script
stringlengths 113
767k
|
|---|
#
# Version Updates
#
# Train Test Split for better Evaluation
# Transfer Learning with EfficinetNetB0
#
#
# ## Setup
import pandas as pd
import numpy as np
import cv2
import os
import random
import matplotlib.pyplot as plt
import seaborn as sns
from tensorflow.keras.applications import EfficientNetB0
from tensorflow.keras.layers import GlobalAveragePooling2D
from tensorflow.keras.models import Model
import tensorflow as tf
import tensorflow_hub as hub
os.environ["TF_CPP_MIN_LOG_LEVEL"] = "3"
from keras.models import Sequential
from keras.layers import Dense, Flatten, Conv2D, MaxPool2D
from keras.layers import Input, Activation, LeakyReLU, Dropout
from keras.losses import BinaryCrossentropy
try:
from keras.optimizer import Adam
except:
from tensorflow.keras.optimizers import Adam
from tensorflow.keras.callbacks import TensorBoard
from tensorflow.keras.preprocessing.image import ImageDataGenerator
from sklearn.metrics import classification_report, confusion_matrix
# ## About Brain Tumor
# A brain tumor is a collection, or mass, of abnormal cells in your brain. Your skull, which encloses your brain, is very rigid. Any growth inside such a restricted space can cause problems.
# Brain tumors can be cancerous (malignant) or noncancerous (benign). When benign or malignant tumors grow, they can cause the pressure inside your skull to increase. This can cause brain damage, and it can be life-threatening.
# ### **How is Brain Tumor diagnosed?**
# ### **Magnetic resonance imaging (MRI)**
# An MRI uses magnetic fields to produce detailed images of the body. MRI can be used to measure the tumor’s size. A special dye called a contrast medium is given before the scan to create a clearer picture. This dye can be injected into a patient’s vein or given as a pill or liquid to swallow. MRIs create more detailed pictures than CT scans (see below) and are the preferred way to diagnose a brain tumor. The MRI may be of the brain, spinal cord, or both, depending on the type of tumor suspected and the likelihood that it will spread in the CNS. There are different types of MRI. The results of a neuro-examination, done by the internist or neurologist, helps determine which type of MRI to use.
# Here we have 253 samples of Brain MRI images, 98 of which show no tumor and 155 show brain tumor. Using a Convolutional Neural Network, we try to **classify whether the sample shows tumor or not**
#
import warnings
warnings.filterwarnings("ignore")
MAIN_DIR = (
"/kaggle/input/brain-mri-images-for-brain-tumor-detection/brain_tumor_dataset/"
)
SEED = 40
# ## Preparing the Image Data
os.listdir(MAIN_DIR)
for dirpath, dirnames, filenames in os.walk(MAIN_DIR):
print(f"{len(dirnames)} directories and {len(filenames)} images in {dirpath}")
# Inspect the raw data before preprocessing
def view_random_image():
subdirs = ["yes/", "no/"]
subdir = np.random.choice(subdirs)
target_folder = MAIN_DIR + subdir
random_image = random.sample(os.listdir(target_folder), 1)
img = cv2.imread(target_folder + random_image[0])
plt.imshow(img, cmap="gray")
plt.axis(False)
plt.title(img.shape)
plt.show()
# View Random Image
view_random_image()
# ### Image Data Generator
IMG_SHAPE = (128, 128)
BATCH_SIZE = 32
datagen = ImageDataGenerator(rescale=1 / 255.0, validation_split=0.5)
train_data = datagen.flow_from_directory(
MAIN_DIR,
target_size=IMG_SHAPE,
batch_size=BATCH_SIZE,
class_mode="binary",
shuffle=True,
subset="training",
)
test_data = datagen.flow_from_directory(
MAIN_DIR,
target_size=IMG_SHAPE,
batch_size=BATCH_SIZE,
class_mode="binary",
shuffle=True,
subset="validation",
)
len(train_data), len(test_data)
# ## Convolutional Neural Network
tf.random.set_seed(SEED)
model = Sequential(
[
Conv2D(filters=64, kernel_size=3, activation="relu"),
Conv2D(32, 3, activation="relu"),
MaxPool2D(pool_size=2),
Conv2D(32, 3, activation="relu"),
Conv2D(16, 3, activation="relu"),
MaxPool2D(2, padding="same"),
Flatten(),
Dense(1, activation="sigmoid"),
]
)
# Compile the model
model.compile(loss=BinaryCrossentropy(), optimizer=Adam(), metrics=["accuracy"])
# Fit the model
history = model.fit(
train_data,
epochs=10,
steps_per_epoch=len(train_data),
validation_data=test_data,
validation_steps=len(test_data),
)
# ## Evaluation
# Function to plot loss curves
def plot_curves(history):
"""
Returns separate loss and accuracy curves
"""
import matplotlib.pyplot as plt
loss = history.history["loss"]
val_loss = history.history["val_loss"]
accuracy = history.history["accuracy"]
val_accuracy = history.history["val_accuracy"]
epochs = range(len(history.history["loss"]))
plt.plot(epochs, loss, label="training_loss")
plt.plot(epochs, val_loss, label="val_loss")
plt.title("loss")
plt.xlabel("epochs")
plt.legend()
plt.figure()
plt.plot(epochs, accuracy, label="training_accuracy")
plt.plot(epochs, val_accuracy, label="val_accuracy")
plt.title("accuracy")
plt.xlabel("epochs")
plt.legend()
plot_curves(history)
result = model.evaluate(test_data, verbose=0)
print(f"Accuracy on Evaluation: {result[1]*100:.2f}%\nLoss: {result[0]:.4f}")
# Set up data generators
train_dir = "/kaggle/input/brain-mri-images-for-brain-tumor-detection/"
valid_dir = "/kaggle/input/brain-mri-images-for-brain-tumor-detection/"
img_size = (224, 224)
batch_size = 32
train_datagen = ImageDataGenerator(
rescale=1.0 / 255,
rotation_range=20,
shear_range=0.2,
zoom_range=0.2,
horizontal_flip=True,
)
valid_datagen = ImageDataGenerator(rescale=1.0 / 255)
train_generator = train_datagen.flow_from_directory(
train_dir, target_size=img_size, batch_size=batch_size, class_mode="binary"
)
valid_generator = valid_datagen.flow_from_directory(
valid_dir, target_size=img_size, batch_size=batch_size, class_mode="binary"
)
# Load EfficientNetB0 model
base_model = EfficientNetB0(
include_top=False, input_shape=(img_size[0], img_size[1], 3)
)
# Add new classification layers
x = GlobalAveragePooling2D()(base_model.output)
x = Dense(128, activation="relu")(x)
x = Dense(1, activation="sigmoid")(x)
model = Model(inputs=base_model.input, outputs=x)
# Freeze base model layers
for layer in base_model.layers:
layer.trainable = False
# Compile model
model.compile(optimizer="adam", loss="binary_crossentropy", metrics=["accuracy"])
# Train model
epochs = 10
history = model.fit(train_generator, epochs=epochs, validation_data=valid_generator)
model.save("model.h5")
from tensorflow.keras.preprocessing import image
# Load the model
model = tf.keras.models.load_model("model.h5")
# Load an example image and preprocess it
img_path = "/kaggle/input/brain-mri-images-for-brain-tumor-detection/no/1 no.jpeg"
img = image.load_img(img_path, target_size=(224, 224))
img_array = image.img_to_array(img)
img_array = np.expand_dims(img_array, axis=0)
img_array = img_array / 255.0
# Make a prediction on the image
prediction = model.predict(img_array)
# Convert the prediction to a label (0 for no tumor, 1 for tumor)
label = int(np.round(prediction)[0][0])
if label == 0:
print("No tumor detected.")
else:
print("Tumor detected.")
# ## Transfer Learning with EfficientNetB0
# EfficientNetB0 Feature Vector
effnet_url = "https://tfhub.dev/tensorflow/efficientnet/b0/feature-vector/1"
# Feature Layer
effnet_layer = hub.KerasLayer(
effnet_url, trainable=False, name="feature_extraction_layer"
)
# Create Sequential model
effnet_model = Sequential([effnet_layer, Dense(1, activation="sigmoid")])
# Compile the model
effnet_model.compile(loss=BinaryCrossentropy(), optimizer=Adam(), metrics=["accuracy"])
# Fit the model
effnet_history = effnet_model.fit(
train_data,
epochs=10,
steps_per_epoch=len(train_data),
validation_data=test_data,
validation_steps=len(test_data),
)
plot_curves(effnet_history)
result = effnet_model.evaluate(test_data, verbose=0)
print(f"Accuracy on Evaluation: {result[1]*100:.2f}%\nLoss: {result[0]:.4f}")
|
# # CNN (Convolutional Neural Network) on Sign Language Digits Dataset
# **Introduction**
#
# In this kernel I try to guess to pictures which are sign language correctly by using CNN. I have used Keras Library.
# **Content**
# [Import Data](#import)
# [Visualize](#visualize)
# [Train-Test Split](#train-test_split)
# [Reshaping](#reshaping)
# [Creating Model](#creating_model)
# [Building Model](#building_model)
# [Defining and Compiling of Optimizer](#optimizer)
# [Accuracy](#accuracy)
# [Conclusion](#conclusion)
#
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
#
# **Import Data**
# Import data
data_x = np.load(
"../input/sign-language-digits-dataset/Sign-language-digits-dataset/X.npy"
)
data_y = np.load(
"../input/sign-language-digits-dataset/Sign-language-digits-dataset/Y.npy"
)
#
# **Visualize**
#
# Visualize to samples
img_size = 64
plt.subplot(1, 3, 1)
plt.imshow(data_x[200].reshape(img_size, img_size))
plt.axis("off")
plt.subplot(1, 3, 2)
plt.imshow(data_x[800].reshape(img_size, img_size))
plt.axis("off")
plt.subplot(1, 3, 3)
plt.imshow(data_x[600].reshape(img_size, img_size))
plt.axis("off")
#
# **Train-Test Split**
# Train-Test Split
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(
data_x, data_y, test_size=0.2, random_state=42
)
# Size of elements of train_test_split methods
print("x train shape: {}".format(x_train.shape))
print("y train shape: {}".format(y_train.shape))
print("x test shape: {}".format(x_test.shape))
print("y test shape: {}".format(y_test.shape))
#
# **Reshaping**
# Reshaping. We reshape x_train and x_test because Keras requires 3 dimention.
x_train = x_train.reshape(-1, 64, 64, 1)
x_test = x_test.reshape(-1, 64, 64, 1)
# New size of x_train and x_shape
print("x train shape: {}".format(x_train.shape))
print("x test shape: {}".format(x_test.shape))
#
# **Creating Model**
# Creating Model
from sklearn.metrics import confusion_matrix
import itertools
from keras.utils.np_utils import to_categorical # convert to one-hot-encoding
from keras.models import Sequential
from keras.layers import Dense, Dropout, Flatten, Conv2D, MaxPool2D
from keras.optimizers import RMSprop, Adam
from keras.preprocessing.image import ImageDataGenerator
from keras.callbacks import ReduceLROnPlateau
#
# **Building Model**
# building of our model
model = Sequential()
# we add convolutional layer, count of filter = 64, kernel_size means that dimension of filter.
model.add(
Conv2D(
filters=64,
kernel_size=(5, 5),
padding="Same",
activation="relu",
input_shape=(64, 64, 1),
)
)
# dimension of (64,64,1) is 3 because kernel requires 3 dimensions. Number "1" shows that it is used as gray scale.
model.add(MaxPool2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
# we rewrite the top one. We don't have to write input shape because these are things that are connected to each other like chains.
model.add(Conv2D(filters=128, kernel_size=(3, 3), padding="Same", activation="relu"))
model.add(MaxPool2D(pool_size=(2, 2), strides=(2, 2)))
model.add(Dropout(0.25))
model.add(Conv2D(filters=128, kernel_size=(3, 3), padding="Same", activation="relu"))
model.add(MaxPool2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
# fully connected
model.add(Flatten())
model.add(Dense(256, activation="relu"))
model.add(Dense(512, activation="relu"))
model.add(Dense(256, activation="relu"))
model.add(Dense(10, activation="softmax"))
# although sigma function is used for binary classification, softmax is a version of sigma function which is used for multi-output classification.
model.summary()
#
# **Defining and Compiling of Optimizer**
#
# defining optimizer
optimizer = Adam(lr=0.001, beta_1=0.9, beta_2=0.999)
# compiling optimizer
model.compile(
optimizer=optimizer, loss="categorical_crossentropy", metrics=["accuracy"]
)
# fitting
history = model.fit(x_train, y_train, epochs=100, validation_data=(x_test, y_test))
#
# **Accuracy**
scores = model.evaluate(x_test, y_test, verbose=0)
print("{}: {:.2f}%".format("accuracy", scores[1] * 100))
|
# Big Five Personality Test
import pandas as pd
import numpy as np
from sklearn.cluster import KMeans
import matplotlib.pyplot as plt
data = pd.read_csv("E:/ML projects/data-final.csv")
x = data.iloc[:, :50]
x.head()
# check null values there
x.isnull().sum()
# fill the values with 0 (1 million>> 1783)
x = x.fillna(0)
x.isnull().sum()
kmc = KMeans(n_clusters=10)
kmc.fit(x)
len(kmc.labels_)
kmc.labels_
# untagging the clusters seperately into 10
one, two, three, four, five, six, seven, eight, nine, ten = kmc.cluster_centers_
one
# observing the distribution of 5 traits in each cluster
# can't take the 1st 10 avg since there are positive and negative statements
oneScores = {}
oneScores["EXT_SCORE"] = (
one[0]
- one[1]
+ one[2]
- one[3]
+ one[4]
- one[5]
+ one[6]
- one[7]
+ one[8]
- one[9]
)
oneScores["EST_SCORE"] = (
one[10]
- one[11]
+ one[12]
- one[13]
+ one[14]
+ one[15]
+ one[16]
+ one[17]
+ one[18]
+ one[19]
)
oneScores["AGR_SCORE"] = (
-one[20]
+ one[21]
- one[22]
+ one[23]
- one[24]
+ one[25]
- one[26]
+ one[27]
+ one[28]
+ one[29]
)
oneScores["CSN_SCORE"] = (
one[30]
- one[31]
+ one[32]
- one[33]
+ one[34]
- one[35]
+ one[36]
- one[37]
+ one[38]
+ one[39]
)
oneScores["OPN_SCORE"] = (
one[40]
- one[41]
+ one[42]
- one[43]
+ one[44]
- one[45]
+ one[46]
- one[47]
+ one[48]
+ one[49]
)
oneScores
types = {
"one": one,
"two": two,
"three": three,
"four": four,
"five": five,
"six": six,
"seven": seven,
"eight": eight,
"nine": nine,
"ten": ten,
}
types.items()
typeScores = {}
for cluster, traitType in types.items():
peronsalScore = {}
peronsalScore["EXT_SCORE"] = (
traitType[0]
- traitType[1]
+ traitType[2]
- traitType[3]
+ traitType[4]
- traitType[5]
+ traitType[6]
- traitType[7]
+ traitType[8]
- traitType[9]
)
peronsalScore["EST_SCORE"] = (
traitType[10]
- traitType[11]
+ traitType[12]
- traitType[13]
+ traitType[14]
+ traitType[15]
+ traitType[16]
+ traitType[17]
+ traitType[18]
+ traitType[19]
)
peronsalScore["AGR_SCORE"] = (
-traitType[20]
+ traitType[21]
- traitType[22]
+ traitType[23]
- traitType[24]
+ traitType[25]
- traitType[26]
+ traitType[27]
+ traitType[28]
+ traitType[29]
)
peronsalScore["CSN_SCORE"] = (
traitType[30]
- traitType[31]
+ traitType[32]
- traitType[33]
+ traitType[34]
- traitType[35]
+ traitType[36]
- traitType[37]
+ traitType[38]
+ traitType[39]
)
peronsalScore["OPN_SCORE"] = (
traitType[40]
- traitType[41]
+ traitType[42]
- traitType[43]
+ traitType[44]
- traitType[45]
+ traitType[46]
- traitType[47]
+ traitType[48]
+ traitType[49]
)
typeScores[cluster] = peronsalScore
typeScores
# since there are negative values, we use min max array to get all positive
# first we have to take the sum of differnt traits of each cluster
totalEXT = []
totalEST = []
totalAGR = []
totalCSN = []
totalOPN = []
for cluster, traitType in typeScores.items():
totalEXT.append(traitType["EXT_SCORE"])
totalEST.append(traitType["EST_SCORE"])
totalAGR.append(traitType["AGR_SCORE"])
totalCSN.append(traitType["CSN_SCORE"])
totalOPN.append(traitType["OPN_SCORE"])
totalEXT
minMaxArray = lambda L: (np.array(L) - np.array(L).min()) / (
np.array(L).max() - np.array(L).min()
)
normExtro = list(minMaxArray(totalEXT))
normEST = list(minMaxArray(totalEST))
normARG = list(minMaxArray(totalAGR))
normCSN = list(minMaxArray(totalCSN))
normOPN = list(minMaxArray(totalOPN))
normCSN
normEST
idx = 0
normTypeScores = {}
for personalityType, personalityScore in typeScores.items():
normPersonScore = {}
normPersonScore["EXT_SCORE"] = normExtro[idx]
normPersonScore["EST_SCORE"] = normEST[idx]
normPersonScore["ARG_SCORE"] = normARG[idx]
normPersonScore["CSN_SCORE"] = normCSN[idx]
normPersonScore["OPN_SCORE"] = normOPN[idx]
normTypeScores[personalityType] = normPersonScore
idx = idx + 1
normTypeScores
# let's see the diffrent traits in cluster 1
plt.bar(list(normTypeScores["one"].keys()), normTypeScores["one"].values())
# let's see the diffrent traits in cluster 7
plt.bar(list(normTypeScores["seven"].keys()), normTypeScores["seven"].values())
|
# Personal Info: [blog](mostafatouny.github.io/), [twitter](@Mostafa_Touny), [email]([email protected])
# ### Preface
# If you wish to see only results without any technical details, hit the above _Run All_ button, skip coding and look at only graphs and tables. At the end there's a summary of the whole notebook's results if you do not like scrolling. Even-though data science methods used here are extremely primitive, I believe there're insights which could be extracted. So, share us your thoughts, as a gamer in interpreting these data. Certainly, they are going to be helpful as these results are just the outset of a long journey I am about to tackle in artifical emotional intellligence in games (still a beginner, anyway).
# ### Goal
# - Top games got high ratings from professional critics but not from community of users
# - Top games got high ratings from community of users but not from progessional critics
# - Graph of percentage of games whose disparity between critics and users are low, moderate, or high
# - Do above steps on four platforms, namely, PS4, Xbox One, Switch, and PC. Then we compare them
# 3rd-party libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
# read input from kaggle into a dataframe
# a map from each platform to its corresponding dataframe
platform_df = {}
# platform names and their corresonding data file names
platformsNames = ["ps4", "xbox", "switch", "pc"]
filesNames = ["ps4.csv", "xbox.csv", "switch.csv", "pc.csv"]
# for each platform, then
for name in platformsNames:
# parse it as pandas dataframe, then map platform name to it
platform_df[name] = pd.read_csv(
"/kaggle/input/metacritic-best-2019-video-games/" + name + ".csv"
)
# take a look at a dataframe
platform_df["ps4"]
platform_df["ps4"].dtypes
# drop unneeded columns and re-organize them
for name in platformsNames:
platform_df[name] = platform_df[name][["title", "user_rating", "critic_rating"]]
# take a look at a dataframe, again
platform_df["xbox"]
# ### compute disparity (difference) between users and critics
# for each platform
for name in platform_df:
# get dataframe of the platform
df = platform_df[name]
# for each record, compute distance between user and critic ratings, then set result to a new column
df["userCritic_difference"] = df.apply(
lambda x: abs(x["user_rating"] - x["critic_rating"]), axis=1
)
# assign updates back to our dataframe
platform_df[name] = df
platform_df["pc"]
# ### discretize disparity computed earlier into categories
# define categories and their intervals
def numToCat(row):
# equal or greater than 30
if row["userCritic_difference"] >= 30:
return "high"
# equal or greater than 20 and less than 30
elif row["userCritic_difference"] >= 20:
return "moderate"
# less than 20
else:
return "low"
# compute categories as defined earlier
# loop on platforms
for platformName in platform_df:
# get dataframe of the platform
df = platform_df[platformName]
# add category based on difference just defined
df["difference_category"] = df.apply(lambda x: numToCat(x), axis=1)
# let categories be recognized by pandas
df["difference_category"] = df["difference_category"].astype("category")
# re-order categories
df["difference_category"] = df["difference_category"].cat.set_categories(
["low", "moderate", "high"]
)
# assign back to our dataframe
platform_df[platformName] = df
# take a look after our new columns added
platform_df["switch"]
# ### sort according to disparity between users and critics
# for each platform
for platformName in platform_df:
# get platform dataframe
df = platform_df[platformName]
# sort it by userCritic_difference
df = df.sort_values(axis=0, by="userCritic_difference", ascending=False)
# assign sorted dataframe back to our dataframe
platform_df[platformName] = df
# ### maximum disparity between users and critics ratings
platform_df["ps4"].head(20)
platform_df["xbox"].head(20)
platform_df["pc"].head(20)
platform_df["switch"].head(20)
# NBA, Fifa, Madden, COD: modern warefare games are on top of nearly all platforms lists
# ### Minimum disparity between users and critics
platform_df["ps4"].tail(20)
# _Star Wars Jedi: Fallen Order_ got zero disparity
# ### Games which got ratings from users higher than from professional critics
# filter only records whose user ratings is greater than critics ratings
def higherUserRatings(platform_in):
return platform_df[platform_in][
platform_df[platform_in]["user_rating"]
> platform_df[platform_in]["critic_rating"]
].head(10)
higherUserRatings("pc")
higherUserRatings("ps4")
higherUserRatings("xbox")
higherUserRatings("switch")
# _Left Alive_ is the most praised game by the community not appreciated by professional critics
# ### Pie graph on disparity between users and critics
plt.close("all")
# for each platform dataframe
for platformName in platform_df:
print("\non platform ", platformName)
# count categories among all records
categories_count = platform_df[platformName].groupby("difference_category").size()
# construct a series based on it
pie_series = pd.Series(categories_count, name="categories percentages")
# plot a pie chart
pie_series.plot.pie(figsize=(6, 6))
plt.show()
# Switch games got much lower percentage of high and moderate disparity
# ### Basic stats on disparity between users and critics
# for each platform
for platformName in platform_df:
# print platform name
print("\n", "on ", platformName)
# show basic stat
print(platform_df[platformName]["userCritic_difference"].describe())
|
import pandas as pd # 导入csv文件的库
import numpy as np # 进行矩阵运算的库
import matplotlib.pyplot as plt # 作图的库
import torch # 一个深度学习的库Pytorch
import torch.nn as nn # neural network,神经网络
from torch.autograd import Variable # 从自动求导中引入变量
import torch.optim as optim # 一个实现了各种优化算法的库
import torch.nn.functional as F # 神经网络函数库
import os # 与操作系统交互,处理文件和目录、管理进程、获取环境变量
from PIL import Image, ImageOps, ImageFilter, ImageEnhance # PIL是图像处理库
import torchvision.transforms as transforms # 图像、视频、文本增强和预处理的库
import zipfile # 对ZIP格式的归档文件进行读取、写入和操作
import warnings # 避免一些可以忽略的报错
warnings.filterwarnings("ignore") # filterwarnings()方法是用于设置警告过滤器的方法,它可以控制警告信息的输出方式和级别。
os.listdir("../input/dogs-vs-cats-redux-kernels-edition") # 返回指定路径下的文件和目录名的列表
zip_files = ["test", "train"]
for zip_file in zip_files:
with zipfile.ZipFile(
"../input/dogs-vs-cats-redux-kernels-edition/{}.zip".format(zip_file), "r"
) as z:
z.extractall(".") # 解压
print("{} unzipped".format(zip_file))
Train_Folder_Path = "../working/train"
Test_Folder_Path = "../working/test"
train_file_names = os.listdir(Train_Folder_Path)
test_file_names = os.listdir(Test_Folder_Path)
targets = []
full_paths = []
for file_name in train_file_names:
target = file_name.split(".")[0]
full_path = os.path.join(Train_Folder_Path, file_name)
full_paths.append(full_path)
targets.append(target)
train_df = pd.DataFrame()
train_df["image_path"] = full_paths
train_df["target"] = targets
full_paths = []
for file_name in test_file_names:
full_path = os.path.join(Test_Folder_Path, file_name)
full_paths.append(full_path)
test_df = pd.DataFrame()
test_df["image_path"] = full_paths
# 定义正则化的标准,3个颜色通道的均值和方差,这是从ImageNet数据集上得出的数值
normalize = transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225])
transform = transforms.Compose(
[
# transforms.RandomRotation(30), #在[-30,30]的范围内随机旋转一个角度
transforms.RandomResizedCrop(
size=128,
scale=(0.8, 1.2),
interpolation=transforms.functional.InterpolationMode.BILINEAR,
), # 先将图像随机进行缩放操作,然后再将图像变成128*128的图像
# transforms.RandomHorizontalFlip(), # 随机水平翻转
# transforms.ColorJitter(brightness=0.5, contrast=0.5, saturation=0.5), # 随机颜色变换
transforms.ToTensor(), # 转换成张量形式
normalize, # 标准化操作
]
)
imgcnt = 25000
data = []
label = []
for i in range(imgcnt):
img = Image.open(train_df["image_path"][i]) # 读取图片
new_img = transform(img)
data.append(new_img.detach().cpu().numpy())
if train_df["target"][i] == "cat":
label.append(0) # 猫是0,狗是1
else:
label.append(1)
if i % 500 == 0:
print(i)
# 划分训练集和测试集
data = np.array(data)
label = np.array(label)
num = [i for i in range(imgcnt)]
np.random.shuffle(num)
data = data[num]
label = label[num]
train_X = data[: 8 * imgcnt // 10]
train_y = label[: 8 * imgcnt // 10]
test_X = data[8 * imgcnt // 10 :]
test_y = label[8 * imgcnt // 10 :]
class CNN(nn.Module):
def __init__(self):
super(CNN, self).__init__()
self.model = nn.Sequential()
# 3*128*128
self.model.add_module("conv1_1", nn.Conv2d(3, 32, 3, 1, 0, bias=False))
# 32*126*126
self.model.add_module("relu1_1", nn.ReLU())
self.model.add_module("maxpool1", nn.MaxPool2d(2, 2, 0))
# 32*63*63
self.model.add_module("batch1", nn.BatchNorm2d(32))
self.model.add_module("conv2_1", nn.Conv2d(32, 64, 3, 1, 0, bias=False))
# 64*61*61
self.model.add_module("SiLU2_1", nn.SiLU())
self.model.add_module("maxpool2", nn.MaxPool2d(2, 2, 0))
# 64*30*30
# 全连接网络层,输入64*30*30,输出10个类别的概率
self.model.add_module("linear1", nn.Linear(64 * 30 * 30, 256))
self.model.add_module("dropout1", nn.Dropout(0.5))
self.model.add_module("Leakyrelu3_1", nn.LeakyReLU())
self.model.add_module("linear2", nn.Linear(256, 2))
self.model.add_module("Tanh3_2", nn.Tanh())
def forward(self, input):
output = input
for name, module in self.model.named_children():
if name == "linear1":
output = output.view(-1, 64 * 30 * 30)
output = module(output)
return F.softmax(output, dim=1)
def weight_init(m):
# 获取对象所属的类的名称
class_name = m.__class__.__name__
# 当对象的name中出现"conv",也就是卷积操作
if class_name.find("conv") != -1:
# 对卷积核按照正态分布的均值和标准差随机初始化
m.weight.data.normal_(0, 0.02)
# 初始化神经网络
netC = CNN()
netC.apply(weight_init)
print(netC)
# 优化器
optimizer = optim.Adam(netC.parameters(), lr=0.0002, betas=(0.5, 0.999))
# 损失函数
criterion = nn.NLLLoss() # 负对数似然损失函数,也是交叉熵损失函数的一种
# 训练周期为20次
num_epochs = 20
for epoch in range(num_epochs):
num = [i for i in range(len(train_X))]
np.random.shuffle(num)
train_X = train_X[num]
train_y = train_y[num]
for i in range(16):
image = []
label = []
for j in range(500):
image.append(train_X[1000 * i + j].reshape((3, 128, 128)))
label.append(train_y[1000 * i + j])
# 将数据转换成可以处理的张量格式
image = torch.Tensor(image) # .to(device)
label = torch.Tensor(label).long() # .to(device)
# 训练
netC.train()
# 将数据放进去训练
output = netC(image) # .to(device)
# 计算每次的损失函数
error = criterion(output, label) # .to(device)
# 反向传播
error.backward()
print(i)
# 优化器进行优化(梯度下降,降低误差)
optimizer.step()
# 将梯度清空
optimizer.zero_grad()
print(epoch, error)
pred_y = []
for i in range(len(train_X)):
pred = netC(torch.Tensor(train_X[i].reshape((1, 3, 128, 128))))[0] # .to(device)
pred = np.argmax(pred.detach().cpu().numpy())
pred_y.append(pred)
if i % 1000 == 0:
print(i)
print("训练集的准确率:", np.sum(pred_y == train_y) / len(train_y), "训练集总数为:", len(train_y))
pred_y = []
for i in range(len(test_X)):
pred = netC(torch.Tensor(test_X[i].reshape((1, 3, 128, 128))))[0]
pred = np.argmax(pred.detach().cpu().numpy())
pred_y.append(pred)
if i % 1000 == 0:
print(i)
print("测试集的准确率:", np.sum(pred_y == test_y) / len(test_y), "测试集总数为:", len(test_y))
torch.save(netC, "catsdogs.pth")
print(
test_df["image_path"][i],
)
imgcnt = 12500
data = []
for i in range(imgcnt):
img = Image.open(test_df["image_path"][i])
new_img = transform(img)
data.append(new_img.detach().cpu().numpy())
if i % 500 == 0:
print(i)
data = np.array(data)
pred_y = [0 for i in range(len(data))]
for i in range(len(data)):
pred = netC(torch.Tensor(data[i].reshape((1, 3, 128, 128))))[0] # .to(device))[0]
pred = np.argmax(pred.detach().cpu().numpy())
t = test_df["image_path"][i].split("/")
t = t[len(t) - 1]
t = int(t[: len(t) - 4])
pred_y[t - 1] = pred
if i % 1000 == 0:
print(i)
# 写入文件
import csv
head = ["id", "label"]
data = []
for i in range(len(pred_y)):
data.append([(i + 1), pred_y[i]])
with open("answer.csv", "w", newline="") as f:
writer = csv.writer(f)
writer.writerow(head)
writer.writerows(data)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
from sklearn.model_selection import train_test_split
from sklearn.svm import LinearSVC
from sklearn.feature_extraction import DictVectorizer
from sklearn.metrics import classification_report
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
df = pd.read_csv("/kaggle/input/titanic/train.csv")
# print(df.head())
useful_df = df[["Survived", "Pclass", "Sex", "Age", "SibSp", "Parch", "Embarked"]]
useful_df.dropna(inplace=True)
train_data, val_data = train_test_split(useful_df, test_size=0.3, random_state=1)
print(train_data.head())
def extract_train_features_and_labels(train_data):
train_labels = list(train_data["Survived"])
train_features = []
pclass = list(train_data["Pclass"])
sex = list(train_data["Sex"])
age = list(train_data["Age"])
sibsp = list(train_data["SibSp"])
parch = list(train_data["Parch"])
embarked = list(train_data["Embarked"])
for i in range(len(train_data)):
feature_dict = {
"pclass": pclass[i],
"sex": sex[i],
"age": age[i],
"sibsp": sibsp[i],
"parch": parch[i],
"embarked": embarked[i],
}
train_features.append(feature_dict)
return train_features, train_labels
# extract_train_features_and_labels(train_data)
def extract_val_features_and_labels(val_data):
val_labels = list(val_data["Survived"])
val_features = []
pclass = list(val_data["Pclass"])
sex = list(val_data["Sex"])
age = list(val_data["Age"])
sibsp = list(val_data["SibSp"])
parch = list(val_data["Parch"])
embarked = list(val_data["Embarked"])
for i in range(len(val_data)):
feature_dict = {
"pclass": pclass[i],
"sex": sex[i],
"age": age[i],
"sibsp": sibsp[i],
"parch": parch[i],
"embarked": embarked[i],
}
val_features.append(feature_dict)
return val_features, val_labels
# extract_val_features_and_labels(val_data)
def create_classifier(train_features, train_labels):
vec = DictVectorizer()
vec_train_features = vec.fit_transform(train_features)
model = LinearSVC()
fitted_model = model.fit(vec_train_features, train_labels)
return vec, fitted_model
def classify_data(vec, fitted_model, val_features):
vec_val_features = vec.transform(val_features)
prediction = fitted_model.predict(vec_val_features)
return prediction
def evaluation(val_labels, prediction):
report = classification_report(val_labels, prediction)
print(report)
return report
train_features, train_labels = extract_train_features_and_labels(train_data)
val_features, val_labels = extract_val_features_and_labels(val_data)
vec, fitted_model = create_classifier(train_features, train_labels)
prediction = classify_data(vec, fitted_model, val_features)
evaluation(val_labels, prediction)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Sentimental Analysis
# Will use 2 main approaches to sentimental analysis
# - VADER (Valence Aware Dictionary sEntiment Reasoner) - Bag of words approach
# - ROBERTA (Transformer) by 🤗
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
plt.style.use("ggplot")
import nltk
import warnings
warnings.filterwarnings("ignore")
df = pd.read_csv("../input/amazon-fine-food-reviews/Reviews.csv", index_col="Id")
df
df.Text.iloc[0]
df.shape
df_subset = df.head(500)
# # EDA
df.Score.value_counts().sort_index().plot(kind="bar", title="Reviews Score Count")
plt.xlabel("Score")
plt.ylabel("Count")
plt.show()
# ProductId and Count of Score type
temp = df_subset.groupby(["ProductId", "Score"]).aggregate({"Score": "count"})
temp.columns = ["count"]
temp.reset_index(inplace=True)
# for sorting
tempmask = df_subset.groupby(["ProductId"])["ProductId"].count()
# Top 10 Products by count
top10_Products = tempmask.sort_values(ascending=False).head(10).index
# Retrieve Top 10 Products
temp["TotalCount"] = temp["ProductId"].map(tempmask)
temp.sort_values("TotalCount", ascending=False, inplace=True)
toptemp = temp[temp["ProductId"].isin(top10_Products)].drop("TotalCount", axis=1)
# Grouped Bar plot
plt.figure(figsize=(15, 5))
sns.barplot(
x="ProductId",
y="value",
hue="Score",
data=pd.melt(toptemp, id_vars=["ProductId", "Score"]),
palette="Paired",
)
plt.title("Top 10 Products Grouped by Score")
plt.show()
# **Top 10 Products**
# - Most of the products are dominated by higher score of 5
# - Product B001ELL608 has the highest frequency of 1-scored reviews
# # NLTK (Python Natural Language Toolkit)
example = df_subset.Text.iloc[50]
example
# ### Tokenize
ex_token = nltk.word_tokenize(example)
np.array(ex_token)
# **The tokenizer is not just split the words by spaces, instead it is extracts real words. Ex: "don't" >> "do", "n't"**
# **This is called Stemming**
# ## Stop-words
from nltk.corpus import stopwords
stop_words = set(nltk.corpus.stopwords.words("english"))
filtered_sentence = [word for word in ex_token if not word.lower() in stop_words]
filtered_sentence
# ### Part Of Speech Tagging
ex_tagged = nltk.pos_tag(ex_token)
ex_tagged
# ref: https://www.educba.com/nltk-pos-tag/
ex_chunked = nltk.chunk.ne_chunk(ex_tagged)
ex_chunked.pprint()
# # VADER Sentiment scoring
# We'll use NLTK's `SentimentIntensityAnalyzer` to get the positive/negative/neutral scores of a text
# - Uses Bag of words:
# - Stop words are removed
# - Each word is stored and combined for a final score
#
# Remember it will score each word individually and then combines the scoring to check wheather the statement is positive or otherwise. Becuase of independent scoring, it does not take into account the relationship between words (context)
from nltk.sentiment import SentimentIntensityAnalyzer
sia = SentimentIntensityAnalyzer()
# Get scores of sentences
print(sia.polarity_scores("I have a happy and hugging face!"))
print(sia.polarity_scores("This couldn't be more awful"))
print(sia.polarity_scores("The product is good but couldn't have been better"))
# **The above statements is more of a;**
# - positive
# - negative
# - neutral
# sentiment resp. (compound shows overall score in range -1 to +1)
print(example)
ex_score = sia.polarity_scores(example)
ex_score
# Get scores for entire dataset
from tqdm.notebook import tqdm # for a beautiful progress bar
result_score = {}
for i, row in tqdm(df_subset.iterrows(), total=len(df_subset)):
result_score[i] = sia.polarity_scores(row["Text"])
vaders = pd.DataFrame(result_score).T
vaders.head()
# Merge to original dataset
df_vader = df_subset.merge(vaders, how="left", left_index=True, right_index=True)
df_vader.head()
# # Plot Vader
# If we assume the Review Score of 5 is more like positive sentiment than negative, we want the Compound score also to be higher.
# **low Score has lower Compound and Higher Score has higher Compound**
sns.barplot(
data=df_vader, x="Score", y="compound", label="Compound score by Review Score"
)
plt.show()
# We can see it is inline with our assumption
fig, axs = plt.subplots(1, 3, figsize=(15, 5))
sns.barplot(data=df_vader, x="Score", y="neg", ax=axs[0])
sns.barplot(data=df_vader, x="Score", y="neu", ax=axs[1])
sns.barplot(data=df_vader, x="Score", y="pos", ax=axs[2])
axs[0].set_title("Score v Neg")
axs[1].set_title("Score v Neu")
axs[2].set_title("Score v Pos")
plt.show()
sia.polarity_scores("You should cut yourself some slack")
# # Roberta - Pretrained Model
# - Roberta is a Transformer
# - Trained on Large corpus of data
# - Transformer models not only account for words, but also for the context of a statement
from transformers import AutoTokenizer # for tokenizing
from transformers import AutoModelForSequenceClassification # for classification
from scipy.special import softmax # for normalizing the output
# Compile Pre-Trained Model
MODEL = "cardiffnlp/twitter-roberta-base-sentiment"
tokenizer = AutoTokenizer.from_pretrained(MODEL)
model = AutoModelForSequenceClassification.from_pretrained(MODEL)
# Encode text
encoded_text = tokenizer(example, return_tensors="pt")
encoded_text
# Run the model
ex_score_roberta = model(**encoded_text)
ex_score_roberta
# Apply softmax
ex_score_roberta = ex_score_roberta[0][0].detach().numpy()
ex_score_roberta = softmax(ex_score_roberta)
ex_score_roberta = {
"roberta_neg": ex_score_roberta[0],
"roberta_neu": ex_score_roberta[1],
"roberta_pos": ex_score_roberta[2],
}
ex_score_roberta
print(example)
print("Score by VADER:", ex_score)
print("Score by Roberta:", ex_score_roberta)
# ### ** We can see Roberta accounts for context and is much more powerful than simple models like VADER **
# # Final Model Combine
def polarity_scores_roberta(text):
encoded_text = tokenizer(text, return_tensors="pt")
score = model(**encoded_text)
score = score[0][0].detach().numpy()
score = softmax(score)
score = {
"roberta_neg": score[0],
"roberta_neu": score[1],
"roberta_pos": score[2],
}
return score
result_score = {}
for i, row in tqdm(df_subset.iterrows(), total=len(df_subset)):
try:
# VADER
vader_score = sia.polarity_scores(row["Text"])
new_vader_score = {}
for key, value in vader_score.items():
new_vader_score[f"vader_{key}"] = value
# Roberta
roberta_score = polarity_scores_roberta(row["Text"])
combine_score = {**new_vader_score, **roberta_score}
result_score[i] = combine_score
except:
print(f"Broke for Id {i} - can't be handled by Roberta")
# Join DataFrame with Scores
result_df = pd.DataFrame(result_score).T
final_df = df_subset.merge(result_df, how="left", left_index=True, right_index=True)
final_df.head()
# # Inferences
sns.pairplot(
data=final_df,
vars=[
"vader_neg",
"vader_neu",
"vader_pos",
"roberta_neg",
"roberta_neu",
"roberta_pos",
],
hue="Score",
)
plt.show()
# **Roberta model exhibited a high kurtosis when compared to the VADER model. This indicates that the Roberta model is more confident in classifying sentiments.**
false_positives = final_df[
(final_df.Score == 1) & (final_df.roberta_pos > 0.5)
].sort_values("roberta_pos", ascending=False)[
["Score", "Text", "vader_pos", "roberta_pos"]
]
false_positives
for text in false_positives.Text:
print(text, "\n")
# **Findings:**
# - All comments even though has 5 stars, is actually saracsting and negative than positive, so Roberta has managed to classified all of them correctly.
false_negatives = final_df[
(final_df.Score == 5) & (final_df.roberta_neg > 0.5)
].sort_values("roberta_neg", ascending=False)[
["Score", "Text", "vader_neg", "roberta_neg"]
]
false_negatives
for text in false_negatives.Text:
print(text, "\n")
# **Findings:**
# - Both comments even though have 1 star, is more on the positive side, but Roberta failed to classify it correctly, and that too with a huge confident error of 96%. Here VADER has performed better in both situation.
True_positives = (
final_df[(final_df.Score == 5) & (final_df.roberta_pos > 0.5)]
.sort_values("roberta_pos", ascending=False)[
[
"Score",
"Text",
"vader_neg",
"vader_neu",
"vader_pos",
"roberta_neg",
"roberta_neu",
"roberta_pos",
]
]
.head(5)
)
True_positives
for text in True_positives.Text:
print(text, "\n")
# **Findings:**
# - Even though all comments are full of positivity, the VADER's bag of words has failed to capure it.
True_negative = (
final_df[(final_df.Score == 1) & (final_df.roberta_neg < 0.5)]
.sort_values("roberta_neg", ascending=False)[
[
"Score",
"Text",
"vader_neg",
"vader_neu",
"vader_pos",
"roberta_neg",
"roberta_neu",
"roberta_pos",
]
]
.head(5)
)
True_negative
for text in True_negative.Text:
print(text, "\n")
# **Findings:**
# - All comments are Negative, Both Roberta and VADER were inclined to classify it as neutral sentiments instead, while VADER being highly confident.
# # Evaluation of Models
temp = final_df[["Score", "vader_neg", "vader_pos", "roberta_neg", "roberta_pos"]]
temp
# Assuming lower scoring as a negative sentiment.
temp["isPosScore"] = temp["Score"].apply(lambda x: 1 if x > 3 else 0)
temp["isPosVader"] = temp["vader_pos"].apply(lambda x: 1 if x > 0.5 else 0)
temp["isPosRoberta"] = temp["roberta_pos"].apply(lambda x: 1 if x > 0.5 else 0)
temp
from sklearn.metrics import confusion_matrix, classification_report
print("VADER\n")
print(confusion_matrix(temp["isPosScore"], temp["isPosVader"]))
print(classification_report(temp["isPosScore"], temp["isPosVader"]))
print("ROBERTA\n")
print(confusion_matrix(temp["isPosScore"], temp["isPosRoberta"]))
print(classification_report(temp["isPosScore"], temp["isPosRoberta"]))
# #### The Accuracy of Roberta model is 87%, which is significantly higher compared to Vader model with merely 19% of accuracy
# # Using Pipelines
from transformers import pipeline
sentiment_pipeline = pipeline("sentiment-analysis")
sentiment_pipeline("How can I be good, if you are great?")
|
# # Big dataset in .csv or .tsv processing with [Dask](http://https://docs.dask.org/en/latest/) library
# ### Motivation
# Data in the form of tables has become a standard in Data Science. It is easy to process and analyse it.
# **But what if you have really a huge data set for every day work, and you need to process tons of files?
# **
# Imagine, you have 5 million records generated every day and you need to process them for production. You may also need to work with them throughout the day - to formulate new hypotheses or analysis.
# This task was set for me to pass one of the stages of the interview - a test task.
# I need to open file in the .tsv format (analogous to .csv) and carry out the following operations:
# 1. Scale test data using train data. Process some columns.
# 2. Find the maximum index value and write the index in a new column.
# To simplify I will show results of my experiments immediately
# # 1. Huge csv file generation.
# ## Skip this stage and use files in the Kaggle input folder unless you want to repeate my experiment
# Here we are preparing a test file (random.csv) of size 9G. This is 5 million records of random numbers (from 100 to 1 million) with 257 columns. Finally we get a table of 5 million for 257 random integers = 1 billion 285 million values.
# **Creating a DataFrame took 5 seconds **, uploading it to .csv format using Pandas (to_csv) - 14 minutes!
# **
# An attempt to open this file using the system or Pandas failed. ( So use files in Kaggle input directory to run the code.
import numpy as np
import pandas as pd
from dask_ml.preprocessing import StandardScaler
import gc
import time
import dask.dataframe as dask
from dask.distributed import Client, progress
# ## You can run LOCALY Dask Dashboard to track the perfomance
# set workers
client = Client(n_workers=2, threads_per_worker=2, memory_limit="2GB")
client # work locally only
# setting the number of rows for the CSV file
start_time = time.time()
N = 5_000_000
columns = 257
# create DF
df = pd.DataFrame(
np.random.randint(999, 999999, size=(N, columns)),
columns=["level_%s" % i for i in range(0, columns)],
)
print("%s seconds" % (time.time() - start_time))
display(df.head(2))
print(f"shape of generated data is {df.shape}")
# # 2. Uncomment and run you want to wait more that 15 minutes.
# # Strongly recommend to Use files in Kaggle input directory!
# # save df to csv
# start_time = time.time()
# df.to_csv('random.csv', sep=',')
# print('%s seconds' % (time.time() - start_time)) # 877.5422155857086 seconds, 8.9 G
# ## Set our small size files path
test = "../input/test.tsv"
train = "../input/train.tsv"
# # 3. Class to load, transform data with Dask
class LoadBigCsvFile:
"""load data from tsv, transform, scale, add two columns
Input .csv, .tsv files
Output transformed file ready to save in .csv, .tsv format
"""
def __init__(self, train, test, scaler=StandardScaler(copy=False)):
self.train = train
self.test = test
self.scaler = (
scaler # here we use StandartScaler of Dask. We can use sklearn one
)
def read_data(self):
# use dask and load with smallest possible format - int16 using 'C'
try:
data_train = dask.read_csv(
self.train, dtype={n: "int16" for n in range(1, 300)}, engine="c"
).reset_index()
data_test = dask.read_csv(
self.test, dtype={n: "int16" for n in range(1, 300)}, engine="c"
).reset_index()
except:
(IOError, OSError), "can not open file"
# if any data?
assert len(data_test) != 0 and len(data_train) != 0, "No data in files"
# fit train and transform test
self.scaler.fit(data_train.iloc[:, 1:])
del data_train # del file that we do not need
test_transformed = self.scaler.transform(data_test.iloc[:, 1:])
# compute values and add columns
test_transformed["max_feature_2_abs_mean_diff"] = abs(
test_transformed.mean(axis=1) - test_transformed.max(axis=1)
)
test_transformed["max_feature_2_index"] = test_transformed.idxmin(axis=1)
test_transformed["job_id"] = data_test.iloc[
:, 0
] # add first column (it is not numerical)
del data_test # del file that we do not need
return test_transformed
# # 4. Let run class instance and track the time
start_time = time.time()
data = LoadBigCsvFile(train, test).read_data()
gc.collect()
print("class loaded in %s seconds" % (time.time() - start_time))
# # 5. Data is ready to save. But what format to choose?
# ## Here we use hdf format - why? please the final test results
# save to hdf for later use or modification
start_time = time.time()
data.to_hdf("test_proc.hdf", key="df1")
print("file saved in hdf in %s seconds" % (time.time() - start_time))
# # 6. Load created file again into the system.
start_time = time.time()
hdf_read = dask.read_hdf("test_proc.hdf", key="df1", mode="r", chunksize=10000)
print("file load into system in %s seconds" % (time.time() - start_time))
display(hdf_read.head(3))
|
# This Python 3 Analysis of NUFORC's database contains 80,0000 rows of reported sightings.
# I'm so glad this one came with Lat/Long,s for me to play with.
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Me: [LinkedIn](https://www.linkedin.com/in/dontadaya/)
# Product for [Mr. Tim Ventura](https://www.youtube.com/channel/UCl1ewEFZwBOTU2I_f0XiSUA) & UAP enthusiasts.
# Feel free to fork, cite, manipulate, scrutinize, comment, and provide insights you deem worthy.
# # Let's load NUFORC's 80K dataset as a csv.
# ## Let's all say thanks to [NUFORC](https://nuforc.org/databank/) for sharing the data!
# ### And most especially a tremendous applause to [Mr. Omariba Collins](https://www.linkedin.com/in/omariba-collins-b28b841b9/) for bringing this phat csv into Kags.
# I hope this serves you something delightful.
# ### What does the data look like?
df = pd.read_csv("/kaggle/input/ufo-sightings/ufos.csv", header=0)
df.datetime
df.head(25)
# Fix datetime column.
# df[''] = pd.to_datetime(df['datetime'], format='%m%b%Y:%H:%M:%S.%f', errors='coerce')
# df['datetime'] = pd.to_datetime(df['datetime'], format='%m/%d/%y %H:%M')
# import datetime as dt
# df['datetime'] = df['datetime'].apply(lambda x: dt.datetime.strptime(x,'%m/%d/%Y %H:%M'))
# ### Duration, Lat, Lon - numerical insight.
#
df.describe()
# #### Average duration of sighting: 9.016889e+03 seconds, or 150.28148333 mins.
# #### The Lat/Lon: 72.700000, 178.441900 - may or may not be significant. We'll see later.
# ## Map it oat.
import pandas as pd
from shapely.geometry import Point
import geopandas as gpd
from geopandas import GeoDataFrame
geometry = [Point(xy) for xy in zip(df["longitude"], df["latitude"])]
gdf = GeoDataFrame(df, geometry=geometry)
# This is a simple map that goes with geopandas
world = gpd.read_file(gpd.datasets.get_path("naturalearth_lowres"))
gdf.plot(ax=world.plot(figsize=(50, 25)), marker="o", color="darkred", markersize=15)
|
# # **Online Shop Customer Sales Data**
# 
# The dataset we have is a collection of online shopping transaction records, containing information about the age, gender, time spent on the website, payment method used, browser, newsletter subscription, and the total purchase value.
# **The aim of this dataset is to identify any patterns and insights that can be gleaned from this data to better understand consumer behavior and to help companies tailor their marketing and sales strategies accordingly.**
# In this analysis, we will be performing exploratory data analysis to understand the distribution of the data, identifying any outliers and missing values, and visualizing the relationships between different variables in the dataset.
# We will also be performing statistical analysis to understand the impact of different variables on the total purchase value, which will help us identify key factors that can be optimized to improve revenue.
# Importing required librries
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
data = pd.read_csv(
"/kaggle/input/online-shop-customer-sales-data/Online Shop Customer Sales Data.csv"
)
# # **Understanding Dataset**
data.columns
# Here is a brief description of each variable:
# - **Customer_id:** A unique identifier for each customer.
# - **Age:** The age of the customer.
# - **Gender:** A binary variable where 0 represents male and 1 represents female.
# - **Revenue_Total:** Total sales revenue by the customer.
# - **N_Purchases:** The number of purchases made by the customer to date.
# - **Purchase_DATE:** The date of the latest purchase made by the customer.
# - **Purchase_VALUE:** The value of the latest purchase made by the customer in euros.
# - **Pay_Method:** A categorical variable indicating the payment method used by the customer. The - - categories are digital wallets, card, PayPal, and other.
# - **Time_Spent:** The time spent by the customer on the website in seconds.
# - **Browser:** A categorical variable indicating the browser used by the customer. The categories are Chrome, Safari, Edge, and other.
# - **Newsletter:** A binary variable indicating whether the customer is subscribed to the newsletter or not.
# - **Voucher:** A binary variable indicating whether the customer has used a voucher or not.
data.head()
data.tail()
# # **Revenue Percentage by Age Group**
print(data["Age"].dtype)
print(data["Age"].describe())
# * The dataset has a total of **65,796** customers with ages ranging from **16 to 63**.
# * The mean age of the customers is **39.59**, and the median age (50th percentile) is **40**.
# * The minimum age of customers in the dataset is **16**, while the maximum age is **63**.
sns.histplot(data=data, x="Age", bins=20)
plt.title("Age Distribution")
plt.xlabel("Age")
plt.ylabel("Frequency")
plt.show()
# Create age bins
age_bins = [15, 25, 35, 45, 65]
age_labels = ["16-25", "26-35", "36-45", "46-63"]
data["Age_Group"] = pd.cut(data["Age"], bins=age_bins, labels=age_labels)
# Group the data by age group and calculate total revenue
age_revenue = data.groupby("Age_Group")["Revenue_Total"].sum().reset_index()
# Calculate the percentage of total revenue generated by each age group
age_revenue["Revenue_Percentage"] = (
age_revenue["Revenue_Total"] / age_revenue["Revenue_Total"].sum() * 100
)
# Print the results
print(age_revenue)
import matplotlib.pyplot as plt
# Set up the figure and axis
fig, ax = plt.subplots()
# Create the pie chart
ax.pie(
age_revenue["Revenue_Percentage"],
labels=age_revenue["Age_Group"],
autopct="%1.1f%%",
startangle=90,
)
# Add a title
ax.set_title("Revenue Percentage by Age Group")
# Show the plot
plt.show()
# * Based on the analysis of revenue by age group, we can see that the highest revenue generation is from customers between **the age group of 46-63, with a revenue percentage of 37.77%.**
# * The revenue percentage for the other age groups is relatively similar, with 20.58% for 16-25, 20.63% for 26-35, and 21.01% for 36-45.
#
# * This indicates that the online shop is catering to a wide range of age groups and has a relatively even distribution of revenue among them, with the exception of the age group 46-63, which generates the highest revenue. This information can be useful for targeted marketing and advertising efforts towards the age group that generates the highest revenue.
# # **Distribution of Customers by Gender**
gender_counts = data["Gender"].value_counts()
total_customers = gender_counts.sum()
male_percent = (gender_counts[0] / total_customers) * 100
female_percent = (gender_counts[1] / total_customers) * 100
print(f"Male percentage: {male_percent:.2f}%")
print(f"Female percentage: {female_percent:.2f}%")
import matplotlib.pyplot as plt
gender_counts = data["Gender"].value_counts()
labels = ["Male", "Female"]
plt.pie(gender_counts, labels=labels, autopct="%1.1f%%")
plt.title("Distribution of Customers by Gender")
plt.show()
# Based on the pie chart of the gender distribution, we can see that around **67.1% of the customers in this dataset are female**, while 32.9% are male.
# **This indicates that the online shop is more popular among female customers.**
# # **Total Revenue by Gender**
gender_revenue = data.groupby("Gender")["Revenue_Total"].sum().reset_index()
gender_revenue["Revenue_Percentage"] = (
gender_revenue["Revenue_Total"] / gender_revenue["Revenue_Total"].sum() * 100
)
print(gender_revenue)
plt.pie(gender_revenue["Revenue_Total"], labels=["Male", "Female"], autopct="%1.1f%%")
plt.title("Total Revenue by Gender")
plt.show()
# The insights we can gather from the gender and total revenue percentage data are:
# * The total revenue generated from female customers is higher than that generated from male customers, with a revenue percentage of 67.11% for females and 32.89% for males.
# * This could be an indication that the online shop has a larger female customer base than male.
# # **No of purchases**
print(data["N_Purchases"].describe())
# * The mean number of purchases made by customers is around 4.
# * The standard deviation is relatively low, suggesting that most customers make a similar number of purchases.
# * The minimum number of purchases is 1, which means there are customers who have made only one purchase.
# * The maximum number of purchases is 7, suggesting that there are no customers who have made more than 7 purchases.
# * The median number of purchases is 4, indicating that half of the customers have made 4 or fewer purchases, and the other half have made 4 or more purchases.
# group the data by age group and aggregate the max and min values of N_Purchases
age_purchase = data.groupby("Age_Group")["N_Purchases"].agg(["min", "max"])
# print the result
print(age_purchase)
# # **Total Revenue by Month**
print(data["Purchase_DATE"].dtype)
print(data["Purchase_DATE"].describe())
data["Purchase_DATE"] = pd.to_datetime(data["Purchase_DATE"], format="%d.%m.%y")
print(data["Purchase_DATE"].dtype)
print(data["Purchase_DATE"].describe())
import pandas as pd
import matplotlib.pyplot as plt
# Load data
data = pd.read_csv(
"/kaggle/input/online-shop-customer-sales-data/Online Shop Customer Sales Data.csv"
)
# Convert purchase date to datetime format
data["Purchase_DATE"] = pd.to_datetime(data["Purchase_DATE"], format="%d.%m.%y")
# Aggregate data by month and calculate total revenue
monthly_revenue = data.groupby(pd.Grouper(key="Purchase_DATE", freq="M"))[
"Revenue_Total"
].sum()
# Plot monthly revenue
fig, ax = plt.subplots(figsize=(10, 6))
monthly_revenue.plot(kind="line", ax=ax)
ax.set_xlabel("Month")
ax.set_ylabel("Total Revenue (€)")
ax.set_title("Total Revenue by Month")
plt.show()
monthly_revenue
# Insights from the monthly revenue:
# * The highest monthly revenue was achieved in December 2021 with a total of 157438.0 units of revenue.
# * The lowest monthly revenue was achieved in January 2022 with a total of 4890.5 units of revenue.
# * The revenue seems to fluctuate over the year but with some general stability.
monthly_revenue.median()
# **The median monthly revenue for the given period is 151661.3.**
# This means that 50% of the monthly revenues are below this value and 50% of the monthly revenues are above this value. It is a useful metric to understand the central tendency of the data, as it is less affected by extreme values than the mean.
# checking starting and end date
print(data["Purchase_DATE"].min())
print(data["Purchase_DATE"].max())
# # **Purchase_VALUE**
print(data["Purchase_VALUE"].dtype)
print(data["Purchase_VALUE"].describe())
# The minimum purchase value is 0.005 and the maximum purchase value is 59.90.
# # **Payment Method Distribution**
print(data["Pay_Method"].dtype)
data["Pay_Method"].unique()
# * 0: Digital Wallets
# * 1: Card
# * 2: PayPal
# * 3: Other
# Count the number of each payment method
payment_counts = data["Pay_Method"].value_counts()
# Convert the counts to percentages
payment_percents = payment_counts / len(data) * 100
payment_percents
import matplotlib.pyplot as plt
# count the number of occurrences of each payment method
counts = data["Pay_Method"].value_counts()
# create a pie chart with labels for each payment method
labels = ["Digital Wallets", "Card", "PayPal", "Other"]
plt.pie(counts, labels=labels, autopct="%1.1f%%")
# add a title
plt.title("Payment Method Distribution")
# display the chart
plt.show()
# * Based on the data, the most commonly used payment method is **card**, accounting for **30.02%** of all purchases.
# * The next most popular method is digital wallets, accounting for 28.99% of purchases.
# * PayPal is the third most common method, accounting for 22.12% of purchases.
# * Other payment methods, which may include cash, bank transfers, or other types of payment, account for 18.87% of purchases.
# **It's important for businesses to understand the preferred payment methods of their customers in order to provide the best possible customer experience and increase sales.**
print(data["Time_Spent"].describe())
# * Time_Spent = time spent (in sec) on website
# * The average time spent is approximately **599 seconds (or around 10 minutes)** per purchase.
# * The standard deviation of the time spent is 278 seconds (or around 4.6 minutes) per purchase.
# * The minimum time spent is **120 seconds (or 2 minutes)** per purchase.
# * The 25th percentile of time spent is 358 seconds (or around 6 minutes) per purchase.
# * The median (50th percentile) of time spent is 598 seconds (or around 10 minutes) per purchase.
# * The 75th percentile of time spent is 840 seconds (or around 14 minutes) per purchase.
# * The maximum time spent is **1080 seconds (or 18 minutes)** per purchase.
# Create age bins
age_bins = [15, 25, 35, 45, 65]
age_labels = ["16-25", "26-35", "36-45", "46-63"]
data["Age_Group"] = pd.cut(data["Age"], bins=age_bins, labels=age_labels)
sns.boxplot(x="Age_Group", y="Time_Spent", data=data)
# **Browser**
# * 0: Chrome
# * 1: Safari
# * 2: Edge
# * 3: Other
# # **most commonly used browser**
browser_count = data["Browser"].value_counts()
print(browser_count)
import matplotlib.pyplot as plt
# Count the number of occurrences of each browser
browser_counts = data["Browser"].value_counts()
# Create a pie chart
labels = ["Chrome", "Safari", "Edge", "Other"]
sizes = [
browser_counts.get(0, 0),
browser_counts.get(1, 0),
browser_counts.get(2, 0),
browser_counts.get(3, 0),
]
colors = ["orange", "lightblue", "green", "pink"]
explode = (0.05, 0.05, 0.05, 0.05)
plt.pie(
sizes,
labels=labels,
colors=colors,
explode=explode,
autopct="%1.1f%%",
startangle=90,
)
plt.axis("equal")
# Add a title
plt.title("Percentage of Browsers Used")
# Show the chart
plt.show()
browser_counts
# * **Chrome is the most commonly used browser, followed by Safari, Other and Edge.**
# * It appears that Chrome is significantly more popular than the other browsers, as it has over three times as many users as Safari, and over twelve times as many users as Edge.
# **Newsletter**
# * 0: not subscribed
# * 1: subscribed
# # **Percentage of Newsletter Subscribers**
newsletter_counts = data["Newsletter"].value_counts()
newsletter_percents = newsletter_counts / newsletter_counts.sum() * 100
labels = ["Not Subscribed", "Subscribed"]
plt.pie(newsletter_percents, labels=labels, autopct="%1.1f%%")
plt.title("Percentage of Newsletter Subscribers")
plt.show()
newsletter_percents
# **84.9% of the customers have not subscribed** to the newsletter, while only 15.1% have subscribed.
# # **Percentage of Customers who have Used Vouchers**#
# * 0: not used
# * 1: used
# Get the voucher counts
voucher_counts = data["Voucher"].value_counts()
voucher_counts
voucher_percents = voucher_counts / voucher_counts.sum() * 100
labels = ["No Voucher", "Used Voucher"]
colors = ["#ff9999", "#66b3ff"]
explode = (0.05, 0)
plt.pie(
voucher_percents,
explode=explode,
labels=labels,
colors=colors,
autopct="%1.1f%%",
startangle=90,
)
plt.axis("equal")
plt.title("Percentage of Customers who have Used Vouchers")
plt.show()
# we can see that there are 75% customers who have not used any voucher, and 25% customers who have used at least one voucher.
# **Newsletter subscription does have any significant impact on voucher usage?**
voucher_pivot = pd.pivot_table(
data, values="Voucher", index="Newsletter", aggfunc=np.mean
)
voucher_pivot
plt.bar(voucher_pivot.index, voucher_pivot["Voucher"] * 100)
plt.xticks([0, 1], ["Not Subscribed", "Subscribed"])
plt.ylabel("Percentage of Voucher Users")
plt.show()
# the percentage of customers who used vouchers is almost the same for both groups, with 0.249485 (24.94%) of customers who didn't subscribe to the newsletter using vouchers and 0.254604 (25.46%) of customers who subscribed to the newsletter using vouchers.
# **Therefore, we can conclude that newsletter subscription does not have a significant impact on voucher usage.**
# # **Analyzing the impact of the browser on newsletter subscription**
import seaborn as sns
browser_newsletter_pivot = pd.pivot_table(
data,
values="Customer_id",
index="Browser",
columns="Newsletter",
aggfunc=lambda x: len(x.unique()),
)
sns.heatmap(browser_newsletter_pivot, annot=True, fmt="g", cmap="Blues")
browser_newsletter_pivot
# The table represents the number of newsletter subscribers and non-subscribers segmented by browser type. It shows that the majority of subscribers and non-subscribers used **Chrome browser.** The second most popular browser for both groups is Safari.
# However, there are differences in the proportion of newsletter subscribers to non-subscribers across browsers.
# For example, the proportion of newsletter subscribers is relatively higher among Safari users compared to non-subscribers. On the other hand, the proportion of non-subscribers is higher among Edge and Other browser users compared to newsletter subscribers.
# This suggests that the impact of newsletters on voucher usage may vary depending on the browser type.
# # **Newsletter subscription impact on Revenue**
import seaborn as sns
import matplotlib.pyplot as plt
sns.boxplot(x="Newsletter", y="Revenue_Total", data=data)
plt.show()
newsletter_revenue = data.groupby("Newsletter")["Revenue_Total"].agg(["mean", "std"])
print(newsletter_revenue)
# The insights from this output are:
# * On average, customers who subscribed to the newsletter **(Newsletter = 1)** have a slightly higher mean Revenue_Total than those who did not **(Newsletter = 0)**, **but the difference is not significant.**
# * The standard deviation of Revenue_Total is almost the same for both groups, indicating that there is not much variation in the Revenue_Total values within each group.
# * **Overall, the Newsletter subscription status does not seem to have a strong impact on Revenue_Total.**
# # **Payment Method by Age Group**
import matplotlib.pyplot as plt
# group the data by age group and payment method
age_pay = data.groupby(["Age_Group", "Pay_Method"])["Customer_id"].count().unstack()
# plot the stacked bar chart
age_pay.plot(kind="bar", stacked=True)
# add labels and title
plt.title("Payment Method by Age Group")
plt.xlabel("Age Group")
plt.ylabel("Count")
plt.xticks(rotation=0)
# show the plot
plt.show()
age_pay
# For all age groups, the most common payment method is **method 1 (card)**, followed by method 2 (PayPal), method 0 (digital wallets), and method 3 (other). We can also see that the number of payments made using method 0 is fairly consistent across all age groups.
# However, the number of payments made using method 1 (card) and method 2 (PayPal) is **highest** in the age group **46-63**.
import matplotlib.pyplot as plt
# Convert Date column to datetime format
data["Purchase_DATE"] = pd.to_datetime(data["Purchase_DATE"])
# Group data by Date and calculate the mean of Time_Spent
time_spent = data.groupby("Purchase_DATE")["Time_Spent"].mean()
# Create lineplot
plt.figure(figsize=(10, 6))
plt.plot(time_spent.index, time_spent.values)
plt.title("Time Spent over Time")
plt.xlabel("Date")
plt.ylabel("Time Spent (seconds)")
plt.show()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
ratings_df = pd.read_csv("/kaggle/input/movie-lens-small-latest-dataset/ratings.csv")[
["movieId", "userId", "rating"]
]
movies_df = pd.read_csv("/kaggle/input/movie-lens-small-latest-dataset/movies.csv")
movieids = list(ratings_df["movieId"].value_counts()[:100].keys())
newdf = ratings_df[ratings_df.movieId.isin(movieids) == True]
d_movie_id = {}
for i in range(100):
d_movie_id[movieids[i]] = i
ids = newdf["movieId"].values
newids = [d_movie_id[i] for i in ids]
newdf["Index"] = newids
newdf
oldid_newid = [
[list(d_movie_id.keys())[i], list(d_movie_id.values())[i]] for i in range(100)
]
rows = [0] * 100
Matrix = [rows.copy() for i in range(592)]
Matrix = np.array(Matrix)
Matrix.shape
# 592 : Number of users
# 100 : Number of movies
# 592 vectors, each vector of length 100, denoting 100 movies rating given by them
# if say row 0 , column 7 has value 6, then user denoted by index 0(row) has given movie denoted by index 7 ( column ) a rating of 6.
# This will form a list of vectors
# * : user 1
# * : user 2
# * ..
# * ..
# * : user 592
# Now if we train autoencoders on these 592 vectors , where it reconstructs the input , it will learn similarities in rating patterns.
# Now if a user inputs a new data then the network tries to predict what will his rating record in future be , as learnt from the 592 users. ie a a single vector with length 100.
# ....
#
userids = list(newdf["userId"].value_counts().keys())
d_user_id = {}
for i in range(592):
d_user_id[userids[i]] = i
userids = newdf["userId"].values
newids = [d_user_id[i] for i in userids]
newdf["userIndex"] = newids
newdf
x = newdf.iloc[:, 2:].values
for i in x:
rating = i[0]
movie = i[1]
user = i[2]
Matrix[int(user)][int(movie)] = rating
colsum = [0] * 100
colcount = [0] * 100
for i in range(592):
for j in range(100):
colsum[j] += Matrix[i][j]
if Matrix[i][j] != 0:
colcount[j] += 1
colavg = np.array([colsum[i] / colcount[i] for i in range(100)])
rowsum = [0] * 592
rowcount = [1] * 592
for i in range(592):
for j in range(100):
rowsum[i] += Matrix[i][j]
if Matrix[i][j] != 0:
rowcount[i] += 1
rowavg = np.array([rowsum[i] / rowcount[i] for i in range(592)])
for i in range(592):
for j in range(100):
if Matrix[i][j] == 0:
Matrix[i][j] = round((rowavg[i] + colavg[j]) / 2, 2)
np.save("MovieLens.npy", Matrix)
nameids = []
moviename = movies_df.iloc[:, :-1].values
for i in oldid_newid:
movid = i[0]
for j in moviename:
if j[0] == movid:
i.append(j[1])
nameids.append(i)
indexes = [x[1] for x in nameids]
names = [x[2] for x in nameids]
df = pd.DataFrame()
df["Index"] = indexes
df["Names"] = names
df.set_index("Index", inplace=True)
df.to_csv("Index_name.csv")
df
# Question : How many of you know about tensorflow?
import tensorflow as tf
# As we saw, an autoencoder model have three main layers. So, let's start building the first layer i.e. the input layer.
# Okay, now how do we arrive at the shape of the input layer??
Matrix.shape
# Columns are the features, right? So, what should be the shape?
input_layer = tf.keras.layers.Input(shape=(100,))
# What is keras and how is it different from tensorflow?
# The next layer is hidden layer. We saw that our data has 100 features which means input layer has 100 neurons. Let's compress it now. Here, we are going to compress to 50 neurons.
hidden_layer = tf.keras.layers.Dense(50, activation="relu")(input_layer)
# What is activation and why did we choose relu?
# Do you know about Dense or a fully connected layer?
# Next and the final layer is output layer. We need the output in the same size as the input.
output_layer = tf.keras.layers.Dense(100, activation="sigmoid")(hidden_layer)
# Again, why sigmoid?
scaled_output_layer = tf.keras.layers.Lambda(lambda x: x * 4 + 1)(
output_layer
) # Scale output to range from 1 to 5
# As we saw, sigmoid gives output in the range of 0 to 1. We are dealing with ratings right? So, we have to scale it so that it remains in the range of 1 to 5.
# Now that we have built our layers, let's put them together to create a model that takes an input and produces an output.
autoencoder = tf.keras.models.Model(inputs=input_layer, outputs=scaled_output_layer)
# Next step is to compile our model and why should we do that??
# This is where we specify about the optimizers, the loss function we are trying to minimize etc.
# This step is necessary for training.
autoencoder.compile(optimizer="adam", loss="mse")
# Now we are going to train our model with our dataset. We use the fit function for that. Before that, is it wise to train our model on the whole data??
# We can train our model on 80 percent of the data and keep the rest 20 percent for testing.
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(Matrix, Matrix, test_size=0.2)
# Okay, now we have to train. What we are gonna do is, we are going to divide our data into small batches and train. Why??
# Let's keep the batch size as 32 , which means that at each pass/update 32 samples are taken.
# Epoch is a pass through the entire training dataset. So, assume we have 100 samples in our data and our batch size is 32. How many batches will an epoch contain??
autoencoder.fit(Matrix, Matrix, epochs=100, batch_size=32)
# As we can see the model is performing so terrible. We are working on real time data and that is not our concern now right? There are several ways to improve the performance of the model, which we encourage you guys to read on and update us. Due to time constraint, let's predict with our model and print the top 10 movies.
# Let's pass the test data to our model and let it predict.
pred = autoencoder.predict(x_test)
print(pred)
# What is our model giving as output??
# We do not need ratings for all the 100 movies for each user right? We only need the top 10. So, let's sort it in descending order and print the top 10 of each user.
# Our output is a numpy array of numpy arrays and we have to sort each numpy array.
# So, we are going to use apply_along_axis and then argsort function.
top_values = np.apply_along_axis(lambda x: np.argsort(x)[-10:], 1, pred)
# This returns the indices of the top 10 movies that a particular user will like. We store it in top_values.
top_values[0] # first user
# Now that we know the indices, we are printing the corresponding movie titles for a particular user.
for i in top_values[0]:
print(df.iloc[i]["Names"])
|
# # SARS-Cov2 seasonality and adaptation are driven by solar activity B
import numpy as np
import pandas as pd
import multiprocessing as mp
import matplotlib.pyplot as plt
import differint.differint as df
import matplotlib.gridspec as gridspec
from PIL import Image
from scipy.signal import find_peaks
from mpl_toolkits.basemap import Basemap
from matplotlib.backends.backend_agg import FigureCanvasAgg as FigureCanvas
###############################################################################
# Visualization functions
###############################################################################
fontsize = 16
def PlotStyle(Axes):
"""
Parameters
----------
Axes : Matplotlib axes object
Applies a general style to the matplotlib object
Returns
-------
None.
"""
Axes.spines["top"].set_visible(False)
Axes.spines["bottom"].set_visible(True)
Axes.spines["left"].set_visible(True)
Axes.spines["right"].set_visible(False)
Axes.xaxis.set_tick_params(labelsize=fontsize)
Axes.yaxis.set_tick_params(labelsize=fontsize)
# # COVID-19 pandemic dynamics follows the fractional sunshine duration rate of change
###############################################################################
# Loading packages
###############################################################################
def GetDayLenght(J, lat):
# CERES model Ecological Modelling 80 (1995) 87-95
phi = 0.4093 * np.sin(0.0172 * (J - 82.2))
coef = (-np.sin(np.pi * lat / 180) * np.sin(phi) - 0.1047) / (
np.cos(np.pi * lat / 180) * np.cos(phi)
)
ha = 7.639 * np.arccos(np.max([-0.87, coef]))
return ha
def GetYearLengths(lat):
return np.array([GetDayLenght(j, lat) for j in range(368)])
def GetDictsBylat(lat, function):
Dict = {}
Dict_n = {}
days = function(lat)
days_n = (days - days.min()) / (days.max() - days.min())
inDict = {}
inDict_n = {}
for k, _ in enumerate(days):
inDict[k] = days[k]
inDict_n[k] = days_n[k]
Dict[0] = inDict
Dict_n[0] = inDict_n
for j in range(1, 4):
localdf = df.GL(j / 3, days, num_points=len(days))
localdf_n = (localdf - localdf.min()) / (localdf.max() - localdf.min())
inDict = {}
inDict_n = {}
for i, _ in enumerate(localdf):
inDict[i] = localdf[i]
inDict_n[i] = localdf_n[i]
Dict[j] = inDict
Dict_n[j] = inDict_n
return Dict, Dict_n
MaxCPUCount = int(0.85 * mp.cpu_count())
def GetDictsBylatDL(lat):
return GetDictsBylat(lat, GetYearLengths)
# Wraper function for parallelization
def GetDataParallel(data, Function):
localPool = mp.Pool(MaxCPUCount)
mData = localPool.map(Function, [val for val in data])
localPool.close()
return mData
###############################################################################
# Loading packages
###############################################################################
dataspots = pd.read_csv("/kaggle/input/sunspots/sunspots.csv")
dataspots["date"] = pd.to_datetime(dataspots["date"])
rollingavgspots = dataspots.groupby("date")["dailysunspots"].mean()
wldata = pd.read_csv(
"/kaggle/input/solar-radiation-spectrum-2018-2023/solarcurrent.csv"
)
wldata = wldata[wldata["irradiance"] > 0]
wbins = [200, 290, 320, 400, 700, 1000, 2500]
wlnames = ["UVA", "UVB", "UVC", "Vis", "NIR", "SWIR"]
sdata = (
wldata.groupby(["date", pd.cut(wldata["wavelength"], wbins)])["irradiance"]
.mean()
.unstack()
)
sdata.columns = wlnames
sdata = sdata.reset_index()
sdata["date"] = pd.to_datetime(sdata["date"])
sdata = sdata.set_index("date")
for val in wlnames:
mean = sdata[val].mean()
std = sdata[val].std()
sdata[val] = [sal if np.abs((sal - mean) / std) < 4 else mean for sal in sdata[val]]
###############################################################################
# Loading packages
###############################################################################
cases_columns = [
"date",
"cases",
"country",
"qry",
"lat",
"long",
"dayofyear",
"year",
"lengthofday",
]
dataam = pd.read_csv("/kaggle/input/covid19-in-the-american-continent/continental2.csv")
dataeu = pd.read_csv("/kaggle/input/covid19-cases/europe.csv")
dataam = dataam[cases_columns]
dataeu = dataeu[cases_columns]
data = pd.concat([dataam, dataeu], axis=0)
data["date"] = pd.to_datetime(data["date"])
data["spots"] = np.array(rollingavgspots.loc[data["date"]])
data["refdate"] = ["2019-12-01 00:00:00" for val in data["date"]]
data["refdate"] = pd.to_datetime(data["refdate"])
data["days_since_initialcase"] = (data["date"] - data["refdate"]).dt.days
lats = data["lat"].unique()
localdicts = GetDataParallel(lats, GetDictsBylatDL)
qryToDL = {}
qryToDL_n = {}
for val, sal in zip(lats, localdicts):
qryToDL[val] = sal[0]
qryToDL_n[val] = sal[1]
data["lengthofdayd03"] = [
qryToDL[val][1][sal] for val, sal in zip(data["lat"], data["dayofyear"])
]
data["lengthofdayd06"] = [
qryToDL[val][2][sal] for val, sal in zip(data["lat"], data["dayofyear"])
]
data["lengthofdayd10"] = [
qryToDL[val][3][sal] for val, sal in zip(data["lat"], data["dayofyear"])
]
data["normspots"] = data["spots"] / data["lengthofday"]
###############################################################################
# Loading packages
###############################################################################
mainindex = sdata.index.intersection(data["date"])
correctqrys = [val for val in data["qry"].unique() if val != "lat==0.0 & long==0.0"]
newdata = data[data["qry"].isin(correctqrys)]
counts = newdata["qry"].value_counts()
highcounts = counts[counts > 700].index
newdata = newdata[newdata["qry"].isin(highcounts)]
newdata = newdata[newdata["date"].isin(mainindex)].copy()
finalqrys = newdata["qry"].unique()
def MakeFeatures(dataset):
for val in wlnames:
dataset[val] = np.array(sdata[val].loc[dataset["date"]])
for val in wlnames:
dataset["norm" + val] = dataset[val] / dataset["lengthofday"]
return dataset
newdata = MakeFeatures(newdata)
###############################################################################
# Loading packages
###############################################################################
largerolling_n = 7 * 8
shortrolling_n = 7 * 2
container = []
dataframes = []
coefs = []
selectedlats = []
dataqrys = []
for k, val in enumerate(finalqrys):
selected = newdata[newdata["qry"] == val]
largerolling = (
selected.groupby("date").mean(numeric_only=True).rolling(largerolling_n).mean()
)
disc = largerolling.corr()["cases"].abs()
nans = largerolling.isna().sum()
for inx in nans.index:
if nans[inx] > int(1.15 * largerolling_n):
disc[inx] = 0
cLat = selected["lat"].mean()
cQry = selected["qry"].iloc[0]
shortrolling = selected.groupby("dayofyear")["cases"].apply(np.median)
shortrolling = shortrolling.to_frame()
lengths = [GetDayLenght(J, cLat) for J in range(0, 366)]
disc_range = [k for k in range(172, 192)]
for i in range(5, 20):
corder = i / 20
diff_dl = df.GL(corder, lengths, num_points=len(lengths))
shortrolling["fractional"] = diff_dl[shortrolling.index - 1]
shortrolling["norm_fractional"] = (
shortrolling["fractional"] - shortrolling["fractional"].min()
) / (shortrolling["fractional"].max() - shortrolling["fractional"].min())
short_disc = (
shortrolling.groupby("norm_fractional")["cases"]
.mean()
.rolling(shortrolling_n)
.mean()
.argmax()
)
if short_disc in disc_range:
coefs.append(corder)
selectedlats.append(cLat)
dataframes.append(shortrolling)
container.append(disc)
dataqrys.append(cQry)
break
###############################################################################
# Loading packages
###############################################################################
fig = plt.figure(figsize=(25, 15))
gs = gridspec.GridSpec(nrows=8, ncols=10)
axs = [fig.add_subplot(gs[0, k]) for k in range(10)]
locationdata = newdata[newdata["qry"].isin(dataqrys)]
gr_locationdata = locationdata.groupby("qry").mean(numeric_only=True)
exampledata = locationdata[locationdata["qry"] == "lat==19.482945 & long==-99.113471"]
exampledata = exampledata.groupby("dayofyear").mean(numeric_only=True)
ex_lengths = [GetDayLenght(J, exampledata["lat"].mean()) for J in range(0, 366)]
ex_colors = [plt.cm.Blues(val) for val in np.linspace(0.5, 1, num=10)]
for k in range(10):
ex_dl = df.GL((k + 1) / 10, ex_lengths, num_points=len(lengths))
exampledata["fractional"] = ex_dl[exampledata.index - 1]
exampledata["norm_fractional"] = (
exampledata["fractional"] - exampledata["fractional"].min()
) / (exampledata["fractional"].max() - exampledata["fractional"].min())
exrolling = (
exampledata.groupby("norm_fractional")["cases"]
.mean()
.rolling(shortrolling_n)
.mean()
)
exrolling.plot(ax=axs[k], color=ex_colors[k], label=str((k + 1) / 10) + "SD")
axs[k].set_ylim([0, 1100])
axs[k].set_xlabel("Normalized \n Fractional Order", fontsize=14)
axs[k].legend(loc=1)
PlotStyle(axs[k])
axs[0].text(
0.01,
0.99,
"A",
size=25,
color="black",
ha="left",
va="top",
transform=axs[0].transAxes,
)
axmp = fig.add_subplot(gs[1:6, 0:7])
m = Basemap(
projection="cyl",
llcrnrlat=-65,
urcrnrlat=80,
llcrnrlon=-180,
urcrnrlon=180,
ax=axmp,
)
m.drawcoastlines(color="gray")
m.fillcontinents(color="gainsboro")
m.drawcountries(color="gray")
parallels = np.arange(-80, 80, 10.0)
# labels = [left,right,top,bottom]
m.drawparallels(parallels, labels=[False, True, True, False], alpha=0.5)
sctr = m.scatter(
gr_locationdata["long"].values,
gr_locationdata["lat"].values,
c=coefs,
s=gr_locationdata["cases"].values / 5,
cmap="Blues_r",
)
axmp.set_frame_on(False)
cbar = plt.colorbar(sctr, location="bottom", aspect=50)
cbar.set_label("Fractional Order", fontsize=fontsize)
axmp.text(
0.01,
0.99,
"B",
size=25,
color="black",
ha="left",
va="top",
transform=axmp.transAxes,
)
axlats = fig.add_subplot(gs[2:8, 7:10])
lat_colors = [plt.cm.Blues(val) for val in np.linspace(0.5, 1, num=len(selectedlats))]
for i, (lt, dfr) in enumerate(zip(selectedlats, dataframes)):
localdf = (
dfr.groupby("norm_fractional")["cases"].mean().rolling(shortrolling_n).mean()
)
localdf = (localdf - localdf.min()) / (localdf.max() - localdf.min())
axlats.plot(
np.array(localdf.index),
(lt + 15 * np.array(localdf)),
alpha=0.05,
color=lat_colors[i],
)
axlats.set_ylabel("Normalized Cases By Latitude", fontsize=14)
axlats.set_xlabel("Normalized \n Fractional Order", fontsize=14)
axlats.text(
0.01,
0.99,
"D",
size=25,
color="black",
ha="left",
va="top",
transform=axlats.transAxes,
)
PlotStyle(axlats)
fulldata = pd.concat(dataframes)
groupdata = fulldata.groupby(pd.cut(fulldata["norm_fractional"], 100)).mean()
inx = [val.right for val in groupdata.index]
axmean = fig.add_subplot(gs[1, 7:10])
axmean.plot(inx, np.array(groupdata["cases"]), color="navy", label="Mean Cases")
axmean.set_ylabel("Normalized \n Cases", fontsize=14)
axmean.set_xlabel("Normalized \n Fractional Order", fontsize=14)
axmean.legend()
axmean.text(
0.01,
0.99,
"C",
size=25,
color="black",
ha="left",
va="top",
transform=axmean.transAxes,
)
PlotStyle(axmean)
ndf = pd.concat(container, axis=1)
ndf.columns = ["qry_" + str(k) for k in range(len(container))]
ndfmean = ndf.T.mean()
datacolumns = [
"dayofyear",
"lengthofday",
"lengthofdayd03",
"lengthofdayd06",
"lengthofdayd10",
"spots",
"UVA",
"UVB",
"UVC",
"Vis",
"NIR",
"SWIR",
"normspots",
"normUVC",
"normVis",
"normSWIR",
]
datanames = [
"DOY",
"SD",
"0.3SD",
"0.6SD",
"SDRC",
"NS",
"UVA",
"UVB",
"UVC",
"Vis",
"NIR",
"SWIR",
"NrNS",
"NrUVC",
"NrVis",
"NrSWIR",
]
cmap = plt.cm.Blues
histaxs = [fig.add_subplot(gs[6 + kk, ii]) for ii in range(7) for kk in range(2)]
for k, cl in enumerate(histaxs):
cnts, values, bars = histaxs[k].hist(
ndf.T[datacolumns[k]].values, bins=50, label=datanames[k]
)
histaxs[k].set_ylim([0, 350])
histaxs[k].set_xlim([0, 1])
alp = values[np.argmax(cnts)]
if (1.25) * alp >= 1:
alp = 1
else:
alp = 1.25 * alp
for i, (cnt, value, bar) in enumerate(zip(cnts, values, bars)):
bar.set_facecolor(cmap(cnt / cnts.max(), alpha=alp))
histaxs[k].set_xlabel("Pearson \n correlation", fontsize=14)
histaxs[k].set_ylabel("Frequency", fontsize=14)
histaxs[k].legend()
PlotStyle(histaxs[k])
histaxs[0].text(
0.01,
0.99,
"E",
size=25,
color="black",
ha="left",
va="top",
transform=histaxs[0].transAxes,
)
plt.tight_layout()
|
# All data are taken at 250 Hz
import time as t
a_s = t.time()
# Importing Libraries Requird for the Preprocessing
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import scipy.io
from math import pi
from scipy.fftpack import fft
import scipy.signal as signal
import os
import math
# It will load the data into matrix
def load_data():
f = []
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
f.append(os.path.join(dirname, filename))
return f
# Calling the load function
f = load_data()
# showing the data
for k in f:
print(k)
# this contain two datatset
# Datatset 1 contain sos, stop, and medicine
# Dataset 3 contain come here and washroom
# In extract raw data function we seprated them into Mdata_1 and Mdata_3
def extract_raw_data():
Mdata_1 = []
Mdata_3 = []
for i in range(len(f)):
mat = scipy.io.loadmat(f[i])
col = mat["labels"]
data = mat["data"]
col = [j for j in col]
# pdata is a dataframe
pdata = pd.DataFrame(data, columns=col)
if i < 13:
Mdata_1.append(pdata)
else:
Mdata_3.append(pdata)
return Mdata_1, Mdata_3
# Calling function to extract dataframe into Mdata_1 and Mdata_3
# Mdata_1 contains::
# + SOS - SOS - SOS + STOP - STOP - STOP - + MEDICINE - MEDICINE - MEDICINE
# +: 5sec, words:2sec, blank: 3sec
# Mdata_3 contains::
# + COME_HERE - COME_HERE - COME_HERE + WASHROOM - WASHROOM - WASHROOM
# +: 5sec, words:2sec, blanks:3sec
Mdata_1, Mdata_3 = extract_raw_data()
del Mdata_1[0]
# showing shape of dataframe of each subject(13 subjects)
# for Mdata_1 :
for i in Mdata_1:
print(i.shape)
from numpy import sin, linspace, pi
from pylab import plot, show, title, xlabel, ylabel, subplot
from scipy import fft, arange
def plotSpectrum(y, Fs):
n = len(y) # length of the signal
k = arange(n)
T = n / Fs
frq = k / T # two sides frequency range
frq = frq[range(n // 2)] # one side frequency range
Y = fft(y) / n # fft computing and normalization
Y = Y[range(n // 2)]
plot(frq, 20 * np.log10(abs(Y)), "r") # plotting the spectrum
xlabel("Freq (Hz)")
ylabel("|Y(freq)|")
plt.rcParams["figure.figsize"] = (20, 20)
"""
def notch_filter(x):
fs = 250.0 # Sample frequency (Hz)
f0 = 50.0 # Frequency to be removed from signal (Hz)
Q = 5 # Quality factor
w0 = f0 / (fs / 2 ) # Normalized Frequency
b, a = signal.iirnotch( w0, Q )
# Look at frequency response
w, h = signal.freqz( b, a )
freq = w * fs / ( 2 * np.pi )
#commentrd earlier
#plt.plot( freq, 20*np.log10( abs( h ) ) )
x_fil=signal.lfilter(b,a,x)
plotSpectrum(x_fil,Fs)
"""
def butter_bandstop_filter(data, lowcut, highcut, fs, order):
col = list(Mdata_1[0].columns)
nyq = 0.5 * fs
low = lowcut / nyq
high = highcut / nyq
i, u = signal.butter(order, [low, high], btype="bandstop")
w, h = signal.freqz(i, u)
freq = w * fs / (2 * np.pi)
# commentrd earlier
# plt.plot( freq, 20*np.log10( abs( h ) ) )
y = signal.lfilter(i, u, data)
# y=signal.filtfilt(i,u,data)
# print(type(y))
return y
# plotSpectrum(x_fil,250)
col = list(Mdata_1[0].columns)
# make dataframe of equal size of Mdata_1
for i in range(len(Mdata_1)):
Mdata_1[i] = Mdata_1[i][0:13500]
print(Mdata_1[i].shape)
# showing shape of dataframe of each subject(13 subjects)
# for Mdata_3 :
for i in Mdata_3:
print(i.shape)
# making dataframe of equal size Mdata_3
for i in range(len(Mdata_3)):
Mdata_3[i] = Mdata_3[i][0:8500]
print(Mdata_3[i].shape)
# Collecting the useful data where subject were thinking the words
# for Mdata_1
def seg_data1():
f = 250
u = []
for n in range(len(Mdata_1)):
u1 = pd.DataFrame()
a = 5 * f
for i in range(3):
if i == 1:
a += 2 * f
if i == 2:
a += 5 * f
for j in range(3):
h = Mdata_1[n][a : a + 2 * f]
u1 = pd.concat([u1, h])
a += 5 * f
# print(len(h),len(u1))
u.append(u1)
return u
# Collecting the useful data where subject were thinking the words
# for Mdata_3
def seg_data3():
f = 250
u = []
for n in range(len(Mdata_3)):
u1 = pd.DataFrame()
a = 5 * f
for i in range(2):
if i == 1:
a += 2 * f
for j in range(3):
h = Mdata_3[n][a : a + 2 * f]
u1 = pd.concat([u1, h])
a += 5 * f
u.append(u1)
return u
data1 = seg_data1()
data3 = seg_data3()
"""
org_data11=[]
org_data33=[]
for i in data1:
temp=i.copy()
org_data11.append(temp)
for i in data3:
temp=i.copy()
org_data33.append(temp)
"""
for i in range(len(data1)):
df1 = data1[i]
for j in col:
df1[j] = (df1[j] - df1[j].mean()) / df1[j].std()
org_data1 = []
org_data3 = []
for i in data1:
temp = i.copy()
org_data1.append(temp)
for i in data3:
temp = i.copy()
org_data3.append(temp)
plt.rcParams["figure.figsize"] = (20, 150)
a, b = plt.subplots(30)
loopi = 0
for y in list(data1[0].columns.values):
b[loopi].plot([i for i in range(len(data1[0][y]))], data1[0][y])
loopi += 1
# frequency spectrum plot of subject1
plt.rcParams["figure.figsize"] = (20, 20)
plotSpectrum(data1[0]["ExG1"], 250)
# filter segmented data to remove 50hz noise
import time as t
d_s = t.time()
for i in range(len(data1)):
for j in range(len(col)):
data1[i][col[j]] = butter_bandstop_filter(data1[i][col[j]], 49, 51, 250, 2)
data1[i][col[j]] = butter_bandstop_filter(data1[i][col[j]], 99, 101, 250, 2)
for i in range(len(data3)):
for j in range(len(col)):
data3[i][col[j]] = butter_bandstop_filter(data3[i][col[j]], 49, 51, 250, 2)
data3[i][col[j]] = butter_bandstop_filter(data3[i][col[j]], 99, 101, 250, 2)
plt.rcParams["figure.figsize"] = (20, 20)
plotSpectrum(data1[0]["ExG1"], 250)
import matplotlib.pyplot as plt
import pywt
import sys
# Data format:
# Raw data should be in a .txt file with two columns, separated by tabs:
# - The first column should be a time-series index
# - The second column should contain the data to be filtered
# Get data:
# file_object = open('./Biosignals.txt', 'r')
# raw_data = file_object.readlines()
def waveletdenoising_(y):
raw_data = y
index = [i for i in range(len(raw_data))]
data = raw_data
# Create wavelet object and define parameters
w = pywt.Wavelet("sym4")
maxlev = pywt.dwt_max_level(len(data), w.dec_len)
# maxlev = 2 # Override if desired
# print("maximum level is " + str(maxlev))
threshold = 0.04 # Threshold for filtering
# Decompose into wavelet components, to the level selected:
coeffs = pywt.wavedec(data, "sym4", level=maxlev)
# cA = pywt.threshold(cA, threshold*max(cA))
# plt.figure()
"""
for i in range(1, len(coeffs)):
plt.subplot(maxlev, 1, i)
plt.plot(coeffs[i])
coeffs[i] = pywt.threshold(coeffs[i], threshold*max(coeffs[i]))
plt.plot(coeffs[i])
"""
datarec = pywt.waverec(coeffs, "sym4")
# plt.figure()
# plt.subplot(2, 1, 1)
# plt.plot(index[mintime:maxtime], data[mintime:maxtime])
"""
plt.plot(index,data)
plt.xlabel('time (s)')
plt.ylabel('microvolts (uV)')
plt.title("Raw signal")
plt.subplot(2, 1, 2)
#plt.plot(index[mintime:maxtime], datarec[mintime:maxtime])
plt.plot(index, datarec)
plt.xlabel('time (s)')
plt.ylabel('microvolts (uV)')
plt.title("De-noised signal using wavelet techniques")
plt.tight_layout()
plt.show()
#fdata= datarec[mintime:maxtime];
"""
return datarec
# fs = 500
# f, t, Sxx = signal.spectrogram(data, fs)
# plt.pcolormesh(t, f, Sxx)
# plt.ylabel('Frequency [Hz]')
# plt.xlabel('Time [sec]')
# plt.ylim(0, 50)
# plt.show()
# wavelet denoising
for i in range(len(data1)):
for j in range(len(col)):
data1[i][col[j]] = waveletdenoising_(data1[i][col[j]])
for i in range(len(data3)):
for j in range(len(col)):
data3[i][col[j]] = waveletdenoising_(data3[i][col[j]])
d_e = t.time()
print("Denoising time ", (d_e - d_s))
plotSpectrum(data1[0]["ExG1"], 250)
plt.rcParams["figure.figsize"] = (20, 10)
plt.subplot(2, 1, 1)
plt.plot([i for i in range(len(org_data1[0]["ExG1"]))], org_data1[0]["ExG1"])
plt.subplot(2, 1, 2)
plt.plot([i for i in range(len(data1[0]["ExG1"]))], data1[0]["ExG1"])
MSE = []
SNR = []
for i in range(len(data1)):
g = []
h = []
for c in col:
ch = (((data1[i][c].subtract(org_data1[i][c])).values ** 2).sum(axis=0)) / len(
data1[i]
)
bh = (data1[i][c].values ** 2).sum(axis=0) / (
(((data1[i][c].subtract(org_data1[i][c])).values ** 2).sum(axis=0))
)
bh = 20 * np.log10(bh)
g.append(ch)
h.append(bh)
MSE.append(g)
SNR.append(h)
mse_file = pd.DataFrame(data=MSE, columns=col)
snr_file = pd.DataFrame(data=SNR, columns=col)
snr_file.to_csv("SNR.csv", header=False, index=False)
mse_file.to_csv("MSE.csv", header=False, index=False)
x_sos = np.array([[0] * 15 for i in range(12)])
x_stop = np.array([[0] * 15 for i in range(12)])
x_medicine = np.array([[0] * 15 for i in range(12)])
# Applying pca
from sklearn.decomposition import PCA
pca = PCA(n_components=5)
rx = []
for d in data1:
p_c = pca.fit_transform(data1[i])
tem = pd.DataFrame(data=p_c, columns=["pc1", "pc2", "pc3", "pc4", "pc5"])
rx.append(tem)
c_s = t.time()
sub = 0
rc = list(rx[0].columns)
for d in rx:
k = 0
for j in rc:
x_sos[sub][k] = d[j][0:1500].mean()
x_stop[sub][k] = d[j][1500:3000].mean()
x_medicine[sub][k] = d[j][3000:4500].mean()
x_sos[sub][k + 1] = d[j][0:1500].std()
x_stop[sub][k + 1] = d[j][1500:3000].std()
x_medicine[sub][k + 1] = d[j][3000:4500].std()
x_sos[sub][k + 2] = (d[j][0:1500].values ** 2).sum(axis=0)
x_stop[sub][k + 2] = (d[j][1500:3000].values ** 2).sum(axis=0)
x_medicine[sub][k + 2] = (d[j][3000:4500].values ** 2).sum(axis=0)
k += 3
sub += 1
X = np.concatenate((x_sos, x_stop), axis=0)
X = np.concatenate((X, x_medicine), axis=0)
X.shape
rx[0].shape
Y = [1] * 12 + [2] * 12 + [3] * 12
Y = np.array(Y)
Y = Y.reshape(-1, 1) # Convert data to a single column
from sklearn.preprocessing import OneHotEncoder
encoder = OneHotEncoder(sparse=False)
Y = encoder.fit_transform(Y)
from keras.models import Sequential
from keras.layers import Dense
from sklearn.model_selection import train_test_split
from keras.optimizers import Adam
model = Sequential()
X_train, X_test, Y_train, Y_test = train_test_split(
X, Y, test_size=0.60, random_state=50
)
model.add(Dense(18, input_shape=(15,), activation="relu"))
model.add(Dense(5, activation="relu"))
model.add(Dense(5, activation="relu"))
model.add(Dense(5, activation="relu"))
model.add(Dense(5, activation="relu"))
model.add(Dense(3, activation="softmax"))
optimizer = Adam(lr=0.001)
model.compile(optimizer, loss="categorical_crossentropy", metrics=["accuracy"])
print("Neural Network Model Summary: ")
print(model.summary())
# Train the model
model.fit(X_train, Y_train, verbose=2, batch_size=5, epochs=10)
c_e = t.time()
print("classification time is ", (c_e - c_s))
# Test on unseen data
results = model.evaluate(X_test, Y_test)
print("Final test set loss: {:4f}".format(results[0]))
print("Final test set accuracy: {:4f}".format(results[1]))
a_e = t.time()
print("total time is ", (a_e - a_s))
|
# Load the Required Library
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
# Loading the dataset
df_train = pd.read_csv("/kaggle/input/playground-series-s3e12/train.csv")
df_test = pd.read_csv("/kaggle/input/playground-series-s3e12/test.csv")
df_sub = pd.read_csv("/kaggle/input/playground-series-s3e12/sample_submission.csv")
df_train
df_train.info(), df_test.info(), df_sub.info()
# Checking the information on data to know wht kind of dataset is...or what is the target
df_train = df_train.drop(["id"], axis=1) # Dropping unecessary columns
df_test = df_test.drop(["id"], axis=1)
Y = df_train["target"] # target which is contous in nature mind it
df_train = df_train.drop(["target"], axis=1) # Drop it from actual data
import xgboost as xgb
from sklearn.model_selection import ShuffleSplit
from sklearn.model_selection import KFold, train_test_split
# define the ShuffleSplit cross-validation strategy
cv = ShuffleSplit(n_splits=5, test_size=0.2, random_state=42)
# Define K-fold cross-validation
kf = KFold(n_splits=5, shuffle=True, random_state=42)
# set XGBoost parameters
params = {
"n_estimators": 1000,
"learning_rate": 0.05,
"max_depth": 7,
"subsample": 1.0,
"colsample_bytree": 1.0,
"n_jobs": -1,
"eval_metric": "logloss",
"objective": "binary:logistic",
"verbosity": 0,
"random_state": 0,
}
model = xgb.XGBClassifier(n_estimators=1000, random_state=42)
# initialize the XGBoost model
model1 = xgb.XGBRegressor(**params)
# train and evaluate the model using cross-validation
scores = []
for train_index, test_index in cv.split(df_train):
X_train, X_test = df_train.iloc[train_index], df_train.iloc[test_index]
Y_train, Y_test = Y.iloc[train_index], Y.iloc[test_index]
model1.fit(X_train, Y_train)
score = model1.score(X_test, Y_test)
scores.append(score)
# train and evaluate the model using cross-validation
scores1 = []
for train_index, test_index in kf.split(df_train):
X_train, X_test = df_train.iloc[train_index], df_train.iloc[test_index]
Y_train, Y_test = Y.iloc[train_index], Y.iloc[test_index]
model.fit(X_train, Y_train)
score = model.score(X_test, Y_test)
scores1.append(score)
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import roc_auc_score
# Define the random forest classifier
rfc = RandomForestClassifier(n_estimators=1000, random_state=42)
# Define K-fold cross-validation
kf = KFold(n_splits=5, shuffle=True, random_state=42)
# Define early stopping criteria
best_score = 0
best_iter = 0
val_preds = np.zeros(len(Y))
for i, (train_idx, val_idx) in enumerate(kf.split(df_train, Y)):
X_train, y_train = df_train.iloc[train_idx], Y.iloc[train_idx]
X_val, y_val = df_train.iloc[val_idx], Y.iloc[val_idx]
rfc.fit(X_train, y_train)
y_pred_proba_train = rfc.predict_proba(X_train)[:, 1]
y_pred_proba_val = rfc.predict_proba(X_val)[:, 1]
# Calculate the AUC score on the training and validation sets
train_score = roc_auc_score(y_train, y_pred_proba_train)
val_score = roc_auc_score(y_val, y_pred_proba_val)
print(
f"Fold {i+1}: Training AUC score = {train_score:.3f}, Validation AUC score = {val_score:.3f}"
)
if val_score > best_score:
best_score = val_score
best_iter = i
# Hold all validation predictions for computing overall AUC score
val_preds[val_idx] = y_pred_proba_val
# Train the final model on the full training set with the best number of iterations
rfc.set_params(
n_estimators=(best_iter + 1) * 1000
) # Multiply by 1000 to get the total number of trees
rfc.fit(X_train, y_train)
# Make predictions on the test set
y_pred = rfc.predict(X_test)
y_pred_proba = rfc.predict_proba(X_test)[:, 1]
# Compute overall AUC score on the validation set
val_auc_score = roc_auc_score(Y, val_preds)
print(f"Overall validation AUC score = {val_auc_score:.3f}")
# make predictions on the test data
Y_pred = model.predict_proba(df_test)
Y_pred1 = rfc.predict_proba(df_test)
m = Y_pred * 0.5 + Y_pred1 * 0.5
df_sub.target = m
df_sub.to_csv("submission.csv", index=False)
pd.read_csv("submission.csv")
|
# ### Video Walkthrough:
# - [video](https://youtube.com/shorts/k2jG-ejRt0c?feature=share)
# ## Contents:
# * [Passenger Class (Ordinal Categorical Feature)](#2)
# * [Label Encoding](#21)
# * [One Hot Encoding](#22)
# * [Plot loss vs iterations](#23)
# * [Surname (Nominal Categorical Feature)](#3)
# * [One Hot Encoding](#31)
# * [Optimal Partition](#32)
# * [Plot loss vs iterations](#33)
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
for dirname, _, filenames in os.walk("/kaggle/output"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# ## 0. Import packages
import matplotlib.pyplot as plt
import lightgbm as lgb
import os
import random
# seed everything
SEED = 0
random.seed(SEED)
np.random.seed(SEED)
os.environ["PYTHONHASHSEED"] = str(SEED)
# ## 1. Read data
df_train = pd.read_csv("/kaggle/input/titanic/train.csv")
# ## 2. Passenger class (Ordinal categorical feature)
# ### 2.1 Label encoding
dtrain = lgb.Dataset(
df_train[["Pclass"]],
label=df_train["Survived"],
feature_name=["Pclass"],
init_score=[0] * df_train.shape[0],
)
params = {
"objective": "binary",
"metric": "binary_logloss",
"verbose": 0,
"force_col_wise": True,
"seed": SEED,
"early_stopping_round": 5,
"max_cat_to_onehot": 4,
}
eval_history_pclass_le = lgb.cv(
params, dtrain, nfold=5, eval_train_metric=True, return_cvbooster=True
)
model_pclass_le = eval_history_pclass_le["cvbooster"].boosters[0]
tree_pclass_le = model_pclass_le.trees_to_dataframe()
lgb.create_tree_digraph(
model_pclass_le,
tree_index=0,
show_info=[
"split_gain",
"internal_value",
"internal_count",
"internal_weight",
"leaf_count",
"leaf_weight",
"data_percentage",
],
)
# ### 2.2 One hot encoding
df_train = df_train.assign(Pclass=df_train.Pclass.astype("category"))
dtrain = lgb.Dataset(
df_train[["Pclass"]],
label=df_train["Survived"],
feature_name=["Pclass"],
init_score=[0] * df_train.shape[0],
categorical_feature=["Pclass"],
)
params = {
"objective": "binary",
"metric": "binary_logloss",
"verbose": 0,
"force_col_wise": True,
"seed": SEED,
"early_stopping_round": 5,
"max_cat_to_onehot": 4,
}
eval_history_pclass_ohe = lgb.cv(
params, dtrain, nfold=5, eval_train_metric=True, return_cvbooster=True
)
model_pclass_ohe = eval_history_pclass_ohe["cvbooster"].boosters[0]
tree_pclass_ohe = model_pclass_ohe.trees_to_dataframe()
lgb.create_tree_digraph(
model_pclass_le,
tree_index=0,
show_info=[
"split_gain",
"internal_value",
"internal_count",
"internal_weight",
"leaf_count",
"leaf_weight",
"data_percentage",
],
)
# ### 2.3 Plot Logloss vs iterations
plt.plot(
eval_history_pclass_le["train binary_logloss-mean"],
color="C0",
linestyle="--",
marker="v",
label="le train",
)
plt.plot(
eval_history_pclass_le["valid binary_logloss-mean"],
color="C1",
linestyle="--",
marker="v",
label="le validation",
)
plt.plot(
eval_history_pclass_ohe["train binary_logloss-mean"],
color="C2",
marker="^",
label="ohe train",
)
plt.plot(
eval_history_pclass_ohe["valid binary_logloss-mean"],
color="C3",
marker="^",
label="ohe validation",
)
plt.ylabel("Logloss")
plt.xlabel("Iteration")
plt.legend()
# ## 3. Surname (Nominal categorical feature)
df_train = df_train.assign(
Surname=df_train.Name.str.split(",")
.apply(lambda x: x[0].strip())
.fillna("NA")
.astype("category")
)
print("Number of unique Surnames =", df_train.Surname.nunique())
surname_mapping = dict(zip(df_train["Surname"].cat.codes, df_train["Surname"]))
print(
"Mapping of Surname:",
[
(k, surname_mapping[k])
for i, k in enumerate(sorted(surname_mapping.keys()))
if k < 10
],
)
# ### 3.1 One hot encoding
max_onehot = 700
dtrain = lgb.Dataset(
df_train[["Surname"]],
label=df_train["Survived"],
feature_name=["Surname"],
init_score=[0] * df_train.shape[0],
categorical_feature=["Surname"],
)
params = {
"objective": "binary",
"metric": "binary_logloss",
"verbose": 0,
"force_col_wise": True,
"seed": SEED,
"early_stopping_round": 5,
"max_cat_to_onehot": max_onehot,
"min_data_in_leaf": 1,
}
eval_history_surname_ohe = lgb.cv(
params, dtrain, nfold=5, eval_train_metric=True, return_cvbooster=True
)
model_surname_ohe = eval_history_surname_ohe["cvbooster"].boosters[0]
tree_surname_ohe = model_surname_ohe.trees_to_dataframe()
lgb.create_tree_digraph(
model_surname_ohe,
tree_index=0,
show_info=[
"split_gain",
"internal_value",
"internal_count",
"internal_weight",
"leaf_count",
"leaf_weight",
"data_percentage",
],
)
# ### 3.2 Optimal Partitioning
# https://xgboost.readthedocs.io/en/stable/tutorials/categorical.html#optimal-partitioning
max_onehot = 4
dtrain = lgb.Dataset(
df_train[["Surname"]],
label=df_train["Survived"],
feature_name=["Surname"],
init_score=[0] * df_train.shape[0],
categorical_feature=["Surname"],
)
params = {
"objective": "binary",
"metric": "binary_logloss",
"verbose": 0,
"force_col_wise": True,
"seed": SEED,
"early_stopping_round": 5,
"max_cat_to_onehot": max_onehot,
"min_data_in_leaf": 1,
"min_data_per_group": 1,
"max_cat_threshold": 255,
"cat_l2": 0.0,
"cat_smooth": 3,
}
eval_history_surname_opt = lgb.cv(
params, dtrain, nfold=5, eval_train_metric=True, return_cvbooster=True
)
model_surname_opt = eval_history_surname_opt["cvbooster"].boosters[0]
tree_surname_opt = model_surname_opt.trees_to_dataframe()
lgb.create_tree_digraph(
model_surname_opt,
tree_index=0,
show_info=[
"split_gain",
"internal_value",
"internal_count",
"internal_weight",
"leaf_count",
"leaf_weight",
"data_percentage",
],
)
# ### 3.3 Plot Logloss vs iterations
plt.plot(
eval_history_surname_ohe["train binary_logloss-mean"],
color="C0",
linestyle="--",
marker="v",
label="ohe train",
)
plt.plot(
eval_history_surname_ohe["valid binary_logloss-mean"],
color="C1",
linestyle="--",
marker="v",
label="ohe validation",
)
plt.plot(
eval_history_surname_opt["train binary_logloss-mean"],
color="C2",
marker="^",
label="gb train",
)
plt.plot(
eval_history_surname_opt["valid binary_logloss-mean"],
color="C3",
marker="^",
label="gb validation",
)
plt.ylabel("Logloss")
plt.xlabel("Iteration")
plt.legend()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)+
import matplotlib.pyplot as plt
import re
import re
from sklearn.ensemble import RandomForestClassifier
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
train_dataset = pd.read_csv("/kaggle/input/titanic/train.csv")
train_dataset.head()
test_dataset = pd.read_csv("/kaggle/input/titanic/test.csv")
test_dataset.head()
women = train_dataset.loc[train_dataset.Sex == "female"]["Survived"]
rate_women = sum(women) / len(women)
print("% of women who survived:", rate_women)
men = train_dataset.loc[train_dataset.Sex == "male"]["Survived"]
rate_men = sum(men) / len(men)
print("% of men who survived:", rate_men)
# Import the needed referances
import pandas as pd
import numpy as np
import csv as csv
from sklearn.model_selection import cross_val_score
from sklearn.linear_model import LogisticRegression
from sklearn.svm import SVC, LinearSVC
from sklearn.ensemble import RandomForestClassifier
# Shuffle the datasets
from sklearn.utils import shuffle
# Learning curve
import matplotlib.pyplot as plt
from sklearn.model_selection import learning_curve
from sklearn.model_selection import ShuffleSplit
# import seaborn as sns
# Output plots in notebook
# %matplotlib inline
addpoly = True
plot_lc = 0 # 1--display learning curve/ 0 -- don't display
print(
"train dataset: %s, test dataset %s"
% (str(train_dataset.shape), str(test_dataset.shape))
)
train_dataset.head()
print("Id is unique.") if train_dataset.PassengerId.nunique() == train_dataset.shape[
0
] else print("oops")
print("Train and test sets are distinct.") if len(
np.intersect1d(train_dataset.PassengerId.values, test_dataset.PassengerId.values)
) == 0 else print("oops")
# print('We do not need to worry about missing values.') if train_dataset.count().min() == train_dataset.shape[0] and test_dataset.count().min() == test_dataset.shape[0] else print('oops we have nan')
datasetHasNan = False
if (
train_dataset.count().min() == train_dataset.shape[0]
and test_dataset.count().min() == test_dataset.shape[0]
):
print("We do not need to worry about missing values.")
else:
datasetHasNan = True
print("oops we have nan")
print("----train dataset column types information-------")
dtype_df = train_dataset.dtypes.reset_index()
dtype_df.columns = ["Count", "Column Type"]
dtype_df.groupby("Column Type").aggregate("count").reset_index()
print("----train dataset information-------")
dtype_df
# Check for missing data & list them
if datasetHasNan == True:
nas = pd.concat(
[train_dataset.isnull().sum(), test_dataset.isnull().sum()],
axis=1,
keys=["Train Dataset", "Test Dataset"],
)
print("Nan in the data sets")
print(nas[nas.sum(axis=1) > 0])
# Class vs Survived
print(
train_dataset[["Pclass", "Survived"]]
.groupby(["Pclass"], as_index=False)
.mean()
.sort_values(by="Survived", ascending=False)
)
# sex vs Survived
print(
train_dataset[["Sex", "Survived"]]
.groupby(["Sex"], as_index=False)
.mean()
.sort_values(by="Survived", ascending=False)
)
# SibSp vs Survived
# Sibling = brother, sister, stepbrother, stepsister
# Spouse = husband, wife (mistresses and fiancés were ignored)
print(
train_dataset[["SibSp", "Survived"]]
.groupby(["SibSp"], as_index=False)
.mean()
.sort_values(by="Survived", ascending=False)
)
# Parch vs Survived
# Parent = mother, father
# Child = daughter, son, stepdaughter, stepson
# Some children travelled only with a nanny, therefore parch=0 for them.
print(
train_dataset[["Parch", "Survived"]]
.groupby(["Parch"], as_index=False)
.mean()
.sort_values(by="Survived", ascending=False)
)
# Data sets cleaing, fill nan (null) where needed and delete not needed columns
print("----Strat data cleaning ------------")
# train_dataset['IsMinor'] = 0
# train_dataset.loc[(train_dataset['Age'] < 14) & ((train_dataset['Pclass'] == 1) | (train_dataset['Pclass'] == 2) ), 'IsMinor'] = 1
# test_dataset['IsMinor'] = 0
# test_dataset.loc[(test_dataset['Age'] < 14) & ((test_dataset['Pclass'] == 1 ) | (test_dataset['Pclass'] == 2 )), 'IsMinor'] = 1
# manage Age
train_random_ages = np.random.randint(
train_dataset["Age"].mean() - train_dataset["Age"].std(),
train_dataset["Age"].mean() + train_dataset["Age"].std(),
size=train_dataset["Age"].isnull().sum(),
)
test_random_ages = np.random.randint(
test_dataset["Age"].mean() - test_dataset["Age"].std(),
test_dataset["Age"].mean() + test_dataset["Age"].std(),
size=test_dataset["Age"].isnull().sum(),
)
train_dataset["Age"][np.isnan(train_dataset["Age"])] = train_random_ages
test_dataset["Age"][np.isnan(test_dataset["Age"])] = test_random_ages
train_dataset["Age"] = train_dataset["Age"].astype(int)
test_dataset["Age"] = test_dataset["Age"].astype(int)
# Embarked
train_dataset["Embarked"].fillna("S", inplace=True)
test_dataset["Embarked"].fillna("S", inplace=True)
train_dataset["Port"] = (
train_dataset["Embarked"].map({"S": 0, "C": 1, "Q": 2}).astype(int)
)
test_dataset["Port"] = (
test_dataset["Embarked"].map({"S": 0, "C": 1, "Q": 2}).astype(int)
)
del train_dataset["Embarked"]
del test_dataset["Embarked"]
# Fare
test_dataset["Fare"].fillna(test_dataset["Fare"].median(), inplace=True)
# Feature that tells whether a passenger had a cabin on the Titanic
train_dataset["Has_Cabin"] = train_dataset["Cabin"].apply(
lambda x: 0 if type(x) == float else 1
)
test_dataset["Has_Cabin"] = test_dataset["Cabin"].apply(
lambda x: 0 if type(x) == float else 1
)
# engineer a new Title feature
# group them
full_dataset = [train_dataset, test_dataset]
##engineer the family size feature
for dataset in full_dataset:
dataset["FamilySize"] = dataset["SibSp"] + dataset["Parch"] + 1
### new try
# Create new feature IsAlone from FamilySize
for dataset in full_dataset:
dataset["IsAlone"] = 0
dataset.loc[dataset["FamilySize"] == 1, "IsAlone"] = 1
##############################
# Get titles from the names
train_dataset["Title"] = train_dataset.Name.str.extract(" ([A-Za-z]+)\.", expand=False)
test_dataset["Title"] = test_dataset.Name.str.extract(" ([A-Za-z]+)\.", expand=False)
for dataset in full_dataset:
dataset["Title"] = dataset["Title"].replace(
[
"Lady",
"Countess",
"Capt",
"Col",
"Don",
"Dr",
"Major",
"Rev",
"Sir",
"Jonkheer",
"Dona",
],
"Rare",
)
dataset["Title"] = dataset["Title"].replace("Mlle", "Miss")
dataset["Title"] = dataset["Title"].replace("Ms", "Miss")
dataset["Title"] = dataset["Title"].replace("Mme", "Mrs")
## Create new column "FamilySizeGroup" and assign "Alone", "Small" and "Big"
for dataset in full_dataset:
dataset["FamilySizeGroup"] = "Small"
dataset.loc[dataset["FamilySize"] == 1, "FamilySizeGroup"] = "Alone"
dataset.loc[dataset["FamilySize"] >= 5, "FamilySizeGroup"] = "Big"
## Get the average survival rate of different FamilySizes
train_dataset[["FamilySize", "Survived"]].groupby(["FamilySize"], as_index=False).mean()
for dataset in full_dataset:
dataset["Sex"] = dataset["Sex"].map({"female": 1, "male": 0}).astype(int)
for dataset in full_dataset:
dataset.loc[dataset["Age"] <= 14, "Age"] = 0
dataset.loc[(dataset["Age"] > 14) & (dataset["Age"] <= 32), "Age"] = 1
dataset.loc[(dataset["Age"] > 32) & (dataset["Age"] <= 48), "Age"] = 2
dataset.loc[(dataset["Age"] > 48) & (dataset["Age"] <= 64), "Age"] = 3
dataset.loc[dataset["Age"] > 64, "Age"] = 4
for dataset in full_dataset:
dataset.loc[dataset["Fare"] <= 7.91, "Fare"] = 0
dataset.loc[(dataset["Fare"] > 7.91) & (dataset["Fare"] <= 14.454), "Fare"] = 1
dataset.loc[(dataset["Fare"] > 14.454) & (dataset["Fare"] <= 31), "Fare"] = 2
dataset.loc[dataset["Fare"] > 31, "Fare"] = 3
dataset["Fare"] = dataset["Fare"].astype(int)
# map the new features
title_mapping = {"Mr": 1, "Miss": 2, "Mrs": 3, "Master": 4, "Rare": 5}
family_mapping = {"Small": 0, "Alone": 1, "Big": 2}
for dataset in full_dataset:
dataset["Title"] = dataset["Title"].map(title_mapping)
dataset["FamilySizeGroup"] = dataset["FamilySizeGroup"].map(family_mapping)
# engineer a new features
for dataset in full_dataset:
dataset["IsChildandRich"] = 0
dataset.loc[(dataset["Age"] <= 0) & (dataset["Pclass"] == 1), "IsChildandRich"] = 1
dataset.loc[(dataset["Age"] <= 0) & (dataset["Pclass"] == 2), "IsChildandRich"] = 1
# for dataset in full_dataset:
# dataset['Age*Class'] = dataset.Age * dataset.Pclass
# for dataset in full_dataset:
# dataset['Sex*Class'] = dataset.Sex * dataset.Pclass
# for dataset in full_dataset:
# dataset['Sex*Age'] = dataset.Sex * dataset.Age
# for dataset in full_dataset:
# dataset['Age*Class*Sex'] = (dataset.Age * dataset.Pclass) + dataset.Sex
for data in full_dataset:
# classify Cabin by fare
data["Cabin"] = data["Cabin"].fillna("X")
data["Cabin"] = data["Cabin"].apply(lambda x: str(x)[0])
data["Cabin"] = data["Cabin"].replace(["A", "D", "E", "T"], "M")
data["Cabin"] = data["Cabin"].replace(["B", "C"], "H")
data["Cabin"] = data["Cabin"].replace(["F", "G"], "L")
data["Cabin"] = data["Cabin"].map({"X": 0, "L": 1, "M": 2, "H": 3}).astype(int)
# data['Cabin'].loc[~data['Cabin'].isnull()] = 1
# data['Cabin'].loc[data['Cabin'].isnull()] = 0
# Delete Name column from datasets (No need for them in the analysis)
del train_dataset["Name"]
del test_dataset["Name"]
del train_dataset["SibSp"]
del test_dataset["SibSp"]
del train_dataset["Parch"]
del test_dataset["Parch"]
del train_dataset["FamilySize"]
del test_dataset["FamilySize"]
# del train_dataset['FamilySizeGroup']
# del test_dataset['FamilySizeGroup']
del train_dataset["Cabin"]
del test_dataset["Cabin"]
# Delete Ticket column from datasets (No need for them in the analysis)
del train_dataset["Ticket"]
del test_dataset["Ticket"]
del train_dataset["Port"]
del test_dataset["Port"]
# Cabin has a lot of nan values, so i will remove it
# del train_dataset['Cabin']
# del test_dataset['Cabin']
##title_dummies_titanic = pd.get_dummies(train_dataset['Title'])
##train_dataset = train_dataset.join(title_dummies_titanic)
##
##title_dummies_titanic = pd.get_dummies(test_dataset['Title'])
##test_dataset = test_dataset.join(title_dummies_titanic)
##
### Drop
##train_dataset.drop(['Title'], axis=1,inplace=True)
##test_dataset.drop(['Title'], axis=1,inplace=True)
print(
"train dataset: %s, test dataset %s"
% (str(train_dataset.shape), str(test_dataset.shape))
)
train_dataset.head()
del train_dataset["PassengerId"]
# X_train = train_dataset.drop("Survived",axis=1).as_matrix()
# Y_train = train_dataset["Survived"].as_matrix()
# X_test = test_dataset.drop("PassengerId",axis=1).copy().as_matrix()
X_train = train_dataset.drop("Survived", axis=1)
Y_train = train_dataset["Survived"]
X_test = test_dataset.drop("PassengerId", axis=1).copy()
print(X_train.shape)
print(Y_train.shape)
print(X_test.shape)
random_forest = RandomForestClassifier(
criterion="gini",
n_estimators=1000,
min_samples_split=10,
min_samples_leaf=1,
max_features="auto",
oob_score=True,
random_state=1,
n_jobs=-1,
)
seed = 42
random_forest = RandomForestClassifier(
n_estimators=1000,
criterion="entropy",
max_depth=5,
min_samples_split=2,
min_samples_leaf=1,
max_features="auto",
bootstrap=False,
oob_score=False,
n_jobs=1,
random_state=seed,
verbose=0,
)
random_forest.fit(X_train, Y_train)
Y_pred = random_forest.predict(X_test)
result_train = random_forest.score(X_train, Y_train)
result_val = cross_val_score(random_forest, X_train, Y_train, cv=5).mean()
print("taring score = %s , while validation score = %s" % (result_train, result_val))
submission = pd.DataFrame(
{"PassengerId": test_dataset["PassengerId"], "Survived": Y_pred}
)
submission.to_csv("titanic.csv", index=False)
print("Done done done done done!")
|
# **Importing Packages**
import pandas as pd
# For Algorithm
from xgboost import XGBClassifier
# For Internal Evaluation, Hyperparameter Selection and Feature Selection
from sklearn.feature_selection import VarianceThreshold
from sklearn.model_selection import GridSearchCV, train_test_split
from sklearn.metrics import f1_score
# **Reading Data**
# Importing Data
train_rawdata = pd.read_csv("/kaggle/input/generated-data/generated_data_train.csv")
test_rawdata = pd.read_csv("/kaggle/input/generated-data/generated_data_test.csv")
X_train = train_rawdata
Y_train = train_rawdata["label"]
X_train = X_train.drop(["label"], axis=1)
X_test = test_rawdata
# **Replacing Null Values**
# Replacing Null With Mean
X_train = X_train.fillna(X_train.mean())
X_test = X_test.fillna(X_test.mean())
# **Variance Threshold**
# Create VarianceThreshold object with the desired threshold
thresh = 10.0 # set your desired variance threshold
selector = VarianceThreshold(threshold=thresh)
# Fit the selector to the data and transform the data
df_selected = selector.fit_transform(X_train)
# Get the selected feature indices
selected_indices = selector.get_support(indices=True)
# Create a new DataFrame with only the selected features
X_train_new = X_train.iloc[:, selected_indices]
X_test_new = X_test.iloc[:, selected_indices]
# **Splitting Data**
train_X, test_X, train_y, test_y = train_test_split(
X_train_new, Y_train, random_state=0, test_size=0.3
)
# **Using GridSearchCV to find best Hyperparameters (One Time Run)**
# Define the XGBClassifier model
# model = XGBClassifier()
# Define the hyperparameters to be tuned
# param_grid = {
# 'max_depth': [3, 5, 7],
# 'learning_rate': [0.1, 0.2, 0.3],
# 'n_estimators': [50, 100, 150],
# 'min_child_weight': [1, 3, 5]
# }
# Create a GridSearchCV object with 5-fold cross validation
# grid_search = GridSearchCV(
# estimator=model,
# param_grid=param_grid,
# cv=5,
# scoring='accuracy',
# n_jobs=-1
# )
# Fit the GridSearchCV object to the training data
# Y_train_binary = Y_train.map({1: 0, 2: 1})
# grid_search.fit(X_train_new, Y_train_binary)
# Print the best hyperparameters and the corresponding accuracy score
# print(f"Best hyperparameters: {grid_search.best_params_}")
# print(f"Accuracy score: {grid_search.best_score_}")
# Best hyperparameters: {'learning_rate': 0.3, 'max_depth': 7, 'min_child_weight': 3, 'n_estimators': 150}
# Accuracy score: 0.9090378112617017
# **Tweaking the hyperparameters of XGBClassifier with respect to "Best Hyperparameters" to find best F1 Macro Score**
train_y_binary = train_y.map({1: 0, 2: 1})
xgb = XGBClassifier(
learning_rate=0.3, max_depth=7, min_child_weight=3, n_estimators=600
).fit(train_X, train_y_binary)
Y_prediction = xgb.predict(test_X)
Y_prediction_original = pd.Series(Y_prediction).map({0: 1, 1: 2})
macro_score = f1_score(test_y, Y_prediction_original, average="macro")
print(macro_score)
# **Running model based on above hyperparameter**
# XGBoost
Y_train_binary = Y_train.map({1: 0, 2: 1})
xgb = XGBClassifier(
learning_rate=0.3, max_depth=7, min_child_weight=3, n_estimators=600
).fit(X_train_new, Y_train_binary)
Y_prediction = xgb.predict(X_test_new)
# Map predicted values back to original format
Y_prediction_original = pd.Series(Y_prediction).map({0: 1, 1: 2})
submission_file = pd.read_csv(
"/kaggle/input/the-toxicity-prediction-challenge-ii/sample_submission.csv"
)
submission_file["Predicted"] = Y_prediction_original
submission_file.to_csv("/kaggle/working/XGB_Parameter3_VT_10.0.csv.csv", index=False)
print("CSV File Generated")
# **Code To Generate "generated_data_train.csv" and "generated_data_train.csv" (ONE TIME RUN)**
# import warnings
# warnings.simplefilter(action='ignore', category=FutureWarning)
# import numpy as np
# import pandas as pd
# from rdkit import Chem
# import matplotlib.pyplot as plt
# train_data = pd.read_csv("/kaggle/input/the-toxicity-prediction-challenge-ii/train_II.csv")
# print(train_data)
# Extract assay Id and chemical from Id
# def extract_assay_Id_and_chemical(data):
# chemical_compound = []
# assay_ids = []
# for columns in data["Id"]:
# d = columns.split(";")
# chemical = d[0]
# assay_id = int(d[-1])
# chemical_compound.append(chemical)
# assay_ids.append(assay_id)
# return assay_ids, chemical_compound
# assay_ids, chemical_compound = extract_assay_Id_and_chemical(train_data)
# train_data["Chemical_Compound"] = chemical_compound
# train_data["Assay_Id"] = assay_ids
# print(train_data)
# train_data = train_data.drop("Id",axis=1)
# print(train_data)
# train_data.to_csv("/kaggle/input/generated-data/extracted_compounds.csv",index=False)
# data = pd.read_csv("/kaggle/input/generated-data/extracted_compounds.csv")
# print(data)
# count_of_assId = data.Assay_Id.value_counts()
# using RDKit to get molicular descriptions
# from rdkit.Chem import AllChem
# from rdkit import Chem
# from rdkit.Chem import Descriptors
# from rdkit.ML.Descriptors import MoleculeDescriptors
# Function written by referring the following Github repository https://github.com/gashawmg/molecular-descriptors
# calc = MoleculeDescriptors.MolecularDescriptorCalculator([x[0] for x in Descriptors._descList])
# desc_names = calc.GetDescriptorNames()
# generated_data = pd.DataFrame(columns=desc_names)
# mol = Chem.MolFromSmiles(data["Chemical_Compound"][1])
# print(mol)
# smile = Chem.MolToSmiles(mol)
# Calculate Each molecule description and add to column
# i=0
# error_set = []
# for SMILE in data["Chemical_Compound"]:
# if(i%10==0):
# print("Calcualting",i)
# calc = MoleculeDescriptors.MolecularDescriptorCalculator([x[0] for x in Descriptors._descList])
# # Converting molicule to molicular form
# mols = Chem.MolFromSmiles(SMILE)
# # Comverting molecule to canocial form
# description = list(calc.CalcDescriptors(mols))
# generated_data.loc[i] = description
# i = i+1
# Saving data for further detail
# generated_data["label"] = train_data["Expected"]
# generated_data["Assay_id"] = data.Assay_Id
# generated_data.to_csv("/kaggle/input/generated-data/generated_data_train.csv",index=False)
# print(data["Chemical_Compound"][10140:10150])
# test_data = pd.read_csv("/kaggle/input/generated-data/test_II.csv")
# print(test_data)
# def extract_assay_Id_and_chemical_for_test(data):
# chemical_compound = []
# assay_ids = []
# for columns in data.x:
# d = columns.split(";")
# chemical = d[0]
# assay_id = int(d[-1])
# chemical_compound.append(chemical)
# assay_ids.append(assay_id)
# return assay_ids, chemical_compound
# assay_ids, chemical_compound = extract_assay_Id_and_chemical_for_test(test_data)
# test_data["Assay_id"] = assay_ids
# test_data["Chemical_Compound"] = chemical_compound
# test_data = test_data.drop("x",axis=1)
# test_data.to_csv("/kaggle/input/generated-data/test_data_new.csv",index=False)
# test_data = pd.read_csv("/kaggle/input/generated-data/test_data_new.csv")
# print(test_data)
# generated_data_test = pd.DataFrame(columns=desc_names)
# i = 0
# error_set = []
# for SMILE in test_data["Chemical_Compound"]:
# if(i%10 == 0):
# print("Calculating",i)
# calc = MoleculeDescriptors.MolecularDescriptorCalculator([x[0] for x in Descriptors._descList])
# Converting molicule to molicular form
# mols = Chem.MolFromSmiles(SMILE)
# Comverting molecule to canocial form
# description = list(calc.CalcDescriptors(mols))
# generated_data_test.loc[i] = description
# i = i+1
# generated_data_test["Assay_id"] = assay_ids
# generated_data_test.to_csv("/kaggle/input/generated-data/generated_data_test.csv",index=False)
|
# # 1. Tổng quan đề tài
# ## Vấn đề
# - Challenge: [Spaceship Titanic](https://www.kaggle.com/competitions/spaceship-titanic)
# - Dự đoán hành khách nào được vận chuyển đến một chiều không gian khác
# - Bối cảnh: Một con tàu vũ trụ Titanic bắt đầu thực hiện chuyến hành trình đầu tiên vận chuyển những người di cư từ hệ mặt trời của chúng ta đến ba hệ hành tinh mới. Titanic đã bị va chạm với một vật thể, và gặp sự cố. Mặc dù con tàu vẫn còn nguyên vẹn, nhưng một số hành khách đã bị chuyển đến một chiều không gian khác. Bộ dữ liệu train.csv bao gồm thông tin các hành khách mà con tàu biết được tình trạng và gửi về trạm, một số còn lại, test.csv, là chưa rõ có bị dịch chuyển hay không. Yêu cầu dự đoán tình trạng các hành khách còn lại đó.
# ## Tại sao lại chọn đề tài này?
# * Chủ đề này thú vị: tàu vũ trụ, mất tích không gian...
# * Phù hợp các yêu cầu của đồ án môn học này
# ## Mục tiêu
# * Mid-term: Tiền xử lý dữ liệu, phân tích, hiểu được dữ liệu, tìm những mô hình khả thi đối với đề tài này, áp dụng thử một mô hình
# * Final: Áp dụng và đánh giá (các) mô hình, tìm mô hình phù hợp và tối ưu nó
# ## Mô tả dataset:
# Bộ dữ liệu gồm: 2 files: train.csv, test.csv
# * train.csv: Để huấn luyện và đánh giá model. Gồm 8693 mẫu với 14 thuộc tính:
# - 13 thuộc tính mô tả và 1 thuộc tính phân lớp
# * test.csv: Để thực hiện dự đoán kết quả.
# ## Đóng góp
# | MSSV | Họ và tên | Email |
# | --- | --- | --- |
# | 19127597 | Trần Khả Trí | [email protected] |
# | 19127607 | Trần Nguyên Trung | [email protected] |
# | Thời gian | Nhiệm vụ | Thực hiện | Kết quả
# | --- | --- | --- | --- |
# | 26-02-2023 | Research Dataset | Trung, Trí | 100% |
# | 01-03-2023 | Data Description | Trung | 100% |
# | | Data Preprocessing, Data Statistical | Trí | 100% |
# | 05-03-2023 | Data Visualization, Model Hypothesis | Trung | 100% |
# | | Model Implementation(Basic) | Trí | 100% |
# | *13-03-2023* | *Midterm Presentation* | *Trung, Trí* | *100%* |
# | 20-03-2023 | Research Model for prediction | Trung, Trí | 100% |
# | 27-03-2023 | Implement RandomForest, LGBM, XGBoost | Trí | 100% |
# | | Implement CatBoost | Trung | 100% |
# | 01-04-2023 | Implement Neuron Network | Trí | 100% |
# | 04-04-2023 | Evaluate Model | Trung | 100% |
# | | Improve Model | Trí | 100% |
# | 08-04-2023 | Predict Data | Trung | 100% |
# | *17-04-2023* | *Final Presentation* | *Trung, Trí* | *100%*|
# # 2. Notebook configuration
# ## 2.1 Librairies loading
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import matplotlib.style as style
import seaborn as sns
from sklearn.preprocessing import LabelEncoder
# ## 2.2 Data description
train_df = pd.read_csv("train.csv")
train_df.head()
train_df.info()
train_df.shape
# Dữ liệu gồm:
# - 8693 dòng và 14 cột
# - Các thuộc tính gồm có:
# - PassengerId: mã số hành khách
# - HomePlanet: Hành tinh xuất phát
# - CryoSleep: Ngủ đông hay không
# - Cabin: Mã số cabin
# - Destination: Hành tinh đến
# - Age: Tuổi
# - VIP: Thuộc VIP hay không
# - RoomService, FoodCourt, ShoppingMall, Spa, VRDeck: Số tiền chi cho các dịch vụ cao cấp
# - Name: Tên
# - Transported: Đã dịch chuyển vào không gian khác chưa (Thuộc tính lớp)
train_df.describe().T
train_df.dtypes.value_counts()
# # 3. Data analysis
total = (
train_df.isnull()
.sum()
.sort_values(ascending=False)[
train_df.isnull().sum().sort_values(ascending=False) != 0
]
)
percent = (train_df.isnull().sum() / train_df.isnull().count()).sort_values(
ascending=False
)[
(train_df.isnull().sum() / train_df.isnull().count()).sort_values(ascending=False)
!= 0
]
missing = pd.concat([total, percent], axis=1, keys=["Total", "Percent"])
print(missing)
# Các cột bị thiếu dữ liệu tương đối đồng đều nhau (2%-2.4%)
missing = train_df.isnull().sum()
missing = missing[missing > 0]
style.use("seaborn-darkgrid")
missing.sort_values(inplace=True)
missing.plot.bar()
# ## 3.1 Visualization
labels = train_df["HomePlanet"].value_counts().index
values = train_df["HomePlanet"].value_counts().values
plt.pie(values, labels=labels, autopct="%.0f%%")
plt.show()
# Hành khách thuộc 'Earth' chiếm tới 54% lượng hành khách
labels = train_df["VIP"].value_counts().index
values = train_df["VIP"].value_counts().values
plt.pie(values, labels=labels, autopct="%.0f%%")
plt.show()
# Số lượng hành khách VIP trên tàu chỉ chiếm 2%
labels = train_df["Destination"].value_counts().index
values = train_df["Destination"].value_counts().values
plt.pie(values, labels=labels, autopct="%.0f%%")
plt.show()
# 69% lượng hành khách xuống ở `TRAPPIST-1e`
labels = train_df["Transported"].value_counts().index
values = train_df["Transported"].value_counts().values
plt.pie(values, labels=labels, autopct="%.0f%%")
plt.show()
# Tỉ lệ bị dịch chuyển ở tất cả hành khách là 50/50
train_df.corr()["Transported"].sort_values(ascending=False)
sns.heatmap(train_df.corr(), annot=True)
continuous_features = ["Age", "Expenses"]
# Combine all expenses into one column
expenses = ["RoomService", "FoodCourt", "ShoppingMall", "Spa", "VRDeck"]
train_df["Expenses"] = train_df[expenses].sum(axis=1)
# train_df.drop(expenses, axis=1, inplace=True)
# Plot continuous features
fig, axs = plt.subplots(nrows=len(continuous_features), figsize=(15, 15))
# Age
sns.histplot(data=train_df, x="Expenses", hue="Transported", bins=30, ax=axs[0])
axs[0].set_title("Expenses Distribution")
axs[0].set_xlabel("Total Expenses")
axs[0].set_yscale("log")
# Expenses
sns.histplot(data=train_df, x="Age", hue="Transported", binwidth=1, kde=True, ax=axs[1])
axs[1].set_title("Age Distribution")
axs[1].set_xlabel("Age")
plt.tight_layout()
plt.show()
# Các khoản chi Expense không theo 1 quy chuẩn nào cả
# Trẻ em <5 tuổi dường như dễ bị dịch chuyển hơn
# Người lớn dường như ít bị dịch chuyển hơn
categorical_features = ["VIP", "CryoSleep", "HomePlanet", "Destination"]
fig = plt.figure(figsize=(10, 16))
for i, feature in enumerate(categorical_features):
ax = fig.add_subplot(4, 2, i + 1)
sns.countplot(data=train_df, x=feature, hue="Transported", axes=ax)
ax.set_title(feature)
plt.tight_layout()
plt.show()
# VIP dường như không liên quan đến khả năng bị dịch chuyển
# CryoSleep có lẽ có liên quan tới khả năng bị dịch chuyển
# ## 3.2 Data preprocessing
#
train_df.info()
train_df.isnull().sum()
# Điền missing value:
# - theo số lần xuất hiện (đối với Destination,HomePlanet )
# - giá trị liền trước (Đối với Cabin, vì khả năng 2 người liền kề có thể là chung Cabin)
# - trung vị cho Age
train_df["Cabin"].fillna(method="ffill", inplace=True)
train_df["Destination"].fillna(train_df["Destination"].mode()[0], inplace=True)
train_df["HomePlanet"].fillna(train_df["HomePlanet"].mode()[0], inplace=True)
train_df["Age"].fillna(train_df["Age"].median(), inplace=True)
# VIP : fill na to False value
train_df["VIP"].fillna(False, inplace=True)
# PassengerId - gggg_pp - gggg là mã nhóm => có thể tách ra
train_df["Pass_group"] = train_df.PassengerId.str.split("_").str[0]
train_df.Pass_group = train_df.Pass_group.astype(float)
# Extract group from PassengerId
train_df["GroupSize"] = train_df["Pass_group"].map(
train_df["Pass_group"].value_counts()
)
plt.subplot(1, 2, 2)
sns.countplot(data=train_df, x="GroupSize", hue="Transported")
plt.title("Group size")
fig.tight_layout()
# Cabin - dạng boong/num/side, side có thể là P hoặc S => có thể tách ra 3 columns
train_df[["Deck", "Cab_num", "Deck_side"]] = train_df.Cabin.str.split("/", expand=True)
train_df.Cab_num = train_df.Cab_num.astype(float)
plt.figure(figsize=(10, 6))
sns.countplot(data=train_df, x="CryoSleep", hue="Transported")
plt.title("CryoSleep Distribution")
plt.show()
# Age - numerical -> categorical
# Split into 4 groups: https://www.statcan.gc.ca/en/concepts/definitions/age2
labels = ["Child", "Youth", "Adult", "Senior"]
bins = [0, 14, 24, 65, 80]
train_df["Age_Group"] = pd.cut(train_df["Age"], bins=bins, labels=labels)
train_df.head()
# Các khoảng chi tiêu cho các dịch vụ sẽ bằng 0 nếu hành khách đang trong trạng thái ngủ đông
train_df.loc[:, "Expenses"] = train_df.apply(
lambda x: 0 if x.CryoSleep == True else x, axis=1
)
train_df.loc[:, "RoomService"] = train_df.apply(
lambda x: 0 if x.CryoSleep == True else x, axis=1
)
train_df.loc[:, "FoodCourt"] = train_df.apply(
lambda x: 0 if x.CryoSleep == True else x, axis=1
)
train_df.loc[:, "ShoppingMall"] = train_df.apply(
lambda x: 0 if x.CryoSleep == True else x, axis=1
)
train_df.loc[:, "Spa"] = train_df.apply(
lambda x: 0 if x.CryoSleep == True else x, axis=1
)
train_df.loc[:, "VRDeck"] = train_df.apply(
lambda x: 0 if x.CryoSleep == True else x, axis=1
)
# Lấy giá trị trung bình các khoảng chi tiêu
RoomService_mean = train_df.RoomService.mean()
FoodCourt_mean = train_df.FoodCourt.mean()
ShoppingMall_mean = train_df.ShoppingMall.mean()
Spa_mean = train_df.Spa.mean()
VRDeck_mean = train_df.VRDeck.mean()
# Trong trường hợp hành khách là Child (0-13 tuổi) và bị mất dữ liệu ở các khoảng chi tiêu,
# thì các khoảng chi tiêu dịch vụ sẽ bằng 0
# Các hành khách còn lại, các giá trị chi tiêu bị thiếu sẽ điền giá trị trung bình
train_df.loc[train_df["Age_Group"] == "Child", "RoomService"].fillna(0)
train_df.RoomService = train_df.RoomService.fillna(RoomService_mean)
train_df.loc[train_df["Age_Group"] == "Child", "FoodCourt"].fillna(0)
train_df.FoodCourt = train_df.FoodCourt.fillna(FoodCourt_mean)
train_df.loc[train_df["Age_Group"] == "Child", "ShoppingMall"].fillna(0)
train_df.ShoppingMall = train_df.ShoppingMall.fillna(ShoppingMall_mean)
train_df.loc[train_df["Age_Group"] == "Child", "Spa"].fillna(0)
train_df.Spa = train_df.Spa.fillna(Spa_mean)
train_df.loc[train_df["Age_Group"] == "Child", "VRDeck"].fillna(0)
train_df.VRDeck = train_df.VRDeck.fillna(VRDeck_mean)
# Tính tổng chi tiêu cho từng hành khách
train_df["Expenses"] = train_df[expenses].sum(axis=1)
train_df.head(5)
train_df = train_df.drop(["Name", "PassengerId", "Cabin", "Age"], axis=1)
# encode categorical features
lbe = LabelEncoder()
categorical_vars = [
"HomePlanet",
"CryoSleep",
"Destination",
"VIP",
"Deck",
"Cab_num",
"Deck_side",
"Age_Group",
"Transported",
]
train_df[categorical_vars] = train_df[categorical_vars].apply(lbe.fit_transform)
train_df.head(5)
# # 4. Implement Model & Predict
# ## 4.1 Split Data
from sklearn.model_selection import train_test_split, StratifiedKFold
label = train_df["Transported"]
feature = train_df.drop("Transported", axis=1)
# Split the 'features' and 'income' data into training and testing sets
X_train, X_test, y_train, y_test = train_test_split(
feature, label, test_size=0.2, random_state=0
)
# Show the results of the split
print("Training set has {} samples.".format(X_train.shape[0]))
print("Testing set has {} samples.".format(X_test.shape[0]))
# ## 4.2 Implementation: Supervised Learning Models
#
from lazypredict.Supervised import LazyClassifier
clf = LazyClassifier(verbose=0, ignore_warnings=True, custom_metric=None)
models, predictions = clf.fit(X_train, X_test, y_train, y_test)
models
# Độ đo được sử dụng trong model trên:
# - **`Accuracy`**: Độ chính xác của mô hình, tính bằng tỉ lệ phần trăm các dự đoán đúng trên tổng số dữ liệu được sử dụng để đánh giá mô hình.
# - `Balanced Accuracy`: Độ chính xác cân bằng của mô hình, tính toán tỉ lệ phần trăm dự đoán đúng của cả hai lớp (positive và negative) và lấy trung bình. Độ chính xác cân bằng thường được sử dụng khi tỷ lệ các lớp trong dữ liệu khác nhau.
# - `ROC AUC`: Đây là độ đo đánh giá khả năng phân loại của mô hình. Nó đo lường khả năng của mô hình phân biệt giữa hai lớp. Đường cong ROC biểu thị tỷ lệ giữa độ nhạy và độ chính xác của mô hình tại một ngưỡng cụ thể.
# - `F1 Score`: Đây là trung bình điểm số của độ chính xác và độ phủ của mô hình. Nó là một độ đo tốt để đánh giá mô hình với một số lượng dữ liệu không cân bằng.
# - **`Time Taken Model`**: Thời gian mô hình hoạt động để đưa ra kết quả cho dữ liệu mới. Thời gian mô hình hoạt động được đo bằng đơn vị thời gian như giây hoặc phút.
# Dựa vào kết quả trên, chúng ta chọn ra được 3 model có độ chính xác cao nhất:
# * `RandomForestClassifier`
# * `LGBMClassifier`
# * `XGBClassifier`
# Ngoài ra có một model được sử dụng cho ra kết quả khá cao trong quá trình tìm hiểu model: `CatBoostClassifier`
# |Thuật toán| Ưu điểm | Nhược điểm | Ý tưởng | Bài toán thích hợp
# |--| --- | --- | ----------------- | --------------------- |
# | RandomForestClassifier | - Tốc độ huấn luyện nhanh hơn so với nhiều thuật toán khác - Có thể xử lý các tập dữ liệu lớn và có nhiều tính năng - Không yêu cầu phải chuẩn hóa dữ liệu trước - Có thể đánh giá tính quan trọng của các tính năng trong tập dữ liệu | - Có thể gặp hiện tượng quá khớp với dữ liệu huấn luyện - Không thể xử lý các giá trị bị khuyết trong tập dữ liệu - Khó khăn khi xử lý các tập dữ liệu có đặc trưng tương quan cao - Không thể áp dụng cho dữ liệu dạng chuỗi hoặc dữ liệu dạng liên tục | Tạo ra nhiều cây quyết định, mỗi cây dự đoán một phần của dữ liệu, và kết quả cuối cùng được dựa trên đa số phiếu bầu của các cây. Các cây được xây dựng bằng cách chọn một số lượng ngẫu nhiên các tính năng cho mỗi nốt trong cây | Phân loại các điểm dữ liệu hoặc dự đoán giá trị đầu ra của chúng dựa trên các tính năng (feature) được cung cấp trong tập dữ liệu |
# | LGBMClassifier | - Có thể xử lý các tập dữ liệu lớn và có nhiều tính năng - Tốc độ huấn luyện nhanh hơn so với nhiều thuật toán khác - Có thể sử dụng cho các tập dữ liệu dạng chuỗi - Hiệu suất tốt hơn so với các thuật toán gradient boosting khác - Có thể xử lý dữ liệu bị khuyết trong tập dữ liệu | - Có thể gặp hiện tượng quá khớp với dữ liệu huấn luyện - Không thể đánh giá tính quan trọng của các tính năng trong tập dữ liệu | Sử dụng các cây quyết định và tối ưu hóa quá trình huấn luyện bằng cách giảm thiểu hàm mất mát thông qua việc sử dụng các thuật toán tối| - Phân loại dữ liệu lớn, phân tích dữ liệu với nhiều features và data points - Bài toán phân loại với nhiều lớp - Phân tích dữ liệu với nhiều missing values - Phân tích dữ liệu trong lĩnh vực y tế, tài chính, marketing, công nghệ |
# |XGBClassifier|- Tốc độ huấn luyện nhanh hơn so với các thuật toán gradient boosting khác. - Hiệu quả trong việc xử lý dữ liệu có tính tương quan cao. - Có thể xử lý dữ liệu bị thiếu giá trị.|- Khó cấu hình đối với các siêu tham số. - Dễ bị overfitting với các tập dữ liệu nhỏ. - Không có tính toán song song. |- Xây dựng mô hình dựa trên nhiều cây quyết định liên tiếp. - Sử dụng kỹ thuật tăng cường gradient cho việc xác định trọng số của cây quyết định. | - Phân loại nhị phân hoặc đa lớp, dữ liệu lớn, số lượng mẫu lớn, số chiều đặc trưng lớn.|
# |CatBoostClassifier|- Tính toán song song giúp tăng tốc độ huấn luyện. - Tự động xử lý dữ liệu bị thiếu giá trị. - Được thiết kế để giảm thiểu overfitting.| - Tốn nhiều bộ nhớ và có thể chậm trong việc dự đoán trên dữ liệu lớn. - Dễ bị overfitting với các tập dữ liệu nhỏ.| - Sử dụng kỹ thuật gradient boosting trên các cây quyết định phân tách theo hạng mục. - Sử dụng kiến trúc dữ liệu đặc trưng thông minh giúp xác định độ ưu tiên của các đặc trưng.| - Phân loại nhị phân hoặc đa lớp, dữ liệu lớn, số lượng mẫu lớn, số chiều đặc trưng lớn.|
#
from time import time
from sklearn.ensemble import (
RandomForestClassifier,
GradientBoostingClassifier,
AdaBoostClassifier,
)
from lightgbm import LGBMClassifier
from xgboost import XGBClassifier
from sklearn.metrics import accuracy_score
from catboost import CatBoostClassifier
from sklearn.model_selection import cross_val_score, train_test_split, GridSearchCV
def predict_and_acc(model, X_train, y_train, X_test, y_test):
result = {}
model = model()
start = time() # Get start time
model.fit(X_train, y_train)
end = time() # Get end time
# Calculate the training time
result["train_time"] = end - start
start = time() # Get start time
predict = model.predict(X_test)
end = time() # Get end time
# Calculate the prediction time
result["pred_time"] = end - start
# accuracy score
result["accuracy"] = float(accuracy_score(y_test, predict))
# cross validation
cvs = cross_val_score(model, X_train, y_train)
result["cross-mean"] = cvs.mean()
print(
f"The accuracy score of {str(model.__class__.__name__)} is {float(accuracy_score(y_test, predict))}"
)
print(
f"The cross validation of {str(model.__class__.__name__)} is:{cvs} with mean of {cvs.mean()}"
)
return result
results = {}
results["RandomForestClassifier"] = predict_and_acc(
RandomForestClassifier, X_train, y_train, X_test, y_test
)
results["LGBMClassifier"] = predict_and_acc(
LGBMClassifier, X_train, y_train, X_test, y_test
)
results["XGBClassifier"] = predict_and_acc(
XGBClassifier, X_train, y_train, X_test, y_test
)
results["CatBoostClassifier"] = predict_and_acc(
CatBoostClassifier, X_train, y_train, X_test, y_test
)
# ## 4.3 Implementation: Neuron Network Models
# Chia dữ liệu thành tập huấn luyện và tập validation
X_train, X_val, y_train, y_val = train_test_split(
feature, label, test_size=0.2, random_state=0
)
# Chuyển đổi y_train thành một mảng 2D
y_train_2d = y_train.values.reshape(-1, 1)
nepochs = 1000
# ### Multi-layer Perceptron
from tensorflow import keras
from tensorflow.keras import layers
# Tạo mô hình
MLP_model = keras.Sequential(
[
layers.Dense(64, activation="relu", input_shape=(16,)),
layers.Dense(64, activation="relu"),
layers.Dense(1, activation="sigmoid"),
]
)
# Compile mô hình
MLP_model.compile(optimizer="adam", loss="binary_crossentropy", metrics=["accuracy"])
# Huấn luyện mô hình
MLP_model.fit(
X_train, y_train_2d, epochs=nepochs, batch_size=32, validation_data=(X_val, y_val)
)
import tensorflow as tf
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Dense, Conv1D, MaxPooling1D, Flatten
CNN_model = Sequential()
CNN_model.add(Conv1D(filters=32, kernel_size=3, activation="relu", input_shape=(16, 1)))
CNN_model.add(MaxPooling1D(pool_size=2))
CNN_model.add(Conv1D(filters=64, kernel_size=3, activation="relu"))
CNN_model.add(MaxPooling1D(pool_size=2))
CNN_model.add(Flatten())
CNN_model.add(Dense(64, activation="relu"))
CNN_model.add(Dense(1, activation="sigmoid"))
CNN_model.compile(loss="binary_crossentropy", optimizer="adam", metrics=["accuracy"])
CNN_model.fit(
X_train, y_train, epochs=nepochs, batch_size=32, validation_data=(X_val, y_val)
)
# ## 4.4 Đánh giá mô hình
# ### Supervised learning
metrics_frame = pd.DataFrame(data=results).transpose().reset_index()
metrics_frame = metrics_frame.rename(columns={"index": "models"})
metrics_frame
# show the visulization
# create shape(4,2) grouped bar plots, it displays metrics of both train and test on each row.
fig, ax = plt.subplots(2, 2, figsize=(15, 10))
# column list for metrics
metrics_col = list(metrics_frame.columns[1:])
i = 0
j = 0
for col in range(int(len(metrics_col) / 2)):
sns.barplot(x="models", y=metrics_col[2 * col], data=metrics_frame, ax=ax[i, j])
j += 1
sns.barplot(x="models", y=metrics_col[2 * col + 1], data=metrics_frame, ax=ax[i, j])
i += 1
j -= 1
if i == 2 and j == 0:
break
# set ylim(0,1) for the three metrics(accuracy, fbeta_score, roc_au_score)
ax[i, j].set_ylim((0, 1))
ax[i, j + 1].set_ylim((0, 1))
plt.suptitle("Performance Metrics for Models", fontsize=25, y=1.10)
plt.tight_layout()
# Nhận xét:
# * Từ bảng này, ta có thể thấy rằng tất cả các mô hình đều đạt được một mức độ chính xác khá cao với `accuracy` dao động từ 0.8 đến 0.81. Tuy nhiên, khi so sánh `cross-validation mean score`, thì `LGBM` và `CatBoost` đều đạt được điểm số cao nhất là 0.81, trong khi đó, `Random Forest` và `XGBoost` đạt điểm số lần lượt là 0.79 và 0.80.
# * Về thời gian huấn luyện và thời gian dự đoán của mỗi mô hình. Ta có thể thấy rằng `LGBM` và `XGBoost` có thời gian huấn luyện và dự đoán ngắn nhất, trong khi đó `CatBoost` có thời gian huấn luyện lâu nhất. Tuy nhiên, điều này có thể được chấp nhận nếu mô hình có hiệu suất tốt hơn.
# * Trong số các mô hình này, ta có thể chọn `LGBM` hoặc `CatBoost` để sử dụng trong ứng dụng thực tế, vì chúng đều có `cross-validation mean score` tốt nhất và thời gian huấn luyện và dự đoán khá ngắn.
# Do đó sử dụng `catboost` là hoàn toàn phù hợp với dự án này.
# ### Deep Learning Model: MLP vs CNN
fig, ax = plt.subplots(1, 2, figsize=(10, 4))
# visualize loss with MLP
ax[0].plot(range(nepochs), MLP_model.history.history["loss"], "r", label="train")
ax[0].plot(range(nepochs), MLP_model.history.history["val_loss"], "b", label="valid")
ax[0].legend()
ax[0].set_xlabel("epochs")
ax[0].set_ylabel("loss")
ax[0].set_title("Loss in MLP")
# visualize loss with CNN
ax[1].plot(range(nepochs), CNN_model.history.history["loss"], "r", label="train")
ax[1].plot(range(nepochs), CNN_model.history.history["val_loss"], "b", label="valid")
ax[1].legend()
ax[1].set_xlabel("epochs")
ax[1].set_ylabel("loss")
ax[1].set_title("Loss in CNN")
plt.show()
# * Đối với MLP: trong quá trình huấn luyện, loss ban đầu giảm khá nhanh, tuy nhiên sau khoảng 20 epochs thì loss bắt đầu giảm chậm hơn và dao động ở mức thấp. Nhìn chung MLP có kết quả loss thấp và giảm dần theo thời gian đào tạo.
# * Đối với CNN: trong quá trình huấn luyện, loss giảm khá nhanh và liên tục, và sau khoảng 20 epochs thì loss dao động ở mức thấp và không có dấu hiệu của overfitting. Kết quả trên tập validation có vẻ ổn định và tương đối tốt. CNN cho kết quả loss giảm dần theo thời gian đào tạo, và giá trị thấp hơn so với MLP.
# Như vậy, CNN cho kết quả loss tốt hơn và có vẻ ổn định hơn so với MLP trong bài toán này.
fig = plt.figure(figsize=(5, 4))
plt.plot(range(nepochs), MLP_model.history.history["accuracy"], "r", label="mlp")
plt.plot(range(nepochs), CNN_model.history.history["accuracy"], "b", label="cnn")
plt.legend()
plt.xlabel("epochs")
plt.ylabel("loss")
plt.title("Accuracy in MLP vs CNN ")
# Nhìn chung cả MLP và CNN đều đạt được accuracy khá cao, ở giai đoạn epoch<200, CNN đạt độ hiệu quả cao hơn MLP, tuy nhiên khi được huấn luyện càng nhiều MLP đạt độ hiệu quả cao hơn rõ rệt so với CNN.
# Như vậy, mặc dù MLP có nhược điểm rất tốn thời gian nếu muốn đạt được một mô hình dự đoán tốt, tuy nhiên với độ hiệu quả khá cao thì chọn MLP là hợp lý với bài toán đang giải quyết.
# ## 4.5 Preprocessing `test.csv`
test_data = pd.read_csv("test.csv")
test_data
test_data.info()
submission = pd.DataFrame({"PassengerId": test_data["PassengerId"]})
test_data.isna().sum()
test_data["Expenses"] = test_data[expenses].sum(axis=1)
# Điền missing value:
# - theo số lần xuất hiện (đối với Destination,HomePlanet )
# - giá trị liền trước (Đối với Cabin, vì khả năng 2 người liền kề có thể là chung Cabin)
# - trung vị cho Age
test_data["Cabin"].fillna(method="ffill", inplace=True)
test_data["Destination"].fillna(test_data["Destination"].mode()[0], inplace=True)
test_data["HomePlanet"].fillna(test_data["HomePlanet"].mode()[0], inplace=True)
test_data["Age"].fillna(test_data["Age"].median(), inplace=True)
# VIP : fill na to False value
test_data["VIP"].fillna(False, inplace=True)
# PassengerId - gggg_pp - gggg là mã nhóm => có thể tách ra
test_data["Pass_group"] = test_data.PassengerId.str.split("_").str[0]
test_data.Pass_group = test_data.Pass_group.astype(float)
# Extract group from PassengerId
test_data["GroupSize"] = test_data["Pass_group"].map(
test_data["Pass_group"].value_counts()
)
# Cabin - dạng boong/num/side, side có thể là P hoặc S => có thể tách ra 3 columns
test_data[["Deck", "Cab_num", "Deck_side"]] = test_data.Cabin.str.split(
"/", expand=True
)
test_data.Cab_num = test_data.Cab_num.astype(float)
# Age - numerical -> categorical
# Split into 4 groups: https://www.statcan.gc.ca/en/concepts/definitions/age2
labels = ["Child", "Youth", "Adult", "Senior"]
bins = [0, 14, 24, 65, 80]
test_data["Age_Group"] = pd.cut(test_data["Age"], bins=bins, labels=labels)
test_data.head()
# Các khoảng chi tiêu cho các dịch vụ sẽ bằng 0 nếu hành khách đang trong trạng thái ngủ đông
test_data.loc[:, "Expenses"] = test_data.apply(
lambda x: 0 if x.CryoSleep == True else x.Expenses, axis=1
)
test_data.loc[:, "RoomService"] = test_data.apply(
lambda x: 0 if x.CryoSleep == True else x.RoomService, axis=1
)
test_data.loc[:, "FoodCourt"] = test_data.apply(
lambda x: 0 if x.CryoSleep == True else x.FoodCourt, axis=1
)
test_data.loc[:, "ShoppingMall"] = test_data.apply(
lambda x: 0 if x.CryoSleep == True else x.ShoppingMall, axis=1
)
test_data.loc[:, "Spa"] = test_data.apply(
lambda x: 0 if x.CryoSleep == True else x.Spa, axis=1
)
test_data.loc[:, "VRDeck"] = test_data.apply(
lambda x: 0 if x.CryoSleep == True else x.VRDeck, axis=1
)
# Lấy giá trị trung bình các khoảng chi tiêu
RoomService_mean = test_data.RoomService.mean()
FoodCourt_mean = test_data.FoodCourt.mean()
ShoppingMall_mean = test_data.ShoppingMall.mean()
Spa_mean = test_data.Spa.mean()
VRDeck_mean = test_data.VRDeck.mean()
# Trong trường hợp hành khách là Child (0-13 tuổi) và bị mất dữ liệu ở các khoảng chi tiêu,
# thì các khoảng chi tiêu dịch vụ sẽ bằng 0
# Các hành khách còn lại, các giá trị chi tiêu bị thiếu sẽ điền giá trị trung bình
test_data.loc[test_data["Age_Group"] == "Child", "RoomService"].fillna(0)
test_data.RoomService = test_data.RoomService.fillna(RoomService_mean)
test_data.loc[test_data["Age_Group"] == "Child", "FoodCourt"].fillna(0)
test_data.FoodCourt = test_data.FoodCourt.fillna(FoodCourt_mean)
test_data.loc[test_data["Age_Group"] == "Child", "ShoppingMall"].fillna(0)
test_data.ShoppingMall = test_data.ShoppingMall.fillna(ShoppingMall_mean)
test_data.loc[test_data["Age_Group"] == "Child", "Spa"].fillna(0)
test_data.Spa = test_data.Spa.fillna(Spa_mean)
test_data.loc[test_data["Age_Group"] == "Child", "VRDeck"].fillna(0)
test_data.VRDeck = test_data.VRDeck.fillna(VRDeck_mean)
# Tính tổng chi tiêu cho từng hành khách
test_data["Expenses"] = test_data[expenses].sum(axis=1)
test_data = test_data.drop(["Name", "PassengerId", "Cabin", "Age"], axis=1)
# encode categorical features
lbe = LabelEncoder()
categorical_vars = [
"HomePlanet",
"CryoSleep",
"Destination",
"VIP",
"Deck",
"Cab_num",
"Deck_side",
"Age_Group",
]
test_data[categorical_vars] = test_data[categorical_vars].apply(lbe.fit_transform)
test_data.head()
# ## 4.6 Predict submission
test_data.head()
# ### Catboost Classifier
# define the parameter grid to search over
param_grid = {
"learning_rate": [0.01, 0.05, 0.1],
"max_depth": [3, 5, 7],
"n_estimators": [50, 100, 200],
}
# create an instance of the CatBoostClassifier
model = CatBoostClassifier()
# create a GridSearchCV object
grid_search = GridSearchCV(model, param_grid=param_grid, cv=5, verbose=2)
# fit the GridSearchCV object to the data
grid_search.fit(X_train, y_train)
# Train model with optimal hyperparameters
model = CatBoostClassifier(**grid_search.best_params_)
model.fit(X_train, y_train)
y_predict_test = model.predict(test_data)
# print the best parameter combination found
print("Best parameters: ", grid_search.best_params_)
# ### Multi-Layer Perceptron
MLP_predict_test = MLP_model.predict(test_data)
MLP_predict_rounded = np.round(MLP_predict_test)
MLP_predict_binary = MLP_predict_rounded.astype(int)
# ### Export submission
submission["Transported"] = MLP_predict_binary.astype("bool")
submission.to_csv("submission.csv", index=False)
submission["Transported"].value_counts()
|
# data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
# # importing neccessary libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
# # reading CSV file and printing the shape of DataFrame
data = pd.read_csv("../input/salary/Salary.csv")
data.shape
# # printing first 5 rows of the DataFrame
data.head(5)
# # renaming the column names
data.rename(columns={"YearsExperience": "yexp", "Salary": "salary"}, inplace=True)
# # Scatter plot representing relation between
# ## experience v/s Salaray
plt.scatter(data["yexp"], data["salary"], color="g")
plt.xlabel("EXPERIENCE")
plt.ylabel("SALARY")
# # reshaping the series in to 2D array using reshape method
x = np.array(data["yexp"]).reshape(-1, 1)
y = np.array(data["salary"]).reshape(-1, 1)
from sklearn.linear_model import LinearRegression
from sklearn.model_selection import train_test_split
from sklearn.metrics import r2_score, mean_squared_error
# # Training Model with training datasets
reg = LinearRegression()
xtrain, xtest, ytrain, ytest = train_test_split(x, y, test_size=0.4, random_state=1)
reg.fit(xtrain, ytrain)
pred = reg.predict(xtest)
# # r_square value is pretty_good 96%
print(r2_score(pred, ytest))
# # MSE is worst ,its due to underfitting ..in plain english less number of features (training examples)
print(mean_squared_error(pred, ytest))
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
# # Part 1: build dataset and visualize
import sys
import time
import math
import gc
import cv2
import torch
import torch.nn as nn
from torch.optim import lr_scheduler
from torch.utils.data import Dataset, DataLoader
import torchvision
import torchvision.transforms as T
import matplotlib.pyplot as plt
import sklearn.metrics
def timeSince(since):
now = time.time()
s = now - since
m = math.floor(s / 60)
s -= m * 60
return "%dm %ds" % (m, s)
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
print(device)
root_train = "/kaggle/input/bengaliaicv19feather"
root_test = "/kaggle/input/bengaliai-cv19"
path = "simpleCNN.pth"
start = time.time()
img_arrs = np.concatenate(
(
pd.read_feather(os.path.join(root_train, "train_image_data_0.feather"))
.drop(columns=["image_id"])
.values,
pd.read_feather(os.path.join(root_train, "train_image_data_1.feather"))
.drop(columns=["image_id"])
.values,
pd.read_feather(os.path.join(root_train, "train_image_data_2.feather"))
.drop(columns=["image_id"])
.values,
pd.read_feather(os.path.join(root_train, "train_image_data_3.feather"))
.drop(columns=["image_id"])
.values,
),
axis=0,
)
print(timeSince(start))
print(img_arrs.shape)
idx = 30
threshold = 0.5
size = 128
img_arr = img_arrs[idx] / 255.0
fig = plt.figure(figsize=(20, 6))
ax = fig.add_subplot(2, 3, 1, xticks=[], yticks=[])
original_img = img_arr.reshape(137, 236)
ax.imshow(original_img, cmap="gray")
ax = fig.add_subplot(2, 3, 2, xticks=[], yticks=[])
cropped_img = original_img[:, 30:190]
ax.imshow(cropped_img, cmap="gray")
ax = fig.add_subplot(2, 3, 3, xticks=[], yticks=[])
resized = cv2.resize(cropped_img, (size, size))
ax.imshow(resized, cmap="gray")
ax = fig.add_subplot(2, 3, 4, xticks=[], yticks=[])
rotate = cv2.rotate(resized, cv2.ROTATE_90_CLOCKWISE)
ax.imshow(rotate, cmap="gray")
ax = fig.add_subplot(2, 3, 5, xticks=[], yticks=[])
rotate = cv2.rotate(resized, cv2.ROTATE_90_COUNTERCLOCKWISE)
ax.imshow(rotate, cmap="gray")
ax = fig.add_subplot(2, 3, 6, xticks=[], yticks=[])
rotate = cv2.rotate(resized, cv2.ROTATE_180)
ax.imshow(rotate, cmap="gray")
plt.show()
# transforms = T.Compose([T.ToPILImage(), T.Resize((size,size)), T.ToTensor()])
transforms = T.Compose([T.ToPILImage(), T.CenterCrop(size), T.ToTensor()])
img_arr = (
255 - img_arrs[idx]
) # flip black and white, so the default padding value (0) could match
original_img = img_arr.reshape(137, 236, 1)
img_tensor = transforms(original_img)
new_tensor = torch.where(
img_tensor <= threshold, torch.zeros_like(img_tensor), img_tensor
)
print(new_tensor.size())
plt.imshow(new_tensor[0], cmap="gray")
plt.show()
class graphemeDataset(Dataset):
def __init__(self, img_arrs, target_file=None, size=128, threshold=0.5):
self.img_arrs = img_arrs
self.target_file = target_file
self.size = size
self.threshold = threshold
if target_file is None:
self.transforms = T.Compose(
[T.ToPILImage(), T.CenterCrop(size), T.ToTensor()]
)
else:
self.transforms = T.Compose(
[T.ToPILImage(), T.CenterCrop(size), T.ToTensor()]
)
# add targets for training
target_df = pd.read_csv(target_file)
self.grapheme = target_df["grapheme_root"].values
self.vowel = target_df["vowel_diacritic"].values
self.consonant = target_df["consonant_diacritic"].values
del target_df
gc.collect()
def __getitem__(self, idx):
img_arr = (
255 - self.img_arrs[idx]
) # flip black and white, so the default padding value (0) could match
img_tensor = self.transforms(img_arr.reshape(137, 236, 1))
new_tensor = torch.where(
img_tensor <= self.threshold, torch.zeros_like(img_tensor), img_tensor
)
if self.target_file is None:
return new_tensor
else:
grapheme_tensor = torch.tensor(self.grapheme[idx], dtype=torch.long)
vowel_tensor = torch.tensor(self.vowel[idx], dtype=torch.long)
consonant_tensor = torch.tensor(self.consonant[idx], dtype=torch.long)
return new_tensor, grapheme_tensor, vowel_tensor, consonant_tensor
def __len__(self):
return len(self.img_arrs)
dataset = graphemeDataset(
img_arrs, target_file="/kaggle/input/bengaliai-cv19/train.csv"
)
print(dataset.__len__())
batch_size = 32
data_loader = DataLoader(dataset, batch_size=batch_size, shuffle=True, drop_last=True)
print(dataset.__len__() / batch_size)
dataiter = iter(data_loader)
img_tensor, grapheme_tensor, vowel_tensor, consonant_tensor = next(dataiter)
print(img_tensor.size())
print(grapheme_tensor.size())
print(vowel_tensor.size())
print(consonant_tensor.size())
fig = plt.figure(figsize=(25, 8))
plot_size = 32
for idx in np.arange(plot_size):
ax = fig.add_subplot(4, plot_size / 4, idx + 1, xticks=[], yticks=[])
ax.imshow(img_tensor[idx][0], cmap="gray")
# # Part 2: design model architecture
class simpleCNN(nn.Module):
def __init__(self, hidden_dim=32):
super(simpleCNN, self).__init__()
self.hidden_dim = hidden_dim
self.features = nn.Sequential(
nn.Conv2d(1, hidden_dim, kernel_size=3),
nn.ReLU(inplace=True),
nn.Conv2d(hidden_dim, hidden_dim, kernel_size=3),
nn.ReLU(inplace=True),
nn.BatchNorm2d(hidden_dim),
nn.MaxPool2d(2),
nn.Conv2d(hidden_dim, hidden_dim, kernel_size=5),
nn.ReLU(inplace=True),
nn.Dropout(p=0.3),
nn.Conv2d(hidden_dim, hidden_dim * 2, kernel_size=3),
nn.ReLU(inplace=True),
nn.Conv2d(hidden_dim * 2, hidden_dim * 2, kernel_size=3),
nn.ReLU(inplace=True),
nn.BatchNorm2d(hidden_dim * 2),
nn.MaxPool2d(2),
nn.Conv2d(hidden_dim * 2, hidden_dim * 2, kernel_size=5),
nn.BatchNorm2d(hidden_dim * 2),
nn.Dropout(p=0.3),
nn.Conv2d(hidden_dim * 2, hidden_dim * 4, kernel_size=3),
nn.ReLU(inplace=True),
nn.Conv2d(hidden_dim * 4, hidden_dim * 4, kernel_size=3),
nn.ReLU(inplace=True),
nn.BatchNorm2d(hidden_dim * 4),
nn.MaxPool2d(2),
nn.Conv2d(hidden_dim * 4, hidden_dim * 4, kernel_size=5),
nn.BatchNorm2d(hidden_dim * 4),
nn.Dropout(p=0.3),
nn.Conv2d(hidden_dim * 4, hidden_dim * 8, kernel_size=3),
nn.ReLU(inplace=True),
nn.Conv2d(hidden_dim * 8, hidden_dim * 8, kernel_size=3),
nn.ReLU(inplace=True),
nn.BatchNorm2d(hidden_dim * 8),
nn.Dropout(p=0.3),
nn.Flatten(),
)
self.grapheme_classifier = nn.Sequential(
nn.Linear(self.hidden_dim * 8, 168), nn.LogSoftmax(dim=1)
)
self.vowel_classifier = nn.Sequential(
nn.Linear(self.hidden_dim * 8, self.hidden_dim * 4),
nn.ReLU(inplace=True),
nn.Linear(self.hidden_dim * 4, 11),
nn.LogSoftmax(dim=1),
)
self.consonant_classifier = nn.Sequential(
nn.Linear(self.hidden_dim * 8, self.hidden_dim * 4),
nn.ReLU(inplace=True),
nn.Linear(self.hidden_dim * 4, 7),
nn.LogSoftmax(dim=1),
)
def forward(self, x):
x = self.features(x)
c1 = self.grapheme_classifier(x)
c2 = self.vowel_classifier(x)
c3 = self.consonant_classifier(x)
return c1, c2, c3
# return x
# model1 = simpleCNN(32)
# out = model1(img_tensor)
# print(out.size())
# plt.imshow(out[0][21].detach().numpy(), cmap='gray')
# plt.show()
# c1, c2, c3 = model1(img_tensor)
# print(c1.size())
# print(c2.size())
# print(c3.size())
# # Part 3: train model
def model_training(
model, dataset, path, batch_size=32, epoches=1, print_every=10, init_lr=0.0005
):
start = time.time()
criterion = nn.NLLLoss()
data_loader = DataLoader(
dataset, batch_size=batch_size, shuffle=True, drop_last=True
)
model.train()
optimizer = torch.optim.Adam(model.parameters(), lr=init_lr)
exp_lr_scheduler = lr_scheduler.StepLR(optimizer, step_size=7, gamma=0.1)
loss_hold = 999
for e in range(epoches):
for counter, (
img_tensor,
grapheme_tensor,
vowel_tensor,
consonant_tensor,
) in enumerate(data_loader):
img_tensor = img_tensor.to(device)
grapheme_tensor = grapheme_tensor.to(device)
vowel_tensor = vowel_tensor.to(device)
consonant_tensor = consonant_tensor.to(device)
c1, c2, c3 = model(img_tensor)
l1 = criterion(c1, grapheme_tensor)
l2 = criterion(c2, vowel_tensor)
l3 = criterion(c3, consonant_tensor)
loss = 0.5 * l1 + 0.25 * l2 + 0.25 * l3
optimizer.zero_grad()
loss.backward()
optimizer.step()
if counter % print_every == 0:
print(
"Epoch {}/{}({}) {}...loss {:6.4f}...grapheme {:6.4f}...vowel {:6.4f}...consonant {:6.4f}".format(
e + 1,
epoches,
counter,
timeSince(start),
loss.item(),
l1.item(),
l2.item(),
l3.item(),
)
)
exp_lr_scheduler.step()
if loss.item() < loss_hold:
torch.save(model.state_dict(), path)
loss_hold = loss.item()
print("Check point saved...")
model_training(
simpleCNN(32).to(device), dataset, path, batch_size=128, epoches=15, print_every=500
)
model = simpleCNN(32)
model.load_state_dict(torch.load(path))
model = model.to(device)
start = time.time()
loader = DataLoader(dataset, batch_size=32, shuffle=False)
grapheme_pred = []
vowel_pred = []
consonant_pred = []
grapheme_true = []
vowel_true = []
consonant_true = []
with torch.no_grad():
for img_tensor, grapheme_tensor, vowel_tensor, consonant_tensor in loader:
img_tensor = img_tensor.to(device)
c1, c2, c3 = model(img_tensor)
grapheme_pred.extend(c1.argmax(1).cpu().tolist())
grapheme_true.extend(grapheme_tensor.tolist())
vowel_pred.extend(c2.argmax(1).cpu().tolist())
vowel_true.extend(vowel_tensor.tolist())
consonant_pred.extend(c3.argmax(1).cpu().tolist())
consonant_true.extend(consonant_tensor.tolist())
scores = [
sklearn.metrics.recall_score(grapheme_true, grapheme_pred, average="macro"),
sklearn.metrics.recall_score(vowel_true, vowel_pred, average="macro"),
sklearn.metrics.recall_score(consonant_true, consonant_pred, average="macro"),
]
final_score = np.average(scores, weights=[2, 1, 1])
print("train acc: {:6.2f} %".format(100 * final_score))
print(timeSince(start))
del img_arrs
gc.collect()
# # Part 4: Predict
# 1. use trained model to predict
# 2. make the results into correct sub format
row_id = []
target = []
for i in range(4):
# load testing data
start = time.time()
img_df = pd.read_parquet(
os.path.join(root_test, "test_image_data_" + str(i) + ".parquet")
)
print(timeSince(start))
img_id = []
img_id.extend(img_df.image_id.tolist())
img_arrs = img_df.iloc[:, 1:].values
dataset = graphemeDataset(img_arrs)
print(dataset.__len__())
loader = DataLoader(dataset, batch_size=32, shuffle=False)
# make predictions
grapheme_pred = []
vowel_pred = []
consonant_pred = []
with torch.no_grad():
for img_tensor in loader:
img_tensor = img_tensor.to(device)
c1, c2, c3 = model(img_tensor)
grapheme_pred.extend(c1.argmax(1).cpu().tolist())
vowel_pred.extend(c2.argmax(1).cpu().tolist())
consonant_pred.extend(c3.argmax(1).cpu().tolist())
# format the results
for idx, g, v, c in zip(img_id, grapheme_pred, vowel_pred, consonant_pred):
row_id.append(idx + "_grapheme_root")
row_id.append(idx + "_vowel_diacritic")
row_id.append(idx + "_consonant_diacritic")
target.append(g)
target.append(v)
target.append(c)
# clean up
del img_arrs, img_df
gc.collect()
pred = pd.DataFrame({"row_id": row_id, "target": target})
pred.head()
pred.to_csv("submission.csv", index=False)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.preprocessing import LabelEncoder, StandardScaler
from sklearn.metrics import roc_auc_score, accuracy_score
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import ExtraTreesClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.metrics import accuracy_score, r2_score
sns.set_style("darkgrid")
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
df = pd.read_csv(
"/kaggle/input/ibm-hr-analytics-attrition-dataset/WA_Fn-UseC_-HR-Employee-Attrition.csv"
)
df.head()
df.info()
sns.countplot(df["Gender"])
sns.countplot(df["OverTime"])
sns.countplot(df["MaritalStatus"])
plt.figure(figsize=(15, 7))
sns.countplot(df["JobRole"])
plt.figure(figsize=(20, 7))
sns.countplot(df["EducationField"])
plt.figure(figsize=(20, 10))
sns.countplot(df["TotalWorkingYears"])
pd.crosstab(df["Attrition"], df["DistanceFromHome"]).plot.bar(figsize=(18, 5))
# sns.countplot(df.DistanceFromHome)
sns.countplot(df["Attrition"])
en = LabelEncoder()
df["Age"] = en.fit_transform(df["Age"])
df["BusinessTravel"] = en.fit_transform(df["BusinessTravel"])
df["DailyRate"] = en.fit_transform(df["DailyRate"])
df["Department"] = en.fit_transform(df["Department"])
df["EducationField"] = en.fit_transform(df["EducationField"])
df["JobRole"] = en.fit_transform(df["JobRole"])
df["MaritalStatus"] = en.fit_transform(df["MaritalStatus"])
df["Attrition"] = en.fit_transform(df["Attrition"])
df["OverTime"] = en.fit_transform(df["OverTime"])
df["Over18"] = en.fit_transform(df["Over18"])
df["Gender"] = en.fit_transform(df["Gender"])
df.head()
X = df.drop("Attrition", axis=1)
y = df["Attrition"]
X = StandardScaler().fit_transform(X)
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.33, random_state=42
)
model = []
log = LogisticRegression()
log.fit(X_train, y_train)
y_predict = log.predict(X_test)
model.append(["LogisticRegression", accuracy_score(y_predict, y_test)])
log = DecisionTreeClassifier()
log.fit(X_train, y_train)
y_predict = log.predict(X_test)
model.append(["DecisionTreeClassifier", accuracy_score(y_predict, y_test)])
log = ExtraTreesClassifier()
log.fit(X_train, y_train)
y_predict = log.predict(X_test)
model.append(["ExtraTreesClassifier", accuracy_score(y_predict, y_test)])
log = KNeighborsClassifier()
log.fit(X_train, y_train)
y_predict = log.predict(X_test)
model.append(["KNeighborsClassifier", accuracy_score(y_predict, y_test)])
log = GaussianNB()
log.fit(X_train, y_train)
y_predict = log.predict(X_test)
model.append(["GaussianNB", accuracy_score(y_predict, y_test)])
model = pd.DataFrame(model)
model.columns = ["Name", "Score"]
model.sort_values(by="Score", ascending=False)
|
# # Project 5 Gaming Industry
# This project is for "BigStream" online Game store, which sells computer games all over the world. Historical game sales data, user and expert ratings, genres and platforms (such as Xbox or PlayStation) are available from public sources.
# The task is to identify the patterns that determine the success of the game. This will allow you to bid on a potentially popular product and plan advertising campaigns. You have data up to 2016. Let's say it's December 2016 and you're planning a campaign for 2017. It is necessary to work out the principle of working with data.
# The abbreviation ESRB (Entertainment Software Rating Board) comes across in the data set - this is an association that determines the age rating of computer games. The ESRB evaluates game content and assigns it to an appropriate age rating, such as Mature, Toddler, or Teen. Data for 2016 may not be complete.
# ### Data Fields:
# * Name - the name of the game
# * Platform - platform
# * Year_of_Release - year of release
# * Genre - game genre
# * NA_sales - sales in North America (millions of copies sold)
# * EU_sales - sales in Europe (millions of copies sold)
# * JP_sales - sales in Japan (millions of copies sold)
# * Other_sales - sales in other countries (millions of copies sold)
# * Critic_Score - Critics score (max 100)
# * User_Score - user score (maximum 10)
# * Rating - rating from the ESRB (Entertainment Software Rating Board). This association determines the rating of computer games and assigns them an appropriate age category.
#
# import libraries and methods
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import math
from scipy import stats as st
# ## Step 1. Open the data file and study the general information
#
try:
data_row = pd.read_csv(
"/Users/dmitry/Documents/Python DS/Modul 1/datasets_modul1/datasets_sumup_games/games.csv"
)
except:
data_row = pd.read_csv("/datasets/games.csv")
data_row.info()
display(data_row.describe())
display(data_row.head(20))
# Let's look at the duplicates and remove the explicit duplicates and duplicates by name and platform
# Check the explicit duplicates
if data_row.duplicated().sum() == 0:
print("\n No explicit duplicates")
else:
print("\n Detected", data.duplicated().sum(), "duplicates")
# Let's check duplicates by name and platform
rows_duplicated = data_row[["Name", "Platform"]].duplicated(keep=False).sort_values()
display(data_row.loc[rows_duplicated].head(20))
# Calculate the number of NaN's
print("Total sales of Nan:")
sum_of_nan = sum(
data_row.loc[659, ["NA_sales", "EU_sales", "JP_sales", "Other_sales"]]
) + sum(data_row.loc[14244, ["NA_sales", "EU_sales", "JP_sales", "Other_sales"]])
print(sum_of_nan)
total_sales_raw = sum(
data_row["NA_sales"]
+ data_row["EU_sales"]
+ data_row["JP_sales"]
+ data_row["Other_sales"]
)
print("Total % sales of Nan in total sales:")
print(sum_of_nan / total_sales_raw)
# Let's delete duplicates by name and platform. I deleted Madden NFL 13, Sonic the Hedgehog and Nan as a duplicates. But Need for Speed: Most Wanted has different years 2005 and 2012. So this is a remaster, let's keep it.
#
data_row = data_row.drop(index=[1591, 659, 4127, 14244, 16230]).reset_index(drop=True)
display(data_row.loc[data_row["Name"] == "Need for Speed: Most Wanted"].head(20))
# Conclusion step 1:
# * there are no explicit duplicates detected
# * There is 'Nan' data for two items. I checked if it was possible to process it. There are <<1% passed values, so we can delete them
# * I discovered that:
# ** year of release is float type, but int type fitts better
# ** there are a lot of miss values in Critic_Score, User_Score, Rating
# ** The age rating is for games in the USA and Canada. If the game didn't release there, then most likely they didn't have a rating either. There could be missed values from here.
# ## Step 2: Data preprocessing
# Let's:
# * Convert data to the distinguish types. Describe in which columns the data type was replaced and why;
# * Process the allowances if necessary
# * Explain why you filled in the gaps in a certain way or why you did not do it
# * Describe the reasons that could lead to omissions
# * Note the abbreviation 'tbd' in the user rating column. Separately analyze this value and describe how to process it
# * Calculate the total sales in all regions and record them in a separate column.
# I'm replacing column names and bringing them to lowercase
# create a new dataset
data = data_row
# let's bring the columns to the snake register
data.columns = data.columns.str.lower()
# let's check if there are any duplicates in the platforms due to different case. if there are, then we will bring it to lowercase
print("\n genre \n", data["genre"].value_counts())
print("\n platform \n", data["platform"].value_counts())
print("\n rating \n", data["rating"].value_counts())
# Conclusion: there are no duplicates in genre and platform, so I'll leave the names unchanged
# rating K-A is the old version for category E. Let's Replace these values.
data["rating"] = data["rating"].replace("K-A", "E")
# There are too many values in the user_score and critic_score columns to check them only with the value_counts method. For them, we call the unique() method
print(data["user_score"].value_counts())
print(data["user_score"].unique())
print("\ncritic_score")
print(data["critic_score"].value_counts())
print(data["critic_score"].unique())
# Conclusion Step 2.1:
# * There is a tbd value in the user_score column
# * In half of the values where user_score == tbd, we know critic_score. Let's try to estimate their correlation
# ## Step 2.2 Checking the missing values male data according to
# Let's check how much of the missing values are in each of the missing columns
# Let's create a function that outputs the number of gaps in the dataframe column
def isna_data_count(df):
for i in df.columns:
isna_count = df[i].isna().mean()
print("% of missing values", i, f"{isna_count:.2%}")
return 0
isna_data_count(data)
# Conclusion:
# * there are also missing values in the name column, but they are not visible from the operation of the isna_data_count function, but can be seen by the describe method
# Let's check the possibility to restore the value of user_score via critic_score or vice versa
# Let's check if these values can be replaced - to do this, check the average of the available data
rows_scores = ~((data["user_score"].isna()) | (data["critic_score"].isna()))
data_scores = data.loc[rows_scores].reset_index(drop=True)
# I am checking the values where user_score is present and critic_score is missing
rows = (~data["user_score"].isna()) & (data["critic_score"].isna())
display(data.loc[rows].shape)
display(data.loc[rows].head(50))
data_scores.info()
# let's evaluate the correlation of user_score and critic_score so that it is possible to replace them
# to do this, I will bring the indicators to a digital format
data_scores["user_score"] = data_scores["user_score"].replace("tbd", np.nan)
data_scores["user_score"] = data_scores["user_score"].fillna(-1)
data_scores["user_score_int"] = round(
data_scores["user_score"].astype(float), 0
).astype(int)
data_scores = data_scores.reset_index(drop=True)
data_scores["user_score"] = data_scores["user_score"].astype(float)
# I am removing the values -1 from the dataset in order to calculate the correlation only by known parameters
data_scores = data_scores.loc[data_scores["user_score"] > 0]
data_scores.plot(kind="scatter", x="critic_score", y="user_score")
plt.show()
display(data_scores.corr())
data["user_score"] = data["user_score"].replace("tbd", np.nan)
# Conclusion:
# * the correlation between user_score and critic_score = 0.58. No linear dependence was found either on the graph or in the function. So we can't replace the average value from one column and put it in another
# * it will not be possible to pick up values from another column, so we will replace the values with np.nan
# I will bring the year to int type, to replace the passes, we will choose the platform average
display(data.loc[data["year_of_release"].isna()].head(30))
data["year_of_release"] = data["year_of_release"].fillna(999)
display(data.loc[183])
# I am changing the data types for user_score - value and year_of_release - floating
data["user_score"] = data["user_score"].astype(float)
data["year_of_release"] = data["year_of_release"].astype(int)
display(data)
data.info()
# Let's see what the critic_score and user_score distributions are
# let's write a function to output estimates
def prnt_score_func(df):
df["critic_score"].hist(bins=100)
plt.xlabel("critic_score")
plt.ylabel("Frequency")
plt.title("Frequency of distribution of critic_score")
plt.show()
df["user_score"].hist(bins=100)
plt.xlabel("user_score")
plt.ylabel("Frequency")
plt.title("Frequency of distribution of user_score")
plt.show()
return 0
# let's output the initial array
print("Before replacing missing values")
prnt_score_func(data)
# we will remove the value in name and genre, since there are few of them, and with the rest we will replace it with a suitable "Unknown". For numeric values, replace the values with -1
data = data.dropna(subset=["name", "genre"]).reset_index(drop=True)
data["critic_score"] = data["critic_score"].fillna(-1)
data["user_score"] = data["user_score"].fillna(-1)
data["rating"] = data["rating"].fillna("Unknown")
print("After replacing the missing values")
prnt_score_func(data)
# Let's add a column with total sales
data["total_sales"] = (
data["na_sales"] + data["eu_sales"] + data["jp_sales"] + data["other_sales"]
)
display(data.head(50))
# Check that all values have been replaced
data.info()
# Conclusion:
# * all missing values are processed correctly
# ### Step 3. Conducting a research analysis of the data
# * Let's
# * See how many games were released among years. Is the data for all periods important?
# * See how sales have changed across platforms. Select the platforms with the highest total sales and build a distribution by year. For what characteristic period of time do new platforms appear and old ones disappear?
# * Take data for the relevant current period. Determine the current period yourself as a result of the study of previous questions. The main factor is that these data will help to build a forecast for 2017.
# * Do not take into account data from previous years in your work
# * Build a "box with a mustache" chart for global game sales by platform. Describe the result.
# * See how the reviews of users and critics affect sales within one popular platform. Build a scatter plot and calculate the correlation between reviews and sales. Formulate conclusions.
# * Correlate the findings with the sales of games on other platforms.
# * Look at the overall distribution of games by genre. What can be said about the most profitable genres? Do genres with high and low sales stand out?
# * Check which platforms are leading in sales, are they growing or falling? Let's select several potentially profitable platforms
# Let's look at the distribution of game sales for each year
data_sales_by_year = data.pivot_table(
index="year_of_release", values=["name", "total_sales"], aggfunc=["count", "sum"]
)
print(data_sales_by_year)
# Conclusion:
# * interestig to notice there has been a rise in sales of classic games since 1996
# * I will use data sterting from 1996 for analysis
print(
data.pivot_table(index="platform", values="total_sales", aggfunc="sum").sort_values(
by="total_sales"
)
)
data = data.loc[data["year_of_release"] >= 1996]
# Conclusion:
# * PS2 has about 1.26 billion copies sold. This is the first place among all platforms.
# * The top three platforms are PS2, Wii and X360
# Let's see how sales by platform have been changing. I selecting the platforms with the highest total sales and build a distribution by year.
# Let's see for what characteristic period of time do new platforms appear and old ones disappear?
#
data_sales_by_platform = data.pivot_table(
index="platform", values="total_sales", aggfunc="sum"
).sort_values(by="total_sales")
# Let's choose platforms with sales of more than 130 million
data_sales_by_platform = data_sales_by_platform.loc[
data_sales_by_platform["total_sales"] > 130
]
platform_to_analize = data_sales_by_platform.index
platform_to_analize = pd.Series(platform_to_analize)
print(platform_to_analize)
data = data.query("platform in @platform_to_analize")
for i in platform_to_analize:
print(i, "\n")
data_to_plot = data.loc[data["platform"] == i]
data_to_plot.pivot_table(
index="year_of_release", values="total_sales", aggfunc="sum"
).plot(kind="bar", figsize=(10, 5), xlim=(1996, 2016))
plt.show()
# Conclusion:
# * on average, platforms live from 4 to 7 years. The longest-lived pc. Games on it were released for the entire time of the study
# * it is necessary to invest in platforms for which games are still being released: XOne, PC, 3ds, PS4, PS3, Wii, X360
# * it is better to choose platforms according to 2 criteria: current sales and growth rate - for example, 2017 is the year when there is a generation change. The ps3 is still relevant and the ps4 has already been released. Similar situation for x360 and Xone. The old generation is "cash cows", and 2 is "stars" according to the BCG matrix.
platform_to_research = ["XOne", "PC", "3DS", "PS4", "PS3", "Wii", "X360"]
platform_to_research = pd.Series(platform_to_research)
data = data.query("platform in @platform_to_research").reset_index(drop=True)
display(data.query("platform == '3DS'").head(50))
# let's find the minimum year for 3DS console release
print(
"The year when consoles started",
data.loc[data["platform"] != "PC"]["year_of_release"].min(),
)
# Conclusion:
# * If we continue to look at the data from 1996 to 2016, we will capture the period of trend change in the video game market, plus we see from the graphs above that many platforms have managed to lose their popularity during this time.
# * Let's limit the data since 2005
data = data.loc[data["year_of_release"] >= 2005].reset_index(drop=True)
print(data.count())
# Let's make a box plot chart for global game sales by platform
data_to_plot = data.pivot_table(
index="name", columns="platform", values="total_sales", aggfunc="sum"
).boxplot(figsize=(16, 12), grid=True)
plt.ylim(-1, 15)
plt.show()
# Conclusion:
# * PS3, PS4, X360 and XOne have similar indicators on average (about 500 copies) and 75% quantile (up to 1 million sales)
data_to_plot = data.pivot_table(
index="name", columns="platform", values="total_sales", aggfunc="sum"
).boxplot(figsize=(16, 12), grid=True)
plt.ylim(-1, 2)
plt.show()
# Let's see how the users and critics scores affect sales within one platform
#
# let's filter data by value, where we know both critic_score and user_score
data = data.loc[data["critic_score"] != -1]
data = data.loc[data["user_score"] != -1].reset_index(drop=True)
display(data.head(50))
for i in platform_to_research:
print(i, "\n")
data_to_plot = data.loc[data["platform"] == i]
data_to_plot = data_to_plot[["total_sales", "critic_score", "user_score"]]
print(data_to_plot.corr())
print("user_score")
data_to_plot.plot(
kind="scatter",
x="user_score",
y="total_sales",
figsize=(12, 6),
alpha=0.5,
xlim=(0, 10),
)
plt.show()
print("critic_score")
data_to_plot.plot(
kind="scatter",
x="critic_score",
y="total_sales",
figsize=(12, 6),
alpha=0.5,
xlim=(0, 100),
)
plt.show()
# Conclusion:
# * the dependence on all data is rather the right branch of the polynomial function (example below). The higher the score of critics and players, the more likely it is that the game will sell a large circulation
# ### пример зависимости
# 
# Let's check the dependence on the horizon of 75% + 3 std deviation
for i in platform_to_research:
print(i, "\n")
data_to_plot = data.loc[data["platform"] == i]
data_to_plot = data_to_plot[["total_sales", "critic_score", "user_score"]]
print(data_to_plot.corr())
print("user_score")
data_to_plot.plot(
kind="scatter",
x="user_score",
y="total_sales",
figsize=(12, 6),
alpha=0.5,
xlim=(0, 10),
ylim=(0, 2),
)
plt.show()
print("critic_score")
data_to_plot.plot(
kind="scatter",
x="critic_score",
y="total_sales",
figsize=(12, 6),
alpha=0.5,
xlim=(0, 100),
ylim=(0, 2),
)
plt.show()
# Conclusion
# * the dependence can be traced on values up to 2 million copies, but a simpler correlation is not visible
#
#
# Let's look at the general distribution of games by genre
print(
data.pivot_table(
index="genre", values="total_sales", aggfunc=["sum", "count", "mean"]
)
)
# Conclusion:
# * Top-3 sales genres are: Action, Shooters and Sports. You should invest in their promotion. The smallest two genres are: Puzzle and Strategy.
# * There are 4 genres among the median sales of more than 1 million copies. 2 of them are leading in total sales.
# ## Step 4 Creating a portrait of the user of each region
# * Let's determine for the user of each region (NA, EU, JP):
# * The most popular platforms (top 5)
# * The most popular genres (top 5)
# * Let's check if the ESRB rating affect sales in a particular region?
regions = ["na_sales", "eu_sales", "jp_sales"]
# let's look at the total sales in the regions
for i in regions:
print(i, round(data[i].sum(), 1))
# Conclusion:
# * Sales in North America are 50% times higher than sales in Europe and 10 times higher than sales in Japan. The North American market is the most promising
# Let's look at popular platforms
for i in regions:
print(
data.pivot_table(index="platform", values=i, aggfunc="sum").sort_values(
i, ascending=False
)
)
# Conclusion:
# * wii, ps3 are in the top 3 platforms among all regions. Let's focus on their promotion
# * We must remember that a new generation of consoles has recently been released and continue capturingthe market. We need to invest in their promotion.
for i in regions:
print(
data.pivot_table(index="genre", values=i, aggfunc="sum").sort_values(
i, ascending=False
)
)
# Conclusion:
# * 4 of the 5 most popular genres in all regions are the same. It's worth investing in such games
# * The most popular genre in Japan is Role-Play
# ESRB raiting
for i in regions:
print(
data.pivot_table(index="rating", values=i, aggfunc="sum").sort_values(
i, ascending=False
)
)
"""
For info:
E - Everyone
T - Teen
M - Mature
E10+ - Everyone 10 and older
EC - Early Childhood
RP - Rating Pending
K-A - Kids to Adult
AO - Adults Only 18+
"""
# Conclusion:
# * the distribution structure of sales by category is the same for NA and EU.
# * Games for an audience of 17+ are in the first place - so this is the most actively developing segment
# * Further, the rating is essentially a funnel by age of consumers. The lower the age limit, the more willing people will be able to buy the game. The exception is EC games up to 3 years old. Children do not buy games, but their parents do it for them.
# * Most games in Japan do not have rating, since rating system is advisory and not mandatory.
# * We need to focus on Games for a wide audience or for an audience over 17 years old
# ## Step 5. Checking the hypotheses
# Let's looking if:
# * Average user ratings of Xbox One and PC platforms are the same
# * Average user ratings of genres Action and Sports are different
# * Explain:
# * How did you formulate the null and alternative hypotheses
# * What criteria were used to test hypotheses and why
# ### Hypothesis 1: The average user ratings of the Xbox One and PC platforms are the same
# * H0 Average user ratings of Xbox One and PC platforms are the same
# * H1 Average user ratings of Xbox One and PC platforms differ
alpha = 0.05 # statistical significance level
results = st.ttest_ind(
data.loc[data["platform"] == "PC"]["user_score"],
data.loc[data["platform"] == "XOne"]["user_score"],
equal_var=True,
)
# the test is two-sided: the p-value will be two times less# the test is two-sided: the p-value will be two times less
print("p-value:", results.pvalue)
# we reject the hypothesis only when the sample mean is significantly less than the assumed value
if results.pvalue < alpha:
print("We reject the null hypothesis")
else:
print("We do not reject the null hypothesis")
# Conclusion:
# * the null hypothesis is rejected, the average user ratings of the Xbox One and PC platforms differ
# ### Hypothesis 2: Average user ratings of the genres Action and Sports are different
# * H0 Average user ratings of the Action and Sports platforms are the same
# * H1 Average user ratings of the Action and Sports platforms differ
alpha = 0.05 # statistical significance level
results = st.ttest_ind(
data.loc[data["genre"] == "Action"]["user_score"],
data.loc[data["genre"] == "Sports"]["user_score"],
)
# the test is two-sided: the p-value will be two times less
print("p-value:", results.pvalue)
# the test is one-sided to the left:
# we reject the hypothesis only when the sample mean is significantly less than the assumed value
if results.pvalue < alpha:
print("We reject the null hypothesis")
else:
print("We do not reject the null hypothesis")
|
import numpy as np
import pandas as pd
import cv2
import matplotlib.pyplot as plt
from PIL import Image
import torch
IMG_SIZE = 240
MARGIN = 10
df = pd.read_csv("../input/deepfake-detection-challenge/sample_submission.csv")
video_names = df["filename"]
video_paths = [
"../input/deepfake-detection-challenge/test_videos/" + n for n in video_names
]
video_paths[:10]
sample_video = video_paths[0]
# ## Read all frames
# 1. Opencv Read Video
v_cap = cv2.VideoCapture(sample_video)
v_int = int(v_cap.get(cv2.CAP_PROP_FRAME_COUNT))
all_frames = []
for i in range(v_int):
ret, frame = v_cap.read()
frame = cv2.cvtColor(frame, cv2.COLOR_BGR2RGB)
all_frames.append(frame)
print(np.array(all_frames).shape)
# 2. Image_ffmpeg
import imageio
reader = imageio.get_reader(sample_video, "ffmpeg")
v_int = reader.count_frames()
meta = reader.get_meta_data()
all_frames = []
for i in range(v_int):
img = reader.get_data(i)
all_frames.append(img)
print(np.array(all_frames).shape)
# ## read specific frames
frames_per_video = 72
# 1. v_cap.grab()
def read_frames1(frames_per_video=60):
v_cap = cv2.VideoCapture(sample_video)
v_int = int(v_cap.get(cv2.CAP_PROP_FRAME_COUNT))
all_frames = []
sample_idx = np.linspace(
0, v_int - 1, frames_per_video, endpoint=True, dtype=np.int
)
for i in range(v_int):
# speed reading without decode unwanted frame
ret = v_cap.grab()
if ret is None:
print("The {} cannot be read".format(i))
continue
# # the frame we want
if i in sample_idx:
ret, frame = v_cap.retrieve()
if ret is None or frame is None:
continue
frame = cv2.cvtColor(frame, cv2.COLOR_BGR2RGB)
all_frames.append(frame)
return all_frames
result = read_frames1(frames_per_video)
np.array(result).shape
# 2. v_cap.read()
def read_frames2(frames_per_video=60):
v_cap = cv2.VideoCapture(sample_video)
v_int = int(v_cap.get(cv2.CAP_PROP_FRAME_COUNT))
all_frames = []
sample_idx = np.linspace(
0, v_int - 1, frames_per_video, endpoint=True, dtype=np.int
)
for i in range(v_int):
ret, frame = v_cap.read()
if ret is None or frame is None:
continue
if i in sample_idx:
frame = cv2.cvtColor(frame, cv2.COLOR_BGR2RGB)
all_frames.append(frame)
return all_frames
result = read_frames2(frames_per_video)
np.array(result).shape
# 3. imageio_ffmpeg
def read_frames3(frames_per_video):
reader = imageio.get_reader(sample_video, "ffmpeg")
v_int = reader.count_frames()
all_frames = []
sample_idx = np.linspace(
0, v_int - 1, frames_per_video, endpoint=True, dtype=np.int
)
for i in sample_idx:
img = reader.get_data(i)
all_frames.append(img)
return all_frames
result = read_frames3(frames_per_video)
np.array(result).shape
# 4. v_cap.cat
def read_frames4(frames_per_video=72):
v_cap = cv2.VideoCapture(sample_video)
v_int = int(v_cap.get(cv2.CAP_PROP_FRAME_COUNT))
all_frames = []
sample_idx = np.linspace(
0, v_int - 1, frames_per_video, endpoint=True, dtype=np.int
)
for i in sample_idx:
v_cap.set(cv2.CAP_PROP_POS_FRAMES, i)
ret, frame = v_cap.read()
frame = cv2.cvtColor(frame, cv2.COLOR_BGR2RGB)
all_frames.append(frame)
return all_frames
result = read_frames4(frames_per_video)
np.array(result).shape
# # MTCNN_PACKAGE (SLOW!!!)
from mtcnn import MTCNN
detector = MTCNN()
def detect_face(img):
img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)
final = []
detected_faces_raw = detector.detect_faces(img)
if detected_faces_raw == []:
# print('no faces found')
return []
confidences = []
for n in detected_faces_raw:
x, y, w, h = n["box"]
final.append([x, y, w, h])
confidences.append(n["confidence"])
if max(confidences) < 0.9:
return []
max_conf_coord = final[confidences.index(max(confidences))]
# return final
return max_conf_coord
def crop(img, x, y, w, h, margin, img_shape):
x -= margin
y -= margin
w += margin * 2
h += margin * 2
if x < 0:
x = 0
if y <= 0:
y = 0
return cv2.cvtColor(
cv2.resize(img[y : y + h, x : x + w], img_shape), cv2.COLOR_BGR2RGB
)
v_cap = cv2.VideoCapture(sample_video)
v_int = int(v_cap.get(cv2.CAP_PROP_FRAME_COUNT))
faces = []
for i in range(v_int):
ret, frame = v_cap.read()
frame = cv2.cvtColor(frame, cv2.COLOR_BGR2RGB)
bounding_box = detect_face(frame)
if bounding_box == []:
continue
else:
x, y, w, h = bounding_box
face = crop(frame, x, y, w, h, MARGIN, (IMG_SIZE, IMG_SIZE))
faces.append(face)
i += 1
np.array(faces).shape
# # Facenet_pytorch
from facenet_pytorch import MTCNN
mtcnn = MTCNN(image_size=IMG_SIZE, margin=MARGIN)
v_cap = cv2.VideoCapture(sample_video)
v_int = int(v_cap.get(cv2.CAP_PROP_FRAME_COUNT))
total_faces = []
for i in range(v_int):
ret, frame = v_cap.read()
frame = cv2.cvtColor(frame, cv2.COLOR_BGR2RGB)
faces = mtcnn(Image.fromarray(frame))
if faces is None:
continue
else:
total_faces.append(faces.numpy()[0])
i += 1
np.array(total_faces).shape
# # BlazeFace
import sys
sys.path.insert(0, "/kaggle/input/blazeface-pytorch")
from blazeface import BlazeFace
net = BlazeFace()
net.load_weights("/kaggle/input/blazeface-pytorch/blazeface.pth")
net.load_anchors("/kaggle/input/blazeface-pytorch/anchors.npy")
# Optionally change the thresholds:
net.min_score_thresh = 0.75
net.min_suppression_threshold = 0.3
# BlazeFace github repo offical version
import matplotlib.pyplot as plt
import matplotlib.patches as patches
def plot_detections(img, detections, with_keypoints=True):
# show the image
fig, ax = plt.subplots(1, figsize=(10, 10))
ax.grid(False)
ax.imshow(img)
if isinstance(detections, torch.Tensor):
detections = detections.cpu().numpy()
if detections.ndim == 1: # normally 2 (1, 17)
detections = np.expand_dims(detections, axis=0)
print("Found %d faces" % detections.shape[0])
for i in range(detections.shape[0]):
ymin = detections[i, 0] * img.shape[0]
xmin = detections[i, 1] * img.shape[1]
ymax = detections[i, 2] * img.shape[0]
xmax = detections[i, 3] * img.shape[1]
rect = patches.Rectangle(
(xmin, ymin),
xmax - xmin,
ymax - ymin,
linewidth=1,
edgecolor="r",
facecolor="none",
alpha=detections[i, 16],
)
ax.add_patch(rect)
if with_keypoints:
for k in range(6):
kp_x = detections[i, 4 + k * 2] * img.shape[1]
kp_y = detections[i, 4 + k * 2 + 1] * img.shape[0]
circle = patches.Circle(
(kp_x, kp_y),
radius=0.5,
linewidth=1,
edgecolor="lightskyblue",
facecolor="none",
alpha=detections[i, 16],
)
ax.add_patch(circle)
plt.show()
def tile_frames(frames, target_size):
num_frames, H, W, _ = frames.shape
split_size = min(H, W)
x_step = (W - split_size) // 2
y_step = (H - split_size) // 2
num_v = 1
num_h = 3 if W > H else 1
splits = np.zeros(
(num_frames * num_v * num_h, target_size[1], target_size[0], 3), dtype=np.uint8
)
i = 0
for f in range(num_frames):
y = 0
for v in range(num_v):
x = 0
for h in range(num_h):
crop = frames[f, y : y + split_size, x : x + split_size, :]
splits[i] = cv2.resize(crop, target_size, interpolation=cv2.INTER_AREA)
x += x_step
i += 1
y += y_step
resize_info = [split_size / target_size[0], split_size / target_size[1], 0, 0]
return splits, resize_info
# 1-1 without ***tile image***, some faces cannot be detectd
# see how many faces are detected
reader = imageio.read(sample_video, "ffmpeg")
v_int = reader.count_frames()
total_faces = 0
# show first frame
img = reader.get_data(0)
img = cv2.resize(img, net.input_size)
plt.imshow(img)
for i in range(v_int):
frame = reader.get_data(i)
frame = cv2.resize(frame, net.input_size)
detections = net.predict_on_image(frame)
# cannot detect
if len(detections) == 0:
continue
else:
total_faces += 1
print("There are {} faces are detectd from {} frames".format(total_faces, v_int))
# 2-1 show detected result without tile image
reader = imageio.read(sample_video, "ffmpeg")
v_int = reader.count_frames()
for i in range(v_int):
frame = reader.get_data(i)
frame = cv2.resize(frame, net.input_size)
detections = net.predict_on_image(frame)
# cannot detect
if len(detections) == 0:
continue
else:
print("Frame idx {}".format(i))
plot_detections(frame, detections)
break
# 1-2 see how many faces are detected with ***tile image***
reader = imageio.read(sample_video, "ffmpeg")
v_int = reader.count_frames()
total_faces = 0
all_frames = []
for i in range(v_int):
frame = reader.get_data(i)
all_frames.append(frame)
plt.imshow(frame)
detections = net.predict_on_image(frame)
print(detections)
break
np.array(total_faces).shape
# # Yolo v2
# source : https://www.kaggle.com/drjerk/detect-faces-using-yolo
def load_mobilenetv2_224_075_detector(path):
input_tensor = Input(shape=(224, 224, 3))
output_tensor = MobileNetV2(
weights=None, include_top=False, input_tensor=input_tensor, alpha=0.75
).output
output_tensor = ZeroPadding2D()(output_tensor)
output_tensor = Conv2D(kernel_size=(3, 3), filters=5)(output_tensor)
model = Model(inputs=input_tensor, outputs=output_tensor)
model.load_weights(path)
return model
# mobilenetv2 = load_mobilenetv2_224_075_detector("../input/facedetection-mobilenetv2/facedetection-mobilenetv2-size224-alpha0.75.h5")
# mobilenetv2.summary()
|
# # Importing and Seeding
# Seeding is important if you want to make a reproducible code. Though there will still be some variations, seeding here will minimize the randomness of each rerun of the kernel.
import numpy as np
import pandas as pd
import random
import time
import gc
from keras.models import Sequential
from keras.layers import Dense, Dropout, BatchNormalization
from keras.optimizers import Adam
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
seed = random.randint(10, 10000)
print("This run's seed is:", seed)
np.random.seed(seed)
random.seed(seed)
# # Model Creation
# The model to be used is a simple regressor model using a Sequential model from Keras. It has three hidden Dense layer of sizes 128, 64 and 16 with activators sigmoid, relu and relu respectively. I also applied Batch Normalization and Dropout to avoid overfitting.
# The loss function to be used is decided to be Mean Square Logarithmic Error or MSLE so that the Y_hat or predicted_value can be minimized. The metrics to be monitored is set to be the Mean Squared Error or MSE of the function. The two are similar but one is on the Logarithmic side while the other is not.
def create_model(input_shape):
model = Sequential()
model.add(Dense(128, input_dim=input_shape, activation="sigmoid"))
model.add(BatchNormalization())
model.add(Dropout(0.5))
model.add(Dense(64, input_dim=input_shape, activation="relu"))
model.add(BatchNormalization())
model.add(Dropout(0.35))
model.add(Dense(16, activation="relu"))
model.add(BatchNormalization())
model.add(Dropout(0.25))
model.add(Dense(1))
optimizer = Adam(lr=0.005, decay=0.0)
model.compile(optimizer=optimizer, loss="msle", metrics=["mse"])
return model
# # Feature Expansion
# To further simplify learning from the data, we would need to do some tweaks to it. One is to fill the N/A parts of the dataset. This can be done mostly by fitting Zero, Mean or Median of the column in its place. For this one, I chose the median for it's versatility in minimizing the amount of extremities.
# To expand the features, I also did a simple boolean columns of form key>N where N is a number between the min and the max of the column.
def data_correction_numeric(keys):
# We will access the global num_df for this
for key in keys:
mn, mx = abs(int(num_df[key].min())), abs(int(num_df[key].max()))
if mx < mn:
mn, mx = mx, mn
print("Min:", mn, "Max:", mx)
try:
for suf in range(mn, mx, int((mx - mn) / 3)):
num_df[key + ">" + str(suf)] = num_df[key].map(lambda x: x > suf)
num_df[key + "<" + str(suf)] = num_df[key].map(lambda x: x < suf)
except:
print("ERROR for %s" % key)
x_val = num_df[key].median()
num_df[key] = num_df[key].fillna(x_val)
num_df[key] = num_df[key] - x_val
def data_correction_category(df, keys):
for key in keys:
x_val = 0 # df[key].value_counts().median()
df[key] = df[key].fillna(x_val)
return df
# # Feature Engineering
# Here, we will be fitting our data correction functions to the full train and test data.
# Read the input data
train = pd.read_csv("../input/house-prices-advanced-regression-techniques/train.csv")
test = pd.read_csv("../input/house-prices-advanced-regression-techniques/test.csv")
print(train.shape, test.shape)
train = train.set_index("Id")
test = test.set_index("Id")
test_index = test.index
# Clean the train and test data
combined = pd.concat([train, test], axis=0, sort=False)
print(combined.columns)
# Free some memory
del train, test
# Get the log(y) to minimize values
Y = combined[combined["SalePrice"].notnull()]["SalePrice"].sort_index().values
log_Y = np.log(Y)
del Y
gc.collect()
numeric_val_list = [
"OverallQual",
"GrLivArea",
"YearBuilt",
"MSSubClass",
"OverallCond",
"GarageCars",
"LotArea",
"Fireplaces",
"LotFrontage",
"TotRmsAbvGrd",
"KitchenAbvGr",
"FullBath",
]
categorical_val_list = [
"BsmtExposure",
"BsmtFinType1",
"Neighborhood",
"BsmtQual",
"MSZoning",
"BsmtCond",
"Exterior1st",
"KitchenQual",
"Exterior2nd",
"SaleCondition",
"HouseStyle",
"LotConfig",
"GarageFinish",
"MasVnrType",
"RoofStyle",
]
num_df = combined[numeric_val_list]
cat_df = combined[categorical_val_list]
# Cleaning the data
data_correction_numeric(numeric_val_list)
cat_df = data_correction_category(cat_df, categorical_val_list)
cat_df = pd.get_dummies(cat_df)
num_df.columns
cat_df.columns
# # Final Adjustments
# Here, we will be adjusting the values of the train and test data once more by fitting them to a scaler.
# Split Data to train and test
train_c = cat_df[cat_df.index <= 1460]
test_c = cat_df[cat_df.index > 1460]
train_n = num_df[num_df.index <= 1460]
test_n = num_df[num_df.index > 1460]
del num_df, cat_df
scale = StandardScaler()
train_n = scale.fit_transform(train_n)
test_n = scale.fit_transform(test_n)
train = np.concatenate((train_n, train_c.values), axis=1)
test = np.concatenate((test_n, test_c.values), axis=1)
del train_c, train_n, test_c, test_n
gc.collect()
# We will also define a plotter so that we can see the train vs validation learning values.
# summarize history for loss
import matplotlib.pyplot as plt
def plotter(history, n):
plt.plot(history.history["mse"])
plt.plot(history.history["val_mse"])
plt.title("model loss")
plt.ylabel("mse")
plt.xlabel("epoch")
plt.legend(["train", "test"], loc="upper right")
plt.ylim(top=0.1, bottom=0.01)
plt.savefig("history_mse_{}.png".format(n))
plt.show()
# # Training
# Now, we fit the training model. I also use some callbacks: ReduceLROnPlateau for slow cooking, and EarlyStopping for, well, early stopping of the training. The patience values are decided after much trial and error, to ensure that there's enough room for adjustment.
# After that, we train and predict the data over a ten-fold repetition of computation. This will minimize the overfitting of the data and will, hopefully, increase the accuracy of the prediction.
# Callbacks
from tensorflow.keras.callbacks import ReduceLROnPlateau, EarlyStopping
lrr = ReduceLROnPlateau(
monitor="val_mse", patience=200, verbose=1, factor=0.75, min_lr=1e-6
)
es = EarlyStopping(
monitor="val_loss", mode="min", verbose=1, patience=1000, restore_best_weights=True
)
print("Shape of Train:", train.shape)
predictions = []
last_mse = []
folds = 10
for x in range(1, folds + 1):
# Separate train data into train and validation data
X_train, X_val, Y_train, Y_val = train_test_split(
train, log_Y, test_size=0.2, shuffle=True, random_state=seed
)
print("#" * 72)
print("Current RERUN: #%i" % (x))
# Design the Model
model = create_model(X_train.shape[1])
# Start the training
history = model.fit(
X_train,
Y_train,
validation_data=(X_val, Y_val),
epochs=10000,
batch_size=128,
verbose=0,
callbacks=[es, lrr],
)
# Predicting
predict = model.predict(test)
try:
predictions = np.concatenate([predictions, predict], axis=1)
except:
predictions = predict
# Show the MSE Plot
plotter(history, x)
loss, mse = model.evaluate(X_val, Y_val)
print("Loss:", loss, "\tMSE:", mse)
last_mse.append(mse)
# Clear some Memory
del X_train, X_val, Y_train, Y_val, model, history, predict, loss, mse
gc.collect()
# # Prediction
# Finally, we will be saving our prediction. As we did a Ten-Fold prediction, we will be doing a weighted combination based on the MSE evaluation of the validation set for each fold of the training, which was save to the variable `last_mse`. Since the metrics we used is the `error` or the predictions, then, the larger the error, the smaller the effect, thus, I used the equation `(total - x)/((n-1)*total)` so as to reverse the relationship.
def ensemble(preds, metrics):
over = sum(metrics)
n = len(metrics)
return [
sum((over - metrics[x]) * preds[i, x] / ((n - 1) * over) for x in range(n))
for i in range(len(preds))
]
print("Predicting the Test data...")
prediction = ensemble(predictions, last_mse)
prediction = np.exp(prediction)
submission = pd.DataFrame()
submission["Id"] = test_index
submission["SalePrice"] = prediction
print("Saving prediction to output...")
submission.to_csv("prediction_regression.csv", index=False)
print("Done.")
print(submission)
x = np.mean(last_mse)
print(x, x**0.5)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
import tensorflow as tf
import matplotlib.pyplot as plt
import glob
import numpy as np
keras = tf.keras
layers = keras.layers
EPOCH = 200
BATCH_SIZE = 16
DATASET_SIZE = 16465
TEST_SIZE = 7382
AUTO = tf.data.experimental.AUTOTUNE
ignore_order = tf.data.Options()
ignore_order.experimental_deterministic = False
print(f"tensorflow version : {tf.__version__}")
dataset_filenames = glob.glob(
r"/kaggle/input/flower-classification-with-tpus/tfrecords-jpeg-512x512/*a*/*.tfrec"
)
test_filenames = glob.glob(
r"/kaggle/input/flower-classification-with-tpus/tfrecords-jpeg-512x512/test/*.tfrec"
)
dataset = tf.data.TFRecordDataset(dataset_filenames)
dataset = dataset.with_options(ignore_order)
dataset_test = tf.data.TFRecordDataset(test_filenames)
dataset_test = dataset_test.with_options(ignore_order)
feature_description = {
"id": tf.io.FixedLenFeature([], tf.string),
"class": tf.io.FixedLenFeature([], tf.int64),
"image": tf.io.FixedLenFeature([], tf.string),
}
test_feature_description = {
"id": tf.io.FixedLenFeature([], tf.string),
"image": tf.io.FixedLenFeature([], tf.string),
}
def dataset_decode(data):
decode_data = tf.io.parse_single_example(data, feature_description)
label = decode_data["class"]
image = tf.image.decode_jpeg(decode_data["image"], channels=3)
image = tf.image.resize_with_pad(image, 512, 512)
image = tf.cast(image, tf.float32)
image = (image - 127.5) / 127.5
return image, label
def test_dataset_decode(data):
decode_data = tf.io.parse_single_example(data, test_feature_description)
ID = decode_data["id"]
image = tf.image.decode_jpeg(decode_data["image"], channels=3)
image = tf.image.resize_with_pad(image, 512, 512)
image = tf.cast(image, tf.float32)
image = (image - 127.5) / 127.5
return ID, image
dataset = dataset.map(dataset_decode)
dataset_test = dataset_test.map(test_dataset_decode)
dataset = dataset.shuffle(DATASET_SIZE).repeat().batch(BATCH_SIZE).prefetch(AUTO)
dataset_test = dataset_test.batch(BATCH_SIZE).prefetch(AUTO)
base_network = keras.applications.InceptionResNetV2(
include_top=False, input_shape=[512, 512, 3]
)
network = keras.Sequential(
[
base_network,
layers.MaxPool2D(),
layers.Conv2D(2048, 3, padding="same"),
layers.BatchNormalization(),
layers.LeakyReLU(),
layers.Conv2D(2048, 3, padding="same"),
layers.BatchNormalization(),
layers.LeakyReLU(),
layers.GlobalAveragePooling2D(),
layers.Dense(2048),
layers.BatchNormalization(),
layers.LeakyReLU(),
layers.Dense(1024),
layers.BatchNormalization(),
layers.LeakyReLU(),
layers.Dense(256),
layers.BatchNormalization(),
layers.LeakyReLU(),
layers.Dense(104, activation="softmax"),
]
)
network.summary()
optimizer = keras.optimizers.Adam()
loss = keras.losses.SparseCategoricalCrossentropy()
metrics = keras.metrics.SparseCategoricalAccuracy()
lr_callback = keras.callbacks.ReduceLROnPlateau(
monitor="train_loss", factor=0.5, patience=15, min_lr=1e-6
)
network.compile(optimizer=optimizer, loss=loss, metrics=[metrics])
network.fit(
dataset,
epochs=EPOCH,
steps_per_epoch=DATASET_SIZE // BATCH_SIZE,
callbacks=[lr_callback],
)
network.save(r"./Xception.h5")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import cv2
import os
from fastai.vision import *
from fastai.metrics import error_rate
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
train = pd.read_csv("../input/digit-recognizer/train.csv", encoding="latin1")
test = pd.read_csv("../input/digit-recognizer/test.csv")
sub = pd.read_csv("../input/digit-recognizer/sample_submission.csv")
# Take a look at the dataset
train.tail()
# Create an image from ds
class CustomImageList(ImageList):
def open(self, fn):
img = fn.reshape(28, 28)
# img = np.stack((img,)*3, axis=-1)
return Image(pil2tensor(img, dtype=np.float32))
@classmethod
def from_csv_custom(
cls,
path: PathOrStr,
csv_name: str,
imgIdx: int = 1,
header: str = "infer",
**kwargs,
) -> "ItemList":
df = pd.read_csv(Path(path) / csv_name, header=header)
res = super().from_df(df, path=path, cols=0, **kwargs)
res.items = df.iloc[:, imgIdx:].apply(lambda x: x.values / 255.0, axis=1).values
return res
# **Dataloader**
tfms = get_transforms(do_flip=False)
data = (
CustomImageList.from_csv_custom(
path="../input/digit-recognizer/",
csv_name="train.csv",
imgIdx=1,
convert_mode="binary",
)
.split_by_rand_pct(0.2)
.label_from_df(cols="label")
.add_test(test)
.transform(tfms)
.databunch(bs=128, num_workers=0)
.normalize(imagenet_stats)
)
# **Shows example**
data.show_batch(rows=3, figsize=(5, 5))
# **Create a learner**
learn = cnn_learner(
data, models.resnet50, metrics=accuracy, model_dir="/kaggle/working/models"
)
learn.lr_find()
learn.recorder.plot(suggestion=True)
# **Train the last layers**
learn.fit_one_cycle(1, 2e-2)
learn.save("one_epoch")
# Let's try to have a bigger accuracy
learn.fit_one_cycle(10, slice(1e-3, 1e-2))
learn.save("second_epoch")
# **Train all the layers**
learn.unfreeze()
learn.lr_find()
learn.recorder.plot()
learn.load("second_epoch")
learn.fit_one_cycle(8, max_lr=slice(1e-6, 1e-4))
learn.save("third_step")
# Show images in top_losses along with their prediction, actual, loss, and probability of actual class.
preds, y, losses = learn.get_preds(with_loss=True)
interp = ClassificationInterpretation(learn, preds, y, losses)
interp.plot_top_losses(9, figsize=(7, 7))
# it's easy to understand why the computer had a dificuly to distinguish between the numbers,
# to increase the next step would be to erase this images and retrain the model to make it more accurate
# **Submission**
# get the predictions
predictions, *_ = learn.get_preds(DatasetType.Test)
labels = np.argmax(predictions, 1)
# output to a file
submission_df = pd.DataFrame(
{"ImageId": list(range(1, len(labels) + 1)), "Label": labels}
)
submission_df.to_csv(f"submission.csv", index=False)
|
import numpy as np
import pandas as pd
TRAIN_PATH = "/kaggle/input/titanic/train.csv"
train = pd.read_csv(TRAIN_PATH)
train.head()
train_class_sex_survived = pd.pivot_table(
train,
index=["Pclass", "Sex"],
columns="Survived",
values=["Age", "Fare"],
aggfunc=["min", "max", "mean", "median"],
)
train_class_sex_survived
train_sex_pclass_survived = pd.pivot_table(
train,
index=["Sex", "Pclass"],
columns="Survived",
values=["Age", "Fare"],
aggfunc=["min", "max", "mean", "median"],
)
train_sex_pclass_survived
|
# # Machine Learning Series - Lecture 3
# ***
# ## Linear regression for a non-linear features-target relationship
# If the parametrization of linear models is not natively adapted to the problem at hand, it is still possible to make linear models more expressive by engineering additional features.
# A machine learning pipeline that combines a non-linear feature engineering step followed by a linear regression step can therefore be considered non-linear regression model as a whole.
# To illustrate these concepts, we will use a dataset where the matrix `data` and the vector `target` do not have a linear link.
import numpy as np
rng = np.random.RandomState(0)
n_sample = 100
data_max, data_min = 1.4, -1.4
len_data = data_max - data_min
# sort the data to make plotting easier later
data = np.sort(rng.rand(n_sample) * len_data - len_data / 2)
noise = rng.randn(n_sample) * 0.3
target = data**3 - 0.5 * data**2 + noise
#
# Note
# To ease the plotting, we will create a pandas dataframe containing the data
# and target:
#
import pandas as pd
full_data = pd.DataFrame({"input_feature": data, "target": target})
import seaborn as sns
_ = sns.scatterplot(
data=full_data, x="input_feature", y="target", color="black", alpha=0.5
)
# We will highlight the limitations of fitting a linear regression model.
# Warning
# In scikit-learn, by convention data (also called X in the scikit-learn
# documentation) should be a 2D matrix of shape (n_samples, n_features).
# If data is a 1D vector, you need to reshape it into a matrix with a
# single column if the vector represents a feature or a single row if the
# vector represents a sample.
#
# X should be 2D for sklearn: (n_samples, n_features)
data = data.reshape((-1, 1))
data.shape
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_squared_error
linear_regression = LinearRegression()
linear_regression.fit(data, target)
target_predicted = linear_regression.predict(data)
mse = mean_squared_error(target, target_predicted)
ax = sns.scatterplot(
data=full_data, x="input_feature", y="target", color="black", alpha=0.5
)
ax.plot(data, target_predicted)
_ = ax.set_title(f"Mean squared error = {mse:.2f}")
#
# Here the coefficient and intercept learnt by `LinearRegression` define the
# best "straight line" that fits the data. We can inspect the coefficients
# using the attributes of the model learnt as follows:
print(
f"weight: {linear_regression.coef_[0]:.2f}, "
f"intercept: {linear_regression.intercept_:.2f}"
)
#
# It is important to note that the learnt model will not be able to handle the
# non-linear relationship between `data` and `target` since linear models
# assume the relationship between `data` and `target` to be linear.
# Indeed, there are 3 possibilities to solve this issue:
# 1. choose a model that can natively deal with non-linearity,
# 2. engineer a richer set of features by including expert knowledge which can
# be directly used by a simple linear model, or
# 3. use a "kernel" to have a locally-based decision function instead of a
# global linear decision function.
# Let's illustrate quickly the first point by using a decision tree regressor
# which can natively handle non-linearity.
from sklearn.tree import DecisionTreeRegressor
tree = DecisionTreeRegressor(max_depth=3).fit(data, target)
target_predicted = tree.predict(data)
mse = mean_squared_error(target, target_predicted)
ax = sns.scatterplot(
data=full_data, x="input_feature", y="target", color="black", alpha=0.5
)
ax.plot(data, target_predicted)
_ = ax.set_title(f"Mean squared error = {mse:.2f}")
#
# Instead of having a model which can natively deal with non-linearity, we
# could also modify our data: we could create new features, derived from the
# original features, using some expert knowledge. In this example, we know that
# we have a cubic and squared relationship between `data` and `target` (because
# we generated the data).
# Indeed, we could create two new features (`data ** 2` and `data ** 3`) using
# this information as follows. This kind of transformation is called a
# polynomial feature expansion:
data.shape
data_expanded = np.concatenate([data, data**2, data**3], axis=1)
data_expanded.shape
linear_regression.fit(data_expanded, target)
target_predicted = linear_regression.predict(data_expanded)
mse = mean_squared_error(target, target_predicted)
ax = sns.scatterplot(
data=full_data, x="input_feature", y="target", color="black", alpha=0.5
)
ax.plot(data, target_predicted)
_ = ax.set_title(f"Mean squared error = {mse:.2f}")
# Let's check the new coefficients and intercept learnt by `LinearRegression` after removing non linearity between features. We can inspect the coefficients using the attributes of the model learnt as follows:
print(
f"data feature weight: {linear_regression.coef_[0]:.2f}, "
f"data^2 feature weight: {linear_regression.coef_[1]:.2f}, "
f"data^3 feature weight: {linear_regression.coef_[2]:.2f}, "
f"intercept: {linear_regression.intercept_:.2f}"
)
# > Weight for feature `data` is **-0.16**, which is approximately **zero**.
# > Weight for feature `data^2` is **-0.52**, which is approximately **-0.5**.
# > Weight for feature `data^3` is **1.07**, which is approximately **1**.
# This highly resembles the actual equation of the `target` as a function in `data` feature: `target = (1) * data^3 + (-0.5) * data^2 + (0) * data`
# We can see that even with a linear model, we can overcome the linearity
# limitation of the model by adding the non-linear components in the design of
# additional features. Here, we created new features by knowing the way the
# target was generated.
# Instead of manually creating such polynomial features one could directly use
# [sklearn.preprocessing.PolynomialFeatures](https://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.PolynomialFeatures.html).
# To demonstrate the use of the `PolynomialFeatures` class, we use a
# scikit-learn pipeline which first transforms the features and then fit the
# regression model.
from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import PolynomialFeatures
polynomial_regression = make_pipeline(
PolynomialFeatures(degree=3),
LinearRegression(),
)
polynomial_regression.fit(data, target)
target_predicted = polynomial_regression.predict(data)
mse = mean_squared_error(target, target_predicted)
ax = sns.scatterplot(
data=full_data, x="input_feature", y="target", color="black", alpha=0.5
)
ax.plot(data, target_predicted)
_ = ax.set_title(f"Mean squared error = {mse:.2f}")
#
# As expected, we observe that the predictions of the this `PolynomialFeatures`
# pipeline match the predictions of the linear model fit on manually engineered
# features.
# ***
# ## Linear Regrssion on US Housing Price
# In statistics, linear regression is a linear approach for modeling the relationship between a scalar dependent variable y and one or more explanatory variables (or independent variables) denoted X. The case of one explanatory variable is called simple linear regression. For more than one explanatory variable, the process is called multiple linear regression.
# Loading data
housing_data = pd.read_csv("../input/usa-housing/USA_Housing.csv")
housing_data.head()
# ### Feature and variable sets
# Make a list of data frame column names.
l_column = list(housing_data.columns) # Making a list out of column names
len_feature = len(l_column) # Length of column vector list
l_column
# Put all the numerical features in X and Price in y, ignore Address which is string for linear regression.
X = housing_data[l_column[0 : len_feature - 2]]
y = housing_data[l_column[len_feature - 2]]
print("Feature set size:", X.shape)
print("Variable set size:", y.shape)
X.head()
y.head()
# ### Test-train split
# Let's split the data into training and testing sets. Then we'll train the model on the training set.
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=123
)
# Check the size and shape of train/test splits (it should be in the ratio as per test_size parameter above)
print("Training feature set size:", X_train.shape)
print("Test feature set size:", X_test.shape)
print("Training variable set size:", y_train.shape)
print("Test variable set size:", y_test.shape)
# ### Model fit and training
lm = LinearRegression() # Creating a Linear Regression object 'lm'
lm.fit(
X_train, y_train
) # Fit the linear model on to the 'lm' object itself i.e. no need to set this to another variable
# Making predictions
train_pred = lm.predict(X_train)
test_pred = lm.predict(X_test)
# ### Evaluate the model
# [R2 Score Documentation](https://scikit-learn.org/stable/modules/generated/sklearn.metrics.r2_score.html)
# Calculate the accuracy
from sklearn.metrics import r2_score
print(
"R-squared value of training:", round(r2_score(y_train, train_pred), 3) * 100, "%"
)
print("R-squared value of testing:", round(r2_score(y_test, test_pred), 3) * 100, "%")
# Scatter plot of predicted price and y_test set to see if the data fall on a 45 degree straight line.
import matplotlib.pyplot as plt
plt.figure(figsize=(10, 7))
plt.title("Actual vs. predicted house prices", fontsize=25)
plt.xlabel("Actual test set house prices", fontsize=18)
plt.ylabel("Predicted house prices", fontsize=18)
plt.scatter(x=y_test, y=test_pred)
# Check the intercept and coefficients and put them in a DataFrame
print("The intercept term of the linear model:", lm.intercept_)
print("The coefficients of the linear model:", lm.coef_)
cdf = pd.DataFrame(data=lm.coef_, index=X_train.columns, columns=["Coefficients"])
cdf
# ***
# ## Let's convert our problem to a Logitic Regression one
housing_class = housing_data.drop("Address", axis=1)
housing_class.loc[
housing_data.Price > housing_data["Price"].mean(), "Price"
] = "High Price"
housing_class.loc[
housing_data.Price <= housing_data["Price"].mean(), "Price"
] = "Low Price"
housing_class
target = housing_class["Price"]
features = housing_class.drop("Price", axis=1)
# Scaling the features to be able to plot them later.
# Import sklearn.preprocessing.StandardScaler
from sklearn.preprocessing import StandardScaler
# Initialize a scaler, then apply it to the features
scaler = StandardScaler() # default=(0, 1)
features = pd.DataFrame(scaler.fit_transform(features), columns=features.columns)
# Show an example of a record with scaling applied
features.head()
# ### Fitting the Logistic model
from sklearn.linear_model import LogisticRegression
logistic_regression = LogisticRegression()
logistic_regression.fit(features, target)
pred_class = logistic_regression.predict(features)
pred_class
# ### Evaluating the Logistic model
# It should have **close** accuracy to the Linear Regression. [F1 Score Documentation](https://scikit-learn.org/stable/modules/generated/sklearn.metrics.f1_score.html)
from sklearn.metrics import f1_score
from sklearn.preprocessing import LabelEncoder
encoded_target = LabelEncoder().fit_transform(target)
encoded_predictions = LabelEncoder().fit_transform(pred_class)
print(
"F1-score value of this fit:",
round(f1_score(encoded_target, encoded_predictions), 1) * 100,
"%",
)
# ### Let's try to reduce the feature for visualization
# Will make feature selection based on correlation between features and target. Next we will make some basic plotting and visualization on the data set.
# Pairplots using seaborn
sns.pairplot(housing_data)
# Correlation matrix and heatmap
housing_data.corr()
plt.figure(figsize=(14, 8))
sns.heatmap(housing_data.corr(), annot=True, linewidths=2)
# Based on the correlation values we will select the two features `Avg. Area Income` and `Avg. Area House Age`.
features_reduced = features[["Avg. Area Income", "Avg. Area House Age"]]
features_reduced.head()
# ### Let's fit the Logistic model again!
logistic_regression = LogisticRegression()
logistic_regression.fit(features_reduced, target)
pred_class = logistic_regression.predict(features_reduced)
# ### Let's evaluate the reduced Logistic model
encoded_target = LabelEncoder().fit_transform(target)
encoded_predictions = LabelEncoder().fit_transform(pred_class)
print(
"F1-score value of this fit:",
round(f1_score(encoded_target, encoded_predictions), 1) * 100,
"%",
)
# This **emphasize** that the selected features are highly correlated to the target `price` class.
# ### Let's visualize the Logistic model
def plot_decision_function(fitted_classifier, range_features, ax=None):
"""Plot the boundary of the decision function of a classifier."""
feature_names = list(range_features.keys())
# create a grid to evaluate all possible samples
plot_step = 0.02
xx, yy = np.meshgrid(
np.arange(*range_features[feature_names[0]], plot_step),
np.arange(*range_features[feature_names[1]], plot_step),
)
# compute the associated prediction
Z = fitted_classifier.predict(np.c_[xx.ravel(), yy.ravel()])
Z = LabelEncoder().fit_transform(Z)
Z = Z.reshape(xx.shape)
# make the plot of the boundary and the data samples
if ax is None:
_, ax = plt.subplots()
ax.contourf(xx, yy, Z, alpha=0.4, cmap="RdBu_r")
return ax
range_features = {
feature_name: (
features_reduced[feature_name].min() - 1,
features_reduced[feature_name].max() + 1,
)
for feature_name in features_reduced.columns
}
ax = sns.scatterplot(
data=features_reduced.join(target),
x="Avg. Area Income",
y="Avg. Area House Age",
hue="Price",
palette=["tab:red", "tab:blue"],
)
_ = plot_decision_function(logistic_regression, range_features, ax=ax)
# Check the intercept and coefficients.
print("The intercept term of the logistic model:", logistic_regression.intercept_)
print("The coefficients of the logistic model:", logistic_regression.coef_)
# ### What if Linear Regression model would fit this data?!
# Define the classifier, and fit it to the data
model = LinearRegression()
model.fit(
features_reduced[["Avg. Area Income"]], features_reduced[["Avg. Area House Age"]]
)
# Making predictions
predictions = model.predict(features_reduced[["Avg. Area Income"]])
ax = sns.scatterplot(
data=features_reduced.join(target),
x="Avg. Area Income",
y="Avg. Area House Age",
hue="Price",
palette=["tab:red", "tab:blue"],
)
ax.plot(features_reduced[["Avg. Area Income"]], predictions, color="black")
# It seems that we used a **wrong hypothesis**!
# ***
# ## Multi-Class Logistic Regression
# Let's split the data into three classes: low, medium and high.
min = housing_data["Price"].min()
mean = housing_data["Price"].mean()
max = housing_data["Price"].max()
lower_mean = (min + mean) / 2
upper_mean = (mean + max) / 2
housing_multiclass = housing_data.drop("Address", axis=1)
housing_multiclass.loc[housing_data.Price < lower_mean, "Price"] = "Low Price"
housing_multiclass.loc[
(housing_data.Price >= lower_mean) & (housing_data.Price <= upper_mean), "Price"
] = "Medium Price"
housing_multiclass.loc[housing_data.Price > upper_mean, "Price"] = "High Price"
housing_multiclass
housing_multiclass.loc[housing_data.Price < lower_mean]
housing_multiclass.loc[housing_data.Price > upper_mean]
target = housing_multiclass["Price"]
print(np.array(target)[1500:1600])
# ### Fitting the multi-class Logistic model
logistic_regression = LogisticRegression()
logistic_regression.fit(features, target)
# Check the intercept and coefficients for the multi-class model.
print(
"The intercept term of the multi-class logistic model:",
logistic_regression.intercept_,
)
print(
"The coefficients of the multi-class logistic model:\n", logistic_regression.coef_
)
|
# # Modeling
# **Input**: Data stored in PostgreSQL (selected features & targets)
# **Output**: Trained model and its performance metrics
# Finally we are ready to create our model.
# The steps we want to follow are:
# - Get the pivot Dataframe containing the targets and the features we have selected
# - Shift the target
# - Standardize data (in order to account for different value ranges across features)
# - Apply PCA to further reduce the amount of features (speed up training while preserving information)
# - Use cross validation to split data in multiple train-test sets in order to evaluate model performance
# - Train model
# - Test model & evaluate performance metrics
# We want then to compare our "time agnostic" model with other "non-time agnostic" models that use also the time feature, like ARIMA/SARIMA/SARIMAX.
# Our model tries to use only Exogenous (external) features, while other models use only Endogenous (internal) features (ARIMA/SARIMA) or a combination of Endogenous + Exogenous (SARIMAX).
# - Target = Gold price
# - Endogenous features = trend of gold price + seasonality of gold price
# - Exogenous features = gdp growth + M3 money supply + unemployment + ...
# Our thought process is that we want to understand how external features impact the return on an asset class, not how that asset class prices move with time. (We know that given enough time the nominal price rises, mainly due to inflation)
# But it could be that prediction in the short term would be dependent also in endogenous features, so we are interested in knowing if that's the case.
# Another way to try and remove non-stationarity would be indeed to adjust (in some way) all relevant features/measures to inflation. We didn't try this in this project, but would be an interesting following step to follow.
# ### 1. Get pivot Dataframe
from portfolio_optimization_helper import get_df_from_table
import pandas as pd
df = get_df_from_table("pivot")
df["date"] = pd.to_datetime(df["date"])
df = df.sort_values(by="date").set_index("date")
df = df.apply(pd.to_numeric)
df = df.asfreq("MS")
df_diff = df.diff()
df_diff.tail()
# ### 2. Shift the target
# We want the model to predict future values of the target, so we need to "shift the targets".
# This means that while now we have for each row in the Dataframe all the data corrisponding to that specific date, we want to shift the target so that, for the target column, in row n we have the value originally in row n+shift.
# Let's choose a single target for the examples in this notebook. target266 represents gold futures prices.
# Eg. (shift = 1)
target = "target266"
y = df_diff[[target]].dropna()
y.tail()
target_df = y.shift(periods=-1)
target_df.tail()
# ### 3. Standardize data
# We build a Dataframe X that contains the features data and than for each target we build a series y with the target shifted (we'll use shift = 1 in this notebook).
# For Dataframe X we need to standardize data using StandardScaler from sklearn. This substitute each value with its z-score. That is (value - mean) / standard deviation.
feature_columns = [col for col in df_diff.columns if "feature" in col]
X = df_diff[feature_columns].dropna()
X.tail()
from sklearn.preprocessing import StandardScaler
def standardize_features(df):
scaled_features = StandardScaler().fit_transform(df.values)
return pd.DataFrame(scaled_features, index=df.index, columns=feature_columns)
X = standardize_features(X)
X.tail()
# ### 4. Apply PCA (Principal Component Analysis)
# Principal Component Analysis (or PCA) uses linear algebra to transform the dataset into a compressed form.
# Generally this is called a data reduction technique. A property of PCA is that you can choose the number of dimensions or principal component in the transformed result.
# This is used to reduce the number of features in our model, while maintining their predictive power.
# You can specify the number of components (features) you get at the end, but we don't want to compress too much as you can lose information if you do. You can use cross validation to find out the best number of components, but for this example we'll just use n_components = half the original number of features
X.tail()
from sklearn.decomposition import PCA
pca_components = int(len(feature_columns) / 2)
def apply_pca(df, pca_components):
idx = df.index
cols = [f"featurePCA{i + 1}" for i in range(pca_components)]
df = PCA(n_components=pca_components).fit_transform(df)
df = pd.DataFrame(df, columns=cols)
df.index = idx
return df
X = apply_pca(X, pca_components)
X.tail()
# ### 5. CV (cross validation)
# To test a model the basic way is to just split the dataset in 2 (a large training set and a smaller test set). You train your model on the training set and that test its performance on the unseed test set.
# A better way is to use cross validation, that is create multiple pairs of training+test and analyse the performance of the model in all the different pairings.
# Usually this train-test split is just done randomly, but for time series data the process is less straighforward as we need to maintain the chronological order between the samples.
# This can be done with a "rolling" training set and a test set coming chronologically after that training set. It's easier to look at a picture.
# 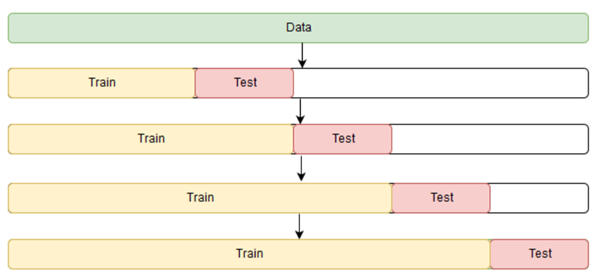
# Below an example using 5 splits and test size of 12.
from sklearn.model_selection import TimeSeriesSplit
def get_X(df, cols):
X = df[cols].dropna()
X = standardize_features(X)
pca_components = int(len(feature_columns) / 2)
X = apply_pca(X, pca_components)
return X
tscv = TimeSeriesSplit(n_splits=5, test_size=12)
split = 1
X = get_X(df_diff, feature_columns)
for train_index, test_index in tscv.split(X):
X_train, X_test = X.iloc[train_index, :], X.iloc[test_index, :]
print(f"train {split}:", min(X_train.index), " - ", max(X_train.index))
print(f"test {split}:", min(X_test.index), " - ", max(X_test.index))
print()
split += 1
# ### 6. Train model
# For each step in the cross validation process we train the model on the train set and test it on the test set.
# With testing we mean using the fitted model (trained) to predict values using the features in the test set. And then compare these predicted values with the actual values of the target in the test set.
# To make sense of the result data and plot something meaningful we need to retransform the data into the original non-stationary form. We do this by transform the prediction values (which are the differences from the previous value) into a cumulative sum (now each value is the difference from the first value - not the previous) and add the first value.
# Let's see an example using a LinearRegression model with the our example target (gold futures price).
from sklearn.linear_model import LinearRegression
import matplotlib.pyplot as plt
target = "target266"
# Features
X = get_X(df_diff, feature_columns)
# Non-stationary target
y_original = df[target].shift(-1).dropna()
# Stationary target (shifted)
y = df_diff[target].shift(-1).dropna()
# Align X, y and y_original in order to have the same rows without nulls
y, X = y.align(X, join="inner", axis=0)
y_original, y = y_original.align(y, join="inner", axis=0)
# Prepare our plot
fig = plt.figure(figsize=(17, 10))
fig.suptitle("2-years Rolling predictions (GOLD)")
subplot_idx = 1
# Function to add subplot, it plots actual values, fitted values and predicted values
def prediction_subplot(y, y_train, y_test, y_fit, y_hat, rows, columns, idx):
ax = fig.add_subplot(rows, columns, idx)
plt.plot(y.index, y, color="blue", label="actual")
plt.plot(y_train.index, y_fit, color="red", label="fit")
plt.plot(y_test.index, y_hat, color="red", linestyle=":", label="predict")
plt.legend()
# CV with 6 splits, each test set is 2-years long
tscv = TimeSeriesSplit(n_splits=6, test_size=24)
# let's store a list of predictions and tests to analyse performance later
y_hat_list = []
y_test_list = []
for train_index, test_index in tscv.split(X):
X_train, X_test = X.iloc[train_index, :], X.iloc[test_index, :]
y_train, y_test = y.iloc[train_index], y.iloc[test_index]
y_original_train, y_original_test = (
y_original.iloc[train_index],
y_original.iloc[test_index],
)
# Create and train the model
model = LinearRegression()
model.fit(X_train, y_train)
# Get non-stationary fitted values
y_fit = model.predict(X_train)
y_fit = y_fit.cumsum()
y_fit += y_original_train[0]
# Get non-stationary predicted values
y_hat = model.predict(X_test)
y_hat = y_hat.cumsum()
y_hat += y_original_train[-1]
# Store prediction and test to analyse performance later
y_hat_list.append(y_hat)
y_test_list.append(y_original_test)
prediction_subplot(y_original, y_train, y_test, y_fit, y_hat, 3, 2, subplot_idx)
subplot_idx += 1
plt.show()
# ### 7. Evaluate model performance
# There are many different metrics we can use to measure the goodness of a time series prediction model, such as:
# - MPE: Mean Percentage Error, measures the average prediction error in % terms
# - MAPE: Mean Absolute Percentage Error, as MPE but absolute values
# - ME: Mean Error, measures the average prediction error in the same unit of measure as the target
# - MAE: Mean Absolute Error, as ME but absolute values
# - RMSE: Root Mean Squared Error, squared root of the average squared prediction error
# - CORR: Pearson correlation between predicted values and actual values
# Let's see an example using the values we have from the last loop of the CV we did in the cell above.
import numpy as np
def forecast_accuracy(forecast, actual):
mae = np.mean(np.abs(forecast - actual)) # MAE
rmse = np.mean((forecast - actual) ** 2) ** 0.5 # RMSE
mape = np.mean(np.abs(forecast - actual) / np.abs(actual)) # MAPE
me = np.mean(forecast - actual) # ME
mpe = np.mean((forecast - actual) / actual) # MPE
corr = np.corrcoef(forecast, actual)[0, 1] # corr
return {"mae": mae, "rmse": rmse, "mape": mape, "me": me, "mpe": mpe, "corr": corr}
def cv_metrics(y_hat_list, y_test_list):
performance_df = []
for i in range(len(y_hat_list)):
performance_df.append(forecast_accuracy(y_hat_list[i], y_test_list[i]))
performance_df = pd.DataFrame(performance_df)
return performance_df
cv_metrics(y_hat_list, y_test_list)
# Each cross validation set has its own performance metrics. Let's compute the average and standard deviation to have an overall performance and robustness of the model.
cv_metrics(y_hat_list, y_test_list).agg(["mean", "std"])
# ### Let's now put all together!
# We want to train and test a model for each target, so we need to cycle through them and collect performance metrics.
# In this example we'll cycle through a couple targets.
from sklearn.preprocessing import StandardScaler
from sklearn.pipeline import Pipeline
feature_columns = [col for col in df_diff.columns if "feature" in col]
target_columns = [col for col in df_diff.columns if "target" in col]
# Limit to 2 targets for this example
target_columns = target_columns[:2]
pca_components = int(len(feature_columns) / 2)
# list to store model performance
performance = []
for target in target_columns:
X = df_diff[feature_columns].dropna()
y = df_diff[target].shift(-1).dropna()
y_original = df[target].shift(-1).dropna()
y, X = y.align(X, join="inner", axis=0)
y_original, y = y_original.align(y, join="inner", axis=0)
for train_index, test_index in tscv.split(X):
X_train, X_test = X.iloc[train_index, :], X.iloc[test_index, :]
y_train, y_test = y.iloc[train_index], y.iloc[test_index]
y_original_train, y_original_test = (
y_original.iloc[train_index],
y_original.iloc[test_index],
)
# a Pipeline object allows us to specify several steps to be executed sequentially
# in this case we want to standardize features and apply PCA
# last step in a pipeline must be an estimator
pipe = Pipeline(
steps=[
("scaler", StandardScaler()),
("pca", PCA(n_components=pca_components)),
("model", LinearRegression()),
]
)
pipe.fit(X_train, y_train)
y_hat = pipe.predict(X_test)
y_hat = y_hat.cumsum()
y_hat += y_original_train[-1]
p_ = forecast_accuracy(y_hat, y_original_test)
p_["target"] = target
performance.append(p_)
p_df = pd.DataFrame(performance)
p_df
# To compare results across different features our best bet are the metrics that compute % error (MAPE, MPE) or the correlation metric.
# What we could now do is trying different regression models (instead of the basic LinearRegression), and also use cross-validation for hyperparameter-tuning. Each model has many hyperparameters that can be tuned and a way to do this is to try different combinations of hyperparameters and compare the performance.
# There are sklearn objects designed specifically with this goal in mind, like GridSearchCV and many others.
# https://scikit-learn.org/stable/modules/classes.html#hyper-parameter-optimizers
# This is a pretty much a big chore, where the goal line is dependent just on computing power and patience.
# We'll just give an example of how to do it below, without entering in too much detail.
def hyperparameter_tuning(X, y):
pipe = Pipeline(
steps=[
("scaler", StandardScaler()),
("pca", PCA(n_components=pca_components)),
("model", LinearRegression()),
]
)
param_grid = [
{
"scaler": [StandardScaler(), "passthrough"],
"pca": [PCA(n_components=5), PCA(n_components=10), "passthrough"],
"model": [SGDRegressor()],
"model__learning_rate": ["invscaling", "adaptive"],
"model__l1_ratio": [0.15, 0.5],
},
{
"scaler": [StandardScaler(), "passthrough"],
"pca": [PCA(n_components=5), PCA(n_components=10), "passthrough"],
"model": [MLPRegressor()],
"model__hidden_layer_sizes": [
(100,),
(10,),
(
50,
3,
),
],
"model__solver": ["lbfgs", "sgd", "adam"],
},
...,
]
cv = GridSearchCV(pipe, param_grid, cv=TimeSeriesSplit(n_splits=6, test_size=24))
cv.fit(X, y)
return pd.DataFrame(cv.cv_results_)
# What we are really interested in is the performance difference of our model with other "time-dependent" models, that make use of the time variable to help them predict.
# ## ARIMA
# ARIMA stands for AutoRegressive Integrated Moving Average. It is a model that uses past values to predict future values.
# ARIMA Components:
# - AR (Autoregression), model uses lag values (past values) to forecast
# - I (Integrated), model apply differencing to make data stationary
# - MA (Moving average), model uses lagged forecast errors to forecast
# ARIMA Parameters:
# - p, order of AR, number of lags of Y to be used as features
# - q, order of MA, number of lagged forecast errors to be used as features
# - d, differencing period, number of differencing needed to make data stationary
# Let's start analysing data to find the correct parameters for our ARIMA model.
target = "target266" # Let's continue using Gold
y = df[[target]].dropna()
y.tail()
# To find d we need to choose how many times we need to differentiate our data in order for it to become stationary.
# We can use the ADFuller test we have already seen in the EDA step.
from statsmodels.tsa.stattools import adfuller
def d_arima(series):
d = 0
tmp = series.copy()
while d < 5:
# take p-value from ADFuller test
p_value = adfuller(tmp)[1]
# if data is stationary, stop
if p_value < 0.05:
return d
# else take another differencing step
else:
tmp = tmp.diff().dropna()
d += 1
return d
d_arima(y)
# We get d = 1, one differencing is enough to make data stationary.
# We can find the value for q parameter by inspecting the ACF plot.
# The autocorrelation function ACF computes the correlation between the actual values and the lagged values. So for lag=1, the correlation between the value and the previous one. For lag=2, the correlation between the value and the value 2 months before, and so on.
# We choose q as the last significant autocorrelated lag.
import statsmodels.api as sm
sm.graphics.tsa.plot_acf(y.diff().dropna(), lags=10)
plt.show()
# In this case no lag is significant, so we choose q = 0. We could make a case for q=1 because the point is pretty much at the significance level, but when in doubt better choose the lower value.
# We can find the value for p parameter by inspecting the PACF plot.
# The difference between autocorrelation function ACF and partial PACF is that in ACF we have the correlation between the actual value with every lag, while in PACF we have only the "incremental autocorrelation".
# The partial autocorrelation for a lag is the correlation that cannot be explained by shorter lags.
# We choose p as the last significant partial autocorrelated lag.
import statsmodels.api as sm
sm.graphics.tsa.plot_pacf(y.diff().dropna(), lags=10, method="ywm")
plt.show()
# In this case no lag is significant, so we choose p = 0. We could make a case for p=1 because the point is pretty much at the significance level, but when in doubt better choose the lower value.
# ARIMA, being only based on the target data and no other exogenous features can predict out-of-sample. Let's try this.
from statsmodels.tsa.arima.model import ARIMA
p = 0
d = 1
q = 0
model = ARIMA(y, order=(p, d, q), trend=[0, 1]).fit()
fc = model.forecast(24)
plt.title("ARIMA 0,1,0 (GOLD)")
plt.plot(y.index, y, color="blue", label="actual")
plt.plot(y.index, model.fittedvalues, color="red", linewidth=0.5, label="fit")
plt.plot(fc.index, fc, color="red", linestyle=":", label="predict")
plt.legend()
plt.show()
fc.tail()
# An easier way to compute the parameters for ARIMA is to use another library that has already a method to automatically compute the optimal parameters based on the data.
# Let's see how.
import pmdarima as pm
model = pm.auto_arima(y, test="adf", seasonal=False)
order = model.get_params()["order"]
print(f"p:{order[0]}, d:{order[1]}, q:{order[2]}")
# Auto ARIMA choose q=1, the only difference with our "manual" parameters.
# Let's use the model to predict.
fc = model.predict(n_periods=24)
plt.title("Auto-ARIMA (GOLD)")
plt.plot(y.index, y, color="blue", label="actual")
plt.plot(y.index, model.fittedvalues(), color="red", linewidth=0.5, label="fit")
plt.plot(fc.index, fc, color="red", linestyle=":", label="predict")
plt.legend()
plt.show()
fc.tail()
# We get similar predicted values.
# Let's see the performance metrics for this model (we have to do cross validation in order to do that and compare with our previous model).
# CV with 6 splits, each test set is 2-years long
tscv = TimeSeriesSplit(n_splits=6, test_size=24)
# Prepare our plot
fig = plt.figure(figsize=(17, 10))
fig.suptitle("ARIMA Predictions (GOLD)")
subplot_idx = 1
# let's store a list of predictions and tests to analyse performance later
y_hat_list = []
y_test_list = []
for train_index, test_index in tscv.split(y):
y_train, y_test = y.iloc[train_index], y.iloc[test_index]
model = pm.auto_arima(y_train, test="adf", seasonal=False)
y_hat = model.predict(n_periods=24)
# Store prediction and test to analyse performance later
y_hat_list.append(y_hat)
y_test_list.append(y_test.iloc[:, 0])
prediction_subplot(
y, y_train, y_test, model.fittedvalues(), y_hat, 3, 2, subplot_idx
)
subplot_idx += 1
plt.show()
cv_metrics(y_hat_list, y_test_list).agg(["mean", "std"])
# This is very similar to our original model, at least for this specific target.
# Let's try to add seasonality to the model to see if we can improve performance.
# ### SARIMA
# SARIMA stands for **Seasonal** AutoRegressive Integrated Moving Average.
# It uses the same concepts as ARIMA (uses past values to forecast future values), but also takes into account any seasonality patterns.
# So, it is more powerful than ARIMA in forecasting complex data spaces containing cycles.
# SARIMA has the same p,q,d parameters as ARIMA, plus 4 additional ones.
# SARIMA additional parameters:
# - m, number of time steps for a single seasonal period
# - P, order of seasonal AR, number of (lags * m) of Y to be used as features
# - Q, order of MA, number of (lagged forecast errors * m) to be used as features
# - D, differencing period, number of differencing needed to make data stationary (shift=m)
# We can use auto_arima to quickly build the model, passing Seasonal=True and a value for m that reflects the use case.
# In this case we use m=12, as we assume an annual seasonality.
# CV with 6 splits, each test set is 2-years long
tscv = TimeSeriesSplit(n_splits=6, test_size=24)
# Prepare our plot
fig = plt.figure(figsize=(17, 10))
fig.suptitle("SARIMA Predictions (GOLD)")
subplot_idx = 1
# let's store a list of predictions and tests to analyse performance later
y_hat_list = []
y_test_list = []
for train_index, test_index in tscv.split(y):
y_train, y_test = y.iloc[train_index], y.iloc[test_index]
model = pm.auto_arima(y_train, test="adf", m=12, seasonal=True)
y_hat = model.predict(n_periods=24)
# Store prediction and test to analyse performance later
y_hat_list.append(y_hat)
y_test_list.append(y_test.iloc[:, 0])
prediction_subplot(
y, y_train, y_test, model.fittedvalues(), y_hat, 3, 2, subplot_idx
)
subplot_idx += 1
plt.show()
cv_metrics(y_hat_list, y_test_list).agg(["mean", "std"])
# We get similar results as ARIMA, due to the fact that probably there is no annual seasonality for gold.
# A way to make ARIMA more powerful is to add exogenous features, with SARIMAX.
# ### SARIMAX
# SARIMAX stands for Seasonal AutoRegressive Integrated Moving Average with **Exogenous** factors.
# It is similar to SARIMA with the difference that we can also pass Exogenous features to the model.
# Let's see it in action.
X = get_X(df, feature_columns)
target = "target266" # Let's continue using Gold
y = df[[target]].dropna()
y, X = y.align(X, join="inner", axis=0)
# CV with 6 splits, each test set is 2-years long
tscv = TimeSeriesSplit(n_splits=6, test_size=24)
# Prepare our plot
fig = plt.figure(figsize=(17, 10))
fig.suptitle("SARIMAX Predictions (GOLD)")
subplot_idx = 1
# let's store a list of predictions and tests to analyse performance later
y_hat_list = []
y_test_list = []
for train_index, test_index in tscv.split(y):
X_train, X_test = X.iloc[train_index, :], X.iloc[test_index, :]
y_train, y_test = y.iloc[train_index], y.iloc[test_index]
model = pm.auto_arima(y_train, X=X_train, test="adf", m=12, seasonal=True)
y_hat = model.predict(n_periods=24, X=X_test)
# Store prediction and test to analyse performance later
y_hat_list.append(y_hat)
y_test_list.append(y_test.iloc[:, 0])
prediction_subplot(
y, y_train, y_test, model.fittedvalues(), y_hat, 3, 2, subplot_idx
)
subplot_idx += 1
plt.show()
cv_metrics(y_hat_list, y_test_list).agg(["mean", "std"])
# Results are slightly better if you take into consideration exogenous variables and overall a little better than our model than did not take into consideration endogenous features (aka past values of the target).
# Of course our model could also be better tuned with hyperparameters optimization.
# While it looks like most of the prediction power comes from exogenous features (like we first thought), it seems that also endogenous features are useful in term of improving our prediction accuracy.
# Let's try to build a new model based on this new information.
# ## Hybrid Models
# We want now to integrate endogenous features into our model (aka past values), so we better start by defining the components of a time series.
# There are 4 main components of a time series:
# - Trend - The trend component refers to the overall direction of the data over time. This can be an increasing, decreasing or a stationary trend
# - Seasonality - Seasonality refers to the periodic fluctuations in the data that occur at regular intervals. These fluctuations could be daily, weekly, monthly, quarterly or yearly
# - Cyclical - The cyclical component of a time series refers to the long-term, non-seasonal fluctuations that occur over an extended period of time. These fluctuations are not periodic, but they tend to be related to business cycles, economic cycles or other long-term trends
# - Random - The random component, also known as the residual, refers to the unpredictable fluctuations in the data that are not explained by the other components
# Different models are better at identifying specific components of a time series, so we can build a hybrid model that makes 2 or models work together to give us the best prediction.
# Let's start with the trend component.
# Here we have a choice to make. The polynomial order of the trend line to fit. The higher the polynomial, the better fit we will get on training data, but the more likely to overfit the model and get poor results on test data.
# For this example we choose 2.
from statsmodels.tsa.deterministic import DeterministicProcess
target = "target266" # Let's continue using Gold
y = df[[target]].dropna()
dp = DeterministicProcess(index=y.index, constant=True, order=2, drop=True)
X_dp = dp.in_sample()
X_dp
# DeterministicProcess build a features Dataframe entirely based on the time variable, with the polynomial order we specify.
# Let's use this simple features to predict.
# CV with 6 splits, each test set is 2-years long
tscv = TimeSeriesSplit(n_splits=6, test_size=24)
# Prepare our plot
fig = plt.figure(figsize=(17, 10))
fig.suptitle("Trend component (GOLD)")
subplot_idx = 1
# let's store a list of predictions and tests to analyse performance later
y_hat_list = []
y_test_list = []
for train_index, test_index in tscv.split(y):
X_train, X_test = X_dp.iloc[train_index, :], X_dp.iloc[test_index, :]
y_train, y_test = y.iloc[train_index], y.iloc[test_index]
model = LinearRegression()
model.fit(X_train, y_train)
y_hat = model.predict(X_test)
# Store prediction and test to analyse performance later
y_hat_list.append(y_hat.ravel())
y_test_list.append(y_test.iloc[:, 0])
prediction_subplot(
y, y_train, y_test, model.predict(X_train), y_hat, 3, 2, subplot_idx
)
subplot_idx += 1
plt.show()
cv_metrics(y_hat_list, y_test_list).agg(["mean", "std"])
# As you can see the model is pretty basic at this point, we still need to add seasonality and cycle components.
# Let's now add seasonality component.
dp = DeterministicProcess(
index=y.index,
constant=True,
order=2,
drop=True,
seasonal=True,
)
X_dp = dp.in_sample()
X_dp
# This add new dummy features representing seasonality.
# Let's try to predict now.
# CV with 6 splits, each test set is 2-years long
tscv = TimeSeriesSplit(n_splits=6, test_size=24)
# Prepare our plot
fig = plt.figure(figsize=(17, 10))
fig.suptitle("Trend + Seasonality components (GOLD)")
subplot_idx = 1
# let's store a list of predictions and tests to analyse performance later
y_hat_trend_list = []
y_fit_trend_list = []
y_test_list = []
for train_index, test_index in tscv.split(y):
X_train, X_test = X_dp.iloc[train_index, :], X_dp.iloc[test_index, :]
y_train, y_test = y.iloc[train_index], y.iloc[test_index]
model = LinearRegression()
model.fit(X_train, y_train)
y_fit = model.predict(X_train)
y_hat = model.predict(X_test)
# Store prediction and test to analyse performance later
y_fit_trend_list.append(y_fit.ravel())
y_hat_trend_list.append(y_hat.ravel())
y_test_list.append(y_test.iloc[:, 0])
prediction_subplot(
y, y_train, y_test, model.predict(X_train), y_hat, 3, 2, subplot_idx
)
subplot_idx += 1
plt.show()
cv_metrics(y_hat_trend_list, y_test_list).agg(["mean", "std"])
# This add little information because as we saw before our data has little seasonality.
# Finally let's add cyclical component using a second model (in this case we'll use a different model - GradientBoostingRegressor, but we could also use another LinearRegression model).
# We want the target to be the residuals of the previous model, because we want to fit this second model just on the errors of the first one. This is the way to combine the strengths of the 2 models.
# The features in this case will be the shifted target (in this case we call this lag because we are shifting in the opposite direction). For example, with lag=1 the row for march 2022 will contain the target value on feb 2022 as a feature.
# First thing we need to choose how many lags to use for our target, and we can do this in a similar way as we did for choosing the ARIMA parameters.
# Plotting the PACF we can see how many lags add information about autocorrelation.
import statsmodels.api as sm
sm.graphics.tsa.plot_pacf(y.dropna(), lags=10, method="ywm")
plt.show()
# We can see that pretty much all autocorrelation is already explained with the first lags, and the subsequential ones add little to no information.
# So for this target we can use lags = 1
from sklearn.ensemble import GradientBoostingRegressor
def make_lags(df, lags):
res = df.copy()
old_cols = df.columns
for i in range(1, lags + 1):
for col in old_cols:
res[f"{col}_lag_{i}"] = res[col].shift(i)
return res.drop(old_cols, axis=1)
lags = 1
X_lags = make_lags(y, lags=lags)
X_lags = X_lags.fillna(0.0)
target = "target266" # Let's continue using Gold
y = df[[target]].dropna()
# CV with 6 splits, each test set is 2-years long
tscv = TimeSeriesSplit(n_splits=6, test_size=24)
# Prepare our plot
fig = plt.figure(figsize=(17, 10))
fig.suptitle("Trend + Seasonality + Cyclical components (GOLD)")
subplot_idx = 1
# let's store a list of predictions and tests to analyse performance later
y_hat_list = []
y_test_list = []
for i, (train_index, test_index) in enumerate(tscv.split(y)):
X_train, X_test = X_lags.iloc[train_index, :], X_lags.iloc[test_index, :]
y_train, y_test = y.iloc[train_index], y.iloc[test_index]
y_residuals = y_train - pd.DataFrame(
y_fit_trend_list[i], index=y_train.index, columns=y_train.columns
)
model = GradientBoostingRegressor()
model.fit(X_train, y_residuals.values.ravel())
y_hat = model.predict(X_test) + y_hat_trend_list[i]
# Store prediction and test to analyse performance later
y_hat_list.append(y_hat)
y_test_list.append(y_test.iloc[:, 0])
prediction_subplot(
y,
y_train,
y_test,
model.predict(X_train) + y_fit_trend_list[i],
y_hat,
3,
2,
subplot_idx,
)
subplot_idx += 1
plt.show()
cv_metrics(y_hat_list, y_test_list).agg(["mean", "std"])
# This is what we can get using only endogenous features, let's try to add exogenous features to our cycle fitting step to further improve the model predictive power.
# We also lag these features with the same lags steps as the target.
# We need to also retrain the model for trend & seasonality components because by adding exogenous features we have to remove nans and doing so reduces also the data points we have available for y.
lags = 1
X = get_X(df, feature_columns)
target = "target266" # Let's continue using Gold
y = df[[target]].dropna()
y, X = y.align(X, join="inner", axis=0)
dp = DeterministicProcess(
index=y.index,
constant=True,
order=2,
drop=True,
seasonal=True,
)
X_dp = dp.in_sample()
X_lags = make_lags(y, lags=lags)
X_lags = pd.concat([X_lags, make_lags(X, lags=lags)], axis=1)
X_lags = X_lags.fillna(0.0)
# CV with 6 splits, each test set is 2-years long
tscv = TimeSeriesSplit(n_splits=6, test_size=24)
# Prepare our plot
fig = plt.figure(figsize=(17, 10))
fig.suptitle("Trend + Seasonality components (GOLD)")
subplot_idx = 1
# let's store a list of predictions and tests to analyse performance later
y_hat_list = []
y_test_list = []
for i, (train_index, test_index) in enumerate(tscv.split(y)):
# Fit trend and seasonality
X_train, X_test = X_dp.iloc[train_index, :], X_dp.iloc[test_index, :]
y_train, y_test = y.iloc[train_index], y.iloc[test_index]
model = LinearRegression()
model.fit(X_train, y_train)
y_fit = pd.DataFrame(
model.predict(X_train), index=y_train.index, columns=y_train.columns
)
y_hat = pd.DataFrame(
model.predict(X_test), index=y_test.index, columns=y_test.columns
)
# Fit cyclical on residuals (with exogenous features)
X_train, X_test = X_lags.iloc[train_index, :], X_lags.iloc[test_index, :]
y_train, y_test = y.iloc[train_index], y.iloc[test_index]
y_residuals = y_train - pd.DataFrame(
y_fit, index=y_train.index, columns=y_train.columns
)
model = GradientBoostingRegressor()
model.fit(X_train, y_residuals.values.ravel())
y_fit += pd.DataFrame(
model.predict(X_train), index=y_train.index, columns=y_train.columns
)
y_hat += pd.DataFrame(
model.predict(X_test), index=y_test.index, columns=y_test.columns
)
# Store prediction and test to analyse performance later
y_hat_list.append(y_hat.iloc[:, 0])
y_test_list.append(y_test.iloc[:, 0])
prediction_subplot(y, y_train, y_test, y_fit, y_hat, 3, 2, subplot_idx)
subplot_idx += 1
plt.show()
cv_metrics(y_hat_list, y_test_list).agg(["mean", "std"])
|
USE_SAMPLE = False
USE_ONLY_SELECTED_FOLDS = True # WARNING - if this is false no folds would me made at all lol, need to fix this!
selected_folds = [0, 1, 2]
WEIGHTS = "dynamic softmax"
ROUND = True
USE_LGB = True
USE_3SPLIT_BERT = False
USE_TEST_SAFETY_ADJUSTMENTS = True
beta = 15
import sys
sys.path.insert(0, "../input/tokenizers0011/")
import pandas as pd
import numpy as np
import lightgbm as lgb
from sklearn.model_selection import KFold, StratifiedKFold
from sklearn.model_selection import GroupKFold
import matplotlib.pyplot as plt
from tqdm.notebook import tqdm
import psutil
import gc
# import tensorflow_hub as hub
import tensorflow as tf
# import bert_tokenization as tokenization
import tensorflow.keras.backend as K
import os
from scipy.stats import spearmanr
from math import floor, ceil
from transformers import *
import nltk
np.set_printoptions(suppress=True)
print(tf.__version__)
pd.set_option("display.max_colwidth", 500)
pd.set_option("display.max_rows", 500)
pd.set_option("display.max_columns", 500)
import matplotlib
import pickle
COLOR = "white"
matplotlib.rcParams["text.color"] = COLOR
matplotlib.rcParams["axes.labelcolor"] = COLOR
matplotlib.rcParams["xtick.color"] = COLOR
matplotlib.rcParams["ytick.color"] = COLOR
pd.set_option("display.max_colwidth", 500)
pd.set_option("display.max_rows", 500)
pd.set_option("display.max_columns", 500)
# #### 1. Read data and tokenizer
# Read tokenizer and data, as well as defining the maximum sequence length that will be used for the input to Bert (maximum is usually 512 tokens)
PATH = "../input/google-quest-challenge/"
BERT_PATH = "../input/bert-base-uncased-huggingface-transformer/"
GPT2_PATH = "../input/gpt2-hugginface-pretrained/"
XLNET_PATH = "../input/xlnet-huggingface-pretrained/"
MAX_SEQUENCE_LENGTH = 512
df_train = pd.read_csv(PATH + "train.csv")
df_test = pd.read_csv(PATH + "test.csv")
df_sub = pd.read_csv(PATH + "sample_submission.csv")
print("train shape =", df_train.shape)
print("test shape =", df_test.shape)
output_categories = list(df_train.columns[11:])
input_categories = list(df_train.columns[[1, 2, 5]])
print("\noutput categories:\n\t", output_categories)
print("\ninput categories:\n\t", input_categories)
if USE_SAMPLE:
df_train = df_train.iloc[0 : round(0.05 * df_train.shape[0]), :]
df_test = df_test.iloc[0 : round(0.1 * df_test.shape[0]), :]
# #### 2. Preprocessing functions
# These are some functions that will be used to preprocess the raw text data into useable Bert inputs.
# *update 4:* credits to [Minh](https://www.kaggle.com/dathudeptrai) for this implementation. If I'm not mistaken, it could be used directly with other Huggingface transformers too! Note that due to the 2 x 512 input, it will require significantly more memory when finetuning BERT.
def save_file(var, name):
pickle.dump(var, open(f"/kaggle/working/{name}.p", "wb"))
def _convert_to_transformer_inputs(
title, question, answer, tokenizer, max_sequence_length
):
"""Converts tokenized input to ids, masks and segments for transformer (including bert)"""
def return_id(str1, str2, truncation_strategy, length):
inputs = tokenizer.encode_plus(
str1,
str2,
add_special_tokens=True,
max_length=length,
truncation_strategy=truncation_strategy,
)
input_ids = inputs["input_ids"]
input_masks = [1] * len(input_ids)
input_segments = inputs["token_type_ids"]
padding_length = length - len(input_ids)
padding_id = tokenizer.pad_token_id if USING_PAD_TOKEN else 0
input_ids = input_ids + ([padding_id] * padding_length)
input_masks = input_masks + ([0] * padding_length)
input_segments = input_segments + ([0] * padding_length)
return [input_ids, input_masks, input_segments]
input_ids_q, input_masks_q, input_segments_q = return_id(
title, question, "longest_first", max_sequence_length
)
input_ids_qa, input_masks_qa, input_segments_qa = return_id(
title + " " + question, answer, "longest_first", max_sequence_length
)
input_ids_a, input_masks_a, input_segments_a = return_id(
answer, None, "longest_first", max_sequence_length
)
return [
input_ids_q,
input_masks_q,
input_segments_q,
input_ids_qa,
input_masks_qa,
input_segments_qa,
input_ids_a,
input_masks_a,
input_segments_a,
]
def compute_input_arrays(df, columns, tokenizer, max_sequence_length):
input_ids_q, input_masks_q, input_segments_q = [], [], []
input_ids_qa, input_masks_qa, input_segments_qa = [], [], []
input_ids_a, input_masks_a, input_segments_a = [], [], []
for _, instance in tqdm(df[columns].iterrows()):
t, q, a = instance.question_title, instance.question_body, instance.answer
(
ids_q,
masks_q,
segments_q,
ids_qa,
masks_qa,
segments_qa,
ids_a,
masks_a,
segments_a,
) = _convert_to_transformer_inputs(t, q, a, tokenizer, max_sequence_length)
# ids_q, masks_q, segments_q, ids_qa, masks_qa, segments_qa = _convert_to_transformer_inputs(t, q, a, tokenizer, max_sequence_length)
input_ids_q.append(ids_q)
input_masks_q.append(masks_q)
input_segments_q.append(segments_q)
input_ids_qa.append(ids_qa)
input_masks_qa.append(masks_qa)
input_segments_qa.append(segments_qa)
input_ids_a.append(ids_a)
input_masks_a.append(masks_a)
input_segments_a.append(segments_a)
return [
np.asarray(input_ids_q, dtype=np.int32),
np.asarray(input_masks_q, dtype=np.int32),
np.asarray(input_segments_q, dtype=np.int32),
np.asarray(input_ids_qa, dtype=np.int32),
np.asarray(input_masks_qa, dtype=np.int32),
np.asarray(input_segments_qa, dtype=np.int32),
np.asarray(input_ids_a, dtype=np.int32),
np.asarray(input_masks_a, dtype=np.int32),
np.asarray(input_segments_a, dtype=np.int32),
]
def compute_output_arrays(df, columns):
return np.asarray(df[columns])
# #### 3. Create model
# `compute_spearmanr()` is used to compute the competition metric for the validation set
# `create_model()` contains the actual architecture that will be used to finetune BERT to our dataset.
#
xlnetcfg = {
"architectures": ["XLNetLMHeadModel"],
"attn_type": "bi",
"bi_data": False,
"bos_token_id": 0,
"clamp_len": -1,
"d_head": 64,
"d_inner": 3072,
"d_model": 768,
"do_sample": False,
"dropout": 0.1,
"end_n_top": 5,
"eos_token_ids": 0,
"ff_activation": "gelu",
"finetuning_task": None,
"id2label": {0: "LABEL_0", 1: "LABEL_1"},
"initializer_range": 0.02,
"is_decoder": False,
"label2id": {"LABEL_0": 0, "LABEL_1": 1},
"layer_norm_eps": 1e-12,
"length_penalty": 1.0,
"max_length": 20,
"mem_len": None,
"model_type": "xlnet",
"n_head": 12,
"n_layer": 12,
"num_beams": 1,
"num_labels": 2,
"num_return_sequences": 1,
"output_attentions": False,
"output_hidden_states": False,
"output_past": True,
"pad_token_id": 0,
"pruned_heads": {},
"repetition_penalty": 1.0,
"reuse_len": None,
"same_length": False,
"start_n_top": 5,
"summary_activation": "tanh",
"summary_last_dropout": 0.1,
"summary_type": "last",
"summary_use_proj": True,
"temperature": 1.0,
"top_k": 50,
"top_p": 1.0,
"torchscript": False,
"untie_r": True,
"use_bfloat16": False,
"vocab_size": 32000,
}
def compute_spearmanr_ignore_nan(trues, preds):
rhos = []
for tcol, pcol in zip(np.transpose(trues), np.transpose(preds)):
rhos.append(spearmanr(tcol, pcol).correlation)
return np.nanmean(rhos)
def create_nn_model(output_len, model_type):
q_id = tf.keras.layers.Input((MAX_SEQUENCE_LENGTH,), dtype=tf.int32)
q_mask = tf.keras.layers.Input((MAX_SEQUENCE_LENGTH,), dtype=tf.int32)
q_atn = tf.keras.layers.Input((MAX_SEQUENCE_LENGTH,), dtype=tf.int32)
if model_type == "BERT":
config = BertConfig() # print(config) to see settings
config.output_hidden_states = False # Set to True to obtain hidden states
# caution: when using e.g. XLNet, XLNetConfig() will automatically use xlnet-large config
# normally ".from_pretrained('bert-base-uncased')", but because of no internet, the
# pretrained model has been downloaded manually and uploaded to kaggle.
bert_model = TFBertModel.from_pretrained(
BERT_PATH + "bert-base-uncased-tf_model.h5", config=config
)
# if config.output_hidden_states = True, obtain hidden states via bert_model(...)[-1]
q_embedding = bert_model(q_id, attention_mask=q_mask, token_type_ids=q_atn)[0]
elif model_type == "GPT2":
# config = GPT2Config.from_pretrained(GPT2_PATH+'gpt2-tf_model.h5')
config = GPT2Config()
# caution: when using e.g. XLNet, XLNetConfig() will automatically use xlnet-large config
# normally ".from_pretrained('bert-base-uncased')", but because of no internet, the
# pretrained model has been downloaded manually and uploaded to kaggle.
gpt2_model = TFGPT2Model.from_pretrained(
GPT2_PATH + "gpt2-tf_model.h5", config=config
)
# if config.output_hidden_states = True, obtain hidden states via bert_model(...)[-1]
q_embedding = gpt2_model(q_id)[0]
elif model_type == "XLNET":
config = XLNetConfig.from_dict(xlnetcfg)
# config = XLNetConfig.from_pretrained(XLNET_PATH+'xlnet-vocab.json')
# config = XLNetConfig()
xlnet_model = TFXLNetModel.from_pretrained(
XLNET_PATH + "xlnet-tf_model.h5", config=config
)
q_embedding = xlnet_model(q_id, attention_mask=q_mask, token_type_ids=q_atn)[0]
x = tf.keras.layers.GlobalAveragePooling1D()(q_embedding)
x = tf.keras.layers.Dense(output_len, activation="sigmoid")(x)
model = tf.keras.models.Model(inputs=[q_id, q_mask, q_atn], outputs=x)
return model
# ### some extra preprocessing
answer_and_question = [
"answer_level_of_information",
"answer_helpful",
"answer_plausible",
"answer_relevance",
"answer_satisfaction",
]
answer_only = [
"answer_type_instructions",
"answer_type_procedure",
"answer_type_reason_explanation",
"answer_well_written",
]
AQ_and_AO = answer_and_question + answer_only
gkf10 = GroupKFold(n_splits=10).split(
X=df_train.question_body, groups=df_train.question_body
)
gkf5 = GroupKFold(n_splits=5).split(
X=df_train.question_body, groups=df_train.question_body
)
common_validation_idx = []
val10 = []
val5 = []
val10_fold0 = None
for fold, (train_idx, valid_idx) in enumerate(gkf10):
if fold in selected_folds:
val10 += list(valid_idx)
if fold == 0:
val10_fold0 = valid_idx
for fold, (train_idx, valid_idx) in enumerate(gkf5):
if fold in selected_folds:
val5 += list(valid_idx)
common_validation_idx = np.array(list(set(val5).intersection(set(val10))))
np.array(list(set(val5).intersection(set(val10)))).shape
def predict_nn(train_data, valid_data, test_data, weights, model_type):
K.clear_session()
model = create_nn_model(train_data[1].shape[1], model_type)
model.load_weights(weights)
# for i in range(len(model.layers)):
# if str.startswith(model.layers[i].name, 'bn'):
# model.layers[i].trainable=True
# trn_preds = model.predict(train_data[0])
trn_preds = np.zeros(train_data[1].shape)
val_preds = model.predict(valid_data[0])
print(f"Lengths of test list is {len(test_data)}")
test_preds = model.predict(test_data) if test_data is not None else None
rho_trn = compute_spearmanr_ignore_nan(train_data[1], trn_preds)
rho_val = compute_spearmanr_ignore_nan(valid_data[1], val_preds)
print(f"Score train {rho_trn}; Score validation {rho_val}")
return trn_preds, val_preds, test_preds
def get_cross_fold_preds_nn(input_idx, model_type):
all_fold_trn_preds, all_fold_val_preds, all_fold_test_preds = [], [], []
all_fold_trn_outputs, all_fold_val_outputs = [], []
gkf = GroupKFold(n_splits=n_splits).split(
X=df_train.question_body, groups=df_train.question_body
)
for fold, (train_idx, valid_idx) in enumerate(gkf):
if USE_ONLY_SELECTED_FOLDS & (fold in selected_folds):
if MOD_DATA_STUCTURE == "2 split":
if input_idx == [0, 1, 2]:
output_idx = [
i for i, z in enumerate(output_categories) if z in question_only
]
if input_idx == [3, 4, 5]:
output_idx = [
i for i, z in enumerate(output_categories) if z in AQ_and_AO
]
if MOD_DATA_STUCTURE == "3 split":
if input_idx == [0, 1, 2]:
output_idx = [
i for i, z in enumerate(output_categories) if z in question_only
]
if input_idx == [3, 4, 5]:
output_idx = [
i
for i, z in enumerate(output_categories)
if z in answer_and_question
]
if input_idx == [6, 7, 8]:
output_idx = [
i for i, z in enumerate(output_categories) if z in answer_only
]
train_inputs = [inputs[i][train_idx] for i in input_idx]
train_outputs = outputs[np.array(train_idx)[:, None], output_idx]
all_fold_trn_outputs.append(train_outputs)
valid_inputs = [inputs[i][valid_idx] for i in input_idx]
valid_outputs = outputs[np.array(valid_idx)[:, None], output_idx]
all_fold_val_outputs.append(valid_outputs)
current_test_input = [test_inputs[i] for i in input_idx]
print(f"Fold {fold}")
input_type = None
if (input_idx == [3, 4, 5]) & (model_type == "XLNET"):
# weights_path = f"../input/gq-xlnet-pretrained/XLNET_question_answer_fold_{fold}.h5"
weights_path = (
f"../input/gq-xlnet-pretrained/XLNET_question_only_fold_{fold}.h5"
)
input_type = "question and answer"
elif (input_idx == [0, 1, 2]) & (model_type == "XLNET"):
# weights_path = f"../input/gq-xlnet-pretrained/XLNET_question_only_fold_{fold}.h5"
weights_path = (
f"../input/gq-xlnet-pretrained/XLNET_question_answer_fold_{fold}.h5"
)
input_type = "question only"
if (input_idx == [3, 4, 5]) & (model_type != "XLNET"):
print(
f"Using weights for BERT fold {fold}, question and answer modification"
)
weights_path = f"../input/{model_roor_dir}/{model_type}_question_answer_fold_{fold}.h5"
input_type = "question and answer"
elif (input_idx == [0, 1, 2]) & (model_type != "XLNET"):
weights_path = f"../input/{model_roor_dir}/{model_type}_question_only_fold_{fold}.h5"
input_type = "question only"
elif (input_idx == [6, 7, 8]) & (model_type != "XLNET"):
weights_path = (
f"../input/{model_roor_dir}/{model_type}_answer_only_fold_{fold}.h5"
)
input_type = "answer only"
trn_preds, val_preds, test_preds = predict_nn(
(train_inputs, train_outputs),
(valid_inputs, valid_outputs),
current_test_input,
weights_path,
model_type,
)
all_fold_trn_preds.append(trn_preds)
all_fold_val_preds.append(val_preds)
all_fold_test_preds.append(test_preds)
trn_preds, val_preds = np.concatenate(all_fold_trn_preds), np.concatenate(
all_fold_val_preds
)
trn_out, val_out = np.concatenate(all_fold_trn_outputs), np.concatenate(
all_fold_val_outputs
)
test_preds = np.stack(all_fold_test_preds, axis=2)
test_preds = np.mean(test_preds, axis=2)
print(f"Finished all folds for {model_type} {input_type}")
print(
test_preds.shape,
trn_out.shape,
val_preds.shape,
val_out.shape,
test_preds.shape,
)
return (trn_preds, trn_out), (val_preds, val_out), test_preds
def get_nn_all_outputs(model_type):
print("Getting all folds for QUESTION ONLY")
qonly_trn, qonly_val, qonly_tst = get_cross_fold_preds_nn([0, 1, 2], model_type)
print("Getting all folds for QUESTION ANSWER")
qa_trn, qa_val, qa_tst = get_cross_fold_preds_nn([3, 4, 5], model_type)
if MOD_DATA_STUCTURE == "3 split":
print("Getting all folds for ANSWER ONLY")
ao_trn, ao_val, ao_tst = get_cross_fold_preds_nn([6, 7, 8], model_type)
trn = (
np.concatenate((qonly_trn[0], qa_trn[0], ao_trn[0]), axis=1),
np.concatenate((qonly_trn[1], qa_trn[1], ao_trn[1]), axis=1),
)
val = (
np.concatenate((qonly_val[0], qa_val[0], ao_val[0]), axis=1),
np.concatenate((qonly_val[1], qa_val[1], ao_val[1]), axis=1),
)
tst = np.concatenate((qonly_tst, qa_tst, ao_tst), axis=1)
if MOD_DATA_STUCTURE == "2 split":
trn = (
np.concatenate((qonly_trn[0], qa_trn[0]), axis=1),
np.concatenate((qonly_trn[1], qa_trn[1]), axis=1),
)
val = (
np.concatenate((qonly_val[0], qa_val[0]), axis=1),
np.concatenate((qonly_val[1], qa_val[1]), axis=1),
)
tst = np.concatenate((qonly_tst, qa_tst), axis=1)
print(f"Finsihed entire dataset (qonly and qa) for {model_type}")
print(trn[0].shape, trn[1].shape, val[0].shape, val[1].shape, tst.shape)
save_file(trn, f"{model_type}_trn")
save_file(val, f"{model_type}_val")
save_file(tst, f"{model_type}_tst")
return trn, val, tst
print(psutil.cpu_percent())
print(dict(psutil.virtual_memory()._asdict()))
gc.collect()
tokenizer = XLNetTokenizer.from_pretrained(
"../input/gq-manual-uploads/xlnet tokenizer from colab/"
)
USING_PAD_TOKEN = False
outputs = compute_output_arrays(df_train, output_categories)
inputs = compute_input_arrays(
df_train, input_categories, tokenizer, MAX_SEQUENCE_LENGTH
)
test_inputs = compute_input_arrays(
df_test, input_categories, tokenizer, MAX_SEQUENCE_LENGTH
)
save_file(outputs, "XLNET_outputs")
save_file(inputs, "XLNET_inputs")
save_file(test_inputs, "XLNET_test_inputs")
model_roor_dir = "gq-xlnet-pretrained"
MOD_DATA_STUCTURE = "2 split"
n_splits = 10
xlnet_trn, xlnet_val, xlnet_tst = get_nn_all_outputs("XLNET")
print(psutil.cpu_percent())
print(dict(psutil.virtual_memory()._asdict()))
gc.collect()
tokenizer = BertTokenizer.from_pretrained(BERT_PATH + "bert-base-uncased-vocab.txt")
USING_PAD_TOKEN = True
outputs = compute_output_arrays(df_train, output_categories)
inputs = compute_input_arrays(
df_train, input_categories, tokenizer, MAX_SEQUENCE_LENGTH
)
test_inputs = compute_input_arrays(
df_test, input_categories, tokenizer, MAX_SEQUENCE_LENGTH
)
save_file(outputs, "BERT_outputs")
save_file(inputs, "BERT_inputs")
save_file(test_inputs, "BERT_test_inputs")
model_roor_dir = "gq-bert-pretrained"
MOD_DATA_STUCTURE = "2 split"
n_splits = 10
bert_trn, bert_val, bert_tst = get_nn_all_outputs("BERT")
model_roor_dir = "3rd-training-2nd-gen-bert-download-from-gdrive"
MOD_DATA_STUCTURE = "3 split"
n_splits = 10
selected_folds = [0]
bert_trn_3split, bert_val_3split, bert_tst_3split = get_nn_all_outputs("BERT")
selected_folds = [0, 1, 2]
model_roor_dir = "2nd-training-1st-gen-bert-download-from-gdrive"
MOD_DATA_STUCTURE = "2 split"
n_splits = 5
bert_trn_5fold, bert_val_5fold, bert_tst_5fold = get_nn_all_outputs("BERT")
print(psutil.cpu_percent())
print(dict(psutil.virtual_memory()._asdict()))
gc.collect()
# tokenizer = GPT2Tokenizer.from_pretrained(GPT2_PATH+'gpt2-vocab.json')
tokenizer = GPT2Tokenizer.from_pretrained(
"../input/gq-manual-uploads/gpt2 config from colab/"
)
USING_PAD_TOKEN = False
outputs = compute_output_arrays(df_train, output_categories)
inputs = compute_input_arrays(
df_train, input_categories, tokenizer, MAX_SEQUENCE_LENGTH
)
test_inputs = compute_input_arrays(
df_test, input_categories, tokenizer, MAX_SEQUENCE_LENGTH
)
save_file(outputs, "GPT2_outputs")
save_file(inputs, "GPT2_inputs")
save_file(test_inputs, "GPT2_test_inputs")
model_roor_dir = "gq-gpt2-pretrained"
MOD_DATA_STUCTURE = "2 split"
n_splits = 10
gpt2_trn, gpt2_val, gpt2_tst = get_nn_all_outputs("GPT2")
model_roor_dir = "2nd-training-1st-gen-gpt-download-from-gdrive"
MOD_DATA_STUCTURE = "2 split"
n_splits = 5
gpt2_trn_5fold, gpt2_val_5fold, gpt2_tst_5fold = get_nn_all_outputs("GPT2")
print(psutil.cpu_percent())
print(dict(psutil.virtual_memory()._asdict()))
gc.collect()
# #### LGB
if USE_LGB:
def remove_articles(df):
for i in ["question_title", "question_body", "answer"]:
df.loc[:, f"{i}_orig"] = df.loc[:, i]
for i in ["question_title", "question_body", "answer"]:
df.loc[:, i] = df.loc[:, i].apply(
lambda x: x.replace(" the ", " ")
.replace(" a ", " ")
.replace(" an ", " ")
)
return df
df_train = remove_articles(df_train)
df_test = remove_articles(df_test)
if USE_LGB:
df_train.loc[:, "q_users_host"] = df_train.apply(
lambda x: x.question_user_name + x.host, axis=1
)
df_train.loc[:, "a_users_host"] = df_train.apply(
lambda x: x.answer_user_name + x.host, axis=1
)
df_test.loc[:, "q_users_host"] = df_test.apply(
lambda x: x.question_user_name + x.host, axis=1
)
df_test.loc[:, "a_users_host"] = df_test.apply(
lambda x: x.answer_user_name + x.host, axis=1
)
q_users_train = dict(df_train.q_users_host.value_counts())
q_users_test = dict(df_test.q_users_host.value_counts())
a_users_train = dict(df_train.a_users_host.value_counts())
a_users_test = dict(df_test.a_users_host.value_counts())
q_users = q_users_train
for i in q_users:
if i in q_users_test:
q_users_train[i] += q_users_test[i]
for i in q_users_test:
if i not in q_users:
q_users[i] = q_users_test[i]
a_users = a_users_train
for i in a_users:
if i in a_users_test:
a_users_train[i] += a_users_test[i]
for i in a_users_test:
if i not in a_users:
a_users[i] = a_users_test[i]
if USE_LGB:
word_categories = [
"adjectives",
"verbs",
"nouns",
"list_maker",
"digits",
"modals",
"posessives",
"persionals",
"interjection",
"direction",
"past_verb",
]
adjectives = ["JJ", "JJR", "JJS", "RB", "RBR", "RBS"]
verbs = ["VB", "VBD", "VBG", "VBN", "VBP", "VBZ"]
nouns = ["NN", "NNS", "NNP", "NNPS"]
list_maker = ["LS"]
digits = ["CD"]
modals = ["MD"]
posessives = ["PPR$", "POS"]
persionals = ["PRP"]
interjection = ["UH"]
direction = ["TO"]
past_verb = ["VBD", "VBN"]
def get_string_stats(df, var):
df.loc[:, f"{var}_numchars"] = df[var].apply(lambda x: len(x))
df.loc[:, f"{var}_numwords"] = df[var].apply(lambda x: len(x.split()))
df.loc[:, f"{var}_exclam_count"] = df[var].apply(lambda x: x.count("!"))
df.loc[:, f"{var}_question_count"] = df[var].apply(lambda x: x.count("?"))
df.loc[:, f"{var}_coma_count"] = df[var].apply(lambda x: x.count(","))
df.loc[:, f"{var}_dot_count"] = df[var].apply(lambda x: x.count("."))
df.loc[:, f"{var}_all_punct_count"] = (
df[f"{var}_question_count"]
+ df[f"{var}_coma_count"]
+ df[f"{var}_exclam_count"]
)
df.loc[:, f"{var}_all_punct_to_sentences"] = (
df.loc[:, f"{var}_all_punct_count"] / df.loc[:, f"{var}_dot_count"]
)
df.loc[:, f"{var}_questions_to_sentences"] = (
df.loc[:, f"{var}_question_count"] / df.loc[:, f"{var}_dot_count"]
)
df.loc[:, f"{var}_questions_to_words"] = (
df.loc[:, f"{var}_question_count"] / df.loc[:, f"{var}_numwords"]
)
df.loc[:, f"{var}_average_word_len"] = (
df[f"{var}_numchars"] / df[f"{var}_numwords"]
)
df.loc[:, f"{var}_capital_count"] = df[var].apply(
lambda x: sum(1 for c in x if c.isupper())
)
df.loc[:, f"{var}_capital_prop"] = (
df[f"{var}_capital_count"] / df[f"{var}_numwords"]
)
df.loc[:, f"{var}_other_ref"] = df[var].apply(
lambda x: sum([x.count(i) for i in [" it ", " they ", " it's ", " their "]])
)
df.loc[:, f"{var}_self_ref"] = df[var].apply(
lambda x: sum([x.count(i) for i in [" I ", " me ", " mine ", " my "]])
)
df.loc[:, f"{var}_total_ref"] = df[f"{var}_self_ref"] + df[f"{var}_other_ref"]
df.loc[:, f"{var}_total_ref_prop"] = (
df.loc[:, f"{var}_total_ref"] / df.loc[:, f"{var}_numwords"]
)
df.loc[:, f"{var}_self_ref_prop"] = (
df[f"{var}_self_ref"] / df[f"{var}_total_ref"]
)
df.loc[:, f"{var}_other_ref_prop"] = (
df[f"{var}_other_ref"] / df[f"{var}_total_ref"]
)
df.loc[:, f"{var}_words_per_sentence"] = (
df[f"{var}_numwords"] / df[f"{var}_dot_count"]
)
df.loc[:, f"{var}_unique_words"] = df[f"{var}"].apply(
lambda x: len(set(str(x).split()))
)
df.loc[:, f"{var}_unique_words_prop"] = (
df.loc[:, f"{var}_unique_words"] / df.loc[:, f"{var}_numwords"]
)
new_cols = [
f"{var}_total_ref_prop",
f"{var}_questions_to_words",
f"{var}_questions_to_sentences",
f"{var}_all_punct_to_sentences",
f"{var}_unique_words_prop",
f"{var}_unique_words",
f"{var}_numchars",
f"{var}_numwords",
f"{var}_exclam_count",
f"{var}_question_count",
f"{var}_coma_count",
f"{var}_dot_count",
f"{var}_all_punct_count",
f"{var}_average_word_len",
f"{var}_capital_count",
f"{var}_capital_prop",
f"{var}_other_ref",
f"{var}_self_ref",
f"{var}_total_ref",
f"{var}_self_ref_prop",
f"{var}_other_ref_prop",
f"{var}_words_per_sentence",
]
for category in word_categories:
df.loc[:, f"{var}_{category}"] = 0
new_cols.append(f"{var}_{category}")
for idx in tqdm(range(df.shape[0]), total=df.shape[0]):
tokens = nltk.word_tokenize(df.loc[idx, var])
tags = nltk.pos_tag(tokens)
tags = [i[1] for i in tags]
# print(idx)
# print(tags)
for category in word_categories:
count = 0
for tag_name in globals()[category]:
count += tags.count(tag_name)
# print(count)
df.loc[idx, f"{var}_{category}"] = (
count / df.loc[idx, f"{var}_numwords"]
if df.loc[idx, f"{var}_numwords"] != 0
else 0
)
# print(df.loc[idx, f'{var}_numwords'])
# break
return df, new_cols
def get_extra_features_and_map(df):
df, nc1 = get_string_stats(df, "question_title")
df, nc2 = get_string_stats(df, "question_body")
df, nc3 = get_string_stats(df, "answer")
df.loc[:, "q_user_q_count"] = df.q_users_host.apply(
lambda x: q_users[x] if x in q_users else 0
)
df.loc[:, "q_user_a_count"] = df.q_users_host.apply(
lambda x: a_users[x] if x in a_users else 0
)
df.loc[:, "a_user_a_count"] = df.a_users_host.apply(
lambda x: a_users[x] if x in a_users else 0
)
df.loc[:, "a_user_q_count"] = df.a_users_host.apply(
lambda x: q_users[x] if x in q_users else 0
)
df.loc[:, "q_user_both_count"] = (
df.loc[:, "q_user_q_count"] + df.loc[:, "q_user_a_count"]
)
df.loc[:, "a_user_both_count"] = (
df.loc[:, "a_user_a_count"] + df.loc[:, "a_user_q_count"]
)
other_features = []
df.loc[:, "q_to_a_all_punct_count"] = (
df.loc[:, "question_body_all_punct_count"]
+ df.loc[:, "question_title_all_punct_count"]
) / df.loc[:, "answer_all_punct_count"]
df.loc[:, "q_to_a_numwords"] = (
df.loc[:, "question_body_numwords"] + df.loc[:, "question_title_numwords"]
) / df.loc[:, "answer_numwords"]
df.loc[:, "q_to_a_capital_count"] = (
df.loc[:, "question_body_capital_count"]
+ df.loc[:, "question_title_capital_count"]
) / df.loc[:, "answer_capital_count"]
df.loc[:, "q_to_a_unique_words_prop"] = (
df.loc[:, "question_body_unique_words_prop"]
+ df.loc[:, "question_title_unique_words_prop"]
) / df.loc[:, "answer_unique_words_prop"]
df.loc[:, "q_to_a_total_ref"] = (
df.loc[:, "question_body_total_ref"] + df.loc[:, "question_title_total_ref"]
) / df.loc[:, "answer_total_ref"]
df.loc[:, "common_words"] = df.apply(
lambda x: len(
set(x.question_body.split()).intersection(set(x.answer.split()))
),
axis=1,
)
other_features += [
"q_to_a_all_punct_count",
"q_to_a_numwords",
"q_to_a_capital_count",
"common_words",
"q_to_a_unique_words_prop",
"q_to_a_total_ref",
]
for category in word_categories:
df.loc[:, f"q_to_a_{category}"] = (
df.loc[:, f"question_body_{category}"] / df.loc[:, f"answer_{category}"]
)
other_features.append(f"q_to_a_{category}")
df.loc[:, "spell_words"] = df.loc[:, "question_title"].apply(
lambda x: sum(
1
for i in x.lower().split()
if i
in [
"spell",
"spelled",
"spelt",
"spelling",
"write",
"wrote",
"written",
]
)
)
df.loc[:, "spell_words"] += df.loc[:, "question_body"].apply(
lambda x: sum(
1
for i in x.lower().split()
if i
in [
"spell",
"spelled",
"spelt",
"spelling",
"write",
"wrote",
"written",
]
)
)
df.loc[:, "compare_words"] = df.loc[:, "question_title"].apply(
lambda x: sum(
1
for i in x.lower().split()
if i in ["better", "best", "worse", "nicer"]
)
)
df.loc[:, "compare_words"] += df.loc[:, "question_body"].apply(
lambda x: sum(
1
for i in x.lower().split()
if i in ["better", "best", "worse", "nicer"]
)
)
df.loc[:, "consequence_words"] = df.loc[:, "question_title"].apply(
lambda x: sum(
1 for i in x.lower().split() if i in ["if", "when", "will", "would"]
)
)
df.loc[:, "consequence_words"] += df.loc[:, "question_body"].apply(
lambda x: sum(
1 for i in x.lower().split() if i in ["if", "when", "will", "would"]
)
)
other_features.append("spell_words")
other_features.append("compare_words")
other_features.append("consequence_words")
onehots = []
for i in df.loc[:, "category"].unique():
df.loc[:, f"{i}_onehot"] = 0
df.loc[df.loc[:, "category"] == i, f"{i}_onehot"] = 1
onehots.append(f"{i}_onehot")
for i in df.loc[:, "host"].unique():
df.loc[:, f"{i}_H_onehot"] = 0
df.loc[df.loc[:, "host"] == i, f"{i}_H_onehot"] = 1
onehots.append(f"{i}_H_onehot")
other_features = (
other_features
+ nc1
+ nc2
+ nc3
+ onehots
+ [
"q_user_q_count",
"q_user_a_count",
"a_user_a_count",
"a_user_q_count",
"q_user_both_count",
"a_user_both_count",
]
)
return df, other_features
df_train, other_features_train = get_extra_features_and_map(df_train)
df_test, other_features = get_extra_features_and_map(df_test)
for i in [a for a in other_features if a not in other_features_train]:
df_train.loc[:, i] = np.zeros(df_train.shape[0])
if USE_LGB:
def get_uids_all(df):
df.loc[:, "answer_uid"] = df.loc[:, "answer_user_page"].apply(
lambda x: int(x.split("/")[-1])
)
df.loc[:, "question_uid"] = df.loc[:, "question_user_page"].apply(
lambda x: int(x.split("/")[-1])
)
for idx in range(df.shape[0]):
split = [i for i in df.loc[idx, "url"].split("/") if i.isdigit()]
df.loc[idx, "url_uid"] = int(split[-1]) if len(split) > 0 else -1
return df
df_train = get_uids_all(df_train)
df_test = get_uids_all(df_test)
if USE_LGB:
se_path = "../input/stackexchange-data"
se_posts = pd.read_parquet(
se_path + "/stackexchange_posts.parquet.gzip", engine="fastparquet"
)
def get_post_info_se(df):
new_other_features = []
new_features = [
"Score",
"ViewCount",
"AnswerCount",
"CommentCount",
"FavoriteCount",
"Tags",
]
df = df.merge(
se_posts.loc[:, ["Id", "host", "AcceptedAnswerId"] + new_features],
how="left",
left_on=["url_uid", "host"],
right_on=["Id", "host"],
sort=False,
)
df.rename({i: "SE_QP_" + i for i in new_features}, inplace=True, axis=1)
new_other_features += ["SE_QP_" + i for i in new_features]
return df, new_other_features
df_train, new_other_features = get_post_info_se(df_train)
df_test, _ = get_post_info_se(df_test)
del se_posts
gc.collect()
other_features += new_other_features
other_features.remove("SE_QP_Tags")
# -----------
all_tags = []
for i in range(df_train.shape[0]):
if (df_train.SE_QP_Tags.iloc[i] == None) or (
pd.isna(df_train.SE_QP_Tags.iloc[i])
):
continue
tags = df_train.SE_QP_Tags.iloc[i].replace("<", "").split(">")
all_tags += [t for t in tags if len(t) > 0]
top_tags = list(pd.DataFrame(all_tags).iloc[:, 0].value_counts()[0:50].index)
other_features += [f"tag_{i}" for i in top_tags]
# --------
for t in top_tags:
df_train.loc[:, f"tag_{t}"] = 0
df_test.loc[:, f"tag_{t}"] = 0
def parse_tags(df):
for i in range(df.shape[0]):
if (df.SE_QP_Tags.iloc[i] == None) or (pd.isna(df.SE_QP_Tags.iloc[i])):
continue
tags = df.SE_QP_Tags.iloc[i].replace("<", "").split(">")
tags = [t for t in tags if (len(t) > 0) & (t in top_tags)]
for t in tags:
df.loc[i, f"tag_{t}"] = 1
return df
df_train = parse_tags(df_train)
df_test = parse_tags(df_test)
# -------
se_path = "../input/stackexchange-data"
se_users = pd.read_parquet(
se_path + "/stackexchange_users.parquet.gzip", engine="fastparquet"
)
def get_user_info_se(df):
new_other_features = []
new_features = ["Reputation", "Views", "Upvotes", "Downvotes"]
df = df.merge(
se_users.loc[:, ["Id", "host"] + new_features],
how="left",
left_on=["question_uid", "host"],
right_on=["Id", "host"],
sort=False,
)
df.rename({i: "SE_Q_" + i for i in new_features}, inplace=True, axis=1)
new_other_features += ["SE_Q_" + i for i in new_features]
new_features = ["Reputation", "Views", "Upvotes", "Downvotes"]
df = df.merge(
se_users.loc[:, ["Id", "host"] + new_features],
how="left",
left_on=["answer_uid", "host"],
right_on=["Id", "host"],
sort=False,
)
df.rename({i: "SE_A_" + i for i in new_features}, inplace=True, axis=1)
new_other_features += ["SE_A_" + i for i in new_features]
return df, new_other_features
df_train, new_other_features = get_user_info_se(df_train)
df_test, _ = get_user_info_se(df_test)
del se_users
gc.collect()
other_features += new_other_features
if USE_LGB:
import bq_helper
from bq_helper import BigQueryHelper
# https://www.kaggle.com/sohier/introduction-to-the-bq-helper-package
stackOverflow = bq_helper.BigQueryHelper(
active_project="bigquery-public-data", dataset_name="stackoverflow"
)
def get_user_info_stackoverflow(df):
print(df.shape)
new_other_features = []
all_q_uids = tuple(
df.loc[df.host == "stackoverflow.com", "answer_uid"].unique()
)
print(len(all_q_uids))
q_users = f"""SELECT id, display_name, reputation, up_votes, down_votes, views from `bigquery-public-data.stackoverflow.users` WHERE id IN {all_q_uids}"""
q_users_df = stackOverflow.query_to_pandas_safe(q_users)
print(q_users_df.shape)
new_features = ["reputation", "up_votes", "down_votes", "views"]
df = df.merge(
q_users_df, left_on="answer_uid", right_on="id", how="left", sort=False
)
df.rename({i: "A_" + i for i in new_features}, inplace=True, axis=1)
new_other_features += ["A_" + i for i in new_features]
all_q_uids = tuple(
df.loc[df.host == "stackoverflow.com", "question_uid"].unique()
)
print(len(all_q_uids))
q_users = f"""SELECT id, display_name, reputation, up_votes, down_votes, views from `bigquery-public-data.stackoverflow.users` WHERE id IN {all_q_uids}"""
q_users_df = stackOverflow.query_to_pandas_safe(q_users)
print(q_users_df.shape)
new_features = ["reputation", "up_votes", "down_votes", "views"]
df = df.merge(
q_users_df, left_on="question_uid", right_on="id", how="left", sort=False
)
df.rename({i: "Q_" + i for i in new_features}, inplace=True, axis=1)
new_other_features += ["Q_" + i for i in new_features]
print(df.shape)
return df, new_other_features
df_train, new_other_features = get_user_info_stackoverflow(df_train)
df_test, _ = get_user_info_stackoverflow(df_test)
other_features += new_other_features
def get_question_info_stackoverflow(df):
print(df.shape)
new_other_features = []
uids_selection = tuple(
df.loc[df.host == "stackoverflow.com", "url_uid"].dropna().unique()
)
print(len(uids_selection))
query = f"""SELECT id, accepted_answer_id, answer_count, comment_count, favorite_count, score, view_count from `bigquery-public-data.stackoverflow.stackoverflow_posts` WHERE id IN {uids_selection}"""
query_as_df = stackOverflow.query_to_pandas_safe(query)
print(query_as_df.shape)
new_features = [
"accepted_answer_id",
"answer_count",
"comment_count",
"favorite_count",
"score",
"view_count",
]
df = df.merge(
query_as_df, left_on="url_uid", right_on="id", how="left", sort=False
)
df.rename({i: "QPAGE_" + i for i in new_features}, inplace=True, axis=1)
new_other_features += ["QPAGE_" + i for i in new_features]
print(df.shape)
return df, new_other_features
df_train, new_other_features = get_question_info_stackoverflow(df_train)
df_test, _ = get_question_info_stackoverflow(df_test)
other_features += new_other_features
if USE_LGB:
def is_answer_accepted(df):
for i in df.loc[
(df.host == "stackoverflow.com") & ~(df.QPAGE_accepted_answer_id.isna()), :
].index.values:
df.loc[i, "answer_accepted"] = (
1
if df.loc[i, "answer_uid"] == df.loc[i, "QPAGE_accepted_answer_id"]
else 0
)
for i in df.loc[
(df.host != "stackoverflow.com") & ~(df.AcceptedAnswerId.isna()), :
].index.values:
df.loc[i, "answer_accepted"] = (
1 if df.loc[i, "answer_uid"] == df.loc[i, "AcceptedAnswerId"] else 0
)
return df
df_train = is_answer_accepted(df_train)
df_test = is_answer_accepted(df_test)
other_features.append("answer_accepted")
if USE_LGB:
class Base_Model(object):
def __init__(
self,
train_df,
test_df,
features,
categoricals=[],
n_splits=5,
verbose=True,
target=None,
predict_test=True,
):
self.train_df = train_df
self.test_df = test_df
self.features = features
self.n_splits = 10
self.categoricals = categoricals
self.target = target
self.cv = self.get_cv()
self.verbose = verbose
self.params = self.get_params()
self.predict_test = predict_test
self.saved_models_dir = "../input/gq-lgb"
(
self.tst_pred,
self.score,
self.model,
self.val_ys,
self.val_preds,
) = self.fit()
def train_model(self, train_set, val_set):
raise NotImplementedError
def get_cv(self):
cv = GroupKFold(n_splits=self.n_splits)
return cv.split(
X=self.train_df.question_body_orig,
groups=self.train_df.question_body_orig,
)
def get_params(self):
raise NotImplementedError
def convert_dataset(self, x_train, y_train, x_val, y_val):
raise NotImplementedError
def convert_x(self, x):
return x
def fit(self):
# oof_pred = np.zeros((len(unseen_valid), )) if MIX_UP else np.zeros((len(self.train_df), ))
oof_pred = []
oof_ys_all = []
y_pred = np.zeros((len(self.test_df),))
for fold, (train_idx, val_idx) in enumerate(self.cv):
if fold < ACTUAL_FOLDS:
x_train, x_val = (
self.train_df[self.features].iloc[train_idx],
self.train_df[self.features].iloc[val_idx],
)
y_train, y_val = (
self.train_df[self.target][train_idx],
self.train_df[self.target][val_idx],
)
train_set, val_set = self.convert_dataset(
x_train, y_train, x_val, y_val
)
model = self.load_model(fold)
conv_x_val = self.convert_x(x_val.reset_index(drop=True))
preds_all = model.predict(conv_x_val)
preds_all = 1 / (1 + np.exp(-preds_all))
# preds_all = np.round(preds_all, ROUND_PLACES) if ROUND else preds_all
oof_pred += preds_all.tolist()
if self.predict_test:
x_test = self.convert_x(self.test_df[self.features])
current_test_preds = model.predict(x_test).reshape(y_pred.shape)
current_test_preds = 1 / (1 + np.exp(-current_test_preds))
current_test_preds = current_test_preds / ACTUAL_FOLDS
y_pred += current_test_preds # no to list as this is stored as single numpy array
if self.verbose:
print(
"Partial score (all) of fold {} is: {}".format(
fold, spearmanr(y_val, preds_all).correlation
)
)
oof_ys_all += list(y_val.reset_index(drop=True).values)
loss_score = spearmanr(oof_ys_all, oof_pred).correlation
if self.verbose:
print("Our oof cohen kappa score (all) is: ", loss_score)
return y_pred, loss_score, model, np.array(oof_ys_all), np.array(oof_pred)
class Lgb_Model(Base_Model):
def train_model(self, train_set, val_set):
verbosity = 100 if self.verbose else 0
return lgb.train(
self.params,
train_set,
valid_sets=[train_set, val_set],
verbose_eval=verbosity,
)
def load_model(self, fold):
model = pickle.load(
open(
f"{self.saved_models_dir}/{self.target}/{self.target}_{fold}.p",
"rb",
)
)
return model
def convert_dataset(self, x_train, y_train, x_val, y_val):
train_set = lgb.Dataset(
x_train, y_train, categorical_feature=self.categoricals
)
val_set = lgb.Dataset(x_val, y_val, categorical_feature=self.categoricals)
return train_set, val_set
def get_params(self):
params = {
"n_estimators": 5000,
"boosting_type": "gbdt",
"objective": "cross_entropy_lambda",
# 'is_unbalance': 'true',
# 'metric': 'huber',
"subsample": 0.75,
"subsample_freq": 1,
"learning_rate": 0.01,
"feature_fraction": 0.8,
"max_depth": 150, # was 15
"num_leaves": 50,
"lambda_l1": 0.1,
"lambda_l2": 0.1,
"early_stopping_rounds": 300,
"min_data_in_leaf": 1,
"min_gain_to_split": 0.01,
"max_bin": 400,
}
return params
print(psutil.cpu_percent())
print(dict(psutil.virtual_memory()._asdict()))
gc.collect()
if USE_LGB:
one_lgb_model = pickle.load(
open(
f"../input/gq-lgb/question_opinion_seeking/question_opinion_seeking_0.p",
"rb",
)
)
lgb_pretrained_features = one_lgb_model.feature_name()
for i in lgb_pretrained_features:
if i not in other_features:
print(f"{i} not in other features here, adding zeros")
df_train.loc[:, i] = np.zeros(df_train.shape[0])
df_test.loc[:, i] = np.zeros(df_test.shape[0])
if USE_LGB:
ACTUAL_FOLDS = 3
lgb_val_scores = []
n_output_categories = len(output_categories)
lgb_val_outputs_all = []
lgb_val_preds_all = []
lgb_tst_preds_all = []
for idx, i in enumerate(output_categories, 1):
lgb_model = Lgb_Model(
df_train, df_test, lgb_pretrained_features, target=i, verbose=False
)
lgb_val_outputs_all.append(lgb_model.val_ys)
lgb_val_preds_all.append(lgb_model.val_preds)
lgb_tst_preds_all.append(lgb_model.tst_pred)
lgb_val_scores.append(lgb_model.score)
print(f"{idx}/{n_output_categories}", i, lgb_model.score)
save_file(lgb_val_outputs_all, "lgb_val_outputs_all")
save_file(lgb_val_preds_all, "lgb_val_preds_all")
save_file(lgb_tst_preds_all, "lgb_tst_preds_all")
save_file(df_train, "df_train")
save_file(df_test, "df_test")
else:
lgb_val_outputs_all = [0] * 30
lgb_val_preds_all = [0] * 30
lgb_tst_preds_all = [0] * 30
print(psutil.cpu_percent())
print(dict(psutil.virtual_memory()._asdict()))
gc.collect()
# #### Compose
def get_rounding(ys, preds):
rounding_types = ["Normal", "Ceil", "Floor"]
rounding_funcs = [np.round, np.ceil, np.floor]
dec_places = [1, 2, 3, 4, 5]
score = spearmanr(ys, preds).correlation
if np.isnan(score):
score = -100
best_result = {"Type": "No rounding", "DP": 0, "func": None}
for r_type, r_func in zip(rounding_types, rounding_funcs):
for dp in dec_places:
if r_type == "Normal":
rounded_preds = r_func(preds, dp)
cur_score = spearmanr(ys, rounded_preds).correlation
else:
rounded_preds = r_func(preds * (10**dp)) / (10**dp)
cur_score = spearmanr(ys, rounded_preds).correlation
if np.isnan(cur_score):
cur_score = 0
if cur_score > score:
score = cur_score
best_result["Type"] = r_type
best_result["DP"] = dp
best_result["func"] = r_func
return score, best_result
from scipy.optimize import minimize
from functools import partial
def inverse_spearman_r(weights, ys, preds):
mixed_val_preds = np.array([i * w for i, w in zip(preds, weights)]).sum(axis=0)
# print(mixed_val_preds.shape)
# print(ys.shape)
score = spearmanr(ys, mixed_val_preds).correlation
if np.isnan(score):
score = -100
return -score
def optimize_mixing_weights(ys, preds):
naive_mix = np.array(preds).mean(axis=0)
score = spearmanr(ys, naive_mix).correlation
if np.isnan(score):
score = -100
c_dict = {"type": "eq", "fun": lambda x: 1 - np.sum(x)}
optim_func = partial(inverse_spearman_r, ys=ys, preds=preds)
x0 = np.array([1 / len(preds)] * len(preds))
res = minimize(optim_func, x0, method="SLSQP", constraints=c_dict)
print(f"Best score {res.fun}; weights {res.x}")
return res.x
from itertools import combinations_with_replacement
def optimize_mixing_weights_guessing(ys, preds):
# variants = [0.1,0.2,0.3,0.4,0.5,0.6,0.7,0.8,0.9]
# variants = [0.0, 0.01, 0.04, 0.1, 0.12, 0.2, 0.25, 0.3, 0.35, 0.4, 0.45, 0.5, 0.55, 0.6, 0.65, 0.7, 0.75, 0.8, 0.85, 0.9, 0.95]
variants = [
0.0,
0.01,
0.04,
0.1,
0.12,
0.16,
0.18,
0.2,
0.22,
0.25,
0.3,
0.35,
0.4,
0.45,
0.5,
0.55,
0.6,
0.65,
0.7,
0.75,
0.8,
0.82,
0.85,
0.88,
0.9,
0.94,
0.95,
]
models_n = len(preds)
weights = [1 / models_n] * models_n
best_weights = weights
best_score = 0
for comb in combinations_with_replacement(variants, models_n):
weights = list(comb)
if np.sum(weights) != 1:
continue
score = spearmanr(
ys, np.array([i * w for i, w in zip(preds, weights)]).sum(axis=0)
).correlation
if np.isnan(score):
score = -100
if score > best_score:
best_weights, best_score = weights.copy(), score
print(f"Best score {best_score}; weights {best_weights}")
return best_weights
# common_validation_idx
val10_common = [np.where(val10 == i)[0][0] for i in common_validation_idx]
val5_common = [np.where(val5 == i)[0][0] for i in common_validation_idx]
val10_fold0_common = [np.where(val10_fold0 == i)[0] for i in common_validation_idx]
val10_fold0_common = [i[0] for i in val10_fold0_common if len(i) > 0]
weights_all = []
weights_for_rounding = []
scores_all = []
df_sub = pd.read_csv(PATH + "sample_submission.csv")
bestonly_raw_scores = []
best_rounded_scores = []
best_weighted_scores = []
if USE_SAMPLE:
df_sub = df_sub.iloc[0 : round(0.1 * df_sub.shape[0]), :]
if USE_LGB:
if len(lgb_val_scores) != 30:
USE_LGB = False
print("Something wrong with LGB, switching it off")
for idx, cat in enumerate(output_categories):
lgb_score = (
spearmanr(xlnet_val[1][:, idx], lgb_val_preds_all[idx]).correlation
if USE_LGB
else np.nan
)
gpt2_score = spearmanr(gpt2_val[1][:, idx], gpt2_val[0][:, idx]).correlation
bert_score = spearmanr(bert_val[1][:, idx], bert_val[0][:, idx]).correlation
xlnet_score = spearmanr(xlnet_val[1][:, idx], xlnet_val[0][:, idx]).correlation
bert_score_3split = (
spearmanr(bert_val_3split[1][:, idx], bert_val_3split[0][:, idx]).correlation
if USE_3SPLIT_BERT
else np.nan
)
bert_score_5fold = spearmanr(
bert_val_5fold[1][:, idx], bert_val_5fold[0][:, idx]
).correlation
gpt2_score_5fold = spearmanr(
gpt2_val_5fold[1][:, idx], gpt2_val_5fold[0][:, idx]
).correlation
scores_all.append(
[
lgb_score,
gpt2_score,
bert_score,
xlnet_score,
bert_score_3split,
bert_score_5fold,
gpt2_score_5fold,
]
)
selected_models_scores = [
i
for i in [
lgb_score,
gpt2_score,
bert_score,
xlnet_score,
bert_score_3split,
bert_score_5fold,
gpt2_score_5fold,
]
if not np.isnan(i)
]
selected_models_val_preds = [
i
for i, s in zip(
[
lgb_val_preds_all[idx],
gpt2_val[0][:, idx],
bert_val[0][:, idx],
xlnet_val[0][:, idx],
bert_val_3split[0][:, idx],
bert_val_5fold[0][:, idx],
gpt2_val_5fold[0][:, idx],
],
[
lgb_score,
gpt2_score,
bert_score,
xlnet_score,
bert_score_3split,
bert_score_5fold,
gpt2_score_5fold,
],
)
if not np.isnan(s)
]
selected_models_tst_preds = [
i
for i, s in zip(
[
lgb_tst_preds_all[idx],
gpt2_tst[:, idx],
bert_tst[:, idx],
xlnet_tst[:, idx],
bert_tst_3split[:, idx],
bert_tst_5fold[:, idx],
gpt2_tst_5fold[:, idx],
],
[
lgb_score,
gpt2_score,
bert_score,
xlnet_score,
bert_score_3split,
bert_score_5fold,
gpt2_score_5fold,
],
)
if not np.isnan(s)
]
current_best = np.max(selected_models_scores)
bestonly_raw_scores.append(current_best)
if USE_LGB:
val_preds_10and5 = [
lgb_val_preds_all[idx][val10_common],
gpt2_val[0][val10_common, idx],
bert_val[0][val10_common, idx],
xlnet_val[0][val10_common, idx],
bert_val_5fold[0][val5_common, idx],
gpt2_val_5fold[0][val5_common, idx],
]
scores10and5 = np.sum(
[
np.exp(i * beta)
for i in [
lgb_score,
gpt2_score,
bert_score,
xlnet_score,
bert_score_5fold,
gpt2_score_5fold,
]
]
)
weights10and5 = [
np.exp(i * beta) / scores10and5
for i in [
lgb_score,
gpt2_score,
bert_score,
xlnet_score,
bert_score_5fold,
gpt2_score_5fold,
]
]
else:
val_preds_10and5 = [
gpt2_val[0][val10_common, idx],
bert_val[0][val10_common, idx],
xlnet_val[0][val10_common, idx],
bert_val_5fold[0][val5_common, idx],
gpt2_val_5fold[0][val5_common, idx],
]
scores10and5 = np.sum(
[
np.exp(i * beta)
for i in [
gpt2_score,
bert_score,
xlnet_score,
bert_score_5fold,
gpt2_score_5fold,
]
]
)
weights10and5 = [
np.exp(i * beta) / scores10and5
for i in [
gpt2_score,
bert_score,
xlnet_score,
bert_score_5fold,
gpt2_score_5fold,
]
]
val_mix = np.array([i * w for i, w in zip(val_preds_10and5, weights10and5)]).sum(
axis=0
)
common_val = xlnet_val[1][val10_common, idx]
weights_for_rounding.append(weights10and5)
if not ROUND:
print(
f"{idx} {cat} scores: LGB: {lgb_score}, GPT2: {gpt2_score}, BERT: {bert_score}, XLNET: {xlnet_score}"
)
if WEIGHTS == "softmax":
all_scores = np.sum(np.exp([i * beta for i in selected_models_scores]))
weights = [np.exp(i * beta) / all_scores for i in selected_models_scores]
weights_all.append(weights)
df_sub.iloc[:, idx + 1] = np.array(
[i * w for i, w in zip(selected_models_tst_preds, weights)]
).sum(axis=0)
if WEIGHTS == "dynamic":
weights = optimize_mixing_weights_guessing(common_val, val_preds_10and5)
weights_all.append(weights)
best_weighted_scores.append(
spearmanr(
common_val,
np.array([i * w for i, w in zip(val_preds_10and5, weights)]).sum(
axis=0
),
).correlation
)
val_mix = np.array([i * w for i, w in zip(val_preds_10and5, weights)]).sum(
axis=0
)
df_sub.iloc[:, idx + 1] = np.array(
[i * w for i, w in zip(selected_models_tst_preds, weights)]
).sum(axis=0)
if WEIGHTS == "dynamic softmax":
ds_score = -100
for b in np.arange(1, 50, 1):
all_scores = np.sum(np.exp([i * b for i in selected_models_scores]))
try_weights = [np.exp(i * b) / all_scores for i in selected_models_scores]
try_mix = np.array(
[i * w for i, w in zip(val_preds_10and5, try_weights)]
).sum(axis=0)
try_score = spearmanr(common_val, try_mix).correlation
if try_score > ds_score:
ds_score = try_score
weights = try_weights.copy()
val_mix = try_mix.copy()
weights_all.append(weights)
best_weighted_scores.append(ds_score)
if WEIGHTS == "bestonly":
df_sub.iloc[:, idx + 1] = selected_models_tst_preds[
np.argmax(selected_models_scores)
]
if ROUND:
best_rounded_score, rounding_method = get_rounding(common_val, val_mix)
best_rounded_scores.append(best_rounded_score)
print(
f"{idx} {cat}: mixed score {best_rounded_score}; {rounding_method['Type']}, {rounding_method['DP']}"
)
unrounded_backup = df_sub.iloc[:, idx + 1].copy()
if rounding_method["Type"] == "Normal":
df_sub.iloc[:, idx + 1] = rounding_method["func"](
df_sub.iloc[:, idx + 1], rounding_method["DP"]
)
elif (rounding_method["Type"] == "Ceil") or (
rounding_method["Type"] == "Floor"
):
df_sub.iloc[:, idx + 1] = rounding_method["func"](
df_sub.iloc[:, idx + 1] * (10 ** rounding_method["DP"])
) / (10 ** rounding_method["DP"])
if USE_TEST_SAFETY_ADJUSTMENTS:
if df_sub.iloc[:, idx + 1].var() == 0:
print("Test predictions are STILL homogenous, reverting to softmax weights")
all_scores = np.sum(np.exp([i for i in selected_models_scores]))
softmax_weights = [np.exp(i) / all_scores for i in selected_models_scores]
df_sub.iloc[:, idx + 1] = np.array(
[i * w for i, w in zip(selected_models_tst_preds, softmax_weights)]
).sum(axis=0)
if df_sub.iloc[:, idx + 1].var() == 0:
raise Exception("var = 0")
if df_sub.iloc[:, idx + 1].min() < 0:
raise Exception("<0")
if df_sub.iloc[:, idx + 1].max() > 1:
raise Exception(">1")
# df_sub.iloc[:,idx+1] *= 0.999998
# df_sub.iloc[:,idx+1] += 0.000001
if df_sub.isna().sum().sum() != 0:
raise Exception("na in sub")
df_sub.to_csv("submission.csv", index=False)
save_file(weights_all, "weights_all")
save_file(weights_for_rounding, "weights_all")
save_file(scores_all, "weights_all")
np.max(df_sub.iloc[:, 1:].to_numpy().flatten()), np.min(
df_sub.iloc[:, 1:].to_numpy().flatten()
), df_sub.shape
np.mean(bestonly_raw_scores), np.min(bestonly_raw_scores), np.max(
bestonly_raw_scores
), len(bestonly_raw_scores)
np.mean(best_weighted_scores), np.min(best_weighted_scores), np.max(
best_weighted_scores
), len(best_weighted_scores)
np.mean(best_rounded_scores), np.min(best_rounded_scores), np.max(
best_rounded_scores
), len(best_rounded_scores)
from matplotlib.pyplot import figure
figure(num=None, figsize=(16, 6), dpi=80, facecolor="black", edgecolor="k")
plt.boxplot(weights_all)
plt.show()
figure(num=None, figsize=(16, 6), dpi=80, facecolor="black", edgecolor="k")
plt.boxplot(weights_for_rounding)
plt.show()
figure(num=None, figsize=(16, 6), dpi=80, facecolor="black", edgecolor="k")
scores_all = [np.array(x) for x in scores_all]
plt.boxplot([x[~np.isnan(x)] for x in scores_all])
plt.show()
[(c, np.round(s, 3)) for c, s in zip(output_categories, scores_all)]
[(c, np.round(s, 3)) for c, s in zip(output_categories, weights_all)]
[(c, np.round(s, 3)) for c, s in zip(output_categories, weights_for_rounding)]
for i in output_categories:
fig, ax = plt.subplots(1, 2, figsize=(10, 3))
ax[0].hist(df_sub[i])
ax[1].hist(df_train[i])
plt.show()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import numpy as np
from scipy.stats import norm
from sklearn.preprocessing import StandardScaler
from scipy import stats
import warnings
warnings.filterwarnings("ignore")
train_df = pd.read_csv(
"/kaggle/input/house-prices-advanced-regression-techniques/train.csv"
)
train_df.columns
train_df["SalePrice"].describe()
sns.displot(train_df["SalePrice"])
print("Skewness: %f" % train_df["SalePrice"].skew())
print("Kurtosis: %f" % train_df["SalePrice"].kurt())
# Relationship with numerical variables
# GrLivArea
# TotalBsmtSF
var = "GrLivArea"
data = pd.concat([train_df["SalePrice"], train_df[var]], axis=1)
sns.scatterplot(x=var, y="SalePrice", data=data)
plt.show()
var = "TotalBsmtSF"
data = pd.concat([train_df["SalePrice"], train_df[var]], axis=1)
sns.scatterplot(x=var, y="SalePrice", data=data)
plt.show()
# Relationship with categorical features
# OverallQual
# YearBuilt
var = "OverallQual"
data = pd.concat([train_df[var], train_df["SalePrice"]], axis=1)
sns.boxplot(x=var, y="SalePrice", data=data)
plt.figure(figsize=(10, 8))
plt.show()
var = "YearBuilt"
data = pd.concat([train_df["SalePrice"], train_df[var]], axis=1)
f, ax = plt.subplots(figsize=(10, 8))
fig = sns.boxplot(x=var, y="SalePrice", data=data)
fig.axis(ymin=0, ymax=800000)
plt.xticks(rotation=90)
# corelation matrix
corrmat = train_df.corr()
plt.figure(figsize=(16, 8))
sns.heatmap(corrmat, vmax=0.9, square="true")
plt.show()
import seaborn as sns
import matplotlib.pyplot as plt
corrmat = train_df.corr()
plt.figure(figsize=(14, 6))
k = 10
cols = corrmat.nlargest(k, "SalePrice")["SalePrice"].index
cm = np.corrcoef(train_df[cols].values.T)
sns.set(font_scale=1.25)
sns.heatmap(
cm,
cbar=True,
annot=True,
square=True,
fmt=".2f",
annot_kws={"size": 12},
yticklabels=cols.values,
xticklabels=cols.values,
)
plt.title("Top 10 Features Correlated with SalePrice", fontsize=16)
plt.show()
# scatterplot
sns.set()
cols = [
"SalePrice",
"OverallQual",
"GrLivArea",
"GarageCars",
"TotalBsmtSF",
"FullBath",
"YearBuilt",
]
sns.pairplot(train_df[cols], size=3)
plt.show()
|
from fastai.imports import *
from fastai.structured import *
from pandas_summary import DataFrameSummary
from sklearn.ensemble import RandomForestRegressor, RandomForestClassifier
from IPython.display import display
from sklearn import metrics
def display_all(df):
with pd.option_context("display.max_rows", 1000, "display.max_columns", 1000):
display(df)
test = pd.read_csv("../input/house-prices-advanced-regression-techniques/test.csv")
train = pd.read_csv("../input/house-prices-advanced-regression-techniques/train.csv")
train_cats(train)
df, y, nas = proc_df(train, "SalePrice")
m = RandomForestRegressor(n_jobs=-1, random_state=0)
m.fit(df, y)
m.score(df, y)
def split_vals(a, n):
return a[:n], a[n:]
n_valid = 2000
n_trn = len(df) - n_valid
X_train, X_valid = split_vals(df, n_trn)
y_train, y_valid = split_vals(y, n_trn)
raw_train, raw_valid = split_vals(train, n_trn)
def rmse(x, y):
return math.sqrt(((x - y) ** 2).mean())
def print_score(m):
res = [
rmse(m.predict(X_train), y_train),
rmse(m.predict(X_valid), y_valid),
m.score(X_train, y_train),
m.score(X_valid, y_valid),
]
if hasattr(m, "oob_score_"):
res.append(m.oob_score_)
print(res)
m = RandomForestRegressor(
n_estimators=40, min_samples_leaf=3, max_features=0.5, n_jobs=-1, oob_score=True
)
m.fit(X_train, y_train)
print_score(m)
fi = rf_feat_importance(m, df)
fi[:30]
|
import numpy as np
import pandas as pd
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from sklearn.metrics import accuracy_score
from sklearn.linear_model import LogisticRegression
from sklearn import svm
from sklearn.naive_bayes import MultinomialNB
from sklearn.ensemble import RandomForestClassifier
# *1. Load your dataset from the “internet_connection_data.csv” csv file.*
data = pd.read_csv("/kaggle/input/connection-data/internet_connection_data.csv")
data.head()
# *2. Explore your dataset and list the name of the columns.*
column_names = data.columns.values.tolist()
print(column_names)
data.shape
data.describe()
data.info()
# *3. Explore your dataset and check if there is any column with missing values.*
data.isnull().values.any()
data.isna().sum()
# *4. Select your input variables and ourput variable.*
x = data["Category"]
x
x.value_counts()
y = data.drop(columns=["Category"])
y
# *5. Split your dataset as %80 training and %20 testing.*
y_train, y_test, x_train, x_test = train_test_split(
y, x, random_state=666, test_size=0.2
)
scaler = StandardScaler()
y_scaled = scaler.fit_transform(y)
y_train_s, y_test_s, x_train_s, x_test_s = train_test_split(
y_scaled, x, random_state=666, test_size=0.2
)
y_train
# *6. Implement four classification model based on Logistic Regression, Support Vector Machine, Multinomial Naive Bayes and Random Forest classifiers.*
models = [
LogisticRegression(multi_class="auto", solver="liblinear", max_iter=10000, C=10),
svm.SVC(),
MultinomialNB(),
RandomForestClassifier(n_estimators=200, max_depth=30),
]
# *7. Train (fit) your network.
# 8. Report the accuracies (by percentage) of the models for the test datasets.*
scores = {}
for i in range(len(models)):
model = models[i]
if i == 2:
model.fit(y_train, x_train)
y_pred = model.predict(y_test)
accuracy = accuracy_score(x_test, x_pred)
else:
model.fit(y_train_s, x_train_s)
x_pred = model.predict(y_test_s)
accuracy = accuracy_score(x_test_s, x_pred)
scores[i] = accuracy * 100
print(f"Accuracy of the model {models[i]}: {accuracy * 100}")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
df = pd.read_csv("/kaggle/input/marketing-campaigns-of-portuguese-bank/bank.csv")
df.head()
df.tail()
df.info()
df.columns
df.describe()
import pandas as pd
df = pd.read_csv(
"/kaggle/input/marketing-campaigns-of-portuguese-bank/bank.csv", delimiter=";"
)
df.head
df.shape
import matplotlib.pyplot as plt
def bar_plot(variable):
var_value = df[variable].value_counts()
plt.figure(figsize=(20, 7))
plt.bar(var_value.index, var_value, color=["blue", "red", "green"])
plt.xticks(var_value.index, var_value.index.values)
plt.ylabel("Frequency")
plt.title(variable)
plt.show()
print("{}: \n {}".format(variable, var_value))
# s = pd.Series(['apple', 'banana', 'banana', 'orange', 'orange', 'orange'])
# orange 3
# banana 2
# apple 1
# dtype: int64 (value.count)
# plt.xticks(var_value.index, var_value.index.values)
# import pandas as pd
# s = pd.Series([10, 20, 30], index=['a', 'b', 'c'])
# plt.bar(s.index, s.values)
# plt.xticks(s.index, s.index.values)
# plt.show()
category = [
"job",
"marital",
"education",
"housing",
"loan",
"contact",
"day",
"month",
"duration",
"campaign",
"pdays",
"previous",
"poutcome",
"y",
]
for c in category:
bar_plot(c)
def plot_hist(variable):
plt.figure(figsize=(9, 6))
plt.hist(df[variable], bins=30, color="gray")
plt.xlabel(variable)
plt.ylabel("Frequency")
plt.title("{} Distribution with Hist".format(variable))
plt.show()
# bins = interval
numeric_vars = ["age", "campaign", "duration"]
for var in numeric_vars:
plot_hist(var)
pd.crosstab(df.age, df.campaign).plot(kind="area", figsize=(15, 7))
plt.title("Age Distribution")
plt.xlabel("Age")
plt.ylabel("Frequency")
plt.show()
# crosstab - Crosstab or Cross-tabulation is a way to represent the relationship between two categorical variables.
# It is a summary table that shows the distribution of one variable in rows and the distribution of another variable in columns.
# relationship it describe
pd.crosstab(df.job, df.marital).plot(kind="barh", figsize=(15, 7))
plt.title("Deposit Age Distribution")
plt.xlabel("Frequency")
plt.ylabel("Job")
plt.show()
# it compares between mariage state & job
# counter - to count the frequency of each index in the outlier_indices list.
from collections import Counter
import numpy as np
def detect_outliers(df, features):
outlier_indices = []
for c in features:
# 1st quartile
Q1 = np.percentile(df[c], 25)
# 3rd quartile
Q3 = np.percentile(df[c], 75)
# IQR
IQR = Q3 - Q1
# Outlier step
outlier_step = IQR * 1.5
# detect outlier and their indices
outlier_list_col = df[
(df[c] < Q1 - outlier_step) | (df[c] > Q3 + outlier_step)
].index
# store indices
outlier_indices.extend(outlier_list_col)
outlier_indices = Counter(outlier_indices)
multiple_outliers = [i for i, v in outlier_indices.items() if v > 1]
return multiple_outliers
df.loc[detect_outliers(df, ["age", "day", "duration", "campaign", "previous"])]
df.isnull().sum()
import seaborn as sns
fig, ax = plt.subplots(figsize=(13, 13)) # it checks the relation between 2 varaible
sns.heatmap(df.corr(), annot=True, linewidths=0.5, ax=ax)
# No correlaiton r=0
# Very weak correlation: r<20
# Weak correlation: between 0.20-0.49
# Moderate correlation: between 0.5-0.79
# Strong correlation: between 0.8-0.99
# Perfect correlation: r=1
df = df.drop(["duration"], axis=1)
df.head()
# one hot encoding catergoical - Gender: Male, Female, Other 1 - male 0 - female other - 2
# color red yellow green
# red 1 0 0
# green 0 0 1
# yellow
from sklearn.preprocessing import StandardScaler, OneHotEncoder
from sklearn.compose import ColumnTransformer
categorical_cols = ["contact", "month", "poutcome", "y"]
preprocessor = ColumnTransformer(
transformers=[("cat", OneHotEncoder(), categorical_cols)], remainder="passthrough"
)
X = preprocessor.fit_transform(
df.iloc[:, :-3]
) # All columns except for the last three columns
Y = df.iloc[:, -1] # Last column 'pdays2' as the target variable
# Perform standard scaling on features
scaler = StandardScaler()
X = scaler.fit_transform(X)
# Print the scaled features
print(X)
# Print the target variable
print(Y)
df.info()
from sklearn.model_selection import train_test_split
from sklearn.metrics import cohen_kappa_score
from sklearn.metrics import classification_report, confusion_matrix
from sklearn.metrics import accuracy_score
from sklearn.metrics import ConfusionMatrixDisplay
from sklearn.metrics import f1_score
X = preprocessor.fit_transform(
df.iloc[:, :-1]
) # All columns except for the last three columns
Y = df.iloc[:, -1]
X_train, X_test, y_train, y_test = train_test_split(
X, Y, test_size=0.2
) # provide a value for test_size
accuracies = {}
kappaScores = {}
f1scores = {}
def pdayswork(pdays):
if pdays == -1:
return 0
elif pdays >= 0:
return 1
df["pdays2"] = df["pdays"].apply(pdayswork)
# if the pdays = 0, it indicates that it has not been contacted before
# if the pdays = 1, it indicates that it was contacted earlier
df.head()
# print('deposit_no' in df.columns)
# print('deposit_yes' in df.columns)
# df = df.drop(['deposit_no', 'deposit_yes'], axis=1)
# def deposit1(deposit):
# if deposit == 'yes':
# return 1
# elif deposit == 'no':
# return 0
# else:
# return None # Add a return statement for cases when deposit value doesn't match 'yes' or 'no'
# df['depositNew'] = df['deposit'].apply(deposit1)
# df = df.drop(['deposit'], axis=1)
from sklearn.preprocessing import StandardScaler, OneHotEncoder
from sklearn.compose import ColumnTransformer
categorical_cols = ["contact", "month", "poutcome", "y"]
preprocessor = ColumnTransformer(
transformers=[("cat", OneHotEncoder(), categorical_cols)], remainder="passthrough"
)
X = preprocessor.fit_transform(
df.iloc[:, :-3]
) # All columns except for the last three columns
Y = df.iloc[:, -1] # Last column 'pdays2' as the target variable
# Perform standard scaling on features
scaler = StandardScaler()
X = scaler.fit_transform(X)
# Print the scaled features
print(X)
# Print the target variable
print(Y)
from sklearn.model_selection import train_test_split
from sklearn.metrics import cohen_kappa_score
from sklearn.metrics import classification_report, confusion_matrix
from sklearn.metrics import accuracy_score
from sklearn.metrics import ConfusionMatrixDisplay
from sklearn.metrics import f1_score
X = preprocessor.fit_transform(
df.iloc[:, :-1]
) # All columns except for the last three columns
Y = df.iloc[:, -1]
X_train, X_test, y_train, y_test = train_test_split(
X, Y, test_size=0.2
) # provide a value for test_size
accuracies = {}
kappaScores = {}
f1scores = {}
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import (
classification_report,
accuracy_score,
f1_score,
cohen_kappa_score,
)
X = preprocessor.fit_transform(
df.iloc[:, :-1]
) # All columns except for the last three columns
Y = df.iloc[:, -1]
lr = LogisticRegression(
random_state=101, multi_class="ovr", solver="liblinear"
) # add a closing parenthesis
lr.fit(X_train, y_train) # add a line break before fit()
prediction = lr.predict(X_test)
print(classification_report(y_test, prediction))
acc = accuracy_score(y_test, prediction) * 100
print("Logistic Regression accuracy:", acc)
accuracies["Logistic Regression"] = acc
f1 = f1_score(y_test, prediction) * 100
print("F1-Score: ", f1)
f1scores["Logistic Regression"] = f1
cohen_kappa = cohen_kappa_score(y_test, prediction) * 100
print("Cohen Kappa score: ", cohen_kappa)
kappaScores["Logistic Regression"] = cohen_kappa
from sklearn.linear_model import LogisticRegression
lr = LogisticRegression(
random_state=101, multi_class="ovr", solver="liblinear"
) # add a closing parenthesis
lr.fit(X_train, y_train)
prediction = lr.predict(X_test)
print(classification_report(y_test, prediction))
acc = accuracy_score(y_test, prediction) * 100
print("Logistic Regression accuracy:", acc)
accuracies["Logistic Regression"] = acc
f1 = f1_score(y_test, prediction) * 100
print("F1-Score: ", f1)
f1scores["Logistic Regression"] = f1
cohen_kappa = cohen_kappa_score(y_test, prediction) * 100
print("Cohen Kappa score: ", cohen_kappa)
kappaScores["Logistic Regression"] = cohen_kappa
score = round(accuracy_score(y_test, prediction), 3)
cm = confusion_matrix(y_test, prediction)
sns.heatmap(cm, annot=True, fmt=".1f", linewidths=3, square=True, cmap="Blues")
plt.ylabel("actual label")
plt.xlabel("predicted label")
plt.title("accuracy score: {0}".format(score), size=12)
plt.show()
from sklearn.ensemble import RandomForestClassifier
clf = RandomForestClassifier(n_estimators=100, max_depth=12, random_state=50)
clf.fit(X_train, y_train)
prediction = clf.predict(X_test)
acc = accuracy_score(y_test, prediction) * 100
print("Random Forest accuracy:", acc)
accuracies["Random Forest"] = acc
f1 = f1_score(y_test, prediction) * 100
print("F1-Score: ", f1)
f1scores["Random Forest"] = f1
cohen_kappa = cohen_kappa_score(y_test, prediction) * 100
print("Cohen Kappa score: ", cohen_kappa)
kappaScores["Random Forest"] = cohen_kappa
from sklearn.naive_bayes import GaussianNB
nb = GaussianNB()
nb.fit(X_train, y_train)
naiveb = nb.predict(X_test)
prediction = nb.predict(X_test)
acc = accuracy_score(y_test, prediction) * 100
print("Naive Bayes accuracy:", acc)
accuracies["Naive Bayes"] = acc
f1 = f1_score(y_test, prediction) * 100
print("F1-Score: ", f1)
f1scores["Naive Bayes"] = f1
cohen_kappa = cohen_kappa_score(y_test, prediction) * 100
print("Cohen Kappa score: ", cohen_kappa)
kappaScores["Naive Bayes"] = cohen_kappa
from sklearn.tree import DecisionTreeClassifier
dtree = DecisionTreeClassifier(criterion="gini", max_depth=10, random_state=None)
dtree.fit(X_train, y_train)
prediction = dtree.predict(X_test)
acc = accuracy_score(y_test, prediction) * 100
print("Decision Tree accuracy:", acc)
accuracies["Decision Tree"] = acc
f1 = f1_score(y_test, prediction) * 100
print("F1-Score: ", f1)
f1scores["Decision Tree"] = f1
cohen_kappa = cohen_kappa_score(y_test, prediction) * 100
print("Cohen Kappa score: ", cohen_kappa)
kappaScores["Decision Tree"] = cohen_kappa
|
# Hello, thanks for checking this out! This is my first time working on Kaggle, so please tell me how I can improve!
list.files(path="../input")
library(tidyverse)
ufc_data < -read.csv("../input/ufcdata/data.csv")
|
from matplotlib import pyplot as plt
import numpy as np
import pandas as pd
import PIL.Image as Image, PIL.ImageDraw as ImageDraw, PIL.ImageFont as ImageFont
import random
import os
import cv2
import gc
from tqdm.auto import tqdm
import numpy as np
import keras
from tensorflow.keras.models import Sequential, clone_model
from tensorflow.keras.layers import (
BatchNormalization,
LeakyReLU,
Dense,
Dropout,
Flatten,
Conv2D,
MaxPooling2D,
)
from tensorflow.keras.optimizers import SGD
from sklearn.model_selection import train_test_split
from tensorflow.keras.callbacks import ReduceLROnPlateau
from tensorflow.keras.preprocessing.image import ImageDataGenerator
from tensorflow.keras.utils import plot_model
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
import numpy as np
import datetime as dt
import tensorflow.compat.v1 as tf
tf.disable_v2_behavior()
# for dirname, _, filenames in os.walk('../input/bengaliai-cv19/'):
# for filename in filenames:
# print(os.path.join(dirname, filename))
train_data = pd.read_csv("../input/bengaliai-cv19/train.csv")
def resize(df, size=64, need_progress_bar=True):
resized = {}
for i in range(df.shape[0]):
image = cv2.resize(df.loc[df.index[i]].values.reshape(137, 236), (size, size))
resized[df.index[i]] = image.reshape(-1)
resized = pd.DataFrame(resized).T
return resized
inputs = keras.Input(shape=(64, 64, 1))
# CONV 1A STARTS
model = layers.Conv2D(
filters=32, kernel_size=(3, 3), padding="SAME", activation="relu"
)(inputs)
model = tf.nn.leaky_relu(model, alpha=0.01, name="Leaky_ReLU")
model = layers.Conv2D(
filters=32, kernel_size=(3, 3), padding="SAME", activation="relu"
)(model)
model = tf.nn.leaky_relu(model, alpha=0.01, name="Leaky_ReLU")
model = layers.BatchNormalization(momentum=0.15)(model)
model = layers.MaxPool2D(pool_size=(2, 2))(model)
model = layers.Conv2D(
filters=32, kernel_size=(5, 5), padding="SAME", activation="relu"
)(model)
model = tf.nn.leaky_relu(model, alpha=0.01, name="Leaky_ReLU")
model = layers.Dropout(rate=0.3)(model)
# CONV 1B STARTS
model = layers.Conv2D(
filters=32, kernel_size=(3, 3), padding="SAME", activation="relu"
)(model)
model = tf.nn.leaky_relu(model, alpha=0.01, name="Leaky_ReLU")
model = layers.Conv2D(
filters=32, kernel_size=(3, 3), padding="SAME", activation="relu"
)(model)
model = tf.nn.leaky_relu(model, alpha=0.01, name="Leaky_ReLU")
model = layers.BatchNormalization(momentum=0.15)(model)
model = layers.MaxPool2D(pool_size=(2, 2))(model)
model = layers.Conv2D(
filters=32, kernel_size=(5, 5), padding="SAME", activation="relu"
)(model)
model = tf.nn.leaky_relu(model, alpha=0.01, name="Leaky_ReLU")
model = layers.Dropout(rate=0.3)(model)
# CONV 2A STARTS
model = layers.Conv2D(
filters=64, kernel_size=(3, 3), padding="SAME", activation="relu"
)(model)
model = tf.nn.leaky_relu(model, alpha=0.01, name="Leaky_ReLU")
model = layers.BatchNormalization(momentum=0.15)(model)
model = layers.Dropout(rate=0.3)(model)
# CONV 2B STARTS
model = layers.Conv2D(
filters=64, kernel_size=(3, 3), padding="SAME", activation="relu"
)(model)
model = tf.nn.leaky_relu(model, alpha=0.01, name="Leaky_ReLU")
model = layers.BatchNormalization(momentum=0.15)(model)
model = layers.Dropout(rate=0.3)(model)
# CONV 3A STARTS
model = layers.Conv2D(
filters=64, kernel_size=(3, 3), padding="SAME", activation="relu"
)(model)
model = tf.nn.leaky_relu(model, alpha=0.01, name="Leaky_ReLU")
model = layers.Conv2D(
filters=64, kernel_size=(3, 3), padding="SAME", activation="relu"
)(model)
model = tf.nn.leaky_relu(model, alpha=0.01, name="Leaky_ReLU")
model = layers.BatchNormalization(momentum=0.15)(model)
model = layers.MaxPool2D(pool_size=(2, 2))(model)
# CONV 3B STARTS
model = layers.Conv2D(
filters=64, kernel_size=(5, 5), padding="SAME", activation="relu"
)(model)
model = tf.nn.leaky_relu(model, alpha=0.01, name="Leaky_ReLU")
model = layers.BatchNormalization(momentum=0.15)(model)
model = layers.Dropout(rate=0.3)(model)
# FLATTEN STARTS
model = layers.Flatten()(model)
model = layers.Dense(1024, activation="relu")(model)
model = layers.Dropout(rate=0.3)(model)
dense = layers.Dense(512, activation="relu")(model)
# CLASS DECSISION STARTS
out_root = layers.Dense(168, activation="softmax", name="root_out")(dense)
out_vowel = layers.Dense(11, activation="softmax", name="vowel_out")(dense)
out_consonant = layers.Dense(7, activation="softmax", name="consonant_out")(dense)
# FINAL MODEL
cnn1model = keras.Model(inputs=inputs, outputs=[out_root, out_vowel, out_consonant])
plot_model(cnn1model, to_file="mode4.png")
# print(cnn1model.summary())
cnn1model.compile(
optimizer="adam",
loss=[
"categorical_crossentropy",
"categorical_crossentropy",
"categorical_crossentropy",
],
metrics=["accuracy"],
)
mc = keras.callbacks.ModelCheckpoint(
"weights{epoch:08d}.h5", save_weights_only=True, period=10
)
batch_size = 128
epochs = 30
history_list = []
paraquets_used = 4
class MultiOutputDataGenerator(keras.preprocessing.image.ImageDataGenerator):
def flow(
self,
x,
y=None,
batch_size=32,
shuffle=True,
sample_weight=None,
seed=None,
save_to_dir=None,
save_prefix="",
save_format="png",
subset=None,
):
targets = None
target_lengths = {}
ordered_outputs = []
for output, target in y.items():
if targets is None:
targets = target
else:
targets = np.concatenate((targets, target), axis=1)
target_lengths[output] = target.shape[1]
ordered_outputs.append(output)
for flowx, flowy in super().flow(
x, targets, batch_size=batch_size, shuffle=shuffle
):
target_dict = {}
i = 0
for output in ordered_outputs:
target_length = target_lengths[output]
target_dict[output] = flowy[:, i : i + target_length]
i += target_length
yield flowx, target_dict
for i in range(paraquets_used):
b_train_data = pd.merge(
pd.read_parquet(f"../input/bengaliai-cv19/train_image_data_{i}.parquet"),
train_data,
on="image_id",
).drop(["image_id"], axis=1)
print("Data Loaded")
(
x_train,
x_test,
y_train_root,
y_test_root,
y_train_vowel,
y_test_vowel,
y_train_consonant,
y_test_consonant,
) = train_test_split(
(
resize(
b_train_data.drop(
[
"grapheme_root",
"vowel_diacritic",
"consonant_diacritic",
"grapheme",
],
axis=1,
)
)
/ 255
).values.reshape(-1, 64, 64, 1),
pd.get_dummies(b_train_data["grapheme_root"]).values,
pd.get_dummies(b_train_data["vowel_diacritic"]).values,
pd.get_dummies(b_train_data["consonant_diacritic"]).values,
test_size=0.08,
random_state=666,
)
del b_train_data
gc.collect()
# del Y_train_root, Y_train_vowel, Y_train_consonant
indeX = str(i)
print("Run " + indeX + " starts")
datagen = MultiOutputDataGenerator(
featurewise_center=False, # set input mean to 0 over the dataset
samplewise_center=False, # set each sample mean to 0
featurewise_std_normalization=False, # divide inputs by std of the dataset
samplewise_std_normalization=False, # divide each input by its std
zca_whitening=False, # apply ZCA whitening
rotation_range=8, # randomly rotate images in the range (degrees, 0 to 180)
zoom_range=0.20, # Randomly zoom image
width_shift_range=0.20, # randomly shift images horizontally (fraction of total width)
height_shift_range=0.20, # randomly shift images vertically (fraction of total height)
horizontal_flip=False, # randomly flip images
vertical_flip=False,
) # randomly flip images
# This will just calculate parameters required to augment the given data. This won't perform any augmentations
datagen.fit(x_train)
history = cnn1model.fit_generator(
datagen.flow(
x_train,
{
"root_out": y_train_root,
"vowel_out": y_train_vowel,
"consonant_out": y_train_consonant,
},
batch_size=batch_size,
),
epochs=epochs,
validation_data=(x_test, [y_test_root, y_test_vowel, y_test_consonant]),
steps_per_epoch=x_train.shape[0] // batch_size,
callbacks=[mc],
)
history_list.append(history)
del datagen
del x_train
del x_test
del y_train_root
del y_test_root
del y_train_vowel
del y_test_vowel
del y_train_consonant
del y_test_consonant
print("Run " + indeX + " ends")
gc.collect()
del train_data
gc.collect()
def plot_loss(his, epoch, title):
plt.figure()
plt.plot(np.arange(0, epoch), his.history["loss"], label="train_loss")
plt.plot(np.arange(0, epoch), his.history["root_out_loss"], label="train_root_loss")
plt.plot(
np.arange(0, epoch), his.history["vowel_out_loss"], label="train_vowel_loss"
)
plt.plot(
np.arange(0, epoch),
his.history["consonant_out_loss"],
label="train_consonant_loss",
)
plt.plot(
np.arange(0, epoch),
his.history["val_root_out_loss"],
label="val_train_root_loss",
)
plt.plot(
np.arange(0, epoch),
his.history["val_vowel_out_loss"],
label="val_train_vowel_loss",
)
plt.plot(
np.arange(0, epoch),
his.history["val_consonant_out_loss"],
label="val_train_consonant_loss",
)
plt.title(title)
plt.xlabel("Epoch #")
plt.ylabel("Loss")
plt.legend(loc="upper right")
plt.show()
def plot_acc(his, epoch, title):
plt.figure()
plt.plot(np.arange(0, epoch), his.history["root_out_acc"], label="train_root_acc")
plt.plot(np.arange(0, epoch), his.history["vowel_out_acc"], label="train_vowel_acc")
plt.plot(
np.arange(0, epoch),
his.history["consonant_out_acc"],
label="train_consonant_acc",
)
plt.plot(np.arange(0, epoch), his.history["val_root_out_acc"], label="val_root_acc")
plt.plot(
np.arange(0, epoch), his.history["val_vowel_out_acc"], label="val_vowel_acc"
)
plt.plot(
np.arange(0, epoch),
his.history["val_consonant_out_acc"],
label="val_consonant_acc",
)
plt.title(title)
plt.xlabel("Epoch #")
plt.ylabel("Accuracy")
plt.legend(loc="upper right")
plt.show()
for dataset in range(paraquets_used):
plot_loss(history_list[dataset], epochs, f"Training Dataset: {dataset}")
plot_acc(history_list[dataset], epochs, f"Training Dataset: {dataset}")
preds_dict = {"grapheme_root": [], "vowel_diacritic": [], "consonant_diacritic": []}
components = ["consonant_diacritic", "grapheme_root", "vowel_diacritic"]
target = [] # model predictions placeholder
row_id = [] # row_id place holder
for i in range(4):
df_test_img = pd.read_parquet(
"../input/bengaliai-cv19/test_image_data_{}.parquet".format(i)
)
df_test_img.set_index("image_id", inplace=True)
X_test = resize(df_test_img, need_progress_bar=False) / 255
X_test = X_test.values.reshape(-1, 64, 64, 1)
preds = cnn1model.predict(X_test)
for i, p in enumerate(preds_dict):
preds_dict[p] = np.argmax(preds[i], axis=1)
for k, id in enumerate(df_test_img.index.values):
for i, comp in enumerate(components):
id_sample = id + "_" + comp
row_id.append(id_sample)
target.append(preds_dict[comp][k])
del df_test_img
del X_test
gc.collect()
df_sample = pd.DataFrame(
{"row_id": row_id, "target": target}, columns=["row_id", "target"]
)
df_sample.to_csv("submission.csv", index=False)
df_sample.head()
|
import torch
import torch.nn as nn
import torch.optim as optim
import torchvision.models as models
import torchvision.transforms as transforms
from torch.utils.data import DataLoader
from torchvision.datasets import ImageFolder
from sklearn.metrics import roc_auc_score, confusion_matrix, precision_recall_curve, auc
import matplotlib.pyplot as plt
import numpy as np
import cv2
import os
import numpy as np
import pandas as pd
from glob import glob
from PIL import Image
import seaborn as sns
import matplotlib.pyplot as plt
from skimage.io import imread
from sklearn.model_selection import train_test_split
# Pytorch Libraries
import torch
import torchvision
import torch.nn as nn
import torch.optim as optim
from torch.optim import lr_scheduler
from torchvision import models, transforms
from torch.utils.data import TensorDataset, DataLoader, Dataset
meta_data = pd.read_csv(
"/kaggle/input/histopathologic-cancer-detection/train_labels.csv"
)
meta_data = meta_data.groupby("label", group_keys=False).apply(lambda x: x.sample(5000))
meta_data.reset_index(drop=True, inplace=True)
meta_data.shape
meta_data.head()
base_dir = os.path.join("..", "input/histopathologic-cancer-detection/train")
imageid_path_dict = {
os.path.splitext(os.path.basename(x))[0]: x
for x in glob(os.path.join(base_dir, "*.tif"))
}
meta_data["path"] = meta_data["id"].map(imageid_path_dict.get)
meta_data.head()
n_samples = 6
n_classes = len(meta_data["label"].unique())
df = meta_data.sort_values(["label"]).groupby("label")
fig, axs = plt.subplots(n_classes, n_samples, figsize=(10, 4))
for ax, (type_, rows) in zip(axs, df):
ax[0].set_title("Class: " + str(type_), fontsize=15)
for sub_ax, (_, subset) in zip(ax, rows.sample(n_samples).iterrows()):
img = imread(subset["path"])
sub_ax.imshow(img)
sub_ax.axis("off")
data = []
for path in meta_data["path"]:
img = cv2.imread(path)
avgR = np.mean(img[:, :, 2])
avgG = np.mean(img[:, :, 1])
avgB = np.mean(img[:, :, 0])
data.append([avgR, avgG, avgB])
rgb = pd.DataFrame(
data, columns=["Red Channel Mean", "Green Channel Mean", "Blue Channel Mean"]
)
meta_data = pd.concat([meta_data.reset_index(drop=True), rgb], axis=1)
g = sns.pairplot(
meta_data[["Red Channel Mean", "Green Channel Mean", "Blue Channel Mean", "label"]],
hue="label",
plot_kws={"alpha": 0.3},
)
g.fig.set_size_inches(12, 8)
extremely_low_pxl_img = meta_data[
(meta_data["Red Channel Mean"] <= 50)
& (meta_data["Blue Channel Mean"] <= 50)
& (meta_data["Green Channel Mean"] <= 50)
]
extremely_high_pxl_img = meta_data[
(meta_data["Red Channel Mean"] >= 250)
& (meta_data["Blue Channel Mean"] >= 250)
& (meta_data["Green Channel Mean"] >= 250)
]
extremely_high_pxl_img
extremely_low_pxl_img
class CustomDataset(Dataset):
def __init__(self, data_df, transform=None):
self.data_df = data_df
self.transform = transform
def __len__(self):
return len(self.data_df)
def __getitem__(self, index):
img_path = self.data_df.iloc[index]["path"]
label = self.data_df.iloc[index]["label"]
# Load image from file
img = Image.open(img_path).convert("RGB")
if self.transform is not None:
img = self.transform(img)
return img, label
# Split data into train and test sets
train_df, test_df = train_test_split(meta_data, test_size=0.2, random_state=42)
# Define the batch size
batch_size = 32
# Define the data transforms for training and testing datasets
train_transforms = transforms.Compose(
[
transforms.RandomResizedCrop(size=256, scale=(0.8, 1.0)),
transforms.RandomRotation(degrees=15),
transforms.RandomHorizontalFlip(),
transforms.CenterCrop(size=224),
transforms.ToTensor(),
transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),
]
)
test_transforms = transforms.Compose(
[
transforms.Resize(size=256),
transforms.CenterCrop(size=224),
transforms.ToTensor(),
transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),
]
)
# Load the data
train_dataset = CustomDataset(train_df, transform=train_transforms)
train_loader = DataLoader(train_dataset, batch_size=batch_size, shuffle=True)
test_dataset = CustomDataset(test_df, transform=test_transforms)
test_loader = DataLoader(test_dataset, batch_size=batch_size)
# Define the models
models_dict = {
"ResNet50": models.resnet50(pretrained=True),
"VGG19": models.vgg19(pretrained=True),
"Inception": models.inception_v3(pretrained=True),
}
# Define the evaluation function
def evaluate(model, test_loader):
y_true, y_pred, y_prob = [], [], []
model.eval()
with torch.no_grad():
for inputs, labels in test_loader:
inputs = inputs.to(device)
labels = labels.to(device)
outputs = model(inputs)
probs = nn.functional.softmax(outputs, dim=1)
_, preds = torch.max(outputs, 1)
y_true.extend(labels.cpu().numpy())
y_pred.extend(preds.cpu().numpy())
y_prob.extend(probs[:, 1].cpu().numpy())
return y_true, y_pred, y_prob
# Define the device
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
# Define the metrics to evaluate
metrics_dict = {
"AUC ROC": roc_auc_score,
"Confusion Matrix": confusion_matrix,
"Precision-Recall": precision_recall_curve,
}
from sklearn.metrics import precision_score
from sklearn.metrics import recall_score
from sklearn.metrics import f1_score
# Initialize the figure and axes objects
fig, axes = plt.subplots(2, 2, figsize=(10, 8))
# Loop through the models
for i, (model_name, model) in enumerate(models_dict.items()):
# Freeze the weights of the convolutional layers
for name, param in model.named_parameters():
if name.startswith("fc"):
param.requires_grad = True
else:
param.requires_grad = True
# Replace the last fully connected layer
model.fc = nn.Linear(in_features=2048, out_features=2)
# Define the optimizer and loss function
optimizer = optim.Adam(model.fc.parameters())
criterion = nn.CrossEntropyLoss()
num_epoch = 10
# Train the model
for epoch in range(num_epoch):
# Set the model to train mode
model.train()
model.to(device)
for inputs, labels in train_loader:
inputs = inputs.to(device)
labels = labels.to(device)
optimizer.zero_grad()
outputs = model(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
# Evaluate the model
y_true, y_pred, y_prob = evaluate(model, test_loader)
# Compute the metrics
auc_roc = roc_auc_score(y_true, y_prob)
print(confusion_matrix(y_true, y_pred).ravel())
tn, fp, fn, tp = confusion_matrix(y_true, y_pred).ravel()
precision, recall, thresholds = precision_recall_curve(y_true, y_prob)
pr_auc = auc(recall, precision)
# Plot the metrics
ax = axes[i // 2, i % 2]
ax.set_title(model_name)
ax.plot(
recall, precision, label="Precision-Recall Curve (AUC = {:.2f})".format(pr_auc)
)
ax.plot([0, 1], [1, 0], "r--", label="Random Guess")
ax.set_xlabel("Recall")
ax.set_ylabel("Precision")
ax.set_xlim([0, 1])
ax.set_ylim([0, 1])
ax.legend()
# Print the metrics
print("{} Metrics:".format(model_name))
print("AUC ROC: {:.2f}".format(auc_roc))
print("Confusion Matrix: \n", np.array([[tn, fp], [fn, tp]]))
print("Precision: {:.2f}".format(precision_score(y_true, y_pred)))
print("Recall: {:.2f}".format(recall_score(y_true, y_pred)))
print("F1 Score: {:.2f}".format(f1_score(y_true, y_pred)))
print("\n")
# Adjust the spacing between the subplots
fig.tight_layout()
# Show the plot
plt.show()
|
# # Bike Share Challenge:
# ## Part I:
# **Goal of challenge**:
# You must predict the total count of bikes rented during each hour covered by the test set, using only information available prior to the rental period.
#
# As we have to simply provide a prediction on the total, we need to predict the number of casual rides + the number of registered rides.
#
# **The dataset**:
# You are provided hourly rental data spanning two years. For this competition, the training set is comprised of the first 19 days of each month, while the test set is the 20th to the end of the month.
# **Data fields:**
# - datetime - hourly date + timestamp
# - season - 1 = spring, 2 = summer, 3 = fall, 4 = winter
# - holiday - whether the day is considered a holiday
# - workingday - whether the day is neither a weekend nor holiday
# - weather - 1: Clear, Few clouds, Partly cloudy, Partly cloudy
# 2: Mist + Cloudy, Mist + Broken clouds, Mist + Few clouds, Mist
# 3: Light Snow, Light Rain + Thunderstorm + Scattered clouds, Light Rain + Scattered clouds
# 4: Heavy Rain + Ice Pallets + Thunderstorm + Mist, Snow + Fog
# - temp - temperature in Celsius
# - atemp - "feels like" temperature in Celsius
# - humidity - relative humidity
# - windspeed - wind speed
# - casual - number of non-registered user rentals initiated
# - registered - number of registered user rentals initiated
# - count - number of total rentals
# **Initial thoughts:**
# - Bike share demand is usually higher on weekends (free floating); however we are only provided with the first 19 days of the month so we can create a variable which says how many weekends in the month provided;
# - Bike share demand is very correlated with good weather (our good weather indicators are: season, weather (1-2-3-4), temp, atemp, humidity, windspeed
# - Casual users: they usually use more on the weekend; they tend to be higher during tourist seasons
# - Registered users: they tend to use more always
# - Total rentals over seasons: usually cyclical over the seasons
# - Holidays: higher number of riders
# **Load relevant libraries:**
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
plt.rc("font", size=18)
sns.set(style="white")
sns.set(style="whitegrid", color_codes=True)
train = pd.read_csv("train.csv")
test = pd.read_csv("test.csv")
train.head()
# **Unique values per variable:**
unique_values = {}
for i in range(1, len(train.columns) - 3):
unique_values[train.columns[i]] = train[train.columns[i]].unique()
unique_values
train.describe()
train.isnull().sum()
# ## Part II: EDA and Visualisation
# **Preparing Out Data For Vis:**
# Datetime:
datasets = [train, test]
for dataset in datasets:
dataset["datetime"] = pd.to_datetime(dataset.datetime)
dataset["hour"] = dataset["datetime"].apply(lambda x: x.hour)
dataset["day"] = dataset["datetime"].apply(lambda x: x.day)
dataset["weekday"] = dataset["datetime"].apply(lambda x: x.weekday())
dataset["month"] = dataset["datetime"].apply(lambda x: x.month)
dataset["year"] = dataset["datetime"].apply(lambda x: x.year)
train.head(2)
test.head(2)
# Names for categorical data:
train_c = train.copy()
train_c["weather"] = train_c["weather"].map(
{1: "Good", 2: "Medium", 3: "Bad", 4: "Very Bad"}
)
train_c["weekday"] = train_c["weekday"].map(
{0: "Mon", 1: "Tue", 2: "Wed", 3: "Thur", 4: "Fri", 5: "Sat", 6: "Sun"}
)
train_c["month"] = train_c["month"].map(
{
1: "Jan",
2: "Feb",
3: "Mar",
4: "Apr",
5: "May",
6: "Jun",
7: "July",
8: "Aug",
9: "Sept",
10: "Oct",
11: "Nov",
12: "Dec",
}
)
train_c["season"] = train_c["season"].map(
{1: "Spring", 2: "Summer", 3: "Fall", 4: "Winter"}
)
train_c["workingday"] = train_c["workingday"].map({0: "No", 1: "Yes"})
train_c["holiday"] = train_c["holiday"].map({0: "No", 1: "Yes"})
# **Season EDA:**|
from numpy import mean
fig, ax = plt.subplots(nrows=2, ncols=2, figsize=(12, 8))
sns.barplot(
x="season",
y="count",
data=train_c,
ci=None,
color="salmon",
hue="year",
estimator=mean,
ax=ax[0, 0],
)
ax[0, 0].set_title("Mean Count by Season hue: Year")
sns.barplot(
x="season",
y="count",
data=train_c,
ci=None,
color="salmon",
hue="weather",
estimator=mean,
ax=ax[0, 1],
)
ax[0, 1].set_title("Mean Count by Season hue: Weather")
sns.barplot(
x="month",
y="count",
data=train_c,
ci=None,
color="indigo",
hue="year",
estimator=mean,
ax=ax[1, 0],
)
ax[1, 0].set_title("Mean Count by Month hue: Year")
sns.barplot(
x="month",
y="count",
data=train_c,
ci=None,
color="indigo",
hue="weather",
estimator=mean,
ax=ax[1, 1],
)
ax[1, 1].set_title("Mean Count by Season hue: Weather")
plt.tight_layout()
# Preliminary observation:
# * We can see a big shift up from 2011 to 2012
# * The very bad weather is associated with January, and yet this is associated with Spring
# (The classification of the winter months as Spring is interesting).
# * We can see that the months / seasons with worse weather indicators have a lower count of rides
# **Humidity, Temperature:**
fig, ax = plt.subplots(nrows=1, ncols=4, figsize=(16, 5))
sns.distplot(train_c["windspeed"], ax=ax[0])
ax[0].set_title("Distplot windspeed")
sns.distplot(train_c["temp"], ax=ax[1])
ax[1].set_title("Distplot temperature")
sns.distplot(train_c["atemp"], ax=ax[2])
ax[2].set_title("Distplot atemperature")
sns.distplot(train_c["humidity"], ax=ax[3])
ax[3].set_title("Distplot humidity")
plt.tight_layout()
# Comments:
# * For atemp('feels like temperature') we can see some spikes around the 30 celcius mark;
# * For temp we can see spikes around the 16 degrees marks
# * Other than that the two displots for temperature would show a *relatively* normal distribution;
# * For windspeed we would see a normal distribution except for the spike at 0 which seems to indicate to be an outlier; let's look into these distributions by looking at their outliers a bit closer.
fig, ax = plt.subplots(nrows=4, ncols=1, figsize=(12, 12))
sns.boxplot(
x="season", y="windspeed", hue="weather", data=train_c, palette="winter", ax=ax[0]
)
ax[0].set_title("Boxplot Wincdspeed by Season: Hue Weather")
sns.boxplot(
x="season", y="temp", hue="weather", data=train_c, palette="winter", ax=ax[1]
)
ax[1].set_title("Boxplot Temperature by Season: Hue Weather")
sns.boxplot(
x="season", y="atemp", hue="weather", data=train_c, palette="winter", ax=ax[2]
)
ax[2].set_title("Boxplot ATemperature by Season: Hue Weather")
sns.boxplot(
x="season", y="humidity", hue="weather", data=train_c, palette="winter", ax=ax[3]
)
ax[3].set_title("Boxplot Humidity by Season: Hue Weather")
plt.tight_layout()
# *Comments:*
# - What we can see here is that out of whisker bounds instances tend to be the lower end for humidity are fall under the occasions of bad weather; particularly prevalent are the Summer and Fall seasons;
# - Winter, for temp atemp and humidity seems to have the least amount of outliers;
# - For temperature we see highest amount of outliers with Fall season for good weather; same with atemp;
# **Day of week and times:**
fig, ax = plt.subplots(1, figsize=(12, 8))
grouped_hours = pd.DataFrame(
train_c.groupby(["hour"], sort=True)["casual", "registered", "count"].mean()
)
grouped_hours.plot(ax=ax)
ax.set_xticks(grouped_hours.index.to_list())
ax.set_xticklabels(grouped_hours.index)
plt.xticks(rotation=45)
plt.title("Avg Count by Hour")
# *Preliminary observations:*
# - We can see that registered users follow commuter patterns, whilst casual users do not - they have higher peaks during the afternoon (potentially from weekend use);
# **Let's look at by day of week:**
fig, ax = plt.subplots(1, figsize=(12, 8))
sns.barplot(
x="weekday", y="count", data=train_c, ci=None, color="indigo", estimator=mean, ax=ax
)
ax.set_title("Avg Count by Weekday")
# * Similar usage overall indicating that higher usage during weekends will compensate for commuter usage during weekdays
# **Can we see a commuter trend?**
fig, ax = plt.subplots(nrows=1, ncols=2, figsize=(15, 8))
workingday = train_c.loc[train_c.workingday == "Yes"]
not_workingday = train_c.loc[train_c.workingday == "No"]
grouped_workingday = pd.DataFrame(
workingday.groupby(["hour"], sort=True)["count"].mean()
)
grouped_notworkingday = pd.DataFrame(
not_workingday.groupby(["hour"], sort=True)["count"].mean()
)
grouped_workingday.plot(ax=ax[0])
ax[0].set_xticks(grouped_workingday.index.to_list())
ax[0].set_xticklabels(grouped_workingday.index)
ax[0].tick_params(labelrotation=45)
ax[0].set_title("Avg Count by Hour - Working Day")
grouped_notworkingday.plot(ax=ax[1])
ax[1].set_xticks(grouped_notworkingday.index.to_list())
ax[1].set_xticklabels(grouped_notworkingday.index)
ax[1].tick_params(labelrotation=45)
ax[1].set_title("Avg Count by Hour - Not Working Day")
# *Preliminary observations:*
# - We can see that the different patterns are very clear here: commuter for working days and leisure for non working days meaning that most usage is during the aft (esp. since weekend)
# ## Part III: Any Outliers?
sns.set(style="ticks")
sns.pairplot(
data=train_c, y_vars=["count"], x_vars=["temp", "atemp", "humidity", "windspeed"]
)
sns.pairplot(
data=train_c,
y_vars=["registered"],
x_vars=["temp", "atemp", "humidity", "windspeed"],
)
sns.pairplot(
data=train_c, y_vars=["casual"], x_vars=["temp", "atemp", "humidity", "windspeed"]
)
# In general we can see that with the casual users temperature seems to be a bigger driver than registered ones.
# **Creating a train without outliers train set:**
Q1 = train.quantile(0.25)
Q3 = train.quantile(0.75)
IQR = Q3 - Q1
train = train.drop(["datetime"], axis=1)
train_without_outliers = train[
~((train < (Q1 - 1.5 * IQR)) | (train > (Q3 + 1.5 * IQR))).any(axis=1)
]
print("train original shape", train.shape[0])
print("train_without_outliers observations", train_without_outliers.shape[0])
# Now let's review some of the outliers we saw with our boxplots in the previous analysis of temperature, windspeed and humidity:
fig, ax = plt.subplots(nrows=4, ncols=2, figsize=(12, 12))
sns.boxplot(x="season", y="windspeed", data=train, palette="winter", ax=ax[0, 0])
ax[0, 0].set_title("Boxplot Wincdspeed by Season WITH OUTLIER")
sns.boxplot(
x="season",
y="windspeed",
data=train_without_outliers,
palette="winter",
ax=ax[0, 1],
)
ax[0, 1].set_title("Boxplot Wincdspeed by Season WITHOUT OUTLIER")
sns.boxplot(x="season", y="temp", data=train, palette="winter", ax=ax[1, 0])
ax[1, 0].set_title("Boxplot Temperature by Season WITH OUTLIERS")
sns.boxplot(
x="season", y="temp", data=train_without_outliers, palette="winter", ax=ax[1, 1]
)
ax[1, 1].set_title("Boxplot Temperature by Season WITHOUT OUTLIERS")
sns.boxplot(x="season", y="atemp", data=train, palette="winter", ax=ax[2, 0])
ax[2, 0].set_title("Boxplot ATemperature WITH OUTLIERS")
sns.boxplot(
x="season", y="atemp", data=train_without_outliers, palette="winter", ax=ax[2, 1]
)
ax[2, 1].set_title("Boxplot ATemperature by Season WITHOUT OUTLIERES")
sns.boxplot(x="season", y="humidity", data=train, palette="winter", ax=ax[3, 0])
ax[3, 0].set_title("Boxplot Humidity by Season WITH OUTLIERS")
sns.boxplot(
x="season", y="humidity", data=train_without_outliers, palette="winter", ax=ax[3, 1]
)
ax[3, 1].set_title("Boxplot Humidity by Season WITHOUT OUTLIERS")
plt.tight_layout()
# **Comments:**
# * Here we can see that the simple method for removing outliers has worked well wiuth windspeed;
# * Its effect on temperature and atemperature however have not been very successful;
# * This comes down to the fact that *whilst on an aggregate level of, say, windspeed we are able to remove outliers, this may not result on a more granular level (of looking at it on a season by season basis)*
# * Note: arguably, if one were to apply a removing outliers method, it would make sense to do this on a season by season basis if we are looking at variables such as weather;
# * We will therefore go ahead with the regression analysis without the non-outlier set first, but then consider it later to see if it makes any great change.
# **How to deal with outliers?**
# https://scikit-learn.org/stable/auto_examples/preprocessing/plot_all_scaling.html
# Summary of scalers:
# from sklearn.preprocessing import MinMaxScaler
# from sklearn.preprocessing import minmax_scale
# from sklearn.preprocessing import MaxAbsScaler
# from sklearn.preprocessing import StandardScaler
# from sklearn.preprocessing import RobustScaler
# from sklearn.preprocessing import Normalizer
# from sklearn.preprocessing import QuantileTransformer
# from sklearn.preprocessing import PowerTransformer
# **MinMaxScaler:**
# - Rescales the data set such that all feature values are in the range [0, 1]
# - MinMaxScaler is very sensitive to the presence of outliers.
# **MaxAbScaler:**
# - Differs from the previous scaler such that the absolute values are mapped in the range [0, 1]. On positive only data, this scaler behaves similarly to MinMaxScaler and therefore also suffers from the presence of large outliers.
# **RobustScaler:**
# - The centering and scaling statistics of this scaler are based on percentiles and are therefore not influenced by a few number of very large marginal outliers.
# **PowerTransformer:**
# - PowerTransformer applies a power transformation to each feature to make the data more Gaussian-like
# - Currently, PowerTransformer implements the Yeo-Johnson and Box-Cox transforms.
# - The power transform finds the optimal scaling factor to stabilize variance and mimimize skewness through maximum likelihood estimation.
# - By default, PowerTransformer also applies zero-mean, unit variance normalization to the transformed output. Note that Box-Cox can only be applied to strictly positive data. Income and number of households happen to be strictly positive, but if negative values are present the Yeo-Johnson transformed is to be preferred.
# **QuantileTransformer:**
# - has an additional output_distribution parameter allowing to match a Gaussian distribution instead of a uniform distribution. Note that this non-parametetric transformer introduces saturation artifacts for extreme values.
# **QuantileTransformer (uniform output):**
# - QuantileTransformer applies a non-linear transformation such that the probability density function of each feature will be mapped to a uniform distribution. In this case, all the data will be mapped in the range [0, 1], even the outliers which cannot be distinguished anymore from the inliers.
# - As RobustScaler, QuantileTransformer is robust to outliers in the sense that adding or removing outliers in the training set will yield approximately the same transformation on held out data. But contrary to RobustScaler, QuantileTransformer will also automatically collapse any outlier by setting them to the a priori defined range boundaries (0 and 1).
# **Normalizer:**
# - The Normalizer rescales the vector for each sample to have unit norm, independently of the distribution of the samples.
# For this project, given the presence of outliers, we will consider the use of RobustScaler().
# ### Conclusions from overall EDA:
# - Casual users tend to be non working day users
# - Non working day users do not use it for commuter times, rather usage is high in the early afternoon not commuter hours
# - Weather is an important factor for usage but plays a stronger role on casual users
# - Usage by weekday stays roughly the same
# - It changes though by season wiht Spring - which has the most intense weather - reporting to be the season with the least rides
# ## Part IV: Correlations
train.corr()
mask = np.array(train.corr())
mask[np.tril_indices_from(mask)] = False
fig, ax = plt.subplots()
fig.set_size_inches(30, 15)
sns.heatmap(
train.corr(),
mask=mask,
vmax=0.8,
square=True,
annot=True,
center=0,
cmap="RdBu_r",
linewidths=0.5,
)
# * For casual and registered: weather seems to be the most contributing factor
# * With casual we also see strong associations with working day (negative)
# ### Identifying the Most Important Factor:
from sklearn.ensemble import RandomForestRegressor
X = train.drop(["count", "casual", "registered"], axis=1)
y = train["count"]
X_train, X_test, Y_train, Y_test = train_test_split(X, y, test_size=0.3, random_state=5)
rf = RandomForestRegressor(n_estimators=100, random_state=2)
rf.fit(X_train, Y_train)
# **Graphical representation of most important factors:**
import matplotlib as mp
plt.subplots(figsize=(15, 10))
core_variables = pd.Series(rf.feature_importances_, index=X.columns)
core_variables = core_variables.nlargest(8)
# Colorize the graph based on likeability:
likeability_scores = np.array(core_variables)
data_normalizer = mp.colors.Normalize()
color_map = mp.colors.LinearSegmentedColormap(
"my_map",
{
"red": [(0, 1.0, 1.0), (1.0, 0.5, 0.5)],
"green": [(0, 0.5, 0.5), (1.0, 0, 0)],
"blue": [(0, 0.50, 0.5), (1.0, 0, 0)],
},
)
plt.title("Most Important Features")
# make the plot
core_variables.plot(kind="barh", color=color_map(data_normalizer(likeability_scores)))
# **Now let's do it with the without outliers dataset:**
continuous_features = ["temp", "atemp", "humidity", "windspeed"]
data = [train_without_outliers]
for dataset in data:
for col in continuous_features:
transf = dataset[col].values.reshape(-1, 1)
scaler = preprocessing.StandardScaler().fit(transf)
dataset[col] = scaler.transform(transf)
train_without_outliers.reset_index()
X = train_without_outliers.drop(["count", "casual", "registered"], axis=1)
y = train_without_outliers["count"]
X_train, X_test, Y_train, Y_test = train_test_split(X, y, test_size=0.3, random_state=5)
rf_without_outliers = RandomForestRegressor(n_estimators=100, random_state=2)
rf_without_outliers.fit(X_train, Y_train)
# **Feature Importance Without Outliers:**
plt.subplots(figsize=(15, 10))
core_variables_without_outliers = pd.Series(
rf_without_outliers.feature_importances_, index=X.columns
)
core_variables_without_outliers = core_variables_without_outliers.nlargest(8)
# Colorize the graph based on likeability:
likeability_scores = np.array(core_variables)
data_normalizer = mp.colors.Normalize()
color_map = mp.colors.LinearSegmentedColormap(
"my_map",
{
"red": [(0, 1.0, 1.0), (1.0, 0.5, 0.5)],
"green": [(0, 0.5, 0.5), (1.0, 0, 0)],
"blue": [(0, 0.50, 0.5), (1.0, 0, 0)],
},
)
# make the plot
core_variables_without_outliers.plot(
kind="barh", color=color_map(data_normalizer(likeability_scores))
)
# **Now let's compare the two plots:**
fig, ax = plt.subplots(nrows=1, ncols=2, figsize=(20, 10))
core_variables.plot(
kind="barh", color=color_map(data_normalizer(likeability_scores)), ax=ax[0]
)
ax[0].set_title("With outliers significance plot")
core_variables_without_outliers.plot(
kind="barh", color=color_map(data_normalizer(likeability_scores)), ax=ax[1]
)
ax[1].set_title("Without outliers significance plot")
# **Conclusion:**
# * With this random forest regressor feature importance we can see that with or without the outliers trai dataset the most import feature by far is *time*.
# * Notable change would be the difference between weekday and working day in importance - with the latter being more important for the original dataset.
# * For this reason we will continue with the normal, complete dataset; however, in our training below we will select only the most important variables.
# ### Random Forest Regressor: Applying Standard Scalers
# **Using Robust Scaler:**
from sklearn.preprocessing import RobustScaler
X = train.drop(["count", "casual", "registered"], axis=1)
y = train["count"]
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=2)
transformer = RobustScaler().fit(X_train)
rescaled_X_train = transformer.transform(X_train)
transformer = RobustScaler().fit(X_test)
rescaled_X_test = transformer.transform(X_test)
y_train = y_train.values.reshape(-1, 1)
y_test = y_test.values.reshape(-1, 1)
transformer = RobustScaler().fit(y_train)
rescaled_y_train = transformer.transform(y_train)
transformer = RobustScaler().fit(y_test)
rescaled_y_test = transformer.transform(y_test)
rf = RandomForestRegressor(n_estimators=100)
rf.fit(rescaled_X_train, rescaled_y_train)
from sklearn.metrics import mean_squared_error
from sklearn import metrics
rf_prediction = rf.predict(rescaled_X_test)
print("MSE:", metrics.mean_squared_error(rescaled_y_test, rf_prediction))
plt.scatter(rescaled_y_test, rf_prediction)
# **Using MinMax Scaler:**
from sklearn.preprocessing import MinMaxScaler
train = train.drop(["datetime"], axis=1)
X = train.drop(["count", "casual", "registered"], axis=1)
y = train["count"]
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=2)
y_train = y_train.values.reshape(-1, 1)
y_test = y_test.values.reshape(-1, 1)
sc_X = MinMaxScaler()
sc_y = MinMaxScaler()
X_train = sc_X.fit_transform(X_train)
X_test = sc_X.fit_transform(X_test)
y_train = sc_y.fit_transform(y_train)
y_test = sc_y.fit_transform(y_test)
from sklearn.ensemble import RandomForestRegressor
rf = RandomForestRegressor(n_estimators=100)
rf.fit(X_train, y_train)
rf_prediction = rf.predict(X_test)
print("MSE:", metrics.mean_squared_error(y_test, rf_prediction))
plt.scatter(y_test, rf_prediction)
# **Using Standard Scaler**:
from sklearn.preprocessing import StandardScaler
train.head(2)
continuous_features = ["temp", "atemp", "humidity", "windspeed", "count"]
train_copy = train.copy()
for col in continuous_features:
transf = train_copy[col].values.reshape(-1, 1)
scaler = preprocessing.StandardScaler().fit(transf)
train_copy[col] = scaler.transform(transf)
X = train_copy.drop(["count", "casual", "registered"], axis=1)
y = train_copy["count"]
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=2)
transformer = StandardScaler().fit(X_train)
standard_X_train = transformer.transform(X_train)
transformer = StandardScaler().fit(X_test)
standard_X_test = transformer.transform(X_test)
y_train = y_train.values.reshape(-1, 1)
y_test = y_test.values.reshape(-1, 1)
transformer = StandardScaler().fit(y_train)
standard_y_train = transformer.transform(y_train)
transformer = StandardScaler().fit(y_test)
standard_y_test = transformer.transform(y_test)
rf = RandomForestRegressor(n_estimators=100)
rf.fit(X_train, y_train)
rf_prediction = rf.predict(standard_X_test)
print("MSE:", metrics.mean_squared_error(standard_y_test, rf_prediction))
plt.scatter(standard_y_test, rf_prediction)
# ## Submission 1:
# For this, therefore, we will use the MinMax() as that performed the best in terms of MSE and the shape of the scatter plot. However, let's try this time using the without outliers dataset:
X = train_without_outliers[
[
"season",
"holiday",
"workingday",
"weather",
"temp",
"atemp",
"humidity",
"year",
"month",
"day",
"hour",
"weekday",
"windspeed",
]
]
y = train_without_outliers["count"]
# Let's also decrease the test size:
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.1)
y_train = y_train.values.reshape(-1, 1)
y_test = y_test.values.reshape(-1, 1)
sc_X = MinMaxScaler()
sc_y = MinMaxScaler()
X_train = sc_X.fit_transform(X_train)
X_test = sc_X.fit_transform(X_test)
y_train = sc_X.fit_transform(y_train)
y_test = sc_y.fit_transform(y_test)
rf = RandomForestRegressor(n_estimators=100)
rf.fit(X_train, y_train)
rf_prediction = rf.predict(X_test)
print("MSE:", metrics.mean_squared_error(y_test, rf_prediction))
test[
[
"season",
"holiday",
"workingday",
"weather",
"temp",
"atemp",
"humidity",
"year",
"month",
"day",
"hour",
"weekday",
"windspeed",
]
] = sc_X.fit_transform(
test[
[
"season",
"holiday",
"workingday",
"weather",
"temp",
"atemp",
"humidity",
"year",
"month",
"day",
"hour",
"weekday",
"windspeed",
]
]
)
test_pred = rf.predict(
test[
[
"season",
"holiday",
"workingday",
"weather",
"temp",
"atemp",
"humidity",
"year",
"month",
"day",
"hour",
"weekday",
"windspeed",
]
]
)
test_pred = test_pred.reshape(-1, 1)
test_pred.shape
test_pred
test_pred = sc_y.inverse_transform(test_pred)
test_pred
test_pred = pd.DataFrame(test_pred, columns=["count"])
submission1 = pd.concat([test["datetime"], test_pred], axis=1)
submission1.head()
submission1.dtypes
submission1["count"] = submission1["count"].astype("int")
submission1.to_csv("submission1.csv", index=False)
# **Score:**
# Score: (private leaderboard for now): 0.49759
# ## Submission 1:
X = train_without_outliers[
[
"season",
"holiday",
"workingday",
"weather",
"temp",
"atemp",
"humidity",
"year",
"month",
"day",
"hour",
"weekday",
"windspeed",
]
]
y = train_without_outliers["count"]
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.1)
rescaled_X_train = RobustScaler().fit_transform(X_train)
rescaled_X_test = RobustScaler().fit_transform(X_test)
y_train = y_train.values.reshape(-1, 1)
y_test = y_test.values.reshape(-1, 1)
rescaled_y_train = RobustScaler().fit_transform(y_train)
rescaled_y_test = RobustScaler().fit_transform(y_test)
rf = RandomForestRegressor(n_estimators=100)
rf.fit(rescaled_X_train, rescaled_y_train)
rf_prediction = rf.predict(rescaled_X_test)
test[
[
"season",
"holiday",
"workingday",
"weather",
"temp",
"atemp",
"humidity",
"year",
"month",
"day",
"hour",
"weekday",
"windspeed",
]
] = sc_X.fit_transform(
test[
[
"season",
"holiday",
"workingday",
"weather",
"temp",
"atemp",
"humidity",
"year",
"month",
"day",
"hour",
"weekday",
"windspeed",
]
]
)
test_pred = rf.predict(
test[
[
"season",
"holiday",
"workingday",
"weather",
"temp",
"atemp",
"humidity",
"year",
"month",
"day",
"hour",
"weekday",
"windspeed",
]
]
)
test_pred = test_pred.reshape(-1, 1)
test_pred.shape
test_pred = transformer.inverse_transform(test_pred)
test_pred
|
# 
# # # IMport
import warnings
warnings.filterwarnings("ignore")
import os
import numpy as np
import pandas as pd
import json
import plotly.express as px
import seaborn as sns
import matplotlib.pyplot as plt
import matplotlib as mpl
from tqdm.notebook import tqdm
from pathlib import Path
class Cfg:
INPUT_ROOT = Path("/kaggle/input/fathomnet-out-of-sample-detection")
OUTPUT_ROOT = Path("/kaggle/working/")
TRAIN_OUTPUT_IMAGE_ROOT = Path("/kaggle/working/images/train")
EVAL_OUTPUT_IMAGE_ROOT = Path("/kaggle/working/images/eval")
TRAIN_FILE = INPUT_ROOT / "multilabel_classification/train.csv"
CATEGORY_KEY_FILE = INPUT_ROOT / "category_key.csv"
SAMPLE_SUBMISSION_FILE = INPUT_ROOT / "sample_submission.csv"
EVAL_JSON_FILE = INPUT_ROOT / "object_detection/eval.json"
TRAIN_JSON_FILE = INPUT_ROOT / "object_detection/train.json"
ANNOTATION_FILE = OUTPUT_ROOT / "annotation.csv"
TRAIN_IMAGE_DATA_FILE = OUTPUT_ROOT / "train_image_data.csv"
EVAL_IMAGE_DATA_FILE = OUTPUT_ROOT / "eval_image_data.csv"
INDEX = "id"
def read_train_data(file=Cfg.TRAIN_FILE, index_col=Cfg.INDEX):
return pd.read_csv(file).set_index(Cfg.INDEX)
def read_category_keys(file=Cfg.CATEGORY_KEY_FILE, index_col=Cfg.INDEX):
return pd.read_csv(file).set_index(Cfg.INDEX)
def read_sample_submission(file=Cfg.SAMPLE_SUBMISSION_FILE, index_col=Cfg.INDEX):
return pd.read_csv(file).set_index(Cfg.INDEX)
def read_json(file):
"""Read a json file."""
f = open(file)
data = json.load(f)
f.close()
return data
def read_object_detection_train():
return read_json(Cfg.TRAIN_JSON_FILE)
def read_object_detection_eval():
return read_json(Cfg.EVAL_JSON_FILE)
train_data = read_train_data()
sample_submission = read_sample_submission()
category_keys = read_category_keys()
category_keys["all_items"] = "items"
f = open("../input/fathomnet-out-of-sample-detection/object_detection/train.json")
obj_detect_train = json.load(f)
f = open("../input/fathomnet-out-of-sample-detection/object_detection/eval.json")
obj_detect_eval = json.load(f)
del (
obj_detect_train["info"],
obj_detect_train["licenses"],
obj_detect_train["categories"],
)
del obj_detect_eval["info"], obj_detect_eval["licenses"], obj_detect_eval["categories"]
assert obj_detect_train.keys() == obj_detect_eval.keys()
fig = px.sunburst(
category_keys[category_keys.all_items == "items"],
path=["all_items", "supercat", "name"],
color_continuous_scale="ice",
)
fig.update_layout(height=600, margin_r=0)
fig.show()
# # # Exploring Data
def get_info(json_data):
return pd.json_normalize(json_data["info"])
def get_licenses(json_data):
return pd.json_normalize(json_data["licenses"])
def get_image_data(json_data):
df = pd.json_normalize(json_data["images"])
return df.set_index(Cfg.INDEX)
def get_annotations(json_data):
df = pd.json_normalize(json_data["annotations"])
return df.set_index(Cfg.INDEX)
def get_categories(json_data):
df = pd.json_normalize(json_data["categories"])
return df.set_index(Cfg.INDEX)
# # # # Train Data
train_data["num_categories"] = train_data["categories"].apply(
lambda x: len(json.loads(x))
)
fig, ax = plt.subplots(nrows=1, ncols=1, figsize=(8, 4))
sns.countplot(
data=train_data,
x="num_categories",
color="g",
alpha=0.8,
ax=ax,
edgecolor="b",
linestyle=":",
linewidth=1.5,
)
ax.set_title("Distribution of the number of categories per image")
plt.tight_layout()
plt.show()
newdata = (
category_keys.groupby("supercat")
.size()
.rename_axis("supercat")
.reset_index(name="Count")
)
sns.barplot(
x=newdata.supercat,
y=newdata.Count,
color="r",
edgecolor="k",
linestyle=":",
linewidth=1.5,
alpha=0.8,
)
plt.xticks(rotation=90)
plt.show()
category_count = pd.DataFrame(category_keys["supercat"].value_counts()).reset_index()
category_count.columns = ["supercat", "count"]
category_count["%"] = np.round(category_count["count"] / len(category_keys), 4) * 100
category_count.set_index("supercat", inplace=True)
fig, ax = plt.subplots(nrows=1, ncols=1, figsize=(12, 5))
df = category_count["count"]
sns.barplot(
x=df.values,
y=df.index,
color="y",
alpha=0.8,
ax=ax,
edgecolor="r",
linestyle=":",
linewidth=1.5,
)
ax.set_title("Distribution in Category Key Table (supercat)")
plt.tight_layout()
plt.show()
object_detection_train = read_object_detection_train()
display(get_info(object_detection_train))
display(get_licenses(object_detection_train))
# # # Train Image Data
train_image_data = get_image_data(object_detection_train)
display(train_image_data)
annotation_data = get_annotations(object_detection_train)
display(annotation_data)
plt.figure(figsize=(7, 3))
np.log10(annotation_data.area[annotation_data.area != 0]).plot(
kind="hist", bins=100, color="r"
)
plt.title("area - log_10 scale")
plt.grid()
plt.show()
train_image_data["num_objects"] = annotation_data.groupby(by="image_id").count()[
"category_id"
]
fig, ax = plt.subplots(nrows=1, ncols=1, figsize=(12, 4))
df = train_image_data[["num_objects"]].head(25)
sns.countplot(
data=train_image_data, x="num_objects", palette=blues_palette, alpha=0.8, ax=ax
)
ax.set_title("Number of Objects per Image")
ax.set_xlim(-1, 20)
plt.tight_layout()
plt.show()
# # # Categories
newdata = (
categories.groupby("supercategory")
.size()
.rename_axis("supercategory")
.reset_index(name="Count")
)
newdata.head()
labels = newdata.supercategory
values = newdata.Count
plt.figure(figsize=(5, 5))
sns.set()
my_circle = plt.Circle((0, 0), 0.7, color="white")
plt.rcParams["text.color"] = "red"
plt.pie(values, labels=labels, wedgeprops={"linewidth": 7, "edgecolor": "white"})
# plt.legend(loc='upper right')
p = plt.gcf()
p.gca().add_artist(my_circle)
plt.show()
# # # # Eval Image Data
object_detection_eval = read_object_detection_eval()
eval_image_data = get_image_data(object_detection_eval)
display(eval_image_data)
sns.displot(eval_image_data.height, kde=True)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import matplotlib.pyplot as plt
import seaborn as sns
df = pd.read_csv("/kaggle/input/bangladesh-weather-dataset/Temp_and_rain.csv")
df.info()
# # ***Monthly Average Rainfall***
df.groupby("Month").rain.mean()
plt.figure(figsize=(12, 5))
sns.barplot(x="Month", y="rain", data=df)
# ****July is the highest Rainfall Month ****
# # Monthly Average temperature
df.groupby("Month").tem.mean().sort_values()
plt.figure(figsize=(12, 5))
sns.barplot(x="Month", y="tem", data=df)
df
# # Average Rainfall in per Year
r_df = df.groupby("Year").rain.mean()
r_df
plt.figure(figsize=(16, 6))
sns.lineplot(x="Year", y="tem", data=df, marker="o")
print("It seen a upward trend in average temperature with the changes of time")
df
|
import os
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
pd.set_option("max_columns", None)
import matplotlib.pyplot as plt
import seaborn as sns
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
data_2019 = pd.read_csv("/kaggle/input/flight-delay-prediction/Jan_2019_ontime.csv")
data_2020 = pd.read_csv("/kaggle/input/flight-delay-prediction/Jan_2020_ontime.csv")
print(data_2019.info(), data_2020.info())
# Join both datasets 2019 and 2020
data = pd.concat([data_2019, data_2020])
data.head()
data.shape
# rename the categories in categorical columns
data["DEP_DEL15"] = np.where(data["DEP_DEL15"] == 0.0, "NO", "YES")
data["CANCELLED"] = np.where(data["CANCELLED"] == 0.0, "NO", "YES")
data["DIVERTED"] = np.where(data["DIVERTED"] == 0.0, "NO", "YES")
data["ARR_DEL15"] = np.where(data["ARR_DEL15"] == 0.0, "NO", "YES")
# Since there is many categories in the ORIGIN and DEST column, I combined them to a single column and extracted the 50 most used routes.
# Combine ORIGIN and DEST into a single column
data["ORIGIN-DEST"] = data["ORIGIN"] + "-" + data["DEST"]
# get the count of each combination into a dataframe
org_dest = data["ORIGIN-DEST"].value_counts().to_frame()
# check the number of observation in the most frequent 50 to check whether the sample size is enough for the analysis
org_dest[:50]["ORIGIN-DEST"].sum()
# extract the data from original dataframe
org_dest_list = org_dest[:50].index.tolist()
data = data[data["ORIGIN-DEST"].isin(org_dest_list)]
# Distance variable is categorized into three to simplify the analysis.
print(
"max distance: ",
data["DISTANCE"].max(),
"\n",
"min distance: ",
data["DISTANCE"].min(),
)
data["DIST_GROUP"] = "SHORT"
data.loc[
(data["DISTANCE"] > 928.0) & (data["DISTANCE"] <= 1757.0), "DIST_GROUP"
] = "MEDIUM"
data.loc[(data["DISTANCE"] > 1757.0), "DIST_GROUP"] = "LONG"
# extract the necessary columns
data = data[
[
"DAY_OF_MONTH",
"DAY_OF_WEEK",
"OP_UNIQUE_CARRIER",
"ORIGIN-DEST",
"DEP_DEL15",
"CANCELLED",
"DIVERTED",
"DIST_GROUP",
"ARR_DEL15",
]
]
data = data.reset_index().drop("index", axis=1)
data.shape
# check for missing values
data.isnull().sum()
data.head()
# # Exploratory Analysis
var = ["DEP_DEL15", "CANCELLED", "DIVERTED"]
fig, axes = plt.subplots(nrows=1, ncols=3, figsize=(20, 10))
for k, ax in zip(range(3), axes.flatten()):
sns.countplot(data=data, x=f"{var[k]}", hue="ARR_DEL15", ax=ax)
ax.set_title(f"Arrival delay vs {var[k]}")
for container in ax.containers:
ax.bar_label(container)
fig, ax = plt.subplots(figsize=(20, 5))
sns.countplot(
data=data,
x="ORIGIN-DEST",
hue="ARR_DEL15",
)
plt.title("Arrival delay with ORIGIN-DESTINATION")
plt.xticks(rotation=45, ha="right")
fig, ax = plt.subplots(figsize=(20, 5))
sns.countplot(
data=data,
x="OP_UNIQUE_CARRIER",
hue="ARR_DEL15",
)
plt.title("Arrival delay with OP_UNIQUE_CARRIER")
fig, ax = plt.subplots(figsize=(20, 10))
sns.countplot(
data=data,
x="DIST_GROUP",
hue="ARR_DEL15",
)
plt.title("Arrival delay with DISTANCE GROUP")
for container in ax.containers:
ax.bar_label(container)
sns.histplot(data=data, x="DAY_OF_WEEK", hue="ARR_DEL15", multiple="dodge", shrink=6)
sns.histplot(data=data, x="DAY_OF_MONTH", hue="ARR_DEL15", multiple="dodge", shrink=0.8)
sns.set(rc={"figure.figsize": (5, 5)})
sns.set_style(rc={"axes.facecolor": "#FFFFFF"})
# plt.figure(figsize=(20,5))
# df.corr()['Churn'].sort_values(ascending = False).plot(kind='bar')
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# ### load the required libraries
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import email
import warnings
warnings.filterwarnings("ignore")
import parser
# read the dataset
df = pd.read_csv("/kaggle/input/enron-email-dataset/emails.csv")
# ## EDA
# display first 5 rows of the dataset using head function
df.head()
# check the shape
df.shape
# get message value
print(df.loc[1]["file"])
print("\n")
print(df.loc[1]["message"])
# transform the email into correct form
message = df.loc[1]["message"]
emails = email.message_from_string(message)
emails.items()
# get email body
emails.get_payload()
emails.get("Date")
# feature we are extract from the email
# 1. date 2. X-From 3. X-To 4. Subject 5. X-Folder
# extract feature from the email for all datasets
def extract_data(feature, df):
column = []
for row in df:
e = email.message_from_string(row)
column.append(e.get(feature))
return column
df["Date"] = extract_data("Date", df["message"])
df["Subject"] = extract_data("Subject", df["message"])
df["X-From"] = extract_data("X-From", df["message"])
df["X-To"] = extract_data("X-To", df["message"])
df["X-Folder"] = extract_data("X-Folder", df["message"])
# extract email body from email message
def get_email_body(data):
column = []
for msg in data:
e = email.message_from_string(msg)
column.append(e.get_payload())
return column
df["body"] = get_email_body(df["message"])
df.head()
# Employee names
def emp_name(data):
column = []
for msg in data:
column.append(msg.split("/")[0])
return column
df["Employee"] = emp_name(df["file"])
df.head(3)
df["Date"]
# find the unique folder
print("number of folder :", df.shape[0])
print("number of unique folder :", df["X-Folder"].unique().shape[0])
unique_emails = pd.DataFrame(df["X-Folder"].value_counts())
unique_emails.reset_index(inplace=True)
# show top 20 folder highest counts
unique_emails.columns = ["Folder_name", "Count"]
unique_emails.iloc[:20, :]
# visualize top 20 folder name
plt.figure(figsize=(10, 6))
sns.barplot(x="Count", y="Folder_name", data=unique_emails.iloc[:20, :])
plt.title("Top 20 Folder")
plt.xlabel("Count")
plt.ylabel("Folder name ")
plt.show()
# top email sender employees
emp_data = pd.DataFrame(df["Employee"].value_counts())
emp_data.reset_index(inplace=True)
emp_data.columns = ["Employee Name", "Count"]
emp_data.iloc[:20, :]
# visualize top 20 emails sender employee
plt.figure(figsize=(10, 6))
sns.barplot(x="Count", y="Employee Name", data=emp_data.iloc[:20, :])
plt.title("Top 20 Emails Sender Emails")
plt.xlabel("Count")
plt.ylabel("Emplyree Name")
plt.show()
df.head(3)
# ### data cleaning and transformation
# Date columns
from datetime import datetime
from dateutil import parser
# this is sample example
x = parser.parse("Fri, 4 May 2001 13:51:00 -0700 (PDT)")
print(x.strftime("%d-%m-%Y %H:%M:%S"))
def change_date_type(data):
column = []
for date in data:
column.append(parser.parse(date).strftime("%d-%m-%Y %H:%M:%S"))
return column
df["Date"] = change_date_type(df["Date"])
# column X-Folder
x_value = df.loc[1, "X-Folder"]
# extract last folder name
folder_name = x_value.split("\\")[-1]
folder_name
def process_folder_name(folders):
column = []
for folder in folders:
if folder is None or folder == "":
column.append(np.nan)
else:
column.append(folder.split("\\")[-1].lower())
return column
df["X-Folder"] = process_folder_name(df["X-Folder"])
df.head(3)
# found unique folder
print("Lenghts of unique folder : ", len(df["X-Folder"].unique()))
# fetch some folder name
df["X-Folder"].unique()[:20]
# see subject empty string
df[df["Subject"] == ""]
# replace empty missing value in subject with np.nan
def replace_empty_with_nan(subject):
column = []
for sub in subject:
if sub == "":
column.append(np.nan)
else:
column.append(sub)
return column
df["Subject"] = replace_empty_with_nan(df["Subject"])
df["X-To"] = replace_empty_with_nan(df["X-To"])
# check missing value in the dataset
df.isnull().sum()
# calculate the missing value percentage
missing_value = df.isnull().sum()
miss = missing_value[missing_value > 0]
miss_percen = miss / df.shape[0]
miss_percen
# drop missing value rows from the dataset
df.dropna(axis=0, inplace=True)
df.isnull().sum()
df.head(3)
# now need to drop some columns which is not necessary for the model
drop_column_names = ["file", "message", "Date", "X-From", "X-To", "Employee"]
df.drop(columns=drop_column_names, axis=1, inplace=True)
df.columns
df.head()
df.to_csv("cleaned_dataset.csv", index=False)
|
# Imports
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
from sklearn.tree import DecisionTreeClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.model_selection import cross_val_score
from sklearn.neural_network import MLPClassifier
from sklearn import ensemble
from sklearn import svm
# Read in data
big_df = pd.read_csv("../input/titanic-cleaned-data/train_clean.csv")
big_df.info()
# util functions
def drop_columns(df, columns_to_drop):
for col in columns_to_drop:
del df[col]
def split_test_train(train_size, all_data):
msk = np.random.rand(len(all_data)) < train_size
train_df = all_data[msk]
test_df = all_data[~msk]
train_y = train_df["Survived"]
train_x = train_df.drop("Survived", axis=1)
test_y = test_df["Survived"]
test_x = test_df.drop("Survived", axis=1)
return (train_x, train_y, test_x, test_y)
def cross_validate(all_data, model):
depth = []
all_y = all_data["Survived"]
all_x = all_data.drop("Survived", axis=1)
for i in range(2, 10):
# Perform n-fold cross validation
scores = cross_val_score(estimator=model, X=all_x, y=all_y, cv=i, n_jobs=4)
# print("i scores for cv: ", scores)
depth.append((i, scores.mean()))
# print(depth)
return depth
def train_and_test(all_data, model):
test_scores = []
train_scores = []
for i in range(1, 10):
(train_x, train_y, test_x, test_y) = split_test_train(0.1 * i, big_df)
# print("len test: ", len(test_x), ", len train: ", len(train_x))
model.fit(train_x, train_y)
pred_test_y = model.predict(test_x) # TODO add wallclock time
test_score = round(model.score(test_x, test_y) * 100, 2)
pred_train_y = model.predict(train_x)
train_score = round(model.score(train_x, train_y) * 100, 2)
test_scores.append(test_score)
train_scores.append(train_score)
return (test_scores, train_scores)
def evaluate_model(all_data, model, model_id):
(test_scores, train_scores) = train_and_test(all_data, model)
cv_scores = cross_validate(all_data, model)
print("{0} test set scores: {1} ".format(model_id, test_scores))
print("{0} train set scores: {1}".format(model_id, train_scores))
print("{0} cross validation set scores: {1}".format(model_id, cv_scores))
# TODO come up with graphing function that takes in two arrays of test and train and plots them
columns_to_drop = [
"Cabin",
"Name",
"Ticket",
"Parch",
"Embarked",
"Title",
"PassengerId",
] # TODO include reasoning for dropping these
drop_columns(big_df, columns_to_drop)
is_male = {"male": 1, "female": 0}
big_df["Sex"].replace(is_male, inplace=True)
# Decision Tree
decision_tree = DecisionTreeClassifier()
evaluate_model(big_df, decision_tree, "decision tree")
# (tree_test_scores, tree_train_scores) = train_and_test(big_df, decision_tree)
# tree_cv_scores = cross_validate(big_df, decision_tree)
# print("decision tree test set scores: ", tree_test_scores)
# print("decision tree train set scores: ", tree_train_scores)
# print("decision tree cross validation set scores: ", tree_cv_scores)
knn_classifier = KNeighborsClassifier(n_neighbors=3)
(knn_test_scores, knn_train_scores) = train_and_test(big_df, knn_classifier)
knn_cv_scores = cross_validate(big_df, knn_classifier)
print("knn test set scores: ", knn_test_scores)
print("knn train set scores: ", knn_train_scores)
print("knn cross validation set scores: ", knn_cv_scores)
neural_net_classifier = MLPClassifier(
solver="lbfgs", alpha=1e-5, hidden_layer_sizes=(8, 6), random_state=1
)
(neural_net_test_scores, neural_net_train_scores) = train_and_test(
big_df, neural_net_classifier
)
neural_net_cv_scores = cross_validate(big_df, neural_net_classifier)
print("neural_net test set scores: ", neural_net_test_scores)
print("neural_net train set scores: ", neural_net_train_scores)
print("neural_net cross validation set scores: ", neural_net_cv_scores)
# boosting_params = {'n_estimators': 500, 'max_depth': 5, 'min_samples_split': 2,
# 'learning_rate': 0.01, 'loss': 'ls'}
# boosting_classifier = ensemble.GradientBoostingRegressor(**boosting_params)
boosting_classifier = ensemble.AdaBoostClassifier(n_estimators=100, random_state=0)
(boosting_test_scores, boosting_train_scores) = train_and_test(
big_df, boosting_classifier
)
boosting_cv_scores = cross_validate(big_df, boosting_classifier)
print("boosting test set scores: ", boosting_test_scores)
print("boosting train set scores: ", boosting_train_scores)
print("boosting cross validation set scores: ", boosting_cv_scores)
# svm_classifier = svm.SVC(kernel='linear')
# (svm_test_scores, svm_train_scores) = train_and_test(big_df, svm_classifier)
# svm_cv_scores = cross_validate(big_df, svm_classifier)
# print("svm test set scores: ", svm_test_scores)
# print("svm train set scores: ", svm_train_scores)
# print("svm cross validation set scores: ", svm_cv_scores)
rbf_svc = svm.SVC(kernel="rbf", gamma=0.013, C=1)
evaluate_model(big_df, rbf_svc, "rbf_svc")
|
# # Table of Contents
# * #### [About Data](#1)
# * #### [Import Libraries](#2)
# * #### [Load and Check Data](#3)
# * #### [Numerical Variables and Categorical Variables](#4)
# * #### [Outlier Detection](#5)
# * #### [Correlation Matrix](#6)
# * #### [Data Visualization](#7)
# * ##### [Price Range Visualization](#8)
# * ##### [Numerical Feature Visualization](#9)
# * ##### [Categorical Feature Visualization](#10)
# * #### [Feature Engineering](#11)
# * ##### [Categorical Features](#12)
# * ##### [Numerical Features](#13)
# * #### [Modeling](#14)
# * ##### [Feature Data and Price Range Values](#15)
# * ##### [Feature Selection](#16)
# * ##### [Train and Test Data](#17)
# * ##### [Model Training and Parameter Tuning](#18)
# * ##### [Model Evaluation](#19)
#
# # About Data
# Bob is the owner of a mobile company but he doesn't know how to estimate the price of the mobiles his company creates. To solve this problem, he collected sales data of mobile phones from various companies. Bob wants to find a relationship between the features of a mobile phone (such as RAM, Internal Memory, etc.) and its selling price. However, he is not proficient in machine learning, so he needs help to solve this problem. In this problem, you do not have to predict the actual price, but rather a price range that indicates how high the price is based on the given data.
#
# Feature Descriptions
#
#
# battery_power = Total energy a battery can store in one time measured in mAh.
#
#
# blue = Has bluetooth or not. | 1: has, 0: doesn't have
#
#
# clock_speed = Speed at which microprocessor executes instructions.
#
#
# dual_sim = Has dual sim support or not | 1: support, 0: doesn't support
#
#
# fc = Front Camera mega pixels.
#
#
# four_g = Has 4G or not. | 1: has , 0: doesn't have
#
#
# int_memory = Internal memory in Gigabytes.
#
#
# m_dep = Mobile Depth in cm.
#
#
# mobile_wt = Weight of mobile phone.
#
#
# n_cores = Number of cores of processor.
#
#
# pc = Primary Camera mega pixels.
#
#
# px_height = Pixel Resolution Height.
#
#
# px_width = Pixel Resolution Width.
#
#
# ram = Random Access Memory in Mega Bytes.
#
#
# sc_h = Screen Height of mobile in cm.
#
#
# sc_w = Screen Width of mobile in cm.
#
#
# talk_time = Longest time that a single battery charge will last when you are.
#
#
# three_g = Has 3G or not. | 1: has, 0: doesn't have
#
#
# touch_screen = Has touch screen or not. | 1: Has, 0:Doesnt have
#
#
# wifi = Has wifi or not. | 1: has, 0: doesn't have
#
#
# price_range = This is the target variable. | 0:Low Cost, 1:Medium Cost, 2:High Cost, 3:Very High Cost
#
#
# # Import Libraries
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
from collections import Counter
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split, GridSearchCV, StratifiedKFold
from sklearn.linear_model import LogisticRegression
from sklearn.svm import SVC
from sklearn.ensemble import RandomForestClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.metrics import (
f1_score,
accuracy_score,
plot_confusion_matrix,
plot_roc_curve,
classification_report,
)
from sklearn.feature_selection import SelectKBest
from sklearn.feature_selection import f_classif # or any other statistical test
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
#
# # Load and Check Data
# Read data from CSV file and create a dataframe
data_train = pd.read_csv("/kaggle/input/mobile-price-classification/train.csv")
# Display the first few rows of the dataframe
data_train.head()
# Display the column names in the dataframe
data_train.columns
# Display the dimensions of the dataframe
data_train.shape
# Display information about the dataframe
data_train.info()
# Check for null values in each column of dataframe
data_train.isnull().sum()
# > * The train dataset contains 21 features and 2000 entries.
# > * There is no missing value in the dataset.
#
# # Numerical Variables and Categorical Variables
# > * We have 21 variables including 20 independent variables and 1 dependent variable: price_range.
# > * We have 7 categorical variables: blue, dual_sim, four_g, three_g, touch_screen, wifi, n_cores
# > * We have 13 numeric variables: battery_power, clock_speed, fc, int_memory, m_dep, mobile_wt, pc, px_height, px_width, ram, sc_h, sc_w, talk_time
# Define lists of categorical features
categorical = [
"blue",
"dual_sim",
"four_g",
"n_cores",
"three_g",
"touch_screen",
"wifi",
]
# Convert the selected columns in the training data to string type
categorical_tr = data_train[categorical].astype(str)
# Calculate the number of unique values for each column
unique_counts = categorical_tr.nunique()
# Find the unique values in each column
unique_values = categorical_tr.apply(lambda x: x.unique())
# Create a pandas DataFrame to display the results
pd.DataFrame({"Number of Unique Values": unique_counts, "Unique Values": unique_values})
# Define lists of numerical features
numerical = [
"battery_power",
"clock_speed",
"fc",
"int_memory",
"m_dep",
"mobile_wt",
"pc",
"px_height",
"px_width",
"ram",
"sc_h",
"sc_w",
"talk_time",
]
# Calculate basic statistics for each column in dataframe
data_train[numerical].describe().T
#
# # Outlier Detection
# Function to detect outliers in dataset
def detect_outliers(df, features):
outlier_indices = [] # Initialize list of outlier indices
# Iterate over features
for i in features:
# Calculate 1st, 3rd quartiles and IQR
Q1 = np.percentile(df[i], 25)
Q3 = np.percentile(df[i], 75)
IQR = Q3 - Q1
# Compute outlier step
outlier_step = IQR * 1.5
# Determine indices of outliers in current feature
outlier_list_col = df[
(df[i] < Q1 - outlier_step) | (df[i] > Q3 + outlier_step)
].index
# Append outlier indices to list
outlier_indices.extend(outlier_list_col)
# Count number of outlier indices for each data point
outlier_indices = Counter(outlier_indices)
# Select rows with more than two outlier indices
multiple_outliers = list(k for k, v in outlier_indices.items() if v > 2)
# Return list of outlier indices
return multiple_outliers
# Use function to find outliers in dataset
outlier_indices = detect_outliers(data_train, numerical)
print("Outlier indices:", outlier_indices)
# > There are no more than two outliers in the dataset.
#
# # Correlation Matrix
# Calculate correlation matrix for dataset
corr = data_train.corr()
# Create heatmap to visualize correlation matrix
plt.figure(figsize=(20, 10))
sns.heatmap(corr, annot=True, cmap="Blues")
# Display heatmap
plt.show()
# > The correlations given indicate the degree of linear association between pairs of variables. A correlation coefficient of 0.64 between pc and fc suggests a moderate positive relationship. Similarly, a correlation coefficient of 0.58 between three_g and four_g implies a moderate positive relationship. A correlation coefficient of 0.51 between px_width and px_height, and between sc_w and sc_h suggests a moderate positive relationship between these variables. Finally, a correlation coefficient of 0.92 between price_range and ram indicates a strong positive relationship between these variables.
#
# # Data Visualization
#
# ## Price Range Visualization
# Visualize number of price range values in dataset
plt.figure(figsize=(15, 5))
# Create bar plot showing count of price range value
plt.subplot(1, 2, 1)
sns.countplot(x="price_range", data=data_train, palette="Set2")
plt.title("Number of Price Range")
# Create pie chart showing proportion of price range value
plt.subplot(1, 2, 2)
plt.pie(
data_train["price_range"].value_counts(),
labels=["low cost", "medium cost", "high cost", "very high cost"],
shadow=True,
autopct="%1.1f%%",
colors=["#B7C3F3", "#8EB897"],
)
# Display plots
plt.show()
# > Mobile phones are evenly divided into 4 price range classes with equal frequency. Therefore, the dataset is completely balanced.
#
# ## Numerical Feature Visualization
# Visualize distributions of numerical features by price range value
i = 0
plt.figure(figsize=(15, 60))
# Create two plots for each numerical feature
for numerical_feature in numerical:
i += 1
plt.subplot(13, 2, i)
# Create histogram showing distribution of feature values by price range value
sns.histplot(
x=data_train[numerical_feature], data=data_train, kde=True, hue="price_range"
)
i += 1
plt.subplot(13, 2, i)
# Create boxplot showing distribution of feature values in general
sns.boxplot(x=data_train[numerical_feature], data=data_train)
# Display plots
plt.show()
# > * It can be said that there is a balanced distribution among the features here. If we are talking about the balance between features, it is not possible to say the same thing for sc_w, px_height, and fc.
# > * It was observed through the boxplot that there are some outliers, but in order to avoid reducing the sample size unnecessarily, data trimming was applied in cases where more than two outliers were detected, and this was already discussed in the section on outlier detection.
#
# ## Categorical Feature Visualization
# Visualize distributions of categorical features by price range value
i = 0
plt.figure(figsize=(15, 25))
# Create plot for each categorical feature
for categorical_feature in categorical:
i += 1
plt.subplot(4, 2, i)
# Create countplot showing count of each category by price range value
sns.countplot(
x=data_train[categorical_feature],
data=data_train,
hue="price_range",
palette="Set2",
)
# Display plots
plt.show()
# > There is a significant difference between having or not having 3G, i.e., in the three_g class. The other classes are balanced.
#
# # Feature Engineering
#
# ## Categorical Features
# Encode categorical features using the get_dummies function
data_train = pd.get_dummies(data_train, columns=categorical)
# > We converted categorical data to binary vector representations.
#
# ## Numerical Features
# Scale numerical features using the StandardScaler function
scaler = StandardScaler()
data_train[numerical] = scaler.fit_transform(data_train[numerical])
# > The numeric values in the dataset were transformed to have a mean of 0 and a variance of 1.
#
# # Modeling
#
# ## Feature Data and Price Range Values
# Create variables for feature data and price range values
X = data_train.drop(["price_range"], axis=1) # feature data train
y = data_train["price_range"].values # price range values train
#
# ## Feature Selection
# Create a SelectKBest object with f_classif as the scoring function and k=25 as the number of top features to select
selector = SelectKBest(score_func=f_classif, k=25)
# Reshape your feature matrix (if necessary)
X_new = selector.fit_transform(X, y)
# Get the indices of the selected features
selected_features = selector.get_support(indices=True)
# Print the names of the selected features
print(X.columns[selected_features])
# Create a new DataFrame with only the selected features
X_new_df = pd.DataFrame(X_new, columns=X.columns[selected_features])
#
# ## Train and Test Data
# Split dataset into training and testing data
X_train, X_test, y_train, y_test = train_test_split(
X_new_df, y, test_size=0.3, random_state=128
) # %70 train data, %30 test data
# Print lengths of training and testing datasets for each feature and target variable
print("X_train: ", len(X_train))
print("X_test: ", len(X_test))
print("y_train: ", len(y_train))
print("y_test: ", len(y_test))
# > A test set was created from the train.csv file to check the accuracy since the price_range data is not available in the test.csv file.
#
# ## Model Training and Parameter Tuning
# Set the random seed
random_state = 128
# Define the machine learning models
model = [
LogisticRegression(random_state=random_state),
DecisionTreeClassifier(random_state=random_state),
SVC(random_state=random_state, probability=True),
RandomForestClassifier(random_state=random_state),
KNeighborsClassifier(),
GaussianNB(),
]
# Define the list of class names
model_names = [
"LogisticRegression",
"DecisionTreeClassifier",
"SVM",
"RandomForestClassifier",
"KNeighborsClassifier",
"NaiveBayes",
]
# Define the hyperparameter grids for each model
lr_params = {
"C": [0.1, 0.3, 0.5, 0.7, 0.9, 1.0, 1.5, 2],
"penalty": ["l1", "l2", "elasticnet"],
"solver": ["liblinear"],
}
dt_params = {"min_samples_split": range(1, 20, 1), "max_depth": range(1, 20, 1)}
svm_params = {
"kernel": ["rbf"],
"gamma": [0.001, 0.01, 0.1],
"C": [1, 10, 50, 100, 200],
}
rf_params = {
"n_estimators": [300, 400, 500, 600],
"max_depth": [10, 20, 30],
"min_samples_split": [2, 5, 10],
}
knn_params = {
"n_neighbors": [30, 35, 40, 45, 50, 60, 70, 80],
"weights": ["uniform", "distance"],
"metric": ["euclidean", "manhattan"],
}
nb_params = {"var_smoothing": np.logspace(0, -9, num=100)}
# Define a list of hyperparameter grids
param_grids = [lr_params, dt_params, svm_params, rf_params, knn_params, nb_params]
# Define two list to store the results and estimators
cv_result = []
best_estimators = []
# Iterate over the models and their hyperparameter grids
for i in range(len(model)):
# Define the stratified K-fold cross-validation scheme
cv = StratifiedKFold(n_splits=10)
# Define the grid search strategy
clf = GridSearchCV(
model[i],
param_grid=param_grids[i],
cv=cv,
scoring="accuracy",
n_jobs=-1,
verbose=1,
)
# Print the cross validation score, best parameters and the train and test accuracy of the model
clf.fit(X_train, y_train)
cv_result.append(clf.best_score_)
best_estimators.append(clf.best_estimator_)
print("Machine Learning Models:", model_names[i])
print("Cross Validation Scores:", cv_result[i])
print("Best Parameters:", best_estimators[i])
print("\n-----------------------------------------------------\n")
print(
"Train Accuracy of Classifier: ", accuracy_score(clf.predict(X_train), y_train)
)
print("Test Accuracy of Classifier: ", accuracy_score(clf.predict(X_test), y_test))
print("\n-----------------------------------------------------\n")
# Print the classification report and plot the ROC curve and confusion matrix
print(
"Classification Report: \n", classification_report(y_test, clf.predict(X_test))
)
print("\n-----------------------------------------------------\n")
print("Confusion Matrix: \n")
plot_confusion_matrix(clf, X_test, y_test)
plt.show()
print("\n****************************************************************\n")
#
# ## Model Evaluation
# Create dataframe with mean accuracy scores for each model
cv_results = pd.DataFrame(
{
"Cross Validation Means": cv_result,
"Model": [
"LogisticRegression",
"DecisionTreeClassifier",
"SVM",
"RandomForestClassifier",
"KNeighborsClassifier",
"NaiveBayes",
],
}
).sort_values(by="Cross Validation Means")
# Create barplot to visualize mean accuracy scores for each model
plt.figure(figsize=(12, 7))
g = sns.barplot(x="Cross Validation Means", y="Model", data=cv_results)
# Set x-axis label and plot title
g.set_xlabel("Mean Accuracy")
g.set_title("Cross Validation Scores")
# Display plot
plt.show()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
from tqdm.auto import tqdm
from glob import glob
import time, gc
import cv2
from torch import nn
import torch
from torch.utils.data import Dataset, DataLoader
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
class Model(nn.Module):
def __init__(self):
super(Model, self).__init__()
self.layer1 = nn.Sequential(
nn.Conv2d(in_channels=1, out_channels=32, kernel_size=(3, 3), padding=1),
nn.ReLU(True),
nn.Conv2d(in_channels=32, out_channels=32, kernel_size=(3, 3), padding=1),
nn.ReLU(True),
nn.Conv2d(in_channels=32, out_channels=32, kernel_size=(3, 3), padding=1),
nn.ReLU(True),
nn.Conv2d(in_channels=32, out_channels=32, kernel_size=(3, 3), padding=1),
nn.ReLU(True),
nn.BatchNorm2d(num_features=32),
nn.MaxPool2d(kernel_size=(2, 2)),
nn.Conv2d(in_channels=32, out_channels=32, kernel_size=(5, 5), padding=2),
nn.ReLU(True),
nn.Dropout2d(p=0.3),
)
self.layer2 = nn.Sequential(
nn.Conv2d(in_channels=32, out_channels=64, kernel_size=(3, 3), padding=1),
nn.ReLU(True),
nn.Conv2d(in_channels=64, out_channels=64, kernel_size=(3, 3), padding=1),
nn.ReLU(True),
nn.Conv2d(in_channels=64, out_channels=64, kernel_size=(3, 3), padding=1),
nn.ReLU(True),
nn.Conv2d(in_channels=64, out_channels=64, kernel_size=(3, 3), padding=1),
nn.BatchNorm2d(num_features=64),
nn.MaxPool2d(kernel_size=(2, 2)),
nn.Conv2d(in_channels=64, out_channels=64, kernel_size=(5, 5), padding=2),
nn.ReLU(True),
nn.Dropout2d(p=0.3),
)
self.layer3 = nn.Sequential(
nn.Conv2d(in_channels=64, out_channels=128, kernel_size=(3, 3), padding=1),
nn.ReLU(True),
nn.Conv2d(in_channels=128, out_channels=128, kernel_size=(3, 3), padding=1),
nn.ReLU(True),
nn.Conv2d(in_channels=128, out_channels=128, kernel_size=(3, 3), padding=1),
nn.ReLU(True),
nn.Conv2d(in_channels=128, out_channels=128, kernel_size=(3, 3), padding=1),
nn.ReLU(True),
nn.BatchNorm2d(num_features=128),
nn.MaxPool2d(kernel_size=(2, 2)),
nn.Conv2d(in_channels=128, out_channels=128, kernel_size=(5, 5), padding=2),
nn.ReLU(True),
nn.Dropout2d(p=0.3),
)
self.layer4 = nn.Sequential(
nn.Conv2d(in_channels=128, out_channels=256, kernel_size=(3, 3), padding=1),
nn.ReLU(True),
nn.Conv2d(in_channels=256, out_channels=256, kernel_size=(3, 3), padding=1),
nn.ReLU(True),
nn.Conv2d(in_channels=256, out_channels=256, kernel_size=(3, 3), padding=1),
nn.ReLU(True),
nn.Conv2d(in_channels=256, out_channels=256, kernel_size=(3, 3), padding=1),
nn.ReLU(True),
nn.BatchNorm2d(num_features=256),
nn.MaxPool2d(kernel_size=(2, 2)),
nn.Conv2d(in_channels=256, out_channels=256, kernel_size=(5, 5), padding=2),
nn.ReLU(True),
nn.Dropout2d(p=0.3),
)
self.flatten = nn.Flatten()
self.fc1 = nn.Linear(in_features=4096, out_features=1024)
self.fc1_dropout = nn.Dropout2d(p=0.3)
self.fc2 = nn.Linear(in_features=1024, out_features=512)
self.fc3 = nn.Linear(in_features=512, out_features=168)
self.fc4 = nn.Linear(in_features=512, out_features=11)
self.fc5 = nn.Linear(in_features=512, out_features=7)
def forward(self, X):
output = self.layer1(X)
output = self.layer2(output)
output = self.layer3(output)
output = self.layer4(output)
output = self.flatten(output)
output = self.fc1(output)
output = self.fc1_dropout(output)
output = self.fc2(output)
output_root = self.fc3(output)
output_vowel = self.fc4(output)
output_consonant = self.fc5(output)
return output_root, output_vowel, output_consonant
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
model = Model()
model = nn.DataParallel(model)
model.load_state_dict(
torch.load("/kaggle/input/40epochspt/40epochs_state_dict.pt", map_location=device)
)
model.eval()
print()
def resize(df, size=64, need_progress_bar=True):
resized = {}
resize_size = 64
if need_progress_bar:
for i in tqdm(range(df.shape[0])):
image = df.loc[df.index[i]].values.reshape(137, 236)
_, thresh = cv2.threshold(
image, 30, 255, cv2.THRESH_BINARY_INV + cv2.THRESH_OTSU
)
contours, _ = cv2.findContours(
thresh, cv2.RETR_LIST, cv2.CHAIN_APPROX_SIMPLE
)[-2:]
idx = 0
ls_xmin = []
ls_ymin = []
ls_xmax = []
ls_ymax = []
for cnt in contours:
idx += 1
x, y, w, h = cv2.boundingRect(cnt)
ls_xmin.append(x)
ls_ymin.append(y)
ls_xmax.append(x + w)
ls_ymax.append(y + h)
xmin = min(ls_xmin)
ymin = min(ls_ymin)
xmax = max(ls_xmax)
ymax = max(ls_ymax)
roi = image[ymin:ymax, xmin:xmax]
resized_roi = cv2.resize(
roi, (resize_size, resize_size), interpolation=cv2.INTER_AREA
)
resized[df.index[i]] = resized_roi.reshape(-1)
else:
for i in range(df.shape[0]):
# image = cv2.resize(df.loc[df.index[i]].values.reshape(137,236),(size,size),None,fx=0.5,fy=0.5,interpolation=cv2.INTER_AREA)
image = df.loc[df.index[i]].values.reshape(137, 236)
_, thresh = cv2.threshold(
image, 30, 255, cv2.THRESH_BINARY_INV + cv2.THRESH_OTSU
)
contours, _ = cv2.findContours(
thresh, cv2.RETR_LIST, cv2.CHAIN_APPROX_SIMPLE
)[-2:]
idx = 0
ls_xmin = []
ls_ymin = []
ls_xmax = []
ls_ymax = []
for cnt in contours:
idx += 1
x, y, w, h = cv2.boundingRect(cnt)
ls_xmin.append(x)
ls_ymin.append(y)
ls_xmax.append(x + w)
ls_ymax.append(y + h)
xmin = min(ls_xmin)
ymin = min(ls_ymin)
xmax = max(ls_xmax)
ymax = max(ls_ymax)
roi = image[ymin:ymax, xmin:xmax]
resized_roi = cv2.resize(
roi, (resize_size, resize_size), interpolation=cv2.INTER_AREA
)
resized[df.index[i]] = resized_roi.reshape(-1)
resized = pd.DataFrame(resized).T
return resized
IMG_SIZE = 64
N_CHANNELS = 1
class GraphemeDataset(Dataset):
def __init__(self, df, _type="train"):
self.df = df
def __len__(self):
return len(self.df)
def __getitem__(self, idx):
return self.df.iloc[idx].values.reshape(64, 64).astype(float)
preds_dict = {"grapheme_root": [], "vowel_diacritic": [], "consonant_diacritic": []}
components = ["consonant_diacritic", "grapheme_root", "vowel_diacritic"]
target = [] # model predictions placeholder
row_id = [] # row_id place holder
test_data = [
"test_image_data_0.parquet",
"test_image_data_1.parquet",
"test_image_data_2.parquet",
"test_image_data_3.parquet",
]
predictions = []
batch_size = 1
for fname in test_data:
data = pd.read_parquet(f"/kaggle/input/bengaliai-cv19/{fname}")
data.set_index("image_id", inplace=True)
data = resize(data, need_progress_bar=False).astype(np.float32) / 255
test_image = GraphemeDataset(data)
test_loader = torch.utils.data.DataLoader(test_image, batch_size=3, shuffle=False)
with torch.no_grad():
for idx, (inputs) in enumerate(test_loader):
inputs.to(device)
preds = model(inputs.unsqueeze(1).float())
for i, p in enumerate(preds_dict):
preds_dict[p] = np.argmax(preds[i], axis=1)
for k, id in enumerate(data.index.values):
for i, comp in enumerate(components):
id_sample = id + "_" + comp
row_id.append(id_sample)
target.append(preds_dict[comp][k].item())
df_sample = pd.DataFrame(
{"row_id": row_id, "target": target}, columns=["row_id", "target"]
)
df_sample.to_csv("submission.csv", index=False)
df_sample.head(20)
|
import os
import glob
import numpy as np
import scipy.signal as sc
import matplotlib.pyplot as plt
import pandas as pd
from IPython.display import display, clear_output
# Define the sampling rate
fs = 100
# Define the Training and Test folders paths
training_path = (
"/kaggle/input/physiological-signals-processing-challenge-2223/Training/"
)
test_path = "/kaggle/input/physiological-signals-processing-challenge-2223/Test/"
# Load the Training CSV file with Pandas
training_csv = pd.read_csv(
"/kaggle/input/physiological-signals-processing-challenge-2223/Training.csv"
)
# Print some rows of the CSV
print(training_csv.head())
print("Number of episodes: ", len(training_csv))
np.random.seed(1947)
# Let's load the data and store it in a list (this process can take a while).
training_signals = []
for i in training_csv["Id"]:
training_signals.append(np.loadtxt(training_path + str(i) + ".txt", delimiter=","))
# Check the number of records (should be 101)
print("The number of loaded records is " + str(len(training_signals)))
# ## Validation Set
# Next, we are going to select 10 patients as a validation dataset in order to test our new features.
#
idx_val = np.random.randint(0, len(training_signals), size=10, dtype=int)
print(idx_val.shape)
idx_val = np.sort(idx_val)
# now lets create idx_subtraining
idx_sub_training = []
for i in range(len(training_signals)):
if i in idx_val:
continue
idx_sub_training.append(i)
print(idx_val)
print(idx_sub_training)
# split training data set into sub_train and validation
sub_training_signals = [training_signals[i] for i in idx_sub_training]
sub_training_labels = training_csv.iloc[idx_sub_training]
val_signals = [training_signals[i] for i in idx_val]
val_labels = training_csv.iloc[idx_val]
# print(val_labels)
# print(val_signals)
# ## Use validation set
# Once you have two set, sub_training and validation, you should work as follow:
# 1. Figure out a feature that you think it would be usefull
# 2. Fit the NB classifier on the sub_training dataset
# 3. Find the predictions of the NB classifier on the sub_traininig dataset and compute the MCCs
# 4. Compute the predictions on validation dataset and compute MCCs using the bootstrap approach to measure the improvement using this new feature
def bootstrap_metric(y_test, y_hat, B=5000, plot=0):
np.random.seed(1947)
matthew_corr_coef_boot = []
for i in range(B):
# bootstrap using id
id_row = np.arange(len(y_test))
ids_boot = np.random.choice(id_row, len(id_row))
# get a bootstrap sampling of the solution
boot_sol = y_test[ids_boot]
# get the boostrap sampling of the submission
boot_sub = y_hat[ids_boot]
# append the matthews_corrcoef
matthew_corr_coef_boot.append(matthews_corrcoef(boot_sol, boot_sub))
# compute confidence interval
mean = np.mean(matthew_corr_coef_boot)
standard_deviation = np.std(matthew_corr_coef_boot)
conf_interval = np.percentile(matthew_corr_coef_boot, [2.5, 97.5])
if plot == 1:
plt.figure()
plt.hist(matthew_corr_coef_boot, bins=20, density=True)
plt.axvline(conf_interval[0], linewidth=3, color="r")
plt.axvline(conf_interval[1], linewidth=3, color="r")
return mean, standard_deviation
from sklearn.metrics import matthews_corrcoef
# 1. New feature
# 2. Fit the NB classifier on sub_training data set
# 3. Predictions on sub_training, let's suppose the new feature leads to the following predicionts
# vector
y_hat_sub_training = np.random.randint(0, 2, size=len(sub_training_labels))
# Compute the MCCS
mcc_sub_training = matthews_corrcoef(
sub_training_labels["Category"].values, y_hat_sub_training
)
print(mcc_sub_training)
# 4. Compute predictions on validation
y_hat_val = np.random.randint(0, 2, size=len(val_labels))
mcc_val = matthews_corrcoef(val_labels["Category"].values, y_hat_val)
print(mcc_val)
# create a bootstrap estimation
mcc_m, mcc_std = bootstrap_metric(
val_labels["Category"].values, y_hat_val, B=2000, plot=1
)
print(mcc_m)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
# The idea is to create a minimal RNN that will provide a baseline model for more complex algorithms,to gain a low level understanding of the working of RNN.
# This kernel was inspired by
# 1. Andrej Karpathy https://gist.github.com/karpathy/d4dee566867f8291f086: Minimal character-level language model with a Vanilla Recurrent Neural Network, in Python/numpy. And the blog http://karpathy.github.io/2015/05/21/rnn-effectiveness/.
# 2. The deep learning book by Michael Nielsen particularly http://neuralnetworksanddeeplearning.com/chap6.html
# 3. Andrew ng Deep learning course (Course 5) on Coursera
#
train_df = pd.read_csv("/kaggle/input/nlp-getting-started/train.csv")
test_df = pd.read_csv("/kaggle/input/nlp-getting-started/test.csv")
train_df.head()
# A negative and a positive text for a disaster
train_df = train_df.sample(frac=1)
print(train_df[train_df["target"] == 0]["text"].values[0])
print(train_df[train_df["target"] == 1]["text"].values[0])
"""Preprocessing and
creating a dictionary that has an index for each of the unique words"""
train_df["text"] = train_df["text"].str.lower()
test_df["text"] = test_df["text"].str.lower()
# removing all the words starting with http and @
train_df["text"] = train_df["text"].map(
lambda x: (
" ".join(word for word in x.split(" ") if not word.startswith(("http", "@")))
)
)
test_df["text"] = test_df["text"].map(
lambda x: (
" ".join(word for word in x.split(" ") if not word.startswith(("http", "@")))
)
)
# removing any non-alphanumeric characters
train_df["text"] = train_df["text"].str.replace("[^a-z A-Z]", "")
test_df["text"] = test_df["text"].str.replace("[^a-z A-Z]", "")
words = list(
train_df["text"].str.split(" ", expand=True).stack().unique()
) # getting the list of unique words
vocabulary_size = len(words)
print("The number of unique words is:%d " % (vocabulary_size))
print(
"The total_number of words is : %d"
% (len(list(train_df["text"].str.split(" ", expand=True).stack())))
)
# creating a dictionary for indexing the words
words_idx = {word: i for i, word in enumerate(words)}
train_df.head()
# words_idx
"""Converting a single training example into the index retrieved from the dictionary """
example = train_df["text"].str.split().values[1]
inputs = [words_idx[i] for i in example]
targets = train_df["target"].values[1]
print(example)
print(inputs)
print(targets)
# hyperparameters
learning_rate = 0.01
n_h = hidden_size = 100
n_x = vocabulary_size
n_y = 2
# model_parameters
Whh = np.random.randn(hidden_size, hidden_size) * 0.1
Whx = np.random.randn(hidden_size, vocabulary_size) * 0.1
Wyh = np.random.randn(2, hidden_size) * 0.1
by = np.zeros((n_y, 1))
bh = np.zeros((n_h, 1))
# """loading the saved model"""
# import pickle
# filename = '/kaggle/input/pkl-model/rnn_model_v2.pkl'
# with open(filename, "rb") as f:
# Whh, Whx, bh, by, Wyh = pickle.load(f)
# validation and training set
copy_df = train_df
train_df = train_df.iloc[:7000]
validation_df = copy_df.iloc[7000:]
print("The training set examples: %d" % (len(train_df)))
print("The validation set examples: %d" % (len(validation_df)))
def feedforward(
inputs,
): # takes in the index of words in a example tweet and return the prediction
xs, hs = [], np.zeros((n_h, 1))
for t in range(len(inputs)):
xs.append(np.zeros((n_x, 1))) # encode in 1-of-k representation
xs[t][inputs[t]] = 1
hs = np.tanh(np.dot(Whh, hs) + np.dot(Whx, xs[t]) + bh) # hidden state
ys = np.dot(Wyh, hs) + by # unnormalized log probabilities for next chars
ps = np.exp(ys) / np.sum(
np.exp(ys), axis=0
) # softmax probabiltity of non -disaster / disaster tweets
prediction = np.argmax(ps)
return prediction
"""Generating a function that takes in one training example, feedforward into the network, calculate the cost which is the function of
predicted y vs actual y. Then we perform a backward propagation and find the gradient of all the parameters and return it"""
def loss_func(inputs, targets):
# input is the list of index in a training example (shape= (1,T_x))
# targets (0 or 1) for a training example[0,1]
xs, hs = [], [] # creating a cache of xs, hs for each unit of propagation
hs.append(np.zeros((n_h, 1)))
loss = 0
# feedforward propagation
for t in range(len(inputs)):
xs.append(np.zeros((n_x, 1)))
xs[t][inputs[t]] = 1
hs.append(np.tanh(np.dot(Whh, hs[t]) + np.dot(Whx, xs[t]) + bh))
ys = np.dot(Wyh, hs[-1]) + by
ps = np.exp(ys) / np.sum(np.exp(ys), axis=0)
# cost
y = np.zeros((2, 1))
y[targets] = 1
loss = -np.log(np.sum(ps * y, axis=0)) # cross_entropy loss
# backward_propagation through time
"""gradient of cost with respect to model parameters """
dWhh, dWyh, dWhx = np.zeros_like(Whh), np.zeros_like(Wyh), np.zeros_like(Whx)
dby, dbh = np.zeros_like(by), np.zeros_like(bh)
dy = ps - y
dWyh = np.dot(dy, hs[-1].transpose())
dby = np.copy(dy)
dh = np.dot(Wyh.transpose(), dy)
dh_raw = (1 - hs[-1] * hs[-1]) * dh
dWhx = np.dot(dh_raw, xs[-1].transpose())
dWhh = np.dot(dh_raw, hs[-2].transpose())
dbh = np.copy(dh_raw)
dh_prev = np.dot(Whh.transpose(), dh_raw)
for t in reversed(range(len(inputs) - 1)):
dh = np.copy(dh_prev)
dh_raw = (1 - hs[t] * hs[t]) * dh
dWhx += np.dot(dh_raw, xs[-1].transpose())
dWhh += np.dot(dh_raw, hs[-2].transpose())
dbh += np.copy(dh_raw)
dh_prev = np.dot(Whh.transpose(), dh_raw)
for dparams in [
dWhh,
dWhx,
dbh,
dby,
dWyh,
]: # clipping to avoid exploding gradients
np.clip(dparams, -5, 5, out=dparams)
return loss, dWhh, dWhx, dbh, dby, dWyh
"""Feeding into the network to retrive the gradient and using Adagrad optimizer to perform the gradient descent.
Then we repeat this for all the training examples and for n epochs."""
num_iterations = 35000
mWhh, mWyh, mWhx = np.zeros_like(Whh), np.zeros_like(Wyh), np.zeros_like(Whx)
mby, mbh = np.zeros_like(by), np.zeros_like(bh) # memory variables for Adagrad
for j in range(num_iterations):
idx = j % len(train_df)
example = train_df["text"].str.split().values[idx]
inputs = [words_idx[i] for i in example]
targets = train_df["target"].values[idx]
loss, dWhh, dWhx, dbh, dby, dWyh = loss_func(inputs, targets)
# Adagrad optimizer
# perform parameter update with Adagrad
for param, dparam, mem in zip(
[Whx, Whh, Wyh, bh, by],
[dWhx, dWhh, dWyh, dbh, dby],
[mWhx, mWhh, mWyh, mbh, mby],
):
mem += dparam * dparam
param += -learning_rate * dparam / np.sqrt(mem + 1e-8) # adagrad update
# validation accuracy
# using for loop instead of vectorization
if j % 700 == 0:
predictions = []
count = 0
actual_targets = validation_df["target"].tolist()
for i in range(len(validation_df)):
example = validation_df["text"].str.split().values[i]
inputs = [words_idx[l] for l in example]
predictions.append(feedforward(inputs))
for y, y_hat in zip(actual_targets, predictions):
if y == y_hat:
count += 1
print(
"The validation_accuracy after iterations:%d is %d"
% (j, (count / len(validation_df)) * 100)
)
# print(predictions[:20])
# print(actual_targets[:20])
# predictions in the test set
test_predictions = []
for i in range(len(test_df)):
example = test_df["text"].str.split().values[i]
inputs = []
for l in example:
if l in words_idx:
inputs.append(words_idx[l])
test_predictions.append(feedforward(inputs))
test_df["target"] = test_predictions
test_df = test_df[["id", "target"]].set_index("id")
test_df.to_csv("submission.csv")
# saving the model
import pickle
filename = "rnn_model_v2.pkl"
with open(filename, "wb") as f:
pickle.dump((Whh, Whx, bh, by, Wyh), f)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
CoronaV = pd.read_csv(
"/kaggle/input/novel-corona-virus-2019-dataset/2019_nCoV_data.csv"
)
print(CoronaV.head(10))
print("\n")
print(CoronaV.info())
# normalize the dataset
CoronaV = CoronaV.drop("Sno", axis=1)
CoronaV.columns = ["State", "Country", "Date", "Confirmed", "Deaths", "Recovered"]
CoronaV["Date"] = CoronaV["Date"].apply(pd.to_datetime).dt.normalize()
CoronaV.info()
CoronaV[["State", "Country", "Date", "Confirmed"]].drop_duplicates().shape[
0
] == CoronaV.shape[0]
CoronaV.describe(include="all")
CoronaV[["Country", "State"]][CoronaV["State"].isnull()].drop_duplicates()
CoronaV[
CoronaV["Country"].isin(
list(
CoronaV[["Country", "State"]][CoronaV["State"].isnull()]["Country"].unique()
)
)
]["State"].unique()
CoronaV.State.unique()
CoronaV.Country.unique()
print(
CoronaV[CoronaV["Country"].isin(["China", "Mainland China"])]
.groupby("Country")["State"]
.unique()
)
print(
CoronaV[CoronaV["Country"].isin(["China", "Mainland China"])]
.groupby("Country")["Date"]
.unique()
)
CoronaV["Country"] = CoronaV["Country"].replace(
["Mainland China"], "China"
) # set 'Mainland China' to 'China'
sorted(CoronaV.Country.unique())
print(CoronaV.head())
china = CoronaV[CoronaV["Country"] == "China"]
china.head()
import matplotlib.pyplot as plt
plt.rcParams["figure.figsize"] = (12, 9)
ax1 = china[["Date", "Confirmed"]].groupby(["Date"]).sum().plot()
ax1.set_ylabel("Total Number of Confirmed Cases")
ax1.set_xlabel("Date")
ax2 = china[["Date", "Deaths", "Recovered"]].groupby(["Date"]).sum().plot()
ax2.set_ylabel("Total N")
ax2.set_xlabel("Date")
import plotly.graph_objects as go
from plotly.subplots import make_subplots
fig = make_subplots(
rows=1,
cols=3,
specs=[[{"type": "pie"}, {"type": "pie"}, {"type": "pie"}]],
subplot_titles=("number of provience in countries", "Deaths", "Recovers"),
)
fig.add_trace(
go.Pie(
labels=CoronaV.groupby("Country")["State"]
.nunique()
.sort_values(ascending=False)[:10]
.index,
values=CoronaV.groupby("Country")["State"]
.nunique()
.sort_values(ascending=False)[:10]
.values,
),
row=1,
col=1,
)
fig.add_trace(
go.Pie(
labels=CoronaV[CoronaV.Deaths > 0].groupby("Country")["Deaths"].sum().index,
values=CoronaV[CoronaV.Deaths > 0].groupby("Country")["Deaths"].sum().values,
),
row=1,
col=2,
)
fig.add_trace(
go.Pie(
labels=CoronaV.groupby("Country")["Recovered"]
.sum()
.sort_values(ascending=False)
.index[:4],
values=CoronaV.groupby("Country")["Recovered"]
.sum()
.sort_values(ascending=False)
.values[:4],
),
row=1,
col=3,
)
fig.update_layout(height=400, showlegend=True)
fig.show()
CoronaV["Date"] = pd.to_datetime(CoronaV["Date"])
CoronaV["Day"] = CoronaV["Date"].apply(lambda x: x.day)
CoronaV["Hour"] = CoronaV["Date"].apply(lambda x: x.hour)
CoronaV = CoronaV[CoronaV["Confirmed"] != 0]
CoronaV
global_case = (
CoronaV.groupby("Country")["Confirmed", "Deaths", "Recovered"].sum().reset_index()
)
global_case.head()
global_case
CoronaV.groupby(["Date", "Country"]).agg(
{
"Confirmed": pd.Series.nunique,
}
).reset_index().pivot(index="Date", columns="Country", values="Confirmed").plot.barh(
stacked=True, figsize=(26, 10), colormap="gist_rainbow"
)
import warnings
warnings.simplefilter(action="ignore", category=FutureWarning)
import seaborn as sns
plt.rcParams["figure.figsize"] = (17, 10)
nums = (
china.groupby(["State"])["Confirmed"]
.aggregate(sum)
.reset_index()
.sort_values("Confirmed", ascending=False)
)
ax = sns.barplot(x="Confirmed", y="State", order=nums["State"], data=china, ci=None)
ax.set_xlabel("Total Confirmed Cases")
def get_ci(N, p):
lci = (p - 1.96 * (((p * (1 - p)) / N) ** 0.5)) * 100
uci = (p + 1.96 * (((p * (1 - p)) / N) ** 0.5)) * 100
return str(np.round(lci, 3)) + "% - " + str(np.round(uci, 3)) + "%"
final = CoronaV[CoronaV.Date == np.max(CoronaV.Date)]
final = final.copy()
final["CFR"] = np.round((final.Deaths.values / final.Confirmed.values) * 100, 3)
final["CFR 95% CI"] = final.apply(
lambda row: get_ci(row["Confirmed"], row["CFR"] / 100), axis=1
)
global_cfr = np.round(
np.sum(final.Deaths.values) / np.sum(final.Confirmed.values) * 100, 3
)
final.sort_values("CFR", ascending=False).head(10)
tops = final.sort_values("CFR", ascending=False)
tops = tops[tops.CFR > 0]
df = final[final["CFR"] != 0]
plt.rcParams["figure.figsize"] = (10, 5)
ax = sns.barplot(y="CFR", x="State", order=tops["State"], data=df, ci=None)
ax.axhline(global_cfr, alpha=0.5, color="r", linestyle="dashed")
ax.set_title("Case Fatality Rates (CFR) as of 30 Jan 2020")
ax.set_ylabel("CFR %")
print("Average CFR % = " + str(global_cfr))
from sklearn.preprocessing import StandardScaler
from sklearn.neighbors import LocalOutlierFactor
scaler = StandardScaler()
scd = scaler.fit_transform(final[["Confirmed", "Deaths", "Recovered"]])
clf = LocalOutlierFactor(
n_neighbors=20, contamination=0.1
) # LOF is very sensitive to the choice of n_neighbors. Generally, n_neighbors = 20 works better
clf.fit(scd)
lofs = clf.negative_outlier_factor_ * -1
final["LOF Score"] = lofs
tops = final.sort_values("LOF Score", ascending=False)
plt.rcParams["figure.figsize"] = (20, 12)
ax = sns.barplot(x="LOF Score", y="State", order=tops["State"], data=final, ci=None)
ax.axvline(1, alpha=0.5, color="g", linestyle="dashed")
ax.axvline(np.median(lofs), alpha=0.5, color="b", linestyle="dashed")
ax.axvline(np.mean(lofs) + 3 * np.std(lofs), alpha=0.5, color="r", linestyle="dashed")
final.sort_values("LOF Score", ascending=False)
from sklearn.cluster import KMeans
plt.rcParams["figure.figsize"] = (5, 5)
wcss = []
for i in range(1, 11):
kmeans = KMeans(
n_clusters=i, init="k-means++", max_iter=300, n_init=10, random_state=1897
)
kmeans.fit(scd)
wcss.append(kmeans.inertia_)
plt.plot(range(1, 11), wcss)
plt.title("Elbow Method")
plt.xlabel("Number of clusters")
plt.ylabel("Within Cluster Sum of Squares")
plt.show()
kmeans = KMeans(
n_clusters=2, init="k-means++", max_iter=300, n_init=10, random_state=1897
)
clusters = np.where(kmeans.fit_predict(scd) == 0, "Cluster 1", "Cluster 2")
clusters
from sklearn import decomposition
pca = decomposition.PCA(n_components=3)
pca.fit(scd)
X = pca.transform(scd)
print(pca.explained_variance_ratio_.cumsum())
plt.rcParams["figure.figsize"] = (7, 7)
ax = sns.scatterplot(X[:, 0], X[:, 1], marker="X", s=80, hue=clusters)
ax.set_title("K-Means Clusters of States")
ax.set_xlabel("Principal Component 1")
ax.set_ylabel("Principal Component 2")
pd.DataFrame(final.State.values, clusters)
X = CoronaV["Deaths"].values.reshape(-1, 1)
y = CoronaV["Recovered"].values.reshape(-1, 1)
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=0)
from sklearn.linear_model import LinearRegression
regressor = LinearRegression()
regressor.fit(X_train, y_train) # training the algorithm
# To retrieve the intercept:
print(regressor.intercept_) # For retrieving the slope:
print(regressor.coef_)
y_pred = regressor.predict(X_test)
import matplotlib.pyplot as plt
import seaborn as sns
import seaborn as seabornInstance
plt.figure(figsize=(15, 10))
plt.tight_layout()
seabornInstance.distplot(CoronaV["Recovered"])
df = pd.DataFrame({"Actual": y_test.flatten(), "Predicted": y_pred.flatten()})
df
df1 = df.head(25)
df1.plot(kind="bar", figsize=(16, 10))
plt.grid(which="major", linestyle="-", linewidth="0.5", color="green")
plt.grid(which="minor", linestyle=":", linewidth="0.5", color="black")
plt.show()
# transform data
from sklearn.preprocessing import MinMaxScaler
scaler = MinMaxScaler()
X_train_scale = scaler.fit_transform(X_train)
X_test_scale = scaler.transform(X_test)
# split training feature and target sets into training and validation subsets
from sklearn.model_selection import train_test_split
X_train_sub, X_validation_sub, y_train_sub, y_validation_sub = train_test_split(
X_train_scale, y_train, random_state=0
)
# import machine learning algorithms
from sklearn.ensemble import GradientBoostingClassifier
from sklearn.metrics import classification_report, confusion_matrix, roc_curve, auc
from sklearn import preprocessing
min_max_scaler = preprocessing.MinMaxScaler()
X_scale = min_max_scaler.fit_transform(X)
X_scale
from sklearn.model_selection import train_test_split
X_train, X_val_and_test, Y_train, Y_val_and_test = train_test_split(
X_scale, y, test_size=0.3
)
X_val, X_test, Y_val, Y_test = train_test_split(
X_val_and_test, Y_val_and_test, test_size=0.5
)
print(
X_train.shape, X_val.shape, X_test.shape, Y_train.shape, Y_val.shape, Y_test.shape
)
from keras.models import Sequential
from keras.layers import Dense
model = Sequential(
[
Dense(32, activation="relu", input_shape=(1,)),
Dense(32, activation="relu"),
Dense(1, activation="sigmoid"),
]
)
model.compile(optimizer="sgd", loss="binary_crossentropy", metrics=["accuracy"])
hist = model.fit(
X_train, Y_train, batch_size=32, epochs=500, validation_data=(X_val, Y_val)
)
model.evaluate(X_test, Y_test)[1]
import matplotlib.pyplot as plt
plt.plot(hist.history["loss"])
plt.plot(hist.history["val_loss"])
plt.title("Model loss")
plt.ylabel("Loss")
plt.xlabel("Epoch")
plt.legend(["Train", "Val"], loc="upper right")
plt.show()
plt.plot(hist.history["accuracy"])
plt.plot(hist.history["val_accuracy"])
plt.title("Model accuracy")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(["Train", "Val"], loc="lower right")
plt.show()
model_2 = Sequential(
[
Dense(1000, activation="relu", input_shape=(1,)),
Dense(1000, activation="relu"),
Dense(1000, activation="relu"),
Dense(1000, activation="relu"),
Dense(1, activation="sigmoid"),
]
)
model_2.compile(optimizer="adam", loss="binary_crossentropy", metrics=["accuracy"])
hist_2 = model_2.fit(
X_train, Y_train, batch_size=32, epochs=100, validation_data=(X_val, Y_val)
)
plt.plot(hist_2.history["accuracy"])
plt.plot(hist_2.history["val_accuracy"])
plt.title("Model accuracy")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(["Train", "Val"], loc="lower right")
plt.show()
from keras.layers import Dropout
from keras import regularizers
model_3 = Sequential(
[
Dense(
1000,
activation="relu",
kernel_regularizer=regularizers.l2(0.01),
input_shape=(1,),
),
Dropout(0.3),
Dense(1000, activation="relu", kernel_regularizer=regularizers.l2(0.01)),
Dropout(0.3),
Dense(1000, activation="relu", kernel_regularizer=regularizers.l2(0.01)),
Dropout(0.3),
Dense(1000, activation="relu", kernel_regularizer=regularizers.l2(0.01)),
Dropout(0.3),
Dense(1, activation="sigmoid", kernel_regularizer=regularizers.l2(0.01)),
]
)
model_3.compile(optimizer="adam", loss="binary_crossentropy", metrics=["accuracy"])
hist_3 = model_3.fit(
X_train, Y_train, batch_size=32, epochs=100, validation_data=(X_val, Y_val)
)
plt.plot(hist_3.history["loss"])
plt.plot(hist_3.history["val_loss"])
plt.title("Model loss")
plt.ylabel("Loss")
plt.xlabel("Epoch")
plt.legend(["Train", "Val"], loc="upper right")
plt.ylim(top=1.2, bottom=0)
plt.show()
plt.plot(hist_3.history["accuracy"])
plt.plot(hist_3.history["val_accuracy"])
plt.title("Model accuracy")
plt.ylabel("Accuracy")
plt.xlabel("Epoch")
plt.legend(["Train", "Val"], loc="lower right")
plt.show()
model.evaluate(X_test, Y_test)[1]
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.tree import DecisionTreeClassifier # Import Decision Tree Classifier
from sklearn.model_selection import train_test_split # Import train_test_split function
from sklearn import (
metrics,
) # Import scikit-learn metrics module for accuracy calculation
from sklearn.model_selection import train_test_split, cross_val_score
import seaborn as sns
from sklearn.tree import export_graphviz
from sklearn.externals.six import StringIO
from IPython.display import Image
from sklearn import tree
from sklearn import metrics
from sklearn.neighbors import KNeighborsClassifier
import pandas as pd
test = pd.read_csv("../input/data-science-london-scikit-learn/test.csv", header=None)
train = pd.read_csv("../input/data-science-london-scikit-learn/train.csv", header=None)
trainLabels = pd.read_csv(
"../input/data-science-london-scikit-learn/trainLabels.csv", header=None
)
## finding size of all data
print("Train Shape:", train.shape)
print("Test Shape:", test.shape)
print("Labels Shape:", trainLabels.shape)
##getting first 5 rows of train
train.head()
trainLabels.columns = ["Target"]
pd.crosstab(
index=trainLabels["Target"].astype("category"), columns="count" # Make a crosstab
)
train.iloc[:, 0:10].describe()
Full_Data = pd.concat([train, trainLabels], axis=1)
Full_Data
Mean_Sum = Full_Data.groupby("Target").agg("mean")
Mean_Sum["Type"] = "Mean"
Sum_Sum = Full_Data.groupby("Target").agg("sum")
Sum_Sum["Type"] = "Sum"
Summ_By_Target = pd.concat([Mean_Sum, Sum_Sum])
Summ_By_Target
Full_Data[Full_Data["Target"] == 0].describe()
Full_Data[Full_Data["Target"] == 1].describe()
sns.lmplot(
x="12", y="28", data=Full_Data.rename(columns=lambda x: str(x)), col="Target"
)
plt.show()
sns.lmplot(x="12", y="22", data=Full_Data.rename(columns=lambda x: str(x)))
plt.show()
# Splitting Data
#
##trying to combine predictor(x) and traget(y), as both are store in differnt varible and combing both will give entire
##training data which will also include target variable.
X, y = train, np.ravel(trainLabels)
##spliting training data into train set and test set. train set has 70% of data while test set has 30% of data.
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3)
# Buliding Decision Tree Model
#
# Create Decision Tree classifer object
clf = DecisionTreeClassifier(max_depth=8)
# Train Decision Tree Classifer
clf = clf.fit(X_train, y_train)
# Predict the response for test dataset
y_pred = clf.predict(X_test)
# Evaluating Model
# Model Accuracy, how often is the classifier correct?
print("Test Accuracy:", metrics.accuracy_score(y_test, y_pred))
# Building RandomForest Model
# Import Random Forest Model
from sklearn.ensemble import RandomForestClassifier
# Create a Gaussian Classifier
clf = RandomForestClassifier(n_estimators=100)
# Train the model using the training sets y_pred=clf.predict(X_test)
clf = clf.fit(X_train, y_train)
y_pred = clf.predict(X_test)
# Evaluating Model
# Model Accuracy, how often is the classifier correct?
print("Accuracy:", metrics.accuracy_score(y_test, y_pred))
# Support Vector Machine
# Import svm model
from sklearn import svm
# Create a svm Classifier
clf = svm.SVC(kernel="linear") # Linear Kernel
# Train the model using the training sets
clf.fit(X_train, y_train)
# Predict the response for test dataset
y_pred = clf.predict(X_test)
# Evaluating Model
# Model Accuracy: how often is the classifier correct?
print("Accuracy:", metrics.accuracy_score(y_test, y_pred))
# KNN means
neighbor = np.arange(1, 26)
k_fold = 10
train_acc = []
valid_acc = []
best_k = 0
trainLabels = np.ravel(trainLabels)
for i, k in enumerate(neighbor):
knn = KNeighborsClassifier(n_neighbors=k)
knn.fit(X_train, y_train)
train_acc.append(knn.score(X_train, y_train))
valid_acc.append(np.mean(cross_val_score(knn, train, trainLabels, cv=k_fold)))
best_k = np.argmax(valid_acc)
print("Best k: ", best_k)
# Evaluating Model
final_model = KNeighborsClassifier(n_neighbors=2)
final_model.fit(train, trainLabels)
y_pred_knn = final_model.predict(X_test)
print("Training final: ", final_model.score(train, trainLabels))
print("Accuracy:", metrics.accuracy_score(y_test, y_pred_knn))
pred_test = final_model.predict(test)
pred_test[:5]
pred_test.shape
submission = pd.DataFrame(pred_test)
submission.columns = ["Solution"]
submission["Id"] = np.arange(1, submission.shape[0] + 1)
submission = submission[["Id", "Solution"]]
submission.head()
filename = "London_Example.csv"
submission.to_csv(filename, index=False)
print("Saved file: " + filename)
|
from matplotlib import pyplot as plt
import pandas as pd # data processing
import numpy as np # statistics
import matplotlib.pyplot as plt # visualization
import seaborn as sns # visualization
data = pd.read_csv("/kaggle/input/co2-csvpy/co2_csv.py")
# Replace all spaces in the 'Country' column with underscores
data["Country"] = data["Country"].str.replace(" ", "")
# Replace all /km² in the 'density(km2) column with empty''
data["Density(km2)"] = data["Density(km2)"].str.replace("/km²", "")
# Write the modified DataFrame back to the CSV file
data.to_csv(
"C:\\Users\\Giorgi\\PycharmProjects\\pythonProject3\\CO2 Emission by countries Year wise (1750-2022)_project\\co2_csv.py",
index=False,
)
print(data.head())
plt.figure(figsize=(12, 8))
yearly_world_emission_data = data.groupby("Year").sum()
plt.plot(yearly_world_emission_data.index, yearly_world_emission_data["CO2Emission"])
plt.title("World yearly CO2 emission")
print(
"""World Yearly CO2 Emission (Cumulative): Based on the data,
there has been a significant increase in CO2 emissions over time.
The cumulative CO2 emissions for the entire world from 1750 to 2023 show a clear upward trend,
with the most rapid increase occurring in the last few decades."""
)
top20_emission_data = (
data[(data.Year > 2012) & (data["Year"] < 2023)]
.groupby("Country")[["CO2Emission"]]
.sum()
.sort_values(by=["CO2Emission"], ascending=False)
.head(20)
)
plt.figure(figsize=(12, 8))
sns.set_style("whitegrid")
sns.barplot(
data=top20_emission_data,
x="CO2Emission",
y=top20_emission_data.index,
palette="bright",
)
plt.title("Top 20 CO2 emitting countries from 2012 - 2022")
plt.xlabel("CO2 emission in tons")
plt.ylabel("Countries")
print(
"""Top 20 CO2 Emitting Countries from 2012-2022:
The top 20 CO2 emitting countries from 2012-2022 account
for a significant proportion of global CO2 emissions.
The list is dominated by large industrialized countries,
including China, the United States, and India."""
)
bottom20_emission_data = (
data[(data.Year > 2012) & (data["Year"] < 2023)]
.groupby("Country")[["CO2Emission"]]
.sum()
.sort_values(by=["CO2Emission"])
.head(20)
)
plt.figure(figsize=(12, 8))
sns.set_style("whitegrid")
sns.barplot(
data=bottom20_emission_data,
x="CO2Emission",
y=bottom20_emission_data.index,
palette="bright",
)
plt.title("Bottom 20 CO2 emitting countries from 2012 - 2022")
plt.xlabel("CO2 emission in tons")
plt.ylabel("Countries")
print(
"""Bottom 20 CO2 Emitting Countries from 2012-2022:
The bottom 20 CO2 emitting countries from 2012-2022 account
for a much smaller proportion of global CO2 emissions.
The list is dominated by small island nations and developing
countries with low levels of industrialization."""
)
# finding the year with the greatest co2 emission from 1750-2020
print(
data.groupby("Year")[["CO2Emission"]]
.sum()
.sort_values(by="CO2Emission", ascending=False)
.head(1)
)
# emission data in Georgia
co2_georgia_data = data[data["Country"] == "Georgia"]
print(co2_georgia_data.head())
plt.figure(figsize=(10, 6))
sns.set_style("whitegrid")
plt.plot(co2_georgia_data["Year"], co2_georgia_data["CO2Emission"])
plt.annotate(
"""Independance from USSR,
War in Abkhazia and Crisis""",
xy=(1991, 4.89e08),
xytext=(1991, 4.89e08),
)
plt.title("Yearly CO2 Emission in Georgia")
print(
"""Yearly CO2 Emission in Georgia (Country):
The data shows that CO2 emissions in Georgia have
increased over time, with a significant increase
occurring in the last few decades. However,
the absolute level of CO2 emissions in Georgia
is relatively low compared to many other countries."""
)
# Emission data in Czech Republic
co_2_cz_data = data[data["Country"] == "Czechia"]
print(co_2_cz_data.head())
plt.figure(figsize=(10, 6))
sns.set_style("whitegrid")
plt.plot(co_2_cz_data["Year"], co_2_cz_data["CO2Emission"])
plt.title("Yearly CO2 Emission in Czech Republic")
print(
"""Yearly CO2 Emission in Czechia:
The data shows that CO2 emissions in
Czechia have fluctuated over time, with significant
increases occurring in the mid-20th century and again
in the last few decades. However, the absolute level of
CO2 emissions in Czechia is relatively low compared to many other countries."""
)
co_2_usa_data = data[data["Country"] == "United_States"]
print(co_2_usa_data.head())
plt.figure(figsize=(10, 6))
sns.set_style("whitegrid")
plt.plot(co_2_usa_data["Year"], co_2_usa_data["CO2Emission"])
plt.title("Yearly CO2 Emission in USA")
print(
"""Yearly CO2 Emission in USA: The data shows that the United States
is one of the largest CO2 emitting countries in the world.
CO2 emissions in the US have increased steadily over time,
with a significant increase occurring in the mid-20th century and again in the last few decades."""
)
# time series emission of top 10 country 1750-2022
top10_data = data[
(data["Country"] == "India")
| (data["Country"] == "China")
| (data["Country"] == "Russia")
| (data["Country"] == "Germany")
| (data["Country"] == "United_Kingdom")
| (data["Country"] == "Poland")
| (data["Country"] == "Ukraine")
| (data["Country"] == "France")
| (data["Country"] == "Japan")
| (data["Country"] == "United_States")
]
top10_data.head()
sns.relplot(
x="Year",
y="CO2Emission",
data=top10_data,
hue="Country",
kind="line",
markers=True,
dashes=True,
height=5,
aspect=2,
style="Country",
)
plt.title("Time progression for top 5 co2 emitting countries")
plt.ylabel("co2 emission in tons")
print(
"""Time Progression for Top 10 CO2 Emitting Countries from 1750 to 2023:
The data shows that the top 10 CO2 emitting countries from 1750 to 2023 are all
large industrialized nations. There has been a significant increase in CO2 emissions
for all countries in this list, with the most rapid increase occurring in the last few decades."""
)
|
# # Let's find out the netflix movies!!!
# ## And how they are formed!!
# ## Import Libraries & Font & File
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import matplotlib as mpl
import seaborn as sns
import plotly.express as px
import plotly.graph_objects as go
# Only for publishing
import warnings
warnings.filterwarnings("ignore")
plt.rc("font", family="Arial Unicode MS") # For MacOS
mpl.rcParams["axes.unicode_minus"] = False
# Bring file into the notebook
nf = pd.read_csv("/kaggle/input/netflix-shows/netflix_titles.csv")
# ## Observe & Ananyze Data
1 # Analyze the heads of the data
nf.head()
# do not abbreviate the columns
pd.set_option("display.max_columns", None)
nf.head(5)
# make new dataframe with this nf['listed_in'].str.split(',').explode().str.strip().str.replace('TV Shows', '').str.replace('Movies', '').str.replace('TV', '').str.strip().replace('', np.nan).dropna().value_counts()
nf_genres = (
nf["listed_in"]
.str.split(",")
.explode()
.str.strip()
.str.replace("TV Shows", "")
.str.replace("Movies", "")
.str.replace("TV", "")
.str.strip()
.replace("", np.nan)
.dropna()
.value_counts()
.to_frame()
.reset_index()
)
nf_genres
# if movie or tv show has genre, it will be 1, if not, it will be 0
nf["listed_in"].str.split(",").explode().str.strip().str.replace(
"TV Shows", ""
).str.replace("Movies", "").str.replace("TV", "").str.strip().replace(
"", np.nan
).dropna().unique()
# make new column for each genre and make new dataframe
for genre in (
nf["listed_in"]
.str.split(",")
.explode()
.str.strip()
.str.replace("TV Shows", "")
.str.replace("Movies", "")
.str.replace("TV", "")
.str.strip()
.replace("", np.nan)
.dropna()
.unique()
):
nf[genre] = nf["listed_in"].str.contains(genre).astype(int)
nf.head(5)
# divide date_added into year by comma
nf["year_added"] = nf["date_added"].str.split(",").str[1].str.strip()
nf.head(5)
# release year desc
nf["release_year"].value_counts().sort_index(ascending=False)
nf.info()
# ## Cleaning Data
# fill in the missing values
nf["director"].fillna("Not Specified", inplace=True)
nf["cast"].fillna("Not Specified", inplace=True)
nf["country"].fillna("Not Specified", inplace=True)
nf["date_added"].fillna("Not Specified", inplace=True)
nf["rating"].fillna("No Rating", inplace=True)
nf.info()
# check duplicates
nf.duplicated("title")
# ## Netflix Colors
# ### I will use Netflix colors to make the plots more attractive
# netflix color palette
netflix_palette = ["#E50914", "#303030", "#FFFFFF", "#000000", "#831010"]
# show netflix color palette
sns.palplot(netflix_palette)
# ## Movies vs TV Shows
# find the number of movies and tv shows
nf["type"].value_counts()
# find the number of movies and tv shows pie chart use plotly, color manually
fig = px.pie(
nf,
values=nf["type"].value_counts(),
names=nf["type"].value_counts().index,
color_discrete_sequence=netflix_palette,
)
fig.update_traces(textposition="inside", textinfo="percent+label")
fig.update_layout(title_text="Number of Movies and TV Shows")
fig.show()
# Almost 70% of the content is movies and 30% is TV Shows
# all added movies and tvshows by year
nf_r_all = nf["release_year"].value_counts().sort_index(ascending=False)
# yearly added movies by year, only movies in type
nf_r_movies = nf[nf["type"] == "Movie"]
nf_r_movies["release_year"].value_counts().sort_index(ascending=False)
# yearly added tv shows by year, only movies in type
nf_r_tv = nf[nf["type"] == "TV Show"]
nf_r_tv["release_year"].value_counts().sort_index(ascending=False)
# make graph by nf_r_all, nf_movies and nf_tv by line graph
fig = go.Figure()
fig.add_trace(
go.Scatter(
x=nf_r_all.index,
y=nf_r_all.values,
name="All",
line=dict(color=netflix_palette[0], width=4),
)
)
fig.add_trace(
go.Scatter(
x=nf_r_movies["release_year"].value_counts().sort_index(ascending=False).index,
y=nf_r_movies["release_year"].value_counts().sort_index(ascending=False).values,
name="Movies",
line=dict(color=netflix_palette[1], width=4),
)
)
fig.add_trace(
go.Scatter(
x=nf_r_tv["release_year"].value_counts().sort_index(ascending=False).index,
y=nf_r_tv["release_year"].value_counts().sort_index(ascending=False).values,
name="TV Shows",
line=dict(color=netflix_palette[4], width=4),
)
)
fig.update_layout(title_text="Number of Movies and TV Shows by Release Year")
fig.update_layout(plot_bgcolor="rgba(0,0,0,0)")
fig.update_xaxes(title_text="Year")
fig.update_yaxes(title_text="Number of Movies and TV Shows")
fig.show()
# 2019 and 2020 was peak year of release of content
# I think this is because of the pandemic, which suppressed people from making Movies and TV Shows
# number of movies added each year, number of tv shows added each year, number of movies and tv shows added each year
fig = go.Figure()
fig.add_trace(
go.Scatter(
x=nf[nf["type"] == "Movie"]
.groupby("year_added")
.count()
.reset_index()["year_added"],
y=nf[nf["type"] == "Movie"]
.groupby("year_added")
.count()
.reset_index()["title"],
name="Movies Added",
line_color=netflix_palette[0],
)
)
fig.add_trace(
go.Scatter(
x=nf[nf["type"] == "TV Show"]
.groupby("year_added")
.count()
.reset_index()["year_added"],
y=nf[nf["type"] == "TV Show"]
.groupby("year_added")
.count()
.reset_index()["title"],
name="TV Shows Added",
line_color=netflix_palette[1],
)
)
fig.add_trace(
go.Scatter(
x=nf.groupby("year_added").count().reset_index()["year_added"],
y=nf.groupby("year_added").count().reset_index()["title"],
name="Total Added",
line_color=netflix_palette[4],
)
)
fig.update_layout(title_text="Number of Movies and TV Shows Added by Year")
# remove background
fig.update_layout(plot_bgcolor="rgba(0,0,0,0)")
# x axis name
fig.update_xaxes(title_text="Year Added")
# y axis name
fig.update_yaxes(title_text="Number of Movies and TV Shows Added")
fig.show()
# Also, added contents are similar with released contents.
# ## Genre
# find genres of movies and tv shows
nf["listed_in"].value_counts()
# with the commma, we can split the genres
nf["listed_in"].str.split(",").explode().value_counts()
# find the number of movies and tv shows by genre
nf["listed_in"].str.split(",").explode().value_counts()
# remove the space in front of the genre
nf["listed_in"].str.split(",").explode().str.strip().value_counts()
# find the number of movies and tv shows by genre
nf["listed_in"].str.split(",").explode().str.strip().value_counts()
# remove if there is TV Shows, Movies, Tv in the genre
nf["listed_in"].str.split(",").explode().str.strip().str.replace(
"TV Shows", ""
).str.replace("Movies", "").str.replace("TV", "").value_counts()
# remove space in front of the genre
nf["listed_in"].str.split(",").explode().str.strip().str.replace(
"TV Shows", ""
).str.replace("Movies", "").str.replace("TV", "").str.strip().value_counts()
# drop blank genre
nf["listed_in"].str.split(",").explode().str.strip().str.replace(
"TV Shows", ""
).str.replace("Movies", "").str.replace("TV", "").str.strip().replace(
"", np.nan
).dropna().value_counts()
# make graph of the number of movies and tv shows by genre with plotly
# opposite color
fig = px.bar(
nf["listed_in"]
.str.split(",")
.explode()
.str.strip()
.str.replace("TV Shows", "")
.str.replace("Movies", "")
.str.replace("TV", "")
.str.strip()
.replace("", np.nan)
.dropna()
.value_counts(),
color=nf["listed_in"]
.str.split(",")
.explode()
.str.strip()
.str.replace("TV Shows", "")
.str.replace("Movies", "")
.str.replace("TV", "")
.str.strip()
.replace("", np.nan)
.dropna()
.value_counts(),
color_continuous_scale=netflix_palette[::-1],
)
fig.update_layout(title_text="Number of Movies and TV Shows by Genre")
fig.update_layout(plot_bgcolor="rgba(0,0,0,0)")
# x axis label
fig.update_xaxes(title_text="Genre")
fig.show()
# International Movies are the most popular genre
# graph of different genres by release year
fig = px.line(
nf.groupby("release_year").sum().reset_index(),
x="release_year",
y=nf.groupby("release_year").sum().reset_index().columns[1:],
)
fig.update_layout(title_text="Number of Movies and TV Shows by Release Year by Genre")
# remove background
fig.update_layout(plot_bgcolor="rgba(0,0,0,0)")
# x axis name
fig.update_xaxes(title_text="Release Year")
# y axis name
fig.update_yaxes(title_text="Number of Movies and TV Shows")
fig.show()
# Similar trend of release and added contents
# ## Find Genre trend
# group by release year and find the different genres count
nf.groupby("release_year").sum().reset_index()
# group by release year and find the average genre
nf.groupby("release_year").sum().reset_index()
# make graph of the average genre by release year just 10 years
fig = px.line(
nf.groupby("release_year").sum().reset_index().iloc[-10:],
x="release_year",
y=nf.groupby("release_year").sum().reset_index().iloc[-10:].columns[1:],
)
fig.update_layout(title_text="Sum of Genre by Release Year")
# remove background
fig.update_layout(plot_bgcolor="rgba(0,0,0,0)")
# x axis name
fig.update_xaxes(title_text="Release Year")
# y axis name
fig.update_yaxes(title_text="Sum of Genre")
fig.show()
# International Movies are most popular genre in 10 years, never changed
# group by year added and find the average genre exclude release year
nf.groupby("year_added").sum().reset_index().drop("release_year", axis=1)
# make graph of the average genre by year added just 10 years
fig = px.line(
nf.groupby("year_added")
.sum()
.reset_index()
.iloc[-10:]
.drop("release_year", axis=1),
x="year_added",
y=nf.groupby("year_added")
.sum()
.reset_index()
.iloc[-10:]
.drop("release_year", axis=1)
.columns[1:],
)
fig.update_layout(title_text="Sum of Genre by Year Added")
# remove background
fig.update_layout(plot_bgcolor="rgba(0,0,0,0)")
# x axis name
fig.update_xaxes(title_text="Year Added")
# y axis name
fig.update_yaxes(title_text="Sum of Genre")
fig.show()
# Sum of Genres by add year is also similar, but seems they updated their contents a lot after 2015
# (Maybe their databse is after 2015)
# ## Rating Categories
# count the number of ratings
nf["rating"].value_counts()
# excclude last 3 ratings
nf["rating"].value_counts()[:-3]
# make graph of the number of ratings
fig = px.bar(
nf["rating"].value_counts()[:-3],
color=nf["rating"].value_counts()[:-3],
color_continuous_scale=netflix_palette[::-1],
)
fig.update_layout(title_text="Number of Ratings")
fig.update_layout(plot_bgcolor="rgba(0,0,0,0)")
# x axis label
fig.update_xaxes(title_text="Rating")
fig.show()
# ## Find rating trend
# make column TV-MA', 'TV-14', 'TV-PG', 'R', 'PG-13', 'TV-Y7', 'TV-Y', 'PG', 'TV-G','NR', 'G', 'TV-Y7-FV', 'No Rating', 'NC-17', 'UR' and if is same with the rating, then 1, else 0
nf["TV-MA"] = np.where(nf["rating"] == "TV-MA", 1, 0)
nf["TV-14"] = np.where(nf["rating"] == "TV-14", 1, 0)
nf["TV-PG"] = np.where(nf["rating"] == "TV-PG", 1, 0)
nf["R"] = np.where(nf["rating"] == "R", 1, 0)
nf["PG-13"] = np.where(nf["rating"] == "PG-13", 1, 0)
nf["TV-Y7"] = np.where(nf["rating"] == "TV-Y7", 1, 0)
nf["TV-Y"] = np.where(nf["rating"] == "TV-Y", 1, 0)
nf["PG"] = np.where(nf["rating"] == "PG", 1, 0)
nf["TV-G"] = np.where(nf["rating"] == "TV-G", 1, 0)
nf["NR"] = np.where(nf["rating"] == "NR", 1, 0)
nf["G"] = np.where(nf["rating"] == "G", 1, 0)
nf["TV-Y7-FV"] = np.where(nf["rating"] == "TV-Y7-FV", 1, 0)
nf["No Rating"] = np.where(nf["rating"] == "No Rating", 1, 0)
nf["NC-17"] = np.where(nf["rating"] == "NC-17", 1, 0)
nf["UR"] = np.where(nf["rating"] == "UR", 1, 0)
nf.head(5)
# yearly count of each rating, TV-MA', 'TV-14', 'TV-PG', 'R', 'PG-13', 'TV-Y7', 'TV-Y', 'PG', 'TV-G','NR', 'G', 'TV-Y7-FV', 'No Rating', 'NC-17', 'UR'
nf_rating_ya = (
nf.groupby("year_added")[
"TV-MA",
"TV-14",
"TV-PG",
"R",
"PG-13",
"TV-Y7",
"TV-Y",
"PG",
"TV-G",
"NR",
"G",
"TV-Y7-FV",
"No Rating",
"NC-17",
"UR",
]
.sum()
.reset_index()
)
nf_rating_ry = (
nf.groupby("release_year")[
"TV-MA",
"TV-14",
"TV-PG",
"R",
"PG-13",
"TV-Y7",
"TV-Y",
"PG",
"TV-G",
"NR",
"G",
"TV-Y7-FV",
"No Rating",
"NC-17",
"UR",
]
.sum()
.reset_index()
)
nf_rating_ry
# make graph of the average rating by release year just 10 years by nf_rating_ry
fig = px.line(
nf_rating_ry.iloc[-10:], x="release_year", y=nf_rating_ry.iloc[-10:].columns[1:]
)
fig.update_layout(title_text="Sum of Rating by Release Year")
# remove background
fig.update_layout(plot_bgcolor="rgba(0,0,0,0)")
# x axis name
fig.update_xaxes(title_text="Release Year")
# y axis name
fig.update_yaxes(title_text="Average Rating")
fig.show()
# make graph of the average rating by release year just 10 years by nf_rating
fig = px.line(
nf_rating_ya.iloc[-10:], x="year_added", y=nf_rating_ya.iloc[-10:].columns[1:]
)
fig.update_layout(title_text="Sum of Rating by Year Added")
# remove background
fig.update_layout(plot_bgcolor="rgba(0,0,0,0)")
# x axis name
fig.update_xaxes(title_text="Year Added")
# y axis name
fig.update_yaxes(title_text="Average Rating")
fig.show()
|
# # Exploratory Data Analysis (EDA)
# **Input**: Clean data stored in PostgreSQL
# **Output**: At the end of this phase we will have explored the data and have the knowledge for selecting useful features in the next step.
# First thing, we are going to answer the following questions:
# 1. How is data distributed?
# 2. How should we manage outliers?
# 3. Should we transform data?
# 4. How are features correlated?
# ## How is data distributed?
# We retrieve data from PostgreSQL created in the "Data Cleaning" step and pivot it to make it easy to analyse and plot.
# To do this we use a helper function that simply requests data from DB with a filter on dates between 1960-01-01 and 2022-12-31.
import psycopg2
from kaggle_secrets import UserSecretsClient
user_secrets = UserSecretsClient()
pg_db = user_secrets.get_secret("pg_db")
pg_host = user_secrets.get_secret("pg_host")
pg_pass = user_secrets.get_secret("pg_pass")
pg_user = user_secrets.get_secret("pg_user")
def get_connection():
"""Connect to the PostgreSQL database server"""
conn = None
try:
# connection parameters
params = {
"database": pg_db,
"user": pg_user,
"password": pg_pass,
"host": pg_host,
}
# connect to the PostgreSQL server
conn = psycopg2.connect(**params)
return conn
except (Exception, psycopg2.DatabaseError) as error:
print("CONNECTION ERROR: ", error)
if conn is not None:
conn.close()
def get_df_from_table(tablename, where=";"):
"""Read table from DB and convert it to pandas DataFrame"""
conn = get_connection()
cur = conn.cursor()
q = f"""SELECT * FROM {tablename} {where}"""
cur.execute(q)
data = cur.fetchall()
cols = []
for elt in cur.description:
cols.append(elt[0])
df = pd.DataFrame(data=data, columns=cols)
cur.close()
return df
# We load the table "indicator" containing cleaned data and the table "indicator_name" containing reference to features and target names.
# This allows us to create aliases for titles and legends in charts.
from datetime import date
import pandas as pd
def get_df():
# We get dataFrame from table "indicator"
df = get_df_from_table(
"indicator",
f"where date between '{date(1960, 1, 1)}' and '{date(2022, 12, 31)}'",
)
df = df.drop("id", axis=1)
# We get dataFrame from table "indicator_name",
# we do this to get labels for features and target
name_df = get_df_from_table("indicator_name")
df = pd.merge(df, name_df, left_on=["name"], right_on=["indicator"], how="inner")
df = df.drop("indicator", axis=1)
name_df = name_df.set_index("id")
# We select which indicators are features and which are targets
df["target_feature"] = "feature"
df.loc[
df["source"].isin(["yahoo_finance", "investing"]), "target_feature"
] = "target"
df.loc[
df["name"].isin(
[
"USSTHPI",
"Short-term interest rates | Total | % per annum",
"Long-term interest rates | Total | % per annum",
"Housing prices | Nominal house prices | 2015=100",
]
),
"target_feature",
] = "target"
df["column_name"] = df["target_feature"] + df["id"].astype(str)
# Converting dataFrame values to datetime and numeric
df["date"] = pd.to_datetime(df["date"])
df["value"] = pd.to_numeric(df["value"])
# Pivot df in order to have features and targets as columns and rows as dates.
df = df.pivot_table(
index="date", columns="column_name", values="value", aggfunc="sum"
).reset_index()
df = df.set_index("date")
return df, name_df
df, name_df = get_df()
df
# We now have a dataframe with 217 columns representing all features and targets.
# To answer the question **How is data distributed?** we are going to plot data in three different ways:
# - A line chart showing actual data across time
# - A histogram showing data distribution
# - A box and whisker chart showing the same data distribution with outliers
# Import plot library
import matplotlib.pyplot as plt
def plot_column(col):
# retrieve title from column name
id = int(col.replace("target", "").replace("feature", ""))
title = name_df.loc[id, "indicator"]
# get series description (count, mean, std and quartiles)
d = df[col].describe()
# To plot everything together we use a figure with 3 columns and 1 row.
fig = plt.figure(figsize=(17, 6))
fig.suptitle(title)
# In the first (left) chart we plot the raw data, and how is distributed in time.
line = fig.add_subplot(131)
line = df[col].plot(kind="line", grid=False)
# In the second (center) chart we want to see an histogram representing the distribution of values.
# to better identify key values we draw key lines on this chart.
hist = fig.add_subplot(132)
hist = df[col].plot(kind="hist", grid=False)
hist.axvline(x=d["mean"], color="r", linestyle="--", lw=2)
hist.axvline(x=d["50%"], color="g", linestyle="--", lw=2)
hist.axvline(x=d["mean"] - d["std"], color="b", linestyle="--", lw=1)
hist.axvline(x=d["mean"] + d["std"], color="b", linestyle="--", lw=1)
hist.axvline(x=d["mean"] - 2 * d["std"], color="b", linestyle="--", lw=2)
hist.axvline(x=d["mean"] + 2 * d["std"], color="b", linestyle="--", lw=2)
# Then in the third (right) chart we want to see a box and whisker that help use identify outliers.
box = fig.add_subplot(133)
box = df[col].plot(kind="box", grid=False)
print(d)
# selecting column 0 as an example, we will plot every column to see how data is distributed.
plot_column(df.columns[0])
plt.show()
# ## How should we manage outliers?
# To loop through columns faster, we decided to only plot those whose data have a certain % of outliers.
# We define an outlier with the following conditions:
# - values that are lower than first quartile minus the interquartile range multiplied by 1.5
# - values that are greater than third quartile plus the interquartile range multiplied by 1.5
def count_outliers(col):
d = df[col].describe()
interquartile_range = d["75%"] - d["25%"]
min_threshold = d["25%"] - interquartile_range * 1.5
max_threshold = d["75%"] + interquartile_range * 1.5
negative_outliers_count = df[df[col] < min_threshold][col].count()
negative_percentage = round(negative_outliers_count / d["count"] * 100, 2)
positive_outliers_count = df[df[col] > max_threshold][col].count()
positive_percentage = round(positive_outliers_count / d["count"] * 100, 2)
print(f"NEGATIVE OUTLIERS: {negative_outliers_count} ({negative_percentage}%)")
print(f"POSITIVE OUTLIERS: {positive_outliers_count} ({positive_percentage}%)")
count_outliers(df.columns[1])
# We analized all series with at least 5% of "positive" or "negative" outliers and took the following considerations:
# - To remove TOTBORR (feature92) *Total Borrowings of Depository Institutions from the Federal Reserve* because it is almost always 0 except for 2 time periods. https://fred.stlouisfed.org/series/TOTBORR
# - To remove BORROW (feature165) *Total Borrowings from the Federal Reserve*, for the same reason. https://fred.stlouisfed.org/series/BORROW
# - To remove NONBORRES (feature169) *Reserves of Depository Institutions, Nonborrowed*. Values are 0 until 2008. https://fred.stlouisfed.org/series/NONBORRES
# - To remove TOTRESNS (feature199) *Reserves of Depository Institutions: Total*, for the same reason. https://fred.stlouisfed.org/series/TOTRESNS
# - To remove TREASURY (feature78) *Treasury Deposits with Federal Reserve Banks*, for the same reason. https://fred.stlouisfed.org/series/TREASURY
# - To remove series with missing data before 1995, like USCI (feature262) and other series
# - To remove series with no data, like M0263AUSM500NNBR (feature85) *Existing Home Mortgage Applications for United States*, https://fred.stlouisfed.org/series/M0263AUSM500NNBR and other series.
# Below some examples with charts of the mentioned series.
f = "feature92"
plot_column(f)
count_outliers(f)
plt.show()
f = "feature169"
plot_column(f)
count_outliers(f)
plt.show()
# ## Should we transform data?
# Looping through all series we also identified some that need specific transformation:
# - Business confidence index (feature10), Consumer confidence index (feature13) from OECD need to be normalized/standardized.
# - JHGDPBRINDX (feature107) *GDP-Based Recession Indicator Index* needs to be normalized/standardized too. https://fred.stlouisfed.org/series/JHGDPBRINDX
# - Normalize/standardized series with "spiky" series, like MTSDS133FMS (feature121) *Federal Surplus or Deficit*. https://fred.stlouisfed.org/series/MTSDS133FMS, and other series.
# In general it seems that features needs to be normalized/standardized as they have very different value ranges, we will approach this in the next step "Feature Selection".
f = "feature10"
plot_column(f)
count_outliers(f)
plt.show()
f = "feature121"
plot_column(f)
count_outliers(f)
plt.show()
# ## How are features correlated?
# Correlated feature aren't very useful since they give the same information and slow down the training process.
# Hence we decided to identify correlated series and mark which one to remove. In general we keep the ones that are higher correlated with targets (and have more data).
# We loop through all features, calculate the correlation, then if the correlation is greater than a certain threshold we decide which one to remove by counting the number of datapoints and evaluating the correlation with all the targets.
# First we update the get_df method to remove the outliers
def get_df(remove_outliers=True):
# We get dataFrame from table "indicator"
df = get_df_from_table(
"indicator",
f"where date between '{date(1960, 1, 1)}' and '{date(2022, 12, 31)}'",
)
df = df.drop("id", axis=1)
# We get dataFrame from table "indicator_name", we do this to get labels for features and target
name_df = get_df_from_table("indicator_name")
df = pd.merge(df, name_df, left_on=["name"], right_on=["indicator"], how="inner")
df = df.drop("indicator", axis=1)
name_df = name_df.set_index("id")
# This new code remove the series with outliers we identified previously
if remove_outliers:
df = df[
~df["name"].isin(
[
"TOTBORR",
"BORROW ",
"TOTRESNS",
"NONBORRES ",
"^DJUSRE",
"^DJCI",
"USCI",
"CL=F",
"GC=F",
"^SP500BDT",
"TREASURY",
"DDDM03USA156NWDB",
"M0263AUSM500NNBR",
"M14062USM027NNBR",
"M0264AUSM500NNBR",
"M1490AUSM157SNBR",
"Quarterly GDP | Total | Percentage change",
"Quarterly GDP | Total | Percentage change",
"Q09084USQ507NNBR",
"M0263AUSM500NNBR",
"M0264AUSM500NNBR",
"M1490AUSM157SNBR",
"M14062USM027NNBR",
"M09075USM476NNBR",
"Q09084USQ507NNBR",
"M09086USM156NNBR",
"DDDM03USA156NWDB",
"DDDM01USA156NWDB",
"DDEM01USA156NWDB",
"LABSHPUSA156NRUG",
"RTFPNAUSA632NRUG",
"SIPOVGINIUSA",
"DDDI06USA156NWDB",
"ITNETUSERP2USA",
"Electricity generation | Total | Gigawatt-hours",
]
)
]
# We select which indicators are features and which are targets
df["target_feature"] = "feature"
df.loc[
df["source"].isin(["yahoo_finance", "investing"]), "target_feature"
] = "target"
df.loc[
df["name"].isin(
[
"USSTHPI",
"Short-term interest rates | Total | % per annum",
"Long-term interest rates | Total | % per annum",
"Housing prices | Nominal house prices | 2015=100",
]
),
"target_feature",
] = "target"
df["column_name"] = df["target_feature"] + df["id"].astype(str)
# Convert dataFrame values to datetime and numeric
df["date"] = pd.to_datetime(df["date"])
df["value"] = pd.to_numeric(df["value"])
# Pivot df in order to have features and targets as columns and rows as dates.
df = df.pivot_table(
index="date", columns="column_name", values="value", aggfunc="sum"
).reset_index()
df = df.set_index("date")
return df, name_df
df, name_df = get_df()
df
# ### Stationarity
# Time series forecasting requires extra preprocessing steps, unlike ordinary machine learning problems.
# Most ML algorithms expect a static relationship between the input features and the output, a static relationship requires inputs and outputs with constant parameters such as mean, median, and variance. In other words, algorithms perform best when the inputs and outputs are stationary.
# Time series are non-stationary data. Meaning, distributions change over time and can have properties such as seasonality and trend. These, cause the mean and variance of the series to change over time, making it hard to model their behaviour.
# First we are going to test non-stationarity of our series. We can do this with some tests. The one we have chosen is *Augmented Dickey-Fuller unit root test*.
# **Null hypothesis**: the series is non-stationary, time-dependent (it has a unit root).
# **Alternative hypothesis**: the series is already stationary, not time-dependent (can't be represented by a unit root)
# The ADFuller test will return a p-value for the time series. If it is smaller than a critical threshold of 0.05 or 0.01, we reject the null hypothesis and conclude that the series is stationary. Otherwise, we fail to reject the null hypothesis and conclude the series is non-stationary.
from statsmodels.tsa.stattools import adfuller
def get_stationarity(df, col):
col_df = df[col].dropna()
id = int(col.replace("target", "").replace("feature", ""))
title = name_df.loc[id, "indicator"]
# rolling statistics
# if rolling mean and/or std move over time (line is not horizontal) data is non-stationary.
# non-stationary = value does depend on date
rolling_mean = col_df.rolling(window=12).mean()
rolling_std = col_df.rolling(window=12).std()
# Dickey–Fuller test
# if p-value is > 0.05 we can conclude data is non-stationary.
# non-stationary = value does depend on date
result = adfuller(col_df)
print("Adfuller p-value: {}".format(result[1]))
# rolling statistics plot
fig = plt.figure(figsize=(12, 6))
fig.suptitle(title)
original = plt.plot(col_df, color="blue", label="Original")
plt.xlabel("date")
mean = plt.plot(rolling_mean, color="red", label="Rolling Mean")
std = plt.plot(rolling_std, color="black", label="Rolling Std")
plt.legend(loc="best")
# We use as example feature77 that represents GDP
col = "feature77"
get_stationarity(df, col)
plt.show()
# We can see that p-value is 1, hence this series is surely time-dependent.
# To make it stationary we are going to use a simple method.
# Differencing data of a time series we can reach stationarity. Meaning we subtract for each data point the previous value. Doing so we will have for each date the difference from the previous date instead of the actual value.
# Pandas dataframe has a function to do exactly that.
stationary_df = df.diff(periods=1)
stationary_df
# Again we use as example feature77 that represents GDP, to see if differencing reduced p-value and gave us a stationary series
col = "feature77"
get_stationarity(stationary_df, col)
plt.show()
# As we can see from the p-value, differencing data with the previous period give us a stationary series.
# With this new dataframe we are going look for correlation among features.
# This step is important since we want features that are truly correlated between them. Otherwise, we could get high correlation because both features are highly correlated with a 3rd variable (time). Removing the time variable was necessary to avoid this situation.
# Below an example of how we calculate the correlation between GDP (feature77) and Saving rate (feature1).
# To see how results change first we are going to check correlation before and after the stationarity trasformation.
from sklearn.linear_model import LinearRegression
from matplotlib.offsetbox import AnchoredText
def get_correlation(df, col1, col2, ax):
title1 = name_df.loc[
int(col1.replace("target", "").replace("feature", "")), "indicator"
]
title2 = name_df.loc[
int(col2.replace("target", "").replace("feature", "")), "indicator"
]
col_df = df[[col1, col2]].dropna()
# To evaluate correlation between the two features
corr = col_df[col1].corr(col_df[col2])
# Get line of best fit
linear = LinearRegression()
linear.fit(col_df[[col1]], col_df[[col2]])
ax.scatter(col_df[col1], col_df[col2])
ax.plot(col_df[col1], linear.predict(col_df[[col1]]), color="red")
ax.axvline(x=0, ymin=-1, ymax=1, linestyle="dashed", color="gray")
ax.axhline(y=0, xmin=-1, xmax=1, linestyle="dashed", color="gray")
ax.set_xlabel(title1)
ax.set_ylabel(title2)
at = AnchoredText(
f"{corr:.0%}",
prop=dict(size="large"),
frameon=True,
loc="lower right",
)
at.patch.set_boxstyle("square, pad=0.0")
ax.add_artist(at)
col1 = "feature77" # GDP
col2 = "feature1" # Saving rate
fig = plt.figure(figsize=(12, 6))
fig.suptitle("GDP - Saving rate Correlation")
ax1 = fig.add_subplot(121)
get_correlation(df, col1, col2, ax1)
ax2 = fig.add_subplot(122)
get_correlation(stationary_df, col1, col2, ax2)
plt.show()
# On the left chart, features are highly correlated (-70%). But this is due to the fact that both are correlated with time.
# On the right chart, we removed the time dependency and we can see that they are not correlated at all (4%).
# To idenfity all possible correlated feature pairs we looped through all features in the dataframe.
# Then, when a pair with high correlation is found (we used 80% as threshold with satisfying results), we evaluate the number of data points and the correlation with the target seriess.
# Features correlation are saved in MongoDB within a new document called *feature_correlation_0.8* in the *feature_selection* collection.
# This document contains one element for each pair with correlation higher than 80%, containing the number of data points for each of 2 features and the absolute mean correlation with the targets for each of the 2 features. To calculate the absolute mean correlation with the target we took the correlation of that feature with each target, and calculate the mean of the absolute values (we are interest in the magnitude of the correlation, not the sign).
# We have a total of 139 feature pairs.
feature_correlation = {
"_id": "feature_correlation_0.8",
"data": [
{
"col_1": "feature100",
"col_2": "feature143",
"corr": 0.9208957065068195,
"c_1": 750,
"c_2": 750,
"corr_1": 0.11212307319417723,
"corr_2": 0.11630831606931778,
},
{
"col_1": "feature102",
"col_2": "feature118",
"corr": 0.9258819460661312,
"c_1": 755,
"c_2": 755,
"corr_1": 0.07906746378736886,
"corr_2": 0.07908686817741475,
},
...,
],
}
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import seaborn as sns
import matplotlib.pyplot as plt
from matplotlib.ticker import EngFormatter
import warnings
warnings.filterwarnings("ignore")
pd.set_option("display.max_columns", 100)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Dataset History
# A retail company “ABC Private Limited” wants to understand the customer purchase behavior (specifically, purchase amount) against various products of different categories. They have shared purchase summaries of various customers for selected high-volume products from last month.
# The data set also contains customer demographics (age, gender, marital status, city type, stayincurrentcity), product details (productid and product category), and Total purchase amount from last month.
# Now, they want to build a model to predict the purchase amount of customers against various products which will help them to create a personalized offer for customers against different products.
sales_data = pd.read_csv("/kaggle/input/black-friday-sale/train.csv")
sales_data.shape
sales_data.head()
sales_data_test = pd.read_csv("/kaggle/input/black-friday/test.csv")
# ## EDA
# * Look for numeric and categorical features
# * Univariate analysis of categorical features
# * Bivariate analysis with respect to target variable (Purchase)
#
sales_data.info()
# **numeric features:**
# > 1. Purchase (target)
# **categorical features:**
# > 1. Gender
# 2. Age
# 3. Occupation
# 4. City_Category
# 5. Stay_In_Current_City_Years
# 6. Marital_Status
#
fmt_obj = EngFormatter()
sales_data.describe()
sns.set_theme(style="whitegrid")
fig, ax = plt.subplots(figsize=(15, 8))
sns.boxplot(data=sales_data, x="Purchase")
"""Shape of DataFrame"""
print(
f"""Number of rows: {sales_data.shape[0]:,}
Number of columns: {sales_data.shape[1]:,} """
)
sales_data.columns
categorical_columns = [
"Gender",
"Age",
"Occupation",
"City_Category",
"Stay_In_Current_City_Years",
"Marital_Status",
"Product_Category_1",
"Product_Category_2",
"Product_Category_3",
]
# Univariate analysis : Distribution of categorical data in their corresponding buckets.
for cat_col in categorical_columns:
fig, ax = plt.subplots(figsize=(15, 8))
sns.countplot(data=sales_data, x=cat_col, ax=ax).set(title=f"{cat_col} buckets")
ax.yaxis.set_major_formatter(fmt_obj)
""" Null values in the dataset"""
sns.heatmap(sales_data.isnull(), yticklabels=False, cbar=True)
sales_data.isnull().sum().reset_index()
""" Null values in the dataset"""
fig, ax = plt.subplots(figsize=(15, 8))
sns.barplot(data=sales_data.isnull().sum().reset_index(), x="index", y=0)
ax.yaxis.set_major_formatter(fmt_obj)
sales_data.groupby("Age").agg({"Purchase": sum}).reset_index()
# Bivariate analysis
for cat_col in categorical_columns:
fig, ax = plt.subplots(figsize=(15, 8))
temp = sales_data.groupby(cat_col).agg({"Purchase": sum}).reset_index()
sns.barplot(data=temp, x=cat_col, y="Purchase", ax=ax).set(
title=f"{cat_col} Vs Purchase"
)
ax.yaxis.set_major_formatter(fmt_obj)
# **OBSERVATIONS**
# > 1. Number of different segment people visited the store is directly proportional to purchase amount
# 2. Ex: No:of men visited the store is greater than women hence the total purchased value is also greater than women.
# 3. This observation holds true for all the segments of people visited the store.
sales_data.columns
"""Commparision of different categorical variables with product_category purchased"""
for cat_col in categorical_columns:
fig, ax = plt.subplots(figsize=(17, 10))
sns.countplot(data=sales_data, x=cat_col, hue="Product_Category_1")
ax.yaxis.set_major_formatter(fmt_obj)
# **OBSERVATIONS**
# > 1. In all the segments the product category that was sold highest is evenly distributed .
# 2. This implies that the product categories that got sold are all equally scaled leading to category 1,5, and 8 as the top 3 sold in all .
# ## Feature Engineering
# **Feature Engineering Techniques for Machine Learning**
# > 1. Imputation
# > > * Numerical Imputation
# > > * Categorical Imputation
# 2. Handling Outliers
# 3. Log Transform
# 4. Encoding
# > > * label
# > > * one-hot
# 5. Scaling
# > > * Normalization
# > > * Standardization
import sklearn.preprocessing as sk_preprocess
# ### sampling to create validation set
sales_data
""" Remove unwanted columns"""
sales_data.drop(columns=["User_ID", "Product_ID"], inplace=True)
validation = sales_data.sample(frac=0.3, replace=False, random_state=1)
x_train = sales_data.drop(validation.index, axis=0)
x_train.reset_index(inplace=True, drop=True)
x_val = validation.reset_index(drop=True)
y_train = x_train["Purchase"]
y_val = x_val["Purchase"]
x_train.drop(columns=["Purchase"], inplace=True)
x_val.drop(columns=["Purchase"], inplace=True)
print(f"Shape of x_train {x_train.shape[0]:,} rows ,{x_train.shape[1]} cols")
print(f"Shape of y_train {y_train.shape[0]:,} rows")
print(f"Shape of x_val {x_val.shape[0]:,} rows,{x_val.shape[1]} cols")
print(f"Shape of y_val {y_val.shape[0]:,} rows")
class feature_engineering:
def __init__(self, training_data=x_train):
self.prod_cat2_mode = x_train["Product_Category_2"].mode().item()
self.prod_cat3_mode = x_train["Product_Category_3"].mode().item()
self.std_scaler = sk_preprocess.StandardScaler()
# ENCODING
def do_encoding(self, df):
"""Encoding Gender"""
df["Gender"] = df["Gender"].map({"F": 0, "M": 1})
""" Encoding Age"""
df["Age"] = df["Age"].map(
{
"0-17": 1,
"18-25": 2,
"26-35": 3,
"36-45": 4,
"46-50": 5,
"51-55": 6,
"55+": 7,
}
)
"""Encoding City_Category"""
city_cats = pd.get_dummies(df["City_Category"], drop_first=True)
df = pd.concat([df, city_cats], axis=1)
df.drop(columns=["City_Category"], inplace=True)
"""Handling Stay_In_Current_City_Years """
df["Stay_In_Current_City_Years"].unique()
df["Stay_In_Current_City_Years"] = (
df["Stay_In_Current_City_Years"].replace("4+", "4").astype(int)
)
return df
# NULL IMPUTATION
def fillna_with_mode(self, df):
df["Product_Category_2"] = df["Product_Category_2"].fillna(self.prod_cat2_mode)
df["Product_Category_3"] = df["Product_Category_3"].fillna(self.prod_cat3_mode)
return df
# FEATURE SCALING
def scaling(self, df, train):
cols = df.columns
if train:
print("Fitting train for scaling")
self.std_scaler.fit(df)
df = self.std_scaler.transform(df)
df = pd.DataFrame(df, columns=cols)
return df
# MAIN_FUNCTION
def do_all(self, df, train=False):
df = self.do_encoding(df=df.copy())
df = self.fillna_with_mode(df=df.copy())
df_ = self.scaling(df, train=train)
if train:
return df, df_
return df
def before_vs_after_scaling(df, df_scaled):
df_scaled.columns = df.columns
no_of_columns = df.shape[1]
fig, ax = plt.subplots(no_of_columns, 2, figsize=(30, 60))
col_list = df.columns
for col, col_name in zip(range(no_of_columns), col_list):
sns.histplot(data=df[col_name], kde=True, ax=ax[col, 0])
ax[col, 0].set_title(f"Non scaled {col_name} distribution")
sns.histplot(data=df[col_name], kde=True, ax=ax[col, 1])
ax[col, 1].set_title(f"Scaled {col_name} distribution")
feat_eng = feature_engineering(training_data=x_train.copy())
non_scaled_x_train, x_train = feat_eng.do_all(df=x_train.copy(), train=True)
x_val = feat_eng.do_all(df=x_val.copy())
""" COMPARISION BETWEEN NON SCALED AND SCALED COLUMNS"""
before_vs_after_scaling(df=non_scaled_x_train, df_scaled=x_train)
x_train.corr()
correlation_matrix = pd.concat([x_train, y_train], axis=1).corr()
# #### Heatmap to get an overview of features correlating
fig, ax = plt.subplots(figsize=(15, 10))
sns.heatmap(data=correlation_matrix, annot=True, cmap="hsv")
# ### Modelling:
# 1. LR
# 2. LR - ridge
# 3. descision tree regression
# 4. Random forest regression
# 5. Knn regression
# 6. Ada boost reg
# 7. gradient boost reg
# 8. Xgb reg
# 9. light bgm reg
from sklearn.linear_model import LinearRegression
from sklearn.metrics import mean_squared_error
lr_obj = LinearRegression()
lr_obj.fit(x_train, y_train)
lr_obj.coef_
lr_obj.intercept_
lr_obj.get_params()
train_pred = lr_obj.predict(x_train)
# Train score
print(f"Regular R2 score on train data is : {lr_obj.score(x_train,y_train):%}")
print(f"Regular R2 score on validation data is : {lr_obj.score(x_val,y_val):%}")
mean_squared_error(y_train, train_pred)
from sklearn.linear_model import Ridge
|
import pandas as pd
import progressbar as pb
import random
data = pd.read_csv("/kaggle/input/movie-reviews/train.csv")
data = data[:10_000]
data.head()
c_data = pd.DataFrame()
movies = []
c_data["id"] = 0
for movie in pb.progressbar(data["movie"]):
if movie not in movies:
movies.append(movie)
c_data[movie] = 0
users = []
for user in pb.progressbar(data["user"]):
if user not in users:
users.append(user)
append_dic = {"id": user}
for column in c_data.columns:
if column != "id":
append_dic[column] = 0
c_data = c_data.append(pd.DataFrame([append_dic]))
c_data = c_data.set_index("id")
for index in pb.progressbar(range(len(data) - 1)):
c_data.loc[data["user"][index], data["movie"][index]] = data["rating"][index]
c_data
c_data.to_csv("converted.csv", index=None, header=True)
|
# # Import Libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from wordcloud import WordCloud, STOPWORDS
from collections import Counter
# # Data Loading
train = pd.read_csv(
"/kaggle/input/jigsaw-toxic-comment-classification-challenge/train.csv.zip"
)
test = pd.read_csv(
"/kaggle/input/jigsaw-toxic-comment-classification-challenge/test.csv.zip"
)
test_y = pd.read_csv(
"/kaggle/input/jigsaw-toxic-comment-classification-challenge/test_labels.csv.zip"
)
print("train shape:", train.shape)
train.head(10)
# * There seems to be comments that doesn't have any tags. They may be clean comments
# * we can see that the 6th message belongs to 4 classes
print("test shape:", test.shape)
test.head()
test_y.head()
# # Exploratory data analysis
# Since there are many comments that are not labelled in any of the six categories, lets mark it as "clean"
# marking comments without any tags as "clean"
tag_sums = train.iloc[:, 2:].sum(axis=1)
train["clean"] = tag_sums == 0
print("Check for missing values in Train dataset")
null_train = train.isnull().sum()
print(null_train)
print("Check for missing values in Train dataset")
null_test = test.isnull().sum()
print(null_test)
# Ok, No null values
# Now lets see some of the comments...
# example of clean comment
train["comment_text"][0]
# example of toxic comment
train[train.toxic == 1].iloc[1, 1]
# example of identity_hate comment
train[train.identity_hate == 1].iloc[1, 1]
# just a random comment
train["comment_text"][157718]
# Yuck, so much hate
# ## Class Distribution
label_count = train[train.columns[2:]].sum()
label_count
plt.figure(figsize=(8, 4))
sns.barplot(
x=label_count.index, y=label_count.values, palette=sns.color_palette("Reds")
)
plt.xticks(rotation=90)
plt.title("Class distribution", fontsize=12)
plt.show()
# Clearly there is class imbalance. We can see that **clean** has the most observations while **threat** has least observations. Also there are comments that belong to multiple classes.
# ## Length Distribution
comment_len = train.comment_text.str.len()
# plot the distribution of comment lengths
plt.figure(figsize=(8, 4))
sns.histplot(comment_len, kde=False, bins=50, color="red")
plt.xlabel("Comment Length (Number of words)", fontsize=12)
plt.ylabel("Number of Comments", fontsize=12)
plt.title("Distribution of comment Lengths", fontsize=12)
# Most of the comments fall between the range of 0-500. We have few comments that go beyond 500 words.
# Let's see the distribution of the comment lengths by each labels.
labels = ["toxic", "severe_toxic", "obscene", "threat", "insult", "identity_hate"]
fig, ax = plt.subplots(nrows=3, ncols=2, figsize=(15, 10), sharex=True)
axes = ax.ravel()
for i in range(6):
comments = train.loc[train[labels[i]] == 1, :]
comment_len = [len(comment.split()) for comment in comments["comment_text"]]
sns.histplot(comment_len, ax=axes[i], bins=50, color="red")
axes[i].title.set_text(labels[i])
# ## Wordclouds
# Represent text data in which the size of each word indicates its frequency or importance.
# clean words
subset = train[train.clean == True]
text = " ".join(i for i in subset.comment_text)
stopwords = set(STOPWORDS)
wordcloud = WordCloud(stopwords=stopwords, colormap="Greens").generate(text)
plt.figure(figsize=(8, 4))
plt.imshow(wordcloud, interpolation="bilinear")
plt.axis("off")
plt.title("Words frequented in Clean Comments", fontsize=20)
plt.show()
stopwords = set(STOPWORDS)
for l in labels:
subset = train[train[l] == 1]
text = " ".join(i for i in subset.comment_text)
wordcloud = WordCloud(
stopwords=stopwords, max_words=200, max_font_size=100, colormap="Reds"
).generate(text)
fig = plt.figure(figsize=(8, 4))
plt.title(l)
plt.imshow(wordcloud)
plt.axis("off")
plt.show()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv("/kaggle/input/titantic/train.csv")
display(df)
display(df.describe(include=[object]))
display(df.describe(include=[np.number]))
df["Age"].fillna(29.7, inplace=True)
df["Embarked"].fillna("S", inplace=True)
new = df.drop(columns=["Cabin", "Ticket", "PassengerId", "Name"])
missing_val_count_by_column = new.isnull().sum()
print(missing_val_count_by_column)
display(new)
from sklearn.preprocessing import MinMaxScaler
new[["Fare"]] = MinMaxScaler().fit_transform(new[["Fare"]])
display(new)
new["Gender"] = new["Sex"].replace({"male": 1, "female": 0})
display(new)
new1 = new.drop(columns=["Sex"])
display(new1)
new1["Depart"] = new1["Embarked"].replace({"S": 0, "C": 1, "Q": 2})
display(new1)
from sklearn.preprocessing import MinMaxScaler
new1[["Parch"]] = MinMaxScaler().fit_transform(new1[["Parch"]])
new2 = new1.drop(columns=["Embarked"])
display(new2)
new2[["Depart"]] = MinMaxScaler().fit_transform(new2[["Depart"]])
new2[["Pclass"]] = MinMaxScaler().fit_transform(new2[["Pclass"]])
new2[["Age"]] = MinMaxScaler().fit_transform(new2[["Age"]])
display(new2)
from sklearn.preprocessing import MinMaxScaler
new2[["SibSp"]] = MinMaxScaler().fit_transform(new2[["SibSp"]])
display(new2.describe(include=[np.number]))
from sklearn.model_selection import train_test_split
train, test = train_test_split(
new2, test_size=0.25, stratify=new2["Survived"], random_state=42
)
X_train = train[["Pclass", "Age", "SibSp", "Parch", "Fare", "Gender", "Depart"]]
Y_train = train.Survived
X_test = test[["Pclass", "Age", "SibSp", "Parch", "Fare", "Gender", "Depart"]]
Y_test = test.Survived
from sklearn.naive_bayes import MultinomialNB
from sklearn.neighbors import KNeighborsClassifier
from sklearn.svm import LinearSVC
from sklearn.tree import DecisionTreeClassifier
model_names = ["Naive Bayes", "K-Nearist Neighbors", "SVM", "Decision Tree"]
models = [
MultinomialNB(),
KNeighborsClassifier(),
LinearSVC(),
DecisionTreeClassifier(),
]
model_dict = dict(zip(model_names, models))
df = pd.DataFrame(columns=["Model", "accuracy", "precision", "recall", "f1"])
for modelkey, modelvalue in model_dict.items():
model = modelvalue.fit(X_train, Y_train)
Y_pred = model.predict(X_test)
metrics_list = compute_metrics(Y_pred, Y_test)
results = pd.DataFrame(
metrics_list, columns=["accuracy", "precision", "recall", "f1"], index=[0]
)
results["Type"] = "Classical Machine Learning"
results["Model"] = modelkey
results = results[["Type", "Model", "accuracy", "precision", "recall", "f1"]]
df = pd.concat([ML_results, results])
display(df)
|
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
from scipy import stats
from sklearn.metrics import (
explained_variance_score,
mean_absolute_error,
mean_squared_error,
median_absolute_error,
r2_score,
)
from sklearn.model_selection import train_test_split
import warnings
warnings.filterwarnings("ignore")
data = pd.read_excel("/kaggle/input/photovoltatic/Pv.xlsx", "Sayfa1")
data
x = data.iloc[:, :-1].values
y = data.iloc[:, -1].values
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.3, random_state=0)
print(stats.describe(x_train, axis=0))
print(np.std(x_train, axis=0))
print(stats.describe(x_train, axis=0))
print(np.std(x_test, axis=0))
print(stats.describe(y_train, axis=0))
print(np.std(y_train, axis=0))
print(stats.describe(y_train, axis=0))
print(np.std(y_test, axis=0))
# pip install pytorch_forecasting -U
from sklearn.preprocessing import MinMaxScaler
scaler = MinMaxScaler()
x_train = scaler.fit_transform(x_test)
x_test = scaler.fit_transform(x_test)
y_train = scaler.fit_transform(y_test.reshape(-1, 1))
y_test = scaler.fit_transform(y_test.reshape(-1, 1))
x.shape, x_train.shape, x_test.shape, x.ndim, y.ndim, x_train.ndim
y.shape, y_train.shape, y_test.shape
from sklearn.linear_model import LinearRegression
model_Regresyon = LinearRegression()
model_Regresyon.fit(x_test, y_train)
y_pred = model_Regresyon.predict(x_test)
import statsmodels.api as sm
model_regresyon_OLS = sm.OLS(endog=y_test, exog=x_train).fit()
model_regresyon_OLS.summary()
print("MAE=%0.2f" % mean_absolute_error(y_test, y_pred))
print("MSE=%0.2f" % mean_squared_error(y_test, y_pred))
print("MedAE=%0.2f" % mean_absolute_error(y_test, y_pred))
print("Belirleme Katysayısı(R2)=%0.2f" % r2_score(y_test, y_pred))
print("RMSE=%0.2f" % np.sqrt(mean_squared_error(y_test, y_pred)))
|
# **epoch**
# **batch size**
# **number of iterations**
import torch
import torchvision
from torch.utils.data import DataLoader
import numpy as np
import math
class Churn_dataset:
def __init__(self):
# constructor
xy = np.loadtxt(
"/kaggle/input/churn-numeric/Churn.csv",
delimiter=",",
dtype=np.float32,
skiprows=1,
)
# convert from numpy to torch
self.x = torch.from_numpy(xy[:, :2])
self.y = torch.from_numpy(xy[:, 2])
self.n_samples = xy.shape[0]
def __getitem__(self, index):
# allows object of a class to use indexing operator
return self.x[index], self.y[index]
def __len__(self):
return self.n_samples
dataset = Churn_dataset()
first_data = dataset[0]
features, labels = first_data
print(features, labels)
dataloader = DataLoader(dataset=dataset, batch_size=10, shuffle=True, num_workers=2)
dataiter = iter(dataloader)
data = next(dataiter)
features, labels = data
print(features, labels)
# Training loop
num_epochs = 3
total_samples = len(dataset)
n_iterations = total_samples / 10
print(total_samples, n_iterations)
for epoch in range(num_epochs):
for i, (features, labels) in enumerate(dataloader):
if (i + 1) % 200 == 0:
print(
f"epoch {epoch+1}/{num_epochs}, step {i+1}/{n_iterations}, features {features.shape}"
)
# **model accepts only numerical features**
import pandas as pd
df = pd.read_csv("/kaggle/input/churn-modelling/Churn_Modelling.csv")
df.columns
df.dtypes
df = df.select_dtypes(include=np.number)
df.head()
df = df.drop("CustomerId", axis=1)
df.dtypes
df.to_csv("churn_modelling_numerical.csv")
|
# ## Hello, world!
# [Read our blog about how & why this notebook was written.](https://medium.com/coinmonks/3-simple-moving-averages-trade-strategy-performance-test-2194f423d36b?source=user_profile---------1----------------------------)
from IPython.display import clear_output
from dateutil.tz import tzlocal
import matplotlib.pyplot as plt
from datetime import datetime
import pandas_ta as ta # https://github.com/twopirllc/pandas-ta
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import time
import warnings # feel free to remove these
warnings.filterwarnings("ignore")
# performance metrics package
# https://vectorbt.dev
import vectorbt as vbt
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
clear_output()
# Three Moving Averages Strategy + VectorBT's Performance Metrics Package
## For backtesting combinations of three simple moving averages
# Upload a couple of historical cryptocurrency data.
btc = pd.read_csv("../input/top-8-crypto-currency-data/BTC-EUR.csv")
eth = pd.read_csv("../input/top-8-crypto-currency-data/ETH-EUR.csv")
# look at last 500 - Ethereum closePrice - over time
eth.Close.plot(figsize=(20, 7))
plt.title("ETH-EUR")
# Pandas_TA has a tsignals function that allows us to set boolean entities as our "entry" & "exit" signals. You might find the algorithm below to be a bit strange, but my goal was to try and determine the best Three-Moving-Averages sma combinations. I thought it would be interesting to reiterate through the multiplicative combinations of trading against thee (3) simple moving averages.
# https://github.com/twopirllc/pandas-ta#performance-metrics
# define True/False signal parameters for 3 SMAs
def tsignals(df, a, b, c):
# Create the "Golden Cross" conditions
# three simple moving average with their periods set as variables
smaA = df.ta.sma(a, append=True)
smaB = df.ta.sma(b, append=True)
smaC = df.ta.sma(c, append=True)
close = df.Close[len(df) - 1]
# when the smaA cross above smaB and smaC, we give a "True" signal for entries
# IMPORTANT part of assumed straegy
# df["GC"] = (smaA > smaB) & (smaB > smaC)
df["GC"] = (close > smaA) & (smaA > smaB) & (smaB > smaC)
golden = df.ta.tsignals(df.GC, asbool=True, append=True)
# Create the Signals Portfolio Performance Assessment report
pf = vbt.Portfolio.from_signals(
df.Close,
entries=golden.TS_Entries,
exits=golden.TS_Exits,
freq="D",
# set initial principal and other params, docs fuond in vectorbt
init_cash=100,
fees=0.0025,
slippage=0.0025,
)
# values from report we care about
ev = pf.stats()["End Value"]
re = pf.stats()["Total Return [%]"]
return pd.Series([ev, re, a, b, c]), df, pf, golden
# range of curious movAvg values & initialize results frame
# after many trials, I decided the best ranges for a,b,c are as follows:
sma_a, sma_b, sma_c, results = (
list(range(7, 40)),
list(range(41, 120)),
list(range(121, 135)),
[],
)
# for the sake of computation efficiency...
df = eth
# re(re(iteration))
# this will take a while to run,
# it evaluates profit/loss outcomes
# by trying each sma(a,b,c) combinations.
thresh = 0 # 15 # hoping to display only results that yield above 15% profit
for c in sma_c:
for b in sma_b:
for a in sma_a:
if tsignals(df, a, b, c)[0][1] > thresh:
try:
print(f"\n*Testing: sma_{a}, sma_{b}, sma_{c}*\n")
print(f"*Score: {tsignals(df,a,b,c)[0][1]}*\n")
results.append(tsignals(df, a, b, c)[0])
clear_output()
except:
print(f"No results yielding over {thresh}.")
res = pd.DataFrame(results)
best_res = (
res.sort_values(res.columns[1], ascending=False).head(3).reset_index(drop=True)
)
print("Best resolution:\nEnd Principal, % Return, SMAa, SMAb, SMAc")
print(best_res)
# highest yielding sma_value
a = best_res[2][0]
b = best_res[3][0]
c = best_res[4][0]
print(f"\nBest SMA Throuple: {a}, {b}, {c}")
print(f"Number of results evaluated: {len(results)}")
########################################################
# le winner
df, pf = tsignals(df, a, b, c)[1], tsignals(df, a, b, c)[2].stats()
golden = tsignals(df, a, b, c)[3]
# params
fig, ax = plt.subplots(figsize=(20, 8))
# plot
ax.plot(df.Close, color="black", label="closePrice")
ax.plot(df.ta.sma(a), color="orange", label=f"sma-{a}")
ax.plot(df.ta.sma(b), color="red", label=f"sma-{b}")
ax.plot(df.ta.sma(c), color="purple", label=f"sma-{c}")
# signals
xcoords_buy, xcoords_sell = (
golden[golden.TS_Entries == True].index,
golden[golden.TS_Exits == True].index,
)
for xc in xcoords_sell:
plt.axvline(x=xc, c="r")
for xc in xcoords_buy:
plt.axvline(x=xc, c="g")
# plot outputs
ax.legend(loc="upper left")
plt.show()
# not very definitive of the absolute BEST combination. but it does yield some pretty neat outputs.
|
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns
import warnings
import xgboost
from sklearn import model_selection
from xgboost import XGBClassifier
from sklearn.metrics import accuracy_score
from sklearn.model_selection import train_test_split
warnings.filterwarnings("ignore")
import tensorflow as tf
# Set columns names
col_Name = [
"Age",
"Work_Class",
"Fnlwgt",
"Education",
"Education_Num",
"Marital_Status",
"Occupation",
"Relationship",
"Race",
"Sex",
"Capital_Gain",
"Capital_Loss",
"Hours_Per_Week",
"Native_Country",
"Salary",
]
train = pd.read_csv("../input/adult.data.csv", sep=",", header=None)
train.columns = col_Name
test1 = pd.read_csv("../input/adult.test.csv", sep=",", header=None)
test1.columns = col_Name
# train null value and info
train.info()
print(train.isnull().sum())
print()
"""
尽管显示空值为0,但是经过观察有很多值为?,也要视为空值,并尝试填充
"""
# Get all data and clean
data_cleaner = [train, test1]
train["Salary"] = train["Salary"].apply(lambda x: 1 if x == " >50K" else 0)
test1["Salary"] = test1["Salary"].apply(lambda x: 1 if x == " >50K." else 0)
train.Salary.value_counts()
train1 = train.copy()
def cc(x):
return sum(x == " ?")
train.apply(cc)
train.loc[train["Work_Class"] == " ?"].apply(cc)
## EDA
# Age versus Salary
train["Age"].describe()
# Age Distribution
bins = np.arange(0, 90, 5)
sns.distplot(train["Age"], bins=bins)
plt.title("Age Distribution")
plt.show()
"""
Age为一个左尾正态分布,从20到30上升过大
"""
# Age versus Salary
AgePlot = sns.FacetGrid(data=train, hue="Salary", aspect=3)
AgePlot.map(sns.distplot, "Age", bins=bins)
AgePlot.set(ylim=(0, 0.05))
AgePlot.set(xlim=(10, 90))
AgePlot.add_legend()
plt.show()
"""
明显Age和Salary会有关系,没钱的更集中在25岁左右,
"""
# 工作与工资比例
plt.figure(figsize=(14, 7))
sns.barplot(y="Salary", x="Work_Class", data=train)
"""
Without-pay, Never-worked没有工资,Self-emp-inc 和 Federal-gov工资比例最高,
"""
# 发现Work_Class变量前面有空格,先删除
for all_data in data_cleaner:
all_data["Work_Class"] = all_data["Work_Class"].apply(lambda x: x.strip())
# Work_Class分布图
plt.figure(figsize=(10, 7))
plt.hist(
x=[
train[train["Salary"] == 0]["Work_Class"],
train[train["Salary"] == 1]["Work_Class"],
],
stacked=True,
color=["g", "r"],
label=["<50K", ">50K"],
)
plt.legend()
"""
绝大多数人为Private职业,但是工资1占比不高,withou-pay 和 Never-worked的人太少,而且工资都为0
"""
# Work_class数量统计
print("Workclass总数:", "\n", train["Work_Class"].value_counts())
print("-" * 50)
print("工资为0数目:", "\n", train[train["Salary"] == 0]["Work_Class"].value_counts())
# Work_Class中?很多, 看一下他在年龄中的分布情况
bins = np.arange(0, 95, 1)
work_age = train[train["Work_Class"] == "?"]
sns.distplot(work_age["Age"], bins=bins)
plt.title("Missing Values in Work Class with Age")
"""
看的出来年龄在24和62左右空值最多,按经验来看24可能是学生为Never-worked团体,而60岁以上为退休集体可能Without-pay
"""
print(train[train["Work_Class"] == "Never-worked"]["Age"].describe())
"""
可以看到'never-worked'人群平均年龄只有20,那么Work-Class可考虑填充为Never-worked
"""
print(train[train["Work_Class"] == "Without-pay"]["Age"])
plt.hist(train[train["Work_Class"] == "Without-pay"]["Age"])
"""
without-pay大部分也是20左右年轻人和60以上老年人,考虑用without-pay进行填补"""
# 尝试填补年轻人Work_Class再观察
train1 = train.copy()
train1.loc[
(train1["Age"] <= 24) & (train1["Work_Class"] == "?"), "Work_Class"
] = "Never-worked"
# 再看分布图
bins = np.arange(0, 95, 1)
age_work = train1[train1["Work_Class"] == "?"]
sns.distplot(age_work["Age"], bins=bins)
plt.xlim(24, 95)
"""
超过60岁还有缺失值,按常理来说应该很多是Without-pay"""
# 观察有无工作与年龄的关系
age_with_work = (
train1[train1["Work_Class"] != "?"].groupby("Age").count().max(1)
/ train1["Age"].value_counts()
)
age_without_work = (
train1[train1["Work_Class"] == "?"].groupby("Age").count().max(1)
/ train1["Age"].value_counts()
)
age_with_work = (age_with_work - age_with_work.mean()) / age_with_work.std()
age_without_work = (age_without_work - age_without_work.mean()) / age_without_work.std()
age_with_work = age_with_work.fillna(0)
age_without_work = age_without_work.fillna(0)
diff = age_with_work - age_without_work
diff.loc[(85, 86, 88),] = 0
age = np.arange(17, 90, 1)
plt.bar(age, diff)
"""
在考虑了年龄的分布后,可以看出大于60岁后,Work_class为空值的都比非空的多,所以可以将60岁以上Work-class为空的设定为retired"""
train1.loc[
(train1["Work_Class"] == "?") & (train1["Age"] >= 60), "Work_Class"
] = "Retired"
bins = np.arange(0, 95, 1)
work_age1 = train1[train1["Work_Class"] == "?"]
sns.distplot(work_age1["Age"], bins=bins)
# Occupation vs salary
plt.figure(figsize=(18, 7))
sns.barplot(x="Occupation", y="Salary", data=train1)
# Education与Salary
print(train1["Education"].value_counts())
print(train1["Education_Num"].value_counts())
"""
这2个为相同的东西,删去后一个变量"""
for all_data in data_cleaner:
all_data["Education"] = all_data["Education"].apply(lambda x: x.strip())
train1 = train.copy()
train1["Education"] = train1["Education"].apply(lambda x: x.strip())
salary_ratio = []
for x in np.unique(train1["Education"]):
salary_ratio.append(
train1[(train1["Education"] == x) & (train1["Salary"] == 1)].count().max()
/ train1[train1["Education"] == x].count().max()
)
salary_class_ratio = {
"Education": np.unique(train1["Education"]),
"Salary": salary_ratio,
}
salary_class_ratio = pd.DataFrame(salary_class_ratio)
salary_class_ratio = salary_class_ratio.sort_values(["Salary"]).reset_index(drop=True)
# 各个Education
plt.figure(figsize=(18, 7))
salary_class_ratio["Salary"] = salary_class_ratio["Salary"].apply(
lambda x: np.round(x, 4)
)
ax = sns.barplot(y="Salary", x="Education", data=salary_class_ratio)
for i, v in enumerate(salary_class_ratio["Salary"].iteritems()):
ax.text(i, v[1], "{:,}".format(v[1]), color="m", va="bottom", rotation=45)
plt.tight_layout()
plt.title("Salary with Education")
plt.show()
"""
看的出来文化水平越高工资比例越高"""
plt.figure(figsize=(18, 7))
Education = pd.DataFrame(train1["Education"].value_counts()).sort_values(by="Education")
sns.barplot(x=Education.index, y=Education["Education"])
plt.ylabel("counts")
plt.title("Education Bar Plot")
plt.show()
"""
可以看到高中毕业最多,考虑将小学生, 高中生分别合并"""
# Marital_Status与Salary
train1["Marital_Status"].value_counts()
plt.figure(figsize=(13, 8))
sns.barplot(y="Salary", x="Marital_Status", data=train1)
plt.title("Salary with Marital_Status")
"""
结婚了的工资相对较高
"""
# Salary vs Marital_status
plt.figure(figsize=(11, 7))
plt.hist(
x=[
train1[train1["Salary"] == 0]["Marital_Status"],
train1[train1["Salary"] == 1]["Marital_Status"],
],
color=["g", "r"],
label=["<50K", ">50K"],
)
plt.legend()
"""
大部分人未婚,但是他们工资比例较低,一般稳定结婚了的比例更高,将MCVS和MAFS结合一起,结婚后只剩一个人的也可考虑结合在一起"""
for all_data in data_cleaner:
all_data["Marital_Status"] = all_data["Marital_Status"].apply(lambda x: x.strip())
# 查看Never-married和年龄的关系
train1 = train.copy()
Marital_age = train1[train1["Marital_Status"] == "Never-married"]
ax = sns.FacetGrid(data=Marital_age, hue="Salary", aspect=3)
ax.map(sns.distplot, "Age")
ax.add_legend()
plt.show()
"""
显示出来大部分年轻人没有结婚,工资也低,而38岁左右不结婚也会有比较高工资
"""
# Occupation缺失值查和Work_class的差不多,看看哪里不同
train.ix[((train["Work_Class"] == "?"))]
print(train[(train["Work_Class"] == "?") & (train["Occupation"] == " ?")].count().max())
"""
Work_class为空的时候Occupation也为空
"""
print(train[(train["Work_Class"] != "?") & (train["Occupation"] == " ?")])
"""
Occupation为空时,Work_class都为Never-Worked,其他都处理方式和Work_class相同
"""
# relationship vs salry
train1["Relationship"].value_counts()
plt.figure(figsize=(14, 8))
sns.barplot(y="Salary", x="Relationship", data=train1)
"""
结了婚工资普遍偏高"""
# relationshion vs age
h = sns.FacetGrid(data=train1, col="Relationship")
h.map(sns.distplot, "Age", color="r")
h.add_legend()
"""
这个数据有些不科学,own-child居然是20岁左右最多,而unmaried居然平均在40左右,由于有sex变量考虑将husband 和 wife合并,他们基本相同分布,并且Salary比例也很相似"""
# 看是否有同性恋
train1.loc[(train1["Relationship"] == " Husband") & (train1["Sex"] == " Female"),]
"""应该是错误数据,删去"""
train = train.drop(index=7109)
# race vs salary(
plt.figure(figsize=(10, 7))
sns.barplot(x="Race", y="Salary", data=train1)
Race_dist = train1.groupby("Race").count().max(1).sort_values()
Race_dist.plot(kind="bar")
"""
亚洲人Salary的比例最高,白人人数最多工资工资比例也高"""
# Sex vs Salary
sns.barplot(x="Sex", y="Salary", data=train1)
"""男性工资普遍比女性高"""
# Native_Country vs Salary
salary_ratio1 = []
for x in np.unique(train1["Native_Country"]):
salary_ratio1.append(
train1[(train1["Native_Country"] == x) & (train1["Salary"] == 1)].count().max()
/ train1[train1["Native_Country"] == x].count().max()
)
salary_country_ratio = {
"Native_Country": np.unique(train1["Native_Country"]),
"Salary": salary_ratio1,
}
salary_country_ratio = pd.DataFrame(salary_country_ratio)
salary_country_ratio = salary_country_ratio.sort_values(
["Salary"], ascending=0
).reset_index(drop=True)
plt.figure(figsize=(18, 7))
salary_country_ratio["Salary"] = salary_country_ratio["Salary"].apply(
lambda x: np.round(x, 4)
)
ax = sns.barplot(x="Salary", y="Native_Country", data=salary_country_ratio)
plt.tight_layout()
plt.title("Native Country with Salary")
plt.show()
# plt.figure(figsize=(10,15))
# ax = sns.barplot(y = 'Native_Country', x = 'Salary', data = train1)
train1["Native_Country"].value_counts().plot(kind="bar")
"""
美国占的数目太多,考虑利用发达国家,发展中国家和贫穷国家进行分割,数据中还有很多空值,但是很难通过其他变量来进行填补"""
train1["Native_Country"] = train1["Native_Country"].apply(lambda x: x.strip())
developed_country = [
"United-States",
"Germany",
"Canada",
"England",
"Italy",
"Japan",
"Taiwan",
"Portugal",
"Greece",
"France",
"Hong",
"Yugoslavia",
"Scotland",
]
developing_country = [
"Mexico",
"Philippines",
"India",
"Cuba",
"China",
"Poland",
"Ecuador",
"Ireland",
"Iran",
"Thailand",
"?",
"Hungary",
]
poor_country = [
"Puerto-Rico",
"El-Salvador",
"Jamaica",
"South",
"Dominican-Republic",
"Vietnam",
"Guatemala",
"Columbia",
"Haiti",
"Nicaragua",
"Peru",
"Cambodia",
"Trinadad&Tobago",
"Laos",
"Outlying-US(Guam-USVI-etc)",
"Honduras",
"Holand-Netherlands",
]
train2 = train1.copy()
train2.loc[
train2["Native_Country"].isin(developed_country), "Native_Country"
] = "Developed_country"
train2.loc[
train2["Native_Country"].isin(developing_country), "Native_Country"
] = "Developing_country"
train2.loc[
train2["Native_Country"].isin(poor_country), "Native_Country"
] = "poor_country"
sns.barplot(y="Salary", x="Native_Country", data=train2)
"""
现在再来看Salary分布比较平均,也比较符合实际"""
# Hours_Per_Week
plt.figure(figsize=(18, 7))
sns.barplot(train1["Hours_Per_Week"], train1["Salary"])
plt.title("Hours Per Week with Salary")
"""
时长越高工资越高"""
plt.figure(figsize=(14, 7))
sns.distplot(train1["Hours_Per_Week"])
"""
分布基本平均在40小时两边,考虑切割为3部分大于40,小于40,等于40,或者利用fulltime,parttime进行分类"""
train1 = train.copy()
train1["Hours_Per_Week"] = train1["Hours_Per_Week"].apply(
lambda x: "<40" if x < 40 else ("=40" if x == 40 else (">40"))
)
# 分割完之后
fig, (axis1, axis2) = plt.subplots(1, 2, figsize=(14, 7))
train1["Hours_Per_Week"].value_counts().plot(kind="bar", ax=axis1)
axis1.set_title("Hourse Per Week distribution")
sns.barplot(x="Hours_Per_Week", y="Salary", data=train1, ax=axis2)
axis2.set_title("Salary vs House_Per_Week")
"""
分割完之后分布更加平稳,也更显示出了Salary与Hours_per_week的关系"""
# Capital_Gain&Capital_Loss
plt.figure(figsize=(14, 7))
plt.subplot(121)
sns.distplot(train1["Capital_Gain"])
plt.title("Capital Gain Distribution")
plt.subplot(122)
sns.distplot(train1["Capital_Loss"])
plt.title("Capital Loss Distribution")
# scatter plot between Gain and Loss
sns.scatterplot(x="Capital_Gain", y="Capital_Loss", data=train1)
plt.title("Gain & Loss Scatter Plot")
"""
基本2者没有同时非零点则考虑将2个变量改为0,1变量"""
train1.loc[(train1["Capital_Gain"] > 0) & (train1["Capital_Loss"] > 0),].count().max()
"""
确实没有既有Gain又有Loss的值"""
##Feature Engineer
df = train.copy()
test = test1.copy()
# 消除所有object变量之前的空格
data_Cleaner = [df, test]
for all_data in data_Cleaner:
for i in col_Name:
if all_data[i].dtype == "object":
all_data[i] = all_data[i].apply(lambda x: x.strip())
# 空值填补
for all_data in data_Cleaner:
all_data.loc[
(all_data["Work_Class"] == "?") & (all_data["Age"] >= 60), "Work_Class"
] = "Retired"
all_data.loc[
(all_data["Age"] <= 24) & (all_data["Work_Class"] == "?"), "Work_Class"
] = "Never-worked"
all_data.loc[
(all_data["Work_Class"] == "Never-worked") & (all_data["Occupation"] == "?"),
"Occupation",
] = "None"
all_data.loc[
(all_data["Occupation"] == "?") & (all_data["Age"] >= 60), "Occupation"
] = "Retired"
all_data.loc[all_data["Work_Class"] == "?", "Work_Class"] = "Unknown"
all_data.loc[all_data["Occupation"] == "?", "Occupation"] = "Unknown"
all_data.loc[all_data["Native_Country"] == "?", "Native_Country"] = "Unknown"
# Work_Class
for all_data in data_Cleaner:
all_data.loc[
all_data["Work_Class"].isin(["Never-worked", "Without-pay"]), "Work_Class"
] = "Others"
# Education
Primary = ["1st-4th", "5th-6th", "Preschool"]
Secondary = ["7th-8th", "9th", "10th", "11th", "12th"]
Teriary = ["HS-grad", "Some-college", "Assoc-voc", "Assoc-acdm"]
Quaternary = ["Prof-school", "Doctorate"]
for all_data in data_Cleaner:
all_data.loc[all_data["Education"].isin(Primary), "Education"] = "Primary"
all_data.loc[all_data["Education"].isin(Secondary), "Education"] = "Secondary"
all_data.loc[all_data["Education"].isin(Teriary), "Education"] = "Teriary"
all_data.loc[all_data["Education"].isin(Quaternary), "Education"] = "Quaternary"
# Marital_Status
Married = ["Married-civ-spouse", "Married-AF-spouse"]
Solo = ["Divorced", "Separated", "Widowed", "Married-spouse-absent"]
for all_data in data_Cleaner:
all_data.loc[all_data["Marital_Status"].isin(Married), "Marital_Status"] = "Married"
all_data.loc[all_data["Marital_Status"].isin(Solo), "Marital_Status"] = "Solo"
# Relationship
Family = ["Husband", "Wife"]
for all_data in data_Cleaner:
all_data.loc[all_data["Relationship"].isin(Family), "Relationship"] = "Family"
# Native_Country
for all_data in data_Cleaner:
all_data.loc[
all_data["Native_Country"].isin(developed_country), "Native_Country"
] = "Developed_country"
all_data.loc[
all_data["Native_Country"].isin(developing_country), "Native_Country"
] = "Developing_country"
all_data.loc[
all_data["Native_Country"].isin(poor_country), "Native_Country"
] = "poor_country"
# Hours_Per_Week
for all_data in data_Cleaner:
all_data["Hours_Per_Week"] = all_data["Hours_Per_Week"].apply(
lambda x: "Part_Time"
if x <= 35
else (
"Full_Time"
if (x > 35) & (x <= 40)
else ("Much_Work" if (x > 40) & (x <= 50) else ("Over_Work"))
)
)
Remove_Columns = ["Fnlwgt", "Education_Num"]
for all_data in data_Cleaner:
all_data.drop(Remove_Columns, axis=1, inplace=True)
df1 = df.copy()
test2 = test.copy()
data_Cleaner1 = [df1, test2]
from sklearn import preprocessing
for all_data in data_Cleaner1:
for column in all_data:
le = preprocessing.LabelEncoder()
all_data[column] = le.fit_transform(all_data[column])
from sklearn.ensemble import RandomForestClassifier
select = RandomForestClassifier()
train_select = df1.drop(columns="Salary", axis=1)
test_select = df1.Salary
param_grid = {
"criterion": ["gini", "entropy"],
"max_depth": [4, 6, 8, 10],
"n_estimators": [50, 100, 200, 300],
}
select1 = model_selection.GridSearchCV(
select, param_grid=param_grid, cv=5, scoring="accuracy"
)
select1.fit(train_select, test_select)
select1.best_params_
selected = RandomForestClassifier(criterion="gini", max_depth=10, n_estimators=100)
selected.fit(train_select, test_select)
importances = selected.feature_importances_
indices = np.argsort(importances)[::-1]
features = train_select.columns
for f in range(train_select.shape[1]):
print(("%2d) %-*s %f" % (f + 1, 30, features[indices[f]], importances[indices[f]])))
# Capital_Gain&Capital_Loss
gain = train1.loc[train1["Capital_Gain"] != 0, "Capital_Gain"]
loss = train1.loc[train1["Capital_Loss"] != 0, "Capital_Loss"]
print("Gain quantile(0.3):", gain.quantile(0.3))
print("Gain quantile(0.7):", gain.quantile(0.7))
print("Loss quantile(0.3):", loss.quantile(0.3))
print("Loss quantile(0.7):", loss.quantile(0.7))
for all_data in data_Cleaner:
all_data["Capital_Total"] = "Zero"
all_data["Capital_Gain"] = all_data["Capital_Gain"].apply(
lambda x: "Low_Gain"
if (x > 0) & (x <= 3942)
else (
"Med_Gain"
if (x > 3942) & (x <= 8614)
else ("High_Gain" if x > 8614 else ("Zero"))
)
)
all_data["Capital_Loss"] = all_data["Capital_Loss"].apply(
lambda x: "Low_Loss"
if (x > 0) & (x <= 1740)
else (
"Med_Loss"
if (x > 1740) & (x <= 1977)
else ("High_Loss" if x > 1977 else ("Zero"))
)
)
all_data["Capital_Total"].loc[all_data["Capital_Gain"] != "Zero"] = all_data[
"Capital_Gain"
]
all_data["Capital_Total"].loc[all_data["Capital_Loss"] != "Zero"] = all_data[
"Capital_Loss"
]
Remove_Columns = ["Capital_Gain", "Capital_Loss"]
for all_data in data_Cleaner:
all_data.drop(Remove_Columns, axis=1, inplace=True)
Dummy = [
"Work_Class",
"Education",
"Marital_Status",
"Occupation",
"Relationship",
"Race",
"Sex",
"Hours_Per_Week",
"Native_Country",
"Capital_Total",
]
dummies1 = pd.get_dummies(df[Dummy], prefix=Dummy)
df = pd.concat([df, dummies1], axis=1)
dummies2 = pd.get_dummies(test[Dummy], prefix=Dummy)
test = pd.concat([test, dummies2], axis=1)
Drop_Coluns = [
"Work_Class",
"Education",
"Marital_Status",
"Occupation",
"Relationship",
"Race",
"Sex",
"Hours_Per_Week",
"Native_Country",
"Capital_Total",
]
df.drop(Drop_Coluns, axis=1, inplace=True)
test.drop(Drop_Coluns, axis=1, inplace=True)
test.to_csv("D_test.csv")
target = ["Salary"]
X = df.drop(target, axis=1)
Y = df[target]
# XGBoost
cv_split = model_selection.ShuffleSplit(
n_splits=3, test_size=0.3, train_size=0.7, random_state=0
)
Xgboost = XGBClassifier(max_depth=7, leaning_rate=0.1)
base_Result = model_selection.cross_validate(
Xgboost, X, Y, cv=cv_split, return_train_score=True
)
# train&test score with no params change xgboost
print("xgboost parameters:", Xgboost.get_params())
print("train score mean:{:.4f}".format(base_Result["train_score"].mean()))
print("test score mean:{:.4f}".format(base_Result["test_score"].mean()))
param_Grid = {"max_depth": [5, 7, 9, 12], "leaning_rate": [0.1, 0.15, 0.2]}
tune_Model = model_selection.GridSearchCV(
XGBClassifier(), param_grid=param_Grid, scoring="roc_auc", cv=cv_split
)
tune_Model.fit(X, Y)
tune_Model.best_params_
Xgboost = XGBClassifier(max_depth=5, leaning_rate=0.1)
Xgboost.fit(X, Y)
importance = Xgboost.feature_importances_
indices = np.argsort(importance)[::-1]
features = X.columns
for f in range(X.shape[1]):
print(("%2d) %-*s %f" % (f + 1, 30, features[indices[f]], importance[indices[f]])))
test_x = test.drop(target, axis=1)
predictions1 = Xgboost.predict(test_x)
accuracy_score(test[target], predictions1)
# random forest
from sklearn.ensemble import RandomForestClassifier
rf = RandomForestClassifier(oob_score=True)
param_Grid1 = {
"criterion": ["gini", "entropy"],
"max_depth": [4, 6, 8, 10],
"n_estimators": [50, 100, 200, 300],
}
tune_Model1 = model_selection.GridSearchCV(
rf, param_grid=param_Grid1, scoring="accuracy", cv=cv_split
)
tune_Model1.fit(X, Y)
tune_Model1.best_params_
rf1 = RandomForestClassifier(criterion="gini", max_depth=10, n_estimators=200)
rf1.fit(X, Y)
predictions2 = rf1.predict(test_x)
accuracy_score(test[target], predictions2)
rf1
# random forest importances
indices = np.argsort(importance)[::-1]
features = X.columns
for f in range(X.shape[1]):
print(("%2d) %-*s %f" % (f + 1, 30, features[indices[f]], importance[indices[f]])))
# svm
import scipy
from sklearn.svm import SVC
sv = SVC()
param_Grid2 = {"C": [0.5, 1, 1.5], "gamma": [0.1, "auto"]}
tune_Model2 = model_selection.GridSearchCV(
sv, param_grid=param_Grid2, cv=cv_split, scoring="accuracy"
)
tune_Model2.fit(X, Y)
tune_Model2.best_params_
sv1
sv1 = SVC(C=1.5, gamma=0.1)
sv1.fit(X, Y)
predictions3 = sv1.predict(test_x)
accuracy_score(test_y, predictions3)
# gdbt
from sklearn.ensemble import GradientBoostingClassifier
gdbt = GradientBoostingClassifier()
gdbt.fit(X, Y)
predictions4 = gdbt.predict(test_x)
accuracy_score(test_y, predictions4)
gdbt
from vecstack import stacking
# stacking
clfs = [
XGBClassifier(max_depth=7, leaning_rate=0.1),
svm.SVC(C=1.5, gamma=0.1),
RandomForestClassifier(criterion="gini", max_depth=10, n_estimators=100),
GradientBoostingClassifier(),
]
X_1, X_2, y_1, y_2 = train_test_split(X, Y, test_size=0.33, random_state=2019)
S_train, S_test = stacking(
clfs,
X,
Y,
test_x,
regression=False,
mode="oof_pred_bag",
needs_proba=False,
save_dir=None,
metric=accuracy_score,
n_folds=5,
stratified=True,
shuffle=True,
verbose=2,
random_state=2019,
)
model = XGBClassifier()
model.fit(S_train, Y)
y_pred = model.predict(S_test)
accuracy_score(test_y, y_pred)
# dnn
test_y = test[target]
print(X.shape[1], Y.shape[1])
# adam optimizer
training_epochs = 1500
learning_rate = 0.01
hidden_layers = X.shape[1] - 1
x = tf.placeholder(tf.float32, [None, 63])
y = tf.placeholder(tf.float32, [None, 1])
is_training = tf.Variable(True, dtype=tf.bool)
initializer = tf.contrib.layers.xavier_initializer()
h0 = tf.layers.dense(x, 120, activation=tf.nn.relu, kernel_initializer=initializer)
h1 = tf.layers.dense(h0, 120, activation=tf.nn.relu)
h2 = tf.layers.dense(h1, 1, activation=None)
cross_entropy = tf.nn.sigmoid_cross_entropy_with_logits(labels=y, logits=h2)
cost = tf.reduce_mean(cross_entropy)
# Momentum = tf.train.MomentumOptimizer(learning_rate=learning_rate, momentum=0.9).minimize(cost)
# RMSprop=tf.train.RMSPropOptimizer(learning_rate=learning_rate, decay=0.9).minimize(cost)
adam = tf.train.AdamOptimizer(learning_rate=learning_rate).minimize(cost)
# gradient=tf.train.GradientDescentOptimizer(learning_rate=learning_rate).minimize(cost)
predicted = tf.nn.sigmoid(h2)
correct_pred = tf.equal(tf.round(predicted), y)
accuracy = tf.reduce_mean(tf.cast(correct_pred, tf.float32))
with tf.Session() as sess:
sess.run(tf.global_variables_initializer())
cost_history = np.empty(shape=1, dtype=float)
for step in range(training_epochs + 1):
sess.run(gradient, feed_dict={x: X, y: Y})
loss, _, acc = sess.run([cost, gradient, accuracy], feed_dict={x: X, y: Y})
Cost_History = np.append(Cost_History, acc)
if step % 200 == 0:
print("Step: {:5}\tLoss: {:.3f}\tAcc: {:.2%}".format(step, loss, acc))
print(
"Test Accuracy by gradient:",
sess.run([accuracy, tf.round(predicted)], feed_dict={x: test_x, y: test_y}),
)
sess.run(tf.global_variables_initializer())
cost_history = np.empty(shape=1, dtype=float)
for step in range(training_epochs + 1):
sess.run(Momentum, feed_dict={x: X, y: Y})
loss, _, acc = sess.run([cost, Momentum, accuracy], feed_dict={x: X, y: Y})
Cost_History = np.append(Cost_History, acc)
if step % 200 == 0:
print("Step: {:5}\tLoss: {:.3f}\tAcc: {:.2%}".format(step, loss, acc))
print(
"Test Accuracy by momentum:",
sess.run([accuracy, tf.round(predicted)], feed_dict={x: test_x, y: test_y}),
)
sess.run(tf.global_variables_initializer())
cost_history = np.empty(shape=1, dtype=float)
for step in range(training_epochs + 1):
sess.run(RMSprop, feed_dict={x: X, y: Y})
loss, _, acc = sess.run([cost, RMSprop, accuracy], feed_dict={x: X, y: Y})
Cost_History = np.append(Cost_History, acc)
if step % 200 == 0:
print("Step: {:5}\tLoss: {:.3f}\tAcc: {:.2%}".format(step, loss, acc))
print(
"Test Accuracy by RMSprop:",
sess.run([accuracy, tf.round(predicted)], feed_dict={x: test_x, y: test_y}),
)
sess.run(tf.global_variables_initializer())
cost_history = np.empty(shape=1, dtype=float)
for step in range(training_epochs + 1):
sess.run(adam, feed_dict={x: X, y: Y})
loss, _, acc = sess.run([cost, adam, accuracy], feed_dict={x: X, y: Y})
Cost_History = np.append(Cost_History, acc)
if step % 200 == 0:
print("Step: {:5}\tLoss: {:.3f}\tAcc: {:.2%}".format(step, loss, acc))
print(
"Test Accuracy by adm:",
sess.run([accuracy, tf.round(predicted)], feed_dict={x: test_x, y: test_y}),
)
with tf.Session() as sess:
sess.run(tf.global_variables_initializer())
cost_history = np.empty(shape=1, dtype=float)
for step in range(training_epochs + 1):
sess.run(adam, feed_dict={x: X, y: Y})
loss, _, acc = sess.run([cost, adam, accuracy], feed_dict={x: X, y: Y})
cost_history = np.append(cost_history, acc)
if step % 200 == 0:
print("Step: {:5}\tLoss: {:.3f}\tAcc: {:.2%}".format(step, loss, acc))
summary_writer = tf.summary.FileWriter("./log/", sess.graph)
print(
"Test Accuracy by adm:",
sess.run([accuracy, tf.round(predicted)], feed_dict={x: test_x, y: test_y}),
)
table = {
"Test Accuracy": [86.364, 86.204, 86.167, 85.831, 85.805, 85.547],
"Model": ["Stacking", "XGBoost", "GDBT", "SVM", "RF", "ANN"],
}
d = ["86.364%", "86.204%", "86.167%", "85.831%", "85.805%", "85.547%"]
sns.barplot(y=table["Model"], x=table["Test Accuracy"])
plt.xlim(85, 86.5)
for i in range(6):
plt.text(x=table["Test Accuracy"][i], y=i, s=d[i])
|
# # 1. Importing of dataset and data preprocessing
# ## 1.1 Import libraries
# Basic Libraries
import numpy as np
import pandas as pd
import seaborn as sb
import matplotlib.pyplot as plt
sb.set() # set the default Seaborn style for graphics
from sklearn.model_selection import train_test_split
import chart_studio
chart_studio.tools.set_credentials_file(
username="LIANGJING", api_key="KeSQYwdxfNybb8zzzMj9"
)
import chart_studio.plotly as py
import plotly.io as pio
pio.templates.default = "none"
import plotly.graph_objects as go
# ## 1.2 Import dataset on town's information
# townName refers to the dataset containing both town code and town name as it has the complete set with proper format. Hence, we will always use back the town name from this dataset instead of the town name from which ever dataset we are using.
townName = pd.read_csv("../input/frenchemployment/name_geographic_information.csv")
townName = townName.drop(
[
"latitude",
"longitude",
"European Union Circonscription",
"region code",
"region",
"administrative center",
"department number",
"department",
"prefecture",
"num of the circumpscription",
"postal code",
"distance",
],
axis=1,
)
townName.sort_values(by=["town code"])
townName["town code"] = townName["town code"].astype(int)
townName = townName.set_index("town code")
# ## 1.3 Import dataset on town's population
# *townDemographics* contains the town's demographics like the town's age category, gender, cohabitation mode. **However, not all town's information is captured, only until town code:13062**. *townSexPopulation* contains the information on the gender demographics for each town. *highestWomenRatioTable* contains the top 10 towns with the highest females vs male ratio. *highestMenRatioTable* contains the top 10 towns with the highest male vs female ratio. *cohabitationTable* contains the information on the cohabitation demographics for each town
# AgeTable contains the information on the age demographics for each town
townDemographics = pd.read_csv("../input/frenchemployment/population.csv")
townSexPopulation = townDemographics.drop(
["age category", "cohabitation mode", "geographic level"], axis=1
)
townSexPopulation = townSexPopulation.pivot_table(
index="town code", columns="sex", aggfunc=sum
)
townSexPopulation.loc["Total", :] = townSexPopulation.sum(axis=0) # Total row
townSexPopulation
highestWomenRatio_col = ["men", "women", "men percentage", "women percentage"]
highestWomenRatioTable = pd.DataFrame(columns=highestWomenRatio_col)
highestWomenRatioTable["women percentage"] = townSexPopulation[
"num of people", "women"
] / (
townSexPopulation["num of people", "women"]
+ townSexPopulation["num of people", "men"]
)
highestWomenRatioTable["men percentage"] = townSexPopulation["num of people", "men"] / (
townSexPopulation["num of people", "women"]
+ townSexPopulation["num of people", "men"]
)
highestWomenRatioTable["men"] = townSexPopulation["num of people", "men"]
highestWomenRatioTable["women"] = townSexPopulation["num of people", "women"]
highestWomenRatioTable = highestWomenRatioTable.nlargest(10, "women percentage")
highestWomenRatioTable = highestWomenRatioTable.join(townName)
highestWomenRatioTable = highestWomenRatioTable.sort_values(
"women percentage", ascending=False
)
highestWomenRatioTable
highestMenRatio_col = ["men", "women", "men percentage", "women percentage"]
highestMenRatioTable = pd.DataFrame(columns=highestMenRatio_col)
highestMenRatioTable["women percentage"] = townSexPopulation[
"num of people", "women"
] / (
townSexPopulation["num of people", "women"]
+ townSexPopulation["num of people", "men"]
)
highestMenRatioTable["men percentage"] = townSexPopulation["num of people", "men"] / (
townSexPopulation["num of people", "women"]
+ townSexPopulation["num of people", "men"]
)
highestMenRatioTable["men"] = townSexPopulation["num of people", "men"]
highestMenRatioTable["women"] = townSexPopulation["num of people", "women"]
highestMenRatioTable = highestMenRatioTable.nlargest(10, "men percentage")
highestMenRatioTable = highestMenRatioTable.join(townName)
highestMenRatioTable = highestMenRatioTable.sort_values(
"men percentage", ascending=False
)
highestMenRatioTable
cohabitationTable = townDemographics.drop(
["age category", "sex", "geographic level", "town name"], axis=1
)
cohabitationTable["num of people"] = cohabitationTable["num of people"].astype(int)
cohabitationTable = cohabitationTable.pivot_table(
index="town code", columns="cohabitation mode", aggfunc=sum
)
cohabitationTable.loc["Total", :] = cohabitationTable.sum(axis=0) # Total row
cohabitationTable
AgeTable = townDemographics.drop(
["cohabitation mode", "sex", "geographic level", "town name"], axis=1
)
AgeTable["num of people"] = AgeTable["num of people"].astype(int)
AgeTable = AgeTable.pivot_table(index="town code", columns="age category", aggfunc=sum)
AgeTable.loc["Total", :] = AgeTable.sum(axis=0) # Total row
AgeTable
# ## 1.4 Import dataset on town's salary information
# *salary* contains salary information in relation with resident's age category, gender, cohabitation mode.
# *salaryGenderType* contains the resident's salary information in relation with the gender demographics for each town.
# *highestSalary* contains the top 15 'mean net salary' disparity among males and females.
# *highestSalaryExecutive* contains the top 15 salary disparity among male and female executives.
# *highestSalaryWorker* contains the top 15 salary disparity among male and female workers.
salary = pd.read_csv("../input/frenchemployment/net_salary_per_town_categories.csv")
salaryGenderType = salary.drop(
[
"town name",
"18-25 yo",
"26-50 yo",
">50 years old",
"women 18-25 yo",
"women 26-50 yo",
"women >50 yo",
"men 18-25 yo",
"men 26-50 yo",
"men >50 yo",
],
axis=1,
)
salaryGenderType = pd.merge(salaryGenderType, townName, on="town code", how="inner")
salaryGenderType
Salary_col = [
"town code",
"town name",
"mean net salary",
"women",
"man",
"pay disparity",
]
Salary = pd.DataFrame(columns=Salary_col)
Salary["town code"] = salaryGenderType["town code"]
Salary["town name"] = salaryGenderType["town name"]
Salary["mean net salary"] = salaryGenderType["mean net salary"]
Salary["women"] = salaryGenderType["women"]
Salary["man"] = salaryGenderType["man"]
Salary["pay disparity"] = Salary["man"] - Salary["women"]
highestSalary = Salary.nlargest(15, "pay disparity")
highestSalary = highestSalary.set_index("town code")
highestSalary
SalaryManager_col = [
"town code",
"town name",
"manager",
"manager (w)",
"manager (m)",
"pay disparity",
]
SalaryManager = pd.DataFrame(columns=SalaryManager_col)
SalaryManager["town code"] = salaryGenderType["town code"]
SalaryManager["town name"] = salaryGenderType["town name"]
SalaryManager["manager"] = salaryGenderType["manager"]
SalaryManager["manager (w)"] = salaryGenderType["manager (w)"]
SalaryManager["manager (m)"] = salaryGenderType["manager (m)"]
SalaryManager["pay disparity"] = (
SalaryManager["manager (m)"] - SalaryManager["manager (w)"]
)
highestSalaryManager = SalaryManager.nlargest(15, "pay disparity")
highestSalaryManager = highestSalaryManager.set_index("town code")
highestSalaryManager
SalaryEmployee_col = [
"town code",
"town name",
"employee",
"employee (w)",
"employee (m)",
"pay disparity",
]
SalaryEmployee = pd.DataFrame(columns=SalaryEmployee_col)
SalaryEmployee["town code"] = salaryGenderType["town code"]
SalaryEmployee["town name"] = salaryGenderType["town name"]
SalaryEmployee["employee"] = salaryGenderType["employee"]
SalaryEmployee["employee (w)"] = salaryGenderType["employee (w)"]
SalaryEmployee["employee (m)"] = salaryGenderType["employee (m)"]
SalaryEmployee["pay disparity"] = (
SalaryEmployee["employee (m)"] - SalaryEmployee["employee (w)"]
)
highestSalaryEmployee = SalaryEmployee.nlargest(15, "pay disparity")
highestSalaryEmployee = highestSalaryEmployee.set_index("town code")
highestSalaryEmployee
SalaryExecutive_col = [
"town code",
"town name",
"executive",
"executive (w)",
"executive (m)",
"pay disparity",
]
SalaryExecutive = pd.DataFrame(columns=SalaryExecutive_col)
SalaryExecutive["town code"] = salaryGenderType["town code"]
SalaryExecutive["town name"] = salaryGenderType["town name"]
SalaryExecutive["executive"] = salaryGenderType["executive"]
SalaryExecutive["executive (w)"] = salaryGenderType["executive (w)"]
SalaryExecutive["executive (m)"] = salaryGenderType["executive (m)"]
SalaryExecutive["pay disparity"] = (
SalaryExecutive["executive (m)"] - SalaryExecutive["executive (w)"]
)
highestSalaryExecutive = SalaryExecutive.nlargest(15, "pay disparity")
highestSalaryExecutive = highestSalaryExecutive.set_index("town code")
highestSalaryExecutive
SalaryWorker_col = [
"town code",
"town name",
"worker",
"worker (w)",
"worker (m)",
"pay disparity",
]
SalaryWorker = pd.DataFrame(columns=SalaryWorker_col)
SalaryWorker["town code"] = salaryGenderType["town code"]
SalaryWorker["town name"] = salaryGenderType["town name"]
SalaryWorker["worker"] = salaryGenderType["worker"]
SalaryWorker["worker (w)"] = salaryGenderType["worker (w)"]
SalaryWorker["worker (m)"] = salaryGenderType["worker (m)"]
SalaryWorker["pay disparity"] = SalaryWorker["worker (m)"] - SalaryWorker["worker (w)"]
highestSalaryWorker = SalaryWorker.nlargest(15, "pay disparity")
highestSalaryWorker = highestSalaryWorker.set_index("town code")
highestSalaryWorker
# # 2: Overall Demographics
# ## 2.1: Gender Demographics
total = townSexPopulation.loc["Total"].tolist()
import matplotlib.pyplot as plt
# Pie chart
labels = ["Men", "Women"]
sizes = total
# colors
colors = ["#66b3ff", "#ff9999"]
fig1, ax1 = plt.subplots()
patches, texts, autotexts = ax1.pie(
sizes, colors=colors, labels=labels, autopct="%1.1f%%", startangle=90
)
for text in texts:
text.set_color("grey")
for autotext in autotexts:
autotext.set_color("grey")
# Equal aspect ratio ensures that pie is drawn as a circle
ax1.axis("equal")
plt.tight_layout()
plt.show()
# ### 2.1.1: Towns with highest ratio of female vs male
import plotly.graph_objects as go
town = highestWomenRatioTable["town name"].values.tolist()
fig = go.Figure()
fig.add_trace(
go.Bar(
x=town,
y=highestWomenRatioTable["men"].values.tolist(),
name="men",
marker_color="lightblue",
)
)
fig.add_trace(
go.Bar(
x=town,
y=highestWomenRatioTable["women"].values.tolist(),
name="women",
marker_color="pink",
)
)
# Here we modify the tickangle of the xaxis, resulting in rotated labels.
fig.update_layout(
title="Top 10 towns with highest ratio of female vs male",
barmode="group",
xaxis_tickangle=-45,
)
fig.show()
# ### 2.1.2: Towns with highest ratio of male vs female
import plotly.graph_objects as go
town = highestMenRatioTable["town name"].values.tolist()
fig = go.Figure()
fig.add_trace(
go.Bar(
x=town,
y=highestMenRatioTable["men"].values.tolist(),
name="men",
marker_color="lightblue",
)
)
fig.add_trace(
go.Bar(
x=town,
y=highestMenRatioTable["women"].values.tolist(),
name="women",
marker_color="pink",
)
)
# Here we modify the tickangle of the xaxis, resulting in rotated labels.
fig.update_layout(
title="Top 10 towns with highest ratio of male vs female",
barmode="group",
xaxis_tickangle=-45,
)
fig.show()
# ## 2.2: Cohabitation Mode Demographics
import plotly.graph_objects as go
x = [
"couple w/o children",
"couple with children",
"children with parents",
"living alone",
"children with single parent",
"single parent with children",
"not living with family",
]
y = cohabitationTable.loc["Total"].tolist()
y.sort(reverse=True)
# Use the hovertext kw argument for hover text
fig = go.Figure(data=[go.Bar(x=x, y=y)])
# Customize aspect
fig.update_traces(
marker_color="#8ac6d1",
marker_line_color="#8ac6d1",
marker_line_width=1.5,
opacity=0.6,
)
fig.update_layout(title_text="Overall demographic: cohabitation mode")
fig.show()
# ## 2.3: Age Category Demographics
import plotly.graph_objects as go
y = [
"0-4 ",
"5-9 ",
"10-14 ",
"15-19 ",
"20-24 ",
"25-29 ",
"30-34 ",
"35-39 ",
"40-44 ",
"45-49 ",
"50-54 ",
"55-59 ",
"60-64 ",
"65-69 ",
"70-74 ",
"75-79 ",
]
x = AgeTable.loc["Total"].tolist()
fig = go.Figure()
fig.add_trace(
go.Bar(
y=y,
x=x,
name="overall cohabitation mode",
orientation="h",
marker=dict(color="#beebe9", line=dict(color="#beebe9", width=1)),
)
)
fig.update_layout(barmode="stack")
fig.update_layout(title_text="Overall demographic: age category")
fig.show()
# # 3: Salary Disparity
# ## 3.1: Overview of salary distribution
# It can be seen from the box plot that the salary difference between female executive and female manager is huge. The range of female salary is quite small compared to the male salary. The maximum of female salary is 35.5 while the maximum of male salary is 93.4. All of the data shown have a lot of outliers, which is understandable as the data is huge and it includes thousands of different values.
import plotly.graph_objects as go
import numpy as np
mean = Salary["women"].values.tolist()
executive = SalaryExecutive["executive (w)"].values.tolist()
manager = SalaryManager["manager (w)"].values.tolist()
employee = SalaryEmployee["employee (w)"].values.tolist()
worker = SalaryWorker["worker (w)"].values.tolist()
fig = go.Figure()
fig.add_trace(go.Box(y=mean, name="mean", marker_color="#f7d695"))
fig.add_trace(go.Box(y=executive, name="executive", marker_color="#ff80b0"))
fig.add_trace(go.Box(y=manager, name="manager", marker_color="#ff80b0"))
fig.add_trace(go.Box(y=employee, name="employee", marker_color="#ff80b0"))
fig.add_trace(go.Box(y=worker, name="worker", marker_color="#ff80b0"))
fig.update_layout(title="Overview distribution on female salary")
fig.show()
import plotly.graph_objects as go
import numpy as np
mean = Salary["man"].values.tolist()
executive = SalaryExecutive["executive (m)"].values.tolist()
manager = SalaryManager["manager (m)"].values.tolist()
employee = SalaryEmployee["employee (m)"].values.tolist()
worker = SalaryWorker["worker (m)"].values.tolist()
fig = go.Figure()
fig.add_trace(go.Box(y=mean, name="mean", marker_color="#f7d695"))
fig.add_trace(go.Box(y=executive, name="executive", marker_color="#88e1f2"))
fig.add_trace(go.Box(y=manager, name="manager", marker_color="#88e1f2"))
fig.add_trace(go.Box(y=employee, name="employee", marker_color="#88e1f2"))
fig.add_trace(go.Box(y=worker, name="worker", marker_color="#88e1f2"))
fig.update_layout(title="Overview distribution on male salary")
fig.show()
# ## 3.2: Top 15 Salary Disparity (mean net salary)
# This graph shows the top 15 salary disparity between males and females. It can be seen that the town's average is following the trend of the males instead of the females. Mainly, because there are more male working compared to females, as shown in the 2.1: gender demographics.
highestTownName = highestSalary["town name"].values.tolist()
highestMean = highestSalary["mean net salary"].values.tolist()
highestWomen = highestSalary["women"].values.tolist()
highestMen = highestSalary["man"].values.tolist()
import plotly.express as px
data = px.data.gapminder()
fig = go.Figure()
fig.add_trace(
go.Scatter(
x=highestWomen,
y=highestTownName,
marker=dict(color="pink", size=12),
mode="markers",
name="Women",
)
)
fig.add_trace(
go.Scatter(
x=highestMen,
y=highestTownName,
marker=dict(color="lightblue", size=12),
mode="markers",
name="Men",
)
)
fig.add_trace(
go.Scatter(
x=highestMean,
y=highestTownName,
marker=dict(color="beige", size=12),
mode="markers",
name="Town's average",
)
)
fig.update_layout(
title="Top 15 Gender Earnings Disparity", xaxis_title="Mean Net Salary"
)
fig.show()
# ## 3.3: Top 15 Salary Disparity among executives
# This graph shows the top 15 salary disparity between male and female executives. It can be seen that even though executive is the highest position, the salary disparity still stays the same.
highestTownName = highestSalaryExecutive["town name"].values.tolist()
highestMean = highestSalaryExecutive["executive"].values.tolist()
highestWomen = highestSalaryExecutive["executive (w)"].values.tolist()
highestMen = highestSalaryExecutive["executive (m)"].values.tolist()
fig = go.Figure()
fig.add_trace(
go.Scatter(
x=highestWomen,
y=highestTownName,
marker=dict(color="pink", size=12),
mode="markers",
name="female executive",
)
)
fig.add_trace(
go.Scatter(
x=highestMen,
y=highestTownName,
marker=dict(color="lightblue", size=12),
mode="markers",
name="male executive",
)
)
fig.add_trace(
go.Scatter(
x=highestMean,
y=highestTownName,
marker=dict(color="beige", size=12),
mode="markers",
name="Town's average",
)
)
fig.update_layout(
title="Top 15 Gender Earnings Disparity (executives)",
xaxis_title="Executive Salary",
)
fig.show()
# ## 3.4: Top 15 Salary Disparity among workers
# This graph shows the top 15 salary disparity between male and female workers. It can be seen that **Fourqueux**'s pay disparity among the male and female workers is huge compared to the others. It can be seen that Fourqueux male worker's salary is even higher than Fourqueux male executive (from the previous graph-3.2 Top 15 Salary Disparity among executives) which is a little odd.
highestTownName = highestSalaryWorker["town name"].values.tolist()
highestMean = highestSalaryWorker["worker"].values.tolist()
highestWomen = highestSalaryWorker["worker (w)"].values.tolist()
highestMen = highestSalaryWorker["worker (m)"].values.tolist()
fig = go.Figure()
fig.add_trace(
go.Scatter(
x=highestWomen,
y=highestTownName,
marker=dict(color="pink", size=12),
mode="markers",
name="female worker",
)
)
fig.add_trace(
go.Scatter(
x=highestMen,
y=highestTownName,
marker=dict(color="lightblue", size=12),
mode="markers",
name="male worker",
)
)
fig.add_trace(
go.Scatter(
x=highestMean,
y=highestTownName,
marker=dict(color="beige", size=12),
mode="markers",
name="Town's average",
)
)
fig.update_layout(
title="Top 15 Gender Earnings Disparity (workers)", xaxis_title="Worker Salary"
)
fig.show()
# # 4: Fourqueux
# The result shown in the previous graphs are a little odd. Hence, we place the data together so we can have a clearer picture of the data. It would have been interesting to find out more about Fourqueux, like the age, gender and cohabitation demographics. However, due to the missing data in *population.csv*, Fourqueux demographic is not available to analyse.
fig = go.Figure()
fig.add_trace(
go.Bar(
y=["mean", "worker", "executive"],
x=[23.4, 18.0, 31.0],
name="female",
orientation="h",
marker=dict(color="pink", line=dict(color="rgba(246, 78, 139, 1.0)", width=1)),
)
)
fig.add_trace(
go.Bar(
y=["mean", "worker", "executive"],
x=[46.9, 53.2, 51.4],
name="male",
orientation="h",
marker=dict(
color="lightblue", line=dict(color="rgba(58, 71, 80, 1.0)", width=1)
),
)
)
fig.update_layout(title="Fourqueux: Gender Earnings Disparity")
fig.show()
|
# Suppress warnings
import warnings
warnings.filterwarnings("ignore")
import pandas as pd
import numpy as np
import nltk
from nltk.corpus import stopwords
import re
import string
from sklearn.feature_extraction.text import CountVectorizer
from sklearn import model_selection
from sklearn.linear_model import LogisticRegression
# This notebook comes as a second part to the **Getting started with NLP Notebooks** that I am writing. I could have included this stuff in my previous notebook but CountVectorizers deserve a notebook of their own.
train = pd.read_csv("../input/nlp-getting-started/train.csv")
test = pd.read_csv("../input/nlp-getting-started/test.csv")
train.head()
# # What is a Countvectorizer
# The CountVectorizer provides a simple way to both tokenize a collection of text documents and build a vocabulary of known words, but also to encode new documents using that vocabulary.
# 
# We take a dataset and convert it into a corpus. Then we create a vocabulary of all the unique words in the corpus. Using this vocabulary, we can then create a feature vector of the count of the words. Let's see this through a simple example. Let's say we have a corpus containing two sentences as follows
sentences = ["The weather is sunny", "The weather is partly sunny and partly cloudy."]
from sklearn.feature_extraction.text import CountVectorizer
vectorizer = CountVectorizer()
vectorizer.fit(sentences)
vectorizer.vocabulary_
vectorizer.transform(sentences).toarray()
# ## 1. Creating a Baseline Model using Basic Countvectorizer
## Basic CountVectorizer with no parameters
count_vectorizer = CountVectorizer()
count_vectorizer.fit(train["text"])
train_vectors = count_vectorizer.transform(train["text"])
test_vectors = count_vectorizer.transform(test["text"])
list(count_vectorizer.vocabulary_)[:10]
train_vectors.shape
from sklearn.linear_model import LogisticRegression
clf = LogisticRegression(C=1.0)
scores = model_selection.cross_val_score(
clf, train_vectors, train["target"], cv=5, scoring="f1"
)
scores
# our assumption will score roughly 0.70 on the leaderboard
clf.fit(train_vectors, train["target"])
# ### Submission
sample_submission = pd.read_csv("../input/nlp-getting-started/sample_submission.csv")
sample_submission["target"] = clf.predict(test_vectors)
sample_submission.to_csv("submission.csv", index=False)
# ## 2. Countvectorizer with Stopword Parameter
# 
# Sometimes, some extremely common words which would appear to be of little value in helping select documents matching a user need are excluded from the vocabulary entirely. These words are called stop words.
# If `stop_word` parameter is specified with a list of stopwords, they will be removed from the vocabulary. Here I'll use the stopwords from NLTK but we can also specify custom stopwords too.
#
stopwords = stopwords.words("english")
count_vectorizer = CountVectorizer(stop_words=stopwords)
count_vectorizer.fit(train["text"])
train_vectors = count_vectorizer.transform(train["text"])
test_vectors = count_vectorizer.transform(test["text"])
train_vectors.shape
# See how the columns have reduced from 21637 to 21498. This is because some of the stopwords were removed.
# ### Eliminating Stopwords using MIN_DF and MAX_DF parameter
# `MIN_DF` lets you ignore those terms that appear rarely in a corpus. In other words, if `MIN_df`is 2, it means that a word has to occur at least two documents to be considered useful.
# `MAX_DF` on the other hand, ignores terms that have a document frequency strictly higher than the given threshold.These will be words which appear a lot of documents.
# This means we can eliminate those words that are either rare or appear too frequently in a corpus.
# When mentioned in absolute values i.e 1,2, etc, the value means if the word appears in 1 or 2 documents. However, when given in float, eg 30%, it means it appears in 30% of the documents.
count_vectorizer = CountVectorizer(stop_words=stopwords, min_df=2)
count_vectorizer.fit(train["text"])
train_vectors = count_vectorizer.transform(train["text"])
test_vectors = count_vectorizer.transform(test["text"])
train_vectors.shape
# The shape drastically reduces on eliminating rare terms. Now, let's see how we can apply the `MAX_DF` parameter.
count_vectorizer = CountVectorizer(stop_words=stopwords, max_df=0.8)
count_vectorizer.fit(train["text"])
train_vectors = count_vectorizer.transform(train["text"])
test_vectors = count_vectorizer.transform(test["text"])
# ## 3. CountVectorizer with Preprocessing parameter
# 
# We can also preprocess the text by passing it as an argument to countvectorizer. The following options are avialable:
# - strip_accents - This removes any accents from the text during the preprocessing step.
# - lowercase - which is default set as true but can be set to False if lowercasing isnot desired
# - preprocessor - we can create our custom preprocessor and set this argument to that.
#
# Creating a custom preprocessor that lowercases, removes special characters, removes hyperlinks and punctuation
def custom_preprocessor(text):
"""
Make text lowercase, remove text in square brackets,remove links,remove special characters
and remove words containing numbers.
"""
text = text.lower()
text = re.sub("\[.*?\]", "", text)
text = re.sub("\\W", " ", text) # remove special chars
text = re.sub("https?://\S+|www\.\S+", "", text)
text = re.sub("<.*?>+", "", text)
text = re.sub("[%s]" % re.escape(string.punctuation), "", text)
text = re.sub("\n", "", text)
text = re.sub("\w*\d\w*", "", text)
return text
count_vectorizer = CountVectorizer(
list(train["text"]), preprocessor=custom_preprocessor
)
train_vectors = count_vectorizer.fit_transform(list(train["text"]))
test_vectors = count_vectorizer.transform(list(test["text"]))
# ## 4. CountVectorizer with Tokenization parameter
# 
def custom_tokenizer(text):
tokenizer = nltk.tokenize.RegexpTokenizer(r"\w+")
text = tokenizer.tokenize(text)
return text
count_vectorizer = CountVectorizer(list(train["text"]), tokenizer=custom_tokenizer)
train_vectors = count_vectorizer.fit_transform(list(train["text"]))
test_vectors = count_vectorizer.transform(list(test["text"]))
print(list(count_vectorizer.vocabulary_)[:20])
# ## 5. CountVectorizer with N-Grams and analyzer parameter
# 
#
# World level Bigrams
count_vectorizer = CountVectorizer(
list(train["text"]), preprocessor=custom_preprocessor, ngram_range=(2, 2)
)
train_vectors = count_vectorizer.fit_transform(list(train["text"]))
test_vectors = count_vectorizer.transform(list(test["text"]))
list(count_vectorizer.vocabulary_)[:10]
# World level unigrams and bigrams
count_vectorizer = CountVectorizer(
list(train["text"]), preprocessor=custom_preprocessor, ngram_range=(1, 2)
)
train_vectors = count_vectorizer.fit_transform(list(train["text"]))
test_vectors = count_vectorizer.transform(list(test["text"]))
list(count_vectorizer.vocabulary_)[:10]
# World level trigrams
count_vectorizer = CountVectorizer(
list(train["text"]), preprocessor=custom_preprocessor, ngram_range=(3, 3)
)
train_vectors = count_vectorizer.fit_transform(list(train["text"]))
test_vectors = count_vectorizer.transform(list(test["text"]))
list(count_vectorizer.vocabulary_)[:20]
# character level bigrams
count_vectorizer = CountVectorizer(
list(train["text"]),
preprocessor=custom_preprocessor,
ngram_range=(2, 2),
analyzer="char_wb",
)
train_vectors = count_vectorizer.fit_transform(list(train["text"]))
test_vectors = count_vectorizer.transform(list(test["text"]))
print(list(count_vectorizer.vocabulary_)[:20])
|
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import plotly.express as px # Visualization
import warnings
warnings.filterwarnings("ignore")
# # About Data
# - Dataset from - goodcarbadcar.net website.
# - Data collected from 2019 to 2022.
# - Total of 4 files, each file has 13 columns with
# + Model: which content car brand and model.
# + Columns Jan - Dec: number of car sold each months.
#
# Load data
sale2019 = pd.read_csv("/kaggle/input/us-auto-sales/carsales2019.csv")
sale2020 = pd.read_csv("/kaggle/input/us-auto-sales/carsales2020.csv")
sale2021 = pd.read_csv("/kaggle/input/us-auto-sales/carsales2021.csv")
sale2022 = pd.read_csv("/kaggle/input/us-auto-sales/carsales2022.csv")
# Preview data
sale2019.head(3)
# Checking datatype and missing data
sale2019.info()
sale2020.info()
sale2021.info()
sale2022.info()
# Next step is to add column Year in each files and combine into 1 dataset
# Add column Year in each corresponding dataframes
sale2019["Year"] = 2019
sale2020["Year"] = 2020
sale2021["Year"] = 2021
sale2022["Year"] = 2022
# Combine all data together
frames = [sale2019, sale2020, sale2021, sale2022]
cardata = pd.concat(frames)
cardata.head()
# Preview data
cardata.info()
# # Data Cleanning
# 1. The months columns are object type need to change the datatype to interger. before that we need to
# - Replace values with '-' indicate no car sold for that month to 0 .
# - Also need to take get rid of the hundreds separator ','
# 2. Add column Total to sum all car sold thoughout the year.
# 3. Split column Model to Make and Model
# 1. Change data type in colums(Jan - Dec) to interger
replace = {",": "", "-": 0}
cardata.iloc[:, 1:13] = cardata.iloc[:, 1:13].replace(replace, regex=True).astype(int)
# 2. Add column Total to sum all car sale though out the year
cardata["Total"] = cardata.iloc[:, 1:13].sum(axis=1)
cardata.head()
# 3. Split column Model to Make and Model
cardata[["Make", "Model"]] = cardata["Model"].str.split(" ", expand=True, n=1)
# Reorder columns
cardata = cardata[
[
"Make",
"Model",
"Year",
"Jan",
"Feb",
"Mar",
"Apr",
"May",
"Jun",
"Jul",
"Aug",
"Sep",
"Oct",
"Nov",
"Dec",
"Total",
]
]
cardata.head()
cardata.info()
# * Dataset has total of 16 columns and 1228 rows.
# * No missing value in data
# The dataset is clean and ready for analysis and visualization
# ## Analysis & Visualization
# 1. Overall analysis
# - Total and average car sold count
# - Overall car sold by Brand distribution
# - Top 5 branch sold
# - Top 5 car brand model distrubition
# - The most and least car brand sold
#
#
# 2. Yearly analysis
# - Total car sold each year
# - Top 5 most and least car brand sold each year
#
#
# 3. Model analysis
# - Top 10 model sold
# - The most Model sold in each Brand
#
#
# 4. Monthly analysis
# - Monthly Cars Sold and Distribution
# # 1. Overall analysis
# total number of cars sold in 4 years
total_count = cardata["Total"].sum()
# avg car sold in 4 years
avg = total_count / 4
overall = pd.DataFrame(
{
"Total Car Sold": [total_count],
"Average Car Sold per Year": [avg],
}
)
overall["Total Car Sold"] = overall["Total Car Sold"].map("{:,}".format)
overall["Average Car Sold per Year"] = overall["Average Car Sold per Year"].map(
"{:,}".format
)
overall
# Overall car sold by Brand
overall_data = cardata.groupby("Make")["Total"].sum().reset_index()
fig = px.pie(
overall_data,
values="Total",
names="Make",
hole=0.4,
title="Cars brands distribution ",
)
fig.show()
# Overall top 5 brands sold
top_brand = cardata.groupby("Make")["Total"].sum().reset_index()
top_5_brand = top_brand.nlargest(5, "Total")
fig = px.bar(
top_5_brand,
x="Make",
y="Total",
color="Make",
title="Overall Top 5 brands ",
width=500,
height=500,
)
fig.show()
# top 5 car brand model distribution
data_selected = cardata.loc[:, ["Make", "Model", "Total"]]
model_group_df = data_selected.groupby(["Make", "Model"]).sum()["Total"].reset_index()
top_5_brand_name = top_5_brand["Make"].tolist()
top5_brand_model = model_group_df.loc[model_group_df["Make"].isin(top_5_brand_name)]
fix = px.treemap(
top5_brand_model,
path=["Make", "Model"],
values="Total",
title="Top 5 car brands model distribution",
)
fix.show()
# The most and least car brand sold
most_brand = top_brand.nlargest(1, "Total")
least_brand = top_brand.nsmallest(1, "Total")
most_least = pd.concat([most_brand, least_brand])
most_least = most_least.rename(index={10: "Most", 18: "Least"})
most_least["Total"] = most_least["Total"].map("{:,}".format)
most_least
# # 2. Yearly analysis
# - Total car sold each year
# - Top 5 most and least car brand sold each year
# Total car sold each year
yearly_sold = cardata.groupby("Year").sum()["Total"].reset_index()
fig = px.pie(
yearly_sold, values="Total", names="Year", hole=0.4, title=" % Total sold Count "
)
fig.show()
# Top 5 brand sold each year
# Group data by Year and Make and sum its Total sold count
select_df = cardata.loc[:, ["Year", "Make", "Model", "Total"]]
group_df = select_df.groupby(["Year", "Make"]).sum()["Total"].reset_index()
# select 2019 data
# pull out top 5 most sold in 2019 and its percentage
data_2019 = group_df[(group_df["Year"] == 2019)]
top5_2019 = data_2019.nlargest(5, "Total")
# repeat step above for each year
data_2020 = group_df[(group_df["Year"] == 2020)]
top5_2020 = data_2020.nlargest(5, "Total")
data_2021 = group_df[(group_df["Year"] == 2021)]
top5_2021 = data_2021.nlargest(5, "Total")
data_2022 = group_df[(group_df["Year"] == 2022)]
top5_2022 = data_2022.nlargest(5, "Total")
df_frames = [top5_2019, top5_2020, top5_2021, top5_2022]
yearly_most_sold = pd.concat(df_frames)
fix = px.treemap(
yearly_most_sold,
path=["Year", "Make"],
values="Total",
title="Yearly 5 most sold cars brand distribution",
)
fix.show()
# The least car brand sold each year
tail_2019 = data_2019.nsmallest(5, "Total")
tail_2020 = data_2020.nsmallest(5, "Total")
tail_2021 = data_2021.nsmallest(5, "Total")
tail_2022 = data_2022.nsmallest(5, "Total")
df_frames = [tail_2019, tail_2020, tail_2021, tail_2022]
yearly_least_sold = pd.concat(df_frames)
fix = px.treemap(
yearly_least_sold,
path=["Year", "Make"],
values="Total",
title="Yearly 5 least sold cars brand distribution",
)
fix.show()
# # 3. Model analysis
# - Top 10 model sold
# - The most Model Sold in each Brand
# Top 10 Models sold
top10 = model_group_df.nlargest(10, "Total")
fig = px.bar(
top10,
x="Model",
y="Total",
color="Make",
title="Top 10 Models sold",
width=800,
height=500,
)
fig.show()
# The most Model sold in each Brand
max_model_m = model_group_df.groupby(["Make"]).agg({"Total": "max"}).reset_index()
max_model = pd.merge(max_model_m, model_group_df, on=["Make", "Total"], how="inner")
max_model = max_model[["Make", "Model", "Total"]]
max_model
# # 4. Monthly analysis
# - Monthly Cars Sold and Distribution.
# Monthly cars sold
month_df = cardata[
["Jan", "Feb", "Mar", "Apr", "May", "Jun", "Jul", "Aug", "Sep", "Oct", "Nov", "Dec"]
]
month_agg = month_df.agg("sum").reset_index()
month_agg_df = month_agg.rename(columns={"index": "Month", 0: "Total Cars Sold"})
fig = px.line(
month_agg_df,
x="Month",
y="Total Cars Sold",
markers=True,
title="Monthly Cars Sold",
width=800,
height=600,
)
fig.show()
# Monthly Cars Sold Distribution
fig = px.pie(
month_agg_df,
values="Total Cars Sold",
names="Month",
title="Monthly Cars Sold Distribution",
hole=0.2,
)
fig.update_traces(textinfo="label+percent", insidetextfont=dict(color="white"))
fig.show()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
full_name = os.path.join(dirname, filename)
if "train.csv" in full_name:
train_raw_path = full_name
elif "test.csv" in full_name:
test_raw_path = full_name
# # Reading Dataset & Analysis
# - csv 파일을 읽고, 데이터의 기본적인 특성을 분석합니다.
import csv
import matplotlib.pyplot as plt
"""
Read Dataset
"""
def read_dataset(train_fname, test_fname):
train_data, test_data = {}, {}
positive_counter = 0
length_counter = []
all_toks = {}
# Data reading
for setname in [train_fname, test_fname]:
column_title = None
with open(setname, newline="") as csvfile:
reader = csv.reader(csvfile)
for row in reader:
if column_title is None: # First row
column_title = row
continue
sent1, sent2 = [int(el) for el in row[1].split()], [
int(el) for el in row[2].split()
]
id_ = int(row[0])
if len(column_title) == 4: # Train
label = int(row[3])
positive_counter += label
train_data[id_] = {"sent1": sent1, "sent2": sent2, "label": label}
length_counter.extend([len(sent1), len(sent2)])
for tok in sent1 + sent2:
assert isinstance(tok, int)
if tok not in all_toks:
all_toks[tok] = 0
all_toks[tok] += 1
else: # Test
test_data[id_] = {"sent1": sent1, "sent2": sent2}
return train_data, test_data, positive_counter, length_counter, all_toks
train_data, test_data, positive_counter, length_counter, all_toks = read_dataset(
train_raw_path, test_raw_path
)
"""
General Statistic
"""
print("[Dataset size]")
print("Train data size: {}".format(len(train_data)))
print("Test data size: {}".format(len(test_data)))
print("\n[Label Distribution]")
print("Pos/Neg: {}/{}".format(positive_counter, len(train_data) - positive_counter))
print("\n[Sentence Length]")
print("Median: {}".format(sorted(length_counter)[len(length_counter) // 2]))
print("Average: {}".format(sum(length_counter) / len(length_counter)))
print("Max, Min: {} {}".format(max(length_counter), min(length_counter)))
print("\n[Vocab Size]")
print("Unique words count: {}".format(len(list(all_toks.keys()))))
import operator
sorted_all_toks = sorted(all_toks.items(), key=operator.itemgetter(1), reverse=True)
plt.figure(figsize=(15, 10))
plt.hist([el[1] for el in sorted_all_toks], label="train", normed=True)
plt.title("Word frequency")
plt.xlabel("Number of word")
plt.xlabel("Frequency")
plt.legend()
plt.show()
dupl_counter = {} # 'sentence': count
for key, item in train_data.items():
sent1, sent2 = " ".join([str(el) for el in item["sent1"]]), " ".join(
[str(el) for el in item["sent2"]]
)
if sent1 in dupl_counter:
dupl_counter[sent1] += 1
else:
dupl_counter[sent1] = 1
if sent2 in dupl_counter:
dupl_counter[sent2] += 1
else:
dupl_counter[sent2] = 1
sentence_frequency = list(dupl_counter.values())
plt.figure(figsize=(12, 5))
plt.hist(sentence_frequency, label="train", normed=True)
plt.title("Histogram of sentence frequency")
plt.legend()
plt.show()
# # Build Vocabulary
# - 이후 모듈들에서 사용할 단어장 클래스를 구축합니다.
class Config:
def __init__(self):
self.w2v_dim = 200
self.vocab_size = 8200
config = Config()
class Vocab:
def __init__(self):
self.word2id, self.id2word = {}, {}
def __len__(self):
assert len(self.word2id) == len(self.id2word)
return len(self.word2id)
def build_vocab(self, all_toks):
# Only with train data, without test data
assert len(self.word2id) == 0
for idx, word in enumerate(all_toks):
self.word2id[word[0]] = idx
assert len(self.word2id) == len(all_toks) == config.vocab_size - 1
self.unk_id = len(self.word2id)
self.word2id["<unk>"] = self.unk_id
for k, v in self.word2id.items():
self.id2word[v] = k
assert len(self.word2id) == len(self.id2word)
print("Vocab size is: {}".format(len(self.word2id)))
self.vocab_size = len(self.word2id)
assert self.vocab_size == config.vocab_size
def sent2ids(self, sent):
assert all([isinstance(tok, int) for tok in sent]) and isinstance(sent, list)
return [
self.word2id[tok] if tok in self.word2id else self.unk_id for tok in sent
]
# # Preprocessing
# - Tokenizing을 거친 데이터가 주어졌기 때문에, 이외에 추가적인 전처리는 진행하지 않습니다.
# - 데이터셋의 word index를 전체 데이터셋의 단어 개수에 맞게 정리합니다.
import pickle
kaggle_path = "/kaggle/working/"
train_bin_fname = kaggle_path + "train_bin.pck"
test_bin_fname = kaggle_path + "test_bin.pck"
vocab_bin_fname = kaggle_path + "vocab_bin.pck"
def preprocess(train_data, test_data, all_toks):
"""
Save the data with replacing the old vocab into own.
"""
# Build Vocab
vocab = Vocab()
vocab.build_vocab(all_toks[: config.vocab_size - 1])
with open(vocab_bin_fname, "wb") as f:
pickle.dump(vocab, f)
my_train_data, my_test_data = {}, {}
for id_, item in train_data.items():
train_data[id_]["sent1"] = vocab.sent2ids(item["sent1"])
train_data[id_]["sent2"] = vocab.sent2ids(item["sent2"])
with open(train_bin_fname, "wb") as f:
pickle.dump(train_data, f)
for id_, item in test_data.items():
test_data[id_]["sent1"] = vocab.sent2ids(item["sent1"])
test_data[id_]["sent2"] = vocab.sent2ids(item["sent2"])
with open(test_bin_fname, "wb") as f:
pickle.dump(test_data, f)
return train_data, test_data, vocab
train_data, test_data, vocab = preprocess(train_data, test_data, sorted_all_toks)
# # Utility Functions
# - 아래 3개의 함수 및 변수는 Feature-based classifier를 구축하는데에 필요한 함수입니다.
# - `get_ngram()`: 두 문장 사이의 n-gram overlap을 측정하는 함수입니다.
# - `get_w2v_model()`: Train set에 있는 데이터를 기반으로 훈련한 word2vec 모델을 반환합니다.
# - `idf_values`: Train set에 있는 단어들의 inverted document frquency를 반환합니다.
"""
Utility functions
"""
def get_ngram(sent, gram=1):
"""
Args:
sent: A list of integers
Return:
result: set of n-grams for the given sent
"""
assert isinstance(sent, list) and all([isinstance(el, int) for el in sent])
result = []
for idx, val in enumerate(sent):
if idx == len(sent) - gram + 1:
break
result.append(" ".join([str(el) for el in sent[idx : idx + gram]]))
return set(result)
# Test
a = [1, 2, 3, 4, 5]
print(get_ngram(a, 1))
print(get_ngram(a, 2))
print(get_ngram(a, 3))
from gensim.models import Word2Vec
"""
Train and load the w2v model
"""
print(vocab.unk_id)
def get_w2v_model():
kaggle_path = "/kaggle/working/"
embedding_path = os.path.join(kaggle_path, "w2v.bin")
sentences = []
all_tok = []
for k, v in train_data.items():
sentences.append(
[str(el) for el in v["sent1"]] + [str(el) for el in v["sent2"]]
)
all_tok.extend(sentences[-1])
model = Word2Vec(sentences, size=config.w2v_dim, window=3, min_count=1)
model.save(embedding_path)
print("8199" in all_tok)
return model
model = get_w2v_model()
print(model)
kaggle_path = "/kaggle/working/"
embedding_path = os.path.join(kaggle_path, "w2v.bin")
"""
Build idf matrix
"""
# Build IDF Matrix
inverted_index = {tok: [] for tok in range(len((vocab.word2id.keys())))}
for idx, sample in train_data.items():
for word_id in sample["sent1"]:
assert isinstance(word_id, int)
inverted_index[word_id].append(2 * idx)
for word_id in sample["sent2"]:
assert isinstance(word_id, int)
inverted_index[word_id].append(2 * idx + 1)
for k, v in inverted_index.items():
inverted_index[k] = set(v)
idf_values = {
k: np.log(2 * len(list(train_data.keys())) / (len(v) + 1))
for k, v in inverted_index.items()
}
# # Feature Engineering
# -`FeatureEngineer()` 클래스는 두 문장이 가진 특징을 추출하고, 이를 classifier에게 전달하는 것을 목표로 합니다. 추출한 feature는 다음과 같습니다.
# ## Sentence-pair features
# - `ngram-overlap`: 두 문장 사이에 겹치는 n-gram을 카운트합니다.
# - `mt-based`: 두 문장 사이의 BLEU score를 측정합니다.
# ## Sentence features
# - `bag-of-word with IDF`: 각 문장에 등장한 단어들을 IDF score로 weighting합니다.
# - `word2vec`: Training 셋에서 훈련한 word2vec 모델로부터 각 단어들의 word embedding 값을 얻고, 이를 min/max/avg pooling하여 문장에 대한 벡터를 얻습니다.
# - 이 때, bag-of-word 및 word2vec의 차원 수가 크기 때문에, 이를 축소하기 위해 `kernel_for_single_sents()`에서 정의한 여러 종류의 kernel을 사용합니다.
"""
A class for traditional NLP feature engineering
"""
from nltk.translate.bleu_score import sentence_bleu, SmoothingFunction
from numpy import dot
from numpy.linalg import norm
from scipy.spatial.distance import cdist
from scipy.stats import spearmanr
from sklearn.metrics.pairwise import rbf_kernel, polynomial_kernel, laplacian_kernel
from sklearn.preprocessing import minmax_scale
# from nltk.translate.nist_score import sentence_nist
class FeatureEngineer:
def __init__(self, idf_values):
self.bleu_smoother = SmoothingFunction()
self.w2v = get_w2v_model()
self.idf_values = idf_values
def get_feature(self, sent1, sent2):
pair_feature = self.get_sentence_pair_feature(sent1, sent2)
single_feature = self.get_sentence_feature(sent1, sent2)
feature = np.concatenate((pair_feature, single_feature), axis=0)
return feature
def get_sentence_pair_feature(self, sent1, sent2):
ngram = self.get_ngram_overlap(sent1, sent2)
mt = self.get_mt_feature(sent1, sent2)
return np.concatenate((ngram, mt), axis=0)
def get_sentence_feature(self, sent1, sent2):
s1_bow, s2_bow = self.get_bow_feature(sent1), self.get_bow_feature(sent2)
s1_emb, s2_emb = self.get_embedding_feature(sent1), self.get_embedding_feature(
sent2
)
bow_feature = self.kernal_for_single_sents(s1_bow, s2_bow)
emb_feature = self.kernal_for_single_sents(s1_emb, s2_emb)
return np.concatenate((bow_feature, emb_feature), axis=0)
def get_bow_feature(self, sent):
bow = [0 for _ in range(vocab.vocab_size)]
for wordid in sent:
bow[wordid] += self.idf_values[wordid] # Weighted by idf
return np.array(bow)
def get_embedding_feature(self, sent):
"""
For a given sentence, make w2v feature with min/max/avg pooling.
"""
embedding_stack = np.asarray([self.w2v.wv[str(tok)] for tok in sent])
min_pool, max_pool, avg_pool = (
embedding_stack.min(0),
embedding_stack.max(0),
np.average(embedding_stack, 0),
)
return np.concatenate((min_pool, max_pool, avg_pool))
def get_mt_feature(self, sent1, sent2):
bleu = sentence_bleu(
[sent1], sent2, smoothing_function=self.bleu_smoother.method3
)
# nist = sentence_nist([sent1], sent2)
# return [float(bleu), float(nist)]
return np.array([float(bleu)])
def get_ngram_overlap(self, sent1, sent2):
# Original Ref: https://www.aclweb.org/anthology/S12-1060.pdf
overlaps = [0, 0, 0]
for n in range(3):
sent1_gram = get_ngram(sent1, n + 1)
sent2_gram = get_ngram(sent2, n + 1)
len_sum = max(1, len(sent1_gram) + len(sent2_gram))
overlaps[n] = 2 / len_sum * len(sent1_gram & sent2_gram)
return np.array(overlaps)
def kernal_for_single_sents(self, feature1, feature2):
# To reduce the dimension of features of two sentences.
cosine = 1 - dot(feature1, feature2) / (norm(feature1) * norm(feature2))
manhanttan = 1 - cdist([feature1], [feature2], metric="cityblock")[0][0]
euclidean = np.linalg.norm(feature1 - feature2)
spearman = spearmanr(feature1, feature2)[0]
sigmoid = 1 / (1 + np.exp(-dot(feature1, feature2)))
rbf = rbf_kernel(np.array([feature1]) - np.array([feature2]), gamma=1)[0][0]
polynomial = polynomial_kernel(np.array([feature1]), np.array([feature2]))[0][0]
laplacian = laplacian_kernel(np.array([feature1]), np.array([feature2]))[0][0]
return np.array(
[
cosine,
manhanttan,
euclidean,
spearman,
sigmoid,
rbf,
polynomial,
laplacian,
]
)
engineer = FeatureEngineer(idf_values)
# Test
sent1, sent2 = [1, 2, 3, 4, 5], [4, 5, 6, 7, 8]
print(engineer.get_feature(sent1, sent2))
def scaling_feature(X):
"""
Scaling each feature into 0-1.
"""
return minmax_scale(X, axis=0, copy=True)
# # Feature Extraction Script
# - 앞서 제작한 `FeatureEngineer` 클래스를 이용하여 데이터셋을 처리하는 스크립트입니다.
from sklearn.model_selection import train_test_split
save_path = os.path.join(kaggle_path, "processed/")
if not os.path.exists(save_path):
os.makedirs(save_path)
serialized_fname = os.path.join(save_path, "processed_everything.pck")
if os.path.exists(serialized_fname):
with open(serialized_fname, "rb") as f:
X_train, X_dev, y_train, y_dev, test_X = pickle.load(f)
else:
X, Y = [], []
for cid, el in train_data.items():
if cid % 1000 == 0:
print(cid)
Y.append(el["label"])
X.append(engineer.get_feature(el["sent1"], el["sent2"]))
X_train, X_dev, y_train, y_dev = train_test_split(
np.array(X), np.array(Y), test_size=0.1, random_state=1515
)
test_X = []
for cid, el in test_data.items():
test_X.append(engineer.get_feature(el["sent1"], el["sent2"]))
test_X = np.array(test_X)
# Serialize
with open(os.path.join(save_path, "processed_everything.pck"), "wb") as f:
pickle.dump([X_train, X_dev, y_train, y_dev, test_X], f)
"""Normalize each feature"""
scaled_X_train, scaled_X_dev = scaling_feature(X_train), scaling_feature(X_dev)
scaled_X_test = scaling_feature(test_X)
# # Vizualize feature space
# - 이렇게 표현된 sentence pair의 feature space를 T-SNE를 통해 시각화한 결과는 아래와 같습니다.
import matplotlib.pyplot as plt
from sklearn.manifold import TSNE
model = TSNE(learning_rate=100)
transformed = model.fit_transform(scaled_X_train)
xs = transformed[:, 0]
ys = transformed[:, 1]
plt.scatter(xs, ys, c=Y[:36000], s=3)
plt.show()
# # Feature-based Classifiers
# - 앞서 추출한 feature를 기반으로 similar여부를 판단하는 모델들입니다.
# - 본 프로젝트에서는 총 5개의 Classifier (Random Forest, Gradient Boosting, SVM, XGBoost, SGD)를 실험하였습니다.
from sklearn.ensemble import RandomForestClassifier, GradientBoostingClassifier
from sklearn import svm
from sklearn.linear_model import SGDClassifier
from sklearn.metrics import accuracy_score
from xgboost import XGBClassifier
class FeatureRegressor:
def __init__(self):
pass
def predict(self, X, Y):
print("## Classifier Prediction Begin ##")
self.rf_pred, self.gb_pred, self.svm_pred, self.xgb_pred, self.sgd_pred = (
self.rf.predict(X),
self.gb.predict(X),
self.svm.predict(X),
self.xgb.predict(X),
self.sgd.predict(X),
)
rf_acc, gb_acc, svm_acc, xgb_acc, sgd_acc = (
accuracy_score(Y, self.rf_pred),
accuracy_score(Y, self.gb_pred),
accuracy_score(Y, self.svm_pred),
accuracy_score(Y, self.xgb_pred),
accuracy_score(Y, self.sgd_pred),
)
print("## Individual Classifier Accuracy ##")
print("Random Forest: {}".format(rf_acc))
print("Gradient Boosting: {}".format(gb_acc))
print("SVM: {}".format(svm_acc))
print("XGBoost: {}".format(xgb_acc))
print("SGD: {}".format(sgd_acc))
print("#" * 15)
def train_classifiers(self, X, Y):
print("## Classifier Train Begin ##")
self.RandomForest(X, Y)
self.GradientBoosting(X, Y)
self.SVM(X, Y)
self.XGBoost(X, Y)
self.SGD(X, Y)
def RandomForest(self, X_train, Y_train):
self.rf = RandomForestClassifier(n_estimators=100, oob_score=True)
self.rf.fit(X_train, Y_train)
def GradientBoosting(self, X_train, Y_train):
self.gb = GradientBoostingClassifier(n_estimators=100, learning_rate=0.1)
self.gb.fit(X_train, Y_train)
def SVM(self, X_train, Y_train):
self.svm = svm.SVC()
self.svm.fit(X_train, Y_train)
def XGBoost(self, X_train, Y_train):
## Best Performace 0.77
self.xgb = XGBClassifier(
n_estimators=100, gamma=3, min_child_weight=1, max_depth=5
)
self.xgb.fit(X_train, Y_train)
def SGD(self, X_train, Y_train):
self.sgd = SGDClassifier(max_iter=500)
self.sgd.fit(X_train, Y_train)
regressor = FeatureRegressor()
# - XGBoost 모델의 hyperparameter tuning을 위한 스크립트입니다.
# Grid Hyperparmeter search
from sklearn.model_selection import KFold, GridSearchCV
from xgboost import XGBClassifier
if False: # Set as True for tuning
model = XGBClassifier()
parameter_grid = {
"booster": ["gbtree"],
"silent": [True],
"max_depth": [5, 8, 10],
"min_child_weight": [1, 3, 5],
"gamma": [0, 1, 2, 3],
"n_estimators": [100],
}
cv = KFold(n_splits=5, random_state=1)
gcv = GridSearchCV(model, param_grid=parameter_grid, cv=cv, scoring="f1", n_jobs=5)
gcv.fit(
np.concatenate((scaled_X_train, scaled_X_dev), 0),
np.concatenate((y_train, y_dev), 0),
)
print("final_params: ", gcv.best_params_)
print("best score", gcv.best_score_)
# - 각 모델들을 훈련하고 결과를 저장합니다. 각 모델들의 결과를 정리하면 아래와 같습니다.
# ```
# ## Individual Classifier Accuracy ##
# Random Forest: 0.6785
# Gradient Boosting: 0.68325
# SVM: 0.68825
# XGBoost: 0.6875
# SGD: 0.69275
# ###############
# ```
TRAIN_WITH_FULL_DATA = (
True # True when the model is trained with full train data (True for submission)
)
if TRAIN_WITH_FULL_DATA:
regressor.train_classifiers(
np.concatenate((scaled_X_train, scaled_X_dev), 0),
np.concatenate((y_train, y_dev), 0),
)
else:
regressor.train_classifiers(scaled_X_train, y_train)
regressor.predict(scaled_X_dev, y_dev)
regressor_prediction = [regressor.xgb.predict(scaled_X_test)]
# regressor_prediction = [regressor.rf.predict(scaled_X_test), regressor.gb.predict(scaled_X_test), regressor.svm.predict(scaled_X_test), regressor.xgb.predict(scaled_X_test), regressor.sgd.predict(scaled_X_test)]
# # Deep Learning based Approach
# - 이후는 딥러닝 기반의 sentence-pair classifier를 구축하기 위한 코드입니다.
# - 모델은 Natural Language Inference task에서 높은 성능을 보였던 ESIM(Enhanced LSTM for Natural Language Inference)을 사용하였습니다. [Paper](https://arxiv.org/pdf/1609.06038.pdf)
# - 모델의 구현은 다음의 링크를 참고하였습니다: [Link](https://github.com/coetaur0/ESIM)
#
import string
import torch
from collections import Counter
from torch.utils.data import Dataset
"""
Data loader class for model
"""
class STSDataset(Dataset):
def __init__(self, data, max_len=None, pad_idx=vocab.vocab_size, is_train=True):
self.sent1_len = [len(el["sent1"]) for k, el in data.items()]
self.sent2_len = [len(el["sent2"]) for k, el in data.items()]
self.max_sent1_len, self.max_sent2_len = max(self.sent1_len), max(
self.sent2_len
)
self.is_train = is_train
self.data_num = len(self.sent1_len)
self.data = {
"ids": [],
"sent1": torch.ones((self.data_num, self.max_sent1_len), dtype=torch.long)
* pad_idx,
"sent2": torch.ones((self.data_num, self.max_sent2_len), dtype=torch.long)
* pad_idx,
}
if self.is_train:
self.data["labels"] = torch.tensor(
[el["label"] for k, el in data.items()], dtype=torch.long
)
for idx, item in enumerate(data.values()):
self.data["ids"].append(idx)
final_pivot = min(len(item["sent1"]), self.max_sent1_len)
self.data["sent1"][idx][:final_pivot] = torch.tensor(
item["sent1"][:final_pivot]
)
final_pivot = min(len(item["sent2"]), self.max_sent1_len)
self.data["sent2"][idx][:final_pivot] = torch.tensor(
item["sent2"][:final_pivot]
)
def __len__(self):
return self.data_num
def __getitem__(self, index):
if self.is_train:
return {
"ids": self.data["ids"][index],
"sent1": self.data["sent1"][index],
"sent2": self.data["sent2"][index],
"sent1_len": min(self.sent1_len[index], self.max_sent1_len),
"sent2_len": min(self.sent2_len[index], self.max_sent2_len),
"label": self.data["labels"][index],
}
else:
return {
"ids": self.data["ids"][index],
"sent1": self.data["sent1"][index],
"sent2": self.data["sent2"][index],
"sent1_len": min(self.sent1_len[index], self.max_sent1_len),
"sent2_len": min(self.sent2_len[index], self.max_sent2_len),
}
# Hyperparameters
class DLConfig:
def __init__(self):
self.batch_size = 32
self.epoch = 15
self.lr = 3e-4
self.keep_rate = 0.5
self.num_class = 2
self.max_grad_norm = 8
self.hidden_dim = 200
self.embed_dim = 200
self.model_path = "model/{}/"
config = DLConfig()
import torch.nn as nn
def sort_by_seq_lens(batch, sequences_lengths, descending=True):
sorted_seq_lens, sorting_index = sequences_lengths.sort(0, descending=descending)
sorted_batch = batch.index_select(0, sorting_index)
idx_range = sequences_lengths.new_tensor(torch.arange(0, len(sequences_lengths)))
_, reverse_mapping = sorting_index.sort(0, descending=False)
restoration_index = idx_range.index_select(0, reverse_mapping)
return sorted_batch, sorted_seq_lens, sorting_index, restoration_index
def get_mask(sequences_batch, sequences_lengths):
batch_size = sequences_batch.size()[0]
max_length = torch.max(sequences_lengths)
mask = torch.ones(batch_size, max_length, dtype=torch.float)
mask[sequences_batch[:, :max_length] == 0] = 0.0
return mask
def masked_softmax(tensor, mask):
tensor_shape = tensor.size()
reshaped_tensor = tensor.view(-1, tensor_shape[-1])
# Reshape the mask so it matches the size of the input tensor.
while mask.dim() < tensor.dim():
mask = mask.unsqueeze(1)
mask = mask.expand_as(tensor).contiguous().float()
reshaped_mask = mask.view(-1, mask.size()[-1])
result = nn.functional.softmax(reshaped_tensor * reshaped_mask, dim=-1)
result = result * reshaped_mask
# 1e-13 is added to avoid divisions by zero.
result = result / (result.sum(dim=-1, keepdim=True) + 1e-13)
return result.view(*tensor_shape)
def weighted_sum(tensor, weights, mask):
weighted_sum = weights.bmm(tensor)
while mask.dim() < weighted_sum.dim():
mask = mask.unsqueeze(1)
mask = mask.transpose(-1, -2)
mask = mask.expand_as(weighted_sum).contiguous().float()
return weighted_sum * mask
def replace_masked(tensor, mask, value):
mask = mask.unsqueeze(1).transpose(2, 1)
reverse_mask = 1.0 - mask
values_to_add = value * reverse_mask
return tensor * mask + values_to_add
def check_prediction(prob, target):
_, out_classes = prob.max(dim=1)
correct = (out_classes == target).sum()
return correct.item()
class RNNDropout(nn.Dropout):
def forward(self, batch):
ones = batch.new_ones(batch.shape[0], batch.shape[-1])
mask = nn.functional.dropout(ones, self.p, self.training, inplace=False)
return mask.unsqueeze(1) * batch
class SeqEncoder(nn.Module):
def __init__(
self, rnn_type, input_dim, hidden_dim, layer_num, dropout, bidirectional=True
):
super(SeqEncoder, self).__init__()
self.rnn_type = rnn_type
self.input_dim = input_dim
self.hidden_dim = hidden_dim
self.layer_num = layer_num
self.dropout = dropout
self.bidirectional = bidirectional
self._encoder = rnn_type(
input_dim,
hidden_dim,
num_layers=layer_num,
bias=True,
batch_first=True,
dropout=dropout,
bidirectional=bidirectional,
)
def forward(self, batch, lens):
sorted_batch, sorted_lens, _, restoration_idx = sort_by_seq_lens(batch, lens)
packed_batch = nn.utils.rnn.pack_padded_sequence(
sorted_batch, sorted_lens, batch_first=True
)
outputs, _ = self._encoder(packed_batch, None)
outputs, _ = nn.utils.rnn.pad_packed_sequence(outputs, batch_first=True)
reordered_outputs = outputs.index_select(0, restoration_idx)
return reordered_outputs
class Attention(nn.Module):
def forward(self, sent1_batch, sent1_mask, sent2_batch, sent2_mask):
matrix = sent1_batch.bmm(sent2_batch.transpose(2, 1).contiguous())
sent1_attn = masked_softmax(matrix, sent2_mask)
sent2_attn = masked_softmax(matrix.transpose(1, 2).contiguous(), sent1_mask)
attned_sent1 = weighted_sum(sent2_batch, sent1_attn, sent1_mask)
attned_sent2 = weighted_sum(sent1_batch, sent2_attn, sent2_mask)
return attned_sent1, attned_sent2
class ESIM(nn.Module):
def __init__(
self,
vocab_size=vocab.vocab_size + 1,
embedding_dim=config.embed_dim,
hidden_dim=config.hidden_dim,
embeddings=None,
padding_idx=vocab.vocab_size,
keep_rate=config.keep_rate,
num_classes=2,
device="cpu",
):
super(ESIM, self).__init__()
self.vocab_size = vocab_size
self.embedding_dim = embedding_dim
self.hidden_dim = hidden_dim
self.num_classes = num_classes
self.dropout = 1 - keep_rate
self.device = device
self.word_embedding = nn.Embedding(
self.vocab_size, self.embedding_dim, padding_idx=padding_idx
)
if embeddings is not None:
self.word_embedding.weight = nn.Parameter(embeddings)
self.rnn_dropout = RNNDropout(p=self.dropout)
self.encoding = SeqEncoder(
nn.LSTM, self.embedding_dim, self.hidden_dim, 1, self.dropout
)
self.attention = Attention()
self.projection = nn.Sequential(
nn.Linear(8 * self.hidden_dim, self.hidden_dim), nn.ReLU()
)
self.composition = SeqEncoder(
nn.LSTM, self.hidden_dim, self.hidden_dim, 1, self.dropout
)
self.fcn = nn.Sequential(
nn.Dropout(p=self.dropout),
nn.Linear(8 * self.hidden_dim, self.hidden_dim),
nn.Tanh(),
nn.Dropout(p=self.dropout),
nn.Linear(self.hidden_dim, self.num_classes),
)
self.apply(_init_model_weights)
def forward(self, sent1, sent1_lens, sent2, sent2_lens):
sent1_mask = get_mask(sent1, sent1_lens).to(self.device)
sent2_mask = get_mask(sent2, sent2_lens).to(self.device)
emb_sent1 = self.word_embedding(sent1)
emb_sent2 = self.word_embedding(sent2)
emb_sent1 = self.rnn_dropout(emb_sent1)
emb_sent2 = self.rnn_dropout(emb_sent2)
enc_sent1 = self.encoding(emb_sent1, sent1_lens)
enc_sent2 = self.encoding(emb_sent2, sent2_lens)
attn_sent1, attn_sent2 = self.attention(
enc_sent1, sent1_mask, enc_sent2, sent2_mask
)
rich_sent1 = torch.cat(
[enc_sent1, attn_sent1, enc_sent1 - attn_sent1, enc_sent1 * attn_sent1],
dim=-1,
)
rich_sent2 = torch.cat(
[enc_sent2, attn_sent2, enc_sent2 - attn_sent2, enc_sent2 * attn_sent2],
dim=-1,
)
projected_sent1 = self.rnn_dropout(self.projection(rich_sent1))
projected_sent2 = self.rnn_dropout(self.projection(rich_sent2))
var_sent1 = self.composition(projected_sent1, sent1_lens)
var_sent2 = self.composition(projected_sent2, sent2_lens)
var_sent1_avg = torch.sum(
var_sent1 * sent1_mask.unsqueeze(1).transpose(2, 1), dim=1
) / torch.sum(sent1_mask, dim=1, keepdim=True)
var_sent2_avg = torch.sum(
var_sent2 * sent2_mask.unsqueeze(1).transpose(2, 1), dim=1
) / torch.sum(sent2_mask, dim=1, keepdim=True)
var_sent1_max, _ = replace_masked(var_sent1, sent1_mask, -1e7).max(dim=1)
var_sent2_max, _ = replace_masked(var_sent2, sent2_mask, -1e7).max(dim=1)
v = torch.cat(
[var_sent1_avg, var_sent1_max, var_sent2_avg, var_sent2_max], dim=-1
)
logits = self.fcn(v)
prob = nn.functional.softmax(logits, dim=-1)
return logits, prob
def _init_model_weights(module):
if isinstance(module, nn.Linear):
nn.init.xavier_uniform_(module.weight.data)
nn.init.constant_(module.bias.data, 0.0)
elif isinstance(module, nn.LSTM):
nn.init.xavier_uniform_(module.weight_ih_l0.data)
nn.init.orthogonal_(module.weight_hh_l0.data)
nn.init.constant_(module.bias_ih_l0.data, 0.0)
nn.init.constant_(module.bias_hh_l0.data, 0.0)
hidden_size = module.bias_hh_l0.data.shape[0] // 4
module.bias_hh_l0.data[hidden_size : (2 * hidden_size)] = 1.0
if module.bidirectional:
nn.init.xavier_uniform_(module.weight_ih_l0_reverse.data)
nn.init.orthogonal_(module.weight_hh_l0_reverse.data)
nn.init.constant_(module.bias_ih_l0_reverse.data, 0.0)
nn.init.constant_(module.bias_hh_l0_reverse.data, 0.0)
module.bias_hh_l0_reverse.data[hidden_size : (2 * hidden_size)] = 1.0
# - 아래는 환경 구축, Training 및 Testing을 위한 함수입니다.
import shutil
from random import shuffle
import matplotlib.pyplot as plt
from torch.utils.data import DataLoader
from tqdm import tqdm
from tensorboardX import SummaryWriter
def build_env(train_, test_, config):
train_dataset = STSDataset(esim_train_data)
train_loader = DataLoader(train_dataset, shuffle=True, batch_size=config.batch_size)
test_dataset = STSDataset(esim_test_data)
test_loader = DataLoader(test_dataset, shuffle=True, batch_size=config.batch_size)
evaluation_dataset = STSDataset(test_data, is_train=False)
evaluation_loader = DataLoader(evaluation_dataset, shuffle=False, batch_size=1)
config.model_path = "model/{}/".format(config.exp_name)
config.save_path, config.log_path = os.path.join(
config.model_path, "save"
), os.path.join(config.model_path, "logdir")
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
return (
train_dataset,
train_loader,
test_dataset,
test_loader,
evaluation_dataset,
evaluation_loader,
config,
device,
)
"""
Test Script
"""
def test_script(model, evaluation_loader, config, restore_epoch=13):
config.batch_size = 1
assert os.path.exists(config.save_path)
model.eval()
checkpoint = torch.load(
os.path.join(config.save_path, "esim_{}.pth.tar".format(restore_epoch))
)
model.load_state_dict(checkpoint["model"])
esim_result = []
with torch.no_grad():
for batch in evaluation_loader:
sent1, sent1_len, sent2, sent2_len = (
batch["sent1"].to(device),
batch["sent1_len"].to(device),
batch["sent2"].to(device),
batch["sent2_len"].to(device),
)
_, prob = model(sent1, sent1_len, sent2, sent2_len)
esim_result.append(prob)
return esim_result
"""
Train Script
"""
def train(model, train_loader, test_loader, config):
# save path and summary writer
if os.path.exists(config.model_path):
shutil.rmtree(config.model_path)
os.makedirs(config.save_path)
os.makedirs(config.log_path)
criterion = nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(model.parameters(), lr=config.lr)
# Begin training
print("\n", "#" * 30, "TRAINING BEGIN", "#" * 30)
step = 0
train_epoch_loss, train_epoch_acc, valid_epoch_loss, valid_epoch_acc = [
[] for _ in range(4)
]
for epoch in range(config.epoch):
model.train()
train_loss, valid_loss = 0, 0
train_accuracy, valid_accuracy = 0, 0
batch_iterator = train_loader
# Training
for batch_index, batch in enumerate(batch_iterator):
sent1, sent1_len, sent2, sent2_len, label = (
batch["sent1"].to(device),
batch["sent1_len"].to(device),
batch["sent2"].to(device),
batch["sent2_len"].to(device),
batch["label"].to(device),
)
optimizer.zero_grad()
logit, prob = model(sent1, sent1_len, sent2, sent2_len)
loss = criterion(logit, label)
accuracy = check_prediction(prob, label)
loss.backward()
nn.utils.clip_grad_norm_(model.parameters(), config.max_grad_norm)
optimizer.step()
train_loss += loss.item()
train_accuracy += accuracy
step += 1
print(
"-> {} epoch {} step Loss {:.4f} Train Accuracy{:.4f}%".format(
epoch,
step,
loss.item() / config.batch_size,
accuracy / config.batch_size,
)
)
train_loss /= len(train_loader)
train_accuracy /= len(train_loader)
# Validation
with torch.no_grad():
for batch in test_loader:
sent1, sent1_len, sent2, sent2_len, label = (
batch["sent1"].to(device),
batch["sent1_len"].to(device),
batch["sent2"].to(device),
batch["sent2_len"].to(device),
batch["label"].to(device),
)
logit, prob = model(sent1, sent1_len, sent2, sent2_len)
loss = criterion(logit, label)
accuracy = check_prediction(prob, label)
valid_loss += loss.item()
valid_accuracy += accuracy
valid_loss /= len(test_loader)
valid_accuracy /= len(test_loader)
# Save the model at every epoch.
torch.save(
{
"epoch": epoch,
"model": model.state_dict(),
"train_loss": train_loss,
"valid_loss": valid_loss,
"train_acc": train_accuracy,
"valid_acc": valid_accuracy,
},
os.path.join(config.save_path, "esim_{}.pth.tar".format(epoch)),
)
train_epoch_loss.append(train_loss)
train_epoch_acc.append(train_accuracy / config.batch_size * 100)
valid_epoch_loss.append(valid_loss)
valid_epoch_acc.append(valid_accuracy / config.batch_size * 100)
# Draw the plot
plt.figure()
epoch_list = [_ for _ in range(config.epoch)]
plt.plot(epoch_list, train_epoch_loss, "-r")
plt.plot(epoch_list, valid_epoch_loss, "-b")
plt.xlabel("epoch")
plt.ylabel("loss")
plt.legend(["Training loss", "Validation loss"])
plt.title("Cross entropy loss")
plt.show()
plt.savefig(os.path.join(config.model_path, "loss.png"))
# Draw the plot
plt.figure()
plt.plot(epoch_list, train_epoch_acc, "-r")
plt.plot(epoch_list, valid_epoch_acc, "-b")
plt.xlabel("epoch")
plt.ylabel("Accuracy(%)")
plt.legend(["Training loss", "Validation loss"])
plt.title("Accuracy")
plt.show()
plt.savefig(os.path.join(config.model_path, "accuracy.png"))
print("SAVEPATH: {}".format(os.path.join(config.model_path, "accuracy.png")))
print([round(el, 2) for el in valid_epoch_acc])
print(
"#" * 30,
"Validation Best Accuracy: {} at {} epoch".format(
max(valid_epoch_acc), np.argmax(valid_epoch_acc)
),
)
# - 위에서 구현한 모델 및 함수를 기반으로 한 스크립트 파일은 아래와 같습니다.
# - Training set에 대해 10-fold cross validation을 통해 모델을 검증하고, 총 10개의 모델을 Ensemble하여 testing set의 결과를 예측하였습니다.
from pprint import pprint
"""
MAIN SCRIPT
"""
key_list = [key for key, val in train_data.items()]
shuffle(key_list)
esim_aggregation = []
exp_name = "esim_{}_10fold"
for fold_index in range(10):
# Dataset split
esim_train_data, esim_test_data = {}, {}
for k, v in train_data.items():
if str(k)[-1] == str(fold_index):
esim_test_data[k] = v
else:
esim_train_data[k] = v
print("#" * 10, "{} of 10 fold cross-validation begin".format(fold_index))
exp_name = "esim_{}_10fold".format(fold_index)
config.exp_name = exp_name
(
train_dataset,
train_loader,
test_dataset,
test_loader,
evaluation_dataset,
evaluation_loader,
config,
device,
) = build_env(esim_train_data, esim_test_data, config)
w2v_model = Word2Vec.load("/kaggle/working/w2v.bin")
w2v_embedding = torch.tensor(
np.array(
[w2v_model.wv[str(el)] for el in range(vocab.vocab_size)]
+ [np.array([0.0 for _ in range(config.embed_dim)])]
),
dtype=torch.float,
).to(device)
print("Experiment name: {}".format(config.exp_name))
print("Save path: {}".format(config.save_path))
print("Model path: {}".format(config.model_path))
print("Device is {}".format(device))
print("TRAINING START")
model = ESIM(embeddings=w2v_embedding, device=device).to(device)
train(model, train_loader, test_loader, config)
print("EVAL START")
model = ESIM(keep_rate=1.0, device=device).to(device)
esim_aggregation.append(test_script(model, evaluation_loader, config))
print("###" * 10)
print("ESIM aggregeation len: {}".format(len(esim_aggregation)))
print("ESIM aggregeation element len: {}".format(len(esim_aggregation[-1])))
# Aggregating ESIM CV results
AGGREGATION_BY_PROB = False
prob_results = [[0.0, 0.0] for _ in range(10000)]
if AGGREGATION_BY_PROB:
for one_fold in esim_aggregation:
for idx, el in enumerate(one_fold):
el = el.cpu().numpy()[0]
assert len(el) == 2
prob_results[idx][0] += el[0]
prob_results[idx][1] += el[1]
else:
for one_fold in esim_aggregation:
for idx, el in enumerate(one_fold):
el = el.cpu().numpy()[0]
assert len(el) == 2
if el[0] > el[1]:
prob_results[idx][0] += 1
else:
prob_results[idx][1] += 1
esim_voting_result = [0 if el[0] > el[1] else 1 for el in prob_results]
# # Final Voting
# - 본 프로젝트에서는 두 개의 서로 다른 페러다임에 기반한 모델들을 구축하였습니다 (1. Hand-crafted feature based classifier 2. deep-learning based classifier). 각 모델들의 예측 결과를 다수결에 기반하여 aggregation하는 코드는 아래와 같습니다.
# - `SELECTED_APPROACH`를 통해 ensemble approach를 선택할 수 있습니다.
"""
Final Voting
"""
SELECTED_APPROACH = "esim" # One of ['full_ensemble', 'feature', 'esim', 'xgb']
SUBMISSION_FNAME = (
"esim_10fold" #'esim_only'#'ensemble_full_data_esim_no_tuning_xgb_gb_esim'
)
def ensemble(pred):
"""
Args:
pred(list): List of prediction result for each classifier.
shape of [n_classifier, np.array(n_test_size)]
"""
prediction = np.transpose(
np.array(pred)
) # Shape of [test_sample_num, classifier_num]
result = []
for preds in prediction:
if sum(preds) >= len(preds) / 2: # positive is majority case
result.append(1)
else:
result.append(0)
return np.array(result)
if SELECTED_APPROACH == "full_ensemble":
regressor_prediction.extend(esim_aggregation)
voting_result = ensemble(regressor_prediction)
elif SELECTED_APPROACH == "esim":
voting_result = ensemble([esim_voting_result])
elif SELECTED_APPROACH == "feature":
voting_result = ensemble(regressor_prediction)
elif SELECTED_APPROACH == "xgb":
assert len(regressor_prediction) == 1
voting_result = ensemble(regressor_prediction)
def make_submission(result, name=""):
submission_dir = os.path.join(kaggle_path, "submission_dir")
if not os.path.exists(submission_dir):
os.makedirs(submission_dir)
fname = os.path.join(submission_dir, "submission_chaehun_{}.csv".format(name))
with open(fname, "w", newline="") as f:
writer = csv.writer(f)
writer.writerow(["id", "label"])
for idx, item in enumerate(result):
writer.writerow([40001 + idx, item])
make_submission(voting_result, name=SUBMISSION_FNAME)
# To get the label distribution of test set
# make_submission([1 for _ in range(10000)], name='all_1')
# make_submission([0 for _ in range(10000)], name='all_0')
|
# The goal of this notebook is to explore the data of both the Men's and Women's competitions with the following goals;
# * How did the game evolved over the years?
# * What stats are most useful to predict the outcome of a game?
# * What are the differences between the Men's and Women's competitions?
# To do so, we need to produce some aggregated statistics. The next few hidden cells have all the function to do just that.
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import explore_data as exp
pd.set_option("max_columns", 300)
def process_details(data):
df = data.copy()
stats = [
"Score",
"FGM",
"FGA",
"FGM3",
"FGA3",
"FTM",
"FTA",
"OR",
"DR",
"Ast",
"TO",
"Stl",
"Blk",
"PF",
"FGM2",
"FGA2",
"Tot_Reb",
"FGM_no_ast",
"Def_effort",
"Reb_opp",
"possessions",
"off_rating",
"def_rating",
"scoring_opp",
"TO_perposs",
"impact",
]
for prefix in ["W", "L"]:
df[prefix + "FG_perc"] = df[prefix + "FGM"] / df[prefix + "FGA"]
df[prefix + "FGM2"] = df[prefix + "FGM"] - df[prefix + "FGM3"]
df[prefix + "FGA2"] = df[prefix + "FGA"] - df[prefix + "FGA3"]
df[prefix + "FG2_perc"] = df[prefix + "FGM2"] / df[prefix + "FGA2"]
df[prefix + "FG3_perc"] = df[prefix + "FGM3"] / df[prefix + "FGA3"]
df[prefix + "FT_perc"] = df[prefix + "FTM"] / df[prefix + "FTA"]
df[prefix + "Tot_Reb"] = df[prefix + "OR"] + df[prefix + "DR"]
df[prefix + "FGM_no_ast"] = df[prefix + "FGM"] - df[prefix + "Ast"]
df[prefix + "FGM_no_ast_perc"] = df[prefix + "FGM_no_ast"] / df[prefix + "FGM"]
df[prefix + "possessions"] = (
df[prefix + "FGA"]
- df[prefix + "OR"]
+ df[prefix + "TO"]
- 0.475 * df[prefix + "FTA"]
)
df[prefix + "off_rating"] = (
df[prefix + "Score"] / df[prefix + "possessions"] * 100
)
df[prefix + "scoring_opp"] = (
df[prefix + "FGA"] + 0.475 * df[prefix + "FTA"]
) / df[prefix + "possessions"]
df[prefix + "TO_perposs"] = df[prefix + "TO"] / df[prefix + "possessions"]
df[prefix + "IE_temp"] = (
df[prefix + "Score"]
+ df[prefix + "FTM"]
+ df[prefix + "FGM"]
+ df[prefix + "DR"]
+ 0.5 * df[prefix + "OR"]
- df[prefix + "FTA"]
- df[prefix + "FGA"]
+ df[prefix + "Ast"]
+ df[prefix + "Stl"]
+ 0.5 * df[prefix + "Blk"]
- df[prefix + "PF"]
)
df["Wdef_rating"] = df["Loff_rating"]
df["Ldef_rating"] = df["Woff_rating"]
df["Wimpact"] = df["WIE_temp"] / (df["WIE_temp"] + df["LIE_temp"])
df["Limpact"] = df["LIE_temp"] / (df["WIE_temp"] + df["LIE_temp"])
del df["WIE_temp"]
del df["LIE_temp"]
df[[col for col in df.columns if "perc" in col]] = df[
[col for col in df.columns if "perc" in col]
].fillna(0)
df["WReb_opp"] = df["WDR"] / (df["LFGA"] - df["LFGM"])
df["LReb_opp"] = df["LDR"] / (df["WFGA"] - df["WFGM"])
return df
def full_stats(data):
df = data.copy()
to_select = [col for col in df.columns if "W" in col and "_perc" not in col]
df_W = df[["Season", "DayNum", "NumOT"] + to_select].copy()
df_W.columns = df_W.columns.str.replace("W", "")
df_W["N_wins"] = 1
to_select = [col for col in df.columns if "L" in col and "_perc" not in col]
df_L = df[["Season", "DayNum", "NumOT"] + to_select].copy()
df_L.columns = df_L.columns.str.replace("L", "")
df_L = df_L.rename(columns={"Woc": "Loc"})
df_L["N_wins"] = 0
df = pd.concat([df_W, df_L])
del df["DayNum"]
del df["Loc"]
to_use = [col for col in df.columns if col != "NumOT"]
means = df[to_use].groupby(["Season", "TeamID"], as_index=False).mean()
sums = df[to_use].groupby(["Season", "TeamID"], as_index=False).sum()
sums["FGM_perc"] = sums.FGM / sums.FGA
sums["FGM2_perc"] = sums.FGM2 / sums.FGA2
sums["FGM3_perc"] = sums.FGM3 / sums.FGA3
sums["FT_perc"] = sums.FTM / sums.FTA
sums["FGM_no_ast_perc"] = sums.FGM_no_ast / sums.FGM
to_use = [
"Season",
"TeamID",
"FGM_perc",
"FGM2_perc",
"FGM3_perc",
"FT_perc",
"FGM_no_ast_perc",
]
sums = sums[to_use].fillna(0)
stats_tot = pd.merge(means, sums, on=["Season", "TeamID"])
return stats_tot
def add_seed(seed_location, total):
seed_data = pd.read_csv(seed_location)
seed_data["Seed"] = seed_data["Seed"].apply(lambda x: int(x[1:3]))
total = pd.merge(total, seed_data, how="left", on=["TeamID", "Season"])
return total
def make_teams_target(data, league):
if league == "men":
limit = 2003
else:
limit = 2010
df = data[data.Season >= limit].copy()
df["Team1"] = np.where((df.WTeamID < df.LTeamID), df.WTeamID, df.LTeamID)
df["Team2"] = np.where((df.WTeamID > df.LTeamID), df.WTeamID, df.LTeamID)
df["target"] = np.where((df["WTeamID"] < df["LTeamID"]), 1, 0)
df["target_points"] = np.where(
(df["WTeamID"] < df["LTeamID"]), df.WScore - df.LScore, df.LScore - df.WScore
)
df.loc[df.WLoc == "N", "LLoc"] = "N"
df.loc[df.WLoc == "H", "LLoc"] = "A"
df.loc[df.WLoc == "A", "LLoc"] = "H"
df["T1_Loc"] = np.where((df.WTeamID < df.LTeamID), df.WLoc, df.LLoc)
df["T2_Loc"] = np.where((df.WTeamID > df.LTeamID), df.WLoc, df.LLoc)
df["T1_Loc"] = df["T1_Loc"].map({"H": 1, "A": -1, "N": 0})
df["T2_Loc"] = df["T2_Loc"].map({"H": 1, "A": -1, "N": 0})
reverse = data[data.Season >= limit].copy()
reverse["Team1"] = np.where(
(reverse.WTeamID > reverse.LTeamID), reverse.WTeamID, reverse.LTeamID
)
reverse["Team2"] = np.where(
(reverse.WTeamID < reverse.LTeamID), reverse.WTeamID, reverse.LTeamID
)
reverse["target"] = np.where((reverse["WTeamID"] > reverse["LTeamID"]), 1, 0)
reverse["target_points"] = np.where(
(reverse["WTeamID"] > reverse["LTeamID"]),
reverse.WScore - reverse.LScore,
reverse.LScore - reverse.WScore,
)
reverse.loc[reverse.WLoc == "N", "LLoc"] = "N"
reverse.loc[reverse.WLoc == "H", "LLoc"] = "A"
reverse.loc[reverse.WLoc == "A", "LLoc"] = "H"
reverse["T1_Loc"] = np.where(
(reverse.WTeamID > reverse.LTeamID), reverse.WLoc, reverse.LLoc
)
reverse["T2_Loc"] = np.where(
(reverse.WTeamID < reverse.LTeamID), reverse.WLoc, reverse.LLoc
)
reverse["T1_Loc"] = reverse["T1_Loc"].map({"H": 1, "A": -1, "N": 0})
reverse["T2_Loc"] = reverse["T2_Loc"].map({"H": 1, "A": -1, "N": 0})
df = pd.concat([df, reverse], ignore_index=True)
to_drop = ["WScore", "WTeamID", "LTeamID", "LScore", "WLoc", "LLoc", "NumOT"]
for col in to_drop:
del df[col]
df.loc[:, "ID"] = (
df.Season.astype(str) + "_" + df.Team1.astype(str) + "_" + df.Team2.astype(str)
)
return df
def make_training_data(details, targets):
tmp = details.copy()
tmp.columns = ["Season", "Team1"] + [
"T1_" + col for col in tmp.columns if col not in ["Season", "TeamID"]
]
total = pd.merge(targets, tmp, on=["Season", "Team1"], how="left")
tmp = details.copy()
tmp.columns = ["Season", "Team2"] + [
"T2_" + col for col in tmp.columns if col not in ["Season", "TeamID"]
]
total = pd.merge(total, tmp, on=["Season", "Team2"], how="left")
if total.isnull().any().any():
raise ValueError("Something went wrong")
stats = [col[3:] for col in total.columns if "T1_" in col]
for stat in stats:
total["delta_" + stat] = total["T1_" + stat] - total["T2_" + stat]
return total
def prepare_data(league):
save_loc = "processed_data/" + league + "/"
if league == "women":
main_loc = "../input/google-cloud-ncaa-march-madness-2020-division-1-womens-tournament/"
regular_season = (
main_loc + "WDataFiles_Stage1/WRegularSeasonDetailedResults.csv"
)
playoff = main_loc + "WDataFiles_Stage1/WNCAATourneyDetailedResults.csv"
playoff_compact = main_loc + "WDataFiles_Stage1/WNCAATourneyCompactResults.csv"
seed = main_loc + "WDataFiles_Stage1/WNCAATourneySeeds.csv"
else:
main_loc = (
"../input/google-cloud-ncaa-march-madness-2020-division-1-mens-tournament/"
)
regular_season = (
main_loc + "MDataFiles_Stage1/MRegularSeasonDetailedResults.csv"
)
playoff = main_loc + "MDataFiles_Stage1/MNCAATourneyDetailedResults.csv"
playoff_compact = main_loc + "MDataFiles_Stage1/MNCAATourneyCompactResults.csv"
seed = main_loc + "MDataFiles_Stage1/MNCAATourneySeeds.csv"
# Season stats
reg = pd.read_csv(regular_season)
reg = process_details(reg)
regular_stats = full_stats(reg)
regular_stats = add_seed(seed, regular_stats)
# Playoff stats
play = pd.read_csv(playoff)
play = process_details(play)
playoff_stats = full_stats(play)
playoff_stats = add_seed(seed, playoff_stats)
# Target data generation
target_data = pd.read_csv(playoff_compact)
target_data = make_teams_target(target_data, league)
all_reg = make_training_data(regular_stats, target_data)
return all_reg, regular_stats, playoff_stats
men_train, men_reg, men_play = prepare_data("men")
men_reg.head()
women_train, women_reg, women_play = prepare_data("women")
women_reg.head()
# # Teams over the years
# It must first be noticed that both competitions saw an increase in the number of teams participating, showing a somewhat similar pattern in the period from 2010 to last year.
fig, ax = plt.subplots(1, 2, figsize=(15, 6))
fig.suptitle("Number of teams competing", fontsize=18)
men_reg.groupby("Season").TeamID.nunique().plot(ax=ax[0])
women_reg.groupby("Season").TeamID.nunique().plot(ax=ax[1])
ax[0].set_title("Men's competition", fontsize=14)
ax[1].set_title("Women's competition", fontsize=14)
ax[0].axvline(2010, color="r", linestyle="--")
plt.show()
# We can then look at how the game has changed over the years by looking at how, on average, the stats are changing season by season
stats = [
"Score",
"FGA",
"FGM",
"FGM_perc",
"FGA3",
"FGM3",
"FGM3_perc",
"FT_perc",
"DR",
"OR",
"Ast",
"TO",
"Stl",
"Blk",
"possessions",
]
for col in stats:
fig, ax = plt.subplots(1, 2, figsize=(15, 6), sharey=True)
fig.suptitle(col, fontsize=18)
men_reg.groupby("Season")[col].mean().plot(ax=ax[0], label="Men")
women_reg.groupby("Season")[col].mean().plot(ax=ax[0], label="Women")
men_play.groupby("Season")[col].mean().plot(ax=ax[1], label="Men")
women_play.groupby("Season")[col].mean().plot(ax=ax[1], label="Women")
ax[0].set_title("Regular Season", fontsize=14)
ax[1].set_title("NCAA Tourney", fontsize=14)
ax[0].axvline(2010, color="r", linestyle="--")
ax[1].axvline(2010, color="r", linestyle="--")
ax[0].legend()
ax[1].legend()
plt.show()
# From these plots, we can see that
# * Men tend to score more than Women, although the Men score less during the playoff while the Women have score a similar amount of points both during regular season and playoffs.
# * Both Men and Women are attempting more and more shots. In particular the number of shots attempted in a Men's game increase quite dramatically in 2016.
# * Men are more accurate when shooting and the accuracy remained stable over the years. We again see a drop in shooting accuracy during the playoff, probably due to a more fierce defence.
# * The increased number of shot attempted seems to be coming from behind the 3 points line. We can also see that this did not affected the accuracy. This is probably telling us that the game, very much as in the NBA and WNBA, has changed towards, focusing its action along the perimeter more and more.
# * The free throw percentage of Men and Women is very similar and it has been slightly increasing
# * Women used to get way more defensive rebounds, but since 2016 Men have caught them up and the difference, both in regular season and in the playoffs, is minimal. This might be related to the larger number of shots taken from the perimeter, which are a low percentage shots and thus increase the opportunities for a defensive rebound.
# * Similarly, the offensive rebounds have been consistently decreasing, with the Women having the upper hand.
# * If in the regular season the number of assists is staying the same for Men and Women (with the now usual jump in 2016 for the Men), we observe opposite trends in the NCAA Tourney. Here, Women are getting more and more assists while Men show a more individualistic game.
# * Women have more turnover but the number, for both Men and Women, is decreasing. Almost as a mirror image, Women steal the ball more.
# * The number of blocks is very similar between Men and Women and it is remaining reasonably stable.
# * Women have more possessions per game but Men are catching up since 2016
# The main takeaway, once again, is that something seems to have changed in the Men's game since 2016.
# # Turning up the heat
# It is no mistery that teams want to bring their A-game in the playoff. This section will explore if this is always the case. We aggregate by seed in order to see if the team quality is somewhat influencing any pattern when we compare regular season and playoff statistics.
men_tot = pd.merge(men_reg, men_play, on=["Season", "TeamID", "Seed"], how="inner")
women_tot = pd.merge(
women_reg, women_play, on=["Season", "TeamID", "Seed"], how="inner"
)
stats = [
"Score",
"FGA",
"FGM",
"FGM_perc",
"FGA3",
"FGM3",
"FGM3_perc",
"FT_perc",
"DR",
"OR",
"Ast",
"FGM_no_ast",
"TO",
"Stl",
"Blk",
"possessions",
]
for stat in stats:
fig, ax = plt.subplots(1, 2, figsize=(15, 8), sharey=True)
fig.suptitle(stat, fontsize=18)
tmp = men_tot[["Seed", f"{stat}_x", f"{stat}_y"]].copy()
tmp.rename(columns={f"{stat}_x": "Regular", f"{stat}_y": "Playoff"}, inplace=True)
tmp.groupby("Seed").mean().sort_values("Seed", ascending=False).plot(
kind="barh", ax=ax[0]
)
tmp = women_tot[["Seed", f"{stat}_x", f"{stat}_y"]].copy()
tmp.rename(columns={f"{stat}_x": "Regular", f"{stat}_y": "Playoff"}, inplace=True)
tmp.groupby("Seed").mean().sort_values("Seed", ascending=False).plot(
kind="barh", ax=ax[1]
)
ax[0].set_title("Men's Competition", fontsize=14)
ax[1].set_title("Women's Competition", fontsize=14)
plt.show()
# Nothing particulary suprising, but we can call out a few things:
# * All the team generally score less during the playoff, but this is more evident for lower seed teams. In particular, in the Women's competition the drop is very large for low ranked teams. This is very understandable, considering that Seed 1 and Seed 16 teams generally meet in the very first round.
# * The Field Goals Attempted, however, tend to go higher during the playoffs
# * The Free Throws percentage is not changing much during the playoffs, but higher ranked teams are stepping up their game
# * Similarly to scoring, defensive rebounds are a key statistic for higher ranking teams
# * Both Men and Women tend to share the ball less during the playoffs but the gap between high and low seeds is more evident in the Women's tournament
# * The number of turnovers stays roughly the same but the number of steals decreases for both Men and Women
# * The same can be said for the number of Blocked shots, with the exception of Seed 1 Women's teams that stay consistent with their regular season performance
# * The number of possessions goes up during the playoffs, in particular for low ranked teams.
# # What makes a winner?
# We turn our attention to the Turney games and on how the regular season statistics can help us to predict their outcome.
# First let's see what features most correlate with the point difference for the **Men's competition**
men_corr = high_corr = exp.plot_correlations(
men_train, target="target_points", limit=12, annot=True
)
# Ignoring the feature `target` as it is obviously very correlated, we see how the Seed of the competing teams is the most correlated feature, either as a difference between the two teams or simply as individual entry. This is very correlated with pretty much all the remaining top features. We can see this correlation even further if we plot these features against the point difference.
exp.corr_target(men_train, "target_points", list(men_corr[2:].index), x_estimator=None)
# Which is suggesting that rather than the individual statistics of Team 1 and 2, it might be a good idea to focus on the differences between Team 1 and Team 2.
# Similarly, for the **Women's competition** we get
women_corr = high_corr = exp.plot_correlations(
women_train, target="target_points", limit=12, annot=True
)
# We notice that **the correlations are much stronger**, confirming the importance of the Seed in predicting the outcome of the games.
# Further investigation confirms this insight
exp.corr_target(
women_train, "target_points", list(women_corr[2:].index), x_estimator=None
)
# Showing a much sharper trend than in the Men's counterpart.
# This invites a further investigation: **how frequently an advantage in a statistic translates into a victory?**
# *Note: the statistics are considering the performance during the regular season as a whole, we are not looking at game statistics vs outcome of the same game*
men_delta = men_train[
["Season", "target", "target_points"]
+ [col for col in men_train if "delta_" in col and "Loc" not in col]
].copy()
women_delta = women_train[
["Season", "target"]
+ [col for col in women_train if "delta_" in col and "Loc" not in col]
].copy()
men_scores = []
men_feats = []
women_scores = []
women_feats = []
for col in [col for col in men_delta if "delta_" in col]:
men_delta[col] = np.sign(men_delta[col])
women_delta[col] = np.sign(women_delta[col])
if "Seed" in col or col == "delta_TO":
men_delta[col] = -men_delta[col]
women_delta[col] = -women_delta[col]
try:
men_scores.append(men_delta.groupby(col)["target"].mean()[1])
men_feats.append(col)
except KeyError:
pass
try:
women_scores.append(women_delta.groupby(col)["target"].mean()[1])
women_feats.append(col)
except KeyError:
pass
men_prob = pd.DataFrame({"feat": men_feats, "Men": men_scores})
women_prob = pd.DataFrame({"feat": women_feats, "Women": women_scores})
tot_prob = pd.merge(men_prob, women_prob, on="feat").sort_values("Men", ascending=False)
tot_prob["feat"] = tot_prob.feat.str.replace("delta_", "")
fig, ax = plt.subplots(1, figsize=(15, 6))
tot_prob.plot(x="feat", kind="bar", ax=ax)
ax.set_xticklabels(ax.get_xticklabels(), fontsize=12)
ax.set_xlabel("")
ax.set_ylim((0, 1))
ax.set_title("Percentage of wins given the stat advantage", fontsize=16)
ax.grid(axis="y")
ax.set_yticklabels(["{:,.0%}".format(x) for x in ax.get_yticks()])
plt.show()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Libraries to be installed
# pyarrow latest version does not work with setfit so pyarrow8.0.0 must be installed.
import setfit
from sentence_transformers.losses import CosineSimilarityLoss
from setfit import SetFitModel, SetFitTrainer, sample_dataset
import pandas as pd
import numpy as np
from datasets import load_dataset
from datasets import Dataset
import tensorflow as tf
from sklearn.model_selection import train_test_split
import tensorflow as tf
import seaborn as sns
from sklearn.metrics import confusion_matrix
import matplotlib.pyplot as plt
# #Did not help
# import torch
# torch.cuda.empty_cache()
# #pip install accelerate
# For setfit to function data must be in a specific format
# 1. Data must be a dataset
# 2. column names of dataset must be text->containing the data to train on , labels: data to predict
# 3. Dataset must be small to ensure cuda does not runs out of space.
# 4. Labels in categorical format must be converted to numerical data
train = pd.read_csv("/kaggle/input/book-genre-prediction/data.csv")
print(train.head())
print(train["genre"].unique())
print(train["genre"].nunique())
fig, ax = plt.subplots(figsize=(10, 7))
ax.hist(train["genre"])
# This cell is where we preprocess data and convert it in a format suitable for setfit
genre_encode = dict(zip(train["genre"].unique(), [*range(0, train["genre"].nunique())]))
train["genre"].replace(genre_encode.keys(), genre_encode.values(), inplace=True)
genre_encode
train = train[["genre", "summary"]]
train = train.sample(frac=1)
train = train.iloc[:100, :]
print(train.head())
print(train["genre"].unique())
print(train["genre"].nunique())
fig, ax = plt.subplots(figsize=(10, 7))
ax.hist(train["genre"])
X_train, X_test, y_train, y_test = train_test_split(
train["summary"], train["genre"], train_size=0.5, test_size=0.5, random_state=0
)
train_ds_dict = {"text": X_train, "label": y_train}
train_ds = Dataset.from_dict(train_ds_dict)
test_ds_dict = {"text": X_test, "label": y_test}
test_ds = Dataset.from_dict(test_ds_dict)
# This is the basic model referred from the following link
# https://huggingface.co/blog/setfit
model = SetFitModel.from_pretrained("sentence-transformers/paraphrase-mpnet-base-v2")
trainer = SetFitTrainer(
model=model,
train_dataset=train_ds,
eval_dataset=test_ds,
loss_class=CosineSimilarityLoss,
batch_size=128,
num_iterations=1, # Number of text pairs to generate for contrastive learning
num_epochs=1, # Number of epochs to use for contrastive learning
column_mapping={"text": "text", "label": "label"},
)
trainer.train()
# ****Experiments with Ray****
# Trials with ray in order to allocate memory and clusters in a better way so the system doesnot run out of memory. Pickle library gave an error and was not resolved by installing a suitable version.
import ray
import time
from datetime import timedelta
import psutil
import pickle5 as pickle
num_cpus = psutil.cpu_count(logical=True)
ray.init(num_cpus=num_cpus, ignore_reinit_error=True)
@ray.remote
def setfit_train():
model = SetFitModel.from_pretrained(
"sentence-transformers/paraphrase-mpnet-base-v2"
)
trainer = SetFitTrainer(
model=model,
train_dataset=train_ds,
eval_dataset=test_ds,
loss_class=CosineSimilarityLoss,
batch_size=128,
num_iterations=1, # Number of text pairs to generate for contrastive learning
num_epochs=1, # Number of epochs to use for contrastive learning
column_mapping={"text": "text", "label": "label"},
)
trainer.train()
return trainer
start = time.time()
trainer = ray.get(setfit_train.remote())
end = time.time()
# trainer=setfit_train().remote()
ray.shutdown()
# Using hyperparameters refering to another link the setfit was successfuly run: https://www.kaggle.com/code/talmanr/easy-few-shot-imdb-prediction-with-setfit
# load model,make sure that you active GPU for reasonable run time
model = SetFitModel.from_pretrained("sentence-transformers/paraphrase-mpnet-base-v2")
# Create trainer
trainer = SetFitTrainer(
model=model,
train_dataset=train_ds,
eval_dataset=test_ds,
loss_class=CosineSimilarityLoss,
batch_size=8,
learning_rate=2e-05,
num_iterations=40, # Number of text pairs to generate for contrastive learning
num_epochs=1, # Number of epochs to use for contrastive learning
)
# train and evaluate :
# train should take ~10 min
trainer.train()
metrics = trainer.evaluate()
metrics
train_ds
test_pred = trainer.model.predict(train_ds["text"])
sns.heatmap(confusion_matrix(train_ds["label"], test_pred), annot=True)
plt.ylabel("label")
plt.xlabel("prediction")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Load the Fake and True news data frames
fake_news_df = pd.read_csv("/kaggle/input/rue-and-fake-news/Fake.csv")
true_news_df = pd.read_csv("/kaggle/input/rue-and-fake-news/True.csv")
# **4. Concatenate the Fake and True news data frames column-wise**
# Concatenate the data frames column-wise
concatenated_df = pd.concat([fake_news_df, true_news_df], axis=1)
concatenated_df
# **5. Reindex the concatenated data frame by resetting the index**
concatenated_df = concatenated_df.reset_index(drop=True)
concatenated_df
# **6. Convert the title and text column to lower case letters**
# Convert 'title' and 'text' columns to lowercase
# concatenated_df[('Title', '')] = concatenated_df[('Title', '')].str.lower()
# concatenated_df[('Text', '')] = concatenated_df[('Text', '')].str.lower()
|
# ### Импорты
import pandas as pd
import numpy as np
import random
import pathlib
import collections
import pickle
import gc
import functools
import networkx as nx
import itertools
import operator
import os
from tqdm import tqdm
tqdm.pandas()
# ### Константы
USE_CLEANED_CACHE = True
USE_PAIRS_CACHE = True
USE_TARGET_CACHE = True
USE_METRICS_CACHE = True
USE_EDGE_FEATURES_CACHE = False
TEST_SIZE = 0.1
RANDOM_SEED = 42
CACHE_OUT_FOLDER = pathlib.Path("../working/cache")
CACHE_IN_FOLDER = pathlib.Path("../input/social-task-v1/cache")
# Создаем директорию для кэшей и задаем сид для воспроизводимости
if not os.path.exists(CACHE_OUT_FOLDER):
os.makedirs(CACHE_OUT_FOLDER)
random.seed(RANDOM_SEED)
np.random.seed(RANDOM_SEED)
if not USE_CLEANED_CACHE:
path = pathlib.Path("../input/socialtaskfriends/friends_dataset.csv")
df = pd.read_csv(path, names=["id1", "id2", "time", "inten"])
print(df[["time", "inten"]].describe())
df[:2]
# ### Очистка
# Взглянем на связи в нашем графе:
# Посчитаем число ребер между каждой парой вершины, которые имеют ребра.
#
# Оказывается, граф у нас направленный, и по-видимому, друзьями являются пользователи (вершины) между которыми есть ребра в обоих направлениях, для каждого направления указываются activity (они могут быть разными). Немного не понятно тогда, какая связь у пользователей, между которыми одно ребро (один - подписчик другого?) и больше 2х ребер (тут вообще никакого объяснения, кроме как ошибки при сборе данных не напрашивается). Исходя из этого, удалим пары пользователей у которых число связей != 2.
CLEANED_FILENAME = "cleaned.csv"
if USE_CLEANED_CACHE:
print(f"Trying to read from {str(CACHE_IN_FOLDER / CLEANED_FILENAME)}")
cleaned = pd.read_csv(CACHE_IN_FOLDER / CLEANED_FILENAME)
else:
data = collections.defaultdict(list)
def find_intersections(row):
id1 = row["id1"]
id2 = row["id2"]
higher = max(id1, id2)
lower = min(id1, id2)
data[(higher, lower)].append(row.name)
df[["id1", "id2"]].progress_apply(find_intersections, axis=1)
# take a look at the data
lens = list(map(len, data.values()))
srs = pd.Series(lens, dtype="int8")
srs.name = "Number of edges"
print("Description:\n", srs.describe())
print("\nUnique values:\n", srs.value_counts())
# drop the edges, that have no connection in the opposite direction or have multiple connections between two nodes
outliers = [val for val in data.values() if len(val) != 2]
outliers = np.concatenate(outliers).astype("int32")
cleaned = df.drop(outliers, axis=0).reset_index(drop=True)
# save to cache anyway, to keep it persistent between Kaggle Kernel relaunch
print(f"Saving")
cleaned.to_csv(CACHE_OUT_FOLDER / CLEANED_FILENAME, index=False)
print(f"Saved to {str(CACHE_OUT_FOLDER / CLEANED_FILENAME)}")
# #### Построение графа и нахождение всех пар вершин, на расстоянии <= 2
# Граф и пары вершин нужны только когда мы рассчитываем метрики в первый раз, в других случаях, мы используем закэшированные результаты
g = nx.from_pandas_edgelist(
cleaned.rename(columns={"time": 1, "inten": 2}),
source="id1",
target="id2",
edge_attr=[1, 2],
create_using=nx.DiGraph,
)
del cleaned
gc.collect()
if USE_PAIRS_CACHE:
print("Trying to read adjancencies")
with open(CACHE_IN_FOLDER / "adjs.npy", "rb") as f:
pairs_np = np.load(f)
else:
print("Calculating adjancencies")
all_adjs = {}
for idx, node in enumerate(tqdm(sorted(g.nodes))):
adjs = g[node]
l2_adjs = set()
for adj_node in adjs:
l2_adjs = l2_adjs | set(g[adj_node])
l2_adjs = l2_adjs - set([node])
all_adjs[node] = sorted(l2_adjs)
print("Calculating pairs")
pairs = []
for idx, (start, finish) in enumerate(tqdm(all_adjs.items())):
first_col = np.full((len(finish)), start, dtype="int32")
second_col = np.asarray(finish, dtype="int32")
pairs.append(np.column_stack((first_col, second_col)))
pairs_np = np.concatenate(pairs, axis=0)
print("Saving adjacency pairs")
with open(CACHE_OUT_FOLDER / "adjs.npy", "wb") as f:
np.save(f, pairs_np)
print("Saved")
@functools.lru_cache()
def are_friends(g, node1, node2):
return node2 in g[node1]
if USE_TARGET_CACHE:
print("Reading target")
with open(CACHE_IN_FOLDER / "target.npy", "rb") as f:
target = np.load(f)
else:
target = np.array(
[
are_friends(g, row[0], row[1])
for row in tqdm(pairs_np, mininterval=1, maxinterval=60)
],
dtype="bool",
)
# save to cache anyway, to keep it persistent between Kaggle Kernel relaunch
print("Saving target")
with open(CACHE_OUT_FOLDER / "target.npy", "wb") as f:
np.save(f, target)
print("Saved")
def compute_metrics(g, metrics, node_pair):
node1, node2 = node_pair
result = np.zeros(len(metrics), dtype="float32")
for i, metric in enumerate(metrics):
result[i] = np.round(metric(g, node1, node2), 3)
return result
def compute_features(g, feature_agg, node_pair):
node1, node2 = node_pair
res = [agg(feature(g, node1, node2)) for feature, agg in feature_agg]
return res
def common_neighbors_score(g, node1, node2):
common_n = _common_neighbors(g, node1, node2)
return common_n.shape[0]
@functools.lru_cache()
def _common_neighbors(g, node1, node2):
node1_n = list(g[node1])
node2_n = list(g[node2])
common_n = np.intersect1d(node1_n, node2_n, assume_unique=True)
return common_n
def adamic_adar_score(g, node1, node2):
common_n = _common_neighbors(g, node1, node2)
degrees = _common_degree(g, common_n)
inv_log = np.divide(1.0, np.log(degrees + 1e-2))
inv_log[inv_log < 0] = 0
return np.sum(inv_log)
def _common_degree(g, common):
N = common.shape[0]
degrees = np.zeros(N, dtype=np.int)
degrees[:] = [len(g[node]) for node in common]
return degrees
def res_allocation(g, node1, node2):
common_n = _common_neighbors(g, node1, node2)
degrees = _common_degree(g, common_n)
score = np.sum(np.divide(1.0, degrees + 1e-2))
return score
@functools.lru_cache(maxsize=10)
def common_times(g, node1, node2):
common = _common_neighbors(g, node1, node2)
times = np.array(
# get the friendship time for each of common friends
[(g[node1][cf][1], g[node2][cf][1]) for cf in common]
)
return times
@functools.lru_cache(maxsize=10)
def common_forward_intensities(g, node1, node2):
common = _common_neighbors(g, node1, node2)
forward_inten = np.array([(g[node1][cf][2], g[cf][node2][2]) for cf in common])
return forward_inten
METRICS = [
common_neighbors_score,
adamic_adar_score,
res_allocation,
]
EDGE_FEATURES = [
(common_times, functools.partial(np.mean, axis=0)),
(common_times, functools.partial(np.min, axis=0)),
(common_times, functools.partial(np.median, axis=0)),
(common_forward_intensities, functools.partial(np.mean, axis=0)),
(common_forward_intensities, functools.partial(np.max, axis=0)),
(common_forward_intensities, functools.partial(np.median, axis=0)),
]
if USE_EDGE_FEATURES_CACHE:
print("Reading edge features")
with open(CACHE_IN_FOLDER / "edge_features.npy", "rb") as f:
edge_features = np.load(f)
else:
edge_features = np.array(
[
compute_features(g, EDGE_FEATURES, row)
for row in tqdm(pairs_np, mininterval=30, maxinterval=60)
],
dtype="float32",
).reshape(-1, len(EDGE_FEATURES) * 2)
print("Saving edge features")
with open(CACHE_OUT_FOLDER / "edge_features.npy", "wb") as f:
np.save(f, edge_features)
print("Saved")
# edge_features = np.random.random((len(pairs_np), len(EDGE_FEATURES) * 2))
if USE_METRICS_CACHE:
print("Reading metrics")
with open(CACHE_IN_FOLDER / "metrics.npy", "rb") as f:
metrics = np.load(f)
else:
metrics = np.array(
[
compute_metrics(g, METRICS, row)
for row in tqdm(pairs_np, mininterval=30, maxinterval=60)
],
dtype="float32",
)
# save to cache anyway, to keep it persistent between Kaggle Kernel relaunch
print("Saving metrics")
with open(CACHE_OUT_FOLDER / "metrics.npy", "wb") as f:
np.save(f, metrics)
print("Saved")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import os.path as osp
from glob import glob
import random
import time
import cv2
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import plotly.express as px
from sklearn.model_selection import train_test_split
from sklearn.model_selection import StratifiedKFold
from sklearn.metrics import accuracy_score
import torch
import torch.nn as nn
import torch.nn.functional as F
import torch.utils.data as data
import torch.optim as optim
import albumentations as A
from albumentations.pytorch import ToTensorV2
data_path = "/kaggle/input/state-farm-distracted-driver-detection/"
imgs_list = pd.read_csv(data_path + "driver_imgs_list.csv")
submission = pd.read_csv(data_path + "sample_submission.csv")
submission.head()
class_map = {
"c0": "Safe driving",
"c1": "Texting - right",
"c2": "Talking on the phone - right",
"c3": "Texting - left",
"c4": "Talking on the phone - left",
"c5": "Operating the radio",
"c6": "Drinking",
"c7": "Reaching behind",
"c8": "Hair and makeup",
"c9": "Talking to passenger",
}
from efficientnet_pytorch import EfficientNet
# 사전 훈련된 efficientnet-b7 모델 불러오기
model = EfficientNet.from_pretrained("efficientnet-b0", num_classes=10)
# model = EfficientNet.from_pretrained('efficientnet-b7', num_classes=10)
# 自分で実装
# テストデータの推論を行う関数
def inference(model, dataloader, device):
model.to(device)
model.eval()
preds = []
for i, (images, labels) in enumerate(dataloader):
images = images.to(device)
with torch.no_grad():
outputs = model(images)
preds += [outputs.detach().cpu().softmax(dim=1).numpy()]
if i % 10 == 0:
print(f"[test][{i+1}/{len(dataloader)}]")
preds = np.concatenate(preds)
return preds
# k個のモデルに対して推論を行い,アンサンブル
def inference_k_fold(df_test):
test_dataset = Dataset(df_test, phase="val", transform=DataTransform())
test_dataloader = data.DataLoader(
test_dataset, batch_size=args.batch_size, shuffle=False
)
model = EfficientNet.from_pretrained(args.model_name, num_classes=args.num_classes)
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
for fold in range(args.folds):
print(f"\n\nFOLD: {fold}")
print("-" * 50)
"/kaggle/input/DriverCassificationModel/"
model.load_state_dict(
torch.load(
f"/kaggle/input/efficientnet-b0-20230405/{args.model_name}_fold_{fold}.pth"
)["model"]
)
# model.load_state_dict(torch.load(f"{args.model_name}_fold_{fold}.pth")['model'])
df_test.loc[:, class_map.keys()] += (
inference(model, test_dataloader, device) / args.folds
)
class args:
model_name = "efficientnet-b0"
num_classes = 10
batch_size = 128
folds = 5
debug = True
train = True
class Dataset(data.Dataset):
def __init__(self, df, phase, transform=None):
super().__init__()
self.df = df
self.phase = phase
self.transform = transform
def __len__(self):
return len(self.df)
def __getitem__(self, index):
label = self.df.iloc[index]["class_num"]
image_path = self.df.iloc[index]["file_path"]
image = cv2.imread(image_path)
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB).astype(np.float32)
image /= 255.0
if self.transform is not None:
image = self.transform(self.phase, image)
return image, label
class DataTransform:
def __init__(self):
self.data_transform = {
"train": A.Compose(
[
A.Crop(x_min=40, y_min=0, x_max=520, y_max=480, p=1.0),
A.Resize(224, 224),
A.Rotate(-10, 10, p=0.5),
A.RandomBrightnessContrast(
brightness_limit=0.1, contrast_limit=0.1, p=0.3
),
A.OneOf([A.Emboss(p=1), A.Sharpen(p=1), A.Blur(p=1)], p=0.3),
ToTensorV2(), # 텐서 변환
]
),
"val": A.Compose(
[
A.Crop(x_min=40, y_min=0, x_max=520, y_max=480, p=1.0),
A.Resize(224, 224),
ToTensorV2(),
]
),
}
def __call__(self, phase, image):
# phase : 'train' or 'val'
transformed = self.data_transform[phase](image=image)
return transformed["image"]
"""
df_test = pd.read_csv(osp.join(data_path, 'sample_submission.csv'))
df_test['file_path'] = df_test.apply(lambda row: osp.join(data_path, f'imgs/test/{row.img}'), axis=1)
df_test['class_num'] = 0
df_test.loc[:, class_map.keys()] = 0
test_dataset = Dataset(df_test, phase="val", transform=DataTransform(
img_w=args.input_width, img_h=args.input_height, color_mean=args.color_mean, color_std=args.color_std))
test_dataloader = data.DataLoader(test_dataset, batch_size=args.batch_size, shuffle=False)
model = EfficientNet.from_pretrained(args.model_name, num_classes=args.num_classes)
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
model.load_state_dict(torch.load("/kaggle/input/efficientnet-test/efficientnet-b0_fold_0.pth")['model'])
df_test.loc[:, class_map.keys()] = inference(model, test_dataloader, device)
results = df_test.drop(['file_path', 'class_num'], axis=1)
results.iloc[:, 1:] = results.iloc[:, 1:].clip(0, 1)
results.to_csv('result.csv', index=False)
"""
df_test = pd.read_csv(osp.join(data_path, "sample_submission.csv"))
df_test["file_path"] = df_test.apply(
lambda row: osp.join(data_path, f"imgs/test/{row.img}"), axis=1
)
df_test["class_num"] = 0
df_test.loc[:, class_map.keys()] = 0
inference_k_fold(df_test)
results = df_test.drop(["file_path", "class_num"], axis=1)
results.iloc[:, 1:] = results.iloc[:, 1:].clip(0, 1)
results.to_csv("result.csv", index=False)
results.head()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
from sklearn.ensemble import (
RandomForestRegressor,
) ##Use machine learning model and import random forest module
# Read training set and test set
train_df = pd.read_csv("/kaggle/input/us-college-completion-rate-analysis/train.csv")
test_df = pd.read_csv("/kaggle/input/us-college-completion-rate-analysis/x_test.csv")
# Select the characteristics and target factors of random forest
features = [
"Tuition_in_state",
"Tuition_out_state",
"Faculty_salary",
"Pell_grant_rate",
"SAT_average",
"ACT_50thPercentile",
"pct_White",
"pct_Black",
"pct_Hispanic",
"pct_Asian",
"Parents_middlesch",
"Parents_highsch",
"Parents_college",
]
target = "Completion_rate"
# Extracting features and target variables from training and test sets
X_train = train_df[features]
y_train = train_df[target]
X_test = test_df[features]
from sklearn.preprocessing import StandardScaler
# Standardize data
scaler = StandardScaler()
X_train_scaled = scaler.fit_transform(X_train)
X_test_scaled = scaler.transform(X_test)
# Performing GridSearchCV on Random Forest Regression and PCA
from sklearn.model_selection import GridSearchCV
from sklearn.decomposition import PCA
pca = PCA()
X_train_pca = pca.fit_transform(X_train_scaled)
X_test_pca = pca.transform(X_test_scaled)
# Parametric search with GridSearchCV
rf_param_grid = {
"n_estimators": [100, 500, 1000],
"max_depth": [None, 10, 20],
"random_state": [42],
}
rf = RandomForestRegressor()
rf_grid_search = GridSearchCV(rf, rf_param_grid, cv=5, scoring="neg_mean_squared_error")
rf_grid_search.fit(X_train_pca, y_train.ravel())
best_rf = rf_grid_search.best_estimator_
X_test_pca = pca.transform(X_test_scaled)
y_pred_rf = best_rf.predict(X_test_pca)
print(y_pred_rf)
# Output CSV file (add ID manually and delete last blank line)
name = ["Completion_rate"]
df = pd.DataFrame(columns=name, data=y_pred_rf)
print(df)
df.to_csv("submission.csv", index=True, index_label="id")
|
# # Question:1
# importing libraries
import pkg_resources
import cmapPy
pkg_resources.get_distribution("cmapPy").version
# importing libraries
import pandas as pd
import numpy as np
from sklearn.decomposition import PCA
import matplotlib.pyplot as plt
import seaborn as sns
# parsing the gct data using cmappy
from cmapPy.pandasGEXpress.parse import parse
data_row = parse(
"PAAD.gct",
).data_df # expressional data
data_col = parse("PAAD.gct", col_meta_only=True).T # informational data
# setting index and visualize the top 5 data row of expressional data
data_row = data_row.set_index(data_row.axes[0])
data_row.head(5)
# visualize the top 5 data row of informational data
data_col.head(5)
# shape of expressional data
data_col.shape
# checking null value and counting
data_row.isnull().sum().sum()
# dropping null values
data_row = data_row.dropna()
# checking null value and counting after dropping null values
data_row.isnull().sum().sum()
# Slicing two informational columns
data_col = data_col.loc[["histological_type", "histological_type_other"]]
# visuslising sliced informational column
data_col.head()
# checking null value and counting row wise
for i in range(len(data_col.index)):
print("Nan in row ", i, " : ", data_col.iloc[i].isnull().sum())
# taking most occurence value for imputing in place of null
data_col.loc["histological_type"].mode()
# filling null values by most occurence value
data_col.loc["histological_type"].fillna(
"pancreas-adenocarcinoma ductal type", inplace=True
)
data_col.loc["histological_type_other"].fillna(
"pancreas-adenocarcinoma ductal type", inplace=True
)
# checking null value and counting
data_col.isnull().sum().sum()
# visuslising sliced informational column
data_col.head()
# checking unique values in data
data_col.iloc[1,].unique()
dataset = data_row.transpose()
# visuslising expressional column
dataset.head()
# Fitting the PCA algorithm with our Data
pca = PCA().fit(dataset)
# Plotting the Cumulative Summation of the Explained Variance
plt.figure()
plt.plot(np.cumsum(pca.explained_variance_ratio_))
plt.grid()
plt.xlabel("Number of Components")
plt.ylabel("Variance (%)") # for each component
plt.title("Plotting for comonents")
plt.show()
# applying pca to dataset
n_components = 125
pca = PCA(n_components)
df = pca.fit_transform(dataset)
pca.explained_variance_ratio_.cumsum()
df.shape
# placing column name and generating dataframe
df1 = pd.DataFrame(
df, columns=["PCA" + str(i) for i in range(1, n_components + 1)], index=None
)
# visualize the top 5 row
df1.head(5)
df1.describe()
# scatter plot pca1 and pca2
plt.figure(figsize=(8, 5))
plt.scatter(df1["PCA1"], df1["PCA2"])
plt.xlabel("PCA1")
plt.ylabel("PCA2")
plt.title("scatter plot pca1 and pca2")
plt.grid()
plt.show()
df2 = df1.copy()
# appending expressinal label to data
df2["label1"] = list(data_col.iloc[0])
df2["label2"] = list(data_col.iloc[1])
df2.head(5)
plt.figure(figsize=(8, 5))
ax = sns.scatterplot(df2["PCA1"], df2["PCA2"], hue=df2["label2"])
plt.xlabel("PCA1")
plt.ylabel("PCA2")
plt.title("scatter plot pca1 and pca2")
ax.legend(loc="center left", bbox_to_anchor=(1, 0.5))
plt.grid()
plt.show()
g = sns.FacetGrid(df2, hue="label2", height=9)
g.map(plt.scatter, "PCA1", "PCA2").add_legend()
plt.show()
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
y = le.fit_transform(df2["label2"])
def myplot(score, coeff, labels, y):
xs = score[:, 0]
ys = score[:, 1]
n = coeff.shape[0]
scalex = 1.0 / (xs.max() - xs.min())
scaley = 1.0 / (ys.max() - ys.min())
from matplotlib.pyplot import figure
figure(num=None, figsize=(12, 8), dpi=80, facecolor="w", edgecolor="k")
plt.scatter(xs * scalex, ys * scaley, c=y)
for i in range(n):
plt.arrow(0, 0, coeff[i, 0], coeff[i, 1], color="r", alpha=0.5)
if labels is None:
plt.text(
coeff[i, 0] * 1.15,
coeff[i, 1] * 1.15,
"Var" + str(i + 1),
color="g",
ha="center",
va="center",
)
else:
plt.text(
coeff[i, 0] * 1.15,
coeff[i, 1] * 1.15,
labels[i],
color="g",
ha="center",
va="center",
)
plt.xlim(-1, 1)
plt.ylim(-1, 1)
plt.xlabel("PC{}".format(1))
plt.ylabel("PC{}".format(2))
plt.grid()
myplot(df[:, 0:2], np.transpose(pca.components_[0:2, :]), df2["label2"].values, y)
plt.show()
# # What does the analysis say about the general behaviour of the different samples?
# # Ans:- Adenocarcinoma sample are forming dense cluster while other one is showing greater variation like Neuroendocrine showing low denser.
# # Are the neuroendocrine tumors clearly separable from the adenocarcinoma tumors?
# # Ans:- some neuroendocrine tumors are clrearly separable from adenocarcinoma tumors but very few are overlapped with adenocarcinoma tumors.
# # What can be said about the variance of the PCA
# # Ans:- 96% variance is captured on eigen vector value 125.
# # Question 2
# reading Type 1 IFN signature
ifn = pd.read_csv("type1_IFN.txt", header=None)
ifn[0].values
# concat informational column and expressional column by condition
new_data = pd.concat([data_row.loc[ifn[0]], data_col.drop("histological_type_other")])
# drop NaN value
hx = new_data.dropna()
hx.head(5)
hx.tail(5)
# transposing
X = hx.T
# slicing the values
X.iloc[:, 1:].values
# taking groupby on histological_type
x1 = X.groupby("histological_type").sum() / X.groupby("histological_type").count()
x1.head(5)
# setting up the dataframe
x2 = pd.DataFrame(x1, index=None, columns=None).T
# visualize the top 5 row
x2.head()
# plotting bar plot of each inferons present in histological type
font = {"family": "normal", "weight": "bold", "size": 24}
plt.rc("font", **font)
ax = x2.plot.bar(figsize=(40, 20))
# for p in ax.patches:
# ax.annotate(str(p.get_height()), (p.get_x() * 1.005, p.get_height() * 1.005))
plt.xlabel("Type 1 IFN signature")
plt.ylabel("score on each histological type")
ax.legend(
loc="upper center", bbox_to_anchor=(0.5, -0.05), fancybox=True, shadow=True, ncol=5
)
plt.tight_layout()
plt.show()
# plotting bar plot of histological type and their correspnding inferons score
ax = x2.T.plot.bar(figsize=(40, 20), rot=0)
# for p in ax.patches:
# ax.annotate(str(p.get_height()), (p.get_x() * 1.005, p.get_height() * 1.005))
plt.xlabel("Histological type")
plt.ylabel("Score on each histological type")
plt.tight_layout()
plt.show()
|
import numpy as np
import pandas as pd
import os
import cv2
labels = os.listdir("/kaggle/input/drowsiness-dataset/train")
labels
import matplotlib.pyplot as plt
plt.imshow(plt.imread("/kaggle/input/drowsiness-dataset/train/Closed/_107.jpg"))
plt.imshow(plt.imread("/kaggle/input/drowsiness-dataset/train/yawn/10.jpg"))
# # for yawn and not_yawn. Take only face
def face_for_yawn(
direc="/kaggle/input/drowsiness-dataset/train",
face_cas_path="/kaggle/input/haarcasacades/haarcascade_frontalface_alt.xml",
):
yaw_no = []
IMG_SIZE = 224
categories = ["yawn", "no_yawn"]
for category in categories:
path_link = os.path.join(direc, category)
class_num1 = categories.index(category)
print(class_num1)
for image in os.listdir(path_link):
image_array = cv2.imread(os.path.join(path_link, image), cv2.IMREAD_COLOR)
face_cascade = cv2.CascadeClassifier(face_cas_path)
faces = face_cascade.detectMultiScale(image_array, 1.3, 5)
for x, y, w, h in faces:
img = cv2.rectangle(image_array, (x, y), (x + w, y + h), (0, 255, 0), 2)
roi_color = img[y : y + h, x : x + w]
resized_array = cv2.resize(roi_color, (IMG_SIZE, IMG_SIZE))
yaw_no.append([resized_array, class_num1])
return yaw_no
yawn_no_yawn = face_for_yawn()
# # for closed and open eye
def get_data(dir_path="/kaggle/input/drowsiness-dataset/train"):
labels = ["Closed", "Open"]
IMG_SIZE = 224
data = []
for label in labels:
path = os.path.join(dir_path, label)
class_num = labels.index(label)
class_num += 2
print(class_num)
for img in os.listdir(path):
try:
img_array = cv2.imread(os.path.join(path, img), cv2.IMREAD_COLOR)
resized_array = cv2.resize(img_array, (IMG_SIZE, IMG_SIZE))
data.append([resized_array, class_num])
except Exception as e:
print(e)
return data
data_train = get_data()
# # extend data and convert array
def append_data():
yaw_no = face_for_yawn()
data = get_data()
yaw_no.extend(data)
return np.array(yaw_no)
# # new variable to store
all_data = append_data()
# # separate label and features
X = []
y = []
for feature, labelss in all_data:
X.append(feature)
y.append(labelss)
# # reshape the array
X = np.array(X)
X = X.reshape(-1, 224, 224, 3)
# # LabelBinarizer
from sklearn.preprocessing import LabelBinarizer
label_bin = LabelBinarizer()
y = label_bin.fit_transform(y)
# # label array
y = np.array(y)
from sklearn.model_selection import train_test_split
seed = 42
test_size = 0.30
X_train, X_test, y_train, y_test = train_test_split(
X, y, random_state=seed, test_size=test_size
)
# # import some dependencies
from keras.layers import Input, Lambda, Dense, Flatten, Conv2D, MaxPooling2D, Dropout
from keras.models import Model
from keras.models import Sequential
from keras.preprocessing.image import ImageDataGenerator
import tensorflow as tf
# # Data Augmentation
train_generator = ImageDataGenerator(
rescale=1 / 255, zoom_range=0.2, horizontal_flip=True, rotation_range=30
)
test_generator = ImageDataGenerator(rescale=1 / 255)
train_generator = train_generator.flow(np.array(X_train), y_train, shuffle=False)
test_generator = test_generator.flow(np.array(X_test), y_test, shuffle=False)
# # Model
model = Sequential()
model.add(Conv2D(256, (3, 3), activation="relu", input_shape=X_train.shape[1:]))
model.add(MaxPooling2D(2, 2))
model.add(Conv2D(128, (3, 3), activation="relu"))
model.add(MaxPooling2D(2, 2))
model.add(Conv2D(64, (3, 3), activation="relu"))
model.add(MaxPooling2D(2, 2))
model.add(Conv2D(32, (3, 3), activation="relu"))
model.add(MaxPooling2D(2, 2))
model.add(Flatten())
model.add(Dropout(0.5))
model.add(Dense(64, activation="relu"))
model.add(Dense(4, activation="softmax"))
model.compile(loss="categorical_crossentropy", metrics=["accuracy"], optimizer="adam")
model.summary()
r = model.fit(
train_generator,
epochs=50,
validation_data=test_generator,
shuffle=True,
validation_steps=len(test_generator),
)
# # history
# loss
plt.figure(figsize=(4, 2))
plt.plot(r.history["loss"], label="train loss")
plt.plot(r.history["val_loss"], label="val loss")
plt.legend()
plt.show()
# accuracies
plt.figure(figsize=(4, 2))
plt.plot(r.history["accuracy"], label="train acc")
plt.plot(r.history["val_accuracy"], label="val acc")
plt.legend()
plt.show()
# # save model
model.save("drowiness_new6.h5")
# # Prediction
prediction = model.predict(X_test)
classes_predicted = np.argmax(prediction, axis=1)
classes_predicted
# # classification report
labels_new = ["yawn", "no_yawn", "Closed", "Open"]
from sklearn.metrics import classification_report
print(
classification_report(
np.argmax(y_test, axis=1), classes_predicted, target_names=labels_new
)
)
# # predicting function
IMG_SIZE = 224
def prepare_yawn(
filepath, face_cas="../input/haarcasacades/haarcascade_frontalface_alt.xml"
):
img_array = cv2.imread(filepath, cv2.IMREAD_COLOR)
img_array = img_array / 255
resized_array = cv2.resize(img_array, (IMG_SIZE, IMG_SIZE))
return resized_array.reshape(-1, IMG_SIZE, IMG_SIZE, 3)
def prepare_eye(
filepath, face_cas="../input/haarcasacades/haarcascade_frontalface_alt.xml"
):
img_array = cv2.imread(filepath, cv2.IMREAD_COLOR)
img_array = img_array / 255
resized_array = cv2.resize(img_array, (IMG_SIZE, IMG_SIZE))
return resized_array.reshape(-1, IMG_SIZE, IMG_SIZE, 3)
model = tf.keras.models.load_model("./drowiness_new6.h5")
IMG_SIZE = 224
def prepare_yawn(
filepath, face_cas="../input/haarcasacades/haarcascade_frontalface_alt.xml"
):
img_array = cv2.imread(filepath, cv2.IMREAD_COLOR)
face_cascade = cv2.CascadeClassifier(face_cas_path)
faces = face_cascade.detectMultiScale(image_array, 1.3, 5)
for x, y, w, h in faces:
img = cv2.rectangle(image_array, (x, y), (x + w, y + h), (0, 255, 0), 2)
roi_color = img[y : y + h, x : x + w]
resized_array = cv2.resize(roi_color, (IMG_SIZE, IMG_SIZE))
# # Prediction
# ## 0-yawn, 1-no_yawn, 2-Closed, 3-Open
prediction = model.predict(
[prepare("../input/d/it136shrriyagoyal/prediction-images/IMG_20230408_204442.jpg")]
)
plt.imshow(
plt.imread("../input/d/it136shrriyagoyal/prediction-images/IMG_20230408_204442.jpg")
)
if np.argmax(prediction) == 0 or np.argmax(prediction) == 2:
print("DROWSY ALERT!!!!")
else:
print("DRIVER IS ACTIVE")
print(np.argmax(prediction))
prediction = model.predict(
[prepare("../input/d/it136shrriyagoyal/prediction-images/IMG_20230408_204505.jpg")]
)
plt.imshow(
plt.imread("../input/d/it136shrriyagoyal/prediction-images/IMG_20230408_204505.jpg")
)
if np.argmax(prediction) == 0 or np.argmax(prediction) == 2:
print("DROWSY ALERT!!!!")
else:
print("DRIVER IS ACTIVE")
print(np.argmax(prediction))
prediction = model.predict(
[prepare("../input/d/it136shrriyagoyal/prediction-images/IMG_20230408_204515.jpg")]
)
plt.imshow(
plt.imread("../input/d/it136shrriyagoyal/prediction-images/IMG_20230408_204515.jpg")
)
if np.argmax(prediction) == 0 or np.argmax(prediction) == 2:
print("DROWSY ALERT!!!!")
else:
print("DRIVER IS ACTIVE")
print(np.argmax(prediction))
prediction = model.predict(
[prepare("../input/d/it136shrriyagoyal/prediction-images/IMG_20230408_204530.jpg")]
)
plt.imshow(
plt.imread("../input/d/it136shrriyagoyal/prediction-images/IMG_20230408_204530.jpg")
)
if np.argmax(prediction) == 0 or np.argmax(prediction) == 2:
print("DROWSY ALERT!!!!")
else:
print("DRIVER IS ACTIVE")
print(np.argmax(prediction))
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import matplotlib.pyplot as plt
from matplotlib.pyplot import imshow
from tensorflow.keras.utils import image_dataset_from_directory
import tensorflow as tf
from tensorflow.keras import callbacks
from tensorflow.keras.models import Sequential
from tensorflow.keras import layers
from tensorflow.keras.applications import EfficientNetB3, ResNet50
from tensorflow.keras.utils import image_dataset_from_directory
from tensorflow.keras import optimizers
from tensorflow.keras import losses
from tensorflow.keras import callbacks
BATCH_SIZE = 32
IMAGE_HEIGHT = 256
IMAGE_WIDTH = 256
NUM_CLASSES = 2
TRAIN_PATH = "../input/diabetic-foot-ulcer-dfu/DFU/Patches"
TEST_PATH = "../input/diabetic-foot-ulcer-dfu/DFU/TestSet"
fpath = TRAIN_PATH + r"/Abnormal(Ulcer)/1.jpg"
img = plt.imread(fpath)
print(img.shape)
imshow(img)
# Load the Training and Validation Dataset
train_ds = image_dataset_from_directory(
TRAIN_PATH,
labels="inferred",
validation_split=0.2,
batch_size=BATCH_SIZE,
image_size=(IMAGE_WIDTH, IMAGE_HEIGHT),
subset="training",
seed=0,
)
validation_ds = image_dataset_from_directory(
TRAIN_PATH,
labels="inferred",
validation_split=0.2,
batch_size=BATCH_SIZE,
image_size=(IMAGE_WIDTH, IMAGE_HEIGHT),
subset="validation",
seed=0,
)
# Load Test Dataset
def decode_img(img):
# Convert the compressed string to a 3D uint8 tensor
img = tf.io.decode_jpeg(img, channels=3)
# Resize the image to the desired size
return tf.image.resize(img, [IMAGE_HEIGHT, IMAGE_WIDTH])
def process_path(file_path):
# Load the raw data from the file as a string
img = tf.io.read_file(file_path)
img = decode_img(img)
return img
test_ds = tf.data.Dataset.list_files(str(TEST_PATH + "/*"), shuffle=False)
# test_ds = test_ds.shuffle(len(os.listdir(TEST_PATH)), reshuffle_each_iteration=False)
test_ds = test_ds.map(process_path, num_parallel_calls=tf.data.AUTOTUNE)
# Get the class names
class_names = train_ds.class_names
print(class_names)
# Visualize the training dataset
plt.figure(figsize=(7, 7))
for images, labels in train_ds.take(1):
for i in range(9):
ax = plt.subplot(3, 3, i + 1)
plt.imshow(images[i].numpy().astype("uint8"))
plt.title(class_names[labels[i]])
plt.axis("off")
# Visualize the testing dataset
plt.figure(figsize=(7, 7))
i = 0
for images in test_ds.take(6):
ax = plt.subplot(3, 3, i + 1)
plt.imshow(images.numpy().astype("uint8"))
plt.axis("off")
i += 1
# Layers of Augmentation
img_augmentation = Sequential(
[
layers.Resizing(IMAGE_WIDTH, IMAGE_HEIGHT),
layers.RandomRotation(factor=0.15),
layers.RandomTranslation(height_factor=0.1, width_factor=0.1),
layers.RandomFlip(),
layers.GaussianNoise(stddev=0.09),
layers.RandomContrast(factor=0.1),
],
name="img_augmentation",
)
# Caching and Prefetching (Optimization)
AUTOTUNE = tf.data.AUTOTUNE
train_ds = train_ds.cache().prefetch(buffer_size=AUTOTUNE)
validation_ds = validation_ds.cache().prefetch(buffer_size=AUTOTUNE)
# Define Callbacks
lr_callback = callbacks.ReduceLROnPlateau(
monitor="val_accuracy", factor=0.1, patience=5
)
stop_callback = callbacks.EarlyStopping(monitor="val_accuracy", patience=8)
# Initialize Model
def build_model():
inputs = layers.Input(shape=(IMAGE_WIDTH, IMAGE_HEIGHT, 3))
x = img_augmentation(inputs)
model = EfficientNetB3(include_top=False, weights="imagenet", input_tensor=x)
# Rebuild top
x = layers.GlobalAveragePooling2D(name="avg_pool")(model.output)
x = layers.BatchNormalization()(x)
top_dropout_rate = 0.45
x = layers.Dropout(top_dropout_rate, seed=123, name="top_dropout")(x)
outputs = layers.Dense(NUM_CLASSES, activation="sigmoid", name="pred")(x)
# Compile
model = tf.keras.Model(inputs, outputs, name="EfficientNet")
optimizer = optimizers.Adam(learning_rate=1e-3)
loss = losses.SparseCategoricalCrossentropy()
model.compile(optimizer=optimizer, loss=loss, metrics=["accuracy"])
model.summary()
return model
model = build_model()
history = model.fit(
train_ds,
validation_data=validation_ds,
callbacks=[lr_callback, stop_callback],
epochs=20,
)
# Testing on Test Set
plt.figure(figsize=(30, 30))
plt.tight_layout()
i = 0
for images in test_ds.take(15):
ax = plt.subplot(5, 5, i + 1)
prediction = np.argmax(model.predict(np.array([images])), axis=1)
plt.imshow(images.numpy().astype("uint8"))
plt.title(class_names[prediction[0]])
plt.axis("off")
i += 1
def plot_hist(hist):
plt.plot(hist.history["accuracy"])
plt.plot(hist.history["val_accuracy"])
plt.title("model accuracy")
plt.ylabel("accuracy")
plt.xlabel("epoch")
plt.legend(["train", "validation"], loc="upper left")
plt.show()
plot_hist(history)
model.save("/kaggle/working/model.h5")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
import os
import numpy as np
import pandas as pd
import random
import cv2
import matplotlib.pyplot as plt
import keras.backend as K
import tensorflow as tf
import warnings
from random import shuffle
from tqdm import tqdm
from PIL import Image
from keras.models import Model, Sequential
from keras.layers import (
Input,
Dense,
Flatten,
Dropout,
BatchNormalization,
Conv2D,
SeparableConv2D,
MaxPool2D,
LeakyReLU,
Activation,
ReLU,
)
from keras.optimizers import Adam
from keras.preprocessing.image import ImageDataGenerator
from keras.callbacks import ModelCheckpoint, ReduceLROnPlateau, EarlyStopping
from sklearn.metrics import accuracy_score, confusion_matrix
from sklearn.preprocessing import MinMaxScaler
from sklearn import svm
from glob import glob
from matplotlib import pyplot as plt
path_train = "../input/chest-xray-pneumonia/chest_xray/train"
path_val = "../input/chest-xray-pneumonia/chest_xray/val"
path_test = "../input/chest-xray-pneumonia/chest_xray/test"
img = glob(path_train + "/PNEUMONIA/*.jpeg")
plt.imshow(np.asarray(plt.imread(img[0])))
# # Augment Data
classes = ["NORMAL", "PNEUMONIA"]
img_height = 299
img_width = 299
train_data = glob(path_train + "/NORMAL/*.jpeg")
train_data += glob(path_train + "/PNEUMONIA/*.jpeg")
train_datagen = ImageDataGenerator(
rescale=1.0 / 255, shear_range=0.2, zoom_range=0.2, horizontal_flip=True
) # set validation split
train_generator = train_datagen.flow_from_directory(
path_train,
target_size=(img_height, img_width),
batch_size=24,
classes=classes,
class_mode="categorical",
) # set as training data
validation_generator = train_datagen.flow_from_directory(
path_val, # same directory as training data
target_size=(img_height, img_width),
batch_size=24,
classes=classes,
class_mode="categorical",
) # set as validation data
test_generator = train_datagen.flow_from_directory(
path_test, # same directory as training data
target_size=(img_height, img_width),
batch_size=24,
classes=classes,
class_mode="categorical",
) # set as validation data
train_generator.image_shape
# # Visualize Some Images
import skimage
from skimage.transform import resize
def plotter(i):
Pimages = os.listdir(path_train + "/PNEUMONIA")
Nimages = os.listdir(path_train + "/NORMAL")
imagep1 = cv2.imread(path_train + "/PNEUMONIA/" + Pimages[i])
imagep1 = skimage.transform.resize(imagep1, (150, 150, 3), mode="reflect")
imagen1 = cv2.imread(path_train + "/NORMAL/" + Nimages[i])
imagen1 = skimage.transform.resize(imagen1, (150, 150, 3))
pair = np.concatenate((imagen1, imagep1), axis=1)
print("(Left) - No Pneumonia Vs (Right) - Pneumonia")
print(
"-----------------------------------------------------------------------------------------------------------------------------------"
)
plt.figure(figsize=(10, 5))
plt.imshow(pair)
plt.show()
for i in range(5, 10):
plotter(i)
from keras.applications.resnet50 import ResNet50
base_model = ResNet50(
weights="imagenet",
include_top=False,
input_tensor=Input(shape=(299, 299, 3)),
input_shape=(299, 299, 3),
)
model = Flatten(name="flatten")(base_model.output)
model = Dense(1024, activation="relu")(model)
model = Dropout(0.7, name="dropout1")(model)
model = Dense(512, activation="relu")(model)
model = Dropout(0.5, name="dropout2")(model)
predictions = Dense(2, activation="softmax")(model)
conv_model = Model(inputs=base_model.input, outputs=predictions)
opt = Adam(lr=0.0001, decay=1e-5)
conv_model.compile(loss="binary_crossentropy", metrics=["accuracy"], optimizer=opt)
print(conv_model.summary())
for layer in conv_model.layers[:-6]:
layer.trainable = False
conv_model.compile(loss="binary_crossentropy", metrics=["accuracy"], optimizer=opt)
history = conv_model.fit_generator(
epochs=5,
shuffle=True,
validation_data=validation_generator,
generator=train_generator,
steps_per_epoch=500,
validation_steps=10,
verbose=2,
)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv(
"/kaggle/input/breast-cancer-gene-expression-profiles-metabric/METABRIC_RNA_Mutation.csv",
delimiter=",",
)
# ilk kısım clinic verilere göre değerlendirme
nRow, nCol = df.shape
print(f"toplam {nRow} satır ve {nCol} sütun var.")
print(df.head(5))
# df'de bulunan verilerden kaç tanesinde nan değer olduğu bilgisi gösteriliyor.
toplam = df.isnull().sum().sort_values(ascending=False)
yuzde = (df.isnull().sum() / df.isnull().count()).sort_values(ascending=False)
missin_data = pd.concat([toplam, yuzde], axis=1, keys=["toplam_NAN", "yüzde_NAN"])
# klinik özellikler ve sonuçları arasındaki ilişki için yeni bir data frame oluşturuluyor.
clinical_features_to_drop = df.columns[31:] # klink olmayan attribute'ler
clinical_df = df.drop(clinical_features_to_drop, axis=1)
print(clinical_df.info())
def to_standard(df):
num_df = df[
df.select_dtypes(include=np.number).columns.tolist()
] # sayı içeren kolonların listesi
ss = StandardScaler()
std = ss.fit_transform(num_df)
std_df = pd.DataFrame(std, index=num_df.index, columns=num_df.columns)
return std_df
clinical_df_standart = to_standard(clinical_df)
# standartlaştırılmış olan aşağıdaki 3 feature çzidiriliyor
sns.kdeplot(data=clinical_df_standart["age_at_diagnosis"])
sns.kdeplot(data=clinical_df_standart["mutation_count"])
sns.kdeplot(data=clinical_df_standart["tumor_size"])
# verideki outlier'lar tespit ediliyor.
ax, fig = plt.subplots(1, 1, figsize=(15, 5))
plt.title("DataFrame'deki clinic verilerin dağılımı", fontsize=20)
sns.boxplot(
y="variable", x="value", data=pd.melt(clinical_df_standart), palette="Spectral"
)
plt.xlabel("Standart hale geldikten sonra veri aralığı", size=16)
plt.ylabel("Klinik veriler", size=16)
# tek tek box plot inceleme
sns.boxplot(clinical_df_standart["mutation_count"])
# sns.boxplot(clinical_df_standart['tumor_size'])
# %% Hayatta kalanlar ve ölenler arasında klinik verilere göre pdf dağılımı.
fig = plt.figure(figsize=(20, 25))
j = 0
num_clinical_columns = [
"age_at_diagnosis",
"lymph_nodes_examined_positive",
"mutation_count",
"nottingham_prognostic_index",
"overall_survival_months",
"tumor_size",
]
for i in clinical_df[num_clinical_columns].columns:
plt.subplot(6, 4, j + 1)
j += 1
sns.distplot(
clinical_df[i][clinical_df["overall_survival"] == 1],
color="g",
label="survived",
)
sns.distplot(
clinical_df[i][clinical_df["overall_survival"] == 0], color="r", label="died"
)
plt.legend(loc="best")
fig.suptitle("Klinik Data Analiz")
fig.subplots_adjust(top=0.95)
plt.show()
# %% Hayatta kalan ve ölenlerin bilgileri
died = clinical_df[clinical_df["overall_survival"] == 0]
survived = clinical_df[clinical_df["overall_survival"] == 1]
# yaş cinsinden bir kişinin hayatta kalama durumunu gösterme
fig, ax = plt.subplots(ncols=1, figsize=(15, 3), sharey=True)
yil = clinical_df["overall_survival_months"] / 12
c = clinical_df["overall_survival"]
data = pd.concat([yil, c], axis=1, keys=["overall_survival_months", "overall_survival"])
sns.boxplot(
x="age_at_diagnosis",
y="overall_survival",
orient="h",
data=clinical_df,
ax=ax,
saturation=0.90,
)
fig.suptitle(
"Bu grafik yaş cinsinden bir kişinin hayatta kalama durumunu gösterir.", fontsize=18
)
ax.set_ylabel("Hayatta kalma durumu")
ax.set_xlabel("Yaş tanısı")
plt.show()
# Tümörün boyutu ve evresine göre hayatta kalma durumu
ig, ax = plt.subplots(figsize=(12, 6))
ax = sns.boxplot(
x="tumor_size",
y="tumor_stage",
data=clinical_df,
orient="h",
hue="overall_survival",
)
ax.set_ylabel("Tümor'ün evresi")
ax.set_xlabel("Tümor'ün boyutu")
fig.suptitle("Tumor stage vs. Tumor size and overall_survival", fontsize=20)
plt.show()
# Alınan tedaviye göre sağ kalanlar ve ölenlerin histogram dağılımı
fig, ax = plt.subplots(ncols=3, figsize=(15, 3))
color = "Spectral"
fig, ax = plt.subplots(ncols=3, figsize=(15, 3))
fig.suptitle("Tedaviye göre sağ kalanlar ve ölenlerin dağılımı", fontsize=18)
sns.countplot(
x=died["chemotherapy"],
color=sns.color_palette(color)[0],
label="Ölü",
ax=ax[0],
saturation=0.90,
)
sns.countplot(
x=survived["chemotherapy"],
color=sns.color_palette(color)[5],
label="Hayatta",
ax=ax[0],
saturation=0.90,
)
ax[0].legend()
ax[0].set(xticklabels=["Hayır", "Evet"])
sns.countplot(
x=died["hormone_therapy"],
color=sns.color_palette(color)[0],
label="Ölü",
ax=ax[1],
saturation=0.90,
)
sns.countplot(
x=survived["hormone_therapy"],
color=sns.color_palette(color)[5],
label="Hayatta",
ax=ax[1],
saturation=0.90,
)
ax[1].legend()
ax[1].set(xticklabels=["Hayır", "Evet"])
sns.countplot(
x=died["radio_therapy"],
color=sns.color_palette(color)[0],
label="Ölü",
ax=ax[2],
saturation=0.90,
)
sns.countplot(
x=survived["radio_therapy"],
color=sns.color_palette(color)[5],
label="Hayatta",
ax=ax[2],
saturation=0.90,
)
ax[2].legend()
ax[2].set(xticklabels=["Hayır", "Evet"])
ax[0].set_xlabel("Kemoterapi")
ax[0].set_ylabel("Hastaların sayısı")
ax[1].set_xlabel("Hormonal terapi")
ax[1].set_ylabel("")
ax[2].set_xlabel("Radyo terapi")
ax[2].set_ylabel("")
plt.show()
# kategorik olmayan sütunlar kategorik hale getirliyor.
categorical_columns = clinical_df.select_dtypes(include=["object"]).columns.tolist()
unwanted_columns = ["patient_id", "death_from_cancer"]
categorical_columns = [
ele for ele in categorical_columns if ele not in unwanted_columns
]
no_id_clinical_df = pd.get_dummies(
clinical_df.drop("patient_id", axis=1), columns=categorical_columns
)
# feature'lar arasında yüksek korelasyon olan sütunlara bakıyoruz.
Corr_survival = no_id_clinical_df.corr()["overall_survival"].sort_values(
ascending=False
) # hayattka kalma ile diğer sütunlar arasındaki ilişkiye bakılıyor.
Corr_df = pd.DataFrame({"Correlation(overall_survival)": Corr_survival})
print(Corr_df.head(10))
print(Corr_df.tail(10))
num_clinical_columns = [
"age_at_diagnosis",
"lymph_nodes_examined_positive",
"mutation_count",
"nottingham_prognostic_index",
"overall_survival_months",
"tumor_size",
]
istatsitik = clinical_df[num_clinical_columns].describe().T
print(istatsitik)
# Tedavi durumuna göre hayatta kalıp kalmama durumu
no_treatment = clinical_df[
(clinical_df["chemotherapy"] == 0)
& (clinical_df["hormone_therapy"] == 0)
& (clinical_df["radio_therapy"] == 0)
]
yes_treatment = clinical_df[
(clinical_df["chemotherapy"] == 1)
& (clinical_df["hormone_therapy"] == 1)
& (clinical_df["radio_therapy"] == 1)
]
print("Tedavi olmayan hasta sayısı: ", no_treatment.shape[0])
print(
"Tedavi olmayıp'ta hayatta kalma oranı: ",
("%.3f" % np.mean(no_treatment["overall_survival"])),
)
print(
"Tedavi olup'ta hayatta kalma oranı: ",
("%.3f" % np.mean(yes_treatment["overall_survival"])),
)
print(
"Genel hayatta kalma oranı: ", ("%.3f" % np.mean(clinical_df["overall_survival"]))
)
print(
"Genel hayatta kalma oranı: "
+ "%.3f"
% (
clinical_df["overall_survival"].value_counts()
/ clinical_df["overall_survival"].count()
).iloc[1]
)
# populastonda yer alan üyelerin ortalama karakteristiği
print("Yaş ortalaması: " + "%.3f" % np.mean(clinical_df["age_at_diagnosis"]))
print(
"En çok görülen tümör evresi: ",
stats.mode(clinical_df["tumor_stage"])[0][0].astype(int),
)
print(
"En çok görülen histopathological tip: ",
stats.mode(clinical_df["neoplasm_histologic_grade"])[0][0].astype(int),
)
print("Ortalama Tümör çapı: " + "%.3f" % np.mean(clinical_df["tumor_size"]))
# Klinik verilerdeki outlier sayıları
Q1 = clinical_df.quantile(0.25)
Q3 = clinical_df.quantile(0.75)
IQR = Q3 - Q1
alt_sinir = Q1 - 1.5 * IQR
ust_sinir = Q3 + 1.5 * IQR
outliers = (
((clinical_df < alt_sinir) | (clinical_df > ust_sinir))
.sum()
.sort_values(ascending=False)
)
print(outliers)
# bundan sonraki kısım genetik verilere göre değerlendirme
# genetik verilere göre değerlendirme
# mutasyonlar siliniyor
genetic_features_to_drop = df.columns[520:]
genetic_df = df.drop(genetic_features_to_drop, axis=1)
# klinik veriler siliniyor.
genetic_features_to_drop = genetic_df.columns[4:35]
genetic_df = genetic_df.drop(genetic_features_to_drop, axis=1)
genetic_df = genetic_df.drop(
["age_at_diagnosis", "type_of_breast_surgery", "cancer_type"], axis=1
)
genetic_df = genetic_df.iloc[:, :-174]
genetic_df["overall_survival"] = df["overall_survival"]
genetic_df.head()
# Her bir kolon için maksimum değer ve standart sapma değeri azalan sıralanıyor.
max_values = genetic_df.max()
std = genetic_df.std(axis=0, skipna=True)
max_data = pd.concat([max_values, std], axis=1, keys=["max_values", "std"])
max_data.sort_values(by="max_values", ascending=False).head()
# Her bir kolon için minimum değer ve standart sapma değeri azalan sıralanıyor.
min_values = genetic_df.min()
std = genetic_df.std(axis=0, skipna=True)
min_data = pd.concat([min_values, std], axis=1, keys=["min_values", "std"])
min_data.sort_values(by="min_values", ascending=True).head()
# Visualizing the mRNA values in a heatmap.
fig, axs = plt.subplots(figsize=(17, 10))
sns.heatmap(
genetic_df.drop(["patient_id", "overall_survival"], axis=1),
ax=axs,
cmap=sns.diverging_palette(180, 10, as_cmap=True),
)
plt.title("Gene Expression Heatmap")
# fix for mpl bug that cuts off top/bottom of seaborn viz
b, t = plt.ylim() # discover the values for bottom and top
b += 0.5 # Add 0.5 to the bottom
t -= 0.5 # Subtract 0.5 from the top
plt.ylim(b, t) # update the ylim(bottom, top) values
plt.show()
# %% listede verilen genlere göre hayatta kalan ve ölenlerin pdf grafiği
fig = plt.figure(figsize=(20, 25))
j = 0
gene_list = [
"rab25",
"eif5a2",
"pik3ca",
"kit",
"fgf1",
"myc",
"egfr",
"notch3",
"kras",
"akt1",
"erbb2",
"pik3r1",
"ccne1",
"akt2",
"aurka",
]
for i in genetic_df.drop(["patient_id"], axis=1).loc[:, gene_list].columns:
plt.subplot(6, 4, j + 1)
j += 1
sns.distplot(
genetic_df[i][genetic_df["overall_survival"] == 0], color="g", label="survived"
)
sns.distplot(
genetic_df[i][genetic_df["overall_survival"] == 1], color="r", label="died"
)
plt.legend(loc="best")
fig.suptitle("Clinical Data Analysis")
fig.tight_layout()
fig.subplots_adjust(top=0.95)
plt.show()
# verideki outlier'lar tespit ediliyor.
sns.boxplot(genetic_df["erbb2"])
# sns.boxplot(genetic_df['dll3'])
# sns.boxplot(genetic_df['mmp1'])
# sns.boxplot(genetic_df['mmp12'])
# sns.boxplot(genetic_df['stat1'])
# Herbir kolondaki outlier'ların sayısı
Q1 = genetic_df.quantile(0.25)
Q3 = genetic_df.quantile(0.75)
IQR = Q3 - Q1
outliers_gen = (
((genetic_df < (Q1 - 1.5 * IQR)) | (genetic_df > (Q3 + 1.5 * IQR)))
.sum()
.sort_values(ascending=False)
)
# Genetik özellikler ve sonuçları arasındaki ilişki
# Genlerin ölüm ile ilişkisi
fig, ax = plt.subplots(figsize=(10, 4))
corrs = []
for col in genetic_df.drop(["patient_id"], axis=1).columns:
corr = genetic_df[[col, "overall_survival"]].corr()["overall_survival"][col]
corrs.append(corr)
corrs.pop(-1)
ax.hist(corrs, bins=25, color="red", edgecolor="black")
ax.set_xlabel("Korelasyon")
ax.set_ylabel("Genlerin sayısı")
ax.set_title("Genlerin hayatta kalma ile ilişkisi", size=16)
plt.show()
# maksimum korelasyona sahip geni bulma
ma = max(corrs)
ma_index = corrs.index(ma)
ma_gen = genetic_df.iloc[0:0, ma_index + 1]
mak_gen_corr = genetic_df[[ma_gen.name, "overall_survival"]].corr()[ma_gen.name][1]
print(ma_gen.name, ":", mak_gen_corr)
# minimum korelasyona sahip geni bulma
mi = min(corrs)
mi_index = corrs.index(mi)
mi_gen = genetic_df.iloc[0:1, mi_index + 1]
min_gen_corr = genetic_df[[mi_gen.name, "overall_survival"]].corr()[mi_gen.name][1]
print(mi_gen.name, ":", min_gen_corr)
# Mutasyona uğrayan genler 1 olacak şekilde yeni bir dataframe oluşturuluyor.
# klinik ve genetik veriler siliniyor.
mutation_features_to_drop = df.columns[4:520]
mutation_df = df.drop(mutation_features_to_drop, axis=1)
mutation_df = mutation_df.drop(
["age_at_diagnosis", "type_of_breast_surgery", "cancer_type"], axis=1
)
# Eğer gen mutasyona uğramışssa 1 uğramamışssa 0 değerini yazıyoruz.
for column in mutation_df.columns[1:]:
mutation_df[column] = (
pd.to_numeric(mutation_df[column], errors="coerce").fillna(1).astype(int)
)
mutation_df.insert(loc=1, column="overall_survival", value=df["overall_survival"])
mutation_df.head()
# mutasyona uğrayan genlerin ölüm ile ilişkisi
fig, ax = plt.subplots(figsize=(10, 4))
corrs = []
for col in mutation_df.drop(["patient_id"], axis=1).columns:
corr = mutation_df[[col, "overall_survival"]].corr()["overall_survival"][col]
corrs.append(corr)
corrs.pop(0)
ax.hist(corrs, bins=25, color="red", edgecolor="black")
ax.set_xlabel("Korelasyon")
ax.set_ylabel("Gen Sayısı")
ax.set_title("Mutasyona uğrayan genlerin ölüm ile ilişkisi ", size=16)
plt.show()
# %% mutasyonlu hücrelerde
# maksimum korelasyona sahip geni bulma
ma = max(corrs)
ma_index = corrs.index(ma)
ma_gen = mutation_df.iloc[0:1, ma_index + 2]
mak_gen_corr = mutation_df[[ma_gen.name, "overall_survival"]].corr()[ma_gen.name][1]
print(ma_gen.name, ":", mak_gen_corr)
# minimum korelasyona sahip geni bulma
mi = min(corrs)
mi_index = corrs.index(mi)
mi_gen = mutation_df.iloc[0:1, mi_index + 2]
min_gen_corr = mutation_df[[mi_gen.name, "overall_survival"]].corr()[mi_gen.name][1]
print(mi_gen.name, ":", min_gen_corr)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
# Importing Necessary libraries
import re
import nltk
import spacy
# Loading Data
train = pd.read_csv("/kaggle/input/nlp-getting-started/train.csv")
train.head()
train.isnull().sum()
# Here we will remove keyword and location variables. They donot have much of a value here. In addition to that, there are many NaN values. So it is better to remove those variables.
train = train.drop(columns=["keyword", "location"])
train.head()
# Now we have everything required for us to get started. Here id is the reference like an index. 'text' is our asset. We will be working on that. 'target' is the target variable.
import string
train["text"] = train["text"].str.lower()
train.head()
# In the earlier preview, you can find a mixture of upper and lower case letters. Now, you can see that whole of the text is in lower case. This forms the first step of text preprocessing. Let us now split our dataset for the target and text variables.
target = train["target"]
target.head()
text = train["text"]
text.head()
# The above data has punctuation with it and they do not have any semantic meaning in our data. So we will remoce it. The following is a better way of removing it.
def remove_punctuation(text):
return text.translate(str.maketrans("", "", string.punctuation))
text_clean = text.apply(lambda text: remove_punctuation(text))
text_clean.head()
# You can find that the functuations are removed. Now we will remove the so called stopwords. They are highly repetitive words in the text but do not posses a greater value for their presence. So we will remove it.
from nltk.corpus import stopwords
STOPWORDS = set(stopwords.words("english"))
def stopwords_(text):
return " ".join([word for word in str(text).split() if word not in STOPWORDS])
text_clean = text_clean.apply(lambda text: stopwords_(text))
text_clean.head()
# You can find ityourself right. The use of removing these words. Want to know what are those words. Take a look at it.
", ".join(stopwords.words("english"))
# Yeah. you have now completed the first phase of the text preprocessing. Now let us proceed to the next one.
# Lemmatization is the process of reducing the words to their roots. Let us take a look at an example for better understanding.
from nltk.stem import WordNetLemmatizer
lemmatizer = WordNetLemmatizer()
def lemmatizer_(text):
return " ".join([lemmatizer.lemmatize(word) for word in text.split()])
lemmatizer.lemmatize("wrote", "v")
lemmatizer.lemmatize("written", "v")
# Do I need to explain further. Hahaha. Not at all necesary. It is self explanatory. But if you have any doubts donot hesitate to comment in the comment section.
# Let us apply this to our text.
text_clean = text_clean.apply(lambda text: lemmatizer_(text))
text_clean.head()
# All of these can also be done by in-built packages. But it is a good parctice in the beginning to understand our data better.
# Now for the fun part, we will look at the most used words in the cleaned text. We will use wordcloud library for that.
from wordcloud import WordCloud
import matplotlib.pyplot as plt
fig, (ax1) = plt.subplots(1, figsize=[12, 12])
wordcloud = WordCloud(background_color="white", width=600, height=400).generate(
" ".join(text_clean)
)
ax1.imshow(wordcloud)
ax1.axis("off")
ax1.set_title("Frequent Words", fontsize=16)
|
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
from sklearn.linear_model import LinearRegression
from xgboost import XGBRegressor
from sklearn.model_selection import train_test_split
from sklearn.metrics import mean_absolute_error, mean_squared_error, accuracy_score
import scipy.stats as stats
df = pd.read_csv(
r"D:\Personal Data\School\IT5006 Fundamentals of Data Analytics\Group Assignment\Milestone 1\CollegeScorecard_Raw_Data_09012022\MERGED2020_21_PP.csv"
)
df
working = df[
[
"UGDS_WHITE",
"UGDS_BLACK",
"UGDS_HISP",
"UGDS_ASIAN",
"NPT41_PUB",
"NPT42_PUB",
"NPT43_PUB",
"NPT44_PUB",
"NPT45_PUB",
"NPT41_PRIV",
"NPT42_PRIV",
"NPT43_PRIV",
"NPT44_PRIV",
"NPT45_PRIV",
"ACTMT25",
]
]
working
working.isna().sum()
# Drop variable if completely NA
working = working.dropna(subset="ACTMT25")
working = working.dropna(
subset=[
"NPT41_PUB",
"NPT42_PUB",
"NPT43_PUB",
"NPT44_PUB",
"NPT45_PUB",
"NPT41_PRIV",
"NPT42_PRIV",
"NPT43_PRIV",
"NPT44_PRIV",
"NPT45_PRIV",
],
how="all",
)
working.isna().sum()
working
working.columns
# Sepearating public and private, duplicate columns
public = working[
[
"UGDS_WHITE",
"UGDS_BLACK",
"UGDS_HISP",
"UGDS_ASIAN",
"NPT41_PUB",
"NPT42_PUB",
"NPT43_PUB",
"NPT44_PUB",
"NPT45_PUB",
"ACTMT25",
]
]
private = working[
[
"UGDS_WHITE",
"UGDS_BLACK",
"UGDS_HISP",
"UGDS_ASIAN",
"NPT41_PRIV",
"NPT42_PRIV",
"NPT43_PRIV",
"NPT44_PRIV",
"NPT45_PRIV",
"ACTMT25",
]
]
# # Public School
public = public.dropna(
subset=["NPT41_PUB", "NPT42_PUB", "NPT43_PUB", "NPT44_PUB", "NPT45_PUB"], how="all"
)
public.isna().sum()
public
public.describe()
# ###### Fillna with mean values of column
public["NPT43_PUB"] = public["NPT43_PUB"].fillna(public["NPT43_PUB"].mean())
public["NPT44_PUB"] = public["NPT44_PUB"].fillna(public["NPT44_PUB"].mean())
public["NPT45_PUB"] = public["NPT45_PUB"].fillna(public["NPT45_PUB"].mean())
publicv1 = public.copy()
# ###### Regression models for predicting 25th percentile ACT math score
# XGBoost Regression
x_trn, x_tst = train_test_split(publicv1, test_size=0.20, random_state=1)
y_trn = x_trn["ACTMT25"]
y_tst = x_tst["ACTMT25"]
x_trn = x_trn.drop(["ACTMT25"], axis=1)
x_tst = x_tst.drop(["ACTMT25"], axis=1)
model = XGBRegressor(subsample=0.5, learning_rate=0.07)
model.fit(x_trn, y_trn)
print("Model Accuracy on Training:", model.score(x_trn, y_trn), "\n")
print(
pd.DataFrame(
model.feature_importances_, index=x_trn.columns, columns=["Variables"]
).sort_values(by=["Variables"], ascending=False),
"\n",
)
y_pred = model.predict(x_tst)
print(
"Accuracy:",
model.score(x_tst, y_tst),
"/ RMSE:",
np.sqrt(mean_squared_error(y_pred, y_tst)),
)
# Linear Regression
x_trn, x_tst = train_test_split(publicv1, test_size=0.20, random_state=1)
y_trn = x_trn["ACTMT25"]
y_tst = x_tst["ACTMT25"]
x_trn = x_trn.drop(["ACTMT25"], axis=1)
x_tst = x_tst.drop(["ACTMT25"], axis=1)
model = LinearRegression()
model.fit(x_trn, y_trn)
print("Model Accuracy on Training:", model.score(x_trn, y_trn), "\n")
# print (pd.DataFrame(model.feature_importances_,index = x_trn.columns,columns =['Variables']).sort_values(by=['Variables'],ascending=False),'\n')
y_pred = model.predict(x_tst)
print(
"Accuracy:",
model.score(x_tst, y_tst),
"/ RMSE:",
np.sqrt(mean_squared_error(y_pred, y_tst)),
)
# ###### Remove outliers
plt.boxplot(public["NPT41_PUB"])
outliers1 = (
public["NPT41_PUB"].quantile(0.25)
- (public["NPT41_PUB"].quantile(0.75) - public["NPT41_PUB"].quantile(0.25)) * 1.5
)
outliers2 = (
public["NPT41_PUB"].quantile(0.75)
+ (public["NPT41_PUB"].quantile(0.75) - public["NPT41_PUB"].quantile(0.25)) * 1.5
)
public = public.drop(public[public["NPT41_PUB"] < outliers1].index)
public = public.drop(public[public["NPT41_PUB"] > outliers2].index)
plt.boxplot(public["NPT42_PUB"])
outliers1 = (
public["NPT42_PUB"].quantile(0.25)
- (public["NPT42_PUB"].quantile(0.75) - public["NPT42_PUB"].quantile(0.25)) * 1.5
)
outliers2 = (
public["NPT42_PUB"].quantile(0.75)
+ (public["NPT42_PUB"].quantile(0.75) - public["NPT42_PUB"].quantile(0.25)) * 1.5
)
public = public.drop(public[public["NPT42_PUB"] < outliers1].index)
public = public.drop(public[public["NPT42_PUB"] > outliers2].index)
plt.boxplot(public["NPT43_PUB"])
outliers1 = (
public["NPT43_PUB"].quantile(0.25)
- (public["NPT43_PUB"].quantile(0.75) - public["NPT43_PUB"].quantile(0.25)) * 1.5
)
outliers2 = (
public["NPT43_PUB"].quantile(0.75)
+ (public["NPT43_PUB"].quantile(0.75) - public["NPT43_PUB"].quantile(0.25)) * 1.5
)
public = public.drop(public[public["NPT43_PUB"] < outliers1].index)
public = public.drop(public[public["NPT43_PUB"] > outliers2].index)
plt.boxplot(public["NPT44_PUB"])
outliers1 = (
public["NPT44_PUB"].quantile(0.25)
- (public["NPT44_PUB"].quantile(0.75) - public["NPT44_PUB"].quantile(0.25)) * 1.5
)
outliers2 = (
public["NPT44_PUB"].quantile(0.75)
+ (public["NPT44_PUB"].quantile(0.75) - public["NPT44_PUB"].quantile(0.25)) * 1.5
)
public = public.drop(public[public["NPT44_PUB"] < outliers1].index)
public = public.drop(public[public["NPT44_PUB"] > outliers2].index)
plt.boxplot(public["NPT45_PUB"])
outliers1 = (
public["NPT45_PUB"].quantile(0.25)
- (public["NPT45_PUB"].quantile(0.75) - public["NPT45_PUB"].quantile(0.25)) * 1.5
)
outliers2 = (
public["NPT45_PUB"].quantile(0.75)
+ (public["NPT45_PUB"].quantile(0.75) - public["NPT45_PUB"].quantile(0.25)) * 1.5
)
public = public.drop(public[public["NPT45_PUB"] < outliers1].index)
public = public.drop(public[public["NPT45_PUB"] > outliers2].index)
public = public.reset_index(drop=True)
public
# ###### Regression models for predicting 25th percentile ACT math score
# XGBoost Regression
x_trn, x_tst = train_test_split(public, test_size=0.20, random_state=1)
y_trn = x_trn["ACTMT25"]
y_tst = x_tst["ACTMT25"]
x_trn = x_trn.drop(["ACTMT25"], axis=1)
x_tst = x_tst.drop(["ACTMT25"], axis=1)
model = XGBRegressor(subsample=0.5, learning_rate=0.07)
model.fit(x_trn, y_trn)
print("Model Accuracy on Training:", model.score(x_trn, y_trn), "\n")
print(
pd.DataFrame(
model.feature_importances_, index=x_trn.columns, columns=["Variables"]
).sort_values(by=["Variables"], ascending=False),
"\n",
)
y_pred = model.predict(x_tst)
print(
"Accuracy:",
model.score(x_tst, y_tst),
"/ RMSE:",
np.sqrt(mean_squared_error(y_pred, y_tst)),
)
# Linear Regression
x_trn, x_tst = train_test_split(public, test_size=0.20, random_state=1)
y_trn = x_trn["ACTMT25"]
y_tst = x_tst["ACTMT25"]
x_trn = x_trn.drop(["ACTMT25"], axis=1)
x_tst = x_tst.drop(["ACTMT25"], axis=1)
model = LinearRegression()
model.fit(x_trn, y_trn)
print("Model Accuracy on Training:", model.score(x_trn, y_trn), "\n")
# print (pd.DataFrame(model.feature_importances_,index = x_trn.columns,columns =['Variables']).sort_values(by=['Variables'],ascending=False),'\n')
y_pred = model.predict(x_tst)
print(
"Accuracy:",
model.score(x_tst, y_tst),
"/ RMSE:",
np.sqrt(mean_squared_error(y_pred, y_tst)),
)
# # Private School
private = private.dropna(
subset=["NPT41_PRIV", "NPT42_PRIV", "NPT43_PRIV", "NPT44_PRIV", "NPT45_PRIV"],
how="all",
)
private.isna().sum()
private
private.describe()
private[private["NPT41_PRIV"] < 100]
private[private["NPT42_PRIV"] < 100]
private = private.drop([194, 1465, 2341])
private.describe()
# ###### Fillna with mean values of column
private["NPT41_PRIV"] = private["NPT41_PRIV"].fillna(private["NPT41_PRIV"].mean())
private["NPT42_PRIV"] = private["NPT42_PRIV"].fillna(private["NPT42_PRIV"].mean())
private["NPT43_PRIV"] = private["NPT43_PRIV"].fillna(private["NPT43_PRIV"].mean())
private["NPT44_PRIV"] = private["NPT44_PRIV"].fillna(private["NPT44_PRIV"].mean())
private["NPT45_PRIV"] = private["NPT45_PRIV"].fillna(private["NPT45_PRIV"].mean())
privatev1 = private.copy()
# ###### Regression models for predicting 25th percentile ACT math score
# XGBoost Regression
x_trn, x_tst = train_test_split(privatev1, test_size=0.20, random_state=1)
y_trn = x_trn["ACTMT25"]
y_tst = x_tst["ACTMT25"]
x_trn = x_trn.drop(["ACTMT25"], axis=1)
x_tst = x_tst.drop(["ACTMT25"], axis=1)
model = XGBRegressor(tree_method="approx", enable_categorical=True)
model.fit(x_trn, y_trn)
print("Model Accuracy on Training:", model.score(x_trn, y_trn), "\n")
print(
pd.DataFrame(
model.feature_importances_, index=x_trn.columns, columns=["Variables"]
).sort_values(by=["Variables"], ascending=False),
"\n",
)
y_pred = model.predict(x_tst)
print(
"Accuracy:",
model.score(x_tst, y_tst),
"/ RMSE:",
np.sqrt(mean_squared_error(y_pred, y_tst)),
)
# Linear Regression
x_trn, x_tst = train_test_split(privatev1, test_size=0.20, random_state=1)
y_trn = x_trn["ACTMT25"]
y_tst = x_tst["ACTMT25"]
x_trn = x_trn.drop(["ACTMT25"], axis=1)
x_tst = x_tst.drop(["ACTMT25"], axis=1)
model = LinearRegression()
model.fit(x_trn, y_trn)
print("Model Accuracy on Training:", model.score(x_trn, y_trn), "\n")
# print (pd.DataFrame(model.feature_importances_,index = x_trn.columns,columns =['Variables']).sort_values(by=['Variables'],ascending=False),'\n')
y_pred = model.predict(x_tst)
print(
"Accuracy:",
model.score(x_tst, y_tst),
"/ RMSE:",
np.sqrt(mean_squared_error(y_pred, y_tst)),
)
# ###### Removing outliers
plt.boxplot(private["NPT41_PRIV"])
outliers1 = (
private["NPT41_PRIV"].quantile(0.25)
- (private["NPT41_PRIV"].quantile(0.75) - private["NPT41_PRIV"].quantile(0.25))
* 1.5
)
outliers2 = (
private["NPT41_PRIV"].quantile(0.75)
+ (private["NPT41_PRIV"].quantile(0.75) - private["NPT41_PRIV"].quantile(0.25))
* 1.5
)
private = private.drop(private[private["NPT41_PRIV"] < outliers1].index)
private = private.drop(private[private["NPT41_PRIV"] > outliers2].index)
plt.boxplot(private["NPT42_PRIV"])
outliers1 = (
private["NPT42_PRIV"].quantile(0.25)
- (private["NPT42_PRIV"].quantile(0.75) - private["NPT42_PRIV"].quantile(0.25))
* 1.5
)
outliers2 = (
private["NPT42_PRIV"].quantile(0.75)
+ (private["NPT42_PRIV"].quantile(0.75) - private["NPT42_PRIV"].quantile(0.25))
* 1.5
)
private = private.drop(private[private["NPT42_PRIV"] < outliers1].index)
private = private.drop(private[private["NPT42_PRIV"] > outliers2].index)
plt.boxplot(private["NPT43_PRIV"])
outliers1 = (
private["NPT43_PRIV"].quantile(0.25)
- (private["NPT43_PRIV"].quantile(0.75) - private["NPT43_PRIV"].quantile(0.25))
* 1.5
)
outliers2 = (
private["NPT43_PRIV"].quantile(0.75)
+ (private["NPT43_PRIV"].quantile(0.75) - private["NPT43_PRIV"].quantile(0.25))
* 1.5
)
private = private.drop(private[private["NPT43_PRIV"] < outliers1].index)
private = private.drop(private[private["NPT43_PRIV"] > outliers2].index)
plt.boxplot(private["NPT44_PRIV"])
outliers1 = (
private["NPT44_PRIV"].quantile(0.25)
- (private["NPT44_PRIV"].quantile(0.75) - private["NPT44_PRIV"].quantile(0.25))
* 1.5
)
outliers2 = (
private["NPT44_PRIV"].quantile(0.75)
+ (private["NPT44_PRIV"].quantile(0.75) - private["NPT44_PRIV"].quantile(0.25))
* 1.5
)
private = private.drop(private[private["NPT44_PRIV"] < outliers1].index)
private = private.drop(private[private["NPT44_PRIV"] > outliers2].index)
plt.boxplot(private["NPT45_PRIV"])
outliers1 = (
private["NPT45_PRIV"].quantile(0.25)
- (private["NPT45_PRIV"].quantile(0.75) - private["NPT45_PRIV"].quantile(0.25))
* 1.5
)
outliers2 = (
private["NPT45_PRIV"].quantile(0.75)
+ (private["NPT45_PRIV"].quantile(0.75) - private["NPT45_PRIV"].quantile(0.25))
* 1.5
)
private = private.drop(private[private["NPT45_PRIV"] < outliers1].index)
private = private.drop(private[private["NPT45_PRIV"] > outliers2].index)
private = private.reset_index(drop=True)
private
# ###### Regression models for predicting 25th percentile ACT math score
# XGBoost Regression
x_trn, x_tst = train_test_split(private, test_size=0.20, random_state=1)
y_trn = x_trn["ACTMT25"]
y_tst = x_tst["ACTMT25"]
x_trn = x_trn.drop(["ACTMT25"], axis=1)
x_tst = x_tst.drop(["ACTMT25"], axis=1)
model = XGBRegressor(tree_method="approx", enable_categorical=True)
model.fit(x_trn, y_trn)
print("Model Accuracy on Training:", model.score(x_trn, y_trn), "\n")
print(
pd.DataFrame(
model.feature_importances_, index=x_trn.columns, columns=["Variables"]
).sort_values(by=["Variables"], ascending=False),
"\n",
)
y_pred = model.predict(x_tst)
print(
"Accuracy:",
model.score(x_tst, y_tst),
"/ RMSE:",
np.sqrt(mean_squared_error(y_pred, y_tst)),
)
x_trn, x_tst = train_test_split(private, test_size=0.20, random_state=1)
y_trn = x_trn["ACTMT25"]
y_tst = x_tst["ACTMT25"]
x_trn = x_trn.drop(["ACTMT25"], axis=1)
x_tst = x_tst.drop(["ACTMT25"], axis=1)
model = LinearRegression()
model.fit(x_trn, y_trn)
print("Model Accuracy on Training:", model.score(x_trn, y_trn), "\n")
# print (pd.DataFrame(model.feature_importances_,index = x_trn.columns,columns =['Variables']).sort_values(by=['Variables'],ascending=False),'\n')
y_pred = model.predict(x_tst)
print(
"Accuracy:",
model.score(x_tst, y_tst),
"/ RMSE:",
np.sqrt(mean_squared_error(y_pred, y_tst)),
)
# # Public vs Private
# ###### Combine, to see if it improves the prediction
private.columns
publicv1["sector"] = 0
publicv1 = publicv1.rename(
columns={
"NPT41_PUB": "NPT41",
"NPT42_PUB": "NPT42",
"NPT43_PUB": "NPT43",
"NPT44_PUB": "NPT44",
"NPT45_PUB": "NPT45",
}
)
privatev1["sector"] = 1
privatev1 = privatev1.rename(
columns={
"NPT41_PRIV": "NPT41",
"NPT42_PRIV": "NPT42",
"NPT43_PRIV": "NPT43",
"NPT44_PRIV": "NPT44",
"NPT45_PRIV": "NPT45",
}
)
PPv1 = pd.concat([publicv1, privatev1], ignore_index=True)
public["sector"] = 0
public = public.rename(
columns={
"NPT41_PUB": "NPT41",
"NPT42_PUB": "NPT42",
"NPT43_PUB": "NPT43",
"NPT44_PUB": "NPT44",
"NPT45_PUB": "NPT45",
}
)
private["sector"] = 1
private = private.rename(
columns={
"NPT41_PRIV": "NPT41",
"NPT42_PRIV": "NPT42",
"NPT43_PRIV": "NPT43",
"NPT44_PRIV": "NPT44",
"NPT45_PRIV": "NPT45",
}
)
PP = pd.concat([public, private], ignore_index=True)
PPv1
PPv1.sector.value_counts()
PP.sector.value_counts()
variables = [
"UGDS_WHITE",
"UGDS_BLACK",
"UGDS_HISP",
"UGDS_ASIAN",
"NPT41",
"NPT42",
"NPT43",
"NPT44",
"NPT45",
"ACTMT25",
]
# ###### Regression models for predicting 25th percentile ACT math score
# ###### Before removing outliers
x_trn, x_tst = train_test_split(PPv1, test_size=0.20, random_state=1)
y_trn = x_trn["ACTMT25"]
y_tst = x_tst["ACTMT25"]
x_trn = x_trn.drop(["ACTMT25"], axis=1)
x_tst = x_tst.drop(["ACTMT25"], axis=1)
model = XGBRegressor(subsample=0.5, learning_rate=0.08)
model.fit(x_trn, y_trn)
print("Model Accuracy on Training:", model.score(x_trn, y_trn), "\n")
print(
pd.DataFrame(
model.feature_importances_, index=x_trn.columns, columns=["Variables"]
).sort_values(by=["Variables"], ascending=False),
"\n",
)
y_pred = model.predict(x_tst)
print(
"Accuracy:",
model.score(x_tst, y_tst),
"/ RMSE:",
np.sqrt(mean_squared_error(y_pred, y_tst)),
)
x_trn, x_tst = train_test_split(PPv1, test_size=0.20, random_state=1)
y_trn = x_trn["ACTMT25"]
y_tst = x_tst["ACTMT25"]
x_trn = x_trn.drop(["ACTMT25"], axis=1)
x_tst = x_tst.drop(["ACTMT25"], axis=1)
model = LinearRegression()
model.fit(x_trn, y_trn)
print("Model Accuracy on Training:", model.score(x_trn, y_trn), "\n")
# print (pd.DataFrame(model.feature_importances_,index = x_trn.columns,columns =['Variables']).sort_values(by=['Variables'],ascending=False),'\n')
y_pred = model.predict(x_tst)
print(
"Accuracy:",
model.score(x_tst, y_tst),
"/ RMSE:",
np.sqrt(mean_squared_error(y_pred, y_tst)),
)
# ###### Two tailed t-test between the variables in 2 classes - Public and Private
for i in variables:
batch_1 = PPv1[PPv1["sector"] == 0][i]
batch_2 = PPv1[PPv1["sector"] == 1][i]
stat, p_value = stats.ttest_ind(batch_1, batch_2)
print(i)
# Level of significance
alpha = 0.05
# conclusion
if p_value < alpha / 2:
print(
"Reject Null Hypothesis (Significant difference between two samples)", "\n"
)
else:
print(
"Do not Reject Null Hypothesis (No significant difference between two samples)",
"\n",
)
# ###### After removing outliers
x_trn, x_tst = train_test_split(PP, test_size=0.20, random_state=1)
y_trn = x_trn["ACTMT25"]
y_tst = x_tst["ACTMT25"]
x_trn = x_trn.drop(["ACTMT25"], axis=1)
x_tst = x_tst.drop(["ACTMT25"], axis=1)
model = XGBRegressor(subsample=0.5, learning_rate=0.08)
model.fit(x_trn, y_trn)
print("Model Accuracy on Training:", model.score(x_trn, y_trn), "\n")
print(
pd.DataFrame(
model.feature_importances_, index=x_trn.columns, columns=["Variables"]
).sort_values(by=["Variables"], ascending=False),
"\n",
)
y_pred = model.predict(x_tst)
print(
"Accuracy:",
model.score(x_tst, y_tst),
"/ RMSE:",
np.sqrt(mean_squared_error(y_pred, y_tst)),
)
x_trn, x_tst = train_test_split(PP, test_size=0.20, random_state=1)
y_trn = x_trn["ACTMT25"]
y_tst = x_tst["ACTMT25"]
x_trn = x_trn.drop(["ACTMT25"], axis=1)
x_tst = x_tst.drop(["ACTMT25"], axis=1)
model = LinearRegression()
model.fit(x_trn, y_trn)
print("Model Accuracy on Training:", model.score(x_trn, y_trn), "\n")
# print (pd.DataFrame(model.feature_importances_,index = x_trn.columns,columns =['Variables']).sort_values(by=['Variables'],ascending=False),'\n')
y_pred = model.predict(x_tst)
print(
"Accuracy:",
model.score(x_tst, y_tst),
"/ RMSE:",
np.sqrt(mean_squared_error(y_pred, y_tst)),
)
# ###### Two tailed t-test between the variables in 2 classes - Public and Private
for i in variables:
batch_1 = PP[PP["sector"] == 0][i]
batch_2 = PP[PP["sector"] == 1][i]
stat, p_value = stats.ttest_ind(batch_1, batch_2)
print(i)
# Level of significance
alpha = 0.05
# conclusion
if p_value < alpha / 2:
print(
"Reject Null Hypothesis (Significant difference between two samples)", "\n"
)
else:
print(
"Do not Reject Null Hypothesis (No significant difference between two samples)",
"\n",
)
|
# # Analysis of Crime in Oakland of the datasets 2011-2016
# ## Background information
# A small project to find details of crimes in Oakland, CA, over the datasets that span from 2011-2016.
# As a bit of background information, there are some names of crimes within the dataset that may not be clear to people outside of the United States (such as myself), so it is useful to research the terminology within the datasets. For e.g. definitions for '*priority 1 & 2*' crimes are:
# *Priority 1 crime is said to be an urgent crime, for e.g. lights and sirens authorised, armed robbery, officer down etc.*
# *Priority 2 crime is said to be of less urgency, for e.g. lights and sirens authorised, but follow basic traffic rules.*
# Whilst evaluating the data I had also found points where I have diregarded for being unnecessary. These were things such as - while analysing priority crimes, a *prirority 0* had appeared in only 3 datasets of which had a count of less than 10 crimes. Where as *priority 1 & 2* had a count of a much larger number for e.g. 25000.
# A big point to make - the data set for 2016 is inconclusive and the last entry is in the middle of the year.
# ### Importing libraries
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from pandas import Timestamp
from datetime import date
from dateutil.relativedelta import relativedelta
from sklearn.linear_model import LinearRegression
# #### Import files
df_2011 = pd.read_csv(
"../input/oakland-crime-statistics-2011-to-2016/records-for-2011.csv",
parse_dates=["Create Time", "Closed Time"],
)
df_2012 = pd.read_csv(
"../input/oakland-crime-statistics-2011-to-2016/records-for-2012.csv",
parse_dates=["Create Time", "Closed Time"],
)
df_2013 = pd.read_csv(
"../input/oakland-crime-statistics-2011-to-2016/records-for-2013.csv",
parse_dates=["Create Time", "Closed Time"],
)
df_2014 = pd.read_csv(
"../input/oakland-crime-statistics-2011-to-2016/records-for-2014.csv",
parse_dates=["Create Time", "Closed Time"],
)
df_2015 = pd.read_csv(
"../input/oakland-crime-statistics-2011-to-2016/records-for-2015.csv",
parse_dates=["Create Time", "Closed Time"],
)
df_2016 = pd.read_csv(
"../input/oakland-crime-statistics-2011-to-2016/records-for-2016.csv",
parse_dates=["Create Time", "Closed Time"],
)
list_dfs = [df_2011, df_2012, df_2013, df_2014, df_2015, df_2016]
# ### First few rows of data for all data sets.
def shapes():
x = 0
for i in list_dfs:
print(f"Shape of dataset for {x+2011} is {i.shape}")
x += 1
shapes()
df_2011.head()
df_2012.head()
df_2013.head()
df_2014.head()
df_2015.head()
df_2016.head()
# I have decided to focus on the Priority column within all datasets, and compare with other columns for analysis.
# > ### Priority Analysis
# Amount of Priority crimes for all years observed:
# Code to show count of priority crimes per year.
a = 0
for i in list_dfs:
print(
i[i["Priority"] != 0]
.groupby(["Priority"])
.size()
.reset_index(name=str(f"Count in {a + 2011}"))
)
a += 1
print(" ")
# Bar charts for comparing priority type crimes
df = pd.DataFrame(
[
[1, 36699, 41926, 43171, 42773, 42418, 24555],
[2, 143314, 145504, 144859, 144707, 150162, 86272],
],
columns=["Priority"] + [f"Count in {x}" for x in range(2011, 2017)],
)
df.plot.bar(x="Priority", subplots=True, layout=(2, 3), figsize=(15, 7))
pri1_2011 = 36699
pri2_2011 = 143314
total_2011 = pri1_2011 + pri2_2011
print(
f"Priority 1 crimes amounted to {round((pri1_2011/total_2011)*100, 3)}%, priority 2 crimes amounted to {round((pri2_2011/total_2011)*100, 3)}% in 2011."
)
print(
"-----------------------------------------------------------------------------------------------------------------------------------------"
)
pri1_2012 = 41926
pri2_2012 = 145504
total_2012 = pri1_2012 + pri2_2012
print(
f"Priority 1 crimes amounted to {round((pri1_2012/total_2012)*100, 3)}%, priority 2 crimes amounted to {round((pri2_2012/total_2012)*100, 3)}% in 2012."
)
print(
"-----------------------------------------------------------------------------------------------------------------------------------------"
)
pri1_2013 = 43171
pri2_2013 = 144859
total_2013 = pri1_2013 + pri2_2013
print(
f"Priority 1 crimes amounted to {round((pri1_2013/total_2013)*100, 3)}%, priority 2 crimes amounted to {round((pri2_2013/total_2013)*100, 3)}% in 2013."
)
print(
"-----------------------------------------------------------------------------------------------------------------------------------------"
)
pri1_2014 = 42773
pri2_2014 = 144707
total_2014 = pri1_2014 + pri2_2014
print(
f"Priority 1 crimes amounted to {round((pri1_2014/total_2014)*100, 3)}% priority 2 crimes amounted to {round((pri2_2014/total_2014)*100, 3)}% in 2014."
)
print(
"-----------------------------------------------------------------------------------------------------------------------------------------"
)
pri1_2015 = 42418
pri2_2015 = 150162
total_2015 = pri1_2015 + pri2_2015
print(
f"Priority 1 crimes amounted to {round((pri1_2015/total_2015)*100, 3)}%, priority 2 crimes amounted to {round((pri2_2015/total_2015)*100, 3)}% in 2015."
)
print(
"-----------------------------------------------------------------------------------------------------------------------------------------"
)
pri1_2016 = 24555
pri2_2016 = 86272
total_2016 = pri1_2016 + pri2_2016
print(
f"Priority 1 crimes amounted to {round((pri1_2016/total_2016)*100, 3)}% and priority 2 crimes amounted to {round((pri2_2016/total_2016)*100, 3)}%, for the first half of 2016."
)
print(
"-----------------------------------------------------------------------------------------------------------------------------------------"
)
# Crime seems to be at a stable rate throughout the datasets. The margin of difference in percentage is only slight throughout the 6 years observed.
# ### Area ID analysis.
# Mean Priority count per Area/Location/Beat
def areaid_groupby():
for i in list_dfs:
print(i[i["Priority"] != 0].groupby(["Area Id", "Priority"]).size())
print(" ")
areaid_groupby()
fig, axes = plt.subplots(2, 3)
for i, d in enumerate(list_dfs):
ax = axes.flatten()[i]
dplot = d[["Area Id", "Priority"]].pivot_table(
index="Area Id", columns=["Priority"], aggfunc=len
)
dplot = (
dplot.assign(total=lambda x: x.sum(axis=1))
.sort_values("total", ascending=False)
.head(10)
.drop("total", axis=1)
)
dplot.plot.bar(ax=ax, figsize=(15, 7), stacked=True)
ax.set_title(f"Plot of Priority 1 and 2 crimes within Area Id for {i+2011}")
plt.tight_layout()
# The Area Id's for each dataset have not been consistent with their category. To see the amount of crimes for each year split by priority, check below:
# Summing the amount of Priority 1 and 2 crimes per dataset we can see that there is an increase in both crimes.
# ### Beat Analysis
# Value count for beats displayed by priority
for i in list_dfs:
print(i[i["Priority"] != 0].groupby(["Beat", "Priority"]).size())
print(" ")
fig, axes = plt.subplots(2, 3)
for i, d in enumerate(list_dfs):
ax = axes.flatten()[i]
dplot = d[["Beat", "Priority"]].pivot_table(
index="Beat", columns=["Priority"], aggfunc=len
)
dplot = (
dplot.assign(total=lambda x: x.sum(axis=1))
.sort_values("total", ascending=False)
.head(10)
.drop("total", axis=1)
)
dplot.plot.bar(ax=ax, figsize=(15, 7), stacked=True)
ax.set_title(f"Top 10 Beats for {i+ 2011}")
plt.tight_layout()
# ### Incident type description (Incident type id) analysis
# Top 20 most popular crimes across the data sets
df1 = df_2011["Incident Type Description"].value_counts()[:10]
df2 = df_2012["Incident Type Description"].value_counts()[:10]
df3 = df_2013["Incident Type Description"].value_counts()[:10]
df4 = df_2014["Incident Type Description"].value_counts()[:10]
df5 = df_2015["Incident Type Description"].value_counts()[:10]
df6 = df_2016["Incident Type Description"].value_counts()[:10]
list_df = [df1, df2, df3, df4, df5, df6]
fig, axes = plt.subplots(2, 3)
for d, i in zip(list_df, range(6)):
ax = axes.ravel()[i]
ax.set_title(f"Top 20 crimes in {i+2011}")
d.plot.barh(ax=ax, figsize=(15, 7))
plt.tight_layout()
fig, axes = plt.subplots(2, 3)
for i, d in enumerate(list_dfs):
ax = axes.flatten()[i]
dplot = d[["Incident Type Id", "Priority"]].pivot_table(
index="Incident Type Id", columns="Priority", aggfunc=len
)
dplot = (
dplot.assign(total=lambda x: x.sum(axis=1))
.sort_values("total", ascending=False)
.head(10)
.drop("total", axis=1)
)
dplot.plot.barh(ax=ax, figsize=(15, 7), stacked=True)
ax.set_title(f"Plot of Top 10 Incidents in {i+2011}")
plt.tight_layout()
# Two graphs to show the 'Indcident Type Decription' as well as it's Id. The first graph shows that 'Alarm Ringer' is by far the most reported crime, however in graph 2 we can see that only a small percentage of that is *priority 1*. All through the 6 datasets we can see that 'Battery/242' is the highest reported *priority 1* crime.
# ### Time analysis
# Total amount of pripority crimes per month
pri_count_list = [
df_2011.groupby(
["Priority", df_2011["Create Time"].dt.to_period("m")]
).Priority.count(),
df_2012.groupby(
["Priority", df_2012["Create Time"].dt.to_period("m")]
).Priority.count(),
df_2013.groupby(
["Priority", df_2013["Create Time"].dt.to_period("m")]
).Priority.count(),
df_2014.groupby(
["Priority", df_2014["Create Time"].dt.to_period("m")]
).Priority.count(),
df_2015.groupby(
["Priority", df_2015["Create Time"].dt.to_period("m")]
).Priority.count(),
df_2016.groupby(
["Priority", df_2016["Create Time"].dt.to_period("m")]
).Priority.count(),
]
fig, axes = plt.subplots(2, 3)
for d, ax in zip(pri_count_list, axes.ravel()):
plot_df1 = d.unstack("Priority").loc[:, 1]
plot_df2 = d.unstack("Priority").loc[:, 2]
plot_df1.index = pd.PeriodIndex(plot_df1.index.tolist(), freq="m")
plot_df2.index = pd.PeriodIndex(plot_df2.index.tolist(), freq="m")
plt.suptitle("Visualisation of priorities by the year")
plot_df1.plot(ax=ax, legend=True, figsize=(15, 7))
plot_df2.plot(ax=ax, legend=True, figsize=(15, 7))
# The visualisation shows us that within each year, Priority 2 crimes seem to peak around July/August time. Apart from in 2014 where there seemed to be a drop.The plot for 2016 shows an inconclusive graph since the dataset was only a 7 month long time span.
count = 2011
x = []
for i in list_dfs:
i["Difference in hours"] = i["Closed Time"] - i["Create Time"]
i["Difference in hours"] = i["Difference in hours"] / np.timedelta64(1, "h")
mean_hours = round(i["Difference in hours"].mean(), 3)
x.append(mean_hours)
print(
f"Difference in hours for {count} is {mean_hours} with a reported {i.shape[0]} crimes."
)
count += 1
|
# #### 1. Train data set is visualized in several ways using swarmplot in seaborn to provide guidelines for feature engineering followed by one-hot encoding. A total of 6 features are used:
# #### (1) Pclass and Sex are used without any modification,
# #### (2) Fare is binned into 4 groups with 'unknown' and by using bin edges 10.5 and 75,
# #### (3) SibSp and Parch are added together and binned to unkown, 0 (travel alone), 4 below, and 4 and above to form a new feature,
# #### (4) Persons with 'Master' in Name are identified and form a new feature,
# #### (5) Female in Pclass 3 with Fare > 24 is identified and forms a new feature,
# #### 2. Eight models with hyper-parameter tuning are constructed for predictions: logistic regression, random forest, gradient boosting, XGBoost, multinomial naive Bayes, k nearest neighbors, stack, and majority vote. The stack model uses all the first 6 models above as the 1st-level models and random forest as the 2nd-level model.
# #### 3. In summary, gradient boost and stack models have the highest mean cross-validation scores (both 0.842), followed by random forest and XGBoost (0.837 and 0.835, respectively), followed by logistic regression and k nearest neighbors (0.828 and 0.827, respectively), and multinomial naive Bayes has the lowest score of 0.780.
# #### However, random forest, together with stack, achieve the highest public scores of 0.799, followed by logistic regression, gradient boost, and XGBoost (all 0.794), followed by k nearest neighbors with 0.789, and multinomial naive Bayes has the lowest public score of 0.746. The majority vote also achieves the highest public score of 0.799.
# #### It is found that model performance (model's public score) may be highly dependent on the number of features chosen and the ways the features are enginnered.
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import operator
import glob
from scipy.stats import uniform, norm
from sklearn.metrics import classification_report, confusion_matrix
from sklearn.preprocessing import OneHotEncoder
from sklearn.model_selection import (
train_test_split,
GridSearchCV,
RandomizedSearchCV,
cross_val_score,
KFold,
StratifiedKFold,
)
from sklearn.linear_model import LogisticRegression
from sklearn.naive_bayes import MultinomialNB
from sklearn.ensemble import RandomForestClassifier, GradientBoostingClassifier
from sklearn.neighbors import KNeighborsClassifier
import xgboost
from xgboost import XGBClassifier
import sklearn
# ## Data visualization
data_train = pd.read_csv("../input/titanic/train.csv")
data_train.info()
data_train.head()
sns.set(style="darkgrid")
colors = ["darkred", "darkseagreen"]
# ### 1. Visualize the survival chance of persons with different fare
data_train[data_train["Fare"] > 300]
fig, ax = plt.subplots(figsize=(17, 12))
ax.set_ylim(0, 300)
ax.set_yticks(np.arange(0, 300, 10))
sns.swarmplot(
y=data_train["Fare"],
x=[""] * len(data_train),
size=4,
hue=data_train["Survived"],
palette=colors,
)
# #### The plot above shows that persons with fare above 75 had a relatively good chance of survival, and for those with fare below about 10.5 the chance was quite bad, and those with fare in between seems to have a chance somewhere in the middle.
# ### 2. Add 'SibSp' and 'Parch' together and visualize the chance of survival
df_try = data_train.copy()
df_try["SibSp_Parch"] = df_try["SibSp"] + df_try["Parch"]
df_try.groupby("SibSp_Parch")["Survived"].value_counts()
fig, ax = plt.subplots(figsize=(17, 7))
sns.swarmplot(
y=df_try["SibSp_Parch"],
x=[""] * len(df_try),
size=4,
hue=df_try["Survived"],
palette=colors,
)
# #### The plot above shows that persons with 4 relatives or above had a relatively small chance of survival, and the same is true (to a lesser extent) with persons who traveled alone with 0 relatives. In contrast, persons with 1 to 3 relatives had a better chance of survival.
# ### 3. Visualize chance of survival in plots combining sex, age, and Pclass
g1 = sns.FacetGrid(
data_train, col="Pclass", hue="Survived", palette=colors, size=5, aspect=1
)
g1 = g1.map(sns.swarmplot, "Sex", "Age", order=["male", "female"], size=5)
g1.add_legend()
# #### It can be seen from the plot above that male with age less than about 12 years old had a better chance of survival compared to male older than this age. We will later create a new feature to reflect this.
mask_master = pd.Series("Master" in i for i in data_train["Name"])
data_train[mask_master].sort_values("Age", ascending=False).head(10)
# #### From the table above it can be seen that if a person has 'Master' in 'Name' then this person is a male with age less than or equal to 12 years old.
fig, ax = plt.subplots(figsize=(17, 7))
sns.swarmplot(
x="Sex",
y="Fare",
data=data_train[data_train["Pclass"] == 3],
size=4,
hue="Survived",
palette=colors,
)
# #### It can be seen from the plot above that female in Pclass 3 with fare greater than about 24 almost all did not make it. We will also later create a new feature to reflect this.
# ## Data cleaning and preprocessing
y = data_train["Survived"]
X = data_train.drop("Survived", axis=1)
X.head()
def combine_Sib_Par(df):
"""Sum the two columns SibSp and Parch together."""
df["SibSp_Parch"] = df["SibSp"] + df["Parch"]
def add_name_master_feature(df):
"""Create a new feature: if Master in Name, then Yes, otherwise, No."""
mask_master = pd.Series("Master" in i for i in df["Name"])
df1 = df["Name"].mask(mask_master, "Yes")
df["Name_Master"] = df1.where(mask_master, "No")
def add_female_pclass_3_high_fare_feature(df):
"""Create a new feature: if female, in Pclass 3, and Fare > 24, Yes, otherwise, No."""
df_temp = df[((df["Pclass"] == 3) & (df["Sex"] == "female")) & (df["Fare"] > 24.0)]
mask = df.index.isin(df_temp.index)
df["Fem_Hfare_Pcl3"] = pd.Series(range(df.shape[0])).mask(mask, "Yes")
df["Fem_Hfare_Pcl3"] = df["Fem_Hfare_Pcl3"].where(mask, "No")
def drop_feature(df):
df.drop(
["PassengerId", "Name", "Age", "SibSp", "Parch", "Ticket", "Cabin", "Embarked"],
axis=1,
inplace=True,
)
def fill_feature(df):
"""Fill all NaN values."""
df["Pclass"] = df["Pclass"].fillna(-1)
df["Sex"] = df["Sex"].fillna("Unknown")
df["SibSp_Parch"] = df["SibSp_Parch"].fillna(-1)
df["Fare"] = df["Fare"].fillna(-0.5)
def bin_fare_and_SibSpParch(df):
"""Bin Fare and SibSp_Parch based on previous visualization results."""
bins = (-1, 0, 10.5, 75, 1500)
group_names = ["Unknown", "10.5_below", "10.5_to_75", "75_above"]
df["Fare"] = pd.cut(df["Fare"], bins, labels=group_names, right=False)
bins = (-1, -0.1, 0.1, 4, 50)
group_names = ["Unknown", "0", "4_below", "4_above"]
df["SibSp_Parch"] = pd.cut(df["SibSp_Parch"], bins, labels=group_names, right=False)
def data_transform(df):
combine_Sib_Par(df)
add_name_master_feature(df)
add_female_pclass_3_high_fare_feature(df)
drop_feature(df)
fill_feature(df)
bin_fare_and_SibSpParch(df)
data_transform(X)
X.head(10)
X.info()
ohe = OneHotEncoder(handle_unknown="ignore")
X_1 = ohe.fit_transform(X).toarray()
list(X_1)[:5]
ohe.categories_
x_ax = ohe.get_feature_names(
["Pclass", "Sex", "Fare", "SibSp_Parch", "Name_Master", "Fem_Hfare_Pcl3"]
)
x_ax
# Create a DataFrame for correlation plot
X_1_frame = pd.DataFrame(X_1, columns=x_ax)
X_1_frame.head()
plt.figure(figsize=(12, 12))
plt.title("Corelation Matrix", size=8)
sns.heatmap(
X_1_frame.corr(),
linewidths=0.1,
vmax=1.0,
square=True,
cmap=plt.cm.RdBu,
linecolor="white",
annot=True,
)
plt.show()
# ## Data training and parameter tuning
# define a cv splitter
cv_splitter = StratifiedKFold(n_splits=5, random_state=42)
# ### 1. First Model: Logistic Regressionfrom
#
logis = LogisticRegression(solver="liblinear", random_state=42)
C_param = sorted(10 ** np.random.uniform(-2, 0, size=200))
# log-uniform distrbution from 0.01 to 1
# Since if there are multiple parameter combinations rank first, GridSearchCV will choose the
# first encountered one as the best result, sort the array so the smallest possible C can be
# picked.
parameter_grid = {"C": C_param, "class_weight": ["balanced", None]}
grid_logis = GridSearchCV(logis, parameter_grid, cv=cv_splitter, refit=True)
grid_logis.fit(X_1, y)
logis_best_param = grid_logis.best_params_
logis_best_param
# best parameter values to be used in the stack model
results = pd.DataFrame(grid_logis.cv_results_)
results.iloc[:, 4:].sort_values("rank_test_score")
x_ax = ohe.get_feature_names(
["Pclass", "Sex", "Fare", "SibSp_Parch", "Name_Master", "Fem_Hfare_Pcl3"]
)
x_ax
fig, ax = plt.subplots(figsize=(35, 8))
ax.bar(x_ax, grid_logis.best_estimator_.coef_[0])
ax.grid
scores_logis = cross_val_score(
grid_logis.best_estimator_, X_1, y, cv=cv_splitter, n_jobs=-1
)
print(scores_logis)
print("Mean (logis): " + str(scores_logis.mean()))
print("SD (logis): " + str(scores_logis.std()))
# ### 2. Second Model: Random Forest
# set max_features normal distribution sample array
num_feature = X_1.shape[1]
max_feature = norm.rvs(np.sqrt(num_feature), 2, size=200, random_state=42).astype(int)
max_feature[max_feature <= 0] = 1
max_feature[max_feature > num_feature] = num_feature
max_feature
# set min_samples_split normal distribution sample array
min_sample_split = norm.rvs(4, 2, size=200, random_state=42).astype(int)
min_sample_split[min_sample_split <= 1] = 2
min_sample_split
rf = RandomForestClassifier(random_state=42)
parameter_grid = {
"n_estimators": np.arange(50, 800, step=5),
"max_features": max_feature,
"min_samples_split": min_sample_split,
"min_samples_leaf": np.arange(1, 5, 1),
"bootstrap": [True, False],
}
grid_random = RandomizedSearchCV(
rf,
parameter_grid,
n_iter=100,
cv=cv_splitter,
random_state=42,
refit=True,
n_jobs=-1,
)
grid_random.fit(X_1, y)
random_forest_best_param = grid_random.best_params_
random_forest_best_param
# best parameter values to be used in the stack model
grid_random.n_splits_
grid_random.best_estimator_.get_params
fig, ax = plt.subplots(figsize=(35, 8))
ax.bar(x_ax, grid_random.best_estimator_.feature_importances_)
ax.grid
scores_random = cross_val_score(
grid_random.best_estimator_, X_1, y, cv=cv_splitter, n_jobs=-1
)
print(scores_random)
print("Mean (random): " + str(scores_random.mean()))
print("SD (random): " + str(scores_random.std()))
# ### 3. Third Model: Gradient Boosting
# #### 1. Tune learning_rate and n_estimators
gb = GradientBoostingClassifier(
learning_rate=0.1,
n_estimators=100,
max_features="sqrt",
subsample=0.8,
random_state=42,
)
parameter_grid = {
"learning_rate": np.arange(0.001, 0.003, 0.0005),
"n_estimators": np.arange(1000, 3000, 500),
}
grid_gradient = GridSearchCV(gb, parameter_grid, cv=cv_splitter, n_jobs=-1)
grid_gradient.fit(X_1, y)
gradient_best_param = grid_gradient.best_params_
gradient_best_param
# best parameter values to be used in the stack model
# update gb with the optimal parameters
gb.set_params(**gradient_best_param)
# #### 2. Tune max_depth and min_sample_split
parameter_grid = {"max_depth": np.arange(1, 5), "min_samples_split": np.arange(2, 6, 1)}
grid_gradient = GridSearchCV(gb, parameter_grid, cv=cv_splitter, n_jobs=-1)
grid_gradient.fit(X_1, y)
grid_gradient.best_params_
gradient_best_param.update(grid_gradient.best_params_)
gradient_best_param
# update best parameter values to be used in the stack model
# update gb with the optimal parameters
gb.set_params(**gradient_best_param)
# #### 3. Tune max_features and subsample
parameter_grid = {
"max_features": np.arange(2, 6),
"subsample": np.arange(0.4, 0.8, step=0.1),
}
grid_gradient = GridSearchCV(gb, parameter_grid, cv=cv_splitter, n_jobs=-1)
grid_gradient.fit(X_1, y)
grid_gradient.best_params_
gradient_best_param.update(grid_gradient.best_params_)
gradient_best_param
# update best parameter values to be used in the stack model
grid_gradient.best_estimator_
fig, ax = plt.subplots(figsize=(35, 8))
ax.bar(x_ax, grid_gradient.best_estimator_.feature_importances_)
ax.grid
scores_gradient = cross_val_score(
grid_gradient.best_estimator_, X_1, y, cv=cv_splitter, n_jobs=-1
)
print(scores_gradient)
print("Mean (gradient): " + str(scores_gradient.mean()))
print("SD (gradient): " + str(scores_gradient.std()))
# ### 4. Fourth Model: XGBoost
# #### (The tuning steps can be found in the article by Aarshay Jain at https://www.analyticsvidhya.com/blog/2016/03/complete-guide-parameter-tuning-xgboost-with-codes-python/)
# #### 1. Fix learning rate at 0.01 and find the optimal number of trees (n_estimators)
xgtrain = xgboost.DMatrix(X_1, label=y.values)
xgb = XGBClassifier(
learning_rate=0.01,
n_estimators=1000,
max_depth=5,
min_child_weight=1,
gamma=0,
subsample=0.8,
colsample_bytree=0.8,
n_jobs=-1,
random_state=42,
)
xgb_param = xgb.get_xgb_params()
xgb_param
cvresult = xgboost.cv(
xgb_param,
xgtrain,
num_boost_round=xgb.get_params()["n_estimators"],
nfold=5,
metrics="auc",
early_stopping_rounds=50,
seed=42,
)
cvresult.head()
cvresult.shape
xgb_best_param = {"n_estimators": cvresult.shape[0]}
xgb_best_param
# best n_estimators value to be used in the stack model
# update xgb with the optimal n_estimators
xgb.set_params(**xgb_best_param)
# #### 2. Tune max_depth and min_child_weight
parameter_grid = {"max_depth": np.arange(2, 4), "min_child_weight": np.arange(1, 4)}
grid_xgb = GridSearchCV(xgb, parameter_grid, cv=cv_splitter, n_jobs=-1)
grid_xgb.fit(X_1, y)
grid_xgb.best_params_
xgb_best_param.update(grid_xgb.best_params_)
xgb_best_param
# best parameter values to be used in the stack model
xgb.set_params(**xgb_best_param)
# update xgb parameters
# #### 3. Tune gamma
10 ** np.random.uniform(-3, 0, size=10)
parameter_grid = {"gamma": 10 ** np.random.uniform(-3, 0, size=10)}
grid_xgb = GridSearchCV(xgb, parameter_grid, cv=cv_splitter, n_jobs=-1)
grid_xgb.fit(X_1, y)
grid_xgb.best_params_
results = pd.DataFrame(grid_xgb.cv_results_)
results.iloc[:, 4:].sort_values("rank_test_score")
xgb_best_param.update(grid_xgb.best_params_)
xgb_best_param
# best parameter values to be used in the stack model
# update xgb with the optimal gamma
xgb.set_params(**xgb_best_param)
# #### 4. Tune subsample and colsample_bytree
parameter_grid = {
"subsample": np.arange(0.6, 1.0, 0.1),
"colsample_bytree": np.arange(0.6, 1.0, 0.1),
}
grid_xgb = GridSearchCV(xgb, parameter_grid, cv=cv_splitter, n_jobs=-1)
grid_xgb.fit(X_1, y)
grid_xgb.best_params_
xgb_best_param.update(grid_xgb.best_params_)
xgb_best_param
# best parameter values to be used in the stack model
# update xgb with the optimal parameters
xgb.set_params(**xgb_best_param)
# #### 5. Finally tune reg_alpha
parameter_grid = {
"reg_alpha": np.arange(0.0, 0.005, 0.001),
}
grid_xgb = GridSearchCV(xgb, parameter_grid, cv=cv_splitter, n_jobs=-1)
grid_xgb.fit(X_1, y)
grid_xgb.best_params_
results = pd.DataFrame(grid_xgb.cv_results_)
results.iloc[:, 4:].sort_values("rank_test_score")
xgb_best_param.update(grid_xgb.best_params_)
xgb_best_param
# best parameter values to be used in the stack model
grid_xgb.best_estimator_
# #### 6. Visualize the feature importance of the final model
fig, ax = plt.subplots(figsize=(35, 8))
ax.bar(x_ax, grid_xgb.best_estimator_.feature_importances_)
ax.grid
scores_xgb = cross_val_score(
grid_xgb.best_estimator_, X_1, y, cv=cv_splitter, n_jobs=-1
)
print(scores_xgb)
print("Mean (xgb): " + str(scores_xgb.mean()))
print("SD (xgb): " + str(scores_xgb.std()))
# ### 5. Fifth Model: Multinomial Naive Bayes
mnb = MultinomialNB()
parameters = {"alpha": np.arange(0.1, 1, 0.1)}
grid_mnb = GridSearchCV(mnb, param_grid=parameters, cv=cv_splitter, n_jobs=-1)
grid_mnb.fit(X_1, y)
grid_mnb.best_params_
pd.DataFrame(grid_mnb.cv_results_)
mnb_best_param = grid_mnb.best_params_
mnb_best_param
grid_mnb.best_estimator_
scores_mnb = cross_val_score(
grid_mnb.best_estimator_, X_1, y, cv=cv_splitter, n_jobs=-1
)
print(scores_mnb)
print("Mean (mnb): " + str(scores_mnb.mean()))
print("SD (mnb): " + str(scores_mnb.std()))
# ### 6. Sixth Model: K Nearest Neighbor
knn = KNeighborsClassifier()
parameter_grid = {
"n_neighbors": np.arange(9, 19, 2),
"weights": ["uniform", "distance"],
"metric": ["minkowski", "manhattan"],
"leaf_size": np.arange(10, 60, 10),
}
grid_knn = GridSearchCV(knn, parameter_grid, cv=cv_splitter, n_jobs=-1)
grid_knn.fit(X_1, y)
grid_knn.best_params_
results = pd.DataFrame(grid_knn.cv_results_)
results.iloc[:, 4:].sort_values("rank_test_score")
knn_best_param = grid_knn.best_params_
knn_best_param
grid_knn.best_estimator_
scores_knn = cross_val_score(
grid_knn.best_estimator_, X_1, y, cv=cv_splitter, n_jobs=-1
)
print(scores_knn)
print("Mean (knn): " + str(scores_knn.mean()))
print("SD (knn): " + str(scores_knn.std()))
# ### 7. Compare Results of the above 5 Models
cross_val_results = pd.Series(
[scores_logis, scores_random, scores_gradient, scores_xgb, scores_mnb, scores_knn],
index=[
"Logistic",
"Random Forrest",
"Gradirnt Boost",
"XGBoost",
"MN Bayes",
"Knn",
],
)
fig, ax = plt.subplots(figsize=(10, 5))
sns.barplot(x=cross_val_results.index, y=cross_val_results.apply(np.mean))
ax.set_ylim(0.77, 0.85)
ax.set_yticks(np.arange(0.77, 0.85, step=0.01))
fig.suptitle("Mean of Cross-Validation Scores")
fig, ax = plt.subplots(figsize=(10, 5))
sns.barplot(x=cross_val_results.index, y=cross_val_results.apply(np.std))
ax.set_ylim(0.01, 0.06)
ax.set_yticks(np.arange(0.01, 0.06, step=0.005))
fig.suptitle("Standard Deviation of Cross-Validation Scores")
# ### 8. Seventh Model: Stackiing
# #### (1) In stacking, kFold cross-validated predictions of 1st-level models are used as input (where the 1st-level models become the new features) for training by a 2nd-level model.
# #### (2) Cross-validated 1st-level models are also used to predict (not train) on the test data set and the outcome (with 1st-level models as the new features) are used as input to the final prediction by the 2nd-level model.
# #### (3) Some discussions and code can be found at https://www.kaggle.com/arthurtok/introduction-to-ensembling-stacking-in-python, https://www.kdnuggets.com/2017/02/stacking-models-imropved-predictions.html.
# #### 1. First test data need to be processed and encoded in the same way as train data (Cross-validated 1st-level models will predict (not train) on the data and the outcome becomes input to the final prediction of the 2nd-level model).
data_test = pd.read_csv("../input/titanic/test.csv")
passenger_id = data_test["PassengerId"]
num_row_test = data_test.shape[0]
data_test.head()
data_test.info()
data_transform(data_test)
data_test.head()
X_test = ohe.transform(data_test).toarray()
list(X_test)[:5]
# #### 2. Define 1st-level models (using the optimal hyperparameters previously)
logis = LogisticRegression(solver="liblinear", random_state=42)
logis.set_params(**logis_best_param)
rf = RandomForestClassifier(random_state=42)
rf.set_params(**random_forest_best_param)
gb = GradientBoostingClassifier(random_state=42)
gb.set_params(**gradient_best_param)
xgb = XGBClassifier(learning_rate=0.01, n_jobs=-1, random_state=42)
xgb.set_params(**xgb_best_param)
mnb = MultinomialNB()
mnb.set_params(**mnb_best_param)
knn = KNeighborsClassifier()
knn.set_params(**knn_best_param)
# #### 3. Prepare 2nd-level training and test data
folds = 5
skf = StratifiedKFold(n_splits=folds, random_state=42)
def get_oof(clf_, X_1_, y_, X_test_, folds_):
"""Obtain out-of-fold predictions of a model on train and test data sets.
Parameters:
----------
clf_: sklearn classifier
a 1st-level model
X_1_: numpy array
train data with shape (891, 15)
y_: pandas series
train data labels with a shape (891,)
X_test_: numpy array
test data with shape (418, 15)
folds: int
number of folds in stratified K-fold cross validator
Returns:
--------
X_train_oof: numpy array
2nd-level train data: out-of-fold predictions of clf_ with shape (891,)
X_test_oof: numpy array
2nd-level test data: mean of out-of-fold predictions of clf_ on test data with
shape (418,)
"""
X_train_oof = np.zeros((X_1_.shape[0],)) # 2nd-level train data with shape (891,)
X_test_oof = np.zeros((X_test_.shape[0],)) # 2nd-level test data with shape (418,)
X_test_oof_folds = np.zeros((folds_, X_test_.shape[0]))
# with shape (5, 418), a temporary array holding out-of-fold predictions of clf_ on test data
for i, (train_index, valid_index) in enumerate(skf.split(X_1_, y_)):
# i: out-of-fold group index (e.g. 0)
# train_index: numpy array holding all train X_1_ and y_ row indices (e.g. from 179-890)
# valid_index: numpy array holding all valid X_1_ and y_ row indices (e.g. from 1-178)
X_train_folds = X_1_[train_index] # select data for train folds
y_train_folds = y_[train_index] # select labels for train folds
clf_.fit(X_train_folds, y_train_folds) # train clf_ on train folds
X_train_valid_fold = X_1_[
valid_index
] # select data for valid (out-of-fold) fold
X_train_oof[valid_index] = clf_.predict(X_train_valid_fold)
# clf_ predicts on valid fold and save to 2nd-level train data
X_test_oof_folds[i, :] = clf_.predict(X_test_)
# clf_ predicts on the entire set of test data and save the results to the i-th row in the
# temporary array X_test_oof_folds
X_test_oof = X_test_oof_folds.mean(axis=0)
# calculate the mean of out-of-fold predcitons by collapsing in the 0-th axid (with 5 rows)
return X_train_oof, X_test_oof
# construct 2nd-level train and test data
clfs = [logis, rf, gb, xgb, mnb, knn]
X_train_oof_final = np.zeros((X_1.shape[0], len(clfs))) # with shape (891, 5)
X_test_oof_final = np.zeros((X_test.shape[0], len(clfs))) # with shape (418, 5)
for i, clf in enumerate(clfs):
clf_train_off, clf_test_off = get_oof(clf, X_1, y, X_test, folds)
X_train_oof_final[:, i] = clf_train_off
X_test_oof_final[:, i] = clf_test_off
X_train_oof_final.shape
X_train_oof_final[:, 0]
X_test_oof_final[:, 0]
# #### 4. Perform the final level 2 modeling using random forest
# set max_features normal distribution sample array
num_feature = X_train_oof_final.shape[1] # 6 features
max_feature = norm.rvs(np.sqrt(num_feature), 2, size=200, random_state=42).astype(int)
max_feature[max_feature <= 0] = 1
max_feature[max_feature > num_feature] = num_feature
max_feature
# set min_samples_split normal distribution sample array
min_sample_split = norm.rvs(4, 2, size=200, random_state=42).astype(int)
min_sample_split[min_sample_split <= 1] = 2
min_sample_split
# Use the 2 previously defined normal distribution sample arrays 'max_feature' and
# 'min_sample_split'
rf = RandomForestClassifier(random_state=42)
parameter_grid = {
"n_estimators": np.arange(50, 800, step=5),
"max_features": max_feature,
"min_samples_split": min_sample_split,
"min_samples_leaf": np.arange(1, 5, 1),
"bootstrap": [True, False],
}
grid_random_stack = RandomizedSearchCV(
rf,
parameter_grid,
n_iter=100,
cv=cv_splitter,
random_state=42,
refit=True,
n_jobs=-1,
)
grid_random_stack.fit(X_train_oof_final, y)
random_forest_stack_best_param = grid_random_stack.best_params_
random_forest_stack_best_param
grid_random_stack.n_splits_
grid_random_stack.best_estimator_
x_clfs = ["Logistic", "Random Forrest", "Gradirnt Boost", "XGBoost", "MN Bayes", "Knn"]
grid_random_stack.best_estimator_.feature_importances_
fig, ax = plt.subplots(figsize=(10, 4))
ax.bar(x_clfs, grid_random_stack.best_estimator_.feature_importances_)
ax.grid
fig.suptitle("Feature Importance in Stack Model")
scores_random_stack = cross_val_score(
grid_random_stack.best_estimator_, X_train_oof_final, y, cv=cv_splitter, n_jobs=-1
)
print(scores_random_stack)
print("Mean (random): " + str(scores_random_stack.mean()))
print("SD (random): " + str(scores_random_stack.std()))
# ### 9. Compare results of all 7 models
cross_val_results_all = pd.Series(
[
scores_logis,
scores_random,
scores_gradient,
scores_xgb,
scores_mnb,
scores_knn,
scores_random_stack,
],
index=[
"Logistic",
"Random Forrest",
"Gradirnt Boost",
"XGBoost",
"MN Bayes",
"Knn",
"Stack",
],
)
fig, ax = plt.subplots(figsize=(11, 5))
sns.barplot(x=cross_val_results_all.index, y=cross_val_results_all.apply(np.mean))
ax.set_ylim(0.77, 0.85)
ax.set_yticks(np.arange(0.77, 0.85, step=0.01))
fig.suptitle("Mean of Cross-Validation Scores")
fig, ax = plt.subplots(figsize=(11, 5))
sns.barplot(x=cross_val_results_all.index, y=cross_val_results_all.apply(np.std))
ax.set_ylim(0.01, 0.06)
ax.set_yticks(np.arange(0.01, 0.06, step=0.005))
fig.suptitle("Standard Deviation of Cross-Validation Scores")
# ## Model Predictions
# ### 1. Logistic regression
y_test_predict_logis = grid_logis.predict(X_test)
submission_logis_2 = pd.DataFrame(
{"PassengerId": passenger_id, "Survived": y_test_predict_logis}
)
existing_file = glob.glob("submission_logis_2.csv")
assert not existing_file, "File already existed."
submission_logis_2.to_csv("submission_logis_2.csv", index=False)
# (This submission got a public score of 0.794)
# ### 2. Random forrest
y_test_predict_random = grid_random.predict(X_test)
submission_random_2 = pd.DataFrame(
{"PassengerId": passenger_id, "Survived": y_test_predict_random}
)
existing_file = glob.glob("submission_random_2.csv")
assert not existing_file, "File already existed."
submission_random_2.to_csv("submission_random_2.csv", index=False)
# (This submission got a public score of 0.799)
# ### 3. Gradient boosting
y_test_predict_gradient = grid_gradient.predict(X_test)
submission_gradient_2 = pd.DataFrame(
{"PassengerId": passenger_id, "Survived": y_test_predict_gradient}
)
existing_file = glob.glob("submission_gradient_2.csv")
assert not existing_file, "File already existed."
submission_gradient_2.to_csv("submission_gradient_2.csv", index=False)
# (This submission got a public score of 0.794)
# ### 4. XGboost
y_test_predict_xgb = grid_xgb.predict(X_test)
submission_xgb_2 = pd.DataFrame(
{"PassengerId": passenger_id, "Survived": y_test_predict_xgb}
)
existing_file = glob.glob("submission_xgb_2.csv")
assert not existing_file, "File already existed."
submission_xgb_2.to_csv("submission_xgb_2.csv", index=False)
# (This submission got a public score of 0.794)
# ### 5. Multinomial Naive Bayes
y_test_predict_mnb = grid_mnb.predict(X_test)
submission_mnb_2 = pd.DataFrame(
{"PassengerId": passenger_id, "Survived": y_test_predict_mnb}
)
existing_file = glob.glob("submission_mnb_2.csv")
assert not existing_file, "File already existed."
submission_mnb_2.to_csv("submission_mnb_2.csv", index=False)
# (This submission got a public score of 0.746)
# ### 6. K Neareat Neighbors
y_test_predict_knn = grid_knn.predict(X_test)
submission_knn_2 = pd.DataFrame(
{"PassengerId": passenger_id, "Survived": y_test_predict_knn}
)
existing_file = glob.glob("submission_knn_2.csv")
assert not existing_file, "File already existed."
submission_knn_2.to_csv("submission_knn_2.csv", index=False)
# (This submission got a public score of 0.789)
# ### 7. Stack
y_test_predict_random_stack = grid_random_stack.predict(X_test_oof_final)
submission_stack_2 = pd.DataFrame(
{"PassengerId": passenger_id, "Survived": y_test_predict_random_stack}
)
existing_file = glob.glob("submission_stack_2.csv")
assert not existing_file, "File already existed."
submission_stack_2.to_csv("submission_stack_2.csv", index=False)
# (This submission got a public score of 0.799)
# ### 8. Finally perform a majority vote using all 7 model predictions
predict_array = np.array(
[
y_test_predict_logis,
y_test_predict_random,
y_test_predict_gradient,
y_test_predict_xgb,
y_test_predict_mnb,
y_test_predict_knn,
y_test_predict_random_stack,
]
)
vote_df = pd.DataFrame(
predict_array,
index=[
"Logistic",
"Random Forrest",
"Gradirnt Boost",
"XGBoost",
"MN Bayes",
"Knn",
"Stack",
],
)
y_test_predict_vote = np.array(vote_df.mode(axis=0))[0]
submission_vote_2 = pd.DataFrame(
{"PassengerId": passenger_id, "Survived": y_test_predict_vote}
)
existing_file = glob.glob("submission_vote_2.csv")
assert not existing_file, "File already existed."
submission_vote_2.to_csv("submission_vote_2.csv", index=False)
# (This submission got a public score of 0.799)
# ### 9. Public score comparison
fig, ax = plt.subplots(figsize=(13, 5))
ax.set_ylim(0.735, 0.805)
ax.set_yticks(np.arange(0.735, 0.805, step=0.01))
sns.barplot(
x=[
"Logistic",
"Random Forrest",
"Gradirnt Boost",
"XGBoost",
"MN Bayes",
"Knn",
"Stack",
"Vote",
],
y=[0.794, 0.799, 0.794, 0.794, 0.746, 0.789, 0.799, 0.799],
)
|
# # 2020 Democratic Primary Endorsements
# **Remark** I have no domain knoledge, thus let me know if I am missing something useful or misusing/misunderstanding the data.
# * [Data Cleaning](#cleaning)
# * [Endorsee Analysis](#endorsee)
# - [nCoV in the World](#world)
# - [Confirmed/Deaths/Recovered over Time](#scatter)
# * [Tweets Analysis](#tweets)
# - [Sentiment Distribution](#sentiment)
# - [WordCloud](#wordcloud)
# - [Hashtags](#hashtags)
# # Data Cleaning
# Load libraries and dataframe
import pandas as pd
pd.set_option("display.max_columns", 100)
pd.set_option("display.max_rows", 100)
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import scipy.stats as stats
import plotly.express as px
import plotly.graph_objs as go
from plotly.offline import iplot, init_notebook_mode
import plotly.figure_factory as ff
from plotly import subplots
from plotly.subplots import make_subplots
init_notebook_mode(connected=True)
from datetime import date, datetime, timedelta
import time, re, os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
def resumetable(df):
print(f"Dataset Shape: {df.shape}")
summary = pd.DataFrame(df.dtypes, columns=["dtypes"])
summary = summary.reset_index()
summary["Name"] = summary["index"]
summary = summary[["Name", "dtypes"]]
summary["Missing"] = df.isnull().sum().values
summary["Uniques"] = df.nunique().values
return summary
df = pd.read_csv(
"/kaggle/input/2020-democratic-primary-endorsements/endorsements-2020.csv"
)
df.head(10)
# Let's rename *endorser party* column which is spaced and see some initial statistics.
df.columns = [c.replace(" ", "_") for c in list(df)]
resumetable(df)
# We can already see that each *Endorser* is unique in the dataframe, however most of them (~75%) do not have an *Endorsee*.
# Other columns with a high number of missing values are *city*, *body*, *order*, *district*, *date* and *source*.
percent_missing = np.round(df.isnull().sum() * 100 / len(df), 2)
missing_value_df = pd.DataFrame(
{"column_name": df.columns, "percent_missing": percent_missing}
).sort_values("percent_missing", ascending=False)
fig = go.Figure()
fig.add_trace(
go.Bar(
x=missing_value_df["column_name"],
y=missing_value_df["percent_missing"],
opacity=0.9,
text=missing_value_df["percent_missing"],
textposition="inside",
marker={"color": "indianred"},
)
)
fig.update_layout(
title={
"text": "Percentage Missing by Column",
"y": 0.95,
"x": 0.5,
"xanchor": "center",
"yanchor": "top",
},
showlegend=False,
xaxis_title_text="Columns",
yaxis_title_text="Percentage",
bargap=0.1,
)
fig.show()
# Let's drop the *city*, *body*, *order* and *district* column due to their very high number of missing values - which I don't know how to fill.
# Furthermore, we can see that the triplets *date*, *endorsee* and *source* are either all missing or all populated, in fact:[](http://)
df.drop(["city", "body", "order", "district"], axis=1, inplace=True)
((df[["source", "date", "endorsee"]].isnull()).astype(int).sum(axis=1)).value_counts()
# Let's do some preprocess for the *source* and *endorsee* columns
df.rename(columns={"source": "raw_source"}, inplace=True)
df["raw_source"] = df.loc[:, "raw_source"].fillna("other")
df["source"] = "other"
keys = ["twitter", "politico", "youtube", "4president", "cnn", "apnews"]
for k in keys:
df["source"] = np.where(df["raw_source"].str.contains(k), k, df["source"])
df.drop("raw_source", axis=1, inplace=True)
df["endorsee"] = df.loc[:, "endorsee"].fillna("no_endorsee")
df["endorser_party"] = df.loc[:, "endorser_party"].fillna("None")
resumetable(df)
#
# # Endorsee Anlysis
# Let's start to analyze and confront endorsees.
endorsee_df = df[df["endorsee"] != "no_endorsee"]
endorsee_df["endorsee"] = (
endorsee_df["endorsee"].str.split(" ").apply(lambda r: r[-1])
) # .apply(lambda r: r[0][0] + '. ' + r[-1])
endorsee_df.head(10)
endorsee_df.endorser_party.unique()
end_df = endorsee_df.groupby("endorsee").agg({"endorser": "count", "points": "sum"})
end_df.rename(
columns={"endorser": "n_endorsements", "points": "tot_points"}, inplace=True
)
end_df["points_endorser_ratio"] = np.round(
np.divide(end_df["tot_points"].to_numpy(), end_df["n_endorsements"].to_numpy()), 2
)
end_df.reset_index(inplace=True)
end_df
# Let's first check the
import plotly.express as px
fig = px.scatter(
end_df,
x="n_endorsements",
y="tot_points",
size=end_df["tot_points"] * 5,
color="points_endorser_ratio",
text="endorsee",
opacity=0.7,
log_x=True,
log_y=True,
)
fig.update_traces(textposition="top center", textfont={"size": 10})
fig.update_layout(
title={
"text": "Total Points per Number of Endorsers",
"y": 0.95,
"x": 0.5,
"xanchor": "center",
"yanchor": "top",
},
showlegend=False,
xaxis_title_text="Number of Endorsers",
yaxis_title_text="Total Points",
)
fig.show()
# Add Bubble chart over time!
endorsee_df[endorsee_df["endorsee"] == "Bloomberg"]
test = endorsee_df[endorsee_df["endorsee"] == "Bloomberg"]
fig = make_subplots(
rows=2, cols=2, specs=[[{}, {}], [{}, {}]], subplot_titles=("11", "12", "21", "22")
)
fig.add_trace(
go.Histogram(
x=test["position"],
# y=missing_value_df['percent_missing'],
opacity=0.9,
# text=missing_value_df['percent_missing'],
# textposition='inside',
marker={"color": "indianred"},
),
row=1,
col=1,
)
fig.add_trace(
go.Histogram(
x=test["endorser_party"],
# y=missing_value_df['percent_missing'],
opacity=0.9,
# text=missing_value_df['percent_missing'],
# textposition='inside',
marker={"color": "indianred"},
),
row=1,
col=2,
)
fig.add_trace(
go.Histogram(
x=test["source"],
# y=missing_value_df['percent_missing'],
opacity=0.9,
# text=missing_value_df['percent_missing'],
# textposition='inside',
marker={"color": "indianred"},
),
row=2,
col=1,
)
fig.add_trace(
go.Histogram(
x=test["category"],
# y=missing_value_df['percent_missing'],
opacity=0.9,
# text=missing_value_df['percent_missing'],
# textposition='inside',
marker={"color": "indianred"},
),
row=2,
col=2,
)
fig.update_layout(
title={
"text": "Endorsee Summary",
"y": 0.95,
"x": 0.5,
"xanchor": "center",
"yanchor": "top",
},
showlegend=False,
# xaxis_title_text='Columns',
# yaxis_title_text='Percentage',
bargap=0.1,
)
fig.show()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
import torch
import torch.nn.functional as F
import matplotlib.pyplot as plt
from torch import nn
data = pd.read_csv("/kaggle/input/380000-lyrics-from-metrolyrics/lyrics.csv")
# 10 first rows
data[:10]
data.describe()
# lyrics containing 'love' keyword
# na=False to avoid errors while copping
data = data[data["song"].str.lower().str.contains("love", na=False)].copy()
data[:10]
# no ponctuation
data["lyrics"] = data["lyrics"].replace(to_replace=[",", "."], value=["", ""])
# Drop NaN values
data = data.dropna(subset=["lyrics"])
# 10 random rows
data.sample(n=10)
data.describe()
# **Neural Network**
# lyrics to text
# lyrics=''
lyrics = " ".join(data.lyrics)
# lyrics=data['lyrics']
len(lyrics)
print("lyrics has " + str(len(lyrics)) + " caracter")
# **Tokenization**
# In the cells, below, I'm creating a couple dictionaries to convert the characters to and from integers. Encoding the characters as integers makes it easier to use as input in the network.
# encode the text and map each character to an integer and vice versa
# we create two dictionaries:
# 1. int2char, which maps integers to characters
# 2. char2int, which maps characters to unique integers
chars = tuple(set(lyrics))
int2char = dict(enumerate(chars))
char2int = {ch: ii for ii, ch in int2char.items()}
# encode the text
encoded = np.array([char2int[ch] for ch in lyrics])
# And we can see those same characters from above, encoded as integers
encoded[:100]
# Pre-processing the data
# As you can see in our char-RNN image above, our LSTM expects an input that is one-hot encoded meaning that each character is converted into an integer (via our created dictionary) and then converted into a column vector where only it's corresponding integer index will have the value of 1 and the rest of the vector will be filled with 0's. Since we're one-hot encoding the data, let's make a function to do that!
def one_hot_encode(arr, n_labels):
# Initialize the encoded array
one_hot = np.zeros((arr.size, n_labels), dtype=np.float32)
# Fill the appropriate elements with ones
one_hot[np.arange(one_hot.shape[0]), arr.flatten()] = 1.0
# Finally reshape it to get back to the original array
one_hot = one_hot.reshape((*arr.shape, n_labels))
return one_hot
# check that the function works as expected
test_seq = np.array([[3, 5, 1]])
one_hot = one_hot_encode(test_seq, 8)
print(one_hot)
# ## Making training mini-batches
# To train on this data, we also want to create mini-batches for training. Remember that we want our batches to be multiple sequences of some desired number of sequence steps. Considering a simple example, our batches would look like this:
# In this example, we'll take the encoded characters (passed in as the `arr` parameter) and split them into multiple sequences, given by `batch_size`. Each of our sequences will be `seq_length` long.
# ### Creating Batches
# **1. The first thing we need to do is discard some of the text so we only have completely full mini-batches. **
# Each batch contains $N \times M$ characters, where $N$ is the batch size (the number of sequences in a batch) and $M$ is the seq_length or number of time steps in a sequence. Then, to get the total number of batches, $K$, that we can make from the array `arr`, you divide the length of `arr` by the number of characters per batch. Once you know the number of batches, you can get the total number of characters to keep from `arr`, $N * M * K$.
# **2. After that, we need to split `arr` into $N$ batches. **
# You can do this using `arr.reshape(size)` where `size` is a tuple containing the dimensions sizes of the reshaped array. We know we want $N$ sequences in a batch, so let's make that the size of the first dimension. For the second dimension, you can use `-1` as a placeholder in the size, it'll fill up the array with the appropriate data for you. After this, you should have an array that is $N \times (M * K)$.
# **3. Now that we have this array, we can iterate through it to get our mini-batches. **
# The idea is each batch is a $N \times M$ window on the $N \times (M * K)$ array. For each subsequent batch, the window moves over by `seq_length`. We also want to create both the input and target arrays. Remember that the targets are just the inputs shifted over by one character. The way I like to do this window is use `range` to take steps of size `n_steps` from $0$ to `arr.shape[1]`, the total number of tokens in each sequence. That way, the integers you get from `range` always point to the start of a batch, and each window is `seq_length` wide.
# > **TODO:** Write the code for creating batches in the function below. The exercises in this notebook _will not be easy_. I've provided a notebook with solutions alongside this notebook. If you get stuck, checkout the solutions. The most important thing is that you don't copy and paste the code into here, **type out the solution code yourself.**
def get_batches(arr, batch_size, seq_length):
"""Create a generator that returns batches of size
batch_size x seq_length from arr.
Arguments
---------
arr: Array you want to make batches from
batch_size: Batch size, the number of sequences per batch
seq_length: Number of encoded chars in a sequence
"""
batch_size_total = batch_size * seq_length
# total number of batches we can make
n_batches = len(arr) // batch_size_total
# Keep only enough characters to make full batches
arr = arr[: n_batches * batch_size_total]
# Reshape into batch_size rows
arr = arr.reshape((batch_size, -1))
# iterate through the array, one sequence at a time
for n in range(0, arr.shape[1], seq_length):
# The features
x = arr[:, n : n + seq_length]
# The targets, shifted by one
y = np.zeros_like(x)
try:
y[:, :-1], y[:, -1] = x[:, 1:], arr[:, n + seq_length]
except IndexError:
y[:, :-1], y[:, -1] = x[:, 1:], arr[:, 0]
yield x, y
# ### Test Your Implementation
# Now I'll make some data sets and we can check out what's going on as we batch data. Here, as an example, I'm going to use a batch size of 8 and 50 sequence steps.
batches = get_batches(encoded, 8, 50)
x, y = next(batches)
# printing out the first 10 items in a sequence
print("x\n", x[:10, :10])
print("\ny\n", y[:10, :10])
# check if GPU is available
train_on_gpu = torch.cuda.is_available()
if train_on_gpu:
print("Training on GPU!")
else:
print("No GPU available, training on CPU; consider making n_epochs very small.")
class CharRNN(nn.Module):
def __init__(self, tokens, n_hidden=256, n_layers=2, drop_prob=0.5, lr=0.001):
super().__init__()
self.drop_prob = drop_prob
self.n_layers = n_layers
self.n_hidden = n_hidden
self.lr = lr
# creating character dictionaries
self.chars = tokens
self.int2char = dict(enumerate(self.chars))
self.char2int = {ch: ii for ii, ch in self.int2char.items()}
## TODO: define the LSTM
self.lstm = nn.LSTM(
len(self.chars), n_hidden, n_layers, dropout=drop_prob, batch_first=True
)
## TODO: define a dropout layer
self.dropout = nn.Dropout(drop_prob)
## TODO: define the final, fully-connected output layer
self.fc = nn.Linear(n_hidden, len(self.chars))
def forward(self, x, hidden):
"""Forward pass through the network.
These inputs are x, and the hidden/cell state `hidden`."""
## TODO: Get the outputs and the new hidden state from the lstm
r_output, hidden = self.lstm(x, hidden)
## TODO: pass through a dropout layer
out = self.dropout(r_output)
# Stack up LSTM outputs using view
# you may need to use contiguous to reshape the output
out = out.contiguous().view(-1, self.n_hidden)
## TODO: put x through the fully-connected layer
out = self.fc(out)
# return the final output and the hidden state
return out, hidden
def init_hidden(self, batch_size):
"""Initializes hidden state"""
# Create two new tensors with sizes n_layers x batch_size x n_hidden,
# initialized to zero, for hidden state and cell state of LSTM
weight = next(self.parameters()).data
if train_on_gpu:
hidden = (
weight.new(self.n_layers, batch_size, self.n_hidden).zero_().cuda(),
weight.new(self.n_layers, batch_size, self.n_hidden).zero_().cuda(),
)
else:
hidden = (
weight.new(self.n_layers, batch_size, self.n_hidden).zero_(),
weight.new(self.n_layers, batch_size, self.n_hidden).zero_(),
)
return hidden
# ## Time to train
# The train function gives us the ability to set the number of epochs, the learning rate, and other parameters.
# Below we're using an Adam optimizer and cross entropy loss since we are looking at character class scores as output. We calculate the loss and perform backpropagation, as usual!
# A couple of details about training:
# >* Within the batch loop, we detach the hidden state from its history; this time setting it equal to a new *tuple* variable because an LSTM has a hidden state that is a tuple of the hidden and cell states.
# * We use [`clip_grad_norm_`](https://pytorch.org/docs/stable/_modules/torch/nn/utils/clip_grad.html) to help prevent exploding gradients.
def train(
net,
data,
epochs=10,
batch_size=10,
seq_length=50,
lr=0.001,
clip=5,
val_frac=0.1,
print_every=10,
):
"""Training a network
Arguments
---------
net: CharRNN network
data: text data to train the network
epochs: Number of epochs to train
batch_size: Number of mini-sequences per mini-batch, aka batch size
seq_length: Number of character steps per mini-batch
lr: learning rate
clip: gradient clipping
val_frac: Fraction of data to hold out for validation
print_every: Number of steps for printing training and validation loss
"""
net.train()
opt = torch.optim.Adam(net.parameters(), lr=lr)
criterion = nn.CrossEntropyLoss()
# create training and validation data
val_idx = int(len(data) * (1 - val_frac))
data, val_data = data[:val_idx], data[val_idx:]
if train_on_gpu:
net.cuda()
counter = 0
n_chars = len(net.chars)
for e in range(epochs):
# initialize hidden state
h = net.init_hidden(batch_size)
for x, y in get_batches(data, batch_size, seq_length):
counter += 1
# One-hot encode our data and make them Torch tensors
x = one_hot_encode(x, n_chars)
inputs, targets = torch.from_numpy(x), torch.from_numpy(y)
if train_on_gpu:
inputs, targets = inputs.cuda(), targets.cuda()
# Creating new variables for the hidden state, otherwise
# we'd backprop through the entire training history
h = tuple([each.data for each in h])
# zero accumulated gradients
net.zero_grad()
# get the output from the model
output, h = net(inputs, h)
# calculate the loss and perform backprop
loss = criterion(output, targets.view(batch_size * seq_length).long())
loss.backward()
# `clip_grad_norm` helps prevent the exploding gradient problem in RNNs / LSTMs.
nn.utils.clip_grad_norm_(net.parameters(), clip)
opt.step()
# loss stats
if counter % print_every == 0:
# Get validation loss
val_h = net.init_hidden(batch_size)
val_losses = []
net.eval()
for x, y in get_batches(val_data, batch_size, seq_length):
# One-hot encode our data and make them Torch tensors
x = one_hot_encode(x, n_chars)
x, y = torch.from_numpy(x), torch.from_numpy(y)
# Creating new variables for the hidden state, otherwise
# we'd backprop through the entire training history
val_h = tuple([each.data for each in val_h])
inputs, targets = x, y
if train_on_gpu:
inputs, targets = inputs.cuda(), targets.cuda()
output, val_h = net(inputs, val_h)
val_loss = criterion(
output, targets.view(batch_size * seq_length).long()
)
val_losses.append(val_loss.item())
net.train() # reset to train mode after iterationg through validation data
print(
"Epoch: {}/{}...".format(e + 1, epochs),
"Step: {}...".format(counter),
"Loss: {:.4f}...".format(loss.item()),
"Val Loss: {:.4f}".format(np.mean(val_losses)),
)
# Instantiating the model
# Now we can actually train the network. First we'll create the network itself, with some given hyperparameters. Then, define the mini-batches sizes, and start training!
# define and print the net
n_hidden = 512
n_layers = 2
net = CharRNN(chars, n_hidden, n_layers)
print(net)
batch_size = 128
seq_length = 100
n_epochs = 20 # start smaller if you are just testing initial behavior
# train the model
train(
net,
encoded,
epochs=n_epochs,
batch_size=batch_size,
seq_length=seq_length,
lr=0.001,
print_every=10,
)
# change the name, for saving multiple files
model_name = "rnn_20_epoch.net"
checkpoint = {
"n_hidden": net.n_hidden,
"n_layers": net.n_layers,
"state_dict": net.state_dict(),
"tokens": net.chars,
}
with open(model_name, "wb") as f:
torch.save(checkpoint, f)
# ---
# ## Making Predictions
# Now that the model is trained, we'll want to sample from it and make predictions about next characters! To sample, we pass in a character and have the network predict the next character. Then we take that character, pass it back in, and get another predicted character. Just keep doing this and you'll generate a bunch of text!
# ### A note on the `predict` function
# The output of our RNN is from a fully-connected layer and it outputs a **distribution of next-character scores**.
# > To actually get the next character, we apply a softmax function, which gives us a *probability* distribution that we can then sample to predict the next character.
# ### Top K sampling
# Our predictions come from a categorical probability distribution over all the possible characters. We can make the sample text and make it more reasonable to handle (with less variables) by only considering some $K$ most probable characters. This will prevent the network from giving us completely absurd characters while allowing it to introduce some noise and randomness into the sampled text. Read more about [topk, here](https://pytorch.org/docs/stable/torch.html#torch.topk).
#
def predict(net, char, h=None, top_k=None):
"""Given a character, predict the next character.
Returns the predicted character and the hidden state.
"""
# tensor inputs
x = np.array([[net.char2int[char]]])
x = one_hot_encode(x, len(net.chars))
inputs = torch.from_numpy(x)
if train_on_gpu:
inputs = inputs.cuda()
# detach hidden state from history
h = tuple([each.data for each in h])
# get the output of the model
out, h = net(inputs, h)
# get the character probabilities
p = F.softmax(out, dim=1).data
if train_on_gpu:
p = p.cpu() # move to cpu
# get top characters
if top_k is None:
top_ch = np.arange(len(net.chars))
else:
p, top_ch = p.topk(top_k)
top_ch = top_ch.numpy().squeeze()
# select the likely next character with some element of randomness
p = p.numpy().squeeze()
char = np.random.choice(top_ch, p=p / p.sum())
# return the encoded value of the predicted char and the hidden state
return net.int2char[char], h
# ### Priming and generating text
# Typically you'll want to prime the network so you can build up a hidden state. Otherwise the network will start out generating characters at random. In general the first bunch of characters will be a little rough since it hasn't built up a long history of characters to predict from.
def sample(net, size, prime="The", top_k=None):
if train_on_gpu:
net.cuda()
else:
net.cpu()
net.eval() # eval mode
# First off, run through the prime characters
chars = [ch for ch in prime]
h = net.init_hidden(1)
for ch in prime:
char, h = predict(net, ch, h, top_k=top_k)
chars.append(char)
# Now pass in the previous character and get a new one
for ii in range(size):
char, h = predict(net, chars[-1], h, top_k=top_k)
chars.append(char)
return "".join(chars)
print(sample(net, 1000, prime="sunset", top_k=5))
|
# **Objective**
# This Data Science for Good Competition intends to use remote sensing techniques to understand Environmental Emissions. Since the whole concept of Satellite Imagery and can be a little overwhelming, this is just an introductory kernel, where I try to explain the various terms and datasets related to satellite Imagery.
# **Problem Statement: Measuring Emissions factors from Satellite Data**
# Air Quality Management is an important area and influences a lot of decisions taken by countries. But how does one ascertain the Air quality of a place? This is done by calculating the Emissions Factor of that area.
# **What is the Emission factor? **
# An emission intensity is the emission rate of a given pollutant relative to the intensity of a specific activity, or an industrial production process; for example grams of carbon dioxide released per megajoule of energy produced, or the ratio of greenhouse gas emissions produced to gross domestic product. **
# * **What is Remote Sensing??**
# Remote sensing is the acquisition of information about an object or phenomenon without making physical contact with the object and thus in contrast to on-site observation, especially the Earth.
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
# **Importing necessary libraries**
## Importing necessary libraries
import numpy as np
import pandas as pd
from skimage.io import imread
import tifffile as tiff
import matplotlib.pyplot as plt
import seaborn as sns
sns.set(style="white", palette="muted", color_codes=True)
from ast import literal_eval
# Analysing datetime
import datetime as dt
from datetime import datetime
# Plotting geographical data
import ee
import folium
from folium import plugins
import rasterio as rio
# File system manangement
import os, random
# **Data Loading**
data = pd.read_csv(
"/kaggle/input/ds4g-environmental-insights-explorer/eie_data/gppd/gppd_120_pr.csv"
)
# **Analyize The Data**
data.columns
data.head(3)
data.shape
data.info()
# Columns with only 0 or NaN values
to_drop = [
"generation_gwh_2013",
"generation_gwh_2014",
"generation_gwh_2015",
"generation_gwh_2016",
"generation_gwh_2017",
"other_fuel1",
"other_fuel2",
"other_fuel3",
"year_of_capacity_data",
]
gpp = data.drop(to_drop, axis=1)
gpp.head(3)
# Columns with all same values
to_drop = ["country", "country_long", "geolocation_source"]
globalPowerPlant = data.drop(to_drop, axis=1)
globalPowerPlant.head(3)
fig, ax = plt.subplots(1, 2, figsize=(10, 5))
sns.countplot(x="primary_fuel", data=globalPowerPlant, ax=ax[0])
sns.catplot(
x="primary_fuel", y="capacity_mw", data=globalPowerPlant, jitter=False, ax=ax[1]
)
plt.close(2) # catplot is a figure-level function, close empty plt
plt.tight_layout()
sns.barplot(
x="primary_fuel",
y="capacity_mw",
data=globalPowerPlant.groupby(["primary_fuel"]).sum().reset_index(),
)
sns.barplot(
x="primary_fuel",
y="estimated_generation_gwh",
data=globalPowerPlant.groupby(["primary_fuel"]).sum().reset_index(),
)
data_path = "/kaggle/input/ds4g-environmental-insights-explorer"
image = "/eie_data/s5p_no2/s5p_no2_20190629T174803_20190705T194117.tif"
data = tiff.imread(data_path + image)
print("Data shape:", data.shape)
print(
"{:.0f}% of the dataset is null".format(
np.isnan(data[:, :, 0]).sum() / np.multiply(*data[:, :, 0].shape) * 100
)
)
print(
"Last measurement at y index {}".format(
np.argwhere(np.isnan(data[0, :, 0])).min() - 1
)
)
# **Detail Analysis**
# NO2
titles = [
"NO2_column_number_density",
"tropospheric_NO2_column_number_density",
"stratospheric_NO2_column_number_density",
"NO2_slant_column_number_density",
]
f = plt.figure()
f.set_size_inches(8, 8)
for i in range(4):
plt.subplot(2, 2, i + 1)
sns.heatmap(data[:, :, i], cbar=False)
f
ax = plt.gca()
ax.get_xaxis().set_visible(False)
ax.get_yaxis().set_visible(False)
plt.title(titles[i], fontsize=10)
# https://seaborn.pydata.org/generated/seaborn.heatmap.html
# **For Wheather**
titles = ["tropopause_pressure", "absorbing_aerosol_index", "cloud_fraction"]
f = plt.figure()
f.set_size_inches(12, 4)
for i in range(3):
plt.subplot(1, 3, i + 1)
sns.heatmap(data[:, :, 4 + i], cbar=False)
f
ax = plt.gca()
ax.get_xaxis().set_visible(False)
ax.get_yaxis().set_visible(False)
plt.title(titles[i], fontsize=16)
# **Satellite Information**
titles = [
"sensor_altitude",
"sensor_azimuth_angle",
"sensor_zenith_angle",
"solar_azimuth_angle",
"solar_zenith_angle",
]
f = plt.figure()
f.set_size_inches(12, 8)
for i in range(5):
plt.subplot(2, 3, i + 1)
sns.heatmap(data[:, :, 7 + i], cbar=False)
f
ax = plt.gca()
ax.get_xaxis().set_visible(False)
ax.get_yaxis().set_visible(False)
plt.title(titles[i], fontsize=16)
for i in range(5):
print("{}: {:.2f}".format(titles[i], np.nanmean(data[:, :, i + 7])))
|
# 1. 导入函数&载入数据集:
import operator
from functools import reduce
import jieba
import gensim
import json
import numpy as np
# from keras.layers import Input
from keras.models import Model, Sequential
from keras.layers import (
InputLayer,
Embedding,
LSTM,
Dense,
TimeDistributed,
SimpleRNN,
Input,
)
from keras.optimizers import SGD, Adam, Adadelta, RMSprop
with open("../input/input11/minipaperoutq1.txt", "r") as lyrics:
raw_text_w2v = lyrics.readlines()
# print(load_file)
# songs = json.load(load_file)
raw_text_w2v = [i.strip("\n") for i in raw_text_w2v]
lyrics = jieba.lcut_for_search(str(raw_text_w2v))
# print(raw_text_w2v)
# raw_text_w2v = ' '.join(raw_text_w2v)#用空格分开
"""
batch_size = 64
epochs = 9
latent_dim = 256
embedding_size = 128
file_name = '../input/poetry.txt'
"""
# 数据初始化——去除文本中的乱码并把文本分解成单独的字:
raw_text = []
raw_text_w2v = []
for lyric in lyrics:
if "、" in lyric:
continue
if "□" in lyric:
continue
# lyric = lyric + '\n'
raw_text.extend([word for word in lyric])
raw_text_w2v.append([word for word in lyric])
# print(l[:1])
"""
l=raw_text_w2v
word=reduce(operator.add,reduce(operator.add,l))
word=word.replace(',',',')
word=word.split(',')
#word=l[0]
print(word[:10])
"""
"""
for i in range(len(word)+1):
if i == len(word)-1:
break
word[i]=word[i]+','+word[i+1]
print(word[:5])
"""
# print(word[:3])
# print(type(raw_text_w2v[1]))
# for line in raw_text_w2v:
# print(line)
"""
input_texts = []
target_texts = []
input_vocab = set()
target_vocab = set()
#with open(file_name, 'r', encoding='utf-8') as f:
# lines = f.readlines()
for line in word:
# 将诗句用逗号分开
line_sp = line.strip().split(',')
# 如果诗中不含逗号,这句诗我们就不用了
if len(line_sp) < 2:
continue
# 上句为input_text,下句为target_text
input_text, target_text = line_sp[0], line_sp[1]
# 在下句前后开始字符和结束字符
target_text = '\t' + target_text[:-1] + '\n'
input_texts.append(input_text)
target_texts.append(target_text)
# 统计输入侧的词汇表和输出侧的词汇表
for ch in input_text:
if ch not in input_vocab:
input_vocab.add(ch)
for ch in target_text:
if ch not in target_vocab:
target_vocab.add(ch)
# 建立字典和反向字典
input_vocab = dict([(char, i) for i, char in enumerate(input_vocab)])
target_vocab = dict([(char, i) for i, char in enumerate(target_vocab)])
reverse_input_char_index = dict((i, char) for char, i in input_vocab.items())
reverse_target_char_index = dict((i, char) for char, i in target_vocab.items())
# 输入侧词汇表大小
encoder_vocab_size = len(input_vocab)
# 最长输入句子长度
encoder_len = max([len(sentence) for sentence in input_texts])
# 输出侧词汇表大小
decoder_vocab_size = len(target_vocab)
# 最长输出句子长度
decoder_len = max([len(sentence) for sentence in target_texts])
print(encoder_vocab_size)
print(encoder_len)
print(decoder_vocab_size)
print(decoder_len)
print(input_vocab)
print(input_texts[:100])
"""
# print(raw_text_w2v[0:100])
# 将拆分完的数据送给word2vec模型训练词向量:
modelx = gensim.models.word2vec.Word2Vec(raw_text_w2v, size=300, min_count=1)
modelx.save("w2v.model")
modelx = gensim.models.Word2Vec.load("./" + "w2v.model")
all_word_vector = modelx[modelx.wv.vocab]
all_word_vector = np.append(all_word_vector, [np.zeros(300)], axis=0)
print(all_word_vector.shape)
modelx.most_similar("好", topn=10)
vocab_inv = list(modelx.wv.vocab)
vocab = {x: index for index, x in enumerate(vocab_inv)}
# 定义一个函数,制作训练数据——训练数据是一个输入输出对,根据我们对语言模型的理解,输入是 (wi,wi+1,…,wi+length) 的话,输出是 (wi+1,wi+2,…,wi+length+1)
def build_matrix(text, vocab, length, step):
M = []
for word in text:
index = vocab.get(word)
if index is None:
M.append(len(vocab))
else:
M.append(index)
num_sentences = len(M) // length
M = M[: num_sentences * length]
M = np.array(M)
X = []
Y = []
for i in range(0, len(M) - length, step):
X.append(M[i : i + length])
Y.append([[x] for x in M[i + 1 : i + length + 1]])
return np.array(X), np.array(Y)
seq_length = 4
X, Y = build_matrix(raw_text, vocab, seq_length, seq_length)
print("第150个输入矩阵:", X[150])
print("第150个输出矩阵:\n", Y[150])
# 搭建神经网络模型:
model = Sequential()
model.add(InputLayer(input_shape=(None,)))
model.add(
Embedding(
input_dim=len(vocab) + 1,
output_dim=300,
trainable=True,
weights=[all_word_vector],
)
)
# 词嵌入层,input_dim表示词典的大小,每个词对应一个output_dim维的词向量,weights为先前训练好的词向量
model.add(LSTM(units=300, return_sequences=True))
# LSTM层,语言处理层,输出形状为(seq_length,300)
model.add(TimeDistributed(Dense(units=len(vocab) + 1, activation="softmax")))
# 输出的300维向量需要经过一次线性变换(也就是Dense层)转化为len(vocab)+1维的向量,用softmax变换将其转化为概率分布,第i维表示下一个时刻的词是i号单词的概率
model.compile(optimizer=Adam(lr=0.001), loss="sparse_categorical_crossentropy")
# 优化器为Adam,损失函数为交叉熵
model.summary()
"""
encoder_input_data = np.zeros((len(input_texts), encoder_len), dtype='int')
decoder_input_data = np.zeros((len(input_texts), decoder_len), dtype='int')
decoder_target_data = np.zeros((len(input_texts), decoder_len, 1), dtype='int')
for i, (input_text, target_text) in enumerate(zip(input_texts, target_texts)):
for t, char in enumerate(input_text):
encoder_input_data[i, t] = input_vocab[char]
for t, char in enumerate(target_text):
decoder_input_data[i, t] = target_vocab[char]
if t > 0:
decoder_target_data[i, t - 1, 0] = target_vocab[char]
print(encoder_input_data.shape)
print(decoder_input_data.shape)
print(encoder_input_data[:100])
print(decoder_input_data.shape)
print(decoder_target_data.shape)
"""
"""
# 编码器输入层
encoder_inputs = Input(shape=(None,))
# 编码器词嵌入层
encoder_embedding = Embedding(input_dim=encoder_vocab_size, output_dim=embedding_size, trainable=True)(encoder_inputs)
# 编码器长短期记忆网络层
encoder = LSTM(latent_dim, return_state=True)
# 编码器长短期记忆网络输出是一个三元组(encoder_outputs, state_h, state_c)
# encoder_outputs是长短期记忆网络每个时刻的输出构成的序列
# state_h和state_c是长短期记忆网络最后一个时刻的隐状态和细胞状态
encoder_outputs, state_h, state_c = encoder(encoder_embedding)
# 我们会把state_h和state_c作为解码器长短期记忆网络的初始状态,之前我们所说的状态向量的传递就是这样实现的
encoder_states = [state_h, state_c]
# 解码器网络建构
# 解码器输入层
decoder_inputs = Input(shape=(None,))
# 解码器词嵌入层
decoder_embedding = Embedding(input_dim=decoder_vocab_size, output_dim=embedding_size, trainable=True)(decoder_inputs)
# 解码器长短期记忆网络层
decoder_lstm = LSTM(latent_dim, return_sequences=True, return_state=True)
# 解码器长短期记忆网络的输出也是三元组,但我们只关心三元组的第一维,同时我们在这里设置了解码器长短期记忆网络的初始状态
decoder_outputs, _, _ = decoder_lstm(decoder_embedding, initial_state=encoder_states)
# 解码器输出经过一个隐层softmax变换转换为对各类别的概率估计
decoder_dense = Dense(decoder_vocab_size, activation='softmax')
# 解码器输出层
decoder_outputs = decoder_dense(decoder_outputs)
# 总模型,接受编码器和解码器输入,得到解码器长短期记忆网络输出
model = Model([encoder_inputs, decoder_inputs], decoder_outputs)
model.compile(optimizer=Adam(lr=0.001), loss='sparse_categorical_crossentropy')
model.summary()
"""
# 训练神经网络:批次大小:512;训练轮数:40
model.fit(X, Y, batch_size=512, epochs=100, verbose=1)
# model.fit([encoder_input_data, decoder_input_data],decoder_target_data, batch_size=64, epochs=200, validation_split=0.2)
model.save("model.h5")
# 歌词生成:
st = "与你无关,"
print(st, end="")
vocab_inv.append("")
i = 0
while i < 200:
X_sample = np.array([[vocab.get(x, len(vocab)) for x in st]])
out = model.predict(X_sample)
out_2 = out[:, -1:, :]
out_3 = out_2[0][0]
out_4 = (-out_3).argsort()
pdt = out_4[:3]
pb = [out_3[index] for index in pdt]
if vocab[","] in pdt:
ch = ","
else:
ch = vocab_inv[np.random.choice(pdt, p=pb / sum(pb))]
print(ch, end="")
st = st + ch
if vocab[ch] != len(vocab) and ch != "," and ch != "。" and ch != "\n":
i += 1
"""
# 第一个黑盒,编码器,给定encoder_inputs,得到encoder的状态
encoder_model = Model(encoder_inputs, encoder_states)
# 第二个黑盒,解码器
# 解码器接受三个输入,两个是初始状态,一个是之前已经生成的文本
decoder_state_input_h = Input(shape=(latent_dim,))
decoder_state_input_c = Input(shape=(latent_dim,))
decoder_states_inputs = [decoder_state_input_h, decoder_state_input_c]
# 解码器产生三个输出,两个当前状态,一个是每个时刻的输出,其中最后一个时刻的输出可以用来计算下一个字
decoder_outputs, state_h, state_c = decoder_lstm(decoder_embedding, initial_state=decoder_states_inputs)
decoder_states = [state_h, state_c]
decoder_outputs = decoder_dense(decoder_outputs)
decoder_model = Model([decoder_inputs] + decoder_states_inputs, [decoder_outputs] + decoder_states)
"""
"""
def decode_sequence(input_seq):
# 先把上句输入编码器得到编码的中间向量,这个中间向量将是解码器的初始状态向量
states_value = encoder_model.predict(input_seq)
# 初始的解码器输入是开始符'\t'
target_seq = np.zeros((1, 1))
target_seq[0, 0] = target_vocab['\t']
stop_condition = False
decoded_sentence = ''
# 迭代解码
while not stop_condition:
# 把当前的解码器输入和当前的解码器状态向量送进解码器
# 得到对下一个时刻的预测和新的解码器状态向量
output_tokens, h, c = decoder_model.predict([target_seq] + states_value)
# 采样出概率最大的那个字作为下一个时刻的输入
sampled_token_index = np.argmax(output_tokens[0, -1, :])
sampled_char = reverse_target_char_index[sampled_token_index]
decoded_sentence += sampled_char
# 如果采样到了结束符或者生成的句子长度超过了decoder_len,就停止生成
#if (sampled_char == '\n' ):
# stop_condition = True
# 否则我们更新下一个时刻的解码器输入和解码器状态向量
target_seq = np.zeros((1, 1))
target_seq[0, 0] = sampled_token_index
states_value = [h, c]
return decoded_sentence
"""
"""
for seq_index in range(200, 300):
input_seq = encoder_input_data[seq_index: seq_index + 1]
decoded_sentence = decode_sequence(input_seq)
print('-')
print('Input sentence:', input_texts[seq_index])
print('Decoded sentence:', decoded_sentence)
"""
# print(encoder_input_data)
# num=45
# num = encoder_input_data[num: num + 1]
# for a in range(20):
# word=decode_sequence(num)
# print(word)
# num=np.array([[input_vocab[i] for i in word]])
# print(num)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import matplotlib.pyplot as plt
import seaborn as sns
train = pd.read_csv(r"/kaggle/input/playground-series-s3e12/train.csv")
test = pd.read_csv(r"/kaggle/input/playground-series-s3e12/test.csv")
submission = pd.read_csv(r"/kaggle/input/playground-series-s3e12/sample_submission.csv")
org = pd.read_csv(
r"/kaggle/input/kidney-stone-prediction-based-on-urine-analysis/kindey stone urine analysis.csv"
)
train.head()
org.head()
print("The shape of the train data is:", train.shape)
print("The shape of the test data is:", test.shape)
print("The shape of the original train data is:", org.shape)
train.info()
test.info()
org.info()
train.isnull().sum()
test.isnull().sum()
org.isnull().sum()
train.duplicated().sum()
test.duplicated().sum()
org.duplicated().sum()
train.describe()
test.describe()
org.describe()
train.drop("id", axis=1, inplace=True)
test.drop("id", axis=1, inplace=True)
plt.figure(figsize=(12, 10))
plt.subplot(3, 2, 1)
sns.kdeplot(train["gravity"])
plt.subplot(3, 2, 2)
sns.kdeplot(train["ph"])
plt.subplot(3, 2, 3)
sns.kdeplot(train["osmo"])
plt.subplot(3, 2, 4)
sns.kdeplot(train["cond"])
plt.subplot(3, 2, 5)
sns.kdeplot(train["urea"])
plt.subplot(3, 2, 6)
sns.kdeplot(train["calc"])
plt.show()
plt.figure(figsize=(12, 10))
plt.subplot(3, 2, 1)
sns.kdeplot(test["gravity"])
plt.subplot(3, 2, 2)
sns.kdeplot(test["ph"])
plt.subplot(3, 2, 3)
sns.kdeplot(test["osmo"])
plt.subplot(3, 2, 4)
sns.kdeplot(test["cond"])
plt.subplot(3, 2, 5)
sns.kdeplot(test["urea"])
plt.subplot(3, 2, 6)
sns.kdeplot(test["calc"])
plt.show()
plt.figure(figsize=(12, 10))
plt.subplot(3, 2, 1)
sns.kdeplot(org["gravity"])
plt.subplot(3, 2, 2)
sns.kdeplot(org["ph"])
plt.subplot(3, 2, 3)
sns.kdeplot(org["osmo"])
plt.subplot(3, 2, 4)
sns.kdeplot(org["cond"])
plt.subplot(3, 2, 5)
sns.kdeplot(org["urea"])
plt.subplot(3, 2, 6)
sns.kdeplot(org["calc"])
plt.show()
columns = test.columns.tolist()
fig, ax = plt.subplots(3, 2, figsize=(10, 10), dpi=300)
ax = ax.flatten()
for i in columns:
sns.kdeplot(train[i], ax=ax[columns.index(i)])
sns.kdeplot(test[i], ax=ax[columns.index(i)])
sns.kdeplot(org[i], ax=ax[columns.index(i)])
ax[columns.index(i)].set_title(f"{i} Distribution")
fig.suptitle("Distribution of Features per Dataset", fontsize=12, fontweight="bold")
fig.legend(["Train", "Test", "Original"])
plt.tight_layout()
train["target"].value_counts().plot(kind="bar")
plt.show()
train.info()
sns.pairplot(train, hue="target")
sns.heatmap(train.corr(), annot=True)
new_train = pd.concat([train, org])
from sklearn.model_selection import train_test_split
x = new_train.drop("target", axis=1)
y = new_train["target"]
X_train, X_test, y_train, y_test = train_test_split(x, y, test_size=30, random_state=1)
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
X_train = scaler.fit_transform(X_train)
X_test = scaler.transform(X_test)
from xgboost import XGBClassifier
model = XGBClassifier()
model.fit(X_train, y_train)
from sklearn.metrics import roc_auc_score, accuracy_score
predict = model.predict(X_test)
score = roc_auc_score(y_test, predict)
score1 = accuracy_score(y_test, predict)
print(score)
# For Kaggle Submission
x_kaggle = new_train.drop("target", axis=1)
y_kaggle = new_train["target"]
xb = XGBClassifier()
xb.fit(x_kaggle, y_kaggle)
xbResult = xb.predict(test)
submission["target"] = xbResult
submission.to_csv("submission1.csv", index=False)
|
# ###### Notebook created by: Arnav Chavan (@[carnav0400](https://www.kaggle.com/carnav0400)), Udbhav Bamba (@[ubamba98](https://www.kaggle.com/ubamba98))
# ## NOTE: Turn on the Internet and GPU for this kernal before starting
# # How to add dataset to the kernal
# * Click on "Add Data"
# * Search "CLabsCVcomp"
# * Click on "Add"
# * Done
# # Importing all Libraries
# PS - FastAI imports all necessary libraries for you
from fastai import *
from fastai.vision import *
from sklearn.metrics import f1_score
# # Seed everything for reproducibility
# You may like to read more about it at [link](https://medium.com/@ODSC/properly-setting-the-random-seed-in-ml-experiments-not-as-simple-as-you-might-imagine-219969c84752).
def seed_everything(seed):
random.seed(seed)
os.environ["PYTHONHASHSEED"] = str(seed)
np.random.seed(seed)
torch.manual_seed(seed)
torch.cuda.manual_seed(seed)
torch.backends.cudnn.deterministic = True
seed_everything(43)
# # EDTA
# ## Reading CSV
train = pd.read_csv("../input/clabscvcomp/data/train.csv")
test_df = pd.read_csv("../input/clabscvcomp/data/sample_submission.csv")
train.head() ## Shows the first five rows of data frame
sorted(train.genres.unique()) ## Shows all classes in the dataframe
train.genres.value_counts(normalize=False) ## Distribution of dataset
# Dataset looks very imbalanced. Try to read more about it. This blog post might be a good read [link](https://www.analyticsvidhya.com/blog/2017/03/imbalanced-classification-problem/)
# # Defining DataBunch for FastAI
# Read more about it [here](https://docs.fast.ai/vision.data.html#ImageDataBunch.from_df)
sz = 512 ## Image rescaled into (128 128 3)
bs = 32 ## Batch size
tfms = get_transforms( ## Transformation to apply on Train data
do_flip=True, ## Horizontal flip
flip_vert=False, ## Vertical flip
max_rotate=45, ## Rotation
max_zoom=2.0, ## Center zoom
max_lighting=0.8, ## lighting
)
data = (
ImageList.from_df(
df=train,
path="",
folder="../input/clabscvcomp/data/train_data/",
cols="id",
suffix=".jpg",
) ## define data path
.split_by_rand_pct(valid_pct=0.2) ## validation split
.label_from_df(cols="genres") ## load labels from
.transform(tfms, size=sz)
.databunch(bs=bs, num_workers=6)
.normalize(imagenet_stats)
)
# Now lets add test data to the DataBunch
test_data = ImageList.from_df(
test_df, path="../input/clabscvcomp/data/test_data/", cols="id", suffix=".jpg"
)
data.add_test(test_data)
# # Visualizing dataset
data.show_batch(2)
# # Define F1 metric
def F1(y_pred, y):
y_pred = y_pred.softmax(dim=1)
y_pred = y_pred.argmax(dim=1)
return torch.tensor(
f1_score(y.cpu(), y_pred.cpu(), labels=list(range(10)), average="weighted"),
device="cuda:0",
)
# # Defining FastAI's Learner
# Learner is an integration of DataBunch + Model + callbacks
# More about it can be found [here](https://docs.fast.ai/vision.learner.html)
learn = cnn_learner(
data, ## DataBunch
models.resnet50, ## Resnet50
metrics=[F1, accuracy], ## Matrices
callback_fns=ShowGraph, ## Allows us to visualize training
)
# # Lets start training!!
# ###### Freeze all layers but last layer and training some epochs with one-cycle policy
# Read more: [1-cycle policy basics](https://sgugger.github.io/the-1cycle-policy.html), [Documentation](https://docs.fast.ai/callbacks.one_cycle.html)
learn.freeze()
learn.fit_one_cycle(6) ##no. of epochs
# ###### Unfreeze all layers and find best learning rate
learn.unfreeze()
learn.lr_find()
learn.recorder.plot(suggestion=True)
# ###### Continue training
learn.fit_one_cycle(9, max_lr=slice(1e-5, 1e-4))
# ## Predicting for test data
preds = learn.get_preds(ds_type=DatasetType.Test) ## get prediction in test data
preds = np.argmax(preds[0].numpy(), axis=1) ##gives index of maximum value
categories = sorted(train.genres.unique().astype("str"))
final_preds = []
for idx in preds:
final_preds.append(categories[idx])
final_submit = pd.read_csv("../input/clabscvcomp/data/sample_submission.csv")
final_submit.genres = final_preds
final_submit.head()
final_submit.to_csv("submission.csv", index=False)
|
# ### Short introduction to Gradient descent.
# In this coding example we explain how gradient descent would work for a single layer preceptron with linear output.
import numpy as np
import matplotlib.pyplot as plt
import pandas
import math
import os
import tensorflow.compat.v1 as tf1
# For Compatibility with older code.
tf1.disable_eager_execution()
# ### Video Links
# https://www.youtube.com/watch?v=sDv4f4s2SB8
# https://www.youtube.com/watch?v=DjF3GiCazwc&list=LLkGG_GT2lhbfvl5e9bZpJ8g&index=2&t=0s
# ### Case 1, Using the aglortihm to solve for a single parameter.
# For a line fitting task, we are provided with one of the parameters "m" which is used scale the line being fit. Using the optimization technique of gradient descent we will find the bias term needed to fit the curve.
# ### Given the data, X & Y
X = np.array([0.5, 2.3, 2.9])
Y = np.array([1.4, 1.9, 3.2])
# ### Using pre-calculated slope m = 0.64
# The fit line with bias term set at 0, results in model with high Sum of squared error.
# $$Y = mx + b$$
# $$SSE = \sum(Y - \hat{Y})^2 = \sum(Y - mx) \ \ \text{Given b = 0}$$
# $$SSE = 3.15$$
plt.scatter(X, Y)
W = 0.64
plt.plot(
range(math.ceil(np.max(X)) + 1), [W * i for i in range(math.ceil(np.max(X)) + 1)]
)
plt.show()
# We will use gradient descent to find the parameter 'm' for the optimum value that minimizes SSE our cost function.
def ss_loss_1(B):
# Using the X&Y decalred at the top
return np.sum(((Y - (W * X + B))) ** 2)
plt.plot(
[p / 1000 for p in range(-10000, 10000)],
[ss_loss_1(i) for i in [p / 1000 for p in range(-10000, 10000)]],
)
plt.axvline(x=0, c="r")
plt.axhline(y=0, c="r")
plt.xlim([-1, 5])
plt.ylim([-1, 10])
# Looking at the curve for SSE vs different alphas, it can be seen that the optimum value of $\alpha$ should be around 1, which minimizes SSE.
# $$ loss = \sum(Y - \hat{Y})^ 2 $$
# $$m = 0.64$$
# $$\hat{y} = b + mx$$
# $$SSE = \sum (y_i - (b + mx_i))^2$$
# $$ SSE = (1.4 - (b + 0.32))^2 + (1.9 - (b + 1.4))^2 + (3.2 - (b + 1.8))^2 $$
# $$SSE = f(x_1) + f(x_2) + f(x_3) $$
# The derivative of SSE will give us an indicator of minimum, as when d/db SSE =0; we will get optimum alpha value
# $$ \frac{d}{db} \ SSE = f'(x_1) + f'(x_2) + f'(x_3) $$
# $$-2(-b + 1.08) - 2(-b + 0.42) - 2(-b + 1.3)$$
# $$\frac{d}{db} \ SSE = 6b - 5.7$$
# $$slope(b) = \frac{d}{db} \ SSE = 6b - 5.7$$
# ### High level overview of gradient descent.
# Given a random starting point in our cost curve (SEE), we use the derivative along with a learning rate C to understand which direction to move. An example is shown below.
# ### Lets initialize gradient descent at $b = 0$
# $$Slope(0) = -5.7$$
# $$Step \ Size(b) = Slope(b) \times C$$
# $$Let \ C = 0.1$$
# $$Step \ Size(Slope(0)) = -5.7 \times 0.1 = -0.57$$
# $$b_{new} = 0 - Step \ Size(Slope(0)) $$
# $$ b_{new} = 0 + 0.57 = 0.57$$
# $$slope(0.57) = -2.3$$
# $$\text{We move closer to 0}$$
# $$sse(0.57) = 1$$
# ### From the previous iteration $b = 0.57$
# $$Slope(0.57) = -2.3$$
# $$Step Size(Slope(0)) = -2.3 \times 0.1 = -0.23$$
# $$b_{new} = 0.57 - Step Size(Slope(0.57)) = 0.57 + 0.23 = 0.8$$
# $$sse(0.8) = 0.5$$
# To clear the defined variables and operations of the previous cell
tf1.reset_default_graph()
# We used a fixed value for m
m = tf1.constant(0.64)
# We initialize b at 0
b = tf1.Variable(0.0)
error = 0
for x, y in zip(X, Y):
y_hat = m * x + b
error += (y - y_hat) ** 2
optimizer = tf1.train.GradientDescentOptimizer(learning_rate=0.1)
train = optimizer.minimize(error)
init = tf1.global_variables_initializer()
with tf1.Session() as sess:
sess.run(init)
training_steps = 1
writer = tf1.summary.FileWriter("logs/", sess.graph)
for i in range(training_steps):
sess.run(train)
slope, intercept = sess.run([m, b])
# ### The values of m,c after a single iteration of gradient descent.
print(" Slope = ", slope, " Intercept = ", intercept)
f, ax = plt.subplots(figsize=(20, 6))
plt.plot(
[p / 1000 for p in range(-10000, 10000)],
[ss_loss_1(i) for i in [p / 1000 for p in range(-10000, 10000)]],
)
plt.axhline(y=0)
with tf1.Session() as sess:
sess.run(init)
training_steps = 10
err = sess.run(error)
slope, intercept = sess.run([m, b])
plt.scatter(intercept, err, c="r", s=150)
for i in range(training_steps):
sess.run(train)
err = sess.run(error)
slope, intercept = sess.run([m, b])
plt.scatter(intercept, err)
plt.scatter(intercept, err, c="g", s=150)
plt.axvline(x=0, c="r")
plt.axhline(y=0, c="r")
plt.xlim([-1, 5])
plt.ylim([-1, 10])
plt.show()
print("Converged Value ", intercept)
# After initializing at m = 0, new alphas move closer and closer to the optimum.
f, ax = plt.subplots(figsize=(20, 6))
with tf1.Session() as sess:
sess.run(init)
training_steps = 10
err = sess.run(error)
slope, intercept = sess.run([m, b])
plt.plot(range(4), [slope * i + intercept for i in range(4)])
for i in range(training_steps):
sess.run(train)
err = sess.run(error)
slope, intercept = sess.run([m, b])
plt.plot(range(4), [slope * i + intercept for i in range(4)])
plt.scatter(X, Y, s=100)
plt.show()
# ### Case 2, Gradient Descent to find both parameters
# Lets use gradient descent to find both parameters alpha and w. Refer to the video links above to understand the calculations behind the equation.
# This time we randomize both variables needed, also we enclose them inside an array
params = tf1.Variable([[8.0, 8.0]], dtype=tf1.float32)
Xmat = np.vstack([X, np.ones(len(X))])
Xmat = Xmat.astype(np.float32)
y_hat = tf1.matmul(params, Xmat)
error = tf1.reduce_sum((Y - y_hat) ** 2)
optimizer = tf1.train.GradientDescentOptimizer(learning_rate=0.01)
train = optimizer.minimize(error)
init = tf1.global_variables_initializer()
from mpl_toolkits.mplot3d import axes3d
def viz_loss(x1, x2):
return (
(1.4 - (x1 + x2 * 0.32)) ** 2
+ (1.9 - (x1 + x2 * 1.4)) ** 2
+ (3.2 - (x1 + x2 * 1.8)) ** 2
)
a1 = np.linspace(-8, 8)
a2 = np.linspace(-8, 8)
A1, A2 = np.meshgrid(a1, a2)
Z = viz_loss(A1, A2)
fig = plt.figure(figsize=(15, 10))
ax = fig.add_subplot(111, projection="3d")
with tf1.Session() as sess:
sess.run(init)
training_steps = 100
for i in range(training_steps):
sess.run(train)
slope, intercept = sess.run(params)[0]
SumSq = sess.run(error)
ax.scatter(slope, intercept, SumSq, c="red")
ax.plot_surface(A1, A2, Z, lw=10, cmap="coolwarm", rstride=1, cstride=1, alpha=0.8)
ax.contour(A1, A2, Z, 10, cmap="coolwarm", linestyles="solid", offset=-1, alpha=0.1)
ax.contour(A1, A2, Z, 10, colors="k", linestyles="solid", alpha=0.1)
ax.view_init(0, 120)
plt.show()
|
# # 11.6 Momentum
# 在 [Section 11.4](https://d2l.ai/chapter_optimization/sgd.html#sec-sgd) 中,我们提到,目标函数有关自变量的梯度代表了目标函数在自变量当前位置下降最快的方向。因此,梯度下降也叫作最陡下降(steepest descent)。在每次迭代中,梯度下降根据自变量当前位置,沿着当前位置的梯度更新自变量。然而,如果自变量的迭代方向仅仅取决于自变量当前位置,这可能会带来一些问题。对于noisy gradient,我们需要谨慎的选取学习率和batch size, 来控制梯度方差和收敛的结果。
# ## An ill-conditioned Problem
# 让我们考虑一个输入和输出分别为二维向量$\boldsymbol{x} = [x_1, x_2]^\top$和标量的目标函数:
# $$f(\boldsymbol{x})=0.1x_1^2+2x_2^2$$
# 与[Section 11.4](https://d2l.ai/chapter_optimization/sgd.html#sec-sgd)一节中不同,这里将$x_1^2$系数从$1$减小到了$0.1$。下面实现基于这个目标函数的梯度下降,并演示使用学习率为$0.4$时自变量的迭代轨迹。
import os
if os.getcwd().split("/")[-1] != "OptimizationCode":
os.chdir(
"/kaggle/input/boyu-d2l-optimization-advance-dataset/OptimizationAdvanceKaggle/Code/OptimizationCode"
)
import sys
sys.path.append("..")
import d2lzh_pytorch as d2l
import torch
eta = 0.4
def f_2d(x1, x2):
return 0.1 * x1**2 + 2 * x2**2
def gd_2d(x1, x2, s1, s2):
return (x1 - eta * 0.2 * x1, x2 - eta * 4 * x2, 0, 0)
d2l.show_trace_2d(f_2d, d2l.train_2d(gd_2d))
# 可以看到,同一位置上,目标函数在竖直方向($x_2$轴方向)比在水平方向($x_1$轴方向)的斜率的绝对值更大。因此,给定学习率,梯度下降迭代自变量时会使自变量在竖直方向比在水平方向移动幅度更大。那么,我们需要一个较小的学习率从而避免自变量在竖直方向上越过目标函数最优解。然而,这会造成自变量在水平方向上朝最优解移动变慢。
# 下面我们试着将学习率调得稍大一点,此时自变量在竖直方向不断越过最优解并逐渐发散。
eta = 0.6
d2l.show_trace_2d(f_2d, d2l.train_2d(gd_2d))
# ## Momentum Algorithm
# 动量法的提出是为了解决梯度下降的上述问题。设时间步$t$的自变量为$\boldsymbol{x}_t$,学习率为$\eta_t$。
# 在时间步 $t=0$,动量法创建速度变量$\boldsymbol{v}_0$,并将其元素初始化成 0。在时间步$t>0$,动量法对每次迭代的步骤做如下修改:
# $$
# \begin{aligned}
# \boldsymbol{v}_t &\leftarrow \gamma \boldsymbol{v}_{t-1} + \eta_t \boldsymbol{g}_t, \\
# \boldsymbol{x}_t &\leftarrow \boldsymbol{x}_{t-1} - \boldsymbol{v}_t,
# \end{aligned}
# $$
# 其中,动量超参数$\gamma$满足$0 \leq \gamma < 1$。当$\gamma=0$时,动量法等价于小批量随机梯度下降。
# 在解释动量法的数学原理前,让我们先从实验中观察梯度下降在使用动量法后的迭代轨迹。
def momentum_2d(x1, x2, v1, v2):
v1 = gamma * v1 + eta * 0.2 * x1
v2 = gamma * v2 + eta * 4 * x2
return x1 - v1, x2 - v2, v1, v2
eta, gamma = 0.4, 0.5
d2l.show_trace_2d(f_2d, d2l.train_2d(momentum_2d))
# 可以看到使用较小的学习率$\eta=0.4$和动量超参数$\gamma=0.5$时,动量法在竖直方向上的移动更加平滑,且在水平方向上更快逼近最优解。下面使用较大的学习率$\eta=0.6$,此时自变量也不再发散。
eta = 0.6
d2l.show_trace_2d(f_2d, d2l.train_2d(momentum_2d))
# ### Exponential Moving Average
# 为了从数学上理解动量法,让我们先解释一下指数加权移动平均(exponential moving average)。给定超参数$0 \leq \gamma < 1$,当前时间步$t$的变量$y_t$是上一时间步$t-1$的变量$y_{t-1}$和当前时间步另一变量$x_t$的线性组合:
# $$y_t = \gamma y_{t-1} + (1-\gamma) x_t.$$
# 我们可以对$y_t$展开:
# $$
# \begin{aligned}
# y_t &= (1-\gamma) x_t + \gamma y_{t-1}\\
# &= (1-\gamma)x_t + (1-\gamma) \cdot \gamma x_{t-1} + \gamma^2y_{t-2}\\
# &= (1-\gamma)x_t + (1-\gamma) \cdot \gamma x_{t-1} + (1-\gamma) \cdot \gamma^2x_{t-2} + \gamma^3y_{t-3}\\
# &= (1-\gamma) \sum_{i=0}^{t} \gamma^{i}x_{t-i}
# \end{aligned}
# $$
# 令$n = 1/(1-\gamma)$,那么 $\left(1-1/n\right)^n = \gamma^{1/(1-\gamma)}$。因为
# $$ \lim_{n \rightarrow \infty} \left(1-\frac{1}{n}\right)^n = \exp(-1) \approx 0.3679,$$
# 所以当$\gamma \rightarrow 1$时,$\gamma^{1/(1-\gamma)}=\exp(-1)$,如$0.95^{20} \approx \exp(-1)$。如果把$\exp(-1)$当作一个比较小的数,我们可以在近似中忽略所有含$\gamma^{1/(1-\gamma)}$和比$\gamma^{1/(1-\gamma)}$更高阶的系数的项。例如,当$\gamma=0.95$时,
# $$y_t \approx 0.05 \sum_{i=0}^{19} 0.95^i x_{t-i}.$$
# 因此,在实际中,我们常常将$y_t$看作是对最近$1/(1-\gamma)$个时间步的$x_t$值的加权平均。例如,当$\gamma = 0.95$时,$y_t$可以被看作对最近20个时间步的$x_t$值的加权平均;当$\gamma = 0.9$时,$y_t$可以看作是对最近10个时间步的$x_t$值的加权平均。而且,离当前时间步$t$越近的$x_t$值获得的权重越大(越接近1)。
# ### 由指数加权移动平均理解动量法
# 现在,我们对动量法的速度变量做变形:
# $$\boldsymbol{v}_t \leftarrow \gamma \boldsymbol{v}_{t-1} + (1 - \gamma) \left(\frac{\eta_t}{1 - \gamma} \boldsymbol{g}_t\right). $$
# 由指数加权移动平均的形式可得,速度变量$\boldsymbol{v}_t$实际上对序列$\{\eta_{t-i}\boldsymbol{g}_{t-i} /(1-\gamma):i=0,\ldots,1/(1-\gamma)-1\}$做了指数加权移动平均。换句话说,相比于小批量随机梯度下降,动量法在每个时间步的自变量更新量近似于将前者对应的最近$1/(1-\gamma)$个时间步的更新量做了指数加权移动平均后再除以$1-\gamma$。所以,在动量法中,自变量在各个方向上的移动幅度不仅取决当前梯度,还取决于过去的各个梯度在各个方向上是否一致。在本节之前示例的优化问题中,所有梯度在水平方向上为正(向右),而在竖直方向上时正(向上)时负(向下)。这样,我们就可以使用较大的学习率,从而使自变量向最优解更快移动。
# ## Implement
# 相对于小批量随机梯度下降,动量法需要对每一个自变量维护一个同它一样形状的速度变量,且超参数里多了动量超参数。实现中,我们将速度变量用更广义的状态变量`states`表示。
features, labels = d2l.get_data_ch7()
def init_momentum_states():
v_w = torch.zeros((features.shape[1], 1), dtype=torch.float32)
v_b = torch.zeros(1, dtype=torch.float32)
return (v_w, v_b)
def sgd_momentum(params, states, hyperparams):
for p, v in zip(params, states):
v.data = hyperparams["momentum"] * v.data + hyperparams["lr"] * p.grad.data
p.data -= v.data
# 我们先将动量超参数`momentum`设0.5
d2l.train_ch7(
sgd_momentum,
init_momentum_states(),
{"lr": 0.02, "momentum": 0.5},
features,
labels,
)
# 将动量超参数`momentum`增大到0.9
d2l.train_ch7(
sgd_momentum,
init_momentum_states(),
{"lr": 0.02, "momentum": 0.9},
features,
labels,
)
# 可见目标函数值在后期迭代过程中的变化不够平滑。直觉上,10倍小批量梯度比2倍小批量梯度大了5倍,我们可以试着将学习率减小到原来的1/5。此时目标函数值在下降了一段时间后变化更加平滑。
d2l.train_ch7(
sgd_momentum,
init_momentum_states(),
{"lr": 0.004, "momentum": 0.9},
features,
labels,
)
# ## Pytorch Class
# 在Pytorch中,```torch.optim.SGD```已实现了Momentum。
d2l.train_pytorch_ch7(torch.optim.SGD, {"lr": 0.004, "momentum": 0.9}, features, labels)
# # 11.7 AdaGrad
# 在之前介绍过的优化算法中,目标函数自变量的每一个元素在相同时间步都使用同一个学习率来自我迭代。举个例子,假设目标函数为$f$,自变量为一个二维向量$[x_1, x_2]^\top$,该向量中每一个元素在迭代时都使用相同的学习率。例如,在学习率为$\eta$的梯度下降中,元素$x_1$和$x_2$都使用相同的学习率$\eta$来自我迭代:
# $$
# x_1 \leftarrow x_1 - \eta \frac{\partial{f}}{\partial{x_1}}, \quad
# x_2 \leftarrow x_2 - \eta \frac{\partial{f}}{\partial{x_2}}.
# $$
# 在[“动量法”](./momentum.ipynb)一节里我们看到当$x_1$和$x_2$的梯度值有较大差别时,需要选择足够小的学习率使得自变量在梯度值较大的维度上不发散。但这样会导致自变量在梯度值较小的维度上迭代过慢。动量法依赖指数加权移动平均使得自变量的更新方向更加一致,从而降低发散的可能。本节我们介绍AdaGrad算法,它根据自变量在每个维度的梯度值的大小来调整各个维度上的学习率,从而避免统一的学习率难以适应所有维度的问题 [1]。
# ## Algorithm
# AdaGrad算法会使用一个小批量随机梯度$\boldsymbol{g}_t$按元素平方的累加变量$\boldsymbol{s}_t$。在时间步0,AdaGrad将$\boldsymbol{s}_0$中每个元素初始化为0。在时间步$t$,首先将小批量随机梯度$\boldsymbol{g}_t$按元素平方后累加到变量$\boldsymbol{s}_t$:
# $$\boldsymbol{s}_t \leftarrow \boldsymbol{s}_{t-1} + \boldsymbol{g}_t \odot \boldsymbol{g}_t,$$
# 其中$\odot$是按元素相乘。接着,我们将目标函数自变量中每个元素的学习率通过按元素运算重新调整一下:
# $$\boldsymbol{x}_t \leftarrow \boldsymbol{x}_{t-1} - \frac{\eta}{\sqrt{\boldsymbol{s}_t + \epsilon}} \odot \boldsymbol{g}_t,$$
# 其中$\eta$是学习率,$\epsilon$是为了维持数值稳定性而添加的常数,如$10^{-6}$。这里开方、除法和乘法的运算都是按元素运算的。这些按元素运算使得目标函数自变量中每个元素都分别拥有自己的学习率。
# ## Feature
# 需要强调的是,小批量随机梯度按元素平方的累加变量$\boldsymbol{s}_t$出现在学习率的分母项中。因此,如果目标函数有关自变量中某个元素的偏导数一直都较大,那么该元素的学习率将下降较快;反之,如果目标函数有关自变量中某个元素的偏导数一直都较小,那么该元素的学习率将下降较慢。然而,由于$\boldsymbol{s}_t$一直在累加按元素平方的梯度,自变量中每个元素的学习率在迭代过程中一直在降低(或不变)。所以,当学习率在迭代早期降得较快且当前解依然不佳时,AdaGrad算法在迭代后期由于学习率过小,可能较难找到一个有用的解。
# 下面我们仍然以目标函数$f(\boldsymbol{x})=0.1x_1^2+2x_2^2$为例观察AdaGrad算法对自变量的迭代轨迹。我们实现AdaGrad算法并使用和上一节实验中相同的学习率0.4。可以看到,自变量的迭代轨迹较平滑。但由于$\boldsymbol{s}_t$的累加效果使学习率不断衰减,自变量在迭代后期的移动幅度较小。
import os
if os.getcwd().split("/")[-1] != "OptimizationCode":
os.chdir(
"/kaggle/input/boyu-d2l-optimization-advance-dataset/OptimizationAdvanceKaggle/Code/OptimizationCode"
)
import math
import torch
import sys
sys.path.append("..")
import d2lzh_pytorch as d2l
def adagrad_2d(x1, x2, s1, s2):
g1, g2, eps = 0.2 * x1, 4 * x2, 1e-6 # 前两项为自变量梯度
s1 += g1**2
s2 += g2**2
x1 -= eta / math.sqrt(s1 + eps) * g1
x2 -= eta / math.sqrt(s2 + eps) * g2
return x1, x2, s1, s2
def f_2d(x1, x2):
return 0.1 * x1**2 + 2 * x2**2
eta = 0.4
d2l.show_trace_2d(f_2d, d2l.train_2d(adagrad_2d))
# 下面将学习率增大到2。可以看到自变量更为迅速地逼近了最优解。
eta = 2
d2l.show_trace_2d(f_2d, d2l.train_2d(adagrad_2d))
# ## Implement
# 同动量法一样,AdaGrad算法需要对每个自变量维护同它一样形状的状态变量。我们根据AdaGrad算法中的公式实现该算法。
features, labels = d2l.get_data_ch7()
def init_adagrad_states():
s_w = torch.zeros((features.shape[1], 1), dtype=torch.float32)
s_b = torch.zeros(1, dtype=torch.float32)
return (s_w, s_b)
def adagrad(params, states, hyperparams):
eps = 1e-6
for p, s in zip(params, states):
s.data += p.grad.data**2
p.data -= hyperparams["lr"] * p.grad.data / torch.sqrt(s + eps)
# 使用更大的学习率来训练模型。
d2l.train_ch7(adagrad, init_adagrad_states(), {"lr": 0.1}, features, labels)
# ## Pytorch Class
# 通过名称为“adagrad”的`Trainer`实例,我们便可使用Pytorch提供的AdaGrad算法来训练模型。
d2l.train_pytorch_ch7(torch.optim.Adagrad, {"lr": 0.1}, features, labels)
# # 11.8 RMSProp
# 我们在[“AdaGrad算法”](adagrad.ipynb)一节中提到,因为调整学习率时分母上的变量$\boldsymbol{s}_t$一直在累加按元素平方的小批量随机梯度,所以目标函数自变量每个元素的学习率在迭代过程中一直在降低(或不变)。因此,当学习率在迭代早期降得较快且当前解依然不佳时,AdaGrad算法在迭代后期由于学习率过小,可能较难找到一个有用的解。为了解决这一问题,RMSProp算法对AdaGrad算法做了修改。该算法源自Coursera上的一门课程,即“机器学习的神经网络”。
# ## Algorithm
# 我们在[“动量法”](momentum.ipynb)一节里介绍过指数加权移动平均。不同于AdaGrad算法里状态变量$\boldsymbol{s}_t$是截至时间步$t$所有小批量随机梯度$\boldsymbol{g}_t$按元素平方和,RMSProp算法将这些梯度按元素平方做指数加权移动平均。具体来说,给定超参数$0 \leq \gamma 0$计算
# $$\boldsymbol{s}_t \leftarrow \gamma \boldsymbol{s}_{t-1} + (1 - \gamma) \boldsymbol{g}_t \odot \boldsymbol{g}_t. $$
# 和AdaGrad算法一样,RMSProp算法将目标函数自变量中每个元素的学习率通过按元素运算重新调整,然后更新自变量
# $$\boldsymbol{x}_t \leftarrow \boldsymbol{x}_{t-1} - \frac{\eta}{\sqrt{\boldsymbol{s}_t + \epsilon}} \odot \boldsymbol{g}_t, $$
# 其中$\eta$是学习率,$\epsilon$是为了维持数值稳定性而添加的常数,如$10^{-6}$。因为RMSProp算法的状态变量$\boldsymbol{s}_t$是对平方项$\boldsymbol{g}_t \odot \boldsymbol{g}_t$的指数加权移动平均,所以可以看作是最近$1/(1-\gamma)$个时间步的小批量随机梯度平方项的加权平均。如此一来,自变量每个元素的学习率在迭代过程中就不再一直降低(或不变)。
# 照例,让我们先观察RMSProp算法对目标函数$f(\boldsymbol{x})=0.1x_1^2+2x_2^2$中自变量的迭代轨迹。回忆在[“AdaGrad算法”](adagrad.ipynb)一节使用的学习率为0.4的AdaGrad算法,自变量在迭代后期的移动幅度较小。但在同样的学习率下,RMSProp算法可以更快逼近最优解。
import os
if os.getcwd().split("/")[-1] != "OptimizationCode":
os.chdir(
"/kaggle/input/boyu-d2l-optimization-advance-dataset/OptimizationAdvanceKaggle/Code/OptimizationCode"
)
import math
import torch
import sys
sys.path.append("..")
import d2lzh_pytorch as d2l
def rmsprop_2d(x1, x2, s1, s2):
g1, g2, eps = 0.2 * x1, 4 * x2, 1e-6
s1 = gamma * s1 + (1 - gamma) * g1**2
s2 = gamma * s2 + (1 - gamma) * g2**2
x1 -= eta / math.sqrt(s1 + eps) * g1
x2 -= eta / math.sqrt(s2 + eps) * g2
return x1, x2, s1, s2
def f_2d(x1, x2):
return 0.1 * x1**2 + 2 * x2**2
eta, gamma = 0.4, 0.9
d2l.show_trace_2d(f_2d, d2l.train_2d(rmsprop_2d))
# ## Implement
# 接下来按照RMSProp算法中的公式实现该算法。
features, labels = d2l.get_data_ch7()
def init_rmsprop_states():
s_w = torch.zeros((features.shape[1], 1), dtype=torch.float32)
s_b = torch.zeros(1, dtype=torch.float32)
return (s_w, s_b)
def rmsprop(params, states, hyperparams):
gamma, eps = hyperparams["gamma"], 1e-6
for p, s in zip(params, states):
s.data = gamma * s.data + (1 - gamma) * (p.grad.data) ** 2
p.data -= hyperparams["lr"] * p.grad.data / torch.sqrt(s + eps)
# 我们将初始学习率设为0.01,并将超参数$\gamma$设为0.9。此时,变量$\boldsymbol{s}_t$可看作是最近$1/(1-0.9) = 10$个时间步的平方项$\boldsymbol{g}_t \odot \boldsymbol{g}_t$的加权平均。
d2l.train_ch7(
rmsprop, init_rmsprop_states(), {"lr": 0.01, "gamma": 0.9}, features, labels
)
# ## Pytorch Class
# 通过名称为“rmsprop”的`Trainer`实例,我们便可使用Gluon提供的RMSProp算法来训练模型。注意,超参数$\gamma$通过`gamma1`指定。
d2l.train_ch7(adagrad, init_adagrad_states(), {"lr": 0.1}, features, labels)
# # 11.9 AdaDelta
# 除了RMSProp算法以外,另一个常用优化算法AdaDelta算法也针对AdaGrad算法在迭代后期可能较难找到有用解的问题做了改进 [1]。有意思的是,AdaDelta算法没有学习率这一超参数。
# ## Algorithm
# AdaDelta算法也像RMSProp算法一样,使用了小批量随机梯度$\boldsymbol{g}_t$按元素平方的指数加权移动平均变量$\boldsymbol{s}_t$。在时间步0,它的所有元素被初始化为0。给定超参数$0 \leq \rho 0$,同RMSProp算法一样计算
# $$\boldsymbol{s}_t \leftarrow \rho \boldsymbol{s}_{t-1} + (1 - \rho) \boldsymbol{g}_t \odot \boldsymbol{g}_t. $$
# 与RMSProp算法不同的是,AdaDelta算法还维护一个额外的状态变量$\Delta\boldsymbol{x}_t$,其元素同样在时间步0时被初始化为0。我们使用$\Delta\boldsymbol{x}_{t-1}$来计算自变量的变化量:
# $$ \boldsymbol{g}_t' \leftarrow \sqrt{\frac{\Delta\boldsymbol{x}_{t-1} + \epsilon}{\boldsymbol{s}_t + \epsilon}} \odot \boldsymbol{g}_t, $$
# 其中$\epsilon$是为了维持数值稳定性而添加的常数,如$10^{-5}$。接着更新自变量:
# $$\boldsymbol{x}_t \leftarrow \boldsymbol{x}_{t-1} - \boldsymbol{g}'_t. $$
# 最后,我们使用$\Delta\boldsymbol{x}_t$来记录自变量变化量$\boldsymbol{g}'_t$按元素平方的指数加权移动平均:
# $$\Delta\boldsymbol{x}_t \leftarrow \rho \Delta\boldsymbol{x}_{t-1} + (1 - \rho) \boldsymbol{g}'_t \odot \boldsymbol{g}'_t. $$
# 可以看到,如不考虑$\epsilon$的影响,AdaDelta算法与RMSProp算法的不同之处在于使用$\sqrt{\Delta\boldsymbol{x}_{t-1}}$来替代超参数$\eta$。
# ## Implement
# AdaDelta算法需要对每个自变量维护两个状态变量,即$\boldsymbol{s}_t$和$\Delta\boldsymbol{x}_t$。我们按AdaDelta算法中的公式实现该算法。
def init_adadelta_states():
s_w, s_b = torch.zeros((features.shape[1], 1), dtype=torch.float32), torch.zeros(
1, dtype=torch.float32
)
delta_w, delta_b = torch.zeros(
(features.shape[1], 1), dtype=torch.float32
), torch.zeros(1, dtype=torch.float32)
return ((s_w, delta_w), (s_b, delta_b))
def adadelta(params, states, hyperparams):
rho, eps = hyperparams["rho"], 1e-5
for p, (s, delta) in zip(params, states):
s[:] = rho * s + (1 - rho) * (p.grad.data**2)
g = p.grad.data * torch.sqrt((delta + eps) / (s + eps))
p.data -= g
delta[:] = rho * delta + (1 - rho) * g * g
d2l.train_ch7(adadelta, init_adadelta_states(), {"rho": 0.9}, features, labels)
# ## Pytorch Class
# 通过名称为“adadelta”的`Trainer`实例,我们便可使用pytorch提供的AdaDelta算法。它的超参数可以通过`rho`来指定。
d2l.train_pytorch_ch7(torch.optim.Adadelta, {"rho": 0.9}, features, labels)
# # 11.10 Adam
# Adam算法在RMSProp算法基础上对小批量随机梯度也做了指数加权移动平均 [1]。下面我们来介绍这个算法。
# ## Algorithm
# Adam算法使用了动量变量$\boldsymbol{v}_t$和RMSProp算法中小批量随机梯度按元素平方的指数加权移动平均变量$\boldsymbol{s}_t$,并在时间步0将它们中每个元素初始化为0。给定超参数$0 \leq \beta_1 < 1$(算法作者建议设为0.9),时间步$t$的动量变量$\boldsymbol{v}_t$即小批量随机梯度$\boldsymbol{g}_t$的指数加权移动平均:
# $$\boldsymbol{v}_t \leftarrow \beta_1 \boldsymbol{v}_{t-1} + (1 - \beta_1) \boldsymbol{g}_t. $$
# 和RMSProp算法中一样,给定超参数$0 \leq \beta_2 < 1$(算法作者建议设为0.999),
# 将小批量随机梯度按元素平方后的项$\boldsymbol{g}_t \odot \boldsymbol{g}_t$做指数加权移动平均得到$\boldsymbol{s}_t$:
# $$\boldsymbol{s}_t \leftarrow \beta_2 \boldsymbol{s}_{t-1} + (1 - \beta_2) \boldsymbol{g}_t \odot \boldsymbol{g}_t. $$
# 由于我们将$\boldsymbol{v}_0$和$\boldsymbol{s}_0$中的元素都初始化为0,
# 在时间步$t$我们得到$\boldsymbol{v}_t = (1-\beta_1) \sum_{i=1}^t \beta_1^{t-i} \boldsymbol{g}_i$。将过去各时间步小批量随机梯度的权值相加,得到 $(1-\beta_1) \sum_{i=1}^t \beta_1^{t-i} = 1 - \beta_1^t$。需要注意的是,当$t$较小时,过去各时间步小批量随机梯度权值之和会较小。例如,当$\beta_1 = 0.9$时,$\boldsymbol{v}_1 = 0.1\boldsymbol{g}_1$。为了消除这样的影响,对于任意时间步$t$,我们可以将$\boldsymbol{v}_t$再除以$1 - \beta_1^t$,从而使过去各时间步小批量随机梯度权值之和为1。这也叫作偏差修正。在Adam算法中,我们对变量$\boldsymbol{v}_t$和$\boldsymbol{s}_t$均作偏差修正:
# $$\hat{\boldsymbol{v}}_t \leftarrow \frac{\boldsymbol{v}_t}{1 - \beta_1^t}, $$
# $$\hat{\boldsymbol{s}}_t \leftarrow \frac{\boldsymbol{s}_t}{1 - \beta_2^t}. $$
# 接下来,Adam算法使用以上偏差修正后的变量$\hat{\boldsymbol{v}}_t$和$\hat{\boldsymbol{s}}_t$,将模型参数中每个元素的学习率通过按元素运算重新调整:
# $$\boldsymbol{g}_t' \leftarrow \frac{\eta \hat{\boldsymbol{v}}_t}{\sqrt{\hat{\boldsymbol{s}}_t} + \epsilon},$$
# 其中$\eta$是学习率,$\epsilon$是为了维持数值稳定性而添加的常数,如$10^{-8}$。和AdaGrad算法、RMSProp算法以及AdaDelta算法一样,目标函数自变量中每个元素都分别拥有自己的学习率。最后,使用$\boldsymbol{g}_t'$迭代自变量:
# $$\boldsymbol{x}_t \leftarrow \boldsymbol{x}_{t-1} - \boldsymbol{g}_t'. $$
# ## Implement
# 我们按照Adam算法中的公式实现该算法。其中时间步$t$通过`hyperparams`参数传入`adam`函数。
import os
if os.getcwd().split("/")[-1] != "OptimizationCode":
os.chdir(
"/kaggle/input/boyu-d2l-optimization-advance-dataset/OptimizationAdvanceKaggle/Code/OptimizationCode"
)
import torch
import sys
sys.path.append("..")
import d2lzh_pytorch as d2l
features, labels = d2l.get_data_ch7()
def init_adam_states():
v_w, v_b = torch.zeros((features.shape[1], 1), dtype=torch.float32), torch.zeros(
1, dtype=torch.float32
)
s_w, s_b = torch.zeros((features.shape[1], 1), dtype=torch.float32), torch.zeros(
1, dtype=torch.float32
)
return ((v_w, s_w), (v_b, s_b))
def adam(params, states, hyperparams):
beta1, beta2, eps = 0.9, 0.999, 1e-6
for p, (v, s) in zip(params, states):
v[:] = beta1 * v + (1 - beta1) * p.grad.data
s[:] = beta2 * s + (1 - beta2) * p.grad.data**2
v_bias_corr = v / (1 - beta1 ** hyperparams["t"])
s_bias_corr = s / (1 - beta2 ** hyperparams["t"])
p.data -= hyperparams["lr"] * v_bias_corr / (torch.sqrt(s_bias_corr) + eps)
hyperparams["t"] += 1
d2l.train_ch7(adam, init_adam_states(), {"lr": 0.01, "t": 1}, features, labels)
# ## Pytorch Class
d2l.train_pytorch_ch7(torch.optim.Adam, {"lr": 0.01}, features, labels)
|
# # Animated EDA and Tweets Analysis
# **Remark**: Many great kernels have already been posted. My goal is to explore the data using the *Plotly* animation feature in scatter and geo plots!
# **Update**: I recently gathered some tweets following the *coronavirus* hashtag and trying to analyze them.
# For the moment this kernel has no predictions.
# * [EDA](#eda)
# - [nCoV in Asia](#asia)
# - [nCoV in the World](#world)
# - [Confirmed/Deaths/Recovered over Time](#scatter)
# * [Tweets Analysis](#tweets)
# - [Sentiment Distribution](#sentiment)
# - [WordCloud](#wordcloud)
# - [Hashtags](#hashtags)
# # (Geographic) EDA
# Load libraries and the dataset.
import pandas as pd
pd.set_option("display.max_columns", 100)
pd.set_option("display.max_rows", 100)
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import scipy.stats as stats
import plotly.express as px
import plotly.graph_objs as go
from plotly.offline import iplot, init_notebook_mode
import plotly.figure_factory as ff
from plotly import subplots
from plotly.subplots import make_subplots
init_notebook_mode(connected=True)
from datetime import date, datetime, timedelta
import time
from textblob import TextBlob
from textblob.sentiments import NaiveBayesAnalyzer
from wordcloud import WordCloud
from collections import Counter
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
def resumetable(df):
print(f"Dataset Shape: {df.shape}")
summary = pd.DataFrame(df.dtypes, columns=["dtypes"])
summary = summary.reset_index()
summary["Name"] = summary["index"]
summary = summary[["Name", "dtypes"]]
summary["Missing"] = df.isnull().sum().values
summary["Uniques"] = df.nunique().values
return summary
df = pd.read_csv(
"../input/novel-corona-virus-2019-dataset/2019_nCoV_data.csv",
)
resumetable(df)
# Let's rename columns, change datetime to date format, drop rows with (0,0,0) triplets.
df.rename(columns={"Last Update": "LastUpdate", "Province/State": "PS"}, inplace=True)
df["Date"] = pd.to_datetime(df["Date"]).dt.date
virus_cols = ["Confirmed", "Deaths", "Recovered"]
df = df[df[virus_cols].sum(axis=1) != 0]
resumetable(df)
# We see that there are lots of missing values in the Province/State column, let's fill with Country value if there are no other Province/State, and drop the remaining 2 rows.
df.loc[
(df["PS"].isnull()) & (df.groupby("Country")["PS"].transform("nunique") == 0), "PS"
] = df.loc[
(df["PS"].isnull()) & (df.groupby("Country")["PS"].transform("nunique") == 0),
"Country",
].to_numpy()
df["Country"] = np.where(df["Country"] == "Mainland China", "China", df["Country"])
df.dropna(inplace=True)
resumetable(df)
# Retrieve latitute and longitude for each Country-Province pair using the time series dataset.
# (Remark, previously I was using the geopy package).
#
usecols = ["Province/State", "Country/Region", "Lat", "Long"]
path = "../input/novel-corona-virus-2019-dataset/time_series_2019_ncov_"
csvs = ["confirmed.csv", "deaths.csv", "recovered.csv"]
coords_df = pd.concat([pd.read_csv(path + csv, usecols=usecols) for csv in csvs])
coords_df.rename(
columns={"Country/Region": "Country", "Province/State": "PS"}, inplace=True
)
coords_df["Country"] = np.where(
coords_df["Country"] == "Mainland China", "China", coords_df["Country"]
)
coords_df = coords_df.drop_duplicates()
df = pd.merge(df, coords_df, on=["Country", "PS"], how="left")
df
# import time
# import geopy
# locator = geopy.Nominatim(user_agent='uagent')
#
# pairs = df[['Country', 'PS']].drop_duplicates().to_numpy()
##d={}
# for p in pairs:
# if p[0] + ', ' + p[1] not in d:
# l = p[0] + ', ' + p[1] if p[0]!=p[1] else p[0]
# location = locator.geocode(l)
#
# d[l] = [location.latitude, location.longitude]
# print(l, location.latitude, location.longitude)
# time.sleep(1)
# def coords(row):
#
# k = row['Country'] +', '+ row['PS'] if row['Country'] != row['PS'] else row['Country']
# row['lat'] = d[k][0]
# row['lon'] = d[k][1]
# return row
#
# df = df.apply(coords, axis=1)
# df.head(10)
df = (
df.groupby(["PS", "Country", "Date"])
.agg(
{
"Confirmed": "sum",
"Deaths": "sum",
"Recovered": "sum",
"Lat": "max",
"Long": "max",
}
)
.reset_index()
)
df = df[df["Date"] > date(2020, 1, 20)]
# Let's plot the virus spreading in Asia and in the rest of the world over time.
# * Size is proportional to number of confirmed cases.
# * Colorscale depends upon the number of deaths.
# ### Asia Scattergeo
dates = np.sort(df["Date"].unique())
data = [
go.Scattergeo(
locationmode="country names",
lon=df.loc[df["Date"] == dt, "Long"],
lat=df.loc[df["Date"] == dt, "Lat"],
text=df.loc[df["Date"] == dt, "Country"]
+ ", "
+ df.loc[df["Date"] == dt, "PS"]
+ "-> Deaths: "
+ df.loc[df["Date"] == dt, "Deaths"].astype(str)
+ " Confirmed: "
+ df.loc[df["Date"] == dt, "Confirmed"].astype(str),
mode="markers",
marker=dict(
size=(df.loc[df["Date"] == dt, "Confirmed"]) ** (1 / 2.7) + 3,
opacity=0.6,
reversescale=True,
autocolorscale=False,
line=dict(width=0.5, color="rgba(0, 0, 0)"),
cmin=0,
color=df.loc[df["Date"] == dt, "Deaths"],
cmax=df["Deaths"].max(),
colorbar_title="Number of Deaths",
),
)
for dt in dates
]
fig = go.Figure(
data=data[0],
layout=go.Layout(
title={
"text": f"Corona Virus spreading in Asia, {dates[0]}",
"y": 0.98,
"x": 0.5,
"xanchor": "center",
"yanchor": "top",
},
geo=dict(
scope="asia",
projection_type="robinson",
showland=True,
landcolor="rgb(252, 240, 220)",
showcountries=True,
showocean=True,
oceancolor="rgb(219, 245, 255)",
countrycolor="rgb(128, 128, 128)",
lakecolor="rgb(219, 245, 255)",
showrivers=True,
showlakes=True,
showcoastlines=True,
countrywidth=1,
),
updatemenus=[
dict(
type="buttons",
buttons=[dict(label="Play", method="animate", args=[None])],
)
],
),
frames=[
go.Frame(
data=dt,
layout=go.Layout(
title={
"text": f"Corona Virus spreading in Asia, {date}",
"y": 0.98,
"x": 0.5,
"xanchor": "center",
"yanchor": "top",
}
),
)
for dt, date in zip(data[1:], dates[1:])
],
)
fig.show()
#
# ### World Scattergeo
dates = np.sort(df["Date"].unique())
data = [
go.Scattergeo(
locationmode="country names",
lon=df.loc[df["Date"] == dt, "Long"],
lat=df.loc[df["Date"] == dt, "Lat"],
text=df.loc[df["Date"] == dt, "Country"]
+ ", "
+ df.loc[df["Date"] == dt, "PS"]
+ "-> Deaths: "
+ df.loc[df["Date"] == dt, "Deaths"].astype(str)
+ " Confirmed: "
+ df.loc[df["Date"] == dt, "Confirmed"].astype(str),
mode="markers",
marker=dict(
size=(df.loc[df["Date"] == dt, "Confirmed"]) ** (1 / 2.7) + 3,
opacity=0.6,
reversescale=True,
autocolorscale=False,
line=dict(width=0.5, color="rgba(0, 0, 0)"),
# colorscale='rdgy', #'jet',rdylbu, 'oryel',
cmin=0,
color=df.loc[df["Date"] == dt, "Deaths"],
cmax=df["Deaths"].max(),
colorbar_title="Number of Deaths",
),
)
for dt in dates
]
fig = go.Figure(
data=data[0],
layout=go.Layout(
title={
"text": f"Corona Virus, {dates[0]}",
"y": 0.98,
"x": 0.5,
"xanchor": "center",
"yanchor": "top",
},
geo=dict(
scope="world",
projection_type="robinson",
showland=True,
landcolor="rgb(252, 240, 220)",
showcountries=True,
showocean=True,
oceancolor="rgb(219, 245, 255)",
countrycolor="rgb(128, 128, 128)",
lakecolor="rgb(219, 245, 255)",
showrivers=True,
showlakes=True,
showcoastlines=True,
countrywidth=1,
),
updatemenus=[
dict(
type="buttons",
buttons=[dict(label="Play", method="animate", args=[None])],
)
],
),
frames=[
go.Frame(
data=dt,
layout=go.Layout(
title={
"text": f"Corona Virus, {date}",
"y": 0.98,
"x": 0.5,
"xanchor": "center",
"yanchor": "top",
}
),
)
for dt, date in zip(data[1:], dates[1:])
],
)
fig.show()
#
# ### Confirmed/Deaths/Recovered over Time
# Also let's check how number of confirmed, deaths and recovered evolve over time, in China and the rest of the world.
# **Take care**, y-scales are very different!
china = df.loc[df["Country"] == "China"]
hubei = china.loc[china["PS"] == "Hubei"]
rest_of_china = china.loc[china["PS"] != "Hubei"].groupby("Date").sum().reset_index()
china = china.groupby("Date").sum().reset_index()
agg_df = df.groupby(["Country", "Date"]).sum().reset_index()
rest_df = agg_df.loc[agg_df["Country"] != "China"].groupby("Date").sum().reset_index()
dates = np.sort(df["Date"].unique())
dt_range = [np.min(dates) - timedelta(days=1), np.max(dates) + timedelta(days=1)]
# Row 1
frames_hubei = [
go.Scatter(
x=hubei["Date"],
y=hubei.loc[hubei["Date"] <= dt, "Confirmed"],
name="Hubei, Confirmed",
legendgroup="21",
)
for dt in dates
]
frames_rchina = [
go.Scatter(
x=rest_of_china["Date"],
y=rest_of_china.loc[rest_of_china["Date"] <= dt, "Confirmed"],
name="Rest of China, Confirmed",
legendgroup="21",
)
for dt in dates
]
frames_world = [
go.Scatter(
x=rest_df["Date"],
y=rest_df.loc[rest_df["Date"] <= dt, "Confirmed"],
name="Rest of the World, Confirmed",
legendgroup="22",
)
for dt in dates
]
# Row 2
frames_china_d = [
go.Scatter(
x=china["Date"],
y=china.loc[china["Date"] <= dt, "Deaths"],
name="China, Deaths",
legendgroup="31",
)
for dt in dates
]
frames_china_r = [
go.Scatter(
x=china["Date"],
y=china.loc[china["Date"] <= dt, "Recovered"],
name="China, Recovered",
legendgroup="31",
)
for dt in dates
]
frames_world_d = [
go.Scatter(
x=rest_df["Date"],
y=rest_df.loc[rest_df["Date"] <= dt, "Deaths"],
name="Rest of World, Deaths",
legendgroup="32",
)
for dt in dates
]
frames_world_r = [
go.Scatter(
x=rest_df["Date"],
y=rest_df.loc[rest_df["Date"] <= dt, "Recovered"],
name="Rest of World, Recovered",
legendgroup="32",
)
for dt in dates
]
fig = make_subplots(
rows=2,
cols=2,
specs=[[{}, {}], [{}, {}]],
subplot_titles=(
"China, Confirmed",
"Rest of the World, Confirmed",
"China, Deaths & Recovered",
"Rest of the World, Deaths & Recovered",
),
)
# Row 1: Confirmed
fig.add_trace(frames_hubei[0], row=1, col=1)
fig.add_trace(frames_rchina[0], row=1, col=1)
fig.add_trace(frames_world[0], row=1, col=2)
# Row 2: Deaths & Recovered
fig.add_trace(frames_china_d[0], row=2, col=1)
fig.add_trace(frames_china_r[0], row=2, col=1)
fig.add_trace(frames_world_d[0], row=2, col=2)
fig.add_trace(frames_world_r[0], row=2, col=2)
# Add Layout
fig.update_xaxes(showgrid=False)
fig.update_layout(
title={
"text": "Corona Virus: Confirmed, Deaths & Recovered",
"y": 0.98,
"x": 0.5,
"xanchor": "center",
"yanchor": "top",
},
height=820,
legend_orientation="h",
# legend=dict(x=1, y=0.4),
xaxis1=dict(range=dt_range, autorange=False),
yaxis1=dict(range=[-10, hubei["Confirmed"].max() * 1.1], autorange=False),
xaxis2=dict(range=dt_range, autorange=False),
yaxis2=dict(range=[-10, rest_df["Confirmed"].max() * 1.1], autorange=False),
xaxis3=dict(range=dt_range, autorange=False),
yaxis3=dict(
range=[-10, np.max([china["Recovered"].max(), china["Deaths"].max()]) * 1.1],
autorange=False,
),
xaxis4=dict(range=dt_range, autorange=False),
yaxis4=dict(
range=[
-0.5,
np.max([rest_df["Recovered"].max(), rest_df["Deaths"].max()]) * 1.1,
],
autorange=False,
),
)
frames = [
dict(
name=str(dt),
data=[
frames_hubei[i],
frames_rchina[i],
frames_world[i],
frames_china_d[i],
frames_china_r[i],
frames_world_d[i],
frames_world_r[i],
],
traces=[0, 1, 2, 3, 4, 5, 6, 7],
)
for i, dt in enumerate(dates)
]
updatemenus = [
dict(
type="buttons",
buttons=[
dict(
label="Play",
method="animate",
args=[
[str(dt) for dt in dates[1:]],
dict(
frame=dict(duration=500, redraw=False),
transition=dict(duration=0),
easing="linear",
fromcurrent=True,
mode="immediate",
),
],
)
],
direction="left",
pad=dict(r=10, t=85),
showactive=True,
x=0.6,
y=-0.1,
xanchor="right",
yanchor="top",
)
]
sliders = [
{
"yanchor": "top",
"xanchor": "left",
"currentvalue": {
"font": {"size": 16},
"prefix": "Date: ",
"visible": True,
"xanchor": "right",
},
"transition": {"duration": 500.0, "easing": "linear"},
"pad": {"b": 10, "t": 50},
"len": 0.9,
"x": 0.1,
"y": -0.2,
"steps": [
{
"args": [
[str(dt)],
{
"frame": {
"duration": 500.0,
"easing": "linear",
"redraw": False,
},
"transition": {"duration": 0, "easing": "linear"},
},
],
"label": str(dt),
"method": "animate",
}
for dt in dates
],
}
]
fig.update(frames=frames),
fig.update_layout(updatemenus=updatemenus, sliders=sliders)
fig.show()
#
# # Tweets Analysis
# I tried to retrieve some tweets with the *coronavirus* hashtag during the last day, you can find the dataset among my inputs.
# The csv has already filtered out all the retweets.
# ### Load data and Clean Text
# Preprocess each tweet, removing some patterns as *http*, *https*, *@[..]* and others..
df_tweets = pd.read_csv("../input/tweets/nCoV_tweets.csv", index_col=0)
df_tweets.rename(columns={"txt": "tweets", "dt": "date"}, inplace=True)
import re
def tweet_parser(text, pattern_regex):
for pr in pattern_regex:
text = re.sub(pr, " ", text)
return text.strip()
pattern_regex = [
"\n",
"\t",
":",
",",
";",
"\.",
'"',
"''",
"@.*?\s+",
"RT.*?\s+",
"http.*?\s+",
"https.*?\s+",
]
df_tweets["tidy_tweets"] = df_tweets.apply(
lambda r: tweet_parser(r["tweets"], pattern_regex), axis=1
)
df_tweets["date"] = pd.to_datetime(df_tweets["date"]).dt.date
df_tweets.head()
# Let's use TextBlob library to infer tweet sentiments, and later categorize them into *Negative*, *Neutral* and *Positive*.
df_tweets["sentiment"] = df_tweets.apply(
lambda r: TextBlob(r["tidy_tweets"]).sentiment.polarity, axis=1
)
df_tweets["sent_adj"] = np.where(
df_tweets["sentiment"] < 0,
"Negative",
np.where(df_tweets["sentiment"] > 0, "Positive", "Neutral"),
)
df_tweets["sent_adj"] = df_tweets["sent_adj"].astype("category")
sizes = df_tweets.groupby("sent_adj").size()
df_tweets.head()
#
# ### Raw Sentiment Distribution and Adjusted Sentiment Histogram
fig = ff.create_distplot(
[df_tweets["sentiment"]],
group_labels=["sentiment"],
bin_size=[0.05],
colors=["indianred"],
)
fig.update_layout(
title={
"text": "Sentiment Distribution",
"y": 0.95,
"x": 0.5,
"xanchor": "center",
"yanchor": "top",
},
showlegend=False,
)
fig.show()
fig = go.Figure()
fig.add_trace(
go.Bar(
x=sizes.index,
y=sizes.values,
opacity=0.9,
text=sizes.values,
textposition="outside",
marker={"color": "indianred"},
)
)
fig.update_layout(
title={
"text": "Sentiment Adjusted Histogram",
"y": 0.95,
"x": 0.5,
"xanchor": "center",
"yanchor": "top",
},
showlegend=False,
xaxis_title_text="Sentiment",
yaxis_title_text="Count",
bargap=0.3,
)
fig.show()
#
# ### Tweets WordCloud
def render_wordcloud(df, sent="Positive"):
color = {"Positive": "Set2", "Negative": "RdGy", "Neutral": "Accent_r"}
words = " ".join([text for text in df.loc[df["sent_adj"] == sent, "tidy_tweets"]])
wordcloud = WordCloud(
width=800,
height=500,
background_color="black",
max_font_size=100,
relative_scaling=0.1,
colormap=color[sent],
).generate(words)
plt.figure(figsize=(14, 10))
plt.imshow(wordcloud, interpolation="bilinear")
plt.title(sent + " Wordcloud", fontsize=20)
plt.axis("off")
plt.show()
for s in ["Positive", "Negative", "Neutral"]:
render_wordcloud(df_tweets, s)
# At least at first sight, all wordclouds look pretty similar, and I cannot see huge differences in the words displayed.
# Let's see if there are differences among the hashtags used.
#
#
# ### Hashtag Analysis
# Let's check hashtags counts, both overall and split by sentiment. From the hashtags I am excluding the #coronavirus one, since it's my tweet research key.
def get_hashtag(series):
s = series.str.lower()
s = s.str.extractall(r"(\#\w*)")[0].value_counts()
return pd.DataFrame(data={"hashtag": s.index, "count": s.values})
def get_hashtag_by_sent(df):
d = {}
for s in df["sent_adj"].unique():
tmp = get_hashtag(df.loc[df["sent_adj"] == s, "tidy_tweets"])
d[s] = tmp[
(tmp["hashtag"].str.len() > 2)
& (~tmp["hashtag"].str.contains("coronavirus"))
]
return d
all_hashtag = get_hashtag(df_tweets["tidy_tweets"])
all_hashtag = all_hashtag[
(all_hashtag["hashtag"].str.len() > 2)
& (~all_hashtag["hashtag"].str.contains("coronavirus"))
]
d = get_hashtag_by_sent(df_tweets)
fig = make_subplots(
rows=2,
cols=3,
specs=[[{"colspan": 3}, None, None], [{}, {}, {}]],
subplot_titles=(
"Overall Most Frequent Hashtags",
"Positive Hashtags",
"Neutral Hashtags",
"Negative Hashtags",
),
)
fig.add_trace(
go.Bar(
x=all_hashtag.loc[:20, "hashtag"].to_numpy(),
y=all_hashtag.loc[:20, "count"].to_numpy(),
opacity=0.8,
orientation="v",
),
row=1,
col=1,
)
for i, k in enumerate(["Positive", "Negative", "Neutral"]):
fig.add_trace(
go.Bar(
x=d[k].loc[:10, "hashtag"].to_numpy(),
y=d[k].loc[:10, "count"].to_numpy(),
opacity=0.8,
orientation="v",
),
row=2,
col=i + 1,
)
fig.update_layout(
title={
"text": "Most Frequent Hashtags",
"y": 1,
"x": 0.5,
"xanchor": "center",
"yanchor": "top",
},
height=1000,
showlegend=False,
)
fig.show()
|
# # E-Commerce Case Study. Sales data analysis.
# **Importing the libraries**
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
# # **Reading the data using read_csv**
# **Checking the data using head(), tail(), shape & info()**
sales_data = pd.read_csv("/kaggle/input/e-commerce-sales-data/all_data.csv")
sales_data
sales_data.head(10)
sales_data.tail(10)
sales_data.info()
sales_data.shape
# # Data Preparation & Cleaning
# **After seeing the head, we can see one null value, so we will use is .isna().sum to get the total null values**
sales_data.isna().sum()
# **Here we can see there are 545 null values in all the 6 columns of the data.**
# **So we will drop all the null values, since 545 in 186k is negligible**
# **We will use dropna() function**
sales_data.dropna(inplace=True)
# inplace = True is used to make the changes permenent
sales_data.isnull().sum()
# here we can see all the 545 null values are removed permenently from the dataset.
# **Now we will check the shape of the data again**
sales_data.shape
# we can see now there are only 186305 rows
# # Finding out the best month for sales and visualizing it in pyplot.
# **For this we are going to create an extra column Month by using split function**
# here we can see the "order date" coloum with data like "09/01/19 22:14"
# here 09 is the month, this can be extracted from this using split
"09/01/19 22:14".split("/")[0] # here we are extracting 0th index after splitting
# **We will define a function and what ever oder date we receive, we will aply the above logic on it and retun the month.**
def Month(x):
return x.split("/")[0]
# **now we will use this funtion in " Order Date"**
sales_data["Month"] = sales_data["Order Date"].apply(Month)
sales_data.head()
# here we can see and extra column "month" extracted from the "Order date"
sales_data.head(5)
# **Now we will create a Sales/Month column**
# **here, sales is basically "Quantity Ordered" * "Price Each"**
# **So, we will check the data type of all the columns as we need mutiplications to get Sales/Month. The object data type needed to be converted to float and int for calculations**
sales_data.dtypes
# **Here we can see Quantity ordered, Price Each, Month, are in Object dtype. Need to convert them to int and float**
# while Trying to convert "Month" dtype to "int" using code, "sales_data["Month"].astype(int)" we got error,
# so we are cheching the unique values of "Month"
sales_data["Month"].unique()
# **We can see "Order Date" comming between the months, so we need to drop that value, in order to convert it to "int" Datatype**
# **We are using Filter operater to do this**
#
# Along with the logical operators AND and OR, there is also an operation called “negation” or ~ .It changes the logical meaning to the opposite. In programming, negation corresponds to the unary operator not:
filter = sales_data["Month"] == "Order Date"
sales_data = sales_data[~filter]
sales_data.head()
# **Now we can easily conver Month into Int datatpe**
sales_data["Month"] = sales_data["Month"].astype(int)
sales_data.dtypes
# **Now we will convert Quantity Ordered to int and Price Each to float datatype**
sales_data["Quantity Ordered"] = sales_data["Quantity Ordered"].astype(int)
sales_data["Price Each"] = sales_data["Price Each"].astype(float)
sales_data.dtypes
# here all the necessay column datatypes are converted to int and flaot for further use
# **Creating new column "Sales per Month"**
sales_data["Sales per Month"] = (
sales_data["Quantity Ordered"] * sales_data["Price Each"]
)
sales_data.head()
# **Now doing the analysis of best month for sales**
# using groupby function to analyse sum of month wise sales
sales_data.groupby("Month")["Sales per Month"].sum()
# **Visualizing the Month wise Sales using pyplot**
months = range(1, 13)
plt.bar(months, sales_data.groupby("Month")["Sales per Month"].sum())
plt.xticks(months)
plt.xlabel("Month")
plt.ylabel("Sales in USD")
# **We can conclude that december and october are the best months for sales**
# # Finding Out the city with Maximum Order
sales_data.head(3)
# **Since there is no column with City, we need to extract the city from the "purchase Address" Coloum**
# **Here we are using the split function to extract the data from Address**
"917 1st St, Dallas, TX 75001".split(",")[1]
# we wil get splited values in form of list
# defining a function City and returning the index 1 of splitted list to it and saving it in a column in Sales_data as "City"
def city(x):
return x.split(",")[1]
sales_data["City"] = sales_data["Purchase Address"].apply(city)
sales_data.head(5)
# **Using Groupby on the basis of City, and counting it based on the number of orders**
sales_data.groupby("City")["City"].count()
# we can see the cities with their maximum orders
# **Plotting the data in barchart**
sales_data.groupby("City")["City"].count().plot.bar()
plt.ylabel("No of Orders")
|
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
import seaborn as sns
from sklearn.linear_model import LinearRegression
from sklearn.model_selection import train_test_split
from sklearn.metrics import mean_squared_error, r2_score
from sklearn.linear_model import ElasticNet
from sklearn.linear_model import Ridge, Lasso
df = pd.read_csv("/kaggle/input/kc-housesales-data/kc_house_data.csv")
df.head()
df.info()
df.tail()
df.corr() # Pandas dataframe.corr(), Python'daki Pandas Dataframe'deki tüm sütunların ikili korelasyonunu bulmak için kullanılır
df.describe() # DataFrame’in özet raporunu almaya yarar
df.shape
df.isnull().sum()
abs(
df.corr()["price"].sort_values(ascending=False)
) # Bu kod, bir veri çerçevesinde ("df") bulunan "price" sütununun diğer sütunlarla olan korelasyonunu hesaplar ve bu korelasyonları mutlak değerlerine göre büyükten küçüğe doğru sıralar.ascending false ile en yuksek deger en basa gelir
sns.countplot(
df["bedrooms"]
) # Bu kod, seaborn (sns) veri görselleştirme kütüphanesi kullanılarak bir veri çerçevesindeki ("df") "bedrooms" sütunundaki verilerin sayısını görselleştirir. "countplot" fonksiyonu, her farklı değere sahip olan öğelerin sayısını gösteren bir çubuk grafik oluşturur.
sns.displot(
df["price"].sort_values()
) # Bu kod, seaborn (sns) veri görselleştirme kütüphanesi kullanılarak bir veri çerçevesindeki ("df") "price" sütunundaki verilerin dağılımını görselleştirir. "displot" fonksiyonu, bir veri setinin dağılımını göstermek için kullanılan bir histogram grafiği oluşturur.
sns.displot(
df["sqft_living"]
) ##Bu kod, seaborn (sns) veri görselleştirme kütüphanesi kullanılarak bir veri çerçevesindeki ("df") "sqft_living" sütunundaki verilerin dağılımını görselleştirir. "displot" fonksiyonu, bir veri setinin dağılımını göstermek için kullanılan bir histogram grafiği oluşturur.
sns.countplot(df["bedrooms"])
plt.xticks(
rotation=90
) # Bu kod, matplotlib (plt) veri görselleştirme kütüphanesi kullanılarak bir çizgi grafiğindeki x eksenindeki etiketlerin yönünü değiştirir. "xticks" fonksiyonu, x eksenindeki etiketlerin ayarlanmasına yardımcı olur. "rotation" parametresi, etiketlerin yönünü belirler ve derece cinsinden açı değeri alır.
sns.displot(df["sqft_above"]) # Aynı şiyi farklı stunda denemiş mal ayla
sns.scatterplot(
x="sqft_living", y="price", data=df, color="purple"
) # Bu kodda, "sqft_living" ve "price" değişkenleri arasındaki ilişkiyi gösteren bir dağılım grafiği oluşturulur.
sns.scatterplot(
x="sqft_basement", y="price", data=df, color="orange"
) # Yukardakinin aynısı farklı tablolar
sns.scatterplot(
x="lat", y="price", data=df, color="green"
) # Yukardakinin aynısı farklı tablolar
plt.figure(figsize=(8, 6))
sns.boxplot(
x="waterfront", y="price", data=df
) # "boxplot" fonksiyonu, bir veri setinin dağılımını göstermek için kullanılan bir grafiğidir. Bu grafiği kullanarak, bir kategorik değişkenin farklı değerleri için sayısal bir değişkenin dağılımını ve istatistiksel özelliklerini (minimum, maksimum, medyan, çeyreklikler, aykırı değerler vb.) görselleştirebiliriz.
sns.boxplot(df["sqft_living"]) # Yukardakinin aynısı farklı tablolar
sns.boxplot(df["sqft_living15"]) # Yukardakinin aynısı farklı tablolar
sns.boxplot(df["sqft_basement"]) # Yukardakinin aynısı farklı tablolar
plt.subplots(figsize=(14, 12))
sns.heatmap(
df.corr(), annot=True
) # Bu grafikte, sayısal veriler arasındaki korelasyon değerleri, renk skalası kullanılarak görselleştirilir. Pozitif korelasyonlar daha koyu renklerle, negatif korelasyonlar daha açık renklerle gösterilir.
# "annot=True" parametresi, grafiğin içine korelasyon değerlerini yazdırmak için kullanılır.
x = df[
[
"bedrooms",
"bathrooms",
"sqft_living",
"floors",
"grade",
"waterfront",
"view",
"sqft_above",
"lat",
"sqft_living15",
"sqft_basement",
]
] # bu stunların olduğu tablo olustur
x.head()
x.describe()
y = df[["price"]]
y.shape
y.head()
df["zipcode"] = df["zipcode"].astype("category")
x = pd.get_dummies(
x, drop_first=True
) # Bu kodlar, veri çerçevesinde ("df") yer alan "zipcode" sütununu kategorik bir değişkene dönüştürür ve "get_dummies" fonksiyonu kullanılarak kategorik değişkenin ikili (binary) kodlamasını yapar.İlk satırda, "astype" fonksiyonu kullanılarak "zipcode" sütunu kategorik bir değişkene dönüştürülür.
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.20, random_state=13
)
x_train.shape, x_test.shape, y_train.shape, y_test.shape
lr = (
LinearRegression()
) # İlk satırda, "LinearRegression()" sınıfı bir doğrusal regresyon modeli nesnesi oluşturulur ve "lr" değişkenine atılır.
regmodel = lr.fit(
x_train, y_train
) # İkinci satırda, "fit" fonksiyonu kullanılarak oluşturulan doğrusal regresyon modeli, eğitim verileri olan "x_train" ve "y_train" verilerine uygulanır. Bu işlem, modelin eğitim verilerindeki bağımlı değişken ("y_train") ile bağımsız değişkenlerin ("x_train") arasındaki ilişkiyi öğrenmesini sağlar.
y_pretest = regmodel.predict(
x_test
) # "predict" fonksiyonu, doğrusal regresyon modeli tarafından öğrenilen katsayıları ve özellikle "x_test" verilerini kullanarak, bağımlı değişken ("y") için tahminler yapar
y_pretest
from sklearn.metrics import mean_squared_error, r2_score
(mean_squared_error)(
y_test, y_pretest
) ** 0.5 # Bu kod, "mean_squared_error" fonksiyonunu kullanarak, test verilerindeki gerçek bağımlı değişken ("y_test") değerleri ile tahmin edilen bağımlı değişken ("y_pretest") değerleri arasındaki hata karelerinin ortalamasını hesaplar. Ardından, bu değerin karekökünü alarak, hata değerlerinin ortalama mutlak hatasını hesaplar
r2_score(
y_test, y_pretest
) # Bu kod, "r2_score" fonksiyonunu kullanarak, test verilerindeki gerçek bağımlı değişken ("y_test") değerleri ile tahmin edilen bağımlı değişken ("y_pretest") değerleri arasındaki ilişkiyi (determinasyon katsayısı) ölçer.
df.describe()
df = df[
df["bedrooms"] < 10
] # Bu kod, "df" veri çerçevesindeki "bedrooms" sütunundaki değerleri 10'dan küçük olan satırların sadece dahil edilmesini sağlar ve veri çerçevesindeki diğer tüm satırları atar.
df = df[df["bathrooms"] < 10]
df = df[df["sqft_living"] < 4000]
df = df[df["sqft_living15"] < 3500]
df = df[df["sqft_above"] < 4000]
df = df[df["view"] == 0]
df = df[df["waterfront"] == 0] # Yukardaki aynısı
df["bedrooms"] = (
df["bedrooms"] ** 2
) # Bu kod, "df" veri çerçevesindeki "bedrooms" sütunundaki tüm değerleri karesine yükseltir ve "bedrooms" sütununu günceller.
df["bathrooms"] = df["bathrooms"] ** 2
df["grade"] = df["grade"] / 2
df["age"] = 2021 - df["yr_built"]
df["sqft"] = (
df["sqft_living"] + df["sqft_living15"] + df["sqft_basement"] + df["sqft_above"]
) # Yukardakinin farklı versiyonu
df["sqft"] = df["sqft"] ** 2
x, y = (
df[
[
"bedrooms",
"bathrooms",
"sqft",
"floors",
"grade",
"waterfront",
"view",
"lat",
"zipcode",
"age",
]
],
df[["price"]],
) # dataframe oluştur seçili stunlardan
x = pd.get_dummies(
x, drop_first=True
) # Bu kod, "x" veri çerçevesindeki kategorik değişkenleri (örneğin, "zipcode") ikili değişkenlere dönüştürür ve her kategorik değişken için ikili sütunlar oluşturur. "drop_first=True" parametresi, her değişken için ilk sütunu düşürerek çoklu doğrusal bağımsızlık durumunu önler.
x.head()
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.20, random_state=13
)
lr = LinearRegression()
regmodel = lr.fit(x_train, y_train)
y_pretest = regmodel.predict(x_test) # Üst kısımlarda yazdım
y_pretest
(mean_squared_error)(y_test, y_pretest) ** 0.5 # Üst kısımlarda yazdım
r2_score(y_test, y_pretest)
# Üst kısımlarda yazdım
R = Ridge()
R.fit(
x_train, y_train
) # Bu kodlar, Ridge regresyon modelinin oluşturulmasını ve eğitilmesini sağlar. "x_train" ve "y_train" verileri kullanılarak Ridge modeli eğitilir ve ardından "x_test" verilerine göre tahminler yapılır. Bu tahminler, "y_pred_ridge" adlı değişkende saklanır.
y_pred_ridge = R.predict(x_test)
y_pred_ridge
(mean_squared_error)(y_test, y_pred_ridge) ** 0.5 # Üst kısımlarda yazdım
r2_score(y_test, y_pred_ridge) # Üst kısımlarda yazdım
L = Lasso()
L.fit(
x_train, y_train
) # Bu kodlar, Lasso regresyon modelinin oluşturulmasını ve eğitilmesini sağlar. "x_train" ve "y_train" verileri kullanılarak Lasso modeli eğitilir ve ardından "x_test" verilerine göre tahminler yapılır. Bu tahminler, "y_pred_lasso" adlı değişkende saklanır.
y_pred_lasso = L.predict(x_test)
(mean_squared_error)(y_test, y_pred_lasso) ** 0.5 # Üst kısımlarda yazdım
r2_score(y_test, y_pred_lasso) # Üst kısımlarda yazdım
E = ElasticNet(l1_ratio=0.8)
E.fit(
x_train, y_train
) # Bu kodlar, Elastic Net regresyon modelinin oluşturulmasını ve eğitilmesini sağlar. "x_train" ve "y_train" verileri kullanılarak Elastic Net modeli eğitilir ve ardından "x_test" verilerine göre tahminler yapılır. Bu tahminler, "y_pred_elastic" adlı değişkende saklanır.
y_pred_elastic = E.predict(x_test)
(mean_squared_error)(y_test, y_pred_elastic) ** 0.5 # Üst kısımlarda yazdım
r2_score(y_test, y_pred_elastic) # Üst kısımlarda yazdım
df.describe() # Üst kısımlarda yazdım
|
# # First of all, let me make things clear.
# KAGGLE IS A GOOSE, NOT A CHICKEN!!
from transformers import *
import numpy as np
tokenizer = BertTokenizer.from_pretrained("bert-base-uncased")
bert_model = TFBertForMaskedLM.from_pretrained("bert-base-uncased")
# DEFINE SENTENCE
str = "[CLS] Data science is [MASK] . [SEP]"
indices = tokenizer.encode(str, add_special_tokens=False, return_tensors="tf")
# PREDICT MISSING WORDS
pred = bert_model(indices)
masked_indices = np.where(indices == 103)[1]
# DISPLAY MISSING WORDS
predicted_words = np.argmax(np.asarray(pred[0][0])[masked_indices, :], axis=1)
print(tokenizer.decode(predicted_words))
str = "[CLS] BERT is the [MASK] . [SEP]"
indices = tokenizer.encode(str, add_special_tokens=False, return_tensors="tf")
# PREDICT MISSING WORDS
pred = bert_model(indices)
masked_indices = np.where(indices == 103)[1]
# DISPLAY MISSING WORDS
predicted_words = np.argmax(np.asarray(pred[0][0])[masked_indices, :], axis=1)
print(tokenizer.decode(predicted_words))
str = "[CLS] Apple really [MASK] good those [MASK] . [SEP]"
indices = tokenizer.encode(str, add_special_tokens=False, return_tensors="tf")
# PREDICT MISSING WORDS
pred = bert_model(indices)
masked_indices = np.where(indices == 103)[1]
# DISPLAY MISSING WORDS
predicted_words = np.argmax(np.asarray(pred[0][0])[masked_indices, :], axis=1)
print(tokenizer.decode(predicted_words))
str = "[CLS] Kaggle is definitely not [MASK] . [SEP]"
indices = tokenizer.encode(str, add_special_tokens=False, return_tensors="tf")
# PREDICT MISSING WORDS
pred = bert_model(indices)
masked_indices = np.where(indices == 103)[1]
# DISPLAY MISSING WORDS
predicted_words = np.argmax(np.asarray(pred[0][0])[masked_indices, :], axis=1)
print(tokenizer.decode(predicted_words))
str = "[CLS] Google will dominate the [MASK] . [SEP]"
indices = tokenizer.encode(str, add_special_tokens=False, return_tensors="tf")
# PREDICT MISSING WORDS
pred = bert_model(indices)
masked_indices = np.where(indices == 103)[1]
# DISPLAY MISSING WORDS
predicted_words = np.argmax(np.asarray(pred[0][0])[masked_indices, :], axis=1)
print(tokenizer.decode(predicted_words))
str = "[CLS] Microsoft will languish in the [MASK] . [SEP]"
indices = tokenizer.encode(str, add_special_tokens=False, return_tensors="tf")
# PREDICT MISSING WORDS
pred = bert_model(indices)
masked_indices = np.where(indices == 103)[1]
# DISPLAY MISSING WORDS
predicted_words = np.argmax(np.asarray(pred[0][0])[masked_indices, :], axis=1)
print(tokenizer.decode(predicted_words))
str = "[CLS] The king of NLP will always [MASK] [MASK] . [SEP]"
indices = tokenizer.encode(str, add_special_tokens=False, return_tensors="tf")
# PREDICT MISSING WORDS
pred = bert_model(indices)
masked_indices = np.where(indices == 103)[1]
# DISPLAY MISSING WORDS
predicted_words = np.argmax(np.asarray(pred[0][0])[masked_indices, :], axis=1)
print(tokenizer.decode(predicted_words))
str = "[CLS] Kaggle is [MASK] . [SEP]"
indices = tokenizer.encode(str, add_special_tokens=False, return_tensors="tf")
# PREDICT MISSING WORDS
pred = bert_model(indices)
masked_indices = np.where(indices == 103)[1]
# DISPLAY MISSING WORDS
predicted_words = np.argmax(np.asarray(pred[0][0])[masked_indices, :], axis=1)
print(tokenizer.decode(predicted_words))
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
train = pd.read_json("/kaggle/input/ml1920-whats-cooking/cooking_train.json")
print("recipes: ", train.shape[0], "\n")
print("cuisines:\n", sorted(set(train.cuisine)), "\n")
print("Head:\n", train.head(), "\n")
print("Sample Ingredients:\n", train.ingredients[:10])
print(train.shape)
from sklearn.feature_extraction.text import CountVectorizer
def preprocessor(line):
tabbed = " ".join(line).lower()
return tabbed
recipes = train.ingredients[:4]
for r in recipes:
print(preprocessor(r))
print()
from sklearn.preprocessing import PolynomialFeatures
from sklearn.feature_extraction.text import TfidfVectorizer
vect = TfidfVectorizer(preprocessor=preprocessor, strip_accents="unicode")
data = pd.DataFrame(
data=vect.fit_transform(train.ingredients).todense(),
columns=sorted(vect.vocabulary_),
)
data.head()
cuisine_occurances = train.cuisine.value_counts()
cuisine_occurances.plot.bar(figsize=(20, 7), title="Cuisines' occurances in train data")
cuisine = train.cuisine
cuisine.rename("original_cuisine", inplace=True)
data = pd.concat([data, cuisine], axis=1)
data = data.groupby("original_cuisine").agg(np.mean)
def get_freq(row):
return row[row["most_frequent"]]
data["most_frequent"] = data.idxmax(axis=1)
data["freq"] = data.apply(get_freq, axis=1)
data[["most_frequent", "freq"]]
labels = sorted(train["cuisine"].unique())
freqs = data.freq.to_numpy()
x = np.arange(len(labels)) # the label locations
width = 0.8 # the width of the bars
fig, ax = plt.subplots(figsize=(20, 7), dpi=256)
rects1 = ax.bar(x, freqs, width)
# Add some text for labels, title and custom x-axis tick labels
ax.set_ylabel("Frequence")
ax.set_title("Most frequent words for each cuisine (according to tfidf formula)")
ax.set_xticks(x)
ax.set_xticklabels(labels)
def autolabel(rects):
"""Attach a text label above each bar in *rects*, displaying connected word"""
for i in range(0, len(rects)):
height = data.most_frequent.to_numpy()[i]
rect = rects[i]
ax.annotate(height, xy=(rect.get_x(), rect.get_height() * 1.02))
autolabel(rects1)
fig.tight_layout()
plt.show()
from nltk.stem.snowball import EnglishStemmer
from nltk.stem import WordNetLemmatizer
stemmer = EnglishStemmer()
lemmatizer = WordNetLemmatizer()
count_analyzer = CountVectorizer().build_analyzer()
def stem(doc):
line = str.lower(" ".join(doc))
return (stemmer.stem(w) for w in count_analyzer(line))
def lemmatize(doc):
line = str.lower(" ".join(doc))
return (lemmatizer.lemmatize(w) for w in count_analyzer(line))
stopwords = [
"a",
"the",
"in",
"of",
"off",
"up",
"down",
"fresh",
"virgin",
"sliced",
"leaf",
"leaves",
"chopped",
"cooked",
"baby",
"yellow",
"red",
"blue",
"white",
"black",
"purple",
"violet",
"over",
]
from sklearn.svm import LinearSVC
from sklearn.model_selection import cross_val_score
from sklearn.preprocessing import PolynomialFeatures
from sklearn.pipeline import Pipeline
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.feature_extraction.text import HashingVectorizer
from sklearn.feature_extraction.text import TfidfTransformer
X_train = train["ingredients"]
y_train = train["cuisine"]
vectorization = Pipeline(
[
(
"vectorizer",
CountVectorizer(
analyzer=lemmatize, strip_accents="unicode", stop_words=stopwords
),
),
("tfidf", TfidfTransformer()),
]
)
preprocessing = Pipeline(
[("vectorization", vectorization), ("poly", PolynomialFeatures(degree=2))]
)
pipeline = Pipeline(
[
("preprocessing", preprocessing),
("model", LinearSVC(random_state=0, C=1.2, max_iter=4000)),
]
)
scores = cross_val_score(pipeline, X_train, y_train, cv=5)
print(np.mean(scores))
ngrams_pipeline = Pipeline(
[
(
"vectorizer",
TfidfVectorizer(analyzer=stem, stop_words="english", ngram_range=(1, 2)),
),
("model", LinearSVC(random_state=0, C=1, max_iter=4000)),
]
)
print(np.mean(cross_val_score(ngrams_pipeline, X_train, y_train, cv=5)))
from sklearn.ensemble import ExtraTreesClassifier
extra_trees_pipeline = Pipeline(
[
("preprocessing", vectorization),
("model", ExtraTreesClassifier(n_estimators=200, random_state=0)),
]
)
scores = cross_val_score(extra_trees_pipeline, X_train, y_train, cv=5)
print(np.mean(scores))
from sklearn.ensemble import RandomForestClassifier
rf = RandomForestClassifier(n_estimators=200, random_state=0)
rf_pipeline = Pipeline([("preprocessing", vectorization), ("model", rf)])
scores = cross_val_score(rf_pipeline, X_train, y_train, cv=5)
print(np.mean(scores))
from sklearn.ensemble import VotingClassifier
from sklearn.ensemble import ExtraTreesClassifier
vc = VotingClassifier(
estimators=[
(
"linearSVC",
Pipeline(
[
("poly", PolynomialFeatures(degree=2)),
("svc", LinearSVC(random_state=0, C=1, max_iter=4000)),
]
),
),
("extraTrees", ExtraTreesClassifier(n_estimators=150, random_state=0)),
("rf", RandomForestClassifier(n_estimators=150, random_state=0)),
],
voting="hard",
)
vc_pipeline = Pipeline([("preprocessing", vectorization), ("model", vc)])
scores = cross_val_score(vc_pipeline, X_train, y_train, cv=5)
print(np.mean(scores))
test = pd.read_json("/kaggle/input/ml1920-whats-cooking/cooking_test.json")
X_test = test["ingredients"]
pipeline.fit(X_train, y_train)
prediction = pipeline.predict(X_test)
submission = test.copy()
submission["cuisine"] = prediction
submission.to_csv("et_submission.csv", index=False, columns=["id", "cuisine"])
|
# Original Codes from :
# https://www.kaggle.com/fatmakursun/titanic-classification-regression
# https://www.kaggle.com/startupsci/titanic-data-science-solutions
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
print(os.getcwd())
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import time
import os
from IPython.display import clear_output
import tensorflow as tf
print("tensorflow version: {}".format(tf.__version__))
print("GPU 사용 가능 여부: {}".format(tf.test.is_gpu_available()))
print(tf.config.list_physical_devices("GPU"))
train_data = pd.read_csv("../input/titanic/train.csv")
sns.heatmap(train_data.isnull(), yticklabels=False, cbar=False, cmap="viridis")
train_data.isnull().sum().sort_values(ascending=False)
sns.set_style("whitegrid")
sns.countplot(x="Survived", data=train_data, palette="RdBu_r")
sns.set_style("whitegrid")
sns.countplot(x="Survived", hue="Sex", data=train_data, palette="RdBu_r")
sns.set_style("whitegrid")
sns.countplot(x="Survived", hue="Pclass", data=train_data, palette="rainbow")
train_data["Age"].hist(bins=30, color="darkred", alpha=0.7)
sns.countplot(x="SibSp", data=train_data)
sns.countplot(x="Parch", data=train_data)
train_data["Fare"].hist(color="green", bins=40, figsize=(8, 4))
plt.figure(figsize=(12, 7))
sns.boxplot(x="Pclass", y="Age", data=train_data, palette="winter")
train_data[["Pclass", "Age"]].groupby(["Pclass"], as_index=False).mean().sort_values(
by="Age", ascending=False
)
def impute_age(cols):
Age = cols[0]
Pclass = cols[1]
if pd.isnull(Age):
if Pclass == 1:
return 38
elif Pclass == 2:
return 30
else:
return 25
else:
return Age
train_data["Age"] = train_data[["Age", "Pclass"]].apply(impute_age, axis=1)
sns.heatmap(train_data.isnull(), yticklabels=False, cbar=False, cmap="viridis")
sns.countplot(x="Embarked", data=train_data)
train_data["Embarked"] = train_data["Embarked"].fillna("S")
sns.heatmap(train_data.isnull(), yticklabels=False, cbar=False, cmap="viridis")
train_data.isnull().sum().sort_values(ascending=False)
train_data.drop(["Name"], axis=1)
train_data["Sex"] = train_data["Sex"].map({"female": 1, "male": 0}).astype(int)
train_data = train_data.drop(["Name", "Ticket", "Cabin"], axis=1)
train_data.info()
train_data["Embarked"] = (
train_data["Embarked"].map({"S": 0, "C": 1, "Q": 2}).astype(int)
)
train_data.head()
def make_dataset(pandas_data):
# 입력, 타겟(라벨) 데이터 생성
sequences = pandas_data.drop(["PassengerId", "Survived"], axis=1)
labels = pandas_data["Survived"]
return np.array(sequences), np.array(labels)
train_sequences, train_labels = make_dataset(train_data)
print(train_sequences)
print(len(train_sequences))
# Input pipeline 만들기
# tf.data.Dadaset 포멧의 데이터로 변환
print(train_sequences, train_sequences.shape, len(train_sequences))
print(train_labels.shape)
batch_size = 6
max_epochs = 130
learning_rate = 0.00001
num_classes = 2
BUFFER_SIZE = len(train_sequences)
train_dataset = tf.data.Dataset.from_tensor_slices((train_sequences, train_labels))
train_dataset = train_dataset.shuffle(BUFFER_SIZE)
train_dataset = train_dataset.batch(128)
print(train_dataset)
class DNNModel(tf.keras.Model):
def __init__(self, num_classes):
super(DNNModel, self).__init__()
## 코드 시작 ##
self.dense1 = tf.keras.layers.Dense(512, activation="relu")
self.bn1 = tf.keras.layers.BatchNormalization()
# self.relu1 = tf.keras.layers.Activation(tf.keras.activations.relu)
self.relu1 = tf.keras.layers.ReLU()
self.dense2 = tf.keras.layers.Dense(200, activation="relu")
self.bn2 = tf.keras.layers.BatchNormalization()
self.relu2 = tf.keras.layers.ReLU()
self.dense3 = tf.keras.layers.Dense(10, activation="relu")
self.bn3 = tf.keras.layers.BatchNormalization()
self.relu3 = tf.keras.layers.ReLU()
self.dense4 = tf.keras.layers.Dense(num_classes)
## 코드 종료 ##
def call(self, inputs, training=False):
"""Run the model."""
## 코드 시작 ##
dense1_out = self.dense1(inputs)
bn1_out = self.bn1(dense1_out, training=training)
relu1_out = self.relu1(bn1_out)
dense2_out = self.dense2(relu1_out)
bn2_out = self.bn2(dense2_out, training=training)
relu2_out = self.relu2(bn2_out)
dense3_out = self.dense2(relu2_out)
bn3_out = self.bn2(dense3_out, training=training)
relu3_out = self.relu2(bn3_out)
dense4_out = self.dense3(relu3_out)
## 코드 종료 ##
return dense4_out
model = DNNModel(num_classes)
for person, labels in train_dataset.take(1):
print(person[0:3])
print("predictions: ", model(person[0:3]))
model.summary()
loss_object = tf.keras.losses.CategoricalCrossentropy()
optimizer = tf.keras.optimizers.Adam(learning_rate=learning_rate)
mean_accuracy = tf.keras.metrics.Accuracy("accuracy")
mean_loss = tf.keras.metrics.Mean("loss")
mean_accuracy_ex = tf.keras.metrics.Accuracy("example_accuracy")
print(mean_accuracy_ex([0, 1, 2, 4], [0, 1, 3, 4])) # 0.75 = 3/4
print(mean_accuracy_ex([5, 1, 7, 3], [5, 0, 2, 7])) # 0.5 = 4/8 위의 계산에서 부터 누적
mean_ex = tf.keras.metrics.Mean("example_mean")
# mean(values) 형태로 사용합니다.
print(mean_ex([0, 1, 2, 3])) # 1.5 = (0 + 1 + 2 + 3) / 4
print(mean_ex([4, 5])) # 2.5 = (0 + 1 + 2 + 3 + 4 + 5) / 6
num_batches_per_epoch = len(list(train_dataset))
for epoch in range(max_epochs):
for step, (persons, labels) in enumerate(train_dataset):
start_time = time.time()
with tf.GradientTape() as tape:
## 코드 시작 ##
predictions = model(persons) # 위의 설명 1. 을 참고하여 None을 채우세요.
labels_onehot = tf.one_hot(labels, num_classes)
loss_value = loss_object(
y_true=labels_onehot, y_pred=predictions
) # 위의 설명 2. 를 참고하여 None을 채우세요.
gradients = tape.gradient(
loss_value, model.trainable_variables
) # 위의 설명 3. 을 참고하여 None을 채우세요.
optimizer.apply_gradients(
zip(gradients, model.trainable_variables)
) # 위의 설명 4. 를 참고하여 None을 채우세요.
## 코드 종료 ##
mean_loss(loss_value)
mean_accuracy(labels, tf.argmax(predictions, axis=1))
if (step + 1) % 5 == 0:
# clear_output(wait=True)
epochs = epoch + step / float(num_batches_per_epoch)
duration = time.time() - start_time
examples_per_sec = batch_size / float(duration)
print(
"Epoch [{}/{}], Step [{}/{}], Loss: {:.4f}, Accuracy: {:.2f}%, ({:.2f} examples/sec; {:.3f} sec/batch)".format(
epoch + 1,
max_epochs,
step + 1,
num_batches_per_epoch,
mean_loss.result(),
mean_accuracy.result() * 100,
examples_per_sec,
duration,
)
)
# clear the history
mean_loss.reset_states()
mean_accuracy.reset_states()
print("training done!")
test_batch_size = 25
batch_index = np.random.choice(len(train_data), size=test_batch_size, replace=False)
batch_xs = train_sequences[batch_index]
batch_ys = train_labels[batch_index]
y_pred_ = model(batch_xs, training=False)
print(batch_ys[:])
print(y_pred_[:, :])
def save_model(model, epoch, train_dir):
model_name = "my_model_" + str(epoch)
model.save_weights(os.path.join(train_dir, model_name))
model.save_weights(os.path.join("./model_checkpoint/", "my_model_loss_0.3824"))
test_data = pd.read_csv("../input/titanic/test.csv")
test_data["Age"] = test_data[["Age", "Pclass"]].apply(impute_age, axis=1)
test_data["Embarked"] = test_data["Embarked"].fillna("S")
test_data.drop(["Name"], axis=1)
test_data["Sex"] = test_data["Sex"].map({"female": 1, "male": 0}).astype(int)
test_data = test_data.drop(["Name", "Ticket", "Cabin"], axis=1)
test_data["Embarked"] = test_data["Embarked"].map({"S": 0, "C": 1, "Q": 2}).astype(int)
test_data = test_data.drop(["PassengerId"], axis=1)
test_input = np.array(test_data)
test_result = model(test_input)
test_result_merge = np.zeros((1309 - 892 + 1, 2))
for i, j in zip(range(0, (1309 - 892 + 1)), range(892, 1309 + 1)):
test_result_merge[i, 0] = j
test_result_merge[i, 1] = np.argmax(test_result[i])
test_result_merge = test_result_merge.astype(int)
test_result_pandas = pd.DataFrame(
test_result_merge, columns=["PassengerId", "Survived"]
)
print(test_result_pandas)
test_result_pandas.to_csv("predictions_loss_03769.csv", index=False)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
def smooze_output(x):
# ここをかきかえてね ひきすうにしてもいいかも
div_num = 90
nums = []
one_cut = 1 / div_num
z = 0
while z < 1:
nums.append(z)
z = z + one_cut
nums.append(1)
for i in range(len(nums)):
if x <= nums[i]:
if i == 0:
x = 0
break
diff_first = abs(x - nums[i]) # i and i-1 is compared(i is bigger)
diff_second = abs(x - nums[i - 1])
if diff_first >= diff_second:
x = nums[i - 1]
break
if diff_second >= diff_first:
x = nums[i]
break
return x
print(smooze_output(0.234))
print(smooze_output(0.223))
df = pd.DataFrame(
{"a": [1, 0.2, 0.3], "b": [0.5, 0.3423, 0.15], "c": [0.545, 0.3434, 0.3423]},
index=["A1", "B1", "C1"],
)
df.applymap(smooze_output)
df.head
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
import pandas as pd
import seaborn as sns
import numpy as np
import matplotlib.pyplot as plt
train = pd.read_csv(
"/kaggle/input/house-prices-advanced-regression-techniques/train.csv"
)
test = pd.read_csv("/kaggle/input/house-prices-advanced-regression-techniques/test.csv")
train.describe()
y_train = train["SalePrice"]
x_train = train.drop("SalePrice", axis=1)
# x_train = train.drop("LotFrontage",axis=1)
x_train = train.drop("Id", axis=1)
x_train.dtypes.sample(10)
plt.hist(np.log(train["LotFrontage"]))
mean_LotFrontage = x_train["LotFrontage"].mean()
std_LotFrontage = x_train["LotFrontage"].std()
x_train.fillna(
np.random.randint(
mean_LotFrontage - std_LotFrontage, mean_LotFrontage + std_LotFrontage
),
inplace=True,
)
x_train["LotFrontage"] = np.log(x_train["LotFrontage"])
x_train["LotArea"] = np.log(x_train["LotArea"])
one_hot_encoded_training_predictors = pd.get_dummies(x_train)
one_hot_encoded_training_predictors
one_hot_encoded_training_predictors
test = pd.read_csv("/kaggle/input/house-prices-advanced-regression-techniques/test.csv")
id = test["Id"]
test = test.drop("Id", axis=1)
# plt.hist(train["LotFrontage"])
mean_LotFrontage = test["LotFrontage"].mean()
std_LotFrontage = test["LotFrontage"].std()
test.fillna(
np.random.randint(
mean_LotFrontage - std_LotFrontage, mean_LotFrontage + std_LotFrontage
),
inplace=True,
)
test["logLotFrontage"] = np.log(test["LotFrontage"])
test["LotArea"] = np.log(test["LotArea"])
one_hot_test = pd.get_dummies(test)
one_hot_test
from sklearn.decomposition import PCA
from sklearn.preprocessing import StandardScaler
pca = PCA(n_components=2)
sc = StandardScaler()
X_train_pca = pca.fit_transform(one_hot_encoded_training_predictors)
X_test_pca = pca.fit_transform(one_hot_test)
data_train_std = sc.fit_transform(X_train_pca)
data_test_std = sc.fit_transform(X_test_pca)
from sklearn.linear_model import LogisticRegression
clf = LogisticRegression(penalty="l2", solver="sag", random_state=0)
clf.fit(data_train_std, y_train)
clf.predict(data_test_std)
from sklearn.ensemble import RandomForestRegressor
regr = RandomForestRegressor(n_estimators=1000, random_state=0, n_jobs=1)
regr.fit(data_train_std, y_train)
print(regr.predict(data_test_std))
predict = regr.predict(data_test_std)
plt.hist(np.log(regr.predict(data_test_std)))
output = pd.DataFrame({"Id": id, "SalePrice": predict})
output.to_csv("submission.csv", index=False)
|
import numpy as np
import pandas as pd
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
import matplotlib.pyplot as plt
import seaborn as sns
import plotly.graph_objects as go
import warnings
warnings.filterwarnings("ignore")
from scipy.stats import shapiro
from sklearn.preprocessing import StandardScaler
from sklearn.utils import resample
import xgboost as xgb
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import (
accuracy_score,
confusion_matrix,
classification_report,
roc_auc_score,
)
from sklearn.model_selection import GridSearchCV
from scipy.stats import chi2_contingency
from sklearn.model_selection import RepeatedStratifiedKFold, KFold, StratifiedKFold
import statsmodels.api as sm
from sklearn.base import BaseEstimator, ClassifierMixin
from sklearn.pipeline import Pipeline
from sklearn.metrics import make_scorer
from sklearn.metrics import log_loss
from sklearn.model_selection import cross_val_predict
df_ts = pd.read_csv("/kaggle/input/playground-series-s3e12/test.csv")
df_tr = pd.read_csv("/kaggle/input/playground-series-s3e12/train.csv")
# # Background:
# Literature review shows top features for stone ML prediction include saturations of uric acid, calcium phosphate, and urinary ammonium. LR used in other models to predict stone type classification with success. Osmolality found as a predictor with pediatric population -> Lower osmo consistent with "stone forming" children. 2-3000 mOsm/kg H20. For adults, there is some evidence that >501 and >577 mmol/kg is associated with risk of calcium stone formation. pH identified as good predictor but not the best. One study found specific gravity as a "good" predictor using null hypothesis testing. Urea and Calcuim may be good predictors for specific stone type formation prediction, but may help with binary classification as well.
# Abraham A, Kavoussi NL, Sui W, Bejan C, Capra JA, Hsi R. Machine Learning Prediction of Kidney Stone Composition Using Electronic Health Record-Derived Features. J Endourol. 2022 Feb;36(2):243-250. doi: 10.1089/end.2021.0211. PMID: 34314237; PMCID: PMC8861926.
# Kavoussi NL, Floyd C, Abraham A, Sui W, Bejan C, Capra JA, Hsi R. Machine Learning Models to Predict 24 Hour Urinary Abnormalities for Kidney Stone Disease. Urology. 2022 Nov;169:52-57. doi: 10.1016/j.urology.2022.07.008. Epub 2022 Jul 16. PMID: 35853510.
# Moreira DM, Friedlander JI, Hartman C, Elsamra SE, Smith AD, Okeke Z. Using 24-hour urinalysis to predict stone type. J Urol. 2013 Dec;190(6):2106-11. doi: 10.1016/j.juro.2013.05.115. Epub 2013 Jun 11. PMID: 23764079.
# Porowski T, Kirejczyk JK, Mrozek P, Protas P, Kozerska A, Łabieniec Ł, Szymański K, Wasilewska A. Upper metastable limit osmolality of urine as a predictor of kidney stone formation in children. Urolithiasis. 2019 Apr;47(2):155-163. doi: 10.1007/s00240-018-1041-2. Epub 2018 Jan 22. PMID: 29356875; PMCID: PMC6420897.
# Silverio AA, Chung WY, Cheng C, Wang HL, Kung CM, Chen J, Tsai VF. The potential of at-home prediction of the formation of urolithiasis by simple multi-frequency electrical conductivity of the urine and the comparison of its performance with urine ion-related indices, color and specific gravity. Urolithiasis. 2016 Apr;44(2):127-34. doi: 10.1007/s00240-015-0812-2. Epub 2015 Aug 13. PMID: 26271351.
# Torricelli FC, De S, Liu X, Calle J, Gebreselassie S, Monga M. Can 24-hour urine stone risk profiles predict urinary stone composition? J Endourol. 2014 Jun;28(6):735-8. doi: 10.1089/end.2013.0769. Epub 2014 Feb 14. PMID: 24460026.
# # EDA
# 1. Univaraiate Analysis (centrality, dispersion, distribution)
# 2. Bivariate Analysis (correlation, covariance)
# 3. Multivariate Analysis (correlations matrix, covariance matrix)
# ## Univariate Analysis
print(df_ts.info())
# # Test Dataset Info
# ## 276 urine specimens
# * id = unique ID
# * gravity = specific gravity, the density of the urine relative to water
# * ph = fixed int, pH, the negative logarithm of the hydrogen ion
# * osmo = osmolarity (mOsm), a unit used in biology and medicine but not in physical chemistry. Osmolarity is proportional to the concentration of molecules in solution
# * cond = conductivity (mMho milliMho). One Mho is one reciprocal Ohm. Conductivity is proportional to the concentration of charged ions in solution
# * urea = urea concentration per millimoles per liter
# * calc = calcium concentration in millimoles liter
# ## Null
# * No null values
print(df_tr.info())
# # Train Dataset Info
# ## 414 urine specimens
# * id = unique ID
# * gravity = specific gravity, the density of the urine relative to water
# * ph = fixed int, pH, the negative logarithm of the hydrogen ion
# * osmo = osmolarity (mOsm), a unit used in biology and medicine but not in physical chemistry. Osmolarity is proportional to the concentration of molecules in solution
# * cond = conductivity (mMho milliMho). One Mho is one reciprocal Ohm. Conductivity is proportional to the concentration of charged ions in solution
# * urea = urea concentration per millimoles per liter
# * calc = calcium concentration in millimoles liter
# * target = binary, 0 no kidney stone, 1 kidney stone
# ## Null
# * No null values
# List features and target variables.
features = ["gravity", "ph", "osmo", "cond", "urea", "calc"]
target = ["target"]
# ## Centrality and Dispersion
df_tr[features].describe().T.style.bar(subset=["mean", "std", "min", "max"])
df_ts[features].describe().T.style.bar(subset=["mean", "std", "min", "max"])
# ## Shape of Distribution
plt.style.use("dark_background")
fig, ax = plt.subplots(2, 3, figsize=(20, 10))
for i, feature in enumerate(features):
sns.distplot(df_tr[feature], ax=ax[i // 3, i % 3])
plt.show()
# **Distribution Analysis**
# - gravity has a narrow dispersion, somewhat bimodal, not normal
# - ph has larger dispersion within ph scale, peaked distribution at 5.5, right tail
# - osmo wide dispersion, nearly bimodal, large std
# - cond wide dispersion, peak at ~25, left tail
# - urea wide dispersion, bimodal in presentation, right tail
# - calc skewed towards ~1, right tail, large std
# Are the feature distributions normal? Graphical representation does not appear normally distributed.
# Therefore, test for normality with Shapiro-Wilk test.
for feature in features:
stat, p = shapiro(df_tr[feature])
print("Statistics=%.3f, p=%.3f" % (stat, p))
if p > 0.05:
print("Training Sample looks Gaussian (fail to reject H0)")
else:
print("Training Sample does not look Gaussian (reject H0)")
print("")
for feature in features:
stat, p = shapiro(df_ts[feature])
print("Statistics=%.3f, p=%.3f" % (stat, p))
if p > 0.05:
print("Test Sample looks Gaussian (fail to reject H0)")
else:
print("Test Sample does not look Gaussian (reject H0)")
print("")
# # Univariate Analysis Complete
# * All features will need to be normalized
# # Bivariate Analysis and Multivariate Anaylsis
# 1. Correlation
# 2. Covariance
display(df_tr[features].corr().style.background_gradient(cmap="coolwarm"))
display(df_tr[features].cov().style.background_gradient(cmap="coolwarm"))
# Perform multivariate analysis on correlation matrix of features in train set
corr = df_tr[features].corr()
mask = np.triu(np.ones_like(corr, dtype=bool))
f, ax = plt.subplots(figsize=(11, 9))
cmap = sns.diverging_palette(230, 20, as_cmap=True)
sns.heatmap(
corr,
mask=mask,
cmap=cmap,
vmax=1,
center=0,
square=True,
linewidths=0.5,
cbar_kws={"shrink": 0.5},
)
cov = df_tr[features].cov()
mask = np.triu(np.ones_like(cov, dtype=bool))
f, ax = plt.subplots(figsize=(11, 9))
cmap = sns.diverging_palette(230, 20, as_cmap=True)
sns.heatmap(
cov,
mask=mask,
cmap=cmap,
vmax=0,
center=0,
square=True,
linewidths=0.5,
cbar_kws={"shrink": 0.5},
)
# # EDA Results
# - Train and test data contain similar values, adequate for training and testing with ML methods
# - Data will need to be normalized prior to predictive modeling for some methods
# - High correlation (with all features) and covariance (with pH) within osmo variable. Consider isolating osmo as univariate predictor or removing from feature set and test for predictive change.
# - Due to lit review revealing osmo as a good predictor, this warrants including in the data set.
# - The impact to the models may be irrelevant for prediction purposes but warrants consideration
# # Analysis of target variable
df_tr[target].value_counts()
fig, ax = plt.subplots(2, 3, figsize=(20, 10))
for i, feature in enumerate(features):
sns.boxplot(x=df_tr["target"], y=df_tr[feature], ax=ax[i // 3, i % 3])
plt.show()
df_tr[features + ["target"]].corr().style.background_gradient(cmap="coolwarm")
# # Chi-Square Analyis for model inclusion
signif_level_1 = 0.05
signif_level_2 = 0.20
result_table = []
for target_col in ["target"]:
for columns in features:
crosstab = pd.crosstab(df_tr[columns], df_tr["target"])
chi2, p, dof, ex = chi2_contingency(crosstab)
if p <= signif_level_1:
result_table.append([columns, target_col, chi2, p, dof])
elif p <= signif_level_2:
result_table.append([columns, target_col, chi2, p, dof])
if not result_table:
print("No p-value less than or equal to 0.20 in the data")
else:
result_table = pd.DataFrame(
result_table,
columns=[
"Column Name",
"Target",
"Chi-Square",
"P-Value",
"Degrees of Freedom",
],
)
display(result_table)
# # Correlation and Chi-Square Analysis
# - It appears that calc has higher correlation than other variables on the target
# - gravity, osmo, cond, urea, and calc meet criteria for model inclusion. omit ph
# create X and y variables
X_train = df_tr[["gravity", "osmo", "cond", "urea", "calc"]]
y_train = df_tr["target"]
X_test = df_ts.drop(columns=["id"])
X_train = sm.add_constant(X_train)
model = sm.OLS(y_train, X_train).fit()
predictions = model.predict(X_train)
model.summary()
# # LR model summary
# - Calc is the only significant predictor in LR model.
# # Predictive Modeling
# ## Data Transformation
# Rebalance Sample
from imblearn.over_sampling import SMOTE
sm = SMOTE(random_state=41889)
X_train, y_train = sm.fit_resample(X_train, y_train)
y_train.value_counts()
# Scale X values
scaler = StandardScaler()
X_train_scaled = scaler.fit_transform(X_train)
X_test_scaled = scaler.transform(X_test)
# Previous model testing and parameter tuning
# Results for SVM
# Best hyperparameters: {'C': 100, 'gamma': 0.001, 'kernel': 'rbf'}
# Best score: 0.7422596153846154
# Results for Random Forest
# Best hyperparameters: {'max_depth': None, 'max_features': 'sqrt', 'n_estimators': 100}
# Best score: 0.7391826923076923
# Results for Gradient Boosting
# Best hyperparameters: {'learning_rate': 1.0, 'max_depth': 10, 'max_features': 'log2', 'n_estimators': 300}
# Best score: 0.7454326923076924
# Results for Logistic Regression
# Best hyperparameters: {'C': 0.1, 'penalty': 'l2'}
# Best score: 0.7235576923076923
# Accuracy on testing set: 0.7681159420289855
# Confusion matrix:
# [[54 14]
# [18 52]]
# Classification report:
# precision recall f1-score support
# 0 0.75 0.79 0.77 68
# 1 0.79 0.74 0.76 70
# accuracy 0.77 138
# macro avg 0.77 0.77 0.77 138
# weighted avg 0.77 0.77 0.77 138
# Cross-validation scores: [0.70769231 0.72307692 0.78125 0.6875 0.71875 ]
# Mean cross-validation score: 0.72
# AUC: 0.83
def fit_model(X_train, y_train, X_val, y_val, model, eval):
if eval:
model.fit(
X_train,
y_train,
eval_set=[(X_val, y_val)],
early_stopping_rounds=1000,
verbose=False,
)
else:
model.fit(X_train, y_train)
y_pred_proba_train = model.predict_proba(X_train)[:, 1]
y_pred_proba_val = model.predict_proba(X_val)[:, 1]
test_preds = model.predict_proba(X_test)[:, 1]
return y_pred_proba_train, y_pred_proba_val, test_preds
rfc_params = {
"n_estimators": [100, 500, 1000],
"max_depth": [None, 10, 20],
"min_samples_split": [2, 5, 10],
}
xgb_params = {
"n_estimators": [100, 500, 1000],
"learning_rate": [0.01, 0.1, 0.2],
"max_depth": [3, 6, 9],
}
rfc = RandomForestClassifier(random_state=41889)
xgb_clf = xgb.XGBClassifier(random_state=41889)
rfc_grid = GridSearchCV(rfc, rfc_params, cv=5, scoring="roc_auc", n_jobs=-1)
xgb_grid = GridSearchCV(xgb_clf, xgb_params, cv=5, scoring="roc_auc", n_jobs=-1)
rfc_grid.fit(X_train, y_train)
xgb_grid.fit(X_train, y_train)
rfc_best_params = rfc_grid.best_params_
xgb_best_params = xgb_grid.best_params_
display(rfc.set_params(**rfc_best_params))
display(xgb_clf.set_params(**xgb_best_params))
class EnsembleClassifier(BaseEstimator, ClassifierMixin):
def __init__(self, rfc, xgb_clf, rfc_weight=0.5):
self.rfc = rfc
self.xgb_clf = xgb_clf
self.rfc_weight = rfc_weight
def fit(self, X, y):
self.rfc.fit(X, y)
self.xgb_clf.fit(X, y)
self.classes_ = self.rfc.classes_
return self
def predict_proba(self, X):
rfc_proba = self.rfc.predict_proba(X)
xgb_proba = self.xgb_clf.predict_proba(X)
ensemble_proba = (rfc_proba * self.rfc_weight) + (
xgb_proba * (1 - self.rfc_weight)
)
return ensemble_proba
def custom_auc_score(y_true, y_pred_proba):
return roc_auc_score(y_true, y_pred_proba)
ensemble_clf = EnsembleClassifier(rfc, xgb_clf)
ensemble_params = {"rfc_weight": [i / 10 for i in range(0, 11)]}
custom_scorer = make_scorer(custom_auc_score, greater_is_better=True, needs_proba=True)
ensemble_grid = GridSearchCV(
ensemble_clf, ensemble_params, cv=5, scoring=custom_scorer, n_jobs=-1
)
ensemble_grid.fit(X_train_scaled, y_train)
ensemble_best_params = ensemble_grid.best_params_
ensemble_clf.set_params(**ensemble_best_params)
splits = 5
kf = StratifiedKFold(n_splits=splits, shuffle=True, random_state=42)
val_preds = np.zeros(len(y_train))
test_preds = np.zeros(len(X_test))
for i, (train_idx, val_idx) in enumerate(kf.split(X_train_scaled, y_train)):
X_train_fold, y_train_fold = X_train_scaled[train_idx], y_train.iloc[train_idx]
X_val_fold, y_val_fold = X_train_scaled[val_idx], y_train.iloc[val_idx]
ensemble_clf.fit(X_train_fold, y_train_fold)
ensemble_y_pred_proba_train = ensemble_clf.predict_proba(X_train_fold)[:, 1]
ensemble_y_pred_proba_val = ensemble_clf.predict_proba(X_val_fold)[:, 1]
ensemble_test_predictions = ensemble_clf.predict_proba(X_test_scaled)[:, 1]
# Hold all validation predictions for computing overall AUC score
val_preds[val_idx] = ensemble_y_pred_proba_val
test_preds += ensemble_test_predictions
# Calculate the AUC score on the training and validation sets
train_score = roc_auc_score(y_train_fold, ensemble_y_pred_proba_train)
val_score = roc_auc_score(y_val_fold, ensemble_y_pred_proba_val)
# Print AUC scores for each fold
print_auc_scores(i + 1, train_score, val_score)
# Compute overall AUC score on the validation set
val_auc_score = roc_auc_score(y_train, val_preds)
print(f"Overall validation AUC score = {val_auc_score:.3f}")
# Initialize the StratifiedKFold cross-validator
cv = StratifiedKFold(n_splits=5, shuffle=True, random_state=42)
# Initialize an empty list to store log loss values for each fold
log_losses = []
# Loop through the cross-validation splits
for i, (train_idx, val_idx) in enumerate(cv.split(X_train_scaled, y_train)):
X_train_fold, y_train_fold = X_train_scaled[train_idx], y_train[train_idx]
X_val_fold, y_val_fold = X_train_scaled[val_idx], y_train[val_idx]
# Fit the ensemble classifier on the training data and predict probabilities on the validation data
ensemble_clf.fit(X_train_fold, y_train_fold)
predicted_proba = ensemble_clf.predict_proba(X_val_fold)
# Compute log loss for the current fold
loss = log_loss(y_val_fold, predicted_proba)
log_losses.append(loss)
print(f"Fold {i + 1}: Log loss = {loss:.3f}")
# Create a Plotly bar chart to visualize the log loss for each fold
fig = go.Figure(data=[go.Bar(x=list(range(1, len(log_losses) + 1)), y=log_losses)])
fig.update_layout(
title="Log Loss during Cross-Validation", xaxis_title="Fold", yaxis_title="Log Loss"
)
fig.show()
# Folds 1-4 were consistent and looked to perform well but fold 5 is different... will investigate
# Initialize lists to store data for analysis
fold_data = []
# Loop through the cross-validation splits
for i, (train_idx, val_idx) in enumerate(cv.split(X_train_scaled, y_train)):
X_val_fold, y_val_fold = X_train_scaled[val_idx], y_train[val_idx]
# Create a DataFrame with features and target variable for the current fold
val_fold_df = pd.DataFrame(X_val_fold, columns=X_train.columns)
val_fold_df["target"] = y_val_fold
val_fold_df["fold"] = i + 1
# Append the DataFrame to the fold_data list
fold_data.append(val_fold_df)
# Combine data from all folds into a single DataFrame
all_folds_df = pd.concat(fold_data)
# Plot the distribution of the target variable for each fold
sns.countplot(data=all_folds_df, x="fold", hue="target")
plt.title("Distribution of Target Variable in Each Fold")
plt.show()
# Plot the distribution of feature values for each fold
for feature in X_train.columns:
sns.boxplot(data=all_folds_df, x="fold", y=feature)
plt.title(f"Distribution of {feature} in Each Fold")
plt.show()
# There's less stones in fold 5 and there is also a higher calc variable in fold 5. Since the regression analysis found calc as a significant predictor for stones, it makes sense that the log loss would degrade with less target =1 in the 5th fold.
# # Prepare prediciton submission
test_preds /= splits
df_ts["target"] = test_preds
df_ts[["id", "target"]].to_csv("submission.csv", index=False)
df_ts[["id", "target"]]
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Forecasting Type of the Network Attacks
# Applied Machine Learning - Lab 2 - Fatih Gurbuz 65223
# 1. Load your dataset from the “internet_connection_data.csv” csv file.
data = pd.read_csv("/kaggle/input/connection-data/internet_connection_data.csv")
data
# 2. Explore your dataset and list the name of the columns.
data.shape
data.head(10)
data.describe()
data.columns
data.info()
# 3. Explore your dataset and check if there is any column with missing values.
data.isna().sum()
# 4. Select your input variables and output variable (hint: output column is the one you want to
# predict. In this case it is the last column which shows the type of network attack).
y = data["Category"]
y
y.value_counts()
X = data.drop(columns=["Category"])
X
# 5. Split your dataset as %80 training and %20 testing.
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from sklearn.metrics import accuracy_score
from sklearn.linear_model import LogisticRegression
from sklearn import svm
from sklearn.naive_bayes import MultinomialNB
from sklearn.ensemble import RandomForestClassifier
X_train, X_test, y_train, y_test = train_test_split(
X, y, random_state=666, test_size=0.2
)
# and this below is for scaled data
scaler = StandardScaler()
X_scaled = scaler.fit_transform(X)
X_train_s, X_test_s, y_train_s, y_test_s = train_test_split(
X_scaled, y, random_state=666, test_size=0.2
)
X_train
# 6. Implement four classification model based on Logistic Regression, Support Vector Machine,
# Multinomial Naive Bayes and Random Forest classifiers.
models = [
LogisticRegression(multi_class="auto", solver="liblinear", max_iter=10000, C=10),
svm.SVC(),
MultinomialNB(),
RandomForestClassifier(n_estimators=200, max_depth=30),
]
# 7. Train (fit) your network.
# 8. Report the accuracies (by percentage) of the models for the test datasets.
scores = {}
for i in range(len(models)):
model = models[i]
if i == 2: # this is because MultinomialNB cannot take values below 0
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
accuracy = accuracy_score(y_test, y_pred)
else: # however, others can use scaled data
model.fit(X_train_s, y_train_s)
y_pred = model.predict(X_test_s)
accuracy = accuracy_score(y_test_s, y_pred)
scores[i] = accuracy * 100
print(f"Accuracy of the model {models[i]}: {accuracy * 100}")
|
#
# ---
# ---
# ### **TMNIST | Neural Network Type Classification**
# **Shreya Jaiswal**
# ---
# ---
# ABSTRACT
# ---
# ABOUT THE DATA
# > The **MNIST database (Modified National Institute of Standards and Technology database)** is a large database of handwritten digits that is commonly used for training various image processing systems. The database is also widely used for training and testing in the field of machine learning.
# > It is an extremely good database for people who want to try machine learning techniques and pattern recognition methods on real-world data while spending minimal time and effort on data preprocessing and formatting. Its simplicity and ease of use are what make this dataset so widely used and deeply understood.
# >dataset origin : https://www.kaggle.com/code/sheshngupta/tminst-character-recognition-94-4-accuracy/input
# ---
# ---
# ---
# IMPORT LIBRARIES
# ---
# ---
#
import numpy as np
import pandas as pd
import re
from sklearn.model_selection import train_test_split
from sklearn.utils import shuffle
import tensorflow as tf
from tensorflow.keras.utils import to_categorical
import matplotlib.pyplot as plt
import warnings
warnings.filterwarnings("ignore")
#
# ---
# READING DATA
# ---
#
# Reading Data
df_tmnist = pd.read_csv(
"/kaggle/input/tmnist-alphabet-94-characters/94_character_TMNIST.csv"
)
df_tmnist.head()
#
# ---
# ---
# FUNCTIONS
# ---
# ---
# ***Function 1*** : *to define and extract regex patterns to check for alphabets, digits and special symbols*
# function to define and extract regex patterns to check for alphabets, digits and special symbols
def regex_pattern(pattern_type, all_labels):
if pattern_type == "uppercase":
pattern_uc = re.compile(r"[A-Z]")
upper_case = pattern_uc.findall(str(all_labels))
return upper_case
elif pattern_type == "lowercase":
pattern_lc = re.compile(r"[a-z]")
lower_case = pattern_lc.findall(str(all_labels))
return lower_case
elif pattern_type == "numbers":
pattern_numbers = re.compile(r"[0-9]")
numbers = pattern_numbers.findall(str(all_labels))
return numbers
elif pattern_type == "symbols":
pattern_symbols = re.compile(r"[\W]|[\_\,]")
symbols = list(set(pattern_symbols.findall(str(all_labels))))
symbols.pop(27)
return symbols
# ***Function 2*** : *Function to Plot Model after Evaluation*
# Function for Plotting
def Plott(data):
fig, ax = plt.subplots(1, 2, figsize=(20, 7))
# summarize history for accuracy
ax[0].plot(data.history["accuracy"])
ax[0].plot(data.history["val_accuracy"])
ax[0].set_title("model accuracy")
ax[0].legend(["train", "test"], loc="upper left")
# summarize history for loss
ax[1].plot(data.history["loss"], label=["loss"])
ax[1].plot(data.history["val_loss"], label=["val_loss"])
ax[1].set_title("model loss")
ax[1].legend(["train", "test"], loc="upper left")
plt.show()
#
# ---
# ---
# Splitting the Input labels to define groups for clasification
# ---
# ---
#
all_labels = list(df_tmnist["labels"].unique())
# all_labels
#
# ---
# Call the regex_pattern function to extract the necessary pattern
# ---
#
# Extracting Pattern
lower_case = regex_pattern("lowercase", all_labels)
upper_case = regex_pattern("uppercase", all_labels)
numbers = regex_pattern("numbers", all_labels)
symbols = regex_pattern("symbols", all_labels)
# ---
# Group the Labels as follows
# ---
# > **Group1** : *lowercase*
# > **Group2** : *uppercase*
# > **Group3** : *numbers*
# > **Group4** : *symbols*
# Creating Groups
group = 1
for label_list in (lower_case, upper_case, numbers, symbols):
df_tmnist.loc[df_tmnist["labels"].isin(label_list), "group"] = str(group)
group += 1
df_tmnist
#
# ---
# Defining X and y for Training
#
# ---
#
# defining X and y for training
X = df_tmnist.iloc[:, 2:-1].astype("float32")
X
# defining X and y for training
y = df_tmnist[["labels"]]
y
# preping the data to create dictionary
labels = y["labels"].unique()
labels
values = [num for num in range(len(df_tmnist["labels"].unique()))]
values
# ---
# Creating Data Dictionary for the Labels
#
# ---
#
# Creating Dictionary
label_dict = dict(zip(labels, values))
label_dict
label_dict_inv = dict(zip(values, labels))
label_dict_inv
# Maping Values
y["labels"].replace(label_dict, inplace=True)
# Checking the mappings
print(label_dict)
#
# ---
# ---
# Creating Train Test Split of 80% and 20%
# ---
# ---
#
# creating test train split of 80% and 20%
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
#
# ---
# ---
# Pre-processing the data for Training
# ---
# ---
#
# Defining the length and height
Length, Height = 28, 28
NCl = y_train.nunique()[0]
NCl
# N of images 28x28
X_train = np.reshape(X_train.values, (X_train.shape[0], Length, Height))
X_test = np.reshape(X_test.values, (X_test.shape[0], Length, Height))
# Target into Categorical Values
y_train = to_categorical(y_train, NCl, dtype="int")
y_test = to_categorical(y_test, NCl, dtype="int")
print(f"X:Train, Test data shape:{X_train.shape},{X_test.shape}")
print(f"Y:Train, Test data shape:{y_train.shape},{y_test.shape}")
#
# ---
# ---
# Displaying Random Characters from our Dataset as samples
# ---
# ---
#
# displaying the charecters of our dataset
random = shuffle(X_train[:500]) # Randomly shuffle
fig, ax = plt.subplots(3, 4, figsize=(10, 10))
axes = ax.flatten()
for i in range(12):
img = np.reshape(random[i], (28, 28)) # reshaping it for displaying
axes[i].imshow(img, cmap="Greys")
img_final = np.reshape(
img, (1, 28, 28, 1)
) # reshapng it for passing into model for prediction
axes[i].grid()
#
# ---
# ---
# IMPLEMENTING NEURAL NETWORKS
# ---
# ---
# ---
# NEURAL NETWORKS
#
# ---
# > 1. Neural Networks (NN) are a type of machine learning model inspired by the structure and function of the human brain. They consist of interconnected nodes, called neurons, organized in layers, where each neuron is a computational unit that processes input data and passes the result to other neurons.
# >2. The most common type of neural network is the feedforward neural network, where the data flows in one direction from the input layer through the hidden layers to the output layer.
# >3. Another type of neural network is the recurrent neural network (RNN), which allows the data to flow in loops, making it well suited for processing sequential data, such as time-series or natural language data.
# >4. There are also several other types of neural networks, such as convolutional neural networks (CNNs) and generative adversarial networks (GANs), that are specialized for specific tasks, such as image recognition and generative modeling, respectively.
# >5. Neural networks have shown remarkable success in various applications, including computer vision, natural language processing, speech recognition, and many others. They have become an important tool in the field of machine learning, and their capabilities are continuously expanding with the development of new architectures and techniques.
# ---
# CONVOLUTION NEURAL NETWORKS (CNN)
# ---
# > 1. CNN stands for Convolutional Neural Network, which is a type of neural network commonly used in computer vision tasks, such as image and video recognition, segmentation, and detection.
# >2. CNNs typically consist of multiple layers, each with a specific function. The input layer receives the raw data, such as an image, and the subsequent layers apply a series of transformations to it. These layers include convolutional layers, pooling layers, and fully connected layers.
# >3. In the convolutional layer, a set of filters slides over the image to extract features, and the output of each filter is called a feature map. In the pooling layer, the size of the feature maps is reduced by selecting the most important features. Finally, the fully connected layer takes the flattened feature maps and applies a traditional neural network on top of it to classify the input image.
# >4. CNNs are typically trained using supervised learning with labeled data. The goal is to optimize the network's parameters, such as the weights and biases, to minimize the difference between the predicted output and the true output. This process is done using an optimizer and a loss function, which measures the difference between the predicted output and the true output.
# ---
# CNN ARCHITECTURE
# ---
# 
# >1. **Input Layer:** This layer takes in the raw image data and passes it on to the next layer.
# >2. **Convolutional Layer:** This layer applies a set of filters (small matrices) to the input image in order to extract features from it. Each filter produces a feature map, which highlights different aspects of the image.
# >3. **Activation Layer:** This layer applies an activation function (such as ReLU) to each element of the feature maps in order to introduce nonlinearity into the network.
# >4. **Pooling Layer:** This layer reduces the spatial dimensionality of the feature maps by performing down-sampling, which helps to reduce the number of parameters in the network and improve its robustness to translation.
# >5. **Fully-Connected Layer:** This layer connects every neuron in the previous layer to every neuron in the next layer, just like in a traditional neural network. This layer is responsible for performing the final classification decision.
# >6. **Output Layer:** This layer produces the final output of the network, which is typically a probability distribution over the different classes.
# ---
# IMPORTING LIBRARIES FOR NEURAL NETWORK
#
# ---
#
# importing libraries for building neural netwrok
from tensorflow.keras.models import Sequential, load_model
from tensorflow.keras.layers import (
Dense,
Conv2D,
MaxPool2D,
Flatten,
Dropout,
BatchNormalization,
)
from tensorflow.keras.optimizers import SGD, Adam, RMSprop
from tensorflow.keras.callbacks import EarlyStopping
#
# ---
# ---
# PRE-PROCESSING THE INPUT FOR CNN
# ---
# ---
#
RGB = 1 # In this case only one instead of 3 because we dont have Color images
X_train = X_train.reshape(X_train.shape[0], X_train.shape[1], X_train.shape[2], RGB)
X_test = X_test.reshape(X_test.shape[0], X_test.shape[1], X_test.shape[2], RGB)
# normalizing the image data
X_train = X_train / 255
X_test = X_test / 255
print(f"Train, Test shapes: {X_train.shape},{X_test.shape}")
#
# ---
# Modeling CNN Architecture
#
# ---
# ---
# Notes
# ---
# >1. `epoch` :In machine learning, an epoch is a complete pass through the entire training dataset during the training process. In other words, it is one iteration over the entire dataset, where the model updates its weights based on the loss calculated on the training set.
# > 2. `params`: This refers to the number of learnable parameters (weights and biases) in the network. The number of parameters in a CNN can be used as an indicator of its complexity and capacity to learn.
model = Sequential()
# 4 Conv with Maxpool and Dropout [25%] -> Flatten - > Dense -> Dense -> output
model.add(
Conv2D(
filters=32,
kernel_size=(3, 3),
input_shape=(Length, Height, RGB),
padding="same",
)
)
model.add(BatchNormalization())
model.add(tf.keras.layers.Activation("relu"))
model.add(MaxPool2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Conv2D(filters=64, kernel_size=(3, 3), padding="same"))
model.add(BatchNormalization())
model.add(tf.keras.layers.Activation("relu"))
model.add(MaxPool2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Conv2D(filters=128, kernel_size=(3, 3), padding="same"))
model.add(BatchNormalization())
model.add(tf.keras.layers.Activation("relu"))
model.add(MaxPool2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Conv2D(filters=256, kernel_size=(3, 3), padding="same"))
model.add(BatchNormalization())
model.add(tf.keras.layers.Activation("relu"))
model.add(MaxPool2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Flatten())
model.add(Dense(350))
model.add(BatchNormalization())
model.add(tf.keras.layers.Activation("relu"))
model.add(Dropout(0.25))
model.add(Dense(NCl, activation="softmax"))
model.summary()
# ---
# Explanation:
#
# ---
# Basic steps while modeling CNN are as below:
# 1. Batch normalization: It is a technique used to normalize the input of each layer, which helps in reducing the internal covariate shift problem. This technique is used to speed up the training of a neural network, as it reduces the number of epochs required to reach convergence.
# 2. Activation: It is a function used to introduce non-linearity in the output of a neuron. In CNN, the Rectified Linear Unit (ReLU) activation function is commonly used, as it is computationally efficient and allows the model to learn complex features.
# 3. Max pooling: Max pooling is a technique used in a type of machine learning algorithm called a convolutional neural network (CNN). It is used to make the neural network more efficient by reducing the amount of information it needs to process.
# 4. Dropout: It is a regularization technique used to prevent overfitting in a neural network. It randomly drops out a fraction of the neurons during training, which reduces the dependence of the network on any one neuron and encourages it to learn more robust features. This technique helps to reduce the variance of the model, which in turn improves its generalization performance.
# ---
# > `model = Sequential()` : This creates a new Sequential model object.
# >`model.add(Conv2D(filters=32, kernel_size=(3,3), input_shape=(Length, Height, RGB), padding='same'))
# `:This adds the first convolutional layer to the model with 32 filters, a 3x3 kernel size, and an input shape of (Length, Height, RGB).
# > `model.add(BatchNormalization())`: This adds a batch normalization layer to the model.
# > `model.add(tf.keras.layers.Activation('relu'))`:This adds a ReLU activation layer to the model.
# >`model.add(MaxPool2D(pool_size=(2,2)))`: model.add(MaxPool2D(pool_size=(2,2)))
# > `model.add(Dropout(0.25))` : This adds a dropout layer to the model with a rate of 25%.
# ---
# defining parameters for training
optimizer = Adam(learning_rate=0.01)
callback = EarlyStopping(monitor="loss", patience=5)
Batch_ = 64
Epochs_ = 100
model.compile(
loss="categorical_crossentropy", optimizer=optimizer, metrics=["accuracy"]
)
# ---
# Explanation:
#
# ---
# > `optimizer = Adam(learning_rate=0.01)` : an optimizer is defined using the Adam algorithm with a learning rate of 0.01. The optimizer is responsible for updating the weights and biases of the model during training in order to minimize the loss function.
# > `callback =EarlyStopping(monitor='loss', patience=5)`: EarlyStopping callback is defined. This callback monitors the loss function during training and stops training early if the loss has not improved for a certain number of epochs (in this case, 5).
# > `Batch_` variable is set to 64, which means that during training, the model will process 64 samples at a time before updating the weights and biases.
# > `Epochs_` variable is set to 100, which means that the model will be trained for 100 epochs (passes through the training data).
# >`model.compile(loss = 'categorical_crossentropy', optimizer = optimizer, metrics = ['accuracy'])` : the model is compiled using categorical cross-entropy as the loss function, the previously defined optimizer, and accuracy as the metric to monitor during training. Compiling the model prepares it for training by setting up the necessary computations and data structures.
# ---
# ---
# MODEL TRAINING
# ---
# ---
#
# Training
history = model.fit(
X_train,
y_train,
validation_data=(X_test, y_test),
batch_size=Batch_,
epochs=Epochs_,
verbose=1,
)
#
# ---
# ---
# MODEL EVALUATION
# ---
# ---
#
# Evaluating model accuracy on test data
score = model.evaluate(X_test, y_test, batch_size=Batch_, verbose=0)
print(f"Test Accuracy:{round(score[1],4)*100}%")
#
# ---
# ---
# MODEL PERFORMANCE: VISUALIZATION
# ---
# ---
#
Plott(history)
# ---
# Explanation:
#
# ---
# > 1. It can be observed that the accuracy of the model for both the training and test datasets exhibits a similar trend, although the test data demonstrates slightly more fluctuations than the training dataset.
# > 2. Furthermore, the model loss for both the training and test datasets shows a comparable decreasing trend.
# ---
# ---
# FINAL PREDICTIONS OBTAINED FROM TRAINING MODEL
# ---
# ---
#
# predicting the characters using trained model
fig, axes = plt.subplots(3, 4, figsize=(8, 12))
axes = axes.flatten()
for i, ax in enumerate(axes):
img = np.reshape(X_test[i], (28, 28)) # reshaping it for displaying
ax.imshow(img, cmap="Greys")
img_final = np.reshape(
img, (1, 28, 28, 1)
) # reshapng it for passing into model for prediction
pred = label_dict_inv[np.argmax(model.predict(img_final))]
ax.set_title("Prediction: " + pred)
ax.grid()
|
import numpy as np, pandas as pd
iris_dataset = pd.read_csv("/kaggle/input/iris/Iris.csv", index_col="Id")
# cancer_dataset = pd.read_csv("/kaggle/input/breast-cancer-wisconsin-data/data.csv")
# heart_dataset = pd.read_csv("/kaggle/input/heart-attack-analysis-prediction-dataset/heart.csv")
iris_dataset
iris_dataset.info()
# cancer_dataset.info()
# heart_dataset.info()
# cancer_dataset.drop(['id','Unnamed: 32'], axis = 1, inplace = True)
# heart_dataset.drop_duplicates(inplace=True)
from sklearn.model_selection import train_test_split
# X Data
X = iris_dataset.iloc[:, :-1]
# X = cancer_dataset.drop(['diagnosis'],axis = 1)
# X = heart_dataset.iloc[:,:-1]
# y Data
y = iris_dataset.iloc[:, -1]
# y = cancer_dataset['diagnosis']
# y = heart_dataset.iloc[:,-1]
# Splitting data
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.33, random_state=0
)
# Splitted Data
print("X_train shape is ", X_train.shape)
print("y_train shape is ", y_train.shape)
print("--------------------------")
print("X_test shape is ", X_test.shape)
print("y_test shape is ", y_test.shape)
# ## k-nearest neighbor
# Applying KNeighborsClassifier Model
"""
sklearn.neighbors.KNeighborsClassifier(n_neighbors=5, weights='uniform’, algorithm=’auto’, leaf_size=30,
p=2, metric='minkowski’, metric_params=None,n_jobs=None)
"""
# import library
from sklearn.neighbors import KNeighborsClassifier
from sklearn.metrics import accuracy_score
# Define object
KNNClassifierModel = KNeighborsClassifier(n_neighbors=7)
# Fit object to data (Training)
KNNClassifierModel.fit(X_train, y_train)
# Calculating Prediction
y_pred = KNNClassifierModel.predict(X_test)
# Calculating Details
print("k-nearest neighbor")
print("--------------------------------------------")
print("Train Score is:", KNNClassifierModel.score(X_train, y_train))
print("Test Score is :", KNNClassifierModel.score(X_test, y_test))
print("--------------------------------------------")
print("Predicted Value for KNN Classifier: ")
print(
pd.DataFrame(
{"y_test": y_test, "y_predict": y_pred, "Match": y_test == y_pred}
).head(X_test.shape[0])
)
print("--------------------------------------------")
# Calculating Accuracy Score : ((TP + TN) / float(TP + TN + FP + FN))
AccScore = accuracy_score(y_test, y_pred, normalize=False)
print("Accuracy Score is: ", AccScore, "out of", X_test.shape[0])
# ## Random Forest
# Applying RandomForestClassifier Model
"""
ensemble.RandomForestClassifier(n_estimators='warn’, criterion=’gini’, max_depth=None,
min_samples_split=2, min_samples_leaf=1,min_weight_fraction_leaf=0.0,
max_features='auto’,max_leaf_nodes=None,min_impurity_decrease=0.0,
min_impurity_split=None, bootstrap=True,oob_score=False, n_jobs=None,
random_state=None, verbose=0,warm_start=False, class_weight=None)
"""
# import library
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import accuracy_score
# Define object
RandomForestClassifierModel = RandomForestClassifier(
criterion="entropy", n_estimators=100, max_depth=1000, random_state=30
)
# Fit object to data (Training)
RandomForestClassifierModel.fit(X_train, y_train)
# Calculating Prediction
y_pred = RandomForestClassifierModel.predict(X_test)
# Calculating Details
print("Random Forest Classifier")
print("--------------------------------------------")
print("Train Score is:", RandomForestClassifierModel.score(X_train, y_train))
print("Test Score is :", RandomForestClassifierModel.score(X_test, y_test))
# print('RandomForestClassifierModel features importances are : ' , RandomForestClassifierModel.feature_importances_)
print("--------------------------------------------")
print("Predicted Value for RandomForestClassifier: ")
print(
pd.DataFrame(
{"y_test": y_test, "y_predict": y_pred, "Match": y_test == y_pred}
).head(X_test.shape[0])
)
print("--------------------------------------------")
# Calculating Accuracy Score : ((TP + TN) / float(TP + TN + FP + FN))
AccScore = accuracy_score(y_test, y_pred, normalize=False)
print("Accuracy Score is: ", AccScore, "out of", X_test.shape[0])
# ## Gradient Boosting
# Applying GradientBoostingClassifier Model
"""
ensemble.GradientBoostingClassifier(loss='deviance’, learning_rate=0.1,n_estimators=100, subsample=1.0,
criterion='friedman_mse’,min_samples_split=2,min_samples_leaf=1,
min_weight_fraction_leaf=0.0,max_depth=3,min_impurity_decrease=0.0,
min_impurity_split=None,init=None, random_state=None,max_features=None,
verbose=0, max_leaf_nodes=None,warm_start=False, presort='auto’,
validation_fraction=0.1,n_iter_no_change=None, tol=0.0001)
"""
# import library
from sklearn.ensemble import GradientBoostingClassifier
from sklearn.metrics import accuracy_score
# Define object
GBCModel = GradientBoostingClassifier(n_estimators=500, max_depth=100, random_state=33)
# Fit object to data (Training)
GBCModel.fit(X_train, y_train)
# Calculating Prediction
y_pred = GBCModel.predict(X_test)
# Calculating Details
print("Gradient Boosting Classifier")
print("--------------------------------------------")
print("Train Score is: ", GBCModel.score(X_train, y_train))
print("Test Score is : ", GBCModel.score(X_test, y_test))
# print('GBCModel features importances are : ' , GBCModel.feature_importances_)
print("--------------------------------------------")
print("Predicted Value for GBC: ")
print(
pd.DataFrame(
{"y_test": y_test, "y_predict": y_pred, "Match": y_test == y_pred}
).head(X_test.shape[0])
)
print("--------------------------------------------")
# Calculating Accuracy Score : ((TP + TN) / float(TP + TN + FP + FN))
AccScore = accuracy_score(y_test, y_pred, normalize=False)
print("Accuracy Score is: ", AccScore, "out of", X_test.shape[0])
# ## Logistic Regression
# Applying LogisticRegression Model
"""
linear_model.LogisticRegression(penalty='l2’,dual=False,tol=0.0001,C=1.0,fit_intercept=True,intercept_scaling=1,
class_weight=None,random_state=None,solver='warn’,max_iter=100,
multi_class='warn’, verbose=0,warm_start=False, n_jobs=None)
"""
# import library
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import accuracy_score
# Define object
LogisticRegressionModel = LogisticRegression(
penalty="l2", solver="sag", C=1.0, random_state=33, max_iter=10000
)
# Fit object to data (Training)
LogisticRegressionModel.fit(X_train, y_train)
# Calculating Prediction
y_pred = LogisticRegressionModel.predict(X_test)
# Calculating Details
print("Logistic Regression")
print("--------------------------------------------")
print("Train Score is: ", LogisticRegressionModel.score(X_train, y_train))
print("Test Score is : ", LogisticRegressionModel.score(X_test, y_test))
# print('Classes are : ' , LogisticRegressionModel.classes_)
# print('No. of iteratios is : ' , LogisticRegressionModel.n_iter_)
print("--------------------------------------------")
print("Predicted Value for LogisticRegression: ")
print(
pd.DataFrame(
{"y_test": y_test, "y_predict": y_pred, "Match": y_test == y_pred}
).head(X_test.shape[0])
)
print("--------------------------------------------")
# Calculating Accuracy Score : ((TP + TN) / float(TP + TN + FP + FN))
AccScore = accuracy_score(y_test, y_pred, normalize=False)
print("Accuracy Score is: ", AccScore, "out of", X_test.shape[0])
# ## Stochastic Gradient Descent
# Applying SGDClassifier Model
"""
sklearn.linear_model.SGDClassifier(loss='hinge’, penalty=’l2’, alpha=0.0001,l1_ratio=0.15, fit_intercept=True,
max_iter=None,tol=None, shuffle=True, verbose=0, epsilon=0.1,n_jobs=None,
random_state=None, learning_rate='optimal’, eta0=0.0, power_t=0.5,
early_stopping=False, validation_fraction=0.1,n_iter_no_change=5,
class_weight=None,warm_start=False, average=False, n_iter=None)
"""
# import library
from sklearn.linear_model import SGDClassifier
from sklearn.metrics import accuracy_score
# Define object
SGDClassifierModel = SGDClassifier(
penalty="l2", loss="squared_error", learning_rate="optimal", random_state=30
)
# Fit object to data (Training)
SGDClassifierModel.fit(X_train, y_train)
# Calculating Prediction
y_pred = LogisticRegressionModel.predict(X_test)
# Calculating Details
print("Stochastic Gradient Descent")
print("--------------------------------------------")
print("Train Score is: ", SGDClassifierModel.score(X_train, y_train))
print("Test Score is : ", SGDClassifierModel.score(X_test, y_test))
# print('loss function is : ' , SGDClassifierModel.loss_function_)
# print('No. of iteratios is : ' , SGDClassifierModel.n_iter_)
print("--------------------------------------------")
print("Predicted Values: ")
print(
pd.DataFrame(
{"y_test": y_test, "y_predict": y_pred, "Match": y_test == y_pred}
).head(X_test.shape[0])
)
print("--------------------------------------------")
# Calculating Accuracy Score : ((TP + TN) / float(TP + TN + FP + FN))
AccScore = accuracy_score(y_test, y_pred, normalize=False)
print("Accuracy Score is: ", AccScore, "out of", X_test.shape[0])
# ## Support Vector Machine
# Applying SVC Model
"""
sklearn.svm.SVC(C=1.0, kernel=’rbf’, degree=3, gamma=’auto_deprecated’, coef0=0.0, shrinking=True,
probability=False, tol=0.001, cache_size=200, class_weight=None,verbose=False,
max_iter=-1, decision_function_shape='ovr’, random_state=None)
"""
# import library
from sklearn.svm import SVC
from sklearn.metrics import accuracy_score
# Define object
SVCModel = SVC(
kernel="rbf", # it can be also linear,poly,sigmoid,precomputed
max_iter=1000,
C=1.0,
gamma="auto",
)
# Fit object to data (Training)
SVCModel.fit(X_train, y_train)
# Calculating Prediction
y_pred = SVCModel.predict(X_test)
# Calculating Details
print("Support Vector Machine")
print("--------------------------------------------")
print("Train Score is : ", SVCModel.score(X_train, y_train))
print("Test Score is : ", SVCModel.score(X_test, y_test))
print("--------------------------------------------")
print("Predicted Values: ")
print(
pd.DataFrame(
{"y_test": y_test, "y_predict": y_pred, "Match": y_test == y_pred}
).head(X_test.shape[0])
)
print("--------------------------------------------")
# Calculating Accuracy Score : ((TP + TN) / float(TP + TN + FP + FN))
AccScore = accuracy_score(y_test, y_pred, normalize=False)
print("Accuracy Score is: ", AccScore, "out of", X_test.shape[0])
# ## Decision Tree
# Applying DecisionTreeClassifier Model
"""
sklearn.tree.DecisionTreeClassifier(criterion='gini’, splitter=’best’, max_depth=None,min_samples_split=2,
min_samples_leaf=1,min_weight_fraction_leaf=0.0,max_features=None,
random_state=None, max_leaf_nodes=None,min_impurity_decrease=0.0,
min_impurity_split=None, class_weight=None,presort=False)
"""
# import library
from sklearn.tree import DecisionTreeClassifier
from sklearn.metrics import accuracy_score
# Define object
DecisionTreeClassifierModel = DecisionTreeClassifier(
criterion="gini", max_depth=10, random_state=33
) # criterion can be entropy
# Fit object to data (Training)
DecisionTreeClassifierModel.fit(X_train, y_train)
# Calculating Prediction
y_pred = DecisionTreeClassifierModel.predict(X_test)
# Calculating Details
print("Decision Tree Classifier")
print("--------------------------------------------")
print("Train Score is: ", DecisionTreeClassifierModel.score(X_train, y_train))
print("Test Score is : ", DecisionTreeClassifierModel.score(X_test, y_test))
# print('Classes are : ' , DecisionTreeClassifierModel.classes_)
# print('feature importances are : ' , DecisionTreeClassifierModel.feature_importances_)
print("--------------------------------------------")
print("Predicted Value for SVC: ")
print(
pd.DataFrame(
{"y_test": y_test, "y_predict": y_pred, "Match": y_test == y_pred}
).head(X_test.shape[0])
)
print("--------------------------------------------")
# Calculating Accuracy Score : ((TP + TN) / float(TP + TN + FP + FN))
AccScore_SVM = accuracy_score(y_test, y_pred, normalize=False)
print("Accuracy Score is: ", AccScore_SVM, "out of", X_test.shape[0])
# ## Linear Discriminant Analysis
# Applying LDA Model
"""
sklearn.discriminant_analysis.LinearDiscriminantAnalysis(solver='svd’,shrinkage=None,priors=None,
n_components=None,store_covariance=False,tol=0.0001)
"""
# import library
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
from sklearn.metrics import accuracy_score
# Define object
LDAModel = LinearDiscriminantAnalysis(n_components=1, solver="svd", tol=0.0001)
# Fit object to data (Training)
LDAModel.fit(X_train, y_train)
# Calculating Prediction
y_pred = LDAModel.predict(X_test)
# Calculating Details
print("Linear Discriminant Analysis")
print("--------------------------------------------")
print("Train Score is: ", LDAModel.score(X_train, y_train))
print("Test Score is : ", LDAModel.score(X_test, y_test))
# print('LDAModel means are : ' , LDAModel.means_)
# print('LDAModel classes are : ' , LDAModel.classes_)
print("--------------------------------------------")
print("Predicted Value for SVC: ")
print(
pd.DataFrame(
{"y_test": y_test, "y_predict": y_pred, "Match": y_test == y_pred}
).head(X_test.shape[0])
)
print("--------------------------------------------")
# Calculating Accuracy Score : ((TP + TN) / float(TP + TN + FP + FN))
AccScore = accuracy_score(y_test, y_pred, normalize=False)
print("Accuracy Score is: ", AccScore, "out of", X_test.shape[0])
# ## Quadratic Discriminant Analysis
# Applying QDA Model
"""
sklearn.discriminant_analysis.QuadraticDiscriminantAnalysis(priors=None,reg_param=0.0,store_covariance=False,
tol=0.0001,store_covariances=None)
"""
# import library
from sklearn.discriminant_analysis import QuadraticDiscriminantAnalysis
from sklearn.metrics import accuracy_score
# Define object
QDAModel = QuadraticDiscriminantAnalysis(tol=0.0001)
# Fit object to data (Training)
QDAModel.fit(X_train, y_train)
# Calculating Prediction
y_pred = QDAModel.predict(X_test)
# Calculating Details
print("Quadratic Discriminant Analysis")
print("--------------------------------------------")
print("Train Score is: ", QDAModel.score(X_train, y_train))
print("Test Score is : ", QDAModel.score(X_test, y_test))
# print('QDAModel means are : ' , QDAModel.means_)
print("--------------------------------------------")
print("Predicted Values: ")
print(
pd.DataFrame(
{"y_test": y_test, "y_predict": y_pred, "Match": y_test == y_pred}
).head(X_test.shape[0])
)
print("--------------------------------------------")
# Calculating Accuracy Score : ((TP + TN) / float(TP + TN + FP + FN))
AccScore = accuracy_score(y_test, y_pred, normalize=False)
print("Accuracy Score is: ", AccScore, "out of", X_test.shape[0])
|
# 
import pandas as pd
pd.pandas.set_option("display.max_columns", None)
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
colors = [
"#FF5F00",
"#AD7BE9",
"#03C988",
"#0081B4",
"#F48484",
"#03C988",
"#CC3636",
"#2DCDDF",
"#FFB200",
"#D5CEA3",
"#54B435",
"#EA047E",
"#FFF80A",
"#1D1CE5",
"#FF0032",
]
df_train = pd.read_csv(
"/kaggle/input/house-prices-advanced-regression-techniques/train.csv"
)
df_train.sample(10)
df_train.shape
df_train.columns
df_train.info()
df_train.duplicated().sum()
df_train.isnull().sum()
plt.figure(figsize=(20, 6))
sns.heatmap(df_train.isnull())
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.