script
stringlengths 113
767k
|
|---|
# # Keşifsel Veri Analizi (EDA)
# Keşifsel Veri Analizi veriyle ilgili yapılan tüm çalışmalarda her zaman olması gereken bir ön aşamadır. Doğru business kararlarını alabilmek, iyi modeller üretebilmek öncesinde yapılan detaylı bir EDA çalışmasına bağlıdır.
# Peki nedir bunun aşamaları?
# 1) **Hangi verilere ihtiyacınız olduğunu belirleyin**
# 2) **Verilerinizi toplayın ve yükleyin**
# 3) **Veri tiplerinizi kontrol edin**
# 4) **Veri setiniz hakkında genel bilgiler toplayın**
# 5) **Tekrar eden verileri kaldırın**
# 6) **Sütunlardaki değerleri gözden geçirin**
# 7) **Verilerinizin dağılımını inceleyin**
# 8) **Boş değerleri tespit edin ve gerekli düzenlemeleri yapın**
# 9) **Daha detaylı analizler için verilerinizi filtreleyin**
# 10) **Görsellerden yararlanarak analizinizi daha da derinleştirin**
# 11) **Sonuçlarınızı sunmak için kullanacağınız görselleri hazırlayın**
# ## Veri Setini Tanıyalım
# 1896 Atina Olimpiyatları'ndan 2016 Rio Olimpiyatları'na kadar uzanan ve oyunların kayıtlarına sahip olan Olimpiyat Oyunlarının kapsamlı bir veri kümesidir.
# Her örnek, bireysel bir Olimpik etkinlikte (sporcu etkinlikleri) yarışan bireysel bir sporcuya karşılık gelir. Kullanılan veri setinin metadatası için: [Metadata](https://www.kaggle.com/datasets/heesoo37/120-years-of-olympic-history-athletes-and-results)
# İlk işimiz kütüphanelerimizi çağırmak :)
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
df1 = pd.read_csv(
"/kaggle/input/120-years-of-olympic-history-athletes-and-results/athlete_events.csv",
index_col=1,
)
df1
df1.info()
# >**SORU:** Weight sütununun float dönmesini beklemiştik ama neden object olarak gelmiş?
# İlerleyen satırlarda bu durumu inceleyelim...
df2 = pd.read_csv(
"/kaggle/input/120-years-of-olympic-history-athletes-and-results/noc_regions.csv"
)
df2
df2.info()
# Veri setlerini birleştirmek için `pandas.merge()` fonksiyonunu kullanabiliriz.
df = pd.merge(df1, df2, on="NOC")
df
# Bu projemizin odak noktası **yalnızca yaz olimpiyatları** olacağı için, veri kümemizden tüm kış olimpiyatlarını filtreleyelim ve veri kümemiz üzerinde bazı temel analizler yapalım.
df = df[df["Season"] == "Summer"]
df
# Öncelikle boş değerlerimize bakalım.
df.isnull().sum()
# Her spor dalı için yalnızca üç kazanan olabileceği gerçeğinden dolayı madalya sütunun birçok NAN değerine sahip olduğuna inanıyorum. Ancak, yüzlerce katılımcı bu üç yer için yer almakta ve yarışmaktadır.
df.Medal.value_counts(normalize=True) # Değerlerin yüzdesine bakalım
missing_percentage = 100 * (df.isna().sum().sort_values(ascending=False) / len(df))
missing_percentage
plt.figure(figsize=(15, 6))
plt.title("Percentage Of Missing Values Per Column")
sns.barplot(x=missing_percentage.index, y=missing_percentage.values, color="Red")
# Görseller oluştururken ihtiyacımız olmayan bilgileri görselimizde bulundurmanın bizim için hiçbir yararı yok. Sebep olacağı tek şey karmaşıklık!
plt.figure(figsize=(8, 5))
plt.title("Percentage Of Missing Values Per Column")
sns.barplot(
x=missing_percentage[:5].index, y=missing_percentage[:5].values, color="Red"
)
# **Notes** sütunun bu EDA projemiz için bir faydası olmayacağını düşündüğümüz için kaldıralım.
df.drop(["notes"], axis=1, inplace=True)
# **Weight** sütunumuzun veri tipinde bir sorun vardı, şimdi bu durumu daha detaylı inceleyelim...
df["Weight"].value_counts()
# Aa ne bulduk... Peki ya bu durumu göremeseydik?
df["Weight"] = df["Weight"].astype(float)
# İlk aşamada boş değer yok gibi görünmesine rağmen "?" olarak girilen değerler var, o zaman bunları da boş değer olarak değiştirmemiz lazım.
df["Weight"] = df["Weight"].replace(
"?", np.nan
) # "?" olarak girilmiş ifadeleri NaN olarak değiştirelim
df.isnull().sum() # Tekrar kontrol edelim boş değer varlığını
# **Age**, **Height** ve **Weight** sütunlarındaki tüm boş değerleri aritmetik ortalamayla dolduralım:
df["Age"].fillna(value=df["Age"].mean(), inplace=True)
df["Height"].fillna(value=df["Height"].mean(), inplace=True)
df["Weight"].fillna(value=df["Weight"].mean(), inplace=True)
# **Weight** sütunu için neden hata aldık beraber inceleyelim...
df["Weight"] = df["Weight"].astype(
float
) # İlk halinde "?" string olduğu için tüm sütunu string algılıyordu, şimdi float tipine çevirelim
df["Weight"].fillna(value=df["Weight"].mean(), inplace=True)
df.info()
# Peki **region** sütunu içindeki boş değerleri ne yapabiliriz, birlikte düşünelim...
df["region"].unique()
df.region.dropna(
inplace=True
) # Sadece 21 tane olduğu için kaldırmamız çok bir sorun yaratmaz
# Yalnızca kendi etkinliklerinin galipleri bir Altın, Gümüş veya Bronz madalyaya sahip olacağından, veri setinde **Medal** sütunundaki tüm boş değerleri **Medal Not Won** olarak doldurabiliriz.
df["Medal"].fillna(value="Medal Not Won", inplace=True)
df.isnull().sum()
# Peki tekrar eden değerler?
df[df.duplicated()]
df.duplicated().sum()
df.drop_duplicates(inplace=True) # tekrar eden değerleri kaldıralım
df.duplicated().sum()
df
# Şimdi ise elimizde veriye bazı sorular soralım ve sorulardan elde ettiğimiz cevapları görseller aracılığıyla karşı tarafla paylaşalım.
# * Yaz Olimpiyatlarına en çok sporcu gönderen ülke hangisidir?
# * Sporcu, ülke ve etkinlik sayısı zaman içinde arttı mı yoksa azaldı mı?
# * Hangi ülkeler en çok madalyayı eve götürdü?
# * Erkek ve kadın sporcuların zaman korelasyonuna katılımı
# * United States'ın Madalya Kazandığı Tüm Sporlar?
# * ......
# ## Yaz Olimpiyatlarına en çok sporcu gönderen ülke hangisi?
athlete_count = df.Team.value_counts()
athlete_count
plt.figure(figsize=[18, 8])
sns.barplot(x=athlete_count.index, y=athlete_count.values)
# Maalesef bu görseli kullanamayız!
plt.figure(figsize=[18, 8])
sns.barplot(x=athlete_count[:10].index, y=athlete_count[:10])
plt.title("Countries Send the Most Athletes to the Olympics")
plt.xlabel("Countries")
plt.ylabel("Athlete Count")
# Sporcuların çoğu Amerika Birleşik Devletleri, İngiltere, Fransa, Almanya, İtalya, Japonya, Kanada ve Hollanda gibi birinci dünya ülkelerini temsil etmiştir.
# ## Hangi ülkeler eve en çok madalyayı götürdü?
df[df.Medal != "Medal Not Won"]
df_filtered = df[df.Medal != "Medal Not Won"]
medals_by_country = (
df_filtered.groupby("Team")["Medal"].count().sort_values(ascending=False)
)
medals_by_country
plt.figure(figsize=[18, 8])
plt.xticks(rotation=45) # Ülkelerin isminin açılı yazılmasını sağlıyor
sns.barplot(
x=medals_by_country[:10].index, y=medals_by_country[:10], palette="YlOrBr_r"
)
plt.title("Countries Won the Most Medals in the Olympics")
# En çok madalya United States temsil eden takımlar tarafından kazanılmış.
# ## Ülkeler gönderdikleri sporcu sayılarına göre madalya kazanmakta ne kadar başarılı?
filtered_athlete = athlete_count[athlete_count > 1000]
country_success = (
(medals_by_country / filtered_athlete).sort_values(ascending=False).dropna()
)
country_success
plt.figure(figsize=[15, 10])
sns.barplot(
x=country_success.values * 100, y=country_success.index, palette="coolwarm_r"
)
plt.title("Countries Medal Won Percentage in the Olympics")
plt.xlabel("Percentage (%)")
# ## Sporcuların Yaşının Zamana Göre Değişimi.
plt.figure(figsize=[20, 10])
sns.boxplot(x="Year", y="Age", data=df)
plt.title("Variation Of Athletes Age Over Time")
# **Box Plot**, sporcuların medyan yaşının 120 yıllık bir süre boyunca nispeten sabit kaldığını ve **25 - 26 yıl** civarında gezindiğini gösteriyor. Bu eğilim, çoğu insanın 20 ila 30 yaşları arasında atletik zirvede olduğu ve ardından vücutlarında geri dönüşü olmayan değişiklikler yaşamaya başladığı gerçeğiyle bilimsel olarak desteklenmektedir.
# ## Yıllar içinde zamanla katılımcı sayılarımız nasıl değişmiş?
athlete_by_year = df.groupby("Year")["Name"].count()
athlete_by_year
plt.figure(figsize=[18, 8])
plt.xticks(np.linspace(1896, 2016, 13)) # eksenlerin aralıklarını ayarlar
plt.grid() # kareli defter gibi bir arkaplan oluşturuyor
sns.lineplot(x=athlete_by_year.index, y=athlete_by_year.values)
plt.title("Change in the Number of Athletes Over the Years")
plt.ylabel("Athlete Count")
# Yıllar geçtikçe, Olimpiyatlardaki yarışmacı sayısı genel olarak giderek artış göstermiş. Ancak, üç durumda (1932, 1956 ve 1980) katılımcı sayısında önemli bir düşüş var.
# > **EK BİLGİ:** Bu durumların sebebi 2 kısımda değerlendirilebilir:
# * 1932 Olimpiyatları Büyük Buhran sırasında gerçekleştiğinden, çok az ülke yarışmak için takım gönderdi.
# * 1956 ve 1980 Olimpiyatlarına katılım, jeopolitik nedenlerle çeşitli ulus gruplarının toptan boykotları nedeniyle oldukça kısıtlandı. (ALAN BİLGİSİ)
plt.figure(figsize=[18, 8])
plt.xticks(np.linspace(1896, 2016, 13))
plt.grid()
sns.lineplot(x=athlete_by_year.index, y=athlete_by_year.values)
plt.title("Change in the Number of Athletes Over the Years")
plt.ylabel("Athlete Count")
plt.text(x=1932, y=1500, s="!", weight="bold", fontsize=35, color="red")
plt.text(x=1956, y=4000, s="!", weight="bold", fontsize=35, color="red")
plt.text(x=1980, y=6000, s="!", weight="bold", fontsize=35, color="red")
|
# # Fun with Variational Autoencoders
# This is a starter kernel to use **Labelled Faces in the Wild (LFW) Dataset** in order to maintain knowledge about main Autoencoder principles. PyTorch will be used for modelling.
# This kernel will not update for a while for the purpose of training by yourself.
# ### **Fork it and give it an upvote.**
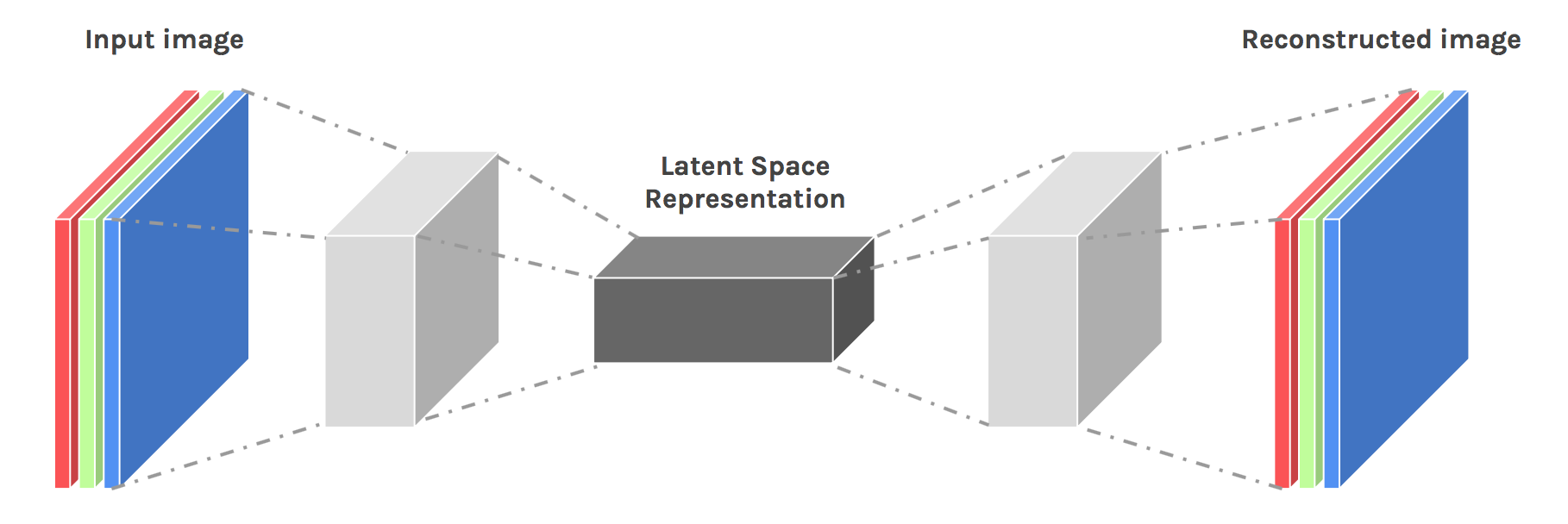
# 
# Useful links:
# * [Building Autoencoders in Keras](https://blog.keras.io/building-autoencoders-in-keras.html)
# * [Conditional VAE (Russian)](https://habr.com/ru/post/331664/)
# * [Tutorial on Variational Autoencoders](https://arxiv.org/abs/1606.05908)
# * [Introducing Variational Autoencoders (in Prose and Code)](https://blog.fastforwardlabs.com/2016/08/12/introducing-variational-autoencoders-in-prose-and.html)
# * [How Autoencoders work - Understanding the math and implementation (Notebook)](https://www.kaggle.com/shivamb/how-autoencoders-work-intro-and-usecases)
# # A bit of theory
# "Autoencoding" is a data compression algorithm where the compression and decompression functions are 1) data-specific, 2) lossy, and 3) learned automatically from examples rather than engineered by a human. Additionally, in almost all contexts where the term "autoencoder" is used, the compression and decompression functions are implemented with neural networks.
# 1) Autoencoders are data-specific, which means that they will only be able to compress data similar to what they have been trained on. This is different from, say, the MPEG-2 Audio Layer III (MP3) compression algorithm, which only holds assumptions about "sound" in general, but not about specific types of sounds. An autoencoder trained on pictures of faces would do a rather poor job of compressing pictures of trees, because the features it would learn would be face-specific.
# 2) Autoencoders are lossy, which means that the decompressed outputs will be degraded compared to the original inputs (similar to MP3 or JPEG compression). This differs from lossless arithmetic compression.
# 3) Autoencoders are learned automatically from data examples, which is a useful property: it means that it is easy to train specialized instances of the algorithm that will perform well on a specific type of input. It doesn't require any new engineering, just appropriate training data.
# source: https://blog.keras.io/building-autoencoders-in-keras.html
#
import matplotlib.pyplot as plt
import os
import glob
import pandas as pd
import random
import numpy as np
import cv2
import base64
import imageio
import torch
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
import torch.utils.data as data_utils
from copy import deepcopy
from torch.autograd import Variable
from tqdm import tqdm
from pprint import pprint
from PIL import Image
from sklearn.model_selection import train_test_split
import os
DATASET_PATH = "/kaggle/input/lfw-dataset/lfw-deepfunneled/lfw-deepfunneled/"
ATTRIBUTES_PATH = "/kaggle/input/lfw-attributes/lfw_attributes.txt"
DEVICE = torch.device("cuda")
# # Explore the data
dataset = []
for path in glob.iglob(os.path.join(DATASET_PATH, "**", "*.jpg")):
person = path.split("/")[-2]
dataset.append({"person": person, "path": path})
dataset = pd.DataFrame(dataset)
# too much Bush
dataset = dataset.groupby("person").filter(lambda x: len(x) < 25)
dataset.head(10)
dataset.groupby("person").count()[:200].plot(kind="bar", figsize=(20, 5))
plt.figure(figsize=(20, 10))
for i in range(20):
idx = random.randint(0, len(dataset))
img = plt.imread(dataset.path.iloc[idx])
plt.subplot(4, 5, i + 1)
plt.imshow(img)
plt.title(dataset.person.iloc[idx])
plt.xticks([])
plt.yticks([])
plt.tight_layout()
plt.show()
# # Prepare the dataset
def fetch_dataset(dx=80, dy=80, dimx=45, dimy=45):
df_attrs = pd.read_csv(
ATTRIBUTES_PATH,
sep="\t",
skiprows=1,
)
df_attrs = pd.DataFrame(df_attrs.iloc[:, :-1].values, columns=df_attrs.columns[1:])
photo_ids = []
for dirpath, dirnames, filenames in os.walk(DATASET_PATH):
for fname in filenames:
if fname.endswith(".jpg"):
fpath = os.path.join(dirpath, fname)
photo_id = fname[:-4].replace("_", " ").split()
person_id = " ".join(photo_id[:-1])
photo_number = int(photo_id[-1])
photo_ids.append(
{"person": person_id, "imagenum": photo_number, "photo_path": fpath}
)
photo_ids = pd.DataFrame(photo_ids)
df = pd.merge(df_attrs, photo_ids, on=("person", "imagenum"))
assert len(df) == len(df_attrs), "lost some data when merging dataframes"
all_photos = (
df["photo_path"]
.apply(imageio.imread)
.apply(lambda img: img[dy:-dy, dx:-dx])
.apply(lambda img: np.array(Image.fromarray(img).resize([dimx, dimy])))
)
all_photos = np.stack(all_photos.values).astype("uint8")
all_attrs = df.drop(["photo_path", "person", "imagenum"], axis=1)
return all_photos, all_attrs
data, attrs = fetch_dataset()
# 45,45
IMAGE_H = data.shape[1]
IMAGE_W = data.shape[2]
N_CHANNELS = 3
data = np.array(data / 255, dtype="float32")
X_train, X_val = train_test_split(data, test_size=0.2, random_state=42)
X_train = torch.FloatTensor(X_train)
X_val = torch.FloatTensor(X_val)
# # Building simple autoencoder
dim_z = 100
X_train.shape
class Autoencoder(nn.Module):
def __init__(self):
super().__init__()
self.encoder = nn.Sequential(
nn.Linear(45 * 45 * 3, 1500),
nn.BatchNorm1d(1500),
nn.ReLU(),
nn.Linear(1500, 1000),
nn.BatchNorm1d(1000),
nn.ReLU(),
# nn.Linear(1000,500),
# nn.ReLU(),
nn.Linear(1000, dim_z),
nn.BatchNorm1d(dim_z),
nn.ReLU(),
)
self.decoder = nn.Sequential(
nn.Linear(dim_z, 1000),
nn.BatchNorm1d(1000),
nn.ReLU(),
# nn.Linear(500,1000),
# nn.ReLU(),
nn.Linear(1000, 1500),
nn.BatchNorm1d(1500),
nn.ReLU(),
nn.Linear(1500, 45 * 45 * 3),
)
def encode(self, x):
return self.encoder(x)
def decode(self, z):
return self.decoder(z)
def forward(self, x):
# print("shape",x.shape)
# x = x.view(-1,45*45*3)
encoded = self.encode(x)
# latent_view = self.latent_view(encoder)
# decoder = self.decoder(latent_view).resize_((45,45,3))
decoded = self.decode(encoded)
return encoded, decoded
class Autoencoder_cnn(nn.Module):
def __init__(self):
super().__init__()
self.encoder = nn.Sequential(
nn.Conv2d(in_channels=3, out_channels=16, kernel_size=3),
nn.ReLU(),
nn.MaxPool2d(kernel_size=2),
nn.Conv2d(in_channels=16, out_channels=8, kernel_size=3),
nn.ReLU(),
nn.MaxPool2d(kernel_size=2),
)
self.decoder = nn.Sequential(
nn.ConvTranspose2d(in_channels=8, out_channels=16, kernel_size=5, stride=2),
nn.ReLU(),
nn.ConvTranspose2d(in_channels=16, out_channels=3, kernel_size=5, stride=2),
# nn.ReLU(),
# nn.ConvTranspose2d(in_channels=8, out_channels=8, kernel_size=3, stride=2),
# nn.ReLU(),
# nn.ConvTranspose2d(in_channels=8, out_channels=3, kernel_size=5, stride=2)
)
def decode(self, z):
return self.decoder(z)
def forward(self, x):
# print("shape",x.shape)
# torch.Size([64, 45, 45, 3])
# x = x.view(-1,45*45*3)
# change to
x = x.permute(0, 3, 1, 2)
encoded = self.encoder(x)
# torch.Size([64, 8, 9, 9])
# print(encoded.shape)
# latent_view = self.latent_view(encoder)
# decoder = self.decoder(latent_view).resize_((45,45,3))
decoded = self.decode(encoded)
return encoded, decoded
model_cnn = Autoencoder_cnn().cuda()
model_auto = Autoencoder().to(DEVICE)
print(model_auto)
print(model_cnn)
# cnn
# torch.Size([64, 45, 45, 3]) -> permute to (64, 3, 45, 45)
# decoder shape: torch.Size([64, 3, 13, 13])
# # Train autoencoder
def get_batch(data, batch_size=64):
total_len = data.shape[0]
for i in range(0, total_len, batch_size):
yield data[i : min(i + batch_size, total_len)]
def plot_gallery(images, h, w, n_row=3, n_col=6, with_title=False, titles=[]):
plt.figure(figsize=(1.5 * n_col, 1.7 * n_row))
plt.subplots_adjust(bottom=0, left=0.01, right=0.99, top=0.90, hspace=0.35)
for i in range(n_row * n_col):
plt.subplot(n_row, n_col, i + 1)
try:
plt.imshow(
images[i].reshape((h, w, 3)),
cmap=plt.cm.gray,
vmin=-1,
vmax=1,
interpolation="nearest",
)
if with_title:
plt.title(titles[i])
plt.xticks(())
plt.yticks(())
except:
pass
def fit_epoch(model, train_x, criterion, optimizer, batch_size, is_cnn=False):
running_loss = 0.0
processed_data = 0
for inputs in get_batch(train_x, batch_size):
if not is_cnn:
inputs = inputs.view(-1, 45 * 45 * 3)
inputs = inputs.to(DEVICE)
optimizer.zero_grad()
encoder, decoder = model(inputs)
# print('decoder shape: ', decoder.shape)
if not is_cnn:
outputs = decoder.view(-1, 45 * 45 * 3)
else:
outputs = decoder.permute(0, 2, 3, 1)
loss = criterion(outputs, inputs)
loss.backward()
optimizer.step()
running_loss += loss.item() * inputs.shape[0]
processed_data += inputs.shape[0]
train_loss = running_loss / processed_data
return train_loss
def eval_epoch(model, x_val, criterion, is_cnn=False):
running_loss = 0.0
processed_data = 0
model.eval()
for inputs in get_batch(x_val):
if not is_cnn:
inputs = inputs.view(-1, 45 * 45 * 3)
inputs = inputs.to(DEVICE)
with torch.set_grad_enabled(False):
encoder, decoder = model(inputs)
if not is_cnn:
outputs = decoder.view(-1, 45 * 45 * 3)
else:
outputs = decoder.permute(0, 2, 3, 1)
loss = criterion(outputs, inputs)
running_loss += loss.item() * inputs.shape[0]
processed_data += inputs.shape[0]
val_loss = running_loss / processed_data
# draw
with torch.set_grad_enabled(False):
pic = x_val[3]
if not is_cnn:
pic_input = pic.view(-1, 45 * 45 * 3)
else:
# print(pic.shape)
pic_input = torch.FloatTensor(pic.unsqueeze(0))
# print(pic.shape)
pic_input = pic_input.to(DEVICE)
encoder, decoder = model(pic_input)
if not is_cnn:
pic_output = decoder.view(-1, 45 * 45 * 3).squeeze()
else:
# print('decoder shape eval: ', decoder.shape)
pic_output = decoder.permute(0, 2, 3, 1)
pic_output = pic_output.to("cpu")
# pic_output = pic_output.view(-1, 45,45,3)
# print(pic)
# print(pic_output)
pic_input = pic_input.to("cpu")
# pics = pic_input + pic_output
# plot_gallery(pics,45,45,10,2)
plot_gallery([pic_input, pic_output], 45, 45, 1, 2)
return val_loss
def train(train_x, val_x, model, epochs=10, batch_size=32, is_cnn=False):
criterion = nn.MSELoss()
optimizer = torch.optim.Adam(model.parameters(), lr=0.001)
history = []
log_template = (
"\nEpoch {ep:03d} train_loss: {t_loss:0.4f} val_loss: {val_loss:0.4f}"
)
with tqdm(desc="epoch", total=epochs) as pbar_outer:
for epoch in range(epochs):
train_loss = fit_epoch(
model, train_x, criterion, optimizer, batch_size, is_cnn
)
val_loss = eval_epoch(model, val_x, criterion, is_cnn)
print("loss: ", train_loss)
history.append((train_loss, val_loss))
pbar_outer.update(1)
tqdm.write(
log_template.format(ep=epoch + 1, t_loss=train_loss, val_loss=val_loss)
)
return history
# history_cnn = train(X_train, X_val, model_cnn, epochs=30, batch_size=64, is_cnn=True)
history = train(X_train, X_val, model_auto, epochs=50, batch_size=64)
train_loss, val_loss = zip(*history)
plt.figure(figsize=(15, 10))
plt.plot(train_loss, label="Train loss")
plt.plot(val_loss, label="Val loss")
plt.legend(loc="best")
plt.xlabel("epochs")
plt.ylabel("loss")
plt.plot()
# # Sampling
# Let's generate some samples from random vectors
z = np.random.randn(25, dim_z)
print(z.shape)
with torch.no_grad():
inputs = torch.FloatTensor(z)
inputs = inputs.to(DEVICE)
model_auto.eval()
output = model_auto.decode(inputs)
# print(output)
plot_gallery(output.data.cpu().numpy(), IMAGE_H, IMAGE_W, n_row=5, n_col=5)
# Convolution autoencoder sampling
# z = torch.rand((25,8,9,9))
#
# with torch.no_grad():
# inputs = torch.FloatTensor(z)
#
# inputs = inputs.to(DEVICE)
# model_cnn.eval()
# output = model_cnn.decode(inputs)
# print(output.shape)
# output = output.permute(0,2,3,1)
# print(output.shape)
#
# #print(output)
#
# plot_gallery(output.data.cpu().numpy(), IMAGE_H, IMAGE_W, n_row=5, n_col=5)
# Attributes
# # Adding smile and glasses
# Let's find some attributes like smiles or glasses on the photo and try to add it to the photos which don't have it. We will use the second dataset for it. It contains a bunch of such attributes.
attrs.head()
attrs.columns
smile_ids = attrs["Smiling"].sort_values(ascending=False).iloc[100:125].index.values
smile_data = data[smile_ids]
no_smile_ids = attrs["Smiling"].sort_values(ascending=True).head(25).index.values
no_smile_data = data[no_smile_ids]
eyeglasses_ids = attrs["Eyeglasses"].sort_values(ascending=False).head(25).index.values
eyeglasses_data = data[eyeglasses_ids]
sunglasses_ids = attrs["Sunglasses"].sort_values(ascending=False).head(25).index.values
sunglasses_data = data[sunglasses_ids]
plot_gallery(
smile_data, IMAGE_H, IMAGE_W, n_row=5, n_col=5, with_title=True, titles=smile_ids
)
plot_gallery(
no_smile_data,
IMAGE_H,
IMAGE_W,
n_row=5,
n_col=5,
with_title=True,
titles=no_smile_ids,
)
plot_gallery(
eyeglasses_data,
IMAGE_H,
IMAGE_W,
n_row=5,
n_col=5,
with_title=True,
titles=eyeglasses_ids,
)
plot_gallery(
sunglasses_data,
IMAGE_H,
IMAGE_W,
n_row=5,
n_col=5,
with_title=True,
titles=sunglasses_ids,
)
# Calculating latent space vector for the selected images.
def to_latent(pic):
with torch.no_grad():
inputs = torch.FloatTensor(pic.reshape(-1, 45 * 45 * 3))
inputs = inputs.to(DEVICE)
model_auto.eval()
output = model_auto.encode(inputs)
return output
def from_latent(vec):
with torch.no_grad():
inputs = vec.to(DEVICE)
model_auto.eval()
output = model_auto.decode(inputs)
return output
smile_data[0].reshape(-1, 45 * 45 * 3).shape
smile_latent = to_latent(smile_data).mean(axis=0)
no_smile_latent = to_latent(no_smile_data).mean(axis=0)
sunglasses_latent = to_latent(sunglasses_data).mean(axis=0)
smile_vec = smile_latent - no_smile_latent
sunglasses_vec = sunglasses_latent - smile_latent
def make_me_smile(ids):
for id in ids:
pic = data[id : id + 1]
latent_vec = to_latent(pic)
latent_vec[0] += smile_vec
pic_output = from_latent(latent_vec)
pic_output = pic_output.view(-1, 45, 45, 3).cpu()
plot_gallery([pic, pic_output], IMAGE_H, IMAGE_W, n_row=1, n_col=2)
def give_me_sunglasses(ids):
for id in ids:
pic = data[id : id + 1]
latent_vec = to_latent(pic)
latent_vec[0] += sunglasses_vec
pic_output = from_latent(latent_vec)
pic_output = pic_output.view(-1, 45, 45, 3).cpu()
plot_gallery([pic, pic_output], IMAGE_H, IMAGE_W, n_row=1, n_col=2)
make_me_smile(no_smile_ids)
give_me_sunglasses(smile_ids)
# While the concept is pretty straightforward the simple autoencoder have some disadvantages. Let's explore them and try to do better.
# # Variational autoencoder
# So far we have trained our encoder to reconstruct the very same image that we've transfered to latent space. That means that when we're trying to **generate** new image from the point decoder never met we're getting _the best image it can produce_, but the quelity is not good enough.
# > **In other words the encoded vectors may not be continuous in the latent space.**
# In other hand Variational Autoencoders makes not only one encoded vector but **two**:
# - vector of means, μ;
# - vector of standard deviations, σ.
# 
# > picture from https://towardsdatascience.com/intuitively-understanding-variational-autoencoders-1bfe67eb5daf
#
dim_z = 256
class VAE(nn.Module):
def __init__(self):
super().__init__()
self.enc_fc1 = nn.Linear(1000, dim_z)
self.enc_fc2 = nn.Linear(1000, dim_z)
self.encoder = nn.Sequential(
nn.Linear(45 * 45 * 3, 1500),
# nn.BatchNorm1d(1500),
nn.ReLU(),
nn.Linear(1500, 1000),
# nn.BatchNorm1d(1000),
nn.ReLU(),
)
self.decoder = nn.Sequential(
nn.Linear(dim_z, 1000),
# nn.BatchNorm1d(1000),
nn.ReLU(),
nn.Linear(1000, 1500),
# nn.BatchNorm1d(1500),
nn.ReLU(),
nn.Linear(1500, 45 * 45 * 3),
)
def encode(self, x):
x = self.encoder(x)
return self.enc_fc1(x), self.enc_fc2(x)
def reparametrize(self, mu, logvar):
##if self.training:
# std = logsigma.exp_()
# eps = Variable(std.data.new(std.size()).normal_())
# return eps.mul(std).add_(mu)
##else:
##return mu
std = logvar.mul(0.5).exp_()
eps = torch.cuda.FloatTensor(std.size()).normal_()
eps = Variable(eps)
return eps.mul(std).add_(mu)
def decode(self, z):
return self.decoder(z)
def forward(self, x):
mu, logvar = self.encode(x)
encoded = self.reparametrize(mu, logvar)
decoded = self.decode(encoded)
return mu, logvar, decoded
# -----------------------------------------------------------------
class VAE2(nn.Module):
def __init__(self):
super(VAE2, self).__init__()
# self.training = True
self.fc1 = nn.Linear(45 * 45 * 3, 1500)
self.fc21 = nn.Linear(1500, 200)
self.fc22 = nn.Linear(1500, 200)
self.fc3 = nn.Linear(200, 1500)
self.fc4 = nn.Linear(1500, 45 * 45 * 3)
self.relu = nn.LeakyReLU()
def encode(self, x):
x = self.relu(self.fc1(x))
return self.fc21(x), self.fc22(x)
def reparametrize(self, mu, logvar):
std = logvar.mul(0.5).exp_()
if torch.cuda.is_available():
eps = torch.cuda.FloatTensor(std.size()).normal_()
else:
eps = torch.FloatTensor(std.size()).normal_()
eps = Variable(eps)
zz = eps.mul(std).add_(mu)
# print(zz.shape)
return zz
def reparametrize2(self, mu, logvar):
if self.training:
std = logvar.exp_()
eps = Variable(std.data.new(std.size()).normal_())
return eps.mul(std).add_(mu)
else:
print("training is off")
return mu
def reparametrize3(self, mu, logvar):
std = logvar.exp_()
eps = Variable(std.data.new(std.size()).normal_())
return eps.mul(std).add_(mu)
# --new---------------------------------------
def reparameterize4(self, mu, logvar):
std = torch.exp(0.5 * logvar)
eps = torch.randn_like(std)
return eps.mul(std).add_(mu)
# ---------------------------------------
def decode(self, z):
z = self.relu(self.fc3(z)) # 1500
return torch.sigmoid(self.fc4(z))
def forward(self, x):
mu, logvar = self.encode(x)
z = self.reparameterize4(mu, logvar)
z = self.decode(z)
return z, mu, logvar
def KL_divergence(mu, logsigma):
return -(1 / 2) * (1 + 2 * logsigma - mu**2 - torch.exp(logsigma) ** 2).sum(
dim=-1
)
def log_likelihood(x, reconstruction):
loss = nn.BCELoss()
loss_val = loss(reconstruction, x)
a = loss_val
return a
def loss_vae(x, mu, logsigma, reconstruction):
# kl = -KL_divergence(mu, logsigma)
# like = log_likelihood(x, reconstruction)
# print('kl ',kl)
# print('like', like)
# return -(kl + like).mean()
# return -(-KL_divergence(mu, logsigma) + log_likelihood(x, reconstruction)).mean()
# print(nn.BCELoss(reduction = 'sum')(reconstruction, x))
BCE = nn.BCELoss(reduction="sum")(reconstruction, x)
KLD = -0.5 * (1 + logsigma - mu.pow(2) - logsigma.exp().pow(2)).sum()
print("BCE ", BCE)
print("KLD ", KLD)
return BCE + KLD
def loss_vae2(x, mu, logvar, reconstruction):
loss = nn.BCELoss()
BCE = loss(reconstruction, x)
KLD_element = mu.pow(2).add_(logvar.exp()).mul_(-1).add_(1).add_(logvar)
KLD = torch.sum(KLD_element).mul_(-0.5)
# KL divergence
return BCE + KLD
# return -(-KL_divergence(mu, logsigma) + log_likelihood(x, reconstruction)).mean()
def loss_vae_fn(x, recon_x, mu, logvar):
# print('recon_x ',recon_x.shape)
# print('x ',x.shape)
BCE = F.binary_cross_entropy(recon_x, x, reduction="sum")
# see Appendix B from VAE paper:
# Kingma and Welling. Auto-Encoding Variational Bayes. ICLR, 2014
# https://arxiv.org/abs/1312.6114
# 0.5 * sum(1 + log(sigma^2) - mu^2 - sigma^2)
KLD = -0.5 * torch.sum(1 + logvar - mu.pow(2) - logvar.exp())
return BCE + KLD
# model_vae = VAE().cuda()
model_vae = VAE2().to(DEVICE)
def fit_epoch_vae(model, train_x, optimizer, batch_size, is_cnn=False):
running_loss = 0.0
processed_data = 0
for inputs in get_batch(train_x, batch_size):
inputs = inputs.view(-1, 45 * 45 * 3)
inputs = inputs.to(DEVICE)
optimizer.zero_grad()
(
decoded,
mu,
logvar,
) = model(inputs)
# print('decoded shape: ', decoded.shape)
outputs = decoded.view(-1, 45 * 45 * 3)
outputs = outputs.to(DEVICE)
loss = loss_vae_fn(inputs, outputs, mu, logvar)
loss.backward()
optimizer.step()
running_loss += loss.item() * inputs.shape[0]
processed_data += inputs.shape[0]
train_loss = running_loss / processed_data
return train_loss
def eval_epoch_vae(model, x_val, batch_size):
running_loss = 0.0
processed_data = 0
model.eval()
for inputs in get_batch(x_val, batch_size=batch_size):
inputs = inputs.view(-1, 45 * 45 * 3)
inputs = inputs.to(DEVICE)
with torch.set_grad_enabled(False):
decoded, mu, logvar = model(inputs)
# print('decoder shape: ', decoder.shape)
outputs = decoded.view(-1, 45 * 45 * 3)
loss = loss_vae_fn(inputs, outputs, mu, logvar)
running_loss += loss.item() * inputs.shape[0]
processed_data += inputs.shape[0]
val_loss = running_loss / processed_data
# draw
with torch.set_grad_enabled(False):
pic = x_val[3]
pic_input = pic.view(-1, 45 * 45 * 3)
pic_input = pic_input.to(DEVICE)
decoded, mu, logvar = model(inputs)
# model.training = False
# print('decoder shape: ', decoded.shape)
pic_output = decoded[0].view(-1, 45 * 45 * 3).squeeze()
# pic_output2 = decoder[1].view(-1, 45*45*3).squeeze()
# pic_output3 = decoder[2].view(-1, 45*45*3).squeeze()
# print(pic_input)
# print('input shape ', pic_input.shape)
# print('outout shape ', pic_output.shape)
pic_output = pic_output.to("cpu")
# pic_output2 = pic_output2.to("cpu")
# pic_output3 = pic_output3.to("cpu")
pic_input = pic_input.to("cpu")
# print(pic_input)
plot_gallery([pic_input, pic_output], 45, 45, 1, 2)
return val_loss
def train_vae(train_x, val_x, model, epochs=10, batch_size=32, lr=0.001):
optimizer = torch.optim.Adam(model.parameters(), lr=lr)
history = []
log_template = (
"\nEpoch {ep:03d} train_loss: {t_loss:0.4f} val_loss: {val_loss:0.4f}"
)
with tqdm(desc="epoch", total=epochs) as pbar_outer:
for epoch in range(epochs):
train_loss = fit_epoch_vae(model, train_x, optimizer, batch_size)
val_loss = eval_epoch_vae(model, val_x, batch_size)
print("loss: ", train_loss)
history.append((train_loss, val_loss))
pbar_outer.update(1)
tqdm.write(
log_template.format(ep=epoch + 1, t_loss=train_loss, val_loss=val_loss)
)
return history
history_vae = train_vae(X_train, X_val, model_vae, epochs=20, batch_size=64, lr=0.0005)
train_loss, val_loss = zip(*history_vae)
plt.figure(figsize=(15, 10))
plt.plot(train_loss, label="Train loss")
plt.plot(val_loss, label="Val loss")
plt.legend(loc="best")
plt.xlabel("epochs")
plt.ylabel("loss")
plt.plot()
|
# # Covid-19 CT ECKP 2023 - Evan
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.metrics import classification_report
import tensorflow as tf
from tensorflow.keras.preprocessing import image_dataset_from_directory
from tensorflow.keras.applications import ResNet50
from tensorflow.keras.models import Sequential, load_model
from tensorflow.keras.layers import (
Dense,
Conv2D,
MaxPooling2D,
InputLayer,
Flatten,
Dropout,
)
def plot_scores(train):
accuracy = train.history["accuracy"]
val_accuracy = train.history["val_accuracy"]
epochs = range(len(accuracy))
plt.plot(epochs, accuracy, "b", label="Score apprentissage")
plt.plot(epochs, val_accuracy, "r", label="Score validation")
plt.title("Scores")
plt.legend()
plt.show()
data_root = "/kaggle/input/large-covid19-ct-slice-dataset/curated_data/curated_data"
random_seed = 1075
batch_size = 32
validation_split = 0.2
image_size = (256, 256)
train_dataset = image_dataset_from_directory(
data_root,
validation_split=validation_split,
subset="training",
seed=random_seed,
image_size=image_size,
batch_size=batch_size,
label_mode="categorical",
)
val_dataset = image_dataset_from_directory(
data_root,
validation_split=validation_split,
subset="validation",
seed=random_seed,
image_size=image_size,
batch_size=batch_size,
label_mode="categorical",
)
resnet50 = ResNet50(include_top=False, input_shape=(256, 256, 3), classes=3)
model = Sequential()
model.add(resnet50)
model.add(Flatten())
model.add(Dense(64, activation="relu"))
model.add(Dense(3, activation="softmax"))
model.summary()
model.compile(
loss="categorical_crossentropy",
optimizer=tf.keras.optimizers.SGD(1e-2),
metrics=["accuracy"],
)
history = model.fit(train_dataset, validation_data=val_dataset, epochs=10, verbose=1)
plot_scores(history)
y_val = np.array([])
y_hat = np.array([])
for x, y in val_dataset:
y_val = np.concatenate([y_val, np.argmax(y.numpy(), axis=-1)])
y_hat = np.concatenate([y_hat, np.argmax(model.predict(x), axis=1)])
print(classification_report(y_val, y_hat, target_names=val_dataset.class_names))
meta_covid = pd.read_csv(
"/kaggle/input/large-covid19-ct-slice-dataset/meta_data_covid.csv",
encoding="windows-1252",
)
meta_cap = pd.read_csv(
"/kaggle/input/large-covid19-ct-slice-dataset/meta_data_cap.csv",
encoding="windows-1252",
)
meta_normal = pd.read_csv(
"/kaggle/input/large-covid19-ct-slice-dataset/meta_data_normal.csv",
encoding="windows-1252",
)
print(f"meta_covid: {meta_covid.shape}")
print(f"meta_cap: {meta_cap.shape}")
print(f"meta_normal: {meta_normal.shape}")
print(f"meta_covid: {meta_covid.columns}")
print(f"meta_normal: {meta_normal.columns}")
print(f"meta_cap: {meta_cap.columns}")
meta_covid.head()
meta_covid.count()
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import xgboost as xgb
import seaborn as sns
import time
from math import sqrt
from numpy import loadtxt
from itertools import product
from tqdm import tqdm
from numpy import loadtxt
import gc
from sklearn import preprocessing
from sklearn.model_selection import cross_val_score
from sklearn.metrics import mean_squared_error, f1_score
from sklearn.metrics import accuracy_score
from sklearn.model_selection import KFold
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import LabelEncoder
from catboost import CatBoostClassifier
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
def reduce_mem_usage(df):
"""
iterate through all the columns of a dataframe and
modify the data type to reduce memory usage.
"""
start_mem = df.memory_usage().sum() / 1024**2
print(("Memory usage of dataframe is {:.2f}" "MB").format(start_mem))
for col in df.columns:
col_type = df[col].dtype
print(str(col_type))
if str(col_type) == "datetime64[ns]":
continue
if col_type != object:
c_min = df[col].min()
c_max = df[col].max()
if str(col_type)[:3] == "int":
if c_min > np.iinfo(np.int8).min and c_max < np.iinfo(np.int8).max:
df[col] = df[col].astype(np.int8)
elif c_min > np.iinfo(np.int16).min and c_max < np.iinfo(np.int16).max:
df[col] = df[col].astype(np.int16)
elif c_min > np.iinfo(np.int32).min and c_max < np.iinfo(np.int32).max:
df[col] = df[col].astype(np.int32)
elif c_min > np.iinfo(np.int64).min and c_max < np.iinfo(np.int64).max:
df[col] = df[col].astype(np.int64)
elif str(col_type) != "Timestamp":
if (
c_min > np.finfo(np.float16).min
and c_max < np.finfo(np.float16).max
):
df[col] = df[col].astype(np.float16)
elif (
c_min > np.finfo(np.float32).min
and c_max < np.finfo(np.float32).max
):
df[col] = df[col].astype(np.float32)
else:
df[col] = df[col].astype(np.float64)
else:
df[col] = df[col].astype("category")
end_mem = df.memory_usage().sum() / 1024**2
print(("Memory usage after optimization is: {:.2f}" "MB").format(end_mem))
print("Decreased by {:.1f}%".format(100 * (start_mem - end_mem) / start_mem))
return df
def df_info(df):
print("----------Top-5- Record----------")
print(df.head(5))
print("-----------Information-----------")
print(df.info())
print("-----------Data Types-----------")
print(df.dtypes)
print("----------Missing value-----------")
print(df.isnull().sum())
print("----------Null value-----------")
print(df.isna().sum())
print("----------Shape of Data----------")
print(df.shape)
print("----------description of Data----------")
print(df.describe())
print("----------Uniques of Data----------")
print(df.nunique())
print("------------Columns in data---------")
print(df.columns)
def downcast_dtypes(df):
float_cols = [c for c in df if df[c].dtype == "float64"]
int_cols = [c for c in df if df[c].dtype in ["int64", "int32"]]
df[float_cols] = df[float_cols].astype(np.float32)
df[int_cols] = df[int_cols].astype(np.int16)
return df
def model_performance_sc_plot(predictions, labels, title):
min_val = max(max(predictions), max(labels))
max_val = min(min(predictions), min(labels))
performance_df = pd.DataFrame({"Labels": labels})
performance_df["Predictions"] = predictions
sns.jointplot(y="Labels", x="Predictions", data=performance_df, kind="reg")
plt.plot([min_val, max_val], [min_val, max_val], "m--")
plt.title(title)
plt.show()
def calculate_counts(column_name, ref_sets, extension_set):
ref_sets_ids = [set(ref[column_name]) for ref in ref_sets]
ext_ids = set(extension_set[column_name])
refs_union = reduce(lambda s1, s2: s1 | s2, ref_sets_ids)
ref_counts = [len(ref) for ref in ref_sets_ids]
ext_count = len(ext_ids)
union_count = len(refs_union)
intersection_count = len(ext_ids & refs_union)
all_counts = ref_counts + [union_count, ext_count, intersection_count]
res_index = ["Ref {}".format(i) for i in range(1, len(ref_sets) + 1)] + [
"Refs Union",
"Extension",
"Union x Extension",
]
return pd.DataFrame({"Count": all_counts}, index=res_index)
train_path = "/kaggle/input/airplane-accidents-severity-dataset/train.csv"
test_path = "/kaggle/input/airplane-accidents-severity-dataset/test.csv"
submission_path = (
"/kaggle/input/airplane-accidents-severity-dataset/sample_submission.csv"
)
train = pd.read_csv(train_path)
df_info(train)
object_cols = ["Severity"]
lb = LabelEncoder()
lb.fit(train[object_cols])
train[object_cols] = lb.transform(train[object_cols])
train["Control_Metric"] = train["Control_Metric"].clip(25, 100)
train.head()
# Accident type code mean wrt control metric, Severity, Turbulence, Maxelevation, Adverse Weatehr
means_accident_code = train.groupby(["Accident_Type_Code"]).agg(
{
"Severity": "mean",
"Safety_Score": "mean",
"Days_Since_Inspection": "mean",
"Total_Safety_Complaints": "mean",
"Control_Metric": "mean",
"Turbulence_In_gforces": "mean",
"Cabin_Temperature": "mean",
"Max_Elevation": "mean",
"Violations": "mean",
"Adverse_Weather_Metric": "mean",
}
)
means_accident_code = means_accident_code.reset_index()
cols = list(means_accident_code.columns)
for i in range(1, len(cols)):
cols[i] = cols[i] + "_mean"
print(cols)
means_accident_code.columns = cols
train = train.merge(means_accident_code, on=["Accident_Type_Code"], how="left")
df = train.groupby(["Accident_Type_Code", "Severity"])["Severity"].agg({"no": "count"})
mask = df.groupby(level=0).agg("idxmax")
df_count = df.loc[mask["no"]]
df_count.drop(["no"], axis=1, inplace=True)
df_count = df_count.reset_index()
df_count.columns = ["Accident_Type_Code", "Severity_max"]
train = train.merge(df_count, on=["Accident_Type_Code"], how="left")
train.drop(["Accident_Type_Code"], axis=1, inplace=True)
train.head()
X = train.drop(["Severity", "Accident_ID"], axis=1)
Y = train["Severity"].values
x_train, x_val, y_train, y_val = train_test_split(X, Y, test_size=0.25)
x_val1, x_val2, y_val1, y_val2 = train_test_split(x_val, y_val, test_size=0.25)
test = pd.read_csv(test_path)
test.head().T
test = test.merge(means_accident_code, on=["Accident_Type_Code"], how="left")
test = test.merge(df_count, on=["Accident_Type_Code"], how="left")
test.drop(["Accident_Type_Code"], axis=1, inplace=True)
x_test = test.drop(["Accident_ID"], axis=1)
cb = CatBoostClassifier(
iterations=1000,
max_ctr_complexity=10,
random_seed=0,
od_type="Iter",
od_wait=50,
verbose=100,
depth=12,
)
from xgboost import XGBClassifier
xgbRegressor = XGBClassifier(
max_depth=10, eta=0.2, n_estimators=500, colsample_bytree=0.7, subsample=0.7, seed=0
)
from sklearn.model_selection import KFold
kf = KFold(n_splits=5, shuffle=True)
y_pred = np.zeros((Y.shape[0], 1))
for train_index, test_index in kf.split(X):
x_train, y_train = X.loc[train_index, :], Y[train_index]
x_val, y_val = X.loc[test_index, :], Y[test_index]
cb.fit(x_train, y_train)
y_pred_cb = cb.predict(x_val)
y_pred[test_index] = y_pred_cb
print("F1_Score val1 : ", f1_score(y_val, y_pred_cb, average="weighted"))
from sklearn.model_selection import KFold
kf = KFold(n_splits=5, shuffle=True)
y_pred_xg = np.zeros((Y.shape[0], 1))
for train_index, test_index in kf.split(X):
x_train, y_train = X.loc[train_index, :], Y[train_index]
x_val, y_val = X.loc[test_index, :], Y[test_index]
xgbRegressor.fit(x_train, y_train, verbose=100)
y_pred_x = xgbRegressor.predict(x_val)
y_pred_xg[test_index] = y_pred_x.reshape(-1, 1)
print("F1_Score val1 : ", f1_score(y_val, y_pred_x, average="weighted"))
cb.fit(X, Y)
test_pred_cb = cb.predict(x_test)
xgbRegressor.fit(X, Y)
test_pred_xg = xgbRegressor.predict(x_test)
first_level = pd.DataFrame(y_pred, columns=["catboost"])
first_level["XGB"] = y_pred_xg
first_level_test = pd.DataFrame(test_pred_cb, columns=["catboost"])
first_level_test["XGB"] = test_pred_xg
first_level_train, first_level_val, y_train, y_val = train_test_split(
first_level, Y, test_size=0.3
)
metamodel = XGBClassifier(
max_depth=8, eta=0.2, n_estimators=500, colsample_bytree=0.7, subsample=0.7, seed=0
)
metamodel.fit(
first_level_train,
y_train,
eval_set=[(first_level_train, y_train), (first_level_val, y_val)],
verbose=20,
early_stopping_rounds=120,
)
ensemble_pred = metamodel.predict(first_level_val)
test_pred = metamodel.predict(first_level_test)
print("Val F1-Score :", f1_score(y_val, ensemble_pred, average="weighted"))
model_performance_sc_plot(ensemble_pred, y_val, "Validation")
# xgbRegressor.fit(X, Y, eval_set = [(X,Y),(x_val1, y_val1)], verbose = 20, early_stopping_rounds = 120)
# test_pred = xgbRegressor.predict(x_test)
y_pred = test_pred
y_pred = y_pred.astype(np.int32)
y_pred = lb.inverse_transform(y_pred)
ids = []
for i in range(test.shape[0]):
ids.append(test.loc[i, "Accident_ID"])
sub = pd.DataFrame(ids, columns=["Accident_ID"])
sub["Severity"] = y_pred
sub.head()
sub.to_csv("Submission.csv", index=False)
|
# 
# # Developer's Survey Data Analysis
# ####We have imported the Developers Survey Dataset which was already available on Kaggle Datasets. This data was collected from the users of Stack Overflow in 2020. We will analyze the survey data and try to find some important insites from the dataset.
# * We will check our directory for the required .csv files from the list of files.
# checking the directory path
import os
os.listdir("/kaggle/input/stack-overflow-developer-survey-2020/developer_survey_2020")
# ####There are four files in our folder, but we need only need files with Survey Questions and Survey Submissions
# importing required libraries
import pandas as pd
import numpy as np
# ####We will import the required files with the help of the pandas therefore we will install the current required libraries for the analysis. The files will be imported as a dataframe with help of *read_csv()* of pandas.
# importing Data
raw_df = pd.read_csv(
"/kaggle/input/stack-overflow-developer-survey-2020/developer_survey_2020/survey_results_public.csv"
)
raw_question_df = pd.read_csv(
"/kaggle/input/stack-overflow-developer-survey-2020/developer_survey_2020/survey_results_schema.csv"
)
# ####Observing the basic structure of the dataset
# viewing data frame
raw_df.head(2)
# observing the columns of the dataset
raw_df.columns
# ####As we can observe that there are many unnecessary columns as per our requirement for the analysis, so we will create a list of columns that we need for the analysis.
# create a list of column that we need for the analysis
selected_columns = [
"Age",
"Age1stCode",
"Employment",
"Gender",
"DevType",
"YearsCode",
"YearsCodePro",
"LanguageDesireNextYear",
"LanguageWorkedWith",
"EdLevel",
"UndergradMajor",
"WorkWeekHrs",
"Hobbyist",
"Country",
]
# ####We will create a new Dataframe from the *raw_df* with only the selected columns, this will considerably reduce the size of dataset used in analysis and it will help us with keeping our focus on important field.
survey_df = raw_df[selected_columns].copy()
# observing new dataframe
survey_df
# # Cleaning the Dataset
# ####We check each column's data for any kind of inconsistency and if found, we will remove that data. Firstly we will check all the columns with the numeric values for any kind of inconsistency.
# checking the numerical columns for inconsistency
survey_df.Age1stCode.unique()
survey_df.Age.unique()
survey_df.YearsCode.unique()
survey_df.YearsCodePro.unique()
# ####We found some string values in those numeric columns therefore we will use *to_numeric* function of pandas to convert all non numeric data to **NAN**.
survey_df["Age1stCode"] = pd.to_numeric(survey_df.Age1stCode, errors="coerce")
survey_df["Age"] = pd.to_numeric(survey_df.Age, errors="coerce")
survey_df["YearsCode"] = pd.to_numeric(survey_df.YearsCode, errors="coerce")
survey_df["YearsCodePro"] = pd.to_numeric(survey_df.YearsCodePro, errors="coerce")
# ####Cheking the numeric columns after the correction.
survey_df.YearsCodePro.unique()
# ####Performing same cleaning procedures for the second table of Questions.
# check the scema table
survey_question_df = raw_question_df.copy()
# setting index to column name for selected number of column
survey_question_df.set_index("Column", inplace=False)
# ####We will change the index to the Columns for filtering the required questions only.
# using transpose and defining the locations
survey_question = survey_question_df.transpose()
survey_question.columns = survey_question.iloc[0]
survey_question = survey_question.iloc[1:]
# Checking the new dataset with selected questions only
survey_question = survey_question[selected_columns]
survey_question
# ####Describing the survey dataset for basic view of data.
survey_df.describe()
# ####We can clearly observe that there are multiple outliers, we will remove these outlier and keep the data authentic.
# * An **outlier** is an observation that lies an abnormal distance from other values in a random sample from a population.
# * In age column we can see that minimum age is **1** year for a programmer and maximum age is **279** years.
# * In Age1stCode column we can see that minimum age is **5** year for a programmer and maximum age is **85** years
# * In WorkWeekHrs column we can see that minimum Work hour is **1** hour and maximum work hour is **475** hours
# deleting the outliers
# creating a new dataframe from old data frame after droping outlier data
survey_df1 = survey_df.drop(survey_df[survey_df["Age"] < 10].index, inplace=False)
# now using newly created dataframe for further process
survey_df1 = survey_df1.drop(survey_df1[survey_df1["Age"] > 80].index, inplace=False)
survey_df1 = survey_df1.drop(
survey_df1[survey_df1["Age1stCode"] > 80].index, inplace=False
)
survey_df1 = survey_df1.drop(
survey_df1[survey_df1["Age1stCode"] < 10].index, inplace=False
)
survey_df1 = survey_df1.drop(
survey_df1[survey_df1["WorkWeekHrs"] > 140].index, inplace=False
)
survey_df1 = survey_df1.drop(
survey_df1[survey_df1["WorkWeekHrs"] < 10].index, inplace=False
)
# observing the dataset
survey_df1.describe()
# ####We will now clean non-numeric data in dataset.
# checking the other columns for any uncertainity
survey_df1.Gender.value_counts()
# ####We can clearly observe that there are multiple gender values which seems to be typing mistakes.
# * We will keep only 3 types of all values and others will be excluded.
# we need only three gender types
survey_cleaned_df = survey_df1.drop(
survey_df1[survey_df1.Gender.str.contains(";", na=False)].index, inplace=False
)
# observing the new gender column
survey_cleaned_df.Gender.value_counts()
# ####We will check other colums.
survey_cleaned_df.Employment.value_counts()
survey_cleaned_df.Hobbyist.value_counts()
# # Analyzing the Dataset and Creating some user friendly Visualizations.
# ####Importing all the required libraries for the Visualization.
# the numerical data is clean, Now analysing and visualizing the filtered data
import seaborn as sns
import matplotlib
import matplotlib.pyplot as plt
# for keeping the background of the chart darker
sns.set_style("darkgrid")
# ####We will find the highest number of users as per countries.
# first analysing data as per geographical location
survey_df1.Country.value_counts()
countries_programmer = survey_cleaned_df.Country.value_counts().head(20)
# ####Getting the list of top **20** countries with most Programers.
countries_programmer
# plotting the graph for top 20 countries
plt.figure(figsize=(12, 12))
plt.title("Countries with Most Users")
plt.xlabel("Number of users as per survey")
plt.ylabel("Countries")
ax = sns.barplot(x=countries_programmer, y=countries_programmer.index)
# annotating the numerical value of data set on graph
n = 0
for p in ax.patches:
x = p.get_x() + p.get_width() + 0.2
y = p.get_y() + p.get_height() / 2 + 0.1
ax.annotate(countries_programmer[n], (x, y))
n = n + 1
plt.show()
# ####We can observe that **United States** and **India** have Largest user base of programmers.
# let us find some trends based on age
plt.figure(figsize=(12, 12))
plt.title("First Code Age Vs Age")
sns.scatterplot(
x=survey_cleaned_df.Age,
y=survey_cleaned_df.Age1stCode,
hue="Gender",
data=survey_cleaned_df,
)
# ####From above scatter plot we can conclude that
# * Multiple people had exposure to programming after **40** years of age, Even through most popular age for first code is between **10** to **30** years.
# ####We will find the distribution of age of programmers in current senario.
plt.figure(figsize=(9, 9))
plt.title("Programmers Age Distribution")
plt.xlabel("Age")
plt.ylabel("Number of users as per survey")
plt.hist(x=survey_cleaned_df.Age, bins=np.arange(10, 80, 5), color="orange")
# ####From above histogram we can conclude that
# * Maximum number of programmers are between age **20 - 45** years.
survey_cleaned_df.head(2)
# ####We will find the Employment status of programmers as per this survey.
# finding some trends related to the employment type
Employment_df = survey_cleaned_df.Employment.value_counts()
Employment_df
plt.figure(figsize=(9, 6))
plt.title("Employment distribution of People")
plt.xlabel("Number of users as per survey")
plt.ylabel("Employment status")
ax = sns.barplot(x=Employment_df, y=Employment_df.index)
total = len(survey_cleaned_df)
for p in ax.patches:
percentage = "{:.1f}%".format(100 * p.get_width() / total)
x = p.get_x() + p.get_width() + 0.2
y = p.get_y() + p.get_height() / 2
ax.annotate(percentage, (x, y))
plt.show()
# ####From above data we can conclude that
# * Most number of Respondents are **Employed full-time**.
# * A significant amount of people are **Students** and **Freelancers**.
# ####We will identify the Educational status of the Respondent Programmers
# Educational level trends
educational_df = survey_df.EdLevel.value_counts()
educational_df
plt.figure(figsize=(9, 6))
plt.title("Programmers Education Status")
plt.xlabel("Number of users as per survey")
plt.ylabel("Education level")
ax = sns.barplot(x=educational_df, y=educational_df.index)
total = len(survey_cleaned_df)
for p in ax.patches:
percentage = "{:.1f}%".format(100 * p.get_width() / total)
x = p.get_x() + p.get_width() + 0.02
y = p.get_y() + p.get_height() / 2
ax.annotate(percentage, (x, y))
plt.show()
# ####From above data we can conclue that
# * Most of programmers have **Educational** background.
# * Most common Educational level is **Bachelors** and **Masters**.
# ####We will find the most common field of study of programers
# Finding the majors of the programmers
majors_list = survey_cleaned_df.UndergradMajor.value_counts()
majors_list
plt.figure(figsize=(9, 6))
plt.title("Programmers Education Major Status")
plt.xlabel("Number of users as per survey")
plt.ylabel("Education Major")
ax = sns.barplot(x=majors_list, y=majors_list.index)
total = len(survey_cleaned_df)
for p in ax.patches:
percentage = "{:.1f}%".format(100 * p.get_width() / total)
x = p.get_x() + p.get_width() + 0.02
y = p.get_y() + p.get_height() / 2
ax.annotate(percentage, (x, y))
plt.show()
# ####We got some important insites from above data they are as follow
# * Maximum number of programmers have there education in the field of **CS**,**CE** or **SE**.
# * Almost **Half** of the programmers are not from **Computer Science** background, they had there primary education in some other Fields.
# ####We will compare the **Working Hours** of programmers of different nation.
work_week_hours_countries = survey_cleaned_df.groupby("Country")
# Top 40
work_week_hours_countries_df = (
round(work_week_hours_countries["WorkWeekHrs"].mean(), 2)
.sort_values(ascending=False)
.head(40)
)
# All countries
work_week_hours_countries_df_all = round(
work_week_hours_countries["WorkWeekHrs"].mean(), 2
).sort_values(ascending=False)
work_week_hours_countries_df
work_week_hours_countries_df_all.describe()
plt.figure(figsize=(12, 12))
plt.title("Programmers Weekly Working Hours")
plt.xlabel("Working Hours")
plt.ylabel("Countries")
ax = sns.barplot(x=work_week_hours_countries_df, y=work_week_hours_countries_df.index)
n = 0
for p in ax.patches:
x = p.get_x() + p.get_width() + 0.2
y = p.get_y() + p.get_height() / 2 + 0.25
ax.annotate(work_week_hours_countries_df[n], (x, y))
n = n + 1
plt.show()
# ####From above data we can conclude that
# * **Asian** and **African** nations have higher Working hours in Week
# * Average worldwide working hours in a week is **41.6 hours**
# ####We will find the most used language in current senario
survey_cleaned_df.head(2)
# ####As we can observe above the **LanguageWorkedWith** column have multiple language responses as a programmers works on multiple programming languages in their lifetime.
# * We will create function which will convert our language worked with column in a dataframe with languages as there columns and boolean value **True** for used and **False** for not used in there respective cells as Rows will have distinct users.
# finding most popular programming language
# As a single programmer can use multiple languages we need to take all of it in account
def split_multicolumn(series):
result = series.to_frame()
options = []
for idx, value in series[series.notnull()].iteritems():
for option in value.split(";"):
if option not in result:
options.append(option)
result[option] = False
result.at[idx, option] = True
return result[options]
# ####We will use the above created function for finding getting the required dataframe for analysis.
# Creating the required dataframe
Language_Worked_With_df = split_multicolumn(survey_cleaned_df.LanguageWorkedWith)
# Observing the dataframe
Language_Worked_With_df
# calculating the percentage of programers prefering a perticular language
language_currently_used_percentage = round(
(100 * Language_Worked_With_df.sum() / Language_Worked_With_df.count()).sort_values(
ascending=False
),
2,
)
language_currently_used_percentage
plt.figure(figsize=(12, 12))
plt.title("Most used current language")
plt.xlabel("Percentage")
plt.ylabel("Language")
ax = sns.barplot(
x=language_currently_used_percentage, y=language_currently_used_percentage.index
)
n = 0
for p in ax.patches:
x = p.get_x() + p.get_width() + 0.2
y = p.get_y() + p.get_height() / 2
ax.annotate(language_currently_used_percentage[n], (x, y))
n = n + 1
plt.show()
# ####From above data we can conclude that
# * **JavaScript, HTML/CSS ,SQL ,Python ,Java** are the top five most used languages by the respondents
#
Language_Want_to_Work_df = split_multicolumn(survey_cleaned_df.LanguageDesireNextYear)
language_most_loved_percentage = round(
(
100 * Language_Want_to_Work_df.sum() / Language_Want_to_Work_df.count()
).sort_values(ascending=False),
2,
)
language_most_loved_percentage
# ploting the language that are most preffered for future
plt.figure(figsize=(12, 12))
plt.title("Most loved programming language for future")
plt.xlabel("Percentage")
plt.ylabel("Language")
ax = sns.barplot(
x=language_most_loved_percentage, y=language_most_loved_percentage.index
)
n = 0
for p in ax.patches:
x = p.get_x() + p.get_width() + 0.2
y = p.get_y() + p.get_height() / 2
ax.annotate(language_most_loved_percentage[n], (x, y))
n = n + 1
plt.show()
# ####From above data we can observe that
# * **Python** and **JavaScript** are the most preferred language
# * Some new Programming languages like **Rust**, **Go** and **TypeScript** are getting popular
# ####Finding the Gender proportionality of the programmer community
gender_percentage = survey_cleaned_df.Gender.value_counts()
gender_percentage
plt.title("Gender proportion of Programming community")
plt.pie(x=gender_percentage, labels=gender_percentage.index, autopct="%1.1f%%")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
from sklearn.ensemble import RandomForestRegressor
from sklearn.model_selection import GridSearchCV
from sklearn.neural_network import MLPRegressor
# Read training set and test set
train_df = pd.read_csv("/kaggle/input/us-college-completion-rate-analysis/train.csv")
test_df = pd.read_csv("/kaggle/input/us-college-completion-rate-analysis/x_test.csv")
# Select the characteristics and target factors of random forest
features = [
"Tuition_in_state",
"Tuition_out_state",
"Faculty_salary",
"Pell_grant_rate",
"SAT_average",
"ACT_50thPercentile",
"pct_White",
"pct_Black",
"pct_Hispanic",
"pct_Asian",
"Parents_middlesch",
"Parents_highsch",
"Parents_college",
]
target = "Completion_rate"
# Extracting features and target variables from training and test sets
X_train = train_df[features]
y_train = train_df[target]
X_test = test_df[features]
from sklearn.ensemble import ExtraTreesRegressor, BaggingRegressor
# Create the Bagging model and specify the base estimator as Extra Trees Regression
model = BaggingRegressor(
base_estimator=ExtraTreesRegressor(n_estimators=100, random_state=42),
n_estimators=10,
random_state=42,
)
# Fitting the model on the training set
model.fit(X_train, y_train)
# Predicting the results of the test set
y_pred = model.predict(X_test)
# Output CSV file (add ID manually and delete last blank line)
name = ["Completion_rate"]
df = pd.DataFrame(columns=name, data=y_pred_mlp)
print(df)
df.to_csv("submission.csv", index=True, index_label="id")
|
# Importo las librerias
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import os
from glob import glob
import seaborn as sns
from PIL import Image
np.random.seed(123)
from sklearn.preprocessing import label_binarize
from sklearn.metrics import confusion_matrix
import itertools
import keras
from keras.utils.np_utils import (
to_categorical,
) # utilizada para convertir etiquetas a codificación en
from keras.models import Sequential
from keras.layers import Dense, Dropout, Flatten, Conv2D, MaxPool2D
from keras import backend as K
import itertools
from keras.layers.normalization import BatchNormalization
from keras.utils.np_utils import to_categorical
from keras.optimizers import Adam
from keras.preprocessing.image import ImageDataGenerator
from keras.callbacks import ReduceLROnPlateau
from sklearn.model_selection import train_test_split
# aqui grafico el modelo de perddias y el modelo de presision
def plot_model_history(model_history):
fig, axs = plt.subplots(1, 2, figsize=(15, 5))
# acc=accurancy val_acc=valor de precision
axs[0].plot(
range(1, len(model_history.history["acc"]) + 1), model_history.history["acc"]
)
axs[0].plot(
range(1, len(model_history.history["val_acc"]) + 1),
model_history.history["val_acc"],
)
axs[0].set_title("Modelo de precision")
axs[0].set_ylabel("Precision")
axs[0].set_xlabel("Epoch") # epocas
axs[0].set_xticks(
np.arange(1, len(model_history.history["acc"]) + 1),
len(model_history.history["acc"]) / 10,
)
axs[0].legend(["train", "val"], loc="best")
axs[1].plot(
range(1, len(model_history.history["loss"]) + 1), model_history.history["loss"]
)
axs[1].plot(
range(1, len(model_history.history["val_loss"]) + 1),
model_history.history["val_loss"],
)
axs[1].set_title("Modelo de perdidas")
axs[1].set_ylabel("Perdidas")
axs[1].set_xlabel("Epoch") # epocas
axs[1].set_xticks(
np.arange(1, len(model_history.history["loss"]) + 1),
len(model_history.history["loss"]) / 10,
)
axs[1].legend(["train", "val"], loc="best")
plt.show()
# aquise hace un diccionario de ruta de imagenes para poder unir las carpetas HAM10000_images_part1 y part2 en la carpeta base_skin_dir
base_skin_dir = os.path.join("..", "input")
# Fusionar imágenes de ambas carpetas HAM10000_images_part1 y part2 en un diccionario
imageid_path_dict = {
os.path.splitext(os.path.basename(x))[0]: x
for x in glob(os.path.join(base_skin_dir, "*", "*.jpg"))
}
# aqui cambio las etiquetas que se muestran por el nombre completo
lesion_type_dict = {
"nv": "Melanocytic nevi",
"mel": "Melanoma",
"bkl": "Benign keratosis-like lesions ",
"bcc": "Basal cell carcinoma",
"akiec": "Actinic keratoses",
"vasc": "Vascular lesions",
"df": "Dermatofibroma",
}
##aqui se hace la lectura de los datos y se procesan los datos
# Leemos el dataset HAM10000_metadata.csvuniendolo a la ruta de la carpeta de imagenes base_skin_di que hice arriba
skin_df = pd.read_csv(os.path.join(base_skin_dir, "HAM10000_metadata.csv"))
# Aqui creo nuevs columnas para poder tener los datos image_id, cell_type tiene el nombre corto de la etiquta del tipo de lesión y
# cell_type_idx que asigna una categoria a cada tipo de lesión va del 0 al 6
skin_df["path"] = skin_df["image_id"].map(imageid_path_dict.get)
skin_df["cell_type"] = skin_df["dx"].map(lesion_type_dict.get)
skin_df["cell_type_idx"] = pd.Categorical(skin_df["cell_type"]).codes
# para ver como esta el datset modificado
skin_df.head()
# # Limpieza de datos
# veo los valores nulos qeu tienen las columnas del dataset
skin_df.isnull().sum()
# como en la columna edad hay datos vacios y no son muchos entonves voya llenarlos con la media
skin_df["age"].fillna((skin_df["age"].mean()), inplace=True)
# veo si se hizo el cambio
skin_df.isnull().sum()
# veo que tipo d e dato maneja cada columna
print(skin_df.dtypes)
# ## Analisis Exploratorio de los Datos
# aqui grafico los tipos de cancer para ver como estan distribuidos los datos
fig, ax1 = plt.subplots(1, 1, figsize=(10, 5))
skin_df["cell_type"].value_counts().plot(kind="bar", ax=ax1)
# nos indica la forma en la fue diagnosticado el cáncer
skin_df["dx_type"].value_counts().plot(kind="bar")
# Grafica de donde esta unicado
skin_df["localization"].value_counts().plot(kind="bar")
# veo la distribucion de la edad
skin_df["age"].hist(bins=40)
# para ver si son hombres o ujeres los paicentes
skin_df["sex"].value_counts().plot(kind="bar")
# aqui cargo y cambio el tamañod elas imagenes cambio el tamaño por que las dimensiones son de 450 x 600 x3 y tensorflow no tranaja con ese tamaño el tamaño debe ser 100*75
# las imagenes se cargan en la caolumna path(sera la ruta de la imagen) en image se guarda el codigod e la imagen en color rgb
skin_df["image"] = skin_df["path"].map(
lambda x: np.asarray(Image.open(x).resize((100, 75)))
)
# veo dataset
skin_df.head()
# aqui veo las imagenes 5 por cada tipo de lesion
n_samples = 5
fig, m_axs = plt.subplots(7, n_samples, figsize=(4 * n_samples, 3 * 7))
for n_axs, (type_name, type_rows) in zip(
m_axs, skin_df.sort_values(["cell_type"]).groupby("cell_type")
):
n_axs[0].set_title(type_name)
for c_ax, (_, c_row) in zip(
n_axs, type_rows.sample(n_samples, random_state=1234).iterrows()
):
c_ax.imshow(c_row["image"])
c_ax.axis("off")
fig.savefig("category_samples.png", dpi=300)
# Comprobando la distribución del tamaño de la imagen
skin_df["image"].map(lambda x: x.shape).value_counts()
features = skin_df.drop(columns=["cell_type_idx"], axis=1)
target = skin_df["cell_type_idx"]
# ## Datos para prueba y entrenamiento
# vamos a dividos los datos ccon 80:20
x_train_o, x_test_o, y_train_o, y_test_o = train_test_split(
features, target, test_size=0.20, random_state=1234
)
# # Normalizacion
# normalizamos los datos de x_train, x_test restando de sus valores medios y luego dividiendo por su desviación estándar.
x_train = np.asarray(x_train_o["image"].tolist())
x_test = np.asarray(x_test_o["image"].tolist())
x_train_mean = np.mean(x_train)
x_train_std = np.std(x_train)
x_test_mean = np.mean(x_test)
x_test_std = np.std(x_test)
x_train = (x_train - x_train_mean) / x_train_std
x_test = (x_test - x_test_mean) / x_test_std
# La capa de salida también tendrá dos nodos, por lo que necesitamos alimentar nuestra serie de etiquetas en un
# marcador de posición para un escalar y luego convertir esos valores en un vector caliente.
y_train = to_categorical(y_train_o, num_classes=7)
y_test = to_categorical(y_test_o, num_classes=7)
# aqui se hace la divicion de datos para el entrenamiento y para la valizacion
# se usara 90:10 90 para entrenar y 10 para validadicones
x_train, x_validate, y_train, y_validate = train_test_split(
x_train, y_train, test_size=0.1, random_state=2
)
# reshape reforma la imagen debe tener 3 dimenciones (alto=75 px,ancho=100 px,canal=3)
x_train = x_train.reshape(x_train.shape[0], *(75, 100, 3))
x_test = x_test.reshape(x_test.shape[0], *(75, 100, 3))
x_validate = x_validate.reshape(x_validate.shape[0], *(75, 100, 3))
# ## Construcción del modelo CNN
# para construir el modelo uso API secuencial de Keras por que aqui solo se tien que agregar una capa a la vez, comenzando desde la entrada.
# convolucional (Conv2D) es como un conjunto de filtros que se pueden aprender. 32 filtros para las dos primeras capas conv2D, 64 filtros para las dos últimas cada filtro que se usa transforma una parte de la imagen (definida por el tamaño del núcleo) utilizando el filtro del núcleo. La matriz de filtro del núcleo se aplica a toda la imagen.
# agrupación (MaxPool2D) esta capa simplemente actúa como un filtro de disminución de resolución. Mira los 2 píxeles vecinos y selecciona el valor máximo. Estos se utilizan para reducir el costo computacional y, en cierta medida, también reducen el sobreajuste.
# La deserción (Dropout) es un método de regularización, donde una proporción de nodos en la capa se ignora aleatoriamente para cada muestra de entrenamiento. Esto deja caer al azar una promoción de la red y obliga a la red a aprender características de forma distribuida. Esta técnica también mejora la generalización y reduce el sobreajuste.
# 'relu' es el rectificador (función de activación max (0, x). La función de activación del rectificador se utiliza para agregar no linealidad a la red.
# La capa Flatten se usa para convertir los mapas de entidades finales en un solo vector 1D. Este paso de aplanamiento es necesario para que pueda utilizar capas completamente conectadas después de algunas capas convolucionales / maxpool. Combina todas las características locales encontradas de las capas convolucionales anteriores.
# 2 Capas Completamente conectadas (densas) que es simplemente un clasificador artificial de redes neuronales (ANN). En la última capa (Denso (10, activación = "softmax")) la distribución de probabilidad de salidas netas de cada clase.
# modelo
input_shape = (75, 100, 3)
num_classes = 7
model = Sequential()
model.add(
Conv2D(
32,
kernel_size=(3, 3),
activation="relu",
padding="Same",
input_shape=input_shape,
)
)
model.add(
Conv2D(
32,
kernel_size=(3, 3),
activation="relu",
padding="Same",
)
)
model.add(MaxPool2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Conv2D(64, (3, 3), activation="relu", padding="Same"))
model.add(Conv2D(64, (3, 3), activation="relu", padding="Same"))
model.add(MaxPool2D(pool_size=(2, 2)))
model.add(Dropout(0.40))
model.add(Flatten())
model.add(Dense(128, activation="relu"))
model.add(Dropout(0.5))
model.add(Dense(num_classes, activation="softmax"))
model.summary()
# defino el optimizador
# Adam Optimizer trata de solventar el problema con la fijación de el ratio de aprendizaje para ello adapta el ratio de aprendizaje en función de cómo estén distribuidos los parámetros.
# Si los parámetros están muy dispersos el ratio de aprendizaje aumentará.
optimizer = Adam(
lr=0.001, beta_1=0.9, beta_2=0.999, epsilon=None, decay=0.0, amsgrad=False
)
# Compilo el modelo
model.compile(
optimizer=optimizer, loss="categorical_crossentropy", metrics=["accuracy"]
)
# para que el optimizador converja más rápido y más cercano al mínimo global de la función de pérdida, se usa el metodo de recocido de la tasa de aprendizaje (LR)
# El LR es el paso por el cual el optimizador recorre el 'panorama de pérdidas'. Cuanto más alto es el LR, más grandes son los pasos y más rápida es la convergencia.
# Sin embargo, el muestreo es muy pobre con un alto LR y el optimizador probablemente podría caer en un mínimo local.
# Con la función ReduceLROnPlateau de Keras.callbacks, se reduce el LR a la mitad si la precisión no mejora después de 3 épocas.
learning_rate_reduction = ReduceLROnPlateau(
monitor="val_acc", patience=3, verbose=1, factor=0.5, min_lr=0.00001 # condicon
)
# se hace un aumento de imagen para evitar el sobreajuste, expandimos el dataset ham
# Para evitar problemas de sobreajuste, necesitamos expandir artificialmente nuestro conjunto de datos HAM 10000. Podemos hacer que su conjunto de datos existente sea aún más grande.
# La idea es alterar los datos de entrenamiento con pequeñas transformaciones para reproducir las variaciones. esto puede duplicar o triplicar el número de ejemplos de entrenamiento y crear un modelo muy robusto.
# solo s ehace al 10%
datagen = ImageDataGenerator(
featurewise_center=False, # establece lamedia d eerada a 0 sobre el datase
samplewise_center=False, # establece cada media de muestra en 0
featurewise_std_normalization=False, # divide las entradas por std del dataset
samplewise_std_normalization=False, # divide cada entrada por su estandar
zca_whitening=False, # aplica blanqueador ZCA este por que hace que la imagen s esiga pareciendo a la original
rotation_range=10, # rota imagen al azar en el rango (grados, 0 a 180)
zoom_range=0.1, # amplia imagen aleatoriamnete
width_shift_range=0.1, # cambio aleatoriamnet ls imagenes a horizontal
height_shift_range=0.1, # cambo aleatorimente las imagens a vertical
horizontal_flip=False, # voltea imagens al azar horizontamnte
vertical_flip=False,
) # voltea imagens al azar verticalemne
datagen.fit(x_train)
# Ajusto el modelo
epochs = 50 # epocas
batch_size = 10 # lotes
history = model.fit_generator(
datagen.flow(x_train, y_train, batch_size=batch_size),
epochs=epochs,
validation_data=(x_validate, y_validate),
verbose=1,
steps_per_epoch=x_train.shape[0] // batch_size,
callbacks=[learning_rate_reduction],
)
# Evaluacion modelo
# aqui se verifica la presicion,se valida el modelo
loss, accuracy = model.evaluate(x_test, y_test, verbose=1)
loss_v, accuracy_v = model.evaluate(x_validate, y_validate, verbose=1)
print("Validation: accuracy = %f ; loss_v = %f" % (accuracy_v, loss_v))
print("Test: accuracy = %f ; loss = %f" % (accuracy, loss))
model.save("model.h5")
plot_model_history(history)
# Function to plot confusion matrix
def plot_confusion_matrix(
cm, classes, normalize=False, title="Confusion matrix", cmap=plt.cm.Blues
):
plt.imshow(cm, interpolation="nearest", cmap=cmap)
plt.title(title)
plt.colorbar()
tick_marks = np.arange(len(classes))
plt.xticks(tick_marks, classes, rotation=45)
plt.yticks(tick_marks, classes)
if normalize:
cm = cm.astype("float") / cm.sum(axis=1)[:, np.newaxis]
thresh = cm.max() / 2.0
for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):
plt.text(
j,
i,
cm[i, j],
horizontalalignment="center",
color="white" if cm[i, j] > thresh else "black",
)
plt.tight_layout()
plt.ylabel("True label")
plt.xlabel("Predicted label")
# Predecir los valores del conjunto de datos de validación
Y_pred = model.predict(x_validate)
# Convierte las clases de predicciones en vectores calientes
Y_pred_classes = np.argmax(Y_pred, axis=1)
# Convertir observaciones de validación a vectores calientes
Y_true = np.argmax(y_validate, axis=1)
# calcular la matriz de confusión
confusion_mtx = confusion_matrix(Y_true, Y_pred_classes)
# grafico dema matriz de confusion
plot_confusion_matrix(confusion_mtx, classes=range(7))
# en esta grafica vemos que categoria tiene mas predicciones incrrectas
label_frac_error = 1 - np.diag(confusion_mtx) / np.sum(confusion_mtx, axis=1)
plt.bar(np.arange(7), label_frac_error)
plt.xlabel("True Label")
plt.ylabel("Fraction classified incorrectly")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
# # 1 Get AutoGluon
import autogluon as ag
# # 2 Data Processing
import pandas as pd
import numpy as np
from sklearn.linear_model import LogisticRegression
import matplotlib.pyplot as plt
from sklearn.utils import shuffle
from sklearn.model_selection import GridSearchCV
tourney_result = pd.read_csv(
"../input/google-cloud-ncaa-march-madness-2020-division-1-mens-tournament/MDataFiles_Stage1/MNCAATourneyCompactResults.csv"
)
tourney_seed = pd.read_csv(
"../input/google-cloud-ncaa-march-madness-2020-division-1-mens-tournament/MDataFiles_Stage1/MNCAATourneySeeds.csv"
)
# deleting unnecessary columns
tourney_result = tourney_result.drop(
["DayNum", "WScore", "LScore", "WLoc", "NumOT"], axis=1
)
tourney_result
tourney_result = pd.merge(
tourney_result,
tourney_seed,
left_on=["Season", "WTeamID"],
right_on=["Season", "TeamID"],
how="left",
)
tourney_result.rename(columns={"Seed": "WSeed"}, inplace=True)
tourney_result = tourney_result.drop("TeamID", axis=1)
tourney_result = pd.merge(
tourney_result,
tourney_seed,
left_on=["Season", "LTeamID"],
right_on=["Season", "TeamID"],
how="left",
)
tourney_result.rename(columns={"Seed": "LSeed"}, inplace=True)
tourney_result = tourney_result.drop("TeamID", axis=1)
tourney_result
def get_seed(x):
return int(x[1:3])
tourney_result["WSeed"] = tourney_result["WSeed"].map(lambda x: get_seed(x))
tourney_result["LSeed"] = tourney_result["LSeed"].map(lambda x: get_seed(x))
tourney_result
season_result = pd.read_csv(
"../input/google-cloud-ncaa-march-madness-2020-division-1-mens-tournament/MDataFiles_Stage1/MRegularSeasonCompactResults.csv"
)
season_win_result = season_result[["Season", "WTeamID", "WScore"]]
season_lose_result = season_result[["Season", "LTeamID", "LScore"]]
season_win_result.rename(columns={"WTeamID": "TeamID", "WScore": "Score"}, inplace=True)
season_lose_result.rename(
columns={"LTeamID": "TeamID", "LScore": "Score"}, inplace=True
)
season_result = pd.concat((season_win_result, season_lose_result)).reset_index(
drop=True
)
season_result
season_score = season_result.groupby(["Season", "TeamID"])["Score"].sum().reset_index()
season_score
tourney_result = pd.merge(
tourney_result,
season_score,
left_on=["Season", "WTeamID"],
right_on=["Season", "TeamID"],
how="left",
)
tourney_result.rename(columns={"Score": "WScoreT"}, inplace=True)
tourney_result = tourney_result.drop("TeamID", axis=1)
tourney_result = pd.merge(
tourney_result,
season_score,
left_on=["Season", "LTeamID"],
right_on=["Season", "TeamID"],
how="left",
)
tourney_result.rename(columns={"Score": "LScoreT"}, inplace=True)
tourney_result = tourney_result.drop("TeamID", axis=1)
tourney_result
tourney_win_result = tourney_result.drop(["Season", "WTeamID", "LTeamID"], axis=1)
tourney_win_result.rename(
columns={
"WSeed": "Seed1",
"LSeed": "Seed2",
"WScoreT": "ScoreT1",
"LScoreT": "ScoreT2",
},
inplace=True,
)
tourney_win_result
tourney_lose_result = tourney_win_result.copy()
tourney_lose_result["Seed1"] = tourney_win_result["Seed2"]
tourney_lose_result["Seed2"] = tourney_win_result["Seed1"]
tourney_lose_result["ScoreT1"] = tourney_win_result["ScoreT2"]
tourney_lose_result["ScoreT2"] = tourney_win_result["ScoreT1"]
tourney_lose_result
tourney_win_result["Seed_diff"] = (
tourney_win_result["Seed1"] - tourney_win_result["Seed2"]
)
tourney_win_result["ScoreT_diff"] = (
tourney_win_result["ScoreT1"] - tourney_win_result["ScoreT2"]
)
tourney_lose_result["Seed_diff"] = (
tourney_lose_result["Seed1"] - tourney_lose_result["Seed2"]
)
tourney_lose_result["ScoreT_diff"] = (
tourney_lose_result["ScoreT1"] - tourney_lose_result["ScoreT2"]
)
tourney_win_result["result"] = 1
tourney_lose_result["result"] = 0
tourney_result = pd.concat((tourney_win_result, tourney_lose_result)).reset_index(
drop=True
)
tourney_result
# X_train = tourney_result.drop('result', axis=1)
# y_train = tourney_result.result
# X_train, y_train = shuffle(X_train, y_train)
# Use autogluon
from autogluon import TabularPrediction as task
train_data = task.Dataset(df=tourney_result)
label_column = "result"
eval_metric = "roc_auc"
output_directory = "./"
predictor = task.fit(
train_data=train_data,
label=label_column,
output_directory=output_directory,
eval_metric=eval_metric,
hyperparameter_tune=False,
verbosity=3,
stack_ensemble_levels=2,
)
# num_bagging_folds=10) # delete last 2 arguments to reduce runtime
results = predictor.fit_summary()
# logreg = LogisticRegression()
# params = {'C': np.logspace(start=-5, stop=3, num=9)}
# clf = GridSearchCV(logreg, params, scoring='neg_log_loss', refit=True)
# clf.fit(X_train, y_train)
# print('Best log_loss: {:.4}, with best C: {}'.format(clf.best_score_, clf.best_params_['C']))
test_df = pd.read_csv(
"../input/google-cloud-ncaa-march-madness-2020-division-1-mens-tournament/MSampleSubmissionStage1_2020.csv"
)
test_df["Season"] = test_df["ID"].map(lambda x: int(x[:4]))
test_df["WTeamID"] = test_df["ID"].map(lambda x: int(x[5:9]))
test_df["LTeamID"] = test_df["ID"].map(lambda x: int(x[10:14]))
test_df
test_df = pd.merge(
test_df,
tourney_seed,
left_on=["Season", "WTeamID"],
right_on=["Season", "TeamID"],
how="left",
)
test_df.rename(columns={"Seed": "Seed1"}, inplace=True)
test_df = test_df.drop("TeamID", axis=1)
test_df = pd.merge(
test_df,
tourney_seed,
left_on=["Season", "LTeamID"],
right_on=["Season", "TeamID"],
how="left",
)
test_df.rename(columns={"Seed": "Seed2"}, inplace=True)
test_df = test_df.drop("TeamID", axis=1)
test_df = pd.merge(
test_df,
season_score,
left_on=["Season", "WTeamID"],
right_on=["Season", "TeamID"],
how="left",
)
test_df.rename(columns={"Score": "ScoreT1"}, inplace=True)
test_df = test_df.drop("TeamID", axis=1)
test_df = pd.merge(
test_df,
season_score,
left_on=["Season", "LTeamID"],
right_on=["Season", "TeamID"],
how="left",
)
test_df.rename(columns={"Score": "ScoreT2"}, inplace=True)
test_df = test_df.drop("TeamID", axis=1)
test_df
test_df["Seed1"] = test_df["Seed1"].map(lambda x: get_seed(x))
test_df["Seed2"] = test_df["Seed2"].map(lambda x: get_seed(x))
test_df["Seed_diff"] = test_df["Seed1"] - test_df["Seed2"]
test_df["ScoreT_diff"] = test_df["ScoreT1"] - test_df["ScoreT2"]
test_df = test_df.drop(["ID", "Pred", "Season", "WTeamID", "LTeamID"], axis=1)
test_df
test_data = task.Dataset(df=test_df)
y_predproba = predictor.predict_proba(test_data)
len(y_predproba), len(test_df)
submission_df = pd.read_csv(
"../input/google-cloud-ncaa-march-madness-2020-division-1-mens-tournament/MSampleSubmissionStage1_2020.csv"
)
submission_df["Pred"] = y_predproba
submission_df
submission_df["Pred"].hist()
submission_df.to_csv("submission.csv", index=False)
|
# # importing libraries
import pandas as pd
import numpy as np
import random as rnd
# visualization
import seaborn as sns
import matplotlib.pyplot as plt
# machine learning models
from sklearn.linear_model import LogisticRegression
from sklearn.svm import SVC, LinearSVC
from sklearn.ensemble import RandomForestClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.linear_model import Perceptron
from sklearn.linear_model import SGDClassifier
from sklearn.tree import DecisionTreeClassifier
import warnings
warnings.filterwarnings("ignore")
# import data
df = pd.read_csv("/kaggle/input/titanic3/titanic3.csv")
df.head()
df.info()
print("Null Values: ", df.isnull().sum().sum())
df.columns.values
df.describe()
df.describe(include=["O"])
# ## Analyze by pivoting features
df[["pclass", "survived"]].groupby(["pclass"], as_index=False).mean().sort_values(
by="survived", ascending=False
)
df[["sex", "survived"]].groupby(["sex"], as_index=False).mean().sort_values(
by="survived", ascending=False
)
df[["sibsp", "survived"]].groupby(["sibsp"], as_index=False).mean().sort_values(
by="survived", ascending=False
)
df[["parch", "survived"]].groupby(["parch"], as_index=False).mean().sort_values(
by="survived", ascending=False
)
# ## Analyze by visualizing data
g = sns.FacetGrid(df, col="survived")
g.map(plt.hist, "age", bins=20)
# grid = sns.FacetGrid(train_df, col='Pclass', hue='Survived')
grid = sns.FacetGrid(df, col="survived", row="pclass", aspect=1.6)
grid.map(plt.hist, "age", alpha=0.5, bins=20)
grid.add_legend()
grid = sns.FacetGrid(df, row="embarked", col="survived", aspect=1.6)
grid.map(sns.barplot, "sex", "fare", alpha=0.5, ci=None)
grid.add_legend()
## deleting unwanted columns
df = df.drop(["ticket", "cabin", "home.dest", "boat"], axis=1)
#'pclass', 'survived', 'name', 'sex', 'age', 'sibsp', 'parch',
# 'ticket', 'fare', 'cabin', 'embarked', 'boat', 'home.dest'],
# Get titles
df["title"] = df["name"].str.split(", ", expand=True)[1].str.split(".", expand=True)[0]
# Print title counts
print(df["title"].value_counts())
# Title mapping ==>
# Mr : 0
# Miss : 1
# Mrs : 2
# Others : 3
for dataset in [df]:
dataset["title"] = dataset["title"].replace(
[
"Lady",
"Countess",
"Capt",
"Col",
"Don",
"Dr",
"Major",
"Rev",
"Sir",
"Jonkheer",
"Dona",
],
"Rare",
)
dataset["title"] = dataset["title"].replace("Mlle", "Miss")
dataset["title"] = dataset["title"].replace("Ms", "Miss")
dataset["title"] = dataset["title"].replace("Mme", "Mrs")
df[["title", "survived"]].groupby(["title"], as_index=False).mean()
# convert the categorical titles to ordinal.
title_mapping = {"Mr": 1, "Miss": 2, "Mrs": 3, "Master": 4, "Rare": 5}
for dataset in [df]:
dataset["title"] = dataset["title"].map(title_mapping)
dataset["title"] = dataset["title"].fillna(0)
df.head()
df = df.drop(["name"], axis=1)
## Converting a categorical feature
for dataset in [df]:
dataset["sex"] = dataset["sex"].map({"female": 1, "male": 0}).astype(int)
df.head()
grid = sns.FacetGrid(df, row="pclass", col="sex", aspect=1.6)
grid.map(plt.hist, "age", alpha=0.5, bins=20)
grid.add_legend()
guess_ages = np.zeros((2, 3))
guess_ages
# iterate over Sex (0 or 1) and Pclass (1, 2, 3) to calculate guessed values of Age for the six combinations.
for dataset in [df]:
for i in range(0, 2):
for j in range(0, 3):
guess_df = dataset[(dataset["sex"] == i) & (dataset["pclass"] == j + 1)][
"age"
].dropna()
age_guess = guess_df.median()
# Convert random age float to nearest .5 age
guess_ages[i, j] = int(age_guess / 0.5 + 0.5) * 0.5
for i in range(0, 2):
for j in range(0, 3):
dataset.loc[
(dataset.age.isnull()) & (dataset.sex == i) & (dataset.pclass == j + 1),
"age",
] = guess_ages[i, j]
dataset["age"] = dataset["age"].astype(int)
df.head()
# create Age bands and determine correlations with Survived.
df["AgeBand"] = pd.cut(df["age"], 5)
df[["AgeBand", "survived"]].groupby(["AgeBand"], as_index=False).mean().sort_values(
by="AgeBand", ascending=True
)
# replace Age with ordinals based on these bands.
for dataset in [df]:
dataset.loc[dataset["age"] <= 16, "age"] = 0
dataset.loc[(dataset["age"] > 16) & (dataset["age"] <= 32), "age"] = 1
dataset.loc[(dataset["age"] > 32) & (dataset["age"] <= 48), "age"] = 2
dataset.loc[(dataset["age"] > 48) & (dataset["age"] <= 64), "age"] = 3
dataset.loc[dataset["age"] > 64, "age"] = 4
df.head()
df = df.drop(["AgeBand"], axis=1)
df.head()
for dataset in [df]:
dataset["FamilySize"] = dataset["sibsp"] + dataset["parch"] + 1
df[["FamilySize", "survived"]].groupby(
["FamilySize"], as_index=False
).mean().sort_values(by="survived", ascending=False)
# We can create another feature called IsAlone.
for dataset in [df]:
dataset["IsAlone"] = 0
dataset.loc[dataset["FamilySize"] == 1, "IsAlone"] = 1
df[["IsAlone", "survived"]].groupby(["IsAlone"], as_index=False).mean()
df = df.drop(["parch", "sibsp", "FamilySize"], axis=1)
df.head()
freq_port = df.embarked.dropna().mode()[0]
freq_port
for dataset in [df]:
dataset["embarked"] = dataset["embarked"].fillna(freq_port)
df[["embarked", "survived"]].groupby(["embarked"], as_index=False).mean().sort_values(
by="survived", ascending=False
)
# Converting categorical feature to numeric
for dataset in [df]:
dataset["embarked"] = dataset["embarked"].map({"S": 0, "C": 1, "Q": 2}).astype(int)
df.head()
df["fare"].fillna(df["fare"].dropna().median(), inplace=True)
df.head()
df["FareBand"] = pd.qcut(df["fare"], 4)
df[["FareBand", "survived"]].groupby(["FareBand"], as_index=False).mean().sort_values(
by="FareBand", ascending=True
)
# Convert the Fare feature to ordinal values based on the FareBand.
for dataset in [df]:
dataset.loc[dataset["fare"] <= 7.91, "fare"] = 0
dataset.loc[(dataset["fare"] > 7.91) & (dataset["fare"] <= 14.454), "fare"] = 1
dataset.loc[(dataset["fare"] > 14.454) & (dataset["fare"] <= 31), "fare"] = 2
dataset.loc[dataset["fare"] > 31, "fare"] = 3
dataset["fare"] = dataset["fare"].astype(int)
df = df.drop(["FareBand"], axis=1)
df.head(10)
# # Model and predict
X = df.drop("survived", axis=1)
Y = df["survived"]
from sklearn.model_selection import train_test_split
# split the data into training and testing sets
X_train, X_test, y_train, y_test = train_test_split(
X, Y, test_size=0.2, random_state=42
)
# KNN Model
knn = KNeighborsClassifier(n_neighbors=7)
knn.fit(X_train, y_train)
Y_pred = knn.predict(X_test)
acc_knn = round(knn.score(X_train, y_train) * 100, 2)
print("accuracy", acc_knn)
# Random Forest Model
random_forest = RandomForestClassifier(n_estimators=200)
random_forest.fit(X_train, y_train)
Y_pred = random_forest.predict(X_test)
random_forest.score(X_train, y_train)
acc_random_forest = round(random_forest.score(X_train, y_train) * 100, 2)
print("accuracy: ", acc_random_forest)
# calculate confusion matrix
from sklearn.metrics import confusion_matrix
cm = confusion_matrix(y_test, Y_pred)
# plot confusion matrix using heatmap
sns.heatmap(cm, annot=True, cmap="Blues")
|
# importing needed libraries to complete notebook
import matplotlib.pylab as plt
import numpy as np
import pandas as pd
from keras import models, layers, optimizers
from keras import applications
from keras.preprocessing.image import ImageDataGenerator
import os
# checking what models are available for me to use from the keras-pretrained-models dataset
from os import listdir, makedirs
from os.path import join, exists, expanduser
cache_dir = expanduser(join("~", ".keras"))
if not exists(cache_dir):
makedirs(cache_dir)
models_dir = join(cache_dir, "models")
if not exists(models_dir):
makedirs(models_dir)
print("Available Pretrained Models:")
# prints the number of categories, and what each of the categories are labelled as
monkey_species = os.listdir("../input/10-monkey-species/training/training")
print("Number of Categories:", len(monkey_species))
print("Categories: ", monkey_species)
# sets target height/width of images
img_width, img_height = 224, 224
# sets paths to data
train_data_dir = "../input/10-monkey-species/training/training"
validation_data_dir = "../input/10-monkey-species/validation/validation"
batch_size = 4
# train and validation datagens for testing
train_datagen = ImageDataGenerator(
rotation_range=30,
rescale=1.0 / 255,
shear_range=0.2,
zoom_range=0.2,
horizontal_flip=True,
)
test_datagen = ImageDataGenerator(rescale=1.0 / 255)
train_generator = train_datagen.flow_from_directory(
train_data_dir,
target_size=(img_height, img_width),
batch_size=batch_size,
class_mode="categorical",
)
validation_generator = test_datagen.flow_from_directory(
validation_data_dir,
target_size=(img_height, img_width),
batch_size=batch_size,
class_mode="categorical",
)
# gets number of samples in train and validation
nb_train_samples = len(train_generator.classes)
nb_validation_samples = len(validation_generator.classes)
# makes dataframes of training and testing data
training_data = pd.DataFrame(train_generator.classes, columns=["classes"])
testing_data = pd.DataFrame(validation_generator.classes, columns=["classes"])
# import inception with pre-trained weights. do not include fully #connected layers
Xception_base = applications.Xception(weights="imagenet", include_top=False)
# add a global spatial average pooling layer
x = Xception_base.output
x = layers.GlobalAveragePooling2D()(x)
# add a fully-connected layer
x = layers.Dense(512, activation="relu")(x)
# and a fully connected output/classification layer
predictions = layers.Dense(
int(len(train_generator.class_indices.keys())), activation="softmax"
)(x)
# create the full network so we can train on it
Xception_transfer = models.Model(inputs=Xception_base.input, outputs=predictions)
Xception_transfer.compile(
loss="categorical_crossentropy",
optimizer=optimizers.SGD(learning_rate=1e-4, momentum=0.9),
metrics=["accuracy"],
)
import tensorflow as tf
with tf.device("/device:GPU:0"):
history = Xception_transfer.fit(
train_generator,
epochs=10,
shuffle=True,
verbose=1,
validation_data=validation_generator,
)
|
# ###
# Table of Contents
# [1. Notebook Versions](#1)
# [2. Loading Libraries](#2)
# [3. Reading Data Files](#3)
# [4. Data Description](#4)
# [5. Data Exploration](#5)
# [6. Baseline Modeling 1.0](#6)
# - [6.1 Logistic Regression](#6.1)
# - [6.2 Random Forest](#6.2)
# - [6.3 Extra Trees](#6.3)
# - [6.4 GradientBoosting](#6.4)
# - [6.5 HistGradientBoosting](#6.5)
# - [6.6 LightGBM](#6.6)
# - [6.7 XGBoost](#6.7)
# - [6.8 CatBoost](#6.8)
# - [6.9 Model Comparison](#6.9)
# [7. Baseline Modeling 2.0](#7)
# # Notebook Versions
# 1. Version 1
# * EDA
# * Data Exploration
# * Baseline modeling 1.0
# 2. Version 2
# * Updating baseline modeling 1.0
#
# 3. Version 3
# * Adding baseline modeling 2.0
# 4. Version 4
# * Updating baseline modeling 2.0 submission
#
# 5. Version 5
# * Updating heatmaps
# # Loading Libraries
import pandas as pd
pd.set_option("display.max_columns", 100)
import numpy as np
from tqdm import tqdm
from functools import partial
import scipy as sp
import matplotlib.pyplot as plt
plt.style.use("ggplot")
import seaborn as sns
import plotly.express as px
from sklearn.tree import DecisionTreeClassifier, plot_tree
from sklearn.preprocessing import MinMaxScaler
from sklearn.model_selection import (
KFold,
StratifiedKFold,
train_test_split,
GridSearchCV,
)
from sklearn.metrics import (
roc_auc_score,
roc_curve,
RocCurveDisplay,
cohen_kappa_score,
log_loss,
)
from sklearn.neighbors import KNeighborsClassifier
from sklearn.feature_selection import RFE, RFECV
from sklearn.isotonic import IsotonicRegression
from sklearn.calibration import CalibrationDisplay
from sklearn.inspection import PartialDependenceDisplay
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import (
RandomForestClassifier,
HistGradientBoostingClassifier,
GradientBoostingClassifier,
ExtraTreesClassifier,
)
from sklearn.svm import SVC
from lightgbm import LGBMClassifier
from xgboost import XGBClassifier
from catboost import CatBoostClassifier
from pygam import LogisticGAM, s, f, l, te
#
# # Reading Data Files
train = pd.read_csv("../input/playground-series-s3e12/train.csv")
test = pd.read_csv("../input/playground-series-s3e12/test.csv")
submission = pd.read_csv("../input/playground-series-s3e12/sample_submission.csv")
print("The dimession of the train dataset is:", train.shape)
print("The dimession of the test dataset is:", test.shape)
train.info()
train.head()
train.describe()
test.head()
test.describe()
# >
# 💡 There are no missing values neither in the train nor test datasets. Also, by a quick eye-ball comparison of the summary statistics of the train and test datasets, they seem to have similar distributions.
# # Data Description
# This is a synthetic dataset generated from the [Kidney Stone Prediction based on Urine Analysis](https://www.kaggle.com/datasets/vuppalaadithyasairam/kidney-stone-prediction-based-on-urine-analysis) dataset. These are the descriptions of the variables in this dataset:
# id: id of the obsevation.
# gravity: specific gravity, the density of the urine relative to water.
# ph: he negative logarithm of the hydrogen ion.
# osmo: osmolarity. Osmolarity is proportional to the concentration of
# molecules in solution.
# cond: conductivity (mMho milliMho). One Mho is one
# reciprocal Ohm. Conductivity is proportional to the concentration of charged
# ions in solution.
# urea: urea concentration in millimoles per litre.
# calc: calcium
# concentration (CALC) in millimolesllitre.
# target: 0- absence of stone 1- presence of stone.
# # Data Exploration
round(100 * train["target"].value_counts() / train.shape[0], 2)
sns.countplot(x="target", hue="target", data=train)
# >
# 💡 From the above chart, we see that the data is pretty balanced (56%-44%).
# We next proceed to explore the relationship between the input and target variables.
fig, axes = plt.subplots(1, 2, figsize=(22, 8))
sns.boxplot(ax=axes[0], x="target", y="gravity", hue="target", data=train)
sns.boxplot(ax=axes[1], x="target", y="ph", hue="target", data=train)
# >
# 💡 From the above boxplots, we see the median of target=1 of gravity is larger than target=0.
fig, axes = plt.subplots(1, 2, figsize=(22, 8))
sns.boxplot(ax=axes[0], x="target", y="osmo", hue="target", data=train)
sns.boxplot(ax=axes[1], x="target", y="cond", hue="target", data=train)
# >
# 💡 In the left panel, we see the median of target=1 of osmo is larger than target=0. On the other hand, in the right panel, we see that the median of target=1 is slighlty larger than target=0
fig, axes = plt.subplots(1, 2, figsize=(22, 8))
sns.boxplot(ax=axes[0], x="target", y="urea", hue="target", data=train)
sns.boxplot(ax=axes[1], x="target", y="calc", hue="target", data=train)
# >
# 💡 From the above boxplots, we see the median of target=1 of urea and calc is larger than target=0.
# We next proceed to explore correlations among the input features.
corr_mat_train = train.drop(columns=["id", "target"], axis=1).corr()
corr_mat_test = test.drop(columns=["id"], axis=1).corr()
train_mask = np.triu(np.ones_like(corr_mat_train, dtype=bool))
test_mask = np.triu(np.ones_like(corr_mat_test, dtype=bool))
cmap = sns.diverging_palette(100, 7, s=75, l=40, n=5, center="light", as_cmap=True)
fig, axes = plt.subplots(1, 2, figsize=(22, 8))
sns.heatmap(
corr_mat_train,
annot=True,
cmap=cmap,
fmt=".2f",
center=0,
annot_kws={"size": 12},
ax=axes[0],
mask=train_mask,
).set_title("Correlations Among Features (in Train)")
sns.heatmap(
corr_mat_test,
annot=True,
cmap=cmap,
fmt=".2f",
center=0,
annot_kws={"size": 12},
ax=axes[1],
mask=test_mask,
).set_title("Correlations Among Features (in Test)")
# >
# 💡 From the above heatmaps, we see that the correlation among the features are almost the same in the train and test datasets.
# We next proceed to compare the distribution of the features in the train and test datasets.
train_vis = train.drop(columns=["id", "target"], axis=1).reset_index(drop=True).copy()
test_vis = test.drop(columns=["id"], axis=1).reset_index(drop=True).copy()
train_vis["Dataset"] = "Train"
test_vis["Dataset"] = "Test"
data_tot = pd.concat([train_vis, test_vis], axis=0).reset_index(drop=True)
fig, axes = plt.subplots(2, 3, figsize=(25, 15))
sns.kdeplot(ax=axes[0, 0], x="gravity", hue="Dataset", data=data_tot, fill=True)
sns.kdeplot(ax=axes[0, 1], x="ph", hue="Dataset", data=data_tot, fill=True)
sns.kdeplot(ax=axes[0, 2], x="osmo", hue="Dataset", data=data_tot, fill=True)
sns.kdeplot(ax=axes[1, 0], x="cond", hue="Dataset", data=data_tot, fill=True)
sns.kdeplot(ax=axes[1, 1], x="urea", hue="Dataset", data=data_tot, fill=True)
sns.kdeplot(ax=axes[1, 2], x="calc", hue="Dataset", data=data_tot, fill=True)
# >
# 💡 From the above density plots, we see that the distributions of the features are very similar in the train and test datasets.
# We next proceed to check for duplicates.
print("There are ", train.shape[0], " observations in the train datset")
print(
"There are ",
train.drop(columns=["id", "target"], axis=1).drop_duplicates().shape[0],
" unique observations in the train datset",
)
print("There are ", test.shape[0], " observations in the test datset")
print(
"There are ",
test.drop(columns=["id"], axis=1).drop_duplicates().shape[0],
" unique observations in the test datset",
)
to_consider = train.drop(columns=["id", "target"], axis=1).columns.tolist()
train_dup = train.drop(columns=["id", "target"], axis=1).drop_duplicates()
test_dup = test.drop(columns=["id"], axis=1).drop_duplicates()
duplicates = pd.merge(train_dup, test_dup, on=to_consider)
print(
"There are ",
duplicates.shape[0],
" rows that appear in the train and test dataset.\n",
)
# We next proceed to explore the relationship of the features with the highest correlations.
fig, axes = plt.subplots(1, 3, figsize=(25, 7))
sns.scatterplot(ax=axes[0], data=train, x="osmo", y="urea", hue="target")
sns.scatterplot(ax=axes[1], data=train, x="osmo", y="cond", hue="target")
sns.scatterplot(ax=axes[2], data=train, x="urea", y="gravity", hue="target")
# >
# 💡 From the above scatter-plots, we see that there is not a clear pattern.
# # Baseline Modeling 1.0
# In this section, we first start by defining the evaluation metric. The ROC curve is created by plotting the true positive rate (TPR) against the false positive rate (FPR) at various threshold settings.
# 
# # Logistic Regression
X = train.drop(columns=["id", "target"], axis=1)
Y = train["target"]
test_baseline = test.drop(columns=["id"], axis=1)
cv_scores, roc_auc_scores = list(), list()
preds = list()
skf = StratifiedKFold(n_splits=10, random_state=42, shuffle=True)
for i, (train_idx, test_idx) in enumerate(skf.split(X, Y)):
## Splitting the data
X_train, X_test = X.iloc[train_idx], X.iloc[test_idx]
Y_train, Y_test = Y.iloc[train_idx], Y.iloc[test_idx]
## Building logistic model
model = LogisticRegression(penalty="l2", C=0.01, max_iter=10000).fit(
X_train, Y_train
)
## Predicting on X_test and test
model_pred_1 = model.predict_proba(X_test)[:, 1]
model_pred_2 = model.predict_proba(test_baseline)[:, 1]
## Computing roc-auc score
score = roc_auc_score(Y_test, model_pred_1)
roc_auc_scores.append(score)
preds.append(model_pred_2)
print("Fold", i, "==> roc-auc-score is ==>", score)
cv_scores.append(np.mean(roc_auc_scores))
logit_cv_score = np.mean(cv_scores)
print(
"The oof average roc-auc-score of the Logistic Regression model over 10-folds is:",
logit_cv_score,
)
logit_preds_test = pd.DataFrame(preds).apply(np.mean, axis=0)
plt.figure(figsize=(8, 6))
sns.histplot(data=logit_preds_test, kde=True, stat="density", color="steelblue").set(
title="Logitistic Predicted Likelihood in Test"
)
submission["target"] = logit_preds_test
submission.to_csv("logit_baseline_submission.csv", index=False)
#
# # Random Forest
X = train.drop(columns=["id", "target"], axis=1)
Y = train["target"]
test_baseline = test.drop(columns=["id"], axis=1)
cv_scores, roc_auc_scores = list(), list()
preds = list()
skf = StratifiedKFold(n_splits=10, random_state=42, shuffle=True)
for i, (train_idx, test_idx) in enumerate(skf.split(X, Y)):
## Splitting the data
X_train, X_test = X.iloc[train_idx], X.iloc[test_idx]
Y_train, Y_test = Y.iloc[train_idx], Y.iloc[test_idx]
## Building RF model
model = RandomForestClassifier(
n_estimators=500,
max_depth=4,
min_samples_split=15,
min_samples_leaf=5,
random_state=i,
).fit(X_train, Y_train)
## Predicting on X_test and test
model_pred_1 = model.predict_proba(X_test)[:, 1]
model_pred_2 = model.predict_proba(test_baseline)[:, 1]
## Computing roc-auc score
score = roc_auc_score(Y_test, model_pred_1)
roc_auc_scores.append(score)
preds.append(model_pred_2)
print("Fold", i, "==> roc-auc-score is ==>", score)
cv_scores.append(np.mean(roc_auc_scores))
RF_cv_score = np.mean(cv_scores)
print(
"The oof average roc-auc-score of the Random Forest model over 10-folds is:",
RF_cv_score,
)
RF_preds_test = pd.DataFrame(preds).apply(np.mean, axis=0)
plt.figure(figsize=(8, 6))
sns.histplot(data=RF_preds_test, kde=True, stat="density", color="steelblue").set(
title="RF Predicted Likelihood in Test"
)
submission["target"] = RF_preds_test
submission.to_csv("RF_baseline_submission.csv", index=False)
#
# # Extra Trees
X = train.drop(columns=["id", "target"], axis=1)
Y = train["target"]
test_baseline = test.drop(columns=["id"], axis=1)
cv_scores, roc_auc_scores = list(), list()
preds = list()
skf = StratifiedKFold(n_splits=10, random_state=42, shuffle=True)
for i, (train_idx, test_idx) in enumerate(skf.split(X, Y)):
## Splitting the data
X_train, X_test = X.iloc[train_idx], X.iloc[test_idx]
Y_train, Y_test = Y.iloc[train_idx], Y.iloc[test_idx]
## Building ExtraTrees model
model = ExtraTreesClassifier(
n_estimators=500, max_depth=4, min_samples_split=15, min_samples_leaf=5
).fit(X_train, Y_train)
## Predicting on X_test and test
model_pred_1 = model.predict_proba(X_test)[:, 1]
model_pred_2 = model.predict_proba(test_baseline)[:, 1]
## Computing roc-auc score
score = roc_auc_score(Y_test, model_pred_1)
roc_auc_scores.append(score)
preds.append(model_pred_2)
print("Fold", i, "==> roc-auc-score is ==>", score)
cv_scores.append(np.mean(roc_auc_scores))
extra_cv_score = np.mean(cv_scores)
print(
"The oof average roc-auc-score of the Extra Trees model over 10-folds is:",
extra_cv_score,
)
extra_preds_test = pd.DataFrame(preds).apply(np.mean, axis=0)
plt.figure(figsize=(8, 6))
sns.histplot(data=extra_preds_test, kde=True, stat="density", color="steelblue").set(
title="Extra Trees Predicted Likelihood in Test"
)
submission["target"] = extra_preds_test
submission.to_csv("extra_baseline_submission.csv", index=False)
#
# # GradientBoosting
X = train.drop(columns=["id", "target"], axis=1)
Y = train["target"]
test_baseline = test.drop(columns=["id"], axis=1)
cv_scores, roc_auc_scores = list(), list()
preds = list()
skf = StratifiedKFold(n_splits=10, random_state=42, shuffle=True)
for i, (train_idx, test_idx) in enumerate(skf.split(X, Y)):
## Splitting the data
X_train, X_test = X.iloc[train_idx], X.iloc[test_idx]
Y_train, Y_test = Y.iloc[train_idx], Y.iloc[test_idx]
## Building GradientBoosting model
model = GradientBoostingClassifier(
n_estimators=500,
learning_rate=0.01,
max_depth=4,
min_samples_split=15,
min_samples_leaf=5,
).fit(X_train, Y_train)
## Predicting on X_test and test
model_pred_1 = model.predict_proba(X_test)[:, 1]
model_pred_2 = model.predict_proba(test_baseline)[:, 1]
## Computing roc-auc score
score = roc_auc_score(Y_test, model_pred_1)
roc_auc_scores.append(score)
preds.append(model_pred_2)
print("Fold", i, "==> roc-auc-score is ==>", score)
cv_scores.append(np.mean(roc_auc_scores))
gradient_cv_score = np.mean(cv_scores)
print(
"The oof average roc-auc-score of the Gradient Boosting model over 10-folds is:",
gradient_cv_score,
)
gradient_preds_test = pd.DataFrame(preds).apply(np.mean, axis=0)
plt.figure(figsize=(8, 6))
sns.histplot(data=gradient_preds_test, kde=True, stat="density", color="steelblue").set(
title="Gradient Boosting Predicted Likelihood in Test"
)
submission["target"] = gradient_preds_test
submission.to_csv("gradient_baseline_submission.csv", index=False)
#
# # HistGradientBoosting
X = train.drop(columns=["id", "target"], axis=1)
Y = train["target"]
test_baseline = test.drop(columns=["id"], axis=1)
cv_scores, roc_auc_scores = list(), list()
preds = list()
skf = StratifiedKFold(n_splits=10, random_state=42, shuffle=True)
for i, (train_idx, test_idx) in enumerate(skf.split(X, Y)):
## Splitting the data
X_train, X_test = X.iloc[train_idx], X.iloc[test_idx]
Y_train, Y_test = Y.iloc[train_idx], Y.iloc[test_idx]
## Building HistGradient model
model = HistGradientBoostingClassifier(
l2_regularization=0.01,
early_stopping=False,
learning_rate=0.01,
max_iter=500,
max_depth=3,
max_bins=255,
min_samples_leaf=5,
max_leaf_nodes=5,
).fit(X_train, Y_train)
## Predicting on X_test and test
model_pred_1 = model.predict_proba(X_test)[:, 1]
model_pred_2 = model.predict_proba(test_baseline)[:, 1]
## Computing roc-auc score
score = roc_auc_score(Y_test, model_pred_1)
roc_auc_scores.append(score)
preds.append(model_pred_2)
print("Fold", i, "==> roc-auc-score is ==>", score)
cv_scores.append(np.mean(roc_auc_scores))
hist_cv_score = np.mean(cv_scores)
print(
"The oof average roc-auc-score of the HistGradient model over 10-folds is:",
hist_cv_score,
)
hist_preds_test = pd.DataFrame(preds).apply(np.mean, axis=0)
plt.figure(figsize=(8, 6))
sns.histplot(data=hist_preds_test, kde=True, stat="density", color="steelblue").set(
title="HistGradientBoosting Predicted Likelihood in Test"
)
submission["target"] = hist_preds_test
submission.to_csv("hist_baseline_submission.csv", index=False)
#
# # LightGBM
X = train.drop(columns=["id", "target"], axis=1)
Y = train["target"]
test_baseline = test.drop(columns=["id"], axis=1)
cv_scores, roc_auc_scores = list(), list()
preds = list()
skf = StratifiedKFold(n_splits=10, random_state=42, shuffle=True)
for i, (train_idx, test_idx) in enumerate(skf.split(X, Y)):
## Splitting the data
X_train, X_test = X.iloc[train_idx], X.iloc[test_idx]
Y_train, Y_test = Y.iloc[train_idx], Y.iloc[test_idx]
## Building LightGBM model
model = LGBMClassifier(
n_estimators=500,
max_depth=4,
learning_rate=0.01,
num_leaves=20,
reg_alpha=3,
reg_lambda=3,
subsample=0.7,
colsample_bytree=0.7,
).fit(X_train, Y_train)
## Predicting on X_test and test
model_pred_1 = model.predict_proba(X_test)[:, 1]
model_pred_2 = model.predict_proba(test_baseline)[:, 1]
## Computing roc-auc score
score = roc_auc_score(Y_test, model_pred_1)
roc_auc_scores.append(score)
preds.append(model_pred_2)
print("Fold", i, "==> roc-auc-score is ==>", score)
cv_scores.append(np.mean(roc_auc_scores))
lgb_cv_score = np.mean(cv_scores)
print(
"The oof average roc-auc-score of the LightGBM model over 10-folds is:",
lgb_cv_score,
)
lgb_preds_test = pd.DataFrame(preds).apply(np.mean, axis=0)
plt.figure(figsize=(8, 6))
sns.histplot(data=lgb_preds_test, kde=True, stat="density", color="steelblue").set(
title="LightGBM Predicted Likelihood in Test"
)
submission["target"] = lgb_preds_test
submission.to_csv("lgb_baseline_submission.csv", index=False)
#
# # XGBoost
X = train.drop(columns=["id", "target"], axis=1)
Y = train["target"]
test_baseline = test.drop(columns=["id"], axis=1)
cv_scores, roc_auc_scores = list(), list()
preds = list()
skf = StratifiedKFold(n_splits=10, random_state=42, shuffle=True)
for i, (train_idx, test_idx) in enumerate(skf.split(X, Y)):
## Splitting the data
X_train, X_test = X.iloc[train_idx], X.iloc[test_idx]
Y_train, Y_test = Y.iloc[train_idx], Y.iloc[test_idx]
## Building XGBoost model
model = XGBClassifier(
tree_method="hist",
colsample_bytree=0.7,
gamma=2,
learning_rate=0.01,
max_depth=4,
min_child_weight=10,
n_estimators=500,
subsample=0.7,
).fit(X_train, Y_train)
## Predicting on X_test and test
model_pred_1 = model.predict_proba(X_test)[:, 1]
model_pred_2 = model.predict_proba(test_baseline)[:, 1]
## Computing roc-auc score
score = roc_auc_score(Y_test, model_pred_1)
roc_auc_scores.append(score)
preds.append(model_pred_2)
print("Fold", i, "==> roc-auc-score is ==>", score)
cv_scores.append(np.mean(roc_auc_scores))
xgb_cv_score = np.mean(cv_scores)
print(
"The oof average roc-auc-score of the XGBoost model over 10-folds is:", xgb_cv_score
)
xgb_preds_test = pd.DataFrame(preds).apply(np.mean, axis=0)
plt.figure(figsize=(8, 6))
sns.histplot(data=xgb_preds_test, kde=True, stat="density", color="steelblue").set(
title="XGBoost Predicted Likelihood in Test"
)
submission["target"] = xgb_preds_test
submission.to_csv("xgb_baseline_submission.csv", index=False)
#
# # CatBoost
X = train.drop(columns=["id", "target"], axis=1)
Y = train["target"]
test_baseline = test.drop(columns=["id"], axis=1)
cv_scores, roc_auc_scores = list(), list()
preds = list()
skf = StratifiedKFold(n_splits=10, random_state=42, shuffle=True)
for i, (train_idx, test_idx) in enumerate(skf.split(X, Y)):
## Splitting the data
X_train, X_test = X.iloc[train_idx], X.iloc[test_idx]
Y_train, Y_test = Y.iloc[train_idx], Y.iloc[test_idx]
## Building XGBoost model
model = CatBoostClassifier(
loss_function="Logloss",
iterations=500,
learning_rate=0.01,
depth=4,
random_strength=0.5,
bagging_temperature=0.7,
border_count=30,
l2_leaf_reg=5,
verbose=False,
task_type="CPU",
).fit(X_train, Y_train)
## Predicting on X_test and test
model_pred_1 = model.predict_proba(X_test)[:, 1]
model_pred_2 = model.predict_proba(test_baseline)[:, 1]
## Computing roc-auc score
score = roc_auc_score(Y_test, model_pred_1)
roc_auc_scores.append(score)
preds.append(model_pred_2)
print("Fold", i, "==> roc-auc-score is ==>", score)
cv_scores.append(np.mean(roc_auc_scores))
cat_cv_score = np.mean(cv_scores)
print(
"The oof average roc-auc-score of the CatBoost model over 10-folds is:",
cat_cv_score,
)
cat_preds_test = pd.DataFrame(preds).apply(np.mean, axis=0)
plt.figure(figsize=(8, 6))
sns.histplot(data=cat_preds_test, kde=True, stat="density", color="steelblue").set(
title="CatBoost Predicted Likelihood in Test"
)
submission["target"] = cat_preds_test
submission.to_csv("cat_baseline_submission.csv", index=False)
#
# # Model Comparison
# The below table summarizes the initial baseline modeling results. Notice that these results are based on using raw data without any transformation.
models_perf = pd.DataFrame(
{
"Model": [
"Logistic",
"RF",
"ExtraTrees",
"GradientBoosting",
"HistGradient",
"LightGBM",
"XGBoost",
"CatBoost",
],
"CV-Score": [
logit_cv_score,
RF_cv_score,
extra_cv_score,
gradient_cv_score,
hist_cv_score,
lgb_cv_score,
xgb_cv_score,
cat_cv_score,
],
}
)
models_perf
md_preds = pd.DataFrame(
{
"Logistic": logit_preds_test,
"RF": RF_preds_test,
"ExtraTrees": extra_preds_test,
"GradientBoosting": gradient_preds_test,
"HistGradient": hist_preds_test,
"LightGBM": lgb_preds_test,
"XGBoost": xgb_preds_test,
"CatBoost": cat_preds_test,
}
)
corr_mat_pred = md_preds.corr()
corr_mask = np.triu(np.ones_like(corr_mat_pred, dtype=bool))
cmap = sns.diverging_palette(100, 7, s=75, l=40, n=10, center="light", as_cmap=True)
fig, axes = plt.subplots(1, 2, figsize=(22, 9))
sns.barplot(ax=axes[0], data=models_perf, x="CV-Score", y="Model", color="steelblue")
sns.heatmap(
corr_mat_pred,
annot=True,
cmap=cmap,
fmt=".2f",
center=0,
annot_kws={"size": 12},
ax=axes[1],
mask=corr_mask,
).set_title("Correlations Among Model Predictions")
ens_preds_test = (
0.25 * RF_preds_test
+ 0.15 * gradient_preds_test
+ 0.2 * lgb_preds_test
+ 0.2 * xgb_preds_test
+ 0.2 * cat_preds_test
)
plt.figure(figsize=(8, 6))
sns.histplot(data=ens_preds_test, kde=True, stat="density", color="steelblue").set(
title="Ensemble Predicted Likelihood in Test"
)
submission["target"] = ens_preds_test
submission.to_csv("ensemble_baseline_submission.csv", index=False)
#
# # Baseline Modeling 2.0
# Based on the results from the previous section, random forest model is the model with the highest cv-score. In this section, we will take a closer look at this model by:
# - Taking a closer look at the partial dependency plots
# - Engineering features
X = train.drop(columns=["id", "target"], axis=1)
Y = train["target"]
model = RandomForestClassifier(
n_estimators=500, max_depth=4, min_samples_split=15, min_samples_leaf=5
).fit(X, Y)
fig, axs = plt.subplots(2, 3, figsize=(15, 10))
plt.suptitle("Partial Dependence Plots", y=1.0)
PartialDependenceDisplay.from_estimator(
model,
X,
X.columns.tolist(),
pd_line_kw={"color": "red"},
ice_lines_kw={"color": "steelblue"},
kind="both",
ax=axs.ravel()[: len(X.columns.tolist())],
)
plt.tight_layout(h_pad=0.5, w_pad=0.5)
plt.show()
# Based on partial dependency plots, we see that:
# - `calc` is an important feature when it comes to predicting the likelihood of kidney stone. We also see when `calc` > 8 the line is pretty flat, which means that the likelihood of kidney stone doesn't increase when `calc` > 8.
# - `gravity` is another important feature. From that plot, we see the likehood of kidney stone increases when `gravity` goes from 1.01 to 1.03, after that, the likelhood stays constant.
# - The partial dependency plots associated to `ph` and `osmo` are pretty flat.
# We next proceed to re-train the random forest model with `gravity`, `cond`, `urea`, and `calc` as predictors. Also we engineer a couple of ratio features (this idea comes from this [post](https://www.kaggle.com/competitions/playground-series-s3e12/discussion/399441).
X = train[["gravity", "ph", "cond", "urea", "calc"]].copy()
Y = train["target"]
X["calc"] = X.calc.clip(None, 8)
X["gravity"] = X.gravity.clip(None, 1.03)
X["calc_gravity_ratio"] = X["calc"] / X["gravity"]
X["calc_urea_ratio"] = X["calc"] / X["urea"]
X["gravity_cond_ratio"] = X["gravity"] / X["cond"]
test_baseline = test[["gravity", "ph", "cond", "urea", "calc"]].copy()
test_baseline["calc"] = test_baseline.calc.clip(None, 8)
test_baseline["gravity"] = test_baseline.gravity.clip(None, 1.03)
test_baseline["calc_gravity_ratio"] = test_baseline["calc"] / test_baseline["gravity"]
test_baseline["calc_urea_ratio"] = test_baseline["calc"] / test_baseline["urea"]
test_baseline["gravity_cond_ratio"] = test_baseline["gravity"] / test_baseline["cond"]
cv_scores, roc_auc_scores = list(), list()
preds = list()
skf = StratifiedKFold(n_splits=10, random_state=42, shuffle=True)
for i, (train_idx, test_idx) in enumerate(skf.split(X, Y)):
## Splitting the data
X_train, X_test = X.iloc[train_idx], X.iloc[test_idx]
Y_train, Y_test = Y.iloc[train_idx], Y.iloc[test_idx]
## Building RF model
model = RandomForestClassifier(
n_estimators=1000,
max_depth=4,
min_samples_split=15,
min_samples_leaf=8,
max_features=3,
random_state=i,
).fit(X_train, Y_train)
## Predicting on X_test and test
model_pred_1 = model.predict_proba(X_test)[:, 1]
model_pred_2 = model.predict_proba(test_baseline)[:, 1]
## Computing roc-auc score
score = roc_auc_score(Y_test, model_pred_1)
roc_auc_scores.append(score)
preds.append(model_pred_2)
print("Fold", i, "==> roc-auc-score is ==>", score)
cv_scores.append(np.mean(roc_auc_scores))
RF_cv_score = np.mean(cv_scores)
print(
"The oof average roc-auc-score of the Random Forest model over 10-folds is:",
RF_cv_score,
)
# From the above, we see an improvement in the cv-score. We next select the oof prediction with `roc-auc-score > 0.8` to ensemble.
to_ensemble = list()
for i in range(0, len(roc_auc_scores)):
if roc_auc_scores[i] > 0.80:
to_ensemble.append(preds[i])
RF_selected_to_ensemble = pd.DataFrame(to_ensemble).apply(np.mean, axis=0)
# Next, we consider GAM as a model to predict the likelihood the having kidney stones.
X = train[["gravity", "cond", "urea", "calc"]].copy()
Y = train["target"]
X["calc_gravity_ratio"] = X["calc"] / X["gravity"]
X["calc_urea_ratio"] = X["calc"] / X["urea"]
X["gravity_cond_ratio"] = X["gravity"] / X["cond"]
test_baseline = test[["gravity", "cond", "urea", "calc"]].copy()
test_baseline["calc_gravity_ratio"] = test_baseline["calc"] / test_baseline["gravity"]
test_baseline["calc_urea_ratio"] = test_baseline["calc"] / test_baseline["urea"]
test_baseline["gravity_cond_ratio"] = test_baseline["gravity"] / test_baseline["cond"]
cv_scores, roc_auc_scores = list(), list()
preds = list()
skf = StratifiedKFold(n_splits=10, random_state=42, shuffle=True)
for i, (train_idx, test_idx) in enumerate(skf.split(X, Y)):
## Splitting the data
X_train, X_test = X.iloc[train_idx], X.iloc[test_idx]
Y_train, Y_test = Y.iloc[train_idx], Y.iloc[test_idx]
## Building GAM
model = LogisticGAM(
s(0, n_splines=10)
+ s(1, n_splines=20)
+ s(2, n_splines=20)
+ s(3, n_splines=20)
+ s(4, n_splines=20)
+ s(5, n_splines=5)
+ s(6, n_splines=5)
).fit(X_train, Y_train)
## Predicting on X_test and test
model_pred_1 = model.predict_proba(X_test)
model_pred_2 = model.predict_proba(test_baseline)
## Computing roc-auc score
score = roc_auc_score(Y_test, model_pred_1)
roc_auc_scores.append(score)
preds.append(model_pred_2)
print("Fold", i, "==> roc-auc-score is ==>", score)
cv_scores.append(np.mean(roc_auc_scores))
gam_cv_score = np.mean(cv_scores)
print("The oof average roc-auc-score of the GAM model over 10-folds is:", gam_cv_score)
# From the above, we see that the cv-score of GAM is better than the random forest model. We next select the oof prediction with `roc-auc-score > 0.8` to ensemble.
to_ensemble = list()
for i in range(0, len(roc_auc_scores)):
if roc_auc_scores[i] > 0.80:
to_ensemble.append(preds[i])
GAM_selected_to_ensemble = pd.DataFrame(to_ensemble).apply(np.mean, axis=0)
submission["target"] = GAM_selected_to_ensemble
submission.to_csv("GAM_submission.csv", index=False)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
from sklearn import svm
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import OrdinalEncoder
from sklearn.neighbors import KNeighborsClassifier
from sklearn.metrics import classification_report, confusion_matrix
df = pd.read_csv("/kaggle/input/breast-cancer-wisconsin-data/data.csv")
df
df["diagnosis"].value_counts()
categorical_cols = ["diagnosis"]
ordinal_encoder = OrdinalEncoder()
df["diagnosis"] = ordinal_encoder.fit_transform(df.loc[:, "diagnosis":"diagnosis"])
df
df["diagnosis"].value_counts()
df.isna().sum()
X_columns = df.columns
X_columns = [col for col in X_columns if col not in ["id", "diagnosis", "Unnamed: 32"]]
y_columns = ["diagnosis"]
X = df.loc[:, X_columns]
y = df.loc[:, y_columns]
X
y
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.30, random_state=0
)
y_train.shape, y_train.values.reshape(-1).shape
clf = svm.SVC()
clf.fit(X_train, y_train.values.reshape(-1))
y_pred = clf.predict(X_test)
y_pred
print(confusion_matrix(y_test, y_pred))
print(classification_report(y_test, y_pred))
y_test["diagnosis"].value_counts()
# SVC is not a good classifier here maybe because the problem is not linearly separable
clf = KNeighborsClassifier(n_neighbors=2)
clf.fit(X_train, y_train.values.reshape(-1))
y_pred = clf.predict(X_test)
y_pred
print(confusion_matrix(y_test, y_pred))
print(classification_report(y_test, y_pred))
|
# Suppress warnings
import warnings
warnings.filterwarnings("ignore")
import pandas as pd
import numpy as np
import nltk
from nltk.corpus import stopwords
import re
import string
from sklearn.feature_extraction.text import CountVectorizer, TfidfVectorizer
from sklearn import model_selection
from sklearn.linear_model import LogisticRegression
# Getting started with Natural Language Processing
# CountVectorizers | TFIDF | Hashing Vectorizer
# Key Objectives:This notebook comes as a second part to the Getting started with NLP Notebooks that I am writing.In this notebook we shall study the various ways of vectorizing text data.Vectorization converts text data into feature vectors.
# Notebooks in this series
#
# Part 1: Getting started with NLP : A General Introduction
# Part 2: Getting started with NLP(2)- CountVectorizer
# ## Importing the dataset
train = pd.read_csv("../input/nlp-getting-started/train.csv")
test = pd.read_csv("../input/nlp-getting-started/test.csv")
train.head()
# # Text Vectorization Methods
# There are many methods to vctorize text, but in this notebook I shall discuss few of them:
# ## 1.Countvectorizer
# The [Scikit-Learn's CountVectorizer](https://scikit-learn.org/stable/modules/generated/sklearn.feature_extraction.text.CountVectorizer.html) provides a simple way to both tokenize a collection of text documents and build a vocabulary of known words, but also to encode new documents using that vocabulary.
# 
# We take a dataset and convert it into a corpus. Then we create a vocabulary of all the unique words in the corpus. Using this vocabulary, we can then create a feature vector of the count of the words. Let's see this through a simple example. Let's say we have a corpus containing two sentences as follows
sentences = ["The weather is sunny", "The weather is partly sunny and partly cloudy."]
from sklearn.feature_extraction.text import CountVectorizer
vectorizer = CountVectorizer()
vectorizer.fit(sentences)
vectorizer.vocabulary_
# Converting all the sentences to arrays
vectorizer.transform(sentences).toarray()
# By default, a scikit learn Count vectorizer can perform the following opertions over a text corpus:
# - Encoding via utf-8
# - converts text to lowercase
# - Tokenizes text using word level tokenization
# CountVectorizer has a number of parameters. Let's look at some of them :
# ### 1.1 Stopword
# Sometimes, some extremely common words which would appear to be of little value in helping select documents matching a user need are excluded from the vocabulary entirely. These words are called stop words. If `stop_word` parameter is specified with a list of stopwords, they will be removed from the vocabulary. Here I'll use the stopwords from NLTK but we can also specify custom stopwords too.
#
stopwords = stopwords.words("english")
count_vectorizer = CountVectorizer(stop_words=stopwords)
count_vectorizer.fit(train["text"])
train_vectors = count_vectorizer.transform(train["text"])
test_vectors = count_vectorizer.transform(test["text"])
train_vectors.shape
# See how the columns have reduced from 21637 to 21498. This is because some of the stopwords were removed.
# ### 1.2 MIN_DF and MAX_DF parameter
# `MIN_DF` lets you ignore those terms that appear rarely in a corpus. In other words, if `MIN_df`is 2, it means that a word has to occur at least two documents to be considered useful.
# `MAX_DF` on the other hand, ignores terms that have a document frequency strictly higher than the given threshold.These will be words which appear a lot of documents.
# This means we can eliminate those words that are either rare or appear too frequently in a corpus.
# When mentioned in absolute values i.e 1,2, etc, the value means if the word appears in 1 or 2 documents. However, when given in float, eg 30%, it means it appears in 30% of the documents.
count_vectorizer = CountVectorizer(stop_words=stopwords, min_df=2, max_df=0.8)
count_vectorizer.fit(train["text"])
train_vectors = count_vectorizer.transform(train["text"])
test_vectors = count_vectorizer.transform(test["text"])
# ### 1.3.Custom Preprocesser
# We can also preprocess the text by passing it as an argument to countvectorizer. The following options are avialable:
# - strip_accents - This removes any accents from the text during the preprocessing step.
# - lowercase - which is default set as true but can be set to False if lowercasing isnot desired
# - preprocessor - we can create our custom preprocessor and set this argument to that.
#
# Creating a custom preprocessor that lowercases, removes special characters, removes hyperlinks and punctuation
def custom_preprocessor(text):
"""
Make text lowercase, remove text in square brackets,remove links,remove special characters
and remove words containing numbers.
"""
text = text.lower()
text = re.sub("\[.*?\]", "", text)
text = re.sub("\\W", " ", text) # remove special chars
text = re.sub("https?://\S+|www\.\S+", "", text)
text = re.sub("<.*?>+", "", text)
text = re.sub("[%s]" % re.escape(string.punctuation), "", text)
text = re.sub("\n", "", text)
text = re.sub("\w*\d\w*", "", text)
return text
count_vectorizer = CountVectorizer(
list(train["text"]), preprocessor=custom_preprocessor
)
train_vectors = count_vectorizer.fit_transform(list(train["text"]))
test_vectors = count_vectorizer.transform(list(test["text"]))
# ### 1.4. N-Grams and analyzer parameter
# This paramneter specifies the upper and lower limit for the range of words/characters to be extracted from text. The following n-grams range stand for:
# (1,1) - unigrams eg 'United'
# (1,2) - unigrams and bigrams eg - 'United', 'United States'
# (2, 2)- only bigrams etc eg 'United States)
#
# World level unigrams and bigrams
count_vectorizer = CountVectorizer(
list(train["text"]), preprocessor=custom_preprocessor, ngram_range=(1, 2)
)
train_vectors = count_vectorizer.fit_transform(list(train["text"]))
test_vectors = count_vectorizer.transform(list(test["text"]))
list(count_vectorizer.vocabulary_)[:10]
# character level bigrams
count_vectorizer = CountVectorizer(
list(train["text"]),
preprocessor=custom_preprocessor,
ngram_range=(2, 2),
analyzer="char_wb",
)
train_vectors = count_vectorizer.fit_transform(list(train["text"]))
test_vectors = count_vectorizer.transform(list(test["text"]))
print(list(count_vectorizer.vocabulary_)[:20])
# ### Creating a Baseline Model using Countvectorizer
count_vectorizer = CountVectorizer(
token_pattern=r"\w{1,}",
ngram_range=(1, 2),
stop_words=stopwords,
preprocessor=custom_preprocessor,
)
count_vectorizer.fit(train["text"])
train_vectors = count_vectorizer.transform(train["text"])
test_vectors = count_vectorizer.transform(test["text"])
from sklearn.linear_model import LogisticRegression
clf = LogisticRegression(C=1.0)
scores = model_selection.cross_val_score(
clf, train_vectors, train["target"], cv=5, scoring="f1"
)
scores
# Fitting a simple Logistic Regression on Counts
clf.fit(train_vectors, train["target"])
# Submission
sample_submission = pd.read_csv("../input/nlp-getting-started/sample_submission.csv")
sample_submission["target"] = clf.predict(test_vectors)
sample_submission.to_csv("submission.csv", index=False)
# This gets me a score of 0.80777 on the Public LB, which isn't bad with simple Logistic Regression model.
# ## 2.TF-IDF Vectorizer
# 
# In the CountVectorizer, we use the counts of the words, in TFIDF we take the relative importance of that term in the entire corpus. TFIDF is composed of two words: TF and IDF.
# **TF** stands for the normalized term frequency. Term Frequency is a scoring of the frequency of the word in the current document.`TF = (Number of times term t appears in a document)/(Number of terms in the document)`
# **IDF** or Inverse Document Frequency: is a scoring of how rare the word is across documents. `IDF = 1+log(N/n)`, where N is the number of documents and n is the number of documents a term t has appeared in.TF-IDF weight is often used in information retrieval and text mining. This weight is a statistical measure used to evaluate how important a word is to a document in a collection or corpus
# *Because the ratio of the id f log function is greater or equal to 1, the TF–IDF score is
# always greater than or equal to zero. We interpret the score to mean that the closer the
# TF–IDF score of a term is to 1, the more informative that term is to that document.
# The closer the score is to zero, the less informative that term is.*
# from : [Applied Text Analysis with Python](https://www.amazon.in/Applied-Text-Analysis-Python-Language-Aware/dp/9352137434/ref=asc_df_9352137434/?tag=googleshopdes-21&linkCode=df0&hvadid=396988721232&hvpos=1o1&hvnetw=g&hvrand=11704105753328600061&hvpone=&hvptwo=&hvqmt=&hvdev=c&hvdvcmdl=&hvlocint=&hvlocphy=9062140&hvtargid=pla-838697427991&psc=1&ext_vrnc=hi)
# TFIDF can be generated at word, character or even N gram level.
# word level
tfidf = TfidfVectorizer(analyzer="word", token_pattern=r"\w{1,}", max_features=5000)
train_tfidf = tfidf.fit_transform(train["text"])
test_tfidf = tfidf.transform(test["text"])
# ngram level
tfidf = TfidfVectorizer(
analyzer="word", ngram_range=(2, 3), token_pattern=r"\w{1,}", max_features=5000
)
train_tfidf = tfidf.fit_transform(train["text"])
test_tfidf = tfidf.transform(test["text"])
# characters level
tfidf = TfidfVectorizer(
analyzer="char", ngram_range=(2, 3), token_pattern=r"\w{1,}", max_features=5000
)
train_tfidf = tfidf.fit_transform(train["text"])
test_tfidf = tfidf.transform(test["text"])
# ### Creating a Baseline Model using TFIDF
tfidf_vectorizer = TfidfVectorizer(
min_df=3,
max_features=None,
analyzer="word",
token_pattern=r"\w{1,}",
ngram_range=(1, 3),
use_idf=1,
smooth_idf=1,
sublinear_tf=1,
stop_words=stopwords,
)
train_tfidf = tfidf.fit_transform(train["text"])
test_tfidf = tfidf.transform(test["text"])
from sklearn.linear_model import LogisticRegression
clf = LogisticRegression(C=1.0)
scores = model_selection.cross_val_score(
clf, train_tfidf, train["target"], cv=5, scoring="f1"
)
scores
# Fitting a simple Logistic Regression on TFIDF
clf.fit(train_tfidf, train["target"])
|
# GPU details
# CPU details
# suppress warnings
import warnings
warnings.filterwarnings("ignore")
import os # for basic operations like folder creation, directory validation etc
import glob # finds file according to wildcard given
import shutil # for moving files
import matplotlib.pyplot as plt # for plotting graphs and viewing images
from numpy import uint8
import numpy as np
import base64
# sbox formation using affine transform
from numpy import uint8
def make_sbox():
s = []
for i in range(0, 256):
q = uint8(i)
x = (
q
^ leftrotater(q, 1)
^ leftrotater(q, 2)
^ leftrotater(q, 3)
^ leftrotater(q, 4)
^ 99
)
x = hex(x)
f = x.split("x")[1]
if len(f) == 1:
f = "0" + f
s.append(f)
return s
sbox = make_sbox()
print(sbox)
# circular left shift in array
def leftrotater(x, shift):
x = (x << shift) | (x >> (8 - shift))
return uint8(x)
# function to convert float values to decimal
def floatdec_convert(my_number, places=18):
res = []
my_whole, my_dec = str(my_number).split(".")
my_whole = int(my_whole)
my_dec = int(my_dec)
if my_dec == 0:
res = "0.000000000000000000"
else:
res = bin(my_whole).lstrip("0b") + "."
rang = 1
while (my_dec != 0) and (rang <= places):
my_whole, my_dec = str((my_decimal_converter(my_dec)) * 2).split(".")
my_dec = int(my_dec)
res += my_whole
rang += 1
while rang <= places:
res += "0"
rang += 1
return res
def my_decimal_converter(num):
while num > 1:
num /= 10
return num
# function to recieve only binary values after decimal point
def arr2bin(a=[]):
lst = []
for i in range(0, len(a)):
print("{0:.9f}".format(float(a[i])))
bin_val = floatdec_convert(str(a[i]))
c = bin_val.split(".")[1]
lst += c
return lst
def appendingzeros(b): ## to make length of list as multiple of 8
lnth = len(b)
if lnth % 8 != 0:
c = 8 - (lnth % 8)
while c > 0:
b.append("0")
c -= 1
return b
def getcode(b): ###to get code word generated from sbox and binary values of features
lnth = len(b)
code = ""
for i in range(0, lnth, 8):
d_val = (
128 * int(b[i])
+ 64 * int(b[i + 1])
+ 32 * int(b[i + 2])
+ 16 * int(b[i + 3])
+ 8 * int(b[i + 4])
+ 4 * int(b[i + 5])
+ 2 * int(b[i + 6])
+ 1 * int(b[i + 7])
)
code += sbox[d_val]
###for extra security base64 encoding of code
data = code
keyword = base64.b64encode(data.encode("utf-8"))
keyword1 = str(keyword, "utf-8")
return keyword1
def callforcode(a):
a /= 100 ####normalizing input
b = arr2bin(a)
c = appendingzeros(b)
word = getcode(c)
return word
a = np.array([256.5625, 6.500, 0.0123]) ###specify input array
word = callforcode(a)
print(word)
print(len(word))
import cv2 as cv
import numpy as np
from matplotlib import pyplot as plt
img = cv.imread("/kaggle/input/fingerprint-database/fvc2006SETA/train/1/1_3.bmp", 0)
print(type(img))
# Otsu's thresholding
ret, thresh_img = cv.threshold(img, 0, 255, cv.THRESH_BINARY + cv.THRESH_OTSU)
def intarray(binstring):
"""Change a 2D matrix of 01 chars into a list of lists of ints"""
return [
[1 if ch == "1" else 0 for ch in line] for line in binstring.strip().split()
]
def chararray(intmatrix):
"""Change a 2d list of lists of 1/0 ints into lines of 1/0 chars"""
return "\n".join("".join(str(p) for p in row) for row in intmatrix)
def toTxt(intmatrix):
"""Change a 2d list of lists of 1/0 ints into lines of '#' and '.' chars"""
return "\n".join("".join(("#" if p else ".") for p in row) for row in intmatrix)
def neighbours(x, y, image):
"""Return 8-neighbours of point p1 of picture, in order"""
i = image
x1, y1, x_1, y_1 = x + 1, y - 1, x - 1, y + 1
# print ((x,y))
return [
i[y1][x],
i[y1][x1],
i[y][x1],
i[y_1][x1], # P2,P3,P4,P5
i[y_1][x],
i[y_1][x_1],
i[y][x_1],
i[y1][x_1],
] # P6,P7,P8,P9
def transitions(neighbours):
n = neighbours + neighbours[0:1] # P2, ... P9, P2
return sum((n1, n2) == (0, 1) for n1, n2 in zip(n, n[1:]))
def zhangSuen(image):
changing1 = changing2 = [(-1, -1)]
while changing1 or changing2:
# Step 1
changing1 = []
for y in range(1, len(image) - 1):
for x in range(1, len(image[0]) - 1):
P2, P3, P4, P5, P6, P7, P8, P9 = n = neighbours(x, y, image)
if (
image[y][x] == 1
and P4 * P6 * P8 == 0 # (Condition 0)
and P2 * P4 * P6 == 0 # Condition 4
and transitions(n) == 1 # Condition 3
and 2 <= sum(n) <= 6 # Condition 2
): # Condition 1
changing1.append((x, y))
for x, y in changing1:
image[y][x] = 0
# Step 2
changing2 = []
for y in range(1, len(image) - 1):
for x in range(1, len(image[0]) - 1):
P2, P3, P4, P5, P6, P7, P8, P9 = n = neighbours(x, y, image)
if (
image[y][x] == 1
and P2 * P6 * P8 == 0 # (Condition 0)
and P2 * P4 * P8 == 0 # Condition 4
and transitions(n) == 1 # Condition 3
and 2 <= sum(n) <= 6 # Condition 2
): # Condition 1
changing2.append((x, y))
for x, y in changing2:
image[y][x] = 0
# print changing1
# print changing2
return image
invert = cv.bitwise_not(thresh_img / 255)
skeleton = zhangSuen(invert)
plt.figure(figsize=(10, 10), dpi=100)
plt.subplot(3, 1, 1)
plt.imshow(img, "gray")
plt.subplot(3, 1, 2)
plt.imshow(thresh_img, "gray")
plt.subplot(3, 1, 3)
plt.imshow(skeleton, "gray")
plt.show()
import cv2 as cv
def intarray(binstring):
"""Change a 2D matrix of 01 chars into a list of lists of ints"""
return [
[1 if ch == "1" else 0 for ch in line] for line in binstring.strip().split()
]
def chararray(intmatrix):
"""Change a 2d list of lists of 1/0 ints into lines of 1/0 chars"""
return "\n".join("".join(str(p) for p in row) for row in intmatrix)
def toTxt(intmatrix):
"""Change a 2d list of lists of 1/0 ints into lines of '#' and '.' chars"""
return "\n".join("".join(("#" if p else ".") for p in row) for row in intmatrix)
def neighbours(x, y, image):
"""Return 8-neighbours of point p1 of picture, in order"""
i = image
x1, y1, x_1, y_1 = x + 1, y - 1, x - 1, y + 1
# print ((x,y))
return [
i[y1][x],
i[y1][x1],
i[y][x1],
i[y_1][x1], # P2,P3,P4,P5
i[y_1][x],
i[y_1][x_1],
i[y][x_1],
i[y1][x_1],
] # P6,P7,P8,P9
def transitions(neighbours):
n = neighbours + neighbours[0:1] # P2, ... P9, P2
return sum((n1, n2) == (0, 1) for n1, n2 in zip(n, n[1:]))
def zhangSuen(image):
changing1 = changing2 = [(-1, -1)]
while changing1 or changing2:
# Step 1
changing1 = []
for y in range(1, len(image) - 1):
for x in range(1, len(image[0]) - 1):
P2, P3, P4, P5, P6, P7, P8, P9 = n = neighbours(x, y, image)
if (
image[y][x] == 1
and P4 * P6 * P8 == 0 # (Condition 0)
and P2 * P4 * P6 == 0 # Condition 4
and transitions(n) == 1 # Condition 3
and 2 <= sum(n) <= 6 # Condition 2
): # Condition 1
changing1.append((x, y))
for x, y in changing1:
image[y][x] = 0
# Step 2
changing2 = []
for y in range(1, len(image) - 1):
for x in range(1, len(image[0]) - 1):
P2, P3, P4, P5, P6, P7, P8, P9 = n = neighbours(x, y, image)
if (
image[y][x] == 1
and P2 * P6 * P8 == 0 # (Condition 0)
and P2 * P4 * P8 == 0 # Condition 4
and transitions(n) == 1 # Condition 3
and 2 <= sum(n) <= 6 # Condition 2
): # Condition 1
changing2.append((x, y))
for x, y in changing2:
image[y][x] = 0
# print changing1
# print changing2
return image
def fingerprint_preprocess(image):
"""
this function accepts a numpy array and return the same
"""
# img = cv.imread(image, 0)
# print(type(image))
# gray = cv.normalize(src=image, dst=None, alpha=0, beta=255, norm_type=cv.NORM_MINMAX, dtype=cv.CV_8UC1)
gray = cv.cvtColor(image, cv.COLOR_RGB2GRAY)
print(type(gray), gray.shape)
# Otsu's thresholding
ret, thresh_img = cv.threshold(gray, 0, 255, cv.THRESH_BINARY + cv.THRESH_OTSU)
# invert image color
invert = cv.bitwise_not(thresh_img / 255)
# skeletonize the fingerprint
skeleton = zhangSuen(invert)
return skeleton
from keras.models import Model
from keras import backend as K
from keras.layers import Input
from keras.layers import Dense, GlobalAveragePooling2D, Dropout, MaxPool1D
from keras.applications.inception_v3 import InceptionV3
from keras.preprocessing.image import ImageDataGenerator
from keras.callbacks import CSVLogger, ModelCheckpoint
from keras.optimizers import Adam, RMSprop, SGD
img_width, img_height = 96, 96
train_data_dir = "/kaggle/input/fingerprint-database/fvc2006SETA/train"
validation_data_dir = "/kaggle/input/fingerprint-database/fvc2006SETA/train/"
nb_train_samples = 3024
nb_validation_samples = 2016
batch_size = 64
epochs = 100
if K.image_data_format() == "channels_last":
input_tensor = Input(shape=(img_width, img_height, 3))
else:
input_tensor = Input(shape=(3, img_width, img_height))
train_datagen = ImageDataGenerator(
rescale=1.0 / 255,
rotation_range=30,
# horizontal_flip=True,
# width_shift_range=0.2,
# height_shift_range=0.2,
# preprocessing_function=fingerprint_preprocess,
validation_split=0.40,
)
train_generator = train_datagen.flow_from_directory(
train_data_dir,
target_size=(img_width, img_height),
batch_size=batch_size,
class_mode="categorical",
subset="training",
)
validation_generator = train_datagen.flow_from_directory(
train_data_dir,
target_size=(img_width, img_height),
batch_size=batch_size,
class_mode="categorical",
subset="validation",
)
# imports the inceptionv3 pretained model
inception_model = InceptionV3(
input_tensor=input_tensor, weights=None, include_top=False
)
x = inception_model.output
x = GlobalAveragePooling2D()(x)
x = Dense(1024, activation="relu")(x)
# x = Dense(512, activation='relu')(x)
x = Dense(256, activation="relu")(x)
# x = Dropout(0.20)(x)
predictions = Dense(140, activation="softmax")(x)
model = Model(inputs=inception_model.input, outputs=predictions)
# functions to calculate f1score
def recall_m(y_true, y_pred):
true_positives = K.sum(K.round(K.clip(y_true * y_pred, 0, 1)))
possible_positives = K.sum(K.round(K.clip(y_true, 0, 1)))
recall = true_positives / (possible_positives + K.epsilon())
return recall
def precision_m(y_true, y_pred):
true_positives = K.sum(K.round(K.clip(y_true * y_pred, 0, 1)))
predicted_positives = K.sum(K.round(K.clip(y_pred, 0, 1)))
precision = true_positives / (predicted_positives + K.epsilon())
return precision
def f1score(y_true, y_pred):
precision = precision_m(y_true, y_pred)
recall = recall_m(y_true, y_pred)
return 2 * ((precision * recall) / (precision + recall + K.epsilon()))
optimizer = Adam(lr=0.01)
model.compile(
optimizer=optimizer,
loss="categorical_crossentropy",
metrics=["accuracy", f1score, recall_m, precision_m],
)
# model.summary()
filepath_accuracy = "accuracy_weights.{epoch:02d}-{val_accuracy:.2f}.hdf5"
filepath_f1score = "f1score_weights.{epoch:02d}-{val_f1score:.2f}.hdf5"
csv_logger = CSVLogger("training.log")
accuracy_checkpoint = ModelCheckpoint(
filepath_accuracy,
monitor="val_accuracy",
verbose=0,
save_best_only=True,
save_weights_only=True,
mode="auto",
period=1,
)
f1score_checkpoint = ModelCheckpoint(
filepath_f1score,
monitor="val_f1score",
verbose=0,
save_best_only=True,
save_weights_only=True,
mode="auto",
period=1,
)
history = model.fit_generator(
train_generator,
steps_per_epoch=nb_train_samples // batch_size,
epochs=epochs,
shuffle=True,
validation_data=validation_generator,
validation_steps=nb_validation_samples // batch_size,
callbacks=[csv_logger, accuracy_checkpoint],
)
model.save("inceptionv3.h5")
print(history.history.keys())
# summarize history for accuracy
plt.plot(history.history["accuracy"])
plt.plot(history.history["val_accuracy"])
plt.title("model accuracy")
plt.ylabel("accuracy")
plt.xlabel("epoch")
plt.legend(["train", "test"], loc="upper left")
plt.show()
# summarize history for loss
plt.plot(history.history["loss"])
plt.plot(history.history["val_loss"])
plt.title("model loss")
plt.ylabel("loss")
plt.xlabel("epoch")
plt.legend(["train", "test"], loc="upper left")
plt.show()
# summarize history for recall
plt.plot(history.history["recall_m"])
plt.plot(history.history["val_recall_m"])
plt.title("model recall_m")
plt.ylabel("recall_m")
plt.xlabel("epoch")
plt.legend(["train", "test"], loc="upper left")
plt.show()
# summarize history for precision
plt.plot(history.history["precision_m"])
plt.plot(history.history["val_precision_m"])
plt.title("model precision_m")
plt.ylabel("precision_m")
plt.xlabel("epoch")
plt.legend(["train", "test"], loc="upper left")
plt.show()
# summarize history for f1score
plt.plot(history.history["f1score"])
plt.plot(history.history["val_f1score"])
plt.title("model f1score")
plt.ylabel("f1score")
plt.xlabel("epoch")
plt.legend(["train", "test"], loc="upper left")
plt.show()
|
# # Linear Regression Model for Real Estate Prices in the USA
# The real estate market is one of the most important sectors of the United States economy. With a wide variety of factors that affect the price of a property, it is crucial for investors and buyers to understand which variables have the greatest impact on price. In this notebook, we will explore a dataset of real estate properties in the USA and use statistical modeling techniques to create a linear regression model that establishes the relationship between the explanatory variables and real estate prices.
# The aim of this study is to use linear regression to identify the variables that most influence real estate prices in the USA and thus help investors and buyers make more informed decisions. To achieve this, we will use the R language and its statistical libraries to analyze the dataset, create the linear regression model, and evaluate its effectiveness.
# ## First Step: Ensuring Data Accuracy by Checking the Working Directory and Software Version
# One of the first steps in any data analysis project is to check the working directory where the report is being created and the version of the software being used. This is important to ensure that the analysis is being performed on the correct data and that any updates or changes to the software are taken into account.
# Checking the working directory involves verifying that the files being used for the analysis are located in the correct folder or directory. This helps to avoid errors caused by accessing the wrong files and ensures that the analysis is being performed on the intended data.
# Verifying the software version is also critical to ensure that the analysis is accurate and consistent. Software updates can introduce changes to the algorithms used for data analysis, so it is important to be aware of any updates or changes that have been made to the software since the last analysis was performed.
# Overall, checking the working directory and software version are essential first steps in any data analysis project, helping to ensure that the analysis is based on the correct data and performed accurately and consistently.
getwd()
sessionInfo()
library(dplyr)
# ## Second Step: Ensuring Accurate Analysis with ETL and Data Validation
# The second critical step in data analysis is the ETL (Extraction, Transformation, and Loading) process. This stage is essential to ensure that the data is ready for analysis and can be reliable for making accurate decisions.
# A fundamental aspect of the ETL process is to understand the database. This involves understanding how the data was collected, what variables are present, the scale of measurement, possible missing values, and other important information. Additionally, it is necessary to evaluate the reliability of the data and if it is complete. This means that the data should be error-free and be within an expected range.
# Another critical step in the ETL process is to ensure that all data is being "read" correctly by the data analysis software. This includes checking if the software is recognizing the variables correctly and if the data is being imported correctly. Data reading errors can lead to inaccurate results and making wrong decisions.
# In summary, understanding the database and evaluating its reliability and completeness, as well as ensuring that all data is being read correctly by the software, is crucial in the ETL process. This will help ensure that the data is ready for analysis and assist in making accurate decisions based on that data.
data < -read.csv("/kaggle/input/house-prices-advanced-regression-techniques/train.csv")
head(data, 5)
sprintf(
"The database has a total of %s columns and %s rows.", dim(data)[2], dim(data)[1]
)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
# Part 1: Deriving Bayes theorem from conditional probability
# **Conditional probability**
# I covered conditional probability in more depth here.
# Conditional probability tells us the probability of an event occurring, given another event.
# **P(A|B) = P(A ∩ B) / P(B) is the probability A occurs, in cases where we knowB occurs. It’s calculated as the probability that both A and B occur, divided by the probability that B occurs.
# But what if we wanted to find the reverse, the probability of B, in cases where A occurs?**
# **sometimes it’s easier to use Bayes theorem.**
# **Deriving Bayes theorem**
# **We start with the formula for conditional probability which can be written either, “A given B” or “B given A”.**
# **We’ll start with the 1st formula, P(A|B)= P(A∩B) / P(B).
# Multiple both sides by P(B). This will cancel out the P(B) denominator on the right, leaving us with below.**
# **What we can now see (more easily if we swapped the left and right sides) is that P(A∩B)= P(A|B) * P(B) . We’ll plug this back into our 2nd original formula**
# 
# Part 2: Predicting if an SMS message is spam
# Bayesian inference has a long history in spam detection. We’ll get into the basics here with some real data.
# 
# **In our case, the probability an SMS is spam, given some word, is equal to the probability of the word, given it is in a spam SMS, multiplied by the probability of spam, all divided by the probability of the word.**
# inspect tje dataframe
dir = "../input/sms-spam-collection-dataset/spam.csv"
import pandas as pd
df = pd.read_csv(dir, encoding="ISO-8859-1")
df.head()
# The columns in the original CSV don’t make sense. So we’ll move the useful information into 2 new columns, one of which is a boolean indicating if the SMS is spam.
# FYI, “ham” means “not spam”.
import pandas as pd
df["sms"] = df["v2"]
df["spam"] = np.where(df["v1"] == "spam", 1, 0)
df.head()
# now drop the old columns
df = df[["sms", "spam"]]
df.head()
# check the no fo records
len(df)
# That’s a lot. Let’s work with a sample of 25% of the original data.
sample_df = df.sample(frac=0.25)
len(sample_df)
# Now split the data into 2 separate dataframes, one for spam and one for ham.
spam_df = sample_df.loc[df["spam"] == 1]
ham_df = sample_df.loc[df["spam"] == 0]
print(len(spam_df))
print(len(ham_df))
# We’ll use sklearn’s TFIDF vectorizer to eyeball some words important in the spam messages, and pick one to plug into our formula.
from sklearn.feature_extraction.text import TfidfVectorizer
vectorizer_spam = TfidfVectorizer(stop_words="english", max_features=30)
vectorizer_spam.fit(spam_df["sms"])
vectorizer_spam.vocabulary_
# We need to pick a word to use in our formula so I’m going to choose the word “win”, though it would be interesting to try this for other words as well.
# P(W|S) = probability of the word “win” being in a spam message
# P(S) = probability of a spam message overall
# P(W) = probability of the word “win” in a message overall
word = "win"
# Calculate P(W|S)
word = "win"
spam_count = 0
spam_with_word_count = 0
for idx, row in spam_df.iterrows():
spam_count += 1
if word in row.sms:
spam_with_word_count += 1
probability_of_word_given_spam = spam_count / spam_with_word_count
print(probability_of_word_given_spam)
# Calculate P(S)
probability_of_spam = len(spam_df) / (len(sample_df))
print(probability_of_spam)
# Calculate P(W)
sms_count = 0
word_in_sms_count = 0
for idx, row in sample_df.iterrows():
sms_count += 1
if word in row.sms:
word_in_sms_count += 1
probability_of_word = word_in_sms_count / sms_count
print(probability_of_word)
# Now putting it all together
(probability_of_word_given_spam * probability_of_spam) / probability_of_word
|
import pandas as pd
iris = pd.read_csv("/kaggle/input/iris/Iris.csv")
iris
# # Analyzing the data
iris.isnull().sum()
iris.Species.unique()
# # Treating the data
iris.drop(columns=["Id"], inplace=True)
iris.Species = iris.Species.replace("Iris-setosa", 0)
iris.Species = iris.Species.replace("Iris-versicolor", 1)
iris.Species = iris.Species.replace("Iris-virginica", 2)
iris
X = iris.drop(columns=["Species"])
y = iris.Species
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
stdscl = StandardScaler()
X = stdscl.fit_transform(X)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=0)
import optuna
import tensorflow as tf
def objective(trial):
# Define the hyperparameters to optimize
n_layers = trial.suggest_int("n_layers", 1, 5)
n_units = trial.suggest_int("n_units", 16, 256)
dropout_rate = trial.suggest_uniform("dropout_rate", 0.0, 0.5)
# Define the model architecture
model = tf.keras.Sequential()
for i in range(n_layers):
model.add(tf.keras.layers.Dense(units=n_units, activation="relu"))
model.add(tf.keras.layers.Dropout(dropout_rate))
model.add(tf.keras.layers.Dense(units=3, activation="softmax"))
# Compile the model with the hyperparameters
model.compile(
optimizer="Adam", loss="sparse_categorical_crossentropy", metrics=["accuracy"]
)
# Train the model and compute the validation accuracy
model.fit(X_train, y_train, epochs=10, batch_size=32, verbose=0)
score = model.evaluate(X_test, y_test, verbose=0)[1]
# Return the score to optimize
return score
# Create the study object and optimize the objective function
study = optuna.create_study(direction="maximize")
study.optimize(objective, n_trials=100)
# Print the best set of hyperparameters and the corresponding score
print("Best score:", study.best_value)
print("Best parameters:", study.best_params)
optuna.visualization.plot_optimization_history(study)
optuna.visualization.plot_contour(study)
optuna.visualization.plot_param_importances(study)
model_optimized = tf.keras.Sequential(
[
tf.keras.layers.Dense(215, input_shape=(X_train.shape[1],), activation="relu"),
tf.keras.layers.Dropout(0.01798081887567679),
tf.keras.layers.Dense(215, input_shape=(X_train.shape[1],), activation="relu"),
tf.keras.layers.Dropout(0.01798081887567679),
tf.keras.layers.Dense(215, input_shape=(X_train.shape[1],), activation="relu"),
tf.keras.layers.Dropout(0.01798081887567679),
tf.keras.layers.Dense(215, input_shape=(X_train.shape[1],), activation="relu"),
tf.keras.layers.Dropout(0.01798081887567679),
tf.keras.layers.Dense(215, input_shape=(X_train.shape[1],), activation="relu"),
tf.keras.layers.Dropout(0.01798081887567679),
tf.keras.layers.Dense(3, activation="sigmoid"),
]
)
import tensorflow as tf
from sklearn.metrics import accuracy_score, precision_score, recall_score, f1_score
model_optimized.compile(
optimizer="adam", loss="sparse_categorical_crossentropy", metrics=["accuracy"]
)
model_optimized.fit(X_train, y_train, epochs=10, batch_size=32)
model_optimized.evaluate(X_test, y_test)
|
# # 1 | Encoding Techniques In Natural Language Processing
# Word encoding techniques in natural language processing (NLP) refer to the process of representing words as numerical values that can be processed by machine learning models. One of the most popular techniques is one-hot encoding, where each word is represented as a vector with a value of 1 in its corresponding index and 0 in all other indices. Another technique is word embedding, which represents words as dense, continuous vectors that capture semantic relationships between words. These techniques enable NLP models to process and analyze large volumes of text data efficiently and effectively.
# # 1.1 | Bag Of Words
# Bag of Words (BoW) is a language modeling technique used in natural language processing (NLP) to represent a text document as a bag of its words, disregarding grammar and word order, but keeping track of their frequency.
# The basic idea behind BoW is to represent a document as a set of its constituent words, and count how many times each word appears in the document. This results in a sparse vector representation of the document, where the vector is as long as the vocabulary size, and each dimension corresponds to a unique word in the vocabulary.
# The BoW technique is often used as a feature extraction method in various NLP tasks, such as text classification, sentiment analysis, and information retrieval. However, it suffers from some limitations, such as not capturing the semantic relationships between words and the context in which they appear. This has led to the development of more advanced techniques, such as word embeddings and deep learning models, that attempt to overcome these limitations.
# ****
# **Note**
# * This notebook is higly inspired by
# * * **[YouTube](https://www.youtube.com/)=>[ritvikmath](https://www.youtube.com/@ritvikmath)=>[Bag Of Word: Natural Language Processing](https://youtu.be/irzVuSO8o4g)**
# * * **[AskPython](https://www.askpython.com/)=>[Python](https://www.askpython.com/python)=>[Examples](https://www.askpython.com/python/examples)=>[Bag Of Words From Scratch](https://www.askpython.com/python/examples/bag-of-words-model-from-scratch)**
import numpy as np
from collections import defaultdict
import nltk
nltk.download("punkt")
# Lets assume we have this sentence
# * She loves pizza, pizza is delicious.
# * She is a good person.
# * good people are the best.
data = [
"She loves pizza, pizza is delicious.",
"She is a good person.",
"good people are the best.",
]
# Its a small case, so lets just manually type the unique words in the combined sentences into a dictionary
identifiers = {
"she",
"loves",
"pizza",
"is",
"delicious",
"a",
"good",
"person",
"people",
"are",
"the",
"best",
}
# Now we will try to break the words. For example our sentence is `She loves pizza, pizza is delicious.`. What we want is `["She" , "loves" , "pizza" , "piazza" , "is" , "delicious"]`.
# For that lets assume we first have this sentecne instead `Swag Like Ohio`, and we want `["Swag" , "Like" , "Ohio"]`
# Our first step should be to break this sentence first
nltk.tokenize.word_tokenize("Swag Like Ohio")
# and it was pretty straight forward
# Now what if the sentence was `[Dang Like Ohio , Swag Like Ohio]` and the expected output was `["Dang" , "Like" , "Ohio" , "Swag" , "Like" , "Ohio"]` You would say, just tokenize them again
nltk.tokenize.word_tokenize("Dang Like Ohio , Swag Like Ohio")
# Ooops, we got an extra `,` here. But we dont want comas. what we can do is to only select letters that are `alpha` using the `str.alpha`.
[
word.lower()
for word in nltk.tokenize.word_tokenize("Dang Like Ohio , Swag Like Ohio")
if word.isalpha()
]
# And that is what something we wanted. But we need to do this for all sentences.
sentences = []
vocab = []
for sent in data:
sentence = [w.lower() for w in nltk.tokenize.word_tokenize(sent) if w.isalpha()]
sentences.append(sentence)
for word in sentence:
if word not in vocab:
vocab.append(word)
vocab
sentences
# Now we need to assign `index` to each word in a dictionary. So that we can use that later
index_word = {}
i = 0
for word in vocab:
index_word[word] = i
i += 1
# Now we just need to define a function that adds value into a vector
def bag_of_words(sent):
count_dict = defaultdict(int)
vec = np.zeros(len(vocab))
for item in sent:
count_dict[item] += 1
for key, item in count_dict.items():
vec[index_word[key]] = item
return vec
# And we have made our `Bag Of Words`.
# Now we just need to make all of this into a global function for better usage
# # 1.1.1 Bag Of Words Final Source Code
def BagOfWords(data):
sentences = []
vocab = []
for sent in data:
sentence = [w.lower() for w in nltk.tokenize.word_tokenize(sent) if w.isalpha()]
sentences.append(sentence)
for word in sentence:
if word not in vocab:
vocab.append(word)
index_word = {}
i = 0
for word in vocab:
index_word[word] = i
i += 1
count_dict = defaultdict(int)
vec = np.zeros(len(vocab))
for item in sent:
count_dict[item] += 1
for key, item in count_dict.items():
vec[index_word[key]] = item
return vec
# # 1.2 | TF-IDF(Term Frequency - Inverse Documentory Frqeuency)
# TF-IDF stands for "term frequency-inverse document frequency," a statistical measure used to evaluate the relevance of a term in a document or corpus.
# TF-IDF is a widely used method in information retrieval and text mining, and it calculates the importance of a word in a document by taking into account both the frequency of the word in the document and the frequency of the word in the entire corpus of documents.
# The "term frequency" component of TF-IDF refers to the number of times a word appears in a particular document. The "inverse document frequency" component refers to how rare or common a word is across all documents in the corpus. This component helps to down-weight the importance of words that appear frequently across all documents in the corpus and boost the importance of words that appear less frequently.
# The TF-IDF score of a term in a document is the product of its term frequency and inverse document frequency. A higher TF-IDF score indicates that a word is more important or relevant to the document.
# TF-IDF is commonly used in information retrieval tasks, such as search engines, document clustering, and text classification, to identify the most relevant documents for a given query.
# ****
# **Note**
# * This notebook is higly inspired by
# * * **[AskPython](https://www.askpython.com/)=>[Python](https://www.askpython.com/python)=>[Examples](https://www.askpython.com/python/examples)=>[Creating a TF-IDF Model from Scratch](https://www.askpython.com/python/examples/tf-idf-model-from-scratch)**
# * * **[YouTube](https://www.youtube.com/)=>[@ritvikmath](https://www.youtube.com/@ritvikmath)=>[TFIDF : Data Science Concepts](https://www.youtube.com/watch?v=OymqCnh-APA)**
# ****
# $$TF = \frac {Number_-of_-times_-a_-word_-"X"_-apprears_-in_-a_-Document}{Number_-Of_-Words_-present_-in_-the_-Document}$$
# $$IDF = log(\frac{Number_-of_-Documents_-present_-in_-a_-Corpus}{Number_-of_-Documents_-where_-word_-"X"_-has_-appeared})$$
# Lets assume you have a $3$ very huge books titles as
# * **The Love Story Of Panda And Capybara**
# * **Capybara Left Panda For Frog**
# * **Panda And Frog Unites (Homies !!!)**
# And you want to know the important words in the books respectively. One way of doing this is to use **Bag Of Words** and count the unique characters in the whole book,or the formula will be like
# $$\frac {Number_-of_-times_-a_-word_-"X"_-apprears_-in_-a_-Document}{Number_-Of_-Words_-present_-in_-the_-Document}$$
# Lets give this a fancy name **Term Frequency**,
# But there is an issue here, Common words like `the` , `and` , `is` and much more, will be having a lot more appreances than the actual words in all of the books,
# One way is to just ignore all of these kind of words and move on with life. But that will be indirectly using a huge dictionary, and finding if the word exists in the dictionary or not.
# Another way is to use this formula
# $$log(\frac{Number_-of_-Documents_-present_-in_-a_-Corpus}{Number_-of_-Documents_-where_-word_-"X"_-has_-appeared})$$
# Lets give this one a more fancy name **Inverse Document Frequency**
# So how is this formula actually solving the porblem we were facing before...?
# Lets assume we have the word `the` and we have $3$, this word appear in all the $3$ documents, so it is not that important. Lets try to plug the values in the $IDF$ formula to get some numrical values
# $$log(\frac{Number_-of_-Documents_-present_-in_-a_-Corpus}{Number_-of_-Documents_-where_-word_-"X"_-has_-appeared})$$
# $$= log \frac {3}{3} => log \frac {1}{1} => log(1) => 0$$
# So now this shows that the word `the` is not that important.
# Now lets unite our $2$ formulas $TF$ and $IDF$ and call it $TF-IDF$, We gave it this name because it was haunting.
# So now our formula is $$TF-IDF = \frac {Number_-of_-times_-a_-word_-"X"_-apprears_-in_-a_-Document}{Number_-Of_-Words_-present_-in_-the_-Document} X log(\frac{Number_-of_-Documents_-present_-in_-a_-Corpus}{Number_-of_-Documents_-where_-word_-"X"_-has_-appeared})$$
# Lets make this a short formula $$TF-IDF = \frac {X_n}{n}log\frac{X_{n_{unique}}}{D_n}$$
# Lets assume we have this data
data = [
"Topic sentences are similar to mini thesis statements. Like a thesis statement, a topic sentence has a specific main point. Whereas the thesis is the main point of the essay",
"the topic sentence is the main point of the paragraph. Like the thesis statement, a topic sentence has a unifying function. But a thesis statement or topic sentence alone doesn’t guarantee unity.",
"An essay is unified if all the paragraphs relate to the thesis, whereas a paragraph is unified if all the sentences relate to the topic sentence.",
]
# We have already created a vocab, so now we can directly apply that here. If you forgot how to make a vocab, so scroll upward a little bit and you will find something magical
vocab = []
for _ in range(len(data)):
k = [j.lower() for j in nltk.word_tokenize(data[0]) if j.isalpha()]
data.append(k)
data.pop(0)
for word in k:
if word not in vocab:
vocab.append(word)
# Now we will try to make a dictionary with this as we did before
indices = {}
i = 0
for word in vocab:
indices[word] = i
i += 1
# Lets just try to take a view at the dictionary we just created
indices
# Now we want to count the number of appearances of a word and store into a dictionary
def count_dict(data):
word_count = {}
for word in vocab:
word_count[word] = 0
for sent in data:
if word in sent:
word_count[word] += 1
return word_count
word_count = count_dict(data)
word_count
# Applying the formaula for `Term Frequency` , $$TF = \frac {Number_-of_-times_-a_-word_-"X"_-apprears_-in_-a_-Document}{Number_-Of_-Words_-present_-in_-the_-Document} = \frac {X_n}{n}$$
def termfreq(document, word):
N = len(document)
occurance = len([token for token in document if token == word])
return occurance / N
# Now applying the formula for `Inverse Document Frequency` $$IDF = log(\frac{Number_-of_-Documents_-present_-in_-a_-Corpus}{Number_-of_-Documents_-where_-word_-"X"_-has_-appeared}) = log\frac{X_{n_{unique}}}{D_n}$$
def inverse_doc_freq(word):
try:
word_occurance = word_count[word] + 1
except:
word_occurance = 1
return np.log(len(data) / word_occurance)
# Combining both of the formulas we get
def tf_idf(sentence):
tf_idf_vec = np.zeros((len(vocab),))
for word in sentence:
tf = termfreq(sentence, word)
idf = inverse_doc_freq(word)
value = tf * idf
tf_idf_vec[indices[word]] = value
return tf_idf_vec
# Now we just need to apply the functions
vectors = []
for sent in data:
vec = tf_idf(sent)
vectors.append(vec)
vectors
# **AND WE JUST MADE OUR TF-IDF, YAYYYYYYYYYYYYYYYYY :)**
# Now lets put all of this into a particular function for better usage
# # 1.2.1 | Final TF-IDF Source Code
import numpy as np
from nltk.tokenize import word_tokenize
import nltk
nltk.download("punkt")
def TF_IDF(data):
vocab = []
for _ in range(len(data)):
k = [j.lower() for j in word_tokenize(data[0]) if j.isalpha()]
data.append(k)
data.pop(0)
for word in k:
if word not in vocab:
vocab.append(word)
indices = {}
i = 0
for word in vocab:
indices[word] = i
i += 1
def count_dict(data):
word_count = {}
for word in vocab:
word_count[word] = 0
for sent in data:
if word in sent:
word_count[word] += 1
return word_count
word_count = count_dict(data)
def termfreq(document, word):
N = len(document)
occurance = len([token for token in document if token == word])
return occurance / N
def inverse_doc_freq(word):
try:
word_occurance = word_count[word] + 1
except:
word_occurance = 1
return np.log(len(data) / word_occurance)
def tf_idf(sentence):
tf_idf_vec = np.zeros((len(vocab),))
for word in sentence:
tf = termfreq(sentence, word)
idf = inverse_doc_freq(word)
value = tf * idf
tf_idf_vec[indices[word]] = value
return tf_idf_vec
vectors = []
for sent in data:
vec = tf_idf(sent)
vectors.append(vec)
return vectors
|
# ## Credit Risk Analytics
# **Definition of Target and Outcome Window:**
# One of the leading banks would like to predict bad customer while customer applying for loan. This model also called as PD models (Probability of Default)
# #### Data PreProcessing:
# - Missing values Treatment - Numerical (Mean/Median imputation) and Categorical (separate missing category or merging)
# - Univariate Analysis - Outlier and Frequency analysis
# #### Data Exploratory Analysis
# - Bivariate Analysis - Numeric(ttest) and Categorical (Chisquare)
# - Bivariate Analysis - Visualization
# - Variable Transformation - P- value based selection
# - Variable Transformation - Bucketing/ Binning for numerical variables and Dummy for categorical variables.
# - Variable Reduction 0 IV/ Somers D
# - Variable Reduction - Multicollinearity.
# #### Model build and Model Diagnostics
# - Train and Test split
# - Significance of each variable
# - Gini and ROC / Concordance analysis - Rank ordering
# - Classfication Table Analysis - Accuracy
# #### Model Validation
# - OOS validation - p-value and sign testing for the model coefficients.
# - Diagnostics check to remain similar to Training model build
# - BootStrapping, if necessary
# #### Model interpretation for its properties
# - Interferencing for finding the most important contributors.
# - Prediction of risk and proactive prevention by targeting segments of the population
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
import statsmodels.formula.api as sm
import scipy.stats as stats
import pandas_profiling
plt.rcParams["figure.figsize"] = 10, 7.5
plt.rcParams["axes.grid"] = True
plt.gray()
from matplotlib.backends.backend_pdf import PdfPages
from sklearn.model_selection import train_test_split
from sklearn import metrics
from sklearn.linear_model import LogisticRegression
from statsmodels.stats.outliers_influence import variance_inflation_factor
from patsy import dmatrices
bankloans = pd.read_csv("../input/bankloans.csv")
bankloans.columns
bankloans.info()
# pandas_profiling.ProfileReport(bankloans)
numeric_var_names = [
key
for key, val in bankloans.dtypes.items()
if val in ["float64", "int64", "float32", "int32"]
]
cat_var_names = [key for key, val in bankloans.dtypes.items() if val in ["object"]]
print(numeric_var_names)
print(cat_var_names)
bankloans_num = bankloans[numeric_var_names]
bankloans_num.head()
bankloans_cat = bankloans[cat_var_names]
bankloans_cat.head()
# ### Creating Data Audit report
# Use a general function that returns multiple values
def var_summary(x):
return pd.Series(
[
x.count(),
x.isnull().sum(),
x.sum(),
x.mean(),
x.median(),
x.std(),
x.var(),
x.min(),
x.dropna().quantile(0.01),
x.dropna().quantile(0.05),
x.dropna().quantile(0.10),
x.dropna().quantile(0.25),
x.dropna().quantile(0.50),
x.dropna().quantile(0.75),
x.dropna().quantile(0.90),
x.dropna().quantile(0.95),
x.dropna().quantile(0.99),
x.max(),
],
index=[
"N",
"NMISS",
"SUM",
"MEAN",
"MEDIAN",
"STD",
"VAR",
"MIN",
"P1",
"P5",
"P10",
"P25",
"P50",
"P75",
"P90",
"P95",
"P99",
"MAX",
],
)
num_summary = bankloans_num.apply(lambda x: var_summary(x)).T
num_summary
bankloans_existing = bankloans_num[bankloans_num.default.isnull() == 0]
bankloans_new = bankloans_num[bankloans_num.default.isnull() == 1]
# ### Handling outliers
def outlier_capping(x):
x = x.clip_upper(x.quantile(0.99))
x = x.clip_lower(x.quantile(0.01))
return x
bankloans_existing = bankloans_existing.apply(lambda x: outlier_capping(x))
bankloans_existing.corr()
# ### Visualize the correlation matrix in seaborn using heatmap
sns.heatmap(bankloans_existing.corr())
plt.show()
# ### Data Exploratory Analysis
# - Variable Transformation: (i) Bucketing
bankloans_existing.columns.difference(["default"])
bp = PdfPages("Transformation plots.pdf")
for num_variable in bankloans_existing.columns.difference(["default"]):
binned = pd.cut(
bankloans_existing[num_variable], bins=10, labels=list(range(1, 11))
)
odds = bankloans_existing.groupby(binned)["default"].sum() / (
bankloans_existing.groupby(binned)["default"].count()
- bankloans_existing.groupby(binned)["default"].sum()
)
log_odds = np.log(odds)
fig, axes = plt.subplots(figsize=(10, 4))
sns.barplot(x=log_odds.index, y=log_odds)
plt.title(
str(
"Logit plot for identifying if the bucketing is required or not for varibale -- "
)
+ str(num_variable)
)
bp.savefig(fig)
bp.close()
# ### 2.5 Data Exploratory Analysis
# - Variable reduction using Somer's D values
bankloans_existing.columns
logreg_model = sm.logit("default~address", data=bankloans_existing).fit()
p = logreg_model.predict(bankloans_existing)
metrics.roc_auc_score(bankloans_existing["default"], p)
2 * metrics.roc_auc_score(bankloans_existing["default"], p) - 1
somersd_df = pd.DataFrame()
for num_variable in bankloans_existing.columns.difference(["default"]):
logreg = sm.logit(formula="default~" + str(num_variable), data=bankloans_existing)
result = logreg.fit()
y_score = pd.DataFrame(result.predict())
y_score.columns = ["Score"]
somers_d = 2 * metrics.roc_auc_score(bankloans_existing["default"], y_score) - 1
temp = pd.DataFrame([num_variable, somers_d]).T
temp.columns = ["Variable Name", "SomersD"]
somersd_df = pd.concat([somersd_df, temp], axis=0)
somersd_df
somersd_df.sort_values(by="SomersD", ascending=False)
# ### Variance Inflation Factor assessment
X = pd.concat(
[
bankloans_existing[bankloans_existing.columns.difference(["default"])],
bankloans_existing["default"],
],
axis=1,
)
features = "+".join(bankloans_existing.columns.difference(["default"]))
X.head()
features
a, b = dmatrices(formula_like="default~" + features, data=X, return_type="dataframe")
vif = pd.DataFrame()
vif["VIF Factor"] = [variance_inflation_factor(b.values, i) for i in range(b.shape[1])]
vif["features"] = b.columns
print(vif)
# ### Train and Test split
train, test = train_test_split(bankloans_existing, test_size=0.3, random_state=43)
train.columns
# ### Building logistic Regression
logreg = sm.logit(formula="default ~ address+age+creddebt+debtinc+employ", data=train)
result = logreg.fit()
print(result.summary2())
# - LE = -2.1527 -0.0865\*address+0.0432\*age+0.6576\*creddebt+0.1021\*debtinc-0.2917\*employ
# - p(Default=1) = exp(LE)/(1+exp(LE))
train_gini = 2 * metrics.roc_auc_score(train["default"], result.predict(train)) - 1
print("The Gini Index for the model built on the Train Data is : ", train_gini)
test_gini = 2 * metrics.roc_auc_score(test["default"], result.predict(test)) - 1
print("The Gini Index for the model built on the Test Data is :", test_gini)
train_auc = metrics.roc_auc_score(train["default"], result.predict(train))
test_auc = metrics.roc_auc_score(test["default"], result.predict(test))
print("The AUC for the model built on the Train Data is:", train_auc)
print("The AUC for the model built on the Test Data is:", test_auc)
# ### Intuition behind ROC curve - predicted probability as a tool for separating the 1's and 0's
train_predicted_prob = pd.DataFrame(result.predict(train))
train_predicted_prob.columns = ["prob"]
train_actual = train["default"]
# ### making a DataFrame with actual and prob columns
train_predict = pd.concat([train_actual, train_predicted_prob], axis=1)
train_predict.columns = ["actual", "prob"]
train_predict.head()
# intuition behind ROC curve - predicted probability as a tool for separating the 1's and 0's
test_predicted_prob = pd.DataFrame(result.predict(test))
test_predicted_prob.columns = ["prob"]
test_actual = test["default"]
# making a DataFrame with actual and prob columns
test_predict = pd.concat([test_actual, test_predicted_prob], axis=1)
test_predict.columns = ["actual", "prob"]
test_predict.head()
np.linspace(0, 1, 50)
train_predict.head()
train_predict["predicted"] = train_predict["prob"].apply(
lambda x: 0.0 if x < 0.2 else 1.0
)
train_predict.head()
train_predict["tp"] = train_predict.apply(
lambda x: 1.0 if x["actual"] == 1.0 and x["predicted"] == 1 else 0.0, axis=1
)
train_predict["fp"] = train_predict.apply(
lambda x: 1.0 if x["actual"] == 0.0 and x["predicted"] == 1 else 0.0, axis=1
)
train_predict["tn"] = train_predict.apply(
lambda x: 1.0 if x["actual"] == 0.0 and x["predicted"] == 0 else 0.0, axis=1
)
train_predict["fn"] = train_predict.apply(
lambda x: 1.0 if x["actual"] == 1.0 and x["predicted"] == 0 else 0.0, axis=1
)
train_predict.head()
accuracy = (train_predict.tp.sum() + train_predict.tn.sum()) / (
train_predict.tp.sum()
+ train_predict.tn.sum()
+ train_predict.fp.sum()
+ train_predict.fn.sum()
)
accuracy
Sensitivity = (train_predict.tp.sum()) / (
train_predict.tp.sum() + train_predict.fn.sum()
)
Sensitivity
# **Intuition behind ROC curve - confusion matrix for each different cut-off shows trade off in sensitivity and specificity**
roc_like_df = pd.DataFrame()
train_temp = train_predict.copy()
for cut_off in np.linspace(0, 1, 50):
train_temp["cut_off"] = cut_off
train_temp["predicted"] = train_temp["prob"].apply(
lambda x: 0.0 if x < cut_off else 1.0
)
train_temp["tp"] = train_temp.apply(
lambda x: 1.0 if x["actual"] == 1.0 and x["predicted"] == 1 else 0.0, axis=1
)
train_temp["fp"] = train_temp.apply(
lambda x: 1.0 if x["actual"] == 0.0 and x["predicted"] == 1 else 0.0, axis=1
)
train_temp["tn"] = train_temp.apply(
lambda x: 1.0 if x["actual"] == 0.0 and x["predicted"] == 0 else 0.0, axis=1
)
train_temp["fn"] = train_temp.apply(
lambda x: 1.0 if x["actual"] == 1.0 and x["predicted"] == 0 else 0.0, axis=1
)
sensitivity = train_temp["tp"].sum() / (
train_temp["tp"].sum() + train_temp["fn"].sum()
)
specificity = train_temp["tn"].sum() / (
train_temp["tn"].sum() + train_temp["fp"].sum()
)
accuracy = (train_temp["tp"].sum() + train_temp["tn"].sum()) / (
train_temp["tp"].sum()
+ train_temp["fn"].sum()
+ train_temp["tn"].sum()
+ train_temp["fp"].sum()
)
roc_like_table = pd.DataFrame([cut_off, sensitivity, specificity, accuracy]).T
roc_like_table.columns = ["cutoff", "sensitivity", "specificity", "accuracy"]
roc_like_df = pd.concat([roc_like_df, roc_like_table], axis=0)
roc_like_df.head()
# finding ideal cut-off for checking if this remains same in OOS validation
roc_like_df["total"] = roc_like_df["sensitivity"] + roc_like_df["specificity"]
roc_like_df.head()
# Cut-off based on highest sum(sensitivity+ specificity) - common way of identifying cut-off
roc_like_df[roc_like_df["total"] == roc_like_df["total"].max()]
# Cut off based on highest sensitivity
roc_like_df[roc_like_df["sensitivity"] == roc_like_df["sensitivity"].max()]
# #Choosen Best Cut-off is 0.20 based on highest (sensitivity+specicity)
test_predict["predicted"] = test_predict["prob"].apply(lambda x: 1 if x > 0.2 else 0)
train_predict["predicted"] = train_predict["prob"].apply(lambda x: 1 if x > 0.2 else 0)
pd.crosstab(train_predict["actual"], train_predict["predicted"])
pd.crosstab(test_predict["actual"], test_predict["predicted"])
print(
"The overall accuracy score for the Train Data is : ",
metrics.accuracy_score(train_predict.actual, train_predict.predicted),
)
print(
"The overall accuracy score for the Test Data is : ",
metrics.accuracy_score(test_predict.actual, test_predict.predicted),
)
print(metrics.classification_report(train_predict.actual, train_predict.predicted))
print(metrics.classification_report(test_predict.actual, test_predict.predicted))
# **Decile Analysis**
# - Top- two deciles - High Risk customers - will reject applications
# - 3rd, 4th, 5th decile - medium risk customers - will accept application with proper scrutiny.
# - 6th decile onwards - Low risk customers - accept the applications
train_predict["Deciles"] = pd.qcut(train_predict["prob"], 10, labels=False)
train_predict.head()
test_predict["Deciles"] = pd.qcut(test_predict["prob"], 10, labels=False)
test_predict.head()
# ### Decile Analysis for train data
no_1s = (
train_predict[["Deciles", "actual"]]
.groupby(train_predict.Deciles)
.sum()
.sort_index(ascending=False)["actual"]
)
no_total = (
train_predict[["Deciles", "actual"]]
.groupby(train_predict.Deciles)
.count()
.sort_index(ascending=False)["actual"]
)
max_prob = (
train_predict[["Deciles", "prob"]]
.groupby(train_predict.Deciles)
.max()
.sort_index(ascending=False)["prob"]
)
min_prob = (
train_predict[["Deciles", "prob"]]
.groupby(train_predict.Deciles)
.min()
.sort_index(ascending=False)["prob"]
)
Decile_analysis_train = pd.concat(
[min_prob, max_prob, no_1s, no_total - no_1s, no_total], axis=1
)
Decile_analysis_train.columns = ["Min_prob", "Max_prob", "#1", "#0", "Total"]
Decile_analysis_train
# ### Decile Analysis for test data
no_1s = (
test_predict[["Deciles", "actual"]]
.groupby(test_predict.Deciles)
.sum()
.sort_index(ascending=False)["actual"]
)
no_total = (
test_predict[["Deciles", "actual"]]
.groupby(test_predict.Deciles)
.count()
.sort_index(ascending=False)["actual"]
)
max_prob = (
test_predict[["Deciles", "prob"]]
.groupby(test_predict.Deciles)
.max()
.sort_index(ascending=False)["prob"]
)
min_prob = (
test_predict[["Deciles", "prob"]]
.groupby(test_predict.Deciles)
.min()
.sort_index(ascending=False)["prob"]
)
Decile_analysis_test = pd.concat(
[min_prob, max_prob, no_1s, no_total - no_1s, no_total], axis=1
)
Decile_analysis_test.columns = ["Min_prob", "Max_prob", "#1", "#0", "Total"]
Decile_analysis_test
# ## Predicting new customers (Implementation of Model on new data)
bankloans_new.head()
# before scoring new customers, you need to process the data using the sames steps you followed while building the model
#
bankloans_new["prob"] = result.predict(bankloans_new)
bankloans_new.head()
bankloans_new["default"] = bankloans_new["prob"].apply(lambda x: 1 if x > 0.20 else 0)
bankloans_new.head()
bankloans_new.default.value_counts()
|
import pickle
import pandas as pd
import numpy as np
import spacy
from sentence_transformers import SentenceTransformer
from datetime import datetime as dt
### Function help to separate text to list of sentences
def text_to_sent_list(
text,
nlp=spacy.load("en_core_web_lg"),
embedder=SentenceTransformer("distilbert-base-nli-mean-tokens"),
min_len=2,
):
# convert to list of sentences
text = nlp(text)
sents = list(text.sents)
# remove newline
sents_clean = [sentence.replace("\n", " ") for sentence in sents]
# remove entries with empty list
sents_clean = [sentence.text for sentence in sents_clean if len(sentence) != 0]
# remove entries with only white space
sents_clean = [sentence.text for sentence in sents_clean if sentence != " "]
# embed sentences. We only use this step for adding "label" attribute.
sents_embedding = np.array(embedder.encode(sents_clean, convert_to_tensor=True))
return sents_clean, sents_embedding
# Read data
data_business = pd.read_csv("../input/bbcnewstocsv/businessDataset.csv")
data_entertainment = pd.read_csv("../input/bbcnewstocsv/entertainmentDataset.csv")
data_politics = pd.read_csv("../input/bbcnewstocsv/politicsDataset.csv")
data_tech = pd.read_csv("../input/bbcnewstocsv/techDataset.csv")
data_sport = pd.read_csv(
"../input/bbcnewstocsv/sportDataset.csv", encoding="unicode_escape"
)
# Drop unused column
data_business = data_business.drop(["ID"], axis=1)
data_business = data_business.reset_index(drop=True)
data_entertainment = data_entertainment.drop(["ID"], axis=1)
data_entertainment = data_entertainment.reset_index(drop=True)
data_politics = data_politics.drop(["ID"], axis=1)
data_politics = data_politics.reset_index(drop=True)
data_tech = data_tech.drop(["ID"], axis=1)
data_tech = data_tech.reset_index(drop=True)
data_sport = data_sport.drop(["ID"], axis=1)
data_sport = data_sport.reset_index(drop=True)
# make dictionary of datasets by category
datasets = {
"business": data_business,
"entertainment": data_entertainment,
"politics": data_politics,
"sport": data_sport,
"tech": data_tech,
}
datasets["business"].head()
for key in datasets:
dataset = datasets[key]
output_file = "train_df_" + key + ".pickle"
# load nlp and embedder
nlp = spacy.load("en_core_web_lg")
embedder = SentenceTransformer("distilbert-base-nli-mean-tokens")
# extract clean sentence list and sentence embedding for each article's text
f = lambda text: text_to_sent_list(text, nlp=nlp, embedder=embedder, min_len=2)
s_interim_tuple = dataset["Text"].apply(f)
dataset["text_clean"] = s_interim_tuple.apply(lambda x: x[0])
dataset["text_embedding"] = s_interim_tuple.apply(lambda x: x[1])
# extract clean sentence list and sentence embedding for each article's summary
f = lambda summ: text_to_sent_list(summ, nlp=nlp, embedder=embedder, min_len=0)
s_interim_tuple = dataset["Summary"].apply(f)
dataset["summary_clean"] = s_interim_tuple.apply(lambda x: x[0])
dataset["summary_embedding"] = s_interim_tuple.apply(lambda x: x[1])
with open(output_file, "wb") as handle:
pickle.dump(dataset, handle)
import pickle
import numpy as np
import pandas as pd
from sklearn.metrics.pairwise import cosine_similarity
from datetime import datetime as dt
### Helper Functions
# Calculate cosine between two vector for checking similarity.
def find_sim_single_summary(summary_sentence_embed, doc_emedding):
cos_sim_mat = cosine_similarity(doc_emedding, summary_sentence_embed)
# Pick most similar sentences.
idx_arr = np.argmax(cos_sim_mat, axis=0)
return idx_arr
# Adding attribute "label" for data.
def label_sent_in_summary(s_text, s_summary):
doc_num = s_text.shape[0]
# initialize zeros. All sentences is labeled by "0"
labels = [np.zeros(doc.shape[0]) for doc in s_text.tolist()]
# find index of summary-picked sentences. Check every pair sentences of text and summary.
idx_list = [
np.sort(find_sim_single_summary(s_summary[j], s_text[j]))
for j in range(doc_num)
]
# Change label to "1" for summary-picked sentences
for j in range(doc_num):
labels[j][idx_list[j]] = 1
return idx_list, labels
df = pd.read_pickle("../input/bbcpreprocess/train_df_tech.pickle")
df.head()
datasets_key = ["sport", "politics", "business", "tech", "entertainment"]
for key in datasets_key:
output_file = "train_df_label_" + key + ".pickle"
df = pd.read_pickle("../input/bbcpreprocess/train_df_" + key + ".pickle")
# get index list and target labels
idx_list, labels = label_sent_in_summary(df.text_embedding, df.summary_embedding)
# wrap in dataframe
df["labels"] = labels
df["labels_idx_list"] = idx_list
# save to pickle
with open(output_file, "wb") as handle:
pickle.dump(df, handle)
|
import os
import cv2
import gc
import numpy as np
import pandas as pd
import itertools
from tqdm.autonotebook import tqdm
import albumentations as A
import matplotlib.pyplot as plt
import torch
from torch import nn
import torch.nn.functional as F
import timm
from transformers import DistilBertModel, DistilBertConfig, DistilBertTokenizer
import os
import collections
import json
import numpy as np
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
import tensorflow_hub as hub
import tensorflow_text as text
import tensorflow_addons as tfa
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
from tqdm import tqdm
import pandas as pd
from transformers import DistilBertModel, DistilBertConfig, DistilBertTokenizer
import torch
from torch import nn
import torch.nn.functional as F
# Suppressing tf.hub warnings
tf.get_logger().setLevel("ERROR")
tfk = tf.keras
tfkl = tf.keras.layers
df = pd.read_csv(
"../input/flickr-image-dataset/flickr30k_images/results.csv", delimiter="|"
)
df.columns = ["image", "caption_number", "caption"]
df["caption"] = df["caption"].str.lstrip()
df["caption_number"] = df["caption_number"].str.lstrip()
df.loc[19999, "caption_number"] = "4"
df.loc[19999, "caption"] = "A dog runs across the grass ."
ids = [id_ for id_ in range(len(df) // 5) for i in range(5)]
df["id"] = ids
df.to_csv("captions.csv", index=False)
df.head()
class CFG:
debug = False
image_path = "../input/flickr-image-dataset/flickr30k_images/flickr30k_images"
captions_path = "."
batch_size = 32
num_workers = 4
head_lr = 1e-3
image_encoder_lr = 1e-4
text_encoder_lr = 1e-5
weight_decay = 1e-3
patience = 1
factor = 0.8
epochs = 2
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
model_name = "resnet50"
image_embedding = 2048
text_encoder_model = "distilbert-base-uncased"
text_embedding = 768
text_tokenizer = "distilbert-base-uncased"
max_length = 200
pretrained = True # for both image encoder and text encoder
trainable = True # for both image encoder and text encoder
temperature = 1.0
# image size
size = 224
# for projection head; used for both image and text encoders
num_projection_layers = 1
projection_dim = 256
dropout = 0.1
class AvgMeter:
def __init__(self, name="Metric"):
self.name = name
self.reset()
def reset(self):
self.avg, self.sum, self.count = [0] * 3
def update(self, val, count=1):
self.count += count
self.sum += val * count
self.avg = self.sum / self.count
def __repr__(self):
text = f"{self.name}: {self.avg:.4f}"
return text
def get_lr(optimizer):
for param_group in optimizer.param_groups:
return param_group["lr"]
class CLIPDataset(torch.utils.data.Dataset):
def __init__(self, image_filenames, captions, tokenizer, transforms):
"""
image_filenames and cpations must have the same length; so, if there are
multiple captions for each image, the image_filenames must have repetitive
file names
"""
self.image_filenames = image_filenames
self.captions = list(captions)
self.encoded_captions = tokenizer(
list(captions), padding=True, truncation=True, max_length=CFG.max_length
)
self.transforms = transforms
def __getitem__(self, idx):
item = {
key: torch.tensor(values[idx])
for key, values in self.encoded_captions.items()
}
image = cv2.imread(f"{CFG.image_path}/{self.image_filenames[idx]}")
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
image = self.transforms(image=image)["image"]
item["image"] = image
item["caption"] = self.captions[idx]
return item
def __len__(self):
return len(self.captions)
def get_transforms(mode="train"):
if mode == "train":
return A.Compose(
[
A.Resize(CFG.size, CFG.size, always_apply=True),
A.Normalize(max_pixel_value=255.0, always_apply=True),
]
)
else:
return A.Compose(
[
A.Resize(CFG.size, CFG.size, always_apply=True),
A.Normalize(max_pixel_value=255.0, always_apply=True),
]
)
def make_train_valid_dfs():
dataframe = pd.read_csv(f"{CFG.captions_path}/captions.csv")
max_id = dataframe["id"].max() + 1 if not CFG.debug else 100
image_ids = np.arange(0, max_id)
np.random.seed(42)
valid_ids = np.random.choice(
image_ids, size=int(0.2 * len(image_ids)), replace=False
)
train_ids = [id_ for id_ in image_ids if id_ not in valid_ids]
train_dataframe = dataframe[dataframe["id"].isin(train_ids)].reset_index(drop=True)
valid_dataframe = dataframe[dataframe["id"].isin(valid_ids)].reset_index(drop=True)
return train_dataframe, valid_dataframe
def build_loaders(dataframe, tokenizer, mode):
transforms = get_transforms(mode=mode)
dataset = CLIPDataset(
dataframe["image"].values,
dataframe["caption"].values,
tokenizer=tokenizer,
transforms=transforms,
)
dataloader = torch.utils.data.DataLoader(
dataset,
batch_size=CFG.batch_size,
num_workers=CFG.num_workers,
shuffle=True if mode == "train" else False,
)
return dataloader
train_df, valid_df = make_train_valid_dfs()
def create_vision_encoder(trainable=False):
# Load the pre-trained Xception model to be used as the base encoder.
resnet = keras.applications.ResNet50(
include_top=False, weights="imagenet", pooling="avg"
)
# Set the trainability of the base encoder.
for layer in resnet.layers:
layer.trainable = trainable
# Receive the images as inputs.
inputs = layers.Input(shape=(224, 224, 3), name="image_input")
# Preprocess the input image.
resnet_input = tf.keras.applications.resnet.preprocess_input(inputs)
# Generate the embeddings for the images using the xception model.
embeddings = resnet(resnet_input)
# Create the vision encoder model.
return keras.Model(inputs, embeddings, name="vision_encoder")
def create_text_encoder(
model_name=CFG.text_encoder_model,
pretrained=CFG.pretrained,
trainable=CFG.trainable,
):
if pretrained:
model = DistilBertModel.from_pretrained(model_name)
else:
model = DistilBertModel(config=DistilBertConfig())
for p in model.parameters():
p.requires_grad = trainable
return model
def project_embeddings(
embeddings,
num_projection_layers=1,
projection_dim=CFG.projection_dim,
dropout_rate=CFG.dropout,
):
projected_embeddings = layers.Dense(units=projection_dim)(embeddings)
for _ in range(num_projection_layers):
x = tf.nn.gelu(projected_embeddings)
x = layers.Dense(projection_dim)(x)
x = layers.Dropout(dropout_rate)(x)
x = layers.Add()([projected_embeddings, x])
projected_embeddings = layers.LayerNormalization()(x)
return projected_embeddings
class DualEncoder(tfk.Model):
def __init__(
self,
text_encoder,
image_encoder,
temperature=CFG.temperature,
image_embedding=CFG.image_embedding,
text_embedding=CFG.text_embedding,
**kwargs,
):
super(DualEncoder, self).__init__(**kwargs)
self.text_encoder = text_encoder
self.image_encoder = image_encoder
self.temperature = temperature
def call(self, image, input_ids, attention_mask):
# Getting Image and Text inputs
image_features = self.image_encoder(image)
input_ids = torch.from_numpy(input_ids)
attention_mask = torch.from_numpy(attention_mask)
text_features = self.text_encoder(
input_ids=input_ids, attention_mask=attention_mask
)
# Getting Image and Text Embeddings (with same dimension)
last_hidden_state = text_features.last_hidden_state
text_features = last_hidden_state[:, 0, :]
text_features = text_features.detach().numpy()
image_embeddings = project_embeddings(image_features)
text_embeddings = project_embeddings(text_features)
return image_embeddings, text_embeddings
import tensorflow as tf
from tensorflow.keras.losses import CategoricalCrossentropy
from tensorflow.keras.activations import softmax
def cross_entropy(preds, targets, reduction="none"):
log_softmax_output = tf.nn.log_softmax(preds, axis=-1)
loss = CategoricalCrossentropy(reduction=reduction)(targets, log_softmax_output)
return loss
def compute_loss(text_embeddings, image_embeddings):
# Calculating the Loss
logits = tf.matmul(text_embeddings, image_embeddings, transpose_b=True) / 1.0
images_similarity = tf.matmul(image_embeddings, image_embeddings, transpose_b=True)
texts_similarity = tf.matmul(text_embeddings, text_embeddings, transpose_b=True)
targets = softmax((images_similarity + texts_similarity) / 2 * 1.0, axis=-1)
texts_loss = cross_entropy(logits, targets, reduction="none")
images_loss = cross_entropy(
tf.transpose(logits), tf.transpose(targets), reduction="none"
)
loss = (images_loss + texts_loss) / 2.0 # shape: (batch_size)
return tf.reduce_mean(loss)
def make_train_valid_dfs():
dataframe = pd.read_csv(f"{CFG.captions_path}/captions.csv")
max_id = dataframe["id"].max() + 1 if not CFG.debug else 100
image_ids = np.arange(0, max_id)
np.random.seed(42)
valid_ids = np.random.choice(
image_ids, size=int(0.2 * len(image_ids)), replace=False
)
train_ids = [id_ for id_ in image_ids if id_ not in valid_ids]
train_dataframe = dataframe[dataframe["id"].isin(train_ids)].reset_index(drop=True)
valid_dataframe = dataframe[dataframe["id"].isin(valid_ids)].reset_index(drop=True)
return train_dataframe, valid_dataframe
def build_loaders(dataframe, tokenizer, mode):
transforms = get_transforms(mode=mode)
dataset = CLIPDataset(
dataframe["image"].values,
dataframe["caption"].values,
tokenizer=tokenizer,
transforms=transforms,
)
dataloader = torch.utils.data.DataLoader(
dataset,
batch_size=CFG.batch_size,
num_workers=CFG.num_workers,
shuffle=True if mode == "train" else False,
)
return dataloader
train_df, valid_df = make_train_valid_dfs()
tokenizer = DistilBertTokenizer.from_pretrained(CFG.text_tokenizer)
train_loader = build_loaders(train_df, tokenizer, mode="train")
valid_loader = build_loaders(valid_df, tokenizer, mode="valid")
vision_encoder = create_vision_encoder()
text_encoder = create_text_encoder()
model = DualEncoder(text_encoder, vision_encoder)
model.compile(optimizer=tf.optimizers.Adam(learning_rate=0.01))
loss_meter = AvgMeter()
tqdm_object = tqdm(train_loader, total=len(train_loader))
optimizer = tf.optimizers.Adam(learning_rate=0.01)
step = 0
for epoch in range(10):
for batch in tqdm_object:
X = batch["image"].numpy()
Y = batch["input_ids"].numpy()
Z = batch["attention_mask"].numpy()
with tf.GradientTape() as tape:
image_embeddings, text_embeddings = model(X, Y, Z)
image_embeddings, text_embeddings = (
image_embeddings.numpy(),
text_embeddings.numpy(),
)
loss_value = compute_loss(text_embeddings, image_embeddings)
grads = tape.gradient(loss_value, model.trainable_weights)
# Run one step of gradient descent by updating
# the value of the variables to minimize the loss.
optimizer.apply_gradients(zip(grads, model.trainable_weights))
step += 1
if step % 10 == 0:
print(loss_value)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv(
"/kaggle/input/health-care-data-set-on-heart-attack-possibility/heart.csv"
)
df.shape
df.head(3)
import matplotlib.pyplot as plt
import seaborn as sns
plt.scatter(df.age, df.chol)
plt.xlabel("Age")
plt.ylabel("Cholestrol")
plt.show()
g = sns.catplot(
x="sex",
y="trestbps",
hue="target",
col="cp",
data=df,
kind="bar",
height=4,
aspect=0.7,
)
h = sns.catplot(
x="sex", y="chol", hue="target", col="cp", data=df, kind="bar", height=4, aspect=0.7
)
df["slope"].unique()
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(
df.drop("target", axis=1), df["target"], test_size=0.2, random_state=42
)
from catboost import CatBoostClassifier
model = CatBoostClassifier(loss_function="Logloss", iterations=1000)
grid = {
"learning_rate": list(np.arange(0, 1, 0.01)),
"depth": list(np.arange(3, 7)),
"l2_leaf_reg": list(np.arange(1, 50, 5)),
}
grid_search_result = model.grid_search(grid, X=x_train, y=y_train)
grid_search_result["params"]
model = CatBoostClassifier(
loss_function="Logloss", iterations=1000, depth=4, l2_leaf_reg=6, learning_rate=0.59
)
model.fit(x_train, y_train)
y_pred = model.predict(x_test)
from sklearn.metrics import f1_score
f1_score(y_test, y_pred, average="macro")
|
# # Machine Learning Series - Lecture 4
# ***
# ## Analyzing Model Performance for Boston Housing Dataset
# The dataset for this exercise originates from the [UCI Machine Learning Repository](https://archive.ics.uci.edu/ml/datasets/Housing). The Boston housing data was collected in 1978 and each of the 506 entries represent aggregated data about 14 features for homes from various suburbs in Boston, Massachusetts. For the purposes of this project, the following preprocessing steps have been made to the dataset:
# - 16 data points have an 'MEDV' value of 50.0. These data points likely contain **missing or censored values** and have been removed.
# - 1 data point has an 'RM' value of 8.78. This data point can be considered an **outlier** and has been removed.
# - The features 'RM', 'LSTAT', 'PTRATIO', and 'MEDV' are essential. The remaining **non-relevant features** have been excluded.
# - The feature 'MEDV' has been **multiplicatively scaled** to account for 35 years of market inflation.
# Run the code cells below to initialize some necessary Python libraries required for visualization and exploration of data and load the Boston housing dataset. You will know the dataset loaded successfully if the size of the dataset is reported.
# Import 'pandas'
import pandas as pd
# Import 'numpy'
import numpy as np
# Import 'matplotlib.pyplot'
import matplotlib.pyplot as plt
# Import supplementary visualizations code diagnostics.py
import diagnostics as dg
# Pretty display for notebooks
# Load the Boston housing dataset
data = pd.read_csv("../input/housing/housing.csv")
prices = data["MEDV"]
features = data.drop("MEDV", axis=1)
# Success
print(
"Boston housing dataset has {} data points with {} variables each.".format(
*data.shape
)
)
# Success - Display the first five records
display(data.head())
# ## Learning Curves
# The following code cell produces four graphs for a decision tree model with different maximum depths. Each graph visualizes the learning curves of the model for both training and testing as the size of the training set is increased. Note that the shaded region of a learning curve denotes the uncertainty of that curve (measured as the standard deviation). The model is scored on both the training and testing sets using R2, the coefficient of determination.
# Run the code cell below and use these graphs to answer the following question.
# Produce learning curves for varying training set sizes and maximum depths
dg.ModelLearning(features, prices)
# ## Complexity Curves
# The following code cell produces a graph for a decision tree model that has been trained and validated on the training data using different maximum depths. The graph produces two complexity curves — one for training and one for validation. Similar to the **learning curves**, the shaded regions of both the complexity curves denote the uncertainty in those curves, and the model is scored on both the training and validation sets using the performance_metric function.
# Run the code cell below to plot the complexity curve and check for the **best maximum depth** value.
# Plot complexity curve for varying maximum depths
dg.ModelComplexity(features, prices)
# ***
# ## Support Vector Machine with Diagnostics
# ## Project: Driving Behavior Prediction
# In this project, we will implement the machine learning algorithm to get a model able to detect the driver's mental state, whether: Focused, De-Focused or Drowsy, from his EEG brain signals readings, using a headset during driving.
# ## Getting Started
# In the next code cells, we will define some functions along with a few of the necessary Python libraries required for visualization and exploration of data, model training and testing, etc in this project.
# Import 'accuracy_score'
from sklearn.metrics import accuracy_score
def performance_metric(y_true, y_predict):
"""Calculates and returns the performance score between
true and predicted values based on the metric chosen."""
# Calculate the performance score between 'y_true' and 'y_predict'
score = accuracy_score(y_true, y_predict)
# Return the score
return score
# Import 'make_scorer', 'SVC', 'GridSearchCV', and 'ShuffleSplit'
from sklearn.metrics import make_scorer
from sklearn.svm import SVC
from sklearn.model_selection import GridSearchCV
from sklearn.model_selection import ShuffleSplit
def fit_model(X, y):
"""Performs grid search over the 'max_depth' parameter for a
decision tree regressor trained on the input data [X, y]."""
# Create cross-validation sets from the training data
cv_sets = ShuffleSplit(n_splits=10, test_size=0.20, random_state=0)
# Create a linear discriminant analysis object
clf = SVC(kernel="rbf", class_weight="balanced")
# Create a dictionary for the parameters
params = {
"C": [1e3, 5e3, 1e4, 5e4, 1e5],
"gamma": [0.0001, 0.0005, 0.001, 0.005, 0.01, 0.1],
}
# Transform 'performance_metric' into a scoring function using 'make_scorer'
scoring_fnc = make_scorer(performance_metric)
# Create the grid search cv object --> GridSearchCV()
# Make sure to include the right parameters in the object:
# (estimator, param_grid, scoring, cv) which have values 'clf', 'params', 'scoring_fnc', and 'cv_sets' respectively.
grid = GridSearchCV(clf, params, scoring=scoring_fnc, cv=cv_sets)
# Fit the grid search object to the data to compute the optimal model
grid = grid.fit(X, y)
# Return the optimal model after fitting the data
return grid.best_estimator_
# Import PCA
from sklearn.decomposition import PCA
def pca_projection(good_data, n_components):
# Apply PCA by fitting the good data with only two dimensions
pca = PCA(n_components=n_components).fit(good_data)
# Transform the good data using the PCA fit above
reduced_data = pca.transform(good_data)
# Create a DataFrame for the reduced data
columns = ["Dimension 1", "Dimension 2", "Dimension 3"]
return pd.DataFrame(reduced_data, columns=columns[:n_components]), pca
# ## Exploring the Data
# In the next section, we will load the data and split it into features and target label. The data consists of 70 features and one target label the `state`. The feature vector for each data entry is about the five frequency bands values for each channel from the 14, concatenated in one vector performing the features. The state classes are represented in a numerical form, where: ***1*** is `Focused`, ***2*** is `De-Focused` and **3** is `Drowsy`.
# The dataset contains **3600** data entries collected from one subject with sampling rate of **1** second over a complete hour. Each third of the data entries belong to one mental state, where each state class has **1200** samples. We can take more samples to increase our dataset size, however **1200** samples are enough to reach a satisfactory performance score.
# The next code cell will display the first fifteen entries, which belong to the third mental state **"Drowsy"**.
# Allows the use of display() for DataFrames
from IPython.display import display
# Read the data.
raw_data = pd.read_csv("../input/eeg-subject/eeg_subject.csv")
# Split the data into features and target label
target_raw = raw_data[raw_data.columns[-1]]
features_raw = raw_data.drop(raw_data.columns[-1], axis=1)
# Print data shape.
print("The shape of the data: {}".format(raw_data.shape))
# Success - Display the first fifteen records
display(raw_data.head(n=15))
# ----
# ## Preparing the Data
# Before data can be used as input for machine learning algorithms, it often must be cleaned, formatted, and restructured — this is typically known as **preprocessing**. Fortunately, for this dataset, there are no invalid or missing entries we must deal with, however, there are some qualities about certain features that must be adjusted. This preprocessing can help tremendously with the outcome and predictive power of nearly all learning algorithms.
# ### Transforming Skewed Continuous Features
# A dataset may sometimes contain at least one feature whose values tend to lie near a single number, but will also have a non-trivial number of vastly larger or smaller values than that single number. Algorithms can be sensitive to such distributions of values and can underperform if the range is not properly normalized. With our dataset for example, feature such as: `'delta ch(2)'`, fit this description.
# The code cell below will plot a histogram of this feature. Note the range of the values present and how they are distributed.
# Visualize skewed continuous features of original data
dg.distribution(raw_data, 6)
# For highly-skewed feature distributions such as `'delta ch(2)'`, it is common practice to apply a logarithmic transformation on the data so that the very large and very small values do not negatively affect the performance of a learning algorithm. Using a logarithmic transformation significantly reduces the range of values caused by outliers. Care must be taken when applying this transformation however: The logarithm of `0` is undefined, so we must translate the values by a small amount above `0` to apply the the logarithm successfully.
# The code cell below will perform a transformation on the data and visualize the results. Again, note the range of values and how they are distributed.
# Log-transform the skewed features
features_log_transformed = features_raw.apply(lambda x: np.log(x + 1))
# Visualize the new log distributions
dg.distribution(features_log_transformed, 6, transformed=True)
# ### Outlier Detection
# Detecting outliers in the data is extremely important in the data preprocessing step of any analysis. The presence of outliers can often skew results which take into consideration these data points. There are many "rules of thumb" for what constitutes an outlier in a dataset. Here, we will use [Tukey's Method for identfying outliers](https://www.stat.cmu.edu/~cshalizi/statcomp/13/labs/05/lab-05.pdf): An **outlier step** is calculated as 1.5 times the interquartile range (IQR). A data point with a feature that is beyond an outlier step outside of the IQR for that feature is considered abnormal.
# Calculate Q1 (25th quantile of the data) for all features.
Q1 = features_log_transformed.quantile(0.25)
# Calculate Q3 (75th quantile of the data) for all features.
Q3 = features_log_transformed.quantile(0.75)
# Use the interquartile range to calculate an outlier step (1.5 times the interquartile range).
IQR = Q3 - Q1
step = 1.5 * IQR
# Remove the outliers from the dataset.
features_log_transformed_out = features_log_transformed[
~(
(features_log_transformed < (Q1 - step))
| (features_log_transformed > (Q3 + step))
).any(axis=1)
]
# Join the features and the target after removing outliers.
preprocessed_data_out = features_log_transformed_out.join(target_raw)
target_raw_out = preprocessed_data_out[preprocessed_data_out.columns[-1]]
# Print data shape after removing outliers.
print(
"The shape of the data after removing outliers: {}".format(
preprocessed_data_out.shape
)
)
# Success - Display the first ten records
display(preprocessed_data_out.head(n=10))
# ### Normalizing Numerical Features
# In addition to performing transformations on features that are highly skewed, it is often good practice to perform some type of scaling on numerical features. Applying a scaling to the data does not change the shape of each feature's distribution (such as `'delta ch(2)'` above); however, normalization ensures that each feature is treated equally when applying supervised learners. Note that once scaling is applied, observing the data in its raw form will no longer have the same original meaning, as exampled below.
# Run the code cell below to normalize each numerical feature. We will use [`sklearn.preprocessing.MinMaxScaler`](http://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.MinMaxScaler.html) for this.
# Import sklearn.preprocessing.StandardScaler
from sklearn.preprocessing import MinMaxScaler
# Initialize a scaler, then apply it to the features
scaler = MinMaxScaler() # default=(0, 1)
features_log_minmax_transform_out = pd.DataFrame(
scaler.fit_transform(features_log_transformed_out), columns=features_raw.columns
)
# Show an example of a record with scaling applied
display(features_log_minmax_transform_out.head())
# ----
# ## Developing a Model
# In this section of the project, we will develop the tools and techniques necessary for a model to make a prediction. Being able to make accurate evaluations of each model's performance through the use of these tools and techniques helps to greatly reinforce the confidence in the model's predictions.
# ### Shuffle and Split Data
# Now that the data has been scaled to a more normal distribution and has had any necessary outliers removed. As always, we will now split the data (both features and their labels) into training and test sets. 80% of the data will be used for training and 20% for testing.
# Import 'train_test_split'
from sklearn.model_selection import train_test_split
# Assign preprocessed data frame to 'good_data'.
good_data = features_log_minmax_transform_out
# Assign the features to the variable Bands, and the labels to the variable state.
Bands = np.array(good_data)
state = np.array(target_raw_out)
# Shuffle and split the data into training and testing subsets.
X_train, X_test, y_train, y_test = train_test_split(
Bands, state, test_size=0.2, random_state=42, shuffle=True
)
# Success
print("Training and testing split was successful.")
# ### PCA Transformation
# We can now apply PCA to the data for dimensionality reduction, to fit the relatively small size of the data compared to its features number, and also to discover which dimensions about the data best maximize the variance of features involved. In addition to finding these dimensions, PCA will also report the *explained variance ratio* of each dimension — how much variance within the data is explained by that dimension alone. Note that a component (dimension) from PCA can be considered a new "feature" of the space, however it is a composition of the original features present in the data.
# Next we will use the PCA to visualize the data in **two** and **three** dimension.
# Project data on two dimensions
reduced_data, pca = pca_projection(good_data, 2)
# Create a biplot
dg.biplot(good_data, reduced_data, target_raw_out, pca)
# Project data on three dimensions
reduced_data, pca = pca_projection(good_data, 3)
# Enabling the `widget` backend.
# Create a triplot
dg.triplot(reduced_data, target_raw_out, pca)
# In the next code cell, we will plot the Cumulative Summation of the Explained Variance, to help us decide the number of principal components to be used in the transformation.
# Fitting the PCA algorithm with our training data.
pca = PCA().fit(X_train)
# Plotting the Cumulative Summation of the Explained Variance.
plt.figure(figsize=(14, 7))
plt.plot(np.cumsum(pca.explained_variance_ratio_))
plt.xlabel("Number of Components")
plt.ylabel("Variance (%)") # For each component.
plt.title("Pulsar Dataset Explained Variance")
plt.show()
# This plot tells us that selecting **20** components we can preserve something around 89% or 90% of the total variance of the data. It makes sense, we’ll not use 100% of our variance, because it denotes all components, and we want only the principal ones.
# With this information in our hands, we can implement the PCA for **20** best components by using the next snippet of code.
# Import time
from time import time
# From the Explained Variance graph.
n_components = 20
print(
"Extracting the top %d eigenfaces from %d faces" % (n_components, X_train.shape[0])
)
t0 = time()
# Create an instance of PCA, initializing with n_components=n_components and whiten=True
pca = PCA(n_components=n_components)
# Pass the training dataset (X_train) to pca's 'fit()' method
pca = pca.fit(X_train)
X_train_pca = pca.transform(X_train)
X_test_pca = pca.transform(X_test)
print("Explained variance ratios:", pca.explained_variance_ratio_ * 100)
print(
"Cumulative explained variance ratio: %0.3f %%"
% (sum(pca.explained_variance_ratio_) * 100)
)
print("done in %0.3fs" % (time() - t0))
# ### Fitting The Model
# Our final implementation requires that we bring everything together and train a model using the **support vector machine algorithm**. To ensure that we are producing an optimized model, we will train the model using the grid search technique to optimize the `'gamma'` and `'C'` parameters for the **svm classifier**. [SVM Documentation](https://scikit-learn.org/stable/modules/generated/sklearn.svm.SVC.html)
# In addition, the implementation is using `ShuffleSplit()` for an alternative form of cross-validation (see the `'cv_sets'` variable). While it is not the K-Fold cross-validation technique, this type of cross-validation technique is just as useful!. The `ShuffleSplit()` implementation will create 10 (`'n_splits'`) shuffled sets, and for each shuffle, 20% (`'test_size'`) of the data will be used as the *validation set*.
# Fit the training data to the model using grid search
model = fit_model(X_train_pca, y_train)
# Produce the value for 'gamma' and 'C'
print(
"Parameter 'gamma' is {} for the optimal model.".format(model.get_params()["gamma"])
)
print("Parameter 'C' is {} for the optimal model.".format(model.get_params()["C"]))
# ### Making Predictions
# Once the model has been trained on a given set of data, it can now be used to make predictions on new sets of input data. In the case of a **support vector machine**, the model has learned **how the frequency bands values relate to the mental state of the driver**, and can respond with a detection of the current **mental state**. We can use these predictions to gain information about data where the value of the target variable is unknown — such as data the model was not trained on (i.e testing data or future readings of the driver's brain signals).
# Make predictions. Store them in the variable y_pred.
y_pred = model.predict(X_test_pca)
# Label states class.
states_class = ["Focused", "De-Focused", "Drowsy"]
# Show predictions
for i, state in enumerate(y_pred[0:20]):
print(
"Predicted mental state for test {}'s bands: {}".format(
i + 1, states_class[state - 1]
)
)
# ## Final Model Evaluation
# In this final section, we will run some evaluation metrics to check the performance of the optimized model on the testing data.
# In the next code cell, we will calculate the `'F1-Score'` of the model on testing data perdictions.
# Import 'f1_score'
from sklearn.metrics import f1_score
# Calculate the f1 score and assign it to the variable score.
score = f1_score(y_test, y_pred, average="micro")
# Print score.
print("F1 score: %0.1f %%" % (score * 100))
# Run the next cell, to calculate and plot the confusion matrix of the **3** state classes.
# Import 'confusion_matrix' and 'plot_confusion_matrix' from 'mlxtend'.
from mlxtend.evaluate import confusion_matrix
from mlxtend.plotting import plot_confusion_matrix
# Calculate the confusion matrix and assign it to the variable matrix.
matrix = confusion_matrix(y_test, y_pred)
# Plot the confusion matrix.
fig, ax = plot_confusion_matrix(matrix)
plt.show()
# Finally, we make a classification report to get a detailed performance evaluation, with scores for the model on each **mental state** class.
# Import 'classification_report'
from sklearn.metrics import classification_report
# Calculate the classification report and assign it to the variable report.
report = classification_report(y_test, y_pred)
# Print the classification report.
print(report)
# Run the code cell below to plot the learning curve for the SVM model.
# Plot learning curve for the SVM model
dg.Svm_Learning_Curve(good_data, target_raw_out, model)
# Save model, pca transformer and feature scaler.
# Import 'pickle'
import pickle
# Save the model to disk.
pickle.dump(model, open("model.sav", "wb"))
# Save PCA transformation.
pickle.dump(pca, open("pca.pkl", "wb"))
# Save scaler mapping values.
pickle.dump(scaler, open("scaler.sav", "wb"))
|
# # GPT (Generative Pre-training)
# **GPT** is the earliest version of the **ChatGPT** which uses currently **GPT 3.5** as angine. **GPT** is named as **openai-gpt** and so-called **GPT-1**.
from transformers import pipeline, set_seed
# ## Sentiment Analysis
set_seed(42)
model_name = "openai-gpt"
# task_pipe = pipeline(‘task_name’,model =’model_name’, tokenizer )
task_pipe = pipeline(
"sentiment-analysis", model=model_name
) # sentiment-analysis, text-generation
task_pipe(["I love you", "I hate you"])
# Following code uses the default model ("**distilbert-base-uncased**") for sentiment analysis to analyze the list of texts data. (https://huggingface.co/distilbert-base-uncased-finetuned-sst-2-english)
#!pip install -q transformers
from transformers import pipeline
sentiment_pipeline = pipeline("sentiment-analysis")
data = ["I love you", "I hate you"]
sentiment_pipeline(data)
# ## Text generation
model_name = "openai-gpt"
task_pipe = pipeline(
"text-generation", model=model_name
) # sentiment-analysis, text-generation
task_pipe("Hi, I like deep learning", max_length=30, num_return_sequences=5)
# ## Question Answering
context = r"""
ChatGPT[a] is an artificial intelligence (AI) chatbot developed by OpenAI and released in November 2022.
It is built on top of OpenAI's GPT-3.5 and GPT-4 families of large language models (LLMs) and
has been fine-tuned (an approach to transfer learning) using both supervised and reinforcement learning techniques.
ChatGPT launched as a prototype on November 30, 2022 and garnered attention for its detailed responses
and articulate answers across many domains of knowledge.[3] Its uneven factual accuracy,
however, has been identified as a significant drawback.[4] In 2023, following the release of ChatGPT,
OpenAI's valuation was estimated at US$29 billion.[5]
The original release of ChatGPT was based on GPT-3.5. A version based on GPT-4,
the newest OpenAI model, was released on March 14, 2023, and is available for paid subscribers on a limited basis."""
# Calling pipeline:
task_pipe = pipeline(
"question-answering"
) # it uses (https://huggingface.co/distilbert-base-cased-distilled-squad) model
result = task_pipe(question="Who developed ChatGPT?", context=context)
print(result)
print(result["answer"])
result = task_pipe(question="When did ChatGPT Launched?", context=context)
print(result)
print(result["answer"])
result = task_pipe(question="Which techniques does ChatGPT use?", context=context)
print(result)
print(result["answer"])
# ## Text Summarization
summarizer = pipeline(
"summarization", model="t5-base", tokenizer="t5-base", framework="tf"
)
summary = summarizer(context, max_length=130, min_length=60)
print(summary)
# ## Language Translation
from transformers import AutoTokenizer, AutoModelForSeq2SeqLM, Pipeline
tokenizer = AutoTokenizer.from_pretrained("Helsinki-NLP/opus-mt-en-ru")
model = AutoModelForSeq2SeqLM.from_pretrained("Helsinki-NLP/opus-mt-en-ru")
# model for "Helsinki-NLP/opus-mt-en-ru" English to Russian translation.
translator = pipeline("translation", model=model, tokenizer=tokenizer)
translated = translator("How are you?")[0].get("translation_text")
print(translated)
|
# The dataset was created using the [wikihowunofficialapi](https://pypi.org/project/wikihowunofficialapi/). So, we can use it to read and use the data.
import sys
import pandas as pd
import wikihowunofficialapi as wha
data = pd.read_pickle("/kaggle/input/wikihow-raw-data/wikihow.pickle")
data.head()
article = data.loc[0, "info"]
print(article.url) # Print Article's URL
print(article.title) # Print Article's Title
print(article.intro) # Print Article's Introduction
print(article.n_methods) # Print number of methods in an Article
print(article.methods) # Print a list of methods in an Article
print(article.num_votes) # Print number of votes given to an Article
print(article.percent_helpful) # Print percentage of helpful votes given to an Article
print(article.is_expert) # Print True if the Article is written by an expert
print(article.last_updated) # Print date when the Article was last updated
print(article.views) # Print the number of views recieved by Article
print(article.co_authors) # Print the number of co-authors of an Article
print(article.references) # Print the number of references in an Article
print(article.summary) # Print Article's summary
print(article.warnings) # Print Article's warnings
print(article.tips) # Print Article's tips
first_method = article.methods[0]
first_step = first_method.steps[0]
print(first_step) # Print Article's first step of the first method
print(first_step.title) # Print the title of Article's first step of the first method
print(
first_step.description
) # Print the description of Article's first step of the first method
|
import pandas as pd
import numpy as np
from sklearn.cluster import KMeans
import matplotlib.pyplot as plt
from sklearn.preprocessing import StandardScaler
# Customer clustering
# Dataset
url = "https://raw.githubusercontent.com/anvarnarz/praktikum_datasets/main/customer_segmentation.csv"
df = pd.read_csv(url)
df.head()
# Data cleaning
# **The Address** column is *text*. We cannot use this column for distance calculation, so we will **discard** this column.
df.drop("Address", axis=1, inplace=True)
df.head()
# Normalization
# We normalize all columns except the **Customer Id** column.
X = df.values[:, 1:]
X = np.nan_to_num(X)
norm_data = StandardScaler().fit_transform(X)
norm_data
# Model
k = 3
k_means = KMeans(n_clusters=k)
k_means.fit(norm_data)
print(k_means.labels_)
df["cluter"] = k_means.labels_
df.head()
# We can get information about clusters by finding the centroid of each cluster. For this, we use groupby and mean functions
df.groupby("cluter").mean()
# What can be inferred about the customers in each cluster?
# For example, if we look at **age**, cluster *0* is older youth (43+), cluster *1* is middle age (33+), cluster *2* is young (up to 31).
# The columns that clearly **distinguish** each cluster are *Income* and *Year Employed*.
# So we can define each cluster as:
# * Cluster 0 - Young, less experienced and lower income customers.
# * Cluster 1 - Older, long-term, high-income customers.
# * Cluster 2 - Middle-aged, 7+ years of experience, low-income customers.
area = np.pi * (X[:, 1]) ** 2
plt.scatter(X[:, 0], X[:, 3], s=area, c=k_means.labels_.astype(float), alpha=0.5)
plt.xlabel("Age", fontsize=18)
plt.ylabel("Income", fontsize=16)
plt.show()
from mpl_toolkits.mplot3d import Axes3D
fig = plt.figure(1, figsize=(8, 6))
plt.clf()
ax = Axes3D(fig, rect=[0, 0, 0.95, 1], elev=48, azim=120)
plt.cla()
# plt.ylabel('Age', fontsize=18)
# plt.xlabel('Income', fontsize=16)
# plt.zlabel('Education', fontsize=16)
ax.set_xlabel("Years Employed")
ax.set_ylabel("Age")
ax.set_zlabel("Income")
ax.scatter(X[:, 2], X[:, 0], X[:, 3], c=k_means.labels_.astype(float))
|
# # Important moment
# WaRP Segmentation. Unsupervised segmentation using CAM methods.
# Serviceability was checked in Google Colab.
# Set the resolution on which the classification model used was trained.
resolution_model = 224
# Download the weights of the classification model to be used as an example.
# # Connect to Drive and prepare Data
# You should use gpu to get results quickly.
# You need to have the following structure so that you don't have to change anything in the code for working with data.
from google.colab import drive
drive.mount("/content/gdrive")
# move data
# rename
# The following cell should be executed if you want to save the result to Google drive.
# # Download the libraries and import them
import torch
import os
import cv2
import numpy as np
import sys
import json
from collections import defaultdict
from torch.utils.data import Dataset, DataLoader
from torch import nn
from torch.nn import functional as F
from torchvision.models import resnet18 as resnet18
from torchvision import models, transforms
import matplotlib.pyplot as plt
from google.colab.patches import cv2_imshow
import timm
from PIL import Image
import random
import pickle
import io
from torch.autograd import Variable
# # CAMERAS
# In the line self.inputResolutions = list(range(**224**, 1000, 100)) - change the underlined number to the one that the classification model was trained for (must match what is set in "Important moment").
# Copy the code below and paste it into the file that is located on the path: /content/CAMERAS.py
import copy
import torch
from torch.nn import functional as F
class CAMERAS:
def __init__(self, model, targetLayerName, inputResolutions=None):
self.model = model
self.inputResolutions = inputResolutions
if self.inputResolutions is None:
self.inputResolutions = list(range(224, 1000, 100)) # Update this line
self.classDict = {}
self.probsDict = {}
self.featureDict = {}
self.gradientsDict = {}
self.targetLayerName = targetLayerName
def _recordActivationsAndGradients(
self, inputResolution, image, classOfInterest=None
):
def forward_hook(module, input, output):
self.featureDict[inputResolution] = copy.deepcopy(
output.clone().detach().cpu()
)
def backward_hook(module, grad_input, grad_output):
self.gradientsDict[inputResolution] = copy.deepcopy(
grad_output[0].clone().detach().cpu()
)
for name, module in self.model.named_modules():
if name == self.targetLayerName:
forwardHandle = module.register_forward_hook(forward_hook)
backwardHandle = module.register_backward_hook(backward_hook)
logits = self.model(image)
softMaxScore = F.softmax(logits, dim=1)
probs, classes = softMaxScore.sort(dim=1, descending=True)
if classOfInterest is None:
ids = classes[:, [0]]
else:
ids = torch.tensor(classOfInterest).unsqueeze(dim=0).unsqueeze(dim=0).cuda()
self.classDict[inputResolution] = ids.clone().detach().item()
self.probsDict[inputResolution] = probs[0, 0].clone().detach().item()
one_hot = torch.zeros_like(logits)
one_hot.scatter_(1, ids, 1.0)
self.model.zero_grad()
logits.backward(gradient=one_hot, retain_graph=False)
forwardHandle.remove()
backwardHandle.remove()
del forward_hook
del backward_hook
def _estimateSaliencyMap(self, classOfInterest):
saveResolution = self.inputResolutions[0]
groundTruthClass = self.classDict[saveResolution]
meanScaledFeatures = None
meanScaledGradients = None
count = 0
for resolution in self.inputResolutions:
if (
groundTruthClass == self.classDict[resolution]
or self.classDict[resolution] == classOfInterest
):
count += 1
upSampledFeatures = F.interpolate(
self.featureDict[resolution].cuda(),
(saveResolution, saveResolution),
mode="bilinear",
align_corners=False,
)
upSampledGradients = F.interpolate(
self.gradientsDict[resolution].cuda(),
(saveResolution, saveResolution),
mode="bilinear",
align_corners=False,
)
if meanScaledFeatures is None:
meanScaledFeatures = upSampledFeatures
else:
meanScaledFeatures += upSampledFeatures
if meanScaledGradients is None:
meanScaledGradients = upSampledGradients
else:
meanScaledGradients += upSampledGradients
meanScaledFeatures /= count
meanScaledGradients /= count
fmaps = meanScaledFeatures
grads = meanScaledGradients
saliencyMap = torch.mul(fmaps, grads).sum(dim=1, keepdim=True)
saliencyMap = F.relu(saliencyMap)
B, C, H, W = saliencyMap.shape
saliencyMap = saliencyMap.view(B, -1)
saliencyMap -= saliencyMap.min(dim=1, keepdim=True)[0]
saliencyMap /= saliencyMap.max(dim=1, keepdim=True)[0]
saliencyMap = saliencyMap.view(B, C, H, W)
saliencyMap = torch.squeeze(torch.squeeze(saliencyMap, dim=0), dim=0)
return saliencyMap
def run(self, image, classOfInterest=None):
for index, inputResolution in enumerate(self.inputResolutions):
if index == 0:
upSampledImage = image.cuda()
else:
upSampledImage = F.interpolate(
image,
(inputResolution, inputResolution),
mode="bicubic",
align_corners=False,
).cuda()
self._recordActivationsAndGradients(
inputResolution, upSampledImage, classOfInterest=classOfInterest
)
saliencyMap = self._estimateSaliencyMap(classOfInterest=classOfInterest)
return saliencyMap, self.classDict, self.probsDict
# The next cell does not need to be copied, just run
import cv2
import torch
import torchvision.models as models
import matplotlib.cm as cm
from CAMERAS import CAMERAS
from torchvision import transforms
import numpy as np
normalizeTransform = transforms.Normalize(
mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]
)
normalizeImageTransform = transforms.Compose(
[transforms.ToTensor(), normalizeTransform]
)
def loadImage(imagePath, imageSize):
rawImage = cv2.imread(imagePath)
rawImage = cv2.resize(
rawImage, (resolution_model,) * 2, interpolation=cv2.INTER_LINEAR
)
rawImage = cv2.resize(rawImage, (imageSize,) * 2, interpolation=cv2.INTER_LINEAR)
image = normalizeImageTransform(rawImage[..., ::-1].copy())
return image, rawImage
def saveMapWithColorMap(filename, map, image):
cmap = cv2.applyColorMap(
np.uint8(255 * map.detach().numpy().squeeze()), cv2.COLORMAP_JET
)
map = (cmap.astype(np.float) + image.astype(np.float)) / 2
cv2.imwrite(filename, np.uint8(map))
def computeAndSaveMaps(model, image_name):
cameras = CAMERAS(model, targetLayerName="layer4") # classic CAMERAS
image, rawImage = loadImage(image_name, imageSize=resolution_model)
image = torch.unsqueeze(image, dim=0)
saliencyMap, predicted_class, probs_predict = cameras.run(image)
saliencyMap = saliencyMap.cpu()
return saliencyMap
# # Implementation of all CAM methods used in the article, except CAMERAS and CCAM
# ## mGrad-CAM
# The next cells does not need to be copied, just run
from pytorch_grad_cam.utils.image import (
show_cam_on_image,
deprocess_image,
preprocess_image,
)
from pytorch_grad_cam.utils.model_targets import ClassifierOutputTarget
import numpy as np
import torch
import ttach as tta
from typing import Callable, List, Tuple
from pytorch_grad_cam.activations_and_gradients import ActivationsAndGradients
from pytorch_grad_cam.utils.svd_on_activations import get_2d_projection
from pytorch_grad_cam.utils.image import scale_cam_image
from pytorch_grad_cam.utils.model_targets import ClassifierOutputTarget
class BaseCAM:
def __init__(
self,
model: torch.nn.Module,
target_layers: List[torch.nn.Module],
use_cuda: bool = False,
reshape_transform: Callable = None,
compute_input_gradient: bool = False,
uses_gradients: bool = True,
) -> None:
self.model = model.eval()
self.target_layers = target_layers
self.cuda = use_cuda
if self.cuda:
self.model = model.cuda()
self.reshape_transform = reshape_transform
self.compute_input_gradient = compute_input_gradient
self.uses_gradients = uses_gradients
self.activations_and_grads = ActivationsAndGradients(
self.model, target_layers, reshape_transform
)
""" Get a vector of weights for every channel in the target layer.
Methods that return weights channels,
will typically need to only implement this function. """
def get_cam_weights(
self,
input_tensor: torch.Tensor,
target_layers: List[torch.nn.Module],
targets: List[torch.nn.Module],
activations: torch.Tensor,
grads: torch.Tensor,
) -> np.ndarray:
raise Exception("Not Implemented")
def get_cam_image(
self,
input_tensor: torch.Tensor,
target_layer: torch.nn.Module,
targets: List[torch.nn.Module],
activations: torch.Tensor,
grads: torch.Tensor,
eigen_smooth: bool = False,
) -> np.ndarray:
weights = self.get_cam_weights(
input_tensor, target_layer, targets, activations, grads
)
weighted_activations = weights * activations
if eigen_smooth:
cam = get_2d_projection(weighted_activations)
else:
cam = weighted_activations.sum(axis=1)
return cam
def forward(
self,
input_tensor: torch.Tensor,
targets: List[torch.nn.Module],
eigen_smooth: bool = False,
) -> np.ndarray:
if self.cuda:
input_tensor = input_tensor.cuda()
if self.compute_input_gradient:
input_tensor = torch.autograd.Variable(input_tensor, requires_grad=True)
outputs = self.activations_and_grads(input_tensor)
if targets is None:
target_categories = np.argmax(outputs.cpu().data.numpy(), axis=-1)
targets = [
ClassifierOutputTarget(category) for category in target_categories
]
if self.uses_gradients:
self.model.zero_grad()
loss = sum([target(output) for target, output in zip(targets, outputs)])
loss.backward(retain_graph=True)
# In most of the saliency attribution papers, the saliency is
# computed with a single target layer.
# Commonly it is the last convolutional layer.
# Here we support passing a list with multiple target layers.
# It will compute the saliency image for every image,
# and then aggregate them (with a default mean aggregation).
# This gives you more flexibility in case you just want to
# use all conv layers for example, all Batchnorm layers,
# or something else.
cam_per_layer = self.compute_cam_per_layer(input_tensor, targets, eigen_smooth)
return self.aggregate_multi_layers(cam_per_layer)
def get_target_width_height(self, input_tensor: torch.Tensor) -> Tuple[int, int]:
width, height = input_tensor.size(-1), input_tensor.size(-2)
return width, height
def compute_cam_per_layer(
self,
input_tensor: torch.Tensor,
targets: List[torch.nn.Module],
eigen_smooth: bool,
) -> np.ndarray:
activations_list = [
a.cpu().data.numpy() for a in self.activations_and_grads.activations
]
grads_list = [
g.cpu().data.numpy() for g in self.activations_and_grads.gradients
]
target_size = self.get_target_width_height(input_tensor)
cam_per_target_layer = []
# Loop over the saliency image from every layer
for i in range(len(self.target_layers)):
target_layer = self.target_layers[i]
layer_activations = None
layer_grads = None
if i < len(activations_list):
layer_activations = activations_list[i]
if i < len(grads_list):
layer_grads = grads_list[i]
cam = self.get_cam_image(
input_tensor,
target_layer,
targets,
layer_activations,
layer_grads,
eigen_smooth,
)
cam = np.maximum(cam, 0)
scaled = scale_cam_image(cam, target_size)
cam_per_target_layer.append(scaled[:, None, :])
return cam_per_target_layer
def aggregate_multi_layers(self, cam_per_target_layer: np.ndarray) -> np.ndarray:
cam_per_target_layer = np.concatenate(cam_per_target_layer, axis=1)
cam_per_target_layer = np.maximum(cam_per_target_layer, 0)
result = np.mean(cam_per_target_layer, axis=1)
return scale_cam_image(result)
def forward_augmentation_smoothing(
self,
input_tensor: torch.Tensor,
targets: List[torch.nn.Module],
eigen_smooth: bool = False,
) -> np.ndarray:
transforms = tta.Compose(
[
tta.HorizontalFlip(),
tta.Multiply(factors=[0.9, 1, 1.1]),
]
)
cams = []
for transform in transforms:
augmented_tensor = transform.augment_image(input_tensor)
cam = self.forward(augmented_tensor, targets, eigen_smooth)
# The ttach library expects a tensor of size BxCxHxW
cam = cam[:, None, :, :]
cam = torch.from_numpy(cam)
cam = transform.deaugment_mask(cam)
# Back to numpy float32, HxW
cam = cam.numpy()
cam = cam[:, 0, :, :]
cams.append(cam)
cam = np.mean(np.float32(cams), axis=0)
return cam
def __call__(
self,
input_tensor: torch.Tensor,
targets: List[torch.nn.Module] = None,
aug_smooth: bool = False,
eigen_smooth: bool = False,
) -> np.ndarray:
# Smooth the CAM result with test time augmentation
if aug_smooth is True:
return self.forward_augmentation_smoothing(
input_tensor, targets, eigen_smooth
)
return self.forward(input_tensor, targets, eigen_smooth)
def __del__(self):
self.activations_and_grads.release()
def __enter__(self):
return self
def __exit__(self, exc_type, exc_value, exc_tb):
self.activations_and_grads.release()
if isinstance(exc_value, IndexError):
# Handle IndexError here...
print(
f"An exception occurred in CAM with block: {exc_type}. Message: {exc_value}"
)
return True
import numpy as np
class GradCAM(BaseCAM):
def __init__(self, model, target_layers, use_cuda=False, reshape_transform=None):
super(GradCAM, self).__init__(model, target_layers, use_cuda, reshape_transform)
def get_cam_weights(
self, input_tensor, target_layer, target_category, activations, grads
):
return grads
# ## LayerCAM, Grad-CAM, FullGrad and others
# The next cell does not need to be copied, just run
from pytorch_grad_cam import (
GradCAM,
ScoreCAM,
GradCAMPlusPlus,
AblationCAM,
XGradCAM,
EigenCAM,
LayerCAM,
FullGrad,
)
from pytorch_grad_cam.utils.image import (
show_cam_on_image,
deprocess_image,
preprocess_image,
)
from pytorch_grad_cam.utils.model_targets import ClassifierOutputTarget
# # Unsupervised segmentation pipeline
# CAMERAS:
# To use the CAMERAS method, you need to follow all the instructions from the text and activate all the cells in the section "CAMERAS".
# ---
# Grad-CAM, mGrad-CAM and etc:
# To use mGrad-CAM -> first go to the section "Implementation of all CAM methods used in the article, except CAMERAS and CCAM", go to the subsection "mGrad-CAM" and activate all the cells, then go back to this section and check in the function "get_saliency_map" set GradCAM.
# ---
# To use other methods (the exceptions are CAMERAS and mGrad-CAM) -> first go to the section "Implementation of all CAM methods used in the article, except CAMERAS and CCAM", go to the subsection "LayerCAM, Grad-CAM, FullGrad and others" execute a single cell, then return to this section and set in the function "get_saliency_map" which CAM will we use.
# ---
# If you change models every time, for example in this sequence::
# 1. Grad-CAM;
# 2. mGrad-CAM;
# 3. Layer-CAM.
# Then be sure to do what is described above every time and for full confidence you can restart the environment.
# If there is no mGrad-CAM in the sequence, then it is enough to do it once and then change the CAM method in get_saliency_map.
# model.layerN[-1] - the last layer in the block;
# model.layerN[-2] - the first layer in the block.
def get_saliency_map(model, image_name):
# From which layers will the class activation maps be taken
target_layers = [model.layer3[-1], model.layer4[-2]]
normalize = transforms.Normalize(
mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]
)
preprocess = transforms.Compose(
[
transforms.Resize((resolution_model, resolution_model)),
transforms.ToTensor(),
normalize,
]
)
# load test image
img_pil = Image.open(image_name).convert("RGB")
img_tensor = preprocess(img_pil)
img_variable = Variable(img_tensor.unsqueeze(0))
input_tensor = img_variable.cuda()
# This specifies which CAM method will be used
cam = GradCAM(model=model, target_layers=target_layers, use_cuda=True)
grayscale_cam = cam(input_tensor=input_tensor)
# In this example grayscale_cam has only one image in the batch:
grayscale_cam = grayscale_cam[0, :]
return grayscale_cam
def iou_pytorch(outputs: torch.Tensor, labels: torch.Tensor):
outputs = outputs.int()
labels = labels.int()
SMOOTH = 1e-8
intersection = (
(outputs & labels).float().sum((1, 2))
) # Will be zero if Truth=0 or Prediction=0
union = (outputs | labels).float().sum((1, 2)) # Will be zero if both are 0
iou = (torch.sum(intersection) + SMOOTH) / (
torch.sum(union) + SMOOTH
) # We smooth(epsilon) our devision to avoid 0/0
return iou
def get_masks(
saliency_map, path_to_image, coefficient=10, resolution=224, visualization=False
):
if not isinstance(saliency_map, np.ndarray):
saliency_map = cv2.applyColorMap(
np.uint8(255 * saliency_map.detach().numpy().squeeze()), cv2.COLORMAP_JET
)
else:
saliency_map = cv2.applyColorMap(np.uint8(255 * saliency_map), cv2.COLORMAP_JET)
hsv = cv2.cvtColor(saliency_map, cv2.COLOR_BGR2HSV)
# Here we define the blue color range in HSV
lower_blue = np.array([coefficient, 50, 50])
upper_blue = np.array([130, 255, 255])
# This method creates a blue mask of the objects found in the frame
mask = cv2.inRange(hsv, lower_blue, upper_blue)
# Inverting the mask
for i in range(len(mask)):
for j in range(len(mask[i])):
if mask[i][j] == 255:
mask[i][j] = 0
else:
mask[i][j] = 255
if visualization == True:
image = cv2.imread(path_to_image)
img = image.copy()
img[mask == 255] = 0
color_channeled_image = cv2.cvtColor(mask, cv2.COLOR_GRAY2BGR)
result = color_channeled_image * 0.4 + img * 0.7
mask_to_seg = saliency_map * 0.5 + image * 0.5
fig, (ax1, ax2) = plt.subplots(ncols=2, figsize=(16, 16))
ax1.set_title("Original picture")
ax2.set_title("The mask was created using CAM methods")
ax1.axis("off")
ax2.axis("off")
_ = ax1.imshow(cv2.cvtColor(mask_to_seg.astype("uint8"), cv2.COLOR_BGR2RGB))
_ = ax2.imshow(cv2.cvtColor(result.astype("uint8"), cv2.COLOR_BGR2RGB))
file_name = path_to_image.split("/")[-1]
# Second argument - gives Grey Scale Image
real_mask = cv2.imread(
"data/mask/"
+ path_to_image.split("/")[2]
+ "/"
+ file_name.split(".")[0]
+ ".png",
0,
)
real_mask = cv2.resize(
real_mask, (resolution, resolution), interpolation=cv2.INTER_NEAREST
)
for i in range(len(real_mask)):
for j in range(len(real_mask[i])):
if real_mask[i][j] > 0:
real_mask[i][j] = 255
else:
real_mask[i][j] = 0
real_mask = torch.Tensor(real_mask)
mask_tensor = torch.Tensor(mask)
return real_mask, mask_tensor
def classes_list():
return [
"bottle-blue-full",
"bottle-transp-full",
"bottle-dark-full",
"bottle-green-full",
"bottle-multicolorv-full",
"bottle-blue5l-full",
"bottle-milk-full",
"bottle-oil-full",
"glass-transp",
"glass-dark",
"glass-green",
"bottle-blue5l",
"bottle-blue",
"bottle-green",
"bottle-dark",
"bottle-milk",
"bottle-transp",
"bottle-multicolor",
"bottle-oil",
"bottle-yogurt",
"juice-cardboard",
"milk-cardboard",
"cans",
"canister",
"detergent-color",
"detergent-transparent",
"detergent-box",
"detergent-white",
]
def get_iou_category(category, model, coefficient):
gt_list = []
pred_list = []
root = ""
dir_name = "data/images/" + category
for root, dirs, files in os.walk(os.path.join(root, dir_name)):
for file in files:
path_to_image = os.path.join(root, file)
# If CAMERAS, then use computeAndSaveMaps() function, the rest CAM - get_saliency_map()
map_to_mask = get_saliency_map(model, path_to_image)
# map_to_mask = computeAndSaveMaps(model, path_to_image)
gt_one, pred_one = get_masks(
map_to_mask,
path_to_image,
coefficient=coefficient,
resolution=resolution_model,
)
gt_list.append(gt_one)
pred_list.append(pred_one)
gt_batch = torch.stack(gt_list)
pred_batch = torch.stack(pred_list)
iou = iou_pytorch(pred_batch, gt_batch).item()
return iou
# When coefficient > 120, regardless of the method, it returns a mask where the entire area is filled in.
# Select the coefficient using the for loop in the code below.
# cam_method_name - the variable responsible for the name of the method, **it must be changed**, since a dictionary with the values IoU and Coefficient is formed from this name.
# Change the CAM class and the layers that will be rendered in the function get_saliency_map(..), which is located at the beginning of the section "Unsupervised segmentation pipeline".
# Changes for CAMERAS need to be made to the function computeAndSaveMap(), which is located in the section "CAMERAS".
# Example from cam_method_name = 'mGradCAM-3.2and4.1'
cam_method_name = "mGradCAM-3.2and4.1"
# Load the pre-trained model
model = resnet18(pretrained=True)
num_ftrs = model.fc.in_features
model.fc = nn.Linear(num_ftrs, 28) # second parameter - number of classes
torch.nn.init.xavier_normal_(model.fc.weight)
model.load_state_dict(
torch.load("ResNet18_Unfreeze3&4_4aug140epoch_model.pth")
) # path to weights
model.eval()
model = model.cuda()
category_classes = classes_list()
category_best_iou = {}
category_best_coefficient = {}
for class_name in category_classes:
category_best_iou[class_name] = 0
category_best_coefficient[class_name] = 0
for coefficient in range(1, 100):
print("Coefficient: ", coefficient)
for class_name in category_classes:
value_category_iou = get_iou_category(
class_name, model, coefficient=coefficient
)
if category_best_iou.get(class_name) < value_category_iou:
category_best_iou[class_name] = value_category_iou
category_best_coefficient[class_name] = coefficient
# Saving the results
with open(cam_method_name + "_best_iou", "w") as f:
json.dump(category_best_iou, f)
with open(cam_method_name + "_best_coefficient", "w") as f:
json.dump(category_best_coefficient, f)
# Execute the following cell if you want to save to google drive
cmd = "cp " + cam_method_name + "_best_iou" + " gdrive/MyDrive/model_ecology/CAM_IoU/"
os.system(cmd)
cmd = (
"cp "
+ cam_method_name
+ "_best_coefficient"
+ " gdrive/MyDrive/model_ecology/CAM_Coefficient/"
)
os.system(cmd)
# ## Creating masks with the best coefficients
# If you received the results in the last session and want to get their masks, transfer the data from Google drive using the following cell
def classes_list():
return [
"bottle-blue-full",
"bottle-transp-full",
"bottle-dark-full",
"bottle-green-full",
"bottle-multicolorv-full",
"bottle-blue5l-full",
"bottle-milk-full",
"bottle-oil-full",
"glass-transp",
"glass-dark",
"glass-green",
"bottle-blue5l",
"bottle-blue",
"bottle-green",
"bottle-dark",
"bottle-milk",
"bottle-transp",
"bottle-multicolor",
"bottle-oil",
"bottle-yogurt",
"juice-cardboard",
"milk-cardboard",
"cans",
"canister",
"detergent-color",
"detergent-transparent",
"detergent-box",
"detergent-white",
]
def get_masks(
saliency_map,
path_to_image,
coefficient=10,
resolution=224,
path_to_dir="",
visualization=False,
):
if not isinstance(saliency_map, np.ndarray):
saliency_map = cv2.applyColorMap(
np.uint8(255 * saliency_map.detach().numpy().squeeze()), cv2.COLORMAP_JET
)
else:
saliency_map = cv2.applyColorMap(np.uint8(255 * saliency_map), cv2.COLORMAP_JET)
hsv = cv2.cvtColor(saliency_map, cv2.COLOR_BGR2HSV)
# Here we define the blue color range in HSV
lower_blue = np.array([coefficient, 50, 50])
upper_blue = np.array([130, 255, 255])
# This method creates a blue mask of the objects found in the frame
mask = cv2.inRange(hsv, lower_blue, upper_blue)
# Inverting the mask
for i in range(len(mask)):
for j in range(len(mask[i])):
if mask[i][j] == 255:
mask[i][j] = 0
else:
mask[i][j] = 255
if visualization == True:
image = cv2.imread(path_to_image)
img = image.copy()
img[mask == 255] = 0
color_channeled_image = cv2.cvtColor(mask, cv2.COLOR_GRAY2BGR)
result = color_channeled_image * 0.4 + img * 0.7
mask_to_seg = saliency_map * 0.5 + image * 0.5
fig, (ax1, ax2) = plt.subplots(ncols=2, figsize=(16, 16))
ax1.set_title("Original picture")
ax2.set_title("The mask was created using CAM methods")
ax1.axis("off")
ax2.axis("off")
_ = ax1.imshow(cv2.cvtColor(mask_to_seg.astype("uint8"), cv2.COLOR_BGR2RGB))
_ = ax2.imshow(cv2.cvtColor(result.astype("uint8"), cv2.COLOR_BGR2RGB))
file_name = path_to_image.split("/")[-1]
# Second argument - gives Grey Scale Image
real_mask = cv2.imread(
"data/mask/"
+ path_to_image.split("/")[2]
+ "/"
+ file_name.split(".")[0]
+ ".png",
0,
)
real_mask = cv2.resize(
real_mask, (resolution, resolution), interpolation=cv2.INTER_NEAREST
)
for i in range(len(real_mask)):
for j in range(len(real_mask[i])):
if real_mask[i][j] > 0:
real_mask[i][j] = 255
else:
real_mask[i][j] = 0
path_for_class_dir = path_to_dir + "/" + file_name.split("_")[0]
try:
os.mkdir(path_for_class_dir)
except:
pass
cv2.imwrite(path_for_class_dir + "/pred_" + file_name.split(".")[0] + ".png", mask)
cv2.imwrite(
path_for_class_dir + "/gt_" + file_name.split(".")[0] + ".png", real_mask
)
cv2.imwrite(
path_for_class_dir + "/map_" + file_name.split(".")[0] + ".png", saliency_map
)
real_mask = torch.Tensor(real_mask)
mask_tensor = torch.Tensor(mask)
return real_mask, mask_tensor
def get_saliency_map(model, image_name, cam_method_name):
if cam_method_name.split("-")[1] == "4.1and4.2":
target_layers = [model.layer4[-2], model.layer4[-1]]
elif cam_method_name.split("-")[1] == "4.2":
target_layers = [model.layer4[-1]]
elif cam_method_name.split("-")[1] == "3.2and4.1":
target_layers = [model.layer3[-1], model.layer4[-2]]
elif cam_method_name.split("-")[1] == "3.1and3.2and4.1and4.2":
target_layers = [
model.layer3[-2],
model.layer3[-1],
model.layer4[-2],
model.layer4[-1],
]
elif cam_method_name.split("-")[1] == "3.2and4.2":
target_layers = [model.layer3[-1], model.layer4[-1]]
elif cam_method_name.split("-")[1] == "3.2and4.1and4.2":
target_layers = [model.layer3[-1], model.layer4[-2], model.layer4[-1]]
elif cam_method_name.split("-")[1] == "3.1and4.2":
target_layers = [model.layer3[-2], model.layer4[-1]]
elif cam_method_name.split("-")[1] == "3.1and4.1":
target_layers = [model.layer3[-2], model.layer4[-2]]
normalize = transforms.Normalize(
mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]
)
preprocess = transforms.Compose(
[
transforms.Resize((resolution_model, resolution_model)),
transforms.ToTensor(),
normalize,
]
)
# load test image
img_pil = Image.open(image_name).convert("RGB")
img_tensor = preprocess(img_pil)
img_variable = Variable(img_tensor.unsqueeze(0))
input_tensor = img_variable.cuda()
# This specifies which CAM method will be used
if (
cam_method_name.split("-")[0] == "GradCAM"
or cam_method_name.split("-")[0] == "mGradCAM"
):
cam = GradCAM(model=model, target_layers=target_layers, use_cuda=True)
elif cam_method_name.split("-")[0] == "LayerCAM":
cam = LayerCAM(model=model, target_layers=target_layers, use_cuda=True)
grayscale_cam = cam(input_tensor=input_tensor)
# In this example grayscale_cam has only one image in the batch:
grayscale_cam = grayscale_cam[0, :]
return grayscale_cam
def get_iou_category(category, model, coefficient, cam_method_name, path_to_dir):
root = ""
dir_name = "data/images/" + category
for root, dirs, files in os.walk(os.path.join(root, dir_name)):
for file in files:
path_to_image = os.path.join(root, file)
# If CAMERAS, then use computeAndSaveMaps() function, the rest CAM - get_saliency_map()
# map_to_mask = get_saliency_map(model, path_to_image)
map_to_mask = computeAndSaveMaps(model, path_to_image)
gt_one, pred_one = get_masks(
map_to_mask,
path_to_image,
coefficient=coefficient,
resolution=resolution_model,
path_to_dir=path_to_dir,
)
import os
absolute_path_dir = "CAM_Coefficient/"
absolute_dir_names = os.listdir(absolute_path_dir)
print(absolute_dir_names)
# GradCAM and LayerCAM
# name_dict_list = ['GradCAM-4.1and4.2_best_coefficient', 'LayerCAM-4.1and4.2_best_coefficient', 'GradCAM-4.2_best_coefficient', 'LayerCAM-3.2and4.1_best_coefficient', 'LayerCAM-3.1and3.2and4.1and4.2_best_coefficient', 'LayerCAM-4.2_best_coefficient', 'GradCAM-3.2and4.2_best_coefficient', 'LayerCAM-3.2and4.1and4.2_best_coefficient', 'LayerCAM-3.2and4.2_best_coefficient', 'LayerCAM-3.1and4.2_best_coefficient', 'LayerCAM-3.1and4.1_best_coefficient']
# mGradCAM
# name_dict_list = ['mGradCAM-4.1and4.2_best_coefficient', 'mGradCAM-3.1and3.2and4.1and4.2_best_coefficient', 'mGradCAM-3.1and4.1_best_coefficient', 'mGradCAM-3.2and4.1_best_coefficient', 'mGradCAM-3.1and4.2_best_coefficient', 'mGradCAM-3.2and4.2_best_coefficient']
# CAMERAS classic
# name_dict_list = ['CAMERAS-classic_best_coefficient']
# CAMERAS custom
name_dict_list = ["CAMERAS-custom-res_best_coefficient"]
# Load the pre-trained model
model = resnet18(pretrained=True)
num_ftrs = model.fc.in_features
model.fc = nn.Linear(num_ftrs, 28) # second parameter - number of classes
torch.nn.init.xavier_normal_(model.fc.weight)
model.load_state_dict(
torch.load("ResNet18_Unfreeze3&4_4aug140epoch_model.pth")
) # path to weights
model.eval()
model = model.cuda()
cam_method_list = []
category_classes = classes_list()
for name_dict in name_dict_list:
cam_method_list.append(name_dict.split("_")[0])
for cam_method_name in cam_method_list:
path_to_cam_method_name = "paper/" + cam_method_name
with open("CAM_Coefficient/" + cam_method_name + "_best_coefficient", "r") as f:
category_best_coefficient = json.load(f)
try:
os.mkdir(path_to_cam_method_name)
except:
pass
for class_name in category_classes:
coefficient = category_best_coefficient.get(class_name)
value_category_iou = get_iou_category(
class_name,
model,
coefficient=coefficient,
cam_method_name=cam_method_name,
path_to_dir=path_to_cam_method_name,
)
|
# ***INRODUCTION***
# 
# For Telco companies it is key to attract new customers and at the same time avoid contract terminations (=churn) to grow their revenue generating base. Looking at churn, different reasons trigger customers to terminate their contracts, for example better price offers, more interesting packages, bad service experiences or change of customers’ personal situations.
# Churn analytics provides valuable capabilities to predict customer churn and also define the underlying reasons that drive it. The churn metric is mostly shown as the percentage of customers that cancel a product or service within a given period (mostly months). If a Telco company had 10 Mio. customers on the 1st of January and received 500K contract terminations until the 31st of January the monthly churn for January would be 5%.
# Telcos apply machine learning models to predict churn on an individual customer basis and take counter measures such as discounts, special offers or other gratifications to keep their customers. A customer churn analysis is a typical classification problem within the domain of supervised learning.
# In this example, a basic machine learning pipeline based on a sample data set from Kaggle is build and performance of different model types is compared. The pipeline used for this example consists of 8 steps:
# Step 1: Problem Definition
# Step 2: Data Collection
# Step 3: Exploratory Data Analysis (EDA)
# Step 4: Feature Engineering
# Step 5: Train/Test Split
# Step 6: Model Evaluation Metrics Definition
# Step 7: Model Selection, Training, Prediction and Assessment
# Step 8: Hyperparameter Tuning/Model Improvement
#
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Importing libraries
import matplotlib.pyplot as plt
import seaborn as sns
# # Data Processing
df = pd.read_excel(
r"/kaggle/input/telecom-churn-dataset/Telecom Churn Rate Dataset.xlsx"
)
df.head()
df.tail()
df.info()
df.describe()
df = df.drop(["Senior_Citizen", "Partner", "Dependents"], axis=1)
df = df.drop(
["Online_Security", "Online_Backup", "Device_Protection", "Tech_Support"], axis=1
)
df = df.drop(
[
"Streaming_TV",
"Streaming_Movies",
"Contract",
"Paper_less_Billing",
"Payment_Method",
],
axis=1,
)
df.drop(["Monthly_Charges"], axis=1)
# # EDA
ax = sns.countplot(x="Gender", data=df)
for bars in ax.containers:
ax.bar_label(bars)
sns.barplot(x="Yearly_Charge", y="Monthly_Charges", data=df)
# # Importing Label Encoder
from sklearn import preprocessing
le = preprocessing.LabelEncoder()
df["Gender"] = le.fit_transform(df["Gender"])
df["Phone_Service"] = le.fit_transform(df["Phone_Service"])
df["Multiple_Lines"] = le.fit_transform(df["Multiple_Lines"])
df["Internet_Service"] = le.fit_transform(df["Internet_Service"])
df["Churn"] = le.fit_transform(df["Churn"])
X = df.drop(["Churn"], axis=1)
y = df["Churn"]
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=42
)
X_train.head()
y_train.head()
# # Importing Machine learning
from sklearn.linear_model import LogisticRegression
clf = LogisticRegression()
clf.fit(X_train, y_train)
y_pred = clf.predict(X_test)
accuracy = clf.score(X_test, y_test)
print(accuracy)
# # KNN
from sklearn.neighbors import KNeighborsClassifier
clf_knn = KNeighborsClassifier()
clf_knn.fit(X_train, y_train)
y_knn_pred = clf_knn.predict(X_test)
accracy = clf_knn.score(X_test, y_test)
print(accracy)
# # Support Vector Classifier
from sklearn.svm import SVC
clf_svm = SVC()
clf_svm.fit(X_train, y_train)
y_svc_pred = clf_svm.predict(X_test)
accuracy = clf_svm.score(X_test, y_test)
print(accuracy)
# # Decision Tree
from sklearn.tree import DecisionTreeClassifier
from sklearn.tree import export_text
dt = DecisionTreeClassifier()
dt.fit(X_train, y_train)
y_dt_pred = dt.predict(X_test)
accuracy = dt.score(X_test, y_test)
accuracy
# # Nayes Bias
from sklearn.naive_bayes import GaussianNB
NB = GaussianNB()
NB.fit(X_train, y_train)
y_nb_pred = NB.predict(X_test)
accuracy = NB.score(X_test, y_test)
print(accuracy)
# # Random Forest
from sklearn.ensemble import RandomForestClassifier
from sklearn.datasets import make_classification
Rf = RandomForestClassifier(n_estimators=4)
Rf.fit(X_train, y_train)
y_rf_pred = Rf.predict(X_test)
accuracy = Rf.score(X_test, y_test)
print(accuracy)
# # Confusion matrix
from sklearn.metrics import confusion_matrix
cm = confusion_matrix(y_test, y_rf_pred)
print(cm)
# # Boosting technique
# # Adaboost
from sklearn.ensemble import AdaBoostClassifier
from sklearn.datasets import make_classification
ada = AdaBoostClassifier()
ada.fit(X_train, y_train)
y_ada_pred = ada.predict(X_test)
accuracy = ada.score(X_test, y_test)
print(accuracy)
# # Gradient Boost
from sklearn.ensemble import GradientBoostingClassifier
xgb = GradientBoostingClassifier()
xgb.fit(X_train, y_train)
y_xgb_pred = xgb.predict(X_test)
accuracy = xgb.score(X_test, y_test)
print(accuracy)
# # Overall Calculation
print("Logistic Regression:", clf.score(X_test, y_test))
print("KNN:", clf_knn.score(X_test, y_test))
print("Support Vector Meachine:", clf_svm.score(X_test, y_test))
print("Decission Tree:", dt.score(X_test, y_test))
print("Naive Bayes:", NB.score(X_test, y_test))
print("Ensemble- Random Forest:", Rf.score(X_test, y_test))
print("Ensemble- adaboost:", ada.score(X_test, y_test))
print("Ensemble- XGBoost:", xgb.score(X_test, y_test))
from sklearn.metrics import f1_score
f1_score(y_test, y_ada_pred, average="macro")
from sklearn.metrics import f1_score
f1_score(y_test, y_xgb_pred, average="macro")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
os.path.join(dirname, filename)
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
os.listdir("/kaggle/input/skin-cancer-malignant-vs-benign/data/")
from tensorflow.keras.preprocessing.image import ImageDataGenerator
datagen = ImageDataGenerator(rescale=1.0 / 255, zoom_range=0.5)
testgen = ImageDataGenerator()
train_path = "/kaggle/input/skin-cancer-malignant-vs-benign/data/train"
test_path = "/kaggle/input/skin-cancer-malignant-vs-benign/data/test"
image_size = (150, 150)
batch_size = 200
train_generator = datagen.flow_from_directory(
train_path,
target_size=image_size,
batch_size=batch_size,
class_mode="binary",
)
test_generator = datagen.flow_from_directory(
test_path, target_size=image_size, batch_size=batch_size, class_mode="binary"
)
from tqdm import tqdm
train_generator.reset()
X_train, y_train = next(train_generator)
for i in tqdm(
range(int(len(train_generator) - 1))
): # 1st batch is already fetched before the for loop.
img, label = next(train_generator)
X_train = np.append(X_train, img, axis=0)
y_train = np.append(y_train, label, axis=0)
print(X_train.shape, y_train.shape)
from tqdm import tqdm
train_generator.reset()
X_test, y_test = next(test_generator)
for i in tqdm(
range(int(len(test_generator) - 1))
): # 1st batch is already fetched before the for loop.
img, label = next(test_generator)
X_test = np.append(X_test, img, axis=0)
y_test = np.append(y_test, label, axis=0)
print(X_test.shape, y_test.shape)
import tensorflow as tf
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Conv2D, MaxPooling2D, Dense, Flatten, Dropout
model = Sequential()
model.add(Conv2D(32, (3, 3), 1, activation="relu", input_shape=(150, 150, 3)))
model.add(MaxPooling2D())
model.add(Conv2D(32, (3, 3), 1, activation="relu"))
model.add(MaxPooling2D())
model.add(Conv2D(32, (3, 3), 1, activation="relu"))
model.add(MaxPooling2D())
model.add(Flatten())
model.add(Dense(150, activation="relu"))
model.add(Dense(1, activation="sigmoid"))
model.compile("adam", loss=tf.losses.BinaryCrossentropy(), metrics=["accuracy"])
model.summary()
history = model.fit(
X_train,
y_train,
validation_split=0.1,
epochs=10,
)
model.evaluate(X_test, y_test)
import matplotlib.pyplot as plt
fig = plt.figure()
plt.plot(history.history["loss"], color="teal", label="loss")
plt.plot(history.history["val_loss"], color="orange", label="val_loss")
fig.suptitle("Loss", fontsize=20)
plt.legend(loc="upper left")
plt.show()
fig = plt.figure()
plt.plot(history.history["accuracy"], color="teal", label="accuracy")
plt.plot(history.history["val_accuracy"], color="orange", label="val_accuracy")
fig.suptitle("Accuracy", fontsize=20)
plt.legend(loc="upper left")
plt.show()
predictions = model.predict(X_test)
# Results
fig = plt.figure()
for i in range(9):
plt.subplot(3, 3, i + 1)
plt.tight_layout()
plt.imshow(X_test[i], cmap="gray", interpolation="none")
title = round(predictions[i][0])
plt.title(title)
plt.xticks([])
plt.yticks([])
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
# for dirname, _, filenames in os.walk('/kaggle/input'):
# for filename in filenames:
# print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import tensorflow as tf
tf.__version__
from PIL import Image
import glob
import os
from tqdm import tqdm
import matplotlib.pyplot as plt
train_data_dir = "/kaggle/input/25-indian-bird-species-with-226k-images/training_set"
IMG_HEIGHT = 299
IMG_WIDHT = 299
batch_size = 64
# sizes = []
# for path in tqdm(glob.glob(train_data_dir+'/*/*')):
# if os.path.isfile(path):
# sizes.append(Image.open(path).size)
# df_sizes = pd.DataFrame(set(sizes))
# df_sizes.describe()
train_image_generator = tf.keras.preprocessing.image.ImageDataGenerator(
rotation_range=0.2,
width_shift_range=0.2,
height_shift_range=0.2,
shear_range=0.2,
zoom_range=0.2,
horizontal_flip=True,
vertical_flip=True,
rescale=1.0 / 255,
data_format="channels_last",
validation_split=0.2,
)
train_generator = train_image_generator.flow_from_directory(
train_data_dir,
target_size=(IMG_HEIGHT, IMG_WIDHT),
batch_size=batch_size,
subset="training",
) # set as training data
validation_generator = train_image_generator.flow_from_directory(
train_data_dir, # same directory as training data
target_size=(IMG_HEIGHT, IMG_WIDHT),
batch_size=batch_size,
subset="validation",
) # set as validation data
def plotImages(images_arr, probabilities=False):
fig, axes = plt.subplots(len(images_arr), 1, figsize=(5, len(images_arr) * 3))
if probabilities is False:
for img, ax in zip(images_arr, axes):
ax.imshow(img)
ax.axis("off")
else:
for img, probability, ax in zip(images_arr, probabilities, axes):
ax.imshow(img)
ax.axis("off")
if probability > 0.5:
ax.set_title("%.2f" % (probability * 100) + "% dog")
else:
ax.set_title("%.2f" % ((1 - probability) * 100) + "% cat")
plt.show()
sample_training_images, _ = next(train_generator)
plotImages(sample_training_images[:5])
# AUTOTUNE = tf.data.AUTOTUNE
# train_ds = train_generator.cache().shuffle(1000).prefetch(buffer_size=AUTOTUNE)
# val_ds = validation_generator.cache().prefetch(buffer_size=AUTOTUNE)
strategy = tf.distribute.MultiWorkerMirroredStrategy()
with strategy.scope():
base_model = tf.keras.applications.inception_resnet_v2.InceptionResNetV2(
include_top=False, input_shape=train_generator.image_shape
)
# add a global spatial average pooling layer
x = base_model.output
x = tf.keras.layers.GlobalAveragePooling2D()(x)
# let's add a fully-connected layer
x = tf.keras.layers.Dense(1024, activation="swish")(x)
# and a logistic layer -- let's say we have 200 classes
predictions = tf.keras.layers.Dense(
len(set(train_generator.classes)), activation="softmax"
)(x)
# this is the model we will train
model = tf.keras.Model(inputs=base_model.input, outputs=predictions)
# first: train only the top layers (which were randomly initialized)
# i.e. freeze all convolutional layers
for layer in base_model.layers:
layer.trainable = False
# compile the model (should be done *after* setting layers to non-trainable)
model.compile(
optimizer="adam",
loss="categorical_crossentropy",
metrics=["accuracy", tf.keras.metrics.FalseNegatives()],
)
early_stopping = tf.keras.callbacks.EarlyStopping(
monitor="val_loss", patience=10, mode="min", restore_best_weights=True
)
reduce_learning = tf.keras.callbacks.ReduceLROnPlateau(
monitor="val_loss", patience=5, mode="min"
)
# train the model on the new data for a few epochs
# model.fit(...)
train_history = model.fit(
train_generator,
epochs=200,
validation_data=validation_generator,
callbacks=[early_stopping, reduce_learning],
)
model.save("model.h5")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import numpy as np
df = pd.read_csv(
"../input/edgeiiotset-cyber-security-dataset-of-iot-iiot/Edge-IIoTset dataset/Selected dataset for ML and DL/DNN-EdgeIIoT-dataset.csv",
low_memory=False,
)
from sklearn.utils import shuffle
drop_columns = [
"frame.time",
"ip.src_host",
"ip.dst_host",
"arp.src.proto_ipv4",
"arp.dst.proto_ipv4",
"http.file_data",
"http.request.full_uri",
"icmp.transmit_timestamp",
"http.request.uri.query",
"tcp.options",
"tcp.payload",
"tcp.srcport",
"tcp.dstport",
"udp.port",
"mqtt.msg",
]
df.drop(drop_columns, axis=1, inplace=True)
df.dropna(axis=0, how="any", inplace=True)
df.drop_duplicates(subset=None, keep="first", inplace=True)
df = shuffle(df)
df.isna().sum()
print(df["Attack_type"].value_counts())
import numpy as np
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from sklearn import preprocessing
def encode_text_dummy(df, name):
dummies = pd.get_dummies(df[name])
for x in dummies.columns:
dummy_name = f"{name}-{x}"
df[dummy_name] = dummies[x]
df.drop(name, axis=1, inplace=True)
encode_text_dummy(df, "http.request.method")
encode_text_dummy(df, "http.referer")
encode_text_dummy(df, "http.request.version")
encode_text_dummy(df, "dns.qry.name.len")
encode_text_dummy(df, "mqtt.conack.flags")
encode_text_dummy(df, "mqtt.protoname")
encode_text_dummy(df, "mqtt.topic")
df.to_csv("preprocessed_DNN.csv", encoding="utf-8", index=False)
import pandas as pd
import numpy as np
from matplotlib import pyplot as plt
import seaborn as sns
df = pd.read_csv("./preprocessed_DNN.csv", low_memory=False)
df
df["Attack_type"].value_counts()
df.info()
feat_cols = list(df.columns)
label_col = "Attack_type"
feat_cols.remove(label_col)
# feat_cols
empty_cols = [col for col in df.columns if df[col].isnull().all()]
empty_cols
skip_list = ["icmp.unused", "http.tls_port", "dns.qry.type", "mqtt.msg_decoded_as"]
df[skip_list[3]].value_counts()
fig, (ax1, ax2) = plt.subplots(nrows=1, ncols=2, figsize=(12, 8))
explode = list(
(
np.array(list(df[label_col].dropna().value_counts()))
/ sum(list(df[label_col].dropna().value_counts()))
)[::-1]
)[:]
labels = list(df[label_col].dropna().unique())[:]
sizes = df[label_col].value_counts()[:]
ax2.pie(
sizes,
explode=explode,
startangle=60,
labels=labels,
autopct="%1.0f%%",
pctdistance=0.8,
)
ax2.add_artist(plt.Circle((0, 0), 0.4, fc="white"))
sns.countplot(y=label_col, data=df, ax=ax1)
ax1.set_title("Count of each Attack type")
ax2.set_title("Percentage of each Attack type")
plt.show()
X = df.drop([label_col], axis=1)
y = df[label_col]
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=1, stratify=y
)
del X
del y
from sklearn.preprocessing import LabelEncoder
label_encoder = LabelEncoder()
y_train = label_encoder.fit_transform(y_train)
y_test = label_encoder.transform(y_test)
label_encoder.classes_
from sklearn.preprocessing import MinMaxScaler
min_max_scaler = MinMaxScaler()
X_train = min_max_scaler.fit_transform(X_train)
X_test = min_max_scaler.transform(X_test)
X_train = X_train.reshape(X_train.shape[0], X_train.shape[1], 1)
X_test = X_test.reshape(X_test.shape[0], X_test.shape[1], 1)
print(X_train.shape)
print(X_test.shape)
input_shape = X_train.shape[1:]
print(X_train.shape, X_test.shape)
print(input_shape)
num_classes = len(np.unique(y_train))
num_classes
from tensorflow.keras.utils import to_categorical
y_train = to_categorical(y_train, num_classes=num_classes)
y_test = to_categorical(y_test, num_classes=num_classes)
print(y_train.shape, y_test.shape)
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Conv1D, MaxPooling1D, Flatten, Dense
model = Sequential()
model.add(Conv1D(32, 3, activation="relu", input_shape=(input_shape)))
model.add(MaxPooling1D(2))
# model.add(Conv1D(64, 3, activation='relu', input_shape=(input_shape)))
# model.add(MaxPooling1D(2))
# model.add(Conv1D(64, 3, activation='relu', input_shape=(input_shape)))
# model.add(MaxPooling1D(2))
model.add(Conv1D(64, 3, activation="relu", input_shape=(input_shape)))
model.add(MaxPooling1D(2))
model.add(Conv1D(128, 3, activation="relu"))
model.add(MaxPooling1D(2))
model.add(Flatten())
model.add(Dense(64, activation="relu"))
model.add(Dense(num_classes, activation="softmax"))
model.summary()
from tensorflow.keras.datasets import mnist
from tensorflow.keras.utils import to_categorical
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Conv1D, Input, ZeroPadding1D
from tensorflow.keras.layers import MaxPooling1D, Add, AveragePooling1D
from tensorflow.keras.layers import Dense, BatchNormalization, Activation
from tensorflow.keras.layers import Flatten
from tensorflow.keras.models import Model
from keras.initializers import glorot_uniform
import keras.backend as K
import tensorflow as tf
from tensorflow.keras.optimizers import Adam # input the model before this cell
# model = build_model(num_classes, input_shape=input_shape)
opt = Adam(learning_rate=0.001)
model.compile(
optimizer=opt, loss=tf.keras.metrics.categorical_crossentropy, metrics=["accuracy"]
)
# !pip install gdown
# !gdown --id 1s-gqNpqkI5GZhHM9hEYSCZmjaZfEIQ_D
# hashable_arr = tuple(np.unique(y_train))
# print(hashable_arr)
# from sklearn.utils import class_weight
# class_weights = class_weight.compute_class_weight('balanced',
# classes=np.unique(y_train),
# y =y_train)
# class_weights = dict(enumerate(class_weights))
from keras.callbacks import ModelCheckpoint, EarlyStopping, ReduceLROnPlateau
# from livelossplot import PlotLossesKeras
# model_weights_file_path = "model_best_weights.h5"
# checkpoint = ModelCheckpoint(filepath=model_weights_file_path, monitor="val_loss", verbose=1, save_best_only=True, mode="min", save_weights_only=True)
# checkpoint = ModelCheckpoint(filepath=model_weights_file_path, monitor="val_loss", verbose=1, save_best_only=True, mode="min", save_weights_only=True)
early_stopping = EarlyStopping(monitor="val_loss", mode="min", verbose=1, patience=10)
lr_reduce = ReduceLROnPlateau(
monitor="val_loss", factor=0.5, patience=5, mode="min", verbose=1, min_lr=0
)
# plotlosses = PlotLossesKeras()
call_backs = [early_stopping, lr_reduce]
EPOCHS = 15
BATCH_SIZE = 256
call_backs = [early_stopping, lr_reduce]
history = model.fit(
X_train,
y_train,
validation_data=(X_test, y_test),
validation_split=0.1,
epochs=EPOCHS,
batch_size=BATCH_SIZE,
callbacks=call_backs,
# class_weight=class_weights,
verbose=1,
)
# from keras.callbacks import ModelCheckpoint, EarlyStopping, ReduceLROnPlateau
# from livelossplot import PlotLossesKeras
# model_weights_file_path = "model_best_weights.h5"
# checkpoint = ModelCheckpoint(filepath=model_weights_file_path, monitor="val_loss", verbose=1, save_best_only=True, mode="min", save_weights_only=True)
# early_stopping = EarlyStopping(monitor="val_loss", mode="min", verbose=1, patience=10)
# lr_reduce = ReduceLROnPlateau(monitor='val_loss', factor=0.5, patience=5, mode="min", verbose=1, min_lr=0)
# plotlosses = PlotLossesKeras()
# call_backs = [checkpoint, early_stopping, lr_reduce, plotlosses]
history_df = pd.DataFrame(history.history)
history_df.loc[:, ["loss", "val_loss"]].plot()
print("Minimum validation loss: {}".format(history_df["val_loss"].min()))
print(y_train.shape)
#!pip install gdown
y_hat = model.predict(X_test)
y_hat = np.argmax(y_hat, axis=1)
print(y_hat)
y_true = np.argmax(y_test, axis=1)
from sklearn.metrics import accuracy_score
def print_score(y_pred, y_real, label_encoder):
print("Accuracy: ", accuracy_score(y_real, y_pred))
print_score(y_hat, y_true, label_encoder)
from sklearn.metrics import (
accuracy_score,
precision_recall_fscore_support,
confusion_matrix,
classification_report,
precision_score,
recall_score,
)
from sklearn.metrics import f1_score as f1_score_rep
# print(confusion_matrix(y_true, y_hat).ravel())
# tn, fp, fn, tp = confusion_matrix(y_true, y_hat).ravel()
# print(tn, fp, fn, tp)
# false_positive_rate = fp / (fp + tn)
# false_negative_rate = fn / (fn + tp)
tn, fp, fn, tp = confusion_matrix(y_true, y_hat).ravel()
tpr = tp / (tp + fn)
fpr = fp / (fp + tn)
|
# * We have used RSNA official datset.It has dcm files to store mammograms.
# * Have to convert to png
# * below code converts train,test dcm sets to png sets
# import libraries
import os
import cv2
import glob
import gdcm
import pydicom
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from tqdm.notebook import tqdm
from joblib import Parallel, delayed
import tensorflow as tf
import tensorflow.keras.backend as K
import matplotlib.image as mpimg
pd.options.display.max_columns = 50
# ### **Convert test data DCM to PNG**
# Save the processed image
def process(f, size=512, save_folder="", extension="png"):
patient = f.split("/")[-2]
image = f.split("/")[-1][:-4]
dicom = pydicom.dcmread(f)
img = dicom.pixel_array
img = (img - img.min()) / (img.max() - img.min())
if dicom.PhotometricInterpretation == "MONOCHROME1":
img = 1 - img
img = cv2.resize(img, (size, size))
cv2.imwrite(
save_folder + f"{patient}_{image}.{extension}", (img * 255).astype(np.uint8)
)
def create_pngs_from_dcms(dcm_path, SAVE_FOLDER, SIZE, EXTENSION):
train_images = glob.glob(dcm_path)
len(train_images) # 54706
print("...........Images loaded")
os.makedirs(SAVE_FOLDER, exist_ok=True)
# an empty directory called train_output in kaggle/working path is created
print("...........New folder created")
_ = Parallel(n_jobs=4)(
delayed(process)(uid, size=SIZE, save_folder=SAVE_FOLDER, extension=EXTENSION)
for uid in tqdm(train_images)
)
print("............Finished")
dcm_path = "/kaggle/input/rsna-breast-cancer-detection/test_images/*/*.dcm"
SAVE_FOLDER = "test_output/"
SIZE = 512
EXTENSION = "png"
create_pngs_from_dcms(dcm_path, SAVE_FOLDER, SIZE, EXTENSION)
# ## **To visulaise data that we are working with**
train_images = glob.glob("/kaggle/input/rsna-breast-cancer-512-pngs/*.png")
train_images[0]
for f in tqdm(train_images[-4:]):
img = cv2.imread(f)
plt.figure(figsize=(15, 15))
plt.imshow(img, cmap="gray")
plt.show()
# ## **Load train and test datasets**
train_df = pd.read_csv("/kaggle/input/rsna-breast-cancer-detection/train.csv")
test_df = pd.read_csv("/kaggle/input/rsna-breast-cancer-detection/test.csv")
train_df.head()
train_df.info()
# ## **Add image paths to train Dataframe**
# row = train_df[train_df['']]
train_images = glob.glob("/kaggle/input/rsna-breast-cancer-512-pngs/*.png")
for path in tqdm(train_images):
name = path.split("/")[-1]
chunks = name.split(".")[0]
patient_id = chunks.split("_")[0]
image_id = chunks.split("_")[1]
idx = (train_df["patient_id"] == int(patient_id)) & (
train_df["image_id"] == int(image_id)
)
train_df.loc[idx, "img_path"] = path
train_df[["patient_id", "image_id", "img_path"]].head()
train_df["img_path"][0]
# ## **Add image paths to test Dataframe**
test_images = glob.glob("/kaggle/working/test_output/*.png")
for path in tqdm(test_images):
name = path.split("/")[-1]
chunks = name.split(".")[0]
patient_id = chunks.split("_")[0]
image_id = chunks.split("_")[1]
idx = (test_df["patient_id"] == int(patient_id)) & (
test_df["image_id"] == int(image_id)
)
test_df.loc[idx, "img_path"] = path
test_df[["patient_id", "image_id", "img_path"]].head()
# # Exploration and Feature Engineering
# * 'site_id'
# * 'patient_id'
# * 'image_id'
# * 'laterality'
# * 'view' = the craniocaudal (CC) view and the mediolateral oblique (MLO) view.
# * 'age'
# * 'cancer'
# * 'biopsy'
# * 'invasive'
# * 'BIRADS'
# * 'implant'
# * 'density'
# * 'machine_id'
# * 'difficult_negative_case'
# * 'img_path'
cols = ["image_id", "age", "machine_id", "img_path"]
for i in list(train_df.drop(cols, axis=1).columns):
print(i)
print(train_df[i].value_counts())
print("----------------\n")
# ## **Make a data frame to reduce bias in dataset non-cancer cases**
# cancer cases
# cancer
# 0 53548
# 1 1158
# Name: cancer, dtype: int64
# ----------------
df = train_df.copy()
# imgs of cancer
canc_count = df.loc[df["cancer"] == 1].shape[0]
canc_count
# pick as many non canc cases as canc cases
df2 = df.loc[df["cancer"] == 0][:canc_count]
# use rest of imgs for testing model
df_test_1 = df.loc[df["cancer"] == 0][canc_count:]
# see how is the split
df2["cancer"].value_counts()
# cancat both categ cases
df3 = df.loc[df["cancer"] == 1]
df4 = pd.concat([df2, df3], axis=0)
# look at the split
df4["cancer"].value_counts()
# df4 = The balanced data set
# make split - train&val : 90% , test : 10%
perc_90 = int(1158 * 0.9)
noncanc_df = df4.loc[df4["cancer"] == 0]
# take 90% non canc cases
noncanc_df_train = noncanc_df[:perc_90]
# take 10% non canc cases
noncanc_df_test = noncanc_df[perc_90:]
canc_df = df4.loc[df4["cancer"] == 1]
# take 90% canc cases
canc_df_train = canc_df[:perc_90]
# take 10% canc cases
canc_df_test = canc_df[perc_90:]
train_df_new = pd.concat([noncanc_df_train, canc_df_train], axis=0)
test_df_new = pd.concat([noncanc_df_test, canc_df_test], axis=0)
train_df_new["cancer"].value_counts()
test_df_new["cancer"].value_counts()
train_datagen = tf.keras.preprocessing.image.ImageDataGenerator(
rescale=1.0 / 255,
horizontal_flip=True,
vertical_flip=True,
rotation_range=90,
validation_split=0.20,
)
test_datagen = tf.keras.preprocessing.image.ImageDataGenerator(rescale=1.0 / 255)
# The value for class_mode in flow_from_dataframe MUST be 'raw' if you are attempting to do multilabel classification.
train_gen = train_datagen.flow_from_dataframe(
train_df_new,
x_col="img_path",
y_col="cancer",
target_size=(128, 128),
class_mode="raw",
batch_size=16,
subset="training",
shuffle=True,
seed=42,
)
val_gen = train_datagen.flow_from_dataframe(
train_df_new,
x_col="img_path",
y_col="cancer",
target_size=(128, 128),
class_mode="raw",
subset="validation",
batch_size=8,
seed=42,
)
test_gen = train_datagen.flow_from_dataframe(
test_df_new,
x_col="img_path",
y_col="cancer",
target_size=(128, 128),
class_mode="raw",
batch_size=8,
seed=42,
)
# # Visualizing Augmentation
# Load some sample images
img_path = "/kaggle/input/rsna-breast-cancer-512-pngs/10006_462822612.png"
img = tf.keras.preprocessing.image.load_img(img_path, target_size=(256, 256))
# Convert image to numpy array
img_array = tf.keras.preprocessing.image.img_to_array(img)
# Add a batch dimension to the array
img_array = np.expand_dims(img_array, axis=0)
# Generate augmented images using the flow() method
aug_iter = train_datagen.flow(img_array, batch_size=1)
# Visualize the augmented images
fig, ax = plt.subplots(1, 5, figsize=(15, 15))
for i in range(5):
aug_img = next(aug_iter)[0]
ax[i].imshow(aug_img)
plt.show()
# # Modeling
def UNet(inputs):
# First convolution block
x = tf.keras.layers.Conv2D(
64, 3, activation="relu", padding="same", kernel_initializer="he_normal"
)(inputs)
d1_con = tf.keras.layers.Conv2D(
64, 3, activation="relu", padding="same", kernel_initializer="he_normal"
)(x)
d1 = tf.keras.layers.MaxPool2D(pool_size=2, strides=2)(d1_con)
# Second convolution block
d2 = tf.keras.layers.Conv2D(
128, 3, activation="relu", padding="same", kernel_initializer="he_normal"
)(d1)
d2_con = tf.keras.layers.Conv2D(
128, 3, activation="relu", padding="same", kernel_initializer="he_normal"
)(d2)
d2 = tf.keras.layers.MaxPool2D(pool_size=2, strides=2)(d2_con)
# Third convolution block
d3 = tf.keras.layers.Conv2D(
256, 3, activation="relu", padding="same", kernel_initializer="he_normal"
)(d2)
d3_con = tf.keras.layers.Conv2D(
256, 3, activation="relu", padding="same", kernel_initializer="he_normal"
)(d3)
d3 = tf.keras.layers.MaxPool2D(pool_size=2, strides=2)(d3_con)
# Fourth convolution block
d4 = tf.keras.layers.Conv2D(
512, 3, activation="relu", padding="same", kernel_initializer="he_normal"
)(d3)
d4_con = tf.keras.layers.Conv2D(
512, 3, activation="relu", padding="same", kernel_initializer="he_normal"
)(d4)
d4 = tf.keras.layers.MaxPool2D(pool_size=2, strides=2)(d4_con)
# Bottleneck layer
b = tf.keras.layers.Conv2D(
1024, 3, activation="relu", padding="same", kernel_initializer="he_normal"
)(d4)
b = tf.keras.layers.Conv2D(
1024, 3, activation="relu", padding="same", kernel_initializer="he_normal"
)(b)
# First upsampling block
u1 = tf.keras.layers.Conv2DTranspose(512, 3, strides=(2, 2), padding="same")(b)
u1 = tf.keras.layers.Concatenate(axis=3)([u1, d4_con])
u1 = tf.keras.layers.Conv2D(
512, 3, activation="relu", padding="same", kernel_initializer="he_normal"
)(u1)
u1 = tf.keras.layers.Conv2D(
512, 3, activation="relu", padding="same", kernel_initializer="he_normal"
)(u1)
# Second upsampling block
u2 = tf.keras.layers.Conv2DTranspose(256, 3, strides=(2, 2), padding="same")(u1)
u2 = tf.keras.layers.Concatenate(axis=3)([u2, d3_con])
u2 = tf.keras.layers.Conv2D(
256, 3, activation="relu", padding="same", kernel_initializer="he_normal"
)(u2)
u2 = tf.keras.layers.Conv2D(
256, 3, activation="relu", padding="same", kernel_initializer="he_normal"
)(u2)
# Third upsampling block
u3 = tf.keras.layers.Conv2DTranspose(128, 3, strides=(2, 2), padding="same")(u2)
u3 = tf.keras.layers.Concatenate(axis=3)([u3, d2_con])
u3 = tf.keras.layers.Conv2D(
128, 3, activation="relu", padding="same", kernel_initializer="he_normal"
)(u3)
u3 = tf.keras.layers.Conv2D(
128, 3, activation="relu", padding="same", kernel_initializer="he_normal"
)(u3)
# Fourth upsampling block
u4 = tf.keras.layers.Conv2DTranspose(64, 3, strides=(2, 2), padding="same")(u3)
u4 = tf.keras.layers.Concatenate(axis=3)([u4, d1_con])
u4 = tf.keras.layers.Conv2D(
64, 3, activation="relu", padding="same", kernel_initializer="he_normal"
)(u4)
u4 = tf.keras.layers.Conv2D(
64, 3, activation="relu", padding="same", kernel_initializer="he_normal"
)(u4)
# Flatten and output
flat = tf.keras.layers.Flatten()(u4)
out = tf.keras.layers.Dense(units=1, activation="softmax")(flat)
model = tf.keras.Model(inputs=[inputs], outputs=[out])
return model
from keras.metrics import Accuracy
auc = tf.keras.metrics.AUC(multi_label=True, thresholds=[0, 1])
aucpr = tf.keras.metrics.AUC(curve="PR", multi_label=True, thresholds=[0, 1])
inputs = tf.keras.layers.Input(shape=(128, 128, 3))
unet = UNet(inputs)
unet.compile(
optimizer="adam", loss="binary_crossentropy", metrics=["accuracy", auc, aucpr]
)
unet.summary()
model_history = unet.fit(train_gen, epochs=10, validation_data=val_gen)
# ## **Plot model loss**
plt.plot(model_history.history["loss"])
plt.plot(model_history.history["val_loss"])
plt.title("model loss")
plt.ylabel("loss")
plt.xlabel("epoch")
plt.legend(["train", "test"], loc="upper left")
plt.show()
# ## **Save Model**
import os
import h5py
# Save model and weights
filename = "Breast_cancer_UNet"
model_name = filename + ".h5" #'Emotion_Model_aug.h5'
save_dir = os.path.join(os.getcwd(), "saved_models")
if not os.path.isdir(save_dir):
os.makedirs(save_dir)
model_path = os.path.join(save_dir, model_name)
unet.save(model_path)
print("Save model and weights at %s " % model_path)
# Save the model to disk
model_json = unet.to_json()
with open(filename + ".json", "w") as json_file:
json_file.write(model_json)
print("file saved to system")
# ## **Evaluating model**
# Evaluate model on test data
evaluation_metrics = unet.evaluate(val_gen)
evaluation_metrics
# for train_gen : loss: 0.6638 - accuracy: 0.6247 - auc_10: 0.5000 - auc_11: 0.6247
# for val_gen : loss: 1.1809 - accuracy: 0.0000e+00 - auc_10: 0.0000e+00 - auc_11: 0.0000e+00
# for test_gen : loss: 0.7860 - accuracy: 0.5000 - auc_10: 0.5000 - auc_11: 0.5000
unet.evaluate(test_gen)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import seaborn as sns
import matplotlib.pyplot as plt
train_data = pd.read_csv("/kaggle/input/titanic/train.csv")
df_train = pd.read_csv("/kaggle/input/titanic/train.csv")
train_data.head()
test_data = pd.read_csv("/kaggle/input/titanic/test.csv")
test_data.head()
test = pd.read_csv("/kaggle/input/titanic/test.csv")
train = pd.read_csv("/kaggle/input/titanic/train.csv")
print("Train columns: ", train.columns.tolist())
print("Test columns: ", test.columns.tolist())
# The tolist() method is used in the code to convert the
# column names of the pandas DataFrame into a Python list.
gender = pd.read_csv("/kaggle/input/titanic/gender_submission.csv")
gender.head()
tested = pd.merge(test_data, gender, on="PassengerId")
tested.head(10)
PassengerId = test["PassengerId"]
train["Ticket_type"] = train["Ticket"].apply(lambda x: x[0:3])
# by taking the first three characters of each value in the "Ticket" column.
train["Ticket_type"] = train["Ticket_type"].astype("category")
train["Ticket_type"] = train["Ticket_type"].cat.codes
# Next, the "cat.codes" method is used to assign a unique code to each unique value in the "Ticket_type"
# column. The codes are integers starting from 0, and each unique value in the column is
# assigned a different code. This is useful for converting categorical data into a numerical format
# that can be used in machine learning algorithms.
test["Ticket_type"] = train["Ticket"].apply(lambda x: x[0:3])
test["Ticket_type"] = test["Ticket_type"].astype("category")
test["Ticket_type"] = test["Ticket_type"].cat.codes
# This line of code is taking the 'Survived' column from the 'train' dataframe and converting it into
# a one-dimensional numpy array using the ravel() method.
y_train = train["Survived"].ravel()
train = train.drop(["Survived"], axis=1)
X_train = train.values
X_test = test.values
|
import pandas as pd
import numpy as np
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from tensorflow import keras
from tensorflow.keras import layers
# Load the data
df = pd.read_csv("/kaggle/input/co2-emissions/CO2 Emissions.csv")
# Drop rows with missing values
df = df.dropna()
# Define the input and output columns
input_cols = [
"Engine Size(L)",
"Cylinders",
"Fuel Consumption City (L/100 km)",
"Fuel Consumption Hwy (L/100 km)",
"Fuel Consumption Comb (L/100 km)",
"Fuel Consumption Comb (mpg)",
]
output_col = "CO2 Emissions(g/km)"
# Split the data into training and test sets
X_train, X_test, y_train, y_test = train_test_split(
df[input_cols], df[output_col], test_size=0.2, random_state=1
)
# Scale the input data
scaler = StandardScaler()
X_train = scaler.fit_transform(X_train)
X_test = scaler.transform(X_test)
# Define the Keras model
model = keras.Sequential(
[
layers.Dense(20, activation="relu", input_shape=[X_train.shape[1]]),
layers.Dense(10, activation="relu"),
layers.Dense(1),
]
)
# Compile the model
model.compile(
optimizer="adam", loss="mean_squared_error", metrics=["mean_absolute_error"]
)
# Train the model
history = model.fit(X_train, y_train, validation_split=0.2, epochs=100, batch_size=32)
# Evaluate the model
test_loss, test_mae = model.evaluate(X_test, y_test)
print("Test loss:", test_loss)
print("Test MAE:", test_mae)
# Make predictions
input_data = np.array([[5.6, 8, 17.5, 12, 15, 19]])
scaled_input_data = scaler.transform(input_data)
prediction = model.predict(scaled_input_data)
print("Predicted CO2 Emissions: ", prediction[0][0])
|
# ## Overview
# The "MLflow PySpark Pipeline for Diabetes Prediction" - Python Notebook is a comprehensive example of how to use the MLflow library to build a machine learning pipeline to predict diabetes in patients. The notebook demonstrates how to use various PySpark libraries, such as Pipeline, PipelineModel, Logistic Regression, BinaryClassificationEvaluator, and MulticlassClassificationEvaluator to build and evaluate the machine learning model.
# **This Python Notebook can be run only in Azure/GCP/AWS Databricks.**
# The first step is to set the MLflow experiment for diabetes prediction. The next step is to load the diabetes dataset, which is split into training and test sets using the randomSplit function. The hyperparameters for the logistic regression model are set, and the model is trained using the pipeline function.
# The categorical features in the dataset are converted to numeric using StringIndexer, and the features vector is created using VectorAssembler. The logistic regression model is defined, and the pipeline is created using these components. The hyperparameters are logged, and the model is trained.
# After training the model, the predictions are made on the test data, and the model is evaluated using BinaryClassificationEvaluator and MulticlassClassificationEvaluator. The evaluation metrics, such as area under the ROC curve and accuracy, are logged using the mlflow.log_metric function.
# The model is then logged using the mlflow.spark.log_model function, which saves the model to the MLflow registry. Finally, the production version of the model is transitioned using the MlflowClient function. Then, The model is then loaded and used to make predictions on new data.
# This notebook is written in **Python** so the default cell type is Python.
import mlflow
from pyspark.ml import Pipeline, PipelineModel
from pyspark.ml.classification import LogisticRegression
from pyspark.ml.evaluation import (
BinaryClassificationEvaluator,
MulticlassClassificationEvaluator,
)
from pyspark.ml.feature import StringIndexer, VectorAssembler
from pyspark.sql import SparkSession
from pyspark.sql.functions import col
from pyspark.sql.types import DoubleType
# Set the MLflow experiment
mlflow.set_experiment("/diabetes-prediction")
# Load the diabetes data
data = spark.read.csv(
"/FileStore/tables/diabetes_prediction_dataset.csv", inferSchema=True, header=True
)
# Convert the diabetes label to double
data = data.withColumn("diabetes", col("diabetes").cast(DoubleType()))
# Split the data into training and test sets
(trainingData, testData) = data.randomSplit([0.7, 0.3], seed=1234)
# Set the hyperparameters for the logistic regression model
lr_params = {"regParam": 0.01, "elasticNetParam": 0.0, "maxIter": 100}
# Train the model and make predictions on the test data
with mlflow.start_run():
# Convert categorical features to numeric using StringIndexer
GenderIndexer = StringIndexer(inputCol="gender", outputCol="genderIndex")
SmokeHistIndexer = StringIndexer(
inputCol="smoking_history", outputCol="smoking_statusIndex"
)
# Create the features vector using VectorAssembler
F_assembler = VectorAssembler(
inputCols=[
"genderIndex",
"age",
"hypertension",
"heart_disease",
"smoking_statusIndex",
"bmi",
"HbA1c_level",
"blood_glucose_level",
],
outputCol="features",
)
# Define the logistic regression model
lr = LogisticRegression(featuresCol="features", labelCol="diabetes")
# Define the pipeline
pipeline = Pipeline(stages=[GenderIndexer, SmokeHistIndexer, F_assembler, lr])
# Log the hyperparameters
mlflow.log_params(lr_params)
# Train the model
models = pipeline.fit(trainingData)
# Make predictions on the test data
predictions = models.transform(testData)
# Evaluate the model
auc_evaluator = BinaryClassificationEvaluator(
labelCol="diabetes", rawPredictionCol="prediction", metricName="areaUnderROC"
)
areaUnderROC = auc_evaluator.evaluate(predictions)
print("Area under ROC curve: {}".format(areaUnderROC))
acc_evaluator = MulticlassClassificationEvaluator(
predictionCol="prediction", labelCol="diabetes", metricName="accuracy"
)
accuracy = acc_evaluator.evaluate(predictions)
print("Accuracy: {:.2f}%".format(accuracy * 100))
# Log the metrics
mlflow.log_metric("Accuracy", accuracy * 100)
mlflow.log_metric("AUC", areaUnderROC)
# Log the model
mlflow.spark.log_model(models, "model")
# Get the ID of the current run
runid = mlflow.active_run().info.run_id
runid # runid contains the unique identifier of the active MLflow run
model_uri = (
"runs:/" + runid + "/model"
) # Here we are constructing the model URI using the run ID of the active run and the 'model' endpoint
model_name = "Diabetes_prediction_model" # Set the name for the registered model
model_details = mlflow.register_model(
model_uri=model_uri, name=model_name
) # Register the trained model with the name and URI specified
from mlflow.tracking.client import MlflowClient # Import the Mlflow client library
client = (
MlflowClient()
) # Create a client instance for accessing the Mlflow tracking server
client.transition_model_version_stage(
name=model_details.name, version=model_details.version, stage="Production"
) # Staging the registered model version to the production stage
import mlflow
# Load the saved MLflow model
loaded_model = mlflow.spark.load_model(model_uri)
# Create a new test dataset as a Spark dataframe
test = spark.createDataFrame(
[("Female", 39.0, 1, 0, "No Info", 79, 8.8, 145)],
[
"gender",
"age",
"hypertension",
"heart_disease",
"smoking_history",
"bmi",
"HbA1c_level",
"blood_glucose_level",
],
)
# Use the loaded model to make predictions on the new dataset
new_predictions = loaded_model.transform(test)
# Display the predicted output
new_predictions.show()
|
# Context
# Cardiovascular diseases (CVDs) are the number 1 cause of death globally, taking an estimated 17.9 million lives each year, which accounts for 31% of all deaths worldwide. Four out of 5CVD deaths are due to heart attacks and strokes, and one-third of these deaths occur prematurely in people under 70 years of age. Heart failure is a common event caused by CVDs and this dataset contains 11 features that can be used to predict a possible heart disease.
# People with cardiovascular disease or who are at high cardiovascular risk (due to the presence of one or more risk factors such as hypertension, diabetes, hyperlipidaemia or already established disease) need early detection and management wherein a machine learning model can be of great help.
# Attribute Information
# **Age:** age of the patient (years)
# **Sex:** sex of the patient (M: Male, F: Female)
# **ChestPainType:** chest pain type (TA: Typical Angina, ATA: Atypical Angina, NAP: Non-Anginal Pain, ASY: Asymptomatic)
# **RestingBP:** Resting blood pressure (mm Hg)
# **Cholesterol:** serum cholesterol (mm/dl)
# **FastingBS:** fasting blood sugar (1: if FastingBS > 120 mg/dl, 0: otherwise)
# **RestingECG:** resting electrocardiogram results (Normal: Normal, ST: having ST-T wave abnormality (T wave inversions and/or ST elevation or depression of > 0.05 mV), LVH: showing probable or definite left ventricular hypertrophy by Estes' criteria)
# **MaxHR:** maximum heart rate achieved (Numeric value between 60 and 202)
# **ExerciseAngina:** exercise-induced angina (Y: Yes, N: No)
# **Oldpeak:** oldpeak = ST (Numeric value measured in depression)
# **ST_Slope:** the slope of the peak exercise ST segment (Up: upsloping, Flat: flat, Down: downsloping)
# **HeartDisease:** output class (1: heart disease, 0: Normal)
# Table of content:
# [1. Importing The Dependencies..](#1)
# [2. Read Data from csv.](#2)
# [3. Data Stucture Analysis.](#3)
# * [3.1 Dataset shape and data type](#3.1)
# * [3.2 Numerical data describe](#3.2)
# * [3.3 Correlation between all features.](#3.3)
# [4. Handling null values and outliers.](#4)
# * [4.1 Null Values ](#4.1)
# * [4.2 Handle if there are any outliers](#4.2)
# [5. Data Visualization.](#5)
# * [5.1 Box plot of all features.](#5.1)
# * [5.2 Distribution of Numerical Features.](#5.2)
# * [5.3 Histogram of Numerical Features.](#5.3)
# * [5.4 Pair plot of Numerical Features.](#5.4)
# [6. Data preproessing.](#6)
# * [6.1 Checking whether data is unbalanced or not.](#6.1)
# * [6.2 Handling categorical variables.](#6.2)
# * [6.3 Splitting the Dataset.](#6.3)
# [7. Building the Models.](#7)
# * [7.1 Confusion Matrix drawing function.](#7.1)
# * [7.2 Decision Tree.](#7.2)
# * [7.3 Random Forest.](#7.3)
# * [7.4 XGBoost.](#7.4)
# ## 1 | Importing The Dependencies.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.model_selection import train_test_split
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
from xgboost import XGBClassifier
from sklearn.model_selection import GridSearchCV
from sklearn import metrics
from sklearn.metrics import accuracy_score
from sklearn.metrics import precision_recall_fscore_support
# ## 2 | Read Data from csv.
data = pd.read_csv("/kaggle/input/heart-failure-prediction/heart.csv")
data.head()
data.tail()
# ## 3 | Data Stucture Analysis
# Now we need to analyze data structure because data structure analysis plays an important role in various aspects of the modeling process, including data preprocessing, feature engineering, and model evaluation.
#
# During data preprocessing, data structure analysis can help in identifying and handling missing values, outliers, and other issues that may affect the quality of the data. By analyzing the data structure, developers can determine the appropriate techniques to use for data cleaning, normalization, and transformation, which can improve the accuracy and reliability of the model.
# 3.1 | Dataset shape and data type
data.shape
data.dtypes
data.info()
data.columns
# Note: We can see that our data has 918 rows and 12 columns. Sex,ChestPainType,RestingECG, ExerciseAngina, and ST_Slope are object time means that are ordinal categorical data others are numbers and our target variable is HeartDisease.
# 3.2 | Numerical data describe
# The describe() method in Pandas is used to get a quick statistical summary of a DataFrame. It calculates several summary statistics for each column in the DataFrame, including the count, mean, standard deviation, minimum value, 25th percentile, median (50th percentile), 75th percentile, and maximum value.
data.describe()
#
# Now we take a look at what describe method shows to us.
# Age: Maximum age is 77 and the minimum age is 28, the mean is 53.51 and 75 percent of the age is below 60.
# RestingBP: Maximum value of RestingBP is 200 and the minimum is 0 and 75 percent of the Bp is below 140.
# Cholesterol: Maximum value of Cholesterol is 603 and the minimum is 0 and 75 percent of the Cholesterol is below 267.
# FastingBS: Maximum value of FastingBSis is 1 and the minimum is 0 and 75 percent of the FastingBS is below 0.
# MaxHR: Maximum value of MaxHR is 202 and the minimum is 60 and 75 percent of the MaxHR is below 156.
# Oldpeak: Maximum value of Oldpeak is 6.2 and the minimum is -2.6 and 75 percent of the Oldpeak is below 1.5.
# 3.3 | Correlation between all features.
# Correlation is a statistical technique that measures the strength and direction of the relationship between two variables. In other words, it helps us understand how much one variable is related to another variable.
# A correlation could be positive, meaning both variables move in the same direction, or negative, meaning that when one variable’s value increases, the other variables’ values decrease. Correlation can also be neutral or zero, meaning that the variables are unrelated.
# Positive Correlation: both variables change in the same direction.
#
# Neutral Correlation: No relationship in the change of the variables.
#
# Negative Correlation: variables change in opposite directions.
#
corr = data.corr()
plt.figure(figsize=(15, 8))
mask = np.triu(np.ones_like(corr, dtype=bool))
sns.heatmap(
corr,
mask=mask,
cmap="BrBG",
vmin=-1,
vmax=1,
center=0,
annot=True,
linewidth=0.5,
square=False,
)
# Correlation between all features with heart disease.
plt.figure(figsize=(15, 8))
sns.heatmap(
data.corr()[["HeartDisease"]].sort_values(by="HeartDisease", ascending=False),
cmap="BrBG",
vmin=-1,
vmax=1,
center=0,
annot=True,
linewidth=0.5,
square=False,
)
# Note: We can see that Age, Oldpeak, RestingBP, and FastingBS all are positively correlated with heart disease, and Cholesterol, and MaxHR is negatively correlated.
# ## 4 | Handling null values and outliers.
# 4.1 | Null Values
data.isnull().sum()
#
# Note: We can see that there are no null values so we do not need to handle it.
# 4.2 | Handle if there are any outliers
# Including outliers in data driven models could be risky. The existence of an extreme single misleading value has the potential to change the conclusion implied by the model. It is therefore important to manage that kind of risk.
# There are many outlier detection algorithms available, including Z-Score, Modified Z-Score, Tukey's Method, and the standard deviation method.
#
# To use the standard deviation method for detecting outliers, you would typically follow these steps:
#
# 1. Calculate the mean (average) of the data set.
#
# 2. Calculate the standard deviation of the data set.
#
# 3. Determine a threshold for identifying outliers. One common approach is to use a multiple of the standard deviation, such as 2 or 3 standard deviations from the mean.
#
# 4. Identify any observations that are more than the threshold distance from the mean as outliers.
#
data.shape
means = data.mean()
std_devs = data.std()
# Define a threshold for identifying outliers (in this case, 2 standard deviations from the mean)
threshold = 4 * std_devs
# Identify any values that are more than the threshold distance from the mean as outliers
outliers = data[(np.abs(data - means) > threshold).any(axis=1)]
data = data.drop(outliers.index)
data.shape
# Histogram without outliers
column = ["Age", "RestingBP", "Cholesterol", "FastingBS", "MaxHR", "Oldpeak"]
plt.figure(figsize=(15, 12))
plt.title("Numerical Data with box plot")
for i, category in enumerate(column):
plt.subplot(2, 3, i + 1)
sns.scatterplot(data=data[category], color="#ffa64d")
plt.title("Histogram of {} without outliers".format(category))
plt.tight_layout()
plt.show()
# Histogram of outliers
column = ["Age", "RestingBP", "Cholesterol", "FastingBS", "MaxHR", "Oldpeak"]
plt.figure(figsize=(15, 12))
plt.title("Numerical Data with box plot")
for i, category in enumerate(column):
plt.subplot(2, 3, i + 1)
sns.scatterplot(data=outliers[category], color="#ffa64d")
plt.title("Histogram of {} outliers".format(category))
plt.tight_layout()
plt.show()
# ## 5 | Data Visualization.
# 5.1 | Box plot of all features.
column = ["Age", "RestingBP", "Cholesterol", "FastingBS", "MaxHR", "Oldpeak"]
plt.figure(figsize=(15, 12))
plt.title("Numerical Data with box plot")
for i, category in enumerate(column):
plt.subplot(2, 3, i + 1)
sns.boxplot(data=data[category], color="#ffa64d", orient="h")
plt.title(category)
plt.show()
# 5.2 | Distribution of Numerical Features.
column = ["Age", "RestingBP", "Cholesterol", "FastingBS", "MaxHR", "Oldpeak"]
plt.figure(figsize=(15, 12))
for i, category in enumerate(column):
plt.subplot(2, 3, i + 1)
sns.kdeplot(data=data, x=data[category], hue="HeartDisease", multiple="stack")
plt.title(category)
plt.show()
# 5.3 | Histogram of Numerical Features.
column = ["Age", "RestingBP", "Cholesterol", "FastingBS", "MaxHR", "Oldpeak"]
plt.figure(figsize=(15, 12))
for i, category in enumerate(column):
plt.subplot(2, 3, i + 1)
sns.histplot(data=data, x=data[category], hue="HeartDisease", multiple="stack")
plt.title(category)
plt.show()
# 5.4 | Pair plot of Numerical Features.
column = [
"Age",
"RestingBP",
"Cholesterol",
"FastingBS",
"MaxHR",
"Oldpeak",
"HeartDisease",
]
sns.pairplot(data=data[column], hue="HeartDisease", corner=True, diag_kind="hist")
plt.suptitle("Pairplot: Numerical Features ", fontsize=24)
plt.show()
# ## 6 | Data preproessing.
# 6.1 | Checking whether data is unbalanced or not.
heart_disease = data[data.HeartDisease == 1]
normal = data[data.HeartDisease == 0]
heart_disease_prcentence = []
heart_disease_prcentence.append((len(heart_disease) / len(data)) * 100)
heart_disease_prcentence.append((len(normal) / len(data)) * 100)
heart_disease_label = ["Heart Disease", "Normal"]
plt.figure(figsize=(5, 5))
plt.pie(
heart_disease_prcentence, labels=heart_disease_label, autopct="%1.1f%%", shadow=True
)
# Note: We can see that the data is balanced so we can leave it as it is.
# 6.2 | Handling categorical variables.
# We have some categorical variables available now we need to handle them. Now there is a function to take care of all categorical variables with pandas that is one hot encoding. one-hot encoding aims to transform a categorical variable with n outputs into n binary variables.
data = pd.get_dummies(data)
data.head()
# Note: Now we can see that categorical variables are converted to a numerical form.
# 6.3 | Splitting the Dataset.
# We need to choose input features and also target variable.
features = data.drop("HeartDisease", axis=1)
features.head()
target_variable = data["HeartDisease"]
target_variable.head()
# Splitting into train and validation set: By using similar data for training and testing, we can minimize the effects of data discrepancies and better understand the characteristics of the model. After a model has been processed by using the training set, we test the model by making predictions against the test set.
# So in this section, we will split our dataset into train and test datasets. We will use the function train_test_split from Scikit-learn.
X_train, X_val, y_train, y_val = train_test_split(
features, target_variable, train_size=0.8, random_state=55
)
print(f"train samples: {len(X_train)}")
print(f"validation samples: {len(X_val)}")
#
# ## 7 | Building the Models.
# 7.1 | Confusion Matrix drawing function.
def draw_confusion_matrix(y_test, test_predict):
confusion_matrix = metrics.confusion_matrix(y_test, test_predict)
cm_display = metrics.ConfusionMatrixDisplay(confusion_matrix=confusion_matrix)
cm_display.plot()
plt.show()
# 7.2 | Decision Tree.
# Decision Tree is the most powerful and popular tool for classification and prediction. A Decision tree is a flowchart-like tree structure, where each internal node denotes a test on an attribute, each branch represents an outcome of the test, and each leaf node (terminal node) holds a class label.
# There are several hyperparameters in the Decision Tree object from Scikit-learn.
# The hyperparameters we will use and investigate here are:
# criterion: The function to measure the quality of a split. Supported criteria are “gini” for the Gini impurity and “log_loss” and “entropy” both for the Shannon information gain.
# min_samples_split: The minimum number of samples required to split an internal node.
# Choosing a higher min_samples_split can reduce the number of splits and may help to reduce overfitting.
# max_depth: The maximum depth of the tree.
# Choosing a lower max_depth can reduce the number of splits and may help to reduce overfitting.
model = DecisionTreeClassifier(random_state=55)
# Find Best Parameter With Gridsearchcv
search_space_dtc = {
"criterion": ["gini", "entropy"],
"max_depth": [1, 2, 3, 4, 8, 16, 32, 64, None],
"min_samples_split": [2, 10, 30, 50, 100, 200, 300, 700],
}
gs = GridSearchCV(
estimator=model,
param_grid=search_space_dtc,
scoring=["r2", "neg_root_mean_squared_error"],
refit="r2",
cv=5,
)
gs.fit(X_train, y_train)
gs.best_params_
X_train_prediction = gs.best_estimator_.predict(X_train)
training_data_accuracy = accuracy_score(X_train_prediction, y_train)
print("Accuracy on Training data : ", training_data_accuracy)
X_test_prediction = gs.best_estimator_.predict(X_val)
test_data_accuracy = accuracy_score(X_test_prediction, y_val)
print("Accuracy score on Test Data : ", test_data_accuracy)
draw_confusion_matrix(y_val, X_test_prediction)
precision_recall_fscore_support(y_val, X_test_prediction, average="macro")
# 7.3 | Random Forest.
# The random forest is a classification algorithm consisting of many decisions trees. It uses bagging and feature randomness when building each individual tree to try to create an uncorrelated forest of trees whose prediction by committee is more accurate than that of any individual tree.
# Now let's try the Random Forest algorithm also, using the Scikit-learn implementation.
# All of the hyperparameters found in the decision tree model will also exist in this algorithm, since a random forest is an ensemble of many Decision Trees.
# One additional hyperparameter for Random Forest is called n_estimators which is the number of Decision Trees that make up the Random Forest.
random_forest_model = RandomForestClassifier()
# Find Best Parameter With Gridsearchcv
search_space_rfc = {
"n_estimators": [100, 200, 300],
"max_depth": [2, 3, 4, 5, 6],
"min_samples_split": [2, 10, 30, 50],
}
gs_rfc = GridSearchCV(
estimator=random_forest_model,
param_grid=search_space_rfc,
scoring=["r2", "neg_root_mean_squared_error"],
refit="r2",
cv=5,
)
gs_rfc.fit(X_train, y_train)
gs_rfc.best_params_
X_train_prediction = gs_rfc.best_estimator_.predict(X_train)
training_data_accuracy = accuracy_score(X_train_prediction, y_train)
print("Accuracy on Training data : ", training_data_accuracy)
X_test_prediction = gs_rfc.best_estimator_.predict(X_val)
test_data_accuracy = accuracy_score(X_test_prediction, y_val)
print("Accuracy score on Test Data : ", test_data_accuracy)
draw_confusion_matrix(y_val, X_test_prediction)
precision_recall_fscore_support(y_val, X_test_prediction, average="macro")
# 7.3 | XGBoost.
# Next is the Gradient Boosting model, called XGBoost. The boosting methods train several trees, but instead of them being uncorrelated to each other, now the trees are fit one after the other in order to minimize the error.
# The model has the same parameters as a decision tree, plus the learning rate.
# The learning rate is the size of the step on the Gradient Descent method that the XGBoost uses internally to minimize the error on each train step.
# One interesting thing about the XGBoost is that during fitting, it can take in an evaluation dataset of the form (X_val,y_val).
xgb_model = XGBClassifier(
n_estimators=500, learning_rate=0.1, verbosity=1, random_state=55
)
xgb_model.fit(X_train, y_train, eval_set=[(X_val, y_val)], early_stopping_rounds=10)
# Even though we initialized the model to allow up to 500 estimators, the algorithm only fit 45 estimators.
# To see why, let's look for the round of training that had the best performance (lowest evaluation metric). We can either view the validation log loss metrics that were output above, or view the model's .best_iteration attribute:
xgb_model.best_iteration
# The best round of training was round 35, with a log loss of 0.30799.
X_train_prediction = xgb_model.predict(X_train)
training_data_accuracy = accuracy_score(X_train_prediction, y_train)
print("Accuracy on Training data : ", training_data_accuracy)
X_test_prediction = xgb_model.predict(X_val)
test_data_accuracy = accuracy_score(X_test_prediction, y_val)
print("Accuracy score on Test Data : ", test_data_accuracy)
draw_confusion_matrix(y_val, X_test_prediction)
precision_recall_fscore_support(y_val, X_test_prediction, average="macro")
|
# # Clustering Mall Customers into socio-economic classes using K-Means
# ### Arkaprabha Poddar
# #### Importing Libraries
# for data manipulation
import numpy as np
import pandas as pd
# for model making
from sklearn.cluster import KMeans
# for data visualisation
import matplotlib.pyplot as plt
import seaborn as sns
import plotly as py
import plotly.graph_objs as go
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# #### Loading the dataset
df = pd.read_csv(
"/kaggle/input/customer-segmentation-tutorial-in-python/Mall_Customers.csv"
)
print(df.shape)
df.head()
# #### Data summary, descriptive statistics and checking null values
df.info()
df.describe()
df.isnull().sum()
# #### Checking the distribution of numerical variables
df.columns
cols = ["Age", "Annual Income (k$)", "Spending Score (1-100)"]
plt.style.use("fivethirtyeight")
plt.figure(figsize=(15, 7))
n = 0
for x in cols:
n = n + 1
plt.subplot(1, 3, n)
sns.distplot(df[x], bins=20)
plt.title("Distplot of {}".format(x))
plt.show()
# #### Plotting number of male and female customers
sns.countplot(y="Gender", data=df)
# #### Plotting relationship betweem numerical variables
plt.figure(1, figsize=(15, 7))
n = 0
for x in cols:
for y in cols:
n = n + 1
plt.subplot(3, 3, n)
plt.subplots_adjust(hspace=0.5, wspace=0.5)
sns.regplot(x=x, y=y, data=df)
plt.show()
# #### Plotting relationship between numerical variables with Gender
# Plotting Age vs Annual Income w.r.t Gender
plt.figure(1, figsize=(15, 6))
for gender in ["Male", "Female"]:
plt.scatter(
x="Age",
y="Annual Income (k$)",
data=df[df["Gender"] == gender],
s=100,
alpha=0.5,
label=gender,
)
plt.xlabel("Age"), plt.ylabel("Annual Income (k$)")
plt.title("Age vs Annual Income w.r.t Gender")
plt.legend()
plt.show()
# Plotting Annual Income vs Spending Score w.r.t Gender
plt.figure(1, figsize=(15, 6))
for gender in ["Male", "Female"]:
plt.scatter(
x="Annual Income (k$)",
y="Spending Score (1-100)",
data=df[df["Gender"] == gender],
s=200,
alpha=0.5,
label=gender,
)
plt.xlabel("Annual Income (k$)"), plt.ylabel("Spending Score (1-100)")
plt.title("Annual Income vs Spending Score w.r.t Gender")
plt.legend()
plt.show()
# #### Choosing the variables Annual Income and Spending Score to cluster the data
X = df.iloc[:, [3, 4]].values
X
# #### Using the elbow method to determine the number of clusters
wcss = []
for i in range(1, 11):
kmeans = KMeans(n_clusters=i, init="k-means++", random_state=42)
kmeans.fit(X)
wcss.append(kmeans.inertia_)
plt.plot(range(1, 11), wcss)
plt.title("The Elbow Method")
plt.xlabel("Number of clusters")
plt.ylabel("WCSS")
plt.show()
# ***Here, after 5 clusters, there is no significant decrease in the WCSS of the datapoints. Hence, we ccan assume value of K to be 5.***
# #### Fitting K-means model with 5 clusters
kmeans = KMeans(n_clusters=5, init="k-means++", random_state=42)
y_kmeans = kmeans.fit_predict(X)
plt.figure(figsize=(10, 7))
plt.scatter(
X[y_kmeans == 0, 0],
X[y_kmeans == 0, 1],
s=100,
c="darkorange",
label="Middle Class",
)
plt.scatter(
X[y_kmeans == 1, 0],
X[y_kmeans == 1, 1],
s=100,
c="mediumseagreen",
label="Frugal Upper Class",
)
plt.scatter(
X[y_kmeans == 2, 0],
X[y_kmeans == 2, 1],
s=100,
c="gold",
label="Lower Middle Classs",
)
plt.scatter(
X[y_kmeans == 3, 0],
X[y_kmeans == 3, 1],
s=100,
c="darkturquoise",
label="Thrifty Lower Middle Class",
)
plt.scatter(
X[y_kmeans == 4, 0], X[y_kmeans == 4, 1], s=100, c="navy", label="Upper Class"
)
plt.scatter(
kmeans.cluster_centers_[:, 0],
kmeans.cluster_centers_[:, 1],
s=150,
c="darkred",
label="Centroids",
)
plt.title("Clusters of Customers")
plt.xlabel("Annual Income (k$)")
plt.ylabel("Spending Score (1-100)")
plt.legend()
plt.show()
|
# ## 1. Постановка задачи
#
#
# Вам предстоит решить настоящую задачу машинного обучения, направленную на автоматизацию бизнес процессов. Мы построим модель, которая будет предсказывать общую продолжительность поездки такси в Нью-Йорке.
# Представьте вы заказываете такси из одной точки Нью-Йорка в другую, причем не обязательно конечная точка должна находиться в пределах города. Сколько вы должны будете за нее заплатить? Известно, что стоимость такси в США рассчитывается на основе фиксированной ставки + тарифная стоимость, величина которой зависит от времени и расстояния. Тарифы варьируются в зависимости от города.
# В свою очередь время поездки зависит от множества факторов таких как, откуда и куда вы едете, в какое время суток вы совершаете вашу поездку, погодных условий и так далее.
# Таким образом, если мы разработаем алгоритм, способный определять длительность поездки, мы сможем прогнозировать ее стоимость самым тривиальным образом, например, просто умножая стоимость на заданный тариф.
# Сервисы такси хранят огромные объёмы информации о поездках, включая такие данные как конечная, начальная точка маршрута, дата поездки и ее длительность. Эти данные можно использовать для того, чтобы прогнозировать длительность поездки в автоматическом режиме с привлечением искусственного интеллекта.
# **Бизнес-задача:** определить характеристики и с их помощью спрогнозировать длительность поездки такси.
# **Техническая задача для вас как для специалиста в Data Science:** построить модель машинного обучения, которая на основе предложенных характеристик клиента будет предсказывать числовой признак - время поездки такси. То есть решить задачу регрессии.
# **Основные цели проекта:**
# 1. Сформировать набор данных на основе нескольких источников информации
# 2. Спроектировать новые признаки с помощью Feature Engineering и выявить наиболее значимые при построении модели
# 3. Исследовать предоставленные данные и выявить закономерности
# 4. Построить несколько моделей и выбрать из них наилучшую по заданной метрике
# 5. Спроектировать процесс предсказания времени длительности поездки для новых данных
# Загрузить свое решение на платформу Kaggle, тем самым поучаствовав в настоящем Data Science соревновании.
# Во время выполнения проекта вы отработаете навыки работы с несколькими источниками данных, генерации признаков, разведывательного анализа и визуализации данных, отбора признаков и, конечно же, построения моделей машинного обучения!
# ## 2. Знакомство с данными, базовый анализ и расширение данных
# Начнём наше исследование со знакомства с предоставленными данными. А также подгрузим дополнительные источники данных и расширим наш исходный датасет.
# Заранее импортируем модули, которые нам понадобятся для решения задачи:
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import plotly.express as px
import warnings
from scipy import stats
from sklearn import linear_model
from sklearn import preprocessing
from sklearn import model_selection
from sklearn import tree
from sklearn import ensemble
from sklearn import metrics
from sklearn import cluster
from sklearn import feature_selection
plt.style.use("seaborn")
warnings.filterwarnings(action="ignore")
# Прочитаем наш файл с исходными данными:
taxi_data = pd.read_csv("Documents/taxi_train_data.csv")
print("Train data shape: {}".format(taxi_data.shape))
taxi_data.head()
# Итак, у нас с вами есть данные о почти 1.5 миллионах поездок и 11 характеристиках, которые описывают каждую из поездок.
# Мы условно разделили признаки нескольких групп. Каждой из групп мы в дальнейшем уделим отдельное внимание.
# **Данные о клиенте и таксопарке:**
# * id - уникальный идентификатор поездки
# * vendor_id - уникальный идентификатор поставщика (таксопарка), связанного с записью поездки
# **Временные характеристики:**
# * pickup_datetime - дата и время, когда был включен счетчик поездки
# * dropoff_datetime - дата и время, когда счетчик был отключен
# **Географическая информация:**
# * pickup_longitude - долгота, на которой был включен счетчик
# * pickup_latitude - широта, на которой был включен счетчик
# * dropoff_longitude - долгота, на которой счетчик был отключен
# * dropoff_latitude - широта, на которой счетчик был отключен
# **Прочие признаки:**
# * passenger_count - количество пассажиров в транспортном средстве (введенное водителем значение)
# * store_and_fwd_flag - флаг, который указывает, сохранилась ли запись о поездке в памяти транспортного средства перед отправкой поставщику. Y - хранить и пересылать, N - не хранить и не пересылать поездку.
# **Целевой признак:**
# * trip_duration - продолжительность поездки в секундах
# Для начала мы проведем базовый анализ того, насколько данные готовы к дальнейшей предобработке и анализу.
# ### Задание 2.1
# Для начала посмотрим на временные рамки, в которых мы работаем с данными.
# Переведите признак pickup_datetime в тип данных datetime с форматом год-месяц-день час:минута:секунда (в функции pd.to_datetime() параметр format='%Y-%m-%d %H:%M:%S').
# Определите временные рамки (без учета времени), за которые представлены данные.
# преобразовываем признаки в формат datetime для удобной работы
taxi_data["pickup_datetime"] = pd.to_datetime(
taxi_data["pickup_datetime"], format="%Y-%m-%d %H:%M:%S"
)
taxi_data["dropoff_datetime"] = pd.to_datetime(
taxi_data["dropoff_datetime"], format="%Y-%m-%d %H:%M:%S"
)
print(taxi_data["pickup_datetime"].min())
print(taxi_data["pickup_datetime"].max())
# ### Задание 2.2
# Посмотрим на пропуски.
# Сколько пропущенных значений присутствует в данных (суммарно по всем столбцам таблицы)?
taxi_data.isnull().sum()
# ### Задание 2.3
# Посмотрим на статистические характеристики некоторых признаков.
# а) Сколько уникальных таксопарков присутствует в данных?
# б) Каково максимальное количество пассажиров?
# в) Чему равна средняя и медианная длительность поездки? Ответ приведите в секундах и округлите до целого.
# г) Чему равно минимальное и максимальное время поездки (в секундах)?
#
round(taxi_data.describe())
# Займемся расширением исходного набора данных как с помощью внешних источников, так и с помощью манипуляций над имеющимися в данных признаками.
# ### Задание 2.4
# Реализуйте функцию add_datetime_features(), которая принимает на вход таблицу с данными о поездках (DataFrame) и возвращает ту же таблицу с добавленными в нее 3 столбцами:
# * pickup_date - дата включения счетчика - начала поездки (без времени);
# * pickup_hour - час дня включения счетчика;
# * pickup_day_of_week - наименование дня недели, в который был включен счетчик.
# а) Сколько поездок было совершено в субботу?
# б) Сколько поездок в среднем совершается в день? Ответ округлите до целого
# реализуем функцию
def add_datetime_features(df):
"""
Функция добавляющая в таблицу с данными о поездках:
- дату включения счётчика
- час влключеня счётчика
- день недели включения счётчика
Args:
df (DataFrame): исходный набор данных
Returns:
df (DataFrame): дополненный набор данных
"""
df["pickup_date"] = df["pickup_datetime"].dt.date
df["pickup_hour"] = df["pickup_datetime"].dt.hour
df["pickup_day_of_week"] = df["pickup_datetime"].dt.dayofweek
return df
# добавляем данные
taxi_data = add_datetime_features(taxi_data)
# рассчитываем показатели
mask = taxi_data["pickup_day_of_week"] == 5
print(taxi_data[mask].shape[0], "поездок было совершено в субботу")
mean_per_day = taxi_data.shape[0] / taxi_data["pickup_date"].nunique()
print(round(mean_per_day), "поездок в среднем совершается в день")
# ### Задание 2.5
# Реализуйте функцию add_holiday_features(), которая принимает на вход две таблицы:
# * таблицу с данными о поездках;
# * таблицу с данными о праздничных днях;
# и возвращает обновленную таблицу с данными о поездках с добавленным в нее столбцом pickup_holiday - бинарным признаком того, начата ли поездка в праздничный день или нет (1 - да, 0 - нет).
# Чему равна медианная длительность поездки на такси в праздничные дни? Ответ приведите в секундах, округлив до целого.
#
# загружаем файл с праздничными датами
holiday_data = pd.read_csv("Documents/holiday_data.csv", sep=";")
# реализуем функцию
def add_holiday_features(df_taxi, df_holiday):
"""
Функция добавляющая в таблицу с данными о поездках
бинарный признак - начата ли поездка в праздничный день.
Args:
df_taxi (DataFrame): таблица с данными о поездках
df_holiday (DataFrame): таблица с праздничными датами
Returns:
df_taxi (DataFrame): дополненный набор данных
"""
df_taxi["pickup_holiday"] = df_taxi["pickup_date"].apply(
lambda x: 1 if str(x) in df_holiday["date"].values else 0
)
return df_taxi
# добавляем данные
taxi_data = add_holiday_features(taxi_data, holiday_data)
# вычисляем длительность поездки
mask = taxi_data["pickup_holiday"] == 1
print(
"Медианная длительность поезди в праздничные дни:",
taxi_data[mask]["trip_duration"].median(),
)
# ### Задание 2.6
# Реализуйте функцию add_osrm_features(), которая принимает на вход две таблицы:
# * таблицу с данными о поездках;
# * таблицу с данными из OSRM;
# и возвращает обновленную таблицу с данными о поездках с добавленными в нее 3 столбцами:
# * total_distance;
# * total_travel_time;
# * number_of_steps.
# а) Чему равна разница (в секундах) между медианной длительностью поездки в данных и медианной длительностью поездки, полученной из OSRM?
# В результате объединения таблиц у вас должны были получиться пропуски в столбцах с информацией из OSRM API. Это связано с тем, что для некоторых поездок не удалось выгрузить данные из веб источника.
# б) Сколько пропусков содержится в столбцах с информацией из OSRM API после объединения таблиц?
# загружаем файл с праздничными датами
osrm_data = pd.read_csv("Documents/osrm_data_train.csv")
# реализуем функцию
def add_osrm_features(df_taxi, df_osrm):
"""
Функция добавляющая в таблицу с данными о поездках данные из OSRM:
- кратчайшее дорожное расстояние
- наименьшее время поездки
- количество дискретных шагов
Args:
df_taxi (DataFrame): таблица с данными о поездках
df_osrm (DataFrame): данные из OSRM (Open Source Routing Machine)
Returns:
df_taxi (DataFrame): дополненный набор данных
"""
df_osrm = df_osrm[["id", "total_distance", "total_travel_time", "number_of_steps"]]
return df_taxi.merge(df_osrm, on="id", how="left")
# добавляем данные
taxi_data = add_osrm_features(taxi_data, osrm_data)
# рассчитываем показатели
median_time_delta = (
taxi_data["trip_duration"].median() - taxi_data["total_travel_time"].median()
)
print("Разница в секундах:", round(median_time_delta))
cols_null_count = taxi_data.isnull().sum()
cols_with_null = cols_null_count[cols_null_count > 0]
print("\n", "Количество пропусков по колонкам:\n", cols_with_null)
# убедимся, что все 3 пропуска находятся в 1 строке
mask = taxi_data["total_distance"].isnull()
display(taxi_data[mask])
def get_haversine_distance(lat1, lng1, lat2, lng2):
"""
Функция для вычисления расстояния Хаверсина (в километрах)
Args:
lat1 и lng1 (array): векторы-столбцы с широтой и долготой первой точки
lat2 и lng2 (array): векторы-столбцы с широтой и долготой второй точки
Returns:
h (float): расстояние между двумя точками на сфере (в километрах)
"""
# переводим углы в радианы
lat1, lng1, lat2, lng2 = map(np.radians, (lat1, lng1, lat2, lng2))
# радиус земли в километрах
EARTH_RADIUS = 6371
# считаем кратчайшее расстояние h по формуле Хаверсина
lat_delta = lat2 - lat1
lng_delta = lng2 - lng1
d = (
np.sin(lat_delta * 0.5) ** 2
+ np.cos(lat1) * np.cos(lat2) * np.sin(lng_delta * 0.5) ** 2
)
h = 2 * EARTH_RADIUS * np.arcsin(np.sqrt(d))
return h
def get_angle_direction(lat1, lng1, lat2, lng2):
"""
Функция для вычисления угла направления движения (в градусах)
Args:
lat1 и lng1 (array): векторы-столбцы с широтой и долготой первой точки
lat2 и lng2 (array): векторы-столбцы с широтой и долготой второй точки
Returns:
alpha (float): угол направления движения от первой точки ко второй
"""
# переводим углы в радианы
lat1, lng1, lat2, lng2 = map(np.radians, (lat1, lng1, lat2, lng2))
# считаем угол направления движения alpha по формуле угла пеленга
lng_delta_rad = lng2 - lng1
y = np.sin(lng_delta_rad) * np.cos(lat2)
x = np.cos(lat1) * np.sin(lat2) - np.sin(lat1) * np.cos(lat2) * np.cos(
lng_delta_rad
)
alpha = np.degrees(np.arctan2(y, x))
return alpha
# ### Задание 2.7.
# Реализуйте функцию add_geographical_features(), которая принимает на вход таблицу с данными о поездках и возвращает обновленную таблицу с добавленными в нее 2 столбцами:
# * haversine_distance - расстояние Хаверсина между точкой, в которой был включен счетчик, и точкой, в которой счетчик был выключен;
# * direction - направление движения из точки, в которой был включен счетчик, в точку, в которой счетчик был выключен.
# Чему равно медианное расстояние Хаверсина поездок (в киллометрах)? Ответ округлите до сотых.
#
# реализуем функцию
def add_geographical_features(df):
"""
Функция добавляющая в таблицу с данными о поездках:
- расстояние Хаверсина
- направление движения
Args:
df (DataFrame): исходный набор данных
Returns:
df (DataFrame): дополненный набор данных
"""
df["haversine_distance"] = get_haversine_distance(
lat1=df["pickup_latitude"],
lng1=df["pickup_longitude"],
lat2=df["dropoff_latitude"],
lng2=df["dropoff_longitude"],
)
df["direction"] = get_angle_direction(
lat1=df["pickup_latitude"],
lng1=df["pickup_longitude"],
lat2=df["dropoff_latitude"],
lng2=df["dropoff_longitude"],
)
return df
taxi_data = add_geographical_features(taxi_data)
print(
"Медианное расстояние Хаверсина:",
round(taxi_data["haversine_distance"].median(), 2),
"км",
)
# ### Задание 2.8.
# Реализуйте функцию add_cluster_features(), которая принимает на вход таблицу с данными о поездках и обученный алгоритм кластеризации. Функция должна возвращать обновленную таблицу с добавленными в нее столбцом geo_cluster - географический кластер, к которому относится поездка.
# Сколько поездок содержится в наименьшем по размеру географическом кластере?
#
# создаем обучающую выборку из географических координат всех точек
coords = np.hstack(
(
taxi_data[["pickup_latitude", "pickup_longitude"]],
taxi_data[["dropoff_latitude", "dropoff_longitude"]],
)
)
# обучаем алгоритм кластеризации
kmeans = cluster.KMeans(n_clusters=10, random_state=42)
kmeans.fit(coords)
# реализуем функцию
def add_cluster_features(df, cluster):
"""
Функция добавляющая в таблицу с данными о поездках
географический кластер, к которому относится поездка.
Args:
df (DataFrame): исходный набор данных
cluster (KMeans): обученный алгоритм кластеризации
Returns:
df (DataFrame): дополненный набор данных
"""
df["geo_cluster"] = cluster.predict(
df[
[
"pickup_latitude",
"pickup_longitude",
"dropoff_latitude",
"dropoff_longitude",
]
]
)
return df
# генерируем новый признак
taxi_data = add_cluster_features(taxi_data, kmeans)
# проверяем получившиеся кластеры
taxi_data["geo_cluster"].value_counts().sort_values(ascending=False)
# ### Задание 2.9.
# Реализуйте функцию add_weather_features(), которая принимает на вход две таблицы:
# * таблицу с данными о поездках;
# * таблицу с данными о погодных условиях на каждый час;
# и возвращает обновленную таблицу с данными о поездках с добавленными в нее 5 столбцами:
# * temperature - температура;
# * visibility - видимость;
# * wind speed - средняя скорость ветра;
# * precip - количество осадков;
# * events - погодные явления.
# а) Сколько поездок было совершено в снежную погоду?
# В результате объединения у вас должны получиться записи, для которых в столбцах temperature, visibility, wind speed, precip, и events будут пропуски. Это связано с тем, что в таблице с данными о погодных условиях отсутствуют измерения для некоторых моментов времени, в которых включался счетчик поездки.
# б) Сколько процентов от общего количества наблюдений в таблице с данными о поездках занимают пропуски в столбцах с погодными условиями? Ответ приведите с точностью до сотых процента.
#
# загружаем данные о погоде
weather_data = pd.read_csv("Documents/weather_data.csv")
# реализуем функцию
def add_weather_features(df_taxi, df_weather):
"""
Функция добавляющая в таблицу с данными о поездках информацию о погоде.
Args:
df_taxi (DataFrame): таблица с данными о поездках
df_weather (DataFrame): таблица с данными о погодных условиях на каждый час
Returns:
df_new (DataFrame): дополненный набор данных
"""
# извлекаем дату и час, оставляем только нужные данные о погоде
df_weather["time"] = pd.to_datetime(df_weather["time"], format="%Y-%m-%d %H:%M:%S")
df_weather["date"] = df_weather["time"].dt.date
df_weather["hour"] = df_weather["time"].dt.hour
df_weather = df_weather[
["temperature", "visibility", "wind speed", "precip", "events", "date", "hour"]
]
# объединяем данные
df_merged = df_taxi.merge(
df_weather,
how="left",
left_on=["pickup_date", "pickup_hour"],
right_on=["date", "hour"],
)
df_new = df_merged.drop(["date", "hour"], axis=1)
return df_new
# применяем полученную функцию к таблице
taxi_data = add_weather_features(taxi_data, weather_data)
mask = taxi_data["events"] == "Snow"
print(taxi_data[mask].shape[0], "поездок было совершено в снежную погоду")
print("-" * 100)
cols_null_perc = taxi_data.isnull().mean() * 100
cols_with_null = cols_null_perc[cols_null_perc > 0].sort_values(ascending=False)
print("Пропуски в процентах:\n", round(cols_with_null, 2))
# ### Задание 2.10.
# Реализуйте функцию fill_null_weather_data(), которая принимает на вход которая принимает на вход таблицу с данными о поездках. Функция должна заполнять пропущенные значения в столбцах.
# Пропуски в столбцах с погодными условиями - temperature, visibility, wind speed, precip заполните медианным значением температуры, влажности, скорости ветра и видимости в зависимости от даты начала поездки. Для этого сгруппируйте данные по столбцу pickup_date и рассчитайте медиану в каждой группе, после чего с помощью комбинации методов transform() и fillna() заполните пропуски.
# Пропуски в столбце events заполните строкой 'None' - символом отсутствия погодных явлений (снега/дождя/тумана).
# Пропуски в столбцах с информацией из OSRM API - total_distance, total_travel_time и number_of_steps заполните медианным значением по столбцам.
# Чему равна медиана в столбце temperature после заполнения пропусков? Ответ округлите до десятых.
#
# реализуем функцию
def fill_null_weather_data(df):
"""
Функция заполняет пропущенные значения в столбцах.
Args:
df (DataFrame): таблица с данными о поездках
Returns:
df (DataFrame): таблица с заполненными пропусками
"""
# заполняем пропуски в столбцах с погодными условиями медианным значением
weather_cols = ["temperature", "visibility", "wind speed", "precip"]
for col in weather_cols:
df[col] = df[col].fillna(df.groupby("pickup_date")[col].transform("median"))
# заполняем пропуски в столбце 'events' строкой 'None'
df["events"] = df["events"].fillna("None")
# заполняем пропуски в столбцах с информацией из OSRM медианным значением
values = {
"total_distance": df["total_distance"].median(),
"total_travel_time": df["total_travel_time"].median(),
"number_of_steps": df["number_of_steps"].median(),
}
df = df.fillna(values)
return df
# применяем созданную функцию к таблице с данными о поездках
taxi_data = fill_null_weather_data(taxi_data)
print("Медиана в столбце temperature:", round(taxi_data["temperature"].median(), 1))
# В завершение первой части найдем очевидные выбросы в целевой переменной - длительности поездки.
# Проще всего найти слишком продолжительные поездки. Давайте условимся, что выбросами будут считаться поездки, длительность которых превышает 24 часа.
# Чуть сложнее с анализом поездок, длительность которых слишком мала. Потому что к ним относятся действительно реальные поездки на короткие расстояния, поездки, которые были отменены через секунду после того как включился счетчик, а также “телепортации” - перемещение на большие расстояния за считанные секунды.
# Условимся, что мы будем считать выбросами только последнюю группу. Как же нам их обнаружить наиболее простым способом?
# Можно воспользоваться информацией о кратчайшем расстоянии, которое проезжает такси. Вычислить среднюю скорость автомобиля на кратчайшем пути следующим образом:
# $$avg\_speed= \frac{total\_distance}{1000*trip\_duration}*3600$$
# Если мы построим диаграмму рассеяния средней скорости движения автомобилей, мы увидим следующую картину:
#
avg_speed = taxi_data["total_distance"] / taxi_data["trip_duration"] * 3.6
fig, ax = plt.subplots(figsize=(10, 5))
sns.scatterplot(x=avg_speed.index, y=avg_speed, ax=ax)
ax.set_xlabel("Index")
ax.set_ylabel("Average speed")
# Как раз отсюда мы видим, что у нас есть “поездки-телепортации”, для которых средняя скорость более 1000 км/ч. Даже есть такая, средняя скорость которой составляла более 12000 км/ч!
# Давайте условимся, что предельная средняя скорость, которую могут развивать таксисты будет 300 км/ч.
# ### Задание 2.11.
# Найдите поездки, длительность которых превышает 24 часа. И удалите их из набора данных.
# а) Сколько выбросов по признаку длительности поездки вам удалось найти?
# Найдите поездки, средняя скорость которых по кратчайшему пути превышает 300 км/ч и удалите их из данных.
# б) Сколько выбросов по признаку скорости вам удалось найти?
print(
"Количество выбросов по признаку длительности поездки:",
taxi_data[taxi_data["trip_duration"] > 86400].shape[0],
)
print("Количество выбросов по признаку скорости:", avg_speed[avg_speed > 300].shape[0])
mask_1 = taxi_data["trip_duration"] <= 86400
mask_2 = (taxi_data["total_distance"] / taxi_data["trip_duration"] * 3.6) <= 300
taxi_data = taxi_data[mask_1 & mask_2]
# ## 3. Разведывательный анализ данных (EDA)
# В этой части нашего проекта мы с вами:
# * Исследуем сформированный набор данных;
# * Попробуем найти закономерности, позволяющие сформулировать предварительные гипотезы относительно того, какие факторы являются решающими в определении длительности поездки;
# * Дополним наш анализ визуализациями, иллюстрирующими; исследование. Постарайтесь оформлять диаграммы с душой, а не «для галочки»: навыки визуализации полученных выводов обязательно пригодятся вам в будущем.
# Начинаем с целевого признака. Забегая вперед, скажем, что основной метрикой качества решения поставленной задачи будет RMSLE - Root Mean Squared Log Error, которая вычисляется на основе целевой переменной в логарифмическом масштабе. В таком случае целесообразно сразу логарифмировать признак длительности поездки и рассматривать при анализе логарифм в качестве целевого признака:
# $$trip\_duration\_log = log(trip\_duration+1),$$
# где под символом log подразумевается натуральный логарифм.
#
taxi_data["trip_duration_log"] = np.log(taxi_data["trip_duration"] + 1)
# ### Задание 3.1.
# Постройте гистограмму и коробчатую диаграмму длительности поездок в логарифмическом масштабе (trip_duration_log).
# Исходя из визуализации, сделайте предположение, является ли полученное распределение нормальным?
# Проверьте свою гипотезу с помощью теста Д’Агостино при уровне значимости $\alpha=0.05$.
# а) Чему равен вычисленный p-value? Ответ округлите до сотых.
# б) Является ли распределение длительности поездок в логарифмическом масштабе нормальным?
# строим гистограмму и коробчатую диаграмму
fig, axes = plt.subplots(nrows=1, ncols=2, figsize=(15, 4))
histplot = sns.histplot(data=taxi_data, x="trip_duration_log", ax=axes[0])
histplot.set_title("Trip Duration Logarithmic Distribution")
boxplot = sns.boxplot(data=taxi_data, x="trip_duration_log", ax=axes[1])
boxplot.set_title("Trip Duration Logarithmic Boxplot")
# формируем нулевую и альтернативную гипотезы для проверки данных на нормальность
H0 = "Данные распределены нормально"
Ha = "Распределение отлично от нормального"
# устанавливаем уровень значимости
alpha = 0.05
# тест Д'Агостино (возвращает двустороннюю вероятность для проверки гипотезы)
_, p = stats.normaltest(taxi_data["trip_duration_log"])
print(f"P-value = {round(p, 2)}")
# интерпритация полученного результата
if (
p > alpha / 2
): # p-value рассчитано для двусторонней гипотезы, поэтому уровень значимости делим на 2
print(H0)
else:
print(Ha)
# ### Задание 3.2.
# Постройте визуализацию, которая позволит сравнить распределение длительности поездки в логарифмическом масштабе (trip_duration_log) в зависимости от таксопарка (vendor_id).
# Сравните два распределения между собой.
# преобразуем признак vendor_id в категориальный тип данных
taxi_data["vendor_id"] = taxi_data["vendor_id"].astype("category")
# строим гистограмму и коробчатую диаграмму
fig, axes = plt.subplots(nrows=2, ncols=1, figsize=(15, 4))
histplot = sns.histplot(
data=taxi_data, x="trip_duration_log", y="vendor_id", ax=axes[0]
)
histplot.set_title("Logarithmic Trip Duration Dependence on Vendor ID")
boxplot = sns.boxplot(data=taxi_data, x="trip_duration_log", y="vendor_id", ax=axes[1])
# #### Выводы
# * Распределения в группах практически не отличаются, признак vendor_id не имеет значения при определении длительности поездки.
# ### Задание 3.3.
# Постройте визуализацию, которая позволит сравнить распределение длительности поездки в логарифмическом масштабе (trip_duration_log) в зависимости от признака отправки сообщения поставщику (store_and_fwd_flag).
# Сравните два распределения между собой.
# на этот раз вместо boxplot построим violinplot
fig, axes = plt.subplots(nrows=2, ncols=1, figsize=(15, 4))
histplot = sns.histplot(
data=taxi_data, x="trip_duration_log", y="store_and_fwd_flag", ax=axes[0]
)
histplot.set_title("Logarithmic Trip Duration Dependence on Store and fwd Flag")
violinplot = sns.violinplot(
data=taxi_data, x="trip_duration_log", y="store_and_fwd_flag", ax=axes[1]
)
# #### Выводы
# * Распределения в группах значительно отличаются, прзнак store_and_fwd_flag имеет значение при определении длительности поездки.
# * Наиболее длительные поездки принадлежат к группе N (не хранить и не пересылать поездку).
# ### Задание 3.4.
# Постройте две визуализации:
# * Распределение количества поездок в зависимости от часа дня;
# * Зависимость медианной длительности поездки от часа дня.
# На основе построенных графиков ответьте на следующие вопросы:
# а) В какое время суток такси заказывают реже всего?
# б) В какое время суток наблюдается пик медианной длительности поездок?
# группируем данные по часу и рассчитываем медианную длительность поездок
duration_by_hour = taxi_data.groupby("pickup_hour")["trip_duration"].median()
# строим графики
fig, axes = plt.subplots(nrows=1, ncols=2, figsize=(15, 4))
countplot = sns.countplot(data=taxi_data, x="pickup_hour", ax=axes[0])
countplot.set_title("Trip Duration Count by Hour")
lineplot = sns.lineplot(data=duration_by_hour, ax=axes[1])
lineplot.set_title("Trip Duration Median by Hour")
lineplot.xaxis.set_ticks(duration_by_hour.index)
lineplot.set_ylabel("median")
# #### Выводы
# * Реже всего такси заказывают с 4:00 до 6:00.
# * Пик медианной длительности поездок наблюдается с 11:00 до 16:00.
# ### Задание 3.5.
# Постройте две визуализации:
# * Распределение количества поездок в зависимости от дня недели;
# * Зависимость медианной длительности поездки от дня недели.
# На основе построенных графиков ответьте на следующие вопросы:
# а) В какой день недели совершается больше всего поездок?
# б) В какой день недели медианная длительность поездок наименьшая?
#
# группируем данные по дню недели и рассчитываем медианную длительность поездок
duration_by_day = taxi_data.groupby("pickup_day_of_week")["trip_duration"].median()
# строим графики
fig, axes = plt.subplots(nrows=1, ncols=2, figsize=(15, 4))
countplot = sns.countplot(data=taxi_data, x="pickup_day_of_week", ax=axes[0])
countplot.set_title("Trip Duration Count by Day of Week")
lineplot = sns.lineplot(data=duration_by_day, ax=axes[1])
lineplot.set_title("Trip Duration Median by Day of Week")
lineplot.set_ylabel("median")
# #### Выводы
# * В пятницу совершается больше всего поездок.
# * В воскресенье медианная длительность поездок наименьшая.
# ### Задание 3.6.
# Посмотрим на обе временные характеристики одновременно.
# Постройте сводную таблицу, по строкам которой отложены часы (pickup_hour), по столбцам - дни недели (pickup_day_of_week), а в ячейках - медианная длительность поездки (trip_duration).
# Визуализируйте полученную сводную таблицу с помощью тепловой карты (рекомендуемая палитра - coolwarm).
# строим сводную таблицу
pivot = taxi_data.pivot_table(
values="trip_duration",
index="pickup_hour",
columns="pickup_day_of_week",
aggfunc="median",
)
# визуализируем полученную таблицу
fig = plt.figure(figsize=(10, 7))
heatmap = sns.heatmap(data=pivot, cmap="coolwarm")
heatmap.set_title("Длительности поездки в зависимости от временных характеристих")
# #### Выводы
# * Самые прдолжительные поездки (в медианном смысле) наблюдаются с понедельника по пятницу в промежутке с 8 до 18 часов.
# * Наибольшая медианная длительность поездки наблюдалась в четверг в 14 часов дня.
# ### Задание 3.7.
# Постройте две диаграммы рассеяния (scatter-диаграммы):
# * первая должна иллюстрировать географическое расположение точек начала поездок (pickup_longitude, pickup_latitude)
# * вторая должна географическое расположение точек завершения поездок (dropoff_longitude, dropoff_latitude).
# Для этого на диаграммах по оси абсцисс отложите широту (longitude), а по оси ординат - долготу (latitude).
# Включите в визуализацию только те точки, которые находятся в пределах Нью-Йорка - добавьте следующие ограничения на границы осей абсцисс и ординат:
#
# city_long_border = (-74.03, -73.75)
# city_lat_border = (40.63, 40.85)
# Добавьте на диаграммы расцветку по десяти географическим кластерам (geo_cluster), которые мы сгенерировали ранее.
# **Рекомендация:** для наглядности уменьшите размер точек на диаграмме рассеяния.
#
# ограничения по осям
city_long_border = (-74.03, -73.75)
city_lat_border = (40.63, 40.85)
# строим диаграммы рассеяния
fig, axes = plt.subplots(nrows=1, ncols=2, figsize=(15, 8))
scatter_pickup = sns.scatterplot(
data=taxi_data,
x="pickup_longitude",
y="pickup_latitude",
hue="geo_cluster",
s=1,
ax=axes[0],
)
scatter_pickup.set_title("Координаты начала поездок")
scatter_pickup.set_xlim(city_long_border)
scatter_pickup.set_ylim(city_lat_border)
scatter_dropoff = sns.scatterplot(
data=taxi_data,
x="dropoff_longitude",
y="dropoff_latitude",
hue="geo_cluster",
s=1,
ax=axes[1],
)
scatter_dropoff.set_title("Координаты завершения поездок")
scatter_dropoff.set_xlim(city_long_border)
scatter_dropoff.set_ylim(city_lat_border)
# #### Выводы
# * Три кластера (из десяти) не попало на диаграммы (находятся за границами Нью-Йорка).
# ## 4. Отбор и преобразование признаков
# Перед тем как перейти к построению модели, осталось сделать ещё несколько шагов.
# * Следует помнить, что многие алгоритмы машинного обучения не могут обрабатывать категориальные признаки в их обычном виде. Поэтому нам необходимо их закодировать;
# * Надо отобрать признаки, которые мы будем использовать для обучения модели;
# * Необходимо масштабировать и трансформировать некоторые признаки для того, чтобы улучшить сходимость моделей, в основе которых лежат численные методы.
#
print("Shape of data: {}".format(taxi_data.shape))
print("Columns: {}".format(taxi_data.columns))
# Для удобства работы сделаем копию исходной таблицы с поездками:
train_data = taxi_data.copy()
train_data.head()
# ### Задание 4.1.
# Сразу позаботимся об очевидных неинформативных и избыточных признаках.
# а) Какой из признаков является уникальным для каждой поездки и не несет полезной информации в определении ее продолжительности?
# б) Утечка данных (data leak) - это…
# в) Подумайте, наличие какого из признаков в обучающем наборе данных создает утечку данных?
# г) Исключите выбранные в пунктах а) и в) признаки из исходной таблицы с данными. Сколько столбцов в таблице у вас осталось?
# #### Ответы
# а) Уникальными для каждой поездки является её идентификатор - столбец id. Он никак не связан с длительностью поездки и не имеет значения при её прогнозировании.
# б) ... ситуация, в которой данные используемые для обучения модели, содержат прямую или косвенную информацию о целевой переменной, но эти данные недоступны в реальных условиях.
# в) В наших данных утечку создаёт признак dropoff_datetime - зафиксированное время остановки счётчика. Начиная поездку, мы никак не можем точно знать, когда она закончится, ведь мы как раз и пытаемся предсказать время окончания поездки.
# исключаем признаки id и dropoff_time из исходной таблицы с данными
train_data = train_data.drop(["id", "dropoff_datetime"], axis=1)
print(f"В таблице осталось {train_data.shape[1]} столбцов")
# Ранее мы извлекли всю необходимую для нас информацию из даты начала поездки, теперь мы можем избавиться от этих признаков, так как они нам больше не понадобятся:
#
drop_columns = ["pickup_datetime", "pickup_date"]
train_data = train_data.drop(drop_columns, axis=1)
print("Shape of data: {}".format(train_data.shape))
# ### Задание 4.2.
# Закодируйте признак vendor_id в таблице train_data таким образом, чтобы он был равен 1, если идентификатор таксопарка равен 0, и 1 в противном случае.
# Закодируйте признак store_and_fwd_flag в таблице train_data таким образом, чтобы он был равен 0, если флаг выставлен в значение "N", и 1 в противном случае.
# а) Рассчитайте среднее по закодированному столбцу vendor_id. Ответ приведите с точностью до сотых.
# б) Рассчитайте среднее по закодированному столбцу store_and_fwd_flag. Ответ приведите с точностью до тысячных.
#
# кодируем признак vendor_id
train_data["vendor_id"] = (
train_data["vendor_id"].apply(lambda x: 0 if x == 1 else 1).astype("int")
)
# кодируем признак store_and_fwd_flag
train_data["store_and_fwd_flag"] = train_data["store_and_fwd_flag"].apply(
lambda x: 0 if x == "N" else 1
)
print("Среднее по столбцу vendor_id:", round(train_data["vendor_id"].mean(), 2))
print(
"Среднее по столбцу store_and_fwd_flag:",
round(train_data["store_and_fwd_flag"].mean(), 3),
)
# ### Задание 4.3.
# Создайте таблицу data_onehot из закодированных однократным кодированием признаков pickup_day_of_week, geo_cluster и events в таблице train_data с помощью OneHotEndoder из библиотеки sklearn. Параметр drop выставите в значение 'first', чтобы удалять первый бинарный столбец, тем самым не создавая излишних признаков.
# В результате работы OneHotEncoder вы получите безымянный numpy-массив, который нам будет необходимо преобразовать обратно в DataFrame, для более удобной работы в дальнейшем. Чтобы получить имена закодированных столбцов у объекта типа OneHotEncoder есть специальный метод get_feature_names_out(). Он возвращает список новых закодированных имен столбцов в формате _.
# Пример использования:
# ``` python
# # Получаем закодированные имена столбцов
# column_names = one_hot_encoder.get_feature_names_out()
# # Составляем DataFrame из закодированных признаков
# data_onehot = pd.DataFrame(data_onehot, columns=column_names)
# ```
# В этом псевдокоде:
# * one_hot_encoder - объект класса OneHotEncoder
# * data_onehot - numpy-массив, полученный в результате трансформации кодировщиком
# В результате выполнения задания у вас должен быть образован DataFrame `data_onehot`, который содержит кодированные категориальные признаки pickup_day_of_week, geo_cluster и events.
# Сколько бинарных столбцов у вас получилось сгенерировать с помощью однократного кодирования?
#
# загружаем библиотеку
one_hot_encoder = preprocessing.OneHotEncoder(drop="first")
# колонки к преобразованию
columns_to_change = ["pickup_day_of_week", "geo_cluster", "events"]
# 'учим' кодировщик и сразу кодируем данные
data_onehot = one_hot_encoder.fit_transform(
train_data[columns_to_change]
).toarray() # результат переводим в массив
# получаем закодированные имена столбцов
column_names = one_hot_encoder.get_feature_names_out()
# составляем DataFrame из закодированных признаков
data_onehot = pd.DataFrame(data_onehot, columns=column_names)
print("Количество новых бинарных столбцов:", data_onehot.shape[1])
# Добавим полученную таблицу с закодированными признаками:
train_data = pd.concat(
[train_data.reset_index(drop=True).drop(columns_to_change, axis=1), data_onehot],
axis=1,
)
print("Shape of data: {}".format(train_data.shape))
# Теперь, когда категориальные признаки предобработаны, сформируем матрицу наблюдений X, вектор целевой переменной y и его логарифм y_log. В матрицу наблюдений войдут все столбцы из таблицы с поездками за исключением целевого признака trip_duration и его логарифмированной версии trip_duration_log:
#
X = train_data.drop(["trip_duration", "trip_duration_log"], axis=1)
y = train_data["trip_duration"]
y_log = train_data["trip_duration_log"]
# Все наши модели мы будем обучать на логарифмированной версии y_log.
# Выбранный тип валидации - hold-out. Разобьем выборку на обучающую и валидационную в соотношении 67/33:
X_train, X_valid, y_train_log, y_valid_log = model_selection.train_test_split(
X, y_log, test_size=0.33, random_state=42
)
# На данный момент у нас достаточно много признаков: скорее всего, не все из них будут важны. Давайте оставим лишь те, которые сильнее всего связаны с целевой переменной и точно будут вносить вклад в повышение качества модели.
# ### Задание 4.4.
# С помощью SelectKBest отберите 25 признаков, наилучшим образом подходящих для предсказания целевой переменной в логарифмическом масштабе. Отбор реализуйте по обучающей выборке, используя параметр score_func = f_regression.
# Укажите признаки, которые вошли в список отобранных
#
# отбираем признаки
selector = feature_selection.SelectKBest(
score_func=feature_selection.f_regression, k=25
)
selector.fit(X_train, y_train_log)
# оставляем только полученные 25 признаков
best_features = selector.get_feature_names_out()
X_train = X_train[best_features]
X_valid = X_valid[best_features]
print("Признаки, которые вошли в список отобранных:\n", best_features)
# Так как мы будем использовать различные модели, в том числе внутри которых заложены численные методы оптимизации, то давайте заранее позаботимся о масштабировании факторов.
# ### Задание 4.5.
# Нормализуйте предикторы в обучающей и валидационной выборках с помощью MinMaxScaler из библиотеки sklearn. Помните, что обучение нормализатора производится на обучающей выборке, а трансформация на обучающей и валидационной!
# Рассчитайте среднее арифметическое для первого предиктора (т. е. для первого столбца матрицы) из валидационной выборки. Ответ округлите до сотых.
#
# инициализируем нормализатор MinMaxScaler
mm_scaler = preprocessing.MinMaxScaler()
# преобразовываем данные
X_train = mm_scaler.fit_transform(X_train)
X_valid = mm_scaler.transform(X_valid)
print(
"Среднее арифметическое для первого предиктора из валидационной выборки:",
np.round(np.mean(X_valid[:, 0]), 2),
)
# составляем DataFrame из массивов
X_train = pd.DataFrame(X_train, columns=best_features)
X_valid = pd.DataFrame(X_valid, columns=best_features)
# ## 5. Решение задачи регрессии: линейная регрессия и деревья решений
# Определим метрику, по которой мы будем измерять качество наших моделей. Мы будем следовать канонам исходного соревнования на Kaggle и в качестве метрики использовать RMSLE (Root Mean Squared Log Error), которая вычисляется как:
# $$RMSLE = \sqrt{\frac{1}{n}\sum_{i=1}^n(log(y_i+1)-log(\hat{y_i}+1))^2},$$
# где:
# * $y_i$ - истинная длительность i-ой поездки на такси (trip_duration)
# * $\hat{y_i}$- предсказанная моделью длительность i-ой поездки на такси
# Заметим, что логарифмирование целевого признака мы уже провели заранее, поэтому нам будет достаточно вычислить метрику RMSE для модели, обученной прогнозировать длительность поездки такси в логарифмическом масштабе:
# $$z_i=log(y_i+1),$$
# $$RMSLE = \sqrt{\frac{1}{n}\sum_{i=1}^n(z_i-\hat{z_i})^2}=\sqrt{MSE(z_i,\hat{z_i})}$$
# ### Задание 5.1.
# Постройте модель линейной регрессии на обучающей выборке (факторы должны быть нормализованы, целевую переменную используйте в логарифмическом масштабе). Все параметры оставьте по умолчанию.
# Для полученной модели рассчитайте метрику RMSLE на тренировочной и валидационной выборках. Ответ округлите до сотых.
#
# создаём объект класса LinearRegression
lr_model = linear_model.LinearRegression()
# обучаем модель - ищем параметры МНК
lr_model.fit(X_train, y_train_log)
# делаем предсказания
y_train_pred = lr_model.predict(X_train)
y_valid_pred = lr_model.predict(X_valid)
# рассчитываем RMSLE
print(
"RMSE на тренировочной выборке:",
round(np.sqrt(metrics.mean_squared_error(y_train_log, y_train_pred)), 2),
)
print(
"RMSE на валидационной выборке:",
round(np.sqrt(metrics.mean_squared_error(y_valid_log, y_valid_pred)), 2),
)
# ### Задание 5.2.
# Сгенерируйте полиномиальные признаки 2-ой степени с помощью PolynomialFeatures из библиотеки sklearn. Параметр include_bias выставите в значение False.
# Постройте модель полиномиальной регрессии 2-ой степени на обучающей выборке (факторы должны быть нормализованы, целевую переменную используйте в логарифмическом масштабе). Все параметры оставьте по умолчанию.
# а) Для полученной модели рассчитайте метрику RMSLE на тренировочной и валидационной выборках. Ответ округлите до сотых.
# б) Наблюдаются ли у вашей модели признаки переобучения?
#
# создаём генератор полиномиальных признаков
poly = preprocessing.PolynomialFeatures(degree=2, include_bias=False)
poly.fit(X_train)
# генерируем полиномиальные признаки
X_train_poly = poly.transform(X_train)
X_valid_poly = poly.transform(X_valid)
# строим модель полиномиальной регрессии
lr_poly = linear_model.LinearRegression()
lr_poly.fit(X_train_poly, y_train_log)
y_train_pred = lr_poly.predict(X_train_poly)
y_valid_pred = lr_poly.predict(X_valid_poly)
# рассчитываем RMSLE
print(
"RMSE на тренировочной выборке:",
round(np.sqrt(metrics.mean_squared_error(y_train_log, y_train_pred)), 2),
)
print(
"RMSE на валидационной выборке:",
round(np.sqrt(metrics.mean_squared_error(y_valid_log, y_valid_pred)), 2),
)
# #### Ответ
# б) Да, у модели наблюдаются признаки переобучения.
# ### Задание 5.3.
# Постройте модель полиномиальной регрессии 2-ой степени с L2-регуляризацией (регуляризация по Тихонову) на обучающей выборке (факторы должны быть нормализованы, целевую переменную используйте в логарифмическом масштабе). Коэффициент регуляризации $\alpha установите равным 1, остальные параметры оставьте по умолчанию.
# Для полученной модели рассчитайте метрику RMSLE на тренировочной и валидационной выборках. Ответ округлите до сотых.
#
# создаём объект класса линейной регрессии с L2-регуляризацией
lr_ridge = linear_model.Ridge(alpha=1)
# обучаем модель
lr_ridge.fit(X_train_poly, y_train_log)
# делаем предсказания
y_train_pred = lr_ridge.predict(X_train_poly)
y_valid_pred = lr_ridge.predict(X_valid_poly)
# рассчитываем RMSLE
print(
"RMSE на тренировочной выборке:",
round(np.sqrt(metrics.mean_squared_error(y_train_log, y_train_pred)), 2),
)
print(
"RMSE на валидационной выборке:",
round(np.sqrt(metrics.mean_squared_error(y_valid_log, y_valid_pred)), 2),
)
# ### Задание 5.4.
# Постройте модель дерева решений (DecisionTreeRegressor) на обучающей выборке (факторы должны быть нормализованы, целевую переменную используйте в логарифмическом масштабе). Все параметры оставьте по умолчанию.
# а) Для полученной модели рассчитайте метрику RMSLE на тренировочной и валидационной выборках. Ответ округлите до сотых.
# б) Наблюдаются ли у вашей модели признаки переобучения?
#
# строим модель дерева решений
dt_reg = tree.DecisionTreeRegressor(random_state=42)
dt_reg.fit(X_train, y_train_log)
# делаем предсказания
y_train_pred = dt_reg.predict(X_train)
y_valid_pred = dt_reg.predict(X_valid)
# рассчитываем RMSLE
print(
"RMSE на тренировочной выборке:",
round(np.sqrt(metrics.mean_squared_error(y_train_log, y_train_pred)), 2),
)
print(
"RMSE на валидационной выборке:",
round(np.sqrt(metrics.mean_squared_error(y_valid_log, y_valid_pred)), 2),
)
# #### Ответ
# б) Да, у модели наблюдаются признаки переобучения.
# ### Задание 5.5.
# Переберите все возможные варианты глубины дерева решений в диапазоне от 7 до 20:
# max_depths = range(7, 20)
# Постройте линейные графики изменения метрики RMSE на тренировочной и валидационной выборках в зависимости от значения параметра глубины дерева решений.
# а) Найдите оптимальное значение максимальной глубины дерева, для которой будет наблюдаться минимальное значение RMSLE на обучающей выборке, но при этом еще не будет наблюдаться переобучение (валидационная кривая еще не начинает возрастать).
# б) Чему равно значение метрик RMSLE на тренировочной и валидационной выборках для дерева решений с выбранной оптимальной глубиной? Ответ округлите до сотых.
#
# создаём список из 13 возможных значений от 7 до 20
max_depths = range(7, 20)
# создаём пустые списки, в которые будем добавлять результаты
train_scores = []
valid_scores = []
for depth in max_depths:
# создаём объект класса DecisionTreeRegressor
dt_reg = tree.DecisionTreeRegressor(max_depth=depth, random_state=42)
# обучаем модель
dt_reg.fit(X_train, y_train_log)
# делаем предсказание для тренировочной выборки
y_train_pred = dt_reg.predict(X_train)
# делаем предсказание для валидационной выборки
y_valid_pred = dt_reg.predict(X_valid)
# рассчитываем RMSLE для двух выборок и добавляем их в списки
train_scores.append(np.sqrt(metrics.mean_squared_error(y_train_log, y_train_pred)))
valid_scores.append(np.sqrt(metrics.mean_squared_error(y_valid_log, y_valid_pred)))
# визуализируем изменения RMSLE в зависимости от max_depth
fig, ax = plt.subplots(figsize=(12, 4)) # фигура + координатная плоскость
ax.plot(
max_depths, train_scores, label="Train"
) # линейный график для тренировочной выборки
ax.plot(
max_depths, valid_scores, label="Valid"
) # линейный график для валидационной выборки
ax.set_xlabel("max_depth") # название оси абсцисс
ax.set_ylabel("RMSLE") # название оси ординат
ax.set_xticks(max_depths) # метки на оси абсцисс
ax.legend()
# отображение легенды
# извлекаем индекс лучшего RMSLE на валидационной выборке
best_index = valid_scores.index(min(valid_scores))
print("Оптимальная глубина дерева решений:", max_depths[best_index])
print("RMSLE на тренировочной выборке:", round(train_scores[best_index], 2))
print("RMSLE на валидационной выборке:", round(valid_scores[best_index], 2))
# ## 6. Решение задачи регрессии: ансамблевые методы и построение прогноза
# Переходим к тяжелой артиллерии: ансамблевым алгоритмам.
# ### Задание 6.1.
# Постройте модель случайного леса на обучающей выборке (факторы должны быть нормализованы, целевую переменную используйте в логарифмическом масштабе). В качестве гиперпараметров укажите следующие:
# * n_estimators=200,
# * max_depth=12,
# * criterion='squared_error',
# * min_samples_split=20,
# * random_state=42
# Для полученной модели рассчитайте метрику RMSLE на тренировочной и валидационной выборках. Ответ округлите до сотых.
#
# создаём объект класса RandomForestRegressor
rf_reg = ensemble.RandomForestRegressor(
n_estimators=200,
max_depth=12,
criterion="squared_error",
min_samples_split=20,
random_state=42,
n_jobs=-1,
)
# обучаем модель
rf_reg.fit(X_train, y_train_log)
# делаем предсказания
y_train_pred = rf_reg.predict(X_train)
y_valid_pred = rf_reg.predict(X_valid)
# рассчитываем RMSLE
print(
"RMSLE на тренировочной выборке:",
round(np.sqrt(metrics.mean_squared_error(y_train_log, y_train_pred)), 2),
)
print(
"RMSLE на валидационной выборке:",
round(np.sqrt(metrics.mean_squared_error(y_valid_log, y_valid_pred)), 2),
)
# ### Задание 6.2.
# Постройте модель градиентного бустинга над деревьями решений (GradientBoostingRegressor) на обучающей выборке (факторы должны быть нормализованы, целевую переменную используйте в логарифмическом масштабе). В качестве гиперпараметров укажите следующие:
# * learning_rate=0.5,
# * n_estimators=100,
# * max_depth=6,
# * min_samples_split=30,
# * random_state=42
# Для полученной модели рассчитайте метрику RMSLE на тренировочной и валидационной выборках. Ответ округлите до сотых.
#
# создаём объект класса GradientBoostingRegressor
gb_reg = ensemble.GradientBoostingRegressor(
learning_rate=0.5,
n_estimators=100,
max_depth=6,
min_samples_split=30,
random_state=42,
)
# обучаем модель
gb_reg.fit(X_train, y_train_log)
# делаем предсказания
y_train_pred = gb_reg.predict(X_train)
y_valid_pred = gb_reg.predict(X_valid)
# рассчитываем RMSLE
print(
"RMSLE на тренировочной выборке:",
round(np.sqrt(metrics.mean_squared_error(y_train_log, y_train_pred)), 2),
)
print(
"RMSLE на валидационной выборке:",
round(np.sqrt(metrics.mean_squared_error(y_valid_log, y_valid_pred)), 2),
)
# ### Задание 6.3.
# Какая из построенных вами моделей показала наилучший результат (наименьшее значение RMSLE на валидационной выборке)?
# * Линейная регрессия
# * Полиномиальная регрессия 2ой степени
# * Дерево решений
# * Случайный лес
# * Градиентный бустинг над деревьями решений
# ### Задание 6.4.
# Постройте столбчатую диаграмму коэффициентов значимости каждого из факторов.
# Укажите топ-3 наиболее значимых для предсказания целевого признака - длительности поездки в логарифмическом масштабе - факторов.
#
fig, ax = plt.subplots(figsize=(13, 5)) # фигура + координатная плоскость
features = X_train.columns # признаки
importances = gb_reg.feature_importances_ # важность признаков
# строим столбчатую диаграмму
sns.barplot(x=features, y=importances, ax=ax)
ax.set_title("Bar plot feature importances") # название графика
ax.set_ylabel("Importance") # название оси ординат
ax.xaxis.set_tick_params(rotation=90) # поворот меток на оси абсцисс
# ### Задание 6.5.
# Для лучшей из построенных моделей рассчитайте медианную абсолютную ошибку (MeAE - в sklearn функция median_absolute_error) предсказания длительности поездки такси на валидационной выборке:
# $$ MeAE = median(|y_i-\hat{y_i}|)$$
# Значение метрики MeAE переведите в минуты и округлите до десятых.
#
# переводим из логарифимического маштаба в изначальный
y_valid = np.exp(y_valid_log) - 1
y_valid_pred_exp = np.exp(y_valid_pred) - 1
# рассчитываем медианную абсолютную ошибку
meae_valid = metrics.median_absolute_error(y_valid, y_valid_pred_exp)
# переводим значение метрики в минуты и округляем
print("MeAE на валидационной выборке:", round(meae_valid / 60, 1), "мин")
# Финальный шаг - сделать submit - предсказание для отложенного тестового набора данных.
# Прочитаем тестовые данные и заранее выделим столбец с идентификаторами поездок из тестового набора данных. Он нам еще пригодится:
#
test_data = pd.read_csv("Documents/Project5_test_data.csv")
osrm_data_test = pd.read_csv("Documents/Project5_osrm_data_test.csv")
test_id = test_data["id"]
# Перед созданием прогноза для тестовой выборки необходимо произвести все манипуляции с данными, которые мы производили с тренировочной выборкой, а именно:
# * Перевести признак pickup_datetime в формат datetime;
# * Добавить новые признаки (временные, географические, погодные и другие факторы);
# * Произвести очистку данных от пропусков;
# * Произвести кодировку категориальных признаков:
# * Закодировать бинарные признаки;
# * Закодировать номинальные признаки с помощью обученного на тренировочной выборке OneHotEncoder’а;
# * Сформировать матрицу наблюдений, оставив в таблице только те признаки, которые были отобраны с помощью SelectKBest;
# * Нормализовать данные с помощью обученного на тренировочной выборке MinMaxScaler’а.
#
test_data["pickup_datetime"] = pd.to_datetime(
test_data["pickup_datetime"], format="%Y-%m-%d %H:%M:%S"
)
test_data = add_datetime_features(test_data)
test_data = add_holiday_features(test_data, holiday_data)
test_data = add_osrm_features(test_data, osrm_data_test)
test_data = add_geographical_features(test_data)
test_data = add_cluster_features(test_data, kmeans)
test_data = add_weather_features(test_data, weather_data)
test_data = fill_null_weather_data(test_data)
test_data["vendor_id"] = test_data["vendor_id"].apply(lambda x: 0 if x == 1 else 1)
test_data["store_and_fwd_flag"] = test_data["store_and_fwd_flag"].apply(
lambda x: 0 if x == "N" else 1
)
test_data_onehot = one_hot_encoder.fit_transform(test_data[columns_to_change]).toarray()
column_names = one_hot_encoder.get_feature_names_out(columns_to_change)
test_data_onehot = pd.DataFrame(test_data_onehot, columns=column_names)
test_data = pd.concat(
[
test_data.reset_index(drop=True).drop(columns_to_change, axis=1),
test_data_onehot,
],
axis=1,
)
X_test = test_data[best_features]
X_test_scaled = mm_scaler.transform(X_test)
print("Shape of data: {}".format(X_test.shape))
# Только после выполнения всех этих шагов можно сделать предсказание длительности поездки для тестовой выборки. Не забудьте перевести предсказания из логарифмического масштаба в истинный, используя формулу:
# $$y_i=exp(z_i)-1$$
# После того, как вы сформируете предсказание длительности поездок на тестовой выборке вам необходимо будет создать submission-файл в формате csv, отправить его на платформу Kaggle и посмотреть на результирующее значение метрики RMSLE на тестовой выборке.
# Код для создания submission-файла:
#
# делаем предсказание для тестовой выборки
y_test_pred_log = gb_reg.predict(X_test_scaled)
# переводим предсказания из логарифмического маштаба в истинный
y_test_predict = np.exp(y_test_pred_log) - 1
# создаём submission-файл
submission = pd.DataFrame({"id": test_id, "trip_duration": y_test_predict})
submission.to_csv("Documents/submission_gb.csv", index=False)
# ### **В качестве бонуса**
# В завершение по ансамблевым мы предлагаем вам попробовать улучшить свое предсказание, воспользовавшись моделью экстремального градиентного бустинга (XGBoost) из библиотеки xgboost.
# **XGBoost** - современная модель машинного обучения, которая является продолжением идеи градиентного бустинга Фридмана. У нее есть несколько преимуществ по сравнению с классической моделью градиентного бустинга из библиотеки sklearn: повышенная производительность путем параллелизации процесса обучения, повышенное качество решения за счет усовершенствования алгоритма бустинга, меньшая склонность к переобучению и широкий функционал возможности управления параметрами модели.
# Для ее использования необходимо для начала установить пакет xgboost:
#!pip install xgboost
# После чего модуль можно импортировать:
import xgboost as xgb
# Перед обучением модели необходимо перевести наборы данных в тип данных xgboost.DMatrix:
# Создание матриц наблюдений в формате DMatrix
dtrain = xgb.DMatrix(X_train, label=y_train_log, feature_names=best_features)
dvalid = xgb.DMatrix(X_valid, label=y_valid_log, feature_names=best_features)
dtest = xgb.DMatrix(X_test_scaled, feature_names=best_features)
# Обучение модели XGBoost происходит с помощью метода train, в который необходимо передать параметры модели, набор данных, количество базовых моделей в ансамбле, а также дополнительные параметры:
#
# Гиперпараметры модели
xgb_pars = {
"min_child_weight": 20,
"eta": 0.1,
"colsample_bytree": 0.9,
"max_depth": 6,
"subsample": 0.9,
"lambda": 1,
"nthread": -1,
"booster": "gbtree",
"eval_metric": "rmse",
"objective": "reg:squarederror",
}
# Тренировочная и валидационная выборка
watchlist = [(dtrain, "train"), (dvalid, "valid")]
# Обучаем модель XGBoost
model = xgb.train(
params=xgb_pars, # гиперпараметры модели
dtrain=dtrain, # обучающая выборка
num_boost_round=300, # количество моделей в ансамбле
evals=watchlist, # выборки, на которых считается матрица
early_stopping_rounds=20, # раняя остановка
maximize=False, # смена поиска максимума на минимум
verbose_eval=10, # шаг, через который происходит отображение метрик
)
# Предсказать целевой признак на новых данных можно с помощью метода predict():
# Делаем предсказание на тестовом наборе данных
y_test_predict = np.exp(model.predict(dtest)) - 1
print("Modeling RMSLE %.5f" % model.best_score)
# создаём submission-файл
submission_xgb = pd.DataFrame({"id": test_id, "trip_duration": y_test_predict})
submission_xgb.to_csv("Documents/submission_xgb.csv", index=False)
# Также как и все модели, основанные на использовании деревьев решений в качестве базовых моделей, XGBoost имеет возможность определения коэффициентов важности факторов. Более того, в библиотеку встроена возможность визуализации важность факторов в виде столбчатой диаграммы. За эту возможность отвечает функция plot_importance():
#
fig, ax = plt.subplots(figsize=(15, 15))
xgb.plot_importance(model, ax=ax, height=0.5)
|
# # Intorduction
# Setup
import os
import cv2
import numpy as np
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
import matplotlib.pyplot as plt
# Avoid OMM Error
physical_devices = tf.config.experimental.list_physical_devices("GPU")
if len(physical_devices) > 0:
tf.config.experimental.set_memory_growth(physical_devices[0], True)
# check the GPU
physical_devices
# define the images directory
data_dir = "/kaggle/input/shai-level-2-training-2023"
print(os.listdir(data_dir))
# let`s make EDA for the dataset
data_splits = os.listdir(data_dir)[3:]
for data_kind in data_splits:
classes = os.listdir(os.path.join(data_dir, data_kind))
print(f"*We Process the {data_kind}")
for class_ in classes:
path = os.path.join(os.path.join(data_dir, data_kind), class_)
if os.path.isdir(path):
print(f" we have a {len(os.listdir(path))} images from class {class_}")
print("")
# # Genrate a Dataset and Visualize
# load data with tf.utils.image_dataset_from_directory
IMAGE_SIZE = (256, 256)
BATCH_SIZE = 16
SEED = 42
# make train-testing-validation datasets
def load_data(data_dir):
data = []
for class_d in os.listdir(data_dir)[3:]:
data.append(
tf.keras.utils.image_dataset_from_directory(
os.path.join(data_dir, class_d),
image_size=IMAGE_SIZE,
batch_size=BATCH_SIZE,
seed=SEED,
)
)
return data
# apllying the Load dataset
train_ds = load_data(data_dir)[0]
# plot Visualize
plt.figure(figsize=(10, 10))
for image, label in train_ds.take(1):
for i in range(9):
plt.subplot(3, 3, i + 1)
plt.imshow(image[i].numpy().astype("uint8"))
plt.title(train_ds.class_names[label[i]])
plt.axis("off")
class_names = train_ds.class_names
print(class_names)
# ## Standardize the data
# standardize values to be in the [0,1] range
normalization_layer = tf.keras.layers.Rescaling(1.0 / 255)
# using thr Dataset.map to apply this layer
normalized_ds = train_ds.map(lambda x, y: (normalization_layer(x), y))
# check pixels after normalization
images_batch, label_batch = next(iter(normalized_ds))
random_image = images_batch[4]
print(f" min value = {np.min(random_image)} , max value = {np.max(random_image)}")
# # Configure the dataset for performance
#
# make sure to use buffered prefetching , yield the data without having I/O become bloking
def Configure(dataset):
dataset = dataset.cache().prefetch(buffer_size=tf.data.AUTOTUNE)
return dataset
# apply the function
train_ds = Configure(train_ds)
# # Build a Model
NUM_CLASSES = 3
INPUT_SHAPE = IMAGE_SIZE + (3,)
# Preprocessing input layers
preprocess_input = tf.keras.applications.vgg19.preprocess_input
# Base Model `VGG19`
base_model = tf.keras.applications.VGG19(
input_shape=INPUT_SHAPE, include_top=False, weights="imagenet"
)
# Unfreeze some layers of the base model for fine-tuning
for layer in base_model.layers[:-4]:
layer.trainable = False
# Feature extractor layers
global_average_layer = tf.keras.layers.GlobalAveragePooling2D()
dense_layer_1 = tf.keras.layers.Dense(1024, activation="relu")
dropout_layer_1 = tf.keras.layers.Dropout(0.5)
dense_layer_2 = tf.keras.layers.Dense(512, activation="relu")
dropout_layer_2 = tf.keras.layers.Dropout(0.4)
prediction_layer = tf.keras.layers.Dense(NUM_CLASSES, activation="softmax")
# Build the model
inputs = tf.keras.Input(shape=INPUT_SHAPE)
x = preprocess_input(inputs)
x = base_model(x, training=True)
x = global_average_layer(x)
x = dense_layer_1(x)
x = dropout_layer_1(x)
x = dense_layer_2(x)
x = dropout_layer_2(x)
outputs = prediction_layer(x)
Mido = tf.keras.Model(inputs, outputs, name="Mido")
# Compile the model
Mido.compile(
optimizer=tf.keras.optimizers.Adam(learning_rate=0.0001),
loss="sparse_categorical_crossentropy",
metrics=["accuracy"],
)
# Print model summary
Mido.summary()
# plot the model
keras.utils.plot_model(Mido, show_shapes=True)
# Define Callback
from tensorflow.keras.callbacks import ModelCheckpoint
# Define the checkpoint filepath and callback
checkpoint_filepath = "best_model_weights.h5"
model_checkpoint_callback = ModelCheckpoint(
filepath=checkpoint_filepath,
save_weights_only=True,
monitor="val_accuracy",
mode="max",
save_best_only=True,
)
# split the dataset
length = len(train_ds)
train_ds_split = train_ds.take(int(length * 0.80))
val_ds_spilt = train_ds.skip(int(length * 0.20)).take(int(length * 0.20))
# check the length
print(" train dataset length", len(train_ds_split))
print(" validation dataset length", len(val_ds_spilt))
# Train the model with the callback
num_epochs = 15
history = Mido.fit(
train_ds_split,
epochs=num_epochs,
validation_data=val_ds_spilt,
callbacks=[model_checkpoint_callback],
)
# # Model Evaluation
# list all data in history
print(history.history.keys())
print("")
plt.figure(figsize=(10, 10))
plt.subplot(2, 1, 1)
# summarize history for accuracy
plt.plot(history.history["accuracy"])
plt.plot(history.history["val_accuracy"])
plt.title("model accuracy")
plt.ylabel("accuracy")
plt.legend(["train", "test"], loc="upper left")
# summarize history for loss
plt.subplot(2, 1, 2)
plt.plot(history.history["loss"])
plt.plot(history.history["val_loss"])
plt.title("model loss")
plt.ylabel("loss")
plt.xlabel("epoch")
plt.legend(["train", "test"], loc="upper left")
plt.show()
# Load the best model weights from the checkpoint
Mido.load_weights(checkpoint_filepath)
# Evaluate the best model
test_loss, test_accuracy = Mido.evaluate(val_ds_spilt)
print(f"Test Loss: {test_loss:.2f}")
print(f"Test Accuracy: {test_accuracy*100:.2f}%")
# # Test the Model and make Sample_submission
# make a prediction
import csv
# Path to the directory containing the images
images_dir = "/kaggle/input/shai-level-2-training-2023/test"
# List to hold the predictions
predictions = []
# Loop over the images in the directory
for image_file in os.listdir(images_dir):
# Load the image
image_path = os.path.join(images_dir, image_file)
image = cv2.imread(image_path)
image = cv2.resize(image, IMAGE_SIZE)
# Make a prediction on the image
prediction = Mido.predict(np.expand_dims(image, axis=0))
predicted_class = class_names[np.argmax(prediction)]
# Append the prediction to the list
predictions.append((image_file, predicted_class))
# Save the predictions to a CSV file
with open("submission.csv", "w", newline="") as csvfile:
writer = csv.writer(csvfile)
writer.writerow(["Image", "Label"])
for prediction in predictions:
writer.writerow(prediction)
import pandas as pd
pred = pd.read_csv("/kaggle/working/submission.csv")
pred.head()
pred.Label.value_counts()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
import sklearn
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Read the DataSet.
df = pd.read_csv("/kaggle/input/co2-emission-by-vehicles/CO2 Emissions_Canada.csv")
# ## Get the head of DataFrame.
df.head()
# ## Check for NaN.
df.isna().sum()
# ## Get info about DataFrame.
df.info()
df["Make"].value_counts()
# ## Data visualization
plt.figure(figsize=(25, 25))
sns.countplot(x=df["Make"])
plt.show()
sns.displot(x=df["CO2 Emissions(g/km)"], kind="kde")
plt.figure(figsize=(25, 25))
sns.countplot(x=df["Vehicle Class"])
df.hist()
# ## Get columns with object values.
target = []
for i in df.columns:
col = df[i]
if type(col[0]) == str:
target.append(i)
print(i)
# ## Converting object values to int.
from sklearn.preprocessing import LabelEncoder
for i in target:
encoder = LabelEncoder()
encoder.fit(list(df[i]))
df[i] = encoder.fit_transform(df[i])
# ## Features selection.
df.columns
X = df.drop("CO2 Emissions(g/km)", axis=1)
y = df["CO2 Emissions(g/km)"]
# ## Data splitting.
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(
X, y, test_size=1 / 5, random_state=41
)
# ## Linear regression.
from sklearn.linear_model import LinearRegression
LR = LinearRegression().fit(x_train, y_train)
# ## Get the prediction.
y_predLR = LR.predict(x_test)
# ## Get r2_score of the model.
from sklearn.metrics import r2_score
accLR = r2_score(y_test, y_predLR)
accLR
# ## RandomForest
from sklearn.ensemble import RandomForestRegressor
RF = RandomForestRegressor(
n_estimators=50, max_depth=15, random_state=55, n_jobs=-1
).fit(x_train, y_train)
# ## Get prediction
y_predRF = RF.predict(x_test)
# ## Get accuracy of RF
accRF = r2_score(y_test, y_predRF)
accRF
# ## KNN.
from sklearn.neighbors import KNeighborsRegressor
KNN = KNeighborsRegressor(n_neighbors=7).fit(x_train, y_train)
# ## Get prediction.
y_predKNN = KNN.predict(x_test)
# ## Get accuracy of KNN.
accKNN = r2_score(y_test, y_predKNN)
accKNN
|
import pandas as pd
import numpy as np
dataset = pd.read_csv(
"/kaggle/input/ipl-complete-dataset-20082020/IPL Matches 2008-2020.csv"
)
dataset
dataset.info()
# format= 1
dataset["date"] = pd.to_datetime(dataset["date"])
dataset.info()
start = "2008-01-01"
end = "2010-12-31"
# dateformat -1
data = dataset.loc[(dataset["date"] >= start) & (dataset["date"] <= end)]
data["date"].unique()
data["date"].value_counts()
data
data.info()
dataset["id"] = pd.to_numeric(dataset["id"])
dataset["date"] = pd.to_datetime(dataset["date"])
dataset.info()
# date format 2
dataset2 = dataset.loc[dataset["date"].between("2012-01-01", "2016-12-31")]
dataset2
# date format 3
dataset1 = dataset[
dataset["date"].isin(pd.date_range(start="2018-01-01", end="2018-12-31"))
]
dataset1
# #date format 4
# df2 = df[df["InsertedDates"].isin(pd.date_range("2021-11-15", "2021-11-17"))]
# print(df2)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
pass
# print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import os
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import cv2
from PIL import Image
import tensorflow as tf
import glob
# # Reading All Images
benign = "/kaggle/input/breast-ultrasound-images-dataset/Dataset_BUSI_with_GT/benign"
malignant = (
"/kaggle/input/breast-ultrasound-images-dataset/Dataset_BUSI_with_GT/malignant"
)
normal = "/kaggle/input/breast-ultrasound-images-dataset/Dataset_BUSI_with_GT/normal"
count_benign = len(os.listdir(benign))
count_malignant = len(os.listdir(malignant))
count_normal = len(os.listdir(normal))
print((count_benign))
print(count_malignant)
print(count_normal)
# Get the list of all the images
bening_cases = glob.glob(benign + "/*")
malignant_cases = glob.glob(malignant + "/*")
normal_cases = glob.glob(normal + "/*")
# An empty list. We will insert the data into this list in (img_path, label) format
exclude = []
train_data_mask = []
train_data_img = []
# Go through all the normal cases. The label for these cases will be 0
for img in normal_cases:
if img.endswith("_mask.png"):
train_data_mask.append(img)
elif img.endswith("_mask_1.png") or img.endswith("_mask_2.png"):
exclude.append(img)
else:
train_data_img.append(img)
# Go through all the bening cases. The label for these cases will be 1
for img in bening_cases:
if img.endswith("_mask.png"):
train_data_mask.append(img)
elif img.endswith("_mask_1.png") or img.endswith("_mask_2.png"):
exclude.append(img)
else:
train_data_img.append(img)
# Go through all the malignant cases. The label for these cases will be 1
for img in malignant_cases:
if img.endswith("_mask.png"):
train_data_mask.append(img)
elif img.endswith("_mask_1.png") or img.endswith("_mask_2.png"):
exclude.append(img)
else:
train_data_img.append(img)
# printing Length of all the images
len(train_data_img), len(train_data_mask), len(exclude)
# Sorting Images
train_data_img = sorted(train_data_img)
train_data_mask = sorted(train_data_mask)
# # Resizing
from skimage import exposure
images = []
masks = []
size_x = 256
size_y = 256
for every_img_path in train_data_img:
img = cv2.imread(every_img_path, cv2.IMREAD_COLOR)
img = cv2.resize(img, (size_y, size_x))
images.append(img)
for every_mask_path in train_data_mask:
mask = cv2.imread(every_mask_path, cv2.IMREAD_GRAYSCALE)
mask = cv2.resize(mask, (size_y, size_x))
masks.append(mask)
images = np.array(images)
masks = np.array(masks)
print(np.shape(images))
print(np.shape(masks))
# checking
if images.shape[0] != masks.shape[0]:
print("Error: number of images and masks do not match.")
else:
print("Number of images and masks match.")
# converting values between 0-1
x = images / 255
y = masks / 255
import random
from skimage.io import imshow
########## Displaying random image from X_train and Y_train #########
random_num = random.randint(0, 516)
print("image index: ", random_num)
imshow(x[random_num])
plt.show()
imshow(y[random_num])
plt.show()
test_img = x[random_num]
test_img2 = y[random_num]
print(test_img.min(), test_img.max())
print(test_img.shape)
print(test_img2.min(), test_img2.max())
print(test_img2.shape)
# # Splitting the images into Training, Testing and Validation
from sklearn.model_selection import train_test_split
# Split the data into training and testing sets
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.2, random_state=42
)
# Split the training data into training and validation sets
x_train, x_val, y_train, y_val = train_test_split(
x_train, y_train, test_size=0.1, random_state=42
)
num_train = x_train.shape[0]
print(
"training images: ",
num_train,
" and the shape of training images numpy array: ",
np.shape(x_train),
)
num_train2 = y_train.shape[0]
print(
"training masks: ",
num_train2,
" and the shape of training masks numpy array: ",
np.shape(y_train),
)
num_test = x_test.shape[0]
print(
"testing images: ",
num_test,
" and the shape of testing images numpy array: ",
np.shape(x_test),
)
num_test2 = y_test.shape[0]
print(
"testing masks: ",
num_test2,
" and the shape of testing masks numpy array: ",
np.shape(y_test),
)
num_val = x_val.shape[0]
print(
"validation images: ",
num_val,
" and the shape of validation images numpy array: ",
np.shape(x_val),
)
num_val2 = y_val.shape[0]
print(
"validation masks: ",
num_val2,
" and the shape of validation masks numpy array: ",
np.shape(y_val),
)
# # Building the Model
from keras.layers import (
Input,
Conv2D,
MaxPooling2D,
UpSampling2D,
concatenate,
Conv2DTranspose,
BatchNormalization,
Dropout,
Lambda,
Add,
Multiply,
Layer,
Activation,
)
from keras.models import Model
IMG_WIDTH = 256
IMG_HEIGHT = 256
IMG_CHANNELS = 3
def conv_block(inputs, num_filters, dropout_rate=0.2):
x = Conv2D(
num_filters,
3,
activation="relu",
padding="same",
kernel_initializer="he_normal",
)(inputs)
x = BatchNormalization()(x)
x = Dropout(dropout_rate)(x)
x = Conv2D(
num_filters,
3,
activation="relu",
padding="same",
kernel_initializer="he_normal",
)(x)
x = BatchNormalization()(x)
return x
# residual convolutional block
def res_conv_block(x, filters, dropout=0.2, kernelsize=3, batchnorm=True):
conv1 = Conv2D(
filters,
(kernelsize, kernelsize),
kernel_initializer="he_normal",
padding="same",
)(x)
if batchnorm is True:
conv1 = BatchNormalization(axis=3)(conv1)
conv1 = Activation("relu")(conv1)
conv2 = Conv2D(
filters,
(kernelsize, kernelsize),
kernel_initializer="he_normal",
padding="same",
)(conv1)
if batchnorm is True:
conv2 = BatchNormalization(axis=3)(conv2)
conv2 = Activation("relu")(conv2)
if dropout > 0:
conv2 = Dropout(dropout)(conv2)
# skip connection
shortcut = Conv2D(
filters, kernel_size=(1, 1), kernel_initializer="he_normal", padding="same"
)(x)
if batchnorm is True:
shortcut = BatchNormalization(axis=3)(shortcut)
res_path = Add()([shortcut, conv2])
res_path = Activation("relu")(res_path)
return res_path
def attention_block(input_encoder, input_decoder, num_filters):
g = Conv2D(num_filters, 1, strides=1, padding="same")(input_encoder)
x = Conv2D(num_filters, 1, strides=1, padding="same")(input_decoder)
x = Add()([g, x])
x = Activation("relu")(x)
x = Conv2D(1, 1, strides=1, padding="same")(x)
x = BatchNormalization()(x)
x = Activation("sigmoid")(x)
x = Multiply()([input_decoder, x])
return x
def AttFrac(IMG_HEIGHT, IMG_WIDTH, IMG_CHANNELS, num_classes=1, num_filters=8):
inputs = Input((IMG_HEIGHT, IMG_WIDTH, IMG_CHANNELS))
# Contracting Path
c0 = res_conv_block(inputs, num_filters)
p0 = MaxPooling2D(pool_size=(2, 2))(c0)
c1 = res_conv_block(p0, num_filters * 2)
p1 = MaxPooling2D(pool_size=(2, 2))(c1)
c2 = res_conv_block(p1, num_filters * 4)
p2 = MaxPooling2D(pool_size=(2, 2))(c2)
c3 = res_conv_block(p2, num_filters * 8)
p3 = MaxPooling2D(pool_size=(2, 2))(c3)
c4 = res_conv_block(p3, num_filters * 16)
p4 = MaxPooling2D(pool_size=(2, 2))(c4)
# bottom
c5 = res_conv_block(p4, num_filters * 32)
# Expansive Path
u6 = Conv2DTranspose(num_filters * 16, (3, 3), strides=(2, 2), padding="same")(c5)
u6 = concatenate([u6, c4])
c6 = res_conv_block(u6, num_filters * 16)
u7 = Conv2DTranspose(num_filters * 8, (3, 3), strides=(2, 2), padding="same")(c6)
u7 = concatenate([u7, c3])
c7 = res_conv_block(u7, num_filters * 8)
u8 = Conv2DTranspose(num_filters * 4, (3, 3), strides=(2, 2), padding="same")(c7)
u8 = concatenate([u8, c2])
c8 = res_conv_block(u8, num_filters * 4)
u9 = Conv2DTranspose(num_filters * 2, (3, 3), strides=(2, 2), padding="same")(c8)
u9 = concatenate([u9, c1])
c9 = res_conv_block(u9, num_filters * 2)
# Contracting Path2
p9 = MaxPooling2D(pool_size=(2, 2))(c9)
p9 = concatenate([p9, c8])
c10 = res_conv_block(p9, num_filters * 4)
p10 = MaxPooling2D(pool_size=(2, 2))(c10)
p10 = concatenate([p10, c7])
c11 = res_conv_block(p10, num_filters * 8)
p11 = MaxPooling2D(pool_size=(2, 2))(c11)
p11 = concatenate([p11, c6])
c12 = res_conv_block(p11, num_filters * 16)
p12 = MaxPooling2D(pool_size=(2, 2))(c12)
p12 = concatenate([p12, c5])
# bottom
c13 = res_conv_block(p12, num_filters * 32)
# Expansive Path2
u14 = Conv2DTranspose(num_filters * 16, (3, 3), strides=(2, 2), padding="same")(c13)
u14 = concatenate([u14, c12])
a1 = attention_block(c12, u14, num_filters * 16) # Attention Gates
c14 = res_conv_block(u14, num_filters * 16)
u15 = Conv2DTranspose(num_filters * 8, (3, 3), strides=(2, 2), padding="same")(c14)
u15 = concatenate([u15, c11])
a1 = attention_block(c11, u15, num_filters * 8) # Attention Gates
c15 = res_conv_block(u15, num_filters * 8)
u16 = Conv2DTranspose(num_filters * 4, (3, 3), strides=(2, 2), padding="same")(c15)
u16 = concatenate([u16, c10])
a1 = attention_block(c10, u16, num_filters * 4) # Attention Gates
c16 = res_conv_block(u16, num_filters * 4)
u17 = Conv2DTranspose(num_filters * 2, (3, 3), strides=(2, 2), padding="same")(c16)
u17 = concatenate([u17, c9])
a1 = attention_block(c9, u17, num_filters * 2) # Attention Gates
c17 = res_conv_block(u17, num_filters * 2)
u18 = Conv2DTranspose(num_filters, (3, 3), strides=(2, 2), padding="same")(c17)
u18 = concatenate([u18, c0])
a1 = attention_block(c0, u18, num_filters * 16) # Attention Gates
c18 = res_conv_block(u18, num_filters)
# Output
outputs = tf.keras.layers.Conv2D(1, (1, 1), activation="sigmoid")(c18)
model = Model(inputs=inputs, outputs=outputs)
return model
model = AttFrac(IMG_HEIGHT, IMG_WIDTH, IMG_CHANNELS)
model.summary()
# # Defining Matrices
from tensorflow.keras.optimizers import Adam
from tensorflow.keras import backend as K
def dice_coeff(y_true, y_pred):
smooth = 1.0
# Flatten
y_true_f = tf.reshape(y_true, [-1])
y_pred_f = tf.reshape(y_pred, [-1])
# y_true_f = tf.y_true_f
# y_pred_f = tf.y_pred_f
intersection = tf.reduce_sum(tf.multiply(y_true_f, y_pred_f))
score = (2.0 * intersection) / (
tf.reduce_sum(y_true_f) + tf.reduce_sum(y_pred_f) + smooth
)
return score
def iou_coef(y_true, y_pred, smooth=1):
intersection = K.sum(K.abs(y_true * y_pred), axis=[1, 2])
union = K.sum(y_true, [1, 2]) + K.sum(y_pred, [1, 2]) - intersection
iou = K.mean((intersection + smooth) / (union + smooth), axis=0)
return iou
def recall(y_true, y_pred):
# y_true_f = K.flatten(y_true)
# y_pred_f = K.flatten(y_pred)
y_true_f = tf.reshape(y_true, [-1])
y_pred_f = tf.reshape(y_pred, [-1])
y_true_f = tf.round(y_true_f)
y_pred_f = tf.round(y_pred_f)
intersection = tf.reduce_sum(tf.multiply(y_true_f, y_pred_f))
re = intersection / (tf.reduce_sum(y_true_f) + 1)
return re
def precision(y_true, y_pred):
# y_true_f = K.flatten(y_true)
# y_pred_f = K.flatten(y_pred)
y_true_f = tf.reshape(y_true, [-1])
y_pred_f = tf.reshape(y_pred, [-1])
y_true_f = tf.round(y_true_f)
y_pred_f = tf.round(y_pred_f)
intersection = tf.reduce_sum(tf.multiply(y_true_f, y_pred_f))
# print()
pr = intersection / (tf.reduce_sum(y_pred_f) + 1)
return pr
def f1_score(y_true, y_pred):
y_true_f = tf.reshape(y_true, [-1])
y_pred_f = tf.reshape(y_pred, [-1])
y_true_f = tf.round(y_true_f)
y_pred_f = tf.round(y_pred_f)
# TP (true positives) is the number of pixels that are correctly classified as positive (belonging to the object of interest)
# FP (false positives) is the number of pixels that are incorrectly classified as positive
# FN (false negatives) is the number of pixels that are incorrectly classified as negative (not belonging to the object of interest)
TP = tf.reduce_sum(tf.multiply(y_true_f, y_pred_f))
FP = tf.reduce_sum(tf.subtract(y_pred_f, tf.multiply(y_true_f, y_pred_f)))
FN = tf.reduce_sum(tf.subtract(y_true_f, tf.multiply(y_true_f, y_pred_f)))
# epsilon() is added to the denominator to avoid division by zero.
precision = TP / (TP + FP + tf.keras.backend.epsilon())
recall = TP / (TP + FN + tf.keras.backend.epsilon())
f1 = 2 * (precision * recall) / (precision + recall + tf.keras.backend.epsilon())
return f1
model.compile(
optimizer=Adam(lr=1e-3),
loss="binary_crossentropy",
metrics=["accuracy", "mse", dice_coeff, iou_coef, recall, precision, f1_score],
)
from keras.callbacks import EarlyStopping, ReduceLROnPlateau
# Define early stopping and reduce learning rate on plateau callbacks
early_stop = EarlyStopping(
monitor="val_loss", patience=4, verbose=1, mode="auto", restore_best_weights=True
)
checkpointer = tf.keras.callbacks.ModelCheckpoint(
"model.h5", verbose=1, save_best_only=True
)
reduce_lr = ReduceLROnPlateau(
monitor="val_loss", factor=0.1, patience=4, verbose=1, mode="auto"
)
results = model.fit(
x_train,
y_train,
batch_size=16,
epochs=50,
callbacks=[reduce_lr, checkpointer],
verbose=1,
validation_data=(x_val, y_val),
)
results.history.keys()
loss, accuracy, mse = (
results.history["loss"],
results.history["accuracy"],
results.history["mse"],
)
val_loss, val_accuracy, val_mse = (
results.history["val_loss"],
results.history["val_accuracy"],
results.history["val_mse"],
)
dice_coeff, iou_coef, recall, precision, f1_score = (
results.history["dice_coeff"],
results.history["iou_coef"],
results.history["recall"],
results.history["precision"],
results.history["f1_score"],
)
val_dice_coeff, val_iou_coef, val_recall, val_precision, val_f1_score = (
results.history["val_dice_coeff"],
results.history["val_iou_coef"],
results.history["val_recall"],
results.history["val_precision"],
results.history["val_f1_score"],
)
# # Graph Visualization
plt.figure(figsize=(20, 8))
plt.subplot(1, 5, 1)
plt.title("Model Loss")
plt.plot(loss, label="Training")
plt.plot(val_loss, label="Validation")
plt.legend()
plt.grid()
plt.subplot(1, 5, 2)
plt.title("Model Accuracy")
plt.plot(accuracy, label="Training")
plt.plot(val_accuracy, label="Validation")
plt.legend()
plt.grid()
plt.subplot(1, 5, 3)
plt.title("Model Dice_Coefficient")
plt.plot(dice_coeff, label="Training")
plt.plot(val_dice_coeff, label="Validation")
plt.legend()
plt.grid()
plt.subplot(1, 5, 4)
plt.title("Model IoU")
plt.plot(iou_coef, label="Training")
plt.plot(val_iou_coef, label="Validation")
plt.legend()
plt.grid()
plt.subplot(1, 5, 5)
plt.title("Model F1_Score")
plt.plot(f1_score, label="Training")
plt.plot(val_f1_score, label="Validation")
plt.legend()
plt.grid()
plt.show()
# # Image Visualization
def show_image(image, title=None, cmap=None, alpha=1):
plt.imshow(image, cmap=cmap, alpha=alpha)
if title is not None:
plt.title(title)
plt.axis("off")
def show_mask(image, mask, cmap=None, alpha=0.4):
plt.imshow(image)
plt.imshow(tf.squeeze(mask), cmap=cmap, alpha=alpha)
plt.axis("off")
plt.figure(figsize=(20, 25))
n = 0
indices = [50, 7, 60, 61, 64] # fixed set of 5 image indices
for i in range(1, (5 * 3) + 1):
plt.subplot(5, 3, i)
if n == 0:
id = indices[i // 3] # select the corresponding fixed index
image = images[id]
mask = masks[id]
pred_mask = model.predict(image[np.newaxis, ...])
plt.title("Original Mask")
show_mask(image, mask)
n += 1
elif n == 1:
plt.title("Predicted Mask")
show_mask(image, pred_mask)
n += 1
elif n == 2:
pred_mask = (pred_mask > 0.5).astype("float")
plt.title("Processed Mask")
show_mask(image, pred_mask)
n = 0
plt.tight_layout()
plt.show()
# # Testing
# Make predictions on the test images
predictions = model.predict(x_test)
# Evaluate the model on the test set
metrics = model.evaluate(x_test, y_test, verbose=0)
# Print the accuracy and MSE
print("Loss on test set: {:.3f}".format(metrics[0]))
print("Accuracy on test set: {:.3f}".format(metrics[1]))
print("MSE on test set: {:.3f}".format(metrics[2]))
# Compute the mean IoU metric on the test set
mean_iou = tf.keras.metrics.MeanIoU(num_classes=2)
mean_iou.update_state(y_test, predictions)
print("Mean IoU on test set: {:.3f}".format(mean_iou.result()))
# Calculate Dice coefficient from IoU
dice_coefficient = 2 * mean_iou.result().numpy() / (1 + mean_iou.result().numpy())
print("Mean Dice coefficient on test set: {:.3f}".format(dice_coefficient))
# Compute the precision and recall on the test set
precision = tf.keras.metrics.Precision()
recall = tf.keras.metrics.Recall()
precision.update_state(y_test, predictions)
recall.update_state(y_test, predictions)
print("precision on test set: {:.3f}".format(precision.result()))
print("recall on test set: {:.3f}".format(recall.result()))
# Compute the F1 score on the test set
f1_score = (
2 * (precision.result() * recall.result()) / (precision.result() + recall.result())
)
print("F1 score on test set: {:.3f}".format(f1_score))
# Reset the metrics
mean_iou.reset_states()
precision.reset_states()
recall.reset_states()
# # ROC Curve
from sklearn.metrics import roc_curve, auc
y_pred = predictions.ravel()
y_test = y_test.ravel()
y_test_binary = np.where(y_test > 0.5, 1, 0)
fpr, tpr, _ = roc_curve(y_test_binary, y_pred)
roc_auc = auc(fpr, tpr)
plt.figure()
plt.plot(fpr, tpr, color="darkorange", lw=2, label="ROC curve (area = %0.2f)" % roc_auc)
plt.plot([0, 1], [0, 1], color="navy", lw=2, linestyle="--")
plt.xlim([0.0, 1.0])
plt.ylim([0.0, 1.05])
plt.xlabel("False Positive Rate")
plt.ylabel("True Positive Rate")
plt.title("Receiver operating characteristic (ROC) curve")
plt.legend(loc="lower right")
plt.show()
|
import numpy as np
import pandas as pd
import os
import json
from pathlib import Path
import matplotlib.pyplot as plt
from matplotlib import colors
import numpy as np
for dirname, _, filenames in os.walk("/kaggle/input"):
print(dirname)
from pathlib import Path
data_path = Path("/kaggle/input/abstraction-and-reasoning-challenge/")
training_path = data_path / "training"
evaluation_path = data_path / "evaluation"
test_path = data_path / "test"
training_tasks = sorted(os.listdir(training_path))
evaluation_tasks = sorted(os.listdir(evaluation_path))
test_tasks = sorted(os.listdir(test_path))
print(training_tasks[:3])
print(len(training_tasks), len(evaluation_tasks), len(test_tasks))
def plot_task(task):
n = len(task["train"]) + len(task["test"])
cmap = colors.ListedColormap(
[
"#000000",
"#0074D9",
"#FF4136",
"#2ECC40",
"#FFDC00",
"#AAAAAA",
"#F012BE",
"#FF851B",
"#7FDBFF",
"#870C25",
]
)
norm = colors.Normalize(vmin=0, vmax=9)
fig, axs = plt.subplots(1, n * 2, figsize=(25, 15), dpi=50)
fig_num = 0
for i, t in enumerate(task["train"]):
axs[fig_num].imshow(t["input"], cmap=cmap, norm=norm)
axs[fig_num].set_title(f"Train-{i} in")
axs[fig_num + 1].imshow(t["output"], cmap=cmap, norm=norm)
axs[fig_num + 1].set_title(f"Train-{i} out")
fig_num += 2
for i, t in enumerate(task["test"]):
axs[fig_num].imshow(t["input"], cmap=cmap, norm=norm)
axs[fig_num].set_title(f"Test-{i} in")
axs[fig_num + 1].imshow(t["output"], cmap=cmap, norm=norm)
axs[fig_num + 1].set_title(f"Test-{i} out")
fig_num += 2
plt.tight_layout()
plt.show()
# # Training Data
for i, json_path in enumerate(training_tasks):
task_file = str(training_path / json_path)
with open(task_file, "r") as f:
task = json.load(f)
print(f"{i:03d}", task_file)
plot_task(task)
# # Evaluation Data
for i, json_path in enumerate(evaluation_tasks):
task_file = str(evaluation_path / json_path)
with open(task_file, "r") as f:
task = json.load(f)
print(f"{i:03d}", task_file)
plot_task(task)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
import pandas_profiling
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
sales = pd.read_csv("/kaggle/input/supermarket-sales/supermarket_sales - Sheet1.csv")
sales.head()
# # Data Pre-processing
sales.info()
sales.describe()
sales.shape
len(sales)
len(sales.columns)
sales.columns
sales.isna().sum()
sales.quantile(q=0.5)
sales["Tax 5%"].quantile(q=0.25)
# # Exploratory Data Analysis
# Pandas Profiler's Interactive Report
# help(pandas_profiling)
sales.profile_report(title="Sales-Report", progress_bar=False)
corr = sales.corr()
corr
plt.figure(figsize=(10, 10))
sns.heatmap(corr, annot=True, fmt="0.2f", cmap="Greens")
|
# ### Анализ игр
# Перед нами стоит задача провести анализ продаж по играм и сформулировать рекомендации, на чем фокусироваться в следующем 2017 году, возможно найти какие-то тренды.
# Из открытых источников доступны исторические данные о продажах игр, оценки пользователей и экспертов, жанры и платформы.
# Причешем эти данные, чтобы с ними можно было работать, посмотрим на основные тенденции рынка и сформулируем две гипотезы, которые далее и проверим.
# ### Предобработка данных
# Начнем с того, что загрузим массив данных и посмотрим на него
import pandas as pd
import numpy as np
import matplotlib as mpl
import matplotlib.pyplot as plt
from scipy import stats as st
try:
data = pd.read_csv("games.csv")
except:
data = pd.read_csv("/datasets/games.csv")
data.head()
# Разделители были запятой, поэтому данные нормально загрузились в датафрейм. Теперь посмотрим информацию о том, какое у нас количество данных и как они хранятся
data.info()
# Из явных проблем - это пропуски в **годах выпуска** и неверный тип данных для **пользовательской оценки**.
# Пропуски в годах критичны, потому что мы строим прогноз, а без информации о дате релиза данные станут для нас бесполезны.
# Пользовательская оценка - это численное значение, на основании которого мы будем смотреть рейтинги и строить гипотезы. Если они хранятся в типе object, это нам не позволит делать математические вычисления с параметром.
# Из неявных проблем - **год релиза** распознан как float. Мешать это не будет, но это испортит эстетическую часть анализа данных. Если заказчик видит графики, где год обозначен, как "2002.0", это вводит в заблуждение.
# Еще одна неявная проблема - это **названия столбцов**. Для комфортной работы приведем названия к нижнему регистру.
# Посмотрим еще на распределения данных. Возможно, графики покажут нам какие-то явные особенности в данных.
data.hist(figsize=(15, 20))
# Явных проблем с данными не выявлено. Приступим к устранению проблем в данных.
# #### Устранение пропусков и ошибок формата данных
# Переименуем названия столбцов
# data = data.rename(columns={'Platform':'platform', 'Year_of_Release':'year_of_release', 'Genre':'genre', 'NA_sales':'na_sales', 'EU_sales':'eu_sales', 'JP_sales':'jp_sales', 'Other_sales':'other_sales', 'Critic_Score':'critic_score', 'User_Score':'user_score', 'Rating':'rating', 'Name':'name'})
data.columns = data.columns.str.lower()
data.info()
# Теперь разберемся с проблемой в типе данных пользовательской оценки. Посмотрим, какие оценки выставляли пользователи и из-за чего pandas посчитала этот тип данных строкой.
data["user_score"].unique()
# Видим значение "tbd", переводится как to be defined/dterminated (будет определено позже). Значит, что с рейтингом этих игр пользователи еще не определились. По сути, это такой же пропуск, как и nan. Заменим tbd на nan. Самих рейтингов мы не узнаем, но это позволит нам работать с переменной, как с float.
data["user_score"] = data["user_score"].replace("tbd", np.NaN)
data["user_score"] = data["user_score"].astype("float64")
data.info()
# Теперь поработаем с удалением пропусков.
# Убедимся, что в датафрейме отсутствуют явные дубли и неявные дубли в названиях платформ
data.duplicated().sum()
# Явные дубли отсутствуют. Посмотрим на платформы.
data["platform"].sort_values().unique()
# Тут тоже неявных дублей не выявлено.
# Помимо пропусков в годах, есть проуски в названиях жанров, названиях игр. Посмотрим на пропуски в названиях жанров.
data.loc[data["genre"].isna()]
# Видим, что эти же два пропуска в названиях игр.
# Суммарно, если мы удалим эти пустые строки, мы потеряем менее 10% данных. Это допустимо, поэтому удаляем.
data = data.loc[
~(data["genre"].isna()) & ~(data["name"].isna()) & ~(data["year_of_release"].isna())
]
data.info()
# Теперь осталось перевести тип данных года в целочисленные.
data["year_of_release"] = data["year_of_release"].astype("Int64")
data.info()
# Пропуски устранены.
# Давайте посмотрим на уникальные значения в оценках критиков.
data["critic_score"].sort_values().unique()
# Выглядит так, что оценки являются целочисленными, но из-за nan формат данных определен неверно.
# Переведем в int
data["critic_score"] = data["critic_score"].astype("Int64")
data.info()
# Теперь можно приступать к анализу данных.
# ### Исследовательский анализ
# #### Общее распределение
# Для начала, посчитаем глобальные продажи, чтобы можно было по этому параметру смотреть общие тенденции рынка
# data['total_sales'] = data['na_sales'] + data['eu_sales'] + data['jp_sales'] + data['other_sales']
data["total_sales"] = data[["na_sales", "eu_sales", "jp_sales", "other_sales"]].sum(
axis=1
)
# Теперь посмотрим на график продаж по годам.
data.pivot_table(
index="year_of_release", values="total_sales", aggfunc="sum"
).sort_values(by="year_of_release").plot(kind="bar", figsize=(15, 7))
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.xlabel("Годы", fontsize=14)
plt.ylabel("Количество продаж в млн. копий", fontsize=14)
plt.title("Диаграмма продаж игр по годам", fontsize=16)
plt.show()
# Мы видим, что продажи имеют волнообразный характер, при этом, крупный пик приходится на 2007-2008 годы. После этого идет спад.
# Явный спад в 2016 году объясняется, скорее всего, тем, что у нас пока отсутствуют данные за 4 квартал (еще не отчитались все о продажах), а может быть и за пол года. Тем не менее, это необходимо учитывать, при анализе данных. Чтобы выявить тенденции роста на падающем рынке, не достаточно смотреть на абсолютные значения.
# Посмотрим на распределение продаж по платформам и выявим лидеров продаж.
# #### Выберем топ 6 платформ
data.pivot_table(index="platform", values="total_sales", aggfunc="sum").sort_values(
by="total_sales"
).plot(kind="bar", figsize=(15, 7))
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.xlabel("Годы", fontsize=14)
plt.ylabel("Количество продаж в млн. копий", fontsize=14)
plt.title("Диаграмма международных продаж игр по платформам", fontsize=16)
plt.show()
# Янвыми лидерами являются PS, PS2, PS3, DS, Wii и X-box 360.
# Разница между устаревшей PS и новой PS4 практически в 2 раза, поэтому для анализа тенденций роста, остановимся на них.
# Посмотрим на диаграммы для каждой из платформ
data.loc[data["platform"] == "PS"].pivot_table(
index="year_of_release", values="total_sales", aggfunc="sum"
).sort_values(by="year_of_release").plot(kind="bar", figsize=(15, 7))
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.xlabel("Годы", fontsize=14)
plt.ylabel("Количество продаж в млн. копий", fontsize=14)
plt.title("Диаграмма продаж игр по годам для PS", fontsize=16)
plt.show()
data.loc[data["platform"] == "DS"].pivot_table(
index="year_of_release", values="total_sales", aggfunc="sum"
).sort_values(by="year_of_release").plot(kind="bar", figsize=(15, 7))
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.xlabel("Годы", fontsize=14)
plt.ylabel("Количество продаж в млн. копий", fontsize=14)
plt.title("Диаграмма продаж игр по годам для DS", fontsize=16)
plt.show()
data.loc[data["platform"] == "PS3"].pivot_table(
index="year_of_release", values="total_sales", aggfunc="sum"
).sort_values(by="year_of_release").plot(kind="bar", figsize=(15, 7))
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.xlabel("Годы", fontsize=14)
plt.ylabel("Количество продаж в млн. копий", fontsize=14)
plt.title("Диаграмма продаж игр по годам для PS3", fontsize=16)
plt.show()
data.loc[data["platform"] == "Wii"].pivot_table(
index="year_of_release", values="total_sales", aggfunc="sum"
).sort_values(by="year_of_release").plot(kind="bar", figsize=(15, 7))
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.xlabel("Годы", fontsize=14)
plt.ylabel("Количество продаж в млн. копий", fontsize=14)
plt.title("Диаграмма продаж игр по годам для Wii", fontsize=16)
plt.show()
data.loc[data["platform"] == "X360"].pivot_table(
index="year_of_release", values="total_sales", aggfunc="sum"
).sort_values(by="year_of_release").plot(kind="bar", figsize=(15, 7))
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.xlabel("Годы", fontsize=14)
plt.ylabel("Количество продаж в млн. копий", fontsize=14)
plt.title("Диаграмма продаж игр по годам для X-box 360", fontsize=16)
plt.show()
data.loc[data["platform"] == "PS2"].pivot_table(
index="year_of_release", values="total_sales", aggfunc="sum"
).sort_values(by="year_of_release").plot(kind="bar", figsize=(15, 7))
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.xlabel("Годы", fontsize=14)
plt.ylabel("Количество продаж в млн. копий", fontsize=14)
plt.title("Диаграмма продаж игр по годам для PS2", fontsize=16)
plt.show()
# Из этих диаграмм видно, что характероное время "жизни" платформы составляет 8 лет - 4 года роста и 4 года падения.
# Это значит, что если мы смотрим на тенденцию за последние 4 года, то динамика продаж должна быть положительной на протяжении всего этого периода. Если динамика положительная за весь период, то мы находим стабильную по продажам, но убывающую платформу. Если динамика положительная только за 2 года, то мы находим предположительно новую "звезду".
# Дополнительно к этому можно заметить особенности продаж для одгого производителя. Явный спад предыдущей консоли (PS) совпадает со стартом следующего поколения (PS2).
# Посмотрим на распределение продаж по годам с 2012 года.
data.loc[data["year_of_release"] >= 2012].pivot_table(
index="year_of_release", columns="platform", values="total_sales", aggfunc="sum"
).plot(figsize=(15, 10))
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.xlabel("Годы", fontsize=14)
plt.ylabel("Количество продаж в млн. копий", fontsize=14)
plt.title("График продаж игр по годам для платформ", fontsize=16)
plt.show()
# Явный рост наблюдается для PS4 и X-box One. При этом, 2016 год все портит. Либо идет реальный спад по этим платформам, либо сказывается неполный период 2016 года.
# Попробуем нормализовать данные и посмотрим на них еще раз
for i in data["year_of_release"].unique():
data.loc[data["year_of_release"] == i, "norm_sales"] = (
data.loc[data["year_of_release"] == i, "total_sales"]
/ data.loc[data["year_of_release"] == i, "total_sales"].sum()
)
data.loc[data["year_of_release"] >= 2012].pivot_table(
index="year_of_release", columns="platform", values="norm_sales", aggfunc="sum"
).plot(figsize=(15, 10))
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.xlabel("Годы", fontsize=14)
plt.ylabel("Количество продаж в млн. копий", fontsize=14)
plt.title("График продаж игр по годам для платформ", fontsize=16)
plt.show()
# Теперь мы видим явный рост PS4 и X-box One. Причем, как и в предыдущие разы, смена поколений консолей явно порождает спад продаж предыдущего поколения и рост продаж актуального.
# Можно определить PS4 и X-box One как потенциально прибыльные платформы.
# Посмотрим на распределение продаж по платформам на предмет явных выбросов данных.
data.loc[data["year_of_release"] >= 2012].pivot_table(
index="year_of_release", columns="platform", values="total_sales", aggfunc="sum"
).boxplot(figsize=(15, 7))
# Имеются незначительные выбросы для некоторых платформ. Явно эти года были для них крайне удачными или крайне неудачными. А в остальном мы видим, что большинство продаж по платформ находятся в пределах нормального распределения.
# Медианное значение явно впереди у PS4
# #### Определим популрные жанры
data.loc[data["year_of_release"] >= 2012].pivot_table(
index="year_of_release", columns="genre", values="total_sales", aggfunc="sum"
).plot(figsize=(15, 10))
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.xlabel("Годы", fontsize=14)
plt.ylabel("Количество продаж в млн. копий", fontsize=14)
plt.title("График продаж игр по годам для разных жанров", fontsize=16)
plt.show()
# К сожалению, на спаде не получается выявить актуальных тенденций рынка. Попробуем посмотреть медианные значения продаж по жанрам.
data.loc[data["year_of_release"] >= 2012].pivot_table(
index="year_of_release", columns="genre", values="total_sales", aggfunc="median"
).plot(figsize=(15, 10))
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.xlabel("Годы", fontsize=14)
plt.ylabel("Медиана продаж в млн. копий", fontsize=14)
plt.title("График продаж игр по годам для разных жанров", fontsize=16)
plt.show()
# Не забываем о том, что данные за 2016 год не полные.
# Теперь отчетливо видно, что в лидеры выбиваются игры жанра Action и Shooter.
# #### Проверим, есть ли зависимость продаж от рейтингов
# Для анализа возьмем две восходящие звезды - PS4 и Xbox One
# #### PS4
# Для начала посмотрим на корреляцию продаж от рейтингов критиков.
data.loc[data["platform"] == "PS4"].plot(
x="total_sales", y="critic_score", kind="scatter", alpha=0.5, figsize=(15, 7)
)
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.xlabel("Количество продаж", fontsize=14)
plt.ylabel("Оценки критиков", fontsize=14)
plt.title("Корреляция количества продаж от оценок критиков", fontsize=16)
plt.show()
data.loc[data["platform"] == "PS4", "total_sales"].corr(
data.loc[data["platform"] == "PS4", "critic_score"].astype("float")
)
# Явной корреляции не наблюдается, но можно обратить внимание на то, что хиты продаж чаще встречаются среди игр с высоким рейтингом.
# Теперь посмотрим на зависимость продаж от рейтинга пользователей
data.loc[data["platform"] == "PS4"].plot(
x="total_sales", y="user_score", kind="scatter", alpha=0.5, figsize=(15, 7)
)
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.xlabel("Количество продаж", fontsize=14)
plt.ylabel("Оценки пользователей", fontsize=14)
plt.title("Корреляция количества продаж от оценок пользователей", fontsize=16)
plt.show()
data.loc[data["platform"] == "PS4", "total_sales"].corr(
data.loc[data["platform"] == "PS4", "user_score"]
)
# Аналогично, прямой корреляции нет, но тут уже чаще можно встретить хиты продаж с не самыми высокими рейтингами.
# Тут могут быть две ситуации:
# - пользователи покупают игру по отзывам критиков, а она им "не зашла"
# - есть нишевые игры, которые покупают фанаты, несмотря на низкие оценки
# #### Xbox One
# Теперь посмотрим, есть ли такая зависимость у конкурента
data.loc[data["platform"] == "XOne"].plot(
x="total_sales", y="critic_score", kind="scatter", alpha=0.5, figsize=(15, 7)
)
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.xlabel("Количество продаж", fontsize=14)
plt.ylabel("Оценки критиков", fontsize=14)
plt.title("Корреляция количества продаж от оценок критиков", fontsize=16)
plt.show()
data.loc[data["platform"] == "XOne", "critic_score"].astype("float").corr(
data.loc[data["platform"] == "XOne", "total_sales"]
)
# Аналогично PS4 прямой зависимости нет, но игры с высоким рейтингом критиков чаще становятся бестселлерами
data.loc[data["platform"] == "XOne"].plot(
x="total_sales", y="user_score", kind="scatter", alpha=0.5, figsize=(15, 7)
)
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.xlabel("Количество продаж", fontsize=14)
plt.ylabel("Оценки пользователей", fontsize=14)
plt.title("Корреляция количества продаж от оценок пользователей", fontsize=16)
plt.show()
data.loc[data["platform"] == "XOne", "total_sales"].corr(
data.loc[data["platform"] == "XOne", "user_score"]
)
# Тут еще больше разброс. На рейтинг пользователей ориентироваться точно не стоит.
# #### Посмотрим на особенности региональных продаж
data.loc[data["year_of_release"] >= 2012].pivot_table(
index="platform", values="eu_sales", aggfunc="sum"
).sort_values(by="eu_sales").plot(kind="bar", figsize=(15, 7))
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.xlabel("Платформы", fontsize=14)
plt.ylabel("Количество продаж в млн. копий", fontsize=14)
plt.title("Диаграмма продаж игр в европейском регионе по платформам", fontsize=16)
plt.show()
# В европе дико популярны игры на PS (3 и 4). Xbox наступают на пятки
data.loc[data["year_of_release"] >= 2012].pivot_table(
index="platform", values="jp_sales", aggfunc="sum"
).sort_values(by="jp_sales").plot(kind="bar", figsize=(15, 7))
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.xlabel("Платформы", fontsize=14)
plt.ylabel("Количество продаж в млн. копий", fontsize=14)
plt.title("Диаграмма продаж игр в японском регионе по платформам", fontsize=16)
plt.show()
# В Японском регионе на компьютере продаж практически нет, при этом значительно выделяется 3DS в топе. Возможно, это связано с тем, что это портативная платформа и особенность менталитета стимулирует покупать игры на платформе, в которе можно играть "на ходу".
# Естественно, в японском регионе скученность японских платформ выше. Игры на Xbox тут практически не покупают
data.loc[data["year_of_release"] >= 2012].pivot_table(
index="platform", values="na_sales", aggfunc="sum"
).sort_values(by="na_sales").plot(kind="bar", figsize=(15, 7))
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.xlabel("Платформы", fontsize=14)
plt.ylabel("Количество продаж в млн. копий", fontsize=14)
plt.title(
"Диаграмма продаж игр в северо-американском регионе по платформам", fontsize=16
)
plt.show()
# В Северо-американском регионе основная доля рынка приходится на Xbox One/360 и PS3/PS4. При том, что нет явных тенденций, кажется разумным ориентироваться на игры, которые поддерживаются консолями предыдущих поколений.
data.loc[data["year_of_release"] >= 2012].pivot_table(
index="platform", values="other_sales", aggfunc="sum"
).sort_values(by="other_sales").plot(kind="bar", figsize=(15, 7))
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.xlabel("Платформы", fontsize=14)
plt.ylabel("Количество продаж в млн. копий", fontsize=14)
plt.title("Диаграмма продаж игр в остальных регионах по платформам", fontsize=16)
plt.show()
# В остальных регионах явный топ у PS4 и PS3.
# Основную долю в продажи вносит северо-американский и еропейские регионы. Япония и остальные регионы в 2 раза меньше по продажам.
# Если ориентироваться на рынок продаж, то стоит обратить внимание на эти регионы.
# #### Теперь посмотрим жанровые предпочтения по регионам
# Проверим, есть ли региональная зависимость у жанра продаваемых игр.
data.loc[data["year_of_release"] >= 2012].pivot_table(
index="genre", values="eu_sales", aggfunc="sum"
).sort_values(by="eu_sales").plot(kind="bar", figsize=(15, 7))
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.xlabel("Жанры", fontsize=14)
plt.ylabel("Количество продаж в млн. копий", fontsize=14)
plt.title("Диаграмма продаж игр в европейском регионе по жанрам", fontsize=16)
plt.show()
# В европейском регионе предпочитают экшн, шутеры и спортивные игры. Это соответствует мировым тенденциям
data.loc[data["year_of_release"] >= 2012].pivot_table(
index="genre", values="jp_sales", aggfunc="sum"
).sort_values(by="jp_sales").plot(kind="bar", figsize=(15, 7))
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.xlabel("Жанры", fontsize=14)
plt.ylabel("Количество продаж в млн. копий", fontsize=14)
plt.title("Диаграмма продаж игр в японском регионе по жанрам", fontsize=16)
plt.show()
# Японский регион явно имеет перекос на ролевые и экшн игры. В случае, если магазин будет ориентироваться на японский регион, следует учитывать эту особенность
data.loc[data["year_of_release"] >= 2012].pivot_table(
index="genre", values="na_sales", aggfunc="sum"
).sort_values(by="na_sales").plot(kind="bar", figsize=(15, 7))
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.xlabel("Жанры", fontsize=14)
plt.ylabel("Количество продаж в млн. копий", fontsize=14)
plt.title("Диаграмма продаж игр в северо-американском регионе по жанрам", fontsize=16)
plt.show()
# В северо-американском регионе, как и в европейском, без каких-то отклонений от общемировых трендов (или правильнее будет говорить, что они и задают мировые тренды)
data.loc[data["year_of_release"] >= 2012].pivot_table(
index="genre", values="other_sales", aggfunc="sum"
).sort_values(by="other_sales").plot(kind="bar", figsize=(15, 7))
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.xlabel("Жанры", fontsize=14)
plt.ylabel("Количество продаж в млн. копий", fontsize=14)
plt.title("Диаграмма продаж игр в остальных регионах по жанрам", fontsize=16)
plt.show()
# В оатсльных регионах такая же картина.
# Вывод: самые популярные жанры - экшн, спортивные игры и шутеры. В японском регионе стоит обратить внимание на ролевые игры.
# #### Есть ли зависимость продаж от возрастных рейтингов
# Давайте посмотрим на это в разрезе региональных продаж.
data.loc[data["year_of_release"] >= 2012].pivot_table(
index="rating", values="eu_sales", aggfunc="sum"
).sort_values(by="eu_sales").plot(kind="bar", figsize=(15, 7))
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.xlabel("Рейтинг", fontsize=14)
plt.ylabel("Количество продаж в млн. копий", fontsize=14)
plt.title("Диаграмма продаж игр в европейских регионах по рейтингу", fontsize=16)
plt.show()
# В европейских регионах преобладают продажи игр с рейтингом 18+, далее - "Для всех".
#
data.loc[data["year_of_release"] >= 2012].pivot_table(
index="rating", values="jp_sales", aggfunc="sum"
).sort_values(by="jp_sales").plot(kind="bar", figsize=(15, 7))
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.xlabel("Рейтинг", fontsize=14)
plt.ylabel("Количество продаж в млн. копий", fontsize=14)
plt.title("Диаграмма продаж игр в японском регионе по рейтингу", fontsize=16)
plt.show()
# В японском регионе основные продажи приходятся на игры с рейтингом "Для всех", "Для подростков" и на третьем месте игры с рейтингом 18+
data.loc[data["year_of_release"] >= 2012].pivot_table(
index="rating", values="na_sales", aggfunc="sum"
).sort_values(by="na_sales").plot(kind="bar", figsize=(15, 7))
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.xlabel("Рейтинг", fontsize=14)
plt.ylabel("Количество продаж в млн. копий", fontsize=14)
plt.title("Диаграмма продаж игр в северо-американском регионе по рейтингу", fontsize=16)
plt.show()
# У Американского региона порядок такой же, как у Европейского. Основные продажи приходятся на игры 18+ и "Для всех"
data.loc[data["year_of_release"] >= 2012].pivot_table(
index="rating", values="other_sales", aggfunc="sum"
).sort_values(by="other_sales").plot(kind="bar", figsize=(15, 7))
plt.xticks(fontsize=14)
plt.yticks(fontsize=14)
plt.xlabel("Рейтинг", fontsize=14)
plt.ylabel("Количество продаж в млн. копий", fontsize=14)
plt.title("Диаграмма продаж игр в остальных регионах по рейтингу", fontsize=16)
plt.show()
# В остальных регионах рейтинги продаж аналогичны европейскому региону. Самые популярные игры 18+, потом "Для всех"
# Выводы: В японском регионе стоит ориентироваться на рейтинг Е, Т и М, тогда как в остальных регионах преобладает интерес к играм рейтинга М, а затем уже Е.
# ### Проверка гипотез
# Сформулируем две гипотезы и проверим их.
# #### Гипотеза 1
# Средние пользовательские рейтинги платформ Xbox One и PC одинаковые.
# Нулевая гипотеза - пользовательские рейтинги платформы Xbox One равны пользовательским рейтингам платформы PC
# Альтернативная гипотеза - пользовательские рейтинги платформы Xbox One не равны пользовательским рейтингам платформы PC
# Определим точность поргового значения в 5%.
# Нет оснований пологать, что вариативность оценок и взаимосвязь между рейтингами устойчивая
results = st.ttest_ind(
data.loc[
(data["year_of_release"] >= 2012)
& (data["platform"] == "XOne")
& ~(data["user_score"].isna()),
"user_score",
],
data.loc[
(data["year_of_release"] >= 2012)
& (data["platform"] == "PC")
& ~(data["user_score"].isna()),
"user_score",
],
equal_var=False,
)
alpha = 0.05
print(results.pvalue)
if results.pvalue < alpha:
print("Отвергаем нулевую гипотезу")
else:
print("Не получилось отвергнуть нулевую гипотезу")
# Не получилось отвергнуть гипотезу о том, что пользовательские рейтинги платформы Xbox One равны пользовательским рейтингам платформы PC
# #### Гиеотеза 2
# Средние пользовательские рейтинги жанров Action и Sports разные
# Нулевая гипотеза - пользовательские рейтинги жанра Action равны пользовательским рейтингам жанра Sports
# Альтернативная гипотеза - пользовательские рейтинги жанра Action не равны пользовательским рейтингам жанра Sports
# Определим точность поргового значения в 5%.
# Нет оснований пологать, что вариативность оценок и взаимосвязь между рейтингами устойчивая
results = st.ttest_ind(
data.loc[
(data["year_of_release"] >= 2012)
& (data["genre"] == "Action")
& ~(data["user_score"].isna()),
"user_score",
],
data.loc[
(data["year_of_release"] >= 2012)
& (data["genre"] == "Sports")
& ~(data["user_score"].isna()),
"user_score",
],
equal_var=False,
)
alpha = 0.05
print(results.pvalue)
if results.pvalue < alpha:
print("Отвергаем нулевую гипотезу")
else:
print("Не получилось отвергнуть нулевую гипотезу")
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import re
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
df11 = pd.read_csv(
"/kaggle/input/the-world-university-rankings-2011-2023/2011_rankings.csv"
)
df11.info()
df11["location"].value_counts()
df11["scores_overall"].plot(kind="box", vert=False)
df11.describe()
df11["aliases"]
df23 = pd.read_csv(
"/kaggle/input/the-world-university-rankings-2011-2023/2023_rankings.csv"
)
df23.info()
subdf23 = df23[
[
"name",
"stats_number_students",
"stats_student_staff_ratio",
"stats_pc_intl_students",
"stats_female_male_ratio",
]
]
subdf23.head()
# load the datasets of top university
df22 = pd.read_csv(
"/kaggle/input/the-world-university-rankings-2011-2023/2022_rankings.csv"
)
df21 = pd.read_csv(
"/kaggle/input/the-world-university-rankings-2011-2023/2021_rankings.csv"
)
df20 = pd.read_csv(
"/kaggle/input/the-world-university-rankings-2011-2023/2020_rankings.csv"
)
df19 = pd.read_csv(
"/kaggle/input/the-world-university-rankings-2011-2023/2019_rankings.csv"
)
df22.info()
df23["location"].value_counts()[:10]
# ### Viz ideas
# 1. Top Uni rankings over a period of past 5 years
# 2. Top Uni with rankings change in over past 5 years in all measures
# 3. radar chart of of scores by individual uni.
# 4. University students count over a period a last 5 years
# ### 1. Top university ranking over past 5 years
def ranking_by_year(df, year):
rank_df = df[["name", "rank"]][:10]
rank_df["year"] = year
return rank_df
rank_19df = ranking_by_year(df19, 2019)
rank_20df = ranking_by_year(df20, 2020)
rank_21df = ranking_by_year(df21, 2021)
rank_22df = ranking_by_year(df22, 2022)
rank_23df = ranking_by_year(df23, 2023)
combined_df = pd.concat([rank_19df, rank_20df, rank_21df, rank_22df, rank_23df])
combined_df["rank"] = combined_df["rank"].apply(
lambda x: "".join([re.sub("^=", "", s) for s in x])
)
combined_df["year"] = combined_df["year"].astype("int64")
# plot the the rankings of each over past 5 years
plt.figure(figsize=(10, 7))
sns.lineplot(x="year", y="rank", hue="name", data=combined_df)
plt.xticks(ticks=[2019, 2020, 2021, 2022, 2023], rotation=30)
plt.show()
# ### 2. Top university teaching and research scores over a past 5 years
def tr_score(df, year):
score_df = df[["name", "scores_teaching", "scores_research"]][:10]
score_df["year"] = year
return score_df
scores_df19 = tr_score(df19, 2019)
scores_df20 = tr_score(df20, 2020)
scores_df21 = tr_score(df21, 2021)
scores_df22 = tr_score(df22, 2022)
scores_df23 = tr_score(df23, 2023)
# combined dataframes for data viz
combined_score_df = pd.concat(
[scores_df19, scores_df20, scores_df21, scores_df22, scores_df23]
)
# plot the viz
plt.figure(figsize=(10, 5))
sns.lineplot(
x="year",
y="scores_teaching",
hue="name",
data=combined_score_df,
legend="full",
markers=True,
)
plt.xticks(ticks=[2019, 2020, 2021, 2022, 2023], rotation=60)
plt.show()
# ### 3. Radar charts of University and their scores
df23[
[
"name",
"scores_teaching",
"scores_research",
"scores_citations",
"scores_industry_income",
"scores_international_outlook",
]
]
# Define the variables and their values for each category
categories = [
"scores_teaching",
"scores_research",
"scores_citations",
"scores_industry_income",
"scores_international_outlook",
]
values = [92.3, 99.7, 99.3, 74.9, 96.2]
width = [1, 1, 1, 1, 1]
colors = ["#ed5f18", "#edb118", "#ccc612", "#adcc12", "#60bf08"]
# Create an array of angles for the radar chart
angles = np.linspace(0, 2 * np.pi, len(categories), endpoint=False)
# Create a figure and polar axes
ax = plt.subplot(projection="polar")
ax.bar(angles, values, width=width, bottom=0.0, alpha=0.8, color=colors)
ax.set_thetagrids(angles * 180 / np.pi, categories)
plt.title("Harvard University Scores Chart")
# Show the plot
plt.show()
|
# # **Description**
# Ce projet est réalisé dans le cadre d'un test technique à ENLILIA. La description est donnée ci-dessous:
# Développer une petite interface web qui prend en entrée un texte et classifie le texte en deux classes positive/négative et affiche le résultat.
# Le classifieur positif/négatif peut être développer sur un des dataset français du Kaggle.
# **Dataset**
# * Dataset choisi : Allocine french movie reviews
# * Lien : [Allocine french movie reviews](https://www.kaggle.com/datasets/djilax/allocine-french-movie-reviews)
# * Description : L'ensemble de données contient des commentaires en français pour des films et séries récupérés sur **Allociné**
#
# Dézipper dans un dossier 'reviews/'
# !unzip '/content/Allocine french movie reviews.zip' -d 'reviews'
import transformers
import tensorflow as tf
import tensorflow.keras as keras
import tensorflow.keras.layers as layers
import matplotlib.pyplot as plt
import seaborn as sns
import numpy as np
import pandas as pd
import os
import glob
# **Data importation and EDA**
# L'ensemble de données est contenu dans reviews et est déjà séparé en train, test, val. Nous allons garder cette séparation.
train_df = pd.read_csv("/kaggle/input/allocine-french-movie-reviews/train.csv").drop(
"Unnamed: 0", axis=1
)
val_df = pd.read_csv("/kaggle/input/allocine-french-movie-reviews/valid.csv").drop(
"Unnamed: 0", axis=1
)
test_df = pd.read_csv("/kaggle/input/allocine-french-movie-reviews/test.csv").drop(
"Unnamed: 0", axis=1
)
train_df.head(5)
full_df = pd.concat([train_df, val_df, test_df], axis=0, ignore_index=True)
print("# lignes dans train", train_df.shape[0])
print("# lignes dans test", test_df.shape[0])
print("# lignes dans val", val_df.shape[0])
columns_with_null = [col for col in full_df.columns if full_df[col].isnull().sum() > 0]
print("Columns contenant des valeurs nulles", columns_with_null)
# Aucune colonne ne contient de valeur nulle.
sns.countplot(data=full_df, x="polarity")
# Le jeu de données est parfaitement équilibré.
# **Preprocessing**
BATCH_SIZE = 8
X_train, y_train = train_df["review"], train_df["polarity"]
train_ds = (
tf.data.Dataset.from_tensor_slices((X_train, y_train))
.batch(BATCH_SIZE)
.cache()
.shuffle(1000)
.prefetch(tf.data.AUTOTUNE)
)
X_test, y_test = test_df["review"], test_df["polarity"]
test_ds = (
tf.data.Dataset.from_tensor_slices((X_test, y_test))
.batch(BATCH_SIZE)
.cache()
.prefetch(tf.data.AUTOTUNE)
)
X_val, y_val = val_df["review"], val_df["polarity"]
val_ds = (
tf.data.Dataset.from_tensor_slices((X_val, y_val))
.batch(BATCH_SIZE)
.cache()
.prefetch(tf.data.AUTOTUNE)
)
# Text encoder
VOCAB_SIZE = 10000
encoder = layers.TextVectorization(max_tokens=VOCAB_SIZE)
encoder.adapt(train_ds.map(lambda text, label: text))
vocab = np.array(encoder.get_vocabulary())
vocab[:20]
encoded_example = encoder("Bonjour, c'est un test. Merci!").numpy()
encoded_example
# **Model**
def create_model():
embedding_dim = 64
inputs = layers.Input(shape=(), dtype=tf.string, name="text")
encoded = encoder(inputs)
embeddings = layers.Embedding(
input_dim=VOCAB_SIZE + 1, output_dim=embedding_dim, mask_zero=True
)(encoded)
x = layers.Bidirectional(tf.keras.layers.LSTM(64, return_sequences=True))(
embeddings
)
x = layers.Bidirectional(tf.keras.layers.LSTM(32))(x)
x = layers.Dense(64, activation="relu")(x)
output = layers.Dense(2, activation="softmax")(x)
return keras.models.Model(inputs=inputs, outputs=output)
model = create_model()
optimizer = keras.optimizers.Adam(learning_rate=1e-4)
loss = keras.losses.SparseCategoricalCrossentropy(from_logits=False)
epochs = 10
model.compile(optimizer=optimizer, loss=loss, metrics="accuracy")
model.summary()
# Fit model
history = model.fit(train_ds, epochs=epochs, validation_data=val_ds)
model.evaluate(test_ds)
def plot_graphs(history, metric):
plt.plot(history.history[metric])
plt.plot(history.history["val_" + metric], "")
plt.xlabel("Epochs")
plt.ylabel(metric)
plt.legend([metric, "val_" + metric])
# Accuracy plot
plot_graphs(history, "accuracy")
# Loss plot
plot_graphs(history, "loss")
# Enregistrement du model
model.save("saved_model/model")
# model.save('saved_model/classifier.h5', save_format='tf')
# Conversion modèle tf lite
# converter = tf.lite.TFLiteConverter.from_saved_model('saved_model/model')
converter = tf.lite.TFLiteConverter.from_keras_model(model)
tflite_model = converter.convert()
with open("model.tflite", "wb") as f:
f.write(tflite_model)
loaded_model = keras.models.load_model("saved_model/model")
loaded_model.summary()
# ## ****BERT****
# **Preprocessing**
# Import generic wrappers
from transformers import AutoModel, AutoTokenizer
# Define the model repo
# model_name = "claudelkros/bert-base-french"
model_name = "dbmdz/bert-base-french-europeana-cased"
# Download pytorch model
model = AutoModel.from_pretrained(model_name)
tokenizer = AutoTokenizer.from_pretrained(model_name)
# Transform input tokens
inputs = tokenizer("Hello world!", return_tensors="tf")
print(inputs)
MAX_LENGTH = 512
def map_to_dict(input_ids, attention_masks, token_type_ids, label):
"""
Convertir les inputs en un dictionnaire valid pour le model en tf
"""
return {
"input_ids": input_ids,
"token_type_ids": token_type_ids,
"attention_mask": attention_masks,
}, label
def encode_examples(df, tokenizer, return_ds=True):
"""
Encoder tous les exempls
:params
return_ds Retourner un Dataset tf
"""
reviews = df["review"].to_list()
labels = df["polarity"].to_list()
inputs = tokenizer.batch_encode_plus(
reviews,
add_special_tokens=True,
# max_length = MAX_LENGTH,
# pad_to_max_length = True,
padding="longest",
truncation=True,
return_attention_mask=True,
)
if not return_ds:
return inputs, labels
return tf.data.Dataset.from_tensor_slices(
(
inputs["input_ids"],
inputs["attention_mask"],
inputs["token_type_ids"],
labels,
)
).map(map_to_dict)
examples_df = encode_examples(train_df.iloc[:5, :], tokenizer, return_ds=True)
print(examples_df)
class CustomDataGen(tf.keras.utils.Sequence):
def __init__(self, df, batch_size, tokenizer, shuffle=True):
self.df = df.copy()
self.batch_size = batch_size
self.shuffle = shuffle
self.tokenizer = tokenizer
self.n = len(self.df)
def on_epoch_end(self):
if self.shuffle:
self.df = self.df.sample(frac=1).reset_index(drop=True)
def __getitem__(self, index):
low = index * self.batch_size
# Cap upper bound at array length; the last batch may be smaller
# if the total number of items is not a multiple of batch size.
high = min(low + self.batch_size, len(self.df))
batch_df = self.df.iloc[low:high, :]
encoded_batch, labels = encode_examples(batch_df, tokenizer, return_ds=False)
inputs = []
# for ids, mask, itype, label in zip(encoded_batch['input_ids'], encoded_batch['attention_mask'], encoded_batch['token_type_ids'], labels):
# example = {
# 'input_ids': ids,
# 'attention_mask': mask,
# 'token_type_ids': itype
# }, label
# inputs.append(example)
return (
tf.convert_to_tensor(encoded_batch["input_ids"]),
tf.convert_to_tensor(encoded_batch["attention_mask"]),
tf.convert_to_tensor(encoded_batch["token_type_ids"]),
), tf.convert_to_tensor(labels)
# return inputs
def __len__(self):
return self.n // self.batch_size
ex_data_gen = CustomDataGen(df=train_df.iloc[:10, :], batch_size=3, tokenizer=tokenizer)
print(len(ex_data_gen))
it = iter(ex_data_gen)
print(next(it))
BATCH_SIZE = 32
# train_ds = encode_examples(train_df, tokenizer, return_ds=True)
# train_ds = train_ds.batch(BATCH_SIZE).cache().shuffle(1000).prefetch(tf.data.AUTOTUNE)
# test_ds = encode_examples(test_df, tokenizer, return_ds=True)
# test_ds = test_ds.batch(BATCH_SIZE).cache().prefetch(tf.data.AUTOTUNE)
# val_ds = encode_examples(val_df, tokenizer, return_ds=True)
# val_ds = val_ds.batch(BATCH_SIZE).cache().prefetch(tf.data.AUTOTUNE)
BATCH_SIZE = 8
train_data_gen = CustomDataGen(
df=train_df, batch_size=BATCH_SIZE, tokenizer=tokenizer, shuffle=True
)
test_data_gen = CustomDataGen(df=test_df, batch_size=BATCH_SIZE, tokenizer=tokenizer)
val_data_gen = CustomDataGen(df=val_df, batch_size=BATCH_SIZE, tokenizer=tokenizer)
# **Model**
from transformers import TFAutoModelForSequenceClassification
model = TFAutoModelForSequenceClassification.from_pretrained(model_name)
epochs = 1
optimizer = keras.optimizers.AdamW(learning_rate=2e-5)
model.compile(
optimizer=optimizer, loss="sparse_categorical_crossentropy", metrics=["accuracy"]
)
history = model.fit(train_data_gen, epochs=epochs, validation_data=val_data_gen)
print(model.evaluate(test_data_gen))
# inputs = tokenizer("Hello world!", return_tensors="tf")
# outputs = model(**inputs)
# print(outputs)
|
# # Introduction
# One of my friends suggested that I look into this dataset to see if we can predict the rating of an app with given data. In addition, I wanted to do a simple hypothesis testing to see if games perform better than other categories of apps. To try and predict the ratings, I decided to use simple regressions. Also, since the full version of the data has more data points, I ended up using it over the 32k data.
# In the end, what I found was that the given data is not enough. General data such as the category or the number of reviews are not good predictors. I assume that better predictors would be data that evaluates user experience. For example, average number of times the user opens the app a day or a week, average number of minutes spent in the app per day, etc. When it comes to the hypothesis testing, I found that games do not really perform better than games.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from scipy import stats
from statsmodels.stats import weightstats as stests
from sklearn.preprocessing import LabelEncoder
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.preprocessing import PolynomialFeatures
from sklearn.metrics import r2_score, mean_absolute_error, mean_squared_error
data = pd.read_csv("/kaggle/input/google-playstore-apps/Google-Playstore-Full.csv")
data.head()
# # Initial data cleaning
# I dropped columns that I thought were unnecessary and renamed 'Content Rating' to 'CR,' trying to avoid having spaces in the names of the columns.
# The name of the app might play a role in the rating, but I assume it was minimal, hence why I removed it. The same logic applies to when the app was last updated and min/max versions required.
data = data.drop(
columns=[
"App Name",
"Unnamed: 11",
"Unnamed: 12",
"Unnamed: 13",
"Unnamed: 14",
"Last Updated",
"Minimum Version",
"Latest Version",
]
)
data = data.rename(columns={"Content Rating": "CR"})
# I did basic exploration of the values for Installs, Price, Reviews, and Rating. I found that with Installs that I probably should just convert it to an int value. In Price, I found couple of string that I got rid of; I also decided to treat the Price as 'Free' for 0 and 'Paid' for 1. With Reviews, I just converted the values to float. With Rating, I found that there were about 10-15 values that basically were words, so I only got values with numbers in them.
data = data[data.Size.str.contains("\d")]
data.Size[data.Size.str.contains("k")] = "0." + data.Size[
data.Size.str.contains("k")
].str.replace(".", "")
data.Size = data.Size.str.replace("k", "")
data.Size = data.Size.str.replace("M", "")
data.Size = data.Size.str.replace(",", "")
data.Size = data.Size.str.replace("+", "")
data.Size = data.Size.astype(float)
data = data[data.Installs.str.contains("\+")]
data.Installs = data.Installs.str.replace("+", "")
data.Installs = data.Installs.str.replace(",", "")
data.Installs.astype(int)
data.Price = data.Price.str.contains("1|2|3|4|5|7|8|9").replace(False, 0)
data = data[data.applymap(np.isreal).Reviews]
data.Reviews = data.Reviews.astype(float)
data = data[data.Rating.str.contains("\d") == True]
data.Rating = data.Rating.astype(float)
# When looking at categories provided initially, I immediately noticed that categories were basically divided into Games and Non-Games. Thus, I decided to split the dataset into two.
data.Category.unique()
data.Category = data.Category.fillna("Unknown")
games = data[data.Category.str.contains("GAME", regex=False)]
other = data[~data.Category.str.contains("GAME", regex=False)]
# Basic cleaning of outliers.
z_Rating = np.abs(stats.zscore(games.Rating))
games = games[z_Rating < 3]
z_Reviews = np.abs(stats.zscore(games.Reviews))
games = games[z_Reviews < 3]
z_Rating2 = np.abs(stats.zscore(other.Rating))
other = other[z_Rating2 < 3]
z_Reviews2 = np.abs(stats.zscore(other.Reviews))
other = other[z_Reviews2 < 3]
# Initial hypothesis testing before doing more exploration. As I mentioned above, I wanted to see which categories perform better than the others. It seems that games generally perform worse than other categories.
games_mean = np.mean(games.Rating)
games_std = np.std(games.Rating)
other_mean = np.mean(other.Rating)
other_std = np.std(games.Rating)
print("Games mean and std: ", games_mean, games_std)
print("Other categories mean and std: ", other_mean, other_std)
ztest, pval = stests.ztest(
games.Rating, other.Rating, usevar="pooled", value=0, alternative="smaller"
)
print("p-value: ", pval)
# # EDA
# ## Games
# By doing basic EDA and plotting simple graphs, the first thought that I had was that there is not much of a correlation between Rating and other independent variables. In the vase of Reviews vs Rating, we can see that, generally, number of reviews does not really affect the rating.
# Something interesting that I noticed was that there are not a lot of Casino games, but they perform on average better than other categories. Also, it seems that not only puzzles are popular, but also people rate puzzles higher than other games.
f, ax = plt.subplots(3, 2, figsize=(10, 15))
games.Category.value_counts().plot(kind="bar", ax=ax[0, 0])
ax[0, 0].set_title("Frequency of Games per Category")
ax[0, 1].scatter(
games.Reviews[games.Reviews < 100000], games.Rating[games.Reviews < 100000]
)
ax[0, 1].set_title("Reviews vs Rating")
ax[0, 1].set_xlabel("# of Reviews")
ax[0, 1].set_ylabel("Rating")
ax[1, 0].hist(games.Rating, range=(3, 5))
ax[1, 0].set_title("Ratings Histogram")
ax[1, 0].set_xlabel("Ratings")
d = games.groupby("Category")["Rating"].mean().reset_index()
ax[1, 1].scatter(d.Category, d.Rating)
ax[1, 1].set_xticklabels(d.Category.unique(), rotation=90)
ax[1, 1].set_title("Mean Rating per Category")
ax[2, 0].hist(games.Size, range=(0, 100), bins=10, label="Size")
ax[2, 0].set_title("Size Histogram")
ax[2, 0].set_xlabel("Size")
games.CR.value_counts().plot(kind="bar", ax=ax[2, 1])
ax[2, 1].set_title("Frequency of Games per Content Rating")
f.tight_layout()
# Some categorical encoding.
games_dum = pd.get_dummies(games, columns=["Category", "CR", "Price"])
# I got this code from [here](https://www.geeksforgeeks.org/exploring-correlation-in-python/). As I predicted above, we can see that there is no correlation between Rating and other independent variables.
corrmat = games_dum.corr()
f, ax = plt.subplots(figsize=(9, 8))
sns.heatmap(corrmat, ax=ax, cmap="YlGnBu", linewidths=0.1)
# ## Other Categories
# After doing some EDA for Games, I figured that I should do the same for other categories. I decided that I get top 17 categories here, since in games I only had 17 types. I went through the same process for EDA here, so I think there is not much of an explanation needed here.
other = other[other.Category.map(other.Category.value_counts() > 3500)]
f, ax = plt.subplots(3, 2, figsize=(10, 15))
other.Category.value_counts().plot(kind="bar", ax=ax[0, 0])
ax[0, 0].set_title("Frequency of Others per Category")
ax[0, 1].scatter(
other.Reviews[other.Reviews < 100000], other.Rating[other.Reviews < 100000]
)
ax[0, 1].set_title("Reviews vs Rating")
ax[0, 1].set_xlabel("# of Reviews")
ax[0, 1].set_ylabel("Rating")
ax[1, 0].hist(other.Rating, range=(3, 5))
ax[1, 0].set_title("Ratings Histogram")
ax[1, 0].set_xlabel("Ratings")
d = other.groupby("Category")["Rating"].mean().reset_index()
ax[1, 1].scatter(d.Category, d.Rating)
ax[1, 1].set_xticklabels(d.Category.unique(), rotation=90)
ax[1, 1].set_title("Mean Rating per Category")
ax[2, 0].hist(other.Size, range=(0, 100), bins=10, label="Size")
ax[2, 0].set_title("Size Histogram")
ax[2, 0].set_xlabel("Size")
other.CR.value_counts().plot(kind="bar", ax=ax[2, 1])
ax[2, 1].set_title("Frequency of Others per Content Rating")
f.tight_layout()
other_dum = pd.get_dummies(other, columns=["Category", "CR", "Price"])
corrmat = other_dum.corr()
f, ax = plt.subplots(figsize=(9, 8))
sns.heatmap(corrmat, ax=ax, cmap="YlGnBu", linewidths=0.1)
# # Regression
# Even if I found that there is no correlation between Rating and other independent variables, I still decided to run regressions, just for the sake of it.
y = games_dum.Rating
X = games_dum.drop(columns=["Rating"])
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.25, random_state=1
)
print("Train", X_train.shape, y_train.shape)
print("Test", X_test.shape, y_test.shape)
# By looking at the R squared value, we see that running a linear regression is basically not helpful at all.
reg = LinearRegression()
reg.fit(X_train, y_train)
pred = reg.predict(X_test)
mae = mean_absolute_error(y_test, pred)
mse = mean_squared_error(y_test, pred)
r2 = r2_score(y_test, pred)
print("MAE: ", mae)
print("RMSE: ", np.sqrt(mse))
print("R2: ", r2)
# Same with polynomial degrees. I was curious to see if it would give me better results.
d = range(4)
for degree in d:
poly = PolynomialFeatures(degree=degree)
Xpoly = poly.fit_transform(X)
Xpoly_test = poly.fit_transform(X_test)
polyreg = LinearRegression()
polyreg.fit(Xpoly, y)
predpoly = polyreg.predict(Xpoly_test)
mae2 = mean_absolute_error(y_test, predpoly)
mse2 = mean_squared_error(y_test, predpoly)
r2poly = r2_score(y_test, pred)
print("Degree: ", degree)
print("MAE: ", mae2)
print("RMSE: ", np.sqrt(mse2))
print("R2: ", r2poly)
|
from keras.datasets import mnist
from keras import models, layers
from keras.utils import to_categorical
(train_data, train_labels), (test_data, test_labels) = mnist.load_data()
print(train_data.shape)
print(len(train_data))
model = models.Sequential()
model.add(layers.Dense(512, activation="relu", input_shape=(28 * 28,)))
model.add(layers.Dense(10, activation="softmax"))
model.compile(
optimizer="rmsprop", loss="categorical_crossentropy", metrics=["accuracy"]
)
train_data = train_data.reshape(60000, 28 * 28)
train_data = train_data.astype("float32") / 255
test_data = test_data.reshape(10000, 28 * 28)
test_data = test_data.astype("float32") / 255
train_labels = to_categorical(train_labels)
test_labels = to_categorical(test_labels)
model.fit(train_data, train_labels, epochs=5, batch_size=128)
(loss_value, accuracy_value) = model.evaluate(test_data, test_labels)
print("accuracy_value: ", accuracy_value * 100)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
import matplotlib.pyplot as plt
import seaborn as sns
import tensorflow as tf
from sklearn.model_selection import train_test_split
from sklearn.metrics import confusion_matrix
from tensorflow.keras.utils import to_categorical
from tensorflow.keras.preprocessing.image import ImageDataGenerator
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import (
Dense,
Conv2D,
Flatten,
MaxPool2D,
Dropout,
BatchNormalization,
)
from tensorflow.keras.optimizers import RMSprop, Adam
from tensorflow.keras.callbacks import ReduceLROnPlateau
train = pd.read_csv("/kaggle/input/Kannada-MNIST/train.csv")
test = pd.read_csv("/kaggle/input/Kannada-MNIST/test.csv")
sample_sub = pd.read_csv("/kaggle/input/Kannada-MNIST/sample_submission.csv")
test.head(3)
test = test.drop("id", axis=1)
X_train = train.drop("label", axis=1)
Y_train = train.label
X_train = X_train / 255
test = test / 255
X_train = X_train.values.reshape(-1, 28, 28, 1)
test = test.values.reshape(-1, 28, 28, 1)
Y_train = to_categorical(Y_train)
X_train, X_test, y_train, y_test = train_test_split(
X_train, Y_train, random_state=42, test_size=0.15
)
datagen = ImageDataGenerator(
featurewise_center=False, # set input mean to 0 over the dataset
samplewise_center=False, # set each sample mean to 0
featurewise_std_normalization=False, # divide inputs by std of the dataset
samplewise_std_normalization=False, # divide each input by its std
zca_whitening=False, # apply ZCA whitening
rotation_range=10, # randomly rotate images in the range (degrees, 0 to 180)
zoom_range=0.1, # Randomly zoom image
width_shift_range=0.1, # randomly shift images horizontally (fraction of total width)
height_shift_range=0.1, # randomly shift images vertically (fraction of total height)
horizontal_flip=False, # randomly flip images
vertical_flip=False,
) # randomly flip images
datagen.fit(X_train)
model = Sequential()
model.add(
Conv2D(
filters=12,
kernel_size=(5, 5),
padding="Same",
activation="relu",
input_shape=(28, 28, 1),
)
)
model.add(Conv2D(filters=12, kernel_size=(5, 5), padding="Same", activation="relu"))
# model.add(BatchNormalization(momentum=.15))
model.add(MaxPool2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Conv2D(filters=24, kernel_size=(3, 3), padding="Same", activation="relu"))
model.add(Conv2D(filters=24, kernel_size=(3, 3), padding="Same", activation="relu"))
# model.add(BatchNormalization(momentum=0.15))
model.add(MaxPool2D(pool_size=(2, 2), strides=(2, 2)))
model.add(Dropout(0.25))
model.add(
Conv2D(
filters=8,
kernel_size=(5, 5),
padding="Same",
activation="relu",
input_shape=(28, 28, 1),
)
)
model.add(Conv2D(filters=8, kernel_size=(5, 5), padding="Same", activation="relu"))
# model.add(BatchNormalization(momentum=.15))
model.add(MaxPool2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Flatten())
model.add(Dense(256, activation="relu", kernel_initializer="he_normal"))
model.add(Dropout(0.37))
model.add(Dense(10, activation="softmax"))
model.summary()
optimizer = Adam(learning_rate=0.001, beta_1=0.9, beta_2=0.999)
model.compile(
optimizer=optimizer, loss=["categorical_crossentropy"], metrics=["accuracy"]
)
learning_rate_reduction = ReduceLROnPlateau(
monitor="val_acc", patience=3, verbose=1, factor=0.5, min_lr=0.00001
)
epochs = 5 # change this to 30 if you need to get better score
batch_size = 128
history = model.fit_generator(
datagen.flow(X_train, y_train, batch_size=batch_size),
epochs=epochs,
validation_data=(X_test, y_test),
verbose=2,
steps_per_epoch=X_train.shape[0] // batch_size,
callbacks=[learning_rate_reduction],
)
fig, ax = plt.subplots(2, 1)
fig.set
x = range(1, 1 + epochs)
ax[0].plot(x, history.history["loss"], color="red")
ax[0].plot(x, history.history["val_loss"], color="blue")
ax[1].plot(x, history.history["accuracy"], color="red")
ax[1].plot(x, history.history["val_accuracy"], color="blue")
ax[0].legend(["trainng loss", "validation loss"])
ax[1].legend(["trainng acc", "validation acc"])
plt.xlabel("Number of epochs")
plt.ylabel("accuracy")
test = pd.read_csv("/kaggle/input/Kannada-MNIST/test.csv")
test_id = test.id
test = test.drop("id", axis=1)
test = test / 255
test = test.values.reshape(-1, 28, 28, 1)
y_pre = model.predict(test)
y_pre = np.argmax(y_pre, axis=1)
sample_sub["label"] = y_pre
sample_sub.to_csv("submission.csv", index=False)
|
# #KERNELS STEP;
# CONTENT;
# 1. [Import Libraries](#1)
# 2. [READ DATA](#2)
# # 1.IMPORT LIBRARIES
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# plotly
# import plotly.plotly as py
from plotly.offline import init_notebook_mode, iplot, plot
import plotly as py
init_notebook_mode(connected=True)
import plotly.graph_objs as go
# word cloud library
from wordcloud import WordCloud
# matplotlib
import matplotlib.pyplot as plt
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list the files in the input directory
import os
print(os.listdir("../input"))
# Any results you write to the current directory are saved as output.
#
# # 2.READ DATA
Data = pd.read_csv("../input/tesla-stock-data-from-2010-to-2020/TSLA.csv")
Data.info()
# I got 7 columns , this also mean I got 7 different attribute as shown above. I show you head five row for you can get it better .
# 1.HEAD DATA
Data.head(10)
# When you look graphic , you can review on which got different number ,but that's not a problem . Now we will split up to month into graphic.
June = Data[:2].copy()
sum_of_June_for_open = sum([June.Open])
sum_of_June_for_High = sum([June.High])
sum_of_June_for_Low = sum([June.Low])
sum_of_June_for_Close = sum([June.Close])
sum_of_June_for_Volume = sum([June.Volume])
July = Data[2:23].copy()
sum_of_July_for_open = sum([July.Open])
sum_of_July_for_High = sum([July.High])
sum_of_July_for_Low = sum([July.Low])
sum_of_July_for_Close = sum([July.Close])
sum_of_July_for_Volume = sum([July.Volume])
August = Data[24:45].copy()
sum_of_August_for_open = sum([August.Open])
sum_of_August_for_High = sum([August.High])
sum_of_August_for_Low = sum([August.Low])
sum_of_August_for_Close = sum([August.Close])
sum_of_August_for_Volume = sum([August.Volume])
September = Data[46:66].copy()
sum_of_September_for_open = sum([September.Open])
sum_of_September_for_High = sum([September.High])
sum_of_September_for_Low = sum([September.Low])
sum_of_September_for_Close = sum([September.Close])
sum_of_September_for_Volume = sum([September.Volume])
October = Data[66:87].copy()
sum_of_October_for_open = sum([October.Open])
sum_of_October_for_High = sum([October.High])
sum_of_October_for_Low = sum([October.Low])
sum_of_October_for_Close = sum([October.Close])
sum_of_October_for_Volume = sum([October.Volume])
November = Data[87:100].copy()
sum_of_November_for_open = sum([November.Open])
sum_of_November_for_High = sum([November.High])
sum_of_November_for_Low = sum([November.Low])
sum_of_November_for_Close = sum([November.Close])
sum_of_November_for_Volume = sum([November.Volume])
print(sum_of_July_for_open)
# I splited up for monthly then we got summarize for each column from 2010 as shown above
JUNE_OPEN = go.Scatter(
x=June.Date,
y=sum_of_June_for_open,
mode="lines",
name="OPENING PRİCE ",
marker=dict(color="rgba(255,0,0,0.8)"),
text=June["Date"],
)
JUNE_HİGH = go.Scatter(
x=June.Date,
y=sum_of_June_for_High,
mode="lines",
name="HIGH PRİCE ",
marker=dict(color="rgba(0,255,0,0.8)"),
text=June["Date"],
)
JUNE_LOW = go.Scatter(
x=June.Date,
y=sum_of_June_for_Low,
mode="lines",
name="LOW PRICE",
marker=dict(color="rgba(0,0,255,0.8)"),
text=June["Date"],
)
JUNE_CLOSE = go.Scatter(
x=June.Date,
y=sum_of_June_for_Close,
mode="lines",
name="CLOSE PRICE ",
marker=dict(color="rgba(255,255,0,0.8)"),
text=June["Date"],
)
JUNE_VOLUME = go.Scatter(
x=June.Date,
y=sum_of_June_for_Volume,
mode="lines",
name="VOLUME PRICE ",
marker=dict(color="rgba(255,0,255,0.8)"),
text=June["Date"],
)
# ---------------------------------------------------------------------------------------------------------------------------------------------------------------------
JULY_OPEN = go.Scatter(
x=July.Date,
y=sum_of_July_for_open,
mode="lines",
name="",
marker=dict(color="rgba(255,0,0,0.8)"),
text=July["Date"],
)
JULY_HIGH = go.Scatter(
x=July.Date,
y=sum_of_July_for_High,
mode="lines",
name="",
marker=dict(color="rgba(0,255,0,0.8)"),
text=July["Date"],
)
JULY_LOW = go.Scatter(
x=July.Date,
y=sum_of_July_for_Low,
mode="lines",
name="",
marker=dict(color="rgba(0,0,255,0.8)"),
text=July["Date"],
)
JULY_CLOSE = go.Scatter(
x=July.Date,
y=sum_of_July_for_Close,
mode="lines",
name="",
marker=dict(color="rgba(255,255,0,0.8)"),
text=July["Date"],
)
JULY_VOLUME = go.Scatter(
x=July.Date,
y=sum_of_July_for_Volume,
mode="lines",
name="",
marker=dict(color="rgba(255,0,255,0.8)"),
text=July["Date"],
)
# ---------------------------------------------------------------------------------------------------------------------------------------------------------------------
AUGUST_OPEN = go.Scatter(
x=August.Date,
y=sum_of_August_for_open,
mode="lines",
name="",
marker=dict(color="rgba(255,0,0,0.8)"),
text=August["Date"],
)
AUGUST_HIGH = go.Scatter(
x=August.Date,
y=sum_of_August_for_High,
mode="lines",
name="",
marker=dict(color="rgba(0,255,0,0.8)"),
text=August["Date"],
)
AUGUST_LOW = go.Scatter(
x=August.Date,
y=sum_of_August_for_Low,
mode="lines",
name="",
marker=dict(color="rgba(0,0,255,0.8)"),
text=August["Date"],
)
AUGUST_CLOSE = go.Scatter(
x=August.Date,
y=sum_of_August_for_Close,
mode="lines",
name="",
marker=dict(color="rgba(255,255,0,0.8)"),
text=August["Date"],
)
AUGUST_VOLUME = go.Scatter(
x=August.Date,
y=sum_of_August_for_Volume,
mode="lines",
name="",
marker=dict(color="rgba(255,0,255,0.8)"),
text=August["Date"],
)
# ---------------------------------------------------------------------------------------------------------------------------------------------------------------------
SEPTEMBER_OPEN = go.Scatter(
x=September.Date,
y=sum_of_September_for_open,
mode="lines",
name="",
marker=dict(color="rgba(255,0,0,0.8)"),
text=September["Date"],
)
SEPTEMBER_HIGH = go.Scatter(
x=September.Date,
y=sum_of_September_for_High,
mode="lines",
name="",
marker=dict(color="rgba(0,255,0,0.8)"),
text=September["Date"],
)
SEPTEMBER_LOW = go.Scatter(
x=September.Date,
y=sum_of_September_for_Low,
mode="lines",
name="",
marker=dict(color="rgba(0,0,255,0.8)"),
text=September["Date"],
)
SEPTEMBER_CLOSE = go.Scatter(
x=September.Date,
y=sum_of_September_for_Close,
mode="lines",
name="",
marker=dict(color="rgba(255,255,0,0.8)"),
text=September["Date"],
)
SEPTEMBER_VOLUME = go.Scatter(
x=September.Date,
y=sum_of_September_for_Volume,
mode="lines",
name="",
marker=dict(color="rgba(255,0,255,0.8)"),
text=September["Date"],
)
# ---------------------------------------------------------------------------------------------------------------------------------------------------------------------
OCTOBER_OPEN = go.Scatter(
x=October.Date,
y=sum_of_October_for_open,
mode="lines",
name="",
marker=dict(color="rgba(255,0,0,0.8)"),
text=October["Date"],
)
OCTOBER_HIGH = go.Scatter(
x=October.Date,
y=sum_of_October_for_High,
mode="lines",
name=" ",
marker=dict(color="rgba(0,255,0,0.8)"),
text=October["Date"],
)
OCTOBER_LOW = go.Scatter(
x=October.Date,
y=sum_of_October_for_Low,
mode="lines",
name=" ",
marker=dict(color="rgba(0,0,255,0.8)"),
text=October["Date"],
)
OCTOBER_CLOSE = go.Scatter(
x=October.Date,
y=sum_of_October_for_Close,
mode="lines",
name=" ",
marker=dict(color="rgba(255,255,0,0.8)"),
text=October["Date"],
)
OCTOBER_VOLUME = go.Scatter(
x=October.Date,
y=sum_of_October_for_Volume,
mode="lines",
name=" ",
marker=dict(color="rgba(255,0,255,0.8)"),
text=October["Date"],
)
# ---------------------------------------------------------------------------------------------------------------------------------------------------------------------
NOVEMBER_OPEN = go.Scatter(
x=November.Date,
y=sum_of_November_for_open,
mode="lines",
name="",
marker=dict(color="rgba(255,0,0,0.8)"),
text=October["Date"],
)
NOVEMBER_HIGH = go.Scatter(
x=November.Date,
y=sum_of_November_for_High,
mode="lines",
name="",
marker=dict(color="rgba(0,255,0,0.8)"),
text=October["Date"],
)
NOVEMBER_LOW = go.Scatter(
x=November.Date,
y=sum_of_November_for_Low,
mode="lines",
name="",
marker=dict(color="rgba(0,0,255,0.8)"),
text=October["Date"],
)
NOVEMBER_CLOSE = go.Scatter(
x=November.Date,
y=sum_of_November_for_Close,
mode="lines",
name="",
marker=dict(color="rgba(255,255,0,0.8)"),
text=October["Date"],
)
NOVEMBER_VOLUME = go.Scatter(
x=November.Date,
y=sum_of_November_for_Volume,
mode="lines",
name="",
marker=dict(color="rgba(255,0,255,0.8)"),
text=October["Date"],
)
# ---------------------------------------------------------------------------------------------------------------------------------------------------------------------
data = [
JUNE_OPEN,
JUNE_LOW,
JUNE_HİGH,
JUNE_CLOSE,
JULY_OPEN,
JULY_HIGH,
JULY_LOW,
JULY_CLOSE,
AUGUST_OPEN,
AUGUST_HIGH,
AUGUST_CLOSE,
SEPTEMBER_OPEN,
SEPTEMBER_HIGH,
SEPTEMBER_LOW,
OCTOBER_OPEN,
OCTOBER_HIGH,
OCTOBER_LOW,
OCTOBER_CLOSE,
NOVEMBER_OPEN,
NOVEMBER_HIGH,
NOVEMBER_LOW,
NOVEMBER_CLOSE,
]
layout = dict(
title=" REWİEV ON PRİCE FOR MOUNTH FROM TESLA ",
xaxis=dict(title="date", ticklen=5, zeroline=False),
)
fig = dict(data=data, layout=layout)
iplot(fig)
# I 'm remainidng you I got four feature such as OPEN , HIGH , CLOSE , LOW ,VOLUME .I didn't use to volume feature due to VOLUME got least values .
# ----------------********** **SCATTER CHART******** --------------------
JUNE_OPEN = go.Scatter(
x=June.Date,
y=sum_of_June_for_open,
mode="markers",
name="OPENING PRİCE ",
marker=dict(color="rgba(255,0,0,0.8)"),
text=June["Date"],
)
JUNE_HİGH = go.Scatter(
x=June.Date,
y=sum_of_June_for_High,
mode="markers",
name="HIGH PRİCE ",
marker=dict(color="rgba(0,255,0,0.8)"),
text=June["Date"],
)
JUNE_LOW = go.Scatter(
x=June.Date,
y=sum_of_June_for_Low,
mode="markers",
name="LOW PRICE",
marker=dict(color="rgba(0,0,255,0.8)"),
text=June["Date"],
)
JUNE_CLOSE = go.Scatter(
x=June.Date,
y=sum_of_June_for_Close,
mode="markers",
name="CLOSE PRICE ",
marker=dict(color="rgba(255,255,0,0.8)"),
text=June["Date"],
)
JUNE_VOLUME = go.Scatter(
x=June.Date,
y=sum_of_June_for_Volume,
mode="markers",
name="VOLUME PRICE ",
marker=dict(color="rgba(255,0,255,0.8)"),
text=June["Date"],
)
# -------------------------------------------------------------------------------------------------------------------------------------------------
JULY_OPEN = go.Scatter(
x=July.Date,
y=sum_of_July_for_open,
mode="markers",
name="",
marker=dict(color="rgba(255,0,0,0.8)"),
text=July["Date"],
)
JULY_HIGH = go.Scatter(
x=July.Date,
y=sum_of_July_for_High,
mode="markers",
name="",
marker=dict(color="rgba(0,255,0,0.8)"),
text=July["Date"],
)
JULY_LOW = go.Scatter(
x=July.Date,
y=sum_of_July_for_Low,
mode="markers",
name="",
marker=dict(color="rgba(0,0,255,0.8)"),
text=July["Date"],
)
JULY_CLOSE = go.Scatter(
x=July.Date,
y=sum_of_July_for_Close,
mode="markers",
name="",
marker=dict(color="rgba(255,255,0,0.8)"),
text=July["Date"],
)
JULY_VOLUME = go.Scatter(
x=July.Date,
y=sum_of_July_for_Volume,
mode="markers",
name="",
marker=dict(color="rgba(255,0,255,0.8)"),
text=July["Date"],
)
# -------------------------------------------------------------------------------------------------------------------------------------------------
AUGUST_OPEN = go.Scatter(
x=August.Date,
y=sum_of_August_for_open,
mode="markers",
name="",
marker=dict(color="rgba(255,0,0,0.8)"),
text=August["Date"],
)
AUGUST_HIGH = go.Scatter(
x=August.Date,
y=sum_of_August_for_High,
mode="markers",
name="",
marker=dict(color="rgba(0,255,0,0.8)"),
text=August["Date"],
)
AUGUST_LOW = go.Scatter(
x=August.Date,
y=sum_of_August_for_Low,
mode="markers",
name="",
marker=dict(color="rgba(0,0,255,0.8)"),
text=August["Date"],
)
AUGUST_CLOSE = go.Scatter(
x=August.Date,
y=sum_of_August_for_Close,
mode="markers",
name="",
marker=dict(color="rgba(255,255,0,0.8)"),
text=August["Date"],
)
AUGUST_VOLUME = go.Scatter(
x=August.Date,
y=sum_of_August_for_Volume,
mode="markers",
name="",
marker=dict(color="rgba(255,0,255,0.8)"),
text=August["Date"],
)
# -------------------------------------------------------------------------------------------------------------------------------------------------
SEPTEMBER_OPEN = go.Scatter(
x=September.Date,
y=sum_of_September_for_open,
mode="markers",
name="",
marker=dict(color="rgba(255,0,0,0.8)"),
text=September["Date"],
)
SEPTEMBER_HIGH = go.Scatter(
x=September.Date,
y=sum_of_September_for_High,
mode="markers",
name="",
marker=dict(color="rgba(0,255,0,0.8)"),
text=September["Date"],
)
SEPTEMBER_LOW = go.Scatter(
x=September.Date,
y=sum_of_September_for_Low,
mode="markers",
name="",
marker=dict(color="rgba(0,0,255,0.8)"),
text=September["Date"],
)
SEPTEMBER_CLOSE = go.Scatter(
x=September.Date,
y=sum_of_September_for_Close,
mode="markers",
name="",
marker=dict(color="rgba(255,255,0,0.8)"),
text=September["Date"],
)
SEPTEMBER_VOLUME = go.Scatter(
x=September.Date,
y=sum_of_September_for_Volume,
mode="markers",
name="",
marker=dict(color="rgba(255,0,255,0.8)"),
text=September["Date"],
)
# -------------------------------------------------------------------------------------------------------------------------------------------------
OCTOBER_OPEN = go.Scatter(
x=October.Date,
y=sum_of_October_for_open,
mode="markers",
name="",
marker=dict(color="rgba(255,0,0,0.8)"),
text=October["Date"],
)
OCTOBER_HIGH = go.Scatter(
x=October.Date,
y=sum_of_October_for_High,
mode="markers",
name=" ",
marker=dict(color="rgba(0,255,0,0.8)"),
text=October["Date"],
)
OCTOBER_LOW = go.Scatter(
x=October.Date,
y=sum_of_October_for_Low,
mode="markers",
name=" ",
marker=dict(color="rgba(0,0,255,0.8)"),
text=October["Date"],
)
OCTOBER_CLOSE = go.Scatter(
x=October.Date,
y=sum_of_October_for_Close,
mode="markers",
name=" ",
marker=dict(color="rgba(255,255,0,0.8)"),
text=October["Date"],
)
OCTOBER_VOLUME = go.Scatter(
x=October.Date,
y=sum_of_October_for_Volume,
mode="markers",
name=" ",
marker=dict(color="rgba(255,0,255,0.8)"),
text=October["Date"],
)
# -------------------------------------------------------------------------------------------------------------------------------------------------
NOVEMBER_OPEN = go.Scatter(
x=November.Date,
y=sum_of_November_for_open,
mode="markers",
name="",
marker=dict(color="rgba(255,0,0,0.8)"),
text=October["Date"],
)
NOVEMBER_HIGH = go.Scatter(
x=November.Date,
y=sum_of_November_for_High,
mode="markers",
name="",
marker=dict(color="rgba(0,255,0,0.8)"),
text=October["Date"],
)
NOVEMBER_LOW = go.Scatter(
x=November.Date,
y=sum_of_November_for_Low,
mode="markers",
name="",
marker=dict(color="rgba(0,0,255,0.8)"),
text=October["Date"],
)
NOVEMBER_CLOSE = go.Scatter(
x=November.Date,
y=sum_of_November_for_Close,
mode="markers",
name="",
marker=dict(color="rgba(255,255,0,0.8)"),
text=October["Date"],
)
NOVEMBER_VOLUME = go.Scatter(
x=November.Date,
y=sum_of_November_for_Volume,
mode="markers",
name="",
marker=dict(color="rgba(255,0,255,0.8)"),
text=October["Date"],
)
# -------------------------------------------------------------------------------------------------------------------------------------------------
data = [
JUNE_OPEN,
JUNE_HİGH,
JUNE_LOW,
JUNE_CLOSE,
JULY_OPEN,
JULY_HIGH,
JULY_LOW,
JULY_CLOSE,
AUGUST_OPEN,
AUGUST_HIGH,
AUGUST_LOW,
AUGUST_CLOSE,
SEPTEMBER_OPEN,
SEPTEMBER_HIGH,
SEPTEMBER_LOW,
SEPTEMBER_CLOSE,
OCTOBER_OPEN,
OCTOBER_HIGH,
OCTOBER_LOW,
OCTOBER_CLOSE,
NOVEMBER_OPEN,
NOVEMBER_HIGH,
NOVEMBER_LOW,
NOVEMBER_CLOSE,
]
layout = dict(
title="PRICE 'S SPLITTED UP FOR MOUNTH SUCH AS BELOW (OPENING,HIGH , LOW ,CLOSE),",
xaxis=dict(title="MONTHS", ticklen=5, zeroline=False),
yaxis=dict(title="AMOUNT OF PRICE(K)", ticklen=5, zeroline=False),
)
fig = dict(data=data, layout=layout)
iplot(fig)
# ------****BAR CHART****---------
JUNE_OPEN = go.Bar(
x=June.Date,
y=sum_of_June_for_open,
name="OPENING PRİCE ",
marker=dict(color="rgba(255,0,0,0.8)", line=dict(color="rgb(0,0,0)", width=3)),
text=June["Date"],
)
JUNE_HİGH = go.Bar(
x=June.Date,
y=sum_of_June_for_High,
name="OPENING PRİCE ",
marker=dict(color="rgba(0,255,0,0.8)", line=dict(color="rgb(0,0,0)", width=3)),
text=June["Date"],
)
JUNE_LOW = go.Bar(
x=June.Date,
y=sum_of_June_for_Low,
name="OPENING PRİCE ",
marker=dict(color="rgba(0,0,255,0.8)", line=dict(color="rgb(0,0,0)", width=3)),
text=June["Date"],
)
JUNE_CLOSE = go.Bar(
x=June.Date,
y=sum_of_June_for_Close,
name="OPENING PRİCE ",
marker=dict(color="rgba(255,255,0,0.8)", line=dict(color="rgb(0,0,0)", width=3)),
text=June["Date"],
)
JUNE_VOLUME = go.Bar(
x=June.Date,
y=sum_of_June_for_Volume,
name="OPENING PRİCE ",
marker=dict(color="rgba(255,255,0,0.8)", line=dict(color="rgb(0,0,0)", width=3)),
text=June["Date"],
)
# ----------------------------------------------------------------------------------------------------------------------------
JULY_OPEN = go.Bar(
x=July.Date,
y=sum_of_July_for_open,
name="",
marker=dict(color="rgba(255,0,0,0.8)"),
text=July["Date"],
)
JULY_HIGH = go.Bar(
x=July.Date,
y=sum_of_July_for_High,
name="",
marker=dict(color="rgba(0,255,0,0.8)", line=dict(color="rgb(0,0,0)", width=3)),
text=July["Date"],
)
JULY_LOW = go.Bar(
x=July.Date,
y=sum_of_July_for_Low,
name="",
marker=dict(color="rgba(0,0,255,0.8)", line=dict(color="rgb(0,0,0)", width=3)),
text=July["Date"],
)
JULY_CLOSE = go.Bar(
x=July.Date,
y=sum_of_July_for_Close,
name="",
marker=dict(color="rgba(255,255,0,0.8)", line=dict(color="rgb(0,0,0)", width=3)),
text=July["Date"],
)
JULY_VOLUME = go.Bar(
x=July.Date,
y=sum_of_July_for_Volume,
name="",
marker=dict(color="rgba(255,0,255,0.8)", line=dict(color="rgb(0,0,0)", width=3)),
text=July["Date"],
)
# ----------------------------------------------------------------------------------------------------------------------------
AUGUST_OPEN = go.Bar(
x=August.Date,
y=sum_of_August_for_open,
name="",
marker=dict(color="rgba(255,0,0,0.8)", line=dict(color="rgb(0,0,0)", width=3)),
text=August["Date"],
)
AUGUST_HIGH = go.Bar(
x=August.Date,
y=sum_of_August_for_High,
name="",
marker=dict(color="rgba(0,255,0,0.8)", line=dict(color="rgb(0,0,0)", width=3)),
text=August["Date"],
)
AUGUST_LOW = go.Bar(
x=August.Date,
y=sum_of_August_for_Low,
name="",
marker=dict(color="rgba(0,0,255,0.8)", line=dict(color="rgb(0,0,0)", width=3)),
text=August["Date"],
)
AUGUST_CLOSE = go.Bar(
x=August.Date,
y=sum_of_August_for_Close,
name="",
marker=dict(color="rgba(255,255,0,0.8)", line=dict(color="rgb(0,0,0)", width=3)),
text=August["Date"],
)
AUGUST_VOLUME = go.Bar(
x=August.Date,
y=sum_of_August_for_Volume,
name="",
marker=dict(color="rgba(255,0,255,0.8)", line=dict(color="rgb(0,0,0)", width=3)),
text=August["Date"],
)
# ----------------------------------------------------------------------------------------------------------------------------
SEPTEMBER_OPEN = go.Bar(
x=September.Date,
y=sum_of_September_for_open,
name="",
marker=dict(color="rgba(255,0,0,0.8)"),
text=September["Date"],
)
SEPTEMBER_HIGH = go.Bar(
x=September.Date,
y=sum_of_September_for_High,
name="",
marker=dict(color="rgba(0,255,0,0.8)", line=dict(color="rgb(0,0,0)", width=3)),
text=September["Date"],
)
SEPTEMBER_LOW = go.Bar(
x=September.Date,
y=sum_of_September_for_Low,
name="",
marker=dict(color="rgba(0,0,255,0.8)", line=dict(color="rgb(0,0,0)", width=3)),
text=September["Date"],
)
SEPTEMBER_CLOSE = go.Bar(
x=September.Date,
y=sum_of_September_for_Close,
name="",
marker=dict(color="rgba(255,255,0,0.8)", line=dict(color="rgb(0,0,0)", width=3)),
text=September["Date"],
)
SEPTEMBER_VOLUME = go.Bar(
x=September.Date,
y=sum_of_September_for_Volume,
name="",
marker=dict(color="rgba(255,0,255,0.8)", line=dict(color="rgb(0,0,0)", width=3)),
text=September["Date"],
)
# ----------------------------------------------------------------------------------------------------------------------------
OCTOBER_OPEN = go.Bar(
x=October.Date,
y=sum_of_October_for_open,
name="",
marker=dict(color="rgba(255,0,0,0.8)"),
text=October["Date"],
)
OCTOBER_HIGH = go.Bar(
x=October.Date,
y=sum_of_October_for_High,
name="",
marker=dict(color="rgba(0,255,0,0.8)", line=dict(color="rgb(0,0,0)", width=3)),
text=October["Date"],
)
OCTOBER_LOW = go.Bar(
x=October.Date,
y=sum_of_October_for_Low,
name="",
marker=dict(color="rgba(0,0,255,0.8)", line=dict(color="rgb(0,0,0)", width=3)),
text=October["Date"],
)
OCTOBER_CLOSE = go.Bar(
x=October.Date,
y=sum_of_October_for_Close,
name="",
marker=dict(color="rgba(255,255,0,0.8)", line=dict(color="rgb(0,0,0)", width=3)),
text=October["Date"],
)
OCTOBER_VOLUME = go.Bar(
x=October.Date,
y=sum_of_October_for_Volume,
name="",
marker=dict(color="rgba(255,0,255,0.8)", line=dict(color="rgb(0,0,0)", width=3)),
text=October["Date"],
)
# ----------------------------------------------------------------------------------------------------------------------------
NOVEMBER_OPEN = go.Bar(
x=November.Date,
y=sum_of_November_for_open,
name="",
marker=dict(color="rgba(255,0,0,0.8)"),
text=November["Date"],
)
NOVEMBER_HIGH = go.Bar(
x=November.Date,
y=sum_of_November_for_High,
name="",
marker=dict(color="rgba(0,255,0,0.8)", line=dict(color="rgb(0,0,0)", width=3)),
text=November["Date"],
)
NOVEMBER_LOW = go.Bar(
x=November.Date,
y=sum_of_November_for_Low,
name="",
marker=dict(color="rgba(0,0,255,0.8)", line=dict(color="rgb(0,0,0)", width=3)),
text=November["Date"],
)
NOVEMBER_CLOSE = go.Bar(
x=November.Date,
y=sum_of_November_for_Close,
name="",
marker=dict(color="rgba(255,255,0,0.8)", line=dict(color="rgb(0,0,0)", width=3)),
text=November["Date"],
)
NOVEMBER_VOLUME = go.Bar(
x=November.Date,
y=sum_of_November_for_Volume,
name="",
marker=dict(color="rgba(255,0,255,0.8)", line=dict(color="rgb(0,0,0)", width=3)),
text=November["Date"],
)
# ----------------------------------------------------------------------------------------------------------------------------
data = [
JUNE_OPEN,
JUNE_HİGH,
JUNE_LOW,
JUNE_CLOSE,
JULY_OPEN,
JULY_HIGH,
JULY_LOW,
JULY_CLOSE,
AUGUST_OPEN,
AUGUST_HIGH,
AUGUST_LOW,
AUGUST_CLOSE,
SEPTEMBER_OPEN,
SEPTEMBER_HIGH,
SEPTEMBER_LOW,
SEPTEMBER_CLOSE,
OCTOBER_OPEN,
OCTOBER_HIGH,
OCTOBER_LOW,
OCTOBER_CLOSE,
NOVEMBER_OPEN,
NOVEMBER_HIGH,
NOVEMBER_LOW,
NOVEMBER_CLOSE,
]
layout = go.Layout(barmode="group")
fig = go.Figure(data=data, layout=layout)
iplot(fig)
import pandas as pd
TSLA = pd.read_csv("../input/tesla-stock-data-from-2010-to-2020/TSLA.csv")
|
import os
from pathlib import Path
from joblib import Parallel, delayed
from tqdm import tqdm
import glob
import pandas as pd
import torch
import torch.nn as nn
import torchaudio
import timm
# # Paste from [Data] notebook
MIN_WINDOW = 32_000 * 5
def maybe_pad(audio):
if len(audio) < MIN_WINDOW:
return torch.concat([audio, torch.zeros(MIN_WINDOW - len(audio))])
return audio
compute_melspec = torchaudio.transforms.MelSpectrogram(
sample_rate=32_000,
n_mels=128,
n_fft=2048,
hop_length=512,
f_min=0,
f_max=32_000 // 2,
)
power_to_db = torchaudio.transforms.AmplitudeToDB(
stype="power",
top_db=80.0,
)
def normalize_to_uint8(X):
mean, std = X.mean(), X.std()
if std < 1e-6:
raise Exception("std is too low")
X = (X - mean) / std
_min, _max = X.min(), X.max()
return (255 * (X - _min) / (_max - _min)).type(torch.uint8)
# # Paste from [Train] notebook
def make_model(out_features, pretrained):
model = timm.create_model("tf_efficientnet_b0_ns", pretrained=pretrained)
model.classifier = nn.Linear(
in_features=model.classifier.in_features, out_features=out_features
)
return model
# # Load stuff
df = pd.read_csv("/kaggle/input/birdclef-2023/train_metadata.csv")
index_to_label = sorted(df.primary_label.unique())
model = make_model(len(index_to_label), pretrained=False)
model.load_state_dict(
torch.load(
"/kaggle/input/bc23-train-melspecs/models/model__0__13__tensor(1_3455).pt",
map_location="cpu",
)["model_state_dict"]
)
model.eval()
filepaths = glob.glob("/kaggle/input/birdclef-2023/test_soundscapes/*.ogg")
# filepaths = [filepaths[0] for i in range(200)] # simulate submission
# # Inference
def process(filepath):
all_predictions = []
name = Path(filepath).stem
audio = torchaudio.load(filepath)[0][0]
for i in range(0, 120):
crop = audio[i * MIN_WINDOW : (i + 1) * MIN_WINDOW]
data = normalize_to_uint8(power_to_db(compute_melspec(maybe_pad(crop))))
data = data / 127 - 1 # normalize to float
data = torch.stack([data, data, data])
with torch.no_grad():
pred = torch.sigmoid(model(data[None])[0])
t = (i + 1) * 5
all_predictions.append({"row_id": f"{name}_{t}", "predictions": pred})
return all_predictions
all_predictions = Parallel(n_jobs=os.cpu_count())(
delayed(process)(filepath) for filepath in tqdm(filepaths, "Processing files")
)
all_predictions = [p2 for p in all_predictions for p2 in p] # flatten
df = pd.concat(
[
pd.DataFrame({"row_id": [p["row_id"] for p in all_predictions]}),
pd.DataFrame(
torch.stack([p["predictions"] for p in all_predictions]).numpy(),
columns=index_to_label,
),
],
axis=1,
)
df
df.to_csv("submission.csv", index=False)
|
# ## Word2vec Example
# Word Embedding: https://www.kaggle.com/ilhamfp31/word2vec-100-indonesian
# More details read https://radimrehurek.com/gensim/models/word2vec.html
import gensim
DIR_DATA_MISC = "../input/word2vec-100-indonesian"
path = "{}/idwiki_word2vec_100.model".format(DIR_DATA_MISC)
id_w2v = gensim.models.word2vec.Word2Vec.load(path)
# ### Embedding for each word
id_w2v["makan"]
# ### Word to index
word = "makan" # for any word in model
index = id_w2v.wv.vocab.get(word).index
print(index)
# ### Index to word
id_w2v.wv.index2word[255]
id_w2v.wv.index2word[index] == word
# ### Most similar
print(id_w2v.most_similar("makan"))
|
# # ***Library import***
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
# # ***Read csv***
train = pd.read_csv("train.csv")
test = pd.read_csv("test.csv")
test.isnull().any().sum(), train.isnull().any().sum()
train.dtypes.value_counts(), test.dtypes.value_counts()
# # ***EDA***
def do(rock):
lets = pd.DataFrame(rock.dtypes, columns=["data types"])
lets["MISSING"] = rock.isnull().sum()
did = rock.describe()
lets["uninque"] = did.nunique()
lets["max"] = did.max()
lets["min"] = did.min()
lets["std"] = did.std()
lets["count"] = did.count()
return lets
do(train).style.background_gradient(cmap="seismic")
def never(show):
love = pd.DataFrame(show.dtypes, columns=["data types"])
love["MISSING"] = show.isnull().sum()
areyou = show.describe()
love["unique"] = areyou.nunique()
love["min"] = areyou.min()
love["max"] = areyou.max()
love["std"] = areyou.std()
love["count"] = areyou.count()
return love
never(test).style.background_gradient(cmap="YlOrBr")
#
plt.figure(facecolor="#d49d31")
sns.regplot(data=train, x="spend", y="Id")
plt.figure(figsize=(16, 15), facecolor="#d49d31")
sns.heatmap(train.corr(), cmap="ocean")
# # ***Preprocessing***
from sklearn.preprocessing import LabelEncoder
L = LabelEncoder()
label = LabelEncoder()
n = train[["X51", "X109"]]
m = test[["X51", "X109"]]
for i in n:
n[i] = L.fit_transform(n[i])
for o in m:
m[o] = label.fit_transform(m[o])
train[["X51", "X109"]] = n
train.dtypes.value_counts().plot(kind="pie")
test[["X51", "X109"]] = m
test.dtypes.value_counts()
# # ***Auto ML***
from pycaret.regression import *
# # ***Setup Model***
setup(train, target="spend", ignore_features="Id", remove_outliers=True)
# # ***List of Model***
models()
# # ***Compare Model***
compare_models()
# # ***Create Model***
best = create_model("omp")
# # ***Pipeline***
finalize_model(best)
# # ***Plots***
plot_model(best)
# # ***Prediction***
predictions = best.predict(test.drop(["Id"], axis=1))
# # ***MAE MSE RMSE R2 RMSLE MAPE***
predict_model(best).head()
# # ***Submission***
submission = pd.DataFrame({"Id": test.Id, "Expected": predictions})
submission.tail()
submission.to_csv("submission.csv", index=False)
|
# # Modelo de Predição da Qualidade de Vinhos utilizando Perceptron
# * **Centro Federal de Educação Tecnológica - CEFET/RJ, UnED Petrópolis**
# * **Sistemas Inteligentes - 2023.1**
# * **Eduardo Paes Silva**
# Importação das bibliotecas e funções que serão utilizadas
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.linear_model import Perceptron
from sklearn.preprocessing import MinMaxScaler, Binarizer
# # Preparação dos Dados
# Carregamento, tratamento e divisão dos dados em conjunto de treino e testes.
# ## Passos realizados
# - Carregamento do CSV;
# - Normalização dos dados utilizando a técnica MinMaxScaler;
# - Separação dos dados de alvo (coluna _'quality'_);
# - Divisão dos dados entre treino e teste (70% e 30%);
# - Ajuste dos valores dos conjuntos alvo para 0 e 1 (Binário);
# - Transposição dos vetores alvo.
# Carregamento dos dados num DataFrame
df = pd.read_csv("/kaggle/input/cefet-si-winequality-red/winequality-red.csv", sep=";")
# Utilizando MinMaxScaler para normalização dos dados
scaler = MinMaxScaler()
# Normalizando todos os dados do DataFrame
df_norm = scaler.fit_transform(df)
# Transformando a matriz normalizada em um novo DataFrame
df_norm = pd.DataFrame(df_norm, columns=df.columns)
# Separação dos dados de alvo
X = df_norm.drop("quality", axis=1).values
y = df_norm["quality"].values
# Divisão dos conjuntos de treino e teste
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=84
)
# Transformando os valores dos conjuntos de alvo em binários usando o limiar 0.5
binarizer = Binarizer(threshold=0.5)
y_train = binarizer.fit_transform(y_train.reshape(-1, 1))
y_test = binarizer.fit_transform(y_test.reshape(-1, 1))
# Transformando os vetores de saída de array unidimensional para vetor coluna
y_train = y_train.ravel()
y_test = y_test.ravel()
# Treinando o Perceptron com o conjunto de dados de treino
perceptron = Perceptron(max_iter=1000, random_state=42)
perceptron.fit(X_train, y_train)
# Avaliando a performance do modelo
score = perceptron.score(X_test, y_test)
# Retorna o score total do modelo
score
##### # Previsões do modelo
# y_pred = perceptron.predict(X_test)
# # Plot dos dados de treino
# plt.scatter(X_train[:, 0], X_train[:, 1], c=y_train, cmap='bwr')
# plt.title('Dados de Treino')
# plt.xlabel('X1')
# plt.ylabel('X2')
# plt.show()
# # Plot dos dados de teste
# plt.scatter(X_test[:, 0], X_test[:, 1], c=y_test, cmap='bwr')
# plt.title('Dados de Teste')
# plt.xlabel('X1')
# plt.ylabel('X2')
# plt.show()
# # Plot dos dados de treino e teste
# plt.scatter(X_train[:, 0], X_train[:, 1], c=y_train, cmap='coolwarm', label='Treino')
# plt.plot(X_test[:, 0], X_test[:, 1], '.', label='Teste')
# # Traçando a linha de separação
# w = perceptron.coef_
# b = perceptron.intercept_
# x1 = np.linspace(0, 1, 100)
# x2 = -(w[0]/w[1])*x1 - (b/w[1])
# plt.plot(x1, x2, 'k--', label='Linha de separação')
# # Legendas e exibição do plot
# plt.xlabel('Característica 1')
# plt.ylabel('Característica 2')
# plt.title('Perceptron para classificação de vinhos')
# plt.legend()
# plt.show()
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
df11 = pd.read_csv(
"/kaggle/input/the-world-university-rankings-2011-2023/2011_rankings.csv"
)
df11.info()
df11["location"].value_counts()
df11["scores_overall"].plot(kind="box", vert=False)
|
# VINCENT CHEN
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
train_data = pd.read_csv("/kaggle/input/titanic/train.csv")
train_data.head()
test_data = pd.read_csv("/kaggle/input/titanic/test.csv")
test_data.head()
women = train_data.loc[train_data.Sex == "female"]["Survived"]
rate_women = sum(women) / len(women)
print("% of women who survived:", rate_women)
men = train_data.loc[train_data.Sex == "male"]["Survived"]
rate_men = sum(men) / len(men)
print("% of men who survived:", rate_men)
# Vincent Chen
from xgboost import XGBRegressor
from sklearn.model_selection import train_test_split
from sklearn.impute import SimpleImputer
from sklearn.impute import SimpleImputer
import pandas as pd
train_data = pd.read_csv("/kaggle/input/titanic/train.csv")
test_data = pd.read_csv("/kaggle/input/titanic/test.csv")
y = train_data["Survived"]
# Columns used
features = ["Pclass", "Sex", "SibSp", "Parch", "Age", "Fare", "Embark"]
X = pd.get_dummies(train_data[features])
X_train, X_test, y_train, y_test = train_test_split(
X, y, train_size=0.8, test_size=0.2, random_state=0
)
X_test_2 = pd.get_dummies(test_data[features])
# Imputer
my_imputer = SimpleImputer()
imputed_X_train = pd.DataFrame(my_imputer.fit_transform(X_train))
imputed_X_test = pd.DataFrame(my_imputer.transform(X_test))
imputed_X_train.columns = X_train.columns
imputed_X_test.columns = X_test.columns
# XGB
model = XGBRegressor(n_esitmators=100, learning_rate=0.01)
model.fit(X, y, early_stopping_rounds=40, eval_set=[(X_test, y_test)])
predictions = model.predict(X_test_2)
list_preds_2 = []
for i in range(len(predictions)):
temp = round(predictions[i])
list_preds_2.append((int)(temp))
print(list_preds_2)
output = pd.DataFrame({"PassengerId": test_data.PassengerId, "Survived": list_preds_2})
output.to_csv("my_submission.csv", index=False)
print("Your submission was successfully saved!")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Euclidean distance
# calculating euclidean distance between vectors
from math import sqrt
def euclidean_distance(a, b):
return sqrt(sum((e1 - e2) ** 2 for e1, e2 in zip(a, b)))
row1 = [10, 20, 15, 10, 5]
row2 = [12, 24, 18, 8, 7]
dist = euclidean_distance(row1, row2)
print(dist)
# euclidean() function from SciPy
from scipy.spatial.distance import euclidean
row1 = [10, 20, 15, 10, 5]
row2 = [12, 24, 18, 8, 7]
dist = euclidean(row1, row2)
print(dist)
# Manhattan distance
# calculating manhattan distance between vectors
from math import sqrt
def manhattan_distance(a, b):
return sum(abs(e1 - e2) for e1, e2 in zip(a, b))
row1 = [10, 20, 15, 10, 5]
row2 = [12, 24, 18, 8, 7]
dist = manhattan_distance(row1, row2)
print(dist)
# using the cityblock() function from SciPy
from scipy.spatial.distance import cityblock
row1 = [10, 20, 15, 10, 5]
row2 = [12, 24, 18, 8, 7]
dist = cityblock(row1, row2)
print(dist)
|
import os
import warnings
warnings.filterwarnings("ignore")
from pathlib import Path
import numpy as np
import pandas as pd
import torch
import timm
from fastai.vision.all import *
from fastcore.parallel import *
# Let's set the hardware to gpu
torch.cuda.set_device = 2
set_seed(42)
# Paths of the training and testing datasets
data_path = "/kaggle/input/leukemia-classification/C-NMC_Leukemia"
trn_path = "/kaggle/input/leukemia-classification/C-NMC_Leukemia/training_data"
valid_path = "/kaggle/input/leukemia-classification/C-NMC_Leukemia/validation_data"
# # Using fastai
# ### Looking at the data
# Get images form the path
files = get_image_files(trn_path)
files[3000]
# Get the parent path
file_path = files[3000]
path = Path(file_path)
print(path.parent.absolute())
# Function to return the label of an image
def get_label(x):
"""It uses the images parent folder name as a label"""
path = Path(x)
return 1 if str(path.parent.absolute()).split("/")[-1] == "all" else 0
get_label(path)
# ## Creating a DataBlock
# We can also use parent folder name as a label in datablock
# to use parent folder name we use: "get_y = parent_label"
dblock = DataBlock(
blocks=(ImageBlock, CategoryBlock),
get_items=get_image_files,
get_x=lambda x: x,
get_y=get_label,
splitter=RandomSplitter(valid_pct=0.2),
item_tfms=Resize(192),
batch_tfms=aug_transforms(size=192, min_scale=0.3),
)
dblock
# Creating a Dataset
dsets = dblock.datasets(trn_path)
# Length of train and valid data
print(f"Length training dataset: {len(dsets.train)}")
print(f"Length validation dataset: {len(dsets.valid)}")
# ### Creating a DataLoader from Datablock
# Creating a DataLoader
dls = dblock.dataloaders(source=trn_path)
# Inspecting the train data
dls.train.show_batch(max_n=6)
# Inspecting Validation data
dls.valid.show_batch(max_n=6)
# ## Using Transfer Learning
# Using pretrained model
arch = "convnext_small_in22k"
# Function to train the vision_learner
def train_model(dblock, arch, item, batch, epochs=5):
dls = dblock.dataloaders(trn_path, arch, item=item, batch=batch)
learn = cnn_learner(dls, arch, pretrained=True, metrics=[error_rate, accuracy])
learn.fine_tune(epochs, 0.01)
return learn
# Training the model
learn = train_model(
dblock, arch, item=Resize(192), batch=aug_transforms(128, min_scale=0.5), epochs=12
)
# Classifying validation data
valid = learn.dls.valid
preds, targets = learn.get_preds(dl=valid)
# Checking the error rate
error_rate(preds, targets)
learn.dls.train.show_batch(max_n=6, unique=True)
# Confusion Matrix
interp = ClassificationInterpretation.from_learner(learn)
interp.plot_confusion_matrix()
# Classifying images from Test Data
tta_preds, _ = learn.tta(dl=valid)
# Calculating the error rate on predictions
# error_rate = 1 - accuracy
error_rate(tta_preds, targets)
# ``` This is a huge improvement on the prediction error_rate ```
# ### Saving the model
# Saving the model as pickel file
learn.save("model.pkl")
# ### Predicting of Test dataset
# Getting test images
test_images = get_image_files(valid_path)
test_dl = learn.dls.test_dl(test_images)
# Classifying test images
preds, _ = learn.get_preds(dl=test_dl)
# Predictions
preds
test_df = pd.read_csv(
"/kaggle/input/leukemia-classification/C-NMC_Leukemia/validation_data/C-NMC_test_prelim_phase_data_labels.csv"
)
test_df["predictions"] = preds[0].argmax(dim=1)
test_df.head()
# Calculating the Accuracy
accuracy = (test_df["labels"] == test_df["predictions"]).value_counts(normalize=True)[1]
print(f"Accuracy of the model: {accuracy}")
|
# # Import Libraries
import gc
import os
import time
import random
import itertools
import numpy as np
import pandas as pd
from pathlib import Path
import matplotlib.pyplot as plt
import seaborn as sns
import plotly.offline as py
py.init_notebook_mode(connected=True)
import plotly.graph_objs as go
import plotly.tools as tls
import lightgbm as lgb
from sklearn.ensemble import (
RandomForestClassifier,
RandomForestRegressor,
ExtraTreesClassifier,
)
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score, roc_auc_score, confusion_matrix
import torch
from torch import nn, cuda
import torchvision.models as models
from torch.utils.data import DataLoader, Dataset
from torch.optim import Adam, SGD, Optimizer
from torch.optim.lr_scheduler import ReduceLROnPlateau
plt.style.use("fivethirtyeight")
pd.set_option("display.max_columns", 100)
pd.set_option("display.max_rows", 100)
import warnings
warnings.filterwarnings("ignore")
use_mlp = True
# # Helper Functions
def seed_everything(seed):
random.seed(seed)
os.environ["PYTHONHASHSEED"] = str(seed)
np.random.seed(seed)
torch.manual_seed(seed)
torch.cuda.manual_seed(seed)
torch.backends.cudnn.deterministic = True
# LABEL ENCODE
def encode_LE(col, train, test, verbose=True):
df_comb = pd.concat([train[col], train[col]], axis=0)
df_comb, _ = df_comb.factorize(sort=True)
nm = col + "_encoded"
if df_comb.max() > 32000:
train[nm] = df_comb[: len(train)].astype("int32")
test[nm] = df_comb[len(train) :].astype("int32")
else:
train[nm] = df_comb[: len(train)].astype("int16")
test[nm] = df_comb[len(train) :].astype("int16")
del df_comb
x = gc.collect()
if verbose:
print(nm)
def SMA(df, target, num_windows=3):
arr = np.array([])
for code in df["code"].unique():
temp_df = df.loc[df["code"] == code]
arr = np.concatenate(
(arr, temp_df[target].rolling(window=num_windows).mean().values)
)
return arr
def EMA(df, target, span_num=3):
arr = np.array([])
for code in df["code"].unique():
temp_df = df.loc[df["code"] == code]
arr = np.concatenate((arr, temp_df[target].ewm(span=span_num).mean().values))
return arr
def to_binary(preds, threshold=0.5):
return np.where(preds >= threshold, 1, 0)
def plot_confusion_matrix(
cm, target_names, title="Validation Confusion matrix", cmap=None, normalize=True
):
accuracy = np.trace(cm) / float(np.sum(cm))
misclass = 1 - accuracy
if cmap is None:
cmap = plt.get_cmap("OrRd_r")
plt.figure(figsize=(8, 6))
plt.imshow(cm, interpolation="nearest", cmap=cmap)
plt.title(title)
plt.colorbar()
if target_names is not None:
tick_marks = np.arange(len(target_names))
plt.xticks(tick_marks, target_names, rotation=45)
plt.yticks(tick_marks, target_names)
if normalize:
cm = cm.astype("float") / cm.sum(axis=1)[:, np.newaxis]
thresh = cm.max() / 1.5 if normalize else cm.max() / 2
for i, j in itertools.product(range(cm.shape[0]), range(cm.shape[1])):
if normalize:
plt.text(
j,
i,
"{:0.4f}".format(cm[i, j]),
horizontalalignment="center",
color="black" if cm[i, j] > thresh else "white",
)
else:
plt.text(
j,
i,
"{:,}".format(cm[i, j]),
horizontalalignment="center",
color="black" if cm[i, j] > thresh else "white",
)
plt.tight_layout()
plt.ylabel("True label")
plt.xlabel(
"Predicted label\naccuracy={:0.4f}; misclass={:0.4f}".format(accuracy, misclass)
)
plt.show()
def make_dict(train_df, test_df, feat_cols, target):
dataset_dict = {}
dataset_dict["X_train1"] = train_df.query("td <= 172")[feat_cols].values
dataset_dict["y_train1"] = train_df.query("td <= 172")[target].values
dataset_dict["X_valid1"] = train_df.query("td > 172 & td <= 206")[feat_cols].values
dataset_dict["y_valid1"] = train_df.query("td > 172 & td <= 206")[target].values
dataset_dict["X_train2"] = train_df.query("td <= 206")[feat_cols].values
dataset_dict["y_train2"] = train_df.query("td <= 206")[target].values
dataset_dict["X_valid2"] = train_df.query("td > 206 & td <= 240")[feat_cols].values
dataset_dict["y_valid2"] = train_df.query("td > 206 & td <= 240")[target].values
dataset_dict["X_train3"] = train_df.query("td <= 240")[feat_cols].values
dataset_dict["y_train3"] = train_df.query("td <= 240")[target].values
dataset_dict["X_valid3"] = train_df.query("td > 240 & td <= 274")[feat_cols].values
dataset_dict["y_valid3"] = train_df.query("td > 240 & td <= 274")[target].values
dataset_dict["x_test"] = test_df[feat_cols].values
return dataset_dict
def make_predictions(
dataset_dict,
df,
feat_cols,
lgb_params,
valid_type="hold_out",
plot_importance=True,
plot_confusion=True,
):
x_test = dataset_dict["x_test"]
if valid_type == "hold_out":
X_train = dataset_dict["X_train3"]
y_train = dataset_dict["y_train3"]
X_valid = dataset_dict["X_valid3"]
y_valid = dataset_dict["y_valid3"]
trn_data = lgb.Dataset(X_train, label=y_train)
val_data = lgb.Dataset(X_valid, label=y_valid)
clf = lgb.train(
lgb_params,
trn_data,
valid_sets=[trn_data, val_data],
verbose_eval=200,
early_stopping_rounds=100,
)
valid_preds = clf.predict(X_valid)
preds = clf.predict(x_test)
print()
print("roc_auc_score: {:.4f}".format(roc_auc_score(y_valid, valid_preds)))
print(
"accuracy_score: {:.4f}".format(
accuracy_score(y_valid, to_binary(valid_preds, 0.5))
)
)
if plot_importance:
feature_importance_df = pd.DataFrame()
feature_importance_df["Feature"] = feat_cols
feature_importance_df["Importance"] = clf.feature_importance()
cols = (
feature_importance_df[["Feature", "Importance"]]
.groupby("Feature")
.mean()
.sort_values(by="Importance", ascending=False)
.index
)
best_features = feature_importance_df.loc[
feature_importance_df.Feature.isin(cols)
]
plt.figure(figsize=(14, 10))
sns.barplot(
x="Importance",
y="Feature",
data=best_features.sort_values(by="Importance", ascending=False)[:20],
ci=None,
)
plt.title("LightGBM Feature Importance", fontsize=20)
plt.tight_layout()
if plot_confusion:
plot_confusion_matrix(
confusion_matrix(y_valid, to_binary(valid_preds, 0.5)),
normalize=False,
target_names=["pos", "neg"],
title="Confusion Matrix",
)
elif valid_type == "sliding_window":
window_num = 3
acc = 0
auc = 0
for num in range(1, window_num + 1):
print(f"num {num} dataset training starts")
preds = np.zeros(len(x_test))
X_train = dataset_dict[f"X_train{num}"]
y_train = dataset_dict[f"y_train{num}"]
X_valid = dataset_dict[f"X_valid{num}"]
y_valid = dataset_dict[f"y_valid{num}"]
trn_data = lgb.Dataset(X_train, label=y_train)
val_data = lgb.Dataset(X_valid, label=y_valid)
clf = lgb.train(
lgb_params,
trn_data,
valid_sets=[trn_data, val_data],
verbose_eval=200,
early_stopping_rounds=100,
)
valid_preds = clf.predict(X_valid)
preds += clf.predict(x_test) / window_num
acc += accuracy_score(y_valid, to_binary(valid_preds, 0.5))
auc += roc_auc_score(y_valid, valid_preds) / 3
print()
print("roc_auc_score: {:.4f}".format(auc))
print("acc_score: {:.4f}".format(acc))
return preds
# # Dataset
seed = 42
seed_everything(seed)
DATASET_PATH = "../input/stock-price"
X_train = pd.read_csv(os.path.join(DATASET_PATH, "train_data.csv")) # 훈련 데이터
Y_train = pd.read_csv(
os.path.join(DATASET_PATH, "train_target.csv")
) # 훈련 데이터에 대한 정답데이터 for regression
Y2_train = pd.read_csv(
os.path.join(DATASET_PATH, "train_target2.csv")
) # 훈련 데이터에 대한 정답데이터 for classification
test_df = pd.read_csv(os.path.join(DATASET_PATH, "test_data.csv")) # 테스트 데이터
X_train = X_train.set_index(["td", "code"])
Y_train = Y_train.set_index(["td", "code"])
Y2_train = Y2_train.set_index(["td", "code"])
# 시각화 및 전처리부터 모델링까지 보다 편하게 수행하기 위해 새로운 데이터셋을 생성
Y2_train = Y2_train.rename(columns={"target": "binned_target"})
train_df = pd.merge(X_train, Y_train["target"], how="left", on=["td", "code"])
train_df = pd.merge(train_df, Y2_train["binned_target"], how="left", on=["td", "code"])
train_df["binary_target"] = train_df["target"].apply(lambda x: 1 if x >= 0 else 0)
train_df = train_df.reset_index()
train_df["td"] = train_df["td"].str[1:].astype("int")
test_df["td"] = test_df["td"].str[1:].astype("int")
train_df.head()
# # Feature Engineering
# encode_LE('code', train_df, test_df)
# train_df['code_encoded'] = train_df['code_encoded'].astype('category')
# train_df['nan_count'] = train_df.isnull().sum(axis=1)
# test_df['nan_count'] = test_df.isnull().sum(axis=1)
train_df = train_df.sort_values(by=["code", "td"]).reset_index(drop=True)
F_cols = [col for col in train_df.columns if col.startswith("F")]
for feat_col in F_cols:
train_df[f"{feat_col}_EMA_3"] = EMA(train_df, feat_col, 3)
test_df[f"{feat_col}_EMA_3"] = EMA(test_df, feat_col, 3)
# train_df[f'{feat_col}_EMA_7'] = EMA(train_df, feat_col, 7)
# test_df[f'{feat_col}_EMA_7'] = EMA(test_df, feat_col, 7)
target_cols = [
col
for col in train_df.columns
if col in ["target", "binned_target", "binary_target"]
]
remove_cols = ["td", "code"]
feat_cols = [col for col in train_df.columns if col not in target_cols + remove_cols]
# feat_cols += ['code_encoded']
if use_mlp:
scaler = StandardScaler()
scaler.fit(train_df[feat_cols])
train_df[feat_cols] = scaler.transform(train_df[feat_cols])
test_df[feat_cols] = scaler.transform(test_df[feat_cols])
# # Validation Strategy
train_df.fillna(-99, inplace=True)
test_df.fillna(-99, inplace=True)
# check NaN
train_df[feat_cols].isnull().sum()
# # Modeling
# ## Random Forest
# https://arxiv.org/abs/1605.00003v1
# 우선 Baeline 모델로 위 논문을 참조하여 앙상블 기법 중 하나인 Random Forest를 적용하여 예측을 해볼 수 있다
# 논문에 의하면 연속값을 예측하는 regression 문제보다, 분류 문제로 접근했을 시에 더 나은
# performance를 보였기 때문에 필자도 **이진 분류 문제**로 접근하고자 한다.
# 모델에서 나온 예측 결과는 주식 시장에서 투자를 하는 의사 결정권자를 support 할 수 있을 것이다.
# 위 논문에서는 예측을 위한 지표로서 많이 활용되는 Relative Strength Index, Stochastic Oscillator와 같은
# Techinical Indicators들을 exponential smoothing을 통해 전처리를 하였고 Random Forest 모델을 활용하여 예측을 하였다.
# Random Forest 모델은 Bootstrap aggregating 기법을 활용하여 Decision Tree의 단점인 오버피팅을 완화시키는 방향으로 학습을 진행한다.
# Boostrapping 기법을 활용하기 떄문에 Validation set을 굳이 나눠도 되냐는 질문이 있을 수 있지만 다음과 같은 이유 때문에 데이터셋을 나누고 진행하기로 한다.
# - 다른 모델과의 보다 정확한 비교
# - OOB (Out-of-Bag) 에러를 활용한 hyper-parameter 최적화
# ## LightGBM
lgb_params = {
"num_leaves": 50,
"num_round": 10000,
"min_data_in_leaf": 30,
"objective": "binary",
"max_depth": 12,
"learning_rate": 0.001,
"min_child_samples": 20,
"boosting": "gbdt",
"feature_fraction": 0.9,
"bagging_freq": 1,
"bagging_fraction": 0.9,
"bagging_seed": 17,
"metric": "auc",
"lambda_l1": 0.1,
"verbosity": -1,
}
dataset_dict = make_dict(train_df, test_df, feat_cols, "binary_target")
# ## Hold-Out-Set
# Validation Split 방법 중에서 가장 간단한 방법이다.
# train set을 시간으로 정렬하여 마지막 부분을 떼어내 validation set으로 활용할 수 있다.
preds = make_predictions(
dataset_dict, train_df, feat_cols, lgb_params, valid_type="hold_out"
)
# ## Sliding Window
# 
# 출처: Predicting the direction of stock market prices using random forest (https://arxiv.org/abs/1605.00003v1)
preds = make_predictions(
dataset_dict, train_df, feat_cols, lgb_params, valid_type="sliding_window"
)
# # MLP Model
class MLP(nn.Module):
def __init__(self, num_classes=1, num_feats=47):
super(MLP, self).__init__()
self.num_classes = num_classes
self.mlp_layers = nn.Sequential(
nn.Linear(num_feats, 1024),
nn.PReLU(),
nn.BatchNorm1d(1024),
nn.Linear(1024, 512),
nn.PReLU(),
nn.BatchNorm1d(512),
nn.Linear(512, 256),
nn.PReLU(),
nn.BatchNorm1d(256),
nn.Dropout(0.3),
nn.Linear(256, 128),
nn.PReLU(),
nn.BatchNorm1d(128),
nn.Dropout(0.2),
nn.Linear(128, self.num_classes),
)
self._initialize_weights()
def forward(self, x):
out = self.mlp_layers(x)
return out
def _initialize_weights(self):
for m in self.modules():
if isinstance(m, nn.Linear):
nn.init.normal_(m.weight, 0, 0.01)
nn.init.constant_(m.bias, 0)
class Stock_dataset(Dataset):
def __init__(self, X, Y):
self.X = X
self.Y = Y
self.X_dataset = []
self.Y_dataset = []
for x in X:
self.X_dataset.append(torch.FloatTensor(x))
try:
for y in Y:
self.Y_dataset.append(torch.tensor(y))
except:
print("no label")
def __len__(self):
return len(self.X)
def __getitem__(self, index):
inputs = self.X_dataset[index]
try:
target = self.Y_dataset[index]
return inputs, target
except:
return inputs
def build_dataloader(X, Y, batch_size, shuffle=False):
dataset = Stock_dataset(X, Y)
dataloader = DataLoader(
dataset, batch_size=batch_size, shuffle=shuffle, num_workers=2
)
return dataloader
def build_model(device, model_name="MLP", num_classes=1, num_feats=47):
if model_name == "MLP":
model = MLP(num_classes, num_feats)
else:
raise NotImplementedError
model.to(device)
return model
def validation(model, criterion, valid_loader, use_cuda):
model.eval()
valid_preds = []
valid_targets = []
val_loss = 0.0
with torch.no_grad():
for batch_idx, (inputs, target) in enumerate(train_loader):
target = target.reshape(-1, 1).float()
valid_targets.append(target.numpy().copy())
if use_cuda:
inputs = inputs.to(device)
target = target.to(device)
output = model(inputs)
loss = criterion(output, target)
valid_preds.append(output.detach().cpu().numpy())
val_loss += loss.item() / len(valid_loader)
valid_preds = np.concatenate(valid_preds)
valid_targets = np.concatenate(valid_targets)
acc_score = accuracy_score(valid_targets, np.where(valid_preds >= 0.5, 1, 0))
auc_score = roc_auc_score(valid_targets, valid_preds)
return val_loss, acc_score, auc_score
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
use_cuda = True if device.type == "cuda" else False
batch_size = 256
train_loader = build_dataloader(X_train, y_train, batch_size, shuffle=True)
valid_loader = build_dataloader(X_val, y_val, batch_size, shuffle=False)
num_classes = 1
num_feats = len(feat_cols)
model = build_model(device, "MLP", num_classes, num_feats)
num_epochs = 1000
lr = 0.00025
criterion = nn.BCEWithLogitsLoss()
optimizer = Adam(model.parameters(), lr, weight_decay=0.000025)
# output path
output_dir = Path("./", "output")
output_dir.mkdir(exist_ok=True, parents=True)
best_valid_acc = 0.0
best_epoch = 0
count = 0
for epoch in range(1, num_epochs + 1):
start_time = time.time()
model.train()
optimizer.zero_grad()
train_loss = 0.0
for batch_idx, (inputs, target) in enumerate(train_loader):
target = target.reshape(-1, 1).float()
if use_cuda:
inputs = inputs.to(device)
target = target.to(device)
output = model(inputs)
loss = criterion(output, target)
loss.backward()
optimizer.step()
optimizer.zero_grad()
train_loss += loss.item() / len(train_loader)
val_loss, acc_score, auc_score = validation(
model, criterion, valid_loader, use_cuda
)
elapsed = time.time() - start_time
lr = [_["lr"] for _ in optimizer.param_groups]
if epoch % 1 == 0:
print(
"Epoch {} / {} train Loss: {:.4f} val_loss: {:.4f} val_acc: {:.4f} val_auc: {:.4f} lr: {:.5f} elapsed: {:.0f}m {:.0f}s".format(
epoch,
num_epochs,
train_loss,
val_loss,
acc_score,
auc_score,
lr[0],
elapsed // 60,
elapsed % 60,
)
)
model_path = output_dir / "best_model.pt"
if acc_score > best_valid_acc:
best_valid_acc = acc_score
best_epoch = epoch
torch.save(model.state_dict(), model_path)
# 30 epoch 동안 개선이 없으면 학습 강제 종료
if count == 20:
print("not improved for 20 epochs")
break
if acc_score < best_valid_acc:
count += 1
else:
count = 0
# scheduler.step(val_score)
best_epoch, best_valid_acc
class MyDataset(Dataset):
def __init__(self, data, window):
self.data = data
self.window = window
def __getitem__(self, index):
x = self.data[index : index + self.window]
return x
def __len__(self):
return len(self.data) - self.window
### Demo Dataset class
class DemoDatasetLSTM(Data.Dataset):
"""
Support class for the loading and batching of sequences of samples
Args:
dataset (Tensor): Tensor containing all the samples
sequence_length (int): length of the analyzed sequence by the LSTM
transforms (object torchvision.transform): Pytorch's transforms used to process the data
"""
## Constructor
def __init__(self, dataset, sequence_length=1, transforms=None):
self.dataset = dataset
self.seq_length = sequence_length
self.transforms = transforms
## Override total dataset's length getter
def __len__(self):
return self.dataset.__len__()
## Override single items' getter
def __getitem__(self, idx):
if idx + self.seq_length > self.__len__():
if self.transforms is not None:
item = torch.zeros(self.seq_length, self.dataset[0].__len__())
item[: self.__len__() - idx] = self.transforms(self.dataset[idx:])
return item, item
else:
item = []
item[: self.__len__() - idx] = self.dataset[idx:]
return item, item
else:
if self.transforms is not None:
return self.transforms(
self.dataset[idx : idx + self.seq_length]
), self.transforms(self.dataset[idx : idx + self.seq_length])
else:
return (
self.dataset[idx : idx + self.seq_length],
self.dataset[idx : idx + self.seq_length],
)
### Helper for transforming the data from a list to Tensor
def listToTensor(list):
tensor = torch.empty(list.__len__(), list[0].__len__())
for i in range(list.__len__()):
tensor[i, :] = torch.FloatTensor(list[i])
return tensor
### Dataloader instantiation
# Parameters
seq_length = 2
batch_size = 2
data_transform = transforms.Lambda(lambda x: listToTensor(x))
dataset = DemoDatasetLSTM(data_from_csv, seq_length, transforms=data_transform)
data_loader = Data.DataLoader(dataset, batch_size, shuffle=False, num_workers=2)
for data in data_loader:
x, _ = data
print(x)
|
# # Bayesian vs Frequentist methods in A/B testing
# >Ville Puuska
# When I was refreshing my knowledge of significance testing and trying to learn about A/B testing, I came across two lectures talking about using Bayesian methods for A/B testing in a business setting that completely sold me on the idea. If you have never heard of or used Bayesian methods I highly recommend watching these lectures: [Talking Bayes to Business: A/B Testing Use Case | Shopify](https://www.youtube.com/watch?v=J6kqvWnUE2Q) and [Thomas Wiecki - Solving Real-World Business Problems with Bayesian Modeling | PyData London 2022](https://www.youtube.com/watch?v=twpZhNqVExc)
# The main selling point that convinced me can be summarized as follows:
# - Imagine we have two versions of an online advertisement, A and B, and we wish to figure out which version has a higher click through rate; we'll denote click through rates by $\theta$.
# - To find this out, we collect data from both versions and analyze the data.
# - A frequentist analysis spits out a p-value that tells us how likely the data would be if the click through rates were equal, i.e. $P(\text{data} \mid \theta_A = \theta_B)$.
# - But this is not what we wanted! We wanted to know $P(\theta_B > \theta_A \mid \text{data})$.
# - Bayesian analysis gives us exactly this, and more!
# In this notebook we'll work through a simulated click through rate A/B test using both standard frequentist methods and Bayesian methods. I'll show how to actually implement both methods and how the different methods perform when the variables in the simulated test change, e.g. what happens to statistical power when changing sample size or the actual difference between versions A and B. The main point is to illustrate how much more intuitive information we can get from our data by using Bayesian methods.
import numpy as np
import pandas as pd
import pymc as pm
import pymc.sampling_jax
import scipy.stats as stats
import matplotlib as mpl
import matplotlib.pyplot as plt
import time
mpl.style.use("ggplot")
np.random.seed(2023)
# # Simulations
# First, let's define our simulated experiment. A single simulated experiment returns two randomly drawn lists of 0's and 1's from two Bernoulli random variables representing no-clicks and clicks for versions A and B.
# To keep later code cleaner, let's start by defining a function to generate simulated tests for us.
def experiment(
n_experiments: int = 1,
n_samples: int = 1000,
n_samples_A: int = None,
n_samples_B: int = None,
theta_A: float = 0.5,
theta_B: float = 0.5,
):
if n_samples_A is None:
n_samples_A = n_samples
if n_samples_B is None:
n_samples_B = n_samples
result = []
for _ in range(n_experiments):
A = np.random.binomial(1, theta_A, size=n_samples_A)
B = np.random.binomial(1, theta_B, size=n_samples_B)
result.append([A, B])
return result
# To illustrate this, here is a sample of three simulated experiments where version A was shown to 50 people with a 50 % probability of clicking, and version B was shown to 75 people with a 75 % probability of clicking.
n = 3
fig, ax = plt.subplots(2 * n, 1, figsize=(10, 2 * n))
arr = experiment(n_experiments=n, n_samples_A=50, n_samples_B=75, theta_B=0.75)
for i in range(n):
ax[2 * i].set_title(f"Experiment #{i+1}")
for j in range(2):
ax[2 * i + j].scatter(range(len(arr[i][j])), arr[i][j])
ax[2 * i + j].set_xlabel("group A" if j == 0 else "group B")
ax[2 * i + j].set_xticks([])
plt.tight_layout()
# # Frequentist tests
# In this section, we look at the standard frequentist analysis of A/B tests. In other words, we'll compute p-values and some confidence intervals. Let's start by simulating tests with four different sample sizes and fixed click through rates and check:
# - How often do the 95 % confidence intervals actually contain the true difference?
# - How are the p-values distributed?
# The answer to first question is obvious so why did I even add it? The simple reason is that I wanted to learn how to compute confidence intervals via bootstrapping in Python and to make sure the intervals actually work as they should. To compute p-values, we'll use Fisher's exact test.
# Since bootstrapping confidence intervals is somewhat computationally expensive, we'll compute only a few hundred simulations first. We fix $\theta_A = 0.10$ and $\theta_B = 0.13$, where $\theta$'s denote the click through rates.
def mean_diff(x, y, axis=0):
return y.mean(axis=axis) - x.mean(axis=axis)
theta_A = 0.10
theta_B = 0.13
results = []
results_ci = []
ns = [100, 250, 1000, 2000]
for n in ns:
print(f"Experiments for n = {n}", flush=True)
results.append([])
results_ci.append([])
arr = experiment(n_experiments=100, n_samples=n, theta_A=theta_A, theta_B=theta_B)
for i, simulated_experiment in enumerate(arr):
A = simulated_experiment[0]
B = simulated_experiment[1]
test_res = stats.fisher_exact(
((sum(A), sum(B)), (len(A) - sum(A), len(B) - sum(B))), alternative="less"
)
results[-1].append(test_res[1])
ci_res = stats.bootstrap((A, B), mean_diff, method="basic")
results_ci[-1].append(
(ci_res.confidence_interval.low, ci_res.confidence_interval.high)
)
print(f"Computed {i+1} experiments", end="\r", flush=True)
print()
results[-1] = np.array(results[-1])
results_ci[-1] = np.array(results_ci[-1])
for i in range(len(ns)):
print("n =", ns[i])
print(
f"Percentage of p-values < 0.05: {100*sum(results[i] < 0.05)/len(results[i]):.0f} %"
)
cis_containing_diff = (
100
* sum(
(results_ci[i][:, 0] <= theta_B - theta_A)
* (results_ci[i][:, 1] >= theta_B - theta_A)
)
/ len(results_ci[i])
)
print(
f"Percentage of 95 % confidence intervals containing the correct difference: {cis_containing_diff:.0f} %"
)
print()
fig, ax = plt.subplots(1, len(ns), figsize=(16, 4))
plt.suptitle("Distributions of p-values", fontsize=16)
for i in range(len(ns)):
ax[i].hist(results[i], bins=np.linspace(0, 1, num=21))
ax[i].set_xlim(0, 1)
ax[i].set_xlabel("p-value")
ax[i].set_title(f"{ns[i]} samples per simulation")
plt.tight_layout()
# The code for confidence intervals seems to be working and the distribution of p-values gets closer to 0 with larger sample sizes as you would obviously expect.
# Let's then forget about the confidence intervals and compute 200 000 simulations with varying click through rates to see how the p-values are distributed and what the false discovery rates/statistical powers are across different click through rates. We'll fix $\theta_A = 0.10$ and vary $\theta_B \in [0.06, 0.17)$.
theta_A = 0.10
theta_Bs = np.linspace(0.06, 0.17, num=200)
n_samples = 2000
results = {}
for theta_B in theta_Bs:
results[theta_B] = []
arr = experiment(
n_experiments=1000, n_samples=n_samples, theta_A=theta_A, theta_B=theta_B
)
for simulated_experiment in arr:
A = simulated_experiment[0]
B = simulated_experiment[1]
test_res = stats.fisher_exact(
((sum(A), sum(B)), (len(A) - sum(A), len(B) - sum(B))), alternative="less"
)
results[theta_B].append(test_res[1])
results[theta_B] = np.array(results[theta_B])
plt.figure(figsize=(15, 5))
y = []
for theta_B in theta_Bs:
y.append(stats.mstats.mquantiles(results[theta_B], prob=[0.05, 0.5, 0.95]))
y = np.array(y)
plt.fill_between(
theta_Bs - theta_A, y[:, 0], y[:, 2], alpha=0.4, label="90 % band of p-values"
)
plt.plot(theta_Bs - theta_A, y[:, 1], label="Median p-value")
plt.hlines(
0.05,
min(theta_Bs) - theta_A,
max(theta_Bs) - theta_A,
label="p = 0.05",
color="black",
linestyle="--",
)
plt.legend()
plt.xlim(min(theta_Bs) - theta_A, max(theta_Bs) - theta_A)
plt.ylim(-0.01, 1.01)
plt.suptitle(
rf"""Observed distribution of p-values across different values of $\theta_B - \theta_A$,
number of samples per simulation = {n_samples}""",
fontsize=16,
)
plt.xlabel(r"Actual difference $\theta_B - \theta_A$")
plt.ylabel("p-values of t-tests on simulations")
plt.figure(figsize=(15, 5))
fdr = []
stat_power = []
for theta_B in theta_Bs:
if theta_B <= theta_A:
fdr.append(sum(results[theta_B] < 0.05) / len(results[theta_B]))
else:
stat_power.append(sum(results[theta_B] < 0.05) / len(results[theta_B]))
plt.plot((theta_Bs - theta_A)[: len(fdr)], fdr, label="False discovery rate")
plt.plot((theta_Bs - theta_A)[len(fdr) :], stat_power, label="Statistical power")
plt.scatter(0, 0.05, s=25, color="black", marker="x")
plt.annotate("y = 0.05", (0.0, 0.05 + 0.02), horizontalalignment="right")
plt.legend()
plt.xlim(min(theta_Bs) - theta_A, max(theta_Bs) - theta_A)
plt.ylim(-0.01, 1.01)
plt.suptitle(
rf"""Observed false discovery rate and statistical power,
number of samples per simulation = {n_samples}""",
fontsize=16,
)
plt.xlabel(r"Actual difference $\theta_B - \theta_A$")
# Next, let's look at the effect of sample sizes on statistical power. We'll run simulations across 30 different sample sizes with fixed $\theta_A = 0.10$ and $\theta_B = 0.13$. Then we'll check how the p-values are distributed and what the statistical power is across different sample sizes.
theta_A = 0.10
theta_B = 0.13
sample_sizes = range(100, 3001, 100)
results = {}
for n_samples in sample_sizes:
results[n_samples] = []
arr = experiment(
n_experiments=1000, n_samples=n_samples, theta_A=theta_A, theta_B=theta_B
)
for simulated_experiment in arr:
A = simulated_experiment[0]
B = simulated_experiment[1]
test_res = stats.fisher_exact(
((sum(A), sum(B)), (len(A) - sum(A), len(B) - sum(B))), alternative="less"
)
results[n_samples].append(test_res[1])
results[n_samples] = np.array(results[n_samples])
plt.figure(figsize=(15, 5))
y = []
for n_samples in sample_sizes:
y.append(stats.mstats.mquantiles(results[n_samples], prob=[0.00, 0.5, 0.80]))
y = np.array(y)
plt.fill_between(
sample_sizes, y[:, 0], y[:, 2], alpha=0.4, label="bottom 80 % of p-values"
)
plt.plot(sample_sizes, y[:, 1], label="Median p-value")
plt.hlines(
0.05,
min(sample_sizes),
max(sample_sizes),
label="p = 0.05",
color="black",
linestyle="--",
)
plt.legend()
plt.xlim(min(sample_sizes), max(sample_sizes))
plt.ylim(-0.01, 1.01)
plt.suptitle(
r"""Observed distribution of p-values across different sample sizes,
$\theta_A = 0.10, \theta_B = 0.13$""",
fontsize=16,
)
plt.xlabel("Sample size")
plt.ylabel("p-values of t-tests on simulations")
plt.figure(figsize=(15, 5))
stat_power = []
for n_samples in sample_sizes:
stat_power.append(sum(results[n_samples] < 0.05) / len(results[n_samples]))
plt.plot(sample_sizes, stat_power, label="Statistical power")
plt.xlim(min(sample_sizes), max(sample_sizes))
plt.ylim(-0.01, 1.01)
plt.suptitle(
r"""Observed statistical power across different sample sizes,
$\theta_A = 0.10, \theta_B = 0.13$""",
fontsize=16,
)
plt.xlabel("Sample size")
# # Bayesian inference
# Now we'll turn to the much more exciting Bayesian methods. As mentioned in the introduction, what we actually want to know is the probability that version B has a higher click through rate than version A, i.e. $P(\theta_B > \theta_A \mid \text{test data})$. Computing this directly can be impossible, but if we can first figure out the distributions $P(\theta_B \mid \text{group B test data})$ and $P(\theta_A \mid \text{group A test data})$ we can then use simple Monte Carlo to figure out the probability we care about. But how do we compute these two distributions?
# Bayes' rule tells us that
# $$P(\theta \mid \text{data}) = \frac{P(\text{data} \mid \theta)P(\theta)}{P(\text{data})}.$$
# The distribution $P(\theta \mid \text{data})$ is called the [_posterior_](https://en.wikipedia.org/wiki/Posterior_probability). In this formula, $P(\text{data} \mid \theta)$ is already known and we can at least numerically compute $P(\text{data})$ by integrating $P(\text{data} \mid \theta)P(\theta)$ over the parameter space. The unknown we're left with is the [_prior_](https://en.wikipedia.org/wiki/Prior_probability) $P(\theta)$.
# In Bayesian inference, choosing the prior is the job of the statistician. If we have some solid idea of what $\theta$ should be, we can use our best guess; if not, we use an uninformative prior, e.g. a uniform distribution over all legal values for $\theta$. If you have never come across priors, this explanation will almost surely not convince you that simply choosing this distribution is safe or even sane. If this is the case, you will simply have to trust me that this works.
# So how do we actually carry out the computation? There are two ways:
# - [conjugate priors](https://en.wikipedia.org/wiki/Conjugate_prior),
# - [probabilistic programming](https://en.wikipedia.org/wiki/Probabilistic_programming)/[Markov chain Monte Carlo methods (MCMC)](https://en.wikipedia.org/wiki/Markov_chain_Monte_Carlo).
# Conjugate priors are essentially an analytic solution to Bayes' rule. Depending on the distributions you have, conjugate priors are not always available. Probabilistic programming is much much more flexible as it does not require any analytical solution.
# Let's start with probabilistic programming.
# # Probabilistic programming using PyMC
# We will use the probabilistic programming language PyMC to compute posteriors. Let's start with a model for a single experiment. We will consider our experiments to be binomial by adding up the Bernoulli trials; this just makes the model a bit nicer in my opinion. I wont explain how this language works beyond surface level details; for a good introduction to using PyMC I'd recommend [Bayesian Methods for Hackers](https://github.com/CamDavidsonPilon/Probabilistic-Programming-and-Bayesian-Methods-for-Hackers) as this is what I learned with or [Bayesian Modeling and Computation in Python](https://bayesiancomputationbook.com/welcome.html), both are free.
# Before defining models, let's get 10 simulated experiments to use throughout this section.
n = 10
arr = experiment(n_experiments=n, theta_A=0.10, theta_B=0.13)
# In the next cell we define a model with two variables, theta_A and theta_B, with unifrom priors from 0 to 1. The model also has two binomial random variables, experiment_A and experiment_B, with probabilities theta_A and theta_B and observed data given by our simulation. Then, we run the default MCMC sampler to compute the posteriors for theta_A and theta_B.
with pm.Model() as model:
t_A = pm.Uniform("theta_A", 0, 1)
t_B = pm.Uniform("theta_B", 0, 1)
experiment_A = pm.Binomial(
"experiment_A", n=len(arr[0][0]), p=t_A, observed=sum(arr[0][0])
)
experiment_B = pm.Binomial(
"experiment_B", n=len(arr[0][1]), p=t_B, observed=sum(arr[0][1])
)
trace = pm.sample(draws=10000, chains=2)
A_trace = trace.posterior.theta_A.to_numpy().flatten()
B_trace = trace.posterior.theta_B.to_numpy().flatten()
pm.model_to_graphviz(model)
# The sampler outputs the following data:
trace
fig, ax = plt.subplots(1, 2, figsize=(15, 3))
plt.suptitle(r"Posteriors of $\theta_A$ and $\theta_B$", fontsize=16)
ax[0].hist(A_trace, bins=50)
ax[0].axvline(0.10, color="black", linestyle="--", label=r"true $\theta_A$")
ax[0].set_xlim(0.06, 0.14)
ax[0].set_yticks([])
ax[0].legend()
ax[0].set_title(r"$\theta_A$")
ax[1].hist(B_trace, bins=50)
ax[1].axvline(0.13, color="black", linestyle="--", label=r"true $\theta_B$")
ax[1].set_xlim(0.09, 0.17)
ax[1].set_yticks([])
ax[1].legend()
ax[1].set_title(r"$\theta_B$")
plt.tight_layout()
# We want to run this simulation multiple times like we did in the frequentist section. We could do this by simply looping and creating a new model for each iteration of the loop. This is very inefficient though as the model needs to be recompiled every iteration unnecessarily. Instead, we can essentially stack these models as shown in the next cell.
starttime = time.time()
with pm.Model() as model:
t_A = pm.Uniform("theta_A", 0, 1, shape=len(arr))
t_B = pm.Uniform("theta_B", 0, 1, shape=len(arr))
experiment_arr = np.transpose(np.array(arr).sum(axis=2))
experiment_A = pm.Binomial(
"experiment_A", n=len(arr[0][0]), p=t_A, observed=experiment_arr[0]
)
experiment_B = pm.Binomial(
"experiment_B", n=len(arr[0][1]), p=t_B, observed=experiment_arr[1]
)
trace = pm.sample(draws=10000, chains=2)
print(
f"Compiling the model and sampling with {n} experiments took {time.time()-starttime:.2f} seconds."
)
pm.model_to_graphviz(model)
show_n = 3
fig, ax = plt.subplots(show_n, 2, figsize=(15, show_n * 3))
ax = ax.flatten()
plt.suptitle(
rf"Posteriors of $\theta_A$ and $\theta_B$ across {show_n} simulations", fontsize=16
)
A_traces = trace.posterior.theta_A
B_traces = trace.posterior.theta_B
for i in range(show_n):
ax[2 * i].hist(A_traces[:, :, i].to_numpy().flatten(), bins=50)
ax[2 * i].axvline(0.10, color="black", linestyle="--", label=r"true $\theta_A$")
ax[2 * i].set_xlim(0.06, 0.14)
ax[2 * i].set_yticks([])
ax[2 * i].legend()
ax[2 * i].set_title(r"$\theta_A$")
ax[2 * i + 1].hist(B_traces[:, :, i].to_numpy().flatten(), bins=50)
ax[2 * i + 1].axvline(0.13, color="black", linestyle="--", label=r"true $\theta_B$")
ax[2 * i + 1].set_xlim(0.09, 0.17)
ax[2 * i + 1].set_yticks([])
ax[2 * i + 1].legend()
ax[2 * i + 1].set_title(r"$\theta_B$")
plt.tight_layout()
# Finally, we can change our sampler from the default sampler in PyMC to something faster. In the next cell we use a NUTS sampler from the numpyro package. This offers a significant speedup.
starttime = time.time()
with pm.Model() as model:
t_A = pm.Uniform("theta_A", 0, 1, shape=len(arr))
t_B = pm.Uniform("theta_B", 0, 1, shape=len(arr))
experiment_arr = np.transpose(np.array(arr).sum(axis=2))
experiment_A = pm.Binomial(
"experiment_A", n=len(arr[0][0]), p=t_A, observed=experiment_arr[0]
)
experiment_B = pm.Binomial(
"experiment_B", n=len(arr[0][1]), p=t_B, observed=experiment_arr[1]
)
trace = pm.sampling_jax.sample_numpyro_nuts(draws=10000, chains=2)
print(
f"\n\nWith the numpyro NUTS sampler, compiling the model and sampling with {n} experiments took {time.time()-starttime:.2f} seconds."
)
fig, ax = plt.subplots(show_n, 2, figsize=(15, show_n * 3))
ax = ax.flatten()
plt.suptitle(
rf"Posteriors of $\theta_A$ and $\theta_B$ across {show_n} simulations", fontsize=16
)
A_traces = trace.posterior.theta_A
B_traces = trace.posterior.theta_B
for i in range(show_n):
ax[2 * i].hist(A_traces[:, :, i].to_numpy().flatten(), bins=50)
ax[2 * i].axvline(0.10, color="black", linestyle="--", label=r"true $\theta_A$")
ax[2 * i].set_xlim(0.06, 0.14)
ax[2 * i].set_yticks([])
ax[2 * i].legend()
ax[2 * i].set_title(r"$\theta_A$")
ax[2 * i + 1].hist(B_traces[:, :, i].to_numpy().flatten(), bins=50)
ax[2 * i + 1].axvline(0.13, color="black", linestyle="--", label=r"true $\theta_B$")
ax[2 * i + 1].set_xlim(0.09, 0.17)
ax[2 * i + 1].set_yticks([])
ax[2 * i + 1].legend()
ax[2 * i + 1].set_title(r"$\theta_B$")
plt.tight_layout()
# # Conjugate priors
# If we had real data from a single A/B test and we wanted to do inference on it, PyMC or any other probabilistic programming language/package would work perfectly. However, since we want to run hundreds of thousands of simulations to see how the test itself performs, probabilistic programming is just far too computationally expensive. For example, above we saw that even with a fast sampler it took us more than 10 seconds to run inference on 10 simulated experiments and sampling took over 5 seconds.
# Luckily, in our setting of A/B testing click through rate, we have a mathematical solution for the posterior!
# > **Fact:** When the prior is a beta distribution and the likelihood is a binomial distribution, the posterior is a beta distribution.
# In a situation like this where the posterior and prior distributions are from the same family, the prior is called a [conjugate prior](https://en.wikipedia.org/wiki/Conjugate_prior) for the likelihood. For more complex models this of course will almost never be the case which is where probabilistic programming has to be used. But in our setting this conjugate prior saves us from very expensive computations.
# Now, the prior we used above was the uniform distribution. That is actually a beta distribution, namely $\beta(1, 1)$. Getting the posterior is extremely simple: if our experiment gave us $a$ ones and $b$ zeros, then the posterior is simply $\beta(1+a, 1+b)$.
# If we wanted to use some more informative priors — e.g. maybe we had an expert tell us they thought that $\theta$ should be around 0.10 — we could find a more informative beta to fit our prior beliefs. Let's plot some example beta distributions to get an idea.
plt.figure(figsize=(15, 5))
x = np.linspace(0, 1, num=1000)
for ones, zeros in [(1, 1), (0.5, 0.5), (10, 5), (3, 20)]:
plt.plot(x, stats.beta(ones, zeros).pdf(x), label=rf"$\beta({ones}, {zeros})$")
plt.xlim(0, 1)
plt.ylim(0, 6.5)
plt.suptitle("Examples of beta distributions", fontsize=16)
plt.legend()
# If we believed our expert we could choose $\beta(3, 20)$ as our prior. To keep our example consistent though, we will stick to the uniform distribution, i.e. $\beta(1, 1)$.
# To make sure that these conjugate priors really work (and that we haven't messed up above with our probabilistic model), let's plot beta posteriors over the final examples in the previous section.
fig, ax = plt.subplots(show_n, 2, figsize=(15, show_n * 3))
ax = ax.flatten()
plt.suptitle(
rf"Posteriors of $\theta_A$ and $\theta_B$ across {show_n} simulations", fontsize=16
)
A_traces = trace.posterior.theta_A
B_traces = trace.posterior.theta_B
for i in range(show_n):
ax[2 * i].hist(A_traces[:, :, i].to_numpy().flatten(), bins=50, density=True)
ax[2 * i].axvline(0.10, color="black", linestyle="--", label=r"true $\theta_A$")
x = np.linspace(0.06, 0.14, num=100)
zeros, ones = len(arr[i][0]) - sum(arr[i][0]), sum(arr[i][0])
ax[2 * i].plot(
x,
stats.beta(1 + ones, 1 + zeros).pdf(x),
label=rf"$\beta(1+{ones}, 1+{zeros})$",
color="blue",
)
ax[2 * i].set_xlim(0.06, 0.14)
ax[2 * i].set_yticks([])
ax[2 * i].legend()
ax[2 * i].set_title(r"$\theta_A$")
ax[2 * i + 1].hist(B_traces[:, :, i].to_numpy().flatten(), bins=50, density=True)
ax[2 * i + 1].axvline(0.13, color="black", linestyle="--", label=r"true $\theta_B$")
x = np.linspace(0.09, 0.17, num=100)
zeros, ones = len(arr[i][1]) - sum(arr[i][1]), sum(arr[i][1])
ax[2 * i + 1].plot(
x,
stats.beta(1 + ones, 1 + zeros).pdf(x),
label=rf"$\beta(1+{ones}, 1+{zeros})$",
color="blue",
)
ax[2 * i + 1].set_xlim(0.09, 0.17)
ax[2 * i + 1].set_yticks([])
ax[2 * i + 1].legend()
ax[2 * i + 1].set_title(r"$\theta_B$")
plt.tight_layout()
# Perfect!
# ## A Bayesian test
# Now that we have the posteriors $P(\theta_A \mid \text{group A test data})$ and $P(\theta_B \mid \text{group B test data})$, we sample values for $\theta_A$ and $\theta_B$ to see what the distribution $P(\theta_B - \theta_A \mid \text{test data})$ looks like. From this distribution we can then easily figure out $P(\theta_B > \theta_A \mid \text{test data})$ and other interesting values.
# People often misinterpret the p-value null hypothesis rejection when p < 0.05 to mean that the alternative hypothesis is true with probability at least 95 %. This of course is not what the p-value tells us. Armed with Bayesian inference we can now make a test that rejects the null hypothesis if the probability that the alternative hypothesis is true is at least 95 % to match peoples intuition. There are many other ways of coming up with a hypothesis test with Bayesian inference, but this test is easiest to directly compare with the frequentist test considered above. In the remainder of this notebook we refer to this test as the **Bayesian test**.
# Before we run the same simulations for the Bayesian test as we did in the frequentist section for Fisher's exact test, let's illustrate how to actually do this sampling.
# If we computed the prior with PyMC, or more generally MCMC, we already have a sampling of $\theta_A$'s and $\theta_B$'s so we can just use them as we do in the next cell.
probs, expected_diffs, expected_lifts = [], [], []
for i in range(len(arr)):
theta_A_sample = A_traces[:, :, i].to_numpy().flatten()
theta_B_sample = B_traces[:, :, i].to_numpy().flatten()
probs.append(sum(theta_B_sample > theta_A_sample) / len(theta_A_sample))
expected_diffs.append(sum(theta_B_sample - theta_A_sample) / len(theta_A_sample))
expected_lifts.append(
sum((theta_B_sample - theta_A_sample) / theta_A_sample) / len(theta_A_sample)
)
pd.DataFrame(
{
r"$P(\theta_B$ $>$ $\theta_A)$": probs,
r"$E(\theta_B$ $-$ $\theta_A)$": expected_diffs,
"Expected lift": expected_lifts,
}
)
# If we instead used conjugate priors, we now need to sample from the priors as we do in the next cell. 10 000 samples per experiment should suffice.
probs, expected_diffs, expected_lifts = [], [], []
for i in range(len(arr)):
zeros_A, ones_A = len(arr[i][0]) - sum(arr[i][0]), sum(arr[i][0])
theta_A_dist = stats.beta(1 + ones_A, 1 + zeros_A)
zeros_B, ones_B = len(arr[i][1]) - sum(arr[i][1]), sum(arr[i][1])
theta_B_dist = stats.beta(1 + ones_B, 1 + zeros_B)
theta_A_sample = theta_A_dist.rvs(10000)
theta_B_sample = theta_B_dist.rvs(10000)
probs.append(sum(theta_B_sample > theta_A_sample) / len(theta_A_sample))
expected_diffs.append(sum(theta_B_sample - theta_A_sample) / len(theta_A_sample))
expected_lifts.append(
sum((theta_B_sample - theta_A_sample) / theta_A_sample) / len(theta_A_sample)
)
pd.DataFrame(
{
r"$P(\theta_B$ $>$ $\theta_A)$": probs,
r"$E(\theta_B$ $-$ $\theta_A)$": expected_diffs,
"Expected lift": expected_lifts,
}
)
# Now, let's run the simulations. For each simulation, we will compute three values:
# - $P(\theta_B > \theta_A \mid \text{test data})$.
# - $E(\theta_B - \theta_A \mid \text{test data})$.
# - Expected lift, i.e. the expected percentage increase/decrease of $\theta_B$ vs $\theta_A$. Explicitly, this is the value $E\Big(\frac{\theta_B - \theta_A}{\theta_A} \mid \text{test data}\Big)$.
# These simulations will take a good while — around 1.5 hours in a Kaggle CPU kernel — so maybe plan something else to do while running the next cell if you're working through this notebook.
starttime = time.time()
theta_A = 0.10
theta_Bs = np.linspace(0.06, 0.17, num=200)
n_samples = 2000
probs, expected_diffs, expected_lifts = {}, {}, {}
for i, theta_B in enumerate(theta_Bs):
probs[theta_B], expected_diffs[theta_B], expected_lifts[theta_B] = [], [], []
arr = experiment(
n_experiments=1000, n_samples=n_samples, theta_A=theta_A, theta_B=theta_B
)
for simulated_experiment in arr:
A = simulated_experiment[0]
B = simulated_experiment[1]
zeros_A, ones_A = len(A) - sum(A), sum(A)
theta_A_dist = stats.beta(1 + ones_A, 1 + zeros_A)
zeros_B, ones_B = len(B) - sum(B), sum(B)
theta_B_dist = stats.beta(1 + ones_B, 1 + zeros_B)
theta_A_sample = theta_A_dist.rvs(10000)
theta_B_sample = theta_B_dist.rvs(10000)
probs[theta_B].append(
sum(theta_B_sample > theta_A_sample) / len(theta_A_sample)
)
expected_diffs[theta_B].append(
sum(theta_B_sample - theta_A_sample) / len(theta_A_sample)
)
expected_lifts[theta_B].append(
sum((theta_B_sample - theta_A_sample) / theta_A_sample)
/ len(theta_A_sample)
)
probs[theta_B] = np.array(probs[theta_B])
expected_diffs[theta_B] = np.array(expected_diffs[theta_B])
expected_lifts[theta_B] = np.array(expected_lifts[theta_B])
print(
f"Computed {i+1} thetas, {100*(i+1)/len(theta_Bs):.0f} % done, {time.time()-starttime:.0f} sec taken",
end="\r",
flush=True,
)
fix, ax = plt.subplots(3, 1, figsize=(15, 15))
y_probs, y_diffs, y_lifts = [], [], []
for theta_B in theta_Bs:
y_probs.append(stats.mstats.mquantiles(probs[theta_B], prob=[0.05, 0.5, 0.95]))
y_diffs.append(
stats.mstats.mquantiles(expected_diffs[theta_B], prob=[0.05, 0.5, 0.95])
)
y_lifts.append(
stats.mstats.mquantiles(expected_lifts[theta_B], prob=[0.05, 0.5, 0.95])
)
y_probs, y_diffs, y_lifts = np.array(y_probs), np.array(y_diffs), np.array(y_lifts)
ax[0].fill_between(
theta_Bs - theta_A,
y_probs[:, 0],
y_probs[:, 2],
alpha=0.4,
label=r"90 % band of $P(\theta_B > \theta_A)$",
)
ax[0].plot(
theta_Bs - theta_A, y_probs[:, 1], label=r"Median of $P(\theta_B > \theta_A)$"
)
ax[0].hlines(
0.95,
min(theta_Bs) - theta_A,
max(theta_Bs) - theta_A,
label="P = 95 %",
color="black",
linestyle="--",
)
ax[0].legend(loc="lower right")
ax[0].set_xlim(min(theta_Bs) - theta_A, max(theta_Bs) - theta_A)
ax[0].set_ylim(-0.01, 1.01)
ax[0].set_title(
rf"""Observed distribution of $P(\theta_B > \theta_A)$ across different values of $\theta_B - \theta_A$,
number of samples per simulation = {n_samples}""",
fontsize=16,
)
ax[0].set_xlabel(r"Actual difference $\theta_B - \theta_A$")
ax[0].set_ylabel(r"$P(\theta_B > \theta_A)$")
ax[1].fill_between(
theta_Bs - theta_A,
y_diffs[:, 0],
y_diffs[:, 2],
alpha=0.4,
label=r"90 % band of $E(\theta_B - \theta_A)$",
)
ax[1].plot(
theta_Bs - theta_A, y_diffs[:, 1], label=r"Median of $E(\theta_B - \theta_A)$"
)
ax[1].plot(
theta_Bs - theta_A,
theta_Bs - theta_A,
color="black",
linestyle="--",
label=r"True $\theta_B - \theta_A$",
)
ax[1].legend(loc="upper left")
ax[1].set_xlim(min(theta_Bs) - theta_A, max(theta_Bs) - theta_A)
ax[1].set_title(
rf"""Observed distribution of $E(\theta_B - \theta_A)$ across different values of $\theta_B - \theta_A$,
number of samples per simulation = {n_samples}""",
fontsize=16,
)
ax[1].set_xlabel(r"Actual difference $\theta_B - \theta_A$")
ax[1].set_ylabel(r"$E(\theta_B - \theta_A)$")
ax[2].fill_between(
(theta_Bs - theta_A) / theta_A,
y_lifts[:, 0],
y_lifts[:, 2],
alpha=0.4,
label="90 % band of expected lift",
)
ax[2].plot(
(theta_Bs - theta_A) / theta_A, y_lifts[:, 1], label="Median of expected lift"
)
ax[2].plot(
(theta_Bs - theta_A) / theta_A,
(theta_Bs - theta_A) / theta_A,
color="black",
linestyle="--",
label="True lift",
)
ax[2].legend(loc="upper left")
ax[2].set_xlim(min((theta_Bs - theta_A) / theta_A), max((theta_Bs - theta_A) / theta_A))
ax[2].set_title(
rf"""Observed distribution of expected lifts across different values of $\theta_B - \theta_A$,
number of samples per simulation = {n_samples}""",
fontsize=16,
)
ax[2].set_xlabel("Actual lift")
ax[2].set_ylabel("Expected lift")
plt.tight_layout()
plt.figure(figsize=(15, 5))
fdr = []
stat_power = []
for theta_B in theta_Bs:
if theta_B <= theta_A:
fdr.append(sum(probs[theta_B] > 0.95) / len(probs[theta_B]))
else:
stat_power.append(sum(probs[theta_B] > 0.95) / len(probs[theta_B]))
plt.plot((theta_Bs - theta_A)[: len(fdr)], fdr, label="False discovery rate")
plt.plot((theta_Bs - theta_A)[len(fdr) :], stat_power, label="Statistical power")
plt.scatter(0, 0.05, s=25, color="black", marker="x")
plt.annotate("y = 0.05", (0.0, 0.05 + 0.02), horizontalalignment="right")
plt.legend()
plt.xlim(min(theta_Bs) - theta_A, max(theta_Bs) - theta_A)
plt.ylim(-0.01, 1.01)
plt.suptitle(
rf"""Observed false discovery rate and statistical power of the Bayesian test,
number of samples per simulation = {n_samples}""",
fontsize=16,
)
plt.xlabel(r"Actual difference $\theta_B - \theta_A$")
starttime = time.time()
theta_A = 0.10
theta_B = 0.13
sample_sizes = range(100, 3001, 100)
probs = {}
for i, n_samples in enumerate(sample_sizes):
probs[n_samples] = []
arr = experiment(
n_experiments=1000, n_samples=n_samples, theta_A=theta_A, theta_B=theta_B
)
for simulated_experiment in arr:
A = simulated_experiment[0]
B = simulated_experiment[1]
zeros_A, ones_A = len(A) - sum(A), sum(A)
theta_A_dist = stats.beta(1 + ones_A, 1 + zeros_A)
zeros_B, ones_B = len(B) - sum(B), sum(B)
theta_B_dist = stats.beta(1 + ones_B, 1 + zeros_B)
theta_A_sample = theta_A_dist.rvs(10000)
theta_B_sample = theta_B_dist.rvs(10000)
probs[n_samples].append(
sum(theta_B_sample > theta_A_sample) / len(theta_A_sample)
)
probs[n_samples] = np.array(probs[n_samples])
print(
f"Computed {i+1} sample sizes, {100*(i+1)/len(sample_sizes):.0f} % done, {time.time()-starttime:.0f} sec taken",
end="\r",
flush=True,
)
plt.figure(figsize=(15, 5))
y = []
for n_samples in sample_sizes:
y.append(stats.mstats.mquantiles(probs[n_samples], prob=[0.2, 0.5, 1.0]))
y = np.array(y)
plt.fill_between(
sample_sizes,
y[:, 0],
y[:, 2],
alpha=0.4,
label=r"top 80 % of $P(\theta_B > \theta_A)$",
)
plt.plot(sample_sizes, y[:, 1], label=r"Median of $P(\theta_B > \theta_A)$")
plt.hlines(
0.95,
min(sample_sizes),
max(sample_sizes),
label="P = 0.95",
color="black",
linestyle="--",
)
plt.legend(loc="lower right")
plt.xlim(min(sample_sizes), max(sample_sizes))
plt.ylim(-0.01, 1.01)
plt.suptitle(
r"""Observed distribution of $P(\theta_B > \theta_A)$ across different sample sizes,
$\theta_A = 0.10, \theta_B = 0.13$""",
y=1.005,
fontsize=16,
)
plt.xlabel("Sample size")
plt.ylabel(r"$P(\theta_B > \theta_A)$")
plt.figure(figsize=(15, 5))
stat_power = []
for n_samples in sample_sizes:
stat_power.append(sum(probs[n_samples] > 0.95) / len(probs[n_samples]))
plt.plot(sample_sizes, stat_power, label="Statistical power")
plt.xlim(min(sample_sizes), max(sample_sizes))
plt.ylim(-0.01, 1.01)
plt.suptitle(
r"""Observed statistical power of the Bayesian test across different sample sizes,
$\theta_A = 0.10, \theta_B = 0.13$""",
fontsize=16,
)
plt.xlabel("Sample size")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import numpy as np
from sklearn.metrics.pairwise import cosine_similarity
import random
ratings = pd.read_csv(
"/kaggle/input/datasettt/ccai422_lab03_part1_data.csv", index_col="Unnamed: 0"
)
ratings.head(5)
# The total number of data points
print("The number of data points in this dataset: " + str(len(ratings)))
# The number of items (i.e. movies) in the dataset
print(
"The number of items (i.e. movies) in the dataset: "
+ str(ratings["movie_id"].nunique())
)
# The number of users in the dataset
print("The number of users in the dataset: " + str(ratings["user_id"].nunique()))
# The average ratings per user
ratings_per_users = ratings.groupby("user_id").count()
print("The average ratings per user: " + str(round(ratings_per_users.mean()[0], 2)))
# The number of ratings/user
print("The below table shows the number of ratings per user\n")
print(ratings_per_users)
ratings = ratings.pivot_table(values="rating", index="user_id", columns="movie_id")
# Before starting computations, copy the original ratings into a dummy
# ratings matrix which will have all null values imputed to zeros
cos_matrix_dummy = ratings.copy()
# rename the axis of the new matrix
cos_matrix_dummy = cos_matrix_dummy.rename_axis("user_id", axis=1).rename_axis(
None, axis=0
)
# Compute the mean rating per user
cos_matrix_dummy["mean"] = cos_matrix_dummy.mean(axis=1)
# Substract the mean from each item
cos_matrix_dummy.loc[:, cos_matrix_dummy.columns != "mean"] = (
cos_matrix_dummy.loc[:, cos_matrix_dummy.columns != "mean"]
- cos_matrix_dummy["mean"].values[:, None]
)
# Drop the newly added mean column from the data
cos_matrix_dummy.drop(columns="mean", inplace=True)
# Display the data centred around the mean
cos_matrix_dummy.head()
# Fill NaN values with zeros
cos_matrix_dummy.fillna(0, inplace=True)
# Compute the cosine similarity. Notice we take the transpose to compute
# the similarity between the items
cos_matrix_dummy_sim = cosine_similarity(cos_matrix_dummy.T)
# Build dataframe based on the similarity value. Note that the type ofr_matrix_dummy_sim is numpy array
cos_matrix_dummy_sim = pd.DataFrame(
cos_matrix_dummy_sim,
columns=list(cos_matrix_dummy.T.index),
index=list(cos_matrix_dummy.T.index),
)
display(cos_matrix_dummy_sim)
# Optionally set a seed
random.seed(3)
# Select random user
random_user = random.randrange(len(ratings))
print("random user ID is {}".format(random_user))
# Retrieve ratings data for the randomly selected user
random_user_ratings = ratings[ratings.index == random_user]
# etrieve unrated item for that the randomly selected user
unrated_items_for_random_user = random_user_ratings.columns[
random_user_ratings.isnull().all(0)
]
# Randomly select an unrated item whose rating will be predicted using
# item-based methods
random_unrated_item = random.choice(list(unrated_items_for_random_user))
print("Item {} is unrated by user{}".format(random_unrated_item, random_user))
rated_items_for_random_user = random_user_ratings.columns[
random_user_ratings.notnull().all(0)
]
filtered_col_sim = list(rated_items_for_random_user)
filtered_col_sim.append(random_unrated_item)
rated_unrated_sim_random_user = cos_matrix_dummy_sim.loc[
cos_matrix_dummy_sim.columns.isin(filtered_col_sim)
]
topn = rated_unrated_sim_random_user.nlargest(3, random_unrated_item).index.tolist()[1:]
neighbors_item_ratings_random_item = random_user_ratings.loc[:, topn]
neighbors_sim = rated_unrated_sim_random_user[[random_unrated_item]].nlargest(
3, random_unrated_item
)[1:]
print(
"The top neighbors for the item {} that is unrated by user {} are theitems: {}".format(
random_unrated_item, random_user, topn
)
)
print(
"The ratings for the top {} neigbors items are: {}".format(
len(topn), *neighbors_item_ratings_random_item.values
)
)
print(
"The cosine similarities between item {} and items: {} are: {}".format(
random_unrated_item, topn, neighbors_sim.values
)
)
predicted_value_for_unrated_item = (
neighbors_sim.T.dot(neighbors_item_ratings_random_item.T).values[0]
/ neighbors_sim.sum()
)
predicted_value_for_unrated_item
|
# Import necessary libraries
import pandas as pd
from mlxtend.frequent_patterns import apriori
from mlxtend.frequent_patterns import association_rules
# Read the CSV data into a pandas DataFrame
csv_data = pd.read_csv(
"/kaggle/input/allstate-purchase-prediction-challenge/train.csv.zip"
)
# Calculate the correlation matrix
correlation_matrix = csv_data.corr(method="pearson")
# Display the correlation matrix
# print(correlation_matrix)
# Select the categorical columns for analysis
categorical_columns = ["state", "car_value", "A", "B", "C", "D", "E", "F", "G"]
categorical_data = csv_data[categorical_columns]
# Convert categorical variables into one-hot encoded format
one_hot_encoded_data = pd.get_dummies(categorical_data)
# Binarize the one-hot encoded data
binarized_data = one_hot_encoded_data.applymap(lambda x: 1 if x >= 1 else 0)
# Convert the binarized data to boolean type
boolean_data = binarized_data.astype(bool)
# Find frequent itemsets using the Apriori algorithm
frequent_itemsets = apriori(boolean_data, min_support=0.1, use_colnames=True)
# Generate association rules
rules = association_rules(frequent_itemsets, metric="confidence", min_threshold=0.7)
# Print the rules sorted by lift
rules = rules.sort_values(by="lift", ascending=False)
print(rules)
import matplotlib.pyplot as plt
import seaborn as sns
# Create a new 'age' column as the average of 'age_oldest' and 'age_youngest'
csv_data["age"] = (csv_data["age_oldest"] + csv_data["age_youngest"]) / 2
# Create age groups
bins = [0, 10, 20, 30, 40, 50, 60, 70, 80, 90, 100]
labels = [
"0-9",
"10-19",
"20-29",
"30-39",
"40-49",
"50-59",
"60-69",
"70-79",
"80-89",
"90-99",
]
csv_data["age_group"] = pd.cut(csv_data["age"], bins=bins, labels=labels)
# Calculate the mean risk factor for each age group
age_risk_data = csv_data.groupby("age_group")["risk_factor"].mean().reset_index()
plt.figure(figsize=(12, 6))
sns.barplot(x="age_group", y="risk_factor", data=age_risk_data, palette="viridis")
plt.title("Mean Risk Factor by Age Group")
plt.xlabel("Age Group")
plt.ylabel("Mean Risk Factor")
plt.show()
state_policy_counts = csv_data["state"].value_counts().reset_index()
state_policy_counts.columns = ["state", "count"]
plt.figure(figsize=(15, 6))
sns.barplot(x="state", y="count", data=state_policy_counts, palette="viridis")
plt.title("Number of Insurance Policies by State")
plt.xlabel("State")
plt.ylabel("Number of Policies")
plt.xticks(rotation=45)
plt.show()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
# for dirname, _, filenames in os.walk('/kaggle/input'):
# for filename in filenames:
# print(os.path.join(dirname, filename))
ts_data = pd.read_csv("/kaggle/input/nba-enhanced-stats/2012-18_teamBoxScore.csv")
# for str in ts_data.keys():
# print(str)
# Any results you write to the current directory are saved as output.
def convert(s):
# initialization of string to ""
new = ""
# traverse in the string
for x in s:
new += x
# return string
return new
def cutYear(lst):
tmp = convert([lst[0:4]])
tmp1 = int(tmp)
return tmp1
def cutMonth(lst):
tmp = convert([lst[5:7]])
tmp1 = int(tmp)
return tmp1
def intTOstr(t):
return str(t)
def getSeason(date):
year = cutYear(date)
month = cutMonth(date)
ans = year
if month < 10:
ans -= 1
return ans
def getwinner(games):
games["win"] = games["teamPTS"] > games["opptPTS"]
games["winner"] = np.where(games["win"], games["teamAbbr"], games["opptAbbr"])
games["looser"] = np.where(games["win"], games["opptAbbr"], games["teamAbbr"])
games["diff"] = abs(games["teamPTS"] - games["opptPTS"])
return games[["winner", "looser", "diff"]]
games = ts_data[
[
"gmDate",
"seasTyp",
"teamConf",
"teamDiv",
"teamAbbr",
"teamPTS",
"teamLoc",
"opptConf",
"opptDiv",
"opptAbbr",
"opptPTS",
]
]
games = games[games.teamLoc == "Home"]
games = games[games.seasTyp == "Regular"]
games = games[
[
"gmDate",
"seasTyp",
"teamConf",
"teamDiv",
"teamAbbr",
"teamPTS",
"opptConf",
"opptDiv",
"opptAbbr",
"opptPTS",
]
]
games["Season"] = games.gmDate.apply(getSeason)
games = games[
[
"Season",
"teamAbbr",
"teamConf",
"teamDiv",
"teamPTS",
"opptAbbr",
"opptConf",
"opptDiv",
"opptPTS",
]
]
season12_13 = games[games.Season == 2012]
season13_14 = games[games.Season == 2013]
season14_15 = games[games.Season == 2014]
season15_16 = games[games.Season == 2015]
season16_17 = games[games.Season == 2016]
season17_18 = games[games.Season == 2017]
season12_13 = getwinner(
season12_13[
[
"teamAbbr",
"teamConf",
"teamDiv",
"teamPTS",
"opptAbbr",
"opptConf",
"opptDiv",
"opptPTS",
]
]
)
season13_14 = getwinner(
season13_14[
[
"teamAbbr",
"teamConf",
"teamDiv",
"teamPTS",
"opptAbbr",
"opptConf",
"opptDiv",
"opptPTS",
]
]
)
season14_15 = getwinner(
season14_15[
[
"teamAbbr",
"teamConf",
"teamDiv",
"teamPTS",
"opptAbbr",
"opptConf",
"opptDiv",
"opptPTS",
]
]
)
season15_16 = getwinner(
season15_16[
[
"teamAbbr",
"teamConf",
"teamDiv",
"teamPTS",
"opptAbbr",
"opptConf",
"opptDiv",
"opptPTS",
]
]
)
season16_17 = getwinner(
season16_17[
[
"teamAbbr",
"teamConf",
"teamDiv",
"teamPTS",
"opptAbbr",
"opptConf",
"opptDiv",
"opptPTS",
]
]
)
season17_18 = getwinner(
season17_18[
[
"teamAbbr",
"teamConf",
"teamDiv",
"teamPTS",
"opptAbbr",
"opptConf",
"opptDiv",
"opptPTS",
]
]
)
study_set = getwinner(games[games.Season < 2017])
test_set = season17_18
centrality_ratings_table = study_set.groupby(["winner"]).sum()
sorted_centrality_ratings_table = centrality_ratings_table.sort_values(
by=["diff"], ascending=False
)
team_west_or_east_table = (games[["teamAbbr", "teamConf"]]).drop_duplicates()
team_west_or_east_table = team_west_or_east_table.rename(columns={"teamAbbr": "winner"})
sorted_centrality_ratings_table = sorted_centrality_ratings_table.merge(
team_west_or_east_table, on="winner"
)
west_playoff_teams = sorted_centrality_ratings_table[
sorted_centrality_ratings_table["teamConf"] == "West"
][0:8]
east_playoff_teams = sorted_centrality_ratings_table[
sorted_centrality_ratings_table["teamConf"] == "East"
][0:8]
play0ff_teams = pd.concat([west_playoff_teams, east_playoff_teams])
sorted_centrality_ratings_table
play0ff_teams
|
# # The Chernobyl Disaster occurred on 26 April 1986
import numpy as np
import pandas as pd
import os
import folium
from folium import plugins
import seaborn as sns
import matplotlib.pyplot as plt
import plotly.express as px
import plotly.graph_objects as go
from plotly.subplots import make_subplots
data = pd.read_csv(
"/kaggle/input/air-concentration-for-the-chernobyl-disaster/data.csv"
)
data["Date"] = pd.to_datetime(data["Date"])
data = data.iloc[:, 2:9]
display(data)
I = []
for i in range(len(data)):
for j in range(6, 7):
if data.iloc[i, j] in ["?", "L", "N"]:
I += [i]
data = data.drop(I, axis=0)
data.iloc[:, 8:] = data.iloc[:, 8:].astype(float)
data.info()
print(sorted(data["Ville"].unique().tolist()))
villes = sorted(data["Ville"].unique().tolist())
display(data)
data3 = data.groupby(
[
"Date",
"Ville",
],
as_index=False,
).agg({"I_131_(Bq/m3)": "max"})
display(data3)
data3["Ville"].value_counts()
fig = make_subplots(specs=[[{"secondary_y": False}]])
for ville in villes:
fig.add_trace(
go.Scatter(
x=data3["Date"],
y=data3[data3["Ville"] == ville]["I_131_(Bq/m3)"],
name=ville,
),
secondary_y=False,
)
fig.update_layout(autosize=False, width=700, height=500, title_text="I_131")
fig.update_xaxes(title_text="Date")
fig.update_yaxes(title_text="Bq/m3", secondary_y=False)
fig.show()
data3 = (
data[data["Date"] == "1986-04-29"]
.groupby("Ville", as_index=False)
.max()
.sort_values("I_131_(Bq/m3)", ascending=False)
)
fig = px.bar(
data3[0:20],
x="Ville",
y="I_131_(Bq/m3)",
title="I_131_(Bq/m3) Ranknig on 1986-04-29",
)
fig.show()
data3 = (
data[data["Date"] == "1986-05-01"]
.groupby("Ville", as_index=False)
.max()
.sort_values("I_131_(Bq/m3)", ascending=False)
)
fig = px.bar(
data3[0:20],
x="Ville",
y="I_131_(Bq/m3)",
title="I_131_(Bq/m3) Ranknig on 1986-05-01 ",
)
fig.show()
data3 = (
data[data["Date"] == "1986-05-02"]
.groupby("Ville", as_index=False)
.max()
.sort_values("I_131_(Bq/m3)", ascending=False)
)
fig = px.bar(
data3[0:20],
x="Ville",
y="I_131_(Bq/m3)",
title="I_131_(Bq/m3) Ranknig on 1986-05-02 ",
)
fig.show()
data1 = data[data["Ville"] == "ISPRA"][data["Date"] == "1986-05-01"]
fig = make_subplots(specs=[[{"secondary_y": False}]])
fig.add_trace(
go.Scatter(
x=data1["End_of_sampling"], y=data1["I_131_(Bq/m3)"], name="I_131_(Bq/m3)"
),
secondary_y=False,
)
fig.update_layout(
autosize=False,
width=700,
height=500,
title_text="I_131,Cs_134,Cs_137 in ISPRA on 1986-05-01",
)
fig.update_xaxes(title_text="Date")
fig.update_yaxes(title_text="Bq/m3", secondary_y=False)
fig.show()
data1 = data[data["Ville"] == "ISPRA"][data["Date"] == "1986-05-02"]
fig = make_subplots(specs=[[{"secondary_y": False}]])
fig.add_trace(
go.Scatter(
x=data1["End_of_sampling"], y=data1["I_131_(Bq/m3)"], name="I_131_(Bq/m3)"
),
secondary_y=False,
)
fig.update_layout(
autosize=False,
width=700,
height=500,
title_text="I_131,Cs_134,Cs_137 in ISPRA on 1986-05-02",
)
fig.update_xaxes(title_text="Date")
fig.update_yaxes(title_text="Bq/m3", secondary_y=False)
fig.show()
data2 = data[["Y", "X"]].drop_duplicates()
eq_map = folium.Map(
location=[50, 10], tiles="Stamen Terrain", zoom_start=5.0, min_zoom=2.0
)
eq_map.add_child(plugins.HeatMap(data2))
eq_map
|
# # Stack-OverFlow Survey Dataset
import pandas as pd
import numpy as np
df = pd.read_csv(
"/kaggle/input/stack-overflow-developer-survey-results-2019/survey_results_public.csv"
)
schema = pd.read_csv(
"/kaggle/input/stack-overflow-developer-survey-results-2019/survey_results_schema.csv",
index_col="Column",
)
df.head()
df.shape
df.columns
df.ConvertedComp.head(10)
# median salary
df.ConvertedComp.median()
df.median()
df.describe().T
# median salary by country
df.groupby(["Country"])
df["Hobbyist"].value_counts()
df["Hobbyist"].value_counts().plot(kind="pie")
schema
schema.loc["SocialMedia"]
df.SocialMedia.value_counts()
df.SocialMedia.value_counts(normalize=True)
df.Country.value_counts()
country_grp = df.groupby(["Country"])
country_grp.get_group("United States")
usa = df["Country"] == "United States"
df.loc[usa]
# same as get_group
df.loc[usa]["SocialMedia"].value_counts()
df.loc[df.Country == "India"]["SocialMedia"].value_counts()
df.groupby("Country")["SocialMedia"].value_counts().head(50)
df.groupby("Country")["SocialMedia"].value_counts().loc["Chile"]
# Creating a generic function so that we donot have to create filters for each country
# Here We just want to specify the country name in loc
df.groupby("Country")["SocialMedia"].value_counts(normalize=True).loc["Chile"]
df.groupby("Country")["ConvertedComp"].agg(["median", "mean"])
df.groupby("Country")["ConvertedComp"].agg(["median", "mean"]).loc["Canada"]
df[df.Country == "India"]["LanguageWorkedWith"].str.contains("Python").value_counts()
df[df.Country == "India"]["LanguageWorkedWith"].str.contains("Python").sum()
df.groupby("Country")["LanguageWorkedWith"].apply(
lambda x: x.str.contains("Python").sum()
)
df.groupby("Country")["LanguageWorkedWith"].apply(
lambda x: x.str.contains("Python").sum()
).loc["India"]
df.groupby("Country")["LanguageWorkedWith"].apply(
lambda x: x.str.contains("Python").value_counts(normalize=True)
).loc["India"][1]
country = df["Country"].value_counts()
country
python_dev = df.groupby("Country")["LanguageWorkedWith"].apply(
lambda x: x.str.contains("Python").sum()
)
python_dev
python_users = pd.concat([country, python_dev], axis=1)
python_users
python_users.columns = ["respondants", "users"]
python_users
python_users["percentage_of_python_users"] = (
python_users["users"] / python_users["respondants"]
) * 100
python_users
python_users.sort_values(by="percentage_of_python_users", ascending=False).head(50)
|
# Merhaba!
# Bu çalışma sayfasında verilen çeşitli müşteri bilgilerine dayanarak müşterilerin kasko sigortası alıp almayacağını tahmin etmeye çalışacağım.
# Fikir ve yorumlarınız benim için değerlidir.
#
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
train = pd.read_csv("/kaggle/input/carinsurance/carInsurance_train.csv")
train.head()
# Elimizdeki data da Id numaraları dahil 20 değişken var bu değişkenler yaş, medeni durum, eğitim durumu, görüşme başlama ve bitiş zamanları gibi detaylı bilgiler dataset'in açıklamalarında mevcut. Elimizdeki değişkenleri görmek ve bu değişkenler hakkında basit bilgilere sahip olmak için;
train.columns
train.describe()
train.isnull().sum() / len(train)
# Veride en fazla boş satırlar Outcome değişkeninde var model kurmadan önce veriyi daha iyi hale getirmek için aşağıdaki adımları uygulayacağım.
# 1- Önce ayların bulunduğu LastContactMonth değişkenini aylıklardan dönemlere çevireceğim bir şekilde mevsimsellik var mı yok mu hangi mevsimlerde ortalama satış artmış veya azalmış görmek için.
#
# 2- Konuşma başlama ve bitiş sürelerini birbirlerinden çıkarıp bir skala haline getireceğim. Konuşma süresinin kasko sigortasına etkisi var mı yok mu konuşma uzadıkça hedef değişkenim ne oluyor görmek için. Bunu yaptıktan sonra bu iki değişkeni datadan atacağım.
#
# 3- Yaşları aynı şekilde bir skalaya getirip genç, orta yaşlı, yaşlı vs gibi gruplayacağım.
#
# 4- Outcome değişkenini datadan çıkaracağım çünkü %76'sı boş, Job değişkeni boş olanları silebilirim çünkü az miktarda bir veri fakat education için hem yaş hemde meslek bilgilerini kullanarak kişilerin eğitim durumu tahmin etmem daha iyi olur.
#
# 5- Tüm değişkenleri encode edip veriyi bağımlı bağımsız değişken olarak bölüp modellemeye hazır hale getireceğim.
#
# Bu işlmelere geçmeden önce data hakkında daha fazla bilgiye sahip olmak için pairplot grafiğine bakılabilir.
#
#
import seaborn as sns
sns.pairplot(data=train, hue="CarInsurance")
# **AYLAR**
yeni_degiskenler = []
i = 0
while i < 4001:
if train["LastContactMonth"][i] == "jan":
yeni_degiskenler.append(1)
elif train["LastContactMonth"][i] == "feb":
yeni_degiskenler.append(1)
elif train["LastContactMonth"][i] == "mar":
yeni_degiskenler.append(1)
elif train["LastContactMonth"][i] == "apr":
yeni_degiskenler.append(2)
elif train["LastContactMonth"][i] == "may":
yeni_degiskenler.append(2)
elif train["LastContactMonth"][i] == "jun":
yeni_degiskenler.append(2)
elif train["LastContactMonth"][i] == "jul":
yeni_degiskenler.append(3)
elif train["LastContactMonth"][i] == "aug":
yeni_degiskenler.append(3)
elif train["LastContactMonth"][i] == "sep":
yeni_degiskenler.append(3)
elif train["LastContactMonth"][i] == "oct":
yeni_degiskenler.append(4)
elif train["LastContactMonth"][i] == "nov":
yeni_degiskenler.append(4)
elif train["LastContactMonth"][i] == "dec":
yeni_degiskenler.append(4)
i += 1
len(yeni_degiskenler)
# Ayları dönemlere çevirdikten sonra konuşma sürelerini hesaplamak için:
konusma = pd.to_datetime(train["CallEnd"]) - pd.to_datetime(train["CallStart"])
konusma = pd.to_numeric(konusma) / 1000000000
konusma = pd.DataFrame(konusma)
a = pd.concat([train.CarInsurance, konusma], names=("sonuc", "saniye"), axis=1)
sns.pairplot(data=a, hue="CarInsurance")
konusma.loc[(konusma[0] >= 0) & (konusma[0] <= 126), "dakika"] = 1
konusma.loc[(konusma[0] > 126) & (konusma[0] <= 232), "dakika"] = 2
konusma.loc[(konusma[0] > 232) & (konusma[0] <= 460), "dakika"] = 3
konusma.loc[(konusma[0] > 460) & (konusma[0] <= 3253), "dakika"] = 4
# Aslında burası bize önemli bir şey söylüyor. Alttaki grafikte görülen şey eğer konuşma süresi kısa ise hedef değişken cevabı 0 olması daha muhtemel tersi yani konuşma süresi uzun ise cevabın 1 olması daha muhtemel gibi.
dakika = pd.DataFrame(konusma["dakika"])
yeni_degiskenler = pd.DataFrame(yeni_degiskenler, columns=["quarter"])
yeni_degiskenler = pd.concat([yeni_degiskenler, dakika], axis=1)
yeni_degiskenler.head()
# Şimdi de yaşları skala edelim.
i = 0
yas = []
while i < 4000:
if train["Age"][i] > 0 and train["Age"][i] < 26:
yas.append(1)
elif train["Age"][i] >= 26 and train["Age"][i] < 35:
yas.append(2)
elif train["Age"][i] >= 35 and train["Age"][i] < 45:
yas.append(3)
elif train["Age"][i] >= 45:
yas.append(4)
i += 1
yas = pd.DataFrame(yas, columns=["yas"])
yeni_degiskenler = pd.concat([yeni_degiskenler, yas], axis=1)
yeni_degiskenler.head()
# Çıkarılan bu 3 yeni değişkeni train'in içine koyma
train = pd.concat([yeni_degiskenler, train], axis=1)
train.head()
# Education içindeki boş verileri doldurmak için yağtığım şey şu benim elimde kişilerin meslekleri ve yaşları var eğer aynı mesleğe sahip insaları datadan çekersem bunların ağırlıklı olarak hangi eğitim düzeyinde oldukları ve eğitim düzeyinin yaşlara dağılımı gibi önemli bilgileri el edebilirim örneğin işi manager olanların büyük çoğunluğu tertiary seviyesinde eğitime sahip olduklarını varsayılım (ki öyle) fakat bir yandan da yaşı 25-30 olup eğitimi tertiary bile olsa mesleği manager olamayacak (çünkü henüz gençler) insanlar olacaktır veya aynı şekilde yaşı 65 olanlar genellikle tertiary eğitim durumu olsa bile retired olarak karşımıza çıkacaklar bunun için education sütünundaki boşlukları hem meslek hemde yaş bilgileri ile beraber doldurmanın daha doğru olacağına inanıyorum.
management = train.loc[train["Job"] == "management"]
blue_collar = train.loc[train["Job"] == "blue-collar"]
technician = train.loc[train["Job"] == "technician"]
admin = train.loc[train["Job"] == "admin."]
services = train.loc[train["Job"] == "services"]
retired = train.loc[train["Job"] == "retired"]
self_employed = train.loc[train["Job"] == "self-employed"]
student = train.loc[train["Job"] == "student"]
management["Education"].value_counts() # tertiary 751 / secondary 94 / primary 22
blue_collar["Education"].value_counts() # secondary 430 / primary 281 / tertiary 17
technician["Education"].value_counts() # secondary 446 / tertiary 177 / primary 16
admin["Education"].value_counts() # secondary 374 / tertiary 62 / primary 9
services["Education"].value_counts() # secondary 269 / primary 27 / tertiary 25
retired["Education"].value_counts() # secondary 100 / primary 93 / tertiary 37
self_employed["Education"].value_counts() # tertiary 84 / secondary 43 / primary 6
student["Education"].value_counts() # secondary 67 / tertiary 31 / primary 8
management["Age"].mean() # 40
blue_collar["Age"].mean() # 40
technician["Age"].mean() # 39
admin["Age"].mean() # 39
services["Age"].mean() # 38
retired["Age"].mean() # 63
self_employed["Age"].mean() # 41
student["Age"].mean() # 25
train["Education"] = train["Education"].replace(np.nan, "?")
i = 0
while i < 4000:
if (
train["Age"][i] >= 35
and train["Job"][i] == "management"
and train["Education"][i] == "?"
):
train["Education"][i] = train["Education"][i].replace("?", "tertiary")
elif (
train["Age"][i] < 35
and train["Job"][i] == "management"
and train["Education"][i] == "?"
):
train["Education"][i] = train["Education"][i].replace("?", "secondary")
i += 1
i = 0
while i < 4000:
if train["Job"][i] == "admin." and train["Education"][i] == "?":
train["Education"][i] = train["Education"][i].replace("?", "secondary")
i += 1
i = 0
while i < 4000:
if train["Job"][i] == "services" and train["Education"][i] == "?":
train["Education"][i] = train["Education"][i].replace("?", "secondary")
i += 1
i = 0
while i < 4000:
if train["Job"][i] == "services" and train["Education"][i] == "?":
train["Education"][i] = train["Education"][i].replace("?", "secondary")
i += 1
i = 0
while i < 4000:
if (
train["Age"][i] >= 28
and train["Job"][i] == "student"
and train["Education"][i] == "?"
):
train["Education"][i] = train["Education"][i].replace("?", "tertiary")
elif (
train["Age"][i] < 28
and train["Job"][i] == "student"
and train["Education"][i] == "?"
):
train["Education"][i] = train["Education"][i].replace("?", "secondary")
i += 1
train["Education"] = train["Education"].replace("?", "secondary")
train.isnull().sum()
# Communacation değişkenindeki boşları doldurmak için daha basit bir yol izleyeceğim çünkü baktığım zaman şöyle bir sonuç çıkıyor.
# Genel olarak telefon kullanılmıyor hücresel veri daha fazla ve telefonu yaşlılar daha fazla kullanıyor fakat bu kullanım oranı yaşlılarda bile az yaşlıların %60'ından fazlası cellular kullanıyor. Bu yüzden mantıklı bir ayrım bulamadığım için tüm boşluklara cellular yerleştirdim ve job kısmındaki 19 boş satırı ve outcome, ıd gibi geresiz değişkenleri datadan çıkardım.
train["Communication"] = train["Communication"].fillna("cellular")
train.isnull().sum()
train = train.drop("Outcome", axis=1)
train = train.drop("CallEnd", axis=1)
train = train.drop("CallStart", axis=1)
train = train.drop("Id", axis=1)
train = train.dropna()
train.isnull().sum()
# Artık boş veri kalmadı yeni değişkenler bulup bunları dataya ekledik şimdi model kurmak için hazırlıkları yapabilirim.
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
cols = ["Job", "Marital", "Education", "Communication", "LastContactMonth"]
for i in cols:
train[i] = le.fit_transform(train[i])
x = train.iloc[:, 0:17]
y = train.iloc[:, 17:18]
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.33, random_state=42
)
# **MODELLER**
# RANDOM FOREST
from sklearn.metrics import accuracy_score
from sklearn.ensemble import RandomForestClassifier
rf = RandomForestClassifier()
rf.fit(x_train, y_train)
rf_tahmin = rf.predict(x_test)
rf_skor = accuracy_score(y_test, rf_tahmin)
rf_skor
# XGBOOOST
from xgboost import XGBClassifier
xgb = XGBClassifier()
xgb.fit(x_train, y_train)
xgb_tahmin = xgb.predict(x_test)
accuracy_score(y_test, xgb_tahmin)
# LIGHTGBM
from lightgbm import LGBMClassifier
lgb = LGBMClassifier()
lgb.fit(x_train, y_train)
lgb_tahmin = lgb.predict(x_test)
accuracy_score(y_test, lgb_tahmin)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
from learntools.core import binder
binder.bind(globals())
from learntools.time_series.ex6 import *
# Setup notebook
from pathlib import Path
import ipywidgets as widgets
from learntools.time_series.style import * # plot style settings
from learntools.time_series.utils import (
create_multistep_example,
load_multistep_data,
make_lags,
make_multistep_target,
plot_multistep,
)
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.linear_model import LinearRegression
from sklearn.multioutput import RegressorChain
from sklearn.preprocessing import LabelEncoder
from xgboost import XGBRegressor
comp_dir = Path("../input/store-sales-time-series-forecasting")
store_sales = pd.read_csv(
comp_dir / "train.csv",
usecols=["store_nbr", "family", "date", "sales", "onpromotion"],
dtype={
"store_nbr": "category",
"family": "category",
"sales": "float32",
"onpromotion": "uint32",
},
parse_dates=["date"],
infer_datetime_format=True,
)
store_sales["date"] = store_sales.date.dt.to_period("D")
store_sales = store_sales.set_index(["store_nbr", "family", "date"]).sort_index()
family_sales = (
store_sales.groupby(["family", "date"]).mean().unstack("family").loc["2017"]
)
test = pd.read_csv(
comp_dir / "test.csv",
dtype={
"store_nbr": "category",
"family": "category",
"onpromotion": "uint32",
},
parse_dates=["date"],
infer_datetime_format=True,
)
test["date"] = test.date.dt.to_period("D")
test = test.set_index(["store_nbr", "family", "date"]).sort_index()
# Convert date to datetime and set it as the index
train["date"] = pd.to_datetime(train["date"])
train = train.set_index("date")
# Group the sales data by store, family, and date and compute the daily sales
daily_sales = (
train.groupby(["store_nbr", "family", pd.Grouper(freq="D")])["sales"]
.sum()
.reset_index()
)
# Reshape the data to have one row per store, family, and date
daily_sales = daily_sales.pivot(
index=["store_nbr", "date"], columns="family", values="sales"
).fillna(0)
# Add columns for year, month, and day of week
daily_sales["year"] = daily_sales.index.get_level_values("date").year
daily_sales["month"] = daily_sales.index.get_level_values("date").month
daily_sales["day_of_week"] = daily_sales.index.get_level_values("date").dayofweek
# Reset the index
daily_sales = daily_sales.reset_index()
# Print the first 5 rows of the dataset
daily_sales.head()
# EDA
import plotly.express as px
import plotly.graph_objs as go
from plotly.subplots import make_subplots
# we can create a line plot of the daily sales for each family using Plotly:
# Create a line plot of the daily sales for each family using Plotly
fig = make_subplots(rows=1, cols=1)
for col in daily_sales.columns[:-3]:
fig.add_trace(
go.Scatter(x=daily_sales["date"], y=daily_sales[col], mode="lines", name=col)
)
fig.update_layout(
title="Daily Sales by Family", xaxis_title="Date", yaxis_title="Sales"
)
fig.show()
# This will produce a line plot that shows the daily sales for each family over time. From this plot, we can see that there are some families that have higher sales than others, and that there are some families that have seasonal patterns in their sales.
# let's look at some descriptive statistics for the data:
# Compute descriptive statistics for the data
sales_stats = daily_sales.describe()
print(sales_stats)
# This will produce a table that shows the mean, standard deviation, minimum, 25th percentile, median, 75th percentile, and maximum values for each family.
# To identify any outliers or anomalies in the data, we can use box plots:
# Create box plots for each family to identify any outliers or anomalies
fig = make_subplots(rows=1, cols=1)
for col in daily_sales.columns[:-3]:
fig.add_trace(go.Box(y=daily_sales[col], name=col))
fig.update_layout(title="Daily Sales by Family", yaxis_title="Sales")
fig.show()
# This will produce a set of box plots that show the distribution of sales for each family. From these plots, we can see if there are any outliers or anomalies in the data.
# Check for missing data
print(daily_sales.isna().sum())
# This will produce a table that shows the number of missing values for each column in the dataset. We can use this information to determine if any imputation or cleaning is necessary before modeling the data.
# Data Preprocessing:
from learntools.time_series.utils import make_lags
# Preprocessing
lags = [1, 7, 14, 21, 28]
X_train = make_lags(family_sales["sales"].values, lags)
y_train = make_multistep_target(family_sales["sales"].values, n_steps=28)
# Modeling:
# * Train a Decision Tree Regressor model to predict future sales based on the lagged features.
# Modeling
from sklearn.tree import DecisionTreeRegressor
regressor = DecisionTreeRegressor(max_depth=10, random_state=0)
regressor.fit(X_train, y_train)
# Forecasting:
# * Apply the trained model to the test set to make sales forecasts.
# * Plot the forecasts using Plotly to visualize the predicted sales trends over time.
# Forecasting
X_test = make_lags(test_sales["sales"].values, lags)
y_pred = regressor.predict(X_test)
# Plotting
plot_multistep(test_sales.index.values, y_pred)
|
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import MinMaxScaler, LabelEncoder
from sklearn.cluster import KMeans
from sklearn.metrics import silhouette_score
df = pd.read_csv("/kaggle/input/iris/Iris.csv")
df.head()
df.shape
df.drop("Id", axis=1, inplace=True)
scaler = MinMaxScaler()
scaler.fit(df[["PetalWidthCm"]])
df["PetalWidthCm"] = scaler.transform(df[["PetalWidthCm"]])
scaler.fit(df[["SepalLengthCm"]])
df["SepalLengthCm"] = scaler.transform(df[["SepalLengthCm"]])
scaler.fit(df[["SepalWidthCm"]])
df["SepalWidthCm"] = scaler.transform(df[["SepalWidthCm"]])
scaler.fit(df[["PetalLengthCm"]])
df["PetalLengthCm"] = scaler.transform(df[["PetalLengthCm"]])
df.head()
sns.heatmap(df.corr(), annot=True, cbar=False, cmap="coolwarm")
sns.pairplot(df)
sns.scatterplot(x="PetalLengthCm", y="PetalWidthCm", hue="Species", data=df)
plt.legend(loc="best")
encoder = LabelEncoder()
df["Species"] = encoder.fit_transform(df["Species"])
df.head()
df["Species"].unique()
kmeans = KMeans(n_clusters=3)
kmeans.fit(df)
labels = kmeans.predict(df)
labels
plt.scatter(x=df["PetalLengthCm"], y=df["PetalWidthCm"], c=labels)
print(silhouette_score(df, labels) * 100)
kmeans = KMeans(n_clusters=4)
kmeans.fit(df)
labels = kmeans.predict(df)
plt.scatter(x=df["PetalLengthCm"], y=df["PetalWidthCm"], c=labels)
print(silhouette_score(df, labels) * 100)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import confusion_matrix
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
train_data = pd.read_csv("/kaggle/input/titanic/train.csv")
X_test = pd.read_csv("/kaggle/input/titanic/test.csv")
submission_data = pd.read_csv("/kaggle/input/titanic/gender_submission.csv")
print(train_data.info())
print("_" * 50)
print(X_test.info())
print(train_data.isna().sum())
print("_" * 50)
print(X_test.isna().sum())
# ## Cleaning Train Dataset
train_data["Age"].fillna(train_data["Age"].mean(), inplace=True)
train_data["Cabin"].fillna("N0", inplace=True)
train_data["Deck"] = train_data["Cabin"].str.slice(0, 1)
train_data["Embarked"].fillna("S", inplace=True)
sex_dummy = pd.get_dummies(train_data["Sex"], drop_first=True)
class_dummy = pd.get_dummies(train_data["Pclass"], drop_first=True)
embarked_dummy = pd.get_dummies(train_data["Embarked"], drop_first=True)
deck_dummy = pd.get_dummies(train_data["Deck"], drop_first=True)
train_data.drop(
["Pclass", "Name", "Cabin", "Deck", "Sex", "Ticket", "Embarked"],
axis=1,
inplace=True,
)
train_data = pd.concat(
[train_data, sex_dummy, class_dummy, embarked_dummy, deck_dummy], axis=1
)
# ## Cleaning Test Dataset
X_test["Age"].fillna(X_test["Age"].mean(), inplace=True)
X_test["Cabin"].fillna("N0", inplace=True)
X_test["Deck"] = X_test["Cabin"].str.slice(0, 1)
X_test["Fare"].fillna(X_test["Fare"].mean(), inplace=True)
sex_dummy = pd.get_dummies(X_test["Sex"], drop_first=True)
class_dummy = pd.get_dummies(X_test["Pclass"], drop_first=True)
embarked_dummy = pd.get_dummies(X_test["Embarked"], drop_first=True)
deck_dummy = pd.get_dummies(X_test["Deck"], drop_first=True)
X_test.drop(
["Pclass", "Name", "Cabin", "Deck", "Sex", "Ticket", "Embarked"],
axis=1,
inplace=True,
)
X_test = pd.concat([X_test, sex_dummy, class_dummy, embarked_dummy, deck_dummy], axis=1)
X_test["T"] = 0
print(train_data.columns)
print("_" * 50)
print(X_test.columns)
y = train_data["Survived"]
train_data = train_data.drop("Survived", axis=1)
X_train, X_val, y_train, y_val = train_test_split(
train_data, y, test_size=0.1, random_state=0
)
X_train.info()
classifier = LogisticRegression(random_state=0)
classifier.fit(X_train, y_train)
y_pred = classifier.predict(X_val)
cm = confusion_matrix(y_val, y_pred)
print((cm[0, 0] + cm[1, 1]) / cm.sum())
y_submit = classifier.predict(X_test)
results_df = pd.DataFrame()
results_df["PassengerId"] = X_test["PassengerId"]
results_df["Survived"] = y_submit
results_df.to_csv("Predictions", index=False)
|
# Content
# Import Libraries
# Load data
# Data Preparation
# Missing values imputation
# Feature Engineering
# Modeling
# Build the model
# Evaluation
# Model performance
# Feature importance
# Who gets the best performing model?
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
# Modelling Algorithms
from sklearn.svm import SVC, LinearSVC
from sklearn import linear_model
# Modelling Helpers
from sklearn.preprocessing import Imputer, Normalizer, scale
from sklearn.feature_selection import RFECV
# Visualisation
import matplotlib as mpl
import matplotlib.pyplot as plt
import matplotlib.pylab as pylab
import seaborn as sns
from sklearn.utils import shuffle
# get TMDB Box Office Prediction train & test csv files as a DataFrame
train = pd.read_csv("/kaggle/input/tmdb-box-office-prediction/train.csv")
test = pd.read_csv("/kaggle/input/tmdb-box-office-prediction/test.csv")
# Shuffle data
tr_shuffle = shuffle(train, random_state=43).reset_index(drop=True)
# Select features
selected_features = {"budget", "popularity"}
# Split into training and validation data set
data_train = tr_shuffle[0:2499]
data_validate = tr_shuffle[2500:2999]
# Create input and out for training and validation set
data_tr_x = data_train[selected_features]
data_tr_y = data_train[{"revenue"}]
data_val_x = data_validate[selected_features]
data_val_y = data_validate[{"revenue"}]
def plot_correlation_map(df):
corr = train.corr()
_, ax = plt.subplots(figsize=(23, 22))
cmap = sns.diverging_palette(220, 10, as_cmap=True)
_ = sns.heatmap(
corr,
cmap=cmap,
square=True,
cbar_kws={"shrink": 0.9},
ax=ax,
annot=True,
annot_kws={"fontsize": 12},
)
def plot_distribution(df, var, target, **kwargs):
row = kwargs.get("row", None)
col = kwargs.get("col", None)
facet = sns.FacetGrid(df, hue=target, aspect=4, row=row, col=col)
facet.map(sns.kdeplot, var, shade=True)
facet.set(xlim=(0, df[var].max()))
facet.add_legend()
# **Visualization**
train.corr()
# edit cast Show first main caracter in first movie with ID and name
print(train.cast.shape[0])
# for index in train.cast:
# print(int((index.split('\'id\':'))[1].split(',')[0]))
# print((index.split('\'name\': \''))[1].split('\',')[0])
print(int((train.cast[1].split("'id':"))[1].split(",")[0]))
print((train.cast[1].split("'name': '"))[1].split("',")[0])
np.count_nonzero(train.budget)
train.describe()
data = pd.concat([train["budget"], train["revenue"]], axis=1)
data.plot.scatter(x="budget", y="revenue", xlim=(0, 1e7), ylim=(0, 1e8))
# plot_correlation_map(train)
# **Training**
# Splitting into Test and validation data and feature selection
# Selecting features Budget and Popularity
train_mod = train[{"budget", "popularity"}]
# Selecting the first 2001 indices of the training data for training
train_train = train_mod[0:2000]
# Selecting the rest of the training data for validation
train_val = train_mod[2001:2999]
# Obtain labels
train_mod_y = train[{"revenue"}]
train_train_y = train_mod_y[0:2000]
train_val_y = train_mod_y[2001:2999]
train_val_title = train["original_title"][2001:2999]
# Check for NaN
if train_mod.isnull().values.any():
print("Too bad, Nan found...")
else:
print("All right!!! Data ok!")
# Initialize and train a linear regression (Lasso) model
model1 = linear_model.Lasso(alpha=1.1, normalize=False, tol=1e-1000)
model1.fit(train_train, train_train_y.values.ravel())
# Evaluate on the training data
res1 = model1.predict(train_val)
# Obtain R2 score (ordinary least square)
acc_lasso = model1.score(train_val, train_val_y)
acc_lasso_trained = model1.score(train_train, train_train_y)
print(acc_lasso, acc_lasso_trained)
# Create the table for comparing predictions with labels
evaluation = pd.DataFrame(
{
"Title": train_val_title.values.ravel(),
"Prediction": res1.round(),
"Actual revenue": train_val_y.values.ravel(),
"Relative error": res1 / train_val_y.values.ravel(),
}
)
evaluation
# Initialize and train a ridge regression model
model2 = linear_model.Ridge(alpha=100000.0)
model2.fit(train_train, train_train_y.values.ravel())
# Evaluate on the training data
res2 = model2.predict(train_val)
# Obtain R2 score (ordinary least square)
acc_ridge = model2.score(train_val, train_val_y)
acc_ridge_trained = model2.score(train_train, train_train_y)
# Create the table for comparing predictions with labels
evaluation = pd.DataFrame(
{
"Title": train_val_title.values.ravel(),
"Prediction": res2.round(),
"Actual revenue": train_val_y.values.ravel(),
"Relative error": res2 / train_val_y.values.ravel(),
}
)
evaluation
# Initialize and train a elasticNet model
model3 = linear_model.ElasticNet(random_state=0)
model3.fit(train_train, train_train_y.values.ravel())
# Evaluate on the training data
res3 = model3.predict(train_val)
# Obtain R2 score (ordinary least square)
acc_elasticNet = model3.score(train_val, train_val_y)
acc_elasticNet_trained = model3.score(train_train, train_train_y)
# Create the table for comparing predictions with labels
evaluation = pd.DataFrame(
{
"Title": train_val_title.values.ravel(),
"Prediction": res3.round(),
"Actual revenue": train_val_y.values.ravel(),
"Relative error": res3 / train_val_y.values.ravel(),
}
)
evaluation
models = pd.DataFrame(
{
"Model": ["Ridge regression", "Lasso", "ElasticNet"],
"Score": [acc_ridge, acc_lasso, acc_elasticNet],
"Score for training data": [
acc_ridge_trained,
acc_lasso_trained,
acc_elasticNet_trained,
],
}
)
models.sort_values(by="Score", ascending=False)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
jpunemployment0 = pd.read_csv(
"../input/fdsadczgdsatewedvcxz/jpunyear.csv", names=["year", "unemployment rate"]
)
jpunemployment = jpunemployment0.loc[45:]
# print(jpunemployment)
jpunemployment["unemployment change"] = (
jpunemployment["unemployment rate"] / jpunemployment["unemployment rate"][45]
)
interest = pd.read_csv(
"../input/fdsadczgdsatewedvcxz/interest.csv", names=["year", "Interest Rate"]
)
ANA0 = pd.read_csv(
"../input/newdata1/ANA.csv",
names=["year", "Trade and Retail"],
header=None,
usecols=[0, 19],
)
ANA = ANA0.loc[7:]
ANA = ANA.astype(float)
ANA_result0 = pd.merge(ANA, jpunemployment, how="inner", on=["year"])
ANA_result = pd.merge(ANA_result0, interest, how="inner", on=["year"])
# print(ANA_result)
ANA_result["trade and retail"] = (
ANA_result["Trade and Retail"] / ANA_result["Trade and Retail"][1]
)
# print(ANA_result)
ANA_result[["trade and retail", "unemployment change", "Interest Rate"]].plot(
figsize=(12, 8)
)
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
jpal0 = pd.read_csv("../input/newdata1/JapanAirlines.csv")
jpal = jpal0.loc[1:]
interest0 = pd.read_csv(
"../input/fdsadczgdsatewedvcxz/interest.csv", names=["year", "Interest Rate"]
)
interest = interest0.loc[0:6]
jpunemployment0 = pd.read_csv(
"../input/fdsadczgdsatewedvcxz/jpunyear.csv", names=["year", "unemployment rate"]
)
jpunemployment = jpunemployment0.loc[45:]
jpunemployment["unemployment change"] = (
jpunemployment["unemployment rate"] / jpunemployment["unemployment rate"][45]
)
jpal_result0 = pd.merge(jpal, jpunemployment, how="inner", on=["year"])
jpal_result = pd.merge(jpal_result0, interest, how="inner", on=["year"])
jpal_result["aircraft"] = jpal_result["Aircraft"] / jpal_result["Aircraft"][1]
print(jpal_result)
jpal_result[["aircraft", "unemployment change", "Interest Rate"]].plot(figsize=(12, 8))
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
Honda = pd.read_csv("../input/newdata1/Honda.csv")
print(Honda)
interest0 = pd.read_csv(
"../input/fdsadczgdsatewedvcxz/interest.csv", names=["year", "Interest Rate"]
)
interest = interest0.loc[0:6]
jpunemployment0 = pd.read_csv(
"../input/fdsadczgdsatewedvcxz/jpunyear.csv", names=["year", "unemployment rate"]
)
jpunemployment = jpunemployment0.loc[45:]
jpunemployment["unemployment change"] = (
jpunemployment["unemployment rate"] / jpunemployment["unemployment rate"][45]
)
Honda_result0 = pd.merge(Honda, jpunemployment, how="inner", on=["year"])
Honda_result = pd.merge(Honda_result0, interest, how="inner", on=["year"])
Honda_result["Automobile Production Data"] = (
Honda_result["Automobile Production Data"]
/ Honda_result["Automobile Production Data"][1]
)
print(Honda_result)
Honda_result[
["Automobile Production Data", "unemployment change", "Interest Rate"]
].plot(figsize=(12, 8))
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
Subaru0 = pd.read_csv("../input/newdata1/subaru production.csv")
print(Subaru0)
Subaru = Subaru0.astype(float)
interest0 = pd.read_csv(
"../input/fdsadczgdsatewedvcxz/interest.csv", names=["year", "Interest Rate"]
)
interest = interest0.loc[0:6]
jpunemployment0 = pd.read_csv(
"../input/fdsadczgdsatewedvcxz/jpunyear.csv", names=["year", "unemployment rate"]
)
jpunemployment = jpunemployment0.loc[45:]
jpunemployment["unemployment change"] = (
jpunemployment["unemployment rate"] / jpunemployment["unemployment rate"][45]
)
Subaru_result0 = pd.merge(Subaru, jpunemployment, how="inner", on=["year"])
Subaru_result = pd.merge(Subaru_result0, interest, how="inner", on=["year"])
Subaru_result["Production"] = (
Subaru_result["Production"] / Subaru_result["Production"][1]
)
print(Subaru_result)
Subaru_result[["Production", "unemployment change", "Interest Rate"]].plot(
figsize=(12, 8)
)
|
import numpy as np, pandas as pd, matplotlib.pyplot as plt
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# importing the datasets and merging them into one
test = pd.read_csv("/kaggle/input/loan-prediciton-classification/test_AbJTz2l.csv")
train = pd.read_csv("/kaggle/input/loan-prediciton-classification/train_v9rqX0R.csv")
data = pd.concat([train, test], axis=0, keys=["train", "test"])
data
print(data.info()) # identifying the columns and data types
# understanding the basic statistical values of numerical features to form questions
base_stat = data.describe()
base_stat
# Based on min, max, std and the mean, it seems like there **could be** outliers in 'Item_Outlet_Sales' and 'Item_Visibility' . Moreover, 'Item_Outlet_Sales' seems to have a significant number of NULL values followed by 'Item_Weight'.
print(
"The percentage of null values in ",
"%.2f" % (((data["Item_Outlet_Sales"].isnull().sum()) / 14204) * 100),
"%",
)
print(
"The count of null values in Item_Outlet_Sales is",
data["Item_Outlet_Sales"].isnull().sum(),
)
# **Features present in dataset (from the above dataframe info):**
# **1) Categorical :** 'Item_Identifier' , 'Item_Fat_Content' , 'Item_Type' , 'Outlet_Size' , 'Outlet_Location_Type' , 'Outlet_Type'
# **2) Numerical :** Item_Weight' , 'Item_Visibility' , 'Item_MRP' , 'Outlet_Establishment_Year' , 'Item_Outlet_Sales'
# checking just in case for any duplicates in the combined data
print(data.shape)
data.duplicated().shape
# based on previous output, there are no duplicates in the data
data_cat = data.loc[
:,
[
"Item_Identifier",
"Item_Fat_Content",
"Item_Type",
"Outlet_Size",
"Outlet_Location_Type",
"Outlet_Type",
],
]
data_num = data.loc[
:,
[
"Item_Weight",
"Item_Visibility",
"Item_MRP",
"Outlet_Establishment_Year",
"Item_Outlet_Sales",
],
]
data_num
data.describe()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import rebound
import numpy as np
sim = rebound.Simulation()
sim.add(m=0.09) # Star
sim.add(m=1.3771 * 3.00273e-6, e=0.00622, a=0.01154, inc=0, M=0.2)
sim.add(m=1.3105 * 3.00273e-6, e=0.00654, a=0.01580, inc=0, M=4)
sim.add(m=0.3885 * 3.00273e-6, e=0.00837, a=0.02227, inc=0, M=2.6)
sim.add(m=0.693 * 3.00273e-6, e=0.00510, a=0.02925, inc=0, M=1.6)
sim.add(m=1.0411 * 3.00273e-6, e=0.01007, a=0.03849, inc=0, M=3)
sim.add(m=1.3238 * 3.00273e-6, e=0.00208, a=0.04683, inc=0, M=0.4)
sim.add(m=0.3261 * 3.00273e-6, e=0.00567, a=0.06189, inc=0, M=5.6)
op = rebound.OrbitPlot(sim, xlim=(-0.1, 0.1), ylim=(-0.1, 0.1))
def simulation(par):
a, e = par # unpack parameters
sim = rebound.Simulation()
sim.integrator = "whfast"
sim.ri_whfast.safe_mode = 0
sim.dt = 0.00013
sim.add(m=0.0898) # Star
sim.add(m=1.3771 * 3.00273e-6, e=e, a=a, inc=0, M=0.2)
sim.add(m=1.3105 * 3.00273e-6, e=0.00654, a=0.01580, inc=0, M=4)
sim.add(m=0.3885 * 3.00273e-6, e=0.00837, a=0.02227, inc=0, M=2.6)
sim.add(m=0.693 * 3.00273e-6, e=0.00510, a=0.02925, inc=0, M=1.6)
sim.add(m=1.0411 * 3.00273e-6, e=0.01007, a=0.03849, inc=0, M=3)
sim.add(m=1.3238 * 3.00273e-6, e=0.00208, a=0.04683, inc=0, M=0.4)
sim.add(m=0.3261 * 3.00273e-6, e=0.00567, a=0.06189, inc=0, M=5.6)
sim.move_to_com()
sim.init_megno()
sim.exit_max_distance = 0.0
try:
sim.integrate(
500 * 2.0 * np.pi, exact_finish_time=0
) # integrate for 500 years, integrating to the nearest
# timestep for each output to keep the timestep constant and preserve WHFast's symplectic nature
megno = sim.calculate_megno()
lyap = sim.calculate_lyapunov()
print(a, e, megno, lyap)
return megno
except rebound.Escape:
return 10.0 # At least one particle got ejected, returning large MEGNO.
simulation((0.01154, 0.00622))
Ngrid = 5
par_a = [0.01152, 0.01153, 0.01154, 0.01155, 0.01156]
par_e = [0, 0.00318, 0.00622, 0.00926, 0.0123]
parameters = []
for e in par_e:
for a in par_a:
parameters.append((a, e))
# print(parameters)
from rebound.interruptible_pool import InterruptiblePool
pool = InterruptiblePool()
results = pool.map(simulation, parameters)
results2d = np.array(results).reshape(Ngrid, Ngrid)
import matplotlib.pyplot as plt
fig = plt.figure(figsize=(7, 5))
ax = plt.subplot(111)
extent = [min(par_a), max(par_a), min(par_e), max(par_e)]
ax.set_xlim(extent[0], extent[1])
ax.set_xlabel("semi-major axis $a$")
# ax.set_ylim(extent[2],extent[3])
ax.set_ylabel("eccentricity $e$")
im = ax.imshow(
results2d,
interpolation="none",
cmap="RdYlGn_r",
origin="lower",
aspect="auto",
extent=extent,
) # , vmin=1.9, vmax=4
cb = plt.colorbar(im, ax=ax)
cb.set_label("MEGNO $\\langle Y \\rangle$")
|
# Here we will classify wines from the **ScikitLearn wines database**.
# The method used will be **Naive Bayes**, the type (Bernoulli, Multinomial, Gaussian) will be decided based on the data after exploration. We will assume we don't know which feature affects the most the prodiction and don't want to search
import numpy as np # linear algebra
# for visualization
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.datasets import load_wine
wines = load_wine()
print(wines["DESCR"])
print("-" * 80)
print("keys: ", ", ".join(wines.keys()))
# important keys:
# - data: for the features
# - target: for... the target
wines["data"][wines["target"] == 1][:5]
# see the distribution of each feature
nrows, nfeat = wines["data"].shape
fig, axes = plt.subplots((nfeat + 4) // 4, 4, figsize=(32, 16))
fig.suptitle("progression of values")
for i in range(nfeat):
ax = axes[i // 4, i % 4]
sns.histplot(
x=wines["data"][:, i], kde=True, hue=wines["target"], element="step", ax=ax
)
ax.set_title(wines["feature_names"][i])
plt.show()
# preparing the data
from sklearn.naive_bayes import BernoulliNB, MultinomialNB, GaussianNB
from sklearn.model_selection import train_test_split, cross_val_score
X_train, X_test, y_train, y_test = train_test_split(
wines["data"], wines["target"], test_size=0.2, random_state=0
)
# selecting the best naive bayes
best = (None, 0)
models = [BernoulliNB(), MultinomialNB(), GaussianNB()]
for model in models:
print(f"evaluating {model}...", end="")
score = cross_val_score(model, X=X_train, y=y_train).sum()
print(f"(score: {round(score, 2)}; state: ", end="")
if score <= best[1]:
print("REJECTED)")
else:
best = (model, score)
print("CHOSEN)")
best
# training the model and checking its accuracy
from sklearn.metrics import confusion_matrix
model = best[0]
model.fit(X_train, y_train)
y_predict = model.predict(X_test)
confmat = confusion_matrix(y_test, y_predict)
for i in np.unique(y_test):
print(
f"there are {X_test[y_test == i].shape[0]} of class `{wines['target_names'][i]}`."
)
sns.heatmap(confmat, annot=True)
plt.show()
# the **Gaussian Naive Bayes** seems to work fine.
# Gaussian NB is used on data set where has a **normal distribution** (bell-shaped curves on the plot), _and this is what we can notice on the 13 plots above for each type_
# playground
model.predict(
[
[
1.237e01,
9.400e-01,
1.360e00,
1.060e01,
8.800e01,
1.980e00,
5.700e-01,
2.800e-01,
4.200e-01,
1.950e00,
1.050e00,
1.820e00,
5.200e02,
],
[
1.233e01,
1.100e00,
2.280e00,
1.600e01,
1.010e02,
2.050e00,
1.090e00,
6.300e-01,
4.100e-01,
3.270e00,
1.250e00,
1.670e00,
6.800e02,
],
[
1.264e01,
1.360e00,
2.020e00,
1.680e01,
1.000e02,
2.020e00,
1.410e00,
5.300e-01,
6.200e-01,
5.750e00,
9.800e-01,
1.590e00,
4.500e02,
],
[
1.367e01,
1.250e00,
1.920e00,
1.800e01,
9.400e01,
2.100e00,
1.790e00,
3.200e-01,
7.300e-01,
3.800e00,
1.230e00,
2.460e00,
6.300e02,
],
[
1.237e01,
1.130e00,
2.160e00,
1.900e01,
8.700e01,
3.500e00,
3.100e00,
1.900e-01,
1.870e00,
4.450e00,
1.220e00,
2.870e00,
4.200e02,
],
]
)
|
# # Python Booleans
print(200 > 8) # boolean values
print(200 < 8)
print(200 == 8)
# # if Conditional
x = 15
y = 5
if y > x:
print("y is greater than x")
else:
print("y is not greater than x")
x = 15
y = 5
if y < x:
print("x is greater than y")
print(bool("Bonjour")) # Evaluating Values and Variables
print(bool(41))
x = "Bonjour"
y = 41
print(bool(x))
print(bool(y))
# # Most Values Are True
bool("HIJ")
bool(567)
bool(["Chanel", "Gucci", "Prada"])
bool(False) # Some Incorrect Values
bool(None)
bool(0)
# # Functions Can Return a Boolean
def myFunc():
return True
if myFunc():
print("Hi!")
else:
print("Bye!")
g = 50 # isinstance function
print(isinstance(g, int))
y = "Good morning!"
print(isinstance(y, str))
# # Python Arithmetic Operators
x = 8
y = 2
print(x + y)
print(x - y)
print(x * y)
print(x / y)
print(x % y)
print(x // y)
print(x**y)
x = 7
x
x = 6
x += 2
print(x)
x = 6
x -= 2
print(x)
x = 6
x *= 2
print(x)
x = 6
x /= 2
print(x)
x = 6
x %= 2
print(x)
x = 6
x **= 2
print(x)
x = 6
x //= 2
print(x)
# # Python Comparison Operators
x = 6
y = 2
print(x == y)
print(x != y)
print(x < y)
print(x > y)
print(x <= y)
print(x >= y)
# # Python Logical Operators
x = 7
print(x > 5 and x < 12)
x = 7
print(x > 5 or x < 12)
x = 7
print(not (x > 5 and x < 12))
# # Python Identity Operators
h = ["pen", "pencil"]
i = ["pen", "pencil"]
j = h
print(h is j)
print(h is i)
print(h == i)
h = ["pen", "pencil"]
i = ["pen", "pencil"]
j = h
print(h is not j)
print(h is not i)
print(h != i)
|
# ## All Space Missions from 1957 | Data Analysis and Visualization
# Group Members:
# ➡️ Alex Tamboli (202011071)
# ➡️ Ashish Kumar Gupta (202011013)
# ➡️ Chinmay Bhalodia (202011016)
# ➡️ Nishesh Jain (202011050)
# ➡️ Prashant Kumar (202011058)
# ## Importing necessary libraries
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import seaborn as sns
import warnings
import plotly.express as px
import plotly.graph_objects as go
from statsmodels.tsa.arima.model import ARIMA
from statsmodels.tsa.seasonal import seasonal_decompose
from datetime import datetime, timedelta
from collections import OrderedDict
from plotly.offline import init_notebook_mode
init_notebook_mode(connected=True)
warnings.filterwarnings("ignore")
plt.style.use("seaborn-dark")
# ## Analysis of dataset
# Let's know what the data is and how the data is.
# loading the dataset
df = pd.read_csv("/kaggle/input/all-space-missions-from-1957/Space_Corrected.csv")
df.head()
# `Unnamed: 0.1` and `Unnamed: 0` are of no use. We will drop them.
df = df.drop(["Unnamed: 0", "Unnamed: 0.1"], axis=1)
df.head()
# ## Visualization #1: Everything related to the Companies.
# ### 1) Number of Launches by Company
# First, we find the total number of unique companies.
number_of_companies = df["Company Name"].unique()
len(number_of_companies)
# Out of total 56 companies, we will see number of launches by top 10 companies.
company_count = df["Company Name"].value_counts()
top_companies = company_count[:10]
plt.figure(figsize=(20, 4))
plt.scatter(
x=top_companies.index,
y=[1] * len(top_companies),
s=top_companies.values * 10,
c="blue",
alpha=0.5,
)
for i, count in enumerate(top_companies.values):
plt.text(
top_companies.index[i],
1,
str(count),
ha="center",
va="center",
fontsize=12,
color="white",
)
plt.xticks(fontsize=12)
plt.xlim(-0.75)
plt.yticks([])
plt.title("Top 10 Companies by number of Launches.", fontsize=15)
plt.show()
# ### 2) Number of Launches by Company per Year
# We need to modify the dataframe to extract the year of launch from the date.
df["Datum"] = pd.to_datetime(df["Datum"], utc=True)
df["Year"] = df["Datum"].dt.year
df.head()
# Plotting the top 10 companies by number of launches per year.
# ## Visualization #2: USA vs India.
countries_dict = {
"New Mexico": "USA",
"Pacific Missile Range Facility": "USA",
"Gran Canaria": "USA",
}
df["Country"] = df["Location"].str.split(", ").str[-1].replace(countries_dict)
compare = df[(df["Country"] == "India") | (df["Country"] == "USA")]
compare.head()
# We will compare the data since first launch by India. The entry of India in space race is determined by following code.
india = compare[compare["Country"] == "India"]
years = india["Year"].unique()
years.sort()
years
# So we will sort the compare dataframe based on timeline `Year>=1979`.
compare = compare[compare["Year"] >= 1979]
# ### Total number of launches by Country.
# Create data
ds = compare["Country"].value_counts().reset_index()
ds.columns = ["Country", "count"]
# Define colors for the pie chart
colors = px.colors.qualitative.Dark24
# Define font style for title
title_font = dict(size=20, color="#444444", family="Arial")
# Create pie chart
fig = px.pie(
ds,
names="Country",
values="count",
title="Number of Launches by Country",
hole=0.5, # Change hole size
color_discrete_sequence=colors, # Assign custom colors
labels={"count": "Number of Launches"}, # Rename labels
width=700,
height=500,
)
# Modify legend properties
fig.update_traces(textposition="inside", textinfo="percent+label")
# Modify title font style
fig.update_layout(title_font=title_font)
fig.show()
# ### Launches Year by Year
# Group data by year and country and count the number of launches
ds = compare.groupby(["Year", "Country"])["Status Mission"].count().reset_index()
ds.columns = ["Year", "Country", "Launches"]
total = ds
# Set custom color palette
colors = ["rgb(53, 83, 255)", "rgb(255, 128, 0)"]
# Create the plot
fig = px.line(
ds,
x="Year",
y="Launches",
color="Country",
title="USA vs India: Launches Year by Year",
color_discrete_sequence=colors, # Set custom color palette
labels={
"Year": "Year",
"Launches": "Number of Launches",
"Country": "Country",
}, # Rename labels
height=500,
width=800,
)
# Update x-axis labels
fig.update_xaxes(tickfont=dict(size=10))
# Update legend properties
fig.update_layout(
legend=dict(
title=None,
orientation="h",
yanchor="top",
y=1.1,
xanchor="left",
x=0.75,
font=dict(size=12),
)
)
# Show the plot
fig.show()
# ### Percentage Failures Year by Year
# Filter data for failures only
ds = compare[compare["Status Mission"] == "Failure"]
ds = ds.groupby(["Year", "Country"])["Status Mission"].count().reset_index()
ds.columns = ["Year", "Country", "Failures"]
for year in ds["Year"]:
ds["Fail Percent"] = ds["Failures"] / total["Launches"] * 100
mean = ds.groupby("Country")["Fail Percent"].mean()
# Create data
mean_df = pd.DataFrame(mean).reset_index()
mean_df.columns = ["Country", "mean"]
# Define colors for the pie chart
colors = px.colors.qualitative.Dark24
# Define font style for title
title_font = dict(size=20, color="#444444", family="Arial")
# Create pie chart
fig = px.pie(
mean_df,
names="Country",
values="mean",
title="Mean Failure Percentage by Country",
hole=0.5, # Change hole size
color_discrete_sequence=colors, # Assign custom colors
labels={"Country": "Country", "mean": "Mean Failure Percentage"}, # Rename labels
width=700,
height=500,
)
# Modify legend properties
fig.update_traces(textposition="inside", textinfo="percent+label")
# Modify title font style
fig.update_layout(title_font=title_font)
fig.show()
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.linear_model import Perceptron
from sklearn.metrics import accuracy_score
from sklearn.tree import DecisionTreeClassifier
from sklearn.svm import SVC
from sklearn.linear_model import LinearRegression
from sklearn.linear_model import LogisticRegression
from keras.models import Sequential
from keras.layers import Dense
from sklearn.neural_network import MLPClassifier
from sklearn.metrics import r2_score
from sklearn.neural_network import MLPRegressor
dataset = pd.DataFrame({"Age": [67, 69, 73, 74, 76, 77, 79, 81, 83, 85]})
target = np.array([2.3, 1.5, 3, 4.5, 5, 5.5, 6, 7, 8.5, 9])
dataset
model = LinearRegression()
model.fit(dataset, target)
y_pred = model.predict(dataset)
accuracy = model.score(dataset, target)
plt.scatter(dataset, target)
plt.plot(dataset, y_pred, color="red")
plt.xlabel("Age")
plt.ylabel("Target")
plt.title("Linear Regression")
plt.show()
bins = [0, 3, 6, 9]
labels = ["low", "medium", "high"]
target_cat = pd.cut(target, bins=bins, labels=labels)
perceptron = Perceptron(max_iter=1000, tol=1e-3, random_state=42)
perceptron.fit(dataset, target_cat)
y_pred = perceptron.predict(dataset)
accuracy = accuracy_score(target_cat, y_pred)
print("Accuracy:", accuracy)
plt.scatter(dataset, target_cat)
plt.scatter(dataset, perceptron.predict(dataset), color="red")
plt.xlabel("Age")
plt.ylabel("Avg. hospitalizations")
plt.show()
|
import cv2
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
from matplotlib import pyplot as plt
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
data = []
import os
for dirname, _, filenames in os.walk(
"/kaggle/input/diabetic-retinopathy-resized/resized_train_cropped"
):
for filename in filenames:
data.append((os.path.join(dirname, filename)))
data[2]
# Any results you write to the current directory are saved as output.
num = len(data)
img = []
for i in range(20000, 20030):
imgloc = data[i]
img1 = cv2.imread(imgloc, 1)
img1 = cv2.resize(img1, (350, 350))
img.append(cv2.cvtColor(img1, cv2.COLOR_BGR2RGB))
f, arr = plt.subplots(6, 5, figsize=(16, 16))
arr[0, 0].imshow(img[0])
arr[0, 1].imshow(img[1])
arr[0, 2].imshow(img[2])
arr[0, 3].imshow(img[3])
arr[0, 4].imshow(img[4])
arr[1, 0].imshow(img[5])
arr[1, 1].imshow(img[6])
arr[1, 2].imshow(img[7])
arr[1, 3].imshow(img[8])
arr[1, 4].imshow(img[9])
arr[2, 0].imshow(img[10])
arr[2, 1].imshow(img[11])
arr[2, 2].imshow(img[12])
arr[2, 3].imshow(img[13])
arr[2, 4].imshow(img[14])
arr[3, 0].imshow(img[15])
arr[3, 1].imshow(img[16])
arr[3, 2].imshow(img[17])
arr[3, 3].imshow(img[18])
arr[3, 4].imshow(img[19])
arr[4, 0].imshow(img[20])
arr[4, 1].imshow(img[21])
arr[4, 2].imshow(img[22])
arr[4, 3].imshow(img[23])
arr[4, 4].imshow(img[24])
arr[5, 0].imshow(img[25])
arr[5, 1].imshow(img[26])
arr[5, 2].imshow(img[27])
arr[5, 3].imshow(img[28])
arr[5, 4].imshow(img[29])
gre = []
for i in range(0, 30):
img2 = np.array(img[i])
gcImg = img2[:, :, 1]
img2 = gcImg
img2 = cv2.cvtColor(img2, cv2.COLOR_BGR2RGB)
gre.append(img2)
f, arr = plt.subplots(6, 5, figsize=(16, 16))
arr[0, 0].imshow(gre[0])
arr[0, 1].imshow(gre[1])
arr[0, 2].imshow(gre[2])
arr[0, 3].imshow(gre[3])
arr[0, 4].imshow(gre[4])
arr[1, 0].imshow(gre[5])
arr[1, 1].imshow(gre[6])
arr[1, 2].imshow(gre[7])
arr[1, 3].imshow(gre[8])
arr[1, 4].imshow(gre[9])
arr[2, 0].imshow(gre[10])
arr[2, 1].imshow(gre[11])
arr[2, 2].imshow(gre[12])
arr[2, 3].imshow(gre[13])
arr[2, 4].imshow(gre[14])
arr[3, 0].imshow(gre[15])
arr[3, 1].imshow(gre[16])
arr[3, 2].imshow(gre[17])
arr[3, 3].imshow(gre[18])
arr[3, 4].imshow(gre[19])
arr[4, 0].imshow(gre[20])
arr[4, 1].imshow(gre[21])
arr[4, 2].imshow(gre[22])
arr[4, 3].imshow(gre[23])
arr[4, 4].imshow(gre[24])
arr[5, 0].imshow(gre[25])
arr[5, 1].imshow(gre[26])
arr[5, 2].imshow(gre[27])
arr[5, 3].imshow(gre[28])
arr[5, 4].imshow(gre[29])
cla = []
clahe = cv2.createCLAHE(clipLimit=2.0, tileGridSize=(8, 8))
for i in range(0, 30):
img3 = cv2.cvtColor(gre[i], cv2.COLOR_BGR2GRAY)
cl1 = clahe.apply(img3)
cl1 = cv2.cvtColor(cl1, cv2.COLOR_BGR2RGB)
cla.append(cl1)
f, arr = plt.subplots(6, 5, figsize=(16, 16))
arr[0, 0].imshow(cla[0])
arr[0, 1].imshow(cla[1])
arr[0, 2].imshow(cla[2])
arr[0, 3].imshow(cla[3])
arr[0, 4].imshow(cla[4])
arr[1, 0].imshow(cla[5])
arr[1, 1].imshow(cla[6])
arr[1, 2].imshow(cla[7])
arr[1, 3].imshow(cla[8])
arr[1, 4].imshow(cla[9])
arr[2, 0].imshow(cla[10])
arr[2, 1].imshow(cla[11])
arr[2, 2].imshow(cla[12])
arr[2, 3].imshow(cla[13])
arr[2, 4].imshow(cla[14])
arr[3, 0].imshow(cla[15])
arr[3, 1].imshow(cla[16])
arr[3, 2].imshow(cla[17])
arr[3, 3].imshow(cla[18])
arr[3, 4].imshow(cla[19])
arr[4, 0].imshow(cla[20])
arr[4, 1].imshow(cla[21])
arr[4, 2].imshow(cla[22])
arr[4, 3].imshow(cla[23])
arr[4, 4].imshow(cla[24])
arr[5, 0].imshow(cla[25])
arr[5, 1].imshow(cla[26])
arr[5, 2].imshow(cla[27])
arr[5, 3].imshow(cla[28])
arr[5, 4].imshow(cla[29])
mor = []
strEl = cv2.getStructuringElement(cv2.MORPH_ELLIPSE, (6, 6))
for i in range(0, 30):
dilateImg = cv2.dilate(cla[i], strEl)
curImg = dilateImg
curImg = cv2.cvtColor(curImg, cv2.COLOR_BGR2RGB)
mor.append(curImg)
f, arr = plt.subplots(6, 5, figsize=(16, 16))
arr[0, 0].imshow(mor[0])
arr[0, 1].imshow(mor[1])
arr[0, 2].imshow(mor[2])
arr[0, 3].imshow(mor[3])
arr[0, 4].imshow(mor[4])
arr[1, 0].imshow(mor[5])
arr[1, 1].imshow(mor[6])
arr[1, 2].imshow(mor[7])
arr[1, 3].imshow(mor[8])
arr[1, 4].imshow(mor[9])
arr[2, 0].imshow(mor[10])
arr[2, 1].imshow(mor[11])
arr[2, 2].imshow(mor[12])
arr[2, 3].imshow(mor[13])
arr[2, 4].imshow(mor[14])
arr[3, 0].imshow(mor[15])
arr[3, 1].imshow(mor[16])
arr[3, 2].imshow(mor[17])
arr[3, 3].imshow(mor[18])
arr[3, 4].imshow(mor[19])
arr[4, 0].imshow(mor[20])
arr[4, 1].imshow(mor[21])
arr[4, 2].imshow(mor[22])
arr[4, 3].imshow(mor[23])
arr[4, 4].imshow(mor[24])
arr[5, 0].imshow(mor[25])
arr[5, 1].imshow(mor[26])
arr[5, 2].imshow(mor[27])
arr[5, 3].imshow(mor[28])
arr[5, 4].imshow(mor[29])
med = []
for i in range(0, 30):
medianImg = cv2.medianBlur(mor[i], 5)
medianImg = cv2.cvtColor(medianImg, cv2.COLOR_BGR2RGB)
med.append(medianImg)
f, arr = plt.subplots(6, 5, figsize=(16, 16))
arr[0, 0].imshow(med[0])
arr[0, 1].imshow(med[1])
arr[0, 2].imshow(med[2])
arr[0, 3].imshow(med[3])
arr[0, 4].imshow(med[4])
arr[1, 0].imshow(med[5])
arr[1, 1].imshow(med[6])
arr[1, 2].imshow(med[7])
arr[1, 3].imshow(med[8])
arr[1, 4].imshow(med[9])
arr[2, 0].imshow(med[10])
arr[2, 1].imshow(med[11])
arr[2, 2].imshow(med[12])
arr[2, 3].imshow(med[13])
arr[2, 4].imshow(med[14])
arr[3, 0].imshow(med[15])
arr[3, 1].imshow(med[16])
arr[3, 2].imshow(med[17])
arr[3, 3].imshow(med[18])
arr[3, 4].imshow(med[19])
arr[4, 0].imshow(med[20])
arr[4, 1].imshow(med[21])
arr[4, 2].imshow(med[22])
arr[4, 3].imshow(med[23])
arr[4, 4].imshow(med[24])
arr[5, 0].imshow(med[25])
arr[5, 1].imshow(med[26])
arr[5, 2].imshow(med[27])
arr[5, 3].imshow(med[28])
arr[5, 4].imshow(med[29])
exu = []
for i in range(0, 30):
retValue, threshImg = cv2.threshold(med[i], 215, 255, cv2.THRESH_BINARY)
threshImg = cv2.cvtColor(threshImg, cv2.COLOR_BGR2RGB)
exu.append(threshImg)
f, arr = plt.subplots(6, 5, figsize=(16, 16))
arr[0, 0].imshow(exu[0])
arr[0, 1].imshow(exu[1])
arr[0, 2].imshow(exu[2])
arr[0, 3].imshow(exu[3])
arr[0, 4].imshow(exu[4])
arr[1, 0].imshow(exu[5])
arr[1, 1].imshow(exu[6])
arr[1, 2].imshow(exu[7])
arr[1, 3].imshow(exu[8])
arr[1, 4].imshow(exu[9])
arr[2, 0].imshow(exu[10])
arr[2, 1].imshow(exu[11])
arr[2, 2].imshow(exu[12])
arr[2, 3].imshow(exu[13])
arr[2, 4].imshow(exu[14])
arr[3, 0].imshow(exu[15])
arr[3, 1].imshow(exu[16])
arr[3, 2].imshow(exu[17])
arr[3, 3].imshow(exu[18])
arr[3, 4].imshow(exu[19])
arr[4, 0].imshow(exu[20])
arr[4, 1].imshow(exu[21])
arr[4, 2].imshow(exu[22])
arr[4, 3].imshow(exu[23])
arr[4, 4].imshow(exu[24])
arr[5, 0].imshow(exu[25])
arr[5, 1].imshow(exu[26])
arr[5, 2].imshow(exu[27])
arr[5, 3].imshow(exu[28])
arr[5, 4].imshow(exu[29])
|
# ## Bank Telemarketing Campaign Case Study.
# In this case study you’ll be learning Exploratory Data Analytics with the help of a case study on "Bank marketing campaign". This will enable you to understand why EDA is a most important step in the process of Machine Learning.
# #### Problem Statement:
#
# The bank provides financial services/products such as savings accounts, current accounts, debit cards, etc. to its customers. In order to increase its overall revenue, the bank conducts various marketing campaigns for its financial products such as credit cards, term deposits, loans, etc. These campaigns are intended for the bank’s existing customers. However, the marketing campaigns need to be cost-efficient so that the bank not only increases their overall revenues but also the total profit. You need to apply your knowledge of EDA on the given dataset to analyse the patterns and provide inferences/solutions for the future marketing campaign.
# The bank conducted a telemarketing campaign for one of its financial products ‘Term Deposits’ to help foster long-term relationships with existing customers. The dataset contains information about all the customers who were contacted during a particular year to open term deposit accounts.
# **What is the term Deposit?**
# Term deposits also called fixed deposits, are the cash investments made for a specific time period ranging from 1 month to 5 years for predetermined fixed interest rates. The fixed interest rates offered for term deposits are higher than the regular interest rates for savings accounts. The customers receive the total amount (investment plus the interest) at the end of the maturity period. Also, the money can only be withdrawn at the end of the maturity period. Withdrawing money before that will result in an added penalty associated, and the customer will not receive any interest returns.
# Your target is to do end to end EDA on this bank telemarketing campaign data set to infer knowledge that where bank has to put more effort to improve it's positive response rate.
# #### Importing the libraries.
# import the warnings.
import warnings
warnings.filterwarnings("ignore")
# import the python libraries.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
# ## Session- 2, Data Cleaning
# ### Segment- 2, Data Types
# There are multiple types of data types available in the data set. some of them are numerical type and some of categorical type. You are required to get the idea about the data types after reading the data frame.
# Following are the some of the types of variables:
# - **Numeric data type**: banking dataset: salary, balance, duration and age.
# - **Categorical data type**: banking dataset: education, job, marital, poutcome and month etc.
# - **Ordinal data type**: banking dataset: Age group.
# - **Time and date type**
# - **Coordinates type of data**: latitude and longitude type.
# #### Read in the Data set.
# read the data set of "bank telemarketing campaign" in inp0.
inp0 = pd.read_csv(
"/Users/sakshimunde/Downloads/Bank Dataset/bank_marketing_updated_v1.csv"
)
inp0.head()
# Print the head of the data frame.
inp0.head()
# ### Segment- 3, Fixing the Rows and Columns
# Checklist for fixing rows:
# - **Delete summary rows**: Total and Subtotal rows
# - **Delete incorrect rows**: Header row and footer row
# - **Delete extra rows**: Column number, indicators, Blank rows, Page No.
# Checklist for fixing columns:
# - **Merge columns for creating unique identifiers**, if needed, for example, merge the columns State and City into the column Full address.
# - **Split columns to get more data**: Split the Address column to get State and City columns to analyse each separately.
# - **Add column names**: Add column names if missing.
# - **Rename columns consistently**: Abbreviations, encoded columns.
# - **Delete columns**: Delete unnecessary columns.
# - **Align misaligned columns**: The data set may have shifted columns, which you need to align correctly.
# #### Read the file without unnecessary headers.
# read the file in inp0 without first two rows as it is of no use.
inp0 = pd.read_csv(
"/Users/sakshimunde/Downloads/Bank Dataset/bank_marketing_updated_v1.csv",
skiprows=2,
)
# print the head of the data frame.
inp0.head()
# print the information of variables to check their data types.
inp0.dtypes
# #### Dropping customer id column.
# drop the customer id as it is of no use
inp0.drop("customerid", axis=1, inplace=True)
# look at dataframe after dropping customerid column
inp0
# #### Dividing "jobedu" column into job and education categories.
# Extract job in newly created 'job' column from "jobedu" column.
inp0["job"] = inp0.jobedu.apply(lambda x: x.split(",")[0])
inp0
# Extract education in newly created 'education' column from "jobedu" column.
inp0["education"] = inp0.jobedu.apply(lambda x: x.split(",")[1])
inp0
# drop the "jobedu" column from the dataframe.
inp0.drop("jobedu", axis=1, inplace=True)
inp0
# ### Segment- 4, Impute/Remove missing values
# Take aways from the lecture on missing values:
# - **Set values as missing values**: Identify values that indicate missing data, for example, treat blank strings, "NA", "XX", "999", etc., as missing.
# - **Adding is good, exaggerating is bad**: You should try to get information from reliable external sources as much as possible, but if you can’t, then it is better to retain missing values rather than exaggerating the existing rows/columns.
# - **Delete rows and columns**: Rows can be deleted if the number of missing values is insignificant, as this would not impact the overall analysis results. Columns can be removed if the missing values are quite significant in number.
# - **Fill partial missing values using business judgement**: Such values include missing time zone, century, etc. These values can be identified easily.
# Types of missing values:
# - **MCAR**: It stands for Missing completely at random (the reason behind the missing value is not dependent on any other feature).
# - **MAR**: It stands for Missing at random (the reason behind the missing value may be associated with some other features).
# - **MNAR**: It stands for Missing not at random (there is a specific reason behind the missing value).
# #### handling missing values in age column.
inp0.isnull().sum()
# count the missing values in age column.
inp0.age.isnull().sum()
# pring the shape of dataframe inp0
inp0.shape
# calculate the percentage of missing values in age column.
float(100.0 * 20 / 45211)
# #### 4% values are missng
# Drop the records with age missing.
# drop the records with age missing in inp0 and copy in inp1 dataframe.
# SHIFT+TAB gives information of this rows
inp1 = inp0[~inp0.age.isnull()].copy()
inp1.shape
# #### handling missing values in month column
# count the missing values in month column in inp1.
inp1.month.isnull().sum()
# print the percentage of each month in the data frame inp1.
# normalize is used to get percentage
inp1.month.value_counts(normalize=True)
# find the mode of month in inp1
month_mode = inp1.month.mode()[0]
month_mode
# fill the missing values with mode value of month in inp1.
inp1.month.fillna(month_mode, inplace=True)
inp1.month.value_counts()
# or inp1.month.fillna(inp1.month.mode()[0])
inp1.month.value_counts(normalize=True)
# let's see the null values in the month column.
inp1.month.isnull().sum()
# #### handling missing values in response column
# count the missing values in response column in inp1.
inp1.response.isnull().sum()
# calculate the percentage of missing values in response column.
100.0 * 30 / 45211
# Target variable is better of not imputed.
# - Drop the records with missing values.
# drop the records with response missings in inp1
inp1 = inp1[~inp1.response.isnull()].copy()
# calculate the missing values in each column of data frame: inp1.
inp1.isnull().sum()
# #### handling pdays column.
# describe the pdays column of inp1.
inp1.pdays.describe()
# -1 indicates the missing values.
# Missing value does not always be present as null.
# How to handle it:
# Objective is:
# - you should ignore the missing values in the calculations
# - simply make it missing - replace -1 with NaN.
# - all summary statistics- mean, median etc. we will ignore the missing values of pdays.
# describe the pdays column with considering the -1 values.
inp1.loc[inp1.pdays < 0, "pdays"] = np.NAN
inp1.pdays.describe()
# ### Segment- 5, Handling Outliers
# Major approaches to the treat outliers:
#
# - **Imputation**
# - **Deletion of outliers**
# - **Binning of values**
# - **Cap the outlier**
# #### Age variable
inp1.head()
# describe the age variable in inp1.
inp1.age.describe()
# plot the histogram of age variable
plt.hist(inp1.age)
plt.show()
# plot the boxplot of age variable.
sns.boxplot(inp1.age)
plt.show()
# #### Salary variable
# describe the salary variable of inp1.
inp1.salary.describe()
# plot the boxplot of salary variable.
sns.boxplot(inp1.salary)
# #### Balance variable
# describe the balance variable of inp1.
inp1.balance.describe()
# plot the boxplot of balance variable.
sns.boxplot(inp1.balance)
# plot the boxplot of balance variable after scaling in 8:2.
plt.figure(figsize=[8, 2])
sns.boxplot(inp1.balance)
# print the quantile (0.5, 0.7, 0.9, 0.95 and 0.99) of balance variable
inp1.balance.quantile([0.5, 0.7, 0.9, 0.95, 0.99])
(inp1.balance > 15000).describe()
inp1[inp1.balance > 15000].describe()
inp1
# ### Segment- 6, Standardising values
# Checklist for data standardization exercises:
# - **Standardise units**: Ensure all observations under one variable are expressed in a common and consistent unit, e.g., convert lbs to kg, miles/hr to km/hr, etc.
# - **Scale values if required**: Make sure all the observations under one variable have a common scale.
# - **Standardise precision** for better presentation of data, e.g., change 4.5312341 kg to 4.53 kg.
# - **Remove extra characters** such as common prefixes/suffixes, leading/trailing/multiple spaces, etc. These are irrelevant to analysis.
# - **Standardise case**: String variables may take various casing styles, e.g., UPPERCASE, lowercase, Title Case, Sentence case, etc.
# - **Standardise format**: It is important to standardise the format of other elements such as date, name, etce.g., change 23/10/16 to 2016/10/23, “Modi, Narendra” to “Narendra Modi", etc.
# #### Duration variable
# describe the duration variable of inp1
inp1.duration.describe()
# convert the duration variable into single unit i.e. minutes. and remove the sec or min prefix.
inp1.duration = inp1.duration.apply(
lambda x: float(x.split()[0]) / 60 if x.find("sec") > 0 else float(x.split()[0])
)
# describe the duration variable
inp1.duration.describe()
inp1
# ## Session- 3, Univariate Analysis
# ### Segment- 2, Categorical unordered univariate analysis
# Unordered data do not have the notion of high-low, more-less etc. Example:
# - Type of loan taken by a person = home, personal, auto etc.
# - Organisation of a person = Sales, marketing, HR etc.
# - Job category of persone.
# - Marital status of any one.
# #### Marital status
# calculate the percentage of each marital status category.
inp1.marital.value_counts(normalize=True)
# plot the bar graph of percentage marital status categories
inp1.marital.value_counts(normalize=True).plot.bar()
# #### Job
inp1.job.value_counts()
# calculate the percentage of each job status category.
inp1.job.value_counts(normalize=True)
# plot the bar graph of percentage job categories
inp1.job.value_counts(normalize=True).plot.bar()
# ### Segment- 3, Categorical ordered univariate analysis
# Ordered variables have some kind of ordering. Some examples of bank marketing dataset are:
# - Age group= <30, 30-40, 40-50 and so on.
# - Month = Jan-Feb-Mar etc.
# - Education = primary, secondary and so on.
# #### Education
# calculate the percentage of each education category.
inp1.education.value_counts(normalize=True)
# plot the pie chart of education categories
inp1.education.value_counts(normalize=True).plot.pie()
# #### poutcome
inp1.poutcome.value_counts(normalize=True)
# calculate the percentage of each poutcome category.
inp1[inp1.poutcome != "unknown"].poutcome.value_counts(normalize=True).plot.bar()
# #### Response the target variable
# calculate the percentage of each response category.
inp1.response.value_counts()
inp1.response.value_counts(normalize=True) * 100
# plot the pie chart of response categories
(inp1.response.value_counts(normalize=True) * 100).plot.bar()
inp1.response.value_counts(normalize=True).plot.pie()
# ## Session- 4, Bivariate and Multivariate Analysis
# ### Segment-2, Numeric- numeric analysis
# There are three ways to analyse the numeric- numeric data types simultaneously.
# - **Scatter plot**: describes the pattern that how one variable is varying with other variable.
# - **Correlation matrix**: to describe the linearity of two numeric variables.
# - **Pair plot**: group of scatter plots of all numeric variables in the data frame.
# plot the scatter plot of balance and salary variable in inp1
# dependent variable should be on x axis
# independent variable should be on y axis
# here balance is dependent on salary
plt.scatter(data=inp1, x="salary", y="balance")
# plot the scatter plot of balance and age variable in inp1
plt.scatter(data=inp1, x="age", y="balance")
plt.show()
# plot the pair plot of salary, balance and age in inp1 dataframe.
sns.pairplot(data=inp1, vars=["salary", "balance", "age"])
plt.show()
# #### Correlation heat map
# plot the correlation matrix of salary, balance and age in inp1 dataframe.
inp1[["age", "balance", "salary"]].corr()
sns.heatmap(inp1[["age", "balance", "salary"]].corr(), cmap="Reds")
sns.heatmap(inp1[["age", "salary", "balance"]].corr(), annot=True, cmap="Greens")
# diagonally high relation 1.rest all are equal to 0 so no relation
# ### Segment- 4, Numerical categorical variable
# #### Salary vs response
# groupby the response to find the mean of the salary with response no & yes seperatly.
inp1.groupby("response")["salary"].mean()
# groupby the response to find the median of the salary with response no & yes seperatly.
inp1.groupby("response")["salary"].median()
# plot the box plot of salary for yes & no responses.
sns.boxplot(data=inp1, x="response", y="salary")
# #### Balance vs response
# plot the box plot of balance for yes & no responses.
sns.boxplot(data=inp1, y="balance", x="response")
# groupby the response to find the mean of the balance with response no & yes seperatly.
inp1.groupby("response")["balance"].mean()
# groupby the response to find the median of the balance with response no & yes seperatly.
inp1.groupby("response")["balance"].median()
# ##### 75th percentile
# function to find the 75th percentile.
def p75(n):
return np.quantile(n, 0.75)
# calculate the mean, median and 75th percentile of balance with response
inp1.groupby("response")["balance"].aggregate(["mean", "median", p75])
# people with high salary around 75% balance said yes in higher number and less number of people with high salary said no
#
def p99(x):
return np.quantile(x, 0.99)
inp1.groupby("response")["salary"].aggregate(["mean", "median", p99])
# plot the bar graph of balance's mean an median with response.
inp1.groupby("response")["balance"].aggregate(["mean", "median"]).plot.bar()
# #### Education vs salary
# groupby the education to find the mean of the salary education category.
inp1.groupby("education")["salary"].mean()
# groupby the education to find the median of the salary for each education category.
inp1.groupby("education")["salary"].median()
# #### Job vs salary
# groupby the job to find the mean of the salary for each job category.
inp1.groupby("job")["salary"].mean()
# job with entrepreneur and management have higher salary and students have the lowest salary
# ### Segment- 5, Categorical categorical variable
# inp1['response_flag1']=inp1.response.apply(lambda x : 1 if x=='yes' else 0)
# inp1['response_flag1'].value_counts()
# OR
# create response_flag of numerical data type where response "yes"= 1, "no"= 0
inp1["response_flag"] = np.where(inp1.response == "yes", 1, 0)
inp1["response_flag"].value_counts()
# #### Education vs response rate
# calculate the mean of response_flag with different education categories.
inp1.groupby("education")["response_flag"].mean() * 100
# #### Marital vs response rate
# calculate the mean of response_flag with different marital status categories.
inp1.groupby("marital")["response_flag"].mean()
# plot the bar graph of marital status with average value of response_flag
inp1.groupby("marital")["response_flag"].mean().plot.bar()
inp1.groupby(["marital", "loan"])["response_flag"].mean() * 100
inp1.groupby(["marital", "loan"])["response_flag"].mean().plot.bar()
# #### Loans vs response rate
# plot the bar graph of personal loan status with average value of response_flag
inp1.groupby("loan")["response_flag"].mean() * 100
inp1.groupby("loan")["response_flag"].mean().plot.bar()
# #### Housing loans vs response rate
# plot the bar graph of housing loan status with average value of response_flag
inp1.groupby("housing")["response_flag"].mean()
inp1.groupby("housing")["response_flag"].mean().plot.bar()
# #### Age vs response
# plot the boxplot of age with response_flag
sns.boxplot(data=inp1, x="response", y="age")
# ##### making buckets from age columns
# create the buckets of <30, 30-40, 40-50 50-60 and 60+ from age column.
inp1["age_group"] = pd.cut(
inp1.age, [0, 30, 40, 50, 60, 200], labels=["<30", "30-40", "40-50", "50-60", "60+"]
)
inp1["age_group"]
inp1
inp1["age_group"].value_counts()
inp1["age_group"].value_counts(normalize=True).plot()
inp1.groupby("response_flag")["age"].mean()
inp1.groupby("age_group")["response_flag"].mean()
inp1.groupby("age_group")["response_flag"].mean().plot()
# plot the percentage of each buckets and average values of response_flag in each buckets. plot in subplots.
inp1["age_group"].value_counts(normalize=True).plot()
inp1.groupby("age_group")["response_flag"].mean().plot()
plt.figure(figsize=[10, 5])
plt.subplot(1, 2, 2)
inp1.age_group.value_counts(normalize=True).plot()
plt.subplot(1, 2, 1)
inp1.groupby("age_group")["response_flag"].mean().plot()
plt.figure(figsize=[10, 5])
plt.subplot(1, 2, 1)
inp1.age_group.value_counts(normalize=True).plot.bar()
plt.subplot(1, 2, 2)
inp1.groupby(["age_group"])["response_flag"].mean().plot.bar()
# plot the bar graph of job categories with response_flag mean value.
(inp1.groupby("job")["response_flag"].mean() * 100)
inp1.groupby("job")["response_flag"].mean().plot.bar()
# ### Segment-6, Multivariate analysis
# #### Education vs marital vs response
result = pd.pivot_table(
data=inp1,
index="marital",
columns="education",
values="response_flag",
aggfunc="mean",
)
result
# create heat map of education vs marital vs response_flag
sns.heatmap(result, annot=True, cmap="Greens")
# single tertiary are giving high positive rsponse to deposit and married people all are not much interested to deposit.
# primary eduaction:people are less interested to deposit with primary eduaction
# secondary education: people who are single are interested than married and divorced
# tertiary : all are interested more single people are interested
inp1.response_flag.describe()
# #### mean is 0.117
sns.heatmap(result, annot=True, cmap="RdYlGn", center=0.117)
# #### Job vs marital vs response
res1 = pd.pivot_table(data=inp1, index="job", columns="marital", values="response_flag")
res1
# create the heat map of Job vs marital vs response_flag
sns.heatmap(res1, annot=True, cmap="Greens")
# #### Education vs poutcome vs response
res2 = pd.pivot_table(
data=inp1, index="education", columns="poutcome", values="response_flag"
)
res2
# create the heat map of education vs poutcome vs response_flag.
sns.heatmap(res2, annot=True, cmap="Greens")
# #### lets find avg of responseFlag so that will be our center
inp1[inp1.pdays > 0].response_flag.mean()
sns.heatmap(res2, annot=True, cmap="RdYlGn", center=0.23077)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
df = pd.read_csv("../input/titanic/train.csv")
df = df[["Sex", "Embarked"]]
df.dropna(axis=0, inplace=True)
df.head()
# ## Label Encode
# it is best for categorical data with ordinal value
# with clear order of hierarchy
from sklearn.preprocessing import LabelEncoder
enc = LabelEncoder()
df["Embarked"] = enc.fit_transform(df["Embarked"])
df.head()
# ## Label Binerizer
# it will binerize the values into 1 or 0
# yes/no -> 1/0
# true/false -> 1/0
# from sklearn.preprocessing import LabelBinarizer
# lb = LabelBinarizer()
# lb.fit_transform(['yes', 'no', 'no', 'yes'])
# array([[1],
# [0],
# [0],
# [1]])
# Binarize labels in a one-vs-all fashion
# ## One Hot Encoding
# pd.get_dummies(df['Sex'], prefix='gender', drop_first=True)
dummies = pd.get_dummies(df["Sex"])
df = pd.concat([df, dummies], axis=1)
df.head()
|
import gc
import numpy as np
import pandas as pd
import time
import torch
import torch.nn as nn
import torch.nn.functional as F
import argparse
import pyarrow.parquet as pq
import cv2
import glob
import os
import sys
sys.path.append("/kaggle/input/utilities/")
import SeResNeXt
from torch.utils.data import Dataset, DataLoader
from tqdm import tqdm
def crop_image_only_outside(img, tol=0):
mask = img > tol
m, n = img.shape
mask0, mask1 = mask.any(0), mask.any(1)
col_start, col_end = mask0.argmax(), n - mask0[::-1].argmax()
row_start, row_end = mask1.argmax(), m - mask1[::-1].argmax()
return img[row_start:row_end, col_start:col_end]
DATA_PATH = "/kaggle/input/bengaliai-cv19/"
sample_submission = pd.read_csv("../input/bengaliai-cv19/sample_submission.csv")
num_samples = sample_submission.shape[0] // 3
TARGET_SIZE = 64
BATCH_SIZE = 96
N_WORKERS = 4
HEIGHT = 137
WIDTH = 236
class GraphemeValidationDataset(Dataset):
def __init__(self, num_samples):
self.num_samples = num_samples
self.images = torch.zeros(
num_samples, TARGET_SIZE, TARGET_SIZE, dtype=torch.uint8
)
img_id = 0
for i in tqdm(range(4)):
datafile = DATA_PATH + "/test_image_data_{}.parquet".format(i)
parq = pq.read_pandas(
datafile, columns=[str(x) for x in range(32332)]
).to_pandas()
parq = 255 - parq.iloc[:, :].values.reshape(-1, HEIGHT, WIDTH).astype(
np.uint8
)
for idx, image in enumerate(parq):
image = (image * (255.0 / image.max())).astype(np.uint8)
image = crop_image_only_outside(image, 80)
image = cv2.resize(image, (TARGET_SIZE, TARGET_SIZE))
self.images[img_id, :, :] = torch.from_numpy(image.astype(np.uint8))
img_id = img_id + 1
del parq
def __len__(self):
return self.num_samples
def __getitem__(self, idx):
return self.images[idx].unsqueeze(0)
class Identity(torch.nn.Module):
def __init__(self):
super(Identity, self).__init__()
def forward(self, x):
return x
class MyNet(nn.Module):
def __init__(self):
super(MyNet, self).__init__()
self.backbone = SeResNeXt.se_resnext101()
self.backbone.conv1 = nn.Conv2d(
1, 64, kernel_size=7, stride=2, padding=3, bias=False
)
self.backbone.fc = Identity()
self.fc1 = nn.Linear(2048, 11) # vowel_diacritic
self.fc2 = nn.Linear(2048, 168) # grapheme_root
self.fc3 = nn.Linear(2048, 7) # consonant_diacritic
def forward(self, x):
x = x / 255.0
x = self.backbone(x)
x = x.view(x.size(0), -1)
vowel_diacritic = self.fc1(x)
grapheme_root = self.fc2(x)
consonant_diacritic = self.fc3(x)
return vowel_diacritic, grapheme_root, consonant_diacritic
bengali_dataset = GraphemeValidationDataset(num_samples=num_samples)
data_loader_test = DataLoader(
bengali_dataset, batch_size=BATCH_SIZE, num_workers=N_WORKERS, shuffle=False
)
import time
import os
from tqdm.notebook import tqdm
model = MyNet()
MODEL_PATH = "/kaggle/input/submission10/0597.pth"
device = torch.device("cuda:0")
checkpoint = torch.load(MODEL_PATH, map_location=device)
if isinstance(checkpoint, dict) and "state_dict" in checkpoint:
model.load_state_dict(checkpoint["state_dict"])
else:
model.load_state_dict(checkpoint)
model.eval()
model.to(device)
del checkpoint
print("start inference")
results = np.zeros((3, num_samples), dtype=np.int)
tic = time.perf_counter()
for batch_idx, images in tqdm(
enumerate(data_loader_test), total=num_samples // BATCH_SIZE
):
images = images.float().to(device)
with torch.no_grad():
vowel_diacritic, grapheme_root, consonant_diacritic = model(images)
start = batch_idx * BATCH_SIZE
end = min((batch_idx + 1) * BATCH_SIZE, num_samples)
results[0, start:end] = consonant_diacritic.argmax(1).cpu().detach().numpy()
results[1, start:end] = grapheme_root.argmax(1).cpu().detach().numpy()
results[2, start:end] = vowel_diacritic.argmax(1).cpu().detach().numpy()
del images
del vowel_diacritic, grapheme_root, consonant_diacritic
del model
print("finish inference in {:.2f} sec.".format(time.perf_counter() - tic))
result_reshape = results.reshape(3 * num_samples, order="F")
sample_submission = pd.read_csv("../input/bengaliai-cv19/sample_submission.csv")
sample_submission.target = np.hstack(result_reshape)
sample_submission.to_csv("submission.csv", index=False)
|
import os
os.chdir("gluon-cv")
os.chdir("..")
import zipfile
def zipdir(path, ziph):
# ziph is zipfile handle
for root, dirs, files in os.walk(path):
for file in files:
ziph.write(os.path.join(root, file))
zipf = zipfile.ZipFile("/kaggle/working/weights.zip", "w", zipfile.ZIP_DEFLATED)
zipdir("/kaggle/working/results/", zipf)
zipf.close()
|
import numpy as np
import pandas as pd
from keras.models import *
from keras.layers import *
from keras.preprocessing import image
from PIL import Image
import matplotlib.pyplot as plt
from tensorflow import keras
from tensorflow.keras.applications import ResNet50V2
import tensorflow as tf
from keras.preprocessing.image import ImageDataGenerator
import os
main_dir = "/kaggle/input/chest-ctscan-images/Data"
train_data = ImageDataGenerator(rescale=1.0 / 255)
test_data = ImageDataGenerator(rescale=1.0 / 255)
valid_data = ImageDataGenerator(rescale=1.0 / 255)
train_dir = os.path.join(main_dir, "train")
train_set = train_data.flow_from_directory(
train_dir, target_size=(460, 460), batch_size=32, class_mode="categorical"
)
test_dir = os.path.join(main_dir, "test")
test_set = test_data.flow_from_directory(
test_dir, target_size=(460, 460), batch_size=32, class_mode="categorical"
)
valid_dir = os.path.join(main_dir, "valid")
valid_set = test_data.flow_from_directory(
valid_dir, target_size=(460, 460), batch_size=32, class_mode="categorical"
)
base_model = ResNet50V2(include_top=False, weights="imagenet")
x = base_model.output
x = GlobalAveragePooling2D()(x)
x = Dense(512, activation="relu")(x)
x = Dense(128, activation="relu")(x)
predictions = Dense(4, activation="softmax")(x)
model = Model(inputs=base_model.input, outputs=predictions)
for layer in base_model.layers:
layer.trainable = False
model.compile(
loss=keras.losses.categorical_crossentropy, optimizer="adam", metrics=["accuracy"]
)
results = model.fit(
train_set, epochs=10, validation_data=[valid_set], verbose=1, batch_size=32
)
pred = model.evaluate(test_set)
plt.plot(results.history["accuracy"])
plt.plot(results.history["val_accuracy"])
plt.plot(results.history["loss"])
plt.plot(results.history["val_loss"])
|
# **importing the libarries**
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.datasets import load_breast_cancer
from sklearn.model_selection import train_test_split
# **Data collection and processing**
breast_cancer = load_breast_cancer()
breast_cancer
breast_cancer_data = pd.DataFrame(
breast_cancer.data, columns=breast_cancer.feature_names
)
breast_cancer_data["target"] = breast_cancer.target
breast_cancer_data
breast_cancer_data["target"].value_counts()
breast_cancer_data.shape
breast_cancer_data.info()
breast_cancer_data.isnull().sum()
breast_cancer_data.describe()
breast_cancer_data.groupby("target").mean()
# **Seperating the features and target**
x = breast_cancer_data.drop("target", axis=1)
y = breast_cancer_data["target"]
print(x)
print(y)
# **Splitting the data into training and testing dataset**
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.2, random_state=10
)
print(x.shape, x_train.shape, x_test.shape)
from sklearn.preprocessing import StandardScaler
scalar = StandardScaler()
x_train_std = scalar.fit_transform(x_train)
x_test_std = scalar.transform(x_test)
# importing tensorflow and keras
import tensorflow as tf
tf.random.set_seed(3)
from tensorflow import keras
# setting up the layers of the neural networks
model = keras.Sequential(
[
keras.layers.Flatten(input_shape=(30,)),
keras.layers.Dense(20, activation="relu"),
keras.layers.Dense(2, activation="sigmoid"),
]
)
# compiling the neural network
model.compile(
optimizer="adam", loss="sparse_categorical_crossentropy", metrics=["accuracy"]
)
# training the neural network
history = model.fit(x_train_std, y_train, validation_split=0.1, epochs=10)
# **Visualizing accuracy and loss**
plt.plot(history.history["accuracy"])
plt.plot(history.history["val_accuracy"])
plt.title("model accuracy")
plt.ylabel("accuracy")
plt.xlabel("epoch")
plt.legend(["training data", "validation data"], loc="lower right")
plt.plot(history.history["loss"])
plt.plot(history.history["val_loss"])
plt.title("model loss")
plt.ylabel("loss")
plt.xlabel("epoch")
plt.legend(["training data", "validation data"], loc="upper right")
# **Accuracy of model on Data**
loss, accuracy = model.evaluate(x_test_std, y_test)
print(accuracy)
print(x_test_std.shape)
print(x_test_std[0])
y_pred = model.predict(x_test_std)
print(y_pred.shape)
print(y_pred[0])
print(x_test_std)
print(y_pred)
# **model.predict() gives the prediction probability of each class for that data point**
my_list = [0.25, 0.56]
index_of_max_value = np.argmax(my_list)
print(my_list)
print(index_of_max_value)
# converting the prediction probability to class labels
y_pred_labels = [np.argmax(i) for i in y_pred]
print(y_pred_labels)
# **Building the predictive system**
input_data = (
11.76,
21.6,
74.72,
427.9,
0.08637,
0.04966,
0.01657,
0.01115,
0.1495,
0.05888,
0.4062,
1.21,
2.635,
28.47,
0.005857,
0.009758,
0.01168,
0.007445,
0.02406,
0.001769,
12.98,
25.72,
82.98,
516.5,
0.1085,
0.08615,
0.05523,
0.03715,
0.2433,
0.06563,
)
# change the input_data to a numpy array
input_data_as_numpy_array = np.asarray(input_data)
# reshape the numpy array as we are predicting for one data point
input_data_reshaped = input_data_as_numpy_array.reshape(1, -1)
# standardizing the input data
from sklearn.preprocessing import StandardScaler
input_data_std = scalar.transform(input_data_reshaped)
prediction = model.predict(input_data_std)
print(prediction)
prediction_label = [np.argmax(prediction)]
print(prediction_label)
if prediction_label[0] == 0:
print("The tumor is Malignant")
else:
print("The tumor is Benign")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
train_data = pd.read_csv("/kaggle/input/ieee-fraud-detection/test_identity.csv")
train_data.head(n=5)
train_data.tail()
train_data.shape
test_data = pd.read_csv("/kaggle/input/ieee-fraud-detection/test_identity.csv")
test_data.head()
test_data.shape
# # to use the matplotlib library, import matplotlib
import matplotlib.pyplot as plt
# # Load the data
train_df = pd.read_csv("/kaggle/input/ieee-fraud-detection/test_transaction.csv")
# # Convert TransactionDT to a datetime object
train_df["TransactionDT"] = pd.to_datetime(train_df["TransactionDT"], unit="s")
# # Create a new feature for the hour of the day
train_df["TransactionHour"] = train_df["TransactionDT"].dt.hour
# # Plot the distribution of transaction hour
plt.hist(train_df["TransactionHour"], bins=24)
plt.xlabel("Hour of the Day")
plt.ylabel("Count")
plt.show()
# this converted the TransactionDT feature to a datetime object & created a new feature TransactionHour to represent the hour of the day for each transaction, then it plots a histogram of the distribution of transaction hours.
# # Plot the distribution of card types
card_types = train_df["card4"].value_counts()
plt.bar(card_types.index, card_types.values)
plt.xlabel("Card Type")
plt.ylabel("Count")
plt.show()
# above histogram, shows the counts of the number of occurrences, of each card-type in the 'card4' feature and plots a bar chart of the distribution of card types.
# # Select two features to compute the correlation between
import seaborn as sns
feat1 = "TransactionAmt"
feat2 = "card1"
# # Compute the correlation between the two features
corr = train_df[[feat1, feat2]].corr()
# # Plot the correlation matrix
sns.heatmap(corr, annot=True, cmap="coolwarm")
plt.show()
|
import torch
import torch.nn as nn
import torch.optim as optim
import torch.nn.functional as F
from torch.optim.lr_scheduler import CosineAnnealingLR
from torch.utils.data import Dataset, DataLoader
from glob import glob
import numpy as np
from PIL import Image
import torchvision
import torchvision.transforms as transforms
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from tqdm import tqdm
import random
data_root = "/kaggle/input/art-price-dataset/artDataset"
import os
# images = glob(f"{data_root}/*.png")
images = list(os.walk("/kaggle/input/art-price-dataset/artDataset"))[0][2]
# targets = pd.read_csv('artDataset.csv')['price']
targets = pd.read_csv("/kaggle/input/art-price-dataset/artDataset.csv")["price"]
targets = targets.apply(lambda item: int(item.strip(" USD").replace(".", "")))
class ArtDataset(Dataset):
def __init__(self, targets: list, transform, image_indexes: list):
self.targets = targets
self.transform = transform
self.image_indexes: list = image_indexes # индексы картинок, которые попали в датасет при текущем разбиении
def __getitem__(self, index):
data = Image.open(
f"{data_root}/image_{self.image_indexes[index] + 1}.png"
).convert("RGB")
target = self.targets[self.image_indexes[index]]
return {
"data": self.transform(data),
"target": target.astype(np.float32),
}
def __len__(self):
return len(self.targets)
train_transform = transforms.Compose(
[
transforms.ToTensor(),
transforms.Resize((228, 228)),
transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5)),
transforms.RandomRotation((-5, 5)),
]
)
test_transform = transforms.Compose(
[
transforms.ToTensor(),
transforms.Resize((228, 228)),
transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5)),
]
)
np.array(range(1, train_size))
train_size: int = 601
train_dataset = ArtDataset(
targets[:train_size], train_transform, image_indexes=np.array(range(1, train_size))
)
test_dataset = ArtDataset(
targets[train_size:],
test_transform,
image_indexes=np.array(range(train_size, len(targets) + 1)),
)
train_loader = DataLoader(train_dataset, batch_size=32, shuffle=True)
test_loader = DataLoader(test_dataset, batch_size=32, shuffle=False)
plt.figure(figsize=(5, 3))
sns.histplot(x=np.log(targets), bins=10)
plt.show()
def myshow(img):
img = img * 0.3 + 0.3 # умножаем на std, прибавляем mean - в Normalize всё наоборот
npimg = img.detach().numpy()
fig = plt.figure(figsize=(10, 10))
plt.imshow(npimg.transpose(1, 2, 0))
trainiter = iter(train_loader)
batch = trainiter.next()
images, labels = batch["data"], batch["target"]
myshow(torchvision.utils.make_grid(images))
class Model(nn.Module):
def __init__(
self,
input_dim: int,
n_classes: int,
):
super().__init__()
self.conv_block = nn.Sequential(
nn.Conv2d(in_channels=3, out_channels=64, kernel_size=3),
nn.LeakyReLU(),
nn.Conv2d(in_channels=64, out_channels=128, kernel_size=3),
nn.LeakyReLU(),
)
self.gap = nn.AdaptiveAvgPool2d((15, 15))
self.fc = nn.Sequential(nn.Linear(128 * 15 * 15, n_classes))
def forward(self, x):
x = self.conv_block(x)
x = self.gap(x)
x = x.view(x.shape[0], -1)
return self.fc(x)
model = Model(3, 1)
from sklearn.metrics import mean_squared_log_error, mean_squared_error
y_pred = [targets.mean() for _ in range(754)]
print(mean_squared_log_error(targets, y_pred, squared=True))
print(mean_squared_error(targets, y_pred, squared=False))
for batch in train_loader:
data, target = batch["data"], batch["target"]
print(data.shape)
print(model(data).shape)
print(torch.max(model(data).data, 1)[1])
break
# def train(model: nn.Module,
# train_loader: DataLoader,
# val_loader: DataLoader,
# criterion: nn.Module,
# optimizer: torch.optim.Optimizer,
# scheduler,
# device: str):
# model.train()
# train_loss = 0
# for batch in tqdm(train_loader):
# data, targets = batch['data'].to(device), batch['target'].to(device)
# predicted = model(data).view(-1)
# loss = criterion(predicted, targets)
# train_loss += loss.item()
# optimizer.zero_grad()
# loss.backward()
# optimizer.step()
# scheduler.step()
# model.eval()
# return train_loss / len(train_loader)
def train(
model,
optimizer,
scheduler,
criterion,
n_epoch,
train_loader,
test_loader,
device: str,
):
for epoch in range(n_epoch):
# обучение
model.train()
train_loss = 0
for batch in tqdm(train_loader):
data, targets = batch["data"].to(device), batch["target"].to(device)
optimizer.zero_grad()
targets_pred = model(data).view(-1)
loss = criterion(targets_pred, targets)
train_loss += loss.item()
loss.backward()
optimizer.step()
scheduler.step()
wandb.log({"train RMSLE": torch.sqrt(loss)}) # todo
# валидация
test_loss = 0
test_size = 0
model.eval()
with torch.no_grad():
for batch in tqdm(test_loader):
data, targets = batch["data"].to(device), batch["target"].to(device)
targets_pred = model(data)
loss = criterion(targets_pred, targets)
# todo
test_loss += loss.item() * y_pred.size(0)
test_size += y_pred.size(0)
# вывод метрик
print(
f"Epoch: {epoch}, RMSLE: {torch.sqrt(torch.tensor(test_loss / test_size))}"
)
wandb.log({"test RMSE": torch.sqrt(torch.tensor(test_loss / test_size))})
class RMSLELoss(nn.Module):
def __init__(self):
super().__init__()
self.mse = nn.MSELoss()
def forward(self, pred, actual):
# return torch.sqrt(self.mse(torch.log(pred + 1), torch.log(actual + 1)))
return self.mse(torch.log(pred + 1), torch.log(actual + 1))
def set_random_seed(seed):
torch.backends.cudnn.deterministic = True
torch.manual_seed(seed)
torch.cuda.manual_seed(seed)
np.random.seed(seed)
random.seed(seed)
import wandb
wandb.login()
set_random_seed(123)
device = "cuda" if torch.cuda.is_available() else "cpu"
model = Model(3, 1)
model.to(device)
num_epochs: int = 2
optimizer = optim.Adam(model.parameters(), lr=3e-4)
criterion = RMSLELoss()
scheduler = CosineAnnealingLR(
optimizer, T_max=int(len(train_dataset) / 32 + 1) * num_epochs
)
wandb.init(
project="artExpertBootcamp",
name="test",
config={"epochs": 1, "batch_size": 32, "model": model, "optimizer": optimizer},
)
wandb.watch(model)
train(
model=model,
optimizer=optimizer,
scheduler=scheduler,
criterion=criterion,
n_epoch=num_epochs,
train_loader=train_loader,
test_loader=test_loader,
device=device,
)
# train(model, optimizer, criterion, 30, train_loader, test_loader, device)
# def fit(model: nn.Module,
# train_loader: DataLoader,
# val_loader: DataLoader,
# criterion,
# optimizer,
# scheduler,
# epochs: int
# ):
# device = 'cuda' if torch.cuda.is_available() else 'cpu'
# model = model.to(device)
# train_losses = []
# for epoch in range(1, epochs + 1):
# train_loss = train(
# model=model,
# optimizer=optimizer,
# scheduler=scheduler,
# criterion=criterion,
# n_epoch=epochs,
# train_loader=train_loader,
# test_loader=val_loader,
# device=device
# )
# # print(f'Epoch: {epoch}, Train Loss: {train_loss:.3f}', )
# train_losses.append(train_loss)
# return train_losses
num_epochs = 1
class RMSLELoss(nn.Module):
def __init__(self):
super().__init__()
self.mse = nn.MSELoss()
def forward(self, pred, actual):
return torch.sqrt(self.mse(torch.log(pred + 1), torch.log(actual + 1)))
model = Model(3, 1)
optimizer = optim.Adam(model.parameters(), lr=3e-4)
# criterion = nn.MSELoss()
criterion = RMSLELoss()
scheduler = CosineAnnealingLR(
optimizer, T_max=int(len(train_dataset) / 32 + 1) * num_epochs
)
train_losses = fit(
model,
train_loader,
train_loader,
criterion,
optimizer,
scheduler,
num_epochs,
)
np.sqrt(260266104)
device = "cpu"
preds = []
with torch.no_grads():
for batch in tqdm(train_loader):
data, targets = batch["data"].to(device), batch["target"].to(device)
predicted = model(data).view(-1)
preds.append(predicted)
preds
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
plt.style.use("bmh") # matplotlib style gallery
sns.set(font_scale=2.5)
import missingno as msno
import warnings
warnings.filterwarnings("ignore")
# 새로운 윈도우 창으로 보여지지 않도록 해줌.
# - feature은 column과 동일하다.
# 1. column engineering을 한다(전처리 하면서 유의미한 변수)
# 2. 그 다음 model을 만든다.
df_train = pd.read_csv("/kaggle/input/titanic/train.csv")
df_test = pd.read_csv("/kaggle/input/titanic/test.csv")
df_train.head()
import pandas as pd
gender_submission = pd.read_csv("../input/titanic/gender_submission.csv")
test = pd.read_csv("../input/titanic/test.csv")
train = pd.read_csv("../input/titanic/train.csv")
df_train.head()
# feature에 대한 특징 파악
df_train.describe()
# numerical한 변수들의 통계적 기본 특징
df_test.describe()
# 테스트 데이터에 대한 설명
# 이 데이터프레임은 Survived를 예측해야 되기 때문에 하나의 feature가 적게 보인다.
df_train.columns
# df_train에 있는 컬럼들을 모두 나타낸다.
# 이것을 for문을 적용
for col in df_train.columns:
msg = "column: {:>10}\t Percent of NaN value: {:.2f}%".format(
col, 100 * (df_train[col].isnull().sum() / df_train[col].shape[0])
)
print(msg)
# null 데이터가 얼마나 있는지
# 셀에서 F 키를 누른 상태로 df_train과 df_train를 바꿔줄 수 있다.
for col in df_test.columns:
msg = "column: {:>10}\t Percent of NaN value: {:.2f}%".format(
col, 100 * (df_test[col].isnull().sum() / df_test[col].shape[0])
)
print(msg)
msno.matrix(df=df_train.iloc[:, :], figsize=(8, 8), color=(0.5, 0.8, 0.1))
# color는 RGB 숫자를 의미한다.
# 그래프상 빈칸은 null값을 의미한다.
msno.bar(df=df_train.iloc[:, :], figsize=(8, 8), color=(0.5, 0.8, 0.1))
# 위와 같이 null data를 파악하는 것인데, 시각화 막대그래프 형태
# # 2. Target label 확인
f, ax = plt.subplots(1, 2, figsize=(18, 8))
df_train["Survived"].value_counts().plot.pie(
explode=[0, 0.1], autopct="%1.1f%%", ax=ax[0], shadow=True
)
# ax는 0 죽은사람과 1 산사람을 나타낸다.
ax[0].set_title("Pie plot - Survived") # 제목 달아주기
ax[0].set_ylabel("") # y축의 라벨 - 파이그래프 왼쪽에 나타난다.
sns.countplot("Survived", data=df_train, ax=ax[1])
ax[1].set_title("Count plot - Survived")
plt.show()
# 생존과 죽음을 비율로 나타낸 파이그래프
# 카운트한 막대그래프 두가지로 나타내보기
df_train.shape
# 인덱스 제외하고 총 11개의 feature가 있다.
# 어떤 feature이 중요할까?
# ## 2.1 Pclass
df_train[["Pclass", "Survived"]].groupby(["Pclass"], as_index=True).count()
df_train[["Pclass", "Survived"]].groupby(["Pclass"], as_index=True).sum()
pd.crosstab(
df_train["Pclass"], df_train["Survived"], margins=True
).style.background_gradient(cmap="summer_r")
# margins는 'All'값을 의미한다.
# cmap은 검색에 'color example code' 검색
df_train[["Pclass", "Survived"]].groupby(["Pclass"], as_index=True).mean().sort_values(
by="Survived", ascending=False
).plot.bar()
# 생존율을 나타내는 것. 0은 죽음, 1은 생존이라면 이것들의 (0 * x + 1 * y) / N를 나타냄
y_position = 1.02
f, ax = plt.subplots(1, 2, figsize=(18, 8))
df_train["Pclass"].value_counts().plot.bar(
color=["#CD7F32", "#FFDF00", "#D3D3D3"], ax=ax[0]
)
ax[0].set_title("Number of Passengers By Pclass", y=y_position)
ax[0].set_ylabel("Count")
sns.countplot("Pclass", hue="Survived", data=df_train, ax=ax[1])
ax[1].set_title("Pclass: Survived vs Dead", y=y_position)
plt.show()
# ## 2.2 Sex
# p.s 셀에서 M을 누르면 마크다운으로 되고, Y를 누르면 스크립트가 된다.
f, ax = plt.subplots(1, 2, figsize=(18, 8))
df_train[["Sex", "Survived"]].groupby(["Sex"], as_index=True).mean().plot.bar(ax=ax[0])
# as_index는 컬럼을 'Sex'로 만드는 것 ??
ax[0].set_title("Survived vs Sex")
sns.countplot("Sex", hue="Survived", data=df_train, ax=ax[1])
ax[1].set_title("Sex: Survived vs Dead")
plt.show()
df_train[["Sex", "Survived"]].groupby(["Sex"], as_index=False).mean().sort_values(
by="Survived", ascending=False
)
pd.crosstab(
df_train["Sex"], df_train["Survived"], margins=True
).style.background_gradient(cmap="summer_r")
# ## 2.3 Both Sex and Pclass
sns.factorplot("Pclass", "Survived", hue="Sex", data=df_train, size=6, aspect=1.5)
# 아래 두가지 중 위의 것이 참고용
# 아래 중 윗쪽은 어느 클래스든 여성이 생존율이 높다. 특히 Pclass의 기울기가 더 크므로.
# 아래 중 아래쪽은 클래스별의 차이를 보여주기에 용이한듯
sns.factorplot(
x="Sex",
y="Survived",
col="Pclass",
data=df_train,
satureation=0.5,
size=9,
aspect=1,
)
sns.factorplot(
x="Pclass",
y="Survived",
col="Sex",
data=df_train,
satureation=0.5,
size=9,
aspect=1,
)
# ## 2.4 Age
print("제일 나이 많은 탑승객 : {:.1f} Years".format(df_train["Age"].max()))
print("제일 어린 탑승객 : {:.1f} Years".format(df_train["Age"].min()))
print("탑승객 평균 나이 : {:.1f} Years".format(df_train["Age"].mean()))
# 히스토그램에 대한 설명
df_train[df_train["Survived"] == 1]["Age"].hist()
# 생존, 사망별 Age의 분포(Kde plot)
fig, ax = plt.subplots(1, 1, figsize=(9, 5))
sns.kdeplot(df_train[df_train["Survived"] == 1]["Age"], ax=ax)
# boolean을 indexing하는 방법을 잘 숙지해야 한다.
# df_train['Survived'] == 1 이라는 것 중 True인 row만 반환해서 'Age'컬럼을 뽑아내는 것
sns.kdeplot(df_train[df_train["Survived"] == 0]["Age"], ax=ax)
plt.legend(["Survived == 1", "Survived == 0"])
plt.show()
# ### 나이가 어릴수록 생존확률이 높다
# Age distribution withing classes
plt.figure(figsize=(8, 6)) # figsize는 도화지를 의미한다.
df_train["Age"][df_train["Pclass"] == 1].plot(kind="kde")
df_train["Age"][df_train["Pclass"] == 2].plot(kind="kde")
df_train["Age"][df_train["Pclass"] == 3].plot(kind="kde")
# kind를 hist로 하면 그래프가 겹쳐서 안보인다.
plt.xlabel("Age")
plt.title("Age Distribution within classes")
plt.legend(["1st Class", "2nd Class", "3rd Class"])
# Class가 높을수록 나이의 평균치가 올라간다.
# 여기에는 Age별로 Class의 분포만 보여주는 것이고, 생존과는 연관이 없다.
# 설명엔 나타나있지 않은 것
# 위에선 생존에 관한 kde를 볼 수 없으므로, boolean을 이용해서 생존자별 나이대의 분포를 알아본다.
# Class별로 생존자 나이대의 분포를 살펴보자.
fig, ax = plt.subplots(1, 1, figsize=(9, 5))
sns.kdeplot(
df_train[(df_train["Survived"] == 0) & (df_train["Pclass"] == 1)]["Age"], ax=ax
)
sns.kdeplot(
df_train[(df_train["Survived"] == 1) & (df_train["Pclass"] == 1)]["Age"], ax=ax
)
plt.legend(["Survived == 0", "Survived == 1"])
plt.title("1st Class")
plt.show()
# 젊은 사람일수록 생존 확률이 높다 (1st Class에서)
fig, ax = plt.subplots(1, 1, figsize=(9, 5))
sns.kdeplot(
df_train[(df_train["Survived"] == 0) & (df_train["Pclass"] == 2)]["Age"], ax=ax
)
sns.kdeplot(
df_train[(df_train["Survived"] == 1) & (df_train["Pclass"] == 2)]["Age"], ax=ax
)
plt.legend(["Survived == 0", "Survived == 1"])
plt.title("2st Class")
plt.show()
fig, ax = plt.subplots(1, 1, figsize=(9, 5))
sns.kdeplot(
df_train[(df_train["Survived"] == 0) & (df_train["Pclass"] == 3)]["Age"], ax=ax
)
sns.kdeplot(
df_train[(df_train["Survived"] == 1) & (df_train["Pclass"] == 3)]["Age"], ax=ax
)
plt.legend(["Survived == 0", "Survived == 1"])
plt.title("3st Class")
plt.show()
# 나이대별로 생존률이 어떻게 되는가?
cummulate_survival_ratio = []
for i in range(1, 80):
cummulate_survival_ratio.append(
df_train[df_train["Age"] < i]["Survived"].sum()
/ len(df_train[df_train["Age"] < i]["Survived"])
)
# 전체 인원 수 중에서(len) survived한 사람들을 (1) sum한 것들의 비율을 반환
plt.figure(figsize=(7, 7))
plt.plot(cummulate_survival_ratio)
plt.title("Survival rate change depending on range of Age", y=1.02)
plt.ylabel("Survival rate")
plt.xlabel("Range of Age(0~x)")
plt.show()
|
# ### The source code is from https://www.kaggle.com/code/estrelia/painter
# ### The source code is also belong to me, but created 2 years ago. I might refer others' code at that time, and can't find now.
from __future__ import division
from torchvision import models
from torchvision import transforms
from PIL import Image
import argparse
import torch
import torchvision
import torch.nn as nn
import numpy as np
import torchvision.utils as vutils
import matplotlib.pyplot as plt
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
def load_image(image_path, transform=None, max_size=None, shape=None):
image = Image.open(image_path)
if max_size:
scale = max_size / max(image.size)
size = np.array(image.size) * scale
image = image.resize(size.astype(int), Image.ANTIALIAS)
if shape:
image = image.resize(shape, Image.LANCZOS)
if transform:
image = transform(image).unsqueeze(0)
# print(image.shape)
return image.to(device)
transform = transforms.Compose(
[
transforms.ToTensor(),
transforms.Normalize(mean=[0.485, 0.456, 0.406], std=[0.229, 0.224, 0.225]),
]
)
content = load_image(
"/kaggle/input/monet-for-test/0a497f768d.jpg", transform, max_size=400
)
style = load_image(
"/kaggle/input/gan-getting-started/monet_jpg/000c1e3bff.jpg",
transform,
shape=[content.size(2), content.size(3)],
)
print(content.shape, style.shape)
unloader = transforms.ToPILImage() # reconvert into PIL image
plt.ion()
def imshow(tensor, title=None):
image = tensor.cpu().clone() # we clone the tensor to not do changes on it
image = image.squeeze(0) # remove the fake batch dimension
image = unloader(image)
plt.imshow(image)
if title is not None:
plt.title(title)
plt.pause(0.001) # pause a bit so that plots are updated
plt.figure()
imshow(style[0], title="Style")
imshow(content[0], title="Image")
class VGGNet(nn.Module):
def __init__(self):
super(VGGNet, self).__init__()
self.select = ["0", "5", "10", "19", "28"]
self.vgg = models.vgg19(
pretrained=True
).features # 这个地方是拿出vgg19的前面部分, 后面的分类器舍弃掉
def forward(self, x):
features = []
for name, layer in self.vgg._modules.items():
x = layer(x)
if name in self.select:
features.append(x)
print(features[0].size(), features[4].size())
return features
target = content.clone().requires_grad_(True)
optimizer = torch.optim.Adam([target], lr=0.003, betas=[0.5, 0.999])
vgg = VGGNet().to(device).eval() # 注意, 这个VGG已经是不训练状态了
print(vgg.modules)
total_step = 2000
style_weight = 100.0
for step in range(total_step):
target_features = vgg(target)
content_features = vgg(content)
style_features = vgg(style)
style_loss = 0
content_loss = 0
for f1, f2, f3 in zip(
target_features, content_features, style_features
): # 对于每一层特征, 下面开始计算损失
content_loss += torch.mean((f1 - f2) ** 2) # 内容损失
_, c, h, w = f1.size() # [c, h, w]
f1 = f1.view(c, h * w) # [c, h*w]
f3 = f3.view(c, h * w) # [c, h*w]
# 计算gram matrix
f1 = torch.mm(f1, f1.t()) # [c, c]
f3 = torch.mm(f3, f3.t()) # [c, c]
style_loss += torch.mean((f1 - f3) ** 2) / (c * h * w) # 风格损失
loss = content_loss + style_weight * style_loss # 总的损失
# 更新target
optimizer.zero_grad()
loss.backward()
optimizer.step()
if step % 100 == 0:
print(
"Step [{}/{}], Content Loss: {:.4f}, Style Loss: {:.4f}".format(
step, total_step, content_loss.item(), style_loss.item()
)
)
denorm = transforms.Normalize((-2.12, -2.04, -1.80), (4.37, 4.46, 4.44)) # 反归一化
img = target.clone().squeeze()
img = denorm(img).clamp_(0, 1) # 控制到0-1
plt.figure()
imshow(img, title="Target Image")
plt.savefig("/kaggle/working/0a49020ae5.png")
from torchsummaryX import summary
import torch
import torchvision.models as models
model = VGGNet()
summary(model, torch.zeros((1, 3, 256, 256)))
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# ## Predicting Normal and Hate Speech in Tweets using Gloves vectors
# Social media has become an integral part of our daily lives, with Twitter being one of the most popular platforms used to share opinions. Unfourtunately, the increased use of abusive language and hate speech represents a well-known problem on online platforms.
# In this Jupyter notebook, I will explore a machine learning approach to classify tweets into normal and hate speech categories using GloVe vectors. GloVe (Global Vectors for Word Representation) is a pre-trained unsupervised method for generating vector representations for words that capture their meanings and relationships in a high-dimensional space.
# I will use a dataset of labeled tweets containing normal and hate speech, and train a logistic regression model on the GloVe vectors to predict the class of new tweets. The performance of the model will be evaluated using metrics such as accuracy, precision, recall, and F1-score.
# This notebook aims to provide a practical guide to using GloVe vectors for text classification, and to demonstrate the effectiveness of machine learning in detecting hate speech on social media.
import pandas as pd
import numpy as np
import re
import nltk
import emoji
from gensim.scripts.glove2word2vec import glove2word2vec
from nltk.tokenize import word_tokenize
from nltk.corpus import stopwords
from sklearn.feature_extraction.text import CountVectorizer, TfidfVectorizer
from sklearn.metrics import accuracy_score, classification_report
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.svm import SVC
from sklearn.naive_bayes import MultinomialNB
from sklearn.ensemble import RandomForestClassifier
from sklearn.pipeline import Pipeline
from sklearn.metrics import confusion_matrix
from nltk.stem import WordNetLemmatizer
from nltk.tokenize import TweetTokenizer
from nltk.stem.porter import PorterStemmer
from sklearn.metrics import plot_confusion_matrix
from sklearn.metrics import precision_recall_curve, auc, roc_curve
from scipy.sparse import hstack
from sklearn.model_selection import GridSearchCV
from gensim.models import Word2Vec
from gensim.models import KeyedVectors
from gensim.test.utils import datapath
import matplotlib.pyplot as plt
print("done!")
# **Uploading all the required text
# First of all, this model will only consider normal and hate speech as a binary variable. To prepare each instance with its label I will proceed at uploading and concatenating the relevant files
# Get all instances of normal speech
normal_1 = pd.read_csv(
"/kaggle/input/normal-hate-and-offensive-speeches/Normal_Speeches_1.csv"
)
normal_2 = pd.read_csv(
"/kaggle/input/normal-hate-and-offensive-speeches/Normal_Speeches_2.csv"
)
normal_3 = pd.read_csv(
"/kaggle/input/normal-hate-and-offensive-speeches/Normal_Speeches_3.csv"
)
normal_4 = pd.read_csv(
"/kaggle/input/normal-hate-and-offensive-speeches/Normal_Speeches_4.csv"
)
df_normal = pd.concat(
[normal_1, normal_2, normal_3, normal_4], axis=0
) # concatenate all normal speech labeled tweets
df_normal.shape
# get all instances of hate speech
hate_1 = pd.read_csv(
"/kaggle/input/normal-hate-and-offensive-speeches/Hate_Speeches_1.csv"
)
hate_2 = pd.read_csv(
"/kaggle/input/normal-hate-and-offensive-speeches/Hate_Speeches_2.csv"
)
hate_3 = pd.read_csv(
"/kaggle/input/normal-hate-and-offensive-speeches/Hate_Speeches_3.csv"
)
hate_4 = pd.read_csv(
"/kaggle/input/normal-hate-and-offensive-speeches/Hate_Speeches_4.csv"
)
df_hate = pd.concat([hate_1, hate_2, hate_3, hate_4], axis=0)
df_hate.shape
# The next step entails creating the labels for each type of speech. Since ML models work on numbers only, I will assign 0 to normal and 1 to hate speech.
X_hate = df_hate["full_text"]
y_hate = [1] * (len(X_hate)) # code hate as 1
X_normal = df_normal["full_text"]
y_normal = [0] * (len(X_normal))
# It is important to check label sizes during the preprocessing of a machine learning (ML) model because it can provide useful insights into the balance of the dataset. For this reason, I will check how many istances of the same category are present by visualising a simple bar-plot.
# exploratory analysis, check label sizes
normal_count = len(X_normal)
hate_count = len(X_hate)
label_counts = [normal_count, hate_count]
label_names = ["Normal Speech", "Hate Speech"]
plt.bar(label_names, label_counts)
plt.title("Occurences of Normal and Hate Speech")
plt.ylabel("Occurences")
plt.show()
# The label sizes are very similar. This model is expected to perform well when properly trained
# After having completed the preprocessing, it is now time to create X and y. X is the dependent variable and will consist of both normal and hate speech text, whereas the variable y, which the model will attempt to predict, will be constituted by the sum of all labeled instances.
# create the X label
X_raw = pd.concat([X_normal, X_hate])
print(X_raw)
# create the y label
y = y_normal + y_hate
# However, the text needs to be further simplified to be turned into numerical vectors. There are a lot of approaches that could be adopted,but, especially in the case of tweets, it is important to strip http tags, emoji and symbols like @ and #, and simplify words that have repeated cahracters. Other steps that are often employed are tokenisation and the removal of stopwords. First, I define a function that performs all these operations and then I apply it to the text
stop_words = set(
stopwords.words("english")
) # gets the stopword list from the dedicated library and saves them
tk = TweetTokenizer() # defines the object, whose method is called in the function
def preprocess_text(text):
# Remove URLs
text = re.sub(r"http\S+", "", text)
# Remove user mentions
text = re.sub(r"@\S+", "", text)
# Remove hashtags
text = re.sub(r"#\S+", "", text)
# Remove emojis
text = emoji.emojize(text, variant="emoji_type")
# Remove repeated characters (e.g. "soooooo" -> "so")
text = re.sub(r"(.)\1+", r"\1", text)
# Lowercase the text
text = text.lower()
# Tokenize the text
words = tk.tokenize(text)
# Remove stop words
words = [w for w in words if w not in stop_words]
# Join the words back into a string
return " ".join(words)
# Apply the preprocess_text function to the 'text' column
X = X_raw.apply(preprocess_text)
X.head()
# The following step entails dividing the whole dataset into training and test set
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=42
)
# Now, it is time to build the model. In this case, I have downloaded word2vec and glove, which are pretrained neural network that assigns to each word a numerical vector. This values will be used convert the tweets into integer strings that will be used to classify the documents. This approach was chosen because word embeddings can capture semantic and syntactic relationships between words, and can be used to create distributed representations of text that can be fed into machine learning models. GloVe is a popular pre-trained word embedding model that has been trained on large amounts of text data and can provide high-quality vector representations of words.
# The GloVe file is in a specific format that is not compatible with the gensim library's implementation of KeyedVectors. Converting the file to word2vec format allows the pre-trained embeddings to be loaded and used in a gensim-based machine learning pipeline.
from gensim.scripts.glove2word2vec import glove2word2vec
from gensim.models import KeyedVectors
# Load GloVe embeddings
# Convert GloVe file to word2vec format
glove_file = "/kaggle/input/glove6b100dtxt/glove.6B.100d.txt"
word2vec_file = "/kaggle/working/glove.6B.100d.word2vec"
glove2word2vec(glove_file, word2vec_file)
# Load word2vec file as KeyedVectors
from gensim.models import KeyedVectors
word_vectors = KeyedVectors.load_word2vec_format(word2vec_file, binary=False)
# After having uploaded the related libraries is now time to transform our tokenised tweets into vectors.
# The function tweet_to_vec takes a tweet as input, tokenizes it into individual words, looks up the vector representation of each word in pre-trained GloVe embeddings, calculates the mean vector of all the word vectors in the tweet, and returns it as the tweet's vector representation.
# Convert tweets to GloVe vectors
def tweet_to_vec(tweet):
tokens = tweet.split()
vecs = []
for token in tokens:
try:
vec = word_vectors.get_vector(token)
vecs.append(vec)
except:
pass # if the word is not present in the vector list, it is just just skipped
if (
len(vecs) == 0
): # this is to handle tweets that have no words that are included in the vector lists
return np.zeros((100,)) # it will return a vector list of 0s
else:
return np.mean(
vecs, axis=0
) # if the tweet has a list of vector, it will compute the mean
# apply the function to the train and test set
X_train_vec = np.stack(X_train.apply(tweet_to_vec))
X_test_vec = np.stack(X_test.apply(tweet_to_vec))
# After having defined this function, it is important to convert the vectorised sets from a pandas series object (resulting from apply) thanks a NumPy function that stacks a sequence of arrays along a new axis, resulting in a new array with one more dimension (2 dimensions in this case). Thus, the resulting array X_train_vec has shape (n_samples, 100), where n_samples is the number of tweets in the training dataset, and 100 is the length of the GloVe word vectors used to represent each tweet.
# apply the function to the train and test set
X_train_vec = np.stack(X_train.apply(tweet_to_vec))
X_test_vec = np.stack(X_test.apply(tweet_to_vec))
print(X_train_vec, 1)
# I have chosen to apply logistic regression because it is suited for binary classification.
# In order to tune the hyperparameters, I will define a function that evaluates the accuracy of the model for several values of C and display it in a plot, so that I can choose the value that performs the best. C is the regularization parameter which controls the trade-off between fitting the training data well and keeping the model simple. The solver is set to lbfgs, but there was no substantial difference between the score of different solvers.
def plot_regularization_performance(X_train, y_train, X_test, y_test, C_values):
train_scores = []
test_scores = []
for C in C_values:
model = LogisticRegression(C=C, solver="lbfgs", max_iter=1000)
model.fit(X_train, y_train)
y_train_pred = model.predict(X_train)
y_test_pred = model.predict(X_test)
train_acc = accuracy_score(y_train, y_train_pred)
test_acc = accuracy_score(y_test, y_test_pred)
train_scores.append(train_acc)
test_scores.append(test_acc)
plt.plot(C_values, train_scores, label="Training Accuracy")
plt.plot(C_values, test_scores, label="Test Accuracy")
plt.xlabel("Regularization Parameter (C)")
plt.ylabel("Accuracy")
plt.legend()
plt.show()
C_values = [0.0001, 0.001, 0.01, 0.1, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 20, 30]
plot_regularization_performance(X_train_vec, y_train, X_test_vec, y_test, C_values)
# 5 seems to perform well, for this reason I will use this value for C when applying Log Regression to the datasets
# Define the logistic regression model
lr_model = LogisticRegression(C=5, solver="lbfgs", max_iter=1000)
# Fit the model on the training data
lr_model.fit(X_train_vec, y_train)
# Predict on the testing data
y_pred = lr_model.predict(X_test_vec)
# Print the accuracy score and classification report
print("Accuracy:", accuracy_score(y_test, y_pred))
print(classification_report(y_test, y_pred))
# With an accuracy score of 0.89, the model performs well. Now I will plot the confusion matrix to visualise the ratio of false negatives and positives.
labs = ["Normal", "Hate"]
plot_confusion_matrix(lr_model, X_test_vec, y_test, display_labels=labs)
plt.text(-0.4, 0.2, "True Negative", fontsize=12)
plt.text(-0.4, 1.2, "False Positive", fontsize=12)
plt.text(0.8, 0.2, "False Negative", fontsize=12)
plt.text(0.8, 1.2, "True Positive", fontsize=12)
plt.show()
|
# # Importing Libraries
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
import plotly.express as px
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
from lightgbm import LGBMRegressor
from imblearn.over_sampling import SMOTE
from imblearn.under_sampling import RandomUnderSampler
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Load the CSV file into a pandas dataframe
df = pd.read_csv("/kaggle/input/epl-soccer-data/EPL_Soccer_MLR_LR.csv")
# Print the first few rows of the dataframe to get an idea of the data
df.head()
# # Exploratory Data Analysis
# Print the number of rows and columns in the dataframe
print(f"Dataframe rows: {df.shape[0]}, columns: {df.shape[1]}")
# Print some basic statistics about the numeric columns in the dataframe
print(df.describe())
# Check for missing values in the dataframe
print(df.isnull().sum())
# Check the data types of the columns in the dataframe
print(df.dtypes)
# Visualize the distribution of a numeric column (e.g. column named 'Goals')
fig = px.histogram(df, x="Goals", nbins=50, color_discrete_sequence=["green"])
fig.update_layout(title="Distribution of Goals")
fig.show()
# Visualize the relationship between two numeric columns (e.g. columns named 'DistanceCovered(InKms)' and 'Score')
fig = px.scatter(
df,
x="DistanceCovered(InKms)",
y="Score",
color="BMI",
size="Cost",
color_continuous_scale="RdYlGn",
)
fig.update_layout(title="Relationship between Distance Covered and Score")
fig.show()
# Visualize the correlation between all numeric columns in the dataframe
fig = px.imshow(df.corr(), color_continuous_scale="RdYlGn")
fig.update_layout(title="Correlation between Numeric Columns")
fig.show()
# Visualize the distribution of each numeric column in the dataframe
for col in df.select_dtypes(include="number"):
fig = px.histogram(df, x=col, nbins=50, color_discrete_sequence=["green"])
fig.update_layout(title=f"Distribution of {col}")
fig.show()
# Visualize the relationship between ShotsPerGame and Goals
fig = px.scatter(
df,
x="ShotsPerGame",
y="Goals",
color="BMI",
size="Cost",
color_continuous_scale="RdYlGn",
)
fig.update_layout(title="Relationship between Shots Per Game and Goals")
fig.show()
# Scatter plot of target variable (Goals) against each numeric variable
numeric_cols = [
"DistanceCovered(InKms)",
"Goals",
"MinutestoGoalRatio",
"ShotsPerGame",
"AgentCharges",
"BMI",
"Cost",
"PreviousClubCost",
"Height",
"Weight",
"Score",
]
for col in numeric_cols:
fig = px.scatter(
df, x=col, y="Goals", trendline="ols", hover_data=["PlayerName", "Club"]
)
fig.show()
# # Data Preprocessing
def one_hot_encode(df):
"""
One-hot encode all object-type columns in a given pandas DataFrame.
Args:
df (pandas.DataFrame): The DataFrame to be one-hot encoded.
Returns:
pandas.DataFrame: The one-hot encoded DataFrame.
"""
obj_cols = df.select_dtypes(include=["object"]).columns
df_encoded = pd.get_dummies(df, columns=obj_cols)
return df_encoded
def log_transform(df):
"""
Perform log transformation on skewed numerical features in a pandas dataframe
Parameters:
df (pandas.DataFrame): Input dataframe
Returns:
pandas.DataFrame: Transformed dataframe
"""
# Get list of numerical columns
numeric_cols = df.select_dtypes(include=[np.number]).columns.tolist()
# Get list of skewed columns
skewed_cols = df[numeric_cols].skew().sort_values(ascending=False).index.tolist()
# Apply log transformation to skewed columns
for col in skewed_cols:
if df[col].skew() > 1:
print(f"Column {col} is skewed. Transforming ...")
# Use np.log1p instead of np.log to handle zero and negative values
df[col] = np.log1p(df[col])
return df
# As we don't want player name to affect our prediction, we want to drop it (predictions more generic)
df.drop(columns=["PlayerName"], inplace=True)
# #### Feature engineering could be applied like finding a way to measure Player Experience. This could be done by combining information from the 'Club' and 'PreviousClubCost' columns, as players who have been transferred for high costs or between high-profile clubs might be more experienced and therefore more likely to have higher scores.
# #### Also as data are only available for three teams then the prediction will be applied also for those teams. If more teams are provided, a ranking could be provided to the dictionary according to e.g. their budgets.
# Assuming data provided are recent, we will rank the teams experience according to their last year standing.
"""
LIV -> 2nd
CHE -> 3rd
MUN -> 6th
Experience level will be -> (total teams participated in 2021-2022) - position of club
eg. LIV = 20-2 = 18
MUN = 20-6 = 14
"""
# Create a dictionary to map club names to experience levels
club_experience_levels = {
"MUN": 14,
"CHE": 17,
"LIV": 18,
# add other clubs and corresponding experience levels
# ...
}
# Map the 'Club' column to the corresponding experience level
df["ExperienceLevel"] = df["Club"].map(club_experience_levels)
# Create a new feature based on the ratio between 'PreviousClubCost' and 'Cost'
df["ExperienceCostRatio"] = df["PreviousClubCost"] / df["Cost"]
# Multiply the 'ExperienceLevel' and 'ExperienceCostRatio' features to get the final 'ExperienceScore' feature
df["ExperienceScore"] = df["ExperienceLevel"] * df["ExperienceCostRatio"]
# # Experiments
def calculate_metrics(y_true, y_pred):
"""
Calculates the Mean Absolute Percentage Error (MAPE), Symmetric Mean Absolute Percentage Error (SMAPE),
and Root Mean Squared Error (RMSE) between the true values and predicted values.
Args:
y_true (array-like): The true values.
y_pred (array-like): The predicted values.
Returns:
float: The MAPE value.
float: The SMAPE value.
float: The RMSE value.
"""
y_true, y_pred = np.array(y_true), np.array(y_pred)
# Calculate MAPE
mape = np.mean(np.abs((y_true - y_pred) / y_true)) * 100
# Calculate SMAPE
smape = (
np.mean(np.abs(y_pred - y_true) / ((np.abs(y_true) + np.abs(y_pred)) / 2)) * 100
)
# Calculate RMSE
rmse = np.sqrt(np.mean((y_true - y_pred) ** 2))
return mape, smape, rmse
def train_models(df, target_col, sampling_mode=None):
"""
Train Linear Regression and LGBM Regression models and return the models and their predictions.
If sampling_mode is 'smote', perform SMOTE on the training data before training the models.
If sampling_mode is 'rus', perform Random Under Sampling on the training data before training the models.
Parameters:
df (pandas.DataFrame): The input dataframe containing the features and target variable.
target_col (str): The name of the target variable column.
sampling_mode (str, optional): The sampling mode to use for balancing the training data. Can be 'smote' or 'rus'.
Returns:
tuple: A tuple containing the trained Linear Regression and LGBM Regression models, as well as their predictions on the test set.
"""
df = one_hot_encode(df)
# Split data into training and test sets
X_train, X_test, y_train, y_test = train_test_split(
df.drop(columns=[target_col]), df[target_col], test_size=0.2, random_state=42
)
if sampling_mode == "smote":
# # Apply SMOTE to balance the training data
# print(f'Applying SMOTE on training data...')
# # Due to constraint of library need to concat and then re-split
# data_to_smote = pd.concat([X_train,y_train], axis=1)
# print(data_to_smote)
# # Perform smote on whole dataframe
# data_to_smote = smogn.smoter(data=data_to_smote, y=target_col, rel_method = 'auto', rel_coef = 0.50)
# # Resplit data
# y_train = data_to_smote[target_col]
# X_train = data_to_smote.drop(columns=[target_col])
pass
elif sampling_mode == "rus":
# # Apply Random Under Sampling to balance the training data
# print(f'Applying Rus on training data...')
# rus = RandomUnderSampler(random_state=42)
# X_train, y_train = rus.fit_resample(X_train, y_train)
pass
if sampling_mode == "weight":
sample_weights = [10 / y for y in y_train]
else:
sample_weights = [1 for y in y_train]
# Train linear regression model
lin_reg = LinearRegression()
lin_reg.fit(X_train, y_train, sample_weight=sample_weights)
# Train LGBM regression model
lgbm_reg = LGBMRegressor()
lgbm_reg.fit(X_train, y_train, sample_weight=sample_weights)
# Make predictions on test set
lin_reg_pred = lin_reg.predict(X_test)
lgbm_reg_pred = lgbm_reg.predict(X_test)
# Calculate evaluation metrics
mape, smape, rmse = calculate_metrics(y_test, lin_reg_pred)
# Print evaluation metrics
print("Linear Regression MAPE:", mape)
print("Linear Regression SMAPE:", smape)
print("Linear Regression RMSE:", rmse)
# Calculate evaluation metrics
mape, smape, rmse = calculate_metrics(y_test, lgbm_reg_pred)
# Print evaluation metrics
print("LGBM Regression MAPE:", mape)
print("LGBM Regression SMAPE:", smape)
print("LGBM Regression RMSE:", rmse)
# Return models and predictions
return lin_reg, lgbm_reg, lin_reg_pred, lgbm_reg_pred
lin_reg, lgbm_reg, lin_reg_pred, lgbm_reg_pred = train_models(df, "Score")
#### Applying log transformation to skewed data features
df_log = log_transform(df.copy())
lin_reg, lgbm_reg, lin_reg_pred, lgbm_reg_pred = train_models(df_log, "Score")
# #### TODO
# #### As the data in the target variable are imbalanced, we can try performing data augmentation (e.g. using SMOTE). Undersampling could also be tested and weighting of the data is also an alternative.
# #### Add weighting by applying 10/y_value sample weight to the data. Similar to invert probability but instead of 1/value is 10/value
# With Smote and log transformations
lin_reg, lgbm_reg, lin_reg_pred, lgbm_reg_pred = train_models(
df_log, "Score", sampling_mode="weight"
)
|
# ## 🧠 Alzheimer's Disease Classification
# ## The data in total consists of 6400 images.
# ## Using Enhanced CNN Models namely,
# ## * Inception Net (V3)
# ## * Resnet 50
# ### Libraries
import numpy as np
import pandas as pd
import seaborn as sns
import tensorflow as tf
from tensorflow import keras
import matplotlib.pyplot as plt
import os
from distutils.dir_util import copy_tree, remove_tree
from PIL import Image
from random import randint
from tensorflow.keras.preprocessing import image, image_dataset_from_directory
from tensorflow.keras.preprocessing.image import ImageDataGenerator
from imblearn.over_sampling import SMOTE
from sklearn.model_selection import train_test_split
from sklearn.metrics import matthews_corrcoef as MCC
from sklearn.metrics import balanced_accuracy_score as BAS
from sklearn.metrics import classification_report, confusion_matrix
from tensorflow.keras.models import Model
import tensorflow_addons as tfa
from keras.utils.vis_utils import plot_model
from tensorflow.keras import Sequential, Input
from tensorflow.keras.layers import Dense, Dropout
from tensorflow.keras.layers import Conv2D, Flatten
from tensorflow.keras.callbacks import ReduceLROnPlateau
from tensorflow.keras.applications.inception_v3 import InceptionV3
from tensorflow.keras.applications.resnet50 import ResNet50
from tensorflow.keras.preprocessing.image import ImageDataGenerator as IDG
from tensorflow.keras.layers import (
SeparableConv2D,
BatchNormalization,
GlobalAveragePooling2D,
)
# ## Inception net V3 Model
# ### Data Pre-Processing
base_dir = "/kaggle/input/alzheimers-dataset-4-class-of-images/Alzheimer_s Dataset/"
root_dir = "./"
test_dir = base_dir + "test/"
train_dir = base_dir + "train/"
work_dir = root_dir + "dataset/"
if os.path.exists(work_dir):
remove_tree(work_dir)
os.mkdir(work_dir)
copy_tree(train_dir, work_dir)
copy_tree(test_dir, work_dir)
print("Working Directory Contents:", os.listdir(work_dir))
WORK_DIR = "./dataset/"
CLASSES = ["NonDemented", "VeryMildDemented", "MildDemented", "ModerateDemented"]
IMG_SIZE = 176
IMAGE_SIZE = [176, 176]
DIM = (IMG_SIZE, IMG_SIZE)
# ### Performing Image Augmentation to have more data samples
ZOOM = [0.99, 1.01]
BRIGHT_RANGE = [0.8, 1.2]
HORZ_FLIP = True
FILL_MODE = "constant"
DATA_FORMAT = "channels_last"
work_dr = IDG(
rescale=1.0 / 255,
brightness_range=BRIGHT_RANGE,
zoom_range=ZOOM,
data_format=DATA_FORMAT,
fill_mode=FILL_MODE,
horizontal_flip=HORZ_FLIP,
)
train_data_gen = work_dr.flow_from_directory(
directory=WORK_DIR, target_size=DIM, batch_size=6500, shuffle=False
)
# ### Data Visualization
def show_images(generator, y_pred=None):
"""
Input: An image generator,predicted labels (optional)
Output: Displays a grid of 9 images with lables
"""
# get image lables
labels = dict(zip([0, 1, 2, 3], CLASSES))
# get a batch of images
x, y = generator.next()
# display a grid of 9 images
plt.figure(figsize=(10, 10))
if y_pred is None:
for i in range(9):
ax = plt.subplot(3, 3, i + 1)
idx = randint(0, 6400)
plt.imshow(x[idx])
plt.axis("off")
plt.title("Class:{}".format(labels[np.argmax(y[idx])]))
else:
for i in range(9):
ax = plt.subplot(3, 3, i + 1)
plt.imshow(x[i])
plt.axis("off")
plt.title(
"Actual:{} \nPredicted:{}".format(
labels[np.argmax(y[i])], labels[y_pred[i]]
)
)
# Display Train Images
show_images(train_data_gen)
# ### Retrieving the data from the ImageDataGenerator iterator
train_data, train_labels = train_data_gen.next()
print(train_data.shape, train_labels.shape)
# ### Performing over-sampling of the data, since the classes are imbalanced
sm = SMOTE(random_state=42)
train_data, train_labels = sm.fit_resample(
train_data.reshape(-1, IMG_SIZE * IMG_SIZE * 3), train_labels
)
train_data = train_data.reshape(-1, IMG_SIZE, IMG_SIZE, 3)
print(train_data.shape, train_labels.shape)
# ### Splitting the data into train, test, and validation sets
train_data, test_data, train_labels, test_labels = train_test_split(
train_data, train_labels, test_size=0.2, random_state=42
)
train_data, val_data, train_labels, val_labels = train_test_split(
train_data, train_labels, test_size=0.2, random_state=42
)
# ### Using the InceptionV3 model as a base model for the task
inception_model = InceptionV3(
input_shape=(176, 176, 3), include_top=False, weights="imagenet"
)
# ### Freezing the pretrained layers for further training
for layer in inception_model.layers:
layer.trainable = False
# ### Further Model Building
custom_inception_model = Sequential(
[
inception_model,
Dropout(0.5),
GlobalAveragePooling2D(),
Flatten(),
BatchNormalization(),
Dense(512, activation="relu"),
BatchNormalization(),
Dropout(0.5),
Dense(256, activation="relu"),
BatchNormalization(),
Dropout(0.5),
Dense(128, activation="relu"),
BatchNormalization(),
Dropout(0.5),
Dense(64, activation="relu"),
Dropout(0.5),
BatchNormalization(),
Dense(4, activation="softmax"),
],
name="inception_cnn_model",
)
# ### Defining a custom callback function to stop training our model when accuracy goes above 99%
class MyCallback(tf.keras.callbacks.Callback):
def on_epoch_end(self, epoch, logs={}):
if logs.get("acc") > 0.99:
print("\nReached accuracy threshold! Terminating training.")
self.model.stop_training = True
my_callback = MyCallback()
# ReduceLROnPlateau to stabilize the training process of the model
rop_callback = ReduceLROnPlateau(monitor="val_loss", patience=3)
METRICS = [
tf.keras.metrics.CategoricalAccuracy(name="acc"),
tf.keras.metrics.AUC(name="auc"),
tfa.metrics.F1Score(num_classes=4),
]
CALLBACKS = [my_callback, rop_callback]
custom_inception_model.compile(
optimizer="rmsprop", loss=tf.losses.CategoricalCrossentropy(), metrics=METRICS
)
custom_inception_model.summary()
# ### Fitting the Model
EPOCHS = 20
history = custom_inception_model.fit(
train_data,
train_labels,
validation_data=(val_data, val_labels),
callbacks=CALLBACKS,
epochs=EPOCHS,
)
# ### Tabulating the Results of our custom InceptionV3 model
# ### Plotting the trend of the metrics during training
fig, ax = plt.subplots(1, 3, figsize=(30, 5))
ax = ax.ravel()
for i, metric in enumerate(["acc", "auc", "loss"]):
ax[i].plot(history.history[metric])
ax[i].plot(history.history["val_" + metric])
ax[i].set_title("Model {}".format(metric))
ax[i].set_xlabel("Epochs")
ax[i].set_ylabel(metric)
ax[i].legend(["train", "val"])
test_scores = custom_inception_model.evaluate(test_data, test_labels)
print("Testing Accuracy: %.2f%%" % (test_scores[1] * 100))
# ### Predicting the test data
pred_labels = custom_inception_model.predict(test_data)
# ### Print the classification report of the tested data
def roundoff(arr):
"""To round off according to the argmax of each predicted label array."""
arr[np.argwhere(arr != arr.max())] = 0
arr[np.argwhere(arr == arr.max())] = 1
return arr
for labels in pred_labels:
labels = roundoff(labels)
print(classification_report(test_labels, pred_labels, target_names=CLASSES))
# ### Plot the confusion matrix to understand the classification in detail
pred_ls = np.argmax(pred_labels, axis=1)
test_ls = np.argmax(test_labels, axis=1)
conf_arr = confusion_matrix(test_ls, pred_ls)
plt.figure(figsize=(8, 6), dpi=80, facecolor="w", edgecolor="k")
ax = sns.heatmap(
conf_arr,
cmap="Greens",
annot=True,
fmt="d",
xticklabels=CLASSES,
yticklabels=CLASSES,
)
plt.title("Alzheimer's Disease Diagnosis")
plt.xlabel("Prediction")
plt.ylabel("Truth")
plt.show(ax)
# ## RESNET 50 Model
# ### Loading Dataset
image_generator = ImageDataGenerator(
rescale=1 / 255, validation_split=0
) # shear_range =.25, zoom_range =.2, horizontal_flip = True, rotation_range=20)
train_dataset = image_generator.flow_from_directory(
batch_size=16,
directory="/kaggle/input/alzheimers-dataset-4-class-of-images/Alzheimer_s Dataset/train",
shuffle=True,
target_size=(224, 224),
subset="training",
class_mode="categorical",
)
image_generator = ImageDataGenerator(rescale=1 / 255, validation_split=0.2)
validation_dataset = image_generator.flow_from_directory(
batch_size=16,
directory="/kaggle/input/alzheimers-dataset-4-class-of-images/Alzheimer_s Dataset/test",
shuffle=True,
target_size=(224, 224),
class_mode="categorical",
)
submission = image_generator.flow_from_directory(
directory="/kaggle/input/alzheimers-dataset-4-class-of-images/Alzheimer_s Dataset/test",
shuffle=False,
subset="validation",
target_size=(224, 224),
class_mode="categorical",
)
# ### Data and label visualization
batch_1_img = train_dataset[0]
for i in range(0, 2):
img = batch_1_img[0][i]
lab = batch_1_img[1][i]
plt.imshow(img)
plt.title(lab)
plt.axis("off")
plt.show()
# ### Using Transfer Learning importing the pretrained model and Fitting the model (RESNET 50)
rn = ResNet50(input_shape=(224, 224, 3), weights="imagenet", include_top=False)
for layer in rn.layers:
layer.trainable = False
x = Flatten()(rn.output)
prediction = Dense(4, activation="softmax")(x)
modelvgg = Model(inputs=rn.input, outputs=prediction)
modelvgg.compile(
optimizer="adam",
loss=tf.losses.CategoricalCrossentropy(),
metrics=[keras.metrics.AUC(name="auc"), "acc"],
)
callback = keras.callbacks.EarlyStopping(
monitor="val_loss", patience=8, restore_best_weights=True
)
modelvgg.fit(
train_dataset, epochs=20, validation_data=validation_dataset, callbacks=callback
)
# ## Sort
def sortit(x, y):
n = len(x)
for i in range(n - 1):
for j in range(n - i - 1):
if x[j] > x[j + 1]:
x[j], x[j + 1] = x[j + 1], x[j]
y[j], y[j + 1] = y[j + 1], y[j]
return x, y
# ### Model's Accuracy (Validation Set)
accuracy = modelvgg.evaluate(validation_dataset)
print("Loss: ", accuracy[0])
print("Accuracy: ", accuracy[1])
# ### Model's Accuracy (Test Set)
accuracy = modelvgg.evaluate(submission)
print("Loss: ", accuracy[0])
print("Accuracy: ", accuracy[1])
# ## Compiling Accuracies of all the previously applied models
import plotly.graph_objects as go
schools = [
"SVM",
"KNN",
"ANN",
"CNN",
"MobileNet",
"MobileNetV2",
"VGG16",
"VGG19",
"DensetNet121",
"DensetNet169",
"ResNet50",
"InceptionNetV3",
]
fig = go.Figure()
fig.add_trace(
go.Scatter(
x=[68, 41, 75.01, 90, 79.35, 78.47, 99.29, 90.51, 93.67, 88.71, 88.97, 89],
y=schools,
marker=dict(color="crimson", size=12),
mode="markers",
name="Accuracy",
)
)
fig.add_trace(
go.Scatter(
x=[
0.30,
0.19,
0.5001,
0.8997,
0.46,
0.47,
0.0727,
0.867,
0.94,
1.039,
0.9890,
0,
],
y=schools,
marker=dict(color="gold", size=12),
mode="markers",
name="F1 Score/Loss Function",
)
)
fig.update_layout(
title="Alzheimer Disease Detection",
xaxis_title="Accuracy (in %,Red) & F1 Score/Loss Val (Yellow)",
yaxis_title="Model Applied",
)
fig.show()
fig = plt.figure(figsize=(18, 7))
y = [
"SVM",
"KNN",
"ANN",
"CNN",
"MobileNet",
"MobileNetV2",
"VGG16",
"VGG19",
"DensetNet121",
"DensetNet169",
"ResNet50",
"InceptionNetV3",
]
x = [68, 41, 75.01, 90, 79.35, 78.47, 99.29, 90.51, 93.67, 88.71, 88.97, 89]
x, y = sortit(x, y)
# Horizontal Bar Plot
plt.bar(y, x)
plt.ylabel("Test Accuracies")
plt.xlabel("Models Used")
# Show Plot
plt.show()
fig = plt.figure(figsize=(18, 7))
y = [
"SVM",
"KNN",
"ANN",
"CNN",
"MobileNet",
"MobileNetV2",
"VGG16",
"VGG19",
"DensetNet121",
"DensetNet169",
"ResNet50",
"InceptionNetV3",
]
x = [
0.30,
0.19,
0.5001,
0.8997,
0.46,
0.47,
0.0727,
0.867,
0.3026,
0.4890,
0.9094,
0.2919,
]
# Horizontal Bar Plot
x, y = sortit(x, y)
plt.bar(y, x)
plt.ylabel("Loss Val")
plt.xlabel("Models Used")
# Show Plot
plt.show()
|
# [Предыдущее занятие](https://www.kaggle.com/zohrab/rus-python-part-3-type-conversion/edit/run/27395984)
# # Структуры данных в Python
# # `list()` - cписки
# Списки в Python - упорядоченные изменяемые коллекции объектов произвольных типов (очень похож на массив данных, только в отличие от массива, списки могут содержать элементы разных типов).
# Физически список представляет собой массив указателей (адресов) на его элементы. С точки зрения производительности списки имеют следующие особенности.
# 1. Время доступа к элементу есть величина постоянная и не зависит от размера списка.
# 2. Время на добавление одного элемента в конец списка есть величина постоянная.
# 3. Время на вставку зависит от того, сколько элементов находится справа от него, т.е. чем ближе элемент к концу списка, тем быстрее идет его вставка.
# 4. Удаление элемента происходит так же, как и в пункте 3.
# 5. Время, необходимое на реверс списка, пропорционально его размеру — O(n).
# 6. Время, необходимое на сортировку, зависит логарифмически от размера списка - — O(log(n))
# ### Создание списка. Способ 1
lst_1 = []
type(lst_1)
# ### Создание списка. Способ 2
lst_2 = list()
type(lst_2)
# ### `id(object)` - встроенная функция, которая возвращает целое число, являющееся "адресом" объекта. Это целое число гарантированно будет уникальным и постоянным для данного объекта в течение срока его существования.
id(lst_2)
# ### Создание списка с предустановленным набором элементов
lst_2 = ["жара", 912, 0.7, True]
lst_2
# Как мы видим, все элементы списка являются разного типа
# ### Меняется `id` списка, хотя его название не изменилось
id(lst_2)
# ### `is` и `is not` - операторы тождественности, которые сравнивают id объектов
id(lst_1)
lst_1 is lst_2
# ### `not` - логический оператор отрицания
lst_1 is not lst_2
# ### Разница между `[ ]` и `list()`
# ### `list(iterable=())` - встроенная функция, которая создает список из любого итерируемого объекта
lst_1 = ["молоко"]
print(lst_1)
lst_1 = list("молоко")
print(lst_1)
# ### При приравнивании списка к другому списку, второй список получает такой же id, как и первый список. Копирования объектов не происходит, а происходит копирование ссылки на один и тот же объект
print(lst_2)
print(id(lst_2))
lst_3 = lst_2
print(lst_3)
print(id(lst_3))
lst_2[0] = 11
print(lst_2)
print(lst_3)
lst_2[0] = "жара"
# ### Полная копия списка. Способ 1
print(lst_2)
print(id(lst_2))
lst_3 = list(lst_2)
print(lst_3)
print(id(lst_3))
lst_2[0] = 11
print(lst_2)
print(lst_3)
lst_2[0] = "жара"
# ### Вывод элемента списка по его индексу
# 
# Нумерация элементов списка начинается с `0`
lst_3[0]
# ### Если мы хотим узнать значение элемента, которого нет в списке, то появится исключение `IndexError`
lst_3[4]
# ### Индексацию можно производить с конца списка, при этом индекс последнего элемента будет равен `-1`
lst_3[-1]
lst_3[-2]
# ### Slice - cрез. При слайсинге мы выбираем диапазон элементов из списка с заданным шагом по следующей формуле:
# ### `[start_index=0 : stop_index=-1 : step=1]`
lst_3
lst_3[0:2]
lst_3[2:4]
# ### Можем делать слайсинг с начала списка или до конца списка
lst_3[:2]
lst_3[2:]
# ### Можно проводить слайсинг с определенным шагом
lst_3[::2]
# ### Шаг может быть отрицательным
lst_3[::-2]
# ### `slice(start=0, end=-1, step=1)` - встроенная функция, которая возвращает объект срез
x = slice(0, 2, 1)
type(x)
lst_3[x]
# ### Вывести весь список
lst_3[:]
# ### При создании среза создается новый объект списка
lst_3 is lst_3[:]
# ### Полная копия списка. Способ 2
print(lst_2)
print(id(lst_2))
lst_3 = lst_2[:]
print(lst_3)
print(id(lst_3))
lst_2[0] = 11
print(lst_2)
print(lst_3)
lst_2[0] = "жара"
|
# # Introduction
# Libraries loading
import pandas as pd
import os
from datetime import datetime, timedelta
import warnings
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
color = sns.color_palette()
sns.set_style("darkgrid")
from scipy import stats
from scipy.stats import norm, skew # statistics for normality and skewness
import numpy as np
ip = get_ipython()
ibe = ip.configurables[-1]
ibe.figure_formats = {"pdf", "png"}
warnings.filterwarnings("ignore")
DATA_DIR = "/kaggle/input/atp-and-wta-tennis-data"
df_atp = pd.read_csv(os.path.join(DATA_DIR, "df_atp.csv"), index_col=0)
df_atp["Date"] = df_atp.Date.apply(lambda x: datetime.strptime(x, "%Y-%m-%d"))
df_atp.head()
print("Total number of matches : " + str(len(df_atp)))
print(list(df_atp.columns))
# We drop the ATP column (Tournament number (men)) because it does seem intuitively unimportant specially for the prediction phase and it might be even contagious to our model if we forget it in the training phase (Example : it might add some leakage into our model)
# verifying the shape of the dataset before droping the 'ATP' column.
print("Shape of the Dataset before droping the 'ATP' Column : {} ".format(df_atp.shape))
# Saving the column (maybe for later use ?)
df_atp_ID = df_atp["ATP"]
# Droping the column
df_atp.drop("ATP", axis=1, inplace=True)
# verifying the shape of the dataset after droping the 'ATP' column
print(
"\nShape of the Dataset after droping the 'ATP' Column : {} ".format(df_atp.shape)
)
# ## Quick glumpse at the data and answering some questions
# ** 1. Who are the three ATP players with the most wins ? **
df_atp["Winner"].describe()
df_atp["Winner"].value_counts()[0:3]
# Return a Series containing counts of unique values in descending order so that the first element is the most frequently-occurring element.
df_atp["Loser"].describe()
df_atp["Loser"].value_counts()[0:3]
# We can see that 'Federer R.' is by far the player with most victories in tournaments being ahead of the second most winner in tournament by 230 matches. While 'Lopez F.' being the player with most losses in Tournaments is only ahead the second most loser one after him with only 14 matches.
# Let's see how these 3 player we were talking about have done in both victories and losses.
print(
"'Federer R.' have won "
+ str(len(df_atp[df_atp["Winner"] == "Federer R."]))
+ " and lost "
+ str(len(df_atp[df_atp["Loser"] == "Federer R."]))
)
print(
"'Nadal R.' have won "
+ str(len(df_atp[df_atp["Winner"] == "Nadal R."]))
+ " and lost "
+ str(len(df_atp[df_atp["Loser"] == "Nadal R."]))
)
print(
"'Djokovic N.' have won "
+ str(len(df_atp[df_atp["Winner"] == "Djokovic N."]))
+ " and lost "
+ str(len(df_atp[df_atp["Loser"] == "Djokovic N."]))
)
print(
"'Lopez F.' have won "
+ str(len(df_atp[df_atp["Winner"] == "Lopez F."]))
+ " and lost "
+ str(len(df_atp[df_atp["Loser"] == "Lopez F."]))
)
print(
"'Youzhny M.' have won "
+ str(len(df_atp[df_atp["Winner"] == "Youzhny M."]))
+ " and lost "
+ str(len(df_atp[df_atp["Loser"] == "Youzhny M."]))
)
print(
"'Verdasco F.' have won "
+ str(len(df_atp[df_atp["Winner"] == "Verdasco F."]))
+ " and lost "
+ str(len(df_atp[df_atp["Loser"] == "Verdasco F."]))
)
# We can see that the top 3 performers: 'Federer R.', 'Nadal R.' and 'Djokovic N.' have won many matches but lost at most less than the quarter of that number in matches. While the top player who lost the most matches : 'Lopez F.', 'Youzhny M.' and 'Verdasco F.' have only won about the same number of matches.
# **2. How many sets did the player “Federer R.” win in total ?**
df_atp["Lsets"] = pd.to_numeric(
df_atp["Lsets"], errors="coerce"
) # tranforming str to numeric values and replcing with nan when we can't
N_sets = (
df_atp["Wsets"][df_atp["Winner"] == "Federer R."].sum()
+ df_atp["Lsets"][df_atp["Loser"] == "Federer R."].sum()
)
print("\nPlayer “Federer R.” won a total of : " + str(N_sets) + " sets.\n")
# ** 3. How many sets did the player “Federer R.” win during the years 2016 and 2017 ?**
# Number of sets the player 'Federer R.' won in 2016 alone:
beg = datetime(2016, 1, 1)
end = datetime(2017, 1, 1)
df_atp_2016 = df_atp[(df_atp["Date"] >= beg) & (df_atp["Date"] < end)]
(
df_atp_2016["Wsets"][df_atp_2016["Winner"] == "Federer R."].sum()
+ df_atp_2016["Wsets"][df_atp_2016["Loser"] == "Federer R."].sum()
)
# Number of sets the player 'Federer R.' won in 2017 alone:
beg = datetime(2017, 1, 1)
end = datetime(2018, 1, 1)
df_atp_2017 = df_atp[(df_atp["Date"] >= beg) & (df_atp["Date"] < end)]
(
df_atp_2017["Wsets"][df_atp_2017["Winner"] == "Federer R."].sum()
+ df_atp_2017["Wsets"][df_atp_2017["Loser"] == "Federer R."].sum()
)
# Number of sets the player 'Federer R.' won during 2016 and 2017: 68+131 = 199 or :
beg = datetime(2016, 1, 1)
end = datetime(2018, 1, 1)
df_atp_2017 = df_atp[(df_atp["Date"] >= beg) & (df_atp["Date"] <= end)]
(
df_atp_2017["Wsets"][df_atp_2017["Winner"] == "Federer R."].sum()
+ df_atp_2017["Wsets"][df_atp_2017["Loser"] == "Federer R."].sum()
)
# **4. For each match, what is the percentage of victories of the winner in the past ?**
unique_player_index_and_score = {}
# Dictionary containing the player name as a key and the tuple (player_unique_index,x,y)
# x : number_of_matches_won
# y : number_of_matches played
# x and y are intiated 0 in the bigining but as we go through the data set we increment x and y by 1 if the player wins a match
# or we increment only y with 1 if the player loses a matches
i = 0
for player in df_atp["Winner"].unique():
if player not in unique_player_index_and_score.keys():
unique_player_index_and_score[player] = (i, 0, 0)
i += 1
for player in df_atp["Loser"].unique():
if player not in unique_player_index_and_score.keys():
unique_player_index_and_score[player] = (i, 0, 0)
i += 1
print("Number of unqiue player names : ", i)
winner_loser_score_tracking_vector = np.zeros((len(df_atp), 2))
# two columns one to track the winner percetage and the other for the loser percentage
# Sorting dataset by date so we can perform our calculation of the player prior win poucentage coorectly by looping one time trough the dataset
df_atp = df_atp.sort_values(by="Date")
for c, row in enumerate(df_atp[["Winner", "Loser"]].values):
score_winner = unique_player_index_and_score[
row[0]
] # Winner up-to date score tracking from the dictionary
score_loser = unique_player_index_and_score[
row[1]
] # Loser up-to date score tracking from the dictionary
# we consider new player that haven't yet played 5 matches as the have 20% of winning in the past
# (kind of a fair approach as they worked hard to get to play in the tournement:))
if score_winner[2] < 5:
winner_loser_score_tracking_vector[c, 0] = 0.2
else:
winner_loser_score_tracking_vector[c, 0] = score_winner[1] / score_winner[2]
if score_loser[2] < 5:
winner_loser_score_tracking_vector[c, 1] = 0.2
else:
winner_loser_score_tracking_vector[c, 1] = score_loser[1] / score_loser[2]
# updating the dictionary based on the new outcome of the current match
unique_player_index_and_score[row[0]] = (
score_winner[0],
score_winner[1] + 1,
score_winner[2] + 1,
) # Winner
unique_player_index_and_score[row[1]] = (
score_loser[0],
score_loser[1],
score_loser[2] + 1,
) # loser
df_atp["Winner_percentage"] = winner_loser_score_tracking_vector[:, 0]
df_atp["Loser_percentage"] = winner_loser_score_tracking_vector[:, 1]
df_atp["Winner_percentage"].describe()
df_atp["Loser_percentage"].describe()
sns.distplot(df_atp["Winner_percentage"], label="Winners")
sns.distplot(df_atp["Loser_percentage"], label="Losers")
plt.ylabel("Frequency")
plt.xlabel("Winners and Losers prior win probabilityt")
plt.title("Winners and Losers prior win probability Distributionn")
plt.legend()
# As we say earlier, the are some winner (the top 3 or 4 performers maybe) have a prior win probability that is very high in the range of [0.75,0.82]. We can see also that Winners tend to win and loser tend to lose from the intersection of the two histograms.
sns.distplot(df_atp["Winner_percentage"], fit=norm)
# Récupèrer les paramètres ajustés utilisés par la fonction
(mu, sigma) = norm.fit(df_atp["Winner_percentage"])
# Tracer la ditribution
plt.legend(
["Normal dist. ($\mu=$ {:.2f} and $\sigma=$ {:.2f} )".format(mu, sigma)], loc="best"
)
plt.ylabel("Frequency")
plt.title("Winner percentage Distribution")
fig = plt.figure()
res = stats.probplot(df_atp["Winner_percentage"], plot=plt)
sns.distplot(df_atp["Loser_percentage"], fit=norm)
# Récupèrer les paramètres ajustés utilisés par la fonction
(mu, sigma) = norm.fit(df_atp["Loser_percentage"])
# Tracer la ditribution
plt.legend(
["Normal dist. ($\mu=$ {:.2f} and $\sigma=$ {:.2f} )".format(mu, sigma)], loc="best"
)
plt.ylabel("Frequency")
plt.title("Loser pourcentage Distribution")
fig = plt.figure()
res = stats.probplot(df_atp["Loser_percentage"], plot=plt)
# the distribution of prior win probability for winner haveextremes values near 0.8 and the ditribution for losers have many values in 0.2 (these are for the playes that have not yet played 5 matches in in tournement in their lifetime). Beside that, both distributions follow an almost normal ditribution Loking at their histogram plot and probability plot expet in the range of
# ## Exploratory Data analisys and Data processing:
# We'll start by the amount of missing data:
train_na = (df_atp.isnull().sum() / len(df_atp)) * 100
train_na = train_na.drop(train_na[train_na == 0].index).sort_values(ascending=False)
missing_data = pd.DataFrame({"Pourcentage of missing values": train_na})
missing_data
# With a visiualisation:
f, ax = plt.subplots(figsize=(15, 12))
plt.xticks(rotation="90")
sns.barplot(x=train_na.index, y=train_na)
plt.xlabel("Columns", fontsize=15)
plt.ylabel("Pourcentage of missing values", fontsize=15)
plt.title("Pourcentage of missing values by variables", fontsize=15)
# Some explications about this missing values:
# - Most of the columns with big missing values are from the odds of betting sites: we shall remove all these columns and keep the columns of the three betting sites : Bet365, EX and PS. First we are going to train a model with data containing these values and also another one with data not containing these values to see its effects on our score. We will be using thoses to also calculate how much we'll win or lose in we used a very simple betting stategy based on the prediction of our model :)
# - The columns L5,L4,W4,W5 (Number of games won in 5th\4th set by match winner\loser respectivly) have missing values because some matches have only 'best out of 3sets' rule while some have 'best out of 5sets' rules.
# - We can think intuitively that we can't use either the columns L1 trough L5 or W5 trought W1 in our predictive modeling phase training data as those variable are set after the matches finishes and we'll be using a discriminative approach in our modeling. So we'll remove those columns too. We can maybe use them if we want to calculate a modified wining set prior probability of each player (in a more advanced modelisation we can think of predicting those values).
# The following columns:
# - MaxW= Maximum odds of match winner (as shown by Oddsportal.com)
# - MaxL= Maximum odds of match loser (as shown by Oddsportal.com)
# - AvgW= Average odds of match winner (as shown by Oddsportal.com)
# - AvgL= Average odds of match loser (as shown by Oddsportal.com)
# maybe having missing as there are mssing values in some matches from all the betting site ? We shall remove them as more than 60 of these column vlues are missing.
# - The columns Lstes and Wsets wich mean Number of sets won by match loser/winner can't be used in as entry data to our model as they are too know after the moatches have finished. But, we we'll keep them to make another variable witch will be the prior probability of winning a sets in the past. We will replace the nan values by mean or median after we explore these columns.
# - As for Lrank or WRank columns, I can't imagine another senario as the one of a new player that just got for the first time in the tournements. we will replace those too by mean or median after we explore those columns too.
# The same applies to the following columns :
# - WPts = ATP Entry points of the match winner as of the start of the tournament
# - LPts = ATP Entry points of the match loser as of the start of the tournament
#
# Drop the columns with missing values and that we won't be using:
for column in train_na.index[:26]:
df_atp.drop(column, axis=1, inplace=True)
# With a visiualisation:
train_na = (df_atp.isnull().sum() / len(df_atp)) * 100
train_na = train_na.drop(train_na[train_na == 0].index).sort_values(ascending=False)
missing_data = pd.DataFrame({"Pourcentage of missing values": train_na})
f, ax = plt.subplots(figsize=(15, 12))
plt.xticks(rotation="90")
sns.barplot(x=train_na.index, y=train_na)
plt.xlabel("Columns", fontsize=15)
plt.ylabel("Pourcentage of missing values", fontsize=15)
plt.title("Pourcentage of missing values by variables", fontsize=15)
# - We also drop the columns W1,L1,W2,L2 as they are intuitivly useless as explained earlier.
#
df_atp.drop("W1", axis=1, inplace=True)
df_atp.drop("L1", axis=1, inplace=True)
df_atp.drop("W2", axis=1, inplace=True)
df_atp.drop("L2", axis=1, inplace=True)
# Let"s explore the remaining columns to see what the best possible values to replace the nan values.
sns.distplot(df_atp["WPts"].dropna(), label="Winners")
sns.distplot(df_atp["LPts"].dropna(), label="Losers")
plt.ylabel("Frequency")
plt.xlabel("Pourcentage of victory in the past")
plt.title("Winners and Losers pourcentage Distribution")
plt.legend()
# seems legit to replace it by mode
sns.distplot(df_atp["PSW"].dropna(), label="Winners")
sns.distplot(df_atp["PSL"].dropna(), label="Losers")
plt.ylabel("Frequency")
plt.xlabel("Pourcentage of victory in the past")
plt.title("Winners and Losers pourcentage Distribution")
plt.legend()
# also mode
df_atp["EXW"] = pd.to_numeric(df_atp["EXW"], errors="coerce")
df_atp["EXW"] = pd.to_numeric(df_atp["EXW"], errors="coerce")
sns.distplot(df_atp["EXW"].dropna(), label="Winners")
sns.distplot(df_atp["EXL"].dropna(), label="Losers")
plt.ylabel("Frequency")
plt.xlabel("Pourcentage of victory in the past")
plt.title("Winners and Losers pourcentage Distribution")
plt.legend()
df_atp["B365W"] = pd.to_numeric(df_atp["B365W"], errors="coerce")
sns.distplot(df_atp["B365W"].dropna(), label="Winners")
sns.distplot(df_atp["B365L"].dropna(), label="Losers")
plt.ylabel("Frequency")
plt.xlabel("Pourcentage of victory in the past")
plt.title("Winners and Losers pourcentage Distribution")
plt.legend()
# also mode
df_atp["Wsets"] = pd.to_numeric(df_atp["Wsets"], errors="coerce")
# df_atp['Lsets']=pd.to_numeric(df_atp['Lsets'],errors='coerce')
# df_atp['Wsets'].replace('scott', np.nan, inplace=True)
sns.distplot(df_atp["Wsets"].dropna(), label="Winners", kde=False)
sns.distplot(df_atp["Lsets"].dropna(), label="Losers")
plt.ylabel("Frequency")
plt.xlabel("Pourcentage of victory in the past")
plt.title("Winners and Losers pourcentage Distribution")
plt.legend()
df_atp["LRank"] = pd.to_numeric(df_atp["LRank"], errors="coerce")
df_atp["WRank"] = pd.to_numeric(df_atp["WRank"], errors="coerce")
sns.distplot(df_atp["LRank"].dropna(), label="Winners")
sns.distplot(df_atp["WRank"].dropna(), label="Losers")
plt.ylabel("Frequency")
plt.xlabel("Pourcentage of victory in the past")
plt.title("Winners and Losers pourcentage Distribution")
plt.legend()
# replacring the value by the mode or mean then plotting poucentage of missing again.
columns = [
"WPts",
"LPts",
"PSW",
"PSL",
"EXW",
"EXL",
"B365W",
"B365L",
"Lsets",
"Wsets",
"LRank",
"WRank",
]
for column in columns:
df_atp[column] = df_atp[column].fillna(float(df_atp[column].mode()[0]))
train_na = (df_atp.isnull().sum() / len(df_atp)) * 100
train_na = train_na.drop(train_na[train_na == 0].index).sort_values(ascending=False)
missing_data = pd.DataFrame({"Pourcentage of missing values": train_na})
missing_data
# No more missing values
# ## More data processing for our datasets so we can use it for prediction and EDA
df_atp.columns
# Description of the rest of the Data:
# - 'B365L', 'B365W','EXL', 'EXW','PSL', 'PSW' : Just betting odds for wiiner and loser repsctivly.
# - 'Bestof' : Maximum number of sets playable in match.
# - 'Comment' :Comment on the match (Completed, won through retirement of loser, or via Walkover)
# - 'Court' : Type of court (outdoors or indoors)
# - 'Date' : date of the match obviously.
# - 'WPts','LPts' : ATP Entry points of the match winner/loser as of the start of the tournament repectivly.
# - 'Location' : Venue of tournament.
# - 'Winner','Loser' : Name of winner/loser respectively.
# - 'Wsets','Lsets' : Number of sets won by match winner/loser respectively
# - 'Round' : Round of match
# - 'Series' : Name of ATP tennis series (Grand Slam, Masters, International or International Gold)
# - 'Surface' : Type of surface (clay, hard, carpet or grass)
# - 'Tournament' : me of tounament (including sponsor if relevant)
# - 'WRank','LRank' : ATP Entry ranking of the match winner/loser respectively as of the start of the tournament
# - 'Winner_percentage','Loser_percentage': ...
# As we said earlier, the column 'B365L', 'B365W','EXL', 'EXW','PSL', 'PSW' will be used in a our modelisationat first and we develop also the same models without those columns. We can think of these columns as some already calculated features given to us by the betting company.
# - The 'comment' column is an event that happend after the matches is finisehd so we'll not be using it too.
# - 'Wsets' and 'Lsets' columns are that reprensents events that happens after the matche is finnished so we'll not be usig it in our model but we'll make of it a new column that reprensets the winner/loser percentage of winning a sets in the past. But we'll drop them after we do some feature engineering with that won't include them having doing some leakage of the target variable; we'll use the calclculate the probability of a player to win a set in the past.
# - All the rest of the columns will be used except maybe fot the tournement name and Location that we'll need to explore them first to see.
df_atp.drop("Comment", axis=1, inplace=True)
df_atp.columns
df_atp.Tournament.describe()
df_atp.Location.describe()
len(df_atp)
# Both columns have only 214,115 unique in a 52298 Data set. thinking about also removing the 2018 and 2017 matches (the later for the tests) and using a cross validation function. I'm not sure if they have any predcitve power seing the few samples for each class. but we'll be keeping them as predictive variables.
df_atp.describe()
# ###### Calculating the player prior win set probability :
# winner prior sets winns pourcentage column
unique_player_index_and_score = {}
# Dictionary containing the player name as a key and the tuple (player_unique_index,x,y)
# x : number_of_set_won
# y : number_of_sets_played
# x and y are intiated 0 in the bigining but as we go through the data set we increment x Wsets(or Lsets) witch are the number of
# won by matches winner(orloser) and we increment y by Wsets+Lsets wich is the number of stes played in that match
i = 0
for player in df_atp["Winner"].unique():
if player not in unique_player_index_and_score.keys():
unique_player_index_and_score[player] = (i, 0, 0)
i += 1
for player in df_atp["Loser"].unique():
if player not in unique_player_index_and_score.keys():
unique_player_index_and_score[player] = (i, 0, 0)
i += 1
print("Number of unqiue player names : ", i)
winner_loser_score_tracking_vector = np.zeros((len(df_atp), 2))
# two columns one to track the winner percetage and the other for the loser percentage
df_atp = df_atp.sort_values(by="Date")
for i in range(len(df_atp)):
row = [df_atp.Winner[i], df_atp.Loser[i]]
score_winner = unique_player_index_and_score[
row[0]
] # Winner up-to date set win score tracking from the dictionary
score_loser = unique_player_index_and_score[
row[1]
] # Loser up-to date set win score tracking from the dictionary
# we consider new player that haven't yet had 15 sets yet as they had a 20% of winning in the past
# (kind of a fair optimist approach as the worked hard to get to play in the tournement:))
if int(score_winner[2]) < 15:
winner_loser_score_tracking_vector[i, 0] = 0.2
else:
winner_loser_score_tracking_vector[i, 0] = score_winner[1] / score_winner[2]
if score_loser[2] < 15:
winner_loser_score_tracking_vector[i, 1] = 0.2
else:
winner_loser_score_tracking_vector[i, 1] = score_loser[1] / score_loser[2]
# updating the dictionary based on the new outcome of the current match
unique_player_index_and_score[row[0]] = (
score_winner[0],
score_winner[1] + float(df_atp.Wsets[i]),
score_winner[2] + float(df_atp.Wsets[i] + df_atp.Lsets[i]),
) # Winner
unique_player_index_and_score[row[1]] = (
score_loser[0],
score_loser[1] + float(df_atp.Lsets[i]),
score_loser[2] + float(df_atp.Wsets[i] + df_atp.Lsets[i]),
) # loser
df_atp["Winner_set_percentage"] = winner_loser_score_tracking_vector[:, 0]
df_atp["Loser_set_percentage"] = winner_loser_score_tracking_vector[:, 1]
df_atp["Winner_set_percentage"].describe()
sns.distplot(df_atp["Winner_set_percentage"])
df_atp["Loser_set_percentage"].describe()
sns.distplot(df_atp["Loser_set_percentage"])
# Again we can remark that winners tend to win and loser tend to lose.
# ###### Calculating the Elo ranking features :
# The Elo rating system is a method for calculating the relative skill levels of players in zero-sum games such as chess. It is named after its creator Arpad Elo, a Hungarian-American physics professor.
# The difference in the ratings between two players serves as a predictor of the outcome of a match. Two players with equal ratings who play against each other are expected to score an equal number of wins. A player whose rating is 100 points greater than their opponent's is expected to score 64%; if the difference is 200 points, then the expected score for the stronger player is 76%.
# Not mine, I took from the internet But i got a full understanding of it :)
def compute_elo_rankings(data):
"""
Given the list on matches in chronological order, for each match, computes
the elo ranking of the 2 players at the beginning of the match
"""
print("Elo rankings computing...")
players = list(pd.Series(list(data.Winner) + list(data.Loser)).value_counts().index)
elo = pd.Series(np.ones(len(players)) * 1500, index=players)
ranking_elo = [(1500, 1500)]
for i in range(1, len(data)):
w = data.iloc[i - 1, :].Winner
l = data.iloc[i - 1, :].Loser
elow = elo[w]
elol = elo[l]
pwin = 1 / (1 + 10 ** ((elol - elow) / 400))
K_win = 32
K_los = 32
new_elow = elow + K_win * (1 - pwin)
new_elol = elol - K_los * (1 - pwin)
elo[w] = new_elow
elo[l] = new_elol
ranking_elo.append((elo[data.iloc[i, :].Winner], elo[data.iloc[i, :].Loser]))
if i % 5000 == 0:
print(str(i) + " matches computed...")
ranking_elo = pd.DataFrame(ranking_elo, columns=["elo_winner", "elo_loser"])
ranking_elo["proba_elo"] = 1 / (
1 + 10 ** ((ranking_elo["elo_loser"] - ranking_elo["elo_winner"]) / 400)
)
return ranking_elo
Elo = compute_elo_rankings(df_atp)
df_atp["Elo_Winner"] = Elo["elo_winner"]
df_atp["Elo_Loser"] = Elo["elo_loser"]
df_atp["Proba_Elo"] = Elo["proba_elo"]
sns.distplot(df_atp["Elo_Winner"], label="Winners")
sns.distplot(df_atp["Elo_Loser"], label="Losers")
plt.legend()
# again winners tend to win biger elo rating and loser tend to have a smaller one.
sns.distplot(df_atp["Proba_Elo"], fit=norm)
# Our distribution of probability is little skewwed trough the right.
# * Let's now drop those 'Wsets' and 'Lsets' columns as they reprensents events that happens after the matche is finished.
#
df_atp.drop(["Wsets", "Lsets"], axis=1, inplace=True)
# One last transformation of our data to augmented it and in the mean time to make it ready for modelisation and to explore it more too.
# -For each row describing a match we'll be having two resulting row. One with target variable 1 and we keep the columns as they are and one with the target variable 0 we put the invert the column of the wiinner with that of the loser and the column of the loser with that the wiinner and we keep the rest of the columsn as they are. That will double the amount of our traning data and will also transform our problem to a binary classification problem.
target_1 = np.ones(len(df_atp))
target_2 = np.zeros(len(df_atp))
target_1 = pd.DataFrame(target_1, columns=["label"])
target_2 = pd.DataFrame(target_2, columns=["label"])
from sklearn import preprocessing
from sklearn.preprocessing import OneHotEncoder
import category_encoders as ce
print(df_atp.columns)
features_categorical = df_atp[
["Series", "Court", "Surface", "Round", "Best of", "Tournament"]
].copy()
features_onehot = pd.get_dummies(features_categorical)
# tournaments_encoded = features_tournaments_encoding(df_atp)
# features_binary = pd.concat([features_categorical_encoded,tournaments_encoded],1)
## For the moment we have one row per match.
## We "duplicate" each row to have one row for each outcome of each match.
## Of course it isn't a simple duplication of each row, we need to "invert" some features
# Elo data
elo_rankings = df_atp[["Elo_Winner", "Elo_Loser", "Proba_Elo"]]
elo_1 = elo_rankings
elo_2 = elo_1[["Elo_Loser", "Elo_Winner", "Proba_Elo"]]
elo_2.columns = ["Elo_Winner", "Elo_Loser", "Proba_Elo"]
elo_2.Proba_Elo = 1 - elo_2.Proba_Elo
# Player prior win probability
win_pourcentage = df_atp[["Winner_percentage", "Loser_percentage"]]
win_1 = win_pourcentage
win_2 = win_1[["Loser_percentage", "Winner_percentage"]]
win_2.columns = ["Winner_percentage", "Loser_percentage"]
# Player prior win set probability
set_win_pourcentage = df_atp[["Winner_set_percentage", "Loser_set_percentage"]]
set_1 = set_win_pourcentage
set_2 = set_1[["Loser_set_percentage", "Winner_set_percentage"]]
set_2.columns = ["Winner_set_percentage", "Loser_set_percentage"]
# Player entry points
Pts = df_atp[["WPts", "LPts"]]
Pts_1 = Pts
Pts_2 = Pts_1[["LPts", "WPts"]]
Pts_2.columns = ["WPts", "LPts"]
# Player Entry Ranking
Rank = df_atp[["WRank", "LRank"]]
Rank_1 = Rank
Rank_2 = Rank_1[["LRank", "WRank"]]
Rank_2.columns = ["LRank", "WRank"]
# Player Odds for winning
Odds = df_atp[["EXW", "EXL", "PSW", "PSL", "B365W", "B365L"]]
Odds_1 = Odds
Odds_2 = Odds_1[["EXL", "EXW", "PSL", "PSW", "B365L", "B365W"]]
Odds_2.columns = ["EXW", "EXL", "PSW", "PSL", "B365W", "B365L"]
# Date
Date_1 = df_atp.Date
Date_2 = df_atp.Date
elo_2.index = range(1, 2 * len(elo_1), 2)
elo_1.index = range(0, 2 * len(elo_1), 2)
win_1.index = range(0, 2 * len(win_1), 2)
win_2.index = range(0, 2 * len(win_1), 2)
set_1.index = range(0, 2 * len(set_1), 2)
set_2.index = range(0, 2 * len(set_1), 2)
Pts_1.index = range(0, 2 * len(Pts_1), 2)
Pts_2.index = range(0, 2 * len(Pts_1), 2)
Rank_1.index = range(0, 2 * len(Rank_1), 2)
Rank_2.index = range(0, 2 * len(Rank_1), 2)
Odds_1.index = range(0, 2 * len(Odds_1), 2)
Odds_2.index = range(0, 2 * len(Odds_1), 2)
Date_1.index = range(0, 2 * len(Date_1), 2)
Date_2.index = range(0, 2 * len(Date_1), 2)
target_1.index = range(0, 2 * len(target_1), 2)
target_2.index = range(0, 2 * len(target_1), 2)
features_elo_ranking = pd.concat([elo_1, elo_2]).sort_index(kind="merge")
features_win_pourcentage = pd.concat([win_1, win_2]).sort_index(kind="merge")
features_set_pourcentage = pd.concat([set_1, set_2]).sort_index(kind="merge")
features_Pts = pd.concat([Pts_1, Pts_2]).sort_index(kind="merge")
features_Rank = pd.concat([Rank_1, Rank_2]).sort_index(kind="merge")
features_Odds = pd.concat([Odds_1, Odds_2]).sort_index(kind="merge")
target = pd.concat([target_1, target_2]).sort_index(kind="merge")
Date = pd.concat([Date_1, Date_2]).sort_index(kind="merge").to_frame()
"""
features_Odds.reset_index(drop=True, inplace=True)
features_elo_ranking.reset_index(drop=True, inplace=True)
#features_onehot.reset_index(drop=True, inplace=True)
features_win_pourcentage.reset_index(drop=True, inplace=True)
features_set_pourcentage.reset_index(drop=True, inplace=True)
features_set_pourcentage.reset_index(drop=True, inplace=True)
features_Pts.reset_index(drop=True, inplace=True)
features_Rank.reset_index(drop=True, inplace=True)
features_Odds.reset_index(drop=True, inplace=True)
target.reset_index(drop=True, inplace=True)
"""
features_onehot = pd.DataFrame(
np.repeat(features_onehot.values, 2, axis=0), columns=features_onehot.columns
)
features_onehot.set_index(pd.Series(range(0, 2 * len(df_atp))), inplace=True)
features_Odds.set_index(pd.Series(range(0, 2 * len(df_atp))), inplace=True)
features_elo_ranking.set_index(pd.Series(range(0, 2 * len(df_atp))), inplace=True)
features_win_pourcentage.set_index(pd.Series(range(0, 2 * len(df_atp))), inplace=True)
features_set_pourcentage.set_index(pd.Series(range(0, 2 * len(df_atp))), inplace=True)
features_Pts.set_index(pd.Series(range(0, 2 * len(df_atp))), inplace=True)
features_Rank.set_index(pd.Series(range(0, 2 * len(df_atp))), inplace=True)
features_Odds.set_index(pd.Series(range(0, 2 * len(df_atp))), inplace=True)
target.set_index(pd.Series(range(0, 2 * len(df_atp))), inplace=True)
Date.set_index(pd.Series(range(0, 2 * len(df_atp))), inplace=True)
### Building of the pre final dataset
# We can remove some features to see the effect on our model
features = pd.concat(
[
features_win_pourcentage,
features_set_pourcentage,
features_elo_ranking,
features_Pts,
features_Rank,
features_Odds,
features_onehot,
Date,
target,
],
1,
)
# Setting the 2019 matches as the test dataset.
# beg = datetime(2016,1,1)
end_train = datetime(2019, 1, 1)
beg_test = datetime(2019, 1, 1)
end_test = datetime(2020, 1, 1)
train = features[features["Date"] < end_train]
test = features[(features["Date"] >= beg_test) & (features["Date"] < end_test)]
# For saving the features
# features.to_csv("df_atp_features.csv",index=False)
# loading after saveing
# features = pd.read_csv('df_atp_features.csv')
print(len(train))
print(len(test))
# # Modeling
# Let's see what might be our most important Features at first try. .
from sklearn.ensemble import RandomForestClassifier
df = features.drop(columns=["Date", "label"])
feat_forest = RandomForestClassifier(n_jobs=-1)
feat_forest.fit(X=df, y=features["label"])
plt.figure(figsize=(10, 10))
feat_imp = feat_forest.feature_importances_
feat_imp, cols = zip(*sorted(zip(feat_imp, df.columns)))
feat_imp = np.array(feat_imp)[-30:]
cols = np.array(cols)[-30:]
d = {"feat_name": cols, "feat_imp": feat_imp}
importance = pd.DataFrame(data=d)
sns.barplot(x=importance["feat_imp"], y=importance["feat_name"])
plt.yticks(range(len(cols[-30:])), cols[-30:])
plt.title("Features Relevance for Classification")
plt.xlabel("Relevance Percentage")
# I will try to build models that are not relying on betting odds,because I've tested models relying on betting odds and the performance were great almost 98% for the best model. Let's remove those columns from the train and test set and see what we'll get; the concerned columns that needs to be removed are : 'EXW','EXL','PSW','PSL','B365W' and 'B365L'
# ## Modeling and performance evaluation:
# We will use a StratifiedKFold as our cross validation strategy(with k=10) to evaluate our models performance on the training phase. After that, we assess our models on the testing data as well and see if those score are signitifictly closer to each other or that we're over-fiting.
from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import RobustScaler
from sklearn.model_selection import (
KFold,
cross_val_score,
train_test_split,
StratifiedKFold,
)
from sklearn.metrics import (
accuracy_score,
precision_score,
recall_score,
f1_score,
make_scorer,
)
# We will be using the accuracy, precision,recall and the f1 as scores to asses our model performence
# Importing most important alogorithms
from sklearn.linear_model import LogisticRegression
from sklearn.neural_network import MLPClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.svm import (
SVC,
) # we will not be using SVM due tot he huge training time required on our dataset.
from sklearn.gaussian_process import GaussianProcessClassifier
from sklearn.gaussian_process.kernels import RBF
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier, AdaBoostClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.discriminant_analysis import (
QuadraticDiscriminantAnalysis,
LinearDiscriminantAnalysis,
)
from sklearn import model_selection # Cross-validation multiple scoring function
# features.drop(Odds_1.columns,axis=1,inplace=True)
X = train.drop(columns=["Date", "label", "EXW", "EXL", "PSW", "PSL", "B365W", "B365L"])
Y = train["label"]
# prepare configuration for cross validation test harness
seed = 42
# prepare models
models = []
models.append(("LR", LogisticRegression()))
models.append(("LDA", LinearDiscriminantAnalysis()))
models.append(("QDA", QuadraticDiscriminantAnalysis()))
models.append(("KNN", KNeighborsClassifier(5, n_jobs=-1)))
models.append(("CART", DecisionTreeClassifier(max_depth=10)))
models.append(("NB", GaussianNB()))
# models.append(('SVM_linear', SVC(kernel="linear", C=0.025)))
# models.append(('SVM_',SVC(gamma=2, C=1)))
models.append(("RandomForest", RandomForestClassifier(n_estimators=100, n_jobs=-1)))
models.append(("MLP", MLPClassifier(alpha=0.0001)))
models.append(("ADABoost", AdaBoostClassifier()))
# evaluate each model in turn
results = []
scoring = {
"accuracy": make_scorer(accuracy_score),
"precision_score": make_scorer(precision_score),
"recall_score": make_scorer(recall_score),
"f1_score": make_scorer(f1_score),
}
names = []
for name, model in models:
stratifiedKFold = model_selection.StratifiedKFold(n_splits=10, random_state=seed)
cv_results = model_selection.cross_validate(
model, X, Y, cv=stratifiedKFold, scoring=scoring
)
results.append(cv_results)
names.append(name)
msg = "-------------------------------------------------------------------------------------------------------------\n"
msg = "Model : %s \n" % (name)
msg = msg + "\n"
msg = msg + "Accuracy : %f (%f)\n" % (
cv_results["test_accuracy"].mean(),
cv_results["test_accuracy"].std(),
)
msg = msg + "Precision score : %f (%f)\n" % (
cv_results["test_precision_score"].mean(),
cv_results["test_precision_score"].std(),
)
msg = msg + "Recall score : %f (%f)\n" % (
cv_results["test_recall_score"].mean(),
cv_results["test_recall_score"].std(),
)
msg = msg + "F1 score : %f (%f)\n" % (
cv_results["test_f1_score"].mean(),
cv_results["test_f1_score"].std(),
)
msg = (
msg
+ "------------------------------------------------------------------------------------------------------------\n"
)
print(msg)
Accuracy = []
Precision = []
Recall = []
F1 = []
for idx, scores in enumerate(results):
Accuracy.append(scores["test_accuracy"])
Precision.append(scores["test_precision_score"])
Recall.append(scores["test_recall_score"])
F1.append(scores["test_f1_score"])
fig = plt.figure(figsize=(14, 12))
fig.suptitle("Algorithms Comparison")
ax = fig.add_subplot(221)
plt.boxplot(Accuracy)
plt.title("Accuracy score")
ax.set_xticklabels(names)
ax = fig.add_subplot(222)
plt.boxplot(Precision)
plt.title("Precision Score")
ax.set_xticklabels(names)
ax = fig.add_subplot(223)
plt.boxplot(Recall)
ax.set_xticklabels(names)
plt.title("Recall score")
ax = fig.add_subplot(224)
plt.title("F1 score")
plt.boxplot(F1)
ax.set_xticklabels(names)
plt.show()
# We can see clearly that almoust all our models have accuracy, precision, recaal, f1 score that are somewhat descent for a first try. But the neural network model with 100 hidden layers is the most well performing by far, followed by the CART model and followed suprisingly by K nearst neighbour classifiers. Let's see if this same performance is reflected on our test set too.
# now to test
from time import time
X_test = test.drop(
columns=["Date", "label", "EXW", "EXL", "PSW", "PSL", "B365W", "B365L"]
)
Y_test = test["label"]
y_pred = []
train_time = []
for name, model in models:
tic = time()
model.fit(X, Y)
toc = time()
y_pred.append(model.predict(X_test))
train_time.append(toc - tic)
print(
"Classifier : {} ===> Training duration : {} sec".format(name, train_time[-1])
)
reports = []
metrics = [
"Classifier",
"Accuracy",
"Precision",
"Recall",
"F1-Score",
"Training Duration (seconds)",
]
for idx, y_clf in enumerate(y_pred):
acc = accuracy_score(Y_test, y_clf)
pre = precision_score(Y_test, y_clf)
rec = recall_score(Y_test, y_clf)
f1s = f1_score(Y_test, y_clf)
report = (models[idx][0], acc, pre, rec, f1s, train_time[idx])
reports.append(report)
display(pd.DataFrame.from_records(reports, columns=metrics))
reports = pd.DataFrame.from_records(reports, columns=metrics)
plt.figure(figsize=(10, 10))
plt.plot(reports["Classifier"].values, reports["Accuracy"].values, label="Accuracy")
plt.plot(
reports["Classifier"], reports["Precision"], lw=1, alpha=0.6, label="Precision"
)
plt.plot(reports["Classifier"], reports["Recall"], lw=1, alpha=0.6, label="Recall")
plt.plot(reports["Classifier"], reports["F1-Score"], lw=1, alpha=0.6, label="F1-Score")
plt.xlabel("Algorithm")
plt.ylabel("score")
plt.title("Algorithms comparison on test set")
plt.legend(loc="lower right")
plt.show()
# The scores from the cross-validation strategy are almost close to the ones found on the test set. and as we saw in the cross valisation phase We can see clearly that almoust all our models have accuracy, precision, recaal, f1 score that are somewhat descent for a first try. But the neural network model with 100 hidden layers is the most well performing by far, followed by the Random forest and CART models wich are followed by K nearst neighbour classifiers. Let's see if this same performance is reflected on our test set too.
from sklearn.metrics import roc_curve, auc
from scipy import interp
y_prob = []
for name, model in models:
y_prob.append(model.predict_proba(X_test)[:, 1])
tprs = []
aucs = []
mean_fpr = np.linspace(0, 1, 100)
i = 0
plt.figure(figsize=(10, 10))
for idx, y_clf in enumerate(y_prob):
# Compute ROC curve and area the curve
fpr, tpr, thresholds = roc_curve(Y_test, y_clf)
tprs.append(interp(mean_fpr, fpr, tpr))
tprs[-1][0] = 0.0
roc_auc = auc(fpr, tpr)
aucs.append(roc_auc)
plt.plot(
fpr,
tpr,
lw=1,
alpha=0.6,
label="ROC Model %s (AUC = %0.2f)" % (models[idx][0], roc_auc),
)
i += 1
plt.plot([0, 1], [0, 1], linestyle="--", lw=2, color="r", label="Chance", alpha=0.7)
plt.xlim([-0.05, 1.05])
plt.ylim([-0.05, 1.05])
plt.xlabel("False Positive Rate")
plt.ylabel("True Positive Rate")
plt.title("Receiver operating characteristic")
plt.legend(loc="lower right")
plt.show()
# the neural entwork model with 100 hiddens units has the best trade-off between sensitivity (true positive rate) and specificity (1 – false positive rate ) with Area Under ROC Curve close to 1.
# # Betting
# ## Using our model for betting
# We will try to assess the return on our investement if you relied on those models for our betting decisions.
betting_columns = ["EXL", "EXW", "PSL", "PSW", "B365L", "B365W"]
# Columns containg the Odds
Betting_Odds = test[betting_columns]
# Our Capital will be 1500Euros for each strategy and for each betting site for a single model.
budget_1 = 1500
import random
def rollDice():
roll = random.randint(1, 100)
if roll == 100:
return False
elif roll <= 50:
return False
elif 100 > roll >= 50:
return True
"""
Simple bettor, betting the same amount each time. This will be our baselane.
"""
def simple_bettor(data, y_true, budget):
# return on investement for each betting site
ROI_1 = budget
ROI_2 = budget
ROI_3 = budget
wager = 10
currentWager = 0
for i in range(len(data)):
if rollDice() and y_true.values[i] == 1:
ROI_1 += wager * (data["EXW"].values[i] - 1)
ROI_2 += wager * (data["PSW"].values[i] - 1)
ROI_3 += wager * (data["B365W"].values[i] - 1)
# checking if we are already broke
if ROI_1 <= 0:
ROI_1 = -100000000000000000000
if ROI_2 <= 0:
ROI_2 = -100000000000000000000
if ROI_3 <= 0:
ROI_3 = -100000000000000000000
elif rollDice() and y_true.values[i] == 0:
ROI_1 -= wager
ROI_2 -= wager
ROI_3 -= wager
# checking if we are already broke
if ROI_1 <= 0:
ROI_1 = -100000000000000000000
if ROI_2 <= 0:
ROI_2 = -100000000000000000000
if ROI_3 <= 0:
ROI_3 = -100000000000000000000
elif not rollDice() and y_true.values[i] == 0:
ROI_1 += wager * (data["EXL"].values[i] - 1)
ROI_2 += wager * (data["PSL"].values[i] - 1)
ROI_3 += wager * (data["B365L"].values[i] - 1)
# checking if we are already broke
if ROI_1 <= 0:
ROI_1 = -100000000000000000000
if ROI_2 <= 0:
ROI_2 = -100000000000000000000
if ROI_3 <= 0:
ROI_3 = -100000000000000000000
else:
ROI_1 -= wager
ROI_2 -= wager
ROI_3 -= wager
# checking if we are already broke
if ROI_1 <= 0:
ROI_1 = -100000000000000000000
if ROI_2 <= 0:
ROI_2 = -100000000000000000000
if ROI_3 <= 0:
ROI_3 = -100000000000000000000
if ROI_1 < 0:
ROI_1 = 0
if ROI_2 < 0:
ROI_2 = 0
if ROI_3 < 0:
ROI_3 = 0
return [
(ROI_1 - budget) / budget,
(ROI_2 - budget) / budget,
(ROI_3 - budget) / budget,
]
# If our model predict that a player is going to win, we'll invest 10Euros on that match for that player winning
# and compare it with the real value to see if we won or lost
def strategy_1(data, y_pred, y_true, budget):
"""
If our model predict that a player is going to win, we'll invest 10Euros on that match for that player winning
and compare it with the real value to see if we won or lost
"""
# Retrun on investement for each betting site
ROI_1 = budget
ROI_2 = budget
ROI_3 = budget
for i in range(0, len(test)):
if y_pred[i] == 1 and y_true.values[i] == 1.0:
ROI_1 += 10 * (data["EXW"].values[i] - 1)
ROI_2 += 10 * (data["PSW"].values[i] - 1)
ROI_3 += 10 * (data["B365W"].values[i] - 1)
# checking if we are already broke
if ROI_1 <= 0:
ROI_1 = -100000000000000000000
if ROI_2 <= 0:
ROI_2 = -100000000000000000000
if ROI_3 <= 0:
ROI_3 = -100000000000000000000
elif y_pred[i] == 1 and y_true.values[i] == 0.0:
ROI_1 += -10
ROI_2 += -10
ROI_3 += -10
# checking if we are already broke
if ROI_1 <= 0:
ROI_1 = -100000000000000000000
if ROI_2 <= 0:
ROI_2 = -100000000000000000000
if ROI_3 <= 0:
ROI_3 = -100000000000000000000
elif y_pred[i] == 0 and y_true.values[i] == 1.0:
# checking if we are already broke
ROI_1 += -10
ROI_2 += -10
ROI_3 += -10
if ROI_1 <= 0:
ROI_1 = -100000000000000000000
if ROI_2 <= 0:
ROI_2 = -100000000000000000000
if ROI_3 <= 0:
ROI_3 = -100000000000000000000
else:
ROI_1 += 10 * (data["EXL"].values[i] - 1)
ROI_2 += 10 * (data["PSL"].values[i] - 1)
ROI_3 += 10 * (data["B365L"].values[i] - 1)
# checking if we are already broke
if ROI_1 <= 0:
ROI_1 = -100000000000000000000
if ROI_2 <= 0:
ROI_2 = -100000000000000000000
if ROI_3 <= 0:
ROI_3 = -100000000000000000000
if ROI_1 < 0:
ROI_1 = 0
if ROI_2 < 0:
ROI_2 = 0
if ROI_3 < 0:
ROI_3 = 0
return [
(ROI_1 - budget) / budget,
(ROI_2 - budget) / budget,
(ROI_3 - budget) / budget,
]
def strategy_2(data, y_proba, y_true, budget):
"""
In each match we'll invest 10(probability_player_win)Euros for the player winning, and 10(probability_player_lose)Euros
for the player losing
"""
ROI_1 = budget
ROI_2 = budget
ROI_3 = budget
for i in range(0, len(test)):
if y_true.values[i] == 1.0:
ROI_1 += (
y_proba[i] * 10 * (data["EXW"].values[i] - 1) - (1 - y_proba[i]) * 10
)
ROI_2 += (
y_proba[i] * 10 * (data["PSW"].values[i] - 1) - (1 - y_proba[i]) * 10
)
ROI_3 += (
y_proba[i] * 10 * (data["B365W"].values[i] - 1) - (1 - y_proba[i]) * 10
)
# checking if we are already broke
if ROI_1 <= 0:
ROI_1 = -100000000000000000000
if ROI_2 <= 0:
ROI_2 = -100000000000000000000
if ROI_3 <= 0:
ROI_3 = -100000000000000000000
else:
ROI_1 += (1 - y_proba[i]) * 10 * (data["EXL"].values[i] - 1) - y_proba[
i
] * 10
ROI_2 += (1 - y_proba[i]) * 10 * (data["PSL"].values[i] - 1) - y_proba[
i
] * 10
ROI_3 += (1 - y_proba[i]) * 10 * (data["B365L"].values[i] - 1) - y_proba[
i
] * 10
# checking if we are already broke
if ROI_1 <= 0:
ROI_1 = -100000000000000000000
if ROI_2 <= 0:
ROI_2 = -100000000000000000000
if ROI_3 <= 0:
ROI_3 = -100000000000000000000
if ROI_1 < 0:
ROI_1 = 0
if ROI_2 < 0:
ROI_2 = 0
if ROI_3 < 0:
ROI_3 = 0
return [
(ROI_1 - budget) / budget,
(ROI_2 - budget) / budget,
(ROI_3 - budget) / budget,
]
# P.S: Seing how we contructed the dataset (Each row repeated one time). We'll be actualy investing 20Euros of our capital in each match instead of 10
# Our Capital will be 1500Euros for each strategy and for each betting site for a single model.
reports = []
metrics = [
"Classifier",
"Strat 1 EX",
"Strat 2 EX",
"Strat 1 PS",
"Strat 2 PS",
"Strat 1 B365",
"Strat 2 B365",
"Random EX",
"Random PS",
"Random B365",
]
for idx, y_clf in enumerate(y_pred):
Random = simple_bettor(Betting_Odds, Y_test, budget_1)
strat_1 = strategy_1(Betting_Odds, y_clf, Y_test, budget_1)
strat_2 = strategy_2(Betting_Odds, y_prob[idx], Y_test, budget_1)
report = (
models[idx][0],
strat_1[1],
strat_2[1],
strat_1[1],
strat_2[1],
strat_1[2],
strat_2[2],
Random[0],
Random[1],
Random[2],
)
reports.append(report)
display(pd.DataFrame.from_records(reports, columns=metrics))
reports = pd.DataFrame.from_records(reports, columns=metrics)
plt.figure(figsize=(10, 10))
plt.plot(
reports["Classifier"].values, reports["Strat 1 EX"].values, label="EX : Strategy 1"
)
plt.plot(
reports["Classifier"],
reports["Strat 2 EX"],
lw=1,
alpha=0.6,
label="EX : Strategy 2",
)
plt.plot(
reports["Classifier"],
reports["Strat 1 PS"],
lw=1,
alpha=0.6,
label="PS : Strategy 1",
)
plt.plot(
reports["Classifier"],
reports["Strat 2 PS"],
lw=1,
alpha=0.6,
label="PS : Strategy 2",
)
plt.plot(
reports["Classifier"],
reports["Strat 1 B365"],
lw=1,
alpha=0.6,
label="B365 : Strategy 1",
)
plt.plot(
reports["Classifier"],
reports["Strat 2 B365"],
lw=1,
alpha=0.6,
label="B365 : Strategy 2",
)
plt.xlabel("Algorithm")
plt.ylabel("Return on investement")
plt.title("Algorithms ROI on Test set")
plt.legend(loc="lower right")
plt.show()
|
import numpy as np
import pandas as pd
data = pd.read_csv("/kaggle/input/salary-data-simple-linear-regression/Salary_Data.csv")
data.info()
import statsmodels.api as sm
X = sm.add_constant(data["YearsExperience"])
X.head(5)
y = data["Salary"]
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, train_size=0.7, random_state=42
)
salary_lm = sm.OLS(y_train, X_train).fit()
salary_lm.params
salary_lm.summary2()
import matplotlib.pyplot as plt
import seaborn as sn
from scipy import stats
resid = salary_lm.resid
probplot = sm.ProbPlot(np.array(resid))
plt.figure(figsize=(10, 8))
probplot.ppplot(line="45")
plt.show()
def get_standarized_values(vals):
return (vals - vals.mean()) / vals.std()
plt.scatter(
get_standarized_values(salary_lm.fittedvalues), get_standarized_values(resid)
)
plt.xlabel("Standarized predicted values")
plt.ylabel("Standarized Residuals")
plt.show()
from scipy.stats import zscore
data["z_score_salary"] = zscore(data.Salary)
data[(data.z_score_salary > 3.0) | (data.z_score_salary < -3.0)]
data_influence = salary_lm.get_influence()
(c, p) = data_influence.cooks_distance
plt.stem(np.arange(len(X_train)), np.round(c, 3), markerfmt=",")
plt.xlabel("Row index")
plt.ylabel("Cooks Distance")
from statsmodels.graphics.regressionplots import influence_plot
fig, ax = plt.subplots(figsize=(10, 8))
influence_plot(salary_lm, ax=ax)
plt.show()
y_pred = salary_lm.predict(X_test)
from sklearn.metrics import r2_score, mean_squared_error
r2_score(y_test, y_pred)
# r2_score(y_pred , y_test) slightly different
np.sqrt(mean_squared_error(y_test, y_pred))
|
# # Finding Pulsar Stars
# Here we have a dataset with 15 thousand stars. Our task is to judge whether a star is a pulsar or not based on certain features that the dataset has given to us about them. A pulsar is a rare type of star which produces radio emission that is detectable on Earth.
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
dataset = pd.read_csv("/kaggle/input/predicting-a-pulsar-star/pulsar_stars.csv")[:15000]
X = np.array([np.array(dataset).T[0], np.array(dataset).T[4]])
y = np.array(dataset).T[8]
X = X.astype(np.float64)
y = y.astype(int)
dataset.head()
# This dataset has 8 features which we can use to predict the target_class. We will simply be using two for this classification: 'Mean of the integrated profile' and 'Mean of the DM-SNR curve'.
# # Plotting data
# For visualising data, we will first use a scatter plot, then a bar graph.
import matplotlib.pyplot as plt
import collections
plt.scatter(X[0], X[1], c=y)
plt.title("Stars", fontsize=15)
plt.xlabel("Mean of the integrated profile", fontsize=13)
plt.ylabel("Mean of the DM-SNR curve", fontsize=13)
plt.show()
plt.bar(range(0, 2), [collections.Counter(y)[0], collections.Counter(y)[1]])
plt.title("Stars", fontsize=15)
plt.xlabel("Regular Pulsar", fontsize=13)
plt.ylabel("Number of stars", fontsize=13)
plt.show()
# The datapoints have a relatively 'classifiable' border, as shown in the scatter plot. As for the bar plot, we can see that the pulsar stars are much less in numbers compared to regular stars.
# # Splitting data and creating models
# Next up, we will use scikit-learn's train_test_split to split the data into train and test sets, which will help our model's accuracy. Afterwards, we will create multiple models which will later be used for fitting the train data.
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X.T, y, test_size=0.2, random_state=1
)
from sklearn.neighbors import KNeighborsClassifier as knn
from sklearn.tree import DecisionTreeClassifier as dtc
from sklearn.svm import SVC
from sklearn.ensemble import RandomForestClassifier as rfc
classifiers = {
"Random Forest Classifier": rfc(n_estimators=100),
"K Nearest Neighbors": knn(),
"Support Vector Classifier": SVC(gamma="scale"),
"Decision Tree": dtc(),
}
for i in classifiers.items():
model = i[1]
model.fit(X_train, y_train)
# # Visualising and presenting accuracy
# Now that our algorithms have been fitted, we will visualise our classification and present our accuracy. We will scatter the data using mlxtend's plot_decision_regions, followed by showing the accuracy using cross validation score, model score and mean absolute error.
from mlxtend.plotting import plot_decision_regions
from sklearn.model_selection import cross_val_score as cvs
from sklearn.metrics import mean_absolute_error
from sklearn.metrics import make_scorer
from sklearn.metrics import accuracy_score
for i in classifiers.items():
model = i[1]
scores = cvs(model, X_test, y_test, cv=5).mean()
pred = model.predict(X_test)
mae = mean_absolute_error(y_test, pred)
print(i[0])
print("Cross validation score: " + str(scores))
print("Model score: " + str(model.score(X_test, y_test)))
print("Mean absolute error: " + str(mae))
plot_decision_regions(X.T, y, clf=model)
plt.title(i[0], fontsize=15)
plt.xlabel("Mean of the integrated profile", fontsize=13)
plt.ylabel("Mean of the DM-SNR curve", fontsize=13)
plt.show()
model = list(classifiers.items())[1][1]
pred = model.predict(X_test)
submis = pd.DataFrame({"Real": y_test, "Prediction": pred})
submis.to_csv("submission.csv", index=False)
|
# # EDA for Wuhan Coronavirus, pray for Wuhan, Hubei, China, World
# **About Wuhan**: Wuhan is a big city in Hubei,China, about 9000,000 peoples live in here. It is a important part of China forever.
# **Abount me**: Hi, my name is 何龙, u can call me Nemo, I live in Shenzhen,Guangdong,China, and many of my friends and colleagues are from Wuhan or Hubei, I'm very worried about them, pray for them and hope them well, and I always believed that our government could save us from this disaster, so go Wuhan, go China, go World.
# **NOW**: Confirmed:24377, Death:492, Recovered:901 in Chine, as of 2020/02/05 14:52.
# From WHO:
# > We must remember that these are people, not numbers.
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
import seaborn as sns
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
cov_data = pd.read_csv("../input/novel-corona-virus-2019-dataset/2019_nCoV_data.csv")
cov_data.Date = pd.to_datetime(cov_data.Date)
cov_data["Last Update"] = pd.to_datetime(cov_data["Last Update"])
whole_china = ["China"]
parts_china = ["Mainland China", "Macau", "Hong Kong", "Taiwan"]
data_whole = cov_data[~cov_data.Country.isin(parts_china)].copy()
data_parts = cov_data[~cov_data.Country.isin(whole_china)].copy()
world_coordinates = pd.read_csv("../input/world-coordinates/world_coordinates.csv")
# ## Take a look
# This data is base on province, not city, so Hubei will replace Wuhan in my notebook.
# Hubei has 13522 confirmed, 414 Deaths, 396 Recovered in 2020-02-04.
data_parts.sort_values(by="Confirmed", ascending=False)
# ## Fix 'Last Update' bigger than 2020-02-05
# The 'Last Update' column has some data 'day' and 'month' reversed.
data_parts["LastUpdateFixed"] = data_parts["Last Update"].astype("object")
data_parts = data_parts.drop("Last Update", axis=1)
data_parts["LastUpdateFixed"] = data_parts["LastUpdateFixed"].apply(
lambda lu: lu
if not ("2020-03" in str(lu) or "2020-04" in str(lu))
else (
(str(lu).split(" ")[0].split("-")[0])
+ "-"
+ (str(lu).split(" ")[0].split("-")[2])
+ "-"
+ (str(lu).split(" ")[0].split("-")[1])
)
+ " "
+ str(lu).split(" ")[1]
)
# ## Analysis with Country(and Hong Kong, Taiwan, Macau)
tmp = (
data_parts.drop("Sno", axis=1)
.groupby("Country")
.max()
.sort_values(by="Confirmed", ascending=False)
)
print(
"Confirmed Mainland Chind / World : {%d}/{%d}, about %d%% in Mainland Chine."
% (
tmp.iloc[0]["Confirmed"],
tmp["Confirmed"].sum(),
int(tmp.iloc[0]["Confirmed"] / tmp["Confirmed"].sum() * 100),
)
)
tmp.iloc[:10]
# We also can see that Top 10 countries are all from asia but Germany, so I think that is a good new for the whole world. That means the virus doesn't spread around the world for now.
# ## Analysis with Mainland China
tmp = (
data_parts.query('Country=="Mainland China"')
.drop("Sno", axis=1)
.groupby("Province/State")
.max()
.sort_values(by="Confirmed", ascending=False)
)
print(
"Confirmed Hubei / Mainland China : {%d}/{%d}, about %d%% in Hubei."
% (
tmp.iloc[0]["Confirmed"],
tmp["Confirmed"].sum(),
int(tmp.iloc[0]["Confirmed"] / tmp["Confirmed"].sum() * 100),
)
)
tmp.iloc[:10]
# But when we look at the Mainland China, thing has been changed, only 66% in Hubei, that mean about 7,000 of Confirmed peoples from other provinces in Mainland China, 829 in Zhejiang, 797 in Guangdong(Where I am), etc. Now we know that coronavirus is more infectious than SARS, we have long way to go.
# ## Analysis with Hubei
data_parts["Province"] = data_parts["Province/State"]
tmp = (
data_parts.query('Country=="Mainland China" and Province=="Hubei"')
.drop(["Sno", "Province/State"], axis=1)
.sort_values(by="Date")
)
plt.subplots(figsize=(20, 4))
plt.subplot(1, 4, 1)
plt.xticks(rotation=40)
sns.lineplot(x="Date", y="Confirmed", data=tmp)
plt.subplot(1, 4, 2)
plt.xticks(rotation=40)
sns.lineplot(x="Date", y="Deaths", data=tmp)
plt.subplot(1, 4, 3)
plt.xticks(rotation=40)
sns.lineplot(x="Date", y="Recovered", data=tmp)
plt.subplot(1, 4, 4)
plt.xticks(rotation=40)
tmp["Deaths/Recovered"] = tmp["Deaths"] / tmp["Recovered"]
sns.lineplot(x="Date", y="Deaths/Recovered", data=tmp)
|
# 1. Student Name : Louis(Yu Liu)
#
# 2. Student ID : 19448104
import numpy as np
import pandas as pd
import xlrd
import matplotlib.pyplot as plt
import scipy as sp
import chart_studio
import simfin as sf
import os
import seaborn as sns
sns.set()
import time
import chart_studio.plotly as py
import plotly.graph_objs as go
from statsmodels.graphics.tsaplots import plot_acf
from statsmodels.tsa.stattools import adfuller as ADF
from statsmodels.graphics.tsaplots import plot_pacf
from statsmodels.stats.diagnostic import acorr_ljungbox
from statsmodels.tsa.arima_model import ARIMA
import pandas_datareader as web
import datetime
import matplotlib.pyplot as plt
from matplotlib import ticker as mticker
import mpl_finance as mpl
# download data
def DownloadData(Ticker, FileSavePath):
sf.set_api_key("free")
sf.set_data_dir(FileSavePath)
Income_Statement = sf.load_income(variant="annual")
Cashflow_Statement = sf.load_cashflow(variant="annual", market="us")
Balance_Statement = sf.load_balance(variant="annual", market="us")
DataInformation_Income = Income_Statement.loc[Ticker]
DataInformation_Cashflow = Cashflow_Statement.loc[Ticker]
DataInformation_Balance = Balance_Statement.loc[Ticker]
Income_SavePath = [FileSavePath, r"\\", r"Income.csv"]
Cashflow_SavePath = [FileSavePath, r"\\", r" Cashflow.csv"]
Balance_SavePath = [FileSavePath, r"\\", r" Balance.csv"]
Income_SavePath = "".join(Income_SavePath)
Cashflow_SavePath = "".join(Cashflow_SavePath)
Balance_SavePath = "".join(Balance_SavePath)
DataInformation_Income.to_csv(Income_SavePath)
DataInformation_Cashflow.to_csv(Cashflow_SavePath)
DataInformation_Balance.to_csv(Balance_SavePath)
print("\n")
print(DataInformation_Income.head())
[print(x) for x in DataInformation_Income.columns]
print(DataInformation_Cashflow.head())
[print(x) for x in DataInformation_Cashflow.columns]
print(DataInformation_Balance.head())
[print(x) for x in DataInformation_Balance.columns]
DataInformation_Income.index = DataInformation_Income["Fiscal Year"]
DataInformation_Balance.index = DataInformation_Balance["Fiscal Year"]
DataInformation_Cashflow.index = DataInformation_Cashflow["Fiscal Year"]
return DataInformation_Income, DataInformation_Cashflow, DataInformation_Balance
Ticker = "NVDA"
FileSavePath = os.getcwd()
(
DataInformation_Income,
DataInformation_Cashflow,
DataInformation_Balance,
) = DownloadData(Ticker, FileSavePath)
# read the data
def ReadLoadData(Ticker, FileSavePath):
N = """
def FindOutTheCurrentPath(Path):
Balance_Sheet_Path = ''.join([Path, r'\\', r'Balance.csv'])
CashFlow_Sheet_Path = ''.join([Path, r'\\', r'Cashflow.csv'])
Income_Sheet_Path = ''.join([Path, r'\\', r'Income.csv'])
return Balance_Sheet_Path, CashFlow_Sheet_Path, Income_Sheet_Path
"""
Balance_SavePath, Cashflow_SavePath, Income_SavePath = DownloadData(
Ticker, FileSavePath
)
N = """
DataInformation_Income = pd.read_csv(Income_SavePath)
DataInformation_Cashflow = pd.read_csv(Cashflow_SavePath)
DataInformation_Balance = pd.read_csv(Balance_SavePath)
"""
DataInformation_Income.index = DataInformation_Income["Fiscal Year"]
DataInformation_Balance.index = DataInformation_Balance["Fiscal Year"]
DataInformation_Cashflow.index = DataInformation_Cashflow["Fiscal Year"]
print("Income Statement", "\n", DataInformation_Income.head())
print("The list of Income Statement Columns:")
[print(x) for x in DataInformation_Income.columns]
print("\n", "Cash flow Statement", "\n", DataInformation_Cashflow.head())
print("The list of Cash flow Statement Columns:")
[print(x) for x in DataInformation_Cashflow.columns]
print("\n", "Balance Statement", "\n", DataInformation_Balance.head())
print("The list of Balance Sheet Columns:")
[print(x) for x in DataInformation_Balance.columns]
return DataInformation_Income, DataInformation_Cashflow, DataInformation_Balance
def excel_to_df(excel_sheet):
df = pd.read_excel(excel_sheet)
df.dropna(how="all", inplace=True)
index_PL = int(
df.loc[df["Data provided by SimFin"] == "Profit & Loss statement"].index[0]
)
index_CF = int(
df.loc[df["Data provided by SimFin"] == "Cash Flow statement"].index[0]
)
index_BS = int(df.loc[df["Data provided by SimFin"] == "Balance Sheet"].index[0])
df_PL = df.iloc[index_PL : index_BS - 1, 1:]
df_PL.dropna(how="all", inplace=True)
df_PL.columns = df_PL.iloc[0]
df_PL = df_PL[1:]
df_PL.set_index("in million USD", inplace=True)
(df_PL.fillna(0, inplace=True))
df_BS = df.iloc[index_BS - 1 : index_CF - 2, 1:]
df_BS.dropna(how="all", inplace=True)
df_BS.columns = df_BS.iloc[0]
df_BS = df_BS[1:]
df_BS.set_index("in million USD", inplace=True)
df_BS.fillna(0, inplace=True)
df_CF = df.iloc[index_CF - 2 :, 1:]
df_CF.dropna(how="all", inplace=True)
df_CF.columns = df_CF.iloc[0]
df_CF = df_CF[1:]
df_CF.set_index("in million USD", inplace=True)
df_CF.fillna(0, inplace=True)
df_CF = df_CF.T
df_BS = df_BS.T
df_PL = df_PL.T
return df, df_PL, df_BS, df_CF
def combine_regexes(regexes):
return "(" + ")|(".join(regexes) + ")"
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
import scipy.stats
import re
from xgboost import XGBClassifier
from sklearn.model_selection import GridSearchCV
pd.set_option("display.max_rows", 50, "display.max_columns", 50)
df_train = pd.read_csv("../input/titanic/train.csv")
df_test = pd.read_csv("../input/titanic/test.csv")
PassengerId = df_test["PassengerId"]
sample_subm = pd.read_csv("../input/titanic/gender_submission.csv")
df_train.head()
plt.figure(figsize=(6, 3))
df_train.Survived.value_counts().plot(kind="bar", rot=360)
plt.show()
# We can see a bit of an imbalance for our target variable
# # Feature engineering
# [Titanic Best Working Classfier](https://www.kaggle.com/sinakhorami/titanic-best-working-classifier) : by Sina
full_data = [df_train, df_test]
# Feature that tells whether a passenger had a cabin on the Titanic
df_train["Has_Cabin"] = df_train["Cabin"].apply(lambda x: 0 if type(x) == float else 1)
df_test["Has_Cabin"] = df_test["Cabin"].apply(lambda x: 0 if type(x) == float else 1)
# Feature engineering steps taken from Sina
# Create new feature FamilySize as a combination of SibSp and Parch
for dataset in full_data:
dataset["FamilySize"] = dataset["SibSp"] + dataset["Parch"] + 1
# Create new feature IsAlone from FamilySize
for dataset in full_data:
dataset["IsAlone"] = 0
dataset.loc[dataset["FamilySize"] == 1, "IsAlone"] = 1
# Remove all NULLS in the Embarked column
for dataset in full_data:
dataset["Embarked"] = dataset["Embarked"].fillna("S")
# Remove all NULLS in the Fare column and create a new feature CategoricalFare
for dataset in full_data:
dataset["Fare"] = dataset["Fare"].fillna(df_train["Fare"].median())
df_train["CategoricalFare"] = pd.qcut(df_train["Fare"], 4)
# Create a New feature CategoricalAge
for dataset in full_data:
age_avg = dataset["Age"].mean()
age_std = dataset["Age"].std()
age_null_count = dataset["Age"].isnull().sum()
age_null_random_list = np.random.randint(
age_avg - age_std, age_avg + age_std, size=age_null_count
)
dataset["Age"][np.isnan(dataset["Age"])] = age_null_random_list
dataset["Age"] = dataset["Age"].astype(int)
df_train["CategoricalAge"] = pd.cut(df_train["Age"], 5)
# Define function to extract titles from passenger names
def get_title(name):
title_search = re.search(" ([A-Za-z]+)\.", name)
# If the title exists, extract and return it.
if title_search:
return title_search.group(1)
return ""
# Create a new feature Title, containing the titles of passenger names
for dataset in full_data:
dataset["Title"] = dataset["Name"].apply(get_title)
# Group all non-common titles into one single grouping "Rare"
for dataset in full_data:
dataset["Title"] = dataset["Title"].replace(
[
"Lady",
"Countess",
"Capt",
"Col",
"Don",
"Dr",
"Major",
"Rev",
"Sir",
"Jonkheer",
"Dona",
],
"Rare",
)
dataset["Title"] = dataset["Title"].replace("Mlle", "Miss")
dataset["Title"] = dataset["Title"].replace("Ms", "Miss")
dataset["Title"] = dataset["Title"].replace("Mme", "Mrs")
for dataset in full_data:
# Mapping Sex
dataset["Sex"] = dataset["Sex"].map({"female": 0, "male": 1}).astype(int)
# Mapping titles
title_mapping = {"Mr": 1, "Miss": 2, "Mrs": 3, "Master": 4, "Rare": 5}
dataset["Title"] = dataset["Title"].map(title_mapping)
dataset["Title"] = dataset["Title"].fillna(0)
# Mapping Embarked
dataset["Embarked"] = dataset["Embarked"].map({"S": 0, "C": 1, "Q": 2}).astype(int)
# Mapping Fare
dataset.loc[dataset["Fare"] <= 7.91, "Fare"] = 0
dataset.loc[(dataset["Fare"] > 7.91) & (dataset["Fare"] <= 14.454), "Fare"] = 1
dataset.loc[(dataset["Fare"] > 14.454) & (dataset["Fare"] <= 31), "Fare"] = 2
dataset.loc[dataset["Fare"] > 31, "Fare"] = 3
dataset["Fare"] = dataset["Fare"].astype(int)
# Mapping Age
dataset.loc[dataset["Age"] <= 16, "Age"] = 0
dataset.loc[(dataset["Age"] > 16) & (dataset["Age"] <= 32), "Age"] = 1
dataset.loc[(dataset["Age"] > 32) & (dataset["Age"] <= 48), "Age"] = 2
dataset.loc[(dataset["Age"] > 48) & (dataset["Age"] <= 64), "Age"] = 3
dataset.loc[dataset["Age"] > 64, "Age"] = 4
# Feature selection
drop_elements = ["PassengerId", "Name", "Ticket", "Cabin", "SibSp"]
df_train = df_train.drop(drop_elements, axis=1)
df_train = df_train.drop(["CategoricalAge", "CategoricalFare"], axis=1)
df_test = df_test.drop(drop_elements, axis=1)
# features = ['Pclass', 'Sex', 'Embarked']
features = df_train.columns[1:]
fig = plt.figure(figsize=(15, 13))
for i in range(len(features)):
fig.add_subplot(4, 3, i + 1)
sns.countplot(x=features[i], hue="Survived", data=df_train)
plt.show()
df_train.head()
df_train = pd.get_dummies(df_train, columns=["Title", "FamilySize", "Embarked"])
y_train = df_train.Survived
X_train = df_train.loc[:, features]
X_test = df_test
params = {
"tree_method": ["gpu_hist"],
"n_estimators": [2000],
"n_jobs": [1],
"scale_pos_weight": [1],
"base_score": [0.5],
"learning_rate": [0.1],
"reg_alpha": [0],
"reg_lambda": [1],
"gamma": [0],
"max_delta_step": [0],
"max_depth": [50],
"objective": ["binary:logistic"],
"random_state": [0],
}
xgb_model = XGBClassifier()
clf = GridSearchCV(
estimator=xgb_model,
param_grid=params,
cv=5,
verbose=0,
scoring="accuracy",
return_train_score=True,
)
clf.fit(X_train, y_train)
clf.cv_results_["mean_test_score"][
0
] # 0.81032548, 0.792368125701459, 0.8080808080808081, 0.7968574635241302, 0.7934904601571269, 0.8035914702581369
y_pred = clf.predict(X_test)
submission = pd.DataFrame({"PassengerId": PassengerId, "Survived": y_pred})
submission.to_csv("submission.csv", index=False)
|
# import package
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import missingno as msno
import cufflinks as cf
cf.set_config_file(
theme="ggplot",
# sharing='public',
offline=True,
dimensions=(500, 300),
offline_show_link=False,
)
import plotly.express as px
import plotly.graph_objects as go
from plotly.subplots import make_subplots
from category_encoders.ordinal import OrdinalEncoder
from category_encoders.target_encoder import TargetEncoder
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
#
# 🌟 EDA with Plotly & Target Encoding
#
#
#
# - 1. Import Data
# - 2. Feature exploration and visualization
# - 2.1 Total Distribution
# - 2.2 Binary Feature
# - 2.3 Nominal Feature
# - 2.4 Ordinal Feature
# - 2.5 Day & Month Feature
# - 3. Feature Encoding
# - 3.1 Binary Feature
# - 3.2 Nominal Feature
# - 3.3 Ordinal Feature
# - 3.4 Day & Month Feature
# > I learn a lot from [Subin's kernal](https://www.kaggle.com/subinium/categorical-data-eda-visualization), and use parts of code in his kernels, please give credit his kernal.Really appreciate your feedback.
# ## 1 Import Data
# Top
train = pd.read_csv("../input/cat-in-the-dat-ii/train.csv")
test = pd.read_csv("../input/cat-in-the-dat-ii/test.csv")
train.head()
target, train_id = train["target"], train["id"]
test_id = test["id"]
train.drop(["id"], axis=1, inplace=True)
test.drop(["id"], axis=1, inplace=True)
# **feature list**
# It's important to know what each feature is, because you need to check how you encode or distribute based on the feature.
# - **bin 0~4** : Binary Feature, label encoding
# - **nom 0~9** : Nominal Feature
# - **ord 0~5** : Ordinal Feature
# - **day/month** : Date, cycle encoding
# ## 2 Feature exploration and visualization
# ### 2.1 Total Distribution
# plot the missing values
msno.matrix(train)
# Plot NULL rate of training data
null_rate = [train[i].isna().sum() / len(train) for i in train.columns]
data = {"train_column": train.columns, "null_rate": null_rate}
train_null_rate = pd.DataFrame(data)
fig = px.bar(
train_null_rate,
x="train_column",
y="null_rate",
text="null_rate",
title="Binary Feature Distribution (Train Data)",
)
fig.update_traces(textposition="outside", texttemplate="%{text:.3p}", textfont_size=20)
fig.update_layout(yaxis_tickformat=".2%")
fig.add_shape(
go.layout.Shape(
type="line",
yref="y",
y0=0.03,
y1=0.03,
xref="x",
x0=-1,
x1=23.5,
line=dict(color="DarkOrange", width=1.5, dash="dash"),
)
)
fig.update_layout(
margin=dict(l=25, t=50, b=0), legend_orientation="h", width=500, height=350
) # this size is for kaggle kernal
fig.show()
# Plot NULL rate of training data
null_rate = [train[i].isna().sum() / len(train) for i in train.columns]
data = {"train_column": train.columns, "null_rate": null_rate}
train_null_rate = pd.DataFrame(data).drop(23)
fig = px.bar(
train_null_rate,
x="train_column",
y="null_rate",
text="null_rate",
color="null_rate",
title="Feature Null Rate (Train Data)",
)
fig.update_traces(textposition="outside", texttemplate="%{text:.2p}", textfont_size=20)
fig.update_layout(yaxis_tickformat="%")
fig.add_shape(
go.layout.Shape(
type="line",
yref="y",
y0=0.03,
y1=0.03,
xref="x",
x0=-1,
x1=23.5,
line=dict(color="gray", width=1.5, dash="dash"),
)
)
fig.update(layout_coloraxis_showscale=False) # hide colorscale
fig.update_layout(
margin=dict(l=25, t=50, b=0), legend_orientation="h", width=500, height=350
) # this size is for kaggle kernal
fig.show()
# First of all, you can see that the target ratio is unbalanced, percentage of target 1 is about 18.7%
# Plot Target Distribution
target.iplot(
kind="histogram",
histnorm="probability",
title="Total Target Distribution",
bargap=0.5,
)
# ### 2.2 Binary Feature
# Plot binary feature distribution for Training Data
sub_title = list(
train[[col for col in train.columns if col.startswith("bin_")]].columns
)
fig = make_subplots(rows=1, cols=5, subplot_titles=sub_title)
for i in range(5):
a = train[f"bin_{i}"].value_counts()
fig.add_trace(
go.Bar(
x=a.index,
y=a.values,
width=0.2,
text=a.values,
textposition="outside",
texttemplate="%{text:.2s}",
name=f"bin_{i}",
showlegend=False,
textfont=dict(size=5),
textangle=0,
constraintext="inside",
marker=dict(
color=["#732FB0", "#2FB09D"], line_width=0.5, line_color="gray"
),
),
row=1,
col=i + 1,
)
fig.update_layout(title_text="Binary Feature Distribution (Train Data)")
# Hide the yaxis
fig.update_layout(
yaxis=dict(visible=False),
yaxis2=dict(visible=False),
yaxis3=dict(visible=False),
yaxis4=dict(visible=False),
yaxis5=dict(visible=False),
)
# this size is for kaggle kernal
fig.update_layout(
margin=dict(l=25, t=50, b=0), legend_orientation="h", width=500, height=350
)
# Set the subtitle size
for i in fig["layout"]["annotations"]:
i["font"] = dict(size=10, color="black")
# fig.show(config={"showLink":True})
fig.show()
# Plot binary feature distribution for Training Data
sub_title = list(train[[col for col in test.columns if col.startswith("bin_")]].columns)
fig = make_subplots(rows=1, cols=5, subplot_titles=sub_title)
for i in range(5):
a = test[f"bin_{i}"].value_counts()
fig.add_trace(
go.Bar(
x=a.index,
y=a.values,
width=0.2,
text=a.values,
textposition="outside",
texttemplate="%{text:.2s}",
name=f"bin_{i}",
showlegend=False,
textfont=dict(size=5),
textangle=0,
constraintext="inside",
marker=dict(
color=["#732FB0", "#2FB09D"], line_width=0.5, line_color="gray"
),
),
row=1,
col=i + 1,
)
fig.update_layout(title_text="Binary Feature Distribution (Test Data)")
# Hide the yaxis
fig.update_layout(
yaxis=dict(visible=False),
yaxis2=dict(visible=False),
yaxis3=dict(visible=False),
yaxis4=dict(visible=False),
yaxis5=dict(visible=False),
)
# this size is for kaggle kernal
fig.update_layout(
margin=dict(l=25, t=50, b=0), legend_orientation="h", width=500, height=350
)
# Set the subtitle size
for i in fig["layout"]["annotations"]:
i["font"] = dict(size=10, color="black")
# fig.show(config={"showLink":True})
fig.show()
# Plot Binary Feature Target Distribution for Training Data
sub_title = list(train[[col for col in test.columns if col.startswith("bin_")]].columns)
fig = make_subplots(rows=1, cols=5, subplot_titles=sub_title)
for i in range(5):
a = train.groupby([f"bin_{i}", "target"]).size().to_frame().reset_index()
a_0 = a[a["target"] == 0]
a_1 = a[a["target"] == 1]
if i == 0:
fig.add_trace(
go.Bar(
x=a_0[f"bin_{i}"],
y=a_0[0],
width=0.2,
text=a_0[0],
textposition="outside",
texttemplate="%{text:.2s}",
name="target_0",
legendgroup="target_0",
textfont=dict(size=5),
textangle=0,
constraintext="inside",
marker=dict(
color=["#732FB0", "#732FB0"], line_width=0.5, line_color="gray"
),
),
row=1,
col=i + 1,
)
fig.add_trace(
go.Bar(
x=a_1[f"bin_{i}"],
y=a_1[0],
width=0.2,
text=a_1[0],
textposition="outside",
texttemplate="%{text:.2s}",
name="target_1",
legendgroup="target_1",
textfont=dict(size=5),
textangle=0,
constraintext="inside",
marker=dict(
color=["#2FB09D", "#2FB09D"], line_width=0.8, line_color="gray"
),
),
row=1,
col=i + 1,
)
else:
fig.add_trace(
go.Bar(
x=a_0[f"bin_{i}"],
y=a_0[0],
width=0.2,
text=a_0[0],
textposition="outside",
texttemplate="%{text:.2s}",
# name=f'bin_{i}'+' target_0',
name="target_0",
legendgroup="target_0",
showlegend=False,
textfont=dict(size=5),
textangle=0,
constraintext="inside",
marker=dict(
color=["#732FB0", "#732FB0"], line_width=0.5, line_color="gray"
),
),
row=1,
col=i + 1,
)
fig.add_trace(
go.Bar(
x=a_1[f"bin_{i}"],
y=a_1[0],
width=0.2,
text=a_1[0],
textposition="outside",
texttemplate="%{text:.2s}",
# name=f'bin_{i}'+' target_1',
name="target_1",
legendgroup="target_1",
showlegend=False,
textfont=dict(size=5),
textangle=0,
constraintext="inside",
marker=dict(
color=["#2FB09D", "#2FB09D"], line_width=0.8, line_color="gray"
),
),
row=1,
col=i + 1,
)
fig.update_layout(
yaxis=dict(visible=False),
yaxis2=dict(visible=False),
yaxis3=dict(visible=False),
yaxis4=dict(visible=False),
yaxis5=dict(visible=False),
)
fig.update_layout(
title_text="Binary Feature Target Distribution (Train Data)",
margin=dict(l=25, t=50, b=0),
legend_orientation="h",
width=500,
height=300,
) # this size is for kaggle kernal
# Set the subtitle size
for i in fig["layout"]["annotations"]:
i["font"] = dict(size=10, color="black")
fig.show()
# fig.show(config={"showLink":True})
# ### 2.3 Nominal Feature
# Describe nominal features
train[[col for col in train.columns if col.startswith("nom_")]].describe(include=["O"])
# nom_0 - nom_4 have few unique values, So let's plot the detail distribution
# Plot nominal feature distribution for Training Data
sub_title = list(
train[[col for col in train.columns if col.startswith("nom_")]].columns
)
fig = make_subplots(rows=1, cols=5, subplot_titles=sub_title)
for i in range(5):
a = train[f"nom_{i}"].value_counts()
fig.add_trace(
go.Bar(
x=a.index,
y=a.values,
width=0.2,
text=a.values,
textposition="outside",
texttemplate="%{text:.2s}",
name=f"bin_{i}",
showlegend=False,
textfont=dict(size=15),
marker=dict(
# color=['#732FB0','#2FB09D'],
line_width=0.5,
line_color="gray",
),
),
row=1,
col=i + 1,
)
fig.update_layout(title_text="Nominal Feature (0-4) Distribution (Train Data)")
# Hide the yaxis
fig.update_layout(
yaxis=dict(visible=False),
yaxis2=dict(visible=False),
yaxis3=dict(visible=False),
yaxis4=dict(visible=False),
yaxis5=dict(visible=False),
)
# this size is for kaggle kernal
fig.update_layout(
margin=dict(l=25, t=50, b=0), legend_orientation="h", width=500, height=350
)
# Set the subtitle size
for i in fig["layout"]["annotations"]:
i["font"] = dict(size=10, color="black")
# fig.show(config={"showLink":True})
fig.show()
# Plot nominal feature distribution for Test Data
sub_title = list(test[[col for col in test.columns if col.startswith("nom_")]].columns)
fig = make_subplots(rows=1, cols=5, subplot_titles=sub_title)
for i in range(5):
a = test[f"nom_{i}"].value_counts()
fig.add_trace(
go.Bar(
x=a.index,
y=a.values,
width=0.2,
text=a.values,
textposition="outside",
texttemplate="%{text:.2s}",
name=f"bin_{i}",
showlegend=False,
textfont=dict(size=15),
marker=dict(
# color=['#732FB0','#2FB09D'],
line_width=0.5,
line_color="gray",
),
),
row=1,
col=i + 1,
)
fig.update_layout(title_text="Nominal Feature (0-4) Distribution (Test Data)")
# Hide the yaxis
fig.update_layout(
yaxis=dict(visible=False),
yaxis2=dict(visible=False),
yaxis3=dict(visible=False),
yaxis4=dict(visible=False),
yaxis5=dict(visible=False),
)
# this size is for kaggle kernal
fig.update_layout(
margin=dict(l=25, t=50, b=0), legend_orientation="h", width=500, height=350
)
# Set the subtitle size
for i in fig["layout"]["annotations"]:
i["font"] = dict(size=10, color="black")
# fig.show(config={"showLink":True})
fig.show()
# Plot Nominal Feature Target Distribution for Training Data
sub_title = list(train[[col for col in test.columns if col.startswith("nom_")]].columns)
fig = make_subplots(rows=1, cols=5, subplot_titles=sub_title)
for i in range(5):
a = (
train.groupby([f"nom_{i}", "target"])
.size()
.to_frame()
.reset_index()
.sort_values(0, ascending=False)
)
a_0 = a[a["target"] == 0]
a_1 = a[a["target"] == 1]
if i == 0:
fig.add_trace(
go.Bar(
x=a_0[f"nom_{i}"],
y=a_0[0],
width=0.2,
text=a_0[0],
textposition="outside",
texttemplate="%{text:.2s}",
name="target_0",
legendgroup="target_0",
showlegend=False,
textfont=dict(size=15),
marker=dict(
# color=['#732FB0'] * a_1.shape[0],
line_width=0.5,
line_color="gray",
),
),
row=1,
col=i + 1,
)
fig.add_trace(
go.Bar(
x=a_1[f"nom_{i}"],
y=a_1[0],
width=0.2,
text=a_1[0],
textposition="outside",
texttemplate="%{text:.2s}",
name="target_1",
legendgroup="target_1",
showlegend=False,
textfont=dict(size=15),
marker=dict(
# color=['#2FB09D'] * a_1.shape[0],
line_width=0.8,
line_color="gray",
),
),
row=1,
col=i + 1,
)
else:
fig.add_trace(
go.Bar(
x=a_0[f"nom_{i}"],
y=a_0[0],
width=0.2,
text=a_0[0],
textposition="outside",
texttemplate="%{text:.2s}",
name="target_0",
legendgroup="target_0",
showlegend=False,
textfont=dict(size=15),
marker=dict(
# color=['#732FB0'] * a_1.shape[0],
line_width=0.5,
line_color="gray",
),
),
row=1,
col=i + 1,
)
fig.add_trace(
go.Bar(
x=a_1[f"nom_{i}"],
y=a_1[0],
width=0.2,
text=a_1[0],
textposition="outside",
texttemplate="%{text:.2s}",
name="target_1",
legendgroup="target_1",
showlegend=False,
textfont=dict(size=15),
marker=dict(
# color=['#2FB09D'] * a_1.shape[0],
line_width=0.8,
line_color="gray",
),
),
row=1,
col=i + 1,
)
fig.update_layout(title_text="Nominal Feature (0-4) Target Distribution (Train Data)")
# Hide the yaxis
fig.update_layout(
yaxis=dict(visible=False),
yaxis2=dict(visible=False),
yaxis3=dict(visible=False),
yaxis4=dict(visible=False),
yaxis5=dict(visible=False),
)
# this size is for kaggle kernal
fig.update_layout(
margin=dict(l=25, t=50, b=0), legend_orientation="v", width=500, height=350
)
# Set the subtitle size
for i in fig["layout"]["annotations"]:
i["font"] = dict(size=10, color="black")
# fig.show(config={"showLink":True})
fig.show()
# Nominal Feature (0-4) Target Rate Distribution
for i in range(5):
data = (
train[[f"nom_{i}", "target"]]
.groupby(f"nom_{i}")["target"]
.value_counts()
.unstack()
)
data["rate"] = data[1] / (data[0] + data[1])
data.sort_values(by=["rate"], inplace=True)
display(
data.style.highlight_max(color="lightgreen")
.highlight_min(color="#cd4f39")
.format({"rate": "{:.2%}"})
)
# ### 2.4 Ordinal Feature
train[[col for col in train.columns if col.startswith("ord_")]].describe(include="all")
# Plot Ordinal Feature (0-3) Target Distribution for Training Data
sub_title = list(
train[[col for col in test.columns if col.startswith("ord_")]].columns
)[:4]
fig = make_subplots(rows=1, cols=4, subplot_titles=sub_title)
for i in range(4):
a = (
train.groupby([f"ord_{i}", "target"])
.size()
.to_frame()
.reset_index()
.sort_values(0, ascending=False)
)
a_0 = a[a["target"] == 0]
a_1 = a[a["target"] == 1]
fig.add_trace(
go.Bar(
x=a_0[f"ord_{i}"],
y=a_0[0],
width=0.2,
text=a_0[0],
textposition="outside",
texttemplate="%{text:.2s}",
name=f"ord_{i}" + " target_0",
showlegend=False,
textfont=dict(size=15),
marker=dict(
# color=['#732FB0','#732FB0'],
line_width=0.5,
line_color="gray",
),
),
row=1,
col=i + 1,
)
fig.add_trace(
go.Bar(
x=a_1[f"ord_{i}"],
y=a_1[0],
width=0.2,
text=a_1[0],
textposition="outside",
texttemplate="%{text:.2s}",
name=f"ord_{i}" + " target_1",
showlegend=False,
textfont=dict(size=15),
marker=dict(
# color=['#2FB09D','#2FB09D'],
line_width=0.8,
line_color="gray",
),
),
row=1,
col=i + 1,
)
fig.update_layout(
title_text="Nominal Ordinal (0-3) Target Distribution (Train Data)", barmode="group"
)
# Hide the yaxis
fig.update_layout(
yaxis=dict(visible=False),
yaxis2=dict(visible=False),
yaxis3=dict(visible=False),
yaxis4=dict(visible=False),
yaxis5=dict(visible=False),
)
# this size is for kaggle kernal
fig.update_layout(
margin=dict(l=25, t=50, b=0), legend_orientation="h", width=500, height=350
)
# Set the subtitle size
for i in fig["layout"]["annotations"]:
i["font"] = dict(size=10, color="black")
# fig.show(config={"showLink":True})
fig.show()
# Plot Ordinal Feature (4) Target Distribution for Training Data
sub_title = list(
train[[col for col in test.columns if col.startswith("ord_")]].columns
)[-2:-1]
fig = make_subplots(rows=1, cols=1, subplot_titles=sub_title)
for i in [4]:
a = (
train.groupby([f"ord_{i}", "target"])
.size()
.to_frame()
.reset_index()
.sort_values(0, ascending=False)
)
a_0 = a[a["target"] == 0]
a_1 = a[a["target"] == 1]
fig.add_trace(
go.Bar(
x=a_0[f"ord_{i}"],
y=a_0[0],
width=0.2,
text=a_0[0],
textposition="outside",
texttemplate="%{text:.2s}",
name=f"ord_{i}" + " target_0",
showlegend=False,
textfont=dict(size=5),
textangle=0,
constraintext="inside",
marker=dict(
# color=['#732FB0','#732FB0'],
line_width=0.5,
line_color="gray",
),
),
row=1,
col=1,
)
fig.add_trace(
go.Bar(
x=a_1[f"ord_{i}"],
y=a_1[0],
width=0.2,
text=a_1[0],
textposition="outside",
texttemplate="%{text:.2s}",
name=f"ord_{i}" + " target_1",
showlegend=False,
textfont=dict(size=5),
textangle=0,
constraintext="inside",
marker=dict(
# color=['#2FB09D','#2FB09D'],
line_width=0.8,
line_color="gray",
),
),
row=1,
col=1,
)
fig.update_layout(
title_text="Nominal Ordinal (4) Target Distribution (Train Data)", barmode="group"
)
# Hide the yaxis
fig.update_layout(
yaxis=dict(visible=False),
yaxis2=dict(visible=False),
yaxis3=dict(visible=False),
yaxis4=dict(visible=False),
yaxis5=dict(visible=False),
)
# this size is for kaggle kernal
fig.update_layout(
margin=dict(l=25, t=50, b=0), legend_orientation="h", width=500, height=350
)
# Set the subtitle size
for i in fig["layout"]["annotations"]:
i["font"] = dict(size=10, color="black")
# fig.show(config={"showLink":True})
fig.show()
# Plot Ordinal Feature (5) Target Distribution for Training Data
sub_title = list(
train[[col for col in test.columns if col.startswith("ord_")]].columns
)[-1:]
fig = make_subplots(rows=1, cols=1, subplot_titles=sub_title)
for i in [5]:
a = (
train.groupby([f"ord_{i}", "target"])
.size()
.to_frame()
.reset_index()
.sort_values(0, ascending=False)
)
a_0 = a[a["target"] == 0]
a_1 = a[a["target"] == 1]
fig.add_trace(
go.Bar(
x=a_0[f"ord_{i}"],
y=a_0[0],
width=0.2,
text=a_0[0],
textposition="outside",
texttemplate="%{text:.2s}",
name=f"ord_{i}" + " target_0",
showlegend=False,
textfont=dict(size=5),
textangle=0,
constraintext="inside",
marker=dict(
# color=['#732FB0','#732FB0'],
line_width=0.5,
line_color="gray",
),
),
row=1,
col=1,
)
fig.add_trace(
go.Bar(
x=a_1[f"ord_{i}"],
y=a_1[0],
width=0.2,
text=a_1[0],
textposition="outside",
texttemplate="%{text:.2s}",
name=f"ord_{i}" + " target_1",
showlegend=False,
textfont=dict(size=5),
textangle=0,
constraintext="inside",
marker=dict(
# color=['#2FB09D','#2FB09D'],
line_width=0.8,
line_color="gray",
),
),
row=1,
col=1,
)
fig.update_layout(
title_text="Nominal Ordinal (5) Target Distribution (Train Data)", barmode="group"
)
# Hide the yaxis
fig.update_layout(
yaxis=dict(visible=False),
yaxis2=dict(visible=False),
yaxis3=dict(visible=False),
yaxis4=dict(visible=False),
yaxis5=dict(visible=False),
)
# this size is for kaggle kernal
fig.update_layout(margin=dict(l=25, t=50, b=0), legend_orientation="h", height=350)
# Set the subtitle size
for i in fig["layout"]["annotations"]:
i["font"] = dict(size=10, color="black")
# fig.show(config={"showLink":True})
fig.show()
# Ordinal Feature (0-4) Target Rate Distribution
for i in range(5):
data = (
train[[f"ord_{i}", "target"]]
.groupby(f"ord_{i}")["target"]
.value_counts()
.unstack()
)
data["rate"] = data[1] / (data[0] + data[1])
data.sort_values(by=["rate"], inplace=True)
display(
data.style.highlight_max(color="lightgreen")
.highlight_min(color="#cd4f39")
.format({"rate": "{:.2%}"})
)
# ### 2.5 Day & Month Feature
# day & month Target Rate Distribution
data = train[["day", "target"]].groupby("day")["target"].value_counts().unstack()
data["rate"] = data[1] / (data[0] + data[1])
data.sort_values(by=["rate"], inplace=True)
display(
data.style.highlight_max(color="lightgreen")
.highlight_min(color="#cd4f39")
.format({"rate": "{:.2%}"})
)
data = train[["month", "target"]].groupby("month")["target"].value_counts().unstack()
data["rate"] = data[1] / (data[0] + data[1])
data.sort_values(by=["rate"], inplace=True)
display(
data.style.highlight_max(color="lightgreen")
.highlight_min(color="#cd4f39")
.format({"rate": "{:.2%}"})
)
# ## 3 Feature Encoding
# [Categorical-encoding](https://github.com/scikit-learn-contrib/categorical-encoding) is a library of sklearn compatible categorical variable encoders. It contains a set of scikit-learn-style transformers for encoding categorical variables into numeric by means of different techniques.
# ### 3.1 Binary Feature Encoding
train[[col for col in train.columns if col.startswith("bin_")]].describe(include="all")
# lable Encoding for binary features
for i in range(5):
ord_order_dict = {
i: j
for j, i in enumerate(
sorted(
list(
set(
list(train[f"bin_{i}"].dropna().unique())
+ list(test[f"bin_{i}"].dropna().unique())
)
)
)
)
}
ord_order_dict["NULL"] = len(
train[f"bin_{i}"].dropna().unique()
) # mapping null value
print(ord_order_dict)
bin_encoding = [{"col": f"bin_{i}", "mapping": ord_order_dict}]
label = OrdinalEncoder(mapping=bin_encoding)
train["lable_" + f"bin_{i}"] = label.fit_transform(train[f"bin_{i}"].fillna("NULL"))
test["lable_" + f"bin_{i}"] = label.fit_transform(test[f"bin_{i}"].fillna("NULL"))
# Target Encoding for binary features
for i in range(5):
label = TargetEncoder()
train["target_" + f"bin_{i}"] = label.fit_transform(
train[f"bin_{i}"].fillna("NULL"), target
)
test["target_" + f"bin_{i}"] = label.transform(test[f"bin_{i}"].fillna("NULL"))
# ### 3.2 Nominal Feature Encoding
train[[col for col in train.columns if col.startswith("nom_")]].describe(include="all")
# Lable Encoding for Nominal features
for i in range(10):
ord_order_dict = {
i: j
for j, i in enumerate(
sorted(
list(
set(
list(train[f"nom_{i}"].dropna().unique())
+ list(test[f"nom_{i}"].dropna().unique())
)
)
)
)
}
ord_order_dict["NULL"] = len(
train[f"nom_{i}"].dropna().unique()
) # mapping null value
# print(ord_order_dict)
bin_encoding = [{"col": f"nom_{i}", "mapping": ord_order_dict}]
label = OrdinalEncoder(mapping=bin_encoding)
train["lable_" + f"nom_{i}"] = label.fit_transform(train[f"nom_{i}"].fillna("NULL"))
test["lable_" + f"nom_{i}"] = label.fit_transform(test[f"nom_{i}"].fillna("NULL"))
# Target Encoding for Nominal features
for i in range(10):
label = TargetEncoder()
train["target_" + f"nom_{i}"] = label.fit_transform(
train[f"nom_{i}"].fillna("NULL"), target
)
test["target_" + f"nom_{i}"] = label.transform(test[f"nom_{i}"].fillna("NULL"))
# ### 3.3 Ordinal Feature Encoding
train[[col for col in train.columns if col.startswith("ord_")]].describe(include="all")
# features 'ord_0', 'ord_1', 'ord_2' follow the order below
ord_order = [
[1.0, 2.0, 3.0],
["Novice", "Contributor", "Expert", "Master", "Grandmaster"],
["Freezing", "Cold", "Warm", "Hot", "Boiling Hot", "Lava Hot"],
]
for i in range(0, 3):
ord_order_dict = {i: j for j, i in enumerate(ord_order[i])}
ord_order_dict["NULL"] = len(
train[f"ord_{i}"].dropna().unique()
) # mapping null value
print(ord_order_dict)
bin_encoding = [{"col": f"ord_{i}", "mapping": ord_order_dict}]
label = OrdinalEncoder(mapping=bin_encoding)
train["lable_" + f"ord_{i}"] = label.fit_transform(train[f"ord_{i}"].fillna("NULL"))
test["lable_" + f"ord_{i}"] = label.fit_transform(test[f"ord_{i}"].fillna("NULL"))
# features 'ord_3', 'ord_4', 'ord_5' follow the alphabet order
for i in range(3, 6):
ord_order_dict = {
i: j
for j, i in enumerate(
sorted(
list(
set(
list(train[f"ord_{i}"].dropna().unique())
+ list(test[f"ord_{i}"].dropna().unique())
)
)
)
)
}
ord_order_dict["NULL"] = len(
train[f"ord_{i}"].dropna().unique()
) # mapping null value
# print(ord_order_dict)
bin_encoding = [{"col": f"ord_{i}", "mapping": ord_order_dict}]
label = OrdinalEncoder(mapping=bin_encoding)
train["lable_" + f"ord_{i}"] = label.fit_transform(train[f"ord_{i}"].fillna("NULL"))
test["lable_" + f"ord_{i}"] = label.fit_transform(test[f"ord_{i}"].fillna("NULL"))
# Target Encoding for Ordinal features
for i in range(6):
label = TargetEncoder()
train["target_" + f"ord_{i}"] = label.fit_transform(
train[f"ord_{i}"].fillna("NULL"), target
)
test["target_" + f"ord_{i}"] = label.transform(test[f"ord_{i}"].fillna("NULL"))
# ### 3.4 Day & Month Feature Encoding
# lable Encoding for Day & Month features
ord_order_dict = {
j + 1: i
for j, i in enumerate(
sorted(
list(
set(
list(train["day"].dropna().unique())
+ list(test["day"].dropna().unique())
)
)
)
)
}
ord_order_dict["NULL"] = len(train["day"].unique()) # mapping null value
bin_encoding = [{"col": "day", "mapping": ord_order_dict}]
label = OrdinalEncoder(mapping=bin_encoding)
train["lable_" + "day"] = label.fit_transform(train["day"].fillna("NULL"))
test["lable_" + "day"] = label.fit_transform(test["day"].fillna("NULL"))
ord_order_dict = {
j + 1: i
for j, i in enumerate(
sorted(
list(
set(
list(train["month"].dropna().unique())
+ list(test["month"].dropna().unique())
)
)
)
)
}
ord_order_dict["NULL"] = len(train["month"].unique()) # mapping null value
bin_encoding = [{"col": "month", "mapping": ord_order_dict}]
label = OrdinalEncoder(mapping=bin_encoding)
train["lable_" + "month"] = label.fit_transform(train["month"].fillna("NULL"))
test["lable_" + "month"] = label.fit_transform(test["month"].fillna("NULL"))
# Target Encoding for Day & Month features
label = TargetEncoder()
train["target_" + "day"] = label.fit_transform(train["day"].fillna("NULL"), target)
test["target_" + "day"] = label.transform(test["day"].fillna("NULL"))
label = TargetEncoder()
train["target_" + "month"] = label.fit_transform(
train[f"ord_{i}"].fillna("NULL"), target
)
test["target_" + "month"] = label.transform(test[f"ord_{i}"].fillna("NULL"))
# target features correlations
df_target_feature = train[[col for col in train.columns if col.startswith("target_")]]
sns.set(style="white")
# Compute the correlation matrix
corr = df_target_feature.corr()
# Generate a mask for the upper triangle
mask = np.triu(np.ones_like(corr, dtype=np.bool))
# Set up the matplotlib figure
f, ax = plt.subplots(figsize=(11, 9))
# Generate a custom diverging colormap
cmap = sns.diverging_palette(220, 10, as_cmap=True)
# Draw the heatmap with the mask and correct aspect ratio
sns.heatmap(
corr,
mask=mask,
cmap=cmap,
vmax=0.3,
center=0,
square=True,
linewidths=0.5,
cbar_kws={"shrink": 0.5},
)
ax.set_title("Target Features Correlation")
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.