content
stringlengths 73
1.12M
| license
stringclasses 3
values | path
stringlengths 9
197
| repo_name
stringlengths 7
106
| chain_length
int64 1
144
|
|---|---|---|---|---|
<jupyter_start><jupyter_text># Text Classification Model for Movie Review Sentiment Analysis Using TensorFlow Take 5
### David Lowe
### December 25, 2020
Template Credit: Adapted from a template made available by Dr. Jason Brownlee of Machine Learning Mastery. [https://machinelearningmastery.com/]
SUMMARY: This project aims to construct a text classification model using a neural network and document the end-to-end steps using a template. The Movie Review Sentiment Analysis dataset is a binary classification situation where we attempt to predict one of the two possible outcomes.
Additional Notes: This script is a replication, with some small modifications, of Dr. Jason Brownlee's blog post, Deep Convolutional Neural Network for Sentiment Analysis (https://machinelearningmastery.com/develop-word-embedding-model-predicting-movie-review-sentiment/). I plan to leverage Dr. Brownlee's tutorial and build a TensorFlow-based text classification notebook template for future modeling of similar datasets.
INTRODUCTION: The Movie Review Data is a collection of movie reviews retrieved from the imdb.com website in the early 2000s by Bo Pang and Lillian Lee. The reviews were collected and made available as part of their research on natural language processing. The dataset comprises 1,000 positive and 1,000 negative movie reviews drawn from an archive of the rec.arts.movies.reviews newsgroup hosted at IMDB. The authors refer to this dataset as the 'polarity dataset.'
We will use the last 100 positive reviews and the last 100 negative reviews as a test set (200 reviews) and the remaining 1,800 reviews as the training dataset. This is a 90% train, 10% split of the data.
In iteration Take1, we constructed the necessary code modules to handle the tasks of loading text, cleaning text, and vocabulary development.
In iteration Take2, we constructed a bag-of-words model and analyzed it with a simple multi-layer perceptron network.
In iteration Take3, we fine-tuned the model using different Tokenizer modes in the Keras API.
In iteration Take4, we will fine-tune the model using word embedding to fit a deep learning model.
In this Take5 iteration, we will fine-tune the model using the Word2Vec algorithm for developing a standalone word embedding mechanism.
ANALYSIS: From iteration Take2, the baseline model's performance achieved an average accuracy score of 85.81% after 25 epochs with ten iterations of cross-validation. Furthermore, the final model processed the test dataset with an accuracy measurement of 91.50%.
From iteration Take3, the best model's performance (using the frequency Tokenizer mode) achieved an average accuracy score of 86.06% after 25 epochs with ten iterations of cross-validation. Furthermore, the final model processed the test dataset with an accuracy measurement of 86.50%.
From iteration Take4, the best model's performance (using the word embedding with Keras) achieved an average accuracy score of 84.17% after ten epochs with ten iterations of cross-validation. Furthermore, the final model processed the test dataset with an accuracy measurement of 88.50%.
In this Take5 iteration, the best model's performance (using the Word2Vec word embedding algorithm) achieved an average accuracy score of 52.61% after ten epochs with ten iterations of cross-validation. Furthermore, the final model processed the test dataset with an accuracy measurement of 49.00%.
CONCLUSION: In this iteration, the Word2Vec word embedding algorithm appeared unsuitable for modeling this dataset. We should consider experimenting with other word embedding techniques for further modeling.
Dataset Used: Movie Review Sentiment Analysis Dataset
Dataset ML Model: Binary class text classification with text-oriented features
Dataset Reference: https://www.cs.cornell.edu/home/llee/papers/cutsent.pdf and http://www.cs.cornell.edu/people/pabo/movie-review-data/review_polarity.tar.gz
One potential source of performance benchmarks: https://machinelearningmastery.com/develop-word-embedding-model-predicting-movie-review-sentiment/
A deep-learning text classification project generally can be broken down into five major tasks:
1. Prepare Environment
2. Load and Prepare Text Data
3. Define and Train Models
4. Evaluate and Optimize Models
5. Finalize Model and Make Predictions# Task 1 - Prepare Environment<jupyter_code># # Install the packages to support accessing environment variable and SQL databases
# !pip install python-dotenv PyMySQL boto3
# # Retrieve GPU configuration information from Colab
# gpu_info = !nvidia-smi
# gpu_info = '\n'.join(gpu_info)
# if gpu_info.find('failed') >= 0:
# print('Select the Runtime → "Change runtime type" menu to enable a GPU accelerator, ')
# print('and then re-execute this cell.')
# else:
# print(gpu_info)
# # Retrieve memory configuration information from Colab
# from psutil import virtual_memory
# ram_gb = virtual_memory().total / 1e9
# print('Your runtime has {:.1f} gigabytes of available RAM\n'.format(ram_gb))
# if ram_gb < 20:
# print('To enable a high-RAM runtime, select the Runtime → "Change runtime type"')
# print('menu, and then select High-RAM in the Runtime shape dropdown. Then, ')
# print('re-execute this cell.')
# else:
# print('You are using a high-RAM runtime!')
# # Direct Colab to use TensorFlow v2
# %tensorflow_version 2.x
# Retrieve CPU information from the system
ncpu = !nproc
print("The number of available CPUs is:", ncpu[0])<jupyter_output>The number of available CPUs is: 4
<jupyter_text>## 1.a) Load libraries and modules<jupyter_code># Set the random seed number for reproducible results
seedNum = 888
# Load libraries and packages
import random
random.seed(seedNum)
import numpy as np
np.random.seed(seedNum)
import pandas as pd
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
import os
import sys
import boto3
import shutil
import string
import nltk
import gensim
from nltk.corpus import stopwords
from collections import Counter
from datetime import datetime
from dotenv import load_dotenv
from sklearn.model_selection import RepeatedKFold
from sklearn.metrics import accuracy_score
from sklearn.metrics import confusion_matrix
from sklearn.metrics import classification_report
import tensorflow as tf
tf.random.set_seed(seedNum)
from tensorflow import keras
from tensorflow.keras import layers
from tensorflow.keras.preprocessing.text import Tokenizer
from tensorflow.keras.preprocessing.sequence import pad_sequences
from tensorflow.keras.callbacks import ReduceLROnPlateau
nltk.download('popular')<jupyter_output>[nltk_data] Downloading collection 'popular'
[nltk_data] |
[nltk_data] | Downloading package cmudict to
[nltk_data] | /home/pythonml/nltk_data...
[nltk_data] | Package cmudict is already up-to-date!
[nltk_data] | Downloading package gazetteers to
[nltk_data] | /home/pythonml/nltk_data...
[nltk_data] | Package gazetteers is already up-to-date!
[nltk_data] | Downloading package genesis to
[nltk_data] | /home/pythonml/nltk_data...
[nltk_data] | Package genesis is already up-to-date!
[nltk_data] | Downloading package gutenberg to
[nltk_data] | /home/pythonml/nltk_data...
[nltk_data] | Package gutenberg is already up-to-date!
[nltk_data] | Downloading package inaugural to
[nltk_data] | /home/pythonml/nltk_data...
[nltk_data] | Package inaugural is already up-to-date!
[nltk_data] | Downloading package movie_reviews to
[nltk_data] | /home/pythonml/nltk_data...
[nltk_data] | Package movie_reviews is a[...]<jupyter_text>## 1.b) Set up the controlling parameters and functions<jupyter_code># Begin the timer for the script processing
startTimeScript = datetime.now()
# Set up the number of CPU cores available for multi-thread processing
n_jobs = 2
# Set up the flag to stop sending progress emails (setting to True will send status emails!)
notifyStatus = False
# Set Pandas options
pd.set_option("display.max_rows", None)
pd.set_option("display.max_columns", None)
pd.set_option("display.width", 140)
# Set the percentage sizes for splitting the dataset
test_set_size = 0.2
val_set_size = 0.25
# Set the number of folds for cross validation
n_folds = 5
n_iterations = 1
# Set various default modeling parameters
default_loss = 'binary_crossentropy'
default_metrics = ['accuracy']
default_optimizer = tf.keras.optimizers.Adam(learning_rate=0.001)
default_kernel_init = tf.keras.initializers.RandomNormal(seed=seedNum)
default_epochs = 10
default_batch_size = 16
default_vector_space = 100
default_filters = 128
default_kernel_size = 5
default_pool_size = 2
default_neighbors = 5
# Define the labels to use for graphing the data
train_metric = "accuracy"
validation_metric = "val_accuracy"
train_loss = "loss"
validation_loss = "val_loss"
# Check the number of GPUs accessible through TensorFlow
print('Num GPUs Available:', len(tf.config.list_physical_devices('GPU')))
# Print out the TensorFlow version for confirmation
print('TensorFlow version:', tf.__version__)
# Set up the parent directory location for loading the dotenv files
# useColab = True
# if useColab:
# # Mount Google Drive locally for storing files
# from google.colab import drive
# drive.mount('/content/gdrive')
# gdrivePrefix = '/content/gdrive/My Drive/Colab_Downloads/'
# env_path = '/content/gdrive/My Drive/Colab Notebooks/'
# dotenv_path = env_path + "python_script.env"
# load_dotenv(dotenv_path=dotenv_path)
# Set up the dotenv file for retrieving environment variables
# useLocalPC = True
# if useLocalPC:
# env_path = "/Users/david/PycharmProjects/"
# dotenv_path = env_path + "python_script.env"
# load_dotenv(dotenv_path=dotenv_path)
# Set up the email notification function
def status_notify(msg_text):
access_key = os.environ.get('SNS_ACCESS_KEY')
secret_key = os.environ.get('SNS_SECRET_KEY')
aws_region = os.environ.get('SNS_AWS_REGION')
topic_arn = os.environ.get('SNS_TOPIC_ARN')
if (access_key is None) or (secret_key is None) or (aws_region is None):
sys.exit("Incomplete notification setup info. Script Processing Aborted!!!")
sns = boto3.client('sns', aws_access_key_id=access_key, aws_secret_access_key=secret_key, region_name=aws_region)
response = sns.publish(TopicArn=topic_arn, Message=msg_text)
if response['ResponseMetadata']['HTTPStatusCode'] != 200 :
print('Status notification not OK with HTTP status code:', response['ResponseMetadata']['HTTPStatusCode'])
if notifyStatus: status_notify('(TensorFlow Text Classification) Task 1 - Prepare Environment has begun on ' + datetime.now().strftime('%A %B %d, %Y %I:%M:%S %p'))
# Reset the random number generators
def reset_random(x):
random.seed(x)
np.random.seed(x)
tf.random.set_seed(x)
if notifyStatus: status_notify('(TensorFlow Text Classification) Task 1 - Prepare Environment completed on ' + datetime.now().strftime('%A %B %d, %Y %I:%M:%S %p'))<jupyter_output><empty_output><jupyter_text># Task 2 - Load and Prepare Text Data<jupyter_code>if notifyStatus: status_notify('(TensorFlow Text Classification) Task 2 - Load and Prepare Text Data has begun on ' + datetime.now().strftime('%A %B %d, %Y %I:%M:%S %p'))<jupyter_output><empty_output><jupyter_text>## 2.a) Download Text Data Archive<jupyter_code># Clean up the old files and download directories before receiving new ones
!rm -rf staging/
!rm review_polarity.tar.gz
!rm vocabulary.txt
!wget https://dainesanalytics.com/datasets/cornell-movie-review-polarity/review_polarity.tar.gz<jupyter_output>--2020-12-13 00:26:39-- https://dainesanalytics.com/datasets/cornell-movie-review-polarity/review_polarity.tar.gz
Resolving dainesanalytics.com (dainesanalytics.com)... 13.225.150.103, 13.225.150.58, 13.225.150.31, ...
Connecting to dainesanalytics.com (dainesanalytics.com)|13.225.150.103|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: 3127238 (3.0M) [application/x-gzip]
Saving to: ‘review_polarity.tar.gz’
review_polarity.tar 100%[===================>] 2.98M 11.8MB/s in 0.3s
2020-12-13 00:26:39 (11.8 MB/s) - ‘review_polarity.tar.gz’ saved [3127238/3127238]
<jupyter_text>## 2.b) Splitting Data for Training and Validation<jupyter_code>staging_dir = 'staging/'
!mkdir staging/
!mkdir staging/testing/
!mkdir staging/testing/pos/
!mkdir staging/testing/neg/
local_archive = 'review_polarity.tar.gz'
shutil.unpack_archive(local_archive, staging_dir)
training_dir = 'staging/txt_sentoken/'
testing_dir = 'staging/testing/'
classA_name = 'pos'
classB_name = 'neg'
# Brief listing of training text files for both classes before splitting
training_classA_dir = os.path.join(training_dir, classA_name)
training_classA_files = os.listdir(training_classA_dir)
print('Number of training images for', classA_name, ':', len(training_classA_files))
print('Training samples for', classA_name, ':', training_classA_files[:10])
training_classB_dir = os.path.join(training_dir, classB_name)
training_classB_files = os.listdir(training_classB_dir)
print('Number of training images for', classB_name, ':', len(training_classB_files))
print('Training samples for', classB_name, ':', training_classB_files[:10])
# Move the testing files from the training directories
testing_classA_dir = os.path.join(testing_dir, classA_name)
testing_classB_dir = os.path.join(testing_dir, classB_name)
file_prefix = 'cv9'
i = 0
for text_file in training_classA_files:
if text_file.startswith(file_prefix):
source_file = training_classA_dir + '/' + text_file
dest_file = testing_classA_dir + '/' + text_file
# print('Moving file from', source_file, 'to', dest_file)
shutil.move(source_file, dest_file)
i = i + 1
print('Number of', classA_name, 'files moved:', i, '\n')
i = 0
for text_file in training_classB_files:
if text_file.startswith(file_prefix):
source_file = training_classB_dir + '/' + text_file
dest_file = testing_classB_dir + '/' + text_file
# print('Moving file from', source_file, 'to', dest_file)
shutil.move(source_file, dest_file)
i = i + 1
print('Number of', classB_name, 'files moved:', i)
# Brief listing of training text files for both classes after splitting
training_classA_files = os.listdir(training_classA_dir)
print('Number of training files for', classA_name, ':', len(training_classA_files))
print('Training samples for', classA_name, ':', training_classA_files[:10])
training_classB_files = os.listdir(training_classB_dir)
print('Number of training files for', classB_name, ':', len(training_classB_files))
print('Training samples for', classB_name, ':', training_classB_files[:10])
# Brief listing of testing text files for both classes after splitting
testing_classA_files = os.listdir(testing_classA_dir)
print('Number of testing files for', classA_name, ':', len(testing_classA_files))
print('Test samples for', classA_name, ':', testing_classA_files[:10])
testing_classB_files = os.listdir(testing_classB_dir)
print('Number of testing files for', classB_name, ':', len(testing_classB_files))
print('Test samples for', classB_name, ':', testing_classB_files[:10])<jupyter_output>Number of testing files for pos : 100
Test samples for pos : ['cv900_10331.txt', 'cv901_11017.txt', 'cv902_12256.txt', 'cv903_17822.txt', 'cv904_24353.txt', 'cv905_29114.txt', 'cv906_11491.txt', 'cv907_3541.txt', 'cv908_16009.txt', 'cv909_9960.txt']
Number of testing files for neg : 100
Test samples for neg : ['cv900_10800.txt', 'cv901_11934.txt', 'cv902_13217.txt', 'cv903_18981.txt', 'cv904_25663.txt', 'cv905_28965.txt', 'cv906_12332.txt', 'cv907_3193.txt', 'cv908_17779.txt', 'cv909_9973.txt']
<jupyter_text>## 2.c) Load Document and Build Vocabulary<jupyter_code># load doc into memory
def load_doc(filename):
# open the file as read only
file = open(filename, 'r')
# read all text
text = file.read()
# close the file
file.close()
return text
# turn a doc into clean tokens
def clean_doc(doc):
# split into tokens by white space
tokens = doc.split()
# remove punctuation from each token
table = str.maketrans('', '', string.punctuation)
tokens = [w.translate(table) for w in tokens]
# remove remaining tokens that are not alphabetic
tokens = [word for word in tokens if word.isalpha()]
# filter out stop words
stop_words = set(stopwords.words('english'))
tokens = [w for w in tokens if not w in stop_words]
# filter out short tokens
tokens = [word for word in tokens if len(word) > 1]
return tokens
# load doc and add to vocab
def add_doc_to_vocab(filename, vocab):
# load doc
doc = load_doc(filename)
# clean doc
tokens = clean_doc(doc)
# update counts
vocab.update(tokens)
# load all docs in a directory
def build_vocab_from_docs(directory, vocab):
# walk through all files in the folder
i = 0
print('Processing the text files and showing the first 10...')
for filename in os.listdir(directory):
# skip files that do not have the right extension
if not filename.endswith(".txt"):
continue
# create the full path of the file to open
path = directory + '/' + filename
# add doc to vocab
add_doc_to_vocab(path, vocab)
i = i + 1
if i < 10: print('Loaded %s' % path)
print('Total number of text files loaded:', i, '\n')
# define vocab
vocab = Counter()
# add all docs to vocab
build_vocab_from_docs(training_classA_dir, vocab)
build_vocab_from_docs(training_classB_dir, vocab)
# print the size of the vocab
print('The total number of words in the vocabulary:', len(vocab))
# print the top words in the vocab
top_words = 50
print('The top', top_words, 'words in the vocabulary:\n', vocab.most_common(top_words))
# keep tokens with a min occurrence
min_occurane = 2
tokens = [k for k,c in vocab.items() if c >= min_occurane]
print('The number of words with the minimum appearance:', len(tokens))
# save list to file
def save_list(lines, filename):
# convert lines to a single blob of text
data = '\n'.join(lines)
# open file
file = open(filename, 'w')
# write text
file.write(data)
# close file
file.close()
# save tokens to a vocabulary file
save_list(tokens, 'vocabulary.txt')<jupyter_output><empty_output><jupyter_text>## 2.d) Prepare Word Embedding using Word2Vec Algorithm<jupyter_code># load the vocabulary
vocab_filename = 'vocabulary.txt'
vocab = load_doc(vocab_filename)
vocab = vocab.split()
vocab = set(vocab)
print('Number of tokens in the vocabulary:', len(vocab))
# turn a doc into clean tokens
def doc_to_clean_lines(doc, vocab):
clean_lines = list()
lines = doc.splitlines()
for line in lines:
# split into tokens by white space
tokens = line.split()
# remove punctuation from each token
table = str.maketrans('', '', string.punctuation)
tokens = [w.translate(table) for w in tokens]
# filter out tokens not in vocab
tokens = [w for w in tokens if w in vocab]
clean_lines.append(tokens)
return clean_lines
# load all docs in a directory
def load_lines_from_docs(directory, vocab):
lines = list()
# walk through all files in the folder
for filename in os.listdir(directory):
# create the full path of the file to open
path = directory + '/' + filename
# load and clean the doc
doc = load_doc(path)
doc_lines = doc_to_clean_lines(doc, vocab)
# add lines to list
lines += doc_lines
return lines
# load training data
positive_train_cases = load_lines_from_docs(training_classA_dir, vocab)
print('The number of positive sentences processed:', len(positive_train_cases))
negative_train_cases = load_lines_from_docs(training_classB_dir, vocab)
print('The number of negative sentences processed:', len(negative_train_cases))
train_sentences = negative_train_cases + positive_train_cases
print('Total training sentences: %d' % len(train_sentences))
# train word2vec model
embed_model = gensim.models.Word2Vec(train_sentences, size=default_vector_space, window=default_neighbors, workers=n_jobs, min_count=1)
# summarize vocabulary size in model
words = list(embed_model.wv.vocab)
print('Vocabulary size: %d' % len(words))
# save model in ASCII (word2vec) format
filename = 'embedding_word2vec.txt'
embed_model.wv.save_word2vec_format(filename, binary=False)<jupyter_output><empty_output><jupyter_text>## 2.e) Create Tokenizer and Encode the Input Text<jupyter_code># load doc, clean and return line of tokens
def doc_to_line(filename, vocab):
# load the doc
doc = load_doc(filename)
# clean doc
tokens = clean_doc(doc)
# filter by vocab
tokens = [w for w in tokens if w in vocab]
return ' '.join(tokens)
# load all docs in a directory
def process_docs_to_lines(directory, vocab):
lines = list()
# walk through all files in the folder
for filename in os.listdir(directory):
# create the full path of the file to open
path = directory + '/' + filename
# load and clean the doc
line = doc_to_line(path, vocab)
# add to list
lines.append(line)
return lines
# prepare encoding of docs
def encode_train_data(train_docs, maxlen):
# create the tokenizer
tokenizer = Tokenizer()
# fit the tokenizer on the documents
tokenizer.fit_on_texts(train_docs)
# encode training data set
encoded_docs = tokenizer.texts_to_sequences(train_docs)
# pad sequences
train_encoded = pad_sequences(encoded_docs, maxlen=maxlen, padding='post')
return train_encoded, tokenizer
# Load all training cases
positive_train_cases = process_docs_to_lines(training_classA_dir, vocab)
print('The number of positive reviews processed:', len(positive_train_cases))
negative_train_cases = process_docs_to_lines(training_classB_dir, vocab)
print('The number of negative reviews processed:', len(negative_train_cases))
training_docs = negative_train_cases + positive_train_cases
# Get maximum doc length for padding sequences
max_length = max([len(s.split()) for s in training_docs])
print('The maximum document length:', max_length)
# encode training and validation datasets
X_train, vocab_tokenizer = encode_train_data(training_docs, max_length)
print('The shape of the encoded training dataset:', X_train.shape)
y_train = np.array([0 for _ in range(len(negative_train_cases))] + [1 for _ in range(len(positive_train_cases))])
print('The shape of the encoded training classes:', y_train.shape)
if notifyStatus: status_notify('(TensorFlow Text Classification) Task 2 - Load and Prepare Text Data completed on ' + datetime.now().strftime('%A %B %d, %Y %I:%M:%S %p'))<jupyter_output><empty_output><jupyter_text># Task 3 - Define and Train Models<jupyter_code>if notifyStatus: status_notify('(TensorFlow Text Classification) Task 3 - Define and Train Models has begun on ' + datetime.now().strftime('%A %B %d, %Y %I:%M:%S %p'))
# Define the default numbers of input/output for modeling
num_outputs = 1
# load embedding as a dict
def load_embedding(filename):
# load embedding into memory, skip first line
file = open(filename,'r')
lines = file.readlines()[1:]
file.close()
# create a map of words to vectors
embedding = dict()
for line in lines:
parts = line.split()
# key is string word, value is numpy array for vector
embedding[parts[0]] = np.asarray(parts[1:], dtype='float32')
return embedding
# create a weight matrix for the Embedding layer from a loaded embedding
def get_weight_matrix(embedding, vocab):
# total vocabulary size plus 0 for unknown words
vocab_size = len(vocab) + 1
# define weight matrix dimensions with all 0
weight_matrix = np.zeros((vocab_size, default_vector_space))
# step vocab, store vectors using the Tokenizer's integer mapping
for word, i in vocab.items():
weight_matrix[i] = embedding.get(word)
return weight_matrix
# load embedding from file
raw_embedding = load_embedding('embedding_word2vec.txt')
# get vectors in the right order
embedding_vectors = get_weight_matrix(raw_embedding, vocab_tokenizer.word_index)
# define vocabulary size (largest integer value)
vocab_size = len(vocab_tokenizer.word_index) + 1
print('The maximum vocabulary size is:', vocab_size)
# create the embedding layer
embedding_layer = keras.layers.Embedding(vocab_size, default_vector_space, weights=[embedding_vectors], input_length=max_length, trainable=False)
# Define the baseline model for benchmarking
def create_nn_model(n_inputs=max_length, n_outputs=num_outputs, embed_layer=embedding_layer, conv1_filters=default_filters, conv1_kernels=default_kernel_size,
n_pools=default_pool_size, opt_param=default_optimizer, init_param=default_kernel_init):
nn_model = keras.Sequential([
embed_layer,
layers.Conv1D(filters=conv1_filters, kernel_size=conv1_kernels, activation='relu'),
layers.MaxPooling1D(pool_size=n_pools),
layers.Flatten(),
layers.Dense(n_outputs, activation='sigmoid', kernel_initializer=init_param)
])
nn_model.compile(loss=default_loss, optimizer=opt_param, metrics=default_metrics)
return nn_model
# Initialize the default model and get a baseline result
startTimeModule = datetime.now()
results = list()
iteration = 0
cv = RepeatedKFold(n_splits=n_folds, n_repeats=n_iterations, random_state=seedNum)
for train_ix, val_ix in cv.split(X_train):
feature_train, feature_validation = X_train[train_ix], X_train[val_ix]
target_train, target_validation = y_train[train_ix], y_train[val_ix]
reset_random(seedNum)
baseline_model = create_nn_model()
baseline_model.fit(feature_train, target_train, epochs=default_epochs, batch_size=default_batch_size, verbose=1)
model_metric = baseline_model.evaluate(feature_validation, target_validation, verbose=0)[1]
iteration = iteration + 1
print('Accuracy measurement from iteration %d >>> %.2f%%' % (iteration, model_metric*100),'\n')
results.append(model_metric)
validation_score = np.mean(results)
validation_variance = np.std(results)
print('Average model accuracy from all validation iterations: %.2f%% (%.2f%%)' % (validation_score*100, validation_variance*100))
print('Total time for model fitting and cross validating:', (datetime.now() - startTimeModule))
if notifyStatus: status_notify('(TensorFlow Text Classification) Task 3 - Define and Train Models completed on ' + datetime.now().strftime('%A %B %d, %Y %I:%M:%S %p'))<jupyter_output><empty_output><jupyter_text># Task 4 - Evaluate and Optimize Models<jupyter_code>if notifyStatus: status_notify('(TensorFlow Text Classification) Task 4 - Evaluate and Optimize Models has begun on ' + datetime.now().strftime('%A %B %d, %Y %I:%M:%S %p'))
# Not applicable for this iteration of modeling
if notifyStatus: status_notify('(TensorFlow Text Classification) Task 4 - Evaluate and Optimize Models completed on ' + datetime.now().strftime('%A %B %d, %Y %I:%M:%S %p'))<jupyter_output><empty_output><jupyter_text># Task 5 - Finalize Model and Make Predictions<jupyter_code>if notifyStatus: status_notify('(TensorFlow Text Classification) Task 5 - Finalize Model and Make Predictions has begun on ' + datetime.now().strftime('%A %B %d, %Y %I:%M:%S %p'))
# prepare encoding of docs
def encode_test_data(test_docs, maxlen, tokenizer):
# encode training data set
encoded_docs = tokenizer.texts_to_sequences(test_docs)
# pad sequences
test_encoded = pad_sequences(encoded_docs, maxlen=maxlen, padding='post')
return test_encoded
# load all validation cases
positive_test_cases = process_docs_to_lines(testing_classA_dir, vocab)
print('The number of positive reviews processed:', len(positive_test_cases))
negative_test_cases = process_docs_to_lines(testing_classB_dir, vocab)
print('The number of negative reviews processed:', len(negative_test_cases))
testing_docs = negative_test_cases + positive_test_cases
# encode training and validation datasets
X_test = encode_test_data(testing_docs, max_length, vocab_tokenizer)
print('The shape of the encoded test dataset:', X_test.shape)
y_test = np.array([0 for _ in range(len(negative_test_cases))] + [1 for _ in range(len(positive_test_cases))])
print('The shape of the encoded test classes:', y_test.shape)
# Create the final model for evaluating the test dataset
reset_random(seedNum)
final_model = create_nn_model()
final_model.fit(X_train, y_train, epochs=default_epochs, batch_size=default_batch_size, verbose=1)
# Summarize the final model
final_model.summary()
# test_predictions = final_model.predict(X_test, batch_size=default_batch, verbose=1)
test_predictions = (final_model.predict(X_test) > 0.5).astype("int32").ravel()
print('Accuracy Score:', accuracy_score(y_test, test_predictions))
print(confusion_matrix(y_test, test_predictions))
print(classification_report(y_test, test_predictions))
if notifyStatus: status_notify('(TensorFlow Text Classification) Task 5 - Finalize Model and Make Predictions completed on ' + datetime.now().strftime('%A %B %d, %Y %I:%M:%S %p'))
print ('Total time for the script:',(datetime.now() - startTimeScript))<jupyter_output>Total time for the script: 0:11:48.403900
|
non_permissive
|
/py_nlp_movie_review_sentiment/py_nlp_movie_review_sentiment_take5.ipynb
|
daines-analytics/nlp-projects
| 12 |
<jupyter_start><jupyter_text>### Nacachian Mauro -- 99619
## Simulacion Monte Carlo para ambos tipos de repetidores
Se simula el sistema analogico y digital para 9 etapas y para 5 a 25 dB. Se usan 1000 muestras <jupyter_code>
import matplotlib.pyplot as plt
import numpy as np
from scipy import stats
from copy import copy
# Funcion indicador para el sistema analogico
# devuelve 1 si el simbolo recibido es distinto a lo recibido
# usando la toma de decision en la utlima etapa
#
def etapaDecision(x,yn):
if x > 0 and yn < 0:
return 1
if x < 0 and yn >= 0:
return 1
else:
return 0
def monteCarloDig(xn, x1, k):
sum = 0
for i in range(k):
if xn[i] != x1[i]:
sum = sum + 1
return sum/k
def monteCarloAnalog(yn, x1, k):
sum = 0
for i in range(k):
sum = sum + etapaDecision(x1[i], yn[i])
return sum/k
def probErrorAnalog(snr,n):
return stats.norm.sf(np.sqrt( 1 / (pow((snr+1) / snr,n) - 1) ))
def probErrorDigital(snr,n):
return (1- pow((1 - 2 * stats.norm.sf( np.sqrt(snr) ) ), n) ) / 2
muestras = 1000 ## cantidad de realizaciones
n = 9 ## cantidad de etapas
h = 1
A = 1
snr_db = np.arange(5,25,1) # en dB
snr = 10**((snr_db)/10)
pErrorAnalog = []
pErrorDig = []
for k in range(np.size(snr_db)):
sigma = pow(h, 2) * pow(A, 2) / snr[k]
Wi = np.random.randn(muestras, n) * np.sqrt(sigma)
X1 = np.random.choice([A, -A], muestras, p = [0.5, 0.5])
#######################
## Sistema Analogico ##
#######################
G = np.sqrt(snr[k] / (snr[k] + 1)) / h
ruido = []
for j in range(muestras):
aux = 0
for i in range(n):
aux = aux + Wi[j][i] * pow(G,n-(i+1)) * pow(h,n-(i+1))
ruido.append(aux)
Yn = []
for i in range(muestras):
Yn = np.append(Yn,pow(G,n-1) * pow(h,n) * X1[i] + ruido[i])
pErrorAnalog = np.append(pErrorAnalog, monteCarloAnalog(Yn, X1, muestras))
#####################
## Sistema Digital ##
#####################
#W = np.random.randn(muestras, n) * np.sqrt(sigma)
Ynd = X1 * h + Wi[:,0] # Accedo a las realizaciones de la primer etapa
Xn = copy(X1)
for i in range(muestras):
for j in range(n):
Ynd[i] = Xn[i] * h + Wi[i][j]
# Toma de decision #
if Ynd[i] < 0:
Xn[i] = -A
else:
Xn[i] = A
pErrorDig = np.append(pErrorDig, monteCarloDig(Xn, X1, muestras))
# Probabilidades por formula
#
pErrorDigTeorica = probErrorDigital(snr, n)
pErrorAnalogTeorica = probErrorAnalog(snr, n)
# Graficos comparativos para n = 9
#
plt.figure(figsize = (15,10))
plt.plot(snr_db, pErrorAnalogTeorica, '--o', label = 'Probabilidad teorica', color = 'black', linewidth = 2)
plt.plot(snr_db, pErrorAnalog, '-^', label = 'Probabilidad simulada', color = 'red', linewidth = 2)
#plt.yscale('log')
plt.legend()
plt.xticks(snr_db)
plt.xlabel('SNR [dB]')
plt.ylabel('Probabilidad de error')
plt.grid(b = True, which = 'major', color = 'black', linestyle = '-', linewidth = 0.4)
plt.grid(b = True, which = 'minor', color = 'black', linestyle = '-', linewidth = 0.4)
plt.legend(loc = 'best')
plt.title('Probabilidad de error para el sistema analogica con n = 9')
plt.savefig("comparativa_analog_sim.png")
plt.figure(figsize = (15,10))
plt.plot(snr_db, pErrorDigTeorica, '--o', label = 'Probabilidad teorica', color = 'black', linewidth = 2)
plt.plot(snr_db, pErrorDig, '-^' ,label = 'Probabilidad simulada', color = 'red', linewidth = 2)
#plt.yscale('log')
plt.legend()
plt.xticks(snr_db)
plt.xlabel('SNR [dB]')
plt.ylabel('Probabilidad de error')
plt.grid(b = True, which = 'major', color = 'black', linestyle = '-', linewidth = 0.4)
plt.grid(b = True, which = 'minor', color = 'black', linestyle = '-', linewidth = 0.4)
plt.legend(loc = 'best')
plt.title('Probabilidad de error para el sistema digital con n = 9')
plt.savefig("comparativa_dig_sim.png")
<jupyter_output><empty_output>
|
no_license
|
/tp1 repetidores/tp1Ejercicio4A.ipynb
|
mauronaca/tps-procesos-estocasticos-fiuba
| 1 |
<jupyter_start><jupyter_text># Text core
> Basic function to preprocess text before assembling it in a `DataBunch`.<jupyter_code>#export
import spacy,html
from spacy.symbols import ORTH<jupyter_output><empty_output><jupyter_text>## Preprocessing rulesThe following are rules applied to texts before or after it's tokenized.<jupyter_code>#export
#special tokens
UNK, PAD, BOS, EOS, FLD, TK_REP, TK_WREP, TK_UP, TK_MAJ = "xxunk xxpad xxbos xxeos xxfld xxrep xxwrep xxup xxmaj".split()
#export
_all_ = ["UNK", "PAD", "BOS", "EOS", "FLD", "TK_REP", "TK_WREP", "TK_UP", "TK_MAJ"]
#export
_re_spec = re.compile(r'([/#\\])')
def spec_add_spaces(t):
"Add spaces around / and #"
return _re_spec.sub(r' \1 ', t)
test_eq(spec_add_spaces('#fastai'), ' # fastai')
test_eq(spec_add_spaces('/fastai'), ' / fastai')
test_eq(spec_add_spaces('\\fastai'), ' \\ fastai')
#export
_re_space = re.compile(' {2,}')
def rm_useless_spaces(t):
"Remove multiple spaces"
return _re_space.sub(' ', t)
test_eq(rm_useless_spaces('a b c'), 'a b c')
#export
_re_rep = re.compile(r'(\S)(\1{2,})')
def replace_rep(t):
"Replace repetitions at the character level: cccc -- TK_REP 4 c"
def _replace_rep(m):
c,cc = m.groups()
return f' {TK_REP} {len(cc)+1} {c} '
return _re_rep.sub(_replace_rep, t)<jupyter_output><empty_output><jupyter_text>It starts replacing at 3 repetitions of the same character or more.<jupyter_code>test_eq(replace_rep('aa'), 'aa')
test_eq(replace_rep('aaaa'), f' {TK_REP} 4 a ')
#export
_re_wrep = re.compile(r'(?:\s|^)(\w+)\s+((?:\1\s+)+)\1(\s|\W|$)')
#hide
"""
Matches any word repeated at least four times with spaces between them
(?:\s|^) Non-Capture either a whitespace character or the beginning of text
(\w+) Capture any alphanumeric character
\s+ One or more whitespace
((?:\1\s+)+) Capture a repetition of one or more times \1 followed by one or more whitespace
\1 Occurence of \1
(\s|\W|$) Capture last whitespace, non alphanumeric character or end of text
""";
#export
def replace_wrep(t):
"Replace word repetitions: word word word word -- TK_WREP 4 word"
def _replace_wrep(m):
c,cc,e = m.groups()
return f' {TK_WREP} {len(cc.split())+2} {c} {e}'
return _re_wrep.sub(_replace_wrep, t)<jupyter_output><empty_output><jupyter_text>It starts replacing at 3 repetitions of the same word or more.<jupyter_code>test_eq(replace_wrep('ah ah'), 'ah ah')
test_eq(replace_wrep('ah ah ah'), f' {TK_WREP} 3 ah ')
test_eq(replace_wrep('ah ah ah ah'), f' {TK_WREP} 4 ah ')
test_eq(replace_wrep('ah ah ah ah '), f' {TK_WREP} 4 ah ')
test_eq(replace_wrep('ah ah ah ah.'), f' {TK_WREP} 4 ah .')
test_eq(replace_wrep('ah ah ahi'), f'ah ah ahi')
#export
def fix_html(x):
"Various messy things we've seen in documents"
x = x.replace('#39;', "'").replace('amp;', '&').replace('#146;', "'").replace('nbsp;', ' ').replace(
'#36;', '$').replace('\\n', "\n").replace('quot;', "'").replace('<br />', "\n").replace(
'\\"', '"').replace('<unk>',UNK).replace(' @.@ ','.').replace(' @-@ ','-').replace('...',' …')
return html.unescape(x)
test_eq(fix_html('#39;bli#146;'), "'bli'")
test_eq(fix_html('Sarah amp; Duck...'), 'Sarah & Duck …')
test_eq(fix_html('a nbsp; #36;'), 'a $')
test_eq(fix_html('\\" <unk>'), f'" {UNK}')
test_eq(fix_html('quot; @.@ @-@ '), "' .-")
test_eq(fix_html('<br />text\\n'), '\ntext\n')
#export
_re_all_caps = re.compile(r'(\s|^)([A-Z]+[^a-z\s]*)(?=(\s|$))')
#hide
"""
Catches any word in all caps, even with ' or - inside
(\s|^) Capture either a whitespace or the beginning of text
([A-Z]+ Capture one capitalized letter or more...
[^a-z\s]*) ...followed by anything that's non lowercase or whitespace
(?=(\s|$)) Look ahead for a space or end of text
""";
#export
def replace_all_caps(t):
"Replace tokens in ALL CAPS by their lower version and add `TK_UP` before."
def _replace_all_caps(m):
tok = f'{TK_UP} ' if len(m.groups()[1]) > 1 else ''
return f"{m.groups()[0]}{tok}{m.groups()[1].lower()}"
return _re_all_caps.sub(_replace_all_caps, t)
test_eq(replace_all_caps("I'M SHOUTING"), f"{TK_UP} i'm {TK_UP} shouting")
test_eq(replace_all_caps("I'm speaking normally"), "I'm speaking normally")
test_eq(replace_all_caps("I am speaking normally"), "i am speaking normally")
#export
_re_maj = re.compile(r'(\s|^)([A-Z][^A-Z\s]*)(?=(\s|$))')
#hide
"""
Catches any capitalized word
(\s|^) Capture either a whitespace or the beginning of text
([A-Z] Capture exactly one capitalized letter...
[^A-Z\s]*) ...followed by anything that's not uppercase or whitespace
(?=(\s|$)) Look ahead for a space of end of text
""";
#export
def replace_maj(t):
"Replace tokens in ALL CAPS by their lower version and add `TK_UP` before."
def _replace_maj(m):
tok = f'{TK_MAJ} ' if len(m.groups()[1]) > 1 else ''
return f"{m.groups()[0]}{tok}{m.groups()[1].lower()}"
return _re_maj.sub(_replace_maj, t)
test_eq(replace_maj("Jeremy Howard"), f'{TK_MAJ} jeremy {TK_MAJ} howard')
test_eq(replace_maj("I don't think there is any maj here"), ("i don't think there is any maj here"),)
#export
def lowercase(t, add_bos=True, add_eos=False):
"Converts `t` to lowercase"
return (f'{BOS} ' if add_bos else '') + t.lower().strip() + (f' {EOS}' if add_eos else '')
#export
def replace_space(t):
"Replace embedded spaces in a token with unicode line char to allow for split/join"
return t.replace(' ', '▁')
#export
defaults.text_spec_tok = [UNK, PAD, BOS, EOS, FLD, TK_REP, TK_WREP, TK_UP, TK_MAJ]
defaults.text_proc_rules = [fix_html, replace_rep, replace_wrep, spec_add_spaces, rm_useless_spaces,
replace_all_caps, replace_maj, lowercase]
defaults.text_postproc_rules = [replace_space]<jupyter_output><empty_output><jupyter_text>## TokenizingA tokenizer is a class that must implement a `pipe` method. This `pipe` method receives a generator of texts and must return a generator with their tokenized versions. Here is the most basic example:<jupyter_code>#export
class BaseTokenizer():
"Basic tokenizer that just splits on spaces"
def __init__(self, split_char=' ', **kwargs): self.split_char=split_char
def __call__(self, items): return (t.split(self.split_char) for t in items)
tok = BaseTokenizer()
for t in tok(["This is a text"]): test_eq(t, ["This", "is", "a", "text"])
tok = BaseTokenizer('x')
for t in tok(["This is a text"]): test_eq(t, ["This is a te", "t"])
#export
class SpacyTokenizer():
"Spacy tokenizer for `lang`"
def __init__(self, lang='en', special_toks=None, buf_sz=5000):
special_toks = ifnone(special_toks, defaults.text_spec_tok)
nlp = spacy.blank(lang, disable=["parser", "tagger", "ner"])
for w in special_toks: nlp.tokenizer.add_special_case(w, [{ORTH: w}])
self.pipe,self.buf_sz = nlp.pipe,buf_sz
def __call__(self, items):
return (L(doc).attrgot('text') for doc in self.pipe(items, batch_size=self.buf_sz))
tok = SpacyTokenizer()
inp,exp = "This isn't the easiest text.",["This", "is", "n't", "the", "easiest", "text", "."]
test_eq(L(tok([inp]*5)), [exp]*5)
#export
class TokenizeBatch:
"A wrapper around `tok_func` to apply `rules` and tokenize in parallel"
def __init__(self, tok_func=SpacyTokenizer, rules=None, post_rules=None, **tok_kwargs ):
self.rules = L(ifnone(rules, defaults.text_proc_rules))
self.post_f = compose(*L(ifnone(post_rules, defaults.text_postproc_rules)))
self.tok = tok_func(**tok_kwargs)
def __call__(self, batch):
return (L(o).map(self.post_f) for o in self.tok(maps(*self.rules, batch)))
f = TokenizeBatch()
test_eq(f(["This isn't a problem"]), [[BOS, TK_MAJ, 'this', 'is', "n't", 'a', 'problem']])
f = TokenizeBatch(BaseTokenizer, rules=[], split_char="'")
test_eq(f(["This isn't a problem"]), [['This▁isn', 't▁a▁problem']])<jupyter_output><empty_output><jupyter_text>The main function that will be called during one of the processes handling tokenization. It will create an instance of a tokenizer with `tok_func` and `tok_kwargs` at init, then iterate through the `batch` of texts, apply them `rules` and tokenize them.<jupyter_code>texts = ["this is a text", "this is another text"]
tok = TokenizeBatch(BaseTokenizer, texts.__getitem__)
test_eq([t for t in tok([0,1])],[['this', 'is', 'a', 'text'], ['this', 'is', 'another', 'text']])
#export
def tokenize1(text, tok_func=SpacyTokenizer, rules=None, post_rules=None, **tok_kwargs):
"Tokenize one `text` with an instance of `tok_func` and some `rules`"
return next(iter(TokenizeBatch(tok_func, rules, post_rules, **tok_kwargs)([text])))
test_eq(tokenize1("This isn't a problem"),
[BOS, TK_MAJ, 'this', 'is', "n't", 'a', 'problem'])
test_eq(tokenize1("This isn't a problem", BaseTokenizer, rules=[], split_char="'"),
['This▁isn', 't▁a▁problem'])
#export
def parallel_tokenize(items, tok_func, rules, as_gen=False, n_workers=defaults.cpus, **tok_kwargs):
"Calls a potential setup on `tok_func` before launching `TokenizeBatch` in parallel"
if hasattr(tok_func, 'setup'): tok_kwargs = tok_func(**tok_kwargs).setup(items, rules)
return parallel_gen(TokenizeBatch, items, as_gen=as_gen, tok_func=tok_func,
rules=rules, n_workers=n_workers, **tok_kwargs)<jupyter_output><empty_output><jupyter_text>### Tokenize texts in filesPreprocessing function for texts in filenames. Tokenized texts will be saved in a similar fashion in a directory suffixed with `_tok` in the parent folder of `path` (override with `output_dir`).<jupyter_code>#export
fn_counter_pkl = 'counter.pkl'
#export
def tokenize_folder(path, extensions=None, folders=None, output_dir=None, n_workers=defaults.cpus,
rules=None, tok_func=SpacyTokenizer, **tok_kwargs):
"Tokenize text files in `path` in parallel using `n_workers`"
path,extensions = Path(path),ifnone(extensions, ['.txt'])
fnames = get_files(path, extensions=extensions, recurse=True, folders=folders)
output_dir = Path(ifnone(output_dir, path.parent/f'{path.name}_tok'))
rules = Path.read + L(ifnone(rules, defaults.text_proc_rules.copy()))
counter = Counter()
for i,tok in parallel_tokenize(fnames, tok_func, rules, as_gen=True, n_workers=n_workers, **tok_kwargs):
out = output_dir/fnames[i].relative_to(path)
out.write(' '.join(tok))
out.with_suffix('.len').write(str(len(tok)))
counter.update(tok)
(output_dir/fn_counter_pkl).save(counter)<jupyter_output><empty_output><jupyter_text>The result will be in `output_dir` (defaults to a folder in the same parent directory as `path`, with `_tok` added to `path.name`) with the same structure as in `path`. Tokenized texts for a given file will be in the file having the same name in `output_dir`. Additionally, a file with a .len suffix contains the number of tokens and the count of all words is stored in `output_dir/counter.pkl`.
`extensions` will default to `['.txt']` and all text files in `path` are treated unless you specify a list of folders in `include`. `tok_func` is instantiated in each process with `tok_kwargs`, and `rules` (that defaults to `defaults.text_proc_rules`) are applied to each text before going in the tokenizer.### Tokenize texts in a dataframe<jupyter_code>#export
def _join_texts(df, mark_fields=False):
"Join texts in row `idx` of `df`, marking each field with `FLD` if `mark_fields=True`"
text_col = (f'{FLD} {1} ' if mark_fields else '' ) + df.iloc[:,0].astype(str)
for i in range(1,len(df.columns)):
text_col += (f' {FLD} {i+1} ' if mark_fields else ' ') + df.iloc[:,i].astype(str)
return text_col.values
#hide
texts = [f"This is an example of text {i}" for i in range(10)]
df = pd.DataFrame({'text': texts, 'text1': texts}, columns=['text', 'text1'])
col = _join_texts(df, mark_fields=True)
for i in range(len(df)):
test_eq(col[i], f'{FLD} 1 This is an example of text {i} {FLD} 2 This is an example of text {i}')
#export
def tokenize_df(df, text_cols, n_workers=defaults.cpus, rules=None, mark_fields=None,
tok_func=SpacyTokenizer, **tok_kwargs):
"Tokenize texts in `df[text_cols]` in parallel using `n_workers`"
text_cols = L(text_cols)
#mark_fields defaults to False if there is one column of texts, True if there are multiple
if mark_fields is None: mark_fields = len(text_cols)>1
rules = L(ifnone(rules, defaults.text_proc_rules.copy()))
texts = _join_texts(df[text_cols], mark_fields=mark_fields)
outputs = L(parallel_tokenize(texts, tok_func, rules, n_workers=n_workers, **tok_kwargs)
).sorted().itemgot(1)
other_cols = df.columns[~df.columns.isin(text_cols)]
res = df[other_cols].copy()
res['text'],res['text_lengths'] = outputs,outputs.map(len)
return res,Counter(outputs.concat())<jupyter_output><empty_output><jupyter_text>This function returns a new dataframe with the same non-text columns, a colum named text that contains the tokenized texts and a column named text_lengths that contains their respective length. It also returns a counter of all words see to quickly build a vocabulary afterward.
`tok_func` is instantiated in each process with `tok_kwargs`, and `rules` (that defaults to `defaults.text_proc_rules`) are applied to each text before going in the tokenizer. If `mark_fields` isn't specified, it defaults to `False` when there is a single text column, `True` when there are several. In that case, the texts in each of those columns are joined with `FLD` markes followed by the number of the field.<jupyter_code>#export
def tokenize_csv(fname, text_cols, outname=None, n_workers=4, rules=None, mark_fields=None,
tok_func=SpacyTokenizer, header='infer', chunksize=50000, **tok_kwargs):
"Tokenize texts in the `text_cols` of the csv `fname` in parallel using `n_workers`"
df = pd.read_csv(fname, header=header, chunksize=chunksize)
outname = Path(ifnone(outname, fname.parent/f'{fname.stem}_tok.csv'))
cnt = Counter()
for i,dfp in enumerate(df):
out,c = tokenize_df(dfp, text_cols, n_workers=n_workers, rules=rules,
mark_fields=mark_fields, tok_func=tok_func, **tok_kwargs)
out.text = out.text.str.join(' ')
out.to_csv(outname, header=(None,header)[i==0], index=False, mode=('a','w')[i==0])
cnt.update(c)
outname.with_suffix('.pkl').save(cnt)
#export
def load_tokenized_csv(fname):
"Utility function to quickly load a tokenized csv ans the corresponding counter"
fname = Path(fname)
out = pd.read_csv(fname)
for txt_col in out.columns[1:-1]:
out[txt_col] = out[txt_col].str.split(' ')
return out,fname.with_suffix('.pkl').load()<jupyter_output><empty_output><jupyter_text>The result will be written in a new csv file in `outname` (defaults to the same as `fname` with the suffix `_tok.csv`) and will have the same header as the original file, the same non-text columns, a text and a text_lengths column as described in `tokenize_df`.
`tok_func` is instantiated in each process with `tok_kwargs`, and `rules` (that defaults to `defaults.text_proc_rules`) are applied to each text before going in the tokenizer. If `mark_fields` isn't specified, it defaults to `False` when there is a single text column, `True` when there are several. In that case, the texts in each of those columns are joined with `FLD` markes followed by the number of the field.
The csv file is opened with `header` and optionally with blocks of `chunksize` at a time. If this argument is passed, each chunk is processed independtly and saved in the output file to save memory usage.<jupyter_code>def _prepare_texts(tmp_d):
"Prepare texts in a folder struct in tmp_d, a csv file and returns a dataframe"
path = Path(tmp_d)/'tmp'
path.mkdir()
for d in ['a', 'b', 'c']:
(path/d).mkdir()
for i in range(5):
with open(path/d/f'text{i}.txt', 'w') as f: f.write(f"This is an example of text {d} {i}")
texts = [f"This is an example of text {d} {i}" for i in range(5) for d in ['a', 'b', 'c']]
df = pd.DataFrame({'text': texts, 'label': list(range(15))}, columns=['text', 'label'])
csv_fname = tmp_d/'input.csv'
df.to_csv(csv_fname, index=False)
return path,df,csv_fname
with tempfile.TemporaryDirectory() as tmp_d:
path,df,csv_fname = _prepare_texts(Path(tmp_d))
#Tokenize as folders
tokenize_folder(path)
outp = Path(tmp_d)/'tmp_tok'
for d in ['a', 'b', 'c']:
p = outp/d
for i in range(5):
test_eq((p/f'text{i}.txt').read(), ' '.join([
BOS, TK_MAJ, 'this', 'is', 'an', 'example', 'of', 'text', d, str(i) ]))
test_eq((p/f'text{i}.len').read(), '10')
cnt_a = (outp/fn_counter_pkl).load()
test_eq(cnt_a['this'], 15)
test_eq(cnt_a['a'], 5)
test_eq(cnt_a['0'], 3)
#Tokenize as a dataframe
out,cnt_b = tokenize_df(df, text_cols='text')
test_eq(list(out.columns), ['label', 'text', 'text_lengths'])
test_eq(out['label'].values, df['label'].values)
test_eq(out['text'], [(outp/d/f'text{i}.txt').read().split(' ') for i in range(5) for d in ['a', 'b', 'c']])
test_eq(cnt_a, cnt_b)
#Tokenize as a csv
out_fname = Path(tmp_d)/'output.csv'
tokenize_csv(csv_fname, text_cols='text', outname=out_fname)
test_eq((out,cnt_b), load_tokenized_csv(out_fname))<jupyter_output><empty_output><jupyter_text>## Sentencepiece<jupyter_code>eu_langs = ["bg", "cs", "da", "de", "el", "en", "es", "et", "fi", "fr", "ga", "hr", "hu",
"it","lt","lv","mt","nl","pl","pt","ro","sk","sl","sv"] # all European langs
#export
class SentencePieceTokenizer():#TODO: pass the special tokens symbol to sp
"Spacy tokenizer for `lang`"
def __init__(self, lang='en', special_toks=None, sp_model=None, vocab_sz=None, max_vocab_sz=30000,
model_type='unigram', char_coverage=None, cache_dir='tmp'):
try: from sentencepiece import SentencePieceTrainer,SentencePieceProcessor
except ImportError:
raise Exception('sentencepiece module is missing: run `pip install sentencepiece`')
self.sp_model,self.cache_dir = sp_model,Path(cache_dir)
self.vocab_sz,self.max_vocab_sz,self.model_type = vocab_sz,max_vocab_sz,model_type
self.char_coverage = ifnone(char_coverage, 0.99999 if lang in eu_langs else 0.9998)
self.special_toks = ifnone(special_toks, defaults.text_spec_tok)
if sp_model is None: self.tok = None
else:
self.tok = SentencePieceProcessor()
self.tok.Load(str(sp_model))
os.makedirs(self.cache_dir, exist_ok=True)
def _get_vocab_sz(self, raw_text_path):
cnt = Counter()
with open(raw_text_path, 'r') as f:
for line in f.readlines():
cnt.update(line.split())
if len(cnt)//4 > self.max_vocab_sz: return self.max_vocab_sz
res = len(cnt)//4
while res%8 != 0: res+=1
return res
def train(self, raw_text_path):
"Train a sentencepiece tokenizer on `texts` and save it in `path/tmp_dir`"
from sentencepiece import SentencePieceTrainer
vocab_sz = self._get_vocab_sz(raw_text_path) if self.vocab_sz is None else self.vocab_sz
spec_tokens = ['\u2581'+s for s in self.special_toks]
SentencePieceTrainer.Train(" ".join([
f"--input={raw_text_path} --vocab_size={vocab_sz} --model_prefix={self.cache_dir/'spm'}",
f"--character_coverage={self.char_coverage} --model_type={self.model_type}",
f"--unk_id={len(spec_tokens)} --pad_id=-1 --bos_id=-1 --eos_id=-1",
f"--user_defined_symbols={','.join(spec_tokens)}"]))
raw_text_path.unlink()
return self.cache_dir/'spm.model'
def setup(self, items, rules):
if self.tok is not None: return {'sp_model': self.sp_model}
raw_text_path = self.cache_dir/'texts.out'
with open(raw_text_path, 'w') as f:
for t in progress_bar(maps(*rules, items), total=len(items), leave=False):
f.write(f'{t}\n')
return {'sp_model': self.train(raw_text_path)}
def __call__(self, items):
for t in items: yield self.tok.EncodeAsPieces(t)
texts = [f"This is an example of text {i}" for i in range(10)]
df = pd.DataFrame({'text': texts, 'label': list(range(10))}, columns=['text', 'label'])
out,cnt = tokenize_df(df, text_cols='text', tok_func=SentencePieceTokenizer, vocab_sz=34)<jupyter_output><empty_output><jupyter_text>## Export -<jupyter_code>#hide
from local.notebook.export import notebook2script
notebook2script(all_fs=True)<jupyter_output>Converted 00_test.ipynb.
Converted 01_core.ipynb.
Converted 01a_torch_core.ipynb.
Converted 02_script.ipynb.
Converted 03_dataloader.ipynb.
Converted 04_transform.ipynb.
Converted 05_data_core.ipynb.
Converted 06_data_transforms.ipynb.
Converted 07_vision_core.ipynb.
Converted 08_pets_tutorial.ipynb.
Converted 09_vision_augment.ipynb.
Converted 11_layers.ipynb.
Converted 11a_vision_models_xresnet.ipynb.
Converted 12_optimizer.ipynb.
Converted 13_learner.ipynb.
Converted 14_callback_schedule.ipynb.
Converted 14a_callback_data.ipynb.
Converted 15_callback_hook.ipynb.
Converted 16_callback_progress.ipynb.
Converted 17_callback_tracker.ipynb.
Converted 18_callback_fp16.ipynb.
Converted 19_callback_mixup.ipynb.
Converted 20_metrics.ipynb.
Converted 21_tutorial_imagenette.ipynb.
Converted 22_vision_learner.ipynb.
Converted 23_tutorial_transfer_learning.ipynb.
Converted 30_text_core.ipynb.
Converted 31_text_data.ipynb.
Converted 32_text_models_awdlstm.ipynb.
Converted 33_text_models_core.ipyn[...]
|
non_permissive
|
/dev/30_text_core.ipynb
|
davanstrien/fastai_dev
| 12 |
<jupyter_start><jupyter_text><jupyter_code>Ingest<jupyter_output><empty_output>
|
no_license
|
/Aparna_R.ipynb
|
aparnark-git/dsi_gittingstarted
| 1 |
<jupyter_start><jupyter_text>Capstone project on Segmenting and Clustering Neighborhoods in Toronto_Part1
## Problem 1
Use the Notebook to build the code to scrape the following Wikipedia page, https://en.wikipedia.org/wiki/List_of_postal_codes_of_Canada:_M, in order to obtain the data that is in the table of postal codes and to transform the data into a pandas dataframe.
To create the above dataframe:
- The dataframe will consist of three columns: PostalCode, Borough, and Neighborhood
- Only process the cells that have an assigned borough. Ignore cells with a borough that is Not assigned.
- More than one neighborhood can exist in one postal code area. For example, in the table on the Wikipedia page, you will notice that - M5A is listed twice and has two neighborhoods: Harbourfront and Regent Park. These two rows will be combined into one row with the - neighborhoods separated with a comma as shown in row 11 in the above table.
- If a cell has a borough but a Not assigned neighborhood, then the neighborhood will be the same as the borough. So for the 9th cell in the table on the Wikipedia page, the value of the Borough and the Neighborhood columns will be Queen's Park.
- Clean your Notebook and add Markdown cells to explain your work and any assumptions you are making.
- In the last cell of your notebook, use the .shape method to print the number of rows of your dataframe.
<jupyter_code>import numpy as np # library to handle data in a vectorized manner
import pandas as pd # library for data analsysis
pd.set_option('display.max_columns', None)
pd.set_option('display.max_rows', None)
import json # library to handle JSON files
#!conda install -c conda-forge geopy --yes # uncomment this line if you haven't completed the Foursquare API lab
from geopy.geocoders import Nominatim # convert an address into latitude and longitude values
import requests # library to handle requests
from pandas.io.json import json_normalize # tranform JSON file into a pandas dataframe
# Matplotlib and associated plotting modules
import matplotlib.cm as cm
import matplotlib.colors as colors
# import k-means from clustering stage
from sklearn.cluster import KMeans
from bs4 import BeautifulSoup
#!conda install -c conda-forge folium=0.5.0 --yes # uncomment this line if you haven't completed the Foursquare API lab
import folium # map rendering library
import requests
webUrl = requests.get('https://en.wikipedia.org/wiki/List_of_postal_codes_of_Canada:_M').text
print('Libraries imported.')<jupyter_output>Libraries imported.
<jupyter_text>###### Scrape the List of postal codes of Canada<jupyter_code>soup = BeautifulSoup(webUrl, 'xml')
table=soup.find('table')
#print(table)
#dataframe will consist of three columns: PostalCode, Borough, and Neighborhood
column_names = ['Postalcode','Borough','Neighborhood']
data = pd.DataFrame(columns = column_names)
# Search all the postcode, borough, neighborhood
for tr in table.find_all('tr'):
rowData=[]
for td in tr.find_all('td'):
rowData.append(td.text.strip())
if len(rowData)==3:
data.loc[len(data)] = rowData
data.head(20)<jupyter_output><empty_output><jupyter_text>###### Data Cleaning
remove rows where Borough is 'Not assigned'<jupyter_code>data=data[data['Borough']!='Not assigned']
data.head()
data[data['Neighborhood']=='Not assigned']=data['Borough']
data.head()
temp_data=data.groupby('Postalcode')['Neighborhood'].apply(lambda x: "%s" % ', '.join(x))
temp_data=temp_data.reset_index(drop=False)
temp_data.rename(columns={'Neighborhood':'Neighborhood_joined'},inplace=True)
data_merge = pd.merge(data, temp_data, on='Postalcode')
data_merge.drop(['Neighborhood'],axis=1,inplace=True)
data_merge.drop_duplicates(inplace=True)
data_merge.rename(columns={'Neighborhood_joined':'Neighborhood'},inplace=True)
data_merge.head()
data_merge.shape<jupyter_output><empty_output>
|
no_license
|
/Neighborhoods_in_Toronto_1st.ipynb
|
natfik/Coursera_Capstone
| 3 |
<jupyter_start><jupyter_text># TD3
**Author:** [pavelkolev](https://github.com/PavelKolev)
**Extending the work of:** [amifunny](https://github.com/amifunny)
**Date created:** 2020/09/22
**Last modified:** 2020/09/22
**Description:** Implementing TD3 algorithm on the LunarLanderContinuous-v2.<jupyter_code>import sys, os
if 'google.colab' in sys.modules:
if not os.path.exists('.setup_complete'):
!wget -q https://raw.githubusercontent.com/yandexdataschool/Practical_RL/master/setup_colab.sh -O- | bash
!touch .setup_complete
# This code creates a virtual display to draw game images on.
# It will have no effect if your machine has a monitor.
if type(os.environ.get("DISPLAY")) is not str or len(os.environ.get("DISPLAY")) == 0:
!bash ../xvfb start
os.environ['DISPLAY'] = ':1'
!pip install box2d-py
import tensorflow as tf
from tensorflow.keras import layers
import gym
import numpy as np<jupyter_output><empty_output><jupyter_text>We use [OpenAIGym](http://gym.openai.com/docs) to create the environment.
We will use the `upper_bound` parameter to scale our actions later.<jupyter_code>env = gym.make('LunarLanderContinuous-v2')
env.reset()
dim_state = env.observation_space.shape[0]
print("Size of State Space -> {}".format(dim_state))
dim_action = env.action_space.shape[0]
print("Size of Action Space -> {}".format(dim_action))
lower_bound = env.action_space.low[0]
upper_bound = env.action_space.high[0]
print("Min Value of Action -> {}".format(lower_bound))
print("Max Value of Action -> {}".format(upper_bound))
import matplotlib.pyplot as plt
%matplotlib inline
plt.imshow(env.render('rgb_array'))
plt.show()<jupyter_output><empty_output><jupyter_text>To implement better exploration by the Actor network, we use noisy perturbations, specifically
an **Ornstein-Uhlenbeck process** for generating noise, as described in the paper.
It samples noise from a correlated normal distribution.<jupyter_code>class Gaussian:
def __init__(self, mean, std_deviation, bounds = None):
self.mean = mean
self.std_dev = std_deviation
self.bounds = bounds
def __call__(self):
x = np.random.normal(self.mean, self.std_dev, self.mean.shape)
if not self.bounds == None:
x = np.clip(x, self.bounds[0], self.bounds[1])
return x<jupyter_output><empty_output><jupyter_text>---
TD3 Algorithm Explained:
https://lilianweng.github.io/lil-log/2018/04/08/policy-gradient-algorithms.html#td3
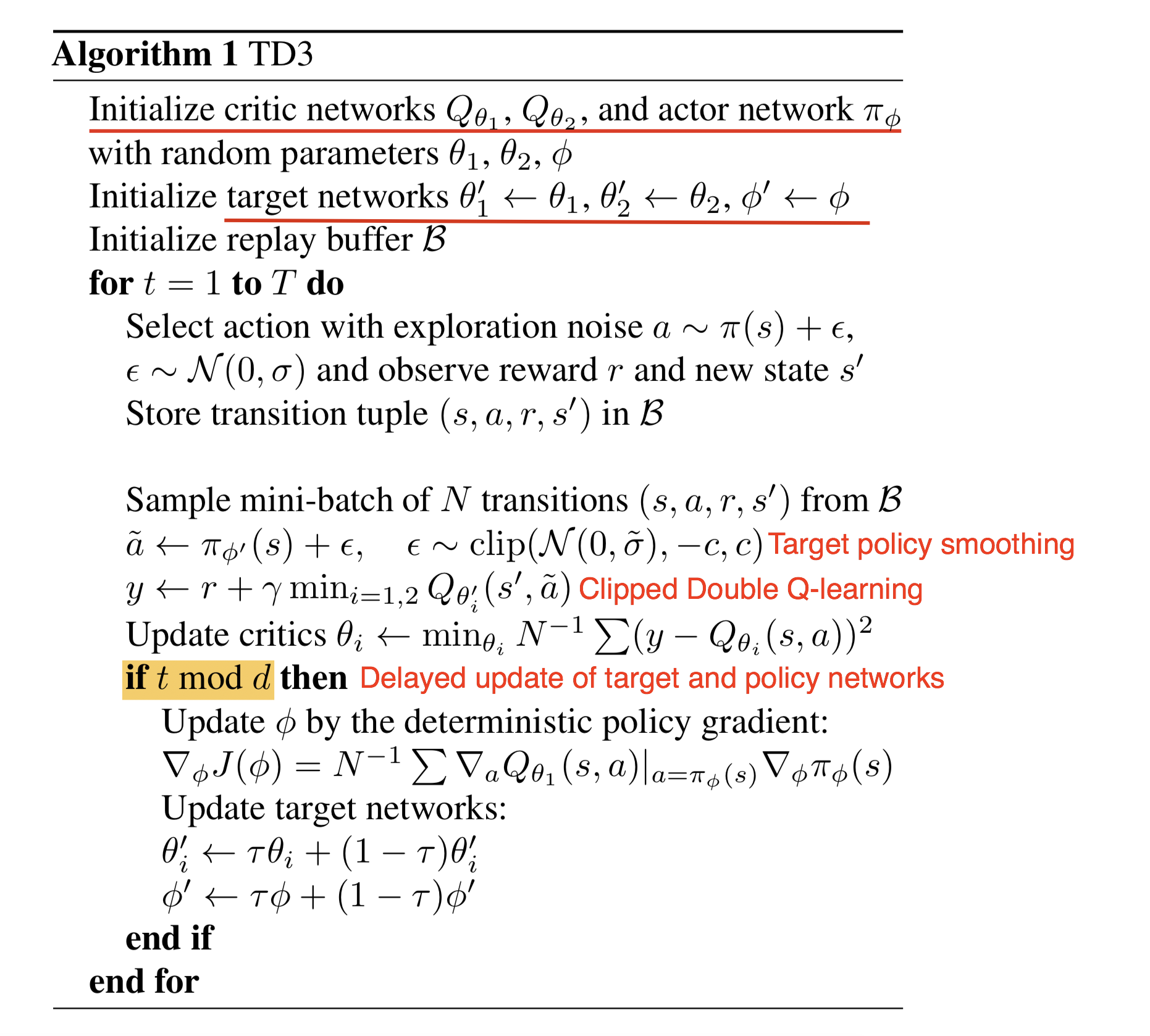
--------------------------------Here we define the Actor and Critic networks. These are basic Dense models
with `ReLU` activation. `BatchNormalization` is used to normalize dimensions across
samples in a mini-batch, as activations can vary a lot due to fluctuating values of input
state and action.
Note: We need the initialization for last layer of the Actor to be between
`-0.003` and `0.003` as this prevents us from getting `1` or `-1` output values in
the initial stages, which would squash our gradients to zero,
as we use the `tanh` activation.<jupyter_code># mu(s) = a
def get_actor():
# Initialize weights between -3e-3 and 3-e3
inputs = layers.Input(shape=(dim_state,))
out = layers.Dense(256, activation="relu")(inputs)
out = layers.Dense(128, activation="relu")(out)
outputs = layers.Dense(dim_action, activation="tanh")(out)
# Scaling: Upper bound
outputs = outputs * upper_bound
model = tf.keras.Model(inputs, outputs)
return model
# Q(s,a)
def get_critic():
# State as input
state_input = layers.Input(shape=(dim_state))
state_out = layers.Dense(32, activation="relu")(state_input)
# Action as input
action_input = layers.Input(shape=(dim_action))
action_out = layers.Dense(32, activation="relu")(action_input)
# Both are passed through seperate layer before concatenating
concat = layers.Concatenate()([state_out, action_out])
out = layers.Dense(256, activation="relu")(concat)
out = layers.Dense(128, activation="relu")(out)
outputs = layers.Dense(1)(out)
# Outputs single value for give state-action
model = tf.keras.Model([state_input, action_input], outputs)
return model<jupyter_output><empty_output><jupyter_text>## Training hyperparameters<jupyter_code># Learning rate for actor-critic models
critic_lr = 0.001
actor_lr = 0.0001
total_episodes = 4000
# Size N
N = 2 ** 16
size_B = 2 ** 6
# Discount factor for future rewards
gamma = 0.99
# Used to update target networks
tau = 0.005
noise_mu = Gaussian(mean = np.zeros(dim_action),
std_deviation = float( 0.1 ) * np.ones(dim_action))
noise_Q = Gaussian(mean = np.zeros(dim_action),
std_deviation = float(0.2) * np.ones(dim_action),
bounds = [-0.5, 0.5])
# Function Approx: mu and Q(s,a)
mu = get_actor()
mu_t = get_actor()
Q1 = get_critic()
Q1_t = get_critic()
Q2 = get_critic()
Q2_t = get_critic()
# Making the weights equal initially
mu_t.set_weights(mu.get_weights())
Q1_t.set_weights(Q1.get_weights())
Q2_t.set_weights(Q2.get_weights())
mu_opt = tf.keras.optimizers.Adam(actor_lr)
Q1_opt = tf.keras.optimizers.Adam(critic_lr)
Q2_opt = tf.keras.optimizers.Adam(critic_lr)<jupyter_output><empty_output><jupyter_text>`policy()` returns an action sampled from our Actor network plus some noise for
exploration.<jupyter_code>def policy(actor, state, noise_object):
# Call "actor" mu
sampled_actions = tf.squeeze( actor(state) )
# Sample Noise
noise = noise_object()
sampled_actions = sampled_actions.numpy() + noise
# Legal action
legal_action = np.clip(sampled_actions, lower_bound, upper_bound)
return legal_action
@tf.function
def policy_symbolic(actor, state, noise_object):
sampled_actions = tf.squeeze( actor(state) )
# Sample Noise
noise = noise_object()
sampled_actions = sampled_actions + noise
# Legal action
legal_action = tf.clip_by_value(sampled_actions, lower_bound, upper_bound)
return legal_action
# Eager execution is turned on by default in TensorFlow 2.
# Decorating with tf.function allows TensorFlow to build a static graph
# out of the logic and computations in our function.
# This provides a large speed up for blocks of code that contain
# many small TensorFlow operations such as this one.
# Input: each has dim [batch,?]
@tf.function
def update_Q1_Q2(state, action, reward, next_state, is_done):
with tf.GradientTape(persistent = True) as tape:
next_action = tf.stop_gradient( policy_symbolic(mu_t, next_state, noise_Q) )
Q1_t_nsa = tf.stop_gradient( Q1_t([next_state, next_action]) )
Q2_t_nsa = tf.stop_gradient( Q2_t([next_state, next_action]) )
y = reward + (1.0 - is_done) * gamma * tf.math.minimum(Q1_t_nsa, Q2_t_nsa)
Q1_nsa = Q1([state, action])
Q2_nsa = Q2([state, action])
diff_1_squared = tf.math.square(y - Q1_nsa)
diff_2_squared = tf.math.square(y - Q2_nsa)
Q1_loss = tf.math.reduce_mean(diff_1_squared)
Q2_loss = tf.math.reduce_mean(diff_2_squared)
Q1_vars = Q1.trainable_variables
Q1_grad = tape.gradient(Q1_loss, Q1_vars)
Q1_opt.apply_gradients(zip(Q1_grad, Q1_vars))
Q2_vars = Q2.trainable_variables
Q2_grad = tape.gradient(Q2_loss, Q2_vars)
Q2_opt.apply_gradients(zip(Q2_grad, Q2_vars))
# Input: each has dim [batch,?]
@tf.function
def update_mu(state, action):
with tf.GradientTape() as tape:
actions = mu(state)
Q1_sa = Q1([state, actions])
# Used "-value" as we want to maximize the value given
# by the critic for our actions
mu_loss = -tf.math.reduce_mean(Q1_sa)
mu_vars = mu.trainable_variables
mu_grad = tape.gradient(mu_loss, mu_vars)
mu_opt.apply_gradients(zip(mu_grad, mu_vars))
# This update target parameters slowly
# Based on rate `tau`, which is much less than one.
@tf.function
def update_target(target_weights, weights, tau):
for (t, w) in zip(target_weights, weights):
t.assign( (1 - tau) * t + tau * w )
class Buffer:
def __init__(self, capacity=2**14, batch_size=2**3):
self.batch_size = batch_size
self.capacity = capacity
self.buffer_index = 0
# capacity
self.state = np.zeros((capacity, dim_state))
self.action = np.zeros((capacity, dim_action))
self.reward = np.zeros((capacity, 1))
self.next_state = np.zeros((capacity, dim_state))
self.is_done = np.zeros((capacity, 1))
# Takes (s,a,r,s',done) obervation tuple
def record(self, obs_tuple):
# Set index to zero if capacity is exceeded,
# replacing old records
index = self.buffer_index % self.capacity
self.state[index] = obs_tuple[0]
self.action[index] = obs_tuple[1]
self.reward[index] = obs_tuple[2]
self.next_state[index] = obs_tuple[3]
self.is_done[index] = obs_tuple[4]
self.buffer_index += 1
# We compute the loss and update parameters
def learn(self, update_mu_target):
# Get sampling range
record_range = min(self.buffer_index, self.capacity)
# Randomly sample indices
batch_indices = np.random.choice(record_range, self.batch_size)
# Convert to tensors
state_batch = tf.convert_to_tensor(self.state[batch_indices])
action_batch = tf.convert_to_tensor(self.action[batch_indices])
reward_batch = tf.convert_to_tensor(self.reward[batch_indices])
reward_batch = tf.cast(reward_batch, dtype=tf.float32)
next_state_batch = tf.convert_to_tensor(self.next_state[batch_indices])
is_done_batch = tf.convert_to_tensor(self.is_done[batch_indices])
is_done_batch = tf.cast(is_done_batch, dtype=tf.float32)
# Update Current Models
deltas = update_Q1_Q2(state_batch, action_batch, reward_batch,
next_state_batch, is_done_batch)
# Train Target Models
if update_mu_target:
# Update policy mu
update_mu(state_batch, action_batch)
# Update All Target models
update_target(Q1_t.variables, Q1.variables, tau)
update_target(Q2_t.variables, Q2.variables, tau)
update_target(mu_t.variables, mu.variables, tau)
<jupyter_output><empty_output><jupyter_text>Now we implement our main training loop, and iterate over episodes.
We sample actions using `policy()` and train with `learn()` at each time step,
along with updating the Target networks at a rate `tau`.<jupyter_code>from IPython.display import clear_output
from tqdm import trange
# To store reward history of each episode
ep_reward_list = []
# To store average reward history of last few episodes
avg_reward_list = []
# Create Buffer
buffer = Buffer(N, size_B)
def save_weights():
# Save the weights
mu.save_weights("mu.h5")
mu_t.save_weights("mu_t.h5")
Q1.save_weights("Q1.h5")
Q1_t.save_weights("Q1_t.h5")
Q2.save_weights("Q2.h5")
Q2_t.save_weights("Q2_t.h5")
total_episodes = 4000
# Takes about 4 min to train
is_train_done = False
curr_iter = 0
for ep in trange(total_episodes + 1):
prev_state = env.reset()
episodic_reward = 0
while True:
# Uncomment this to see the Actor in action
# But not in a python notebook.
# env.render()
tf_prev_state = tf.expand_dims(tf.convert_to_tensor(prev_state), 0)
action = policy(mu, tf_prev_state, noise_mu)
# Recieve state and reward from environment.
state, reward, done, info = env.step(action)
reward = reward / 200.
obs_tuple = (prev_state, action, reward, state, done)
buffer.record(obs_tuple)
episodic_reward += reward
update_mu_target = (curr_iter % 2 == 0 )
buffer.learn(update_mu_target)
# End this episode when `done` is True
if done:
break
prev_state = state
curr_iter += 1
ep_reward_list.append(episodic_reward)
# Mean of last 40 episodes
avg_reward = np.mean(ep_reward_list[-40:])
avg_reward_list.append(200 * avg_reward)
if ep % 50 == 0:
# Plotting graph
# Episodes versus Avg. Rewards
clear_output(True)
plt.plot(avg_reward_list)
plt.xlabel("Episode")
plt.ylabel("Avg. Epsiodic Reward")
plt.show()
if avg_reward >= 200:
print("Pass!")
is_train_done = True
break
if is_train_done:
save_weights()
break
if ep >=1 and ep % 500 == 0:
save_weights()
<jupyter_output><empty_output><jupyter_text>Evaluate Learned Agent<jupyter_code>def det_policy(state):
# Use deterministic actor: "mu"
sampled_actions = tf.squeeze( mu(state) )
# We make sure action is within bounds
legal_action = np.clip(sampled_actions, lower_bound, upper_bound)
return legal_action
def evaluate(env, n_games=1):
"""Plays an a game from start till done, returns per-game rewards """
game_rewards = []
for _ in range(n_games):
# initial observation and memory
obs = env.reset()
total_reward = 0
while True:
action = det_policy(obs[None])
obs, reward, done, info = env.step(action)
total_reward += reward
if done:
break
game_rewards.append(total_reward)
return game_rewards
import gym.wrappers
with gym.wrappers.Monitor(env, directory="videos", force=True) as env_monitor:
final_rewards = evaluate(env_monitor, n_games=10)
print("Final mean reward", np.mean(final_rewards))<jupyter_output>Final mean reward 274.87823239927627
<jupyter_text>-------------------------------------------------<jupyter_code>gpu_info = !nvidia-smi
gpu_info = '\n'.join(gpu_info)
if gpu_info.find('failed') >= 0:
print('Select the Runtime > "Change runtime type" menu to enable a GPU accelerator, ')
print('and then re-execute this cell.')
else:
print(gpu_info)
from psutil import virtual_memory
ram_gb = virtual_memory().total / 1e9
print('Your runtime has {:.1f} gigabytes of available RAM\n'.format(ram_gb))
if ram_gb < 20:
print('To enable a high-RAM runtime, select the Runtime > "Change runtime type"')
print('menu, and then select High-RAM in the Runtime shape dropdown. Then, ')
print('re-execute this cell.')
else:
print('You are using a high-RAM runtime!')
<jupyter_output><empty_output>
|
no_license
|
/TD3_Gaussian/TD3_LLCont.ipynb
|
PavelKolev/rl_algos
| 9 |
<jupyter_start><jupyter_text># Load and Test Model for test set
This notebook loads the model and performs predictions on all the data in the Test set (the split with the least data samples). Once the predictions are completed, a classification plot and confusion matrix is displayed.## Load dependencies<jupyter_code>import matplotlib.pyplot as plt
import numpy as np
from PIL import Image
import torch
from torchvision import transforms
from tqdm import tqdm
import sys
import av
import pandas as pd
import os
from sklearn.metrics import classification_report, confusion_matrix, ConfusionMatrixDisplay<jupyter_output><empty_output><jupyter_text>## Init file paths and dataframes<jupyter_code>data_dir = "dataset"
test_dir = f'../{data_dir}/test'
# All labels
filtered_data = "data"
test_label_df = pd.read_csv(f'../{filtered_data}/test.csv', header=None)
# convert all into hashmap - key = u_vid_name , value = label
test_label = {f"{test_dir}/{k[0]}": k[1] for k in test_label_df.values.tolist()}
# Total label + turkish to english translation
total_label = pd.read_csv(f'../{filtered_data}/filtered_ClassId.csv')
u_len_label = len(total_label['ClassId'].unique())
print("total unique label:", u_len_label)
class_id_to_label = {k[0]: k[2] for k in total_label.values.tolist()}
print(class_id_to_label)<jupyter_output>{0: 'champion', 1: 'glass', 2: 'wrong', 3: 'bad', 4: 'married', 5: 'potato', 6: 'congratulations', 7: 'child', 8: 'inform', 9: 'father'}
<jupyter_text>## Load and process video<jupyter_code>def extract_frames(vid_path, frames_cap, transforms=None):
"""Extract and transform video frames
Parameters:
vid_path (str): path to video file
frames_cap (int): number of frames to extract, evenly spaced
transforms (torchvision.transforms, optional): transformations to apply to frame
Returns:
list of numpy.array: vid_arr
"""
vid_arr = []
with av.open(vid_path) as container:
stream = container.streams.video[0]
n_frames = stream.frames
remainder = n_frames % frames_cap
interval = n_frames // frames_cap
take_frame_idx = 0
for frame_no, frame in enumerate(container.decode(stream)):
if frame_no == take_frame_idx:
img = frame.to_image()
if transforms:
img = transforms(img)
vid_arr.append(np.array(img))
if remainder > 0:
take_frame_idx += 1
remainder -= 1
take_frame_idx += interval
if len(vid_arr) < frames_cap:
raise ValueError(f"video with path '{vid_path}' is too short, please make sure that video has >={frames_cap} frames")
return vid_arr<jupyter_output><empty_output><jupyter_text>## Load and test model<jupyter_code>module_path = os.path.abspath(os.path.join('../'))
if module_path not in sys.path:
sys.path.append(module_path)
from dev_model.models import CNN_LSTM
model = CNN_LSTM(10,
latent_size=512,
n_cnn_layers=6,
n_rnn_layers=1,
n_rnn_hidden_dim=512,
cnn_bn=True,
bidirectional=True,
dropout_rate=0.8,
device="cpu",
attention=True)
checkpoint = torch.load("../../models/final-model.pt", map_location=torch.device("cpu"))
model.load_state_dict(checkpoint['model_state_dict'])
model.eval()
def load_and_test_video(vid_name):
"""Load video from dataset and pass video to model
Parameters:
vid_name (str): video name to display
"""
transforms_compose = transforms.Compose([transforms.Resize(256),
transforms.ToTensor(),
transforms.Normalize(mean=[0.5], std=[0.5])])
vid_color_path = f"{test_dir}/{vid_name}_color.mp4"
rgb_arr = extract_frames(vid_color_path, 30, transforms=transforms_compose)
vid_arr = np.array(rgb_arr)
vid_arr = vid_arr/255
vid_arr = torch.from_numpy(vid_arr).float()
vid_arr = vid_arr.unsqueeze(0)
predict_id = model.forward(vid_arr)
predict_id = torch.max(predict_id, 1)[1].item()
ground_truth_id = test_label[f"{test_dir}/{vid_name}"]
return predict_id, class_id_to_label[predict_id], ground_truth_id, class_id_to_label[ground_truth_id]
# Load the test set and perform predictions on each sample. Add their ground truths and predictions to lists
df = pd.read_csv("../data/test.csv", names=["video_name", "class_id"])
predict_ids = []
predict_labels = []
ground_truth_ids = []
ground_truth_labels = []
for index, row in tqdm(df.iterrows(), total=df.shape[0]):
predict_id, predict_label, ground_truth_id, ground_truth_label = load_and_test_video(row['video_name'])
predict_ids.append(predict_id)
predict_labels.append(predict_label)
ground_truth_ids.append(ground_truth_id)
ground_truth_labels.append(ground_truth_label)
df['predict_id'] = predict_ids
df['predict_label'] = predict_labels
df['ground_truth_id'] = ground_truth_ids
df['ground_truth_label'] = ground_truth_labels
df<jupyter_output><empty_output><jupyter_text>## Analysis of results
- Table of a classification report that details precision, recall, f1-score per class and accuracy
- Confusion matrix of ground truth labels and predictions<jupyter_code>target_names = [class_id_to_label[key] if key in class_id_to_label.keys() else 0 for key in range(10)]
print(classification_report(ground_truth_ids, predict_ids, target_names=target_names))
print(f'Total test data:{len(df)}')
cm = confusion_matrix(ground_truth_ids, predict_ids)
disp = ConfusionMatrixDisplay(confusion_matrix=cm, display_labels=np.array(target_names))
fig, ax = plt.subplots(figsize=(14,14))
disp.plot(ax=ax)<jupyter_output>Total test data:164
|
no_license
|
/src/test_model/load_and_test_model.ipynb
|
penyepesto/signo-lingo
| 5 |
<jupyter_start><jupyter_text># Regression diagnosticsThis example file shows how to use a few of the ``statsmodels`` regression diagnostic tests in a real-life context. You can learn about more tests and find out more information about the tests here on the [Regression Diagnostics page.](https://www.statsmodels.org/stable/diagnostic.html)
Note that most of the tests described here only return a tuple of numbers, without any annotation. A full description of outputs is always included in the docstring and in the online ``statsmodels`` documentation. For presentation purposes, we use the ``zip(name,test)`` construct to pretty-print short descriptions in the examples below.## Estimate a regression model<jupyter_code>%matplotlib inline
from statsmodels.compat import lzip
import numpy as np
import pandas as pd
import statsmodels.formula.api as smf
import statsmodels.stats.api as sms
import matplotlib.pyplot as plt
# Load data
url = 'https://raw.githubusercontent.com/vincentarelbundock/Rdatasets/master/csv/HistData/Guerry.csv'
dat = pd.read_csv(url)
# Fit regression model (using the natural log of one of the regressors)
results = smf.ols('Lottery ~ Literacy + np.log(Pop1831)', data=dat).fit()
# Inspect the results
print(results.summary())<jupyter_output><empty_output><jupyter_text>## Normality of the residualsJarque-Bera test:<jupyter_code>name = ['Jarque-Bera', 'Chi^2 two-tail prob.', 'Skew', 'Kurtosis']
test = sms.jarque_bera(results.resid)
lzip(name, test)<jupyter_output><empty_output><jupyter_text>Omni test:<jupyter_code>name = ['Chi^2', 'Two-tail probability']
test = sms.omni_normtest(results.resid)
lzip(name, test)<jupyter_output><empty_output><jupyter_text>## Influence tests
Once created, an object of class ``OLSInfluence`` holds attributes and methods that allow users to assess the influence of each observation. For example, we can compute and extract the first few rows of DFbetas by:<jupyter_code>from statsmodels.stats.outliers_influence import OLSInfluence
test_class = OLSInfluence(results)
test_class.dfbetas[:5,:]<jupyter_output><empty_output><jupyter_text>Explore other options by typing ``dir(influence_test)``
Useful information on leverage can also be plotted:<jupyter_code>from statsmodels.graphics.regressionplots import plot_leverage_resid2
fig, ax = plt.subplots(figsize=(8,6))
fig = plot_leverage_resid2(results, ax = ax)<jupyter_output><empty_output><jupyter_text>Other plotting options can be found on the [Graphics page.](https://www.statsmodels.org/stable/graphics.html)## Multicollinearity
Condition number:<jupyter_code>np.linalg.cond(results.model.exog)<jupyter_output><empty_output><jupyter_text>## Heteroskedasticity tests
Breush-Pagan test:<jupyter_code>name = ['Lagrange multiplier statistic', 'p-value',
'f-value', 'f p-value']
test = sms.het_breuschpagan(results.resid, results.model.exog)
lzip(name, test)<jupyter_output><empty_output><jupyter_text>Goldfeld-Quandt test<jupyter_code>name = ['F statistic', 'p-value']
test = sms.het_goldfeldquandt(results.resid, results.model.exog)
lzip(name, test)<jupyter_output><empty_output><jupyter_text>## Linearity
Harvey-Collier multiplier test for Null hypothesis that the linear specification is correct:<jupyter_code>name = ['t value', 'p value']
test = sms.linear_harvey_collier(results)
lzip(name, test)<jupyter_output><empty_output>
|
permissive
|
/examples/notebooks/regression_diagnostics.ipynb
|
hsiaoyi0504/statsmodels
| 9 |
<jupyter_start><jupyter_text># Minimizing cost(4)
minimizing cost using tensorflow gradient<jupyter_code>import tensorflow as tf
# tf Graph Input
X = [1, 2, 3]
Y = [1, 2, 3]
# set wrong model weights
W = tf.Variable(5.)
# Linear model
hypothesis = X * W
# Manual gradient
gradient = tf.reduce_mean((W * X - Y) * X) * 2
# cost/loss function
cost = tf.reduce_mean(tf.square(hypothesis - Y))
# Minimize: Gradient Descent Optimizer
optimizer = tf.train.GradientDescentOptimizer(learning_rate=0.01)
# Get gradients
gvs = optimizer.compute_gradients(cost)
# Apply gradients
apply_gradients = optimizer.apply_gradients(gvs)
# Launch the graph in a session.
with tf.Session() as sess:
# Initializes global variables in the graph.
sess.run(tf.global_variables_initializer())
# gradient_val은 gradient값을,
# gvs_val은 gradient값과 그때의 W값을 출력
for step in range(101):
gradient_val, gvs_val, _ = sess.run([gradient, gvs, apply_gradients])
print(step, gradient_val, gvs_val)<jupyter_output>0 37.333332 [(37.333336, 4.6266665)]
1 33.84889 [(33.84889, 4.2881775)]
2 30.689657 [(30.689657, 3.9812808)]
3 27.825287 [(27.825287, 3.703028)]
4 25.228262 [(25.228262, 3.4507453)]
5 22.873621 [(22.873623, 3.2220092)]
6 20.738752 [(20.73875, 3.0146217)]
7 18.803137 [(18.803137, 2.8265903)]
8 17.048176 [(17.048176, 2.6561086)]
9 15.457013 [(15.457014, 2.5015385)]
10 14.014359 [(14.01436, 2.361395)]
11 12.706352 [(12.706352, 2.2343314)]
12 11.520427 [(11.520427, 2.119127)]
13 10.445186 [(10.445185, 2.0146751)]
14 9.470302 [(9.470302, 1.9199722)]
15 8.586407 [(8.586407, 1.8341081)]
16 7.785009 [(7.785009, 1.756258)]
17 7.0584083 [(7.0584083, 1.685674)]
18 6.399624 [(6.399624, 1.6216778)]
19 5.8023257 [(5.8023252, 1.5636545)]
20 5.260776 [(5.260776, 1.5110468)]
21 4.7697697 [(4.7697697, 1.4633491)]
22 4.324591 [(4.324591, 1.4201032)]
23 3.9209633 [(3.9209635, 1.3808936)]
24 3.5550067 [(3.5550067, 1.3453435)]
25 3.2232056 [(3.2232056, 1.3131114)]
26 2.9223735 [(2.9223735, 1.2838877)]
27 2.[...]
|
no_license
|
/online-lecture/sungkim/practice/lab3-04.ipynb
|
stellakang/deeplearning-study
| 1 |
<jupyter_start><jupyter_text>### Equipo de Trabajo:
Kevin Martínez Gallego; Mateo Llano Avendaño; Deiry Sofía Navas### Matrix Multiplication#### Setting up environment<jupyter_code>import findspark
#findspark.init(spark_path)
findspark.init()
from pyspark.sql import SparkSession, SQLContext
from pyspark import SparkContext, SparkConf
environment_to_connect = 'local' # CHANGE IT IF CONNECTION TO A CLUSTER
conf = SparkConf().setAppName('matrix_multiplication_method').setMaster(environment_to_connect)
sc = SparkContext(conf=conf)
spark = SQLContext(sc)
# other useful imports
import numpy as np
import time<jupyter_output><empty_output><jupyter_text>#### Source<jupyter_code>j_max_values = []
def convert_row(line, row): # i : row number
coo_row = []
j_max = 0
for j in range(len(line)):
value = line[j]
if value != 0:
coo_row.append((row,j,value))
j_max = j
global j_max_values
j_max_values.append(j_max)
return(coo_row)
def perform_mult(matrix_A_path, matrix_B_path, num_partitions_matrix_A, num_partitions_matrix_B):
# Read matrices with spark
matrix_A = sc.textFile(matrix_A_path, num_partitions_matrix_A)
matrix_B = sc.textFile(matrix_B_path, num_partitions_matrix_B)
matrix_A_rows = matrix_A.count()
matrix_B_rows = matrix_B.count()
# Tokenize values and convert them into float
matrix_A = matrix_A.map( lambda line: list(map(float, line.split(' '))) )
matrix_B = matrix_B.map( lambda line: list(map(float, line.split(' '))) )
# Convert matrices to coordinates (sparse) format
matrix_A_list = matrix_A.collect()
matrix_B_list = matrix_B.collect()
get_coo_matrix = (lambda matrix_list: [(convert_row(matrix_list[index], index)) for index in range(len(matrix_list))])
coo_matrix_A = get_coo_matrix(matrix_A_list)
global j_max_values
matrix_A_columns = np.max(j_max_values) + 1 # Getting max value of j as number of columns
j_max_values = []
coo_matrix_B = get_coo_matrix(matrix_B_list)
matrix_B_columns = np.max(j_max_values) + 1 # Getting max value of j as number of columns
print('Matrix A --> Rows: ' + str(matrix_A_rows), 'Columns: ' + str(matrix_A_columns))
print('Matrix B --> Rows: ' + str(matrix_B_rows), 'Columns: ' + str(matrix_B_columns))
# Parallelize matrices with sparse format in order to be processed (create RDDs)
coo_matrix_A = sc.parallelize(coo_matrix_A, num_partitions_matrix_A)
coo_matrix_B = sc.parallelize(coo_matrix_B, num_partitions_matrix_B)
# Save RDDs in main memory
coo_matrix_A.cache()
coo_matrix_B.cache()
# MATRIX MULTIPLICATION --> Two Map/Reduce steps
start_time = time.time()
# Produce key, value pairs (j, (i, Aij)) and (j, (k, Bjk))
first_map_matrix_A = coo_matrix_A.flatMap( lambda line: [(row[1], (row[0], row[2])) for row in line] )
first_map_matrix_B = coo_matrix_B.flatMap( lambda line: [(row[0], (row[1], row[2])) for row in line] )
# For each key j: generate a key-value pair, where the key is (i, k) and the value is Aij*Bjk
# Then, apply the identity function.
first_reduce_sec_map = first_map_matrix_A.join(first_map_matrix_B).\
map( lambda line: ((line[1][0][0], line[1][1][0]), line[1][0][1] * line[1][1][1]) )
# Group by key (i,k) and sum the obtained results
second_reduce = first_reduce_sec_map.reduceByKey( lambda x, y: round(x + y, 4) )
# Sort result. The result consists of pairs ((i,k), v) for the output matrix
result = second_reduce.sortByKey()
end_time = time.time()
print("Total execution time for map/reduce steps: {} seconds".format(round(end_time - start_time, 2)))
return(result)<jupyter_output><empty_output><jupyter_text>### Set parameters<jupyter_code>matrix_A_path = 'Matriz_Ejemplo_A.dat'
num_partitions_matrix_A = 4
matrix_B_path = 'Matriz_Ejemplo_B.dat'
num_partitions_matrix_B = 4<jupyter_output><empty_output><jupyter_text>#### Execute multiplication<jupyter_code>result_matrix = perform_mult(matrix_A_path, matrix_B_path, num_partitions_matrix_A, num_partitions_matrix_B)
result_matrix.count()
result_matrix.take(70)
result_matrix.getNumPartitions()
result_matrix.saveAsTextFile("result") # Save result to disk<jupyter_output><empty_output><jupyter_text>---<jupyter_code>output_matrix = sc.textFile("result") # Restore output matrix from generated files
output_matrix
output_matrix.getNumPartitions()
output_matrix.count()
output_matrix.take(70)<jupyter_output><empty_output>
|
no_license
|
/Matrix_Multiplication/matrix_multiplication_method.ipynb
|
kevinmaiden7/Spark
| 5 |
<jupyter_start><jupyter_text># Tuples
In Python tuples are very similar to lists, however, unlike lists they are *immutable* meaning they can not be changed. You would use tuples to present things that shouldn't be changed, such as days of the week, or dates on a calendar.
In this section, we will get a brief overview of the following:
1.) Constructing Tuples
2.) Basic Tuple Methods
3.) Immutability
4.) When to Use Tuples
You'll have an intuition of how to use tuples based on what you've learned about lists. We can treat them very similarly with the major distinction being that tuples are immutable.
## Constructing Tuples
The construction of a tuples use () with elements separated by commas. For example:<jupyter_code># Create a tuple
t = (1,2,3)
# Check len just like a list
len(t)
# Can also mix object types
t = ('one',2)
# Show
t
# Use indexing just like we did in lists
t[0]
# Slicing just like a list
t[-1]<jupyter_output><empty_output><jupyter_text>## Basic Tuple Methods
Tuples have built-in methods, but not as many as lists do. Let's look at two of them:<jupyter_code># Use .index to enter a value and return the index
t.index('one')
# Use .count to count the number of times a value appears
t.count('one')<jupyter_output><empty_output><jupyter_text>## Immutability
It can't be stressed enough that tuples are immutable. To drive that point home:<jupyter_code>t[0]= 'change'<jupyter_output><empty_output><jupyter_text>Because of this immutability, tuples can't grow. Once a tuple is made we can not add to it.<jupyter_code>t.append('nope')<jupyter_output><empty_output>
|
no_license
|
/06Tuples.ipynb
|
udaysurya/Python-Basics
| 4 |
<jupyter_start><jupyter_text># Workshop - 1: K- Means Clustering
This notebook will walk through some of the basics of K-Means Clustering.<jupyter_code>import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
data=pd.read_csv(r"C:\Users\karth\Desktop\Intenship\Task 2\Iris.csv")
data.head()
data.shape
data.info()<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 150 entries, 0 to 149
Data columns (total 6 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Id 150 non-null int64
1 SepalLengthCm 150 non-null float64
2 SepalWidthCm 150 non-null float64
3 PetalLengthCm 150 non-null float64
4 PetalWidthCm 150 non-null float64
5 Species 150 non-null object
dtypes: float64(4), int64(1), object(1)
memory usage: 7.2+ KB
<jupyter_text>### To find optimum number of clusters for K Means<jupyter_code>X=data.drop("Species",axis=1)
X
from sklearn.cluster import KMeans<jupyter_output><empty_output><jupyter_text>#### To find the right cluster we plot elbow plot <jupyter_code>elbow=[]
for i in range(1,11):
kmeans=KMeans(n_clusters=i,max_iter=300,n_init=10,init='k-means++')
kmeans.fit(X)
elbow.append(kmeans.inertia_)
elbow
plt.plot(range(1,11),elbow)
plt.title("elbow Plot Method")
plt.xlabel("The Number of Cluster")
plt.ylabel("elbow Points")
plt.show()<jupyter_output><empty_output><jupyter_text>### From the above grap we can clear see that **3** is right cluster <jupyter_code>kmeans=KMeans(n_clusters=3,max_iter=300,n_init=10,init='k-means++')
kmeans.fit(X)
y_kmeans=kmeans.predict(X)
y_kmeans
plt.scatter(X.iloc[y_kmeans == 0, 0], X.iloc[y_kmeans == 0, 1],
s = 100, c = 'red', label = 'Iris-setosa')
plt.scatter(X.iloc[y_kmeans ==1,0], X.iloc[y_kmeans == 1,1],
s = 100, c ="blue", label = 'Iris-setosa')
plt.scatter(X.iloc[y_kmeans ==2,0], X.iloc[y_kmeans ==2,1],
s = 100, c ="green", label = 'Iris-setosa')
plt.scatter(kmeans.cluster_centers_[:,0],kmeans.cluster_centers_[:,1],s = 100, c = 'black', label = 'Centroids')
plt.show()<jupyter_output><empty_output><jupyter_text>### Ploting the centroids on above Grap<jupyter_code>kmeans.cluster_centers_<jupyter_output><empty_output>
|
no_license
|
/Task 2 complted.ipynb
|
Karthikeyan-Ravichandran22/K--Means-Clustering-The-Sparks-Foundation-Task-2
| 5 |
<jupyter_start><jupyter_text># Problem 1<jupyter_code>t,m,u = np.loadtxt('Wmass_data.txt' , unpack = True)
print(t,m,u)
PDG = 80.379
lista=([])
for i in range (len(t)):
lista.insert(i,PDG)
dof= 7
Eu = (1/np.sum(u))**(1/2)
Wm = np.sum(m*u)/np.sum(u)
chi = np.sum((m-PDG)**2/u**2)
Rchi = chi/dof
pvalue = sit.gammaincc(dof/2, chi/2)
print("Uncertainty Error", Eu, "Weighted Mean", Wm, "chisq", chi, "Reduce chisq", Rchi, "P Value", pvalue)
fig= plt.figure()
ax=fig.add_axes([0.8,0.8,0.8,0.8])
ax.scatter(t,m,marker='o')
ax.plot(t,lista,label='Expected value from PDG')
ax.legend()
ax.set_xlabel('Trial')
ax.set_ylabel('Mass(GeV/c^2)')
ax.errorbar(t,m,u, fmt='.', label = 'Error')
ax.legend();<jupyter_output>Uncertainty Error 1.494035761667992 Weighted Mean 80.39910491071429 chisq 8.707036304862834 Reduce chisq 1.243862329266119 P Value 0.2743795204721061
<jupyter_text># Problem 2
<jupyter_code>tp,mp,up = np.loadtxt('proton_radius_data.txt', unpack = True)
print(tp,mp,up)
PDGp = 0.8751
listp=([])
for i in range (len(tp)):
listp.insert(i,PDGp)
dofp = 13
Eup = (1/np.sum(up))**(1/2)
Wmp = np.sum(mp*up)/np.sum(up)
chip = np.sum((mp-PDGp)**2/up**2)
Rchip = chip/dofp
pvaluep = sit.gammaincc(dofp/2, chip/2)
print("Uncertainty Error", Eup, "Weighted Mean", Wmp, "chisq", chip, "Reduce chisq", Rchip, "P value",pvaluep)
fig2= plt.figure()
ax=fig2.add_axes([0.8,0.8,0.8,0.8])
ax.scatter(tp,mp,marker='o')
ax.plot(tp,listp,label='Expected value from PDG')
ax.legend()
ax.set_xlabel('Trial')
ax.set_ylabel('Mass(GeV/c^2)')
ax.errorbar(tp,mp,up, fmt='.', label = 'Error')
ax.legend();<jupyter_output>Uncertainty Error 2.5627549867848893 Weighted Mean 0.8876640673847367 chisq 10285.48511513286 Reduce chisq 791.1911627025277 P value 0.0
|
no_license
|
/Module02/Assignment_2b_Victoire_Djuidje.ipynb
|
Vhub99/Phys-3511-Fall2021
| 2 |
<jupyter_start><jupyter_text>Made a list of files to manipulate.<jupyter_code>csv_names = os.listdir()[1:]
print(csv_names)<jupyter_output>['assist_ratio.csv', 'defensive_efficiency.csv', 'defensive_rebounding_rate.csv', 'effective_fg_pct.csv', 'offensive_efficiency.csv', 'offensive_rebound_rate.csv', 'pace.csv', 'rebound_rate.csv', 'true_shooting.csv', 'turnover_ratio.csv']
<jupyter_text>Wrote for loop to go through those files, read it in with pandas, drop the first two cols, change the name of the stat cols, then write that to a csv with a "clean" file name.<jupyter_code>clean_dfs = []
#shit python, needs clarification
def make_nice(file):
filename = file[:-4]
filename = pd.read_csv(file)
filename = filename.drop(labels=['Unnamed: 0', 'Rank', 'Last 3', 'Last 1'], axis=1)
filename.columns = ['Team', f'2020_{file[:-4]}', f'Home_{file[:-4]}', f'Away_{file[:-4]}', f'2019_{file[:-4]}']
print(file[:-4])
clean_dfs.append(filename)
return filename.head(3)
for file in csv_names:
make_nice(file)
clean_dfs.append(file)
#shit python
final_df = clean_dfs[0].merge(clean_dfs[1], on='Team').merge(clean_dfs[2], on='Team').merge(clean_dfs[3], on='Team').merge(clean_dfs[4], on='Team').merge(clean_dfs[5], on='Team').merge(clean_dfs[6], on='Team').merge(clean_dfs[7], on='Team').merge(clean_dfs[8], on='Team').merge(clean_dfs[9], on='Team')
final_df.head()
final_df.to_csv('stats_df.csv')
final_df.info()<jupyter_output><class 'pandas.core.frame.DataFrame'>
Int64Index: 30 entries, 0 to 29
Data columns (total 41 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Team 30 non-null object
1 2020_assist_ratio 30 non-null float64
2 Home_assist_ratio 30 non-null float64
3 Away_assist_ratio 30 non-null float64
4 2019_assist_ratio 30 non-null float64
5 2020_defensive_efficiency 30 non-null float64
6 Home_defensive_efficiency 30 non-null float64
7 Away_defensive_efficiency 30 non-null float64
8 2019_defensive_efficiency 30 non-null float64
9 2020_defensive_rebounding_rate 30 non-null object
10 Home_defensive_rebounding_rate 30 non-null object
11 Away_defensive_rebounding_rate 30 non-null object
12 2019_defensive_rebounding_rate 30 non[...]
|
no_license
|
/Merge Dataframes.ipynb
|
sharkoe/NBA
| 2 |
<jupyter_start><jupyter_text># Simple Linear Regression Model with Hyperledger Fabric dataImport modules<jupyter_code>import requests as rqt
import pandas as pd
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LinearRegression
import matplotlib.pyplot as plt<jupyter_output><empty_output><jupyter_text>Get data from Hyperledger Fabric Network<jupyter_code>data = rqt.get("http://localhost:8000/api/getAllAssets")
jsonifiedData = data.json()
jsonifiedData["response"]<jupyter_output><empty_output><jupyter_text>Make pandas dataframe from data.<jupyter_code>dataframe = pd.DataFrame(jsonifiedData["response"])
dataframe = dataframe.sort_values(by = ["force"])
dataframe
X = dataframe.iloc[:, 0].values
Y = dataframe.iloc[:, 1].values<jupyter_output><empty_output><jupyter_text>Generate training and test sets.<jupyter_code>X_train, X_test, Y_train, Y_test = train_test_split(X, Y, test_size = 1/3, random_state = 0)<jupyter_output><empty_output><jupyter_text>Create a LinearRegression instance and fit the model with the training sets.<jupyter_code>regressor = LinearRegression()
regressor.fit(X_train.reshape(-1, 1), Y_train.reshape(-1, 1))<jupyter_output><empty_output><jupyter_text>Predict results of trained model in testing set.<jupyter_code>Y_pred = regressor.predict(X_test.reshape(-1, 1))<jupyter_output><empty_output><jupyter_text>Visualization of model in training set.<jupyter_code>plt.scatter(X_train, Y_train, color = 'red')
plt.plot(X_train, regressor.predict(X_train.reshape(-1, 1)), color = 'blue')
plt.title("Stretching vs Force")
plt.xlabel('Force (N)')
plt.ylabel('Stretching (m)')
plt.show()<jupyter_output><empty_output><jupyter_text>Visualization of model applied in test set.<jupyter_code>plt.scatter(X_test, Y_test, color = 'red')
plt.plot(X, regressor.predict(X.reshape(-1, 1)), color = 'blue')
plt.title("Stretching vs Force")
plt.xlabel('Force (N)')
plt.ylabel('Stretching (m)')
plt.show()
regressor.coef_<jupyter_output><empty_output><jupyter_text>Shows intercept of the model.<jupyter_code>regressor.intercept_<jupyter_output><empty_output><jupyter_text>Shows angular coefficient of the model.<jupyter_code>regressor.predict([[2]])<jupyter_output><empty_output>
|
non_permissive
|
/MLModel/LinearRegression.ipynb
|
lucianoacsilva/fabric-demo
| 10 |
<jupyter_start><jupyter_text># Homework 3
This assignment covers the Harris corner detector, RANSAC and the HOG descriptor for panorama stitching.<jupyter_code># Setup
from __future__ import print_function
import numpy as np
from skimage import filters
from skimage.feature import corner_peaks
from skimage.io import imread
import matplotlib.pyplot as plt
from time import time
%matplotlib inline
plt.rcParams['figure.figsize'] = (15.0, 12.0) # set default size of plots
plt.rcParams['image.interpolation'] = 'nearest'
plt.rcParams['image.cmap'] = 'gray'
# for auto-reloading extenrnal modules
%load_ext autoreload
%autoreload 2<jupyter_output>The autoreload extension is already loaded. To reload it, use:
%reload_ext autoreload
<jupyter_text>## Introduction: Panorama Stitching
Panorama stitching is an early success of computer vision. Matthew Brown and David G. Lowe published a famous [panoramic image stitching paper](http://matthewalunbrown.com/papers/ijcv2007.pdf) in 2007. Since then, automatic panorama stitching technology has been widely adopted in many applications such as Google Street View, panorama photos on smartphones,
and stitching software such as Photosynth and AutoStitch.
In this assignment, we will detect and match keypoints from multiple images to build a single panoramic image. This will involve several tasks:
1. Use Harris corner detector to find keypoints.
2. Build a descriptor to describe each point in an image.
Compare two sets of descriptors coming from two different images and find matching keypoints.
3. Given a list of matching keypoints, use least-squares method to find the affine transformation matrix that maps points in one image to another.
4. Use RANSAC to give a more robust estimate of affine transformation matrix.
Given the transformation matrix, use it to transform the second image and overlay it on the first image, forming a panorama.
5. Implement a different descriptor (HOG descriptor) and get another stitching result.## Part 1 Harris Corner Detector (20 points)
In this section, you are going to implement Harris corner detector for keypoint localization. Review the lecture slides on Harris corner detector to understand how it works. The Harris detection algorithm can be divide into the following steps:
1. Compute $x$ and $y$ derivatives ($I_x, I_y$) of an image
2. Compute products of derivatives ($I_x^2, I_y^2, I_{xy}$) at each pixel
3. Compute matrix $M$ at each pixel, where
$$
M = \sum_{x,y} w(x,y)
\begin{bmatrix}
I_{x}^2 & I_{x}I_{y} \\
I_{x}I_{y} & I_{y}^2
\end{bmatrix}
$$
4. Compute corner response $R=Det(M)-k(Trace(M)^2)$ at each pixel
5. Output corner response map $R(x,y)$
Step 1 is already done for you in the function **`harris_corners`** in `panorama.py`. Complete the function implementation and run the code below.
*-Hint: You may use the function `scipy.ndimage.filters.convolve`, which is already imported in `panoramy.py`*<jupyter_code>from panorama import harris_corners
img = imread('sudoku.png', as_gray=True)
# Compute Harris corner response
response = harris_corners(img)
# Display corner response
plt.subplot(1,2,1)
plt.imshow(response)
plt.axis('off')
plt.title('Harris Corner Response')
plt.subplot(1,2,2)
plt.imshow(imread('solution_harris.png', as_gray=True))
plt.axis('off')
plt.title('Harris Corner Solution')
plt.show()<jupyter_output><empty_output><jupyter_text>Once you implement the Harris detector correctly, you will be able to see small bright blobs around the corners of the sudoku grids and letters in the output corner response image. The function `corner_peaks` from `skimage.feature` performs non-maximum suppression to take local maxima of the response map and localize keypoints.<jupyter_code># Perform non-maximum suppression in response map
# and output corner coordiantes
corners = corner_peaks(response, threshold_rel=0.01)
# Display detected corners
plt.imshow(img)
plt.scatter(corners[:,1], corners[:,0], marker='x')
plt.axis('off')
plt.title('Detected Corners')
plt.show()<jupyter_output><empty_output><jupyter_text>## Part 2 Describing and Matching Keypoints (20 points)
We are now able to localize keypoints in two images by running the Harris corner detector independently on them. Next question is, how do we determine which pair of keypoints come from corresponding locations in those two images? In order to *match* the detected keypoints, we must come up with a way to *describe* the keypoints based on their local appearance. Generally, each region around detected keypoint locations is converted into a fixed-size vectors called *descriptors*.
### Part 2.1 Creating Descriptors (10 points)
In this section, you are going to implement the **`simple_descriptor`** function, where each keypoint is described by the normalized intensity of a small patch around it.
<jupyter_code>from panorama import harris_corners
img1 = imread('uttower1.jpg', as_gray=True)
img2 = imread('uttower2.jpg', as_gray=True)
# Detect keypoints in two images
keypoints1 = corner_peaks(harris_corners(img1, window_size=3),
threshold_rel=0.05,
exclude_border=8)
keypoints2 = corner_peaks(harris_corners(img2, window_size=3),
threshold_rel=0.05,
exclude_border=8)
# Display detected keypoints
plt.subplot(1,2,1)
plt.imshow(img1)
plt.scatter(keypoints1[:,1], keypoints1[:,0], marker='x')
plt.axis('off')
plt.title('Detected Keypoints for Image 1')
plt.subplot(1,2,2)
plt.imshow(img2)
plt.scatter(keypoints2[:,1], keypoints2[:,0], marker='x')
plt.axis('off')
plt.title('Detected Keypoints for Image 2')
plt.show()<jupyter_output><empty_output><jupyter_text>### Part 2.2 Matching Descriptors (10 points)
Next, implement the **`match_descriptors`** function to find good matches in two sets of descriptors. First, calculate Euclidean distance between all pairs of descriptors from image 1 and image 2. Then use this to determine if there is a good match: if the distance to the closest vector is significantly (by a given factor) smaller than the distance to the second-closest, we call it a match. The output of the function is an array where each row holds the indices of one pair of matching descriptors.<jupyter_code>from panorama import simple_descriptor, match_descriptors, describe_keypoints
from utils import plot_matches
patch_size = 5
# Extract features from the corners
desc1 = describe_keypoints(img1, keypoints1,
desc_func=simple_descriptor,
patch_size=patch_size)
desc2 = describe_keypoints(img2, keypoints2,
desc_func=simple_descriptor,
patch_size=patch_size)
# Match descriptors in image1 to those in image2
matches = match_descriptors(desc1, desc2, 0.7)
# Plot matches
fig, ax = plt.subplots(1, 1, figsize=(15, 12))
ax.axis('off')
plot_matches(ax, img1, img2, keypoints1, keypoints2, matches)
plt.show()
plt.imshow(imread('solution_simple_descriptor.png'))
plt.axis('off')
plt.title('Matched Simple Descriptor Solution')
plt.show()<jupyter_output><empty_output><jupyter_text>## Part 3 Transformation Estimation (20 points)
We now have a list of matched keypoints across the two images. We will use this to find a transformation matrix that maps points in the second image to the corresponding coordinates in the first image. In other words, if the point $p_1 = [y_1,x_1]$ in image 1 matches with $p_2=[y_2, x_2]$ in image 2, we need to find an affine transformation matrix $H$ such that
$$
\tilde{p_2}H = \tilde{p_1},
$$
where $\tilde{p_1}$ and $\tilde{p_2}$ are homogenous coordinates of $p_1$ and $p_2$.
Note that it may be impossible to find the transformation $H$ that maps every point in image 2 exactly to the corresponding point in image 1. However, we can estimate the transformation matrix with least squares. Given $N$ matched keypoint pairs, let $X_1$ and $X_2$ be $N \times 3$ matrices whose rows are homogenous coordinates of corresponding keypoints in image 1 and image 2 respectively. Then, we can estimate $H$ by solving the least squares problem,
$$
X_2 H = X_1
$$
Implement **`fit_affine_matrix`** in `panorama.py`
*-Hint: read the [documentation](https://docs.scipy.org/doc/numpy/reference/generated/numpy.linalg.lstsq.html) about np.linalg.lstsq*<jupyter_code>from panorama import fit_affine_matrix
# Sanity check for fit_affine_matrix
# Test inputs
a = np.array([[0.5, 0.1], [0.4, 0.2], [0.8, 0.2]])
b = np.array([[0.3, -0.2], [-0.4, -0.9], [0.1, 0.1]])
H = fit_affine_matrix(b, a)
# Target output
sol = np.array(
[[1.25, 2.5, 0.0],
[-5.75, -4.5, 0.0],
[0.25, -1.0, 1.0]]
)
error = np.sum((H - sol) ** 2)
if error < 1e-20:
print('Implementation correct!')
else:
print('There is something wrong.')<jupyter_output>Implementation correct!
<jupyter_text>After checking that your `fit_affine_matrix` function is running correctly, run the following code to apply it to images.
Images will be warped and image 2 will be mapped to image 1.<jupyter_code>from utils import get_output_space, warp_image
# Extract matched keypoints
p1 = keypoints1[matches[:,0]]
p2 = keypoints2[matches[:,1]]
# Find affine transformation matrix H that maps p2 to p1
H = fit_affine_matrix(p1, p2)
output_shape, offset = get_output_space(img1, [img2], [H])
print("Output shape:", output_shape)
print("Offset:", offset)
# Warp images into output sapce
img1_warped = warp_image(img1, np.eye(3), output_shape, offset)
img1_mask = (img1_warped != -1) # Mask == 1 inside the image
img1_warped[~img1_mask] = 0 # Return background values to 0
img2_warped = warp_image(img2, H, output_shape, offset)
img2_mask = (img2_warped != -1) # Mask == 1 inside the image
img2_warped[~img2_mask] = 0 # Return background values to 0
# Plot warped images
plt.subplot(1,2,1)
plt.imshow(img1_warped)
plt.title('Image 1 warped')
plt.axis('off')
plt.subplot(1,2,2)
plt.imshow(img2_warped)
plt.title('Image 2 warped')
plt.axis('off')
plt.show()<jupyter_output>Output shape: [496 615]
Offset: [-39.37184617 0. ]
<jupyter_text>Next, the two warped images are merged to get a panorama. Your panorama may not look good at this point, but we will later use other techniques to get a better result.<jupyter_code>merged = img1_warped + img2_warped
# Track the overlap by adding the masks together
overlap = (img1_mask * 1.0 + # Multiply by 1.0 for bool -> float conversion
img2_mask)
# Normalize through division by `overlap` - but ensure the minimum is 1
normalized = merged / np.maximum(overlap, 1)
plt.imshow(normalized)
plt.axis('off')
plt.show()<jupyter_output><empty_output><jupyter_text>## Part 4 RANSAC (20 points)
Rather than directly feeding all our keypoint matches into ``fit_affine_matrix`` function, we can instead use RANSAC ("RANdom SAmple Consensus") to select only "inliers" to use for computing the transformation matrix.
The steps of RANSAC are:
1. Select random set of matches
2. Compute affine transformation matrix
3. Find inliers using the given threshold
4. Repeat and keep the largest set of inliers
5. Re-compute least-squares estimate on all of the inliers
Implement **`ransac`** in `panorama.py`, run through the following code to get a panorama. You can see the difference from the result we get without RANSAC.<jupyter_code>from panorama import ransac
# Set seed to compare output against solution image
np.random.seed(131)
H, robust_matches = ransac(keypoints1, keypoints2, matches, threshold=1)
# Visualize robust matches
fig, ax = plt.subplots(1, 1, figsize=(15, 12))
plot_matches(ax, img1, img2, keypoints1, keypoints2, robust_matches)
plt.axis('off')
plt.show()
plt.imshow(imread('solution_ransac.png'))
plt.axis('off')
plt.title('RANSAC Solution')
plt.show()<jupyter_output>41
[[ 1.04721152e+00 4.99175866e-02 0.00000000e+00]
[-3.13753090e-02 1.01292955e+00 0.00000000e+00]
[ 1.22271580e+01 2.55793274e+02 1.00000000e+00]]
<jupyter_text>We can now use the tranformation matrix $H$ computed using the robust matches to warp our images and create a better-looking panorama.<jupyter_code>output_shape, offset = get_output_space(img1, [img2], [H])
# Warp images into output sapce
img1_warped = warp_image(img1, np.eye(3), output_shape, offset)
img1_mask = (img1_warped != -1) # Mask == 1 inside the image
img1_warped[~img1_mask] = 0 # Return background values to 0
img2_warped = warp_image(img2, H, output_shape, offset)
img2_mask = (img2_warped != -1) # Mask == 1 inside the image
img2_warped[~img2_mask] = 0 # Return background values to 0
# Plot warped images
plt.subplot(1,2,1)
plt.imshow(img1_warped)
plt.title('Image 1 warped')
plt.axis('off')
plt.subplot(1,2,2)
plt.imshow(img2_warped)
plt.title('Image 2 warped')
plt.axis('off')
plt.show()
merged = img1_warped + img2_warped
# Track the overlap by adding the masks together
overlap = (img1_mask * 1.0 + # Multiply by 1.0 for bool -> float conversion
img2_mask)
# Normalize through division by `overlap` - but ensure the minimum is 1
normalized = merged / np.maximum(overlap, 1)
plt.imshow(normalized)
plt.axis('off')
plt.show()
plt.imshow(imread('solution_ransac_panorama.png'))
plt.axis('off')
plt.title('RANSAC Panorama Solution')
plt.show()<jupyter_output><empty_output><jupyter_text>## Part 5 Histogram of Oriented Gradients (HOG) (20 points)
In the above code, you are using the `simple_descriptor`, and in this section, you are going to implement a simplified version of HOG descriptor.
HOG stands for Histogram of Oriented Gradients. In HOG descriptor, the distribution ( histograms ) of the directions of gradients ( oriented gradients ) are used as features. Gradients ( x and y derivatives ) of an image are useful because the magnitude of a gradient is large around edges and corners ( regions of abrupt intensity changes ) and we know that edges and corners pack in a lot more information about object shape than flat regions.
The steps of HOG are:
1. Compute the gradient image in x and y directions
Use the sobel filter provided by skimage.filters
2. Compute gradient histograms
Divide image into cells, and calculate histogram of gradients in each cell
3. Flatten block of histograms into feature vector
4. Normalize flattened block
Implement **`hog_descriptor`** in `panorama.py` and run through the following code to get a panorama image.<jupyter_code>from panorama import hog_descriptor
img1 = imread('uttower1.jpg', as_grey=True)
img2 = imread('uttower2.jpg', as_grey=True)
# Detect keypoints in both images
keypoints1 = corner_peaks(harris_corners(img1, window_size=3),
threshold_rel=0.05,
exclude_border=8)
keypoints2 = corner_peaks(harris_corners(img2, window_size=3),
threshold_rel=0.05,
exclude_border=8)
# Extract features from the corners
desc1 = describe_keypoints(img1, keypoints1,
desc_func=hog_descriptor,
patch_size=16)
desc2 = describe_keypoints(img2, keypoints2,
desc_func=hog_descriptor,
patch_size=16)
# Match descriptors in image1 to those in image2
matches = match_descriptors(desc1, desc2, 0.7)
# Plot matches
fig, ax = plt.subplots(1, 1, figsize=(15, 12))
ax.axis('off')
plot_matches(ax, img1, img2, keypoints1, keypoints2, matches)
plt.show()
plt.imshow(imread('solution_hog.png'))
plt.axis('off')
plt.title('HOG descriptor Solution')
plt.show()<jupyter_output><empty_output><jupyter_text>Once we've described our keypoints with the HOG descriptor and have found matches between these keypoints, we can use RANSAC to select robust matches for computing the tranasformtion matrix.
<jupyter_code>from panorama import ransac
# Set seed to compare output against solution image
np.random.seed(131)
H, robust_matches = ransac(keypoints1, keypoints2, matches, threshold=1)
# Plot matches
fig, ax = plt.subplots(1, 1, figsize=(15, 12))
plot_matches(ax, img1, img2, keypoints1, keypoints2, robust_matches)
plt.axis('off')
plt.show()
plt.imshow(imread('solution_hog_ransac.png'))
plt.axis('off')
plt.title('HOG descriptor + RANSAC Solution')
plt.show()<jupyter_output><empty_output><jupyter_text>Now we use the computed transformation matrix $H$ to warp our images and produce our panorama.<jupyter_code>output_shape, offset = get_output_space(img1, [img2], [H])
# Warp images into output sapce
img1_warped = warp_image(img1, np.eye(3), output_shape, offset)
img1_mask = (img1_warped != -1) # Mask == 1 inside the image
img1_warped[~img1_mask] = 0 # Return background values to 0
img2_warped = warp_image(img2, H, output_shape, offset)
img2_mask = (img2_warped != -1) # Mask == 1 inside the image
img2_warped[~img2_mask] = 0 # Return background values to 0
# Plot warped images
plt.subplot(1,2,1)
plt.imshow(img1_warped)
plt.title('Image 1 warped')
plt.axis('off')
plt.subplot(1,2,2)
plt.imshow(img2_warped)
plt.title('Image 2 warped')
plt.axis('off')
plt.show()
merged = img1_warped + img2_warped
# Track the overlap by adding the masks together
overlap = (img1_mask * 1.0 + # Multiply by 1.0 for bool -> float conversion
img2_mask)
# Normalize through division by `overlap` - but ensure the minimum is 1
normalized = merged / np.maximum(overlap, 1)
plt.imshow(normalized)
plt.axis('off')
plt.show()
plt.imshow(imread('solution_hog_panorama.png'))
plt.axis('off')
plt.title('HOG Descriptor Panorama Solution')
plt.show()<jupyter_output><empty_output><jupyter_text>## Extra Credit: Better Image Merging
You will notice the blurry region and unpleasant lines in the middle of the final panoramic image. Using a very simple technique called linear blending, we can smooth out a lot of these artifacts from the panorama.
Currently, all the pixels in the overlapping region are weighted equally. However, since the pixels at the left and right ends of the overlap are very well complemented by the pixels in the other image, they can be made to contribute less to the final panorama.
Linear blending can be done with the following steps:
1. Define left and right margins for blending to occur between
2. Define a weight matrix for image 1 such that:
- From the left of the output space to the left margin the weight is 1
- From the left margin to the right margin, the weight linearly decrements from 1 to 0
3. Define a weight matrix for image 2 such that:
- From the right of the output space to the right margin the weight is 1
- From the left margin to the right margin, the weight linearly increments from 0 to 1
4. Apply the weight matrices to their corresponding images
5. Combine the images
In **`linear_blend`** in `panorama.py` implement the linear blending scheme to make the panorama look more natural. This extra credit can be worth up to 1% of your final grade. <jupyter_code>from panorama import linear_blend
img1 = imread('uttower1.jpg', as_grey=True)
img2 = imread('uttower2.jpg', as_grey=True)
# Set seed to compare output against solution
np.random.seed(131)
# Detect keypoints in both images
keypoints1 = corner_peaks(harris_corners(img1, window_size=3),
threshold_rel=0.05,
exclude_border=8)
keypoints2 = corner_peaks(harris_corners(img2, window_size=3),
threshold_rel=0.05,
exclude_border=8)
# Extract features from the corners
desc1 = describe_keypoints(img1, keypoints1,
desc_func=hog_descriptor,
patch_size=16)
desc2 = describe_keypoints(img2, keypoints2,
desc_func=hog_descriptor,
patch_size=16)
# Match descriptors in image1 to those in image2
matches = match_descriptors(desc1, desc2, 0.7)
H, robust_matches = ransac(keypoints1, keypoints2, matches, threshold=1)
output_shape, offset = get_output_space(img1, [img2], [H])
# Warp images into output sapce
img1_warped = warp_image(img1, np.eye(3), output_shape, offset)
img1_mask = (img1_warped != -1) # Mask == 1 inside the image
img1_warped[~img1_mask] = 0 # Return background values to 0
img2_warped = warp_image(img2, H, output_shape, offset)
img2_mask = (img2_warped != -1) # Mask == 1 inside the image
img2_warped[~img2_mask] = 0 # Return background values to 0
# Merge the warped images using linear blending scheme
merged = linear_blend(img1_warped, img2_warped)
plt.imshow(merged)
plt.axis('off')
plt.show()<jupyter_output><empty_output><jupyter_text>## Extra Credit: Stitching Multiple Images
Implement **`stitch_multiple_images`** in `panorama.py` to stitch together an ordered chain of images. This extra credit can be worth up to 1% of your final grade.
Given a sequence of $m$ images ($I_1, I_2,...,I_m$), take every neighboring pair of images and compute the transformation matrix which converts points from the coordinate frame of $I_{i+1}$ to the frame of $I_{i}$. Then, select a reference image $I_{ref}$, which is in the middle of the chain. We want our final panorama image to be in the coordinate frame of $I_{ref}$.
*-Hint:*
- If you are confused, you may want to review the Linear Algebra slides on how to combine the effects of multiple transformation matrices.
- The inverse of transformation matrix has the reverse effect. Please use [`numpy.linalg.inv`](https://docs.scipy.org/doc/numpy/reference/generated/numpy.linalg.inv.html) function whenever you want to compute matrix inverse.<jupyter_code>from panorama import stitch_multiple_images
# Set seed to compare output against solution
np.random.seed(131)
# Load images to be stitched
img1 = imread('yosemite1.jpg', as_grey=True)
img2 = imread('yosemite2.jpg', as_grey=True)
img3 = imread('yosemite3.jpg', as_grey=True)
img4 = imread('yosemite4.jpg', as_grey=True)
imgs = [img1, img2, img3, img4]
# Stitch images together
panorama = stitch_multiple_images(imgs, desc_func=simple_descriptor, patch_size=5)
# Visualize final panorama image
plt.imshow(panorama)
plt.axis('off')
plt.show()<jupyter_output><empty_output>
|
no_license
|
/hw3_release/hw3.ipynb
|
whu-pzhang/cs131_fall2018
| 15 |
<jupyter_start><jupyter_text># Question 1: Find the mean, median for Sachin, Rahul, India<jupyter_code>import numpy as np
cric_data = np.loadtxt("cric_data.tsv", skiprows=1)
cric_data.shape
cric_data = cric_data[:, [1, 2, 3]]
cric_data.shape
sachin = cric_data[:, 0]
rahul = cric_data[:, 1]
india = cric_data[:, 2]
sachin.shape
def stats(col):
print('Mean', np.mean(col))
print('Median', np.median(col))
stats(sachin)
stats(rahul)
stats(india)
np.mean(cric_data, axis=0)<jupyter_output><empty_output><jupyter_text># Question 2: Find histogram of Sachin's scores with 10 bins<jupyter_code>np.histogram(sachin)<jupyter_output><empty_output><jupyter_text># Question 3: find mean of sachin's scores grouped by 25 matches<jupyter_code>sachin.shape
sachin25 = sachin.reshape(9, 25)
sachin25.shape
np.mean(sachin25, axis=1)<jupyter_output><empty_output><jupyter_text># Question 4: Find mean of Sachin's scores where he has scored a century<jupyter_code>sachin >= 100
sachin[sachin >= 100]
<jupyter_output><empty_output>
|
no_license
|
/cricket.ipynb
|
akashshetty9/Dlithe-internship
| 4 |
<jupyter_start><jupyter_text># Data Science Academy - Python Fundamentos - Capítulo 3
## Download: http://github.com/dsacademybr<jupyter_code># Versão da Linguagem Python
from platform import python_version
print('Versão da Linguagem Python Usada Neste Jupyter Notebook:', python_version())<jupyter_output>Versão da Linguagem Python Usada Neste Jupyter Notebook: 3.8.3
<jupyter_text>### While<jupyter_code># Usando o loop while para imprimir os valores de 0 a 9
counter = 0
while counter < 10:
print(counter)
counter = counter + 1
# Também é possível usar a claúsula else para encerrar o loop while
x = 0
while x < 10:
print ('O valor de x nesta iteração é: ', x)
print (' x ainda é menor que 10, somando 1 a x')
x += 1
else:
print ('Loop concluído!')<jupyter_output>O valor de x nesta iteração é: 0
x ainda é menor que 10, somando 1 a x
O valor de x nesta iteração é: 1
x ainda é menor que 10, somando 1 a x
O valor de x nesta iteração é: 2
x ainda é menor que 10, somando 1 a x
O valor de x nesta iteração é: 3
x ainda é menor que 10, somando 1 a x
O valor de x nesta iteração é: 4
x ainda é menor que 10, somando 1 a x
O valor de x nesta iteração é: 5
x ainda é menor que 10, somando 1 a x
O valor de x nesta iteração é: 6
x ainda é menor que 10, somando 1 a x
O valor de x nesta iteração é: 7
x ainda é menor que 10, somando 1 a x
O valor de x nesta iteração é: 8
x ainda é menor que 10, somando 1 a x
O valor de x nesta iteração é: 9
x ainda é menor que 10, somando 1 a x
Loop concluído!
<jupyter_text>### Pass, Break, Continue<jupyter_code>counter = 0
while counter < 100:
if counter == 4:
break
else:
pass
print(counter)
counter = counter + 1
for verificador in "Python":
if verificador == "h":
continue
print(verificador)<jupyter_output>P
y
t
o
n
<jupyter_text>### While e For juntos<jupyter_code>for i in range(2,30):
j = 2
counter = 0
while j < i:
if i % j == 0:
counter = 1
j = j + 1
else:
j = j + 1
if counter == 0:
print(str(i) + " é um número primo")
counter = 0
else:
counter = 0<jupyter_output>2 é um número primo
3 é um número primo
5 é um número primo
7 é um número primo
11 é um número primo
13 é um número primo
17 é um número primo
19 é um número primo
23 é um número primo
29 é um número primo
|
permissive
|
/PythonFundamentos/Cap03/Notebooks/DSA-Python-Cap03-03-While.ipynb
|
paulohos06/curso-python-dsa
| 4 |
<jupyter_start><jupyter_text># Neural Machine Translation
- Translate a given sentence in one language to another desired language.
#### In this notebook, we aim to build a model which can translate German sentences to English.## Dataset
Dataset is taken from http://www.manythings.org/anki/.
We are considering German – English deu-eng.zip file from the above mentioned website.
In the above zip file there is a file with name **`deu.txt`** that contains **152,820** pairs of English to German phrases, one pair per line with a tab separating the phrases.
For example,
The first 5 lines in deu.txt are as given below.
***
```
Hi. Hallo!
Hi. Grüß Gott!
Run! Lauf!
Wow! Potzdonner!
Wow! Donnerwetter!
```
***
## Problem
### Given a sequence of words in German as input, predict the sequence of words in English.### 1. Prepare Data
The preprocessing of the data involves:
1. Removing punctuation marks from the data.
2. Converting text corpus into lower case characters.
3. Split into Train and Test sets.
4. Shuffling the sentences.
The above tasks are done and full dataset is given as **``english-german-both.pkl``** respectively.
Download dataset files from here: https://drive.google.com/open?id=1gWVk7SuuE93Cf_nT9Lb7GBCiwfAgdBiX
# Character level Machine Translation## Initialize parameters
Run the below code to initialize the variables required for the model.<jupyter_code>batch_size = 64 # Batch size for training.
epochs = 10 # Number of epochs to train for.
latent_dim = 256 # Latent dimensionality of the encoding space.
num_samples = 10000 # Number of samples to train on.
# Path to the data txt file on disk.
data_path = '/content/drive/My Drive/AIML-Residency10/deu.txt'<jupyter_output><empty_output><jupyter_text>### Connect to google drive<jupyter_code>from google.colab import drive
drive.mount('/content/drive/')<jupyter_output>Go to this URL in a browser: https://accounts.google.com/o/oauth2/auth?client_id=947318989803-6bn6qk8qdgf4n4g3pfee6491hc0brc4i.apps.googleusercontent.com&redirect_uri=urn%3Aietf%3Awg%3Aoauth%3A2.0%3Aoob&scope=email%20https%3A%2F%2Fwww.googleapis.com%2Fauth%2Fdocs.test%20https%3A%2F%2Fwww.googleapis.com%2Fauth%2Fdrive%20https%3A%2F%2Fwww.googleapis.com%2Fauth%2Fdrive.photos.readonly%20https%3A%2F%2Fwww.googleapis.com%2Fauth%2Fpeopleapi.readonly&response_type=code
Enter your authorization code:
··········
Mounted at /content/drive/
<jupyter_text>### Give the path for the folder in which the dataset is present in google drive<jupyter_code>project_path = "/content/drive/My Drive/AIML-Residency10/"<jupyter_output><empty_output><jupyter_text>### Change present working directory<jupyter_code>import os
os.chdir(project_path)<jupyter_output><empty_output><jupyter_text>## Load the pickle file (`english-german-both.pkl`) into a variable with name `dataset`
Run the below code to load the .pkl file.<jupyter_code>import pickle
with open(project_path + 'english-german-both.pkl', 'rb') as f:
dataset = pickle.load(f)<jupyter_output><empty_output><jupyter_text>## Check the `dataset` variable at this step. It should be as given below<jupyter_code>dataset<jupyter_output><empty_output><jupyter_text>## Feature set and target set division from the **dataset**
### Run the below code to divide the dataset into feature set(input) and target set(output).
1. We are creating two lists for storing input sentences and output sentences separately.
2. We are storing each character in a list from both input and target sets separately.
3. Print and check `input_texts` and `target_texts`.
4. Print and check `input_characters` and `target_characters`.<jupyter_code># Vectorize the data.
input_texts = []
target_texts = []
input_characters = set()
target_characters = set()
for line in dataset[: min(num_samples, len(dataset) - 1)]:
input_text, target_text = line[0], line[1]
# We use "tab" as the "start sequence" character
# for the targets, and "\n" as "end sequence" character.
target_text = '\t' + target_text + '\n'
input_texts.append(input_text)
target_texts.append(target_text)
for char in input_text:
if char not in input_characters:
input_characters.add(char)
for char in target_text:
if char not in target_characters:
target_characters.add(char)<jupyter_output><empty_output><jupyter_text>### Print input text<jupyter_code>print('Input Text:', input_texts)<jupyter_output>Input Text: ['stay with us', 'she wants him', 'youre strong', 'examine this', 'heres my card', 'tom burped', 'it is no joke', 'tom is a spy', 'im a teenager', 'im not crazy', 'we cant do it', 'i like sweets', 'put it down', 'tom looks fine', 'i am hungarian', 'youre dying', 'go away', 'tom denied this', 'yes of course', 'this is ugly', 'tom cried again', 'ill prove it', 'do as he says', 'please tell me', 'he had ambition', 'look here', 'cuff him', 'is it far away', 'were cousins', 'is tom dreaming', 'i envy tom', 'find a job', 'im the expert', 'just try it', 'theyll find me', 'a car hit tom', 'have tom do it', 'close your eyes', 'come on in here', 'i like this', 'tom is history', 'try it on', 'ill sleep here', 'does that help', 'tom moved away', 'well be ok', 'im on my own', 'toms out cold', 'eat everything', 'you said it', 'he looked young', 'dont insult me', 'im upset', 'you never ask', 'i broke it', 'theyre happy', 'tom seemed sad', 'tom is a loser', 'its up to you', 'are these ours[...]<jupyter_text>### Print target text<jupyter_code>print('Target Text:', target_texts)<jupyter_output>Target Text: ['\tbleib bei uns\n', '\tsie will ihn\n', '\tdu bist stark\n', '\tuntersuchen sie das\n', '\thier ist meine karte\n', '\ttom stie auf\n', '\tdas ist kein witz\n', '\ttom ist ein spion\n', '\tich bin ein teenager\n', '\tich bin nicht verruckt\n', '\twir konnen das nicht\n', '\tich mag sues\n', '\tlegt es hin\n', '\ttom sieht gut aus\n', '\tich bin ungarin\n', '\tdu stirbst\n', '\tverkrumele dich\n', '\ttom hat das bestritten\n', '\tja naturlich\n', '\tdas ist grauslich\n', '\ttom hat wieder geweint\n', '\tich werde es beweisen\n', '\ttu was er sagt\n', '\tbitte sagt es mir\n', '\ter war strebsam\n', '\tschau her\n', '\tlegen sie ihm handschellen an\n', '\tist es weit weg\n', '\twir sind cousins\n', '\tist tom am traumen\n', '\tich beneide tom\n', '\tsuch dir einen job\n', '\tich bin der mann vom fach\n', '\tversuchs doch einfach\n', '\tsie werden mich finden\n', '\ttom wurde von einem auto angefahren\n', '\tlass es tom machen\n', '\tmacht die augen zu\n', '\tkomm herein\n',[...]<jupyter_text>### Print input character<jupyter_code>print('Input Characters:', input_characters)<jupyter_output>Input Characters: {' ', 'y', 'q', 'f', 'l', 'c', 'u', 'j', 'e', 'z', 'p', 'b', 'w', 'm', 'i', 't', 's', 'd', 'v', 'x', 'r', 'a', 'k', 'g', 'n', 'o', 'h'}
<jupyter_text>### Print target character<jupyter_code>print('Target Characters:', target_characters)<jupyter_output>Target Characters: {' ', 'y', 'q', 'f', 'l', 'c', 'u', 'z', 'e', 'j', '\n', 'p', 'b', 'w', 'm', 'i', 't', 's', 'd', '\t', 'v', 'x', 'r', 'a', 'k', 'g', 'n', 'o', 'h'}
<jupyter_text>## Stats from the dataset
### Run the below code to check the stats from the dataset<jupyter_code>input_characters = sorted(list(input_characters))
target_characters = sorted(list(target_characters))
num_encoder_tokens = len(input_characters)
num_decoder_tokens = len(target_characters)
max_encoder_seq_length = max([len(txt) for txt in input_texts])
max_decoder_seq_length = max([len(txt) for txt in target_texts])
print('Number of samples:', len(input_texts))
print('Number of unique input tokens:', num_encoder_tokens)
print('Number of unique output tokens:', num_decoder_tokens)
print('Max sequence length for inputs:', max_encoder_seq_length)
print('Max sequence length for outputs:', max_decoder_seq_length)<jupyter_output>Number of samples: 9999
Number of unique input tokens: 27
Number of unique output tokens: 29
Max sequence length for inputs: 15
Max sequence length for outputs: 51
<jupyter_text>## Build character to index dictionary names `input_token_index` and `target_token_index` for input and target sets respectively.<jupyter_code>input_token_index = dict(
[(char, i) for i, char in enumerate(input_characters)])
target_token_index = dict(
[(char, i) for i, char in enumerate(target_characters)])<jupyter_output><empty_output><jupyter_text>### Print input_index_token<jupyter_code>input_token_index<jupyter_output><empty_output><jupyter_text>### Print target_token_index<jupyter_code>target_token_index<jupyter_output><empty_output><jupyter_text>## Build Model
Initialize the required layers from keras
### Import libraries<jupyter_code>from __future__ import print_function
from keras.models import Model
from keras.layers import Input, LSTM, Dense
import numpy as np<jupyter_output>Using TensorFlow backend.
<jupyter_text>### Run the below code to build one-hot vectors for the characters<jupyter_code>encoder_input_data = np.zeros(
(len(input_texts), max_encoder_seq_length, num_encoder_tokens),
dtype='float32')
decoder_input_data = np.zeros(
(len(input_texts), max_decoder_seq_length, num_decoder_tokens),
dtype='float32')
decoder_target_data = np.zeros(
(len(input_texts), max_decoder_seq_length, num_decoder_tokens),
dtype='float32')
for i, (input_text, target_text) in enumerate(zip(input_texts, target_texts)):
for t, char in enumerate(input_text):
encoder_input_data[i, t, input_token_index[char]] = 1.
for t, char in enumerate(target_text):
# decoder_target_data is ahead of decoder_input_data by one timestep
decoder_input_data[i, t, target_token_index[char]] = 1.
if t > 0:
# decoder_target_data will be ahead by one timestep
# and will not include the start character.
decoder_target_data[i, t - 1, target_token_index[char]] = 1.<jupyter_output><empty_output><jupyter_text>### Build the encoder Model
Define an input sequence and process it.
Discard `encoder_outputs` and only keep the states.<jupyter_code># Define an input sequence and process it.
encoder_inputs = Input(shape=(None, num_encoder_tokens))
encoder = LSTM(latent_dim, return_state=True)
encoder_outputs, state_h, state_c = encoder(encoder_inputs)
# We discard `encoder_outputs` and only keep the states.
encoder_states = [state_h, state_c]
encoder_model = Model(encoder_inputs, encoder_states)<jupyter_output>WARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/tensorflow/python/framework/op_def_library.py:263: colocate_with (from tensorflow.python.framework.ops) is deprecated and will be removed in a future version.
Instructions for updating:
Colocations handled automatically by placer.
<jupyter_text>### Build the decoder Model
Set up the decoder, using `encoder_states` as initial state.
We set up our decoder to return full output sequences, and to return internal states as well. We don't use the return states in the training model, but we will use them in inference.<jupyter_code># Set up the decoder, using `encoder_states` as initial state.
decoder_inputs = Input(shape=(None, num_decoder_tokens))
# We set up our decoder to return full output sequences,
# and to return internal states as well. We don't use the
# return states in the training model, but we will use them in inference.
decoder_lstm = LSTM(latent_dim, return_sequences=True, return_state=True)
decoder_outputs, _, _ = decoder_lstm(decoder_inputs,
initial_state=encoder_states)
decoder_dense = Dense(num_decoder_tokens, activation='softmax')
decoder_outputs = decoder_dense(decoder_outputs)<jupyter_output><empty_output><jupyter_text>### Define Model
Define the model that will turn `encoder_input_data ` & ` decoder_input_data` into `decoder_target_data`<jupyter_code># Define the model that will turn
# `encoder_input_data` & `decoder_input_data` into `decoder_target_data`
model = Model([encoder_inputs, decoder_inputs], decoder_outputs)<jupyter_output><empty_output><jupyter_text>### Compile and fit the model<jupyter_code># Run training
model.compile(optimizer='rmsprop', loss='categorical_crossentropy')
model.fit([encoder_input_data, decoder_input_data], decoder_target_data,
batch_size=batch_size,
epochs=30,
validation_split=0.2)<jupyter_output>WARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/tensorflow/python/ops/math_ops.py:3066: to_int32 (from tensorflow.python.ops.math_ops) is deprecated and will be removed in a future version.
Instructions for updating:
Use tf.cast instead.
WARNING:tensorflow:From /usr/local/lib/python3.6/dist-packages/tensorflow/python/ops/math_grad.py:102: div (from tensorflow.python.ops.math_ops) is deprecated and will be removed in a future version.
Instructions for updating:
Deprecated in favor of operator or tf.math.divide.
Train on 7999 samples, validate on 2000 samples
Epoch 1/30
7999/7999 [==============================] - 14s 2ms/step - loss: 0.9570 - val_loss: 0.8086
Epoch 2/30
7999/7999 [==============================] - 10s 1ms/step - loss: 0.7473 - val_loss: 0.6725
Epoch 3/30
7999/7999 [==============================] - 11s 1ms/step - loss: 0.6431 - val_loss: 0.6009
Epoch 4/30
7999/7999 [==============================] - 11s 1ms/step - loss: 0.5866 - val_loss: 0.5591
Epoch 5/3[...]<jupyter_text>### Save the model<jupyter_code># Save model
model.save('/content/drive/My Drive/AIML-Residency10/s2s.h5')<jupyter_output>/usr/local/lib/python3.6/dist-packages/keras/engine/network.py:877: UserWarning: Layer lstm_2 was passed non-serializable keyword arguments: {'initial_state': [<tf.Tensor 'lstm_1/while/Exit_2:0' shape=(?, 256) dtype=float32>, <tf.Tensor 'lstm_1/while/Exit_3:0' shape=(?, 256) dtype=float32>]}. They will not be included in the serialized model (and thus will be missing at deserialization time).
'. They will not be included '
<jupyter_text>## Run the below code for inferencing the model<jupyter_code>encoder_model = Model(encoder_inputs, encoder_states)
decoder_state_input_h = Input(shape=(latent_dim,))
decoder_state_input_c = Input(shape=(latent_dim,))
decoder_states_inputs = [decoder_state_input_h, decoder_state_input_c]
decoder_outputs, state_h, state_c = decoder_lstm(decoder_inputs, initial_state=decoder_states_inputs)
decoder_states = [state_h, state_c]
decoder_outputs = decoder_dense(decoder_outputs)
decoder_model = Model(
[decoder_inputs] + decoder_states_inputs,
[decoder_outputs] + decoder_states)<jupyter_output><empty_output><jupyter_text>## Reverse-lookup token index to decode sequences back to something readable.<jupyter_code># Reverse-lookup token index to decode sequences back to
# something readable.
reverse_input_char_index = dict(
(i, char) for char, i in input_token_index.items())
reverse_target_char_index = dict(
(i, char) for char, i in target_token_index.items())
print(reverse_input_char_index)
print(reverse_target_char_index)
def decode_sequence(input_seq):
# Encode the input as state vectors.
states_value = encoder_model.predict(input_seq)
# Generate empty target sequence of length 1.
target_seq = np.zeros((1, 1, num_decoder_tokens))
# Populate the first character of target sequence with the start character.
target_seq[0, 0, target_token_index['\t']] = 1.
# Sampling loop for a batch of sequences
# (to simplify, here we assume a batch of size 1).
stop_condition = False
decoded_sentence = ''
while not stop_condition:
output_tokens, h, c = decoder_model.predict(
[target_seq] + states_value)
# Sample a token
sampled_token_index = np.argmax(output_tokens[0, -1, :])
sampled_char = reverse_target_char_index[sampled_token_index]
decoded_sentence += sampled_char
# Exit condition: either hit max length
# or find stop character.
if (sampled_char == '\n' or
len(decoded_sentence) > max_decoder_seq_length):
stop_condition = True
# Update the target sequence (of length 1).
target_seq = np.zeros((1, 1, num_decoder_tokens))
target_seq[0, 0, sampled_token_index] = 1.
# Update states
states_value = [h, c]
return decoded_sentence<jupyter_output><empty_output><jupyter_text>## Run the below code for checking some outputs from the model.<jupyter_code>for seq_index in range(10):
# Take one sequence (part of the training set)
# for trying out decoding.
input_seq = encoder_input_data[seq_index: seq_index + 1]
decoded_sentence = decode_sequence(input_seq)
print('-')
print('Input sentence:', input_texts[seq_index])
print('Decoded sentence:', decoded_sentence)
<jupyter_output><empty_output>
|
no_license
|
/Residency_10/nmt_ger_eng_questions.ipynb
|
GreatLearningAIML1/chennai-aug-batch-Shanmugapriya03
| 25 |
<jupyter_start><jupyter_text># ENGG\*3130 Project: CartPole Game
By: Brandon, Dylan, and Gwyneth## Requirements
To install the requirements please run `pip install -r requirements.txt` inside of this directory. It will install the following dependencies:
- `gym`
- `matplotlib`
- `control`
- `keras`
- `tensorflow`## Introduction
Cartpole is a game provided by OpenAI Gym that is commonly used for evaluating various machine learning algorithms. Gym is a toolkit for developing and comparing reinforcement learning algorithms. There is a library of environments that can be used to test various algorithms in order to complete the objectives of the games. These games range from simple controls to classic arcade games which involve more complicated controls. Cartpole consists of a pole that is attached by an un-actuated joint to a cart, which moves along a frictionless track. The system is controlled by applying a force towards the right or left of the cart. The game starts with the pendulum upright and the goal is to prevent it from falling over. Scoring is determined by awarding a point for each timestep the pole remains upright. The game is finished when the pole is more than 15 degrees from vertical or if the cart moves more than 2.4 units from the center.## Testing
Before starting to create our strategies we must first determine how we are to test the game out. We have decided to let the game run for a set amount of episdoes and then analyze the highest score acheived as well as the average score. We will then plot the scores as well as the strategies graph for it's highest score.
First we will import `BaseStrategy` and `Game` from the `Game.py` file. `BaseStrategy` is a class that get's information about the pole as well as let the pole make actions. `Game` contains code in order to run the game with a given strategy, log all of it's results, and plot those results. <jupyter_code>from Game import Game, BaseStrategy<jupyter_output><empty_output><jupyter_text>We will then determine how many episodes we will like to run and whether or not to render the game. For the purpose of this notebook we will not be rendering the game as it takes too long to complete each episode.<jupyter_code>episodes = 200
render=False<jupyter_output><empty_output><jupyter_text>Next we will create a function `play_game` in order to play the Strategies we will be testing.<jupyter_code>def play_game(strategy, title="", episodes=100):
game = Game(episodes, strategy(), render)
game.play()
game.close()
game.plot(title)<jupyter_output><empty_output><jupyter_text>### Random Strategy
The Random Strategy will be the first strategy we will try to implement. It will randomly select a float from 0 to 1 using the `random` library and determine if the the CartPole will move left or right. <jupyter_code>import random
class RandomStrategy(BaseStrategy):
def calculate(self, observation):
if random.random() < 0.5:
return self.make_move_left_action()
else:
return self.make_move_right_action()
play_game(RandomStrategy, "Random Strategy", episodes)<jupyter_output>High Score: 112, Average Score: 21.84
<jupyter_text>### Brute Force Strategies
For Brute Force strategies we decided on 4 different ones. Position, Angle, Tip Velocity, and Tip Velocity and Position Strategy. These all track their respective component and moves the pole accordingly. #### Position Strategy
If the position of the pole is left of the center, push the pole right or vice versa<jupyter_code>class PositionStrategy(BaseStrategy):
def calculate(self, observation):
if (self.is_pole_positioned_right(observation)):
return self.make_move_left_action()
else:
return self.make_move_right_action()
play_game(PositionStrategy, "Position Strategy", episodes)<jupyter_output>High Score: 65, Average Score: 28.99
<jupyter_text>#### Angle Strategy
If the angle of the pole is left, push the pole left to fix the angle and vice versa<jupyter_code>class AngleStrategy(BaseStrategy):
def calculate(self, observation):
if (self.is_pole_angled_left(observation)):
return self.make_move_left_action()
else:
return self.make_move_right_action()
play_game(AngleStrategy, "Angle Strategy", episodes)<jupyter_output>High Score: 65, Average Score: 41.95
<jupyter_text>#### Tip Velocity Strategy
If the tip of the pole is going left, push the pole left or vice versa<jupyter_code>class TipVelocityStrategy(BaseStrategy):
def calculate(self, observation):
if (self.is_pole_tip_velocity_left(observation)):
return self.make_move_left_action()
else:
return self.make_move_right_action()
play_game(TipVelocityStrategy, "Tip Velocity Strategy", episodes)<jupyter_output>High Score: 340, Average Score: 201.345
<jupyter_text>#### Tip Velocity and Position Strategy
If the tip of the pole is going left, push the pole left or vice versa. If the position of the pole is not centered, push the pole back to the center.<jupyter_code>class TipVelocityAndPositionStrategy(BaseStrategy):
count = 0
def calculate(self, observation):
self.count = self.count + 1
if self.count % 2 == 0:
if (self.is_pole_positioned_right(observation)):
return self.make_move_left_action()
else:
return self.make_move_right_action()
if (self.is_pole_tip_velocity_left(observation)):
return self.make_move_left_action()
else:
return self.make_move_right_action()
play_game(TipVelocityAndPositionStrategy, "Tip Velocity and Position Strategy", episodes)<jupyter_output>High Score: 228, Average Score: 60.23
<jupyter_text>### PID Controller Strategy
This strategy utilizes a PID controller in order to try and keep the pole balanced and not fall outside of the given parameters. <jupyter_code>import control
import math
"""
m: mass of pendulum
M: mass of cart
b: coefficient of friction of the cart (zero in this case)
I: inertia of the pendulum (might be 0?)
l = length of the pendulum
q = (M+m) * (I+m*(l^2))-((m*l)^2)
closed loop transfer_function = Theta(s)/Force(s) = (-m*l*s)/q) / ( (s^3) + ((b*(m(l^2) + I)/q)*(s^2) + (((M + m)*g*m*l)/q)*s + b*m*g*l/q
No significance to values for this model
kd = 1
kp = 1
ki = 1
pid_controller = kd*s + kp + (ki/s)
open_loop_transfer_function = ((-self.m*self.l)/q) / ((s^2) + (((self.M + self.m)*self.g*self.m*self.l)/q))
"""
class PIDStrategy(BaseStrategy):
def __init__(self):
self.g = 9.8
self.m = 0.1
self.M = 1.0
self.l = 1.0 # or 0.5?
self.I = 0
def pid_controller_pendulum (self, observation):
error = (180/math.pi) * self.get_pole_angle(observation)
#amount of deviation from vertical
q = (self.M+self.m) * (self.I+self.m*(self.l**2))-((self.m*self.l)**2)
#q => setting reocurring constants
open_loop_transfer_function = control.TransferFunction([((-self.m*self.l)/q)], [1, 0, (((self.M + self.m)*self.g*self.m*self.l)/q)])
kd = 1000
kp = -100
ki = 0
pid_controller = control.TransferFunction([kd, kp, ki], [1, 0])
#setting up control function
transfer_function = (open_loop_transfer_function/(1 + open_loop_transfer_function*pid_controller))
#closed loop = open/(1 + (open*control func))
t, force_array = control.impulse_response(transfer_function, X0 = error)
#gives an impulse and stores how it responds over time
#IC = starting angle (error), creates a time array (t) and force array, the latter is the actual system output
force = force_array[0]
#only care about first value as it corresponds to our IC
return force
def calculate (self, observation):
force = self.pid_controller_pendulum(observation)
if (force >= 0):
action = self.make_move_left_action()
# sol = self.make_more_....()*force
elif (force < 0):
action = self.make_move_right_action()
return action
play_game(PIDStrategy, "PID Strategy", episodes)<jupyter_output>High Score: 67, Average Score: 41.69
<jupyter_text>### DQN Strategy
DQN is a reinforcement learning technique that can be used to solve CartPole. We will split up the strategy in a few parts. First part is to setup a Network class in order to compute the reinforcement learning. Next we will actually solve the game then plot it. Lastly is to have a different way of playing the game due to the reinforcement learning part.<jupyter_code>from Logger import Logger, ScoreLogger
import gym
from keras.models import Sequential
from keras.layers import Dense, Dropout
from keras.optimizers import rmsprop, Adam
import numpy as np
from collections import deque
from statistics import mean
import h5py
LEARNING_RATE = 1e-3
MAX_MEMORY = 1000000
BATCH_SIZE = 20
GAMMA = 0.95
EXPLORATION_DECAY = 0.995
EXPLORATION_MIN = 0.01
class Network:
def __init__(self, observation_space, action_space):
self.action_space = action_space
self.memory = deque(maxlen=MAX_MEMORY)
self.exploration_rate = 1.0
self.model = Sequential()
self.model.add(Dense(32, input_shape=(observation_space,), activation='relu'))
self.model.add(Dense(32, activation='relu'))
self.model.add(Dense(self.action_space, activation='linear'))
self.model.compile(loss='mse', optimizer=Adam(lr=LEARNING_RATE))
def add_to_memory(self, state, action, reward, next_state, done):
self.memory.append((state, action, reward, next_state, done))
def take_action(self, state):
if np.random.rand() < self.exploration_rate:
return random.randrange(0, self.action_space)
q_values = self.model.predict(state)
return np.argmax(q_values[0])
def experience_replay(self):
if len(self.memory) < BATCH_SIZE:
return
else:
minibatch = random.sample(self.memory, BATCH_SIZE)
for state, action, reward, state_next, done in minibatch:
Q = reward
if not done:
Q = (reward + GAMMA * np.amax(self.model.predict(state_next)[0]))
Q_values = self.model.predict(state)
Q_values[0][action] = Q
self.model.fit(state, Q_values, verbose=0)
self.exploration_rate *= EXPLORATION_DECAY
self.exploration_rate = max(EXPLORATION_MIN, self.exploration_rate)
def get_model(self):
return self.model
class DQNGameSolver(BaseStrategy):
def __init__(self, max_episodes, render=False):
self.max_episodes = max_episodes
self.logger = Logger()
self.scorelogger = ScoreLogger()
self.render = render
self.highscore = -1
self.totalscore = 0
self.score_table = deque(maxlen=400)
self.average_of_last_runs = None
self.model = None
env = gym.make('CartPole-v1')
observation_space = env.observation_space.shape[0]
action_space = env.action_space.n
self.solver = Network(observation_space, action_space)
def log_score(self, score):
self.totalscore += score
self.scorelogger.log(score)
def log_observation(self, observation, logger):
logger.log(
self.get_pole_position(observation),
self.get_pole_velocity(observation),
self.get_pole_angle(observation),
self.get_pole_tip_velocity(observation)
)
def plot(self, title=""):
print("High Score: " + str(self.highscore) + ", Average Score: " + str(self.totalscore/self.max_episodes))
self.scorelogger.plot(title)
self.logger.plot(title)
def print_logs(self):
print(self.logger.get_positions())
print(self.logger.get_velocities())
print(self.logger.get_angles())
print(self.logger.get_tip_velocities())
print(self.scorelogger.get_scores())
def play(self):
env = gym.make('CartPole-v1')
observation_space = env.observation_space.shape[0]
action_space = env.action_space.n
self.model = self.solver.get_model()
episode = 0
while episode < self.max_episodes:
episode += 1
state = env.reset()
state = np.reshape(state, [1, observation_space])
step = 0
logger = Logger()
while True:
step += 1
if self.render:
env.render()
action = self.solver.take_action(state)
state_next, reward, done, info = env.step(action)
self.log_observation(state_next, logger)
if not done:
reward = reward
else:
reward = -reward
state_next = np.reshape(state_next, [1, observation_space])
self.solver.add_to_memory(state, action, reward, state_next, done)
state = state_next
if done:
self.log_score(step)
if step > self.highscore:
self.highscore = step
self.logger = logger
env.close()
break
self.solver.experience_replay()
def play_dqn_game(title="", episodes=100):
game = DQNGameSolver(episodes, render)
game.play()
game.plot(title)
play_dqn_game("DQN Strategy", episodes)<jupyter_output>WARNING:tensorflow:From /opt/conda/lib/python3.6/site-packages/keras/backend/tensorflow_backend.py:422: The name tf.global_variables is deprecated. Please use tf.compat.v1.global_variables instead.
High Score: 500, Average Score: 114.895
|
no_license
|
/jupyter_notebook/.ipynb_checkpoints/cartpole_notebook-checkpoint.ipynb
|
brandonyap/engg3130-finalproject
| 10 |
<jupyter_start><jupyter_text># Data Preparation
-----### Constants and Folder Paths<jupyter_code>import os
dataset_folder_path = os.path.join("..", "files", "dataset")
NUM_SAMPLES = 50<jupyter_output><empty_output><jupyter_text>### Load Data and Split into *Test*, *Train/Valid*<jupyter_code>from data.DataSet import DataSet
dataset = DataSet()
dataset.load(dataset_folder_path, test_set_percentage=0.5, validation_set_percentage=0)
print(len(dataset.train_data))
print(len(dataset.test_data))<jupyter_output>1800
1800
<jupyter_text>### Data Preprocessing<jupyter_code>from utils.preprocessing import *
from functools import partial
dataset.apply(apply_mean_centering)
dataset.apply(apply_unit_distance_normalization)
#dataset.apply(partial(normalize_pressure_value, max_pressure_val=512))
dataset.apply(partial(spline_interpolate_and_resample, num_samples=NUM_SAMPLES))
dataset.expand(reverse_digit_sequence)
# dataset.apply(lambda digit: convert_xy_to_derivative(digit, normalize=False))
#dataset.apply(partial(convert_xy_to_derivative, normalize=True))
print(len(dataset.train_data))
print(len(dataset.test_data))<jupyter_output>3600
3600
<jupyter_text>### Split Dataset into *Train*, *Valid*, and *Test*<jupyter_code>import numpy as np
from sklearn.model_selection import train_test_split
X_train_valid = np.array(dataset.train_data)
X_test = np.array(dataset.test_data)
# Convert labels to numpy array and OneHot encode them
encoder, train_valid_labels, _, Y_test = dataset.onehot_encode_labels()
train_valid_labels = train_valid_labels.astype('float32').todense()
Y_test = Y_test.astype('float32').todense()
# Split Data
X_train, X_valid, Y_train, Y_valid = train_test_split(X_train_valid, train_valid_labels, shuffle=True, stratify=train_valid_labels, random_state=42)<jupyter_output><empty_output><jupyter_text>----------
# Neural Network Setup and Training
----------## **Regularized Naive GRU**### Parameters<jupyter_code>PARAM_NUM_EPOCHS = 30
PARAM_BATCH_SIZE = 300<jupyter_output><empty_output><jupyter_text>### Setup Model<jupyter_code>from models.regularized_gru import NaiveRegularizedGRU
mymodel = NaiveRegularizedGRU(X_train.shape[1:])
mymodel.batch_size = PARAM_BATCH_SIZE
mymodel.num_epochs = PARAM_NUM_EPOCHS
mymodel.initialize()
print(mymodel)<jupyter_output>
----------
Optimizer: <class 'keras.optimizers.Nadam'>
Batch Size: 300
Number of Epochs: 30
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
gru_15 (GRU) (None, 50, 256) 198912
_________________________________________________________________
dropout_15 (Dropout) (None, 50, 256) 0
_________________________________________________________________
gru_16 (GRU) (None, 256) 393984
_________________________________________________________________
dropout_16 (Dropout) (None, 256) 0
_________________________________________________________________
dense_8 (Dense) (None, 10) 2570
_________________________________________________________________
activation_8 (Activation) (None, 10) [...]<jupyter_text>#### Save Model Summary<jupyter_code>mymodel.save_summary(dataset.get_recorded_operations())
mymodel.save_config()<jupyter_output><empty_output><jupyter_text>### Train Model<jupyter_code>mymodel.train(X_train, Y_train, X_valid, Y_valid)<jupyter_output>Train on 2700 samples, validate on 900 samples
Epoch 1/30
2700/2700 [==============================] - 1s 494us/step - loss: 2.1803 - categorical_accuracy: 0.1863 - val_loss: 2.1012 - val_categorical_accuracy: 0.1911
Epoch 00001: val_categorical_accuracy improved from -inf to 0.19111, saving model to checkpoints/1524406933.4725032/naive_regularized_gru-01-0.19.hdf5
Epoch 2/30
2700/2700 [==============================] - 1s 283us/step - loss: 2.0356 - categorical_accuracy: 0.2422 - val_loss: 1.8963 - val_categorical_accuracy: 0.2844
Epoch 00002: val_categorical_accuracy improved from 0.19111 to 0.28444, saving model to checkpoints/1524406933.4725032/naive_regularized_gru-02-0.28.hdf5
Epoch 3/30
2700/2700 [==============================] - 1s 284us/step - loss: 1.9089 - categorical_accuracy: 0.2985 - val_loss: 1.5668 - val_categorical_accuracy: 0.4489
Epoch 00003: val_categorical_accuracy improved from 0.28444 to 0.44889, saving model to checkpoints/1524406933.4725032/naive_regularize[...]<jupyter_text>### Evaluate Model#### Test Set Accuracy<jupyter_code>test_score = tuple(mymodel.model.evaluate(X_test, Y_test))
print("Test Loss: %.3f, Test Acc: %.3f%%" % (test_score[0], test_score[1] * 100))<jupyter_output>3600/3600 [==============================] - 2s 490us/step
Test Loss: 0.294, Test Acc: 93.056%
<jupyter_text>#### Recall, Precision, F1_Score on Validation set<jupyter_code>from utils.evaluation import get_evaluation_metrics, get_confusion_matrix
Y_predicted_valid = mymodel.model.predict_classes(X_valid, verbose=1)
rpf_valid = get_evaluation_metrics(Y_valid, Y_predicted_valid)
rpf_valid<jupyter_output>900/900 [==============================] - 1s 947us/step
<jupyter_text>Average F1 Score for Validation Set<jupyter_code>rpf_valid.mean()<jupyter_output><empty_output><jupyter_text>#### Recall, Precision, F1_Score on Test set<jupyter_code>from utils.evaluation import get_evaluation_metrics, get_confusion_matrix
Y_predicted_test = mymodel.model.predict_classes(X_test, verbose=1)
rpf_test = get_evaluation_metrics(Y_test, Y_predicted_test)
rpf_test<jupyter_output>3600/3600 [==============================] - 2s 481us/step
<jupyter_text>Average F1 Score for Test Set<jupyter_code>rpf_test.mean()<jupyter_output><empty_output><jupyter_text>##### Increase default plotsize for matplotlib<jupyter_code>import matplotlib.pyplot as plt
plt.rcParams['figure.figsize'] = [15, 10]<jupyter_output><empty_output><jupyter_text>#### Plot Validation Set Confusion Matrix<jupyter_code>from utils.evaluation import get_confusion_matrix
# Confusion Matrix
confmat = get_confusion_matrix(Y_valid, Y_predicted_valid, plot=True)<jupyter_output><empty_output><jupyter_text>#### Plot Test Set Confusion Matrix<jupyter_code>from utils.evaluation import get_confusion_matrix
# Confusion Matrix
confmat = get_confusion_matrix(Y_test, Y_predicted_test, plot=True)<jupyter_output><empty_output><jupyter_text># Backup Code
-----<jupyter_code>from keras.models import load_model
mymodel.model = load_model("checkpoints/1523369406.7123575/naive_overfit_gru-30-0.97.hdf5")<jupyter_output><empty_output>
|
non_permissive
|
/notebooks/train_naive_regularized_thesis.ipynb
|
ronnymajani/dynamic-motion-data-tools
| 17 |
<jupyter_start><jupyter_text># Manipulación de DatosEl principal objetivo de esta clase es introducir la biblioteca de análisis de datos __pandas__, la cual agrega mayor flexibilidad y opciones que Numpy, sin embargo el costo de esto se traduce en pérdida de rendimiento y eficiencia. Conocer los elemnos básicos pandas permite manejar desde pocos dados en un arhivo _excel_ hasta miles y millones de registros de una base de datos.## Introducción a pandasDesde de la [página oficial](https://pandas.pydata.org/) Pandas se introduce de la siguiente forma:
_pandas is a fast, powerful, flexible and easy to use open source data analysis and manipulation tool,
built on top of the Python programming language._
Mientras que su [misión](https://pandas.pydata.org/about/):
_pandas aims to be the fundamental high-level building block for doing practical, real world data analysis in Python. Additionally, it has the broader goal of becoming the most powerful and flexible open source data analysis / manipulation tool available in any language._### Principales Características
* A fast and efficient __DataFrame__ object for data manipulation with integrated indexing;
* Tools for __reading and writing data__ between in-memory data structures and different formats: CSV and text files, Microsoft Excel, SQL databases, and the fast HDF5 format;
* Intelligent __data alignment__ and integrated handling of __missing data__: gain automatic label-based alignment in computations and easily manipulate messy data into an orderly form;
* Flexible __reshaping__ and pivoting of data sets;
* Intelligent label-based __slicing__, __fancy indexing__, and __subsetting__ of large data sets;
* Columns can be inserted and deleted from data structures for __size mutability__;
* Aggregating or transforming data with a powerful __group by__ engine allowing split-apply-combine operations on data sets;
* High performance __merging and joining__ of data sets;
* __Hierarchical axis indexing__ provides an intuitive way of working with high-dimensional data in a lower-dimensional data structure;
* __Time series__-functionality: date range generation and frequency conversion, moving window statistics, date shifting and lagging. Even create domain-specific time offsets and join time series without losing data;
* Highly __optimized for performance__, with critical code paths written in Cython or C.
* Python with pandas is in use in a wide variety of __academic and commercial__ domains, including Finance, Neuroscience, Economics, * Statistics, Advertising, Web Analytics, and more.<jupyter_code>import pandas as pd
pd.__version__<jupyter_output><empty_output><jupyter_text>
## SeriesArreglos unidimensionales con etiquetas. Se puede pensar como una generalización de los diccionarios de Python.<jupyter_code># Descomenta y ejecuta para ver la documentación sobre pd.Series
pd.Series<jupyter_output><empty_output><jupyter_text>Para crear una instancia de una serie existen muchas opciones, las más comunes son:
* A partir de una lista.
* A partir de un _numpy.array_.
* A partir de un diccionario.
* A partir de un archivo (por ejemplo un csv).<jupyter_code>my_serie = pd.Series(range(3, 33, 3))
my_serie
type(my_serie)
# Presiona TAB y sorpréndete con la cantidad de métodos y atributos que poseen!
# my_serie.<jupyter_output><empty_output><jupyter_text>Las series son arreglos unidemensionales que constan de _data_ e _index_.<jupyter_code># Data
my_serie.values
type(my_serie.values)
# Index
my_serie.index
type(my_serie.index)<jupyter_output><empty_output><jupyter_text>¿Te fijaste que el index es de otra clase?A diferencia de Numpy, pandas ofrece más flexibilidad para los valores e índices.<jupyter_code>my_serie_2 = pd.Series(range(3, 33, 3), index=list('abcdefghij'))
my_serie_2<jupyter_output><empty_output><jupyter_text>### Acceder a los valores de una<jupyter_code>my_serie_2['b']
my_serie_2.loc['b']
my_serie_2.iloc[1]<jupyter_output><empty_output><jupyter_text>```loc```?? ```iloc```?? ¿Qué es eso?A modo de resumen:
* ```loc``` es un método que hace referencia a las etiquetas (*labels*) del objeto .
* ```iloc``` es un método que hace referencia posicional del objeto.**Consejo**: Si quieres editar valores siempre utiliza ```loc``` y/o ```iloc```.<jupyter_code>my_serie_2.loc['d'] = 1000
my_serie_2<jupyter_output><empty_output><jupyter_text>¿Y si quiero escoger más de un valor?<jupyter_code>my_serie_2.loc["b":"e"] # Incluso retorna el último valor!
my_serie_2.iloc[1:5] # Incluso retorna el último valor!<jupyter_output><empty_output><jupyter_text>Sorpresa! También puedes filtrar según condiciones!En la mayoría de los tutoriales en internet encontrarás algo como lo siguiente:<jupyter_code>my_serie_2[my_serie_2 % 2 == 0]<jupyter_output><empty_output><jupyter_text>Lo siguiente se conoce como _mask_, y se basa en el siguiente hecho:<jupyter_code>my_serie_2 % 2 == 0 # Retorna una serie con valores booleanos pero los mismos index!<jupyter_output><empty_output><jupyter_text>Si es una serie resultante de otra operación, tendrás que guardarla en una variable para así tener el nombre y luego acceder a ella. La siguiente manera puede qeu sea un poco más verboso, pero te otorga más flexibilidad.<jupyter_code>my_serie_2.loc[lambda s: s % 2 == 0]<jupyter_output><empty_output><jupyter_text>Una función lambda es una función pequeña y anónima. Pueden tomar cualquer número de argumentos pero solo tienen una expresión.### Trabajar con fechasPandas incluso permite que los index sean fechas! Por ejemplo, a continuación se crea una serie con fechas y valores de temperatura.<jupyter_code>temperature = pd.read_csv(
"https://raw.githubusercontent.com/jbrownlee/Datasets/master/daily-min-temperatures.csv",
index_col="Date",
squeeze=True
)
temperature.head(10)
temperature.tail(10)
temperature.dtype
temperature.index<jupyter_output><empty_output><jupyter_text>**OJO!** Los valores del Index son _strings_ (_object_ es una generalización). **Solución:** _Parsear_ a elementos de fecha con la función ```pd.to_datetime()```.<jupyter_code>pd.to_datetime
temperature.index = pd.to_datetime(temperature.index, format='%Y-%m-%d')
temperature.index<jupyter_output><empty_output><jupyter_text>Para otros tipos de _parse_ puedes visitar la documentación [aquí](https://docs.python.org/3/library/datetime.html#strftime-and-strptime-behavior).
La idea de los elementos de fecha es poder realizar operaciones que resulten naturales para el ser humano. Por ejemplo:<jupyter_code>temperature.index.min()
temperature.index.max()
temperature.index.max() - temperature.index.min()<jupyter_output><empty_output><jupyter_text>Volviendo a la Serie, podemos trabajar con todos sus elementos, por ejemplo, determinar rápidamente la máxima tendencia.<jupyter_code>max_temperature = temperature.max()
max_temperature<jupyter_output><empty_output><jupyter_text>Para determinar el _index_ correspondiente al valor máximo usualmente se utilizan dos formas:
* Utilizar una máscara (*mask*)
* Utilizar métodos ya implementados<jupyter_code># Mask
temperature[temperature == max_temperature]
# Built-in method
temperature.idxmax()<jupyter_output><empty_output><jupyter_text>
## DataFramesArreglo bidimensional y extensión natural de una serie. Podemos pensarlo como la generalización de un numpy.array.Para motivar utilizaremos los datos de COVID-19 disponibilizados por el Ministerio de Ciencias, Tecnología, Conocimiento e Innovación de Chile. En particular, los casos totales por comuna, puedes leer más detalles en el [repositorio oficial](https://github.com/MinCiencia/Datos-COVID19/tree/master/output/producto1).<jupyter_code>covid = pd.read_csv("https://raw.githubusercontent.com/MinCiencia/Datos-COVID19/master/output/producto1/Covid-19_std.csv")
covid.head()
covid.tail()
covid.query("Region == 'Valparaíso'")
type(covid)
covid.info(memory_usage=True)
covid.dtypes<jupyter_output><empty_output><jupyter_text>Puedes pensar que un dataframe es una colección de series<jupyter_code>covid['Casos confirmados'].head()
type(covid['Casos confirmados'])<jupyter_output><empty_output><jupyter_text>### Exploración <jupyter_code>covid.describe().T
covid.describe(include='all').T
covid.max()<jupyter_output><empty_output><jupyter_text>Para extraer elementos lo más recomendable es el método loc.<jupyter_code>covid.loc[19271, 'Region']<jupyter_output><empty_output><jupyter_text>Evita acceder con doble corchete<jupyter_code>covid[19271]['Region']<jupyter_output><empty_output><jupyter_text>Aunque en ocasiones funcione, no se asegura que sea siempre así. [Más info aquí.](https://pandas.pydata.org/pandas-docs/stable/indexing.html#why-does-assignment-fail-when-using-chained-indexing)<jupyter_code>covid['Region'].value_counts()<jupyter_output><empty_output><jupyter_text>### Valores perdidos/nulosPandas ofrece herramientas para trabajar con valors nulos, pero es necesario conocerlas y saber aplicarlas. Por ejemplo, el método ```isnull()``` entrega un booleano si algún valor es nulo.__Ejemplo:__ ¿Qué registros tienen casos confirmados nulos?<jupyter_code>covid.index.shape
covid.loc[lambda x: x['Casos confirmados'].isnull()]<jupyter_output><empty_output><jupyter_text>Si deseamos encontrar todas las filas que contengan por lo menos un valor nulo.<jupyter_code>covid.isnull()
rows_null_mask = covid.isnull().any(axis=1) # axis=1 hace referencia a las filas.
rows_null_mask.head()
covid[rows_null_mask].head()
covid[rows_null_mask].shape<jupyter_output><empty_output><jupyter_text>Para determinar aquellos que no tienen valors nulos el procedimiento es similar.<jupyter_code>covid.loc[lambda x: x.notnull().all(axis=1)].head()<jupyter_output><empty_output><jupyter_text>Pandas incluso ofrece opciones para eliminar elementos nulos!<jupyter_code># Cualquier registro con null
print(covid.dropna().shape)
# Filas con elementos nulos
print(covid.dropna(axis=0).shape)
# Columnas con elementos nulos
print(covid.dropna(axis=1).shape)<jupyter_output>(18681, 7)
(18681, 7)
(19548, 4)
|
permissive
|
/lessons/M2L03_data_manipulation.ipynb
|
ClaudioFigueroa/mat281_portfolio
| 26 |
<jupyter_start><jupyter_text>
# Optimal Growth II: Accelerating the Code with Numba## Contents
- [Optimal Growth II: Accelerating the Code with Numba](#Optimal-Growth-II:-Accelerating-the-Code-with-Numba)
- [Overview](#Overview)
- [The Model](#The-Model)
- [Computation](#Computation)
- [Exercises](#Exercises)
- [Solutions](#Solutions) In addition to what’s in Anaconda, this lecture will need the following libraries:<jupyter_code>!pip install --upgrade quantecon
!pip install --upgrade interpolation<jupyter_output><empty_output><jupyter_text>## Overview
[Previously](https://python-programming.quantecon.org/optgrowth.html), we studied a stochastic optimal
growth model with one representative agent.
We solved the model using dynamic programming.
In writing our code, we focused on clarity and flexibility.
These are important, but there’s often a trade-off between flexibility and
speed.
The reason is that, when code is less flexible, we can exploit structure more
easily.
(This is true about algorithms and mathematical problems more generally:
more specific problems have more structure, which, with some thought, can be
exploited for better results.)
So, in this lecture, we are going to accept less flexibility while gaining
speed, using just-in-time (JIT) compilation to
accelerate our code.
Let’s start with some imports:<jupyter_code>import numpy as np
import matplotlib.pyplot as plt
from interpolation import interp
from numba import jit, njit, jitclass, prange, float64, int32
from quantecon.optimize.scalar_maximization import brent_max
%matplotlib inline<jupyter_output><empty_output><jupyter_text>We are using an interpolation function from
[interpolation.py](https://github.com/EconForge/interpolation.py) because it
helps us JIT-compile our code.
The function `brent_max` is also designed for embedding in JIT-compiled code.
These are alternatives to similar functions in SciPy (which, unfortunately, are not JIT-aware).## The Model
The model is the same as discussed in our [previous lecture](https://python-programming.quantecon.org/optgrowth.html)
on optimal growth.
We will start with log utility:
$$
u(c) = \ln(c)
$$
We continue to assume that
- $ f(k) = k^{\alpha} $
- $ \phi $ is the distribution of $ \xi := \exp(\mu + s \zeta) $ when $ \zeta $ is standard normal
We will once again use value function iteration to solve the model.
In particular, the algorithm is unchanged, and the only difference is in the implementation itself.
As before, we will be able to compare with the true solutions<jupyter_code>
def v_star(y, α, β, μ):
"""
True value function
"""
c1 = np.log(1 - α * β) / (1 - β)
c2 = (μ + α * np.log(α * β)) / (1 - α)
c3 = 1 / (1 - β)
c4 = 1 / (1 - α * β)
return c1 + c2 * (c3 - c4) + c4 * np.log(y)
def σ_star(y, α, β):
"""
True optimal policy
"""
return (1 - α * β) * y<jupyter_output><empty_output><jupyter_text>## Computation
We will again store the primitives of the optimal growth model in a class.
But now we are going to use [Numba’s](https://python-programming.quantecon.org/numba.html) `@jitclass` decorator to target our class for JIT compilation.
Because we are going to use Numba to compile our class, we need to specify the data types.
You will see this as a list called `opt_growth_data` above our class.
Unlike in the [previous lecture](https://python-programming.quantecon.org/optgrowth.html), we
hardwire the production and utility specifications into the
class.
This is where we sacrifice flexibility in order to gain more speed.<jupyter_code>
opt_growth_data = [
('α', float64), # Production parameter
('β', float64), # Discount factor
('μ', float64), # Shock location parameter
('s', float64), # Shock scale parameter
('grid', float64[:]), # Grid (array)
('shocks', float64[:]) # Shock draws (array)
]
@jitclass(opt_growth_data)
class OptimalGrowthModel:
def __init__(self,
α=0.4,
β=0.96,
μ=0,
s=0.1,
grid_max=4,
grid_size=120,
shock_size=250,
seed=1234):
self.α, self.β, self.μ, self.s = α, β, μ, s
# Set up grid
self.grid = np.linspace(1e-5, grid_max, grid_size)
# Store shocks (with a seed, so results are reproducible)
np.random.seed(seed)
self.shocks = np.exp(μ + s * np.random.randn(shock_size))
def f(self, k):
"The production function"
return k**self.α
def u(self, c):
"The utility function"
return np.log(c)
def f_prime(self, k):
"Derivative of f"
return self.α * (k**(self.α - 1))
def u_prime(self, c):
"Derivative of u"
return 1/c
def u_prime_inv(self, c):
"Inverse of u'"
return 1/c<jupyter_output><empty_output><jupyter_text>The class includes some methods such as `u_prime` that we do not need now
but will use in later lectures.### The Bellman Operator
We will use JIT compilation to accelerate the Bellman operator.
First, here’s a function that returns the value of a particular consumption choice `c`, given state `y`, as per the Bellman equation [(9)](https://python-programming.quantecon.org/optgrowth.html#equation-fpb30).<jupyter_code>@njit
def state_action_value(c, y, v_array, og):
"""
Right hand side of the Bellman equation.
* c is consumption
* y is income
* og is an instance of OptimalGrowthModel
* v_array represents a guess of the value function on the grid
"""
u, f, β, shocks = og.u, og.f, og.β, og.shocks
v = lambda x: interp(og.grid, v_array, x)
return u(c) + β * np.mean(v(f(y - c) * shocks))<jupyter_output><empty_output><jupyter_text>Now we can implement the Bellman operator, which maximizes the right hand side
of the Bellman equation:<jupyter_code>@jit(nopython=True)
def T(v, og):
"""
The Bellman operator.
* og is an instance of OptimalGrowthModel
* v is an array representing a guess of the value function
"""
v_new = np.empty_like(v)
v_greedy = np.empty_like(v)
for i in range(len(og.grid)):
y = og.grid[i]
# Maximize RHS of Bellman equation at state y
result = brent_max(state_action_value, 1e-10, y, args=(y, v, og))
v_greedy[i], v_new[i] = result[0], result[1]
return v_greedy, v_new<jupyter_output><empty_output><jupyter_text>We use the `solve_model` function to perform iteration until convergence.<jupyter_code>def solve_model(og,
tol=1e-4,
max_iter=1000,
verbose=True,
print_skip=25):
"""
Solve model by iterating with the Bellman operator.
"""
# Set up loop
v = og.u(og.grid) # Initial condition
i = 0
error = tol + 1
while i < max_iter and error > tol:
v_greedy, v_new = T(v, og)
error = np.max(np.abs(v - v_new))
i += 1
if verbose and i % print_skip == 0:
print(f"Error at iteration {i} is {error}.")
v = v_new
if i == max_iter:
print("Failed to converge!")
if verbose and i < max_iter:
print(f"\nConverged in {i} iterations.")
return v_greedy, v_new<jupyter_output><empty_output><jupyter_text>Let’s compute the approximate solution at the default parameters.
First we create an instance:<jupyter_code>og = OptimalGrowthModel()<jupyter_output><empty_output><jupyter_text>Now we call `solve_model`, using the `%%time` magic to check how long it
takes.<jupyter_code>%%time
v_greedy, v_solution = solve_model(og)<jupyter_output><empty_output><jupyter_text>You will notice that this is *much* faster than our [original implementation](https://python-programming.quantecon.org/optgrowth.html).
Here is a plot of the resulting policy, compared with the true policy:<jupyter_code>fig, ax = plt.subplots()
ax.plot(og.grid, v_greedy, lw=2,
alpha=0.8, label='approximate policy function')
ax.plot(og.grid, σ_star(og.grid, og.α, og.β), 'k--',
lw=2, alpha=0.8, label='true policy function')
ax.legend()
plt.show()<jupyter_output><empty_output><jupyter_text>Again, the fit is excellent — this is as expected since we have not changed
the algorithm.
The maximal absolute deviation between the two policies is<jupyter_code>np.max(np.abs(v_greedy - σ_star(og.grid, og.α, og.β)))<jupyter_output><empty_output><jupyter_text>## Exercises
### Exercise 1
Time how long it takes to iterate with the Bellman operator
20 times, starting from initial condition $ v(y) = u(y) $.
Use the default parameterization.### Exercise 2
Modify the optimal growth model to use the CRRA utility specification.
$$
u(c) = \frac{c^{1 - \gamma} } {1 - \gamma}
$$
Set `γ = 1.5` as the default value and maintaining other specifications.
(Note that `jitclass` currently does not support inheritance, so you will
have to copy the class and change the relevant parameters and methods.)
Compute an estimate of the optimal policy, plot it and compare visually with
the same plot from the [analogous exercise](https://python-programming.quantecon.org/optgrowth.html#ogex1) in the first optimal
growth lecture.
Compare execution time as well.
### Exercise 3
In this exercise we return to the original log utility specification.
Once an optimal consumption policy $ \sigma $ is given, income follows
$$
y_{t+1} = f(y_t - \sigma(y_t)) \xi_{t+1}
$$
The next figure shows a simulation of 100 elements of this sequence for three
different discount factors (and hence three different policies).
In each sequence, the initial condition is $ y_0 = 0.1 $.
The discount factors are `discount_factors = (0.8, 0.9, 0.98)`.
We have also dialed down the shocks a bit with `s = 0.05`.
Otherwise, the parameters and primitives are the same as the log-linear model discussed earlier in the lecture.
Notice that more patient agents typically have higher wealth.
Replicate the figure modulo randomness.## Solutions### Exercise 1
Let’s set up the initial condition.<jupyter_code>v = og.u(og.grid)<jupyter_output><empty_output><jupyter_text>Here’s the timing:<jupyter_code>%%time
for i in range(20):
v_greedy, v_new = T(v, og)
v = v_new<jupyter_output><empty_output><jupyter_text>Compared with our [timing](https://python-programming.quantecon.org/optgrowth.html#og-ex2) for the non-compiled version of
value function iteration, the JIT-compiled code is usually an order of magnitude faster.### Exercise 2
Here’s our CRRA version of `OptimalGrowthModel`:<jupyter_code>
opt_growth_data = [
('α', float64), # Production parameter
('β', float64), # Discount factor
('μ', float64), # Shock location parameter
('γ', float64), # Preference parameter
('s', float64), # Shock scale parameter
('grid', float64[:]), # Grid (array)
('shocks', float64[:]) # Shock draws (array)
]
@jitclass(opt_growth_data)
class OptimalGrowthModel_CRRA:
def __init__(self,
α=0.4,
β=0.96,
μ=0,
s=0.1,
γ=1.5,
grid_max=4,
grid_size=120,
shock_size=250,
seed=1234):
self.α, self.β, self.γ, self.μ, self.s = α, β, γ, μ, s
# Set up grid
self.grid = np.linspace(1e-5, grid_max, grid_size)
# Store shocks (with a seed, so results are reproducible)
np.random.seed(seed)
self.shocks = np.exp(μ + s * np.random.randn(shock_size))
def f(self, k):
"The production function."
return k**self.α
def u(self, c):
"The utility function."
return c**(1 - self.γ) / (1 - self.γ)
def f_prime(self, k):
"Derivative of f."
return self.α * (k**(self.α - 1))
def u_prime(self, c):
"Derivative of u."
return c**(-self.γ)
def u_prime_inv(c):
return c**(-1 / self.γ)<jupyter_output><empty_output><jupyter_text>Let’s create an instance:<jupyter_code>og_crra = OptimalGrowthModel_CRRA()<jupyter_output><empty_output><jupyter_text>Now we call `solve_model`, using the `%%time` magic to check how long it
takes.<jupyter_code>%%time
v_greedy, v_solution = solve_model(og_crra)<jupyter_output><empty_output><jupyter_text>Here is a plot of the resulting policy:<jupyter_code>fig, ax = plt.subplots()
ax.plot(og.grid, v_greedy, lw=2,
alpha=0.6, label='Approximate value function')
ax.legend(loc='lower right')
plt.show()<jupyter_output><empty_output><jupyter_text>This matches the solution that we obtained in our non-jitted code, [in
the exercises](https://python-programming.quantecon.org/optgrowth.html#ogex1).
Execution time is an order of magnitude faster.### Exercise 3
Here’s one solution:<jupyter_code>def simulate_og(σ_func, og, y0=0.1, ts_length=100):
'''
Compute a time series given consumption policy σ.
'''
y = np.empty(ts_length)
ξ = np.random.randn(ts_length-1)
y[0] = y0
for t in range(ts_length-1):
y[t+1] = (y[t] - σ_func(y[t]))**og.α * np.exp(og.μ + og.s * ξ[t])
return y
fig, ax = plt.subplots()
for β in (0.8, 0.9, 0.98):
og = OptimalGrowthModel(β=β, s=0.05)
v_greedy, v_solution = solve_model(og, verbose=False)
# Define an optimal policy function
σ_func = lambda x: interp(og.grid, v_greedy, x)
y = simulate_og(σ_func, og)
ax.plot(y, lw=2, alpha=0.6, label=rf'$\beta = {β}$')
ax.legend(loc='lower right')
plt.show()<jupyter_output><empty_output>
|
no_license
|
/optgrowth_fast.ipynb
|
lystahi/lecture-python-intro.notebooks
| 18 |
<jupyter_start><jupyter_text># MNIST
MNISTは手書き数字画像60,000枚と、テスト画像10,000枚を集めた、画像データセット。
各画像に正解ラベルがついており、手書き文字の画像から、書かれた数字を当てるのが目的である。
いわゆる多クラス分類の問題である### インポートとデータ読み込み
各画像は28画素x28画素(784ピクセル)の白黒画像となっている。
以下ではランダムに選んだ81画像を表示させている。<jupyter_code># tensorflow 2.x には keras>=2.3.x が必要
!pip install -U keras
import tensorflow as tf
import keras
from keras.models import Sequential
from keras.layers import Dense, Dropout
from keras.optimizers import Adam
from keras.datasets import mnist
from keras.utils.np_utils import to_categorical
import matplotlib.pyplot as plt
%matplotlib inline
print("Tensorflow version " + tf.__version__)
#Kerasの関数でデータの読み込み。データをシャッフルして学習データと訓練データに分割
(x_train, y_train), (x_test, y_test) = mnist.load_data()
#MNISTデータの表示
fig = plt.figure(figsize=(9, 9))
fig.subplots_adjust(left=0, right=1, bottom=0, top=0.5, hspace=0.05, wspace=0.05)
for i in range(81):
ax = fig.add_subplot(9, 9, i + 1, xticks=[], yticks=[])
ax.imshow(x_train[i].reshape((28, 28)), cmap='gray')<jupyter_output>Requirement already up-to-date: keras in /usr/local/lib/python3.6/dist-packages (2.3.1)
Requirement already satisfied, skipping upgrade: h5py in /usr/local/lib/python3.6/dist-packages (from keras) (2.10.0)
Requirement already satisfied, skipping upgrade: keras-preprocessing>=1.0.5 in /usr/local/lib/python3.6/dist-packages (from keras) (1.1.0)
Requirement already satisfied, skipping upgrade: six>=1.9.0 in /usr/local/lib/python3.6/dist-packages (from keras) (1.12.0)
Requirement already satisfied, skipping upgrade: numpy>=1.9.1 in /usr/local/lib/python3.6/dist-packages (from keras) (1.18.2)
Requirement already satisfied, skipping upgrade: scipy>=0.14 in /usr/local/lib/python3.6/dist-packages (from keras) (1.4.1)
Requirement already satisfied, skipping upgrade: pyyaml in /usr/local/lib/python3.6/dist-packages (from keras) (3.13)
Requirement already satisfied, skipping upgrade: keras-applications>=1.0.6 in /usr/local/lib/python3.6/dist-packages (from keras) (1.0.8)
Tensorflow version 2.2.0-[...]<jupyter_text>### データ加工
- 28x28の2次元データを、1次元の大きさ784の数値列に変換する
### ※工夫のポイント
- データの中身は、扱うものによって、さまざまな値をとる(例:テストの点数(0-100)、気温、株価、緯度経度、金額)
- このうち、複数のデータ値を一緒に学習することを考える
- 例:ニューヨークの気温とNYSEの株価
- この際に、気温はおおよそ20-30の範囲、株価は10000前後、のように桁が違うデータを同時にトレーニングすると精度が落ちる
- したがって、すべてのデータを同じ範囲に揃える(たとえば0.0~1.0の間)これを **正規化** と呼ぶ
### 正規化
一般的には、配列 x があったとき、以下のような式で、値を `0.0 - 1.0` に収めることができる
```python
def normalize(x):
max_x = np.max(x, axis=(0, 1), keepdims=True)
min_x = np.min(x, axis=(0, 1), keepdims=True)
return (x - min_x) / (max_x - min_x)
```
画像データの場合、データの値は `0 - 255` の値を取るので、上の式に当てはめると、
`min_x` が 0 となり、もとのデータの値を `255` で割れば良いことがわかる。<jupyter_code># 2次元データを1次元に変換
# (60000, 28, 28) -> (60000, 784)
x_train = x_train.reshape(60000, 784)
x_test = x_test.reshape(10000, 784)
# 型変換
x_train = x_train.astype('float32')
x_test = x_test.astype('float32')
# 255で割ったものを新たに変数とする
x_train /= 255
x_test /= 255<jupyter_output><empty_output><jupyter_text>### 正解ラベルの加工
- 元のデータセットには4, 7, 9, 0, 1, ...のように数値で入っています (これを名義尺度といいます)。
- 名義尺度の数(ここでは名義尺度は 0-9 なので 10)をクラス数といいます。
- 名義尺度を大きさがクラス数のベクトルに直します (これを One-Hot表現といいます)
-(例:`2` -> `[0, 0, 1, 0, 0, 0, 0, 0, 0, 0]`)
- one-hot表現にする理由は、そのほうが精度が上がるから。(精度が上がる理由は各自調べる)<jupyter_code>num_classes = 10
y_train = y_train.astype('int32')
y_test = y_test.astype('int32')
# to_categorical: one-hot encodingを施すためのメソッド
y_train = to_categorical(y_train, num_classes)
y_test = to_categorical(y_test, num_classes)<jupyter_output><empty_output><jupyter_text>### ネットワークの構築
- モデルを定義(作成)します
<jupyter_code># モデル作成
# Sequential: ネットワークを1列に積み上げているシンプルな方法
model = Sequential()
# 入力層では、入力データの大きさ input_shape を指定する。
# (1次元の場合は (xxx, ) と書かないとタプルではなく数値扱いとなってしまう)
# Dense は入力と出力をすべて接続する 全結合 のレイヤー (ここでは出力次元数が 256)
# 活性化関数(activation) はネットワークに「非線形性」を導入するために必要。
# 正規化線形関数 relu は精度が良いとされている活性化関数の一つ
model.add(Dense(256, activation='relu', input_shape=(784,)))
# Dropoutは、モデルの「過学習」を防止するためのテクニックの一つ。
# 指定された割合の値を `0` にしてしまうことで、学習を難しくし、過学習を防止する
# 過学習とは、トレーニング用のデータに特化しすぎたモデルとなってしまい、
# テスト用のデータで性能があがらなくなってしまう状態のこと。
# テスト用のデータでの性能を「汎化性能」と言ったりする
model.add(Dropout(0.2))
# Deep Learningは一般的に 3層以上のニューラルネットワークを使用するものをいう。
# ネットワークは層が多ければ、性能は良くなりやすいが、学習時間が伸びてしまい、リソースも多く必要になる。
# 学習時間の短縮と性能の向上をさまざまなアルゴリズムやテクニックで伸ばしていく。
model.add(Dense(256, activation='relu'))
model.add(Dropout(0.2))
model.add(Dense(256, activation='relu'))
model.add(Dropout(0.2))
model.add(Dense(128, activation='relu'))
model.add(Dropout(0.2))
# 出力層では、クラス数が出力データ数となるようにする。
# 多クラス分類では、出力層の活性化関数に softmax を使うのが一般的
model.add(Dense(num_classes, activation='softmax'))<jupyter_output><empty_output><jupyter_text>### モデルサマリの表示<jupyter_code>model.summary()<jupyter_output>Model: "sequential_2"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
dense_6 (Dense) (None, 256) 200960
_________________________________________________________________
dropout_5 (Dropout) (None, 256) 0
_________________________________________________________________
dense_7 (Dense) (None, 256) 65792
_________________________________________________________________
dropout_6 (Dropout) (None, 256) 0
_________________________________________________________________
dense_8 (Dense) (None, 256) 65792
_________________________________________________________________
dropout_7 (Dropout) (None, 256) 0
______________________________________________________[...]<jupyter_text>## 学習の設定
### 損失関数
- 損失関数(コスト関数) loss は、モデルを学習する際に基準となる関数で、この関数の値が小さくなるようにパラメータを更新していく。
- パラメータを更新するためには、どのようにパラメータを更新すれば、損失関数の値が小さくなるか分かる必要があり、そのために、損失関数は連続で微分可能である必要がある。
- 多クラス分類では、損失関数として categorical_crossentropy がよく使われる(性能がよい)
### 最適化関数
- 最適化関数 optimizer は、パラメータを「どのくらい」更新したらよいかを決める関数。
- Adam はその中でも収束が早い(学習が早い) 最適化関数として知られている
### 学習の指標
ここでは、学習の指標として 正解率 accuracy を使う<jupyter_code>model.compile(loss='categorical_crossentropy',
optimizer=Adam(),
metrics=['accuracy'])<jupyter_output><empty_output><jupyter_text>### 学習とテスト
- バッチサイズとは、一回の計算で同時に処理するデータ数です
- **エポック** とは、トレーニングデータセット(この場合、60000データ)をいったん全部学習することを指します
- 通常、エポックを複数回繰り返すことで、誤差が徐々に小さくなるため、60000データの学習を何セットも繰り返す、ということを行います
### 学習の途中経過の表示
- 1エポック終了ごとに、そこまでの経過を表示します
- loss: トレーニングデータでの誤差の大きさ(小さいほど良い)
- acc: トレーニングデータでの精度(大きいほど良い。1.0=100%)
- エポック終了ごとにテストデータでも精度をみます。
- val_loss: テストデータでの誤差の大きさ(小さいほど良い)
- val_acc: テストデータでの精度(大きいほど良い。1.0=100%)<jupyter_code># バッチサイズ、エポック数
batch_size = 128
epochs = 20
history = model.fit(x_train, y_train,
batch_size=batch_size,
epochs=epochs,
verbose=1,
validation_data=(x_test, y_test))
score = model.evaluate(x_test, y_test, verbose=0)
print('Test loss:', score[0])
print('Test accuracy:', score[1])<jupyter_output>Test loss: 0.06843761885338036
Test accuracy: 0.9818999767303467
<jupyter_text>### 精度と損失関数のグラフ化
- 可視化は大事
### 見るべきポイント
- loss/accともに、トレーニングデータだけ最適化されてテストデータの精度(損失)が著しく悪ければ、それは「過学習」のあらわれ
- 言い換えると、トレーニングデータの精度だけあがっても、テストデータやその他の現実のデータで精度が高くないと現実的に使えるAIにならない
<jupyter_code>#Accuracy
plt.plot(history.history['accuracy'])
plt.plot(history.history['val_accuracy'])
plt.title('model accuracy')
plt.ylabel('accuracy')
plt.xlabel('epoch')
plt.legend(['train', 'test'], loc='upper left')
plt.show()
#loss
plt.plot(history.history['loss'])
plt.plot(history.history['val_loss'])
plt.title('model loss')
plt.ylabel('loss')
plt.xlabel('epoch')
plt.legend(['train', 'test'], loc='upper left')
plt.show()<jupyter_output><empty_output>
|
no_license
|
/実装/FNN_MNIST_for_IT_Arch.ipynb
|
IMOKURI/ML-DL
| 8 |
<jupyter_start><jupyter_text># Machine Learning. Data Analysis 2
# Text generation with an RNN
## Artiom Carabas; Fatih KarabayThis tutorial demonstrates how to generate text using a character-based RNN. You will work with a dataset of Shakespeare's writing from Andrej Karpathy's [The Unreasonable Effectiveness of Recurrent Neural Networks](http://karpathy.github.io/2015/05/21/rnn-effectiveness/). Given a sequence of characters from this data ("Shakespear"), train a model to predict the next character in the sequence ("e"). Longer sequences of text can be generated by calling the model repeatedly.
Note: Enable GPU acceleration to execute this notebook faster. In Colab: *Runtime > Change runtime type > Hardware accelerator > GPU*.
This tutorial includes runnable code implemented using [tf.keras](https://www.tensorflow.org/guide/keras/sequential_model) and [eager execution](https://www.tensorflow.org/guide/eager). The following is the sample output when the model in this tutorial trained for 30 epochs, and started with the prompt "Q":
QUEENE:
I had thought thou hadst a Roman; for the oracle,
Thus by All bids the man against the word,
Which are so weak of care, by old care done;
Your children were in your holy love,
And the precipitation through the bleeding throne.
BISHOP OF ELY:
Marry, and will, my lord, to weep in such a one were prettiest;
Yet now I was adopted heir
Of the world's lamentable day,
To watch the next way with his father with his face?
ESCALUS:
The cause why then we are all resolved more sons.
VOLUMNIA:
O, no, no, no, no, no, no, no, no, no, no, no, no, no, no, no, no, no, no, no, no, it is no sin it should be dead,
And love and pale as any will to that word.
QUEEN ELIZABETH:
But how long have I heard the soul for this world,
And show his hands of life be proved to stand.
PETRUCHIO:
I say he look'd on, if I must be content
To stay him from the fatal of our country's bliss.
His lordship pluck'd from this sentence then for prey,
And then let us twain, being the moon,
were she such a case as fills m
While some of the sentences are grammatical, most do not make sense. The model has not learned the meaning of words, but consider:
* The model is character-based. When training started, the model did not know how to spell an English word, or that words were even a unit of text.
* The structure of the output resembles a play—blocks of text generally begin with a speaker name, in all capital letters similar to the dataset.
* As demonstrated below, the model is trained on small batches of text (100 characters each), and is still able to generate a longer sequence of text with coherent structure.## Setup### Import TensorFlow and other libraries<jupyter_code>import tensorflow as tf
from tensorflow.keras.layers.experimental import preprocessing
import numpy as np
import os
import time<jupyter_output><empty_output><jupyter_text>### Download the Shakespeare dataset
Change the following line to run this code on your own data.<jupyter_code>path_to_file = tf.keras.utils.get_file('pride-and-prejudice.txt', 'https://raw.githubusercontent.com/artemcarabash/machine_learning/main/pride-and-prejudice.txt')<jupyter_output><empty_output><jupyter_text>### Read the data
First, look in the text:<jupyter_code># Read, then decode for py2 compat.
text = open(path_to_file, 'rb').read().decode(encoding='utf-8')
# length of text is the number of characters in it
print(f'Length of text: {len(text)} characters')
# Take a look at the first 250 characters in text
print(text[:250])
# The unique characters in the file
vocab = sorted(set(text))
print(f'{len(vocab)} unique characters')<jupyter_output>76 unique characters
<jupyter_text>## Process the text### Vectorize the text
Before training, you need to convert the strings to a numerical representation.
The `preprocessing.StringLookup` layer can convert each character into a numeric ID. It just needs the text to be split into tokens first.<jupyter_code>example_texts = ['abcdefg', 'xyz']
chars = tf.strings.unicode_split(example_texts, input_encoding='UTF-8')
chars<jupyter_output><empty_output><jupyter_text>Now create the `preprocessing.StringLookup` layer:<jupyter_code>ids_from_chars = preprocessing.StringLookup(
vocabulary=list(vocab))<jupyter_output><empty_output><jupyter_text>It converts form tokens to character IDs, padding with `0`:<jupyter_code>ids = ids_from_chars(chars)
ids<jupyter_output><empty_output><jupyter_text>Since the goal of this tutorial is to generate text, it will also be important to invert this representation and recover human-readable strings from it. For this you can use `preprocessing.StringLookup(..., invert=True)`. Note: Here instead of passing the original vocabulary generated with `sorted(set(text))` use the `get_vocabulary()` method of the `preprocessing.StringLookup` layer so that the padding and `[UNK]` tokens are set the same way.<jupyter_code>chars_from_ids = tf.keras.layers.experimental.preprocessing.StringLookup(
vocabulary=ids_from_chars.get_vocabulary(), invert=True)<jupyter_output><empty_output><jupyter_text>This layer recovers the characters from the vectors of IDs, and returns them as a `tf.RaggedTensor` of characters:<jupyter_code>chars = chars_from_ids(ids)
chars<jupyter_output><empty_output><jupyter_text>You can `tf.strings.reduce_join` to join the characters back into strings. <jupyter_code>tf.strings.reduce_join(chars, axis=-1).numpy()
def text_from_ids(ids):
return tf.strings.reduce_join(chars_from_ids(ids), axis=-1)<jupyter_output><empty_output><jupyter_text>### The prediction taskGiven a character, or a sequence of characters, what is the most probable next character? This is the task you're training the model to perform. The input to the model will be a sequence of characters, and you train the model to predict the output—the following character at each time step.
Since RNNs maintain an internal state that depends on the previously seen elements, given all the characters computed until this moment, what is the next character?
### Create training examples and targets
Next divide the text into example sequences. Each input sequence will contain `seq_length` characters from the text.
For each input sequence, the corresponding targets contain the same length of text, except shifted one character to the right.
So break the text into chunks of `seq_length+1`. For example, say `seq_length` is 4 and our text is "Hello". The input sequence would be "Hell", and the target sequence "ello".
To do this first use the `tf.data.Dataset.from_tensor_slices` function to convert the text vector into a stream of character indices.<jupyter_code>all_ids = ids_from_chars(tf.strings.unicode_split(text, 'UTF-8'))
all_ids
ids_dataset = tf.data.Dataset.from_tensor_slices(all_ids)
for ids in ids_dataset.take(10):
print(chars_from_ids(ids).numpy().decode('utf-8'))
seq_length = 100
examples_per_epoch = len(text)//(seq_length+1)<jupyter_output><empty_output><jupyter_text>The `batch` method lets you easily convert these individual characters to sequences of the desired size.<jupyter_code>sequences = ids_dataset.batch(seq_length+1, drop_remainder=True)
for seq in sequences.take(1):
print(chars_from_ids(seq))<jupyter_output>tf.Tensor(
[b'C' b'h' b'a' b'p' b't' b'e' b'r' b' ' b'1' b'\r' b'\n' b'\r' b'\n'
b'\r' b'\n' b'I' b't' b' ' b'i' b's' b' ' b'a' b' ' b't' b'r' b'u' b't'
b'h' b' ' b'u' b'n' b'i' b'v' b'e' b'r' b's' b'a' b'l' b'l' b'y' b' '
b'a' b'c' b'k' b'n' b'o' b'w' b'l' b'e' b'd' b'g' b'e' b'd' b',' b' '
b't' b'h' b'a' b't' b' ' b'a' b' ' b's' b'i' b'n' b'g' b'l' b'e' b' '
b'm' b'a' b'n' b' ' b'i' b'n' b' ' b'p' b'o' b's' b's' b'e' b's' b's'
b'i' b'o' b'n' b'\r' b'\n' b'o' b'f' b' ' b'a' b' ' b'g' b'o' b'o' b'd'
b' ' b'f' b'o' b'r'], shape=(101,), dtype=string)
<jupyter_text>It's easier to see what this is doing if you join the tokens back into strings:<jupyter_code>for seq in sequences.take(5):
print(text_from_ids(seq).numpy())<jupyter_output>b'Chapter 1\r\n\r\n\r\nIt is a truth universally acknowledged, that a single man in possession\r\nof a good for'
b'tune, must be in want of a wife.\r\n\r\nHowever little known the feelings or views of such a man may be o'
b'n his\r\nfirst entering a neighbourhood, this truth is so well fixed in the minds\r\nof the surrounding f'
b'amilies, that he is considered the rightful property\r\nof some one or other of their daughters.\r\n\r\n"My'
b' dear Mr. Bennet," said his lady to him one day, "have you heard that\r\nNetherfield Park is let at las'
<jupyter_text>For training you'll need a dataset of `(input, label)` pairs. Where `input` and
`label` are sequences. At each time step the input is the current character and the label is the next character.
Here's a function that takes a sequence as input, duplicates, and shifts it to align the input and label for each timestep:<jupyter_code>def split_input_target(sequence):
input_text = sequence[:-1]
target_text = sequence[1:]
return input_text, target_text
split_input_target(list("Tensorflow"))
dataset = sequences.map(split_input_target)
for input_example, target_example in dataset.take(1):
print("Input :", text_from_ids(input_example).numpy())
print("Target:", text_from_ids(target_example).numpy())<jupyter_output>Input : b'Chapter 1\r\n\r\n\r\nIt is a truth universally acknowledged, that a single man in possession\r\nof a good fo'
Target: b'hapter 1\r\n\r\n\r\nIt is a truth universally acknowledged, that a single man in possession\r\nof a good for'
<jupyter_text>### Create training batches
You used `tf.data` to split the text into manageable sequences. But before feeding this data into the model, you need to shuffle the data and pack it into batches.<jupyter_code># Batch size
BATCH_SIZE = 64
# Buffer size to shuffle the dataset
# (TF data is designed to work with possibly infinite sequences,
# so it doesn't attempt to shuffle the entire sequence in memory. Instead,
# it maintains a buffer in which it shuffles elements).
BUFFER_SIZE = 10000
dataset = (
dataset
.shuffle(BUFFER_SIZE)
.batch(BATCH_SIZE, drop_remainder=True)
.prefetch(tf.data.experimental.AUTOTUNE))
dataset<jupyter_output><empty_output><jupyter_text>## Build The ModelThis section defines the model as a `keras.Model` subclass (For details see [Making new Layers and Models via subclassing](https://www.tensorflow.org/guide/keras/custom_layers_and_models)).
This model has three layers:
* `tf.keras.layers.Embedding`: The input layer. A trainable lookup table that will map each character-ID to a vector with `embedding_dim` dimensions;
* `tf.keras.layers.GRU`: A type of RNN with size `units=rnn_units` (You can also use an LSTM layer here.)
* `tf.keras.layers.Dense`: The output layer, with `vocab_size` outputs. It outputs one logit for each character in the vocabulary. These are the log-likelihood of each character according to the model.<jupyter_code># Length of the vocabulary in chars
vocab_size = len(vocab)
# The embedding dimension
embedding_dim = 256
# Number of RNN units
rnn_units = 1024
class MyModel(tf.keras.Model):
def __init__(self, vocab_size, embedding_dim, rnn_units):
super().__init__(self)
self.embedding = tf.keras.layers.Embedding(vocab_size, embedding_dim)
self.gru = tf.keras.layers.GRU(rnn_units,
return_sequences=True,
return_state=True)
self.dense = tf.keras.layers.Dense(vocab_size)
def call(self, inputs, states=None, return_state=False, training=False):
x = inputs
x = self.embedding(x, training=training)
if states is None:
states = self.gru.get_initial_state(x)
x, states = self.gru(x, initial_state=states, training=training)
x = self.dense(x, training=training)
if return_state:
return x, states
else:
return x
model = MyModel(
# Be sure the vocabulary size matches the `StringLookup` layers.
vocab_size=len(ids_from_chars.get_vocabulary()),
embedding_dim=embedding_dim,
rnn_units=rnn_units)<jupyter_output><empty_output><jupyter_text>For each character the model looks up the embedding, runs the GRU one timestep with the embedding as input, and applies the dense layer to generate logits predicting the log-likelihood of the next character:
Note: For training you could use a `keras.Sequential` model here. To generate text later you'll need to manage the RNN's internal state. It's simpler to include the state input and output options upfront, than it is to rearrange the model architecture later. For more details see the [Keras RNN guide](https://www.tensorflow.org/guide/keras/rnn#rnn_state_reuse).## Try the model
Now run the model to see that it behaves as expected.
First check the shape of the output:<jupyter_code>for input_example_batch, target_example_batch in dataset.take(1):
example_batch_predictions = model(input_example_batch)
print(example_batch_predictions.shape, "# (batch_size, sequence_length, vocab_size)")<jupyter_output>(64, 100, 78) # (batch_size, sequence_length, vocab_size)
<jupyter_text>In the above example the sequence length of the input is `100` but the model can be run on inputs of any length:<jupyter_code>model.summary()<jupyter_output>Model: "my_model"
_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
embedding (Embedding) multiple 19968
_________________________________________________________________
gru (GRU) multiple 3938304
_________________________________________________________________
dense (Dense) multiple 79950
=================================================================
Total params: 4,038,222
Trainable params: 4,038,222
Non-trainable params: 0
_________________________________________________________________
<jupyter_text>To get actual predictions from the model you need to sample from the output distribution, to get actual character indices. This distribution is defined by the logits over the character vocabulary.
Note: It is important to _sample_ from this distribution as taking the _argmax_ of the distribution can easily get the model stuck in a loop.
Try it for the first example in the batch:<jupyter_code>sampled_indices = tf.random.categorical(example_batch_predictions[0], num_samples=1)
sampled_indices = tf.squeeze(sampled_indices, axis=-1).numpy()<jupyter_output><empty_output><jupyter_text>This gives us, at each timestep, a prediction of the next character index:<jupyter_code>sampled_indices<jupyter_output><empty_output><jupyter_text>Decode these to see the text predicted by this untrained model:<jupyter_code>print("Input:\n", text_from_ids(input_example_batch[0]).numpy())
print()
print("Next Char Predictions:\n", text_from_ids(sampled_indices).numpy())<jupyter_output>Input:
b' relations, with only\r\none serious, parting look, went away.\r\n\r\nAs he quitted the room, Elizabeth fe'
Next Char Predictions:
b'68_Ek[UNK]P1f\nH3,UWcgG?z9r_a9\'OpnsLx\nr A!*TUE5l;.44\rKpByK""NgRgi"xMp[UNK]_Ka*S(:A-wR\'-BJ09q_Hewh,e42L3qPT"[UNK]V'
<jupyter_text>## Train the modelAt this point the problem can be treated as a standard classification problem. Given the previous RNN state, and the input this time step, predict the class of the next character.### Attach an optimizer, and a loss functionThe standard `tf.keras.losses.sparse_categorical_crossentropy` loss function works in this case because it is applied across the last dimension of the predictions.
Because your model returns logits, you need to set the `from_logits` flag.
<jupyter_code>loss = tf.losses.SparseCategoricalCrossentropy(from_logits=True)
example_batch_loss = loss(target_example_batch, example_batch_predictions)
mean_loss = example_batch_loss.numpy().mean()
print("Prediction shape: ", example_batch_predictions.shape, " # (batch_size, sequence_length, vocab_size)")
print("Mean loss: ", mean_loss)<jupyter_output>Prediction shape: (64, 100, 78) # (batch_size, sequence_length, vocab_size)
Mean loss: 4.356842
<jupyter_text>A newly initialized model shouldn't be too sure of itself, the output logits should all have similar magnitudes. To confirm this you can check that the exponential of the mean loss is approximately equal to the vocabulary size. A much higher loss means the model is sure of its wrong answers, and is badly initialized:<jupyter_code>tf.exp(mean_loss).numpy()<jupyter_output><empty_output><jupyter_text>Configure the training procedure using the `tf.keras.Model.compile` method. Use `tf.keras.optimizers.Adam` with default arguments and the loss function.<jupyter_code>model.compile(optimizer='adam', loss=loss)<jupyter_output><empty_output><jupyter_text>### Configure checkpointsUse a `tf.keras.callbacks.ModelCheckpoint` to ensure that checkpoints are saved during training:<jupyter_code># Directory where the checkpoints will be saved
checkpoint_dir = './training_checkpoints'
# Name of the checkpoint files
checkpoint_prefix = os.path.join(checkpoint_dir, "ckpt_{epoch}")
checkpoint_callback = tf.keras.callbacks.ModelCheckpoint(
filepath=checkpoint_prefix,
save_weights_only=True)<jupyter_output><empty_output><jupyter_text>### Execute the trainingTo keep training time reasonable, use 10 epochs to train the model. In Colab, set the runtime to GPU for faster training.<jupyter_code>EPOCHS = 20
history = model.fit(dataset, epochs=EPOCHS, callbacks=[checkpoint_callback])<jupyter_output>Epoch 1/20
107/107 [==============================] - 8s 57ms/step - loss: 3.4253
Epoch 2/20
107/107 [==============================] - 7s 58ms/step - loss: 2.1914
Epoch 3/20
107/107 [==============================] - 7s 59ms/step - loss: 1.8482
Epoch 4/20
107/107 [==============================] - 7s 59ms/step - loss: 1.5726
Epoch 5/20
107/107 [==============================] - 7s 58ms/step - loss: 1.3956
Epoch 6/20
107/107 [==============================] - 7s 57ms/step - loss: 1.2785
Epoch 7/20
107/107 [==============================] - 7s 57ms/step - loss: 1.2030
Epoch 8/20
107/107 [==============================] - 7s 57ms/step - loss: 1.1471
Epoch 9/20
107/107 [==============================] - 7s 56ms/step - loss: 1.1010
Epoch 10/20
107/107 [==============================] - 7s 56ms/step - loss: 1.0546
Epoch 11/20
107/107 [==============================] - 7s 56ms/step - loss: 1.0147
Epoch 12/20
107/107 [==============================] - 7s 56ms/step - loss: 0.9756
Epoch 13/20
1[...]<jupyter_text>## Generate textThe simplest way to generate text with this model is to run it in a loop, and keep track of the model's internal state as you execute it.

Each time you call the model you pass in some text and an internal state. The model returns a prediction for the next character and its new state. Pass the prediction and state back in to continue generating text.
The following makes a single step prediction:<jupyter_code>class OneStep(tf.keras.Model):
def __init__(self, model, chars_from_ids, ids_from_chars, temperature=1.0):
super().__init__()
self.temperature = temperature
self.model = model
self.chars_from_ids = chars_from_ids
self.ids_from_chars = ids_from_chars
# Create a mask to prevent "" or "[UNK]" from being generated.
skip_ids = self.ids_from_chars(['', '[UNK]'])[:, None]
sparse_mask = tf.SparseTensor(
# Put a -inf at each bad index.
values=[-float('inf')]*len(skip_ids),
indices=skip_ids,
# Match the shape to the vocabulary
dense_shape=[len(ids_from_chars.get_vocabulary())])
self.prediction_mask = tf.sparse.to_dense(sparse_mask)
@tf.function
def generate_one_step(self, inputs, states=None):
# Convert strings to token IDs.
input_chars = tf.strings.unicode_split(inputs, 'UTF-8')
input_ids = self.ids_from_chars(input_chars).to_tensor()
# Run the model.
# predicted_logits.shape is [batch, char, next_char_logits]
predicted_logits, states = self.model(inputs=input_ids, states=states,
return_state=True)
# Only use the last prediction.
predicted_logits = predicted_logits[:, -1, :]
predicted_logits = predicted_logits/self.temperature
# Apply the prediction mask: prevent "" or "[UNK]" from being generated.
predicted_logits = predicted_logits + self.prediction_mask
# Sample the output logits to generate token IDs.
predicted_ids = tf.random.categorical(predicted_logits, num_samples=1)
predicted_ids = tf.squeeze(predicted_ids, axis=-1)
# Convert from token ids to characters
predicted_chars = self.chars_from_ids(predicted_ids)
# Return the characters and model state.
return predicted_chars, states
one_step_model = OneStep(model, chars_from_ids, ids_from_chars)<jupyter_output><empty_output><jupyter_text>Run it in a loop to generate some text. Looking at the generated text, you'll see the model knows when to capitalize, make paragraphs and imitates a Shakespeare-like writing vocabulary. With the small number of training epochs, it has not yet learned to form coherent sentences.<jupyter_code>start = time.time()
states = None
next_char = tf.constant(['Generated Text:'])
result = [next_char]
for n in range(1000):
next_char, states = one_step_model.generate_one_step(next_char, states=states)
result.append(next_char)
result = tf.strings.join(result)
end = time.time()
print(result[0].numpy().decode('utf-8'), '\n\n' + '_'*80)
print('\nRun time:', end - start)<jupyter_output>Generated Text:
Elizabeth saw that of _this_ we are all somewiment over my two most guarded and sigh
of seeing Jane, she added:
"Lizzy, I have," said Jane, "that mine would not ake, could bear such a favourite; one, observing that let de Bourgh, and taking the obligation must
belonging to orrout me dow."
"Oh! by in that variety of this house
un. Neversonly see him in his fear. Shanly a gave way, and gave them together, which
both she added in distressed
constant communication. It ore to the same time. It is every mind,
"It might attention to visit his wife a gentleman did not advantage. But poor Meryton? Oh!
why, in silence with Elizabeth
grieved for his patroness.
They better not very fan by themselves; and I did not listen, they friends and breakfast, took
the necessary terms over. Elizabeth then reached to confirm her. What are her fiture beyond to part over him again.
Half when she left Netherfield believing it. Well, if I have had to see you
at Pemberley, eye,[...]<jupyter_text>The easiest thing you can do to improve the results is to train it for longer (try `EPOCHS = 30`).
You can also experiment with a different start string, try adding another RNN layer to improve the model's accuracy, or adjust the temperature parameter to generate more or less random predictions.If you want the model to generate text *faster* the easiest thing you can do is batch the text generation. In the example below the model generates 5 outputs in about the same time it took to generate 1 above. <jupyter_code>start = time.time()
states = None
next_char = tf.constant(['Generated Text:', 'Generated Text:', 'Generated Text:', 'Generated Text:', 'Generated Text:'])
result = [next_char]
for n in range(1000):
next_char, states = one_step_model.generate_one_step(next_char, states=states)
result.append(next_char)
result = tf.strings.join(result)
end = time.time()
print(result, '\n\n' + '_'*80)
print('\nRun time:', end - start)<jupyter_output>tf.Tensor(
[b'Generated Text:--\r\n\r\n"But your father has no best cordial feelings, and only the first sentence alarming on the perpossible match, or occasion, was\r\nnot seen a very propition which their first object was never so prevailed on\r\none. An end or carriage? As a chill protee that were larger to be the carriage?"\r\n\r\nShe then exclaimed, "but I am very serious like of her curiosity; he was allowed at fir the\r\nentrance of Bingley, and she turned her interest; for who, in a moment which she felt a smile at suspense,\r\nand Mr. Denny and his sonsisting more surprised before the dance with Mr. Wickham, and\r\nintroduction at long, he was lagged before the intention maintains, expressively the\r\nshopled better offered her eetrest disposition.\r\n\r\nIt cannot understand it, I could hardly hulf home. Mrs. Gardiner regained her gown. She lamed at\r\nbr the exercise of tea-ten thousand pounds, you was it difficult on terms.\r\nThis was now patience to accept the dance, and [...]<jupyter_text>## Export the generator
This single-step model can easily be [saved and restored](https://www.tensorflow.org/guide/saved_model), allowing you to use it anywhere a `tf.saved_model` is accepted.<jupyter_code>tf.saved_model.save(one_step_model, 'one_step')
one_step_reloaded = tf.saved_model.load('one_step')
states = None
next_char = tf.constant(['Generated Text:'])
result = [next_char]
for n in range(100):
next_char, states = one_step_reloaded.generate_one_step(next_char, states=states)
result.append(next_char)
print(tf.strings.join(result)[0].numpy().decode("utf-8"))<jupyter_output>WARNING:tensorflow:5 out of the last 5 calls to <function recreate_function.<locals>.restored_function_body at 0x7fee726f6dd0> triggered tf.function retracing. Tracing is expensive and the excessive number of tracings could be due to (1) creating @tf.function repeatedly in a loop, (2) passing tensors with different shapes, (3) passing Python objects instead of tensors. For (1), please define your @tf.function outside of the loop. For (2), @tf.function has experimental_relax_shapes=True option that relaxes argument shapes that can avoid unnecessary retracing. For (3), please refer to https://www.tensorflow.org/guide/function#controlling_retracing and https://www.tensorflow.org/api_docs/python/tf/function for more details.
<jupyter_text>## Advanced: Customized Training
The above training procedure is simple, but does not give you much control.
It uses teacher-forcing which prevents bad predictions from being fed back to the model, so the model never learns to recover from mistakes.
So now that you've seen how to run the model manually next you'll implement the training loop. This gives a starting point if, for example, you want to implement _curriculum learning_ to help stabilize the model's open-loop output.
The most important part of a custom training loop is the train step function.
Use `tf.GradientTape` to track the gradients. You can learn more about this approach by reading the [eager execution guide](https://www.tensorflow.org/guide/eager).
The basic procedure is:
1. Execute the model and calculate the loss under a `tf.GradientTape`.
2. Calculate the updates and apply them to the model using the optimizer.<jupyter_code>class CustomTraining(MyModel):
@tf.function
def train_step(self, inputs):
inputs, labels = inputs
with tf.GradientTape() as tape:
predictions = self(inputs, training=True)
loss = self.loss(labels, predictions)
grads = tape.gradient(loss, model.trainable_variables)
self.optimizer.apply_gradients(zip(grads, model.trainable_variables))
return {'loss': loss}<jupyter_output><empty_output><jupyter_text>The above implementation of the `train_step` method follows [Keras' `train_step` conventions](https://www.tensorflow.org/guide/keras/customizing_what_happens_in_fit). This is optional, but it allows you to change the behavior of the train step and still use keras' `Model.compile` and `Model.fit` methods.<jupyter_code>model = CustomTraining(
vocab_size=len(ids_from_chars.get_vocabulary()),
embedding_dim=embedding_dim,
rnn_units=rnn_units)
model.compile(optimizer = tf.keras.optimizers.Adam(),
loss=tf.keras.losses.SparseCategoricalCrossentropy(from_logits=True))
model.fit(dataset, epochs=1)<jupyter_output>107/107 [==============================] - 13s 106ms/step - loss: 2.8803
<jupyter_text>Or if you need more control, you can write your own complete custom training loop:<jupyter_code>EPOCHS = 10
mean = tf.metrics.Mean()
for epoch in range(EPOCHS):
start = time.time()
mean.reset_states()
for (batch_n, (inp, target)) in enumerate(dataset):
logs = model.train_step([inp, target])
mean.update_state(logs['loss'])
if batch_n % 50 == 0:
template = f"Epoch {epoch+1} Batch {batch_n} Loss {logs['loss']:.4f}"
print(template)
# saving (checkpoint) the model every 5 epochs
if (epoch + 1) % 5 == 0:
model.save_weights(checkpoint_prefix.format(epoch=epoch))
print()
print(f'Epoch {epoch+1} Loss: {mean.result().numpy():.4f}')
print(f'Time taken for 1 epoch {time.time() - start:.2f} sec')
print("_"*80)
model.save_weights(checkpoint_prefix.format(epoch=epoch))<jupyter_output>Epoch 1 Batch 0 Loss 2.2837
Epoch 1 Batch 50 Loss 2.1464
Epoch 1 Batch 100 Loss 1.9971
Epoch 1 Loss: 2.1193
Time taken for 1 epoch 12.23 sec
________________________________________________________________________________
Epoch 2 Batch 0 Loss 1.9429
Epoch 2 Batch 50 Loss 1.8356
Epoch 2 Batch 100 Loss 1.6687
Epoch 2 Loss: 1.7960
Time taken for 1 epoch 11.91 sec
________________________________________________________________________________
Epoch 3 Batch 0 Loss 1.6579
Epoch 3 Batch 50 Loss 1.5514
Epoch 3 Batch 100 Loss 1.4544
Epoch 3 Loss: 1.5508
Time taken for 1 epoch 11.87 sec
________________________________________________________________________________
Epoch 4 Batch 0 Loss 1.4252
Epoch 4 Batch 50 Loss 1.3652
Epoch 4 Batch 100 Loss 1.2843
Epoch 4 Loss: 1.3827
Time taken for 1 epoch 11.81 sec
________________________________________________________________________________
Epoch 5 Batch 0 Loss 1.2892
Epoch 5 Batch 50 Loss 1.2735
Epoch 5 Batch 100 Loss 1.2576
Epoch 5 Loss: 1.2753[...]
|
no_license
|
/text_generation.ipynb
|
artemcarabash/machine_learning
| 32 |
<jupyter_start><jupyter_text>
#**Artificial Intelligence - MSc**
##ET5003 - MACHINE LEARNING APPLICATIONS
###Instructor: Enrique Naredo
###ET5003_KaggleCompetition<jupyter_code>#@title Current Date
Today = '2021-10-15' #@param {type:"date"}
#@markdown ---
#@markdown ### Enter your details here:
Team_Number = "5" #@param {type:"string"}
Student_ID&Name = " 20157347 Robert Barrett" #@param {type:"string"}
Student_ID&Name = " 20157347 Robert Barrett" #@param {type:"string"}
Student_ID&Name = " " #@param {type:"string"}
Student_ID&Name = " " #@param {type:"string"}
Student_ID&Name = " " #@param {type:"string"}
#@markdown ---
#@title Notebook information
Notebook_type = 'Etivity' #@param ["Example", "Lab", "Practice", "Etivity", "Assignment", "Exam"]
Version = Final #@param ["Draft", "Final"] {type:"raw"}
Submission = False #@param {type:"boolean"}<jupyter_output><empty_output><jupyter_text># INTRODUCTIONYour introduction here.***The goal is to use advanced Machine Learning methods to predict House price.***## Imports<jupyter_code># Suppressing Warnings:
import warnings
warnings.filterwarnings("ignore")
# standard libraries
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
# to plot
import matplotlib.colors
from mpl_toolkits.mplot3d import Axes3D
# to generate classification, regression and clustering datasets
import sklearn.datasets as dt
# to create data frames
from pandas import DataFrame
# to generate data from an existing dataset
from sklearn.neighbors import KernelDensity
from sklearn.model_selection import GridSearchCV
# Scikit-learn is an open source machine learning library
# that supports supervised and unsupervised learning
# https://scikit-learn.org/stable/
from sklearn.model_selection import train_test_split
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.naive_bayes import MultinomialNB
from sklearn.metrics import accuracy_score, confusion_matrix
# Regular expression operations
#https://docs.python.org/3/library/re.html
import re
# Natural Language Toolkit
# https://www.nltk.org/install.html
import nltk
# Stemming maps different forms of the same word to a common “stem”
# https://pypi.org/project/snowballstemmer/
from nltk.stem import SnowballStemmer
# https://www.nltk.org/book/ch02.html
from nltk.corpus import stopwords
# https://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.StandardScaler.html
from sklearn.preprocessing import StandardScaler
!pip install gpy
import GPy as GPy
import numpy as np
import pylab as pb
import pymc3 as pm
import arviz as az
# Define the seed so that results can be reproduced
seed = 11
rand_state = 11
# Define the color maps for plots
color_map = plt.cm.get_cmap('RdYlBu')
color_map_discrete = matplotlib.colors.LinearSegmentedColormap.from_list("", ["red","cyan","magenta","blue"])<jupyter_output><empty_output><jupyter_text># DATASETExtract from this [paper](https://ieeexplore.ieee.org/document/9300074):
* House prices are a significant impression of the economy, and its value ranges are of great concerns for the clients and property dealers.
* Housing price escalate every year that eventually reinforced the need of strategy or technique that could predict house prices in future.
* There are certain factors that influence house prices including physical conditions, locations, number of bedrooms and others.
1. [Download the dataset](https://github.com/UL-CS6134/CS6134_SEM1_2021-2/tree/main/Week-5).
2. Upload the dataset into your folder.
<jupyter_code>from google.colab import drive
drive.mount('/content/drive')
path = '/content/sample_data/'
# Due to formatting issues, I found that explicitly defining features of interest and omitting ones that are problematic was useful
train_file = path+'HousePrice_Train.csv'
test_file = path+'HousePrice_Test.csv'
# Import the data, while excluding 'features' and 'description_block' explicitly on import
train_data = pd.read_csv(train_file)
test_data = pd.read_csv(test_file)
print(train_data.shape)
print(test_data.shape)<jupyter_output>(1638, 14)
(702, 13)
<jupyter_text>The challenge is to predict the final price of each house.## Exploratory Data Analysis#### Let's look at samples of the tabular data and review some basic statistics<jupyter_code># Review samples of training data
train_data.head()
# Review samples of test data
test_data.head()
# Generate statistcis for numeric training data
train_data.describe()<jupyter_output><empty_output><jupyter_text>#### Initial Analysis of Training Data Statistics
The above table provides a some key information:
1. **counts** indicate no missing values for these numeric features
2. **mean** and **50% Quartile** are the same or close except for "Surface" and "Price" showing an imbalance in the data .. we may need to deal with this.
3. **min** and **max** indicate ranges that need to be scaled<jupyter_code># Check for missing values in Training data
train_missing = train_data.isnull().sum()*100/len(train_data)
print(train_missing)<jupyter_output>Index 0.000000
ID 0.000000
Location 0.000000
Num_Bathrooms 0.000000
Num_Beds 0.000000
BER_class 15.323565
Description 0.000000
Services 64.774115
Features 0.000000
Latitude 0.000000
Longitude 0.000000
Type 0.000000
Surface 0.000000
Price 0.000000
dtype: float64
<jupyter_text>**Comments** There are two features have missing values. These are categorical features as they did not appear in above statistics report. High % missing features cannot be remediated and should be removed. We should look at 'BER_class' and see if it is an important feature. If it is, then we should look to see if there is a way to handle the missing values.
Approach here will be to
1. Remove 'ID' as it provides no information
2. Remove the feature 'Services'
3. Determine importance of 'BER_class' as a predictor of price. We will use XGBoost and its correspondng "feature_importance" metric.
Reviewing the features for non-uniqueness, there are no features with just one value.<jupyter_code># Let's look at obvious erroneous values - no zero values in the training data
train_data_zeros = (train_data == 0).all()
print(train_data_zeros)<jupyter_output>Index False
ID False
Location False
Num_Bathrooms False
Num_Beds False
BER_class False
Description False
Services False
Features False
Latitude False
Longitude False
Type False
Surface False
Price False
dtype: bool
<jupyter_text>There are no features with zeros so we do not have to analyse this further.<jupyter_code># Let's look for obvious coordinates outside Dublin range
outliers = train_data[(train_data.Longitude > -6) | (train_data.Latitude < 53)]
outliers.head()<jupyter_output><empty_output><jupyter_text>This location has coordinates for a UK address and should be removed.<jupyter_code># Remove location in UK
df_train = train_data.drop(train_data[(train_data.Longitude > -6) | (train_data.Latitude < 53)].index)
# Drop unimportant features
df_train.drop(['ID', 'Services'], axis=1, inplace=True)
# Let's deal with categorical data
# Feature map 'locations'
locations = np.hstack((train_data['Location'], test_data['Location']))
unique_loc = np.unique(locations)
unique_loc_index=[]
# Create a dictionary lookup
i = [i+1 for i in range(len(unique_loc))]
unique_areas_index = dict(zip(unique_loc, i))
# Replace area name with the index from the 'unique_areas_index' dictionary
df_train_1 = df_train.replace({'Location': unique_areas_index})
# Feature map 'BER_class'
ber_stack = np.hstack((train_data['BER_class'], test_data['BER_class'])) # get all BER Classes from Train and Test datasets to ensure we have all classes in each set.
ber_class = [c for c in ber_stack if not(pd.isna(c)) == True] # remove NaN from feature map list
unique_ber = np.unique(ber_class)
unique_ber_index=[]
# Create a dictionary lookup
i = [i+1 for i in range(len(unique_ber))]
unique_ber_index = dict(zip(unique_ber, i))
# Replace BER_class with the index from the 'unique_ber_index' dictionary
df_train_2 = df_train_1.replace({'BER_class': unique_ber_index})
# One-hot Encode 'Type'
df_train_2 = pd.get_dummies(df_train_2, columns=['Type'])
df_train = df_train_2.copy()
# Take a copy of the Location and BER_class column before scaling - this will be used during imputing for NaNs later
loc_ber_train_values = df_train[['Location', 'BER_class']].copy()
<jupyter_output><empty_output><jupyter_text>We will now look at the feature distributions and outliers.<jupyter_code>df_train.columns<jupyter_output><empty_output><jupyter_text>We will look at these key features:
'Location', 'Num_Bathrooms', 'Num_Beds', 'BER_class', 'Latitude', 'Longitude', 'Surface', 'Price'<jupyter_code># First we will look at the distribution of key predictor features
fig = plt.figure(figsize=(20,30))
ax1 = fig.add_subplot(8, 2, 1)
ax1.set_title("Plot of 'Location'")
df_train.boxplot(column=['Location'])
ax2 = fig.add_subplot(8, 2, 2)
ax2.set_title("Histogram of 'Location'")
df_train['Location'].hist(bins=20)
ax3 = fig.add_subplot(8, 2, 3)
ax3.set_title("Plot of 'Num_Bathrooms'")
df_train.boxplot(column=['Num_Bathrooms'])
ax4 = fig.add_subplot(8, 2, 4)
ax4.set_title("Histogram of 'Num_Bathrooms'")
df_train['Num_Bathrooms'].hist(bins=20)
ax5 = fig.add_subplot(8, 2, 5)
ax5.set_title("Plot of 'Num_Beds'")
df_train.boxplot(column=['Num_Beds'])
ax6 = fig.add_subplot(8, 2, 6)
ax6.set_title("Histogram of 'Num_Beds'")
df_train['Num_Beds'].hist(bins=20)
ax7 = fig.add_subplot(8, 2, 7)
ax7.set_title("Plot of 'BER_class'")
df_train.boxplot(column=['BER_class'])
ax8 = fig.add_subplot(8, 2, 8)
ax8.set_title("Histogram of 'BER_class'")
df_train['BER_class'].hist(bins=20)
ax9 = fig.add_subplot(8, 2, 9)
ax9.set_title("Plot of 'Longitude'")
df_train.boxplot(column=['Longitude'])
ax10 = fig.add_subplot(8, 2, 10)
ax10.set_title("Histogram of 'Longitude'")
df_train['Longitude'].hist(bins=20)
ax11 = fig.add_subplot(8, 2, 11)
ax11.set_title("Plot of 'Latitude'")
df_train.boxplot(column=['Latitude'])
ax12 = fig.add_subplot(8, 2, 12)
ax12.set_title("Histogram of 'Latitude'")
df_train['Latitude'].hist(bins=20)
ax13 = fig.add_subplot(8, 2, 13)
ax13.set_title("Plot of 'Surface'")
df_train.boxplot(column=['Surface'])
ax14 = fig.add_subplot(8, 2, 14)
ax14.set_title("Histogram of 'Surface'")
df_train['Surface'].hist(bins=20)
ax15 = fig.add_subplot(8, 2, 15)
ax15.set_title("Plot of 'Price'")
df_train.boxplot(column=['Price'])
ax16 = fig.add_subplot(8, 2, 16)
ax16.set_title("Histogram of 'Price'")
df_train['Price'].hist(bins=20)<jupyter_output><empty_output><jupyter_text>There is an outlier on 'Surface' that needs to be investigated<jupyter_code>df_train[(df_train.Surface > 2000)]<jupyter_output><empty_output><jupyter_text>There is only two values in the range greater than 2,000. Decision to remove it seems valid.<jupyter_code>df_train = df_train.drop(df_train[df_train.Surface > 2000].index)
# Apply scalers to data to resolve distribution and outlier issues
from sklearn.preprocessing import StandardScaler
from sklearn.preprocessing import MinMaxScaler
minmax = MinMaxScaler()
standardscaler = StandardScaler()
# Not a normal distribution so just scale with MinMax [0, 1]
df_train[['Location']] = minmax.fit_transform(df_train[['Location']])
# Deal with outliers with a Log Transform - later we will use Standard Scaler as well on some feature
df_train['Num_Bathrooms'] = np.log(df_train['Num_Bathrooms'])
df_train['Num_Beds'] = np.log(df_train['Num_Beds'])
df_train['Latitude'] = np.log(df_train['Latitude'])
df_train['Surface'] = np.log(df_train['Surface'])
df_train['Price'] = np.log(df_train['Price'])
# Standard scaler for normal distributions
df_train[['BER_class', 'Longitude', 'Latitude', 'Surface', 'Price']] = standardscaler.fit_transform(df_train[['BER_class', 'Longitude', 'Latitude', 'Surface', 'Price']])
<jupyter_output><empty_output><jupyter_text>Do the same scaling to the Test Data<jupyter_code># Check for missing values in Training data - calculate % misisng values
test_missing = test_data.isnull().sum()*100/len(test_data)
print(test_missing)
# Drop unimportant features
test_data.drop(['ID', 'Services'], axis=1, inplace=True)
df_test = test_data.copy()
# Let's deal with categorical data for Test Dataset
# Replace area name with the index from the 'unique_areas_index' dictionary
df_test_1 = df_test.replace({'Location': unique_areas_index})
# Replace BER_class with the index from the 'unique_ber_index' dictionary
df_test_2 = df_test_1.replace({'BER_class': unique_ber_index})
# One-hot Encode 'Type'
df_test_2 = pd.get_dummies(df_test_2, columns=['Type'])
df_test = df_test_2.copy()
# Take a copy of the Location and BER_class column before scaling - this will be used during imputing for NaNs later
loc_ber_test_values = df_test[['Location', 'BER_class']].copy()
# Apply same scalers as for Training dataset
df_test[['Location']] = minmax.fit_transform(df_test[['Location']])
df_test['Num_Bathrooms'] = np.log(df_test['Num_Bathrooms'])
df_test['Num_Beds'] = np.log(df_test['Num_Beds'])
df_test['Latitude'] = np.log(df_test['Latitude'])
df_test['Surface'] = np.log(df_test['Surface'])
# Standard scaler for normal distributions
df_test[['BER_class', 'Longitude', 'Latitude', 'Surface']] = standardscaler.fit_transform(df_test[['BER_class', 'Longitude', 'Latitude', 'Surface']])
<jupyter_output><empty_output><jupyter_text>Now let's look at 'BER_class' and see how important it is for predicting price.
The following approach uses XGBoost Regression algorithm which has some powerful built in functions for Feature Importance.<jupyter_code># Let's take a closer look at 'BER_class' using feature importance on all data set
# We will look at the feature Importance of 'BER_class' with empty features removed from the dataset
# and left in to see check the impact also for imputation.
import xgboost as xgb
from sklearn.model_selection import train_test_split
from sklearn.metrics import mean_squared_error, r2_score
def xgb_model(X, y, msg):
xgb_model = xgb.XGBRegressor(max_depth=7, learning_rate = 0.6, n_jobs=4, objective = 'reg:squarederror', random_state=10, n_estimators=50).fit(X, y)
y_xgb_pred = xgb_model.predict(X)
RMSE_xgb = mean_squared_error(y, y_xgb_pred, squared=False)
r2_xgb = r2_score(y, y_xgb_pred)
print(msg)
print("RMSE_score: %.4f R2_score: %.4f" %(RMSE_xgb, r2_xgb))
return xgb_model, RMSE_xgb, r2_xgb
# Let's look at the Feature Importance with all BER_class values equal to NULL removed
# We will also remove large text features for now.
features = ['Location', 'Num_Bathrooms', 'Num_Beds', 'BER_class',
'Latitude', 'Longitude', 'Surface', 'Type_apartment',
'Type_bungalow', 'Type_detached', 'Type_duplex',
'Type_end-of-terrace', 'Type_semi-detached', 'Type_site', 'Type_studio',
'Type_terraced', 'Type_townhouse', 'Price']
df_train_xgb = df_train[features]
X_train_xgb = df_train_xgb.iloc[:,0:-1].values
y_train_xgb = df_train_xgb.iloc[:,-1].values.reshape(-1,1)
df_train_xgb_partial = df_train_xgb.dropna(how='all', subset=['BER_class'])
X_train_xgb_partial = df_train_xgb_partial.iloc[:,0:-1].values
y_train_xgb_partial = df_train_xgb_partial.iloc[:,-1].values.reshape(-1,1)
# Call XGBoost Model for each dataset
Model_Full, RMSE_xgb_train_full, r2_xgb_train_full = xgb_model(X_train_xgb, y_train_xgb, "Performance Evaluation for Full Training Set")
Model_Partial, RMSE_xgb_train_partial, r2_xgb_train_partial = xgb_model(X_train_xgb_partial, y_train_xgb_partial, "Performance Evaluation for NaNs Removed")
# Let's plot the Feature Importance for each of these solutions
import seaborn as sn
fig = plt.figure(figsize=(25,6))
Model_Full.get_booster().feature_names = features[:-1]
ax1 = fig.add_subplot(1, 2, 1)
ax1.set_title("Plot of Feature Importance for Full Model")
xgb.plot_importance(Model_Full, max_num_features= 17, ax=ax1, importance_type="weight")
Model_Partial.get_booster().feature_names = features[:-1]
ax2 = fig.add_subplot(1, 2, 2)
ax2.set_title("Plot of Feature Importance for Model with NaNs removed.")
xgb.plot_importance(Model_Partial, max_num_features= 17, ax=ax2, importance_type="weight")
plt.show()
<jupyter_output><empty_output><jupyter_text>**Analysis of Plots** It can be seen from the above plots of Training Dataset with NaNs removed (plot on right) and NaNs left (plot on left) in that both plots confirm the importance of BER_class. XGBoost handles NaN values by default but we cannot expect that all models wil do so. We will keep the BER_class and compute imputed values for missing data.
Note also that both models agree on the overall importance of features. The Top 7 Features have a greater influence on model performance. It seems that property 'Type' is not so important.
For this exercise, I will concentrate on the Top 7 key features and remove the 'Type'#### Imputing BER_class value: replace NaNs with average BER by location<jupyter_code># First we will determine the average BER_class for each Location.
# This approach was taken because many locations have a mix of BER ratings (determined from analysis)
# but are, in general, either new development areas or mature development areas and as such
# are likely to have ratings within a range based on maturity of the location as a whole.
# Determine the average BER_class rating by location
feat = ['Location', 'BER_class']
train_feat = df_train_2[feat]
test_feat = df_test_2[feat]
train_test_stack = pd.concat([train_feat, test_feat])
average_vals = train_test_stack.groupby(['Location'], dropna=True)['BER_class'].mean().round()
<jupyter_output><empty_output><jupyter_text>**Comment** The code above creates a lookup of Average BER_class by Location. However, this is not a perfect lookup as there are several locations in the combined Training and Test datasets that have only one property in certain locations and these properties have a NaN value for BER_class. This means that the lookup will not be able to replace these NaNs with and average. Tests have shown that the effect is limited and the BER_class is improved from 15.4% missing values to 0.1% missing values. <jupyter_code># Look for NaNs and replace them with the average for the given location
import math
def impute_ber(data):
for index, row in data.iterrows():
if math.isnan(row['BER_class']):
loc = row['Location']
lookup_ber = average_vals.loc[loc]
data.loc[index, "BER_class"] = lookup_ber
return data
# Impute BER_class on training data
ber_tr_values = impute_ber(loc_ber_train_values)
# Impute BER_class on test data
ber_ts_values = impute_ber(loc_ber_test_values)
# There are still a very small number (2 in both training and test datsets) of NaNs due to single properties in locations that have NaNs and therefore could not be averaged
# We will apply an average over the entire feature for BER_class to populate these
ber_tr_values['BER_class'].fillna((ber_tr_values['BER_class'].mean()), inplace=True)
ber_ts_values['BER_class'].fillna((ber_ts_values['BER_class'].mean()), inplace=True)
# We can apply the standard scaler to each of the training and test set BER_class features
ber_tr_values[['BER_class']] = standardscaler.fit_transform(ber_tr_values[['BER_class']])
ber_ts_values[['BER_class']] = standardscaler.fit_transform(ber_ts_values[['BER_class']])
# Then we insert these columns back into the df_train and df_test datasets
df_train['BER_class'] = ber_tr_values['BER_class']
df_test['BER_class'] = ber_ts_values['BER_class']
# Let's test for any performance changes with the XGBoost Model
# Remove 'Type' from dataset as it does not appear to be too important
features = ['Location', 'Num_Bathrooms', 'Num_Beds', 'BER_class',
'Latitude', 'Longitude', 'Surface','Price']
df_train_xgb = df_train[features]
X_train_xgb = df_train_xgb.iloc[:,0:-1].values
y_train_xgb = df_train_xgb.iloc[:,-1].values.reshape(-1,1)
# Call XGBoost Model for each dataset
Model_Full, RMSE_xgb_train_full, r2_xgb_train_full = xgb_model(X_train_xgb, y_train_xgb, "Performance Evaluation for Full Training Set")
<jupyter_output>Performance Evaluation for Full Training Set
RMSE_score: 0.0410 R2_score: 0.9983
<jupyter_text>**Comment** The performance of the XGBoost Model has not shown much improvement with the imputing of missing BER_values. However, that is expected as XGBoost handles missing values by default. We expect to see no great impact on the model, but no deterioration.<jupyter_code># Plot the Feature Importance to see what the impact of imputing BER_class has been
fig, ax = plt.subplots(figsize=(12, 6))
Model_Full.get_booster().feature_names = features[:-1]
xgb.plot_importance(Model_Full, max_num_features= 7, ax=ax, importance_type="weight")
plt.show()<jupyter_output><empty_output><jupyter_text>**Comment** As mentioned above, we expect to see no great change from in either BER_class feature importance. Key here is the model performs well with the newly processed data and that is confirmed. We want to process the data to perform well with models that do not handle missing data well and that is what we have done here.### Analysis of Exploratory Data Analysis and Feature Engineering/Preprocessing
**Explain the approach, setps and results**
**Discuss the imputing for BER_class**
>>1) Not much impact and
>>2) not expected with XGBoost but will be important for modelling with algorithms that do not handle NaNs as well as XGBoost>
**Discuss the Type feature(s)** and the fact they do not look useful and may be removed from further processing
**The data is fully processed and ready for next stage - NLP Processing**The training and test datasets have been fully processed by removing unnecessary columns removing erroneous records, scaling and imputing values to provide complete datasets ready for further processing.**Comment** key to note here is the scales on all features are within similar ranges and not with large differences.# NATURAL LANGUAGE PROCESSINGNatural language processing ([NLP](https://en.wikipedia.org/wiki/Natural_language_processing)) is a subfield of linguistics, computer science, and artificial intelligence.
* NLP concerned with the interactions between computers and human language.
* In particular how to program computers to process and analyze large amounts of natural language data.
* The goal is a computer capable of "understanding" the contents of documents.
* Including the contextual nuances of the language within them.
* The technology can then accurately extract information and insights contained in the documents.
* As well as categorize and organize the documents themselves. For this section, we will process the two textual features using NLP techniqus. The two textual features are:
1. Description
2. Features
We are looking to see if these features have an impact on price. There maybe some words or phrases (bi- or tri-grams) that may influence the Price paid for the propertes. This is interesting because these are typically used by sellers or estate agents to dscribe the prperty for sale. The use of descriptions like this, as well as more detail on features of the prperty, are likely to influence buyers, so we will test this.### Approach to using NLP to increase the performance of the datasets
There are many options available to using NLP to process text data. For the current exercise, I will look at using NLP to provide additional features that may enhance the results and increase performance of the prediction models.
The approach will take the follow steps
1. Process 'Description' and 'Features' features to extract relevant words or phrases (n-grams). **Note**: It has been observed that these features, especially 'Description' can have similar text content. An initial review has also found descriptions of property features in the 'Description' field and "None" in the 'Features' field. This will be mitigated by combining these textual fields for processing.
2. Combine 'Description' and 'Features' fields and process by removing extraneous content such as numbers, punctuation, stopwords, etc and then Stemming, Lemmatizing, etc. to produce clean textual content.
3. Apply TF-IDF to extract the most important words and phrases across the textual corpus.
4. Order and select the top X number of words and phrases and create new features from them on the datasets, i.e. 'has_Parking', 'has_Garden', 'local_Amenities', etc.
5. Process each property record in both Training and Test datasets to update these features with [0, 1] if any of the words or phrases are present in the text of the record/sample.
6. This will provide additional features to train on - I will train on a full set of features on both Bayes Regression Model and Bayes Nueral Network.
7. I will also see if these features change the Feature Importance selected by XGBoost.
8. Overall performance improvement will be evaluated across models with/without these new features.## Word Counting in Property Corpus<jupyter_code># Set up the various components of the NLP processing code
from nltk.tokenize import word_tokenize
from nltk.stem import WordNetLemmatizer
from nltk.stem.porter import *
nltk.download('punkt')
nltk.download('wordnet')
import nltk
nltk.download('stopwords')
# Define a text preprocessing function
def nlp_preprocess(text):
text = text.lower() # set all text to lower case
string = re.sub('[^a-zA-Z]', ' ', text) # extract only words from text
text = " ".join(string.split()) # split text into words
shortword = re.compile(r'\W*\b\w{1,2}\b') # remove single or double letter words
text = (shortword.sub('', text))
# 1. Tokenization
NLP_token = word_tokenize(text) # Tokenise the words
# 3. Lemmatization
WL = WordNetLemmatizer()
NLP_lemma = []
for word in NLP_token:
#for word in NLP_stem:
NLP_lemma.append(WL.lemmatize(word))
# 4. Stopword
FS = []
NLP_stop = set(stopwords.words("english"))
for w in NLP_lemma:
if w not in NLP_stop:
FS.append(w)
# 5. Punctuation
punctuations = "?:!.,;"
for word in FS:
if word in punctuations:
FS.remove(word)
return FS
# Function to count words in text corpi
def count_words(text):
for word in text:
if word not in list(word_counts.keys()): word_counts.update({word:1})
else: word_counts[word] += 1
return word_counts
# Firstly, let's add back the "Description" and "Features" columns to the df_train and df_test datsets
df_train['Desc_Feat'] = train_data['Description'] + ' ' + train_data['Features']
df_test['Desc_Feat'] = train_data['Description'] +' ' + train_data['Features']
# Generate the list of words sorted by count of word occurrences
word_counts={}
n = df_train.shape[0]
for p in range(n):
text= nlp_preprocess((df_train.iloc[p]['Desc_Feat']))
count_words(text)
sorted_words = dict(sorted(word_counts.items(), key=lambda x: x[1], reverse=True))
# Create a DataFrame from the sorted list of words by word occurrences
# Select just top 120 values
words = list(sorted_words.keys())[0:120]
count = list(sorted_words.values())[0:120]
df_wdcount=pd.DataFrame(words, columns=['Words'])
df_wdcount['Counts'] = count
# Plot the list of words by word accurences acorss all the prperty records
plt.figure(figsize=(20, 8))
ax = sn.barplot(x='Words', y='Counts', data=df_wdcount, order=df_wdcount.sort_values('Counts', ascending=False).Words)
plt.xticks(rotation=90)<jupyter_output><empty_output><jupyter_text>### Word Occurrence Plot
**Analysis** This plot of Word Occurrence in the corpus of text provides us with an view on recurring words in the description+feature text describing the properties. Recurring descriptive words are important as they are used by estate agents to differentiate properties and attract buyers. Many word are typically used by estate agents and they have a lexicon that they use for advertisements.
However, the word occurrece, while important, does not tell the whole story. Recurring words may not differentiate properties in a large list of poperties. Very common words may not attract attention to buy or have an impact on price, which is what we are predicting here. To augment our analysis, we will also look at TF-IDF to dentify important words within the property descriptions.## Term Frequency-Inverse Document Frequency (TF-IDF) in Property Corpus<jupyter_code>df_train.columns
# Test out TF-IDF on the corpus of Description+Features
# Preprocess the data first ... although for TF-IDF it would push common words down the list anyway, but for simplicity we remove them
corpus = []
n = df_train.shape[0]
for p in range(n):
text = nlp_preprocess((df_train.iloc[p]['Desc_Feat']))
corpus.append(" ".join(text))
# Apply TF-IDF to the corpus of text across all property records
# This approach will provide us with another list, but this time rather than just calculating common words
# we will calculate using TF-IDF hwo important words are to the corpus of documents
from sklearn.feature_extraction.text import TfidfTransformer
from sklearn.feature_extraction.text import CountVectorizer
tfidfTransform = TfidfTransformer(use_idf=True)
CV = CountVectorizer()
word_count = CV.fit_transform(corpus)
tf_idf = tfidfTransform.fit_transform(word_count)
df = pd.DataFrame(tf_idf[0].T.todense(), index=CV.get_feature_names(), columns=['tfidf'])
df = df.sort_values('tfidf', ascending=False)
print(df.head(120))
# Reset the index of the dataframe and rename the index to make it easier to plot.
df.reset_index(inplace=True)
df_150=df.loc[1:150] # select 150 words from the dataframe
df_150_words = df_150.rename(columns={"index":"words"})
# Plot ordered list of words by TF-IDF score
plt.figure(figsize=(20, 8))
ax = sn.barplot(x='words', y='tfidf', data=df_150_words, order=df_150_words.sort_values('tfidf', ascending=False).words)
plt.xticks(rotation=90)<jupyter_output><empty_output><jupyter_text>### Term Frequency - Inverse Document Frequency (TF-IDF) Plot
**Analysis** This plot differs from the pure word count plot above. It provides a visual of the words that are most important to the corpus of data. You can see the words order by *how important they are* with words that are very common, i.e. recur very frequenetly in the property descriptions, set to '0' - this can be seen by the suddent drop after the word "accomodation".
A good description of how TF-IDF works can be found in the text by Silge and Robinson (2021) or Chaudhary (2020).
<jupyter_code># Prints word position for analysis below
print (df_150_words[df_150_words['words'] == "amenity"])
print (df_wdcount[df_wdcount['Words'] == "amenity"])<jupyter_output>Empty DataFrame
Columns: [words, tfidf]
Index: []
Words Counts
47 amenity 1577
<jupyter_text>### How do we create new features from this information?
With the Word Occurrence and TF-IDF information we can now apply an heuristic approach to determine the new features.
Neither plots provides us with all the information but given an understanding of what is important for prospective home buyers (or investors as the case may be), we can say:
1. Having a garden is an important feature of a house but not an apartment. The word 'garden' appears a lot (high on Word Occurrence and low (< 7000) on TF-IDF). Given this we can assume that 'garden' is not a maor differentiator in general but it is an important consideration to *generalise* our model
2. Parking appears quite high in both word occurrence and TF-IDF score. It is clearly important for both house and apartment buyers. We will include this as one of our features
3. School also appears on both plots: word occurrence (46) and TF-IDF (115). For families we know that this is important. Families live on both houses and apartments and it can be a deciding factor
4. Public Transport appears as "transport" in TF-IDF and as "bus" in word count. Based on knowledge that public transport is important in Dublin, we can take these values as important features.
5. Local Amenities appear in both lists of words as "amentity", "retaurant", "pharmacy", "supermarket. These are key both from the position on both plots and heuristic domain knowledge.
With these findings, we can now search the individual words in each property record looking for these words and setting [0] if it does not occur and [1] if it does for the following nw features:
1. has_Garden
2. has_Parking
3. local_School (close to schools to include "school", "secondary", "primary")
4. local_Transport (to include "transport", "bus")
5. local_Amenities (to include "amenity", "shop", "shopping", "restaurant", "supermarket")
### Create new features from above rules<jupyter_code># Reload the data to ensure we are working with clean datasets
path = '/content/sample_data/'
# Due to formatting issues, I found that explicitly defining features of interest and omitting ones that are problematic was useful
train_file = path+'HousePrice_Train.csv'
test_file = path+'HousePrice_Test.csv'
# Import the data, while excluding 'features' and 'description_block' explicitly on import
train_data = pd.read_csv(train_file)
test_data = pd.read_csv(test_file)
# Create new features in training and test dataframes
# Go back to original datasets: we will want to split them for performance testing on training and validation sets
df_train = train_data.join(pd.DataFrame(
{
'has_Garden': 0,
'has_Parking': 0,
'local_School': 0,
'local_Transport': 0,
'local_Amenities': 0
}, index=train_data.index
))
# Combine 'Description' and 'Features'
df_train['Desc_Feat'] = df_train['Description'] + ' ' + df_train['Features']
# check if words are in a text
def words_in_text(list, text):
return set(list).intersection(text)
# Process the text descriptions in the new 'Desc_Feat' column for each property
_Garden = ["garden"]
_Parking = ["parking"]
_localSchool = ["school", "primary school", "secondary school"]
_localTransport = ["transport", "bus"]
_localAmenities = ["amenity", "shop", "shopping", "restaurant", "supermarket", "pharmacy"]
for index, row in df_train.iterrows():
descfeat = []
text = nlp_preprocess((df_train.iloc[index]['Desc_Feat']))
descfeat.append(" ".join(text))
# check for key keywords to set the new features
if words_in_text(_Garden, text): df_train.loc[index, "has_Garden"] = 1
if words_in_text(_Parking, text): df_train.loc[index, "has_Parking"] = 1
if words_in_text(_localSchool, text): df_train.loc[index, "local_School"] = 1
if words_in_text(_localTransport, text): df_train.loc[index, "local_Transport"] = 1
if words_in_text(_localAmenities, text): df_train.loc[index, "local_Amenities"] = 1
# Drop these columns from the datasets; they are no longer needed
df_train.drop(columns=['Description', 'Features', 'Desc_Feat'], inplace=True)
df_train.head()<jupyter_output><empty_output><jupyter_text>### Comments on New Features for datasets
Now we have added five new features to the property datasets. Before we take thenext step to train and test ML Models, I will quickly take a look to see if these features have impacted Feature Importance order.### Pipeline Function
Firstly we create two pipeline functions to encapsulate the pre-processing steps before modelling.<jupyter_code># Create a processing pipeline as a function
def drop_outliers(dataset):
# Remove outliers for location that is not in Dublin
df_dataset = dataset.drop(dataset[(dataset.Longitude > -6) | (dataset.Latitude < 53)].index)
# Remove Surface values over 2000.
df_dataset = df_dataset.drop(df_dataset[df_dataset.Surface > 2000].index)
return df_dataset
def process_dataset(dataset, exclude="None"):
# Drop unimportant features
dataset.drop(['ID', 'Index', 'Services'], axis=1, inplace=True)
df_dataset = dataset.copy()
# Replace Locations with index value from 'unique_areas_index' lookup
df_dataset_1 = df_dataset.replace({'Location': unique_areas_index})
# Replace BER_class with index value from 'unique_ber_index'
df_dataset_2 = df_dataset_1.replace({'BER_class': unique_ber_index})
# One-hot-encode 'Type
if exclude == "None":
df_dataset_2 = pd.get_dummies(df_dataset_2, columns=['Type'])
else: df_dataset_2.drop(columns=['Type'], inplace=True)
df_dataset = df_dataset_2.copy()
# Take a copy of the Location and BER_class before applying further scaling
loc_ber_dataset_values = df_dataset[['Location', 'BER_class']].copy()
# Apply scalers to features
# Not a normal distribution so just scale with MinMax [0, 1]
minmax=MinMaxScaler()
standardscaler = StandardScaler()
df_dataset[['Location']] = minmax.fit_transform(df_dataset[['Location']])
# Deal with outliers with a Log Transform - later we will use Standard Scaler as well on some feature
df_dataset['Num_Bathrooms'] = np.log(df_dataset['Num_Bathrooms'].apply(pd.to_numeric))
df_dataset['Num_Beds'] = np.log(df_dataset['Num_Beds'].apply(pd.to_numeric))
df_dataset['Latitude'] = np.log(df_dataset['Latitude'].apply(pd.to_numeric))
df_dataset['Surface'] = np.log(df_dataset['Surface'].apply(pd.to_numeric))
# Standard scaler for normal distributions
df_dataset[['BER_class', 'Longitude', 'Latitude', 'Surface']] = standardscaler.fit_transform(df_dataset[['BER_class', 'Longitude', 'Latitude', 'Surface']])
# Impute BER_class
ber_values = impute_ber(loc_ber_dataset_values)
# There are still a very small number (2 in both training and test datsets) of NaNs due to single properties in locations that have NaNs and therefore could not be averaged
# We will apply an average over the entire feature for BER_class to populate these
ber_values['BER_class'].fillna((ber_values['BER_class'].mean()), inplace=True)
# We can apply the standard scaler to each of the training and test set BER_class features
ber_values[['BER_class']] = standardscaler.fit_transform(ber_values[['BER_class']])
# Then we insert these columns back into the df_train and df_test datasets
df_dataset['BER_class'] = ber_values['BER_class']
return df_dataset
# Create a New Features pipeline as a function
def new_features(data):
df_data = data.join(pd.DataFrame(
{
'has_Garden': 0,
'has_Parking': 0,
'local_School': 0,
'local_Transport': 0,
'local_Amenities': 0
}, index=data.index
))
# Combine 'Description' and 'Features'
df_data['Desc_Feat'] = df_data['Description'] + ' ' + df_data['Features']
# Process the text descriptions in the new 'Desc_Feat' column for each property
_Garden = ["garden"]
_Parking = ["parking"]
_localSchool = ["school", "primary school", "secondary school"]
_localTransport = ["transport", "bus"]
_localAmenities = ["amenity", "shop", "shopping", "restaurant", "supermarket", "pharmacy"]
for idx, row in df_data.iterrows():
descfeat = []
text = nlp_preprocess((df_data.iloc[idx]['Desc_Feat']))
descfeat.append(" ".join(text))
# check for key keywords to set the new features
if words_in_text(_Garden, text): df_data.loc[idx, "has_Garden"] = 1
if words_in_text(_Parking, text): df_data.loc[idx, "has_Parking"] = 1
if words_in_text(_localSchool, text): df_data.loc[idx, "local_School"] = 1
if words_in_text(_localTransport, text): df_data.loc[idx, "local_Transport"] = 1
if words_in_text(_localAmenities, text): df_data.loc[idx, "local_Amenities"] = 1
# Drop these columns from the dataset; they are no longer needed
df_data.drop(columns=['Description', 'Features', 'Desc_Feat'], inplace=True)
return df_data
# Function to calculate MAPE
def MAPE(y_pred, y):
sum = 0
n = len(y_pred)
for i in range(n):
sum = sum + abs(y_pred[i] - y[i])/y[i]
mape = sum/n
return mape
# Reload the data to ensure we are working with clean datsets
path = '/content/sample_data/'
# Due to formatting issues, I found that explicitly defining features of interest and omitting ones that are problematic was useful
train_file = path+'HousePrice_Train.csv'
test_file = path+'HousePrice_Test.csv'
# Import the data, while excluding 'features' and 'description_block' explicitly on import
train_data = pd.read_csv(train_file)
test_data = pd.read_csv(test_file)
df_tr = drop_outliers(train_data)
# Separate the training dataset into Training and Validation Sets
colnames = list(df_tr.columns)
X = df_tr.iloc[:,0:-1].values
y = df_tr.iloc[:,-1].values.reshape(-1,1)
# Split training data into training and validation sets
X_train, X_val, y_train, y_val = train_test_split(X, y, test_size = 0.3, shuffle = False)
# Convert numpy arrays to dataframes before processing
X_df_tr = pd.DataFrame(X_train, columns=colnames[:-1])
X_df_vl = pd.DataFrame(X_val, columns=colnames[:-1])
# save features of the training dataset
features = ['Location', 'Num_Bathrooms', 'Num_Beds', 'BER_class', 'Latitude',
'Longitude', 'Surface', 'has_Garden', 'has_Parking', 'local_School',
'local_Transport', 'local_Amenities', 'Type_apartment', 'Type_bungalow',
'Type_detached', 'Type_duplex', 'Type_end-of-terrace',
'Type_semi-detached', 'Type_studio', 'Type_terraced', 'Type_townhouse']
# Execute preprocessing steps in two calls
# First for training data
X_tr_step_1 = new_features(X_df_tr)
X_trn_ = process_dataset(X_tr_step_1)
X_trn_ = X_trn_[features]
X_trn = X_trn_.values
# Next for validation data
X_vl_step_1 = new_features(X_df_vl)
X_valid_ = process_dataset(X_vl_step_1)
X_valid_ = X_valid_[features]
X_valid = X_valid_.values
# Scale 'Price' if the dataset if Training
ylog = np.log(y_train.astype('float'))
yscaler = standardscaler.fit(ylog)
y_trn = yscaler.transform(ylog)<jupyter_output><empty_output><jupyter_text>**Caution** the following may fail if all columns are not in both training and validation. I fixed this by shuffling instead of random_state for the train/test split. This is caused by the split data not having samples of all 'Types' and thus when one-hot-encoded this feature may be missing in either datset.<jupyter_code># Let's test for any performance changes with the XGBoost Model
# Call XGBoost Model for each dataset
model_processed_data, RMSE_xgb_train_processed, r2_xgb_train_processed = xgb_model(X_trn, y_trn, "Performance Evaluation for Full Training Set with New Features")
# Calculate MAPE scores for training and validation datsets
# Predictions and MAPE score for training data
# X_trn = X_trn_[features].values
y_pred_trn = model_processed_data.predict(X_trn)
y_pred_blr_trn = np.exp(yscaler.inverse_transform(y_pred_trn))
mape_trn = MAPE(y_pred_blr_trn, y_train)
# Predictions and MAPE score for validation data
y_pred_val = model_processed_data.predict(X_valid)
y_pred_blr_val = np.exp(yscaler.inverse_transform(y_pred_val))
mape_val = MAPE(y_pred_blr_val, y_val)
print("MAPE_training: %.4f MAPE_validation: %.4f" %(mape_trn, mape_val))<jupyter_output>MAPE_training: 0.0058 MAPE_validation: 0.1715
<jupyter_text>### Comment on model performance
The performance of the XGBoost on the data is very good. Performance of the model on the validation set (17.1%) with the addition NLP feature is 2% better than without. We will use this model to examine the Feature Importance, now with al features.<jupyter_code># Plot the Feature Importance to see what the impact of imputing BER_class has been
fig, ax = plt.subplots(figsize=(12, 6))
model_processed_data.get_booster().feature_names = features
xgb.plot_importance(model_processed_data, max_num_features= 21, ax=ax, importance_type="weight")
plt.show()<jupyter_output><empty_output><jupyter_text>### **Analysis of new feature importance metrics**
From above plot it can be seen that the addition of the new features has impacted on the feature importance ranking. has_Parking, local_Transport and local_School are all more important that the 'Type' features. Most of the NLP features rank better than the 'Type' features also. These are interesting observations given that the features were embedded in text descriptions. The plot above has proven the value of NLP feature engineering, certainly in this case.## Training, Validation & Test Data<jupyter_code># Reload the data to ensure we are working with clean datasets
path = '/content/sample_data/'
# Due to formatting issues, I found that explicitly defining features of interest and omitting ones that are problematic was useful
train_file = path+'HousePrice_Train.csv'
test_file = path+'HousePrice_Test.csv'
# Import the data, while excluding 'features' and 'description_block' explicitly on import
train_data = pd.read_csv(train_file)
test_data = pd.read_csv(test_file)
df_tr = drop_outliers(train_data)
# Separate the training dataset into Training and Validation Sets
colnames = list(df_tr.columns)
X = df_tr.iloc[:,0:-1].values
y = df_tr.iloc[:,-1].values.reshape(-1,1)
# Split training data into training and validation sets
X_train, X_val, y_train, y_val = train_test_split(X, y, test_size = 0.3, shuffle = False)
# Convert numpy arrays to dataframes before processing
X_df_trn = pd.DataFrame(X_train, columns=colnames[:-1])
X_df_val = pd.DataFrame(X_val, columns=colnames[:-1])
# Execute preprocessing steps for each dataset
# First for training data
X_trn_step_1 = new_features(X_df_trn)
X_trn_ = process_dataset(X_trn_step_1, exclude="Type")
X_trn = X_trn_.values
# Next for validation data
X_val_step_1 = new_features(X_df_val)
X_val_ = process_dataset(X_val_step_1, exclude="Type")
X_val = X_val_.values
# Then for Test data
X_tst_step_1 = new_features(test_data)
X_tst_ = process_dataset(X_tst_step_1, exclude="Type")
X_tst = X_tst_.values
# Scale 'Price' if the dataset if Training
ylog = np.log(y_train.astype('float'))
yscaler = standardscaler.fit(ylog)
y_trn = yscaler.transform(ylog)<jupyter_output><empty_output><jupyter_text>### Train dataset<jupyter_code># show first data frame rows
X_trn_.head(2)
# Generate descriptive statistics
X_trn_.describe()<jupyter_output><empty_output><jupyter_text>### Validation dataset<jupyter_code>X_val_.head(2)
X_val_.describe()<jupyter_output><empty_output><jupyter_text>### Test dataset<jupyter_code># show first data frame rows
X_tst_.head(2)
# Generate descriptive statistics
X_tst_.describe()<jupyter_output><empty_output><jupyter_text>### Baseline Predictions on Test with a Known Model Performance
Let's set a baseline prediction on test data from XGBoost so we can compare with Bayesian Regression and Bayesian Neural Network<jupyter_code># Call XGBoost Model for each dataset
model_processed_data, RMSE_xgb_processed, r2_xgb_processed = xgb_model(X_trn, y_trn, "Performance Evaluation for Full Training Set with New Features")
# Predict on test dataset
y_pred_tst = model_processed_data.predict(X_tst)
y_pred_blr_tst = np.exp(yscaler.inverse_transform(y_pred_tst))
# Record baseline prices
df_prices = test_data['Index']
df_prices_tst = pd.DataFrame(y_pred_blr_tst, columns=['Baseline_price'])
full_test_predictions = pd.concat([df_prices, df_prices_tst], axis=1)
full_test_predictions<jupyter_output><empty_output><jupyter_text>**Comment** These are the baseline predictions from XGBoost with a MAPE of 17% - we expect these to be a good estimate to check other methods against.# PIECEWISE REGRESSION**Piecewise regression**, extract from [Wikipedia](https://en.wikipedia.org/wiki/Segmented_regression):
Segmented regression, also known as piecewise regression or broken-stick regression, is a method in regression analysis in which the independent variable is partitioned into intervals and a separate line segment is fit to each interval.
* Segmented regression analysis can also be performed on
multivariate data by partitioning the various independent variables.
* Segmented regression is useful when the independent variables, clustered into different groups, exhibit different relationships between the variables in these regions.
* The boundaries between the segments are breakpoints.
* Segmented linear regression is segmented regression whereby the relations in the intervals are obtained by linear regression. ### Bayesian Model Price Prediction on dataset with all key features (excluding 'Type' as determined above in EDA Section<jupyter_code># model
with pm.Model() as model:
#prior over the parameters of linear regression
alpha = pm.Normal('alpha', mu=0, sigma=30)
#we have one beta for each column of Xn
beta = pm.Normal('beta', mu=0, sigma=30, shape=X_trn.shape[1])
#prior over the variance of the noise
sigma = pm.HalfCauchy('sigma_n', 5)
#linear regression model in matrix form
mu = alpha + pm.math.dot(beta, X_trn.T)
#likelihood, be sure that observed is a 1d vector
like = pm.Normal('like', mu=mu, sigma=sigma, observed=y_trn[:,0])
#number of iterations of the algorithms
iter = 50000
# run the model
with model:
approximation = pm.fit(iter,method='advi')
# check the convergence
plt.plot(approximation.hist);
# samples from the posterior
posterior = approximation.sample(draws=702)
# prediction
ll=np.mean(posterior['alpha']) + np.dot(np.mean(posterior['beta'],axis=0), X_tst.T)
y_pred_BLR = np.exp(yscaler.inverse_transform(ll.reshape(-1,1)))[:,0]
print(len(y_pred_BLR))
df_prices = full_test_predictions
df_prices_bayes_full = pd.DataFrame(y_pred_BLR, columns=['Bayes_full_price'])
full_test_predictions = pd.concat([full_test_predictions, df_prices_bayes_full], axis=1)
full_test_predictions
mape_bayes_full = MAPE(y_pred_blr_tst, y_pred_BLR)
print("MAPE_validation: %.4f MAPE_bayes_full: %.4f" %(mape_val, mape_bayes_full))<jupyter_output>MAPE_validation: 0.1715 MAPE_bayes_full: 0.1920
<jupyter_text>**Comments** MAPE_bayes_full is the MAPE score when comparing the baseline estimated prices on the test dataset with the estimated prices from the Bayesian Regression Model with all key features. This score of 19.3 % is a good performance if we are confident that the baseline estimates are good enough. Given the performance of the baseline estimation using XGBoost, it is reasonable to take this view.### Price Prediction using Piecewise Bayesian Model (Clustering by Top 2 Key Features)<jupyter_code>X_trn_.columns<jupyter_output><empty_output><jupyter_text>**Comment** from the Feature Importance analysis above, the Longitude and Latitude features rank the highest. We will look at these features for the Piecewise approach.<jupyter_code># clustering by features 4, 5; from list above these are 'longitude' and 'latitude'
ind=[4, 5]
X_ind = np.vstack([X_trn[:,ind],X_tst[:,ind]])
# training gaussian mixture model
from sklearn.mixture import GaussianMixture
gmm = GaussianMixture(n_components=4)
# Gaussian Mixture
gmm.fit(X_ind)
# plot predicted clusters
label = gmm.predict(X_ind)
plt.scatter(X_ind[:,0],X_ind[:,1], c = label)
# centroids: orange dots
plt.scatter(gmm.means_[:,0],gmm.means_[:,1], c = 'orange')<jupyter_output><empty_output><jupyter_text>**Comment** reviewing 3, 4, 5 and 6 clusters, four clusters seems a reasonable choice.#### Prediction on 4 Clusters<jupyter_code># Split training data into training and validation sets
X_train, X_val, y_train, y_val = train_test_split(X, y, test_size = 0.3, shuffle = False)
# Change names to make them more meaningful
X_train = X_trn
X_test = X_tst
# train clusters
clusters_train = gmm.predict(X_train[:,ind])
unique_train, counts_train = np.unique(clusters_train, return_counts=True)
dict(zip(unique_train, counts_train))
# test clusters
clusters_test = gmm.predict(X_test[:,ind])
unique_test, counts_test = np.unique(clusters_test, return_counts=True)
dict(zip(unique_test, counts_test))
# Process data for cluster 0
Xn0 = X_train[clusters_train==0,:]
Xtestn0 = X_test[clusters_test==0,:]
ylog0 = np.log(y_train.astype('float')[clusters_train==0,:])
yscaler0 = StandardScaler().fit(ylog0)
yn0 = yscaler0.transform(ylog0)
# Process data for cluster 1
Xn1 = X_train[clusters_train==1,:]
Xtestn1 = X_test[clusters_test==1,:]
ylog1 = np.log(y_train.astype('float')[clusters_train==1,:])
yscaler1 = StandardScaler().fit(ylog1)
yn1 = yscaler1.transform(ylog1)
# Process data for cluster 2
Xn2 = X_train[clusters_train==2,:]
Xtestn2 = X_test[clusters_test==2,:]
ylog2 = np.log(y_train.astype('float')[clusters_train==2,:])
yscaler2 = StandardScaler().fit(ylog2)
yn2 = yscaler2.transform(ylog2)
# Process data for cluster 3
Xn3 = X_train[clusters_train==3,:]
Xtestn3 = X_test[clusters_test==3,:]
ylog3 = np.log(y_train.astype('float')[clusters_train==3,:])
yscaler3 = StandardScaler().fit(ylog3)
yn3 = yscaler3.transform(ylog3)
# Train model_0
with pm.Model() as model_0:
# prior over the parameters of linear regression
alpha = pm.Normal('alpha', mu=0, sigma=30)
# we have a beta for each column of Xn0
beta = pm.Normal('beta', mu=0, sigma=30, shape=Xn0.shape[1])
# prior over the variance of the noise
sigma = pm.HalfCauchy('sigma_n', 5)
# linear regression relationship
#linear regression model in matrix form
mu = alpha + pm.math.dot(beta, Xn0.T)
# likelihood, be sure that observed is a 1d vector
like = pm.Normal('like', mu=mu, sigma=sigma, observed=yn0[:,0])
with model_0:
# iterations of the algorithm
approximation = pm.fit(40000,method='advi')
# samples from the posterior
posterior0 = approximation.sample(counts_test[0])
# Train model_1
with pm.Model() as model_1:
# prior over the parameters of linear regression
alpha = pm.Normal('alpha', mu=0, sigma=30)
# we have a beta for each column of Xn
beta = pm.Normal('beta', mu=0, sigma=30, shape=Xn1.shape[1])
# prior over the variance of the noise
sigma = pm.HalfCauchy('sigma_n', 5)
# linear regression relationship
#linear regression model in matrix form
mu = alpha + pm.math.dot(beta, Xn1.T)
# likelihood, #
like = pm.Normal('like', mu=mu, sigma=sigma, observed=yn1[:,0])
with model_1:
# iterations of the algorithm
approximation = pm.fit(40000,method='advi')
# samples from the posterior
posterior1 = approximation.sample(counts_test[1])
# Train model_2
with pm.Model() as model_2:
# prior over the parameters of linear regression
alpha = pm.Normal('alpha', mu=0, sigma=30)
# we have a beta for each column of Xn
beta = pm.Normal('beta', mu=0, sigma=30, shape=Xn2.shape[1])
# prior over the variance of the noise
sigma = pm.HalfCauchy('sigma_n', 5)
# linear regression relationship
# linear regression model in matrix form
mu = alpha + pm.math.dot(beta, Xn2.T)
# likelihood, be sure that observed is a 1d vector
like = pm.Normal('like', mu=mu, sigma=sigma, observed=yn2[:,0])
with model_2:
# iterations of the algorithms
approximation = pm.fit(40000,method='advi')
# samples from the posterior
posterior2 = approximation.sample(counts_test[2])
# Train model_3
with pm.Model() as model3:
# prior over the parameters of linear regression
alpha = pm.Normal('alpha', mu=0, sigma=30)
# we have a beta for each column of Xn
beta = pm.Normal('beta', mu=0, sigma=30, shape=Xn3.shape[1])
# prior over the variance of the noise
sigma = pm.HalfCauchy('sigma_n', 5)
# linear regression relationship
mu = alpha + pm.math.dot(beta, Xn3.T)#linear regression model in matrix form
# likelihood, be sure that observed is a 1d vector
like = pm.Normal('like', mu=mu, sigma=sigma, observed=yn3[:,0])
with model3:
# number of iterations of the algorithms
approximation = pm.fit(40000,method='advi')
# samples from the posterior
posterior3 = approximation.sample(counts_test[3])
# Posterior predictive checks (PPCs)
def ppc(alpha,beta,sigma, X, nsamples=500):
#we select nsamples random samples from the posterior
ind = np.random.randint(0,beta.shape[0],size=nsamples)
alphai = alpha[ind]
betai = beta[ind,:]
sigmai = sigma[ind]
Ypred = np.zeros((nsamples,X.shape[0]))
for i in range(X.shape[0]):
#we generate data from linear model
y_pred = alphai + np.dot(betai, X[i:i+1,:].T).T +np.random.randn(len(sigmai))*sigmai
Ypred[:,i]=y_pred[0,:]
return Ypred
# Simulation for cluster 0
# Reverse the scaling for predicted value to get back to the original price scale
Ypred0 = yscaler0.inverse_transform(ppc(posterior0['alpha'],posterior0['beta'],posterior0['sigma_n'],Xn0, nsamples=200))
for i in range(Ypred0.shape[0]):
az.plot_dist( Ypred0[i,:],color='r',plot_kwargs={"linewidth": 0.2})
az.plot_dist(Ypred0[i,:],color='r',plot_kwargs={"linewidth": 0.2}, label="prediction")
#plt.plot(np.linspace(-8,8,100),norm.pdf(np.linspace(-8,8,100),df=np.mean(posterior_1['nu'])))
#plt.xlim([0,10e7])
az.plot_dist(ylog0,label='true observations');
plt.legend()
plt.xlabel("log(y) - output variable")
plt.ylabel("density plot");
#Simulation for cluster 1
# Reverse the scaling for predicted value to get back to the original price scale
Ypred1 = yscaler1.inverse_transform(ppc(posterior1['alpha'],posterior1['beta'],posterior1['sigma_n'],Xn1, nsamples=200))
for i in range(Ypred1.shape[0]):
az.plot_dist( Ypred1[i,:],color='r',plot_kwargs={"linewidth": 0.2})
az.plot_dist(Ypred1[i,:],color='r',plot_kwargs={"linewidth": 0.2}, label="prediction")
#plt.plot(np.linspace(-8,8,100),norm.pdf(np.linspace(-8,8,100),df=np.mean(posterior_1['nu'])))
#plt.xlim([0,10e7])
az.plot_dist(ylog1,label='true observations');
plt.legend()
plt.xlabel("log(y) - output variable")
plt.ylabel("density plot");
#Simulation
Ypred2 = yscaler2.inverse_transform(ppc(posterior2['alpha'],posterior2['beta'],posterior2['sigma_n'],Xn2, nsamples=200))
for i in range(Ypred2.shape[0]):
az.plot_dist( Ypred2[i,:],color='r',plot_kwargs={"linewidth": 0.2})
az.plot_dist(Ypred2[i,:],color='r',plot_kwargs={"linewidth": 0.2}, label="prediction")
#plt.plot(np.linspace(-8,8,100),norm.pdf(np.linspace(-8,8,100),df=np.mean(posterior_1['nu'])))
#plt.xlim([0,10e7])
az.plot_dist(ylog2,label='true observations');
plt.legend()
plt.xlabel("log(y) - output variable")
plt.ylabel("density plot");
#Simulation for Cluster 3
# Reverse the scaling for predicted value to get back to the original price scale
Ypred3 = yscaler3.inverse_transform(ppc(posterior3['alpha'],posterior3['beta'],posterior3['sigma_n'],Xn3, nsamples=200))
for i in range(Ypred3.shape[0]):
az.plot_dist( Ypred3[i,:],color='r',plot_kwargs={"linewidth": 0.2})
az.plot_dist(Ypred3[i,:],color='r',plot_kwargs={"linewidth": 0.2}, label="prediction")
#plt.plot(np.linspace(-8,8,100),norm.pdf(np.linspace(-8,8,100),df=np.mean(posterior_1['nu'])))
#plt.xlim([0,10e7])
az.plot_dist(ylog3,label='true observations');
plt.legend()
plt.xlabel("log(y) - output variable")
plt.ylabel("density plot");
# Using PPC, take 200 samples from the posteriors of each cluster
Ypred0 = ppc(posterior0['alpha'],posterior0['beta'],posterior0['sigma_n'],Xn0, nsamples=200)
Ypred1 = ppc(posterior1['alpha'],posterior1['beta'],posterior1['sigma_n'],Xn1, nsamples=200)
Ypred2 = ppc(posterior2['alpha'],posterior2['beta'],posterior2['sigma_n'],Xn2, nsamples=200)
Ypred3 = ppc(posterior3['alpha'],posterior3['beta'],posterior3['sigma_n'],Xn3, nsamples=200)
# Create a combined predicted value by reversing the scaling done on each cluster prediction
Ypred = np.hstack([ yscaler0.inverse_transform(Ypred0),
yscaler1.inverse_transform(Ypred1),
yscaler2.inverse_transform(Ypred2),
yscaler3.inverse_transform(Ypred3)])
# Plot the distribution for the combined set of samples
for i in range(Ypred.shape[0]):
az.plot_dist( Ypred[i,:],color='r',plot_kwargs={"linewidth": 0.2})
# plot the predicted distributions against the true distribution to see how they are close
az.plot_dist(Ypred[i,:],color='r',plot_kwargs={"linewidth": 0.2}, label="prediction")
ylog=np.vstack([ylog0,ylog1,ylog2,ylog3])
az.plot_dist(ylog,label='true observations');
plt.legend()
plt.xlabel("log(y) - output variable")
plt.ylabel("density plot");
# cluster 0
y_pred_BLR0 = np.exp(yscaler0.inverse_transform(np.mean(posterior0['alpha'])
+ np.dot(np.mean(posterior0['beta'],axis=0), Xtestn0.T)))
# cluster 1
y_pred_BLR1 = np.exp(yscaler1.inverse_transform(np.mean(posterior1['alpha'])
+ np.dot(np.mean(posterior1['beta'],axis=0), Xtestn1.T)))
# cluster 2
y_pred_BLR2 = np.exp(yscaler2.inverse_transform(np.mean(posterior2['alpha'])
+ np.dot(np.mean(posterior2['beta'],axis=0), Xtestn2.T)))
# cluster 3
y_pred_BLR3 = np.exp(yscaler3.inverse_transform(np.mean(posterior3['alpha'])
+ np.dot(np.mean(posterior3['beta'],axis=0), Xtestn3.T)))
joint_y_pred=np.hstack([y_pred_BLR0, y_pred_BLR1, y_pred_BLR2, y_pred_BLR3])
# Save predictions to predictions dataframe
df_1 = full_test_predictions
df_2 = pd.DataFrame(joint_y_pred, columns=['Price_Piecewise'])
full_test_predictions = pd.concat([df_1, df_2], axis=1)
full_test_predictions
mape_bayes_cluster = MAPE(y_pred_blr_tst, joint_y_pred)
print("MAPE_validation: %.4f MAPE_bayes_cluster: %.4f" %(mape_val, mape_bayes_cluster))<jupyter_output>MAPE_validation: 0.1715 MAPE_bayes_cluster: 0.7458
<jupyter_text>**Comments** MAPE_bayes_cluster is the MAPE score when comparing the baseline estimated prices on the test dataset with the estimated prices from the Bayesian Piecewise Regression Model with all key features. This score of 73 % is poor performance and is attributed to the use of only 2 featres, where the other key feature have a significant impact on performance. If the model was trained in clusters of all features, we would likely see a small improvement over just unclustered features.# BAYESIAN NNA [Bayesian network](https://en.wikipedia.org/wiki/Bayesian_network) (also known as a Bayes network, Bayes net, belief network, or decision network) is a probabilistic graphical model that represents a set of variables and their conditional dependencies via a directed acyclic graph (DAG).
* Bayesian networks are ideal for taking an event that occurred and predicting the likelihood that any one of several possible known causes was the contributing factor.
* For example, a Bayesian network could represent the probabilistic relationships between diseases and symptoms.
* Given symptoms, the network can be used to compute the probabilities of the presence of various diseases.Your comments, explanation, and references here.<jupyter_code># Instantiate Kernel
kernel = GPy.kern.RBF(input_dim=1,lengthscale=0.15,variance=0.2)
print(kernel)
# Split training data into training and validation sets
X_train, X_val, y_train, y_val = train_test_split(X, y, test_size = 0.3, shuffle = False)
# Change names to make them more meaningful
X_train = X_trn
X_test = X_tst
# Scale 'Price' if the dataset if Training
ylog = np.log(y_train.astype('float'))
yscaler = standardscaler.fit(ylog)
y_train = yscaler.transform(ylog)
# https://theano-pymc.readthedocs.io/en/latest/
import theano
# add a column of ones to include an intercept in the model
x1 = np.hstack([np.ones((X_train.shape[0],1)), X_train])
floatX = theano.config.floatX
l = 15
# Initialize random weights between each layer
# we do that to help the numerical algorithm that computes the posterior
init_1 = np.random.randn(x1.shape[1], l).astype(floatX)
init_out = np.random.randn(l).astype(floatX)
# pymc3 model as neural_network
with pm.Model() as bayesian_neural_network:
# we convert the data in theano type so we can do dot products with the correct type.
ann_input = pm.Data('ann_input', x1)
ann_output = pm.Data('ann_output', y_train)
# Priors
# Weights from input to hidden layer
weights_in_1 = pm.Normal('w_1', 0, sigma=1,
shape=(x1.shape[1], l), testval=init_1)
# Weights from hidden layer to output
weights_2_out = pm.Normal('w_0', 0, sigma=1,
shape=(l,),testval=init_out)
# Build neural-network using tanh activation function
# Inner layer
act_1 = pm.math.tanh(pm.math.dot(ann_input,weights_in_1))
# Linear layer, like in Linear regression
act_out = pm.Deterministic('act_out',pm.math.dot(act_1, weights_2_out))
# standard deviation of noise
sigma = pm.HalfCauchy('sigma',5)
# Normal likelihood
out = pm.Normal('out',
act_out,
sigma=sigma,
observed=ann_output[:,0])
par_1 = 100
par_2 = 1000
with bayesian_neural_network:
posterior = pm.sample(par_1, tune=par_2, chains=1)
# we can do instead an approximated inference
param3 = 20000 # start with 1000, then use 50000+
VI = 'advi' # 'advi', 'fullrank_advi', 'svgd', 'asvgd', 'nfvi'
OP = pm.adam # pm.adam, pm.sgd, pm.adagrad, pm.adagrad_window, pm.adadelta
LR = 0.001
with bayesian_neural_network:
approx = pm.fit(param3, method=VI, obj_optimizer=pm.adam(learning_rate=LR))
# plot
pb.plot(approx.hist, label='Variational Inference: '+ VI.upper(), alpha=.3)
pb.legend(loc='upper right')
# Evidence Lower Bound (ELBO)
# https://en.wikipedia.org/wiki/Evidence_lower_bound
pb.ylabel('ELBO')
pb.xlabel('iteration');
# draw samples from variational posterior
D = 702
posterior = approx.sample(draws=D)
# add a column of ones to include an intercept in the model
x2 = np.hstack([np.ones((X_test.shape[0],1)), X_test])
y_pred = []
for i in range(posterior['w_1'].shape[0]):
#inner layer
t1 = np.tanh(np.dot(posterior['w_1'][i,:,:].T,x2.T))
#outer layer
y_pred.append(np.dot(posterior['w_0'][i,:],t1))
# predictions
y_pred = np.array(y_pred)
# prediction
y_pred_BNN = np.exp(yscaler.inverse_transform(y_pred))[:,0]
# Let's check how close these are to the Full Mdel Predictions (Piecewise Approach)
df_1 = full_test_predictions.copy()
df_2 = pd.DataFrame(y_pred_BNN, columns=['Price_NN'])
full_test_predictions = pd.concat([df_1, df_2], axis=1)
full_test_predictions
full_test_predictions.drop(columns=['Price_NN','Price_NN_7'], inplace=True)<jupyter_output><empty_output><jupyter_text>**Comments** MAPE_BNN is the MAPE score when comparing the baseline estimated prices on the test dataset with the estimated prices from the Bayesian Neural Network Model with all key features. This score of 38.4% is not great performance. The model requires tuning. Results:
1) No tuning = 38.4%
2) Learning rate 0.001 = 36.0%
3) Learning_rate 0.0001 = 37.8%
4) Addition of NUTS Sampler = 37.1%
Recommend, if time permits, a GridSearch approach to hyperparameter tuning.<jupyter_code># Save this file as a CSV
full_test_predictions.to_csv(path+'ET5003_Kaggle_RobertBarrett_20157347.csv')<jupyter_output><empty_output>
|
permissive
|
/ET5003_Kaggle_RobertBarrett_20157347 (Code Complete).ipynb
|
rcb2021/ET5003_SEM1_2021-3
| 35 |
<jupyter_start><jupyter_text># Inference in PyTorch, what do the wrappers mean? What's best?
> A tour through PyTorch's various context managers, torch script, and comparing performance
- toc: true
- badges: true
- comments: true
- image: images/chart-preview.png
- category: torch---
This blog is also a Jupyter notebook available to run from the top down. There will be code snippets that you can then run in any environment. In this section I will be posting what version of `torch` I am currently running at the time of writing this:
* `torch`: 1.10.2
Benchmarks were ran on a NVIDIA RTX 3070 Laptop GPU
---## Why are we doing this?
Earlier today, [Francesco Pochetti](https://twitter.com/Fra_Pochetti) had pinged on the fastai discord asking if when using inference for a torch model, whether `no_grad` was needed when the model has been scripted. This then got me curious on timings, which as we've seen previously I *love* to do benchmarks like this!
So, I followed along PyTorch's fantastic [inference tutorial using TorchScript](https://pytorch.org/tutorials/recipes/torchscript_inference.html) and went to work!
What we'll explore in this article are the three "modes" for running a torch model:
- Regular
- `no_grad`
- `inference_mode`
How each of them differ in what they do, and overall how the timings for each performed.
For the initial testing, we'll use a resnet18 on three different batch sizes (1, 16, and 64) to see the full effect of our efforts## Okay, so what are these [modes](https://pytorch.org/docs/stable/notes/autograd.html#grad-modes)?
When we simply call `model(torch.tensor([...]))`, a trace of the functions called is stored, along with the results of each layer, so that later gradients can be calculated if needed. This can become a time sink, and greatly increases the memory being used during inference. To speed things up, in pytorch you would typically wrap the call to `model()` with a `no_grad()` context manager. Under this, computations are never recorded in the model as the inference is performed, and looks like so:
```python
with torch.no_grad():
model(torch.tensor([...]))
```
Finally, we get to `inference_mode` which is the **extreme** version of `no_grad`. With this context manager, you should assume that you'll **never** need to have any recordings done in the backwards of the graph (like `no_grad`), *and* any tensors made during inference mode won't be used for any computations touching autograd later. Hence the naming, `inference_mode`.
It looks like so:
```python
with torch.inference_mode():
model(torch.tensor([...]))
```## Making a Baseline
When doing experiments, it's always important to make proper benchmarks! Since we're comparing three modes, as well as two different models (torch scripted vs not), the the initial benchmark will be of our base model and torch scripted model without any context managers.
First let's make our models:<jupyter_code>import torch
from torchvision.models import resnet18
baseline_resnet = resnet18(pretrained=True).eval()
scripted_resnet = torch.jit.script(baseline_resnet).eval()<jupyter_output><empty_output><jupyter_text>Next we'll setup our batches:<jupyter_code>bs = [1,16,64]
batches = [torch.rand(size, 3, 224, 224) for size in bs]<jupyter_output><empty_output><jupyter_text>And finally set them all to CUDA<jupyter_code>baseline_resnet.cuda()
scripted_resnet.cuda()
for i in range(len(batches)):
batches[i] = batches[i].cuda()<jupyter_output><empty_output><jupyter_text>We'll also keep this interesting by keeping track of the allocated memory used by each. First we'll grab the current memory being used:<jupyter_code>def get_mb(key):
"A helpful function to get a readable size of an allocation"
sz = torch.cuda.memory_stats()[key]
return sz // 1024 // 1024
get_mb("allocated_bytes.all.current")<jupyter_output><empty_output><jupyter_text>This is how much our current memory usage is, and then we can track the peak memory usage once we start doing inference!Lastly we'll make some dictionaries to provide quick access to what we need:<jupyter_code>import contextlib
modes = {
"none":contextlib.suppress,
"no_grad":torch.no_grad,
"inference_mode":torch.inference_mode
}
models = {
"baseline":baseline_resnet,
"scripted":scripted_resnet
}
ranges = {
1:1000,
16:100,
64:10
}<jupyter_output><empty_output><jupyter_text>Now we just wrap up our configuration to get us some benchmarks! The latter half of this blog will be looking at the data:<jupyter_code>import time
from prettytable import PrettyTable
def benchmark_modes():
overall_reports = []
for mode in ["none", "no_grad", "inference_mode"]:
for batch in batches:
num_times = ranges[batch.shape[0]]
for model_type in ["baseline", "scripted"]:
total_time = 0
total_memory = 0
for i in range(num_times):
torch.cuda.reset_peak_memory_stats()
initial_memory = get_mb("allocated_bytes.all.current")
start_time = time.perf_counter_ns()
with modes[mode]():
_ = models[model_type](batch)
torch.cuda.synchronize()
total_time += (time.perf_counter_ns() - start_time)/1e6
peak_memory = get_mb("allocated_bytes.all.peak")
total_memory += peak_memory - initial_memory
overall_reports.append(
{
"mode":mode,
"batch_size":batch.shape[0],
"model_type":model_type,
"time":round(total_time/num_times, 2),
"memory_used":round(total_memory/num_times, 2),
}
)
return overall_reports
results = benchmark_modes()<jupyter_output><empty_output><jupyter_text>## Examining the Results
Let's dive deep into our results. First we'll look at everything, based on the context manager and the batch size:<jupyter_code>#hide_input
import copy
print("Experiment Results:")
table = PrettyTable(["Context Mode", "Batch Size", "Model Type", "Average Time Taken (ms)", "Average Total Memory Used (mb)"])
prev_bs = None
prev_mode = None
reports = copy.deepcopy(results)
for report in reports:
if prev_mode == report["mode"]:
report["mode"] = ""
else:
prev_mode = None
if prev_bs == report["batch_size"]:
report["batch_size"] = ""
else:
prev_bs = None
if prev_bs is None: prev_bs = report["batch_size"]
if prev_mode is None: prev_mode = report["mode"]
table.add_row(report.values())
print(table)<jupyter_output>Experiment Results:
+----------------+------------+------------+-------------------------+--------------------------------+
| Context Mode | Batch Size | Model Type | Average Time Taken (ms) | Average Total Memory Used (mb) |
+----------------+------------+------------+-------------------------+--------------------------------+
| none | 1 | baseline | 1.78 | 33.0 |
| | | scripted | 1.39 | 33.0 |
| | 16 | baseline | 7.65 | 345.95 |
| | | scripted | 7.69 | 346.0 |
| | 64 | baseline | 26.51 | 1389.5 |
| | | scripted | 26.53 | 1389.5 |
| no_grad | 1 | baseline |[...]<jupyter_text>We can see that generally the scripted model tends to be slightly faster, independent of the context manager being used. It also uses the same memory footprint as the non-scripted model.
But what if we compare by each context manager themselves?<jupyter_code>#hide
table = PrettyTable(["Model Type", "Batch Size", "Context Mode", "Average Time Taken (ms)", "Average Total Memory Used (mb)"])
prev_bs = None
prev_mode = None
reports = copy.deepcopy(results)
reports = sorted(reports, key=lambda x: (x["model_type"], x["batch_size"]))
for report in reports:
table.add_row([report[key] for key in ["model_type", "batch_size", "mode", "time", "memory_used"]])
#hide_input
for bs, (start,end) in [[1,(0,3)],[16,(3,6)],[64,(6,9)]]:
print(
table.get_string(
sort_key= lambda x: (x["Batch Size"], x["Model Type"]),
fields = list(table.field_names)[2:],
title=f"Baseline Model, Batch Size {bs}",
start=start,
end=end,
hrules=0
)
)<jupyter_output>+---------------------------------------------------------------------------+
| Baseline Model, Batch Size 1 |
+----------------+-------------------------+--------------------------------+
| Context Mode | Average Time Taken (ms) | Average Total Memory Used (mb) |
+----------------+-------------------------+--------------------------------+
| none | 1.78 | 33.0 |
| no_grad | 1.63 | 13.0 |
| inference_mode | 1.58 | 13.0 |
+----------------+-------------------------+--------------------------------+
+---------------------------------------------------------------------------+
| Baseline Model, Batch Size 16 |
+----------------+-------------------------+--------------------------------+
| Context Mode | Average Time Taken (ms) | Average Total Memor[...]<jupyter_text>For our non-scripted model, we find that for a batch size of 1, inference mode does the best! We see an average speedup of 12%!
However, as the batch size increases, this speedup becomes less and less radical, becoming only a fraction of a millisecond.
> Note: Notice the importance of having any context manager vs none in the total memory used. We reduced it from 1.3gb to 392mb being used, which is important!
Does this pattern continue for our scripted model?<jupyter_code>#hide_input
for bs, (start,end) in [[1,(9,12)],[16,(12,15)],[64,(15,18)]]:
print(
table.get_string(
sort_key= lambda x: (x["Batch Size"], x["Model Type"]),
fields = list(table.field_names)[2:],
title=f"Scripted Model, Batch Size {bs}",
start=start,
end=end,
hrules=0
)
)<jupyter_output>+---------------------------------------------------------------------------+
| Scripted Model, Batch Size 1 |
+----------------+-------------------------+--------------------------------+
| Context Mode | Average Time Taken (ms) | Average Total Memory Used (mb) |
+----------------+-------------------------+--------------------------------+
| none | 1.39 | 33.0 |
| no_grad | 1.32 | 13.0 |
| inference_mode | 1.29 | 13.0 |
+----------------+-------------------------+--------------------------------+
+---------------------------------------------------------------------------+
| Scripted Model, Batch Size 16 |
+----------------+-------------------------+--------------------------------+
| Context Mode | Average Time Taken (ms) | Average Total Memor[...]<jupyter_text>Again, we do see this pattern occur even here! But, it looks like we have a time decrease, doesn't it? Our scripted model actually is a decent chunk faster in some cases:<jupyter_code>#hide_input
import operator
for bs, (start,end) in [[1,(18,12)],[16,(12,6)],[64,(6,0)]]:
print(
table.get_string(
sort_key=operator.itemgetter(2,3),
fields = table.field_names[:1] + table.field_names[2:],
reversesort=True,
sortby="Batch Size",
title=f"Scripted Model, Batch Size {bs}",
start=end,
end=start,
hrules=0
)
)<jupyter_output>+----------------------------------------------------------------------------------------+
| Scripted Model, Batch Size 1 |
+------------+----------------+-------------------------+--------------------------------+
| Model Type | Context Mode | Average Time Taken (ms) | Average Total Memory Used (mb) |
+------------+----------------+-------------------------+--------------------------------+
| baseline | none | 1.78 | 33.0 |
| scripted | none | 1.39 | 33.0 |
| baseline | no_grad | 1.63 | 13.0 |
| scripted | no_grad | 1.32 | 13.0 |
| baseline | inference_mode | 1.58 | 13.0 |
| scripted | inference_mode | 1.29 | 13.0 |[...]<jupyter_text>We see it specifically packing the punch when there was a batch size of 1. Otherwise no matter the context manager used, it always added in a few hundredth's of a second of time.
But when does the loss of value happen? Let's find out.
We'll run a fresh set of benchmarks, examining the batch size from 1 to 8:<jupyter_code>for i in range(1,9):
ranges[i] = 100
batches = [torch.rand(i, 3, 224, 224) for i in range(1,9)]
for i in range(len(batches)):
batches[i] = batches[i].cuda()
results = benchmark_modes()<jupyter_output><empty_output><jupyter_text>Next, we'll plot a chart of `batch_size` x `time (ms)`, looking at the distribution based on each kind:<jupyter_code>import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
df = pd.DataFrame(results)
df.columns = 'mode', 'Batch Size', 'model_type', 'Time (ms)', 'memory_used'
fig, ax = plt.subplots()
i = 0
colors = "b","g",'r','c','m','y'
for (key, grp) in df.groupby(['mode']):
for (key2, grp2) in grp.groupby(["model_type"]):
ax = grp2.plot(ax=ax, kind='scatter', x='Batch Size', y='Time (ms)', label=f'{key2}, {key}', c=colors[i])
i += 1
plt.legend(loc='best')
plt.show()<jupyter_output><empty_output><jupyter_text>We see that for a single item and two, it's **extremely** important to use the right context manager, but as we increase our batch size it matters less and less until we hit 8.
For morbid curiosity, I decided to check how 8 to 16 might look, and here's those results:<jupyter_code>for i in range(8,16):
ranges[i] = 100
batches = [torch.rand(i, 3, 224, 224) for i in range(8,16)]
for i in range(len(batches)):
batches[i] = batches[i].cuda()
results = benchmark_modes()
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
df = pd.DataFrame(results)
df.columns = 'mode', 'Batch Size', 'model_type', 'Time (ms)', 'memory_used'
fig, ax = plt.subplots()
i = 0
colors = "b","g",'r','c','m','y'
for (key, grp) in df.groupby(['mode']):
for (key2, grp2) in grp.groupby(["model_type"]):
ax = grp2.plot(ax=ax, kind='scatter', x='Batch Size', y='Time (ms)', label=f'{key2}, {key}', c=colors[i])
i += 1
plt.legend(loc='best')
plt.show()<jupyter_output><empty_output><jupyter_text>We find yet again that the distribution between the different modes is ~ <.1 milliseconds.## Finally, what about CPU?
Here's our experiment again, performed on a CPU:<jupyter_code>#hide
for i in range(1,8):
ranges[i] = 10
batches = [torch.rand(i, 3, 224, 224) for i in range(1,8)]
for i in range(len(batches)):
batches[i] = batches[i].cpu()
for model in models.values():
model.cpu()
results = benchmark_modes()
#hide_input
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
df = pd.DataFrame(results)
df.columns = 'mode', 'Batch Size', 'model_type', 'Time (ms)', 'memory_used'
fig, ax = plt.subplots()
i = 0
colors = "b","g",'r','c','m','y'
for (key, grp) in df.groupby(['mode']):
for (key2, grp2) in grp.groupby(["model_type"]):
ax = grp2.plot(ax=ax, kind='scatter', x='Batch Size', y='Time (ms)', label=f'{key2}, {key}', c=colors[i])
i += 1
plt.legend(loc='best')
plt.show()<jupyter_output><empty_output>
|
permissive
|
/_notebooks/2022-05-14-PyTorchInference.ipynb
|
muellerzr/fastblog
| 14 |
<jupyter_start><jupyter_text># 反応拡散モデルの結果プロット用ノートブック
内容は前回のセルオートマトンのプロット用ノートブックと同じ<jupyter_code>%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
from ipywidgets import interact
# 関数の定義
def plot_RD(data, step, height=100):
"""
data: 反応拡散モデルのの出力結果を読み込んだNumPyの多次元配列
step: ステップ
height: 1ステップのサイズ(何列か)
"""
plt.figure( figsize=(5, 5), dpi=120)
plt.matshow(data[step*height:(step+1)*height], cmap=plt.cm.gray, fignum=1)
plt.show()<jupyter_output><empty_output><jupyter_text>## データの読み込みとプロット<jupyter_code># データ(rd_u.csv)の読み込み
data = np.genfromtxt("./11-2.csv", dtype="float",delimiter=",")
# プロット
height=100
interact(lambda step:plot_RD(data, step, height), step=(0,int(len(data)/height)-1,1))<jupyter_output><empty_output>
|
no_license
|
/11/plot_RD.ipynb
|
ishiharatomomi/CompBio2018Code
| 2 |
<jupyter_start><jupyter_text># BY: Mohamed Ashraf Gaber## The Data.
#### This data is for markets. It contains the markets' sales. The columns: The Product name, its price, The time for each order and the address.## Importing libraries I will need.<jupyter_code>%matplotlib inline
import pandas as pd
import os
import matplotlib.pyplot as plt
import seaborn as sns
from itertools import combinations
from collections import Counter<jupyter_output><empty_output><jupyter_text>### The data contains 12 files for each month, so first I'll concat them all in one single file.<jupyter_code>path = 'Sales_Data' # The path of the files.
# Getting the data files and ignoring the hidden files.
data_files = [file for file in os.listdir(path) if not file.startswith('.')]
all_data = pd.DataFrame() # This data frame will contain all data.
# Looping for each file
for file in data_files:
df_small = pd.read_csv(f'./{path}/{file}') # Reading the file.
all_data = pd.concat([all_data, df_small], axis=0) # Concat this file with the big one.
all_data.info()
all_data.head()<jupyter_output><empty_output><jupyter_text>### Exporting the final data to CSV file.<jupyter_code>all_data.to_csv('./Final_Data/final_data.csv', index=False)<jupyter_output><empty_output><jupyter_text>## Importing the data.<jupyter_code>df = pd.read_csv('./Final_Data/final_data.csv')
df.head()<jupyter_output><empty_output><jupyter_text>## Some exploratory data analysis and data visualization.<jupyter_code>df.info()
df.describe()<jupyter_output><empty_output><jupyter_text>## Checking for null values.<jupyter_code>df.isnull().sum()<jupyter_output><empty_output><jupyter_text>## Visualizing the null values.<jupyter_code>plt.figure(figsize=(12, 8));
sns.heatmap(df.isnull(), cbar=False, yticklabels=False, cmap='viridis');<jupyter_output><empty_output><jupyter_text>#### Dropping the null values.<jupyter_code>df.dropna(inplace=True)
plt.figure(figsize=(12, 8));
sns.heatmap(df.isnull(), cbar=False, yticklabels=False, cmap='viridis');
df.isnull().sum()<jupyter_output><empty_output><jupyter_text>### Printing some duplicates rows that I'll drop from the data.<jupyter_code>df[df['Order Date'].str[:] != 'Order Date'].head()
df = df[df['Order Date'].str[:] != 'Order Date']
df[df['Order Date'].str[:] == 'Order Date']<jupyter_output><empty_output><jupyter_text>#### Converting the Order Data column to datetime type instead of object.<jupyter_code>df['Order Date'] = pd.to_datetime(df['Order Date'])
df.info()<jupyter_output><class 'pandas.core.frame.DataFrame'>
Int64Index: 185950 entries, 0 to 186849
Data columns (total 6 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 Order ID 185950 non-null object
1 Product 185950 non-null object
2 Quantity Ordered 185950 non-null object
3 Price Each 185950 non-null object
4 Order Date 185950 non-null datetime64[ns]
5 Purchase Address 185950 non-null object
dtypes: datetime64[ns](1), object(5)
memory usage: 9.9+ MB
<jupyter_text>### Creating some columns which will help me in the Analysis<jupyter_code>df['month'] = df['Order Date'].dt.month # Creating a column for the month.
df['hour'] = df['Order Date'].dt.hour # Creating a column for the hour.
num_cols = ['Order ID', 'Quantity Ordered', 'Price Each']
# Converting the num_cols to numeric instead of object.
for col in num_cols:
df[col] = pd.to_numeric(df[col])
df.info()
df.head()<jupyter_output><empty_output><jupyter_text>### Creating a column for the final price.<jupyter_code>df['final_price'] = df.apply(lambda x: x['Quantity Ordered'] * x['Price Each'], axis=1)
df.head()<jupyter_output><empty_output><jupyter_text>### Creating a column for City.<jupyter_code>def get_city_state(address):
city = address.split(', ')[1]
state = address.split(', ')[2][:2]
return city + ', ' + state
df['city'] = df['Purchase Address'].apply(get_city_state)
df.head()<jupyter_output><empty_output><jupyter_text>### Creating some helpers functions that will help me in visualization.<jupyter_code>def create_bar(x, height, xlabel, ylabel, title, ticks=None, rotation=None):
plt.figure(figsize=(12, 8));
plt.bar(x=x, height=height);
plt.xlabel(xlabel);
plt.ylabel(ylabel);
plt.title(title);
plt.xticks(ticks=ticks, rotation=rotation);
def create_plot(x, y, xlabel, ylabel, title, ticks=None, rotation=None):
plt.figure(figsize=(12, 8));
plt.plot(x, y);
plt.xlabel(xlabel);
plt.ylabel(ylabel);
plt.title(title);
plt.xticks(ticks=ticks, rotation=rotation);<jupyter_output><empty_output><jupyter_text>## Question 1: What was the best month for sales?<jupyter_code>month_sales = df.groupby('month').sum()['final_price']
month_sales
create_bar(x=month_sales.index, height=month_sales.values,
xlabel='Month', ylabel='Sales', title='Sales for each Month',
ticks=list(range(1, 13)))<jupyter_output><empty_output><jupyter_text>## Answering to Question 1: The best month for sales was December.
## Question 2: What city sold the most products?<jupyter_code>city_sales = df.groupby('city').sum()['final_price'].sort_values()
city_sales
create_bar(x=city_sales.index, height=city_sales.values,
xlabel='City', ylabel='Sales', title='Sales for each City', rotation=15)<jupyter_output><empty_output><jupyter_text>## Answering to Question 2: The city that sold most products was San Francisco, CA.
## Question 3: What was the best hour to display advertisements?<jupyter_code>hour_count = df.groupby('hour').count()['Order ID']
hour_count[:5]
create_plot(x=hour_count.index, y=hour_count.values,
xlabel='Hour', ylabel='Number of Orders', title='Number of Orders every Hour',
ticks=list(range(0, 24)))<jupyter_output><empty_output><jupyter_text>## Answering to Question 3: The best hours to display advertisements were 11 A.M and 7 P.M.
## Question 4: What products are most often sold together?<jupyter_code>df.duplicated(keep=False).sum()
df[df['Order ID'] == 178158]<jupyter_output><empty_output><jupyter_text>### Creating new data frame that will have the Order ID and all its Products.<jupyter_code>df_products = df[df['Order ID'].duplicated(keep=False)]
df_products['Product'] = df_products.groupby('Order ID')['Product'].transform(lambda x: ','.join(x))
df_products = df_products[['Order ID', 'Product']].drop_duplicates()
df_products.head()<jupyter_output>D:\Anaconda\lib\site-packages\ipykernel_launcher.py:3: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
This is separate from the ipykernel package so we can avoid doing imports until
<jupyter_text>### Getting the most 2 products that were sold together.<jupyter_code>count = Counter()
for row in df_products['Product']:
row_products = row.split(',')
count.update(Counter(combinations(row_products, 2)))
for key, value in count.most_common(10):
print(key, value)<jupyter_output>('iPhone', 'Lightning Charging Cable') 1005
('Google Phone', 'USB-C Charging Cable') 987
('iPhone', 'Wired Headphones') 447
('Google Phone', 'Wired Headphones') 414
('Vareebadd Phone', 'USB-C Charging Cable') 361
('iPhone', 'Apple Airpods Headphones') 360
('Google Phone', 'Bose SoundSport Headphones') 220
('USB-C Charging Cable', 'Wired Headphones') 160
('Vareebadd Phone', 'Wired Headphones') 143
('Lightning Charging Cable', 'Wired Headphones') 92
<jupyter_text>### Getting the most 3 products that were sold together.<jupyter_code>count = Counter()
for row in df_products['Product']:
row_products = row.split(',')
count.update(Counter(combinations(row_products, 3)))
for key, value in count.most_common(10):
print(key, value)<jupyter_output>('Google Phone', 'USB-C Charging Cable', 'Wired Headphones') 87
('iPhone', 'Lightning Charging Cable', 'Wired Headphones') 62
('iPhone', 'Lightning Charging Cable', 'Apple Airpods Headphones') 47
('Google Phone', 'USB-C Charging Cable', 'Bose SoundSport Headphones') 35
('Vareebadd Phone', 'USB-C Charging Cable', 'Wired Headphones') 33
('iPhone', 'Apple Airpods Headphones', 'Wired Headphones') 27
('Google Phone', 'Bose SoundSport Headphones', 'Wired Headphones') 24
('Vareebadd Phone', 'USB-C Charging Cable', 'Bose SoundSport Headphones') 16
('USB-C Charging Cable', 'Bose SoundSport Headphones', 'Wired Headphones') 5
('Vareebadd Phone', 'Bose SoundSport Headphones', 'Wired Headphones') 5
<jupyter_text>## Answering to Question 4: The most 2 products were iPhone and Lightning Charging Cable. And the most 3 were Google Phone, USB-C Charging Cable and Wired Headphones.
## Question 5: Which product has the highest profit?<jupyter_code>product_sales = df.groupby('Product')['final_price'].sum().sort_values()
product_sales
create_bar(x=product_sales.index, height=product_sales.values,
xlabel='Product', ylabel='Sales', title='Sales for each Product', rotation=90)<jupyter_output><empty_output><jupyter_text>## Answering to Question 5: It was Macbook Pro Laptop
## Question 6: What product sold the most?<jupyter_code>product_quantities = df.groupby('Product')['Quantity Ordered'].sum().sort_values()
product_quantities
create_bar(x=product_quantities.index, height=product_quantities.values,
xlabel='Product', ylabel='Quantity Ordered', title='Products and its Quantity Ordered',
rotation=90)<jupyter_output><empty_output><jupyter_text>### Plotting the products with its price and its Quantity Ordered.<jupyter_code>prices = df.groupby('Product').mean()['Price Each']
prices = prices.reindex(index=product_quantities.index)
fig, ax1 = plt.subplots(figsize=(12, 8));
ax2 = ax1.twinx();
ax1.bar(x=product_quantities.index, height=product_quantities.values, color='g');
ax1.set_xlabel('Products');
ax1.set_ylabel('Quantity Ordered');
ax1.set_title('Products and its Quantity Ordered');
ax1.set_xticklabels(product_quantities.index, rotation=90);
ax2.plot(product_quantities.index, prices, 'b');
ax2.set_ylabel('Each Price');
prices<jupyter_output><empty_output>
|
no_license
|
/Sales_Analysis.ipynb
|
Mohamed-code-13/Sales-Analysis-for-Markets
| 24 |
<jupyter_start><jupyter_text>## Homework 3: Assignment 2
INSTRUCTIONS
1. States the question you want to ask, and formulates the Null and Alternative hypothesis (remember the confidence level!)
2. Use pandas to read in the CitiBike files, either from the DF, or locally, but you must be able to download them on the spot (so the TA can reproduce your work).
3. Display the top few rows of the DF in your notebook. This table must be rendered.
4. Display the reducted dataframe. This table must be rendered.
5. Plot your data distributions.## IDEA:
Does age of gender affect ridership? (i.e. Are men above age 40 are less likely than women above age 40 to ride a citi bike?)
## NULL HYPOTHESIS:
The ratio of man above age 45 to man age 45 or below riding a bike is the same or greater than the ratio of woman above age 45 to woman age 45 or below riding a bike
$H_0$ : (Women_above45 / Women_45orbelow) <= (Men_above45/Men_45orbelow)
$H_1$ : (Women_above45 / Women_45orbelow) > (Men_above45/Men_45orbelow)
## ALTERNATIVE HYPOTHESIS:
The ratio of man above age 45 to man age 45 or below riding a bike is the smaller than the ratio of woman above age 45 to man age 45 or below riding a bike.
I will use a significance leve $\alpha=0.05$<jupyter_code>from __future__ import print_function, division
import pylab as pl
import pandas as pd
import numpy as np
import pandas as pd
import os
%pylab inline
if os.getenv ('PUI2016') is None:
print ("Must set env variable PUI2016")
if os.getenv ('PUIDATA') is None:
print ("Must set env variable PUIDATA")
os.getenv ('PUI2016')
os.getenv ('PUIDATA')
def getCitiBikeCSV(datestring):
print ("Downloading", datestring)
### First I will check that it is not already there
if not os.path.isfile(os.getenv("PUIDATA") + "/" + datestring + "-citibike-tripdata.csv"):
if os.path.isfile(datestring + "-citibike-tripdata.csv"):
# if in the current dir just move it
if os.system("mv " + datestring + "-citibike-tripdata.csv " + os.getenv("PUIDATA")):
print ("Error moving file!, Please check!")
#otherwise start looking for the zip file
else:
if not os.path.isfile(os.getenv("PUIDATA") + "/" + datestring + "-citibike-tripdata.zip"):
if not os.path.isfile(datestring + "-citibike-tripdata.zip"):
os.system("curl -O https://s3.amazonaws.com/tripdata/" + datestring + "-citibike-tripdata.zip")
### To move it I use the os.system() functions to run bash commands with arguments
os.system("mkdir " + os.getenv("PUIDATA")) ##you have to make a new file directory first
os.system("mv " + datestring + "-citibike-tripdata.zip " + os.getenv("PUIDATA"))
### unzip the csv
os.system("unzip " + os.getenv("PUIDATA") + "/" + datestring + "-citibike-tripdata.zip -d " + os.getenv("PUIDATA"))
## NOTE: old csv citibike data had a different name structure.
if '2014' in datestring:
os.system("mv " + datestring[:4] + '-' + datestring[4:] +
"/ -/ Citi/ Bike/ trip/ data.csv " + datestring + "-citibike-tripdata.csv")
os.system("mv " + datestring + "-citibike-tripdata.csv " + os.getenv("PUIDATA"))
### One final check:
if not os.path.isfile(os.getenv("PUIDATA") + "/" + datestring + "-citibike-tripdata.csv"):
print ("WARNING!!! something is wrong: the file is not there!")
else:
print ("file in place, you can continue")
os.system("rm " + os.getenv("PUIDATA") + "/" + datestring + "-citibike-tripdata.zip")
datestring = '201503'
getCitiBikeCSV(datestring)
df = pd.read_csv(os.getenv("PUIDATA") + "/" +
datestring + '-citibike-tripdata.csv')
df.head()
df.drop(['starttime', 'stoptime', 'start station id',
'start station name', 'start station latitude',
'start station longitude', 'end station id', 'end station name',
'end station latitude', 'end station longitude', 'bikeid'], axis=1, inplace=True)
df.head()
df1 = df[df.usertype != 'Customer']
df1.head()
df_m_above45 = (df1['birth year'][df1['gender'] == 1]).groupby(df1['birth year'] < 1971.0).count()
df_m_above45
error_m = np.sqrt(df_m_above45)
df_w_above45 = (df1['birth year'][df1['gender'] == 2]).groupby(df1['birth year'] < 1971.0).count()
error_w = np.sqrt(df_w_above45)
fig = pl.figure(figsize(10,5))
norm_w = 1
norm_m = 1
ax=((df_m_above45) / norm_m).plot(kind="bar", yerr=[
((error_m) / norm_m, (error_m) / norm_m)], alpha=0.5, rot=0, label='men bikers')
((df_w_above45) / norm_w).plot(kind="bar", color='IndianRed', alpha=0.5, rot=0, yerr=[
((error_w) / norm_w, (error_w) / norm_w)], label='women bikers')
ax.xaxis.set_ticklabels(['age 45 or below','age above 45'], fontsize=15)
ax.set_ylabel ("Number of rides", fontsize=20)
ax.set_xlabel ("Age", fontsize=20)
pl.legend(['men bikers','women bikers'], loc='best', fontsize=15)<jupyter_output><empty_output><jupyter_text>## Figure 1: Distribution of Citibike bikers by Age in March 2015, absolute counts<jupyter_code>fig = pl.figure(figsize(10,5))
norm_m = df_m_above45.sum()
ax=((df_m_above45) / norm_m).plot(kind="bar", color='Blue', yerr=[
((error_m) / norm_m, (error_m) / norm_m)], alpha=0.5, rot=0, label='men bikers')
norm_w = df_w_above45.sum()
((df_w_above45) / norm_w).plot(kind="bar", color='IndianRed', alpha=0.4, rot=0, yerr=[
((error_w) / norm_w, (error_w) / norm_w)], label='women bikers')
ax.xaxis.set_ticklabels(['age 45 or below','age above 45'], fontsize=15)
ax.set_ylabel ("Fractions of rides", fontsize=20)
ax.set_xlabel ("Age", fontsize=20)
pl.legend(['men bikers','women bikers'], loc='best', fontsize=15)<jupyter_output><empty_output><jupyter_text>## Figure 2: Distribution of Citibike bikers by age in March 2015, normalized<jupyter_code>fig = pl.figure(figsize(15,6))
ax1 = fig.add_subplot(121)
ax2 = fig.add_subplot(122)
ax1.set_xticks([])
ax2.set_xticks([])
m_above45 = sum(df_m_above45[1]) * 1.0 / norm_m
m_45orbelow = sum(df_m_above45[0]) * 1.0 / norm_m
eAbove45_m = np.sqrt(sum(error_m[1]**2)) / norm_m
e45orbelow_m = np.sqrt(sum(error_m[0]**2)) / norm_m
w_above45 = sum(df_w_above45[1]) * 1.0 / norm_w
w_45orbelow = sum(df_w_above45[0]) * 1.0 / norm_w
eAbove45_w = np.sqrt(sum(error_w[1]**2)) / norm_w
e45orbelow_w = np.sqrt(sum(error_w[0]**2)) / norm_w
print("Men: above age 45:{0:.3f}, age 45 or below:{1:.3f}, above age 45 error:{2:.3f}, age 45 or below error:{3:.3f}"\
.format(m_above45, m_45orbelow, eAbove45_m, e45orbelow_m))
print("Women: above age 45:{0:.3f}, age 45 or below:{1:.3f}, above age 45 error:{2:.3f}, age 45 or below error:{3:.3f}"\
.format(w_above45, w_45orbelow, eAbove45_w, e45orbelow_w))
ax1.errorbar([0.4], [m_above45], yerr=[eAbove45_m], fmt='o', label='men')
ax1.errorbar([0.2], [w_above45], yerr=[eAbove45_w], fmt='o', label='women')
ax1.set_xlim(0, 0.5)
ax2.errorbar([0.4], [m_45orbelow], yerr=[e45orbelow_m], fmt='o', label='men')
ax2.errorbar([0.2], [w_45orbelow], yerr=[e45orbelow_w], fmt='o', label='women')
ax1.set_xlim(0, 0.5)
ax1.set_title("Above age 45")
ax2.set_title("Age 45 or below")
ax2.set_ylabel("Fraction of normalized rides by age", fontsize = 20)
ax1.set_ylabel("Fraction of normalized rides by age", fontsize = 20)
pl.xlim(-0.5, 1.5)
pl.legend(fontsize = 20)<jupyter_output>Men: above age 45:0.345, age 45 or below:0.655, above age 45 error:0.001, age 45 or below error:0.002
Women: above age 45:0.307, age 45 or below:0.693, above age 45 error:0.002, age 45 or below error:0.003
|
no_license
|
/HW3_fhl204/HW3_2_fhl204.ipynb
|
fhl204/PUI2016_fhl204
| 3 |
<jupyter_start><jupyter_text># Model Training
We are using the basic features derived from the `basic_featureset_constructor.py` script.
For full description of the data and the schema used, please refer to the README.md file in this directory and `basic_featureset_constructor.py`.<jupyter_code># perform imports
import numpy as np
import pandas as pd
# read in the data and establish the headers
headers = [
"label",
"video_url",
"mean_0",
"mean_1",
"mean_2",
"var_0",
"var_1",
"var_2",
"kurt_0",
"kurt_1",
"kurt_2",
"skew_0",
"skew_1",
"skew_2",
"mfcc_0",
"mfcc_1",
"mfcc_2",
"mfcc_3",
"mfcc_4",
"mfcc_5",
"mfcc_6",
"mfcc_7",
"mfcc_8",
"mfcc_9"
]
data_path = "basic_image_mfcc.csv"
df = pd.read_csv(data_path,
names=headers)
df
<jupyter_output><empty_output><jupyter_text>## Standardise by historical mean and variance<jupyter_code>def standardise_hist(row):
'''
helper function to do historical standardisation
The data are split evenly between the classes, however this is not
the case in real life, where the long term average will be totally dominated
by the "No Danger" class (in Co. Leitrim anyway...) so the long term average
values we will experience in the field will be very close to those found
in the "No Danger" class for that particular video.
To resolve this, simply standardise using the mean and variance of the
feature observed from "No Danger" frames from that particular video.
'''
video_url = row[headers[1]] # find video_url column
num_cols = row[headers[2:len(headers)]] # find numeric columns
hist_vals = df.loc[(df[headers[1]]==video_url)&(df[headers[0]]=="No_Danger"),headers[2:len(headers)]]
hist_mean = np.mean(hist_vals, axis=0)
hist_std = np.std(hist_vals, axis=0)
return (num_cols-hist_mean)/hist_std
df_std = df.apply(standardise_hist, axis=1, raw=False)
df_std.describe()
df.describe()
pd.set_option('display.max_rows', 500)
pd.set_option('display.max_columns', 500)
# merge the standardised data with the labels
# should see close to zero mean and one standard deviation for No Danger for each specific video
ml_df= df_std.merge(df[headers[:2]],how="inner",left_index=True, right_index=True).dropna()
ml_df.loc[(ml_df[headers[0]]=="No_Danger")&(ml_df[headers[1]]==df.iloc[0,1]),:].describe()<jupyter_output><empty_output><jupyter_text># Try Out Some Models<jupyter_code># split into training, validation and testing sets
# ensure that no video appears in more than one set so divide the frames by video_url
# find unique video urls
np.random.seed(0)
video_urls = ml_df.loc[:,headers[1]].unique()
# randomly sample
train_prop = 0.5 # save half the videos for validation run
val_prop = 0.25
train_idx = np.random.choice(len(video_urls),int(np.floor(train_prop*len(video_urls))),
replace=False)
val_idx_full = np.setdiff1d(np.arange(len(video_urls)), train_idx)
val_idx = np.random.choice(val_idx_full,int(np.floor(val_prop*len(video_urls))),
replace=False)
test_idx = np.setdiff1d(val_idx_full, val_idx)
# find the videos corresponding to these indices
train_videos = video_urls[train_idx]
val_videos = video_urls[val_idx]
test_videos = video_urls[test_idx]
ml_train = ml_df.loc[ml_df[headers[1]].isin(train_videos),:]
ml_val = ml_df.loc[ml_df[headers[1]].isin(val_videos) ,:]
ml_test = ml_df.loc[ml_df[headers[1]].isin(test_videos) ,:]
print("Train set shape: ", ml_train.shape)
print("Validation set shape: ",ml_val.shape)
print("Test set shape: ",ml_test.shape)
<jupyter_output>Train set shape: (4338, 24)
Validation set shape: (1947, 24)
Test set shape: (1617, 24)
<jupyter_text>## Logistic Regression
### Training<jupyter_code># load scikit learn logistic regression
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import train_test_split, ShuffleSplit
from sklearn.metrics import confusion_matrix
# hide convergence warnings
import warnings
warnings.filterwarnings("ignore")
# show progress
import progressbar
X = ml_train[headers[2:]]
y = np.where(ml_train[headers[0]]=="Danger",1,0) # 1: Danger, 0 No_Danger
def score_model(clf, X_test, y_test):
# get predictions
y_hat = clf.predict(X_test)
# get the confusion matrix
tn, fp, fn, tp = confusion_matrix(y_test, y_hat).ravel()
# get accuracy
acc = (tn+tp)/(tn+fp+fn+tp)
# get sensitivity
sens = tp/(tp+fn)
# get specificity
spec = tn/(tn+fp)
return acc, sens, spec
# perform bootstrapping to observe which coefficients can be dropped
b = 200
n_train = 0.5 # train on half, test on half
params = np.zeros((b,1+len(headers[2:]))) # storage for coefficeints
# storage for accuracy, sensitivity, specificity (for both train and test to check for overfitting)
scores = np.zeros((b, 6))
bs = ShuffleSplit(n_splits = b,
random_state=0,
test_size=0.5)
iter = 0
with progressbar.ProgressBar(max_value=b) as bar:
for train_index, test_index in bs.split(X):
X_train, X_test = X.iloc[train_index,:], X.iloc[test_index,:]
y_train, y_test = y[train_index], y[test_index]
# fit the classifier
clf = LogisticRegression(random_state=0,
solver="sag").fit(X_train,y_train)
# record the model parameters
params[iter,:] = np.squeeze(np.hstack((clf.intercept_[:,None], clf.coef_)))
# score the model on train data first
scores[iter,:3] = score_model(clf, X_train, y_train)
# score the model on test data
scores[iter,3:] = score_model(clf, X_test, y_test)
# iteration counter
bar.update(iter)
iter +=1
# plotting
import matplotlib.pyplot as plt
import seaborn as sns
# boxplot of the parameters
fig = plt.figure(figsize = (8,8))
ax = fig.add_subplot(2, 2, 1)
ax.boxplot(params)
ax.axhline(y=0, c="b")
ax.set_xticklabels(["intercept"]+headers[2:], rotation = 90)
# boxplot of the scorings, grouped by type if possible
# reconfigure so that the
ax = fig.add_subplot(2, 2, 2)
ax.boxplot(scores[:,[0,3]])
ax.set_xticklabels(["Training\nAccuracy", "Testing\nAccuracy"])
ax = fig.add_subplot(2, 2, 3)
ax.boxplot(scores[:,[1,4]])
ax.set_xticklabels(["Training\nSensitivity", "Testing\nSensitivity"])
ax = fig.add_subplot(2, 2, 4)
ax.boxplot(scores[:,[2,5]])
ax.set_xticklabels(["Training\nSpecificity", "Testing\nSpecificity"])
plt.subplots_adjust(hspace=0.3)
plt.savefig("lr.pdf")
plt.show()<jupyter_output><empty_output><jupyter_text>### Threshold Selection
Use the training set for training, and validation set for selecting threshold<jupyter_code>def adjusted_classes(y_scores, t):
"""
This function adjusts class predictions based on the prediction threshold (t).
Will only work for binary classification problems.
"""
return [1 if y >= t else 0 for y in y_scores]
def precision_recall_threshold(p, r, thresholds, t=0.5):
"""
plots the precision recall curve and shows the current value for each
by identifying the classifier's threshold (t).
"""
# generate new class predictions based on the adjusted_classes
# function above and view the resulting confusion matrix.
y_pred_adj = adjusted_classes(y_scores, t)
print(pd.DataFrame(confusion_matrix(y_test, y_pred_adj),
columns=['pred_neg', 'pred_pos'],
index=['neg', 'pos']))
# plot the curve
plt.figure(figsize=(8,8))
plt.title("Precision and Recall curve ^ = current threshold")
plt.step(r, p, color='b', alpha=0.2,
where='post')
plt.fill_between(r, p, step='post', alpha=0.2,
color='b')
plt.ylim([0.5, 1.01]);
plt.xlim([0.5, 1.01]);
plt.xlabel('Recall');
plt.ylabel('Precision');
# plot the current threshold on the line
close_default_clf = np.argmin(np.abs(thresholds - t))
plt.plot(r[close_default_clf], p[close_default_clf], '^', c='k',
markersize=15)
X_train, X_test = ml_train[:,2:], ml_val[:,2:]
y_train, y_test = np.where(ml_train[headers[0]]=="Danger",1,0), np.where(ml_val[headers[0]]=="Danger",1,0)
clf = LogisticRegression(random_state=0,
solver="sag").fit(X_train,y_train)
# get y_hat
y_hat = clf.predict_proba(X_test)[:, 1]
p, r, thresholds = precision_recall_curve(y_test, y_hat)<jupyter_output><empty_output>
|
no_license
|
/common/model/training/n3060_basic/.ipynb_checkpoints/n3060_basic-checkpoint.ipynb
|
mdunlop2/GapWatch
| 5 |
<jupyter_start><jupyter_text>
# Lab 6.4
# *PCA Lab*
**In this lab, we will:**
- Explore how PCA is related to correlation.
- Use PCA to perform dimensionality reduction.### 1. Load Data
Features are computed from a digitized image of a fine needle aspirate (FNA) of a breast mass. They describe characteristics of the cell nuclei present in the image. n the 3-dimensional space is that described in: [K. P. Bennett and O. L. Mangasarian: "Robust Linear Programming Discrimination of Two Linearly Inseparable Sets", Optimization Methods and Software 1, 1992, 23-34].
This database is also available through the UW CS ftp server: ftp ftp.cs.wisc.edu cd math-prog/cpo-dataset/machine-learn/WDBC/
Also can be found on UCI Machine Learning Repository: https://archive.ics.uci.edu/ml/datasets/Breast+Cancer+Wisconsin+%28Diagnostic%29
Attribute Information:
1) ID number 2) Diagnosis (M = malignant, B = benign) 3-32)
Ten real-valued features are computed for each cell nucleus:
a) radius (mean of distances from center to points on the perimeter) b) texture (standard deviation of gray-scale values) c) perimeter d) area e) smoothness (local variation in radius lengths) f) compactness (perimeter^2 / area - 1.0) g) concavity (severity of concave portions of the contour) h) concave points (number of concave portions of the contour) i) symmetry j) fractal dimension ("coastline approximation" - 1)
The mean, standard error and "worst" or largest (mean of the three largest values) of these features were computed for each image, resulting in 30 features. For instance, field 3 is Mean Radius, field 13 is Radius SE, field 23 is Worst Radius.
All feature values are recoded with four significant digits.
Missing attribute values: none
Class distribution: 357 benign, 212 malignant<jupyter_code># IMPORT LABRARIES
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.preprocessing import StandardScaler
%matplotlib inline
df = pd.read_csv('breast_cancer.csv', index_col = 'id')<jupyter_output><empty_output><jupyter_text>### 2. EDA
Explore dataset. Clean data. Find correlation. <jupyter_code>df.info()
df.drop(columns = ['Unnamed: 32'], inplace = True)<jupyter_output><empty_output><jupyter_text>### 3. Subset & Normalize
Subset the data to only include all columns except diagnosis. We will be comparing the principal components to age specifically, so we are leaving age out.<jupyter_code># ANSWER
X = df.copy()
X.drop(columns = ['diagnosis'], inplace = True)
X
features = X.columns
features
scaler = StandardScaler()
X_scaled = scaler.fit_transform(X)
X_scaled = pd.DataFrame(X_scaled, columns = ['radius_mean', 'texture_mean', 'perimeter_mean', 'area_mean',
'smoothness_mean', 'compactness_mean', 'concavity_mean',
'concave points_mean', 'symmetry_mean', 'fractal_dimension_mean',
'radius_se', 'texture_se', 'perimeter_se', 'area_se', 'smoothness_se',
'compactness_se', 'concavity_se', 'concave points_se', 'symmetry_se',
'fractal_dimension_se', 'radius_worst', 'texture_worst',
'perimeter_worst', 'area_worst', 'smoothness_worst',
'compactness_worst', 'concavity_worst', 'concave points_worst',
'symmetry_worst', 'fractal_dimension_worst'])
X_scaled
y = df.diagnosis
y<jupyter_output><empty_output><jupyter_text>### Calculate correlation matrix
We will be using the correlation matrix to calculate the eigenvectors and eigenvalues.<jupyter_code># ANSWER
correlation_matrix = X_scaled.corr()
correlation_matrix<jupyter_output><empty_output><jupyter_text>### 4. Calculate the eigenvalues and eigenvectors from the correlation matrix
numpy has a convenient function to calculate this:
eigenvalues, eigenvectors = np.linalg.eig(correlation_matrix)<jupyter_code># ANSWER
eigenvalues, eigenvectors = np.linalg.eig(correlation_matrix)
print('eigenvalues \n', eigenvalues)
print('eigenvectors \n', eigenvectors)<jupyter_output>eigenvalues
[1.32816077e+01 5.69135461e+00 2.81794898e+00 1.98064047e+00
1.64873055e+00 1.20735661e+00 6.75220114e-01 4.76617140e-01
4.16894812e-01 3.50693457e-01 2.93915696e-01 2.61161370e-01
2.41357496e-01 1.57009724e-01 9.41349650e-02 7.98628010e-02
5.93990378e-02 5.26187835e-02 4.94775918e-02 1.33044823e-04
7.48803097e-04 1.58933787e-03 6.90046388e-03 8.17763986e-03
1.54812714e-02 1.80550070e-02 2.43408378e-02 2.74394025e-02
3.11594025e-02 2.99728939e-02]
eigenvectors
[[ 2.18902444e-01 -2.33857132e-01 -8.53124284e-03 4.14089623e-02
-3.77863538e-02 1.87407904e-02 1.24088340e-01 7.45229622e-03
-2.23109764e-01 9.54864432e-02 4.14714866e-02 5.10674568e-02
1.19672116e-02 -5.95061348e-02 5.11187749e-02 -1.50583883e-01
2.02924255e-01 1.46712338e-01 -2.25384659e-01 -7.02414091e-01
2.11460455e-01 -2.11194013e-01 -1.31526670e-01 1.29476396e-01
1.92264989e-02 -1.82579441e-01 9.85526942e-02 -7.29289034e-02
-4.96986642e-02 6.85700057e-02]
[ 1.03724578e-[...]<jupyter_text>### 5. Calculate and plot the explained variance
A useful measure is the **explained variance**, which is calculated from the eigenvalues.
The explained variance tells us how much information (variance) is captured by each principal component.
### $$ ExpVar_i = \bigg(\frac{eigenvalue_i}{\sum_j^n{eigenvalue_j}}\bigg) * 100$$<jupyter_code>cum_var_exp = []
var_exp = []
for i in eigenvalues:
var_i = (i/np.sum(eigenvalues))*100
var_exp.append(var_i)
cum_var_exp = np.cumsum(var_exp)
cum_var_exp<jupyter_output><empty_output><jupyter_text>def calculate_cum_var_exp(eig_vals):
'''
Calculate Explained Variance from Eigenvalues
Return a list or array containing the cumulative explained variance
'''
var_exp = []
tot = sum(eig_vals)
for i in eig_vals:
var_i = (i/tot)*100
var_exp.append(var_i)
cum_var_exp = np.cumsum(var_exp)
return cum_var_exp
cum_var_exp<jupyter_code>plt.figure(figsize=(9,7))
component_number = [i+1 for i in range(len(cum_var_exp))]
plt.plot(component_number, cum_var_exp, lw=7)
plt.axhline(y=0, linewidth=5, color='grey', ls='dashed')
plt.axhline(y=100, linewidth=3, color='grey', ls='dashed')
ax = plt.gca()
ax.set_xlim([1,30])
ax.set_ylim([-5,105])
ax.set_ylabel('cumulative variance explained', fontsize=16)
ax.set_xlabel('component', fontsize=16)
for tick in ax.xaxis.get_major_ticks():
tick.label.set_fontsize(12)
for tick in ax.yaxis.get_major_ticks():
tick.label.set_fontsize(12)
ax.set_title('component v++s cumulative variance explained\n', fontsize=20)
plt.show()<jupyter_output><empty_output><jupyter_text>### 6. Using sklearn For PCA
from sklearn.decomposition import PCA
- Create an instance of PCA
- Fit X
- Plot the explained variance
- Define n_components
- n_component
- Apply dimensionality reduction to X
- transform
- Create PairPlot of PCA<jupyter_code># ANSWER
from sklearn.decomposition import PCA
# Create an instance of PCA
pca = PCA()
# Fit Xs
pca.fit(X_scaled)
y = 100*pca.explained_variance_ratio_
y
X = np.arange(1,31)
X
# ANSWER
# Plot explained_variance_
plt.plot(X, 100-y)
plt.xlabel('number of components')
plt.ylabel('Explained_variance')
plt.show()
# ANSWER
# Apply dimensionality reduction to Xs using transform
pca = PCA(n_components = 16)
X_pca = pca.fit_transform(X_scaled)
X_pca = pd.DataFrame(X_pca)
X_pca
pca.explained_variance_ratio_.cumsum()
# ANSWER
# Create PairPlot of PCA
sns.pairplot(X_pca)<jupyter_output><empty_output><jupyter_text>### 7. Split Data to 80/20 and Use PCA you gon in 6 as X
Split data 80/20 and Use KNN to find score.<jupyter_code># ANSWER
# Split Data
# ANSWER<jupyter_output><empty_output>
|
no_license
|
/Module 6/IOD_Lab_6_4_Principal_Component_Analysis.ipynb
|
DavidMuwandi/Institue_of_data
| 9 |
<jupyter_start><jupyter_text># Emulating Numeric Types<jupyter_code>from math import hypot
class Vector:
def __init__(self, x=0,y=0):
self.x = x
self.y = y
# String representation, should be unambiguous, if possible,
# match the source code necessary to re-create the object being represented
# it is different from __str__, called by str() and implicitly used by the print function.
# if there is not __str__, Python will call __repr__ instead
def __repr__(self):
return 'Vector(%r, %r)' % (self.x, self.y)
def __abs__(self):
return hypot(self.x, self.y)
# by default, if we don't have this method, Python will call __len__, if not, it always return True
def __bool__(self):
return bool(abs(self))
def __add__(self, other):
x = self.x + other.x
y = self.y + other.y
return Vector(x, y)
def __mul__(self, scalar):
return Vector(self.x * scalar, self.y * scalar)
v1 = Vector(2,4)
v2 = Vector(2,1)
v = v1 + v2
v
print(v)
abs(v)
abs(v * 3)<jupyter_output><empty_output>
|
no_license
|
/Chapter1.ipynb
|
sethips/fluent-python-tutorial
| 1 |
<jupyter_start><jupyter_text><jupyter_code>import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import tensorflow as tf
import os
from PIL import Image
print(os.listdir())
%cd drive/My Drive/chest_xray/
train_folder='train'
test_folder='test'
val_folder='val'
os.listdir(train_folder)
train_n=train_folder+'/NORMAL/'
train_p=train_folder+'/PNEUMONIA/'
val_n=val_folder+'/NORMAL/'
val_p=val_folder+'/PNEUMONIA/'
test_n=test_folder+'/NORMAL/'
test_p=test_folder+'/PNEUMONIA/'
print(len(os.listdir(train_n)))
rand_norm=np.random.randint(0,len(os.listdir(train_n)))
norm_pic = os.listdir(train_n)[rand_norm]
print('normal picture title: ',norm_pic)
norm_pic_address = train_n+norm_pic
print(len(os.listdir(train_p)))
rand_p=np.random.randint(0,len(os.listdir(train_p)))
pneu_pic = os.listdir(train_p)[rand_p]
print('pneumonia picture title: ',pneu_pic)
pneu_pic_address=train_p+pneu_pic
#plotting the images
norm_load = Image.open(norm_pic_address)
pneu_load = Image.open(pneu_pic_address)
f = plt.figure(figsize= (10,6))
a1 = f.add_subplot(1,2,1)
img_plot = plt.imshow(norm_load)
a1.set_title('Normal')
a2 = f.add_subplot(1, 2, 2)
img_plot = plt.imshow(pneu_load)
a2.set_title('Pneumonia')
#building the cnn
model=tf.keras.models.Sequential([tf.keras.layers.Conv2D(32,(3,3),strides=(1,1),padding='valid',activation=tf.nn.relu,input_shape=(300,300,1)),
tf.keras.layers.MaxPool2D(pool_size=(2,2),strides=(2,2),padding='valid'),
tf.keras.layers.Conv2D(64,(3,3),strides=(1,1),padding='valid',activation=tf.nn.relu),
tf.keras.layers.MaxPool2D(pool_size=(2,2),strides=(2,2),padding='valid'),
tf.keras.layers.Conv2D(128,(3,3),strides=(1,1),padding='valid',activation=tf.nn.relu),
tf.keras.layers.MaxPool2D(pool_size=(2,2),strides=(2,2),padding='valid'),
tf.keras.layers.Flatten(),
tf.keras.layers.Dense(units=256,activation=tf.nn.relu),
tf.keras.layers.Dropout(0.1),
tf.keras.layers.Dense(units=1,activation=tf.nn.sigmoid)
])
model.compile(optimizer='adam',
loss='binary_crossentropy',
metrics=['accuracy'])
model.summary()
from keras.preprocessing.image import ImageDataGenerator, load_img
from sklearn.metrics import classification_report, confusion_matrix
train_datagen = ImageDataGenerator(rescale = 1./255,
shear_range = 0.2,
rotation_range=5,
zoom_range = 0.2,
width_shift_range=0.1,
height_shift_range=0.1,
)
test_datagen = ImageDataGenerator(rescale = 1./255) #Image normalization.
training_set = train_datagen.flow_from_directory(train_folder,
target_size = (300, 300),
shuffle=True,
batch_size = 32,
class_mode = 'binary',
color_mode='grayscale')
validation_generator = test_datagen.flow_from_directory(val_folder,
target_size=(300, 300),
batch_size=32,
class_mode='binary',
color_mode='grayscale')
test_set = test_datagen.flow_from_directory(test_folder,
target_size = (300, 300),
batch_size = 32,
class_mode = 'binary',
color_mode='grayscale')
model.fit(training_set,
epochs = 1,
validation_data = validation_generator,
shuffle=False)
test_accu = model.evaluate(test_set,steps=624)
print("The testing accuracy is : ",test_accu*100,'%')
<jupyter_output><empty_output>
|
permissive
|
/Pneumonia.ipynb
|
AritraJana1810/t81_558_deep_learning
| 1 |
<jupyter_start><jupyter_text># What's this PyTorch business?
You've written a lot of code in this assignment to provide a whole host of neural network functionality. Dropout, Batch Norm, and 2D convolutions are some of the workhorses of deep learning in computer vision. You've also worked hard to make your code efficient and vectorized.
For the last part of this assignment, though, we're going to leave behind your beautiful codebase and instead migrate to one of two popular deep learning frameworks: in this instance, PyTorch (or TensorFlow, if you switch over to that notebook).
### What is PyTorch?
PyTorch is a system for executing dynamic computational graphs over Tensor objects that behave similarly as numpy ndarray. It comes with a powerful automatic differentiation engine that removes the need for manual back-propagation.
### Why?
* Our code will now run on GPUs! Much faster training. When using a framework like PyTorch or TensorFlow you can harness the power of the GPU for your own custom neural network architectures without having to write CUDA code directly (which is beyond the scope of this class).
* We want you to be ready to use one of these frameworks for your project so you can experiment more efficiently than if you were writing every feature you want to use by hand.
* We want you to stand on the shoulders of giants! TensorFlow and PyTorch are both excellent frameworks that will make your lives a lot easier, and now that you understand their guts, you are free to use them :)
* We want you to be exposed to the sort of deep learning code you might run into in academia or industry.
### PyTorch versions
This notebook assumes that you are using **PyTorch version 0.4**. Prior to this version, Tensors had to be wrapped in Variable objects to be used in autograd; however Variables have now been deprecated. In addition 0.4 also separates a Tensor's datatype from its device, and uses numpy-style factories for constructing Tensors rather than directly invoking Tensor constructors.## How will I learn PyTorch?
Justin Johnson has made an excellent [tutorial](https://github.com/jcjohnson/pytorch-examples) for PyTorch.
You can also find the detailed [API doc](http://pytorch.org/docs/stable/index.html) here. If you have other questions that are not addressed by the API docs, the [PyTorch forum](https://discuss.pytorch.org/) is a much better place to ask than StackOverflow.
# Table of Contents
This assignment has 5 parts. You will learn PyTorch on different levels of abstractions, which will help you understand it better and prepare you for the final project.
1. Preparation: we will use CIFAR-10 dataset.
2. Barebones PyTorch: we will work directly with the lowest-level PyTorch Tensors.
3. PyTorch Module API: we will use `nn.Module` to define arbitrary neural network architecture.
4. PyTorch Sequential API: we will use `nn.Sequential` to define a linear feed-forward network very conveniently.
5. CIFAR-10 open-ended challenge: please implement your own network to get as high accuracy as possible on CIFAR-10. You can experiment with any layer, optimizer, hyperparameters or other advanced features.
Here is a table of comparison:
| API | Flexibility | Convenience |
|---------------|-------------|-------------|
| Barebone | High | Low |
| `nn.Module` | High | Medium |
| `nn.Sequential` | Low | High |# Part I. Preparation
First, we load the CIFAR-10 dataset. This might take a couple minutes the first time you do it, but the files should stay cached after that.
In previous parts of the assignment we had to write our own code to download the CIFAR-10 dataset, preprocess it, and iterate through it in minibatches; PyTorch provides convenient tools to automate this process for us.<jupyter_code>import torch
import torch.nn as nn
import torch.optim as optim
from torch.utils.data import DataLoader
from torch.utils.data import sampler
import torchvision.datasets as dset
import torchvision.transforms as T
import numpy as np
NUM_TRAIN = 49000
# The torchvision.transforms package provides tools for preprocessing data
# and for performing data augmentation; here we set up a transform to
# preprocess the data by subtracting the mean RGB value and dividing by the
# standard deviation of each RGB value; we've hardcoded the mean and std.
transform = T.Compose([
T.ToTensor(),
T.Normalize((0.4914, 0.4822, 0.4465), (0.2023, 0.1994, 0.2010))
])
# We set up a Dataset object for each split (train / val / test); Datasets load
# training examples one at a time, so we wrap each Dataset in a DataLoader which
# iterates through the Dataset and forms minibatches. We divide the CIFAR-10
# training set into train and val sets by passing a Sampler object to the
# DataLoader telling how it should sample from the underlying Dataset.
cifar10_train = dset.CIFAR10('./cs231n/datasets', train=True, download=True,
transform=transform)
loader_train = DataLoader(cifar10_train, batch_size=64,
sampler=sampler.SubsetRandomSampler(range(NUM_TRAIN)))
cifar10_val = dset.CIFAR10('./cs231n/datasets', train=True, download=True,
transform=transform)
loader_val = DataLoader(cifar10_val, batch_size=64,
sampler=sampler.SubsetRandomSampler(range(NUM_TRAIN, 50000)))
cifar10_test = dset.CIFAR10('./cs231n/datasets', train=False, download=True,
transform=transform)
loader_test = DataLoader(cifar10_test, batch_size=64)<jupyter_output>Files already downloaded and verified
Files already downloaded and verified
Files already downloaded and verified
<jupyter_text>You have an option to **use GPU by setting the flag to True below**. It is not necessary to use GPU for this assignment. Note that if your computer does not have CUDA enabled, `torch.cuda.is_available()` will return False and this notebook will fallback to CPU mode.
The global variables `dtype` and `device` will control the data types throughout this assignment. <jupyter_code>USE_GPU = True
dtype = torch.float32 # we will be using float throughout this tutorial
if USE_GPU and torch.cuda.is_available():
device = torch.device('cuda')
else:
device = torch.device('cpu')
# Constant to control how frequently we print train loss
print_every = 100
print('using device:', device)
# http://pytorch.org/
from os import path
from wheel.pep425tags import get_abbr_impl, get_impl_ver, get_abi_tag
platform = '{}{}-{}'.format(get_abbr_impl(), get_impl_ver(), get_abi_tag())
accelerator = 'cu80' if path.exists('/opt/bin/nvidia-smi') else 'cpu'
!pip install -q http://download.pytorch.org/whl/{accelerator}/torch-0.3.0.post4-{platform}-linux_x86_64.whl torchvision
import torch<jupyter_output><empty_output><jupyter_text># Part II. Barebones PyTorch
PyTorch ships with high-level APIs to help us define model architectures conveniently, which we will cover in Part II of this tutorial. In this section, we will start with the barebone PyTorch elements to understand the autograd engine better. After this exercise, you will come to appreciate the high-level model API more.
We will start with a simple fully-connected ReLU network with two hidden layers and no biases for CIFAR classification.
This implementation computes the forward pass using operations on PyTorch Tensors, and uses PyTorch autograd to compute gradients. It is important that you understand every line, because you will write a harder version after the example.
When we create a PyTorch Tensor with `requires_grad=True`, then operations involving that Tensor will not just compute values; they will also build up a computational graph in the background, allowing us to easily backpropagate through the graph to compute gradients of some Tensors with respect to a downstream loss. Concretely if x is a Tensor with `x.requires_grad == True` then after backpropagation `x.grad` will be another Tensor holding the gradient of x with respect to the scalar loss at the end.### PyTorch Tensors: Flatten Function
A PyTorch Tensor is conceptionally similar to a numpy array: it is an n-dimensional grid of numbers, and like numpy PyTorch provides many functions to efficiently operate on Tensors. As a simple example, we provide a `flatten` function below which reshapes image data for use in a fully-connected neural network.
Recall that image data is typically stored in a Tensor of shape N x C x H x W, where:
* N is the number of datapoints
* C is the number of channels
* H is the height of the intermediate feature map in pixels
* W is the height of the intermediate feature map in pixels
This is the right way to represent the data when we are doing something like a 2D convolution, that needs spatial understanding of where the intermediate features are relative to each other. When we use fully connected affine layers to process the image, however, we want each datapoint to be represented by a single vector -- it's no longer useful to segregate the different channels, rows, and columns of the data. So, we use a "flatten" operation to collapse the `C x H x W` values per representation into a single long vector. The flatten function below first reads in the N, C, H, and W values from a given batch of data, and then returns a "view" of that data. "View" is analogous to numpy's "reshape" method: it reshapes x's dimensions to be N x ??, where ?? is allowed to be anything (in this case, it will be C x H x W, but we don't need to specify that explicitly). <jupyter_code>def flatten(x):
N = x.shape[0] # read in N, C, H, W
return x.view(N, -1) # "flatten" the C * H * W values into a single vector per image
def test_flatten():
x = torch.arange(12).view(2, 1, 3, 2)
print('Before flattening: ', x)
print('After flattening: ', flatten(x))
test_flatten()<jupyter_output>Before flattening: tensor([[[[ 0., 1.],
[ 2., 3.],
[ 4., 5.]]],
[[[ 6., 7.],
[ 8., 9.],
[ 10., 11.]]]])
After flattening: tensor([[ 0., 1., 2., 3., 4., 5.],
[ 6., 7., 8., 9., 10., 11.]])
<jupyter_text>### Barebones PyTorch: Two-Layer Network
Here we define a function `two_layer_fc` which performs the forward pass of a two-layer fully-connected ReLU network on a batch of image data. After defining the forward pass we check that it doesn't crash and that it produces outputs of the right shape by running zeros through the network.
You don't have to write any code here, but it's important that you read and understand the implementation.<jupyter_code>import torch.nn.functional as F # useful stateless functions
def two_layer_fc(x, params):
"""
A fully-connected neural networks; the architecture is:
NN is fully connected -> ReLU -> fully connected layer.
Note that this function only defines the forward pass;
PyTorch will take care of the backward pass for us.
The input to the network will be a minibatch of data, of shape
(N, d1, ..., dM) where d1 * ... * dM = D. The hidden layer will have H units,
and the output layer will produce scores for C classes.
Inputs:
- x: A PyTorch Tensor of shape (N, d1, ..., dM) giving a minibatch of
input data.
- params: A list [w1, w2] of PyTorch Tensors giving weights for the network;
w1 has shape (D, H) and w2 has shape (H, C).
Returns:
- scores: A PyTorch Tensor of shape (N, C) giving classification scores for
the input data x.
"""
# first we flatten the image
x = flatten(x) # shape: [batch_size, C x H x W]
w1, w2 = params
# Forward pass: compute predicted y using operations on Tensors. Since w1 and
# w2 have requires_grad=True, operations involving these Tensors will cause
# PyTorch to build a computational graph, allowing automatic computation of
# gradients. Since we are no longer implementing the backward pass by hand we
# don't need to keep references to intermediate values.
# you can also use `.clamp(min=0)`, equivalent to F.relu()
x = F.relu(x.mm(w1))
x = x.mm(w2)
return x
def two_layer_fc_test():
hidden_layer_size = 42
x = torch.zeros((64, 50), dtype=dtype) # minibatch size 64, feature dimension 50
w1 = torch.zeros((50, hidden_layer_size), dtype=dtype)
w2 = torch.zeros((hidden_layer_size, 10), dtype=dtype)
scores = two_layer_fc(x, [w1, w2])
print(scores.size()) # you should see [64, 10]
two_layer_fc_test()<jupyter_output>torch.Size([64, 10])
<jupyter_text>### Barebones PyTorch: Three-Layer ConvNet
Here you will complete the implementation of the function `three_layer_convnet`, which will perform the forward pass of a three-layer convolutional network. Like above, we can immediately test our implementation by passing zeros through the network. The network should have the following architecture:
1. A convolutional layer (with bias) with `channel_1` filters, each with shape `KW1 x KH1`, and zero-padding of two
2. ReLU nonlinearity
3. A convolutional layer (with bias) with `channel_2` filters, each with shape `KW2 x KH2`, and zero-padding of one
4. ReLU nonlinearity
5. Fully-connected layer with bias, producing scores for C classes.
**HINT**: For convolutions: http://pytorch.org/docs/stable/nn.html#torch.nn.functional.conv2d; pay attention to the shapes of convolutional filters!<jupyter_code>def three_layer_convnet(x, params):
"""
Performs the forward pass of a three-layer convolutional network with the
architecture defined above.
Inputs:
- x: A PyTorch Tensor of shape (N, 3, H, W) giving a minibatch of images
- params: A list of PyTorch Tensors giving the weights and biases for the
network; should contain the following:
- conv_w1: PyTorch Tensor of shape (channel_1, 3, KH1, KW1) giving weights
for the first convolutional layer
- conv_b1: PyTorch Tensor of shape (channel_1,) giving biases for the first
convolutional layer
- conv_w2: PyTorch Tensor of shape (channel_2, channel_1, KH2, KW2) giving
weights for the second convolutional layer
- conv_b2: PyTorch Tensor of shape (channel_2,) giving biases for the second
convolutional layer
- fc_w: PyTorch Tensor giving weights for the fully-connected layer. Can you
figure out what the shape should be?
- fc_b: PyTorch Tensor giving biases for the fully-connected layer. Can you
figure out what the shape should be?
Returns:
- scores: PyTorch Tensor of shape (N, C) giving classification scores for x
"""
conv_w1, conv_b1, conv_w2, conv_b2, fc_w, fc_b = params
scores = None
################################################################################
# TODO: Implement the forward pass for the three-layer ConvNet. #
################################################################################
conv1 = F.conv2d(x,conv_w1,conv_b1,padding=2)
relu1 = F.relu(conv1)
conv2 = F.conv2d(relu1,conv_w2,conv_b2,padding=1)
#print(conv2.shape)
relu2 = F.relu(conv2)
relu2 = flatten(relu2)
scores = relu2.mm(fc_w) + fc_b
################################################################################
# END OF YOUR CODE #
################################################################################
return scores<jupyter_output><empty_output><jupyter_text>After defining the forward pass of the ConvNet above, run the following cell to test your implementation.
When you run this function, scores should have shape (64, 10).<jupyter_code>def three_layer_convnet_test():
x = torch.zeros((64, 3, 32, 32), dtype=dtype) # minibatch size 64, image size [3, 32, 32]
conv_w1 = torch.zeros((6, 3, 5, 5), dtype=dtype) # [out_channel, in_channel, kernel_H, kernel_W]
conv_b1 = torch.zeros((6,)) # out_channel
conv_w2 = torch.zeros((9, 6, 3, 3), dtype=dtype) # [out_channel, in_channel, kernel_H, kernel_W]
conv_b2 = torch.zeros((9,)) # out_channel
# you must calculate the shape of the tensor after two conv layers, before the fully-connected layer
fc_w = torch.zeros((9 * 32 * 32, 10))
fc_b = torch.zeros(10)
scores = three_layer_convnet(x, [conv_w1, conv_b1, conv_w2, conv_b2, fc_w, fc_b])
print(scores.size()) # you should see [64, 10]
three_layer_convnet_test()<jupyter_output>torch.Size([64, 10])
<jupyter_text>### Barebones PyTorch: Initialization
Let's write a couple utility methods to initialize the weight matrices for our models.
- `random_weight(shape)` initializes a weight tensor with the Kaiming normalization method.
- `zero_weight(shape)` initializes a weight tensor with all zeros. Useful for instantiating bias parameters.
The `random_weight` function uses the Kaiming normal initialization method, described in:
He et al, *Delving Deep into Rectifiers: Surpassing Human-Level Performance on ImageNet Classification*, ICCV 2015, https://arxiv.org/abs/1502.01852<jupyter_code>def random_weight(shape):
"""
Create random Tensors for weights; setting requires_grad=True means that we
want to compute gradients for these Tensors during the backward pass.
We use Kaiming normalization: sqrt(2 / fan_in)
"""
if len(shape) == 2: # FC weight
fan_in = shape[0]
else:
fan_in = np.prod(shape[1:]) # conv weight [out_channel, in_channel, kH, kW]
# randn is standard normal distribution generator.
w = torch.randn(shape, device=device, dtype=dtype) * np.sqrt(2. / fan_in)
w.requires_grad = True
return w
def zero_weight(shape):
return torch.zeros(shape, device=device, dtype=dtype, requires_grad=True)
# create a weight of shape [3 x 5]
# you should see the type `torch.cuda.FloatTensor` if you use GPU.
# Otherwise it should be `torch.FloatTensor`
random_weight((3, 5))<jupyter_output><empty_output><jupyter_text>### Barebones PyTorch: Check Accuracy
When training the model we will use the following function to check the accuracy of our model on the training or validation sets.
When checking accuracy we don't need to compute any gradients; as a result we don't need PyTorch to build a computational graph for us when we compute scores. To prevent a graph from being built we scope our computation under a `torch.no_grad()` context manager.<jupyter_code>def check_accuracy_part2(loader, model_fn, params):
"""
Check the accuracy of a classification model.
Inputs:
- loader: A DataLoader for the data split we want to check
- model_fn: A function that performs the forward pass of the model,
with the signature scores = model_fn(x, params)
- params: List of PyTorch Tensors giving parameters of the model
Returns: Nothing, but prints the accuracy of the model
"""
split = 'val' if loader.dataset.train else 'test'
print('Checking accuracy on the %s set' % split)
num_correct, num_samples = 0, 0
with torch.no_grad():
for x, y in loader:
x = x.to(device=device, dtype=dtype) # move to device, e.g. GPU
y = y.to(device=device, dtype=torch.int64)
scores = model_fn(x, params)
_, preds = scores.max(1)
num_correct += (preds == y).sum()
num_samples += preds.size(0)
acc = float(num_correct) / num_samples
print('Got %d / %d correct (%.2f%%)' % (num_correct, num_samples, 100 * acc))<jupyter_output><empty_output><jupyter_text>### BareBones PyTorch: Training Loop
We can now set up a basic training loop to train our network. We will train the model using stochastic gradient descent without momentum. We will use `torch.functional.cross_entropy` to compute the loss; you can [read about it here](http://pytorch.org/docs/stable/nn.html#cross-entropy).
The training loop takes as input the neural network function, a list of initialized parameters (`[w1, w2]` in our example), and learning rate.<jupyter_code>def train_part2(model_fn, params, learning_rate):
"""
Train a model on CIFAR-10.
Inputs:
- model_fn: A Python function that performs the forward pass of the model.
It should have the signature scores = model_fn(x, params) where x is a
PyTorch Tensor of image data, params is a list of PyTorch Tensors giving
model weights, and scores is a PyTorch Tensor of shape (N, C) giving
scores for the elements in x.
- params: List of PyTorch Tensors giving weights for the model
- learning_rate: Python scalar giving the learning rate to use for SGD
Returns: Nothing
"""
for t, (x, y) in enumerate(loader_train):
# Move the data to the proper device (GPU or CPU)
x = x.to(device=device, dtype=dtype)
y = y.to(device=device, dtype=torch.long)
# Forward pass: compute scores and loss
scores = model_fn(x, params)
loss = F.cross_entropy(scores, y)
# Backward pass: PyTorch figures out which Tensors in the computational
# graph has requires_grad=True and uses backpropagation to compute the
# gradient of the loss with respect to these Tensors, and stores the
# gradients in the .grad attribute of each Tensor.
loss.backward()
# Update parameters. We don't want to backpropagate through the
# parameter updates, so we scope the updates under a torch.no_grad()
# context manager to prevent a computational graph from being built.
with torch.no_grad():
for w in params:
w -= learning_rate * w.grad
# Manually zero the gradients after running the backward pass
w.grad.zero_()
if t % print_every == 0:
print('Iteration %d, loss = %.4f' % (t, loss.item()))
check_accuracy_part2(loader_val, model_fn, params)
print()<jupyter_output><empty_output><jupyter_text>### BareBones PyTorch: Train a Two-Layer Network
Now we are ready to run the training loop. We need to explicitly allocate tensors for the fully connected weights, `w1` and `w2`.
Each minibatch of CIFAR has 64 examples, so the tensor shape is `[64, 3, 32, 32]`.
After flattening, `x` shape should be `[64, 3 * 32 * 32]`. This will be the size of the first dimension of `w1`.
The second dimension of `w1` is the hidden layer size, which will also be the first dimension of `w2`.
Finally, the output of the network is a 10-dimensional vector that represents the probability distribution over 10 classes.
You don't need to tune any hyperparameters but you should see accuracies above 40% after training for one epoch.<jupyter_code>hidden_layer_size = 4000
learning_rate = 1e-2
w1 = random_weight((3 * 32 * 32, hidden_layer_size))
w2 = random_weight((hidden_layer_size, 10))
train_part2(two_layer_fc, [w1, w2], learning_rate)<jupyter_output>Iteration 0, loss = 3.8582
Checking accuracy on the val set
Got 172 / 1000 correct (17.20%)
Iteration 100, loss = 2.4662
Checking accuracy on the val set
Got 353 / 1000 correct (35.30%)
Iteration 200, loss = 2.1477
Checking accuracy on the val set
Got 349 / 1000 correct (34.90%)
Iteration 300, loss = 2.0237
Checking accuracy on the val set
Got 365 / 1000 correct (36.50%)
Iteration 400, loss = 1.6529
Checking accuracy on the val set
Got 420 / 1000 correct (42.00%)
Iteration 500, loss = 1.8114
Checking accuracy on the val set
Got 426 / 1000 correct (42.60%)
Iteration 600, loss = 1.9078
Checking accuracy on the val set
Got 405 / 1000 correct (40.50%)
Iteration 700, loss = 1.7006
Checking accuracy on the val set
Got 438 / 1000 correct (43.80%)
<jupyter_text>### BareBones PyTorch: Training a ConvNet
In the below you should use the functions defined above to train a three-layer convolutional network on CIFAR. The network should have the following architecture:
1. Convolutional layer (with bias) with 32 5x5 filters, with zero-padding of 2
2. ReLU
3. Convolutional layer (with bias) with 16 3x3 filters, with zero-padding of 1
4. ReLU
5. Fully-connected layer (with bias) to compute scores for 10 classes
You should initialize your weight matrices using the `random_weight` function defined above, and you should initialize your bias vectors using the `zero_weight` function above.
You don't need to tune any hyperparameters, but if everything works correctly you should achieve an accuracy above 42% after one epoch.<jupyter_code>learning_rate = 3e-3
channel_1 = 32
channel_2 = 16
conv_w1 = None
conv_b1 = None
conv_w2 = None
conv_b2 = None
fc_w = None
fc_b = None
################################################################################
# TODO: Initialize the parameters of a three-layer ConvNet. #
################################################################################
conv_w1 = random_weight((channel_1,3,5,5))
conv_b1 =zero_weight((channel_1,))
conv_w2 = random_weight((channel_2,channel_1,3,3))
conv_b2 = zero_weight((channel_2,))
fc_w = random_weight((channel_2 * 32 * 32,10))
fc_b = zero_weight((10,))
################################################################################
# END OF YOUR CODE #
################################################################################
params = [conv_w1, conv_b1, conv_w2, conv_b2, fc_w, fc_b]
train_part2(three_layer_convnet, params, learning_rate)<jupyter_output>Iteration 0, loss = 3.4028
Checking accuracy on the val set
Got 81 / 1000 correct (8.10%)
Iteration 100, loss = 1.7824
Checking accuracy on the val set
Got 351 / 1000 correct (35.10%)
Iteration 200, loss = 1.9526
Checking accuracy on the val set
Got 401 / 1000 correct (40.10%)
Iteration 300, loss = 1.5768
Checking accuracy on the val set
Got 411 / 1000 correct (41.10%)
Iteration 400, loss = 1.5590
Checking accuracy on the val set
Got 455 / 1000 correct (45.50%)
Iteration 500, loss = 1.8510
Checking accuracy on the val set
Got 464 / 1000 correct (46.40%)
Iteration 600, loss = 1.2943
Checking accuracy on the val set
Got 460 / 1000 correct (46.00%)
Iteration 700, loss = 1.7689
Checking accuracy on the val set
Got 475 / 1000 correct (47.50%)
<jupyter_text># Part III. PyTorch Module API
Barebone PyTorch requires that we track all the parameter tensors by hand. This is fine for small networks with a few tensors, but it would be extremely inconvenient and error-prone to track tens or hundreds of tensors in larger networks.
PyTorch provides the `nn.Module` API for you to define arbitrary network architectures, while tracking every learnable parameters for you. In Part II, we implemented SGD ourselves. PyTorch also provides the `torch.optim` package that implements all the common optimizers, such as RMSProp, Adagrad, and Adam. It even supports approximate second-order methods like L-BFGS! You can refer to the [doc](http://pytorch.org/docs/master/optim.html) for the exact specifications of each optimizer.
To use the Module API, follow the steps below:
1. Subclass `nn.Module`. Give your network class an intuitive name like `TwoLayerFC`.
2. In the constructor `__init__()`, define all the layers you need as class attributes. Layer objects like `nn.Linear` and `nn.Conv2d` are themselves `nn.Module` subclasses and contain learnable parameters, so that you don't have to instantiate the raw tensors yourself. `nn.Module` will track these internal parameters for you. Refer to the [doc](http://pytorch.org/docs/master/nn.html) to learn more about the dozens of builtin layers. **Warning**: don't forget to call the `super().__init__()` first!
3. In the `forward()` method, define the *connectivity* of your network. You should use the attributes defined in `__init__` as function calls that take tensor as input and output the "transformed" tensor. Do *not* create any new layers with learnable parameters in `forward()`! All of them must be declared upfront in `__init__`.
After you define your Module subclass, you can instantiate it as an object and call it just like the NN forward function in part II.
### Module API: Two-Layer Network
Here is a concrete example of a 2-layer fully connected network:<jupyter_code>class TwoLayerFC(nn.Module):
def __init__(self, input_size, hidden_size, num_classes):
super().__init__()
# assign layer objects to class attributes
self.fc1 = nn.Linear(input_size, hidden_size)
# nn.init package contains convenient initialization methods
# http://pytorch.org/docs/master/nn.html#torch-nn-init
nn.init.kaiming_normal_(self.fc1.weight)
self.fc2 = nn.Linear(hidden_size, num_classes)
nn.init.kaiming_normal_(self.fc2.weight)
def forward(self, x):
# forward always defines connectivity
x = flatten(x)
scores = self.fc2(F.relu(self.fc1(x)))
return scores
def test_TwoLayerFC():
input_size = 50
x = torch.zeros((64, input_size), dtype=dtype) # minibatch size 64, feature dimension 50
model = TwoLayerFC(input_size, 42, 10)
scores = model(x)
print(scores.size()) # you should see [64, 10]
test_TwoLayerFC()<jupyter_output>torch.Size([64, 10])
<jupyter_text>### Module API: Three-Layer ConvNet
It's your turn to implement a 3-layer ConvNet followed by a fully connected layer. The network architecture should be the same as in Part II:
1. Convolutional layer with `channel_1` 5x5 filters with zero-padding of 2
2. ReLU
3. Convolutional layer with `channel_2` 3x3 filters with zero-padding of 1
4. ReLU
5. Fully-connected layer to `num_classes` classes
You should initialize the weight matrices of the model using the Kaiming normal initialization method.
**HINT**: http://pytorch.org/docs/stable/nn.html#conv2d
After you implement the three-layer ConvNet, the `test_ThreeLayerConvNet` function will run your implementation; it should print `(64, 10)` for the shape of the output scores.<jupyter_code>class ThreeLayerConvNet(nn.Module):
def __init__(self, in_channel, channel_1, channel_2, num_classes):
super().__init__()
########################################################################
# TODO: Set up the layers you need for a three-layer ConvNet with the #
# architecture defined above. #
########################################################################
self.conv1 = nn.Conv2d(in_channel,channel_1,(5,5),padding=2)
self.conv2 = nn .Conv2d(channel_1,channel_2,(3,3),padding=1)
self.fc = nn.Linear(channel_2*32*32,num_classes)
nn.init.kaiming_normal_(self.conv1.weight)
nn.init.constant_(self.conv1.bias,0)
nn.init.kaiming_normal_(self.conv2.weight)
nn.init.constant_(self.conv2.bias,0)
nn.init.kaiming_normal_(self.fc.weight)
nn.init.constant_(self.fc.bias,0)
########################################################################
# END OF YOUR CODE #
########################################################################
def forward(self, x):
scores = None
########################################################################
# TODO: Implement the forward function for a 3-layer ConvNet. you #
# should use the layers you defined in __init__ and specify the #
# connectivity of those layers in forward() #
########################################################################
layer1 = F.relu(self.conv1(x))
layer2 = F.relu(self.conv2(layer1))
scores = self.fc(flatten(layer2))
########################################################################
# END OF YOUR CODE #
########################################################################
return scores
def test_ThreeLayerConvNet():
x = torch.zeros((64, 3, 32, 32), dtype=dtype) # minibatch size 64, image size [3, 32, 32]
model = ThreeLayerConvNet(in_channel=3, channel_1=12, channel_2=8, num_classes=10)
scores = model(x)
print(scores.size()) # you should see [64, 10]
test_ThreeLayerConvNet()<jupyter_output>torch.Size([64, 10])
<jupyter_text>### Module API: Check Accuracy
Given the validation or test set, we can check the classification accuracy of a neural network.
This version is slightly different from the one in part II. You don't manually pass in the parameters anymore.<jupyter_code>def check_accuracy_part34(loader, model):
if loader.dataset.train:
print('Checking accuracy on validation set')
else:
print('Checking accuracy on test set')
num_correct = 0
num_samples = 0
model.eval() # set model to evaluation mode
with torch.no_grad():
for x, y in loader:
x = x.to(device=device, dtype=dtype) # move to device, e.g. GPU
y = y.to(device=device, dtype=torch.long)
scores = model(x)
_, preds = scores.max(1)
num_correct += (preds == y).sum()
num_samples += preds.size(0)
acc = float(num_correct) / num_samples
print('Got %d / %d correct (%.2f)' % (num_correct, num_samples, 100 * acc))<jupyter_output><empty_output><jupyter_text>### Module API: Training Loop
We also use a slightly different training loop. Rather than updating the values of the weights ourselves, we use an Optimizer object from the `torch.optim` package, which abstract the notion of an optimization algorithm and provides implementations of most of the algorithms commonly used to optimize neural networks.<jupyter_code>def train_part34(model, optimizer, epochs=1):
"""
Train a model on CIFAR-10 using the PyTorch Module API.
Inputs:
- model: A PyTorch Module giving the model to train.
- optimizer: An Optimizer object we will use to train the model
- epochs: (Optional) A Python integer giving the number of epochs to train for
Returns: Nothing, but prints model accuracies during training.
"""
model = model.to(device=device) # move the model parameters to CPU/GPU
for e in range(epochs):
for t, (x, y) in enumerate(loader_train):
model.train() # put model to training mode
x = x.to(device=device, dtype=dtype) # move to device, e.g. GPU
y = y.to(device=device, dtype=torch.long)
scores = model(x)
loss = F.cross_entropy(scores, y)
# Zero out all of the gradients for the variables which the optimizer
# will update.
optimizer.zero_grad()
# This is the backwards pass: compute the gradient of the loss with
# respect to each parameter of the model.
loss.backward()
# Actually update the parameters of the model using the gradients
# computed by the backwards pass.
optimizer.step()
if t % print_every == 0:
print('Iteration %d, loss = %.4f' % (t, loss.item()))
check_accuracy_part34(loader_val, model)
print()<jupyter_output><empty_output><jupyter_text>### Module API: Train a Two-Layer Network
Now we are ready to run the training loop. In contrast to part II, we don't explicitly allocate parameter tensors anymore.
Simply pass the input size, hidden layer size, and number of classes (i.e. output size) to the constructor of `TwoLayerFC`.
You also need to define an optimizer that tracks all the learnable parameters inside `TwoLayerFC`.
You don't need to tune any hyperparameters, but you should see model accuracies above 40% after training for one epoch.<jupyter_code>hidden_layer_size = 4000
learning_rate = 1e-2
model = TwoLayerFC(3 * 32 * 32, hidden_layer_size, 10)
optimizer = optim.SGD(model.parameters(), lr=learning_rate)
train_part34(model, optimizer)<jupyter_output>Iteration 0, loss = 3.1364
Checking accuracy on validation set
Got 125 / 1000 correct (12.50)
Iteration 100, loss = 2.5477
Checking accuracy on validation set
Got 269 / 1000 correct (26.90)
Iteration 200, loss = 2.0673
Checking accuracy on validation set
Got 304 / 1000 correct (30.40)
Iteration 300, loss = 2.2261
Checking accuracy on validation set
Got 389 / 1000 correct (38.90)
Iteration 400, loss = 1.9842
Checking accuracy on validation set
Got 342 / 1000 correct (34.20)
Iteration 500, loss = 1.7411
Checking accuracy on validation set
Got 412 / 1000 correct (41.20)
Iteration 600, loss = 1.8313
Checking accuracy on validation set
Got 402 / 1000 correct (40.20)
Iteration 700, loss = 1.6024
Checking accuracy on validation set
Got 447 / 1000 correct (44.70)
<jupyter_text>### Module API: Train a Three-Layer ConvNet
You should now use the Module API to train a three-layer ConvNet on CIFAR. This should look very similar to training the two-layer network! You don't need to tune any hyperparameters, but you should achieve above above 45% after training for one epoch.
You should train the model using stochastic gradient descent without momentum.<jupyter_code>learning_rate = 3e-3
channel_1 = 32
channel_2 = 16
model = None
optimizer = None
################################################################################
# TODO: Instantiate your ThreeLayerConvNet model and a corresponding optimizer #
################################################################################
model = ThreeLayerConvNet(3,channel_1,channel_2,10)
optimizer =optim.SGD(model.parameters(),lr=learning_rate)
################################################################################
# END OF YOUR CODE
################################################################################
train_part34(model, optimizer)<jupyter_output>Iteration 0, loss = 3.7966
Checking accuracy on validation set
Got 136 / 1000 correct (13.60)
Iteration 100, loss = 1.8770
Checking accuracy on validation set
Got 376 / 1000 correct (37.60)
Iteration 200, loss = 1.5875
Checking accuracy on validation set
Got 405 / 1000 correct (40.50)
Iteration 300, loss = 1.7029
Checking accuracy on validation set
Got 444 / 1000 correct (44.40)
Iteration 400, loss = 1.6423
Checking accuracy on validation set
Got 466 / 1000 correct (46.60)
Iteration 500, loss = 1.6394
Checking accuracy on validation set
Got 480 / 1000 correct (48.00)
Iteration 600, loss = 1.4693
Checking accuracy on validation set
Got 482 / 1000 correct (48.20)
Iteration 700, loss = 1.4205
Checking accuracy on validation set
Got 489 / 1000 correct (48.90)
<jupyter_text># Part IV. PyTorch Sequential API
Part III introduced the PyTorch Module API, which allows you to define arbitrary learnable layers and their connectivity.
For simple models like a stack of feed forward layers, you still need to go through 3 steps: subclass `nn.Module`, assign layers to class attributes in `__init__`, and call each layer one by one in `forward()`. Is there a more convenient way?
Fortunately, PyTorch provides a container Module called `nn.Sequential`, which merges the above steps into one. It is not as flexible as `nn.Module`, because you cannot specify more complex topology than a feed-forward stack, but it's good enough for many use cases.
### Sequential API: Two-Layer Network
Let's see how to rewrite our two-layer fully connected network example with `nn.Sequential`, and train it using the training loop defined above.
Again, you don't need to tune any hyperparameters here, but you shoud achieve above 40% accuracy after one epoch of training.<jupyter_code># We need to wrap `flatten` function in a module in order to stack it
# in nn.Sequential
class Flatten(nn.Module):
def forward(self, x):
return flatten(x)
hidden_layer_size = 4000
learning_rate = 1e-2
model = nn.Sequential(
Flatten(),
nn.Linear(3 * 32 * 32, hidden_layer_size),
nn.ReLU(),
nn.Linear(hidden_layer_size, 10),
)
# you can use Nesterov momentum in optim.SGD
optimizer = optim.SGD(model.parameters(), lr=learning_rate,
momentum=0.9, nesterov=True)
train_part34(model, optimizer)<jupyter_output>Iteration 0, loss = 2.3842
Checking accuracy on validation set
Got 176 / 1000 correct (17.60)
Iteration 100, loss = 1.7823
Checking accuracy on validation set
Got 385 / 1000 correct (38.50)
Iteration 200, loss = 1.8912
Checking accuracy on validation set
Got 404 / 1000 correct (40.40)
Iteration 300, loss = 1.8636
Checking accuracy on validation set
Got 418 / 1000 correct (41.80)
Iteration 400, loss = 1.6576
Checking accuracy on validation set
Got 451 / 1000 correct (45.10)
Iteration 500, loss = 2.0532
Checking accuracy on validation set
Got 455 / 1000 correct (45.50)
Iteration 600, loss = 1.5167
Checking accuracy on validation set
Got 450 / 1000 correct (45.00)
Iteration 700, loss = 1.6628
Checking accuracy on validation set
Got 481 / 1000 correct (48.10)
<jupyter_text>### Sequential API: Three-Layer ConvNet
Here you should use `nn.Sequential` to define and train a three-layer ConvNet with the same architecture we used in Part III:
1. Convolutional layer (with bias) with 32 5x5 filters, with zero-padding of 2
2. ReLU
3. Convolutional layer (with bias) with 16 3x3 filters, with zero-padding of 1
4. ReLU
5. Fully-connected layer (with bias) to compute scores for 10 classes
You should initialize your weight matrices using the `random_weight` function defined above, and you should initialize your bias vectors using the `zero_weight` function above.
You should optimize your model using stochastic gradient descent with Nesterov momentum 0.9.
Again, you don't need to tune any hyperparameters but you should see accuracy above 55% after one epoch of training.<jupyter_code>channel_1 = 32
channel_2 = 16
learning_rate = 1e-2
model = None
optimizer = None
################################################################################
# TODO: Rewrite the 2-layer ConvNet with bias from Part III with the #
# Sequential API. #
################################################################################
model = nn.Sequential(
nn.Conv2d(3,channel_1,(5,5),padding=2),
nn.ReLU(),
nn.Conv2d(channel_1,channel_2,(3,3),padding=1),
nn.ReLU(),
Flatten(),
nn.Linear(32*32*channel_2,10),
)
def my_init(m):
if(type(m)==nn.Conv2d or type(m)==nn.Linear):
print(type(m))
nn.init.kaiming_normal_(m.weight)
nn.init.constant_(m.bias,0)
model.apply(my_init)
optimizer = optim.SGD(model.parameters(),lr=learning_rate,momentum=0.9, nesterov=True)
################################################################################
# END OF YOUR CODE
################################################################################
train_part34(model, optimizer)<jupyter_output><class 'torch.nn.modules.conv.Conv2d'>
<class 'torch.nn.modules.conv.Conv2d'>
<class 'torch.nn.modules.linear.Linear'>
Iteration 0, loss = 2.9956
Checking accuracy on validation set
Got 107 / 1000 correct (10.70)
Iteration 100, loss = 1.8609
Checking accuracy on validation set
Got 321 / 1000 correct (32.10)
Iteration 200, loss = 1.8420
Checking accuracy on validation set
Got 373 / 1000 correct (37.30)
Iteration 300, loss = 1.6415
Checking accuracy on validation set
Got 444 / 1000 correct (44.40)
Iteration 400, loss = 1.7806
Checking accuracy on validation set
Got 481 / 1000 correct (48.10)
Iteration 500, loss = 1.5548
Checking accuracy on validation set
Got 463 / 1000 correct (46.30)
Iteration 600, loss = 1.6234
Checking accuracy on validation set
Got 499 / 1000 correct (49.90)
Iteration 700, loss = 1.3825
Checking accuracy on validation set
Got 504 / 1000 correct (50.40)
<jupyter_text>Well... I can't make it above 55%.
And it's strange that with the same code, sometimes the final accuracy is even below 20%. # Part V. CIFAR-10 open-ended challenge
In this section, you can experiment with whatever ConvNet architecture you'd like on CIFAR-10.
Now it's your job to experiment with architectures, hyperparameters, loss functions, and optimizers to train a model that achieves **at least 70%** accuracy on the CIFAR-10 **validation** set within 10 epochs. You can use the check_accuracy and train functions from above. You can use either `nn.Module` or `nn.Sequential` API.
Describe what you did at the end of this notebook.
Here are the official API documentation for each component. One note: what we call in the class "spatial batch norm" is called "BatchNorm2D" in PyTorch.
* Layers in torch.nn package: http://pytorch.org/docs/stable/nn.html
* Activations: http://pytorch.org/docs/stable/nn.html#non-linear-activations
* Loss functions: http://pytorch.org/docs/stable/nn.html#loss-functions
* Optimizers: http://pytorch.org/docs/stable/optim.html
### Things you might try:
- **Filter size**: Above we used 5x5; would smaller filters be more efficient?
- **Number of filters**: Above we used 32 filters. Do more or fewer do better?
- **Pooling vs Strided Convolution**: Do you use max pooling or just stride convolutions?
- **Batch normalization**: Try adding spatial batch normalization after convolution layers and vanilla batch normalization after affine layers. Do your networks train faster?
- **Network architecture**: The network above has two layers of trainable parameters. Can you do better with a deep network? Good architectures to try include:
- [conv-relu-pool]xN -> [affine]xM -> [softmax or SVM]
- [conv-relu-conv-relu-pool]xN -> [affine]xM -> [softmax or SVM]
- [batchnorm-relu-conv]xN -> [affine]xM -> [softmax or SVM]
- **Global Average Pooling**: Instead of flattening and then having multiple affine layers, perform convolutions until your image gets small (7x7 or so) and then perform an average pooling operation to get to a 1x1 image picture (1, 1 , Filter#), which is then reshaped into a (Filter#) vector. This is used in [Google's Inception Network](https://arxiv.org/abs/1512.00567) (See Table 1 for their architecture).
- **Regularization**: Add l2 weight regularization, or perhaps use Dropout.
### Tips for training
For each network architecture that you try, you should tune the learning rate and other hyperparameters. When doing this there are a couple important things to keep in mind:
- If the parameters are working well, you should see improvement within a few hundred iterations
- Remember the coarse-to-fine approach for hyperparameter tuning: start by testing a large range of hyperparameters for just a few training iterations to find the combinations of parameters that are working at all.
- Once you have found some sets of parameters that seem to work, search more finely around these parameters. You may need to train for more epochs.
- You should use the validation set for hyperparameter search, and save your test set for evaluating your architecture on the best parameters as selected by the validation set.
### Going above and beyond
If you are feeling adventurous there are many other features you can implement to try and improve your performance. You are **not required** to implement any of these, but don't miss the fun if you have time!
- Alternative optimizers: you can try Adam, Adagrad, RMSprop, etc.
- Alternative activation functions such as leaky ReLU, parametric ReLU, ELU, or MaxOut.
- Model ensembles
- Data augmentation
- New Architectures
- [ResNets](https://arxiv.org/abs/1512.03385) where the input from the previous layer is added to the output.
- [DenseNets](https://arxiv.org/abs/1608.06993) where inputs into previous layers are concatenated together.
- [This blog has an in-depth overview](https://chatbotslife.com/resnets-highwaynets-and-densenets-oh-my-9bb15918ee32)
### Have fun and happy training! <jupyter_code>def my_train(model, optimizer, epochs=1):
"""
Train a model on CIFAR-10 using the PyTorch Module API.
Inputs:
- model: A PyTorch Module giving the model to train.
- optimizer: An Optimizer object we will use to train the model
- epochs: (Optional) A Python integer giving the number of epochs to train for
Returns: Nothing, but prints model accuracies during training.
"""
model = model.to(device=device) # move the model parameters to CPU/GPU
for e in range(epochs):
for t, (x, y) in enumerate(loader_train):
model.train() # put model to training mode
x = x.to(device=device, dtype=dtype) # move to device, e.g. GPU
y = y.to(device=device, dtype=torch.long)
scores = model(x)
loss = F.cross_entropy(scores, y)
# Zero out all of the gradients for the variables which the optimizer
# will update.
optimizer.zero_grad()
# This is the backwards pass: compute the gradient of the loss with
# respect to each parameter of the model.
loss.backward()
# Actually update the parameters of the model using the gradients
# computed by the backwards pass.
optimizer.step()
if t % print_every == 0:
print('Iteration %d, loss = %.4f' % (t, loss.item()))
check_accuracy_part34(loader_val, model)
print()
check_accuracy_part34(loader_train,model) #print the final train and val accuracy to avoid over-fitting
check_accuracy_part34(loader_val, model)
def my_init(m):
if(type(m)==nn.Conv2d or type(m)==nn.Linear):
print(type(m))
nn.init.kaiming_normal_(m.weight)
nn.init.constant_(m.bias,0)
learning_rate = 1e-3
model = nn.Sequential(
nn.BatchNorm2d(3),nn.Conv2d(3,64,(3,3),padding=1),nn.ReLU(),
nn.BatchNorm2d(64),nn.Conv2d(64,64,(3,3),padding=1),nn.ReLU(),
nn.MaxPool2d((2,2)),
nn.BatchNorm2d(64),nn.Conv2d(64,128,(3,3),padding=1),nn.ReLU(),
nn.MaxPool2d((2,2)),
nn.BatchNorm2d(128),nn.Conv2d(128,256,(3,3),padding=1),nn.ReLU(),
nn.MaxPool2d((2,2)),
nn.BatchNorm2d(256),
Flatten(),
nn.Linear(256*4*4,512),nn.ReLU(),nn.Dropout(0.5),
nn.Linear(512,10),
)
model.apply(my_init)
optimizer = optim.Adam(model.parameters(),lr=learning_rate)
print_every=100
my_train(model,optimizer,epochs=5)<jupyter_output><class 'torch.nn.modules.conv.Conv2d'>
<class 'torch.nn.modules.conv.Conv2d'>
<class 'torch.nn.modules.conv.Conv2d'>
<class 'torch.nn.modules.conv.Conv2d'>
<class 'torch.nn.modules.linear.Linear'>
<class 'torch.nn.modules.linear.Linear'>
Iteration 0, loss = 2.8770
Checking accuracy on validation set
Got 154 / 1000 correct (15.40)
Iteration 100, loss = 1.4513
Checking accuracy on validation set
Got 497 / 1000 correct (49.70)
Iteration 200, loss = 1.6458
Checking accuracy on validation set
Got 597 / 1000 correct (59.70)
Iteration 300, loss = 1.2044
Checking accuracy on validation set
Got 617 / 1000 correct (61.70)
Iteration 400, loss = 1.1888
Checking accuracy on validation set
Got 639 / 1000 correct (63.90)
Iteration 500, loss = 1.1744
Checking accuracy on validation set
Got 661 / 1000 correct (66.10)
Iteration 600, loss = 0.8904
Checking accuracy on validation set
Got 696 / 1000 correct (69.60)
Iteration 700, loss = 0.9292
Checking accuracy on validation set
Got 697 / 1000 corre[...]<jupyter_text>## Describe what you did
In the cell below you should write an explanation of what you did, any additional features that you implemented, and/or any graphs that you made in the process of training and evaluating your network.First, I tried the recommended architecture 3 with different N and M, but they didn't perform very well.
Actually, the highest val accuracy achieved is lower than 70%.
Then I considered adding Max Pooling layers after Conv layers, and after some trials, the results got much better.
## Test set -- run this only once
Now that we've gotten a result we're happy with, we test our final model on the test set (which you should store in best_model). Think about how this compares to your validation set accuracy.<jupyter_code>best_model = model_3
check_accuracy_part34(loader_test, best_model)<jupyter_output>Checking accuracy on test set
Got 8073 / 10000 correct (80.73)
|
no_license
|
/assignment2/PyTorch.ipynb
|
forestLoop/Learning-CS231n
| 21 |
<jupyter_start><jupyter_text># Batch Normalization
:label:`sec_batch_norm`
Training deep neural nets is difficult.
And getting them to converge in a reasonable amount of time can be tricky.
In this section, we describe batch normalization (BN)
:cite:`Ioffe.Szegedy.2015`, a popular and effective technique
that consistently accelerates the convergence of deep nets.
Together with residual blocks—covered in :numref:`sec_resnet`—BN
has made it possible for practitioners
to routinely train networks with over 100 layers.
## Training Deep Networks
To motivate batch normalization, let us review
a few practical challenges that arise
when training ML models and neural nets in particular.
1. Choices regarding data preprocessing often
make an enormous difference in the final results.
Recall our application of multilayer perceptrons
to predicting house prices (:numref:`sec_kaggle_house`).
Our first step when working with real data
was to standardize our input features
to each have a mean of *zero* and variance of *one*.
Intuitively, this standardization plays nicely with our optimizers
because it puts the parameters a-priori at a similar scale.
2. For a typical MLP or CNN, as we train,
the activations in intermediate layers
may take values with widely varying magnitudes—both
along the layers from the input to the output,
across nodes in the same layer,
and over time due to our updates to the model's parameters.
The inventors of batch normalization postulated informally
that this drift in the distribution of activations
could hamper the convergence of the network.
Intuitively, we might conjecture that if one
layer has activation values that are 100x that of another layer,
this might necessitate compensatory adjustments in the learning rates.
3. Deeper networks are complex and easily capable of overfitting.
This means that regularization becomes more critical.
Batch normalization is applied to individual layers
(optionally, to all of them) and works as follows:
In each training iteration,
we first normalize the inputs (of batch normalization)
by subtracting their mean and
dividing by their standard deviation,
where both are estimated based on the statistics of the current minibatch.
Next, we apply a scaling coefficient and a scaling offset.
It is precisely due to this *normalization* based on *batch* statistics
that *batch normalization* derives its name.
Note that if we tried to apply BN with minibatches of size $1$,
we would not be able to learn anything.
That is because after subtracting the means,
each hidden node would take value $0$!
As you might guess, since we are devoting a whole section to BN,
with large enough minibatches, the approach proves effective and stable.
One takeaway here is that when applying BN,
the choice of minibatch size may be
even more significant than without BN.
Formally, BN transforms the activations at a given layer $\mathbf{x}$
according to the following expression:
$$\mathrm{BN}(\mathbf{x}) = \mathbf{\gamma} \odot \frac{\mathbf{x} - \hat{\mathbf{\mu}}}{\hat\sigma} + \mathbf{\beta}$$
Here, $\hat{\mathbf{\mu}}$ is the minibatch sample mean
and $\hat{\mathbf{\sigma}}$ is the minibatch sample standard deviation.
After applying BN, the resulting minibatch of activations
has zero mean and unit variance.
Because the choice of unit variance
(vs some other magic number) is an arbitrary choice,
we commonly include coordinate-wise
scaling coefficients $\mathbf{\gamma}$ and offsets $\mathbf{\beta}$.
Consequently, the activation magnitudes
for intermediate layers cannot diverge during training
because BN actively centers and rescales them back
to a given mean and size (via $\mathbf{\mu}$ and $\sigma$).
One piece of practitioner's intuition/wisdom
is that BN seems to allows for more aggressive learning rates.
Formally, denoting a particular minibatch by $\mathcal{B}$,
we calculate $\hat{\mathbf{\mu}}_\mathcal{B}$ and $\hat\sigma_\mathcal{B}$ as follows:
$$\hat{\mathbf{\mu}}_\mathcal{B} \leftarrow \frac{1}{|\mathcal{B}|} \sum_{\mathbf{x} \in \mathcal{B}} \mathbf{x}
\text{ and }
\hat{\mathbf{\sigma}}_\mathcal{B}^2 \leftarrow \frac{1}{|\mathcal{B}|} \sum_{\mathbf{x} \in \mathcal{B}} (\mathbf{x} - \mathbf{\mu}_{\mathcal{B}})^2 + \epsilon$$
Note that we add a small constant $\epsilon > 0$
to the variance estimate
to ensure that we never attempt division by zero,
even in cases where the empirical variance estimate might vanish.
The estimates $\hat{\mathbf{\mu}}_\mathcal{B}$
and $\hat{\mathbf{\sigma}}_\mathcal{B}$ counteract the scaling issue
by using noisy estimates of mean and variance.
You might think that this noisiness should be a problem.
As it turns out, this is actually beneficial.
This turns out to be a recurring theme in deep learning.
For reasons that are not yet well-characterized theoretically,
various sources of noise in optimization
often lead to faster training and less overfitting.
While traditional machine learning theorists
might buckle at this characterization,
this variation appears to act as a form of regularization.
In some preliminary research,
:cite:`Teye.Azizpour.Smith.2018` and :cite:`Luo.Wang.Shao.ea.2018`
relate the properties of BN to Bayesian Priors and penalties respectively.
In particular, this sheds some light on the puzzle
of why BN works best for moderate minibatches sizes in the $50$–$100$ range.
Fixing a trained model, you might (rightly) think
that we would prefer to use the entire dataset
to estimate the mean and variance.
Once training is complete, why would we want
the same image to be classified differently,
depending on the batch in which it happens to reside?
During training, such exact calculation is infeasible
because the activations for all data points
change every time we update our model.
However, once the model is trained,
we can calculate the means and variances
of each layer's activations based on the entire dataset.
Indeed this is standard practice for
models employing batch normalization
and thus BN layers function differently
in *training mode* (normalizing by minibatch statistics)
and in *prediction mode* (normalizing by dataset statistics).
We are now ready to take a look at how batch normalization works in practice.
## Batch Normalization Layers
Batch normalization implementations for fully-connected layers
and convolutional layers are slightly different.
We discuss both cases below.
Recall that one key differences between BN and other layers
is that because BN operates on a full minibatch at a time,
we cannot just ignore the batch dimension
as we did before when introducing other layers.
### Fully-Connected Layers
When applying BN to fully-connected layers,
we usually insert BN after the affine transformation
and before the nonlinear activation function.
Denoting the input to the layer by $\mathbf{x}$,
the linear transform (with weights $\theta$) by $f_{\theta}(\cdot)$,
the activation function by $\phi(\cdot)$,
and the BN operation with parameters $\mathbf{\beta}$ and $\mathbf{\gamma}$
by $\mathrm{BN}_{\mathbf{\beta}, \mathbf{\gamma}}$,
we can express the computation of a BN-enabled,
fully-connected layer $\mathbf{h}$ as follows:
$$\mathbf{h} = \phi(\mathrm{BN}_{\mathbf{\beta}, \mathbf{\gamma}}(f_{\mathbf{\theta}}(\mathbf{x}) ) ) $$
Recall that mean and variance are computed
on the *same* minibatch $\mathcal{B}$
on which the transformation is applied.
Also recall that the scaling coefficient $\mathbf{\gamma}$
and the offset $\mathbf{\beta}$ are parameters that need to be learned
jointly with the more familiar parameters $\mathbf{\theta}$.
### Convolutional Layers
Similarly, with convolutional layers,
we typically apply BN after the convolution
and before the nonlinear activation function.
When the convolution has multiple output channels,
we need to carry out batch normalization
for *each* of the outputs of these channels,
and each channel has its own scale and shift parameters,
both of which are scalars.
Assume that our minibatches contain $m$ each
and that for each channel,
the output of the convolution has height $p$ and width $q$.
For convolutional layers, we carry out each batch normalization
over the $m \cdot p \cdot q$ elements per output channel simultaneously.
Thus we collect the values over all spatial locations
when computing the mean and variance
and consequently (within a given channel)
apply the same $\hat{\mathbf{\mu}}$ and $\hat{\mathbf{\sigma}}$
to normalize the values at each spatial location.
### Batch Normalization During Prediction
As we mentioned earlier, BN typically behaves differently
in training mode and prediction mode.
First, the noise in $\mathbf{\mu}$ and $\mathbf{\sigma}$
arising from estimating each on minibatches
are no longer desirable once we have trained the model.
Second, we might not have the luxury
of computing per-batch normalization statistics, e.g.,
we might need to apply our model to make one prediction at a time.
Typically, after training, we use the entire dataset
to compute stable estimates of the activation statistics
and then fix them at prediction time.
Consequently, BN behaves differently during training and at test time.
Recall that dropout also exhibits this characteristic.
## Implementation from Scratch
Below, firstly we get all the relevant libraries needed to implement BatchNorm. After that, we implement a batch normalization layer with NDArrays from scratch:
<jupyter_code>%load ../utils/djl-imports
%load ../utils/plot-utils
%load ../utils/Training.java
%load ../utils/Accumulator.java
import ai.djl.basicdataset.cv.classification.*;
import org.apache.commons.lang3.ArrayUtils;
public NDList batchNormUpdate(NDArray X, NDArray gamma,
NDArray beta, NDArray movingMean, NDArray movingVar,
float eps, float momentum, boolean isTraining) {
// attach moving mean and var to submanager to close intermediate computation values
// at the end to avoid memory leak
try(NDManager subManager = movingMean.getManager().newSubManager()){
movingMean.attach(subManager);
movingVar.attach(subManager);
NDArray xHat;
NDArray mean;
NDArray var;
if (!isTraining) {
// If it is the prediction mode, directly use the mean and variance
// obtained from the incoming moving average
xHat = X.sub(movingMean).div(movingVar.add(eps).sqrt());
} else {
if (X.getShape().dimension() == 2) {
// When using a fully connected layer, calculate the mean and
// variance on the feature dimension
mean = X.mean(new int[]{0}, true);
var = X.sub(mean).pow(2).mean(new int[]{0}, true);
} else {
// When using a two-dimensional convolutional layer, calculate the
// mean and variance on the channel dimension (axis=1). Here we
// need to maintain the shape of `X`, so that the broadcast
// operation can be carried out later
mean = X.mean(new int[]{0, 2, 3}, true);
var = X.sub(mean).pow(2).mean(new int[]{0, 2, 3}, true);
}
// In training mode, the current mean and variance are used for the
// standardization
xHat = X.sub(mean).div(var.add(eps).sqrt());
// Update the mean and variance of the moving average
movingMean = movingMean.mul(momentum).add(mean.mul(1.0f - momentum));
movingVar = movingVar.mul(momentum).add(var.mul(1.0f - momentum));
}
NDArray Y = xHat.mul(gamma).add(beta); // Scale and shift
// attach moving mean and var back to original manager to keep their values
movingMean.attach(subManager.getParentManager());
movingVar.attach(subManager.getParentManager());
return new NDList(Y, movingMean, movingVar);
}
}<jupyter_output><empty_output><jupyter_text>
We can now create a proper `BatchNorm` layer. Our layer will maintain proper parameters corresponding for scale gamma and shift beta, both of which will be updated in the course of training. Additionally, our layer will maintain a moving average of the means and variances for subsequent use during model prediction. The numFeatures parameter required by the BatchNorm instance is the number of outputs for a fully-connected layer and the number of output channels for a convolutional layer. The numDimensions parameter also required by this instance is 2 for a fully-connected layer and 4 for a convolutional layer.
Putting aside the algorithmic details, note the design pattern underlying our implementation of the layer. Typically, we define the math in a separate function, say `batchNormUpdate`. We then integrate this functionality into a custom layer, whose code mostly addresses bookkeeping matters, such as moving data to the right device context, allocating and initializing any required variables, keeping track of running averages (here for mean and variance), etc. This pattern enables a clean separation of math from boilerplate code. Also note that for the sake of convenience we did not worry about automatically inferring the input shape here, thus we need to specify the number of features throughout. Do not worry, the DJL `BatchNorm` layer will care of this for us.
<jupyter_code>public class BatchNormBlock extends AbstractBlock {
private static final byte VERSION = 1;
private NDArray movingMean;
private NDArray movingVar;
private Parameter gamma;
private Parameter beta;
private Shape shape;
// num_features: the number of outputs for a fully-connected layer
// or the number of output channels for a convolutional layer.
// num_dims: 2 for a fully-connected layer and 4 for a convolutional layer.
public BatchNormBlock(int numFeatures, int numDimensions) {
super(VERSION);
if (numDimensions == 2) {
shape = new Shape(1, numFeatures);
} else {
shape = new Shape(1, numFeatures, 1, 1);
}
// The scale parameter and the shift parameter involved in gradient
// finding and iteration are initialized to 0 and 1 respectively
gamma = addParameter(
Parameter.builder()
.setName("gamma")
.setType(Parameter.Type.GAMMA)
.optShape(shape)
.build());
beta = addParameter(
Parameter.builder()
.setName("beta")
.setType(Parameter.Type.BETA)
.optShape(shape)
.build());
// All the variables not involved in gradient finding and iteration are
// initialized to 0. Create a base manager to maintain their values
// throughout the entire training process
NDManager manager = NDManager.newBaseManager();
movingMean = manager.zeros(shape);
movingVar = manager.zeros(shape);
}
@Override
public String toString() {
return "BatchNormBlock()";
}
@Override
protected NDList forwardInternal(
ParameterStore parameterStore,
NDList inputs,
boolean training,
PairList<String, Object> params) {
NDList result = batchNormUpdate(inputs.singletonOrThrow(),
gamma.getArray(), beta.getArray(), this.movingMean, this.movingVar, 1e-12f, 0.9f, training);
// close previous NDArray before assigning new values
if(training){
this.movingMean.close();
this.movingVar.close();
}
// Save the updated `movingMean` and `movingVar`
this.movingMean = result.get(1);
this.movingVar = result.get(2);
return new NDList(result.get(0));
}
@Override
public Shape[] getOutputShapes(Shape[] inputs) {
Shape[] current = inputs;
for (Block block : children.values()) {
current = block.getOutputShapes(current);
}
return current;
}
}<jupyter_output><empty_output><jupyter_text>## Using a Batch Normalization LeNet
To see how to apply `BatchNorm` in context,
below we apply it to a traditional LeNet model (:numref:`sec_lenet`).
Recall that BN is typically applied
after the convolutional layers and fully-connected layers
but before the corresponding activation functions.
<jupyter_code>SequentialBlock net = new SequentialBlock()
.add(
Conv2d.builder()
.setKernelShape(new Shape(5, 5))
.setFilters(6).build())
.add(new BatchNormBlock(6, 4))
.add(Pool.maxPool2dBlock(new Shape(2, 2), new Shape(2, 2)))
.add(
Conv2d.builder()
.setKernelShape(new Shape(5, 5))
.setFilters(16).build())
.add(new BatchNormBlock(16, 4))
.add(Activation::sigmoid)
.add(Pool.maxPool2dBlock(new Shape(2, 2), new Shape(2, 2)))
.add(Blocks.batchFlattenBlock())
.add(Linear.builder().setUnits(120).build())
.add(new BatchNormBlock(120, 2))
.add(Activation::sigmoid)
.add(Blocks.batchFlattenBlock())
.add(Linear.builder().setUnits(84).build())
.add(new BatchNormBlock(84, 2))
.add(Activation::sigmoid)
.add(Linear.builder().setUnits(10).build());<jupyter_output><empty_output><jupyter_text>Let's initialize the batchSize, numEpochs and the relevant arrays to store the data from the training function.<jupyter_code>int batchSize = 256;
int numEpochs = Integer.getInteger("MAX_EPOCH", 10);
double[] trainLoss;
double[] testAccuracy;
double[] epochCount;
double[] trainAccuracy;
epochCount = new double[numEpochs];
for (int i = 0; i < epochCount.length; i++) {
epochCount[i] = i+1;
}<jupyter_output><empty_output><jupyter_text>As before, we will train our network on the Fashion-MNIST dataset.
This code is virtually identical to that when we first trained LeNet (:numref:`sec_lenet`).
The main difference is the considerably larger learning rate.
<jupyter_code>FashionMnist trainIter = FashionMnist.builder()
.optUsage(Dataset.Usage.TRAIN)
.setSampling(batchSize, true)
.optLimit(Long.getLong("DATASET_LIMIT", Long.MAX_VALUE))
.build();
FashionMnist testIter = FashionMnist.builder()
.optUsage(Dataset.Usage.TEST)
.setSampling(batchSize, true)
.optLimit(Long.getLong("DATASET_LIMIT", Long.MAX_VALUE))
.build();
trainIter.prepare();
testIter.prepare();
float lr = 1.0f;
Loss loss = Loss.softmaxCrossEntropyLoss();
Tracker lrt = Tracker.fixed(lr);
Optimizer sgd = Optimizer.sgd().setLearningRateTracker(lrt).build();
DefaultTrainingConfig config = new DefaultTrainingConfig(loss)
.optOptimizer(sgd) // Optimizer (loss function)
.optDevices(Device.getDevices(1)) // single GPU
.addEvaluator(new Accuracy()) // Model Accuracy
.addTrainingListeners(TrainingListener.Defaults.logging()); // Logging
Model model = Model.newInstance(Device.defaultDevice().toString());
model.setBlock(net);
Trainer trainer = model.newTrainer(config);
trainer.initialize(new Shape(1, 1, 28, 28));
Map<String, double[]> evaluatorMetrics = new HashMap<>();
double avgTrainTimePerEpoch = 0;
Training.trainingChapter6(trainIter, testIter, numEpochs, trainer, evaluatorMetrics, avgTrainTimePerEpoch);
trainLoss = evaluatorMetrics.get("train_epoch_SoftmaxCrossEntropyLoss");
trainAccuracy = evaluatorMetrics.get("train_epoch_Accuracy");
testAccuracy = evaluatorMetrics.get("validate_epoch_Accuracy");
System.out.printf("loss %.3f,", trainLoss[numEpochs - 1]);
System.out.printf(" train acc %.3f,", trainAccuracy[numEpochs - 1]);
System.out.printf(" test acc %.3f\n", testAccuracy[numEpochs - 1]);
System.out.printf("%.1f examples/sec", trainIter.size() / (avgTrainTimePerEpoch / Math.pow(10, 9)));
System.out.println();<jupyter_output><empty_output><jupyter_text>Let us have a look at the scale parameter `gamma`
and the shift parameter `beta` learned
from the first batch normalization layer.
<jupyter_code>// Printing the value of gamma and beta in the first BatchNorm layer.
List<Parameter> batchNormFirstParams = net.getChildren().values().get(1).getParameters().values();
System.out.println("gamma " + batchNormFirstParams.get(0).getArray().reshape(-1));
System.out.println("beta " + batchNormFirstParams.get(1).getArray().reshape(-1));<jupyter_output><empty_output><jupyter_text><jupyter_code>String[] lossLabel = new String[trainLoss.length + testAccuracy.length + trainAccuracy.length];
Arrays.fill(lossLabel, 0, trainLoss.length, "train loss");
Arrays.fill(lossLabel, trainAccuracy.length, trainLoss.length + trainAccuracy.length, "train acc");
Arrays.fill(lossLabel, trainLoss.length + trainAccuracy.length,
trainLoss.length + testAccuracy.length + trainAccuracy.length, "test acc");
Table data = Table.create("Data").addColumns(
DoubleColumn.create("epoch", ArrayUtils.addAll(epochCount, ArrayUtils.addAll(epochCount, epochCount))),
DoubleColumn.create("metrics", ArrayUtils.addAll(trainLoss, ArrayUtils.addAll(trainAccuracy, testAccuracy))),
StringColumn.create("lossLabel", lossLabel)
);
render(LinePlot.create("", data, "epoch", "metrics", "lossLabel"),"text/html");<jupyter_output><empty_output><jupyter_text>## Concise Implementation
Compared with the `BatchNorm` class, which we just defined ourselves, the `BatchNorm` class defined by `nn` module in DJL is easier to use. In DJL, we do not have to worry about `numFeatures` or `numDimensions`. Instead, these parameter values will be inferred automatically via delayed initialization. Otherwise, the code looks virtually identical to the application our implementation above.
<jupyter_code>SequentialBlock block = new SequentialBlock()
.add(
Conv2d.builder()
.setKernelShape(new Shape(5, 5))
.setFilters(6).build())
.add(BatchNorm.builder().build())
.add(Pool.maxPool2dBlock(new Shape(2, 2), new Shape(2, 2)))
.add(
Conv2d.builder()
.setKernelShape(new Shape(5, 5))
.setFilters(16).build())
.add(BatchNorm.builder().build())
.add(Activation::sigmoid)
.add(Pool.maxPool2dBlock(new Shape(2, 2), new Shape(2, 2)))
.add(Blocks.batchFlattenBlock())
.add(Linear.builder().setUnits(120).build())
.add(BatchNorm.builder().build())
.add(Activation::sigmoid)
.add(Blocks.batchFlattenBlock())
.add(Linear.builder().setUnits(84).build())
.add(BatchNorm.builder().build())
.add(Activation::sigmoid)
.add(Linear.builder().setUnits(10).build());<jupyter_output><empty_output><jupyter_text>Below, we use the same hyperparameters to train out model.
Note that as usual, the high-level API variant runs much faster
because its code has been compiled to C++/CUDA
while our custom implementation must be interpreted by Python.
<jupyter_code>Loss loss = Loss.softmaxCrossEntropyLoss();
Tracker lrt = Tracker.fixed(1.0f);
Optimizer sgd = Optimizer.sgd().setLearningRateTracker(lrt).build();
Model model = Model.newInstance(Device.defaultDevice().toString());
model.setBlock(block);
DefaultTrainingConfig config = new DefaultTrainingConfig(loss)
.optOptimizer(sgd) // Optimizer (loss function)
.addEvaluator(new Accuracy()) // Model Accuracy
.addTrainingListeners(TrainingListener.Defaults.logging()); // Logging
Trainer trainer = model.newTrainer(config);
trainer.initialize(new Shape(1, 1, 28, 28));
Map<String, double[]> evaluatorMetrics = new HashMap<>();
double avgTrainTimePerEpoch = 0;
Training.trainingChapter6(trainIter, testIter, numEpochs, trainer, evaluatorMetrics, avgTrainTimePerEpoch);
trainLoss = evaluatorMetrics.get("train_epoch_SoftmaxCrossEntropyLoss");
trainAccuracy = evaluatorMetrics.get("train_epoch_Accuracy");
testAccuracy = evaluatorMetrics.get("validate_epoch_Accuracy");
System.out.printf("loss %.3f,", trainLoss[numEpochs - 1]);
System.out.printf(" train acc %.3f,", trainAccuracy[numEpochs - 1]);
System.out.printf(" test acc %.3f\n", testAccuracy[numEpochs - 1]);
System.out.printf("%.1f examples/sec", trainIter.size() / (avgTrainTimePerEpoch / Math.pow(10, 9)));
System.out.println();<jupyter_output><empty_output><jupyter_text><jupyter_code>String[] lossLabel = new String[trainLoss.length + testAccuracy.length + trainAccuracy.length];
Arrays.fill(lossLabel, 0, trainLoss.length, "train loss");
Arrays.fill(lossLabel, trainAccuracy.length, trainLoss.length + trainAccuracy.length, "train acc");
Arrays.fill(lossLabel, trainLoss.length + trainAccuracy.length,
trainLoss.length + testAccuracy.length + trainAccuracy.length, "test acc");
Table data = Table.create("Data").addColumns(
DoubleColumn.create("epoch", ArrayUtils.addAll(epochCount, ArrayUtils.addAll(epochCount, epochCount))),
DoubleColumn.create("metrics", ArrayUtils.addAll(trainLoss, ArrayUtils.addAll(trainAccuracy, testAccuracy))),
StringColumn.create("lossLabel", lossLabel)
);
render(LinePlot.create("", data, "epoch", "metrics", "lossLabel"),"text/html");<jupyter_output><empty_output>
|
non_permissive
|
/chapter_convolutional-modern/batch-norm.ipynb
|
panda-run/d2l-java-zh
| 10 |
<jupyter_start><jupyter_text>## Clean Code#### Helper Functions<jupyter_code>def get_xml(valid_xml_file):
'''
Return a xml object with patent data
:param valid_xml_file: a valid xml object
:return the root object of the xml file, ready to parse
'''
tree = etree.parse(valid_xml_file)
root = tree.getroot()
return root
def process_patent_numbers(raw_patent_num):
'''
Helper function ot transform patent ids into thier final format
:param raw_patent_num: patent number extracted from the raw XML
:return cleaned patent id.
'''
num = re.findall('\d+', raw_patent_num)[0] #get the just numbers in string form
if num[0].startswith("0"):
num = num[1:]
let = re.findall('[a-zA-Z]+', raw_patent_num) #get the letter prefixes
if let:
let = let[0]#list to string
clean_patent_num = let + num
else:
clean_patent_num = num
return clean_patent_num
def process_date(date):
'''
Takes a date formated as 6 numbers and returns it with dashes and days that are 00 replaced with 01
:params date: a date object formatted as 6 numbers
:returns cleaned up date
'''
if date is not None:
if date[6:] != "00":
date = date[:4]+'-'+date[4:6]+'-'+date[6:]
else:
date = date[:4]+'-'+date[4:6]+'-'+'01'
return date
def recursive_children(xml_element, parent_field=""):
'''
:params xml_element: xml object can be nested
:params parent_field: parent of nested xml object
:returns a dictionary of the tags and texts of nested xml object
'''
test_list = []
if len(xml_element)==0:
if parent_field:
test_list.append((parent_field+"-"+xml_element.tag, xml_element.text))
else:
test_list.append((xml_element.tag, xml_element.text))
#print xml_element.tag, xml_element.text
else:
parent_field = xml_element.tag
for element in xml_element:
test_list += recursive_children(element, parent_field)
return test_list<jupyter_output><empty_output><jupyter_text>#### Data Functions<jupyter_code>def get_entity(patent, entity_name, attribute_list=None):
'''
:params patent: take the xml object representing a patent
:params entity_name: a string with the xml tag for an entity with single or multiple entities
:returns a list of default dictionaries with all the data for the entity and processes dates
'''
var_list=[]
xml = patent.findall('.//'+entity_name)
for field in xml:
data={}
if attribute_list:
for attribute in attribute_list:
data[attribute]=field.attrib[attribute]
#recursive function modifies data dictionary defined above
results_list = recursive_children(field)
data.update(dict(results_list))
for key in data.keys():
if 'date' in key:
data[key] = process_date(data[key])
var_list.append(data)
return var_list<jupyter_output><empty_output><jupyter_text>### Processed Entities<jupyter_code>#Single
#number-of-claims
#us-application-series code
#figures
#pct-or-regional-publishing-data
#us-term-of-grant
#pct-or-regional-filing-data
#publication-reference: this gives you the patent data
#application-reference, attribute_list=['appl-type']
#invention-title, attribute_list=['id']
#us-application-series-code
#us-botanic
#Multiple
#us-exemplary-claim
#classifications-ipcr
#classification-national
#inventors
#us-references-cited
#assignees
#'us-applicants', attribute_list=['sequence', 'app-type', 'designation', 'applicant-authority-category'
#'inventors', attribute_list=['sequence', 'designation']
#'agents', attribute_list=['sequence', 'rep-type']
#us_related_docs
<jupyter_output><empty_output><jupyter_text>## To Run Process<jupyter_code>patents = get_xml("D:\Caitlin_PV\Data\ipg171212_clean.xml")<jupyter_output><empty_output><jupyter_text>## Working Area<jupyter_code>patents #is an xml-tree object that has all the patents an information
patents[104] #is the element that represnts the 104th patent
#The main highest level grouings are things like 'abstract','figures', and 'us-bibliographic-data-grant
#'us-bibliographic-data-grant' has most of the fields we are working with
patent = patents[3705]<jupyter_output><empty_output><jupyter_text>### PostProcessing<jupyter_code>#TODO role for examiners
#TODO #classification-national post processing, line 556<jupyter_output><empty_output>
|
no_license
|
/Development/XML_Parser_Dev.ipynb
|
vkelk/PatentsView-DB
| 6 |
<jupyter_start><jupyter_text>[View in Colaboratory](https://colab.research.google.com/github/marcelcerri/Integra-o-ordens-de-rea-o/blob/master/Integrando_equa%C3%A7%C3%B5es_diferenciais_e_ajustando_pontos_experimentais_importando_os_dados.ipynb)**Integrando equações diferenciais e ajuste de pontos experimentais completo**<jupyter_code>#Importação todos os pacotes utilizados
import pandas as pd
from google.colab import files
import io
from scipy import stats
from scipy.interpolate import *
from scipy.integrate import odeint
from scipy.optimize import curve_fit
import numpy as np
import matplotlib.pyplot as plt
#carregar o arquivo em csv
uploaded = files.upload()
#verificação do upload
for fn in uploaded.keys():
print('O arquivo "{name}" com tamanho de {length} bytes'.format(
name=fn, length=len(uploaded[fn])))
#transformando o arquivo csv em data frame
calibracao_df = pd.read_csv(io.StringIO(uploaded['Cultivo_e_coli_1000_rpm_sem_lag.csv'].decode('utf-8')))
#impressão do arquivo em data frame
calibracao_df
#transformando data frame em vetor
calibracao_np = calibracao_df.values
#impressão do vetor
calibracao_np
#vetor do eixo x
t = calibracao_np[:,0]
print(t)
#vetor do eixo y
data_ca = calibracao_np[:,1]
print(data_ca)
def growth(Ca,t, mi):
return mi*Ca
def y(t, mi, Ca0):
y = odeint(growth, Ca0, t, args=(mi,))
return y.ravel()
popt, cov = curve_fit(y, t, data_ca, [0.1, 0.1])
a_opt, y0_opt = popt
print(y)
print("mi = %g" % a_opt)
print("Ca0 = %g" % y0_opt)
import matplotlib.pyplot as plt
plt.plot(t, data_ca, '.',
t, y(t, a_opt, y0_opt), '-')
plt.gcf().set_size_inches(13, 9)
plt.savefig('out.png', dpi=100)
plt.xlabel('Tempo (horas)')
plt.ylabel('Densidade Optica (-)')
plt.title('DO em funcao do tempo')
plt.legend()
plt.show()
files.download( "out.png" )
#y = odeint(growth, y0_opt, t, args=(a_opt,))
#guardando as informações e salvando em txt
data = np.vstack((t, y.T))
data = data.T
print(data)
#salvando em csv
df = pd.DataFrame(data)
df.to_csv("data.csv")
files.download('data.csv')
print(df)<jupyter_output> 0 1
0 0.0 0.203015
1 2.0 0.272297
2 4.0 0.365222
3 6.0 0.489860
4 8.0 0.657031
5 10.0 0.881252
6 12.0 1.181992
7 14.0 1.585364
8 22.0 5.130813
9 24.0 6.881776
|
no_license
|
/Crescimento_microbiano_E_coli_experimental_1000_rpm_23082018.ipynb
|
marcelcerri/Ajuste-de-modelos-n-o-lineares
| 1 |
<jupyter_start><jupyter_text>### Preparar ambiente<jupyter_code>import sys
!{sys.executable} -m pip install PyAthena<jupyter_output>Requirement already satisfied: PyAthena in /home/ec2-user/anaconda3/envs/python3/lib/python3.6/site-packages (2.3.0)
Requirement already satisfied: botocore>=1.5.52 in /home/ec2-user/anaconda3/envs/python3/lib/python3.6/site-packages (from PyAthena) (1.20.79)
Requirement already satisfied: tenacity>=4.1.0 in /home/ec2-user/anaconda3/envs/python3/lib/python3.6/site-packages (from PyAthena) (7.0.0)
Requirement already satisfied: boto3>=1.4.4 in /home/ec2-user/anaconda3/envs/python3/lib/python3.6/site-packages (from PyAthena) (1.17.79)
Requirement already satisfied: s3transfer<0.5.0,>=0.4.0 in /home/ec2-user/anaconda3/envs/python3/lib/python3.6/site-packages (from boto3>=1.4.4->PyAthena) (0.4.2)
Requirement already satisfied: jmespath<1.0.0,>=0.7.1 in /home/ec2-user/anaconda3/envs/python3/lib/python3.6/site-packages (from boto3>=1.4.4->PyAthena) (0.10.0)
Requirement already satisfied: urllib3<1.27,>=1.25.4 in /home/ec2-user/anaconda3/envs/python3/lib/python3.6/site-packages (from botocore[...]<jupyter_text>### Conectar con pyathena<jupyter_code>from pyathena.pandas.cursor import PandasCursor
from pyathena import connect
import pandas as pd
import time
# agregar directorio en el cual se almacenan las tablas que se van a consultar
directorio = 's3://iadbprod-csd-hub-analyticaldata/graphdata-mobility-temporal/athena-results/'
# base de datos
bd = 'graphdata' # Database in Glue
cursor = connect(s3_staging_dir = directorio, region_name = 'us-east-1', schema_name = bd, cursor_class = PandasCursor).cursor()<jupyter_output><empty_output><jupyter_text>### Completar con columnas lat_frecuentes, long_frecuentes y lat_final, long_final<jupyter_code>fecha_inicio = '09/01/2020'
fecha_final = '09/30/2020'
fechas = pd.date_range(fecha_inicio,fecha_final)
for fecha in fechas:
day = str(fecha.day)
if len(day) == 1:
day = f'0{day}'
codigo_fecha = str(fecha.month) + day
print(codigo_fecha)
tabla_usuarios_distancia = f'todos_usuarios_delta_dist_{codigo_fecha}'
# Tabla historica preliminar para capturar latitud y longitud frecuente (mes base: 0 +1)
tabla_historico_usuarios = 'historico_usuarios_preliminar'
query_usuario_freq = f'''CREATE VIEW hogares_frecuentes AS
SELECT t_res.caid,
t_res.iso_country_code,
t_res.first_lat,
t_res.first_long
FROM
(SELECT t_aux.caid AS caid,
t_aux.iso_country_code AS iso_country_code,
FIRST_VALUE(t_aux.lat_hogar) over( partition by t_aux.caid,
t_aux.iso_country_code) AS first_lat,
FIRST_VALUE(t_aux.long_hogar) over( partition by t_aux.caid,
t_aux.iso_country_code) AS first_long
FROM
(SELECT t_hist.caid,
t_hist.iso_country_code,
t_hist.lat_hogar,
t_hist.long_hogar,
COUNT(*) AS count_hogar
FROM {tabla_usuarios_distancia} AS t_ud
LEFT JOIN {tabla_historico_usuarios} AS t_hist
ON t_ud.caid = t_hist.caid
AND t_ud.iso_country_code = t_hist.iso_country_code
GROUP BY t_hist.caid, t_hist.iso_country_code, t_hist.lat_hogar, t_hist.long_hogar
ORDER BY COUNT(*) DESC, SUM(t_hist.n_obs_hogar) DESC) AS t_aux ) AS t_res
GROUP BY t_res.caid, t_res.iso_country_code, t_res.first_lat, t_res.first_long'''
cursor.execute(query_usuario_freq)
time.sleep(6)
while (not cursor.has_result_set):
time.sleep(6)
cursor.close()
tabla_usuarios_distancia_cor = f'todos_usuarios_delta_dist_corregida_{codigo_fecha}'
query = f'''CREATE TABLE {tabla_usuarios_distancia_cor} AS
SELECT t_users.*,
t_hogares.first_lat AS lat_frecuente,
t_hogares.first_long AS long_frecuente,
CASE
WHEN t_users.lat_hogar is NULL THEN
t_hogares.first_lat
ELSE t_users.lat_hogar END as lat_final,
CASE
WHEN t_users.long_hogar is NULL THEN
t_hogares.first_long
ELSE t_users.long_hogar END as long_final
FROM {tabla_usuarios_distancia} AS t_users
LEFT JOIN hogares_frecuentes AS t_hogares
ON t_users.caid = t_hogares.caid
AND t_users.iso_country_code = t_hogares.iso_country_code'''
cursor.execute(query)
cursor.execute("DROP VIEW graphdata.hogares_frecuentes")<jupyter_output>901
902
903
904
905
906
907
908
909
910
911
912
913
914
915
916
917
918
919
920
921
922
923
924
925
926
927
928
929
930
|
no_license
|
/graphdata-mobility-iadb/Fase1/1_Ventana/6_Completar hogar faltantes_Ventana.ipynb
|
datapartnership/covid19-lac
| 3 |
<jupyter_start><jupyter_text>Load data<jupyter_code>repeat_sizes = pd.read_csv('/Users/kibanez/Documents/STRs/VALIDATION/table_validation_simple_withMaxAllele.tsv', sep='\t')
repeat_sizes.head()<jupyter_output><empty_output><jupyter_text>Prepare data for plotting<jupyter_code>repeat_info = {
'AR': {'expected_range': [34, 38], 'unit_len': 3, 'index': 12},
'ATN1': {'expected_range': [46, 48], 'unit_len': 3, 'index': 11},
'ATXN1': {'expected_range': [39, 44], 'unit_len': 3, 'index': 10},
'ATXN2': {'expected_range': [32, 33], 'unit_len': 3, 'index': 9},
'ATXN3': {'expected_range': [45, 60], 'unit_len': 3, 'index': 8},
'ATXN7': {'expected_range': [34, 36], 'unit_len': 3, 'index': 7},
'CACNA1A': {'expected_range': [18, 20], 'unit_len': 3, 'index': 6},
'C9orf72': {'expected_range': [60, 60], 'unit_len': 6, 'index': 5},
'FXN': {'expected_range': [44, 66], 'unit_len': 3, 'index': 4},
'HTT': {'expected_range': [36, 40], 'unit_len': 3, 'index': 3},
'TBP': {'expected_range': [44, 49], 'unit_len': 3, 'index': 2},
'FMR1': {'expected_range': [55, 200], 'unit_len': 3, 'index': 1},
'PPP2R2B': {'expected_range': [32, 51], 'unit_len': 3, 'index': 0}
}
max_repeat_size = 650
fig, ax = plt.subplots(figsize=(15, 7))
for repeat_id, rec in repeat_sizes.groupby(by='loci'):
normal_cutoff, premutation_cutoff = repeat_info[repeat_id]['expected_range']
base_y = repeat_info[repeat_id]['index']
ax.add_patch(patches.Rectangle((0, base_y - 0.5),
normal_cutoff, 1.0,
facecolor='#048732', alpha=0.2, ec='k'))
ax.add_patch(patches.Rectangle((normal_cutoff, base_y - 0.5),
premutation_cutoff - normal_cutoff,
1.0, facecolor='#eda407', alpha=0.2, ec='k'))
ax.add_patch(patches.Rectangle((premutation_cutoff, base_y - 0.5),
max_repeat_size - premutation_cutoff,
1.0, facecolor='#ed0716', alpha=0.2, ec='k'))
#allele_sizes = list(rec.max_EH_allele)
allele_sizes = list(rec.EH_a1) + list(rec.EH_a2)
ys = np.random.normal(base_y, 0.12, len(allele_sizes))
ax.scatter(allele_sizes, ys, c='black')
gene_names = [(gene, rec['index']) for gene, rec in repeat_info.items()]
gene_names = sorted(gene_names, key=lambda k: k[1])
gene_names = [g for g, _ in gene_names]
ax.yaxis.set_ticks(range(0, len(gene_names)))
ax.yaxis.set_ticklabels(gene_names)
ax.margins(x=0, y=0)
ax.set(xlabel='Expansion Hunter\'s estimation - size of the expansion', ylabel='Locus containing the repeat');
# Let's save the plot into a PDF file
fig.savefig("/Users/kibanez/Documents/STRs/GEL_Conference_Autumn2019/figures/repeat-sizes_EHa1_EHa2_average.pdf", bbox_inches='tight')
# Let's save the plot into a high quality PNG file
fig.savefig("/Users/kibanez/Documents/STRs/GEL_Conference_Autumn2019/figures/repeat-sizes_EHa1_EHa2_average.png", bbox_inches='tight', dpi=600)<jupyter_output><empty_output>
|
no_license
|
/plotting_validation_table_repeat_sizes.ipynb
|
kibanez/STR_detection_python3
| 2 |
<jupyter_start><jupyter_text>**Other Callbacks**<jupyter_code>class LR_Find(Callback):
_order = 1
def __init__(self, max_iter=100, min_lr=1e-6,max_lr=10):
self.max_iter, self.min_lr,self.max_lr = max_iter,min_lr,max_lr
self.best_loss =1e9
def begin_batch(self):
if not self.in_train: return
pos = self.n_iter/self.max_iter
lr = self.min_lr * (self.max_lr/self.min_lr) ** pos
for pg in self.opt.param_groups: pg['lr'] = lr
def after_step(self):
if self.n_iter>=self.max_iter or self.loss>self.best_loss*10:
raise CancelTrainException()
if self.loss < self.best_loss: self.best_loss = self.loss
learn = create_learner(get_model, loss_func, data)
run = Runner(cb_funcs=[LR_Find, Recorder])
run.fit(2,learn)
run.recorder.plot(skip_last=5)
!pip install fire
!python notebook2script.py 05b_early_stopping.ipynb<jupyter_output>Converted 05b_early_stopping.ipynb to exp/nb_05b.py
|
no_license
|
/05b_early_stopping.ipynb
|
Hlompho-Dash/first_try_of_fastai
| 1 |
<jupyter_start><jupyter_text>### 4.2.2 过拟合问题
真实应用中想要的不是让模型尽量模拟训练数据的行为,而是希望通过训练出来的模型对未知的数据给出判断。
过拟合:模型过为复杂,可以很好的“记忆”每一个训练数据中随机噪音的部分,而忘记了要去“学习”训练数据中通用的趋势。
为了避免模型出现过拟合,非常常用的方法就是正则化。
### 正则化
思想:在损失函数中加入刻画模型复杂程度的指标。一般来说,模型复杂度只由权重w决定。
L1正则化:$R(w) = ||w||_1 = \sum_{i}{w_i}$
L2正则化:$R(w) = ||w||_2^2 = \sum_{i}{w_i^2}$
L1正则化会让参数变得更稀疏,即会有更多的参数变为0,这样可以达到类似特征选取的功能。L2不会。
L1计算公式不可导,L2可导。因为在优化时,需要计算损失函数的偏导数,所以对L2正则化损失函数的优化更简洁。
实践中,也可将二者同时使用:
$$R(w) = \sum_{i}{a}{|w_i|} + (1-a)w_i^2$$#### 1. 生成模拟数据集。<jupyter_code>import tensorflow as tf
import matplotlib.pyplot as plt
import numpy as np
dataset_size = 200
data = []
label = []
np.random.seed(0)
# 以原点为圆心,半径为1的圆把散点划分成红蓝两部分,并加入随机噪音。
for i in range(dataset_size):
x1 = np.random.uniform(-1,1)
x2 = np.random.uniform(0,2)
if x1**2 + x2**2 <= 1:
data.append([np.random.normal(x1, 0.1),np.random.normal(x2,0.1)])
label.append(0)
else:
data.append([np.random.normal(x1, 0.1), np.random.normal(x2, 0.1)])
label.append(1)
data = np.hstack(data).reshape(-1,2)
label = np.hstack(label).reshape(-1, 1)
plt.scatter(data[:,0], data[:,1], c=label.reshape(200,),
cmap="RdBu", vmin=-.2, vmax=1.2, edgecolor="white")
plt.show()<jupyter_output><empty_output><jupyter_text>#### 2. 定义一个获取权重,并自动加入正则项到损失的函数。一般tf.Variable都是需要训练的权重参数<jupyter_code>def get_weight(shape, var_lambda):
# 生成一个变量
w = tf.Variable(tf.random_normal(shape), dtype=tf.float32)
# 将新生成变量的L2正则化损失加入集合
# losses是集合名,其后是要加入集合的内容
tf.add_to_collection('losses', tf.contrib.layers.l2_regularizer(var_lambda)(w))
return w<jupyter_output><empty_output><jupyter_text>#### 3. 定义神经网络。<jupyter_code>x = tf.placeholder(tf.float32, shape=(None, 2))
y_ = tf.placeholder(tf.float32, shape=(None, 1))
# 定义每层网络中节点的个数
layer_dimension = [2,10,5,3,1]
# 神经网络层数
n_layers = len(layer_dimension)
# 该变量维护前向传播时最深层的节点,开始的时候就是输入层
cur_layer = x
# 当前层的节点个数
in_dimension = layer_dimension[0]
# 通过循环来生成5层全连接的神经网络结构
# 输入层X[None, 2]
# 隐藏层W1[2, 10]、W2[10, 5]、W3[5, 3]
# 输出层Y[None, 1]
for i in range(1, n_layers):
# 下一层的节点个数
out_dimension = layer_dimension[i]
# 生成当前层中权重的变量weight,并将这个变量的L2正则化损失加入计算图上的集合
weight = get_weight([in_dimension, out_dimension], 0.003)
bias = tf.Variable(tf.constant(0.1, shape=[out_dimension]))
# 使用ReLU激活函数
cur_layer = tf.nn.elu(tf.matmul(cur_layer, weight) + bias)
# 进入下一层之前将下一层的节点个数更新为当前层节点个数
in_dimension = layer_dimension[i]
y= cur_layer
# 在神经网络前向传播的同时,已经将所有的L2正则化损失加入了图上的集合
# 这里只需要计算模型在训练数据上表现的损失函数
mse_loss = tf.reduce_sum(tf.pow(y_ - y, 2)) / dataset_size
# 将均方差误差损失函数加入损失集合
tf.add_to_collection('losses', mse_loss)
# get_collection 返回一个列表,这个列表是所有这个集合中的元素。
# 该元素是损失函数的不同部分,将它们加起来就可以得到最终的损失函数
loss = tf.add_n(tf.get_collection('losses'))<jupyter_output><empty_output><jupyter_text>#### 4. 训练不带正则项的损失函数mse_loss。<jupyter_code># 定义训练的目标函数mse_loss,训练次数及训练模型
train_op = tf.train.AdamOptimizer(0.001).minimize(mse_loss)
TRAINING_STEPS = 10000
with tf.Session() as sess:
tf.global_variables_initializer().run()
for i in range(TRAINING_STEPS):
sess.run(train_op, feed_dict={x: data, y_: label})
if i % 1000 == 1000 - 1:
print("After %d steps, mse_loss: %f" % (i,sess.run(mse_loss, feed_dict={x: data, y_: label})))
# 画出训练后的分割曲线
xx, yy = np.mgrid[-1.2:1.2:.01, -0.2:2.2:.01]
grid = np.c_[xx.ravel(), yy.ravel()]
probs = sess.run(y, feed_dict={x:grid})
probs = probs.reshape(xx.shape)
plt.scatter(data[:,0], data[:,1], c=label.reshape(200,),
cmap="RdBu", vmin=-.2, vmax=1.2, edgecolor="white")
plt.contour(xx, yy, probs, levels=[.5], cmap="Greys", vmin=0, vmax=.1)
plt.show()<jupyter_output>After 999 steps, mse_loss: 0.066249
After 1999 steps, mse_loss: 0.060282
After 2999 steps, mse_loss: 0.050951
After 3999 steps, mse_loss: 0.043452
After 4999 steps, mse_loss: 0.037738
After 5999 steps, mse_loss: 0.035935
After 6999 steps, mse_loss: 0.033375
After 7999 steps, mse_loss: 0.027632
After 8999 steps, mse_loss: 0.022587
After 9999 steps, mse_loss: 0.020984
<jupyter_text>#### 5. 训练带正则项的损失函数loss。
这里有点奇怪,一直运行出错:
```
InvalidArgumentError: You must feed a value for placeholder tensor 'Placeholder_3' with dtype float and shape [?,1]
```
我将 x,y_,data,label的shape都打印出来,并没有发现问题。不知道错在何处。<jupyter_code># 定义训练的目标函数loss,训练次数及训练模型
train_op = tf.train.AdamOptimizer(0.001).minimize(loss)
TRAINING_STEPS = 10000
with tf.Session() as sess:
tf.global_variables_initializer().run()
for i in range(TRAINING_STEPS):
print(x.get_shape(), y_.get_shape())
print(data.shape, label.shape)
sess.run(train_op, feed_dict={x: data, y_: label})
if i % 1000 == 1000 - 1:
print("After %d steps, loss: %f" % (i, sess.run(loss, feed_dict={x: data, y_: label})))
# 画出训练后的分割曲线
xx, yy = np.mgrid[-1:1:.01, 0:2:.01]
grid = np.c_[xx.ravel(), yy.ravel()]
probs = sess.run(y, feed_dict={x:grid})
probs = probs.reshape(xx.shape)
plt.scatter(data[:,0], data[:,1], c=label.reshape(200,),
cmap="RdBu", vmin=-.2, vmax=1.2, edgecolor="white")
plt.contour(xx, yy, probs, levels=[.5], cmap="Greys", vmin=0, vmax=.1)
plt.show()<jupyter_output>(?, 2) (?, 1)
(200, 2) (200, 1)
|
no_license
|
/Chapter04/3. 正则化.ipynb
|
xiaqunfeng/TensorFlowCombat_GoogleDeepLearningFramework
| 5 |
<jupyter_start><jupyter_text># Fare exploration
** IT APPEARS THAT FARE IS SHARED IN CASE OF MULTIPLE PEOPLE EMBARKING ON A SINGLE TICKET !!! **
Let's throw this to the bin and start anew.
We need to see
+ how fare is correlated with survival
+ how we can forge a new simpler and more relevant Fare feature
+ if there are missing values we might need to complete## 0 Setup
### 0.1 Loading of data, external libraries and functions.<jupyter_code>library(here)
setwd(here())
print(getwd())
source(file="misc_functions.R")
source("experiments/titanic_challenge/1_feature_engineering.R")
library(gridExtra)
library(lattice)<jupyter_output>[1] "/home/sapristi/Documents/ML"
<jupyter_text>### 0.2 Creation of a dataframe with relevant rows, and forged features (age, title)<jupyter_code>train.raw <- read.csv("datasets/titanic_na/train.csv")
train.ff <- fe$forge_features(train.raw)
train.raw$Survived <- as.factor(train.raw$Survived)
cols.notna <- (!is.na(train.raw$Age))
train.notna <- train.raw[cols.notna,]
train.notna$Pclass <- as.factor(train.raw$Pclass[cols.notna])
train.notna$Title <- train.ff$Title[cols.notna]
train.notna$Age <- train.ff$Age[cols.notna]
train.notna$Age.simple <- train.ff$Age.simple[cols.notna]
train.notna$Age.disc5 <- train.ff$Age.disc5[cols.notna]
train.notna$Fare.disc10 <- sapply(train.notna$Fare, function (f) {as.integer(f/10)})
train.notna$Fare.disc10 <- as.factor(train.notna$Fare.disc10)
train.notna$Fare.sqrtdisc <- sapply(train.notna$Fare, function(f) {as.integer(sqrt(f)/3)})
train.notna$Fare.sqrtdisc <- as.factor(train.notna$Fare.sqrtdisc)
<jupyter_output><empty_output><jupyter_text>## 1 Visualisation
### 1.1 Fare ~ Survived<jupyter_code>options(repr.plot.height= 5)
histogram(~ Fare | Survived , data = train.notna, type="count")<jupyter_output><empty_output><jupyter_text>We can see some difference in surivival between low and high fare, but not very clearly.
### 1.2 Survival | Fare<jupyter_code>options(repr.plot.height= 5, repr.plot.width=10)
h1 <- histogram( ~ Survived | Fare.disc10, data = train.notna, type="count",
lattice.options = list(key = list(cex.title = 50)))
h2 <- histogram( ~ Survived | Fare.disc10, data = train.notna, type="density")
print(h1, split=c(1,1,2,1), more=TRUE)
print(h2, split=c(2,1,2,1))<jupyter_output><empty_output><jupyter_text>### 1.3 Survival | sqrt(Fare) + Title<jupyter_code>h3 <- histogram( ~ Survived | Fare.sqrtdisc + Title, data = train.notna, type="count",
lattice.options = list(key = list(cex.title = 50)))
h4 <- histogram( ~ Survived | Fare.sqrtdisc + Title, data = train.notna, type="density")
print(h3, split=c(1,1,2,1), more=TRUE)
print(h4, split=c(2,1,2,1))<jupyter_output><empty_output><jupyter_text>### 1.4 Survival | log(Fare) + Title<jupyter_code>nonzerofare <- train.notna$Fare != 0
train.nonzerofare <- train.notna[nonzerofare, ]
train.nonzerofare$Fare.logdisc <- sapply(train.nonzerofare$Fare, function(f) {floor(log(f))})
train.nonzerofare$Fare.logdisc <- as.factor(train.nonzerofare$Fare.logdisc)
h5 <- histogram( ~ Survived | Fare.logdisc + Title, data = train.nonzerofare, type="count",
lattice.options = list(key = list(cex.title = 50)))
h6 <- histogram( ~ Survived | Fare.logdisc + Title, data = train.nonzerofare, type="density")
print(h5, split=c(1,1,2,1), more=TRUE)
print(h6, split=c(2,1,2,1))<jupyter_output><empty_output><jupyter_text>### 1.5 Comparison of log and sqrt Fare scales for Survival | Fare + Title<jupyter_code>options(repr.plot.height= 10)
print(h3, split=c(1,1,2,1), more=TRUE)
print(h5, split=c(2,1,2,1))<jupyter_output><empty_output><jupyter_text>## 1.3 Pclass and Fare are obviously related, but how so ?<jupyter_code># h1 <- histogram( ~ Survived | Pclass + Title, data = train.notna, type="density")
h2 <- histogram(~ Fare.sqrtdisc | Pclass, data = train.notna, type="count")
print(h1, split=c(1,1,2,1), more=TRUE)
print(h2, split=c(2,1,2,1))
histogram(~ Fare.sqrtdisc | Pclass + Embarked, data = train.notna, type="percent")
<jupyter_output><empty_output>
|
no_license
|
/experiments/titanic_challenge/fare_exploration_old.ipynb
|
sapristi/ML
| 8 |
<jupyter_start><jupyter_text><jupyter_code>import pandas as pd
import numpy as np
import warnings
warnings.filterwarnings('ignore')
import matplotlib.pyplot as plt
from sklearn import cluster, datasets, metrics
import seaborn as sns
df_country = pd.read_csv('https://raw.githubusercontent.com/sagihaider/CE888_2021/main/Lab_6/Data/Country-data.csv')
df_country
df_country.shape[0]
df_data = pd.read_csv('https://raw.githubusercontent.com/sagihaider/CE888_2021/main/Lab_6/Data/data-dictionary.csv')
df_data
df_country['exports'] = df_country['exports']*df_country['gdpp']/100
df_country['imports'] = df_country['imports']*df_country['gdpp']/100
df_country['health'] = df_country['health']*df_country['gdpp']/100
df_country.head()
#check outliers
fig = plt.figure(figsize = (8,8))
sns.boxplot(data=df_country)
plt.show()
df_country.dtypes
df_country.isnull().sum()
%matplotlib inline
plt.figure(figsize = (10,8))
sns.heatmap(df_country.corr(),annot = True)
plt.show()
from sklearn.preprocessing import StandardScaler
df_c = df_country.drop('country',1) ## Droping string feature country name.
standard_scaler = StandardScaler()
df_c_scaled = standard_scaler.fit_transform(df_c)
df_c_scaled.shape
#fit pca on dataset
from sklearn.decomposition import PCA
pca = PCA()
X_pca = pca.fit_transform(df_c_scaled)
#pca.explained_variance_ratio_
%matplotlib inline
fig = plt.figure(figsize = (8,6))
plt.plot(np.cumsum(pca.explained_variance_ratio_))
plt.xlabel('number of components')
plt.ylabel('cumulative explained variance')
plt.show()
Most of the variance can be explained by 5 features and so a new dataframe is created with only the 5 features.
colnames = df_c.columns.tolist()
pcs = pd.DataFrame({ 'Feature':colnames,'PC1':pca.components_[0],'PC2':pca.components_[1],'PC3':pca.components_[2],
'PC4':pca.components_[3],'PC5':pca.components_[4]})
pcs
from sklearn.decomposition import IncrementalPCA
pca_final = IncrementalPCA(n_components=5
)
df_pca = pca_final.fit_transform(df_c_scaled)
df_pca.shape
pc = np.transpose(df_pca)
pcs_2 = pd.DataFrame({'PC1':pc[0],'PC2':pc[1],'PC3':pc[2],'PC4':pc[3],'PC5':pc[4]})
pcs_2.shape
fig = plt.figure(figsize = (8,6))
sns.boxplot(data=pcs_2)
plt.show()
fig = plt.figure(figsize = (6,4))
sns.scatterplot(x='PC1',y='PC2',data=pcs_2)
plt.show()
from sklearn.cluster import KMeans
km = KMeans(n_clusters = 3, random_state = 50)
km.fit(pcs_2)
km_cluster = pd.concat([df_post_pca , pd.Series(km.labels_)], axis=1)
km_cluster.columns = ['PC1', 'PC2','PC3','PC4','PC5','ClusterID']
km_cluster
km_data['ClusterID'].value_counts()
new_data = pd.merge(df_country,km_data['ClusterID'], left_index=True,right_index=True)
new_data.head()
Clus_gdpp = pd.DataFrame(new_data.groupby(["ClusterID"]).gdpp.mean())
Clus_child_mor = pd.DataFrame(new_data.groupby(["ClusterID"]).child_mort.mean())
Clus_exports = pd.DataFrame(new_data.groupby(["ClusterID"]).exports.mean())
Clus_income = pd.DataFrame(new_data.groupby(["ClusterID"]).income.mean())
Clus_health = pd.DataFrame(new_data.groupby(["ClusterID"]).health.mean())
Clus_imports = pd.DataFrame(new_data.groupby(["ClusterID"]).imports.mean())
Clus_inflation = pd.DataFrame(new_data.groupby(["ClusterID"]).inflation.mean())
Clus_life_expec = pd.DataFrame(new_data.groupby(["ClusterID"]).life_expec.mean())
Clus_total_fer = pd.DataFrame(new_data.groupby(["ClusterID"]).total_fer.mean())
df = pd.concat([Clus_gdpp, Clus_child_mor, Clus_income, Clus_exports, Clus_health,
Clus_imports, Clus_inflation, Clus_life_expec, Clus_total_fer], axis=1)
df.columns = ["gdpp","child_mort","income","exports","health","imports","inflation","life_expec","total_fer"]
df
fig = plt.figure(figsize = (8,6))
df.rename(index={0: 'Developed Countries'},inplace=True)
df.rename(index={1: 'Developing Countries'},inplace=True)
df.rename(index={2: 'Under-developed Countries'},inplace=True)
s=sns.barplot(x=df.index,y='gdpp',data=df)
plt.xlabel('Country Groups', fontsize=10)
plt.ylabel('GDP per Capita', fontsize=10)
plt.title('Country Groups On the basis of GDPP')
plt.show()
####using child mortality
fig = plt.figure(figsize = (10,6))
sns.barplot(x=df.index,y='child_mort',data=df)
plt.xlabel('Country Groups', fontsize=10)
plt.title('Country Groups On the basis of Child mortality Rate')
plt.show()<jupyter_output><empty_output>
|
no_license
|
/Lab6/Lab6_Task7.ipynb
|
rajsumanth19/dsdm-19
| 1 |
<jupyter_start><jupyter_text># Decision Tree Examples
+ Using the Iris dataset
+ Using graphviz for visualization of the tree
### Graphviz installation
1. Download and install from [graphviz site](https://graphviz.gitlab.io/download)
2. Put the executable on the system path
3. Install the python package using command
`conda install -c conda-forge python-graphviz`### Packages<jupyter_code>%matplotlib inline
# data and plotting packages
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import os
# modeling packages
import scipy.stats as stats
from sklearn.datasets import load_iris
from sklearn.tree import DecisionTreeClassifier, DecisionTreeRegressor
from sklearn.tree import export_graphviz
import mlutils
from mlutils import plot_decision_boundary, plot_regression<jupyter_output><empty_output><jupyter_text>### Display Settings<jupyter_code>plt.style.use('seaborn')
plt.rcParams['figure.figsize'] = (20, 10)
plt.rcParams['legend.fontsize'] = 'x-large'
plt.rcParams['axes.labelsize'] = 'x-large'
plt.rcParams['axes.titlesize'] = 'x-large'
plt.rcParams['xtick.labelsize'] = 'x-large'
plt.rcParams['ytick.labelsize'] = 'x-large'<jupyter_output><empty_output><jupyter_text>### Data Loading<jupyter_code># load the iris dataset from sklearn; note sklearn does not use pandas dataframes
iris = load_iris()
# anatomy of the data object
print("feature_names: {}".format(iris.feature_names))
print("feature_shape: {}".format(iris.data.shape))
print("target_names: {}".format(iris.target_names))
print("target_shape: {}".format(iris.target.shape))
#print(iris.DESCR)<jupyter_output>feature_names: ['sepal length (cm)', 'sepal width (cm)', 'petal length (cm)', 'petal width (cm)']
feature_shape: (150, 4)
target_names: ['setosa' 'versicolor' 'virginica']
target_shape: (150,)
<jupyter_text>### Decision Tree Classification
We fit a Decision Tree classifier model using only two attributes.<jupyter_code># Pick the indices of two features and two classes
feat0_idx = 2
feat1_idx = 3
X = iris.data[:, (feat0_idx, feat1_idx)] # choose the features here
y = iris.target
# Create an instance of DecisionTreeClassifier and fit the data.
# Switch off the C hyperparameter
dtree = DecisionTreeClassifier(criterion='gini', max_depth=2, random_state=2)
print(dtree)
dtree.fit(X, y)
y_fit = dtree.predict(X)
n_fail = (y != y_fit).sum()
print("Total points: {0}; Mislabeled points: {1}\n".format(len(y), n_fail))
# export tree graph
fbasename = 'iris-dtree'
export_graphviz(dtree, out_file=fbasename + '.dot',
feature_names=[iris.feature_names[i] for i in [feat0_idx, feat1_idx]],
class_names=iris.target_names, filled=True)
# render the graph (requires graphviz exeecutable on the path)
cmd = 'dot -Tpng ' + fbasename + '.dot -o ' + fbasename + '.png'
os.system(cmd);<jupyter_output>DecisionTreeClassifier(class_weight=None, criterion='gini', max_depth=2,
max_features=None, max_leaf_nodes=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, presort=False, random_state=2,
splitter='best')
Total points: 150; Mislabeled points: 6
<jupyter_text>### Visualize the tree
### Decision Boundaries
<jupyter_code># axes limits for plotting
xlim = [min(X[:, 0]) - 0.5, max(X[:, 0]) + 0.5]
ylim = [min(X[:, 1]) - 0.5, max(X[:, 1]) + 0.5]
axlim = xlim + ylim
# create a plot
fig = plt.figure(figsize=(10, 10))
ax = fig.add_subplot(111)
plot_decision_boundary(dtree, ax, axlim)
# plot the observations
ax.plot(X[:, 0][y==0], X[:, 1][y==0], "yo", label=iris.target_names[0])
ax.plot(X[:, 0][y==1], X[:, 1][y==1], "bs", label=iris.target_names[1])
ax.plot(X[:, 0][y==2], X[:, 1][y==2], "g^", label=iris.target_names[2])
ax.set(xlabel=iris.feature_names[feat0_idx], ylabel=iris.feature_names[feat1_idx])
ax.legend(loc='upper left')<jupyter_output><empty_output><jupyter_text>Prediction probabilities are estimated empirically<jupyter_code>x_in = [3.2, 1.7] # a new observation
y_pred = dtree.predict([x_in])
p_pred = dtree.predict_proba([x_in])
print('Prediction: {}'.format(iris.target_names[y_pred][0]))
print('Probabilities:')
for species, prob in zip(iris.target_names, np.round(p_pred, 3).tolist()[-1]):
print('{0:10s}: {1:5.3f}'.format(species, prob))<jupyter_output>Prediction: versicolor
Probabilities:
setosa : 0.000
versicolor: 0.907
virginica : 0.093
<jupyter_text>### Instability
Suppose we remove one point, the versicolor with the highest petal width.
We refit the tree and plot the decision boundary.<jupyter_code>rem_feat = 1
rem_species = 1
# the maximum rem_feat for rem_species
max_feat = X[:, rem_feat][y == rem_species].max()
keep_idx = (X[:, rem_feat] != max_feat) | (y != rem_species)
# remove the row for rem_species that has the maximum rem_feat
X_mod = X[keep_idx].copy()
y_mod = y[keep_idx].copy()
# fit a tree with one row removed
dmodtree = DecisionTreeClassifier(criterion='gini', max_depth=2, random_state=1)
dmodtree.fit(X_mod, y_mod)
# create a plot
fig1 = plt.figure(figsize=(10, 10))
ax1 = fig1.add_subplot(111)
plot_decision_boundary(dmodtree, ax1, axlim)
# plot the observations
ax1.plot(X_mod[:, 0][y_mod==0], X_mod[:, 1][y_mod==0], "yo", label=iris.target_names[0])
ax1.plot(X_mod[:, 0][y_mod==1], X_mod[:, 1][y_mod==1], "bs", label=iris.target_names[1])
ax1.plot(X_mod[:, 0][y_mod==2], X_mod[:, 1][y_mod==2], "g^", label=iris.target_names[2])
ax1.set(xlabel=iris.feature_names[feat0_idx], ylabel=iris.feature_names[feat1_idx])
ax1.legend(loc='upper left')<jupyter_output><empty_output><jupyter_text>### Decision Tree Regression
Using synthetic data.
We generate quadratic data and fit two regression trees with different depths.<jupyter_code>np.random.seed(0) # fix the seed for reproducibily
n_samples = 1000 # total number of samples
sigma = 0.1 # noise volatility
XX = np.random.rand(n_samples, 1) # from uniform [0, 1] distrobution
yy = 4 * (XX - 0.5) ** 2
yy = yy + sigma * np.random.randn(n_samples, 1)
max_depth1 = 2
regtree1 = DecisionTreeRegressor(max_depth=max_depth1, random_state=42)
regtree1.fit(XX, yy)
max_depth2 = 4
regtree2 = DecisionTreeRegressor(max_depth=max_depth2, random_state=42)
regtree2.fit(XX, yy);
# Display results
fig2 = plt.figure(figsize=(16, 8))
ax2 = fig2.add_subplot(121)
ax3 = fig2.add_subplot(122)
plot_regression(regtree1, XX, yy, ax2, [0, 1, -0.2, 1])
plot_regression(regtree2, XX, yy, ax3, [0, 1, -0.2, 1])
ax2.set(xlabel='$x$', ylabel='$y$')
ax2.set(title='Regression Tree with depth ' + str(max_depth1))
ax3.set(xlabel='$x$', ylabel='$y$')
ax3.set(title='Regression Tree with depth ' + str(max_depth2));<jupyter_output><empty_output>
|
permissive
|
/LectureExamples/Code/L07-DecisionTree-Examples.ipynb
|
bondxue/FRE7773-Machine-Learning-in-Finance
| 8 |
<jupyter_start><jupyter_text>___
___
# Support Vector Machines Project
Welcome to your Support Vector Machine Project! Just follow along with the notebook and instructions below. We will be analyzing the famous iris data set!
## The Data
For this series of lectures, we will be using the famous [Iris flower data set](http://en.wikipedia.org/wiki/Iris_flower_data_set).
The Iris flower data set or Fisher's Iris data set is a multivariate data set introduced by Sir Ronald Fisher in the 1936 as an example of discriminant analysis.
The data set consists of 50 samples from each of three species of Iris (Iris setosa, Iris virginica and Iris versicolor), so 150 total samples. Four features were measured from each sample: the length and the width of the sepals and petals, in centimeters.
Here's a picture of the three different Iris types:<jupyter_code># The Iris Setosa
from IPython.display import Image
url = 'http://upload.wikimedia.org/wikipedia/commons/5/56/Kosaciec_szczecinkowaty_Iris_setosa.jpg'
Image(url,width=300, height=300)
# The Iris Versicolor
from IPython.display import Image
url = 'http://upload.wikimedia.org/wikipedia/commons/4/41/Iris_versicolor_3.jpg'
Image(url,width=300, height=300)
# The Iris Virginica
from IPython.display import Image
url = 'http://upload.wikimedia.org/wikipedia/commons/9/9f/Iris_virginica.jpg'
Image(url,width=300, height=300)<jupyter_output><empty_output><jupyter_text>The iris dataset contains measurements for 150 iris flowers from three different species.
The three classes in the Iris dataset:
Iris-setosa (n=50)
Iris-versicolor (n=50)
Iris-virginica (n=50)
The four features of the Iris dataset:
sepal length in cm
sepal width in cm
petal length in cm
petal width in cm
## Get the data
**Use seaborn to get the iris data by using: iris = sns.load_dataset('iris') **<jupyter_code>import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import cufflinks as cf
cf.go_offline()
%matplotlib inline
iris = sns.load_dataset('iris')<jupyter_output><empty_output><jupyter_text>Let's visualize the data and get you started!
## Exploratory Data Analysis
Time to put your data viz skills to the test! Try to recreate the following plots, make sure to import the libraries you'll need!
**Import some libraries you think you'll need.**<jupyter_code>iris.head()<jupyter_output><empty_output><jupyter_text>** Create a pairplot of the data set. Which flower species seems to be the most separable?**<jupyter_code>sns.pairplot(iris,hue='species',palette='viridis')<jupyter_output><empty_output><jupyter_text>**Create a kde plot of sepal_length versus sepal width for setosa species of flower.**<jupyter_code>setosa = iris[iris['species']=='setosa']
sns.set(style='darkgrid')
sns.kdeplot(data=setosa['sepal_width'],data2=setosa['sepal_length'],cmap='plasma',shade=True,shade_lowest=False)<jupyter_output><empty_output><jupyter_text># Train Test Split
** Split your data into a training set and a testing set.**<jupyter_code>from sklearn.model_selection import train_test_split
y = iris['species']
X = iris.drop('species',axis=1)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3)<jupyter_output><empty_output><jupyter_text># Train a Model
Now its time to train a Support Vector Machine Classifier.
**Call the SVC() model from sklearn and fit the model to the training data.**<jupyter_code>from sklearn.svm import SVC
svm = SVC()
svm.fit(X_train,y_train)<jupyter_output>C:\Users\LENOVO\Anaconda3\lib\site-packages\sklearn\svm\base.py:196: FutureWarning:
The default value of gamma will change from 'auto' to 'scale' in version 0.22 to account better for unscaled features. Set gamma explicitly to 'auto' or 'scale' to avoid this warning.
<jupyter_text>## Model Evaluation
**Now get predictions from the model and create a confusion matrix and a classification report.**<jupyter_code>pred = svm.predict(X_test)
from sklearn.metrics import confusion_matrix,classification_report
print(confusion_matrix(y_test,pred))
print(classification_report(y_test,pred))<jupyter_output> precision recall f1-score support
setosa 1.00 1.00 1.00 17
versicolor 0.88 1.00 0.93 14
virginica 1.00 0.86 0.92 14
micro avg 0.96 0.96 0.96 45
macro avg 0.96 0.95 0.95 45
weighted avg 0.96 0.96 0.96 45
<jupyter_text>Wow! You should have noticed that your model was pretty good! Let's see if we can tune the parameters to try to get even better (unlikely, and you probably would be satisfied with these results in real like because the data set is quite small, but I just want you to practice using GridSearch.## Gridsearch Practice
** Import GridsearchCV from SciKit Learn.**<jupyter_code>from sklearn.model_selection import GridSearchCV<jupyter_output><empty_output><jupyter_text>**Create a dictionary called param_grid and fill out some parameters for C and gamma.**<jupyter_code>param_grid = {'C':[0.1,1,10,100,1000],'gamma':[1,0.1,0.001,0.001,0.0001]}<jupyter_output><empty_output><jupyter_text>** Create a GridSearchCV object and fit it to the training data.**<jupyter_code>grid = GridSearchCV(SVC(),param_grid,refit=True,verbose=2)
grid.fit(X_train,y_train)<jupyter_output>C:\Users\LENOVO\Anaconda3\lib\site-packages\sklearn\model_selection\_split.py:2053: FutureWarning:
You should specify a value for 'cv' instead of relying on the default value. The default value will change from 3 to 5 in version 0.22.
[Parallel(n_jobs=1)]: Using backend SequentialBackend with 1 concurrent workers.
[Parallel(n_jobs=1)]: Done 1 out of 1 | elapsed: 0.0s remaining: 0.0s
<jupyter_text>** Now take that grid model and create some predictions using the test set and create classification reports and confusion matrices for them. Were you able to improve?**<jupyter_code>g_pred = grid.predict(X_test)
print(confusion_matrix(y_test,g_pred))
print(classification_report(y_test,g_pred))<jupyter_output> precision recall f1-score support
setosa 1.00 1.00 1.00 17
versicolor 0.93 0.93 0.93 14
virginica 0.93 0.93 0.93 14
micro avg 0.96 0.96 0.96 45
macro avg 0.95 0.95 0.95 45
weighted avg 0.96 0.96 0.96 45
|
no_license
|
/Support Vector Machines Project.ipynb
|
M-Hammad0/ML
| 12 |
<jupyter_start><jupyter_text># **Regression.**## **Linear Regression.**
### Given housing data build a machine learning model that can predict home prices based on square feet area uisng linear regression.<jupyter_code>import warnings
warnings.filterwarnings("ignore")
import pandas as pd
import numpy as np
from sklearn import linear_model
import matplotlib.pyplot as plt
%matplotlib inline #this line is used to print the plot other plotting wont occur.
df = pd.read_csv('/home/dharmendra/Gamma/datasets/homeprices.csv')
df
# here the area is the independent variable and price is dependent variable.
#as the price of a house depends on the area.
%matplotlib inline
plt.xlabel('area')
plt.ylabel('price')
plt.scatter(df.area,df.price,color='red',marker='+')
#the next step is to design the model for single regression.
area = df['area']
area
area = df[['area']]
area
price = df.price
price
# let's Create a linear regression object
reg = linear_model.LinearRegression()
reg.fit(area,price)
#let's predict the price of home whose area is 3300
reg.predict(3300)
#since we know the equation for regression in single varialble is y=m*x+b
#now for manual testing let's find the co-efficient in regular expresssion which is m in above equation.
reg.coef
#let's find the intercept in regular expresssion which is b in above equation.
reg.intercept_
#this is the price(y) of area(x) of 3300 with value of m and b being inserted.
3300*135.78767123 + 180616.43835616432<jupyter_output><empty_output><jupyter_text>### steps
1. Data Preprocessing
<jupyter_code>import pandas as pd
import numpy as np
import csv
sf = pd.read_csv('/home/dharmendra/Gamma/datasets/Bay area_house_price.csv')
sf.head()
#There are several features that we do not need, such as “info”, “z_address”, “zipcode”(We have “neighborhood” as a location variable), “zipid” and “zestimate”(This is the price estimated by Zillow, we don’t want our model to be affected by this).
#so, we will drop them.
sf.drop(sf.columns[[0, 2, 3, 15, 17, 18]], axis=1, inplace=True)
sf.info()
#The data type of “zindexvalue” should be numeric, so let’s change that.
sf['zindexvalue'] = sf['zindexvalue'].str.replace(',', '')
sf['zindexvalue'] = sf['zindexvalue'].convert_objects(convert_numeric=True)
sf.lastsolddate.min(), sf.lastsolddate.max()<jupyter_output><empty_output><jupyter_text>* The house sold period in the dateset was between January 2013 and December 2015.<jupyter_code>sf.describe()
#The count, mean, min and max rows are self-explanatory.
#The std shows the standard deviation, and the 25%, 50% and 75% rows show the corresponding percentiles.
#To get a feel for the type of data we are dealing with, we plot a histogram for each numeric variable.
%matplotlib inline
import matplotlib.pyplot as plt
sf.hist(bins=50, figsize=(20,15))
plt.savefig("attribute_histogram_plots")
plt.show()
#Let’s create a scatter plot with latitude and longitude to visualize the data:
sf.plot(kind="scatter", x="longitude", y="latitude", alpha=0.2)
plt.savefig('map1.png')
#now let’s color code from the most expensive to the least expensive areas:
sf.plot(kind="scatter", x="longitude", y="latitude", alpha=0.4, figsize=(10,7),
c="lastsoldprice", cmap=plt.get_cmap("jet"), colorbar=True,
sharex=False)<jupyter_output><empty_output><jupyter_text>* This image tells us that the most expensive houses sold were in the north area.
* The **variable** we are going to **predict** is the **“last sold price”.**
* **So let’s look at how much each independent variable correlates with this dependent variable.**<jupyter_code>corr_matrix = sf.corr()
corr_matrix["lastsoldprice"].sort_values(ascending=False)<jupyter_output><empty_output><jupyter_text>* The last sold price tends to increase when the finished sqft and the number of bathrooms go up. You can see a small negative correlation between the year built and the last sold price.
* And finally, coefficients close to zero indicate that there is no linear correlation.<jupyter_code>from pandas.tools.plotting import scatter_matrix
attributes = ["lastsoldprice", "bathrooms","finishedsqft" ,"zindexvalue"]
scatter_matrix(sf[attributes], figsize=(12, 8))
plt.savefig('matrix.png')<jupyter_output><empty_output><jupyter_text>* To visualize the correlation between variables by using Pandas’ **scatter_matrix** function. We will just focus on a few promising variables, that seem the most correlated with the last sold price as showm above.
* The most promising variable for predicting the last sold price is the finished sqft, so let’s zoom in on their correlation scatter plot.<jupyter_code>sf.plot(kind="scatter", x="finishedsqft", y="lastsoldprice", alpha=0.5)
plt.savefig('scatter.png')<jupyter_output><empty_output><jupyter_text>* The correlation is indeed very strong; you can clearly see the upward trend and that the points are not too dispersed.
* Because each house has different square footage and each neighborhood has different home prices, what we really need is the price per sqft. So, we add a new variable “price_per_sqft”. We then check to see how much this new independent variable correlates with the last sold price.
<jupyter_code>sf['price_per_sqft'] = sf['lastsoldprice']/sf['finishedsqft']
corr_matrix = sf.corr()
corr_matrix["lastsoldprice"].sort_values(ascending=False)<jupyter_output><empty_output><jupyter_text>* Unfortunately, the new price_per_sqft variable shows only a very small positive correlation with the last sold price. But we still need this variable for grouping neighborhoods.
* But we still need this variable for grouping neighborhoods.
* There are 71 neighborhoods in the data, and we are going to group them.<jupyter_code>len(sf['neighborhood'].value_counts())
#now let's cluster the neighborhood into three groups:
#1. low price; 2. high price low frequency; 3. high price 4. high frequency.
freq = sf.groupby('neighborhood').count()['address']
mean = sf.groupby('neighborhood').mean()['price_per_sqft']
cluster = pd.concat([freq, mean], axis=1)
cluster['neighborhood'] = cluster.index
cluster.columns = ['freq', 'price_per_sqft','neighborhood']
cluster
cluster.describe()
#low price neighborhoods:
cluster1 = cluster[cluster.price_per_sqft < 756]
cluster1.index
#high price and low frequency neighborhoods.
cluster_temp = cluster[cluster.price_per_sqft >= 756]
cluster2 = cluster_temp[cluster_temp.freq <123]
cluster2.index
#high price and high frequency neighborhoods.
cluster3 = cluster_temp[cluster_temp.freq >=123]
cluster3.index<jupyter_output><empty_output>
|
no_license
|
/Supervised Algorithms/Regression.ipynb
|
Dammonoit/Machine-Learning-Arsenal
| 8 |
<jupyter_start><jupyter_text># Support Vector Machine - SVM
[**SVM**](https://en.wikipedia.org/wiki/Support_vector_machine) is popular and widely use for superviser learning to classification algorithm (pattern recognization)
A good seperation by using [**hyperplane**](https://en.wikipedia.org/wiki/Hyperplane)
* Advantages:
* Can use for regression or classification
* Easier than neural network
* Not influence by noisy data
* Disadvantages:
* Combination of kernel and model parameters
* Slow when input data has large number of features
* Blackbox model therefore it is to hard to undestand the beyond
> For SVM library and dataset, we are using from [*scikit learn*](http://scikit-learn.org/stable/modules/svm.html)
## Example 1: Classifier the sample data
Given the dataset with **blue class** as **0**, **red class** as **1**. Using SVM to train the training dataset then predict the testing sample data.<jupyter_code>import numpy as np
from matplotlib import pyplot as plt
from sklearn import svm
# Blue class as 0
xBlue = np.array([0.3,0.5,1,1.4,1.7,2])
yBlue = np.array([1,4.5,2.3,1.9,8.9,4.1])
# Red class as 1
xRed = np.array([3.3,3.5,4,4.4,5.7,6])
yRed = np.array([7,1.5,6.3,1.9,2.9,7.1])
# Consolidate data
X = np.array([[0.3,1],[0.5,4.5],[1,2.3],[1.4,1.9],[1.7,8.9],[2,4.1],[3.3,7],[3.5,1.5],[4,6.3],[4.4,1.9],[5.7,2.9],[6,7.1]])
y = np.array([0,0,0,0,0,0,1,1,1,1,1,1])
# Sample testing dat
xSample = 3
ySample = 4
# Using SVM to training data
classifier = svm.SVC()
classifier.fit(X, y)
# Classifier the sample data
print "SVM predict the sample data belongs to class", classifier.predict([[xSample, ySample]])
# Ploting data
plt.plot(xBlue, yBlue, 'ro', color='blue')
plt.plot(xRed, yRed, 'ro', color='red')
plt.plot(xSample, ySample, 'ro', color='green', markersize=14)
plt.axis([0, 7, 0, 9.5])
plt.show()<jupyter_output>SVM predict the sample data belongs to class [0]
<jupyter_text>## Example 2: Character Recognition
Using SVM to recognize the digit. The dataset of digit is used in this example available at [scikit load digit](http://scikit-learn.org/stable/modules/generated/sklearn.datasets.load_digits.html)
> We are going to use the svm.SVC, for more information about argurment of [**svm.SVC**](http://scikit-learn.org/stable/modules/generated/sklearn.svm.SVC.html#sklearn.svm.SVC)<jupyter_code>import numpy as np
from matplotlib import pyplot as plt
import matplotlib.cm as cm
from sklearn import datasets
from sklearn import svm
# Load image dataset
numberImages = datasets.load_digits()
print "Dataset information", numberImages.data.shape
# Show first image in dataset
print "Visualize the first image in dataset"
plt.matshow(numberImages.images[0], cmap=cm.Greys_r)
plt.show()
# C: Penalty parameter the error term.
# gamma: Kernel coefficient
model = svm.SVC(gamma=0.0001, C=100, kernel='rbf')
# Create the training dataset (expect the last 5 item use for testing set)
# Training dataset -> *.data
# Training label -> *.target
X_train, y_train = numberImages.data[:-5], numberImages.target[:-5]
# Using SVM to train the dataset
model.fit(X_train, y_train)
# The testing image
predict_Image = numberImages.data[-2]
# Visualize the testing image
print "Visualize the testing image"
plt.matshow(numberImages.images[-2], cmap=cm.Greys_r)
plt.show()
# Predict the testing image
print "The testing image is", model.predict([predict_Image])<jupyter_output>Dataset information (1797L, 64L)
Visualize the first image in dataset
|
no_license
|
/Section 5 - Support Vector Machine SVM/svm.ipynb
|
leduynguyen/Introduction-Machine-Learning
| 2 |
<jupyter_start><jupyter_text>## My Musical Journey
Our beloved spotify provided us with a 2010s decade end by wrapping up one’s listening history throughout the year - in my case this meant from 2016 to 2019.
Formally known as Spotify Wrapped (https://open.spotify.com/genre/2019-page)
These insights have ignited my curiosity to explore beyond my liked and disliked songs, as well as what Spotify showed to us.
So I decided to play around with the data of tracks that made to the top global list in 2016, 2017, 2018, 2019. Overall I thought it was very interesting to try to justify the numbers and results with song knowledge and intuituion.<jupyter_code>#necessary libraries
import json
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import seaborn as sb
%matplotlib inline
import plotly.express as px
import plotly.figure_factory as ff
from sklearn.model_selection import train_test_split
from sklearn.ensemble import RandomForestRegressor
from sklearn import metrics
from sklearn.metrics import classification_report, confusion_matrix, accuracy_score
from sklearn.preprocessing import MinMaxScaler
from sklearn.linear_model import LogisticRegression
from sklearn.decomposition import PCA
import statsmodels.api as sm
from scipy import stats
from scipy.stats import ttest_1samp, ttest_rel, ttest_ind
from IPython.display import display, Image, SVG, Math, YouTubeVideo
#import dataset created in other jupyter notebook
data = pd.read_csv('../data/top_year.csv')
data
data.dtypes
sb.catplot(x = 'Year', y = 'instrumentalness', data = data, kind = 'violin')
sb.catplot(x = 'Year', y = 'loudness', data = data, kind = 'violin')
sb.catplot(x = 'Year', y = 'liveness', data = data, kind = 'violin')<jupyter_output><empty_output><jupyter_text>## Whole Picture Comparison
Unlike the ones above, let’s do another type of comparison for the other features.
- Due to the fact that the features have varying range, I needed to rescale the values to a relatively similar range using min-max scaler.
- That kind of scaler has an advantage over the standard scaler in terms of distortion in original data.<jupyter_code>features = ['danceability', 'energy', 'key','loudness', 'mode', 'acousticness', 'instrumentalness', 'liveness','valence', 'tempo']
from sklearn.preprocessing import MinMaxScaler
# Min-max scaling
df_scaled = pd.DataFrame(MinMaxScaler().fit_transform(data[features]),
columns=data[features].columns)
# KDE plot
fig = ff.create_distplot([df_scaled[c] for c in df_scaled.columns],
df_scaled.columns,
show_hist=False)
fig.show()
#Now all of the data is on ranging between 0 and 1;
image_3 = fig.write_image("../images/image3.png")
#because git doesn't load the graph above
Image(filename='../images/image3.png',width = 1000)
df_scaled['Year'] = data['Year']
df_scaled.head()
# Radar plot
df_radar = df_scaled.groupby('Year').mean().reset_index() \
.melt(id_vars='Year', var_name="features", value_name="avg") \
.sort_values(by=['Year','features']).reset_index(drop=True)
fig = px.line_polar(df_radar,
r="avg",
theta="features",
title='Mean Values of Each Track Features',
color="Year",
line_close=True,
line_shape='spline',
range_r=[0, 0.8],
color_discrete_sequence=px.colors.cyclical.mygbm[:-6])
fig.show()
image_4 = fig.write_image("../images/image4.png")
#because git doesn't load the graph above
Image(filename='../images/image4.png',width = 800)<jupyter_output><empty_output><jupyter_text>###### At a glance, there is seemingly no significant gap among the playlists.
- Looking in closely, the tracks in 2019 are way more acoustic and with higher level of instrumentalness compared to the other editions.
- 2016 has higher level of loudness and energy due to some rock and Eminem trackers there.
- According to Valence I was the happiest in 2016 and the saddest in 2017.
- Mode feature tell us that on average I liked songs with major modes in 2016 vs minor in 2018 and 2019.### PCA
Principal Component Analysis (PCA) is one of the most common linear dimensionality reduction techniques. It emphasizes variation and brings out strong patterns in a dataset.
- The explained variance ratios after projection are 35%, 21%, 15% for PC1, PC2, PC3 respectively. Then I took the absolute value of the eigenvectors for each projection space relative to each original feature.
- The result was acousticness has the most weights in forming the second principal component.<jupyter_code># Transform
pca = PCA(n_components=3, random_state=42)
df_pca = pd.DataFrame(data=pca.fit_transform(df_scaled.iloc[:,:-1]), columns=['PC1','PC2','PC3'])
# Inspect eigenvectors and eigenvalues
pca.components_
pca.explained_variance_ratio_
plt.matshow(pca.components_, cmap='viridis')
plt.yticks([0, 1, 2], ["First component", "Second component", "Third component"])
plt.colorbar()
plt.xticks(range(len(data[features].columns)),data[features], rotation=60, ha='left')
plt.xlabel("Feature")
plt.ylabel("Principal components")
df_pca_fix = df_pca.merge(data, left_index=True, right_index=True)
df_pca_fix = df_pca_fix[['PC1', 'PC2', 'PC3', 'title', 'main_artist', 'acousticness']]
df_pca_fix.head()
# Plot the PCA
px.scatter_3d(df_pca_fix,
x='PC1',
y='PC2',
z='PC3',
title='Principal Component Analysis Projection (3-D)',
color='acousticness',
size=np.ones(len(df_pca_fix)),
size_max=5,
height=600,
hover_name='title',
hover_data=['main_artist'],
color_continuous_scale=px.colors.cyclical.mygbm[:-6])
#because the 3D graph above doesn't load on git:
Image(filename='../images/newplot.png',width = 800)<jupyter_output><empty_output><jupyter_text>#### There we can see each song position and its distance to other songs based on the audio features that have been transformed.
- Most points are concentrated on the green-ish area and others on a pink one.
- The mapping also confirms that acousticness does correlate with PC2 to some extent.
- Salvador Sobral, Rubel and Tim Bernardes are on the opposite side to the way way less acoustic level with Shakira's famous world cup Waka Waka (what happened to me in 2016)
<jupyter_code>data[data['Year']==2019].sort_values(by="acousticness", ascending=False).head(10)
data[data['Year']==2016].sort_values(by="loudness", ascending=False).head(10)
data[data['Year']==2019].sort_values(by="instrumentalness", ascending=False).head(10)<jupyter_output><empty_output><jupyter_text>### LA ROSALÍA. TRA TRA.
Every year is marked by a specific artist (e.g. 2018 - Frank Ocean).
2019 was the year I discovered the Catalan Phenomenon - Rosalía - a singer with strong Flamengo basis that doesn't fear to explore with Pop, Trap, hip hop, etc. Her music is known as very experimental. Should be interesting to check the features on the songs I heard more during 2019:<jupyter_code>data[data['main_artist']== 'ROSALÍA'].mean()
#I would expect Rosalía to have higher level of instrumentalness
#The closer the instrumentalness value is to 1.0, the greater likelihood the track contains no vocal content.
data.mean()
Image(filename='../images/Rosalía.png')<jupyter_output><empty_output><jupyter_text>### From the land of Oz. Tash Sultana.
Let's analyse other of my absolutely favourite artists - a one woman show - Tash Sultana.
https://www.youtube.com/watch?v=bWgWKdqoJmo<jupyter_code>data[data['main_artist']== 'Tash Sultana'].mean()
#lower level of danceability - makes sense
#low acousticness - she's know to be an artist in eletric guitar
#super low level of instrumentalness - weird - she doen't sing in most part of the songs
data.mean()<jupyter_output><empty_output>
|
no_license
|
/code/.ipynb_checkpoints/My Musical Journey -checkpoint.ipynb
|
ollie0317/Spotify-Analysis
| 6 |
<jupyter_start><jupyter_text># [作業目標]
- 利用範例的創建方式, 創建一組資料, 並練習如何取出最大值# [作業重點]
- 練習創立 DataFrame (In[2])
- 如何取出口數最多的國家 (In[3], Out[3])## 練習時間
在小量的資料上,我們用眼睛就可以看得出來程式碼是否有跑出我們理想中的結果
請嘗試想像一個你需要的資料結構 (裡面的值可以是隨機的),然後用上述的方法把它變成 pandas DataFrame
#### Ex: 想像一個 dataframe 有兩個欄位,一個是國家,一個是人口,求人口數最多的國家
### Hints: [隨機產生數值](https://blog.csdn.net/christianashannon/article/details/78867204)<jupyter_code>import pandas as pd
import numpy as np
list(np.random.randint(100000000,size=5))
data = {'國家': ['Taiwan','United States','Japan','Singapore','Korea'],
'人口': list(np.random.randint(100000000,size=5))}
data1= pd.DataFrame(data)
print(data1)
data1['人口'].max()<jupyter_output><empty_output>
|
no_license
|
/homework/Day_005-1_HW.ipynb
|
veronikachn/3rd-ML100Days
| 1 |
<jupyter_start><jupyter_text>### FAKE NEWS DETECTION<jupyter_code>import numpy as np
import pandas as pd
import itertools
from sklearn.model_selection import train_test_split
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.linear_model import PassiveAggressiveClassifier
from sklearn.metrics import accuracy_score, confusion_matrix
# reading data
df=pd.read_csv('news.csv')
#Get shape and head
df.shape
df.head()
#Get the labels
labels=df.label
labels.head()
# Split the dataset
x_train,x_test,y_train,y_test=train_test_split(df['text'], labels, test_size=0.2, random_state=7)
#Initialize a TfidfVectorizer
tfidf_vectorizer=TfidfVectorizer(stop_words='english', max_df=0.7)
#Fit and transform train set, transform test set
tfidf_train=tfidf_vectorizer.fit_transform(x_train)
tfidf_test=tfidf_vectorizer.transform(x_test)
#Initialize a PassiveAggressiveClassifier
pac=PassiveAggressiveClassifier(max_iter=50)
pac.fit(tfidf_train,y_train)
#Predict on the test set and calculate accuracy
y_pred=pac.predict(tfidf_test)
score=accuracy_score(y_test,y_pred)
print(f'Accuracy: {round(score*100,2)}%')
#Build confusion matrix
confusion_matrix(y_test,y_pred, labels=['FAKE','REAL'])
<jupyter_output><empty_output>
|
no_license
|
/Fake News Project.ipynb
|
Nishanto/python-projects
| 1 |
<jupyter_start><jupyter_text># 0. Package Dependency
- [nltk](https://www.nltk.org)
- [sklearn](http://scikit-learn.org/stable/)<jupyter_code># Load packages
import pandas as pd
from sklearn.feature_extraction.text import CountVectorizer
from nltk.tokenize import WordPunctTokenizer
import numpy as np
# Load data
trn_texts = open("trn-reviews.txt").read().strip().split("\n")
trn_labels = open("trn-labels.txt").read().strip().split("\n")
print("Training data ...")
print("%d, %d" % (len(trn_texts), len(trn_labels)))
dev_texts = open("dev-reviews.txt").read().strip().split("\n")
dev_labels = open("dev-labels.txt").read().strip().split("\n")
print("Development data ...")
print("%d, %d" % (len(dev_texts), len(dev_labels)))
glove = pd.read_csv("glove.6b.50d.txt", sep=' ', header=None, quotechar=None, quoting=3)
dict = {}
for i, row in glove.iterrows():
if len(dict)%50000 ==0:
print(len(dict))
dict[row[0]] = np.array(row.drop([0]))
wtrn_data = np.array([WordPunctTokenizer().tokenize(i.lower()) for i in trn_texts])
wdev_data = np.array([WordPunctTokenizer().tokenize(i.lower()) for i in dev_texts])
def txt_rep(data):
new_data = np.array([[0]*50])
k = 0
for i in data:
addition = np.array([0]*50)
count = 0
for j in i:
try:
temp = dict[j]
addition = np.sum([temp, addition], axis=0)
count+=1
except: pass
count = max(1, count)
try:
new_data = np.append(new_data,[addition/count], axis=0)
except:
print(addition, count)
if k%10000 == 0:
print(k)
k+=1
return new_data
gtrn_data = txt_rep(wtrn_data)[1:]
gdev_data = txt_rep(wdev_data)[1:]
print('this block is done')
print(gtrn_data.shape)
from sklearn.linear_model import LogisticRegression
print(gtrn_data.shape)
# Define a LR classifier
classifier = LogisticRegression(solver="liblinear", multi_class="auto")
classifier.fit(gtrn_data, trn_labels)
# Measure the performance on training and dev data
print("Training accuracy = %f" % classifier.score(gtrn_data, trn_labels))
print("Dev accuracy = %f", classifier.score(gdev_data, dev_labels))
choice = 1
if choice == 1:
print("Preprocessing without any feature selection")
vectorizer = CountVectorizer(lowercase=False)
# vocab size 77166
elif choice == 2:
print("Lowercasing all the tokens")
vectorizer = CountVectorizer(lowercase=True)
# vocab size 60610
else:
raise ValueError("Unrecognized value: choice = %d" % choice)
trn_data = vectorizer.fit_transform(trn_texts)
print(trn_data.shape)
dev_data = vectorizer.transform(dev_texts)
print(dev_data.shape)
from scipy.sparse import coo_matrix, hstack, csr_matrix
# dev_data = dev_data.toarray()
print(trn_data.shape, gtrn_data.shape)
# cdev_data = np.concatenate((dev_data.toarray(),gdev_data), axis=1)
# ctrn_data = np.concatenate((trn_data.toarray(),gtrn_data), axis=1)
A = csr_matrix(gtrn_data.astype(float))
ctrn_data = hstack([trn_data,A])
B = csr_matrix(gdev_data.astype(float))
cdev_data = hstack([dev_data,B])
# def comb(data, gdata):
# new_data = np.array([[0]*77216])
# k = 0
# for i,j in zip(data,gdata):
# if k%(n/10) ==0:
# print (k)
# if k == n:
# print(k)
# break
# new_data = np.append(new_data, [np.append(i.toarray(), j )], axis=0)
# k+=1
# return new_data
# ctrn_data = comb(trn_data, gtrn_data)[1:]
# cdev_data = comb(dev_data, gdev_data)[1:]
print(ctrn_data.shape)<jupyter_output>(40000, 77216)
<jupyter_text># 3. Logistic Regression
Please refer to the document of [_LogisticRegression_](https://scikit-learn.org/stable/modules/generated/sklearn.linear_model.LogisticRegression.html) for the parameters of this function. <jupyter_code>from sklearn.linear_model import LogisticRegression
# Define a LR classifier
classifier = LogisticRegression(solver="liblinear", multi_class="auto")
classifier.fit(ctrn_data, trn_labels)
# Measure the performance on training and dev data
print("Training accuracy = %f" % classifier.score(ctrn_data, trn_labels))
print("Dev accuracy = %f", classifier.score(cdev_data, dev_labels))
from sklearn.linear_model import LogisticRegression
# Define a LR classifier
classifier = LogisticRegression(solver="saga", multi_class="auto")
classifier.fit(ctrn_data, trn_labels)
# Measure the performance on training and dev data
print("Training accuracy = %f" % classifier.score(ctrn_data, trn_labels))
print("Dev accuracy = %f", classifier.score(cdev_data, dev_labels))
from sklearn.linear_model import LogisticRegression
# Define a LR classifier
classifier = LogisticRegression(solver="sag", multi_class="auto")
classifier.fit(ctrn_data, trn_labels)
# Measure the performance on training and dev data
print("Training accuracy = %f" % classifier.score(ctrn_data, trn_labels))
print("Dev accuracy = %f", classifier.score(cdev_data, dev_labels))
from sklearn.linear_model import LogisticRegression
# Define a LR classifier
classifier = LogisticRegression(solver="lbfgs", multi_class="auto")
classifier.fit(ctrn_data, trn_labels)
# Measure the performance on training and dev data
print("Training accuracy = %f" % classifier.score(ctrn_data, trn_labels))
print("Dev accuracy = %f", classifier.score(cdev_data, dev_labels))<jupyter_output>c:\users\ssjk\appdata\local\programs\python\python37\lib\site-packages\sklearn\linear_model\logistic.py:758: ConvergenceWarning: lbfgs failed to converge. Increase the number of iterations.
"of iterations.", ConvergenceWarning)
|
no_license
|
/HW4&5/Text-classification/Hw4.ipynb
|
sanatkumarsjk/NLP
| 2 |
<jupyter_start><jupyter_text># Задания
#### Обязательная часть
1. Дан датасет в виде бинарной строки в файле *JEOPARDY_CSV.csv* (~30Mb) с содержанием вопросов викторины Jeopardy!. Прочитайте построчно первые 1 + 10 строк, включая шапку и выведите их в печать вместе с текущей строкой и положением указателя в файле (метод tell для файла), после чего воспользуйтесь f.seek() и переместите указатель считывания на начало и выведите еще 5 строк на экран. В помощь к решению прилагаю считывание файла и конвертацию в юникод строки.
```python
with open('JEOPARDY_CSV.csv', 'rb') as f:
# decode применяется к считываемой строке и перевод из байт-строки в utf-8
head = f.readline().decode('utf-8')
# ВАШ КОД ...
```
Замечание: прерывание цикла *for* можно обеспечить как условием *if* так и оператором *break*:
```python
for i in range(10):
print(i)
if i == 3:
break # выйти из цикла for
```<jupyter_code>with open('JEOPARDY_CSV.csv', 'rb') as f:
# decode применяется к считываемой строке и перевод из байт-строки в utf-8
head = f.readline().decode('utf-8')
print(head)
for i in range(10):
line = f.readline().decode('utf-8')
print(i+1, end=' ')
print(f.tell(), end=': ')
print(line)
f.seek(0)
for i in range(5):
line = f.readline().decode('utf-8')
print(i+1, end=' ')
print(f.tell(), end=': ')
print(line)
f.close()<jupyter_output>Show Number, Air Date, Round, Category, Value, Question, Answer
1 221: 4680,2004-12-31,Jeopardy!,"HISTORY","$200","For the last 8 years of his life, Galileo was under house arrest for espousing this man's theory","Copernicus"
2 412: 4680,2004-12-31,Jeopardy!,"ESPN's TOP 10 ALL-TIME ATHLETES","$200","No. 2: 1912 Olympian; football star at Carlisle Indian School; 6 MLB seasons with the Reds, Giants & Braves","Jim Thorpe"
3 577: 4680,2004-12-31,Jeopardy!,"EVERYBODY TALKS ABOUT IT...","$200","The city of Yuma in this state has a record average of 4,055 hours of sunshine each year","Arizona"
4 732: 4680,2004-12-31,Jeopardy!,"THE COMPANY LINE","$200","In 1963, live on ""The Art Linkletter Show"", this company served its billionth burger","McDonald's"
5 908: 4680,2004-12-31,Jeopardy!,"EPITAPHS & TRIBUTES","$200","Signer of the Dec. of Indep., framer of the Constitution of Mass., second President of the United States","John Adams"
6 1049: 4680,2004-12-31,Jeopardy!,"3-LETTER WORDS",[...]<jupyter_text>После этого заново считайте файл и распарсите его в структуру данных (словарь), ключи в котором будут являться поля *head*, а значениями списки с соответствующими данными, после чего при считывании добавьте парсинг каждой строки и добавление в соответствующий список данных после конвертации:
* Show Number, Value - int
* Air Date - создайте объект `datetime.date` из встроенной библиотеки [datetime](https://docs.python.org/3.7/library/datetime.html)
* Round, Category, Question, Answer - строки
Замечание:
* вы можете воспользоваться встроенными и ранее пройденными методами форматирования строк, а можете воспользоваться [регулярными выражениями](https://habr.com/ru/post/349860/) с помощью библиотеки [re](https://docs.python.org/3.7/library/re.html)
* рекомендую использовать функции как можно больше для форматирования, парсинга и т.д.<jupyter_code>import re
import datetime
with open('JEOPARDY_CSV.csv', 'rb') as f:
lines = f.readlines()
head = lines.pop(0).decode('utf-8')
head = head.replace("\r\n","")
keys = head.split(", ")
#print(keys)
leo_dic=[]
for k in range(len(lines)):
lines[k] = lines[k].decode('utf-8').replace("\r\n","")
values=re.split(',(?=(?:[^\"]*\"[^\"]*\")*(?![^\"]*\"))', lines[k])
#print(values)
leo_dic.append({keys[0]: int(values[0])})
leo_dic[k][keys[1]]= datetime.date.fromisoformat(values[1])
leo_dic[k][keys[4]]= int(values[4].replace('$', '').replace('"', '').replace(',', '').replace('None', '0'))
for i in [2,3,5,6]:
values[i]=re.sub('(^")|("$)','',values[i])
values[i]=values[i].replace('\\"', '"')
values[i]=values[i].replace('""', '"')
leo_dic[k][keys[i]]= values[i]
f.close()
for i in range(5):
print(leo_dic[i])<jupyter_output>{'Show Number': 4680, 'Air Date': datetime.date(2004, 12, 31), 'Value': 200, 'Round': 'Jeopardy!', 'Category': 'HISTORY', 'Question': "For the last 8 years of his life, Galileo was under house arrest for espousing this man's theory", 'Answer': 'Copernicus'}
{'Show Number': 4680, 'Air Date': datetime.date(2004, 12, 31), 'Value': 200, 'Round': 'Jeopardy!', 'Category': "ESPN's TOP 10 ALL-TIME ATHLETES", 'Question': 'No. 2: 1912 Olympian; football star at Carlisle Indian School; 6 MLB seasons with the Reds, Giants & Braves', 'Answer': 'Jim Thorpe'}
{'Show Number': 4680, 'Air Date': datetime.date(2004, 12, 31), 'Value': 200, 'Round': 'Jeopardy!', 'Category': 'EVERYBODY TALKS ABOUT IT...', 'Question': 'The city of Yuma in this state has a record average of 4,055 hours of sunshine each year', 'Answer': 'Arizona'}
{'Show Number': 4680, 'Air Date': datetime.date(2004, 12, 31), 'Value': 200, 'Round': 'Jeopardy!', 'Category': 'THE COMPANY LINE', 'Question': 'In 1963, live on "The Art Linkletter S[...]<jupyter_text>
* Сохраните созданную структуру данных в форматах [JSON](https://docs.python.org/3.7/library/json.html) (библиотека json) и с использованием модуля [pickle](https://docs.python.org/3.7/library/pickle.html).<jupyter_code>import json
class ComplexEncoder(json.JSONEncoder):
def default(self, obj):
if isinstance(obj, datetime.date):
return obj.isoformat()
return json.JSONEncoder.default(self, obj)
json.dumps(leo_dic, cls=ComplexEncoder)[1:1000]
import pickle
pickle.dumps(leo_dic)[1:1000]<jupyter_output><empty_output><jupyter_text>2. Вам дана заготовка скрипта **animal.py**. Необходимо:
* Определить абстрактный класс Animal, в котором будут методы: *voice()*, *say_name()*, *feed(q)*. Класс Animal является родителем для классов Cat, Dog, Cow, которые будут определять поведение наследованных методов.
* Метод *voice()* должен возвращать строку "{animal type} say: {animal_voice}!" (например, "Cow say: mooooo!"). Голоса для животных: Cow - "mooooo!", Dog - "wooof", Cat - "meooow".
* Метод *say_name()* должен возвращать строку "I am {animal type}, my name is {name}"
* Метод *feed(q)* должен покормить животное едой размером q и вернуть строку. Для разных объемов q будут различные варианты возвращаемых строк:
* q = 0 -> "I am still whant to eat"
* q = 1 -> "Not so much food"
* q = 2 -> "I am feeling good"
* q > 2 -> "Is today a holiday?"
* У каждого из классов в заготовке дан также метод, который будет использоваться как опция.
После определения классов вам необходимо будет запускать скрипт "animal.py" через комманду в командной строке (Anaconda Prompt для Windows или Bash Shell для Linux). Например:
`python path_to_folder/animal.py -t "animal_type" -q *some_number* -a *what_to_do"`
В скрипте должен быть реализован следующий сценарий:
- для заданного animal_type вы создаёте животное с любым именем на ваше усмотрение, вызываете `say_name()` и кормите его `feed(q)` с q, который был получен через *-q*.
- для заданного -a сделать либо `voice()`, либо специфичный для каждого класса метод.
<jupyter_code>import argparse
ap = argparse.ArgumentParser()
ap.add_argument("--action", "-a", help='what action animal should do')
ap.add_argument("--quantity", "-q", help='how much times they should do actions')
ap.add_argument("--type", "-t", help='what type of animal should born')
class Animal:
def __init__(self, name):
self.name = name
self.animal_type = self.__class__.__name__
self.food = 0
def voice(self):
return(f"{self.animal_type} say: {self.animal_voice}!")
def say_name(self):
return(f"I am {self.animal_type}, my name is {self.name}")
def feed(self, q):
self.food += q
if (q==0):
return("I am still whant to eat")
elif(q==1):
return("Not so much food")
elif(q==2):
return("I am feeling good")
elif(q>2):
return("Is today a holiday?")
class Dog(Animal):
animal_voice = "wooof"
pass
def play_with_ball(self):
print(f"{self.name.capitalize()} run to a ball and become happy")
class Cat(Animal):
animal_voice = "meooow"
pass
def nightrunner(self):
print(f"Everybody wake up, {self.name} is happy")
class Cow(Animal):
animal_voice = "mooooo!"
pass
def walking_with_bell(self):
print(f"Cow walking on the grass")
def main():
animal_types={"Dog": Dog("Snoopy"),
"Cat": Cat("Ralf"),
"Cow": Cow("Moon"),
}
an = animal_types[args.type]
print(an.say_name())
print(an.feed(int(args.quantity)))
if args.action == "play_with_ball":
an.play_with_ball()
elif args.action == "nightrunner":
an.nightrunner()
elif args.action == "walking_with_bell":
an.walking_with_bell()
else:
print(an.voice())
pass
if __name__ == '__main__':
args = ap.parse_args()
main()
args = ap.parse_args('-a play_with_ball -q 2 -t Dog'.split())
main()
print("")
args = ap.parse_args('-a nightrunner -q 1 -t Cat'.split())
main()
print("")
args = ap.parse_args('-a walking_with_bell -q 10 -t Cow'.split())
main()
print("")
args = ap.parse_args('-a voice -q 0 -t Dog'.split())
main()<jupyter_output>I am Dog, my name is Snoopy
I am feeling good
Snoopy run to a ball and become happy
I am Cat, my name is Ralf
Not so much food
Everybody wake up, Ralf is happy
I am Cow, my name is Moon
Is today a holiday?
Cow walking on the grass
I am Dog, my name is Snoopy
I am still whant to eat
Dog say: wooof!
|
no_license
|
/HW_3.ipynb
|
fizik/pycourse
| 4 |
<jupyter_start><jupyter_text># Station
In this tutorial, the following topics shall be covered:
- What is a station
- How to create it, and work with it
- Snapshot of a station
- Configuring station using a YAML configuration file<jupyter_code># Useful imports:
from pprint import pprint # for pretty-printing python variables like 'dict'
import qcodes
from qcodes.instrument import Parameter
from qcodes.station import Station
from qcodes.tests.instrument_mocks import DummyInstrument<jupyter_output><empty_output><jupyter_text>## What is a Station?
Experimental setups are generally large as they consist of many instruments. Each instrument in a given setup tends to be quite complex due to the fact that each comprises a variety of adjustable parameters and other stateful parts. As a result, it deems useful to have a bucket in which the necessary information concerning the instruments that are going to be used in a particular experiment can be conveniently stored, and accessed.
The concept of a station, in essence, is a programmatical representation of such a bucket. Instruments, parameters, and other "components" can be added to a station. The user gets a station instance that can be referred to in order to access those "components".
A station can be configured from a text file which simplifies the initialisation of the instruments. In particular, in this tutorial, we shall provide an example configuration of a station by using a YAML file.
A special use case of a station in an experiment would be the capturing of the state of an experimental setup, known as a snapshot. We shall devote a subsection for the concept of a snapshot. ## How to create a Station and work with it?
Let us, first, create a dummy parameter, and a dummy instrument which we shall use throughout the tutorial.<jupyter_code># A dummy self-standing parameter
p = Parameter('p', label='Parameter P', unit='kg', set_cmd=None, get_cmd=None)
p.set(123)
# A dummy instrument with three parameters
instr = DummyInstrument('instr', gates=['input', 'output', 'gain'])
instr.gain(42)<jupyter_output><empty_output><jupyter_text>### Creating a StationWe create a ``Station`` object and add previously defined parameter and instrument as its components as follows: <jupyter_code>station = Station()
station.add_component(p)
station.add_component(instr)<jupyter_output><empty_output><jupyter_text>It is straightforward to verify if the station contains our parameter and instrument:<jupyter_code># Now station contains both `p` and `instr`
station.components<jupyter_output><empty_output><jupyter_text>Note that it is also possible to add components to a station via arguments of its constructor:<jupyter_code>station = Station(p, instr)<jupyter_output><empty_output><jupyter_text>### Accessing Station componentsNow that the components have been added to the station, it is possible to access them as its attributes (by using the "dot" notation). With this feature, users can use tab-completion to find the instrument in the station they'd like to access.<jupyter_code># Let's confirm that station's `p` is
# actually the `p` parameter defined above
assert station.p is p<jupyter_output><empty_output><jupyter_text>### Removing components from a StationRemoving components from a station should be done with `remove_component` method - the name of the component that is to be removed should be passed as the argument of the method:<jupyter_code>station.remove_component('p')
# Now station contains only `instr`
station.components<jupyter_output><empty_output><jupyter_text>### Default Station
The `Station` class is designed in such a way that it always contains a reference to a `default` station object (the `Station.default` attribute). The constructor of the station object has a `default` keyword argument that allows to specify whether the resulting instance shall be stored as a default station, or not.
This feature is a convenience. Other objects which consume an instance of `Station` as an argument (for example, `Measurement`) can now implement a logic to resort to `Station.default` in case a `Station` instance was not explicitly given to them.## Snapshot of a Station
The station has a `snapshot` method that allows to create a collective, single snapshot of all the instruments, parameters, and submodules that have been added to it. It would be very time-consuming for the user to manually go through every instrument and parameter, and collect the snapshot data.
For example, the `Measurement` object accepts a station argument exactly for the purpose of storing a snapshot of the whole experimental setup next to the measured data.
Read more about snapshots in general, how to work with them, station's snapshot in particular, and more -- in ["Working with snapshots" example notebook](DataSet/Working with snapshots.ipynb) ([nbviewer.jupyter.org link](https://nbviewer.jupyter.org/github/QCoDeS/Qcodes/tree/master/docs/examples/DataSet/Working with snapshots.ipynb)).## Configuring the Station by using a YAML configuration file
The instantiation of the instruments, setting up the proper initial values of the corresponding parameters and similar pre-specifications of a measurement constitutes the initialization portion of the code. In general, this portion can be quite long and tedious to maintain. For example, consider a case in which a certain instrument is no longer needed. In this case a common practice is commenting out the lines of initialization script which are related to the said instrument, and re-run the initialization script. The latter may easily cause a bloaded code with possible repetitions. In another case, we may want to share initialization scripts among collaborators and fail to do so as it is difficult due to the fact that even similar experiments may require different instantiations.
These (and more) concerns are to be solved by a YAML configuration file of the `Station` object (formerly known as the `StationConfigurator`).
The YAML configuration file allows one to statically and uniformly specify settings of all the instruments (and their parameters) that the measurement setup (the "physical" station) consists of, and load them with those settings on demand. The `Station` object implements convenient methods for this procedure.
The YAML configuration file, if used, is stored in the station as a attribute with the name `config`, and is thus included in the snapshot of the whole station.
Note that we are not obliged to use the YAML configuration file to set up a `Station` (see, for example, the section "How to create a Station and work with it" of this tutorial).
In what follows, we shall discuss:
- The structure of the YAML configuration file
- `Station`s methods related to working with the YAML configuration
- Entries in QCoDeS configuration that are related to `Station`### Example of YAML Station configurationHere, we provide an example YAML station configuration file. All the fields within the configuration file are explained via inline comments that should not be disregarded. A careful inspection of the comments by the reader is strongly recomended for a clear understanding.
In particular, here, we would like to underline the difference between `parameters` and `add_parameters` sections. In this example file for the `QDac` instrument, we define a `Bx` parameter as a new, additional parameter. The `Bx` parameter will have the properties such as `limits`, `scale`, etc. __different__ from its "source" parameter `ch02.v` that it controls. Specifically, this means that when setting `Bx` to `2.0`:
1. the value of `2.0` is being validated against the limits of `Bx` (`0.0, 3.0`),
2. then the raw ("scaled") value of `130.468` (`= 2.0 * 65.234`) is passed to the `ch02.v` parameter,
3. then that value of `130.468` is validated against the limits of `ch02.v` (`0.0, 1.5e+3`),
4. then the raw ("scaled") value of `1.30468` (`= 130.468 * 0.01`) is finally passed to the physical instrument.
We also note that in the exponential represantations of numbers, it is required to provide `+` and `-` signs after `e`, e.g., we write `7.8334e+5` and `2.5e-23`. Refer to YAML file format specification for more information.```yaml
# Example YAML Station configuration file
#
# This file gets snapshotted and can be read back from the JSON
# snapshot for every experiment run.
#
# All fields are optional unless explicitly mentioned otherwise.
#
# As in all YAML files a one-line notation can also be used
# instead of nesting notation.
#
# The file starts with a list of loadable instruments instances,
# i.e. there can be two entries for two instruments of the same
# type if you want to specify two different use cases
# e.g. "dmm1-readout" and "dmm1-calibration".
#
instruments:
# Each instrument is specified by its name.
# This name is what is looked up by the `load_instrument`
# method of `Station`.
# Simulated instruments can also be specified here, just put
# the path to the similation .yaml file as the value of the
# "init"->"visalib" field (see below for an example of the
# "init" section as well as an example of specifying
# a simulated instrument).
qdac:
# Full import path to the python class of the instrument
# driver
type: qcodes.instrument_drivers.QDev.QDac_channels.QDac
# Visa address of the instrument.
# Note that this field can also be specified in the
# "init" section (see below) but the address specified
# here will overrule the address from the "init" section.
# Essentially, specifying address here allows avoiding
# the "init" section completely when address is the only
# neccesary argument that the instrument driver needs.
# For obvious reasons, this field is required for VISA
# instruments.
address: ASRL4::INSTR
# If an instrument with this name is already instantiated,
# and this field is true, then the existing instrument
# instance will be closed before instantiating this new one.
# If this field is false, or left out, closing will not
# happen.
enable_forced_reconnect: true
#
# The "init" section specifies constant arguments that are
# to be passed to the __init__ function of the instrument.
# Note that it is the instrument's driver class that defines
# the allowed arguments, for example, here "update_currents"
# is an argument that is specific to "QDac" driver.
init:
terminator: \n
update_currents: false
#
# Setting up properties of parameters that already exist on
# the instrument.
parameters:
# Each parameter is specified by its name from the
# instrument driver class.
# Note that "dot: notation can be used to specify
# parameters in (sub)channels and submodules.
ch01.v:
# If an alias is specified, the paramater becomes
# accessible under another name, so that you can write
# `qdac.cutter_gate(0.2)` instead of `qdac.ch01.v(0.2)`.
# Note that the parameter instance does not get copied,
# so that `(qdac.ch01.v is qdac.cutter_gate) == True`.
alias: cutter_gate
# Set new label.
label: Cutter Gate Voltage
# Set new unit.
unit: mV
# Set new scale.
scale: 0.001
# Set new post_delay.
post_delay: 0
# Set new inter_delay.
inter_delay: 0.01
# Set new step.
step: 1e-4
# If this field is given, and contains an array of two
# numbers like here, then the parameter
# gets a new `Numbers` validator with these values as
# lower and upper limits, respectively (in this case, it
# is `Numbers(-0.1, 0.1)`).
limits: [-0.1, 0.1]
# Set the parameter to this given initial value upon
# instrument initialization.
# Note that if the current value on the physical
# instrument is different, the parameter will be ramped
# with the delays and step specified in this file.
initial_value: 0.01
# In case this values equals to true, upon loading this
# instrument from this configuration this parameter will
# be appended to the list of parameters that are
# displayed in QCoDeS `Monitor`.
monitor: true
# As in all YAML files a one-line notation can also be
# used, here is an example.
ch02.v: {scale: 0.01, limits: [0.0, 1.5e+3] , label: my label}
ch04.v: {alias: Q1lplg1, monitor: true}
#
# This section allows to add new parameters to the
# instrument instance which are based on existing parameters
# of the instrument. This functionality is based on the use
# of the `DelegateParameter` class.
add_parameters:
# For example, here we define a parameter that represents
# magnetic field control. Setting and getting this
# parameter will actually set/get a specific DAC channel.
# So this new magnetic field parameter is playing a role
# of a convenient proxy - it is much more convenient to
# perform a measurement where "Bx" is changed in tesla as
# opposed to where some channel of some DAC is changed in
# volts and one has to clutter the measurement code with
# the mess of conversion factors and more.
# Every new parameter definition starts with a name of
# the new parameter.
Bx:
# This field specifies the parameter which "getter" and
# "setter" will be used when calling `get`/`set` on this
# new parameter.
# Required field.
source: ch02.v
# Set the label. Otherwise, the one of the source parameter
# will be used.
label: Magnetic Field X-Component
# Set the unit. Otherwise, the one of the source parameter
# will be used.
unit: T
# Other fields have the same purpose and behavior as for
# the entries in the `add_parameter` section.
scale: 65.243
inter_delay: 0.001
post_delay: 0.05
step: 0.001
limits: [0.0, 3.0]
initial_value: 0.0
# For the sake of example, we decided not to monitor this
# parameter in QCoDeS `Monitor`.
#monitor: true
#
# More example instruments, just for the sake of example.
# Note that configuring simulated instruments also works,
# see the use of 'visalib' argument field below
dmm1:
type: qcodes.instrument_drivers.agilent.Agilent_34400A.Agilent_34400A
enable_forced_reconnect: true
address: GPIB::1::65535::INSTR
init:
visalib: 'Agilent_34400A.yaml@sim'
parameters:
volt: {monitor: true}
mock_dac:
type: qcodes.tests.instrument_mocks.DummyInstrument
enable_forced_reconnect: true
init:
# To pass an list of items use {}.
gates: {"ch1", "ch2"}
add_parameters:
Bx: {source: ch1, label: Bx, unit: T,
scale: 28, limits: [-1, 1], monitor: true}
mock_dac2:
type: qcodes.tests.instrument_mocks.DummyInstrument
enable_forced_reconnect: true
```### QCoDeS configuration entries related to StationQCoDeS configuration contains entries that are related to the `Station` and its YAML configuration file. Refer to [the description of the 'station' section in QCoDeS config](http://qcodes.github.io/Qcodes/user/configuration.html?highlight=station#default-config) for more specific information.### Using Station with YAML configuration filesIn this section, we shall briefly describe the usage of `Station` with YAML configuration files. For more details, we refer to the docstrings of the methods of the `Station` class.
A `Station` can be created with or without one or more YAML configuration files. With YAML configuration files, `Station` can be created by passing the file names (or file names with absolute path) to `Station`'s constructor. File names and location resolution also takes into account related entries in the `'station'` section of the QCoDeS configuration. We refer to corresponding documentation for more information.
```python
station = Station(config_file='qutech_station_25.yaml')
```
or
```python
station = Station(config_file=['example_1.yaml', 'example_2.yaml'])
```
Alternatively, `load_config_file` or `load_config_files` methods can be called on an already instantiated Station to load the configuration files.
* `load_config_file` method can be used for loading 1 configuration file.
* `load_config_files` method can be used for loading 1 or more configuration files. If multiple configuration files are provided, this method first merges them into one temporary file and then loads that into the station. Please note that, the merged file is temporary and not saved on the disk.
```python
station = Station()
station.load_config_file=r'Q:\\station_data\\qutech_station_25.yaml'
```
or
```python
station = Station()
station.load_config_files('example_1.yaml', 'example_2.yaml', 'example_3.yaml')
```
In case the configuration is already available as a YAML string, then that configuration can be loaded using `Station`'s `load_config` method. We refer to it's docstring and signature for more information.
Once the YAML configuration is loaded, the `load_instrument` method of the `Station` can be used to instantiate a particular instrument that is described in the YAML configuration. Calling this method not only will return an instance of the instantiated instrument, but will also add it to the station object.
For example, to instantiate the `qdac` instrument from the YAML configuration example from the previous section, it is sufficient to execute the following:
```python
loaded_qdac = station.load_instrument('qdac')
```
Note the `load_instrument`'s `revive_instance` argument, as well as `enable_force_reconnect` setting from the YAML configuration - these define what to do in case an instrument with the given name has already been instantiated in this python session.
There is a more convenient way to load the instruments. Upon load of the YAML configuration, convenient `load_` methods are being generated on the `Station` object. Users can make use of tab-completion in their development environments to list what instruments can be loaded by the station object from the loaded YAML configuration. For example, loading the QDac instrument can, alternatively, be done via:
```python
conveniently_loaded_qdac = station.load_qdac()
```
We note that instruments are instantiated only when `load_*` methods are called. This means that loading a YAML configuration does NOT automatically instantiate anything.
For the instruments that are loaded with the `load_*` methods, it is recommended to use `Station`'s `close_and_remove_instrument` method for closing and removing those from the station.### Setting up Visual Studio Code to work with station files
VSCode can be conveniently used to work with station files. The [YAML extension by Red Hat](https://marketplace.visualstudio.com/items?itemName=redhat.vscode-yaml), allows to use JSON schemas to validate and auto-complete YAML files.

**Quick setup steps**
- install [YAML extension by Red Hat](https://marketplace.visualstudio.com/items?itemName=redhat.vscode-yaml)
- run `qcodes.utils.installation.register_station_schema_with_vscode()`
- make sure your station file has the extension `.station.yaml`**Setup details**
To associate the qcodes station schema with `*.station.yaml` files you can either run<jupyter_code>from qcodes.utils.installation import register_station_schema_with_vscode
import os
import sys
# if statement for CI only
if sys.platform == 'win32' and os.path.exists(os.path.expandvars(os.path.join('%APPDATA%', 'Code', 'User', 'settings.json'))):
register_station_schema_with_vscode()<jupyter_output><empty_output><jupyter_text>or manually add the following to your vscode user/workspace settings:
```
"yaml.schemas": {
"file:///path/to/home/schemas/station.schema.json": "*.station.yaml"
}
```To get autocompletion for instruments from other packages run
```python
import qcodes
import qcodes_contrib_drivers
import zhinst
qcodes.station.update_config_schema(qcodes_contrib_drivers, zhinst)
```where `qcodes_contrib_drivers` and `zhinst` are example packages that are available in the python environment where qcodes is running from.
**Beware** that the generated schema file is not python-environment aware. To circumvent that, you can manually generate a schema file for each environment and register them as a workspace setting in vscode.<jupyter_code>os.path.expandvars(os.path.join('%APPDATA%', 'Code', 'User', 'settings.json'))<jupyter_output><empty_output>
|
non_permissive
|
/docs/examples/Station.ipynb
|
jenshnielsen/Qcodes
| 9 |
<jupyter_start><jupyter_text>#Projeto Pokemón - Análise Estatística
Objetivo: Analisar alguns atributos dos Pokémons fazendo uso de gráficos, dados estatísticos ou análise de correlações e, em seguida, criar um relatório de análise com base no que foi observado.
**Atenção**
Para melhor organização da tabela, resolvemos reduzir o número de pokémons que foram mostrados: de 721 pokémons, foram seleciodos 20 . Como critério de escolha, colocamos a soma das características totais dos pokémons em ordem crescente e pegamos os 20 últimos pokémons da nova tabela.<jupyter_code>import pandas as pd
import numpy as np
from google.colab import drive
import matplotlib.pyplot as plt
%matplotlib inline
pd.read_csv("/content/drive/My Drive/POKEmonio.csv")
df = pd.read_csv("/content/drive/My Drive/POKEmonio.csv")
df.tail(21)
df = df.tail(21)
df
df = df.drop(columns='#')
df<jupyter_output><empty_output><jupyter_text> Efetuamos o cálculo da `variância` de todas as colunas.<jupyter_code>df1 = df.drop(columns='Total')
vari = df1.var()
vari<jupyter_output><empty_output><jupyter_text>.Cálculo do Desvio Padrão com o comando `std`.<jupyter_code>df1 = df.drop(columns='Total')
df1.std()<jupyter_output><empty_output><jupyter_text>.Usamos o comando ``describe``para descrever diferentes formas de medidas de tendência, média e dispersão concentradas<jupyter_code>df1 = df.drop(columns='Total')
df1.describe()<jupyter_output><empty_output><jupyter_text>Inserimos um filtro com duas condições; HP > 80 e Speed > 50.<jupyter_code>df.query("HP > 80 and Speed >= 50")<jupyter_output><empty_output><jupyter_text>Selecionamos os 6 ultimos Pokémons da lista pelo comando:<jupyter_code>df.tail(12)
df = df.tail(12)<jupyter_output><empty_output><jupyter_text>Dessa forma, fizemos um grafico dos HPs dos ultimos 6 Pokémons.<jupyter_code>df["Sp. Atk"]
x = df["Sp. Atk"]
plt.plot(x,'r.-')
plt.title("Especial de Ataque")
plt.xlabel("Pokemons")
plt.ylabel("Ataque")
plt.grid()<jupyter_output><empty_output><jupyter_text>Novamente selecionamos os 6 ultimos Pokémons com o mesmo comando.<jupyter_code>df.tail(12)
df = df.tail(12)<jupyter_output><empty_output><jupyter_text>E por meio do comando anterior, foi plotado um grafico do Especial de Defesa dos pokémons do comando anterior.<jupyter_code>df["Sp. Def"]
y = df['Sp. Def']
plt.plot(y,'b.-')
plt.title("Sp. Def")
plt.xlabel("Pokemons")
plt.ylabel("Especial de Defesa")
plt.grid()<jupyter_output><empty_output><jupyter_text>Organizamos os dois gráficos anteriores juntos de uma forma direta e prática, com o intuito de tornar possivel fazer a comparação entre o HP e a Speed de cada Pokémon.<jupyter_code>plt.plot(x,'r.-')
plt.plot(y,'b.-')
plt.title("Especial de Ataque e Defesa")
plt.xlabel("Pokemons")
plt.ylabel("Ataque e Defesa")
plt.grid()<jupyter_output><empty_output><jupyter_text>Gráfico onde mostra o valor do ataque dos ultimos 6 Pokémons selecionados da tabela.<jupyter_code>df.tail(12)
df = df.tail(12)
df["Attack"]
y2 = df['Attack']
plt.plot(y2,'g.-')
plt.xlabel("Pokemons")
plt.ylabel("Attack")
plt.grid()<jupyter_output><empty_output><jupyter_text>Assim como no gráfico anterior, utilizamos os mesmos Pokémons para a comparação da defesa em relação do Especial de Ataque, Especial de Defesa e o Attack. Nesse Gráfico, mostramos ``APENAS`` a defesa dos mesmos que foram comparados.<jupyter_code>df.tail(12)
df = df.tail(12)
df["Defense"]
x2 = df['Defense']
plt.plot(x2,'r.-')
plt.xlabel("Pokemons")
plt.ylabel("Defesa")
plt.grid()
plt.plot(x2,'g.-')
plt.plot(y2,'r.-')
plt.title("Ataque e Defesa")
plt.xlabel("Pokemons")
plt.ylabel("Ataque e Defesa")
plt.grid()<jupyter_output><empty_output><jupyter_text>No gráfico a seguir, plotamos o Especial de Ataque, Especial de Defesa, Attack e Defesa em um gráfico de barras para observar a força máxima de cada Pokémon em cada categoria.<jupyter_code>grupos = ["Sp. Atk","Sp. Def","Attack","Defense"]
valores = [170,150,160,181]
plt.bar(grupos,valores)
plt.show()<jupyter_output><empty_output>
|
no_license
|
/Projeto_Pokemon.ipynb
|
brunovmouraf/PokemonUFABC
| 13 |
<jupyter_start><jupyter_text>## for presentation<jupyter_code>zip_map = crime2016['zipcode'].value_counts().index
ax = plt.gca()
for z in zip_map:
crime2016[crime2016['zipcode'] == z].plot('x', 'y', kind='scatter', figsize=(20,20),\
alpha=0.7, c=['0.75','0.25','0.25'], ax=ax)
ax.set_axis_bgcolor('white')
crime2016.columns<jupyter_output><empty_output><jupyter_text>## geopandas<jupyter_code>import geopandas as gpd
sf = gpd.read_file('../map_visualization/2010_census_tracts_neighborhoods/census_tracts.shp')
sf.plot()
crime2016[['x', 'y']].head(1)
# y is latitude and x is longitude
plt.scatter(crime2016['x'], crime2016['y'])
#missing person
#assault, sex offenses
#forcible, sex offenses, non forcible
#stabbing: threats against life, aggravated assault with a deadly weapon, aggravated assault with a knife
#aggravated assault with a gun
crime2016[crime2016.category == 'assault']['descript'].value_counts()
#crime2016.category.value_counts()
crime_drug = crime2016[crime2016.category == 'drug/narcotic']
<jupyter_output><empty_output><jupyter_text>import mplleaflet
f = plt.figure(figsize=(15, 8))
ax = f.gca()
plt.scatter(crime_drug['x'], crime_drug['y'])
mplleaflet.display(fig=f)## geopandas<jupyter_code>sf.head()
sf.crs
sf = sf.to_crs({'init': 'epsg:4326'})
sf['geometry'].head()
sf.plot()
import shapely
crime_drug=crime_drug.reset_index(drop=True)
listings2 = gpd.GeoDataFrame(crime_drug, geometry=crime_drug.apply(
lambda srs: shapely.geometry.Point(srs['x'], srs['y']), axis='columns'
))
f = plt.figure(figsize=(5, 5))
ax = f.gca()
sf.plot(ax=ax, alpha=0.1, linewidth=0.25, color='white')
f = plt.figure(figsize=(8, 8))
ax = f.gca()
sf.plot(ax=ax, alpha=0.1, linewidth=0.25, color='white')
sns.kdeplot(data=listings2.apply(lambda srs: pd.Series({'x': srs.geometry.x, 'y': srs.geometry.y}), axis='columns'),
ax=ax, alpha=1)
ax.set_axis_off()
import numpy as np
def assign_census_tract(df):
bools = [geom.contains(df['geometry']) for geom in sf['geometry']]
if True in bools:
return sf.iloc[bools.index(True)]['tractce10']
else:
return np.nan
#listings['census_tract'] = listings.apply(assign_census_tract, axis='columns')
sf.groupby('nhood')['drug'].sum().sort_values(ascending=False).head(20)
crime_drug['census_tract'] = crime_drug.apply(assign_census_tract, axis='columns')
crime_drug['census_tract'].value_counts().head()
sf['drug'] = sf['tractce10'].map(crime_drug['census_tract'].value_counts())
sf = sf.fillna(0)
f = plt.figure(figsize=(8, 8))
ax = f.gca()
kw = dict(column='drug', k=6, cmap='Reds', alpha=1, legend=True, edgecolor='gray', linewidth=0.5)
sf.plot(scheme='QUANTILES', ax=ax, **kw)
ax.set_axis_off()<jupyter_output><empty_output><jupyter_text>In our case we'll fix it by figuring out AirBnB density per square kilometer, not just the raw number, and plot that.
Remember, however, that areas in the latitude-longitude coordinate system are not equal. To get an accurate measurement of area we need to first reproject to what's called an "equal-area" projection (epsg:3395 will do nicely), and then take the areas of those polygons instead. Note also that shapely returns areas in square meters; we divide by a constant to get square kilometers. All this is done by the workhourse one-liner below.<jupyter_code>sf
sf['drugDensity'] = (sf['drug'] / (sf['geometry'].to_crs({'init': 'epsg:4326'}).map(lambda p: p.area*10**5))).fillna(0)
f = plt.figure(figsize=(8, 8))
ax = f.gca()
kw = dict(column='drugDensity', k=6, cmap='PuBu', alpha=1, legend=True, edgecolor='gray', linewidth=0.5)
sf.plot(scheme='Fisher_Jenks',ax=ax, **kw)
ax.set_title('SF Drug Crime Dentsity Map')
ax.set_axis_off()<jupyter_output><empty_output><jupyter_text>## Spatial Weights<jupyter_code>import pysal as ps
qW = ps.queen_from_shapefile('../map_visualization/2010_census_tracts_neighborhoods/census_tracts.shp')<jupyter_output>WARNING: there is one disconnected observation (no neighbors)
('Island id: ', [183])
<jupyter_text>## Spatial Lag<jupyter_code>drug_spatial_lag = ps.lag_spatial(qW, sf['drugDensity'])
spatial_lag_class = ps.Quantiles(drug_spatial_lag, k=5)
drug_spatial_lag
spatial_lag_class
f = plt.figure(figsize=(8,8))
ax = f.gca()
kw = dict(column='spatial_class', k=5, cmap='Reds', alpha=1, legend=True, edgecolor='gray',
linewidth=0.5, categorical=True)
sf.assign(spatial_class=spatial_lag_class.yb).plot(ax=ax, **kw)
ax.set_axis_off()<jupyter_output><empty_output><jupyter_text>This geographic pattern is distinctly not random, strong evidence again that our data is geographically dependent.
How dependent? The tool of choice for this is the a Moran scatterplot. If geography matters our data should show a strong liner structure of some kind: high lag values correspond with high actual values, and low lags with low ones.
A quick plot shows that this is, indeed, the case:<jupyter_code>f, ax = plt.subplots(1, figsize=(6, 6))
plt.plot(sf['drugDensity'], drug_spatial_lag, '.', color='firebrick')
# Calculate and plot a line of best fit.
b,a = np.polyfit(sf['drugDensity'], drug_spatial_lag, 1)
plt.plot(sf['drugDensity'], a + b*sf['drugDensity'], 'r')
plt.title('Moran Scatterplot')
plt.ylabel('SF Drug Density Spatial Lag')
plt.xlabel('SF Drug Density Actual')
ax.set_axis_bgcolor('white')
plt.show()
from math import log
moran = ps.Moran(sf['drugDensity'].values, qW)
moran.I, moran.p_sim<jupyter_output>('WARNING: ', 183, ' is an island (no neighbors)')
<jupyter_text>## Spatial Clustering<jupyter_code>listings2['descript'].value_counts()<jupyter_output><empty_output><jupyter_text>### possession and sale of drug (spatial clustering)<jupyter_code>possession_of_drug = listings2.query('descript == "possession of narcotics paraphernalia" or\
descript == "possession of meth-amphetamine" or\
descript == "possession of marijuana" or\
descript == "possession of heroin" or\
descript == "possession of base/rock cocaine" or\
descript == "possession of controlled substance" or\
descript == "possession of cocaine" or\
descript == "transportation of marijuana"').groupby('census_tract').count()['descript']
sale_of_drug = listings2.query('descript == "possession of base/rock cocaine for sale" or\
descript == "possession of meth-amphetamine for sale" or\
descript == "possession of marijuana for sales" or\
descript == "ppossession of heroin for sales" or\
descript == "sale of base/rock cocaine" or\
descript == "sale of marijuana" or\
descript == "loitering where narcotics are sold/used" or\
descript == "possession of cocaine for sales"').groupby('census_tract').count()['descript']
sf['drugDensity_poss_drug'] = sf['tractce10'].map(possession_of_drug).fillna(0)
sf['drugDensity_sale_drug'] = sf['tractce10'].map(sale_of_drug).fillna(0)
sf_drug_location = sf[['drugDensity', 'drugDensity_poss_drug', 'drugDensity_sale_drug']]
import sklearn.cluster
import sklearn.preprocessing
cl = sklearn.cluster.KMeans(n_clusters=3)
X = sklearn.preprocessing.scale(sf_drug_location.values)
classes = cl.fit(X)
f = plt.figure(figsize=(10, 10))
ax = f.gca()
kw = dict(column='cluster', k=3, cmap='Reds', alpha=1, legend=True, edgecolor='gray', linewidth=0.5, categorical=True)
sf.assign(cluster=classes.labels_).plot(ax=ax, **kw)
ax.set_axis_off()
sf_drug_location.assign(cluster=classes.labels_).groupby('cluster').mean()
from mpl_toolkits.mplot3d import Axes3D
fig = plt.figure()
ax = fig.add_subplot(111, projection='3d')
ax.scatter(sf_drug_location['drugDensity'], sf_drug_location['drugDensity_poss_drug'],
zs=sf_drug_location['drugDensity_sale_drug'], c=classes.labels_, cmap='Reds')
cl = ps.region.maxp.Maxp(qW, sf_drug_location.values, 30, sf['drug'].values[:,None])
cl.
f = plt.figure(figsize=(10, 10))
ax = f.gca()
kw = dict(column='cluster', k=10, cmap='Reds', alpha=1, legend=True, edgecolor='gray', linewidth=0.5, categorical=True)
sf.assign(cluster=cl).plot(ax=ax, **kw)
ax.set_axis_off()
sf_drug_location.assign(cluster=cl.area2region.values()).groupby('cluster').mean()<jupyter_output><empty_output><jupyter_text>## load or save work<jupyter_code>with open('../final_dataset/crime2016_origin_modified_col.pkl') as handle:
crime2016 = pickle.load(handle)
crime2016['category'].value_counts().head(30)
crime2016['descript'].value_counts().head(30)
[col for col in crime2016.columns]
## save stabbing one
#aggravated assault with a deadly weapon, aggravated assault with a knife
#aggravated assault with a gun
stabbing = crime2016[(crime2016.descript == 'aggravated assault with a deadly weapon') | \
(crime2016.descript == 'aggravated assault with a knife') | \
(crime2016.descript == '#aggravated assault with a gun')]
with open('../final_dataset/stabbing.pkl', 'wb') as handle:
pickle.dump(stabbing, handle)
stabbing2 = crime2016[crime2016.descript == 'aggravated assault with a knife']
with open('../final_dataset/stabbing2.pkl', 'wb') as handle:
pickle.dump(stabbing2, handle)<jupyter_output><empty_output>
|
no_license
|
/final_report/map_geo.ipynb
|
sitang/all_projects
| 10 |
<jupyter_start><jupyter_text># Style Transfer
In this notebook we will implement the style transfer technique from ["Image Style Transfer Using Convolutional Neural Networks" (Gatys et al., CVPR 2015)](http://www.cv-foundation.org/openaccess/content_cvpr_2016/papers/Gatys_Image_Style_Transfer_CVPR_2016_paper.pdf).
The general idea is to take two images, and produce a new image that reflects the content of one but the artistic "style" of the other. We will do this by first formulating a loss function that matches the content and style of each respective image in the feature space of a deep network, and then performing gradient descent on the pixels of the image itself.
The deep network we use as a feature extractor is [SqueezeNet](https://arxiv.org/abs/1602.07360), a small model that has been trained on ImageNet. You could use any network, but we chose SqueezeNet here for its small size and efficiency.
Here's an example of the images you'll be able to produce by the end of this notebook:

## Setup<jupyter_code>
%load_ext autoreload
%autoreload 2
from scipy.misc import imread, imresize
import os
import numpy as np
from scipy.misc import imread
import matplotlib.pyplot as plt
import tensorflow as tf
# Helper functions to deal with image preprocessing
from cs231n.image_utils import load_image, preprocess_image, deprocess_image
%matplotlib inline
def get_session():
"""Create a session that dynamically allocates memory."""
# See: https://www.tensorflow.org/tutorials/using_gpu#allowing_gpu_memory_growth
config = tf.ConfigProto()
config.gpu_options.allow_growth = True
session = tf.Session(config=config)
return session
def rel_error(x,y):
return np.max(np.abs(x - y) / (np.maximum(1e-8, np.abs(x) + np.abs(y))))
# Older versions of scipy.misc.imresize yield different results
# from newer versions, so we check to make sure scipy is up to date.
def check_scipy():
import scipy
vnum = int(scipy.__version__.split('.')[1])
assert vnum >= 16, "You must install SciPy >= 0.16.0 to complete this notebook."
check_scipy()<jupyter_output><empty_output><jupyter_text>Load the pretrained SqueezeNet model. This model has been ported from PyTorch, see `cs231n/classifiers/squeezenet.py` for the model architecture.
To use SqueezeNet, you will need to first **download the weights** by changing into the `cs231n/datasets` directory and running `get_squeezenet_tf.sh` . Note that if you ran `get_assignment3_data.sh` then SqueezeNet will already be downloaded.<jupyter_code>from cs231n.classifiers.squeezenet import SqueezeNet
import tensorflow as tf
tf.reset_default_graph() # remove all existing variables in the graph
sess = get_session() # start a new Session
# Load pretrained SqueezeNet model
SAVE_PATH = 'cs231n/datasets/squeezenet.ckpt'
if not os.path.exists(SAVE_PATH):
raise ValueError("You need to download SqueezeNet!")
model = SqueezeNet(save_path=SAVE_PATH, sess=sess)
# Load data for testing
content_img_test = preprocess_image(load_image('styles/tubingen.jpg', size=192))[None]
style_img_test = preprocess_image(load_image('styles/starry_night.jpg', size=192))[None]
answers = np.load('style-transfer-checks-tf.npz')
<jupyter_output>INFO:tensorflow:Restoring parameters from cs231n/datasets/squeezenet.ckpt
<jupyter_text>## Computing Loss
We're going to compute the three components of our loss function now. The loss function is a weighted sum of three terms: content loss + style loss + total variation loss. You'll fill in the functions that compute these weighted terms below.## Content loss
We can generate an image that reflects the content of one image and the style of another by incorporating both in our loss function. We want to penalize deviations from the content of the content image and deviations from the style of the style image. We can then use this hybrid loss function to perform gradient descent **not on the parameters** of the model, but instead **on the pixel values** of our original image.
Let's first write the content loss function. Content loss measures how much the feature map of the generated image differs from the feature map of the source image. We only care about the content representation of one layer of the network (say, layer $\ell$), that has feature maps $A^\ell \in \mathbb{R}^{1 \times C_\ell \times H_\ell \times W_\ell}$. $C_\ell$ is the number of filters/channels in layer $\ell$, $H_\ell$ and $W_\ell$ are the height and width. We will work with reshaped versions of these feature maps that combine all spatial positions into one dimension. Let $F^\ell \in \mathbb{R}^{N_\ell \times M_\ell}$ be the feature map for the current image and $P^\ell \in \mathbb{R}^{N_\ell \times M_\ell}$ be the feature map for the content source image where $M_\ell=H_\ell\times W_\ell$ is the number of elements in each feature map. Each row of $F^\ell$ or $P^\ell$ represents the vectorized activations of a particular filter, convolved over all positions of the image. Finally, let $w_c$ be the weight of the content loss term in the loss function.
Then the content loss is given by:
$L_c = w_c \times \sum_{i,j} (F_{ij}^{\ell} - P_{ij}^{\ell})^2$<jupyter_code>def content_loss(content_weight, content_current, content_original):
"""
Compute the content loss for style transfer.
Inputs:
- content_weight: scalar constant we multiply the content_loss by.
- content_current: features of the current image, Tensor with shape [1, height, width, channels]
- content_target: features of the content image, Tensor with shape [1, height, width, channels]
Returns:
- scalar content loss
"""
shape = tf.shape(content_current)
content_current = tf.transpose(content_current,perm=[3,1,2,0])
content_current = tf.reshape(content_current,[shape[0]*shape[3],shape[1]*shape[2]])
content_original = tf.transpose(content_original,perm=[3,1,2,0])
content_original = tf.reshape(content_original,[shape[0]*shape[3],shape[1]*shape[2]])
loss = tf.reduce_sum(content_weight*tf.squared_difference(content_current,content_original))
return loss
pass
<jupyter_output><empty_output><jupyter_text>Test your content loss. You should see errors less than 0.001.<jupyter_code>def content_loss_test(correct):
content_layer = 3
content_weight = 6e-2
c_feats = sess.run(model.extract_features()[content_layer], {model.image: content_img_test})
bad_img = tf.zeros(content_img_test.shape)
feats = model.extract_features(bad_img)[content_layer]
student_output = sess.run(content_loss(content_weight, c_feats, feats))
error = rel_error(correct, student_output)
print('Maximum error is {:.3f}'.format(error))
content_loss_test(answers['cl_out'])<jupyter_output>Maximum error is 0.000
<jupyter_text>## Style loss
Now we can tackle the style loss. For a given layer $\ell$, the style loss is defined as follows:
First, compute the Gram matrix G which represents the correlations between the responses of each filter, where F is as above. The Gram matrix is an approximation to the covariance matrix -- we want the activation statistics of our generated image to match the activation statistics of our style image, and matching the (approximate) covariance is one way to do that. There are a variety of ways you could do this, but the Gram matrix is nice because it's easy to compute and in practice shows good results.
Given a feature map $F^\ell$ of shape $(1, C_\ell, M_\ell)$, the Gram matrix has shape $(1, C_\ell, C_\ell)$ and its elements are given by:
$$G_{ij}^\ell = \sum_k F^{\ell}_{ik} F^{\ell}_{jk}$$
Assuming $G^\ell$ is the Gram matrix from the feature map of the current image, $A^\ell$ is the Gram Matrix from the feature map of the source style image, and $w_\ell$ a scalar weight term, then the style loss for the layer $\ell$ is simply the weighted Euclidean distance between the two Gram matrices:
$$L_s^\ell = w_\ell \sum_{i, j} \left(G^\ell_{ij} - A^\ell_{ij}\right)^2$$
In practice we usually compute the style loss at a set of layers $\mathcal{L}$ rather than just a single layer $\ell$; then the total style loss is the sum of style losses at each layer:
$$L_s = \sum_{\ell \in \mathcal{L}} L_s^\ell$$
Begin by implementing the Gram matrix computation below:<jupyter_code>def gram_matrix(features, normalize=True):
"""
Compute the Gram matrix from features.
Inputs:
- features: Tensor of shape (1, H, W, C) giving features for
a single image.
- normalize: optional, whether to normalize the Gram matrix
If True, divide the Gram matrix by the number of neurons (H * W * C)
Returns:
- gram: Tensor of shape (C, C) giving the (optionally normalized)
Gram matrices for the input image.
"""
f = tf.transpose(features,perm=[0,3,1,2])
out = tf.shape(f)
f = tf.reshape(f,shape=[out[0]*out[1],out[2]*out[3]])
G = tf.matmul(f,tf.transpose(f))
if normalize:
G = tf.divide(G,tf.cast(out[1]*out[2]*out[3],dtype=tf.float32))
return G
pass
<jupyter_output><empty_output><jupyter_text>Test your Gram matrix code. You should see errors less than 0.001.<jupyter_code>def gram_matrix_test(correct):
gram = gram_matrix(model.extract_features()[5])
student_output = sess.run(gram, {model.image: style_img_test})
error = rel_error(correct, student_output)
print('Maximum error is {:.3f}'.format(error))
gram_matrix_test(answers['gm_out'])<jupyter_output>Maximum error is 0.000
<jupyter_text>Next, implement the style loss:<jupyter_code>def style_loss(feats, style_layers, style_targets, style_weights):
"""
Computes the style loss at a set of layers.
Inputs:
- feats: list of the features at every layer of the current image, as produced by
the extract_features function.
- style_layers: List of layer indices into feats giving the layers to include in the
style loss.
- style_targets: List of the same length as style_layers, where style_targets[i] is
a Tensor giving the Gram matrix the source style image computed at
layer style_layers[i].
- style_weights: List of the same length as style_layers, where style_weights[i]
is a scalar giving the weight for the style loss at layer style_layers[i].
Returns:
- style_loss: A Tensor contataining the scalar style loss.
"""
# Hint: you can do this with one for loop over the style layers, and should
# not be very much code (~5 lines). You will need to use your gram_matrix function.
s_loss = tf.zeros(shape=[1])
for i in range(len(style_layers)):
l = style_layers[i]
gram = gram_matrix(feats[l])
loss = style_weights[i]*tf.reduce_sum(tf.squared_difference(gram,style_targets[i]))
s_loss = s_loss + loss
return s_loss
<jupyter_output><empty_output><jupyter_text>Test your style loss implementation. The error should be less than 0.001.<jupyter_code>def style_loss_test(correct):
style_layers = [1, 4, 6, 7]
style_weights = [300000, 1000, 15, 3]
feats = model.extract_features()
style_target_vars = []
for idx in style_layers:
style_target_vars.append(gram_matrix(feats[idx]))
style_targets = sess.run(style_target_vars,
{model.image: style_img_test})
s_loss = style_loss(feats, style_layers, style_targets, style_weights)
student_output = sess.run(s_loss, {model.image: content_img_test})
error = rel_error(correct, student_output)
print('Error is {:.3f}'.format(error))
style_loss_test(answers['sl_out'])<jupyter_output>Error is 0.000
<jupyter_text>## Total-variation regularization
It turns out that it's helpful to also encourage smoothness in the image. We can do this by adding another term to our loss that penalizes wiggles or "total variation" in the pixel values.
You can compute the "total variation" as the sum of the squares of differences in the pixel values for all pairs of pixels that are next to each other (horizontally or vertically). Here we sum the total-variation regualarization for each of the 3 input channels (RGB), and weight the total summed loss by the total variation weight, $w_t$:
$L_{tv} = w_t \times \sum_{c=1}^3\sum_{i=1}^{H-1} \sum_{j=1}^{W-1} \left( (x_{i,j+1, c} - x_{i,j,c})^2 + (x_{i+1, j,c} - x_{i,j,c})^2 \right)$
In the next cell, fill in the definition for the TV loss term. To receive full credit, your implementation should not have any loops.<jupyter_code>def tv_loss(img, tv_weight):
"""
Compute total variation loss.
Inputs:
- img: Tensor of shape (1, H, W, 3) holding an input image.
- tv_weight: Scalar giving the weight w_t to use for the TV loss.
Returns:
- loss: Tensor holding a scalar giving the total variation loss
for img weighted by tv_weight.
"""
# Your implementation should be vectorized and not require any loops!
out = tf.shape(img)
loss1 = tf.reduce_sum(tf.squared_difference(img[:,0:out[1]-1,:,:],img[:,1:,:,:]))
loss2 = tf.reduce_sum(tf.squared_difference(img[:,:,0:out[2]-1,:],img[:,:,1:,:]))
loss = tv_weight*(loss1 + loss2)
return loss
<jupyter_output><empty_output><jupyter_text>Test your TV loss implementation. Error should be less than 0.001.<jupyter_code>def tv_loss_test(correct):
tv_weight = 2e-2
t_loss = tv_loss(model.image, tv_weight)
student_output = sess.run(t_loss, {model.image: content_img_test})
error = rel_error(correct, student_output)
print('Error is {:.3f}'.format(error))
tv_loss_test(answers['tv_out'])<jupyter_output>Error is 0.000
<jupyter_text>## Style TransferLets put it all together and make some beautiful images! The `style_transfer` function below combines all the losses you coded up above and optimizes for an image that minimizes the total loss.<jupyter_code>def style_transfer(content_image, style_image, image_size, style_size, content_layer, content_weight,
style_layers, style_weights, tv_weight, init_random = False):
"""Run style transfer!
Inputs:
- content_image: filename of content image
- style_image: filename of style image
- image_size: size of smallest image dimension (used for content loss and generated image)
- style_size: size of smallest style image dimension
- content_layer: layer to use for content loss
- content_weight: weighting on content loss
- style_layers: list of layers to use for style loss
- style_weights: list of weights to use for each layer in style_layers
- tv_weight: weight of total variation regularization term
- init_random: initialize the starting image to uniform random noise
"""
# Extract features from the content image
content_img = preprocess_image(load_image(content_image, size=image_size))
feats = model.extract_features(model.image)
content_target = sess.run(feats[content_layer],
{model.image: content_img[None]})
# Extract features from the style image
style_img = preprocess_image(load_image(style_image, size=style_size))
style_feat_vars = [feats[idx] for idx in style_layers]
style_target_vars = []
# Compute list of TensorFlow Gram matrices
for style_feat_var in style_feat_vars:
style_target_vars.append(gram_matrix(style_feat_var))
# Compute list of NumPy Gram matrices by evaluating the TensorFlow graph on the style image
style_targets = sess.run(style_target_vars, {model.image: style_img[None]})
# Initialize generated image to content image
if init_random:
img_var = tf.Variable(tf.random_uniform(content_img[None].shape, 0, 1), name="image")
else:
img_var = tf.Variable(content_img[None], name="image")
# Extract features on generated image
feats = model.extract_features(img_var)
# Compute loss
c_loss = content_loss(content_weight, feats[content_layer], content_target)
s_loss = style_loss(feats, style_layers, style_targets, style_weights)
t_loss = tv_loss(img_var, tv_weight)
loss = c_loss + s_loss + t_loss
# Set up optimization hyperparameters
initial_lr = 3.0
decayed_lr = 0.1
decay_lr_at = 180
max_iter = 1000
# Create and initialize the Adam optimizer
lr_var = tf.Variable(initial_lr, name="lr")
# Create train_op that updates the generated image when run
with tf.variable_scope("optimizer") as opt_scope:
train_op = tf.train.AdamOptimizer(lr_var).minimize(loss, var_list=[img_var])
# Initialize the generated image and optimization variables
opt_vars = tf.get_collection(tf.GraphKeys.GLOBAL_VARIABLES, scope=opt_scope.name)
sess.run(tf.variables_initializer([lr_var, img_var] + opt_vars))
# Create an op that will clamp the image values when run
clamp_image_op = tf.assign(img_var, tf.clip_by_value(img_var, -1.5, 1.5))
f, axarr = plt.subplots(1,2)
axarr[0].axis('off')
axarr[1].axis('off')
axarr[0].set_title('Content Source Img.')
axarr[1].set_title('Style Source Img.')
axarr[0].imshow(deprocess_image(content_img))
axarr[1].imshow(deprocess_image(style_img))
plt.show()
plt.figure()
# Hardcoded handcrafted
for t in range(max_iter):
# Take an optimization step to update img_var
sess.run(train_op)
if t < decay_lr_at:
sess.run(clamp_image_op)
if t == decay_lr_at:
sess.run(tf.assign(lr_var, decayed_lr))
if t % 100 == 0:
print('Iteration {}'.format(t))
img = sess.run(img_var)
plt.imshow(deprocess_image(img[0], rescale=True))
plt.axis('off')
plt.show()
print('Iteration {}'.format(t))
img = sess.run(img_var)
plt.imshow(deprocess_image(img[0], rescale=True))
plt.axis('off')
plt.show()<jupyter_output><empty_output><jupyter_text>## Generate some pretty pictures!
Try out `style_transfer` on the three different parameter sets below. Make sure to run all three cells. Feel free to add your own, but make sure to include the results of style transfer on the third parameter set (starry night) in your submitted notebook.
* The `content_image` is the filename of content image.
* The `style_image` is the filename of style image.
* The `image_size` is the size of smallest image dimension of the content image (used for content loss and generated image).
* The `style_size` is the size of smallest style image dimension.
* The `content_layer` specifies which layer to use for content loss.
* The `content_weight` gives weighting on content loss in the overall loss function. Increasing the value of this parameter will make the final image look more realistic (closer to the original content).
* `style_layers` specifies a list of which layers to use for style loss.
* `style_weights` specifies a list of weights to use for each layer in style_layers (each of which will contribute a term to the overall style loss). We generally use higher weights for the earlier style layers because they describe more local/smaller scale features, which are more important to texture than features over larger receptive fields. In general, increasing these weights will make the resulting image look less like the original content and more distorted towards the appearance of the style image.
* `tv_weight` specifies the weighting of total variation regularization in the overall loss function. Increasing this value makes the resulting image look smoother and less jagged, at the cost of lower fidelity to style and content.
Below the next three cells of code (in which you shouldn't change the hyperparameters), feel free to copy and paste the parameters to play around them and see how the resulting image changes. <jupyter_code># Composition VII + Tubingen
params1 = {
'content_image' : 'styles/tubingen.jpg',
'style_image' : 'styles/composition_vii.jpg',
'image_size' : 192,
'style_size' : 512,
'content_layer' : 3,
'content_weight' : 5e-2,
'style_layers' : (1, 4, 6, 7),
'style_weights' : (20000, 500, 12, 1),
'tv_weight' : 5e-2
}
style_transfer(**params1)
# Scream + Tubingen
params2 = {
'content_image':'styles/tubingen.jpg',
'style_image':'styles/the_scream.jpg',
'image_size':192,
'style_size':224,
'content_layer':3,
'content_weight':3e-2,
'style_layers':[1, 4, 6, 7],
'style_weights':[200000, 800, 12, 1],
'tv_weight':2e-2
}
style_transfer(**params2)
# Starry Night + Tubingen
params3 = {
'content_image' : 'styles/pixel2.jpg',
'style_image' : 'styles/starry_night.jpg',
'image_size' : 192,
'style_size' : 192,
'content_layer' : 3,
'content_weight' : 6e-2,
'style_layers' : [1, 4, 6, 7],
'style_weights' : [300000, 1000, 15, 3],
'tv_weight' : 2e-2
}
style_transfer(**params3)<jupyter_output><empty_output><jupyter_text>## Feature Inversion
The code you've written can do another cool thing. In an attempt to understand the types of features that convolutional networks learn to recognize, a recent paper [1] attempts to reconstruct an image from its feature representation. We can easily implement this idea using image gradients from the pretrained network, which is exactly what we did above (but with two different feature representations).
Now, if you set the style weights to all be 0 and initialize the starting image to random noise instead of the content source image, you'll reconstruct an image from the feature representation of the content source image. You're starting with total noise, but you should end up with something that looks quite a bit like your original image.
(Similarly, you could do "texture synthesis" from scratch if you set the content weight to 0 and initialize the starting image to random noise, but we won't ask you to do that here.)
[1] Aravindh Mahendran, Andrea Vedaldi, "Understanding Deep Image Representations by Inverting them", CVPR 2015
<jupyter_code># Feature Inversion -- Starry Night + Tubingen
params_inv = {
'content_image' : 'styles/tubingen.jpg',
'style_image' : 'styles/starry_night.jpg',
'image_size' : 192,
'style_size' : 192,
'content_layer' : 3,
'content_weight' : 6e-2,
'style_layers' : [1, 4, 6, 7],
'style_weights' : [0, 0, 0, 0], # we discard any contributions from style to the loss
'tv_weight' : 2e-2,
'init_random': True # we want to initialize our image to be random
}
style_transfer(**params_inv)
# Texture Synthesis -- Starry Night + Tubingen
params_inv = {
'content_image' : 'styles/tubingen.jpg',
'style_image' : 'styles/starry_night.jpg',
'image_size' : 192,
'style_size' : 192,
'content_layer' : 3,
'content_weight' : 0,
'style_layers' : [1, 4, 6, 7],# we discard any contributions from content to the loss
'style_weights' : [3000000, 1000, 15, 3],
'tv_weight' : 2e-2,
'init_random': True # we want to initialize our image to be random
}
style_transfer(**params_inv)<jupyter_output><empty_output>
|
permissive
|
/CS231n-master/assignment3/StyleTransfer-TensorFlow.ipynb
|
saiamrit/CS231n
| 13 |
<jupyter_start><jupyter_text># 作業 : (Kaggle)鐵達尼生存預測# [作業目標]
- 試著模仿範例寫法, 在鐵達尼生存預測中, 使用葉編碼並觀察預測效果# [作業重點]
- 仿造範例, 完成葉編碼的寫作 : 使用隨機森林 (In[3], Out[3], In[4], Out[4])
- 仿造範例, 觀察葉編碼搭配邏輯斯迴歸後的效果 (In[5], Out[5], In[6], Out[6]) <jupyter_code># 做完特徵工程前的所有準備
import pandas as pd
import numpy as np
import copy
from sklearn.preprocessing import LabelEncoder, MinMaxScaler
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import RandomForestClassifier
# 因為擬合(fit)與編碼(transform)需要分開, 因此不使用.get_dummy, 而採用 sklearn 的 OneHotEncoder
from sklearn.preprocessing import OneHotEncoder
from sklearn.model_selection import train_test_split
from sklearn.metrics import roc_curve
data_path = 'data/'
df = pd.read_csv(data_path + 'titanic_train.csv')
train_Y = df['Survived']
df = df.drop(['PassengerId', 'Survived'] , axis=1)
df.head()
# 因為需要把類別型與數值型特徵都加入, 故使用最簡版的特徵工程
LEncoder = LabelEncoder()
MMEncoder = MinMaxScaler()
for c in df.columns:
df[c] = df[c].fillna(-1)
if df[c].dtype == 'object':
df[c] = LEncoder.fit_transform(list(df[c].values))
df[c] = MMEncoder.fit_transform(df[c].values.reshape(-1, 1))
df.head()
train_X = df.values
# 因為訓練邏輯斯迴歸時也要資料, 因此將訓練及切成三部分 train / val / test, 採用 test 驗證而非 k-fold 交叉驗證
# train 用來訓練梯度提升樹, val 用來訓練邏輯斯迴歸, test 驗證效果
train_X, test_X, train_Y, test_Y = train_test_split(train_X, train_Y, test_size=0.5)
train_X, val_X, train_Y, val_Y = train_test_split(train_X, train_Y, test_size=0.5)<jupyter_output><empty_output><jupyter_text># 作業1
* 請對照範例,完成隨機森林的鐵達尼生存率預測,以及對應的葉編碼+邏輯斯迴歸<jupyter_code># 隨機森林擬合後, 再將葉編碼 (*.apply) 結果做獨熱 / 邏輯斯迴歸
rf = RandomForestClassifier(n_estimators=20, min_samples_split=10, min_samples_leaf=5,
max_features=4, max_depth=3, bootstrap=True)
onehot = OneHotEncoder()
lr = LogisticRegression(solver='lbfgs', max_iter=1000)
"""
Your Code Here (Hint : 隨機森林的葉編碼(.apply)不需要加上[:, :, 0], 直接用rf.apply()調用即可, 本作業其餘寫法相同)
"""
rf.fit(train_X, train_Y)
onehot.fit(rf.apply(train_X))
lr.fit(onehot.transform(rf.apply(val_X)), val_Y)
# 將隨機森林+葉編碼+邏輯斯迴歸結果輸出
"""
Your Code Here
"""
pred_rf_lr = lr.predict_proba(onehot.transform(rf.apply(test_X)))[:, 1]
fpr_rf_lr, tpr_rf_lr, _ = roc_curve(test_Y, pred_rf_lr)
# 將隨機森林結果輸出
"""
Your Code Here
"""
pred_rf = rf.predict_proba(test_X)[:, 1]
fpr_rf, tpr_rf, _ = roc_curve(test_Y, pred_rf)<jupyter_output><empty_output><jupyter_text># 作業2
* 上述的結果,葉編碼是否有提高預測的正確性呢?
> * Ans: 看起來 加上 LR 的正確率有提升
>
> 下面是比較 ROC 曲線, 而根據說明, ROC 曲線對 X 軸之積分值越高代表預測準確率越高
>
> 由於積分很難計算, 改計算 $\sum\limits_{i = 0}^{n - 1}(x_{i + 1} - x_{i})\frac{(y_{i + 1} + y_{i})}{2}$ 代替.
> 也就是相鄰兩點的平均值乘上相鄰兩點之差代替相鄰兩點之間之積分<jupyter_code>import matplotlib.pyplot as plt
# 將結果繪圖
"""
Your Code Here
"""
plt.plot([0, 1], [0, 1], 'k--')
plt.plot(fpr_rf, tpr_rf, label='RF')
plt.plot(fpr_rf_lr, tpr_rf_lr, label='RF + LR')
plt.xlabel('False positive rate')
plt.ylabel('True positive rate')
plt.title('ROC curve')
plt.legend(loc='best')
plt.show()
def integral(x, y):
dist = x[1:] - x[:-1]
mul = dist * (y[1:] + y[:-1]) / 2
return np.sum(mul)
print(integral(fpr_rf_lr, tpr_rf_lr) - integral(fpr_rf, tpr_rf))<jupyter_output>0.013296041308089479
|
no_license
|
/homework/Day_032_HW.ipynb
|
clmikechan/4th-ML100Days
| 3 |
<jupyter_start><jupyter_text># IntroductionIPython, pandas and matplotlib have a number of useful options you can use to make it easier to view and format your data. This notebook collects a bunch of them in one place. I hope this will be a useful reference.
The original blog posting is on http://pbpython.com/ipython-pandas-display-tips.html## Import modules and some sample dataFirst, do our standard pandas, numpy and matplotlib imports as well as configure inline displays of plots.<jupyter_code>import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline<jupyter_output><empty_output><jupyter_text>One of the simple things we can do is override the default CSS to customize our DataFrame output.
This specific example is from - [Brandon Rhodes' talk at pycon](https://www.youtube.com/watch?v=5JnMutdy6Fw "Pandas From The Ground Up")For the purposes of the notebook, I'm defining CSS as a variable but you could easily read in from a file as well.<jupyter_code>CSS = """
body {
margin: 0;
font-family: Helvetica;
}
table.dataframe {
border-collapse: collapse;
border: none;
}
table.dataframe tr {
border: none;
}
table.dataframe td, table.dataframe th {
margin: 0;
border: 1px solid white;
padding-left: 0.25em;
padding-right: 0.25em;
}
table.dataframe th:not(:empty) {
background-color: #fec;
text-align: left;
font-weight: normal;
}
table.dataframe tr:nth-child(2) th:empty {
border-left: none;
border-right: 1px dashed #888;
}
table.dataframe td {
border: 2px solid #ccf;
background-color: #f4f4ff;
}
"""<jupyter_output><empty_output><jupyter_text>Now add this CSS into the current notebook's HTML.<jupyter_code>from IPython.core.display import HTML
HTML('<style>{}</style>'.format(CSS))
SALES=pd.read_csv("../data/sample-sales-tax.csv", parse_dates='True')
SALES.head()<jupyter_output><empty_output><jupyter_text>You can see how the CSS is now applied to the DataFrame and how you could easily modify it to customize it to your liking.
Jupyter notebooks do a good job of automatically displaying information but sometimes you want to force data to display. Fortunately, ipython provides and option. This is especially useful if you want to display multiple dataframes.<jupyter_code>from IPython.display import display
display(SALES.head(2))
display(SALES.tail(2))
display(SALES.describe())<jupyter_output><empty_output><jupyter_text>## Using pandas settings to control outputPandas has many different options to control how data is displayed.
You can use max_rows to control how many rows are displayed<jupyter_code>pd.set_option("display.max_rows",4)
SALES<jupyter_output><empty_output><jupyter_text>Depending on the data set, you may only want to display a smaller number of columns.<jupyter_code>pd.set_option("display.max_columns",6)
SALES<jupyter_output><empty_output><jupyter_text>You can control how many decimal points of precision to display<jupyter_code>pd.set_option('precision',2)
SALES
pd.set_option('precision',7)
SALES<jupyter_output><empty_output><jupyter_text>You can also format floating point numbers using float_format<jupyter_code>pd.set_option('float_format', '{:.2f}'.format)
SALES<jupyter_output><empty_output><jupyter_text>This does apply to all the data. In our example, applying dollar signs to everything would not be correct for this example.<jupyter_code>pd.set_option('float_format', '${:.2f}'.format)
SALES<jupyter_output><empty_output><jupyter_text>## Third Party PluginsQtopian has a useful plugin called qgrid - https://github.com/quantopian/qgrid
Import it and install it.<jupyter_code>import qgrid
qgrid.nbinstall()<jupyter_output><empty_output><jupyter_text>Showing the data is straighforward.<jupyter_code>qgrid.show_grid(SALES, remote_js=True)<jupyter_output><empty_output><jupyter_text>The plugin is very similar to the capability of an Excel autofilter. It can be handy to quickly filter and sort your data.## Improving your plotsI have mentioned before how the default pandas plots don't look so great. Fortunately, there are style sheets in matplotlib which go a long way towards improving the visualization of your data.
Here is a simple plot with the default values.<jupyter_code>SALES.groupby('name')['quantity'].sum().plot(kind="bar")<jupyter_output><empty_output><jupyter_text>We can use some of the matplolib styles available to us to make this look better.
http://matplotlib.org/users/style_sheets.html<jupyter_code>plt.style.use('ggplot')
SALES.groupby('name')['quantity'].sum().plot(kind="bar")<jupyter_output><empty_output><jupyter_text>You can see all the styles available<jupyter_code>plt.style.available
plt.style.use('bmh')
SALES.groupby('name')['quantity'].sum().plot(kind="bar")
plt.style.use('fivethirtyeight')
SALES.groupby('name')['quantity'].sum().plot(kind="bar")<jupyter_output><empty_output>
|
permissive
|
/notebooks/Ipython-pandas-tips-and-tricks.ipynb
|
blackzw/pbpython
| 14 |
<jupyter_start><jupyter_text>## GPSMap<jupyter_code>point_x_node = pd.read_csv("data/w.tsv", sep=" ", index_col=0)
point_ = point_x_node.index.to_numpy()
node_ = point_x_node.columns.to_numpy()
point_name = point_x_node.index.name
point_x_node = point_x_node.to_numpy()
node_name = "Node"
node_x_node = squareform(pdist(point_x_node.T))
gps_map = kraft.gps_map.GPSMap(
node_name,
node_,
node_x_node,
point_name,
point_,
point_x_node,
)
gps_map.plot()
point_highlight_ = np.asarray(("Feature 6", "Feature 10", "Feature 29"))
for point in point_highlight_:
print(point)
print(gps_map.point_x_dimension[gps_map.point_ == point])
gps_map.plot(point_highlight_=point_highlight_)
score_name = "Dimension Sum"
score_ = gps_map.point_x_dimension.sum(axis=1)
gps_map.plot(score_=score_)
score_[::2] = np.nan
gps_map.plot(score_name=score_name, score_=score_, score_nan_opacity=0.4)
gps_map.set_group("closest_node")
gps_map.plot()
gps_map.set_group(gps_map.point_x_dimension.argmax(axis=1))
gps_map.plot()
gps_map.plot(score_=score_)
new_point_ = point_highlight_
new_point_x_node = np.concatenate(
[point_x_node[point_ == point] for point in point_highlight_]
)
new_point_x_node
gps_map.predict(
"New", new_point_, new_point_x_node, point_trace={"marker": {"size": 32}}
)
node_x_point = pd.read_csv("data/kras_gps_map.h.tsv", sep=" ", index_col=0)
node_ = node_x_point.index.to_numpy()
point_ = node_x_point.columns.to_numpy()
node_name = node_x_point.index.name
point_name = "Cell Line"
point_x_node = node_x_point.to_numpy().T
point_x_node = np.apply_along_axis(kraft.array.normalize, 0, point_x_node, "-0-")
point_x_node = point_x_node.clip(min=-3, max=3)
point_x_node = np.apply_along_axis(kraft.array.normalize, 0, point_x_node, "0-1")
point_x_node **= 2
node_x_node = squareform(pdist(point_x_node.T))
gps_map = kraft.gps_map.GPSMap(
node_name, node_, node_x_node, point_name, point_, point_x_node
)
group_ = (
pd.read_csv("data/kras_gps_map.k_x_h_point.tsv", sep=" ", index_col=0)
.loc["K15", :]
.to_numpy()
)
gps_map.set_group(group_)
gps_map.plot()
every_n = 10
gps_map.predict("New", point_[::every_n], point_x_node[::every_n])
path = os.path.expanduser("~/Downloads/gps_map.pickle.gz")<jupyter_output><empty_output><jupyter_text>## write<jupyter_code>kraft.gps_map.write(path, gps_map)<jupyter_output><empty_output><jupyter_text>## read<jupyter_code>gps_map = kraft.gps_map.read(path)
gps_map.plot()<jupyter_output><empty_output>
|
permissive
|
/notebook/gps_map.ipynb
|
KwatME/ccal
| 3 |
<jupyter_start><jupyter_text># Modeling - Naive Bayes (after minimal data cleaning)<jupyter_code>import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import spacy
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.linear_model import LogisticRegression
from sklearn.pipeline import make_pipeline
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split, GridSearchCV
from sklearn.naive_bayes import MultinomialNB
from sklearn.metrics import plot_confusion_matrix, balanced_accuracy_score
#read in data
postings = pd.read_csv('../datasets/postings_minimally_cleaned.csv')
postings
#define X and y
X = postings['text']
y = np.where(postings['topic'] == 'hiking',1,0)
#checking X
X
#checking y
y
sum(y) #this should be 6986 (number of hiking rows) -- confirmed!<jupyter_output><empty_output><jupyter_text>### Train, test, split<jupyter_code>X_train, X_test, y_train, y_test = train_test_split(X,y, stratify=y, random_state=123)
def input_to_df(model, cleaning, lemmatized):
"""
Appends model information into model summary dataframe for comparison
"""
df = pd.read_csv('../datasets/model_list_summary.csv')
if lemmatized == 'no':
info = [cleaning, #Indicates if data was minimally or deeply cleaned
lemmatized, #Indicates if text was lemmatized or not
model.score(X_train,y_train),
model.score(X_test, y_test),
balanced_accuracy_score(y_train, model.predict(X_train)),
balanced_accuracy_score(y_test, model.predict(X_test)),
model.best_params_
]
elif lemmatized == 'yes':
info = [cleaning, #Indicates if data was minimally or deeply cleaned
lemmatized, #Indicates if text was lemmatized or not
model.score(X_train_lem,y_train),
model.score(X_test_lem, y_test),
balanced_accuracy_score(y_train, model.predict(X_train_lem)),
balanced_accuracy_score(y_test, model.predict(X_test_lem)),
model.best_params_
]
info_series = pd.Series(info, index=df.columns)
df = df.append(info_series, ignore_index=True)
df.to_csv('../datasets/model_list_summary.csv', index=False)<jupyter_output><empty_output><jupyter_text>### Building my pipe and GS<jupyter_code>def model_fit_and_scores(model):
model.fit(X_train,y_train)
print(f' Score on train data: {model.score(X_train,y_train)}')
print(f' Score on test data: {model.score(X_test, y_test)}')
print(f' balanced accuracy score on train data: {balanced_accuracy_score(y_train, model.predict(X_train))}')
print(f' balanced accuracy score on test data: {balanced_accuracy_score(y_test, model.predict(X_test))}')
print(model.best_params_)
pipe = make_pipeline(CountVectorizer(),
MultinomialNB())
params = {
'countvectorizer__stop_words': [None, 'english'],
'countvectorizer__max_features':[500,4000,6000],
'countvectorizer__ngram_range':[(1,1), (2,2)],
'multinomialnb__alpha':[.005,.1, 1, 10]
}
gs = GridSearchCV(pipe,params)
model_fit_and_scores(gs)
pipe2 = make_pipeline(CountVectorizer(),
MultinomialNB())
params2 = {
'countvectorizer__stop_words': [None, 'english'],
'countvectorizer__max_features':[5500,6000,6500],
'countvectorizer__ngram_range':[(1,1), (1,2)],
'multinomialnb__alpha':[.05, .1, .5]
}
gs2 = GridSearchCV(pipe2, params2)
model_fit_and_scores(gs2)
pipe3 = make_pipeline(CountVectorizer(),
MultinomialNB())
params3 = {
'countvectorizer__stop_words': ['english'],
'countvectorizer__max_features':[6500,6600,6700],
'countvectorizer__ngram_range':[(1,1)],
'multinomialnb__alpha':[.09, .1, .12]
}
gs3 = GridSearchCV(pipe3, params3)
model_fit_and_scores(gs3)
pipe4 = make_pipeline(CountVectorizer(),
MultinomialNB())
params4 = {
'countvectorizer__stop_words': ['english'],
'countvectorizer__max_features':[6500,6700,7000],
'countvectorizer__ngram_range':[(1,1)],
'multinomialnb__alpha':[.12, .14, .15]
}
gs4 = GridSearchCV(pipe4, params4)
model_fit_and_scores(gs4)
# Since this is the best so far, I will input this into my model summary
input_to_df(gs4, 'minimal', 'no')<jupyter_output><empty_output>
|
no_license
|
/code/modeling-1-bayes.ipynb
|
auroravictoria/Reddit-Classification-NLP
| 3 |
<jupyter_start><jupyter_text>#### It looks like $\frac{\mathrm{d}E\hat{z}}{\mathrm{d}r_\mathrm{hole}}$ within 6 mm diameter hole sizes are less than 10 ppm of the electric field. So varying this hole up to this size seems pretty negligible. Keep in mind that this derivative appears to decrease quadratically so for larger hole sizes this could be of concern but we are more than likely will not go larger than a 6 mm diameter hole size. This 10 ppm figure could very well change as we vary the plate separation and diameter of the disk<jupyter_code>Ez_center[0]
## Plotting E-field strength as a function of the distance from the center of the flux surface
for i in range(0,len(E_flux_rad[0,:])):
plt.plot(np.arange(500e-6,abs(E_flux_rad[:,i]))
time_elapsed = file['time_elapsed'][()]
time_elapsed<jupyter_output><empty_output>
|
no_license
|
/code/fea_electrodes/python_scripts/calculate_e_field_flux.ipynb
|
dancvan/AlGaAs_electrooptic_effect
| 1 |
<jupyter_start><jupyter_text>#Some Python examples##Load data<jupyter_code>from azureml import Workspace
ws = Workspace(
workspace_id='be61456bd29047529d24a146a9cb6233',
authorization_token='2652318930204669ac2bff77d523c284',
endpoint='https://studioapi.azureml.net'
)
ds = ws.datasets['iris.csv']
import pandas as pd
frame = ds.to_dataframe()
iris=pd.DataFrame(frame,columns=['Sepal.Length','Sepal.Width','Petal.Length','Petal.Width','Species'])
iris<jupyter_output><empty_output><jupyter_text>##Summary data<jupyter_code>print set(iris['Species'])<jupyter_output>set([u'setosa', u'versicolor', u'virginica'])
<jupyter_text>##Plot data<jupyter_code>from pylab import *
cols=['bo','ro','go']
i=0
for spec in set(iris['Species']):
plot(iris.loc[iris['Species']==spec,'Sepal.Length'],iris.loc[iris['Species']==spec,'Sepal.Width'],cols[i])
i=i+1
show()
hist(iris['Sepal.Length'])
show()
import random
iris['Target']=zeros(len(iris["Species"]))
iris.loc[iris.Species=='setosa','Target']=np.random.random_integers(0,1,len(iris['Target']+1))
iris.head()<jupyter_output>/home/nbcommon/env/lib/python2.7/site-packages/pandas/core/internals.py:648: DeprecationWarning: assignment will raise an error in the future, most likely because your index result shape does not match the value array shape. You can use `arr.flat[index] = values` to keep the old behaviour.
values[indexer] = value
<jupyter_text># Random Forest<jupyter_code>from sklearn.cross_validation import cross_val_score
from sklearn.cross_validation import cross_val_score
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import roc_curve, auc
rows = random.sample(iris.index, int(len(iris.index)*0.6))
dat=iris.ix[:,0:3]
training = dat.ix[rows]
validation=dat.drop(rows)
y_train=iris.Target[rows]
y_val=iris.Target.drop(rows)
clf = RandomForestClassifier(max_depth=None, min_samples_split=1,random_state=0)
clf = clf.fit(training, y_train)
tree_probas = clf.predict_proba(validation)
tree_fpr, tree_tpr, tree_thresholds = roc_curve(y_val,
tree_probas[:, 1])
plot(tree_fpr, tree_tpr, label='Tree')
xlabel('False Positive Rate')
ylabel('True Positive Rate')
title('Receiver operating characteristic')
legend(loc="lower right")
show()
res=validation
res['Predict']=clf.predict(validation)
res['Target']=y_val
res
<jupyter_output><empty_output>
|
no_license
|
/Python_training_prework.ipynb
|
pascalgaehler/data_science_for_analytics
| 4 |
<jupyter_start><jupyter_text>## collecting samples from webcam<jupyter_code>pip install --upgrade pip
pip install imgaug
pip install opencv-contrib-python
import cv2
import numpy as np
face_classifier = cv2.CascadeClassifier('C:/Users/PC-LENOVO/anaconda/Lib/site-packages/cv2/data/haarcascade_frontalface_default.xml')
def face_extractor(img): # so the image we work under gray scale not in rgb
gray = cv2.cvtColor(img,cv2.COLOR_BGR2GRAY)
faces = face_classifier.detectMultiScale(gray,1.3,6)
# 1.3 is scale factor
# 5 is minimum numbers of neighbours its value vary between 3 and 6 but the value at 6 or nearer to 6 give good result
if faces is (): # if face is not present
return None
for (x,y,w,h) in faces: # for loop is for cropping frontal faces to see ity
cropped_face = img[y:y+h,x:x+w] # y is for rows and x is for columns
return cropped_face
cap = cv2.VideoCapture(0) # 0 is our by default camera id
count = 0 # this variable is derive to take the number of photos
while True :
ret, frame = cap.read()
if face_extractor(frame) is not None:
count+=1
face = cv2.resize(face_extractor(frame),(200,200)) # we resize the image to 200,200
face = cv2.cvtColor(face,cv2.COLOR_BGR2GRAY) # now convert that img to gray
file_name_path = 'C:/Users/PC-LENOVO/Downloads/facial tutorial/faces/user'+str(count)+'.jpg' # now we save the face value so for that we have to define a path to aparticular file # user we define in path is the name of the samples we collected and count should be depicted in string format in jpg
cv2.imwrite(file_name_path,face) # so it basically saving it for that we pass path and image
cv2.putText(face,str(count),(50,50),cv2.FONT_HERSHEY_COMPLEX,1,(0,255,0),2) # 50,50 is size it detect 0,255,0 define green color rbg 1 defines the scale 2 defines the font size
cv2.imshow('Face Cropper',face) # it shows us current face samples
else:
print("Face not found")
pass
if cv2.waitKey(1)==13 or count==100: # now for closing it 13 is the asci code for enter to stop capturing photo or it will take 100 samples
break
cap.release() # now we have to release the camera
cv2.destroyAllWindows()
print("collecting samples complete!!!")
<jupyter_output>Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
Face not found
collecting samples complete!!!
<jupyter_text># training that model by using those samples<jupyter_code>import cv2
import numpy as np
from os import listdir # this helps in fetching data from current working directory
from os.path import isfile, join
data_path = 'C:/Users/PC-LENOVO/Downloads/facial tutorial/faces/' # so we place / at the end of working directory to fetch the data
onlyfiles = [f for f in listdir(data_path) if isfile(join(data_path,f))] # so it basically join the corrent working directory to fetch the data stored in it
Training_Data, Labels = [],[] # so we have two empty list to insert training data and labels
for i,files in enumerate(onlyfiles): # enumerate provide the number of iteration according to the amount of data saved inside it
image_path = data_path + onlyfiles[i]
images = cv2.imread(image_path,cv2.IMREAD_GRAYSCALE) # so it defines images we are fetching is in gray scale
Training_Data.append(np.asarray(images,dtype=np.uint8)) # uint is unsigned integer
Labels.append(i)
Labels = np.asarray(Labels, dtype=np.int32)
model = cv2.face.LBPHFaceRecognizer_create() # LINEAR BINARY PHASE HISTROGRAM RECOGNIZER so our classification model is built up
model.train(np.asarray(Training_Data),np.asarray(Labels))
print("Model Training is Completed")
<jupyter_output>Model Training is Completed
<jupyter_text>## testing the model<jupyter_code>face_classifier = cv2.CascadeClassifier('C:/Users/PC-LENOVO/anaconda/Lib/site-packages/cv2/data/haarcascade_frontalface_default.xml')
def face_detector(img, size=0.5):
gray = cv2.cvtColor(img, cv2.COLOR_BGR2GRAY) # so it convert img to grayscale
faces = face_classifier.detectMultiScale(gray,1.3,6)
if faces is():# image is not present
return img,[]
for(x,y,w,h) in faces: # we scan each area of faces
cv2.rectangle(img, (x,y),(x+w,y+h),(0,255,0),2) # so it shows rectangle through x,y and x+w,y+h this shows how much increament should be done in the face and (0,255,0) shows the color of the rectangle 2 shows the thickness of the rectangle
roi = img[y:y+h,x:x+w]# region of interest
roi = cv2.resize(roi,(200,200))
return img,roi
cap = cv2.VideoCapture(0)
while True:
ret,frame = cap.read()
image, face = face_detector(frame)
try:
face = cv2.cvtColor(face, cv2.COLOR_BGR2GRAY)
result = model.predict(face)
if result[1] < 500: # if the first value in result is lesser than 500
confidence = int(100*(1-(result[1])/300)) # we find the confidence value which describe how much percentage of faces will match
display_string = str(confidence)+'% Confidence it is user' # it prints the integer value
cv2.putText(image,display_string,(100,120),cv2.FONT_HERSHEY_COMPLEX,1,(250,120,255),2)# (250,120,255) is a color (100,120) is size 1 is scale 2 is font
if confidence >75:
cv2.putText(image,"Unlocked",(250,450),cv2.FONT_HERSHEY_COMPLEX,1,(0,255,0),2)
cv2.imshow('Face Cropper', image)
else:
cv2.putText(image,"Locked",(250,450),cv2.FONT_HERSHEY_COMPLEX,1,(0,0,255),2)
cv2.imshow('Face Cropper', image)
except:
cv2.putText(image,"Face Not Found",(250,450),cv2.FONT_HERSHEY_COMPLEX,1,(255,0,0),2)
cv2.imshow('Face Cropper', image)
pass # it heps in countinous running
if cv2.waitKey(1)==13:
break
cap.release()
cv2.destroyAllWindows()
<jupyter_output><empty_output>
|
no_license
|
/facialrecog/face_recog.ipynb
|
Ravikshdikola/my-projects
| 3 |
<jupyter_start><jupyter_text># AI Lab 8 - Naive Bayes from Scratch
__By Hasnain Naeem (212728), BSCS-7B, NUST__
__Description__: Naive bayes implementation from scratch using Numpy and Pandas. Although binary classification is demonstrated but it works for more than 2 classes. ## Imports<jupyter_code>from tabulate import tabulate
import numpy as np
import pandas as pd<jupyter_output><empty_output><jupyter_text>## Defining Training Set Dataframe<jupyter_code># dataframe columns
column_names=["department", "status", "age", "salary"]
# define dataframe data
age_groups = {"A": "21-25", "B": "26-30", "C": "31-35", "D": "36-40",
"E": "41-45", "F": "46-50"}
salary_groups = {"A": "26-30", "B": "31-35", "C": "36-40", "D": "41-45",
"E": "46-50", "F": "51-55", "G": "56-60", "H": "61-65", "I": "66-70"}
data = {"department": ["sales", "sales", "sales", "systems", "systems",
"systems", "systems", "marketing", "marketing",
"secretary", "secretary"],
"status": ["senior", "junior", "junior", "junior", "senior",
"junior", "senior", "senior", "junior", "senior",
"junior"],
"age": ["C", "B", "C", "A", "C", "B", "E", "D", "C", "F", "B"],
"salary": ["E", "A", "B", "E", "I", "E", "I", "E", "D", "C", "A"]
}
# create dataframe
df = pd.DataFrame(data)
print("Training Set:")
print(tabulate(df))<jupyter_output>Training Set:
-- --------- ------ - -
0 sales senior C E
1 sales junior B A
2 sales junior C B
3 systems junior A E
4 systems senior C I
5 systems junior B E
6 systems senior E I
7 marketing senior D E
8 marketing junior C D
9 secretary senior F C
10 secretary junior B A
-- --------- ------ - -
<jupyter_text>## Naive Bayes Implementation<jupyter_code>def get_feature_table(feature_name, feature_column, label_column, pc=0):
"""
Takes the feature column and label column to calculate the probability tables.
Parameters:
pc: pseudocount constant used for laplace smoothing
"""
# get column names for the feature dataframe
label_values = list(label_column.unique())
feature_df_cols = label_values.copy()
# get list of column names as: feature_name, label values...
feature_df_cols.insert(0, feature_name)
feature_values = feature_column.unique()
feature_table = pd.DataFrame([[feature_val, 0, 0] for feature_val in feature_values], columns=feature_df_cols)
feature_table = feature_table.set_index(feature_name)
# fill the feature table with the counts of each feature value
label_count = {label_value: 0 for label_value in label_values}
# count the values
for i, feature_value in feature_column.iteritems():
feature_table[label_column[i]][feature_value] = feature_table[label_column[i]][feature_value] + 1
label_count[label_column[i]] += 1
# divide to get probabilities
for label_value in label_values:
feature_table[label_value] = (feature_table[label_value] + pc) / (label_count[label_value] + (pc * len(label_values)))
return feature_table
# select the feature to be predicted;
label_class = "status"
# column containing the values of the label class
label_column = df[label_class]
# get the prior probabilities of values of each label class
label_counts = label_column.value_counts()
label_prob = label_counts / label_column.size
label_classes = [label_class for label_class, _ in label_prob.iteritems()]
# get the prior probabilties of features
feature_prior_prob = {column_name:None for column_name in column_names}
for feature_name, feature_val_prior_prob in feature_prior_prob.items():
val_counts = df[feature_name].value_counts()
val_prob = val_counts / label_column.size
feature_prior_prob[feature_name] = val_prob<jupyter_output><empty_output><jupyter_text>### Calculate Feature Tables without Laplace Smoothing<jupyter_code># generate the probability tables for each feature
feature_tables = {}
for col_name, col_data in df.iteritems():
feature_table = get_feature_table(col_name, col_data, label_column)
feature_tables[col_name] = feature_table
print("Feature Tables")
print("_______________\n")
for feature_name, feature_table in feature_tables.items():
print("Feature Name: "+feature_name)
print("Feature Table:")
print(tabulate(feature_table))
print()<jupyter_output>Feature Tables
_______________
Feature Name: department
Feature Table:
--------- --- --------
sales 0.2 0.333333
systems 0.4 0.333333
marketing 0.2 0.166667
secretary 0.2 0.166667
--------- --- --------
Feature Name: status
Feature Table:
------ - -
senior 1 0
junior 0 1
------ - -
Feature Name: age
Feature Table:
- --- --------
C 0.4 0.333333
B 0 0.5
A 0 0.166667
E 0.2 0
D 0.2 0
F 0.2 0
- --- --------
Feature Name: salary
Feature Table:
- --- --------
E 0.4 0.333333
A 0 0.333333
B 0 0.166667
I 0.4 0
D 0 0.166667
C 0.2 0
- --- --------
<jupyter_text>### Calculate Feature Tables with Laplace Smoothing<jupyter_code># generate the probability tables for each feature
feature_tables_with_ls = {}
for col_name, col_data in df.iteritems():
feature_table = get_feature_table(col_name, col_data, label_column, pc=1) # using pseudocount = 1
feature_tables_with_ls[col_name] = feature_table
print("Feature Tables")
print("_______________\n")
for feature_name, feature_table in feature_tables_with_ls.items():
print("Feature Name: "+feature_name)
print("Feature Table:")
print(tabulate(feature_table))
print()<jupyter_output>Feature Tables
_______________
Feature Name: department
Feature Table:
--------- -------- -----
sales 0.285714 0.375
systems 0.428571 0.375
marketing 0.285714 0.25
secretary 0.285714 0.25
--------- -------- -----
Feature Name: status
Feature Table:
------ -------- -----
senior 0.857143 0.125
junior 0.142857 0.875
------ -------- -----
Feature Name: age
Feature Table:
- -------- -----
C 0.428571 0.375
B 0.142857 0.5
A 0.142857 0.25
E 0.285714 0.125
D 0.285714 0.125
F 0.285714 0.125
- -------- -----
Feature Name: salary
Feature Table:
- -------- -----
E 0.428571 0.375
A 0.142857 0.375
B 0.142857 0.25
I 0.428571 0.125
D 0.142857 0.25
C 0.285714 0.125
- -------- -----
<jupyter_text>## Defining Test Set Dataframe<jupyter_code>test_rows = [
["marketing", "C", "E"],
["sales", "C", "I"]
]
test_columns = column_names.copy()
test_columns.remove(label_class)
test_df = pd.DataFrame(test_rows, columns=test_columns)
print("Test Set:")
print(tabulate(test_df))<jupyter_output>Test Set:
- --------- - -
0 marketing C E
1 sales C I
- --------- - -
<jupyter_text>## Making Predictions<jupyter_code>def make_prediction(i, row, feature_tables):
"""
Makes predictions depending on the given feature table.
Parameters:
i: index of sample
row: row of test set dataframe
feature_table: probability tables for each feature
"""
likelihoods = {label_class: 1 for label_class in label_classes}
predict_prior_probs = {label_class: 0 for label_class in label_classes}
predicted_probs = likelihoods.copy()
for feature_name, feature_val in row.iteritems():
for sample_class, class_prior_prob in label_prob.iteritems():
likelihoods[sample_class] *= feature_tables[feature_name].loc[feature_val][sample_class]
predict_prior_probs[sample_class] += feature_prior_prob[feature_name][feature_val]
# Calculate Feature Tables without Laplace Smoothing
for sample_class, class_prior_prob in label_prob.iteritems():
predicted_probs[sample_class] = (likelihoods[sample_class] * label_prob[sample_class]) / predict_prior_probs[sample_class]
print("\tProbability of class '" + sample_class+"': " + str(predicted_probs[sample_class]))
print("\tPredicted class: '" + max(predicted_probs, key=predicted_probs.get)+"'")<jupyter_output><empty_output><jupyter_text>### Making Predictions without Laplace Smoothing<jupyter_code>for i, row in test_df.iterrows():
print("Making prediction on " + str(i) +"th sample:")
make_prediction(i, row, feature_tables)
print()<jupyter_output>Making prediction on 0th sample:
Probability of class 'junior': 0.01111111111111111
Probability of class 'senior': 0.016000000000000004
Predicted class: 'senior'
Making prediction on 1th sample:
Probability of class 'junior': 0.0
Probability of class 'senior': 0.017777777777777785
Predicted class: 'senior'
<jupyter_text>### Making Predictions with Laplace Smoothing<jupyter_code>for i, row in test_df.iterrows():
print("Making prediction on " + str(i) +"th sample:")
make_prediction(i, row, feature_tables_with_ls)
print()<jupyter_output>Making prediction on 0th sample:
Probability of class 'junior': 0.021093749999999998
Probability of class 'senior': 0.02623906705539358
Predicted class: 'senior'
Making prediction on 1th sample:
Probability of class 'junior': 0.011718749999999998
Probability of class 'senior': 0.029154518950437313
Predicted class: 'senior'
|
permissive
|
/Lab 8 - Naive Bayes from Scratch/Lab 8 - Naive Bayes.ipynb
|
hasnainnaeem/Artificial-Intelligence-Coursework
| 9 |
<jupyter_start><jupyter_text># Amortization Example in PandasCode to support article at [Practical Business Python](http://pbpython.com/amortization-model.html)Setup the imports and matplotlib plotting<jupyter_code>import pandas as pd
import numpy as np
from datetime import date
import matplotlib.pyplot as plt
import matplotlib
%matplotlib inline
matplotlib.style.use('ggplot')<jupyter_output><empty_output><jupyter_text>Define a function to build an amortization table/schedule and return the value as a pandas DataFrame<jupyter_code>def amortization_table(interest_rate, years, payments_year, principal, addl_principal=0, start_date=date.today()):
""" Calculate the amortization schedule given the loan details
Args:
interest_rate: The annual interest rate for this loan
years: Number of years for the loan
payments_year: Number of payments in a year
principal: Amount borrowed
addl_principal (optional): Additional payments to be made each period. Assume 0 if nothing provided.
must be a value less then 0, the function will convert a positive value to
negative
start_date (optional): Start date. Will start on first of next month if none provided
Returns:
schedule: Amortization schedule as a pandas dataframe
summary: Pandas dataframe that summarizes the payoff information
"""
# Ensure the additional payments are negative
if addl_principal > 0:
addl_principal = -addl_principal
# Create an index of the payment dates
rng = pd.date_range(start_date, periods=years * payments_year, freq='MS')
rng.name = "Payment_Date"
# Build up the Amortization schedule as a DataFrame
df = pd.DataFrame(index=rng,columns=['Payment', 'Principal', 'Interest',
'Addl_Principal', 'Curr_Balance'], dtype='float')
# Add index by period (start at 1 not 0)
df.reset_index(inplace=True)
df.index += 1
df.index.name = "Period"
# Calculate the payment, principal and interests amounts using built in Numpy functions
per_payment = np.pmt(interest_rate/payments_year, years*payments_year, principal)
df["Payment"] = per_payment
df["Principal"] = np.ppmt(interest_rate/payments_year, df.index, years*payments_year, principal)
df["Interest"] = np.ipmt(interest_rate/payments_year, df.index, years*payments_year, principal)
# Round the values
df = df.round(2)
# Add in the additional principal payments
df["Addl_Principal"] = addl_principal
# Store the Cumulative Principal Payments and ensure it never gets larger than the original principal
df["Cumulative_Principal"] = (df["Principal"] + df["Addl_Principal"]).cumsum()
df["Cumulative_Principal"] = df["Cumulative_Principal"].clip(lower=-principal)
# Calculate the current balance for each period
df["Curr_Balance"] = principal + df["Cumulative_Principal"]
# Determine the last payment date
try:
last_payment = df.query("Curr_Balance <= 0")["Curr_Balance"].idxmax(axis=1, skipna=True)
except ValueError:
last_payment = df.last_valid_index()
last_payment_date = "{:%m-%d-%Y}".format(df.loc[last_payment, "Payment_Date"])
# Truncate the data frame if we have additional principal payments:
if addl_principal != 0:
# Remove the extra payment periods
df = df.ix[0:last_payment].copy()
# Calculate the principal for the last row
df.ix[last_payment, "Principal"] = -(df.ix[last_payment-1, "Curr_Balance"])
# Calculate the total payment for the last row
df.ix[last_payment, "Payment"] = df.ix[last_payment, ["Principal", "Interest"]].sum()
# Zero out the additional principal
df.ix[last_payment, "Addl_Principal"] = 0
# Get the payment info into a DataFrame in column order
payment_info = (df[["Payment", "Principal", "Addl_Principal", "Interest"]]
.sum().to_frame().T)
# Format the Date DataFrame
payment_details = pd.DataFrame.from_items([('payoff_date', [last_payment_date]),
('Interest Rate', [interest_rate]),
('Number of years', [years])
])
# Add a column showing how much we pay each period.
# Combine addl principal with principal for total payment
payment_details["Period_Payment"] = round(per_payment, 2) + addl_principal
payment_summary = pd.concat([payment_details, payment_info], axis=1)
return df, payment_summary
<jupyter_output><empty_output><jupyter_text>## Examples of running the function<jupyter_code>schedule1, stats1 = amortization_table(0.05, 30, 12, 100000, addl_principal=0)<jupyter_output><empty_output><jupyter_text>Take a look at the start and end of the table as well as the summary stats<jupyter_code>schedule1.head()
schedule1.tail()
stats1<jupyter_output><empty_output><jupyter_text>Try running some other scenarios and combining them into a single DataFrame<jupyter_code>schedule2, stats2 = amortization_table(0.05, 30, 12, 100000, addl_principal=-200)
schedule3, stats3 = amortization_table(0.04, 15, 12, 100000, addl_principal=0)
# Combine all the scenarios into 1 view
pd.concat([stats1, stats2, stats3], ignore_index=True)
schedule3.head()<jupyter_output><empty_output><jupyter_text>## Examples of plotting the data<jupyter_code>schedule1.plot(x='Payment_Date', y='Curr_Balance', title="Pay Off Timeline");
fig, ax = plt.subplots(1, 1)
schedule1.plot(x='Payment_Date', y='Curr_Balance', label="Scenario 1", ax=ax)
schedule2.plot(x='Payment_Date', y='Curr_Balance', label="Scenario 2", ax=ax)
schedule3.plot(x='Payment_Date', y='Curr_Balance', label="Scenario 3", ax=ax)
plt.title("Pay Off Timelines");
schedule1["Cum_Interest"] = schedule1["Interest"].abs().cumsum()
schedule2["Cum_Interest"] = schedule2["Interest"].abs().cumsum()
schedule3["Cum_Interest"] = schedule3["Interest"].abs().cumsum()
fig, ax = plt.subplots(1, 1)
schedule1.plot(x='Payment_Date', y='Cum_Interest', label="Scenario 1", ax=ax)
schedule2.plot(x='Payment_Date', y='Cum_Interest', label="Scenario 2", ax=ax, style='+')
schedule3.plot(x='Payment_Date', y='Cum_Interest', label="Scenario 3", ax=ax)
ax.legend(loc="best");
fig, ax = plt.subplots(1, 1)
y1_schedule = schedule1.set_index('Payment_Date').resample("A")["Interest"].sum().abs().reset_index()
y1_schedule["Year"] = y1_schedule["Payment_Date"].dt.year
y1_schedule.plot(kind="bar", x="Year", y="Interest", ax=ax, label="30 Years @ 5%")
plt.title("Interest Payments");<jupyter_output><empty_output>
|
permissive
|
/notebooks/Amortization-Model-Article.ipynb
|
routinezh/pbpython
| 6 |
<jupyter_start><jupyter_text>## [作業重點]
確保你了解隨機森林模型中每個超參數的意義,並觀察調整超參數對結果的影響## 作業
1. 試著調整 RandomForestClassifier(...) 中的參數,並觀察是否會改變結果?
2. 改用其他資料集 (boston, wine),並與回歸模型與決策樹的結果進行比較<jupyter_code>from sklearn import datasets, metrics
from sklearn.ensemble import RandomForestClassifier
from sklearn.ensemble import RandomForestRegressor
from sklearn.model_selection import train_test_split
from sklearn.metrics import mean_squared_error, r2_score
# 忽略警告訊息
%matplotlib inline
import warnings
warnings.filterwarnings('ignore')
wine = datasets.load_wine()
# 切分訓練集/測試集
x_train, x_test, y_train, y_test = train_test_split(wine.data, wine.target, test_size=0.1, random_state=4)
# 建立一個RandomForestClassifier
# n_estimators #決策樹的數量量
# max_features #如何選取 features
clf =RandomForestClassifier(n_estimators=20,
max_depth=5,
min_samples_split=2,
min_samples_leaf=1)
# 將訓練資料丟進去模型訓練
clf.fit(x_train, y_train)
# 將測試資料丟進模型得到預測結果
y_pred = clf.predict(x_test)
acc = metrics.accuracy_score(y_test, y_pred)
print("Accuracy: ", acc)
wine = datasets.load_wine()
# 切分訓練集/測試集
x_train, x_test, y_train, y_test = train_test_split(wine.data, wine.target, test_size=0.1, random_state=4)
# 建立一個RandomForestClassifier
# Criterion: 衡量量資料相似程度的
# n_estimators #決策樹的數量量
# metricMax_depth: 樹能⽣生長的最深限制
# Min_samples_split: ⾄至少要多少樣本以上才進⾏行行切分
# Min_samples_lear: 最終的葉⼦子 (節點) 上⾄至少要有多少樣本
clf =RandomForestClassifier(criterion='gini',
n_estimators=100,
max_depth=10,
min_samples_split=5,
min_samples_leaf=1)
# 將訓練資料丟進去模型訓練
clf.fit(x_train, y_train)
# 將測試資料丟進模型得到預測結果
y_pred = clf.predict(x_test)
acc = metrics.accuracy_score(y_test, y_pred)
print("Accuracy: ", acc)
wine = datasets.load_wine()
# 切分訓練集/測試集
x_train, x_test, y_train, y_test = train_test_split(wine.data, wine.target, test_size=0.1, random_state=4)
# 建立一個RandomForestClassifier
# Criterion: 衡量量資料相似程度的
# n_estimators #決策樹的數量量
# metricMax_depth: 樹能⽣生長的最深限制
# Min_samples_split: ⾄至少要多少樣本以上才進⾏行行切分
# Min_samples_lear: 最終的葉⼦子 (節點) 上⾄至少要有多少樣本
clf =RandomForestRegressor( criterion='mse',
n_estimators=100,
max_depth=10,
min_samples_split=5,
min_samples_leaf=1)
# 將訓練資料丟進去模型訓練
clf.fit(x_train, y_train)
# 將測試資料丟進模型得到預測結果
y_pred = clf.predict(x_test)
# 預測值與實際值的差距,使用 MSE
print("Mean squared error:{0}".format(mean_squared_error(y_test, y_pred)))<jupyter_output>Mean squared error:0.01777505303602922
|
no_license
|
/homework/Day_044_HW.ipynb
|
LiChengJhe/2nd-ML100Days
| 1 |
<jupyter_start><jupyter_text>## Sentiment Analysis
> 1. Merge `Title` & `Content`
> 2. Prep text. I set-up to run in parallel and it's much faster but still takes ~7 minutes on my laptop.
> 3. Perform Sentiment Analysis using `TextBlob`. May take ~150 hours to run in parallel on my machine. A powerful AWS instance with 64 cores will still take ~9 hours to run. The best approach to optimize performance here is to use convert to **C** code using `Cython` and possibly even use distributed computing.
**Next: try optimizing code for efficiency using `Dask` and `Cython`.**<jupyter_code># instantiate `myNLP` object
myNLP = myNLP()
# merge `Topics` and `Content`
merged = merge_2_string_lists(docs['Title'], docs['Content'])
# clean and prep text
parallel_tasks = merged
parallel_func = myNLP.prep_docs_lematize
docs['Processed Text'] = parallelize(parallel_func, merged)
# perform Sentiment Analysis
parallel_tasks = docs['Processed Text']
parallel_func = get_sentiment
docs['Sentiment'] = parallelize(parallel_func, parallel_tasks)
i = 3
text = docs['Processed Text'][i]
content = docs['Content'][i]
print(content)
print(get_sentiment(text))
docs.head()
# save
docs['Sentiment'].to_csv('src/results/sentiment.csv', sep=',')<jupyter_output><empty_output>
|
no_license
|
/NLP_Sentiment_Analysis.ipynb
|
dast1/gdp_prediction
| 1 |
<jupyter_start><jupyter_text>## Parameters<jupyter_code>f = 0.1 #Mining rate
m = 1000 #Number of voting chains
log_epsilon = 20 #Confirmation guarantee
beta_actual = 0.25
<jupyter_output><empty_output><jupyter_text>## Prism Latency Simulation<jupyter_code># Number of adverserial blocks in private at time t
def adverserial(t, beta, fv):
x = np.arange(0,100,1)
mu = fv*t*beta/(1-beta)
return poisson.pmf(x, mu)
#Probability that a vote with depth k will be removed when adversary has Z_t blocks in private
def p_k_t(k, Z_t,beta):
ans = 0
k = int(k)
for i in range(k):
ans += Z_t[i]*np.power(beta/(1-beta), k - i)
ans += np.sum(Z_t[k:])
return ans
def runExp(m, log_epsilon, fv):
events = int(2000*log_epsilon)
time_array = np.zeros_like(beta_array)
fv_effective = fv/(1.0+2*fv) #Forking
exp_random_v = np.random.exponential(1/(m*fv_effective), size=events)
chain_random_v = np.random.randint(m, size=events)
for j, beta in enumerate(beta_array):
# exponent = 0.8*(1-2*beta)/(1-beta)*np.log((1-beta)/beta)
votes, votes_permanence_E, votes_permanence_V = np.zeros(m), np.zeros(m), np.zeros(m)
time, mean, var = 0,0,0
lower_bar = np.zeros_like(exp_random_v)
mined_by_adversary_or_honest_node = np.random.choice(['Honest', 'Adversary'], events, p = [1 - beta_actual, beta_actual])
voter_cast = np.zeros(m)
for i, tDiff in enumerate(exp_random_v):
time += tDiff
# Adding vote on a random chain
rChain = chain_random_v[i]
if mined_by_adversary_or_honest_node[i] == 'Honest':
voter_cast[rChain] = True
if voter_cast[rChain]:
votes[rChain] +=1
#Solidified probability
Z_t = adverserial(time, beta, fv)
p_i = 1 - p_k_t(votes[rChain], Z_t, beta)
# p_i = 1-np.power(np.e, -votes[rChain]*exponent) # Old calculation.
#Updating the mean an variance
mean -= votes_permanence_E[rChain]
var -= votes_permanence_V[rChain]
votes_permanence_E[rChain] = p_i
votes_permanence_V[rChain] = p_i*(1-p_i)
mean += votes_permanence_E[rChain]
var += votes_permanence_V[rChain]
lower_bar[i] = mean#-np.sqrt(2*var*log_epsilon)
# The block got confirmed
if mean-np.sqrt(2*var*log_epsilon) > m/2+1:
time_array[j] = time
lower_bar[i:] = mean#-np.sqrt(2*var*log_epsilon)
break
return time_array
beta_array = np.array([ 0.25, 0.275, .3, .325, .35, .375, 0.4])
prism_ans = np.zeros_like(beta_array)
prism_ans_square = np.zeros_like(beta_array)
no_exp = 5
#T1: Time taken for a unique proposer block to refer the tx_block
#T2: Time taken for a proposer block to get confirmed
for i in range(no_exp):
a = runExp(m, log_epsilon, f)
a += np.power(np.e, 1*f)/f
prism_ans += a
prism_ans_square += a*a
prism_ans /= no_exp
prism_ans_square /= no_exp
prism_ans_std = np.sqrt(prism_ans_square - prism_ans*prism_ans) + np.sqrt(np.power(np.e, 1*f)/f)<jupyter_output><empty_output><jupyter_text>## Bitcoin Latency calculation<jupyter_code>exponent = 0.8*(1-2*beta_array)/(1-beta_array)*np.log((1-beta_array)/beta_array)
bitcoin_ans= (log_epsilon)/(exponent*f)+(1./f)*(beta_actual/1-beta_actual)<jupyter_output><empty_output><jupyter_text>## Plot<jupyter_code>fig = plt.figure(figsize=(15,7))
ax = fig.add_subplot(111)
bitcoin_ans = bitcoin_ans.astype(int)
prism_ans = prism_ans.astype(int)
plt.plot(beta_array, bitcoin_ans, '-o', lw=3, label="Longest chain")
plt.errorbar(beta_array, prism_ans, yerr = 10*prism_ans_std, fmt='-o',lw=3, label="Prism")
for i in range(bitcoin_ans.shape[0]):
ax.annotate(bitcoin_ans[i],xy=(beta_array[i],bitcoin_ans[i]+40), size=20)
ax.annotate(prism_ans[i],xy=(beta_array[i],prism_ans[i]+10), size=20)
plt.ylabel("Confirmation latency (secs)", size=25)
plt.xlabel("Beta", size=25)
plt.tick_params(axis='both', which='major', labelsize=20)
# plt.title("Time to confirm for epsilon = $e$^("+str(-log_epsilon)+") gaurantee with active beta = "+str(beta_actual), size=25)
plt.legend(prop={'size': 25})
plt.show()
<jupyter_output><empty_output>
|
no_license
|
/Confirmation latency - Bitcoin vs Prism-Censorship attack.ipynb
|
bagavi/Prism_simulations
| 4 |
<jupyter_start><jupyter_text># TFDS로 input pipeline 구축하기
다음 colab을 따라한 것임
* https://colab.research.google.com/github/tensorflow/docs/blob/master/site/en/guide/data.ipynb<jupyter_code>import tensorflow as tf
import pathlib
import os
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
np.set_printoptions(precision=4)<jupyter_output><empty_output><jupyter_text>## Basic mechanism
* from_tensor_slice로 Dataset 생성
* for in 이나 next로 개별 data 접근하기
* lambda 함수 적용 예시 (reduce)<jupyter_code>dataset = tf.data.Dataset.from_tensor_slices([1,2,3])
dataset
for elem in dataset:
print(elem.numpy())
it = iter(dataset)
it
print(next(it).numpy())<jupyter_output>1
<jupyter_text>## TFDS.TypeSpec 확인하기<jupyter_code>dataset1 = tf.data.Dataset.from_tensor_slices(tf.random.uniform([4, 10]))
dataset1.element_spec
dataset2 = tf.data.Dataset.from_tensor_slices(
(tf.random.uniform([4]), tf.random.uniform([4,100], maxval=100, dtype=tf.int32)))
dataset2.element_spec
dataset3 = tf.data.Dataset.zip((dataset1, dataset2))
dataset3.element_spec
# with sparse tensor
dataset4 = tf.data.Dataset.from_tensors(
tf.SparseTensor(indices=[[0,0], [1,2]], values=[1,2], dense_shape=[3,4]))
print(dataset4.element_spec)
print(dataset4.element_spec.value_type)<jupyter_output>SparseTensorSpec(TensorShape([3, 4]), tf.int32)
<class 'tensorflow.python.framework.sparse_tensor.SparseTensor'>
<jupyter_text>## Numpy array 로 dataset 생성하기<jupyter_code>train, test = tf.keras.datasets.fashion_mnist.load_data()
images, labels = train
images = images / 255
print(type(images))
dataset = tf.data.Dataset.from_tensor_slices((images, labels))
dataset<jupyter_output><class 'numpy.ndarray'>
<jupyter_text>## Python generator로 Dataset 생성하기 <jupyter_code># generator
def pyfunc_count(stop):
i = 0
while i<stop:
yield i
i +=1
for n in pyfunc_count(3):
print(n)
ds_counter = tf.data.Dataset.from_generator(
pyfunc_count,
args=[3],
output_types=tf.int32,
output_shapes=(), )
for count in ds_counter.take(10): # single element generation
print(count.numpy())
for count_batch in ds_counter.repeat().batch(5).take(3): # mini-batch generation
print(count_batch.numpy())<jupyter_output>[0 1 2 0 1]
[2 0 1 2 0]
[1 2 0 1 2]
<jupyter_text>## 가변 series를 generate 하고, padded batch 만들기<jupyter_code>def gen_series():
i = 0
while True:
size = np.random.randint(0, 5)
yield i, np.random.normal(size=(size,))
i += 1
for i, series in gen_series():
print(i, ":", str(series))
if i > 5:
break
ds_series = tf.data.Dataset.from_generator(
gen_series,
output_types=(tf.int32, tf.float32),
output_shapes=((), (None, ))) # None으로 가변형 표현
ds_series
ds_series_batch = ds_series.padded_batch(5)
ids, seq_batch = next(iter(ds_series_batch))
seq_batch.numpy() # zero-padded 고정 길이형 확인<jupyter_output><empty_output><jupyter_text>## Image Generator로부터 dataset 만들기<jupyter_code>flowers = tf.keras.utils.get_file(
'flower_photos',
'https://storage.googleapis.com/download.tensorflow.org/example_images/flower_photos.tgz',
untar=True)
img_gen = tf.keras.preprocessing.image.ImageDataGenerator(rescale=1./255, rotation_range=20)
images, labels = next(img_gen.flow_from_directory(flowers)) # tf.keras의 generator
print(images.dtype, images.shape)
print(labels.dtype, labels.shape)
ds = tf.data.Dataset.from_generator(
lambda: img_gen.flow_from_directory(flowers),
output_types = (tf.float32, tf.float32),
output_shapes = ([32,256,256,3], [32,5])
)
ds.element_spec
images, labels = next(iter(ds)) # tfds로부터 iteration
print(images.dtype, images.shape)
print(labels.dtype, labels.shape)<jupyter_output>Found 3670 images belonging to 5 classes.
<dtype: 'float32'> (32, 256, 256, 3)
<dtype: 'float32'> (32, 5)
<jupyter_text>## TFRecord 로부터 TFRecordDataset 만들기<jupyter_code>fsns_test_file = tf.keras.utils.get_file("fsns.tfrec", "https://storage.googleapis.com/download.tensorflow.org/data/fsns-20160927/testdata/fsns-00000-of-00001")
dataset = tf.data.TFRecordDataset(filenames=[fsns_test_file])
dataset
raw_example = next(iter(dataset))
parsed = tf.train.Example.FromString(raw_example.numpy())
parsed.features.feature['image/text']<jupyter_output><empty_output><jupyter_text>## Text data로부터 TextLineDataset 만들기<jupyter_code># 텍스트 파일들 가져오기
directory_url = 'https://storage.googleapis.com/download.tensorflow.org/data/illiad/'
file_names = ['cowper.txt', 'derby.txt', 'butler.txt']
file_paths = [
tf.keras.utils.get_file(file_name, directory_url + file_name)
for file_name in file_names
]
dataset = tf.data.TextLineDataset(file_paths)
dataset
line = next(iter(dataset))
print(line.numpy())<jupyter_output>b"\xef\xbb\xbfAchilles sing, O Goddess! Peleus' son;"
<jupyter_text>#### interleave를 사용해서 여러 개의 file로부터 온 line들을 섞기<jupyter_code>files_ds = tf.data.Dataset.from_tensor_slices(file_paths)
lines_ds = files_ds.interleave(tf.data.TextLineDataset, cycle_length=3)
for i, line in enumerate(lines_ds.take(9)):
if i % 3 == 0:
print()
print(line.numpy())<jupyter_output>
b"\xef\xbb\xbfAchilles sing, O Goddess! Peleus' son;"
b"\xef\xbb\xbfOf Peleus' son, Achilles, sing, O Muse,"
b'\xef\xbb\xbfSing, O goddess, the anger of Achilles son of Peleus, that brought'
b'His wrath pernicious, who ten thousand woes'
b'The vengeance, deep and deadly; whence to Greece'
b'countless ills upon the Achaeans. Many a brave soul did it send'
b"Caused to Achaia's host, sent many a soul"
b'Unnumbered ills arose; which many a soul'
b'hurrying down to Hades, and many a hero did it yield a prey to dogs and'
<jupyter_text>#### 불필요한 line skip 및 line filter 조건 <jupyter_code>titanic_file = tf.keras.utils.get_file("train.csv", "https://storage.googleapis.com/tf-datasets/titanic/train.csv")
titanic_lines = tf.data.TextLineDataset(titanic_file)
for line in titanic_lines.take(10):
print(line.numpy())
def survived(line):
return tf.not_equal(tf.strings.substr(line, 0, 1), "0")
survivors = titanic_lines.skip(1).filter(survived)
for line in survivors.take(10):
print(line.numpy())<jupyter_output>b'1,female,38.0,1,0,71.2833,First,C,Cherbourg,n'
b'1,female,26.0,0,0,7.925,Third,unknown,Southampton,y'
b'1,female,35.0,1,0,53.1,First,C,Southampton,n'
b'1,female,27.0,0,2,11.1333,Third,unknown,Southampton,n'
b'1,female,14.0,1,0,30.0708,Second,unknown,Cherbourg,n'
b'1,female,4.0,1,1,16.7,Third,G,Southampton,n'
b'1,male,28.0,0,0,13.0,Second,unknown,Southampton,y'
b'1,female,28.0,0,0,7.225,Third,unknown,Cherbourg,y'
b'1,male,28.0,0,0,35.5,First,A,Southampton,y'
b'1,female,38.0,1,5,31.3875,Third,unknown,Southampton,n'
<jupyter_text>## CSV -> Pandas -> Dataset 만들기<jupyter_code>titanic_file = tf.keras.utils.get_file("train.csv", "https://storage.googleapis.com/tf-datasets/titanic/train.csv")
df = pd.read_csv(titanic_file)
titanic_ds = tf.data.Dataset.from_tensor_slices(dict(df))
feature_batch = next(iter(titanic_ds))
for key, value in feature_batch.items():
print(" {!r:20s}: {}".format(key, value))<jupyter_output> 'survived' : 0
'sex' : b'male'
'age' : 22.0
'n_siblings_spouses': 1
'parch' : 0
'fare' : 7.25
'class' : b'Third'
'deck' : b'unknown'
'embark_town' : b'Southampton'
'alone' : b'n'
<jupyter_text>## CSV 로부터 한번에 Dataset 만들기 (from disk directly)<jupyter_code>titanic_batches = tf.data.experimental.make_csv_dataset(
titanic_file, batch_size=4,
select_columns=['class', 'fare', 'survived'],
label_name="survived"
)
titanic_batches
feature_batch, label_batch = next(iter(titanic_batches))
print("'survived': {}".format(label_batch))
for key, value in feature_batch.items():
print(" {!r:20s}: {}".format(key, value))<jupyter_output>'survived': [1 0 1 0]
'fare' : [ 26.2875 7.0542 146.5208 26.25 ]
'class' : [b'First' b'Third' b'First' b'Second']
<jupyter_text>## CSV 로부터 한번에 CsvDataset 만들기<jupyter_code>titanic_types = [tf.int32, tf.string, tf.float32, tf.int32, tf.int32, tf.float32, tf.string, tf.string, tf.string, tf.string]
dataset = tf.data.experimental.CsvDataset(titanic_file, titanic_types, header=True)
dataset
line = next(iter(dataset))
print( [item.numpy() for item in line] )<jupyter_output>[0, b'male', 22.0, 1, 0, 7.25, b'Third', b'unknown', b'Southampton', b'n']
<jupyter_text>## 복수 파일로부터 dataset 만들기<jupyter_code>flowers_root = tf.keras.utils.get_file(
'flower_photos',
'https://storage.googleapis.com/download.tensorflow.org/example_images/flower_photos.tgz',
untar=True)
flowers_root = pathlib.Path(flowers_root)
for item in flowers_root.glob("*"):
print(item.name) # subdirectory로 개별 클래스 인스턴스 파일들을 가지고 있다.
list_ds = tf.data.Dataset.list_files(str(flowers_root/'*/*'))
for f in list_ds.take(5):
print(f.numpy())
# path에서 label을 추출해서 label을 가진 ds 만들기
def process_path(file_path):
label = tf.strings.split(file_path, os.sep)[-2]
return tf.io.read_file(file_path), label
labeled_ds = list_ds.map(process_path)
image_raw, label_text = next(iter(labeled_ds))
print(repr(image_raw.numpy())[:100])
print()
print(label_text.numpy())
<jupyter_output>b'\xff\xd8\xff\xe0\x00\x10JFIF\x00\x01\x01\x00\x00\x01\x00\x01\x00\x00\xff\xdb\x00C\x00\x03\x02\x02\
b'dandelion'
<jupyter_text>## BatchDataset 만들기<jupyter_code>dataset = tf.data.Dataset.range(100)
dataset = dataset.map(lambda x: tf.fill([tf.cast(x,tf.int32)],x))
for elem in dataset.take(5):
print(elem.numpy())
# padded batch 만들기: Mini-Batch 안의 가장 긴 시퀀스대로 패치한다.
pad_dataset = dataset.padded_batch(batch_size=3, padded_shapes=(None,))
for elem in pad_dataset.take(5):
print(elem.numpy())<jupyter_output>[[0 0]
[1 0]
[2 2]]
[[3 3 3 0 0]
[4 4 4 4 0]
[5 5 5 5 5]]
[[6 6 6 6 6 6 0 0]
[7 7 7 7 7 7 7 0]
[8 8 8 8 8 8 8 8]]
[[ 9 9 9 9 9 9 9 9 9 0 0]
[10 10 10 10 10 10 10 10 10 10 0]
[11 11 11 11 11 11 11 11 11 11 11]]
[[12 12 12 12 12 12 12 12 12 12 12 12 0 0]
[13 13 13 13 13 13 13 13 13 13 13 13 13 0]
[14 14 14 14 14 14 14 14 14 14 14 14 14 14]]
|
no_license
|
/tfds/tfds_input_pipeline.ipynb
|
hoondori/coursera_tensorflow
| 16 |
<jupyter_start><jupyter_text># Machine Learning Engineer Nanodegree
## Unsupervised Learning
## Project: Creating Customer SegmentsWelcome to the third project of the Machine Learning Engineer Nanodegree! In this notebook, some template code has already been provided for you, and it will be your job to implement the additional functionality necessary to successfully complete this project. Sections that begin with **'Implementation'** in the header indicate that the following block of code will require additional functionality which you must provide. Instructions will be provided for each section and the specifics of the implementation are marked in the code block with a `'TODO'` statement. Please be sure to read the instructions carefully!
In addition to implementing code, there will be questions that you must answer which relate to the project and your implementation. Each section where you will answer a question is preceded by a **'Question X'** header. Carefully read each question and provide thorough answers in the following text boxes that begin with **'Answer:'**. Your project submission will be evaluated based on your answers to each of the questions and the implementation you provide.
>**Note:** Code and Markdown cells can be executed using the **Shift + Enter** keyboard shortcut. In addition, Markdown cells can be edited by typically double-clicking the cell to enter edit mode.## Getting Started
In this project, you will analyze a dataset containing data on various customers' annual spending amounts (reported in *monetary units*) of diverse product categories for internal structure. One goal of this project is to best describe the variation in the different types of customers that a wholesale distributor interacts with. Doing so would equip the distributor with insight into how to best structure their delivery service to meet the needs of each customer.
The dataset for this project can be found on the [UCI Machine Learning Repository](https://archive.ics.uci.edu/ml/datasets/Wholesale+customers). For the purposes of this project, the features `'Channel'` and `'Region'` will be excluded in the analysis — with focus instead on the six product categories recorded for customers.
Run the code block below to load the wholesale customers dataset, along with a few of the necessary Python libraries required for this project. You will know the dataset loaded successfully if the size of the dataset is reported.<jupyter_code># Import libraries necessary for this project
import numpy as np
import pandas as pd
from IPython.display import display # Allows the use of display() for DataFrames
# Import supplementary visualizations code visuals.py
import visuals as vs
# Pretty display for notebooks
%matplotlib inline
# Load the wholesale customers dataset
try:
data = pd.read_csv("customers.csv")
data.drop(['Region', 'Channel'], axis = 1, inplace = True)
print("Wholesale customers dataset has {} samples with {} features each.".format(*data.shape))
except:
print("Dataset could not be loaded. Is the dataset missing?")<jupyter_output>Wholesale customers dataset has 440 samples with 6 features each.
<jupyter_text>## Data Exploration
In this section, you will begin exploring the data through visualizations and code to understand how each feature is related to the others. You will observe a statistical description of the dataset, consider the relevance of each feature, and select a few sample data points from the dataset which you will track through the course of this project.
Run the code block below to observe a statistical description of the dataset. Note that the dataset is composed of six important product categories: **'Fresh'**, **'Milk'**, **'Grocery'**, **'Frozen'**, **'Detergents_Paper'**, and **'Delicatessen'**. Consider what each category represents in terms of products you could purchase.<jupyter_code># Display a description of the dataset
display(data.describe())
data.head()<jupyter_output><empty_output><jupyter_text>### Implementation: Selecting Samples
To get a better understanding of the customers and how their data will transform through the analysis, it would be best to select a few sample data points and explore them in more detail. In the code block below, add **three** indices of your choice to the `indices` list which will represent the customers to track. It is suggested to try different sets of samples until you obtain customers that vary significantly from one another.<jupyter_code># TODO: Select three indices of your choice you wish to sample from the dataset
indices = [1,47,240]
# Create a DataFrame of the chosen samples
samples = pd.DataFrame(data.loc[indices], columns = data.keys()).reset_index(drop = True)
print("Chosen samples of wholesale customers dataset:")
display(samples)<jupyter_output>Chosen samples of wholesale customers dataset:
<jupyter_text>### Question 1
Consider the total purchase cost of each product category and the statistical description of the dataset above for your sample customers.
* What kind of establishment (customer) could each of the three samples you've chosen represent?
**Hint:** Examples of establishments include places like markets, cafes, delis, wholesale retailers, among many others. Avoid using names for establishments, such as saying *"McDonalds"* when describing a sample customer as a restaurant. You can use the mean values for reference to compare your samples with. The mean values are as follows:
* Fresh: 12000.2977
* Milk: 5796.2
* Grocery: 7951.2
* Frozen: 3071.9
* Detergents_paper: 2881.4
* Delicatessen: 1524.8
Knowing this, how do your samples compare? Does that help in driving your insight into what kind of establishments they might be?
**Answer:**
- Customer 0: Spending on Fresh, Frozen less than average. Spending on Milk, Grocery, Detergents_paper, Delicatessen more than average.
- Customer 1: Spending on everything (Fresh, Milk, Grocery, Frozen, Detergents_paper, Delicatessen) more than average.
- Customer 2: Spending on Milk, Grocery, Detergents_paper less than average. Spending on Fresh, Frozen, Delicatessen more than average.
It doesnt give me any insight into what kind of establishments they might be.### Implementation: Feature Relevance
One interesting thought to consider is if one (or more) of the six product categories is actually relevant for understanding customer purchasing. That is to say, is it possible to determine whether customers purchasing some amount of one category of products will necessarily purchase some proportional amount of another category of products? We can make this determination quite easily by training a supervised regression learner on a subset of the data with one feature removed, and then score how well that model can predict the removed feature.
In the code block below, you will need to implement the following:
- Assign `new_data` a copy of the data by removing a feature of your choice using the `DataFrame.drop` function.
- Use `sklearn.cross_validation.train_test_split` to split the dataset into training and testing sets.
- Use the removed feature as your target label. Set a `test_size` of `0.25` and set a `random_state`.
- Import a decision tree regressor, set a `random_state`, and fit the learner to the training data.
- Report the prediction score of the testing set using the regressor's `score` function.<jupyter_code>from sklearn.cross_validation import train_test_split
from sklearn.tree import DecisionTreeRegressor
# TODO: Make a copy of the DataFrame, using the 'drop' function to drop the given feature
new_data = data.copy()
new_data.drop('Frozen', axis = 1, inplace = True)
target = data['Frozen']
# TODO: Split the data into training and testing sets(0.25) using the given feature as the target
# Set a random state.
X_train, X_test, y_train, y_test = train_test_split(new_data, target, test_size = 0.25, random_state = 50)
# TODO: Create a decision tree regressor and fit it to the training set
regressor = DecisionTreeRegressor(random_state = 50)
regressor.fit(X_train,y_train)
y_pred = regressor.predict(X_test)
# TODO: Report the score of the prediction using the testing set
score = regressor.score(X_test,y_test)
score<jupyter_output>/opt/conda/lib/python3.6/site-packages/sklearn/cross_validation.py:41: DeprecationWarning: This module was deprecated in version 0.18 in favor of the model_selection module into which all the refactored classes and functions are moved. Also note that the interface of the new CV iterators are different from that of this module. This module will be removed in 0.20.
"This module will be removed in 0.20.", DeprecationWarning)
<jupyter_text>### Question 2
* Which feature did you attempt to predict?
* What was the reported prediction score?
* Is this feature necessary for identifying customers' spending habits?
**Hint:** The coefficient of determination, `R^2`, is scored between 0 and 1, with 1 being a perfect fit. A negative `R^2` implies the model fails to fit the data. If you get a low score for a particular feature, that lends us to beleive that that feature point is hard to predict using the other features, thereby making it an important feature to consider when considering relevance.**Answer:**
* Which feature did you attempt to predict? ---> I tried to predict 'Frozen' features
* What was the reported prediction score? ---> The score is -1.409. The score should be between 0 and 1. A negative score implies the model fails to fit the data. It's hard to predict "Frozen" feature using other features.
* Is this feature necessary for identifying customers' spending habits? ---> Yes.
### Visualize Feature Distributions
To get a better understanding of the dataset, we can construct a scatter matrix of each of the six product features present in the data. If you found that the feature you attempted to predict above is relevant for identifying a specific customer, then the scatter matrix below may not show any correlation between that feature and the others. Conversely, if you believe that feature is not relevant for identifying a specific customer, the scatter matrix might show a correlation between that feature and another feature in the data. Run the code block below to produce a scatter matrix.<jupyter_code># Produce a scatter matrix for each pair of features in the data
pd.scatter_matrix(data, alpha = 0.3, figsize = (14,8), diagonal = 'kde');<jupyter_output>/opt/conda/lib/python3.6/site-packages/ipykernel_launcher.py:2: FutureWarning: pandas.scatter_matrix is deprecated. Use pandas.plotting.scatter_matrix instead
<jupyter_text>### Question 3
* Using the scatter matrix as a reference, discuss the distribution of the dataset, specifically talk about the normality, outliers, large number of data points near 0 among others. If you need to sepearate out some of the plots individually to further accentuate your point, you may do so as well.
* Are there any pairs of features which exhibit some degree of correlation?
* Does this confirm or deny your suspicions about the relevance of the feature you attempted to predict?
* How is the data for those features distributed?
**Hint:** Is the data normally distributed? Where do most of the data points lie? You can use [corr()](https://pandas.pydata.org/pandas-docs/stable/generated/pandas.DataFrame.corr.html) to get the feature correlations and then visualize them using a [heatmap](http://seaborn.pydata.org/generated/seaborn.heatmap.html) (the data that would be fed into the heatmap would be the correlation values, for eg: `data.corr()`) to gain further insight.<jupyter_code>data.corr()
import seaborn as sns
ax = sns.heatmap(data.corr())
import matplotlib.pyplot as plt
g = sns.PairGrid(data,vars = ['Fresh','Milk','Grocery','Frozen','Detergents_Paper','Delicatessen'])
g.map(plt.scatter)
plt.show()
features = ['Fresh','Milk','Grocery','Frozen','Detergents_Paper','Delicatessen']
fig = plt.figure(figsize= (20,15))
i=0
for a in features:
fig.add_subplot(2,3,i+1)
plt.hist(data[a])
plt.title(a)
i += 1<jupyter_output><empty_output><jupyter_text>**Answer:**
* The data is not normally distributed. Most of the data points lie closely to zero.
* Are there any pairs of features which exhibit some degree of correlation? ---> Yes. Grocery vs Detergents_Paper(0.924641), Detergents_Paper vs Milk (0.661816), Grocery vs. Milk(0.728335)
* Does this confirm or deny your suspicions about the relevance of the feature you attempted to predict? ---> Yes, looking at the correlation between Frozen and others, I see there are no correlation.
* How is the data for those features distributed? ---> most of the data is close to zero, and they have big outliers. The data is skewed to the right.
## Data Preprocessing
In this section, you will preprocess the data to create a better representation of customers by performing a scaling on the data and detecting (and optionally removing) outliers. Preprocessing data is often times a critical step in assuring that results you obtain from your analysis are significant and meaningful.### Implementation: Feature Scaling
If data is not normally distributed, especially if the mean and median vary significantly (indicating a large skew), it is most [often appropriate](http://econbrowser.com/archives/2014/02/use-of-logarithms-in-economics) to apply a non-linear scaling — particularly for financial data. One way to achieve this scaling is by using a [Box-Cox test](http://scipy.github.io/devdocs/generated/scipy.stats.boxcox.html), which calculates the best power transformation of the data that reduces skewness. A simpler approach which can work in most cases would be applying the natural logarithm.
In the code block below, you will need to implement the following:
- Assign a copy of the data to `log_data` after applying logarithmic scaling. Use the `np.log` function for this.
- Assign a copy of the sample data to `log_samples` after applying logarithmic scaling. Again, use `np.log`.<jupyter_code># TODO: Scale the data using the natural logarithm
log_data = np.log(data)
# TODO: Scale the sample data using the natural logarithm
log_samples = np.log(samples)
# Produce a scatter matrix for each pair of newly-transformed features
pd.scatter_matrix(log_data, alpha = 0.3, figsize = (14,8), diagonal = 'kde');<jupyter_output>/opt/conda/lib/python3.6/site-packages/ipykernel_launcher.py:8: FutureWarning: pandas.scatter_matrix is deprecated. Use pandas.plotting.scatter_matrix instead
<jupyter_text>### Observation
After applying a natural logarithm scaling to the data, the distribution of each feature should appear much more normal. For any pairs of features you may have identified earlier as being correlated, observe here whether that correlation is still present (and whether it is now stronger or weaker than before).
Run the code below to see how the sample data has changed after having the natural logarithm applied to it.<jupyter_code># Display the log-transformed sample data
display(log_samples)<jupyter_output><empty_output><jupyter_text>### Implementation: Outlier Detection
Detecting outliers in the data is extremely important in the data preprocessing step of any analysis. The presence of outliers can often skew results which take into consideration these data points. There are many "rules of thumb" for what constitutes an outlier in a dataset. Here, we will use [Tukey's Method for identfying outliers](http://datapigtechnologies.com/blog/index.php/highlighting-outliers-in-your-data-with-the-tukey-method/): An *outlier step* is calculated as 1.5 times the interquartile range (IQR). A data point with a feature that is beyond an outlier step outside of the IQR for that feature is considered abnormal.
In the code block below, you will need to implement the following:
- Assign the value of the 25th percentile for the given feature to `Q1`. Use `np.percentile` for this.
- Assign the value of the 75th percentile for the given feature to `Q3`. Again, use `np.percentile`.
- Assign the calculation of an outlier step for the given feature to `step`.
- Optionally remove data points from the dataset by adding indices to the `outliers` list.
**NOTE:** If you choose to remove any outliers, ensure that the sample data does not contain any of these points!
Once you have performed this implementation, the dataset will be stored in the variable `good_data`.<jupyter_code># For each feature find the data points with extreme high or low values
outliers = []
for feature in log_data.keys():
# TODO: Calculate Q1 (25th percentile of the data) for the given feature
Q1 = np.percentile(log_data[feature],25)
# TODO: Calculate Q3 (75th percentile of the data) for the given feature
Q3 = np.percentile(log_data[feature],75)
# TODO: Use the interquartile range to calculate an outlier step (1.5 times the interquartile range)
step = (Q3 - Q1) * 1.5
# Display the outliers
print("Data points considered outliers for the feature '{}':".format(feature))
outliers_data = log_data[~((log_data[feature] >= Q1 - step) & (log_data[feature] <= Q3 + step))]
display(outliers_data)
for each_index in outliers_data.index.values:
outliers.append(each_index)
print ("This is the outliers of the log_data:")
print (outliers)
# I will remove the duplicates in the outliers
def remove_duplicates(outliers):
final_list = []
for each in outliers:
if each not in final_list:
final_list.append(each)
return final_list
final_outliers = remove_duplicates(outliers)
print ("Before we had {} outliers. After removing duplicates, we have {} outliers". format(len(outliers), len(final_outliers)))
# Remove the outliers, if any were specified
good_data = log_data.drop(log_data.index[final_outliers]).reset_index(drop = True)
len(log_data)
len(good_data)<jupyter_output><empty_output><jupyter_text>### Question 4
* Are there any data points considered outliers for more than one feature based on the definition above?
* Should these data points be removed from the dataset?
* If any data points were added to the `outliers` list to be removed, explain why.
** Hint: ** If you have datapoints that are outliers in multiple categories think about why that may be and if they warrant removal. Also note how k-means is affected by outliers and whether or not this plays a factor in your analysis of whether or not to remove them.**Answer:**
* Are there any data points considered outliers for more than one feature based on the definition above? yes. There are 6 of them.
* Should these data points be removed from the dataset? yes, I removed them.
* If any data points were added to the `outliers` list to be removed, explain why. I removed them because the outliers might skew the data. It might lead to wrong insights. Sometimes the outliers are created by mistakes. Removing outliers helps us calculate or create algorithm easier, and we still keep a large portion of the data.
## Feature Transformation
In this section you will use principal component analysis (PCA) to draw conclusions about the underlying structure of the wholesale customer data. Since using PCA on a dataset calculates the dimensions which best maximize variance, we will find which compound combinations of features best describe customers.### Implementation: PCA
Now that the data has been scaled to a more normal distribution and has had any necessary outliers removed, we can now apply PCA to the `good_data` to discover which dimensions about the data best maximize the variance of features involved. In addition to finding these dimensions, PCA will also report the *explained variance ratio* of each dimension — how much variance within the data is explained by that dimension alone. Note that a component (dimension) from PCA can be considered a new "feature" of the space, however it is a composition of the original features present in the data.
In the code block below, you will need to implement the following:
- Import `sklearn.decomposition.PCA` and assign the results of fitting PCA in six dimensions with `good_data` to `pca`.
- Apply a PCA transformation of `log_samples` using `pca.transform`, and assign the results to `pca_samples`.<jupyter_code>from sklearn.decomposition import PCA
# TODO: Apply PCA by fitting the good data with the same number of dimensions as features
pca = PCA(n_components = 6)
pca.fit(good_data)
# TODO: Transform log_samples using the PCA fit above
pca_samples = pca.transform(log_samples)
# Generate PCA results plot
pca_results = vs.pca_results(good_data, pca)<jupyter_output><empty_output><jupyter_text>### Question 5
* How much variance in the data is explained* **in total** *by the first and second principal component?
* How much variance in the data is explained by the first four principal components?
* Using the visualization provided above, talk about each dimension and the cumulative variance explained by each, stressing upon which features are well represented by each dimension(both in terms of positive and negative variance explained). Discuss what the first four dimensions best represent in terms of customer spending.
**Hint:** A positive increase in a specific dimension corresponds with an *increase* of the *positive-weighted* features and a *decrease* of the *negative-weighted* features. The rate of increase or decrease is based on the individual feature weights.**Answer:**
* How much variance in the data is explained* **in total** *by the first and second principal component? the first one is 0.4993, and the second one is 0.2259. Total is 0.7252.
* How much variance in the data is explained by the first four principal components? in the first four principal components, we have total variance 0.9279.
- Dimension 1: has mostly everything in positive (Fresh and Frozen are slightly negative). Detergents_Paper, Milk, and Grocery have the most weight in this dimension.
- Dimension 2: has mostly everything in positive. Different from dimension 1, most weight of this dimension are on Fresh , Frozen, Delicatessen.
- Dimension 3: has Fresh and Detergents_Paper in negative. That makes dimension 3 different from others.
- Dimension 4: has Frozen and Detergents_Paper in negative. That makes dimension 4 different from others.
### Observation
Run the code below to see how the log-transformed sample data has changed after having a PCA transformation applied to it in six dimensions. Observe the numerical value for the first four dimensions of the sample points. Consider if this is consistent with your initial interpretation of the sample points.<jupyter_code># Display sample log-data after having a PCA transformation applied
display(pd.DataFrame(np.round(pca_samples, 4), columns = pca_results.index.values))<jupyter_output><empty_output><jupyter_text>### Implementation: Dimensionality Reduction
When using principal component analysis, one of the main goals is to reduce the dimensionality of the data — in effect, reducing the complexity of the problem. Dimensionality reduction comes at a cost: Fewer dimensions used implies less of the total variance in the data is being explained. Because of this, the *cumulative explained variance ratio* is extremely important for knowing how many dimensions are necessary for the problem. Additionally, if a signifiant amount of variance is explained by only two or three dimensions, the reduced data can be visualized afterwards.
In the code block below, you will need to implement the following:
- Assign the results of fitting PCA in two dimensions with `good_data` to `pca`.
- Apply a PCA transformation of `good_data` using `pca.transform`, and assign the results to `reduced_data`.
- Apply a PCA transformation of `log_samples` using `pca.transform`, and assign the results to `pca_samples`.<jupyter_code># TODO: Apply PCA by fitting the good data with only two dimensions
pca = PCA(n_components = 2)
pca.fit(good_data)
# TODO: Transform the good data using the PCA fit above
reduced_data = pca.transform(good_data)
# TODO: Transform log_samples using the PCA fit above
pca_samples = pca.transform(log_samples)
# Create a DataFrame for the reduced data
reduced_data = pd.DataFrame(reduced_data, columns = ['Dimension 1', 'Dimension 2'])<jupyter_output><empty_output><jupyter_text>### Observation
Run the code below to see how the log-transformed sample data has changed after having a PCA transformation applied to it using only two dimensions. Observe how the values for the first two dimensions remains unchanged when compared to a PCA transformation in six dimensions.<jupyter_code># Display sample log-data after applying PCA transformation in two dimensions
display(pd.DataFrame(np.round(pca_samples, 4), columns = ['Dimension 1', 'Dimension 2']))<jupyter_output><empty_output><jupyter_text>## Visualizing a Biplot
A biplot is a scatterplot where each data point is represented by its scores along the principal components. The axes are the principal components (in this case `Dimension 1` and `Dimension 2`). In addition, the biplot shows the projection of the original features along the components. A biplot can help us interpret the reduced dimensions of the data, and discover relationships between the principal components and original features.
Run the code cell below to produce a biplot of the reduced-dimension data.<jupyter_code># Create a biplot
vs.biplot(good_data, reduced_data, pca)<jupyter_output><empty_output><jupyter_text>### Observation
Once we have the original feature projections (in red), it is easier to interpret the relative position of each data point in the scatterplot. For instance, a point the lower right corner of the figure will likely correspond to a customer that spends a lot on `'Milk'`, `'Grocery'` and `'Detergents_Paper'`, but not so much on the other product categories.
From the biplot, which of the original features are most strongly correlated with the first component? --> Milk, Grocery, and Detergents_paper are most strongly correlated with the first component.
What about those that are associated with the second component? --> Fresh, Frozen, and Delicatessen are most strongly correlated with the second component.
Do these observations agree with the pca_results plot you obtained earlier? --> Yes.## Clustering
In this section, you will choose to use either a K-Means clustering algorithm or a Gaussian Mixture Model clustering algorithm to identify the various customer segments hidden in the data. You will then recover specific data points from the clusters to understand their significance by transforming them back into their original dimension and scale. ### Question 6
* What are the advantages to using a K-Means clustering algorithm?
* What are the advantages to using a Gaussian Mixture Model clustering algorithm?
* Given your observations about the wholesale customer data so far, which of the two algorithms will you use and why?
** Hint: ** Think about the differences between hard clustering and soft clustering and which would be appropriate for our dataset.**Answer:**
What are the advantages to using a K-Means clustering algorithm?
- Easy to implement
- With a large number of variables, K-Means may be computationally faster than hierarchical clustering (if K is small).
- k-Means may produce tighter clusters than hierarchical clustering
- An instance can change cluster (move to another cluster) when the centroids are recomputed.
What are the advantages to using a Gaussian Mixture Model clustering algorithm?
- It is the fastest algorithm for learning mixture models
- It's alot more flexible (different shapes: full, spherical, tied, diag)
- It allows mixed membership which means overlap.
Given your observations about the wholesale customer data so far, which of the two algorithms will you use and why?
- K-means is hard clustering, and Gaussian Mixture Model is soft clustering.
- I choose the Gaussian Mixture model because I believe customer dimensions can be overlapped. ### Implementation: Creating Clusters
Depending on the problem, the number of clusters that you expect to be in the data may already be known. When the number of clusters is not known *a priori*, there is no guarantee that a given number of clusters best segments the data, since it is unclear what structure exists in the data — if any. However, we can quantify the "goodness" of a clustering by calculating each data point's *silhouette coefficient*. The [silhouette coefficient](http://scikit-learn.org/stable/modules/generated/sklearn.metrics.silhouette_score.html) for a data point measures how similar it is to its assigned cluster from -1 (dissimilar) to 1 (similar). Calculating the *mean* silhouette coefficient provides for a simple scoring method of a given clustering.
In the code block below, you will need to implement the following:
- Fit a clustering algorithm to the `reduced_data` and assign it to `clusterer`.
- Predict the cluster for each data point in `reduced_data` using `clusterer.predict` and assign them to `preds`.
- Find the cluster centers using the algorithm's respective attribute and assign them to `centers`.
- Predict the cluster for each sample data point in `pca_samples` and assign them `sample_preds`.
- Import `sklearn.metrics.silhouette_score` and calculate the silhouette score of `reduced_data` against `preds`.
- Assign the silhouette score to `score` and print the result.<jupyter_code>from sklearn.mixture import GaussianMixture as gm
import sklearn.metrics as metrics
# TODO: Apply your clustering algorithm of choice to the reduced data
clusterer = gm(n_components = 2, n_init = 3, random_state = 50)
clusterer = clusterer.fit(reduced_data)
# TODO: Predict the cluster for each data point
preds = clusterer.predict(reduced_data)
# TODO: Find the cluster centers
centers = clusterer.means_
# TODO: Predict the cluster for each transformed sample data point
sample_preds = clusterer.predict(pca_samples)
# TODO: Calculate the mean silhouette coefficient for the number of clusters chosen
score = metrics.silhouette_score(reduced_data,preds)
score<jupyter_output><empty_output><jupyter_text>### Question 7
* Report the silhouette score for several cluster numbers you tried.
* Of these, which number of clusters has the best silhouette score?**Answer:**
Cluster numbers: 2 , silhouette score: 0.4474 <---this is the best silhouette score
Cluster numbers: 3 , silhouette score: 0.2729
Cluster numbers: 4 , silhouette score: 0.3070
Cluster numbers: 5 , silhouette score: 0.2162### Cluster Visualization
Once you've chosen the optimal number of clusters for your clustering algorithm using the scoring metric above, you can now visualize the results by executing the code block below. Note that, for experimentation purposes, you are welcome to adjust the number of clusters for your clustering algorithm to see various visualizations. The final visualization provided should, however, correspond with the optimal number of clusters. <jupyter_code># Display the results of the clustering from implementation
vs.cluster_results(reduced_data, preds, centers, pca_samples)<jupyter_output><empty_output><jupyter_text>### Implementation: Data Recovery
Each cluster present in the visualization above has a central point. These centers (or means) are not specifically data points from the data, but rather the *averages* of all the data points predicted in the respective clusters. For the problem of creating customer segments, a cluster's center point corresponds to *the average customer of that segment*. Since the data is currently reduced in dimension and scaled by a logarithm, we can recover the representative customer spending from these data points by applying the inverse transformations.
In the code block below, you will need to implement the following:
- Apply the inverse transform to `centers` using `pca.inverse_transform` and assign the new centers to `log_centers`.
- Apply the inverse function of `np.log` to `log_centers` using `np.exp` and assign the true centers to `true_centers`.
<jupyter_code># TODO: Inverse transform the centers
log_centers = pca.inverse_transform(centers)
# TODO: Exponentiate the centers
true_centers = np.exp(log_centers)
# Display the true centers
segments = ['Segment {}'.format(i) for i in range(0,len(centers))]
true_centers = pd.DataFrame(np.round(true_centers), columns = data.keys())
true_centers.index = segments
display(true_centers)
# Display a description of the dataset
data_without_outliers = data.drop(data.index[final_outliers]).reset_index(drop = True)
display(data_without_outliers.describe())
<jupyter_output><empty_output><jupyter_text>### Question 8
* Consider the total purchase cost of each product category for the representative data points above, and reference the statistical description of the dataset at the beginning of this project(specifically looking at the mean values for the various feature points). What set of establishments could each of the customer segments represent?
**Hint:** A customer who is assigned to `'Cluster X'` should best identify with the establishments represented by the feature set of `'Segment X'`. Think about what each segment represents in terms their values for the feature points chosen. Reference these values with the mean values to get some perspective into what kind of establishment they represent.**Answer:**
Fresh, Frozen and Delicatessen values from Segment0 and Segment 1 are less than the mean of the population. In order to distinguish segment 0 and segment 1, we look at these features: Milk, Grocery, and Detergents_paper. If the cost in those features are less than the mean, the customer should belong to segment 0. On the other hand, the customer is belong to segment 1 if those feature values are greater than the mean.
### Question 9
* For each sample point, which customer segment from* **Question 8** *best represents it?
* Are the predictions for each sample point consistent with this?*
Run the code block below to find which cluster each sample point is predicted to be.<jupyter_code># Display the predictions
for i, pred in enumerate(sample_preds):
print("Sample point", i, "predicted to be in Cluster", pred)
samples
display(true_centers)<jupyter_output><empty_output><jupyter_text>**Answer:**
The mean of features: Milk, Grocery, and Detergents_Paper are 5486.31, 7504.90,2725.37 respectively. In sample 2, we have Milk(3575), Grocery(7041), and Detergents_Paper(343) values less than the mean. It should be in segment 0. Sample 0 and sample 1 should be in segment 1 because their values are above the mean.
**Comparing between samples and true_centers:**
- Sample 0: Fresh is kinda in the middle between 2 segments. Milk (9810), Grocery (9568), Detergent_paper (3293), and Delicatessen (1776) are close to segment 1. Most of the features are close to segment 1, so sample 0 should belong to segment 1.
- Sample 1: Fresh, Frozen are close to segment 0. Milk, Grocery, Detergents_Paper, Delicatessen are close to segment 1. Most of the features are close to segment 1, so sample 1 should belong to segment 1.
- Sample 2: Fresh, Milk, Frozen, Detergents_Paper are close to segment 0. Grocery, Delicatessen are close to segment 1. Most of the features are close to segment 0, so sample 2 should belong to segment 0.
**Are the predictions for each sample point consistent with this?* <--- Yes**## ConclusionIn this final section, you will investigate ways that you can make use of the clustered data. First, you will consider how the different groups of customers, the ***customer segments***, may be affected differently by a specific delivery scheme. Next, you will consider how giving a label to each customer (which *segment* that customer belongs to) can provide for additional features about the customer data. Finally, you will compare the ***customer segments*** to a hidden variable present in the data, to see whether the clustering identified certain relationships.### Question 10
Companies will often run [A/B tests](https://en.wikipedia.org/wiki/A/B_testing) when making small changes to their products or services to determine whether making that change will affect its customers positively or negatively. The wholesale distributor is considering changing its delivery service from currently 5 days a week to 3 days a week. However, the distributor will only make this change in delivery service for customers that react positively.
* How can the wholesale distributor use the customer segments to determine which customers, if any, would react positively to the change in delivery service?*
**Hint:** Can we assume the change affects all customers equally? How can we determine which group of customers it affects the most?**Answer:**
We assume the change affects all customers equally. We see that segment 0 spends less than segment 1. It makes sense if we delivery to segment 1 more regularly which is 5 days a week. We might apply 3-day-delivery to segment 0.### Question 11
Additional structure is derived from originally unlabeled data when using clustering techniques. Since each customer has a ***customer segment*** it best identifies with (depending on the clustering algorithm applied), we can consider *'customer segment'* as an **engineered feature** for the data. Assume the wholesale distributor recently acquired ten new customers and each provided estimates for anticipated annual spending of each product category. Knowing these estimates, the wholesale distributor wants to classify each new customer to a ***customer segment*** to determine the most appropriate delivery service.
* How can the wholesale distributor label the new customers using only their estimated product spending and the **customer segment** data?
**Hint:** A supervised learner could be used to train on the original customers. What would be the target variable?**Answer:**
The target variables would be Milk, Grocery, and Detergents_Paper. The learner can compare those values with the mean of the population. If those values are less than the means, a new customer should be in segment 0. On the other hand, the customer should be in segment 1 if those values are more than the means.### Visualizing Underlying Distributions
At the beginning of this project, it was discussed that the `'Channel'` and `'Region'` features would be excluded from the dataset so that the customer product categories were emphasized in the analysis. By reintroducing the `'Channel'` feature to the dataset, an interesting structure emerges when considering the same PCA dimensionality reduction applied earlier to the original dataset.
Run the code block below to see how each data point is labeled either `'HoReCa'` (Hotel/Restaurant/Cafe) or `'Retail'` the reduced space. In addition, you will find the sample points are circled in the plot, which will identify their labeling.<jupyter_code># Display the clustering results based on 'Channel' data
vs.channel_results(reduced_data, outliers, pca_samples)
# Display the results of the clustering from implementation
vs.cluster_results(reduced_data, preds, centers, pca_samples)<jupyter_output><empty_output>
|
no_license
|
/customer_segments.ipynb
|
BenjaminDKLuong/Project_customer_segments
| 19 |
<jupyter_start><jupyter_text># Exploring precision and recall
The goal of this second notebook is to understand precision-recall in the context of classifiers.
* Use Amazon review data in its entirety.
* Train a logistic regression model.
* Explore various evaluation metrics: accuracy, confusion matrix, precision, recall.
* Explore how various metrics can be combined to produce a cost of making an error.
* Explore precision and recall curves.
Because we are using the full Amazon review dataset (not a subset of words or reviews), in this assignment we return to using GraphLab Create for its efficiency. As usual, let's start by **firing up GraphLab Create**.
Make sure you have the latest version of GraphLab Create (1.8.3 or later). If you don't find the decision tree module, then you would need to upgrade graphlab-create using
```
pip install graphlab-create --upgrade
```
See [this page](https://dato.com/download/) for detailed instructions on upgrading.<jupyter_code>import graphlab
from __future__ import division
import numpy as np
graphlab.canvas.set_target('ipynb')<jupyter_output><empty_output><jupyter_text># Load amazon review dataset<jupyter_code>products = graphlab.SFrame('amazon_baby.gl/')<jupyter_output><empty_output><jupyter_text># Extract word counts and sentimentsAs in the first assignment of this course, we compute the word counts for individual words and extract positive and negative sentiments from ratings. To summarize, we perform the following:
1. Remove punctuation.
2. Remove reviews with "neutral" sentiment (rating 3).
3. Set reviews with rating 4 or more to be positive and those with 2 or less to be negative.<jupyter_code>def remove_punctuation(text):
import string
return text.translate(None, string.punctuation)
# Remove punctuation.
review_clean = products['review'].apply(remove_punctuation)
# Count words
products['word_count'] = graphlab.text_analytics.count_words(review_clean)
# Drop neutral sentiment reviews.
products = products[products['rating'] != 3]
# Positive sentiment to +1 and negative sentiment to -1
products['sentiment'] = products['rating'].apply(lambda rating : +1 if rating > 3 else -1)<jupyter_output><empty_output><jupyter_text>Now, let's remember what the dataset looks like by taking a quick peek:<jupyter_code>products<jupyter_output><empty_output><jupyter_text>## Split data into training and test sets
We split the data into a 80-20 split where 80% is in the training set and 20% is in the test set.<jupyter_code>train_data, test_data = products.random_split(.8, seed=1)<jupyter_output><empty_output><jupyter_text>## Train a logistic regression classifier
We will now train a logistic regression classifier with **sentiment** as the target and **word_count** as the features. We will set `validation_set=None` to make sure everyone gets exactly the same results.
Remember, even though we now know how to implement logistic regression, we will use GraphLab Create for its efficiency at processing this Amazon dataset in its entirety. The focus of this assignment is instead on the topic of precision and recall.<jupyter_code>model = graphlab.logistic_classifier.create(train_data, target='sentiment',
features=['word_count'],
validation_set=None)<jupyter_output><empty_output><jupyter_text># Model EvaluationWe will explore the advanced model evaluation concepts that were discussed in the lectures.
## Accuracy
One performance metric we will use for our more advanced exploration is accuracy, which we have seen many times in past assignments. Recall that the accuracy is given by
$$
\mbox{accuracy} = \frac{\mbox{# correctly classified data points}}{\mbox{# total data points}}
$$
To obtain the accuracy of our trained models using GraphLab Create, simply pass the option `metric='accuracy'` to the `evaluate` function. We compute the **accuracy** of our logistic regression model on the **test_data** as follows:<jupyter_code>accuracy= model.evaluate(test_data, metric='accuracy')['accuracy']
print "Test Accuracy: %s" % accuracy
recall= model.evaluate(test_data, metric='recall')['recall']
print "Test Recall: %s" % recall<jupyter_output>Test Recall: 0.949955508098
<jupyter_text>## Baseline: Majority class prediction
Recall from an earlier assignment that we used the **majority class classifier** as a baseline (i.e reference) model for a point of comparison with a more sophisticated classifier. The majority classifier model predicts the majority class for all data points.
Typically, a good model should beat the majority class classifier. Since the majority class in this dataset is the positive class (i.e., there are more positive than negative reviews), the accuracy of the majority class classifier can be computed as follows:<jupyter_code>precision= model.evaluate(test_data, metric='precision')['precision']
print "Test Precision: %s" % precision
baseline = len(test_data[test_data['sentiment'] == 1])/len(test_data)
print "Baseline accuracy (majority class classifier): %s" % baseline<jupyter_output>Baseline accuracy (majority class classifier): 0.842782577394
<jupyter_text>** Quiz Question:** Using accuracy as the evaluation metric, was our **logistic regression model** better than the baseline (majority class classifier)?## Confusion Matrix
The accuracy, while convenient, does not tell the whole story. For a fuller picture, we turn to the **confusion matrix**. In the case of binary classification, the confusion matrix is a 2-by-2 matrix laying out correct and incorrect predictions made in each label as follows:
```
+---------------------------------------------+
| Predicted label |
+----------------------+----------------------+
| (+1) | (-1) |
+-------+-----+----------------------+----------------------+
| True |(+1) | # of true positives | # of false negatives |
| label +-----+----------------------+----------------------+
| |(-1) | # of false positives | # of true negatives |
+-------+-----+----------------------+----------------------+
```
To print out the confusion matrix for a classifier, use `metric='confusion_matrix'`:<jupyter_code>confusion_matrix = model.evaluate(test_data, metric='confusion_matrix')['confusion_matrix']
confusion_matrix<jupyter_output><empty_output><jupyter_text>**Quiz Question**: How many predicted values in the **test set** are **false positives**?<jupyter_code>1443<jupyter_output><empty_output><jupyter_text>## Computing the cost of mistakes
Put yourself in the shoes of a manufacturer that sells a baby product on Amazon.com and you want to monitor your product's reviews in order to respond to complaints. Even a few negative reviews may generate a lot of bad publicity about the product. So you don't want to miss any reviews with negative sentiments --- you'd rather put up with false alarms about potentially negative reviews instead of missing negative reviews entirely. In other words, **false positives cost more than false negatives**. (It may be the other way around for other scenarios, but let's stick with the manufacturer's scenario for now.)
Suppose you know the costs involved in each kind of mistake:
1. \$100 for each false positive.
2. \$1 for each false negative.
3. Correctly classified reviews incur no cost.
**Quiz Question**: Given the stipulation, what is the cost associated with the logistic regression classifier's performance on the **test set**?<jupyter_code>1443 * 100 + 1 * 1406<jupyter_output><empty_output><jupyter_text>## Precision and RecallYou may not have exact dollar amounts for each kind of mistake. Instead, you may simply prefer to reduce the percentage of false positives to be less than, say, 3.5% of all positive predictions. This is where **precision** comes in:
$$
[\text{precision}] = \frac{[\text{# positive data points with positive predicitions}]}{\text{[# all data points with positive predictions]}} = \frac{[\text{# true positives}]}{[\text{# true positives}] + [\text{# false positives}]}
$$So to keep the percentage of false positives below 3.5% of positive predictions, we must raise the precision to 96.5% or higher.
**First**, let us compute the precision of the logistic regression classifier on the **test_data**.<jupyter_code>precision = model.evaluate(test_data, metric='precision')['precision']
print "Precision on test data: %s" % precision<jupyter_output>Precision on test data: 0.948706099815
<jupyter_text>**Quiz Question**: Out of all reviews in the **test set** that are predicted to be positive, what fraction of them are **false positives**? (Round to the second decimal place e.g. 0.25)<jupyter_code>.05<jupyter_output><empty_output><jupyter_text>**Quiz Question:** Based on what we learned in lecture, if we wanted to reduce this fraction of false positives to be below 3.5%, we would: (see the quiz)A complementary metric is **recall**, which measures the ratio between the number of true positives and that of (ground-truth) positive reviews:
$$
[\text{recall}] = \frac{[\text{# positive data points with positive predicitions}]}{\text{[# all positive data points]}} = \frac{[\text{# true positives}]}{[\text{# true positives}] + [\text{# false negatives}]}
$$
Let us compute the recall on the **test_data**.<jupyter_code>recall = model.evaluate(test_data, metric='recall')['recall']
print "Recall on test data: %s" % recall<jupyter_output>Recall on test data: 0.949955508098
<jupyter_text>**Quiz Question**: What fraction of the positive reviews in the **test_set** were correctly predicted as positive by the classifier?
**Quiz Question**: What is the recall value for a classifier that predicts **+1** for all data points in the **test_data**?# Precision-recall tradeoff
In this part, we will explore the trade-off between precision and recall discussed in the lecture. We first examine what happens when we use a different threshold value for making class predictions. We then explore a range of threshold values and plot the associated precision-recall curve.
## Varying the threshold
False positives are costly in our example, so we may want to be more conservative about making positive predictions. To achieve this, instead of thresholding class probabilities at 0.5, we can choose a higher threshold.
Write a function called `apply_threshold` that accepts two things
* `probabilities` (an SArray of probability values)
* `threshold` (a float between 0 and 1).
The function should return an array, where each element is set to +1 or -1 depending whether the corresponding probability exceeds `threshold`.<jupyter_code>def threshold_helper(prob, thresh):
if prob >= thresh:
return 1
else:
return -1
def apply_threshold(probabilities, threshold):
### YOUR CODE GOES HERE
#t = probabilities.apply(lambda p: threshold_helper(p, threshold))
t = probabilities.apply(lambda p: 1 if p >= threshold else -1)
return t
<jupyter_output><empty_output><jupyter_text>Run prediction with `output_type='probability'` to get the list of probability values. Then use thresholds set at 0.5 (default) and 0.9 to make predictions from these probability values.<jupyter_code>probabilities = model.predict(test_data, output_type='probability')
predictions_with_default_threshold = apply_threshold(probabilities, 0.5)
predictions_with_high_threshold = apply_threshold(probabilities, 0.9)
print "Number of positive predicted reviews (threshold = 0.5): %s" % (predictions_with_default_threshold == 1).sum()
print "Number of positive predicted reviews (threshold = 0.9): %s" % (predictions_with_high_threshold == 1).sum()<jupyter_output>Number of positive predicted reviews (threshold = 0.9): 25630
<jupyter_text>**Quiz Question**: What happens to the number of positive predicted reviews as the threshold increased from 0.5 to 0.9?## Exploring the associated precision and recall as the threshold variesBy changing the probability threshold, it is possible to influence precision and recall. We can explore this as follows:<jupyter_code># Threshold = 0.5
precision_with_default_threshold = graphlab.evaluation.precision(test_data['sentiment'],
predictions_with_default_threshold)
recall_with_default_threshold = graphlab.evaluation.recall(test_data['sentiment'],
predictions_with_default_threshold)
# Threshold = 0.9
precision_with_high_threshold = graphlab.evaluation.precision(test_data['sentiment'],
predictions_with_high_threshold)
recall_with_high_threshold = graphlab.evaluation.recall(test_data['sentiment'],
predictions_with_high_threshold)
print "Precision (threshold = 0.5): %s" % precision_with_default_threshold
print "Recall (threshold = 0.5) : %s" % recall_with_default_threshold
print "Precision (threshold = 0.9): %s" % precision_with_high_threshold
print "Recall (threshold = 0.9) : %s" % recall_with_high_threshold<jupyter_output>Precision (threshold = 0.9): 0.969527896996
Recall (threshold = 0.9) : 0.884463427656
<jupyter_text>**Quiz Question (variant 1)**: Does the **precision** increase with a higher threshold?
**Quiz Question (variant 2)**: Does the **recall** increase with a higher threshold?## Precision-recall curve
Now, we will explore various different values of tresholds, compute the precision and recall scores, and then plot the precision-recall curve.<jupyter_code>threshold_values = np.linspace(0.5, 1, num=100)
print threshold_values<jupyter_output>[ 0.5 0.50505051 0.51010101 0.51515152 0.52020202 0.52525253
0.53030303 0.53535354 0.54040404 0.54545455 0.55050505 0.55555556
0.56060606 0.56565657 0.57070707 0.57575758 0.58080808 0.58585859
0.59090909 0.5959596 0.6010101 0.60606061 0.61111111 0.61616162
0.62121212 0.62626263 0.63131313 0.63636364 0.64141414 0.64646465
0.65151515 0.65656566 0.66161616 0.66666667 0.67171717 0.67676768
0.68181818 0.68686869 0.69191919 0.6969697 0.7020202 0.70707071
0.71212121 0.71717172 0.72222222 0.72727273 0.73232323 0.73737374
0.74242424 0.74747475 0.75252525 0.75757576 0.76262626 0.76767677
0.77272727 0.77777778 0.78282828 0.78787879 0.79292929 0.7979798
0.8030303 0.80808081 0.81313131 0.81818182 0.82323232 0.82828283
0.83333333 0.83838384 0.84343434 0.84848485 0.85353535 0.85858586
0.86363636 0.86868687 0.87373737 0.87878788 0.88383838 0.88888889
0.89393939 0.8989899 0.9040404 0.90909091 0.[...]<jupyter_text>For each of the values of threshold, we compute the precision and recall scores.<jupyter_code>precision_all = []
recall_all = []
probabilities = model.predict(test_data, output_type='probability')
for threshold in threshold_values:
predictions = apply_threshold(probabilities, threshold)
precision = graphlab.evaluation.precision(test_data['sentiment'], predictions)
recall = graphlab.evaluation.recall(test_data['sentiment'], predictions)
precision_all.append(precision)
recall_all.append(recall)<jupyter_output><empty_output><jupyter_text>Now, let's plot the precision-recall curve to visualize the precision-recall tradeoff as we vary the threshold.<jupyter_code>import matplotlib.pyplot as plt
%matplotlib inline
def plot_pr_curve(precision, recall, title):
plt.rcParams['figure.figsize'] = 7, 5
plt.locator_params(axis = 'x', nbins = 5)
plt.plot(precision, recall, 'b-', linewidth=4.0, color = '#B0017F')
plt.title(title)
plt.xlabel('Precision')
plt.ylabel('Recall')
plt.rcParams.update({'font.size': 16})
plot_pr_curve(precision_all, recall_all, 'Precision recall curve (all)')<jupyter_output><empty_output><jupyter_text>**Quiz Question**: Among all the threshold values tried, what is the **smallest** threshold value that achieves a precision of 96.5% or better? Round your answer to 3 decimal places.<jupyter_code>thresh_idx = np.argmax(np.array(precision_all) > 0.965)
threshold_values[thresh_idx]<jupyter_output><empty_output><jupyter_text>**Quiz Question**: Using `threshold` = 0.98, how many **false negatives** do we get on the **test_data**? (**Hint**: You may use the `graphlab.evaluation.confusion_matrix` function implemented in GraphLab Create.)<jupyter_code>targets = test_data['sentiment']
predictions = apply_threshold(probabilities, .98)
confusion_matrix = graphlab.evaluation.confusion_matrix(targets, predictions)
print confusion_matrix
<jupyter_output>+--------------+-----------------+-------+
| target_label | predicted_label | count |
+--------------+-----------------+-------+
| -1 | 1 | 487 |
| 1 | 1 | 22269 |
| 1 | -1 | 5826 |
| -1 | -1 | 4754 |
+--------------+-----------------+-------+
[4 rows x 3 columns]
<jupyter_text>This is the number of false negatives (i.e the number of reviews to look at when not needed) that we have to deal with using this classifier.# Evaluating specific search termsSo far, we looked at the number of false positives for the **entire test set**. In this section, let's select reviews using a specific search term and optimize the precision on these reviews only. After all, a manufacturer would be interested in tuning the false positive rate just for their products (the reviews they want to read) rather than that of the entire set of products on Amazon.
## Precision-Recall on all baby related items
From the **test set**, select all the reviews for all products with the word 'baby' in them.<jupyter_code>baby_reviews = test_data[test_data['name'].apply(lambda x: 'baby' in x.lower())]<jupyter_output><empty_output><jupyter_text>Now, let's predict the probability of classifying these reviews as positive:<jupyter_code>probabilities = model.predict(baby_reviews, output_type='probability')<jupyter_output><empty_output><jupyter_text>Let's plot the precision-recall curve for the **baby_reviews** dataset.
**First**, let's consider the following `threshold_values` ranging from 0.5 to 1:<jupyter_code>threshold_values = np.linspace(0.5, 1, num=100)<jupyter_output><empty_output><jupyter_text>**Second**, as we did above, let's compute precision and recall for each value in `threshold_values` on the **baby_reviews** dataset. Complete the code block below.<jupyter_code>precision_all = []
recall_all = []
for threshold in threshold_values:
# Make predictions. Use the `apply_threshold` function
## YOUR CODE HERE
predictions = apply_threshold(probabilities, threshold)
confusion_matrix = graphlab.evaluation.confusion_matrix(baby_reviews['sentiment'],
predictions)
# Calculate the precision.
# YOUR CODE HERE
precision = graphlab.evaluation.precision(baby_reviews['sentiment'], predictions)
# precision= model.evaluate(baby_reviews, metric='precision')['precision']
# print "Test Precision: %s" % precision...
# YOUR CODE HERE
recall = graphlab.evaluation.recall(baby_reviews['sentiment'], predictions)
# print "Test Recall: %s" % recall
# Append the precision and recall scores.
precision_all.append(precision)
recall_all.append(recall)<jupyter_output><empty_output><jupyter_text>**Quiz Question**: Among all the threshold values tried, what is the **smallest** threshold value that achieves a precision of 96.5% or better for the reviews of data in **baby_reviews**? Round your answer to 3 decimal places.<jupyter_code>thresh_idx = np.argmax(np.array(precision_all) > 0.965)
threshold_values[thresh_idx]<jupyter_output><empty_output><jupyter_text>**Quiz Question:** Is this threshold value smaller or larger than the threshold used for the entire dataset to achieve the same specified precision of 96.5%?
**Finally**, let's plot the precision recall curve.<jupyter_code>plot_pr_curve(precision_all, recall_all, "Precision-Recall (Baby)")<jupyter_output><empty_output>
|
no_license
|
/module-9-precision-recall-assignment-blank.ipynb
|
JeffADaniels/UWash_Machine_Learning
| 28 |
<jupyter_start><jupyter_text># Introduction to Deep Learning with PyTorch
---
# Using PyTorch on Google Server
Google has made a version of Jupyter Notebook available online for **free** that allow us to use GPUs for faster training time! I do not recommend you use the local installation unless you don't have access to the internet.
Go to https://colab.research.google.com and sign in with your Google account. If you do not have a Google account you can create one. From there you can create a new notebook.# Install using Conda (for local installation - NOT Recommended)
`conda install pytorch torchvision -c pytorch`<jupyter_code># usual suspects
import os
import time
import shutil
import requests
import zipfile
from PIL import Image
from io import BytesIO
import numpy as np
import pandas as pd
from cycler import cycler
# the good stuff
import torch
import torch.nn as nn
from torch.autograd import Variable
import torch.nn.functional as F
from torch.utils.data import Dataset, DataLoader
import torchvision
from torchvision import datasets, models, transforms
# standard sklearn import
from sklearn.metrics import accuracy_score
# minor changes to plotting functions
import matplotlib.pyplot as plt
cmap=plt.cm.tab10
c = cycler('color', cmap(np.linspace(0,1,10)))
plt.rcParams["axes.prop_cycle"] = c
# see if GPU is available
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
%matplotlib inline
# Download all necessary files for the tutorial
data_url = 'https://www.dropbox.com/s/z0ilg0u8rsl2udz/pytorch_data.zip?dl=1'
if not os.path.exists('data'):
# Download the data zip file.
response = requests.get(data_url, stream=True)
zip_path = 'pytorch_data.zip'
with open(zip_path, 'wb') as f:
shutil.copyfileobj(response.raw, f)
# Unzip the file.
with zipfile.ZipFile(zip_path, 'r') as z:
z.extractall()
# Clean up.
os.remove(zip_path)<jupyter_output><empty_output><jupyter_text># PyTorch
What is PyTorch?
* It is a replacement for NumPy to use GPUs
* A deep learning platform built for flexibility and speed
## Tensor Overview
What are tensors?
Tensors are similar to NumPy's `ndarrays`
We normally think of tensors as a generalization of matrices. In fact, matrices are 2-D tensors!

Here is a great visualization of tensors from 1-D to 5-D

As mentioned before, since tensors are generalizations of matrices, we should be able to create them in similar ways. We can also expect *most* operations to stay the same. In particular, addition of tensors is the same as for matrices. Multiplication is a bit different, but we won't have to concern ourselves with that in this lecture.## Autograd - AKA why PyTorch is awesome
Central to all Neural Networks in PyTorch is the `autograd` package.
The `autograd` package provides automatic differentiation for all operations on Tensors. It is a define-by-run framework,
which means that your backprop is defined by how your code is run, and that every single iteration can be different.
Let's look at a basic example of this before returning to our Neural Networks again.<jupyter_code># Create a tensor and set requires_grad=True to track computation with it
x = torch.ones(2,2, requires_grad=True)
print(x)
# Do some operation on said tensor
y = 3*x + 7
print(y)
# Because y was created as a result of an operation, it now has a grad_fn method
y.grad_fn
# We can do more stuff to y (and thus x) and calculate its derivatives
z = 2*y**2
w = z.mean()
print(z, w)
# Backpropagation in one line!
w.backward()
# et voila!
x.grad<jupyter_output><empty_output><jupyter_text>---
# Neural NetsNeural networks (NNs) are special forms of nonlinear regressions where the decision system for which the NN is built mimics the way
the brain is supposed to work (whether it works like a NN is up for grabs of course).
Like many of the algorithms we have seen before, it is a supervised learning technique that can perform complex tasks.## PerceptronsThe basic building block of a neural network is a perceptron. A perceptron is like a neuron in a human brain. It takes inputs
(e.g. sensory in a real brain) and then produces an output signal. An entire network of perceptrons is called a neural net.
In general, a perceptron could have more or fewer inputs.Instead of assigning equal weight to each of the inputs, we can assign real numbers $w_1, w_2, \ldots$ expressing the importance of the respective inputs to the output. The nueron's output, 0 or 1, is determined whether the weighted sum $\sum_j w_j x_j$ is less than or greater than some *threshold value*.
Perceptrons may emit continuous signals or binary $(0,1)$ signals. In the case of a credit card application, the final perceptron is a binary one (approved or denied). Such perceptrons are implemented by means of squashing functions. For example, a really simple squashing function is one that issues a 1 if the function value is positive and a $-1$ if it is negative.To put this in more mathematical terms, let $z = \sum_{j=0}^n w_j x_j$ .
Then the *activation function* $\phi(z)$ is defined as
$$
\phi(z) =
\begin{cases}
-1 & \text{if } z < \theta\\
1 & \text{if } z \geq \theta
\end{cases}
$$The whole point of the perceptron is to mimic how a single nueron in the brain works: it either *fires* or it doesn't. Thus, the
perceptron rule is fairly simple and can be summarized by the following steps.
* Initialize the weights to zero or small random numbers
* For each training sample $\textbf{x}_n$ perform the following steps:
* Compute the output value $y$
* Calculate error in $y$ vs $\hat y$
* Update the weights
Here, the output value is the class label predicted by the activation function that we defined earlier, and the
simultaneous update of weight $w_j$ in the weight vector $\textbf{w}$ can be more formally written as
$$\bar w_j = w_j + \Delta w_j$$
## Fitting a model
Let's go back to our linear perceptron. It has the following parameters:
* $x_i$: inputs
* $y$ : output
* $w_i$: learned weights
What we would like to do is adjust the $w_i$'s until our model has the best fit.
First, initialize the $w_i$'s in some meaningful way (usually they're drawn from a randon uniform distribution).
Then, we put it into our usual **algorithm workflow:**
* calculate prediction $\hat y$
* calculate Loss function $L(y, \hat y)$
* update weights using backpropagation
### Loss function and backpropagation
To figure out how well our prediction was during each epoch, we'll use a basic loss function, mean squared error (MSE):
$L(y,\hat{y}) = ||~ y-\hat{y} ~||^2$,
ultimately trying to find $L_{\rm min}$, defined by the point in parameter space where $\nabla_{w_i} L = 0$.
Per-iteration update:
$ w_i \to w_i - \eta \nabla_{w_i} L $,
where $\eta$ is known as the learning rate; too small and takes very long to converge, too big and you oscillate about the minimum.
## A basic example
We'll build a Rube Goldberg adding machine that will illustrate how neural nets work.<jupyter_code>model = nn.Sequential(
nn.Linear(2, 1, bias=False))
optimizer = torch.optim.Adam(model.parameters())
criterion = nn.MSELoss()
print(model)
total_loss = []
num_samples = 10000
for num in range(1, num_samples+1):
# Progress bar indicator
if num % (num_samples//5) == 0:
print('{0}: %: {1:.3f}'.format(num,num/num_samples * 100))
# data prep
x = 4*torch.rand(2) #generate two random numbers uniformly on (0,4)
data, target = Variable(x), Variable(x[0] + x[1])
# Feed forward through NN
output = model(data)
loss = criterion(output, target)
total_loss.append(loss)
# Backpropagation
optimizer.zero_grad()
loss.backward()
optimizer.step()
fig,ax=plt.subplots(figsize=(11,8))
ax.plot(total_loss,marker='.',ls='',markersize=1.)
ax.set_ylim(0,);ax.set_xlim(0,);ax.grid(alpha=0.2);
ax.set_xlabel('training examples');ax.set_ylabel('mean squared loss');
x,y=np.linspace(-5,8,100),[]
for xx in x:
yy=model(torch.tensor([xx,2])).data.cpu().numpy()[0]
y.append(yy)
y=np.array(y)
fig,ax=plt.subplots(figsize=(11,8))
ax.plot([-5,8],[-3,10],lw=2.0,label='actual',alpha=0.7)
ax.fill_betweenx([-3,10],0,4,alpha=0.2)
ax.scatter(x,y,marker='.',s=3.,label='prediction',color='r')
ax.text(0.2,0,'Where we have \ntraining data')
ax.legend()
ax.set_ylim(-3,10);ax.set_xlim(-5,8);
ax.grid(alpha=0.2);
<jupyter_output><empty_output><jupyter_text>## Feedforward Neural Network

For our case of learning linear relationships, the modification to the linear regression architecture is depicted below:

where
$$\varphi(z) = \frac{1}{1+e^{-z}}$$
is the so-called sigmoid function; this is typically the activation function that is first introduced, I think because of historical reasons. In modern practice, it finds most of its use in transforming single outputs from a NN into a probability. It's worth noting that if your NN will output multiple probabilities, for example, if your NN will categorize between black cats, red cats, white cats, etc., a multi-dimensional generalization of the sigmoid, called the softmax function, is typically used.
The motivation behind adding an activation function is the hope that the NN model may capture non-linear relationships that exist in the data. Below are some commonly used activation functions.

In practice, a lot of architectures use the rectified linear unit (ReLU), along with it's close cousin, the so-called leaky-ReLU. In introducing this idea though, we'll focus on the sigmoid which maps real numbers from $(-\infty,\infty) \to [0,1]$.
Of course our data is linear in the case of a straight line (!) but let's see what happens if we try to force a non-linear activation layer to capture a linear relationship..## Non-linear model for a linear relationship
### Deep Feedforward Network with sigmoid activation<jupyter_code>model = nn.Sequential(
nn.Linear(2, 20),
nn.Sigmoid(),
nn.Linear(20, 20),
nn.Sigmoid(),
nn.Linear(20, 20),
nn.Sigmoid(),
nn.Linear(20, 1))
optimizer = torch.optim.Adam(model.parameters())
criterion = nn.MSELoss()
print(model)
total_loss = []
num_samples = 10000
for num in range(1, num_samples+1):
# Progress bar indicator
if num % (num_samples//5) == 0:
print('{0}: %: {1:.3f}'.format(num,num/num_samples * 100))
# data prep
x = 4*torch.rand(2) #generate two random numbers uniformly on (0,4)
data, target = Variable(x), Variable(x[0] + x[1])
# Feed forward through NN
output = model(data)
loss = criterion(output, target)
total_loss.append(loss)
# Backpropagation
optimizer.zero_grad()
loss.backward()
optimizer.step()
fig,ax=plt.subplots(figsize=(11,8))
ax.plot(total_loss,marker='.',ls='',markersize=.8)
ax.set_ylim(0,);ax.set_xlim(0,);ax.grid(alpha=0.2);
ax.set_xlabel('training examples');ax.set_ylabel('mean squared loss');
x,y=np.linspace(-5,8,100),[]
for xx in x:
yy=model(torch.tensor([xx,2])).data.cpu().numpy()[0]
y.append(yy)
y=np.array(y)
fig,ax=plt.subplots(figsize=(11,8))
ax.plot([-5,8],[-3,10],lw=2.0,label='actual',alpha=0.7)
ax.fill_betweenx([-3,10],0,4,alpha=0.2)
ax.scatter(x,y,marker='.',s=3.,label='prediction',color='r')
ax.text(0.2,0,'Where we have \ntraining data')
ax.legend()
ax.set_ylim(-3,10);ax.set_xlim(-5,8);
ax.grid(alpha=0.2);
ax.set_title('Sigmoid activation function');<jupyter_output><empty_output><jupyter_text>## ReLU activation function<jupyter_code>model = nn.Sequential(
nn.Linear(2, 20),
nn.ReLU(),
nn.Linear(20, 20),
nn.ReLU(),
nn.Linear(20, 20),
nn.ReLU(),
nn.Linear(20, 1))
optimizer = torch.optim.Adam(model.parameters())
criterion = nn.MSELoss()
print(model)
total_loss = []
num_samples = 10000
for num in range(1, num_samples+1):
# Progress bar indicator
if num % (num_samples//5) == 0:
print('{0}: %: {1:.3f}'.format(num,num/num_samples * 100))
# data prep
x = 4*torch.rand(2) #generate two random numbers uniformly on (0,4)
data, target = Variable(x), Variable(x[0] + x[1])
# Feed forward through NN
output = model(data)
loss = criterion(output, target)
total_loss.append(loss)
# Backpropagation
optimizer.zero_grad()
loss.backward()
optimizer.step()
fig,ax=plt.subplots(figsize=(11,8))
ax.plot(total_loss,marker='.',ls='',markersize=.8)
ax.set_ylim(0,);ax.set_xlim(0,);ax.grid(alpha=0.2);
ax.set_xlabel('training examples');ax.set_ylabel('mean squared loss');
x,y=np.linspace(-5,8,100),[]
for xx in x:
yy=model(torch.tensor([xx,2])).data.cpu().numpy()[0]
y.append(yy)
y=np.array(y)
fig,ax=plt.subplots(figsize=(11,8))
ax.plot([-5,8],[-3,10],lw=2.0,label='actual',alpha=0.7)
ax.fill_betweenx([-3,10],0,4,alpha=0.2)
ax.scatter(x,y,marker='.',s=3.,label='prediction',color='r')
ax.text(0.2,0,'Where we have \ntraining data')
ax.legend()
ax.set_ylim(-3,10);ax.set_xlim(-5,8);
ax.grid(alpha=0.2);
ax.set_title('ReLu activation function');<jupyter_output><empty_output><jupyter_text>## Teaching a machine to draw circles
Here the NN learns attempts to learn the 2d rotation matrix, parameterized by the generator of rotations in two dimensions:
$R={\begin{bmatrix}\cos \theta &-\sin \theta \\\sin \theta &\cos \theta \\\end{bmatrix}}$<jupyter_code># First some helper functions for plotting and model training
def train_models(models, optimizers, num_samples=1000, circle_interval=1.0, save_models=False, cuda=torch.cuda.is_available(), recurrent=False):
total_loss = []
for num in range(1, num_samples+1):
# progress indicator
if num % (num_samples//20) ==0:
print('{0}: %: {1:.3f}'.format(num, num/num_samples * 100))
# data calc
# take a random point on the circle of radius 1
x, theta = torch.ones(2), circle_interval*2*np.pi*torch.rand(1)
R = torch.zeros(2,2)
R[0,:] = torch.Tensor([np.cos(theta[0]),-np.sin(theta[0])])
R[1,:] = torch.Tensor([np.sin(theta[0]), np.cos(theta[0])])
data, target = Variable(theta), Variable(torch.mv(R,x))
# Check if GPU can be used
if cuda:
data, target = data.cuda(), target.cuda()
# learning phases
for idx, model in enumerate(models):
loss_iter = []
# forward
if recurrent:
output = model(data,None)
else:
output = model(data)
loss = criterion(output, target)
loss_iter.append(loss.data.item())
# backward
optimizers[idx].zero_grad()
loss.backward()
optimizers[idx].step()
total_loss.append(np.mean(loss_iter))
# save model state
if save_models:
for l,model in enumerate(models):
torch.save(model.state_dict(), 'rotations_{}.pth'.format(l))
return total_loss,theta
def plot_circles(models, offset=0, CI=False):
fig, axes = plt.subplots(figsize=(5*3,3.9),ncols=3)
x = torch.ones(2)
for k,ax in enumerate(axes):
ax.scatter(x[0],x[1], facecolors='none', edgecolors='r')
ax.scatter(x[0],x[1], facecolors='none', edgecolors='b')
x_real, y_real = [],[]
x_mean, y_mean = [],[]
x_std, y_std = [],[]
for theta in np.linspace((k+offset) *2*np.pi,(k+1+offset) *2*np.pi,300):
x_model,y_model = [],[]
# sythetic (real) data
data = Variable(torch.Tensor([theta]))#.cuda()
R = torch.zeros(2,2)
R[0,:] = torch.Tensor([np.cos(theta),-np.sin(theta)])
R[1,:] = torch.Tensor([np.sin(theta), np.cos(theta)])
real = torch.mv(R,x)
x_real.append(real[0].numpy())
y_real.append(real[1].numpy())
# predict w/ all models
for model in models:
if torch.cuda.is_available():
model.cpu()
outputs=model(data).data
xx_model, yy_model = outputs[0],outputs[1]
x_model.append(xx_model.numpy())
y_model.append(yy_model.numpy())
else:
outputs=model(data).data
xx_model, yy_model = outputs[0],outputs[1]
x_model.append(xx_model.numpy())
y_model.append(yy_model.numpy())
# summarize all model predictions
x_mean.append(np.mean(x_model))
y_mean.append(np.mean(y_model))
x_std.append(np.std(x_model))
y_std.append(np.std(y_model))
# plotting data
ax.scatter(x_real,y_real, facecolors='none', edgecolors='r',label='real data',s=2.)
ax.scatter(x_mean,y_mean, facecolors='none', edgecolors='k',label='model data', alpha=0.9,s=2.)
if CI:
ax.fill_betweenx(y_mean,x_mean-3*np.array(x_std),x_mean+3*np.array(x_std), alpha=0.1,color='b')
ax.fill_between(x_mean,y_mean-3*np.array(y_std),y_mean+3*np.array(y_std), alpha=0.1,color='b')
ax.legend()
ax.set_ylim(-2,2);ax.set_xlim(-2,2);ax.grid(alpha=0.3)
ax.set_title(r'${}\pi \leq \theta \leq {}\pi$'.format(2*(k+offset),2*(k+1+offset)),y=1.01);
return x_mean, y_mean, np.array(x_std), np.array(y_std)
def weight_init(m): # so-called xavier normalization https://arxiv.org/abs/1211.5063
if isinstance(m, nn.Linear):
size = m.weight.size()
fan_out = size[0]
fan_in = size[1]
variance = np.sqrt(2.0/(fan_in + fan_out))
m.weight.data.normal_(0.0, variance)
num_nodes=10
# Usually we define neural nets as a class in pytorch
class Rotations(nn.Module):
def __init__(self):
super(Rotations, self).__init__()
self.layer1 = nn.Sequential(
nn.Linear(1,num_nodes),
nn.Sigmoid(),
nn.Linear(num_nodes,2))
def forward(self, x):
out=self.layer1(x)
return out
model = Rotations().to(device)
optimizer = torch.optim.Adam(model.parameters())
criterion = nn.MSELoss()
print(model)
total_loss, theta = train_models([model], [optimizer], num_samples=200000, circle_interval=2.0)
fig,ax = plt.subplots(figsize=(11,8))
ax.plot(total_loss,marker='.',ls='',markersize=.8)
ax.set_ylim(0,);ax.set_xlim(0,);ax.grid(alpha=0.2);
ax.set_xlabel('training examples');ax.set_ylabel('mean squared loss');
output=plot_circles([model],offset=0,CI=False)<jupyter_output><empty_output><jupyter_text>### Will a deeper network perform better?<jupyter_code># Add more layers to the model and see if performance on the test set increases
num_nodes=10
class Rotations(nn.Module):
def __init__(self):
super(Rotations, self).__init__()
self.layer1 = nn.Sequential(
nn.Linear(1,num_nodes),
nn.Sigmoid(),
nn.Linear(num_nodes,num_nodes),
nn.Sigmoid(),
nn.Linear(num_nodes,num_nodes),
nn.Sigmoid(),
nn.Linear(num_nodes,2))
def forward(self, x):
out=self.layer1(x)
return out
model=Rotations().to(device)
optimizer=torch.optim.Adam(model.parameters())
criterion=nn.MSELoss()
print(model)
total_loss,theta=train_models([model],[optimizer],num_samples=150000,circle_interval=2.0)
fig,ax=plt.subplots(figsize=(7,5))
ax.plot(total_loss,marker='.',ls='',markersize=.8)
ax.set_ylim(0,);ax.set_xlim(0,);ax.grid(alpha=0.2);
ax.set_xlabel('training examples');ax.set_ylabel('mean squared loss');
output=plot_circles([model],offset=0,CI=False)<jupyter_output><empty_output><jupyter_text>## Bayesian approachOne simple approach to apply the bayesian method described by Yarin Gal, is to take multiple of the same models and train them independently. This allows each model to take independent paths through parameter space, usually finding their way near some optimal minima. In practice, this allows you to hedge the risk of getting stuck in some local minima and missing out on the global one, if it exists.
A visual way of understanding the situation of training a machine learning model, in general, is by considering a 3D surface plot where the x and y dimensions are two parameters you may modify, with the loss on the z axis, or height, which your aim is to minimize.

The surface that the data carves out in this space is predicated by the data; the aim of the model design is then to build a model flexible and robust enough to find the global minima, but not overly complex enough to overfit and get stuck at a local minima. Also, if your model is too simple, it can skip right over all the minima altogether, and not learn the nuance of the process described by the data. Having too high/low of a learning rate can also make the training process difficult.---
# Image Recognition and Transfer Learning
# Transfer Learning
Transfer learning is one of the most useful discoveries to come out of the computer vision community. Stated simply, transfer learning allows one model that was trained on different types of images, e.g. dogs vs cats, to be used for a different set of images, e.g. planes vs trains, while reducing the training time dramatically. When Google released ImageNet, they stated it took them over 14 **days** to train the model on some of the most powerful GPUs available at the time. Now, with transfer learning, we will train an, albeit smaller, model in less than 5 minutes.
The philosophy behind transfer learning is simple. We keep the "base" layers of a model frozen since the weights have already been tuned to identify patterns such as lines, circles, and other shapes, and insert layers at the end that will be tuned for the specific task at hand.


For our task, let's take a look at the King and Queen of the Miami food scene:
**The Cuban Sandwich**

**The Stuffed Arepa**
<jupyter_code>normalize = transforms.Normalize(mean=[0.485, 0.456, 0.406],
std=[0.229, 0.224, 0.225])
data_transforms = {
'train':
transforms.Compose([
transforms.Resize((224,224)),
transforms.RandomAffine(0, shear=10, scale=(0.8,1.2)),
transforms.RandomHorizontalFlip(),
transforms.ToTensor()]),
'validation':
transforms.Compose([
transforms.Resize((224,224)),
transforms.ToTensor()])}
image_datasets = {
'train':
datasets.ImageFolder('data/cva_data/train', data_transforms['train']),
'validation':
datasets.ImageFolder('data/cva_data/validation', data_transforms['validation'])}
dataloaders = {
'train':
torch.utils.data.DataLoader(
image_datasets['train'],
batch_size=32,
shuffle=True,
num_workers=4),
'validation':
torch.utils.data.DataLoader(
image_datasets['validation'],
batch_size=32,
shuffle=False,
num_workers=4)}
model = models.resnet50(pretrained=True).to(device)
for param in model.parameters():
param.requires_grad = False
# modify the final layer of resnet50 (called 'fc')
# originally model.fc = nn.Linear(2048, 1000) for the 1000 image classes
# we modify it to our specific needs
model.fc = nn.Sequential(
nn.Linear(2048, 128),
# inplace=True is a shortcut to not modify the forward method
nn.ReLU(inplace=True),
nn.Linear(128, 2)).to(device)
criterion = nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(model.fc.parameters())
def train_model(model, criterion, optimizer, num_epochs=3):
for epoch in range(num_epochs):
print(f'Epoch {epoch + 1}/{num_epochs}')
print('-' * 10)
for phase in ['train', 'validation']:
if phase == 'train':
model.train()
else:
model.eval()
running_loss = 0.0
running_corrects = 0
for inputs, labels in dataloaders[phase]:
inputs = inputs.to(device)
labels = labels.to(device)
outputs = model(inputs)
loss = criterion(outputs, labels)
if phase == 'train':
optimizer.zero_grad()
loss.backward()
optimizer.step()
_, preds = torch.max(outputs, 1)
running_loss += loss.item() * inputs.size(0)
running_corrects += torch.sum(preds == labels.data)
epoch_loss = running_loss / len(image_datasets[phase])
epoch_acc = running_corrects.double() / len(image_datasets[phase])
print(f'{phase} loss: {epoch_loss}, acc: {epoch_acc}')
return model
model_trained = train_model(model, criterion, optimizer, num_epochs=3)<jupyter_output><empty_output><jupyter_text>### Saving Models<jupyter_code>torch.save(model_trained.state_dict(),'models/cva_weights.h5')<jupyter_output><empty_output><jupyter_text>### Loading Models<jupyter_code>model = models.resnet50(pretrained=False).to(device)
model.fc = nn.Sequential(
nn.Linear(2048, 128),
nn.ReLU(inplace=True),
nn.Linear(128, 2)).to(device)
model.load_state_dict(torch.load('models/cva_weights.h5'))
model.eval() # stop training model<jupyter_output><empty_output><jupyter_text>### Make predictions on test images<jupyter_code>validation_img_paths = ["data/cva_data/validation/arepas/00000165.jpg",
"data/cva_data/validation/cubanos/00000037.jpg",
"data/cva_data/validation/cubanos/00000061.jpg",
"data/cva_data/validation/arepas/00000003.jpeg"]
img_list = [Image.open(img_path) for img_path in validation_img_paths]
validation_batch = torch.stack([data_transforms['validation'](img).to(device)
for img in img_list])
pred_logits_tensor = model(validation_batch)
pred_probs = F.softmax(pred_logits_tensor, dim=1).cpu().data.numpy()
fig, axs = plt.subplots(1, len(img_list), figsize=(20, 5))
for i, img in enumerate(img_list):
ax = axs[i]
ax.axis('off')
ax.set_title("{:.0f}% Arepa, {:.0f}% Cubano".format(100*pred_probs[i,0],
100*pred_probs[i,1]))
ax.imshow(img)
<jupyter_output><empty_output><jupyter_text># CNN from Scratch### `Datasets` and `Dataloaders`
In PyTorch, you'll usually create or import a `Dataset` subclass to represent your data. Once you've done that, you can use it to instantiate a `Dataloader` object which allows you to easily iterate over your training set in `BATCH_SIZE` chunks.<jupyter_code>image_size = 28
num_classes = 10
num_channels = 1
batch_size = 64
id_to_label = {
0 :'T-shirt/top',
1 :'Trouser',
2 :'Pullover',
3 :'Dress',
4 :'Coat',
5 :'Sandal',
6 :'Shirt',
7 :'Sneaker',
8 :'Bag',
9 :'Ankle boot'}
class FashionDataset(Dataset):
def __init__(self, path,
image_size, num_channels, image_transform=None):
self.num_channels = num_channels
self.image_size = image_size
self.image_transform = image_transform
data_df = pd.read_csv(path)
self.X = data_df.values[:, 1:]
self.X = self.X.reshape(-1, image_size, image_size, num_channels)
self.X = self.X.astype('float32')
self.y = data_df.values[:, 0]
def __getitem__(self, index):
batch_X, batch_y = self.X[index], self.y[index]
if self.image_transform is not None:
batch_X = self.image_transform(batch_X)
return batch_X, batch_y
def __len__(self):
return len(self.X)
# This simple transform coverts the image from an numpy array
# to a PyTorch tensor and remaps its values from 0-255 to 0-1.
# Many other types of transformations are available, and they
# can easily be composed into a pipeline. For more info see:
# https://pytorch.org/docs/stable/torchvision/transforms.html
image_transform = transforms.Compose([transforms.ToTensor()])
train_dataset = FashionDataset(
'fashionmnist/fashion-mnist_train.csv',
image_size,
num_channels,
image_transform)
train_dataloader = DataLoader(
dataset=train_dataset,
batch_size=batch_size,
shuffle=True)
val_dataset = FashionDataset(
'fashionmnist/fashion-mnist_test.csv',
image_size,
num_channels,
image_transform)
val_dataloader = DataLoader(
dataset=train_dataset,
batch_size=batch_size)<jupyter_output><empty_output><jupyter_text>### Show some examples from the training set<jupyter_code>def plot_images(data, labels, image_size):
fig, axes = plt.subplots(
1, data.shape[0], figsize=(16, 4),
subplot_kw={'xticks':[], 'yticks':[]},
gridspec_kw=dict(hspace=0.1, wspace=0.1))
for i, ax in enumerate(axes.flat):
ax.imshow(data[i].reshape(image_size, image_size), cmap='binary')
ax.set_xlabel(labels[i])
images = train_dataset.X[:10]
class_ids = train_dataset.y[:10]
class_labels = [id_to_label[class_id] for class_id in class_ids]
plot_images(images, class_labels, image_size)<jupyter_output><empty_output><jupyter_text>## ModelingThis baseline network uses a single convolution with only 4 filters. Because of the simplicity of our dataset, it still manages to achieve nearly 90% accuracy after only 5 epochs.
Experiment and see if you can increase the accuracy on the validation set to above 95%.
Things you might try:
* Increasing the number of filters per convolution.
* Adding more convolutions.
* Adding a `BatchNorm2d` layer after `Conv2d`.
* Increasing the number of epochs.
* Changing the kernel size.
* Using different types of pooling or using stride > 1 in convolutional layers instead of pooling.
You might also find the [PyTorch API reference](https://pytorch.org/docs/stable/nn.html) useful.<jupyter_code>class FashionModel(nn.Module):
def __init__(self, num_channels, num_classes):
super().__init__()
self.conv_1 = nn.Sequential(
nn.Conv2d(1, 16, kernel_size=3, stride=1, padding=1),
nn.BatchNorm2d(16),
nn.ReLU(inplace=True),
nn.MaxPool2d(kernel_size=2, stride=2, padding=0))
# Add more conv2d layers
# recall that the input size for the new layer is 16!
# Make the outputs 32 and 64 to create 3 total convolution layers
# ------------- CODE GOES HERE ------------- #
# to determine the correct inputs for the last layer you'll need the formulas:
# output_size of conv2d = (image_width - kernel_size + 2*padding)/stride + 1
# output_size of maxpool = (output_size of conv2d - kernel_size + 2*padding)/stride + 1
# then it's the number of size of output, e.g., 16, 32, 64, times the output size of maxpool
# example with first conv layer:
# input_width = 28 (our images are square otherwise we would have to do this for width/height independently)
# output width after convolution = (28 - 3 + 2*1)/1 + 1 = 28
# output width after maxpool = (28 - 2 + 2*0)/2 + 1 = 14
# so since we have 16 output channels, our input to the linear layer is 14 * 14 * 16
self.linear = nn.Linear(14 * 14 * 16, num_classes)
def forward(self, x):
x = self.conv_1(x)
# Uncomment as more convolution layers are added
# x = self.conv_2(x)
# x = self.conv_3(x)
x = x.reshape(x.size(0), -1)
x = self.linear(x)
return x
# Instantiate the model.
model = FashionModel(num_channels, num_classes)
# Send the model's tensors to the GPU (if available).
model = model.to(device) <jupyter_output><empty_output><jupyter_text>## Training<jupyter_code>num_epochs = 10
log_freq = 100
checkpoint_path = 'checkpoint.pickle'
optimizer = torch.optim.Adam(model.parameters())
criterion = nn.CrossEntropyLoss()
for epoch in range(1, num_epochs + 1):
model.train() # Switch to training mode.
print(f'Starting epoch {epoch}.')
epoch_start_time = time.time()
running_loss = 0.0
running_accuracy = 0.0
for batch_id, (batch_X, batch_y) in enumerate(train_dataloader):
batch_X = batch_X.to(device)
batch_y = batch_y.to(device)
output = model(batch_X)
loss = criterion(output, batch_y)
optimizer.zero_grad()
loss.backward()
optimizer.step()
# Periodically print the loss and prediction accuracy.
running_loss += loss.item()
y_pred = output.argmax(dim=-1)
running_accuracy += accuracy_score(batch_y.cpu(), y_pred.cpu())
if batch_id % log_freq == log_freq - 1:
average_loss = running_loss / log_freq
average_accuracy = running_accuracy / log_freq
print(f'Mini-batch: {batch_id + 1}/{len(train_dataloader)} '
f'Loss: {average_loss:.5f} Accuracy: {average_accuracy:.5f}')
running_loss = 0.0
running_accuracy = 0.0
# Log elapsed_time for the epoch.
elapsed_time = time.time() - epoch_start_time
print(f'\nEpoch {epoch} completed in {elapsed_time // 60:.0f} minutes '
f'{elapsed_time % 60:.0f} seconds.')
# Calculate and log loss on validation set.
with torch.no_grad():
model.eval()
running_loss = 0.0
running_accuracy = 0.0
for batch_id, (batch_X, batch_y) in enumerate(val_dataloader):
batch_X = batch_X.to(device)
batch_y = batch_y.to(device)
output = model(batch_X)
loss = criterion(output, batch_y)
running_loss += loss.item()
y_pred = output.argmax(dim=-1)
running_accuracy += accuracy_score(batch_y.cpu(), y_pred.cpu())
average_loss = running_loss / len(val_dataloader)
average_accuracy = running_accuracy / len(val_dataloader)
print(f'Val Loss: {average_loss:.5f} Val Accuracy: {average_accuracy:.5f}\n')<jupyter_output><empty_output><jupyter_text># Semi-Supervised Learning - Style Transfer
Style transfer, to me, is one of the coolest "discoveries" in the computer vision and deep learning community from the past few years. In essence, it allows us to take the "content" from an image (shapes, objects, arrangements) and reproduce a new target that is in the "style" (style, colors, textures) of another.
We'll be taking inspiration from the paper, [Image Style Transfer Using Convolutional Neural Networks, by Gatys](https://www.cv-foundation.org/openaccess/content_cvpr_2016/papers/Gatys_Image_Style_Transfer_CVPR_2016_paper.pdf), and implementing the model in PyTorch.
In the paper, style transfer uses the features found in the 19-layer VGG Network, which are comprised of a series of convolutional and pooling layers, and a few fully-connected layers. (Recall that this is similar to many of the computer vision models discussed earlier)
In the image below, the convolutional layers are named by stack and their order in the stack.
For example, `Conv_1_1` is the first convolutional layer that an image is passed through in the *first* stack. `Conv_2_1` is the first convolutional layer in the *second* stack. The deepest convolutional layer in the network is `Conv_5_4`.
Style transfer relies on separating the content and style of an image. To do so, we aim to create a new **target** image which should contain our desired content and style components.
**Note:**
* objects and their arrangement are similar to that of the **content image**
* style, colors, and textures are similar to that of the **style image**
## Load VGG19
VGG19 is split into two portions:
* `vgg19.features` - contains all the convolutional and pooling layers
* `vgg19.classifier` - contains the three fully connected layers layers at the end
We only need the `features` portion, which we're going to load in and "freeze" the weights of. This is similar to what we did for our transfer learning section.<jupyter_code># get the "features" portion of VGG19
vgg = models.vgg19(pretrained=True).features
# freeze all VGG parameters since we're only optimizing the target image
for param in vgg.parameters():
param.requires_grad_(False)
# move the model to GPU
vgg.to(device)<jupyter_output><empty_output><jupyter_text>### Load in Content and Style Images
Load in any images you want! The code below is a helper function for loading in any type and size of image. The `load_image` function also converts images to normalized Tensors.<jupyter_code>def load_image(img_path, max_size=400, shape=None):
''' Load in and transform an image, making sure the image
is <= 400 pixels in the x-y dims.'''
if "http" in img_path:
response = requests.get(img_path)
image = Image.open(BytesIO(response.content)).convert('RGB')
else:
image = Image.open(img_path).convert('RGB')
# large images will slow down processing
if max(image.size) > max_size:
size = max_size
else:
size = max(image.size)
if shape is not None:
size = shape
in_transform = transforms.Compose([
transforms.Resize(size),
transforms.ToTensor(),
transforms.Normalize((0.485, 0.456, 0.406),
(0.229, 0.224, 0.225))])
# discard the transparent, alpha channel (that's the :3) and add the batch dimension
image = in_transform(image)[:3,:,:].unsqueeze(0)
return image
# load in content and style image
content = load_image('data/img/lion.jpg').to(device)
# Resize style to match content, makes code easier
style = load_image('data/img/magritte-la-belle-captive.jpg', shape=content.shape[-2:]).to(device)
# helper function for un-normalizing an image
# and converting it from a Tensor image to a NumPy image for display
def im_convert(tensor):
""" Display a tensor as an image. """
image = tensor.to("cpu").clone().detach()
image = image.numpy().squeeze()
image = image.transpose(1,2,0)
image = image * np.array((0.229, 0.224, 0.225)) + np.array((0.485, 0.456, 0.406))
image = image.clip(0, 1)
return image
# display the images
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(20, 10))
# content and style ims side-by-side
ax1.imshow(im_convert(content))
ax2.imshow(im_convert(style))<jupyter_output><empty_output><jupyter_text>## VGG19 Layers
To get the content and style representations of an image, we have to pass an image forward throug the VGG19 network until we get to the desired layer(s) and then get the output from that layer.<jupyter_code># print out VGG19 model so you can see the names of all the layers
print(vgg)<jupyter_output><empty_output><jupyter_text>## Content and Style Features
Below, complete the mapping of layer names to the names found in the paper for the _content representation_ and the _style representation_.<jupyter_code>def get_features(image, model, layers=None):
""" Run an image forward through a model and get the features for
a set of layers. Default layers are for VGGNet matching Gatys et al (2016)
"""
## Here we gather the layers need to preserve style and content of an image
if layers is None:
layers = {'0': 'conv1_1',
'5': 'conv2_1',
'10': 'conv3_1',
'19': 'conv4_1',
'21': 'conv4_2', ## content representation
'28': 'conv5_1'}
features = {}
x = image
# model._modules is a dictionary holding each module in the model
for name, layer in model._modules.items():
x = layer(x)
if name in layers:
features[layers[name]] = x
return features<jupyter_output><empty_output><jupyter_text>## Gram Matrix
The output of every convolutional layer is a Tensor with dimensions associated with the `batch_size`, a depth, `d` and some height and width (`h`, `w`). The Gram matrix of a convolutional layer can be calculated as follows:
* Get the depth, height, and width of a tensor using `batch_size, d, h, w = tensor.size`
* Reshape that tensor so that the spatial dimensions are flattened
* Calculate the gram matrix by multiplying the reshaped tensor by it's transpose <jupyter_code>def gram_matrix(tensor):
""" Calculate the Gram Matrix of a given tensor
Gram Matrix: https://en.wikipedia.org/wiki/Gramian_matrix
"""
# get the batch_size, depth, height, and width of the Tensor
_, d, h, w = tensor.size()
# reshape so we're multiplying the features for each channel
tensor = tensor.view(d, h * w)
# calculate the gram matrix
gram = tensor @ tensor.t()
return gram <jupyter_output><empty_output><jupyter_text>## We're Almost there!
Here's what our helper functions do:
* Extract the features for our content and style images from VGG19
* Compute the Gram Matrix for a given convolutional layer
What we need:
* Put it all together!<jupyter_code># get content and style features only once before training
content_features = get_features(content, vgg)
style_features = get_features(style, vgg)
# calculate the gram matrices for each layer of our style representation
style_grams = {layer: gram_matrix(style_features[layer]) for layer in style_features}
# create a third "target" image and prep it for change
# it is a good idea to start off with the target as a copy of our *content* image
# then iteratively change its style
target = content.clone().requires_grad_(True).to(device)<jupyter_output><empty_output><jupyter_text>## Loss and Weights
#### Individual Layer Style Weights
In the script below, we have the option to weight the style representation at each relevant layer. This will allow us to fine tune what effect size we want for each layer - earlier layers have larger style artifacts and later layers place emphasis on smaller features. Remember, each layer is a different size and by combining them we can create multi-scale style representations.
The paper suggests using a range between 0-1 to weight the layers.
#### Content and Style Weight
The paper defines a **style ratio** of $\alpha/\beta$, where $\alpha$ is the `content_weight` and $\beta$ is the `style_weight`. This ratio will affect how _stylized_ the final image is. It's recommended that to leave the content_weight = 1 and set the style_weight to achieve the ratio needed for a desired effect style. Note that this is not exact science, there will be lots of tuning of the ratio and the weights of the layers to get a result we're pleased with.
Remember - the reason this is called "Semi-Supervised" is because there is no right answer. We decide when to stop the training based on intermediate results we plot and stop when we are happy with what we see.<jupyter_code># weights for each style layer
# weighting earlier layers more will result in *larger* style artifacts
# notice we are excluding `conv4_2` our content representation
style_weights = {'conv1_1': 1.,
'conv2_1': 0.75,
'conv3_1': 0.2,
'conv4_1': 0.2,
'conv5_1': 0.2}
content_weight = 1 # alpha
style_weight = 1e6 # beta<jupyter_output><empty_output><jupyter_text>## Update the Target & Calculate Losses
Like in every training loop we've seen before, we need to decide on how many passes we want to do on our model (via gradient descent). The difference here is that we will be changing our **target** image and nothing about the VGG19 model or our original content and style images. Since this is semi-supervised, the number of steps to choose is up to you. Keep in mind that after ~50,000 steps you probably won't see any noticeable differences in the images and by using ~2,000 steps you can see early on whether the style ratio ($\alpha/\beta$) is giving the desired effect.
Experiment with different weights or images to see some really cool effects!
#### Content Loss
The content loss will be the mean squared difference between the target and content features at layer `conv4_2`. This can be calculated as follows: (see paper)
```
content_loss = torch.mean((target_features['conv4_2'] - content_features['conv4_2'])**2)
```
#### Style Loss
The style loss is calculated in a similar way, but we have to iterate through the layers specified by name in our dictionary `style_weights`.
> Calculate the gram matrix for the target image, `target_gram` and style image `style_gram` at each of these layers and compare those gram matrices, calculating the `layer_style_loss`.
#### Total Loss
Finally, the total loss is calculated by adding up the individual style and content losses and weighting them with the specified alpha and beta values chosen.
Intermittently, we'll print out an intermediate image and its loss - don't be alarmed if the loss is very large! It takes some time for an image's style to change and you should focus on the appearance of your target image rather than any loss value, but we should still be seeing the loss go down over time.<jupyter_code># for displaying the target image, intermittently
show_every = 400
# iteration hyperparameters
optimizer = torch.optim.Adam([target], lr=0.003)
steps = 2000 # decide how many iterations to update your image
for step in range(1, steps+1):
# get the features from your target image
target_features = get_features(target, vgg)
# the content loss
content_loss = torch.mean((target_features['conv4_2'] - content_features['conv4_2'])**2)
# the style loss
# initialize the style loss to 0
style_loss = 0
# then add to it for each layer's gram matrix loss
for layer in style_weights:
# get the "target" style representation for the layer
target_feature = target_features[layer]
target_gram = gram_matrix(target_feature)
_, d, h, w = target_feature.shape
# get the "style" style representation
style_gram = style_grams[layer]
# the style loss for one layer, weighted appropriately
layer_style_loss = style_weights[layer] * torch.mean((target_gram - style_gram)**2)
# add to the style loss
style_loss += layer_style_loss / (d * h * w)
# calculate the *total* loss
total_loss = content_weight * content_loss + style_weight * style_loss
# update your target image
optimizer.zero_grad()
total_loss.backward()
optimizer.step()
# display intermediate images and print the loss
if step % show_every == 0:
print('Total loss: ', total_loss.item())
plt.imshow(im_convert(target))
plt.show()<jupyter_output><empty_output><jupyter_text>## Display the Target Image<jupyter_code># display content and target image
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(20, 10))
ax1.imshow(im_convert(content))
ax2.imshow(im_convert(target))
# display style and target image
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(20, 10))
ax1.imshow(im_convert(style))
ax2.imshow(im_convert(target))<jupyter_output><empty_output><jupyter_text>---
# RNN Character Level Generation
Text generation is a fun way to familiarize yourself with Recurrent Neural Nets.
In this notebook, we will deal with **character-level** text generation and why they can be just as useful as word-level text generation.
For this example, we'll be using text files to generate code similar to our input. In other words, if we put in Trump tweets, our generator should output words that sound like Trump.
### Our current understanding of RNNs

### Reminder

#### "RNNs have a hidden state that feeds back into the cell at the next time step"## What is actually going on?
### Example with sequence length 5### What about the backwards pass?## Preprocessing<jupyter_code># install unidecode
!pip install unidecode
# Read in text and change unicode characters to ASCII
import unidecode
import string
import random
import re
all_characters = string.printable
n_characters = len(all_characters)
# read in file to train RNN
file = unidecode.unidecode(open('data/shakespeare.txt').read())
file_len = len(file)
print(f'file_len = {file_len}')<jupyter_output><empty_output><jupyter_text>To give our model inputs from this large string of text, we'll split it up into chunks<jupyter_code>chunk_len = 400
def random_chunk():
start_index = random.randint(0, file_len - chunk_len)
end_index = start_index + chunk_len + 1
return file[start_index:end_index]
print(random_chunk())<jupyter_output><empty_output><jupyter_text>## Build Model
This model will take as input the character for step $t$ and is expected to output the next character for step $t+1$. There are three layers - one linear layer that encodes the input character into an internal state, one GRU layer (which may itself have multiple layers) that operates on that internal state and a hidden state, and a decoder layer that outputs the probability distribution.<jupyter_code>class RNN(nn.Module):
def __init__(self, input_size, hidden_size, output_size, n_layers=1):
super(RNN, self).__init__()
self.input_size = input_size
self.hidden_size = hidden_size
self.output_size = output_size
self.n_layers = n_layers
self.encoder = nn.Embedding(input_size, hidden_size)
self.gru = nn.GRU(hidden_size, hidden_size, n_layers)
self.decoder = nn.Linear(hidden_size, output_size)
def forward(self, input, hidden):
output = self.encoder(input.view(1, -1))
output, hidden = self.gru(output.view(1, 1, -1), hidden)
output = self.decoder(output.view(1, -1))
return output, hidden
def init_hidden(self):
return Variable(torch.randn(self.n_layers, 1, self.hidden_size))<jupyter_output><empty_output><jupyter_text>## Inputs and Targets
Now that we've defined our model, we need to give it both input data, via our chunks, and our target data. Each character is one-hot encoded to the vocab size <jupyter_code>def char2tensor(string):
tensor = torch.zeros(len(string)).long()
for char in range(len(string)):
if string[char] in all_characters:
tensor[char] = all_characters.index(string[char])
else:
tensor[char] = 94 #predict space if character unknown
return Variable(tensor)
# Let's see it in action.
print(char2tensor('Metis0123abczABC'))<jupyter_output><empty_output><jupyter_text>Now that we can generate chunks of data, we can build our inputs and targets.
Our inputs will be all of the chunk except for the last letter.
Our target will be all of the chunk except for the first letter.<jupyter_code>def random_training_set():
chunk = random_chunk()
inp = char2tensor(chunk[:-1])
target = char2tensor(chunk[1:])
return inp, target<jupyter_output><empty_output><jupyter_text>## Evaluating the Model
To evaluate the network we will feed one character at a time, use the outputs of the network as a probability distribution for the next character, and repeat. To start generation we pass a priming string to start building up the hidden state, from which we then generate one character at a time.<jupyter_code>def evaluate(model, prime_str='A', predict_len=100, temperature=0.8):
hidden = model.init_hidden()
prime_input = char2tensor(prime_str)
predicted = prime_str
# use priming string to build up hidden state
for p in range(len(prime_str) - 1):
_, hidden = model(prime_input[p], hidden)
inp = prime_input[-1]
for p in range(predict_len):
output, hidden = model(inp, hidden)
# Sample from the network as a multinomial distribution
output_dist = output.data.view(-1).div(temperature).exp()
top_i = torch.multinomial(output_dist, 1)[0]
# Add predicted character to string and use as next input
predicted_char = all_characters[top_i]
predicted += predicted_char
inp = char2tensor(predicted_char)
return predicted<jupyter_output><empty_output><jupyter_text>## Training<jupyter_code># helper function
import time, math
def time_since(since):
s = time.time() - since
m = math.floor(s/60)
s -= m*60
return '%dm %ds' % (m, s)
# The actual training part
def train(inp, target):
hidden = model.init_hidden()
model.zero_grad()
loss = 0
for char in range(chunk_len):
output, hidden = model(inp[char], hidden)
loss += criterion(output, target[char].unsqueeze(0))
loss.backward()
model_optimizer.step()
return loss.data.item() / chunk_len
# parameters
n_epochs = 1000
print_every = 100
plot_every = 10
hidden_size = 256
n_layers = 2
learning_rate = 0.001
# model declaration
model = RNN(n_characters, hidden_size, n_characters, n_layers)
model_optimizer = torch.optim.Adam(model.parameters(), lr=learning_rate)
criterion = nn.CrossEntropyLoss()
start = time.time()
all_losses = []
loss_avg = 0
for epoch in range(1, n_epochs + 1):
loss = train(*random_training_set())
loss_avg += loss
if epoch % print_every == 0:
print('[%s (%d %d%%) %.4f]' % (time_since(start), epoch, epoch / n_epochs * 100, loss))
print(evaluate(model, 'A ', 100), '\n')
if epoch % plot_every == 0:
all_losses.append(loss_avg / plot_every)
loss_avg = 0
import matplotlib.pyplot as plt
import matplotlib.ticker as ticker
%matplotlib inline
plt.figure()
plt.plot(all_losses)<jupyter_output><empty_output><jupyter_text>### Load pre-trained models<jupyter_code>trump = torch.load('data/models/potus.pt')
billy = torch.load('data/models/shakespeare.pt')
cells = torch.load('data/models/cell.pt')
# Evaluate Trump
print(evaluate(trump, 'covfefe ', predict_len=240, temperature=0.5))
# Evaluate Billy Shakespeare
print(evaluate(billy, 'To be or not to be: ', predict_len=200, temperature=0.5))
# Evaluate NLP model of math biology latex file
print(evaluate(cells, 'Vascular ', predict_len=200, temperature=0.6))<jupyter_output><empty_output>
|
no_license
|
/Deep_Neural_Nets.ipynb
|
lenakchen/pytorch_tutorial
| 32 |
<jupyter_start><jupyter_text>## NFL Punt Analytics Proposal ##
Thirty seven known punt-related concussions occurred during the 2016 and 2017 NFL seasons. By reviewing the evidence from all 6,681 punt plays that are recorded from these two seasons, I seek to show that the following three rule changes will improve player safety while being implementable and preserving the excitement and unpredictability of the great game of football.
* [Proposal #1](#proposal_1): Award a five yard bonus to the receiving team on a fair catch.
* [Proposal #2](#proposal_2): Require single coverage of gunners by the receiving team.
* [Proposal #3](#proposal_3): Install helmet sensors to monitor deceleration.
In support of proposal #1, **I attempt to quantify the risk-reward tradeoff of reducing the number of punt returns and increasing the number of fair catches**. I find that 32% of punt returns conditional on a punt received yield fewer than five yards on the return. In the current framework, punt returners have an incentive to attempt a return whenever they think their expected value of returning is greater than 0 yards. In the new framework, punt returns will have an incentive to make a return only when they believe the expected value of returning is greater than 5 yards. The concussion rate is 10 injuries per 1000 plays for a fielded return versus 1.8 injuries per 1000 plays for a fair catch, so we expect a 80% decrease in concussions for each incremental fair catch that is called. On the other hand, punt returners will continue to be able to try to go for a return and make a play if they see an opportunity to go for it.
In support of proposal #2, **I analyze the injury rate conditioning on both the choice of coverage and on the yards between the line of scrimmage and the end zone, as these factors are not independent of one another**. Teams are more likely to choose double coverage when there is more open field, i.e. greater yards to go between the kicking team and the receiving team's end zone. On the other hand, plays where the field is longer also have higher injury rates, perhaps because the possibility of a touchback or coffin corner is reduced and the coverage team has to run farther to reach the spot of punt reception, resulting in a more open field and higher speed play. I show that even when one conditions on yards-to-go, double coverage generates somewhat higher injury rates than single coverage in these long field situations. On the other hand, double coverage does not appear to help the receiving team make longer punt returns, i.e. the punt returner is able to generate just as much excitement whether or not the gunners were single covered or not. Enforcing single coverage would then appear to reduce injuries without limiting excitement, and so seems like a costless design proposal, although the statistical significance of these findings (recall there are only 37 concussion events in this entire sample) is limited by the small sample size.
In support of proposal #3, **I show that deceleration appears more important than velocity in determining injury, but that the NGS data is inherently limited in its ability to measure deceleration.**. The code below attempts to measure each player's velocity and deceleration at the time of a tackle event. I find that players who are not injured on a play have velocity as great as or greater than players involved in an injury generating tackle. On the other hand, the injured player experiences much greater deceleration than uninjured players. This suggests that deceleration is a more important factor in injury than velocity. The NGS data provides (x, y) coordinates at 100 millisecond resolution, so a player who decelerates from 9.8 m/s to 0.0 m/s would be recorded as having experienced at most 10 g's in deceleration, since 10g = (9.8m/s - 0.0m/s) / 0.1s. In fact the player could have experienced much greater deceleration, e.g. he could have decelerated to 0 in 20 milliseconds, but the resolution of the NGS system is not granular enough to show this. For the purpose of measuring head injury, the deceleration experienced at the head would seem to be a critical indicator of whether injury is likely to have occurred. This data would be collected not to take players out of the game or to limit their playing time, but rather to better study the conditions and plays that result in greater head deceleration and likely injury.
The notebook below provides the analysis and figures that support each of these three proposals. There is an extensive amount of formatting code and data preparation code, which I have attempted to hide, so that the focus can be on the interesting parts - the output and figures.
[Additional observations](#additional_observations) are found at the end.
[Final thoughts](#final_thoughts) concludes.
<jupyter_code>%matplotlib inline
import os
import sys
import re
import pandas as pd
import numpy as np
import glob
import os
import logging
import sys
import re
import matplotlib
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings('ignore')
ngs_files = ['../input/NGS-2016-pre.csv',
'../input/NGS-2016-reg-wk1-6.csv',
'../input/NGS-2016-reg-wk7-12.csv',
'../input/NGS-2016-reg-wk13-17.csv',
'../input/NGS-2016-post.csv',
'../input/NGS-2017-pre.csv',
'../input/NGS-2017-reg-wk1-6.csv',
'../input/NGS-2017-reg-wk7-12.csv',
'../input/NGS-2017-reg-wk13-17.csv',
'../input/NGS-2017-post.csv']
PUNT_TEAM = set(['GL', 'PLW', 'PLT', 'PLG', 'PLS', 'PRG', 'PRT', 'PRW', 'PC',
'PPR', 'P', 'GR'])
RECV_TEAM = set(['VR', 'PDR', 'PDL', 'PLR', 'PLM', 'PLL', 'VL', 'PFB', 'PR'])
<jupyter_output><empty_output><jupyter_text>### **Proposal 1: **Award five yard bonus on fair catches ###
**Fair catches are 80% safer than punt received events.**
The graph below shows that the concussion rate is 10.0 per 1000 returns on punt received events vs. 1.8 on fair catch events. While fair catches are not as exciting as punt returns, they are certainly much safer. Our goal should not be to ban all punt returns wholesale, but rather to reduce the number of dangerous but less exciting (i.e. short) punt returns while preserving the potential for long punt returns (ESPN highlight material). There is always risk in each play (even fair catches), and the analysis below helps us to find the best trade off between risk and reward.
<jupyter_code>plays_df = pd.read_csv('../input/play_information.csv')
def get_return_yards(s):
m = re.search('for ([0-9]+) yards', s)
if m:
return int(m.group(1))
elif re.search('for no gain', s):
return 0
else:
return np.nan
plays_df['Return'] = plays_df['PlayDescription'].map(
lambda x: get_return_yards(x))
video_review = pd.read_csv('../input/video_review.csv')
video_review = video_review.rename(columns={'GSISID': 'InjuredGSISID'})
plays_df= plays_df.merge(video_review, how='left',
on=['Season_Year', 'GameKey', 'PlayID'])
plays_df['InjuryOnPlay'] = 0
plays_df.loc[plays_df['InjuredGSISID'].notnull(), 'InjuryOnPlay'] = 1
plays_df = plays_df[['Season_Year', 'GameKey', 'PlayID', 'Return', 'InjuryOnPlay']]
ngs_df = []
for filename in ngs_files:
df = pd.read_csv(filename, parse_dates=['Time'])
df = df.loc[df['Event'].isin(['fair_catch', 'punt_received'])]
df = pd.concat([df, pd.get_dummies(df['Event'])], axis=1)
df = df.groupby(['Season_Year', 'GameKey', 'PlayID'])[['fair_catch', 'punt_received']].max()
ngs_df.append(df.reset_index())
ngs_df = pd.concat(ngs_df)
plays_df = plays_df.merge(ngs_df, on=['Season_Year', 'GameKey', 'PlayID'])
injury_per_1000_fair_catch = 1000 * plays_df.loc[plays_df['fair_catch']==1,
'InjuryOnPlay'].mean()
injury_per_1000_punt_received = 1000 * plays_df.loc[plays_df['punt_received']==1,
'InjuryOnPlay'].mean()
fig = plt.figure()
ax = plt.subplot2grid((1, 1), (0, 0))
plt.bar([0, 1], [injury_per_1000_fair_catch, injury_per_1000_punt_received])
ax.set_xticks([0, 1])
ax.set_xticklabels(['Fair Catch', 'Punt Received'])
plt.text(0, injury_per_1000_fair_catch+0.2, '{:.1f}'.format(injury_per_1000_fair_catch))
plt.text(1, injury_per_1000_punt_received+0.2, '{:.1f}'.format(injury_per_1000_punt_received))
plt.title("Concussion Rate")
plt.ylabel("Injuries per 1000 Events")
sns.despine(top=True, right=True)
plt.show()<jupyter_output><empty_output><jupyter_text>#### ** 32% of punt returns result in fewer than 5 yards ** ####
The graph below shows that 32% of punt returns yield fewer than 5 yards. These seem like prime candidates for events that we could replace with an awarded bonus of 5 yards, with little effect on field position but with substantial decrease in injury.
**Nudge Theory**
As an additional alternative, we could consider a proposal that makes fair catch the default, and require a returner to make a signal or gesture in order to indicate that he will not choose a fair catch. Evidence in support of setting an optimal default when giving people choices include research by Richard Thaler, Nobel prize-winning economist at the University of Chicago, who has written extensively on the notion of 'nudges.' If we nudge receivers into choosing a fair catch, then they will have to make the conscious decision to elect to return after assessing that the situation presents an opportunity to make a long return.<jupyter_code>x_groups = ['0-3 yds', '3-5 yds', '5-7 yds', '7-9 yds',
'9-12 yds', '12-15 yds', '15-20 yds', '20+ yds']
rec = plays_df.loc[(plays_df['punt_received']==1)
&(plays_df['Return'].notnull())]
y_groups = [sum(rec['Return']<=3) / len(rec),
sum((rec['Return']>3) & (rec['Return']<=5)) / len(rec),
sum((rec['Return']>5) & (rec['Return']<=7)) / len(rec),
sum((rec['Return']>7) & (rec['Return']<=9)) / len(rec),
sum((rec['Return']>9) & (rec['Return']<=12)) / len(rec),
sum((rec['Return']>12) & (rec['Return']<=15)) / len(rec),
sum((rec['Return']>15) & (rec['Return']<=20))/ len(rec),
sum(rec['Return']>20) / len(rec)]
y_bottoms = [0,
sum(rec['Return']<=3) / len(rec),
sum(rec['Return']<=5) / len(rec),
sum(rec['Return']<=7) / len(rec),
sum(rec['Return']<=9) / len(rec),
sum(rec['Return']<=12) / len(rec),
sum(rec['Return']<=15) / len(rec),
sum(rec['Return']<=20) / len(rec)]
fig = plt.figure(figsize=(8.5,4.5))
ax = plt.subplot2grid((1, 1), (0, 0))
plt.bar(range(len(x_groups)), y_groups, bottom=y_bottoms)
ax.set_xticks(range(len(x_groups)))
ax.set_xticklabels(x_groups)
ax.set_yticks([0, 0.2, 0.4, 0.6, 0.8, 1.0])
ax.set_yticklabels(['0%', '20%', '40%', '60%', '80%', '100%'])
for i in range(len(x_groups)):
plt.text(i-0.2, y_bottoms[i]+y_groups[i]+0.02, '{:.0f}%'.format(100*y_groups[i]))
sns.despine(top=True, right=True)
plt.title("Distribution of Punt Returns by Length")
plt.show()<jupyter_output><empty_output><jupyter_text>#### ** Example 1 of a play where the punt returner should probably have called a fair catch ** ####
A NFL punter can kick a punt with hang time of 4.4 seconds on average, which gives NFL level gunners time to advance 40 yards. In practice there is usually a 1-2 second gap between the time of ball snap and the time of the punt, allowing the gunner to advance 50 or 60 yards down the field before the ball is caught That's a lot of time, and sometimes I cringe while watching a punt returner (aka sitting duck) about to get nailed by a gunner - often the punt returner is too busy focusing on the ball to even notice that a player is about to hit him on his blind side. This appears to be one of those plays, although as often happens it's the gunner who suffers the injury and not the returner.<jupyter_code>play_player_role_data = pd.read_csv('../input/play_player_role_data.csv')
gfp = video_review.merge(play_player_role_data, on=['GameKey', 'PlayID', 'Season_Year'])
df_29 = pd.read_csv('../input/NGS-2016-pre.csv', parse_dates=['Time'])
df_29 = df_29.loc[(df_29['GameKey']==29) & (df_29['PlayID']==538)]
df_29 = df_29.merge(gfp, on=['GameKey', 'PlayID', 'Season_Year', 'GSISID'])
df_29 = df_29.sort_values(['GameKey', 'PlayID', 'Season_Year', 'GSISID', 'Time'])
fig = plt.figure(figsize = (10, 4.5))
ax = plt.subplot2grid((1, 1), (0, 0))
line_set = df_29.loc[df_29['Event'].isin(['ball_snap'])]
line_set_time = line_set['Time'].min()
line_of_scrimmage = line_set.loc[line_set['Role'].isin(['PLS', 'PLG', 'PRG']), 'x'].median()
recv_df = df_29.loc[df_29['Event']=='punt_received']
recv_time = recv_df['Time'].min()
event_df = df_29.loc[df_29['Time'] <= recv_time]
event_df = event_df.loc[event_df['Time'] >= recv_time + pd.Timedelta('-2s')]
injured = df_29['InjuredGSISID'].values[0]
partner = float(df_29['Primary_Partner_GSISID'].values[0])
players = event_df['GSISID'].unique()
for player in players:
player_df = event_df.loc[event_df['GSISID'] == player]
role = str(player_df['Role'].values[0])
if re.sub('[io0-9]', '', str(role)) in PUNT_TEAM:
color = '#fdc086'
marker = 'x'
linewidth = 6
else:
color = '#beaed4'
marker = 'o'
linewidth = 6
if player == injured:
marker = '*'
linewidth = 10
linestyle = '-'
color = '#f0027f'
elif player == partner:
marker = '*'
linewidth = 10
linestyle = '-'
color = '#ffff99'
else:
linestyle = '--'
alphas = np.ones(len(player_df))
alphas = alphas.cumsum() / alphas.sum()
px = player_df['x'].values
py = player_df['y'].values
for k in range(len(px)):
plt.plot(px[k:], py[k:], color=color,
linewidth=linewidth*(k+1+4)/(4+len(px)),
linestyle=linestyle,
alpha=(k+1)/len(px))
player_df = player_df.reset_index(drop=True)
x = player_df['x'].iloc[-1]
y = player_df['y'].iloc[-1]
marker = (3, 0, 90 + player_df['o'].iloc[-1])
plt.scatter(player_df['x'].iloc[-1],
player_df['y'].iloc[-1],
marker=marker,
s=linewidth*60,
color=color)
if (role == 'PR'):
circ = plt.Circle((player_df['x'].iloc[-1],
player_df['y'].iloc[-1]),
5, color=color,
fill=False)
ax.set_xlim(0, 120)
ax.set_ylim(0, 53.3)
plt.axvline(x=0, color='w', linewidth=2)
plt.axvline(x=10, color='w', linewidth=2)
plt.axvline(x=15, color='w', linewidth=2)
plt.axvline(x=20, color='w', linewidth=2)
plt.axvline(x=25, color='w', linewidth=2)
plt.axvline(x=30, color='w', linewidth=2)
plt.axvline(x=35, color='w', linewidth=2)
plt.axvline(x=40, color='w', linewidth=2)
plt.axvline(x=45, color='w', linewidth=2)
plt.axvline(x=50, color='w', linewidth=2)
plt.axvline(x=55, color='w', linewidth=2)
plt.axvline(x=60, color='w', linewidth=2)
plt.axvline(x=65, color='w', linewidth=2)
plt.axvline(x=70, color='w', linewidth=2)
plt.axvline(x=75, color='w', linewidth=2)
plt.axvline(x=80, color='w', linewidth=2)
plt.axvline(x=85, color='w', linewidth=2)
plt.axvline(x=90, color='w', linewidth=2)
plt.axvline(x=95, color='w', linewidth=2)
plt.axvline(x=100, color='w', linewidth=2)
plt.axvline(x=105, color='w', linewidth=2)
plt.axvline(x=110, color='w', linewidth=2)
plt.axvline(x=120, color='w', linewidth=2)
plt.axvline(x=line_of_scrimmage, color='y', linewidth=3)
plt.axhline(y=0, color='w', linewidth=2)
plt.axhline(y=53.3, color='w', linewidth=2)
plt.text(x=18, y=2, s= '1', color='w')
plt.text(x=21, y=2, s= '0', color='w')
plt.text(x=28, y=2, s= '2', color='w')
plt.text(x=31, y=2, s= '0', color='w')
plt.text(x=38, y=2, s= '3', color='w')
plt.text(x=41, y=2, s= '0', color='w')
plt.text(x=48, y=2, s= '4', color='w')
plt.text(x=51, y=2, s= '0', color='w')
plt.text(x=58, y=2, s= '5', color='w')
plt.text(x=61, y=2, s= '0', color='w')
plt.text(x=68, y=2, s= '4', color='w')
plt.text(x=71, y=2, s= '0', color='w')
plt.text(x=78, y=2, s= '3', color='w')
plt.text(x=81, y=2, s= '0', color='w')
plt.text(x=88, y=2, s= '2', color='w')
plt.text(x=91, y=2, s= '0', color='w')
plt.text(x=98, y=2, s= '1', color='w')
plt.text(x=101, y=2, s= '0', color='w')
ax.set_xticks([0, 120])
ax.set_yticks([0, 53.3])
ax.set_xticklabels(['', ''])
ax.set_yticklabels(['', ''])
ax.tick_params(axis=u'both', which=u'both', length=0)
ax.add_artist(circ)
ax.set_facecolor("#2ca25f")
plt.title("GR is injured while tackling PR (helmet to body).\nGameKey 29, Play ID 538. NYJ Punting to WAS.")
plt.show() <jupyter_output><empty_output><jupyter_text>#### Example 2 of a play where the punt returner should probably have called a fair catch ####
Here the punt returner isn't even directly involved in the injury, but rather two members of the coverage team collide with each other. When multiple players have a chance to converge at a high rate of speed to the same stationary target, there is increased risk. <jupyter_code>df_296 = pd.read_csv('../input/NGS-2016-reg-wk13-17.csv', parse_dates=['Time'])
df_296 = df_296.loc[(df_296['GameKey']==296) & (df_296['PlayID']==2667)]
df_296 = df_296.merge(gfp, on=['GameKey', 'PlayID', 'Season_Year', 'GSISID'])
df_296 = df_296.sort_values(['GameKey', 'PlayID', 'Season_Year', 'GSISID', 'Time'])
fig = plt.figure(figsize = (10, 4.5))
ax = plt.subplot2grid((1, 1), (0, 0))
line_set = df_296.loc[df_296['Event'].isin(['ball_snap'])]
line_set_time = line_set['Time'].min()
line_of_scrimmage = line_set.loc[line_set['Role'].isin(['PLS', 'PLG', 'PRG']), 'x'].median()
recv_df = df_296.loc[df_296['Event']=='punt_received']
recv_time = recv_df['Time'].min()
event_df = df_296.loc[df_296['Time'] <= recv_time]
event_df = event_df.loc[event_df['Time'] >= recv_time + pd.Timedelta('-2s')]
injured = df_296['InjuredGSISID'].values[0]
partner = float(df_296['Primary_Partner_GSISID'].values[0])
players = event_df['GSISID'].unique()
for player in players:
player_df = event_df.loc[event_df['GSISID'] == player]
role = str(player_df['Role'].values[0])
if re.sub('[io0-9]', '', str(role)) in PUNT_TEAM:
color = '#fdc086'
marker = 'x'
linewidth = 6
else:
color = '#beaed4'
marker = 'o'
linewidth = 6
if player == injured:
marker = '*'
linewidth = 10
linestyle = '-'
color = '#f0027f'
elif player == partner:
marker = '*'
linewidth = 10
linestyle = '-'
color = '#ffff99'
else:
linestyle = '--'
alphas = np.ones(len(player_df))
alphas = alphas.cumsum() / alphas.sum()
px = player_df['x'].values
py = player_df['y'].values
for k in range(len(px)):
plt.plot(px[k:], py[k:], color=color,
linewidth=linewidth*(k+1+4)/(4+len(px)),
linestyle=linestyle,
alpha=(k+1)/len(px))
player_df = player_df.reset_index(drop=True)
x = player_df['x'].iloc[-1]
y = player_df['y'].iloc[-1]
marker = (3, 0, 90 + player_df['o'].iloc[-1])
plt.scatter(player_df['x'].iloc[-1],
player_df['y'].iloc[-1],
marker=marker,
s=linewidth*60,
color=color)
if (role == 'PR'):
circ = plt.Circle((player_df['x'].iloc[-1],
player_df['y'].iloc[-1]),
5, color=color,
fill=False)
ax.set_xlim(0, 120)
ax.set_ylim(0, 53.3)
plt.axvline(x=0, color='w', linewidth=2)
plt.axvline(x=10, color='w', linewidth=2)
plt.axvline(x=15, color='w', linewidth=2)
plt.axvline(x=20, color='w', linewidth=2)
plt.axvline(x=25, color='w', linewidth=2)
plt.axvline(x=30, color='w', linewidth=2)
plt.axvline(x=35, color='w', linewidth=2)
plt.axvline(x=40, color='w', linewidth=2)
plt.axvline(x=45, color='w', linewidth=2)
plt.axvline(x=50, color='w', linewidth=2)
plt.axvline(x=55, color='w', linewidth=2)
plt.axvline(x=60, color='w', linewidth=2)
plt.axvline(x=65, color='w', linewidth=2)
plt.axvline(x=70, color='w', linewidth=2)
plt.axvline(x=75, color='w', linewidth=2)
plt.axvline(x=80, color='w', linewidth=2)
plt.axvline(x=85, color='w', linewidth=2)
plt.axvline(x=90, color='w', linewidth=2)
plt.axvline(x=95, color='w', linewidth=2)
plt.axvline(x=100, color='w', linewidth=2)
plt.axvline(x=105, color='w', linewidth=2)
plt.axvline(x=110, color='w', linewidth=2)
plt.axvline(x=120, color='w', linewidth=2)
plt.axvline(x=line_of_scrimmage, color='y', linewidth=3)
plt.axhline(y=0, color='w', linewidth=2)
plt.axhline(y=53.3, color='w', linewidth=2)
plt.text(x=18, y=2, s= '1', color='w')
plt.text(x=21, y=2, s= '0', color='w')
plt.text(x=28, y=2, s= '2', color='w')
plt.text(x=31, y=2, s= '0', color='w')
plt.text(x=38, y=2, s= '3', color='w')
plt.text(x=41, y=2, s= '0', color='w')
plt.text(x=48, y=2, s= '4', color='w')
plt.text(x=51, y=2, s= '0', color='w')
plt.text(x=58, y=2, s= '5', color='w')
plt.text(x=61, y=2, s= '0', color='w')
plt.text(x=68, y=2, s= '4', color='w')
plt.text(x=71, y=2, s= '0', color='w')
plt.text(x=78, y=2, s= '3', color='w')
plt.text(x=81, y=2, s= '0', color='w')
plt.text(x=88, y=2, s= '2', color='w')
plt.text(x=91, y=2, s= '0', color='w')
plt.text(x=98, y=2, s= '1', color='w')
plt.text(x=101, y=2, s= '0', color='w')
ax.set_xticks([0, 120])
ax.set_yticks([0, 53.3])
ax.set_xticklabels(['', ''])
ax.set_yticklabels(['', ''])
ax.tick_params(axis=u'both', which=u'both', length=0)
ax.add_artist(circ)
ax.set_facecolor("#2ca25f")
plt.title("GL and GR collide, injuring GL (helmet to helmet friendly fire).\nGameKey 296, Play ID 2667. TEN punting to JAX.")
plt.show()<jupyter_output><empty_output><jupyter_text>#### **Going for it on fourth down**
While somewhat out of the scope of this study, there is considerable outside evidence that teams are overly conservative on fourth down, choosing to punt when they would be better off attempting to move the chains. If the fair catch bonus of 5 yards were to encourage more teams to go for it rather than punt, that would seem to a positive side effect.### **Proposal 2** : require single coverage on gunners.
Does single coverage result in fewer injuries? To start to analyze this, we classify plays as having single, hybrid, or double coverage, and see whether or not an injury occurred. Because teams are more likely to opt for double coverage in plays where the field is long (i.e. the punt is less likely to be coffin cornered or result in touchback), we need to bucket our data in two dimensions: (a) choice of coverage and (b) distance from line of scrimmage to the end zone.
<jupyter_code>ppr = pd.read_csv('../input/play_player_role_data.csv')
ppr['Role'] = ppr['Role'].map(lambda x: re.sub('[oi0-9]', '', x))
roles = ppr['Role'].unique()
ppr = pd.concat([ppr, pd.get_dummies(ppr['Role'])], axis=1)
ppr = ppr.groupby(['Season_Year', 'GameKey', 'PlayID'])[roles].sum()
ppr = ppr.reset_index()
vi = pd.read_csv('../input/video_review.csv')
vi = vi[['Season_Year', 'GameKey', 'PlayID', 'GSISID']]
ppr = ppr.merge(vi, on=['Season_Year', 'GameKey', 'PlayID'], how='left')
ppr['Injury'] = 0
ppr['const'] = 1
ppr.loc[ppr['GSISID'].notnull(), 'Injury'] = 1
play_information = pd.read_csv('../input/play_information.csv')
def extract_recv_yards(s):
m = re.search('for ([0-9]+) yards', s)
if m:
return int(m.group(1))
elif re.search('for no gain', s):
return 0
else:
return np.nan
play_information['recv_length'] = play_information['PlayDescription'].map(
lambda x: extract_recv_yards(x))
play_information = play_information[['Season_Year', 'GameKey', 'PlayID', 'YardLine', 'Poss_Team', 'recv_length']]
play_information['yards_to_go'] = play_information['YardLine'].map(lambda x: int(x[-2:]))
play_information['back_half'] = play_information.apply(lambda x:
x['YardLine'].startswith(x['Poss_Team']), axis = 1)
play_information.loc[play_information['back_half']==1, 'yards_to_go'] = 100 - (
play_information.loc[play_information['back_half']==1, 'yards_to_go'])
play_information = play_information[['Season_Year', 'GameKey', 'PlayID', 'yards_to_go', 'recv_length']]
ppr = ppr.merge(play_information, on=['Season_Year', 'GameKey', 'PlayID'], how='inner')
col = 'VR_VL'
ppr[col] = ppr.loc[:, ['VR', 'VL']].sum(axis=1)
ptiles = np.percentile(ppr[col], [0, 25, 50, 75, 100])
ppr['yards'] = ''
ppr['coverage'] = ''
ppr.loc[ppr[col]==2, 'coverage'] = 'single'
ppr.loc[ppr[col]==3, 'coverage'] = 'hybrid'
ppr.loc[ppr[col]==4, 'coverage'] = 'double'
<jupyter_output><empty_output><jupyter_text>This graph below shows that receiving teams choose single coverage 84% of the time when the field of play is effectively short, e.g. there are fewer than 50 yards from the line of scrimmage to the end zone. On the other hand, receiving teams choose double coverage, single coverage, and hybrid coverage with more or less even probability when there are more than 70 yards between the line of scrimmage and the goal line. <jupyter_code>### For graphing, keep track of counts of plays by single, hybrid, or double coverage
### Sort by yards-to-go.
x_single = [0.00, 1.00, 2.00]
x_hybrid = [0.25, 1.25, 2.25]
x_double = [0.50, 1.50, 2.50]
y_single = []
y_hybrid = []
y_double = []
injury_rate = {'Yards to Go': [], '30-50': [], '50-70': [], '70-100': []}
for i in range(2, 5):
if i == 2:
mode = 'Single'
elif i == 3:
mode = 'Hybrid'
elif i == 4:
mode = 'Double'
injury_rate['Yards to Go'].append(mode)
for r in ([30, 50], [50, 70], [70, 100]):
# Edit : condition on punts that are returned,
# ie ignore fair catch
ii = ((ppr['yards_to_go']>=r[0])
&(ppr['yards_to_go']<r[1])
&(ppr['recv_length'].notnull())
&(ppr['recv_length']!=0))
if (r[0]==30) & (r[1]==50):
ppr.loc[ii, 'yards'] = '30 to 50'
elif (r[0]==50) & (r[1]==70):
ppr.loc[ii, 'yards'] = '50 to 70'
elif (r[0]==70) & (r[1]==100):
ppr.loc[ii, 'yards'] = '70 to 100'
pprt = ppr.loc[ii]
if len(pprt) == 0:
pass
iii = (pprt[col] == i)
if sum(iii) == 0:
continue
# Keep track of coverage choice by yards to go
if mode == 'Single':
y_single.append(sum(iii) / sum(ii)) # Ratio of times single coverage is elected
elif mode == 'Hybrid':
y_hybrid.append(sum(iii) / sum(ii)) # Ratio of times hybrid coverage is elected
elif mode == 'Double':
y_double.append(sum(iii) / sum(ii)) # Ratio of times double coverage is elected
if r[0] == 30:
injury_rate['30-50'].append(1000 * pprt.loc[iii, 'Injury'].mean())
elif r[0] == 50:
injury_rate['50-70'].append(1000 * pprt.loc[iii, 'Injury'].mean())
elif r[0] == 70:
injury_rate['70-100'].append(1000 * pprt.loc[iii, 'Injury'].mean())
injury_rate['Yards to Go'].append("All")
injury_rate['30-50'].append(1000 * ppr.loc[
(ppr['yards_to_go']>=30) & (ppr['yards_to_go']<50) &(ppr['recv_length'].notnull())
&(ppr['recv_length']!=0) , 'Injury'].mean())
injury_rate['50-70'].append(1000 * ppr.loc[
(ppr['yards_to_go']>=50) & (ppr['yards_to_go']<70) &(ppr['recv_length'].notnull())
&(ppr['recv_length']!=0), 'Injury'].mean())
injury_rate['70-100'].append(1000 * ppr.loc[
(ppr['yards_to_go']>=70) & (ppr['yards_to_go']<100) &(ppr['recv_length'].notnull())
&(ppr['recv_length']!=0), 'Injury'].mean())
injury_rate = pd.DataFrame(injury_rate)
injury_rate = injury_rate[['Yards to Go', '30-50', '50-70', '70-100']]
fig = plt.figure(figsize = (6.5, 4.5))
ax = plt.subplot2grid((1, 1), (0, 0))
plt.bar(x_single, y_single, color='#7fc97f', width=.25, label='Single Coverage')
plt.bar(x_hybrid, y_hybrid, color='#beaed4', width=.25, label='Hybrid Coverage')
plt.bar(x_double, y_double, color='#fdc086', width=.25, label='Double Coverage')
for i in range(3):
plt.text(x_single[i]-0.06, y_single[i]+0.02, '{:.0f}%'.format(y_single[i] * 100))
plt.text(x_hybrid[i]-0.06, y_hybrid[i]+0.02, '{:.0f}%'.format(y_hybrid[i] * 100))
plt.text(x_double[i]-0.06, y_double[i]+0.02, '{:.0f}%'.format(y_double[i] * 100))
plt.legend()
ax.set_yticks([0, 0.25, 0.5, 0.75, 1.00])
ax.set_yticklabels(['0%', '25%', '50%', '75%', '100%'])
ax.set_xticks([0.25, 1.25, 2.25])
ax.set_xticklabels(['30-50 Yards', '50-70 Yards', '70-100 Yards'])
ax.set_xlabel('Yards from Line of Scrimmage to End Zone')
ax.set_ylabel('Coverage Choice (%)')
sns.despine(top=True, right=True)
plt.title("Coverage Choice vs. Yards between Line of Scrimmage and End Zone\n")
plt.show()<jupyter_output><empty_output><jupyter_text>The injury rate is higher on double coverage and hybrid coverage vs. single coverage, at least in plays with longer fields. E.g. looking at all coverage types (the last row of the table below), there are 3 injuries per 1000 plays with 30-50 yards-to-go versus 9 injuries per 1000 plays when there are 70-100 yards-to-go. Because choice of coverage is correlated with yards-to-go, we need to control for both effects when trying to evaluate whether coverage is related to injury rate.
By subsetting plays on both dimensions, we can see that even when controlling for yards to go, double coverage appears to result in substantially higher injury rates than single and hybrid coverage. That said, the statistical significance of this finding is limited by the small sample size, as there are too few observed injuries in the provided data to draw sharp inferences. Given that we are not strongly confident that double coverage increases the injury rate, we need at least to be sure that prohibiting double coverage does not reduce the quality of game play.
** Table of Injury Rate vs. Yards-to-Go and Coverage Choice **<jupyter_code>injury_rate.style.set_precision(2).hide_index()<jupyter_output><empty_output><jupyter_text>One measure of quality of play is the length of punt return, with longer punt returns being (in my mind) associated with more exciting games. Let us look at the distribution of punt return length for punts received with single, hybrid, and double coverage. Again, we group by yards-to-go, since coverage decisions appear partly conditioned on the length of the playable field. The "violin plots" below show the density and distribution of punt returns, condtioned on coverage choice and yards to go. At first glance, return length appears to be indistinguishable between single, hybrid, and double coverage, suggesting that the return team would do just as well electing single coverage.
** Distribution of Punt Return Length Conditioned on Yards-to-Go and Coveage Choice **<jupyter_code>fig = plt.figure(figsize = (8.5, 5.5))
ax = plt.subplot2grid((1, 1), (0, 0))
pal = {'single': '#7fc97f', 'hybrid': '#beaed4', 'double': '#fdc086'}
ax = sns.violinplot(x="yards", y="recv_length", hue="coverage",
data=ppr, palette=pal,
order=['30 to 50', '50 to 70', '70 to 100'],
hue_order=['single', 'hybrid', 'double'],
cut=0)
ax.legend().remove()
sns.despine(top=True, right=True)
plt.legend()
plt.show()<jupyter_output><empty_output><jupyter_text>### **Proposal 3**: Require helmet monitors to better measure deceleration at time of tackle.
As the plots below show, deceleration is greater for tackles generating injuries than for tackles without injuries. On the other hand, velocities are somewhat higher on non-injury players and plays. Intuitively, there is nothing dangerous about running fast, unless you have to stop suddenly.
Because NGS data is provided only at 100 millisecond resolution, the maximum observable deceleration is approximately 100 m/s/s (assuming a person running flat out at 10 m/s at time t is moving at 0 m/s in the next NGS data point, 100 milli seconds later) , or less than 10g's. On the other hand, it is possible for players to have experienced much greater g-forces, and for those forces to be stronger at the helmet than in the rest of the body. Without helmet sensor data, there is a finite upper bound on the measurable deceleration forces, one that is too coarse to be really useful at distinguishing injury from safe and routine play.
<jupyter_code># Collate data
game_data = pd.read_csv('../input/game_data.csv')
play_information = pd.read_csv('../input/play_information.csv')
player_punt_data = pd.read_csv('../input/player_punt_data.csv')
player_punt_data = player_punt_data.groupby('GSISID').head(1).reset_index()
play_player_role_data = pd.read_csv('../input/play_player_role_data.csv')
video_review = pd.read_csv('../input/video_review.csv')
combined = game_data.merge(play_information.drop(['Game_Date'], axis=1),
on=['GameKey', 'Season_Year', 'Season_Type', 'Week'])
combined = combined.merge(play_player_role_data,
on=['GameKey', 'Season_Year', 'PlayID'])
combined = combined.merge(player_punt_data, on=['GSISID'])
combined = combined.merge(video_review, how='left',
on=['Season_Year', 'GameKey', 'PlayID', 'GSISID'])
combined['injury'] = 0
combined.loc[combined['Player_Activity_Derived'].notnull(), 'injury'] = 1
ngs_files = ['../input/NGS-2016-pre.csv',
'../input/NGS-2016-reg-wk1-6.csv',
'../input/NGS-2016-reg-wk7-12.csv',
'../input/NGS-2016-reg-wk13-17.csv',
'../input/NGS-2016-post.csv',
'../input/NGS-2017-pre.csv',
'../input/NGS-2017-reg-wk1-6.csv',
'../input/NGS-2017-reg-wk7-12.csv',
'../input/NGS-2017-reg-wk13-17.csv',
'../input/NGS-2017-post.csv']
max_decel_df = []
for filename in ngs_files:
logging.info("Loading file " + filename)
group_keys = ['Season_Year', 'GameKey', 'PlayID', 'GSISID']
df = pd.read_csv(filename, parse_dates=['Time'])
logging.info("Read file " + filename)
df = df.sort_values(group_keys + ['Time'])
df['dx'] = df.groupby(group_keys)['x'].diff(1)
df['dy'] = df.groupby(group_keys)['y'].diff(1)
df['dis'] = (df['dx']**2 + df['dy']**2)**0.5
df['dt'] = df.groupby(group_keys)['Time'].diff(1).dt.total_seconds()
df['velocity'] = 0
ii = (df['dis'].notnull() & df['dt'].notnull() & (df['dt']>0))
df.loc[ii, 'velocity'] = df.loc[ii, 'dis'] / df.loc[ii, 'dt']
df['velocity'] *= 0.9144 # Convert yards to meters
df['deceleration'] = -1 * df.groupby(group_keys)['velocity'].diff(1)
df['velocity'] = df.groupby(group_keys)['velocity'].shift(1)
# Only look at the one second window around each tackle
df['Event'] = df.groupby(group_keys)['Event'].ffill(limit=5)
df['Event'] = df.groupby(group_keys)['Event'].bfill(limit=5)
t_df = df.loc[df['Event']=='tackle']
t_max_decel = t_df.loc[t_df.groupby(['Season_Year', 'GameKey', 'PlayID', 'GSISID'])['deceleration'].idxmax()]
t_max_decel = t_max_decel[['Season_Year', 'GameKey', 'PlayID', 'GSISID', 'deceleration']].rename(columns={'deceleration': 'deceleration_at_tackle'})
t_max_velocity = t_df.loc[t_df.groupby(['Season_Year', 'GameKey', 'PlayID', 'GSISID'])['velocity'].idxmax()]
t_max_velocity = t_max_velocity[['Season_Year', 'GameKey', 'PlayID', 'GSISID', 'velocity']].rename(columns={'velocity': 'velocity_at_tackle'})
max_decel = t_max_velocity.merge(t_max_decel, on=['Season_Year', 'GameKey', 'PlayID', 'GSISID'],
how='outer')
max_decel_df.append(max_decel)
max_decel_df = pd.concat(max_decel_df)
combined = combined.merge(max_decel_df, on=['Season_Year', 'GameKey', 'PlayID', 'GSISID'], how='left')
combined['tackle_injury'] = combined['Player_Activity_Derived'].isin(['Tackled', 'Tackling'])
### Original work by Halla Yang<jupyter_output><empty_output><jupyter_text>#### Distribution of Player Velocities and Deceleration at time of Event Tackle, Injury Plays vs. non-Injury Plays <jupyter_code>fig = plt.figure(figsize = (6.5, 5.0))
ax = plt.subplot2grid((1, 1), (0, 0))
inj = combined.loc[(combined['injury']==1)&(combined['velocity_at_tackle'].notnull())
&(combined['deceleration_at_tackle'].notnull())]
ax = sns.kdeplot(inj.velocity_at_tackle, inj.deceleration_at_tackle,
cmap="Reds")
notinj = combined.loc[(combined['injury']==0)&(combined['velocity_at_tackle'].notnull())
&(combined['deceleration_at_tackle'].notnull())]
ax = sns.kdeplot(notinj.velocity_at_tackle, notinj.deceleration_at_tackle,
cmap="Blues")
ax.set_xlim(0, 10)
ax.set_ylim(0, 2)
plt.xlabel("Velocity at Tackle")
plt.ylabel("Deceleration at Tackle")
sns.despine(top=True, right=True)
plt.title("Velocity/Deceleration at Time of Tackle for Injuries shown in Red, Non-Injuries in Blue")
plt.subplots_adjust(top=0.9)
plt.show()
<jupyter_output><empty_output><jupyter_text>We show that players with less than median deceleration (approximately 0.5) have much lower injury rates than players with greater than median deceleration. Similar results can be found using logistic regressions, that deceleration is a significant predictor of injury. If we required players to wear mouthguard or helmet sensors that measured deceleration, we would have a better way to empirically measure whether rule changes, formation changes, etc. reduce the likelihood of injury. <jupyter_code># Multiply by 22 since there are 22 players per play.
ii = (combined['deceleration_at_tackle']>=0.0) & (combined['deceleration_at_tackle']<0.5)
jj = (combined['deceleration_at_tackle']>=0.5) & (combined['deceleration_at_tackle']<100)
rate1 = 22*1000*combined.loc[ii, 'tackle_injury'].mean()
rate2 = 22*1000*combined.loc[jj, 'tackle_injury'].mean()
print("Injury rate per 1000 plays when deceleration at tackle is less than 0.5: {:.2f} (N={:.0f})".format(rate1,
sum(ii)))
print("Injury rate per 1000 plays when deceleration at tackle is greater than 0.5: {:.2f} (N={:.0f})".format(rate2,
sum(jj)))
<jupyter_output><empty_output>
|
no_license
|
/datasets/NFL-Punt-Analytics-Competition/kernels/NULL---hallayang---nfl-punt-analytics-proposal.ipynb
|
mindis/GDS
| 12 |
<jupyter_start><jupyter_text>### Process raw WIPO patent data<jupyter_code>import codecs
import glob
import os
import pandas as pd
import pycountry
from io import StringIO
# find target path for data files, assuming the notebook is in the right place
data_path = _dh[0]
assert data_path.endswith(os.path.join('em-2020','data'))
output_path = os.path.join(data_path, 'WIPO_reshaped.csv')<jupyter_output><empty_output><jupyter_text>The WIPO file format is hideous and needs a custom loader:<jupyter_code>wipo_file = 'patent_1 - Total patent applications (direct and PCT national phase entries)_Total count by filing office_1980_2018.csv'
wipo_raw = codecs.open(os.path.join(data_path, wipo_file), encoding='ISO-8859-1').readlines()
wipo_clean = StringIO()
wipo_clean.write(wipo_raw[7].strip())
wipo_clean.write(',')
wipo_clean.write(wipo_raw[6][3:])
for line in wipo_raw[8:]:
wipo_clean.write(line)
wipo_clean.seek(0)
wipo_data = pd.read_csv(wipo_clean, index_col=False, na_values='', keep_default_na=False)
wipo_data.iloc[-5:,:5]
def map_iso_alpha2(x):
country = pycountry.countries.get(alpha_2=x)
if country is None:
return None
else:
return country.alpha_3
iso_alpha3 = wipo_data['Office (Code)'].map(map_iso_alpha2)
valid = ~iso_alpha3.isnull()
wipo_reshaped = pd.DataFrame(wipo_data.loc[valid]
.set_index(iso_alpha3.loc[valid])
.drop(columns=['Office', 'Office (Code)', 'Origin'])
.stack())
wipo_reshaped.index.names = ['Country', 'Year']
wipo_reshaped.index.set_levels(wipo_reshaped.index.levels[-1].astype(int), level=-1, inplace=True)
wipo_reshaped.columns = ['WIPO patents']
wipo_reshaped.iloc[:5, :5]
wipo_reshaped.to_csv(output_path)<jupyter_output><empty_output>
|
no_license
|
/data/D2_extract WIPO_data.ipynb
|
drakesiard/em-2020
| 2 |
<jupyter_start><jupyter_text># Build Spectral Library<jupyter_code>from dataclasses import dataclass
@dataclass
class spectrum:
"""Class for keeping track of a."""
name: str
wave: np.array #wavelengths (microns)
A: np.array #spectral albedo
def shkuratov(wave, n, k, S, q=0.7):
"""This function computes a spectra with the shkuratov radiative
transfer equations for the specified inputs.
It returns the spectral albedo (reflectance spectra) at the wavelengths
input in wave.
INPUTS:
wave: array: array of wavelengths for corresponding n and k values
n, k: arrays: arrays of n and k values to be use din shkuratov formula
S: number: grain size, in microns
q: default 0.7: volume filling fraction (default is 79%)
OUTPUT:
A: array: spectral albedo values for wavelength in wave
"""
#Fresnell coefficient
r_o = ((n-1)**2)/((n+1)**2)
#Reflection Coefficients
R_i = 1.04 - (1/(n**2))
R_e = r_o + 0.05
R_b = (0.28*n - 0.20)*R_e
R_f = R_e-R_b
#Transmission Coefficients
T_e = 1 - R_e
T_i = 1 - R_i
#Tau: optical density
tau = (4*np.pi*k*S)/wave
#Light Scattering Indicatrices
r_b = R_b + 0.5*T_e*T_i*R_i*np.exp(-2*tau )/(1 - R_i*np.exp(-1*tau))
r_f = R_f + T_e*T_i*np.exp(-1*tau) + 0.5*T_e*T_i*R_i*np.exp(-2*tau )/(1 - R_i*np.exp(-1*tau))
#1D indicatrices
rho_b = q*r_b
rho_f = q*r_f + (1-q)
#Spectral Albedo
return ((1 + rho_b**2 - rho_f**2)/(2*rho_b)) - np.sqrt(((1 + rho_b**2 - rho_f**2)/(2*rho_b))**2 -1)
spec_library = []
#For now, populate spec_library with spectra to be used in linear model
# Mg salts (hydrated)
spec_library.append(spectrum(name='Mg chloride', wave = mg_chloride[:,0], A = mg_chloride[:,1]))
spec_library.append(spectrum(name='Mg chlorate', wave = mg_chlorate[:,0], A = mg_chlorate[:,1]))
spec_library.append(spectrum(name='Mg perchlorate', wave = mg_perchlorate[:,0], A = mg_perchlorate[:,1]))
#Amorphous Water
spec_library.append(spectrum(name='Amorphous 5 micron', wave=amor_nk[:,0],
A = shkuratov(amor_nk[:,0], amor_nk[:,1], amor_nk[:,2], 5., q=0.7)))
spec_library.append(spectrum(name='Amorphous 25 micron', wave=amor_nk[:,0],
A = shkuratov(amor_nk[:,0], amor_nk[:,1], amor_nk[:,2], 25., q=0.7)))
spec_library.append(spectrum(name='Amorphous 200 micron', wave=amor_nk[:,0],
A = shkuratov(amor_nk[:,0], amor_nk[:,1], amor_nk[:,2], 200., q=0.7)))
spec_library.append(spectrum(name='Amorphous 1 mm', wave=amor_nk[:,0],
A = shkuratov(amor_nk[:,0], amor_nk[:,1], amor_nk[:,2], 1000., q=0.7)))
#Crystalline Water
spec_library.append(spectrum(name='Crystalline 25 micron', wave=crys_nk[:,0],
A = shkuratov(crys_nk[:,0], crys_nk[:,1], crys_nk[:,2], 25., q=0.7)))
spec_library.append(spectrum(name='Crystalline 200 micron', wave=crys_nk[:,0],
A = shkuratov(crys_nk[:,0], crys_nk[:,1], crys_nk[:,2], 200., q=0.7)))
spec_library.append(spectrum(name='Crystalline 1 mm', wave=crys_nk[:,0],
A = shkuratov(crys_nk[:,0], crys_nk[:,1], crys_nk[:,2], 1000., q=0.7)))
#Sulfuric Acid Octahydrate
spec_library.append(spectrum(name='Sulfuric Acid Octahydrate', wave=sulfuric_acid[:,0], A = sulfuric_acid[:,1]))
from abc import ABC, abstractmethod
from typing import Callable, List, Tuple, Optional, Sequence
from tqdm.auto import tqdm
from copy import deepcopy
from lmfit import Parameters, Minimizer
def fill_parameters(params: Parameters, values: Sequence[float]) -> Parameters:
assert len(params) == len(values)
params = deepcopy(params)
for p,v in zip(params.values(), values):
p.value = v
return params
class SpectralModel(ABC):
""" An abstract class representing a model which can be fit, and which can
be used to estimate spectral intensity.
"""
@property
@abstractmethod
def parameters(self) -> Parameters:
"""Get a copy of the model parameters. """
pass
@abstractmethod
def set_parameters(self, params: Parameters):
pass
@abstractmethod
def estimate(self, params: Parameters) -> np.ndarray:
pass
def residual_fn(self, b: np.ndarray) -> Callable[[Parameters], np.ndarray]:
"""Get a function which computes residuals when provided parameters.
Used for lmfit optimization.
"""
def _fn(params):
return self.estimate(params) - b
return _fn
def fit(self, b: np.ndarray, parameters=None, *args, **kwargs) -> Tuple[lmfit.minimizer.MinimizerResult, np.ndarray]:
""" Compute a fit, given target values 'b'. Uses the (optional) parameters to start the minimization.
Returns a MinimizerResult as well as an array of residual sum-of-square errors, indexed by iteration."""
errors = []
pbar = tqdm()
def log_errors(params, iter, res):
err = np.linalg.norm(res)**2
pbar.set_description(f'err = {err:.3f}')
errors.append(err)
pbar.update(1)
if parameters is None:
parameters = self.parameters
mini = Minimizer(self.residual_fn(b), parameters, iter_cb=log_errors)
out = mini.minimize(*args, **kwargs)
self.set_parameters(out.params)
return out, np.array(errors)
def fill_parameters(self, values: Sequence[float]):
""" Set the values on the model's parameters. """
self.set_parameters(
fill_parameters(self.parameters, values)
)
class NaiveLinearModel(SpectralModel):
"""A simple linear model, based on known wavelength and spectral library. """
def __init__(self, waves: np.ndarray, spec_library: List[spectrum]):
# initialize model matrix
self.A = np.zeros((len(waves), len(spec_library)))
for i,spec in enumerate(spec_library):
self.A[:,i] = np.interp(waves, spec.wave, spec.A)
# initialize parameters
self._params = Parameters()
for i,spec in enumerate(spec_library):
self._params.add(spec.name.replace(" ", "_"), vary=True, min=0)
def estimate(self, params: Parameters) -> np.ndarray:
params_dict = params.valuesdict()
x = np.array([params_dict[key] for key in params_dict.keys()]).flatten()
return self.A @ x
@property
def parameters(self):
return self._params
def set_parameters(self, params):
self._params = params
# Remove Nan values from VLT spectrum
waves = wave[np.where(~np.isnan(europa_spec))]
b = europa_spec[np.where(~np.isnan(europa_spec))]<jupyter_output><empty_output><jupyter_text># Naive Linear Model: Ax = b<jupyter_code>naive_linear = NaiveLinearModel(waves, spec_library)
naive_linear.fill_parameters(np.random.rand(11)) #Provide random starting conditions to avoid boundary issues
nl_out, nl_errors = naive_linear.fit(b, method='leastsq')
from lmfit import report_fit
report_fit(nl_out)
fig, ax = plt.subplots(2,1, figsize=(15,15),
gridspec_kw={'height_ratios': [1.5, 2]})
plt.title('260W, Non-Icy Region')
ax[0].plot(waves, b, color='b')
ax[0].plot(waves, naive_linear.estimate(nl_out.params), color='r', linewidth=3,
label='Reduced Chi: '+format(nl_out.redchi,'.3e'))
ax[0].set_xlabel('Wavelength (microns)')
ax[0].set_ylabel('Reflectance')
ax[0].legend()
#Plot components
for i,p in enumerate(nl_out.params.values()):
ax[1].plot(waves, naive_linear.A[:,i]*p.value,
label=p.name+': '+format(p.value,'0.4f'))
ax[1].legend()
ax[1].set_xlabel('Wavelength (microns)')
ax[1].set_ylabel('Reflectance')<jupyter_output><empty_output><jupyter_text># Constrained Linear Model: Ax = b where $\sum x_i = 1$<jupyter_code>class ConstrainedLinearModel(SpectralModel):
"""A simple linear model, based on known wavelength and spectral library where the
sum of coefficients are constrained to be 1. """
def __init__(self, waves: np.ndarray, spec_library: List[spectrum]):
# initialize model matrix
self.A = np.zeros((len(waves), len(spec_library)))
for i,spec in enumerate(spec_library):
self.A[:,i] = np.interp(waves, spec.wave, spec.A)
# initialize parameters
self._params = Parameters()
expr = '1'
for spec in spec_library[:-1]:
#Create initial blank paramters to allow expression building
self._params.add(spec.name.replace(" ", "_"), vary=True, min=0)
#Add to constraint expression
expr+= '-'+str(spec.name.replace(" ", "_"))
#Constrain all parameters to sum to 1
self._params.add(spec_library[-1].name.replace(" ", "_"), vary=True, min=0, expr=expr)
def estimate(self, params: Parameters) -> np.ndarray:
params_dict = params.valuesdict()
x = np.array([params_dict[key] for key in params_dict.keys()]).flatten()
return self.A @ x
@property
def parameters(self):
return self._params
def set_parameters(self, params):
self._params = params
constrained_linear = ConstrainedLinearModel(waves, spec_library)
rand = np.random.rand(11)
rand /= np.sum(rand)
constrained_linear.fill_parameters(rand) #Provide random starting conditions (that add to 1) to avoid boundary issues
cl_out, cl_errors = constrained_linear.fit(b, method='leastsq')
report_fit(cl_out)
fig, ax = plt.subplots(2,1, figsize=(15,15),
gridspec_kw={'height_ratios': [1.5, 2]})
plt.title('260W, Non-Icy Region')
ax[0].plot(waves, b, color='b')
ax[0].plot(waves, constrained_linear.estimate(cl_out.params), color='r', linewidth=3,
label='Reduced Chi: '+format(cl_out.redchi, '.3e'))
ax[0].set_xlabel('Wavelength (microns)')
ax[0].set_ylabel('Reflectance')
ax[0].legend()
#Plot components
for i,p in enumerate(cl_out.params.values()):
ax[1].plot(waves, constrained_linear.A[:,i]*p.value,
label=p.name+': '+format(p.value,'0.4f'))
#ax[1].set_yscale('log')
ax[1].legend()
ax[1].set_xlabel('Wavelength (microns)')
ax[1].set_ylabel('Reflectance')<jupyter_output><empty_output><jupyter_text># Linear Model with Slope Variation: $b = \sum x_i A_i *(\lambda + S)$<jupyter_code>class SlopeAdjustedModel(SpectralModel):
"""A simple linear model, based on known wavelength and spectral library with an additional fudge
factor (S) to adjust the spectral slope.
\sum x_i spec_i * (lambda + S)
"""
def __init__(self, waves: np.ndarray, spec_library: List[spectrum]):
# initialize model matrix
self.A = np.zeros((len(waves), len(spec_library)))
for i,spec in enumerate(spec_library):
self.A[:,i] = np.interp(waves, spec.wave, spec.A)
# initialize parameters
self._params = Parameters()
for spec in spec_library:
self._params.add(spec.name.replace(" ", "_"), vary=True, min=0)
# Add paramater for S
self._params.add('S', vary=True, min=0)
def estimate(self, params: Parameters) -> np.ndarray:
x = np.array([p.value for p in params.values() if p.name != 'S']).flatten()
S = params['S'].value
return self.A @ x * (waves+S)
@property
def parameters(self):
return self._params
def set_parameters(self, params):
self._params = params
slope_adjusted = SlopeAdjustedModel(waves, spec_library)
slope_adjusted.fill_parameters(np.concatenate([np.random.rand(11), [1]])) #Provide random starting conditions
sa_out, sa_errors = slope_adjusted.fit(b, method='leastsq')
report_fit(sa_out)
fig, ax = plt.subplots(2,1, figsize=(15,15),
gridspec_kw={'height_ratios': [1.5, 2]})
plt.title('260W, Non-Icy Region')
ax[0].plot(waves, b, color='b')
ax[0].plot(waves, slope_adjusted.estimate(sa_out.params), color='r', linewidth=3,
label='Reduced Chi: '+format(sa_out.redchi,'.3e'))
ax[0].set_xlabel('Wavelength (microns)')
ax[0].set_ylabel('Reflectance')
ax[0].legend()
#Plot components
for i,p in enumerate(sa_out.params.values()):
if p.name != 'S':
ax[1].plot(waves, slope_adjusted.A[:,i]*p.value,
label=p.name+': '+format(p.value,'.2e'))
#else:
# ax[1].plot(waves, S*waves - 15, label='S: '+format(p.value,'0.4f'))
#ax[1].set_yscale('log')
ax[1].legend()
ax[1].set_xlabel('Wavelength (microns)')
ax[1].set_ylabel('Reflectance')<jupyter_output><empty_output><jupyter_text># Constrained Linear Model with Slope Variation: $b = \sum x_i A_i *(\lambda + S)$
# where $\sum x_i = 1$<jupyter_code>class ConstrainedSlopeAdjustedModel(SpectralModel):
"""A simple linear model, based on known wavelength and spectral library with an additional fudge
factor (S) to adjust the spectral slope.
\sum x_i spec_i * (lambda + S)
"""
def __init__(self, waves: np.ndarray, spec_library: List[spectrum]):
# initialize model matrix
self.A = np.zeros((len(waves), len(spec_library)))
for i,spec in enumerate(spec_library):
self.A[:,i] = np.interp(waves, spec.wave, spec.A)
# initialize parameters
self._params = Parameters()
expr = '1'
for spec in spec_library[:-1]:
#Create initial blank paramters to allow expression building
self._params.add(spec.name.replace(" ", "_"), vary=True, min=0)
#Add to constraint expression
expr+= '-'+str(spec.name.replace(" ", "_"))
#Constrain all parameters to sum to 1
self._params.add(spec_library[-1].name.replace(" ", "_"), vary=True, min=0, expr=expr)
# Add paramater for S
self._params.add('S', vary=True, min=0)
def estimate(self, params: Parameters) -> np.ndarray:
x = np.array([p.value for p in params.values() if p.name != 'S']).flatten()
S = params['S'].value
return self.A @ x * (waves+S)
@property
def parameters(self):
return self._params
def set_parameters(self, params):
self._params = params
constrained_slope_adjusted = ConstrainedSlopeAdjustedModel(waves, spec_library)
rand = np.random.rand(11)
rand /= np.sum(rand)
constrained_slope_adjusted.fill_parameters(np.concatenate([rand, [1]])) #Provide random starting conditions
csa_out, csa_errors = constrained_slope_adjusted.fit(b, method='leastsq')
report_fit(csa_out)
fig, ax = plt.subplots(2,1, figsize=(15,15),
gridspec_kw={'height_ratios': [1.5, 2]})
plt.title('260W, Non-Icy Region')
ax[0].plot(waves, b, color='b')
ax[0].plot(waves, constrained_slope_adjusted.estimate(csa_out.params), color='r', linewidth=3,
label='Reduced Chi: '+format(csa_out.redchi,'.3e'))
ax[0].set_xlabel('Wavelength (microns)')
ax[0].set_ylabel('Reflectance')
ax[0].legend()
#Plot components
for i,p in enumerate(csa_out.params.values()):
if p.name != 'S':
ax[1].plot(waves, constrained_slope_adjusted.A[:,i]*p.value,
label=p.name+': '+format(p.value,'0.4f'))
#else:
# ax[1].plot(waves, S*waves - 15, label='S: '+format(p.value,'0.4f'))
#ax[1].set_yscale('log')
ax[1].legend()
ax[1].set_xlabel('Wavelength (microns)')
ax[1].set_ylabel('Reflectance')<jupyter_output><empty_output><jupyter_text># Linear Model with Sloped Base: $b = \sum x_i A_i + (x_m \lambda + x_b)$<jupyter_code>class SlopedBaseModel(SpectralModel):
"""A simple linear model, based on known wavelength and spectral library with an additional slobed base.
\sum x_i spec_i + (x_m lambda + x_b)
"""
def __init__(self, waves: np.ndarray, spec_library: List[spectrum]):
# initialize model matrix
self.A = np.zeros((len(waves), len(spec_library)))
for i,spec in enumerate(spec_library):
self.A[:,i] = np.interp(waves, spec.wave, spec.A)
# initialize parameters
self._params = Parameters()
for spec in spec_library:
self._params.add(spec.name.replace(" ", "_"), vary=True, min=0)
# Add paramater for x_m and x_b
self._params.add('slope', vary=True, min=0)
self._params.add('yint', vary=True, min=0)
def estimate(self, params: Parameters) -> np.ndarray:
x = np.array([p.value for p in params.values() if p.name != 'slope' and p.name != 'yint']).flatten()
m = params['slope'].value
yint = params['yint'].value
return self.A @ x + (m*waves+yint)
@property
def parameters(self):
return self._params
def set_parameters(self, params):
self._params = params
sloped_base = SlopedBaseModel(waves, spec_library)
rand = np.random.rand(11)
sloped_base.fill_parameters(np.concatenate([rand, [1,0]])) #Provide random starting conditions
sb_out, sb_errors = sloped_base.fit(b, method='leastsq')
report_fit(sb_out)
fig, ax = plt.subplots(2,1, figsize=(15,15),
gridspec_kw={'height_ratios': [1.5, 2]})
plt.title('260W, Non-Icy Region')
ax[0].plot(waves, b, color='b')
ax[0].plot(waves, sloped_base.estimate(sb_out.params), color='r', linewidth=3,
label='Reduced Chi: '+format(sb_out.redchi,'.3e'))
ax[0].set_xlabel('Wavelength (microns)')
ax[0].set_ylabel('Reflectance')
ax[0].legend()
#Plot components
for i,p in enumerate(sb_out.params.values()):
if p.name != 'slope' and p.name != 'yint':
ax[1].plot(waves, sloped_base.A[:,i]*p.value,
label=p.name+': '+format(p.value,'0.4f'))
ax[1].plot(waves, sb_out.params['slope']*waves+sb_out.params['yint'] , label='sloped base')
#ax[1].set_yscale('log')
ax[1].legend()
ax[1].set_xlabel('Wavelength (microns)')
ax[1].set_ylabel('Reflectance')<jupyter_output><empty_output><jupyter_text># Constrained Linear Model with Sloped Base: $b = \sum x_i A_i + x_j(x_m \lambda + x_b)$
# where $\sum x_i + x_j = 1$<jupyter_code>class ConstrainedSlopedBaseModel(SpectralModel):
"""A simple linear model, based on known wavelength and spectral library with an additional slobed base
where the coefficients are constrained to sum to 1.
0
\sum x_i spec_i + x_j(x_m lambda + x_b) where \sum(x_i) + x_j = 1
"""
def __init__(self, waves: np.ndarray, spec_library: List[spectrum]):
# initialize model matrix
self.A = np.zeros((len(waves), len(spec_library)))
for i,spec in enumerate(spec_library):
self.A[:,i] = np.interp(waves, spec.wave, spec.A)
# initialize parameters
self._params = Parameters()
# Add paramater for x_j, x_m, and x_b
self._params.add('x_j', vary=True, min=0)
self._params.add('x_m', vary=True, min=0)
self._params.add('x_b', vary=True, min=0)
#Constrain all parameters (x_i and x_j) to sum to 1
expr = '1-x_j'
for spec in spec_library[:-1]:
#Create initial blank paramters to allow expression building
self._params.add(spec.name.replace(" ", "_"), vary=True, min=0)
#Add to constraint expression
expr+= '-'+str(spec.name.replace(" ", "_"))
#Constrain all parameters to sum to 1
self._params.add(spec_library[-1].name.replace(" ", "_"), vary=True, min=0, expr=expr)
def estimate(self, params: Parameters) -> np.ndarray:
x = np.array([p.value for p in params.values() if not 'x_' in p.name]).flatten()
x_j = params['x_j'].value
slope = params['x_m'].value
yint = params['x_b'].value
return self.A @ x + x_j*(slope*waves+yint)
@property
def parameters(self):
return self._params
def set_parameters(self, params):
self._params = params
constrained_sloped_base = ConstrainedSlopedBaseModel(waves, spec_library)
rand = np.random.rand(12)
rand /= np.sum(rand)
constrained_sloped_base.fill_parameters(np.concatenate([[rand[0]], [1,0], rand[1:]])) #Provide random starting conditions
csb_out, csb_errors = constrained_sloped_base.fit(b, method='leastsq')
report_fit(csb_out)
fig, ax = plt.subplots(2,1, figsize=(15,15),
gridspec_kw={'height_ratios': [1.5, 2]})
plt.title('260W, Non-Icy Region')
ax[0].plot(waves, b, color='b')
ax[0].plot(waves, constrained_sloped_base.estimate(csb_out.params), color='r', linewidth=3,
label='Reduced Chi: '+format(csb_out.redchi,'.3e'))
ax[0].set_xlabel('Wavelength (microns)')
ax[0].set_ylabel('Reflectance')
ax[0].legend()
#Plot components
i=0
for p in csb_out.params.values():
if 'x_' not in p.name:
ax[1].plot(waves, constrained_sloped_base.A[:,i]*p.value,
label=p.name+': '+format(p.value,'0.4f'))
i+=1
ax[1].plot(waves, csb_out.params['x_j']*(csb_out.params['x_m']*waves+csb_out.params['x_b'])
, label='sloped base')
#ax[1].set_yscale('log')
ax[1].legend()
ax[1].set_xlabel('Wavelength (microns)')
ax[1].set_ylabel('Reflectance')<jupyter_output><empty_output><jupyter_text># See Mathematically best possible fit for this family of functions:
## Slope Adjusted per Spectra: $b = \sum x_i*A_i*\lambda + A_i*y_i$
<jupyter_code>class SlopedAdjustedPerSpecModel(SpectralModel):
"""Not physically reasonable, but to see "mathematically best possible fit", we let the slope of
each spectrum vary independently
"""
def __init__(self, waves: np.ndarray, spec_library: List[spectrum]):
# initialize model matrix
self.A = np.zeros((len(waves), len(spec_library)))
for i,spec in enumerate(spec_library):
self.A[:,i] = np.interp(waves, spec.wave, spec.A)
# initialize parameters
self._params = Parameters()
for spec in spec_library:
self._params.add(spec.name.replace(" ", "_"), vary=True, min=0)
self._params.add(spec.name.replace(" ", "_")+'_y', vary=True, min=0)
def estimate(self, params: Parameters) -> np.ndarray:
x = np.array([p.value for p in params.values() if '_y' not in p.name]).flatten()
y = np.array([p.value for p in params.values() if '_y' in p.name]).flatten()
return (self.A @ x)*waves + (self.A @ y)
@property
def parameters(self):
return self._params
def set_parameters(self, params):
self._params = params
indiv_slope_adjusted = SlopedAdjustedPerSpecModel(waves, spec_library)
indiv_slope_adjusted.fill_parameters(np.random.rand(22)) #Provide random starting conditions
isa_out, isa_errors = indiv_slope_adjusted.fit(b, method='leastsq')
report_fit(isa_out)
fig, ax = plt.subplots(2,1, figsize=(15,15),
gridspec_kw={'height_ratios': [1.5, 2]})
plt.title('260W, Non-Icy Region')
ax[0].plot(waves, b, color='b')
ax[0].plot(waves, indiv_slope_adjusted.estimate(isa_out.params), color='r', linewidth=3,
label='Reduced Chi: '+format(isa_out.redchi,'.3e'))
ax[0].set_xlabel('Wavelength (microns)')
ax[0].set_ylabel('Reflectance')
ax[0].legend()
#Plot components
i=0
for key in isa_out.params.keys():
if '_y' not in key:
A = indiv_slope_adjusted.A[:,i]
ax[1].plot(waves, isa_out.params[key].value*A*waves + A*isa_out.params[key+'_y'],
label=isa_out.params[key].name)#+': '+format(isa_out.params[key].value,'0.4f'))
i+=1
#ax[1].set_yscale('log')
ax[1].legend()
ax[1].set_xlabel('Wavelength (microns)')
ax[1].set_ylabel('Reflectance')<jupyter_output><empty_output>
|
no_license
|
/LMFit_tutorial.ipynb
|
RyleighFitz/VLT_Sinfoni_Europa
| 8 |
<jupyter_start><jupyter_text># The Prediction Task
We will use the dataset gathered by our scraping script [Github](https://github.com/shkhaksar/book-classification) merged with two datasets from Kaggle. [Dataset1](https://www.kaggle.com/mdhamani/goodreads-books-100k) - [Dataset2](https://www.kaggle.com/sp1thas/book-depository-dataset)
The dataset contains book summaries for 200K books extracted from [Goodreads](https://www.goodreads.com/) and [BookDepository](https://www.bookdepository.com/) websites.
The classification goal is, given a new book, to predict the book belongs to which category of books.#### Install dependencies<jupyter_code>%pip install langdetect nltk<jupyter_output><empty_output><jupyter_text>#### Import Useful packages<jupyter_code>import pyspark
from pyspark.sql import *
from pyspark.sql.types import *
from pyspark.sql.functions import *
from pyspark import SparkContext, SparkConf
from pyspark.sql.functions import isnan
import numpy as np
from numpy import nan
import pandas as pd
%matplotlib inline
import matplotlib.pyplot as plt
import seaborn as sns
RANDOM_SEED=100<jupyter_output><empty_output><jupyter_text>## 1. Data Acquisition
This is the first step we need to accomplish before going any further. The dataset will be downloaded and loaded to DBFS.
##### Download the dataset to the local driver node's /tmp folder using wget<jupyter_code>%sh wget -P /tmp https://github.com/shkhaksar/book-classification/raw/master/classifier/dataset.csv.bz2
%fs ls file:/tmp/<jupyter_output><empty_output><jupyter_text>##### Move the file from local driver node's file system to DBFS (in the directory we created)<jupyter_code>%fs mkdirs book-classifier/datasets/
dbutils.fs.mv("file:/tmp/dataset.csv.bz2", "dbfs:/book-classifier/datasets/dataset.csv.bz2")
#dbutils.fs.rm("dbfs:/book_classifier/datasets/dataset.csv.bz2") remove csv command<jupyter_output><empty_output><jupyter_text>##### Read dataset file into a Spark Dataframe<jupyter_code>book_df = spark.read.load("dbfs:/book-classifier/datasets/dataset.csv.bz2",
format="csv",quote="\"",escape="\"",
sep=",", multiLine=True,
inferSchema="true",
header="true")<jupyter_output><empty_output><jupyter_text>##### Dataset Shape and Schema
The dataset contains 200K+ records of Books; each record, is represented by the following set of 3 columns:
- Title: The books title, although it's not being used in this task.
- Description: A brief description about the book.
- Category: A category of books, consists of 34 distinct categories.##### Check the shape of the loaded dataset, i.e., number of rows and columns<jupyter_code>print("The shape of the dataset is {:d} rows by {:d} columns".format(book_df.count(), len(book_df.columns)))<jupyter_output><empty_output><jupyter_text>##### Print out the schema of the loaded dataset<jupyter_code>book_df.printSchema()<jupyter_output><empty_output><jupyter_text>We drop `Title` column to decrease memory footprint of our training task.<jupyter_code>book_df = book_df.drop('Title')<jupyter_output><empty_output><jupyter_text>##### Display the first 5 rows of the dataset<jupyter_code>book_df.show(n=5,truncate=True)<jupyter_output><empty_output><jupyter_text>##### Check for duplicated Books<jupyter_code>print("The total number of duplicated books are {:d} out of {:d}".format(book_df.count() - book_df.dropDuplicates(['Description']).count(), book_df.count())) <jupyter_output><empty_output><jupyter_text>##### Drop duplicated books<jupyter_code>book_df = book_df.dropDuplicates(['Description'])
print("The total number of unique books is: {:d}".format(book_df.count()))<jupyter_output><empty_output><jupyter_text>##### Check for any missing values<jupyter_code>for c in book_df.columns:
print("N. of missing values of column `{:s}` = {:d}".format(c, book_df.where(col(c).isNull() | isnan(col(c)) | (col(c) == " ")).count()))<jupyter_output><empty_output><jupyter_text>##### Remove the missing values<jupyter_code>book_df = book_df.na.drop()
for c in book_df.columns:
book_df = book_df.filter(~col(c).isNull()).filter(~isnan(col(c))).filter(~(col(c) == " ")) #drop nan values too
for c in book_df.columns:
print("N. of missing values of column `{:s}` = {:d}".format(c, book_df.where(col(c).isNull() | isnan(col(c)) | (col(c) == " ")).count()))<jupyter_output><empty_output><jupyter_text>##### Now we briefly check our categories<jupyter_code>book_df.groupBy("Category").count().orderBy("Count",ascending=False).show(30,truncate=False)<jupyter_output><empty_output><jupyter_text>##### Let's see how many different categories do we have in our dataset<jupyter_code>n_cats = book_df.groupby("Category").count().count()
print("Total Number of categories in our database {}".format(n_cats))<jupyter_output><empty_output><jupyter_text>## 2. Data Exploration/Pre-processing#### Detecting text languages
To train only based on english books, all records which content is not in engish needs to be removed.
We detect language by using [langdetect](https://github.com/Mimino666/langdetect). The result will be a new dataframe with an aditional column `lang`. Finally, we will remove rows not containing `en`.
Below you can view the helper function for detecting languages:<jupyter_code>def language_detect(df, description_col="Description"):
from langdetect import detect
def detector(text)->str:
try:
return detect(text)
except:
return None
detect_udf = udf(lambda desc: detector(desc), StringType())
detected_df = df.withColumn("lang", detect_udf(description_col)).cache()
detected_df.show(10)
return detected_df
book_df = language_detect(book_df)<jupyter_output><empty_output><jupyter_text>Now let's see how many records have `en` as their language.<jupyter_code>book_df.groupBy("lang").count().orderBy("Count",ascending=False).show()<jupyter_output><empty_output><jupyter_text>Now our dataset is tagged in `lang` column. we can now safely remove rows without `en`tag, then drop `lang` column.<jupyter_code>book_df = book_df.filter((col("lang") == "en")).select(["Description","Category"])
print("The shape of the dataset is {:d} rows by {:d} columns".format(book_df.count(), len(book_df.columns)))<jupyter_output><empty_output><jupyter_text>#### Data Visualization
It is a good time to visualize our data. We use matplotlib library to get a sense of possible insights.<jupyter_code>genre_list = [ # this is a refrence for all of possible genres
'Art','Biography','Business & Travel','Children & Comics','Christian','Classics','Cookbooks','Fantasy', 'Science Fiction','Nonfiction','Historical Fiction','Fiction','Graphic Novels','History','Horror','Memoir','Mystery', 'Romance','Science', 'Self Help','Thriller','Young Adult']
p_book_df = book_df.toPandas()<jupyter_output><empty_output><jupyter_text><jupyter_code>fig, axe = plt.subplots(1, 1, figsize=(120,20))
ax = sns.countplot(p_book_df["Category"], ax=axe)
_ = ax.set_xticklabels(ax.get_xticklabels(), rotation=45, horizontalalignment='right')
for tick in ax.xaxis.get_major_ticks():
tick.label.set_fontsize(60)
for tick in ax.yaxis.get_major_ticks():
tick.label.set_fontsize(40)
<jupyter_output><empty_output><jupyter_text>#### We want to have a look a few description and category pairs.<jupyter_code>def print_plot(index):
example = p_book_df[p_book_df.index == index][['Description', 'Category']].values[0]
if len(example) > 0:
print(example[0])
print("---------")
print('Category:', example[1])
print_plot(10)
print("\n")
print_plot(100)<jupyter_output><empty_output><jupyter_text>#### Text Pre-processing and cleaning
As preliminary steps of any NLP task, at least the following pipeline must be executed first:
- Text cleaning:
- Case normalization (lower) -> convert all text to lower case;
- Filter out leading and trailing whitespaces (trim);
- Filter out punctuation symbols (regexp_replace);
- Filter out newlines and tabs (regexp_replace);
- Filter out any internal extra whitespace resulting from the step above (regexp_replace + trim).
- Tokenization (Tokenizer): splitting raw text into a list of individual tokens (i.e., words), typically using whitespace as delimiter
- Stopwords removal (StopWordsRemover): removing so-called stopwords, namely words that do not contribute to the deeper meaning of the document like "the", "a", "me", etc.
- Stemming (SnowballStemmer): reducing each word to its root or base. For example "fishing", "fished", "fisher" all reduce to the stem "fish".<jupyter_code>def clean_text(df, column_name="Description"):
"""
This function takes the raw text data and applies a standard NLP preprocessing pipeline consisting of the following steps:
- Text cleaning
- Tokenization
- Stopwords removal
- Stemming (Snowball stemmer)
parameter: dataframe
returns: the input dataframe along with the `cleaned_content` column as the results of the NLP preprocessing pipeline
"""
from pyspark.sql.functions import udf, col, lower, trim, regexp_replace
from pyspark.ml.feature import Tokenizer, StopWordsRemover
from nltk.stem.snowball import SnowballStemmer # BE SURE NLTK IS INSTALLED ON THE CLUSTER
# Text preprocessing pipeline
print("***** Text Preprocessing Pipeline *****\n")
# 1. Text cleaning
print("# 1. Text Cleaning\n")
# 1.a Case normalization
print("1.a Case normalization:")
lower_case_df = df.withColumn(column_name,lower(col(column_name)).alias(column_name))
lower_case_df.show(10)
# 1.b Trimming
print("1.b Trimming:")
trimmed_df = lower_case_df.withColumn(column_name,trim(col(column_name)).alias(column_name))
trimmed_df.show(10)
# 1.c Filter out punctuation symbols
print("1.c Filter out punctuation:")
no_punct_df = trimmed_df.withColumn(column_name,(regexp_replace(col(column_name), "[^a-zA-Z\\s]", "")).alias(column_name))
no_punct_df.show(10)
# 1.d Filter out any new line
print("1.d Filter out any new line or tab:")
no_new_line_df = no_punct_df.withColumn(column_name,trim(regexp_replace(col(column_name), "\s+", " ")).alias(column_name))
no_new_line_df.show(10)
# 1.d Filter out any internal extra whitespace
print("1.d Filter out extra whitespaces:")
cleaned_df = no_new_line_df.withColumn(column_name,trim(regexp_replace(col(column_name), " +", " ")).alias(column_name))
cleaned_df.show(10)
# 2. Tokenization (split text into tokens)
print("# 2. Tokenization:")
tokenizer = Tokenizer(inputCol=column_name, outputCol="tokens")
tokens_df = tokenizer.transform(cleaned_df).cache()
tokens_df.show(10)
# 3. Stopwords removal
print("# 3. Stopwords removal:")
stopwords_remover = StopWordsRemover(inputCol="tokens", outputCol="terms")
terms_df = stopwords_remover.transform(tokens_df).cache()
terms_df.show(10)
# 4. Stemming (Snowball stemmer)
print("# 4. Stemming:")
stemmer = SnowballStemmer(language="english")
stemmer_udf = udf(lambda tokens: [stemmer.stem(token) for token in tokens], ArrayType(StringType()))
terms_stemmed_df = terms_df.withColumn("terms_stemmed", stemmer_udf("terms")).cache()
terms_stemmed_df.show(10)
return terms_stemmed_df
clean_book_df = clean_text(book_df,"Description")<jupyter_output><empty_output><jupyter_text>We rename column `terms_stemmed` to `terms` for convenience:<jupyter_code>clean_book_df = clean_book_df.select(['Category','Description','terms_stemmed']).withColumnRenamed('terms_stemmed','terms')<jupyter_output><empty_output><jupyter_text>## 3. Learning Pipeline
##### - Logistic Regression (TF-IDF - ParamGrid - K-Flod Cross Validation)
##### - Naive Bayes Model (TF-IDF - ParamGrid - K-Flod Cross Validation)
##### - Random Forest (TF-IDF - ParamGrid - K-Flod Cross Validation)<jupyter_code>VOCAB_SIZE = 20000 # number of words to be retained as vocabulary
MIN_DOC_FREQ = 10 # minimum number of documents a word has to appear in to be included in the vocabulary
N_FEATURES = 20000 # default embedding vector size (if HashingTF or, later, Word2Vec are used)<jupyter_output><empty_output><jupyter_text>As our database is (Almost) balanced we will use Simple Random Spliting to split our train and testset.<jupyter_code>train_df, test_df = clean_book_df.randomSplit([0.7, 0.3], seed=RANDOM_SEED)
print("Training Set count: {}".format(train_df.count()))
print("Test Set count: {}".format(test_df.count()))
def logistic_regression_pipeline(df, terms_feature_col="terms", label_col="Category"):
from pyspark.ml.feature import StringIndexer,HashingTF, IDF
from pyspark.ml.classification import LogisticRegression
from pyspark.ml import Pipeline
from pyspark.ml.evaluation import MulticlassClassificationEvaluator
from pyspark.ml.tuning import ParamGridBuilder, CrossValidator
label_string_idx = StringIndexer(inputCol=label_col, outputCol = "label") #categories => numebers
hashingTF = HashingTF(inputCol=terms_feature_col, outputCol="tf_features", numFeatures=N_FEATURES)
idf = IDF(inputCol="tf_features", outputCol="features")
lr = LogisticRegression(maxIter=20)
pipeline = Pipeline(stages=[label_string_idx, hashingTF, idf, lr])
# Create ParamGrid for Cross Validation
paramGrid = (ParamGridBuilder()
.addGrid(lr.regParam, [0.1, 0.3, 0.5]) # regularization parameter
.addGrid(lr.elasticNetParam, [0.0, 0.1]) # Elastic Net Parameter (Ridge = 0)
.build())
# Create 5-fold CrossValidator
cv = CrossValidator(estimator=pipeline, \
estimatorParamMaps=paramGrid, \
evaluator=MulticlassClassificationEvaluator(), \
numFolds=5, collectSubModels=True)
cv_model = cv.fit(df)
return cv_model
cv_model = logistic_regression_pipeline(train_df ,terms_feature_col="terms", label_col="Category")
def lr_model_evaluation(cv_models, test_dataset):
from pyspark.ml.evaluation import MulticlassClassificationEvaluator
# Make predictions.
predictions = cv_models.transform(test_dataset)
predictions.filter(predictions['prediction'] == 0) \
.select("Description","Category","probability","label","prediction") \
.orderBy("probability", ascending=False) \
.show(n = 10, truncate = 30)
evaluator = MulticlassClassificationEvaluator(predictionCol="prediction")
accuracy= evaluator.evaluate(predictions)
print("""****Test evaluation*****""")
print("Test Accuracy = %g "%(accuracy))
print("""****Test evaluation*****""")
print("-----------")
lrModel = cv_models.bestModel.stages[2]
print(lrModel) # summary only
lr_model_evaluation(cv_model,test_df)
lr_model_evaluation(cv_model,train_df)<jupyter_output><empty_output><jupyter_text>### Naive Bayes Model<jupyter_code>def nb_pipeline(df, terms_feature_col="terms", label_col="Category"):
from pyspark.ml.feature import StringIndexer, HashingTF, IDF
from pyspark.ml import Pipeline
from pyspark.ml.evaluation import MulticlassClassificationEvaluator
from pyspark.ml.tuning import ParamGridBuilder, CrossValidator
from pyspark.ml.classification import NaiveBayes
label_string_idx = StringIndexer(inputCol=label_col, outputCol = "label") #categories => numebers
hashingTF = HashingTF(inputCol=terms_feature_col, outputCol="tf_features", numFeatures=N_FEATURES)
idf = IDF(inputCol="tf_features", outputCol="features")
nb = NaiveBayes(smoothing=1)
pipeline = Pipeline(stages=[label_string_idx, hashingTF, idf, nb])
paramGrid = (ParamGridBuilder()
.addGrid(nb.smoothing, [0.0, 0.5, 1.0])
.build())
# Create 5-fold CrossValidator
cv = CrossValidator(estimator=pipeline, \
estimatorParamMaps=paramGrid, \
evaluator=MulticlassClassificationEvaluator(), \
numFolds=5, collectSubModels=True)
cv_model = cv.fit(df)
return cv_model
nb_model = nb_pipeline(train_df,terms_feature_col="terms", label_col="Category")
def nb_model_evaluation(nb_model,test_dataset):
from pyspark.ml.evaluation import MulticlassClassificationEvaluator
# Make predictions.
predictions = nb_model.transform(test_dataset)
predictions.filter(predictions['prediction'] == 0) \
.select("Description","Category","probability","label","prediction") \
.orderBy("probability", ascending=False) \
.show(n = 10, truncate = 30)
evaluator = MulticlassClassificationEvaluator(predictionCol="prediction")
accuracy = evaluator.evaluate(predictions)
print("Test Accuracy = %g "%(accuracy))
print("-----------")
nb_model = nb_model.bestModel.stages[2]
print(nb_model) # summary only
nb_model_evaluation(nb_model,test_df)
nb_model_evaluation(nb_model,train_df)<jupyter_output><empty_output><jupyter_text># Random Forest<jupyter_code>def rf_pipeline(df, terms_feature_col="terms", label_col="Category"):
from pyspark.ml.feature import StringIndexer,HashingTF, IDF
from pyspark.ml.classification import RandomForestClassifier
from pyspark.ml import Pipeline
from pyspark.ml.evaluation import MulticlassClassificationEvaluator
from pyspark.ml.tuning import ParamGridBuilder, CrossValidator
from pyspark.ml.classification import NaiveBayes
label_string_idx = StringIndexer(inputCol=label_col, outputCol = "label") #categories => numebers
hashingTF = HashingTF(inputCol=terms_feature_col, outputCol="tf_features", numFeatures=N_FEATURES)
idf = IDF(inputCol="tf_features", outputCol="features")
rf = RandomForestClassifier()
pipeline = Pipeline(stages=[label_string_idx, hashingTF, idf, rf])
paramGrid = (ParamGridBuilder()
.addGrid(rf.maxDepth, [3, 8]) \
.addGrid(rf.numTrees, [10,100]) \
.build())
# Create 5-fold CrossValidator
cv = CrossValidator(estimator=pipeline, \
estimatorParamMaps=paramGrid, \
evaluator=MulticlassClassificationEvaluator(), \
numFolds=5, collectSubModels=True)
cv_model = cv.fit(df)
return cv_model
rf_model = rf_pipeline(train_df,terms_feature_col="terms", label_col="Category")
def evaluate_model(predictions):
from pyspark.ml.evaluation import MulticlassClassificationEvaluator
evaluator = MulticlassClassificationEvaluator(predictionCol="prediction")
return evaluator.evaluate(predictions)
def rf_model_evaluation(rf_model,test_dataset):
from pyspark.ml.evaluation import MulticlassClassificationEvaluator
# Make predictions.
predictions = rf_model.transform(test_dataset)
predictions.filter(predictions['prediction'] == 0) \
.select("Description","Category","probability","label","prediction") \
.orderBy("probability", ascending=False) \
.show(n = 10, truncate = 30)
print("***** Test Set *****")
print("Accuracy = ",evaluate_model(predictions))
print(rf_model) # summary only
print("***** Test Set *****")
print("--------------------")
rf_model = rf_model.bestModel.stages[2]
rf_model_evaluation(rf_model,test_df)<jupyter_output><empty_output>
|
no_license
|
/classifier/book-classifier.ipynb
|
shkhaksar/book-classification
| 27 |
<jupyter_start><jupyter_text># 教學目標
主要說明matplotlib 的基礎操作
1. 使用常見的子圖與軸圖來做畫面配置
2. 長條圖
# 範例重點
如何使用亂數, 資料集來操作
# 軸圖進階
但是可以將圖放置在圖中的任何位置。因此,如果要在較大的圖中放置較小的圖,則可以使用軸。
#特別提醒: tick 刻度線定位器
格式正確的刻度線是準備發布的數據的重要組成部分。Matplotlib為滴答提供了一個完全可配置的系統。有刻度線定位器可以指定刻度線應出現的位置,刻度線格式化程序可以為刻度線提供所需的外觀。主刻度線和次刻度線可以相互獨立地定位和格式化。
# 作業: 繪製如下圖示:

<jupyter_code>import matplotlib.pyplot as plt
#決定底框
plt.axes([0.1, 0.1, .5, .5])
#給定刻度, 若不給定值, 圖的周邊無文字
plt.xticks([]), plt.yticks([])
plt.text(0.1, 0.1, 'axes([0.1,0.1,.5,.5])', ha = 'left', va = 'center', size = 16, alpha = .5)
#決定第二層框
plt.axes([0.3, 0.3, .5, .5])
#給定刻度, 若不給定值, 圖的周邊無文字
#plt.xticks([]), plt.yticks([])
plt.text(0.1, 0.1, 'axes([0.3, 0.3, .5, .5])', ha = 'left', size = 16, alpha = .5)
#決定第三層框
plt.axes([0.5, 0.5, .5, .5])
#給定刻度
#plt.xticks([]), plt.yticks([])
plt.text(0.1, 0.1, 'axes([0.5, 0.5, .5, .5])', ha = 'left', size = 16, alpha = .5)
#決定第四層框
plt.axes([0.7, 0.7, .5, .5])
plt.xticks([]), plt.yticks([])
plt.text(0.1, 0.1, 'axes([0.7, 0.7, .5, .5])', ha = 'left', size = 16, alpha = .5)
plt.show()<jupyter_output><empty_output><jupyter_text># 條型圖: Bar Plots
長條圖主要用來呈現兩個維度的資料,一個為X軸另一個則為Y軸(當然這邊指的是二維的狀況,較為常見)
主要用來呈現兩個維度的資料
# 問題: 嘗試通過添加紅色條形標籤重現上側的圖形。
# 在 四個像限的一二像限繪圖<jupyter_code>import numpy as np
import matplotlib.pyplot as plt
#配置12 組 Bar
n = 12
X = np.arange(n)
#給定數學運算式
Y1 = (1-X/float(n)) * np.random.uniform(0.5, 1.0, n)
Y2 = (1-X/float(n)) * np.random.uniform(0.5, 1.0, n)
#指定上半部繪製區域, 給定 Bar 顏色, 邊界顏色
plt.bar(X, +Y1, facecolor = '#9999ff', edgecolor = 'white')
# +Y 指的是 XY 四象限的第一象限
# -Y 指的是 XY 四象限的第二象限
#在此coding
#指定下半部繪製區域, 給定 Bar 顏色, 邊界顏色
#顏色除了用色標外, 也可以用顏色文字描述, red: 紅色
plt.bar(X, -Y2, facecolor = 'red', edgecolor = 'black')
#設定繪圖圖示區間
for x, y in zip(X, Y1):
plt.text(x + 0.4, y + 0.05, '%.2f'%y, ha = 'center', va = 'bottom')
#設定Y軸區間
plt.ylim(-1.25, +1.25)
plt.show()<jupyter_output><empty_output>
|
no_license
|
/20_Homework.ipynb
|
NickChen0417/DS_60days
| 2 |
<jupyter_start><jupyter_text>### Seulement pour le debug / la trame<jupyter_code>plan_coord
add = np.array([in_coord[0],])
in_coord = np.concatenate((in_coord, add))
in_coord
fig = plt.figure()
plt.scatter(in_coord[:, 0], in_coord[:, 1], c='black')
plt.show(plt)
(size, dim) = in_coord.shape
size
for i in range(0, size - 1):
x1 = in_coord[i,0]
y1 = in_coord[i,1]
x2 = in_coord[i+1,0]
y2 = in_coord[i+1,1]
print("x1 {} y1 {} x2 {} y2 {}".format(x1,y1,x2,y2))
print("OK")
plan_coord = np.array([[]])
index = -1
for i in range(0, size - 1):
x1 = in_coord[i,0]
y1 = in_coord[i,1]
x2 = in_coord[i+1,0]
y2 = in_coord[i+1,1]
if index == -1:
plan_coord = np.array([[(int) (x1 + x2)/2, (int) (y1 + y2)/2],])
index = index + 1
else:
add = np.array([[(int) (x1 + x2)/2, (int) (y1 + y2)/2],])
plan_coord = np.concatenate((plan_coord, add))
add = np.array([[x2, y2],])
plan_coord = np.concatenate((plan_coord, add))
add = np.array([plan_coord[0],])
plan_coord = np.concatenate((plan_coord, add))
print("plan_coord --> {}".format(plan_coord))
fig = plt.figure()
plt.scatter(in_coord[:, 0], in_coord[:, 1], s=500, c='black')
plt.scatter(plan_coord[:, 0], plan_coord[:, 1], s=100, c='green')
plt.show(plt)
def bezier(points, steps=10):
n = len(points)
b = [scipy.special.binom(n - 1, i) for i in range(n)]
r = np.arange(n)
#print("points {} binom {} r {}".format(points, b, r))
for t in np.linspace(0, 1, steps):
u = np.power(t, r) * np.power(1 - t, n - r - 1) * b
#print("u {}".format(u))
yield t, u @ points
i = 3
k = 0
curve_coord = np.array([p for _, p in bezier(plan_coord[k:k+i])])
for j in range(0,size-1):
k = k + 2;
add = np.array([p for _, p in bezier(plan_coord[k:k+i])])
curve_coord = np.concatenate((curve_coord, add))
#curve_coord
fig = plt.figure()
plt.scatter(in_coord[:, 0], in_coord[:, 1], s=500, c='black')
plt.scatter(plan_coord[:, 0], plan_coord[:, 1], s=100, c='green')
plt.scatter(curve_coord[:, 0], curve_coord[:, 1], c='red')
plt.show(plt)
<jupyter_output><empty_output>
|
no_license
|
/.ipynb_checkpoints/02_Planning_Bezier-checkpoint.ipynb
|
gr0mph/CodersStrikeBack
| 1 |
<jupyter_start><jupyter_text># Pretty Good Privacy (PGP)
- A data encryption/decryption tool
- Can be used to encrypt and authenticat email, files, etc.
- Created by Phil Zimmermann in 1991
- A practical hybrid system that uses symmetric and asymmetric crypto
image source: wikipedia<jupyter_code>def enc_key(public_key, message):
pass
def dec_key(private_key, ciphertext):
pass
def enc_msg(key, iv, msg):
pass
def dec_msg(key, iv, ciphertext):
pass
def gen_hmac(key, msg):
pass
def verify_hmac(key, msg, sig):
pass
k1 = os.urandom(16)
k2 = os.urandom(16)
iv = os.urandom(16)
msg = b"PyCon2017 Crypto"
cipher = enc_msg(k1, iv, msg)
sig = gen_hmac(k2, msg)
encrypted_key = enc_key(public_key, k1)
decrypted_key = dec_key(private_key, encrypted_key)
plaintext = dec_msg(decrypted_key, iv, cipher)
verify_hmac(k2, plaintext, sig)<jupyter_output><empty_output>
|
permissive
|
/6. Pretty Good Privacy (PGP).ipynb
|
dfeinzeig/pycon2017_crypto_tutorial
| 1 |
<jupyter_start><jupyter_text># Oblig 1 - Sanders<jupyter_code>import time
import math
import matplotlib.pyplot as plt
import numpy as np
import geneticAlgorithm as ga
import exhaustiveSearch as es
import hill
import data
<jupyter_output><empty_output><jupyter_text>My cities are mapped to a list of ints, to make it easier. Under is the mapping shown.<jupyter_code>df = data.data(data.path_to_datafile)
representation = data.get_representation(df)
cities = df.columns
# [0, 1, 9, 4, 5, 2, 6, 8, 3, 7]
for city, city_int in zip(cities, representation):
print(f" City: {city} = {city_int} ")<jupyter_output> City: Barcelona = 0
City: Belgrade = 1
City: Berlin = 2
City: Brussels = 3
City: Bucharest = 4
City: Budapest = 5
City: Copenhagen = 6
City: Dublin = 7
City: Hamburg = 8
City: Istanbul = 9
City: Kiev = 10
City: London = 11
City: Madrid = 12
City: Milan = 13
City: Moscow = 14
City: Munich = 15
City: Paris = 16
City: Prague = 17
City: Rome = 18
City: Saint Petersburg = 19
City: Sofia = 20
City: Stockholm = 21
City: Vienna = 22
City: Warsaw = 23
<jupyter_text>
## Exhaustive Search
<jupyter_code>es_time_data = {} # For timing each iteration
exhaustive_search_data = {} # For staring the best paths and socres
for i in range(2,11): # iterating trough subset sizes
start_time = time.time()
bestPath, bestScore = es.exhaustiveSearch(i) # Running algorithm
end_time = time.time()
# Storing data
exhaustive_search_data[i] = (bestScore, bestPath)
es_time_data[i] = end_time - start_time
for key in es_time_data:
print(f"#Cities: {key}\n time: {es_time_data[key]}, \n Score: {exhaustive_search_data[key][0]},\n Path: {exhaustive_search_data[key][1]}")<jupyter_output>#Cities: 2
time: 0.007389068603515625,
Score: 3056.26,
Path: [0, 1]
#Cities: 3
time: 0.0030007362365722656,
Score: 4024.99,
Path: [0, 1, 2]
#Cities: 4
time: 0.002092123031616211,
Score: 4241.89,
Path: [0, 1, 2, 3]
#Cities: 5
time: 0.0,
Score: 4983.38,
Path: [0, 1, 4, 2, 3]
#Cities: 6
time: 0.01593637466430664,
Score: 5018.8099999999995,
Path: [0, 1, 4, 5, 2, 3]
#Cities: 7
time: 0.051158905029296875,
Score: 5487.89,
Path: [0, 1, 4, 5, 2, 6, 3]
#Cities: 8
time: 0.38487792015075684,
Score: 6667.49,
Path: [0, 1, 4, 5, 2, 6, 3, 7]
#Cities: 9
time: 3.308502674102783,
Score: 6678.55,
Path: [0, 1, 4, 5, 2, 6, 8, 3, 7]
#Cities: 10
time: 33.172536849975586,
Score: 7486.3099999999995,
Path: [0, 1, 9, 4, 5, 2, 6, 8, 3, 7]
<jupyter_text>
We see that for each city we add the time is increased by a substantial amount. From 9 to 10 it is aprox. a factor of 10. But we know gereraly in parmutation problems the increase of complexity is N!.
To calculate a rough estimate of time for 24 cities, we can take the time 10 cities take and multiply it with the facotr 24! / 10!. Wich means 24 cities takes aprox. 31 * 24! / 10! seconds.<jupyter_code>years = ((math.factorial(24)/math.factorial(10))*es_time_data[10])/60/60/24/365.25
print(years)<jupyter_output>179728667880.4142
<jupyter_text>So that is not going to work.## Hill Climbing
<jupyter_code># Hill Climbing
bestScore, worstScore = -1, -1
numCities_list = list(range(2,11)) + [24]
runs = 20
hill_scores = {}
bestScores = {}
worstScores = {}
sdScores = {}
times_run = {}
means = {}
for numCities in numCities_list:
scores = []
times = []
for _ in range(runs):
data_subset = data.data_subset(data.path_to_datafile, numCities)
start_time = time.time()
path, score = hill.hill(data_subset, 1000)
end_time = time.time()
scores.append(score)
times.append(end_time - start_time)
# Best
bestScores[numCities] = min(scores)
# Worst
worstScores[numCities] = max(scores)
# Mean
# ip.embed()
mean = sum(scores) / len(scores)
means[numCities] = mean
# Sd
s = 0
for score in scores:
s += (score - mean)**2
s = (s/len(scores))**(1/2)
sdScores[numCities] = s
# Time
mean_time = sum(times) / len(times)
times_run[numCities] = mean_time
# Report
for numCities in numCities_list:
print(
f"numCities: {numCities}\n" +
f" Best: {bestScores[numCities]},\n" +
f" Worst: {worstScores[numCities]}, \n" +
f" SD: {sdScores[numCities]}, \n" +
f" time: {times_run[numCities]}"
)<jupyter_output>numCities: 2
Best: 3056.26,
Worst: 3056.26,
SD: 9.094947017729282e-13,
time: 0.02123422622680664
numCities: 3
Best: 4024.99,
Worst: 4024.99,
SD: 4.547473508864641e-13,
time: 0.03040180206298828
numCities: 4
Best: 4241.89,
Worst: 4241.89,
SD: 0.0,
time: 0.03973288536071777
numCities: 5
Best: 4983.38,
Worst: 5776.78,
SD: 229.13615651834587,
time: 0.048692739009857176
numCities: 6
Best: 5018.8099999999995,
Worst: 6107.72,
SD: 344.6748576789435,
time: 0.05705734491348267
numCities: 7
Best: 5487.89,
Worst: 7167.74,
SD: 415.5250063507005,
time: 0.06733838319778443
numCities: 8
Best: 7082.219999999999,
Worst: 8921.189999999999,
SD: 524.4511249132275,
time: 0.0767057180404663
numCities: 9
Best: 7539.180000000001,
Worst: 9985.86,
SD: 578.7263467086581,
time: 0.08658030033111572
numCities: 10
Best: 8597.86,
Worst: 11309.64,
SD: 690.3060079240219,
time: 0.09535678625106811
numCities: 24
Best: 24263.2099999[...]<jupyter_text>## Genetic Algorithm
I have chosen to use inversion mutation and pmx for crossover, and with the subset sizes 6, 10, 24.<jupyter_code># Genetic algorithm
print("Genetic Algorithm")
numCities_list = [6,10, 24]
runs = 20
iterations = 1000 # If you want ot run recomened to ether recude iterations or pop size
mutation_prob = 0.05
pop_size = 40
hill_scores = {}
bestScores = {}
worstScores = {}
sdScores = {}
times_run = {}
means = {}
average_scores_run = {}
# r = ga.geneticAlgorithm(
# 10,
# 10000,
# 10,
# 0.05,
# debug=False
# )
for numCities in numCities_list:
local_min_scores = []
scores = []
times = []
for _ in range(runs):
data_subset = data.data_subset(data.path_to_datafile, numCities)
start_time = time.time()
r = ga.geneticAlgorithm(
pop_size,
iterations,
numCities,
mutation_prob,
debug=False
)
score = r["bestScore"]
path = r["bestPath"]
minScores =r["minScores"]
end_time = time.time()
scores.append(score)
times.append(end_time - start_time)
local_min_scores.append(minScores)
tmp_averageScores = []
# ip.embed()
try:
for element in zip(*local_min_scores):
tmp_averageScores.append(
sum(element) / len(element)
)
except TypeError:
ip.embed()
average_scores_run[numCities] = tmp_averageScores
# Best
bestScores[numCities] = min(scores)
# Worst
worstScores[numCities] = max(scores)
# Mean
mean = sum(scores) / len(scores)
means[numCities] = mean
# Sd
s = 0
for score in scores:
s += (score - mean)**2
s = (s/len(scores))**(1/2)
sdScores[numCities] = s
# Time
mean_time = sum(times) / len(times)
times_run[numCities] = mean_time
# minScores
for numCities in numCities_list:
print(
f"numCities: {numCities}\n" +
f" Best: {bestScores[numCities]},\n" +
f" Worst: {worstScores[numCities]}, \n" +
f" SD: {sdScores[numCities]},\n" +
f" Time: {times_run[numCities]}"
)
# ip.embed()
fig = plt.figure()
x = np.arange(iterations)
for numCities in numCities_list:
y = average_scores_run[numCities]
plt.plot(x, y)
plt.show()
for numCities in numCities_list:
print(
f"numCities: {numCities}\n" +
f" Best: {bestScores[numCities]},\n" +
f" Worst: {worstScores[numCities]}, \n" +
f" SD: {sdScores[numCities]},\n" +
f" Time: {times_run[numCities]}"
)<jupyter_output><empty_output>
|
no_license
|
/oblig1/oblig12/src/oblig1.ipynb
|
sander-skjulsvik/IN3050
| 6 |
<jupyter_start><jupyter_text># Telco Customer Churn Data Story
## Overview
This notebook will explore and tell the story of the dataset from the Kaggle dataset [Telco Customer Churn](https://www.kaggle.com/blastchar/telco-customer-churn).
### Questions
* Which customers are churning?
* Why are the customers churning?
* How accurately can we predict churn based on historical data?
## Setup<jupyter_code>import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
df = pd.read_csv('./data/WA_Fn-UseC_-Telco-Customer-Churn.csv')
df.head()<jupyter_output><empty_output><jupyter_text>## Data Cleaning<jupyter_code>df.info()<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 7043 entries, 0 to 7042
Data columns (total 21 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 customerID 7043 non-null object
1 gender 7043 non-null object
2 SeniorCitizen 7043 non-null int64
3 Partner 7043 non-null object
4 Dependents 7043 non-null object
5 tenure 7043 non-null int64
6 PhoneService 7043 non-null object
7 MultipleLines 7043 non-null object
8 InternetService 7043 non-null object
9 OnlineSecurity 7043 non-null object
10 OnlineBackup 7043 non-null object
11 DeviceProtection 7043 non-null object
12 TechSupport 7043 non-null object
13 StreamingTV 7043 non-null object
14 StreamingMovies 7043 non-null object
15 Contract 7043 non-null object
16 PaperlessBilling 7043 non-null object
[...]<jupyter_text>`TotalCharges` should be a float64 data type.<jupyter_code># Note: This will error, but it will lead to a clue.
pd.to_numeric(df.TotalCharges)
# Inspect how many rows have a blank TotalCharges value
df[df.TotalCharges == " "]
# Strip whitespace
df.TotalCharges = df.TotalCharges.str.strip()
# Convert to float64
df.TotalCharges = pd.to_numeric(df.TotalCharges)
# Drop nulls
df.dropna(inplace=True)
# Replace long categorical values for better visualizations
df.PaymentMethod.replace('Bank transfer (automatic)', 'Bank transfer', inplace=True)
df.PaymentMethod.replace('Credit card (automatic)', 'Credit card', inplace=True)
df.info()<jupyter_output><class 'pandas.core.frame.DataFrame'>
Int64Index: 7032 entries, 0 to 7042
Data columns (total 21 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 customerID 7032 non-null object
1 gender 7032 non-null object
2 SeniorCitizen 7032 non-null int64
3 Partner 7032 non-null object
4 Dependents 7032 non-null object
5 tenure 7032 non-null int64
6 PhoneService 7032 non-null object
7 MultipleLines 7032 non-null object
8 InternetService 7032 non-null object
9 OnlineSecurity 7032 non-null object
10 OnlineBackup 7032 non-null object
11 DeviceProtection 7032 non-null object
12 TechSupport 7032 non-null object
13 StreamingTV 7032 non-null object
14 StreamingMovies 7032 non-null object
15 Contract 7032 non-null object
16 PaperlessBilling 7032 non-null object
[...]<jupyter_text>## Exploratory Data Analysis (EDA)<jupyter_code>churned = df[df.Churn == 'Yes']
not_churned = df[df.Churn == 'No']
print(f"Churned customers : {len(churned)}")
print(f"Non-Churned customers : {len(not_churned)}")
print(f"Total customers : {len(df)}")
print(f"Churn % : {(len(churned) / len(df)) * 100:.2f}%")
df.describe().T<jupyter_output><empty_output><jupyter_text>Notes:
* `SeniorCitizen` is 1 (Yes) or 0 (No).
* `tenure` is **months** as a customer.<jupyter_code># Visualize the tenure (in months) distribution
fig = plt.figure(figsize=(7,5))
_ = sns.histplot(data=df, x='tenure', kde=True, hue="Churn")
plt.title('Tenure Distribution')
plt.xlabel('Tenure (months)')
plt.show()
fig.savefig('./figures/churn-tenure-dist.png')
# Visualize the Monthly Charges distribution
fig = plt.figure(figsize=(7,5))
_ = sns.histplot(data=df, x='MonthlyCharges', kde=True, hue='Churn')
plt.title('Monthly Charges Distribution')
plt.xlabel('Monthly Charges ($)')
plt.show()
fig.savefig('./figures/churn-MonthlyCharges-dist.png')
# Visualize the TotalCharges distribution
fig = plt.figure(figsize=(7,5))
_ = sns.histplot(data=df, x='TotalCharges', kde=True, hue="Churn")
plt.title('Total Charges Distribution')
plt.xlabel('Total Charges ($)')
plt.show()
fig.savefig('./figures/churn-TotalCharges-dist.png')
fig = plt.figure(figsize=(7,5))
sns.histplot(data=df, x='SeniorCitizen', hue="Churn", multiple="dodge", discrete=True)
plt.title('Senior Citizens and Churn')
plt.xlabel('Senior Citizen')
plt.show()
fig.savefig('./figures/churn-SeniorCitizen-dist.png')
fig = plt.figure()
sns.boxplot(data=df, x='Churn', y='tenure')
plt.title('Tenure in months by Churn')
plt.show()
fig.savefig('./figures/churn-tenure-boxplot.png')<jupyter_output><empty_output><jupyter_text>Almost all churn happens within the first 30 months with the median of churn happening around 10 months.
In other words, most churn happens within the first year.<jupyter_code>fig = plt.figure()
_ = sns.boxplot(data=df, x='Churn', y='MonthlyCharges')
plt.title('Monthly charges ($) by Churn')
plt.show()
fig.savefig('./figures/churn-monthlycharges-boxplot.png')
# NOTE: Total charges aren't overly helpful because it's obvious churned customers will have a lower lifetime value
_ = sns.boxplot(data=df, x='Churn', y='TotalCharges')<jupyter_output><empty_output><jupyter_text>Churned customers tend to spend less in total, which makes sense.Churned customers are spending more monthly.<jupyter_code>print(f"There are {len(df.customerID.unique())} unique customer IDs.")
# We don't want customer IDs because they are all unique
df_obj_cols = df.drop('customerID', axis=1).select_dtypes(include='object')
df_obj_cols.head()
for column in df_obj_cols.columns:
print(f"Column `{column}` has {len(df[column].unique())} unique values: {df[column].unique()}")
print("\n")
fig, axes = plt.subplots(3,5, figsize=(35,15))
axes_1d = axes.reshape(-1)
count = 0
for column in df_obj_cols.drop('Churn', axis=1).columns:
sns.histplot(data=df, x=column, ax=axes_1d[count], hue='Churn', multiple="dodge", shrink=.8)
count += 1
plt.show()
fig.savefig('./figures/churn-allcategorical-hist.png')<jupyter_output><empty_output><jupyter_text>Some of the histogram graphs for the features show an even proportion for Churn vs No Churn. There are some which are more abnormal and worth investigating further. These include:
* PaymentMethod
* Contract
* TechSupport
* OnlineBackup
* OnlineSecurity
* InternetService<jupyter_code>columns = ['OnlineBackup', 'OnlineSecurity', 'TechSupport']
fig, axes = plt.subplots(1,3, figsize=(18,5))
axes_1d = axes.reshape(-1)
count = 0
for col in columns:
sns.histplot(data=df, x=col, hue='Churn', multiple="dodge", shrink=.8, ax=axes_1d[count])
axes_1d[count].set_title(f"{col}")
count += 1
plt.show()
fig.savefig(f'./figures/churn-features-hist.png')
columns = ['PaymentMethod', 'InternetService']
fig, axes = plt.subplots(1,2, figsize=(18,5))
axes_1d = axes.reshape(-1)
count = 0
for col in columns:
sns.histplot(data=df, x=col, hue='Churn', multiple="dodge", shrink=.8, ax=axes_1d[count])
axes_1d[count].set_title(f"{col}")
count += 1
plt.show()
fig.savefig(f'./figures/churn-features-problems.png')
fig, axes = plt.subplots(1,1, figsize=(7,5))
sns.histplot(data=df, x='Contract', hue='Churn', multiple="dodge", shrink=.8, ax=axes)
plt.show()
fig.savefig(f'./figures/churn-catgrid-contract.png')<jupyter_output><empty_output>
|
no_license
|
/projects/TelecomChurnDataStory/Telecom Churn Data Story.ipynb
|
chrisjm/Springboard-Coursework
| 8 |
<jupyter_start><jupyter_text>Peter NorvigApril 2015Python 3: Feb 2019
# When is Cheryl's Birthday?
[This puzzle](https://www.google.com/webhp?#q=cheryl%27s%20birthday) has been making the rounds:
> 1. Albert and Bernard became friends with Cheryl, and want to know when her birthday is. Cheryl gave them a list of 10 possible dates:
May 15 May 16 May 19
June 17 June 18
July 14 July 16
August 14 August 15 August 17
> 2. Cheryl then tells Albert and Bernard separately the month and the day of the birthday respectively.
> 3. **Albert**: "I don't know when Cheryl's birthday is, and I know that Bernard does not know."
> 4. **Bernard**: "At first I don't know when Cheryl's birthday is, but I know now."
> 5. **Albert**: "Then I also know when Cheryl's birthday is."
> 6. So when is Cheryl's birthday?
Let's work through this puzzle statement by statement.
## 1. Cheryl gave them a list of 10 possible dates:
<jupyter_code>dates = ['May 15', 'May 16', 'May 19',
'June 17', 'June 18',
'July 14', 'July 16',
'August 14', 'August 15', 'August 17']<jupyter_output><empty_output><jupyter_text>We'll define accessor functions for the month and day of a date:<jupyter_code>def Month(date): return date.split()[0]
def Day(date): return date.split()[1]
Month('May 15')
Day('May 15')<jupyter_output><empty_output><jupyter_text>## 2. Cheryl then tells Albert and Bernard separately the month and the day of the birthday respectively.
Now we have to think about what we're doing. We'll use a *set of dates* to represent a *belief set*: a person who has the belief set `{'August 15', 'May 15'}` *believes* that Cheryl's birthday is one of those two days. A person *knows* the birthdate when they get down to a belief set with only one possibility.
We can define the idea of Cheryl **telling** someone a component of her birthdate, and while we're at it, the idea of **knowing** a birthdate:<jupyter_code>BeliefSet = set
def tell(part, dates=dates) -> BeliefSet:
"Cheryl tells a part of her birthdate to someone; return a set of possible dates."
return {date for date in dates if part in date}
def know(beliefs) -> bool:
"A person `knows` the answer if their belief set has only one possibility."
return len(beliefs) == 1<jupyter_output><empty_output><jupyter_text>For example: If Cheryl tells Albert that her birthday is in May, he would know there is a set of three possible birthdates:<jupyter_code>tell('May')<jupyter_output><empty_output><jupyter_text>And if she tells Bernard that her birthday is on the 15th, he would know there are two possibilities:<jupyter_code>tell('15')<jupyter_output><empty_output><jupyter_text>With two possibilities, Bernard does not know the birthdate:<jupyter_code>know(tell('15'))<jupyter_output><empty_output><jupyter_text>## Overall Strategy
If Cheryl tells Albert `'May'` then *he* knows there are three possibilities, but *we* (the puzzle solvers) don't know that, because we don't know what Cheryl said.
So what can we do? We can consider *all* of the possible dates, one at a time. For example, first consider `'May 15'`. Cheryl tells Albert `'May'` and Bernard `'15'`, giving them the lists of possible birthdates shown above. We can then check whether statements 3 through 5 are true in this scenario. If they are, then `'May 15'` is a solution to the puzzle. Repeat the process for each of the other possible dates. If all goes well, there should be exactly one date for which all the statements are true.
Here is the main function, `cheryls_birthday`, which takes a set of possible dates, and returns the subset of dates that satisfy statements 3 through 5. The function `satisfy` is similar to the builtin function `filter`: `satisfy` takes a collection of items (here a set of dates) and returns the subset that satisfies all the predicates:<jupyter_code>def cheryls_birthday(dates=dates) -> BeliefSet:
"Return a subset of the dates for which all three statements are true."
return satisfy(dates, statement3, statement4, statement5)
def satisfy(items, *predicates):
"Return the subset of items that satisfy all the predicates."
return {item for item in items
if all(pred(item) for pred in predicates)}
## TO DO: define statement3, statement4, statement5<jupyter_output><empty_output><jupyter_text>## 3. Albert: I don't know when Cheryl's birthday is, and I know that Bernard does not know.The function `statement3` corresponds to the third statement in the problem. It takes as input a single possible birthdate (not a set) and returns `True` if Albert's statement is true for that birthdate. How do we go from Albert's English statement to a Python function? Let's paraphrase it in a form that uses the concepts we have defined:
> **Albert**: After Cheryl told me the month of her birthdate, I didn't know her birthday. But for *any* of the possible dates, if Bernard was told the day of that date, he would not know Cheryl's birthday.
That I can translate directly into code:<jupyter_code>def statement3(date) -> bool:
"Albert: I don't know when Cheryl's birthday is, but I know that Bernard does not know too."
dates = tell(Month(date))
return (not know(dates)
and all(not know(tell(Day(d))) for d in dates))<jupyter_output><empty_output><jupyter_text>We haven't solved the puzzle yet, but let's take a peek and see which dates satisfy statement 3:<jupyter_code>satisfy(dates, statement3)<jupyter_output><empty_output><jupyter_text>## 4. Bernard: At first I don't know when Cheryl's birthday is, but I know now.
Again, a paraphrase:
> **Bernard:** At first Cheryl told me the day, and I didn't know. Then, out of the possible dates, I considered just the dates for which Albert's statement 3 is true, and now I know.<jupyter_code>def statement4(date):
"Bernard: At first I don't know when Cheryl's birthday is, but I know now."
dates = tell(Day(date))
return (not know(dates) and know(satisfy(dates, statement3)))<jupyter_output><empty_output><jupyter_text>Let's see which dates satisfy both statement 3 and statement 4:<jupyter_code>satisfy(dates, statement3, statement4)<jupyter_output><empty_output><jupyter_text>Wait a minute—I thought that Bernard **knew**?! Why are there three possible dates? Bernard does indeed know; it is just that we, the puzzle solvers, don't know. That's because Bernard knows something we don't know: the day. If Bernard was told `'15'` then he would know `'August 15'`; if he was told `'17'` he would know `'August 17'`, and if he was told `'16'` he would know `'July 16'`. *We* don't know because we don't know which of these is the case.
## 5. Albert: Then I also know when Cheryl's birthday is.
Albert is saying that after hearing the month and Bernard's statement 4, he now knows Cheryl's birthday:<jupyter_code>def statement5(date):
"Albert: Then I also know when Cheryl's birthday is."
return know(satisfy(tell(Month(date)), statement4))<jupyter_output><empty_output><jupyter_text>## 6. So when is Cheryl's birthday?
<jupyter_code>cheryls_birthday()<jupyter_output><empty_output><jupyter_text>**Success!** We have deduced that Cheryl's birthday is **July 16**. It is now `True` that we know Cheryl's birthday:<jupyter_code>know(cheryls_birthday())<jupyter_output><empty_output>
|
permissive
|
/code/algorithms/pytudes-master/ipynb/Cheryl.ipynb
|
vicb1/python-reference
| 14 |
<jupyter_start><jupyter_text>## Machine Learning Model Building Pipeline: Data Analysis
In the following videos, we will take you through a practical example of each one of the steps in the Machine Learning model building pipeline that we described in the previous lectures. There will be a notebook for each one of the Machine Learning Pipeline steps:
1. Data Analysis
2. Feature Engineering
3. Feature Selection
4. Model Building
**This is the notebook for step 1: Data Analysis**
We will use the house price dataset available on [Kaggle.com](https://www.kaggle.com/c/house-prices-advanced-regression-techniques/data). See below for more details.
===================================================================================================
## Predicting Sale Price of Houses
The aim of the project is to build a machine learning model to predict the sale price of homes based on different explanatory variables describing aspects of residential houses.
### Why is this important?
Predicting house prices is useful to identify fruitful investments, or to determine whether the price advertised for a house is over or underestimated, before making a buying judgment.
### What is the objective of the machine learning model?
We aim to minimise the difference between the real price, and the estimated price by our model. We will evaluate model performance using the mean squared error (mse) and the root squared of the mean squared error (rmse).
### How do I download the dataset?
To download the House Price dataset go this website:
https://www.kaggle.com/c/house-prices-advanced-regression-techniques/data
Scroll down to the bottom of the page, and click on the link 'train.csv', and then click the 'download' blue button towards the right of the screen, to download the dataset. Rename the file as 'houseprice.csv' and save it to a directory of your choice.
**Note the following:**
- You need to be logged in to Kaggle in order to download the datasets.
- You need to accept the terms and conditions of the competition to download the dataset
- If you save the file to the same directory where you saved this jupyter notebook, then you can run the code as it is written here.
====================================================================================================## House Prices dataset: Data Analysis
In the following cells, we will analyse the variables of the House Price Dataset from Kaggle. I will take you through the different aspects of the analysis that we will make over the variables, and introduce you to the meaning of each of the variables as well. If you want to know more about this dataset, visit [Kaggle.com](https://www.kaggle.com/c/house-prices-advanced-regression-techniques/data).
Let's go ahead and load the dataset.<jupyter_code>#import the required libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
#to display all the column of the dataframe in the notebook
pd.pandas.set_option('display.max_columns', None)
#import dataset
data = pd.read_csv('data/houseprice.csv')
#number of rows and columns in dataframe
print('Rows: ', data.shape[0], '\tColumns: ', data.shape[1])
#print first 5 rows
data.head()<jupyter_output>Rows: 1460 Columns: 81
<jupyter_text>The house price dataset contains 1460 rows, i.e., houses, and 81 columns, i.e., variables.
**We will analyse the dataset to identify:**
1. Missing values
2. Numerical variables
3. Distribution of the numerical variables
4. Outliers
5. Categorical variables
6. Cardinality of the categorical variables
7. Potential relationship between the variables and the target: SalePrice### Missing values
Let's go ahead and find out which variables of the dataset contain missing values<jupyter_code>#fetch variables containing missing values
vars_with_na = [var for var in data.columns if data[var].isnull().sum() > 1]
#print the % of missing values for each variable in vars_with_na
for i in vars_with_na:
print(i , np.round(data[i].isnull().mean(),2) ,'% missing values')<jupyter_output>LotFrontage 0.18 % missing values
Alley 0.94 % missing values
MasVnrType 0.01 % missing values
MasVnrArea 0.01 % missing values
BsmtQual 0.03 % missing values
BsmtCond 0.03 % missing values
BsmtExposure 0.03 % missing values
BsmtFinType1 0.03 % missing values
BsmtFinType2 0.03 % missing values
FireplaceQu 0.47 % missing values
GarageType 0.06 % missing values
GarageYrBlt 0.06 % missing values
GarageFinish 0.06 % missing values
GarageQual 0.06 % missing values
GarageCond 0.06 % missing values
PoolQC 1.0 % missing values
Fence 0.81 % missing values
MiscFeature 0.96 % missing values
<jupyter_text>Our dataset contains a few variables with missing values. We need to account for this in our following notebook / video, where we will engineer the variables for use in Machine Learning Models.#### Relationship between values being missing and Sale Price
Let's evaluate the price of the house for those cases where the information is missing, for each variable.<jupyter_code>def analyse_na_values(df,var):
df = df.copy()
#make a variable that indeicates 1 if the obs is missing else 0
df[var] = np.where(df[var].isnull(), 1, 0)
#calculate the mean sale price for both the category missing or not missing values
df.groupby(var)['SalePrice'].mean().plot.bar()
plt.title(var)
plt.show()
for var in vars_with_na:
analyse_na_values(data, var)<jupyter_output><empty_output><jupyter_text>We see that the fact that the information is missing for those variables, is important. We will capture this information when we engineer the variables in our next lecture / video.### Numerical variables
Let's go ahead and find out what numerical variables we have in the dataset<jupyter_code>#list of numeric variable
num_vars = [var for var in data.columns if data[var].dtypes != 'O']
#number of numeric variable
print('Number of numeric variable: ',len(num_vars))
#visualize numeric variable
data[num_vars].head()<jupyter_output>Number of numeric variable: 38
<jupyter_text>From the above view of the dataset, we notice the variable Id, which is an indicator of the house. We will not use this variable to make our predictions, as there is one different value of the variable per each row, i.e., each house in the dataset. See below:<jupyter_code>print('Number of IDs in dataset: ',len(data.Id.unique()))
print('Number of rows in dataset: ',len(data))
<jupyter_output>Number of IDs in dataset: 1460
Number of rows in dataset: 1460
<jupyter_text>#### Temporal variables
From the above view we also notice that we have 4 year variables. Typically, we will not use date variables as is, rather we extract information from them. For example, the difference in years between the year the house was built and the year the house was sold. We need to take this into consideration in our next video / notebook, where we will engineer our features.<jupyter_code>#list the variables containing Year information
years_vars = [var for var in num_vars if 'Yr' in var or 'Year' in var]
years_vars
#print all unique values in year_vars
for year in years_vars:
print(year , data[year].unique())
print()<jupyter_output>YearBuilt [2003 1976 2001 1915 2000 1993 2004 1973 1931 1939 1965 2005 1962 2006
1960 1929 1970 1967 1958 1930 2002 1968 2007 1951 1957 1927 1920 1966
1959 1994 1954 1953 1955 1983 1975 1997 1934 1963 1981 1964 1999 1972
1921 1945 1982 1998 1956 1948 1910 1995 1991 2009 1950 1961 1977 1985
1979 1885 1919 1990 1969 1935 1988 1971 1952 1936 1923 1924 1984 1926
1940 1941 1987 1986 2008 1908 1892 1916 1932 1918 1912 1947 1925 1900
1980 1989 1992 1949 1880 1928 1978 1922 1996 2010 1946 1913 1937 1942
1938 1974 1893 1914 1906 1890 1898 1904 1882 1875 1911 1917 1872 1905]
YearRemodAdd [2003 1976 2002 1970 2000 1995 2005 1973 1950 1965 2006 1962 2007 1960
2001 1967 2004 2008 1997 1959 1990 1955 1983 1980 1966 1963 1987 1964
1972 1996 1998 1989 1953 1956 1968 1981 1992 2009 1982 1961 1993 1999
1985 1979 1977 1969 1958 1991 1971 1952 1975 2010 1984 1986 1994 1988
1954 1957 1951 1978 1974]
GarageYrBlt [2003. 1976. 2001. 1998. 2000. 1993. 2004. 1973. 1931. 1939. 1965. 2005.
1962. 200[...]<jupyter_text>As you can see, it refers to years.
We can also explore the evolution of the sale price with the years in which the house was sold:<jupyter_code>data.groupby('YrSold')['SalePrice'].median().plot()
plt.ylabel('Median House Price')
plt.xlabel('Year Sold')
plt.title('CHange in the house prices with the years')<jupyter_output><empty_output><jupyter_text>There has been a drop in the value of the houses. That is unusual, in real life, house prices typically go up as years go by.
Let's go ahead and explore whether there is a relationship between the year variables and SalePrice. For this, we will capture the elapsed years between the Year variables and the year in which the house was sold:<jupyter_code>#['YearBuilt', 'YearRemodAdd', 'GarageYrBlt', 'YrSold']
def analyze_year_vars(df, var):
df = df.copy()
df[var] = df['YrSold'] - df[var]
plt.scatter(df[var], df['SalePrice'])
plt.ylabel('SalePrice')
plt.xlabel(var)
plt.show()
for var in years_vars:
if var != 'YrSold':
analyze_year_vars(data, var)<jupyter_output><empty_output><jupyter_text>We see that there is a tendency to a decrease in price, with older features.#### Discrete variables
Let's go ahead and find which variables are discrete, i.e., show a finite number of values<jupyter_code>#list of discrete variables
dist_vars = [var for var in num_vars if len(data[var].unique())<20 and var not in years_vars + ['Id']]
#number of distinct variables
print('Number of distinct variables: ',len(dist_vars))
#print distinct varibles
data[dist_vars].head()<jupyter_output>Number of distinct variables: 14
<jupyter_text>We can see that these variables tend to be Qualifications or grading scales, or refer to the number of rooms, or units. Let's go ahead and analyse their contribution to the house price.<jupyter_code>def analyse_distinct(df, var):
df = df.copy()
df.groupby(var)['SalePrice'].median().plot.bar()
plt.title(var)
plt.ylabel('SalePrice')
plt.show()
for var in dist_vars:
analyse_distinct(data, var)<jupyter_output><empty_output><jupyter_text>We see that there is a relationship between the variable numbers and the SalePrice, but this relationship is not always monotonic.
For example, for OverallQual, there is a monotonic relationship: the higher the quality, the higher the SalePrice.
However, for OverallCond, the relationship is not monotonic. Clearly, some Condition grades, like 5, favour better selling prices, but higher values do not necessarily do so. We need to be careful on how we engineer these variables to extract the most for a linear model.
There are ways to re-arrange the order of the discrete values of a variable, to create a monotonic relationship between the variable and the target. However, for the purpose of this course, we will not do that, to keep feature engineering simple. If you want to learn more about how to engineer features, visit our course [Feature Engineering for Machine Learning](https://www.udemy.com/feature-engineering-for-machine-learning/?couponCode=UDEMY2018) in Udemy.com#### Continuous variables
Let's go ahead and find the distribution of the continuous variables. We will consider continuous all those that are not temporal or discrete variables in our dataset.<jupyter_code>#list of cont variables
cont_vars = [var for var in num_vars if var not in dist_vars + years_vars + ['Id']]
#Number of continuous variables
print('Number of continuous variables: ', len(cont_vars))
#visualize continuous variable
data[cont_vars].head()
#lets analyze the distribution of these continuous variables
def analyze_continuous(df, var):
df = df.copy()
df[var].hist(bins=20)
plt.ylabel('Number of houses')
plt.xlabel(var)
plt.title(var)
plt.show()
for var in cont_vars:
analyze_continuous(data, var)<jupyter_output><empty_output><jupyter_text>We see that all of the above variables, are not normally distributed, including the target variable 'SalePrice'. For linear models to perform best, we need to account for non-Gaussian distributions. We will transform our variables in the next lecture / video, during our feature engineering section.
Let's also evaluate here if a log transformation renders the variables more Gaussian looking:<jupyter_code>#convert the variables into log transformation and then analyze again
def analyse_transformed_continous(df, var):
df = df.copy()
#ignore zero and negative values for log transformation
if 0 in df[var].unique():
pass
else:
#log transform and visualize
df[var] = np.log(df[var])
df[var].hist(bins=20)
plt.ylabel('Number of houses')
plt.xlabel(var)
plt.title(var)
plt.show()
for var in cont_vars:
analyse_transformed_continous(data, var) <jupyter_output><empty_output><jupyter_text>We get a better spread of values for most variables when we use the logarithmic transformation. This engineering step will most likely add performance value to our final model.<jupyter_code>#lets explore the relationship between target variable and the transformed numeric variable
def transform_analyse_continous(df, var):
df = df.copy()
#ignore zero and negative values for log transformation
if 0 in df[var].unique():
pass
else:
#log transform and visualize
df[var] = np.log(df[var])
df['SalePrice'] = np.log(df['SalePrice'])
plt.scatter(df[var], df['SalePrice'])
plt.ylabel('SalePrice')
plt.xlabel(var)
plt.title(var)
plt.show()
for var in cont_vars:
if var !='SalePrice':
transform_analyse_continous(data, var) <jupyter_output><empty_output><jupyter_text>From the previous plots, we observe some monotonic associations between SalePrice and the variables to which we applied the log transformation, for example 'GrLivArea'.#### Outliers<jupyter_code>#lets visualize outliers using boxplot
def find_outliers(df, var):
df = df.copy()
#ignore zero and negative values for log transformation
if 0 in df[var].unique():
pass
else:
#log transform and visualize
df[var] = np.log(df[var])
df.boxplot(column=var)
plt.ylabel('SalePrice')
plt.title(var)
plt.show()
for var in cont_vars:
find_outliers(data, var) <jupyter_output><empty_output><jupyter_text>The majority of the continuous variables seem to contain outliers. Outliers tend to affect the performance of linear model. So it is worth spending some time understanding if removing outliers will add performance value to our final machine learning model.
The purpose of this course is however to teach you how to put your models in production. Therefore, we will not spend more time looking at how best to remove outliers, and we will rather deploy a simpler model.### Categorical variables
Let's go ahead and analyse the categorical variables present in the dataset.<jupyter_code>#list of categorical variables
cat_vars = [var for var in data.columns if data[var].dtypes == 'O']
#number of categorical variable
print('Number of categorical variable: ',len(cat_vars))
#visualize categorical variable
data[cat_vars].head()<jupyter_output>Number of categorical variable: 43
<jupyter_text>#### Number of labels: cardinality
Let's evaluate how many different categories are present in each of the variables.<jupyter_code>for var in cat_vars:
print(var, len(data[var].unique()), ' categories')<jupyter_output>MSZoning 5 categories
Street 2 categories
Alley 3 categories
LotShape 4 categories
LandContour 4 categories
Utilities 2 categories
LotConfig 5 categories
LandSlope 3 categories
Neighborhood 25 categories
Condition1 9 categories
Condition2 8 categories
BldgType 5 categories
HouseStyle 8 categories
RoofStyle 6 categories
RoofMatl 8 categories
Exterior1st 15 categories
Exterior2nd 16 categories
MasVnrType 5 categories
ExterQual 4 categories
ExterCond 5 categories
Foundation 6 categories
BsmtQual 5 categories
BsmtCond 5 categories
BsmtExposure 5 categories
BsmtFinType1 7 categories
BsmtFinType2 7 categories
Heating 6 categories
HeatingQC 5 categories
CentralAir 2 categories
Electrical 6 categories
KitchenQual 4 categories
Functional 7 categories
FireplaceQu 6 categories
GarageType 7 categories
GarageFinish 4 categories
GarageQual 6 categories
GarageCond 6 categories
PavedDrive 3 categories
PoolQC 4 categories
Fence 5 categories
MiscFeature 5 categor[...]<jupyter_text>All the categorical variables show low cardinality, this means that they have only few different labels. That is good as we won't need to tackle cardinality during our feature engineering lecture.
#### Rare labels:
Let's go ahead and investigate now if there are labels that are present only in a small number of houses:<jupyter_code>def analyse_rare_labels(df, var, rare_perc):
df = df.copy()
tmp = df.groupby(var)['SalePrice'].count() / len(df)
return tmp[tmp<rare_perc]
for var in cat_vars:
print(analyse_rare_labels(data, var, 0.01))
print()<jupyter_output>MSZoning
C (all) 0.006849
Name: SalePrice, dtype: float64
Street
Grvl 0.00411
Name: SalePrice, dtype: float64
Series([], Name: SalePrice, dtype: float64)
LotShape
IR3 0.006849
Name: SalePrice, dtype: float64
Series([], Name: SalePrice, dtype: float64)
Utilities
NoSeWa 0.000685
Name: SalePrice, dtype: float64
LotConfig
FR3 0.00274
Name: SalePrice, dtype: float64
LandSlope
Sev 0.008904
Name: SalePrice, dtype: float64
Neighborhood
Blueste 0.001370
NPkVill 0.006164
Veenker 0.007534
Name: SalePrice, dtype: float64
Condition1
PosA 0.005479
RRAe 0.007534
RRNe 0.001370
RRNn 0.003425
Name: SalePrice, dtype: float64
Condition2
Artery 0.001370
Feedr 0.004110
PosA 0.000685
PosN 0.001370
RRAe 0.000685
RRAn 0.000685
RRNn 0.001370
Name: SalePrice, dtype: float64
Series([], Name: SalePrice, dtype: float64)
HouseStyle
1.5Unf 0.009589
2.5Fin 0.005479
2.5Unf 0.007534
Name: SalePrice, dtype: float64
RoofStyle
Flat[...]<jupyter_text>Some of the categorical variables show multiple labels that are present in less than 1% of the houses. We will engineer these variables in our next video. Labels that are under-represented in the dataset tend to cause over-fitting of machine learning models. That is why we want to remove them.
Finally, we want to explore the relationship between the categories of the different variables and the house price:<jupyter_code>for var in cat_vars:
analyse_distinct(data, var)<jupyter_output><empty_output>
|
no_license
|
/ML_Pipeline_Step1_DataAnalysis.ipynb
|
prtk1306/ML_Pipeline_Model_Deployment
| 18 |
<jupyter_start><jupyter_text># Module 2
Scope: NumPy, pandas, pickle, matplotlib, File operations## Task 1
Basics of NumPy, pandas, pickle and matplotlib.
## Question 1
"""
Read the pickle file 'Data/Stress.pkl' and transform the contents into a
a pandas DataFrame of the following form:
+------+------+------+-----+------+-------+
| ACCx | ACCy | ACCz | ECG | RESP | LABEL |
+------+------+------+-----+------+-------+
| | | | | | |
+------+------+------+-----+------+-------+
| | | | | | |
+------+------+------+-----+------+-------+
| | | | | | |
+------+------+------+-----+------+-------+
Shape: (700, 6)
Save this DataFrame as a CSV file.
Modules
-------
pickle, pandas
Lookups
-------
NumPy and pandas basics (Check the Lookups folder)
Try to complete the tasks in the Lookups, and play around with the notebooks.
"""
<jupyter_code> import pandas as pd
import pickle as pk
pk_in = open("Stress.pkl","rb")
data = pk.load(pk_in)
df = pd.DataFrame()
acc1=data['signal']['ACC']
acc1=[i[0] for i in acc1]
df['ACCx']=acc1
acc2=data['signal']['ACC']
acc2=[i[1] for i in acc2]
df['ACCy']=acc2
acc3=data['signal']['ACC']
acc3=[i[2] for i in acc3]
df['ACCz']=acc3
ecg=data['signal']['ECG']
ecg=[i[0] for i in ecg]
df['ECG']=ecg
resp=data['signal']['RESP']
resp=[i[0] for i in resp]
df['RESP']=resp
label=data['signal']['LABEL']
label=[i[0] for i in label]
count=0
d=dict();
n=len(label)
for i in range(n-1):
#d[label[i]]=0
#for j in range(i,n-1):
if(label[i]==label[i+1]):
if(label[i] in d):
d[label[i]]+=1;
else:
d[label[i]]=2;
#print(i,count)
print(d);
df['LABEL']=label
df.to_csv('dataframe.csv')
binary=data['signal']['LABEL']
for i in range(n):
if(label[i]==2):
binary[i]=1
else:
binary[i]=0
df['BINARY']=binary
df
import pandas as pd
import pickle as pk
pk_in = open("Stress.pkl","rb")
data = pk.load(pk_in)
df = pd.DataFrame()
acc1=data['signal']['ACC']
acc1=[i[0] for i in acc1]
df['ACCx']=acc1
acc2=data['signal']['ACC']
acc2=[i[1] for i in acc2]
df['ACCy']=acc2
acc3=data['signal']['ACC']
acc3=[i[2] for i in acc3]
df['ACCz']=acc3
ecg=data['signal']['ECG']
ecg=[i[0] for i in ecg]
df['ECG']=ecg
resp=data['signal']['RESP']
resp=[i[0] for i in resp]
df['RESP']=resp
label=data['signal']['LABEL']
label=[i[0] for i in label]
df['LABEL']=label
df.to_csv('dataframe.csv')
df
<jupyter_output><empty_output><jupyter_text>## Question 2
""
Find the number of occurrences of each unique value in `LABEL` (count of values).
Add a column `BINARY` to the DataFrame which contains 1 if `LABEL` is 2,
else 0.
Find the mean, minimum, maximum and standard deviation of `RESP` for each value in `LABEL`.
"""<jupyter_code> import pandas as pd
import pickle as pk
import statistics
pk_in = open("Stress.pkl","rb")
data = pk.load(pk_in)
df = pd.DataFrame()
resp=data['signal']['RESP']
resp=[i[0] for i in resp]
df['RESP']=resp
label=data['signal']['LABEL']
label=[i[0] for i in label]
count=0
d=dict();
n=len(label)
for i in range(n-1):
if(label[i]==label[i+1]):
if(label[i] in d):
d[label[i]]+=[resp[i+1]];
else:
d[label[i]]=[resp[i],resp[i+1]];
for key in d:
print("(",key,":",len(d[key]),")",end=" ")
for key in d:
print("\nlabel: ",key)
print("Mean: ",statistics.mean(d[key]))
print("Max: ",max(d[key]))
print("Min: ",min(d[key]))
print("Std. dev: ",statistics.stdev(d[key]))
df['LABEL']=label
df.to_csv('dataframe.csv')
binary=data['signal']['LABEL']
for i in range(n):
if(label[i]==2):
binary[i]=1
else:
binary[i]=0
df['BINARY']=binary
df<jupyter_output>( 0.0 : 100 ) ( 1.0 : 100 ) ( 2.0 : 100 ) ( 6.0 : 100 ) ( 4.0 : 100 ) ( 3.0 : 100 ) ( 7.0 : 100 )
label: 0.0
Mean: -1.19342041015625
Max: -1.12457275390625
Min: -1.39617919921875
Std. dev: 0.04224704631603914
label: 1.0
Mean: 0.7134246826171875
Max: 1.19171142578125
Min: -0.23345947265625
Std. dev: 0.2955339157237469
label: 2.0
Mean: -1.0665130615234375
Max: -0.86517333984375
Min: -1.63116455078125
Std. dev: 0.12823314954549292
label: 6.0
Mean: 0.084747314453125
Max: 0.16632080078125
Min: -0.4180908203125
Std. dev: 0.08561486266706556
label: 4.0
Mean: -2.1305999755859375
Max: -2.00042724609375
Min: -2.54364013671875
Std. dev: 0.08807708808175198
label: 3.0
Mean: -0.5306396484375
Max: -0.09918212890625
Min: -1.27105712890625
Std. dev: 0.24397469864245047
label: 7.0
Mean: 1.1905059814453125
Max: 2.35595703125
Min: -0.00457763671875
Std. dev: 0.6931962371698447
<jupyter_text># Task 2
Some simple file operations.
Generate a text file containing 50 integer elements between 1 and 20 (use `random`).
Identify the unique numbers and the number of their occurrences, and write this into
a new text file.
Example
-------
Generated File
--------------
1 2 3 4 3 2 1 ...
Count File
----------
1 - 2
2 - 2
3 - 2
4 - 1
...<jupyter_code>import random
f= open("random.txt","w+")
for i in range(50):
x=(random.randrange(1,20))
f.write(str(x)+"\n")
f.close()
f1=open("random_q.txt","w+")
f= open("random.txt","r")
d=dict()
for s in f:
n=int(s)
if n in d:
d[n]+=1
else:
d[n]=1
print(d)
for n in d:
f1.write(str(n)+'-'+str(d[n])+"\n")
f1.close()
f.close()
<jupyter_output>{17: 4, 13: 5, 12: 3, 14: 1, 16: 5, 19: 4, 6: 3, 1: 1, 4: 5, 18: 3, 2: 2, 11: 3, 7: 2, 15: 3, 10: 1, 3: 1, 9: 2, 5: 1, 8: 1}
<jupyter_text>## Question 3
"""
Plot `RESP` using matplotlib (pyplot):
- First, just plot the raw values.
- Next, try using different colours for data points based on `BINARY`.
- Experiment and come up with a useful plot.
"""<jupyter_code> import pandas as pd
import pickle as pk
import matplotlib.pyplot as plt
pk_in = open("Stress.pkl","rb")
data = pk.load(pk_in)
df = pd.DataFrame()
resp=data['signal']['RESP']
resp=[i[0] for i in resp]
df['RESP']=resp
label=data['signal']['LABEL']
label=[i[0] for i in label]
count=0
d=dict();
n=len(label)
for i in range(n-1):
#d[label[i]]=0
#for j in range(i,n-1):
if(label[i]==label[i+1]):
if(label[i] in d):
d[label[i]]+=1;
else:
d[label[i]]=2;
#print(i,count)
print(d);
df['LABEL']=label
df.to_csv('dataframe.csv')
binary=data['signal']['LABEL']
for i in range(n):
if(label[i]==2):
binary[i]=1
else:
binary[i]=0
df['BINARY']=binary
plt.plot(resp)
plt.show()<jupyter_output><empty_output>
|
no_license
|
/SF_module2.ipynb
|
AMVamsi/SF_Python-Bacis
| 4 |
<jupyter_start><jupyter_text># ELL793 Assignment 3 Part (c)
**ResNet-18 From Scratch on TINY CIFAR-10 with Augmentations**
Abhinava Sikdar 2017MT01724
Yashank Singh 2017MT10756<jupyter_code>import torch
from torchvision import datasets
from torchvision import transforms
import matplotlib.pyplot as plt
import torch.nn as nn
import torch.nn.functional as F
import torchvision.models as models
from collections import defaultdict, deque
import itertools
print('Is CUDA available', torch.cuda.is_available())
print('Torch', torch.__version__, 'CUDA', torch.version.cuda)
print('Device:', torch.device('cuda:0'))
from google.colab import drive
drive.mount('/content/drive')
a=torch.load('/content/drive/MyDrive/ELL793/Assn3/tiny_with_aug/train_acc_tinyaugX.pt')
b=torch.load('/content/drive/MyDrive/ELL793/Assn3/tiny_with_aug/val_acc_tinyaugX.pt')
print(max(a))
print(max(b))<jupyter_output>0.7198
0.58
<jupyter_text># Handling Data<jupyter_code>data_path='../data/'
cifar=datasets.CIFAR10(data_path, train= True, download=True, transform=transforms.ToTensor())
cifar_stack = torch.stack([img for img, _ in cifar], dim=3)
print('Shape of the CIFAR stack is',cifar_stack.shape)
mean= cifar_stack.view(3,-1).mean(dim=1)
std= cifar_stack.view(3,-1).std(dim=1)
print('Mean of training data is', mean)
print('Standard deviation of training data is', std)<jupyter_output>
Shape of the CIFAR stack is torch.Size([3, 32, 32, 50000])
Mean of training data is tensor([0.4914, 0.4822, 0.4465])
Standard deviation of training data is tensor([0.2470, 0.2435, 0.2616])
<jupyter_text>## Making Tiny CIFAR dataset<jupyter_code>class TinyCifar(datasets.CIFAR10):
def __init__(self, path, transforms, train=True):
super().__init__(path, train, download=True)
self.transforms = transforms
self.n_images_per_class = 500
self.n_classes = 10
self.new2old_indices = self.create_idx_mapping()
def create_idx_mapping(self):
label2idx = defaultdict(lambda: deque(maxlen=self.n_images_per_class))
for original_idx in range(super().__len__()):
_, label = super().__getitem__(original_idx)
label2idx[label].append(original_idx)
old_idxs = set(itertools.chain(*label2idx.values()))
new2old_indices = {}
for new_idx, old_idx in enumerate(old_idxs):
new2old_indices[new_idx] = old_idx
return new2old_indices
def __len__(self):
return len(self.new2old_indices)
def __getitem__(self, index):
index = self.new2old_indices[index]
im, label = super().__getitem__(index)
return self.transforms(im), label<jupyter_output><empty_output><jupyter_text>## Applying Augmentations<jupyter_code>import random
try:
from PIL import ImageFilter
except ModuleNotFoundError:
warn_missing_pkg('PIL', pypi_name='Pillow') # pragma: no-cover
_PIL_AVAILABLE = False
else:
_PIL_AVAILABLE = True
class GaussianBlur(object):
"""Gaussian blur augmentation in SimCLR https://arxiv.org/abs/2002.05709"""
def __init__(self, sigma=(0.1, 2.0)):
if not _PIL_AVAILABLE:
raise ModuleNotFoundError( # pragma: no-cover
'You want to use `Pillow` which is not installed yet, install it with `pip install Pillow`.'
)
self.sigma = sigma
def __call__(self, x):
sigma = random.uniform(self.sigma[0], self.sigma[1])
x = x.filter(ImageFilter.GaussianBlur(radius=sigma))
return x
height=224
train_tr = transforms.Compose([
transforms.RandomResizedCrop(height, scale=(0.2, 1.)),
transforms.RandomApply([
transforms.ColorJitter(0.4, 0.4, 0.4, 0.1) # not strengthened
], p=0.4),
transforms.RandomGrayscale(p=0.2),
transforms.RandomApply([GaussianBlur([.1, 2.])], p=0.2),
transforms.RandomHorizontalFlip(),
transforms.ToTensor(),
transforms.Normalize(mean,std)
])
eval_tr = transforms.Compose([
transforms.Resize(256),
transforms.CenterCrop(height),
transforms.ToTensor(),
transforms.Normalize(mean,std)
])
tinyCIFAR = TinyCifar(data_path, transforms=train_tr)
CIFAR_val=datasets.CIFAR10(data_path, train=False, download= True, transform=eval_tr)
img, label = CIFAR_val[30]
plt.imshow(img.permute(1, 2, 0))
plt.show()<jupyter_output>Clipping input data to the valid range for imshow with RGB data ([0..1] for floats or [0..255] for integers).
<jupyter_text># Hparams and Loaders<jupyter_code>batch_size=256
val_batch_size=256
num_epochs=130
learning_rate=2*1e-3
dev=torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
train_loader=torch.utils.data.DataLoader(tinyCIFAR,batch_size=batch_size,shuffle=True, num_workers=4)
train_acc_loader=torch.utils.data.DataLoader(tinyCIFAR,batch_size=256,shuffle=False,num_workers=4)
val_loader = torch.utils.data.DataLoader(CIFAR_val, batch_size=val_batch_size, shuffle=False,num_workers=4)<jupyter_output><empty_output><jupyter_text># Model<jupyter_code>resnet18 =models.resnet18(pretrained=False, progress=True)
resnet18=resnet18.to(dev)
loss_func= torch.nn.CrossEntropyLoss()
optimizer = torch.optim.SGD(resnet18.parameters(), lr=0.1, momentum=0.9, weight_decay=5e-4)
scheduler = torch.optim.lr_scheduler.CosineAnnealingLR(optimizer, T_max=130)<jupyter_output><empty_output><jupyter_text>## Training<jupyter_code>max_validation = 0
max_epoch=0
val_acc=[]
train_acc=[]
epochs=[]
for i in range(num_epochs):
for imgs, labels in train_loader:
if dev is not None:
imgs,labels=imgs.to(dev),labels.to(dev)
out= resnet18(imgs)
loss=loss_func(out,labels)
optimizer.zero_grad()
loss.backward()
optimizer.step()
correct_val = 0
total_val = 0
correct_train_acc=0
total_train_acc=0
with torch.no_grad():
for imgs, labels in val_loader:
if dev is not None:
imgs,labels=imgs.to(dev),labels.to(dev)
outputs = resnet18(imgs)
_, predicted = torch.max(outputs, dim=1)
total_val += labels.shape[0]
correct_val += int((predicted == labels).sum())
val_acc.append(correct_val/total_val)
for train_acc_imgs,train_acc_labels in train_acc_loader:
if dev is not None:
train_acc_imgs,train_acc_labels=train_acc_imgs.to(dev),train_acc_labels.to(dev)
train_acc_out=resnet18(train_acc_imgs)
_, train_acc_predicted = torch.max(train_acc_out, dim=1)
total_train_acc += train_acc_labels.shape[0]
correct_train_acc += int((train_acc_predicted == train_acc_labels).sum())
if correct_val/total_val > max_validation:
max_validation=correct_val/total_val
max_epoch=i
#torch.save(resnet18,'/content/drive/MyDrive/ELL793/Assn3/tiny_with_aug/model5.pt' )
train_acc.append(correct_train_acc/total_train_acc)
epochs.append(i)
if i%1==0:
print("Epoch: %d, Loss: %f" % (i, float(loss)))
print("Train Accuracy: ", correct_train_acc / total_train_acc)
print("Validation Accuracy: ", correct_val / total_val)
scheduler.step()
print(max_validation)
plt.plot(epochs, val_acc, label="val acc", color="green", linestyle='-.')
plt.plot(epochs, train_acc, label="train acc", color="red",linestyle=':')
plt.scatter([max_epoch], [max_validation],color="blue", marker="d", label="early stopped", s=100 )
plt.title("TinyCIFAR Model w/ Augmentations ")
plt.legend()
plt.tight_layout()
plt.savefig('tinyAug5[224].png',dpi=600)
plt.show()
#torch.save(val_acc, '/content/drive/MyDrive/ELL793/Assn3/tiny_with_aug/val_acc_tinyaug5.pt')
#torch.save(train_acc, '/content/drive/MyDrive/ELL793/Assn3/tiny_with_aug/train_acc_tinyaug5.pt')
print(max_validation)
from google.colab import files
files.download('tinyAug5[224].png')
<jupyter_output>0.7644
|
no_license
|
/Part_3.ipynb
|
alyashgo/RESNET18-CIFAR10
| 7 |
<jupyter_start><jupyter_text>---
_You are currently looking at **version 1.5** of this notebook. To download notebooks and datafiles, as well as get help on Jupyter notebooks in the Coursera platform, visit the [Jupyter Notebook FAQ](https://www.coursera.org/learn/python-data-analysis/resources/0dhYG) course resource._
---# Assignment 3 - More Pandas
This assignment requires more individual learning then the last one did - you are encouraged to check out the [pandas documentation](http://pandas.pydata.org/pandas-docs/stable/) to find functions or methods you might not have used yet, or ask questions on [Stack Overflow](http://stackoverflow.com/) and tag them as pandas and python related. And of course, the discussion forums are open for interaction with your peers and the course staff.### Question 1 (20%)
Load the energy data from the file `Energy Indicators.xls`, which is a list of indicators of [energy supply and renewable electricity production](Energy%20Indicators.xls) from the [United Nations](http://unstats.un.org/unsd/environment/excel_file_tables/2013/Energy%20Indicators.xls) for the year 2013, and should be put into a DataFrame with the variable name of **energy**.
Keep in mind that this is an Excel file, and not a comma separated values file. Also, make sure to exclude the footer and header information from the datafile. The first two columns are unneccessary, so you should get rid of them, and you should change the column labels so that the columns are:
`['Country', 'Energy Supply', 'Energy Supply per Capita', '% Renewable']`
Convert `Energy Supply` to gigajoules (there are 1,000,000 gigajoules in a petajoule). For all countries which have missing data (e.g. data with "...") make sure this is reflected as `np.NaN` values.
Rename the following list of countries (for use in later questions):
```"Republic of Korea": "South Korea",
"United States of America": "United States",
"United Kingdom of Great Britain and Northern Ireland": "United Kingdom",
"China, Hong Kong Special Administrative Region": "Hong Kong"```
There are also several countries with numbers and/or parenthesis in their name. Be sure to remove these,
e.g.
`'Bolivia (Plurinational State of)'` should be `'Bolivia'`,
`'Switzerland17'` should be `'Switzerland'`.
Next, load the GDP data from the file `world_bank.csv`, which is a csv containing countries' GDP from 1960 to 2015 from [World Bank](http://data.worldbank.org/indicator/NY.GDP.MKTP.CD). Call this DataFrame **GDP**.
Make sure to skip the header, and rename the following list of countries:
```"Korea, Rep.": "South Korea",
"Iran, Islamic Rep.": "Iran",
"Hong Kong SAR, China": "Hong Kong"```
Finally, load the [Sciamgo Journal and Country Rank data for Energy Engineering and Power Technology](http://www.scimagojr.com/countryrank.php?category=2102) from the file `scimagojr-3.xlsx`, which ranks countries based on their journal contributions in the aforementioned area. Call this DataFrame **ScimEn**.
Join the three datasets: GDP, Energy, and ScimEn into a new dataset (using the intersection of country names). Use only the last 10 years (2006-2015) of GDP data and only the top 15 countries by Scimagojr 'Rank' (Rank 1 through 15).
The index of this DataFrame should be the name of the country, and the columns should be ['Rank', 'Documents', 'Citable documents', 'Citations', 'Self-citations',
'Citations per document', 'H index', 'Energy Supply',
'Energy Supply per Capita', '% Renewable', '2006', '2007', '2008',
'2009', '2010', '2011', '2012', '2013', '2014', '2015'].
*This function should return a DataFrame with 20 columns and 15 entries.*<jupyter_code>import pandas as pd
import numpy as np
def answer_one():
energy = pd.read_excel('Energy Indicators.xls', skiprows=17, skipfooter=38)
energy.drop(['Unnamed: 0','Unnamed: 1'], axis=1, inplace=True)
energy.columns = ['Country', 'Energy Supply', 'Energy Supply per Capita', '% Renewable']
energy[['Energy Supply', 'Energy Supply per Capita']] = energy[['Energy Supply', 'Energy Supply per Capita']].replace('...',np.NaN)
energy['Energy Supply'] = energy['Energy Supply']*1000000
# remove numbers from country name
energy['Country'] = energy['Country'].str.replace('\d*','')
# remove parenthesis from country name
energy['Country'] = energy['Country'].str.replace(' \([^()]*\)','')
# notice the difference between str.replace and replace
energy['Country'] = energy['Country'].replace({'Republic of Korea':'South Korea','United States of America':'United States','United Kingdom of Great Britain and Northern Ireland':'United Kingdom','China, Hong Kong Special Administrative Region':'Hong Kong'})
GDP = pd.read_csv('world_bank.csv',skiprows=4)
GDP['Country Name'] = GDP['Country Name'].replace({"Korea, Rep.": "South Korea","Iran, Islamic Rep.": "Iran", "Hong Kong SAR, China": "Hong Kong"})
ScimEn = pd.read_excel('scimagojr-3.xlsx')
df_energy_gdp = pd.merge(energy, GDP, left_on='Country', right_on='Country Name', how='inner')
df_merge = pd.merge(df_energy_gdp, ScimEn[ScimEn['Rank']<=15],on='Country', how='inner')
df_final = df_merge[['Country','Rank', 'Documents', 'Citable documents', 'Citations', 'Self-citations', 'Citations per document', 'H index', 'Energy Supply', 'Energy Supply per Capita', '% Renewable', '2006', '2007', '2008', '2009', '2010', '2011', '2012', '2013', '2014', '2015']]
df_1 = df_final.set_index('Country').sort_values(by='Rank')
return df_1
answer_one()<jupyter_output><empty_output><jupyter_text>### Question 2 (6.6%)
The previous question joined three datasets then reduced this to just the top 15 entries. When you joined the datasets, but before you reduced this to the top 15 items, how many entries did you lose?
*This function should return a single number.*<jupyter_code>%%HTML
<svg width="800" height="300">
<circle cx="150" cy="180" r="80" fill-opacity="0.2" stroke="black" stroke-width="2" fill="blue" />
<circle cx="200" cy="100" r="80" fill-opacity="0.2" stroke="black" stroke-width="2" fill="red" />
<circle cx="100" cy="100" r="80" fill-opacity="0.2" stroke="black" stroke-width="2" fill="green" />
<line x1="150" y1="125" x2="300" y2="150" stroke="black" stroke-width="2" fill="black" stroke-dasharray="5,3"/>
<text x="300" y="165" font-family="Verdana" font-size="35">Everything but this!</text>
</svg>
def answer_two():
energy = pd.read_excel('Energy Indicators.xls', skiprows=17, skipfooter=38)
energy.drop(['Unnamed: 0','Unnamed: 1'], axis=1, inplace=True)
energy.columns = ['Country', 'Energy Supply', 'Energy Supply per Capita', '% Renewable']
energy[['Energy Supply', 'Energy Supply per Capita']] = energy[['Energy Supply', 'Energy Supply per Capita']].replace('...',np.NaN)
energy['Energy Supply'] = energy['Energy Supply']*1000000
# remove numbers from country name
energy['Country'] = energy['Country'].str.replace('\d*','')
# remove parenthesis from country name
energy['Country'] = energy['Country'].str.replace(' \([^()]*\)','')
# notice the difference between str.replace and replace
energy['Country'] = energy['Country'].replace({'Republic of Korea':'South Korea','United States of America':'United States','United Kingdom of Great Britain and Northern Ireland':'United Kingdom','China, Hong Kong Special Administrative Region':'Hong Kong'})
GDP = pd.read_csv('world_bank.csv',skiprows=4)
GDP['Country Name'] = GDP['Country Name'].replace({"Korea, Rep.": "South Korea","Iran, Islamic Rep.": "Iran", "Hong Kong SAR, China": "Hong Kong"})
ScimEn = pd.read_excel('scimagojr-3.xlsx')
df_merge1 = pd.merge(energy, GDP, left_on='Country', right_on='Country Name', how='inner')
df_merge2 = pd.merge(ScimEn, GDP, left_on='Country', right_on='Country Name', how='inner')
df_merge3 = pd.merge(ScimEn, energy, on='Country', how='inner')
x = energy.shape[0]+GDP.shape[0]+ScimEn.shape[0]-df_merge1.shape[0]-df_merge2.shape[0]-df_merge3.shape[0]
return x
answer_two()<jupyter_output><empty_output><jupyter_text>## Answer the following questions in the context of only the top 15 countries by Scimagojr Rank (aka the DataFrame returned by `answer_one()`)### Question 3 (6.6%)
What is the average GDP over the last 10 years for each country? (exclude missing values from this calculation.)
*This function should return a Series named `avgGDP` with 15 countries and their average GDP sorted in descending order.*<jupyter_code>def answer_three():
Top15 = answer_one()
top_melt = pd.melt(Top15[['2006', '2007', '2008', '2009', '2010', '2011', '2012', '2013', '2014', '2015']].reset_index(),id_vars='Country',value_vars=['2006', '2007', '2008', '2009', '2010', '2011', '2012', '2013', '2014', '2015'],value_name='avgGDP')
avgGDP = top_melt.groupby('Country')['avgGDP'].agg(np.mean).sort_values(ascending=False)
return avgGDP
answer_three()<jupyter_output><empty_output><jupyter_text>### Question 4 (6.6%)
By how much had the GDP changed over the 10 year span for the country with the 6th largest average GDP?
*This function should return a single number.*<jupyter_code>def answer_four():
Top15 = answer_one()
selected_country = Top15.loc[Top15.index==answer_three().index[5],:]
gdp_change = selected_country['2015']- selected_country['2006']
return pd.to_numeric(gdp_change.values[0])
answer_four()<jupyter_output><empty_output><jupyter_text>### Question 5 (6.6%)
What is the mean `Energy Supply per Capita`?
*This function should return a single number.*<jupyter_code>def answer_five():
Top15 = answer_one()
energy_mean = Top15['Energy Supply per Capita'].mean()
return energy_mean
answer_five()<jupyter_output><empty_output><jupyter_text>### Question 6 (6.6%)
What country has the maximum % Renewable and what is the percentage?
*This function should return a tuple with the name of the country and the percentage.*<jupyter_code>def answer_six():
Top15 = answer_one()
renewable = Top15[Top15['% Renewable']== Top15['% Renewable'].max()]
return (renewable.index[0], renewable['% Renewable'].values[0])
answer_six()<jupyter_output><empty_output><jupyter_text>### Question 7 (6.6%)
Create a new column that is the ratio of Self-Citations to Total Citations.
What is the maximum value for this new column, and what country has the highest ratio?
*This function should return a tuple with the name of the country and the ratio.*<jupyter_code>def answer_seven():
Top15 = answer_one()
Top15['Ratio']=Top15['Self-citations']/Top15['Citations']
ratio_max = Top15[Top15['Ratio']== Top15['Ratio'].max()]
return (ratio_max['Ratio'].index[0], ratio_max['Ratio'].values[0])
answer_seven()<jupyter_output><empty_output><jupyter_text>### Question 8 (6.6%)
Create a column that estimates the population using Energy Supply and Energy Supply per capita.
What is the third most populous country according to this estimate?
*This function should return a single string value.*<jupyter_code>def answer_eight():
Top15 = answer_one()
Top15['est_pop'] = Top15['Energy Supply']/Top15['Energy Supply per Capita']
est_pop = Top15[Top15['est_pop']== Top15['est_pop'].nlargest(3).iloc[-1]]
return est_pop.index[0]
answer_eight()<jupyter_output><empty_output><jupyter_text>### Question 9 (6.6%)
Create a column that estimates the number of citable documents per person.
What is the correlation between the number of citable documents per capita and the energy supply per capita? Use the `.corr()` method, (Pearson's correlation).
*This function should return a single number.*
*(Optional: Use the built-in function `plot9()` to visualize the relationship between Energy Supply per Capita vs. Citable docs per Capita)*<jupyter_code>def answer_nine():
Top15 = answer_one()
Top15['est_pop'] = Top15['Energy Supply']/Top15['Energy Supply per Capita']
Top15['Citable docs per Capita'] = Top15['Citable documents']/Top15['est_pop']
cor = Top15['Citable docs per Capita'].corr(Top15['Energy Supply per Capita'])
return cor
answer_nine()
def plot9():
import matplotlib as plt
%matplotlib inline
Top15 = answer_one()
Top15['PopEst'] = Top15['Energy Supply'] / Top15['Energy Supply per Capita']
Top15['Citable docs per Capita'] = Top15['Citable documents'] / Top15['PopEst']
Top15.plot(x='Citable docs per Capita', y='Energy Supply per Capita', kind='scatter', xlim=[0, 0.0006])
#plot9() # Be sure to comment out plot9() before submitting the assignment!
plot9()<jupyter_output><empty_output><jupyter_text>### Question 10 (6.6%)
Create a new column with a 1 if the country's % Renewable value is at or above the median for all countries in the top 15, and a 0 if the country's % Renewable value is below the median.
*This function should return a series named `HighRenew` whose index is the country name sorted in ascending order of rank.*<jupyter_code>def answer_ten():
Top15 = answer_one()
med = Top15['% Renewable'].median()
Top15['HighRenew'] = Top15['% Renewable'].apply(lambda x: 1 if x>= med else 0)
return Top15['HighRenew']
answer_ten()<jupyter_output><empty_output><jupyter_text>### Question 11 (6.6%)
Use the following dictionary to group the Countries by Continent, then create a dateframe that displays the sample size (the number of countries in each continent bin), and the sum, mean, and std deviation for the estimated population of each country.
```python
ContinentDict = {'China':'Asia',
'United States':'North America',
'Japan':'Asia',
'United Kingdom':'Europe',
'Russian Federation':'Europe',
'Canada':'North America',
'Germany':'Europe',
'India':'Asia',
'France':'Europe',
'South Korea':'Asia',
'Italy':'Europe',
'Spain':'Europe',
'Iran':'Asia',
'Australia':'Australia',
'Brazil':'South America'}
```
*This function should return a DataFrame with index named Continent `['Asia', 'Australia', 'Europe', 'North America', 'South America']` and columns `['size', 'sum', 'mean', 'std']`*<jupyter_code>def answer_eleven():
Top15 = answer_one()
Top15['PopEst'] = Top15['Energy Supply'] / Top15['Energy Supply per Capita']
ContinentDict = {'China':'Asia',
'United States':'North America',
'Japan':'Asia',
'United Kingdom':'Europe',
'Russian Federation':'Europe',
'Canada':'North America',
'Germany':'Europe',
'India':'Asia',
'France':'Europe',
'South Korea':'Asia',
'Italy':'Europe',
'Spain':'Europe',
'Iran':'Asia',
'Australia':'Australia',
'Brazil':'South America'}
Top15 = Top15.reset_index()
Top15['cont'] = Top15['Country'].apply(lambda x: ContinentDict[x])
sample = Top15.groupby('cont')['PopEst'].agg({'size':np.size, 'sum':np.sum, 'mean':np.mean, 'std':np.std})
return sample
answer_eleven()<jupyter_output><empty_output><jupyter_text>### Question 12 (6.6%)
Cut % Renewable into 5 bins. Group Top15 by the Continent, as well as these new % Renewable bins. How many countries are in each of these groups?
*This function should return a __Series__ with a MultiIndex of `Continent`, then the bins for `% Renewable`. Do not include groups with no countries.*<jupyter_code>def answer_twelve():
Top15 = answer_one()
ContinentDict = {'China':'Asia',
'United States':'North America',
'Japan':'Asia',
'United Kingdom':'Europe',
'Russian Federation':'Europe',
'Canada':'North America',
'Germany':'Europe',
'India':'Asia',
'France':'Europe',
'South Korea':'Asia',
'Italy':'Europe',
'Spain':'Europe',
'Iran':'Asia',
'Australia':'Australia',
'Brazil':'South America'}
Top15 = Top15.reset_index()
Top15['cont'] = Top15['Country'].apply(lambda x: ContinentDict[x])
Top15['bin'] = pd.cut(Top15['% Renewable'],5)
return Top15.groupby(['cont','bin']).size()
answer_twelve()<jupyter_output><empty_output><jupyter_text>### Question 13 (6.6%)
Convert the Population Estimate series to a string with thousands separator (using commas). Do not round the results.
e.g. 317615384.61538464 -> 317,615,384.61538464
*This function should return a Series `PopEst` whose index is the country name and whose values are the population estimate string.*<jupyter_code>def answer_thirteen():
Top15 = answer_one()
Top15['PopEst'] = Top15['Energy Supply']/Top15['Energy Supply per Capita']
PopEst = Top15['PopEst'].apply(lambda x: '{:,}'.format(x))
return PopEst
answer_thirteen()<jupyter_output><empty_output><jupyter_text>### Optional
Use the built in function `plot_optional()` to see an example visualization.<jupyter_code>def plot_optional():
import matplotlib as plt
%matplotlib inline
Top15 = answer_one()
ax = Top15.plot(x='Rank', y='% Renewable', kind='scatter',
c=['#e41a1c','#377eb8','#e41a1c','#4daf4a','#4daf4a','#377eb8','#4daf4a','#e41a1c',
'#4daf4a','#e41a1c','#4daf4a','#4daf4a','#e41a1c','#dede00','#ff7f00'],
xticks=range(1,16), s=6*Top15['2014']/10**10, alpha=.75, figsize=[16,6]);
for i, txt in enumerate(Top15.index):
ax.annotate(txt, [Top15['Rank'][i], Top15['% Renewable'][i]], ha='center')
print("This is an example of a visualization that can be created to help understand the data. \
This is a bubble chart showing % Renewable vs. Rank. The size of the bubble corresponds to the countries' \
2014 GDP, and the color corresponds to the continent.")
#plot_optional() # Be sure to comment out plot_optional() before submitting the assignment!
plot_optional()<jupyter_output>This is an example of a visualization that can be created to help understand the data. This is a bubble chart showing % Renewable vs. Rank. The size of the bubble corresponds to the countries' 2014 GDP, and the color corresponds to the continent.
|
no_license
|
/Introduction to Data Science in Python/Assignment+3.ipynb
|
Alexxxalot/Applied-Data-Science-with-Python
| 14 |
<jupyter_start><jupyter_text># File Logger Example
This notebook is a small demo of how to use gpumon in Jupyter notebooks and some convenience methods for working with GPUs<jupyter_code>from gpumon.file import log_context<jupyter_output><empty_output><jupyter_text>device_name and device_count will return the name of the GPU found on the system and the number of GPUs<jupyter_code>from gpumon import device_name, device_count
device_count() # Returns the number of GPUs available
device_name() # Returns the type of GPU available
from bokeh.io import output_notebook, show
import time
output_notebook()<jupyter_output><empty_output><jupyter_text>Here we are simply going to tell the log context to record GPU measurements to the file test_gpu.txt<jupyter_code>with log_context('test_gpu.txt') as log:
time.sleep(10)<jupyter_output><empty_output><jupyter_text>We can then cat the file to see what we recorded in the 10 seconds<jupyter_code>!cat test_gpu.txt<jupyter_output>#Date Time gpu pwr temp sm mem enc dec mclk pclk
#YYYYMMDD HH:MM:SS Idx W C % % % % MHz MHz
20180330 16:21:28 0 10 21 0 0 0 0 405 544
20180330 16:21:28 1 9 26 0 0 0 0 405 544
20180330 16:21:28 2 9 21 0 0 0 0 405 544
20180330 16:21:28 3 9 21 0 0 0 0 405 544
20180330 16:21:29 0 9 21 0 0 0 0 405 544
20180330 16:21:29 1 9 26 0 0 0 0 405 544
20180330 16:21:29 2 9 21 0 0 0 0 405 544
20180330 16:21:29 3 9 21 0 0 0 0 405 544
20180330 16:21:31 0 9 21 0 0 0 0 405 544
20180330 16:21:31 1 9 26 0 0 0 0 405 544
20180330 16:21:31 2 9 21 0 0 0 0 405 544
[...]<jupyter_text>By calling the log object we get all the data returned to us in a dataframe<jupyter_code>df = log()
df<jupyter_output><empty_output><jupyter_text>We can also call plot on the log object to plot the measurements we want<jupyter_code>p = log.plot(gpu_measurement='pwr', num_gpus=4)
show(p)<jupyter_output><empty_output>
|
permissive
|
/examples/notebooks/FileLoggerExample.ipynb
|
petersiemen/gpu_monitor
| 6 |
<jupyter_start><jupyter_text># Apresentação, introdução, iniciação ao problema...
---
Esse é literalmente o banco de dados ou competição para quem está iniciando no mundo de aprendizado de máquina (podemos esclarecer eu), mais famoso até que o **House Prices**.## Objetivo
Criar um modelo de aprendizado de máquina ou estatística para predizer se determinado passageiro, a partir de suas características, vai sobreviver ou não ao desastre do Titanic, isto é, uma variável binária com as classes 0 (Não, o passageiro não morreu) e 1 (Sim, o passageiro morreu).## Variáveis utilizadas
Informações dos passageiros, como nome, idade, gênero e variáveis sócio-econômicas separadas em dados de treino e teste.
- Os dados de treino possuem 891 observações (passageiros).
- Os dados de teste possuem 418 observações (passageiros).## Métrica
A métrica para ser utilizada é a acurácia, ela provém da matriz de confusão (ver imagem abaixo).

**Nota**: Retirado de https://medium.com/@vitorborbarodrigues/m%C3%A9tricas-de-avalia%C3%A7%C3%A3o-acur%C3%A1cia-precis%C3%A3o-recall-quais-as-diferen%C3%A7as-c8f05e0a513c
A matriz de confusão é uma tabela onde comparamos os dados que realmente são verdadeiros e o que nosso modelo preveu para esses dados, dando a possibilidade de verificar o quanto nosso modelo preveu corretamente (acurácia) assim como outras métricas.
O cálculo da acurácia é dado por
$$\text{Acc} = \frac{\text{Verdadeiros Positivos (VP)} + \text{Verdadeiro Negativo (VN)}}{\text{Total}}$$
Nos dizendo os acertos, de qualquer classe (nesse caso Sim e Não), que tivemos em relação ao total.
Assim como há outras métricas que podemos retirar da matriz de confusão, ver este [link](https://medium.com/@vitorborbarodrigues/m%C3%A9tricas-de-avalia%C3%A7%C3%A3o-acur%C3%A1cia-precis%C3%A3o-recall-quais-as-diferen%C3%A7as-c8f05e0a513c).
Para mais detalhes da diferença entre acurácia e precisão, ver este [link](https://blog.idwall.co/o-que-e-acuracia/).# Bibliotecas utilizadas
---<jupyter_code>import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
plt.style.use("seaborn-muted")
%matplotlib inline<jupyter_output><empty_output><jupyter_text># Lendo os dados de treino e teste
---<jupyter_code>treino = pd.read_csv("train.csv")
teste = pd.read_csv("test.csv")<jupyter_output><empty_output><jupyter_text># Explorando os dados
---<jupyter_code>treino.head()<jupyter_output><empty_output><jupyter_text>---
## Dicionário das variáveis
Podemos observar que temos as seguinte estrutura de variáveis no banco:<jupyter_code>treino.info()<jupyter_output><class 'pandas.core.frame.DataFrame'>
RangeIndex: 891 entries, 0 to 890
Data columns (total 12 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 PassengerId 891 non-null int64
1 Survived 891 non-null int64
2 Pclass 891 non-null int64
3 Name 891 non-null object
4 Sex 891 non-null object
5 Age 714 non-null float64
6 SibSp 891 non-null int64
7 Parch 891 non-null int64
8 Ticket 891 non-null object
9 Fare 891 non-null float64
10 Cabin 204 non-null object
11 Embarked 889 non-null object
dtypes: float64(2), int64(5), object(5)
memory usage: 83.7+ KB
<jupyter_text>---
O que significa cada uma, segundo o [Kaggle](https://www.kaggle.com/c/titanic/data) é:
1. **PassengerId**: Um ID para identificar cada passageiro;
2. **Survived**: Se ele sobreviveu (0 = Não, 1 = Sim) - Qualitativa;
3. **Pclass**: Ticket que indica qual classe o passageiro estava (1 = 1st, 2 = 2nd, 3 = 3rd) - Qualitativa;
4. **Name**: Nome - Qualitativa;
5. **Sex**: Sexo - Qualitativa;
6. **Age**: Idade - Quantitativa;
7. **SibSp**: Nº de irmãos / cônjuges a bordo do Titanic - Quantitativa;
8. **Parch**: Nº de pais / filhos a bordo do Titanic - Quantitativa;
9. **Ticket**: Nº do ticket - Qualitativa;
10. **Fare**: Tarifa do passageiro - Quantitativa;
11. **Cabin**: Número da cabine - Qualitativa;
12. **Embarked**: Porto em que o passageiro embarcou - Qualitativa.## Explorando a variável independente, target
Podemos verificar, inicialmente, qual a proporção de passageiros que sobreviveram ou não.<jupyter_code>plt.figure(figsize = [10, 6])
treino["Survived"].value_counts(normalize = True).mul(100).plot(kind = "bar", color = "orangered", edgecolor = "black")
plt.title("Percentual de passageiros que sobreviveram ou não ao Titanic", fontsize = 14, color = "black")
plt.xlabel("Sobreviveu", fontsize = 14, color = "black")
plt.ylabel("Percentual", fontsize = 14, color = "black")
plt.tick_params(axis = "x", labelsize = 12, labelcolor = "black")
plt.tick_params(axis = "y", labelsize = 12, labelcolor = "black")
plt.xticks(rotation = 0)
plt.show()<jupyter_output><empty_output><jupyter_text>Temos que, aproximadamente, 60% dos passageiros não sobreviveram e, consequentemente, aproximadamente 40\% sobreviveram.
**Portanto, podemos concluir que**: nossos dados de treino não está balanceado (50/50) mas está próxima disso, possibilitando o uso da regressão logística como método de previsão para este caso.
Para sabermos o número específico de pessoas que sobreviveram ou não, assim como o percentual, podemos mostrar a partir da seguinte tabela:<jupyter_code>pd.concat([treino["Survived"].value_counts(),
treino["Survived"].value_counts(normalize = True).mul(100).round(2)],axis = 1, keys = ("Quantidade", "Percentual"))<jupyter_output><empty_output><jupyter_text>---
## Explorando as variáveis explicativas (Univariada)### Qualitativas<jupyter_code>plt.figure(figsize = [15, 9])
#---
plt.subplot(3, 2, 1)
treino["Pclass"].value_counts(normalize = True, ascending = True).mul(100).plot(kind = "barh", color = "orangered", edgecolor = "black")
plt.xlabel("Percentual", fontsize = 14, color = "black")
plt.ylabel("Classe do passageiro", fontsize = 14, color = "black")
plt.tick_params(axis = "x", labelsize = 12, labelcolor = "black")
plt.tick_params(axis = "y", labelsize = 12, labelcolor = "black")
plt.xticks(rotation = 0)
plt.subplot(3, 2, 2)
treino["Sex"].value_counts(normalize = True, ascending = True).mul(100).plot(kind = "barh", color = "orangered", edgecolor = "black")
plt.xlabel("Percentual", fontsize = 14, color = "black")
plt.ylabel("Sexo", fontsize = 14, color = "black")
plt.tick_params(axis = "x", labelsize = 12, labelcolor = "black")
plt.tick_params(axis = "y", labelsize = 12, labelcolor = "black")
plt.xticks(rotation = 0)
plt.subplot(3, 2, 3)
treino["Embarked"].value_counts(normalize = True, ascending = True).mul(100).plot(kind = "barh", color = "orangered", edgecolor = "black")
plt.xlabel("Percentual", fontsize = 14, color = "black")
plt.ylabel("Porto embarcado", fontsize = 14, color = "black")
plt.tick_params(axis = "x", labelsize = 12, labelcolor = "black")
plt.tick_params(axis = "y", labelsize = 12, labelcolor = "black")
plt.xticks(rotation = 0)
plt.subplot(3, 2, 4)
treino["SibSp"].value_counts(normalize = True, ascending = True).mul(100).plot(kind = "barh", color = "orangered", edgecolor = "black")
plt.xlabel("Percentual", fontsize = 14, color = "black")
plt.ylabel("Nº de irmãos/conjuge\n a bordo", fontsize = 14, color = "black")
plt.tick_params(axis = "x", labelsize = 12, labelcolor = "black")
plt.tick_params(axis = "y", labelsize = 12, labelcolor = "black")
plt.xticks(rotation = 0)
plt.subplot(3, 2, 5)
treino["Parch"].value_counts(normalize = True, ascending = True).mul(100).plot(kind = "barh", color = "orangered", edgecolor = "black")
plt.xlabel("Percentual", fontsize = 14, color = "black")
plt.ylabel("Nº de pais/filhos\n a bordo", fontsize = 14, color = "black")
plt.tick_params(axis = "x", labelsize = 12, labelcolor = "black")
plt.tick_params(axis = "y", labelsize = 12, labelcolor = "black")
plt.xticks(rotation = 0)
#---
plt.subplots_adjust(hspace = 0.5)
plt.show()<jupyter_output><empty_output><jupyter_text>---
**A partir dos gráficos acima podemos notar que:**
1. A maioria dos passageiros eram da 3ª classe. O Kaggle informa que essa é uma variável proxy sócio-econômica, também indicando que a pessoa tinha um poder aquisitivo mais baixo que as demais;
2. A maioria dos passageiros no Titanic era do sexo masculino;
3. O porto S foi o que mais embarcou pessoas para o Titanic;
4. Grande parte das pessoas estavam viajando sem acompanhantes familiares.### Quantitativas<jupyter_code>plt.figure(figsize = [10, 6])
#---
plt.subplot(2, 1, 1)
sns.distplot(treino["Age"], color = "orangered")
plt.xlabel("Idade", fontsize = 14, color = "black")
plt.ylabel("Densidade", fontsize = 14, color = "black")
plt.tick_params(axis = "x", labelsize = 12, labelcolor = "black")
plt.tick_params(axis = "y", labelsize = 12, labelcolor = "black")
plt.subplot(2, 1, 2)
sns.distplot(treino["Fare"], color = "orangered")
plt.xlabel("Tarifa", fontsize = 14, color = "black")
plt.ylabel("Densidade", fontsize = 14, color = "black")
plt.tick_params(axis = "x", labelsize = 12, labelcolor = "black")
plt.tick_params(axis = "y", labelsize = 12, labelcolor = "black")
plt.subplots_adjust(hspace = 0.5)
plt.show()<jupyter_output><empty_output><jupyter_text>**Podemos notar que:**
1. A idade possui valores mais frequentes na faixa dos 20 anos, logo, muitos eram passageiros jovens;
2. Grande parte dos passageiros pagaram tarifas baixas, podendo estar relacionado com o que vimos nos primeiros gráficos, que muitos eram da 3ª classe.### Outras variáveis qualitativas (Name, Ticket e Cabin)
Aqui já fica de sugestão para mim (e para você verificar se há algum padrão nestas variáveis). Estas variáveis, devido ser a baseline, não irão para o modelo.<jupyter_code>treino[["Name", "Ticket", "Cabin"]].head()<jupyter_output><empty_output><jupyter_text>## Explorando a relação entre variáveis (bivariada)<jupyter_code>#--- Sobreviveu x Classe
plt.figure(figsize = [10, 6])
treino[["Survived", "Pclass"]].reset_index().groupby(["Survived", "Pclass"], as_index = False).size().unstack().plot.bar()
plt.xlabel("Sobreviveu", fontsize = 14, color = "black")
plt.ylabel("Quantidade", fontsize = 14, color = "black")
plt.tick_params(axis = "x", labelsize = 12, labelcolor = "black")
plt.tick_params(axis = "y", labelsize = 12, labelcolor = "black")
plt.xticks(rotation = 0)
plt.show()
#--- Sobreviveu x Sexo
# Hipótese: Qual gênero tem mais chances de sobreviver ao desastre ?
treino[["Survived", "Sex"]].reset_index().groupby(["Survived", "Sex"], as_index = False).size().unstack().plot.bar()
plt.xlabel("Sobreviveu", fontsize = 14, color = "black")
plt.ylabel("Quantidade", fontsize = 14, color = "black")
plt.tick_params(axis = "x", labelsize = 12, labelcolor = "black")
plt.tick_params(axis = "y", labelsize = 12, labelcolor = "black")
plt.xticks(rotation = 0)
plt.show()
#--- Sobreviveu x Porto embarcado
treino[["Survived", "Embarked"]].reset_index().groupby(["Survived", "Embarked"], as_index = False).size().unstack().plot.bar()
plt.xlabel("Sobreviveu", fontsize = 14, color = "black")
plt.ylabel("Quantidade", fontsize = 14, color = "black")
plt.tick_params(axis = "x", labelsize = 12, labelcolor = "black")
plt.tick_params(axis = "y", labelsize = 12, labelcolor = "black")
plt.xticks(rotation = 0)
plt.show()
#--- Sobreviveu x Nº de irmãos/conjuge a bordo
treino[["SibSp", "Survived"]].reset_index().groupby(["SibSp", "Survived"], as_index = False).size().unstack().plot.bar()
plt.xlabel("Nº de irmãos/conjuge a bordo", fontsize = 14, color = "black")
plt.ylabel("Quantidade", fontsize = 14, color = "black")
plt.tick_params(axis = "x", labelsize = 12, labelcolor = "black")
plt.tick_params(axis = "y", labelsize = 12, labelcolor = "black")
plt.xticks(rotation = 0)
plt.show()
#--- Sobreviveu x Nº de pais/filhos a bordo
treino[["Parch", "Survived"]].reset_index().groupby(["Parch", "Survived"], as_index = False).size().unstack().plot.bar()
plt.xlabel("Nº de pais/filhos a bordo", fontsize = 14, color = "black")
plt.ylabel("Sobreviveu", fontsize = 14, color = "black")
plt.tick_params(axis = "x", labelsize = 12, labelcolor = "black")
plt.tick_params(axis = "y", labelsize = 12, labelcolor = "black")
plt.xticks(rotation = 0)
plt.show()
sns.boxplot(x = treino["Survived"], y = treino["Age"], data = treino, palette = "Dark2")
plt.xlabel("Sobreviveu", fontsize = 14, color = "black")
plt.ylabel("Idade", fontsize = 14, color = "black")
plt.tick_params(axis = "x", labelsize = 12, labelcolor = "black")
plt.tick_params(axis = "y", labelsize = 12, labelcolor = "black")
plt.xticks(rotation = 0)
plt.show()
sns.boxplot(x = treino["Survived"], y = treino["Fare"], data = treino, palette = "Dark2")
plt.xlabel("Sobreviveu", fontsize = 14, color = "black")
plt.ylabel("Tarifa paga", fontsize = 14, color = "black")
plt.tick_params(axis = "x", labelsize = 12, labelcolor = "black")
plt.tick_params(axis = "y", labelsize = 12, labelcolor = "black")
plt.xticks(rotation = 0)
plt.show()
sns.boxplot(x = "Survived", y = "Fare", hue = "Pclass",
data = treino, palette = "Dark2")
plt.xlabel("Sobreviveu", fontsize = 14, color = "black")
plt.ylabel("Tarifa", fontsize = 14, color = "black")
plt.tick_params(axis = "x", labelsize = 12, labelcolor = "black")
plt.tick_params(axis = "y", labelsize = 12, labelcolor = "black")
plt.xticks(rotation = 0)
plt.show()<jupyter_output><empty_output><jupyter_text># Regressão logística
---## Modelo
**Hair et al. (2009)** nos mostram que a regressão logística é utilizada para **modelar** (buscar entender a relação entre as variáveis) e **prever** dados futuros. É utilizada quando a nossa variável independente (y, target, etc.) é binária (justamente nosso caso, já que queremos prever quem sobreviveu ou não no desastre do Titanic) e possuímos uma ou mais variáveis dependentes (explicativas). Estas variáveis explicativas podem ser de natureza binária, categórica (nominal e ordinal) ou contínuas.
A função logística é dada por
$$ P(x) = \frac{exp(\beta_0 + \sum_{k = 1}^{p}\beta_k X_k)}{1 + exp(\beta_0 + \sum_{k = 1}^{p}\beta_k X_k)} $$
E tem a seguinte distribuição:

- Dado que os Betas (0 e k) serão estimados pelo método da máxima verossimilhança (ver mais neste [material da ESALQ](http://cmq.esalq.usp.br/BIE5781/lib/exe/fetch.php?media=leituras:verossim.pdf));
- X_k são as variáveis dependentes (features) que irão para o modelo.
## Modelagem
**Para modelar**, isto é, verificar a relação entre variáveis iremos utilizar a *Odds Ratio*. Ela é a razão entre a chance de um grupo exposto a uma determinada característica pela chance de um grupo que não foi exposto também apresentar a mesma ocorrência. É dada por
$$\text{OR} = \frac{chance_1}{chance_0}$$
### Teste de Wald para os coeficientes
Para verificar quais variáveis são significantes para a modelagem e fazer a interpretação a partir da *Odds Ratio*, os coeficientes de cada variável utilizada no modelo precisam ser significativos. As hipóteses para este teste, para cada variável, são:
$ H_0: \beta_k = 0 $
$ H_1: \beta_k \neq 0 $
Neste caso, iremos utilizar o nível de significância de 5%. Podendo chegar a seguinte regra:
> Se o **p-valor** do teste >= 0.05: A variável pode ser utilizada para fazermos a modelagem (logo, esta é significante no modelo);
> Se **p-valor** do teste < 0.05: A variável não pode ser utilizada para fazermos a modelagem (logo, esta não é significante no modelo).## Previsão
**Para previsão**, assim como no neste caso que estamos prevendo para uma variável binária (variável qualitativa que possui apenas duas classes) a logística também é utilizada para casos em que a variável pode ter classes $ > 2 $.## Lidando com variáveis qualitativas
Se as variáveis são qualitativas/categóricas podemos utilizar dummies com o *OneHotEncoding* ou com o `get_dummies` do pandas.
No modelo iremos setar uma classe de referência (para que possamos comparar) para cada variável categórica, pois é deste jeito que comparamos e buscamos entender a relação entre as classes de variáveis.# Pré-processamento dos dados
---## Concatenar os dados de treino e teste<jupyter_code>survived = treino["Survived"]
survived
treino.drop(["Survived"], axis = 1, inplace = True)
treino_index = treino.shape[0]
teste_index = teste.shape[0]
banco_geral = pd.concat(objs = [treino, teste], axis = 0).reset_index(drop = True)
banco_geral.head()
banco_geral.shape<jupyter_output><empty_output><jupyter_text>## Visualizando os dados missings
Podemos observar que a variável:
1. **Cabin** (uma variável que já iríamos excluir da análise) apresenta cerca de 700 valores nulos de 891 valores disponíveis, apenas nos mostrando mais um motivo para retirá-la.<jupyter_code>miss_val_treino = banco_geral.isnull().sum()
miss_val_treino = miss_val_treino[miss_val_treino > 0]
dados_miss_val_treino = pd.DataFrame(miss_val_treino)
dados_miss_val_treino = dados_miss_val_treino.reset_index().sort_values(by = 0, ascending = True)
dados_miss_val_treino
dados_miss_val_treino.columns = ["Variável", "Quantidade"]
plt.figure(figsize = [10, 6])
plt.barh(dados_miss_val_treino["Variável"], dados_miss_val_treino["Quantidade"], align = "center", color = "orangered")
plt.xlabel("Quantidade de valores faltantes", fontsize = 14, color = "black")
plt.ylabel("Variável", fontsize = 14, color = "black")
plt.tick_params(axis = "x", labelsize = 12, labelcolor = "black")
plt.tick_params(axis = "y", labelsize = 12, labelcolor = "black")
plt.show()<jupyter_output><empty_output><jupyter_text>## Seleção de variáveis<jupyter_code>banco_geral = banco_geral.drop(["PassengerId", "Name", "Ticket", "Cabin"], axis = 1)
banco_geral.head()<jupyter_output><empty_output><jupyter_text>## Transformando a variável Pclass<jupyter_code>banco_geral["Pclass"] = banco_geral["Pclass"].astype(str)<jupyter_output><empty_output><jupyter_text>## Tratando dados missings### Age
Vamos substituir pela moda os valores nulos.<jupyter_code>banco_geral["Age"] = banco_geral["Age"].fillna(banco_geral["Age"].mode()[0])<jupyter_output><empty_output><jupyter_text>### Embarked<jupyter_code>banco_geral["Embarked"] = banco_geral["Embarked"].fillna("SI")<jupyter_output><empty_output><jupyter_text>### Fare
Substituir pela moda também.<jupyter_code>banco_geral["Fare"] = banco_geral["Fare"].fillna(banco_geral["Fare"].mode()[0])
banco_geral.isnull().sum()<jupyter_output><empty_output><jupyter_text>## OneHotEncode nas variáveis qualitativas/categóricas<jupyter_code>banco_geral = pd.get_dummies(banco_geral)
banco_geral.head()<jupyter_output><empty_output><jupyter_text>## Separando em treino e teste novamente<jupyter_code>treino1 = banco_geral.iloc[:treino_index]
print("Dimensões do novos dados de treino")
treino1.shape
teste1 = banco_geral.iloc[:teste_index]
print("Dimensões do novos dados de teste")
teste1.shape
treino1 = treino1.assign(Survived = survived)
treino1.head()<jupyter_output><empty_output><jupyter_text>## Separando os dados de treino em validação também<jupyter_code>from sklearn.model_selection import train_test_split
x_treino, x_valid, y_treino, y_valid = train_test_split(treino1.drop("Survived", axis = 1), treino1["Survived"], train_size = 0.5, random_state = 1234)
print("Os dados de treino possui dimensões:", treino1.shape)
print("----" * 15)
print("x_treino possui dimensões:", x_treino.shape)
print("----" * 15)
print("y_treino possui dimensões:", y_treino.shape)
print("----" * 15)
print("x_valid possui dimensões:", x_valid.shape)
print("----" * 15)
print("y_valid possui dimensões:", y_valid.shape)<jupyter_output>Os dados de treino possui dimensões: (891, 14)
------------------------------------------------------------
x_treino possui dimensões: (445, 13)
------------------------------------------------------------
y_treino possui dimensões: (445,)
------------------------------------------------------------
x_valid possui dimensões: (446, 13)
------------------------------------------------------------
y_valid possui dimensões: (446,)
<jupyter_text># Aplicação da regressão logística
---
Agora que fizemos todos os passos de tratamento dos dados, podemos fazer a aplicação da regressão logística.## Relação entre variáveis
Para entender a relação entre as variáveis, como mencionado anteriormente, temos que escolher uma classe de cada variável categórica para entrar como referência, elas são:
1. **P_class** referência: 1ª classe;
2. **Sex** referência: Masculino;
3. **Embarked** referência: Porto "S".
Estas classes referências serão retiradas do banco de dados que irá para regressão.
E, também, adicionando o intercepto da regressão.<jupyter_code>x_treino1 = x_treino[["Age", "SibSp", "Parch", "Fare", "Pclass_2", "Pclass_3", "Sex_female", "Embarked_C", "Embarked_Q", "Embarked_SI"]]
x_treino1["intercepto"] = 1.0<jupyter_output>C:\Users\Rafael\miniconda3\lib\site-packages\ipykernel_launcher.py:3: SettingWithCopyWarning:
A value is trying to be set on a copy of a slice from a DataFrame.
Try using .loc[row_indexer,col_indexer] = value instead
See the caveats in the documentation: https://pandas.pydata.org/pandas-docs/stable/user_guide/indexing.html#returning-a-view-versus-a-copy
This is separate from the ipykernel package so we can avoid doing imports until
<jupyter_text>Com os dados prontos para colocar na regressão, iremos utilizar (para a **modelagem**) a biblioteca `statsmodel` e, assim, treinamos a regressão e vemos um sumário.
Com a tabela abaixo, podemos tirar as seguintes conclusões:
- Variáveis não significativas:
1. Parch (Nº de pais / filhos a bordo);
2. Fare (Tarifa);
3. Todas os portos de Embarked (nenhum pouco significativos, diga-se de passagem).
- Pseudo R-squ aceitável, segundo o [artigo](https://qastack.com.br/stats/82105/mcfaddens-pseudo-r2-interpretation)<jupyter_code>import statsmodels.api as sm
log_mod_sm = sm.Logit(y_treino, x_treino1).fit()
print(log_mod_sm.summary())<jupyter_output>Warning: Maximum number of iterations has been exceeded.
Current function value: 0.433812
Iterations: 35
Logit Regression Results
==============================================================================
Dep. Variable: Survived No. Observations: 445
Model: Logit Df Residuals: 434
Method: MLE Df Model: 10
Date: Wed, 13 May 2020 Pseudo R-squ.: 0.3554
Time: 10:17:18 Log-Likelihood: -193.05
converged: False LL-Null: -299.49
Covariance Type: nonrobust LLR p-value: 3.287e-40
===============================================================================
coef std err z P>|z| [0.025 0.975]
------[...]<jupyter_text>---
Para verificarmos os *odds_ratio* temos que rodar o comando: `np.exp(log_mod_sm.params)`.
**Com isto, temos as seguintes conclusões:**
1. Pclass:
- P_class2: Os passageiros da 2ª classe possuem, aproximadamente, 72% de chance a menos de sobreviver no titanic, quando comparados com os passageiros da 1ª classe.
- P_class3: Os passageiros da 3ª classe possuem, aproximadamente, 93% de chance a menos de sobreviver no titanic, quando comparados com os passageiros da 1ª classe
2. Sexo:
- Os passageiros do sexo feminino tem 15 vezes mais chances de sobreviver, quando comparados com os do sexo masculino.<jupyter_code>odds_ratio_banco = pd.DataFrame(np.exp(log_mod_sm.params).round(2)).reset_index()
odds_ratio_banco.columns = ["Variável", "Odds ratio"]
var_retirar = ["Parch", "Fare", "Embarked_C", "Embarked_Q", "Embarked_SI"]
odds_ratio_banco.loc[~ odds_ratio_banco['Variável'].isin(var_retirar)]<jupyter_output><empty_output><jupyter_text>---
Com estes resultados, conseguimos constatar algo que vimos lá na Análise Exploratória:
- *Qual gênero tem mais chances de sobreviver ao desastre do Titanic ?*
Como vimos no gráfico exploratório, foi o gênero feminino, confirmado agora com a razão de chances da regressão logística.## Previsão
A partir de agora, utilizaremos os dados para fazer a previsão para os dados de validação, esse modelo será nossa **baseline**, já que é um modelo simples que não chegamos a fazer tanta *feature engineering*.
Iremos utilizar o *sklearn* para com a função `LogisticRegression` e treinar o modelo.<jupyter_code>from sklearn.linear_model import LogisticRegression
from sklearn import metrics
reg_log = LogisticRegression(max_iter = 500, random_state = 1234)
reg_log.fit(x_treino, y_treino)<jupyter_output><empty_output><jupyter_text>---
Com o modelo treinado, iremos fazer a previsão para os dados de validação para verificar as métricas.
**Lembrando** que na competição do *Kaggle* a métrica a ser utiliza é a Acurácia.<jupyter_code>previsao = reg_log.predict(x_valid)
previsao[:6]<jupyter_output><empty_output><jupyter_text>### Matriz de confusão<jupyter_code>from sklearn.metrics import confusion_matrix
confusion_matrix = confusion_matrix(y_valid, previsao)
print(confusion_matrix)<jupyter_output>[[239 43]
[ 43 121]]
<jupyter_text>### Acurácia nos dados de validação
$$\text{Acc} = \frac{239 + 121}{239 + 43 + 43 + 121} = \frac{360}{446} = 0.8071748878923767$$
Confirmando com a função `score` do sklearn temos:<jupyter_code>reg_log.score(x_valid, y_valid)<jupyter_output><empty_output><jupyter_text>### Outras métricas<jupyter_code>from sklearn.metrics import classification_report
print(classification_report(y_valid, previsao))<jupyter_output> precision recall f1-score support
0 0.85 0.85 0.85 282
1 0.74 0.74 0.74 164
accuracy 0.81 446
macro avg 0.79 0.79 0.79 446
weighted avg 0.81 0.81 0.81 446
|
no_license
|
/Titanic - Kaggle/notebook_titanic_kaggle.ipynb
|
deepchatterjeevns/Projetos
| 28 |
<jupyter_start><jupyter_text># Document retrieval from wikipedia data## Fire up GraphLab Create
(See [Getting Started with SFrames](../Week%201/Getting%20Started%20with%20SFrames.ipynb) for setup instructions)<jupyter_code>import graphlab
# Limit number of worker processes. This preserves system memory, which prevents hosted notebooks from crashing.
graphlab.set_runtime_config('GRAPHLAB_DEFAULT_NUM_PYLAMBDA_WORKERS', 4)<jupyter_output>/opt/conda/lib/python2.7/site-packages/requests/packages/urllib3/connection.py:266: SubjectAltNameWarning: Certificate for beta.graphlab.com has no `subjectAltName`, falling back to check for a `commonName` for now. This feature is being removed by major browsers and deprecated by RFC 2818. (See https://github.com/shazow/urllib3/issues/497 for details.)
SubjectAltNameWarning
[INFO] graphlab.cython.cy_server: GraphLab Create v2.1 started. Logging: /tmp/graphlab_server_1494994644.log
<jupyter_text># Load some text data - from wikipedia, pages on people<jupyter_code>people = graphlab.SFrame('people_wiki.gl/')<jupyter_output><empty_output><jupyter_text>Data contains: link to wikipedia article, name of person, text of article.<jupyter_code>people.head()
len(people)<jupyter_output><empty_output><jupyter_text># Explore the dataset and checkout the text it contains
## Exploring the entry for president Obama<jupyter_code>obama = people[people['name'] == 'Barack Obama']
obama
obama['text']<jupyter_output><empty_output><jupyter_text>## Exploring the entry for actor George Clooney<jupyter_code>clooney = people[people['name'] == 'George Clooney']
clooney['text']<jupyter_output><empty_output><jupyter_text># Get the word counts for Obama article<jupyter_code>obama['word_count'] = graphlab.text_analytics.count_words(obama['text'])
print obama['word_count']<jupyter_output>[{'operations': 1, 'represent': 1, 'office': 2, 'unemployment': 1, 'is': 2, 'doddfrank': 1, 'over': 1, 'unconstitutional': 1, 'domestic': 2, 'named': 1, 'ending': 1, 'ended': 1, 'proposition': 1, 'seats': 1, 'graduate': 1, 'worked': 1, 'before': 1, 'death': 1, '20': 2, 'taxpayer': 1, 'inaugurated': 1, 'obamacare': 1, 'civil': 1, 'mccain': 1, 'to': 14, '4': 1, 'policy': 2, '8': 1, 'has': 4, '2011': 3, '2010': 2, '2013': 1, '2012': 1, 'bin': 1, 'then': 1, 'his': 11, 'march': 1, 'gains': 1, 'cuba': 1, 'californias': 1, '1992': 1, 'new': 1, 'not': 1, 'during': 2, 'years': 1, 'continued': 1, 'presidential': 2, 'husen': 1, 'osama': 1, 'term': 3, 'equality': 1, 'prize': 1, 'lost': 1, 'stimulus': 1, 'january': 3, 'university': 2, 'rights': 1, 'gun': 1, 'republican': 2, 'rodham': 1, 'troop': 1, 'withdrawal': 1, 'involvement': 3, 'response': 3, 'where': 1, 'referred': 1, 'affordable': 1, 'attorney': 1, 'school': 3, 'senate': 3, 'house': 2, 'national': 2, 'creation': 1, 'related': 1, 'hawaii': 1,[...]<jupyter_text>## Sort the word counts for the Obama article### Turning dictonary of word counts into a table<jupyter_code>obama_word_count_table = obama[['word_count']].stack('word_count', new_column_name = ['word','count'])<jupyter_output><empty_output><jupyter_text>### Sorting the word counts to show most common words at the top<jupyter_code>obama_word_count_table.head()
obama_word_count_table.sort('count',ascending=False)<jupyter_output><empty_output><jupyter_text>Most common words include uninformative words like "the", "in", "and",...# Compute TF-IDF for the corpus
To give more weight to informative words, we weigh them by their TF-IDF scores.<jupyter_code>people['word_count'] = graphlab.text_analytics.count_words(people['text'])
people.head()
tfidf = graphlab.text_analytics.tf_idf(people['word_count'])
# Earlier versions of GraphLab Create returned an SFrame rather than a single SArray
# This notebook was created using Graphlab Create version 1.7.1
if graphlab.version <= '1.6.1':
tfidf = tfidf['docs']
tfidf
people['tfidf'] = tfidf
people.head()
<jupyter_output><empty_output><jupyter_text>## Examine the TF-IDF for the Obama article<jupyter_code>obama = people[people['name'] == 'Barack Obama']
obama[['tfidf']].stack('tfidf',new_column_name=['word','tfidf']).sort('tfidf',ascending=False)<jupyter_output><empty_output><jupyter_text>Words with highest TF-IDF are much more informative.# Manually compute distances between a few people
Let's manually compare the distances between the articles for a few famous people. <jupyter_code>clinton = people[people['name'] == 'Bill Clinton']
beckham = people[people['name'] == 'David Beckham']
obama['tfidf'][0]
<jupyter_output><empty_output><jupyter_text>## Is Obama closer to Clinton than to Beckham?
We will use cosine distance, which is given by
(1-cosine_similarity)
and find that the article about president Obama is closer to the one about former president Clinton than that of footballer David Beckham.<jupyter_code>graphlab.distances.cosine(obama['tfidf'][0],clinton['tfidf'][0])
graphlab.distances.cosine(obama['tfidf'][0],beckham['tfidf'][0])<jupyter_output><empty_output><jupyter_text># Build a nearest neighbor model for document retrieval
We now create a nearest-neighbors model and apply it to document retrieval. <jupyter_code>knn_model = graphlab.nearest_neighbors.create(people,features=['tfidf'],label='name')<jupyter_output><empty_output><jupyter_text># Applying the nearest-neighbors model for retrieval## Who is closest to Obama?<jupyter_code>knn_model.query(obama)<jupyter_output><empty_output><jupyter_text>As we can see, president Obama's article is closest to the one about his vice-president Biden, and those of other politicians. ## Other examples of document retrieval<jupyter_code>swift = people[people['name'] == 'Taylor Swift']
knn_model.query(swift)
jolie = people[people['name'] == 'Angelina Jolie']
knn_model.query(jolie)
arnold = people[people['name'] == 'Arnold Schwarzenegger']
knn_model.query(arnold)
elton = people[people['name'] == 'Elton John']
elton_word_count_table = elton[['word_count']].stack('word_count', new_column_name = ['word','count'])
elton_word_count_table.sort('count', ascending =False)[0:3]
elton_tfidf_table = elton[['tfidf']].stack('tfidf', new_column_name = ['word', 'tfidf']).sort('tfidf', ascending = False)
elton_tfidf_table
paul = people[people['name'] == 'Paul McCartney']
victoria = people[people['name'] == 'Victoria Beckham']
graphlab.distances.cosine(elton['tfidf'][0], paul['tfidf'][0])
graphlab.distances.cosine(elton['tfidf'][0], victoria['tfidf'][0])
one_model = graphlab.nearest_neighbors.create(people, features = ['word_count'], label = 'name', distance = 'cosine')
second_model = graphlab.nearest_neighbors.create(people, features = ['tfidf'], label = 'name', distance = 'cosine')
one_model.query(elton)
second_model.query(elton)
one_model.query(victoria)
second_model.query(victoria)<jupyter_output><empty_output>
|
no_license
|
/UW-machine/ML-Case Studies/Week 4/Document+Retrieval.ipynb
|
mhassa22/Coursera
| 15 |
<jupyter_start><jupyter_text>Fitting 10 folds for each of 45 candidates, totalling 450 fits
[Parallel(n_jobs=-1)]: Done 42 tasks | elapsed: 47.2min
[Parallel(n_jobs=-1)]: Done 192 tasks | elapsed: 273.3min
[Parallel(n_jobs=-1)]: Done 442 tasks | elapsed: 631.1min
[Parallel(n_jobs=-1)]: Done 450 out of 450 | elapsed: 646.4min finished<jupyter_code>print 'Best parameter set: %s' % result.best_params_
print 'CV accuracy %0.3f' % result.best_score_
clf = result.best_estimator_<jupyter_output><empty_output><jupyter_text>Best parameter set: {'clf__max_depth': 8, 'clf__learning_rate': 0.1, 'clf__n_estimators': 200}
CV accuracy -1.303<jupyter_code>predictions = clf.predict(text_test)
print 'macro f1:', f1_score(encoded_Y_test, predictions, average='macro')<jupyter_output><empty_output>
|
no_license
|
/whisper_data_science/XGBoost_jupyter.ipynb
|
oliverlewis/Data-Science-Projects
| 2 |
<jupyter_start><jupyter_text># UW Data Science Winter 2017
# Dave Wine 8430191
# Term Project - Refugees, Terrorism, and AirstrikesPerspectives
1. You are a US customs official with a long line of refugees. You need to decide whether each is a potential
based on their sex, country of origin, and religion.
2. You are a terrorist mastermind trying to recruit people who will not arouse US customs supervision.
3. You are the President of the United States trying to choose an entry policy.
4. You are a citizen of the United States that is voting for a President that is claiming extreme danger
from foreign terrorists posing as refugees.
5. You are a citizen of one of those countries, which is currently subject to airstrikes by the United
States. How safe are you compared to the US citizen?
6. You are a policy wonk in the US government, trying to decide whether to add additional screening to
a refugee admission process.<jupyter_code># Import packages
require(ggplot2)
require(car)
require(plyr)
require(dplyr)
# File read function
read.file = function(file = f){
file.data <- read.csv(file, header=TRUE, stringsAsFactors=FALSE)
# numcols <- c('price','peak.rpm')
# auto.data[, numcols]<-lapply(auto.data[,numcols], as.numeric)
#factcols <- c('fuel.type','aspiration','drive.wheels','body.style')
#auto.data[, factcols]<-lapply(auto.data[,factcols], as.factor)
file.data[complete.cases(file.data),]
}
# Histogram Plot Function
plot.t <- function(a, b, plotvar,a.name, b.name,cols = c(a.name,b.name), nbins = 20){
maxs = max(c(max(a), max(b)))
mins = min(c(min(a), min(b)))
breaks = seq(maxs, mins, length.out = (nbins + 1))
par(mfrow = c(2, 1))
hist(a, breaks = breaks, main = paste('Histogram of', cols[1]), xlab = plotvar)
abline(v = mean(a), lwd = 4, col = 'red')
hist(b, breaks = breaks, main = paste('Histogram of', cols[2]), xlab = plotvar)
abline(v = mean(b), lwd = 4, col = 'red')
par(mfrow = c(1, 1))
}
# ANOVA Plot Function
ANOVA.plot <- function (df){
df$group = factor(df$group) # Make sure your groups are a factor (for further analysis below)
boxplot(df$val ~ df$group)
df_aov = aov(val ~ group, data = df)
summary(df_aov)
print(df_aov)
tukey_aov = TukeyHSD(df_aov) # Tukey's Range test:
plot(tukey_aov)
print(tukey_aov)
}
####
# Main Code
####
#set working directory
setwd("~/GitHub/UW-MDA-2017-DW/Test")
# Read data in
GTD <- 'globalterrorismdb_0616dist.csv'
GTD.data = read.file(GTD)
# View dataset and summary statistics
str(GTD.data)
summary(GTD.data)
####<jupyter_output>'data.frame': 0 obs. of 137 variables:
$ eventid : num
$ iyear : int
$ imonth : int
$ iday : int
$ approxdate : chr
$ extended : int
$ resolution : chr
$ country : int
$ country_txt : chr
$ region : int
$ region_txt : chr
$ provstate : chr
$ city : chr
$ latitude : num
$ longitude : num
$ specificity : int
$ vicinity : int
$ location : chr
$ summary : chr
$ crit1 : int
$ crit2 : int
$ crit3 : int
$ doubtterr : int
$ alternative : int
$ alternative_txt : chr
$ multiple : int
$ success : int
$ suicide : int
$ attacktype1 : int
$ attacktype1_txt : chr
$ attacktype2 : int
$ attacktype2_txt : chr
$ attacktype3 : int
$ attacktype3_txt : chr
$ targt[...]
|
no_license
|
/Term Project - Dave Wine.ipynb
|
dwine66/UW-MDA-2017-DW
| 1 |
<jupyter_start><jupyter_text># Train ATF2 and make predictions for HepG2
---
Takes something like 10-20 minutes, and uses 50GB!
----<jupyter_code>import time
import pandas as pd
import numpy as np
import subprocess
from sklearn.metrics import roc_auc_score
from sklearn.linear_model import SGDClassifier
TF='ATF2'
TRAIN_CELL_TYPES=['GM12878', 'H1-hESC']
VALID_CELL_TYPE='MCF-7'
TEST_CELL_TYPE='HepG2'
DATA_DIR='/mnt/vdisk/data/synapse/'<jupyter_output><empty_output><jupyter_text>### Load data<jupyter_code>start=time.time()
# load fold coverage tables for all train cell lines
fc_train=[pd.read_hdf(DATA_DIR+'fold_cov_data/'+cl+'_dnase_fold_cov.hdf',
'dnase_fold_cov')
for cl in TRAIN_CELL_TYPES ]
#load fold coverage table for valid cell line
fc_valid=pd.read_hdf(DATA_DIR+'fold_cov_data/'+VALID_CELL_TYPE+'_dnase_fold_cov.hdf',
'dnase_fold_cov')
#load fold coverage table for the final submission cell line
fc_test=pd.read_hdf(DATA_DIR+'fold_cov_data/'+TEST_CELL_TYPE+'_dnase_fold_cov.hdf',
'dnase_fold_cov')
#load motif table for the transciption factor
motif=pd.read_hdf(DATA_DIR+'motif_data/'+TF+'_motif.hdf','motif')
#load labels for the transcription factor
labels=pd.read_hdf(DATA_DIR+'extended_labels/'+TF+'_labels.hdf','labels')
print (time.time()-start),'s'
start=time.time()
# fix index not set before
motif.set_index(['chr1','600','800'],inplace=True)
print (time.time()-start),'s'<jupyter_output>25.8023970127 s
<jupyter_text>### Create train,valid,test data<jupyter_code>start=time.time()
# packing columns into the train dataset
x_train=[fc_train[0].values, #cell line specific fc column
motif.values ] + [ #motif scores
x.values for x in fc_train[1:]] + [ #other cell line fc values
labels[tf].values for tf in TRAIN_CELL_TYPES[1:] ] #other cell line labels
x_train=np.column_stack(x_train)
y_train=labels[TRAIN_CELL_TYPES[0]].values.astype('int')
print (time.time()-start),'s'
start=time.time()
x_valid=np.array(x_train)
x_valid[:,0]=fc_valid.values.flatten()
y_valid=labels[VALID_CELL_TYPE].values
print (time.time()-start),'s'<jupyter_output>6.9238550663 s
<jupyter_text>### Train SGD logistic reg
- it doesn't seem to use the 12 cores for most of the time<jupyter_code>start=time.time()
clf = SGDClassifier(loss='log', class_weight='balanced', n_jobs=12)
clf.fit(x_train,y_train)
print (time.time()-start),'s'<jupyter_output>169.176910877 s
<jupyter_text>### Evaluate<jupyter_code>start=time.time()
print 'auc:',roc_auc_score(y_valid,clf.predict_proba(x_valid)[:,1])
print (time.time()-start),'s'<jupyter_output>auc: 0.644590469777
33.3002209663 s
<jupyter_text>### Predict<jupyter_code>start=time.time()
x_test=np.array(x_train)
x_test[:,0]=fc_test.values.flatten()
print (time.time()-start),'s'
start=time.time()
y_test_pred=clf.predict_proba(x_test)[:,1]
print (time.time()-start),'s'<jupyter_output>4.03232192993 s
<jupyter_text>### Annotate predictions with the test regions
- It has to be in the exact order of the test regions
- https://www.synapse.org/#!Synapse:syn6131484/wiki/402044
- I missed the very frst line when creating the tables, I just add a 0 there<jupyter_code>start=time.time()
#load index
idx=pd.read_hdf(DATA_DIR+'/annotations/test_regions.hdf',
'test_regions').index
print (time.time()-start),'s'
start=time.time()
res_df=pd.DataFrame(np.concatenate([[0],y_test_pred]),index=idx)
print (time.time()-start),'s'<jupyter_output>0.28665804863 s
<jupyter_text>### The slowest part of the whole process is to write the tsv, so now i just make it in hdf<jupyter_code>start=time.time()
#slooooooooooow
#res_df.to_csv(
# TF+'_test.h',sep='\t',header=False,compression='gzip')
res_df.to_hdf(TF+'_'+TEST_CELL_TYPE+'_test.hdf','preds')
print (time.time()-start),'s'<jupyter_output>2.23854184151 s
<jupyter_text>### Join the results using shell commands
- Not too elegant but faster<jupyter_code>start=time.time()
np.savetxt(TF+'_'+TEST_CELL_TYPE+'_test.txt',np.concatenate([[0],y_test_pred]))
print (time.time()-start),'s'
start=time.time()
cmd = ' paste '
cmd+= ' <(zcat '+DATA_DIR+'/annotations/test_regions.blacklistfiltered.bed.gz ) '
cmd+= TF+'_'+TEST_CELL_TYPE +'_test.txt '
cmd+= ' | gzip -c -1 > '+'F.'+TF+'.'+TEST_CELL_TYPE+'.tab.gz'
print cmd
subprocess.check_output(cmd,shell=True, executable='/bin/bash')
print (time.time()-start),'s'<jupyter_output> paste <(zcat /mnt/vdisk/data/synapse//annotations/test_regions.blacklistfiltered.bed.gz ) ATF2_HepG2_test.txt | gzip -c -1 > F.ATF2.HepG2.tab.gz
46.6792120934 s
|
no_license
|
/deadline_09_30/final_subs_09_30/train_v0_ATF2_HepG2.ipynb
|
riblidezso/dream_encode_challenge
| 9 |
<jupyter_start><jupyter_text>Table of Contents
1 Beta Variational Autoencoder Using Keras data Dsprites1.1 Data1.1.1 Description of the dataset1.1.2 Generating the Training and test data1.2 Building the Beta-VAE1.2.1 Beta-VAE Summary1.3 Training1.4 Analysis1.4.1 Traversals# Beta Variational Autoencoder Using Keras data Dsprites
The code is based on the book [Generative Deep Learning by David Foster](https://www.oreilly.com/library/view/generative-deep-learning/9781492041931/).## DataThe data is taken from [Dsprites dataset](https://github.com/deepmind/dsprites-dataset).
For loading the dataset we used this [notebook](https://github.com/deepmind/dsprites-dataset/blob/master/dsprites_reloading_example.ipynb).
The datafiles are on the input folder.<jupyter_code>from __future__ import absolute_import
from __future__ import division
from __future__ import print_function
from __future__ import unicode_literals
from matplotlib import pyplot as plt
import numpy as np
import seaborn as sns
import os
# Change figure aesthetics
%matplotlib inline
sns.set_context('paper', font_scale=1.2, rc={'lines.linewidth': 1.5})
path = os.getcwd()
dataset_zip = np.load(path+'/input/dsprites/dsprites_ndarray_co1sh3sc6or40x32y32_64x64.npz'
,encoding='bytes', allow_pickle=True)
print('Keys in the dataset:', dataset_zip.keys())
imgs = dataset_zip['imgs']
latents_values = dataset_zip['latents_values']
latents_classes = dataset_zip['latents_classes']
metadata = dataset_zip['metadata'][()]
print('Metadata: \n', metadata)<jupyter_output>Keys in the dataset: KeysView(<numpy.lib.npyio.NpzFile object at 0x7f502574eb38>)
Metadata:
{b'date': b'April 2017', b'description': b'Disentanglement test Sprites dataset.Procedurally generated 2D shapes, from 6 disentangled latent factors.This dataset uses 6 latents, controlling the color, shape, scale, rotation and position of a sprite. All possible variations of the latents are present. Ordering along dimension 1 is fixed and can be mapped back to the exact latent values that generated that image.We made sure that the pixel outputs are different. No noise added.', b'version': 1, b'latents_names': (b'color', b'shape', b'scale', b'orientation', b'posX', b'posY'), b'latents_possible_values': {b'orientation': array([0. , 0.16110732, 0.32221463, 0.48332195, 0.64442926,
0.80553658, 0.96664389, 1.12775121, 1.28885852, 1.44996584,
1.61107316, 1.77218047, 1.93328779, 2.0943951 , 2.25550242,
2.41660973, 2.57771705, 2.73882436, 2.89993168, 3.061039 ,
3.22[...]<jupyter_text>### Description of the dataset
From the metadata we have the description of the dataset and how it was generated:
'Disentanglement test Sprites dataset.Procedurally generated 2D shapes, from 6 disentangled latent factors.This dataset uses 6 latents, controlling the color, shape, scale, rotation and position of a sprite. All possible variations of the latents are present. Ordering along dimension 1 is fixed and can be mapped back to the exact latent values that generated that image.We made sure that the pixel outputs are different. No noise added.'
The latent space size for generating the dataset is:
- 'latents_sizes': array([ 1, 3, 6, 40, 32, 32]),
Each latent affect only one attribute of the figure, the atributes are:
- 'latents_names': ('color', 'shape', 'scale', 'orientation', 'posX', 'posY')
**Therefore, we have only one color, 3 shapes, 6 types of scales, 40 points regarding rotations, 32 points for the posX (or posY).**<jupyter_code># Define number of values per latents and functions to convert to indices
latents_sizes = metadata[b'latents_sizes']
latents_bases = np.concatenate((latents_sizes[::-1].cumprod()[::-1][1:],
np.array([1,])))<jupyter_output><empty_output><jupyter_text>Generating helper functions to handle the data<jupyter_code>def latent_to_index(latents):
return np.dot(latents, latents_bases).astype(int)
def ytest_to_index(latents):
"""Creating ytest index for posX feature.
Parameters:
-----------------------------------------
latents(list): Latents.
"""
return np.dot(latents,[0,0,0,0,1,0]).astype(int)
def sample_latent(size=1):
"""Sample the latent space.
Parameters:
-----------------------------------------
size(integer): Number of samples.
"""
samples = np.zeros((size, latents_sizes.size))
for lat_i, lat_size in enumerate(latents_sizes):
samples[:, lat_i] = np.random.randint(lat_size, size=size)
return samples<jupyter_output><empty_output><jupyter_text>### Generating the Training and test data
Here we generate the dataset randomly.<jupyter_code># Sample latents randomly
latents_sampled = sample_latent(size=5000)
# Select images
indices_sampled = latent_to_index(latents_sampled)
#To use as an index for the scatterplot (posX)
y_test = ytest_to_index(latents_sampled)
imgs_sampled = imgs[indices_sampled]
x_test = imgs[indices_sampled]
x_test = x_test.reshape(x_test.shape + (1,))
# Sample latents randomly
latents_sampled_train = sample_latent(size=30000)
# Select images
indices_sampled_train = latent_to_index(latents_sampled_train)
imgs_sampled = imgs[indices_sampled]
x_train = imgs[indices_sampled_train]
x_train = x_train.reshape(x_train.shape + (1,))
print('Analysing the shapes:\n'+
' X train shape: {} \n Y test shape: {} \n X test shape: {} \n'.format(x_train.shape,
y_test.shape,
x_test.shape))<jupyter_output>Analysing the shapes:
X train shape: (30000, 64, 64, 1)
Y test shape: (5000,)
X test shape: (5000, 64, 64, 1)
<jupyter_text>## Building the Beta-VAE*In this notebook I will ignore Furure and Deprecation Warnings*<jupyter_code>import warnings
warnings.simplefilter(action='ignore', category=FutureWarning)
warnings.filterwarnings('ignore', category=DeprecationWarning)
from Models.VAE_Keras import VariationalAutoencoder_Keras<jupyter_output>Using TensorFlow backend.
<jupyter_text>**Choose 'build' if you want to train the VAE, if you want to reload weights type 'reconstruct'.**<jupyter_code>import os
# run params
SECTION = 'bvae'
RUN_ID = '0001'
DATA_NAME = 'D'
RUN_FOLDER = 'run/{}/'.format(SECTION)
RUN_FOLDER += '_'.join([RUN_ID, DATA_NAME])
if not os.path.exists(RUN_FOLDER):
os.makedirs(RUN_FOLDER)
os.mkdir(os.path.join(RUN_FOLDER, 'viz'))
os.mkdir(os.path.join(RUN_FOLDER, 'images'))
os.mkdir(os.path.join(RUN_FOLDER, 'weights'))
mode = 'build'
#Latent size
ZDIM = 10
bvae = VariationalAutoencoder_Keras(
input_dim = (64,64,1)
, encoder_conv_filters = [32,64,64,64]#, 64]
, encoder_conv_kernel_size = [3,3,3,3]#,3]
, encoder_conv_strides = [1,2,2,1]#,1]
, decoder_conv_t_filters = [64,64,32,1]
, decoder_conv_t_kernel_size = [3,3,3,3]
, decoder_conv_t_strides = [1,2,2,1]
, z_dim = ZDIM
)
if mode == 'build':
bvae.save(RUN_FOLDER)
else:
bvae.load_weights(os.path.join(RUN_FOLDER, 'weights/weights.h5'))<jupyter_output><empty_output><jupyter_text>### Beta-VAE Summary<jupyter_code>bvae.encoder.summary()
bvae.decoder.summary()<jupyter_output>_________________________________________________________________
Layer (type) Output Shape Param #
=================================================================
decoder_input (InputLayer) (None, 10) 0
_________________________________________________________________
dense_2 (Dense) (None, 16384) 180224
_________________________________________________________________
reshape_2 (Reshape) (None, 16, 16, 64) 0
_________________________________________________________________
decoder_conv_t_0 (Conv2DTran (None, 16, 16, 64) 36928
_________________________________________________________________
leaky_re_lu_12 (LeakyReLU) (None, 16, 16, 64) 0
_________________________________________________________________
decoder_conv_t_1 (Conv2DTran (None, 32, 32, 64) 36928
_________________________________________________________________
leaky_re_l[...]<jupyter_text>## Training<jupyter_code>#Compile Parameters (These parameters depend on a fine tuning)
LEARNING_RATE = 0.0005
R_LOSS_FACTOR = 5000
BETA = 10
bvae.compile(LEARNING_RATE, R_LOSS_FACTOR, BETA)
# Training Parameters
BATCH_SIZE = 126
EPOCHS = 20
PRINT_EVERY_N_BATCHES = 100
INITIAL_EPOCH = 0
bvae.train(
x_train
, batch_size = BATCH_SIZE
, epochs = EPOCHS
, run_folder = RUN_FOLDER
, print_every_n_batches = PRINT_EVERY_N_BATCHES
, initial_epoch = INITIAL_EPOCH
)<jupyter_output>Epoch 1/20
30000/30000 [==============================] - 510s 17ms/step - loss: 140.6081 - vae_r_loss: 101.5043 - vae_kl_loss: 3.9104
Epoch 00001: saving model to run/bvae/0001_D/weights/weights-001-140.61.h5
Epoch 00001: saving model to run/bvae/0001_D/weights/weights.h5
Epoch 2/20
30000/30000 [==============================] - 508s 17ms/step - loss: 126.2686 - vae_r_loss: 78.4650 - vae_kl_loss: 4.7804
Epoch 00002: saving model to run/bvae/0001_D/weights/weights-002-126.27.h5
Epoch 00002: saving model to run/bvae/0001_D/weights/weights.h5
Epoch 3/20
30000/30000 [==============================] - 509s 17ms/step - loss: 123.2918 - vae_r_loss: 74.7729 - vae_kl_loss: 4.8519
Epoch 00003: saving model to run/bvae/0001_D/weights/weights-003-123.29.h5
Epoch 00003: saving model to run/bvae/0001_D/weights/weights.h5
Epoch 4/20
30000/30000 [==============================] - 511s 17ms/step - loss: 120.1351 - vae_r_loss: 71.3680 - vae_kl_loss: 4.8767
Epoch 00004: saving model to run/bvae/0[...]<jupyter_text>## Analysis### TraversalsWe choose a random point on the latent space that corresponds to an image.<jupyter_code>n_to_show = 1
example_idx = np.random.choice(range(len(x_test)), n_to_show)
example_images = x_test[example_idx]
z_points = bvae.encoder.predict(example_images)
reconst_images = bvae.decoder.predict(z_points)
def traverse(pos,traverse_value,latent_vector):
"""Make the traverse on pos at the latent
vector
Parameters
------------------------------------------
pos(int): Position at the latent vector that
you want to traverse.
latent_vector(list): Latent vector.
"""
latent_vector[0][pos] = traverse_value
return latent_vector
from copy import copy
#number of imgs
n_traverse = 10
#minimum value of traversal
min_t = -3
#maximum value of traversal
max_t = 3
plt.imshow(reconst_images[0,:,:,0], cmap='gray')
plt.title("Original \n Latent Vector: {}".format(z_points[0]))
for j in range(ZDIM):
fig = plt.figure(figsize=(18,5))
fig.subplots_adjust(hspace=0.4,wspace=0.1)
for i,val in enumerate(np.linspace(min_t,max_t,n_traverse)):
#Use copy in order to not overwrite z_points
Latent_T = traverse(j,val,copy(z_points))
reconst_image = bvae.decoder.predict(Latent_T)
ax = fig.add_subplot(1, n_traverse, i+1)
ax.imshow(reconst_image[0,:,:,0], cmap='gray')
ax.axis('off')
ax.set_title("{:.3f}".format(val))
fig.suptitle('Changing Latent Position {}'.format(j),x=0.5,y=0.75, fontsize=16)
plt.show()<jupyter_output><empty_output>
|
no_license
|
/BETA-VAE with Keras Dsprites - Traversals.ipynb
|
nahumsa/Variational-Autoencoder
| 9 |
<jupyter_start><jupyter_text>In this notebook I will try and train a U-net to output the masks of synthetic bat-like calls that I've made spectrograms + masks of. The SNRs, call durations and shape of the calls in the spectrograms vary. Please refer to the previous notebook 'Generating masks for synthetic call data' for further reference.
Most of the code here is taken directly from https://www.depends-on-the-definition.com/unet-keras-segmenting-images/<jupyter_code>import keras
from keras.models import Model, load_model
from keras.layers import Input, BatchNormalization, Activation, Dense, Dropout
from keras.layers.core import Lambda, RepeatVector, Reshape
from keras.layers.convolutional import Conv2D, Conv2DTranspose
from keras.layers.pooling import MaxPooling2D, GlobalMaxPool2D
from keras.layers.merge import concatenate, add
from keras.callbacks import EarlyStopping, ModelCheckpoint, ReduceLROnPlateau
from keras.optimizers import Adam
from keras.preprocessing.image import ImageDataGenerator, array_to_img, img_to_array, load_img
import matplotlib.pyplot as plt
plt.rcParams['agg.path.chunksize'] = 10000
import pickle
import numpy as np
from skimage.io import imread, imshow, concatenate_images
from skimage.transform import resize
from skimage.morphology import label
# from sklearn.model_selection import train_test_split --> don't need this anyway as it's already separated
import tensorflow as tf
# let's load the data and make it into tensors :
training_file = open('sound_spec_mask//training_data.pkl', 'rb')
training = pickle.load(training_file)
training_file.close()
testing_file = open('sound_spec_mask//test_data.pkl', 'rb')
testing = pickle.load(testing_file)
testing_file.close()
validn_file = open('sound_spec_mask//validation_data.pkl', 'rb')
validation = pickle.load(validn_file)
validn_file.close()
# reshape everything as it needs to be for the U-net
def reshape_and_normalise(data_container):
filenames = data_container.keys()
num_files = len(filenames)
img_rows, img_cols = data_container[filenames[0]]['specgm'].shape
num_channels = 1
fin_rows, fin_cols = 128, 128
image_tensor = np.zeros((num_files,fin_rows, fin_cols, num_channels))
mask_tensor = np.zeros((num_files, fin_rows, fin_cols, num_channels))
for i, each_file in enumerate(filenames):
img_raw = data_container[each_file]['specgm']
norm_img = img_raw/np.max(img_raw)
norm_img = np.reshape(norm_img, (img_rows, img_cols, 1))
resized_normimg = resize(norm_img, (128, 128, 1), mode='constant', preserve_range=True)
mask_img_raw = data_container[each_file]['mask_specgm']
mask_img = np.reshape(mask_img_raw, (img_rows, img_cols, 1))
resized_mask_img = resize(mask_img, (128, 128, 1), mode='constant', preserve_range=True)
threshold = np.percentile(resized_mask_img.flatten(), 99)
resized_mask_img[resized_mask_img >= threshold] = 1
resized_mask_img[resized_mask_img < threshold] = 0
image_tensor[i,:,:,:] = resized_normimg
mask_tensor[i,:,:,:] = resized_mask_img
return(image_tensor, mask_tensor)
training_img, training_mask = reshape_and_normalise(training)
validation_img, validation_mask = reshape_and_normalise(validation)
test_img, test_mask = reshape_and_normalise(testing)
%matplotlib notebook
def conv2d_block(input_tensor, n_filters, kernel_size=3, batchnorm=True):
# first layer
x = Conv2D(filters=n_filters, kernel_size=(kernel_size, kernel_size), kernel_initializer="he_normal",
padding="same")(input_tensor)
if batchnorm:
x = BatchNormalization()(x)
x = Activation("relu")(x)
# second layer
x = Conv2D(filters=n_filters, kernel_size=(kernel_size, kernel_size), kernel_initializer="he_normal",
padding="same")(x)
if batchnorm:
x = BatchNormalization()(x)
x = Activation("relu")(x)
return x
def get_unet(input_img, n_filters=16, dropout=0.5, batchnorm=True):
# contracting path
c1 = conv2d_block(input_img, n_filters=n_filters*1, kernel_size=3, batchnorm=batchnorm)
p1 = MaxPooling2D((2, 2)) (c1)
p1 = Dropout(dropout*0.5)(p1)
c2 = conv2d_block(p1, n_filters=n_filters*2, kernel_size=3, batchnorm=batchnorm)
p2 = MaxPooling2D((2, 2)) (c2)
p2 = Dropout(dropout)(p2)
c3 = conv2d_block(p2, n_filters=n_filters*4, kernel_size=3, batchnorm=batchnorm)
p3 = MaxPooling2D((2, 2)) (c3)
p3 = Dropout(dropout)(p3)
c4 = conv2d_block(p3, n_filters=n_filters*8, kernel_size=3, batchnorm=batchnorm)
p4 = MaxPooling2D(pool_size=(2, 2)) (c4)
p4 = Dropout(dropout)(p4)
c5 = conv2d_block(p4, n_filters=n_filters*16, kernel_size=3, batchnorm=batchnorm)
# expansive path
u6 = Conv2DTranspose(n_filters*8, (3, 3), strides=(2, 2), padding='same') (c5)
u6 = concatenate([u6, c4])
u6 = Dropout(dropout)(u6)
c6 = conv2d_block(u6, n_filters=n_filters*8, kernel_size=3, batchnorm=batchnorm)
u7 = Conv2DTranspose(n_filters*4, (3, 3), strides=(2, 2), padding='same') (c6)
u7 = concatenate([u7, c3])
u7 = Dropout(dropout)(u7)
c7 = conv2d_block(u7, n_filters=n_filters*4, kernel_size=3, batchnorm=batchnorm)
u8 = Conv2DTranspose(n_filters*2, (3, 3), strides=(2, 2), padding='same') (c7)
u8 = concatenate([u8, c2])
u8 = Dropout(dropout)(u8)
c8 = conv2d_block(u8, n_filters=n_filters*2, kernel_size=3, batchnorm=batchnorm)
u9 = Conv2DTranspose(n_filters*1, (3, 3), strides=(2, 2), padding='same') (c8)
u9 = concatenate([u9, c1], axis=3)
u9 = Dropout(dropout)(u9)
c9 = conv2d_block(u9, n_filters=n_filters*1, kernel_size=3, batchnorm=batchnorm)
outputs = Conv2D(1, (1, 1), activation='sigmoid') (c9)
model = Model(inputs=[input_img], outputs=[outputs])
return model
im_height = 128
im_width = 128
input_img = Input((im_height, im_width, 1), name='img')
model = get_unet(input_img, n_filters=16, dropout=0.05, batchnorm=True)
model.compile(optimizer=Adam(), loss="binary_crossentropy", metrics=["accuracy"])
model.summary()
# serialize model to JSON
model_json = model.to_json()
with open("batca;;_unet.json", "w") as json_file:
json_file.write(model_json)
i = 50
plt.figure()
plt.subplot(211)
plt.title('Original resized spectrogram')
plt.imshow(training_img[i,:,:,:].reshape(128,128), aspect='auto')
plt.subplot(212)
plt.title('Resized mask')
plt.imshow(training_mask[i,:,:,:].reshape(128,128), aspect='auto')
callbacks = [
EarlyStopping(patience=10, verbose=1),
ReduceLROnPlateau(factor=0.1, patience=3, min_lr=0.00001, verbose=1),
ModelCheckpoint('model-tgs-salt.h5', verbose=1, save_best_only=True, save_weights_only=True)
]
results = model.fit(training_img, training_mask, batch_size=32, epochs=10, callbacks=callbacks,
validation_data=(validation_img, validation_mask))
#copy pasted code from the post :
plt.figure(figsize=(8, 8))
plt.title("Learning curve")
plt.plot(results.history["loss"], label="loss")
plt.plot(results.history["val_loss"], label="val_loss")
plt.plot( np.argmin(results.history["val_loss"]), np.min(results.history["val_loss"]), marker="x", color="r", label="best model")
plt.xlabel("Epochs")
plt.ylabel("log_loss")
plt.legend();<jupyter_output><empty_output><jupyter_text>And now let's try out the performance of the model:<jupyter_code>preds_val = model.predict(test_img, verbose=1)
predicted_masks_file = open('sound_spec_mask/predicted_test_masks.pkl', 'wb')
pickle.dump(preds_val, predicted_masks_file)
predicted_masks_file.close()
img_ind = np.random.choice(test_img.shape[0],1)
plt.figure(figsize=(8,8))
plt.subplot(311)
plt.title(str(img_ind))
plt.imshow(test_img[img_ind,:,:,:].reshape(128,128), aspect='auto')
plt.subplot(312)
plt.title('actual mask')
plt.imshow(test_mask[img_ind,:,:,:].reshape(128,128), aspect='auto')
plt.subplot(313)
plt.title('predicted mask')
plt.imshow(preds_val[img_ind,:,:,:].reshape(128,128), aspect='auto')<jupyter_output><empty_output>
|
no_license
|
/Training a U-net to output bat call maks.ipynb
|
thejasvibr/bat_call_detection_and_segmentation
| 2 |
<jupyter_start><jupyter_text>## Decision Tree### Loading Data<jupyter_code>import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.model_selection import train_test_split
from sklearn.feature_extraction.text import CountVectorizer, TfidfVectorizer
from gensim.models import KeyedVectors, Word2Vec
import pickle
from sklearn.metrics import confusion_matrix, roc_curve, auc, roc_auc_score
from tqdm import tqdm_notebook as tqdm
import os
from pathlib import Path
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import Normalizer
from scipy.sparse import hstack
from wordcloud import WordCloud
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import make_scorer, roc_auc_score, confusion_matrix
from sklearn.tree import DecisionTreeClassifier
import plotly.offline as offline
import plotly.graph_objs as go
offline.init_notebook_mode()
import numpy as np
%matplotlib inline
df = pd.read_csv('data/preprocessed_df.csv')
print(df.shape)
df.head()
y = df['project_is_approved'].values
X = df.drop(['project_is_approved'], axis=1)
X.head()<jupyter_output><empty_output><jupyter_text>### Splitting data into Train and cross validation(or test): Stratified Sampling<jupyter_code>X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, stratify=y)
X_train, X_cv, y_train, y_cv = train_test_split(X_train, y_train, test_size=0.2, stratify=y_train)
print(X_train.shape, y_train.shape), print(X_cv.shape, y_cv.shape), print(X_test.shape, y_test.shape)
<jupyter_output><empty_output><jupyter_text>## Make Data Model Ready: encoding eassay, and project_title<jupyter_code>tfidf_all_features = []
tfidf_w2v_all_features = []<jupyter_output><empty_output><jupyter_text>#### Essay<jupyter_code>vectorizer_tfidf = TfidfVectorizer(min_df=10, ngram_range=(1,4), max_features=20000)
X_train_essay_tfidf = vectorizer_tfidf.fit_transform(X_train['essay'].values)
X_cv_essay_tfidf = vectorizer_tfidf.transform(X_cv['essay'].values)
X_test_essay_tfidf = vectorizer_tfidf.transform(X_test['essay'].values)
print(X_train_essay_tfidf.shape, y_train.shape)
print(X_cv_essay_tfidf.shape, y_cv.shape)
print(X_test_essay_tfidf.shape, y_test.shape)
print("="*100)
tfidf_all_features.extend(vectorizer_tfidf.get_feature_names())
list_of_sentence_train=[]
for sentence in X_train['essay']:
list_of_sentence_train.append(sentence.split())
w2v_model = Word2Vec(list_of_sentence_train, min_count=10, size=50, workers=4)
w2v_words = list(w2v_model.wv.vocab)
tf_idf_vect = TfidfVectorizer(min_df=10, ngram_range=(1,4), max_features=20000)
tf_idf_matrix = tf_idf_vect.fit_transform(X_train['essay'])
tfidf_feat = tf_idf_vect.get_feature_names()
dictionary = dict(zip(tf_idf_vect.get_feature_names(), list(tf_idf_vect.idf_)))
tfidf_w2v_train = []
row = 0
for sentence in tqdm(list_of_sentence_train):
sentence_vec = np.zeros(50)
weight_sum = 0
for word in sentence:
if word in w2v_words and word in tfidf_feat:
vec = w2v_model.wv[word]
tf_idf = dictionary[word] * (sentence.count(word) / len(sentence))
sentence_vec += (vec * tf_idf)
weight_sum += tf_idf
if weight_sum != 0:
sentence_vec /= weight_sum
tfidf_w2v_train.append(sentence_vec)
row += 1
list_of_sentence_cv = []
for sentence in X_cv['essay']:
list_of_sentence_cv.append(sentence.split())
tfidf_w2v_cv = []
row = 0
for sentence in tqdm(list_of_sentence_cv):
sentence_vec = np.zeros(50)
weight_sum = 0
for word in sentence:
if word in w2v_words and word in tfidf_feat:
vec = w2v_model.wv[word]
tf_idf = dictionary[word] * (sentence.count(word) / len(sentence))
sentence_vec += (vec * tf_idf)
weight_sum += tf_idf
if weight_sum != 0:
sentence_vec /= weight_sum
tfidf_w2v_cv.append(sentence_vec)
row += 1
list_of_sentence_test = []
for sentence in X_test['essay']:
list_of_sentence_test.append(sentence.split())
tfidf_w2v_test = []
row = 0
for sentence in tqdm(list_of_sentence_test):
sentence_vec = np.zeros(50)
weight_sum = 0
for word in sentence:
if word in w2v_words and word in tfidf_feat:
vec = w2v_model.wv[word]
tf_idf = dictionary[word] * (sentence.count(word) / len(sentence))
sentence_vec += (vec * tf_idf)
weight_sum += tf_idf
if weight_sum != 0:
sentence_vec /= weight_sum
tfidf_w2v_test.append(sentence_vec)
row += 1
tfidf_w2v = np.array([tfidf_w2v_train, tfidf_w2v_cv, tfidf_w2v_test])
tfidf = np.array([X_train_essay_tfidf, X_cv_essay_tfidf, X_test_essay_tfidf])
with open('tmp/vectorizer_tfidf', 'wb') as f:
pickle.dump(vectorizer_tfidf.get_feature_names(), f)
tfidf_w2v_all_features.extend(w2v_words)<jupyter_output><empty_output><jupyter_text>### Title<jupyter_code>vectorizer_tfidf_item = TfidfVectorizer(min_df=10, ngram_range=(1,4), max_features=5000)
X_train_title_tfidf = vectorizer_tfidf_item.fit_transform(X_train['project_title'].values.astype('U'))
X_cv_title_tfidf = vectorizer_tfidf_item.transform(X_cv['project_title'].values.astype('U'))
X_test_title_tfidf = vectorizer_tfidf_item.transform(X_test['project_title'].values.astype('U'))
print(X_train_title_tfidf.shape, y_train.shape)
print(X_cv_title_tfidf.shape, y_cv.shape)
print(X_test_title_tfidf.shape, y_test.shape)
print("="*100)
tfidf_all_features.extend(vectorizer_tfidf_item.get_feature_names())
X_train['project_title'].values.astype('U')
list_of_title_train=[]
for sentence in X_train['project_title'].values.astype('U'):
list_of_title_train.append(sentence.split())
w2v_model = Word2Vec(list_of_title_train, min_count=10, size=50, workers=4)
w2v_words = list(w2v_model.wv.vocab)
tf_idf_vect = TfidfVectorizer(min_df=10, ngram_range=(1,4), max_features=20000)
tf_idf_matrix = tf_idf_vect.fit_transform(X_train['project_title'].values.astype('U'))
tfidf_feat = tf_idf_vect.get_feature_names()
dictionary = dict(zip(tf_idf_vect.get_feature_names(), list(tf_idf_vect.idf_)))
tfidf_w2v_title_train = []
row = 0
for sentence in tqdm(list_of_title_train):
sentence_vec = np.zeros(50)
weight_sum = 0
for word in sentence:
if word in w2v_words and word in tfidf_feat:
vec = w2v_model.wv[word]
tf_idf = dictionary[word] * (sentence.count(word) / len(sentence))
sentence_vec += (vec * tf_idf)
weight_sum += tf_idf
if weight_sum != 0:
sentence_vec /= weight_sum
tfidf_w2v_title_train.append(sentence_vec)
row += 1
list_of_title_cv = []
for sentence in X_cv['project_title'].values.astype('U'):
list_of_title_cv.append(sentence.split())
tfidf_w2v_title_cv = []
row = 0
for sentence in tqdm(list_of_title_cv):
sentence_vec = np.zeros(50)
weight_sum = 0
for word in sentence:
if word in w2v_words and word in tfidf_feat:
vec = w2v_model.wv[word]
tf_idf = dictionary[word] * (sentence.count(word) / len(sentence))
sentence_vec += (vec * tf_idf)
weight_sum += tf_idf
if weight_sum != 0:
sentence_vec /= weight_sum
tfidf_w2v_title_cv.append(sentence_vec)
row += 1
list_of_title_test = []
for sentence in X_test['project_title'].values.astype('U'):
list_of_title_test.append(sentence.split())
tfidf_w2v_title_test = []
row = 0
for sentence in tqdm(list_of_title_test):
sentence_vec = np.zeros(50)
weight_sum = 0
for word in sentence:
if word in w2v_words and word in tfidf_feat:
vec = w2v_model.wv[word]
tf_idf = dictionary[word] * (sentence.count(word) / len(sentence))
sentence_vec += (vec * tf_idf)
weight_sum += tf_idf
if weight_sum != 0:
sentence_vec /= weight_sum
tfidf_w2v_title_test.append(sentence_vec)
row += 1
tfidf_w2v_all_features.extend(w2v_words)
title_tfidf_w2v = np.array([tfidf_w2v_title_train, tfidf_w2v_title_cv, tfidf_w2v_title_test])
title_tfidf = np.array([X_train_title_tfidf, X_cv_title_tfidf, X_test_title_tfidf])
# with open('tmp/vectorizer_tfidf_item', 'wb') as f:
# pickle.dump(vectorizer_tfidf_item.get_feature_names(), f)<jupyter_output><empty_output><jupyter_text>### Encoding categorical variable school state<jupyter_code>vectorizer = CountVectorizer()
vectorizer.fit(X_train['school_state'].values)
X_train_state_ohe = vectorizer.transform(X_train['school_state'].values)
X_cv_state_ohe = vectorizer.transform(X_cv['school_state'].values)
X_test_state_ohe = vectorizer.transform(X_test['school_state'].values)
print(X_train_state_ohe.shape, y_train.shape)
print(X_cv_state_ohe.shape, y_cv.shape)
print(X_test_state_ohe.shape, y_test.shape)
print(vectorizer.get_feature_names())
print("="*100)
tfidf_all_features.extend(vectorizer.get_feature_names())
tfidf_w2v_all_features.extend(vectorizer.get_feature_names())<jupyter_output><empty_output><jupyter_text>### encoding categorical features: teacher_prefix<jupyter_code>vectorizer = CountVectorizer()
vectorizer.fit(X_train['teacher_prefix'].values)
X_train_teacher_ohe = vectorizer.transform(X_train['teacher_prefix'].values)
X_cv_teacher_ohe = vectorizer.transform(X_cv['teacher_prefix'].values)
X_test_teacher_ohe = vectorizer.transform(X_test['teacher_prefix'].values)
print(X_train_teacher_ohe.shape, y_train.shape)
print(X_cv_teacher_ohe.shape, y_cv.shape)
print(X_test_teacher_ohe.shape, y_test.shape)
print(vectorizer.get_feature_names())
print("="*100)
tfidf_all_features.extend(vectorizer.get_feature_names())
tfidf_w2v_all_features.extend(vectorizer.get_feature_names())
vectorizer = CountVectorizer()
vectorizer.fit(X_train['project_grade_category'].values)
X_train_project_ohe = vectorizer.transform(X_train['project_grade_category'].values)
X_cv_project_ohe = vectorizer.transform(X_cv['project_grade_category'].values)
X_test_project_ohe = vectorizer.transform(X_test['project_grade_category'].values)
print(X_train_project_ohe.shape, y_train.shape)
print(X_cv_project_ohe.shape, y_cv.shape)
print(X_test_project_ohe.shape, y_test.shape)
print(vectorizer.get_feature_names())
print("="*100)
tfidf_all_features.extend(vectorizer.get_feature_names())
tfidf_w2v_all_features.extend(vectorizer.get_feature_names())<jupyter_output><empty_output><jupyter_text>### Normalize price<jupyter_code>normalizer = Normalizer()
normalizer.fit(X_train['price'].values.reshape(-1, 1))
X_train_price_norm = normalizer.transform(X_train['price'].values.reshape(-1, 1))
X_cv_price_norm = normalizer.transform(X_cv['price'].values.reshape(-1, 1))
X_test_price_norm = normalizer.transform(X_test['price'].values.reshape(-1, 1))
print(X_train_price_norm.shape, y_train.shape)
print(X_cv_price_norm.shape, y_cv.shape)
print(X_test_price_norm.shape, y_test.shape)
print("="*100)
tfidf_all_features.extend(vectorizer.get_feature_names())
tfidf_w2v_all_features.extend(vectorizer.get_feature_names())
normalizer = Normalizer()
normalizer.fit(X_train['teacher_number_of_previously_posted_projects'].values.reshape(1, -1))
X_train_prev_proj_norm = normalizer.transform(X_train['teacher_number_of_previously_posted_projects'].values.reshape(-1, 1))
X_cv_prev_proj_norm = normalizer.transform(X_cv['teacher_number_of_previously_posted_projects'].values.reshape(-1, 1))
X_test_prev_proj_norm = normalizer.transform(X_test['teacher_number_of_previously_posted_projects'].values.reshape(-1, 1))
print(X_train_prev_proj_norm.shape, y_train.shape)
print(X_cv_prev_proj_norm.shape, y_cv.shape)
print(X_test_prev_proj_norm.shape, y_test.shape)
print("="*100)
tfidf_all_features.extend(vectorizer.get_feature_names())
tfidf_w2v_all_features.extend(vectorizer.get_feature_names())<jupyter_output><empty_output><jupyter_text>## Preparing data for TFIDF<jupyter_code>X_train_tfidf = hstack((X_train_essay_tfidf, X_train_title_tfidf, X_train_state_ohe, X_train_teacher_ohe, X_train_project_ohe, X_train_price_norm, X_train_prev_proj_norm)).tocsr()
X_cv_tfidf = hstack((X_cv_essay_tfidf, X_cv_title_tfidf, X_cv_state_ohe, X_cv_teacher_ohe, X_cv_project_ohe, X_cv_price_norm, X_cv_prev_proj_norm)).tocsr()
X_test_tfidf = hstack((X_test_essay_tfidf, X_test_title_tfidf, X_test_state_ohe, X_test_teacher_ohe, X_test_project_ohe, X_test_price_norm, X_test_prev_proj_norm)).tocsr()
print("Final Data matrix")
print(X_train_tfidf.shape, y_train.shape)
print(X_cv_tfidf.shape, y_cv.shape)
print(X_test_tfidf.shape, y_test.shape)
print("="*100)<jupyter_output><empty_output><jupyter_text>## Preparing data for TFIDF-W2V<jupyter_code>X_train_tfidf_w2v = hstack((tfidf_w2v_train, tfidf_w2v_title_train, X_train_state_ohe, X_train_teacher_ohe, X_train_project_ohe, X_train_price_norm, X_train_prev_proj_norm)).tocsr()
X_cv_tfidf_w2v = hstack((tfidf_w2v_cv, tfidf_w2v_title_cv, X_cv_state_ohe, X_cv_teacher_ohe, X_cv_project_ohe, X_cv_price_norm, X_cv_prev_proj_norm)).tocsr()
X_test_tfidf_w2v = hstack((tfidf_w2v_test, tfidf_w2v_title_test, X_test_state_ohe, X_test_teacher_ohe, X_test_project_ohe, X_test_price_norm, X_test_prev_proj_norm)).tocsr()
print("Final Data matrix")
print(X_train_tfidf_w2v.shape, y_train.shape)
print(X_cv_tfidf_w2v.shape, y_cv.shape)
print(X_test_tfidf_w2v.shape, y_test.shape)
print("="*100)<jupyter_output><empty_output><jupyter_text>## Applying Decision tree on processed Data<jupyter_code>def cross_validate_model(X_train, Y_train):
parameters = { 'max_depth': [1, 5, 10, 50], 'min_samples_split': [5, 10, 100, 500] }
model = DecisionTreeClassifier()
clf = GridSearchCV(model, parameters, cv=3, scoring=make_scorer(roc_auc_score), return_train_score=True, verbose=1, n_jobs=4)
clf.fit(X_train, y_train)
train_auc_mean = clf.cv_results_['mean_train_score']
train_auc_std = clf.cv_results_['std_train_score']
cv_auc_mean = clf.cv_results_['mean_test_score']
cv_auc_std = clf.cv_results_['std_test_score']
optimal_depth = clf.best_params_['max_depth']
optimal_split = clf.best_params_['min_samples_split']
return parameters['max_depth'], parameters['min_samples_split'], optimal_depth, optimal_split, train_auc_mean, train_auc_std, cv_auc_mean, cv_auc_std
def plot_cross_validate(depth, split, train_auc_mean, train_auc_std, cv_auc_mean, cv_auc_std):
# this code is refered from: https://stackoverflow.com/a/48803361/4084039
plt.figure(figsize=(20, 15))
# pdb.set_trace();
trace1 = go.Scatter3d(x=split,y=depth,z=train_auc_mean, name = 'train')
trace2 = go.Scatter3d(x=split,y=depth,z=cv_auc_mean, name = 'Cross validation')
data = [trace1, trace2]
layout = go.Layout(scene = dict(
xaxis = dict(title='n_estimators'),
yaxis = dict(title='max_depth'),
zaxis = dict(title='AUC'),))
fig = go.Figure(data=data, layout=layout)
offline.iplot(fig, filename='3d-scatter-colorscale')
def plot_cross_validate_hm(depth, split, train_auc_mean, train_auc_std, cv_auc_mean, cv_auc_std):
n = len(depth)
m = len(split)
df_train = pd.DataFrame(data=train_auc_mean.reshape(n, m), index=depth, columns=split)
df_test = pd.DataFrame(data=cv_auc_mean.reshape(n, m), index=depth, columns=split)
fig, ax = plt.subplots(1, 2, figsize=(20, 12))
sns.heatmap(ttt, annot = True, fmt='.4g', ax=ax[0])
sns.heatmap(ttt, annot = True, fmt='.4g', ax=ax[1])
def model_auc_roc_score(X_train, y_train, X_test, y_test, optimal_depth, optimal_split):
model = DecisionTreeClassifier(max_depth=optimal_depth, min_samples_split=optimal_split)
model.fit(X_train, y_train)
y_train_proba = model.predict_proba(X_train)[:,1]
y_test_proba = model.predict_proba(X_test)[:, 1]
train_fpr, train_tpr, train_thresholds = roc_curve(y_train, y_train_proba)
test_fpr, test_tpr, test_thresholds = roc_curve(y_test, y_test_proba)
return model, y_train_proba, train_fpr, train_tpr, train_thresholds, y_test_proba, test_fpr, test_tpr, test_thresholds
def plot_auc_roc_score(train_fpr, train_tpr, test_fpr, test_tpr):
plt.figure(figsize=(15, 12))
plt.plot(train_fpr, train_tpr, label=f"Train AUC = {auc(train_fpr, train_tpr)}")
plt.plot(test_fpr, test_tpr, label=f"Test AUC = {auc(test_fpr, test_tpr)}")
plt.plot([0, 1], [0, 1], 'g--')
plt.legend()
plt.xlabel("False Positive Rate(FPR)")
plt.ylabel("True Positive Rate(TPR)")
plt.title("AUC")
plt.grid(color='black', linestyle='-', linewidth=0.5)
plt.show()
def predict(proba, thresholds, tpr, fpr):
pred = []
best_thres = thresholds[np.argmax(tpr * (1 - fpr))]
for prob in proba:
if prob >= best_thres:
pred.append(1)
else:
pred.append(0)
return best_thres, pred
def plot_confusion_matrix(y_test, y_pred):
cm = confusion_matrix(y_test, y_pred)
df = pd.DataFrame(cm)
plt.figure(figsize=(10, 7))
sns.heatmap(df, annot=True, fmt='d')
plt.title("Confusion Matrix for test data")
plt.xlabel("Predicted Label")
plt.ylabel("True Label")
plt.show()
def get_top_and_worst_features(model, all_features, num_of_features=20):
pos_indx = model.feature_importances_.argsort()[::-1][:num_of_features]
neg_indx = model.feature_importances_.argsort()[:num_of_features]
top_features = []
worst_features = []
for i in list(pos_indx[:num_of_features]):
top_features.append(all_features[i])
for i in list(neg_indx[:num_of_features]):
worst_features.append(all_features[i])
return top_features, worst_features
def build_word_cloud(features):
features = ' '.join(features)
wordcloud = WordCloud(width = 800, height = 800, background_color ='white').generate(features)
plt.figure(figsize = (6, 6), facecolor = None)
plt.imshow(wordcloud, interpolation='bilinear')
plt.axis("off")
plt.show()
def get_false_positives(y_pred, y_test, feature=[]):
fp_index = []
for i in range(len(y_test)) :
if (y_test[i] == 0) & (y_pred[i] == 1) :
fp_index.append(i)
fp_names = []
for i in fp_index :
fp_names.append(X_test['essay'].values[i])
return fp_names, fp_index
def build_false_positive_price(fp_index):
pdb.set_trace();
cols = X_test.columns
X_test_falsePos1 = pd.DataFrame(columns=cols)
for i in fp_index:
X_test_falsePos1 = X_test_falsePos1.append(X_test.filter(items=[i], axis=0))
sns.boxplot(y='price', data=X_test_falsePos1)
return X_test_falsePos1
def false_positive_pdf(X_test_falsePos):
plt.figure(figsize=(8,5))
counts, bin_edges = np.histogram(X_test_falsePos['teacher_number_of_previously_posted_projects'],bins='auto', density=True)
pdf = counts/sum(counts)
cdf = np.cumsum(pdf)
pdfP, = plt.plot(bin_edges[1:], pdf)
cdfP, = plt.plot(bin_edges[1:], cdf)
plt.legend([pdfP, cdfP], ["PDF", "CDF"])
plt.xlabel('teacher_number_of_previously_posted_projects')
plt.show()
def build_model(X_train, y_train, X_test, y_test, all_features, num_of_features=20):
depth, split, optimal_depth, optimal_split, train_auc_mean, train_auc_std, cv_auc_mean, cv_auc_std = cross_validate_model(X_train, y_train)
plot_cross_validate(depth, split, train_auc_mean, train_auc_std, cv_auc_mean, cv_auc_std)
model, y_train_proba, train_fpr, train_tpr, train_thresholds, y_test_proba, test_fpr, test_tpr, test_thresholds = model_auc_roc_score(X_train, y_train, X_test, y_test, optimal_depth, optimal_split)
plot_auc_roc_score(train_fpr, train_tpr, test_fpr, test_tpr)
thresh, y_pred = predict(y_test_proba, test_thresholds, test_tpr, test_fpr)
print(f'The best threshold value:: {thresh}')
plot_confusion_matrix(y_test, y_pred)
fp_names, fp_index = get_false_positives(y_pred, y_test)
X_test_falsePos = build_false_positive_price(fp_index)
false_positive_pdf(X_test_falsePos)
# top_features, worst_features = get_top_and_worst_features(model, all_features, num_of_features)
# build_word_cloud(top_features)
# build_word_cloud(worst_features)
return optimal_depth, optimal_split, train_fpr, train_tpr, test_fpr, test_tpr, y_pred
[1, 5], [1, 5]
depth, split
train_auc_mean
ttt.head()
depth, split, optimal_depth, optimal_split, train_auc_mean, train_auc_std, cv_auc_mean, cv_auc_std = cross_validate_model(X_train_tfidf, y_train)
plot_cross_validate(depth, split, train_auc_mean, train_auc_std, cv_auc_mean, cv_auc_std)
model, y_train_proba, train_fpr, train_tpr, train_thresholds, y_test_proba, test_fpr, test_tpr, test_thresholds = model_auc_roc_score(X_train_tfidf, y_train, X_test_tfidf, y_test, optimal_depth, optimal_split)
plot_auc_roc_score(train_fpr, train_tpr, test_fpr, test_tpr)
thresh, y_pred = predict(y_test_proba, test_thresholds, test_tpr, test_fpr)
plot_confusion_matrix(y_test, y_pred)
fp_names, fp_index = get_false_positives(y_pred, y_test)
build_word_cloud(fp_names)
X_test_falsePos = build_false_positive_price(fp_index)
false_positive_pdf(X_test_falsePos)
fp_names, fp_index = get_false_positives(y_pred, y_test)
X_test_falsePos = build_false_positive_price(fp_index)
false_positive_pdf(X_test_falsePos)
parameters = { 'max_depth': [1, 5], 'min_samples_split': [5, 10] }
model = DecisionTreeClassifier()
clf = GridSearchCV(model, parameters, cv=3, scoring=make_scorer(roc_auc_score), return_train_score=True, verbose=1, n_jobs=4)
clf.fit(X_train_tfidf, y_train)
max_scores = pd.DataFrame(clf.cv_results_).groupby(['param_min_samples_split', 'param_max_depth']).max().unstack()[['mean_test_score', 'mean_train_score']]
max_scores.head()
fig, ax = plt.subplots(1,2, figsize=(20,6))
sns.heatmap(max_scores.mean_train_score, annot = True, fmt='.4g', ax=ax[0])
sns.heatmap(max_scores.mean_test_score, annot = True, fmt='.4g', ax=ax[1])
ax[0].set_title('Train Set')
ax[1].set_title('CV Set')
plt.show()
from sklearn.externals.six import StringIO
from IPython.display import Image
from sklearn.tree import export_graphviz
import pydotplus
#Analysis on the False positives
fpi = []
for i in range(len(y_test)) :
if (y_test[i] == 0) & (y_pred_tfidf[i] == 1) :
fpi.append(i)
fp_essay1 = []
for i in fpi :
fp_essay1.append(X_test['essay'].values[i])
from wordcloud import WordCloud, STOPWORDS
comment_words = ' '
stopwords = set(STOPWORDS)
for val in fp_essay1 :
val = str(val)
tokens = val.split()
for i in range(len(tokens)):
tokens[i] = tokens[i].lower()
for words in tokens :
comment_words = comment_words + words + ' '
wordcloud = WordCloud(width = 800, height = 800, background_color ='white', stopwords = stopwords,min_font_size = 10).generate(comment_words)
plt.figure(figsize = (6, 6), facecolor = None)
plt.imshow(wordcloud)
plt.axis("off")
plt.tight_layout(pad = 0)
plt.show()
cols = X_test.columns
X_test_falsePos1 = pd.DataFrame(columns=cols)
# get the data of the false pisitives
for i in fpi : # (in fpi all the false positives data points indexes)
X_test_falsePos1 = X_test_falsePos1.append(X_test.filter(items=[i], axis=0))
sns.boxplot(y='price', data=X_test_falsePos1)
plt.figure(figsize=(8,5))
counts, bin_edges = np.histogram(X_test_falsePos1['teacher_number_of_previously_posted_projects'],bins='auto', density=True)
pdf = counts/sum(counts)
cdf = np.cumsum(pdf)
pdfP, = plt.plot(bin_edges[1:], pdf)
cdfP, = plt.plot(bin_edges[1:], cdf)
plt.legend([pdfP, cdfP], ["PDF", "CDF"])
plt.xlabel('teacher_number_of_previously_posted_projects')
plt.show()
from prettytable import PrettyTable
x = PrettyTable()
x.field_names = ["Vectorizer", "Model", "Hyperparameter: Max Depth", "Hyperparameter: Split", "Train AUC", "Test AUC"]
# auc_train_bow = auc(train_fpr_bow, train_tpr_bow)
# auc_test_bow = auc(test_fpr_bow, test_tpr_bow)
auc_train_tfidf = auc(train_fpr_tfidf, train_tpr_tfidf)
auc_test_tfidf = auc(test_fpr_tfidf, test_tpr_tfidf)
# x.add_row(["BOW", "Multinomial Naive Bayes", optimal_alpha_bow, round(auc_train_bow, 2),round(auc_test_bow, 2)])
x.add_row(["TF-IDF", "Multinomial Naive Bayes", optimal_depth_tfidf, optimal_split_tfidf, round(auc_train_tfidf, 2),round(auc_test_tfidf, 2)])
print(x)<jupyter_output>+------------+-------------------------+---------------------------+-----------------------+-----------+----------+
| Vectorizer | Model | Hyperparameter: Max Depth | Hyperparameter: Split | Train AUC | Test AUC |
+------------+-------------------------+---------------------------+-----------------------+-----------+----------+
| TF-IDF | Multinomial Naive Bayes | 5 | 10 | 0.61 | 0.61 |
+------------+-------------------------+---------------------------+-----------------------+-----------+----------+
|
no_license
|
/.ipynb_checkpoints/decision_tree-checkpoint.ipynb
|
lalitzz/Donor_Choose
| 11 |
<jupyter_start><jupyter_text># Spark read and write on remote Object Stores
By loading the right libraries, Spark is able to both read and write to external Object Stores in a distributed way.
SQL Server Server Big Data Clusters ships libraries to access S3 and ADLS Gen2 protocols.
Libraries are updated with each cumulative update, please make sure to list the available libraries. To list S3 protocol libraries use the following:
```
kubectl -n exec sparkhead-0 -- bash -c "ls /opt/hadoop/share/hadoop/tools/lib/*aws*"
```
If your scenario requires a library either unavailable or version incompatible with what is shipped with Big Data Clusters, you have some options:
1. Use a session based configure cell with dynamic library loading on Notebooks or Jobs.
2. Copy the additional libraries to a known HDFS on BDC and reference that at session configuration.
These two scenarios are described in detail in the [Manage libraries](https://docs.microsoft.com/en-us/sql/big-data-cluster/spark-install-packages?view=sql-server-ver15) and [Submit Spark jobs by using command-line tools](https://docs.microsoft.com/en-us/sql/big-data-cluster/spark-submit-job-command-line?view=sql-server-ver15) articles.
## Step 1 - Configure access to the remote storage
In this example we will access a remote S3 protocol object store.
The example considers a [MinIO](https://min.io/) object store service, but would would work with other S3 protocol providers.
Please check your S3 object store provider documentation to understand which libraries are required.
With that information at hand, configure your notebook session or job to use the right library like the example bellow.<jupyter_code>%%configure -f \
{
"conf": {
"spark.driver.extraClassPath": "/opt/hadoop/share/hadoop/tools/lib/aws-java-sdk-bundle-1.11.271.jar:/opt/hadoop/share/hadoop/tools/lib/hadoop-aws-3.1.168513.jar",
"spark.executor.extraClassPath": "/opt/hadoop/share/hadoop/tools/lib/aws-java-sdk-bundle-1.11.271.jar:/opt/hadoop/share/hadoop/tools/lib/hadoop-aws-3.1.168513.jar",
"spark.hadoop.fs.s3a.buffer.dir": "/var/opt/yarnuser"
}
}
spark<jupyter_output><empty_output><jupyter_text>## Step 2 - Add in access tokens to access the remote storage dynamically
Follow your S3 provider security documentation to change the following cells to correctly configure Spark to connect to the endpoint.<jupyter_code>access_key="YOUR_ACCESS_KEY"
secret="YOUR_SECRET"
spark._jsc.hadoopConfiguration().set("fs.s3a.aws.credentials.provider", "org.apache.hadoop.fs.s3a.SimpleAWSCredentialsProvider")
spark._jsc.hadoopConfiguration().set("fs.s3a.access.key", access_key)
spark._jsc.hadoopConfiguration().set("fs.s3a.secret.key", secret)
spark._jsc.hadoopConfiguration().set("fs.s3a.endpoint", "YOUR_ENDPOINT")
spark._jsc.hadoopConfiguration().set("spark.hadoop.fs.s3a.buffer.dir", "/var/opt/yarnuser") # Temp dir for writes back to S3
spark._jsc.hadoopConfiguration().set("fs.s3a.connection.ssl.enabled", "false")<jupyter_output><empty_output><jupyter_text>## Spark read and write patterns
Use the following examples to cover a range of read and write scenarios to remote object stores.
### Read from external S3 and write to BDC HDFS as table<jupyter_code>df = spark.read.csv("s3a://NYC-Cab/fhv_tripdata_2015-01.csv", header=True)
df.count()
df.write.format("parquet").save("/securelake/fhv_tripdata_2015-01")
%%sql
DROP TABLE tripdata
%%sql
CREATE TABLE tripdata
USING parquet
LOCATION '/securelake/fhv_tripdata_2015-01'
%%sql
select count(*) from tripdata
%%sql
select * from tripdata limit 10<jupyter_output><empty_output><jupyter_text>### Write back to S3 as parquet<jupyter_code>df.write.format("parquet").save("s3a://NYC-Cab/fhv_tripdata_2015-01-3")<jupyter_output><empty_output><jupyter_text>### Create external table on S3
This example virtualizes a folder on external object store as a Hive table.<jupyter_code>%%sql
DROP TABLE tripdata_s3
%%sql
CREATE TABLE tripdata_s3
USING parquet
LOCATION 's3a://NYC-Cab/fhv_tripdata_2015-01-3'
%%sql
select count(*) from tripdata_s3
%%sql
select * from tripdata_s3 limit 10<jupyter_output><empty_output>
|
permissive
|
/samples/features/sql-big-data-cluster/spark/data-virtualization/spark_external_object_store.ipynb
|
microsoft/sql-server-samples
| 5 |
<jupyter_start><jupyter_text># Visualizing Earnings Based on College MajorsThis dataset is from [American Community Survey]() about the job outcomes of students who graduated from college between 2010 and 2012. There's a cleaned version from [FiveThirtyEight]() which were released on their Github repo.
Some of the column names are not that intuitive nor self-explanatory, so I'll jot them here for easy reference:
* Rank - Rank by median earnings (the dataset is ordered by this column).
* Major_code - Major code.
* Major - Major description.
* Major_category - Category of major.
* Total - Total number of people with major.
* Sample_size - Sample size (unweighted) of full-time.
* Men - Male graduates.
* Women - Female graduates.
* ShareWomen - Women as share of total.
* Employed - Number employed.
* Median - Median salary of full-time, year-round workers.
* Low_wage_jobs - Number in low-wage service jobs.
* Full_time - Number employed 35 hours or more.
* Part_time - Number employed less than 35 hours.
<jupyter_code>import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
recent_grads = pd.read_csv("recent-grads.csv")
recent_grads.iloc[0]
recent_grads.head()
recent_grads.tail()
recent_grads.describe()
raw_data_count = recent_grads.shape[0]
raw_data_count
recent_grads.dropna(inplace = True)
cleaned_data_count = recent_grads.shape[0]
cleaned_data_count<jupyter_output><empty_output><jupyter_text>## Pandas, Scatter Plots<jupyter_code>recent_grads.plot(x = 'Sample_size', y = 'Median' , kind = 'scatter')
recent_grads.plot(x = 'Sample_size', y = 'Unemployment_rate' , kind = 'scatter')
recent_grads.plot(x = 'Full_time', y = 'Median' , kind = 'scatter')
recent_grads.plot(x = 'ShareWomen', y = 'Unemployment_rate' , kind = 'scatter')
recent_grads.plot(x = 'Men', y = 'Median' , kind = 'scatter')
recent_grads.plot(x = 'Women', y = 'Median' , kind = 'scatter')<jupyter_output><empty_output><jupyter_text>Using the plots above to explore the following questions:
1. Do students in more popular majors make more money?
2. Do students that majored in subjects that were majority female make more money?
3. Is there any link between the number of full-time employees and median salary?## Observation:
1. Do students in more popular majors make more money?
We don’t have a “popularity” column. I suggest that, as an approximation for this, you use the column Total. Plot this against Median (the median salary of full-time, year-round workers.)<jupyter_code>recent_grads.plot(x='Total', y='Median', kind='scatter')<jupyter_output><empty_output><jupyter_text>It can be inferred from the plot that as 'Total' increases, 'Median' decreases. If one finds it hard to analyze the results from this plot, one could use `.set_xlim()` to zoom in on this plot. Lets set the x limit from 0 to 100000 as most of the values reside within this range.<jupyter_code>ax = recent_grads.plot(x='Total', y='Median', kind='scatter')
ax.set_xlim(0,100000)
ax.set_title("Popularity VS Median")<jupyter_output><empty_output><jupyter_text>2. Do students that majored in subjects that were majority female make more money?
TO know about thus relationship, we will make a scatter plot of the variables `ShareWomen` and `Median`.<jupyter_code>recent_grads.plot(x = "ShareWomen", y = "Median", kind = 'scatter')<jupyter_output><empty_output><jupyter_text>It is quite evident from the above plot that as the share of women in a major increases, the median income shows a declining trend3. Is there any link between the number of full-time employees and median salary?<jupyter_code>recent_grads.plot(x = "Full_time", y = "Median", kind = 'scatter')<jupyter_output><empty_output><jupyter_text>there’s no significant relation between the number of full time employees and the median salary.When Median grows, noting really happens to the number of full time employees## Pandas, HistogramUsing histogram to explore the distributions of the following columns:
1. Sample_size
2. Median
3. Employed
4. Full_time
5. ShareWomen
6. Unemploment_rate
7. Men
8. Women
Using the plots below, lets explore the following questions:
* What percent of majors are predominatly male? Predominatly female?
* what's the most common median salary range?<jupyter_code>recent_grads['Sample_size'].hist()
recent_grads['Median'].hist(range=(0,100000)).set_xlabel("Median")<jupyter_output><empty_output><jupyter_text>Obseravtion: using a bin of 10
Common Median Salary Range is 30000-40000<jupyter_code>recent_grads['Employed'].hist()
recent_grads['Full_time'].plot(kind='hist')
recent_grads['ShareWomen'].hist(bins= 2,range = (0,1))<jupyter_output><empty_output><jupyter_text>Observations:
I can infer from the histogram that more than half of the majors in the dataset are predominately female.<jupyter_code>recent_grads['Unemployment_rate'].plot(kind='hist')
recent_grads['Men'].plot(kind='hist')
recent_grads['Women'].plot(kind='hist')
plt.show()<jupyter_output><empty_output><jupyter_text>## Scatter Matrix Plot<jupyter_code>from pandas.plotting import scatter_matrix
scatter_matrix(recent_grads[['Sample_size','Median']], figsize = (10,10))
plt.show()
scatter_matrix(recent_grads[['Sample_size','Median','Unemployment_rate']], figsize = (10,10))
plt.show()<jupyter_output><empty_output><jupyter_text>## Panas, Bar Plots<jupyter_code>recent_grads[:10].plot.bar(x = 'Major', y = 'ShareWomen')
recent_grads[-10:].plot.bar(x = 'Major', y = 'ShareWomen')
recent_grads[:10].plot.bar(x = 'Major', y = 'Unemployment_rate')
recent_grads[-10:].plot.bar(x = 'Major', y = 'Unemployment_rate')<jupyter_output><empty_output><jupyter_text>## Using boxplot<jupyter_code>recent_grads["Median"].plot(kind = 'box').set_ylim(recent_grads['Median'].min(),80000)
plt.show()<jupyter_output><empty_output>
|
no_license
|
/Visualizing Earnings Based On College Majors.ipynb
|
doyinsolamiolaoye/DataQuest_Projects
| 12 |
<jupyter_start><jupyter_text><jupyter_code>from google.colab import files
uploaded = files.upload()
import pandas as pd
import io
data = pd.read_csv(io.BytesIO(uploaded['amazon_vfl_reviews.csv']))
data.head()
x=data['review']
y=data['rating']
x, y = data.review.fillna(' '), data.rating
data.isnull().sum()
data=data.dropna()
data=data.reset_index(drop=True)
data.isnull().sum()
import string
punct=string.punctuation
punct
from spacy.lang.en.stop_words import STOP_WORDS
stopwords=list(STOP_WORDS)
import spacy
nlp=spacy.load('en_core_web_sm')
def text_data_cleaning(sentence):
doc=nlp(sentence)
tokens=[]
for token in doc:
if token.lemma !="-PRON-":
temp=token.lemma_.lower().strip()
else:
temp=token.lower_
tokens.append(temp)
cleaned_tokens=[]
for token in tokens:
if token not in stopwords and token not in punct:
cleaned_tokens.append(token)
return cleaned_tokens
from sklearn.svm import LinearSVC
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.pipeline import Pipeline
tfidf=TfidfVectorizer(tokenizer=text_data_cleaning)
classifier=LinearSVC()
from sklearn.model_selection import train_test_split
x_train,x_test,y_train,y_test= train_test_split(x, y,test_size=0.2,random_state=0)
x_train.shape,x_test.shape
x_train.head()
clf=Pipeline([('tfidf',tfidf), ('clf',classifier)])
clf.fit(x_train,y_train)
from sklearn.metrics import accuracy_score,confusion_matrix,classification_report
y_pred=clf.predict(x_test)
confusion_matrix(y_pred,y_test)
accuracy_score(y_pred,y_test)
print(classification_report(y_pred,y_test))
clf.predict(["wonderfull product"])
import joblib
joblib.dump(clf,"Email_class")
!pip install streamlit
!pip install pyngrok==4.1.1 --quiet
from pyngrok import ngrok
%%writefile app.py
import streamlit as st
import joblib
model=joblib.load("Email_class")
st.title('review classifier')
ip=st.text_input('enter your review')
op=model.predict([ip])
if st.button('Predict'):
st.title(op[0])
!nohup streamlit run app.py &
url=ngrok.connect(port='8501')
url
!pip install pipreqs
!pipreqs /content<jupyter_output>INFO: Successfully saved requirements file in /content/requirements.txt
|
no_license
|
/MajorProject.ipynb
|
shrutee2000/Major-Project
| 1 |
<jupyter_start><jupyter_text># Setup Notebook for Exercises##### IMPORTANT: Only modify cells which have the following comment:
```python
# Modify this cell
```
##### Do not add any new cells when you submit the homework## Creating the Spark Context<jupyter_code>import findspark
findspark.init()
from pyspark import SparkContext
sc = SparkContext(master="local[4]")
<jupyter_output><empty_output><jupyter_text>## Importing necessary libraries<jupyter_code>import os
import sys
from pyspark.sql import SQLContext
from pyspark.sql.types import Row, StructField, StructType, StringType, IntegerType
from pyspark.sql.functions import *
import Tester.SparkSQL as SparkSQL
pickleFile="Tester/SparkSQL.pkl"<jupyter_output><empty_output><jupyter_text>## Creating the SQL Context<jupyter_code># Just like using Spark requires having a SparkContext, using SQL requires an SQLContext
sqlContext = SQLContext(sc)<jupyter_output><empty_output><jupyter_text>## Dataframes
Dataframes are a special type of RDDs. They are similar to, but not the same as, pandas dataframes. They are used to store two dimensional data, similar to the type of data stored in a spreadsheet. Each column in a dataframe can have a different type and each row contains a `record`.
Spark DataFrames are similar to `pandas` DataFrames. With the important difference that spark DataFrames are **distributed** data structures, based on RDDs.# Exercises## Exercise 1 -- Creating and transforming dataframes from JSON files
[JSON](http://www.json.org/) is a very popular readable file format for storing structured data.
Among it's many uses are **twitter**, `javascript` communication packets, and many others. In fact this notebook file (with the extension `.ipynb` is in json format. JSON can also be used to store tabular data and can be easily loaded into a dataframe.
In this exercise, you will do the following:
* Read the dataset from a json file and store it in a dataframe
* Filter the rows which has the column "make_is_common" equal to 1
* Group the rows by make_country column and compute the count for each country
* Return the list of countries which have count greater than n
###### Sample Input:
```python
example.json has following contents
{"make_id":"acura","make_display":"Acura","make_is_common":"0","make_country":"USA"}
{"make_id":"alpina","make_display":"Alpina","make_is_common":"1","make_country":"UK"}
{"make_id":"aston-martin","make_display":"Aston Martin","make_is_common":"1","make_country":"UK"}
country_list = get_country_list("example.json", 1, sqlContext)
```
###### Sample Output:
country_list = ['UK']
<jupyter_code>def get_country_list(json_filepath, n, sqlContext):
makes_df = sqlContext.read.json(json_filepath)
# check the schema of the json file, uncomment the next line to see the schema
#makes_df.printSchema()
# The scheme should look like the one below
#root
# |-- make_country: string (nullable = true)
# |-- make_display: string (nullable = true)
# |-- make_id: string (nullable = true)
# |-- make_is_common: string (nullable = true)
sqlContext.registerDataFrameAsTable(makes_df, "makes_table")
query = "SELECT make_country, COUNT(make_country) as count \
FROM makes_table WHERE make_is_common == '1'\
GROUP BY make_country HAVING count > " + str(n)
country_rdd = sqlContext.sql(query).rdd.map(lambda row: row.make_country)
country_list = country_rdd.collect()
return country_list
import Tester.SparkSQL as SparkSQL
SparkSQL.exercise_1(sqlContext, pickleFile, get_country_list)<jupyter_output>Correct Output: ['Germany', 'Italy', 'USA', 'UK', 'Japan']
Great Job!
<jupyter_text>## Exercise 2 -- Creating and transforming dataframes from Parquet files
[Parquet](http://parquet.apache.org/) is a columnar format that is supported by many other data processing systems. Spark SQL provides support for both reading and writing Parquet files that automatically preserves the schema of the original data.
In this exercise, you will do the following:
* Read the dataset from a parquet file and store it in a dataframe
* Write a SQL query to group the rows by make_country and compute the count for each make_country
* Sort the make_country based on the count in descending order
* Return the list of tuples (country, count) of top "n" make_country
###### Sample Input:
```python
example.parquet has contents similar to the following json data
{"make_id":"a","make_display":"A","make_is_common":"0","make_country":"USA"}
{"make_id":"b","make_display":"B","make_is_common":"1","make_country":"UK"}
{"make_id":"c","make_display":"C","make_is_common":"1","make_country":"UK"}
{"make_id":"d","make_display":"D","make_is_common":"1","make_country":"USA"}
{"make_id":"e","make_display":"E","make_is_common":"0","make_country":"Germany"}
{"make_id":"f","make_display":"F","make_is_common":"0","make_country":"UK"}
top_n_country_list = get_top_n_country_list("example.parquet", 2, sqlContext)
```
###### Sample Output:
top_n_country_list = [ ('UK', 3), ('USA', 2)]<jupyter_code># Modify this cell
def get_top_n_country_list(parquet_path, n, sqlContext):
makes_df = sqlContext.read.load(parquet_path)
# The scheme should look like the one below
#root
# |-- make_country: string (nullable = true)
# |-- make_display: string (nullable = true)
# |-- make_id: string (nullable = true)
# |-- make_is_common: string (nullable = true)
makes_df.registerTempTable("makes_table")
query = "SELECT make_country, COUNT(make_country) as make_count \
FROM makes_table GROUP BY make_country \
ORDER BY make_count DESC"
query_result_df = sqlContext.sql(query)
country_rdd = query_result_df.rdd.map(lambda row: (row.make_country, row.make_count))
top_n_country_list = country_rdd.collect()[:n]
return top_n_country_list
import Tester.SparkSQL as SparkSQL
SparkSQL.exercise_2(sqlContext, pickleFile, get_top_n_country_list)<jupyter_output>Correct Output: [('USA', 16), ('Japan', 10), ('UK', 8)]
Great Job!
<jupyter_text>## Exercise 3 -- Creating and transforming dataframes from CSV files
In this exercise, you will do the following:
* Read the dataset from a csv file and store it in a dataframe
* Filter the rows which has the word "city" in the first column of csv file - "name" and return the count
###### Sample Input:
```python
example.csv has contents similar to the following csv data
name, country, subcountry, geonameid
logan city,australia,queensland,7281838
carindale,australia,queensland,7281839
city_count = get_city_count("example.csv", sqlContext)
```
###### Sample Output:
city_count = 1<jupyter_code>def get_city_count(csv_filepath, sqlContext):
city_df = sqlContext.read.csv(csv_filepath)
count = city_df.filter("_c0 like \'%city%\'").count()
return count
import Tester.SparkSQL as SparkSQL
SparkSQL.exercise_3(sqlContext, pickleFile, get_city_count)<jupyter_output>Correct Output: 15
Great Job!
|
permissive
|
/tutorials/spark-da-cse255/006_Spark_SQL.ipynb
|
superkley/spark-playground
| 6 |
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.