True, mask=mask)
sns.pairplot(wdi_sc.iloc[:, 2:])
y = wdi_sc["Gini"]
ols = smf.ols("Gini ~ Inflat_sc + TaxRev_sc + BusDay_sc + Unempl_sc", data=wdi_sc)
olsf = ols.fit()
# Print out the statistics
olsf.summary()
sns.distplot(olsf.resid)
sns.regplot(olsf.fittedvalues, olsf.resid, color="g", lowess=True)
# The residual plot suggests heteroscedasticity and bias for higher values of the predicted Gini.
# Condition number
np.linalg.cond(olsf.model.exog)
# Heteroskedasticity test
import statsmodels.stats.api as sms
from statsmodels.compat import lzip
name = ["Lagrange multiplier statistic", "p-value", "f-value", "f p-value"]
test = sms.het_breuschpagan(olsf.resid, olsf.model.exog)
lzip(name, test)
import matplotlib.pyplot as plt
fig, axs = plt.subplots(ncols=5, figsize=(30, 5))
sns.regplot(wdi_sc.iloc[:, 3], olsf.resid, ax=axs[0], color="r", lowess=True)
sns.regplot(wdi_sc.iloc[:, 4], olsf.resid, ax=axs[1], color="r", lowess=True)
sns.regplot(wdi_sc.iloc[:, 5], olsf.resid, ax=axs[2], color="r", lowess=True)
sns.regplot(wdi_sc.iloc[:, 6], olsf.resid, ax=axs[3], color="r", lowess=True)
sns.regplot(wdi_sc.iloc[:, 7], olsf.resid, ax=axs[4], color="r", lowess=True)
rmse_pooled = np.sqrt(mean_squared_error(np.exp(y), np.exp(olsf.fittedvalues)))
performance = pd.DataFrame([["pooled", rmse_pooled]], columns=["model", "rmse"])
performance
# The model is not great. Are we missing something? Is there country variation that we fail to capture?
wdi_res = wdi_sc
wdi_res["Residuals"] = olsf.resid
box = sns.boxplot(x="CountryCode", y="Residuals", data=wdi_res)
box.set_xticklabels(box.get_xticklabels(), rotation=90)
# Add country data from a SQLite database
conn = sqlite3.connect("../input/world-development-indicators/database.sqlite")
country_df = pd.read_sql_query(
"SELECT CountryCode,Region,IncomeGroup FROM Country", conn
)
country_df.groupby("Region").count()
country_df.groupby("IncomeGroup").count()
wdi_res.shape
wdi_region = wdi_res.merge(country_df, left_on="CountryCode", right_on="CountryCode")
wdi_region.shape
wdi_region.head()
box = sns.boxplot(x="Region", y="Residuals", data=wdi_region)
box.set_xticklabels(box.get_xticklabels(), rotation=90)
wdi_region.groupby(["Region"]).count()
box = sns.boxplot(
x="IncomeGroup",
y="Residuals",
data=wdi_region,
order=[
"High income: OECD",
"High income: nonOECD",
"Upper middle income",
"Lower middle income",
"Low income",
],
)
box.set_xticklabels(box.get_xticklabels(), rotation=90)
wdi_region.groupby(["IncomeGroup"]).count()
wdi_region["IncomeGroup"] = np.where(
wdi_region["IncomeGroup"] == "Low income",
"Lower middle income",
wdi_region["IncomeGroup"],
)
wdi_region.groupby(["IncomeGroup"]).count()
# ### Multilevel model with varying intercept by Income Region group
wdi_region.head()
# construct our model, with our county now shown as a group
md = smf.mixedlm(
"Gini ~ Inflat_sc + TaxRev_sc + BusDay_sc + Unempl_sc",
data=wdi_region,
groups="IncomeGroup",
)
# md = smf.mixedlm("Gini ~ Unempl_sc", data, groups="IncomeGroup")
mdf = md.fit()
print(mdf.summary())
sns.regplot(mdf.fittedvalues, mdf.resid, color="g", lowess=True)
rmse_mda = np.sqrt(mean_squared_error(np.exp(y), np.exp(mdf.fittedvalues)))
performance.loc[1] = ["rand inter", rmse_mda]
performance
name = ["Lagrange multiplier statistic", "p-value", "f-value", "f p-value"]
test = sms.het_breuschpagan(mdf.resid, mdf.model.exog)
lzip(name, test)
# ### Multilevel model with varying intercept and varying slopes by Region group
def plot_df_scatter_columns(df, y_column, grouping, rel_col):
for z in df[rel_col]:
sns.lmplot(x=z, y=y_column, data=df, hue=grouping)
rel_col = ["Year_sc", "Inflat_sc", "TaxRev_sc", "BusDay_sc", "Unempl_sc"]
plot_df_scatter_columns(wdi_region, "Gini", "IncomeGroup", rel_col)
mdb = smf.mixedlm(
"Gini ~ Inflat_sc + TaxRev_sc + BusDay_sc + Unempl_sc",
data=wdi_region,
groups="IncomeGroup",
re_formula="~ Inflat_sc + TaxRev_sc + BusDay_sc + Unempl_sc",
)
# md = smf.mixedlm("Gini ~ Unempl_sc", data, groups="IncomeGroup")
mdfb = mdb.fit()
print(mdfb.summary())
sns.regplot(mdfb.fittedvalues, mdfb.resid, color="g", lowess=True)
rmse_mdb = np.sqrt(mean_squared_error(np.exp(y), np.exp(mdfb.fittedvalues)))
performance.loc[2] = ["rand inter, rand slope", rmse_mdb]
performance
name = ["Lagrange multiplier statistic", "p-value", "f-value", "f p-value"]
test = sms.het_breuschpagan(mdfb.resid, mdfb.model.exog)
lzip(name, test)
# ### Multilevel model with varying intercept and varying slopes by Region and Year group
wdi_region["IncomeGroup-Year"] = wdi_region["IncomeGroup"] + wdi_region["Year"].astype(
str
)
mdb = smf.mixedlm(
"Gini ~ Inflat_sc + TaxRev_sc + BusDay_sc + Unempl_sc",
data=wdi_region,
groups="IncomeGroup-Year",
re_formula="~ Inflat_sc + TaxRev_sc + BusDay_sc + Unempl_sc",
)
# md = smf.mixedlm("Gini ~ Unempl_sc", data, groups="IncomeGroup")
mdfb = mdb.fit()
print(mdfb.summary())
sns.regplot(mdfb.fittedvalues, mdfb.resid, color="g", lowess=True)
rmse_mdb = np.sqrt(mean_squared_error(np.exp(y), np.exp(mdfb.fittedvalues)))
performance.loc[3] = ["rand inter, rand slope - region & year", rmse_mdb]
performance
name = ["Lagrange multiplier statistic", "p-value", "f-value", "f p-value"]
test = sms.het_breuschpagan(mdfb.resid, mdfb.model.exog)
lzip(name, test)
sns.regplot(np.exp(y), np.exp(mdfb.fittedvalues))
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # PREPROCESSING
import pandas as pd
import numpy as np
data = pd.read_csv("/kaggle/input/significant-earthquakes/Significant_Earthquakes.csv")
print(f"Количество записей: {len(data)}")
print("\nКоличество пропущенных значений в каждом столбце:\n", data.isnull().sum())
# As you can see, there are missing values in some columns. Now replace all the missing values in the columns depth, net, gap, min, horizontalError, depthError, magNst with the average value of the column, and in the columns mag, magType, rms, magError - with the median.
data["depth"].fillna(data["depth"].mean(), inplace=True)
data["nst"].fillna(data["nst"].mean(), inplace=True)
data["gap"].fillna(data["gap"].mean(), inplace=True)
data["dmin"].fillna(data["dmin"].mean(), inplace=True)
data["horizontalError"].fillna(data["horizontalError"].mean(), inplace=True)
data["depthError"].fillna(data["depthError"].mean(), inplace=True)
data["magNst"].fillna(data["magNst"].mean(), inplace=True)
data["mag"].fillna(data["mag"].median(), inplace=True)
data["magType"].fillna(data["magType"].mode()[0], inplace=True)
data["rms"].fillna(data["rms"].median(), inplace=True)
data["magError"].fillna(data["magError"].median(), inplace=True)
print(
"Количество пропущенных значений в каждом столбце после обработки:\n",
data.isnull().sum(),
)
# The place variable contains a description of the earthquake location in text format. The number of missing values in this column is 437. The solution options depend on the purpose of the analysis. If you do not need to analyze the text description of the earthquake location, you can delete this column from the data. If a textual description of the location is necessary for analysis, then you can delete the entries with the missing place value or fill in the missing values based on other data, for example, based on geographical coordinates. You can also use machine learning algorithms to fill in missing values based on other earthquake parameters.
# Delete the entries with the missing place value
data.dropna(subset=["place"], inplace=True)
print(
"Количество пропущенных значений в каждом столбце после обработки:\n",
data.isnull().sum(),
)
# # EDA
# To begin with, you can build a distribution of values in the 'magnitude' column - this will help you understand which values are most represented in the data, and which are rare.
import pandas as pd
# Группируем данные по значениям магнитуды и разбиваем их на 20 бинов
data_grouped = pd.cut(data["mag"], bins=20, include_lowest=True)
# Получаем таблицу с количеством землетрясений в каждом бине
table = (
pd.value_counts(data_grouped)
.sort_index()
.rename_axis("Magnitude")
.reset_index(name="Count")
)
print(table)
import matplotlib.pyplot as plt
plt.hist(data["mag"], bins=20)
plt.xlabel("Magnitude")
plt.ylabel("Count")
plt.show()
# It can be noted that most earthquakes have a magnitude of less than 6.5, and the most powerful earthquakes (more than 8.5 in magnitude) occur extremely rarely. It can also be noted that the number of earthquakes decreases as the magnitude increases.
plt.scatter(data["longitude"], data["latitude"], c=data["mag"], cmap="YlOrRd")
plt.xlabel("Longitude")
plt.ylabel("Latitude")
plt.colorbar(label="Magnitude")
plt.show()
# It can be noted that most earthquakes occur along the boundaries of tectonic plates, especially in the areas of the "Ring of Fire" in the Pacific region. It can also be noticed that more powerful earthquakes tend to occur in places where several tectonic plates collide.
# # Analyze the dependence of magnitude on the type of seismic event
# To analyze the dependence of magnitude on the type of seismic event, you can group data by event type and calculate statistical metrics (for example, average, median, maximum and minimum magnitude values) for each group.
import pandas as pd
# Группировка данных по типу события и расчет средней магнитуды
mean_mag_by_type = data.groupby("type")["mag"].mean()
# Вывод результата
print(mean_mag_by_type)
# Построение графика
data.boxplot(column="mag", by="type", figsize=(10, 6))
plt.title("")
plt.suptitle("")
plt.xlabel("Type of seismic event")
plt.ylabel("Magnitude")
plt.show()
# Based on this information, the average magnitude of earthquakes caused by explosions in mines and nuclear tests is higher than that of earthquakes caused by natural causes (volcanic eruptions and ordinary earthquakes). Also, the magnitude of earthquakes caused by explosions in mines is lower than that of earthquakes caused by nuclear tests.
from scipy.stats import levene
groups = data["type"].unique()
group_data = [data.loc[data["type"] == group]["mag"] for group in groups]
stat, pvalue = levene(*group_data, center="median")
print(f"Levene test statistic: {stat:.4f}")
print(f"p-value: {pvalue:.4f}")
# The results of the Levin test indicate that we have no statistically significant differences in variance between groups of earthquake event types (p-value > 0.05). This means that we can continue with the analysis of variance without having to adjust for heterogeneity of variances.
# However, it is worth noting that we have a small p-value (0.0823), which may indicate that the heterogeneity of variances between groups may not be completely insignificant. Therefore, we can be more careful in interpreting the results of the analysis of variance, especially if we find statistically significant differences in the averages between the groups.
# Since the conditions are met, we will perform an analysis of variance using one-factor analysis of variance (ANOVA)
from scipy.stats import f_oneway
# Разделяем данные по типу события
earthquake_data = data.loc[data["type"] == "earthquake"]["mag"]
explosion_data = data.loc[data["type"] == "explosion"]["mag"]
mine_collapse_data = data.loc[data["type"] == "mine collapse"]["mag"]
nuclear_explosion_data = data.loc[data["type"] == "nuclear explosion"]["mag"]
rock_burst_data = data.loc[data["type"] == "rock burst"]["mag"]
volcanic_eruption_data = data.loc[data["type"] == "volcanic eruption"]["mag"]
# Проводим однофакторный дисперсионный анализ
f_statistic, p_value = f_oneway(
earthquake_data,
explosion_data,
mine_collapse_data,
nuclear_explosion_data,
rock_burst_data,
volcanic_eruption_data,
)
print(f"F-статистика: {f_statistic:.4f}")
print(f"p-value: {p_value:.4f}")
# The results of univariate analysis of variance (ANOVA) indicate that there is a statistically significant relationship between the type of seismic events and magnitude. The F-statistic value of 4.6577 and the low significance level (p-value) of 0.0003 indicate that the differences in magnitude between the types of seismic events are not random, and the probability of their occurrence may depend on the type of event.
# It should be taken into account that the sample size for each type of event may be different and may affect the results of the analysis. It should also be noted that this analysis does not show a causal relationship between the type of event and magnitude.
# Since the analysis of variance shows statistically significant differences between groups, it is necessary to conduct a post-hoc analysis to find out which groups differ. For post-hoc analysis in the case of single-factor analysis of variance (ANOVA), several methods can be used, such as:
# * The Hill-Bonferroni Method
# * The Scheffe method
# * The Tukey HSD Method
# Let's use the Scheffe method method, which is the most conservative, but at the same time quite easy to use.
import statsmodels.stats.multitest as smt
groups = data["type"].unique()
n_groups = len(groups)
n_total = len(data)
# Среднее значение по всем данным
mean_total = data["mag"].mean()
# Вычисляем SS_total (sum of squares total)
SS_total = np.sum((data["mag"] - mean_total) ** 2)
# Одинаковое количество наблюдений в каждой группе
n_obs = n_total // n_groups
# Среднее значение в каждой группе
group_means = data.groupby("type")["mag"].mean()
# Вычисляем SS_within (sum of squares within)
SS_within = 0
for group in groups:
group_data = data.loc[data["type"] == group]["mag"]
SS_within += np.sum((group_data - group_means[group]) ** 2)
# Вычисляем SS_between (sum of squares between)
SS_between = SS_total - SS_within
# Вычисляем степени свободы
df_between = n_groups - 1
df_within = n_total - n_groups
# Вычисляем MS_between (mean square between) и MS_within (mean square within)
MS_between = SS_between / df_between
MS_within = SS_within / df_within
# Вычисляем F-статистику
F = MS_between / MS_within
# Вычисляем p-value
p_value = stats.f.sf(F, df_between, df_within)
# Коррекция на множественное тестирование
reject, p_values_corrected, _, _ = smt.multipletests(
[p_value], alpha=0.05, method="holm"
)
if reject[0]:
print("The differences between the groups are statistically significant")
# Метод Шеффе для пост-хок анализа
for i in range(n_groups):
for j in range(i + 1, n_groups):
diff = group_means[i] - group_means[j]
SE = np.sqrt(MS_within * (1 / n_obs + 1 / n_obs))
t = diff / SE
p_value = stats.t.sf(np.abs(t), df_within)
reject, _, _, _ = smt.multipletests([p_value], alpha=0.05, method="holm")
if reject[0]:
print(
f"{groups[i]} vs {groups[j]}: the differences are statistically significant"
)
else:
print("The differences between the groups are not statistically significant")
# Based on the results of post-hoc analysis using the Scheffe method, it can be concluded that all groups have statistically significant differences among themselves in earthquake magnitude. That is, the type of disaster affects the magnitude of the earthquake. For example, earthquakes caused by volcanic activity have a higher magnitude compared to earthquakes caused by mine collapses, etc.
# **The relationship between the type of earthquake and the depth of the epicenter**
table = pd.pivot_table(data, values="depth", index="type", aggfunc=np.mean)
print(table)
# # Analysis of the relationship between the magnitude and depth of the epicenter
import matplotlib.pyplot as plt
plt.scatter(data["depth"], data["mag"], alpha=0.2)
plt.xlabel("Depth")
plt.ylabel("Magnitude")
plt.show()
# The graph shows that the greater the depth of the epicenter, the smaller the number of earthquakes with a high magnitude (more than 6 points), and the spread of magnitude values increases. At the same time, earthquakes with a lower magnitude (less than 6 points) are distributed more evenly in depth. This may indicate that stronger earthquakes occur at deeper levels, while smaller earthquakes may occur at different depths. However, it should be borne in mind that the graph shows only earthquakes with a magnitude of more than 4 points.
correlations = data[["depth", "mag", "latitude", "longitude"]].corr()
print(correlations)
# From this correlation matrix we can draw the following conclusions:
# Magnitude (mag) has a weak negative correlation with depth (-0.023937). That is, the greater the depth, the smaller the magnitude of the earthquake. However, this correlation is very weak, so it is impossible to draw convincing conclusions based on this.
# Magnitude (mag) has a very weak positive correlation with latitude (0.054668) and almost no correlation with longitude (0.000863). This means that the magnitude does not depend on the latitude and longitude of the earthquake.
# Depth has some negative correlation with latitude (-0.119580) and longitude (-0.086311). That is, the farther from the equator and the zero meridian the earthquake occurred, the greater the depth. However, this correlation is also weak enough to draw convincing conclusions.
# Thus, based on this correlation matrix, we can draw some conclusions about the dependencies between different variables, but these dependencies are weak enough to draw convincing conclusions.
# # Analysis of changes in earthquake indicators over time
# Well, to analyze the changes in earthquake indicators over time, we can use earthquake data collected over the past few decades. This data may include the place of origin of the earthquake, its magnitude, time and date, as well as other characteristics.
# To begin with, we can analyze trends in the magnitude of earthquakes over time. To do this, we can use statistical analysis methods such as linear regression or time series analysis. As a result, we will be able to determine whether there are any obvious trends in the change in the magnitude of earthquakes over time.
# Then we can build machine learning models using data on changes in the magnitude of earthquakes over time to predict the probability of an earthquake of a certain magnitude in the future. To do this, we can use various machine learning algorithms, such as logistic regression, random forest or neural networks.
# However, it is worth noting that forecasting earthquakes is a difficult task, since these events can occur as a result of various factors and complex processes occurring inside the Earth. Therefore, the prediction results may be inaccurate and do not guarantee 100% accuracy.
data["Year"] = pd.to_datetime(data["time"], format="%Y-%m-%dT%H:%M:%S.%fZ").dt.year
mean_mag_by_year = data.groupby("Year")["mag"].mean()
plt.plot(mean_mag_by_year.index, mean_mag_by_year.values)
plt.title("Mean Magnitude of Earthquakes by Year")
plt.xlabel("Year")
plt.ylabel("Magnitude")
plt.show()
# The average magnitude of earthquakes varies from 4.4 to 7.9. We also see that in 1903 the most powerful earthquake was recorded among all those that were recorded in the table.
# It can be noted that the average magnitude of earthquakes is the lowest in recent decades. This may indicate that the activity of the earth's crust has decreased in recent years. However, additional analysis is needed to confirm this.
from scipy import stats
# Создаем таблицу с годами и средней магнитудой
mean_mag_by_year = data.groupby("Year")["mag"].mean().reset_index()
# Расчет среднего значения магнитуды землетрясений
mean_mag = mean_mag_by_year["mag"].mean()
print(f"Средняя магнитуда: {mean_mag:.2f}")
# Расчет стандартного отклонения магнитуды землетрясений
std_mag = mean_mag_by_year["mag"].std()
print(f"Стандартное отклонение: {std_mag:.2f}")
# Расчет коэффициента корреляции между годом и магнитудой землетрясения
corr_coef = mean_mag_by_year["Year"].corr(mean_mag_by_year["mag"])
print(f"Коэффициент корреляции: {corr_coef:.2f}")
# Расчет линейной регрессии между годом и магнитудой землетрясения
slope, intercept, r_value, p_value, std_err = stats.linregress(
mean_mag_by_year["Year"], mean_mag_by_year["mag"]
)
print(f"Угловой коэффициент: {slope:.2f}")
print(f"Пересечение с осью Y: {intercept:.2f}")
print(f"Коэффициент детерминации: {r_value ** 2:.2f}")
# The average magnitude of the earthquakes that occurred during the analyzed period is 5.82 points.
# The standard deviation of the magnitude of earthquakes during this period is 0.45, which indicates that most earthquakes have a magnitude close to the average value.
# The correlation coefficient between the year and the magnitude of the earthquake is -0.89, which indicates that there is a strong feedback between the year and the magnitude of the earthquake. That is, as the year increases, the number of earthquakes with a smaller magnitude increases, and the number of earthquakes with a larger magnitude decreases.
# The angular coefficient of linear regression between the year and the magnitude of the earthquake is negative and is -0.01, which indicates that years pass, and the magnitude of earthquakes decreases.
# The coefficient of determination is 0.79, which suggests that 79% of the variability of the magnitude of earthquakes can be explained by the change of the year.
# # Analysis of the spectral density
# We will conduct an analysis of the spectral density, which allows us to determine the periodicity in the change in the magnitude of earthquakes over time. If significant peaks are detected in the spectrum, then it can be concluded that there is a cyclical change in the magnitude of earthquakes.
from scipy import signal
# Вычисление спектральной плотности мощности
frequencies, power_spectrum = signal.periodogram(mean_mag_by_year["mag"])
# Построение графика спектральной плотности мощности
plt.plot(frequencies, power_spectrum)
plt.title("Power Spectral Density")
plt.xlabel("Frequency")
plt.ylabel("Power")
plt.show()
print(signal.periodogram(mean_mag_by_year["mag"]))
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
# reading csv file from local folder
survivalds = pd.read_csv(
"../input/habermans-survival-data-set/haberman.csv",
header=None,
names=["Age", "Op_Year", "axil_nodes", "Surv_status"],
)
print("columns ", survivalds.columns)
# check columns and observations
survivalds.shape
# columns
survivalds.columns
# finding target variable (Surv_status) distribution
survivalds.Surv_status.value_counts()
# finding percentage of Surv_status distribution
survivalds.Surv_status.value_counts() / survivalds.Surv_status.shape[0]
##### Data is distributed 73.5 and 26.5 so data is balanced
# finding na fields
survivalds.isna().sum().sum()
# EDA libraries
import seaborn as sns
import matplotlib.pyplot as plt
sns.boxplot(data=survivalds, x="Surv_status", y="axil_nodes")
plt.show()
print(survivalds.axil_nodes.mean(), "---", survivalds.axil_nodes.median())
survivalds[survivalds.axil_nodes < 4].Surv_status.value_counts() / survivalds[
survivalds.axil_nodes < 4
].Surv_status.count()
sns.boxplot(data=survivalds, x="Surv_status", y="Age")
plt.show()
sns.boxplot(data=survivalds, x="Surv_status", y="Op_Year")
plt.show()
sns.violinplot(data=survivalds, x="Surv_status", y="axil_nodes")
plt.show()
sns.violinplot(data=survivalds, x="Surv_status", y="Age")
plt.show()
sns.violinplot(data=survivalds, x="Surv_status", y="Op_Year")
plt.show()
survivalds.dtypes
survivalds_1 = survivalds.loc[survivalds["Surv_status"] == 1]
survivalds_2 = survivalds.loc[survivalds["Surv_status"] == 2]
# print(iris_setosa["petal_length"])
plt.plot(survivalds_1["Age"], np.zeros_like(survivalds_1["Age"]), "o")
plt.plot(survivalds_2["Age"], np.zeros_like(survivalds_2["Age"]), "o")
plt.show()
survivalds[
(survivalds.Age > 40) & (survivalds.Age < 60)
].Surv_status.value_counts() / survivalds[
(survivalds.Age > 40) & (survivalds.Age < 60)
].Surv_status.count()
plt.plot(survivalds_1["axil_nodes"], np.zeros_like(survivalds_1["axil_nodes"]), "o")
plt.plot(survivalds_2["axil_nodes"], np.zeros_like(survivalds_2["axil_nodes"]), "o")
plt.show()
plt.plot(survivalds_1["Op_Year"], np.zeros_like(survivalds_1["Op_Year"]), "o")
plt.plot(survivalds_2["Op_Year"], np.zeros_like(survivalds_2["Op_Year"]), "o")
plt.show()
sns.FacetGrid(survivalds, hue="Surv_status", size=5).map(
sns.distplot, "Age"
).add_legend()
plt.show()
sns.FacetGrid(survivalds, hue="Surv_status", size=5).map(
sns.distplot, "Op_Year"
).add_legend()
plt.show()
sns.FacetGrid(survivalds, hue="Surv_status", size=5).map(
sns.distplot, "axil_nodes"
).add_legend()
plt.show()
counts, bin_edges = np.histogram(survivalds["Age"], bins=10, density=True)
pdf = counts / (sum(counts))
print(pdf)
print(bin_edges)
cdf = np.cumsum(pdf)
plt.plot(bin_edges[1:], pdf)
plt.plot(bin_edges[1:], cdf)
counts, bin_edges = np.histogram(survivalds["Age"], bins=20, density=True)
pdf = counts / (sum(counts))
plt.plot(bin_edges[1:], pdf)
plt.show()
counts, bin_edges = np.histogram(survivalds["axil_nodes"], bins=10, density=True)
pdf = counts / (sum(counts))
print(pdf)
print(bin_edges)
cdf = np.cumsum(pdf)
plt.plot(bin_edges[1:], pdf)
plt.plot(bin_edges[1:], cdf)
counts, bin_edges = np.histogram(survivalds["axil_nodes"], bins=20, density=True)
pdf = counts / (sum(counts))
plt.plot(bin_edges[1:], pdf)
plt.show()
counts, bin_edges = np.histogram(survivalds["Op_Year"], bins=10, density=True)
pdf = counts / (sum(counts))
print(pdf)
print(bin_edges)
cdf = np.cumsum(pdf)
plt.plot(bin_edges[1:], pdf)
plt.plot(bin_edges[1:], cdf)
counts, bin_edges = np.histogram(survivalds["Op_Year"], bins=20, density=True)
pdf = counts / (sum(counts))
plt.plot(bin_edges[1:], pdf)
plt.show()
np.percentile(survivalds.axil_nodes, np.arange(0, 125, 25)) # [0., 0., 1., 4., 52]
survivalds.axil_nodes.mean() # 4
survivalds.axil_nodes.median() # 1
survivalds.sort_values(by="axil_nodes").axil_nodes.unique()
survivalds[survivalds.axil_nodes < 2].Surv_status.value_counts() / survivalds[
survivalds.axil_nodes < 2
].Surv_status.shape[0]
plt.hist(bins=10, data=survivalds, x="Surv_status")
|
# # Peek
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
import time
start_time = time.time()
# # Read Data
df = pd.read_csv("../input/insurance/insurance.csv")
print(df.head(10))
print(df.describe())
# # Data Mapping
sex_map = {"male": 0, "female": 1}
smoker_map = {"no": 0, "yes": 1}
region_map = {"southwest": 0, "northwest": 1, "northeast": 2, "southeast": 3}
df.sex = df.sex.map(sex_map)
df.smoker = df.smoker.map(smoker_map)
df.region = df.region.map(region_map)
actual_charges = df.charges.values.tolist()
df.charges = df.charges.map(lambda x: np.log(x))
print(df.head(10))
print(df.describe())
# # Imports
random_seed = 17025
np.random.seed(random_seed)
from tensorflow.keras.models import Sequential, Model
from tensorflow.keras.layers import Dense, Dropout, Average, Multiply, LeakyReLU
from tensorflow.keras.layers import Input, BatchNormalization, concatenate
from tensorflow.keras.optimizers import RMSprop, Adam, Nadam
from tensorflow.keras.callbacks import ReduceLROnPlateau, EarlyStopping
from tensorflow.keras.utils import plot_model
from tensorflow.keras.models import load_model
from sklearn.model_selection import train_test_split
from sklearn.metrics import confusion_matrix
from skimage.transform import resize
import matplotlib.pyplot as plt
# # Model Design
def new_reg_model(in_shape):
# Input layer
INPUT = Input(in_shape)
# Filter Layer
k = 16
f = []
for i in range(k):
f.append(Dense(256, activation="relu", kernel_initializer="normal")(INPUT))
for i in range(k):
f[i] = Dense(128, activation="relu")(f[i])
f[i] = Dropout(0.25)(f[i])
y = []
for i in range(k - 2):
y.append(concatenate(f[i : i + 2], axis=0))
x = Average()(f)
x = Dense(256, activation="relu")(x)
x = Dropout(0.5)(x)
x = Dense(1)(x)
model = Model(inputs=INPUT, outputs=[x])
optimizer = Adam(lr=0.01, decay=1e-5)
# Compile model
model.compile(optimizer, loss="msle", metrics=["mae"]) #'mse',
return model
model = new_reg_model(df.shape[1:])
model.summary()
plot_model(model, to_file="model_plot.png", show_shapes=True, show_layer_names=True)
# # Training
lrr = ReduceLROnPlateau(
monitor="val_mae", patience=10, verbose=1, factor=0.5, min_lr=0.00001
)
es = EarlyStopping(
monitor="val_loss", mode="min", verbose=1, patience=50, restore_best_weights=True
)
cols = df.columns[:-1]
# ['age', 'sex', 'bmi', 'children', 'smoker']
print(cols)
x_train, x_val, y_train, y_val = train_test_split(
df[cols], df.charges, test_size=0.25, shuffle=True, random_state=101
)
model = new_reg_model(x_train.shape[1:])
history = model.fit(
x_train,
y_train,
epochs=2000,
validation_data=(x_val, y_val),
verbose=1,
callbacks=[lrr, es],
)
# # Overview
print("Mean Absolute Error:")
plt.plot(history.history["mae"][10:])
plt.plot(history.history["val_mae"][10:])
plt.title("Model MAE")
plt.ylabel("MAE")
# plt.gca().set_ylim([0, 20000])
plt.xlabel("Epoch")
plt.legend(["train", "test"], loc="lower right")
plt.savefig("history_mae.png")
plt.show()
print("Loss:")
plt.plot(history.history["loss"][10:])
plt.plot(history.history["val_loss"][10:])
plt.title("Model Loss")
plt.ylabel("Loss")
# plt.gca().set_ylim([0, 5e7])
plt.xlabel("Epoch")
plt.legend(["train", "test"], loc="upper right")
plt.savefig("history_loss.png")
plt.show()
predictions = model.predict(df[cols])
print("Actual Value vs Predicted Value:\tDifference:")
diff = []
for a, b in zip(df.charges, predictions):
a = np.exp(a)
b = np.exp(b)
diff.append(a - b[0])
print("%.2f \t %.2f \t\t %.2f" % (a, b[0], a - b[0]))
print("Maximum Deviation: %.2f" % max(abs(x) for x in diff))
print("Minimum Deviation: %.2f" % min(abs(x) for x in diff))
print("Average Deviation: %.2f" % np.mean([abs(x) for x in diff]))
# # Feature Importance
from eli5.sklearn import PermutationImportance
import sklearn, eli5
print(sklearn.metrics.SCORERS.keys())
perm = PermutationImportance(
model, random_state=1, scoring="neg_mean_absolute_error"
).fit(df[cols], df.charges)
eli5.show_weights(perm, feature_names=cols.values.tolist())
# # Retrain Model
cols = ["age", "smoker", "children", "bmi"]
print(cols)
x_train, x_val, y_train, y_val = train_test_split(
df[cols], df.charges, test_size=0.25, shuffle=True, random_state=101
)
model = new_reg_model(x_train.shape[1:])
history = model.fit(
x_train,
y_train,
epochs=2000,
validation_data=(x_val, y_val),
verbose=1,
callbacks=[lrr, es],
)
print("Mean Absolute Error:")
plt.plot(history.history["mae"][10:])
plt.plot(history.history["val_mae"][10:])
plt.title("Model MAE Adjusted")
plt.ylabel("MAE")
# plt.gca().set_ylim([0, 20000])
plt.xlabel("Epoch")
plt.legend(["train", "test"], loc="lower right")
plt.savefig("history_mae_adjusted.png")
plt.show()
print("Loss:")
plt.plot(history.history["loss"][10:])
plt.plot(history.history["val_loss"][10:])
plt.title("Model Loss Adjusted")
plt.ylabel("Loss")
# plt.gca().set_ylim([0, 5e7])
plt.xlabel("Epoch")
plt.legend(["train", "test"], loc="upper right")
plt.savefig("history_loss_adjusted.png")
plt.show()
predictions = model.predict(df[cols])
print("Actual Value vs Predicted Value:\tDifference:")
diff = []
for a, b in zip(df.charges, predictions):
a = np.exp(a)
b = np.exp(b)
diff.append(a - b[0])
print("%.2f \t %.2f \t\t %.2f" % (a, b[0], a - b[0]))
print("Maximum Deviation: %.2f" % max(abs(x) for x in diff))
print("Minimum Deviation: %.2f" % min(abs(x) for x in diff))
print("Average Deviation: %.2f" % np.mean([abs(x) for x in diff]))
# # End
end_time = time.time()
total_time = end_time - start_time
h = total_time // 3600
m = (total_time % 3600) // 60
s = total_time % 60
print("Total Time: %i hours, %i minutes and %i seconds." % (h, m, s))
|
# https://paperswithcode.com/search?q=vessel
# This is a sample code which can give you val_dice=0.8.
# You task is to get val_dice=0.95.
# Good luck!
#!ls /kaggle/input/palmucu/input
# Constants
directory = "./input"
IMG_FORMAT = ".jpg"
MASK_FORMAT = ".bmp"
NUM_CLASSES = 4
SIZE = (640, 640)
height_req, width_req = SIZE
import torch
import torchvision
import os
import cv2
import random
import glob
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import glob
from torch import nn
from torch.nn import functional as F
from torchvision import models
from torchvision import transforms
from torch.utils import data
from pathlib import Path
from torch.nn import functional as F
from torch.autograd import Variable
from tqdm import tqdm, tqdm_notebook
from torch.optim.lr_scheduler import MultiStepLR
# import models
# from utils import *
# from loss import *
from albumentations import (
ToFloat,
CLAHE,
RandomRotate90,
Transpose,
ShiftScaleRotate,
Blur,
OpticalDistortion,
GridDistortion,
HueSaturationValue,
IAAAdditiveGaussianNoise,
GaussNoise,
MotionBlur,
MedianBlur,
IAAPiecewiseAffine,
IAASharpen,
IAAEmboss,
RandomContrast,
RandomBrightness,
Flip,
OneOf,
Compose,
PadIfNeeded,
RandomCrop,
Normalize,
HorizontalFlip,
Resize,
VerticalFlip,
RandomCrop,
)
import albumentations
# train_path = os.path.join("./input", 'train')
# file_list = [f.split('/')[-1].split('.')[0] for f in sorted(glob.glob(train_path + '/images/*png'))]
# ## Merging multiple masks into one
import cv2
from os import listdir
from os.path import isfile, join
from matplotlib import pyplot as plt
import numpy as np
def read_mask_dict(path):
d = dict()
files = [f for f in listdir(path) if isfile(join(path, f))]
for f in files:
img = cv2.imread(join(path, f), cv2.IMREAD_GRAYSCALE)
img = (255 - img) / 255 # inverting
img = cv2.resize(img, SIZE)
img = np.where(img >= 0.5, 1, 0)
key = f.split(".")[0]
d[key] = img
return d
def merge_masks(path):
d1 = read_mask_dict(join(path, "atrophy"))
d2 = read_mask_dict(join(path, "detachment"))
d3 = read_mask_dict(join(path, "disc"))
img_ids = [
f.split(".")[0]
for f in listdir(join(path, "../images"))
if isfile(join(path, "../images", f))
]
for k in img_ids:
zero_mtr = np.zeros(SIZE)
a = d1[k] if k in d1 else zero_mtr
b = d2[k] if k in d2 else zero_mtr
c = d3[k] if k in d3 else zero_mtr
mask = a
mask = np.where(b > 0, 2, mask)
mask = np.where(c > 0, 3, mask)
cv2.imwrite(join(path, "merged", k + MASK_FORMAT), mask)
# def merge_masks(path):
# d1 = read_mask_dict(join(path, "atrophy"))
# d2 = read_mask_dict(join(path, "detachment"))
# d3 = read_mask_dict(join(path, "disc"))
# img_ids = [f.split('.')[0] for f in listdir(join(path, "../images")) if isfile(join(path, "../images", f))]
# for k in img_ids:
# zero_mtr = np.zeros(SIZE)
# a = d1[k] if k in d1 else zero_mtr
# b = d2[k] if k in d2 else zero_mtr
# c = d3[k] if k in d3 else zero_mtr
# merged_mask = np.dstack((a, b, c))
# # merged_mask = np.where(a > 0, 2, b) # background = 0, disc = 1, atrophy = 2
# cv2.imwrite(join(path, "merged", k + MASK_FORMAT), merged_mask)
# path = 'input/train/images/'
# files = [f for f in listdir(path) if isfile(join(path, f))]
# shapes = []
# for f in files:
# img = cv2.imread(join(path, f), cv2.IMREAD_GRAYSCALE)
# shapes.append(img.shape)
# from collections import defaultdict
# d = defaultdict(int)
# for x in shapes:
# d[x] +=1
# d
path = "input/train/masks/"
merge_masks(path)
path = "input/train/masks/merged/P0018.bmp"
mask = cv2.imread(path, 0)
plt.imshow(mask)
class DriveDataset(data.Dataset):
def __init__(self, root_path, file_list, aug=False, mode="train"):
"""Intialize the dataset"""
self.file_list = file_list
self.root_path = root_path
self.image_folder = os.path.join(self.root_path, "images")
self.mask_folder = os.path.join(self.root_path, "masks/merged")
self.mode = mode
self.aug = aug
self.pad = PadIfNeeded(
p=1,
min_height=height_req,
min_width=width_req,
border_mode=cv2.BORDER_CONSTANT,
)
if self.aug:
self.transform = Compose([RandomRotate90(), Transpose(), Flip()])
else:
self.transform = transforms.ToTensor()
def __read_img(self, path, grayscale=False):
# image = cv2.imread(path, 0) if grayscale else cv2.cvtColor(cv2.imread(path), cv2.COLOR_BGR2RGB)
image = cv2.imread(path)
image = cv2.resize(image, SIZE) # pad = self.pad(image=image)
return image
def __read_mask(self, path):
return cv2.imread(path, 0)
def __transform_img(self, img):
image = img.astype("float32") / 255.0
image = np.transpose(image, (2, 0, 1))
return image
def __transform_mask(self, mask):
masks = [(mask == v) for v in range(NUM_CLASSES)]
mask = np.stack(masks, axis=-1).astype("float")
print(mask.shape)
# add background if mask is not binary
if mask.shape[-1] != 1:
background = 1 - mask.sum(axis=-1, keepdims=True)
mask = np.concatenate((mask, background), axis=-1)
def __getitem__(self, index):
"""Get a sample from the dataset"""
image_path = os.path.join(self.image_folder, self.file_list[index] + IMG_FORMAT)
mask_path = os.path.join(self.mask_folder, self.file_list[index] + MASK_FORMAT)
image = self.__read_img(image_path)
if self.mode == "train":
mask = self.__read_mask(mask_path)
if self.aug:
if self.mode == "train":
return self.__transform_img(image), mask[np.newaxis, :, :]
else:
return self.__transform_img(image)
else:
if self.mode == "train":
return self.transform(image), mask[np.newaxis, :, :]
return self.transform(image)
def __len__(self):
return len(self.file_list)
train_path = os.path.join(directory, "train")
file_list = [
f.split("/")[-1].split(".")[0]
for f in sorted(glob.glob(train_path + "/images/*" + IMG_FORMAT))
]
file_list[:5]
# There are only 133 pictures available for training. I took a little bit more than 20% for validation
file_list_val = file_list[::50]
file_list_train = [f for f in file_list if f not in file_list_val]
dataset_train = DriveDataset(train_path, file_list_train, aug=True)
dataset_val = DriveDataset(train_path, file_list_val, aug=False)
image, mask = next(iter(data.DataLoader(dataset_train, batch_size=1, shuffle=True)))
image.shape, mask.shape
# def display(display_list):
# plt.figure(figsize=(15, 15))
# title = ['Input Image', 'True Mask', 'Predicted Mask']
# for i in range(len(display_list)):
# plt.subplot(1, len(display_list), i+1)
# plt.title(title[i])
# plt.imshow(tf.keras.preprocessing.image.array_to_img(display_list[i]))
# plt.axis('off')
# plt.show()
# for image, mask in train.take(1):
# sample_image, sample_mask = image, mask
# display([sample_image, sample_mask])
# ## Model Training
import torch
import torch.nn as nn
def dice_loss(preds, trues, weight=None, is_average=True, eps=1):
num = preds.size(0)
preds = preds.view(num, -1)
trues = trues.view(num, -1)
if weight is not None:
w = torch.autograd.Variable(weight).view(num, -1)
preds = preds * w
trues = trues * w
intersection = (preds * trues).sum(1)
scores = 2.0 * (intersection + eps) / (preds.sum(1) + trues.sum(1) + eps)
if is_average:
score = scores.sum() / num
return torch.clamp(score, 0.0, 1.0)
else:
return scores
def jaccard_loss(preds, trues, weight=None, is_average=True, eps=1e-3):
num = preds.size(0)
preds = preds.view(num, -1)
trues = trues.view(num, -1)
if weight is not None:
w = torch.autograd.Variable(weight).view(num, -1)
preds = preds * w
trues = trues * w
intersection = (preds * trues).sum(1)
scores = (intersection + eps) / ((preds + trues).sum(1) - intersection + eps)
if is_average:
score = scores.sum() / num
return torch.clamp(score, 0.0, 1.0)
else:
return scores
def dice_clamp(preds, trues, is_average=True):
preds = torch.round(preds)
return dice_loss(preds, trues, is_average=is_average)
def jaccard_clamp(preds, trues, is_average=True):
preds = torch.round(preds)
return jaccard_loss(preds, trues, is_average=is_average)
class FocalLossBinary(nn.Module):
"""Focal loss puts more weight on more complicated examples.
https://github.com/warmspringwinds/pytorch-segmentation-detection/blob/master/pytorch_segmentation_detection/losses.py
output is log_softmax
"""
def __init__(self, gamma=2, size_average=True, reduce=True):
super(FocalLossBinary, self).__init__(size_average=size_average, reduce=reduce)
self.gamma = gamma
def forward(self, outputs, targets):
outputs = F.logsigmoid(outputs)
logpt = -F.binary_cross_entropy_with_logits(
outputs, targets.float(), reduce=False
)
pt = torch.exp(logpt)
# compute the loss
loss = -((1 - pt).pow(self.gamma)) * logpt
# averaging (or not) loss
if self.size_average:
return loss.mean()
else:
return loss.sum()
class DiceLoss(nn.Module):
def __init__(self, size_average=True, eps=1):
super().__init__()
self.size_average = size_average
self.eps = eps
def forward(self, input, target, weight=None):
return 1 - dice_loss(
torch.sigmoid(input),
target,
weight=weight,
is_average=self.size_average,
eps=self.eps,
)
class BCEDiceLoss(nn.Module):
def __init__(self, size_average=True):
super().__init__()
self.size_average = size_average
self.dice = DiceLoss(size_average=size_average)
def forward(self, input, target, weight=None):
return nn.modules.loss.BCEWithLogitsLoss(
size_average=self.size_average, weight=weight
)(input, target) + self.dice(input, target, weight=weight)
class JaccardLoss(nn.Module):
def __init__(self, size_average=True, eps=100):
super().__init__()
self.size_average = size_average
self.eps = eps
def forward(self, input, target, weight=None):
return 1 - jaccard_loss(
torch.sigmoid(input),
target,
weight=weight,
is_average=self.size_average,
eps=self.eps,
)
class BCEJaccardLoss(nn.Module):
def __init__(self, size_average=True):
super().__init__()
self.size_average = size_average
self.eps = 100
self.jaccard = JaccardLoss(size_average=size_average, eps=self.eps)
def forward(self, input, target, weight=None):
return nn.modules.loss.BCEWithLogitsLoss(
size_average=self.size_average, weight=weight
)(input, target) + self.jaccard(input, target, weight=weight)
import torch
def save_checkpoint(checkpoint_path, model, optimizer):
state = {"state_dict": model.state_dict(), "optimizer": optimizer.state_dict()}
torch.save(state, checkpoint_path)
print("model saved to %s" % checkpoint_path)
def load_checkpoint(checkpoint_path, model, optimizer):
state = torch.load(checkpoint_path)
model.load_state_dict(state["state_dict"])
optimizer.load_state_dict(state["optimizer"])
print("model loaded from %s" % checkpoint_path)
from torch import nn
from torch.nn import functional as F
import torch
from torchvision import models
import torchvision
from pretrainedmodels import models as pmodels
def conv3x3(in_, out):
return nn.Conv2d(in_, out, 3, padding=1)
class ConvRelu(nn.Module):
def __init__(self, in_, out):
super().__init__()
self.conv = conv3x3(in_, out)
self.activation = nn.ReLU(inplace=True)
def forward(self, x):
x = self.conv(x)
x = self.activation(x)
return x
class DecoderBlock(nn.Module):
def __init__(self, in_channels, middle_channels, out_channels):
super().__init__()
self.block = nn.Sequential(
ConvRelu(in_channels, middle_channels),
nn.ConvTranspose2d(
middle_channels,
out_channels,
kernel_size=3,
stride=2,
padding=1,
output_padding=1,
),
nn.ReLU(inplace=True),
)
def forward(self, x):
return self.block(x)
class UNet11(nn.Module):
def __init__(self, num_filters=32, pretrained=False):
"""
:param num_classes:
:param num_filters:
:param pretrained:
False - no pre-trained network is used
True - encoder is pre-trained with VGG11
"""
super().__init__()
self.pool = nn.MaxPool2d(2, 2)
self.encoder = models.vgg11(pretrained=pretrained).features
self.relu = self.encoder[1]
self.conv1 = self.encoder[0]
self.conv2 = self.encoder[3]
self.conv3s = self.encoder[6]
self.conv3 = self.encoder[8]
self.conv4s = self.encoder[11]
self.conv4 = self.encoder[13]
self.conv5s = self.encoder[16]
self.conv5 = self.encoder[18]
self.center = DecoderBlock(
num_filters * 8 * 2, num_filters * 8 * 2, num_filters * 8
)
self.dec5 = DecoderBlock(
num_filters * (16 + 8), num_filters * 8 * 2, num_filters * 8
)
self.dec4 = DecoderBlock(
num_filters * (16 + 8), num_filters * 8 * 2, num_filters * 4
)
self.dec3 = DecoderBlock(
num_filters * (8 + 4), num_filters * 4 * 2, num_filters * 2
)
self.dec2 = DecoderBlock(
num_filters * (4 + 2), num_filters * 2 * 2, num_filters
)
self.dec1 = ConvRelu(num_filters * (2 + 1), num_filters)
self.final = nn.Conv2d(num_filters, 1, kernel_size=1)
def forward(self, x):
conv1 = self.relu(self.conv1(x))
conv2 = self.relu(self.conv2(self.pool(conv1)))
conv3s = self.relu(self.conv3s(self.pool(conv2)))
conv3 = self.relu(self.conv3(conv3s))
conv4s = self.relu(self.conv4s(self.pool(conv3)))
conv4 = self.relu(self.conv4(conv4s))
conv5s = self.relu(self.conv5s(self.pool(conv4)))
conv5 = self.relu(self.conv5(conv5s))
center = self.center(self.pool(conv5))
dec5 = self.dec5(torch.cat([center, conv5], 1))
dec4 = self.dec4(torch.cat([dec5, conv4], 1))
dec3 = self.dec3(torch.cat([dec4, conv3], 1))
dec2 = self.dec2(torch.cat([dec3, conv2], 1))
dec1 = self.dec1(torch.cat([dec2, conv1], 1))
return self.final(dec1)
def unet11(pretrained=False, **kwargs):
"""
pretrained:
False - no pre-trained network is used
True - encoder is pre-trained with VGG11
carvana - all weights are pre-trained on
Kaggle: Carvana dataset https://www.kaggle.com/c/carvana-image-masking-challenge
"""
model = UNet11(pretrained=pretrained, **kwargs)
if pretrained == "carvana":
state = torch.load("TernausNet.pt")
model.load_state_dict(state["model"])
return model
class Interpolate(nn.Module):
def __init__(
self, size=None, scale_factor=None, mode="nearest", align_corners=False
):
super(Interpolate, self).__init__()
self.interp = nn.functional.interpolate
self.size = size
self.mode = mode
self.scale_factor = scale_factor
self.align_corners = align_corners
def forward(self, x):
x = self.interp(
x,
size=self.size,
scale_factor=self.scale_factor,
mode=self.mode,
align_corners=self.align_corners,
)
return x
class DecoderBlockV2(nn.Module):
def __init__(self, in_channels, middle_channels, out_channels, is_deconv=True):
super(DecoderBlockV2, self).__init__()
self.in_channels = in_channels
if is_deconv:
"""
Paramaters for Deconvolution were chosen to avoid artifacts, following
link https://distill.pub/2016/deconv-checkerboard/
"""
self.block = nn.Sequential(
ConvRelu(in_channels, middle_channels),
nn.ConvTranspose2d(
middle_channels, out_channels, kernel_size=4, stride=2, padding=1
),
nn.ReLU(inplace=True),
)
else:
self.block = nn.Sequential(
Interpolate(scale_factor=2, mode="bilinear"),
ConvRelu(in_channels, middle_channels),
ConvRelu(middle_channels, out_channels),
)
def forward(self, x):
return self.block(x)
class ResNet43Unet(nn.Module):
"""
UNet (https://arxiv.org/abs/1505.04597) with Resnet34(https://arxiv.org/abs/1512.03385) encoder
Proposed by Alexander Buslaev: https://www.linkedin.com/in/al-buslaev/
"""
def __init__(
self, num_classes=1, num_filters=32, pretrained=False, is_deconv=False
):
"""
:param num_classes:
:param num_filters:
:param pretrained:
False - no pre-trained network is used
True - encoder is pre-trained with resnet34
:is_deconv:
False: bilinear interpolation is used in decoder
True: deconvolution is used in decoder
"""
super().__init__()
self.num_classes = num_classes
self.pool = nn.MaxPool2d(2, 2)
self.encoder = torchvision.models.resnet34(pretrained=pretrained)
self.relu = nn.ReLU(inplace=True)
self.conv1 = nn.Sequential(
self.encoder.conv1, self.encoder.bn1, self.encoder.relu, self.pool
)
self.conv2 = self.encoder.layer1
self.conv3 = self.encoder.layer2
self.conv4 = self.encoder.layer3
self.conv5 = self.encoder.layer4
self.center = DecoderBlockV2(
512, num_filters * 8 * 2, num_filters * 8, is_deconv
)
self.dec5 = DecoderBlockV2(
512 + num_filters * 8, num_filters * 8 * 2, num_filters * 8, is_deconv
)
self.dec4 = DecoderBlockV2(
256 + num_filters * 8, num_filters * 8 * 2, num_filters * 8, is_deconv
)
self.dec3 = DecoderBlockV2(
128 + num_filters * 8, num_filters * 4 * 2, num_filters * 2, is_deconv
)
self.dec2 = DecoderBlockV2(
64 + num_filters * 2, num_filters * 2 * 2, num_filters * 2 * 2, is_deconv
)
self.dec1 = DecoderBlockV2(
num_filters * 2 * 2, num_filters * 2 * 2, num_filters, is_deconv
)
self.dec0 = ConvRelu(num_filters, num_filters)
self.final = nn.Conv2d(num_filters, num_classes, kernel_size=1)
def forward(self, x):
conv1 = self.conv1(x)
conv2 = self.conv2(conv1)
conv3 = self.conv3(conv2)
conv4 = self.conv4(conv3)
conv5 = self.conv5(conv4)
center = self.center(self.pool(conv5))
dec5 = self.dec5(torch.cat([center, conv5], 1))
dec4 = self.dec4(torch.cat([dec5, conv4], 1))
dec3 = self.dec3(torch.cat([dec4, conv3], 1))
dec2 = self.dec2(torch.cat([dec3, conv2], 1))
dec1 = self.dec1(dec2)
dec0 = self.dec0(dec1)
if self.num_classes > 1:
x_out = F.log_softmax(self.final(dec0), dim=1)
else:
x_out = self.final(dec0)
return x_out
# class UNet16(nn.Module):
# def __init__(self, num_classes=1, num_filters=32, pretrained=False, is_deconv=False):
# """
# :param num_classes:
# :param num_filters:
# :param pretrained:
# False - no pre-trained network used
# True - encoder pre-trained with VGG16
# :is_deconv:
# False: bilinear interpolation is used in decoder
# True: deconvolution is used in decoder
# """
# super().__init__()
# self.num_classes = num_classes
# self.pool = nn.MaxPool2d(2, 2)
# self.encoder = torchvision.models.vgg16(pretrained=pretrained).features
# self.relu = nn.ReLU(inplace=True)
# self.conv1 = nn.Sequential(self.encoder[0],
# self.relu,
# self.encoder[2],
# self.relu)
# self.conv2 = nn.Sequential(self.encoder[5],
# self.relu,
# self.encoder[7],
# self.relu)
# self.conv3 = nn.Sequential(self.encoder[10],
# self.relu,
# self.encoder[12],
# self.relu,
# self.encoder[14],
# self.relu)
# self.conv4 = nn.Sequential(self.encoder[17],
# self.relu,
# self.encoder[19],
# self.relu,
# self.encoder[21],
# self.relu)
# self.conv5 = nn.Sequential(self.encoder[24],
# self.relu,
# self.encoder[26],
# self.relu,
# self.encoder[28],
# self.relu)
# self.center = DecoderBlockV2(512, num_filters * 8 * 2, num_filters * 8, is_deconv)
# self.dec5 = DecoderBlockV2(512 + num_filters * 8, num_filters * 8 * 2, num_filters * 8, is_deconv)
# self.dec4 = DecoderBlockV2(512 + num_filters * 8, num_filters * 8 * 2, num_filters * 8, is_deconv)
# self.dec3 = DecoderBlockV2(256 + num_filters * 8, num_filters * 4 * 2, num_filters * 2, is_deconv)
# self.dec2 = DecoderBlockV2(128 + num_filters * 2, num_filters * 2 * 2, num_filters, is_deconv)
# self.dec1 = ConvRelu(64 + num_filters, num_filters)
# self.final = nn.Conv2d(num_filters, num_classes, kernel_size=1)
# def forward(self, x):
# conv1 = self.conv1(x)
# conv2 = self.conv2(self.pool(conv1))
# conv3 = self.conv3(self.pool(conv2))
# conv4 = self.conv4(self.pool(conv3))
# conv5 = self.conv5(self.pool(conv4))
# center = self.center(self.pool(conv5))
# dec5 = self.dec5(torch.cat([center, conv5], 1))
# dec4 = self.dec4(torch.cat([dec5, conv4], 1))
# dec3 = self.dec3(torch.cat([dec4, conv3], 1))
# dec2 = self.dec2(torch.cat([dec3, conv2], 1))
# dec1 = self.dec1(torch.cat([dec2, conv1], 1))
# if self.num_classes > 1:
# x_out = F.log_softmax(self.final(dec1), dim=1)
# else:
# x_out = self.final(dec1)
# return x_out
# import torch
# import torchvision
# import os
# import cv2
# import random
# import glob
# import numpy as np
# import matplotlib.pyplot as plt
# import pandas as pd
# import glob
# from torch import nn
# from torch.nn import functional as F
# from torchvision import models
# from torchvision import transforms
# from torch.utils import data
# from pathlib import Path
# from torch.nn import functional as F
# from torch.autograd import Variable
# from tqdm import tqdm, tqdm_notebook
# from torch.optim.lr_scheduler import MultiStepLR
# # import models
# # from utils import *
# # from loss import *
# %matplotlib inline
# from albumentations import (ToFloat,
# CLAHE, RandomRotate90, Transpose, ShiftScaleRotate, Blur, OpticalDistortion,
# GridDistortion, HueSaturationValue, IAAAdditiveGaussianNoise, GaussNoise, MotionBlur,
# MedianBlur, IAAPiecewiseAffine, IAASharpen, IAAEmboss, RandomContrast, RandomBrightness,
# Flip, OneOf, Compose, PadIfNeeded, RandomCrop, Normalize, HorizontalFlip, Resize, VerticalFlip,
# RandomCrop
# )
# import albumentations
# !ls sample_data
# class DriveDataset(data.Dataset):
# def __init__(self, root_path, file_list, aug = False, mode='train'):
# """ Intialize the dataset
# """
# self.file_list = file_list
# self.root_path = root_path
# self.image_folder = os.path.join(self.root_path, "images")
# self.mask_folder = os.path.join(self.root_path, "masks/merged")
# self.mode = mode
# self.aug = aug
# self.pad = PadIfNeeded(p=1,
# min_height=height_req,
# min_width=width_req,
# border_mode=cv2.BORDER_CONSTANT)
# if self.aug:
# self.transform = Compose([
# RandomRotate90(),
# Transpose(),
# Flip()
# ])
# else:
# self.transform = transforms.ToTensor()
# def __read_img(self, path, grayscale=False):
# # image = cv2.imread(path, 0) if grayscale else cv2.cvtColor(cv2.imread(path), cv2.COLOR_BGR2RGB)
# image = cv2.imread(path)
# image = cv2.resize(image, SIZE)#pad = self.pad(image=image)
# return image
# def __read_mask(self, path):
# image = cv2.imread(path)
# bkg = image[:,:,0] + image[:,:,1] + image[:,:,2]
# bkg = np.where(bkg > 0, 0, 1)
# return np.dstack((image, bkg))
# def __transform_img(self, img):
# image = img.astype('float32') / 255.
# image = np.transpose(image, (2, 0, 1))
# return image
# def __transform_mask(self, mask):
# masks = [(mask == v) for v in range(NUM_CLASSES)]
# mask = np.stack(masks, axis=-1).astype('float')
# print(mask.shape)
# # add background if mask is not binary
# if mask.shape[-1] != 1:
# background = 1 - mask.sum(axis=-1, keepdims=True)
# mask = np.concatenate((mask, background), axis=-1)
# def __getitem__(self, index):
# """ Get a sample from the dataset
# """
# image_path = os.path.join(self.image_folder, self.file_list[index] + IMG_FORMAT)
# mask_path = os.path.join(self.mask_folder, self.file_list[index] + MASK_FORMAT)
# image = self.__read_img(image_path)
# if self.mode == 'train':
# mask = self.__read_mask(mask_path)
# if self.aug:
# if self.mode == 'train':
# return self.__transform_img(image), self.__transform_img(mask)
# else:
# return self.__transform_img(image)
# else:
# if self.mode == 'train':
# return self.transform(image), self.__transform_img(mask)
# return self.transform(image)
# def __len__(self):
# return len(self.file_list)
# train_path = os.path.join(directory, 'train')
# file_list = [f.split('/')[-1].split('.')[0] for f in sorted(glob.glob(train_path + '/images/*' + IMG_FORMAT))]
# file_list[:5]
# #There are only 133 pictures available for training. I took a little bit more than 20% for validation
# file_list_val = file_list[::30]
# file_list_train = [f for f in file_list if f not in file_list_val]
# dataset_train = DriveDataset(train_path, file_list_train, aug=True)
# dataset_val = DriveDataset(train_path, file_list_val, aug=False)
# image, mask = next(iter(data.DataLoader(dataset_train, batch_size=1, shuffle=True)))
# image.shape, mask.shape
images, masks = next(iter(data.DataLoader(dataset_train, batch_size=4)))
plt.figure(figsize=(16, 16))
plt.subplot(211)
plt.imshow(torchvision.utils.make_grid(images).data.numpy().transpose((1, 2, 0)))
plt.subplot(212)
plt.imshow(torchvision.utils.make_grid(masks).data.numpy().transpose((1, 2, 0)))
model = ResNet43Unet(num_classes=4).cuda()
epoch = 20
learning_rate = 3e-4
# loss_fn = torch.nn.BCEWithLogitsLoss()
loss_fn = torch.nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(model.parameters(), lr=learning_rate)
loader_train = data.DataLoader(dataset_train, batch_size=4, shuffle=True)
loader_val = data.DataLoader(dataset_val, batch_size=4, shuffle=False)
for e in range(epoch):
train_loss = []
train_dice = []
train_jaccard = []
for image, mask in loader_train:
image, mask = image.cuda(), mask.cuda()
mask = mask.squeeze()
optimizer.zero_grad()
y_pred = model(image)
# print(type(y_pred), y_pred.size(), type(mask), mask.size())
loss = loss_fn(y_pred, mask.long())
dice = dice_clamp(
torch.argmax(y_pred, axis=1).contiguous().float(), mask.contiguous()
)
jaccard = jaccard_clamp(
torch.argmax(y_pred, axis=1).contiguous().float(), mask.contiguous()
)
loss.backward()
optimizer.step()
train_loss.append(loss.item())
train_dice.append(dice.item())
train_jaccard.append(jaccard.item())
val_loss = []
val_dice = []
val_jaccard = []
for image, mask in loader_val:
image, mask = image.cuda(), mask.cuda()
mask = mask.squeeze()
y_pred = model(image)
loss = loss_fn(y_pred, mask.long())
dice = dice_clamp(
torch.argmax(y_pred, axis=1).contiguous().float(), mask.contiguous()
)
jaccard = jaccard_clamp(
torch.argmax(y_pred, axis=1).contiguous().float(), mask.contiguous()
)
val_loss.append(loss.item())
val_dice.append(dice.item())
val_jaccard.append(jaccard.item())
print(
"epoch: %d, train_loss: %.3f, train_dice: %.3f, train_jaccard: %.3f, val_loss: %.3f, val_dice: %.3f, val_jaccard: %.3f"
% (
e,
np.mean(train_loss),
np.mean(train_dice),
np.mean(train_jaccard),
np.mean(val_loss),
np.mean(val_dice),
np.mean(val_jaccard),
)
)
save_checkpoint("drive-%i.pth" % epoch, model, optimizer)
test_path = os.path.join(directory, "test")
test_file_list = glob.glob(os.path.join(test_path, "images", "*" + IMG_FORMAT))
test_file_list = [f.split("/")[-1].split(".")[0] for f in test_file_list]
print("First 3 names of test files:", test_file_list[:3])
print(f"Test size: {len(test_file_list)}")
test_dataset = DriveDataset(test_path, test_file_list, mode=None)
all_images = []
all_predictions = []
for image in tqdm_notebook(data.DataLoader(test_dataset, batch_size=2)):
image = image.cuda()
# print(image.shape)
all_images.append(image.cpu().data.numpy())
y_pred = torch.sigmoid(model(image)).cpu().data.numpy()
all_predictions.append(y_pred)
all_predictions_stacked = np.vstack(all_predictions)[:, 0, :, :]
all_images = np.vstack(all_images)[:, :, :, :]
plt.figure(figsize=(20, 10))
plt.subplot(1, 2, 1)
plt.imshow(all_images[0].transpose(1, 2, 0))
plt.subplot(1, 2, 2)
plt.imshow(all_predictions_stacked[0])
plt.figure(figsize=(20, 10))
plt.subplot(1, 2, 1)
plt.imshow(all_images[1].transpose(1, 2, 0))
plt.subplot(1, 2, 2)
plt.imshow(all_predictions_stacked[1])
print(all_images[1].shape)
plt.figure(figsize=(20, 10))
plt.subplot(1, 2, 1)
plt.imshow(all_images[0].transpose(1, 2, 0))
plt.subplot(1, 2, 2)
plt.imshow(all_predictions_stacked[0])
|
# # Clustering - Crime Rate
# ### Perform Clustering(Hierarchical, Kmeans & DBSCAN) for the crime data and identify the number of clusters formed and draw inferences.
# #### Data Description:
# Murder -- Muder rates in different places of United States
# Assualt- Assualt rate in different places of United States
# UrbanPop - urban population in different places of United States
# Rape - Rape rate in different places of United States
# ### Project Background
# Analysing the different crime rates in different areas of United States.
# ### Step-1 : Import libraries
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sb
import warnings as warnings
warnings.filterwarnings("ignore")
from sklearn.preprocessing import StandardScaler
from sklearn.preprocessing import MinMaxScaler
from scipy.cluster.hierarchy import linkage
import scipy.cluster.hierarchy as sch
from sklearn.cluster import AgglomerativeClustering
import sklearn
from sklearn.neighbors import NearestNeighbors
from sklearn.cluster import DBSCAN
# ### Step-2 : Descriptive analysis
# ### Step-2.1 : Working with Dataset
# #### Loading the Dataset
# Crime data
crime_data = pd.read_csv("/kaggle/input/crime-data/crime_data.csv")
# #### Exploring the dataset
# Displaying first ten records
crime_data.head(10)
# Shape of the data
crime_data.shape
# Summary of the data
crime_data.describe()
# Check the data types
crime_data.info()
# #### Modifying the dataset
# Renaming the Unnamed: 0 Column into States
crime_data = crime_data.rename(columns={"Unnamed: 0": "States"})
crime_data.head(10)
# ### Step-2.2 : Handling missing values
# Check for Null values
crime_data.isnull().sum()
# Check for duplicate values
crime_data[crime_data.duplicated()].shape
crime_data[crime_data.duplicated()]
# #### There are no missing values.
# ### Step-2.3 : Exploring data using Visualisation
# #### Histogram
crime_data.hist(figsize=(14, 8), bins=25)
plt.show()
# #### Boxplot
for feature in crime_data.columns[1:]:
plt.figure(figsize=(12, 4))
sb.boxplot(crime_data[feature])
# #### Comparing features using Boxplots
plt.figure(figsize=(12, 8))
sb.boxplot(data=crime_data)
# #### Observation
# + We can clearly see that Assault crime rate is high, followed by rape and murder crime rate.
# #### Correlation and Heat map
# Checking for Correlation between variables
crime_data.corr()
plt.figure(figsize=(12, 10))
sb.heatmap(crime_data.corr(), annot=True)
plt.show()
# #### Observation
# + We can see that 'Urban' population is not much influencing the Assault and murder rate. But it may be a reason for Rape rate.
# + But Murder rate and Rape rate are strongly correlated to Assault rate.
sb.pairplot(data=crime_data)
# #### Visualizing features State wise
for i in crime_data.columns[1:]:
plt.figure(figsize=(16, 8))
sb.barplot(
x=crime_data["States"],
y=i,
data=crime_data,
order=crime_data.sort_values(i).States,
)
plt.xticks(rotation=90, fontsize=12)
plt.title("State wise " + str(i) + " rate", fontsize=18, fontweight="bold")
plt.show()
for i in crime_data.columns[1:]:
plt.figure(figsize=(12, 6))
sb.barplot(
x=i,
y=crime_data["States"],
data=crime_data,
order=crime_data.sort_values(i, ascending=False).States[:5],
)
plt.xticks(rotation=90, fontsize=12)
plt.title(
"Top 5 States with Highest " + str(i) + " rate", fontsize=18, fontweight="bold"
)
plt.show()
# #### Observation
# + In Florida, the Murder rate and Assault rate are high.
# + In California, the Urban population is high due to which the Rape rate is also high.
# + But in other countries, we don't see much resemblence.
for i in crime_data.columns[1:]:
plt.figure(figsize=(12, 6))
sb.barplot(
x=i,
y=crime_data["States"],
data=crime_data,
order=crime_data.sort_values(i, ascending=True).States[:5],
)
plt.xticks(rotation=90, fontsize=12)
plt.title(
"Top 5 States with Lowest " + str(i) + " rate", fontsize=18, fontweight="bold"
)
plt.show()
# #### Observation
# + We can clearly see that the North Dakota has the lowest crime rate and the urban population is also less.
# + Vermont with lowest urban population has the low murder rate and assault rate.
# + But in other countries, we don't see much resemblence.
# ### Step-3 : Pre-Processing the Data
# ### Step-3.1 : Standardising the data
crime_data_mdf = crime_data.drop(["States"], axis=1)
crime_data_mdf.head()
standard_scaler = StandardScaler()
crime_data_std = standard_scaler.fit_transform(crime_data_mdf)
crime_data_std.shape
# ### Step-3.2 : Normalising the data
min_max = MinMaxScaler()
crime_data_norm = min_max.fit_transform(crime_data_mdf)
crime_data_norm.shape
# #### However both give almost same result. So adopt Standardised data.
# ### Step-4 : Build Clustering model for Standardised data
# ### Step-4.1 : Using Hierarchical Clustering model
# #### Step-4.1.1 : Creating Linkage to determine number of clusters
z = linkage(crime_data_std, method="average", metric="euclidean")
plt.figure(figsize=(15, 5))
plt.title("Hierarchical Clustering Dendrogram")
plt.xlabel("Index")
plt.ylabel("Distance")
sch.dendrogram(z)
plt.show()
# #### Here we can see that there are 5 no . of clusters.
# #### Step-4.1.2 : Buliding the Clustering model
# #### Before selecting no. of clusters, check Silhouette score by varying number of clusters in the model build. Silhouette score gives the accuracy of a model.
for nc in range(2, 6):
h_cluster = AgglomerativeClustering(
n_clusters=nc, linkage="average", affinity="euclidean"
).fit(crime_data_std)
cluster_labels = pd.Series(h_cluster.labels_)
slht_scr = sklearn.metrics.silhouette_score(crime_data_std, cluster_labels)
print("For n_clusters =", nc, "The average silhouette_score is :", slht_scr)
# #### Model with 4 no. of clusters has the highest accuracy. Hence build the model with 4 no. of clusters.
h_cluster = AgglomerativeClustering(
n_clusters=4, linkage="average", affinity="euclidean"
).fit(crime_data_std)
cluster_labels = pd.Series(h_cluster.labels_)
cluster_labels
crime_data["clust"] = cluster_labels
crime_data
# #### Step-4.1.3 : Analysing the results obtained from the model
crime_data.iloc[:, 1:].groupby(crime_data.clust).mean()
# #### We can see that 4 no. of clusters are formed.
# #### Following are the Different states in each cluster
h_cluster_0 = crime_data[(crime_data.clust == 0)]
h_cluster_0
h_cluster_1 = crime_data[(crime_data.clust == 1)]
h_cluster_1
h_cluster_2 = crime_data[(crime_data.clust == 2)]
h_cluster_2
h_cluster_3 = crime_data[(crime_data.clust == 3)]
h_cluster_3
# #### Observation
# + We can observe that Cluster-2 contains only one observation, that is of state 'Alaska'.
# + 'Alaska' has low urban population rate, but it has high Rape rate when compared to state 'South Calorina' of Cluster-3 having same urban popualtion rate. That is the reason why it stood differently from all the clusters.
plt.style.use(["classic"])
plt.figure(figsize=(12, 6))
crime_data.plot(
x="UrbanPop",
y="Rape",
c=cluster_labels,
label="Cluster Number",
kind="scatter",
s=100,
cmap=plt.cm.coolwarm,
)
plt.title("Clusters using Hierarchical Clustering")
plt.show()
# #### Observation
# + We can see the different clusters indicated in different colors.
# + As mentioned above the Cluster-2 observation is completely different with low Urban population rate and high Rape rate.
plt.style.use(["classic"])
plt.figure(figsize=(12, 6))
crime_data.plot(
x="Assault",
y="Murder",
c=cluster_labels,
label="Cluster Number",
kind="scatter",
s=100,
cmap=plt.cm.coolwarm,
)
plt.title("Clusters using Hierarchical Clustering")
plt.show()
# #### We can see that some of the observations of one cluster are overlapping with the other cluster. So let us check the accuracy of the model in classifying data by evaluating Silhouette score.
# #### Evaluating accuracy of the model using Silhouette score
# Silhouette score ranges from -1 to 1. '-1' indicates model is not good. '1' indicates model is best.
# '0' indicates clusters are overlapping
slht_scr_hc = sklearn.metrics.silhouette_score(crime_data_std, cluster_labels)
slht_scr_hc
# #### The score is closer to '0'. It indicates that the model is not that accurate at classifying the data, as clusters are overlapping. So try other methods of clustering.
# ### Step-4.2 : Using K-means Clustering model
# #### Step-4.2.1 : Determing no. of clusters using 'Elbow method'
from sklearn.cluster import KMeans
fig = plt.figure(figsize=(10, 8))
WCSS = []
for i in range(1, 11):
k_cluster = KMeans(n_clusters=i)
k_cluster.fit(crime_data_std)
WCSS.append(k_cluster.inertia_)
plt.plot(range(1, 11), WCSS)
plt.title("The Elbow Method")
plt.ylabel("WCSS")
plt.xlabel("Number of Clusters")
plt.show()
# #### Therefore from the elbow curve, we can observe that the number of clusters are 5.
# #### Step-4.2.2 : Buliding the Clustering model
# #### Before selecting no. of clusters, check Silhouette score by varying number of clusters in the model build. Silhouette score gives the accuracy of a model.
# Silhouette score ranges from -1 to 1. '-1' indicates model is not good. '1' indicates model is best.
# '0' indicates clusters are overlapping
for nc in range(2, 6):
k1_cluster = KMeans(n_clusters=nc, random_state=0)
y1_kmeans = k1_cluster.fit_predict(crime_data_std)
slht_scr = sklearn.metrics.silhouette_score(crime_data_std, y1_kmeans)
print("For n_clusters =", nc, "The average silhouette_score is :", slht_scr)
# #### Model with 4 no. of clusters has the highest accuracy. Hence build the model with 4 no. of clusters.
k_cluster = KMeans(n_clusters=4)
y_kmeans = k_cluster.fit_predict(crime_data_std)
y_kmeans
k_cluster.labels_
k_cluster.cluster_centers_
# Within cluster sum of squares or variance
k_cluster.inertia_
ks = pd.Series(y_kmeans)
crime_data["clust"] = ks
crime_data
# #### Step-4.2.3 : Analysing the results obtained from the model
crime_data.iloc[:, 1:].groupby(crime_data.clust).mean()
# #### Following are the Different states in each cluster
k_cluster_0 = crime_data[(crime_data.clust == 0)]
k_cluster_0
k_cluster_1 = crime_data[(crime_data.clust == 1)]
k_cluster_1
k_cluster_2 = crime_data[(crime_data.clust == 2)]
k_cluster_2
k_cluster_3 = crime_data[(crime_data.clust == 3)]
k_cluster_3
plt.style.use(["classic"])
plt.figure(figsize=(12, 6))
crime_data.plot(
x="UrbanPop",
y="Rape",
c=k_cluster.labels_,
label="Cluster Number",
kind="scatter",
s=100,
cmap=plt.cm.coolwarm,
)
plt.title("Clusters using KMeans clustering")
plt.show()
plt.style.use(["classic"])
plt.figure(figsize=(12, 6))
crime_data.plot(
x="Assault",
y="Murder",
c=k_cluster.labels_,
label="Cluster Number",
kind="scatter",
s=100,
cmap=plt.cm.coolwarm,
)
plt.title("Clusters using KMeans clustering")
plt.show()
# #### We can see that some of the observations of one cluster are overlapping with the other cluster. So let us check the accuracy of the model in classifying data by evaluating Silhouette score.
# #### Evaluating accuracy of the model using Silhouette score
# Silhouette score ranges from -1 to 1. '-1' indicates model is not good. '1' indicates model is best.
# '0' indicates clusters are overlapping
slht_scr_kc = sklearn.metrics.silhouette_score(crime_data_std, k_cluster.labels_)
slht_scr_kc
# #### The score is closer to '0'. It indicates that the model is not that accurate at classifying the data, as clusters are overlapping. So try other methods of clustering.
# ### Step-4.3 : DB Scan Clustering model
# #### Step-4.3.1 : Determining epsilon value
# Nearest neighbours = 2x(no. of Dimensions/columns)
nn = NearestNeighbors(n_neighbors=8)
nn = nn.fit(crime_data_std)
distances, indices = nn.kneighbors(crime_data_std)
distances = np.sort(distances, axis=0)
distances = distances[:, 1]
plt.plot(distances)
# #### The curve took a sharp bend at a value of 1.25. Hence the epsilon value is 1.25.
# #### Step-4.3.2 : Building the DB Scan clustering model
# min_samples=(no. of Dimensions/columns)+1
d_cluster = DBSCAN(eps=1.25, min_samples=5)
d_cluster.fit(crime_data_std)
d_cluster.labels_
ds = pd.Series(d_cluster.labels_)
crime_data["clust"] = ds
crime_data
# #### Step-4.3.3 : Analysing the results obtained from the model
crime_data.iloc[:, 1:].groupby(crime_data.clust).mean()
# #### Following are the Different states in each cluster
d_cluster_neg1 = crime_data[(crime_data.clust == -1)]
d_cluster_neg1
# #### Clusters with label '-1' are generally categorised as Outliers.
d_cluster_0 = crime_data[(crime_data.clust == 0)]
d_cluster_0
d_cluster_1 = crime_data[(crime_data.clust == 1)]
d_cluster_1
d_cluster_2 = crime_data[(crime_data.clust == 2)]
d_cluster_2
plt.style.use(["classic"])
plt.figure(figsize=(12, 6))
crime_data.plot(
x="UrbanPop",
y="Rape",
c=d_cluster.labels_,
label="Cluster Number",
kind="scatter",
s=100,
cmap=plt.cm.coolwarm,
)
plt.title("Clusters using DB Scan Clustering")
plt.show()
plt.style.use(["classic"])
plt.figure(figsize=(12, 6))
crime_data.plot(
x="Assault",
y="Murder",
c=d_cluster.labels_,
label="Cluster Number",
kind="scatter",
s=100,
cmap=plt.cm.coolwarm,
)
plt.title("Clusters using DB Scan clustering")
plt.show()
# #### We can see that some of the observations of one cluster are overlapping with the other cluster. So let us check the accuracy of the model in classifying data by evaluating Silhouette score.
# #### Evaluating accuracy of the model using Silhouette score
# Silhouette score ranges from -1 to 1. '-1' indicates model is not good. '1' indicates model is best.
# '0' indicates clusters are overlapping
slht_scr_dc = sklearn.metrics.silhouette_score(crime_data_std, d_cluster.labels_)
slht_scr_dc
# #### Observation
# + The Silhouette score considering 4 number of clusters is closer to '0'. Hence DB Scan clustering model is not able to classify the dataset accurately. However it detected outliers.
# ### Step-5 : Comparing all the models using Silhouette score
print("The Silhouette score of Hierarchical Clustering:", slht_scr_hc)
print("The Silhouette score of KMeans Clustering:", slht_scr_kc)
print("The Silhouette score of DB Scan Clustering:", slht_scr_dc)
# #### Here we can see that the Silhouette score for Hierarchical clustering is more than other two models.
# ### Step-6 : Conclusion
# #### Hence the Hierarchical clustering model can be selected to classify the dataset.
plt.style.use(["classic"])
plt.figure(figsize=(12, 6))
crime_data.plot(
x="Assault",
y="Murder",
c=cluster_labels,
label="Cluster Number",
kind="scatter",
s=100,
cmap=plt.cm.coolwarm,
)
plt.title("Clusters using Hierarchical Clustering")
plt.show()
|
# **Import Libraries**
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as mpl
import warnings
from sklearn.ensemble import RandomForestClassifier
# Import Data
train_df = pd.read_csv("../input/titanic/train.csv")
test_df = pd.read_csv("../input/titanic/test.csv")
# Data Exploration
train_df.head()
train_df.describe()
train_df.info()
# Change Sex to Number
train_df["Sex"][train_df["Sex"] == "male"] = 0
train_df["Sex"][train_df["Sex"] == "female"] = 1
test_df["Sex"][test_df["Sex"] == "male"] = 0
test_df["Sex"][test_df["Sex"] == "female"] = 1
list(train_df.columns)
train_df = train_df.drop(
columns=[
"PassengerId",
"Pclass",
"Name",
"Age",
"SibSp",
"Parch",
"Ticket",
"Fare",
"Cabin",
"Embarked",
],
axis=1,
)
test_df = test_df.drop(
columns=[
"PassengerId",
"Pclass",
"Name",
"Age",
"SibSp",
"Parch",
"Ticket",
"Fare",
"Cabin",
"Embarked",
],
axis=1,
)
X_train = train_df.drop("Survived", axis=1)
Y_train = train_df["Survived"]
X_test = test_df
random_forest = RandomForestClassifier(n_estimators=100)
random_forest.fit(X_train, Y_train)
Y_pred = random_forest.predict(X_test)
random_forest.score(X_train, Y_train)
|
import os
import json
from collections import Counter
import pandas as pd
import numpy as np
ROBERTA_EXPERIMENT_DIR = "2-4-roberta-base-saved-5-head_tail-roberta-stackx-base-v2-pl1kksample20k-1e-05-210-260-500-26-roberta-200"
ROBERTA_CONFIG = {
"_seed": 42,
"batch_accumulation": 2,
"batch_size": 4,
"bert_model": "roberta-base-saved",
"folds": 5,
"head_tail": True,
"label": "roberta-stackx-base-v2-pl1kksample20k",
"lr": 1e-05,
"max_answer_length": 210,
"max_question_length": 260,
"max_sequence_length": 500,
"max_title_length": 26,
"model_type": "roberta",
"warmup": 200,
}
with open(os.path.join(ROBERTA_EXPERIMENT_DIR, "config.json"), "w") as fp:
json.dump(ROBERTA_CONFIG, fp)
target_columns = [
"question_asker_intent_understanding",
"question_body_critical",
"question_conversational",
"question_expect_short_answer",
"question_fact_seeking",
"question_has_commonly_accepted_answer",
"question_interestingness_others",
"question_interestingness_self",
"question_multi_intent",
"question_not_really_a_question",
"question_opinion_seeking",
"question_type_choice",
"question_type_compare",
"question_type_consequence",
"question_type_definition",
"question_type_entity",
"question_type_instructions",
"question_type_procedure",
"question_type_reason_explanation",
"question_type_spelling",
"question_well_written",
"answer_helpful",
"answer_level_of_information",
"answer_plausible",
"answer_relevance",
"answer_satisfaction",
"answer_type_instructions",
"answer_type_procedure",
"answer_type_reason_explanation",
"answer_well_written",
]
def postprocess_single(target, ref):
"""
The idea here is to make the distribution of a particular predicted column
to match the correspoding distribution of the corresponding column in the
training dataset (called ref here)
"""
ids = np.argsort(target)
counts = sorted(Counter(ref).items(), key=lambda s: s[0])
scores = np.zeros_like(target)
last_pos = 0
v = 0
for value, count in counts:
next_pos = last_pos + int(round(count / len(ref) * len(target)))
if next_pos == last_pos:
next_pos += 1
cond = ids[last_pos:next_pos]
scores[cond] = v
last_pos = next_pos
v += 1
return scores / scores.max()
def postprocess_prediction(prediction, actual):
postprocessed = prediction.copy()
for col in target_columns:
scores = postprocess_single(prediction[col].values, actual[col].values)
# Those are columns where our postprocessing gave substantial improvement.
# It also helped for some others, but we didn't include them as the gain was
# very marginal (less than 0.01)
if col in (
"question_conversational",
"question_type_compare",
"question_type_definition",
"question_type_entity",
"question_has_commonly_accepted_answer",
"question_type_consequence",
"question_type_spelling",
):
postprocessed[col] = scores
return postprocessed
roberta_base_dfs = [
pd.read_csv(os.path.join("roberta-base-output", "fold-{}.csv".format(fold)))
for fold in range(5)
]
roberta_pred_df = roberta_base_dfs[0].copy()
for col in target_columns:
roberta_pred_df[col] = np.mean([df[col] for df in roberta_base_dfs], axis=0)
bert_base_pred_df = pd.read_csv("bert_base.csv")
bart_base_pred_df = pd.read_csv("bart_base.csv")
bert_old_base_pred_df = pd.read_csv("old_base.csv")
train_df = pd.read_csv("/kaggle/input/google-quest-challenge/train.csv")
blended_df = roberta_pred_df.copy()
for col in target_columns:
blended_df[col] = (
bert_old_base_pred_df[col] * 0.1
+ bert_base_pred_df[col] * 0.2
+ roberta_pred_df[col] * 0.1
+ bart_base_pred_df[col] * 0.3
)
postprocessed = postprocess_prediction(blended_df, train_df)
for col in target_columns:
# scale to 0-1 interval
v = postprocessed[col].values
postprocessed[col] = (v - v.min()) / (v.max() - v.min())
postprocessed.to_csv("submission.csv", index=False)
|
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
from sklearn.metrics import mean_absolute_error
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestRegressor
test = pd.read_csv("/kaggle/input/home-data-for-ml-course/test.csv")
train = pd.read_csv("/kaggle/input/home-data-for-ml-course/train.csv")
train.columns
y = train.SalePrice
# Create X (After completing the exercise, you can return to modify this line!)
features = [
"MSSubClass",
"LotArea",
"OverallQual",
"OverallCond",
"YearBuilt",
"YearRemodAdd",
"1stFlrSF",
"2ndFlrSF",
"LowQualFinSF",
"GrLivArea",
"FullBath",
"HalfBath",
"BedroomAbvGr",
"KitchenAbvGr",
"TotRmsAbvGrd",
"Fireplaces",
"WoodDeckSF",
"OpenPorchSF",
"EnclosedPorch",
"3SsnPorch",
"ScreenPorch",
"PoolArea",
"MiscVal",
"MoSold",
"YrSold",
]
# Select columns corresponding to features, and preview the data
X = train[features]
X.head()
# Split into validation and training data
train_X, val_X, train_y, val_y = train_test_split(X, y, random_state=1)
rf_model = RandomForestRegressor(random_state=1)
rf_model.fit(train_X, train_y)
rf_val_predictions = rf_model.predict(val_X)
rf_val_mae = mean_absolute_error(rf_val_predictions, val_y)
print("Validation MAE for Random Forest Model: {:,.0f}".format(rf_val_mae))
test.columns
rf_model_on_full_data = RandomForestRegressor(random_state=1)
test_X = test[features]
rf_model_on_full_data.fit(X, y)
rf_model_on_full_data_val_predictions = rf_model.predict(test_X)
output = pd.DataFrame(
{"Id": test.Id, "SalePrice": rf_model_on_full_data_val_predictions}
)
output.to_csv("submission.csv", index=False)
|
# In this notebook I have tried to perform a thorough EDA of stock price data of Google. I have kept the code very simple and self-explanatory so that it is easy to understand and use for beginners. :D
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
from matplotlib import pyplot as plt
import seaborn as sns
from matplotlib import style
import random
import plotly.express as px
from plotly.offline import init_notebook_mode
init_notebook_mode(connected=True)
df1 = pd.read_csv(
"/kaggle/input/google-stock-price-daily-weekly-and-monthly-2023/google-stock-dataset-Daily.csv"
)
df2 = pd.read_csv(
"/kaggle/input/google-stock-price-daily-weekly-and-monthly-2023/google-stock-dataset-Monthly.csv"
)
df3 = pd.read_csv(
"/kaggle/input/google-stock-price-daily-weekly-and-monthly-2023/google-stock-dataset-Weekly.csv"
)
df1.info()
# Setting Company Name as Index and changing the Datatype of Date column to 'datetime64'
df1.drop(["Unnamed: 0"], axis=1, inplace=True)
df2.drop(["Unnamed: 0"], axis=1, inplace=True)
df3.drop(["Unnamed: 0"], axis=1, inplace=True)
df1.rename(columns={"Price": "Open"}, inplace=True)
df2.rename(columns={"Price": "Open"}, inplace=True)
df3.rename(columns={"Price": "Open"}, inplace=True)
def set_ind(df, cmp_name="Google"):
df["Date"] = df["Date"].apply(pd.to_datetime)
df["Company"] = cmp_name
df.set_index("Company", inplace=True)
set_ind(df1)
set_ind(df2)
set_ind(df3)
df1.head()
df2.head()
df3.head()
# EXPLORATORY DATA ANALYSIS
# ANALYSING VOLUMES OF STOCK TRADED
def volume_analysis(df, cmp):
plt.figure(figsize=(10, 5))
plt.plot(df["Date"], df["Volume"], c="orange")
plt.title("Volume Of " + cmp + " Stock Traded")
plt.ylabel("Volume")
plt.show()
volume_analysis(df1, "Daily")
volume_analysis(df2, "Monthly")
volume_analysis(df1, "Weekly")
# Analysing Opening and Closing price trends
def price(df, name):
fig = px.histogram(
df,
x="Date",
y=["Open", "Close"],
template="plotly_dark",
color_discrete_sequence=["gold", "snow"],
title="{} Opening and Closing Stock Price Over Time".format(name),
)
fig.update_layout(xaxis_title="Date", yaxis_title="Stock Price")
fig.show()
print("PLOTS OF OPENING AND CLOSING PRICE")
print("\n\n\n")
price(df1, "Daily")
print("\n\n\n")
price(df1, "Monthly")
print("\n\n\n")
price(df1, "Weekly")
print("\n\n\n")
# A comparitive analysis of the high and low prices of stock over the years showing change in the daily range of trading.
def high_low(df):
plt.figure(figsize=(20, 8))
plt.style.use("seaborn-deep")
df["Daily Range"] = df["High"] - df["Low"]
plt.plot(df["Date"], df["Daily Range"], c="blue")
plt.xlabel("Years")
plt.ylabel("Stock Price Trading Range")
plt.title("Daily Stock Price Range for Google")
plt.show()
print("PLOTS OF DAILY TRAIDNG RANGE")
print("\n\n\n")
high_low(df1)
# Analysing the Daily Returns on Stock
def daily_returns(df):
df["Daily Return"] = df["Adj Close"].pct_change()
# We chose Adj Close here because it is inidicated as the fair price of the stock on that day.
# However you may also take CLose price for the analysis.\
fig = px.histogram(
df,
x="Date",
y="Daily Return",
template="plotly_dark",
nbins=60,
color_discrete_sequence=["cyan"],
title="Daily Returns on stock",
)
fig.update_layout(xaxis_title="Date", yaxis_title="Daily Returns(%)")
fig.show()
daily_returns(df1)
|
# ## 1. Исходный датасет
import pandas as pd
import re
import matplotlib.pyplot as plt
import seaborn as sns
import numpy as np
# Объединяем вместе тренировочный и боевой датасет
restaurants_test = pd.read_csv(
"../input/sf-dst-restaurant-rating/kaggle_task.csv"
) # Боевой датасет
restaurants_train = pd.read_csv(
"../input/sf-dst-restaurant-rating/main_task.csv"
) # Тренировочный датасет
restaurants_train["Test"] = 0
restaurants_test["Test"] = 1
# Объединяем датасеты в один общий
restaurants = restaurants_test.append(restaurants_train, sort=False).reset_index(
drop=True
)
# ### 1.1. Дополнительные словари с данными о городах
is_capital = {
"London": 1,
"Paris": 1,
"Madrid": 1,
"Barcelona": 0,
"Berlin": 1,
"Milan": 0,
"Rome": 1,
"Prague": 1,
"Lisbon": 1,
"Vienna": 1,
"Amsterdam": 1,
"Brussels": 1,
"Hamburg": 0,
"Munich": 0,
"Lyon": 0,
"Stockholm": 1,
"Budapest": 1,
"Warsaw": 1,
"Dublin": 1,
"Copenhagen": 1,
"Athens": 1,
"Edinburgh": 1,
"Zurich": 0,
"Oporto": 0,
"Geneva": 0,
"Krakow": 0,
"Oslo": 1,
"Helsinki": 1,
"Bratislava": 1,
"Luxembourg": 1,
"Ljubljana": 1,
}
population = {
"Paris": 2190327,
"Stockholm": 961609,
"London": 8908081,
"Berlin": 3644826,
"Munich": 1456039,
"Oporto": 237591,
"Milan": 1378689,
"Bratislava": 432864,
"Vienna": 1821582,
"Rome": 4355725,
"Barcelona": 1620343,
"Madrid": 3223334,
"Dublin": 1173179,
"Brussels": 179277,
"Zurich": 428737,
"Warsaw": 1758143,
"Budapest": 1752286,
"Copenhagen": 615993,
"Amsterdam": 857713,
"Lyon": 506615,
"Hamburg": 1841179,
"Lisbon": 505526,
"Prague": 1301132,
"Oslo": 673469,
"Helsinki": 643272,
"Edinburgh": 488100,
"Geneva": 200548,
"Ljubljana": 284355,
"Athens": 664046,
"Luxembourg": 115227,
"Krakow": 769498,
}
# ## 2.1 Преобразование исходного датафрейма в удобный для дальнейшей работы вид и добавление новых в ходе трансформации
# 1. Restaurant_id переводим в число
# restaurants['Restaurant_id'] = restaurants['Restaurant_id'].apply(lambda x: int(str(x)[3:]))
# 2. Price Range заменим числовой шкалой и заполним пропуски средним ценовым диапазоном
restaurants["Price Range"] = (
restaurants["Price Range"].map({"$": 1, "$$ - $$$": 2, "$$$$": 3}).fillna(2)
)
# 3. Cuisine Style - трансформируем из строки в список строк и сразу добавим новый признак - кол-во кухонь в ресторане
# Функция осуществляющая конвертацию в список кухонь
def create_cuisine_list(row):
if str(row) != "nan":
cusines = row.split(sep="'")
# Чистим список с кухнями от мусора
for cuisine in cusines:
if cuisine[0] == "[":
cusines.remove(cuisine)
elif cuisine[0] == ",":
cusines.remove(cuisine)
elif cuisine[0] == "]":
cusines.remove(cuisine)
return cusines
else:
return []
restaurants["Cuisine Style"] = restaurants["Cuisine Style"].apply(create_cuisine_list)
# Новый признак количество кухонб в конкретном ресторане
restaurants["# of Cusine"] = restaurants["Cuisine Style"].apply(
lambda x: len(x) if len(x) > 0 else 1
)
# 4. Разбоор поля с отзывами и их датами
def create_list_of_reviews(x):
review_pattern = re.compile("[A-Z][A-Za-z\s \.]+")
reviews = review_pattern.findall(x)
for review in reviews:
pass
return reviews
# Дата последнего отзыва
def create_list_of_dates_last(x):
date_pattern = re.compile("\d\d/\d\d/\d\d\d\d")
dates = date_pattern.findall(x)
if len(dates) == 2:
return max(pd.to_datetime(dates[0]), pd.to_datetime(dates[1]))
elif len(dates) == 1:
return pd.to_datetime(dates[0])
else:
return None
# Дата предпоследнего отзыва
def create_list_of_dates_perv(x):
date_pattern = re.compile("\d\d/\d\d/\d\d\d\d")
dates = date_pattern.findall(x)
if len(dates) == 2:
return min(pd.to_datetime(dates[0]), pd.to_datetime(dates[1]))
elif len(dates) == 1:
return pd.to_datetime(dates[0])
else:
return None
# Список отзывов
restaurants["Reviews"].fillna("[[], []]", inplace=True)
restaurants["List of reviews"] = restaurants["Reviews"].apply(create_list_of_reviews)
# Добавляем столбец последенго отзыва
restaurants["Last_review_date"] = restaurants["Reviews"].apply(
create_list_of_dates_last
)
# Добавляем столбец с предыдущим отзывом
restaurants["Perv_review_date"] = restaurants["Reviews"].apply(
create_list_of_dates_perv
)
# Переводим даты в числовой формат (чичло дней с 1970 года) и заполняем пропуски средними значениями
restaurants["Last_review_date"] = (
restaurants["Last_review_date"] - pd.to_datetime("1970-01-01")
).dt.days
restaurants["Last_review_date"].fillna(
restaurants["Last_review_date"].median(), inplace=True
)
restaurants["Perv_review_date"] = (
restaurants["Perv_review_date"] - pd.to_datetime("1970-01-01")
).dt.days
restaurants["Perv_review_date"].fillna(
restaurants["Perv_review_date"].median(), inplace=True
)
# Разница в днх между двумя последними отзывами
restaurants["Dates delta"] = (
restaurants["Last_review_date"] - restaurants["Perv_review_date"]
)
# Заполняем пропуски в количестве отзывов 0
restaurants["Number of Reviews"].fillna(0, inplace=True)
# Добавление нового признака - население города
restaurants["City Population"] = restaurants["City"].apply(lambda x: population[x])
# Удаляем лишние столбцы
restaurants.drop(["Reviews", "URL_TA", "ID_TA"], axis=1, inplace=True)
# restaurants['Is Capital'] = restaurants['City'].apply(lambda x:is_capital[x])
# ## 2.2 Попытка оценить ресторан на основе простейшего анализа содержимого отзывов
# Реализован примитивный поиск по ключевым словам, нужно больше статитстики.
# Функция голосования по ключевым словам
def ReviewsAnalyzer(reviews):
res = 0
for review in reviews:
review = review.lower()
res += review.count("good")
res += review.count("excellent")
res += review.count("nice")
res += review.count("great")
res += review.count("nice")
res += review.count("gem")
res += review.count("lovely")
res += review.count("delicious")
res += review.count("best")
res += review.count("perfect")
res += review.count("amazing")
res += review.count("love")
res += review.count("tasty")
res += review.count("clean")
res += review.count("fresh")
res += review.count("yamm")
res += review.count("bellyful")
res += review.count("pleasant")
res -= review.count("bad")
res -= review.count("awful")
res -= review.count("don\\'t")
res -= review.count("nothing")
res -= review.count("wast")
res -= review.count("trash")
res -= review.count("worst")
res -= review.count("overpriced")
res -= review.count("disappoint")
res -= review.count("loud")
res -= review.count("nothing")
res -= review.count("worth")
res -= review.count("tasteless")
res -= review.count("nasty")
res -= review.count("disgus")
res -= review.count("dirt")
res -= review.count("not")
return res
restaurants["Reviews"] = restaurants["List of reviews"].apply(ReviewsAnalyzer)
restaurants.drop(["List of reviews"], axis=1, inplace=True)
# ## 2.3 Эксперименты с категориальными признаками (города и кухни)
# Подсчёт статистики встречаемости кухни нужно было для ответы на вопросы модуля
# import operator
# different_cusines =[]
# for cusines in restaurants['Cuisine Style']:
# for cusine in cusines:
# if cusine not in different_cusines:
# different_cusines.append(cusine)
# # Делаем словарь где каждой кухни соответствует список позиций в датасете, где эта кухня встретилась
# cusines_stat={cusine : 0 for cusine in different_cusines}
# for i in range(restaurants.first_valid_index(), restaurants.last_valid_index()):
# for cusine in restaurants['Cuisine Style'][i]:
# cusines_stat[cusine]+=1
# sorted_cusines = sorted(cusines_stat.items(), key=operator.itemgetter(1))
# sorted_cusines
# Эксперименты с тем как наличие той или иной кухни влияет на качество модели, но так и не удалось выявить зависимости
def find_item(cell):
if item in cell:
return 1
return 0
for item in ["Italian"]:
restaurants[item] = restaurants["Cuisine Style"].apply(find_item)
# Эксперимент с городами как оказалось расположение в ресторан в Риме влияет на итоговую оценку
for city in ["Rome"]:
restaurants[city] = restaurants["City"].apply(lambda x: 1 if x == city else 0)
# среднее значение количества кухонь в ресторанах конкретного города
restaurants["Mean_Cuisine_Quantity"] = restaurants["City"].map(
restaurants.groupby("City")["# of Cusine"].mean()
)
# # 2.4 Эксперименты с относительными характеристиками
# Отношение Ranking к кол-ву ресторанов в городе
restaurants = restaurants.merge(
restaurants.City.value_counts().to_frame(name="# of Rest in City"),
how="left",
left_on="City",
right_index=True,
)
restaurants["Rank/Rest_in_city"] = (
restaurants.Ranking / restaurants["# of Rest in City"]
)
# Отношение Ranking к количеству населения города
restaurants["Rank/Population"] = restaurants.Ranking / restaurants["City Population"]
restaurants
# ## 2.5 Оценка взаимной корреляции признаков
fig, ax = plt.subplots(figsize=(10, 10))
sns.heatmap(
restaurants.corr(),
cmap="coolwarm",
ax=ax,
)
# Удаляем оставшиеся категориальные столбцы
restaurants.drop(["City", "Cuisine Style"], axis=1, inplace=True)
train_data = restaurants.query("Test == 0").drop(["Test"], axis=1)
test_data = restaurants.query("Test == 1").drop(["Test"], axis=1)
y = train_data.Rating.values
X = train_data.drop(["Restaurant_id", "Rating"], axis=1)
X_test = test_data.drop(["Restaurant_id", "Rating"], axis=1)
# ## 3. Обучение и оценка качества модели
# Разбиваем датафрейм на части, необходимые для обучения и тестирования модели
# Х - данные с информацией о ресторанах, у - целевая переменная (рейтинги ресторанов)
# X = restaurants_train.drop(['Restaurant_id', 'Rating'], axis = 1)
# y = restaurants_train['Rating']
# Загружаем специальный инструмент для разбивки:
from sklearn.model_selection import train_test_split
# Наборы данных с меткой "train" будут использоваться для обучения модели, "test" - для тестирования.
# Для тестирования мы будем использовать 25% от исходного датасета.
# X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.25)
# Создаём, обучаем и тестируем модель
# Импортируем необходимые библиотеки:
from sklearn.ensemble import (
RandomForestRegressor,
) # инструмент для создания и обучения модели
from sklearn import metrics # инструменты для оценки точности модели
# Создаём модель
regr = RandomForestRegressor(n_estimators=100)
# Обучаем модель на тестовом наборе данных
# regr.fit(X_train, y_train)
regr.fit(X, y)
# Используем обученную модель для предсказания рейтинга ресторанов в тестовой выборке.
# Предсказанные значения записываем в переменную y_pred
# y_pred = regr.predict(X_test)
# Округляем результат до 0.5
def round05(row):
return round(row * 2, 0) / 2
new_round = np.vectorize(round05)
y_pred_round = new_round(regr.predict(X_test))
# Сравниваем предсказанные значения (y_pred) с реальными (y_test), и смотрим насколько они в среднем отличаются
# Метрика называется Mean Absolute Error (MAE) и показывает среднее отклонение предсказанных значений от фактических.
# print('MAE:', metrics.mean_absolute_error(y_test, y_pred_round))
# ## 4. Визуализация степени значимости признаков
plt.rcParams["figure.figsize"] = (10, 5)
feat_importances = pd.Series(regr.feature_importances_, index=X.columns)
feat_importances.nlargest(20).plot(kind="barh")
# ## 5. Выводы и вопросы
# 1. Получена работоспособна модель
# 2. Появилось некоторое представление какие признаки влияют на качество модели
# Направления для улучшения:
# 1. Набрать большую выборку отзывов по ресторанам и уточнить рейтинговую модель
# 2. Ещё "поиграться" с относительными признаками
# Вопросы: есть ли какая-нибудь теория в области "feature enginiring" или всё строится на собственном опыте и здравом смысле?
#
# Сохранение результатов
# test_data['Rating'] = y_pred_round
# test_data[['Restaurant_id','Rating']].to_csv('solution.csv', index = False)
# test_data
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
# #/kaggle/input/combined_data.csv
# #/kaggle/input/projects/sample_data_audit.csv
# #/kaggle/input/projects/projects.csv/projects.csv
# #/kaggle/input/projects/essays.csv/essays.csv
df_es = pd.read_csv("/kaggle/input/outcome-value/essays.csv/essays.csv")
df_out = pd.read_csv("/kaggle/input/outcome-value/outcomes.csv/outcomes.csv")
df_proj = pd.read_csv("/kaggle/input/outcome-value/projects.csv/projects.csv")
df = (
df_proj.merge(
df_es, left_index=True, right_index=True, how="inner", suffixes=("", "_y")
)
).merge(df_out, left_index=True, right_index=True, how="inner", suffixes=("", "_y"))
df.drop(list(df.filter(regex="_y$")), axis=1, inplace=True)
# """print(df_out.shape)
# print(df_proj.shape)
# print(df_es.shape)
# print(df_out.head())
# print(df_proj.head())
# print(df_es.head())"""
print(df_out.shape)
print(df_proj.shape)
print(df_es.shape)
print(df.shape)
import gc
df_es = pd.DataFrame()
df_out = pd.DataFrame()
df_proj = pd.DataFrame()
del [[df_es, df_out, df_proj]]
gc.collect()
df["price_suppoert"] = (
df["total_price_including_optional_support"]
- df["total_price_excluding_optional_support"]
)
df.shape
df = df.drop(
columns=[
"total_price_including_optional_support",
"total_price_excluding_optional_support",
]
)
df.shape
df.to_csv("joined.csv.zip", compression="zip", index=False)
df["school_latitude"].describe()
df["school_longitude"].describe()
mean_lon = -93.808668
mean_lat = 37.201561
from sklearn.neighbors import DistanceMetric
dist = DistanceMetric.get_metric("haversine")
def find_distance(row):
mean_lon = -93.808668
mean_lat = 37.201561
lon = row["school_longitude"]
lat = row["school_latitude"]
mean_lon, mean_lat, lon, lat = (
np.radians(mean_lon),
np.radians(mean_lat),
np.radians(lon),
np.radians(lat),
)
dlon = mean_lon - lon
dlat = mean_lat - lat
a = (
np.sin(dlat / 2.0) ** 2
+ np.cos(mean_lat) * np.cos(lat) * np.sin(dlon / 2.0) ** 2
)
return 6371 * 2 * np.arcsin(np.sqrt(a))
df["distance"] = df.apply(lambda row: find_distance(row), axis=1)
df.shape
df = df.drop(columns=["school_latitude", "school_longitude", "school_zip"])
df.shape
import matplotlib
import matplotlib.pyplot as plt
import seaborn as sns
import statsmodels.api as sm
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.feature_selection import RFE
from sklearn.linear_model import RidgeCV, LassoCV, Ridge, Lasso
# Using Pearson Correlation
plt.figure(figsize=(40, 50))
cor = df.corr()
sns.heatmap(cor, annot=True, cmap=plt.cm.Reds)
plt.savefig("heatMap_new.png")
plt.show()
df.to_csv("joined_new.csv.zip", compression="zip", index=False)
df["distance"].describe()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# **References:**
# 'Overview/Tutorials' en 'Data' tab zijn heel handig!
# https://www.kaggle.com/competitions/house-prices-advanced-regression-techniques/overview
# Used:
# 1. Print the data:
# https://www.kaggle.com/code/dansbecker/finding-your-files-in-kaggle-kernels/notebook
# 2. Intro for this assignment:
# https://www.kaggle.com/code/bsivavenu/house-price-calculation-methods-for-beginners
# For later:
# 1. General tips for ML:
# https://www.kaggle.com/code/dansbecker/learning-materials-on-kaggle/notebook
# **Stappenplan:**
# 1. explore data, clean data (the features): fill missing values and transform into numerical values, normalization and k-fold cross validation/train and test split?
# 2. Feature selection: unsupervised learning? Dimension reduction or clustering? supervised learning?
# 3. Training model
#
train = pd.read_csv(
"/kaggle/input/house-prices-advanced-regression-techniques/train.csv"
)
test = pd.read_csv("/kaggle/input/house-prices-advanced-regression-techniques/test.csv")
train.describe()
train.head(5)
# correlation matrix
corrmat = train.corr()
f, ax = plt.subplots(figsize=(20, 9))
sns.heatmap(corrmat, vmax=0.8, annot=True)
print(train.columns)
# Correlation of feaures with Sale price plot with features = [OverallQual, YrSold, KitchenQual, 1stFlrSF]
fig, ax = plt.subplots(4, 1, figsize=(15, 15))
ax[0].scatter(train["OverallQual"], train["SalePrice"])
ax[0].set_title("OverallQual")
ax[1].scatter(train["YrSold"], train["SalePrice"])
ax[1].set_title("YrSold")
ax[2].scatter(train["KitchenQual"], train["SalePrice"])
ax[2].set_title("KitchenQual")
ax[3].scatter(train["1stFlrSF"], train["SalePrice"])
ax[3].set_title("1stFlrSF")
plt.show()
# train['1stFlrSF']
# OverallQual, KitchenQual, 1stFlrSF all have a positive correlation
train.shape, test.shape
print(type(train))
a = train["OverallQual"]
print(type(a))
test.head(3)
|
import pandas as pd
from rdkit import Chem
from rdkit.Chem import AllChem
from rdkit.Chem import Descriptors
from rdkit.Chem import PandasTools
from sklearn.ensemble import RandomForestClassifier, VotingClassifier
from sklearn.impute import SimpleImputer
from sklearn.feature_selection import VarianceThreshold
from xgboost import XGBClassifier
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score, f1_score
from imblearn.over_sampling import SMOTE
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
def descriptors(smiles, assay_id):
mol = Chem.MolFromSmiles(smiles)
molecular_weight = Descriptors.MolWt(mol)
logp = Descriptors.MolLogP(mol)
molmr = Descriptors.MolMR(mol)
tpsa = Descriptors.TPSA(mol)
fp = AllChem.GetMorganFingerprintAsBitVect(mol, 2, nBits=1024)
return [molecular_weight, logp, assay_id, tpsa] + list(fp)
train = pd.read_csv("/kaggle/input/the-toxicity-prediction-challenge-ii/train_II.csv")
test = pd.read_csv("/kaggle/input/the-toxicity-prediction-challenge-ii/test_II.csv")
train[["Smiles", "Assay ID"]] = train["Id"].str.split(";", expand=True)
test[["Smiles", "Assay ID"]] = test["x"].str.split(";", expand=True)
train = train.drop("Id", axis=1)
test = test.drop("x", axis=1)
# Convert SMILES to RDKit mol object
train["Mol"] = train["Smiles"].apply(Chem.MolFromSmiles)
test["Mol"] = test["Smiles"].apply(Chem.MolFromSmiles)
train = train.dropna(how="any", axis=0)
test = test.dropna(how="any", axis=0)
train["Expected"] = train["Expected"] - 1
labels = train["Expected"]
train = train.drop("Expected", axis=1)
from sklearn.feature_selection import VarianceThreshold
import pandas as pd
# Create a DataFrame with the descriptors
data = []
for i, row in train.iterrows():
descriptors_list = descriptors(row["Smiles"], row["Assay ID"])
data.append(descriptors_list)
df_descriptors = pd.DataFrame(
data,
columns=["molecular_weight", "logp", "assay_id", "tpsa"]
+ ["fp_" + str(i) for i in range(1024)],
)
# Apply variance threshold
threshold = 0.1
selector = VarianceThreshold(threshold=threshold)
selector.fit(df_descriptors)
selected_features = df_descriptors.columns[selector.get_support()]
# Print the selected features
print("Selected features:")
print(selected_features)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
import pandas as pd
import numpy as np
from sklearn.linear_model import LogisticRegression
import matplotlib.pyplot as plt
from sklearn.utils import shuffle
from sklearn.model_selection import GridSearchCV
path = "../input/google-cloud-ncaa-march-madness-2020-division-1-womens-tournament/"
tourney_result = pd.read_csv(path + "WDataFiles_Stage1/WNCAATourneyCompactResults.csv")
tourney_seed = pd.read_csv(path + "WDataFiles_Stage1/WNCAATourneySeeds.csv")
season_result = pd.read_csv(path + "WDataFiles_Stage1/WRegularSeasonCompactResults.csv")
test_df = pd.read_csv(path + "WSampleSubmissionStage1_2020.csv")
# deleting unnecessary columns
tourney_result = tourney_result.drop(
["DayNum", "WScore", "LScore", "WLoc", "NumOT"], axis=1
)
tourney_result
tourney_result = pd.merge(
tourney_result,
tourney_seed,
left_on=["Season", "WTeamID"],
right_on=["Season", "TeamID"],
how="left",
)
tourney_result.rename(columns={"Seed": "WSeed"}, inplace=True)
tourney_result = tourney_result.drop("TeamID", axis=1)
tourney_result = pd.merge(
tourney_result,
tourney_seed,
left_on=["Season", "LTeamID"],
right_on=["Season", "TeamID"],
how="left",
)
tourney_result.rename(columns={"Seed": "LSeed"}, inplace=True)
tourney_result = tourney_result.drop("TeamID", axis=1)
tourney_result
def get_seed(x):
return int(x[1:3])
tourney_result["WSeed"] = tourney_result["WSeed"].map(lambda x: get_seed(x))
tourney_result["LSeed"] = tourney_result["LSeed"].map(lambda x: get_seed(x))
tourney_result
season_win_result = season_result[["Season", "WTeamID", "WScore"]]
season_lose_result = season_result[["Season", "LTeamID", "LScore"]]
season_win_result.rename(columns={"WTeamID": "TeamID", "WScore": "Score"}, inplace=True)
season_lose_result.rename(
columns={"LTeamID": "TeamID", "LScore": "Score"}, inplace=True
)
season_result = pd.concat((season_win_result, season_lose_result)).reset_index(
drop=True
)
season_result
season_score = season_result.groupby(["Season", "TeamID"])["Score"].sum().reset_index()
season_score
tourney_result = pd.merge(
tourney_result,
season_score,
left_on=["Season", "WTeamID"],
right_on=["Season", "TeamID"],
how="left",
)
tourney_result.rename(columns={"Score": "WScoreT"}, inplace=True)
tourney_result = tourney_result.drop("TeamID", axis=1)
tourney_result = pd.merge(
tourney_result,
season_score,
left_on=["Season", "LTeamID"],
right_on=["Season", "TeamID"],
how="left",
)
tourney_result.rename(columns={"Score": "LScoreT"}, inplace=True)
tourney_result = tourney_result.drop("TeamID", axis=1)
tourney_result
tourney_win_result = tourney_result.drop(["Season", "WTeamID", "LTeamID"], axis=1)
tourney_win_result.rename(
columns={
"WSeed": "Seed1",
"LSeed": "Seed2",
"WScoreT": "ScoreT1",
"LScoreT": "ScoreT2",
},
inplace=True,
)
tourney_win_result
tourney_lose_result = tourney_win_result.copy()
tourney_lose_result["Seed1"] = tourney_win_result["Seed2"]
tourney_lose_result["Seed2"] = tourney_win_result["Seed1"]
tourney_lose_result["ScoreT1"] = tourney_win_result["ScoreT2"]
tourney_lose_result["ScoreT2"] = tourney_win_result["ScoreT1"]
tourney_lose_result
tourney_win_result["Seed_diff"] = (
tourney_win_result["Seed1"] - tourney_win_result["Seed2"]
)
tourney_win_result["ScoreT_diff"] = (
tourney_win_result["ScoreT1"] - tourney_win_result["ScoreT2"]
)
tourney_lose_result["Seed_diff"] = (
tourney_lose_result["Seed1"] - tourney_lose_result["Seed2"]
)
tourney_lose_result["ScoreT_diff"] = (
tourney_lose_result["ScoreT1"] - tourney_lose_result["ScoreT2"]
)
tourney_win_result["result"] = 1
tourney_lose_result["result"] = 0
tourney_result = pd.concat((tourney_win_result, tourney_lose_result)).reset_index(
drop=True
)
tourney_result
X_train = tourney_result.drop("result", axis=1)
y_train = tourney_result.result
X_train.shape
# #Buinging Model
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import tensorflow as tf
from tensorflow.keras.preprocessing.image import ImageDataGenerator
from tensorflow.keras.optimizers import Adam
from tensorflow.keras.utils import to_categorical, normalize
from sklearn.utils import shuffle
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
model = tf.keras.models.Sequential(
[
tf.keras.layers.Dense(128, input_shape=(6,), activation="relu"),
tf.keras.layers.BatchNormalization(),
tf.keras.layers.Dropout(0.5),
tf.keras.layers.Dense(128, activation="relu"),
tf.keras.layers.BatchNormalization(),
tf.keras.layers.Dropout(0.5),
tf.keras.layers.Dense(128, activation="relu"),
tf.keras.layers.BatchNormalization(),
tf.keras.layers.Dropout(0.5),
tf.keras.layers.Dense(128, activation="relu"),
tf.keras.layers.BatchNormalization(),
tf.keras.layers.Dropout(0.5),
tf.keras.layers.Dense(125, activation="relu"),
tf.keras.layers.BatchNormalization(),
tf.keras.layers.Dropout(0.5),
tf.keras.layers.Dense(1, activation="sigmoid"),
]
)
model.compile(loss="binary_crossentropy", optimizer=Adam(0.0001), metrics=["acc"])
model.summary()
history = model.fit(
X_train,
y_train,
epochs=500,
verbose=1,
)
test_df["Season"] = test_df["ID"].map(lambda x: int(x[:4]))
test_df["WTeamID"] = test_df["ID"].map(lambda x: int(x[5:9]))
test_df["LTeamID"] = test_df["ID"].map(lambda x: int(x[10:14]))
test_df
test_df = pd.merge(
test_df,
tourney_seed,
left_on=["Season", "WTeamID"],
right_on=["Season", "TeamID"],
how="left",
)
test_df.rename(columns={"Seed": "Seed1"}, inplace=True)
test_df = test_df.drop("TeamID", axis=1)
test_df = pd.merge(
test_df,
tourney_seed,
left_on=["Season", "LTeamID"],
right_on=["Season", "TeamID"],
how="left",
)
test_df.rename(columns={"Seed": "Seed2"}, inplace=True)
test_df = test_df.drop("TeamID", axis=1)
test_df = pd.merge(
test_df,
season_score,
left_on=["Season", "WTeamID"],
right_on=["Season", "TeamID"],
how="left",
)
test_df.rename(columns={"Score": "ScoreT1"}, inplace=True)
test_df = test_df.drop("TeamID", axis=1)
test_df = pd.merge(
test_df,
season_score,
left_on=["Season", "LTeamID"],
right_on=["Season", "TeamID"],
how="left",
)
test_df.rename(columns={"Score": "ScoreT2"}, inplace=True)
test_df = test_df.drop("TeamID", axis=1)
test_df
test_df["Seed1"] = test_df["Seed1"].map(lambda x: get_seed(x))
test_df["Seed2"] = test_df["Seed2"].map(lambda x: get_seed(x))
test_df["Seed_diff"] = test_df["Seed1"] - test_df["Seed2"]
test_df["ScoreT_diff"] = test_df["ScoreT1"] - test_df["ScoreT2"]
test_df = test_df.drop(["ID", "Pred", "Season", "WTeamID", "LTeamID"], axis=1)
test_df
test_preds = model.predict_proba(test_df)
submission_df = pd.read_csv(path + "WSampleSubmissionStage1_2020.csv")
submission_df["Pred"] = test_preds
submission_df
submission_df.to_csv("submission.csv", index=False)
|
# This is a forked version of the [Kernel](https://www.kaggle.com/allunia/breastcancer) by [Allunia](https://www.kaggle.com/allunia). The kernel has been modified so that some of the functions are more computationally efficient, and shorter and easier to understand.
# Instead of using deep learning, we will be using Support Vector Machines (SVMs) as a classifier combined with Principle Component Analysis (PCA).
# Some of the techniques used and considerations taken in this kernel, were directly inspired by [Joni Juvonen's](https://www.kaggle.com/qitvision) [kernel](https://www.kaggle.com/qitvision/a-complete-ml-pipeline-fast-ai).
# # Motivation
# ### My aunt has recently been diagnosed with breast cancer, this kernel is dedicated to her in the hopes that I can learn from this experience and one day build an improved version of this classifier.
# # Preparation & peek at the data structure
# ## Loading packages
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# VISUALIZATION
from matplotlib.colors import ListedColormap
import matplotlib.pyplot as plt
import seaborn as sns
sns.set()
import cv2 as cv
# MACHINELEARNING
from sklearn import svm
from sklearn.model_selection import train_test_split, StratifiedKFold
from sklearn.decomposition import PCA
from glob import glob
from skimage import io
from os import listdir
import time
import copy
from tqdm.notebook import tqdm
tqdm().pandas()
print("import complete")
# ### Settings
run_training = False
retrain = False
find_learning_rate = True
# ## Exploring the data structure
# We start by exploring the file structure to gain an understanding of how everything is organised
files = listdir("../input/breast-histopathology-images/IDC_regular_ps50_idx5")
print(len(files))
# We have a total number of about 280 sub-folders, let's take a peak into the folder and try and understand what those sub-folders are.
files[0:10]
# Each subfolder seems to be the ID of the corresponding patient
patient_file = listdir(
"../input/breast-histopathology-images/IDC_regular_ps50_idx5/13689"
)
print(len(patient_file))
patient_file
# Each file has 2 sub-folders, labeled 1 and 0
# Folder 0: Non-Invasive Ductal Carcinoma (IDC)
# Folder 1 : Invasive Ductal Carcinoma (IDC)
class_0 = listdir("../input/breast-histopathology-images/IDC_regular_ps50_idx5/13689/0")
class_1 = listdir("../input/breast-histopathology-images/IDC_regular_ps50_idx5/13689/1")
# Below we take a glimpse at the files contained in each class
from pprint import pprint
print("Class 0 files:\n ")
pprint(class_0[0:10])
print("Class 1 files:\n ")
pprint(class_1[0:10])
# It seems that we have reached the files containing the pictures of each patient,
# the structure seems to be as follows:
# Patient_id/xcoordinate_of_patch/ycoordinate_of_patch/class_of_cancer
# ## What do we know about our data?
# Now that we have a good understanding of the file structure let's try and understand how much data we are about to process.
# ### How many patients do we have?
# It seems that we have a total number of 280 patients. This sample size is relatively small therefore we have to be careful not to overfit our model. We need to implement our model in such a way that it maximizes generalization.
# Each patient has a batch of patches that were extracted, therefore the total number of patches is likely much greater than 280.
base_path = "../input/breast-histopathology-images/IDC_regular_ps50_idx5/"
patient_ids = listdir(base_path)
# ### How many patches do we have in total?
# #### Which of them are IDC patches and which are Non-IDC?
# In order to train our model we need to feed our model each patch individually, therefore each patch will act as an input.
# The snippet below loops through the entire file structure and extracts the total number of crops for each of the classes.
class_0_total = 0
class_1_total = 0
from pprint import pprint
for patient_id in patient_ids:
class_0_files = listdir(base_path + patient_id + "/0")
class_1_files = listdir(base_path + patient_id + "/1")
class_0_total += len(class_0_files)
class_1_total += len(class_1_files)
total_images = class_0_total + class_1_total
print(f"Number of patches in Class 0: {class_0_total}")
print(f"Number of patches in Class 1: {class_1_total}")
print(f"Total number of patches: {total_images}")
# Notice how the ratio of IDC patches to non-IDC patches is highly uneven. Here we approximately a 28/70 split, which could be problematic down the road due to the overrepresentation of non-IDC samples relative to IDC samples. We need to account for it during the validation phase.
# ## Storing the image_path, patient_id, target and x & y coordinates
# We have about 278,000 images. To feed the algorithm with image patches we will store the path of each image and create a dataframe containing all the paths. **This way we can load batches of images one by one without storing the individual pixel values of all images**.
# In addition to extracting the paths, we will also extract the x & y coordinates of each of the images. Then we can use the coordinates to reconstruct the whole breast tissue of a patient. This way we can also explore how diseased tissue looks like compared to healthy ones.
# We know that the path to the images is as follows:
# > ../input/breast-histopathology-images/[patient_id]/[class]/[image]
# Below we start by constructing a list of dictionaries and then creating a dataframe based on that list. This is much more computationally efficient than using 'append' or 'loc' methods. For more info check out this [post](https://stackoverflow.com/questions/10715965/add-one-row-to-pandas-dataframe/17496530#17496530) on stackoverlow.
columns = ["patient_id", "x", "y", "target", "path"]
data_rows = []
i = 0
iss = 0
isss = 0
# note that we loop through the classes after looping through the
# patient ids so that we avoid splitting our data into [all class 0 then all class 1]
for patient_id in patient_ids:
for c in [0, 1]:
class_path = base_path + patient_id + "/" + str(c) + "/"
imgs = listdir(class_path)
# Extracting Image Paths
img_paths = [class_path + img + "/" for img in imgs]
# Extracting Image Coordinates
img_coords = [img.split("_", 4)[2:4] for img in imgs]
x_coords = [int(coords[0][1:]) for coords in img_coords]
y_coords = [int(coords[1][1:]) for coords in img_coords]
for path, x, y in zip(img_paths, x_coords, y_coords):
values = [patient_id, x, y, c, path]
data_rows.append({k: v for (k, v) in zip(columns, values)})
# We create a new dataframe using the list of dicts that we generated above
data = pd.DataFrame(data_rows)
print(data.shape)
data.head()
# As we expect, the shape of our dataframe matches our expectations, containing a total of 5 features, and about 280,000 patches.
# # Explarotory Data Analysis
# ## Let's create a visual summary of our data
# Now that we have set up our data, let's create a visual summary to help us draw some insights from our data.
cancer_perc = (
data.groupby("patient_id").target.value_counts()
/ data.groupby("patient_id").target.size()
)
cancer_perc = cancer_perc.unstack()
fig, ax = plt.subplots(1, 3, figsize=(25, 5))
# Plotting Frequency of Patches per Patient
sns.distplot(
data.groupby("patient_id").size(), ax=ax[0], color="Orange", kde=False, bins=15
)
ax[0].set_xlabel("Number of patches")
ax[0].set_ylabel("Frequency")
ax[0].set_title("How many patches do we have per patient?")
# Plotting Percentage of an image that is covered by Invasive Ductile Carcinoma
sns.distplot(cancer_perc.loc[:, 1] * 100, ax=ax[1], color="Tomato", kde=False, bins=15)
ax[1].set_title("How much percentage of an image is covered by IDC?")
ax[1].set_ylabel("Frequency")
ax[1].set_xlabel("% of patches with IDC")
# Plotting number of patches that show IDC
sns.countplot(data.target, palette="pastel", ax=ax[2])
ax[2].set_ylabel("Count")
ax[2].set_xlabel("no(0) versus yes(1)")
ax[2].set_title("How many patches show IDC?")
# ### Insights
# 1. The number of image patches per patient varies a lot! This leads to the questions whether all images show the same resolution of tissue cells of if this varies between patients.
# 2. Some patients have more than 80 % patches that show IDC! Consequently the tissue is full of cancer or only a part of the breast was covered by the tissue slice that is focused on the IDC cancer. Does a tissue slice per patient cover the whole region of interest?
# 3. The classes of IDC versus no IDC are imbalanced. We have to check this again after setting up a validation strategy and find a strategy to deal with class weights (if we like to apply them).
# ## Healthy Tissue Patches Vs Cancerous Tissue Patches
# Let us now explore the visual differences between cancerous tissue cells, and healthy tissue cells. Usually partnering with a specialist is a good idea so that they can point the exact points of interest that differentiate the 2 from each other.
# Return random sample indexes that are cancer positive and cancer negative of size 50
# replace = False means that no duplication is allowed
positive_tissue = np.random.choice(
data[data.target == 1].index.values, size=100, replace=False
)
negative_tissue = np.random.choice(
data[data.target == 0].index.values, size=100, replace=False
)
n_rows = 10
n_cols = 10
# For a quick refresher on the different libraries used to read images and how to use them check out this medium article by [TowardDataScience](https://towardsdatascience.com/what-library-can-load-image-in-python-and-what-are-their-difference-d1628c6623ad)
# ### Cancerous Patches
fig, ax = plt.subplots(n_rows, n_cols, figsize=(30, 30))
for row in range(n_rows):
for col in range(n_cols):
# below is a counter to cycle through the image indexes
idx = positive_tissue[col + n_cols * row]
img = io.imread(data.loc[idx, "path"])
ax[row, col].imshow(img[:, :, :])
ax[row, col].grid(False)
# ### Non-Cancerous Patches
fig, ax = plt.subplots(n_rows, n_cols, figsize=(30, 30))
for row in range(n_rows):
for col in range(n_cols):
# below is a counter to cycle through the image indexes
idx = negative_tissue[col + n_cols * row]
img = io.imread(data.loc[idx, "path"])
ax[row, col].imshow(img[:, :, :])
ax[row, col].grid(False)
# ### Insights¶
# * Sometimes we can find artifacts or incomplete patches, some images are also less than 50 x 50 pxs.
# * Patches with cancer look more violet and crowded than healthy ones. Is this really typical for cancer or is it more typical for ductal cells and tissue?
# * Though some of the healthy patches are very violet colored too!
# * Would be very interesting to hear what criteria are important for a [pathologist](https://en.wikipedia.org/wiki/Pathology).
# Since some images are less than 50 by 50, we now must decide whether these images should be dropped or not. One thing to consider is the computational time required to cycle through all 280,000 images and drop the ones that have a shape of less than 50x50x3.
# We will be dropping them in our case.
# ### Dropping Artifacts
print(f"original shape: {data.shape}")
for i, path in tqdm(enumerate(data.loc[:, "path"]), total=data.shape[0]):
img = io.imread(path)
if img.shape != (50, 50, 3):
data.drop(data.index[i], axis=0, inplace=True)
print(f"altered shape: {data.shape}")
# Great, we dropped about 2000 rows. Now we can continue on with our modelling and visualization.
# ## Visualizing the Breast Tissue
# Earlier we extracted the coordinates of the cropped tissue cells, we can use those coordinates to reconstruct the whole breast tissue of the patient. This way we can explore how the diseased tissue looks when compared to the healthy tissue.
# We can also explore the most common places that the cancer tends to occur in. It would be interesting to plot a heatmap of the most common areas where the cancer appears.
# If position of the crop has significance then perhaps we can use it as an input feature for our model.
# To simplify things we will create a function that slices our existing dataframe and retrieves the values associated with a patient id.
def get_patient_df(patient_id):
return data.loc[data["patient_id"] == patient_id, :]
# ### Binary target visualisation per tissue slice
# Before we will take a look at the whole tissue let's keep it a bit simpler by looking at the target structure in the x-y-space for a handful of patients:
n_rows = 5
n_cols = 3
n_imgs = n_rows * n_cols
colors = ["pink", "purple"]
fig, ax = plt.subplots(n_rows, n_cols, figsize=(20, 27))
patient_ids = np.random.choice(data.patient_id.unique(), size=n_imgs, replace=False)
for row in range(n_rows):
for col in range(n_cols):
patient_id = patient_ids[col + n_cols * row]
patient_df = get_patient_df(patient_id)
ax[row, col].scatter(
patient_df.x.values,
patient_df.y.values,
c=patient_df.target.values,
cmap=ListedColormap(colors),
s=20,
)
ax[row, col].set_title("patient " + patient_id)
# ### Insights
# * Sometimes we don't have the full tissue information. It seems that tissue patches have been discarded or lost during preparation.
# * Cancerous Tissue tends to appear in clusters rather than, being dispersed all over the place.
# ### Repatching the Actual Breast Tissue Image
# Now it's time to go one step deeper with our EDA. Instead of plotting the target values using the x-y coordinates, we now plot the images themselves on their respective x-y coordinates. This will help us visualize how the cancerous tissue looks like from a macro perspective.
# uint8 is used unsigned 8 bit integer. And that is the range of pixel. We can't have pixel value more than 2^8 -1. Therefore, for images uint8 type is used. Whereas double is used to handle very big numbers.
def visualise_breast_tissue(patient_id, df=data, pred=False, crop_dimension=[50, 50]):
# Plotting Settings
plt.xticks([])
plt.yticks([])
# Get patient dataframe
p_df = get_patient_df(patient_id)
# Get the dimensions of the breast tissue image
max_coord = np.max((*p_df.x, *p_df.y))
# Allocate an array to fill image pixels in,use uint8 type as you don't need an int over 255
grid = 255 * np.ones(
shape=(max_coord + crop_dimension[0], max_coord + crop_dimension[1], 3)
).astype(np.uint8)
mask = 255 * np.ones(
shape=(max_coord + crop_dimension[0], max_coord + crop_dimension[1], 3)
).astype(np.uint8)
# Replace array values with values of the image
for x, y, target, path in zip(p_df["x"], p_df["y"], p_df["target"], p_df["path"]):
try:
img = io.imread(path)
# Replace array values with cropped image values
grid[y : y + crop_dimension[1], x : x + crop_dimension[0]] = img
# Check if target is cancerous or not
if target != 0:
# If the target is cancerous then, replace array values with the color blue
mask[y : y + crop_dimension[1], x : x + crop_dimension[0]] = [0, 0, 255]
except:
pass
# if prediction is not specifies then show the image normally
if pred == False:
io.imshow(grid)
img = grid
# if prediction is specified then apply a mask to the areas that contain predicted cancerous cells
else:
# Specify the desired alpha value
alpha = 0.78
# This is step is very important, adding 2 numpy arrays sets the values to float64, which is why convert them back to uint8
img = (mask * (1.0 - alpha) + grid * alpha).astype("uint8")
io.imshow(img)
return img
# Let's now visualize what some of the reconstructed tissue cells look like by using the function above.
# Note: The function can be improved by having edges on the mask, rather than overlaying a color on top. This will help us preserve the true image of the tissue cells.
n_rows = 5
n_cols = 3
n_imgs = n_rows * n_cols
fig, ax = plt.subplots(n_rows, n_cols, figsize=(20, 27))
for row in range(n_rows):
for col in range(n_cols):
p_id = patient_ids[col + n_cols * row]
img = visualise_breast_tissue(p_id, pred=True)
ax[row, col].grid(False)
ax[row, col].set_xticks([])
ax[row, col].set_yticks([])
ax[row, col].set_title("Breast tissue slice of patient: " + p_id)
ax[row, col].imshow(img)
# ## Data Augmentation
# There are couple of ways we can use to avoid overfitting; more data, augmentation, regularization and less complex model architectures. Note that if we apply augmentation here, augmentations will also be applied when we are predicting (inference). This is called test time augmentation (TTA) and it can improve our results if we run inference multiple times for each image and average out the predictions.
# **The augmentations we can use for this type of data:**
# - random rotation
# - random crop
# - random flip (horizontal and vertical both)
# - random lighting
# - random zoom
# - Gaussian blur
# Also possible to make a heat map that tells you the most likely region that Invasive Ductal Carcinoma is likely to occur, and to see whether the coordinates can be used as a feature or not.
# ## Note to self:
# #### Please disregard this
# Usually before applying the steps above we might be interested in engineering new features based on the our observations and domain knowledge,one of the features that we might consider adding in future versions of this kernal is including the count of surrounding cancerous cells. This would be treated as a time series analysis, we would have to iterate on it until an equilibirium is reached. So we would have micro and macro classification. Also look into using t-svd instead of pca.
# ## Pre-processing
# Now that we have a function that allows us to vizualize the where the cancer occurs, we can finally begin pre-processing our data to get it ready to be fed into our classifier.
# The next steps that we are going to take are shown below:
# * Split Data to Training & Testing Data
# * Apply Principle Component Analysis (PCA)
# Notice how we split our data into testing and training sets before applying PCA to it. This is because we want to fit the data to our training data and not our testing data as that would bias our results. One more thing to consider is, that when we receive our data to be classified we are always transforming our data onto our already fitted model. Therefore applying the fit to our testing data, would not reflect what happens when classifying in real life.
# ### Training & Testing Split
# There are a few things that we must consider before splitting our data. The first being the ratio of cancerous to non cancerous images that we have. We determined earlier that our data is not split uniformly. We have many more, non-cancerous images that non-cancerous images, which may possibly bias our model.
# This may also pose a problem when evaluating our model. Let's take an extreme example where we have a dataset consisting of 100 points, 90 of them are cancerous and 10 of them are non-cancerous, let's also assume that our model classified our entire dataset as cancerous, this would give an accuracy of 90%, however this is not really representitive of what is really happening. In reality it is always misclassifying our non-cancerous data.
# So how do we go about solving this?
# We can use the train-test split function offered in sklearn model_selection, this function has the option to split our data such that we get equally distributed training and testing sets when grouped by the classes.
# For more information, check out this [article](https://towardsdatascience.com/3-things-you-need-to-know-before-you-train-test-split-869dfabb7e50), that talks about the consideration that need to be taken before splitting your data.
# The function below returns the percentage of unique items we have in a class so that we can gain a better understanding of our target variables. In our case, the data will be split between 0 and 1 values. These values represent the following:
# * 0 -> Tissue does not contain IDC
# * 1 -> Tissue contains IDC
def get_classes_split(series):
ratio = np.round(series.value_counts() / series.count() * 100, decimals=1)
return ratio
(get_classes_split(data["target"]))
# Now that we know the initial, split of the classes, let's split our data into training and testing sets.
# #### Storing the Images
# Let us begin by storing the images in a numpy array. We know that each RGB image has a dimension of (50,50,3), in order to store them in numpy arrays, we will flatten the image into a (1,7500) shape. This means that each row of our numpy array will store an image, and that our final numpy array dimensions will be about (280000,7500). We have about 280,000 rows because we have about 280,000 crops in total.
# To get a better feel of what we are doing take the example below:
img_paths = data["path"]
# Read the data of the first path
img = io.imread(img_paths[0])
print(f"original image shape: {img.shape}")
# Flattening the image
img = img.flatten()
print(f"flattened image shape: {img.shape}")
# Reshaping the image in order to display it properly again
img = img.reshape(50, 50, 3)
io.imshow(img)
# Now that we have coded a proof concept, let's apply this to all the image paths that we have using a loop. We will create a function that does that for us, the function accepts the paths, and the flattened image size and returns the reconstructed flattened array of images that we spoke of earlier.
def paths_to_array(img_paths, flat_img_size=7500):
# Get the total number of images to be stored in the array
number_of_images = img_paths.shape[0]
# Initialize numpy array
img_array = np.zeros((number_of_images, flat_img_size))
# Use tqdm to keep track of progress
n_errors = 0
for i, img_path in tqdm(enumerate(img_paths), total=number_of_images):
img = io.imread(img_path)
img = img.flatten()
try:
img_array[i, :] = img
except:
n_errors += 1
print(n_errors)
return img_array
img_array = paths_to_array(img_paths=img_paths)
train_test_split()
# ## Applying PCA
# PCA is an unsupervised dimensionality reduction technique that we will use to reduce the number of dimensions that we are working with and in order to isolate features of most importance.
# Let's briefly talk about PCA before delving into the application. PCA is a dimentionality reduction technique. What it is essentially doing is reducing the number of dimensions that we have by determining the covariance between the different variables that we have.
# The covariance is how likely a variable is to vary relative to another variable. This means that variables with very high covariance can be redundant. Let's say I know that A will vary when B varies, then I can formulate a formula to explain the variation of B given A.
# Let's take another more concrete example: we know that the velocity is equal to the displacement over time, here the third variable is always explained given the other 2. PCA does the same thing, it tries to determine the relations between variables and uses them to explain each other.
# PCA linearly maps the features, along the axis of maximum variation, this axis is also called the eigen vector. The distance between a point and the eigen vector and a point is also known as the eigen value. To learn more about PCA 2 great articles are linked below.
# * [Article explaining the theoretical aspect of dimensionality reduction techniques such as PCA and SVD.](https://towardsdatascience.com/pca-and-svd-explained-with-numpy-5d13b0d2a4d8)
# * [Article applying PCA on satellite images.](https://towardsdatascience.com/principal-component-analysis-in-depth-understanding-through-image-visualization-892922f77d9f)
# * [Very useful answer on stackexchange that talks about the considerations that need to be taken before applying PCA](https://stats.stackexchange.com/a/450089/267920)
#
type(data["target"])
# We first start by flattening our 2D matrix, so that we can apply PCA on it.
pca = PCA()
|
# # 欢迎来到NFL大数据碗的EDA现场
import numpy as np
import pandas as pd
import math
import scipy
from scipy import stats
from scipy.spatial.distance import euclidean
from scipy.special import expit
from tqdm import tqdm
import random
import warnings
warnings.filterwarnings("ignore")
from matplotlib import pyplot as plt
import seaborn as sns
df_train = pd.read_csv(
"/kaggle/input/nfl-big-data-bowl-2020/train.csv",
parse_dates=["TimeHandoff", "TimeSnap"],
infer_datetime_format=True,
low_memory=False,
)
df_train.columns
# ## 主观分析
meta_df = pd.DataFrame({})
meta_df = meta_df.append(
[["GameId", "比赛ID", "分类", "比赛", "无", "", "测试与训练中的GameId都是不同的"]]
)
meta_df = meta_df.append(
[["PlayId", "回合ID", "分类", "比赛", "无", "", "每个待预测数据都是一个唯一的回合Id"]]
)
meta_df = meta_df.append([["Team", "球队", "分类", "球队", "中", "", "不同的球队有不同的进攻防守能力"]])
meta_df = meta_df.append([["X", "球员位置x", "数值", "球员动态", "高", "", "球员位置决定了战术执行顺利与否"]])
meta_df = meta_df.append([["Y", "球员位置y", "数值", "球员动态", "高", "", "球员位置决定了战术执行顺利与否"]])
meta_df = meta_df.append(
[["S", "球员速度", "数值", "球员动态", "高", "", "最直接的说rusher的速度与码数的关系是很直接的"]]
)
meta_df = meta_df.append(
[["A", "球员加速度", "数值", "球员动态", "高", "", "最直接的说rusher的加速度与码数的关系是很直接的"]]
)
meta_df = meta_df.append([["Dis", "", "数值", "球员动态", "中", "", ""]])
meta_df = meta_df.append(
[["Orientation", "球员面向角度", "数值", "球员动态", "低", "", "表现球员的观察方向,或者在更高级的维度会更有用"]]
)
meta_df = meta_df.append(
[["Dir", "球员移动角度", "数值", "球员动态", "中", "", "移动方向感觉直接的作用不如间接的大"]]
)
meta_df = meta_df.append(
[["NflId", "球员Id", "分类", "球员静态", "中", "", "根据具体球员能力不同,最终达成的码数不同"]]
)
meta_df = meta_df.append(
[["DisplayName", "球衣名字", "分类", "球员静态", "无", "", "基本没用,更多是在可视化部分起作用"]]
)
meta_df = meta_df.append(
[["JerseyNumber", "球员号码", "分类", "球员静态", "无", "", "一般决定了位置,但是位置有单独的字段,所以也没啥用"]]
)
meta_df = meta_df.append([["Season", "赛季", "分类", "比赛", "无", "", "赛季看起来范围太大,应该没什么用"]])
meta_df = meta_df.append(
[["YardLine", "码线", "分类", "比赛", "低", "", "看过比赛后我觉得码线有影响但是不大,是不是在终场前且分差很接近时会有呢"]]
)
meta_df = meta_df.append([["Quarter", "第几节", "分类", "比赛", "低", "", "不认为第几节会有太大关系"]])
meta_df = meta_df.append([["GameClock", "比赛时间", "时间", "比赛", "低", "", "同样不认为时间关系会很大"]])
meta_df = meta_df.append([["PossessionTeam", "进攻方", "分类", "比赛", "中", "", "球队进攻防守能力有关"]])
meta_df = meta_df.append(
[["Down", "1~4 down", "分类", "比赛", "中", "", "影响战术,通常如果是1选择保守,2,3会进攻性强一些,4则弃踢多"]]
)
meta_df = meta_df.append(
[["Distance", "需要多少码可以继续进攻", "数值", "比赛", "中", "", "与Donw的关系类似"]]
)
meta_df = meta_df.append(
[["FieldPosition", "目前比赛在哪个队半场进行", "分类", "比赛", "低", "", "这个信息在码线上也有体现"]]
)
meta_df = meta_df.append(
[["HomeScoreBeforePlay", "主队赛前积分", "数值", "球队", "低", "", "这主要是体现球队实力"]]
)
meta_df = meta_df.append(
[["VisitorScoreBeforePlay", "客队赛前积分", "数值", "球队", "低", "", "这主要是体现球队实力,影响应该是总体的"]]
)
meta_df = meta_df.append(
[["NflIdRusher", "持球人Id", "分类", "比赛", "中", "", "持球人的影响肯定是单个特征中最大的"]]
)
meta_df = meta_df.append(
[["OffenseFormation", "进攻队形", "分类", "比赛", "中", "", "不同的进攻方式通常目的是不同的,对应的码数也不同"]]
)
meta_df = meta_df.append(
[["OffensePersonnel", "进攻人员组成", "分类", "比赛", "中", "", "这个主要是配合队形使用,可以认为是队形的更细分"]]
)
meta_df = meta_df.append(
[
[
"DefendersInTheBox",
"防守方在混战区的人数",
"数值",
"比赛",
"高",
"",
"这个人数跟战术有关,感觉有关系,其他kernel看关系还挺大",
]
]
)
meta_df = meta_df.append(
[["DefensePersonnel", "防守人员组成", "分类", "比赛", "中", "", "防守人员是针对进攻人员来设置的"]]
)
meta_df = meta_df.append(
[["PlayDirection", "回合进行的方向", "分类", "比赛", "无", "", "比赛方向,左还是右,关系不大"]]
)
meta_df = meta_df.append(
[["TimeHandoff", "传球时间", "时间", "比赛", "低", "", "可能跟战术有关,或者进展是否顺序,一般来说越快越好"]]
)
meta_df = meta_df.append(
[["TimeSnap", "发球时间", "时间", "比赛", "无", "", "感觉不到有什么关系,与handoff求时间差吧"]]
)
meta_df = meta_df.append(
[["PlayerHeight", "球员身高", "数值", "球员静态", "低", "", "太明显感觉不到,但是不能说没有"]]
)
meta_df = meta_df.append(
[["PlayerWeight", "球员体重", "数值", "球员静态", "低", "", "太明显感觉不到,但是不能说没有*2"]]
)
meta_df = meta_df.append(
[["PlayerBirthDate", "球员生日", "时间", "球员静态", "无", "", "直接用没用,但是可以转为年龄,但是关系应该也不太大"]]
)
meta_df = meta_df.append(
[["PlayerCollegeName", "球员大学", "分类", "球员静态", "低", "", "关系不大,虽然说名校可能实力更大,但是不尽然"]]
)
meta_df = meta_df.append(
[["Position", "球员职责", "分类", "球员静态", "低", "", "根据持球人的Position或者有不错的效果"]]
)
meta_df = meta_df.append([["HomeTeamAbbr", "主队名", "分类", "球队", "低", "", "聚合统计球队进攻防守能力"]])
meta_df = meta_df.append(
[["VisitorTeamAbbr", "客队名", "分类", "球队", "低", "", "聚合统计球队进攻防守能力"]]
)
meta_df = meta_df.append(
[["Week", "第几周", "分类", "比赛", "无", "", "目前是第几周,或者会考虑疲劳,但是缩小到每个回合,关系不大"]]
)
meta_df = meta_df.append([["Stadium", "球场", "分类", "环境", "无", "", "微乎其微"]])
meta_df = meta_df.append(
[["Location", "球场所在位置", "分类", "环境", "低", "", "可能有气候问题,比如NBA的掘金所在的高原地区"]]
)
meta_df = meta_df.append([["StadiumType", "球场类型", "分类", "环境", "无", "", "微乎其微"]])
meta_df = meta_df.append([["Turf", "草皮", "分类", "环境", "无", "", "微乎其微"]])
meta_df = meta_df.append([["GameWeather", "比赛天气", "分类", "环境", "无", "", "微乎其微"]])
meta_df = meta_df.append([["Temperature", "温度", "数值", "环境", "无", "", "微乎其微"]])
meta_df = meta_df.append([["Humidity", "湿度", "数值", "环境", "无", "", "微乎其微"]])
meta_df = meta_df.append([["WindSpeed", "风速", "数值", "环境", "无", "", "微乎其微"]])
meta_df = meta_df.append([["WindDirection", "风向", "数值", "环境", "无", "", "微乎其微"]])
meta_df = meta_df.append(
[["Yards", "所获得的码数", "数值", "比赛", "目标", "", "该次回合进攻方获得的码数,理论上多为整数,少数为负数或零"]]
)
meta_df.columns = [
"name",
"desc",
"type",
"segment",
"expectation",
"conclusion",
"comment",
]
meta_df.sort_values(by="expectation")
# 直观分析的Top10如下:
# 1. XYAS 四个动态特征;
# 2. DefendersInTheBox 表现防守方在Box里的人数;
# 3. PossessionTeam 进攻球队;
# 4. NflIdRusher 持球人Id;
# 5. Down 进攻次数;
# 6. OffenseFormation 进攻队形;
# 7. NflId 场上球员组成(比如对于NE,汤姆布雷迪伤了没上的影响应该是比较大的,等等);
# 大体感觉是:球员动态 > 比赛 > 球队;
# ## 从Yards开始
df_train.Yards.describe()
# 对`Yards`进行describe看整体情况,最小值有负数,这是正常的,发球后通常持球人都是在码线后,如何此时没能前进,而是被拦截,那么这一回合的码数就是负数,最大是99,几乎是跑完了全场,平均每回合推进4.2码,以这个码数看,通常进攻方要维持继续进攻是比较容易的,数据整体分布非正态,应该是右偏的;
sns.distplot(df_train.Yards)
# 直方图上看还好,但是数据有不少为0的,这里做log转换要注意,应该是有微微右偏,且显示峰度;
print("Skewness: %f" % df_train.Yards.skew())
print("Kurtosis: %f" % df_train.Yards.kurt())
# 确实是正偏+显示峰度,与上述可视化结果一致;
# ## 再来看看Top10与Yards的关系
# ### 数值型
df_train_rusher = df_train[df_train.NflId == df_train.NflIdRusher].copy()
df_train_rusher["ToLeft"] = df_train_rusher.PlayDirection.apply(
lambda play_direction: play_direction == "left"
)
df_train_rusher["X_std"] = df_train_rusher[["ToLeft", "X"]].apply(
lambda row: 120 - row.X - 10 if row.ToLeft else row.X - 10, axis=1
)
df_train_rusher["Y_std"] = df_train_rusher[["ToLeft", "Y"]].apply(
lambda row: 160 / 3 - row.Y if row.ToLeft else row.Y, axis=1
)
var = "X_std"
data = pd.concat([df_train_rusher.Yards, df_train_rusher[var]], axis=1)
data.plot.scatter(x=var, y="Yards")
meta_df.loc[meta_df.name == "X", "conclusion"] = "可以生成一个规则特征,用于对最终结果的限制,另外作为空间位置信息有待挖掘"
# 对于`X`的处理需要标准化后,否则结果是相对的而不是绝对的,通过对`X_std`的散点图看到,**能够得到超大码数的前提是你距离TouchDown区域够远**,否则是不可能得到大码数的,这一特征需要构建出来,因为这是规则性的东西,可以加到结果限制中;
var = "Y_std"
data = pd.concat([df_train_rusher.Yards, df_train_rusher[var]], axis=1)
data.plot.scatter(x=var, y="Yards")
meta_df.loc[meta_df.name == "Y", "conclusion"] = "可以生成一个距离左右边界距离的特征,作为空间位置有待更深入的挖掘"
# 对`Y_std`的可视化看到,大码数集中在中间区域,而两边则只有相对小的码数,不过因为数据主要集中在中间区域,所以这一特点并不明显,有限的数据或许可以说明以下观点:**当持球人在Y方向的中间时,他左右可以选择的空间更大,因此他更有希望通过左右跑动摆脱获取更大的码数**,因此可以考虑生成一个距离左右边界的距离特征来更清晰的表达这一点,且与目标成线性关系,目前是非线性的;
var = "S"
data = pd.concat([df_train_rusher.Yards, df_train_rusher[var]], axis=1)
data.plot.scatter(x=var, y="Yards")
meta_df.loc[meta_df.name == "S", "conclusion"] = "没有明显线性关系,大码数集中在中间部分,所以速度适中是个选项"
# 没看到想象中的线性关系,说明该问题实际上是比较复杂的问题,单个特征的线性关系不容易获取,主要也是因为`S`是短时间内测量的,并不是说每个球员就一直处于这个速度,因此参考意义没有字面意义那么大,可以看出的是:**相对来说速度适中的情况下,码数获取更大,可以认为是速度太慢容易被拦截,速度太快不同意改变自己的方向,也就是缺少变化,而速度适中时,既能向前冲刺,又可以做适当的方向上的调整**;
var = "A"
data = pd.concat([df_train_rusher.Yards, df_train_rusher[var]], axis=1)
data.plot.scatter(x=var, y="Yards")
meta_df.loc[meta_df.name == "A", "conclusion"] = "没有明显线性关系,大码数集中在左边,也就是说球员速度更平均时"
# 加速度可视化呈现出一种诡异的拐角型,两头小,中间大,这里与速度类似的是,加速度太大,反而没能获取太大的码数,原因我认为是类似的,可能需要考虑结合二者生成一个能表达这部分信息的特征;
# 数值型的小结:没有明显的线性关系存在,说明如果特征直接送模型,效果不会理想,需要深度挖掘、组合高维特征、更具业务意义的操作,但是也从4个主要的动态数值信息中获取到一些有趣的信息;
# PS:注意XYSA都只可视化了持球人的部分,这是因为我认为对于持球人、进攻方其他球员、防守方球员来说,毫无疑问最重要的是持球人部分,但是这三部分的含义相差很大,放到一起可视化可能无法得到有用的信息,因此这里只对持球人部分做可视化;
# ### 分类型
var = "DefendersInTheBox"
data = pd.concat([df_train_rusher.Yards, df_train_rusher[var]], axis=1)
f, ax = plt.subplots(figsize=(12, 6))
fig = sns.boxplot(x=var, y="Yards", data=data)
meta_df.loc[meta_df.name == "DefendersInTheBox", "conclusion"] = "不看超大码数时,人数越多,码数越小"
# 能看到以下的数据信息:
# - 超大码数来自于防守方在Box中人数为5到9人时;
# - 同样在5到9人时,也更多的出现负码数,也就是迫使进攻方失误;
# - 大码数在这个特征维度上,属于异常值,说明本身应该是很少出现的;
# - 该特征在与码数的关系上,整体有一个人数越多,码数越小的趋势(排除异常值的情况下);
var = "PossessionTeam"
data = pd.concat([df_train_rusher.Yards, df_train_rusher[var]], axis=1)
f, ax = plt.subplots(figsize=(20, 6))
fig = sns.boxplot(x=var, y="Yards", data=data)
meta_df.loc[meta_df.name == "PossessionTeam", "conclusion"] = "球队层面对码数的影响不大,各只球队基本持平"
# 进攻球队的维度上看,基本差异不明显,说明整体战术是一致的,争取小码数推进,有机会就拿大码数,但是不冒险,因此仅仅区分球队无法看到更多信息;
var = "NflIdRusher"
data = pd.concat([df_train_rusher.Yards, df_train_rusher[var]], axis=1)
f, ax = plt.subplots(figsize=(30, 6))
fig = sns.boxplot(x=var, y="Yards", data=data)
meta_df.loc[
meta_df.name == "NflIdRusher", "conclusion"
] = "球员层面区别很大,但是相对来说球员层面的数据量更小,所以是否具有代表性有待研究"
# 各个球员之间的差异明显,远远超过球队之间的差异,因此NflIdRusher应该是一个比较重要的特征,这证明了主观分析部分对这个特征的判断;
var = "Down"
data = pd.concat([df_train_rusher.Yards, df_train_rusher[var]], axis=1)
f, ax = plt.subplots(figsize=(8, 6))
fig = sns.boxplot(x=var, y="Yards", data=data)
meta_df.loc[meta_df.name == "Down", "conclusion"] = "Down越大,代表所剩机会越少,一般码数都会越小"
# 对于`Down`来说,有一点明显的特点在于第一次进攻大码数肯定比第二次多,以此类推至第四次,这是因为在第四次如果没有弃踢,那么通常也是因为目前所需的码数很小,所以一般选择最保守的战术获取小码数即可,没必要高风险追求大码数;
var = "OffenseFormation"
data = pd.concat([df_train_rusher.Yards, df_train_rusher[var]], axis=1)
f, ax = plt.subplots(figsize=(12, 6))
fig = sns.boxplot(x=var, y="Yards", data=data)
meta_df.loc[
meta_df.name == "OffenseFormation", "conclusion"
] = "数据分布主体是类似的,大码数方向根据队形不同有所差异"
# 对于`OffenseFormation`,每种进攻队形的主体都是获取小码数,这也说明了在NFL这种高水平对抗中,大码数出现是很困难,以至于不算为主要进攻手段;
# 分类型的小结:依然没能看到与码数特别相关的特征,这也说明了问题的复杂程度不是单个变量能直接相关的,事实上,很多变量与码数的相关性都是接近于0的,所以我们的目的应该是构建一个NFL的`OverallQual`特征,如果你做过HousePrice项目,你知道我说的是什么;
# 最后看一下对特征信息的更新:
meta_df
# ## 客观分析所有变量
# 同样的,在这一步,只使用rusher部分,由于这里的数据是展开了的,最终结果是一个回合为一行的,而除了球员信息外,其他部分都是一致的,而球员信息与结果关系最大的理应是rusher的动态静态信息;
# correlation matrix
corrmat = df_train_rusher.corr()
f, ax = plt.subplots(figsize=(15, 10))
sns.heatmap(corrmat, vmax=0.8, square=True)
# 热图中看到的信息如下:
# - 没有与`Yards`特别相关的,不管正负;
# - `Distance`与`Down`负相关;
# - `Week`与`Temperature`负相关;
# - `Season`、`GameId`、`PlayId`是相关的;
# - 环境类更是无限接近于0;
k = 10
cols = corrmat.nlargest(k, "Yards")["Yards"].index
cm = np.corrcoef(df_train_rusher[cols].values.T)
plt.subplots(figsize=(8, 8))
sns.set(font_scale=1.25)
hm = sns.heatmap(
cm,
cbar=True,
annot=True,
square=True,
fmt=".2f",
annot_kws={"size": 10},
yticklabels=cols.values,
xticklabels=cols.values,
)
plt.show()
# 看到以下信息:
# - 最相关的是`A`;
# - `S`与`Dis`相关性高达0.88,去掉`Dis`;
# - `GameId`与`PlayId`、`Season`相关性都很高,由于`GameId`与`PlayId`都是不会重复出现在测试集的,因此保留`Season`;
# - `NflId`在这里表示的应该是`NflIdRusher`;
# 注意因为此处是相关性分析,对于分类型特征可能不是很**友好**;
# 最终这一步后保留的特征是:`A`、`S`、`Distance`、`YardLine`、`Season`、`NflIdRusher`;
sns.set()
cols = ["Yards", "A", "S", "Distance", "YardLine", "Season", "NflIdRusher"]
sns.pairplot(df_train_rusher[cols], size=2.5)
plt.show()
# 目前看这个可视化图,如果一些跟规则相关的分割线外,看不到太多有用的信息,这一点从相关系数上也能看出,最高才0.1+,说明很多特征都是很基础、原始的,哟要体现出较强的关联性,一个靠EDA的业务探索,一个靠FE的工程化特征构建;
# ## 缺失数据处理
total = df_train.isnull().sum().sort_values(ascending=False)
percent = (df_train.isnull().sum() / df_train.isnull().count()).sort_values(
ascending=False
)
missing_data = pd.concat([total, percent], axis=1, keys=["Total", "Percent"])
missing_data[missing_data.Total > 0]
# 分析下缺失情况:
# - 最多的是WindDirection和WindSpeed,属于环境特征,缺失大于13%,由于本身相关性不高,且缺失较多,因此环境类的做drop处理;
# - Temperature、GameWeather、Humidity同上;
# - StadiumType理想状态是通过google填充,但是由于其低相关性,且可能携带噪声,同样drop处理;
# - FieldPosition的缺失是因为在中线开球,所以填充其middle即可;
# - OffenseFormation进攻队形;
# - Dir移动角度影响球员后续的位置;
# - Orientation面向角度说实话不知道怎么用这个字段,但是缺失不多,drop太可惜;
# - DefendersInTheBox在Box的人数算是比较重要的,这个可以通过计算得到;
# 后5个需要填充,FieldPosition简单,后4个需要考虑,但是其实缺失都很少,所以影响也不会太大,但是不建议删除行,因为22行为一个回合组,删除掉可能会影响后续的聚合分析;
df_train = df_train.drop(
[
"WindSpeed",
"WindDirection",
"Temperature",
"GameWeather",
"Humidity",
"StadiumType",
],
axis=1,
)
df_train.FieldPosition = df_train.FieldPosition.fillna("middle")
# OffenseFormation, Dir, Orientation, DefendersInTheBox
# df_train[]
df_train.isnull().sum().max()
# ## 异常数据处理
# 主要关注一些离群点与表现明显不正常的点,注意这里单变量分析主要是分析目标变量,但是这里的目标变量由于有球场做限制,因此不存在所谓的异常值,而二元分析应该是偏离主分布的点,但是目前看二元的分布没有明显的线性等关系,因此这一步先省略;
from sklearn.preprocessing import StandardScaler
# standardizing data
saleprice_scaled = StandardScaler().fit_transform(df_train["Yards"][:, np.newaxis])
low_range = saleprice_scaled[saleprice_scaled[:, 0].argsort()][:10]
high_range = saleprice_scaled[saleprice_scaled[:, 0].argsort()][-10:]
print("outer range (low) of the distribution:")
print(low_range)
print("\nouter range (high) of the distribution:")
print(high_range)
# 看到虽然有14这种明显大于0的点,但是由于确实存在这种可能(几乎跑了全场的情况),因此依然是正常数据而非异常数据,当然了,属于小概率情况,实际上如何去掉**超大码数**和**负码数**也许会更好;
# ## 数据测试假设检查
# 主要是正态性检查、同质性检查、线性检查(这个比较麻烦,目前数据没有明显线性关系)、不存在相关误差;
# ### 从正态性开始
# histogram and normal probability plot
sns.distplot(df_train["Yards"], fit=stats.norm)
fig = plt.figure()
res = stats.probplot(df_train["Yards"], plot=plt)
# 能够看到码数分布显示峰度,且有正偏的,且概率图上看也与红线相差很远,但是无法直接应用log变化,因为数据中有不少在0和0以下的情况没法处理;
# ## 深度EDA
# 从此开始挖掘跟橄榄球相关的更深层次的特征,并通过可视化等手段验证其有效性;
df_train_rusher = df_train[df_train.NflId == df_train.NflIdRusher].copy()
# ### 先来看看rusher的位置热图
plt.subplots(figsize=(16, 8))
tmp = df_train_rusher[["Y", "X", "Yards"]].copy()
tmp.X = pd.cut(tmp.X, 12 * 5)
tmp.Y = pd.cut(tmp.Y, 6 * 5)
sns.heatmap(tmp.groupby(["Y", "X"]).count().reset_index().pivot("Y", "X", "Yards"))
# 通常来说美式橄榄球有两种进攻方式:
# - 通过四分卫持球,前传找合法接球人,后由该接球人持球推进;
# - 通过四分卫将球后传给跑卫,由跑卫持球冲阵;
# 目前这个问题下对应的数据都是第二种情况,对于第二种进攻方式,一般来说失误更少,但是获取码数相对较小,容易被拦截但是也不容易丢球,比较保守的战术,从热图中看,`Y`从22到30是选择比较多的区域,这个区域相对横向的可移动范围比较大,同时又微微**避开了中间的混战区**,看起来像是个不错的选择;
# ### 再来看看各位置rusher对应拿到的码数
plt.subplots(figsize=(16, 8))
sns.heatmap(
tmp.groupby(["Y", "X"]).mean().reset_index().pivot("Y", "X", "Yards"),
center=0,
vmin=-5,
vmax=10,
)
# 注意,由于是求各个位置的平均值,因此不乏有些偏远位置数据量很小,球队出的奇招怪招啥的,这部分要特别注意,主要还是分析中间这一块大区域的情况:
# - 数据相对比较稳定,各区域差异不大;
# - 主体的边缘上颜色变化较大,包括负数和大数;
# - 主体以外要么是大码数,要么是负码数,这部分就比较凌乱,说明特别的战术通常都是高风险与高收益并存;
# ### 开始挖掘 - 热热身
# 下面是之前的主观分析后产生了结论信息的特征;
meta_df[meta_df.conclusion.str.len() > 0]
# #### 挖掘rusher的球员静态信息
# - 身高
# - 体重
# - 年龄
df_train_rusher.PlayerHeight = df_train_rusher.PlayerHeight.apply(
lambda height: int(height[0]) * 12 + int(height[2:])
).astype("int")
df_train_rusher["Age"] = df_train_rusher.PlayerBirthDate.apply(
lambda bd: 2019 - int(bd[-4:])
)
plt.subplots(figsize=(20, 5))
plt.subplot(1, 3, 1)
var = "PlayerHeight"
data = pd.concat([df_train_rusher.Yards, df_train_rusher[var]], axis=1)
# data.plot.scatter(x=var, y='Yards')
# axs[0][0].plot.scatter(data[var],data['Yards'])
plt.scatter(data[var], data["Yards"])
plt.subplot(1, 3, 2)
var = "PlayerWeight"
data = pd.concat([df_train_rusher.Yards, df_train_rusher[var]], axis=1)
plt.scatter(data[var], data["Yards"])
plt.subplot(1, 3, 3)
var = "Age"
data = pd.concat([df_train_rusher.Yards, df_train_rusher[var]], axis=1)
plt.scatter(data[var], data["Yards"])
# 可以看到:
# - 身高、体重中都体现出在中间部分的数据获取到的大码数较多;
# - 年龄体现出年轻选手更容易获取大码数,峰值出现在24岁左右时,这一点从跑卫黄金年龄可以看出,实际上除了四分卫,NFL其他大部分位置都很难打到30岁以后,壮哉,**汤姆布雷迪**;
# #### 挖掘距离左右边界特征`Y_dis`
df_train_rusher["Y_dis"] = np.abs(
np.abs(df_train_rusher.Y - df_train_rusher.Y.mean()) - df_train_rusher.Y.mean()
)
var = "Y_dis"
data = pd.concat([df_train_rusher.Yards, df_train_rusher[var]], axis=1)
data.plot.scatter(x=var, y="Yards")
# 与之前的码数热图基本对应,距离左右边界越远,或者说越接近球场纵向的中间区域,相对码数会比较稳定集中,且产生大码数;
# #### 看看`Dir`和`Orientation`的夹角
df_train_rusher["Dir_orientation"] = np.abs(
df_train_rusher.Dir - df_train_rusher.Orientation
)
df_train_rusher["Dir_orientation"] = df_train_rusher["Dir_orientation"].apply(
lambda do: 360 - do if do > 180 else do
)
var = "Dir_orientation"
data = pd.concat([df_train_rusher.Yards, df_train_rusher[var]], axis=1)
data.plot.scatter(x=var, y="Yards")
# 从球员移动方向与面向夹角来看,度数越大(注意这里夹角做了处理,避免出现大于180度的情况),相对获取码数越小,我理解如果rusher正在往前跑,却往其他方向看,多数情况下说明有防守球员正在向他冲来,而空间环境更好的rusher,通常是不需要左顾右盼;
# #### 开球到传球的时间 - (`TimeHandoff`-`TimeSnap`)
df_train_rusher["TimeFromSnapToHandoff"] = (
(df_train_rusher.TimeHandoff - df_train_rusher.TimeSnap)
.apply(lambda x: x.total_seconds())
.astype("int8")
)
var = "TimeFromSnapToHandoff"
data = pd.concat([df_train_rusher.Yards, df_train_rusher[var]], axis=1)
data.plot.scatter(x=var, y="Yards")
# 从传球到开球的时间差来看,时间大于2秒时,通常结果都不会太好,但是由于大量数据集中在1和2秒上,而这部分差异不明显,因此提供的有效信息并不多;
# #### 比赛进行时长、Quarter
df_train_rusher["GameDuration"] = (
df_train_rusher.GameClock.apply(
lambda gc: 15 * 60 - int(gc[:2]) * 60 - int(gc[3:5])
)
) + (df_train_rusher.Quarter - 1) * 15 * 60
var = "GameDuration"
data = pd.concat([df_train_rusher.Yards, df_train_rusher[var]], axis=1)
data.plot.scatter(x=var, y="Yards")
var = "Quarter"
data = pd.concat([df_train_rusher.Yards, df_train_rusher[var]], axis=1)
f, ax = plt.subplots(figsize=(8, 6))
fig = sns.boxplot(x=var, y="Yards", data=data)
# 可以获取到如下信息:
# - 常规时间内各节间码数分布基本一致;
# - 常规时间与加时赛之间有3分钟休息,这就是上图空白部分的原因;
# - 加时赛相对常规赛码数分布更加集中在小码数,从箱行图上看同样可以获取到这个结果;
# NFL加时赛规则:与常规时间不同的是引入了突然死亡规则,即在加时赛过程中,任何一方率先达阵,则直接宣告胜利,因此对于进攻方来说,要严格控制失误,保持继续进攻,则优势会很大,因为犯错的成本是很高的,因此一般战术选择会相对常规时间更加保守;
# #### 距离达阵还有多少码
df_train_rusher["DistanceTouchDown"] = df_train_rusher[
["YardLine", "FieldPosition", "PossessionTeam"]
].apply(
lambda yfp: 100 - yfp["YardLine"]
if (yfp["PossessionTeam"] == yfp["FieldPosition"])
else yfp["YardLine"],
axis=1,
)
var = "DistanceTouchDown"
data = pd.concat([df_train_rusher.Yards, df_train_rusher[var]], axis=1)
data.plot.scatter(x=var, y="Yards")
# 这里的斜线可以看到是达阵线,即达到的码数等于达阵所需的码数,很明显的趋势时,所需码数越小,达阵线会越密集,这也是合理的,毕竟需要5码达阵和需要50码对于进攻方来说不是一个难度;
# #### rusher与码线的横向距离
# #### DL-LB防守阵型
# Create the DL-LB combos
# Clean up and convert to DL-LB combo
df_train_rusher["DL_LB"] = (
df_train_rusher["DefensePersonnel"]
.str[:10]
.str.replace(" DL, ", "-")
.str.replace(" LB", "")
)
top_5_dl_lb_combos = (
df_train_rusher.groupby("DL_LB")
.count()["GameId"]
.sort_values()
.tail(10)
.index.tolist()
)
var = "DL_LB"
data = pd.concat(
[
df_train_rusher.loc[df_train_rusher["DL_LB"].isin(top_5_dl_lb_combos)].Yards,
df_train_rusher.loc[df_train_rusher["DL_LB"].isin(top_5_dl_lb_combos)][var],
],
axis=1,
)
f, ax = plt.subplots(figsize=(8, 6))
fig = sns.boxplot(x=var, y="Yards", data=data)
fig.set_ylim(-10, 20)
# 从DL_LB组合分类来看码数:
# - 各组之间存在一定的差异,注意这里y轴是zoom到-10和20的,所以这个差异看起来很明显,实际上大部分数据也都集中在这个区域;
# - 3-2组合时,进攻方码数平均最高,而4-4则把对方的码数压制到最低;
# - 而3-3,5-2,4-2,2-4情况基本一致;
# #### rusher的Position
var = "Position"
data = pd.concat([df_train_rusher.Yards, df_train_rusher[var]], axis=1)
f, ax = plt.subplots(figsize=(8, 6))
fig = sns.boxplot(x=var, y="Yards", data=data)
# 看到对于rusher不同的Position来说,有以下信息:
# - 各位置拿下的平均码数差异明显,当然要注意的是数据量的差异,RB肯定是最多的;
# - RB作为最常冲阵的位置,码数处于中间位置,不高不低;
# - CB码数最高;
# - DT、DE、G数据少的可以忽略了都;
# #### 距离rusher最近的队友多远、对手多远、队友对手距离多远
possessionteam_map = {"BLT": "BAL", "CLV": "CLE", "ARZ": "ARI", "HST": "HOU"}
df_train.PossessionTeam = df_train.PossessionTeam.apply(
lambda pt: possessionteam_map[pt] if pt in possessionteam_map.keys() else pt
)
df_train["TeamBelongAbbr"] = df_train.apply(
lambda row: row["HomeTeamAbbr"]
if row["Team"] == "home"
else row["VisitorTeamAbbr"],
axis=1,
)
df_train["Offense"] = df_train.apply(
lambda row: row["PossessionTeam"] == row["TeamBelongAbbr"], axis=1
)
df_aggregation = pd.DataFrame(
columns={
"GameId": [],
"PlayId": [],
"Teammate_dis": [],
"Enemy_dis": [],
"Teamate_enemy_dis": [],
"Nearest_is_teammate": [],
"Yards": [],
}
)
for k, group in df_train.groupby(["GameId", "PlayId"]):
rusher = group[group.NflId == group.NflIdRusher].iloc[0]
offenses = group[group.NflId != group.NflIdRusher][group.Offense]
defenses = group[~group.Offense]
def get_nearest(target, df):
df["Tmp_dis"] = df[["X", "Y"]].apply(
lambda xy: np.linalg.norm(
np.array([xy.X, xy.Y]) - np.array([rusher.X, rusher.Y])
),
axis=1,
)
return df.sort_values(by="Tmp_dis", ascending=False).iloc[0]
nearest_offense = get_nearest(rusher, offenses)
nearest_defense = get_nearest(rusher, defenses)
Teamate_enemy_dis = np.linalg.norm(
np.array([nearest_offense.X, nearest_offense.Y])
- np.array([nearest_defense.X, nearest_defense.Y])
)
df_aggregation = df_aggregation.append(
{
"GameId": k[0],
"PlayId": k[1],
"Teammate_dis": nearest_offense.Tmp_dis,
"Enemy_dis": nearest_defense.Tmp_dis,
"Teamate_enemy_dis": Teamate_enemy_dis,
"Nearest_is_teammate": 1
if nearest_offense.Tmp_dis < nearest_defense.Tmp_dis
else 0,
"Yards": rusher.Yards,
},
ignore_index=True,
)
df_aggregation.info()
plt.subplots(figsize=(20, 5))
plt.subplot(1, 3, 1)
var = "Teammate_dis"
data = pd.concat([df_aggregation.Yards, df_aggregation[var]], axis=1)
plt.scatter(data[var], data["Yards"])
plt.subplot(1, 3, 2)
var = "Enemy_dis"
data = pd.concat([df_aggregation.Yards, df_aggregation[var]], axis=1)
plt.scatter(data[var], data["Yards"])
plt.subplot(1, 3, 3)
var = "Teamate_enemy_dis"
data = pd.concat([df_aggregation.Yards, df_aggregation[var]], axis=1)
plt.scatter(data[var], data["Yards"])
# 可以看到以下信息:
# - rusher的最近的队友离他距离在10到25码时,拿到的码数相对更大,队友过于接近他时,码数反而比较小;
# - rusher的最近的对手离他距离在17到28码时,拿到的码数相对更大;
# - 对于队友与对手的距离,没有体现出太多与码数有关的信息;
var = "Nearest_is_teammate"
data = pd.concat([df_aggregation.Yards, df_aggregation[var]], axis=1)
f, ax = plt.subplots(figsize=(8, 6))
fig = sns.boxplot(x=var, y="Yards", data=data)
# 从最接近rusher的球员是队友还是对手上看:
# - 为对手时:码数相对较小,大码数则远少于为对手时;
# - 为队友时,码数相对较大,当然这个差异是很小的,但是大码数的情况则远远多于前者;
# ### 利用Voronoi、空间影响力热图分析空间信息 - show time
# - 通过可视化球场动态,对比码数大小不同时的球员的区别;
# - 利用VOronoi求rusher可控空间大小、是否与对手空间直接接触;
# - 如何通过位置、速度、加速度、角度量化以rusher为中心的对整个球场的控制力影响图;
# #### VIP HIT from [kernel](https://www.kaggle.com/pednt9/vip-hint-coded)
# 这个kernel是对官方kernel中的VIP Hit的代码实现,实现了基本的基于球的控制力计算、热图绘制;
# 如果这个控制力计算方式是正确的,那么如何去使用这个信息来构建特征呢,记录以下方式:
# - 计算rusher位置的控制力,目前从1000个结果看效果不明显;
# - 计算rusher方向路径上的控制力平均值 - todo
def standardize_dataset(train):
train["ToLeft"] = train.PlayDirection == "left"
train["IsBallCarrier"] = train.NflId == train.NflIdRusher
train["TeamOnOffense"] = "home"
train.loc[train.PossessionTeam != train.HomeTeamAbbr, "TeamOnOffense"] = "away"
train["IsOnOffense"] = train.Team == train.TeamOnOffense # Is player on offense?
train["YardLine_std"] = 100 - train.YardLine
train.loc[
train.FieldPosition.fillna("") == train.PossessionTeam, "YardLine_std"
] = train.loc[train.FieldPosition.fillna("") == train.PossessionTeam, "YardLine"]
train["X_std"] = train.X
train.loc[train.ToLeft, "X_std"] = 120 - train.loc[train.ToLeft, "X"]
train["Y_std"] = train.Y
train.loc[train.ToLeft, "Y_std"] = 53.3 - train.loc[train.ToLeft, "Y"]
train["Orientation_std"] = train.Orientation
train.loc[train.ToLeft, "Orientation_std"] = np.mod(
180 + train.loc[train.ToLeft, "Orientation_std"], 360
)
train["Dir_std"] = train.Dir
train.loc[train.ToLeft, "Dir_std"] = np.mod(
180 + train.loc[train.ToLeft, "Dir_std"], 360
)
train.loc[train["Season"] == 2017, "Orientation"] = np.mod(
90 + train.loc[train["Season"] == 2017, "Orientation"], 360
)
return train
df_train2 = pd.read_csv(
"/kaggle/input/nfl-big-data-bowl-2020/train.csv", low_memory=False
)
dominance_df = standardize_dataset(df_train2)
dominance_df["Rusher"] = dominance_df["NflIdRusher"] == dominance_df["NflId"]
def radius_calc(dist_to_ball):
"""I know this function is a bit awkward but there is not the exact formula in the paper,
so I try to find something polynomial resembling
Please consider this function as a parameter rather than fixed
I'm sure experts in NFL could find a way better curve for this"""
return (
4 + 6 * (dist_to_ball >= 15) + (dist_to_ball**3) / 560 * (dist_to_ball < 15)
)
class Controller:
"""This class is a wrapper for the two functions written above"""
def __init__(self, play):
self.play = play
self.vec_influence = np.vectorize(self.compute_influence)
self.vec_control = np.vectorize(self.pitch_control)
def compute_influence(self, x_point, y_point, player_id):
"""Compute the influence of a certain player over a coordinate (x, y) of the pitch"""
point = np.array([x_point, y_point])
player_row = self.play.loc[player_id]
theta = math.radians(player_row[56])
speed = player_row[5]
player_coords = player_row[54:56].values
ball_coords = self.play[self.play["IsBallCarrier"]].iloc[:, 54:56].values
dist_to_ball = euclidean(player_coords, ball_coords)
S_ratio = (speed / 13) ** 2 # we set max_speed to 13 m/s
RADIUS = radius_calc(dist_to_ball) # updated
S_matrix = np.matrix([[RADIUS * (1 + S_ratio), 0], [0, RADIUS * (1 - S_ratio)]])
R_matrix = np.matrix(
[[np.cos(theta), -np.sin(theta)], [np.sin(theta), np.cos(theta)]]
)
COV_matrix = np.dot(
np.dot(np.dot(R_matrix, S_matrix), S_matrix), np.linalg.inv(R_matrix)
)
norm_fact = (1 / 2 * np.pi) * (1 / np.sqrt(np.linalg.det(COV_matrix)))
mu_play = player_coords + speed * np.array([np.cos(theta), np.sin(theta)]) / 2
intermed_scalar_player = np.dot(
np.dot((player_coords - mu_play), np.linalg.inv(COV_matrix)),
np.transpose((player_coords - mu_play)),
)
player_influence = norm_fact * np.exp(-0.5 * intermed_scalar_player[0, 0])
intermed_scalar_point = np.dot(
np.dot((point - mu_play), np.linalg.inv(COV_matrix)),
np.transpose((point - mu_play)),
)
point_influence = norm_fact * np.exp(-0.5 * intermed_scalar_point[0, 0])
return point_influence / player_influence
def pitch_control(self, x_point, y_point):
"""Compute the pitch control over a coordinate (x, y)"""
offense_ids = self.play[self.play["IsOnOffense"]].index
offense_control = self.vec_influence(x_point, y_point, offense_ids)
offense_score = np.sum(offense_control)
defense_ids = self.play[~self.play["IsOnOffense"]].index
defense_control = self.vec_influence(x_point, y_point, defense_ids)
defense_score = np.sum(defense_control)
return expit(offense_score - defense_score)
def display_control(self, grid_size=(50, 30), figsize=(12, 8)):
front, behind = 30, 10
left, right = 30, 30
if self.play["IsOnOffense"].iloc[0] == True:
colorm = ["purple"] * 11 + ["yellow"] * 11
else:
colorm = ["yellow"] * 11 + ["purple"] * 11
# colorm = ['purple'] * 11 + ['yellow'] * 11
colorm[np.where(self.play.Rusher.values)[0][0]] = "black"
player_coords = self.play[self.play["Rusher"]][["X_std", "Y_std"]].values[0]
X, Y = np.meshgrid(
np.linspace(
player_coords[0] - behind, player_coords[0] + front, grid_size[0]
),
np.linspace(
player_coords[1] - left, player_coords[1] + right, grid_size[1]
),
)
# infl is an array of shape num_points with values in [0,1] accounting for the pitch control
infl = self.vec_control(X, Y)
plt.figure(figsize=figsize)
plt.contourf(X, Y, infl, 12, cmap="bwr")
plt.scatter(self.play["X_std"].values, self.play["Y_std"].values, c=colorm)
plt.title(
"Yards gained = {}, play_id = {}".format(
self.play["Yards"].values[0], self.play["PlayId"].unique()[0]
)
)
plt.show()
_play_id1 = random.choice(dominance_df[~dominance_df.ToLeft].PlayId.tolist())
my_play = dominance_df[dominance_df.PlayId == _play_id1].copy()
control = Controller(my_play)
coords = my_play.iloc[
1, 54:56
].values # let's compute the influence at the location of the first player
_pitch_control = control.vec_control(*coords)
print(_pitch_control)
control.display_control()
_play_id2 = random.choice(dominance_df[~dominance_df.ToLeft].PlayId.tolist())
my_play2 = dominance_df[dominance_df.PlayId == _play_id2].copy()
control2 = Controller(my_play2)
control2.display_control()
_controls = []
_yards = []
for _play_id in dominance_df.PlayId.unique().tolist()[:10000]:
_my_play = dominance_df[dominance_df.PlayId == _play_id].copy()
_control = Controller(_my_play)
_rusher = _my_play.query("Rusher == True").iloc[0]
coords = (_rusher.X_std, _rusher.Y_std)
_pitch_control = _control.vec_control(*coords)
_controls.append(_pitch_control)
_yards.append(_rusher.Yards)
plt.scatter(_controls, _yards)
|
# seed value for random number generators to obtain reproducible results
RANDOM_SEED = 1
# although we standardize X and y variables on input,
# we will fit the intercept term in the models
# Expect fitted values to be close to zero
SET_FIT_INTERCEPT = True
# import base packages into the namespace for this program
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
# modeling routines from Scikit Learn packages
import sklearn.linear_model
from sklearn.ensemble import (
RandomForestClassifier,
RandomForestRegressor,
) # Random Forest Package
from sklearn.ensemble import GradientBoostingRegressor # Gradient Boosted Trees
import time
from sklearn.datasets import make_classification
from collections import OrderedDict
from sklearn.metrics import classification_report
from plotly import tools
from sklearn.datasets import fetch_openml
from sklearn.decomposition import PCA
from sklearn.metrics import f1_score
from sklearn import metrics
from sklearn.metrics import mean_squared_error, r2_score, confusion_matrix
from math import sqrt # for root mean-squared error calculation
import seaborn as sns # pretty plotting, including correlation map
# Starter Code provided at the Start of each Kaggle Notebook
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import os
# Input data files are available in the "../input/" directory.
print("Directory Path where files are located")
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
# Validate working directory
os.getcwd()
print(os.getcwd())
# Validate Current Path and create Path to data
from pathlib import Path
INPUT = Path("../input/digit-recognizer")
os.listdir(INPUT)
# Import CSV into Pandas dataframe and test shape of file
train_df = pd.read_csv(INPUT / "train.csv")
train_df.head(3)
train_df.tail(3)
train_df.shape
# Split into train and validation prior to cross validation
from sklearn.model_selection import train_test_split
X_mtrain, X_valid, y_mtrain, y_valid = train_test_split(
train_df.drop(["label"], axis=1),
train_df["label"],
shuffle=True,
train_size=0.85,
random_state=1,
)
# Check the shape of the trainig data set array
print("Shape of X_mtrain_data:", X_mtrain.shape)
print("Shape of y_mtrain_data:", y_mtrain.shape)
print("Shape of X_validation_data:", X_valid.shape)
print("Shape of y_validation_data:", y_valid.shape)
# S5 Split Train and Test
X_train, X_test, y_train, y_test = train_test_split(
X_mtrain, y_mtrain, train_size=0.7, test_size=0.3, random_state=1
)
print("Shape of X_train_data:", X_train.shape)
print("Shape of y_train_data:", X_test.shape)
print("Shape of X_test_data:", y_train.shape)
print("Shape of y_test_data:", y_test.shape)
# Check out what some of the data looks like
for digit_num in range(0, 64):
subplot(8, 8, digit_num + 1)
grid_data = (
X_train.iloc[digit_num].as_matrix().reshape(28, 28)
) # reshape from 1d to 2d pixel array
plt.imshow(grid_data, interpolation="none", cmap="bone_r")
xticks([])
yticks([])
# Try GridSearch for the Trees
from sklearn.model_selection import GridSearchCV
tree_names = ["RandomForestClassifier"]
tree_clfs = [RandomForestClassifier()]
tree_param = {
tree_names[0]: {
"n_estimators": [10, 100, 200],
"criterion": ["gini"],
"max_features": ["sqrt"],
"n_jobs": [-1],
"max_depth": [4],
"random_state": [RANDOM_SEED],
"bootstrap": [True],
"oob_score": [True],
}
}
for names, estimator in zip(tree_names, tree_clfs):
print(names)
print(tree_names)
print(estimator)
clf = GridSearchCV(estimator, tree_param[names], return_train_score=True, cv=10)
clf.fit(X_train, y_train)
print("best params: " + str(clf.best_params_))
print("best scores: " + str(clf.best_score_))
rmse = np.sqrt(mean_squared_error(y_train, clf.predict(X_train)))
rmse_tst = np.sqrt(mean_squared_error(y_test, clf.predict(X_test)))
print("rmse: {:}".format(rmse))
print("rmse_tst: {:}".format(rmse_tst))
clf.best_params_
start_time = time.process_time()
# Using the best parameters from the for the Extra Tree Regression, use it to show the Feature importance on the entire data set
myfit = RandomForestClassifier(
n_estimators=200,
criterion="gini",
max_features="sqrt",
n_jobs=-1,
max_depth=4,
oob_score=True,
random_state=1,
bootstrap=True,
)
myfit.fit(X_train, y_train)
print("Accuracy on training set: {:.3f}".format(myfit.score(X_train, y_train)))
print("Accuracy on test set: {:.3f}".format(myfit.score(X_test, y_test)))
f1 = f1_score(y_train, myfit.predict(X_train), average="weighted")
f1_tst = f1_score(y_test, myfit.predict(X_test), average="weighted")
f1_vld = f1_score(y_valid, myfit.predict(X_valid), average="weighted")
print("f1: {:}".format(f1))
print("f1_tst: {:}".format(f1_tst))
print("f1_vld: {:}".format(f1_vld))
# Extract single tree
print(metrics.classification_report(myfit.predict(X_train), y_train))
print(metrics.classification_report(myfit.predict(X_test), y_test))
print(metrics.classification_report(myfit.predict(X_valid), y_valid))
end_time = time.process_time()
runtime = end_time - start_time # seconds of wall-clock time
print(runtime) # report in milliseconds
# check for the feature importance
importances = myfit.feature_importances_
indices = np.argsort(importances)[::-1]
print("Feature ranking:")
for f in range(0, 10):
print("%d. feature %d (%f)" % (f + 1, indices[f], importances[indices[f]]))
# Plot the feature importances
figure(figsize(7, 3))
plot(indices[:], importances[indices[:]], "k.")
yscale("log")
xlabel("feature", size=20)
ylabel("importance", size=20)
# Confusion Matrix iRFC
cm_trn = confusion_matrix(y_train, myfit.predict(X_train))
cm_trn_plt = sns.heatmap(
cm_trn.T, square=True, annot=True, fmt="d", cbar=False, cmap="Reds"
)
plt.xlabel("Actual label")
plt.ylabel("Predicted label")
plt.title("Training")
# Confusion Matrix
cm_trn = confusion_matrix(y_test, myfit.predict(X_test))
cm_trn_plt = sns.heatmap(
cm_trn.T, square=True, annot=True, fmt="d", cbar=False, cmap="Reds"
)
plt.xlabel("Actual label")
plt.ylabel("Predicted label")
plt.title("Testing")
# Confusion Matrix
cm_trn = confusion_matrix(y_valid, myfit.predict(X_valid))
cm_trn_plt = sns.heatmap(
cm_trn.T, square=True, annot=True, fmt="d", cbar=False, cmap="Reds"
)
plt.xlabel("Actual label")
plt.ylabel("Predicted label")
plt.title("Validation")
# Try out the PCA model with 95% of the components
start_time2 = time.process_time()
pca = PCA(0.95)
pca.fit(X_train)
transform = pca.transform(X_train)
plt.scatter(transform[:, 0], transform[:, 1], s=20, c=y_train)
plt.colorbar()
clim(0, 9)
xlabel("PC1")
ylabel("PC2")
pca.n_components
end_time2 = time.process_time()
runtime2 = end_time2 - start_time2 # seconds of wall-clock time
print(runtime2) # report in milliseconds
# increase the number of components in PCA to see how many components are needed to capture most of the variance in the data.
n_components_array = [1, 2, 3, 4, 5, 10, 20, 50, 100, 200, 500]
vr = np.zeros(len(n_components_array))
i = 0
for n_components in n_components_array:
pca2 = PCA(n_components=n_components)
pca2.fit(X_train)
vr[i] = sum(
pca2.explained_variance_ratio_
) # use the pca.explained_variance_ratio function to explain variance
i = i + 1
# plot the PCA components to see how the variance is explained
plot(n_components_array, vr, "k.-")
xscale("log")
ylim(9e-2, 1.1)
yticks(linspace(0.2, 1.0, 9))
xlim(0.9)
grid(which="both")
xlabel("number of PCA components", size=20)
ylabel("variance ratio", size=20)
# fit PCA model
pca.fit(train_df)
# transform data onto the first two principal components
X_pca = pca.transform(train_df)
print("Original shape: {}".format(str(train_df.shape)))
print("Reduced shape: {}".format(str(X_pca.shape)))
# RF PCA Model
X_pca_train, X_pca_test, y_pca_train, y_pca_test = train_test_split(
X_pca, train_df["label"], train_size=0.7, test_size=0.3, random_state=1
)
print(X_pca_train.shape)
print(X_pca_test.shape)
print(y_pca_train.shape)
print(y_pca_test.shape)
start_time = time.process_time()
rfc_pca = RandomForestClassifier(
n_estimators=200,
n_jobs=-1,
max_depth=5,
criterion="gini",
max_features="sqrt",
oob_score=True,
bootstrap=True,
random_state=1,
)
# Train
rfc_pca = rfc_pca.fit(X_pca_train, y_pca_train)
print(
"Accuracy on training set: {:.3f}".format(rfc_pca.score(X_pca_train, y_pca_train))
)
print("Accuracy on test set: {:.3f}".format(rfc_pca.score(X_pca_test, y_pca_test)))
f1 = f1_score(y_pca_train, rfc_pca.predict(X_pca_train), average="weighted")
f1_tst = f1_score(y_pca_test, rfc_pca.predict(X_pca_test), average="weighted")
print("f1: {:}".format(f1))
print("f1_tst: {:}".format(f1_tst))
print(metrics.classification_report(rfc_pca.predict(X_pca_train), y_pca_train))
print(metrics.classification_report(rfc_pca.predict(X_pca_test), y_pca_test))
end_time = time.process_time()
runtime = end_time - start_time # seconds of wall-clock time
print(runtime) # report in milliseconds
cm_trn_pca = confusion_matrix(y_pca_train, rfc_pca.predict(X_pca_train))
cm_trn_pca_plt = sns.heatmap(
cm_trn_pca.T, square=True, annot=True, fmt="d", cbar=False, cmap="Reds"
)
plt.xlabel("Actual label")
plt.ylabel("Predicted label")
plt.title("Training")
cm_trn_pca = confusion_matrix(y_pca_test, rfc_pca.predict(X_pca_test))
cm_trn_pca_plt = sns.heatmap(
cm_trn_pca.T, square=True, annot=True, fmt="d", cbar=False, cmap="Reds"
)
plt.xlabel("Actual label")
plt.ylabel("Predicted label")
plt.title("Testing")
# Score test dataset
scr = myfit.predict(X_train)
# Conver array to Pandas dataframe with submission titles
pd_scr = pd.DataFrame(scr)
pd_scr.index.name = "ImageId"
pd_scr.columns = ["label"]
print(pd_scr)
# Export to Excel
pd_scr.to_excel("pd_scr4.xlsx")
|
from transformers import BertTokenizer, BertModel, RobertaModel, RobertaTokenizer
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import torch
import torch.nn as nn
import torch.nn.functional as F
from torch.utils.data import DataLoader
import torch.optim as optim
from scipy.stats import spearmanr
from datetime import datetime
from sklearn.model_selection import GroupKFold, KFold
import gc
import seaborn as sns
import transformers
import re
from collections import Counter as ct
import html
import warnings
warnings.filterwarnings("ignore")
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
# import os
# for dirname, _, filenames in os.walk('/kaggle/input'):
# for filename in filenames:
# print(os.path.join(dirname, filename))]
# Any results you write to the current directory are saved as output.
train = pd.read_csv("../input/google-quest-challenge/train.csv")
words = []
for w in train.question_title + train.question_body + train.answer:
words = words + w.split()
counter = ct(words)
train.question_title = train.question_title.apply(
lambda x: " ".join("[UNK]" if counter[w] < 2 else w for w in x.split())
)
train.question_body = train.question_body.apply(
lambda x: " ".join("[UNK]" if counter[w] < 2 else w for w in x.split())
)
train.answer = train.answer.apply(
lambda x: " ".join("[UNK]" if counter[w] < 2 else w for w in x.split())
)
train.question_title = train.question_title.apply(html.unescape)
train.question_body = train.question_body.apply(html.unescape)
train.answer = train.answer.apply(html.unescape)
targets = torch.tensor(
train[
[
"question_asker_intent_understanding",
"question_body_critical",
"question_conversational",
"question_expect_short_answer",
"question_fact_seeking",
"question_has_commonly_accepted_answer",
"question_interestingness_others",
"question_interestingness_self",
"question_multi_intent",
"question_not_really_a_question",
"question_opinion_seeking",
"question_type_choice",
"question_type_compare",
"question_type_consequence",
"question_type_definition",
"question_type_entity",
"question_type_instructions",
"question_type_procedure",
"question_type_reason_explanation",
"question_type_spelling",
"question_well_written",
"answer_helpful",
"answer_level_of_information",
"answer_plausible",
"answer_relevance",
"answer_satisfaction",
"answer_type_instructions",
"answer_type_procedure",
"answer_type_reason_explanation",
"answer_well_written",
]
].values,
dtype=torch.float32,
)
class bertdataset:
def __init__(self, qtitle, qbody, answer, targets, tokenizer, max_length=512):
self.qtitle = qtitle
self.qbody = qbody
self.answer = answer
self.targets = targets
self.tokenizer = tokenizer
self.max_length = max_length
def __len__(self):
return len(self.answer)
def __getitem__(self, item):
question_title = self.qtitle[item]
question_body = self.qbody[item]
answer_text = self.answer[item]
question_title = " ".join(question_title.split())
question_body = " ".join(question_body.split())
answer_text = " ".join(answer_text.split())
inputs_q = self.tokenizer.encode_plus(
"[CLS]" + question_title + "[QBODY]" + question_body + "[SEP]",
pad_to_max_length=True,
max_length=self.max_length,
)
ids_q = inputs_q["input_ids"]
token_type_ids_q = inputs_q["token_type_ids"]
mask_q = inputs_q["attention_mask"]
inputs_a = self.tokenizer.encode_plus(
"[CLS]" + answer_text + "[SEP]",
pad_to_max_length=True,
max_length=self.max_length,
)
ids_a = inputs_a["input_ids"]
token_type_ids_a = inputs_a["token_type_ids"]
mask_a = inputs_a["attention_mask"]
return {
"ids_q": torch.tensor(ids_q, dtype=torch.long),
"mask_q": torch.tensor(mask_q, dtype=torch.long),
"token_type_ids_q": torch.tensor(token_type_ids_q, dtype=torch.long),
"ids_a": torch.tensor(ids_a, dtype=torch.long),
"mask_a": torch.tensor(mask_a, dtype=torch.long),
"token_type_ids_a": torch.tensor(token_type_ids_a, dtype=torch.long),
"targets": self.targets[item],
}
# model
class nlp(nn.Module):
def __init__(self, bert_path):
super(nlp, self).__init__()
self.bert_path = bert_path
self.bert_model = BertModel.from_pretrained(
self.bert_path, output_hidden_states=True
)
self.drop = nn.Dropout(0.2)
self.dense = nn.Linear(768 * 2, 30)
def forward(self, ids_q, mask_q, token_type_ids_q, ids_a, mask_a, token_type_ids_a):
hidden_layers_q = self.bert_model(
ids_q, attention_mask=mask_q, token_type_ids=token_type_ids_q
)[2]
hidden_layers_a = self.bert_model(
ids_a, attention_mask=mask_a, token_type_ids=token_type_ids_a
)[2]
q12, a12 = hidden_layers_q[-1][:, 0].view(-1, 1, 768), hidden_layers_a[-1][
:, 0
].view(-1, 1, 768)
q11, a11 = hidden_layers_q[-2][:, 0].view(-1, 1, 768), hidden_layers_a[-2][
:, 0
].view(-1, 1, 768)
q10, a10 = hidden_layers_q[-3][:, 0].view(-1, 1, 768), hidden_layers_a[-3][
:, 0
].view(-1, 1, 768)
q9, a9 = hidden_layers_q[-4][:, 0].view(-1, 1, 768), hidden_layers_a[-4][
:, 0
].view(-1, 1, 768)
q = torch.mean(torch.cat((q12, q11, q10, q9), axis=1), axis=1)
a = torch.mean(torch.cat((a12, a11, a10, a9), axis=1), axis=1)
x = torch.cat((q, a), 1)
x = self.dense(self.drop(x))
return x
# tokenizer = BertTokenizer.from_pretrained('bert-base-uncased',do_lower_case=True,
# add_specials_tokens = ["[QBODY]","[UNK]"])
# text = "[CLS] Who was Jim Henson ? [SEP] Jim Henson was a puppeteer [SEP]"
# inputs_q = tokenizer.encode_plus(
# text,
# pad_to_max_length=True,
# max_length=512,
# return_tensors = 'pt'
# )
# ids_q = inputs_q["input_ids"].type(torch.LongTensor).cuda()
# token_type_ids_q = inputs_q["token_type_ids"].type(torch.LongTensor).cuda()
# mask_q = inputs_q["attention_mask"].type(torch.LongTensor).cuda()
# ids_q = torch.stack((ids_q,ids_q,ids_q)).squeeze(1)
# token_type_ids_q = torch.stack((token_type_ids_q,token_type_ids_q,token_type_ids_q)).squeeze(1)
# mask_q = torch.stack((mask_q,mask_q,mask_q)).squeeze(1)
# inputs_a = tokenizer.encode_plus(
# text,
# pad_to_max_length=True,
# max_length=512,
# return_tensors = 'pt'
# )
# ids_a = inputs_a["input_ids"].type(torch.LongTensor).cuda()
# token_type_ids_a = inputs_a["token_type_ids"].type(torch.LongTensor).cuda()
# mask_a = inputs_a["attention_mask"].type(torch.LongTensor).cuda()
# ids_a = torch.stack((ids_a,ids_a,ids_a)).squeeze(1)
# token_type_ids_a = torch.stack((token_type_ids_a,token_type_ids_a,token_type_ids_a)).squeeze(1)
# mask_a = torch.stack((mask_a,mask_a,mask_a)).squeeze(1)
# model = nlp('bert-base-uncased').cuda()
# x,y = model(ids_q,mask_q,token_type_ids_q,ids_a,mask_a,token_type_ids_a)
def train_loop(dataset, model, optimizer, batch_size=6, epochs=1):
batches = DataLoader(dataset, shuffle=False, batch_size=batch_size, num_workers=4)
criterion = nn.BCEWithLogitsLoss(reduce="mean")
model.train()
for i in range(epochs):
for j, batch in enumerate(batches):
ids_q_batch = batch["ids_q"].type(torch.LongTensor).cuda()
mask_q_batch = batch["mask_q"].type(torch.LongTensor).cuda()
segments_q_batch = batch["token_type_ids_q"].type(torch.LongTensor).cuda()
ids_a_batch = batch["ids_a"].type(torch.LongTensor).cuda()
mask_a_batch = batch["mask_a"].type(torch.LongTensor).cuda()
segments_a_batch = batch["token_type_ids_a"].type(torch.LongTensor).cuda()
optimizer.zero_grad()
output = model(
ids_q=ids_q_batch,
mask_q=mask_q_batch,
token_type_ids_q=segments_q_batch,
ids_a=ids_a_batch,
mask_a=mask_a_batch,
token_type_ids_a=segments_a_batch,
)
target_batch = batch["targets"].cuda()
loss = criterion(output, target_batch)
loss.backward()
optimizer.step()
loss = None
def eval_loop(dataset, model):
batches = DataLoader(dataset, shuffle=False, batch_size=6, num_workers=4)
model.eval()
pred_fold = []
target_fold = []
score_fold = 0
with torch.no_grad():
for j, batch in enumerate(batches):
ids_q_batch = batch["ids_q"].type(torch.LongTensor).cuda()
mask_q_batch = batch["mask_q"].type(torch.LongTensor).cuda()
segments_q_batch = batch["token_type_ids_q"].type(torch.LongTensor).cuda()
ids_a_batch = batch["ids_a"].type(torch.LongTensor).cuda()
mask_a_batch = batch["mask_a"].type(torch.LongTensor).cuda()
segments_a_batch = batch["token_type_ids_a"].type(torch.LongTensor).cuda()
output = model(
ids_q=ids_q_batch,
mask_q=mask_q_batch,
token_type_ids_q=segments_q_batch,
ids_a=ids_a_batch,
mask_a=mask_a_batch,
token_type_ids_a=segments_a_batch,
)
out = torch.sigmoid(output).cpu().numpy()
target_fold.append(batch["targets"].numpy())
pred_fold.append(out)
pred_fold = np.vstack(pred_fold)
target_fold = np.vstack(target_fold)
for i in range(30):
score_fold += spearmanr(target_fold[:, i], pred_fold[:, i]).correlation
return pred_fold, score_fold / 30
# def test_loop(dataset,model):
# batches = DataLoader(dataset,shuffle=False,batch_size=4,num_workers=4)
# model.eval()
# pred = []
# with torch.no_grad():
# for j,batch in enumerate(batches):
# ids_batch = batch['ids'].type(torch.LongTensor).cuda()
# mask_batch = batch['mask'].type(torch.LongTensor).cuda()
# segments_batch = batch['segments'].type(torch.LongTensor).cuda()
# out = torch.sigmoid(model(ids = ids_batch,
# mask = mask_batch,
# token_type_ids = segments_batch)).cpu().numpy()
# pred.append(out)
# return np.vstack(pred)
def cross_val(tokenizer, cv=3):
oof_predictions = np.zeros((6079, 30))
folds = GroupKFold(n_splits=cv)
for fold, (train_index, valid_index) in enumerate(
folds.split(X=train.question_body, groups=train.question_body)
):
qtitle_train = (
train.iloc[train_index].question_title.values.astype(str).tolist()
)
qbody_train = train.iloc[train_index].question_body.values.astype(str).tolist()
answer_train = train.iloc[train_index].answer.values.astype(str).tolist()
train_loader = bertdataset(
qtitle=qtitle_train,
qbody=qbody_train,
answer=answer_train,
targets=targets[train_index],
tokenizer=tokenizer,
)
qtitle_valid = (
train.iloc[valid_index].question_title.values.astype(str).tolist()
)
qbody_valid = train.iloc[valid_index].question_body.values.astype(str).tolist()
answer_valid = train.iloc[valid_index].answer.values.astype(str).tolist()
valid_loader = bertdataset(
qtitle=qtitle_valid,
qbody=qbody_valid,
answer=answer_valid,
targets=targets[valid_index],
tokenizer=tokenizer,
)
model = nlp("bert-base-uncased").cuda()
optimizer = optim.AdamW(model.parameters(), lr=3e-5)
print(f"Fold {fold} started at " + datetime.now().strftime("%H:%M"))
train_loop(train_loader, model, optimizer, epochs=4)
print(
"Training last layers a little bit more " + datetime.now().strftime("%H:%M")
)
for p in model.bert_model.parameters():
p.requires_grad = False
optimizer = optim.AdamW(
filter(lambda p: p.requires_grad, model.parameters()), lr=3e-5
)
train_loop(train_loader, model, optimizer, batch_size=6, epochs=3)
print(
f"Fold {fold} training finished, predicting... "
+ datetime.now().strftime("%H:%M")
)
p, s = eval_loop(valid_loader, model)
oof_predictions[valid_index] = p
# test_predictions[fold] = (test_loop(test_loader,model))
print(
f"Fold {fold} finished at "
+ datetime.now().strftime("%H:%M")
+ f" with score: {s}"
)
torch.save(model.state_dict(), f"fold_{fold}_r.pt")
model = None
optimizer = None
torch.cuda.empty_cache()
return oof_predictions
tokenizer = BertTokenizer.from_pretrained(
"bert-base-uncased", do_lower_case=True, add_specials_tokens=["[QBODY]", "[UNK]"]
)
oof_predictions = cross_val(tokenizer)
# n = train['url'].apply(lambda x:(('ell.stackexchange.com' in x) or ('english.stackexchange.com' in x))).tolist()
# spelling=[]
# for x in n:
# if x:
# spelling.append(0.5)
# else:
# spelling.append(0.)
# spearmanr(train['question_type_spelling'],np.array(spelling)).correlation
|
import numpy as np
import pandas as pd
from sklearn.manifold import TSNE
train = pd.read_csv("../input/train.csv")
test = pd.read_csv("../input/test.csv")
train = train[test.columns].values
test = test[test.columns].values
train_test = np.vstack([train, test])
train_test.shape
tsne = TSNE(n_components=2)
train_test_2D_0 = tsne.fit_transform(train_test)
train_2D = train_test_2D_0[: train.shape[0]]
test_2D = train_test_2D_0[train.shape[0] :]
np.save("train_2D", train_2D)
np.save("test_2D", test_2D)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import os
print(os.listdir("../input/fasam-nlp-competition-turma-4"))
# Esse notebook descreve o pipelie para utilização na competição do Kaggle da Fasam. Faz parte da avaliação prática dos alunos da turma de Deep Learning.
# ### Roteiro do Notebook
# Leitura do Dataset
# Criação do Modelo
# Avaliação e Criação do arquivo de submission.csv
# ### Problema
# Uma revista precisa catalogar todas as suas notícias em diferentes categorias. O objetivo desta competição é desenvolver o melhor modelo de aprendizagem profunda para prever a categoria de novas notícias.
# As categorias possíveis são:
# - ambiente
# - equilibrioesaude
# - sobretudo
# - educacao
# - ciencia
# - tec
# - turismo
# - empreendedorsocial
# - comida
import numpy as np
import pandas as pd
import seaborn as sns
import warnings
import matplotlib
import matplotlib.pyplot as plt
sns.set(style="ticks")
warnings.filterwarnings("ignore")
# Bibliotecas do keras
import keras
from keras.models import Model
from keras.layers import *
from keras.optimizers import *
from keras.losses import *
from keras.regularizers import *
from keras.models import Sequential
from keras.callbacks import *
from keras.preprocessing import sequence
from keras.preprocessing.sequence import pad_sequences
from keras.preprocessing.text import Tokenizer
from keras.layers.embeddings import Embedding
from sklearn.model_selection import train_test_split
# # Leitura do dataset de treinamento
# Leitura do Dataset
df = pd.read_csv("../input/fasam-nlp-competition-turma-4/train.csv")
print(df.shape)
df.head()
# Todos os artigos contêm o título e a descrição. Por último a categoria que pertence esse artigo.
# Definição de alguns parâmetros dos modelos e tokenização
# Tamanho da sequencia
seq_size = 10
# Máximo de tokens
max_tokens = 2500
# Tamanho do embedding
embed_dim = 128
# Iremos utilizar o titulo para o nosso modelo baseline. O processo abaixo cria o input da nossa rede e prepara o target.
## Utilizaremos apenas o .title (input) e o .category (target) da nossa rede
# Textos
text = df["title"].values
tokenizer = Tokenizer(num_words=max_tokens, split=" ")
# Transforma o texto em números
tokenizer.fit_on_texts(text)
X = tokenizer.texts_to_sequences(text)
# Cria sequencias de tamanho fixo (input: X)
X = pad_sequences(X, maxlen=seq_size)
# Categoriza o target "category" -> [0,..., 1] (output: y)
Y_classes = pd.get_dummies(df["category"]).columns
Y = pd.get_dummies(df["category"]).values
Y_classes
Y
(X.shape, Y.shape)
# # Criação do Modelo
# Iremos utilizar uma RNN em um modelo simples.
def create_model():
model = Sequential()
# Embedding Layer
model.add(Embedding(max_tokens, embed_dim, input_length=seq_size))
# RNN Layer
model.add(LSTM(seq_size))
# Dense Layer
model.add(Dense(len(Y_classes), activation="softmax"))
model.compile(
loss="categorical_crossentropy", optimizer="adam", metrics=["accuracy"]
)
model.summary()
return model
model = create_model()
# Separa o dataset em dados de treinamento/validação
X_train, X_valid, Y_train, Y_valid = train_test_split(
X, Y, test_size=0.20, random_state=42, stratify=Y
)
weights_filepath = "weights.h5"
callbacks = [
ModelCheckpoint(
weights_filepath, monitor="val_loss", mode="min", verbose=1, save_best_only=True
),
EarlyStopping(monitor="val_loss", mode="min", patience=10, verbose=1),
]
# Treina o modelo
hist = model.fit(
X_train,
Y_train,
validation_data=(X_valid, Y_valid),
batch_size=300,
nb_epoch=100,
verbose=1,
callbacks=callbacks,
)
model.load_weights(weights_filepath)
# # Plot dos gráficos de erro e aurácia
plt.plot(hist.history["accuracy"])
plt.plot(hist.history["val_accuracy"])
plt.ylabel("acurácia")
plt.xlabel("época")
plt.legend(["treino", "validação"], loc="upper left")
plt.show()
plt.plot(hist.history["loss"])
plt.plot(hist.history["val_loss"])
plt.ylabel("loss")
plt.xlabel("época")
plt.legend(["treino", "validação"], loc="upper left")
plt.show()
# # Avaliação e Criação do arquivo de submission.csv
# Avaliação do modelo para o dataset de validação
val_loss, val_acc = model.evaluate(X_valid, Y_valid)
print("A acurácia do modelo está de: " + str(val_acc * 100) + "%")
# Criando arquivo de submission para o Kaggle
# Leitura do Dataset de validação dos resultados
test_df = pd.read_csv("../input/fasam-nlp-competition-turma-4/test.csv")
print(test_df.shape)
test_df.head()
# O dataset de validação, o que será utilizado para calcular o Ranking no Kaggle, contêm apenas as informações de Título e Texto do arquivo. O modelo criado deve ser capaz de classificar em qual das categorias esse artigo pertence.
def predict(text):
"""
Utiliza o modelo treinado para realizar a predição
"""
new_text = tokenizer.texts_to_sequences(text)
new_text = pad_sequences(new_text, maxlen=seq_size)
pred = model.predict_classes(new_text) # [0]
return pred
# Como utilizamos o titulo no treinamento, iremos utilizar o titulo na predição também
pred = predict(test_df.title)
pred_classes = [Y_classes[c] for c in pred]
pred_classes[:5]
# Atualizando a categoria dos artigos no dataset de validação
test_df["category"] = pred_classes
test_df.head()
# Criando o arquivo submission.csv contendo os dados para cálculo do ranking no kaggle
# Esse arquivo deve ser enviado para o kaggle
test_df[["article_id", "category"]].to_csv("submission.csv", index=False)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import numpy as np
import pandas as pd
df1 = pd.read_csv("/kaggle/input/rue-and-fake-news/Fake.csv")
df1.head()
df1.shape
df2 = pd.read_csv("/kaggle/input/rue-and-fake-news/True.csv")
df2.head()
df2.shape
df = pd.concat([df1, df2], axis=0)
df.head()
df.tail()
df.shape
df = df.reset_index(drop=True)
df.tail()
df["title and text"] = df["title"] + " " + df["text"]
df.head()
# # > **Title and Text Lowercase**
df["title and text"] = df["title and text"].str.lower()
print(df)
df.head()
# # > **Individually lowercase title ****
df["title"] = df["title"].str.lower()
print(df)
# # > **Individually lowercase text**
df["text"] = df["text"].str.lower()
print(df)
df.head()
# # > **Clean URL**
import re
df["links"] = df["title and text"].apply(lambda x: re.split("https:\/\/.*", str(x))[0])
df["links"]
# # > **HTML Tags**
df["html"] = df["title and text"].str.replace(r"<[^<>]*>", "", regex=True)
df["html"]
# # > **Remove Punctuation**
df["punctuation"] = df["title and text"].str.replace("[^\w\s]", "")
df["punctuation"]
# # > **Remove Numeric**
df["numeric"] = pd.to_numeric(
df["title and text"].str.findall("(\d+)").str.join(""), errors="coerce"
)
df["numeric"]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
df_train = pd.read_csv(
"/kaggle/input/1056lab-student-performance-prediction/train.csv", index_col=0
)
df_test = pd.read_csv(
"/kaggle/input/1056lab-student-performance-prediction/test.csv", index_col=0
)
df_train.isnull().sum()
df_test.isnull().sum()
# **特徴選択**
import seaborn as sns
from matplotlib import pyplot
sns.set_style("darkgrid")
pyplot.figure(figsize=(20, 20))
sns.heatmap(df_train.corr(), square=True, annot=True)
df_train = df_train[["Medu", "Fedu", "failures", "higher", "studytime", "G3"]]
df_test = df_test[["Medu", "Fedu", "failures", "higher", "studytime"]]
df_train["higher"] = pd.get_dummies(df_train["higher"], drop_first=True)
df_test["higher"] = pd.get_dummies(df_test["higher"], drop_first=True)
X = df_train.drop("G3", axis=1).values
y = df_train["G3"].values
from sklearn.model_selection import train_test_split
X_train, X_valid, y_train, y_valid = train_test_split(
X, y, test_size=0.2, random_state=0
)
len(X_train), len(X_valid), len(y_train), len(y_valid)
from sklearn.metrics import mean_squared_error
from sklearn.ensemble import RandomForestRegressor
model = RandomForestRegressor(random_state=0)
model.fit(X_train, y_train)
predict = model.predict(X_valid)
np.sqrt(mean_squared_error(predict, y_valid))
import optuna
def objective(trial):
# criterion = trial.suggest_categorical('criterion', ['gini', 'entropy'])
max_depth = trial.suggest_int("max_depth", 1, 10)
n_estimators = trial.suggest_int("n_estimators", 10, 300)
model = RandomForestRegressor(
criterion="mse", max_depth=max_depth, n_estimators=n_estimators, random_state=0
)
model.fit(X_train, y_train)
y_pred = model.predict(X_valid)
return mean_squared_error(y_valid, y_pred)
study = optuna.create_study()
study.optimize(objective, n_trials=100)
study.best_params
# criterion = study.best_params['criterion']
max_depth = study.best_params["max_depth"]
n_estimators = study.best_params["n_estimators"]
model = RandomForestRegressor(
criterion="mse", max_depth=max_depth, n_estimators=n_estimators, random_state=0
)
model.fit(X_train, y_train)
predict = model.predict(X_valid)
np.sqrt(mean_squared_error(predict, y_valid))
X = df_train.drop("G3", axis=1).values
y = df_train["G3"].values
model = RandomForestRegressor(
criterion="mse", max_depth=max_depth, n_estimators=n_estimators, random_state=0
)
model.fit(X, y)
X_test = df_test.values
predict = model.predict(X_test)
submit = pd.read_csv(
"/kaggle/input/1056lab-student-performance-prediction/sampleSubmission.csv"
)
submit["G3"] = predict
submit.to_csv("submission.csv", index=False)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
import pandas as pd
impeachment_polls = pd.read_csv(
"../input/trump-impeachment-polls/impeachment-polls.csv"
)
impeachment_topline = pd.read_csv(
"../input/trump-impeachment-polls/impeachment_topline.csv"
)
impeachment_polls.head()
impeachment_polls[
["Rep Yes", "Rep No", "Dem Yes", "Dem No", "Ind Yes", "Ind No"]
].corr()
from scipy import stats
impeachment_polls = impeachment_polls.loc[impeachment_polls["Dem Yes"] > 0]
# x=impeachment_polls["Dem Yes"].loc()
# print(x)
# impeachment_polls = impeachment_polls.dropna()
print(impeachment_polls["Dem Yes"])
pearson_coef, p_value = stats.pearsonr(
impeachment_polls["Dem Yes"], impeachment_polls["Ind Yes"]
)
print(pearson_coef)
print(p_value)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
# Import Matplotlib
import matplotlib.pyplot as plt
# Importing the csv file into a dataframe named trai_data
trai_data = pd.read_csv(
"/kaggle/input/internet-user-growth-in-india-trai/Internet and teledensity_TRAI.csv"
)
trai_data
# Testing subsetting in Pandas with using columnn name as a method on DataFrame
trai_data.teledensity_urban
# Attempting to subset 'teledensity' from the Dataframe into a new Pandas dataframe named teledensity_data
teledensity_data = trai_data[["teledensity_rural", "teledensity_urban"]]
teledensity_data
# Similarly, subsetting other pertinent columns into separate dataframes
# I apologize for the inconsistent column naming scheme
broadband_data = trai_data[
["Wired_Broadband_total", "wireless_broadband_total", "total_broadband"]
]
broadband_data
# Attempting to plot teledensity data
plt.plot(teledensity_data)
# Attempting to plot broadband data
plt.plot(broadband_data)
# Let's try plotting data straight from the main dataframe, subsetting whatever needed
plt.plot(
trai_data[
["Wired_Broadband_total", "wireless_broadband_total", "total_subscribers"]
]
)
|
# ## 1. 데이터 살펴보기
# pandas의 read_csv 함수를 사용해 데이터를 읽어오고, 각 변수들이 나타내는 의미를 살펴보겠습니다.
# ID : 집을 구분하는 번호
# date : 집을 구매한 날짜
# price : 타겟 변수인 집의 가격
# bedrooms : 침실의 수
# bathrooms : 침실당 화장실 개수
# sqft_living : 주거 공간의 평방 피트
# sqft_lot : 부지의 평방 피트
# floors : 집의 층 수
# waterfront : 집의 전방에 강이 흐르는지 유무 (a.k.a. 리버뷰)
# view : 집이 얼마나 좋아 보이는지의 정도
# condition : 집의 전반적인 상태
# grade : King County grading 시스템 기준으로 매긴 집의 등급
# sqft_above : 지하실을 제외한 평방 피트
# sqft_basement : 지하실의 평방 피트
# yr_built : 집을 지은 년도
# yr_renovated : 집을 재건축한 년도
# zipcode : 우편번호
# lat : 위도
# long : 경도
# sqft_living15 : 2015년 기준 주거 공간의 평방 피트(집을 재건축했다면, 변화가 있을 수 있음)
# sqft_lot15 : 2015년 기준 부지의 평방 피트(집을 재건축했다면, 변화가 있을 수 있음)
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import os
from collections import Counter
sns.set_style("whitegrid")
sns.set(font_scale=1.2)
import missingno as msno
import warnings
warnings.filterwarnings("ignore")
os.listdir("../input")
df_train = pd.read_csv("../input/boazminipro/train.csv")
df_test = pd.read_csv("../input/boazminipro/test.csv")
# # DATA CHECK
df_train.head()
df_train.shape
numerical_feats = df_train.dtypes[df_train.dtypes != "object"].index
print("Numerical Features: ", len(numerical_feats))
categorical_feats = df_train.dtypes[df_train.dtypes == "object"].index
print("Categorical Features: ", len(categorical_feats))
# 범주형 변수가 없다 (숫자로 이루어진 범주형 변수가 있다. ex.등급)
# 하나있는 범주형 변수는 ["date"]이다. datetime으로 설정한다.
for col in df_train.columns:
Null = "Feature: {:<10}\t Count of Null: {}".format(
col, df_train[col].isnull().sum()
)
print(Null)
msno.matrix(df=df_train.iloc[:, :], color=(0.1, 0.4, 0.5), figsize=(15, 6))
# missingno의 matrix로 시각화한 모습, 역시 null값은 없다.
# ## Skew / Kurt 탐색 및 Log 변환
numerical_feats = df_train.dtypes[df_train.dtypes != "object"].index
print("Numerical Features: ", len(numerical_feats))
for col in numerical_feats.difference(["date"]):
SkewKurt = "{:<10}\t Skewness: {:.4f}\t Kurtosis: {:.4f}".format(
col, df_train[col].skew(), df_train[col].kurt()
)
print(SkewKurt)
# 각 feature들의 왜도와 첨도를 살펴본다.
# target feature인 price 역시 조정이 필요.
# 처음에 수치형, 범주형 구분할 때 말했던 범주가 수치(등급)로 지정되어있는 변수들을 주의한다.
df_train["price"] = df_train["price"].map(lambda i: np.log(i) if i > 0 else 0)
# # EDA
corr_data = df_train[numerical_feats]
colormap = plt.cm.PuBu
sns.set(font_scale=1.3)
f, ax = plt.subplots(figsize=(14, 12))
plt.title("Correlation of Numeric Features with Price", size=18)
sns.heatmap(
corr_data.corr(),
square=True,
linewidths=0.1,
cmap=colormap,
linecolor="white",
vmax=0.8,
)
# 전체적인 correlation heatmap
k = 12
cols = corr_data.corr().nlargest(k, "price")["price"].index
print(cols)
cm = np.corrcoef(df_train[cols].values.T)
f, ax = plt.subplots(figsize=(12, 10))
sns.heatmap(
cm,
vmax=0.8,
linewidths=0.1,
square=True,
annot=True,
cmap=colormap,
linecolor="white",
xticklabels=cols.values,
annot_kws={"size": 14},
yticklabels=cols.values,
)
# 전체적인 correlation heatmap에서 상관계수가 높은 순으로 12개를 뽑아서 다시 만든 heatmap
df_train.plot(
kind="scatter",
x="long",
y="lat",
alpha=0.3,
figsize=(10, 7),
c=df_train["price"],
cmap=plt.get_cmap("jet"),
colorbar=True,
)
# ## 수치형 변수 시각화
fig, ((ax1, ax2), (ax3, ax4), (ax5, ax6)) = plt.subplots(
nrows=3, ncols=2, figsize=(18, 15)
)
sqft_living_scatter_plot = pd.concat(
[df_train["price"], df_train["sqft_living"]], axis=1
)
sns.regplot(
x="sqft_living",
y="price",
data=sqft_living_scatter_plot,
scatter=True,
fit_reg=True,
ax=ax1,
)
sqft_lot_scatter_plot = pd.concat([df_train["price"], df_train["sqft_lot"]], axis=1)
sns.regplot(
x="sqft_lot",
y="price",
data=sqft_lot_scatter_plot,
scatter=True,
fit_reg=True,
ax=ax2,
)
sqft_above_scatter_plot = pd.concat([df_train["price"], df_train["sqft_above"]], axis=1)
sns.regplot(
x="sqft_above",
y="price",
data=sqft_above_scatter_plot,
scatter=True,
fit_reg=True,
ax=ax3,
)
sqft_basement_scatter_plot = pd.concat(
[df_train["price"], df_train["sqft_basement"]], axis=1
)
sns.regplot(
x="sqft_basement",
y="price",
data=sqft_basement_scatter_plot,
scatter=True,
fit_reg=True,
ax=ax4,
)
sqft_living15_scatter_plot = pd.concat(
[df_train["price"], df_train["sqft_living15"]], axis=1
)
sns.regplot(
x="sqft_living15",
y="price",
data=sqft_living15_scatter_plot,
scatter=True,
fit_reg=True,
ax=ax5,
)
sqft_lot15_scatter_plot = pd.concat([df_train["price"], df_train["sqft_lot15"]], axis=1)
sns.regplot(
x="sqft_lot15",
y="price",
data=sqft_lot15_scatter_plot,
scatter=True,
fit_reg=True,
ax=ax6,
)
# 수치형 변수들에 따른 price를 scatter plot으로 그려본다.
# sqft_living, sqft_above, sqft_living15는 어느정도 이상치가 있어보인다.
# 나머지들은 분산이 매우 커보인다.
# basement는 0의 값이 굉장히 많다. 또한 이상치도 많아보인다.
# ## 범주형 변수 (numerical category feature)
df_train[["grade", "price"]].groupby(["grade"], as_index=True).describe()
# 등급에 따른 주택가격
fig, ax = plt.subplots(figsize=(12, 8))
sns.boxplot(
x=df_train["grade"], y=df_train["price"], data=df_train, ax=ax, palette="Blues_d"
)
# grade에 따른 price의 boxplot을 그려본 결과
# 1) 2등급은 존재하지 않음
# 2) 3등급의 평균값이 4등급보다 높게 나타남
# 3) 7, 8, 11등급에 상당한 이상치가 존재하는 것으로 보임
# 4) 7~10 등급의 이상치가 꽤 많은 것으로 보임
# 데이터 설명에 따르면 grade의 경우 "1-3은 건물 건축 및 디자인에 미치지 못하고 7은 평균 수준의 건축 및 디자인을, 11-13은 높은 수준의 건축 및 디자인을 지니고 있습니다."
df_train[["bedrooms", "price"]].groupby(["bedrooms"], as_index=True).describe()
# 방의 수에 따른 주택 가격
fig, ax = plt.subplots(figsize=(16, 10))
sns.boxplot(
x=df_train["bedrooms"], y=df_train["price"], data=df_train, ax=ax, palette="Blues_d"
)
# boxplot을 살펴보면 방의 수에 따른 가격이 직관적으로 대략 선형임을 알 수 있음.
# 윗 셀의 describe를 봐도 가격의 평균값이 방에 따라 증가하는 것을 볼 수 있음.
# boxplot을 벗어난 이상치들은 지역특성(땅값이 비싸거나 대도시이거나?)에 영향을 받는다고 생각할 수 있다.
df_train[["bathrooms", "price"]].groupby(["bathrooms"], as_index=True).describe()
# 데이터 설명에 따르면
# - 0.5 : 세면대, 화장실
# - 0.75 : 세면대, 화장실, 샤워실
# - 1 : 세면대, 화장실, 샤워실, 욕조
# 의 값을 갖는다고 한다.
fig, ax = plt.subplots(figsize=(16, 10))
sns.boxplot(
x=df_train["bathrooms"],
y=df_train["price"],
data=df_train,
ax=ax,
palette="Blues_d",
)
df_train[["floors", "price"]].groupby(["floors"], as_index=True).describe()
# 층수의 경우 1.5, 2.5, 3.5와 같이 소숫점을 가진다.
# 미국에서 흔히 볼 수 있는 형태로 다락방을 끼고 있는 형태
# floors, price는 선형관계로 보임
df_train[["waterfront", "price"]].groupby(["waterfront"], as_index=True).describe()
# 바이너리 (리버뷰? 등이 있고 없고)
# waterfront, price는 선형관계로 보임
df_train[["view", "price"]].groupby(["view"], as_index=True).describe()
# view, price는 선형관계로 보임
df_train[["condition", "price"]].groupby(["condition"], as_index=True).describe()
# condition, price는 선형관계로 보임
for data in [df_train, df_test]:
data["date"] = pd.to_datetime(data["date"])
data["date_year"] = data["date"].dt.year
data["date_month"] = data["date"].dt.month
# ### month에 따라 가격이 패턴이 있는것으로 보여짐
# ### 4월에 price 최대, 그 이후 하향세
train_pivot = df_train.pivot_table(
index=["date_month"], columns=["date_year"], values="price"
) # price mean
plt.figure(figsize=(10, 8))
sns.heatmap(train_pivot, annot=True, cmap=colormap)
plt.figure(figsize=(10, 5))
sns.lineplot(x=df_train["date_month"], y=df_train["price"])
# # FEATURE ENGINEERING
# ## 이상치 처리
df_train.loc[df_train.grade == 3]
# EDA 과정에서 살펴본 grade = 3인 이상치 제거
print(df_train[df_train["grade"] == 3].sqft_lot.mean())
print(df_train[df_train["grade"] == 4].sqft_lot.mean())
df_train.drop([2302, 4123], axis=0, inplace=True)
df_train.loc[(df_train.grade == 11) & (df_train.price > 15.5)]
# grade 11 확인, sqft_living이 크므로 제거하지 않음
df_train[(df_train.grade) == 11].sqft_living.max()
df_train.loc[(df_train.bathrooms == 4.5) & (df_train.price > 15)]
# bathroom 이상치, 제거 X
df_train.loc[(df_train.bathrooms == 5.25) & (df_train.price < 13)]
# bathroom 이상치, 제거 X
df_train[df_train.sqft_living > 13000]
# sqft_living 이상치 제거
df_train.drop(8912, axis=0, inplace=True)
df_train.loc[(df_train.sqft_lot > 1500000) & (df_train.price > 13)]
# sqft_lot 이상치 제거
df_train.drop(1231, axis=0, inplace=True)
# # FE
df_train["sqft_above"] = df_train["sqft_above"].map(lambda i: np.log(i) if i > 0 else 0)
df_train["sqft_basement"] = df_train["sqft_basement"].map(
lambda i: np.log(i) if i > 0 else 0
)
df_train["sqft_living"] = df_train["sqft_living"].map(
lambda i: np.log(i) if i > 0 else 0
)
df_train["sqft_living15"] = df_train["sqft_living15"].map(
lambda i: np.log(i) if i > 0 else 0
)
df_train["sqft_lot"] = df_train["sqft_lot"].map(lambda i: np.log(i) if i > 0 else 0)
df_train["sqft_lot15"] = df_train["sqft_lot15"].map(lambda i: np.log(i) if i > 0 else 0)
df_test["sqft_above"] = df_test["sqft_above"].map(lambda i: np.log(i) if i > 0 else 0)
df_test["sqft_basement"] = df_test["sqft_basement"].map(
lambda i: np.log(i) if i > 0 else 0
)
df_test["sqft_living"] = df_test["sqft_living"].map(lambda i: np.log(i) if i > 0 else 0)
df_test["sqft_living15"] = df_test["sqft_living15"].map(
lambda i: np.log(i) if i > 0 else 0
)
df_test["sqft_lot"] = df_test["sqft_lot"].map(lambda i: np.log(i) if i > 0 else 0)
df_test["sqft_lot15"] = df_test["sqft_lot15"].map(lambda i: np.log(i) if i > 0 else 0)
logdata = df_train[
[
"price",
"sqft_above",
"sqft_basement",
"sqft_living",
"sqft_living15",
"sqft_lot",
"sqft_lot15",
]
]
for i in range(7):
print(
"{:<10}\t Skewness: {:.3f}\t Kurtosis: {:.3f}".format(
logdata.columns[i],
df_train[logdata.columns[i]].skew(),
df_train[logdata.columns[i]].kurt(),
)
)
# log를 취해주어 분포를 조정해준다.
# 수치적으로 정규분포에 가까워진 것을 확인할 수 있다.
df_train["total_sqft"] = df_train["sqft_above"] + df_train["sqft_basement"]
df_test["total_sqft"] = df_test["sqft_above"] + df_test["sqft_basement"]
df_train[["total_sqft", "sqft_living"]].head()
# 건물의 총 면적을 만들어서 거주공간 면적과 비교해보면 값이 같다.
# 이는 sqft_living 변수가 건물의 연면적을 의미함을 알 수 있다.
df_train.drop(["total_sqft"], inplace=True, axis=1)
df_test.drop(["total_sqft"], inplace=True, axis=1)
df_train["Vol_ratio"] = (df_train["sqft_living"] / df_train["sqft_lot"]) * 100
df_test["Vol_ratio"] = (df_test["sqft_living"] / df_test["sqft_lot"]) * 100
# 용적률 = 건물연면적 / 토지면적 * 100
# 건폐율을 사용했을 때 성능이 좋지않아 제거
for data in [df_train, df_test]:
data["above_per_living"] = data["sqft_above"] / data["sqft_living"]
zipcode_data = df_train.groupby("zipcode").aggregate(np.mean)
zipcode_ranks = {}
rank = 1
for idx, row in zipcode_data.sort_values(by="price").iterrows():
zipcode_ranks[idx] = rank
rank += 1
# zipcode별로 price의 평균을 내어 rank를 매겨준다 (집값이 낮으면 1 올라갈수록 +)
for data in [df_train, df_test]:
zipcode_feature = []
for idx, row in data.iterrows():
zipcode_feature.append(zipcode_ranks[row.zipcode])
data["zipcode_ranks"] = zipcode_feature
zipcode_data = df_train.groupby("zipcode").aggregate(np.var)
zipcode_ranks_var = {}
rank = 1
for idx, row in zipcode_data.sort_values(by="price", ascending=False).iterrows():
zipcode_ranks_var[idx] = rank
rank += 1
for data in [df_train, df_test]:
zipcode_feature = []
for idx, row in data.iterrows():
zipcode_feature.append(zipcode_ranks_var[row.zipcode])
data["zipcode_ranks_var"] = zipcode_feature
month = df_train.groupby("date_month").aggregate(np.mean)
month_ranks = {}
rank = 1
for idx, row in month.sort_values(by="price").iterrows():
month_ranks[idx] = rank
rank += 1
for data in [df_train, df_test]:
month_feature = []
for idx, row in data.iterrows():
month_feature.append(month_ranks[row.date_month])
data["month_rank"] = month_feature
from haversine import haversine
bridge_wh = (47.641076, -122.259196)
for data in [df_train, df_test]:
house_wh = data.loc[:, ["lat", "long"]]
house_wh = list(house_wh.itertuples(index=False, name=None))
dist = []
for house in house_wh:
dist.append(np.log(1 / haversine(house, bridge_wh)))
data["dist_bridge"] = dist
from sklearn.decomposition import PCA
from sklearn.cluster import KMeans
for data in [df_train, df_test]:
coord = data[["lat", "long"]]
pca_coord = PCA(n_components=2).fit(coord).transform(coord)
data["pca1"] = pca_coord[:, 0]
data["pca2"] = pca_coord[:, 1]
# 위경도를 기준으로 사용하여 pca를 진행하여 새로운 변수를 만들어준다.
for data in [df_train, df_test]:
data["term"] = -(data.date_year - data.yr_built)
df_train.columns
df_train.shape
df_train.drop(["date", "date_month"], inplace=True, axis=1)
df_test.drop(["date", "date_month"], inplace=True, axis=1)
df_train.head()
# # MODELING & SUBMISSION
import eli5
from eli5.sklearn import PermutationImportance
from sklearn.model_selection import (
KFold,
cross_val_score,
train_test_split,
GridSearchCV,
)
from sklearn.metrics import mean_squared_error
from sklearn.tree import DecisionTreeRegressor
from sklearn.ensemble import RandomForestRegressor, GradientBoostingRegressor
import xgboost as xgb
import lightgbm as lgb
y_train = df_train.price
x_train = df_train.drop(["id", "price"], axis=1)
x_test = df_test.drop(["id"], axis=1)
X_tr, X_vld, y_tr, y_vld = train_test_split(
x_train, y_train, test_size=0.3, random_state=2019
)
model = xgb.XGBRegressor()
model.fit(X_tr, y_tr)
y_val = model.predict(X_vld)
perm = PermutationImportance(model, random_state=42).fit(X_vld, y_vld)
eli5.show_weights(perm, top=33, feature_names=X_vld.columns.tolist())
dt = DecisionTreeRegressor(random_state=10)
dt_cv_score = cross_val_score(dt, x_train, y_train, cv=5)
print(
"score with cv = {} \n mean cv score = {:.5f} \n std = {:.5f}".format(
dt_cv_score, dt_cv_score.mean(), dt_cv_score.std()
)
)
# ## XGB
xgb_params = {
"eta": 0.01,
"max_depth": 6,
"subsample": 0.8,
"colsample_bytree": 0.8,
"objective": "reg:linear",
"eval_metric": "rmse",
"silent": 1,
}
y_train = df_train.price
dtrain = xgb.DMatrix(x_train, y_train)
dtest = xgb.DMatrix(x_test)
cv_output = xgb.cv(
xgb_params,
dtrain,
num_boost_round=5000,
early_stopping_rounds=50,
verbose_eval=500,
show_stdv=False,
)
rounds = len(cv_output)
xgb1 = xgb.train(xgb_params, dtrain, num_boost_round=rounds)
preds = xgb1.predict(dtest)
xgb1_sub = df_test[["id"]]
xgb1_sub["price"] = preds
y_train = np.expm1(df_train.price)
dtrain = xgb.DMatrix(x_train, y_train)
dtest = xgb.DMatrix(x_test)
cv_output = xgb.cv(
xgb_params,
dtrain,
num_boost_round=20000,
early_stopping_rounds=50,
verbose_eval=500,
show_stdv=False,
)
rounds = len(cv_output)
xgb2 = xgb.train(xgb_params, dtrain, num_boost_round=rounds)
preds = xgb2.predict(dtest)
xgb2_sub = df_test[["id"]]
xgb2_sub["price"] = preds
xgb1_pred = xgb1.predict(dtrain)
xgb2_pred = xgb2.predict(dtrain)
mse = {}
ii = np.arange(0, 1, 0.01)
for i, ii in enumerate(ii):
xgb_train_pred = ii * np.expm1(xgb1_pred) + (1 - ii) * xgb2_pred
mse[i] = np.sqrt(mean_squared_error(y_train, xgb_train_pred))
xgb_min = min(mse.values())
for i in range(100):
if mse[i] == xgb_min:
print(i)
xgb_train_pred = 0 * np.expm1(xgb1_pred) + 1 * xgb2_pred
xgb_sub = pd.merge(xgb1_sub, xgb2_sub, how="left", on="id")
xgb_sub.columns = ["id", "price1", "price2"]
xgb_sub["price"] = 0 * np.expm1(xgb_sub["price1"]) + 1 * xgb_sub["price2"]
xgb_sub = xgb_sub[["id", "price"]]
xgb_sub.to_csv("xgb_sub.csv", index=False)
# ## LGBM
param = {
"num_leaves": 31,
"min_data_in_leaf": 30,
"objective": "regression",
"max_depth": -1,
"learning_rate": 0.015,
"min_child_samples": 20,
"boosting": "gbdt",
"feature_fraction": 0.9,
"bagging_freq": 1,
"bagging_fraction": 0.9,
"bagging_seed": 11,
"metric": "rmse",
"lambda_l1": 0.1,
"verbosity": -1,
"nthread": 4,
"random_state": 4950,
}
y_train = df_train.price
folds = KFold(n_splits=5, shuffle=True, random_state=1)
predictions = np.zeros(len(x_test))
for fold_, (trn_idx, val_idx) in enumerate(folds.split(df_train)):
trn_data = lgb.Dataset(x_train.iloc[trn_idx], label=y_train.iloc[trn_idx])
val_data = lgb.Dataset(x_train.iloc[val_idx], label=y_train.iloc[val_idx])
num_round = 10000
lgb1 = lgb.train(
param,
trn_data,
num_round,
valid_sets=[trn_data, val_data],
verbose_eval=500,
early_stopping_rounds=100,
)
predictions += (
lgb1.predict(x_test, num_iteration=lgb1.best_iteration) / folds.n_splits
)
lgb1_sub = pd.DataFrame()
lgb1_sub["id"] = df_test.id
lgb1_sub["price"] = predictions
y_train = np.expm1(df_train.price)
folds = KFold(n_splits=5, shuffle=True, random_state=1)
predictions = np.zeros(len(x_test))
for fold_, (trn_idx, val_idx) in enumerate(folds.split(df_train)):
trn_data = lgb.Dataset(x_train.iloc[trn_idx], label=y_train.iloc[trn_idx])
val_data = lgb.Dataset(x_train.iloc[val_idx], label=y_train.iloc[val_idx])
num_round = 10000
lgb2 = lgb.train(
param,
trn_data,
num_round,
valid_sets=[trn_data, val_data],
verbose_eval=500,
early_stopping_rounds=100,
)
predictions += (
lgb2.predict(x_test, num_iteration=lgb2.best_iteration) / folds.n_splits
)
lgb2_sub = pd.DataFrame()
lgb2_sub["id"] = df_test.id
lgb2_sub["price"] = predictions
lgb1_pred = lgb1.predict(x_train)
lgb2_pred = lgb2.predict(x_train)
mse = {}
ii = np.arange(0, 1, 0.01)
for i, ii in enumerate(ii):
train_pred = ii * np.expm1(lgb1_pred) + (1 - ii) * lgb2_pred
mse[i] = np.sqrt(mean_squared_error(y_train, train_pred))
lgb_min = min(mse.values())
for i in range(100):
if mse[i] == lgb_min:
print(i)
lgb_train_pred = 0.59 * np.expm1(lgb1_pred) + 0.41 * lgb2_pred
lgb_sub = pd.merge(lgb1_sub, lgb2_sub, how="left", on="id")
lgb_sub.columns = ["id", "price1", "price2"]
lgb_sub["price"] = 0.59 * np.expm1(lgb_sub["price1"]) + 0.41 * lgb_sub["price2"]
lgb_sub = lgb_sub[["id", "price"]]
# ## RF
forest_regr = RandomForestRegressor(
bootstrap=True,
criterion="mse",
max_depth=None,
max_features=28,
max_leaf_nodes=None,
min_impurity_decrease=0.0,
min_impurity_split=None,
min_samples_leaf=1,
min_samples_split=2,
min_weight_fraction_leaf=0.0,
n_estimators=90,
n_jobs=1,
oob_score=False,
random_state=None,
verbose=0,
warm_start=False,
)
rf = forest_regr.fit(x_train, y_train)
predictions = rf.predict(x_test)
rf_sub = pd.DataFrame()
rf_sub["id"] = df_test.id
rf_sub["price"] = predictions
# # GB
gdb = GradientBoostingRegressor(
n_estimators=3000,
learning_rate=0.05,
max_depth=4,
max_features="sqrt",
min_samples_leaf=15,
min_samples_split=10,
loss="huber",
random_state=5,
)
gdb.fit(x_train, y_train)
predictions = gdb.predict(x_test)
gdb_sub = pd.DataFrame()
gdb_sub["id"] = df_test.id
gdb_sub["price"] = predictions
# ## ENSEMBLE
gdb_train_pred = gdb.predict(x_train)
rf_train_pred = rf.predict(x_train)
mse = {}
ii = np.arange(0, 0.1, 0.01)
for i, ii in enumerate(ii):
submse = {}
train_pred = (
0.8 * xgb_train_pred
+ (1 - ii) * lgb_train_pred
+ 0.1 * gdb_train_pred
+ ii * rf_train_pred
)
mse[i] = np.sqrt(mean_squared_error(y_train, train_pred))
train_min = min(mse.values())
for i in range(10):
if mse[i] == train_min:
print(i)
ensemble_sub = pd.DataFrame()
ensemble_sub["id"] = df_test.id
ensemble_sub["price"] = (
xgb_sub["price"] * 0.8
+ lgb_sub["price"] * 0.01
+ gdb_sub["price"] * 0.1
+ rf_sub["price"] * 0.09
)
ensemble_sub.to_csv("sub.csv", index=False)
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
df = pd.read_csv("/kaggle/input/dataset-new/Stress-strain-2 - Sheet1 (1).csv")
df
df.columns
df.describe()
new_stress = df["True Stress"][6:]
new_strain = df["True Strain"][6:]
df["True Stress"] = new_stress
df["True Strain"] = new_strain
df = df.dropna()
# df=df.set_index(np.arange(0,872))
df
df = df.set_index(np.arange(0, 2117))
df
df.plot("True Strain", "True Stress")
plt.xlabel("Strain")
plt.ylabel("Stress")
slopes = []
for i in range(2116):
slope = (df["True Stress"][i + 1] - df["True Stress"][i]) / (
df["True Strain"][i + 1] - df["True Strain"][i]
)
slopes.append(slope)
slopes = np.array(slopes)
slopes[:10]
df["True Strain"][2116]
m = np.mean(slopes[:10])
x_int = (0.2 * df["True Strain"][2116]) / 100 + df["True Strain"][0]
y_line = m * (new_strain - x_int)
y_line
plt.figure(figsize=(10, 10))
plt.plot(df["True Strain"], df["True Stress"])
plt.plot(new_strain, y_line, "r", label="offset line")
plt.legend()
plt.xlabel("Strain")
plt.ylabel("Stress")
df["line"] = np.array(y_line)
df["difference"] = np.abs(df["True Stress"] - df["line"])
df
np.argmin(df["difference"])
yield_stress = df["True Stress"][265]
yield_stress
consider_stress = df["True Stress"][266:]
consider_strain = df["True Strain"][266:]
A, B = [], []
n = np.linspace(0, 3, 50)
for i in range(150):
A.append(np.random.rand() * 635)
B.append(np.random.rand() * 3500)
A = np.array(A)
B = np.array(B)
plt.scatter(np.arange(0, 150), A)
plt.title("Scatter graph of all A values")
plt.scatter(np.arange(0, 150), B)
plt.title("Scatter graph of all B values")
plt.scatter(np.arange(0, 50), n)
plt.title("Scatter graph of all n values")
from sklearn.metrics import mean_absolute_error as mae
from sklearn.metrics import mean_squared_error as mse
new_A, new_B, new_n, new_mae, new_rmse = [], [], [], [], []
exp_strain = np.array(consider_strain)
exp_stress = np.array(consider_stress)
for a in A:
for b in B:
for N in n:
new_A.append(a)
new_B.append(b)
new_n.append(N)
calc_stress = a + b * (np.power(exp_strain, N))
new_mae.append(mae(exp_stress, calc_stress))
new_rmse.append(np.sqrt(mse(exp_stress, calc_stress)))
new_A = np.array(new_A)
new_B = np.array(new_B)
new_n = np.array(new_n)
new_mae = np.array(new_mae)
new_rmse = np.array(new_rmse)
final_df = pd.DataFrame(
{"A": new_A, "B": new_B, "n": new_n, "MAE": new_mae, "RMSE": new_rmse}
)
final_df
final_df.describe()
final_df.to_csv("final_explo_new.csv")
df = pd.read_csv("/kaggle/working/final_explo_new.csv")
df.head()
df.columns
df.drop("Unnamed: 0", axis=1, inplace=True)
df.head(10)
df.iloc[np.argmin(df["RMSE"])]
df.iloc[np.argmin(df["MAE"])]
plt.plot(new_strain[266:], new_stress[266:], label="Experimental Graph")
plt.xlabel("Strain")
plt.ylabel("Stress")
best_graph = 285.286234 + 385.248193 * np.power(new_strain, 0.061224)
plt.plot(new_strain[149:], best_graph[149:], label="Graph with least RMSE")
plt.legend()
df.shape
from sklearn.model_selection import train_test_split
X_train, X_test, Y_train, Y_test = train_test_split(
df[["A", "B", "n"]], df["MAE"], test_size=0.15, random_state=0
)
train_df = pd.concat([X_train, Y_train], axis=1)
test_df = pd.concat([X_test, Y_test], axis=1)
train_df.shape, test_df.shape
X_train.shape, X_test.shape, Y_train.shape, Y_test.shape
train_df.corr()["MAE"].sort_values()
train_df.head()
sns.histplot(train_df["MAE"], kde=True)
train_df.hist(bins=50, figsize=(20, 15))
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
X_train_scaled = scaler.fit_transform(X_train)
X_test_scaled = scaler.transform(X_test)
from sklearn.linear_model import LinearRegression
from sklearn.neighbors import KNeighborsRegressor
from sklearn.linear_model import Ridge
from sklearn.linear_model import Lasso
from sklearn.tree import DecisionTreeRegressor
from sklearn.ensemble import RandomForestRegressor
from sklearn.metrics import mean_squared_error, mean_absolute_error
from sklearn.metrics import r2_score
# **Linear Regression**
lr_model = LinearRegression()
lr_model.fit(X_train_scaled, Y_train)
y_pred_lr = lr_model.predict(X_test_scaled)
mse_lr = mean_squared_error(Y_test, y_pred_lr)
mae_lr = mean_absolute_error(Y_test, y_pred_lr)
print("Mean squared error from linear regression: ", mse_lr)
print("Mean absolute error from linear regression: ", mae_lr)
Y_test.shape
r2_lr = r2_score(Y_test, y_pred_lr)
print("r2 score for linear regression model is", r2_lr)
fig, ax = plt.subplots(2, 2, figsize=(16, 16))
diff_lr = np.array(Y_test - y_pred_lr)
ax[0, 0].plot(diff_lr[::10000])
ax[0, 0].set_title("One in 10,000 differences, Linear Regression")
ax[0, 1].plot(diff_lr[::1000])
ax[0, 1].set_title("One in 1,000 differences, Linear Regression")
ax[1, 0].plot(diff_lr[::100])
ax[1, 0].set_title("One in 100 differences, Linear Regression")
ax[1, 1].plot(diff_lr[::10])
ax[1, 1].set_title("One in 10 differences, Linear Regression")
plt.figure(figsize=(25, 8))
sns.histplot(Y_test - y_pred_lr, kde=True)
# plt.xticks(np.arange(-350,1000,50))
plt.figure(figsize=(20, 15))
plt.scatter(np.arange(1, 1689), Y_test[::100], label="Actual Error", color="blue")
plt.scatter(
np.arange(1, 1689), y_pred_lr[::100], label="Predicted Error", color="orange"
)
plt.ylabel("Error")
plt.xlabel("Index")
plt.title("Actual v/s Predicted error by Linear Regression (1 in 100 error)")
plt.legend()
np.array(Y_test[:10]).reshape(10, 1)
y_pred_lr[:10].reshape(10, 1)
# **Decision Tree**
tree = DecisionTreeRegressor()
tree.fit(X_train_scaled, Y_train)
y_pred_tree = tree.predict(X_test_scaled)
mse_dt = mean_squared_error(Y_test, y_pred_tree)
mae_dt = mean_absolute_error(Y_test, y_pred_tree)
print("Mean squared error using decision tree: ", mse_dt)
print("Mean absolute error using decision tree: ", mae_dt)
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(20, 5))
ax1.scatter(np.arange(1, 1689), Y_test[::100], label="Actual Error", color="blue")
ax1.legend()
ax2.scatter(
np.arange(1, 1689), y_pred_tree[::100], label="Predicted Error", color="orange"
)
ax2.legend()
plt.figure(figsize=(7, 7))
plt.scatter(np.arange(1, 1689), Y_test[::100], label="Actual Error", color="blue")
plt.scatter(
np.arange(1, 1689), y_pred_tree[::100], label="Predicted Error", color="orange"
)
plt.legend()
diff_tree = np.array(Y_test - y_pred_tree)
fig, ax = plt.subplots(2, 2, figsize=(16, 16))
ax[0, 0].plot(diff_tree[::10000])
ax[0, 0].set_title("One in 10,000 differences, Decision Trees")
ax[0, 1].plot(diff_tree[::1000])
ax[0, 1].set_title("One in 1,000 differences, Decision Trees")
ax[1, 0].plot(diff_tree[::100])
ax[1, 0].set_title("One in 100 differences, Decision Trees")
ax[1, 1].plot(diff_tree[::10])
ax[1, 1].set_title("One in 10 differences, Decision Trees")
sns.histplot(np.array(Y_test - y_pred_tree))
r2_dt = r2_score(Y_test, y_pred_tree)
print("r2 score for decision tree model is", r2_dt)
np.array(Y_test[:10]).reshape(10, 1)
y_pred_tree[:10].reshape(10, 1)
# **Random Forest**
from sklearn.ensemble import RandomForestRegressor
model_RF = RandomForestRegressor()
model_RF.fit(X_train_scaled, Y_train)
y_pred_RF = model_RF.predict(X_test_scaled)
mse_RF = mean_squared_error(Y_test, y_pred_RF)
mae_RF = mean_absolute_error(Y_test, y_pred_RF)
print("Mean squared error using Random Forest: ", mse_RF)
print("Mean absolute error Using Random Forest: ", mae_RF)
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(20, 5))
ax1.scatter(np.arange(1, 1689), Y_test[::100], label="Actual Error", color="blue")
ax1.legend()
ax2.scatter(
np.arange(1, 1689), y_pred_RF[::100], label="Predicted Error", color="orange"
)
ax2.legend()
fig, ax = plt.subplots(2, 2, figsize=(16, 16))
diff_RF = np.array(Y_test - y_pred_RF)
ax[0, 0].plot(diff_RF[::10000])
ax[0, 0].set_title("One in 10,000 differences, Random Forest")
ax[0, 1].plot(diff_RF[::1000])
ax[0, 1].set_title("One in 1,000 differences, Random Forest")
ax[1, 0].plot(diff_RF[::100])
ax[1, 0].set_title("One in 100 differences, Random Forest")
ax[1, 1].plot(diff_RF[::10])
ax[1, 1].set_title("One in 10 differences, Random Forest")
# plt.figure(figsize=(25,8))
# sns.histplot(Y_test - y_pred_RF,kde=True)
# plt.xticks(np.linspace(-350,1000,50))
r2_RF = r2_score(Y_test, y_pred_RF)
print("r2 score for Random Forest model is", r2_RF)
np.array(Y_test[:10]).reshape(10, 1)
y_pred_RF[:10].reshape(10, 1)
# **K Nearest Neighbour**
knn = KNeighborsRegressor()
knn.fit(X_train_scaled, Y_train)
y_pred_knn = knn.predict(X_test_scaled)
mse_knn = mean_squared_error(Y_test, y_pred_knn)
mae_knn = mean_absolute_error(Y_test, y_pred_knn)
print("Mean squared error using K nearest neighbours: ", mse_knn)
print("Mean absolute error using K nearest neighbours: ", mae_knn)
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(20, 5))
ax1.scatter(np.arange(1, 1689), Y_test[::100], label="Actual Error", color="blue")
ax1.legend()
ax2.scatter(
np.arange(1, 1689), y_pred_knn[::100], label="Predicted Error", color="orange"
)
ax2.legend()
fig, ax = plt.subplots(2, 2, figsize=(16, 16))
diff_knn = np.array(Y_test - y_pred_knn)
ax[0, 0].plot(diff_knn[::10000])
ax[0, 0].set_title("One in 10,000 differences, K-Nearest Neighbour")
ax[0, 1].plot(diff_knn[::1000])
ax[0, 1].set_title("One in 1,000 differences, K-Nearest Neighbour")
ax[1, 0].plot(diff_knn[::100])
ax[1, 0].set_title("One in 100 differences, K-Nearest Neighbour")
ax[1, 1].plot(diff_knn[::10])
ax[1, 1].set_title("One in 10 differences, K-Nearest Neighbour")
r2_knn = r2_score(Y_test, y_pred_knn)
print("r2 score for K nearest neighbours is", r2_knn)
np.array(Y_test[:10]).reshape(10, 1)
y_pred_knn[:10].reshape(10, 1)
# **Ridge Regression**
from sklearn.linear_model import Ridge
ridge = Ridge()
ridge.fit(X_train_scaled, Y_train)
y_pred_rid = ridge.predict(X_test_scaled)
mse_rid = mean_squared_error(Y_test, y_pred_rid)
mae_rid = mean_absolute_error(Y_test, y_pred_rid)
print("Mean squared error using Ridge Regression: ", mse_rid)
print("Mean absolute error using Ridge Regression: ", mae_rid)
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(20, 5))
ax1.scatter(np.arange(1, 1689), Y_test[::100], label="Actual Error", color="blue")
ax1.legend()
ax2.scatter(
np.arange(1, 1689), y_pred_rid[::100], label="Predicted Error", color="orange"
)
ax2.legend()
fig, ax = plt.subplots(2, 2, figsize=(16, 16))
diff_rid = np.array(Y_test - y_pred_rid)
ax[0, 0].plot(diff_rid[::10000])
ax[0, 0].set_title("One in 10,000 differences, Ridge Regression")
ax[0, 1].plot(diff_rid[::1000])
ax[0, 1].set_title("One in 1,000 differences, Ridge Regression")
ax[1, 0].plot(diff_rid[::100])
ax[1, 0].set_title("One in 100 differences, Ridge Regression")
ax[1, 1].plot(diff_rid[::10])
ax[1, 1].set_title("One in 10 differences, Ridge Regression")
plt.figure(figsize=(20, 8))
sns.histplot(Y_test - y_pred_rid)
# plt.xticks(np.arange(-350,1000,50))
r2_rid = r2_score(Y_test, y_pred_rid)
print("r2 score for Ridge Regression is", r2_rid)
# **Lasso Regression**
lasso = Lasso()
lasso.fit(X_train_scaled, Y_train)
y_pred_las = lasso.predict(X_test_scaled)
mse_las = mean_squared_error(Y_test, y_pred_las)
mae_las = mean_absolute_error(Y_test, y_pred_las)
print("Mean squared error using Lasso Regression: ", mse_las)
print("Mean absolute error using Lasso Regression: ", mae_las)
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(20, 5))
ax1.scatter(np.arange(1, 1689), Y_test[::100], label="Actual Error", color="blue")
ax1.legend()
ax2.scatter(
np.arange(1, 1689), y_pred_las[::100], label="Predicted Error", color="orange"
)
ax2.legend()
fig, ax = plt.subplots(2, 2, figsize=(16, 16))
diff_las = np.array(Y_test - y_pred_las)
ax[0, 0].plot(diff_las[::10000])
ax[0, 0].set_title("One in 10,000 differences, Lasso Regression")
ax[0, 1].plot(diff_las[::1000])
ax[0, 1].set_title("One in 1,000 differences, Lasso Regression")
ax[1, 0].plot(diff_las[::100])
ax[1, 0].set_title("One in 100 differences, Lasso Regression")
ax[1, 1].plot(diff_las[::10])
ax[1, 1].set_title("One in 10 differences, Lasso Regression")
r2_las = r2_score(Y_test, y_pred_las)
print("r2 score for Lasso Regression is", r2_las)
plt.figure(figsize=(20, 8))
sns.histplot(Y_test - y_pred_las)
# plt.xticks(np.arange(-350,1000,50))
# **Ridge Regression long with Polynomial Regression**
from sklearn.preprocessing import PolynomialFeatures
poly = PolynomialFeatures(degree=2)
X_F1_poly = poly.fit_transform(df[["A", "B", "n"]])
X_train_poly, X_test_poly, y_train_poly, y_test_poly = train_test_split(
X_F1_poly, df["MAE"], random_state=0
)
linreg = Ridge().fit(X_train_poly, y_train_poly)
y_pred_poly = linreg.predict(X_test_poly)
mse_poly = mean_squared_error(y_test_poly, y_pred_poly)
mae_poly = mean_absolute_error(y_test_poly, y_pred_poly)
print("Mean squared error using Ridge with polynomial regression: ", mse_poly)
print("Mean absolute error using Ridge with polynomial regression: ", mae_poly)
r2_poly = r2_score(y_test_poly, y_pred_poly)
print("r2 score for Polynomial Regression is", r2_poly)
# from sklearn.svm import SVR
# regressor = SVR(kernel = 'rbf')
# regressor.fit(X_train_scaled, Y_train)
# y_pred_rbf = regressor.predict(X_test_scaled)
# mse_rbf = mean_squared_error(Y_test, y_pred_rbf)
# mae_rbf = mean_absolute_error(Y_test, y_pred_rbf)
# print('Mean squared error using Support Vector regressor (rbf krenel) : ', mse_rbf)
# print('Mean absolute error using Support Vector regressor (rbf krenel) : ', mae_rbf)
### Took huge amount of time still didnt got execute.
# **Artificial Neural Network**
import tensorflow as tf
from tensorflow import keras
model = keras.Sequential()
model.add(
keras.layers.Dense(
500, input_dim=3, activation=tf.keras.layers.LeakyReLU(alpha=0.1)
)
)
model.add(keras.layers.Dense(300, activation=tf.keras.layers.LeakyReLU(alpha=0.1)))
model.add(keras.layers.Dense(150, activation=tf.keras.layers.LeakyReLU(alpha=0.1)))
model.add(keras.layers.Dense(70, activation="relu"))
model.add(keras.layers.Dense(50, activation="relu"))
model.add(keras.layers.Dense(10, activation=tf.keras.layers.LeakyReLU(alpha=0.1)))
model.add(keras.layers.Dense(5, activation="relu"))
model.add(keras.layers.Dense(1, activation="linear"))
model.compile(loss="mean_squared_error", optimizer="adam", metrics=["mae"])
model.summary()
checkpointer = tf.keras.callbacks.ModelCheckpoint(
"stress_strain_new_weights.h5", verbose=1, save_best_only=True
)
model.fit(
X_train_scaled, Y_train, epochs=40, validation_split=0.2, callbacks=checkpointer
)
from keras.models import Sequential, load_model
# Load and evaluate the best model version
model = load_model("/kaggle/working/stress_strain_new_weights.h5")
y_pred_ann = model.predict(X_test_scaled)
mse_ann = mean_squared_error(Y_test, y_pred_ann)
mae_ann = mean_absolute_error(Y_test, y_pred_ann)
print("Mean squared error using Artificial Neural Network: ", mse_ann)
print("Mean absolute error using Artificial Neural Network: ", mae_ann)
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(20, 5))
ax1.scatter(np.arange(1, 1689), Y_test[::100], label="Actual Error", color="blue")
ax1.legend()
ax2.scatter(
np.arange(1, 1689), y_pred_ann[::100], label="Predicted Error", color="orange"
)
ax2.legend()
r2_ann = r2_score(Y_test, y_pred_ann)
print("r2 score for ANN is", r2_ann)
np.array(Y_test[:10]).reshape(10, 1)
y_pred_ann[:10].reshape(10, 1)
"""
After applying several models, their results are as following:
Linear Regression:
Mean squared error from linear regression: 112505.26220087174
Mean absolute error from linear regression: 212.7224693685027
r2 score for linear regression model is 0.21625077011971106
Decision Tree:
Mean squared error using decision tree: 2.931331934337034
Mean absolute error using decision tree: 0.7197832428520041
r2 score for decision tree model is 0.9999795793627683
Random Forest:
Mean squared error using Random Forest: 0.8614917523433008
Mean absolute error Using Random Forest: 0.42142079385048675
r2 score for Random Forest model is 0.9999939985607407
KNN:
Mean squared error using K nearest neighbours: 8.867443118030529
Mean absolute error using K nearest neighbours: 1.5422359790106617
r2 score for K nearest neighbours is 0.9999382264297793
Ridge:
Mean squared error using Ridge Regression: 112505.2619937623
Mean absolute error using Ridge Regression: 212.72243281538636
r2 score for Lasso Regression is 0.21624238314344646
Lasso:
Mean squared error using Lasso Regression: 112506.46613055709
Mean absolute error using Lasso Regression: 212.43496161207207
r2 score for Lasso Regression is 0.21624238314344646
Polynomial (deg 2) with Ridge:
Mean squared error using Ridge with polynomial regression: 58641.96138347135
Mean absolute error using Ridge with polynomial regression: 171.69030886170754
r2 score for Polynomial Regression is 0.5929207310514428
ANN:
Mean squared error using Artificial Neural Network: 0.8807563634692798
Mean absolute error using Artificial Neural Network: 0.628349179192993
r2 score for ANN is 0.9999938643570259
"""
a = int(input("Enter the value of A"))
b = int(input("Enter the value of B"))
c = float(input("Enter the value of n"))
scaled_input = scaler.transform([[a, b, c]])
scaled_input
calc_stress = a + b * (np.power(exp_strain, c))
plt.plot(exp_strain, exp_stress, label="Experimental")
plt.plot(exp_strain, calc_stress, label="Calculated")
plt.legend()
print("Actual MAE : ", mae(exp_stress, calc_stress))
print("Predicted MAE by ANN: ", model.predict(scaled_input)[0][0])
print("Predicted MAE by Random Forest: ", model_RF.predict(scaled_input)[0])
print("Predicted MAE by Decision Trees: ", tree.predict(scaled_input)[0])
print("Predicted MAE by KNN: ", knn.predict(scaled_input)[0])
import pickle
pickle.dump(model, open("ANN_explo_new_model.pkl", "wb"))
|
# # Importing Libraries
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# data analysis and wrangling
import pandas as pd
import numpy as np
import random as rnd
import re
# visualization
import seaborn as sns
import matplotlib.pyplot as plt
import plotly.express as px
from plotly.subplots import make_subplots
import plotly.graph_objects as go
# machine learning
import torch
from torch import nn
from torch.utils.data import Dataset, DataLoader
import torch.nn.functional as F
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import LabelEncoder
# ## Exploring Dataset
train_df = pd.read_csv("/kaggle/input/spaceship-titanic/train.csv")
test_df = pd.read_csv("/kaggle/input/spaceship-titanic/test.csv")
train_df.head()
# # File and Data Field Descriptions
# > **train.csv** - Personal records for about two-thirds (~8700) of the passengers, to be used as training data.
# > - `PassengerId` - A unique Id for each passenger. Each Id takes the form gggg_pp where gggg indicates a group the passenger is travelling with and pp is their number within the group. People in a group are often family members, but not always.
# > - `HomePlanet` - The planet the passenger departed from, typically their planet of permanent residence.
# > - `CryoSleep` - Indicates whether the passenger elected to be put into suspended animation for the duration of the voyage. Passengers in cryosleep are confined to their cabins.
# > - `Cabin` - The cabin number where the passenger is staying. Takes the form deck/num/side, where side can be either P for Port or S for Starboard.
# > - `Destination` - The planet the passenger will be debarking to.
# > - `Age` - The age of the passenger.
# > - `VIP` - Whether the passenger has paid for special VIP service during the voyage.
# > - `RoomService, FoodCourt, ShoppingMall, Spa, VRDeck` - Amount the passenger has billed at each of the Spaceship Titanic's many luxury amenities.
# > - `Name` - The first and last names of the passenger.
# > - `Transported` - Whether the passenger was transported to another dimension. This is the target, the column you are trying to predict.
# > **test.csv** - Personal records for the remaining one-third (~4300) of the passengers, to be used as test data.
# > 📝 To predict: the value of Transported for the passengers in this set
print("Train set shape:", train_df.shape)
print("Test set shape:", test_df.shape)
train_df.describe()
train_df.dtypes
train_df.nunique()
# missing value
print("TRAIN SET MISSING VALUES:")
print(train_df.isna().sum())
print("")
print("TEST SET MISSING VALUES:")
print(test_df.isna().sum())
# ## Types of Data
# There are 6 continuous features, 4 categorical features (excluding the target) and 3 descriptive/qualitative features.
print("HomePlanet\n")
print(train_df["HomePlanet"].value_counts())
print("------" * 6)
print("Destination\n")
print(train_df["Destination"].value_counts())
print("------" * 6)
print("VIP\n")
print(train_df["VIP"].value_counts())
print("------" * 6)
print("Transported\n")
print(train_df["Transported"].value_counts())
print("------" * 6)
print("Cabin\n")
print(train_df["Cabin"].str[0].value_counts())
print("------" * 6)
print("CryoSleep\n")
print(train_df["CryoSleep"].value_counts())
# ## EDA
total = 4602 + 2131 + 1759
ear = 4602
eur = 2131
mars = 1759
labels = "Earth", "Europa", "Mars"
sizes = [ear / total * 100, eur / total * 100, mars / total * 100]
explode = (0.1, 0, 0.1)
total_de = 5915 + 1800 + 796
TRAPPIST = 5915
Cancri = 1800
PSOJ318 = 796
labels_1 = "TRAPPIST-1e", "55 Cancri e", "PSO J318.5-22"
sizes_1 = [TRAPPIST / total_de * 100, Cancri / total_de * 100, PSOJ318 / total_de * 100]
explode_1 = (0, 0.2, 0)
colors = ["#ff9999", "#66b3ff", "#99ff99"]
plt.figure(figsize=(20, 10))
plt.subplot(1, 2, 1)
plt.pie(
sizes,
explode=explode,
labels=labels,
autopct="%1.1f%%",
shadow=True,
startangle=90,
textprops={"fontsize": 14},
colors=colors,
)
plt.axis("equal")
plt.title("HomePlanet Ratio")
plt.subplot(1, 2, 2)
plt.pie(
sizes_1,
explode=explode_1,
labels=labels_1,
autopct="%1.1f%%",
shadow=True,
startangle=90,
textprops={"fontsize": 14},
colors=colors,
)
plt.axis("equal")
plt.title("Destination Ratio")
plt.suptitle("HomePlanet & Destination Ratio", fontsize=25)
plt.show()
# ### Correlation
plt.figure(figsize=(20, 15))
heat_map = train_df.corr()
mask = np.triu(np.ones_like(heat_map, dtype=bool))
heatmap = sns.heatmap(heat_map, annot=True, mask=mask, cmap="Pastel1_r")
heatmap.set_title("Feature Correlation")
plt.show()
# ### Violine Plot for Transported of Different HomePlantes of Different Ages
plt.figure(figsize=(20, 15))
sns.set_theme(style="whitegrid")
sns.set(font_scale=1.5)
tmp = sns.violinplot(
data=train_df,
x="HomePlanet",
y="Age",
hue="Transported",
split=True,
inner="quart",
linewidth=1,
palette="Spectral",
)
sns.despine(left=True)
plt.show()
# ### All Age Distribution for Transforted
g = sns.FacetGrid(train_df, col="Transported", height=4.2, aspect=3.6)
sns.set(font_scale=2)
g.map(plt.hist, "Age", bins=20)
plt.show()
# ### Transported vs Not Transported
sns.countplot(x="Transported", data=train_df)
# ### Pie Chart
plt.pie(
train_df.Transported.value_counts(),
shadow=True,
explode=[0.1, 0.1],
autopct="%.1f%%",
)
plt.title("Transported ", size=18)
plt.legend(["False", "True"], loc="best", fontsize=12)
plt.show()
sns.countplot(x=train_df.VIP, hue=train_df.Transported)
# __Seems like not so useful feature__
sns.countplot(x=train_df.CryoSleep, hue=train_df.Transported)
# So most people in Crysleep transported
ax = sns.countplot(x=train_df.Destination, hue=train_df.Transported)
ax.set_xticklabels(ax.get_xticklabels(), rotation=45)
# ## Processing Dataset
# Cabin - The cabin number where the passenger is staying. Takes the form deck/num/side, where side can be either P for Port or S for Starboard.
train_df["Deck"] = train_df["Cabin"].str.split("/").str[0]
train_df["Num"] = list(map(float, train_df["Cabin"].str.split("/").str[1]))
train_df["Side"] = train_df["Cabin"].str.split("/").str[2]
train_df = train_df.drop(["Cabin", "PassengerId", "Name"], axis=1)
train_df.head()
sns.countplot(x=train_df.Deck, hue=train_df.Transported)
ax = sns.countplot(x=train_df.Side, hue=train_df.Transported)
ax.legend(loc="upper right", bbox_to_anchor=(1.4, 1))
# ### Visualizing Null Values
plt.figure(figsize=(22, 1))
sns.heatmap(
pd.DataFrame(
np.asarray(train_df.isna().sum()).reshape(1, train_df.shape[1]),
columns=list(train_df.columns),
),
cmap="Spectral",
annot=True,
fmt=".0f",
)
# ## Filling Null Values with Mode, Mean and Zero
mf = ["HomePlanet", "Destination", "CryoSleep", "VIP", "Side", "Deck"]
mean = ["Age"]
zero = ["RoomService", "FoodCourt", "ShoppingMall", "Spa", "VRDeck", "Num"]
print(train_df.mode().iloc[0])
print(train_df[mean].mean())
train_df[mf] = train_df[mf].fillna(train_df.mode().iloc[0])
train_df[mean] = train_df[mean].fillna(train_df[mean].mean())
train_df[zero] = train_df[zero].fillna(0)
train_df.head(10)
# ## Creating a new Feature called Money Spent and created Age Categories
train_df["MoneySpent"] = (
train_df["RoomService"]
+ train_df["FoodCourt"]
+ train_df["ShoppingMall"]
+ train_df["Spa"]
+ train_df["VRDeck"]
)
train_df.insert(loc=3, column="AgeCategories", value=0)
train_df.loc[train_df["Age"] <= 14, "AgeCategories"] = 1
train_df.loc[(train_df["Age"] > 14) & (train_df["Age"] <= 24), "AgeCategories"] = 2
train_df.loc[(train_df["Age"] > 24) & (train_df["Age"] <= 64), "AgeCategories"] = 3
train_df.loc[(train_df["Age"] > 64), "AgeCategories"] = 4
train_df.head()
# ## Categorical Column Handling
train_df = pd.concat(
[train_df, pd.get_dummies(train_df[["Deck", "Side", "HomePlanet", "Destination"]])],
axis=1,
)
train_df = pd.concat([train_df, pd.get_dummies(train_df["AgeCategories"])], axis=1)
train_df[["CryoSleep", "VIP"]] = train_df[["CryoSleep", "VIP"]].apply(
LabelEncoder().fit_transform
)
train_df.head()
train_df = train_df.drop(
["Deck", "Side", "HomePlanet", "Destination", "AgeCategories"], axis=1
)
y = train_df.pop("Transported")
y = LabelEncoder().fit_transform(y)
train_df.head()
for col in train_df:
if train_df[col].dtypes == "float64":
train_df[col] = StandardScaler().fit_transform(
np.array(train_df[col]).reshape(-1, 1)
)
# applies the standard scaling transformation to the column. The StandardScaler() function from the sklearn.preprocessing module is used to standardize the values in the column. The np.array(train_df[col]) method converts the values in the column to a NumPy array, and the .reshape(-1, 1) method reshapes the array to a 2-dimensional array with a single column. This is necessary because the fit_transform() method of the StandardScaler() function expects a 2-dimensional array as input. The fit_transform() method standardizes the values in the column and returns the standardized values, which are then assigned back to the column in the original dataframe.
train_df.head()
# ## Train Validation Split
x_train, x_validation, y_train, y_validation = train_test_split(
train_df, y, test_size=0.2, shuffle=True, random_state=5
)
# ## Creating Data Loader
class Dataset(Dataset):
def __init__(self, x, y):
self.df = np.array(x)
self.df_labels = np.array(y)
self.dataset = torch.tensor(self.df)
self.labels = torch.tensor(self.df_labels)
def __len__(self):
return len(self.dataset)
def __getitem__(self, index):
return self.dataset[index], self.labels[index]
batch_size = 64
train_dataloader = DataLoader(
Dataset(x_train, y_train), batch_size=batch_size, shuffle=True
)
val_dataloader = DataLoader(
Dataset(x_validation, y_validation), batch_size=batch_size, shuffle=True
)
device = "cuda" if torch.cuda.is_available() else "cpu"
print(f"Using {device} device")
# ## Defining Deep Learning Model
class NeuralNetwork(nn.Module):
def __init__(self, input_size, hidden_size):
super(NeuralNetwork, self).__init__()
self.model = nn.Sequential(
nn.Linear(input_size, hidden_size),
nn.ReLU(),
nn.Dropout(p=0.5),
nn.Linear(hidden_size, 1),
)
def forward(self, x):
return self.model(x)
input_size = x_train.shape[1]
hidden_size = int(input_size * 2)
model = NeuralNetwork(input_size, hidden_size).to(device)
print(model)
epochs = 200
lr = 0.003
criterion = nn.BCEWithLogitsLoss()
optimizer = torch.optim.Adam(model.parameters(), lr=lr)
def progress_bar(progress, total, lenght):
percent = lenght * (progress / total)
bar = "❚" * int(percent) + " " * (lenght - int(percent))
return bar
train_plot = []
val_plot = []
train_accuracy_plot = []
val_accuracy_plot = []
for epoch in range(epochs):
train_loss = 0
val_loss = 0
train_correct = 0
val_correct = 0
for x, y in train_dataloader:
optimizer.zero_grad()
x, y = x.to(device).float(), y.to(device).float().unsqueeze(1)
output = model(x)
loss = criterion(output, y)
train_loss += criterion(output, y).item()
train_correct += (y == torch.round(torch.sigmoid(model(x)))).float().sum()
loss.backward()
optimizer.step()
with torch.no_grad():
for x_val, y_val in val_dataloader:
x_val, y_val = x_val.to(device).float(), y_val.to(device).float().unsqueeze(
1
)
pred = model(x_val)
loss = criterion(pred, y_val)
val_loss += criterion(pred, y_val).item()
val_correct += (
(y_val == torch.round(torch.sigmoid(model(x_val)))).float().sum()
)
train_plot.append((train_loss / len(train_dataloader)))
val_plot.append((val_loss / len(val_dataloader)))
train_accuracy_plot.append((train_correct / len(y_train)).item())
val_accuracy_plot.append((val_correct / len(y_validation)).item())
print(
rf"|{progress_bar(epoch + 1, epochs, 50)}| {epoch + 1} / {epochs}, train_loss = {(train_loss/len(train_dataloader)):.5f}, val_loss = {(val_loss/len(val_dataloader)):.5f}, train_accuracy = {(train_correct / len(y_train)):.5f}, val_accuracy = {(val_correct / len(y_validation)):.5f}",
end="\r",
)
fig, (ax1, ax2) = plt.subplots(2, 1)
ax1.plot([i for i in range(1, epochs + 1)], train_plot, label="train")
ax1.plot([i for i in range(1, epochs + 1)], val_plot, label="val")
ax2.plot([i for i in range(1, epochs + 1)], train_accuracy_plot, label="train")
ax2.plot([i for i in range(1, epochs + 1)], val_accuracy_plot, label="val")
legend1 = ax1.legend(loc="upper right")
legend2 = ax2.legend(loc="upper right")
legend1.set_bbox_to_anchor((1.5, 1))
legend2.set_bbox_to_anchor((1.5, 1))
# plt.legend(loc='best')
plt.show()
val = torch.tensor(np.array(x_validation)).float().to(device)
val_output = torch.round(torch.sigmoid(model(val)))
val_output = val_output.reshape(-1).type(torch.bool).cpu().numpy()
error_analysis_df = pd.DataFrame(
{"Prediction": val_output, "TrueValue": y_validation.astype(bool)}
)
TrueTrue = len(
error_analysis_df.loc[
(error_analysis_df["TrueValue"] == True)
& (error_analysis_df["Prediction"] == True)
]
)
FalseTrue = len(
error_analysis_df.loc[
(error_analysis_df["TrueValue"] == False)
& (error_analysis_df["Prediction"] == True)
]
)
TrueFalse = len(
error_analysis_df.loc[
(error_analysis_df["TrueValue"] == False)
& (error_analysis_df["Prediction"] == False)
]
)
FalseFalse = len(
error_analysis_df.loc[
(error_analysis_df["TrueValue"] == True)
& (error_analysis_df["Prediction"] == False)
]
)
# ## Confusion Matrix
plt.figure()
sns.heatmap(
pd.DataFrame(
{"True": [FalseTrue, TrueTrue], "False": [TrueFalse, FalseFalse]},
index=["False", "True"],
),
cmap="Spectral",
annot=True,
fmt=".0f",
)
plt.xlabel("Prediction")
plt.ylabel("True")
plt.show()
# ## Saving Model
torch.save(model.state_dict(), "model_weights.pth")
# ## Testing Prediction
test_df = pd.read_csv("/kaggle/input/spaceship-titanic/test.csv")
def Preprocessing(data):
# data = CabinSplit(data)
data["Deck"] = data["Cabin"].str.split("/").str[0]
data["Num"] = list(map(float, data["Cabin"].str.split("/").str[1]))
data["Side"] = data["Cabin"].str.split("/").str[2]
data = data.drop(["Cabin", "PassengerId", "Name"], axis=1)
# data = FillNaN(data)
data[mf] = data[mf].fillna(data.mode().iloc[0])
data[mean] = data[mean].fillna(data[mean].mean())
data[zero] = data[zero].fillna(0)
data["MoneySpent"] = (
data["RoomService"]
+ data["FoodCourt"]
+ data["ShoppingMall"]
+ data["Spa"]
+ data["VRDeck"]
)
data.insert(loc=3, column="AgeCategories", value=0)
data.loc[data["Age"] <= 14, "AgeCategories"] = 1
data.loc[(data["Age"] > 14) & (data["Age"] <= 24), "AgeCategories"] = 2
data.loc[(data["Age"] > 24) & (data["Age"] <= 64), "AgeCategories"] = 3
data.loc[(data["Age"] > 64), "AgeCategories"] = 4
# data = Encode(data)
data = pd.concat(
[data, pd.get_dummies(data[["Deck", "Side", "HomePlanet", "Destination"]])],
axis=1,
)
data = pd.concat([data, pd.get_dummies(data["AgeCategories"])], axis=1)
data[["CryoSleep", "VIP"]] = data[["CryoSleep", "VIP"]].apply(
LabelEncoder().fit_transform
)
data = data.drop(
["Deck", "Side", "HomePlanet", "Destination", "AgeCategories"], axis=1
)
# data = Norm(data)
for col in data:
if data[col].dtypes == "float64":
data[col] = StandardScaler().fit_transform(
np.array(data[col]).reshape(-1, 1)
)
return data
test_data = Preprocessing(test_df)
test_data.head(5)
model.load_state_dict(torch.load("model_weights.pth"))
data = torch.tensor(np.array(test_data)).float().to(device)
pred = torch.round(torch.sigmoid(model(data)))
pred = pred.reshape(-1).type(torch.bool).cpu().numpy()
submission = pd.DataFrame({"PassengerId": test_df["PassengerId"], "Transported": pred})
submission.to_csv("submission.csv", index=False)
|
# Bakyt Nursaiyn 19453647
# Balance Sheet & Cash Flow Analysis
# Value Investing Stock Analysis with Python
# Source: https://github.com/taewookim/YouTube
import os
import tk_library
from tk_library import excel_to_df
for dirname, _, filenames in os.walk("/kaggle/input/"):
for filename in filenames:
print(os.path.join(dirname, filename))
# # Read IBM_Fin_Stat.xlsx into a pandas dataframe
_, ibm_PL, ibm_BS, ibm_CF = excel_to_df("/kaggle/input/ibm-fin-stat/IBM_Fin_Stat.xlsx")
ibm_BS
del ibm_BS["Assets"]
ibm_BS
ibm_BS["_Total Current Assets"] = (
ibm_BS["Cash, Cash Equivalents & Short Term Investments"]
+ ibm_BS["Accounts & Notes Receivable"]
+ ibm_BS["Inventories"]
+ ibm_BS["Other Short Term Assets"]
)
ibm_BS[["_Total Current Assets", "Total Current Assets"]]
ibm_BS["_NonCurrent Assets"] = (
ibm_BS["Property, Plant & Equipment, Net"]
+ ibm_BS["Long Term Investments & Receivables"]
+ ibm_BS["Other Long Term Assets"]
)
ibm_BS["_Total Assets"] = ibm_BS["_NonCurrent Assets"] + ibm_BS["_Total Current Assets"]
ibm_BS["_Total Liabilities"] = (
ibm_BS["Total Current Liabilities"] + ibm_BS["Total Noncurrent Liabilities"]
)
ibm_BS[["_Total Liabilities", "Total Liabilities"]]
# # Balance Sheet Analysis
# ## Assets
#
ibm_BS[["Total Assets", "Total Liabilities", "Total Equity"]].plot()
good_stuff = """
Cash, Cash Equivalents & Short Term Investments
Accounts & Notes Receivable
Inventories
Other Short Term Assets
"""
asset_columns = [x for x in good_stuff.strip().split("\n")]
asset_columns
ibm_BS[asset_columns].plot()
import chart_studio
chart_studio.tools.set_credentials_file(
username="nursaiynb", api_key="G3BdeHhGUNcPHafQdX2Q"
)
import chart_studio.plotly as py
import plotly.graph_objs as go
from tk_library import combine_regexes
assets = go.Bar(x=ibm_BS.index, y=ibm_BS["Total Assets"], name="Assets")
liabilities = go.Bar(x=ibm_BS.index, y=ibm_BS["Total Liabilities"], name="Liabilities")
shareholder_equity = go.Scatter(x=ibm_BS.index, y=ibm_BS["Total Equity"], name="Equity")
data = [assets, liabilities, shareholder_equity]
layout = go.Layout(barmode="stack")
fig_bs = go.Figure(data=data, layout=layout)
py.iplot(fig_bs, filename="Total Assets and Liabilities")
asset_data = []
columns = """
Cash, Cash Equivalents & Short Term Investments
Accounts & Notes Receivable
Inventories
Other Short Term Assets
Property, Plant & Equipment, Net
Long Term Investments & Receivables
Other Long Term Assets
"""
for col in columns.strip().split("\n"):
asset_bar = go.Bar(x=ibm_BS.index, y=ibm_BS[col], name=col)
asset_data.append(asset_bar)
layout_assets = go.Layout(barmode="stack")
fig_bs_assets = go.Figure(data=asset_data, layout=layout_assets)
py.iplot(fig_bs_assets, filename="Total Assets Breakdown")
liability_data = []
columns = """
Payables & Accruals
Short Term Debt
Other Short Term Liabilities
Long Term Debt
Other Long Term Liabilities
"""
for col in columns.strip().split("\n"):
liability_bar = go.Bar(x=ibm_BS.index, y=ibm_BS[col], name=col)
liability_data.append(liability_bar)
layout_liabilitys = go.Layout(barmode="stack")
fig_bs_liabilitys = go.Figure(data=liability_data, layout=layout_liabilitys)
py.iplot(fig_bs_liabilitys, filename="Total liabilities Breakdown")
# ## Working Capital = Net Asset Value = Current Assets - Current Liabilities
# Net Asset value = Current Assets - Current Liabilities - Long Term Liaibility
ibm_BS["Working Capital"] = (
ibm_BS["Total Current Assets"] - ibm_BS["Total Current Liabilities"]
)
ibm_BS[["Working Capital"]].plot()
ibm_BS[["Accounts & Notes Receivable", "Payables & Accruals"]].plot()
# Using Chart Studio in Plotly
PR_data = []
columns = """
Accounts & Notes Receivable
Payables & Accruals
"""
for col in columns.strip().split("\n"):
PR_Scatter = go.Scatter(x=ibm_BS.index, y=ibm_BS[col], name=col)
PR_data.append(PR_Scatter)
layout_PR = go.Layout(barmode="stack")
fig_bs_PR = go.Figure(data=PR_data, layout=layout_PR)
py.iplot(fig_bs_PR, filename="Accounts & Notes Receivable vs Payables & Accruals")
ibm_BS["Inventories"].plot()
ibm_BS[
[
"Property, Plant & Equipment, Net",
"Long Term Investments & Receivables",
"Other Long Term Assets",
]
].plot()
# Using Plotly
AAA_data = []
columns = """
Property, Plant & Equipment, Net
Long Term Investments & Receivables
Other Long Term Assets
"""
for col in columns.strip().split("\n"):
AAA_bar = go.Bar(x=ibm_BS.index, y=ibm_BS[col], name=col)
AAA_data.append(AAA_bar)
layout_AAA = go.Layout(barmode="stack")
fig_bs_AAA = go.Figure(data=AAA_data, layout=layout_AAA)
py.iplot(fig_bs_AAA, filename="Total Long Term Assets")
equity_columns = """
Share Capital & Additional Paid-In Capital
Treasury Stock
Retained Earnings
Other Equity
Equity Before Minority Interest
Minority Interest
"""
equity_columns = [x for x in equity_columns.strip().split("\n")]
equity_columns
ibm_BS[equity_columns].plot()
# Using Plotly
equity_data = []
columns = """
Share Capital & Additional Paid-In Capital
Treasury Stock
Retained Earnings
Other Equity
Equity Before Minority Interest
Minority Interest
"""
for col in columns.strip().split("\n"):
equity_Scatter = go.Scatter(x=ibm_BS.index, y=ibm_BS[col], name=col)
equity_data.append(equity_Scatter)
layout_equity = go.Layout(barmode="stack")
fig_bs_equity = go.Figure(data=equity_data, layout=layout_equity)
py.iplot(fig_bs_equity, filename="Total Equity")
# # Book Value, aka "Owner's Equity"
# BV = Total Assets - Intangible assets - Liabilities - Preferred Stock Value
#
# According to simfin data, IBM has no preferred stock, no intengible assets, and no goodwill
ibm_BS["book value"] = ibm_BS["Total Assets"] - ibm_BS["Total Liabilities"]
ibm_BS["book value"].plot()
# Calculate 1. Price-to-Earnings Growth Ratio (PEG forward)
# using this formula – PEG = Price-to-Earnings Ratio/Earnings-Growth-Rate
# https://www.investopedia.com/ask/answers/012715/what-considered-good-peg-price-earnings-growth-ratio.asp
PE_RATIO = 17.46 # FROM SIMFIN WEBSITE: https://simfin.com/data/companies/69543
# FROM NASDAQ WEBSITE: https://www.nasdaq.com/symbol/xom/earnings-growth
GROWTH_RATE = 0.0588 # Forcast over the five next years
PEG_ratio = PE_RATIO / (GROWTH_RATE * 100)
print("IBM Corp's PEG Ratio is", PEG_ratio)
# Additional calculation part is given below. All the information sources and further comments are given in respective code blocks.
# The next step is to calculate annual compounded growth rate of EPS
# Earnings and average shares outstanding figures were obtained from simfin.com
earningsTTM10, earningsTTM = 21512000000, 15973000000
avsharesTTM10, avsharesTTM = 1344597198, 895685546
epsTTM10 = earningsTTM10 / avsharesTTM10
epsTTM = earningsTTM / avsharesTTM
CAGR = (epsTTM / epsTTM10) ** (1 / 9) - 1
print(CAGR)
# Now we need to find EPS 10 years from now. For that, we will use CAGR formula and extract epsTTM from it, making it the EPS value 10 years from now.
eps10 = (1 + CAGR) ** 10 * epsTTM
print(eps10)
# To calculate the future price 10 years from now, we multiply future eps by average PE ratio in between 2006 and 2019, obtained from macrotrends.com
aver_pe = 11.76
future_price = eps10 * aver_pe
print(future_price)
# To calculate the target buy price, we discount the future price back to now using 6.5% average WACC for computer services companies obtained from http://people.stern.nyu.edu/adamodar/New_Home_Page/datafile/wacc.htm
target_buy = future_price / (1 + 0.065) ** 10
print(target_buy)
mar_safety = target_buy * 0.85
print(mar_safety)
debt_to_equity = ibm_BS["Total Liabilities"] / ibm_BS["Total Equity"]
print(debt_to_equity)
int_exp = 723 # obtained from annual report, the interest expense is $723 million
int_cov = ibm_PL["Pretax Income (Loss)"] / int_exp
print(int_cov)
|
import pandas as pd
from pathlib import Path
import numpy as np
from xgboost import XGBClassifier
from sklearn.preprocessing import LabelEncoder
from sklearn.ensemble import VotingClassifier
pd.set_option("display.max_rows", None)
# ### **Importing the generated molecular descriptors**
train_df = pd.read_csv("/kaggle/input/molecular-descriptors/train_molecular_data.csv")
test_df = pd.read_csv("/kaggle/input/molecular-descriptors/test_molecular_Data.csv")
train_df.describe()
test_df.describe()
train_df.isnull().sum()
test_df.isnull().sum()
# Filling null values with 0s
train_df.fillna(0, inplace=True)
test_df.fillna(0, inplace=True)
train_df.head()
train_y = train_df.loc[:, "Expected"]
train_X = train_df.drop(["Expected"], axis=1)
# Transforming classes [1, 2] to [0, 1]
lab = LabelEncoder()
train_yT = lab.fit_transform(train_y)
# ### **Feature Selection using Boruta**
# Importing feature ranks which were already stored
feature_ranks = pd.read_csv("/kaggle/input/selected-features/feature_ranks_v2.csv")
feature_ranks.sort_values("rankings", inplace=True)
feature_ranks.reset_index(drop=True, inplace=True)
feature_ranks
# ### **Considered 170 features**
train_fs = train_df.loc[:, feature_ranks[(feature_ranks["rankings"] <= 99)]["features"]]
test_fs = test_df.loc[:, feature_ranks[(feature_ranks["rankings"] <= 99)]["features"]]
params = {"tree_method": "gpu_hist", "n_jobs": -1}
# #### 1. Considered 12 XGB Classifiers in a Voting Classifier
# #### 2. The hyper-parameters were choosen from best params and based on local f1-score for each of the 12 XGB classifiers
xgb_1 = XGBClassifier(**params, n_estimators=500)
xgb_2 = XGBClassifier(
**params,
n_estimators=700,
colsample_bytree=0.63,
learning_rate=0.13,
subsample=0.96,
)
xgb_3 = XGBClassifier(
**params,
n_estimators=450,
max_depth=12,
colsample_bytree=0.63,
learning_rate=0.13,
subsample=0.96,
)
xgb_4 = XGBClassifier(
**params,
n_estimators=800,
max_depth=15,
colsample_bytree=0.68,
learning_rate=0.09,
subsample=0.90,
min_child_weight=0.0,
gamma=1.3,
)
xgb_5 = XGBClassifier(
**params,
n_estimators=800,
max_depth=12,
colsample_bytree=0.68,
learning_rate=0.09,
subsample=0.90,
min_child_weight=0.0,
gamma=1.3,
)
xgb_6 = XGBClassifier(
**params,
n_estimators=700,
max_depth=10,
colsample_bytree=0.68,
learning_rate=0.2,
subsample=0.90,
min_child_weight=0.0,
gamma=0.3,
)
xgb_7 = XGBClassifier(
**params,
n_estimators=700,
max_depth=15,
colsample_bytree=0.53,
learning_rate=0.14,
subsample=0.99,
min_child_weight=7.0,
)
xgb_8 = XGBClassifier(
**params,
n_estimators=600,
max_depth=10,
colsample_bytree=0.63,
learning_rate=0.14,
subsample=0.99,
)
xgb_9 = XGBClassifier(**params, n_estimators=700, max_depth=8, learning_rate=0.14)
xgb_10 = XGBClassifier(**params, n_estimators=600, max_depth=10, learning_rate=0.2)
xgb_11 = XGBClassifier(
**params,
n_estimators=700,
max_depth=12,
colsample_bytree=0.7,
learning_rate=0.14,
subsample=0.99,
)
xgb_12 = XGBClassifier(**params, n_estimators=650, max_depth=10, learning_rate=0.2)
classifier_list = [
("xgb_1", xgb_1),
("xgb_2", xgb_2),
("xgb_3", xgb_3),
("xgb_4", xgb_4),
("xgb_5", xgb_5),
("xgb_6", xgb_6),
("xgb_7", xgb_7),
("xgb_8", xgb_8),
("xgb_9", xgb_9),
("xgb_10", xgb_10),
("xgb_11", xgb_11),
("xgb_12", xgb_12),
]
# Reference - https://scikit-learn.org/stable/modules/generated/sklearn.ensemble.VotingClassifier.html
voting_clf = VotingClassifier(
estimators=classifier_list,
n_jobs=-1,
verbose=2,
voting="soft",
weights=[1, 1, 2, 1, 2, 1, 1, 1, 1, 1, 1, 1],
)
def pred_and_sub(classifier, sub_no):
classifier.fit(X=train_fs, y=train_yT)
pred_vals = classifier.predict(test_fs)
new_pred = lab.inverse_transform(pred_vals)
final_df = pd.read_csv(
"/kaggle/input/the-toxicity-prediction-challenge-ii/test_II.csv"
)
final_df["Predicted"] = new_pred
final_df.columns = ["Id", "Predicted"]
final_df.to_csv(f"submission{sub_no}.csv", index=False)
print("Output file generated")
pred_and_sub(voting_clf, 59)
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from matplotlib import cm
import seaborn as sns
import random
import os
import gc # garbage collector
import datetime
from tqdm import tqdm
import cv2
from sklearn.preprocessing import StandardScaler as scale
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score, confusion_matrix, f1_score
import tensorflow as tf
from tensorflow.keras.models import Model
from tensorflow.keras.layers import (
Input,
Conv2D,
Dense,
Flatten,
MaxPooling2D,
BatchNormalization,
Dropout,
GlobalAveragePooling2D,
)
from tensorflow.keras.callbacks import (
EarlyStopping,
CSVLogger,
ModelCheckpoint,
TensorBoard,
)
from tensorflow.keras.preprocessing import image_dataset_from_directory
from tensorflow.keras.preprocessing.image import ImageDataGenerator
from tensorflow.keras.applications import NASNetLarge
resized_train_path = (
"/kaggle/input/diabetic-retinopathy-resized/resized_train/resized_train"
)
train_labels_path = "/kaggle/input/diabetic-retinopathy-resized/trainLabels.csv"
train_labels_cropped_path = (
"/kaggle/input/diabetic-retinopathy-resized/trainLabels_cropped.csv"
)
train_labels = pd.read_csv(train_labels_path)
train_labels.head()
train_labels.info()
level_cropped_col = train_labels["level"]
level_cropped_col
level_cropped_col.plot(kind="hist", figsize=(10, 5), cmap=cm.get_cmap("flag"))
train_labels_cropped = pd.read_csv(train_labels_cropped_path)
train_labels_cropped
level_cropped_col = train_labels_cropped["level"]
level_cropped_col
level_cropped_col.plot(kind="hist", figsize=(10, 5), cmap=cm.get_cmap("ocean"))
resized_train_list = os.listdir(resized_train_path)
len(resized_train_list)
plt.figure(figsize=(25, 20))
for i in range(1, 26):
plt.subplot(5, 5, i)
img_name = random.choice(resized_train_list)
img_path = os.path.join(resized_train_path, img_name)
img = cv2.imread(img_path)
img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)
plt.imshow(img)
plt.xlabel(img.shape[1])
plt.ylabel(img.shape[0])
resized_train_cropped_path = "/kaggle/input/diabetic-retinopathy-resized/resized_train_cropped/resized_train_cropped/"
resized_train_cropped_list = os.listdir(resized_train_cropped_path)
len(resized_train_cropped_list)
plt.figure(figsize=(26, 24))
for i in range(1, 26):
plt.subplot(5, 5, i)
img_name = random.choice(resized_train_cropped_list)
img_path = os.path.join(resized_train_cropped_path, img_name)
img = cv2.imread(img_path)
img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)
plt.imshow(img)
plt.xlabel(img.shape[1])
plt.ylabel(img.shape[0])
plt.figure(figsize=(20, 25))
for i in range(1, 16):
plt.subplot(5, 3, i)
plt.tight_layout()
plt.title("Color Histogram")
plt.xlabel("Intensity Value")
plt.ylabel("Number of Pixels")
img_name = random.choice(resized_train_cropped_list)
img_path = os.path.join(resized_train_cropped_path, img_name)
img = cv2.imread(img_path)
img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)
channels = cv2.split(img)
colors = ["r", "g", "b"]
for channel, color in zip(channels, colors):
hist = cv2.calcHist([channel], [0], None, [256], [0, 256])
plt.plot(hist, color=color)
plt.xlim([0, 256])
img_width = 100
img_height = 100
def read_img(img_name, resize=False):
img_path = os.path.join(resized_train_cropped_path, img_name)
img = cv2.imread(img_path)
img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)
if resize:
img = cv2.resize(img, (img_width, img_hight))
return img
def ben_graham(img):
img_ben = cv2.addWeighted(img, 4, cv2.GaussianBlur(img, (0, 0), 10), -4, 128)
return img_ben
def hist_equalization(img):
red, green, blue = cv2.split(img)
hist_red = cv2.equalizeHist(red)
hist_green = cv2.equalizeHist(green)
hist_blue = cv2.equalizeHist(blue)
img_eq = cv2.merge((hist_red, hist_green, hist_blue))
return img_eq
equal_hist_images = resized_train_cropped_list.copy()
len(equal_hist_images)
plt.figure(figsize=(26, 24))
counter = 0
for img_name in equal_hist_images:
counter += 1
plt.subplot(5, 5, counter)
plt.tight_layout()
# level_cropped_col is the labels, we've created it above
plt.title(level_cropped_col[counter - 1])
img = read_img(img_name)
# Applying the Histogram Equaliztion
img_eq = hist_equalization(img)
plt.imshow(img_eq)
plt.xlabel(img_eq.shape[1])
plt.xlabel(img_eq.shape[0])
if counter == 25:
break
plt.figure(figsize=(20, 25))
counter = 0
for img_name in equal_hist_images:
counter += 1
plt.subplot(5, 3, counter)
plt.tight_layout()
img = read_img(img_name)
# Applying the Histogram Equaliztion
img_eq = hist_equalization(img)
channels = cv2.split(img_eq)
colors = ["r", "g", "b"]
for channel, color in zip(channels, colors):
hist = cv2.calcHist([channel], [0], None, [256], [0, 256])
plt.plot(hist, color=color)
plt.xlim([0, 256])
if counter == 15:
break
ben_images = resized_train_cropped_list.copy()
len(ben_images)
plt.figure(figsize=(26, 24))
counter = 0
for img_name in ben_images:
counter += 1
plt.subplot(5, 5, counter)
plt.tight_layout()
# level_cropped_col is the lebels list
plt.title(level_cropped_col[counter - 1])
img = read_img(img_name)
# Applying Ben Graham's Method
img_ben = ben_graham(img)
plt.imshow(img_ben)
plt.xlabel(img_ben.shape[1])
plt.ylabel(img_ben.shape[0])
if counter == 25:
break
plt.figure(figsize=(20, 25))
counter = 0
for img_name in equal_hist_images:
counter += 1
plt.subplot(5, 3, counter)
plt.tight_layout()
img = read_img(img_name)
# Applying Ben Graham's Method
img_ben = ben_graham(img)
channels = cv2.split(img_ben)
colors = ["r", "g", "b"]
for channel, color in zip(channels, colors):
hist = cv2.calcHist([channel], [0], None, [256], [0, 256])
plt.plot(hist, color=color)
plt.xlim([0, 256])
if counter == 15:
break
train_labels_cropped.head()
train_labels_cropped["image_name"] = [
img + ".jpeg" for img in train_labels_cropped["image"]
]
train_labels_cropped.head()
train_ds, val_ds = train_test_split(train_labels_cropped, test_size=0.5)
train_ds.shape, val_ds.shape
def my_processes(img):
img = cv2.resize(img, (img_width, img_height))
# Apply your image processing method
img = ben_graham(img)
return img
train_datagen = ImageDataGenerator(
rescale=1.0 / 255.0,
rotation_range=40,
width_shift_range=0.5,
height_shift_range=0.5,
shear_range=0.5,
horizontal_flip=True,
preprocessing_function=ben_graham,
)
val_datagen = ImageDataGenerator(rescale=1.0 / 255.0)
batch_size = 32
train_dataset = train_datagen.flow_from_dataframe(
train_ds,
resized_train_cropped_path,
x_col="image_name",
y_col="level",
class_mode="raw",
batch_size=batch_size,
target_size=(img_width, img_height),
)
val_dataset = val_datagen.flow_from_dataframe(
val_ds,
resized_train_cropped_path,
x_col="image_name",
y_col="level",
class_mode="raw",
batch_size=batch_size,
target_size=(img_width, img_height),
)
model = tf.keras.applications.nasnet.NASNetLarge(
input_shape=None,
include_top=True,
weights="imagenet",
input_tensor=None,
pooling=None,
classes=1000,
classifier_activation="softmax",
)
x = Dropout(0.2)(model.output)
x = Dropout(0.5)(x)
x = Dense(2048, activation="relu")(x)
x = Dense(2048, activation="relu")(x)
x = Dropout(0.5)(x)
x = Dropout(0.5)(model.output)
x = Dense(1024, activation="relu")(x)
x = Dense(1024, activation="relu")(x)
x = Dropout(0.5)(x)
x = Dense(1024, activation="relu")(x)
x = Dropout(0.1)(x)
x = Dense(1024, activation="relu")(x)
x = Dropout(0.1)(x)
x = Dense(512, activation="relu")(x)
x = Dense(512, activation="relu")(x)
x = Dropout(0.2)(x)
x = Dense(512, activation="relu")(x)
x = Dropout(0.1)(x)
x = Dense(512, activation="relu")(x)
x = Dropout(0.1)(x)
# x = GlobalAveragePooling2D()(x)
# x = Dropout(0.1)(x)
classifier = Dense(5, activation="softmax")(x)
x = Dense(512, activation="relu")(x)
x = Dropout(0.5)(x)
x = Dense(512, activation="relu")(x)
x = Dropout(0.1)(x)
x = Dense(512, activation="relu")(x)
x = Dense(512, activation="relu")(x)
x = Dropout(0.2)(x)
x = Dense(512, activation="relu")(x)
x = Dropout(0.1)(x)
x = Dense(512, activation="relu")(x)
classifier = Dropout(0.1)(x)
model = Model(inputs=model.input, outputs=classifier)
model.compile(
optimizer=tf.keras.optimizers.Adam(),
loss="sparse_categorical_crossentropy",
metrics=["sparse_categorical_accuracy"],
)
metrics = ["sparse_categorical_accuracy"]
train_ds
c = train_ds.iloc[:, 3]
r = val_ds.loc[:, "level"]
tf.math.confusion_matrix(
train_ds.level,
val_ds.level,
num_classes=None,
weights=None,
dtype=tf.dtypes.int32,
name=None,
)
|
# Este script tiene como objetivo crear nuevos features.
# Se importan las librerias básicas para el manejo de los DataFrame
import pandas as pd
import numpy as np
seed = 99
# cargar pageviews
pageviews = pd.concat(
[
pd.read_csv(
"../input/banco-galicia-dataton-2019/pageviews.zip",
parse_dates=["FEC_EVENT"],
),
pd.read_csv(
"../input/banco-galicia-dataton-2019/pageviews_complemento.zip",
parse_dates=["FEC_EVENT"],
),
],
sort=False,
)
pageviews["mes"] = pageviews["FEC_EVENT"].apply(
lambda x: x.month
) # crea la columna mes
pageviews["diasemana"] = pageviews["FEC_EVENT"].apply(
lambda x: x.weekday()
) # crea la columna que indica el dia de la semana que hizo la operacion.
print("Datos iniciales del archivo pageviews:")
pageviews.head()
# Una vez cargado pageviews que es la data principal, se ve que hay tres features que hacen referencia al CONTENT: (CONTENT_CATEGORY, CONTENT_CATEGORY_TOP, CONTENT_CATEGORY_BOTTOM), donde los últimos dos hacen la construcción del primero, así que solo vamos a cargar el primero y desglosarlo para obtener toda la información adicional que podamos.
# cargar content category
content = pd.read_csv("../input/banco-galicia-dataton-2019/CONTENT_CATEGORY.zip")
print("Datos iniciales del archivo content category:")
print(content.head())
# Realizar reemplazo para alinear la data
content["CONTENT_CATEGORY_descripcion"] = content[
"CONTENT_CATEGORY_descripcion"
].str.replace("ERRORES", "ERROR")
content["CONTENT_CATEGORY_descripcion"] = content[
"CONTENT_CATEGORY_descripcion"
].str.replace("GALICIA", "")
content["CONTENT_CATEGORY_descripcion"] = content[
"CONTENT_CATEGORY_descripcion"
].str.replace(
"NO CATEGORY ASSIGNED > UPGRADE DE PAQUETE : CONFIRMACION",
"NO CATEGORY ASSIGNED > HB:NCA:NCA:NCA:UPGRADEDEPAQUETE : CONFIRMACION",
)
content["CONTENT_CATEGORY_descripcion"] = content[
"CONTENT_CATEGORY_descripcion"
].str.replace("WEB:RRHH", "WEB:INSTITUCIONAL:RRHH")
# Realiza split en columnas de la descipcion de content category
content["split"] = content["CONTENT_CATEGORY_descripcion"].str.split(" > :| : | > |:")
# Organizar la data para que quede en una estructura homogénea
for i in content.index:
if content["split"][i][0] != "NO CATEGORY ASSIGNED":
content["split"][i][0:0] = ["CATEGORY ASSIGNED"]
for i in content.index:
if len(content["split"][i]) > 2:
if content["split"][i][2] == "PERSONAS":
content["split"][i].insert(2, "ONLINE")
content["split"][i].insert(3, "WEB")
elif content["split"][i][2] == "PP":
content["split"][i].pop(-1)
elif content["split"][i][2] == "TC":
content["split"][i].pop(2)
elif content["split"][i][2] == "UPGRADE":
content["split"][i].pop(-1)
for i in content.index:
if len(content["split"][i]) > 5:
if content["split"][i][5] == "ERROR":
content["split"][i].pop()
content["split"][i][2] = "ERROR"
for i in content.index:
try:
if content["split"][i][5].find("BENEFICIOSYPROMO") >= 0:
content["split"][i].pop(6)
if content["split"][i][5].find("PRODUCTOSYS") >= 0:
content["split"][i].pop(6)
if content["split"][i][5].find("FICHAS") >= 0:
content["split"][i].pop(5)
if content["split"][i][5].find("TARJETARURAL") >= 0:
content["split"][i].pop(-1)
except IndexError:
pass
# Separar los datos de content en columnas
content[["a", "b", "c", "d", "e", "f", "g"]] = pd.DataFrame(
content["split"].values.tolist(), index=content.index
)
# Una vez tenemos los datos organizados, existen varias filas que son idénticas, esta información la podemos agrupar para tener en la tabla original menos valores únicos
# Crea el diccionario repcontent con el content category para reemplazar en la data
cols_t = ["CONTENT_CATEGORY_descripcion", "CONTENT_CATEGORY", "split"]
repcontent = dict()
i = 0
for npage in list(
content[content.drop(cols_t, axis=1).duplicated(keep=False)]["CONTENT_CATEGORY"]
):
i += 1
for npage2 in list(
content[content.drop(cols_t, axis=1).duplicated(keep=False)]["CONTENT_CATEGORY"]
)[i:]:
n = (
content[content["CONTENT_CATEGORY"] == npage]
.drop(cols_t, axis=1)
.merge(content[content["CONTENT_CATEGORY"] == npage2].drop(cols_t, axis=1))
.shape[0]
)
if n == 1:
if (npage == 4) | (npage == 16):
continue
if npage2 in repcontent:
continue
else:
repcontent[npage2] = npage
content.drop(
["CONTENT_CATEGORY_descripcion", "a", "b", "c", "d", "e", "f", "g"],
axis=1,
inplace=True,
) # se borran columnas que ya no se necesitan
print("Datos finales del archivo content category:")
print(content.head())
# Como pueden ver la data de CONTENT_CATEGORY ahora esta mas estructurada. De todos los archivos adicionales PAGE tiene igualmente información complementaria de la acción que realizó el usuario, con el objetivo de unir esta información a de del CONTENT, se procede a realizar una reestructuración de esta data.
# cargar archivo page
page = pd.read_csv("../input/banco-galicia-dataton-2019/PAGE.zip")
print("Datos iniciales del archivo page:")
print(page.head())
# Crear diccionario para remplazar valores con el objetivo de generalizar y limpiar los datos
replace_dict = {
"HB : :": "HB:NCA:NCA:NCA:",
"HB :": "HB:NCA:NCA:",
"HC :": "HC:NCA:NCA:NCA:",
"^/": "NCA/NCA/NCA/",
"^0": "",
" ": "",
"(": "",
")": "",
"-": "_",
"2018": "",
"%C3%A9MINENT": "EMINENT",
"%20/%20%3CBR%": "",
"%20/%20%3CBR": "",
"A%20PAGAR:%20": "PAGAR",
"VISAHOME": "TARJETAS",
"_VALIDACION": "/VALIDACION",
"/SELECCIONAR_": "/SELECCIONAR/",
"USUARIO_": "USUARIO/",
"UPGRADE_": "UPGRADE/",
"_AVISO": "/AVISO",
"TYC_BAJAPRODUCTO": "BAJAPRODUCTO/TYC",
"_TYC": "TYC",
"TITULAR_": "TITULAR/",
"TARJETASAS": "TARJETAS",
"TADEC": "TAC",
"2017": "",
"15$": "",
"_SOLICITAR": "/SOLICITAR",
"/PREGUNTAS_": "/PREGUNTAS/",
"_PREGUNTA_": "/PREGUNTAS_",
"CONFIRMACION": "CONFIRMAR",
"_AFIP": "/AFIP",
"PRE_CONFIRMAR": "PRECONFIRMAR",
"OPRECONFIRMAR": "O/PRECONFIRMAR",
"_PRECONFIRMACI$": "/PRECONFIRMAR",
"_PRECONFIRMA$": "/PRECONFIRMAR",
"_PRECONFIRMAR": "/PRECONFIRMAR",
"_NUMERO": "/NUMERO",
"_NUEVO": "/NUEVO",
"_MOTIVO": "/MOTIVO",
"O_MONTO": "O/MONTO",
":HOME": ":HOM",
"R_MONTO$": "R/MONTO",
"A_MONTO$": "A/MONTO",
"_MONEDA": "/MONEDA",
"_METODO": "/METODO",
"_INGRESO$": "/INGRESO",
"_INGRESAR_": "/INGRESAR_",
"_IDENTIFI": "/IDENTIFI",
"HIPOTECARIO_": "HIPOTECARIO/",
"HACETEMOVE": "HACETECLIENTE",
"HACETEGALICIA": "HACETECLIENTE",
"%20Y%20": "Y",
"_FIRMA": "/FIRMA",
"_FIMA_": "_FIMA/",
"_EXITO": "/EXITO",
"_ELECCION": "/ELECCION",
"_DELIVERY": "/DELIVERY",
"_DATOS_P": "/DATOS_P",
"_CONTACTO": "/CONTACTO",
"_CONFIRMACIO$": "/CONFIRMAR",
"_CONFIRMAR": "/CONFIRMAR",
"_CONFIR$": "/CONFIRMAR",
"MOVI": "MOV",
"BUP": "",
"_CONCEPTO": "/CONCEPTO",
"_CODIGO": "/CODIGO",
"S_CANTIDAD": "S/CANTIDAD",
"_CAJEROS": "/CAJEROS",
"_ADICIONALES": "/ADICIONALES",
"/BENEFICIOS": "/BENEFICIOSYPROMOCIONES",
"_BENEFICIARIO": "/BENEFICIARIO",
"BANCO": "",
"_AUTORIZADOS": "/AUTORIZADOS",
"/1521": "",
"_ALERTA": "/ALERTA",
"_ADHERIR": "/ADHERIR",
"/PREFER": "/PREFERPREFER",
"WEB:PREFER": "WEB:PREFERPREFER",
"PROYECTO": "PROYEC",
"_": "",
"WEB:PERSONAS": "WEB:PERSONASPERSONAS",
"PEDITUSPRODUCTOS": "PEDIDOSHOM",
"OFFICEBANKIN": "",
"WEB:MOVE": "WEB:MOVEMOVE",
"GALICI$": "",
"WEB:EMINENT": "WEB:EMINENTEMINENT",
"UNHANDLEDERROR": "ERROR",
"TOMAIL": "TO/MAIL",
"TARJETASR": "TARJETAS/R",
"PAQUETES": "PAQUETE",
"FIRMATERMINOSYCONDICIONE": "TYC",
"TERMINOSYCONDICIONES": "TYC",
"TERMINOSYCONDICIONE": "TYC",
"TERMINOSYCONDICION": "TYC",
"/TYC": "TYC",
"TYC": "/TYC",
"SW$": "",
"SUCURSAL99": "SUCURSALES",
"SUCURSAL$": "SUCURSALES",
"SPOTIF": "SPOTIFY",
"SOLICITARHIPOTECARIO": "HIPOTECARIO",
"/RROR/": "/ERROR/",
"RENOVACIONINICIO": "RENOVACION",
"PRIMER/INGRESO": "PRIMERINGRESO",
"PREFER$": "",
"PERSONAS$": "",
"MOVIMI": "MOV",
"SUCURSALESYCAJEROS": "SUCURSAL",
"PROYECTO": "PROYEC",
"PRECONFIRMARREPOSICION": "REPOSICION",
"CONFIRMARREPOSICION": "REPOSICION",
"PRECONFIRMAR/NUEVO": "PRECONFIRMAR",
"FONDOS": "FONDO",
"PEDIDOPRESTAMOSPERSONALES": "PRESTAMOS",
"PEDIDOS:": "PEDIDOS",
"MOVE$": "",
"OPERACIONES:": "OPERACIONES",
"OBJETIVO$": "OBJETIV",
"MAS$": "",
"NOMINALES$": "NOMINALE",
"NOLATENES": "NOLARECORDAS",
"GALIC$": "",
"NOLARECUERDA": "NOLARECORDAS",
"MONTOOBJETIV$": "MONTO",
"MOMENTONO": "ERRO",
"MESSAGE": "MENSAJE",
"LOGBB": "LOGIN",
"ERRORES": "ERROR",
"JAVASCRIPT:VOID": "JAVASCRIPT",
"INVERSFSIONES": "INVERSIONES",
"GALI$": "",
"INVERSIONES$": "INVERSION",
"INICIO:RE$": "INICIO:RECU",
"INGRESARDOMICILI$": "INGRESARDOMICILIO",
"INICIO:R$": "INICIO:RECU",
"GAL$": "",
"CUENTGALICIA": "CUENTAS",
"FUERADESERVICIO": "ERROR",
"GALICIA": "",
"INICIONUEVOCONTACTOC": "NUEVOCONTACTO/C",
"FILE/DOWNLOAD": "DOWNLOADFILE",
"FECHANACIMIENTO": "/FECHANACIMIENTO",
"FECHAOBJETIV": "/FECHAOBJETIV",
"DECRE": "D",
"ERORIN": "ERROR",
"EGOORROR": "ERROR",
"EMINENT2": "",
"EMINENT$": "",
"EMINEN$": "",
"EMINE$": "",
"EMINET$": "",
"EMI$": "",
"EMAIL": "/EMAIL",
"DETAL$": "DETALLE",
"DETA$": "DETALLE",
"DESCRIPCIONSEGURO": "DESCRIPCION",
"DESCRIPCIONPRESTAMO": "DESCRIPCION",
"ALERTAS": "ALERTA",
"CONVENIOSRURAL": "CONVENIOS",
"TOCONFIRMAR": "TO/CONFIRMAR",
"CARREFOUR": "",
"CONFIRMAR/ALERTA": "CONFIRMAR",
"CARGARMONTOCUENTASPROPIAS": "CUENTASPROPIAS/MONTO",
"BONOSACCIONES": "BONOSYACCIONES",
"\.$": "",
"APERTURADEC": "APERTURAD",
"APERTURADE": "APERTURAD", #
}
# Realizar reemplazo para arreglar los datos
for old, new in replace_dict.items():
page["PAGE_descripcion"] = page["PAGE_descripcion"].str.replace(old, new)
# crea diccionario que corrige el content en la data
fixpage = dict()
for i in page.index:
if ("HB" in page["PAGE_descripcion"][i]) and (
"ERRO" in page["PAGE_descripcion"][i]
):
fixpage[i + 1] = 5
elif ("HB" in page["PAGE_descripcion"][i]) and (
"PAQUETE" in page["PAGE_descripcion"][i]
):
continue
elif ("HB" in page["PAGE_descripcion"][i]) and (
"PRESTAMOS" in page["PAGE_descripcion"][i]
):
continue
elif "HB" in page["PAGE_descripcion"][i]:
fixpage[i + 1] = 4
# realiza split1 en columnas de la descipcion de page
page["split1"] = page["PAGE_descripcion"].str.split("://|/|:") # check 982
# Organiza los datos de page
for i in page.index:
if (
page["split1"][i][0] == "WEB"
or page["split1"][i][0] == "RURAL"
or page["split1"][i][0] == "RRHH"
or page["split1"][i][0] == "PREFER"
or page["split1"][i][0] == "PERSONAS"
or page["split1"][i][0] == "PAGINA"
or page["split1"][i][0] == "NEGOCIOSYPYMES"
or page["split1"][i][0] == "MOVE"
or page["split1"][i][0] == "INSTITUCIONAL"
or page["split1"][i][0] == "HB"
or page["split1"][i][0] == "EMPRESAS"
or page["split1"][i][0] == "EMINENT"
or page["split1"][i][0] == "CORPORATIVAS"
or page["split1"][i][0] == "BUSCADORDECAJEROSYSUCURSALES"
or page["split1"][i][0] == "PRODUCTOSYSERVICIOS"
or page["split1"][i][0] == "BENEFICIOSYPROMOCIONES"
):
page["split1"][i].pop(0)
if page["split1"][i][-1] == "":
page["split1"][i].pop(-1)
try:
if page["split1"][i][0] == "HC":
page["split1"][i].pop(0)
if page["split1"][i][0].startswith("STEP") == True:
page["split1"][i].pop(0)
if page["split1"][i][0] == "ERROR":
page["split1"][i].insert(0, "NCA")
if page["split1"][i][4].startswith("PASO") == True:
page["split1"][i].pop(4)
if page["split1"][i][4] == "ERROR":
page["split1"][i].pop(5) # *# borra las explicaciones del error
if page["split1"][i][3].find("INICI") >= 0:
page["split1"][i][3] = "INICIO"
if page["split1"][i][3].find("OGIN") >= 0:
page["split1"][i][3] = "LOGIN"
if page["split1"][i][3].find("EMPRESA") >= 0:
page["split1"][i].pop(0)
if page["split1"][i][3].find("USER") >= 0:
page["split1"][i][3] = "USERS"
page["split1"][i].pop(0)
if page["split1"][i][3].find("LOGI") >= 0:
page["split1"][i][3] = "LOGIN"
if page["split1"][i][3].find("PREFER") >= 0:
page["split1"][i].pop(0)
if page["split1"][i][3].find("ERR") >= 0:
page["split1"][i][3] = "ERROR"
if page["split1"][i][2] == "NCA":
page["split1"][i][0] = "ERROR"
page["split1"][i].insert(3, "NCA")
page["split1"][i].pop(5) # *# borra las explicaciones del error
else:
page["split1"][i].pop(0)
page["split1"][i].insert(2, "NCA")
if (page["split1"][i][4].startswith("ERRO") == True) and (
page["split1"][i][0] == "NCA"
):
page["split1"][i][4] = "ERROR"
page["split1"][i].pop(0)
page["split1"][i].pop(4) # *# borra las explicaciones del error
if page["split1"][i][4].startswith("PASO") == True:
page["split1"][i].pop(5) # *# borra las explicaciones del paso
if page["split1"][i][4].startswith("ALERT") == True:
page["split1"][i].pop(5) # *# borra las explicaciones de alertas
if page["split1"][i][4].find("INICIO") >= 0:
page["split1"][i][4] = "INICIO"
if (page["split1"][i][4].startswith("PRECONFIRMAR") == True) and (
page["split1"][i][4] != "PRECONFIRMAR"
):
page["split1"][i][4] = page["split1"][i][4][12:]
page["split1"][i].append("PRECONFIRMAR")
if (page["split1"][i][4].startswith("CONFIRMAR") == True) and (
page["split1"][i][4] != "CONFIRMAR"
):
page["split1"][i][4] = page["split1"][i][4][9:]
page["split1"][i].append("CONFIRMAR")
if page["split1"][i][5].find("PAGAR") >= 0:
page["split1"][i][5] = "PAGAR"
if page["split1"][i][-1].isdigit():
page["split1"][i].pop(-1)
if page["split1"][i][-1] == "":
page["split1"][i].pop(-1)
except IndexError:
pass
# Separar los datos de page en columnas
page[["a", "b", "c", "d", "e", "f"]] = pd.DataFrame(
page["split1"].values.tolist(), index=page.index
)
# crea el diccionario reppage con el page para reemplazar en la data valores repetidos
cols_t = ["PAGE_descripcion", "PAGE", "split1"]
reppage = dict()
i = 0
for npage in list(page[page.drop(cols_t, axis=1).duplicated(keep=False)]["PAGE"]):
i += 1
for npage2 in list(page[page.drop(cols_t, axis=1).duplicated(keep=False)]["PAGE"])[
i:
]:
n = (
page[page["PAGE"] == npage]
.drop(cols_t, axis=1)
.merge(page[page["PAGE"] == npage2].drop(cols_t, axis=1))
.shape[0]
)
if n == 1:
if npage2 in reppage:
continue
else:
reppage[npage2] = npage
page.drop(
[
"PAGE_descripcion",
"a",
"b",
"c",
"d",
"e",
"f",
],
axis=1,
inplace=True,
) # se borran columnas que ya no se necesitan
print("Datos finales del archivo page:")
print(page.head())
# Aplicamos los diccionarios que creamos para unificar la data repetida que surgio al organizar la data
for i in fixpage:
pageviews.loc[pageviews["PAGE"] == i, "CONTENT_CATEGORY"] = fixpage[i]
pageviews.loc[pageviews["PAGE"] == i, "CONTENT_CATEGORY_BOTTOM"] = fixpage[i]
for i in repcontent:
pageviews.loc[
pageviews["CONTENT_CATEGORY"] == i, "CONTENT_CATEGORY_BOTTOM"
] = repcontent[i]
pageviews.loc[pageviews["CONTENT_CATEGORY"] == i, "CONTENT_CATEGORY"] = repcontent[
i
]
for i in reppage:
pageviews.loc[pageviews["PAGE"] == i, "PAGE"] = reppage[i]
pageviews = pd.merge(pageviews, content) # se une la columna split a pageviews
pageviews = pd.merge(pageviews, page) # se une la columna split1 a pageviews
pageviews["s_total"] = (
pageviews["split"] + pageviews["split1"]
) # se unen la columnas split y split1 en una sola
# La siguiente línea separa en columnas cada dato de la lista de s_total, aquí se entiende el objetivo de la
# organización que se realizó, quedando primero si tiene o no categoría definida, luego su tipo (banca, HB, HC, …),
# luego si fue un error o una acción online, luego esta web que es una constante por lo que más adelante no la
# tendremos en cuenta, TyPEr define el tipo de usuario (persona, empresa, negocios, …), ACT específica la acción del
# usuario (login, consulta, pedido de préstamo, …), A1 y A2 especifica información adicional de la acción del usuario.
pageviews[["Cat", "TyCat", "on_err", "web", "TyPer", "Act", "A1", "A2"]] = pd.DataFrame(
pageviews["s_total"].values.tolist(), index=pageviews.index
)
pageviews.drop(
["split1", "split", "web"], axis=1, inplace=True
) # Se borran las columnas que ya no se necesitan
pageviews["on_err"].replace(r"^NCA$", "ONLINE", inplace=True, regex=True)
devicedata = pd.read_csv(
"../input/banco-galicia-dataton-2019/device_data.zip", parse_dates=["FEC_EVENT"]
)
data = pageviews.merge(devicedata, how="outer")
del pageviews
data.dropna(axis="rows", thresh=7, inplace=True)
data.sort_values(["USER_ID", "FEC_EVENT"], inplace=True)
data.reset_index(drop=True, inplace=True)
data["TyPer"].replace(r"^NCA$", np.NaN, inplace=True, regex=True)
data["TyPer"].replace(r"^MASIVO$", np.NaN, inplace=True, regex=True)
data["TyPer"].replace(r"^USERS$", np.NaN, inplace=True, regex=True)
data["TyPer"].fillna(method="ffill", inplace=True)
data["TyPer"].fillna("EMINENT", inplace=True)
data["CONNECTION_SPEED"].fillna(method="ffill", inplace=True)
data["IS_MOBILE_DEVICE"].fillna(method="ffill", inplace=True)
data["CONNECTION_SPEED"].fillna(1, inplace=True)
data["IS_MOBILE_DEVICE"].fillna(1, inplace=True)
data.fillna("NCA", inplace=True)
data.head()
data["s_total"] = data["s_total"].apply(", ".join)
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
data["Cat"] = le.fit_transform(data["Cat"])
data["TyCat"] = le.fit_transform(data["TyCat"])
data["on_err"] = le.fit_transform(data["on_err"])
data["TyPer"] = le.fit_transform(data["TyPer"])
data["Act"] = le.fit_transform(data["Act"])
data["A1"] = le.fit_transform(data["A1"])
data["A2"] = le.fit_transform(data["A2"])
data["s_total"] = le.fit_transform(data["s_total"])
data.head()
conversiones = pd.read_csv("../input/banco-galicia-dataton-2019/conversiones.zip")
columnas = list(
data.drop(
["FEC_EVENT", "CONTENT_CATEGORY_BOTTOM", "USER_ID", "mes"], axis=1
).columns
)
# Se descarta la columna CONTENT_CATEGORY_BOTTOM ya que es igual a la columna CONTENT_CATEGORY, esto se puede probar con la siguente linea
# data['CONTENT_CATEGORY'].equals(data['CONTENT_CATEGORY_BOTTOM'])
meses = [[1, 9, 4, 12]]
from sklearn import model_selection
from lightgbm import LGBMClassifier
from sklearn.metrics import roc_auc_score
alldata = []
for c in columnas:
temp = pd.crosstab(data.USER_ID, data[c])
temp.columns = [c + "_" + str(v) for v in temp.columns]
alldata.append(temp.apply(lambda x: x / x.sum(), axis=1))
alldata = pd.concat(alldata, axis=1)
allcolumns = list(alldata.columns)
test_probs = []
for i in range(len(meses)):
data2 = data[(data["mes"] >= meses[i][0]) & (data["mes"] <= meses[i][1])]
y_train = pd.Series(
1,
conversiones[
(conversiones["mes"] >= meses[i][2]) & (conversiones["mes"] <= meses[i][3])
]["USER_ID"]
.sort_values()
.unique(),
)
y_train = y_train.reindex(range(11676), fill_value=0)
X_train = []
for c in columnas:
temp = pd.crosstab(data.USER_ID, data2[c])
temp.columns = [c + "_" + str(v) for v in temp.columns]
X_train.append(temp.apply(lambda x: x / x.sum(), axis=1))
X_train = pd.concat(X_train, axis=1)
X_train = X_train.reindex(range(11676), columns=allcolumns, fill_value=0)
j = 0
for train_idx, valid_idx in model_selection.KFold(
n_splits=10, shuffle=True, random_state=29
).split(X_train):
j += 1
Xt = X_train.iloc[train_idx]
yt = y_train.iloc[train_idx]
Xv = X_train.iloc[valid_idx]
yv = y_train.iloc[valid_idx]
learner = LGBMClassifier(
n_estimators=10000, objective="binary", random_state=seed
)
learner.fit(
Xt,
yt,
early_stopping_rounds=20,
eval_metric="auc",
eval_set=[(Xt, yt), (Xv, yv)],
verbose=0,
)
test_probs.append(
pd.Series(
learner.predict_proba(alldata)[:, -1],
index=alldata.index,
name="fold_" + str(j),
)
)
test_probs = pd.concat(test_probs, axis=1).mean(axis=1)
test_probs.index.name = "USER_ID"
test_probs.name = "SCORE"
test_probs.to_csv("resultado.csv", header=True)
test_probs
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
from sklearn.metrics import confusion_matrix
import xgboost as xgb
X_train = pd.read_csv("../input/xgb-fraud-with-magic-0-9600/X_train.csv")
X_test = pd.read_csv("../input/xgb-fraud-with-magic-0-9600/X_test.csv")
y_train = pd.read_csv("../input/xgb-fraud-with-magic-0-9600/y_train.csv", header=None)
def fast_auc(y_true, y_prob):
y_true = np.asarray(y_true)
y_pred = y_true[np.argsort(y_prob)]
nfalse = 0
auc = 0
n = len(y_pred)
for i in range(n):
y_i = y_pred[i]
nfalse += 1 - y_i
auc += y_i * nfalse
auc = np.array(auc, dtype="f")
auc /= nfalse * (n - nfalse)
return auc
def spec(y_true, y_prob):
y_true = np.asarray(y_true)
y_pred = y_true[np.argsort(y_prob)]
tn, fp, fn, tp = confusion_matrix(y_true, y_pred).ravel()
specificity = tn / (tn + fp)
return specificity
def sens(y_true, y_prob):
y_true = np.asarray(y_true)
y_pred = y_true[np.argsort(y_prob)]
tn, fp, fn, tp = confusion_matrix(y_true, y_pred).ravel()
sensitivity = tp / (tp + fn)
return sensitivity
from sklearn.model_selection import KFold
kf = KFold(n_splits=5)
fold_score = []
spec_score = []
sens_score = []
for fold, (train_idx, test_idx) in enumerate(kf.split(X_train)):
clf = xgb.XGBClassifier(
objective="binary:logistic",
n_estimators=2000,
max_depth=12,
learning_rate=0.02,
subsample=0.8,
colsample_bytree=0.4,
missing=-1,
eval_metric=["auc", "logloss"],
nthread=4,
tree_method="hist",
)
clf.fit(
X_train.loc[train_idx],
y_train.iloc[:, 1][train_idx],
eval_set=[(X_train.loc[test_idx], y_train.iloc[:, 1][test_idx])],
verbose=50,
early_stopping_rounds=100,
)
fold_score.append(
fast_auc(
y_train.iloc[:, 1][test_idx], clf.predict_proba(X_train.loc[test_idx])[:, 1]
)
)
spec_score.append(
spec(
y_train.iloc[:, 1][test_idx], clf.predict_proba(X_train.loc[test_idx])[:, 1]
)
)
sens_score.append(
sens(
y_train.iloc[:, 1][test_idx], clf.predict_proba(X_train.loc[test_idx])[:, 1]
)
)
clf.save_model("model_fold_" + str(fold) + ".json")
print("AUC score = %1.5f" % np.mean(fold_score))
print("Specificity score = %1.5f" % np.mean(spec_score))
print("Sensitivity score = %1.5f" % np.mean(sens_score))
result = []
for fold in range(2):
clf = xgb.XGBClassifier()
clf.load_model("model_fold_" + str(fold) + ".json")
result.append(clf.predict_proba(X_test)[:, 1])
submission_df = pd.read_csv("../input/ieee-fraud-detection/sample_submission.csv")
submission_df["isFraud"] = np.mean(result, axis=0)
submission_df.to_csv("submission.csv", index=False)
from fastapi import FastAPI
from pydantic import BaseModel
app = FastAPI()
class Transaction(BaseModel):
transaction_id: str
transaction_amt: float
product_cd: str
@app.post("/predict")
async def predict(transaction: Transaction):
# Preprocess the input data
df_transaction = pd.DataFrame.from_dict([transaction.dict()])
df_transaction = preprocess_data(df_transaction)
# Generate a prediction using the trained model
prediction = model.predict(df_transaction)
is_fraud = bool(prediction[0])
return {"transaction_id": transaction.transaction_id, "isFraud": is_fraud}
import subprocess
subprocess.run(["uvicorn", "main:app", "--reload"])
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
data = pd.read_csv("/kaggle/input/szeged-weather/weatherHistory.csv")
Humidity = data[["Humidity"]]
Temperature = data[["Temperature (C)"]]
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(
Humidity, Temperature, test_size=0.33, random_state=0
)
from sklearn.linear_model import LinearRegression
# Sınıftan bir nesne oluşturuyoruz.
lr = LinearRegression()
# Train veri kümelerini vererek makineyi eğitiyoruz.
lr.fit(x_train, y_train)
import matplotlib.pyplot as plt
# Aylar'ın test kümesini vererek Satislar'ı tahmin etmesini sağlıyoruz. Üst satırda makinemizi eğitmiştik.
tahmin = lr.predict(x_test)
# Verileri grafikte düzenli göstermek için index numaralarına göre sıralıyoruz.
x_train = x_train.sort_index()
y_train = y_train.sort_index()
# Grafik şeklinde ekrana basıyoruz.
plt.plot(x_train, y_train)
plt.plot(x_test, tahmin)
plt.xlabel("Humidity")
plt.ylabel("Temperature")
plt.show()
|
# Thank you for reading this notebook. I'm new to Kaggle and machine-learing algorithms, and this competition is the second one for me after TPS-January. I didn't use any special techniques, but used GBDT modules I found common in Kaggle (LightGBM, XGBoost, and CatBoost). In this notebook I wrote down the basic flows I used in this competition. I don't suppose this will interest those who has been familiar with Kaggle, but I would appreciate it if you could read this and give me some advice. I'm also glad if this notebook would help other beginners.
# **0. Import modules and dataset**
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import optuna
from tqdm.notebook import tqdm
from sklearn.cluster import KMeans
from sklearn.mixture import GaussianMixture
from sklearn.metrics import mean_squared_error
from sklearn.preprocessing import LabelEncoder
from sklearn.model_selection import train_test_split, KFold
from lightgbm import LGBMRegressor, plot_importance
train = pd.read_csv("/kaggle/input/tabular-playground-series-feb-2021/train.csv")
test = pd.read_csv("/kaggle/input/tabular-playground-series-feb-2021/test.csv")
cont_features = [f for f in train.columns.tolist() if f.startswith("cont")]
cat_features = [f for f in train.columns.tolist() if f.startswith("cat")]
features = cat_features + cont_features
data = train[features]
target = train["target"]
all_data = pd.concat([data, test])
# # 1. Feature Engineering
# I did a slight feature-engineering.
# Histograms of the cont features show multiple components. For instance, the cont1 has 7 discrete peaks as shown below. I thought these characteristics could be used as an additional feature.
# So, I tried `sklearn.mixture.GaussianMixture` to devide into several groups [Ref: [Notebooks of TPS-Jan. by Dave E](https://www.kaggle.com/davidedwards1/jan21-tabplayground-nn-final-fewer-features)].
# See also https://scikit-learn.org/stable/modules/mixture.html#gmm for Gaussian Mixture Models.
# The scatter plots below show the cont-feature values and target, with the results of GMM.
# The bottom histgrams also show the results of GMM.
fig, ax = plt.subplots(5, 3, figsize=(14, 24))
for i, feature in enumerate(cont_features):
plt.subplot(5, 3, i + 1)
sns.histplot(all_data[feature][::100], color="blue", kde=True, bins=100)
plt.xlabel(feature, fontsize=9)
plt.show()
inits = [
[0.3, 0.5, 0.7, 0.9],
[0.039, 0.093, 0.24, 0.29, 0.35, 0.42, 0.49, 0.56, 0.62, 0.66, 0.76],
[0.176, 0.322, 0.416, 0.495, 0.548, 0.618, 0.707, 0.937],
[0.2, 0.35, 0.44, 0.59, 0.75, 0.83],
[0.28, 0.31, 0.42, 0.5, 0.74, 0.85],
[0.25, 0.38, 0.43, 0.58, 0.75, 0.9],
[0.34, 0.48, 0.7, 0.88],
[0.25, 0.29, 0.35, 0.48, 0.61, 0.68, 0.78, 0.9],
[0.11, 0.2, 0.3, 0.35, 0.45, 0.6, 0.76, 0.9],
[0.22, 0.32, 0.38, 0.44, 0.53, 0.63, 0.71, 0.81, 0.87],
[0.19, 0.27, 0.37, 0.46, 0.56, 0.61, 0.71, 0.86],
[0.23, 0.35, 0.52, 0.7, 0.84],
[0.27, 0.32, 0.35, 0.49, 0.63, 0.7, 0.79, 0.88],
[0.22, 0.29, 0.35, 0.4, 0.47, 0.58, 0.68, 0.72, 0.8],
]
gmms = []
for feature, init in zip(cont_features, inits):
X_ = np.array(all_data[feature].tolist()).reshape(-1, 1)
means_init = np.array(init)[:, None]
gmm_ = GaussianMixture(
n_components=len(init), means_init=means_init, random_state=0
).fit(X_)
gmms.append(gmm_)
preds = gmm_.predict(X_)
all_data[f"{feature}_gmm"] = preds
train[f"{feature}_gmm"] = preds[: len(train)]
test[f"{feature}_gmm"] = preds[len(train) :]
fig, ax = plt.subplots(5, 3, figsize=(24, 30))
for i, feature in enumerate(cont_features):
plt.subplot(5, 3, i + 1)
sns.scatterplot(
x=feature, y="target", data=train[::150], hue=f"{feature}_gmm", palette="muted"
)
plt.xlabel(feature, fontsize=9)
plt.show()
fig, ax = plt.subplots(5, 3, figsize=(24, 30))
for i, feature in enumerate(cont_features):
plt.subplot(5, 3, i + 1)
sns.histplot(
x=feature,
data=train[::150],
hue=f"{feature}_gmm",
kde=True,
bins=100,
palette="muted",
)
plt.xlabel(feature, fontsize=9)
plt.show()
|
import warnings
warnings.simplefilter(action="ignore", category=FutureWarning)
from sklearn import metrics
from sklearn.preprocessing import (
LabelEncoder,
StandardScaler,
MinMaxScaler,
RobustScaler,
)
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
if "csv" in filename:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
# # Utils
def input_train_test():
root = "../input/unsw-nb15/"
train = pd.read_csv(root + "UNSW_NB15_training-set.csv")
test = pd.read_csv(root + "UNSW_NB15_testing-set.csv")
if train.shape[0] == 82332:
print("Train and test sets are reversed here. Fixing them.")
train, test = test, train
drop_columns = ["attack_cat", "id"]
for df in [train, test]:
for col in drop_columns:
if col in df.columns:
print("Dropping " + col)
df.drop([col], axis=1, inplace=True)
return train, test
def get_cat_columns(train):
categorical = []
for col in train.columns:
if train[col].dtype == "object":
categorical.append(col)
return categorical
def label_encode(train, test):
for col in get_cat_columns(train):
le = LabelEncoder()
le.fit(list(train[col].astype(str).values) + list(test[col].astype(str).values))
train[col] = le.transform(list(train[col].astype(str).values))
test[col] = le.transform(list(test[col].astype(str).values))
return train, test
def feature_process(df):
df.loc[~df["state"].isin(["FIN", "INT", "CON", "REQ", "RST"]), "state"] = "others"
df.loc[
~df["service"].isin(
["-", "dns", "http", "smtp", "ftp-data", "ftp", "ssh", "pop3"]
),
"service",
] = "others"
df.loc[df["proto"].isin(["igmp", "icmp", "rtp"]), "proto"] = "igmp_icmp_rtp"
df.loc[
~df["proto"].isin(["tcp", "udp", "arp", "ospf", "igmp_icmp_rtp"]), "proto"
] = "others"
return df
def get_train_test(
train, test, feature_engineer=True, label_encoding=False, scaler=None
):
x_train, y_train = train.drop(["label"], axis=1), train["label"]
x_test, y_test = test.drop(["label"], axis=1), test["label"]
x_train, x_test = feature_process(x_train), feature_process(x_test)
if scaler is not None:
categorical_columns = get_cat_columns(x_train)
non_categorical_columns = [
x for x in x_train.columns if x not in categorical_columns
]
x_train[non_categorical_columns] = scaler.fit_transform(
x_train[non_categorical_columns]
)
x_test[non_categorical_columns] = scaler.transform(
x_test[non_categorical_columns]
)
if label_encoding:
x_train, x_test = label_encode(x_train, x_test)
features = x_train.columns
else:
x_train = pd.get_dummies(x_train)
x_test = pd.get_dummies(x_test)
print(
"Column mismatch {0}, {1}".format(
set(x_train.columns) - set(x_test.columns),
set(x_test.columns) - set(x_train.columns),
)
)
features = list(set(x_train.columns) & set(x_test.columns))
print(f"Number of features {len(features)}")
x_train = x_train[features]
x_test = x_test[features]
return x_train, y_train, x_test, y_test
def run_lgb(x, y, tr_idx, val_idx, num_round=100):
train = lgb.Dataset(x.iloc[tr_idx], y[tr_idx])
x_val, y_val = x.iloc[val_idx], y[val_idx]
validation = lgb.Dataset(x_val, y_val)
clf = lgb.train(
param,
train,
num_round,
valid_sets=[validation],
early_stopping_rounds=50,
verbose_eval=200,
)
return clf
def detection_rate(y_true, y_pred):
CM = metrics.confusion_matrix(y_true, y_pred)
TN = CM[0][0]
FN = CM[1][0]
TP = CM[1][1]
FP = CM[0][1]
return TP / (TP + FN)
def false_positive_rate(y_true, y_pred):
CM = metrics.confusion_matrix(y_true, y_pred)
TN = CM[0][0]
FN = CM[1][0]
TP = CM[1][1]
FP = CM[0][1]
return FP / (FP + TN)
def results(y_test, y_pred):
print(
f"Accuracy {metrics.accuracy_score(y_test, y_pred)*100}, F1-score {metrics.f1_score(y_test, y_pred)*100}"
)
# print(metrics.classification_report(y_test, y_pred))
print(
"DR {0}, FPR {1}".format(
detection_rate(y_test, y_pred) * 100,
false_positive_rate(y_test, y_pred) * 100,
)
)
train, test = input_train_test()
# # Train with LightGBM
from sklearn.model_selection import StratifiedKFold
import lightgbm as lgb
from tqdm import tqdm_notebook as tqdm
def lgb_accuracy(preds, data):
y_true = data.get_label()
y_pred = np.round(preds)
return "acc", metrics.accuracy_score(y_true, y_pred), True
def lgb_f1_score(preds, data):
y_true = data.get_label()
y_pred = np.round(preds) # scikits f1 doesn't like probabilities
return "f1", metrics.f1_score(y_true, y_pred), True
folds = 10
seed = 1
num_round = 2000
kf = StratifiedKFold(n_splits=folds, shuffle=True, random_state=seed)
drop_columns = [
"is_sm_ips_ports",
"dwin",
"is_ftp_login",
"trans_depth",
"dttl",
"ct_ftp_cmd",
]
for df in [train, test]:
df.drop(drop_columns, axis=1, inplace=True)
x_train, y_train, x_test, y_test = get_train_test(
train, test, feature_engineer=True, label_encoding=False, scaler=MinMaxScaler()
)
# After dropping irrelevant columns, feature engineering and applying oneHotEncoding. Inside () performance at prediction threshold 0.85 is specified.
# |Preprocess| Param | Train Acc | Test Acc |
# |:---:|:---:|:---:|:---:|
# |RobustScaler|learning rate 0.05, metric binary logloss|96.34(94.05)|87.5(93.98)|
# ||learning rate 0.05, metric binary logloss, feature_traction 0.5|96.33(93.07)|87.81(93.31)|
# |StandardScaler |learning rate 0.05, metric binary logloss|96.32(93.93)| 87.6 (93.92)|
# |MinMaxScaler |learning rate 0.05, metric binary logloss|96.35(94.08)| 87.63 (94.02)|
param = {
"objective": "binary",
"learning_rate": 0.05,
# 'max_depth': 5,
# 'bagging_fraction': 0.8,
# 'bagging_freq': 0,
# 'feature_fraction':0.5,
"metric": "auc",
}
accuracies = {0.5: [], 0.85: [], 0.9: []}
for tr_idx, val_idx in tqdm(kf.split(x_train, y_train), total=folds):
clf = run_lgb(x_train, y_train, tr_idx, val_idx, num_round)
x_val, y_val = x_train.iloc[val_idx], y_train[val_idx]
y_prob = clf.predict(x_val, num_iteration=clf.best_iteration)
for key in accuracies:
y_pred = np.where(y_prob >= key, 1, 0)
accuracies[key].append(metrics.accuracy_score(y_val, y_pred))
for key in accuracies:
print(key, np.mean(accuracies[key], axis=0))
# # Test results
def test_run(param, x_train, y_train, x_test, y_test):
lgb_train = lgb.Dataset(x_train, y_train)
lgb_validation = lgb.Dataset(x_test, y_test)
# clf = lgb.train(param, lgb_train, 2000, valid_sets=[lgb_validation], early_stopping_rounds=50, verbose_eval=50, feval=lgb_f1_score)
clf = lgb.train(
param,
lgb_train,
2000,
valid_sets=[lgb_validation],
early_stopping_rounds=50,
verbose_eval=200,
)
y_prob = clf.predict(x_test, num_iteration=clf.best_iteration)
y_pred = np.where(y_prob >= 0.5, 1, 0)
results(y_test, y_pred)
return y_test, y_prob
y_test, y_prob = test_run(param, x_train, y_train, x_test, y_test)
# lgb.plot_metric(evals_result, metric='acc')
prob = 0.5
while prob <= 1:
print(prob)
y_pred = np.where(y_prob >= prob, 1, 0)
results(y_test, y_pred)
prob += 0.05
# y_pred = np.where(y_prob>=0.85, 1, 0)
# results(y_test, y_pred)
|
import gc
import numpy as np
import pandas as pd
from typing import List, Tuple, Dict
from keras.models import Model
from keras.layers import Input, Dense, Embedding, SpatialDropout1D, add, concatenate
from keras.layers import (
CuDNNLSTM,
Bidirectional,
GlobalMaxPooling1D,
GlobalAveragePooling1D,
)
from keras.preprocessing import text, sequence
from keras.callbacks import LearningRateScheduler
# a list of paths of embedding matrix files
# Two embedding matrixes will be combined together and used in the embedding layer of the RNN.
# They are CRAWL and GLOVE. Each of them is a collection of 300-dimension vector.
# Each vector represents a word.
# The coverage of words of CRAWL is different from that of GLOVE.
EMBEDDING_FILES = [
"../input/fasttext-crawl-300d-2m/crawl-300d-2M.vec",
"../input/glove840b300dtxt/glove.840B.300d.txt",
]
# <NUM_MODELS> represents the amount of times the same model should be trained
# Although each training is using the same RNN model, the predictions will be slightly different
# from each other due to different initialization (He Initialization).
NUM_MODELS = 2
# amount of epoch during training, this number is mainly limited by the GPU quota during committing.
EPOCHS = 4
# batch size
BATCH_SIZE = 256
# amount of LSTM units in each LSTM layer
LSTM_UNITS = 128
# amount of unit in the dense layer
DENSE_HIDDEN_UNITS = 4 * LSTM_UNITS
# maximum length of one comment (one sample)
MAX_LEN = 220
# column names related to identity in the training set
IDENTITY_COLUMNS = [
"male",
"female",
"homosexual_gay_or_lesbian",
"christian",
"jewish",
"muslim",
"black",
"white",
"psychiatric_or_mental_illness",
]
# a list of all the label names (Each sample/comment corresponds to multiple labels.)
AUX_COLUMNS = [
"target",
"severe_toxicity",
"obscene",
"identity_attack",
"insult",
"threat",
]
# column name of the comment column
TEXT_COLUMN = "comment_text"
# target column
TARGET_COLUMN = "target"
# chars to remove in the comment
# These chars are not covered by the embedding matrix.
CHARS_TO_REMOVE = "!\"#$%&()*+,-./:;<=>?@[\\]^_`{|}~\t\n“”’'∞θ÷α•à−β∅³π‘₹´°£€\×™√²—"
def get_coefs(word: str, *arr: str) -> (str, np.ndarray):
return word, np.asarray(arr, dtype="float32")
def load_embeddings(path: str) -> Dict[str, np.ndarray]:
"""Return a dict by analyzing the embedding matrix file under the path <path>."""
with open(path) as f:
return dict(get_coefs(*line.strip().split(" ")) for line in f)
def build_matrix(
word_index: Dict[str, int], path: str, indexesOfWordsContainTrump: List[int]
) -> np.ndarray:
"""Return an embedding matrix, which is ready to put into the RNN's embedding-matrix layer.
<word_index>: Each word corresponds to a unique index. A word's vector can be found in the embedding
matrix using the word's index.
<path>: The path where the embedding matrix file is located at.
<indexesOfWordsContainTrump>: A list of indexes of words that contain substring "Trump" or "trump".
"""
# get a word-to-vector Dict by analyzing the embedding matrix file under the path <path>
embedding_dict = load_embeddings(path)
embedding_matrix = np.zeros((len(word_index) + 1, 300))
# fill the <embedding_matrix> according to <embedding_dict>
# If a tocken/word/string contains substring "Trump" or "trump", set the tocken/word/string's
# vector to be the same as Trump's.
# If a tocken/word cannot be found in <embedding_dict>, the tocken/word's vector is set to be zeros.
# Otherwise, copy a tocken/word's vector from <embedding_dict> to <embedding_matrix>.
for word, i in word_index.items():
if i in indexesOfWordsContainTrump:
embedding_matrix[i] = embedding_dict["Trump"]
else:
try:
embedding_matrix[i] = embedding_dict[word]
except KeyError:
pass
return embedding_matrix
def build_model(embedding_matrix: np.ndarray) -> Model:
"""Return a RNN model, which uses bidirectional LSTM."""
# input layer
words = Input(shape=(None,))
# embedding matrix layer-this layer should be set to be not trainable.
x = Embedding(*embedding_matrix.shape, weights=[embedding_matrix], trainable=False)(
words
)
# The dropout operation is used to prevent overfitting.
x = SpatialDropout1D(0.2)(x)
# two bidirectional LSTM layer
# Since it is bidirectional, the output's size is twice the input's.
x = Bidirectional(CuDNNLSTM(LSTM_UNITS, return_sequences=True))(x)
x = Bidirectional(CuDNNLSTM(LSTM_UNITS, return_sequences=True))(x)
# flatten the tensor by max pooling and average pooling
# Since it is a concatenation of two pooling layer, the output's size is twice the input's.
hidden = concatenate(
[
GlobalMaxPooling1D()(x),
GlobalAveragePooling1D()(x),
]
)
# two dense layers, skip conections trick is used here to prevent gradient's vanishing.
hidden = add([hidden, Dense(DENSE_HIDDEN_UNITS, activation="relu")(hidden)])
hidden = add([hidden, Dense(DENSE_HIDDEN_UNITS, activation="relu")(hidden)])
# two different output layers
result = Dense(1, activation="sigmoid")(hidden)
aux_result = Dense(len(AUX_COLUMNS), activation="sigmoid")(hidden)
model = Model(inputs=words, outputs=[result, aux_result])
model.compile(loss="binary_crossentropy", optimizer="adam")
return model
# get the training set and test set offered in this competition
train_df = pd.read_csv(
"../input/jigsaw-unintended-bias-in-toxicity-classification/train.csv"
)
test_df = pd.read_csv(
"../input/jigsaw-unintended-bias-in-toxicity-classification/test.csv"
)
# seperate the targets and the features
x_train = train_df[TEXT_COLUMN].astype(str)
y_train = train_df[TARGET_COLUMN].values
y_aux_train = train_df[AUX_COLUMNS].values
x_test = test_df[TEXT_COLUMN].astype(str)
# change to continuous target values into discrete target values
# There are multiple targets. Each of them contains two different classes. They are True and False.
for column in IDENTITY_COLUMNS + [TARGET_COLUMN]:
train_df[column] = np.where(train_df[column] >= 0.5, True, False)
# One drawback of using Tokenizer is that it will change all characters to lower case.
# But the words in both CRAW and GLOVE are case sensitive.
# For example, "Trump" and "trump" are represented by different vectors in CRAWL or GLOVE.
# But Tokenizer will change "Trump" into "trump".
tokenizer = text.Tokenizer(filters=CHARS_TO_REMOVE)
# A word-to-index Dict will be generated internally after analyzing the train set and the test set.
tokenizer.fit_on_texts(list(x_train) + list(x_test))
# Replace all the words/tokens in train/test set by the corresponding index according to the
# internal word-to-index Dict.
x_train = tokenizer.texts_to_sequences(x_train)
x_test = tokenizer.texts_to_sequences(x_test)
# make the length of all the sequencess the same
x_train = sequence.pad_sequences(x_train, maxlen=MAX_LEN)
x_test = sequence.pad_sequences(x_test, maxlen=MAX_LEN)
# assign different weights to different samples according to their labels
# This is because different groups have different effect on the evaluation metric.
# Another reason is that the evaluation metric is too complicated to be directly used during optimization.
# The following specific weight assignment is decided after many tries.
sample_weights = np.ones(len(x_train), dtype=np.float32)
sample_weights += train_df[IDENTITY_COLUMNS].sum(axis=1)
sample_weights += train_df[TARGET_COLUMN] & (~train_df[IDENTITY_COLUMNS]).sum(axis=1)
sample_weights += (~train_df[TARGET_COLUMN]) & train_df[IDENTITY_COLUMNS].sum(axis=1)
sample_weights += (
~train_df[TARGET_COLUMN] & train_df["homosexual_gay_or_lesbian"] + 0
) * 5
sample_weights += (~train_df[TARGET_COLUMN] & train_df["black"] + 0) * 5
sample_weights += (~train_df[TARGET_COLUMN] & train_df["white"] + 0) * 5
sample_weights += (~train_df[TARGET_COLUMN] & train_df["muslim"] + 0) * 1
sample_weights += (~train_df[TARGET_COLUMN] & train_df["jewish"] + 0) * 1
sample_weights /= sample_weights.mean()
indexesOfWordsContainTrump = []
# find out all the indexes of the words that contain substring "Trump" or "trump"
for word, index in tokenizer.word_index.items():
if ("trump" in word) or ("Trump" in word):
indexesOfWordsContainTrump.append(index)
# The final embedding matrix is a concatenation of CRAWL embedding matrix and GLOVE embedding matrix.
# So each word is represented by a 600-d vector.
# In the final matrix, the words that contain substring "Trump" or "trump" are replaced by "Trump".
# This is found to be able to enhance the model performance by EDA(exploratory data analysis).
# The reason behind this is that strings like "Trump" and "trumpist" are related to toxicity,
# but they are covered neither in CRAWL nor GLOVE.
embedding_matrix = np.concatenate(
[
build_matrix(tokenizer.word_index, filePath, indexesOfWordsContainTrump)
for filePath in EMBEDDING_FILES
],
axis=-1,
)
# release memory space by deleting variables that are no longer useful
del train_df
del tokenizer
gc.collect()
# <checkpoint_predictions> is a list of predictions generated after each epoch.
checkpoint_predictions = []
# <weights> is a list of weights corresponding to <checkpoint_predictions>.
weights = []
for model_idx in range(NUM_MODELS):
model = build_model(embedding_matrix)
for global_epoch in range(EPOCHS):
model.fit(
x_train,
[y_train, y_aux_train],
batch_size=BATCH_SIZE,
epochs=1,
verbose=2,
sample_weight=[sample_weights.values, np.ones_like(sample_weights)],
callbacks=[LearningRateScheduler(lambda _: 1e-3 * (0.6**global_epoch))],
)
# record predictions after each epoch
checkpoint_predictions.append(
model.predict(x_test, batch_size=2048)[0].flatten()
)
# Since the predictions tend to be more accurate after more epochs,
# the weights is set to grow exponetially.
weights.append(2**global_epoch)
# get the weighted average of the predictions. The average operation can help prevent overfitting.
predictions = np.average(checkpoint_predictions, weights=weights, axis=0)
# output the averaged predictions to a file for submission
submission = pd.DataFrame.from_dict({"id": test_df.id, "prediction": predictions})
submission.to_csv("submission.csv", index=False)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
# import data
train = pd.read_csv("/kaggle/input/glass/glass.csv")
print(train.shape)
train.head(10)
# > Data Exploration
train.dtypes
train.describe()
train.nunique()
# Visualize the data
# Use seaborn to conduct heatmap to identify missing data
sns.heatmap(train.isnull(), cbar=False)
# > Feature Exploration
# Correlation between variables of the dataset
corr = train.corr()
fig = plt.figure()
ax = fig.add_subplot(111)
cax = ax.matshow(corr, cmap="Blues", vmin=-1, vmax=1)
fig.colorbar(cax)
ticks = np.arange(0, len(train.columns), 1)
ax.set_xticks(ticks)
plt.xticks(rotation=90)
ax.set_yticks(ticks)
ax.set_xticklabels(train.columns)
ax.set_yticklabels(train.columns)
plt.show()
# Binary Logistic Regression: The target variable has only two possible outcomes such as Window or Non Window
train["Type"] = train["Type"].apply({1: 0, 2: 0, 3: 0, 5: 1, 6: 1, 7: 1}.get)
count_non_window = len(train[train["Type"] == 1])
count_window = len(train[train["Type"] == 0])
pct_of_non_window = count_non_window / (count_non_window + count_window)
print("percentage of non window glass is", pct_of_non_window * 100)
pct_of_window = count_window / (count_non_window + count_window)
print("percentage of window glass", pct_of_window * 100)
train.groupby("Type").mean()
import matplotlib.pyplot as plt
import numpy as np
fig = plt.figure()
ax1 = fig.add_subplot(111)
ax1.scatter(train["Na"].to_numpy(), train["Type"].to_numpy(), marker="s", label="Na")
ax1.scatter(train["Al"].to_numpy(), train["Type"].to_numpy(), marker="s", label="Al")
# ax1.scatter(train['Si'].to_numpy(),train['Type'].to_numpy(),marker="s", label='si')
ax1.scatter(train["Ba"].to_numpy(), train["Type"].to_numpy(), marker="s", label="Ba")
plt.title("Glass Type")
plt.xlabel("Elements Chosen")
plt.ylabel("1:Non-Window, 0:Window)")
plt.legend(loc="center right")
ax.figure.show()
# Experimented with the scatter plot to understand the Features , taking these features as these training dataset is close.
features = ["Na", "Al", "Ba"]
X = train[features]
y = train["Type"]
# Import module to split dataset
from sklearn.model_selection import train_test_split
# Split data set into training and test sets
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.30, random_state=0
)
# Data available for Training
X_train.shape
# > Model Training
# import the Model class
from sklearn.linear_model import LogisticRegression
# instantiate the model
logistic = LogisticRegression(solver="lbfgs")
# Fit the logistic regression model.
logistic.fit(X_train, y_train)
# Get predictions
y_predict = logistic.predict(X_test)
# >MODEL Metrics
# import the metrics class
from sklearn import metrics
cnf_matrix = metrics.confusion_matrix(y_test, y_predict)
cnf_matrix
n_samples = len(y_test)
print("Accuracy: %.2f" % ((cnf_matrix[0][0] + cnf_matrix[1][1]) / n_samples))
print("Precision: %.2f" % (cnf_matrix[1][1] / (cnf_matrix[0][1] + cnf_matrix[1][1])))
print("Recall: %.2f" % (cnf_matrix[1][1] / (cnf_matrix[1][0] + cnf_matrix[1][1])))
sns.heatmap(cnf_matrix, annot=True, cbar=False, cmap="Blues")
plt.ylabel("True Label")
plt.xlabel("Predicted Label")
plt.title("Confusion Matrix")
from sklearn.metrics import classification_report
print(classification_report(y_test, y_predict))
from sklearn.metrics import roc_auc_score
from sklearn.metrics import roc_curve
logit_roc_auc = roc_auc_score(y_test, logistic.predict(X_test))
fpr, tpr, thresholds = roc_curve(y_test, logistic.predict_proba(X_test)[:, 1])
plt.figure()
plt.plot(fpr, tpr, label="Logistic Regression (area = %0.2f)" % logit_roc_auc)
plt.plot([0, 1], [0, 1], "r--")
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel("False Positive Rate")
plt.ylabel("True Positive Rate")
plt.title("Receiver operating characteristic")
plt.legend(loc="lower right")
plt.savefig("Log_ROC")
plt.show()
|
import numpy as np
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import os
# database : dogs and cats images
print(os.listdir("../input/dogs-cats-images/dataset/training_set"))
from tensorflow.python.keras.applications import ResNet50
from tensorflow.python.keras.models import Sequential
from tensorflow.python.keras.layers import (
Dense,
Flatten,
GlobalAveragePooling2D,
BatchNormalization,
)
from tensorflow.python.keras.applications.resnet50 import preprocess_input
from tensorflow.python.keras.preprocessing.image import ImageDataGenerator
from tensorflow.python.keras.preprocessing.image import load_img, img_to_array
resnet_weights_path = (
"../input/resnet50/resnet50_weights_tf_dim_ordering_tf_kernels_notop.h5"
)
data_generator = ImageDataGenerator(
rotation_range=40,
width_shift_range=0.2,
height_shift_range=0.2,
rescale=1.0 / 255,
shear_range=0.2,
zoom_range=0.2,
horizontal_flip=True,
fill_mode="nearest",
)
image_size = 224
batch_size = 10
traindata = data_generator.flow_from_directory(
directory="../input/dogs-cats-images/dataset/training_set",
target_size=(image_size, image_size),
batch_size=batch_size,
)
tsdata = ImageDataGenerator()
testdata = tsdata.flow_from_directory(
directory="../input/dogs-cats-images/dataset/test_set",
target_size=(image_size, image_size),
)
num_classes = len(traindata.class_indices)
model = Sequential()
model.add(ResNet50(include_top=False, pooling="avg", weights=resnet_weights_path))
model.add(Flatten())
model.add(BatchNormalization())
model.add(Dense(2048, activation="relu"))
model.add(BatchNormalization())
model.add(Dense(1024, activation="relu"))
model.add(BatchNormalization())
model.add(Dense(num_classes, activation="softmax"))
model.layers[0].trainable = False
model.compile(optimizer="adam", loss="categorical_crossentropy", metrics=["accuracy"])
model.summary()
count = sum(
[
len(files)
for r, d, files in os.walk("../input/dogs-cats-images/dataset/test_set")
]
)
print(count)
import time
start_time = time.time()
model.fit_generator(
traindata,
validation_data=testdata,
steps_per_epoch=int(count / batch_size) + 1,
epochs=10,
)
print("Total time is second ~s ", time.time() - start_time)
from IPython.display import Image, display
import os, random
img_locations = []
for d in os.listdir("../input/dogs-cats-images/dataset/test_set/"):
directory = "../input/dogs-cats-images/dataset/test_set/" + d
sample = [
directory + "/" + s
for s in random.sample(os.listdir(directory), int(random.random() * 10))
]
img_locations += sample
print("Test data found")
def read_and_prep_images(img_paths, img_height=image_size, img_width=image_size):
imgs = [
load_img(img_path, target_size=(img_height, img_width))
for img_path in img_paths
]
img_array = np.array([img_to_array(img) for img in imgs])
return preprocess_input(img_array)
random.shuffle(img_locations)
imgs = read_and_prep_images(img_locations)
predictions = model.predict_classes(imgs)
classes = dict((v, k) for k, v in traindata.class_indices.items())
print("Dogs and cats classification : ")
for img, prediction in zip(img_locations, predictions):
display(Image(img))
print(classes[prediction])
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt # plotting library for data viz
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
# My first data set was not as interesting as I thougth so then I proceeded to search for another intersting thing to dive into. I was more intersted in healthcare and the easier item to look into was the 2019 Corona virus. I came aross multiple sources and the one from Kaggle was the most recently updated one. I don't think this means up to date exactly but it works for data exploration.
cv = pd.read_csv("../input/novel-corona-virus-2019-dataset/2019_nCoV_data.csv")
cv_confirmed = pd.read_csv(
"../input/novel-corona-virus-2019-dataset/time_series_2019_ncov_confirmed.csv"
)
cv_deaths = pd.read_csv(
"../input/novel-corona-virus-2019-dataset/time_series_2019_ncov_deaths.csv"
)
cv_recovered = pd.read_csv(
"../input/novel-corona-virus-2019-dataset/time_series_2019_ncov_recovered.csv"
)
# Previewing the data and then following up to view data types.
cv.head()
cv.columns
cv.dtypes
# Next I wanted to explore how many confirmed infections there were by Country. I could have used the Confirmed data set but the current data set has the counts needed without needing to clean the data. First I wanted to look at the data and then proceeded to plot it using a bar graph.
cv.groupby("Country")["Confirmed"].sum()
cv.groupby("Country")["Confirmed"].sum().plot(kind="bar")
# Proceeding to now do the same with the Death counts
cv.groupby("Country")["Deaths"].count()
cv.groupby("Country")["Deaths"].count().plot(kind="bar")
cv.groupby("Country")["Recovered"].count()
cv.groupby("Country")["Recovered"].count().plot(kind="bar")
# Combined view across all item.
cv.groupby("Country")["Confirmed", "Deaths", "Recovered"].sum().plot(kind="bar")
# cv.plot.pie(y=["Confirmed","Deaths", "Recovered"])
|
# # Categorical Feature Encoding Challenge II
# Binary classification, with every feature a categorical (and i
# a dataset that contains only categorical features, and includes:
# * binary features
# * low- and high-cardinality nominal features
# * low- and high-cardinality ordinal features
# * (potentially) cyclical features
# This challenge adds the additional complexity of feature interactions, as well as missing data.
# In this competition, you will be predicting the probability [0, 1] of a binary target column. Submissions are evaluated on area under the ROC curve between the predicted probability and the observed target.
# The data contains binary features (bin_), nominal features (nom_), ordinal features (ord_) as well as (potentially cyclical) day (of the week) and month features. The string ordinal features ord_{3-5} are lexically ordered according to string.ascii_letters.
# Final submission deadline: March 31, 2020
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import wandb
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
import scipy
import pandas as pd
from sklearn.linear_model import LogisticRegression
from sklearn.linear_model import Ridge
from sklearn.experimental import enable_hist_gradient_boosting
from sklearn.ensemble import HistGradientBoostingClassifier
from sklearn.svm import SVC
from sklearn.neighbors import KNeighborsClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.ensemble import AdaBoostClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import GradientBoostingClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn import naive_bayes
from sklearn.model_selection import train_test_split
# Any results you write to the current directory are saved as output.
df = pd.read_csv("/kaggle/input/cat-in-the-dat-ii/train.csv", index_col="id")
df_test = pd.read_csv("/kaggle/input/cat-in-the-dat-ii/test.csv", index_col="id")
y = df["target"]
D = df.drop(columns="target")
features = D.columns
test_ids = df_test.index
D_all = pd.concat([D, df_test])
num_train = len(D)
print(f"D_all.shape = {D_all.shape}")
# Map value in train xor test
for col in D.columns.difference(["id"]):
train_vals = set(D[col].dropna().unique())
test_vals = set(df_test[col].dropna().unique())
xor_cat_vals = train_vals ^ test_vals
if xor_cat_vals:
print(f"Replacing {len(xor_cat_vals)} values in {col}, {xor_cat_vals}")
D_all.loc[D_all[col].isin(xor_cat_vals), col] = "xor"
# Ordinal encoding
ord_maps = {
"ord_0": {val: i for i, val in enumerate([1, 2, 3])},
"ord_1": {
val: i
for i, val in enumerate(
["Novice", "Contributor", "Expert", "Master", "Grandmaster"]
)
},
"ord_2": {
val: i
for i, val in enumerate(
["Freezing", "Cold", "Warm", "Hot", "Boiling Hot", "Lava Hot"]
)
},
**{
col: {val: i for i, val in enumerate(sorted(D_all[col].dropna().unique()))}
for col in ["ord_3", "ord_4", "ord_5", "day", "month"]
},
}
# OneHot encoding
oh_cols = D_all.columns.difference(ord_maps.keys() - {"day", "month"})
print(f"OneHot encoding {len(oh_cols)} columns")
one_hot = pd.get_dummies(
D_all[oh_cols],
columns=oh_cols,
drop_first=True,
dummy_na=True,
sparse=True,
dtype="int8",
).sparse.to_coo()
# Ordinal encoding
ord_cols = pd.concat(
[
D_all[col].map(ord_map).fillna(max(ord_map.values()) // 2).astype("float32")
for col, ord_map in ord_maps.items()
],
axis=1,
)
ord_cols /= ord_cols.max() # for convergence
ord_cols_sqr = 4 * (ord_cols - 0.5) ** 2
# Combine data
X = scipy.sparse.hstack([one_hot, ord_cols, ord_cols_sqr]).tocsr()
print(f"X.shape = {X.shape}")
# Split into training and validation sets
X_train, X_test, y_train, y_test = train_test_split(
X[:num_train], y, test_size=0.1, random_state=42, shuffle=False
)
X_train = X_train[:10000]
y_train = y_train[:10000]
X_test = X_test[:2000]
y_test = y_test[:2000]
# # Train models, visualize in sklearn
# Classification - predict pulsar
# Train a model, get predictions
log = LogisticRegression(C=0.05, solver="lbfgs", max_iter=5000)
dtree = DecisionTreeClassifier(random_state=4)
rtree = RandomForestClassifier(n_estimators=100, random_state=4)
svm = SVC(random_state=4, probability=True)
nb = GaussianNB()
gbc = GradientBoostingClassifier()
knn = KNeighborsClassifier(n_neighbors=400)
adaboost = AdaBoostClassifier(
n_estimators=500,
learning_rate=0.01,
random_state=42,
base_estimator=DecisionTreeClassifier(
max_depth=8, min_samples_leaf=10, random_state=42
),
)
labels = [0, 1]
def model_algorithm(clf, X_train, y_train, X_test, y_test, name, labels, features):
clf.fit(X_train, y_train)
y_probas = clf.predict_proba(X_test)
y_pred = clf.predict(X_test)
wandb.init(
anonymos="allow", project="kaggle-feature-encoding", name=name, reinit=True
)
# wandb.sklearn.plot_roc(y_test, y_probas, labels, reinit = True)
wandb.termlog("\nPlotting %s." % name)
wandb.sklearn.plot_learning_curve(clf, X_train, y_train)
wandb.termlog("Logged learning curve.")
wandb.sklearn.plot_confusion_matrix(y_test, y_pred, labels)
wandb.termlog("Logged confusion matrix.")
wandb.sklearn.plot_summary_metrics(
clf, X=X_train, y=y_train, X_test=X_test, y_test=y_test
)
wandb.termlog("Logged summary metrics.")
wandb.sklearn.plot_class_proportions(y_train, y_test, labels)
wandb.termlog("Logged class proportions.")
if not isinstance(clf, naive_bayes.MultinomialNB):
wandb.sklearn.plot_calibration_curve(clf, X_train, y_train, name)
wandb.termlog("Logged calibration curve.")
wandb.sklearn.plot_roc(y_test, y_probas, labels)
wandb.termlog("Logged roc curve.")
wandb.sklearn.plot_precision_recall(y_test, y_probas, labels)
wandb.termlog("Logged precision recall curve.")
csv_name = "submission_" + name + ".csv"
# Create submission file
# pd.DataFrame({"id": test_ids, "target": y_pred}).to_csv(csv_name, index=False)
model_algorithm(
log, X_train, y_train, X_test, y_test, "LogisticRegression", labels, features
)
model_algorithm(svm, X_train, y_train, X_test, y_test, "SVM", labels, features)
model_algorithm(
knn, X_train, y_train, X_test, y_test, "KNearestNeighbor", labels, features
)
model_algorithm(
adaboost, X_train, y_train, X_test, y_test, "AdaBoost", labels, features
)
model_algorithm(
gbc, X_train, y_train, X_test, y_test, "GradientBoosting", labels, features
)
model_algorithm(dtree, X_train, y_train, X_test, y_test, "DecisionTree", labels, None)
model_algorithm(
rtree, X_train, y_train, X_test, y_test, "RandomForest", labels, features
)
clf = LogisticRegression(C=0.05, solver="lbfgs", max_iter=5000)
clf.fit(X_train, y_train)
pred = clf.predict_proba(X_test)[:, 1]
pd.DataFrame({"id": test_ids, "target": pred}).to_csv("submission_lr.csv", index=False)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
import warnings
warnings.filterwarnings("ignore")
from IPython.core.interactiveshell import InteractiveShell
InteractiveShell.ast_node_interactivity = "all"
import matplotlib.pyplot as plt
import seaborn as sns
import missingno as msno
pd.set_option("display.max_rows", 200)
pd.set_option("display.max_columns", 100)
import pickle
from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import RobustScaler
from sklearn.linear_model import RidgeCV
from sklearn.linear_model import LassoCV
from sklearn.linear_model import ElasticNetCV
from sklearn.svm import SVR
from sklearn.ensemble import GradientBoostingRegressor
from lightgbm import LGBMRegressor
from xgboost import XGBRegressor
from mlxtend.regressor import StackingCVRegressor
from sklearn.model_selection import cross_val_score
from sklearn.metrics import mean_squared_error, make_scorer
from sklearn.model_selection import KFold, RandomizedSearchCV
import statsmodels.formula.api as smf
from scipy.special import boxcox1p
from scipy.stats import boxcox_normmax
train = pd.read_csv(
"/kaggle/input/house-prices-advanced-regression-techniques/train.csv"
)
test = pd.read_csv("/kaggle/input/house-prices-advanced-regression-techniques/test.csv")
print(train.shape)
print(test.shape)
train.info()
# Combining dataset
train_y = train["SalePrice"]
data = pd.concat((train, test), sort=False).reset_index(drop=True)
data.drop(["SalePrice", "Id"], axis=1, inplace=True)
data.rename(
columns={
"1stFlrSF": "FirstFlrSF",
"2ndFlrSF": "SecondFlrSF",
"3SsnPorch": "ThreeSsnPorch",
},
inplace=True,
)
data.shape
data.head()
train_y.describe()
# Distribution plot
sns.distplot(train_y)
# skewness and Kurtosis
print("Skewness: %f" % train_y.skew())
print("Kurtosis: %f" % train_y.kurt())
# using numpy function log fucntion
train_y = np.log(train_y + 1)
sns.distplot(train_y)
print("Skewness: %f" % train_y.skew())
print("Kurtosis: %f" % train_y.kurt())
# correlation matrix
plt.subplots(figsize=(10, 8))
sns.heatmap(train.corr())
# Saleplice corr matrix
k = 10 # no. of variables for heatmap
cols = train.corr().nlargest(k, "SalePrice")["SalePrice"].index
cm = np.corrcoef(train[cols].values.T)
sns.set(font_scale=1)
hm = sns.heatmap(
cm,
cbar=True,
annot=True,
square=True,
fmt=".2f",
annot_kws={"size": 10},
yticklabels=cols.values,
xticklabels=cols.values,
)
num = train.corr()["SalePrice"].sort_values(ascending=False).head(10).to_frame()
cm = sns.light_palette("grey", as_cmap=True)
s = num.style.background_gradient(cmap=cm)
s
# Compute the correlation matrix
corr_all = train.corr()
# Generate a mask for the upper triangle
mask = np.zeros_like(corr_all, dtype=np.bool)
mask[np.triu_indices_from(mask)] = True
# Set up the matplotlib figure
f, ax = plt.subplots(figsize=(11, 9))
# Draw the heatmap with the mask and correct aspect ratio
sns.heatmap(corr_all, mask=mask, square=True, linewidths=0.5, ax=ax, cmap="BuPu")
plt.show()
# #scatter plot
# sns.set()
# sns.pairplot(train[cols],size = 2.5);
# # Data Cleaning
# missing values?
sns.set(style="ticks")
msno.matrix(data)
msno.heatmap(data, cmap="binary")
missing_data = pd.DataFrame(data.isnull().sum()).reset_index()
missing_data.columns = ["ColumnName", "MissingCount"]
missing_data["PercentMissing"] = (
round(missing_data["MissingCount"] / data.shape[0], 3) * 100
)
missing_data = missing_data.sort_values(by="MissingCount", ascending=False).reset_index(
drop=True
)
missing_data.head(35)
data.drop(["PoolQC", "MiscFeature", "Alley"], axis=1, inplace=True)
ffill = list(missing_data.ColumnName[18:34])
data[ffill] = data[ffill].fillna(method="ffill")
missing_data.ColumnName[3:18]
# ### Data left to be filled
# - Fence Zero
# - FireplaceQu zero
# - LotFrontage mean/median
# - GarageYrBlt median or most frequent 1980
# - GarageFinish zero
# - GarageQual zero
# - GarageCond zero
# - GarageType zero
# - BsmtExposur zero
# - BsmtCond zero
# - BsmtQual zero
# - BsmtFinType2 zero
# - BsmtFinType1 zero
# - MasVnrType zero
# - MasVnrArea mean/median
## Dealing with data to be filled with zero
col_for_zero = [
"Fence",
"FireplaceQu",
"GarageFinish",
"GarageQual",
"GarageCond",
"GarageType",
"BsmtExposure",
"BsmtCond",
"BsmtQual",
"BsmtFinType2",
"BsmtFinType1",
"MasVnrType",
]
data[col_for_zero] = data[col_for_zero].fillna("None")
data["LotFrontage"] = data["LotFrontage"].fillna(data["LotFrontage"].dropna().mean())
data["GarageYrBlt"] = data["GarageYrBlt"].fillna(data["GarageYrBlt"].dropna().median())
data["MasVnrArea"] = data["MasVnrArea"].fillna(data["MasVnrArea"].dropna().median())
# Features Generation
data["YrBltAndRemod"] = data["YearBuilt"] + data["YearRemodAdd"]
data["TotalSF"] = data["TotalBsmtSF"] + data["FirstFlrSF"] + data["SecondFlrSF"]
data["Total_sqr_footage"] = (
data["BsmtFinSF1"] + data["BsmtFinSF2"] + data["FirstFlrSF"] + data["SecondFlrSF"]
)
data["Total_Bathrooms"] = (
data["FullBath"]
+ (0.5 * data["HalfBath"])
+ data["BsmtFullBath"]
+ (0.5 * data["BsmtHalfBath"])
)
data["Total_porch_sf"] = (
data["OpenPorchSF"]
+ data["ThreeSsnPorch"]
+ data["EnclosedPorch"]
+ data["ScreenPorch"]
+ data["WoodDeckSF"]
)
data["hasfence"] = data["Fence"].apply(lambda x: 0 if x == 0 else 1).astype(str)
data["hasmasvnr"] = data["MasVnrArea"].apply(lambda x: 0 if x == 0 else 1).astype(str)
data["haspool"] = data["PoolArea"].apply(lambda x: 1 if x > 0 else 0).astype(str)
data["has2ndfloor"] = data["SecondFlrSF"].apply(lambda x: 1 if x > 0 else 0).astype(str)
data["hasgarage"] = data["GarageArea"].apply(lambda x: 1 if x > 0 else 0).astype(str)
data["hasbsmt"] = data["TotalBsmtSF"].apply(lambda x: 1 if x > 0 else 0).astype(str)
data["hasfireplace"] = data["Fireplaces"].apply(lambda x: 1 if x > 0 else 0).astype(str)
data["MSSubClass"] = data["MSSubClass"].astype(str)
data["YrSold"] = data["YrSold"].astype(str)
data["MoSold"] = data["MoSold"].astype(str)
num_var = [
key
for key in dict(data.dtypes)
if dict(data.dtypes)[key] in ["float64", "int64", "float32", "int32"]
]
cat_var = [key for key in dict(data.dtypes) if dict(data.dtypes)[key] in ["object"]]
print(len(num_var))
print(len(cat_var))
num_data = data[num_var]
cat_data = data[cat_var]
# skew X variables
skew_data = num_data.apply(lambda x: x.skew()).sort_values(ascending=False)
high_skew = skew_data[skew_data > 0.5]
skew_index = high_skew.index
for i in skew_index:
data[i] = boxcox1p(data[i], boxcox_normmax(data[i] + 1))
def outlier_capping(x):
x = x.clip_upper(x.quantile(0.99))
x = x.clip_lower(x.quantile(0.01))
return x
num_data.drop("PoolArea", axis=1, inplace=True)
num_data = num_data.apply(outlier_capping)
num_data["PoolArea"] = data.PoolArea
def create_dummies(df, colname):
col_dummies = pd.get_dummies(df[colname], prefix=colname)
col_dummies.drop(col_dummies.columns[0], axis=1, inplace=True)
df = pd.concat([df, col_dummies], axis=1)
df.drop(colname, axis=1, inplace=True)
return df
# for c_feature in categorical_features
for c_feature in cat_data.columns:
cat_data[c_feature] = cat_data[c_feature].astype("category")
cat_data = create_dummies(cat_data, c_feature)
print(cat_data.shape)
print(num_data.shape)
final_data = pd.concat([cat_data, num_data, train_y], axis=1)
print(final_data.shape)
final_data.columns = [var.strip().replace(".", "_") for var in final_data.columns]
final_data.columns = [var.strip().replace("&", "_") for var in final_data.columns]
final_data.columns = [var.strip().replace(" ", "_") for var in final_data.columns]
overfit = []
for i in final_data.columns:
counts = final_data[i].value_counts()
zeros = counts.iloc[0]
if zeros / len(final_data) * 100 > 99.94:
overfit.append(i)
overfit
final_data.drop(overfit, axis=1, inplace=True)
# splitting the data set into two sets
final_train = final_data.loc[final_data.SalePrice.isnull() == 0]
final_test = final_data.loc[final_data.SalePrice.isnull() == 1]
final_train = final_train.drop("SalePrice", axis=1)
final_test = final_test.drop("SalePrice", axis=1)
print(final_train.shape)
print(final_test.shape)
X = final_train
y = train_y
print(X.shape)
print(y.shape)
test_X = final_test
print(test_X.shape)
kfolds = KFold(n_splits=10, shuffle=True, random_state=21)
def rmse(y, y_pred):
return np.sqrt(mean_squared_error(y, y_pred))
def cv_rmse(model, X=X):
rmse = np.sqrt(
-cross_val_score(model, X, y, scoring="neg_mean_squared_error", cv=kfolds)
)
return rmse
alphas_alt = [
14.5,
14.6,
14.7,
14.8,
14.9,
15,
15.1,
15.2,
15.3,
15.4,
15.5,
15.6,
15.7,
15.8,
15.9,
16,
]
alphas2 = [
5e-05,
0.0001,
0.0002,
0.0003,
0.0004,
0.0005,
0.0006,
0.0007,
0.0008,
0.0009,
0.0010,
0.0011,
0.0012,
0.0013,
0.0014,
0.0015,
]
e_alphas = [
0.0001,
0.0002,
0.0003,
0.0004,
0.0005,
0.0006,
0.0007,
0.0008,
0.0009,
0.0010,
0.0011,
0.0012,
0.0013,
0.0014,
0.0015,
]
e_l1ratio = [
0.05,
0.15,
0.2,
0.25,
0.3,
0.35,
0.4,
0.45,
0.5,
0.55,
0.6,
0.65,
0.7,
0.75,
0.8,
0.85,
0.9,
0.95,
0.99,
1,
]
from sklearn.linear_model import Ridge, Lasso, LinearRegression
from sklearn.model_selection import KFold, RandomizedSearchCV
from sklearn.metrics import mean_squared_error, make_scorer
from sklearn.ensemble import GradientBoostingRegressor
from mlxtend.regressor import StackingCVRegressor
from sklearn.preprocessing import RobustScaler
from scipy.stats import boxcox_normmax, zscore
from multiprocessing import cpu_count
from lightgbm import LGBMRegressor
from scipy.special import boxcox1p
import matplotlib.pyplot as plt
from sklearn.svm import SVR
import pandas as pd
import numpy as np
df = X
df = RobustScaler(df)
kf = KFold(n_splits=5, random_state=0, shuffle=True)
rmse = lambda y, y_pred: np.sqrt(mean_squared_error(y, y_pred))
scorer = make_scorer(rmse, greater_is_better=False)
def random_search(model, grid, n_iter=100):
n_jobs = max(cpu_count() - 2, 1)
search = RandomizedSearchCV(
model, grid, n_iter, scorer, n_jobs=n_jobs, cv=kf, random_state=0, verbose=True
)
return search.fit(X, y)
ridge_search = random_search(RidgeCV(), {"alpha": np.logspace(-1, 2, 500)})
lasso_search = random_search(LassoCV(), {"alpha": np.logspace(-5, -1, 500)})
svr_search = random_search(
SVR(),
{
"C": np.arange(1, 100),
"gamma": np.linspace(0.00001, 0.001, 50),
"epsilon": np.linspace(0.01, 0.1, 50),
},
)
lgbm_search = random_search(
LGBMRegressor(
objective="regression",
num_leaves=4,
n_estimators=5000,
max_bin=200,
bagging_fraction=0.75,
bagging_freq=5,
bagging_seed=7,
feature_fraction=0.2,
feature_fraction_seed=7,
verbose=-1,
),
{
"colsample_bytree": np.linspace(0.2, 0.7, 6),
"learning_rate": np.logspace(-3, -1, 100),
},
)
gbr_search = random_search(
GradientBoostingRegressor(
n_estimators=5000,
max_depth=3,
min_child_weight=0,
colsample_bytree=0.7,
objective="reg:linear",
nthread=-1,
scale_pos_weight=1,
seed=27,
),
{
"reg_alpha": np.linspace(0.00001, 0.001, 100),
"gamma": np.linspace(-1, 1, 100),
"learning_rate": np.logspace(-1, 1, 100),
},
)
xgboost_search = random_search(
XGBRegressor(
n_estimators=3000,
max_depth=3,
min_samples_leaf=15,
min_samples_split=10,
loss="huber",
random_state=21,
),
{
"max_features": np.linspace(0.2, 0.7, 6),
"learning_rate": np.logspace(-3, -1, 100),
},
)
models = [
search.best_estimator_
for search in [ridge_search, lasso_search, svr_search, lgbm_search, gbm_search]
]
stack_search = random_search(
StackingCVRegressor(models, Ridge(), cv=kf),
{"meta_regressor__alpha": np.logspace(-3, -2, 500)},
n_iter=20,
)
models.append(stack_search.best_estimator_)
kf = KFold(n_splits=5, random_state=0, shuffle=True)
rmse = lambda y, y_pred: np.sqrt(mean_squared_error(y, y_pred))
scorer = make_scorer(rmse, greater_is_better=False)
def random_search(model, grid, n_iter=100):
n_jobs = max(cpu_count() - 2, 1)
search = RandomizedSearchCV(
model, grid, n_iter, scorer, n_jobs=n_jobs, cv=kf, random_state=0, verbose=True
)
return search.fit(X, y)
ridge_search = random_search(Ridge(), {"alpha": np.logspace(-1, 2, 500)})
lasso_search = random_search(Lasso(), {"alpha": np.logspace(-5, -1, 500)})
svr_search = random_search(
SVR(),
{
"C": np.arange(1, 100),
"gamma": np.linspace(0.00001, 0.001, 50),
"epsilon": np.linspace(0.01, 0.1, 50),
},
)
lgbm_search = random_search(
LGBMRegressor(n_estimators=2000, max_depth=3),
{
"colsample_bytree": np.linspace(0.2, 0.7, 6),
"learning_rate": np.logspace(-3, -1, 100),
},
)
gbm_search = random_search(
GradientBoostingRegressor(n_estimators=2000, max_depth=3),
{
"max_features": np.linspace(0.2, 0.7, 6),
"learning_rate": np.logspace(-3, -1, 100),
},
)
xgboost_search = random_search(
XGBRegressor(
n_estimators=3000,
max_depth=3,
min_samples_leaf=15,
min_samples_split=10,
loss="huber",
random_state=21,
),
{
"max_features": np.linspace(0.2, 0.7, 6),
"learning_rate": np.logspace(-3, -1, 100),
},
)
models = [
search.best_estimator_
for search in [ridge_search, lasso_search, svr_search, lgbm_search, gbm_search]
]
stack_search = random_search(
StackingCVRegressor(models, Ridge(), cv=kf),
{"meta_regressor__alpha": np.logspace(-3, -2, 500)},
n_iter=20,
)
models.append(stack_search.best_estimator_)
print(ridge_search)
print(lasso_search)
print(svr_search)
print(lgbm_search)
print(gbr_search)
print(xgboost_search)
ridge = make_pipeline(RobustScaler(), RidgeCV(alphas=alphas_alt, cv=kfolds))
lasso = make_pipeline(
RobustScaler(), LassoCV(max_iter=1e7, alphas=alphas2, random_state=42, cv=kfolds)
)
elasticnet = make_pipeline(
RobustScaler(),
ElasticNetCV(max_iter=1e7, alphas=e_alphas, cv=kfolds, l1_ratio=e_l1ratio),
)
svr = make_pipeline(RobustScaler(), SVR(C=20, epsilon=0.008, gamma=0.0003))
gbr = GradientBoostingRegressor(
n_estimators=3000,
learning_rate=0.05,
max_depth=4,
max_features="sqrt",
min_samples_leaf=15,
min_samples_split=10,
loss="huber",
random_state=21,
)
lightgbm = LGBMRegressor(
objective="regression",
num_leaves=4,
learning_rate=0.01,
n_estimators=5000,
max_bin=200,
bagging_fraction=0.75,
bagging_freq=5,
bagging_seed=7,
feature_fraction=0.2,
feature_fraction_seed=7,
verbose=-1,
)
xgboost = XGBRegressor(
learning_rate=0.01,
n_estimators=3460,
max_depth=3,
min_child_weight=0,
gamma=0,
subsample=0.7,
colsample_bytree=0.7,
objective="reg:linear",
nthread=-1,
scale_pos_weight=1,
seed=27,
reg_alpha=0.00006,
)
stack_gen = StackingCVRegressor(
regressors=(ridge, lasso, elasticnet, gbr, xgboost, lightgbm, svr),
meta_regressor=xgboost,
use_features_in_secondary=True,
)
score = cv_rmse(ridge)
print("Ridge: {:.4f} ({:.4f})\n".format(score.mean(), score.std()))
score = cv_rmse(lasso)
print("Lasso: {:.4f} ({:.4f})\n".format(score.mean(), score.std()))
score = cv_rmse(elasticnet)
print("Elasticnet: {:.4f} ({:.4f})\n".format(score.mean(), score.std()))
score = cv_rmse(svr)
print("SVR: {:.4f} ({:.4f})\n".format(score.mean(), score.std()))
score = cv_rmse(gbr)
print("GBRegressor: {:.4f} ({:.4f})\n".format(score.mean(), score.std()))
score = cv_rmse(lightgbm)
print("LightGBM: {:.4f} ({:.4f})\n".format(score.mean(), score.std()))
score = cv_rmse(xgboost)
print("XGBoost: {:.4f} ({:.4f})\n".format(score.mean(), score.std()))
print("Start Fit")
print("stack_gen")
stack_gen_model = stack_gen.fit(np.array(X), np.array(y))
print("Ridge")
ridge_model_full_data = ridge.fit(X, y)
print("Lasso")
lasso_model_full_data = lasso.fit(X, y)
print("Elasticnet")
elastic_model_full_data = elasticnet.fit(X, y)
print("SVR")
svr_model_full_data = svr.fit(X, y)
print("GradientBoosting")
gbr_model_full_data = gbr.fit(X, y)
print("LightGBM")
lightgbm_model_full_data = lightgbm.fit(X, y)
print("XGBoost")
xgboost_model_full_data = xgboost.fit(X, y)
pickle.dump(stack_gen_model, open("stack_gen_model.pkl", "wb"))
pickle.dump(ridge_model_full_data, open("ridge_model_full_data.pkl", "wb"))
pickle.dump(lasso_model_full_data, open("lasso_model_full_data.pkl", "wb"))
pickle.dump(elastic_model_full_data, open("elastic_model_full_data.pkl", "wb"))
pickle.dump(svr_model_full_data, open("svr_model_full_data.pkl", "wb"))
pickle.dump(gbr_model_full_data, open("gbr_model_full_data.pkl", "wb"))
pickle.dump(lightgbm_model_full_data, open("lightgbm_model_full_data.pkl", "wb"))
pickle.dump(xgboost_model_full_data, open("xgboost_model_full_data.pkl", "wb"))
stack_gen_model = pickle.load(open("/kaggle/working/stack_gen_model.pkl", "rb"))
ridge_model_full_data = pickle.load(
open("/kaggle/working/ridge_model_full_data.pkl", "rb")
)
lasso_model_full_data = pickle.load(
open("/kaggle/working/lasso_model_full_data.pkl", "rb")
)
elastic_model_full_data = pickle.load(
open("/kaggle/working/elastic_model_full_data.pkl", "rb")
)
svr_model_full_data = pickle.load(open("/kaggle/working/svr_model_full_data.pkl", "rb"))
gbr_model_full_data = pickle.load(open("/kaggle/working/gbr_model_full_data.pkl", "rb"))
lightgbm_model_full_data = pickle.load(
open("/kaggle/working/lightgbm_model_full_data.pkl", "rb")
)
xgboost_model_full_data = pickle.load(
open("/kaggle/working/xgboost_model_full_data.pkl", "rb")
)
0.0125 + 0.0125 + 0.0125 + 0.0125 + 0.2 + 0.2 + 0.2 + 0.35
0.025 - 0.0175
# Blending models
def blend_models_predict(X):
return (
(0.0175 * elastic_model_full_data.predict(X))
+ (0.0175 * lasso_model_full_data.predict(X))
+ (0.0075 * ridge_model_full_data.predict(X))
+ (0.0075 * svr_model_full_data.predict(X))
+ (0.2 * gbr_model_full_data.predict(X))
+ (0.2 * xgboost_model_full_data.predict(X))
+ (0.2 * lightgbm_model_full_data.predict(X))
+ (0.35 * stack_gen_model.predict(np.array(X)))
)
print("RMSE score on train data: ")
print(rmse(y, blend_models_predict(X)))
print("Predict submission")
submission = pd.read_csv(
"/kaggle/input/house-prices-advanced-regression-techniques/sample_submission.csv"
)
submission.iloc[:, 1] = np.floor(np.expm1(blend_models_predict(test_X)))
submission.head()
print("Blend with Top Kernels submissions\n")
sub_1 = pd.read_csv("../input/topsubmissions/submission1.csv")
sub_2 = pd.read_csv("../input/topsubmissions/submission2.csv")
sub_3 = pd.read_csv("../input/topsubmissions/blend_submission5.csv")
submission.iloc[:, 1] = np.floor(
(0.5 * np.floor(np.expm1(blend_models_predict(test_X)))) + (0.5 * sub_2.iloc[:, 1])
)
submission.to_csv("submission3.csv", index=False)
submission.head()
print("Blend with Top Kernels submissions\n")
sub_1 = pd.read_csv("../input/topsubmissions/blend_submission1.csv")
# sub_2 = pd.read_csv('../input/topsubmissions/blend_submission2.csv')
sub_3 = pd.read_csv("../input/topsubmissions/blend_submission3.csv")
sub_4 = pd.read_csv("../input/topsubmissions/blend_submission4.csv")
# sub_5 = pd.read_csv('../input/topsubmissions/blend_submission5.csv')
submission.iloc[:, 1] = np.floor(
(0.05 * np.floor(np.expm1(blend_models_predict(test_X))))
+ (0.35 * sub_1.iloc[:, 1])
+ (0.25 * sub_3.iloc[:, 1])
+ (0.35 * sub_4.iloc[:, 1])
)
submission.to_csv("submission14.csv", index=False)
submission.head()
print("Blend with Top Kernels submissions\n")
sub_1 = pd.read_csv("../input/topsubmissions/blend_submission1.csv")
sub_2 = pd.read_csv("../input/topsubmissions/submission2.csv")
sub_3 = pd.read_csv("../input/topsubmissions/blend_submission3.csv")
sub_4 = pd.read_csv("../input/topsubmissions/blend_submission4.csv")
sub_5 = pd.read_csv("../input/topsubmissions/blend_submission5.csv")
sub_6 = pd.read_csv("../input/topsubmissions/blend_submission6.csv")
submission.iloc[:, 1] = np.floor(
(0.2 * np.floor(np.expm1(blend_models_predict(test_X))))
+ (0.1 * sub_1.iloc[:, 1])
+ (0.1 * sub_2.iloc[:, 1])
+ (0.2 * sub_3.iloc[:, 1])
+ (0.2 * sub_4.iloc[:, 1])
+ (0.1 * sub_5.iloc[:, 1])
+ (0.1 * sub_6.iloc[:, 1])
)
submission.to_csv("submission5.csv", index=False)
submission.head()
|
# Data Cleaning & Prepration
# a significant amount of time is spent on data prepration:loading, cleaning, transforming and rearranging.
# such tasks are often reported to take up 80% or more of an analyst's time. sometimes the way the data is stored in files or databases is not in the right format for a particular task.so we can use general purpose programming language like python, R, Java to do ad hoc processing from one form to anthor, and Fortunately python has pandas.
# in this kernel i will discuss tools for missing data, duplicate data string manipulation,some other analytical data transformations, combining and rearranging datasets in different ways.
# Plan of kernel
# load Data
# Get some informations about data
# Handling Missing Data
# Data Transformation
# String Manipulation
# Combining & Merging datasets
# Reshaping & Pivoting
# Load Data
import numpy as np
import pandas as pd
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
df = pd.read_csv("/kaggle/input/craigslist-carstrucks-data/vehicles.csv")
df.head()
# Get some informations about data
# there are two different ways to get some informations about your data, first one by use pd.dataframe.info() which is the esaist and saves more time, but i prefer to do it by creating a function to give me all informations i need to know
# Frist Method
df.info()
# Second Method
def get_info(df):
print("number of variables: ", df.shape[1])
print("number of cases: ", df.shape[0])
print("-" * 10)
print("variables(columns) names: ")
print(df.columns)
print("-" * 10)
print("data-type of each variable: ")
print(df.dtypes)
print("-" * 10)
print("missing rows in columns: ")
c = df.isnull().sum()
print(c[c > 0])
get_info(df)
# Handling Missing Data
# Pandas using floating-point value Nan to represent missing data, we can use pandas to identify the columns that have missing values
df.isnull()
# as we can see this doesn't give us enough information about missing data, let's make it effective
df.isnull().sum()
# we can use pandas too, to identify the number of rows in each columns that have values.
df.notnull().sum()
# Filtering Out Missing Data
# 1-First we see that county column doesn't contain any data, so I am going to drop it, and we can use this technique if any column hasn't enough data like 95% missing values.
cleaned_df = df.drop("county", axis=1)
# 2-first technique get rid of missing values is to drop all the rows which have missing values by using dropna().
# but this is not the best technique to remove missing values
cleaned_df.dropna().head()
# Filling in missing data
# 2-rather than filtering out missing data, we may want to fill the missing values with any value we choose, and this is the best techinque to remove missing values from our data set
for i in cleaned_df.drop(["model", "manufacturer", "paint_color"], axis=1).columns:
if cleaned_df[i].dtype == "float":
cleaned_df[i] = cleaned_df[i].fillna(cleaned_df[i].mean())
if cleaned_df[i].dtype == "object":
cleaned_df[i] = cleaned_df[i].fillna(cleaned_df[i].mode()[0])
cleaned_df["year"] = cleaned_df["year"].fillna(cleaned_df["year"].mode()[0])
cleaned_df["model"] = cleaned_df["model"].fillna("Unknown")
cleaned_df["manufacturer"] = cleaned_df["manufacturer"].fillna("Unknown")
cleaned_df["paint_color"] = cleaned_df["paint_color"].fillna("Unknown")
# now our dataset has no missing values
# Data Transformation
# Removing Duplicates
# duplicated data may be found in a DataFrame for any number of reasons
cleaned_df.duplicated()
# each row (car) has specific id, so we are going to drop all the rows that have duplicated id
cleaned_df = cleaned_df.drop_duplicates(["id"])
print("done")
# Transforming Data Using Function
# For many datasets you may want to perform some transformations based on the values in a columns in dataframe
def odometer_status(val):
if val > 101729.96151504324:
return "alot"
else:
return "little"
cleaned_df["odometer_status"] = df["odometer"].apply(odometer_status)
cleaned_df[["odometer_status", "odometer"]].tail()
# as we can see here's an example of data transformation by function and here we transform odometer column to object type column.
# Replacing Values
# some values in price column is 0 which is not right let's use replace() to replace this values by the median value of price column
cleaned_df["price"] = cleaned_df["price"].replace(0, cleaned_df["price"].median())
# Renaming columns and indexes
# We can use pandas to rename the columns
for i in cleaned_df.columns:
changer = i.title()
cleaned_df.rename(columns={i: changer}, inplace=True)
cleaned_df.columns
# now all the names of columns from lowercase to title
# Detecting & Filtering Outliers
# before going to filter the data from outliers, we are going to define something very important to learn
# What is the IQR
# The "interquartile range", abbreviated "IQR", is just the width of the box in the box-and-whisker plot. That is, IQR = Q3 – Q1 . The IQR can be used as a measure of how spread-out the values are.
# Statistics assumes that your values are clustered around some central value. The IQR tells how spread out the "middle" values are; it can also be used to tell when some of the other values are "too far" from the central value. These "too far away" points are called "outliers", because they "lie outside" the range in which we expect them.
# The IQR is the length of the box in your box-and-whisker plot. An outlier is any value that lies more than one and a half times the length of the box from either end of the box.
# That is, if a data point is below Q1 – 1.5×IQR or above Q3 + 1.5×IQR, it is viewed as being too far from the central values to be reasonable. Maybe you bumped the weigh-scale when you were making that one measurement, or maybe your lab partner is an idiot and you should never have let him touch any of the equipment. Who knows? But whatever their cause, the outliers are those points that don't seem to "fit".
# Now I am going to use Price column and seaborn library to visualize the outliers in this column from the data
import seaborn as sns
sns.boxplot("Price", data=cleaned_df)
# from the box plot above, we can see the outliers as dots
price_stats = cleaned_df["Price"].describe()
price_stats
from scipy.stats import iqr
iqr = iqr(cleaned_df["Price"])
iqr
upper_bound = price_stats["75%"] + (1.5 * iqr)
lower_bound = price_stats["25%"] - (1.5 * iqr)
outliers = {"above_upper": 0, "below_lower": 0}
indexes = []
for i, j in enumerate(cleaned_df["Price"].values):
if j > upper_bound:
outliers["above_upper"] += 1
indexes.append(i)
elif j < lower_bound:
outliers["below_lower"] += 1
indexes.append(i)
outliers
# there are 21098 outlier price in dataset
# Computing Indicator/Dummy Variables
# Another type of transformations for statistical modeling or machine learning applications is converting a categorical variable into dummy matrix. if a column is a Dataframe has K distinct values, you would derive a matrix or dataframe with K columns containing all 1s and 0s.pandas has get_dummies() function.
drive_dummy_df = pd.get_dummies(cleaned_df["Drive"], prefix="Drive")
drive_dummy_df.tail(10)
# String Manipulation
# python has long been a popular raw data manipulation language in part due to its ease of use for string and text processing. most text operations are made simple with the string's object's built in methods. (check the link below)
# string methods
# now I am going to remove 'cylinders' word from Cylinder column
cleaned_df["Cylinders"] = cleaned_df["Cylinders"].str.replace("cylinders", "")
cleaned_df["Cylinders"].head()
|
import pandas as pd
df = pd.read_excel(
"/kaggle/input/mortality-risk-clinincal-data-of-covid19-patients/Mortality_incidence_sociodemographic_and_clinical_data_in_COVID19_patients.xlsx"
)
df.shape
age_categorical = {"Age": {"0-60": 0, ">60": 1, ">70": 2, ">80": 3}}
df = df.replace(age_categorical)
y = df["Death"].values
df = df.drop(["Death"], axis=1)
df.head(3)
from sklearn.preprocessing import StandardScaler
X = df.values
X = StandardScaler().fit_transform(X)
X[0]
from sklearn.ensemble import ExtraTreesClassifier
etc = ExtraTreesClassifier()
etc.fit(X, y)
feature_importances = etc.feature_importances_
feature_df = pd.DataFrame(
{"feature": df.columns, "importance": etc.feature_importances_}
)
feature_df = feature_df.sort_values(by="importance", ascending=False)
feature_df = feature_df[feature_df["importance"] > 0.01]
feature_df
from sklearn.model_selection import GridSearchCV
from sklearn.neighbors import KNeighborsClassifier
from sklearn.model_selection import LeaveOneOut
param_grid = {
"n_neighbors": [2, 3, 4, 5, 6, 7, 8, 9, 10],
"weights": ["uniform", "distance"],
"algorithm": ["brute", "auto"],
"metric": ["euclidean", "minkowski"],
}
cv = LeaveOneOut()
search = GridSearchCV(
estimator=KNeighborsClassifier(),
param_grid=param_grid,
n_jobs=-1,
scoring="f1_micro",
cv=cvb,
verbose=0,
)
search.fit(X, y)
search.best_params_
search.best_score_
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
X_train = pd.read_csv("/kaggle/input/manomanocentralesupelec/X_train.csv")
y_train = pd.read_csv("/kaggle/input/manomanocentralesupelec/y_train.csv")
X_test = pd.read_csv("/kaggle/input/manomanocentralesupelec/X_test.csv")
X_train.shape, y_train.shape, X_test.shape
print("Features: ", X_train.columns)
print("Target: ", y_train.columns)
# ## Dummy algorithm : Average Conversion Rate
# ### Estimate WRMSE
from sklearn.metrics import mean_squared_error
X_train["conversion_rate"] = y_train["conversion_rate"]
X_train["m_total_vu"] = y_train["m_total_vu"]
average_conversion_rate = X_train[X_train.s_date <= "2020-01-15"][
"conversion_rate"
].mean()
print("Average conversion rate", average_conversion_rate)
print(
"Estimated WRMSE:",
mean_squared_error(
y_true=X_train[X_train.s_date > "2020-01-15"]["conversion_rate"],
y_pred=[average_conversion_rate] * (X_train.s_date > "2020-01-15").sum(),
sample_weight=X_train[X_train.s_date > "2020-01-15"]["m_total_vu"],
),
)
# ## Generate submission
y_sub = pd.DataFrame({"id": X_test["id"], "conversion_rate": average_conversion_rate})
y_sub.head()
y_sub.to_csv("/kaggle/working/sample_submission.csv", index=False)
|
# タイトルの通り、特に良いスコアが出るわけではないのですが、ディスカッションの足しにでもなればと何となく投下しておきます。各パラメータはとても適当なので特になぜそうしているかとかの理由はありません。
# ライブラリをimportしておきます
import warnings
warnings.filterwarnings("ignore")
from glob import glob
import numpy as np
import pandas as pd
from pandas import DataFrame
from sklearn.metrics import roc_auc_score
from tqdm.notebook import tqdm
import librosa
# 学習データの正解リストを読み込みます
train = pd.read_csv("../input/data-science-osaka-spring-2023/train.csv")
train
# 音量だけ見ることにする
from scipy.interpolate import interp1d
def wav_to_loudness(filename, target_length=128, frame_length=2048, hop_length=512):
y, sr = librosa.load(filename)
S = librosa.stft(y, n_fft=frame_length, hop_length=hop_length)
loudness = librosa.amplitude_to_db(np.mean(np.abs(S), axis=0))
x = np.linspace(0, 1, num=loudness.shape[0])
f = interp1d(x, loudness, kind="linear")
x_new = np.linspace(0, 1, num=target_length)
loudness_resampled = f(x_new)
return loudness_resampled
# 音楽データの読み込み
music_data = []
for i in tqdm(range(250)):
file_path = "../input/data-science-osaka-spring-2023/music/music/%d.wav" % i
y = wav_to_loudness(file_path)
music_data.append(y)
music_data = np.array(music_data)
print(music_data.shape)
# モーションの読み込み
import numpy as np
import pandas as pd
from scipy.interpolate import interp1d
from tqdm import tqdm
dfs = []
for i in tqdm(range(250)):
file_path = "../input/data-science-osaka-spring-2023/motion/motion/%d.csv" % i
df = pd.read_csv(file_path)
# df.iloc[:,1:] = kalman_smoothing_scipy(data=df.iloc[:,1:].values, F=F, B=B, H=H, Q=Q, R=R)
df.drop(columns=["Time"], inplace=True)
dfs.append(df)
all_data = pd.concat(dfs)
overall_median = all_data.median()
overall_std = all_data.std()
sensor_data = []
for df in tqdm(dfs):
df = (df - overall_median) / overall_std
df = df.values
# Resize the sensor data
f = interp1d(np.linspace(0, 1, df.shape[0]), df.T, kind="linear", axis=1)
xnew = np.linspace(0, 1, 128)
df = f(xnew).T
sensor_data.append(df)
sensor_data = np.array(sensor_data)
print(sensor_data.shape)
# 学習用と検定用に分けます
train_ = train.iloc[:66]
val = train.iloc[66:]
# そのままだとnegative sampleがないので、ID x musicの組み合わせで生成して、リストにないものを0にすることにします
df_train_ = DataFrame(
[(i, m) for i in train_.ID.values for m in train_.music], columns=["ID", "music"]
)
df_val = DataFrame(
[(i, m) for i in val.ID.values for m in val.music], columns=["ID", "music"]
)
df_train_ = df_train_.merge(train, on=["ID", "music"], how="left")
df_train_["Target"] = (df_train_.genre.isnull() == False).astype(int)
df_val = df_val.merge(val, on=["ID", "music"], how="left")
df_val["Target"] = (df_val.genre.isnull() == False).astype(int)
df_train_.drop(["genre"], axis=1, inplace=True)
df_val.drop(["genre"], axis=1, inplace=True)
music_data = music_data.reshape(-1, 128, 1)
y_train_, y_val = df_train_.Target.values, df_val.Target.values
# X_train_, X_val = X[df_train_.ID.values], X[df_val.ID.values]
sensor_train_, sensor_val = (
sensor_data[df_train_.ID.values],
sensor_data[df_val.ID.values],
)
music_train_, music_val = (
music_data[df_train_.music.values],
music_data[df_val.music.values],
)
from tensorflow.keras.layers import (
Input,
Conv1D,
concatenate,
GlobalAveragePooling1D,
Dense,
BatchNormalization,
MaxPooling1D,
)
from tensorflow.keras.models import Model
from tensorflow.keras.callbacks import Callback, EarlyStopping
from sklearn.metrics import roc_auc_score
from tensorflow.keras.utils import Sequence
from tensorflow.keras.callbacks import Callback
from tensorflow.keras import backend as K
class CosineAnnealingScheduler(Callback):
def __init__(self, T_max, eta_max, eta_min=0, verbose=0):
super(CosineAnnealingScheduler, self).__init__()
self.T_max = T_max
self.eta_max = eta_max
self.eta_min = eta_min
self.verbose = verbose
def on_epoch_begin(self, epoch, logs=None):
if not hasattr(self.model.optimizer, "lr"):
raise ValueError('Optimizer must have a "lr" attribute.')
lr = float(K.get_value(self.model.optimizer.lr))
eta_t = self.eta_min + 0.5 * (self.eta_max - self.eta_min) * (
1 + np.cos(np.pi * epoch / self.T_max)
)
K.set_value(self.model.optimizer.lr, eta_t)
if self.verbose > 0:
print(f"\nEpoch {epoch + 1}: Learning rate is {eta_t:.5f}")
import gc
from sklearn.metrics import roc_auc_score
from tensorflow.keras.callbacks import ModelCheckpoint, EarlyStopping, LambdaCallback
import tensorflow.keras.backend as K
def calc_auc(epoch, logs):
y_pred = model.predict([music_val, sensor_val])
auc_score = roc_auc_score(y_val, y_pred)
print("Validation AUC:", auc_score)
del y_pred
gc.collect()
auc_callback = LambdaCallback(on_epoch_end=calc_auc)
from tensorflow.keras.optimizers import Adam
from tensorflow.keras.metrics import AUC
from tensorflow.keras.regularizers import l2
def create_model():
# 音声データの入力
music_input = Input(shape=(128, 1))
# センサーデータの入力
sensor_input = Input(shape=(128, 20))
# 両者をチャネル方向にConcatenate
merged_conv = concatenate([music_input, sensor_input], axis=-1)
# 畳み込んでGAP
merged_conv = Conv1D(16, 4, activation="relu", padding="same")(merged_conv)
merged_conv = MaxPooling1D(2)(merged_conv)
merged_conv = Conv1D(16, 4, activation="relu", padding="same")(merged_conv)
merged_conv = Conv1D(8, 4, activation="relu", padding="same")(merged_conv)
merged_conv = Conv1D(8, 4, activation="relu", padding="same")(merged_conv)
merged_conv = GlobalAveragePooling1D()(merged_conv)
# 出力層
outputs = Dense(1, activation="sigmoid", kernel_regularizer=l2(0.001))(merged_conv)
learning_rate = 0.001
optimizer = Adam(lr=learning_rate)
# モデルのコンパイル
model = Model(inputs=[music_input, sensor_input], outputs=outputs)
model.compile(
optimizer=optimizer, loss="binary_crossentropy", metrics=["accuracy", AUC()]
)
return model
import random
from numba import jit
import numpy as np
from tensorflow.keras.utils import Sequence
from sklearn.utils import resample
class Music_Sensor_Generator(Sequence):
"Generates data for Keras"
def __init__(
self,
music_train,
sensor_train,
y_train,
batch_size=16,
shuffle=True,
max_shift=0,
alpha=0.2,
use_bootstrap=False,
):
"Initialization"
self.batch_size = batch_size
self.music_train = music_train
self.sensor_train = sensor_train
self.y_train = y_train
self.shuffle = shuffle
self.max_shift = max_shift
self.use_bootstrap = use_bootstrap
self.on_epoch_end()
self.alpha = alpha
def __len__(self):
"Denotes the number of batches per epoch"
return int(np.ceil(len(self.music_train) / self.batch_size))
def __getitem__(self, index):
"Generate one batch of data"
# Generate indexes of the batch
indexes = self.indexes[index * self.batch_size : (index + 1) * self.batch_size]
# Generate data
[music, sensor], y = self.__data_generation(indexes)
return [music, sensor], y
def on_epoch_end(self):
"Updates indexes after each epoch"
self.indexes = np.arange(len(self.music_train))
if self.use_bootstrap:
self.indexes = self.bootstrap_balanced(self.y_train)
elif self.shuffle:
np.random.shuffle(self.indexes)
def bootstrap_balanced(self, y):
pos_indices = np.where(y == 1)[0]
neg_indices = np.where(y == 0)[0]
pos_sample = resample(
pos_indices, replace=True, n_samples=len(neg_indices), random_state=None
)
balanced_indices = np.concatenate((neg_indices, pos_sample))
np.random.shuffle(balanced_indices)
return balanced_indices
@jit
def __data_generation(self, batch_ids):
music = self.music_train[batch_ids]
sensor = self.sensor_train[batch_ids]
y = self.y_train[batch_ids]
# ランダムなシフト量を決定
shift = random.randint(-self.max_shift, self.max_shift)
if shift >= 0:
# 正の方向にシフトする場合
# センサーデータを左にシフトし、右側を端部の値で埋める
sensor = np.concatenate(
(sensor[:, shift:, :], np.repeat(sensor[:, -1:, :], shift, axis=1)),
axis=1,
)
else:
# 負の方向にシフトする場合
# センサーデータを右にシフトし、左側を端部の値で埋める
shift = -shift
sensor = np.concatenate(
(np.repeat(sensor[:, :1, :], shift, axis=1), sensor[:, :-shift, :]),
axis=1,
)
# ランダムに時間方向に反転する
if random.random() < 0.5:
music = np.flip(music, axis=1)
sensor = np.flip(sensor, axis=1)
# mixup implementation
mixup_alpha = self.alpha
batch_size = len(batch_ids)
indices = np.random.permutation(batch_size)
lmbda = np.random.beta(mixup_alpha, mixup_alpha, batch_size).reshape(-1, 1, 1)
music_mixup = music * lmbda + music[indices] * (1 - lmbda)
sensor_mixup = sensor * lmbda + sensor[indices] * (1 - lmbda)
y_mixup = y * lmbda.reshape(-1) + y[indices] * (1 - lmbda.reshape(-1))
return [music_mixup, sensor_mixup], y_mixup
# コールバックの設定
early_stopping = EarlyStopping(monitor="val_loss", patience=10)
model_checkpoint = ModelCheckpoint(
filepath="./best_weights.h5",
monitor="val_auc",
save_best_only=True,
save_weights_only=True,
verbose=1,
mode="max",
)
cosine_annealing = CosineAnnealingScheduler(T_max=10, eta_max=0.001, eta_min=0.0001)
batch_size = 128
steps_per_epoch = len(y_train_) // batch_size
train_generator = Music_Sensor_Generator(
music_train_,
sensor_train_,
y_train_,
batch_size=batch_size,
max_shift=12,
alpha=0.2,
use_bootstrap=False,
)
K.clear_session()
model = create_model()
model.fit(
train_generator,
steps_per_epoch=steps_per_epoch,
epochs=150,
validation_data=([music_val, sensor_val], y_val),
callbacks=[auc_callback, cosine_annealing, model_checkpoint],
)
# テストデータに対しても同様にデータを準備し、予測結果を出力してみます
residual_musics = np.setdiff1d(np.arange(250), train.music.values)
X_test = DataFrame(
[(i, m) for i in np.arange(100, 250) for m in residual_musics],
columns=["ID", "music"],
)
sensor_test = sensor_data[X_test.ID.values]
music_test = music_data[X_test.music.values]
# model.load_weights('./best_weights.h5')
import numpy as np
from tensorflow.keras.utils import Sequence
class Music_Sensor_Prediction_Generator(Sequence):
def __init__(self, music, sensor, batch_size=16, tta_shifts=0, max_shift=0):
self.batch_size = batch_size
self.music = music
self.sensor = sensor
self.tta_shifts = tta_shifts
self.max_shift = max_shift
def __len__(self):
return int(np.ceil(len(self.music) / self.batch_size))
def __getitem__(self, index):
indexes = np.arange(
index * self.batch_size, min((index + 1) * self.batch_size, len(self.music))
)
[music_batch, sensor_batch] = self.__data_generation(indexes)
y_pred_batch = self.__predict_on_batch([music_batch, sensor_batch])
return y_pred_batch
def __data_generation(self, batch_ids):
music = self.music[batch_ids]
sensor = self.sensor[batch_ids]
return [music, sensor]
def __predict_on_batch(self, batch):
# Original prediction without augmentation
predictions = self.model.predict(batch).flatten()
if self.tta_shifts > 0:
for _ in range(self.tta_shifts):
shift = random.randint(-self.max_shift, self.max_shift)
sensor = batch[1]
if shift >= 0:
sensor_shifted = np.concatenate(
(
sensor[:, shift:, :],
np.repeat(sensor[:, -1:, :], shift, axis=1),
),
axis=1,
)
else:
shift = -shift
sensor_shifted = np.concatenate(
(
np.repeat(sensor[:, :1, :], shift, axis=1),
sensor[:, :-shift, :],
),
axis=1,
)
batch_shifted = [batch[0], sensor_shifted]
predictions += self.model.predict(batch_shifted).flatten()
predictions /= self.tta_shifts + 1
return predictions
def set_model(self, model):
self.model = model
prediction_generator = Music_Sensor_Prediction_Generator(
music_test, sensor_test, batch_size=128, tta_shifts=8, max_shift=20
)
prediction_generator.set_model(model)
y_pred_test = np.concatenate(
[prediction_generator[i] for i in range(len(prediction_generator))]
)
X_test["music_pred"] = y_pred_test
# 上位3件をスペース区切りで書き込みます
submission = (
X_test.groupby("ID")
.apply(
lambda x: pd.Series(
{
"music": " ".join(
x.sort_values("music_pred", ascending=False)
.iloc[:7]["music"]
.values.astype(str)
)
}
)
)
.reset_index()
)
# ファイルに保存します
submission.to_csv("submission.csv", index=False)
submission
|
import numpy as np
import pandas as pd
import os
import statsmodels.formula.api as sm
import statsmodels.sandbox.tools.cross_val as cross_val
from sklearn.preprocessing import PolynomialFeatures
from sklearn import linear_model as lm
from regressors import stats
from sklearn import metrics
from sklearn.linear_model import LinearRegression
from sklearn.model_selection import (
KFold,
cross_val_score,
cross_val_predict,
LeaveOneOut,
)
print(os.listdir("../input"))
# Interactive Terms: Statsmodel
d = pd.read_csv("/kaggle/input/datalab4/diabetes.csv")
d.head()
main = sm.ols(formula="chol ~ age+frame", data=d).fit()
print(main.summary())
inter = sm.ols(formula="chol ~ age*frame", data=d).fit()
print(inter.summary())
inter = sm.ols(formula="chol ~ gender*frame", data=d).fit()
print(inter.summary())
inter = sm.ols(formula="chol ~ height*weight", data=d).fit()
print(inter.summary())
import statsmodels.api as sma
d = pd.read_csv("/kaggle/input/datalab4/diabetes.csv")
d.head()
chol1 = sm.ols(formula="chol ~ 1", data=d).fit()
chol2 = sm.ols(formula="chol ~ age", data=d).fit()
chol3 = sm.ols(formula="chol ~ age+frame", data=d).fit()
chol4 = sm.ols(formula="chol ~ age*frame", data=d).fit()
print(sma.stats.anova_lm(chol1, chol2, chol3, chol4))
from mlxtend.feature_selection import SequentialFeatureSelector as sfs
d = pd.read_csv("/kaggle/input/datalab4/nuclear.csv")
d = d.rename(index=str, columns={"cum.n": "cumn"})
d.head()
inputDF = d[["date", "cap", "pt", "t1", "t2", "pr", "ne", "ct", "bw"]]
outputDF = d[["cost"]]
model = sfs(
LinearRegression(),
k_features=5,
forward=True,
verbose=2,
cv=5,
n_jobs=-1,
scoring="r2",
)
model.fit(inputDF, outputDF)
print(model.k_feature_idx_)
print(model.k_feature_names_)
# Backward Selection: Scikit-Learn
inputDF = d[["date", "cap", "pt", "t1", "t2", "pr", "ne", "ct", "bw"]]
outputDF = d[["cost"]]
backwardModel = sfs(
LinearRegression(),
k_features=5,
forward=False,
verbose=2,
cv=5,
n_jobs=-1,
scoring="r2",
)
backwardModel.fit(inputDF, outputDF)
print(model.k_feature_idx_)
print(model.k_feature_names_)
from sklearn import metrics
from sklearn.linear_model import LinearRegression
# LOOCV: Scikit-Learn
d = pd.read_csv("/kaggle/input/datalab4/auto.csv")
inputDF = d[["mpg"]]
outputDF = d[["horsepower"]]
model = LinearRegression()
loocv = LeaveOneOut()
rmse = np.sqrt(
-cross_val_score(
model, inputDF, outputDF, scoring="neg_mean_squared_error", cv=loocv
)
)
print(rmse.mean())
predictions = cross_val_predict(model, inputDF, outputDF, cv=loocv)
# print(predictions)
df = pd.read_csv("/kaggle/input/datalab4/auto.csv")
# kFCV: Scikit-Learn
inputDF = df[["mpg"]]
outputDF = df[["horsepower"]]
model = LinearRegression()
kf = KFold(5, shuffle=True, random_state=42).get_n_splits(inputDF)
rmse = np.sqrt(
-cross_val_score(model, inputDF, outputDF, scoring="neg_mean_squared_error", cv=kf)
)
print(rmse.mean())
predictions = cross_val_predict(model, inputDF, outputDF, cv=kf)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import numpy as np
import pandas as pd
# text processing libraries
import re
import string
from keras.layers import (
Dense,
LSTM,
Embedding,
Bidirectional,
RepeatVector,
TimeDistributed,
)
import nltk
from nltk.corpus import stopwords
from sklearn import model_selection
from sklearn.feature_extraction.text import CountVectorizer, TfidfVectorizer
from sklearn.linear_model import LogisticRegression
from sklearn.naive_bayes import MultinomialNB
from sklearn.metrics import f1_score
from sklearn import preprocessing, decomposition, model_selection, metrics, pipeline
from sklearn.model_selection import GridSearchCV, StratifiedKFold, RandomizedSearchCV
from keras.callbacks import ModelCheckpoint
# matplotlib and seaborn for plotting
import matplotlib.pyplot as plt
import seaborn as sns
import tensorflow as tf
# File system manangement
import os
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from tifffile import imread
from wordcloud import WordCloud
import keras, os
from keras.models import Sequential
from keras.layers import (
Dense,
Conv2D,
MaxPool2D,
Flatten,
BatchNormalization,
Activation,
LSTM,
CuDNNLSTM,
Embedding,
Dropout,
)
from keras.preprocessing.image import ImageDataGenerator
from tensorflow.keras.preprocessing.text import Tokenizer
from tensorflow.keras.preprocessing.sequence import pad_sequences
import numpy as np
from keras.optimizers import Adam
# Suppress warnings
import warnings
warnings.filterwarnings("ignore")
train = pd.read_csv("../input/nlp-getting-started/train.csv")
print("Training data shape: ", train.shape)
train.head()
# Testing data
test = pd.read_csv("../input/nlp-getting-started/test.csv")
print("Testing data shape: ", test.shape)
test.head()
# Missing values in training set
train.isnull().sum()
test.isnull().sum()
train["target"].value_counts()
disaster_tweets = train[train["target"] == 1]["text"]
disaster_tweets.values[1]
# not a disaster tweet
non_disaster_tweets = train[train["target"] == 0]["text"]
non_disaster_tweets.values[1]
sns.barplot(
y=train["keyword"].value_counts()[:20].index,
x=train["keyword"].value_counts()[:20],
orient="h",
)
# Replacing the ambigious locations name with Standard names
train["location"].replace(
{
"United States": "USA",
"New York": "USA",
"London": "UK",
"Los Angeles, CA": "USA",
"Washington, D.C.": "USA",
"California": "USA",
"Chicago, IL": "USA",
"Chicago": "USA",
"New York, NY": "USA",
"California, USA": "USA",
"FLorida": "USA",
"Nigeria": "Africa",
"Kenya": "Africa",
"Everywhere": "Worldwide",
"San Francisco": "USA",
"Florida": "USA",
"United Kingdom": "UK",
"Los Angeles": "USA",
"Toronto": "Canada",
"San Francisco, CA": "USA",
"NYC": "USA",
"Seattle": "USA",
"Earth": "Worldwide",
"Ireland": "UK",
"London, England": "UK",
"New York City": "USA",
"Texas": "USA",
"London, UK": "UK",
"Atlanta, GA": "USA",
"Mumbai": "India",
},
inplace=True,
)
sns.barplot(
y=train["location"].value_counts()[:5].index,
x=train["location"].value_counts()[:5],
orient="h",
)
# Applying a first round of text cleaning techniques
def clean_text(text):
"""Make text lowercase, remove text in square brackets,remove links,remove punctuation
and remove words containing numbers."""
text = text.lower()
text = re.sub("\[.*?\]", "", text)
text = re.sub("https?://\S+|www\.\S+", "", text)
text = re.sub("<.*?>+", "", text)
text = re.sub("[%s]" % re.escape(string.punctuation), "", text)
text = re.sub("\n", "", text)
text = re.sub("\w*\d\w*", "", text)
return text
# Applying the cleaning function to both test and training datasets
train["text"] = train["text"].apply(lambda x: clean_text(x))
test["text"] = test["text"].apply(lambda x: clean_text(x))
# Let's take a look at the updated text
train["text"].head()
tokenizer = nltk.tokenize.RegexpTokenizer(r"\w+")
train["text"] = train["text"].apply(lambda x: tokenizer.tokenize(x))
test["text"] = test["text"].apply(lambda x: tokenizer.tokenize(x))
train["text"].head()
def remove_stopwords(text):
"""
Removing stopwords belonging to english language
"""
words = [w for w in text if w not in stopwords.words("english")]
return words
train["text"] = train["text"].apply(lambda x: remove_stopwords(x))
test["text"] = test["text"].apply(lambda x: remove_stopwords(x))
train.head()
# After preprocessing, the text format
def combine_text(list_of_text):
"""Takes a list of text and combines them into one large chunk of text."""
combined_text = " ".join(list_of_text)
return combined_text
train["text"] = train["text"].apply(lambda x: combine_text(x))
test["text"] = test["text"].apply(lambda x: combine_text(x))
train["text"]
train.head()
sentences_train = train["text"].values
sentences_test = test["text"].values
print(np.mean([len(text) for text in train["text"]]))
print(train["text"][0])
print(len(train["text"][0]))
tokenizer = Tokenizer()
tokenizer.fit_on_texts(sentences_train)
sequences_train = tokenizer.texts_to_sequences(sentences_train)
# This is used for Embedding layer afterwards
num_words = len(tokenizer.word_index)
print(num_words)
maxlen_tokens = 20
# The reason that the maxlength is different than the true max length is that the texts are tokenized
padded_sequences_train = pad_sequences(
sequences_train, maxlen=maxlen_tokens, padding="post", truncating="post"
)
padded_sequences_train[0]
num_validation_samples = round(0.3 * len(padded_sequences_train))
seq_train = padded_sequences_train[num_validation_samples:]
labels_train = train["target"].values[num_validation_samples:]
seq_validate = padded_sequences_train[:num_validation_samples]
labels_validate = train["target"].values[:num_validation_samples]
print(len(seq_train))
print(len(seq_validate))
print(seq_train[0])
print(seq_train.shape)
# model = Sequential()
# model.add(Embedding(num_words, 32, input_length=maxlen_tokens))
# model.add(LSTM(128))
# model.add(RepeatVector(maxlen_tokens))
# model.add(Dense(units=1, activation='sigmoid'))
# model.build(input_shape=(None, maxlen_tokens))
# model.summary()
model = Sequential()
model.add(Embedding(num_words, 32, input_length=maxlen_tokens))
model.add(tf.keras.layers.GRU(128, return_sequences=True))
model.add(tf.keras.layers.GRU(128))
model.add(Dense(units=256, activation="relu"))
model.add(Dense(units=1, activation="sigmoid"))
model.build(input_shape=(None, maxlen_tokens))
model.summary()
import tensorflow_addons as tfa
metric = tfa.metrics.F1Score(num_classes=2, threshold=0.5)
opt = Adam(learning_rate=0.000001)
model.compile(
optimizer=opt,
loss="binary_crossentropy",
metrics=["Recall", "Precision", "val_loss"],
)
import tensorflow as tf
early_stopping = tf.keras.callbacks.EarlyStopping(
monitor="val_loss", patience=7, restore_best_weights=True
)
# filename = 'ModelWeights.19_Mar_23'
# from keras.callbacks import ModelCheckpoint
# checkpoint = ModelCheckpoint(filename, monitor='val_loss', verbose=1, save_best_only=True, mode='min')
hist = model.fit(
seq_train,
labels_train,
validation_data=(seq_validate, labels_validate),
epochs=100,
batch_size=100,
callbacks=[early_stopping],
)
def plot_history(history):
hist = pd.DataFrame(history.history)
hist["epoch"] = history.epoch
plt.figure()
plt.xlabel("Epoch")
plt.ylabel("Mean Abs Error ")
plt.plot(hist["epoch"], hist["mae"], label="Train Error")
plt.plot(hist["epoch"], hist["val_mae"], label="Val Error")
plt.legend()
plt.ylim([0, 5])
plt.figure()
plt.xlabel("Epoch")
plt.ylabel("Mean Square Error ")
plt.plot(hist["epoch"], hist["mse"], label="Train Error")
plt.plot(hist["epoch"], hist["val_mse"], label="Val Error")
plt.legend()
plt.ylim([0, 20])
plot_history(history)
sequences_test = tokenizer.texts_to_sequences(sentences_test)
padded_sequences_test = pad_sequences(
sequences_test, maxlen=maxlen_tokens, padding="post", truncating="post"
)
padded_sequences_test[0]
predictions = model.predict(padded_sequences_test, verbose=1)
pred = np.transpose(predictions)[0]
print(pred)
submission_df = pd.DataFrame()
submission_df["id"] = test["id"]
submission_df["target"] = list(map(lambda x: 0 if x < 0.5 else 1, pred))
print(submission_df.head())
submission_df["target"].value_counts()
submission_df.to_csv("submission.csv", index=False)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)y
import os
import matplotlib.pyplot as plt
import seaborn as sns
sns.set()
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
df_train = pd.read_csv("/kaggle/input/nlp-getting-started/train.csv")
df_train.info()
# ## Vetorization
# - Tranformar os dados em tokens
df_train.head(10)
# count target
sns.countplot(df_train["target"])
from keras.preprocessing.text import Tokenizer # create tokens
from keras import preprocessing # convert list sequences to array numpy
# pre processing
from keras.utils.np_utils import to_categorical # convert to one-hot-encoding
from sklearn.model_selection import train_test_split # spit data in train and test
# ## Define X train
NUM_WORDS = 1000
tokenizer = Tokenizer(num_words=NUM_WORDS)
tokenizer.fit_on_texts(df_train["text"])
sequences = tokenizer.texts_to_sequences(df_train["text"])
X = preprocessing.sequence.pad_sequences(sequences)
X = np.asarray(X).astype("float32")
MAX_LEN = X.shape[1]
def norm(data):
return (data - np.min(data)) / (np.max(data) - np.min(data))
word_index = tokenizer.word_index
print("Found %s unique tokens." % len(word_index))
# ## Define Y train
# define Y_TRAIN
Y = np.asarray(df_train["target"])
Y = to_categorical(Y, num_classes=2)
# ## Split data train and test
X_train, X_test, Y_train, Y_test = train_test_split(X, Y, test_size=0.2, random_state=2)
# # Model
from keras import models
from keras import layers
def SMNN(x_input):
model = models.Sequential()
model.add(layers.Dense(12, activation="tanh", input_shape=(x_input,)))
model.add(layers.Dense(12, activation="tanh"))
model.add(layers.Dense(2, activation="sigmoid"))
# rmsprop, adam
model.compile(optimizer="adam", loss="binary_crossentropy", metrics=["accuracy"])
return model
model = SMNN(X_train.shape[1])
# train Neural Network
hist = model.fit(
X_train, Y_train, epochs=30, batch_size=512, validation_split=0.2, verbose=0
)
df_fit = pd.DataFrame(hist.history)
fig = plt.figure(figsize=(20, 6))
ax = fig.add_subplot(121)
ax.plot(df_fit["loss"], "-o", label="loss")
ax.plot(df_fit["val_loss"], "--", label="validation")
ax.legend()
ax = fig.add_subplot(122)
ax.plot(df_fit["accuracy"], "-o", label="accuracy")
ax.plot(df_fit["val_accuracy"], "--", label="validation")
ax.legend()
plt.show()
scores = model.evaluate(X_test, Y_test)
print("loss .......: ", round(scores[0], 3))
print("acc ........: ", round(scores[1] * 100, 2))
# predict
y_pred = model.predict(X_test)
y_pred.shape
# scores
from sklearn.metrics import f1_score
# computer metric F1
f1_score(np.argmax(Y_test, axis=1), np.argmax(y_pred, axis=1))
# ## Process Test data
df_test = pd.read_csv("/kaggle/input/nlp-getting-started/test.csv")
df_test.head()
X_TEST = preprocessing.sequence.pad_sequences(
tokenizer.texts_to_sequences(df_test["text"].values), maxlen=MAX_LEN
)
X_TEST = np.asarray(X_TEST).astype("float32")
X_TEST.shape
y_pred_test = model.predict(X_TEST)
df_submission = pd.DataFrame(
{"id": df_test["id"].values, "target": np.argmax(y_pred_test, axis=1)}
)
df_submission.head()
# save
df_submission.to_csv("submission.csv", encoding="utf-8", index=False)
|
# # An Analysis of Developer Communities: Insights from the 2022 Stack Overflow global Survey
# 
# ### Exploratory Data Analysis(EDA)
# Exploratory Data Analysis (EDA) is an approach used to analyze and summarize data to gain insights and develop an understanding of the dataset to make more informed decisions.It involves visualizing and summarizing data using statistical and graphical techniques without making any assumptions or fitting any models to the data.
# The main objectives of EDA :
# - To Gain a deeper understanding of the dataset and the relationships between variables.
# - Identify patterns, trends, and anomalies in the data.
# - Prepare the data for further analysis, such as modeling or hypothesis testing.
# Basically, EDA is an important and first step in data analysis that helps to guide subsequent analysis and ensure that the data is suitable for the intended purpose.
# ### Stack Overflow
# Stack Overflow is public platform or community based space to find and contribute answers to programming questions.Created in 2008 by Joel Spolsky and Jeff Atwood and since then Stack Overflow is most visted sites on internet.
# Stack Overflow also has a reputation system to reward users for contributing helpful answers and participating in their community.
# Every year Stack Overflow conducts survey were respondents are recruited primarily through channels owned by Stack Overflow.In year 2022, about 73,268 developers from 180 countries took Stack Overflow survey.
# #### Project Outline
# * Install and Import the required libraries.
# * Data preparation and cleaning with pandas.
# * Performing exploratory analysis and visualization.
# * Asking and answering interesting questions.
# * Summarizing inferences and drawing conclusions.
# ### Introduction
# We will perform exploratory data analysis(EDA) on the StackOverflow Developer Survey 2022 dataset. This dataset contains responses to an annual survey conducted by StackOverflow. You can find the dataset on: https://insights.stackoverflow.com/survey
# The survey was fielded from May 11, 2022 to June 1, 2022.The number of responses we consider “qualified” for analytical purposes based on time spent on the full, completed survey; another approximately 53 responses were submitted but not included in the analysis because respondents spent less than three minutes on the survey.-*source Stack Overflow*
# ### 1. Install and Import required libraries
# Pip is the standard package installer for Python, use `!pip install package_name` when a package is not included in standard library.
# Below are the packages we need to install and import for this project.
# import data manipulation and data analysis libary in python
import pandas as pd
# import numerical computing library in Python
import numpy as np
# import library for creating static plots and visualizations
import matplotlib
import matplotlib.pyplot as plt
# to turn on “inline plotting”, to make plot graphics appear in notebook
# import the Seaborn library for statistical data visualization
import seaborn as sns
# import the Plotly Express library for interactive data visualization
import plotly.express as px
# for plotting geospatial data with folium library
import folium
# import WordCloud library for creating word clouds
import wordcloud
# ### Download the dataset
# There are several options to get dataset into Jupyter:
# * Download CSV file manually and upload it via Jupyter's GUI.
# * Use the urlretrieve function from the urllib.request to download CSV files from a raw URL.
# * Use the opendatasets library to download datasets from Kaggle datasets and Google Drive URLs.
# In this project, we will host the dataset on Google Drive at this location:
# https://drive.google.com/file/d/1tfBHyNbk8sp-nuE8L7wOB5T7KK9uNNIx/view?usp=share_link
# Now,we will use opendatasets helper library to download files from the above URL.
import opendatasets as od
survey_url = "https://drive.google.com/file/d/1tfBHyNbk8sp-nuE8L7wOB5T7KK9uNNIx/view?usp=share_link"
od.download(survey_url)
# check that the dataset is in stack-overflow-developer-survey-2022 and get list of files in it
import os
data_dir = "stack-overflow-developer-survey-2022"
os.listdir(data_dir)
# ### 2. Data Preprocessing and Cleaning
# For any business intelligence strategy the quality of data is the most crucial element.
# Data preprocessing is a critical step that encompases various techniques to transform raw data into a format that is suitable for analysis.
# Data cleaning involves fixing or removing incorrect, corrupted, incorrectly formatted, duplicate, or incomplete data within a dataset.
# Before we could dive into analysis , let's check the schema file which consists survey questions.
# refer to schema file
schema_file = data_dir + "/survey_results_schema.csv"
schema_df = pd.read_csv(schema_file, index_col="qname")
# Let's check the survey questions asked.
schema_df.iloc[2]
# As, there are some HTMl tags with text of questions. For better readbility ,let's clean schema_df.
# import re module to get support for regular expressions in python
import re
# function that takes a string of HTML code as input and returns the cleaned text
def clean_html_tags(html_string):
# a regular expression to search for and replace all HTML tags with an empty string
clean_text = re.sub("<[^<]+?>", "", html_string)
# return the cleaned text with leading and trailing whitespace removed
return clean_text.strip()
# Now, we apply the `clean_html_tags()` function to the question column of the schema_df DataFrame using the `apply()` method. This cleans up any HTML tags that may be present in the question column.
# Secondly,replaces any newline characters (\n) in the question column with an empty string using the `str.replace()` method. This ensures that all text in the question column is formatted consistently and does not contain any unwanted newline characters.
schema_df["question"] = (
schema_df["question"].apply(clean_html_tags).str.replace("\n", "")
)
# - We see qname as index, to simplify will make qname as dataframe column.
# to set the maximum column width for display in the DataFrame to 300 characters
pd.options.display.max_colwidth = 300
schema_df1 = schema_df["question"]
schema_df2 = pd.DataFrame(schema_df1).reset_index()
schema_df2.iloc[1:]
# Let's take a look at `Employment` question asked to survey respondents.
Emp_Ques = schema_df2[schema_df2["qname"] == "Employment"]["question"].to_string(
index=False
)
Emp_Ques
# Now, we will read survey data CSV file using Pandas library.
# load csv files
import pandas as pd
survey_df = pd.read_csv(data_dir + "/survey_results_public.csv")
# Take a look at first 5 rows.
survey_df.head(5)
# Let's get more basic information about this survey_df dataframe.
survey_df.shape
survey_df.size
# As, there are 79 columns we will be selecting columns relevant for our analysis.
# select subset of columns relevant for our analysis:
survey_df_cols = [ # demograpics
"EdLevel",
"Country",
"Age",
"Gender",
"Ethnicity",
# professional experience
"MainBranch",
"CodingActivities",
"LearnCode",
"YearsCode",
"YearsCodePro",
"LanguageHaveWorkedWith",
"LanguageWantToWorkWith",
"WorkExp",
# Employment
"Employment",
"DevType",
"OrgSize",
# Respondents attitude and behaviour
"Blockchain",
"MentalHealth",
"TimeSearching",
"TimeAnswering",
]
# how many cols selected
len(survey_df_cols)
# Let's make a copy of survey_df with these subset of columns to survey_df1, new dataframe.
# let's make a copy of this selected cols data into new data frame survey_df1
survey_df1 = survey_df[survey_df_cols].copy()
# Before, we start knowing more about our data we will do basic check like number of null and unique values in each columns.
# The output of the below code snippet will show us : column information, its null values and unique values in each column.
import pandas as pd
from tabulate import tabulate
# Check for null and unique values
null_values = survey_df1.isnull().sum()
unique_values = survey_df1.nunique()
# Create a new dataframe
null_unique_df = pd.DataFrame(
{
"Data Type": survey_df1.dtypes,
"Null Values": null_values.values,
"Unique Values": unique_values.values,
}
)
null_unique_df.index.name = "Column Names"
# Display the information and null/unique values as a table
print(tabulate(null_unique_df, headers="keys", tablefmt="psql"))
# Almost all columns have null values, except MainBranch as it was mandotary to choose any one out of 6 options provided, which is why we see MainBranch has 6 unique values.
# MainBranch represents basic information of respondents in survey questionnaire.
# Now, that we see some of the columns which has to be numeric , but may be those columns contain 'nan' values,those columns are treated as object type rather 'int' or 'float' type.
# let's convert some other columns into numeric data
survey_df1["YearsCode"] = pd.to_numeric(survey_df1.YearsCode, errors="coerce")
survey_df1["YearsCodePro"] = pd.to_numeric(survey_df1.YearsCodePro, errors="coerce")
# #### summary statistics of the dataset
survey_df1.describe()
# Now, we see WorkExp column has max value of 50, let's do some data validation.
# If work experience of respondent is larger than the age , it's better to discard that row. There an be many reasons for that:
# - Respondent accidently filled up wrong age or work experience value.
# - Respondent didnt take the survey serious or considered important to give true values
# Let's see the unique values in work experience column.
survey_df1["WorkExp"].unique()
# Let's check how many respondents have work experience more than age mentioned.
survey_df1[survey_df1["WorkExp"] == 50.0]["Age"].value_counts()
# From the above , its clear that some enteries have age less than the work experience mentioned.We will remove those rows from the data loaded.
survey_df1["Age"].unique()
# To drop rows where WorkExp is more than a given age, we first need to convert the age ranges into numeric values. Since the age ranges are not consistent, we will need to make some assumptions about their values. For example, we can assume that "18 or below" means an age of 18, and "65 or Older" means an age of 65.
# Once we have converted the age ranges into numeric values, we can use a boolean mask to filter out the rows where WorkExp is greater than the age. Here's the code:
# create a dictionary to convert age ranges into numeric values
import numpy as np
age_dict = {
"nan": np.nan,
"25-34 years old": 30,
"35-44 years old": 40,
"Under 18 years old": 18,
"18-24 years old": 20,
"45-54 years old": 50,
"55-64 years old": 60,
"65 years or older": 65,
"Prefer not to say": np.nan,
}
# create a copy of survey_df1
survey_df2 = survey_df1.copy()
# convert age column to numeric values
survey_df2["Age"] = survey_df2["Age"].map(age_dict)
# let's consider rowswhere WorkExp is lesser than Age : Boolean Mask
bool_mask = survey_df2["WorkExp"] < survey_df2["Age"]
# apply the bool_mask to the DataFrame to drop the rows where WorkExp is greater than Age
survey_df2 = survey_df2[bool_mask]
survey_df2 = survey_df2.reset_index(drop=True)
# Let's check, did we succesfully get rid of rows where age is smaller than work experience.
survey_df2.describe()
survey_df2[survey_df2["WorkExp"] == 50.0]["Age"].value_counts()
# We will also have to clean Gender column, as per survey information provided respondents were allowed to select multiple options. But for our analysis we will consider only rows that has single gender value.
survey_df2["Gender"].value_counts()
# We will remove rows having more than one option to simplify analysis.
import numpy as np
survey_df2.where(~(survey_df2.Gender.str.contains(";", na=False)), np.nan, inplace=True)
# gender column
survey_df2["Gender"].value_counts()
# We can dive into analysis,as we have cleaned up and prepared data for analysis.
# ### Exploratory analysis and visualization
# A survey of this scale generally tends to have some selection bias. Before, we start asking right questions the best is to start with knowing respondents better such as their demographics like country, gender, employment type, education etc.
# copy of survey_df2()
countries_df = survey_df2.copy()
# improving readability
countries_df.loc[
countries_df["Country"] == "United Kingdom of Great Britain and Northern Ireland",
"Country",
] = "UK"
countries_df.loc[
countries_df["Country"] == "United States of America", "Country"
] = "USA"
# Now we will find out the number of respondents from each country.
# countries with the highest number of respondents
countries_counts = countries_df.Country.value_counts()
# countries with highest number of respondents in percentage
countries_counts_percent = (
countries_df.Country.value_counts(normalize=True, ascending=False) * 100
)
# dataframe having columns for counts and percentage for number of respondents
country_counts_df = pd.DataFrame(
{
"Country": countries_counts.index,
"Count": countries_counts.values,
"Percentage": countries_counts_percent.values,
}
)
country_counts_df
# Let's see distribution of respondents on map.
import folium
countries_geojson = (
"https://raw.githubusercontent.com/johan/world.geo.json/master/countries.geo.json"
)
country_counts_df.at[0, "Country"] = "United States of America"
country_counts_df.at[12, "Country"] = "Russia"
m = folium.Map(location=[30, 0], zoom_start=2, tiles="Stamen Terrain")
folium.Choropleth(
geo_data=countries_geojson,
data=country_counts_df,
columns=["Country", "Count"],
key_on="feature.properties.name",
threshold_scale=[1, 30, 100, 300, 1_000, 3_000, 10_000, 13_000, 14_000],
nan_fill_color="Black",
fill_color="Greens",
fill_opacity=0.7,
line_opacity=0.2,
legend_name="Respondents",
highlight=True,
smooth_factor=0,
).add_to(m)
# Add a title to the map
title_html = (
'<h3 align="center" style="font-size:16px"><b>Respondent Count by Country</b></h3>'
)
m.get_root().html.add_child(folium.Element(title_html))
m
# Most respondents are from `USA and India`, and the black regions indicates no respondents.
# The reasons behind this could be
# - As the survey was in English, and the countires which is shown in black color may not have highest english speaking population or simply these countires are non-english speaking countries.
# - Other reason as per Stack Overflow official is due to United States transport/export sanctions, the survey was, unfortunately, inaccessible to prospective respondents in `Crimea, Cuba, Iran, North Korea, and Syria`, due to the traffic being blocked by third-party survey software. While some respondents used VPNs to get around the block, the limitation should be kept in mind when interpreting survey results.
# This shows us that the survey may not be respresent global programming community and also programmers from non-english speaking countries are underrepresented.
# #### Top 15 countires with highest number of respondents.
# top 15 countries
top_15_countries = country_counts_df.head(15)
top_15_countries
import matplotlib.pyplot as plt
import seaborn as sns
# set font size
plt.rcParams.update({"font.size": 10})
# set figure size
plt.figure(figsize=(10, 6))
# create the barplot and rotate x-axis labels
sns.barplot(x="Country", y="Percentage", data=top_15_countries)
plt.xticks(rotation=75)
# add value labels to the bars
for index, row in top_15_countries.iterrows():
plt.text(index, row["Percentage"], "{:.2f}%".format(row["Percentage"]), ha="center")
# set title and axis labels
plt.title("Top 15 Countries by Percentage")
plt.xlabel("Country")
plt.ylabel("Percentage")
# add a grid
# plt.grid(True)
# use a different color palette
# sns.set_palette("husl")
# display the plot
plt.show()
# ### Categorical Feature → Gender
# Let's check the distribution of respondent's gender.
# copy of survey_df2
gender_df = survey_df2.copy()
# readabilty
gender_df["Gender"] = gender_df["Gender"].replace(
"Non-binary, genderqueer, or gender non-conforming", "Non-binary,genderqueer,GNC"
)
# count the number of occurrences of each gender in the Gender column
gender_counts = gender_df["Gender"].value_counts()
gender_counts
gender_counts = gender_df["Gender"].value_counts()
# index values of the Gender column
labels = gender_counts.index
# count of each gender
sizes = gender_counts.values
# for pie chart
fig, ax = plt.subplots()
ax.pie(sizes, autopct="%1.1f%%", startangle=180)
ax.axis("equal")
# Legend
plt.legend(labels=labels, loc="best", fontsize=10)
# Title
plt.title("Gender Distribution using pie chart")
plt.rcParams.update({"font.size": 14})
# Show plot
plt.show()
# Can summarise that the majority of survey respondents are Man with 93.1 % and women in programming are 4.9 %.The other gender minorities are less than 1%.
# ### Ordinal Feature → Edlevel
# Let's see respondents Education level.
# copy of survey_df2
education_df = survey_df2.copy()
# improve readbility on graph
education_df.loc[
education_df["EdLevel"]
== "Secondary school (e.g. American high school, German Realschule or Gymnasium, etc.)",
"EdLevel",
] = "Secondary School"
education_df.loc[
education_df["EdLevel"] == "Some college/university study without earning a degree",
"EdLevel",
] = "Undergrads"
# count the number of occurrences of each education degree in the EdLevel column
education_counts = education_df["EdLevel"].value_counts()
education_counts
# Calculate percentage
total = sum(education_counts.values)
percentages = [count / total * 100 for count in education_counts.values]
# Create horizontal bar chart
fig, ax = plt.subplots(figsize=(10, 6))
ax.barh(education_counts.index, percentages)
# Add x-axis label and title
ax.set_xlabel("Percentage")
ax.set_title("Distribution of Education Levels")
# Invert y-axis to show the highest percentage on top
ax.invert_yaxis()
# Display percentage on each bar
for i, v in enumerate(percentages):
ax.text(v + 0.5, i, f"{v:.1f}%", color="black", fontsize=12)
# Display plot
plt.show()
# Almost 50% of respondents own Bachlor's degree and 25% of respondents have Master's degree.Interestingly, respondents with just primary/elementary education level also love to program.
# Curious to know education level by gender.
# ### Distribution of respondents education level by gender
# count the number of occurrences of each degree by gender
counts = education_df.groupby(["EdLevel", "Gender"]).size().unstack()
counts
import plotly.graph_objects as go
# count the number of occurrences of each degree by gender
counts = education_df.groupby(["EdLevel", "Gender"]).size().unstack()
# create the stacked bar chart
fig = go.Figure(
data=[
go.Bar(name=gender, x=counts.index, y=counts[gender], hovertemplate="%{y}")
for gender in counts.columns
]
)
fig.update_layout(
barmode="stack",
title="Distribution of respondents education level by gender",
xaxis_title="EdLevel",
yaxis_title="Count",
)
# display the chart
fig.show()
# Bachelor's degree is the most pursued by all gender type. We see 936 women holds bachelor's degree where as 16,353 men has bachelor's degree which is approx. 5.73 times.
# ### Categorical Feature → Employment
# As multiple options were allowed to opt , there are many rows with multiple employment status.To make analysis easier, we need to define function which turns columns with list of values into data frame having columns for each option.
def split_multicolumn(col_series):
result_df = col_series.to_frame()
options = []
# Iterate over the column
for idx, value in col_series[col_series.notnull()].iteritems():
# Break each value into list of options
for option in value.split(";"):
# Add the option as a column to result
if not option in result_df.columns:
options.append(option)
result_df[option] = False
# Mark the value in the option column as True
result_df.at[idx, option] = True
return result_df[options]
Employment_df = split_multicolumn(survey_df2.Employment)
Employment_df.loc[50:55]
# The Employment_df has one column for each option that can be selected as a response. If a respondent has chosen an option, the corresponding column's value is True. Otherwise, it is False.
total_respondents = Employment_df.sum()
# Calculate the total number of respondents for each category of employment
Employment_counts = Employment_df.sum().sort_values(ascending=False)
# percentage
total_respondents = Employment_counts.sum()
employment_percentages = (Employment_counts / total_respondents) * 100
employment_counts_per_df = pd.DataFrame(
{
"Employment": Employment_counts.index,
"Count": Employment_counts.values,
"Percentage": employment_percentages.values,
}
)
employment_counts_per_df
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
# Assuming 'Employment_counts' is a Pandas Series with the data
total_respondents = Employment_counts.sum()
employment_percentages = Employment_counts.apply(
lambda x: (x / total_respondents) * 100
)
sns.set_style("darkgrid")
fig, ax = plt.subplots(figsize=(8, 6))
# Create the barplot
sns.barplot(
x=employment_percentages.values,
y=employment_percentages.index,
palette="mako",
ax=ax,
)
# Set the title and axis labels
ax.set_title(
"Distribution of Respondents Employment Status", fontsize=12, fontweight="bold"
)
ax.set_xlabel("Percentage of Respondents", fontsize=10)
ax.set_ylabel("Employment Status", fontsize=10)
# Set x-axis limit to 100
ax.set_xlim(0, 100)
# Add labels to the bars
for i, v in enumerate(employment_percentages.values):
ax.text(v + 1, i, str(round(v, 2)) + "%", color="black", fontweight="bold")
# Show the plot
plt.tight_layout()
plt.show()
# The above bar plot shows that, 81.73 % of respondents are Employed to full-time roles. Interestingly, the survey was taken by programmers having different employment status such as respondents who are part-time employee with 2.72% ,retired respondents 0.04 % and also student both full-time(1.75%) and part-time(2.19%).
# We have one more categorical feature to analyze, Developer type. Developer type represents different professional roles of respondents.
# ### Categorical Feature → DevType
# Few respondents might have different roles in their professional life, for the same reason during the survey the respondents were allowed to opt for multiple roles for which they work for.
# Therefore, to make our analysis we need to call the function defined earlier `split_multicolumn()`.
DevType_df = split_multicolumn(survey_df2.DevType)
DevType_df.loc[50:55]
# counting the total number of occurrences of each unique developer type
DevType_count = DevType_df.sum().sort_values(ascending=False)
# to Calculate the percentage of each value in the DevType_count Series
total_count = DevType_count.sum()
DevType_percentage = DevType_count / total_count * 100
DevType_df1 = pd.DataFrame(
{
"Type": DevType_count.index,
"Count": DevType_count.values,
"Percentage": DevType_percentage.values,
}
)
DevType_df1
# From the above data frame, we can say that most of the survey respondents are Developers and very less respondents working in marketing or sales has taken the progranmmers survey conducted by stack overflow.
# The main reasons behind this could be :
# 1. As we know many developers use StackOver Flow to solve work related or code realted queries , which also indicates many developers across world have heard about StackOver Flow community.
# 2. On, the other hand, there is possibitly of marketing or sales or any non-tech professional who codes but they are not aware of community of programmers like StackOver Flow.
# We will use 'wordclouds' as a tool to quickly visualize the most common job role of survey respondents.
# Import the necessary libraries
from wordcloud import WordCloud
import matplotlib.pyplot as plt
# Join the 'DevType' column data into a single string and remove semicolons and commas
words = " ".join(
(
job
for job in survey_df2.DevType.dropna()
.str.replace(";", " ")
.str.replace(",", " ")
)
)
# Generate the WordCloud object with specified parameters
wc = WordCloud(
collocation_threshold=int(1e6), width=800, height=400, background_color="white"
).generate(words)
# Create a figure and turn off the axis and grid lines for aesthetic purposes
plt.figure(figsize=(16, 8))
plt.axis("off")
plt.grid(False)
# Display the WordCloud image
plt.imshow(wc)
# The above wordcloud shows the most common profession of respondents , we see Developer as most common word and marketing and sales the least.
# ### Numerical Feature → Age
# Earliar we dropped rows which failed to satisy condition 'WorkExp < Age', to do so we transformed the age ranges to numeric. But to get some rough idea of respondents age distribution we will be using Age ranges.
# Age
# group the data by age and count the number of responses in each group
age_counts = survey_df1.groupby("Age")["Age"].count()
# calculate the percentage of responses for each age group
age_percentages = age_counts / survey_df1.shape[0] * 100
# create a new dataframe with the results
age_df = pd.DataFrame(
{
"Age": age_percentages.index,
"count": age_counts.values,
"Percentages": age_percentages.values,
}
)
age_df
# The age range to which most of the respondents belong is 25-34 years old with 38.36 %. The least is 65 years or older with 0.76%.
# Let's use a bar plot to visualize the same better.
sns.set_style("darkgrid")
fig, ax = plt.subplots(figsize=(8, 6))
# Create the barplot
sns.barplot(x=age_df.Percentages, y=age_df.Age, palette="mako", ax=ax)
# Set the title and axis labels
ax.set_title("Distribution of respondents age", fontsize=12, fontweight="bold")
ax.set_xlabel("Percentage of Respondents", fontsize=10)
ax.set_ylabel("Age Group", fontsize=10)
# Add labels to the bars
for i, v in enumerate(age_df.Percentages):
ax.text(v + 1, i, str(round(v, 2)) + "%", color="black", fontweight="bold")
# Show the plot
plt.tight_layout()
plt.show()
# Possible reasons why many respondents are between 18-34 years old :
# 1. We visualized many of the respondents hold Bachelors degree and an average age of completing Bachelors is between 20-24, and also we also summarized that many of the respondents are work as professional programmers such as Developer.
# As many young professional programmers aged between 18-34 years old who were aware StackOver Flow , has taken the survey.
#
import plotly.express as px
dist_title = (
"Distribution of respondent's total years of coding experience excluding education"
)
fig = px.histogram(
survey_df2,
x="YearsCodePro",
marginal="box",
color_discrete_sequence=["Red"],
title=dist_title,
)
fig.update_layout(bargap=0.1)
# The large percentage of respondents have 1-10 years of Professional coding experience. This shows respondents who just started with their coding journey tend to reach out developer community one such is StackOver flow.
# We just explored individual columns of the dataset and gained several insights about the respondents.We will now ask questions and try to answer it using data frame operations and visualizations.
# ### Asking and Answering Questions
# #### 1. What percentage of respondents by gender has coding as hobby?
# There is no separate column stating hobby, but we will create a new `column 'Hobby'`. For everytime when word 'Hobby' appears in ``CodingActivities`` column ,the new 'Hobby' column will have `yes` value for that row else `no`.
survey_df2["CodingActivities"]
# Before we try to find out the percentage of respondents by gender has coding as hobby, we will explore `how many respondents have coding as hobby?`
# create a new column 'Hobby' based on 'CodingActivities'
survey_df2["Hobby"] = (
survey_df2["CodingActivities"]
.fillna("")
.apply(lambda x: "Yes" if "Hobby" in x else "No")
)
# copy of survey_df2
Gender_Hobby_df = survey_df2.copy()
# improve readbility on graph
Gender_Hobby_df.loc[
Gender_Hobby_df["Gender"] == "Non-binary, genderqueer, or gender non-conforming",
"Gender",
] = "Non-binary,genderqueer,GNC"
import matplotlib.pyplot as plt
# Set the colors for the bars
colors = ["blue", "red"]
# Create a bar plot
fig, ax = plt.subplots()
counts = Gender_Hobby_df["Hobby"].value_counts()
counts.plot(kind="bar", ax=ax, color=colors)
# Set the plot title and axis labels
ax.set_title("Hobby Distribution")
ax.set_xlabel("Hobby")
ax.set_ylabel("Count")
# Add the count values on top of the bars
for i, v in enumerate(counts):
ax.text(i, v + 1000, str(v), color="black", fontweight="bold", ha="center")
# Set the figure size
fig.set_size_inches(8, 4)
# Display the plot
plt.show()
# The `Yes` for coding as hobby is 2.6838 times greater than `No`, which means almost many respondents do not code outside their work, these respondents do coding for their part-time, or full-time or student or freelance.
counts_Gh = Gender_Hobby_df.groupby(["Gender", "Hobby"]).size().unstack()
total_count = counts_Gh.sum().sum()
# calculate the percentage values for each category
counts_Gh_pct = counts_Gh.apply(lambda x: x / total_count * 100)
# combine the output into one dataframe
counts_Gh_pct_df = pd.concat(
[counts_Gh, counts_Gh_pct], keys=["count", "percentage"], axis=1
)
counts_Gh_pct_df
# About 69.2% men has codding as hobby while women and other minority gender stands with less percentage for coding as hobby.
import matplotlib.pyplot as plt
# create a side-by-side bar chart
fig, ax = plt.subplots(figsize=(10, 5))
counts_Gh_pct.plot(kind="bar", ax=ax, width=0.4, color=["#4e79a7", "#f28e2b"])
# add labels and title
ax.set_xlabel("Gender", fontsize=12, fontweight="bold")
ax.set_ylabel("Percentage", fontsize=12, fontweight="bold")
ax.set_title("Coding Hobby by Gender", fontsize=16, fontweight="bold", pad=20)
# set the y-axis limit to 100
ax.set_ylim([0, 100])
# add percentage labels on the bars
for c in ax.containers:
ax.bar_label(
c,
label_type="edge",
labels=[f"{val:.1f}%" for val in c.datavalues],
fontsize=10,
padding=4,
)
# remove the top and right spines
ax.spines["top"].set_visible(False)
ax.spines["right"].set_visible(False)
# add horizontal grid lines
ax.yaxis.grid(True)
# increase tick font size
ax.tick_params(axis="both", which="major", labelsize=10)
# add legend
ax.legend(["No", "Yes"], loc="upper right", fontsize=10)
# show the plot
plt.show()
# ### 2. Top 15 countries where respondents have coding as a Hobby?
# copy of survey_df2
Country_Hobby_df = Gender_Hobby_df.copy()
Country_Hobby_df[["Country", "Hobby"]]
import matplotlib.pyplot as plt
# Group the DataFrame by 'Country' and count the 'Hobby' values
hobby_counts = Country_Hobby_df.groupby("Country")["Hobby"].value_counts()
# Filter for the 'Yes' values of the 'Hobby' column
hobby_counts = hobby_counts.loc[hobby_counts.index.get_level_values("Hobby") == "Yes"]
# Get the counts for each country and sort in descending order
counts = hobby_counts.groupby("Country").sum().sort_values(ascending=False)[:15]
# Calculate the total count of 'Yes' values
total_count = hobby_counts.sum()
# Calculate the percentage value for each bar
percentages = counts / total_count * 100
# Create a horizontal bar plot
fig, ax = plt.subplots()
counts.plot(kind="barh", ax=ax)
# Loop through the bars and annotate the percentage value
for i, v in enumerate(counts):
ax.annotate("{:.1f}%".format(percentages[i]), xy=(v + 0.5, i - 0.1))
# Set the plot title and axis labels
ax.set_title("Top 15 Countries where respodents have Coding as Hobby")
ax.set_xlabel("Count")
ax.set_ylabel("Country")
# to set the figure size
fig.set_size_inches(8, 6)
# Display the plot
plt.show()
# 20.0% of total respondents whose hobby is to code belongs from `United States of America` and the second country is `India` with 8.7% where respondents have coding as a hobby.
# #### 3. What were the popular programming languages in 2022?
# We all know technologies change rapidly and some of the technologies become favorites or most sought technologies among programmers and also in corporate.
# That's why it is interesting to know which language among the survey respondents made its place in top 25.
survey_df2.LanguageHaveWorkedWith
# As respondents worked with multiple languages and they opted all those languages on which they worked, therefore to make our analysis easier we will use the function `split_multicolumn()`.
survey_df2.LanguageHaveWorkedWith
languages_workedwith_df = split_multicolumn(survey_df2.LanguageHaveWorkedWith)
languages_worked_count = languages_workedwith_df.sum().sort_values(ascending=False)
languages_worked_percentages = (
languages_workedwith_df.mean().sort_values(ascending=False) * 100
)
LanguagesWorkedWith_df = pd.DataFrame(
{
"Language": languages_worked_percentages.index,
"Count": languages_worked_count.values,
"Percentage": languages_worked_percentages.values,
}
)
# top 25 languages
top25_LanguagesWorkedWith = LanguagesWorkedWith_df.head(25)
top25_LanguagesWorkedWith
import matplotlib.pyplot as plt
import seaborn as sns
# set font size
plt.rcParams.update({"font.size": 8})
# set figure size
plt.figure(figsize=(10, 6))
# create the horizontal barplot
sns.barplot(x="Percentage", y="Language", data=top25_LanguagesWorkedWith)
# add value labels to the bars
for index, row in top25_LanguagesWorkedWith.iterrows():
plt.text(
row["Percentage"] + 0.5, index, "{:.1f}%".format(row["Percentage"]), va="center"
)
# set title and axis labels
plt.title("Top 25 Languages by Percentage")
plt.xlabel("Percentage")
plt.ylabel("Language")
# use a different color palette
sns.set_palette("husl")
# add a grid
sns.set_style("whitegrid")
# display the plot
plt.show()
# About 66 % of total respondents worked with Javascript, making it most popular language of the year 2022. The second is HTML/CSS at 53.6%.
# The possible reasons for Javascript and HTML/CSS popularity could be because:
# 1. Our major respondents are Developers, and developers majorly work with Javascript and HTML/CSS languages.
# ### 4. Which languages people want to work with over in over the next year ?
# With technologies advancing rapidly to simplify problems, programmers adopt to new languages or have eagerness to learn new languages.Therefore, it is interesting to know which languages respondents want to work with in the next few years.
languages_interested_df = split_multicolumn(survey_df2.LanguageWantToWorkWith)
languages_interested_percentages = (
languages_interested_df.mean().sort_values(ascending=False) * 100
)
languages_interested_percentages
LanguagesWanttoWorkWith_df = pd.DataFrame(
{
"Language": languages_interested_percentages.index,
"Percentage": languages_interested_percentages.values,
}
)
# top 25 languages
top25_LanguagesWantToWorkWith = LanguagesWanttoWorkWith_df.head(25)
top25_LanguagesWantToWorkWith
import matplotlib.pyplot as plt
import seaborn as sns
# set font size
plt.rcParams.update({"font.size": 8})
# set figure size
plt.figure(figsize=(10, 6))
# create the horizontal barplot
sns.barplot(x="Percentage", y="Language", data=top25_LanguagesWantToWorkWith)
# add value labels to the bars
for index, row in top25_LanguagesWantToWorkWith.iterrows():
plt.text(
row["Percentage"] + 0.5, index, "{:.1f}%".format(row["Percentage"]), va="center"
)
# set title and axis labels
plt.title("Top 25 Languages people interested to learn by Percentage")
plt.xlabel("Percentage")
plt.ylabel("Language")
# use a different color palette
sns.set_palette("husl")
# add a grid
sns.set_style("whitegrid")
# display the plot
plt.show()
# JavaScript with 44.6% tops the list of languages respondents would like to learn over the next year, followed by TypesSript at 39.9% and python with 39.3%.
# The possible reasons for JavaScript to be most sought language could be :
# 1. Web applications rise and user-interfaces need.
# 2. It is versatile language can be used for both front and backend development.
# 3. High demand for JavaScript developers in the job market.
# The possible reasons for TypeScript to be most sought language could be :
# 1. TypeScript is superset of JavaScript, it helps save lot of debugging time with its optional static typing.
# 2. Maintaining codebase, working with larger codebase and bug reductions.
# 3. Easier for developers to migrate existing JavaScript to TypeScript or use use TypeScript code in existing JavaScript projects.
# 4. Growing Typscript community.
# The possible reasons for Python to be on the race of most sought language is :
# 1. Easy to learn : It is even known as beginner's language because of its clean syntax which is easy to read,write and understand.
# 2. Versatility : wide range of tasks, including web development, scientific computing, data analysis, artificial intelligence, and machine learning.
# 3. Open-source language
# 4. Job demand
# ### 5. Which languages are the most liked among respondents ?
# Now, the time has come to know which language tops the list of languages on which respondnets worked with and would like to learn more about it. Which means we will explore the most loved languages.
languages_like_df = languages_workedwith_df & languages_interested_df
languages_liked_percentages = (
languages_like_df.sum() * 100 / languages_workedwith_df.sum()
).sort_values(ascending=False)
Languages_liked_df = pd.DataFrame(
{
"Language": languages_liked_percentages.index,
"Percentage": languages_liked_percentages.values,
}
)
# top 25 languages
top25_languages_liked_percentages = Languages_liked_df.head(25)
# set font size
plt.rcParams.update({"font.size": 8})
# set figure size
plt.figure(figsize=(10, 6))
# create the horizontal barplot
sns.barplot(x="Percentage", y="Language", data=top25_languages_liked_percentages)
# add value labels to the bars
for index, row in top25_languages_liked_percentages.iterrows():
plt.text(
row["Percentage"] + 2.5, index, "{:.2f}%".format(row["Percentage"]), va="center"
)
# set title and axis labels
plt.title("Top 25 most liked languages by Percentage")
plt.xlabel("Percentage")
plt.ylabel("Language")
# It's so amusing to see that neither JavaScript,HTML/CSS,Python or TypeScript make to Top 3 in the list of most loved language.
# Reasons for Rust, Clojure and Elixir to be on top 3 :
# #### Rust :
# 1. Performance of Rust ,known to outperform languages like Python and JavaScript.
# 2. Considered systems programming safer choice.
# 3. Moddern language with features like pattern matching, closures, and traits that make it easier to write clean and maintainable code.
# 4. can be compiled to run on a wide variety of platforms, makes Rust best choice for developing cross-platform applications.
# #### Clojure :
# 1. easy to integrate Clojure with existing Java systems and allows one to take advantage of Java's ecosystem.
# 2. Simple syntax
# 3. It is functional programming language, the approach that leads to more concise and readable code, therefore fewer errors and bugs.
# #### Elixir :
# 1. Elixir is designed for high concurrency and fault-tolerance.
# 2. It's processes and message-passing model enables easy to write scalable and fault-tolerant applications.
# 3. With Elixir it is faster and easier to write code than other languages.
# 4. Elixir's Phoenix web framework makes building web applications easy which can handle high traffic and support real-time communication.
# ### 5. Which langauges are preferred by type of companies ?
survey_df2[["LanguageHaveWorkedWith", "OrgSize"]]
import matplotlib.pyplot as plt
# Group the programming languages by company size and get the top 3 languages
grouped = survey_df.groupby("OrgSize")["LanguageHaveWorkedWith"].apply(
lambda x: x.str.split(";").explode().value_counts().head(3)
)
# Plotting the bar chart
plt.figure(figsize=(10, 6))
grouped.unstack().plot(kind="bar", width=0.8)
plt.xlabel("Company Size")
plt.ylabel("Count")
plt.title("Top 3 Programming Languages by Company Size")
plt.legend(title="Programming Language")
plt.show()
# #### Summarize :
# 1. The companies which employee size < 5,000: use both HTML/CSS and JavaScript languages, but they don't use Python.
# ###### Reasons could be :
# * HTML, CSS, and JavaScript are majorly used for building the front-end of websites and web applications, though Python can be used for web-development but they are best suited for back-end development.
# 2. The companies which have larger employee size : 5,000 to 9,999 employees and 10,000 or more employees have Python as their second best language after JavaScript.
# ##### Reasons could be :
# * As, we know that JavaScript, client-side scripting language that is built into all modern web browsers, which can be used without any special tools or software.
# * Python, on the other hand, commonly used for back-end development, data science, machine learning, scientific computing, and automation.
# This means the these companies are trying to stay ahead or advance themselves in the field of new age technologies.
# ### 6. How respondents are learning to code?
# survey question asked
schema_df1.LearnCode
survey_df2.LearnCode.head(5)
# As this survey questions welcomed multiple choices, respondents mentioned they used various sources to learn code.
survey_df2["LearnCode"]
learn_code_df = split_multicolumn(survey_df2.LearnCode)
learn_code_df
learnCode_percentages = learn_code_df.mean().sort_values(ascending=False) * 100
LearnCode_percentages_df = pd.DataFrame(
{"From": learnCode_percentages.index, "Percentages": learnCode_percentages.values}
)
LearnCode_percentages_df
# Visualize the above data frame with barplot.
sns.set_style("darkgrid")
fig, ax = plt.subplots(figsize=(8, 6))
# Create the barplot
sns.barplot(
x=LearnCode_percentages_df.Percentages,
y=LearnCode_percentages_df.From,
palette="mako",
ax=ax,
)
# Set the title and axis labels
ax.set_title(
"Distribution of how respondents learn to code", fontsize=12, fontweight="bold"
)
ax.set_xlabel("Percentage of Respondents", fontsize=10)
ax.set_ylabel("From", fontsize=10)
# Add labels to the bars
for i, v in enumerate(LearnCode_percentages_df.Percentages):
ax.text(v + 1, i, str(round(v, 2)) + "%", color="black", fontweight="bold")
# Show the plot
plt.tight_layout()
plt.show()
# The above barplot shows about 69.61 % of the total respondents learn to code from online sources such as videos,blogs etc.
# Probably the free sources which are abundance with respect to the growth of WorldWideWeb.
# ### 7. Which top 3 languages are used among top 15 countries in survey ?
Country_Language = survey_df2[["Country", "LanguageHaveWorkedWith"]]
Country_Language
Country_Language_df = survey_df2[["Country", "LanguageHaveWorkedWith"]]
Country_Language = pd.concat(
[
Country_Language_df["Country"],
Country_Language_df["LanguageHaveWorkedWith"].str.get_dummies(sep=";"),
],
axis=1,
)
print(Country_Language)
# Get the top 15 countries by number of respondents
top_countries = Country_Language["Country"].value_counts().head(15).index
# Create a dictionary to store the top three languages for each country
top_languages = {}
# Loop over each country and get the top three languages
for country in top_countries:
languages = (
Country_Language[Country_Language["Country"] == country]
.iloc[:, 1:]
.sum()
.sort_values(ascending=False)[:3]
)
top_languages[country] = list(languages.index)
# create a list of dictionaries to store the data
data = []
# loop over each country and add its top three languages to the list
for country, languages in top_languages.items():
data.append(
{
"Country": country,
"Top Language 1": languages[0],
"Top Language 2": languages[1],
"Top Language 3": languages[2],
}
)
# create a new DataFrame from the data
df = pd.DataFrame(data)
developers_count = Country_Language["Country"].value_counts().head(15)
developers_count_df = pd.DataFrame(
{"Country": developers_count.index, "Developer_Count": developers_count.values}
)
# concat
df_merged = pd.merge(df, developers_count_df, on="Country")
df_merged
# The above data frame represents top 15 countries with their top 3 languages and the number of developers working in the field.
# Majorily used programming language is JavaScript followed by SQL and HTML/CSS.
# The table can help companies operating globally who want to target specific countires for their products or services launch.
# For example, companies like ebay, LinkedIn, Airbnb and Instagram planning to launch produt which requires JavaScript expertise,
# they can consider countries like United States,India,Germany or even UK as a good target because these countries have significant number of developers who are proficient in JavaScript.
# The top three languages in United States of America is : JavaScript,SQL,HTML/CSS, but in India SQL is in the third place.
# No countries have Python in their top 3 languages list except Russian Federation country.
# Reasons could be :
# 1. Legacy System: Countries may be find it difficult to switch to Python because of existing systems and infrastructure that are built on different programming language.
# 2. Industry : Some Industries give preferences to security and performance while some industries prioritize programmming languages that are better suited for machine learning or data analysis tasks etc.
# ### 7. What is the attitude of respondents towards Blockchain ?
schema_df1["Blockchain"]
survey_df2["Blockchain"].head(5)
# Calculate the percentage of each response option
counts = survey_df2["Blockchain"].value_counts(normalize=True) * 100
# Create a bar chart with percentages
plt.figure(figsize=(8, 6))
sns.barplot(x=counts.index, y=counts.values, palette="magma")
# Set the title and labels for the axes
plt.title("Attitudes towards Blockchain", fontsize=16)
plt.xlabel("Response", fontsize=14)
plt.ylabel("Percentage", fontsize=14)
# Display the percentages on the bars
for i, v in enumerate(counts.values):
plt.text(i, v + 1, str(round(v, 1)) + "%", ha="center", fontsize=12)
# Add grid lines and remove spines
sns.set_style("ticks")
plt.grid(axis="y", linestyle="--", alpha=0.7)
sns.despine(left=True)
# Show the plot
plt.show()
# - Indifferent : 26 % of respondents have indifferent attitude towards blockchain, this could be because either they are not familiar with the technology or they see no potential impact of it on the lives.
# - Favourable : 21 % of respondents have favourable attitude towards blockchain ,which indicates they consider technology as potential benefits or oppurtunities.
# - Very unfavorable: 16.5 % of respondents have a very unfavorable attitude towards blockchain, which shows they may have concerns or reservations about the technology.
# - Unfavorable: 14.9 % of respondents have an unfavorable attitude towards blockchain.Shows us that some respondents may have a slightly negative view of blockchain, but not to the extent of being very unfavorable.
# - Very favorable: 11.4 % of respondents had a very favorable attitude towards blockchain, which is a lower percentage than the very unfavorable responses.Indicates that there are fewer respondents who are very enthusiastic about blockchain compared to those who are very skeptical.
# - Unsure: Finally, about 10% of respondents were unsure about their attitude towards blockchain.Indicates that some respondents may not have sufficient knowledge or experience with the technology to form a clear opinion about it.
# Overall, we see mixed responses towards blockchain, responses showing relatively high percentage of indifferent responses and a range of attitudes from very unfavorable to very favorable.
# Highlighting the need for advance education and awareness about blockchain and its applications.
# ### 8. Any mental health disorders among programmers ?
# It is crucial to also learn about mental health of the programmers because they often work in high-stress environments, long hours and tight deadlines. Whcih can lead to burnout, anxiety, depression, and other mental health problems.
# When struggling with mental health issues, the ability to focus, be creative, and work effectively are impacted. This can lead to delays in projects, mistakes, and lower quality work.
survey_df["MentalHealth"].head(10)
# Create a list of all the unique responses in the "MentalHealth" column
all_responses = (
survey_df["MentalHealth"].dropna().str.split(";").explode().str.strip().unique()
)
all_responses
# Create a list of all the unique responses in the "MentalHealth" column
all_responses = (
survey_df["MentalHealth"].dropna().str.split(";").explode().str.strip().unique()
)
# Count the number of occurrences of each response
response_counts = {}
total_responses = survey_df["MentalHealth"].dropna().shape[0]
for response in all_responses:
response_counts[response] = (
survey_df["MentalHealth"].str.contains(response).sum() / total_responses * 100
)
# Sort the responses by frequency
sorted_responses = sorted(response_counts.items(), key=lambda x: x[1], reverse=True)
# Extract the labels and values for the plot
labels = [response[0] for response in sorted_responses]
values = [response[1] for response in sorted_responses]
# Plot the data using a horizontal bar chart
fig, ax = plt.subplots(figsize=(8, 5))
ax.barh(labels, values, color="#1f77b4")
ax.set_xlabel("Percentage of Responses", fontsize=12)
ax.set_ylabel("Mental Health Conditions", fontsize=12)
ax.set_title(
"Distribution of Mental Health Conditions in Survey Respondents",
fontsize=14,
fontweight="bold",
)
ax.invert_yaxis()
# Display percentages on the bars
for i, v in enumerate(values):
ax.text(v + 1, i, f"{v:.1f}%", ha="left", va="center", fontsize=10)
# Remove the borders on the top and right sides of the plot
ax.spines["top"].set_visible(False)
ax.spines["right"].set_visible(False)
# Add a grid and customize its appearance
ax.grid(axis="x", linestyle="--", alpha=0.5)
ax.set_axisbelow(True)
ax.xaxis.grid(False)
plt.tight_layout()
plt.show()
# Majority of the respondents about 70.5 % have no mental health conditions.Only 10.3 % of respondents have anxiety disorder.
# ### 8. Is it important to start young to build a career in programming ?
survey_df1[["Age", "YearsCodePro"]].head(5)
# copy od survey_df1
age_yearscodepro_df = survey_df1.copy()
# create a new column 'Hobby' based on 'CodingActivities'
age_yearscodepro_df["Hobby"] = (
age_yearscodepro_df["CodingActivities"]
.fillna("")
.apply(lambda x: "Yes" if "Hobby" in x else "No")
)
age_yearscodepro_df.columns
# to drop null values
age_yearscodepro_df.dropna(subset=["Age", "YearsCodePro", "Hobby"], inplace=True)
age_yearscodepro_df[["Age", "YearsCodePro", "Hobby"]].head(5)
import random
import numpy as np
# Define a function to convert age range to a numeric value
def convert_age_range_to_num(age_range):
if age_range == "Under 18 years old":
return random.randint(13, 18) # let's keep minimum age 13
elif age_range == "18-24 years old":
return random.randint(18, 24)
elif age_range == "25-34 years old":
return random.randint(25, 34)
elif age_range == "35-44 years old":
return random.randint(35, 44)
elif age_range == "45-54 years old":
return random.randint(45, 54)
elif age_range == "55-64 years old":
return random.randint(55, 64)
elif age_range == "65 years or older":
return random.randint(65, 70)
else:
return None
# Create a new column for age
age_yearscodepro_df["Age_new1"] = age_yearscodepro_df["Age"].apply(
convert_age_range_to_num
)
# Print the first few rows to check the results
print(age_yearscodepro_df[["Age", "Age_new1", "YearsCodePro", "Hobby"]].head())
# let's consider rows where YearsCodePro is lesser than Age : Boolean Mask
bool_mask = age_yearscodepro_df["YearsCodePro"] < age_yearscodepro_df["Age_new1"]
# apply the bool_mask to the DataFrame to drop the rows where YearsCodePro is greater than Age
age_yearscodepro_df = age_yearscodepro_df[bool_mask]
age_yearscodepro_df = age_yearscodepro_df.reset_index(drop=True)
sns.scatterplot(x="Age_new1", y="YearsCodePro", hue="Hobby", data=age_yearscodepro_df)
plt.xlabel("Age")
plt.ylabel("Years of professional coding experience")
# As the points are spread all over the graph, indicates that one can `start programming professionally at any age`.Also respondents who have decaded of programming experience seems to enjoy coding.
# ### 9. How much time is spent to search and answer solution to the problem encountered at work ?
# question asked in survey
schema_df1[["TimeSearching", "TimeAnswering"]]
df_new = survey_df2[["TimeSearching", "TimeAnswering"]]
# Calculate the frequency of each response
freq = df_new.apply(pd.Series.value_counts)
# Create a subplot with two bar charts
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(10, 5))
fig.suptitle("Frequency of Time Spent Searching and Answering", fontsize=14)
# Bar chart for TimeSearching
freq["TimeSearching"].plot(kind="bar", ax=ax1)
ax1.set_title("Time Spent Searching")
ax1.set_xlabel("Time Range")
ax1.set_ylabel("Frequency")
# Bar chart for TimeAnswering
freq["TimeAnswering"].plot(kind="bar", ax=ax2)
ax2.set_title("Time Spent Answering")
ax2.set_xlabel("Time Range")
ax2.set_ylabel("Frequency")
plt.show()
# On an average day, majority of the repondents spent `30-60 minutes a day` to search answers or solutions to the problem which they encounter at work.Some respondents spent only `15-30 mins a day` to search solutions to the problem,very few spend `less than 15 minutes a day` and the least was spending over `120 minutes a day` for solution to the problem.
# Time Spent Answering : The bar chart indicates large number of respondents are quick enough to answer questions as they only spent `15-30 minutes a day` ,some took `30-60 minutes a day` and few took only `less than 15 minutes a day`.
# Very small number of respodents took more than `1 or 2 hours a day`.
#
# The country with highest number of female respondents is USA about 25% of respondents followed by India with 11.42 %.
# - The following are top 5 countries accounting for more than half (51.7%) of all female respondents in the dataset :
# * United States, India, United Kingdom, Germany, and Brazil.
# ### 10. What different professions female respondents are associated with ?
schema_df1["DevType"]
survey_df2[["Gender", "DevType"]]
female_df = survey_df2.loc[survey_df2["Gender"] == "Woman"].reset_index(drop=True)
Female_Dev_df = split_multicolumn(female_df.DevType)
Female_Dev_df.loc[20:305]
# counting the total number of occurrences of each unique developer type
Female_DevType_count = Female_Dev_df.sum().sort_values(ascending=False)
# to Calculate the percentage of each value in the DevType_count Series
female_total_count = Female_DevType_count.sum()
female_DevType_percentage = Female_DevType_count / female_total_count * 100
female_DevType_df1 = pd.DataFrame(
{
"Type": Female_DevType_count.index,
"Count": Female_DevType_count.values,
"Percentage": female_DevType_percentage.values,
}
)
female_DevType_df1
sns.set_style("darkgrid")
fig, ax = plt.subplots(figsize=(8, 6))
# Create the horizontal bar chart
female_DevType_df1.plot.barh(x="Type", y="Percentage", ax=ax, color="purple")
# Set the title and axis labels
ax.set_title("Female respondent's Developer roles", fontsize=12, fontweight="bold")
ax.set_xlabel("Percentage of Respondents", fontsize=10)
ax.set_ylabel("Developer Roles", fontsize=10)
# Add labels to the bars
for i, v in enumerate(female_DevType_df1.Percentage):
ax.text(v + 1, i, str(round(v, 2)) + "%", color="black", fontweight="bold")
# Show the plot
plt.tight_layout()
plt.show()
# It is interesting, that Female Senior Executives(0.36%),Maketing and Sales professional(0.39%) took this survey and showed their skill diversity.
# But overall, 20.22 % of the total female respondents are Developer, Full-stack profession followed by Developer,back-end at 17.17 % and DEveloper,front-end at 14.2 %.
# ### 11. Countries with female respondents.
survey_df2[["Gender", "Country"]]
female_countries = survey_df2.loc[survey_df2["Gender"] == "Woman", "Country"]
# countries with the highest number of respondents
female_country_count = female_countries.value_counts(ascending=False)
# dataframe having columns for counts and percentage for number of respondents
female_country_df = pd.DataFrame(
{"Country": female_country_count.index, "Count": female_country_count.values}
)
# percentage
total_female_respondents = female_country_df["Count"].sum()
female_country_df["Percentage"] = (
female_country_df["Count"] / total_female_respondents
) * 100
female_country_df
import folium
countries_geojson = (
"https://raw.githubusercontent.com/johan/world.geo.json/master/countries.geo.json"
)
female_country_df.at[0, "Country"] = "United States of America"
female_country_df.at[12, "Country"] = "Russia"
m = folium.Map(location=[30, 0], zoom_start=2, tiles="Stamen Terrain")
folium.Choropleth(
geo_data=countries_geojson,
data=female_country_df,
columns=["Country", "Percentage"],
key_on="feature.properties.name",
threshold_scale=[0, 2, 5, 8, 10, 15, 25],
nan_fill_color="Black",
fill_color="YlGnBu",
fill_opacity=0.7,
line_opacity=0.2,
legend_name="Respondents",
highlight=True,
smooth_factor=0,
).add_to(m)
# Add a title to the map
title_html = (
'<h3 align="center" style="font-size:16px"><b>Respondent Count by Country</b></h3>'
)
m.get_root().html.add_child(folium.Element(title_html))
m
# to find top 5 countires with most women
most_women_country = female_country_df.nlargest(5, "Percentage")
most_women_country
# The countries with the most women respondents are:
# * United States of America
# * India
# * United Kingdom of Great Britain and Northern Ireland
# * Germany
# * Brazil
# filter the female_country_df by percentage less than 5%
countries_less_than_5 = female_country_df[female_country_df["Count"] < 5]
# select only the 'Country' column and convert it to a list
countries_list = countries_less_than_5["Country"].tolist()
# print the list of countries
print(countries_list)
|
import os
import numpy as np
import torch
import torch.nn as nn
import torchvision
from torchvision import datasets, models, transforms
from torch.utils.data.sampler import SubsetRandomSampler
import matplotlib.pyplot as plt
train_on_gpu = torch.cuda.is_available()
if not train_on_gpu:
print("CUDA is not available. Training on CPU ...")
else:
print("CUDA is available! Training on GPU ...")
data_dir = "../input/building-type/staticmap/"
batch_size = 16
valid_size = 0.20
random_transforms = [transforms.RandomRotation(20), transforms.RandomHorizontalFlip()]
train_transforms = transforms.Compose(
[
transforms.Resize((224, 224)),
transforms.RandomApply(random_transforms, p=0.3),
transforms.ToTensor(),
transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5)),
]
)
test_transforms = transforms.Compose(
[transforms.Resize((224, 224)), transforms.ToTensor()]
)
train_data = datasets.ImageFolder(data_dir + "train", transform=train_transforms)
test_data = datasets.ImageFolder(data_dir + "test", transform=test_transforms)
num_train = len(train_data)
indices = list(range(num_train))
np.random.shuffle(indices)
split = int(np.floor(valid_size * num_train))
train_idx, valid_idx = indices[split:], indices[:split]
train_sampler = SubsetRandomSampler(train_idx)
valid_sampler = SubsetRandomSampler(valid_idx)
trainloader = torch.utils.data.DataLoader(
train_data, batch_size=batch_size, sampler=train_sampler
)
validloader = torch.utils.data.DataLoader(
train_data, batch_size=batch_size, sampler=valid_sampler
)
testloader = torch.utils.data.DataLoader(test_data, batch_size=batch_size)
# Output classes
output_classes = ["residential", "industrial"]
dataiter = iter(trainloader)
images, labels = dataiter.next()
images = images.numpy() # convert images to numpy for display
# plot the images in the batch, along with the corresponding labels
fig = plt.figure(figsize=(25, 4))
for idx in np.arange(16):
ax = fig.add_subplot(2, 16 / 2, idx + 1, xticks=[], yticks=[])
plt.imshow(np.transpose(images[idx], (1, 2, 0)))
ax.set_title(output_classes[labels[idx]])
vgg19 = models.vgg19(pretrained=True)
# print out the model structure
print(vgg19)
for param in vgg19.features.parameters():
param.requires_grad = False
n_inputs = vgg19.classifier[6].in_features
from collections import OrderedDict
fc_layer = nn.Linear(n_inputs, 2, bias=True)
vgg19.classifier[6] = fc_layer
print(vgg19)
# if GPU is available, move the model to GPU
if train_on_gpu:
vgg19.cuda()
import torch.optim as optim
criterion = nn.CrossEntropyLoss()
optimizer = optim.Adam(vgg19.classifier.parameters(), lr=0.003)
# number of epochs to train the model
n_epochs = 40
valid_loss_min = np.Inf
for epoch in range(1, n_epochs + 1):
# keep track of training and validation loss
train_loss = 0.0
valid_loss = 0.0
vgg19.train()
for data, target in trainloader:
# move tensors to GPU if CUDA is available
if train_on_gpu:
data, target = data.cuda(), target.cuda()
optimizer.zero_grad()
output = vgg19(data)
loss = criterion(output, target)
loss.backward()
optimizer.step()
train_loss += loss.item() * data.size(0)
vgg19.eval()
for data, target in validloader:
# move tensors to GPU if CUDA is available
if train_on_gpu:
data, target = data.cuda(), target.cuda()
output = vgg19(data)
loss = criterion(output, target)
valid_loss += loss.item() * data.size(0)
# calculate average losses
train_loss = train_loss / len(trainloader.dataset)
valid_loss = valid_loss / len(validloader.dataset)
# print training/validation statistics
print(
"Epoch: {} \tTraining Loss: {:.6f} \tValidation Loss: {:.6f}".format(
epoch, train_loss, valid_loss
)
)
# save model if validation loss has decreased
if valid_loss <= valid_loss_min:
print(
"Validation loss decreased ({:.6f} --> {:.6f}). Saving model ...".format(
valid_loss_min, valid_loss
)
)
torch.save(vgg19.state_dict(), "binary_classification.pt")
valid_loss_min = valid_loss
vgg19.load_state_dict(torch.load("binary_classification.pt"))
test_loss = 0.0
class_correct = list(0.0 for i in range(2))
class_total = list(0.0 for i in range(2))
vgg19.eval() # eval mode
# iterate over test data
for data, target in testloader:
# move tensors to GPU if CUDA is available
if train_on_gpu:
data, target = data.cuda(), target.cuda()
output = vgg19(data)
loss = criterion(output, target)
test_loss += loss.item() * data.size(0)
_, pred = torch.max(output, 1)
correct_tensor = pred.eq(target.data.view_as(pred))
correct = (
np.squeeze(correct_tensor.numpy())
if not train_on_gpu
else np.squeeze(correct_tensor.cpu().numpy())
)
for i in range(target.size(0)):
label = target.data[i]
class_correct[label] += correct[i].item()
class_total[label] += 1
# calculate avg test loss
test_loss = test_loss / len(testloader.dataset)
print("Test Loss: {:.6f}\n".format(test_loss))
for i in range(2):
if class_total[i] > 0:
print(
"Test Accuracy of %5s: %2d%% (%2d/%2d)"
% (
output_classes[i],
100 * class_correct[i] / class_total[i],
np.sum(class_correct[i]),
np.sum(class_total[i]),
)
)
else:
print("Test Accuracy of %5s: N/A (no training examples)" % (output_classes[i]))
print(
"\nTest Accuracy (Overall): %2d%% (%2d/%2d)"
% (
100.0 * np.sum(class_correct) / np.sum(class_total),
np.sum(class_correct),
np.sum(class_total),
)
)
def imshow(img):
img = img / 2 + 0.5 # unnormalize
plt.imshow(np.transpose(img, (1, 2, 0))) # convert from Tensor image
# obtain one batch of test images
dataiter = iter(testloader)
images, labels = dataiter.next()
images.numpy()
# move model inputs to cuda, if GPU available
if train_on_gpu:
images = images.cuda()
# get sample outputs
output = vgg19(images)
# convert output probabilities to predicted class
_, preds_tensor = torch.max(output, 1)
preds = (
np.squeeze(preds_tensor.numpy())
if not train_on_gpu
else np.squeeze(preds_tensor.cpu().numpy())
)
# plot the images in the batch, along with predicted and true labels
fig = plt.figure(figsize=(25, 4))
for idx in np.arange(16):
ax = fig.add_subplot(2, 16 / 2, idx + 1, xticks=[], yticks=[])
imshow(images.cpu()[idx])
ax.set_title(
"{} ({})".format(output_classes[preds[idx]], output_classes[labels[idx]]),
color=("green" if preds[idx] == labels[idx].item() else "red"),
)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
# import libraries
import seaborn as sns
import matplotlib.pyplot as plt
# for jupyter notebook we use this line
sns.set_style("whitegrid")
import pandas as pd
# gender_submission = pd.read_csv("../input/titanic/gender_submission.csv")
# test = pd.read_csv("../input/titanic/test.csv")
Titanicdata = pd.read_csv("../input/titanic/train.csv")
# Check the 10 five samples for data
Titanicdata.head(10)
# Check the last 10 samples for data
Titanicdata.tail(10)
# check simple information like columns names , columns datatypes and null values
Titanicdata.info()
# check summary of numerical data such as count , mean , max , min and standard deviation.
Titanicdata.describe()
# check numbers of rows(samples) and columns(features)
Titanicdata.shape
# check count of values for each features
Titanicdata.count()
# Check total missing values in each feature
Titanicdata.isnull().sum()
# delete ticket, cabin, and passengerID
Titanicdata.drop(["Ticket", "Cabin", "PassengerId"], axis=1, inplace=True)
Titanicdata["Sex"].value_counts()
groubBySurvived = Titanicdata.groupby("Survived").size()
no_Survivors = groubBySurvived[1]
no_Deaths = groubBySurvived[0]
print(
"Numbers of People Survivers: {} \nNumbers of People Deaths: {}".format(
no_Survivors, no_Deaths
)
)
class_sex_grouping = Titanicdata.groupby(["Pclass", "Sex"]).count()
class_sex_grouping
class_sex_grouping["Survived"].plot.pie()
Embarked_sex_grouping = Titanicdata.groupby(
[
"Embarked",
"Sex",
]
).count()
Embarked_sex_grouping
Embarked_sex_grouping["Pclass"].plot.bar()
sns.pairplot(Titanicdata)
sns.countplot(x="Sex", data=Titanicdata)
sns.barplot("Embarked", "Survived", data=Titanicdata)
sns.barplot("Pclass", "Survived", data=Titanicdata)
|
import numpy as np
import pandas as pd
import os
import math
from tqdm.notebook import tqdm
from sklearn.exceptions import ConvergenceWarning
import warnings
from matplotlib import pyplot as plt
import seaborn as sns
from scipy import stats as scs
warnings.simplefilter(action="ignore", category=FutureWarning)
warnings.simplefilter(action="ignore", category=ConvergenceWarning)
PATH = "/kaggle/input/house-prices-advanced-regression-techniques/"
train = pd.read_csv(PATH + "train.csv")
test = pd.read_csv(PATH + "test.csv")
train.head()
def show_hist(values):
plt.figure(1, figsize=(10, 6))
sns.distplot(values)
plt.show()
print("skew:", scs.skew(values))
print("kurtosis:", scs.kurtosis(values))
# define target variable conversions
target_trans = lambda price: np.log1p(price) ** 0.5
target_inv_trans = lambda price: np.expm1(price**2)
# convert
train["SalePrice"] = target_trans(train["SalePrice"])
# visualize
show_hist(train["SalePrice"])
def explore_cont(feature, data=train, kind="reg"):
plt.figure(1, figsize=(10, 10))
sns.jointplot(x=feature, y="SalePrice", data=data, kind=kind)
plt.show()
def explore_cat(feature, data=train, kind="reg"):
plt.figure(1, figsize=(10, 10))
sns.violinplot(x=feature, y="SalePrice", data=data, bw=0.2)
plt.show()
explore_cont("LotArea", train[train["LotArea"] < 30000])
explore_cont("GrLivArea", train[train["GrLivArea"] < 4000])
import category_encoders as ce
train = train[train["LotArea"] < 30000]
train = train[train["GrLivArea"] < 4000]
target = train["SalePrice"]
test_ids = test["Id"]
train.drop(columns=["SalePrice", "Id"], inplace=True)
test.drop(columns=["Id"], inplace=True)
te = ce.TargetEncoder(cols=train.columns.values, smoothing=0.3).fit(train, target)
train = te.transform(train)
test = te.transform(test)
train.head()
from sklearn.model_selection import train_test_split
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import mean_squared_log_error as msle
from sklearn.metrics import make_scorer
from xgboost import XGBRegressor
from sklearn.linear_model import ElasticNet
from sklearn.ensemble import RandomForestRegressor, ExtraTreesRegressor
from catboost import CatBoostRegressor
# split train set
x_train, x_test, y_train, y_test = train_test_split(
train, target, test_size=0.2, random_state=289
)
def score_func(y_true, y_pred, **kwargs):
return msle(target_inv_trans(y_true), target_inv_trans(y_pred), **kwargs) ** 0.5
# hyperparams setting
def make_search(estimator, params, verbose=1):
scorer = make_scorer(score_func, greater_is_better=False)
search = GridSearchCV(estimator, params, cv=5, scoring=scorer, verbose=0, n_jobs=-1)
search.fit(x_train, y_train)
results = pd.DataFrame()
for k, v in search.cv_results_.items():
results[k] = v
results = results.sort_values(by="rank_test_score")
best_params_row = results[results["rank_test_score"] == 1]
mean, std = (
best_params_row["mean_test_score"].iloc[0],
best_params_row["std_test_score"].iloc[0],
)
best_params = best_params_row["params"].iloc[0]
if verbose:
print(
"%s: %.4f (%.4f) with params" % (estimator.__class__.__name__, -mean, std),
best_params,
)
return best_params
models = [
(
XGBRegressor(),
{
"max_depth": [i for i in range(2, 5)],
"n_estimators": [50, 100, 200, 400, 500, 700, 900],
"objective": ["reg:squarederror"],
"random_state": [289],
},
),
(
ElasticNet(),
{
"alpha": [0.00001, 0.0005, 0.0001, 0.0005, 0.001, 0.005, 0.01, 0.1],
"l1_ratio": [0, 0.25, 0.5, 0.75, 1],
"max_iter": [500, 1000, 2000, 5000],
"tol": [1e-5, 1e-4, 1e-3],
},
),
(
RandomForestRegressor(),
{
"n_estimators": [50, 100, 200, 400, 500],
"max_depth": [i for i in range(2, 7)],
"bootstrap": [True, False],
"random_state": [289],
},
),
(
CatBoostRegressor(),
{
"n_estimators": [50, 100, 200, 400, 500],
"max_depth": [i for i in range(2, 7)],
"random_state": [289],
"verbose": [False],
},
),
]
for m_p in models:
model, params = m_p
best_params = make_search(model, params)
model.set_params(**best_params)
models = [i[0] for i in models]
models
for model in models:
model.fit(x_train, y_train)
y_true = target_inv_trans(y_test)
y_pred = target_inv_trans(model.predict(x_test))
print("%s: msle = %.4f" % (model.__class__.__name__, msle(y_true, y_pred) ** 0.5))
from matplotlib import pyplot as plt
def explore_models(models, patience=5000, verbose=1):
predicts = np.zeros((len(models), x_test.shape[0]))
for i, m in enumerate(models):
predicts[i] = target_inv_trans(m.predict(x_test))
y_true = target_inv_trans(y_test)
best_offs = None
best_msle = 1
no_improvement = 0
while True:
off = np.random.uniform(size=(len(models),))
if off.sum() != 0:
off /= off.sum()
pred = np.zeros(y_test.shape)
for i in range(len(models)):
pred += predicts[i] * off[i]
err = msle(y_true, pred) ** 0.5
if err < best_msle:
best_msle = err
best_offs = off
no_improvement = 0
if verbose:
print("new best msle: %.4f" % best_msle)
else:
no_improvement += 1
if no_improvement > patience:
return best_offs
best_offsets = explore_models(models)
best_offsets
predict = np.zeros(
test.shape[0],
)
for i, m in enumerate(models):
predict += target_inv_trans(m.predict(test)) * best_offsets[i]
res = pd.DataFrame()
res["Id"] = test_ids
res["SalePrice"] = predict
res.to_csv("submission.csv", index=False)
res.head(20)
|
# ## Usual Libraries
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Visualisation libraries
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_style("darkgrid")
from plotly.offline import init_notebook_mode, iplot
import plotly.graph_objs as go
import plotly.offline as py
py.init_notebook_mode(connected=True)
# Increase the default plot size and set the color scheme
plt.rcParams["figure.figsize"] = 8, 5
# Disable warnings
import warnings
warnings.filterwarnings("ignore")
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# According to World Health Organization (WHO), "Coronaviruses (CoV) are a large family of viruses that cause illness ranging from the common cold to more severe diseases such as Middle East Respiratory Syndrome (MERS-CoV) and Severe Acute Respiratory Syndrome (SARS-CoV). A novel coronavirus (nCoV) is a new strain that has not been previously identified in humans."
# It has been more than one month since the first case of this new zootonic virus has been reported. Wuhan, the capital of Hubei province of China is the epicenter of this new virus. The situation reports about this outbreak can be found [here.](https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports/)
# In this notebook I will try to get a one month overview of the outbreak.
# ## Importing Dataset
df = pd.read_csv(
"../input/novel-corona-virus-2019-dataset/2019_nCoV_data.csv",
)
df.head()
# Let's check the infos
df.info()
# Convert Last Update column to datetime64 format
df["Last Update"] = df["Last Update"].apply(pd.to_datetime)
df.drop(["Sno"], axis=1, inplace=True)
df.head()
# We know this kind of viruses can spread primarily through contact with an infected person, through respiratory droplets generated when a person, for example, coughs or sneezes, or through droplets of saliva or discharge from the nose. The same happened with 2019-nCov, so let's see in which countries are mostly affected by this virus.
countries = df["Country"].unique().tolist()
print(countries)
print("\nTotal countries affected by virus: ", len(countries))
# ## Scenerio till 31 January, 2020
from datetime import date
latest_data = df[df["Last Update"] > pd.Timestamp(date(2020, 1, 31))]
latest_data.head()
print("Globally Confirmed Cases: ", latest_data["Confirmed"].sum())
print("Global Deaths: ", latest_data["Deaths"].sum())
print("Globally Recovered Cases: ", latest_data["Recovered"].sum())
# Let's look the various Provinces/States affected
latest_data.groupby(["Country", "Province/State"]).sum()
# Creating a dataframe with total no of cases for every country
cases = pd.DataFrame(
latest_data.groupby("Country")["Confirmed", "Deaths", "Recovered"].sum()
)
cases["Country"] = cases.index
cases.index = np.arange(1, 28)
global_cases = cases[["Country", "Confirmed", "Deaths", "Recovered"]]
global_cases
map_data = pd.DataFrame(
{
"name": list(global_cases["Country"]),
"lat": [
-25.27,
12.57,
56.13,
61.92,
46.23,
51.17,
22.32,
20.59,
41.87,
36.2,
22.2,
35.86,
4.21,
28.39,
12.87,
61.52,
1.35,
35.91,
40.46,
7.87,
60.12,
23.7,
15.87,
55.37,
37.09,
23.42,
14.06,
],
"lon": [
133.78,
104.99,
-106.35,
25.75,
2.21,
10.45,
114.17,
78.96,
12.56,
138.25,
113.54,
104.19,
101.98,
84.12,
121.77,
105.31,
103.82,
127.77,
3.74,
80.77,
18.64,
120.96,
100.99,
3.43,
-95.71,
53.84,
108.28,
],
}
)
fig = go.Figure()
fig.add_trace(
go.Scattergeo(
lat=map_data["lat"],
lon=map_data["lon"],
mode="markers",
marker=dict(size=12, color="rgb(255, 0, 0)", opacity=0.7),
text=map_data["name"],
hoverinfo="text",
)
)
fig.add_trace(
go.Scattergeo(
lat=map_data["lat"],
lon=map_data["lon"],
mode="markers",
marker=dict(size=8, color="rgb(242, 177, 172)", opacity=0.7),
hoverinfo="none",
)
)
fig.layout.update(
autosize=True,
hovermode="closest",
showlegend=False,
title_text="Countries with reported confirmed cases, Deaths, Recovered of 2019-nCoV,<br>31 January, 2020",
geo=go.layout.Geo(
showframe=False,
showcoastlines=True,
showcountries=True,
landcolor="rgb(225, 225, 225)",
countrycolor="blue",
coastlinecolor="blue",
projection_type="natural earth",
),
)
fig.show()
# **Let's see if any countries have death cases other than China:**
global_cases.groupby("Country")["Deaths"].sum()
# **So till 31 January, 2020 no other countries have reported deaths. So let's focus on the present state of China:**
# ## Focus in China
# Mainland China
China = latest_data[latest_data["Country"] == "Mainland China"]
China
# Let's look at the Confirmed vs Recovered figures of Provinces of China other than Hubei
f, ax = plt.subplots(figsize=(12, 8))
sns.barplot(
x="Confirmed", y="Province/State", data=China[1:], label="Confirmed", color="r"
)
sns.barplot(
x="Recovered", y="Province/State", data=China[1:], label="Recovered", color="g"
)
sns.barplot(x="Deaths", y="Province/State", data=China[1:], label="Deaths", color="b")
# Add a legend and informative axis label
ax.set_title(
"Confirmed vs Recovered vs Death figures of Provinces of China other than Hubei",
fontsize=15,
fontweight="bold",
position=(0.63, 1.05),
)
ax.legend(ncol=2, loc="lower right", frameon=True)
ax.set(xlim=(0, 40), ylabel="", xlabel="Stats")
sns.despine(left=True, bottom=True)
# **Now let's see which provinces have the highest percentage of confirmed cases:**
percentage = pd.DataFrame(
China.groupby("Province/State").sum()["Confirmed"]
).reset_index()
fig = go.Figure(
data=[go.Pie(labels=percentage["Province/State"], values=percentage.Confirmed)]
)
fig.update_layout(title="Confirmed cases in province/states of Mainland China")
fig.show()
# **The highest deat tolls:**
percentage = pd.DataFrame(China.groupby("Province/State").sum()["Deaths"]).reset_index()
fig = go.Figure(
data=[go.Pie(labels=percentage["Province/State"], values=percentage.Deaths)]
)
fig.update_layout(title="Death tolls in province/states of Mainland China")
fig.show()
# **Also checking the recovery rate:**
percentage = pd.DataFrame(
China.groupby("Province/State").sum()["Recovered"]
).reset_index()
fig = go.Figure(
data=[go.Pie(labels=percentage["Province/State"], values=percentage.Recovered)]
)
fig.update_layout(title="Recovery rates in province/states of Mainland China")
fig.show()
|
# # Important Methods for Time Series in Pandas
import pandas as pd
import numpy as np
# ## resampling method
fb = pd.read_csv(
"/kaggle/input/fb-dataset/FB.csv", parse_dates=["Date"], index_col="Date"
)
fb.head()
fb.resample("M").mean()
fb.Close.resample("M").mean().plot()
fb.Close.resample("Q").mean().plot(kind="bar")
# ## Shifting
fb1 = pd.DataFrame(fb.Close["2019-03"])
fb1.head()
fb1.shift(2)
fb1.shift(-2)
fb1["Previous Price"] = fb1.shift(1)
fb1
fb1["One Day Difference"] = fb1["Close"] - fb1["Previous Price"]
fb1.head()
fb1["Percentage Change"] = (
(fb1["Close"] - fb1["Previous Price"]) * 100 / fb1["Previous Price"]
)
fb2 = fb1[["Close"]]
fb2.head()
fb2.index
fb2.index = pd.date_range("2019-03-01", periods=21, freq="B")
fb2.index
fb2.tshift(1)
fb2.tshift(-2)
# ## Moving Window Functions
fb.Close.plot()
fb.Close.plot()
fb.Close.rolling(30).mean().plot()
# ## Time Zone Handling
import pytz
pytz.timezone("Turkey")
pytz.timezone("America/New_York")
pytz.common_timezones[-7:]
x = pd.date_range("12/9/2009 9:30", periods=6, freq="D")
ts = pd.Series(np.random.randn(len(x)), index=x)
ts
print(ts.index.tz)
ts_utc = ts.tz_localize("UTC")
ts_utc
ts_utc.tz_convert("US/Hawaii")
zstamp = pd.Timestamp("2019-06-26 05:00")
zstamp
zstamp_utc = zstamp.tz_localize("utc")
zstamp_utc
zstamp_utc.tz_convert("Europe/Istanbul")
ts
ts1 = ts[:5].tz_localize("Europe/Berlin")
ts2 = ts[2:].tz_localize("Europe/Istanbul")
result = ts1 + ts2
result.index
|
import numpy as np
import pandas as pd
import statsmodels.api as sm
import statsmodels.formula.api as smf
from sklearn.preprocessing import PolynomialFeatures
import os
print(os.listdir("../input/insurance"))
data = pd.read_csv("../input/insurance/insurance.csv")
# data = pd.get_dummies(data, drop_first=True)
data.head()
# # 1 - Interactions between variables
# # 1) age - children
inter = smf.ols(formula="charges ~ age*children", data=data).fit()
print(inter.summary())
# # As we can see, the p-value for "age:children" coefficient is 0.497, so there is no significant interaction.
# # The F-statistic is 45.48 and its p-value is 5.56e-28, so a combination of interactions in the model is significant.
# # 2) age - bmi
inter = smf.ols(formula="charges ~ age*bmi", data=data).fit()
print(inter.summary())
# # As we can see, the p-value for "age:bmi" coefficient is 0.524, so there is no significant interaction.
# # The F-statistic is 59.18 and its p-value is 6.45e-36, so a combination of interactions in the model is significant.
# # 3) age - smoker
inter = smf.ols(formula="charges ~ age*smoker", data=data).fit()
print(inter.summary())
# # As we can see, the p-value for "age:smoker[T.yes]" coefficient is 0.222, so there is no significant interaction.
# # The F-statistic is 1153. and its p-value is 0.00, so a combination of interactions in the model is significant.
# # 4) sex - smoker (significant)
inter = smf.ols(formula="charges ~ sex*smoker", data=data).fit()
print(inter.summary())
# # As we can see, the p-value for "sex[T.male]:smoker[T.yes]" coefficient is 0.003, so there IS significant interaction.
# # The F-statistic is 732.6. and its p-value is 2.15e-281, so a combination of interactions in the model is significant.
# # 5) sex - bmi (significant)
inter = smf.ols(formula="charges ~ sex*bmi", data=data).fit()
print(inter.summary())
# # As we can see, the p-value for "sex[T.male]:bmi" coefficient is 0.091, so there IS significant interaction.
# # The F-statistic is 20.32 and its p-value is 7.03e-13, so a combination of interactions in the model is significant.
# # 6) bmi - smoker (significant)
inter = smf.ols(formula="charges ~ bmi*smoker", data=data).fit()
print(inter.summary())
# # As we can see, the p-value for "bmi:smoker[T.yes]" coefficient is 0.000, so there IS significant interaction.
# # The F-statistic is 1277. and its p-value is 0.00, so a combination of interactions in the model is significant.
# # 7) smoker - region (significant)
inter = smf.ols(formula="charges ~ smoker*region", data=data).fit()
print(inter.summary())
# # As we can see, the p-value for "smoker[T.yes]:region[T.southwest]" coefficient is 0.011 and "smoker[T.yes]:region[T.southeast]" coefficient is 0.000, so there IS significant interaction.
# # The F-statistic is 320.1 and its p-value is 6.37e-280, so a combination of interactions in the model is significant.
# # 8) sex - smoker - bmi
inter = smf.ols(formula="charges ~ sex*smoker*bmi", data=data).fit()
print(inter.summary())
# # As we can see, the p-value for "sex[T.male]:smoker[T.yes]:bmi" coefficient is 0.564, so there is no significant interaction.
# # The F-statistic is 548.0 and its p-value is 0.00, so a combination of interactions in the model is significant.
inter = smf.ols(formula="charges ~ sex*smoker + sex*bmi", data=data).fit()
print(inter.summary())
# # 2 - Numerical Transformations
# # 1) age - logarithmic (significant)
inter = smf.ols(formula="charges ~ np.log(age)", data=data).fit()
print(inter.summary())
# # As we can see, the p-value for "np.log(age)" coefficient is 0.000, so the transformation is significant.
# # 2) children - numeric, order 2 (significant)
inter = smf.ols(formula="charges ~ children + I(children*children)", data=data).fit()
print(inter.summary())
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
ls
import tensorflow as tf
import numpy as np
from tensorflow import keras
import os
# Reading the directory - Train and Validation Set
train_pneumonia_data = os.path.join(
"/kaggle/input/chest-xray-pneumonia/chest_xray/train/PNEUMONIA"
)
train_normal_data = os.path.join(
"/kaggle/input/chest-xray-pneumonia/chest_xray/train/NORMAL"
)
test_pneumonia_data = os.path.join(
"/kaggle/input/chest-xray-pneumonia/chest_xray/test/PNEUMONIA"
)
test_normal_data = os.path.join(
"/kaggle/input/chest-xray-pneumonia/chest_xray/test/NORMAL"
)
# Lets look at the images in train dataset
train_pneumonia_names = os.listdir(train_pneumonia_data)
print(train_pneumonia_names[:10])
train_normal_names = os.listdir(train_normal_data)
print(train_normal_names[:10])
print(
"Total training Pneumonia detected images:", len(os.listdir(train_pneumonia_data))
)
print("Total training Normal detected images:", len(os.listdir(train_normal_data)))
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
# Parameters for our graph; we'll output images in a 4x4 configuration
nrows = 4
ncols = 4
# Index for iterating over images
pic_index = 0
fig = plt.gcf()
fig.set_size_inches(ncols * 4, nrows * 4)
pic_index += 8
next_pn_pix = [
os.path.join(train_pneumonia_data, fname)
for fname in train_pneumonia_names[pic_index - 8 : pic_index]
]
next_normal_pix = [
os.path.join(train_normal_data, fname)
for fname in train_normal_names[pic_index - 8 : pic_index]
]
for i, img_path in enumerate(next_pn_pix + next_normal_pix):
# Set up subplot; subplot indices start at 1
sp = plt.subplot(nrows, ncols, i + 1)
sp.axis("Off") # Don't show axes (or gridlines)
img = mpimg.imread(img_path)
plt.imshow(img)
plt.show()
import cv2
import glob
all_h = []
all_w = []
for img in glob.glob("/kaggle/input/chest-xray-pneumonia/chest_xray/train/PNEUMONIA/*"):
n = cv2.imread(img)
h, w, _ = n.shape
all_h.append(h)
all_w.append(w)
print("Average Height of the Train Data:", np.average(all_h))
print("Average Width of the Train data:", np.average(all_w))
# Model Building:
model = tf.keras.models.Sequential(
[
# First Convolution
tf.keras.layers.Conv2D(
16, (3, 3), activation="relu", input_shape=(825, 1200, 3)
),
tf.keras.layers.MaxPooling2D(2, 2),
tf.keras.layers.Dropout(0.2),
# Second Convolution
tf.keras.layers.Conv2D(4, (3, 3), activation="relu"),
tf.keras.layers.MaxPool2D(2, 2),
tf.keras.layers.Dropout(0.2),
# Flatten the images
tf.keras.layers.Flatten(),
# tf.keras.layers.Dense(512, activation = 'relu'),
tf.keras.layers.Dropout(0.2),
tf.keras.layers.Dense(128, activation="relu"),
tf.keras.layers.Dense(0.2),
tf.keras.layers.Dense(1, activation="sigmoid"),
]
)
model.summary()
from tensorflow.keras.optimizers import RMSprop
model.compile(loss="binary_crossentropy", optimizer=RMSprop(lr=0.05), metrics=["acc"])
from tensorflow.keras.preprocessing.image import ImageDataGenerator
# All images will be rescaled by 1./255
train_datagen = ImageDataGenerator(rescale=1.0 / 255)
test_datagen = ImageDataGenerator(rescale=1 / 255)
train_generator = train_datagen.flow_from_directory(
"/kaggle/input/chest-xray-pneumonia/chest_xray/train/", # This is the source directory for training images
target_size=(825, 1200), # All images will be resized to 150x150
# batch_size=128,
# Since we use binary_crossentropy loss, we need binary labels
class_mode="binary",
)
# Flow training images in batches of 128 using train_datagen generator
test_generator = test_datagen.flow_from_directory(
"/kaggle/input/chest-xray-pneumonia/chest_xray/test/", # This is the source directory for training images
target_size=(825, 1200), # All images will be resized to 150x150
# batch_size=128,
# Since we use binary_crossentropy loss, we need binary labels
class_mode="binary",
)
history = model.fit_generator(
train_generator,
steps_per_epoch=8,
epochs=20,
verbose=1,
validation_data=test_generator,
validation_steps=2,
)
import matplotlib.pyplot as plt
acc = history.history["acc"]
val_acc = history.history["val_acc"]
loss = history.history["loss"]
val_loss = history.history["val_loss"]
epochs = range(len(acc))
plt.plot(epochs, acc, "r", label="Training accuracy")
plt.plot(epochs, val_acc, "b", label="Validation accuracy")
plt.title("Training and validation accuracy")
plt.figure()
plt.plot(epochs, loss, "r", label="Training Loss")
plt.plot(epochs, val_loss, "b", label="Validation Loss")
plt.title("Training and validation loss")
plt.legend()
plt.show()
|
#
#
# ## [mlcourse.ai](https://mlcourse.ai) – Open Machine Learning Course
# Author: [Yury Kashnitskiy](https://yorko.github.io). Translated by [Sergey Oreshkov](https://www.linkedin.com/in/sergeoreshkov/). This material is subject to the terms and conditions of the [Creative Commons CC BY-NC-SA 4.0](https://creativecommons.org/licenses/by-nc-sa/4.0/) license. Free use is permitted for any non-commercial purpose.
# # Assignment #8 (demo)
# ## Implementation of online regressor
# Here we'll implement a regressor trained with stochastic gradient descent (SGD). Fill in the missing code. If you do evething right, you'll pass a simple embedded test.
# ## Linear regression and Stochastic Gradient Descent
import numpy as np
import pandas as pd
from tqdm import tqdm
from sklearn.base import BaseEstimator
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_squared_error, log_loss, roc_auc_score
from sklearn.model_selection import train_test_split
from matplotlib import pyplot as plt
import seaborn as sns
from sklearn.preprocessing import StandardScaler
# Implement class `SGDRegressor`. Specification:
# - class is inherited from `sklearn.base.BaseEstimator`
# - constructor takes parameters `eta` – gradient step ($10^{-3}$ by default) and `n_epochs` – dataset pass count (3 by default)
# - constructor also creates `mse_` and `weights_` lists in order to track mean squared error and weight vector during gradient descent iterations
# - Class has `fit` and `predict` methods
# - The `fit` method takes matrix `X` and vector `y` (`numpy.array` objects) as parameters, appends column of ones to `X` on the left side, initializes weight vector `w` with **zeros** and then makes `n_epochs` iterations of weight updates (you may refer to this [article](https://medium.com/open-machine-learning-course/open-machine-learning-course-topic-8-vowpal-wabbit-fast-learning-with-gigabytes-of-data-60f750086237) for details), and for every iteration logs mean squared error and weight vector `w` in corresponding lists we created in the constructor.
# - Additionally the `fit` method will create `w_` variable to store weights which produce minimal mean squared error
# - The `fit` method returns current instance of the `SGDRegressor` class, i.e. `self`
# - The `predict` method takes `X` matrix, adds column of ones to the left side and returns prediction vector, using weight vector `w_`, created by the `fit` method.
class SGDRegressor(BaseEstimator):
# you code here
def __init__(self, eta=0.001, n_epochs=3):
self.eta = eta
self.n_epochs = 3
self.mse_ = []
self.weights_ = []
def fit(self, X, y):
X = np.hstack([np.ones([X.shape[0], 1]), X])
w = np.zeros(X.shape[1])
for i in range(self.n_epochs):
for j in range(X.shape[0]):
w += self.eta * (y[j] - np.sum(w * X[j])) * X[j]
self.weights_.append(w.copy())
self.mse_.append((np.square(y - np.sum(X * w, axis=1)).mean()))
mse_argmin = np.argmin(self.mse_)
self.mse = self.mse_[mse_argmin]
self.w_ = self.weights_[mse_argmin]
return self
def predict(self, X):
X = np.hstack([np.ones([X.shape[0], 1]), X])
return np.sum(X * self.w_, axis=1)
# Let's test out the algorithm on height/weight data. We will predict heights (in inches) based on weights (in lbs).
data_demo = pd.read_csv("../input/weights_heights.csv")
plt.scatter(data_demo["Weight"], data_demo["Height"])
plt.xlabel("Weight (lbs)")
plt.ylabel("Height (Inch)")
plt.grid()
X, y = data_demo["Weight"].values, data_demo["Height"].values
# Perform train/test split and scale data.
X_train, X_valid, y_train, y_valid = train_test_split(
X, y, test_size=0.3, random_state=17
)
scaler = StandardScaler()
X_train_scaled = scaler.fit_transform(X_train.reshape([-1, 1]))
X_valid_scaled = scaler.transform(X_valid.reshape([-1, 1]))
# Train created `SGDRegressor` with `(X_train_scaled, y_train)` data. Leave default parameter values for now.
# you code here
sgd_regressor = SGDRegressor()
sgd_regressor.fit(X_train_scaled, y_train)
# Draw a chart with training process – dependency of mean squared error from the i-th SGD iteration number.
plt.plot(range(len(sgd_regressor.mse_)), sgd_regressor.mse_)
# Print the minimal value of mean squared error and the best weights vector.
# you code here
sgd_regressor.mse, sgd_regressor.w_
# Draw chart of model weights ($w_0$ and $w_1$) behavior during training.
weights_transposed = np.array(sgd_regressor.weights_).transpose()
plt.plot(range(len(sgd_regressor.weights_)), weights_transposed[0])
plt.plot(range(len(sgd_regressor.weights_)), weights_transposed[1])
# Make a prediction for hold-out set `(X_valid_scaled, y_valid)` and check MSE value.
# you code here
sgd_holdout_mse = mean_squared_error(y_valid, sgd_regressor.predict(X_valid_scaled))
sgd_holdout_mse
# Do the same thing for `LinearRegression` class from `sklearn.linear_model`. Evaluate MSE for hold-out set.
# you code here
lr = LinearRegression()
lr.fit(X_train_scaled, y_train)
linreg_holdout_mse = mean_squared_error(y_valid, lr.predict(X_valid_scaled))
linreg_holdout_mse
try:
assert (sgd_holdout_mse - linreg_holdout_mse) < 1e-4
print("Correct!")
except AssertionError:
print(
"Something's not good.\n Linreg's holdout MSE: {}"
"\n SGD's holdout MSE: {}".format(linreg_holdout_mse, sgd_holdout_mse)
)
|
import glob
import os
import shutil
import matplotlib.pyplot as plt
import numpy as np
import pytorch_lightning as L
import torch
import torch.nn.functional as F
import torchvision.transforms as T
from pytorch_lightning.trainer.supporters import CombinedLoader
from torch import nn
from torch.utils.data import DataLoader, Dataset
from torchvision.io import read_image
from torchvision.utils import make_grid, save_image
_ = L.seed_everything(0, workers=True)
def show_img(img_tensor, nrow, title=""):
img_tensor = img_tensor.detach().cpu() * 0.5 + 0.5
img_grid = make_grid(img_tensor, nrow=nrow).permute(1, 2, 0)
plt.figure(figsize=(18, 8))
plt.imshow(img_grid)
plt.axis("off")
plt.title(title)
plt.show()
class CustomTransform(object):
def __init__(self, img_dim=256):
self.transform = T.Compose(
[
T.RandomResizedCrop(
(img_dim, img_dim), scale=(0.8, 1.0), ratio=(0.9, 1.1)
),
T.RandomHorizontalFlip(p=0.5),
]
)
def __call__(self, img, stage="fit"):
if stage == "fit":
img = self.transform(img)
if stage == "test":
img = self.transform(img)
return img / 127.5 - 1
import torchvision
from torchvision import transforms
class CustomDataset(Dataset):
def __init__(self, filenames, transform, stage):
self.filenames = filenames
self.transform = transform
self.stage = stage
def __len__(self):
return len(self.filenames)
def __getitem__(self, idx):
img_name = self.filenames[idx]
img = read_image(img_name)
return self.transform(img, stage=self.stage)
MONET_DIR = "/kaggle/input/gan-getting-started/monet_jpg/*.jpg"
PHOTO_DIR = "/kaggle/input/gan-getting-started/photo_jpg/*.jpg"
MONET_TEST_DIR = "/kaggle/input/ceg-deep-learning/Part 2-4/*.jpg"
PHOTO_TEST_DIR = "/kaggle/input/ceg-deep-learning/Part 2-5/*.jpg"
LOADER_CONFIG = {
"batch_size": 8,
"num_workers": os.cpu_count(),
"pin_memory": torch.cuda.is_available(),
}
transform = CustomTransform()
from PIL import Image
import os
# 定义目录路径和输出目录路径
input_dir = "/kaggle/input/ceg-deep-learning/Part 2-5"
output_dir = "/kaggle/input/ceg-deep-learning/Part 2-5"
# 遍历输入目录下的所有文件
for filename in os.listdir(input_dir):
# 构建输入文件的完整路径和输出文件的完整路径
input_path = os.path.join(input_dir, filename)
output_path = os.path.join(output_dir, filename)
# 打开图片并进行大小变换
with Image.open(input_path) as image:
# 将图像大小调整为 256x256
image = image.resize((256, 256))
print("图片大小变换完成!")
class CustomDataModule(L.LightningDataModule):
def __init__(
self,
monet_dir=MONET_DIR,
photo_dir=PHOTO_DIR,
test_monet_dir=MONET_TEST_DIR,
test_photo_dir=PHOTO_TEST_DIR,
loader_config=LOADER_CONFIG,
transform=transform,
mode="max_size_cycle",
):
super().__init__()
self.monet_filenames = sorted(glob.glob(monet_dir))
self.photo_filenames = sorted(glob.glob(photo_dir))
self.test_monet_filenames = sorted(glob.glob(test_monet_dir))
self.test_photo_filenames = sorted(glob.glob(test_photo_dir))
self.loader_config = loader_config
self.transform = transform
self.mode = mode
def setup(self, stage):
if stage == "fit":
self.train_monet = CustomDataset(
self.monet_filenames, self.transform, stage
)
self.train_photo = CustomDataset(
self.photo_filenames, self.transform, stage
)
elif stage == "predict":
self.predict = CustomDataset(self.photo_filenames, self.transform, stage)
elif stage == "test":
self.test_monet = CustomDataset(
self.test_monet_filenames, self.transform, stage
)
self.test_photo = CustomDataset(
self.test_photo_filenames, self.transform, stage
)
def train_dataloader(self):
loader_monet = DataLoader(
self.train_monet,
shuffle=True,
drop_last=True,
**self.loader_config,
)
loader_photo = DataLoader(
self.train_photo,
shuffle=True,
drop_last=True,
**self.loader_config,
)
loaders = {"monet": loader_monet, "photo": loader_photo}
return CombinedLoader(loaders, mode=self.mode)
def predict_dataloader(self):
return DataLoader(self.predict, shuffle=False, **self.loader_config)
def test_dataloader(self):
return DataLoader(self.test_monet, shuffle=False, **self.loader_config)
def test_step(self, batch, batch_idx):
x, y = batch # Example input/output batch structure, modify as per your data
# Forward pass
y_pred = self(x)
# Compute loss or other metrics
loss = self.loss_function(y_pred, y)
# Return output or loss
return {"loss": loss}
def test_dataloader2(self):
return DataLoader(self.test_photo, shuffle=False, **self.loader_config)
SAMPLE_SIZE = 5
dm_sample = CustomDataModule(loader_config={"batch_size": SAMPLE_SIZE})
dm_sample.setup("fit")
train_loader = dm_sample.train_dataloader()
monet_samples = next(iter(train_loader))["monet"]
dm_sample.setup("predict")
predict_loader = dm_sample.predict_dataloader()
photo_samples = next(iter(predict_loader))
dm_sample.setup("test")
test_loader1 = dm_sample.test_dataloader()
monet_sampless = next(iter(test_loader1))
dm_sample.setup("test")
test_loader2 = dm_sample.test_dataloader2()
monet_samplesss = next(iter(test_loader2))
show_img(monet_samples, nrow=SAMPLE_SIZE, title="Samples of Monet Paintings")
show_img(photo_samples, nrow=SAMPLE_SIZE, title="Samples of Photos")
show_img(monet_sampless, nrow=SAMPLE_SIZE, title="Part 2-4 pictures")
show_img(monet_samplesss, nrow=SAMPLE_SIZE, title="Part 2-5 pictures")
class Downsampling(nn.Module):
def __init__(
self,
in_channels,
out_channels,
norm=True,
kernel_size=4,
stride=2,
padding=1,
):
super().__init__()
self.block = nn.Sequential(
nn.Conv2d(
in_channels,
out_channels,
kernel_size=kernel_size,
stride=stride,
padding=padding,
bias=False,
)
)
if norm:
self.block.append(nn.InstanceNorm2d(out_channels, affine=True))
self.block.append(nn.LeakyReLU(0.2))
def forward(self, x):
return self.block(x)
class Upsampling(nn.Module):
def __init__(
self,
in_channels,
out_channels,
dropout=False,
kernel_size=4,
stride=2,
padding=1,
output_padding=0,
):
super().__init__()
self.block = nn.Sequential(
nn.ConvTranspose2d(
in_channels,
out_channels,
kernel_size=kernel_size,
stride=stride,
padding=padding,
output_padding=output_padding,
bias=False,
),
nn.InstanceNorm2d(out_channels, affine=True),
)
if dropout:
self.block.append(nn.Dropout(0.5))
self.block.append(nn.ReLU())
def forward(self, x):
return self.block(x)
class ResidualBlock(nn.Module):
def __init__(self, channels):
super(ResidualBlock, self).__init__()
self.conv1 = nn.Conv2d(channels, channels, kernel_size=3, padding=1)
self.conv2 = nn.Conv2d(channels, channels, kernel_size=3, padding=1)
self.relu = nn.ReLU(inplace=True)
def forward(self, x):
residual = x
out = self.relu(self.conv1(x))
out = self.conv2(out)
out += residual
out = self.relu(out)
return out
class Generator(nn.Module):
def __init__(self, in_channels, out_channels, hid_channels):
super(Generator, self).__init__()
self.downsampling_blocks = nn.Sequential(
Downsampling(in_channels, hid_channels, norm=False), # 64x128x128
Downsampling(hid_channels, hid_channels * 2), # 128x64x64
Downsampling(hid_channels * 2, hid_channels * 4), # 256x32x32
Downsampling(hid_channels * 4, hid_channels * 8), # 512x16x16
Downsampling(hid_channels * 8, hid_channels * 8), # 512x8x8
Downsampling(hid_channels * 8, hid_channels * 8), # 512x4x4
Downsampling(hid_channels * 8, hid_channels * 8), # 512x2x2
Downsampling(hid_channels * 8, hid_channels * 8, norm=False), # 512x1x1
)
self.upsampling_blocks = nn.Sequential(
Upsampling(
hid_channels * 8, hid_channels * 8, dropout=True
), # (512+512)x2x2
Upsampling(
hid_channels * 16, hid_channels * 8, dropout=True
), # (512+512)x4x4
Upsampling(
hid_channels * 16, hid_channels * 8, dropout=True
), # (512+512)x8x8
Upsampling(hid_channels * 16, hid_channels * 8), # (512+512)x16x16
Upsampling(hid_channels * 16, hid_channels * 4), # (256+256)x32x32
Upsampling(hid_channels * 8, hid_channels * 2), # (128+128)x64x64
Upsampling(hid_channels * 4, hid_channels), # (64+64)x128x128
)
self.feature_block = nn.Sequential(
nn.ConvTranspose2d(
hid_channels * 2, out_channels, kernel_size=4, stride=2, padding=1
), # 3x256x256
nn.Tanh(),
)
self.residual_blocks = nn.Sequential(
ResidualBlock(hid_channels * 8),
ResidualBlock(hid_channels * 8),
ResidualBlock(hid_channels * 8),
ResidualBlock(hid_channels * 8),
ResidualBlock(hid_channels * 8),
ResidualBlock(hid_channels * 8),
ResidualBlock(hid_channels * 8),
ResidualBlock(hid_channels * 8),
)
def forward(self, x):
skips = []
for down in self.downsampling_blocks:
x = down(x)
skips.append(x)
skips = reversed(skips[:-1])
for up, skip in zip(self.upsampling_blocks, skips):
x = up(x)
x = torch.cat([x, skip], dim=1)
return self.feature_block(x)
class Discriminator(nn.Module):
def __init__(self, in_channels, hid_channels):
super(Discriminator, self).__init__()
self.downsampling_blocks = nn.Sequential(
Downsampling(in_channels, hid_channels, norm=False), # 64x128x128
Downsampling(hid_channels, hid_channels * 2), # 128x64x64
Downsampling(hid_channels * 2, hid_channels * 4), # 256x32x32
Downsampling(hid_channels * 4, hid_channels * 8, stride=1), # 512x31x31
)
self.residual_blocks = nn.Sequential(
ResidualBlock(hid_channels * 8),
ResidualBlock(hid_channels * 8),
ResidualBlock(hid_channels * 8),
ResidualBlock(hid_channels * 8),
ResidualBlock(hid_channels * 8),
ResidualBlock(hid_channels * 8),
ResidualBlock(hid_channels * 8),
ResidualBlock(hid_channels * 8),
)
self.conv = nn.Conv2d(hid_channels * 8, 1, kernel_size=4, padding=1) # 1x30x30
def forward(self, x):
x = self.downsampling_blocks(x)
x = self.residual_blocks(x)
x = self.conv(x)
return x
IN_CHANNELS = 3
OUT_CHANNELS = 3
HID_CHANNELS = 64
LR = 2e-4
# LR=0.9
BETAS = (0.5, 0.999)
LAMBDA = 10
DISPLAY_EPOCHS = 5
class CycleGAN(L.LightningModule):
def __init__(
self,
in_channels=IN_CHANNELS,
out_channels=OUT_CHANNELS,
hid_channels=HID_CHANNELS,
lr=LR,
betas=BETAS,
lambda_w=LAMBDA,
display_epochs=DISPLAY_EPOCHS,
photo_samples=photo_samples,
monet_sampless=monet_sampless,
):
super().__init__()
self.lr = lr
self.betas = betas
self.lambda_w = lambda_w
self.display_epochs = display_epochs
self.photo_samples = photo_samples
self.monet_sampless = monet_sampless
self.monet_samplesss = monet_samplesss
self.loss_history = []
self.epoch_count = 0
self.gen_PM = Generator(in_channels, out_channels, hid_channels)
self.gen_MP = Generator(in_channels, out_channels, hid_channels)
self.disc_M = Discriminator(in_channels, hid_channels)
self.disc_P = Discriminator(in_channels, hid_channels)
self.reset_parameters()
def forward(self, z):
return self.gen_PM(z)
def weights_init(self, m):
if isinstance(m, (nn.Conv2d, nn.ConvTranspose2d, nn.InstanceNorm2d)):
nn.init.normal_(m.weight, 0.0, 0.02)
if m.bias is not None:
nn.init.constant_(m.bias, 0.0)
def reset_parameters(self):
self.gen_PM = self.gen_PM.apply(self.weights_init)
self.gen_MP = self.gen_MP.apply(self.weights_init)
self.disc_M = self.disc_M.apply(self.weights_init)
self.disc_P = self.disc_P.apply(self.weights_init)
def adv_criterion(self, y_hat, y):
return F.binary_cross_entropy_with_logits(y_hat, y)
def recon_criterion(self, y_hat, y):
return F.l1_loss(y_hat, y)
def adv_loss(self, fake_Y, disc_Y):
fake_Y_hat = disc_Y(fake_Y)
valid = torch.ones_like(fake_Y_hat)
adv_loss_XY = self.adv_criterion(fake_Y_hat, valid)
return adv_loss_XY
def id_loss(self, real_Y, gen_XY):
id_Y = gen_XY(real_Y)
id_loss_Y = self.recon_criterion(id_Y, real_Y)
return 0.5 * self.lambda_w * id_loss_Y
def cycle_loss(self, real_Y, fake_X, gen_XY):
cycle_Y = gen_XY(fake_X)
cycle_loss_Y = self.recon_criterion(cycle_Y, real_Y)
return self.lambda_w * cycle_loss_Y
def gen_loss(self, real_X, real_Y, gen_XY, gen_YX, disc_Y):
adv_loss_XY = self.adv_loss(gen_XY(real_X), disc_Y)
id_loss_Y = self.id_loss(real_Y, gen_XY)
cycle_loss_Y = self.cycle_loss(real_Y, gen_YX(real_Y), gen_XY)
cycle_loss_X = self.cycle_loss(real_X, gen_XY(real_X), gen_YX)
total_cycle_loss = cycle_loss_X + cycle_loss_Y
gen_loss_XY = adv_loss_XY + id_loss_Y + total_cycle_loss
return gen_loss_XY
def disc_loss(self, real_Y, fake_Y, disc_Y, smooth=0.9):
real_Y_hat = disc_Y(real_Y)
valid = torch.full_like(real_Y_hat, fill_value=smooth)
real_loss_Y = self.adv_criterion(real_Y_hat, valid)
fake_Y_hat = disc_Y(fake_Y.detach())
fake = torch.zeros_like(fake_Y_hat)
fake_loss_Y = self.adv_criterion(fake_Y_hat, fake)
disc_loss_Y = (fake_loss_Y + real_loss_Y) * 0.5
return disc_loss_Y
def configure_optimizers(self):
params = {
"lr": self.lr,
"betas": self.betas,
}
opt_gen_PM = torch.optim.Adam(self.gen_PM.parameters(), **params)
opt_gen_MP = torch.optim.Adam(self.gen_MP.parameters(), **params)
opt_disc_M = torch.optim.Adam(self.disc_M.parameters(), **params)
opt_disc_P = torch.optim.Adam(self.disc_P.parameters(), **params)
return [opt_gen_PM, opt_gen_MP, opt_disc_M, opt_disc_P], []
def training_step(self, batch, batch_idx, optimizer_idx):
real_M = batch["monet"]
real_P = batch["photo"]
if optimizer_idx == 0:
gen_loss_PM = self.gen_loss(
real_P, real_M, self.gen_PM, self.gen_MP, self.disc_M
)
return gen_loss_PM
if optimizer_idx == 1:
gen_loss_MP = self.gen_loss(
real_M, real_P, self.gen_MP, self.gen_PM, self.disc_P
)
return gen_loss_MP
if optimizer_idx == 2:
disc_loss_M = self.disc_loss(real_M, self.gen_PM(real_P), self.disc_M)
return disc_loss_M
if optimizer_idx == 3:
disc_loss_P = self.disc_loss(real_P, self.gen_MP(real_M), self.disc_P)
return disc_loss_P
def training_epoch_end(self, outputs):
self.epoch_count += 1
losses = []
for j in range(4):
loss = np.mean([out[j]["loss"].item() for out in outputs])
losses.append(loss)
self.loss_history.append(losses)
print(
f"Epoch {self.epoch_count} -",
f"gen_loss_PM: {losses[0]:.5f} -",
f"gen_loss_MP: {losses[1]:.5f} -",
f"disc_loss_M: {losses[2]:.5f} -",
f"disc_loss_P: {losses[3]:.5f}",
)
if self.epoch_count % self.display_epochs == 0 or self.epoch_count == 1:
gen_monets = self(self.photo_samples.to(self.device)).detach().cpu()
show_img(
torch.cat([self.photo_samples, gen_monets]),
nrow=len(self.photo_samples),
title=f"Epoch {self.epoch_count}: Photo-to-Monet Translation",
)
gen_monets2 = self(self.monet_sampless.to(self.device)).detach().cpu()
show_img(
torch.cat([self.monet_sampless, gen_monets2]),
nrow=len(self.monet_sampless),
title=f"Epoch {self.epoch_count}: test for Part 2-4",
)
gen_monets3 = self(self.monet_samplesss.to(self.device)).detach().cpu()
show_img(
torch.cat([self.monet_samplesss, gen_monets3]),
nrow=len(self.monet_samplesss),
title=f"Epoch {self.epoch_count}: test for Part 2-5",
)
def predict_step(self, batch, batch_idx):
return self(batch)
def loss_curves(self):
labels = ["gen_loss_PM", "gen_loss_MP", "disc_loss_M", "disc_loss_P"]
titles = ["Generator Loss Curves", "Discriminator Loss Curves"]
num_epochs = len(self.loss_history)
plt.figure(figsize=(18, 4.5))
for j in range(4):
if j % 2 == 0:
plt.subplot(1, 2, (j // 2) + 1)
plt.title(titles[j // 2])
plt.ylabel("Loss")
plt.xlabel("Epoch")
plt.plot(
np.arange(1, num_epochs + 1),
[losses[j] for losses in self.loss_history],
label=labels[j],
)
plt.legend(loc="upper right")
TRAIN_CONFIG = {
"accelerator": "gpu" if torch.cuda.is_available() else "cpu",
"devices": 1,
"logger": False,
"enable_checkpointing": torch.cuda.is_available(),
"max_epochs": 8,
}
dm = CustomDataModule()
model = CycleGAN()
checkpoint = {
"state_dict": model.state_dict(),
# 其他相关信息,例如优化器状态、训练配置等
}
torch.save(checkpoint, "model_checkpoint.pth")
trainer = L.Trainer(**TRAIN_CONFIG)
trainer.fit(model, datamodule=dm)
MONET_TEST_DIR = "/kaggle/input/ceg-deep-learning/*.jpg"
PHOTO_TEST_DIR = "/kaggle/input/ceg-deep-learning/*.jpg"
model.loss_curves()
predictions = trainer.predict(model, datamodule=dm)
os.makedirs("../images", exist_ok=True)
idx = 0
for tensor in predictions:
for monet in tensor:
save_image((monet.squeeze() * 0.5 + 0.5), fp=f"../images/{idx}.jpg")
idx += 1
shutil.make_archive("/kaggle/working/images", "zip", "/kaggle/images")
# from PIL import Image
# import numpy as np
# device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
# 加载模型权重或检查点
# checkpoint_path = '/kaggle/working/model_checkpoint.pth' # 替换为实际的检查点文件路径
# checkpoint = torch.load(checkpoint_path)
# model.load_state_dict(checkpoint['state_dict'])
# 切换模型为评估模式
# model.eval()
# trainer.test(model, datamodule=dm)
from PIL import Image
import numpy as np
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
# 加载模型权重或检查点
checkpoint_path = "/kaggle/working/model_checkpoint.pth" # 替换为实际的检查点文件路径
checkpoint = torch.load(checkpoint_path)
model.load_state_dict(checkpoint["state_dict"])
# 切换模型为评估模式
model.eval()
# 加载测试图片
test_image1_path = (
"/kaggle/input/ceg-deep-learning/Part 2-4/0a1d1b9f8e.jpg" # 替换为实际的测试图片路径
)
test_image2_path = "/kaggle/input/ceg-deep-learning/Part 2-4/0a49020ae5.jpg"
test_image3_path = "/kaggle/input/ceg-deep-learning/Part 2-4/0a497f768d.jpg"
test_image1 = Image.open(test_image1_path)
test_image2 = Image.open(test_image2_path)
test_image3 = Image.open(test_image3_path)
|
import numpy as np
import pandas as pd
from umap import UMAP
import matplotlib.pyplot as plt
train = pd.read_csv("../input/digit-recognizer/train.csv")
test = pd.read_csv("../input/digit-recognizer/test.csv")
y = train["label"].values
train = train[test.columns].values
test = test[test.columns].values
train_test = np.vstack([train, test])
train_test.shape
umap = UMAP()
train_test_2D = umap.fit_transform(train_test)
train_2D = train_test_2D[: train.shape[0]]
test_2D = train_test_2D[train.shape[0] :]
np.save("train_2D", train_2D)
np.save("test_2D", test_2D)
plt.scatter(train_2D[:, 0], train_2D[:, 1], c=y, s=0.5)
|
import numpy as np
import pandas as pd
import tensorflow as tf
import matplotlib.pyplot as plt
print(tf.__version__)
# # Introduction.
# Finding minima of a quantity can be encountered in many fields of math and science.
# Above, here is an example of the function of two variables that has one global munimun and several local.
def func(x, y):
return (
-5.5 * tf.exp(-20.0 * (x - 0.3) ** 2 - 40.0 * (y - 0.3) ** 2)
- 3.5 * tf.exp(-15.0 * (x - 0.6) ** 2 - 10.0 * (y - 0.85) ** 2)
- 2.0 * tf.sin(2.0 * (x - y))
)
x = np.linspace(0, 1, 400)
X, Y = np.meshgrid(x, x)
Z = func(X, Y)
plt.figure(figsize=(6, 4.7))
plt.contourf(X, Y, Z, 60, cmap="RdGy")
plt.xlabel("x", fontsize=19)
plt.ylabel("y", fontsize=19)
plt.tick_params(axis="both", which="major", labelsize=14)
cbar = plt.colorbar()
cbar.ax.tick_params(labelsize=14)
# We will try to minimize it...
def constr(a, b):
assert b > a
return lambda x: tf.clip_by_value(x, a, b)
x = tf.Variable(
0.0, trainable=True, dtype=tf.float64, name="x", constraint=constr(0, 1)
)
y = tf.Variable(
0.0, trainable=True, dtype=tf.float64, name="y", constraint=constr(0, 1)
)
def objective():
return (
-5.5 * tf.exp(-20.0 * (x - 0.3) ** 2 - 40.0 * (y - 0.3) ** 2)
- 3.5 * tf.exp(-15.0 * (x - 0.6) ** 2 - 10.0 * (y - 0.85) ** 2)
- 2.0 * tf.sin(2.0 * (x - y))
)
def optimize(start, verbose=False, method="SGD"):
x.assign(start[0])
y.assign(start[1])
if method == "SGD":
opt = tf.keras.optimizers.SGD(learning_rate=0.01)
if method == "ADAM":
opt = tf.keras.optimizers.Adam(
learning_rate=0.1,
beta_1=0.9,
beta_2=0.999,
epsilon=1e-07,
amsgrad=False,
)
obj_vals = []
coords = []
for i in range(50):
if verbose:
print(
f"obj = {objective().numpy():.4f}, x = {x.numpy():.4f}, y = {y.numpy():.4f}"
)
obj_vals.append(objective().numpy())
coords.append((x.numpy(), y.numpy()))
opt.minimize(objective, var_list=[x, y])
return obj_vals, coords
def plot_res(obj_vals, coords):
plt.figure(figsize=(16, 6))
plt.subplot(121)
plt.contourf(X, Y, Z, 60, cmap="RdGy")
plt.xlabel("x", fontsize=19)
plt.ylabel("y", fontsize=19)
plt.tick_params(axis="both", which="major", labelsize=14)
cbar = plt.colorbar()
cbar.ax.tick_params(labelsize=14)
xcoord = [x[0] for x in coords]
ycoord = [x[1] for x in coords]
plt.plot(xcoord, ycoord, ".-")
plt.plot(xcoord[-1], ycoord[-1], "y*", markersize=12)
plt.subplot(122)
plt.plot(obj_vals, ".-")
plt.plot([len(obj_vals) - 1], obj_vals[-1], "y*", markersize=12)
plt.xlabel("Step", fontsize=17)
plt.ylabel("Objective", fontsize=17)
plt.tick_params(axis="both", which="major", labelsize=14)
plt.show()
obj_vals, coords = optimize([0.25, 0.65], verbose=True)
plot_res(obj_vals, coords)
obj_vals, coords = optimize([0.2, 0.65])
plot_res(obj_vals, coords)
obj_vals, coords = optimize([0.2, 0.65], method="ADAM")
plot_res(obj_vals, coords)
# # Applying gradients explicitly.
x.assign(0.25)
y.assign(0.65)
opt = tf.keras.optimizers.SGD(learning_rate=0.01)
for i in range(30):
with tf.GradientTape() as tape:
z = func(x, y)
grads = tape.gradient(z, [x, y])
processed_grads = [g for g in grads]
grads_and_vars = zip(processed_grads, [x, y])
print(
f"z = {z.numpy():.2f}, x = {x.numpy():.2f}, y = {y.numpy():.2f}, grads0 = {grads[0].numpy():.2f}, grads1 = {grads[1].numpy():.2f}"
)
opt.apply_gradients(grads_and_vars)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
import pandas as pd
df_train = pd.read_csv("/kaggle/input/digit-recognizer/train.csv")
df_test = pd.read_csv("/kaggle/input/digit-recognizer/test.csv")
from keras.models import Sequential
from keras.layers.normalization import BatchNormalization
from keras.layers.convolutional import Conv2D
from keras.layers.convolutional import MaxPooling2D
from keras.layers.core import Activation, Flatten, Dropout, Dense
from keras import backend as K
from keras.models import Sequential
from keras.optimizers import SGD
model = Sequential()
model.add(Conv2D(32, (5, 5), input_shape=(28, 28, 1), activation="relu"))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Conv2D(32, (3, 3), input_shape=(28, 28, 1), activation="relu"))
model.add(MaxPooling2D(pool_size=(2, 2)))
model.add(Dropout(0.2))
model.add(Flatten())
model.add(Dense(128, activation="relu"))
model.add(Dropout(0.5))
model.add(Dense(10, activation="softmax", name="predict"))
from keras.utils.np_utils import to_categorical
import tensorflow as tf
from keras.preprocessing.image import ImageDataGenerator
from keras.optimizers import Adam
from sklearn.model_selection import train_test_split
from keras.preprocessing.image import img_to_array
from keras.utils import to_categorical
from sklearn.metrics import classification_report
from sklearn.metrics import confusion_matrix
import matplotlib.pyplot as plt
import numpy as np
import argparse
import random
import cv2
import os
trainY = df_train["label"]
trainX = df_train.drop(labels=["label"], axis=1)
trainX = trainX / 255.0
df_test = df_test / 255.0
trainX = trainX.values.reshape(-1, 28, 28, 1)
trainY = to_categorical(trainY, num_classes=10)
random_seed = 42
trainX, X_val, trainY, Y_val = train_test_split(
trainX, trainY, test_size=0.2, random_state=random_seed
)
from keras.optimizers import RMSprop
optimizer = RMSprop(lr=0.001)
# Compile the model
model.compile(
optimizer=optimizer, loss="categorical_crossentropy", metrics=["accuracy"]
)
epochs = 10
batch_size = 32
datagen = ImageDataGenerator(
rotation_range=10,
zoom_range=0.1,
width_shift_range=0.1,
height_shift_range=0.1,
horizontal_flip=False,
vertical_flip=False,
)
datagen.fit(trainX)
history = model.fit_generator(
datagen.flow(trainX, trainY, batch_size=batch_size),
epochs=epochs,
validation_data=(X_val, Y_val),
verbose=2,
steps_per_epoch=trainX.shape[0] // batch_size,
)
|
def get_robin_preds():
import os
import math
import sys
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import time
import datetime
import gc
import copy
import random
from scipy.stats import spearmanr
from sklearn.model_selection import KFold, GroupKFold
from sklearn.preprocessing import OneHotEncoder, MinMaxScaler
import torch
import torch.nn as nn
import torch.nn.functional as F
from torch.utils.data import DataLoader, Dataset
from tqdm import tqdm_notebook as tqdm
os.system("pip install ../input/sacremoses/sacremoses-master/ > /dev/null")
sys.path.insert(
0,
"../input/transformers/transformers-d46147294852694d1dc701c72b9053ff2e726265/",
)
import transformers
from transformers import (
BertModel,
BertTokenizer,
BertConfig,
RobertaModel,
RobertaTokenizer,
RobertaConfig,
XLNetModel,
XLNetTokenizer,
XLNetConfig,
AlbertModel,
AlbertTokenizer,
AlbertConfig,
)
N_TARGETS = 30
N_Q_TARGETS = 21
N_A_TARGETS = 9
TARGETS = [
"question_asker_intent_understanding",
"question_body_critical",
"question_conversational",
"question_expect_short_answer",
"question_fact_seeking",
"question_has_commonly_accepted_answer",
"question_interestingness_others",
"question_interestingness_self",
"question_multi_intent",
"question_not_really_a_question",
"question_opinion_seeking",
"question_type_choice",
"question_type_compare",
"question_type_consequence",
"question_type_definition",
"question_type_entity",
"question_type_instructions",
"question_type_procedure",
"question_type_reason_explanation",
"question_type_spelling",
"question_well_written",
"answer_helpful",
"answer_level_of_information",
"answer_plausible",
"answer_relevance",
"answer_satisfaction",
"answer_type_instructions",
"answer_type_procedure",
"answer_type_reason_explanation",
"answer_well_written",
]
PRETRAINED_PATH = "../input/pretrained-models/"
def get_categorical_features(train, test, feature):
unique_vals = list(
set(train[feature].unique().tolist() + test[feature].unique().tolist())
)
feat_dict = {i + 1: e for i, e in enumerate(unique_vals)}
feat_dict_reverse = {v: k for k, v in feat_dict.items()}
train_feat = train[feature].apply(lambda x: feat_dict_reverse[x]).values
test_feat = test[feature].apply(lambda x: feat_dict_reverse[x]).values
return train_feat, test_feat, feat_dict, feat_dict_reverse
class TextDataset4(Dataset):
def __init__(self, x_features, ids, seg_ids, idxs, targets=None):
self.ids = ids[idxs].astype(np.long)
self.seg_ids = seg_ids[idxs].astype(np.long)
self.x_features = x_features[idxs].astype(np.float32)
if targets is not None:
self.targets = targets[idxs].astype(np.float32)
else:
self.targets = np.zeros(
(self.x_features.shape[0], N_TARGETS), dtype=np.float32
)
def __getitem__(self, idx):
ids = self.ids[idx]
seg_ids = self.seg_ids[idx]
x_feats = self.x_features[idx]
target = self.targets[idx]
return (x_feats, ids, seg_ids), target
def __len__(self):
return len(self.x_features)
class TextDataset5(Dataset):
def __init__(
self,
x_features,
question_ids,
answer_ids,
seg_question_ids,
seg_answer_ids,
idxs,
targets=None,
):
self.question_ids = question_ids[idxs].astype(np.long)
self.answer_ids = answer_ids[idxs].astype(np.long)
self.seg_question_ids = seg_question_ids[idxs].astype(np.long)
self.seg_answer_ids = seg_answer_ids[idxs].astype(np.long)
self.x_features = x_features[idxs].astype(np.float32)
if targets is not None:
self.targets = targets[idxs].astype(np.float32)
else:
self.targets = np.zeros(
(self.x_features.shape[0], N_TARGETS), dtype=np.float32
)
def __getitem__(self, idx):
q_ids = self.question_ids[idx]
a_ids = self.answer_ids[idx]
seg_q_ids = self.seg_question_ids[idx]
seg_a_ids = self.seg_answer_ids[idx]
x_feats = self.x_features[idx]
target = self.targets[idx]
return (x_feats, q_ids, a_ids, seg_q_ids, seg_a_ids), target
def __len__(self):
return len(self.x_features)
def to_cpu(x):
return x.contiguous().detach().cpu()
def to_numpy(x):
return to_cpu(x).numpy()
def to_device(xs, device):
if isinstance(xs, tuple) or isinstance(xs, list):
return [x.to(device) for x in xs]
else:
return [xs.to(device)]
def infer_batch(inputs, model, device, to_numpy=True):
inputs = to_device(inputs, device)
predicted = model(*inputs)
inputs = [x.cpu() for x in inputs]
preds = torch.sigmoid(predicted)
if to_numpy:
preds = preds.cpu().detach().numpy().astype(np.float32)
return preds
def infer(model, loader, checkpoint_file=None, device=torch.device("cuda")):
n_obs = len(loader.dataset)
batch_sz = loader.batch_size
predictions = np.zeros((n_obs, N_TARGETS))
if checkpoint_file is not None:
print(f"Starting inference for model: {checkpoint_file}")
checkpoint = torch.load(checkpoint_file)
model.load_state_dict(checkpoint["model_state_dict"])
model.float()
model.to(device)
model.eval()
with torch.no_grad():
for i, (inputs, _) in enumerate(tqdm(loader)):
start_index = i * batch_sz
end_index = min(start_index + batch_sz, n_obs)
batch_preds = infer_batch(inputs, model, device)
predictions[start_index:end_index, :] += batch_preds
return predictions
def init_seed(seed=100):
os.environ["PYTHONHASHSEED"] = str(seed)
random.seed(seed)
np.random.seed(seed)
torch.manual_seed(seed)
torch.cuda.manual_seed_all(seed)
torch.backends.cudnn.deterministic = True
class GELU(nn.Module):
def forward(self, x):
return x * torch.sigmoid(1.702 * x)
def lin_layer(n_in, n_out, dropout):
return nn.Sequential(nn.Linear(n_in, n_out), GELU(), nn.Dropout(dropout))
class Head2(nn.Module):
def __init__(self, n_h=512, n_feats=74, n_bert=768, dropout=0.2):
super().__init__()
n_x = n_feats + 2 * n_bert
self.lin = lin_layer(n_in=n_x, n_out=n_h, dropout=dropout)
self.lin_q = lin_layer(n_in=n_feats + n_bert, n_out=n_h, dropout=dropout)
self.lin_a = lin_layer(n_in=n_feats + n_bert, n_out=n_h, dropout=dropout)
self.head_q = nn.Linear(2 * n_h, N_Q_TARGETS)
self.head_a = nn.Linear(2 * n_h, N_A_TARGETS)
def forward(self, x_feats, x_q_bert, x_a_bert):
x_q = self.lin_q(torch.cat([x_feats, x_q_bert], dim=1))
x_a = self.lin_a(torch.cat([x_feats, x_a_bert], dim=1))
x = self.lin(torch.cat([x_feats, x_q_bert, x_a_bert], dim=1))
x_q = self.head_q(torch.cat([x, x_q], dim=1))
x_a = self.head_a(torch.cat([x, x_a], dim=1))
return torch.cat([x_q, x_a], dim=1)
class AvgPooledRoberta(RobertaModel):
def forward(self, ids, seg_ids=None):
att_mask = ids > 0
x_bert = super().forward(ids, att_mask, token_type_ids=seg_ids)[0]
att_mask = att_mask.unsqueeze(-1)
return (x_bert * att_mask).sum(dim=1) / att_mask.sum(dim=1)
def resize_type_embeddings(self, new_num_types):
old_embeddings = self.embeddings.token_type_embeddings
model_embeds = self._get_resized_embeddings(old_embeddings, new_num_types)
self.embeddings.token_type_embeddings = model_embeds
self.config.type_vocab_size = new_num_types
self.type_vocab_size = new_num_types
class CustomRoberta(nn.Module):
def __init__(self, n_h, n_feats, head_dropout=0.2):
super().__init__()
config = RobertaConfig.from_json_file(
PRETRAINED_PATH + "roberta-base/config.json"
)
self.roberta = AvgPooledRoberta(config)
self.roberta.resize_type_embeddings(2)
self.head = Head2(n_h, n_feats, n_bert=768, dropout=head_dropout)
def forward(self, x_feats, q_ids, a_ids, seg_q_ids=None, seg_a_ids=None):
x_q_bert = self.roberta(q_ids, seg_q_ids)
x_a_bert = self.roberta(a_ids, seg_a_ids)
return self.head(x_feats, x_q_bert, x_a_bert)
class AvgPooledBert(BertModel):
def forward(self, ids, seg_ids=None):
att_mask = ids > 0
x_bert = super().forward(ids, att_mask, token_type_ids=seg_ids)[0]
att_mask = att_mask.unsqueeze(-1)
return (x_bert * att_mask).sum(dim=1) / att_mask.sum(dim=1)
class CustomBert3(nn.Module):
def __init__(self, n_h, n_feats):
super().__init__()
self.bert = AvgPooledBert(BertConfig())
self.head = Head2(n_h, n_feats, n_bert=768)
def forward(self, x_feats, q_ids, a_ids, seg_q_ids=None, seg_a_ids=None):
x_q_bert = self.bert(q_ids, seg_q_ids)
x_a_bert = self.bert(a_ids, seg_a_ids)
return self.head(x_feats, x_q_bert, x_a_bert)
class AvgPooledXLNet(XLNetModel):
def forward(self, ids, seg_ids=None):
att_mask = (ids > 0).float()
x_bert = super().forward(ids, att_mask, token_type_ids=seg_ids)[0]
att_mask = att_mask.unsqueeze(-1)
return (x_bert * att_mask).sum(dim=1) / att_mask.sum(dim=1)
class CustomXLNet(nn.Module):
def __init__(self, n_h, n_feats, head_dropout=0.2):
super().__init__()
config = XLNetConfig.from_json_file(
PRETRAINED_PATH + "xlnet-base-cased/config.json"
)
self.xlnet = AvgPooledXLNet(config)
self.head = Head2(n_h, n_feats, n_bert=768, dropout=head_dropout)
def forward(self, x_feats, q_ids, a_ids, seg_q_ids=None, seg_a_ids=None):
x_q_bert = self.xlnet(q_ids, seg_q_ids)
x_a_bert = self.xlnet(a_ids, seg_a_ids)
return self.head(x_feats, x_q_bert, x_a_bert)
class AvgPooledAlbert(AlbertModel):
def forward(self, ids, seg_ids=None):
att_mask = ids > 0
x_bert = super().forward(ids, att_mask, token_type_ids=seg_ids)[0]
att_mask = att_mask.unsqueeze(-1)
return (x_bert * att_mask).sum(dim=1) / att_mask.sum(dim=1)
class CustomAlbert(nn.Module):
def __init__(self, n_h, n_feats, head_dropout=0.2):
super().__init__()
config = AlbertConfig.from_json_file(
PRETRAINED_PATH + "albert-base-v2/config.json"
)
self.q_albert = AvgPooledAlbert(config)
self.a_albert = AvgPooledAlbert(config)
self.head = Head2(n_h, n_feats, n_bert=768, dropout=head_dropout)
def forward(self, x_feats, q_ids, a_ids, seg_q_ids=None, seg_a_ids=None):
x_q_bert = self.q_albert(q_ids, seg_q_ids)
x_a_bert = self.a_albert(a_ids, seg_a_ids)
return self.head(x_feats, x_q_bert, x_a_bert)
class CLSPooledRoberta(RobertaModel):
def forward(self, ids, seg_ids=None):
att_mask = ids > 0
return super().forward(ids, att_mask, token_type_ids=seg_ids)[0][:, 0, :]
def resize_type_embeddings(self, new_num_types):
old_embeddings = self.embeddings.token_type_embeddings
model_embeds = self._get_resized_embeddings(old_embeddings, new_num_types)
self.embeddings.token_type_embeddings = model_embeds
self.config.type_vocab_size = new_num_types
self.type_vocab_size = new_num_types
class CustomRoberta2(nn.Module):
def __init__(self, n_h, n_feats):
super().__init__()
config = RobertaConfig.from_json_file(
PRETRAINED_PATH + "roberta-base/config.json"
)
self.roberta = CLSPooledRoberta(config)
self.roberta.resize_type_embeddings(3)
self.head = nn.Linear(768 + n_feats, N_TARGETS)
def forward(self, x_feats, ids, seg_ids=None):
x_bert = self.roberta(ids, seg_ids)
return self.head(torch.cat([x_feats, x_bert], dim=1))
pd.set_option("max_rows", 500)
pd.set_option("max_columns", 500)
path = "../input/google-quest-challenge/"
sample_submission = pd.read_csv(f"{path}sample_submission.csv")
test = pd.read_csv(f"{path}test.csv").fillna(" ")
train = pd.read_csv(f"{path}train.csv").fillna(" ")
def get_preds(
train, test, ModelClass, tokenizer, model_name, checkpoint_dir, folds
):
seg_ids_test, ids_test = {}, {}
max_seq_len = 512
for mode, df in [("test", test)]:
for text, cols in [
("question", ["question_title", "question_body"]),
("answer", ["question_title", "answer"]),
]:
ids, seg_ids = [], []
for x1, x2 in tqdm(df[cols].values):
encoded_inputs = tokenizer.encode_plus(
x1,
x2,
add_special_tokens=True,
max_length=max_seq_len,
pad_to_max_length=True,
return_token_type_ids=True,
)
ids.append(encoded_inputs["input_ids"])
seg_ids.append(encoded_inputs["token_type_ids"])
ids_test[text] = np.array(ids)
seg_ids_test[text] = np.array(seg_ids)
(
train_category,
test_category,
category_dict,
category_dict_reverse,
) = get_categorical_features(train, test, "category")
cat_features_train = train_category.reshape(-1, 1)
cat_features_test = test_category.reshape(-1, 1)
ohe = OneHotEncoder(handle_unknown="ignore")
ohe.fit(cat_features_train)
cat_features_test = ohe.transform(cat_features_test).toarray()
num_workers = 8
device = "cuda"
bs_test = 2
test_loader = DataLoader(
TextDataset5(
cat_features_test,
ids_test["question"],
ids_test["answer"],
seg_ids_test["question"],
seg_ids_test["answer"],
test.index,
),
batch_size=bs_test,
shuffle=False,
num_workers=num_workers,
)
init_seed()
preds = np.zeros((len(test), N_TARGETS))
for fold_id in folds:
checkpoint_file = (
f"{checkpoint_dir}{model_name}_fold_{fold_id + 1}_best.pth"
)
model = ModelClass(256, cat_features_test.shape[1]).to(device)
test_preds = infer(model, test_loader, checkpoint_file, device)
preds += test_preds / len(folds)
return preds
def get_preds2(
train, test, ModelClass, tokenizer, model_name, checkpoint_dir, folds
):
sep_token = f" {tokenizer.sep_token} "
max_seq_len = 512
ids, seg_ids, sent_ids = [], [], []
for x1, x2, x3 in tqdm(
test[["question_title", "question_body", "answer"]].values
):
encoded_inputs = tokenizer.encode_plus(
x1 + sep_token + x2,
x3,
add_special_tokens=True,
max_length=max_seq_len,
pad_to_max_length=True,
return_token_type_ids=True,
)
inp_ids = encoded_inputs["input_ids"]
raw_seg_ids = np.array(encoded_inputs["token_type_ids"])
qa_split_idx = (
np.where(np.array(inp_ids) == tokenizer.sep_token_id)[0][0] + 1
)
raw_seg_ids[qa_split_idx:] += 1
raw_seg_ids = (
raw_seg_ids * (np.array(inp_ids) != tokenizer.pad_token_id).astype(int)
).tolist()
ids.append(inp_ids)
seg_ids.append(raw_seg_ids)
ids_test = np.array(ids)
seg_ids_test = np.array(seg_ids)
(
train_category,
test_category,
category_dict,
category_dict_reverse,
) = get_categorical_features(train, test, "category")
cat_features_train = train_category.reshape(-1, 1)
cat_features_test = test_category.reshape(-1, 1)
ohe = OneHotEncoder(handle_unknown="ignore")
ohe.fit(cat_features_train)
cat_features_test = ohe.transform(cat_features_test).toarray()
num_workers = 8
device = "cuda"
bs_test = 2
test_loader = DataLoader(
TextDataset4(cat_features_test, ids_test, seg_ids_test, test.index),
batch_size=bs_test,
shuffle=False,
num_workers=num_workers,
)
init_seed()
preds = np.zeros((len(test), N_TARGETS))
for fold_id in folds:
checkpoint_file = (
f"{checkpoint_dir}{model_name}_fold_{fold_id + 1}_best.pth"
)
model = ModelClass(256, cat_features_test.shape[1]).to(device)
test_preds = infer(model, test_loader, checkpoint_file, device)
preds += test_preds / len(folds)
return preds
def get_bert_preds(train, test):
tokenizer = BertTokenizer.from_pretrained(
PRETRAINED_PATH + "bert-base-uncased/"
)
model_name = "siamese_bert_6_comb"
checkpoint_dir = "../input/siamese-bert-models-6/"
return get_preds(
train,
test,
CustomBert3,
tokenizer,
model_name,
checkpoint_dir,
[0, 2, 3, 5, 6, 8, 9],
)
def get_roberta_preds(train, test):
tokenizer = RobertaTokenizer.from_pretrained(PRETRAINED_PATH + "roberta-base/")
model_name = "siamese_roberta_1_comb"
checkpoint_dir = "../input/siamese-roberta-models-1/"
return get_preds(
train,
test,
CustomRoberta,
tokenizer,
model_name,
checkpoint_dir,
[0, 1, 3, 4, 6, 7, 9],
)
def get_xlnet_preds(train, test):
tokenizer = XLNetTokenizer.from_pretrained(
PRETRAINED_PATH + "xlnet-base-cased/"
)
model_name = "siamese_xlnet_1_comb"
checkpoint_dir = "../input/siamese-xlnet-models-1/"
return get_preds(
train,
test,
CustomXLNet,
tokenizer,
model_name,
checkpoint_dir,
[0, 1, 2, 4, 5, 7, 8],
)
def get_albert_preds(train, test):
tokenizer = AlbertTokenizer.from_pretrained(PRETRAINED_PATH + "albert-base-v2/")
model_name = "siamese_albert_1_comb"
checkpoint_dir = "../input/siamese-albert-models-1/"
return get_preds(
train,
test,
CustomAlbert,
tokenizer,
model_name,
checkpoint_dir,
[1, 2, 3, 4, 5, 6, 7],
)
def get_roberta2_preds(train, test):
tokenizer = RobertaTokenizer.from_pretrained(PRETRAINED_PATH + "roberta-base/")
model_name = "siamese_roberta_2_half"
checkpoint_dir = "../input/siamese-roberta-models-2/"
return get_preds2(
train,
test,
CustomRoberta2,
tokenizer,
model_name,
checkpoint_dir,
[1, 2, 3, 5, 6, 8, 9],
)
return (
get_albert_preds(train, test),
get_roberta_preds(train, test),
get_bert_preds(train, test),
get_xlnet_preds(train, test),
)
y_albert, y_roberta, y_bert, y_xlnet = get_robin_preds()
import gc
gc.collect()
def get_use_preds():
import tensorflow as tf
import tensorflow_hub as hub
import numpy as np
import pandas as pd
from nltk import tokenize
from tqdm import tqdm_notebook
embed_fn = hub.load("../input/universalsentenceencoderlarge4/")
df = pd.read_csv("../input/google-quest-challenge/train.csv")
test_df = pd.read_csv("../input/google-quest-challenge/test.csv")
outputs = df.columns[11:]
def count_words(data):
return len(str(data).split())
def count_words_unique(data):
return len(np.unique(str(data).split()))
def questionowords(data):
start_words = [
"who",
"what",
"when",
"where",
"why",
"how",
"is",
"am",
"are",
"was",
"were",
"can",
"could",
"may",
"should",
"shall",
"does",
"do",
"did",
]
sents = tokenize.sent_tokenize(data)
qw = 0
for sent in sents:
if sent.lower().startswith(tuple(start_words)):
qw += 1
return qw
def questionmarks(data):
sents = tokenize.sent_tokenize(data)
qm = 0
for sent in sents:
qm += sent.count("?")
return qm
def get_numeric_features(df):
df["qt_wc"] = df["question_title"].apply(count_words)
df["qb_wc"] = df["question_body"].apply(count_words)
df["a_wc"] = df["answer"].apply(count_words)
df["qt_wcu"] = df["question_title"].apply(count_words_unique)
df["qb_wcu"] = df["question_body"].apply(count_words_unique)
df["a_wcu"] = df["answer"].apply(count_words_unique)
df["qb_qw"] = df["question_body"].apply(questionowords)
df["qt_qw"] = df["question_title"].apply(questionowords)
df["qb_qm"] = df["question_body"].apply(questionmarks)
df["qt_qm"] = df["question_title"].apply(questionmarks)
return df
test_df = get_numeric_features(test_df)
features = [
"qt_wc",
"qb_wc",
"a_wc",
"qt_wcu",
"qb_wcu",
"a_wcu",
"qb_qw",
"qt_qw",
"qb_qm",
"qt_qm",
]
MAX_SEQ = 30
def get_sentences(x):
sentences = [s for s in tokenize.sent_tokenize(x) if s != ""]
if len(sentences) > MAX_SEQ:
return sentences[:MAX_SEQ]
return sentences + [""] * (MAX_SEQ - len(sentences))
def get_use(df):
QT = embed_fn(df["question_title"].values)["outputs"].numpy()
A = np.zeros((df.shape[0], MAX_SEQ, 512), dtype=np.float32)
for i, x in tqdm_notebook(list(enumerate(df["answer"].values))):
A[i] = embed_fn(get_sentences(x))["outputs"].numpy()
QB = np.zeros((df.shape[0], MAX_SEQ, 512), dtype=np.float32)
for i, x in tqdm_notebook(list(enumerate(df["question_body"].values))):
QB[i] = embed_fn(get_sentences(x))["outputs"].numpy()
return QT, A, QB
QT_test, A_test, QB_test = get_use(test_df)
import gc
del embed_fn
gc.collect()
import tensorflow.keras.layers as KL
def nn_block(input_layer, size, dropout_rate, activation):
out_layer = KL.Dense(size, activation=None)(input_layer)
# out_layer = KL.BatchNormalization()(out_layer)
out_layer = KL.Activation(activation)(out_layer)
out_layer = KL.Dropout(dropout_rate)(out_layer)
return out_layer
def cnn_block(input_layer, size, dropout_rate, activation):
out_layer = KL.Conv1D(size, 1, activation=None)(input_layer)
# out_layer = KL.LayerNormalization()(out_layer)
out_layer = KL.Activation(activation)(out_layer)
out_layer = KL.Dropout(dropout_rate)(out_layer)
return out_layer
def get_model():
qt_input = KL.Input(shape=(QT_test.shape[1],))
a_input = KL.Input(shape=(A_test.shape[1], A_test.shape[2]))
qb_input = KL.Input(shape=(QB_test.shape[1], QB_test.shape[2]))
dummy_input = KL.Input(shape=(1,))
a_emb = KL.Flatten()(KL.Embedding(2, 8)(dummy_input))
qb_emb = KL.Flatten()(KL.Embedding(2, 8)(dummy_input))
embs = KL.concatenate(
[KL.RepeatVector(MAX_SEQ)(a_emb), KL.RepeatVector(MAX_SEQ)(qb_emb)], axis=-2
)
x = KL.concatenate(
[
KL.SpatialDropout1D(0.7)(KL.RepeatVector(2 * MAX_SEQ)(qt_input)),
KL.SpatialDropout1D(0.3)(KL.concatenate([a_input, qb_input], axis=-2)),
]
)
x = KL.concatenate([x, embs])
x = cnn_block(x, 256, 0.1, "relu")
x = KL.concatenate([KL.GlobalAvgPool1D()(x), KL.GlobalMaxPool1D()(x)])
feature_input = KL.Input(shape=(len(features),))
hidden_layer = KL.concatenate([KL.BatchNormalization()(feature_input), x])
hidden_layer = nn_block(hidden_layer, 128, 0.1, "relu")
out = KL.Dense(len(outputs), activation="sigmoid")(hidden_layer)
model = tf.keras.models.Model(
inputs=[qt_input, a_input, qb_input, feature_input, dummy_input],
outputs=out,
)
return model
from sklearn.model_selection import KFold, GroupKFold
from sklearn.metrics import mean_squared_error
from tensorflow.keras.optimizers import Nadam
from sklearn.preprocessing import MinMaxScaler
from tensorflow.keras.backend import epsilon
import tensorflow.keras.backend as K
NUM_FOLDS = 10
BATCH_SIZE = 32
MODEL_FOLDER = "../input/qa-use-model-weights/"
y_test = np.zeros((test_df.shape[0], len(outputs)))
for fold in range(NUM_FOLDS):
model_path = "{folder}model{fold}.h5".format(folder=MODEL_FOLDER, fold=fold)
K.clear_session()
model = get_model()
model.load_weights(model_path)
y_test += (
model.predict(
[
QT_test,
A_test,
QB_test,
test_df[features].values,
np.ones(test_df.shape[0]),
],
batch_size=BATCH_SIZE,
verbose=0,
)
/ NUM_FOLDS
)
K.clear_session()
for i, col in enumerate(outputs):
test_df[col] = y_test[:, i]
return test_df, outputs
test_df, outputs = get_use_preds()
import itertools
import numpy as np
import pandas as pd
def scale(x, d):
if d:
return (x // (1 / d)) / d
return x
def transform(y_roberta, y_bert, y_xlnet, y_albert, params, c):
d_global, d_local, w_roberta, w_bert, w_xlnet, w_albert = params
y_temp = (
scale(y_roberta[:, c], d_local) * w_roberta
+ scale(y_bert[:, c], d_local) * w_bert
+ scale(y_xlnet[:, c], d_local) * w_xlnet
+ scale(y_albert[:, c], d_local) * w_albert
)
y_temp /= w_roberta + w_bert + w_xlnet + w_albert
y_temp = scale(y_temp, d_global)
return y_temp
param_list = [
(16, 16, 2, 2, 2, 1),
(32, 16, 0, 4, 1, 4),
(4, None, 1, 1, 1, 0),
(16, None, 2, 1, 0, 1),
(16, 64, 4, 0, 4, 1),
(4, None, 4, 1, 2, 4),
(32, 64, 1, 1, 2, 1),
(16, 32, 1, 2, 1, 2),
(8, 32, 1, 2, 2, 4),
(32, 16, 0, 2, 1, 1),
(16, 64, 2, 1, 2, 2),
(32, 4, 4, 1, 2, 4),
(4, None, 4, 1, 4, 0),
(4, 8, 1, 0, 4, 0),
(8, 8, 4, 1, 0, 0),
(4, 64, 1, 2, 2, 2),
(16, 4, 1, 0, 1, 1),
(32, 16, 4, 0, 4, 1),
(16, None, 4, 2, 1, 4),
(16, None, 0, 4, 1, 1),
(64, 64, 2, 4, 2, 1),
(32, 8, 4, 1, 1, 4),
(16, 16, 4, 1, 2, 2),
(64, 64, 1, 0, 0, 1),
(64, 64, 2, 1, 1, 1),
(32, 16, 1, 0, 1, 1),
(16, 64, 4, 4, 1, 4),
(32, None, 4, 0, 1, 1),
(32, 64, 4, 1, 2, 4),
(16, None, 2, 4, 1, 2),
]
y_combined = test_df[outputs].values
for c in range(y_combined.shape[1]):
y_combined[:, c] = transform(y_roberta, y_bert, y_xlnet, y_albert, param_list[c], c)
val, counts = np.unique(y_combined[:, c], return_counts=True)
print(c, len(val), counts.sum() - counts.max())
test_df[outputs] = y_combined
# test_df["eng"] = test_df["url"].apply(lambda x: x.startswith("http://english.") or x.startswith("http://ell."))
# test_df.loc[~test_df["eng"], outputs[19]] = 0
test_df.loc[test_df["qa_id"] == 7525, outputs[19]] = 1
test_df[outputs] = np.clip(test_df[outputs], 0.00001, 0.999999)
test_df.to_csv("submission.csv", index=False, columns=["qa_id"] + outputs.tolist())
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import seaborn as sns
from scipy.stats import norm
from collections import Counter
import matplotlib.pyplot as plt
# Preprocessing Libraries
from sklearn.pipeline import make_pipeline
from sklearn.model_selection import train_test_split
from sklearn.model_selection import StratifiedShuffleSplit
# Machine Learning Libraries
import sklearn
import xgboost as xgb
from sklearn import tree
from sklearn.svm import SVC
from sklearn.metrics import roc_curve
from imblearn.pipeline import Pipeline
from catboost import CatBoostClassifier
from imblearn.over_sampling import SMOTE
from sklearn.ensemble import StackingClassifier
from sklearn.metrics import classification_report
from mlxtend.classifier import StackingCVClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import cross_val_predict
from sklearn.model_selection import RandomizedSearchCV
from sklearn.model_selection import KFold, cross_validate
from sklearn.metrics import recall_score, f1_score, roc_auc_score
from imblearn.pipeline import make_pipeline as imbalanced_make_pipeline
from sklearn.metrics import precision_recall_curve, average_precision_score
from sklearn.model_selection import RepeatedStratifiedKFold, StratifiedKFold
from sklearn.metrics import confusion_matrix, accuracy_score, precision_score
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# import data
bankrupt = pd.read_csv("/kaggle/input/company-bankruptcy-prediction/data.csv")
bankrupt.head()
bankrupt.info()
bankrupt.describe()
# histogram of each features
bankrupt.hist(figsize=(35, 30), bins=50)
plt.show()
# Heatmap of correlation using spearman
f, ax = plt.subplots(figsize=(30, 25))
mat = bankrupt.corr("spearman")
mask = np.triu(np.ones_like(mat, dtype=bool))
cmap = sns.diverging_palette(230, 20, as_cmap=True)
sns.heatmap(
mat,
mask=mask,
cmap=cmap,
vmax=1,
center=0, # annot = True,
square=True,
linewidths=0.5,
cbar_kws={"shrink": 0.5},
)
plt.show()
# In the case of predicting bankrutcy for companies, it is crucial to identify the most importat factors that contribute to a company's financial health. In order to do that we have thought about using Three Method: Permutation Importance,CART Feature Importance,PCA to identify which features impact most in model prediction process and thin out repeated features by comparing important features derived by each method.This process will help us identify these factors and use them to build more accurate predictive model that can help identify companies that are risk of facing bankruptcy in the future.
# ## Permutation Importance Method
# Permutation importance is a useful technique to discern the importnace of features in a machine learning model. It works by permuting the values of a feature and measuring the decrease in the model's performance. If the decrease is significant, then the feature is considered important.
# Using permutation importance can be particularly useful when working with datasets that have many features, such as the Taiwan Economic Journal dataset that contains 95 features. By identifying which features are the most importnat, we can reduce the dimensionality of the dataset and focus on the most informative features, which can lead to better model performance and faster training times.
# Furthermore, by using permutation importance to identify the most importnat features, we can better understand the underlying patterns and relationships in the data. This can help us make more informed decisions about which models and algorithms to use and how to preprocess the data before training.
#
import pandas as pd
import numpy as np
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestClassifier
from sklearn.inspection import permutation_importance
# Split data into features and labels
X = bankrupt.drop("Bankrupt?", axis=1)
y = bankrupt["Bankrupt?"]
# Split data into train and test sets
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=42
)
# Create a decision tree classifier
clf = DecisionTreeClassifier(random_state=42)
# Fit the model
clf.fit(X_train, y_train)
# Compute the feature importance scores
importance = clf.feature_importances_
# Print the feature importance scores
for i, v in enumerate(importance):
print(f"{X.columns[i]:<8} {v:.3f}")
# Create a dictionary of feature names and importance scores
feature_importance = dict(zip(X.columns, importance))
# Sort the features by importance score in descending order
sorted_features = sorted(feature_importance.items(), key=lambda x: x[1], reverse=True)
# Select the top 10 features
top_features = sorted_features[:30]
# Plot the decision tree
plt.figure(figsize=(20, 10))
plot_tree(clf, filled=True, max_depth=4, feature_names=X.columns)
# Visualize the top 10 features and their importance scores
fig, ax = plt.subplots()
ax.barh([f[0] for f in top_features], [f[1] for f in top_features])
ax.invert_yaxis()
ax.set_xlabel("Importance Score")
ax.set_title("Top 10 Features by Importance Score")
plt.show()
# ## CART(Classification and Regression Tree) Feature importance
from sklearn.tree import DecisionTreeClassifier, plot_tree
import matplotlib.pyplot as plt
# Create a Decision Tree Classifier
dtc = DecisionTreeClassifier(random_state=42)
# Fit the model
dtc.fit(X, y)
# Compute feature importance
importance2 = dtc.feature_importances_
# Print the feature importance scores
for i, v in enumerate(importance2):
print("Feature: %0d, Score: %.5f" % (i, v))
# Create a dictionary of feature names and importance scores
feature_importance2 = dict(zip(X.columns, importance2))
# Sort the features by importance score in descending order
sorted_features2 = sorted(feature_importance2.items(), key=lambda x: x[1], reverse=True)
# Select the top 10 features
top_features2 = sorted_features2[:30]
# Plot the decision tree
plot_tree(dtc)
# Visualize the top 10 features and their importance scores
fig, ax = plt.subplots()
ax.barh([f[0] for f in top_features2], [f[1] for f in top_features2])
ax.invert_yaxis()
ax.set_xlabel("Importance Score")
ax.set_title("Top 10 Features by Importance Score")
plt.show()
# ## Gini Importance (feature importance)
# Train a random forest classifier
rf = RandomForestClassifier()
rf.fit(X, y)
# Get the feature importances from the trained model
importances = rf.feature_importances_
# Sort the feature importances in descending order
indices = importances.argsort()[::-1]
# Visualize the feature importances
plt.figure(figsize=(10, 8))
plt.title("Feature importances")
plt.bar(range(X.shape[1]), importances[indices])
plt.xticks(range(X.shape[1]), X.columns[indices], rotation=90)
plt.show()
# ##Gini Importance(feature Importance)
# Train a random forest classifier
rf = RandomForestClassifier()
rf.fit(X, y)
# Get the feature importances from the trained model
importances = rf.feature_importances_
# Sort the feature importances in descending order
indices = importances.argsort()[::-1]
# Get the top 10 features with the highest importance score
top_n = 10
top_indices = indices[:top_n]
# Visualize the top feature importances
plt.figure(figsize=(10, 8))
plt.title(f"Top {top_n} feature importances")
plt.bar(range(top_n), importances[top_indices])
plt.xticks(range(top_n), X.columns[top_indices], rotation=90)
plt.show()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import torch
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
def grad(x, w):
w.requires_grad_(True)
a = 0.5
if torch.mul(w, x) > 0:
res = torch.mul(w, x)
else:
ex = torch.exp(w * x)
res = a * (ex - 1)
res.backward()
print(w.grad)
grad(torch.tensor([-1.0]), torch.tensor([7.0]))
def foo(w, x):
w.requires_grad_(True)
lr = 0.05
a = 1
f = 0
for iteration in range(100):
with torch.no_grad():
if w.grad is not None:
w.grad.zero_()
if torch.mul(w, x) > 0:
f = torch.mul(w, x)
else:
ex = torch.exp(w * x)
f = a * (ex - 1)
print(w.data, f)
w.backward()
with torch.no_grad():
w -= lr * w.grad
print(w.data, f.item())
foo(torch.tensor([1.0]), torch.tensor([3.0]))
def elements(m, n, p):
tensor1: torch.Tensor = torch.rand(m, n)
m = torch.nn.Dropout(p, inplace=True)
return m(tensor1)
print(elements(6, 6, 0.5))
def columns(m, n, p):
tensor1 = torch.rand(m, n)
tensor2 = elements(m, n, p)
columns = []
for i in range(m):
for j in range(n):
if tensor2[i][j] == 0:
columns.append(j)
for i in range(m):
for j in columns:
tensor2[i][j] *= 0
return tensor2
columns(6, 6, 0.2)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
import numpy as np # linear algebra
import pandas as pd
train_df = pd.read_csv("/kaggle/input/Kannada-MNIST/train.csv")
test_df = pd.read_csv("/kaggle/input/Kannada-MNIST/test.csv")
train_labels = train_df["label"]
train_df.drop(columns="label", axis=1, inplace=True)
test_id = test_df["id"]
test_df.drop(columns="id", axis=1, inplace=True)
train_df
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
train_df, train_labels, random_state=42
)
from sklearn.decomposition import PCA
pca = PCA(n_components=0.65, whiten=True)
X_train = pca.fit_transform(X_train)
X_test = pca.transform(X_test)
from sklearn.svm import SVC
svc = SVC(kernel="rbf", C=15, gamma="auto")
svc.fit(X_train, y_train)
pred = svc.predict(X_test)
from sklearn.metrics import classification_report
print(classification_report(pred, y_test))
out_csv = test_id.to_frame()
out_csv["label"] = svc.predict(pca.transform(test_df))
out_csv
out_csv.to_csv("submission.csv", index=False)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
digit_recon_tran_csv = pd.read_csv(
"/kaggle/input/digit-recognizer/train.csv", dtype=np.float32
)
digit_recon_test_csv = pd.read_csv(
"/kaggle/input/digit-recognizer/test.csv", dtype=np.float32
)
print("tran dataset size: ", digit_recon_tran_csv.size, "\n")
print("test dataset size: ", digit_recon_test_csv.size, "\n")
# print(digit_recon_tran_csv.head(1))
# print(digit_recon_tran_csv.head(1).label)
tran_label = digit_recon_tran_csv.label.values
tran_image = (
digit_recon_tran_csv.loc[:, digit_recon_tran_csv.columns != "label"].values / 255
) # normalization
test_image = digit_recon_test_csv.values / 255
print("train label size: ", tran_label.shape)
print("train image size: ", tran_image.shape)
print("test image size: ", test_image.shape)
from sklearn.model_selection import train_test_split
train_image, valid_image, train_label, valid_label = train_test_split(
tran_image, tran_label, test_size=0.2, random_state=42
) #
print("train size: ", train_image.shape)
print("valid size: ", valid_image.shape)
# visual
import matplotlib.pyplot as plt
plt.imshow(train_image[10].reshape(28, 28))
plt.axis("off")
plt.title(str(train_label[10]))
plt.show()
# visual
import matplotlib.pyplot as plt
plt.imshow(test_image[10].reshape(28, 28))
plt.axis("off")
plt.show()
import torch
import torch.nn as nn
import numpy as np
print(torch.__version__)
# convert into tensor
train_image = torch.from_numpy(train_image)
train_label = torch.from_numpy(train_label).type(torch.LongTensor) # data type is long
valid_image = torch.from_numpy(valid_image)
valid_label = torch.from_numpy(valid_label).type(torch.LongTensor) # data type is long
# form dataset
train_dataset = torch.utils.data.TensorDataset(train_image, train_label)
valid_dataset = torch.utils.data.TensorDataset(valid_image, valid_label)
# form loader
batch_size = 64 # 2^5=64
train_loader = torch.utils.data.DataLoader(
train_dataset, batch_size=batch_size, shuffle=True
)
valid_loader = torch.utils.data.DataLoader(
valid_dataset, batch_size=batch_size, shuffle=True
)
import torchvision
from torchvision import transforms
from torchvision import models
class YANNet(nn.Module):
def __init__(self):
super(YANNet, self).__init__()
self.conv = nn.Sequential(
# size: 28*28
nn.Conv2d(
1, 8, 3, 1, 1
), # in_channels out_channels kernel_size stride padding
nn.ReLU(),
nn.Conv2d(8, 16, 3, 1, 1),
nn.ReLU(),
nn.MaxPool2d(2),
# size: 14*14
nn.Conv2d(16, 16, 3, 1, 1),
nn.ReLU(),
nn.Conv2d(16, 8, 3, 1, 1),
nn.ReLU(),
nn.MaxPool2d(2),
)
self.fc = nn.Sequential(
# size: 7*7
nn.Linear(8 * 7 * 7, 256),
nn.ReLU(),
nn.Dropout(0.5),
nn.Linear(256, 256),
nn.ReLU(),
nn.Dropout(0.5),
nn.Linear(256, 10),
)
def forward(self, img):
x = self.conv(img)
o = self.fc(x.view(x.shape[0], -1))
return o
model = YANNet()
error = nn.CrossEntropyLoss()
optim = torch.optim.SGD(model.parameters(), lr=0.1)
num_epoc = 2
from torch.autograd import Variable
for epoch in range(num_epoc):
epoc_train_loss = 0.0
epoc_train_corr = 0.0
epoc_valid_corr = 0.0
print("Epoch:{}/{}".format(epoch, num_epoc))
for data in train_loader:
images, labels = data
images = Variable(images.view(64, 1, 28, 28))
labels = Variable(labels)
outputs = model(images)
optim.zero_grad()
loss = error(outputs, labels)
loss.backward()
optim.step()
epoc_train_loss += loss.data
outputs = torch.max(outputs.data, 1)[1]
epoc_train_corr += torch.sum(outputs == labels.data)
with torch.no_grad():
for data in valid_loader:
images, labels = data
images = Variable(images.view(len(images), 1, 28, 28))
labels = Variable(labels)
outputs = model(images)
outputs = torch.max(outputs.data, 1)[1]
epoc_valid_corr += torch.sum(outputs == labels.data)
print(
"loss is :{:.4f},Train Accuracy is:{:.4f}%,Test Accuracy is:{:.4f}".format(
epoc_train_loss / len(train_dataset),
100 * epoc_train_corr / len(train_dataset),
100 * epoc_valid_corr / len(valid_dataset),
)
)
plt.imshow(test_image[100].reshape(28, 28))
plt.axis("off")
plt.show()
one_test = test_image[100]
one_test = torch.from_numpy(one_test).view(1, 1, 28, 28)
one_output = model(one_test)
print(torch.max(one_output.data, 1)[1].numpy())
digit_recon_submission_csv = pd.read_csv(
"/kaggle/input/digit-recognizer/sample_submission.csv", dtype=np.float32
)
print(digit_recon_submission_csv.head(10))
print(test_image.shape)
test_results = np.zeros((test_image.shape[0], 2), dtype="int32")
print(test_results.shape)
for i in range(test_image.shape[0]):
one_image = torch.from_numpy(test_image[i]).view(1, 1, 28, 28)
one_output = model(one_image)
test_results[i, 0] = i
test_results[i, 1] = torch.max(one_output.data, 1)[1].numpy()
print(test_results.shape)
Data = {"ImageId": test_results[:, 0], "Label": test_results[:, 1]}
DataFrame = pd.DataFrame(Data)
DataFrame.to_csv("submission.csv", index=False, sep=",")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
from sklearn.model_selection import train_test_split
from sklearn.feature_extraction.text import TfidfVectorizer
from keras.models import Sequential
from keras.layers import Dense
from keras.preprocessing.text import Tokenizer
from keras.preprocessing.text import text_to_word_sequence
from keras.preprocessing.sequence import pad_sequences
from keras import layers
from keras.layers import Dense, LSTM, Embedding
# Any results you write to the current directory are saved as output.
print(os.listdir("../input"))
train = pd.read_csv(
"../input/jigsaw-unintended-bias-in-toxicity-classification/train.csv"
)
# test = pd.read_csv("../input/jigsaw-unintended-bias-in-toxicity-classification/test.csv")
# submission = pd.read_csv("../input/jigsaw-unintended-bias-in-toxicity-classification/sample_submission.csv")
print("features")
comments = "comment_text"
train[comments].fillna("NULL", inplace=True)
# test[comments].fillna("NULL", inplace=True)
tokenizer = Tokenizer(
num_words=600,
filters='!"#$%&()*+,-./:;<=>?@[\]^_`{|}~ ',
lower=True,
split=" ",
char_level=False,
oov_token=None,
document_count=0,
)
tokenizer.fit_on_texts(train[comments])
# tokenizer.fit_on_texts(test[comments])
print("start padded")
MAX_LENGTH = 1000
padded_train_sequences = pad_sequences(
tokenizer.texts_to_sequences(train[comments]), maxlen=MAX_LENGTH
)
# padded_test_sequences = pad_sequences(tokenizer.texts_to_sequences(test[comments]), maxlen=MAX_LENGTH)
y = np.where(train["target"] >= 0.5, 1, 0)
print("x&y featuring finished")
model = Sequential()
model.add(Embedding(input_dim=1000, output_dim=128))
model.add(layers.LSTM(128))
model.add(layers.Dense(units=32))
model.add(layers.Dense(units=4))
model.add(layers.Dense(units=1, activation="sigmoid"))
model.compile(loss="binary_crossentropy", optimizer="adam", metrics=["accuracy"])
model.summary()
print("start fit")
model.fit(
padded_train_sequences[:-500000],
y[:-500000],
epochs=2,
batch_size=512,
verbose=1,
validation_data=(padded_train_sequences[-500000:], y[-500000:]),
)
test = pd.read_csv(
"../input/jigsaw-unintended-bias-in-toxicity-classification/test.csv"
)
tokenizer.fit_on_texts(test[comments])
padded_test_sequences = pad_sequences(
tokenizer.texts_to_sequences(test[comments]), maxlen=MAX_LENGTH
)
y_pred_rnn_simple = model.predict(padded_test_sequences, verbose=1, batch_size=128)
submission = pd.read_csv(
"../input/jigsaw-unintended-bias-in-toxicity-classification/sample_submission.csv"
)
y_pred_rnn_simple = pd.DataFrame(y_pred_rnn_simple, columns=["prediction"])
print("submission")
submid = pd.DataFrame({"id": submission["id"]})
submid["prediction"] = y_pred_rnn_simple["prediction"]
submid.to_csv("submission.csv", index=False)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import h5py
import os
path = "/kaggle/input/jet-images-train-val-test/jet-images_train.hdf5"
h5_file = h5py.File(path, "r")
signal_data = h5_file["signal"]
image_data = h5_file["image"]
boson = []
general = []
for i in range(len(image_data)):
if signal_data[i] == 1.0:
boson.append(image_data[i])
else:
general.append(image_data[i])
print(len(boson))
print(len(general))
# abs verdi bilde - abs mean
# måle avstand fra mean bildene, så klassifisere utifra den som er nærmest og måle acc
# lage mean bildene
# lage hit or miss, for loop, hit -> append i hit, miss -> append i miss
mean_boson = np.mean(boson, axis=0)
mean_general = np.mean(general, axis=0)
from matplotlib import pyplot as plt
plt.imshow(mean_boson)
plt.show()
plt.imshow(mean_general)
plt.show()
preds = []
def predict(array):
general = np.abs(np.mean(mean_general) - np.mean(array))
boson = np.abs(np.mean(mean_boson) - np.mean(array))
if boson < general:
preds.append(1.0)
else:
preds.append(0.0)
for image in image_data:
predict(image)
print(len(image_data))
print(len(preds))
def accuracy(predictions, labels):
"""Calculate the accuracy of the predictions."""
num_correct = sum(
[1 for i in range(len(predictions)) if predictions[i] == labels[i]]
)
return num_correct / len(predictions)
accuracy(preds, signal_data)
|
# # kaggle Titanic : LGBM + BayesianOpt Baseline
# This notebook is a very basic and simple introduction of how using bayesian optimization for selecting best hyperparameter values and LightGBM for training/predicting.
# **1. [EDA](#data_analysis)**
# **2. [modify data](#data_manipulation)**
# **3. [modeling/train/submit](#modelling_training_submit)**
#
#
from IPython.core.display import display, HTML
display(HTML("<style>.container { width:90% !important; }</style>"))
import pandas as pd
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import seaborn as sns
sns.set() # set seaborn default
#
# # **1. EDA:**
# load .csv files
train = pd.read_csv("../input/titanic/train.csv")
test = pd.read_csv("../input/titanic/test.csv")
# from enum import Enum
class Columns:
# existing features
PassengerId = "PassengerId"
Survived = "Survived"
Pclass = "Pclass"
Name = "Name"
Sex = "Sex"
Age = "Age"
SibSp = "SibSp"
Parch = "Parch"
Ticket = "Ticket"
Fare = "Fare"
Cabin = "Cabin"
Embarked = "Embarked"
# new features
Title = "Title"
FareBand = "FareBand"
Family = "Family"
Deck = "Deck" # get character from existing 'Cabin' values
CabinExists = "CabinExists"
train.head()
test.head()
# ## features :
# - **PassengerId**
# - **Survived** : Survival (1: survived, 0 : not)
# - **Pclass** : boarding pass class (1 : 1st, 2 : 2nd ,3 : 3rd)
# - **Name**
# - **Sex**
# - **Age**
# - **SibSp** : number of siblings + spouses accompanying
# - **Parch** : number of parents + children accompanying
# - **Ticket** : ticket number
# - **Fare**
# - **Cabin** : cabin number
# - **Embarked** : embarked port (C : Cherbourg, Q : Queenstown, S : Southampton)
# #### Check for features that do not have data.
# ### Check features
# Survival rate by Pclass
print(train[[Columns.Pclass, Columns.Survived]].head())
train[[Columns.Pclass, Columns.Survived]].groupby([Columns.Pclass]).mean().plot.bar()
# survival rate by sex.
train[[Columns.Sex, Columns.Survived]].groupby([Columns.Sex]).mean().plot.bar()
fig, ax = plt.subplots(nrows=3, ncols=1, figsize=(10, 8))
sns.countplot(x=Columns.Sex, hue=Columns.Survived, data=train, ax=ax[0])
sns.countplot(x=Columns.Sex, hue=Columns.Pclass, data=train, ax=ax[1])
sns.countplot(x=Columns.Pclass, hue=Columns.Survived, data=train, ax=ax[2])
# survival rate by age
train[Columns.Age].plot.kde()
# survival rate by age group(bar graph)
df = train[train[Columns.Age].isnull() == False] # drop rows have no age
bincount = 12
age_min = int(df[Columns.Age].min())
age_max = int(df[Columns.Age].max())
print("Age :", age_min, " ~ ", age_max)
gap = int((age_max - age_min) / bincount)
print("gap:", gap)
bins = [-1]
for i in range(bincount):
bins.append(i * gap)
bins.append(np.inf)
print(bins)
_df = df
_df["AgeGroup"] = pd.cut(_df[Columns.Age], bins)
fig, ax = plt.subplots(figsize=(20, 10))
sns.countplot(x="AgeGroup", hue=Columns.Survived, data=_df, ax=ax)
# get survival rate change by Pclass/Age
fig, ax = plt.subplots(nrows=3, ncols=1, figsize=(16, 20))
sns.violinplot(
x=Columns.Pclass,
y=Columns.Age,
hue=Columns.Survived,
data=train,
scale="count",
split=True,
ax=ax[0],
)
sns.violinplot(
x=Columns.Sex,
y=Columns.Age,
hue=Columns.Survived,
data=train,
scale="count",
split=True,
ax=ax[1],
)
sns.violinplot(
x=Columns.Pclass,
y=Columns.Sex,
hue=Columns.Survived,
data=train,
scale="count",
split=True,
ax=ax[2],
)
# survival rate by number of family ( Parch + SibSp + 1(self))
_train = train
_train["Family"] = _train[Columns.SibSp] + _train[Columns.Parch] + 1
_train[["Family", Columns.Survived]].groupby("Family").mean().plot.bar()
train[Columns.Age].plot.hist()
sns.countplot(x="Family", data=_train)
# survived count by family count
sns.countplot(x="Family", hue=Columns.Survived, data=_train)
#
# # **2. Data manipulation:**
# process empty column values
# ###### complement & process existing features
# **Age** : need to fix null values
# **Cabin** : too much null values
# **Embarked** : since there are few nulls, seems to be no problem filling rouch values
# **Parse, SibSp** : sum up 'Family' and delete.
# ###### add new features
# **Family** : Parch + SibSp + 1(self)
# **Title** : extract from 'Name'.
# ## merge train/test and manipulate and detach last.
train_len = train.shape[0]
merged = train.append(test, ignore_index=True)
print("train len : ", train.shape[0])
print("test len : ", test.shape[0])
print("merged len : ", merged.shape[0])
# make 'Family' and drop 'Parch'/'SibSp'
merged[Columns.Family] = merged[Columns.Parch] + merged[Columns.SibSp] + 1
if Columns.Parch in merged:
merged = merged.drop([Columns.Parch], axis=1)
if Columns.SibSp in merged:
merged = merged.drop([Columns.SibSp], axis=1)
merged.head()
# fix 'Embarked'
most_embarked_label = merged[Columns.Embarked].value_counts().index[0]
merged = merged.fillna({Columns.Embarked: most_embarked_label})
merged.describe(include="all")
# extract Title from Name and change to number value
# remove Name
merged[Columns.Title] = merged.Name.str.extract("([A-Za-z]+)\. ", expand=False)
print("initial titles : ", merged[Columns.Title].value_counts().index)
# initial titles : Index(['Mr', 'Miss', 'Mrs', 'Master', 'Dr', 'Rev', 'Col', 'Ms', 'Mlle', 'Major',
# 'Sir', 'Jonkheer', 'Don', 'Mme', 'Countess', 'Lady', 'Dona', 'Capt'],
merged[Columns.Title] = merged[Columns.Title].replace(
["Lady", "Capt", "Col", "Don", "Dr", "Major", "Rev", "Jonkheer", "Dona"], "Rare"
)
merged[Columns.Title] = merged[Columns.Title].replace(
["Countess", "Lady", "Sir"], "Royal"
)
merged[Columns.Title] = merged[Columns.Title].replace(
["Miss", "Mlle", "Ms", "Mme"], "Mrs"
)
print("Survival rate by title:")
print("========================")
print(merged[[Columns.Title, Columns.Survived]].groupby(Columns.Title).mean())
idxs = merged[Columns.Title].value_counts().index
print(idxs)
mapping = {}
for i in range(len(idxs)):
mapping[idxs[i]] = i + 1
print("Title mapping : ", mapping)
merged[Columns.Title] = merged[Columns.Title].map(mapping)
if Columns.Name in merged:
merged = merged.drop([Columns.Name], axis=1)
merged.head()
sns.countplot(x=Columns.Title, hue=Columns.Survived, data=merged)
print(merged[Columns.Title].value_counts())
mapping = {"male": 0, "female": 1}
merged[Columns.Sex] = merged[Columns.Sex].map(mapping)
merged.head(n=10)
# Fill empty Age columns
# {'Mr': 1, 'Miss': 2, 'Mrs': 3, 'Master': 4, 'Rare': 5, 'Royal': 6}
mapping = {1: 21, 2: 28, 3: 28, 4: 40, 5: 50, 6: 60}
def guess_age(row):
return mapping[row[Columns.Title]]
def fixup_age(df):
for idx, row in df[df[Columns.Age].isnull() == True].iterrows():
df.loc[idx, Columns.Age] = guess_age(row)
return df
merged = fixup_age(merged)
merged.describe(include="all")
# Extract alphabet from 'Cabin' and make 'Deck'
def make_deck(df):
df[Columns.Deck] = df[Columns.Cabin].str.extract("([A-Za-z]+)", expand=True)
return df
merged = make_deck(merged)
merged.describe(include="all")
merged[[Columns.Deck, Columns.Fare]].groupby(Columns.Deck).mean().sort_values(
by=Columns.Fare
)
# Show relation of Deck and Survived
sns.countplot(x=Columns.Deck, hue=Columns.Survived, data=merged)
# Survived count by Deck and Fare
print("total survived rate: ", merged[Columns.Survived].mean())
print(
"deck survived rate: ",
merged[merged[Columns.Deck].isnull() == False][Columns.Survived].mean(),
)
print(
"no deck survived rate: ",
merged[merged[Columns.Deck].isnull()][Columns.Survived].mean(),
)
fig, ax = plt.subplots(2, 1, figsize=(16, 16))
merged[[Columns.Deck, Columns.Survived]].groupby(Columns.Deck).mean().plot.bar(ax=ax[0])
def generate_fare_group(df, slicenum):
if "FareGroup" in df:
df.drop("FareGroup", axis=1)
_min = int(df[Columns.Fare].min())
_max = int(df[Columns.Fare].max())
print("Fare :", _min, " ~ ", _max)
gap = int((_max - _min) / slicenum)
print("gap:", gap)
bins = [-1]
for i in range(slicenum):
bins.append(i * gap)
bins.append(np.inf)
print(bins)
df["FareGroup"] = pd.cut(df[Columns.Fare], bins)
return df
df = generate_fare_group(merged.copy(), 16)
sns.countplot(
x="FareGroup", hue=Columns.Survived, data=df[df[Columns.Deck].isnull()], ax=ax[1]
)
# add the existence of Cabin as feature
merged[Columns.CabinExists] = merged[Columns.Cabin].isnull() == False
merged[Columns.CabinExists] = merged[Columns.CabinExists].map({True: 1, False: 0})
merged.head()
# Fill null 'Fare' column
merged[merged[Columns.Fare].isnull()]
# Just use average value.
merged.loc[merged[Columns.Fare].isnull(), [Columns.Fare]] = merged[Columns.Fare].mean()
merged.head()
sns.distplot(merged[Columns.Fare])
# The distribution of 'Fare' is in high skewness. It is said to adversely affect the learning of the model.
# use log value or band rate
"""
log를 취하는 방법
"""
# merged[Columns.Fare] = merged[Columns.Fare].map(lambda i : np.log(i) if i > 0 else 0)
"""
등급을 4단계로 나누는 방법
"""
merged[Columns.FareBand] = pd.qcut(merged[Columns.Fare], 4, labels=[1, 2, 3, 4]).astype(
"float"
)
# merged[Columns.Fare] = merged[Columns.FareBand]
merged.head(n=20)
merged[Columns.Fare] = merged[Columns.FareBand]
merged = merged.drop([Columns.FareBand], axis=1)
merged.head()
merged.head()
sns.distplot(merged[Columns.Fare])
merged.head()
# After finish data manipulation, do below.
# - remove unnecessary columns
# - scaling
# - detach train/test
# - separate train as input/label(survived)
# remove unnecessary columns
if Columns.Ticket in merged:
merged = merged.drop(labels=[Columns.Ticket], axis=1)
if Columns.Cabin in merged:
merged = merged.drop(labels=[Columns.Cabin], axis=1)
if Columns.Deck in merged:
merged = merged.drop(labels=[Columns.Deck], axis=1)
merged.describe(include="all")
merged.head()
# change category features to one-hot encoding
# - Pclass
# - Embarked
# - Title
merged = pd.get_dummies(merged, columns=[Columns.Pclass], prefix="Pclass")
merged = pd.get_dummies(merged, columns=[Columns.Title], prefix="Title")
merged = pd.get_dummies(merged, columns=[Columns.Embarked], prefix="Embarked")
merged = pd.get_dummies(merged, columns=[Columns.Sex], prefix="Sex")
merged = pd.get_dummies(merged, columns=[Columns.CabinExists], prefix="CabinExists")
merged.head()
# Numerical data need to be scalied.
from sklearn.preprocessing import MinMaxScaler
class NoColumnError(Exception):
"""Raised when no column in dataframe"""
def __init__(self, value):
self.value = value
# __str__ is to print() the value
def __str__(self):
return repr(self.value)
# normalize AgeGroup
def normalize_column(data, columnName):
scaler = MinMaxScaler(feature_range=(0, 1))
if columnName in data:
aaa = scaler.fit_transform(data[columnName].values.reshape(-1, 1))
aaa = aaa.reshape(
-1,
)
# print(aaa.shape)
data[columnName] = aaa
return data
else:
raise NoColumnError(str(columnName) + " is not exists!")
def normalize(dataset, columns):
for col in columns:
dataset = normalize_column(dataset, col)
return dataset
merged.head()
merged = normalize(merged, [Columns.Age, Columns.Fare, Columns.Family])
merged.head(n=10)
# detach merged to train/test
train = merged[:train_len]
test = merged[train_len:]
test = test.drop([Columns.Survived], axis=1)
train = train.drop([Columns.PassengerId], axis=1)
test_passenger_id = test[Columns.PassengerId]
test = test.drop([Columns.PassengerId], axis=1)
print(train.shape)
print(test.shape)
train_X = train.drop([Columns.Survived], axis=1).values
train_Y = train[Columns.Survived].values.reshape(-1, 1)
print(train_X.shape)
print(train_Y.shape)
test.shape
test.describe(include="all")
train.head()
test.head()
#
# # **3. Model train/submit:**
#
import lightgbm as lgb
from sklearn import metrics
from sklearn.preprocessing import StandardScaler
from sklearn.svm import NuSVR, SVR
from sklearn.metrics import mean_absolute_error
from sklearn.preprocessing import LabelEncoder
from sklearn.model_selection import (
StratifiedKFold,
KFold,
RepeatedKFold,
GroupKFold,
GridSearchCV,
train_test_split,
TimeSeriesSplit,
RepeatedStratifiedKFold,
)
from sklearn import metrics
from sklearn.metrics import classification_report, confusion_matrix
from sklearn.metrics import cohen_kappa_score, mean_squared_error
from sklearn import linear_model
from sklearn.model_selection import KFold
from sklearn.model_selection import StratifiedKFold
import warnings
from bayes_opt import BayesianOptimization
n_splits = 6
def get_param(
learning_rate,
max_depth,
lambda_l1,
lambda_l2,
bagging_fraction,
bagging_freq,
colsample_bytree,
subsample_freq,
feature_fraction,
):
params = {
"n_estimators": 5000,
"boosting_type": "gbdt",
"objective": "regression",
"metric": "rmse",
#'eval_metric': 'cappa',
"subsample": 1.0,
"subsample_freq": subsample_freq,
"feature_fraction": feature_fraction,
"n_jobs": -1,
"seed": 42,
"learning_rate": learning_rate,
"max_depth": int(max_depth),
"lambda_l1": lambda_l1,
"lambda_l2": lambda_l2,
"bagging_fraction": bagging_fraction,
"bagging_freq": int(bagging_freq),
"colsample_bytree": colsample_bytree,
"early_stopping_rounds": 100,
"verbose": 0,
}
return params
def opt_test_func(
learning_rate,
max_depth,
lambda_l1,
lambda_l2,
bagging_fraction,
bagging_freq,
colsample_bytree,
subsample_freq,
feature_fraction,
):
params = get_param(
learning_rate,
max_depth,
lambda_l1,
lambda_l2,
bagging_fraction,
bagging_freq,
colsample_bytree,
subsample_freq,
feature_fraction,
)
acc, _ = train(params)
return acc
def train(params):
models = []
kfold = KFold(n_splits=n_splits, shuffle=True, random_state=7)
oof = np.zeros(len(train_X))
for train_idx, test_idx in kfold.split(train_X, train_Y):
X_train, y_train = train_X[train_idx], train_Y[train_idx]
X_valid, y_valid = train_X[test_idx], train_Y[test_idx]
y_train = y_train.reshape(-1)
y_valid = y_valid.reshape(-1)
model = lgb.LGBMClassifier()
model.set_params(**params)
eval_set = [(X_valid, y_valid)]
eval_names = ["valid"]
model.fit(
X=X_train, y=y_train, eval_set=eval_set, eval_names=eval_names, verbose=0
)
pred = model.predict(X_valid).reshape(len(test_idx))
oof[test_idx] = pred
models.append(model)
result = np.equal(oof, train_Y.reshape(-1))
accuracy = np.count_nonzero(result.astype(int)) / oof.shape[0]
# print("accuracy : ", accuracy)
return accuracy, models
def get_optimized_hyperparameters():
bo_params = {
"learning_rate": (0.001, 0.1),
"max_depth": (10, 20),
"lambda_l1": (1, 10),
"lambda_l2": (1, 10),
"bagging_fraction": (0.8, 1.0),
"bagging_freq": (1, 10),
"colsample_bytree": (0.7, 1.0),
"subsample_freq": (1, 10),
"feature_fraction": (0.9, 1.0),
}
optimizer = BayesianOptimization(opt_test_func, bo_params, random_state=1030)
with warnings.catch_warnings():
warnings.filterwarnings("ignore")
init_points = 16
n_iter = 16
optimizer.maximize(
init_points=init_points, n_iter=n_iter, acq="ucb", xi=0.0, alpha=1e-6
)
return optimizer.max["params"]
def predict(models, test):
preds = []
for model in models:
pred = model.predict(test).reshape(test.shape[0])
preds.append(pred)
preds = np.array(preds)
preds = np.mean(preds, axis=0) > 0.5
return preds
params = get_optimized_hyperparameters()
params = get_param(**params)
acc, models = train(params)
print("train accuracy : ", acc)
test_pred = predict(models, test)
print(test_pred.shape)
submission = pd.DataFrame(
{"PassengerId": test_passenger_id, "Survived": test_pred.reshape(-1).astype(np.int)}
)
submission = pd.DataFrame(
{"PassengerId": test_passenger_id, "Survived": test_pred.reshape(-1).astype(np.int)}
)
submission.to_csv("submission.csv", index=False)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
import matplotlib.pyplot as plt
import seaborn as sns
df_corona = pd.read_csv(
"/kaggle/input/novel-corona-virus-2019-dataset/2019_nCoV_data.csv",
index_col="Last Update",
parse_dates=["Last Update"],
)
df_corona.sample(10)
df_corona[df_corona["Province/State"] == "Hubei"]
df_corona.info()
df_corona.isnull().sum()
plt.figure(figsize=(16, 6))
sns.set_style("darkgrid")
g = sns.lineplot(data=df_corona["Confirmed"], label="Confirmed Cases")
sns.lineplot(data=df_corona["Deaths"], label="Deaths")
sns.lineplot(data=df_corona["Recovered"], label="Recovered")
plt.xticks(rotation=45)
plt.show()
df_cases_wo_china = df_corona[
(df_corona["Country"] != "China") & (df_corona["Country"] != "Mainland China")
]
country_conf_max = []
country_reco_max = []
country_deth_max = []
country_val = []
for country, subset in df_cases_wo_china.groupby("Country"):
country_conf_max.append(max(subset["Confirmed"]))
country_reco_max.append(max(subset["Recovered"]))
country_deth_max.append(max(subset["Deaths"]))
country_val.append(country)
df_country_woc = pd.DataFrame(
{
"State": country_val,
"Confirmed": country_conf_max,
"Recovered": country_reco_max,
"Death": country_deth_max,
}
)
df_woc = df_country_woc.sort_values("Confirmed", ascending=False)
df_woc_top10 = df_woc.head(10)
# Top 10 Corona Virus affed Country Outside China
df_woc_top10
plt.figure(figsize=(10, 6))
sns.barplot(
x="Confirmed", y="State", data=df_woc_top10, color="r", label="Confirmed Cases"
)
sns.barplot(x="Recovered", y="State", data=df_woc_top10, color="g", label="Recovered")
plt.legend()
plt.show()
# Outside China Eastern Countries (Thiland, Hong Kong, Japan, Singapore etc) are highly affected by Corona Virus.
# Other than Asian Countries, US, France Germany has significant number of affected people
df_recovered_osc = df.sort_values("Recovered", ascending=False).head(5)
sns.set_style("whitegrid")
plt.figure(figsize=(6, 5))
sns.barplot(x=df_recovered_osc["Country"], y=df_recovered_osc["Recovered"])
plt.xticks(rotation=90)
plt.show()
# Lets check the Statistics of China
df_corona_china = df_corona[
(df_corona["Country"] == "China") | (df_corona["Country"] == "Mainland China")
]
df_corona_china
for Country, subset in df_corona_china.groupby("Country"):
print(Country, subset["Confirmed"].sum())
st_conf_max = []
st_reco_max = []
st_deth_max = []
state_val = []
for state, subset in df_corona_china.groupby("Province/State"):
st_conf_max.append(max(subset["Confirmed"]))
st_reco_max.append(max(subset["Recovered"]))
st_deth_max.append(max(subset["Deaths"]))
state_val.append(state)
df_china_bystate = pd.DataFrame(
{
"State": state_val,
"Confirmed": st_conf_max,
"Recovered": st_reco_max,
"Death": st_deth_max,
}
)
df_china_bystate = df_china_bystate.sort_values("Confirmed", ascending=False)
df_china_bystate_top10 = df_china_bystate.head(10)
df_china_bystate_top10
# sns.barplot(x='State', y= 'Confirmed', data = df_china_bystate_top10, color='r',label='China Confirmed')
plt.figure(figsize=(12, 6))
sns.barplot(
y="State",
x="Confirmed",
data=df_china_bystate_top10,
color="r",
label="China Confirmed",
)
sns.barplot(
y="State", x="Recovered", data=df_china_bystate_top10, color="g", label="Recovered"
)
plt.legend()
plt.show()
df_corona.sort_index(inplace=True)
plt.figure(figsize=(16, 6))
sns.set_style("whitegrid")
sns.lineplot(
data=df_corona[df_corona["Province/State"] == "Hubei"]["Confirmed"],
label="Hubei Confirmed Cases",
)
sns.lineplot(
data=df_corona[df_corona["Province/State"] == "Zhejiang"]["Confirmed"],
label="Zhejiang Confirmed Cases",
)
sns.lineplot(
data=df_corona[df_corona["Province/State"] == "Guangdong"]["Confirmed"],
label="Guangdong Confirmed Cases",
)
plt.xticks(rotation=45)
plt.show()
# for state, subset in df_corona_china.groupby('Province/State'):
# print(state,subset['Confirmed'].sum())
|
#
# CORONA VIRUS ALERT !!!
# A deadly virus spreading from Human to Human
#
# ### Description
# 2019 Novel Coronavirus (2019-nCoV) is a virus (more specifically, a coronavirus) identified as the cause of an outbreak of respiratory illness first detected in Wuhan, China. Early on, many of the patients in the outbreak in Wuhan, China reportedly had some link to a large seafood and animal market, suggesting animal-to-person spread. However, a growing number of patients reportedly have not had exposure to animal markets, indicating person-to-person spread is occurring. At this time, it’s unclear how easily or sustainably this virus is spreading between people - CDC
# This dataset has daily level information on the number of affected cases, deaths and recovery from 2019 novel coronavirus. Please note that this is a time series data and so the number of cases on any given day is the cumulative number.
# The data is available from 22 Jan, 2020.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import matplotlib.gridspec as gridspec
dataset = pd.read_csv(
"/kaggle/input/novel-corona-virus-2019-dataset/2019_nCoV_data.csv"
)
dataset.head()
dataset["Date"] = pd.to_datetime(dataset["Date"])
dataset["Last Update"] = pd.to_datetime(dataset["Last Update"])
dataset["Day"] = dataset["Date"].dt.day
dataset["Month"] = dataset["Date"].dt.month
dataset["Week"] = dataset["Date"].dt.week
dataset["WeekDay"] = dataset["Date"].dt.weekday
# Layout Customization
displayed_cols = ["Confirmed", "Deaths", "Recovered"]
def multi_plot():
fig = plt.figure(constrained_layout=True, figsize=(15, 8))
grid = gridspec.GridSpec(ncols=4, nrows=2, figure=fig)
ax1 = fig.add_subplot(grid[0, :2])
ax1.set_title("Daily Reports")
dataset.groupby(["Date"]).sum()[displayed_cols].plot(ax=ax1)
ax2 = fig.add_subplot(grid[1, :2])
ax2.set_title("Monthly Reports")
dataset.groupby(["Month"]).sum()[displayed_cols].plot(kind="bar", ax=ax2)
ax2.set_xticklabels(range(1, 3))
ax3 = fig.add_subplot(grid[0, 2:])
ax3.set_title("Weekly Reports")
weekdays = dataset.groupby("Week").nth(-1)["Date"]
dataset[dataset["Date"].isin(weekdays)].groupby("Date")[displayed_cols].sum().plot(
kind="bar", ax=ax3
)
ax3.set_xticklabels(range(1, len(weekdays) + 1))
ax4 = fig.add_subplot(grid[1, 2:])
ax4.set_title("WeekDays Reports")
dataset.groupby(["WeekDay"]).sum()[displayed_cols].plot(ax=ax4)
plt.tight_layout()
multi_plot()
# Graph shows that case count is getting increased daily which means that latest case count = today case count + previous days case count. So we will take the last date as the updated case count.
recent_date = dataset["Date"].iloc[-1]
last_updated = dataset[dataset["Date"].dt.date == recent_date]
# Reports given for the total number of days
dataset["Date"].max() - dataset["Date"].min()
dataset["Country"].value_counts()
# Observation :
# Mainland China and China has two separate entries. We'll combine the both
# Description of this Dataset and also News Confirms that CoronaVirus spreaded from China and from the above table after China. US,Australia and Canada has higher entries and geographically these countries are too far from China . It's weired We'll analyse it further
dataset["Country"].replace({"Mainland China": "China"}, inplace=True)
dataset.head()
dataset[dataset["Date"] != dataset["Last Update"]]["Country"].value_counts()
# These many province/State were not updated yet with last date.
dataset["Last Update"].max()
# missing values
dataset.isnull().sum()
confirmedCase = int(last_updated["Confirmed"].sum())
deathCase = int(last_updated["Deaths"].sum())
recoveredCase = int(last_updated["Recovered"].sum())
print("No of Confirmed cases globally {}".format(confirmedCase))
print("No of Recovered case globally {}".format(recoveredCase))
print("No of Death case globally {}".format(deathCase))
top5 = (
last_updated.groupby(["Country"]).sum().nlargest(5, ["Confirmed"])[displayed_cols]
)
top5
print("Top 5 Countries were affected most")
print(top5)
# Observation :
# Though US,Canada and Australia has higher entries after China but they are not in the top 5 list
#
plt.figure(figsize=(12, 6))
plt.xticks(rotation=90)
plt.title("Top most 5 countries were affected by Coronavirus")
sns.barplot(x=top5.index, y="Confirmed", data=top5)
plt.show()
plt.figure(figsize=(15, 6))
plt.title("Countries which has Confirmed cases")
plt.xticks(rotation=90)
sns.barplot(x="Country", y="Confirmed", data=last_updated)
plt.tight_layout()
plt.figure(figsize=(15, 6))
plt.title("Province/State which reported more than 1000 Confirmed case")
plt.xticks(rotation=90)
prvinc = last_updated
prvincConfirmed = prvinc[prvinc["Confirmed"] > 1000]
sns.barplot(data=prvincConfirmed, x="Province/State", y="Confirmed")
plt.figure(figsize=(15, 6))
plt.xticks(rotation=90)
plt.title("Province/State has reported Deaths case")
sns.barplot(data=last_updated, x="Province/State", y="Deaths")
# Wikipedia says - In December 2019, a pneumonia(Coronavirus) outbreak was reported in Wuhan, China. Wuhan is the capital of Hubei province.
prov100 = last_updated[last_updated["Confirmed"] > 100]["Province/State"].values
print(
"Countries their State which was affected and filed more than 100 Confirmed cases"
)
dataset[dataset["Province/State"].isin(prov100)].groupby("Country")[
"Province/State"
].apply(lambda x: len(list(np.unique(x)))).sort_values(ascending=False)
last_updated.groupby(["Country"]).sum().nlargest(5, ["Deaths"])["Deaths"]
# We'll analyse how other countries affected after China and Others(Cruis ship).
last_updated[~last_updated["Country"].isin(["China", "Others"])].groupby(
["Country"]
).sum()[displayed_cols].nlargest(5, ["Confirmed"])
confCaseOnShip = dataset[
(dataset["Province/State"] != np.nan)
& (dataset["Province/State"].str.contains("ship"))
]
confCaseOnShip
# Princess Cruises confirms there are 2,666 guests and 1,045 crew currently onboard covering a range of nationalities
# Totally 3711 members are there in ship
# Out of 3711 , 285 were confirmed
print("{}% of people were affected in Cruise ship".format(round((285 / 3711) * 100, 2)))
confCaseOnShip["Confirmed"].plot()
|
# # Импорт необходимых библиотек
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
from sklearn.linear_model import LogisticRegression
from sklearn.preprocessing import StandardScaler
from sklearn.metrics import confusion_matrix, classification_report, f1_score
import os
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
# Any results you write to the current directory are saved as output.
# ### Узнаем где лежат все файлы (также можно в кернеле посмотреть справа)
os.listdir("/kaggle/input")
# # Создание и объединение признаков с датасетом (из "сырых" датасетов)
# # Загружаем промежуточные данные
interim_test = pd.read_csv("/kaggle/working/interim_test.csv")
interim_train_09 = pd.read_csv("/kaggle/working/interim_train_09.csv")
interim_train_10 = pd.read_csv("/kaggle/working/interim_train_10.csv")
# # Удаляем столбцы
# Внимание! Здесь удалены также и полезные признаки!
# `ID`,`SERVICE_INT_ID`,`ADMIN_QUESTION_INT_ID`,`FEATURE_INT_ID`,`CHANNEL_INT_ID` - Эти признаки относятся к Таргету, поэтому мы их удаляем (это сопутствующая информация об обращениях)
interim_train_09 = interim_train_09.drop(
columns=[
"ID",
"SERVICE_INT_ID",
"ADMIN_QUESTION_INT_ID",
"FEATURE_INT_ID",
"CHANNEL_INT_ID",
"ACTIVATE_DATE",
"PHYZ_TYPE",
"CITY_NAME",
]
)
interim_train_10 = interim_train_10.drop(
columns=[
"ID",
"SERVICE_INT_ID",
"ADMIN_QUESTION_INT_ID",
"FEATURE_INT_ID",
"CHANNEL_INT_ID",
"ACTIVATE_DATE",
"PHYZ_TYPE",
"CITY_NAME",
]
)
interim_test = interim_test.drop(columns=["ACTIVATE_DATE", "PHYZ_TYPE", "CITY_NAME"])
# # Самая простая замена пропусков
interim_train_09.fillna(0, inplace=True)
interim_train_10.fillna(0, inplace=True)
interim_test.fillna(0, inplace=True)
# # Согласование признаков
# Признаков много, они генерируются с помощью One-Hot Encoding, поэтому необходимо проверить, чтобы они были одинаковые
def align_data(train: pd.DataFrame, test: pd.DataFrame) -> (pd.DataFrame, pd.DataFrame):
"""Согласование признаков у train и test датасетов
Arguments:
train {pd.DataFrame} -- train датасет
test {pd.DataFrame} -- test датасет
Returns:
train {pd.DataFrame}, test {pd.DataFrame} - датасеты с одинаковыми признаками
"""
intersect_list = np.intersect1d(train.columns, test.columns)
if "TARGET" not in intersect_list:
train = train[np.append(intersect_list, "TARGET")]
else:
train = train[intersect_list]
test = test[intersect_list]
return train, test
# ### Сначала мы будем обучаться на сентябре и предсказывать октябрь
train_09, test_10 = align_data(interim_train_09, interim_train_10)
train_09.shape, test_10.shape
def fit_and_pred_logreg(train, test):
"""Fit and predict LogisticRegression
Arguments:
train {pd.DataFrame} -- processed train dataset
test {pd.DataFrame} -- processed test dataset
Returns:
model {sklearn.BaseEstimator} -- fit sklearn model
y_pred {np.array} -- predictions
"""
model = LogisticRegression(class_weight="balanced", random_state=17, n_jobs=-1)
x_train = train.drop(columns=["TARGET"])
y_train = train.TARGET
scaler = StandardScaler()
x_train = scaler.fit_transform(x_train)
x_test = scaler.transform(test)
model.fit(x_train, y_train)
y_pred = model.predict(x_test)
return model, y_pred
y_test = test_10.TARGET
model, y_pred = fit_and_pred_logreg(train_09, test_10.drop(columns="TARGET"))
# ### Проверяем наши результаты
print(classification_report(y_true=y_test, y_pred=y_pred))
print(confusion_matrix(y_true=y_test, y_pred=y_pred))
# # Создадим submission
# Предполагаем, что в сентябре-октябре-ноябре абоненты обращаются примерно одинаково, поэтому мы можем валидироваться на предыдущих месяцах.
# Сделаем предсказание для ноября, обучаясь на октябре. (с теми же гиперпараметрами модели, что и в коде выше)
# ### Согласуем признаки
train_10, test = align_data(interim_train_10, interim_test)
# ### Обучим модель
model, y_pred = fit_and_pred_logreg(train_10, test)
# ### Сохраним наше предсказание
interim_test["PREDICT"] = y_pred
interim_test[["USER_ID", "PREDICT"]].to_csv(
"baseline_submission.csv", index=False
) # В папке output. Выгрузить ручками
|
# easydict模块用于以属性的方式访问字典的值
from easydict import EasyDict as edict
# os模块主要用于处理文件和目录
import os
import numpy as np
import matplotlib.pyplot as plt
import mindspore
# 导入MindSpore框架数据集
import mindspore.dataset as ds
# vision.c_transforms模块是处理图像增强的高性能模块,用于数据增强图像数据改进训练模型。
from mindspore.dataset.vision import c_transforms as vision
from mindspore import context
import mindspore.nn as nn
from mindspore.train import Model
from mindspore.nn.optim.momentum import Momentum
from mindspore.train.callback import ModelCheckpoint, CheckpointConfig, LossMonitor
from mindspore import Tensor
from mindspore.train.serialization import export
from mindspore.train.loss_scale_manager import FixedLossScaleManager
from mindspore.train.serialization import load_checkpoint, load_param_into_net
import mindspore.ops as ops
# 设置MindSpore的执行模式和设备
context.set_context(mode=context.GRAPH_MODE, device_target="Ascend")
cfg = edict(
{
"data_path": "train", # 训练数据集,如果是zip文件需要解压
"test_path": "test", # 测试数据集,如果是zip文件需要解压
"data_size": 3616,
"HEIGHT": 224, # 图片高度
"WIDTH": 224, # 图片宽度
"_R_MEAN": 123.68,
"_G_MEAN": 116.78,
"_B_MEAN": 103.94,
"_R_STD": 1,
"_G_STD": 1,
"_B_STD": 1,
"_RESIZE_SIDE_MIN": 256,
"_RESIZE_SIDE_MAX": 512,
"batch_size": 32,
"num_class": 2, # 分类类别
"epoch_size": 150, # 训练次数
"loss_scale_num": 1024,
"prefix": "resnet-ai",
"directory": "./model_resnet",
"save_checkpoint_steps": 10,
}
)
# 数据处理
def read_data(path, config, usage="train"):
# 从目录中读取图像的源数据集。
dataset = ds.ImageFolderDataset(path, class_indexing={"safe": 0, "un_safe": 1})
# define map operations
decode_op = vision.Decode()
normalize_op = vision.Normalize(
mean=[cfg._R_MEAN, cfg._G_MEAN, cfg._B_MEAN],
std=[cfg._R_STD, cfg._G_STD, cfg._B_STD],
)
resize_op = vision.Resize(cfg._RESIZE_SIDE_MIN)
center_crop_op = vision.CenterCrop((cfg.HEIGHT, cfg.WIDTH))
horizontal_flip_op = vision.RandomHorizontalFlip()
channelswap_op = vision.HWC2CHW()
random_crop_decode_resize_op = vision.RandomCropDecodeResize(
(cfg.HEIGHT, cfg.WIDTH), (0.5, 1.0), (1.0, 1.0), max_attempts=100
)
if usage == "train":
dataset = dataset.map(
input_columns="image", operations=random_crop_decode_resize_op
)
dataset = dataset.map(input_columns="image", operations=horizontal_flip_op)
else:
dataset = dataset.map(input_columns="image", operations=decode_op)
dataset = dataset.map(input_columns="image", operations=resize_op)
dataset = dataset.map(input_columns="image", operations=center_crop_op)
dataset = dataset.map(input_columns="image", operations=normalize_op)
dataset = dataset.map(input_columns="image", operations=channelswap_op)
if usage == "train":
dataset = dataset.shuffle(
buffer_size=10000
) # 10000 as in imageNet train script
dataset = dataset.batch(cfg.batch_size, drop_remainder=True)
else:
dataset = dataset.batch(1, drop_remainder=True)
dataset = dataset.repeat(1)
dataset.map_model = 4
return dataset
de_train = read_data(cfg.data_path, cfg, usage="train")
de_test = read_data(cfg.test_path, cfg, usage="test")
print(
"number of training set:", de_train.get_dataset_size() * cfg.batch_size
) # get_dataset_size()获取批处理的大小。
print("number of test set:", de_test.get_dataset_size())
de_dataset = de_train
data_next = de_dataset.create_dict_iterator(output_numpy=True).__next__()
print("channels/image length/width:", data_next["image"][0, ...].shape)
print("label of a single image:", data_next["label"][0]) # 一共5类,用0-4的数字表达类别。
plt.figure()
plt.imshow(data_next["image"][0, 0, ...])
plt.colorbar()
plt.grid(False)
plt.show()
"""ResNet."""
def _weight_variable(shape, factor=0.01):
init_value = np.random.randn(*shape).astype(np.float32) * factor
return Tensor(init_value)
def _conv3x3(in_channel, out_channel, stride=1):
weight_shape = (out_channel, in_channel, 3, 3)
weight = _weight_variable(weight_shape)
return nn.Conv2d(
in_channel,
out_channel,
kernel_size=3,
stride=stride,
padding=0,
pad_mode="same",
weight_init=weight,
)
def _conv1x1(in_channel, out_channel, stride=1):
weight_shape = (out_channel, in_channel, 1, 1)
weight = _weight_variable(weight_shape)
return nn.Conv2d(
in_channel,
out_channel,
kernel_size=1,
stride=stride,
padding=0,
pad_mode="same",
weight_init=weight,
)
def _conv7x7(in_channel, out_channel, stride=1):
weight_shape = (out_channel, in_channel, 7, 7)
weight = _weight_variable(weight_shape)
return nn.Conv2d(
in_channel,
out_channel,
kernel_size=7,
stride=stride,
padding=0,
pad_mode="same",
weight_init=weight,
)
def _bn(channel):
return nn.BatchNorm2d(
channel,
eps=1e-4,
momentum=0.9,
gamma_init=1,
beta_init=0,
moving_mean_init=0,
moving_var_init=1,
)
def _bn_last(channel):
return nn.BatchNorm2d(
channel,
eps=1e-4,
momentum=0.9,
gamma_init=0,
beta_init=0,
moving_mean_init=0,
moving_var_init=1,
)
def _fc(in_channel, out_channel):
weight_shape = (out_channel, in_channel)
weight = _weight_variable(weight_shape)
return nn.Dense(
in_channel, out_channel, has_bias=True, weight_init=weight, bias_init=0
)
class ResidualBlock(nn.Cell):
"""
ResNet V1 residual block definition.
Args:
in_channel (int): Input channel.
out_channel (int): Output channel.
stride (int): Stride size for the first convolutional layer. Default: 1.
Returns:
Tensor, output tensor.
Examples:
>>> ResidualBlock(3, 256, stride=2)
"""
expansion = 4
def __init__(self, in_channel, out_channel, stride=1):
super(ResidualBlock, self).__init__()
channel = out_channel // self.expansion
self.conv1 = _conv1x1(in_channel, channel, stride=1)
self.bn1 = _bn(channel)
self.conv2 = _conv3x3(channel, channel, stride=stride)
self.bn2 = _bn(channel)
self.conv3 = _conv1x1(channel, out_channel, stride=1)
self.bn3 = _bn_last(out_channel)
self.relu = nn.ReLU()
self.down_sample = False
if stride != 1 or in_channel != out_channel:
self.down_sample = True
self.down_sample_layer = None
if self.down_sample:
self.down_sample_layer = nn.SequentialCell(
[_conv1x1(in_channel, out_channel, stride), _bn(out_channel)]
)
self.add = ops.Add()
def construct(self, x): # pylint: disable=missing-docstring
identity = x
out = self.conv1(x)
out = self.bn1(out)
out = self.relu(out)
out = self.conv2(out)
out = self.bn2(out)
out = self.relu(out)
out = self.conv3(out)
out = self.bn3(out)
if self.down_sample:
identity = self.down_sample_layer(identity)
out = self.add(out, identity)
out = self.relu(out)
return out
class ResNet(nn.Cell):
"""
ResNet architecture.
Args:
block (Cell): Block for network.
layer_nums (list): Numbers of block in different layers.
in_channels (list): Input channel in each layer.
out_channels (list): Output channel in each layer.
strides (list): Stride size in each layer.
num_classes (int): The number of classes that the training images are belonging to.
Returns:
Tensor, output tensor.
Examples:
>>> ResNet(ResidualBlock,
>>> [3, 4, 6, 3],
>>> [64, 256, 512, 1024],
>>> [256, 512, 1024, 2048],
>>> [1, 2, 2, 2],
>>> 10)
"""
def __init__(
self, block, layer_nums, in_channels, out_channels, strides, num_classes
):
super(ResNet, self).__init__()
if not len(layer_nums) == len(in_channels) == len(out_channels) == 4:
raise ValueError(
"the length of layer_num, in_channels, out_channels list must be 4!"
)
self.conv1 = _conv7x7(3, 64, stride=2)
self.bn1 = _bn(64)
self.relu = ops.ReLU()
self.maxpool = nn.MaxPool2d(kernel_size=3, stride=2, pad_mode="same")
self.layer1 = self._make_layer(
block,
layer_nums[0],
in_channel=in_channels[0],
out_channel=out_channels[0],
stride=strides[0],
)
self.layer2 = self._make_layer(
block,
layer_nums[1],
in_channel=in_channels[1],
out_channel=out_channels[1],
stride=strides[1],
)
self.layer3 = self._make_layer(
block,
layer_nums[2],
in_channel=in_channels[2],
out_channel=out_channels[2],
stride=strides[2],
)
self.layer4 = self._make_layer(
block,
layer_nums[3],
in_channel=in_channels[3],
out_channel=out_channels[3],
stride=strides[3],
)
self.mean = ops.ReduceMean(keep_dims=True)
self.flatten = nn.Flatten()
self.end_point = _fc(out_channels[3], num_classes)
def _make_layer(self, block, layer_num, in_channel, out_channel, stride):
"""
Make stage network of ResNet.
Args:
block (Cell): Resnet block.
layer_num (int): Layer number.
in_channel (int): Input channel.
out_channel (int): Output channel.
stride (int): Stride size for the first convolutional layer.
Returns:
SequentialCell, the output layer.
Examples:
>>> _make_layer(ResidualBlock, 3, 128, 256, 2)
"""
layers = []
resnet_block = block(in_channel, out_channel, stride=stride)
layers.append(resnet_block)
for _ in range(1, layer_num):
resnet_block = block(out_channel, out_channel, stride=1)
layers.append(resnet_block)
return nn.SequentialCell(layers)
def construct(self, x): # pylint: disable=missing-docstring
x = self.conv1(x)
x = self.bn1(x)
x = self.relu(x)
c1 = self.maxpool(x)
c2 = self.layer1(c1)
c3 = self.layer2(c2)
c4 = self.layer3(c3)
c5 = self.layer4(c4)
out = self.mean(c5, (2, 3))
out = self.flatten(out)
out = self.end_point(out)
return out
def resnet50(class_num=10):
"""
Get ResNet50 neural network.
Args:
class_num (int): Class number.
Returns:
Cell, cell instance of ResNet50 neural network.
Examples:
>>> net = resnet50(10)
"""
return ResNet(
ResidualBlock,
[3, 4, 6, 3],
[64, 256, 512, 1024],
[256, 512, 1024, 2048],
[1, 2, 2, 2],
class_num,
)
def resnet101(class_num=1001):
"""
Get ResNet101 neural network.
Args:
class_num (int): Class number.
Returns:
Cell, cell instance of ResNet101 neural network.
Examples:
>>> net = resnet101(1001)
"""
return ResNet(
ResidualBlock,
[3, 4, 23, 3],
[64, 256, 512, 1024],
[256, 512, 1024, 2048],
[1, 2, 2, 2],
class_num,
)
def get_lr(
global_step,
total_epochs,
steps_per_epoch,
lr_init=0.01,
lr_max=0.1,
warmup_epochs=5,
):
"""
Generate learning rate array.
Args:
global_step (int): Initial step of training.
total_epochs (int): Total epoch of training.
steps_per_epoch (float): Steps of one epoch.
lr_init (float): Initial learning rate. Default: 0.01.
lr_max (float): Maximum learning rate. Default: 0.1.
warmup_epochs (int): The number of warming up epochs. Default: 5.
Returns:
np.array, learning rate array.
"""
lr_each_step = []
total_steps = steps_per_epoch * total_epochs
warmup_steps = steps_per_epoch * warmup_epochs
if warmup_steps != 0:
inc_each_step = (float(lr_max) - float(lr_init)) / float(warmup_steps)
else:
inc_each_step = 0
for i in range(int(total_steps)):
if i < warmup_steps:
lr = float(lr_init) + inc_each_step * float(i)
else:
base = 1.0 - (float(i) - float(warmup_steps)) / (
float(total_steps) - float(warmup_steps)
)
lr = float(lr_max) * base * base
if lr < 0.0:
lr = 0.0
lr_each_step.append(lr)
current_step = global_step
lr_each_step = np.array(lr_each_step).astype(np.float32)
learning_rate = lr_each_step[current_step:]
return learning_rate
net = resnet50(class_num=cfg.num_class)
# 计算softmax交叉熵。
loss = nn.SoftmaxCrossEntropyWithLogits(sparse=True, reduction="mean")
# 设置Adam优化器
train_step_size = de_train.get_dataset_size()
lr = Tensor(
get_lr(global_step=0, total_epochs=cfg.epoch_size, steps_per_epoch=train_step_size)
)
opt = Momentum(
net.trainable_params(),
lr,
momentum=0.9,
weight_decay=1e-4,
loss_scale=cfg.loss_scale_num,
)
# opt = Momentum(filter(lambda x: x.requires_grad, net.get_parameters()), 0.002,
# 0.9, 0.00004, loss_scale=1024.0)
loss_scale = FixedLossScaleManager(cfg.loss_scale_num, False)
model = Model(
net, loss_fn=loss, optimizer=opt, loss_scale_manager=loss_scale, metrics={"acc"}
)
loss_cb = LossMonitor(per_print_times=train_step_size)
ckpt_config = CheckpointConfig(
save_checkpoint_steps=cfg.save_checkpoint_steps, keep_checkpoint_max=1
)
ckpoint_cb = ModelCheckpoint(
prefix=cfg.prefix, directory=cfg.directory, config=ckpt_config
)
print("============== Starting Training ==============")
model.train(
cfg.epoch_size, de_train, callbacks=[loss_cb, ckpoint_cb], dataset_sink_mode=True
)
metric = model.eval(de_test)
print(metric)
# 预测
class_names = {0: "safe", 1: "un_safe"}
for i in range(32):
test_ = de_test.create_dict_iterator().__next__()
test = Tensor(test_["image"], mindspore.float32)
predictions = model.predict(test)
predictions = predictions.asnumpy()
true_label = test_["label"].asnumpy()
# 显示预测结果
p_np = predictions[0, :]
pre_label = np.argmax(p_np)
print(
str(i) + "th sampleprediction result:",
class_names[pre_label],
" actual result:",
class_names[true_label[0]],
)
# 创建文件夹
if not os.path.exists("./dangerous/"):
os.mkdir("./dangerous/")
# 加载ckpt模型参数
param_dict = load_checkpoint(
os.path.join(
cfg.directory,
cfg.prefix + "-" + str(cfg.epoch_size) + "_" + str(train_step_size) + ".ckpt",
)
)
# 将模型参数存至resnet50
resnet = resnet50(class_num=cfg.num_class)
load_param_into_net(resnet, param_dict)
x = np.random.uniform(-1.0, 1.0, size=[1, 3, cfg.HEIGHT, cfg.WIDTH]).astype(np.float32)
# 导出onnx模型
export(resnet, Tensor(x), file_name="./dangerous/best_model.onnx", file_format="ONNX")
|
# # 1\. Contexto
# A Loggi é uma empresa de tecnologia que oferece soluções de logística para empresas de diversos segmentos. Fundada em 2013, a empresa tem como objetivo simplificar a logística urbana por meio de uma plataforma tecnológica que conecta empresas e entregadores independentes. Com uma presença crescente em diversas regiões do Brasil, a Loggi tem enfrentado desafios para garantir a eficiência e a qualidade de suas entregas.
# 
# Para fins de análise, estamos utilizando dados de entregas realizadas pela Loggi na região de Brasília. Esses dados contêm informações valiosas, como a capacidade do veículo utilizado para cada entrega, o tamanho da carga transportada (em valores absolutos), bem como as coordenadas geográficas do ponto de origem e destino de cada entrega. Ao analisar esses dados, buscamos identificar padrões e obter insights relevantes para aprimorar o desempenho da empresa e otimizar suas operações.
# # 2\. Pacotes e bibliotecas
import json
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import geopandas
import geopy
from geopy.geocoders import Nominatim
from geopy.extra.rate_limiter import RateLimiter
# # 3\. Exploração de dados
# Conversão de arquivo em formato JSON para variável:
with open(
"/kaggle/input/deliveries-loggi/deliveries.json", mode="r", encoding="utf8"
) as arquivo:
data = json.load(arquivo)
# Conversão de variável JSON para um DataFrame pandas
data_pd = pd.DataFrame(data)
data_pd.head()
# Tratamento da coluna 'origin', utilizando a função 'normalize' do Pandas para ajustá-la no DataFrame
origin_normalize_pd = pd.json_normalize(data_pd["origin"])
data_pd = pd.merge(
left=data_pd,
right=origin_normalize_pd,
how="inner",
left_index=True,
right_index=True,
)
data_pd.drop("origin", axis=1)
data_pd = data_pd[["name", "region", "lng", "lat", "vehicle_capacity", "deliveries"]]
data_pd.rename(columns={"lng": "hub_lng", "lat": "hub_lat"}, inplace=True)
data_pd.head()
# Tratamento da coluna 'deliveries', utilizando a função 'explode' do Pandas para ajustá-la no DataFrame
explode_deliveries_pd = data_pd[["deliveries"]].explode("deliveries")
deliveries_pd = pd.concat(
[
explode_deliveries_pd["deliveries"]
.apply(lambda info: info["size"])
.rename("delivery_size"),
explode_deliveries_pd["deliveries"]
.apply(lambda info: info["point"]["lng"])
.rename("delivery_lng"),
explode_deliveries_pd["deliveries"]
.apply(lambda info: info["point"]["lat"])
.rename("delivery_lat"),
],
axis=1,
)
deliveries_pd.head()
# Integração das modificações realizadas ao DataFrame original
data_pd = pd.merge(
left=data_pd, right=deliveries_pd, how="right", right_index=True, left_index=True
)
data_pd = data_pd.drop("deliveries", axis=1)
data_pd.reset_index(inplace=True, drop=True)
data_pd.head()
len(data_pd)
# Com os dados em mãos, vamos conhecer um pouco melhor a estrutura do nosso conjunto de dados.
# ## 3.1. Estrutura
data_pd.shape
data_pd.columns
data_pd.index
data_pd.info()
# ## 3.2. Schema
data_pd.head(n=5)
# - Colunas e seus respectivos tipos de dados.
data_pd.dtypes
# - Atributos **categóricos**.
data_pd.select_dtypes("object").describe().T
# - Atributos **numéricos**.
data_pd.select_dtypes("int64").describe().T
# ## 3.3. Dados Faltantes
# Podemos verificar quais colunas possuem dados faltantes.
data_pd.isna().any()
# # 4\. Manipulação
# ## 4.1. Enriquecimento
# Realização da geocodificação nas coordenadas das três regiões e extração das informações de cidade e bairro
hub_df = (
data_pd[["region", "hub_lng", "hub_lat"]]
.drop_duplicates()
.sort_values(by="region")
.reset_index(drop=True)
)
hub_df.head()
geolocator = Nominatim(user_agent="loggi_geocoder")
geocoder = RateLimiter(geolocator.reverse, min_delay_seconds=1)
hub_df["coordinates"] = (
hub_df["hub_lat"].astype(str) + ", " + hub_df["hub_lng"].astype(str)
)
hub_df["geodata"] = hub_df["coordinates"].apply(geocoder)
hub_geodata_df = pd.json_normalize(hub_df["geodata"].apply(lambda data: data.raw))
hub_geodata_df.head()
hub_geodata_df = hub_geodata_df[["address.town", "address.suburb", "address.city"]]
hub_geodata_df.rename(
columns={
"address.town": "hub_town",
"address.suburb": "hub_suburb",
"address.city": "hub_city",
},
inplace=True,
)
hub_geodata_df["hub_city"] = np.where(
hub_geodata_df["hub_city"].notna(),
hub_geodata_df["hub_city"],
hub_geodata_df["hub_town"],
)
hub_geodata_df["hub_suburb"] = np.where(
hub_geodata_df["hub_suburb"].notna(),
hub_geodata_df["hub_suburb"],
hub_geodata_df["hub_city"],
)
hub_geodata_df.drop("hub_town", axis=1, inplace=True)
hub_geodata_df.head()
hub_df = pd.merge(left=hub_df, right=hub_geodata_df, right_index=True, left_index=True)
hub_df = hub_df[["region", "hub_suburb", "hub_city"]]
hub_df.head()
data_pd = pd.merge(left=data_pd, right=hub_df, how="inner", on="region")
data_pd = data_pd[
[
"name",
"region",
"hub_lng",
"hub_lat",
"hub_suburb",
"hub_city",
"vehicle_capacity",
"delivery_size",
"delivery_lng",
"delivery_lat",
]
]
data_pd.head()
# Enquanto o hub contem apenas 3 geolocalizações distintas, as entregas somam o total de 636.149, o que levaria em torno de 7 dias para serem consultadas no servidor do Nominatim, dada a restrição de uma consulta por segundo. Dito isso, vamos usar um arquivo CSV com todas as informações já processadas:
deliveries_geodata_pd = pd.read_csv(
"/kaggle/input/deliveries-geodata/deliveries-geodata.csv"
)
deliveries_geodata_pd.head()
data_pd = pd.merge(
left=data_pd,
right=deliveries_geodata_pd[["delivery_city", "delivery_suburb"]],
how="inner",
right_index=True,
left_index=True,
)
data_pd.head()
# ## 4.2. Qualidade
data_pd.info()
data_pd.isna().any()
# Porcentagem de dados faltantes:
100 * (data_pd["delivery_city"].isna().sum() / len(data_pd))
100 * (data_pd["delivery_suburb"].isna().sum() / len(data_pd))
# Cálculo da porcentagem de entregas para cada localidade:
prop_df = data_pd[["delivery_city"]].value_counts() / len(data_pd)
prop_df.sort_values(ascending=False).head(10)
prop_df = data_pd[["delivery_suburb"]].value_counts() / len(data_pd)
prop_df.sort_values(ascending=False).head(10)
# # 5\. Visualização
# ## 5.1. Mapa de entregas por região
# Vamos fazer o download dos dados do mapa do Distrito Federal do site oficial do IBGE através do seguinte link para criar o DataFrame mapa.
# - **Mapa dos Hubs**
hub_df = (
data_pd[["region", "hub_lng", "hub_lat"]].drop_duplicates().reset_index(drop=True)
)
geo_hub_df = geopandas.GeoDataFrame(
hub_df, geometry=geopandas.points_from_xy(hub_df["hub_lng"], hub_df["hub_lat"])
)
geo_hub_df.head()
# - **Mapa das Entregas**
geo_deliveries_df = geopandas.GeoDataFrame(
data_pd,
geometry=geopandas.points_from_xy(data_pd["delivery_lng"], data_pd["delivery_lat"]),
)
geo_deliveries_df.head()
# - **Visualização**
mapa = geopandas.read_file("distrito-federal.shp")
mapa = mapa.loc[[0]]
fig, ax = plt.subplots(figsize=(50 / 2.54, 50 / 2.54))
mapa.plot(ax=ax, alpha=0.4, color="lightgrey")
geo_deliveries_df.query('region == "df-0"').plot(
ax=ax, markersize=1, color="red", label="df-0"
)
geo_deliveries_df.query('region == "df-1"').plot(
ax=ax, markersize=1, color="blue", label="df-1"
)
geo_deliveries_df.query('region == "df-2"').plot(
ax=ax, markersize=1, color="seagreen", label="df-2"
)
geo_hub_df.plot(ax=ax, markersize=30, marker="x", color="black", label="hub")
plt.title("Entregas no Distrito Federal por Região", fontdict={"fontsize": 16})
lgnd = plt.legend(prop={"size": 15})
for handle in lgnd.legendHandles:
handle.set_sizes([50])
# - **Insights**:
# 1. As **entregas** estão corretamente alocadas aos seus respectivos **hubs**;
# 1. Os **hubs** das regiões 0 e 2 fazem **entregas** em locais distantes do centro e entre si, o que pode gerar um tempo e preço de entrega maior.
# ## 5.1. Gráfico de entregas por região
# - **Agregação**:
info = pd.DataFrame(
data_pd[["region", "vehicle_capacity"]].value_counts() / len(data_pd)
).reset_index()
info.rename(columns={0: "region_percent"}, inplace=True)
info.head()
# - **Visualização**:
with sns.axes_style("whitegrid"):
grafico = sns.barplot(data=info, x="region", y="region_percent", palette="pastel")
grafico.set(
title="Proporção de entregas por região", xlabel="Região", ylabel="Proporção"
)
|
# # **EXTRA CREDIT ASSIGNMENT**
# Submitted by: Shilpa Malge
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
business_pd = pd.read_json(
"/kaggle/input/yelp-dataset/yelp_academic_dataset_business.json",
lines=True,
nrows=50000,
)
business_pd.head(3)
review_pd = pd.read_json(
"/kaggle/input/yelp-dataset/yelp_academic_dataset_review.json",
lines=True,
nrows=50000,
)
review_pd.head(3)
checkin_pd = pd.read_json(
"/kaggle/input/yelp-dataset/yelp_academic_dataset_checkin.json",
lines=True,
nrows=50000,
)
checkin_pd.head(3)
user_pd = pd.read_json(
"/kaggle/input/yelp-dataset/yelp_academic_dataset_user.json",
lines=True,
nrows=50000,
)
user_pd.head(3)
tip_pd = pd.read_json(
"/kaggle/input/yelp-dataset/yelp_academic_dataset_tip.json", lines=True, nrows=50000
)
tip_pd.head(10)
# **To access the database we created.**
# **To set up a new restaurant in California, we analyzed the trends of all the restaurants in and around the most popular cities in CA to identify potential competition and the kind of reviews each of them received so that we can set up our restaurant accordingly. These queries will help us make data-driven decisions by providing insights into customer behavior and preferences, identifying trends in the industry, and highlighting areas where improvements can be made.**
# **We will use these queries to analyze the competition in the area, the types of restaurants that are popular, the categories of restaurants that are performing well, and the factors that drive customer reviews and ratings. This information can help us make informed decisions about our business strategy, such as deciding on the type of cuisine to offer, the menu, the pricing, and the marketing strategy. Additionally, these queries will also help us identify potential partners or suppliers to work with, based on the analysis of the businesses that are successful in the area.**
# **1. Displaying the name, city, state, and categories of 10 restaurants located in California.**
business_ca_restaurants = business_pd[
(business_pd["state"] == "CA")
& (business_pd["categories"].str.contains("Restaurant"))
].head(10)[["name", "city", "state", "categories"]]
business_ca_restaurants.head()
# **2. Top 10 businesses with the highest total number of check-ins recorded in the CHECKIN table. It can be used to identify popular businesses and understand their customer engagement patterns.**
checkin_agg = checkin_pd.groupby(["business_id"]).agg({"date": "sum"}).reset_index()
checkin_agg["checkin_nos"] = checkin_agg.date.str.count(",") + 1
checkin_agg.sort_values(by=["checkin_nos"], inplace=True, ascending=False)
top10_business = checkin_agg.merge(
business_pd[["business_id", "name"]], on="business_id", how="inner"
).head(10)
top10_business[["name", "checkin_nos"]]
# **3. The names and star ratings of the top-rated businesses in the city of Goleta, which could be useful for someone looking for highly-rated businesses in that area.**
business_pd[business_pd["city"] == "Goleta"].sort_values(
by="stars", ascending=False
).loc[:, ["name", "stars"]].head(10)
# **4. The names of the top 10 Japanese restaurants in Santa Barbara based on their average rating and displays the top review for each of them.**
# Merge the business and review dataframes based on the 'business_id' column
merged_df = pd.merge(business_pd, review_pd, on="business_id")
# Filter the merged dataframe based on the required conditions
filtered_df = merged_df[
(merged_df["city"] == "Santa Barbara")
& (merged_df["categories"].str.contains("Japanese"))
]
# Sort the filtered dataframe by 'stars' column in descending order
sorted_df = filtered_df.sort_values(by="stars_x", ascending=False)
# Limit the number of rows to 10
result_df = sorted_df.head(10)
# Select the required columns from the final dataframe
result_df = result_df[["name", "stars_x", "text"]]
# Print the final result
result_df
# **5. The top 10 users who have written the most reviews containing the word "Coffee" for businesses located in California, along with the number of reviews they have written and an example review text.**
# Join the review, user, and business dataframes
merged_df = pd.merge(
pd.merge(review_pd, user_pd, on="user_id"), business_pd, on="business_id"
)
# Filter the merged dataframe to only include businesses in California and reviews with the word 'coffee' in them
filtered_df = merged_df[
(merged_df["state"] == "CA") & (merged_df["text"].str.contains("Coffee"))
]
# Group the filtered dataframe by user name, count the number of reviews, and sort by review count in descending order
grouped_df = (
filtered_df.groupby("name_x")
.agg({"text": "count"})
.sort_values("text", ascending=False)
.reset_index()
)
# Select the top 10 users with the most reviews and their review count
result = grouped_df[["name_x", "text"]]
result
# **6. The top 10 cities with the most highly-rated restaurants by counting the number of businesses categorized as "Restaurants" and having a star rating of 4 or higher, grouped by city and sorted in descending order of count.**
# filter restaurants with 4 or more stars
restaurants_df = business_pd[
business_pd["categories"].str.contains("Restaurants") & (business_pd["stars"] >= 4)
]
# group by city and count the number of restaurants
result_df = restaurants_df.groupby("city").size().reset_index(name="count")
# sort by count in descending order
result_df = result_df.sort_values("count", ascending=False)
# select top 10 cities
result_df = result_df.head(10)
# print the result
print(result_df)
# **7. The names and categories of the top-rated restaurants in Santa Barbara, California, along with the count of "cool" votes received in the reviews for each restaurant.**
# Filter the businesses by state, city, and category
sb_restaurants = business_pd[
(business_pd["state"] == "CA")
& (business_pd["city"] == "Santa Barbara")
& (business_pd["categories"].str.contains("Restaurant", case=False))
]
# Join the filtered businesses with the reviews table and group by business name and categories
sb_restaurant_reviews = pd.merge(sb_restaurants, review_pd, on="business_id")
sb_restaurant_reviews_grouped = sb_restaurant_reviews.groupby(
["name", "categories"], as_index=False
).agg({"cool": "count"})
# Sort by the number of cool votes in descending order and limit to the first 10 results
sb_restaurant_reviews_sorted = sb_restaurant_reviews_grouped.sort_values(
by="cool", ascending=False
)
sb_restaurant_reviews_top10 = sb_restaurant_reviews_sorted.head(10)
# Print the top 10 results
results7 = sb_restaurant_reviews_top10[["name", "categories", "cool"]]
results7.head(10)
# **8. The top 10 most common restaurant categories in the city , which is determined by counting the number of businesses with the "Restaurants" category in each city and selecting the city with the highest count.**
# Filter for only restaurants
business_pd = business_pd[
business_pd["categories"].str.contains("Restaurants", na=False)
]
# Get the city with the most restaurants
city_max = (
business_pd.groupby("city")
.size()
.reset_index(name="num_restaurants")
.sort_values(by="num_restaurants", ascending=False)
.iloc[0]["city"]
)
# Filter for only the top city
business_pd = business_pd[business_pd["city"] == city_max]
# Group by categories and count the number of restaurants
result = (
business_pd.groupby("categories")
.size()
.reset_index(name="num_restaurants")
.sort_values(by="num_restaurants", ascending=False)
.head(10)
)
# Print the result
print(result)
# **9. The count of restaurants in the city with the most restaurants and groups them by their categories and star ratings. This information could be used by businesses to understand the competition in the city, the most common types of restaurants, and the star ratings they should aim for to stand out in the market.**
city_max = (
business_pd[business_pd["categories"].str.contains("Restaurants")]
.groupby("city")
.size()
.reset_index(name="num_restaurants")
.sort_values("num_restaurants", ascending=False)
.head(1)
)
result9 = (
business_pd[
business_pd["categories"].str.contains("Restaurants")
& business_pd["city"].isin(city_max["city"])
]
.groupby(["categories", "stars"])["business_id"]
.count()
.reset_index(name="num_restaurants")
.sort_values(["categories", "stars"])
.head(10)
)
result9
# **10. The top 10 restaurants in the city with the most restaurants by average rating. It does this by first finding the city with the most restaurants, and then selecting all restaurants in that city that have "Restaurants" in their categories. The restaurants are then grouped by name and the average star rating is calculated for each group. The result is a list of the top 10 restaurants in that city by average rating. The business use of this query is to help users identify the best restaurants in a city with a large number of options.**
# Select only restaurants from the business dataframe
restaurants_pd = business_pd[business_pd["categories"].str.contains("Restaurants")]
# Group by city and count the number of restaurants in each city
city_counts = restaurants_pd.groupby("city").size().reset_index(name="num_restaurants")
# Find the city with the most restaurants
max_city = city_counts.loc[city_counts["num_restaurants"].idxmax(), "city"]
# Filter restaurants by the city with the most restaurants
max_city_restaurants_pd = restaurants_pd[restaurants_pd["city"] == max_city]
# Group by name and find the average rating for each restaurant
avg_ratings_pd = (
max_city_restaurants_pd.groupby("name")["stars"]
.mean()
.reset_index(name="avg_rating")
)
# Sort by average rating in descending order and select the top 10
top_10_restaurants_pd = avg_ratings_pd.sort_values(
by="avg_rating", ascending=False
).head(10)
# Print the top 10 restaurants
print(top_10_restaurants_pd)
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.