script
stringlengths 113
767k
|
|---|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
epl_df = pd.read_csv("../input/english-premier-league202021/EPL_20_21.csv")
epl_df.head()
epl_df.info()
epl_df.describe()
epl_df.isna().sum()
epl_df[" MinsPerMatch"] = (epl_df["Mins"] / epl_df["Matches"]).astype(int)
epl_df[" GoalsPerMatch"] = (epl_df["Goals"] / epl_df["Matches"]).astype(float)
print(epl_df.head())
##total Goal
Total_Goals = epl_df["Goals"].sum()
print(Total_Goals)
Total_PenaltyGoals = epl_df["Penalty_Goals"].sum()
print(Total_PenaltyGoals)
Total_PenaltyAttempts = epl_df["Penalty_Attempted"].sum()
print(Total_PenaltyAttempts)
import os, sys
plt.figure(figsize=(13, 6))
pl_not_scored = epl_df["Penalty_Attempted"].sum() - Total_PenaltyGoals
data = [pl_not_scored, Total_PenaltyGoals]
labels = ["Penalty missed", "Penalties Scored"]
color = sns.color_palette("Set2")
plt.pie(data, labels=labels, colors=color, autopct="%.0f%%")
plt.show()
# unique position
epl_df["Position"].unique()
## total fw
epl_df[epl_df["Position"] == "FW"]
##plyer nation
np.size((epl_df["Nationality"].unique()))
##most player come
nationality = epl_df.groupby("Nationality").size().sort_values(ascending=False)
nationality.head(10).plot(kind="bar", figsize=(12, 6), color=sns.color_palette("magma"))
# max player in sqad
epl_df["Club"].value_counts().nlargest(5).plot(
kind="bar", color=sns.color_palette("viridis")
)
# player
epl_df["Club"].value_counts().nsmallest(5).plot(
kind="bar", color=sns.color_palette("viridis")
)
under20 = epl_df[epl_df["Age"] <= 20]
under20_25 = epl_df[(epl_df["Age"] > 20) & (epl_df["Age"] < 25)]
under25_30 = epl_df[(epl_df["Age"] > 25) & (epl_df["Age"] < 30)]
Above30 = epl_df[epl_df["Age"] > 30]
x = np.array(
[
under20["Name"].count(),
under20_25["Name"].count(),
under25_30["Name"].count(),
Above30["Name"].count(),
]
)
mylabels = ["<=20", ">20 & <=25", ">25 & <=30", ">30"]
plt.title("Total Player With Age", fontsize=20)
plt.pie(x, labels=mylabels, autopct="%.1f%%")
plt.show()
##total under20
players_under20 = epl_df[epl_df["Age"] < 20]
players_under20["Club"].value_counts().plot(
kind="bar", color=sns.color_palette("cubehelix")
)
# under 20 manu
players_under20[players_under20["Club"] == "Manchester United"]
# under 20 chelsi
players_under20[players_under20["Club"] == "Chelsea"]
## avarage age
plt.figure(figsize=(12, 6))
sns.boxenplot(x="Club", y="Age", data=epl_df)
plt.xticks(rotation=90)
num_player = epl_df.groupby("Club").size()
data = (epl_df.groupby("Club")["Age"].sum()) / num_player
data.sort_values(ascending=False)
## total assist
|
# # **Point Couds with zarr**
# Generate Point Clouds while leveraging efficient image loading with zarr. Since we will be able to use all surfaces let's see if we can denoise the point cloud.
# Credit: https://www.kaggle.com/code/brettolsen/efficient-image-loading-with-zarr/notebookimport os
import os
import shutil
from tifffile import tifffile
import time
import numpy as np
import PIL.Image as Image
import matplotlib.pyplot as plt
import matplotlib.patches as patches
from tqdm import tqdm
from ipywidgets import interact, fixed
from IPython.display import HTML, display
import zarr
import open3d as o3
INPUT_FOLDER = "/kaggle/input/vesuvius-challenge-ink-detection"
WORKING_FOLDER = "/kaggle/working/"
TEMP_FOLDER = "kaggle/temp/"
class TimerError(Exception):
pass
class Timer:
def __init__(self, text=None):
if text is not None:
self.text = text + ": {:0.4f} seconds"
else:
self.text = "Elapsed time: {:0.4f} seconds"
def logfunc(x):
print(x)
self.logger = logfunc
self._start_time = None
def start(self):
if self._start_time is not None:
raise TimerError("Timer is already running. Use .stop() to stop it.")
self._start_time = time.time()
def stop(self):
if self._start_time is None:
raise TimerError("Timer is not running. Use .start() to start it.")
elapsed_time = time.time() - self._start_time
self._start_time = None
if self.logger is not None:
self.logger(self.text.format(elapsed_time))
return elapsed_time
def __enter__(self):
self.start()
return self
def __exit__(self, exc_type, exc_value, exc_traceback):
self.stop()
class FragmentImageException(Exception):
pass
class FragmentImageData:
"""A general class that uses persistent zarr objects to store the surface volume data,
binary data mask, and for training sets, the truth data and infrared image of a papyrus
fragment, in a compressed and efficient way.
"""
def __init__(self, sample_type: str, sample_index: str, working: bool = True):
if sample_type not in ("test, train"):
raise FragmentImageException(
f"Invalid sample type f{sample_type}, must be one of 'test' or 'train'"
)
zarrpath = self._zarr_path(sample_type, sample_index, working)
if os.path.exists(zarrpath):
self.zarr = self.load_from_zarr(zarrpath)
else:
dirpath = os.path.join(INPUT_FOLDER, sample_type, sample_index)
if not os.path.exists(dirpath):
raise FragmentImageException(
f"No input data found at f{zarrpath} or f{dirpath}"
)
self.zarr = self.load_from_directory(dirpath, zarrpath)
@property
def surface_volume(self):
return self.zarr.surface_volume
@property
def mask(self):
return self.zarr.mask
@property
def truth(self):
return self.zarr.truth
@property
def infrared(self):
return self.zarr.infrared
@staticmethod
def _zarr_path(sample_type: str, sample_index: str, working: bool = True):
filename = f"{sample_type}-{sample_index}.zarr"
if working:
return os.path.join(WORKING_FOLDER, filename)
else:
return os.path.join(TEMP_FOLDER, filename)
@staticmethod
def clean_zarr(sample_type: str, sample_index: str, working: bool = True):
zarrpath = FragmentImageData._zarr_path(sample_type, sample_index, working)
if os.path.exists(zarrpath):
shutil.rmtree(zarrpath)
@staticmethod
def load_from_zarr(filepath):
with Timer("Loading from existing zarr"):
return zarr.open(filepath, mode="r")
@staticmethod
def load_from_directory(dirpath, zarrpath):
if os.path.exists(zarrpath):
raise FragmentImageException(
f"Trying to overwrite existing zarr at f{zarrpath}"
)
# Initialize the root zarr group and write the file
root = zarr.open_group(zarrpath, mode="w")
# Load in the surface volume tif files
with Timer("Surface volume loading"):
init = True
imgfiles = sorted(
[
imgfile
for imgfile in os.listdir(os.path.join(dirpath, "surface_volume"))
]
)
for imgfile in imgfiles:
print(f"Loading file {imgfile}", end="\r")
# img_data = np.array(
# Image.open(os.path.join(dirpath, "surface_volume", imgfile))
# )
img_data = tifffile.imread(
os.path.join(dirpath, "surface_volume", imgfile)
)
if init:
surface_volume = root.zeros(
name="surface_volume",
shape=(img_data.shape[0], img_data.shape[1], len(imgfiles)),
chunks=(1000, 1000, 4),
dtype=img_data.dtype,
write_empty_chunks=False,
)
init = False
z_index = int(imgfile.split(".")[0])
surface_volume[:, :, z_index] = img_data
# Load in the mask
with Timer("Mask loading"):
img_data = np.array(
Image.open(os.path.join(dirpath, "mask.png")), dtype=bool
)
mask = root.array(
name="mask",
data=img_data,
shape=img_data.shape,
chunks=(1000, 1000),
dtype=img_data.dtype,
write_empty_chunks=False,
)
# Load in the truth set (if it exists)
with Timer("Truth set loading"):
truthfile = os.path.join(dirpath, "inklabels.png")
if os.path.exists(truthfile):
img_data = np.array(Image.open(truthfile), dtype=bool)
truth = root.array(
name="truth",
data=img_data,
shape=img_data.shape,
chunks=(1000, 1000),
dtype=img_data.dtype,
write_empty_chunks=False,
)
# Load in the infrared image (if it exists)
with Timer("Infrared image loading"):
irfile = os.path.join(dirpath, "ir.png")
if os.path.exists(irfile):
img_data = np.array(Image.open(irfile))
infrared = root.array(
name="infrared",
data=img_data,
shape=img_data.shape,
chunks=(1000, 1000),
dtype=img_data.dtype,
write_empty_chunks=False,
)
return root
# # Load data
FragmentImageData.clean_zarr("train", 1)
data = FragmentImageData("train", "1")
print(data.surface_volume.info)
print(data.mask.info)
print(data.truth.info)
print(data.infrared.info)
with Timer():
plt.imshow(data.mask, cmap="gray")
with Timer():
plt.imshow(data.surface_volume[:, :, 20], cmap="gray")
# ### Plot vertical slices of the surface volumes
with Timer():
plt.figure(figsize=(10, 1))
plt.imshow(data.surface_volume[2000, :, :].T, cmap="gray", aspect="auto")
with Timer():
plt.figure(figsize=(10, 1))
plt.imshow(data.surface_volume[:, 2000, :].T, cmap="gray", aspect="auto")
# # Create Point Cloud
# ## Sample from Surface Volumes
ROWS = data.surface_volume.shape[0]
COLS = data.surface_volume.shape[1]
Z_DIM = data.surface_volume.shape[2] # number of volume slices
N_SAMPLES = 10000
with Timer():
# sample from valid regions of surface volume
c = np.ravel(data.mask).cumsum()
samples = np.random.uniform(low=0, high=c[-1], size=(N_SAMPLES, Z_DIM)).astype(int)
# get valid indexes
x, y = np.unravel_index(c.searchsorted(samples), data.mask.shape)
x, y = x[np.newaxis, ...], y[np.newaxis, ...]
# get z dimensions from surface volume locations
z = np.arange(0, Z_DIM)
z = np.tile(z, N_SAMPLES).reshape(N_SAMPLES, -1)[np.newaxis, ...]
# get point cloud
xyz = np.vstack((x, y, z))
xyz.shape
# ### Get Normalized Intensities
intensities = np.zeros((N_SAMPLES, Z_DIM))
with Timer():
for i in range(Z_DIM):
img = data.surface_volume[:, :, i]
intensities[:, i] = img[xyz[0, :, i], xyz[1, :, i]] / 65535.0
intensities = intensities.astype(np.float32)
# #### Sanity Check
print(xyz[:, 20, 1], intensities[20, 1])
print(
xyz.T.reshape((-1, 3))[20 + N_SAMPLES, :],
intensities.T.reshape((-1))[20 + N_SAMPLES],
)
# ### Reshape and Normalize
xyz = xyz.T.reshape((-1, 3))
xyz = xyz / xyz.max(axis=0)
intensities = intensities.T.reshape((-1)).repeat((3)).reshape((-1, 3))
# ## Get Colormap and Convert to Point Cloud
colors = plt.get_cmap("bone") # also use 'cool', 'bone'
colors
pcd = o3.geometry.PointCloud()
pcd.points = o3.utility.Vector3dVector(xyz)
pcd.colors = o3.utility.Vector3dVector(colors(intensities)[:, 0, :3])
pcd
# # Display Point Cloud
o3.visualization.draw_plotly([pcd])
# # Inspect distribution at each layer
# ### Animate intensity Histograms for each surface layer
# Animation code resused from: https://www.kaggle.com/code/leonidkulyk/eda-vc-id-volume-layers-animation
from celluloid import Camera
fig, ax = plt.subplots(1, 1)
camera = Camera(fig) # define the camera that gets the fig we'll plot
for i in range(Z_DIM):
cnts, bins, _ = plt.hist(
np.ravel(data.surface_volume[:, :, i][data.mask]) / 65535.0, bins=100
)
ax.set_title(f"Surfacer Layer: {i}")
ax.text(
0.5,
1.08,
f"Surfacer Layer: {i}",
fontweight="bold",
fontsize=18,
transform=ax.transAxes,
horizontalalignment="center",
)
camera.snap() # the camera takes a snapshot of the plot
plt.close(fig) # close figure
animation = camera.animate() # get plt animation
fix_video_adjust = (
"<style> video {margin: 0px; padding: 0px; width:100%; height:auto;} </style>"
)
display(HTML(fix_video_adjust + animation.to_html5_video())) # displaying the animation
# There seems to be a mix of modes, especially in the lower surface volume layers. There seems to be a single mode around 0.35 for all surface volumes, while the lower volumes contain a mode with a wider spread with a center that seems to move across each layer.
# The 0.35 centered mode is the dominate mode for the upper surface volumes which seem to contain less papyrus according to the volume slice cuts above. Also ccording to these histograms, the majority of the variance comes from the lower surface cuts. This can also be seen in the point cloud and the surface volume cuts.
# Could this dominate mode be noisy data? Let's take a closer look at with a new point cloud.
# # Denoise Point Cloud
# Hypothesis/Idea: The top layers do not contain useful data, they are just nosie.
# In the surface volume slices and point cloud the top layers look like they may not contain much useful data. The histograms may also support this hypothesis, since the dominate mode at 0.35 is the only mode at the top layers.
# Let's use the top surface volume to estimate to estimate the distribution of the noisy data. All we need is the mean and standard deviation.
mu = np.mean(np.ravel(data.surface_volume[:, :, -1][data.mask]) / 65535.0)
sig = np.std(np.ravel(data.surface_volume[:, :, -1][data.mask]) / 65535.0)
mu, sig
# Let's get upper and lower bounds of data to remove. We will just go up to 4 sigma for now to get the lower and upper bounds to remove.
# We should also be weary of just naively truncating data. We will need to investigate this further, but that will be for another time.
lower, upper = mu - 4 * sig, mu + 4 * sig
lower, upper
# get intensity mask
i_mask = (intensities[:, 0] > lower) & (intensities[:, 0] < upper)
# remove 0 intensities?
i_mask = ~i_mask & (intensities[:, 0] != 0)
pcd_2 = o3.geometry.PointCloud()
pcd_2.points = o3.utility.Vector3dVector(xyz[i_mask])
pcd_2.colors = o3.utility.Vector3dVector(colors(intensities[i_mask])[:, 0, :3])
pcd_2
o3.visualization.draw_plotly([pcd_2])
# Let's go ahead and plot plot the mean and standard dev for each surface layer.
# Code from: https://www.kaggle.com/code/brettolsen/fast-efficient-image-storing-and-manipulation
#
with Timer():
zindices = np.arange(Z_DIM)
means = np.zeros_like(zindices)
stdevs = np.zeros_like(zindices)
# mask = data.mask[:,:,0]
for z in zindices:
print(z, end="\r")
array = data.surface_volume[:, :, z][data.mask]
means[z] = array.mean()
stdevs[z] = np.std(array)
plt.figure()
plt.grid()
plt.errorbar(zindices, means, stdevs, marker="o")
plt.plot(zindices, stdevs)
|
# Source Code: https://github.com/rasbt/machine-learning-book with additional explanation
#
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from matplotlib.colors import ListedColormap
# The whole idea behind the perceptron model is to use a simplified approach to mimic how a single neuron in the brain works. The Perceptron Rule is simple, and can be summarized in the following steps:
# * Initialize the weights and bias unit to 0 or small random numbers
# * For each training example, x superscript i:
# * Compute output value y hat
# * Update the weights and bias unit
# Weights can be initialized to 0, however the learning rate would have no effect onn the decision boundary. If all w's are initialized to 0 , then the learning rate parameter affects only the scale oof the weight vector, ot the direction
class Perceptron:
"""
Perceptron Classifier
Parameters:
lr: float
learnning rate 0.0 >= lr <=1.0
n_iter: int
passes over the training dataset
random_state: int
Random number generator seed for random weights initialization
Attributes:
w_: 1d array - weights after fitting
b_: Scalar - bias unit after fitting
errors_: list - Number of missclassification (updates) in epoch
"""
def __init__(self, lr=0.01, n_iter=50, random_state=1):
self.lr = lr
self.n_iter = n_iter
self.random_state = random_state
def fit(self, X, y):
"""
Fit Training Data
Parameters:
X: {array-like}, shape=[n_examples, n_features]
y: {array-like}, shape=[n_examples], Target values
Returns:
self: object
"""
rgen = np.random.RandomState(self.random_state)
# generate random weights
self.w_ = rgen.normal(loc=0.0, scale=0.1, size=X.shape[1])
# self.w_ = np.zeros(X.shape[1])
self.b_ = np.float(0.0)
self.errors_ = []
for _ in range(self.n_iter):
errors = 0
for xi, target in zip(X, y):
update = self.lr * (target - self.predict(xi))
self.w_ += update * xi
self.b_ += update
errors += int(update != 0.0)
self.errors_.append(errors)
return self
def net_input(self, X):
"""Calculate net input"""
return np.dot(X, self.w_) + self.b_
def predict(self, X):
"""Return Class Label after each unit step"""
return np.where(self.net_input(X) > 0.0, 1, 0)
df = pd.read_csv("/kaggle/input/irisdataset/iris.data", header=None, encoding="utf-8")
df
# select setosa and versicolor
y = df.iloc[:100, 4].values
y = np.where(y == "Iris-setosa", 0, 1)
# extract sepal length and petal length
X = df.iloc[0:100, [0, 2]].values
# plot data
plt.scatter(X[:50, 0], X[:50, 1], color="red", marker="o", label="Setosa")
plt.scatter(X[50:100, 0], X[50:100, 1], color="blue", marker="s", label="Versicolor")
plt.xlabel("Sepal length [cm]")
plt.ylabel("Petal length [cm]")
plt.legend(loc="upper left")
plt.show()
# ### ***Full Code***
class Perceptron:
"""
Perceptron Classifier
Parameters:
lr: float
learnning rate 0.0 >= lr <=1.0
n_iter: int
passes over the training dataset
random_state: int
Random number generator seed for random weights initialization
Attributes:
w_: 1d array - weights after fitting
b_: Scalar - bias unit after fitting
errors_: list - Number of missclassification (updates) in epoch
"""
def __init__(self, lr=0.01, n_iter=50, random_state=1):
self.lr = lr
self.n_iter = n_iter
self.random_state = random_state
def fit(self, X, y):
"""
Fit Training Data
Parameters:
X: {array-like}, shape=[n_examples, n_features]
y: {array-like}, shape=[n_examples], Target values
Returns:
self: object
"""
rgen = np.random.RandomState(self.random_state)
# generate random weights
self.w_ = rgen.normal(loc=0.0, scale=0.1, size=X.shape[1])
# self.w_ = np.zeros(X.shape[1])
self.b_ = np.float(0.0)
self.errors_ = []
for _ in range(self.n_iter):
errors = 0
for xi, target in zip(X, y):
update = self.lr * (target - self.predict(xi))
self.w_ += update * xi
self.b_ += update
errors += int(update != 0.0)
self.errors_.append(errors)
return self
def net_input(self, X):
"""Calculate net input"""
return np.dot(X, self.w_) + self.b_
def predict(self, X):
"""Return Class Label after each unit step"""
return np.where(self.net_input(X) > 0.0, 1, 0)
ppn = Perceptron(lr=0.1, n_iter=10)
ppn.fit(X, y)
plt.plot(range(1, len(ppn.errors_) + 1), ppn.errors_, marker="o")
plt.xlabel("Epochs")
plt.ylabel("Number of updates")
plt.show()
def plot_decision_regions(X, y, classifier, resolution=0.02):
# setup marker generator and color map
markers = ("o", "s", "^", "v", "<")
colors = ("red", "blue", "lightgreen", "gray", "cyan")
cmap = ListedColormap(colors[: len(np.unique(y))])
# plot the decision surface
x1_min, x1_max = X[:, 0].min() - 1, X[:, 0].max() + 1
x2_min, x2_max = X[:, 1].min() - 1, X[:, 1].max() + 1
xx1, xx2 = np.meshgrid(
np.arange(x1_min, x1_max, resolution), np.arange(x2_min, x2_max, resolution)
)
lab = classifier.predict(np.array([xx1.ravel(), xx2.ravel()]).T)
lab = lab.reshape(xx1.shape)
plt.contourf(xx1, xx2, lab, alpha=0.3, cmap=cmap)
plt.xlim(xx1.min(), xx1.max())
plt.ylim(xx2.min(), xx2.max())
# plot class examples
for idx, cl in enumerate(np.unique(y)):
plt.scatter(
x=X[y == cl, 0],
y=X[y == cl, 1],
alpha=0.8,
c=colors[idx],
marker=markers[idx],
label=f"Class {cl}",
edgecolor="black",
)
plot_decision_regions(X, y, classifier=ppn)
plt.xlabel("Sepal length [cm]")
plt.ylabel("Petal length [cm]")
plt.legend(loc="upper left")
plt.show()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
train_df = pd.read_csv("/kaggle/input/playground-series-s3e12/train.csv")
train_df.head()
train_df.info()
train_df.describe()
# 1. There is no null value.
import plotly.express as px
import matplotlib
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_style("darkgrid")
matplotlib.rcParams["font.size"] = 14
matplotlib.rcParams["figure.figsize"] = (10, 6)
matplotlib.rcParams["figure.facecolor"] = "#00000000"
px.histogram(train_df, x="target")
px.histogram(train_df, x="gravity", marginal="box")
# 1. Lesser the gravity, lesser the chance of getting stones.
#
px.histogram(train_df, x="ph", marginal="box")
# Low ph cause kidney stones.
px.histogram(train_df, x="cond", marginal="box")
# Majorly conductivity range of 20mMho - 30mMho cause kidney stones.
px.histogram(train_df, x="calc", marginal="box")
# Larger the amount of calcium present in the urine, larger the chance of kidney stones.
train_df.corr()
sns.heatmap(train_df.corr(), annot=True)
# This heatmap proves our previous observation.
# #### Modeling
import xgboost as xgb
from sklearn import metrics
def modelfit(
alg, dtrain, predictors, useTrainCV=True, cv_folds=5, early_stopping_rounds=50
):
if useTrainCV:
xgb_param = alg.get_xgb_params()
xgtrain = xgb.DMatrix(dtrain[predictors].values, label=dtrain["target"].values)
cvresult = xgb.cv(
xgb_param,
xgtrain,
num_boost_round=alg.get_params()["n_estimators"],
nfold=cv_folds,
metrics="auc",
early_stopping_rounds=early_stopping_rounds,
verbose_eval=True,
)
alg.set_params(n_estimators=cvresult.shape[0])
# Fit the algorithm on the data
alg.fit(dtrain[predictors], dtrain["target"], eval_metric="auc")
# Predict training set:
dtrain_predictions = alg.predict(dtrain[predictors])
dtrain_predprob = alg.predict_proba(dtrain[predictors])[:, 1]
# Print model report:
print("\nModel Report")
print(
"Accuracy : %.4g"
% metrics.accuracy_score(dtrain["target"].values, dtrain_predictions)
)
print(
"AUC Score (Train): %f"
% metrics.roc_auc_score(dtrain["target"], dtrain_predprob)
)
predictors = [x for x in train_df.columns if x not in ["target", "id"]]
xgb1 = xgb.XGBClassifier(
learning_rate=0.1,
n_estimators=500,
max_depth=3,
min_child_weight=1,
gamma=0,
subsample=0.8,
colsample_bytree=0.8,
objective="binary:logistic",
seed=27,
)
modelfit(xgb1, train_df, predictors)
from sklearn.model_selection import GridSearchCV
param_test1 = {"max_depth": range(1, 10, 2), "min_child_weight": range(1, 6, 2)}
gsearch1 = GridSearchCV(
estimator=xgb.XGBClassifier(
learning_rate=0.1,
n_estimators=17,
max_depth=3,
min_child_weight=1,
objective="binary:logistic",
seed=27,
),
param_grid=param_test1,
scoring="roc_auc",
n_jobs=4,
cv=5,
)
gsearch1.fit(train_df[predictors], train_df["target"])
gsearch1.best_params_, gsearch1.best_score_
param_test3 = {"gamma": [i / 10.0 for i in range(0, 5)]}
gsearch3 = GridSearchCV(
estimator=xgb.XGBClassifier(
learning_rate=0.1,
n_estimators=17,
max_depth=1,
min_child_weight=1,
gamma=0,
objective="binary:logistic",
seed=27,
),
param_grid=param_test3,
scoring="roc_auc",
n_jobs=4,
cv=5,
)
gsearch3.fit(train_df[predictors], train_df["target"])
gsearch3.best_params_, gsearch3.best_score_
predictors = [x for x in train_df.columns if x not in ["target", "id"]]
xgb1 = xgb.XGBClassifier(
learning_rate=0.1,
n_estimators=500,
max_depth=3,
min_child_weight=1,
gamma=0,
subsample=0.8,
colsample_bytree=0.8,
objective="binary:logistic",
seed=27,
)
modelfit(xgb1, train_df, predictors)
param_test1 = {"max_depth": range(1, 10, 2), "min_child_weight": range(1, 6, 2)}
gsearch1 = GridSearchCV(
estimator=xgb.XGBClassifier(
learning_rate=0.01,
n_estimators=29,
max_depth=3,
min_child_weight=1,
objective="binary:logistic",
seed=27,
),
param_grid=param_test1,
scoring="roc_auc",
n_jobs=4,
cv=5,
)
gsearch1.fit(train_df[predictors], train_df["target"])
gsearch1.best_params_, gsearch1.best_score_
param_test3 = {"gamma": [i / 10.0 for i in range(0, 5)]}
gsearch3 = GridSearchCV(
estimator=xgb.XGBClassifier(
learning_rate=0.01,
n_estimators=29,
max_depth=3,
min_child_weight=1,
gamma=0,
objective="binary:logistic",
seed=27,
),
param_grid=param_test3,
scoring="roc_auc",
n_jobs=4,
cv=5,
)
gsearch3.fit(train_df[predictors], train_df["target"])
gsearch3.best_params_, gsearch3.best_score_
from sklearn.model_selection import train_test_split
X = train_df.drop(["target"], axis=1)
y = train_df["target"]
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.33, random_state=42
)
xgb_classifier1 = xgb.XGBClassifier(
learning_rate=0.1,
n_estimators=17,
max_depth=1,
min_child_weight=1,
)
xgb_classifier1.fit(X_train, y_train)
xgb_classifier1.score(X_train, y_train)
xgb_classifier1.score(X_test, y_test)
# #### Test Prediction
test_df = pd.read_csv("/kaggle/input/playground-series-s3e12/test.csv")
Y_pred = xgb_classifier1.predict_proba(test_df)[:, 1]
submission = pd.DataFrame({"id": test_df["id"], "target": Y_pred})
submission.to_csv("submission.csv", index=False)
|
#
# # 1. Import Libraries
# ([Go to top](#top))
from tensorflow.keras.datasets import imdb
from tensorflow.keras.preprocessing.text import text_to_word_sequence
from tensorflow.keras.preprocessing.text import Tokenizer
from tensorflow.keras.preprocessing.sequence import pad_sequences
from tensorflow.keras import models
from tensorflow.keras import layers
from tensorflow.keras import losses
from tensorflow.keras import metrics
from tensorflow.keras import optimizers
from tensorflow.keras.utils import plot_model
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.model_selection import train_test_split
from collections import Counter
from pathlib import Path
import os
import numpy as np
import re
import string
import nltk
nltk.download("punkt")
from nltk.tokenize import word_tokenize
from nltk.corpus import stopwords
nltk.download("stopwords")
from nltk.stem.porter import PorterStemmer
from nltk.stem import WordNetLemmatizer
nltk.download("wordnet")
from nltk.corpus import wordnet
import unicodedata
import html
stop_words = stopwords.words("english")
#
# # 2. Load Data
# ([Go to top](#top))
#
t_set = pd.read_csv("/kaggle/input/nlp-getting-started/train.csv")
t_set.shape
t_set.head()
# For training our deep learning model, we will use the 'text' column as our input feature or `Train_X`, and the 'target' column as our output or `y_train`. Any other columns that are not required will be dropped as we are only interested in the 'text' column.
Train_X_raw = t_set["text"].values
Train_X_raw.shape
Train_X_raw[5]
_ = list(map(print, Train_X_raw[:5] + "\n"))
y_train = t_set["target"].values
y_train.shape
y_train[:5]
#
# # 3. Data Splitting
# ([Go to top](#top))
# As we see there's a little bit class imbalance, so we'll use `stratify` parameter with `train_test_split`.
_ = sns.countplot(x=y_train)
from sklearn.model_selection import train_test_split
Train_X_raw, Validate_X_raw, y_train, y_validation = train_test_split(
Train_X_raw, y_train, test_size=0.2, stratify=y_train
)
print(Train_X_raw.shape)
print(y_train.shape)
print()
print(Validate_X_raw.shape)
print(y_validation.shape)
#
# # 4. Text Preprocessing
# ([Go to top](#top))
# In this phase, we apply some operations on the text, to make it in the most usable form for the task at hand. Mainly we clean it up to be more appealing to the problem we try to solve. The input is __text__ and the output is a transformed __text__.
def remove_special_chars(text):
recoup = re.compile(r" +")
x1 = (
text.lower()
.replace("#39;", "'")
.replace("amp;", "&")
.replace("#146;", "'")
.replace("nbsp;", " ")
.replace("#36;", "$")
.replace("\\n", "\n")
.replace("quot;", "'")
.replace("<br />", "\n")
.replace('\\"', '"')
.replace("<unk>", "u_n")
.replace(" @.@ ", ".")
.replace(" @-@ ", "-")
.replace("\\", " \\ ")
)
return recoup.sub(" ", html.unescape(x1))
def remove_non_ascii(text):
"""Remove non-ASCII characters from list of tokenized words"""
return (
unicodedata.normalize("NFKD", text)
.encode("ascii", "ignore")
.decode("utf-8", "ignore")
)
def to_lowercase(text):
return text.lower()
def remove_punctuation(text):
"""Remove punctuation from list of tokenized words"""
translator = str.maketrans("", "", string.punctuation)
return text.translate(translator)
def replace_numbers(text):
"""Replace all interger occurrences in list of tokenized words with textual representation"""
return re.sub(r"\d+", "", text)
def remove_whitespaces(text):
return text.strip()
def remove_stopwords(words, stop_words):
"""
:param words:
:type words:
:param stop_words: from sklearn.feature_extraction.stop_words import ENGLISH_STOP_WORDS
or
from spacy.lang.en.stop_words import STOP_WORDS
:type stop_words:
:return:
:rtype:
"""
return [word for word in words if word not in stop_words]
def stem_words(words):
"""Stem words in text"""
stemmer = PorterStemmer()
return [stemmer.stem(word) for word in words]
def lemmatize_words(words):
"""Lemmatize words in text"""
lemmatizer = WordNetLemmatizer()
return [lemmatizer.lemmatize(word) for word in words]
def lemmatize_verbs(words):
"""Lemmatize verbs in text"""
lemmatizer = WordNetLemmatizer()
return " ".join([lemmatizer.lemmatize(word, pos="v") for word in words])
def text2words(text):
return word_tokenize(text)
def normalize_text(text):
text = remove_special_chars(text)
text = remove_non_ascii(text)
text = remove_punctuation(text)
text = to_lowercase(text)
text = replace_numbers(text)
words = text2words(text)
words = remove_stopwords(words, stop_words)
# words = stem_words(words)# Either stem ovocar lemmatize
words = lemmatize_words(words)
words = lemmatize_verbs(words)
return "".join(words)
Train_X_clean = Train_X_raw.copy()
Validate_X_clean = Validate_X_raw.copy()
Train_X_clean = list(map(normalize_text, Train_X_clean))
Validate_X_clean = list(map(normalize_text, Validate_X_clean))
# Train_X_clean
#
# # 5. Model
# ([Go to top](#top))
evaluation_df = pd.DataFrame()
models_dict = {}
#
# ## 5.1 BOW
# ([Go to top](#top))
#
# ##### Text Preparation for BOW
vectorizer = CountVectorizer()
Train_X = vectorizer.fit_transform(Train_X_clean)
Validate_X = vectorizer.transform(Validate_X_clean)
# vectorizer.vocabulary_
Train_X = Train_X.toarray()
Validate_X = Validate_X.toarray()
print(Train_X.shape)
print(Validate_X.shape)
#
# ##### Building Model
model = models.Sequential()
model.add(layers.Dense(16, activation="relu", input_shape=(Train_X.shape[1],)))
model.add(layers.Dense(16, activation="relu"))
model.add(layers.Dense(1, activation="sigmoid"))
model.summary()
from keras.utils import plot_model
plot_model(model)
from keras import optimizers
model.compile(
optimizer=optimizers.RMSprop(learning_rate=0.001),
loss="binary_crossentropy",
metrics=["accuracy"],
)
history = model.fit(
Train_X,
y_train,
epochs=50,
batch_size=512,
validation_data=(Validate_X, y_validation),
)
#
# ##### Training VS Validation
accuracy = history.history["accuracy"]
loss = history.history["loss"]
val_accuracy = history.history["val_accuracy"]
val_loss = history.history["val_loss"]
plt.plot(loss, label="Training loss")
plt.plot(val_loss, label="Validation loss")
plt.title("Training and validation loss")
plt.legend()
plt.show()
plt.plot(accuracy, label="Training accuracy")
plt.plot(val_accuracy, label="Validation accuracy")
plt.title("Training and validation accuracy")
plt.legend()
plt.show()
#
# ##### Save Model
model.save("/kaggle/working/bow.h5")
#
# ##### Save Performance
model_name = "BOW"
models_dict[model_name] = "/kaggle/working/bow.h5"
train_loss, train_accuracy = model.evaluate(Train_X, y_train)
validation_loss, validation_accuracy = model.evaluate(Validate_X, y_validation)
evaluation = pd.DataFrame(
{
"Model": [model_name],
"Train": [train_accuracy],
"Validation": [validation_accuracy],
}
)
evaluation_df = pd.concat([evaluation_df, evaluation], ignore_index=True)
#
# ## 5.2 BOW Vectors
# ([Go to top](#top))
#
# ##### Text Preparation for BOW Vectors
from keras.preprocessing.text import Tokenizer
t = Tokenizer()
t.fit_on_texts(Train_X_clean)
vocab_size = len(t.word_index) + 1
# integer encode the documents
train_encoded_docs = t.texts_to_sequences(Train_X_clean)
validation_encoded_docs = t.texts_to_sequences(Validate_X_clean)
# print(train_encoded_docs)
# Determine the optimal maximum padding length
figure, subplots = plt.subplots(1, 2, figsize=(20, 5))
_ = sns.countplot(x=list(map(len, train_encoded_docs)), ax=subplots[0])
_ = sns.kdeplot(list(map(len, train_encoded_docs)), fill=True, ax=subplots[1])
from statistics import mode
train_encoded_docs_length = list(map(len, train_encoded_docs))
mode(train_encoded_docs_length)
# As we see the most frequent value in the histogram (mode) = 12.
# If we set the max padding length to be equal to the most frequent value in the histogram (mode = 12), let's see how many sentences we can get rid of.
len(
list(
filter(
lambda x: x >= mode(train_encoded_docs_length), train_encoded_docs_length
)
)
)
len(train_encoded_docs_length)
# We will lose around 34% of the data (6090 tweets).
round(
len(
list(
filter(
lambda x: x >= mode(train_encoded_docs_length),
train_encoded_docs_length,
)
)
)
/ len(train_encoded_docs_length)
* 100,
2,
)
# That's why we will set the max padding length to be equal to the length of the longest tweet.
max(train_encoded_docs_length)
max_length = max(train_encoded_docs_length)
train_seq = pad_sequences(train_encoded_docs, maxlen=max_length, padding="post")
validate_seq = pad_sequences(validation_encoded_docs, maxlen=max_length, padding="post")
print(train_seq)
#
# ##### Building Model
c_latent_factors = 32
model = models.Sequential()
model.add(layers.Embedding(vocab_size + 1, c_latent_factors, input_length=max_length))
model.add(layers.Flatten())
model.add(layers.Dense(16, activation="relu"))
model.add(layers.Dense(16, activation="relu"))
model.add(layers.Dense(1, activation="sigmoid"))
model.summary()
from keras.utils import plot_model
plot_model(model)
from keras import optimizers
model.compile(
optimizer=optimizers.RMSprop(learning_rate=0.001),
loss="binary_crossentropy",
metrics=["accuracy"],
)
history = model.fit(
train_seq,
y_train,
epochs=50,
batch_size=512,
validation_data=(validate_seq, y_validation),
)
#
# ##### Training VS Validation
accuracy = history.history["accuracy"]
loss = history.history["loss"]
val_accuracy = history.history["val_accuracy"]
val_loss = history.history["val_loss"]
plt.plot(loss, label="Training loss")
plt.plot(val_loss, label="Validation loss")
plt.title("Training and validation loss")
plt.legend()
plt.show()
plt.plot(accuracy, label="Training accuracy")
plt.plot(val_accuracy, label="Validation accuracy")
plt.title("Training and validation accuracy")
plt.legend()
plt.show()
#
# ##### Save Model
model.save("/kaggle/working/bow_vectors.h5")
#
# ##### Save Performance
model_name = "BOW Vectors"
models_dict[model_name] = "/kaggle/working/bow_vectors.h5"
train_loss, train_accuracy = model.evaluate(train_seq, y_train)
validation_loss, validation_accuracy = model.evaluate(validate_seq, y_validation)
evaluation = pd.DataFrame(
{
"Model": [model_name],
"Train": [train_accuracy],
"Validation": [validation_accuracy],
}
)
evaluation_df = pd.concat([evaluation_df, evaluation], ignore_index=True)
#
# ## 5.3 LSTM
# ([Go to top](#top))
#
# ##### Building Model
c_latent_factors = 32
model = models.Sequential()
model.add(layers.Embedding(vocab_size + 1, c_latent_factors, input_length=max_length))
model.add(layers.LSTM(32, dropout=0.2, recurrent_dropout=0.4))
model.add(layers.Flatten())
model.add(layers.Dense(16, activation="relu"))
model.add(layers.Dense(16, activation="relu"))
model.add(layers.Dense(1, activation="sigmoid"))
model.summary()
from keras.utils import plot_model
plot_model(model)
from keras import optimizers
model.compile(
optimizer=optimizers.RMSprop(learning_rate=0.001),
loss="binary_crossentropy",
metrics=["accuracy"],
)
history = model.fit(
train_seq,
y_train,
epochs=50,
batch_size=512,
validation_data=(validate_seq, y_validation),
)
#
# ##### Training VS Validation
accuracy = history.history["accuracy"]
loss = history.history["loss"]
val_accuracy = history.history["val_accuracy"]
val_loss = history.history["val_loss"]
plt.plot(loss, label="Training loss")
plt.plot(val_loss, label="Validation loss")
plt.title("Training and validation loss")
plt.legend()
plt.show()
plt.plot(accuracy, label="Training accuracy")
plt.plot(val_accuracy, label="Validation accuracy")
plt.title("Training and validation accuracy")
plt.legend()
plt.show()
#
# ##### Save Model
model.save("/kaggle/working/lstm.h5")
#
# ##### Save Performance
model_name = "LSTM"
models_dict[model_name] = "/kaggle/working/lstm.h5"
train_loss, train_accuracy = model.evaluate(train_seq, y_train)
validation_loss, validation_accuracy = model.evaluate(validate_seq, y_validation)
evaluation = pd.DataFrame(
{
"Model": [model_name],
"Train": [train_accuracy],
"Validation": [validation_accuracy],
}
)
evaluation_df = pd.concat([evaluation_df, evaluation], ignore_index=True)
#
# ## 5.4 GRU
# ([Go to top](#top))
#
# ##### Building Model
c_latent_factors = 32
model = models.Sequential()
model.add(layers.Embedding(vocab_size + 1, c_latent_factors, input_length=max_length))
model.add(layers.GRU(32, dropout=0.2, recurrent_dropout=0.4))
model.add(layers.Flatten())
model.add(layers.Dense(16, activation="relu"))
model.add(layers.Dense(16, activation="relu"))
model.add(layers.Dense(1, activation="sigmoid"))
model.summary()
from keras.utils import plot_model
plot_model(model)
from keras import optimizers
model.compile(
optimizer=optimizers.RMSprop(learning_rate=0.001),
loss="binary_crossentropy",
metrics=["accuracy"],
)
history = model.fit(
train_seq,
y_train,
epochs=50,
batch_size=512,
validation_data=(validate_seq, y_validation),
)
#
# ##### Training VS Validation
accuracy = history.history["accuracy"]
loss = history.history["loss"]
val_accuracy = history.history["val_accuracy"]
val_loss = history.history["val_loss"]
plt.plot(loss, label="Training loss")
plt.plot(val_loss, label="Validation loss")
plt.title("Training and validation loss")
plt.legend()
plt.show()
plt.plot(accuracy, label="Training accuracy")
plt.plot(val_accuracy, label="Validation accuracy")
plt.title("Training and validation accuracy")
plt.legend()
plt.show()
#
# ##### Save Model
model.save("/kaggle/working/gru.h5")
#
# ##### Save Performance
model_name = "GRU"
models_dict[model_name] = "/kaggle/working/gru.h5"
train_loss, train_accuracy = model.evaluate(train_seq, y_train)
validation_loss, validation_accuracy = model.evaluate(validate_seq, y_validation)
evaluation = pd.DataFrame(
{
"Model": [model_name],
"Train": [train_accuracy],
"Validation": [validation_accuracy],
}
)
evaluation_df = pd.concat([evaluation_df, evaluation], ignore_index=True)
#
# # 6. Evaluation
# ([Go to top](#top))
evaluation_df
from keras.models import load_model
# Get best model according to validation score.
best_model = evaluation_df[
evaluation_df["Validation"] == evaluation_df["Validation"].max()
]["Model"].values[0]
best_model
model = load_model(models_dict[best_model])
model.summary()
#
# # 7. Submission File Generation
# ([Go to top](#top))
test_data = pd.read_csv("/kaggle/input/nlp-getting-started/test.csv")
test_data.shape
test_data.head()
# Text preprocessing utilized during training
test_data["clean text"] = test_data["text"].apply(normalize_text)
test_data.head()
# Get the appropriate preparation based on model
if best_model == "BOW":
X_test = vectorizer.transform(test_data["clean text"])
X_test = X_test.toarray()
else:
X_test = t.texts_to_sequences(test_data["clean text"])
X_test = pad_sequences(X_test, maxlen=max_length, padding="post")
predictions = model.predict(X_test).round()
submission = pd.read_csv("/kaggle/input/nlp-getting-started/sample_submission.csv")
submission["target"] = np.round(predictions).astype("int")
submission.head()
submission.to_csv("submission.csv", index=False)
|
# Data Types and Data Structures in Python
# 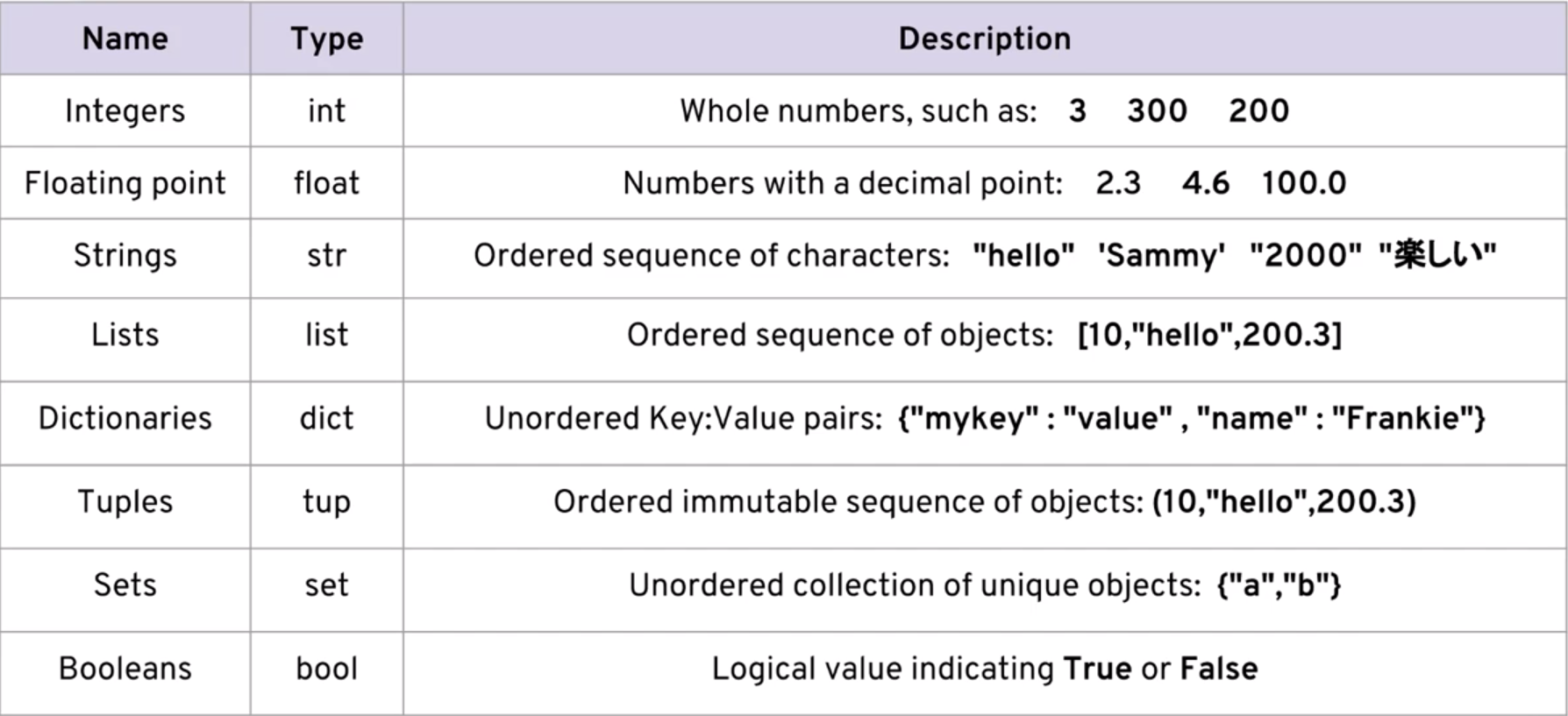
# Data types in Python are the different types of data that can be used to store information in a program. There are several built-in data types in Python, including numeric, string, Boolean, list, tuple, set, and dictionary. Each data type is used for a specific purpose, and they have their own properties and operations.
# * Numeric data types include integers, floats, and complex numbers. Integers are whole numbers, floats are numbers with decimal points, and complex numbers have a real and imaginary part. For example, the length of a rectangle can be stored as an integer, the weight of a person can be stored as a float, and the impedance of a circuit can be stored as a complex number.
# * String data types are used to store text data, such as names, addresses, and messages. For example, a person's name can be stored as a string, a product description can be stored as a string, and a message sent through a chat application can be stored as a string.
# * Boolean data types are used to represent true or false values. They are often used in conditional statements and loops to control the flow of a program. For example, a program that checks whether a user is logged in or not can use a Boolean value to represent the login status.
# * List, tuple, set, and dictionary data types are used to store collections of data. Lists are used to store ordered collections of data, tuples are used to store immutable collections of data, sets are used to store unordered collections of unique data, and dictionaries are used to store key-value pairs. For example, a list can be used to store the grades of a group of students, a tuple can be used to store the coordinates of a point on a map, a set can be used to store the unique values in a dataset, and a dictionary can be used to store the properties of a person, such as their name, age, and address.
# * Data structures in Python are different ways of organizing and storing data in a program. They are used to efficiently store and manipulate large amounts of data. Some common data structures in Python include arrays, linked lists, stacks, queues, trees, and graphs. Each data structure has its own properties and operations, and is used for a specific purpose.
#
# Operations in Numeric Datatypes
x = 10
y = 3.5
z = 2 + 3j
# Arithmetic operations
print(x + y) # 13.5
print(x - y) # 6.5
print(x * y) # 35.0
print(x / y) # 2.857142857142857
print(x // y) # 2
print(x % 3) # 1
print(z.real) # 2.0
print(z.imag) # 3.0
# Operations in Boolean Datatypes
a = True
b = False
# Logical operations
print(a and b) # False
print(a or b) # True
print(not a) # False
|
# ## İş Problemi
# Scout’lar tarafından izlenen futbolcuların özelliklerine verilen puanlara göre, oyuncuların hangi sınıf (average, highlighted) oyuncu olduğunu tahminleme
# ## Veri Seti Hikayesi
# Veri seti Scoutium’dan maçlarda gözlemlenen futbolcuların özelliklerine göre scoutların değerlendirdikleri futbolcuların, maç içerisinde puanlanan özellikleri ve puanlarını içeren bilgilerden oluşmaktadır.
import pandas as pd
import seaborn as sns
import numpy as np
import matplotlib.pyplot as plt
from sklearn.ensemble import RandomForestClassifier, GradientBoostingClassifier
from sklearn.linear_model import LogisticRegression
from sklearn.neighbors import KNeighborsClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.model_selection import (
train_test_split,
GridSearchCV,
cross_validate,
validation_curve,
)
from sklearn.metrics import precision_score, f1_score, recall_score, roc_auc_score
from sklearn.preprocessing import StandardScaler, LabelEncoder
from xgboost import XGBClassifier
from lightgbm import LGBMClassifier
from catboost import CatBoostClassifier
pd.set_option("display.max_columns", None)
pd.set_option("display.float_format", lambda x: "%.3f" % x)
pd.set_option("display.width", 500)
# Veri setlerine genel bakış.
att_df = pd.read_csv(
"/kaggle/input/scoutiumattributes/scoutium_attributes.csv", sep=";"
)
pot_df = pd.read_csv(
"/kaggle/input/scoutiumpotentiallabels/scoutium_potential_labels.csv", sep=";"
)
att_df.head()
pot_df.head()
# Veri setlerini birleştirelim.
df = att_df.merge(
pot_df, on=["task_response_id", "match_id", "evaluator_id", "player_id"]
)
df.head()
# position_id içerisindeki Kaleci (1) sınıfını veri setinden kaldıralım.
df = df.loc[~(df["position_id"] == 1)]
df.head()
# Adım 4: potential_label içerisindeki below_average sınıfını veri setinden kaldıralım.( below_average sınıfı tüm verisetinin %1'ini oluşturur)
df = df.loc[~(df["potential_label"] == "below_average")]
df.head()
# İndekste “player_id”,“position_id” ve “potential_label”, sütunlarda “attribute_id” ve değerlerde scout’ların oyunculara verdiği puan“attribute_value” olacak şekilde pivot table’ı oluşturalım.
pivot_df = pd.pivot_table(
df,
values="attribute_value",
index=["player_id", "position_id", "potential_label"],
columns=["attribute_id"],
)
pivot_df.head()
# İndeksleri değişken olarak atayalım ve “attribute_id” sütunlarının isimlerini stringe çevirelim.
pivot_df = pivot_df.reset_index()
pivot_df.columns = pivot_df.columns.astype("str")
# Label Encoder fonksiyonunu kullanarak “potential_label” kategorilerini (average, highlighted) sayısal olarak ifade edelim.
le = LabelEncoder()
pivot_df["potential_label"] = le.fit_transform(pivot_df["potential_label"])
pivot_df.head()
# Sayısal değişken kolonlarını “num_cols” adıyla bir listeye atayalım.
num_cols = [
col
for col in pivot_df.columns
if pivot_df[col].dtypes != "O" and pivot_df[col].nunique() > 5
]
num_cols = [col for col in num_cols if col not in "player_id"]
num_cols = num_cols[1:]
# Kaydettiğimiz bütün “num_cols” değişkenlerindeki veriyi ölçeklendirmek için StandardScaler uygulayalım.
ss = StandardScaler()
df = pivot_df.copy()
df[num_cols] = ss.fit_transform(df[num_cols])
df.head()
# ## Model
X = df.drop(["potential_label"], axis=1)
y = df["potential_label"]
def base_models(X, y, scoring="roc_auc"):
print("Base Models....")
models = [
("LR", LogisticRegression()),
("KNN", KNeighborsClassifier()),
("CART", DecisionTreeClassifier()),
("RF", RandomForestClassifier()),
("GBM", GradientBoostingClassifier()),
("XGBoost", XGBClassifier(eval_metric="logloss")),
("LightGBM", LGBMClassifier()),
("CatBoost", CatBoostClassifier(verbose=False)),
]
for name, classifier in models:
cv_results = cross_validate(classifier, X, y, cv=3, scoring=scoring)
print(f"{scoring}: {round(cv_results['test_score'].mean(), 4)} ({name}) ")
# roc_auc skorları
base_models(X, y)
# f1 skorları
base_models(X, y, scoring="f1")
# accuracy skorları
base_models(X, y, scoring="accuracy")
# precision skorları
base_models(X, y, scoring="precision")
# Catboost'a göre hiperparametre optimizasyonu yapalım.
catboost_model = CatBoostClassifier(random_state=17, verbose=False)
catboost_params = {
"iterations": [200, 500],
"learning_rate": [0.01, 0.1],
"depth": [3, 6],
}
catboost_grid = GridSearchCV(
catboost_model, catboost_params, cv=5, n_jobs=-1, verbose=False
).fit(X, y)
catboost_final = catboost_model.set_params(
**catboost_grid.best_params_, random_state=17
).fit(X, y)
cv_results = cross_validate(
catboost_final, X, y, cv=5, scoring=["accuracy", "f1", "roc_auc", "precision"]
)
# Hata metriklerimize bakalım.
cv_results["test_accuracy"].mean()
cv_results["test_f1"].mean()
cv_results["test_roc_auc"].mean()
cv_results["test_precision"].mean()
# Değişkenlerin önem düzeyini belirten feature_importance fonksiyonunu kullanarak özelliklerin sıralamasını çizdirelim.
def plot_importance(model, features, num=len(X), save=False):
feature_imp = pd.DataFrame(
{"Value": model.feature_importances_, "Feature": features.columns}
)
plt.figure(figsize=(10, 10))
sns.set(font_scale=1)
sns.barplot(
x="Value",
y="Feature",
data=feature_imp.sort_values(by="Value", ascending=False)[0:num],
)
plt.title("Features")
plt.tight_layout()
plt.show()
if save:
plt.savefig("importances.png")
plot_importance(catboost_final, X)
|
import torch
import torch.nn as nn
import torch.nn.functional as F
from torch.utils.data import DataLoader, random_split
from torchvision import datasets, transforms, models
from torchvision.utils import make_grid
from sklearn.model_selection import train_test_split
from sklearn.metrics import classification_report
from sklearn.metrics import roc_curve, auc
import torch.optim as optim
from tqdm import tqdm
from torchsummary import summary
import os
import random
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.metrics import confusion_matrix
import seaborn as sns
import shutil
import json
from PIL import Image as PilImage
from omnixai.data.image import Image
from omnixai.explainers.vision.specific.gradcam.pytorch.gradcam import GradCAM
import warnings
warnings.filterwarnings("ignore")
BATCH_SIZE = 128
LR = 0.0001
# Define the preprocessing transforms
transform = transforms.Compose(
[
transforms.Resize((224, 224)),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225]),
]
)
train_dataset = datasets.ImageFolder(
root="/kaggle/input/flower-classification-5-classes-roselilyetc/Flower Classification V2/V2/Training Data",
transform=transform,
)
test_dataset = datasets.ImageFolder(
root="/kaggle/input/flower-classification-5-classes-roselilyetc/Flower Classification V2/V2/Testing Data",
transform=transform,
)
val_dataset = datasets.ImageFolder(
root="/kaggle/input/flower-classification-5-classes-roselilyetc/Flower Classification V2/V2/Validation Data",
transform=transform,
)
# Define the dataloaders
train_dataloader = DataLoader(train_dataset, batch_size=BATCH_SIZE, shuffle=True)
val_dataloader = DataLoader(val_dataset, batch_size=BATCH_SIZE, shuffle=False)
test_dataloader = DataLoader(test_dataset, batch_size=BATCH_SIZE, shuffle=False)
class_names = train_dataset.classes
print(class_names)
print(len(class_names))
class VGG16(nn.Module):
def __init__(self, num_classes=5):
super(VGG16, self).__init__()
self.features = models.vgg16(pretrained=False).features
self.classifier = nn.Linear(512 * 7 * 7, num_classes)
def forward(self, x):
x = self.features(x)
x = torch.flatten(x, 1)
x = self.classifier(x)
return x
preprocess = lambda ims: torch.stack([transform(im.to_pil()) for im in ims])
# Set the device
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
device
model = VGG16(num_classes=len(class_names)).to(device)
best_checkpoint = torch.load(
"/kaggle/input/d2-vgg16-model/d2_vgg16_model_checkpoint.pth"
)
print(best_checkpoint.keys())
model.load_state_dict(best_checkpoint["state_dict"])
# load the saved weights from a .pth file
# set the model to evaluation mode
model.eval()
img_path = "/kaggle/input/flower-classification-5-classes-roselilyetc/Flower Classification V2/V2/Testing Data/Aster/Aster-Test (105).jpeg"
img = Image(PilImage.open(img_path).convert("RGB"))
explainer = GradCAM(
model=model, target_layer=model.features[-1], preprocess_function=preprocess
)
explanations = explainer.explain(img)
explanations.ipython_plot(index=0, class_names=class_names)
import os
import matplotlib.pyplot as plt
import numpy as np
test_dir = "/kaggle/input/flower-classification-5-classes-roselilyetc/Flower Classification V2/V2/Training Data/"
classes = [
"Aster",
"Daisy",
"Iris",
"Lavender",
"Lily",
"Marigold",
"Orchid",
"Poppy",
"Rose",
"Sunflower",
]
num_images = 2
for i, cls in enumerate(classes):
cls_dir = os.path.join(test_dir, cls)
img_files = os.listdir(cls_dir)
img_files = np.random.choice(img_files, size=num_images, replace=False)
for j, img_file in enumerate(img_files):
img_path = os.path.join(cls_dir, img_file)
img = Image(PilImage.open(img_path).convert("RGB"))
explainer = GradCAM(
model=model, target_layer=model.features[-1], preprocess_function=preprocess
)
explanations = explainer.explain(img)
explanations.ipython_plot(index=0, class_names=class_names)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Titanic Project Example Walk Through
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sbs
train_data = pd.read_csv("/kaggle/input/train-data/train.csv")
test_data = pd.read_csv("/kaggle/input/train-data/train.csv")
train_data["train_test"] = 1
test_data["train_test"] = 0
test_data["Survived"] = np.NaN
all_data = pd.concat([train_data, test_data])
all_data.columns
train_data.head()
train_data.isnull()
sbs.heatmap(train_data.isnull())
sbs.set_style("whitegrid")
sbs.countplot(x="Survived", data=train_data)
sbs.set_style("whitegrid")
sbs.countplot(x="Survived", hue="Sex", data=train_data)
sbs.set_style("whitegrid")
sbs.countplot(x="Survived", hue="Pclass", data=train_data)
sbs.distplot(train_data["Age"].dropna(), kde=False, bins=10)
sbs.countplot(x="SibSp", data=train_data)
sbs.distplot(train_data["Fare"], bins=20)
sbs.boxplot(x="Pclass", y="Age", data=train_data)
def function(columns):
Age = columns[0]
Pclass = columns[1]
if pd.isnull(Age):
if Pclass == 1:
return 37
elif Pclass == 2:
return 28
else:
return 25
else:
return Age
train_data["Age"] = train_data[["Age", "Pclass"]].apply(function, axis=1)
sbs.heatmap(train_data.isnull())
train_data.drop("Cabin", axis=1, inplace=True)
train_data.head()
# To calculate the required percenatge of women who survived
women = train_data.loc[train_data.Sex == "female"]["Survived"]
rate_women = (sum(women) / len(women)) * 100
print("Percentage of women who survived", rate_women)
# To calculate the required number of men who survived
men = train_data.loc[train_data.Sex == "male"]["Survived"]
rate_men = (sum(men) / len(men)) * 100
print("Percentage of women who survived", rate_men)
train_data.head()
sbs.displot(data=train_data, x="Age", row="Sex", col="Pclass", hue="Survived")
# Graphic Representation to check how many women survived and how many men survived.
labels = ["Women who Survived", "Men who Suvived"]
x = [rate_women, rate_men]
plt.pie(x, labels=labels)
# Through this presentation, we conclude that more percenatge of women survived than men
|
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# #### This is my first machine learning project that I build myself from scratch.
# # IMPORT LIBRARIES
import pandas as pd
import sklearn
from sklearn.feature_selection import RFE
from sklearn.preprocessing import StandardScaler, MinMaxScaler
from sklearn.linear_model import SGDRegressor
# from sklearn.linear_model import Ridge
from sklearn.preprocessing import PolynomialFeatures, OneHotEncoder
# from sklearn.linear_model import LinearRegression
from tensorflow.keras import Sequential
import tensorflow as tf
import numpy as np
from sklearn.metrics import mean_squared_error as mse
from sklearn.model_selection import train_test_split
from sklearn.utils import resample
from sklearn.metrics import mean_squared_log_error
from tensorflow.keras.activations import relu, linear
from tensorflow.keras.layers import Dense
from tensorflow.keras.optimizers import Adam
import matplotlib
# import matplotlib.pyplot as plt
# # IMPORT DATA
holidays_original = pd.read_csv(
"/kaggle/input/store-sales-time-series-forecasting/holidays_events.csv"
)
oil_original = pd.read_csv("/kaggle/input/store-sales-time-series-forecasting/oil.csv")
stores_original = pd.read_csv(
"/kaggle/input/store-sales-time-series-forecasting/stores.csv"
)
train_original = pd.read_csv(
"/kaggle/input/store-sales-time-series-forecasting/train.csv"
)
transactions_original = pd.read_csv(
"/kaggle/input/store-sales-time-series-forecasting/transactions.csv"
)
test_original = pd.read_csv(
"/kaggle/input/store-sales-time-series-forecasting/test.csv"
)
# # DATA WRANGLING
# #### Convert date (object) -> datetime
train_original["date"] = pd.to_datetime(train_original["date"])
test_original["date"] = pd.to_datetime(test_original["date"])
transactions_original["date"] = pd.to_datetime(transactions_original["date"])
holidays_original["date"] = pd.to_datetime(holidays_original["date"])
oil_original["date"] = pd.to_datetime(oil_original["date"])
# ### HOLIDAYS DATASET
# Remove transferred column
holidays = holidays_original.loc[(holidays_original["transferred"] == False)][
["date", "type", "locale", "locale_name"]
]
# Mutate city and state column
holidays["city_of_holidays"] = np.where(
holidays["locale"] == "Local", holidays["locale_name"], np.nan
)
holidays["state_of_holidays"] = np.where(
holidays["locale"] == "Regional", holidays["locale_name"], np.nan
)
# Group the date so we have unique date for each holiday
holidays.loc[holidays["city_of_holidays"].isnull() == False].groupby("date").agg(
np.array
)
# ### OIL DATA
# #### Make oil data for everyday in the period
full_date_oil = pd.DataFrame(
pd.date_range(start="2013-01-01", end="2017-08-31", name="date")
)
oil_price = np.zeros(len(full_date_oil))
oil_index = 0
for i in range(len(oil_price)):
if full_date_oil.iloc[i][0] < oil_original.iloc[oil_index + 1]["date"]:
oil_price[i] = oil_original.iloc[oil_index]["dcoilwtico"]
elif oil_index == len(oil_original) - 1:
break
else:
oil_index += 1
oil_price[i] = oil_original.iloc[oil_index]["dcoilwtico"]
full_date_oil["oil_price"] = oil_price
# Fill NA values
full_date_oil.fillna(method="backfill", inplace=True)
full_date_oil
# ## JOIN DATASETS
train_with_stores = pd.merge(
train_original,
stores_original,
left_on="store_nbr",
right_on="store_nbr",
how="left",
)
train_joined = pd.merge(
train_with_stores, full_date_oil, left_on=["date"], right_on=["date"], how="left"
)
train_joined = pd.merge(
train_joined,
holidays[
[
"date",
"type",
"locale",
"locale_name",
"city_of_holidays",
"state_of_holidays",
]
]
.groupby("date")
.agg(" ".join),
left_on=["date"],
right_on=["date"],
how="left",
)
test_with_stores = pd.merge(
test_original,
stores_original,
left_on="store_nbr",
right_on="store_nbr",
how="left",
)
test_joined = pd.merge(
test_with_stores, full_date_oil, left_on=["date"], right_on=["date"], how="left"
)
test_joined = pd.merge(
test_joined,
holidays[
[
"date",
"type",
"locale",
"locale_name",
"city_of_holidays",
"state_of_holidays",
]
]
.groupby("date")
.agg(" ".join),
left_on=["date"],
right_on=["date"],
how="left",
)
train_joined
# ### One hot for holidays
# Create a column to determine whether the store is located near the holiday or not. If the holiday is national then value equals 1, or the state or the city of the store match with the holidays then the value equals 1. Otherwise the value is 0.
#
def condition(row):
if (
"National" in str(row["locale"])
or str(row["city"]) in str(row["locale_name"])
or str(row["state"]) in str(row["locale_name"])
):
return 1
else:
return 0
train_joined["holidays?"] = train_joined.apply(condition, axis=1)
test_joined["holidays?"] = test_joined.apply(condition, axis=1)
# ### Add salary day
train_joined["month"] = train_joined["date"].dt.month
train_joined["day_of_month"] = train_joined["date"].dt.day
train_joined["year"] = train_joined["date"].dt.year
train_joined["salary_day?"] = train_joined["date"].apply(
lambda x: 1 if (x.is_month_end == True or x.date().day == 15) else 0
)
## For test dataset
test_joined["month"] = test_joined["date"].dt.month
test_joined["day_of_month"] = test_joined["date"].dt.day
test_joined["year"] = test_joined["date"].dt.year
test_joined["salary_day?"] = test_joined["date"].apply(
lambda x: 1 if (x.is_month_end == True or x.date().day == 15) else 0
)
# # RENAME COLUMN FOR BETTER INTEPRETATION
train_joined = train_joined.rename(
columns={"type_x": "store_type", "type_y": "holiday_type"}
)
train_joined
test_joined = test_joined.rename(
columns={"type_x": "store_type", "type_y": "holiday_type"}
)
test_joined
# # RESAMPLE FOR SMALLER TRAINING DATA
train_resampled = resample(train_joined, n_samples=1500000)
train_resampled
# # NORMALIZING DATA
# Select features for training
ohc = OneHotEncoder(sparse=False, drop="first")
one_hot = ohc.fit(
train_resampled[
[
"holidays?",
"salary_day?",
"locale",
"family",
"city",
"state",
"store_type",
"locale_name",
"holiday_type",
]
]
)
train_one_hot = ohc.fit_transform(
train_resampled[
[
"holidays?",
"salary_day?",
"locale",
"family",
"city",
"state",
"store_type",
"locale_name",
"holiday_type",
]
]
)
# train_one_hot=pd.get_dummies(train_resampled[['holidays?','salary_day?','locale','family','city','state','store_type','locale_name','holiday_type']])
train_unscaled_x = train_resampled[
["year", "store_nbr", "onpromotion", "oil_price", "month", "cluster"]
]
poly = PolynomialFeatures(degree=4, include_bias=False)
poly_fit = poly.fit(train_unscaled_x)
item_poly = poly_fit.transform(train_unscaled_x)
# Scale
train_unscaled_y = train_resampled["sales"]
scaler = StandardScaler()
scaler = scaler.fit(item_poly)
item_train = scaler.fit_transform(item_poly)
y_for_train = np.array(train_unscaled_y).reshape(-1, 1)
train_final = np.concatenate((item_train, np.array(train_one_hot)), axis=1)
train_final.shape
pd.DataFrame(train_final).describe()
test_one_hot = pd.get_dummies(
test_joined[
[
"holidays?",
"salary_day?",
"locale",
"family",
"city",
"state",
"store_type",
"locale_name",
"holiday_type",
]
]
)
test_one_hot = ohc.transform(
test_joined[
[
"holidays?",
"salary_day?",
"locale",
"family",
"city",
"state",
"store_type",
"locale_name",
"holiday_type",
]
]
)
test_unscaled = test_joined[
["year", "store_nbr", "onpromotion", "oil_price", "month", "cluster"]
]
test_poly = poly_fit.transform(test_unscaled)
test_scaled = scaler.transform(test_poly)
test_final = np.concatenate((test_scaled, np.array(test_one_hot)), axis=1)
test_final.shape
# # SPLIT DATA FOR TRAINING
X_train, X_val, y_train, y_val = train_test_split(
train_final, y_for_train, test_size=0.10, random_state=1
)
print("X_train.shape", X_train.shape, "y_train.shape", y_train.shape)
print("X_val.shape", X_val.shape, "y_val.shape", y_val.shape)
# # BUILD THE MODEL
# Initialize learning rate decay
initial_learning_rate = 0.001
lr_schedule = tf.keras.optimizers.schedules.ExponentialDecay(
initial_learning_rate, decay_steps=30000, decay_rate=0.9, staircase=True
)
opt = Adam(lr_schedule)
model = Sequential(
[
tf.keras.Input(shape=(342,)),
Dense(
342, activation="relu", activity_regularizer=tf.keras.regularizers.L1(0.01)
),
Dense(
32, activation="relu", activity_regularizer=tf.keras.regularizers.L2(0.02)
),
Dense(1, activation="relu"),
],
name="my_model",
)
model.compile(optimizer=opt, loss=tf.keras.losses.MeanSquaredLogarithmicError())
history = model.fit(X_train, y_train, epochs=80)
model.evaluate(X_val, y_val)
submit = pd.DataFrame(model.predict(test_final))
submit_ = pd.concat((test_joined["id"], submit[0]), axis=1)
submit_ = submit_.rename(columns={0: "sales"})
submit_.to_csv("submit6.csv", index=False)
submit_
# Save weights
model.save_weights("./saved/checkpoints")
# # PLOT THE RESULT
model.predict(X_val)
lim = y_val.max()
matplotlib.pyplot.scatter(y_val, model.predict(X_val), marker="o")
matplotlib.pyplot.xlabel("Ground Truth")
matplotlib.pyplot.ylabel("Predict")
matplotlib.pyplot.xlim(0, lim)
matplotlib.pyplot.ylim(0, lim)
|
# ## Visualization Technique
# For this demonstration I will be using the jointplot and the backbone of it the JointGrid from the seaborn library.
# This plotting technique is a way to see the interaction between two continuous variables. The grid is split into three sections, the joint plot which is in the center and the marginal plots, one of which is on top and the other which is on the right side. The joint plot shows the interaction of the variables, and the marginal plots show the distributions. Using The jointplot we can quickly graph a scatterplot and also see the histograms of those variables in the joint and marginal plots respectively. We can specify different plot types in the jointplot, but once we select the type we want to use in the joint plot we are locked into the choice for the marginal plots. To overcome this, we can use the JointGrid instead. The jointplot is built on top of the JointGrid, using the latter gives us more control but is a bit more involved. "This is intended to be a fairly lightweight wrapper; if you need more flexibility, you should use JointGrid directly."(Seaborn.jointplot — Seaborn 0.11.1 Documentation)
# Plotting with the jointplot and JointGrid interface is a great way to see bivariate data as well as each univariate distribution on the same figure. These techniques can be used with any bivariate and continuous data. Just like a scatter plot we can see the relationship between the variables, and we can even specify a regression plot to fit a regression line. Although this technically works with categorical data and we can make barplots in the JointGrid, at least one of the marginal plots won't make sense and it would be better visualized using another plotting function. This can be seen in Example 9.
# ## Visulaization library
# The seaborn Library is an open source statistical plotting library derived from matplotlib. It is more intuitive and makes quick plotting easy with nice looking default values. Seaborn has a quick and easy way to create this jointplot, which is technically possible to create in matplotlib's pyplot, but it would take a lot of code and testing to get right. Using seaborn to create the jointplot has its limitations. With many of the seaborn functions they can take a matplotlib axis as an argument and most also return that axis for further customization. This is not the case with jointplot. It is built on top of the JointGrid and has access to the axis objects created from the jointgrid for customization but no axis or figure objects can be passed to JointGrid initially. Unlike matplotlib's pyplot, seaborn is declarative not procedural. As stated in the seaborn introduction "Its dataset-oriented, declarative API lets you focus on what the different elements of your plots mean, rather than on the details of how to draw them"(An Introduction to Seaborn — Seaborn 0.9.0 Documentation). The Seaborn library was created by Michael Waskom and can be integrated wherever matplotlib can be which creates a very diverse array of environments where it can be used including the jupyter environment without any alteration.
# ### Installation
# `pip install seaborn`
# or
# `conda install seaborn`
# ### Dependencies
# `numpy`
# `scipy`
# `pandas`
# `matplotlib`
# ### Optional Dependencies
# `statsmodels`
# `fastcluster`
# (Seaborn.jointplot — Seaborn 0.11.1 Documentation)
# ## Visualization demonstration
# For this demonstration I am going to use one of the built-in datasets from the seaborn library. Specifically, the miles per gallon (mpg.csv) dataset. it can be retrieved by `seaborn.load_dataset('mpg')` or by going to https://github.com/mwaskom/seaborn-data.
# ### Import and data cleaning
# first we import our modules
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
# set the pandas DataFrame to display all columns
pd.set_option("display.max_columns", None)
# Load The data, it is from seaborn datasets
data = sns.load_dataset("mpg")
# drop any rows with null values
data.dropna(inplace=True)
# lets look at a the shape and a sample of the data
print("Shape of the dataframe")
print(f"{data.shape[0]} rows")
print(f"{data.shape[1]} columns\n")
print(data.info())
print("\nSample from the DataFrame")
data.sample(5)
# ### Plotting
# #### Example 1
# To create a simple jointplot we specify the data we are working with and the variables to assign to x and y. Seaborn will create a plot with axis labels and does a good job of picking default values including axis labels, tick marks, color, etc.
sns.jointplot(data=data, x="horsepower", y="weight")
# #### Example 2
# We can specify a plot `kind` and the jointplot automatically picks appropriate values and changes the marginal plots appropriately. there are many types of plots that can be used. The built-in ones are scatter, regression, kde, histogram, hex, and residual.
# specify that we are using a regression plot
sns.jointplot(data=data, x="horsepower", y="weight", kind="reg")
# #### Example 3
# here we see the hex plot kind
sns.jointplot(data=data, x="mpg", y="weight", kind="hex")
# #### Example 4
# The Residual plot shows the distribution of the residuals if we were predicting mpg from horsepower. The plot fits a model and plots the residuals.
# to show the residual plot it will take x as the predictor variable and y as the response variabel
sns.jointplot(data=data, x="horsepower", y="mpg", kind="resid", order=2)
# here we had to specify an order parameter it is the order of the
# polynomial fit to determin the residuals where 1 in a linear fit
# this relationship can be seen in Example 5
# #### Example 5
# To see the interaction of a third variable we can specify a hue. A legend will appear, and the marginal plots will again change accordingly. We can control for the height of the figure, though it will always be square.
# adding in a hue and height variable
sns.jointplot(data=data, x="horsepower", y="mpg", hue="origin", height=8)
# #### Example 6
# Although the default values look nice in many cases, sometimes we want to do some kind of customization. We can send optional arguments to the individual plots, either the joint plot or the marginal plots. These optional keyword arguments are passed to the underlying functions so they will change depending on the `kind` specified.
sns.jointplot(
data=data,
x="horsepower",
y="acceleration",
kind="kde",
joint_kws=dict(fill=True, color="cyan"),
marginal_kws=dict(color="green", fill=False, linewidth=3),
)
# #### Example 7
# The jointplot function returns a JointGrid object so we can use its methods to do some additional plotting to combine plot types.
# a seaborn kdeplot ontop of a scatterplot of the same data
# here i only specified the jointplot so the kdeplot is only represented there
# and the marginal plots are unchanged
grid = sns.jointplot(data=data, x="horsepower", y="displacement")
grid.plot_joint(sns.kdeplot)
# #### Example 8
# Using the jointplot is extremely easy and intuitive, but for greater control and more plotting options we can use the JointGrid directly. The JointGrid takes many of the same parameters as jointplot but here there are not optional argument parameters for the individual plots. They must be specified when calling one of the plot functions.
# Here we are using a seaborn scatterplot as the jointplot
# and seaborn violinplots for the marginal plots.
# the color parameter is optional and effects both
grid = sns.JointGrid(data=data, x="displacement", y="mpg")
grid.plot(sns.scatterplot, sns.violinplot, color="lightblue")
# #### Example 9
# This is an example of why categorical data does not work as well. The barplot itself turns out fine and the marginal plot on the right is fine as well, however the top marginal plot is quite meaningless. Since the individual plots cannot be turned off we are better off visualizing the categorical data in another plot type. Or as we will see in example 12 we can use the underlying matplotlib axis to plot or not.
# not ideal for categorical data
grid = sns.JointGrid(data=data, x="cylinders", y="mpg")
grid.plot(sns.barplot, sns.kdeplot, color="red")
# #### Example 10
# We are not bound by the seaborn library for plot types. We can also use matplotlib.pyplot functions if we so choose.
# we can also use matplotlib's pyplot functions to plot
grid = sns.JointGrid(data=data, x="mpg", y="acceleration")
grid.plot(plt.scatter, plt.hist, color="forestgreen")
# as an asside for the marginal plots i was not able to get
# pyplot's boxplot violinplot or plot functions to work.
# It threw an error that 'orient_kw_x' was used before it was
# declared. It also could not be passed through by Jointgrid.plot
# this may work if we use the technique seen in example 12
# #### Example 11
# We can plot the joint and marginal plots separately passing different keyword arguments to each to customize them even more or if there are conflicting parameters to pass to the joint and marginal plots. (fill the joint kde but don’t fill the marginal ones). We can also do some label customization. Seaborn is also great for the vast array of color pallets to choose from to avoid picking individual colors.
# There is a great article here https://medium.com/@morganjonesartist/color-guide-to-seaborn-palettes-da849406d44f that shows an example of each color pallet.
# plot individually then customize axis labels
grid = sns.JointGrid(
data=data,
x="horsepower",
y="mpg",
hue="origin",
space=0,
height=10,
ratio=3,
palette="Set2",
)
grid.plot_joint(sns.kdeplot, fill=True, alpha=0.6)
grid.plot_marginals(sns.histplot, element="poly", linewidth=1.5, fill=False)
grid.set_axis_labels(
xlabel="Horsepower HP", ylabel="Miles per Gallon", fontsize=18, color="steelblue"
)
# #### Example 12
# Finally we can access the individual matplotlib axis variables for even more controle. They can be used for customization after the initial plotting or by using the matplotlib plotting functions them selves. Aditionally we can take the axis and pass them into a seaborn plotting function. however when we are plottingthis way we must make sure that our plotting is consistent. we can easily switch the marginal plots showing the wrong distribution on them as compared to the joint plot.
grid = sns.JointGrid(
data=data, x="mpg", y="acceleration", hue="origin", space=2, height=8, ratio=3
)
# plotting on the jont grid normally
grid.plot_joint(sns.scatterplot)
# using the matplotlib axis to set the tick params
grid.ax_joint.tick_params(axis="both", direction="inout", length=20, color="cyan")
# plotting from the matplotlib axis this must correspond to the data plotted on the y axis in the joint plot
grid.ax_marg_y.hist(data["acceleration"], orientation="horizontal")
# passing the matplotlib axis to a seaborn plotting function
# again this must correspond to the data plotted on the x axis in the joint plot
sns.kdeplot(data=data, x="mpg", legend=False, ax=grid.ax_marg_x)
# adjusting the x marginal plot tick params
grid.ax_marg_x.tick_params(axis="x", color="red", width=4, length=10)
# accessing the figure to add a title
grid.fig.suptitle("Comparing miles per gallon and acceleration", y=1.06, fontsize=18)
# ## Conclusion
# Although there are limitations to the seaborn jointplot and JointGrid they are extremely useful in creating quick, good looking and informative plots of this type. They can create powerful visualizations and inform on the interaction of the variables and the distributions of those variables at the same time. There is potential for all this customization to get confusing and can greatly skew the data ink ratio defined by Tufte making some of these visualizations hard to follow.
# ### Sources
# “Seaborn.jointplot — Seaborn 0.11.1 Documentation.” Seaborn.pydata.org,
# seaborn.pydata.org/generated/seaborn.jointplot.html#seaborn.jointplot.
# Accessed 22 Mar. 2021.
# “An Introduction to Seaborn — Seaborn 0.9.0 Documentation.” Pydata.org,
# 2012, seaborn.pydata.org/introduction.html.
# Accessed 22 Mar. 2021.
# “Installing and Getting Started — Seaborn 0.11.1 Documentation.”
# Seaborn.pydata.org, seaborn.pydata.org/installing.html.
# Accessed 22 Mar. 2021.
#
# ### Documentation
#
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Packages
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
# # Load Data
df_kidney = pd.read_csv(
"/kaggle/input/kidney-stone-prediction-based-on-urine-analysis/kindey stone urine analysis.csv"
)
df_train = pd.read_csv(
"/kaggle/input/playground-series-s3e12/train.csv", index_col="id"
)
df_test = pd.read_csv("/kaggle/input/playground-series-s3e12/test.csv", index_col="id")
df_sample = pd.read_csv("/kaggle/input/playground-series-s3e12/sample_submission.csv")
print(f"df_kidney.shape: {df_kidney.shape}")
print(f"df_train.shape: {df_train.shape}")
print(f"df_test.shape: {df_test.shape}")
print(f"missing values in origional dataset: \n{df_kidney.isna().sum()}")
print("-" * 50)
print(f"missing values in train dataset: \n{df_train.isna().sum()}")
print("-" * 50)
print(f"missing values in test dataset: \n{df_test.isna().sum()}")
# We can see that there are 414 training datapoints and 79 origional datapoints.
# There is no missing data so we do not need to worry about what to do with missing data.
# Here is what the data looks like...
df_train.head()
df_kidney.describe()
df_train.describe()
df_test.describe()
df_kidney.nunique()
df_train.nunique()
df_test.nunique()
print(f"origional duplicates: {df_kidney.duplicated().sum()}")
print(f"train duplicates: {df_train.duplicated().sum()}")
print(f"train duplicates: {df_test.duplicated().sum()}")
print(
f"duplicates when combining origional and train: {pd.concat([df_kidney, df_train]).duplicated().sum()}"
)
# -----
# There are 6 independent variables and a classification target.
# From the [origional dataset](https://www.kaggle.com/datasets/vuppalaadithyasairam/kidney-stone-prediction-based-on-urine-analysis/code) the independ variables are:
# gravity: specific gravity, the density of the urine relative to water
# ph: pH, the negative logarithm of the hydrogen ion.
# osmo: osmolarity (mOsm), a unit used in biology and medicine but not in physical chemistry. Osmolarity is proportional to the concentration of molecules in solution
# cond: conductivity (mMho milliMho), one Mho is one reciprocal Ohm. Conductivity is proportional to the concentration of charged ions in solution
# urea: urea concentration, in millimoles per litre
# calc: calcium concentration (CALC), in millimolesllitre
# The dependent variable is `target` which is whether or not someone has a kidneystone (0 - No, 1 - Yes).
# # Plotting
# ## Vairable Distributions Across Datasets
# create subplots
fig, axs = plt.subplots(nrows=2, ncols=3, figsize=(10, 10))
# loop through the columns and create a line plot for each column
for idx, col in enumerate(df_test.columns):
row_idx = idx // 3
col_idx = idx % 3
ax = axs[row_idx, col_idx]
ax.grid(True)
sns.kdeplot(data=df_train[col], fill=True, ax=ax, label="train")
sns.kdeplot(data=df_test[col], fill=True, ax=ax, label="test")
sns.kdeplot(data=df_kidney[col], fill=True, ax=ax, label="og")
ax.set_title(f"{col}")
ax.legend()
ax.grid(True)
# set the main title and adjust the spacing between subplots
fig.suptitle("Density Plot of Continuous Features", fontsize=16)
plt.tight_layout()
# show the plot
plt.show()
# Some differences between the origional dataset and the generated data.
# The training and test data has some differences but they are small.
# We will only use the generated training set.
# ## Correlation
sns.heatmap(
data=df_train.corr(),
vmin=-1,
vmax=1,
annot=True,
cmap=sns.color_palette("vlag", as_cmap=True),
)
plt.title("Correlation Plot for Train Data")
# # Scatterplots
# create subplots
fig, axs = plt.subplots(nrows=2, ncols=3, figsize=(10, 10))
# loop through the columns and create a scatterplots plot for each column
for idx, col in enumerate(df_test.columns):
row_idx = idx // 3
col_idx = idx % 3
ax = axs[row_idx, col_idx]
ax.grid(True)
sns.regplot(data=df_train, x=col, y="target", ax=ax, lowess=True)
ax.set_title(f"{col}")
ax.legend()
# set the main title and adjust the spacing between subplots
fig.suptitle("Variable Effects on Kidney Stone Target", fontsize=16)
plt.tight_layout()
# ChatGPT is not interpreting my problem correctly. For one of my plots the lowess curve produced has a section of the curve that is 1. I was wondering if there was a way to fix this. Note
# create subplots
fig, axs = plt.subplots(nrows=2, ncols=3, figsize=(10, 10))
# loop through the columns and create a scatterplots plot for each column
for idx, col in enumerate(df_test.columns):
row_idx = idx // 3
col_idx = idx % 3
ax = axs[row_idx, col_idx]
ax.grid(True)
sns.regplot(
data=df_train,
x=col,
y="target",
ax=ax,
lowess=True,
line_kws={"color": "green"},
)
sns.regplot(
data=df_train,
x=col,
y="target",
ax=ax,
logistic=True,
line_kws={"color": "orange"},
scatter_kws={"color": "#4C72B0"},
)
ax.set_title(f"{col}")
ax.legend()
# set the main title and adjust the spacing between subplots
fig.suptitle("Variable Effects on Kidney Stone Target (Train)", fontsize=16)
plt.tight_layout()
# Weird how the lowess curve for gravity starts to decrease after 1.025.
# create subplots
fig, axs = plt.subplots(nrows=2, ncols=3, figsize=(10, 10))
# loop through the columns and create a scatterplots plot for each column
for idx, col in enumerate(df_test.columns):
row_idx = idx // 3
col_idx = idx % 3
ax = axs[row_idx, col_idx]
ax.grid(True)
sns.regplot(
data=df_kidney,
x=col,
y="target",
ax=ax,
lowess=True,
line_kws={"color": "green"},
)
sns.regplot(
data=df_kidney,
x=col,
y="target",
ax=ax,
logistic=True,
line_kws={"color": "orange"},
scatter_kws={"color": "#4C72B0"},
)
ax.set_title(f"{col}")
# set the main title and adjust the spacing between subplots
fig.suptitle("Variable Effects on Kidney Stone Target (Origional)", fontsize=16)
plt.tight_layout()
# Looks like the data generator did not follow the same pattern for usg in the train data that is shown in the origional dataset.
# Will have to see if this carried over into test dataset as well.
# # Violin Plots of the Variables with the Target
# Create violin plot to see the distributions across kidney stones
fig, axs = plt.subplots(nrows=2, ncols=3, figsize=(10, 10))
for idx, col in enumerate(df_test.columns):
row_idx = idx // 3
col_idx = idx % 3
ax = axs[row_idx, col_idx]
ax.grid(True)
sns.violinplot(x="target", y=col, data=df_train, ax=ax)
fig.suptitle("Violin Plots of Kidney Stone Target (Train)", fontsize=16)
plt.tight_layout()
# # Use Paired scatterplot to check for outliers
FEATURES = df_test.columns.to_list()
# Create scatter plots, check for outliers
fig, axs = plt.subplots(nrows=5, ncols=3, figsize=(15, 15))
count = 0
for idx, col1 in enumerate(FEATURES[:-1]):
for col2 in FEATURES[(idx + 1) :]:
row_idx = count // 3
col_idx = count % 3
ax = axs[row_idx, col_idx]
# add grid to plot
ax.grid(True)
# create separate regression plots for each value of the hue variable
for hue_val in df_train["target"].unique():
subset = df_train[df_train["target"] == hue_val]
sns.regplot(
x=col1,
y=col2,
data=subset,
ax=ax,
lowess=True,
scatter_kws={"alpha": 0.5},
label=f"{hue_val}",
)
# remove legend from individual subplot
ax.legend().remove()
count += 1
fig.suptitle("Scatter Plots of the Training Variables (Train)\n\n", fontsize=16)
# Add single legend for the entire figure
handles, labels = axs[0, 0].get_legend_handles_labels()
fig.legend(handles, labels, loc="upper center", ncol=2, bbox_to_anchor=(0.5, 0.96))
plt.tight_layout()
|
# # Clone DiffWave Repo
# upgrade pip
# install torch
# install diffwave
# ! pip install .
# # Rewrite
import numpy as np
import os
import torch
import torch.nn as nn
from torch.nn.parallel import DistributedDataParallel
from torch.utils.tensorboard import SummaryWriter
from tqdm import tqdm
from diffwave.dataset import from_path, from_gtzan
from diffwave.model import DiffWave
from diffwave.params import AttrDict
def _nested_map(struct, map_fn):
if isinstance(struct, tuple):
return tuple(_nested_map(x, map_fn) for x in struct)
if isinstance(struct, list):
return [_nested_map(x, map_fn) for x in struct]
if isinstance(struct, dict):
return {k: _nested_map(v, map_fn) for k, v in struct.items()}
return map_fn(struct)
class DiffWaveLearner:
def __init__(self, model_dir, model, dataset, optimizer, params, *args, **kwargs):
os.makedirs(model_dir, exist_ok=True)
self.model_dir = model_dir
self.model = model
self.dataset = dataset
self.optimizer = optimizer
self.params = params
self.autocast = torch.cuda.amp.autocast(enabled=kwargs.get("fp16", False))
self.scaler = torch.cuda.amp.GradScaler(enabled=kwargs.get("fp16", False))
self.step = 0
self.is_master = True
beta = np.array(self.params.noise_schedule)
noise_level = np.cumprod(1 - beta)
self.noise_level = torch.tensor(noise_level.astype(np.float32))
self.loss_fn = nn.L1Loss()
self.summary_writer = None
def state_dict(self):
if hasattr(self.model, "module") and isinstance(self.model.module, nn.Module):
model_state = self.model.module.state_dict()
else:
model_state = self.model.state_dict()
return {
"step": self.step,
"model": {
k: v.cpu() if isinstance(v, torch.Tensor) else v
for k, v in model_state.items()
},
"optimizer": {
k: v.cpu() if isinstance(v, torch.Tensor) else v
for k, v in self.optimizer.state_dict().items()
},
"params": dict(self.params),
"scaler": self.scaler.state_dict(),
}
def load_state_dict(self, state_dict):
if hasattr(self.model, "module") and isinstance(self.model.module, nn.Module):
self.model.module.load_state_dict(state_dict["model"])
else:
self.model.load_state_dict(state_dict["model"])
self.optimizer.load_state_dict(state_dict["optimizer"])
self.scaler.load_state_dict(state_dict["scaler"])
self.step = state_dict["step"]
def save_to_checkpoint(self, filename="weights"):
save_basename = f"{filename}-{self.step}.pt"
save_name = f"{self.model_dir}/{save_basename}"
link_name = f"{self.model_dir}/{filename}.pt"
torch.save(self.state_dict(), save_name)
if os.name == "nt":
torch.save(self.state_dict(), link_name)
else:
if os.path.islink(link_name):
os.unlink(link_name)
os.symlink(save_basename, link_name)
def restore_from_checkpoint(self, filename="weights"):
try:
checkpoint = torch.load(f"{self.model_dir}/{filename}.pt")
self.load_state_dict(checkpoint)
return True
except FileNotFoundError:
return False
def train(self, max_steps=None):
device = next(self.model.parameters()).device
while True:
for features in (
tqdm(self.dataset, desc=f"Epoch {self.step // len(self.dataset)}")
if self.is_master
else self.dataset
):
if max_steps is not None and self.step >= max_steps:
return
features = _nested_map(
features,
lambda x: x.to(device) if isinstance(x, torch.Tensor) else x,
)
loss = self.train_step(features)
if torch.isnan(loss).any():
raise RuntimeError(f"Detected NaN loss at step {self.step}.")
if self.is_master:
if self.step % 50 == 0:
self._write_summary(self.step, features, loss)
if self.step % len(self.dataset) == 0 and self.step == int(
max_steps - len(self.dataset)
):
self.save_to_checkpoint()
self.step += 1
def train_step(self, features):
for param in self.model.parameters():
param.grad = None
audio = features["audio"]
spectrogram = features["spectrogram"]
N, T = audio.shape
device = audio.device
self.noise_level = self.noise_level.to(device)
with self.autocast:
t = torch.randint(
0, len(self.params.noise_schedule), [N], device=audio.device
)
noise_scale = self.noise_level[t].unsqueeze(1)
noise_scale_sqrt = noise_scale**0.5
noise = torch.randn_like(audio)
noisy_audio = noise_scale_sqrt * audio + (1.0 - noise_scale) ** 0.5 * noise
predicted = self.model(noisy_audio, t, spectrogram)
loss = self.loss_fn(noise, predicted.squeeze(1))
self.scaler.scale(loss).backward()
self.scaler.unscale_(self.optimizer)
self.grad_norm = nn.utils.clip_grad_norm_(
self.model.parameters(), self.params.max_grad_norm or 1e9
)
self.scaler.step(self.optimizer)
self.scaler.update()
return loss
def _write_summary(self, step, features, loss):
writer = self.summary_writer or SummaryWriter(self.model_dir, purge_step=step)
writer.add_audio(
"feature/audio",
features["audio"][0],
step,
sample_rate=self.params.sample_rate,
)
if not self.params.unconditional:
writer.add_image(
"feature/spectrogram",
torch.flip(features["spectrogram"][:1], [1]),
step,
)
writer.add_scalar("train/loss", loss, step)
writer.add_scalar("train/grad_norm", self.grad_norm, step)
writer.flush()
self.summary_writer = writer
def _train_impl(replica_id, model, dataset, args, params):
torch.backends.cudnn.benchmark = True
opt = torch.optim.Adam(model.parameters(), lr=params.learning_rate)
learner = DiffWaveLearner(
args.model_dir, model, dataset, opt, params, fp16=args.fp16
)
learner.is_master = replica_id == 0
learner.restore_from_checkpoint()
learner.train(max_steps=args.max_steps)
def train(args, params):
if args.data_dirs[0] == "gtzan":
dataset = from_gtzan(params)
else:
dataset = from_path(args.data_dirs, params)
model = DiffWave(params).cuda()
_train_impl(0, model, dataset, args, params)
def train_distributed(replica_id, replica_count, port, args, params):
os.environ["MASTER_ADDR"] = "localhost"
os.environ["MASTER_PORT"] = str(port)
torch.distributed.init_process_group(
"nccl", rank=replica_id, world_size=replica_count
)
if args.data_dirs[0] == "gtzan":
dataset = from_gtzan(params, is_distributed=True)
else:
dataset = from_path(args.data_dirs, params, is_distributed=True)
device = torch.device("cuda", replica_id)
torch.cuda.set_device(device)
model = DiffWave(params).to(device)
model = DistributedDataParallel(model, device_ids=[replica_id])
_train_impl(replica_id, model, dataset, args, params)
import numpy as np
class AttrDict(dict):
def __init__(self, *args, **kwargs):
super(AttrDict, self).__init__(*args, **kwargs)
self.__dict__ = self
def override(self, attrs):
if isinstance(attrs, dict):
self.__dict__.update(**attrs)
elif isinstance(attrs, (list, tuple, set)):
for attr in attrs:
self.override(attr)
elif attrs is not None:
raise NotImplementedError
return self
params = AttrDict(
# Training params
batch_size=4,
learning_rate=2e-4,
max_grad_norm=None,
# Data params
sample_rate=44100,
n_mels=80,
n_fft=1024,
hop_samples=256,
crop_mel_frames=62, # Probably an error in paper.
# Model params
residual_layers=30,
residual_channels=64,
dilation_cycle_length=10,
unconditional=False,
noise_schedule=np.linspace(1e-8, 0.05, 50).tolist(),
inference_noise_schedule=[0.0001, 0.001, 0.01, 0.05, 0.2, 0.5],
# unconditional sample len
audio_len=44100 * 3, # unconditional_synthesis_samples
)
# install again
# # Training
# copy the dataset to the working directory
# !cp -r /kaggle/input/the-lj-speech-dataset /kaggle/working/
# #### Immature
# preprocess the immature dataset
# start training
# #### Mature
# preprocess the mature dataset
# ! python -m diffwave.preprocess /kaggle/working/coconut-signals-generation/mature
# start training
# ! python -m diffwave /kaggle/working/models/mature /kaggle/working/coconut-signals-generation/mature --max_steps 1800
# #### Overmature
# preprocess the mature dataset
# ! python -m diffwave.preprocess /kaggle/working/coconut-signals-generation/overmature
# start training
# ! python -m diffwave /kaggle/working/models/overmature /kaggle/working/coconut-signals-generation/overmature --max_steps 3000
# # Inference
# #### Immature
import os
import torchaudio
from diffwave.inference import predict as diffwave_predict
model_path = "/kaggle/working/models/immature/weights-147.pt"
input_folder = "/kaggle/working/coconut-signals-generation/immature"
output_folder = "/kaggle/working/inference/immature"
# create output folder if it doesn't exist
if not os.path.exists(output_folder):
os.makedirs(output_folder)
# iterate over input folder and process each file
for filename in os.listdir(input_folder):
if filename.endswith(".npy"):
input_path = os.path.join(input_folder, filename)
# load spectrogram and reshape if necessary
spectrogram = torch.from_numpy(np.load(input_path))
if len(spectrogram.shape) == 2:
spectrogram = spectrogram.unsqueeze(0)
# generate audio
audio, sample_rate = diffwave_predict(
spectrogram, model_path, fast_sampling=True
)
# move tensor from GPU to CPU and save to output file
audio = audio.cpu().numpy()
output_path = os.path.join(
output_folder, os.path.splitext(filename)[0] + ".wav"
)
torchaudio.save(output_path, torch.tensor(audio), sample_rate)
# #### Mature
# import os
# import subprocess
# from tqdm import tqdm
# model_path = '/kaggle/working/models/mature/weights-1797.pt'
# input_folder = '/kaggle/working/coconut-signals-generation/mature'
# output_folder = '/kaggle/working/inference/mature'
# # create output folder if it doesn't exist
# if not os.path.exists(output_folder):
# os.makedirs(output_folder)
# # iterate over all npy files in the input folder
# for filename in tqdm(os.listdir(input_folder)):
# if filename.endswith('.npy'):
# # construct paths for input and output files
# input_path = os.path.join(input_folder, filename)
# output_path = os.path.join(output_folder, os.path.splitext(filename)[0] + '.wav')
# # construct command-line arguments
# args = ['python', '-m', 'diffwave.inference', '--fast', model_path, input_path, '-o', output_path]
# # run the command for the current file
# subprocess.run(args, check=True)
# #### Overmature
# import os
# import subprocess
# from tqdm import tqdm
# model_path = '/kaggle/working/models/overmature/weights-2995.pt'
# input_folder = '/kaggle/working/coconut-signals-generation/overmature'
# output_folder = '/kaggle/working/inference/overmature'
# # create output folder if it doesn't exist
# if not os.path.exists(output_folder):
# os.makedirs(output_folder)
# # iterate over all npy files in the input folder
# for filename in tqdm(os.listdir(input_folder)):
# if filename.endswith('.npy'):
# # construct paths for input and output files
# input_path = os.path.join(input_folder, filename)
# output_path = os.path.join(output_folder, os.path.splitext(filename)[0] + '.wav')
# # construct command-line arguments
# args = ['python', '-m', 'diffwave.inference', '--fast', model_path, input_path, '-o', output_path]
# # run the command for the current file
# subprocess.run(args, check=True)
# # Remove all other folders
import os
from tqdm import tqdm
# specify the path to the directory containing the folders
directory_path = "/kaggle/working/"
# specify the names of the folders you want to keep
folder_names_to_keep = ["models", "inference"]
# get a list of all the folders in the directory
folders = os.listdir(directory_path)
# loop over all the folders and delete the ones that are not in the list of folders to keep
for folder in tqdm(folders):
if folder not in folder_names_to_keep:
folder_path = os.path.join(directory_path, folder)
if os.path.isdir(folder_path):
print("Deleting folder:", folder_path)
os.rmdir(folder_path)
|
# # Smart Environment Task
# **IBNU BORISMAN FARREL**
# **20/457751/PA/18789**
# We will be examining a collection of data that specifically pertains to the expenses incurred during the treatment of various patients. The amount spent on treatment is influenced by a variety of factors, such as the location of the clinic, the city of residence, the patient's age, and more. Although we lack data on the patients' diagnoses, we possess other details that could aid us in determining their overall health and utilizing regression analysis. In any event, my hope is for your good health. So let's delve into our data. We will be using linear and decision tree
# # Data Collection
# The quantity & quality of your data dictate how accurate our model is
# The outcome of this step is generally a representation of data which we will use for training
# Using pre-collected data, by way of datasets from Kaggle, UCI, etc., still fits into this step
# Load libraries and the dataset
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn import metrics
import warnings
warnings.filterwarnings("ignore")
# # Importing Data
data = pd.read_csv("../input/insurance.csv")
data.head()
data.dtypes # check datatype
# # Data Preparation
# * age: age of primary beneficiary
# * sex: gender
# * bmi: Body mass index, providing an understanding of body, weights that are relatively high or low relative to height, objective index of body weight (kg / m ^ 2) using the ratio of height to weight, ideally 18.5 to 24.9
# * children: Number of children covered by health insurance / Number of dependents
# * smoker: Smoking
# * region: the beneficiary's residential area in the US, northeast, southeast, southwest, northwest.
# * charges: Individual medical costs billed by health insurance
#
data.mean()
data.corr()
data.count()
data.max()
data.std()
data.isnull().sum(axis=0)
# There no missing value which is good.
data["sex"].unique()
data["smoker"].unique()
data["region"].unique()
from sklearn.preprocessing import LabelEncoder
label = LabelEncoder()
data.sex = label_encoder.fit_transform(data.sex)
data.smoker = label_encoder.fit_transform(data.smoker)
data.region = label_encoder.fit_transform(data.region)
data.dtypes
data.head()
# # Checking Correlation
#
data["sex"] = pd.factorize(data["sex"])[0] + 1
data["region"] = pd.factorize(data["region"])[0] + 1
data["smoker"] = pd.factorize(data["smoker"])[0] + 1
corr = data.corr()
corr["charges"].sort_values(ascending=False)
ax = sns.lmplot(x="age", y="charges", data=data, hue="smoker", palette="Set1")
ax = sns.lmplot(x="bmi", y="charges", data=data, hue="smoker", palette="Set2")
ax = sns.lmplot(x="children", y="charges", data=data, hue="smoker", palette="Set3")
# Smoking has the highest impact on medical costs, even though the costs are growing with age, bmi and children.
fig = plt.figure(figsize=(10, 10))
data_ploting = data.corr(method="pearson")
sns.heatmap(data_ploting, cmap="Reds", linecolor="black", linewidths=2)
plt.show()
# # Splitting data
y = data["charges"]
X = data.drop(["charges"], axis=1)
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.20, random_state=0
)
# # Scalling
scaler = StandardScaler()
scaler.fit(X_train)
X_train_scaled = scaler.transform(X_train)
X_test_scaled = scaler.transform(X_test)
# # Linear Regression
from sklearn.model_selection import train_test_split
from sklearn import linear_model
reg = linear_model.LinearRegression()
reg.fit(X_train, y_train)
y_pred = reg.predict(X_test)
y_pred = pd.DataFrame(y_pred)
MAE_li_reg = metrics.mean_absolute_error(y_test, y_pred)
MSE_li_reg = metrics.mean_squared_error(y_test, y_pred)
RMSE_li_reg = np.sqrt(MSE_li_reg)
pd.DataFrame(
[MAE_li_reg, MSE_li_reg, RMSE_li_reg],
index=["MAE_li_reg", "MSE_li_reg", "RMSE_li_reg"],
columns=["Metrics"],
)
scores = cross_val_score(reg, X_train, y_train, cv=5)
print(np.sqrt(scores))
reg.score(X_test, y_test)
# # Decision Tree Regressor
#
from sklearn.tree import DecisionTreeRegressor
regressor = DecisionTreeRegressor(random_state=0)
regressor.fit(X_train, y_train)
y_pred = regressor.predict(X_test_scaled)
y_pred = pd.DataFrame(y_pred)
MAE_tree_reg = metrics.mean_absolute_error(y_test, y_pred)
MSE_tree_reg = metrics.mean_squared_error(y_test, y_pred)
RMSE_tree_reg = np.sqrt(MSE_tree_reg)
pd.DataFrame(
[MAE_tree_reg, MSE_tree_reg, RMSE_tree_reg],
index=["MAE_tree_reg", "MSE_tree_reg", "RMSE_tree_reg"],
columns=["Metrics"],
)
regressor.score(X_test, y_test)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import matplotlib.pyplot as plt
import seaborn as sns
df = pd.read_csv("/kaggle/input/uber-fares-dataset/uber.csv")
df.sample(20)
df.describe()
df.isnull().sum()
df.info()
df.dropna(axis=0, inplace=True)
df.isnull().sum()
df.columns
df.drop(["Unnamed: 0", "key"], axis=1, inplace=True)
df.columns
df.corr()
# haversine distance
from math import radians
from math import *
def dis(longitude1, latitude1, longitude2, latitude2):
dist = []
for i in range(len(longitude1)):
long1, lat1, long2, lat2 = map(
radians, [longitude1[i], latitude1[i], longitude2[i], latitude2[i]]
)
distlong = long2 - long1
distlat = lat2 - lat1
a = sin(distlat / 2) ** 2 + cos(lat1) * cos(lat2) * sin(distlong / 2) ** 2
c = 2 * asin(sqrt(a)) * 6371
dist.append(c)
return dist
df.columns
df["distance_travelled"] = dis(
df["pickup_longitude"].to_numpy(),
df["pickup_latitude"].to_numpy(),
df["dropoff_longitude"].to_numpy(),
df["dropoff_latitude"].to_numpy(),
)
df.isnull().sum()
df["datetime"] = pd.to_datetime(df["pickup_datetime"])
df["year"] = df["datetime"].dt.year
df["month"] = df["datetime"].dt.month
df["weekday"] = df["datetime"].dt.dayofweek
df["weekdayname"] = df["datetime"].dt.day_name()
df["monthname"] = df["datetime"].dt.month_name()
df["hour"] = df["datetime"].dt.hour
df = df.drop(
[
"pickup_datetime",
"pickup_longitude",
"pickup_latitude",
"dropoff_longitude",
"dropoff_latitude",
],
axis=1,
)
df.info()
import plotly.express as px
figure = px.scatter_3d(
data_frame=df, x="distance_travelled", y="fare_amount", z="month"
)
figure.show()
df["hour"].value_counts(normalize=True).plot.bar()
plt.xticks(rotation=0)
plt.scatter(x=df["weekday"], y=df["distance_travelled"])
plt.scatter(x=df["weekday"], y=df["fare_amount"])
plt.scatter(x=df["hour"], y=df["distance_travelled"])
plt.scatter(x=df["hour"], y=df["fare_amount"])
plt.scatter(x=df["month"], y=df["distance_travelled"])
plt.scatter(x=df["month"], y=df["fare_amount"])
plt.scatter(x=df["year"], y=df["distance_travelled"])
plt.scatter(x=df["year"], y=df["fare_amount"])
|
KFOLD_SOURCES = ["train"]
# ограничение на размер группы мутаций
EXCLUDE_CT_UNDER = 18 # 20
import Bio
import biopandas
from biopandas.pdb import PandasPdb
from Bio.PDB.ResidueDepth import ResidueDepth
from Bio.PDB.PDBParser import PDBParser
from scipy.stats import rankdata, spearmanr
import pandas as pd
pd.options.mode.chained_assignment = None
pd.set_option("display.max_columns", None)
import numpy as np
import sklearn
from colorama import Fore, Back, Style
import math
import time
import io
import os
import gc
import matplotlib.pyplot as plt
from tqdm.notebook import tqdm
tqdm.pandas()
import warnings
warnings.filterwarnings("ignore")
def seed_everything(seed: int):
import random, os
import numpy as np
import torch
random.seed(seed)
os.environ["PYTHONHASHSEED"] = str(seed)
np.random.seed(seed)
torch.manual_seed(seed)
torch.cuda.manual_seed(seed)
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = True
seed_everything(42)
print("\n\n... IMPORTS COMPLETE ...\n")
# Добавление признака записи последовательности с мутацией
def get_mutant_seq(row):
row["sequence"] = row["sequence"].strip(
"X"
) # удаление неопределенных аминокислотных остатков
row["mutant_seq"] = (
row["sequence"][: row.position - 1]
+ row.mutation
+ row["sequence"][row.position :]
)
assert len(row["sequence"]) == len(row["mutant_seq"]), row
return row
# загружаем специальный датасет с данными о месте и виде мутации, ddG и dTm
df = pd.read_csv("../input/14656-unique-mutations-voxel-features-pdbs/dataset.csv")
display(df.head())
print("было", len(df))
# оставляем где ddG отрицательно и dT отрицательно или неопределенно
df = df[((df.ddG < 0)) & ((df.dT < 0) | df.dT.isna())].reset_index(drop=True)
# df = df[(df.ddG < 0) | (df.ddG > 0)].reset_index(drop=True)
# df = df[df.ddG.notna()].reset_index(drop=True)
display(df.head())
print("осталось", len(df))
df["position"] = df["seq_position"].values + 1
df = df.rename({"PDB_chain": "PDB", "mutant": "mutation", "dT": "dTm"}, axis=1)
df = df.drop(["seq_position", "wT"], axis=1)
df = df.apply(get_mutant_seq, axis=1)
df["source"] = "kaggle"
df = df[df.columns]
df["source"] = "train"
# сортировка по названию файла pdb природной последовательности и по позиции мутации
df = df.sort_values(["PDB", "position"]).reset_index(drop=True)
print("Конечный вид")
display(df.head())
# ## Feature Engineer
# Расшифровка однобуквенной кодировки аминокислотных остатков
aa_map = {
"VAL": "V",
"PRO": "P",
"ASN": "N",
"GLU": "E",
"ASP": "D",
"ALA": "A",
"THR": "T",
"SER": "S",
"LEU": "L",
"LYS": "K",
"GLY": "G",
"GLN": "Q",
"ILE": "I",
"PHE": "F",
"CYS": "C",
"TRP": "W",
"ARG": "R",
"TYR": "Y",
"HIS": "H",
"MET": "M",
}
volume = {
"A": 67,
"R": 148,
"N": 96,
"D": 91,
"C": 86,
"E": 109,
"Q": 114,
"G": 48,
"H": 118,
"I": 124,
"L": 124,
"K": 135,
"M": 124,
"F": 135,
"P": 90,
"S": 73,
"T": 93,
"W": 163,
"Y": 141,
"V": 105,
}
hydrophobicity = {
"A": 1.8,
"R": -4.5,
"N": -3.5,
"D": -3.5,
"C": 2.5,
"E": -3.5,
"Q": -3.5,
"G": -0.4,
"H": -3.2,
"I": 4.5,
"L": 3.8,
"K": -3.9,
"M": 1.9,
"F": 2.8,
"P": -1.6,
"S": -0.8,
"T": -0.7,
"W": -0.9,
"Y": -1.3,
"V": 4.2,
}
polarity = {
"A": [0, 0, 0, 1],
"R": [1, 0, 0, 0],
"N": [0, 0, 1, 0],
"D": [0, 1, 0, 0],
"C": [0, 0, 1, 0],
"E": [0, 1, 0, 0],
"Q": [0, 0, 1, 0],
"G": [0, 0, 1, 0],
"H": [1, 0, 0, 0],
"I": [0, 0, 0, 1],
"L": [0, 0, 0, 1],
"K": [1, 0, 0, 0],
"M": [0, 0, 0, 1],
"F": [0, 0, 0, 1],
"P": [0, 0, 0, 1],
"S": [0, 0, 1, 0],
"T": [0, 0, 1, 0],
"W": [0, 0, 0, 1],
"Y": [0, 0, 1, 0],
"V": [0, 0, 0, 1],
}
acid_base = {
"A": [0, 0, 1],
"R": [0, 1, 0],
"N": [1, 0, 0],
"D": [0, 0, 1],
"C": [0, 0, 1],
"E": [0, 0, 1],
"Q": [1, 0, 0],
"G": [0, 0, 1],
"H": [0, 1, 0],
"I": [0, 0, 1],
"L": [0, 0, 1],
"K": [0, 1, 0],
"M": [0, 0, 1],
"F": [0, 0, 1],
"P": [0, 0, 1],
"S": [0, 0, 1],
"T": [0, 0, 1],
"W": [0, 0, 1],
"Y": [0, 0, 1],
"V": [0, 0, 1],
}
def get_new_row(atom_df, j, row, k=25, n=25):
##################
# ATOM_DF - PDB
# J - номер
# ROW - строка
# K - число ближайших аминокислотных остатков
# N - число ближайших остатков с дополнительными признаками
##################
dd = None
tmp = atom_df.loc[(atom_df.residue_number == j)].reset_index(drop=True)
prev = atom_df.loc[(atom_df.residue_number == j - 1)].reset_index(drop=True)
post = atom_df.loc[(atom_df.residue_number == j + 1)].reset_index(drop=True)
# FEATURE ENGINEER
if len(tmp) > 0:
atm = ["N", "H", "CA", "O"]
atom_df = atom_df.loc[atom_df.atom_name.isin(atm)]
# Центр остатка получается путем усреднения трехмерных координат ключевых атомов каждого остатка
central_coord = (
atom_df.groupby("residue_number")["x_coord", "y_coord", "z_coord"]
.mean()
.reset_index(drop=True)
)
residue_names = (
atom_df.groupby("residue_number")["residue_name"]
.unique()
.reset_index(drop=True)
)
mutation_coord = central_coord.loc[row["position"] - 1]
distance = np.zeros((len(central_coord)))
distance = np.sqrt(
np.sum((central_coord.values - mutation_coord.values) ** 2, axis=1)
)
dic = {dist: j for j, dist in enumerate(distance)}
dic = dict(sorted(dic.items()))
k_nearest_aa_distance = np.zeros((k))
k_nearest_aa_position = np.zeros((k))
k_angle1 = []
k_angle2 = []
k_nearest_aa_distance = list(dic.keys())[1 : k + 1]
k_nearest_aa_position = list(dic.values())[1 : k + 1]
relative_distance = (
np.array(k_nearest_aa_distance) / np.array(k_nearest_aa_distance).max()
)
dd = {}
all_angles1 = (mutation_coord.z_coord - central_coord.z_coord) / distance
all_angles2 = (mutation_coord.y_coord - central_coord.y_coord) / distance
all_angles3 = (mutation_coord.x_coord - central_coord.x_coord) / distance
all_angles4 = (mutation_coord.y_coord - central_coord.y_coord) / (
mutation_coord.x_coord - central_coord.x_coord
)
all_angles5 = (mutation_coord.x_coord - central_coord.x_coord) / (
mutation_coord.z_coord - central_coord.z_coord
)
all_angles6 = (mutation_coord.z_coord - central_coord.z_coord) / (
mutation_coord.y_coord - central_coord.y_coord
)
for kk in range(k):
# compute angles
angle1 = math.acos(all_angles1[k_nearest_aa_position[kk]]) * 180 / math.pi
angle2 = math.acos(all_angles2[k_nearest_aa_position[kk]]) * 180 / math.pi
angle3 = math.acos(all_angles3[k_nearest_aa_position[kk]]) * 180 / math.pi
angle4 = math.atan(all_angles4[k_nearest_aa_position[kk]]) * 180 / math.pi
angle5 = math.atan(all_angles5[k_nearest_aa_position[kk]]) * 180 / math.pi
angle6 = math.atan(all_angles6[k_nearest_aa_position[kk]]) * 180 / math.pi
# создание данных
dd[f"nearest_dist_{kk}"] = relative_distance[kk]
dd[f"aver_dist"] = np.array(k_nearest_aa_distance).mean()
dd[f"k_angle1_{kk}"] = angle1
dd[f"k_angle2_{kk}"] = angle2
dd[f"k_angle3_{kk}"] = angle3
dd[f"k_angle4_{kk}"] = angle4
dd[f"k_angle5_{kk}"] = angle5
dd[f"k_angle6_{kk}"] = angle6
name = residue_names[k_nearest_aa_position[kk]]
dd[f"volume_{kk}"] = volume[aa_map[name[0]]]
dd[f"hydr_{kk}"] = hydrophobicity[aa_map[name[0]]]
if kk < n:
dd[f"pol_pos_{kk}"] = polarity[aa_map[name[0]]][0]
dd[f"pol_neg_{kk}"] = polarity[aa_map[name[0]]][1]
dd[f"pol_no_{kk}"] = polarity[aa_map[name[0]]][2]
dd[f"nopol_{kk}"] = polarity[aa_map[name[0]]][3]
dd[f"acidic_{kk}"] = acid_base[aa_map[name[0]]][0]
dd[f"basic_{kk}"] = acid_base[aa_map[name[0]]][1]
dd[f"neutral_{kk}"] = acid_base[aa_map[name[0]]][2]
dd["WT"] = row.wildtype
dd["WT_volume"] = volume[row.wildtype]
dd["WT_hydr"] = hydrophobicity[row.wildtype]
# dd['WT_pol_pos'] = polarity[row.wildtype][0]
# dd['WT_pol_neg'] = polarity[row.wildtype][1]
# dd['WT_pol_no'] = polarity[row.wildtype][2]
# dd['WT_nopol'] = polarity[row.wildtype][3]
dd["WT2"] = tmp.residue_name.map(aa_map)[0]
dd["MUT"] = row.mutation
dd["MUT_volume"] = volume[row.mutation]
dd["MUT_hydr"] = hydrophobicity[row.mutation]
# dd['MUT_pol_pos'] = polarity[row.mutation][0]
# dd['MUT_pol_neg'] = polarity[row.mutation][1]
# dd['MUT_pol_no'] = polarity[row.mutation][2]
# dd['MUT_nopol'] = polarity[row.mutation][3]
dd["multi_hydr"] = hydrophobicity[row.wildtype] * hydrophobicity[row.mutation]
dd["position"] = row.position
dd["relative_position"] = row.position / len(row.sequence)
for i in range(4):
for j in range(4):
dd[f"WT-MUT_pol_{i}_{j}"] = (
polarity[row.wildtype][i] * polarity[row.mutation][j]
)
for i in range(3):
for j in range(3):
dd[f"WT-MUT_ABN_{i}_{j}"] = (
acid_base[row.wildtype][i] * acid_base[row.mutation][j]
)
# 3D расположение мутации
atm = ["N", "H", "CA", "O"]
atoms = atom_df.loc[atom_df.atom_name.isin(atm)]
# средние координаты всех особых атомов в молекуле
centroid1 = np.array(
[atoms.x_coord.mean(), atoms.y_coord.mean(), atoms.z_coord.mean()]
)
# центр меняемого аминокислотного остатка
tmp = tmp.loc[tmp.atom_name.isin(atm)]
centroid2 = np.array(
[tmp.x_coord.mean(), tmp.y_coord.mean(), tmp.z_coord.mean()]
)
dist = centroid2 - centroid1
# расстояние между ними
dd["location3d"] = dist.dot(dist)
# TARGETS AND SOURCES
dd["ddG"] = row.ddG
dd["dTm"] = row.dTm
dd["pdb"] = row.PDB
dd["source"] = row.source
return dd
# ## Преобразование тренировочных данных
from Bio.PDB.SASA import ShrakeRupley
pdb = None
rows = []
bad = []
offsets = []
from biopandas.mmcif import PandasMmcif
for index, row in df.iterrows():
if row.PDB != pdb:
pdb = row.PDB
try:
flag = True
pdb_path = f"../input/14656-unique-mutations-voxel-features-pdbs/pdbs/{row.PDB}/{row.PDB}_relaxed.pdb"
atom_df = PandasPdb().read_pdb(pdb_path)
atom_df = atom_df.df["ATOM"]
except:
flag = False
bad.append(row.PDB)
print(f"{row.PDB} is not found.")
if flag:
try:
dd = get_new_row(atom_df, row.pdb_position, row)
if dd is not None:
rows.append(dd)
except:
print(
f"{row.PDB}_{row.wildtype}{row.pdb_position}{row.mutation}_relaxed.pdb is not found"
)
# ## Создание датасета
train = pd.DataFrame(rows)
train = train.loc[train.WT == train.WT2].reset_index(drop=True)
train["ct"] = train.groupby("pdb").WT.transform("count")
train = train.loc[train.ct > EXCLUDE_CT_UNDER].reset_index(drop=True)
train = train.drop(["WT2", "ct"], axis=1)
print("Размер датасета после удаления малых групп мутаций", train.shape)
"""# создание целевой переменной, ранжирующей изменение термостабильности при мутации
train['target'] = 0.5
for g in train.pdb.unique():
target = 'dTm'
tmp = train.loc[train.pdb==g,'dTm']
if tmp.isna().sum()>len(tmp)/2: target = 'ddG' # если нулевых значений dTm больше половины для
# конкретной природной последовательности, то
# ранжировать по ddG
train.loc[train.pdb==g,'target'] =\
rankdata(train.loc[train.pdb==g,target])/len(train.loc[train.pdb==g,target])
train.head()"""
"""train['target'] = 0.0
for g in train.pdb.unique():
target = 'ddG'
#train.loc[train.pdb==g,'target'] =\
#rankdata(train.loc[train.pdb==g,target])/len(train.loc[train.pdb==g,target])
#train.head()
train.loc[train.pdb==g,'target'] =\
(train.loc[train.pdb==g,target] - train.loc[train.pdb==g,target].mean())/train.loc[train.pdb==g,target].std() + 2*rankdata(train.loc[train.pdb==g,target])/len(train.loc[train.pdb==g,target])
train.head(25)"""
train["target"] = 0.0
for g in train.pdb.unique():
target = "ddG"
# train.loc[train.pdb==g,'target'] =\
# rankdata(train.loc[train.pdb==g,target])/len(train.loc[train.pdb==g,target])
# train.head()
train.loc[train.pdb == g, "target"] = (
train.loc[train.pdb == g, target] - train.loc[train.pdb == g, target].mean()
) / train.loc[train.pdb == g, target].std() + rankdata(
train.loc[train.pdb == g, target]
) / len(
train.loc[train.pdb == g, target]
)
train.head(25)
train = train.loc[train.source.isin(KFOLD_SOURCES)].reset_index(drop=True)
train["group"], _ = train.pdb.factorize()
# признаки - геометрические показатели для K соседних остатков
EXCLUDE = [
"WT",
"MUT",
"prev",
"post",
"ddG",
"dTm",
"pdb",
"source",
"target",
"group",
"oof",
"position",
]
FEATURES = [c for c in train.columns if c not in EXCLUDE]
print(f" {len(FEATURES)} признаков для модели:")
print(FEATURES)
# # XGBoost Model
from sklearn.model_selection import GroupKFold
from sklearn.metrics import mean_squared_error
import xgboost as xgb
print("XGB Version", xgb.__version__)
FOLDS = 11
SEED = 123
xgb_parms = {
"max_depth": 8,
"learning_rate": 0.01,
"subsample": 0.6,
"colsample_bytree": 0.35,
#'n_estimators' : 200,
"eval_metric": "rmse",
"objective": "reg:squarederror",
"random_state": SEED,
}
importances = []
importances2 = []
oof = np.zeros(len(train))
os.system("mkdir xgb3_models")
skf = GroupKFold(n_splits=FOLDS)
for fold, (train_idx, valid_idx) in enumerate(
skf.split(train, train.target, train.group)
):
print("#" * 25)
print("### Fold", fold + 1)
print("### Train size", len(train_idx), "Valid size", len(valid_idx))
print("#" * 25)
X_train = train.loc[train_idx, FEATURES]
y_train = train.loc[train_idx, "target"]
X_valid = train.loc[valid_idx, FEATURES]
y_valid = train.loc[valid_idx, "target"]
dtrain = xgb.DMatrix(data=X_train, label=y_train)
dvalid = xgb.DMatrix(data=X_valid, label=y_valid)
model = xgb.train(
xgb_parms,
dtrain=dtrain,
evals=[(dtrain, "train"), (dvalid, "valid")],
num_boost_round=12000,
early_stopping_rounds=50,
verbose_eval=100,
)
print(model)
model.save_model(f"xgb3_models/XGB_fold{fold}.xgb")
# Создание списков значимости параметров
dd = model.get_score(importance_type="weight")
df = pd.DataFrame({"feature": dd.keys(), f"importance_{fold}": dd.values()})
importances.append(df)
dd = model.get_score(importance_type="gain")
df = pd.DataFrame({"feature": dd.keys(), f"importance_{fold}": dd.values()})
importances2.append(df)
oof_preds = model.predict(dvalid)
rmse = mean_squared_error(y_valid.values, oof_preds, squared=False)
print("RMSE =", rmse, "\n")
oof[valid_idx] = oof_preds
del dtrain, X_train, y_train, dd, df
del X_valid, y_valid, dvalid, model
_ = gc.collect()
print("#" * 25)
rsme = mean_squared_error(train.target.values, oof, squared=False)
print("Итоговое значение RSME =", rsme, "\n")
train["oof"] = oof
# ## Feature Importance
df1 = importances[0].copy()
for k in range(1, FOLDS):
df1 = df1.merge(importances[k], on="feature", how="left")
df1["importance"] = df1.iloc[:, 1:].mean(axis=1)
df1 = df1.sort_values("importance", ascending=False)
NUM_FEATURES = 30
plt.figure(figsize=(10, 5 * NUM_FEATURES // 10))
plt.barh(np.arange(NUM_FEATURES, 0, -1), df1.importance.values[:NUM_FEATURES])
plt.yticks(np.arange(NUM_FEATURES, 0, -1), df1.feature.values[:NUM_FEATURES])
plt.title(f"XGB WEIGHT Feature Importance - Top {NUM_FEATURES}")
plt.show()
df2 = importances2[0].copy()
for k in range(1, FOLDS):
df2 = df2.merge(importances2[k], on="feature", how="left")
df2["importance"] = df2.iloc[:, 1:].mean(axis=1)
df2 = df2.sort_values("importance", ascending=False)
NUM_FEATURES = 30
plt.figure(figsize=(10, 5 * NUM_FEATURES // 10))
plt.barh(np.arange(NUM_FEATURES, 0, -1), df2.importance.values[:NUM_FEATURES])
plt.yticks(np.arange(NUM_FEATURES, 0, -1), df2.feature.values[:NUM_FEATURES])
plt.title(f"XGB GAIN Feature Importance - Top {NUM_FEATURES}")
plt.show()
# # Validate OOF (on either dTm or ddG)
sp = []
sp_dtm = []
sp_ddg = []
for p in train.pdb.unique():
tmp = train.loc[train.pdb == p].reset_index(drop=True)
ttarget = "dTm"
if tmp["dTm"].isna().sum() > 0.3 * len(tmp):
ttarget = "ddG"
print("Protein", p, "has mutation count =", len(tmp), "and target =", ttarget)
r = np.abs(
spearmanr(tmp.oof.values, tmp[ttarget].values, nan_policy="omit").correlation
)
print("Spearman Metric =", r)
sp.append(r)
if ttarget == "dTm":
sp_dtm.append(r)
else:
sp_ddg.append(r)
print()
print("#" * 25)
if len(sp_dtm) > 0:
print(f"Overall Spearman Metric (predicting dTm) =", np.nanmean(sp_dtm))
if len(sp_ddg) > 0:
print(f"Overall Spearman Metric (predicting ddG) =", np.nanmean(sp_ddg))
# # Преобразование тестовых данных
base = "VPVNPEPDATSVENVALKTGSGDSQSDPIKADLEVKGQSALPFDVDCWAILCKGAPNVLQRVNEKTKNSNRDRSGANKGPFKDPQKWGIKALPPKNPSWSAQDFKSPEEYAFASSLQGGTNAILAPVNLASQNSQGGVLNGFYSANKVAQFDPSKPQQTKGTWFQITKFTGAAGPYCKALGSNDKSVCDKNKNIAGDWGFDPAKWAYQYDEKNNKFNYVGK"
len(base)
# Загрузка
test = pd.read_csv("../input/novozymes-enzyme-stability-prediction/test.csv")
deletions = test.loc[test.protein_sequence.str.len() == 220, "seq_id"].values
print("Test shape", test.shape)
test.head()
pdb_path = "../input/nesp-test-wildtype-pdb/model.pdb"
atom_df = PandasPdb().read_pdb("../input/nesp-test-wildtype-pdb/model.pdb")
atom_df = atom_df.df["ATOM"]
atom_df.head()
def get_test_mutation(row):
for i, (a, b) in enumerate(zip(row.protein_sequence, base)):
if a != b:
break
row["wildtype"] = base[i]
row["mutation"] = row.protein_sequence[i]
row["position"] = i + 1
return row
test = test.apply(get_test_mutation, axis=1)
test["ddG"] = np.nan
test["dTm"] = np.nan
test["CIF"] = None
test["sequence"] = base
test = test.rename({"protein_sequence": "mutant_seq"}, axis=1)
test["source"] = "test"
test["PDB"] = "kaggle"
rows = []
print(f"Extracting embeddings and feature engineering {len(test)} test rows...")
for index, row in test.iterrows():
if index % 100 == 0:
print(index, ", ", end="")
j = row.position
dd = get_new_row(atom_df, j, row)
rows.append(dd)
test = pd.DataFrame(rows)
test.head(20)
# ## Результат моделей для тестовых данных
# TEST DATA FOR XGB
X_test = test[FEATURES]
dtest = xgb.DMatrix(data=X_test)
# INFER XGB MODELS ON TEST DATA
model = xgb.Booster()
model.load_model(f"xgb3_models/XGB_fold0.xgb")
preds = model.predict(dtest)
for f in range(1, FOLDS):
model.load_model(f"xgb3_models/XGB_fold{f}.xgb")
preds += model.predict(dtest)
preds /= FOLDS
plt.hist(preds, bins=100)
plt.title("Test preds histogram", size=16)
plt.show()
sub = pd.read_csv(
"../input/novozymes-enzyme-stability-prediction/sample_submission.csv"
)
sub.tm = preds
# обучающие данные не содержали примеров удаления остатка,
# предсказанные результаты для таких случаев усредняются
sub.loc[sub.seq_id.isin(deletions), "tm"] = sub.loc[
~sub.seq_id.isin(deletions), "tm"
].mean()
sub.to_csv(f"XGB3_submission.csv", index=False)
sub.head(20)
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import os
import gc
import tensorflow as tf
from tensorflow import keras
from keras import models, layers
from tensorflow.python.ops.numpy_ops import np_config
import warnings
warnings.filterwarnings("ignore")
np_config.enable_numpy_behavior()
import tensorflow as tf
import time
import os
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from tensorflow.keras.preprocessing.text import Tokenizer
from tensorflow.keras.layers import (
Embedding,
MultiHeadAttention,
Dense,
Input,
Dropout,
LayerNormalization,
)
from tensorflow.keras.preprocessing.sequence import pad_sequences
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
en_df = pd.read_csv(
"/kaggle/input/english-to-french/small_vocab_en.csv", header=None, usecols=[0]
)
fr_df = pd.read_csv(
"/kaggle/input/english-to-french/small_vocab_fr.csv", header=None, usecols=[0]
)
en_df.head()
fr_df.head()
# # Use just portion of the dataset
english_sentences = en_df[0].values
french_sentences = fr_df[0].values
print(f"the first sentence in english: {english_sentences[0]}")
print(f"the first sentence in french: {french_sentences[0]}")
for i in range(len(english_sentences)):
english_sentences[i] = "sos " + english_sentences[i] + " eos."
french_sentences[i] = "sos " + french_sentences[i] + " eos."
# # tokenization
num_words = 100000
tokenizer_en = Tokenizer(
num_words=num_words, filters='!#$%&()*+,-/:;<=>@«»""[\\]^_`{|}~\t\n'
)
tokenizer_en.fit_on_texts(english_sentences)
english_sentences = tokenizer_en.texts_to_sequences(english_sentences)
word_index = tokenizer_en.word_index
print(f"The number of words in the English vocabulary: {len(word_index)}")
tokenizer_fr = Tokenizer(
num_words=num_words, filters='!#$%&()*+,-/:;<=>@«»""[\\]^_`{|}~\t\n'
)
tokenizer_fr.fit_on_texts(french_sentences)
french_sentences = tokenizer_fr.texts_to_sequences(french_sentences)
word_index_fr = tokenizer_fr.word_index
print(f"The number of words in the French vocabulary: {len(word_index_fr)}")
english_sentences = pad_sequences(
english_sentences, maxlen=30, padding="post", truncating="post"
)
french_sentences = pad_sequences(
french_sentences, maxlen=30, padding="post", truncating="post"
)
print(f"the first sentence in english: {english_sentences[0]}")
print(f"the first sentence in french: {french_sentences[0]}")
test_en = tokenizer_en.sequences_to_texts(english_sentences[:5])
test_en
def get_angles(pos, i, d_model):
"""
Function to compute the angles for positional encoding.
Returns the angle computed
"""
angle_rates = 1 / np.power(10000, (2 * (i // 2)) / np.float32(d_model))
return pos * angle_rates
def positional_encoding(position, d_model):
"""
Adds positional encoding to the Embeddings to be fed to the Transformer model.
Computes a sin and cos of the angles determined by the get_angles() function
and adds the value computed to an axis of the embeddings.
"""
angle_rads = get_angles(
np.arange(position)[:, np.newaxis], np.arange(d_model)[np.newaxis, :], d_model
)
# apply sin to even indices in the array. ie 2i
angle_rads[:, 0::2] = np.sin(angle_rads[:, 0::2])
# apply cos to odd indices in the array. ie 2i+1
angle_rads[:, 1::2] = np.cos(angle_rads[:, 1::2])
pos_encoding = angle_rads[np.newaxis, ...]
return tf.cast(pos_encoding, dtype=tf.float32)
def scaled_dot_product_attention(q, k, v, mask):
"""
Computes the attention weight for the q, k, v vectors
Attention in the transformer is popularly known as self-attention because the q, k, v vectors are
sourced from the same sequence. Self Attention, also called intra Attention, is an attention mechanism relating
different positions of a single sequence in order to compute a representation of the same sequence.
q, k, v must have leading dimensions - same 'batch_size'
k, v must have matching penultimate dimension, i.e.: seq_len_k = seq_len_v
q - query vectors; shape == (..., seq_len_q, depth)
k - key vectors; shape == (..., seq_len_k, depth)
v - value vectors; shape == (..., seq_len_v, depth_v)
Returns - attention weights, output
"""
matmul_qk = tf.matmul(q, k, transpose_b=True)
dk = tf.cast(tf.shape(k)[-1], dtype=tf.float32)
scaled_dk = tf.math.sqrt(dk)
scaled_attention_logits = matmul_qk / scaled_dk
# add mask
if mask is not None:
scaled_attention_logits += mask * -1e9
# normalize with softmax
attention_weights = tf.nn.softmax(scaled_attention_logits, axis=-1)
output = tf.matmul(attention_weights, v)
return output, attention_weights
class MultiHeadAttention(tf.keras.layers.Layer):
"""Computes the attention for several heads in the transformer"""
def __init__(self, key_dim, num_heads, dropout_rate=0.0):
super(MultiHeadAttention, self).__init__()
self.num_heads = num_heads
self.key_dim = key_dim
assert (
key_dim % num_heads == 0
) # this ensures the dimension of the embedding can be evenly split across attention heads
self.depth = self.key_dim // self.num_heads
self.wq = tf.keras.layers.Dense(key_dim)
self.wk = tf.keras.layers.Dense(key_dim)
self.wv = tf.keras.layers.Dense(key_dim)
self.dropout = tf.keras.layers.Dropout(dropout_rate)
self.dense = tf.keras.layers.Dense(key_dim)
def split_heads(self, x, batch_size):
"""Split the last dimension into (num_heads, depth)
Transpose the result such that the shape is (batch_size, num_heads, seq_len, depth)
"""
x = tf.reshape(x, (batch_size, -1, self.num_heads, self.depth))
return tf.transpose(x, perm=[0, 2, 1, 3])
def call(self, v, k, q, mask=None):
batch_size = tf.shape(q)[0]
# Dense on the q, k, v vectors
q = self.wq(q)
k = self.wk(k)
v = self.wv(v)
# split the heads
q = self.split_heads(q, batch_size)
k = self.split_heads(k, batch_size)
v = self.split_heads(v, batch_size)
# compute attention weights
scaled_attention, attention_weights = scaled_dot_product_attention(
q, k, v, mask
)
scaled_attention = tf.transpose(scaled_attention, perm=[0, 2, 1, 3])
# reshape and add Dense layer
concat_attention = tf.reshape(scaled_attention, (batch_size, -1, self.key_dim))
output = self.dense(concat_attention)
output = self.dropout(output)
return output, attention_weights
def get_config(self):
"""Implement serialization in order to save the model"""
config = super(MultiHeadAttention).get_config()
config.update(
{
"key_dim": self.key_dim,
"num_heads": self.num_heads,
"dropout_rate": self.dropout.rate,
}
)
return config
class EncoderLayer(tf.keras.layers.Layer):
def __init__(self, d_model, num_heads, dff, rate=0.1):
"""Initializes the encoder layer
Args:
d_model: depth of the transformer model
num_heads: number of heads for multi-head attention
dff: depth of the feed forward network
rate: dropout rate for training
"""
super(EncoderLayer, self).__init__()
self.multihead = MultiHeadAttention(d_model, num_heads, rate)
self.layernorm1 = tf.keras.layers.LayerNormalization(epsilon=1e-6)
self.layernorm2 = tf.keras.layers.LayerNormalization(epsilon=1e-6)
self.dropout1 = tf.keras.layers.Dropout(rate)
self.dropout2 = tf.keras.layers.Dropout(rate)
self.ffn = pointwise_feed_forward_network(d_model, dff)
def __call__(self, x, training, mask):
attn_output, _ = self.multihead(x, x, x, mask)
attn_output = self.dropout1(attn_output, training=training)
out1 = self.layernorm1(attn_output + x)
ffn_output = self.ffn(out1)
ffn_output = self.dropout2(ffn_output, training=training)
out2 = self.layernorm2(ffn_output + x)
return out2
def get_config(self):
config = super(EncoderLayer, self).get_config()
class DecoderLayer(tf.keras.layers.Layer):
def __init__(self, d_model, num_heads, dff, rate=0.1):
super(DecoderLayer, self).__init__()
self.multihead1 = MultiHeadAttention(d_model, num_heads, rate)
self.multihead2 = MultiHeadAttention(d_model, num_heads, rate)
self.ffn = pointwise_feed_forward_network(d_model, dff)
self.layernorm1 = layers.LayerNormalization(epsilon=1e-6)
self.layernorm2 = layers.LayerNormalization(epsilon=1e-6)
self.layernorm3 = layers.LayerNormalization(epsilon=1e-6)
# self.dropout1 = layers.Dropout(rate)
# self.dropout2 = layers.Dropout(rate)
self.dropout3 = layers.Dropout(rate)
def __call__(self, x, enc_output, training, look_ahead_mask, padding_mask):
"""
x - query vector for the Decoder
enc_output - a set of attention vectors k and v from the top Encoder layer
training - mode for Dropout
look_ahead_mask/padding_mask - required for MultiHeadAttention
"""
attn1, attn_weights_block1 = self.multihead1(x, x, x, look_ahead_mask)
# attn1 = self.dropout1(attn1, training = training)
out1 = self.layernorm1(attn1 + x)
attn2, attn_weights_block2 = self.multihead2(
enc_output, enc_output, out1, padding_mask
)
# attn2 = self.dropout2(attn2, training = training)
out2 = self.layernorm2(attn2 + out1)
ffn_output = self.ffn(out2)
ffn_output = self.dropout3(ffn_output, training=training)
out3 = self.layernorm3(ffn_output + out2)
return out3, attn_weights_block1, attn_weights_block2
class Encoder(tf.keras.layers.Layer):
def __init__(
self,
num_layers,
d_model,
num_heads,
dff,
input_vocab_size,
maximum_position_encoding,
rate=0.1,
):
super(Encoder, self).__init__()
self.num_layers = num_layers
self.d_model = d_model
self.embedding = layers.Embedding(input_vocab_size, d_model)
self.pos_encoding = positional_encoding(maximum_position_encoding, d_model)
self.enc_layers = [
EncoderLayer(d_model, num_heads, dff, rate) for _ in range(num_layers)
]
self.dropout = layers.Dropout(rate)
def __call__(self, inputs, training, mask):
seq_len = tf.shape(inputs)[1]
# embed and add positonal encoding
inputs = self.embedding(inputs)
inputs *= tf.math.sqrt(
tf.cast(self.d_model, tf.float32)
) # tf.math.sqrt doesn't support int types
inputs += self.pos_encoding[:, :seq_len, :]
inputs = self.dropout(inputs, training=training)
for i in range(self.num_layers):
inputs = self.enc_layers[i](inputs, training, mask)
return inputs
class Decoder(tf.keras.layers.Layer):
def __init__(
self,
num_layers,
d_model,
num_heads,
dff,
target_vocab_size,
maximum_position_encoding,
rate=0.1,
):
super(Decoder, self).__init__()
self.num_layers = num_layers
self.d_model = d_model
self.embedding = layers.Embedding(target_vocab_size, d_model)
self.pos_encoding = positional_encoding(maximum_position_encoding, d_model)
self.dec_layers = [
DecoderLayer(d_model, num_heads, dff, rate=0.1) for _ in range(num_layers)
]
self.dropout = layers.Dropout(rate)
def __call__(self, x, enc_output, training, look_ahead_mask, padding_mask):
seq_len = tf.shape(x)[1]
attention_weights = {}
# add embedding and positional encoding
x = self.embedding(x)
x *= tf.math.sqrt(tf.cast(self.d_model, tf.float32))
x += self.pos_encoding[:, :seq_len, :]
x = self.dropout(x, training=training)
for i in range(self.num_layers):
x, block1, block2 = self.dec_layers[i](
x, enc_output, training, look_ahead_mask, padding_mask
)
attention_weights[f"decoder_layer{i + 1}_block1"] = block1
attention_weights[f"decoder_layer{i + 1}_block2"] = block2
return x, attention_weights
class Transformer(tf.keras.Model):
def __init__(
self,
num_layers,
d_model,
num_heads,
dff,
input_vocab_size,
target_vocab_size,
pe_input,
pe_target,
rate=0.1,
):
super(Transformer, self).__init__()
self.encoder = Encoder(
num_layers, d_model, num_heads, dff, input_vocab_size, pe_input, rate
)
self.decoder = Decoder(
num_layers, d_model, num_heads, dff, input_vocab_size, pe_target, rate
)
self.final_layer = tf.keras.layers.Dense(target_vocab_size)
def get_config(self):
"""Implement serialization in order to save model"""
config = super(Transformer, self).get_config()
config.update(
{
"encoder": self.encoder,
"decoder": self.decoder,
"final_layer": self.final_layer,
}
)
def __call__(
self, inp, tar, training, enc_padding_mask, look_ahead_mask, dec_padding_mask
):
enc_output = self.encoder(inp, training, enc_padding_mask)
dec_output, attention_weights = self.decoder(
tar, enc_output, training, look_ahead_mask, dec_padding_mask
)
final_output = self.final_layer(dec_output)
return final_output, attention_weights
def create_padding_mask(seq):
seq = tf.cast(tf.math.equal(seq, 0), tf.float32)
# add extra dimension to add padding to the attention logits
return seq[:, tf.newaxis, tf.newaxis, :] # (batch_size, 1, 1, seq_len)
def create_look_ahead_mask(size):
mask = 1 - tf.linalg.band_part(tf.ones((size, size)), -1, 0)
return mask # (seq_len, seq_len)
def create_masks(inp, tar):
# encoder padding mask (also necessary for 2nd attention block in the decoder)
enc_padding_mask = create_padding_mask(inp)
# dec padding mask - used in the 2nd attention block in the decoder
dec_padding_mask = create_padding_mask(inp)
# used in the first attention block
# used to pad and mask future tokens in the tokens received by the decoder
look_ahead_mask = create_look_ahead_mask(tf.shape(tar)[1])
dec_target_padding_mask = create_padding_mask(tar)
combined_mask = tf.maximum(dec_target_padding_mask, look_ahead_mask)
return enc_padding_mask, combined_mask, dec_padding_mask
d_model = 128
dff = 512
num_layers = 4
num_heads = 8
dropout_rate = 0.1
input_vocab_size = len(tokenizer_fr.word_index) + 2 # french is the input
target_vocab_size = len(tokenizer_en.word_index) + 2 # english is the target
pe_input = input_vocab_size
pe_target = target_vocab_size
EPOCHS = 10
batch_size = 64
# learning rate
class CustomSchedule(tf.keras.optimizers.schedules.LearningRateSchedule):
def __init__(self, d_model, warmup_steps=4000):
super(CustomSchedule, self).__init__()
self.embedding_dim = tf.cast(d_model, dtype=tf.float32)
self.warmup_steps = tf.cast(warmup_steps, dtype=tf.float32)
def __call__(self, step):
step = tf.cast(step, dtype=tf.float32)
arg1 = tf.math.rsqrt(step)
arg2 = step * (self.warmup_steps**-1.5)
return tf.math.rsqrt(self.embedding_dim) * tf.math.minimum(arg1, arg2)
learning_rate = CustomSchedule(d_model)
def pointwise_feed_forward_network(d_model, dff):
model = tf.keras.Sequential(
[tf.keras.layers.Dense(dff, activation="relu"), tf.keras.layers.Dense(d_model)]
)
return model
transformer = Transformer(
num_layers,
d_model,
num_heads,
dff,
input_vocab_size,
target_vocab_size,
pe_input,
pe_target,
dropout_rate,
)
optimizer = tf.keras.optimizers.Adam(
learning_rate, beta_1=0.9, beta_2=0.98, epsilon=1e-9
)
loss_object = tf.keras.losses.SparseCategoricalCrossentropy(
from_logits=True, reduction="none"
)
def loss_function(real, pred):
mask = tf.math.logical_not(tf.math.equal(real, 0))
loss_ = loss_object(real, pred)
mask = tf.cast(mask, dtype=loss_.dtype)
loss_ *= mask
return tf.reduce_sum(loss_) / tf.reduce_sum(mask)
def accuracy_function(real, pred):
accuracies = tf.equal(real, tf.argmax(pred, axis=2))
mask = tf.math.logical_not(tf.math.equal(real, 0))
accuracies = tf.math.logical_and(mask, accuracies)
accuracies = tf.cast(accuracies, dtype=tf.float32)
mask = tf.cast(mask, dtype=tf.float32)
return tf.reduce_sum(accuracies) / tf.reduce_sum(mask)
train_loss = tf.keras.metrics.Mean(name="train_loss")
train_accuracy = tf.keras.metrics.SparseCategoricalAccuracy(name="train_accuracy")
# checkpointing
checkpoint_path = "./train"
ckpt = tf.train.Checkpoint(transformer=transformer, optimizer=optimizer)
ckpt_manager = tf.train.CheckpointManager(ckpt, checkpoint_path, max_to_keep=3)
# if a checkpoint exists, restore the latest checkpoint.
if ckpt_manager.latest_checkpoint:
ckpt.restore(ckpt_manager.latest_checkpoint)
print("Latest checkpoint restored!!")
# the train function
train_step_signature = [
tf.TensorSpec(shape=(batch_size, 30), dtype=tf.int64),
tf.TensorSpec(shape=(batch_size, 30), dtype=tf.int64),
]
@tf.function(input_signature=train_step_signature)
def train_step(inp, tar):
tar_inp = tar[:, :-1]
tar_real = tar[:, 1:]
enc_padding_mask, combined_mask, dec_padding_mask = create_masks(inp, tar_inp)
with tf.GradientTape() as tape:
predictions, _ = transformer(
inp, tar_inp, True, enc_padding_mask, combined_mask, dec_padding_mask
)
loss = loss_function(tar_real, predictions)
gradients = tape.gradient(loss, transformer.trainable_variables)
optimizer.apply_gradients(zip(gradients, transformer.trainable_variables))
train_loss(loss)
train_accuracy(tar_real, predictions)
# import time
# tf.function(experimental_relax_shapes=True)
# for epoch in range(1):
# start = time.time()
# train_loss.reset_states()
# train_accuracy.reset_states()
# start_pt = 0
# # inp -> french, tar -> english
# for i in range(int(len(english_sentences)/batch_size)):
# inp = tf.convert_to_tensor(np.array(french_sentences[start_pt:start_pt+batch_size]),dtype=tf.int64)
# tar = tf.convert_to_tensor(np.array(english_sentences[start_pt:start_pt+batch_size]),dtype=tf.int64)
# start_pt = start_pt + batch_size
# train_step(inp, tar)
# if i % 100 == 0:
# print ('Epoch {} Batch {} Loss {:.4f} Accuracy {:.4f}'.format(epoch + 1, i, train_loss.result(), train_accuracy.result()))
# if (epoch + 1) % 5 == 0:
# ckpt_save_path = ckpt_manager.save()
# print ('Saving checkpoint for epoch {} at {}'.format(epoch+1, ckpt_save_path))
# print ('Epoch {} Loss {:.4f} Accuracy {:.4f}'.format(epoch + 1, train_loss.result(), train_accuracy.result()))
# print ('Total time taken for that epoch: {} secs\n'.format(time.time() - start))
maxlen = 25
def evaluate(sentence):
sentence = "sos " + sentence[0] + " eos."
sentence = [sentence] # done because of the way TensorFlow's tokenizer
# vectorize and pad the sentence
sentence = tokenizer_fr.texts_to_sequences(sentence)
sentence = pad_sequences(sentence, maxlen=30, padding="post", truncating="post")
inp = tf.convert_to_tensor(
np.array(sentence), dtype=tf.int64
) # convert input to tensors
# tokenize the start of the decoder input & convert to tensor
decoder_input = tokenizer_en.texts_to_sequences(["sos"])
decoder_input = tf.convert_to_tensor(np.array(decoder_input), dtype=tf.int64)
for i in range(maxlen):
enc_padding_mask, combined_mask, dec_padding_mask = create_masks(
inp, decoder_input
)
predictions, _ = transformer(
inp, decoder_input, False, enc_padding_mask, combined_mask, dec_padding_mask
)
# select the last word from the seq_len dimension
predictions = predictions[:, -1:, :]
predicted_id = tf.cast(tf.argmax(predictions, axis=-1), tf.int64)
# return the result if the predicted_id is equal to the end token
if predicted_id == tokenizer_en.texts_to_sequences(["eos"]):
return tf.squeeze(decoder_input, axis=0)
# concatentate the predicted_id to the output which is given to the decoder
# as its input.
decoder_input = tf.concat([decoder_input, predicted_id], axis=1)
return tf.squeeze(decoder_input, axis=0)
def translate(sentence):
sentence = [sentence] # our evaluate function requires lists
print("Input: {}".format(sentence[0]))
print("Please wait while we translate: \n")
result = (evaluate(sentence)).tolist()
predicted_sentence = tokenizer_en.sequences_to_texts(
[
[
i
for i in result
if i != tokenizer_en.texts_to_sequences(["sos"])[0][0]
and i != tokenizer_en.texts_to_sequences(["eos."])[0][0]
]
]
)
print("Predicted translation: {}".format(predicted_sentence[0]))
sentence = "new jersey est parfois calme pendant l' automne"
translate(sentence)
sentence = "we vous banana."
translate(sentence)
# ## Positional Encoding
# **When working with sequence to sequence tasks, the order of the data is crucial. While training RNNs, the input order is preserved automatically. However, when training Transformer networks, all data is input at once, and there's no inherent order information. To overcome this, positional encoding is used to specify the position of each input in the sequence. This encoding is achieved through sine and cosine formulas as follows:**
# $$
# PE_{(pos, 2i)}= sin\left(\frac{pos}{{10000}^{\frac{2i}{d}}}\right)
# \tag{1}$$
# $$PE_{(pos, 2i+1)}= cos\left(\frac{pos}{{10000}^{\frac{2i}{d}}}\right)
# \tag{2}$$
# **Here, $pos$ refers to the position of the input in the sequence, $i$ refers to the index of the dimension in the embedding vector, and $d$ refers to the dimensionality of the model.**
# Generate positional encodings
pos_encodings = positional_encoding(100, 128)
# Visualize the encodings as a heatmap
plt.figure(figsize=(10, 6))
sns.heatmap(pos_encodings[0], cmap="viridis")
plt.xlabel("Embedding Dimension")
plt.ylabel("Position in Sequence")
plt.title("Positional Encodings")
plt.show()
|
import pandas as pd
import numpy as np
import pickle5
import re
import emoji
import csv
import seaborn as sns
import matplotlib as mpl
import matplotlib.pyplot as plt
import sklearn
from sklearn.model_selection import train_test_split
from sklearn.model_selection import cross_val_score
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.neighbors import KNeighborsClassifier
from sklearn.metrics import accuracy_score
from sklearn.metrics import f1_score
from sklearn.metrics import classification_report
df = pd.read_csv("/kaggle/input/assign8/hindi_train_val.csv")
def extract_emojis(s):
return "".join((" " + c + " ") if c in emoji.UNICODE_EMOJI["en"] else c for c in s)
# @title Data Preprocessing
df["text"] = df["text"].apply(extract_emojis)
for ind in df.index:
normal_string = df["text"][ind]
special_characters = ["@", "#", "$", "&", ".", "_", "!", '"']
for idx in special_characters:
normal_string = normal_string.replace(idx, "")
df.at[ind, "text"] = normal_string
for ind in df.index:
special_string = df["text"][ind]
sample_list = []
for idx in range(0, len(special_string)):
if special_string[idx] == "*":
if idx == 0:
if special_string[idx + 1] == "*":
sample_list.append(special_string[idx])
elif idx == len(special_string) - 1:
if special_string[idx - 1] == "*":
sample_list.append(special_string[idx])
elif (special_string[idx - 1] == "*") or (special_string[idx + 1] == "*"):
sample_list.append(special_string[idx])
else:
sample_list.append(special_string[idx])
normal_string = "".join(sample_list)
df.at[ind, "text"] = normal_string
for ind in df.index:
normal_string = df["text"][ind]
str_ = re.sub("[\*]+", " **", normal_string)
df.at[ind, "text"] = str_
# @title TfIdf
df1 = df.copy()
v = TfidfVectorizer()
Fit = v.fit(df1["text"])
x = Fit.transform(df1["text"])
X = pd.DataFrame(x.toarray(), columns=v.get_feature_names_out())
y = df1["label"]
# @title Predict K using cross validation
# k_values = [i for i in range (1,31)]
# scores = []
# for k in k_values:
# knn = KNeighborsClassifier(n_neighbors = k, weights = 'distance')
# score = cross_val_score(knn, X, y, cv = 5)
# scores.append(np.mean(score))
max_k = 8 # scores.index(max(scores)) + 1
# @title KNN
knn = KNeighborsClassifier(n_neighbors=max_k, weights="distance")
knn.fit(X, y)
# @title Save KNN model
pickle5.dump(knn, open("knn.pkl", "wb"))
# rango = ['मादरचोद कौन जाएगा रहेगा आ जाएगा दिन में', 'दीदी कास्ट है घरवाला रेडी नहीं होंगे', 'भोसड़ी के कितना मैंने भी सच्चा प्यार किया']
X_test = pd.read_csv("/kaggle/input/assignment8/hindi_test.csv") # path of testing file
# comp = pd.read_csv('/kaggle/input/assign8/hindi_train_val.csv')
# X_test.columns = ['text']
# X_test.drop(['index'], axis=1)
# print(X_test.info())
X_test["text"] = X_test["text"].apply(extract_emojis)
for ind in X_test.index:
normal_string = X_test["text"][ind]
special_characters = ["@", "#", "$", "&", ".", "_", "!", '"']
for idx in special_characters:
normal_string = normal_string.replace(idx, "")
X_test.at[ind, "text"] = normal_string
for ind in X_test.index:
special_string = X_test["text"][ind]
sample_list = []
for idx in range(0, len(special_string)):
if special_string[idx] == "*":
if idx == 0:
if special_string[idx + 1] == "*":
sample_list.append(special_string[idx])
elif idx == len(special_string) - 1:
if special_string[idx - 1] == "*":
sample_list.append(special_string[idx])
elif (special_string[idx - 1] == "*") or (special_string[idx + 1] == "*"):
sample_list.append(special_string[idx])
else:
sample_list.append(special_string[idx])
normal_string = "".join(sample_list)
X_test.at[ind, "text"] = normal_string
for ind in X_test.index:
normal_string = X_test["text"][ind]
str_ = re.sub("[\*]+", " **", normal_string)
X_test.at[ind, "text"] = str_
X_test1 = X_test.copy()
sin = Fit.transform(X_test1["text"])
X_new = pd.DataFrame(sin.toarray(), columns=v.get_feature_names_out())
# @title Load KNN model
KNNmodel = pickle5.load(open("/kaggle/working/knn.pkl", "rb"))
# @title Predict KNN
pred = KNNmodel.predict(X_new)
CSV_list1 = []
for i in pred:
CSV_list1.append({"label": i})
filename = "KNN_result.csv"
with open(filename, "w") as csvfile:
writer = csv.DictWriter(csvfile, fieldnames=["label"])
writer.writeheader()
writer.writerows(CSV_list1)
accuracy = accuracy_score(X_test["label"], pred)
print("Accuracy:", accuracy)
f1_score_ = f1_score(X_test["label"], pred, average="macro")
print("macro f1-score:", f1_score_)
X_test = pd.read_csv("/kaggle/input/assignment8/hindi_test.csv") # path of testing file
comp = pd.read_csv("/kaggle/input/assign8/hindi_train_val.csv")
# @ Calculate intersection
merge_count = comp.merge(X_test, on="text").shape[0]
print(merge_count)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import os
import seaborn as sns
sns.set()
# Setup feedback system
from learntools.core import binder
binder.bind(globals())
from learntools.computer_vision.ex1 import *
# Imports
import os, warnings
import matplotlib.pyplot as plt
from matplotlib import gridspec
import numpy as np
import tensorflow as tf
from tensorflow.keras.preprocessing import image_dataset_from_directory
# Reproducability
def set_seed(seed=31415):
np.random.seed(seed)
tf.random.set_seed(seed)
os.environ["PYTHONHASHSEED"] = str(seed)
os.environ["TF_DETERMINISTIC_OPS"] = "1"
set_seed()
# Set Matplotlib defaults
plt.rc("figure", autolayout=True)
plt.rc(
"axes",
labelweight="bold",
labelsize="large",
titleweight="bold",
titlesize=18,
titlepad=10,
)
plt.rc("image", cmap="magma")
warnings.filterwarnings("ignore") # to clean up output cells
train_df = image_dataset_from_directory(
"/kaggle/input/code-to-cure-10/Training",
labels="inferred",
label_mode="int",
image_size=[512, 512],
interpolation="nearest",
batch_size=64,
shuffle=True,
)
|
# # Overview
#
# Abstract:
# - This notebook present a beginner approach to episode 12 of season 3 playground series which is about Kidney Stone Prediction based on Urine Analysis.
# - Using StandardScaler works better than MinMaxScaler and gives a better ROC score.
# - Introducing engineered features helped in increasing the LB scores.
# - Outliers were removed from the dataset.
# - Random Forest Classifier seems like the best choice for the auc-roc score metrics and further tunning should be conducted.
# - Using Kfold cross validation in higher values of splits increase the LB score to some extend but computational costs were high.
#
# About Dataset:
# - Original dataset consists of 79 urine specimens, to determine if certain physical characteristics of the urine might be related to the formation of calcium oxalate crystals.
# - The six physical characteristics of the urine are:
# - **(1) specific gravity**, the density of the urine relative to water;
# - **(2) pH**, the negative logarithm of the hydrogen ion;
# - **(3) osmolarity (mOsm)**, a unit used in biology and medicine but not in physical chemistry. Osmolarity is proportional to the concentration of molecules in solution;
# - **(4) conductivity (mMho milliMho)**, One Mho is one reciprocal Ohm. Conductivity is proportional to the concentration of charged ions in solution;
# - **(5) urea concentration in millimoles per litre**;
# - **(6) calcium concentration (CALC) in millimolesllitre**.
# - The data is obtained from @'Physical Characteristics of Urines With and Without Crystals',a chapter from Springer Series in Statistics.
# # Outline
# * [ 1. Import dataset and libraries ](#1)
# * [ 2. EDA](#2)
# * [2.1 Data summary](#2.1)
# * [2.2 Feature distribution](#2.2)
# * [2.3 Target distribution](#2.3)
# * [2.4 Correlation Matrix](#2.4)
# * [2.5 Outliers](#2.5)
# * [ 3. Data Preparation](#3)
# * [ 4. Modeling](#4)
# * [4.1 Baseline Random Forest Classifier Model with all features](#4.1)
# * [4.2 Comparison between different models with All features](#4.2)
# * [4.3 Simple Stacking of classifiers](#4.3)
# * [ 5. Submission](#5)
# # 1. Import dataset and libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import copy
# Data manipulation libraries
from sklearn.preprocessing import MinMaxScaler, StandardScaler
from sklearn.model_selection import (
train_test_split,
cross_val_score,
KFold,
StratifiedKFold,
)
# Model evaluaion libraries
from sklearn.metrics import (
roc_auc_score,
roc_curve,
accuracy_score,
classification_report,
)
# Classiication Models libraries
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import RandomForestClassifier
from xgboost import XGBClassifier, plot_importance
from sklearn.svm import SVC
from sklearn.neighbors import KNeighborsClassifier
from sklearn.naive_bayes import GaussianNB
from catboost import CatBoostClassifier
from mlxtend.classifier import StackingCVClassifier
# Remove warnings
import warnings
warnings.simplefilter(action="ignore", category=FutureWarning)
RANDOM_STATE = 42
# Playground dataset train & test
P_train_df = pd.read_csv("/kaggle/input/playground-series-s3e12/train.csv")
test_df = pd.read_csv("/kaggle/input/playground-series-s3e12/test.csv")
# Original dataset train & test
O_train_df = pd.read_csv(
"/kaggle/input/kidney-stone-prediction-based-on-urine-analysis/kindey stone urine analysis.csv"
)
# # Playground dataset train & test
# P_train_df = pd.read_csv('Data/Input/Playground/train.csv')
# test_df = pd.read_csv('Data/Input/Playground/test.csv')
# # Original dataset train & test
# O_train_df = pd.read_csv('Data/Input/Original/kindey stone urine analysis.csv')
# Combining original and playground datasets
train_df = pd.concat([P_train_df, O_train_df], axis=0)
train_df.drop(columns=["id"], inplace=True)
#
# # 2. EDA
# ## 2.1 Data summary
def summary(df):
print(f"Dataset has {df.shape[1]} features and {df.shape[0]} examples.")
summary = df.describe().T
summary.drop(columns=["count", "25%", "50%", "75%"], inplace=True)
summary["mode"] = df.mode().values[0]
summary["median"] = df.median()
summary["Unique"] = df.nunique().values
summary["Missing"] = df.isnull().sum().values
summary["Duplicated"] = df.duplicated().sum()
summary["Types"] = df.dtypes
# summary.drop(labels="id", inplace=True)
return summary
summary(P_train_df)
summary(O_train_df)
# Combination dataset
summary(train_df)
summary(test_df)
#
# Insights:
# - Train and Test dataset has zero duplicated values.
# - Train and Test dataset has zero Missing values.
# - All of features seem to be continuous and dataset seems to be small.
# - Regarding the number of training examples **"gravity"** and **"cond"** has the lowest unique values.
# - **"gravity"** has the smallest range and "osmo" has the largest range.
# - **"osmo"** and **"urea"** are the only int features of the dataset and other features are float.
# - **"target"** is the output value which is one when paitent has kidney stone and zero when there isn't any stones.
# ## 2.2 Feature distribution
# First of all, It is recommended to evaluate the distribution of the train and test datasets and compare the original and playground datasets. In this step histograms were created to show how each feature was distributed in a given dataset. After that Kernel density function was plotted to see the resemblance more vividly.
# Create multiple plots with a given size
fig = plt.figure(figsize=(15, 12))
features = P_train_df.columns[1:-1]
# Create a countplot to evaluate the distribution
for i, feature in enumerate(features):
ax = plt.subplot(3, 3, i + 1)
sns.histplot(
x=feature,
data=P_train_df,
label="Playground_Train",
color="#800080",
ax=ax,
alpha=0.5,
kde=True,
)
sns.histplot(
x=feature,
data=O_train_df,
label="Original_Train",
color="#FF5733",
ax=ax,
alpha=0.5,
kde=True,
)
sns.histplot(
x=feature,
data=test_df,
label="Test",
color="#006b3c",
ax=ax,
alpha=0.5,
kde=True,
)
# ax.set_xlabel(feature, fontsize=12)
# Create the legend
fig.legend(
labels=["Playground_Train", "Original_Train", "Test"],
loc="upper center",
bbox_to_anchor=(0.5, 0.96),
fontsize=12,
ncol=3,
)
# Adjust the spacing between the subplots and the legend
fig.subplots_adjust(
top=0.90, bottom=0.05, left=0.10, right=0.95, hspace=0.45, wspace=0.45
)
plt.show()
#
# Insights:
# - Trend of distribution in playground and orginal training set it seems quiet similar and test set has a good resemblace to both of them too.
# - Our dataset is quiet small and all of features seem to be continuous.
# - Value of features suggest that scaling will be quiet important in this problem specially for **"calc"**.
# ## 2.3 Target distribution
colorPal = ["#780060", "#4CAF50"]
piee = train_df["target"].value_counts()
mylabels = "True", "False"
myexplode = [0.2, 0]
fig, ax = plt.subplots(figsize=(6, 6), subplot_kw=dict(aspect="equal"))
patches, texts, autotexts = ax.pie(
piee,
autopct="%1.1f%%",
explode=myexplode,
labels=mylabels,
shadow=True,
colors=colorPal,
startangle=90,
radius=0.8,
)
ax.legend(
patches,
mylabels,
title="Kidney stone: ",
loc="lower left",
fontsize=10,
bbox_to_anchor=(1, 0, 0.5, 1),
)
plt.setp(autotexts, size=12, weight="bold")
plt.setp(texts, size=12, weight="bold")
ax.set_title("Target distribution", fontsize=12, weight="bold")
plt.show()
#
# Insights:
#
# - Looking in the distribution of **"target"** we understand that this is a classification problem which has more data of True kidney stone.
# - Pie chart suggests that 55.8 % of patients have kidney stone and 44.2 % don't have kidney stone.
# ## 2.4 Correlation Matrix
corr = train_df.iloc[:, 1:].corr()
# create mask for upper triangle
mask = np.triu(np.ones_like(corr, dtype=bool))
# Create heatmap
plt.figure(figsize=(6, 3))
# set theme to change overall style
sns.set_theme(style="white", font_scale=0.7)
sns.heatmap(
corr, cmap="cool", linewidths=2, mask=mask, vmin=-1, vmax=1, annot=True, fmt=".2f"
)
plt.title("Correlation Matrix", fontsize=12, weight="bold", pad=20)
plt.show()
#
# Insights:
#
# - Correlatin matrix suggests that **"ph"** has a negative relationship with target and **"calc"** and **"gravity"** seems to have a positive relationship.
# - Patients with lower **"ph"** and higher **"calc"** are more likely to have kidney stone.
# - As **"Conductivity"** increases **"osmolarity"** raises too which means it is more likely that patient have kidney stone.
# ## 2.5 Outliers
# Set the figure size
plt.figure(figsize=(15, 10))
sns.set_theme(style="whitegrid")
# Create categorical features subplots
for i, feature in enumerate(features):
ax = plt.subplot(3, 3, i + 1)
sns.boxplot(x="target", y=feature, data=P_train_df, ax=ax, palette=colorPal)
plt.subplots_adjust(top=0.9, bottom=0.1, left=0.1, right=0.9, hspace=0.5, wspace=0.5)
#
# Insights:
#
# - Considering the boxplot of different features there are some outliers in the dataset which should be removed.
# - Typically outliers are points beyond the whiskers of the boxplot and you can see them in **gravity**, **ph** and **Calc**.
# # 3. Data preparation
# # Removing outliers
# # Kudus to @klyushnik
# def outlier_removal(data , i):
# q1 = np.quantile(data[i] ,0.25)
# q3 = np.quantile(data[i] , 0.75)
# iqr = q3-q1
# lower_tail = q1 - 1.5*iqr
# upper_tail = q3 + 1.5*iqr
# data.drop(data[data[i]> upper_tail].index , inplace = True)
# data.drop(data[data[i]< lower_tail].index ,inplace =True)
# outlier_list = ['gravity', 'ph', 'osmo', 'cond', 'urea', 'calc']
# for i in outlier_list:
# outlier_removal(train_df ,i)
def prepXy(df, X_label, y_label=None):
selected_feat = []
data = copy.deepcopy(df)
# Feature Engineering
# Ion product of calcium and urea
data["ion_product"] = data["calc"] * data["urea"]
# Calcium-to-urea ratio
data["calcium_to_urea_ratio"] = data["calc"] / data["urea"]
# Electrolyte balance
data["electrolyte_balance"] = data["cond"] / (10 ** (-data["ph"]))
# Osmolality-to-specific gravity ratio
data["osmolality_to_sg_ratio"] = data["osmo"] / data["gravity"]
# The product of osmolarity and density is created as a new property
data["osmo_density"] = data["osmo"] * data["gravity"]
# ******************************************************************
# Calculate ammonium concentration (assuming ammonium is not directly measured)
data["ammonium_concentration"] = (
(data["gravity"] - 1.010) * (140 - (2 * data["ph"])) * 1.2
)
# Calculate phosphate concentration (assuming phosphate is not directly measured)
data["phosphate_concentration"] = (data["gravity"] - 1.010) * (
32 - (0.06 * data["ph"])
)
# Split into features and target
X = data[X_label].values
if y_label is None:
y = np.zeros(data.shape[0])
else:
y = np.ravel(data[y_label].values)
# Scaling dataset
# scaler = MinMaxScaler()
scaler = StandardScaler()
X = scaler.fit_transform(X)
return data, X, y
features = [c for c in train_df.columns if c != "target"]
# Define engineered features
eng_features = [
"ion_product",
"calcium_to_urea_ratio",
"electrolyte_balance",
"osmolality_to_sg_ratio",
"osmo_density",
"ammonium_concentration",
"phosphate_concentration",
]
features.extend(eng_features)
#
# # 4. Modeling
# ## 4.1 Baseline Random Forest Classifier Model with all features
# Data preparatin
new_train_df, X, y = prepXy(train_df, X_label=features, y_label=["target"])
new_test_df, X_, y_ = prepXy(test_df, X_label=features, y_label=None)
# split data
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=RANDOM_STATE
)
# Train model
clf = RandomForestClassifier(n_estimators=100, random_state=RANDOM_STATE)
clf.fit(X_train, y_train)
# Make predictions using predict() and predict_proba() methods
y_pred = clf.predict(X_test) # predict() method
y_pred_prob = clf.predict_proba(X_test) # predict_proba() method
acc_score = accuracy_score(y_test, y_pred)
auc_score = roc_auc_score(y_test, y_pred)
print(f"Test set Accuracy score: {acc_score}")
print(f"Test set ROC AUC score: {auc_score}")
idx = [c for c in new_train_df.columns if c != "target"]
feature_imp = pd.Series(clf.feature_importances_, index=idx).sort_values(
ascending=False
)
# Creating a bar plot
sns.barplot(x=feature_imp, y=feature_imp.index, palette="plasma")
# Add labels to your graph
plt.xlabel("Feature Importance Score")
plt.ylabel("Features")
plt.title("Visualizing Important Features")
plt.show()
#
# ## 4.2 Comparison between different models with All features
remove_col = [] # add if want to remove features
for i in remove_col:
features.remove(i)
print(features)
# XGBoost parameters
XGBParams = {
"n_estimators": 500,
"learning_rate": 0.01,
"max_depth": 5,
"objective": "binary:logistic",
"eval_metric": "auc",
"tree_method": "auto",
"grow_policy": "lossguide",
"reg_alpha": 1.0,
"reg_lambda": 0.5,
}
# Random Forest parameters
RFParams = {"n_estimators": 550, "max_depth": 5}
CatParams = {
"iterations": 875,
"learning_rate": 0.003,
"depth": 4,
"l2_leaf_reg": 1.852525829263644e-08,
"bootstrap_type": "Bayesian",
"random_strength": 2.468077989304625e-07,
"bagging_temperature": 7.3264062526863345,
"od_type": "IncToDec",
"od_wait": 21,
}
# # Data preparation
# new_train_df,X,y = prepXy(train_df,X_label,y_label)
# new_test_df,X_,y_ = prepXy(test_df,X_label,y_label=None)
# # Split into training and testing sets
# X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=RANDOM_STATE)
# evaluate each model using cross-validation and compute ROC AUC scores
scores = []
Simp_classifiers = {
"Logistic Regression": LogisticRegression(),
"Random Forest Classifier": RandomForestClassifier(
**RFParams, random_state=RANDOM_STATE
),
"XGboost": XGBClassifier(**XGBParams, random_state=RANDOM_STATE),
"CatBoost": CatBoostClassifier(**CatParams, random_state=RANDOM_STATE, verbose=0),
# 'SVC':SVC(probability=True, random_state=RANDOM_STATE),
# 'KNN':KNeighborsClassifier(n_neighbors=30),
# 'Naive Bayes':GaussianNB()
}
# Creating subplots for further evaluation
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(10, 4))
sns.set_theme(style="whitegrid", font_scale=0.8)
for name, model in Simp_classifiers.items():
model.fit(X_train, y_train)
y_pred_prob = model.predict_proba(X_test)[:, 1]
y_predict = model.predict(X_test)
# calc roc
roc_auc = roc_auc_score(y_test, y_pred_prob)
scores.append((name, roc_auc))
# plotting roc curve
fpr, tpr, _ = roc_curve(y_test, y_pred_prob)
ax1.plot(fpr, tpr, label="{} (AUC = {:.4f})".format(name, roc_auc))
# Print classification score of the model
report = classification_report(
y_test,
y_predict,
target_names=["Have Kidney stone", "Dont have Kidney stone"],
labels=[0, 1],
)
print(f"{name}\n{report}\n")
# Create the roc_curve description
ax1.plot([0, 1], [0, 1], linestyle="--", color="gray", label="Random Guessing")
ax1.set_xlabel("False Positive Rate")
ax1.set_ylabel("True Positive Rate")
ax1.set_title(
"Receiver Operating Characteristic (ROC) Curves", fontsize=9, weight="bold", pad=10
)
ax1.legend(loc="lower right", fontsize="x-small")
# Find mean score of models and sort them and print the best model
names, model_scores = zip(*scores)
comp = dict(zip(names, model_scores))
sorted_comp = dict(sorted(comp.items(), key=lambda x: x[1]))
sorted_names = list(sorted_comp.keys())
sorted_model_scores = list(sorted_comp.values())
print(
f"The best model is {sorted_names[-1]} with a mean ROC AUC score of {sorted_model_scores[-1]:.4f}"
)
# Creating a barplot to compare models
ax2.bar(
sorted_names,
sorted_model_scores,
align="center",
alpha=0.9,
ecolor="black",
capsize=10,
)
ax2.set_ylabel("ROC AUC Score")
ax2.set_xticks(range(len(sorted_names)))
ax2.set_xticklabels(sorted_names, rotation=45, ha="right", weight="bold")
ax2.set_title(
"Comparison of Classification Models for Kidney Stone Classifier",
fontsize=9,
weight="bold",
pad=10,
)
plt.show()
#
# ## 4.3 Simple Stacking of classifiers
# create the stacking classifier
sclf = StackingCVClassifier(
classifiers=[Simp_classifiers[classifier] for classifier in Simp_classifiers],
meta_classifier=RandomForestClassifier(**RFParams, random_state=RANDOM_STATE),
cv=5,
use_probas=True,
)
# fit the stacking classifier to the training data
sclf.fit(X_train, y_train)
# Predict the class probabilities
y_proba = sclf.predict_proba(X_test)[:, 1]
# evaluate the performance of the stacking classifier on the test data
accuracy = sclf.score(X_test, y_test)
# Calculate the AUC score
rocauc = roc_auc_score(y_test, y_proba)
print("Accuracy:", accuracy)
print("Roc_Auc:", rocauc)
#
# # 5. Submission
# y_sub=Simp_classifiers['Random Forest Classifier'].predict_proba(X_)[:,1]
y_sub = sclf.predict_proba(X_)[:, 1]
# for classifier in Simp_classifiers:
# y_sub = Simp_classifiers[classifier].predict_proba(X_)[:,1]
Export = np.hstack(
(np.reshape(test_df["id"].values, (-1, 1)), np.reshape(y_sub, (-1, 1)))
)
Submission = pd.DataFrame(Export, columns=["id", "target"])
Submission.to_csv(r"submission.csv", index=False, header=["id", "target"])
Submission.shape
|
# # Step 1 : data gathering and preparation
# Goal: obtain a concatenated dataset with all rlevant raw data, for each time stamp
# **Data sources**
# The price data comes from:
# * ENTSO-E Transparency platform, series 12.1.D, frequency=1hour
# The intermitent renewables data comes from:
# * ENTSO-E Transparency platform, series 14.1.D (solar & wind generation forecast day-ahead + actual generation), frequency=15minutes
# The electricity consumption:
# * ENTSO-E Transparency platform, series 6.1.B (total load forecast day-ahead + actual load), frequency=15minutes
#
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import gc
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# each input file contain 1 year worth of data, so we need to concatenate them:
def yearly_files_concatenation(directory):
i = 0
for dirname, _, filenames in os.walk("/kaggle/input/" + str(directory)):
for filename in filenames:
if i == 0:
out = pd.read_csv(
"/kaggle/input/" + str(directory) + "/" + str(filename)
)
i = 1
else:
df = pd.read_csv(
"/kaggle/input/" + str(directory) + "/" + str(filename)
)
out = pd.concat([df, out])
return out
DE_DA_Prices = yearly_files_concatenation(
"da-prices-germany-ensto-e-transparency-platform"
)
# to check that we have several years worth of data: 7*365*24=61 320 data points for 7 years of data with 1-hour frequency:
len(DE_DA_Prices)
DE_DA_Prices.head()
# time is written as a time window, text format. Let s turn the text into a recognizable timestamp for python, and keep only the begining of the window, in order to sort values
def timestamp_reading(string):
return pd.to_datetime(string.split(" - ")[0], format="%d.%m.%Y %H:%M")
DE_DA_Prices["MTU (CET)"] = DE_DA_Prices["MTU (CET)"].apply(timestamp_reading)
DE_DA_Prices.head()
# the following function sets timestamp as index of the dataframe for easy merging
# due to daylight savings, some timestamps are redundant. The function arbitrarly removes the first occurence of the duplicates
def remove_timestamp_duplicates_and_set_as_index(timestamp_column, df):
dup_df = df[timestamp_column].value_counts().to_frame()
duplicates = dup_df[dup_df[timestamp_column] > 1].index.to_list()
print("the following timestamps are in duplicate", duplicates)
for d in duplicates:
# remove the first occurence:
df.drop(df.index[df[timestamp_column] == d][0], inplace=True)
df.set_index(timestamp_column, drop=True, inplace=True, verify_integrity=True)
df.sort_index(inplace=True)
return df
DE_DA_Prices = remove_timestamp_duplicates_and_set_as_index("MTU (CET)", DE_DA_Prices)
DE_DA_Prices.head()
# last step before merging: turning non numerical values into NaN, and removing missing data at the begining and end of timeranges
def df_value_cleaning(df, columns):
df.dropna(inplace=True)
for column in columns:
df[column] = pd.to_numeric(df[column], errors="coerce")
return df
DE_DA_Prices = df_value_cleaning(DE_DA_Prices, ["Day-ahead Price [EUR/MWh]"])
DE_DA_Prices["Day-ahead Price [EUR/MWh]"].plot(figsize=(20, 5))
# repeating the steps above for other data
# renewables generation data:
RE = yearly_files_concatenation("wind-and-solar-germany-entsoe-transparency")
RE["MTU (CET)"] = RE["MTU (CET)"].apply(timestamp_reading)
RE = remove_timestamp_duplicates_and_set_as_index("MTU (CET)", RE)
RE = df_value_cleaning(RE, RE.columns)
# this function aggregates 15 minutes frequencies into 1-hour rows
def hour_aggregation(df):
df = df.groupby(pd.Grouper(freq="H")).mean()
return df
RE = hour_aggregation(RE)
# removing some empty columns and aggragating onshore and offshore wind:
RE.drop(
columns=[
"Generation - Solar [MW] Current / Germany (DE)",
"Generation - Wind Offshore [MW] Current / Germany (DE)",
"Generation - Wind Onshore [MW] Current / Germany (DE)",
],
inplace=True,
)
RE["Generation - Wind [MW] Day Ahead/ Germany (DE)"] = (
RE["Generation - Wind Offshore [MW] Day Ahead/ Germany (DE)"]
+ RE["Generation - Wind Onshore [MW] Day Ahead/ Germany (DE)"]
)
RE["Generation - Wind [MW] Intraday / Germany (DE)"] = (
RE["Generation - Wind Offshore [MW] Intraday / Germany (DE)"]
+ RE["Generation - Wind Onshore [MW] Intraday / Germany (DE)"]
)
RE.drop(
[
"Generation - Wind Offshore [MW] Day Ahead/ Germany (DE)",
"Generation - Wind Offshore [MW] Intraday / Germany (DE)",
"Generation - Wind Onshore [MW] Day Ahead/ Germany (DE)",
"Generation - Wind Onshore [MW] Intraday / Germany (DE)",
],
axis=1,
inplace=True,
)
RE.head()
# electricity consumption data:
cons = yearly_files_concatenation("load-germany-entso-e-transparency-platform")
cons["Time (CET)"] = cons["Time (CET)"].apply(timestamp_reading)
cons = remove_timestamp_duplicates_and_set_as_index("Time (CET)", cons)
cons = df_value_cleaning(cons, cons.columns)
cons = hour_aggregation(cons)
cons.head()
# adding natural gas TTF index in the data
# data manually read from European Commission quaterly reports on gas markets
DE_DA_Prices["Dutch TTF natural gas price"] = np.nan
DE_DA_Prices.loc[
"2015-01-01 00:00:00":"2015-01-01 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 19
DE_DA_Prices.loc[
"2015-01-02 00:00:00":"2015-01-02 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 22.3
DE_DA_Prices.loc[
"2015-01-03 00:00:00":"2015-01-03 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 21.7
DE_DA_Prices.loc[
"2015-01-04 00:00:00":"2015-01-04 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 21.9
DE_DA_Prices.loc[
"2015-01-05 00:00:00":"2015-01-05 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 20.5
DE_DA_Prices.loc[
"2015-01-06 00:00:00":"2015-01-06 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 20.4
DE_DA_Prices.loc[
"2015-01-07 00:00:00":"2015-01-07 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 20.9
DE_DA_Prices.loc[
"2015-01-08 00:00:00":"2015-01-08 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 19.5
DE_DA_Prices.loc[
"2015-01-09 00:00:00":"2015-01-09 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 19.3
DE_DA_Prices.loc[
"2015-01-10 00:00:00":"2015-01-10 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 18.1
DE_DA_Prices.loc[
"2015-01-11 00:00:00":"2015-01-11 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 17.1
DE_DA_Prices.loc[
"2015-01-12 00:00:00":"2015-01-12 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 15.5
DE_DA_Prices.loc[
"2016-01-01 00:00:00":"2016-01-01 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 13.8
DE_DA_Prices.loc[
"2016-01-02 00:00:00":"2016-01-02 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 12.4
DE_DA_Prices.loc[
"2016-01-03 00:00:00":"2016-01-03 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 12.2
DE_DA_Prices.loc[
"2016-01-04 00:00:00":"2016-01-04 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 12
DE_DA_Prices.loc[
"2016-01-05 00:00:00":"2016-01-05 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 12.8
DE_DA_Prices.loc[
"2016-01-06 00:00:00":"2016-01-06 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 14.4
DE_DA_Prices.loc[
"2016-01-07 00:00:00":"2016-01-07 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 14.1
DE_DA_Prices.loc[
"2016-01-08 00:00:00":"2016-01-08 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 11.9
DE_DA_Prices.loc[
"2016-01-09 00:00:00":"2016-01-09 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 12.3
DE_DA_Prices.loc[
"2016-01-10 00:00:00":"2016-01-10 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 16
DE_DA_Prices.loc[
"2016-01-11 00:00:00":"2016-01-11 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 17.9
DE_DA_Prices.loc[
"2016-01-12 00:00:00":"2016-01-12 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 17.6
DE_DA_Prices.loc[
"2017-01-01 00:00:00":"2017-01-01 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 19.9
DE_DA_Prices.loc[
"2017-01-02 00:00:00":"2017-01-02 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 19.5
DE_DA_Prices.loc[
"2017-01-03 00:00:00":"2017-01-03 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 15.7
DE_DA_Prices.loc[
"2017-01-04 00:00:00":"2017-01-04 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 16.1
DE_DA_Prices.loc[
"2017-01-05 00:00:00":"2017-01-05 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 15.5
DE_DA_Prices.loc[
"2017-01-06 00:00:00":"2017-01-06 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 15.1
DE_DA_Prices.loc[
"2017-01-07 00:00:00":"2017-01-07 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 15
DE_DA_Prices.loc[
"2017-01-08 00:00:00":"2017-01-08 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 15.9
DE_DA_Prices.loc[
"2017-01-09 00:00:00":"2017-01-09 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 17.1
DE_DA_Prices.loc[
"2017-01-10 00:00:00":"2017-01-10 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 17
DE_DA_Prices.loc[
"2017-01-11 00:00:00":"2017-01-11 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 20.3
DE_DA_Prices.loc[
"2017-01-12 00:00:00":"2017-01-12 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 19.6
DE_DA_Prices.loc[
"2018-01-01 00:00:00":"2018-01-01 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 17.9
DE_DA_Prices.loc[
"2018-01-02 00:00:00":"2018-01-02 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 17.4
DE_DA_Prices.loc[
"2018-01-03 00:00:00":"2018-01-03 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 18.2
DE_DA_Prices.loc[
"2018-01-04 00:00:00":"2018-01-04 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 20.6
DE_DA_Prices.loc[
"2018-01-05 00:00:00":"2018-01-05 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 22.6
DE_DA_Prices.loc[
"2018-01-06 00:00:00":"2018-01-06 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 21.9
DE_DA_Prices.loc[
"2018-01-07 00:00:00":"2018-01-07 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 22.1
DE_DA_Prices.loc[
"2018-01-08 00:00:00":"2018-01-08 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 25.8
DE_DA_Prices.loc[
"2018-01-09 00:00:00":"2018-01-09 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 27.3
DE_DA_Prices.loc[
"2018-01-10 00:00:00":"2018-01-10 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 24.3
DE_DA_Prices.loc[
"2018-01-11 00:00:00":"2018-01-11 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 24.7
DE_DA_Prices.loc[
"2018-01-12 00:00:00":"2018-01-12 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 22
DE_DA_Prices.loc[
"2019-01-01 00:00:00":"2019-01-01 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 19.9
DE_DA_Prices.loc[
"2019-01-02 00:00:00":"2019-01-02 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 17.8
DE_DA_Prices.loc[
"2019-01-03 00:00:00":"2019-01-03 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 14.2
DE_DA_Prices.loc[
"2019-01-04 00:00:00":"2019-01-04 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 14.4
DE_DA_Prices.loc[
"2019-01-05 00:00:00":"2019-01-05 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 11.2
DE_DA_Prices.loc[
"2019-01-06 00:00:00":"2019-01-06 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 10.1
DE_DA_Prices.loc[
"2019-01-07 00:00:00":"2019-01-07 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 11.0
DE_DA_Prices.loc[
"2019-01-08 00:00:00":"2019-01-08 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 12.8
DE_DA_Prices.loc[
"2019-01-09 00:00:00":"2019-01-09 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 16.4
DE_DA_Prices.loc[
"2019-01-10 00:00:00":"2019-01-10 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 16.1
DE_DA_Prices.loc[
"2019-01-11 00:00:00":"2019-01-11 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 16.3
DE_DA_Prices.loc[
"2019-01-12 00:00:00":"2019-01-12 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 12.1
DE_DA_Prices.loc[
"2020-01-01 00:00:00":"2020-01-01 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 9.8
DE_DA_Prices.loc[
"2020-01-02 00:00:00":"2020-01-02 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 8.9
DE_DA_Prices.loc[
"2020-01-03 00:00:00":"2020-01-03 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 6.9
DE_DA_Prices.loc[
"2020-01-04 00:00:00":"2020-01-04 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 6.2
DE_DA_Prices.loc[
"2020-01-05 00:00:00":"2020-01-05 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 4.4
DE_DA_Prices.loc[
"2020-01-06 00:00:00":"2020-01-06 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 6.2
DE_DA_Prices.loc[
"2020-01-07 00:00:00":"2020-01-07 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 6.0
DE_DA_Prices.loc[
"2020-01-08 00:00:00":"2020-01-08 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 11.2
DE_DA_Prices.loc[
"2020-01-09 00:00:00":"2020-01-09 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 13.3
DE_DA_Prices.loc[
"2020-01-10 00:00:00":"2020-01-10 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 14.1
DE_DA_Prices.loc[
"2020-01-11 00:00:00":"2020-01-11 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 15.1
DE_DA_Prices.loc[
"2020-01-12 00:00:00":"2020-01-12 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 19.1
DE_DA_Prices.loc[
"2021-01-01 00:00:00":"2021-01-01 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 19.8
DE_DA_Prices.loc[
"2021-01-02 00:00:00":"2021-01-02 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 15.7
DE_DA_Prices.loc[
"2021-01-03 00:00:00":"2021-01-03 00:00:00",
"Dutch TTF natural gas price":"Dutch TTF natural gas price",
] = 16.1
DE_DA_Prices["Dutch TTF natural gas price"] = DE_DA_Prices[
"Dutch TTF natural gas price"
].fillna(method="ffill")
DE_DA_Prices.head()
# finally, merging of the 3 dataframes created earlier
data = DE_DA_Prices.copy(deep=True)
data = data.join(RE)
data = data.join(cons)
data.dropna(inplace=True)
print(len(data))
del RE
del cons
del DE_DA_Prices
gc.collect()
data.head()
# the import column names accidently contain special json characters (invisible when printing), creating issues later on. Let us remove them:
import string
dic = {}
# printable = set(string.printable)
for column in data.columns.values:
# dic[column]=''.join(filter(lambda x: x in printable, column))
# dic[column]=column.encode("ascii", errors="ignore").decode()
dic[column] = str(column).replace("[", "").replace("]", "")
data.rename(columns=dic, inplace=True)
# # Step 2 : EDA, feature engineering
# goal: identify relevant correlations, and transform raw data into better explaing factors
# useful imports:
from sklearn.metrics import mean_squared_error
import matplotlib.pyplot as plt
import lightgbm as lgb
import seaborn as sns
import statsmodels.api as sm
from statsmodels.api import add_constant
# engineering a few features, based on intuition:
data["clean_ignition"] = data["Dutch TTF natural gas price"] / 0.5
data["total intermitent RE forecast"] = (
data["Generation - Solar MW Day Ahead/ Germany (DE)"]
+ data["Generation - Wind MW Day Ahead/ Germany (DE)"]
)
data["% supply by renewables"] = (
data["total intermitent RE forecast"]
/ data["Day-ahead Total Load Forecast MW - Germany (DE)"]
)
# splitting X and Y:
# X=data.drop('Day-ahead Price EUR/MWh',axis=1)
# X=data.drop(['Day-ahead Price EUR/MWh','total intermitent RE forecast'],axis=1)
# X=data.drop(['Day-ahead Price EUR/MWh','total intermitent RE forecast','Generation - Wind MW Intraday / Germany (DE)','Generation - Solar MW Intraday / Germany (DE)','Actual Total Load MW - Germany (DE)'],axis=1)
# X=data.drop(['Day-ahead Price EUR/MWh','total intermitent RE forecast','Generation - Wind MW Intraday / Germany (DE)','Generation - Solar MW Intraday / Germany (DE)','Actual Total Load MW - Germany (DE)','Generation - Wind MW Day Ahead/ Germany (DE)','Generation - Solar MW Day Ahead/ Germany (DE)'],axis=1)
# X=data.drop(['Day-ahead Price EUR/MWh','total intermitent RE forecast','Generation - Solar MW Day Ahead/ Germany (DE)','Generation - Wind MW Day Ahead/ Germany (DE)','Day-ahead Total Load Forecast MW - Germany (DE)','Generation - Wind MW Day Ahead/ Germany (DE)','Generation - Solar MW Day Ahead/ Germany (DE)'],axis=1)
# X=data.drop(['Day-ahead Price EUR/MWh','Generation - Wind MW Intraday / Germany (DE)','Generation - Solar MW Intraday / Germany (DE)','Actual Total Load MW - Germany (DE)','Generation - Wind MW Day Ahead/ Germany (DE)','Generation - Solar MW Day Ahead/ Germany (DE)'],axis=1)
# X=data.drop(['Day-ahead Price EUR/MWh','Generation - Wind MW Intraday / Germany (DE)','Generation - Solar MW Intraday / Germany (DE)','Actual Total Load MW - Germany (DE)','Generation - Wind MW Day Ahead/ Germany (DE)','Generation - Solar MW Day Ahead/ Germany (DE)'],axis=1)
# X=data.drop(['Day-ahead Price EUR/MWh','total intermitent RE forecast','Generation - Wind MW Intraday / Germany (DE)','Generation - Solar MW Intraday / Germany (DE)','Actual Total Load MW - Germany (DE)','Generation - Wind MW Day Ahead/ Germany (DE)','Generation - Solar MW Day Ahead/ Germany (DE)'],axis=1)
X = data.drop(
[
"Day-ahead Price EUR/MWh",
"% supply by renewables",
"Generation - Wind MW Intraday / Germany (DE)",
"Generation - Solar MW Intraday / Germany (DE)",
"Actual Total Load MW - Germany (DE)",
"Generation - Wind MW Day Ahead/ Germany (DE)",
"Generation - Solar MW Day Ahead/ Germany (DE)",
],
axis=1,
)
Y = data["Day-ahead Price EUR/MWh"]
# splitting train and test sets:
X_train = X["2015-01-05 00:00:00":"2019-12-31 23:00:00"]
X_test = X["2020-01-01 00:00:00":]
Y_train = Y["2015-01-05 00:00:00":"2019-12-31 23:00:00"].to_frame()
Y_test = Y["2020-01-01 00:00:00":].to_frame()
# sanity check for erronous values.
sns.heatmap(data)
# Nothing abnormal, since:
# * load time series, appearing very bright, aggregates all the load of the country, and thus are very high compared to other series
# * Solar shows a pattern, corresponding to day/night cycles
# * Wind shows some volatility, as can be expected
# feature importance
model = lgb.LGBMRegressor(importance_type="gain")
model.fit(X_train, Y_train)
df_fimp = pd.DataFrame()
df_fimp["feature"] = X.columns.values
df_fimp["importance"] = model.booster_.feature_importance()
plt.figure(figsize=(14, 7))
sns.barplot(
x="importance",
y="feature",
data=df_fimp.sort_values(by="importance", ascending=False),
)
plt.title("LightGBM Feature Importance")
plt.tight_layout()
# feature importance
data_set = lgb.Dataset(X_train, label=Y_train, free_raw_data=False)
params = {
"objective": "regression",
"boosting": "gbdt",
"num_leaves": 200,
"learning_rate": 0.5,
"feature_fraction": 1.0,
"reg_lambda": 1,
"metric": "rmse",
}
model = lgb.train(params=params, train_set=data_set)
df_fimp = pd.DataFrame()
df_fimp["feature"] = X.columns.values
df_fimp["importance"] = model.feature_importance()
plt.figure(figsize=(14, 7))
sns.barplot(
x="importance",
y="feature",
data=df_fimp.sort_values(by="importance", ascending=False),
)
plt.title("LightGBM Feature Importance")
plt.tight_layout()
data_bis = Y_test.copy()
data_bis["prediction"] = model.predict(X_test)
data_bis[["Day-ahead Price EUR/MWh", "prediction"]].plot(figsize=(20, 10))
print(mean_squared_error(data_bis["Day-ahead Price EUR/MWh"], data_bis["prediction"]))
# accuracy of RE forecast:
# data_bis['wind - relative error']=abs(data_bis['Generation - Wind MW Day Ahead/ Germany (DE)']-data_bis['Generation - Wind MW Intraday / Germany (DE)'])/data_bis['Generation - Wind MW Day Ahead/ Germany (DE)']
# print(data_bis['wind - relative error']['2015-01-01 00:00:00':'2015-01-12 00:00:00'].mean())
# print(data_bis['wind - relative error']['2020-01-01 00:00:00':'2020-01-12 00:00:00'].min())
# print(data_bis['wind - relative error']['2020-01-01 00:00:00':'2020-01-12 00:00:00'].max())
# accuracy heavily depends on year
# Build Model
X_bis = sm.add_constant(X_train)
X_bis_test = sm.add_constant(X_test)
model = sm.OLS(Y_train, X_bis)
results = model.fit()
# results.summary() si on veut voir un tableau plutôt qu’un graphique
def coefplot(results, X, normalization=False):
# Create dataframe of results summary
coef_df = pd.DataFrame(results.summary().tables[1].data)
# Add column names
coef_df.columns = coef_df.iloc[0]
# Drop the extra row with column labels
coef_df = coef_df.drop(0)
# Set index to variable names
coef_df = coef_df.set_index(coef_df.columns[0])
# Change datatype from object to float
coef_df = coef_df.astype(float)
# Get errors; (coef - lower bound of conf interval)
errors = coef_df["coef"] - coef_df["[0.025"]
# Append errors column to dataframe
coef_df["errors"] = errors
# normalization steps:
if normalization:
for var in coef_df.index.values:
if var == "const":
pass
else:
coef_df.loc[var, "coef"] = coef_df.loc[var, "coef"] * X[var].std()
coef_df.loc[var, "errors"] = coef_df.loc[var, "errors"] * X[var].std()
# Sort values by coef descending
coef_df = coef_df.sort_values(by=["coef"], ascending=False)
### Plot Coefficients ###
# x-labels
variables = list(coef_df.index.values)
# Add variables column to dataframe
coef_df["variables"] = variables
# Set sns plot style back to 'poster'
# This will make bars wide on plot
sns.set_context("poster")
# Define figure, axes, and plot
fig, ax = plt.subplots(figsize=(15, 10))
# Error bars for 95% confidence interval
# Can increase capsize to add whiskers
coef_df.plot(
x="variables",
y="coef",
kind="bar",
ax=ax,
color="none",
fontsize=22,
ecolor="steelblue",
capsize=0,
yerr="errors",
legend=False,
)
# Set title & labels
if normalization:
plt.title(
"Normalized coefficients of Features w/ 95% Confidence Intervals",
fontsize=30,
)
else:
plt.title("Coefficients of Features w/ 95% Confidence Intervals", fontsize=30)
ax.set_ylabel("Coefficients", fontsize=22)
ax.set_xlabel("", fontsize=22)
# Coefficients
ax.scatter(
x=pd.np.arange(coef_df.shape[0]),
marker="o",
s=80,
y=coef_df["coef"],
color="steelblue",
)
# Line to define zero on the y-axis
ax.axhline(y=0, linestyle="--", color="red", linewidth=1)
return plt.show()
coefplot(results, X_bis, True)
print(
mean_squared_error(Y_test["Day-ahead Price EUR/MWh"], results.predict(X_bis_test))
)
data_bis = Y_test.copy()
data_bis["prediction"] = results.predict(X_bis_test)
data_bis[["Day-ahead Price EUR/MWh", "prediction"]].plot(figsize=(20, 10))
a = Y_train.mean().to_numpy()[0]
dumb_pred = np.full((len(Y_test), 1), a)
print(mean_squared_error(Y_test["Day-ahead Price EUR/MWh"], dumb_pred))
plot_df = pd.DataFrame(
columns=[
"number of hours with negative prices",
"ratio between intermittent power generation and consumption",
]
)
for y in range(2015, 2021):
plot_df.loc[y, "number of hours with negative prices"] = len(
data[data["Day-ahead Price EUR/MWh"] <= 0].loc[
str(y) + "-01-01 00:00:00" : str(y + 1) + "-01-01 00:00:00",
]
)
plot_df.loc[
y, "ratio between intermittent power generation and consumption"
] = data["total intermitent RE forecast"].loc[
str(y) + "-01-01 00:00:00" : str(y + 1) + "-01-01 00:00:00",
].sum() / (
data["Day-ahead Total Load Forecast MW - Germany (DE)"]
.loc[str(y) + "-01-01 00:00:00" : str(y + 1) + "-01-01 00:00:00",]
.sum()
)
plot_df
fig = plt.figure(figsize=(20, 5))
ax = fig.add_subplot(111)
plt.scatter(
x=plot_df["ratio between intermittent power generation and consumption"],
y=plot_df["number of hours with negative prices"],
)
# plt.axis([0,0.4,0,300])
for y in plot_df.index.values:
ax.annotate(
y,
xy=(
plot_df.loc[
y, "ratio between intermittent power generation and consumption"
],
plot_df.loc[y, "number of hours with negative prices"],
),
)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv(
os.path.join("/kaggle/input/-spotify-tracks-dataset/dataset.csv")
).drop_duplicates(subset="track_name")
df = df.sort_values(by="duration_ms")
df.to_csv("sorted.csv")
df
df = pd.read_csv(os.path.join("sorted.csv"))
df["popularity"] = (df["popularity"] - df["popularity"].min()) / (
df["popularity"].max() - df["popularity"].min()
)
count = 0
for i in df.columns[1:]:
if df[i].dtype.kind in "iufc":
df[i] = (df[i] - df[i].min()) / (df[i].max() - df[i].min())
df["explicit"] = df["explicit"].astype(int)
moderated_df = df.copy()
moderated_df = moderated_df.drop(
[*df.columns[:9], df.columns[13], df.columns[-1]], axis=1
)
moderated_df
df
sal1 = np.array(
[
moderated_df.iloc[15182, :].values,
moderated_df.iloc[22520, :].values,
moderated_df.iloc[26534, :].values,
moderated_df.iloc[22549, :].values,
moderated_df.iloc[38010, :].values,
]
)
sal2 = np.array(
[
moderated_df.iloc[250, :].values,
moderated_df.iloc[321, :].values,
moderated_df.iloc[7, :].values,
moderated_df.iloc[728, :].values,
moderated_df.iloc[421, :].values,
]
)
sal3 = np.array(
[
moderated_df.iloc[254, :].values,
moderated_df.iloc[222, :].values,
moderated_df.iloc[14, :].values,
moderated_df.iloc[728, :].values,
moderated_df.iloc[422, :].values,
]
)
sal4 = np.array(
[
moderated_df.iloc[300, :].values,
moderated_df.iloc[169, :].values,
moderated_df.iloc[16, :].values,
moderated_df.iloc[728, :].values,
moderated_df.iloc[423, :].values,
]
)
sal5 = np.array(
[
moderated_df.iloc[301, :].values,
moderated_df.iloc[196, :].values,
moderated_df.iloc[20, :].values,
moderated_df.iloc[728, :].values,
moderated_df.iloc[424, :].values,
]
)
activities = np.array([sal1, sal2, sal3, sal4, sal5])
moderated_df
def create_gaussian(values):
mean = np.mean(values)
std = np.std(values)
gaussian = np.random.normal(loc=mean, scale=std, size=1000)
q1 = np.percentile(gaussian, 25)
q3 = np.percentile(gaussian, 75)
return np.array([mean, mean - 0.001, mean + 0.001])
def cosine_similarity(a, b):
"""
calculates cosine similarity of two vectors
"""
dot_product = np.dot(a, b)
norm_a = np.linalg.norm(a)
norm_b = np.linalg.norm(b)
similarity = dot_product / (norm_a * norm_b)
return similarity
def calculate_min_cosine(ls):
"""
Enter the list of songs identified by the person for a particular exercise as a numpy array in the format:
ls = [
song1,
song2,
song3
]
returns:
cosine similarity of mean with minimum of (75th quartile of gaussian distribution of song attribute and 25th quartile of the same)
"""
results = []
ls = np.transpose(ls)
for i in ls:
results.append(create_gaussian(i))
results = np.transpose(results)
cos_min_mean = cosine_similarity(results[0], results[1])
cos_max_mean = cosine_similarity(results[0], results[2])
return min(cos_min_mean, cos_max_mean)
def all_act_min_cosine(activities):
"""
calculates minimum cosine generated from all exercises
"""
cosines = []
for activity in activities:
cosines.append(calculate_min_cosine(activity))
return min(cosines)
def return_means_all(activities):
"""
Return mean of the list of songs per exercise
"""
means = []
for activity in activities:
activity = np.transpose(activity)
temp = []
for i in activity:
temp.append(create_gaussian(i))
means.append(np.array(temp))
return means
def calculate_approximate_song_vector(means, exercise_index_realtime, limiter):
"""
inputs:
mean value of songs
realtime list of exercises detected
limiter (window size)
"""
net_value = means[exercise_index_realtime[0]]
exercise_index_realtime = exercise_index_realtime[:limiter]
for i in exercise_index_realtime[1:]:
net_value += means[i]
return net_value / limiter
def return_reference_list(activities):
"""
for a given list of activities, it returns, a reference of mean, q1 and q3 for each activity in that list
"""
ref = []
for i in activities:
ref.append(create_gaussian(i))
return np.array(ref)
def locate_songs(df, start_value, end_value):
temp = df.columns[0]
return df.loc[(df[temp] >= start_value) & (df[temp] <= end_value)].index
def return_combined_songs(column_wise_list, appx_song_with_ranges):
songs = set()
for i in range(len(column_wise_list)):
temp = set(
locate_songs(
column_wise_list[i],
appx_song_with_ranges[i][1],
appx_song_with_ranges[i][2],
)
)
songs = songs | temp
return songs
def recommend_songs(
real_df, normalized_df, activities_songs, workout, limiter, popularity=False
):
"""
returns recommendations in a df format.
real_df: un-adulterated dataframe of songs(all columns)
normalized_df: df where non-important values are thrown out and all values are floats.(the df from which cosines are calculated)
activities_songs: numpy.ndarray where each index specifies an exercise and consistes of a list of appropriate songs for that exercise chosen by user.
workout: list of exercises detected by lstm model. placeholder is an integer. the integer is used to locate song preference for that
activity from activities songs as an index.
limiter: window size(int)
"""
ref = return_means_all(activities_songs)
appx_song_with_ranges = calculate_approximate_song_vector(ref, workout, limiter)
column_wise_list = []
for i in normalized_df.columns:
column_wise_list.append(normalized_df[i].sort_values().to_frame())
combined = return_combined_songs(column_wise_list, appx_song_with_ranges)
final_df = normalized_df.loc[combined]
min_cosine = np.min(all_act_min_cosine(activities_songs))
reference = np.transpose(appx_song_with_ranges)[0]
indices = []
for i in range(len(combined)):
k = cosine_similarity(reference, final_df.iloc[i].tolist())
if k > 0.95:
indices.append([i, k])
indices = list(zip(*sorted(indices, key=lambda x: x[1], reverse=True)))
songs = real_df.iloc[list(indices[0])]
if popularity:
return songs.sort_values(by="popularity", ascending=False).iloc[:5]
else:
return songs.iloc[:15]
temp = recommend_songs(
df.copy(), moderated_df.copy(), activities, [0, 0, 0, 0, 0, 0], 5, popularity=True
)
temp
recommend_songs(df.copy(), moderated_df.copy(), activities, [0, 0, 0, 0, 0, 0], 5)
recommend_songs(
df.copy(), moderated_df.copy(), activities, [0, 0, 0, 0, 0, 0], 5, popularity=True
)
def create_gaussian(values):
mean = np.mean(values)
std = np.std(values)
gaussian = np.random.normal(loc=mean, scale=std, size=1000)
q1 = np.percentile(gaussian, 25)
q3 = np.percentile(gaussian, 75)
# return np.array([mean, mean-0.01, mean+0.01])
return std
activity = np.transpose(activities[0])
print(activities[0].shape)
for i in activity:
print(create_gaussian(i))
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# ## Read Data
# reading Datasets
path = "/kaggle/input/amazon-sales-dataset/amazon.csv"
df = pd.read_csv(path)
df.head()
df.dtypes
# Links wont be useful for me
df.drop(["img_link", "product_link"], inplace=True, axis=1)
df.info()
# ## Null values
df.isnull().sum()
df.loc[df.rating_count.isnull()]
df.drop(index=[282, 324], inplace=True)
df.info()
# ### Changine dtype as some numrical variables are O type
df.actual_price = df.actual_price.str.replace("₹", "")
df.actual_price = df.actual_price.str.replace(",", "")
df.discounted_price = df.discounted_price.str.replace("₹", "")
df.discounted_price = df.discounted_price.str.replace(",", "")
df.discount_percentage = df.discount_percentage.str.replace("%", "")
df.rating_count = df.rating_count.str.replace(",", "")
# df.rating.astype('float64')
df.rating.unique()
df.loc[df.rating == "|"]
df.drop(index=1279, inplace=True)
df.info()
df.discounted_price = df.discounted_price.astype("float64")
df.actual_price = df.actual_price.astype("float64")
df.discount_percentage = df.discount_percentage.astype("float64")
df.rating_count = df.rating_count.astype("float64")
df.rating = df.rating.astype("float64")
df.info()
df.describe()
df.head()
import matplotlib.pyplot as plt
import seaborn as sns
|
# ### 1. Load library
import pandas as pd
import numpy as np
from sklearn.model_selection import train_test_split, GridSearchCV
from sklearn.preprocessing import StandardScaler
from sklearn.ensemble import RandomForestClassifier
from imblearn.over_sampling import SMOTE
import plotly.graph_objects as go
from plotly.subplots import make_subplots
import plotly.express as px
# ### 2. Read data and EDA using plotly
data = pd.read_csv(
"../input/stroke-prediction-dataset/healthcare-dataset-stroke-data.csv"
)
data.isnull().sum(axis=0)
fig = make_subplots(rows=5, cols=2)
fig.add_trace(
go.Bar(
x=data.gender.value_counts().index,
y=data.gender.value_counts().values,
name="Gender",
),
row=1,
col=1,
)
fig.add_trace(go.Histogram(x=data.age, name="Age"), row=1, col=2)
fig.add_trace(
go.Bar(
x=data.hypertension.value_counts().index,
y=data.hypertension.value_counts().values,
name="HyperTension",
),
row=2,
col=1,
)
fig.add_trace(
go.Bar(
x=data.heart_disease.value_counts().index,
y=data.heart_disease.value_counts().values,
name="heart_disease",
),
row=2,
col=2,
)
fig.add_trace(
go.Bar(
x=data.ever_married.value_counts().index,
y=data.ever_married.value_counts().values,
name="ever_married",
),
row=3,
col=1,
)
fig.add_trace(
go.Bar(
x=data.work_type.value_counts().index,
y=data.work_type.value_counts().values,
name="work_type",
),
row=3,
col=2,
)
fig.add_trace(
go.Bar(
x=data.Residence_type.value_counts().index,
y=data.Residence_type.value_counts().values,
name="Residence_type",
),
row=4,
col=1,
)
fig.add_trace(
go.Histogram(x=data.avg_glucose_level, name="avg_glucose_level"), row=4, col=2
)
fig.add_trace(
go.Bar(
x=data.smoking_status.value_counts().index,
y=data.smoking_status.value_counts().values,
name="smoking_status",
),
row=5,
col=1,
)
fig.add_trace(
go.Bar(
x=data.stroke.value_counts().index,
y=data.stroke.value_counts().values,
name="stroke",
),
row=5,
col=2,
)
fig.show()
other_index = data[data["gender"] == "Other"].index
data = data.drop(other_index)
px.histogram(data, x="bmi", color="stroke")
bmi_mean = round(data.bmi.mean(skipna=True), 1)
data["bmi"] = data["bmi"].fillna(bmi_mean)
data.bmi.isnull().any()
# ### 3. Data Preprosessing
# > In general, children is 'never smoked' category.
# > So, If work_types are children, it can be modified from 'Unknown' to 'never smoked'.
px.bar(data, x="smoking_status", color="work_type")
data.loc[
(data["smoking_status"] == "Unknown") & (data["work_type"] == "children"),
"smoking_status",
] = "never smoked"
px.bar(data, x="smoking_status", color="work_type")
gender_dummy = pd.get_dummies(data.gender)
ever_married_dummy = pd.get_dummies(data.ever_married)
work_type_dummy = pd.get_dummies(data.work_type)
Residence_type_dummy = pd.get_dummies(data.Residence_type)
sc = StandardScaler()
sc.fit(data[["age", "avg_glucose_level", "bmi"]])
data[["age", "avg_glucose_level", "bmi"]] = sc.transform(
data[["age", "avg_glucose_level", "bmi"]]
)
data = pd.concat(
[data, gender_dummy, ever_married_dummy, work_type_dummy, Residence_type_dummy],
axis="columns",
)
data = data.drop(
["gender", "ever_married", "work_type", "Residence_type"], axis="columns"
)
# > For data analysis, I will predict 'Unknown' type using randomforest
smoke_train = data.copy()
# gender_dummy = pd.get_dummies(smoke_train.gender)
# ever_married_dummy = pd.get_dummies(smoke_train.ever_married)
# work_type_dummy = pd.get_dummies(smoke_train.work_type)
# Residence_type_dummy = pd.get_dummies(smoke_train.Residence_type)
# sc = StandardScaler()
# sc.fit(smoke_train[['age', 'avg_glucose_level', 'bmi']])
# smoke_train[['age', 'avg_glucose_level', 'bmi']] = sc.transform(smoke_train[['age', 'avg_glucose_level', 'bmi']])
# smoke_train = pd.concat([smoke_train, gender_dummy, ever_married_dummy,work_type_dummy,Residence_type_dummy], axis='columns')
# smoke_train = smoke_train.drop(['id','gender','ever_married','work_type','Residence_type'], axis='columns')
smoke_tr = smoke_train.loc[smoke_train["smoking_status"] != "Unknown", :]
smoke_te = smoke_train.loc[smoke_train["smoking_status"] == "Unknown", :]
smoke_x = smoke_tr.drop(["smoking_status"], axis="columns")
smoke_y = smoke_tr["smoking_status"]
smoke_y.isnull().any()
rf = RandomForestClassifier()
rf.fit(smoke_x, smoke_y)
smoke_te = smoke_te.drop(["smoking_status"], axis="columns")
smoke_pred = rf.predict(smoke_te)
smoke_te["smoking_status"] = smoke_pred
data_pre = pd.concat([smoke_tr, smoke_te], axis="rows")
data_pre.isnull().any()
# ### 4. SMOTE (Unbalancing stroke data)
data_pre["stroke"].value_counts()
smoking_status_dummy = pd.get_dummies(data_pre.smoking_status)
data_pre = pd.concat([data_pre, smoking_status_dummy], axis="columns")
data_pre = data_pre.drop(["smoking_status"], axis="columns")
data_pre
sm = SMOTE(random_state=0)
X_train = data_pre.drop(["stroke"], axis="columns")
y_train = data_pre["stroke"]
X_resampled, y_resampled = sm.fit_resample(X_train, list(y_train))
print("After OverSampling, the shape of train_X: {}".format(X_resampled.shape))
print("After OverSampling, the shape of train_y: {} \n".format(X_resampled.shape))
|
# # import modules
import os
import glob
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
# # Overview
# - **objective** : detect the start and stop of freezing of gait and types of FOG events: Start Hesitation, Turn, and Walking.
# - The competition dataset includes time-series data from 3-D accelerometer on subjects lower-back.
# - The data series include three dataset, collected under distinct circumstances.
# - The tDCS FOG (**tdcsfog**) : Collected in the lab. Subjects completed a FOG-provoking protcol.
# - The DeFOG (**defog**) : Collected in the subject's home. Subject completed a FOG-provoking protcol.
# - The Daily Living (**daily**:) : Comprising one week of continuous 24/7 recordings from sixty-five subjects. Forty-five subjects exhibit FOG symptoms (series in the defog dataset), while the other twenty subjects exhibit no FOG symptoms (no series).
#
# - The tdcsfog and defog dataset include annotation by expert reviewers.
# - The daily dataset are *unannotated*. This dataset may be useful for modeling unsupervised or semi-supervised learning.
# # Explore train data
# ## tdcsfog
# **Memo**
# - Measurement frequency: 128 Hz(128 timesteps per second)
# - The column names **[AccV, AccML, AccAP]** represent the measured values of a 3-axis accelerometer, where V denotes vertical, ML denotes anterior-posterior, and AP denotes medial-lateral directions. The unit is **m/s^2**.
# - The column names **[StartHesitation, Turn, Walking]** indicate the presence of events. 0 (False) indicates FOG and 1 (True) indicates the occurrence of FOG.
pdir = "/kaggle/input/tlvmc-parkinsons-freezing-gait-prediction/"
# ### Anumber of csv files
# - There are 833 csv files for 65 subjects.
# - Subjects' acceleration and event series may have been subdivided.
tdcsfog_train_files = os.listdir(os.path.join(pdir, "train/tdcsfog"))
print(f"a number of files: {len(tdcsfog_train_files)}")
# ### Check a csv file
# - The column "Time" is recorded as integer values.
# - The sampling frequency is 128Hz, so it was assumed that each row represents a time interval of 1/128 seconds, but it may instead represent simple measurement points contrary to my expectations.
csv_fname = tdcsfog_train_files[0]
df_tdcs_train = pd.read_csv(os.path.join(pdir, "train/tdcsfog", csv_fname))
display(df_tdcs_train)
# ### Visualize a DataFrame
# - The visualization example below suggests the occurrence of a FOG event in Turn.
rows = 3
columns = 2
names = ["AccV", "AccML", "AccAP", "StartHesitation", "Turn", "Walking"]
i = 0
fig, ax = plt.subplots(3, 2, figsize=(15, 8))
for c in range(columns):
for r in range(rows):
ax[r, c].plot(df_tdcs_train[names[i]])
ax[r, c].set_title(names[i])
i += 1
fig.suptitle(csv_fname, fontsize=20)
plt.tight_layout(rect=[0, 0, 1, 0.99])
plt.show()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
#
# # Table of content
#
# * [1. Introduction](#1)
# - [Problem statement](#1.1)
# - [Data description](#1.2)
#
# * [2. Import Libraries](#2)
#
# * [3. Basic Exploration](#3)
# - [Read dataset](#3.1)
# - [Some information](#3.2)
# - [Data transformation](#3.3)
# - [Data visualization](#3.4)
# * [4. Machine Learning model](#4)
#
# * [5 Conclusion](#5)
# * [6 Author Message](#6)
# # Introduction
# Problem statement
# Data description
# # Import Libraries
import pandas as pd
import numpy as np
pd.plotting.register_matplotlib_converters()
import matplotlib.pyplot as plt
import seaborn as sns
import plotly.express as px
from pandas.api.types import CategoricalDtype
print("Setup Complete")
#
# # Basic Exploration
# Read dataset
def read_dataset(file_path):
data = pd.read_csv(file_path, index_col=0)
return data
data = read_dataset(
"/kaggle/input/customer-shopping-dataset/customer_shopping_data.csv"
)
#
# Some information
data.head()
data.shape
data.info()
data.nunique()
data.duplicated().any()
#
# Data transformation
# >
# Missing Data Treatment
total_null = data.isnull().sum().sort_values(ascending=False)
percent = ((data.isnull().sum() / data.isnull().count()) * 100).sort_values(
ascending=False
)
print("Total records = ", data.shape[0])
missing_data = pd.concat(
[total_null, percent.round(2)], axis=1, keys=["Total Missing", "In Percent"]
)
missing_data
# >
# Duplicated Data Treatment
duplicated_data = pd.DataFrame(data.loc[data.duplicated()].count())
duplicated_data.columns = ["Total Duplicate"]
duplicated_data
# >
# Clean Data
print("Gender :", data["gender"].unique().tolist())
print("Category :", data["category"].unique().tolist())
print("Payment method :", data["payment_method"].unique().tolist())
print("Shopping mall :", data["shopping_mall"].unique().tolist())
data["age_group"] = pd.cut(
x=data["age"],
bins=[0, 16, 30, 45, 100],
labels=["Child", "Young Adults", "Middle-aged Adults", "Old-aged Adults"],
)
data[["age", "age_group"]].head(10)
data["invoice_date"] = pd.to_datetime(data["invoice_date"])
data["day"] = data["invoice_date"].dt.day
data["month"] = data["invoice_date"].dt.strftime("%b")
data["year"] = data["invoice_date"].dt.year.astype("str")
data["day_of_week"] = data["invoice_date"].dt.day_name()
data[["quantity", "price"]].describe().round(2)
data.head()
#
# Data visualization
plt.style.use("seaborn")
fig = plt.figure(figsize=(10, 5))
colors = ["steelblue", "lightcoral"]
sns.histplot(data=data, x="age", hue="gender", palette=colors, alpha=0.7)
plt.title("Age Distribution by Gender", pad=10, fontsize=15)
plt.ylabel("Quality", labelpad=20)
plt.xlabel("Age", labelpad=20)
plt.legend(title="Gender", loc="upper right")
sns.despine()
plt.tight_layout()
plt.show()
df_age_group = data.groupby("age_group")["customer_id"].count().reset_index()
df_age_group.columns = ["age_group", "quality"]
df_age_group["percent"] = (
df_age_group["quality"] / df_age_group["quality"].sum() * 100
).round(2)
df_category = data.groupby("category")["customer_id"].count().reset_index()
df_category.columns = ["category", "quality"]
df_category["percent"] = (
df_category["quality"] / df_category["quality"].sum() * 100
).round(2)
df_payment_method = data.groupby("payment_method")["customer_id"].count().reset_index()
df_payment_method.columns = ["payment_method", "quality"]
df_payment_method["percent"] = (
df_payment_method["quality"] / df_payment_method["quality"].sum() * 100
).round(2)
df_shopping_mall = data.groupby("shopping_mall")["customer_id"].count().reset_index()
df_shopping_mall.columns = ["shopping_mall", "quality"]
df_shopping_mall["percent"] = (
df_shopping_mall["quality"] / df_shopping_mall["quality"].sum() * 100
).round(2)
fig, ((ax1, ax2), (ax3, ax4)) = plt.subplots(2, 2, figsize=(12, 8))
colors = sns.color_palette("Set2")
ax1.pie(
df_age_group["percent"],
labels=df_age_group["age_group"].tolist(),
colors=colors,
autopct="%1.1f%%",
startangle=90,
shadow=True,
wedgeprops=dict(width=0.5),
)
ax1.set_title("Distribution of Values by Age Group", pad=20, fontsize=15)
ax1.axis("equal")
# ax1.legend(loc="upper right", labels=df_age_group['age_group'].tolist(), bbox_to_anchor=(-0.2, 1), ncol=1)
ax2.pie(
df_category["percent"],
labels=df_category["category"].tolist(),
colors=colors,
autopct="%1.1f%%",
startangle=90,
shadow=True,
wedgeprops=dict(width=0.5),
)
ax2.set_title("Distribution of Values by Category", pad=20, fontsize=15)
ax2.axis("equal")
# ax2.legend(loc="upper right", labels=df_category['category'].tolist(), bbox_to_anchor=(-0.2, 1), ncol=1)
ax3.pie(
df_payment_method["percent"],
labels=df_payment_method["payment_method"].tolist(),
colors=colors,
autopct="%1.1f%%",
startangle=90,
shadow=True,
wedgeprops=dict(width=0.5),
)
ax3.set_title("Distribution of Values by Payment Method", pad=20, fontsize=15)
ax3.axis("equal")
# ax3.legend(loc="upper right", labels=df_payment_method['payment_method'].tolist(), bbox_to_anchor=(-0.2, 1), ncol=1)
ax4.pie(
df_shopping_mall["percent"],
labels=df_shopping_mall["shopping_mall"].tolist(),
colors=colors,
autopct="%1.1f%%",
startangle=90,
shadow=True,
wedgeprops=dict(width=0.5),
)
ax4.set_title("Distribution of Values by Shopping Mall", pad=20, fontsize=15)
ax4.axis("equal")
# ax4.legend(loc="upper right", labels=df_shopping_mall['shopping_mall'].tolist(), bbox_to_anchor=(-0.2, 1), ncol=1)
plt.subplots_adjust(hspace=1.0)
plt.tight_layout()
plt.show()
df_day = data.groupby(["gender", "day"])["quantity"].sum().reset_index()
months = [
"Jan",
"Feb",
"Mar",
"Apr",
"May",
"Jun",
"Jul",
"Aug",
"Sep",
"Oct",
"Nov",
"Dec",
]
month_type = CategoricalDtype(categories=months, ordered=True)
df_month = data.groupby(["gender", "month"])["quantity"].sum().reset_index()
df_month["month"] = df_month["month"].astype(month_type)
df_year = data.groupby(["gender", "year"])["quantity"].sum().reset_index()
cats = ["Monday", "Tuesday", "Wednesday", "Thursday", "Friday", "Saturday", "Sunday"]
cat_type = CategoricalDtype(categories=cats, ordered=True)
df_day_of_week = data.groupby(["gender", "day_of_week"])["quantity"].sum().reset_index()
df_day_of_week["day_of_week"] = df_day_of_week["day_of_week"].astype(cat_type)
fig, axes = plt.subplots(2, 2, figsize=(16, 10), sharey=True)
sns.lineplot(
ax=axes[0, 0], data=df_day, x="day", y="quantity", hue="gender", palette="Dark2"
)
sns.lineplot(
ax=axes[0, 1], data=df_month, x="month", y="quantity", hue="gender", palette="Dark2"
)
sns.lineplot(
ax=axes[1, 0], data=df_year, x="year", y="quantity", hue="gender", palette="Dark2"
)
sns.lineplot(
ax=axes[1, 1],
data=df_day_of_week,
x="day_of_week",
y="quantity",
hue="gender",
palette="Dark2",
)
axes[0, 0].set_title("Quantity by Day", pad=10, fontsize=15)
axes[0, 0].set_ylabel("Number of Products", labelpad=20)
axes[0, 0].set_xlabel("Day", labelpad=20)
axes[0, 1].set_title("Quantity by Month", pad=10, fontsize=15)
axes[0, 1].set_ylabel("Number of Products", labelpad=20)
axes[0, 1].set_xlabel("Month", labelpad=20)
axes[1, 0].set_title("Quantity by Year", pad=10, fontsize=15)
axes[1, 0].set_ylabel("Number of Products", labelpad=20)
axes[1, 0].set_xlabel("Year", labelpad=20)
axes[1, 1].set_title("Quantity by Weekday", pad=10, fontsize=15)
axes[1, 1].set_ylabel("Number of Products", labelpad=20)
axes[1, 1].set_xlabel("Weekday", labelpad=20)
plt.tight_layout()
plt.show()
df_day = data.groupby(["gender", "day"])["price"].sum().reset_index()
months = [
"Jan",
"Feb",
"Mar",
"Apr",
"May",
"Jun",
"Jul",
"Aug",
"Sep",
"Oct",
"Nov",
"Dec",
]
month_type = CategoricalDtype(categories=months, ordered=True)
df_month = data.groupby(["gender", "month"])["price"].sum().reset_index()
df_month["month"] = df_month["month"].astype(month_type)
df_year = data.groupby(["gender", "year"])["price"].sum().reset_index()
cats = ["Monday", "Tuesday", "Wednesday", "Thursday", "Friday", "Saturday", "Sunday"]
cat_type = CategoricalDtype(categories=cats, ordered=True)
df_day_of_week = data.groupby(["gender", "day_of_week"])["price"].sum().reset_index()
df_day_of_week["day_of_week"] = df_day_of_week["day_of_week"].astype(cat_type)
fig, axes = plt.subplots(2, 2, figsize=(16, 10), sharey=True)
sns.lineplot(
ax=axes[0, 0], data=df_day, x="day", y="price", hue="gender", palette="Dark2"
)
sns.lineplot(
ax=axes[0, 1], data=df_month, x="month", y="price", hue="gender", palette="Dark2"
)
sns.lineplot(
ax=axes[1, 0], data=df_year, x="year", y="price", hue="gender", palette="Dark2"
)
sns.lineplot(
ax=axes[1, 1],
data=df_day_of_week,
x="day_of_week",
y="price",
hue="gender",
palette="Dark2",
)
axes[0, 0].set_title("Price by Day", pad=10, fontsize=15)
axes[0, 0].set_ylabel("Number of Products", labelpad=20)
axes[0, 0].set_xlabel("Day", labelpad=20)
axes[0, 1].set_title("Price by Month", pad=10, fontsize=15)
axes[0, 1].set_ylabel("Number of Products", labelpad=20)
axes[0, 1].set_xlabel("Month", labelpad=20)
axes[1, 0].set_title("Price by Year", pad=10, fontsize=15)
axes[1, 0].set_ylabel("Number of Products", labelpad=20)
axes[1, 0].set_xlabel("Year", labelpad=20)
axes[1, 1].set_title("Price by Weekday", pad=10, fontsize=15)
axes[1, 1].set_ylabel("Number of Products", labelpad=20)
axes[1, 1].set_xlabel("Weekday", labelpad=20)
plt.tight_layout()
plt.show()
months = [
"Jan",
"Feb",
"Mar",
"Apr",
"May",
"Jun",
"Jul",
"Aug",
"Sep",
"Oct",
"Nov",
"Dec",
]
month_type = CategoricalDtype(categories=months, ordered=True)
df_month = pd.DataFrame(
data.groupby(["shopping_mall", "month"])["shopping_mall"].count()
)
df_month.columns = ["quality"]
df_month = df_month.reset_index()
df_month["month"] = df_month["month"].astype(month_type)
cats = ["Monday", "Tuesday", "Wednesday", "Thursday", "Friday", "Saturday", "Sunday"]
cat_type = CategoricalDtype(categories=cats, ordered=True)
df_day_of_week = pd.DataFrame(
data.groupby(["shopping_mall", "day_of_week"])["shopping_mall"].count()
)
df_day_of_week.columns = ["quality"]
df_day_of_week = df_day_of_week.reset_index()
df_day_of_week["day_of_week"] = df_day_of_week["day_of_week"].astype(cat_type)
# Set a custom color palette
colors = ["#a6cee3", "#1f78b4", "#b2df8a", "#33a02c", "#fb9a99", "#e31a1c"]
# Create subplots
fig, axes = plt.subplots(1, 2, figsize=(16, 10), sharey=True)
# Plot line charts with custom color palette and linewidth
sns.lineplot(
ax=axes[0],
data=df_month,
x="month",
y="quality",
hue="shopping_mall",
linewidth=2.5,
)
sns.lineplot(
ax=axes[1],
data=df_day_of_week,
x="day_of_week",
y="quality",
hue="shopping_mall",
linewidth=2.5,
)
# Set titles, labels, and legends
axes[0].set_title(
"Shopping Mall Performance by Month", pad=10, fontsize=18, fontweight="bold"
)
axes[0].set_ylabel("Quality", labelpad=20, fontsize=14)
axes[0].set_xlabel("Month", labelpad=20, fontsize=14)
axes[0].legend(loc="upper left", bbox_to_anchor=(0, 1), fontsize=12)
axes[1].set_title(
"Shopping Mall Performance by Weekday", pad=10, fontsize=18, fontweight="bold"
)
axes[1].set_ylabel("Quality", labelpad=20, fontsize=14)
axes[1].set_xlabel("Weekday", labelpad=20, fontsize=14)
axes[1].legend(loc="upper left", bbox_to_anchor=(0, 1), fontsize=12)
# Add gridlines to the plots
for ax in axes:
ax.grid(axis="y", alpha=0.5)
# Add a tight layout and show the plots
plt.tight_layout()
plt.show()
plt.style.use("seaborn")
# Create a pivot table of count of categories by payment method
table = pd.pivot_table(
data, values="quantity", index="category", columns="payment_method", aggfunc="count"
)
# Define custom colors for each payment method
colors = ["steelblue", "limegreen", "gold"]
# Plot a stacked bar chart
ax = table.plot(kind="bar", stacked=True, figsize=(10, 5), color=colors)
# Set chart title and axis labels
ax.set_title("Distribution of Categories by Payment Method", pad=10, fontsize=15)
ax.set_xlabel("Category", labelpad=20, fontsize=12)
ax.set_ylabel("Count", labelpad=20, fontsize=12)
# Add a legend
handles, labels = ax.get_legend_handles_labels()
ax.legend(handles, labels, title="Payment Method", loc="upper right")
# Remove top and right spines
sns.despine()
# Show the chart
plt.show()
plt.style.use("seaborn")
# Create a pivot table of count of categories by payment method
table = pd.pivot_table(
data,
values="quantity",
index="shopping_mall",
columns="payment_method",
aggfunc="count",
)
# Define custom colors for each payment method
colors = ["steelblue", "limegreen", "gold"]
# Plot a stacked bar chart
ax = table.plot(kind="bar", stacked=True, figsize=(10, 5), color=colors)
# Set chart title and axis labels
ax.set_title("Distribution of Shopping mall by Payment Method", pad=10, fontsize=15)
ax.set_xlabel("Shopping mall", labelpad=20, fontsize=12)
ax.set_ylabel("Count", labelpad=20, fontsize=12)
# Add a legend
handles, labels = ax.get_legend_handles_labels()
ax.legend(handles, labels, title="Payment Method", loc="upper right")
# Remove top and right spines
sns.despine()
# Show the chart
plt.show()
plt.style.use("seaborn")
# Create a pivot table of count of categories by payment method
table = pd.pivot_table(
data, values="quantity", index="category", columns="gender", aggfunc="count"
)
# Define custom colors for each payment method
colors = ["steelblue", "limegreen", "gold"]
# Plot a stacked bar chart
ax = table.plot(kind="bar", stacked=True, figsize=(10, 5), color=colors)
# Set chart title and axis labels
ax.set_title("Distribution of Categories by Gender", pad=10, fontsize=15)
ax.set_xlabel("Category", labelpad=20, fontsize=12)
ax.set_ylabel("Count", labelpad=20, fontsize=12)
# Add a legend
handles, labels = ax.get_legend_handles_labels()
ax.legend(handles, labels, title="Gender", loc="upper right")
# Remove top and right spines
sns.despine()
# Show the chart
plt.show()
plt.style.use("seaborn")
# Create a pivot table of count of categories by payment method
table = pd.pivot_table(
data, values="quantity", index="shopping_mall", columns="gender", aggfunc="count"
)
# Define custom colors for each payment method
colors = ["steelblue", "limegreen", "gold"]
# Plot a stacked bar chart
ax = table.plot(kind="bar", stacked=True, figsize=(10, 5), color=colors)
# Set chart title and axis labels
ax.set_title("Distribution of Shopping mall by Gender", pad=10, fontsize=15)
ax.set_xlabel("Shopping mall", labelpad=20, fontsize=12)
ax.set_ylabel("Count", labelpad=20, fontsize=12)
# Add a legend
handles, labels = ax.get_legend_handles_labels()
ax.legend(handles, labels, title="Gender", loc="upper right")
# Remove top and right spines
sns.despine()
# Show the chart
plt.show()
plt.style.use("seaborn")
# Create a pivot table of count of categories by payment method
table = pd.pivot_table(
data, values="quantity", index="category", columns="age_group", aggfunc="count"
)
# Define custom colors for each payment method
colors = ["steelblue", "limegreen", "gold"]
# Plot a stacked bar chart
ax = table.plot(kind="bar", stacked=True, figsize=(10, 5), color=colors)
# Set chart title and axis labels
ax.set_title("Distribution of Categories by Age Group", pad=10, fontsize=15)
ax.set_xlabel("Category", labelpad=20, fontsize=12)
ax.set_ylabel("Count", labelpad=20, fontsize=12)
# Add a legend
handles, labels = ax.get_legend_handles_labels()
ax.legend(handles, labels, title="Age Group", loc="upper right")
# Remove top and right spines
sns.despine()
# Show the chart
plt.show()
plt.style.use("seaborn")
# Create a pivot table of count of categories by payment method
table = pd.pivot_table(
data, values="quantity", index="shopping_mall", columns="age_group", aggfunc="count"
)
# Define custom colors for each payment method
colors = ["steelblue", "limegreen", "gold"]
# Plot a stacked bar chart
ax = table.plot(kind="bar", stacked=True, figsize=(10, 5), color=colors)
# Set chart title and axis labels
ax.set_title("Distribution of Shopping mall by Age Group", pad=10, fontsize=15)
ax.set_xlabel("Shopping mall", labelpad=20, fontsize=12)
ax.set_ylabel("Count", labelpad=20, fontsize=12)
# Add a legend
handles, labels = ax.get_legend_handles_labels()
ax.legend(handles, labels, title="Age Group", loc="upper right")
# Remove top and right spines
sns.despine()
# Show the chart
plt.show()
# Create a box plot of age by category
sns.set(style="ticks", palette="pastel")
plt.figure(figsize=(16, 6))
sns.boxplot(x="category", y="age", data=data)
sns.despine(offset=10, trim=True)
plt.title("Distribution of Age by Product Category", fontsize=16)
plt.xlabel("Category", fontsize=14)
plt.ylabel("Age", fontsize=14)
plt.xticks(fontsize=12)
plt.yticks(fontsize=12)
plt.show()
sns.set_style("whitegrid")
fig, ax = plt.subplots(figsize=(15, 8))
sns.boxplot(x="category", y="price", data=data, palette="pastel", ax=ax)
sns.despine(offset=10, trim=True)
ax.set_title("Distribution of Price by Product Category", fontsize=16)
ax.set_xlabel("Category", fontsize=14)
ax.set_ylabel("Price", fontsize=14)
plt.xticks(fontsize=12)
plt.yticks(fontsize=12)
plt.show()
fig, axes = plt.subplots(1, 2, figsize=(15, 5))
sns.set(style="ticks", palette="pastel")
sns.boxplot(ax=axes[0], x="payment_method", y="age", data=data)
sns.boxplot(ax=axes[1], x="payment_method", y="price", data=data)
for ax_idx, var in enumerate(["age", "price"]):
axes[ax_idx].set_title(
f"Distribution of {var.capitalize()} by Payment Method", fontsize=16
)
axes[ax_idx].set_xlabel("Payment Method", fontsize=14)
axes[ax_idx].set_ylabel(f"{var.capitalize()}", fontsize=14)
sns.despine(offset=10, trim=True)
plt.xticks(fontsize=12)
plt.yticks(fontsize=12)
plt.tight_layout(pad=2)
plt.show()
# Set the size of the figure
plt.figure(figsize=(20, 8))
# Set the style of the plot
sns.set_style("whitegrid")
# Create a violin plot with gender as hue
sns.violinplot(x="shopping_mall", y="price", hue="gender", data=data, palette="Set2")
# Add a title and axis labels
plt.title("Distribution of Price by Shopping Mall", fontsize=18, fontweight="bold")
plt.xlabel("Shopping Mall", fontsize=14)
plt.ylabel("Price", fontsize=14)
# Increase the font size of the tick labels
plt.xticks(fontsize=12)
plt.yticks(fontsize=12)
# Display the plot
plt.show()
# Set the size of the figure
plt.figure(figsize=(10, 5))
# Set the style of the plot
sns.set_style("whitegrid")
# Create a violin plot with gender as hue
sns.violinplot(x="age_group", y="price", hue="gender", data=data, palette="Set2")
# Add a title and axis labels
plt.title("Distribution of Price by Age Group", fontsize=18, fontweight="bold")
plt.xlabel("Age Group", fontsize=14)
plt.ylabel("Price", fontsize=14)
# Increase the font size of the tick labels
plt.xticks(fontsize=12)
plt.yticks(fontsize=12)
plt.tight_layout()
# Display the plot
plt.show()
# create a pivot table to count frequency of each category at each shopping mall
pivot_table = data.pivot_table(
index="shopping_mall", columns="category", values="quantity", aggfunc="count"
)
# create heatmap
fig, ax = plt.subplots(figsize=(10, 8))
sns.heatmap(pivot_table, cmap="Blues", annot=True, fmt="g", linewidths=0.5, ax=ax)
# set plot title and axis labels
ax.set_title("Frequency of Product Categories by Shopping Mall", fontsize=16)
ax.set_xlabel("Category", fontsize=14)
ax.set_ylabel("Shopping Mall", fontsize=14)
plt.show()
|
# # Electronic Structure using Qiskit Nature
# Link : https://qiskit.org/documentation/nature/tutorials/01_electronic_structure.html
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
from qiskit_nature.units import DistanceUnit
from qiskit_nature.second_q.drivers import PySCFDriver
from qiskit_nature.second_q.algorithms import (
GroundStateEigensolver,
NumPyMinimumEigensolverFactory,
)
from qiskit_nature.second_q.mappers import JordanWignerMapper, QubitConverter
driver = PySCFDriver(
atom="H 0 0 0; H 0 0 0.735",
basis="sto3g",
charge=0,
spin=0,
unit=DistanceUnit.ANGSTROM,
)
problem = driver.run()
problem.__dict__
hamiltonian = problem.hamiltonian
coefficients = hamiltonian.electronic_integrals
print(coefficients.alpha)
second_q_op = hamiltonian.second_q_op()
print(second_q_op)
hamiltonian.nuclear_repulsion_energy # NOT included in the second_q_op above
solver = GroundStateEigensolver(
QubitConverter(JordanWignerMapper()),
NumPyMinimumEigensolverFactory(),
)
result = solver.solve(problem)
print(result)
dict_res = result.__dict__
no = 0
for key, value in dict_res.items():
no += 1
print(no, key, " : ", value, "\n")
result.eigenstates
NR = result._nuclear_repulsion_energy
GE = result._computed_energies.item()
Total = GE + NR
print(NR, GE, Total)
# ## Function
def experiment(dist):
# Molecule
molecule = "H 0 0 -{0}; H 0 0 {0}"
driver = PySCFDriver(
atom=molecule.format(dist / 2),
basis="sto3g",
charge=0,
spin=0,
unit=DistanceUnit.ANGSTROM,
)
# Set up driver
problem = driver.run()
# Ground State Eigen Solver
numpy_solver = NumPyMinimumEigensolverFactory()
converter = QubitConverter(JordanWignerMapper())
solver = GroundStateEigensolver(converter, numpy_solver)
# Result
result = solver.solve(problem)
nr_energy = result._nuclear_repulsion_energy
com_energy = result._computed_energies.item()
total_energy = com_energy + nr_energy
return total_energy, nr_energy, com_energy
distances = np.arange(0.2, 3, 0.1)
print("number of step:", len(distances))
total_energies = []
nr_energies = []
elec_energies = []
for i, d in enumerate(distances):
if i % 10 == 0:
print("step-", i, end=" ")
total, nr, elec = experiment(d)
total_energies.append(total)
nr_energies.append(nr)
elec_energies.append(elec)
print("step-", i, end=" ")
print("Run Complete")
minE = min(total_energies)
loc_minE = np.where(total_energies == minE)
dist_minE = distances[loc_minE]
min_nr = nr_energies[loc_minE[0][0]]
min_elec = elec_energies[loc_minE[0][0]]
print("E0:", minE)
print("dist:", dist_minE)
# fig = plt.figure(figsize=(10, 5))
plt.plot(distances, total_energies, label="H2")
plt.plot(round(dist_minE[0], 4), minE, "x")
plt.text(
3,
0,
"min E({:}) = {:.5f}".format(round(dist_minE[0], 1), minE),
ha="right",
va="top",
fontsize=12,
)
plt.xlabel("Interatomic Distance")
plt.ylabel("Energy")
plt.title("Ground State Energy")
# plt.xticks(np.arange(min(distances), max(distances), 0.2))
plt.legend(loc="upper right")
plt.savefig("H2_energidasar.png")
fig = plt.figure(figsize=(16, 4))
plt.subplot(1, 3, 1)
plt.plot(distances, nr_energies)
plt.text(
3,
2.5,
"min E({:}) = {:.5f}".format(round(dist_minE[0], 1), min_nr),
ha="right",
va="top",
fontsize=12,
)
plt.xlabel("Interatomic Distance")
plt.ylabel("Energy")
plt.title("Nuclear Repulsion")
plt.subplot(1, 3, 2)
plt.plot(distances, elec_energies)
plt.text(
3,
-2.4,
"min E({:}) = {:.5f}".format(round(dist_minE[0], 1), min_elec),
ha="right",
va="top",
fontsize=12,
)
plt.xlabel("Interatomic Distance")
plt.ylabel("Energy")
plt.title("Electron Energies")
plt.savefig("H2_energi2.png", bbox_inches="tight", pad_inches=0)
data = {
"r": distances,
"Nuclear Repulsion": nr_energies,
"Electron Energies": elec_energies,
"Ground State Energy": total_energies,
}
hasil_df = pd.DataFrame(data)
hasil_df
hasil_df.to_csv("energidasar.csv")
# ## Hasil Quanttangle Competition
# 
# ## Optimal Bond Length
distances2 = np.arange(0.7, 0.8, 0.01)
print("number of step:", len(distances2))
total_energies2 = []
nr_energies2 = []
elec_energies2 = []
for i, d in enumerate(distances2):
print("step-", i, end=" ")
total, nr, elec = experiment(d)
total_energies2.append(total)
nr_energies2.append(nr)
elec_energies2.append(elec)
minE2 = min(total_energies2)
loc_minE2 = np.where(total_energies2 == minE2)
dist_minE2 = distances2[loc_minE2]
min_nr2 = nr_energies2[loc_minE2[0][0]]
min_elec2 = elec_energies2[loc_minE2[0][0]]
print("E0:", minE2)
print("dist:", dist_minE2)
# fig = plt.figure(figsize=(10, 5))
plt.plot(distances2, total_energies2, label="Hartree-Fock")
plt.plot(round(dist_minE2[0], 4), minE2, "x")
plt.text(
0.7,
-1.1345,
"min E({:}) = {:.5f}".format(round(dist_minE2[0], 4), minE2),
ha="left",
va="top",
fontsize=12,
)
plt.xlabel("Interatomic Distance")
plt.ylabel("Energy")
plt.title("H2 Ground State Energy")
plt.legend(loc="upper right")
# 
# - Link: https://quantaggle.com/competitions/gs-pes/
# ## Function NumpyMinimumEigensolver & VQE
from qiskit.algorithms.minimum_eigensolvers import NumPyMinimumEigensolver
from qiskit.algorithms.optimizers import SLSQP
from qiskit.primitives import Estimator
from qiskit_nature.second_q.algorithms import VQEUCCFactory
from qiskit_nature.second_q.circuit.library import UCCSD
def experiment2(dist):
# Molecule
molecule = "H 0 0 -{0}; H 0 0 {0}"
driver = PySCFDriver(
atom=molecule.format(dist / 2),
basis="sto3g",
charge=0,
spin=0,
unit=DistanceUnit.ANGSTROM,
)
# Run Driver
problem = driver.run()
# Solver
numpy_solver = NumPyMinimumEigensolver()
vqe_solver = VQEUCCFactory(Estimator(), UCCSD(), SLSQP())
# Calculation
converter = QubitConverter(JordanWignerMapper(), two_qubit_reduction=True)
calc_numpy = GroundStateEigensolver(converter, numpy_solver)
calc_vqe = GroundStateEigensolver(converter, vqe_solver)
# Result
res_numpy = calc_numpy.solve(problem)
res_vqe = calc_vqe.solve(problem)
nr_np = res_numpy._nuclear_repulsion_energy
com_np = res_numpy._computed_energies.item()
total_np = com_np + nr_np
nr_vqe = res_vqe._nuclear_repulsion_energy
com_vqe = res_vqe._computed_energies.item()
total_vqe = com_vqe + nr_vqe
return total_np, total_vqe
distances = np.arange(0.2, 3, 0.1)
GE_numpy = []
GE_vqe = []
for i, d in enumerate(distances):
if i % 10 == 0:
print("step-", i, end=" ")
t1, t2 = experiment2(d)
GE_numpy.append(t1)
GE_vqe.append(t2)
plt.plot(distances, GE_numpy, label="Numpy Solver")
plt.plot(distances, GE_vqe, "x", label="VQE Solver")
plt.xlabel("Interatomic Distance")
plt.ylabel("Energy")
plt.title("H2 Ground State Energy")
plt.legend(loc="upper right")
minE_np = min(GE_numpy)
loc_minE_np = np.where(GE_numpy == minE_np)
dist_minE_np = distances[loc_minE_np]
print("E0:", minE_np)
print("dist:", dist_minE_np)
minE_vqe = min(GE_vqe)
loc_minE_vqe = np.where(GE_vqe == minE_vqe)
dist_minE_vqe = distances[loc_minE_vqe]
print("E0:", minE_vqe)
print("dist:", dist_minE_vqe)
def experiment3(dist):
# Molecule
molecule = "He 0 0 -{0}; H 0 0 {0}"
driver = PySCFDriver(
atom=molecule.format(dist / 2),
basis="sto3g",
charge=1,
spin=0,
unit=DistanceUnit.ANGSTROM,
)
# Set up driver
problem = driver.run()
# Ground State Eigen Solver
method = NumPyMinimumEigensolver()
converter = QubitConverter(JordanWignerMapper(), two_qubit_reduction=True)
solver = GroundStateEigensolver(converter, method)
# Result
result = solver.solve(problem)
nr_energy = result._nuclear_repulsion_energy
com_energy = result._computed_energies.item()
total_energy = com_energy + nr_energy
return total_energy, nr_energy, com_energy
distances = np.arange(0.1, 3, 0.1)
print("number of step:", len(distances))
total_energies = []
nr_energies = []
elec_energies = []
for i, d in enumerate(distances):
if i % 10 == 0:
print("step-", i, end=" ")
total, nr, elec = experiment3(d)
total_energies.append(total)
nr_energies.append(nr)
elec_energies.append(elec)
print("step-", i, end=" ")
minE = min(total_energies)
loc_minE = np.where(total_energies == minE)
dist_minE = distances[loc_minE]
min_nr = nr_energies[loc_minE[0][0]]
min_elec = elec_energies[loc_minE[0][0]]
print("E0:", minE)
print("dist:", dist_minE)
# fig = plt.figure(figsize=(10, 5))
plt.plot(distances, total_energies, label="Hartree-Fock")
plt.text(
max(distances),
minE,
"min E({:}) = {:.5f}".format(round(dist_minE[0], 1), minE),
ha="right",
va="top",
fontsize=12,
)
plt.xlabel("Interatomic Distance")
plt.ylabel("Energy")
plt.title("HeHe Ground State Energy")
# plt.xticks(np.arange(min(distances), max(distances), 0.2))
plt.legend(loc="upper right")
fig = plt.figure(figsize=(16, 4))
plt.subplot(1, 3, 1)
plt.plot(distances, nr_energies)
plt.text(
max(distances),
min_nr,
"min E({:}) = {:.5f}".format(round(dist_minE[0], 1), min_nr),
ha="right",
va="top",
fontsize=12,
)
plt.xlabel("Interatomic Distance")
plt.ylabel("Energy")
plt.title("He2 Nuclear Repulsion")
plt.subplot(1, 3, 2)
plt.plot(distances, elec_energies)
plt.text(
0.2,
min_elec,
"min E({:}) = {:.5f}".format(round(dist_minE[0], 1), min_elec),
ha="left",
va="top",
fontsize=12,
)
plt.xlabel("Interatomic Distance")
plt.ylabel("Energy")
plt.title("He2 Electron Energies")
plt.subplot(1, 3, 3)
plt.plot(distances, total_energies)
plt.text(
max(distances),
minE,
"min E({:}) = {:.5f}".format(round(dist_minE[0], 1), minE),
ha="right",
va="top",
fontsize=12,
)
plt.xlabel("Interatomic Distance")
plt.ylabel("Energy")
plt.title("He2 Ground State Energy")
|
# ## Θεωρητικό Μέρος
# α) Σύμφωνα με το paper, η διαδικασία παραγωγής της Γκαουσιανής πυραμίδας ισοδυναμεί με την συνέλιξη της αρχικής εικονάς με ένα σετ ισοδύναμων συναρτήσεων βαρών h. Η συνάρτηση αυτή h μοιάζει όλο και περισσότερο με την συνάρτηση της Γκαουσιανής κατανομής όσο το α γίνεται μικρότερο της μονάδας αλλά καθώς η παράμετρος α προσεγγίζει την μονάδα, το σχήμα της συνάρτησης βαρών h παίρνει πιο τριγωνικές μορφές. Επιπλέον η παράμετρος α καθορίζει το κατά πόσο θα μειωθούν η διακύμανση και η εντροπία των ιστογραμμάτων των εικόνων του κάθε επιπέδου της πυραμίδας.
# b) Η εντροπία είναι ο ελάχιστος αριθμός από bits ανά pixel, που χρειαζόμαστε για να κωδικοποιήσουμε μία εικόνα. Επειδή χρησιμοποιούμε 8 bits για την αναπαράσταση του κάθε pixel σε μία grayscale εικόνα, άρα έχουμε 2^8=256 δυνατά αποτελέσματα, η μέγιστη εντροπία θα είναι:
# $ -\sum \limits _{n=0}^ {255}P(n)\log(P(n)) = -\sum \limits _{n=0}^{255}2^{(-8)}\log(2^{(-8)}) = -\log(2^{(-8)}) = 8 $
# c)
# d)
# ## Εργαστηριακό Μέρος
# ### Α. Υλοποίηση Αλγορίθμου
import numpy as np
from skimage import io
from skimage.transform import resize
from skimage import color
import matplotlib.pyplot as plt
def GKernel(a=0.0):
w_n = np.array(
[(0.25 - a / 2), 0.25, a, 0.25, (0.25 - a / 2)]
) # initializing row vector w(n) with given constraints
w_m = w_n.reshape((5, 1)) # initializing column vector w(m)
w = np.outer(w_m, w_n) # getting the 5x5 kernel
return w
def GReduce(I, h):
if I.ndim < 3: # grayscale image
window = 5
offset = window // 2
row, col = I.shape
if row % 2 == 0:
height = row - offset
else:
height = row - offset - 1
if col % 2 == 0:
width = row - offset
else:
width = row - offset - 1
nextLevel = np.zeros((width // 2 - 1, height // 2 - 1))
for i in range(2, width):
for j in range(2, height):
if j % 2 == 0 and i % 2 == 0:
patch = I[i - offset : i + offset + 1, j - offset : j + offset + 1]
psum = np.dot(patch, h).sum()
nextLevel[(i // 2) - 1, (j // 2) - 1] = psum
return nextLevel
else: # coloured image
window = 5
offset = window // 2
row, col, ch = I.shape
# splitting rgb channels to process seperately
red = I[:, :, 0]
green = I[:, :, 1]
blue = I[:, :, 2]
if row % 2 == 0:
height = row - offset
else:
height = row - offset - 1
if col % 2 == 0:
width = row - offset
else:
width = row - offset - 1
nextRedLevel = np.zeros((width // 2 - 1, height // 2 - 1, ch))
nextGreenLevel = np.zeros((width // 2 - 1, height // 2 - 1, ch))
nextBlueLevel = np.zeros((width // 2 - 1, height // 2 - 1, ch))
for i in range(2, width):
for j in range(2, height):
if j % 2 == 0 and i % 2 == 0:
patch = red[
i - offset : i + offset + 1, j - offset : j + offset + 1
]
psum = np.dot(patch, h).sum()
nextRedLevel[(i // 2) - 1, (j // 2) - 1] = psum
patch = green[
i - offset : i + offset + 1, j - offset : j + offset + 1
]
psum = np.dot(patch, h).sum()
nextGreenLevel[(i // 2) - 1, (j // 2) - 1] = psum
patch = blue[
i - offset : i + offset + 1, j - offset : j + offset + 1
]
psum = np.dot(patch, h).sum()
nextBlueLevel[(i // 2) - 1, (j // 2) - 1] = psum
nextLevel = np.array([nextRedLevel, nextGreenLevel, nextBlueLevel])
return nextRedLevel
img = io.imread("/kaggle/input/lenapng/lena.png")
img = img / 255
gray_img = color.rgb2gray(img)
red = GReduce(img[:, :, 0], GKernel(0.5))
plt.imshow(red, cmap="gray")
plt.show()
green = GReduce(img[:, :, 1], GKernel(0.5))
plt.imshow(green, cmap="gray")
plt.show()
blue = GReduce(img[:, :, 2], GKernel(0.5))
plt.imshow(blue, cmap="gray")
plt.show()
output_array = np.zeros((254, 254, 3))
output_array[:, :, 0] = red
output_array[:, :, 1] = green
output_array[:, :, 2] = blue
plt.imshow(output_array)
plt.show()
|
import pandas as pd
from sklearn.datasets import load_digits
info = load_digits()
info
# data.info() wont work here as load_digit is an pictorial data
dir(info)
# We used dir() because its an image data
import matplotlib.pyplot as plt
plt.gray()
for i in range(4):
plt.matshow(info.images[i])
# data.image becuase its an image based data
df = pd.DataFrame(info.data)
df.head()
# data.data is used because we have to identify Object and features/Atribute, Data is the table also here
# For Example Student.Age Student is Object and Age is the attribute
df["Target"] = info.target
df
# Why target is 0,1,2,3,4.....63 - Pixel distubution is stored
x = df.drop("Target", axis="columns")
y = df.Target
x
y
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(x, y, train_size=0.8)
from sklearn.ensemble import RandomForestClassifier
model = RandomForestClassifier()
model.fit(x_train, y_train)
model.score(x_test, y_test)
# Accuracy Measure is very high, If not required this much of high accuracy then you can tune theparameter
from sklearn.ensemble import RandomForestClassifier
model = RandomForestClassifier(n_estimators=20, max_depth=5)
model.fit(x_train, y_train)
# n_estimators=20 indicates I have taken 20 number of decision tree
# Default parameter=100, default impurity=gini
model.score(x_test, y_test)
from sklearn.ensemble import RandomForestClassifier
model = RandomForestClassifier(n_estimators=20)
model.fit(x_train, y_train)
# Default depth=None
model.score(x_test, y_test)
from sklearn.ensemble import RandomForestClassifier
model = RandomForestClassifier(n_estimators=20, max_depth=5, ccp_alpha=0.05)
model.fit(x_train, y_train)
# We took ccp_alpha=0.05 for tunning
model.score(x_test, y_test)
# We are working on random forest because we saw overfitting problem in decision tree
# So to avoid overfitting we require reduction in high bais
y_predicted = model.predict(x_test)
y_predicted
from sklearn.metrics import confusion_matrix
from sklearn.metrics import plot_confusion_matrix
cm = confusion_matrix(y_test, y_predicted)
cm
plot_confusion_matrix(model, x_test, y_test)
import matplotlib.pyplot as plt
plt.figure(figsize=(10, 7))
import seaborn as sns
sns.heatmap(cm, annot=True)
plt.xlabel("Predicted")
plt.ylabel("Actual")
|
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import datetime as dt
from kmodes.kprototypes import KPrototypes
from sklearn.preprocessing import StandardScaler
df = pd.read_csv(
"/kaggle/input/customer-personality-analysis/marketing_campaign.csv", sep="\t"
)
df.info()
pd.set_option("display.max_columns", None)
df.describe().round()
num_unique_consumerID = df["ID"].nunique()
num_unique_consumerID
df = df.rename(columns={"Response": "AcceptedCmp6"})
# Create a dictionary to map the campaign numbers to the column names
campaign_dict = {
"AcceptedCmp1": 1,
"AcceptedCmp2": 2,
"AcceptedCmp3": 3,
"AcceptedCmp4": 4,
"AcceptedCmp5": 5,
"AcceptedCmp6": 6,
}
# Loop through the rows and find the campaign where the customer accepted the offer
for i in range(len(df)):
for col, campaign in campaign_dict.items():
if df.loc[i, col] == 1:
df.loc[i, "AcceptedCmp"] = campaign
break
else:
df.loc[i, "AcceptedCmp"] = 0
# Delete the original campaign columns
df.drop(columns=campaign_dict.keys(), inplace=True)
df["AcceptedCmp"] = df["AcceptedCmp"].astype("object")
df["Teenhome"] = df["Teenhome"].astype("object")
df["Kidhome"] = df["Kidhome"].astype("object")
df["Complain"] = df["Complain"].astype("object")
# creating a new column age and enrollment_time and deleting year_birth and Dt_customer columns
# calculate age based on Year_Birth
current_year = dt.datetime.now().year
df["Age"] = current_year - df["Year_Birth"]
# convert 'Dt_Customer' to datetime format and calculate enrollment time
df["Dt_Customer"] = pd.to_datetime(df["Dt_Customer"], format="%d-%m-%Y")
df["Enrollment_time"] = ((dt.datetime.now() - df["Dt_Customer"]).dt.days / 12).round(1)
# drop the original columns
df.drop(columns=["Year_Birth", "Dt_Customer"], inplace=True)
# deleting the z_costcontact and Z_revenue columns as they have constant values.
df = df.drop(columns=["Z_CostContact", "Z_Revenue", "ID"])
df.describe().round()
df.head()
df.info()
# select columns with data type 'object'
object_cols = df.select_dtypes(include=["object"]).columns
# get the index of the object columns
object_cols_index = object_cols.to_list()
object_cols_index = [df.columns.get_loc(col) for col in object_cols]
print(object_cols_index) # prints [1, 2]
# select only the non-object columns
non_obj_cols = df.select_dtypes(exclude=["object"]).columns
# apply standardization to the non-object columns
scaler = StandardScaler()
df[non_obj_cols] = scaler.fit_transform(df[non_obj_cols])
df.describe().round()
|
import pandas as pd
import numpy as np
from matplotlib import pyplot as plt
import seaborn as sns
import xgboost as xgb
from boruta import BorutaPy
from sklearn.preprocessing import LabelEncoder, StandardScaler
from sklearn.metrics import accuracy_score
from sklearn.metrics import confusion_matrix
from sklearn.base import BaseEstimator, TransformerMixin
from sklearn.pipeline import Pipeline
import pickle
df_train = pd.read_csv(
"/kaggle/input/yoga82-cluster-nomarlization/Yoga82_cluster_normalization/normalized_train.csv",
index_col=0,
)
le = LabelEncoder()
le.fit(df_train["name"])
df_train["name"] = le.transform(df_train["name"])
# class LabelEncode(BaseEstimator, TransformerMixin):
# def fit(self, X, y=None):
# return self
# def transform(self, X):
# le = LabelEncoder()
# X["label"] = le.fit_transform(X["label"])
# return self
# class Scale(BaseEstimator, TransformerMixin):
# def fit(self, X, y=None):
# return self
# def transform(self, X):
# ss = StandardScaler()
# X["label"] = ss.fit_transform(X["label"])
# return self
def f_selector(gname, X):
df = X[X["group_name"] == gname]
X_train = df.drop(labels=["image_name", "name", "group_name"], axis=1).to_numpy()
y_train = df["name"]
dump_model = xgb.XGBClassifier()
local_encoder = LabelEncoder()
y_train = local_encoder.fit_transform(y_train)
feature_selector = BorutaPy(dump_model, n_estimators="auto", random_state=42)
feature_selector.fit(X_train, y_train)
print("ranking: ")
ranking = feature_selector.ranking_
print(ranking)
feature_used = [
df.columns[index] for index, value in enumerate(ranking) if value == 1
]
return feature_used
# class LabelEncode(BaseEstimator, TransformerMixin):
# def fit(self, X, y=None):
# return self
# def transform(self, X):
# le = LabelEncoder()
# X["label"] = le.fit_transform(X["label"])
# return self
# def train_invidual_group(gname, X):
# df = X[X["group_name"]==gname]
# X_train = df.drop(labels=["image_name","name","group_name"],axis=1)
# y_train = df["name"]
# pipe = Pipeline([
# ('encode', LabelEncoder())
# ])
# local_encoder = LabelEncoder()
# y_train = local_encoder.fit_transform(y_train)
# feature_used = f_selector(gname, X_train, y_train)
# X_train_filtered = X_train.loc[:,feature_used]
# xgb_model = xgb.XGBClassifier()
# xgb_model.fit(X_train_filterd, y_train)
# xgb_model.save_model(gname+".json")
# print(f"{gname} model: Done, saved to {gname}.json")
grouped_name = [
"Split",
"Leg_bend",
"Forward_bend_sitting",
"Side",
"Up_facing_wheel",
"Leg_straight_up",
"Forward_bend_standing",
"Others_standing",
"Side_facing",
"Up_facing_reclining",
"Down_facing_reclining",
"Plank_balance",
"Others_wheel",
"Front",
"Normal1",
"Twist",
"Normal2",
"Straight",
"Side_bend",
]
# note 'Down_facing_wheel' is contain only 1 label => ignored
stored = {}
for gname in grouped_name[15:17]:
feature_selection = f_selector(gname, df_train)
stored[gname] = feature_selection
import json
with open("selection_feature.json", "w") as outfile:
json.dump(stored, outfile)
# **TRAINING**
# xgb_model = xgb.XGBClassifier()
# xgb_model.fit(X_train_filtered, y_train_encoded)
# df_split_test = df_test[df_test["group_name"]=="Split"]
# X_test = df_split_test.drop(labels=["image_name","name","group_name"],axis=1)
# y_test = df_split_test["name"]
# X_demo = X_test.loc[:,["left_ear_z",
# "right_ear_z",
# "right_pinky_1_y",
# "right_hip_y",
# "left_eye_inner+mouth_right+right_shoulder",
# "left_ear+right_knee+right_ankle",
# "right_hip+right_knee+right_ear",
# "right_hip+right_shoulder+left_hip",
# "right_shoulder+right_hip+right_knee",
# "right_shoulder+right_hip+right_ankle",
# "left_knee+right_knee+left_elbow",
# "left_thumb_2+right_pinky_1+left_index_1"]]
# X_demo
# X_test_scaled = ss.transform(X_test)
# X_test_filtered = feature_selector.transform(X_test_scaled)
# y_test_encoded = le.transform(y_test)
# y_pred = xgb_model.predict(X_test_filtered)
# y_pred_label = le.inverse_transform(y_pred)
# accuracy_score(y_test,y_pred_label)
# cm = confusion_matrix(y_test, y_pred_label)
# sns.heatmap(cm, annot=True)
# xgb_model.save_model("split.json")
# import xgboost as xgb
# import pickle
# a = xgb.XGBModel
# bst = xgb.XGBClassifier()
# bst.load_model('/kaggle/input/test-boruta-split/split.json')
# X_test_scaled
# bst.predict(X_test_scaled)
# bst2 = xgb.Booster(model_file="/kaggle/input/test-boruta-split/split.json")
# import pickle
# bst3 = pickle.load(open("/kaggle/input/test-boruta-split/split.json", "rb"))
|
from keras.datasets import mnist
import numpy as np
from matplotlib.pyplot import plot as plt
import sys
def standardize(x):
return (x - np.mean(x)) / np.std(x)
def sigmoid(z):
return 1 / (1 + np.exp(-z))
def predict(X, W, b):
return sigmoid(np.dot(X, W) + b)
def cost(y, y_hat):
return np.mean(
-y * np.log(y_hat + sys.float_info.min)
- (1 - y) * np.log(1 - y_hat + sys.float_info.min)
)
def train(X, y, learning_rate, epochs):
W = np.random.random(X.shape[1])
b = np.random.random()
costs = np.array([])
for i in range(epochs):
y_hat = predict(X, W, b)
dW = np.dot(X.T, (y_hat - y)) / len(X)
db = np.mean(y_hat - y)
W -= learning_rate * dW
b -= learning_rate * db
if i % 100 == 0:
c: float = cost(y, y_hat)
costs = np.append(costs, c)
print(f"Cost at epoch {i}: {c}")
return W, b, costs
def test(X, y, W, b):
y_hat = predict(X, W, b)
y_hat = np.round(y_hat)
return accuracy(y, y_hat)
def k_fold_cross_validation(X, y, k, lr, epochs):
fold_size = len(X) // k
accuracies = []
for i in range(k):
# Divide the data into training and testing sets
X_train = np.concatenate([X[: i * fold_size], X[(i + 1) * fold_size :]])
Y_train = np.concatenate([y[: i * fold_size], y[(i + 1) * fold_size :]])
X_test = X[i * fold_size : (i + 1) * fold_size]
Y_test = y[i * fold_size : (i + 1) * fold_size]
# Train the model
print(f"----------Training at Fold {i + 1} of {k}----------")
w, b, c = train(X_train, Y_train, lr, epochs)
# Test the model
print(f"----------Testing at Fold {i + 1} of {k}----------")
acc = test(X_test, Y_test, w, b)
print(f"Accuracy at Fold {i + 1} of {k}: {acc}")
accuracies.append(acc)
return np.mean(accuracies), c
def accuracy(y, y_hat):
return np.mean(y == y_hat)
trainSet, testSet = mnist.load_data()
xTrain = trainSet[0].astype("float32")
yTrain = trainSet[1].astype("int32")
xTrain = xTrain.reshape(xTrain.shape[0], -1)
xTrain = np.concatenate([xTrain[yTrain == 0], xTrain[yTrain == 1]])
yTrain = np.concatenate([yTrain[yTrain == 0], yTrain[yTrain == 1]])
xTrain = standardize(xTrain)
k = 10
print(f"----------K-Fold Cross Validation with {k} Folds and eta 0.01----------")
average_accuracy, c = k_fold_cross_validation(xTrain, yTrain, k, 0.01, 1000)
print(average_accuracy)
print(c)
# graph the cost
plt(c)
k = 10
print(f"----------K-Fold Cross Validation with {k} Folds and eta 0.001----------")
average_accuracy, c = k_fold_cross_validation(xTrain, yTrain, k, 0.001, 1000)
print(average_accuracy)
print(c)
# graph the cost
plt(c)
|
# Import des Moduls "Panda" mit dem Alias "pd"
import pandas as pd
# Erstellen einer Serie
pd.Series([1, 2, 3, 4, 5])
# Erstellen einer Serie mit Index und Namen
pd.Series(
[30, 35, 40], index=["2015 Sales", "2016 Sales", "2017 Sales"], name="Product A"
)
# ### AUFGABE:
# Erstellen Sie eine Serie, die wie folgt aussieht (die linke Spalte ist der Index):
# Punkte Kandidat 1
# 17
# Punkte Kandidat 2
# 19
# Punkte Kandidat 3
# 12
#
pd.Series(
[17, 19, 12],
index=["Punkte Kandidat 1", "Punkte Kandidat 2", "Punkte Kandidat 3"],
name="Klausurleistung",
)
# Erstellen eines Dataframes mit "Integer"-Einträgen
pd.DataFrame({"Yes": [50, 21], "No": [131, 2]})
# Erstellen eines Dataframes mit "String"-Einträgen
pd.DataFrame(
{"Bob": ["I liked it.", "It was awful."], "Sue": ["Pretty good.", "Bland."]}
)
# Erstellen eines Dataframes mit einem Labelindex für die Zeilen
pd.DataFrame(
{"Bob": ["I liked it.", "It was awful."], "Sue": ["Pretty good.", "Bland."]},
index=["Product A", "Product B"],
)
# ### AUFGABE:
# Erstellen Sie ein Dataframe, das wie folgt aussieht:
# Die linke Spalte und die obere Spalte sind jeweils ein Labelindex.
# Die Werte der mittleren Spalte haben das Format "String".
# Die Werte der rechten Spalte haben das Format "Integer".
#
# KanzlerIn
# Dauer in Jahren
# 1949–1963
# Konrad Adenauer
# 14
# 1963–1966
# Ludwig Erhard
# 3
# 1969–1974
# Willy Brandt
# 5
#
# 2005–2021
# Angela Merkel
# 16
#
#
pd.DataFrame(
{
"Kanzlerin": [
"Konrad Adenauer",
"Ludwig Erhard",
"Willy Brandt",
"Angela Merkel",
],
"Dauer in Jahren": ["14", "3", "5", "16"],
},
index=["1949-1963", "1963-1966", "1969-1974", "2005-2021"],
)
# Einlesen einer csv-Datei aus Kaggle
pd.read_csv(
"/kaggle/input/global-commodity-trade-statistics/commodity_trade_statistics_data.csv",
index_col=0,
)
# Einlesen einer csv-Datei aus einer anderen Quelle: Laden Sie die Datei auf Ihre Festplatte. Klicken Sie dann oben links auf "File" und dann auf "Upload Data".
pd.read_csv(
"/kaggle/input/global-commodity-trade-statistics/commodity_trade_statistics_data.csv"
)
# Ausmaß des Datensatz bestimmen (Anzahl Beobachtungen, Anzahl Variablen)
df.shape
# Zeige die obersten 5 Zeilen des Datensatzes
df.head(5)
# ### AUFGABE:
# Erstellen Sie einen Kaggle-Account und schicken Sie mir Ihren Nickname, damit ich meinen Code für Sie freigeben kann.
# Suchen Sie einen CSV-Datensatz zu einem Thema, das Sie interessant finden. Der Datensatz sollte eine mögliche metrische abhängige Variable und eine mögliche unabhängige Variable beinhalten.
# Laden Sie den Datensatz in dieses Python Notebook.
# Führen Sie eine erste sehr kurze Zusammenhangsanalyse mit Hilfe des df.groupby Befehls oder mit Hilfe des df.corr Befehls durch. Schreiben Sie Ihre Interpretation in den Code (als #Kommentar oder als Markdown-Zelle).
# Teilen Sie dieses Python Nootebook mit mir. Klicken Sie dazu oben rechts auf "File" und dann auf "Share".
#
df[["trade_usd", "weight_kg"]].corr()
# Die Korrelationsmatrix df_corr = df[['weight_kg', 'trade_usd']].corr() zeigt eine positive Korrelation von 0,315879 zwischen dem Handelswert ('trade_usd') und dem Gewicht ('weight_kg') an. Dies deutet darauf hin, dass höhere Handelswerte tendenziell mit höheren Gewichten einhergehen. Der Zusammenhang zwischen Handelswert und Gewicht ist jedoch nicht sehr stark, da die Korrelation nicht bei 1 liegt. Wenn der Korrelationskoeffizient nahe bei 0 liegt, gibt es möglicherweise keine signifikante Korrelation zwischen den Variablen.
df.groupby(["country_or_area", "year"])["trade_usd"].sum().reset_index()
|
# # *Tentukan Library yang ditentukan*
# # fungsi library
# - NumPy adalah library Python yang digunakan untuk bekerja dengan array
# - Pandas digunakan untuk menganalisis data
# - Melalui scikit-learn dapat mengimplementasikan berbagai model pembelajaran mesin untuk regresi, klasifikasi, pengelompokan, dan alat statistik untuk menganalisisnya
# - Pickle untuk menyimpan data codingan dalam format 'sav'
import numpy as np
import pandas as pd
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import train_test_split
from sklearn import svm
from sklearn.metrics import accuracy_score
import pickle
# # METODE CRISP-DM
# # '''1.Business Understanding'''
# Tahap ini bertujuan untuk memahami masalah bisnis, yaitu bagaimana meningkatkan akurasi prediksi tentang suatu penyakit yaitu Hepatitis C agar dapat memberikan informasi yang lebih akurat bagi pasien dan membantu Dokter dalam pengambilan keputusan ketika mendiagnosa pasien tertentu
# # '''2. Data Understanding'''
# Konteks
# Kumpulan data berisi nilai laboratorium donor darah dan pasien Hepatitis C dan nilai demografis seperti usia.
# Isi
# Semua atribut kecuali Kategori dan Jenis Kelamin adalah numerik.
# Atribut 1 sampai 4 mengacu pada data pasien:
# 1) X (KTP/No. Pasien)
# 2) Kategori (diagnosis) (nilai: '0=Donor Darah', '0s=Suspek Donor Darah', '1=Hepatitis', '2=Fibrosis', '3=Sirosis')
# 3) Umur (dalam tahun)
# 4) Jenis Kelamin (f,m)
# Atribut 5 sampai 14 mengacu pada data laboratorium:
# 5) ALB (Albumin Blood Test)
# 6) ALP (Alkaline phosphatase)
# 7) ALT (Alanine Transaminase)
# 8) AST (Aspartate Transaminase)
# 9) BIL (Bilirubin)
# 10) CHE (Acetylcholinesterase)
# 11) CHOL (Cholesterol)
# 12) CREA (Creatinine)
# 13) GGT (Gamma-Glutamyl Transferase)
# 14) PROT (Proteins)
# Atribut target untuk klasifikasi adalah Kategori (2)
# # *Load Dataset*
HepatitisC_Dataset = pd.read_csv("HepatitisCdata.csv")
HepatitisC_Dataset.head(10)
# # '''3.Data Preparation'''
# # a. Melakukan pengecekan kembali pada kebenaran data
# Mengubah Nama-nama Kolom Yang di perlukan
HepatitisC_Dataset.columns = [
"Patient_ID",
"Category",
"Age",
"Gender",
"ALB",
"ALP",
"ALT",
"AST",
"BIL",
"CHE",
"CHOL",
"CREA",
"GGT",
"PROT",
]
HepatitisC_Dataset.head(10)
# Menghapus Columns Patient ID
HepatitisC_Dataset = HepatitisC_Dataset.drop("Patient_ID", axis=1)
HepatitisC_Dataset.head(10)
# Mengganti Category ['0=Blood Donor', '0s=suspect Blood Donor'] menjadi '0'
# dan mengganti Category ['1=Hepatitis', '2=Fibrosis', '3=Cirrhosis'] menjadi '1'
HepatitisC_Dataset["Category"] = HepatitisC_Dataset["Category"].replace(
{
"0=Blood Donor": 0,
"0s=suspect Blood Donor": 0,
"1=Hepatitis": 1,
"2=Fibrosis": 1,
"3=Cirrhosis": 1,
}
)
# Mengganti Gender ['m'] menjadi 0
# dan mengganti Gender ['f'] menjadi 1
HepatitisC_Dataset["Gender"] = HepatitisC_Dataset["Gender"].replace({"m": 0, "f": 1})
HepatitisC_Dataset
# # b.Memberlakukan data missing dan data inkonsistensi
# Handling Duplicate Values
# Mencari data yang sama atau duplikat
# Menghapus data yang duplikat jika ditemukan
HepatitisC_Dataset = HepatitisC_Dataset.drop_duplicates()
# Handling Missing Values
# - Mencari terlebih dahulu kolom yang memiliki data null
print(
HepatitisC_Dataset.isnull().sum()
) # Menampilkan jumlah data null pada suatu kolom
# - Mereplace kolom yang memiliki data kosong dengan rata-rata dari kolom tersebut (mean imputation)
HepatitisC_Dataset["ALB"].fillna(
HepatitisC_Dataset["ALB"].mean(), inplace=True
) # Mengisi data yang kosong di kolom tsb dengan rata-rata dari kolom tsb
HepatitisC_Dataset["ALP"].fillna(
HepatitisC_Dataset["ALP"].mean(), inplace=True
) # Mengisi data yang kosong di kolom tsb dengan rata-rata dari kolom tsb
HepatitisC_Dataset["ALT"].fillna(
HepatitisC_Dataset["ALT"].mean(), inplace=True
) # Mengisi data yang kosong di kolom tsb dengan rata-rata dari kolom tsb
HepatitisC_Dataset["CHOL"].fillna(
HepatitisC_Dataset["CHOL"].mean(), inplace=True
) # Mengisi data yang kosong di kolom tsb dengan rata-rata dari kolom tsb
HepatitisC_Dataset["PROT"].fillna(
HepatitisC_Dataset["PROT"].mean(), inplace=True
) # Mengisi data yang kosong di kolom tsb dengan rata-rata dari kolom tsb
print(
HepatitisC_Dataset.isnull().sum()
) # Menampilkan jumlah data null pada suatu kolom
# Jumlah Dataset yang ada
HepatitisC_Dataset.shape
HepatitisC_Dataset["Category"].value_counts()
# # '''4.Modeling'''
# Memisahkan data dan Label
X = HepatitisC_Dataset.drop(columns="Category", axis=1)
Y = HepatitisC_Dataset["Category"]
print(X)
print(Y)
# # *Standarisasi Data*
scaler = StandardScaler()
scaler.fit(X)
standarized_data = scaler.transform(X)
print(standarized_data)
X = standarized_data
Y = HepatitisC_Dataset["Category"]
print(X)
print(Y)
# # *Memisahkan Data Training dan Data Testing*
X_train, X_test, Y_train, Y_test = train_test_split(
X, Y, test_size=0.2, stratify=Y, random_state=2
)
print(X.shape, X_train.shape, X_test.shape)
# # *Membuat Data Latih Menggunakan Algoritma SVM*
classifier = svm.SVC(kernel="linear")
classifier.fit(X_train, Y_train)
# # '''5.Evaluasi'''
# # *Membuat Model Evaluasi Untuk Mengukur Tingkat akurasi*
X_train_prediction = classifier.predict(X_train)
training_data_accuracy = accuracy_score(X_train_prediction, Y_train)
print("Akurasi data training adalah = ", training_data_accuracy)
X_test_prediction = classifier.predict(X_test)
test_data_accuracy = accuracy_score(X_test_prediction, Y_test)
print("Akurasi data testing adalah = ", test_data_accuracy)
# # *Membuat Model Prediksi*
input_data = (32, 0, 38.5, 52.5, 7.7, 22.1, 7.5, 6.93, 3.23, 106, 12.1, 69)
input_data_as_numpy_array = np.array(input_data)
input_data_reshape = input_data_as_numpy_array.reshape(1, -1)
std_data = scaler.transform(input_data_reshape)
print(std_data)
prediction = classifier.predict(std_data)
print(prediction)
if prediction[0] == 0:
print("Pasien tidak terdiagnosa Hepatitis C")
else:
print("Pasien terdiagnosa Hepatitis C")
input_data = (23, 0, 47, 19.1, 38.9, 164.2, 17, 7.09, 3.2, 79.3, 90.4, 70.1)
input_data_as_numpy_array = np.array(input_data)
input_data_reshape = input_data_as_numpy_array.reshape(1, -1)
std_data = scaler.transform(input_data_reshape)
print(std_data)
prediction = classifier.predict(std_data)
print(prediction)
if prediction[0] == 0:
print("Pasien tidak terdiagnosa Hepatitis C")
else:
print("Pasien terdiagnosa Hepatitis C")
# # '''6.Deployment'''
# # *a.Simpan Model*
filename = "hepatitisC_model.sav"
pickle.dump(classifier, open(filename, "wb"))
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
10
10 + 10
10 * 5
10.10
if 10 > 4:
print("On dörtten büyüktür!")
print("Selam Ali")
type(1)
type(10.10)
type("Yusuf Alsanjari")
print("Merhaba,YUSUF!")
print("Manuel Icardi!")
print("Trabzonspor!")
# En Büyük Galatasaray
# Aşkın Olayım
# Söylenmedi Hiç
print("Hello, Trabzon!")
"""
No Pain No Gain
Never Give Up
Dont Forget Yourself
"""
print("Hello, Trabzon!")
x = 50
y = 55
print(x)
print(y)
x + y
z = x + y
z
d = "Yusuf"
d
"STRING İFADE"
x = 10 # integer, int
x = "10" # string, str
print(x)
# bir değişkenin veri tipini belirtmek için
x = str(8) # x will be '8'
y = int(8) # y will be 8
z = float(8) # z will be 8.0
print(x)
print(y)
print(z)
ornek = "Yuusf Trabzona gidiyor" # ornek objesine string ifadeyi atamış olduk
x = 10
print(type(x))
y = "Yusuf"
print(type(y))
x = "Doğukan"
# ikisi de aynı
x = "Ysusf"
x = 10
X = "Ysusf"
myvar = "Ysusf"
my_var = "Ysusf"
_my_var = "Ysusf"
myVar = "Ysusf"
MYVAR = "Ysusf"
myvar2 = "Ysusf"
myvar = "Ysusf"
myvar = "Ysusf"
myvar = "Ysusf"
benimDegiskenAdim = "Yusuf"
benimDegiskenAdim = "Yusuf"
benim_degisken_adim = "Yusuf"
x, y, z = "Inek", "Kedi", "Kus"
print(x)
print(y)
print(z)
x = y = z = "Kulaklık"
print(x)
print(y)
print(z)
meyveler = ["avakado", "mango", "ananas"]
x, y, z = meyveler
print(x)
print(y)
print(z)
x = "Para is Para"
print(x)
x = "Para"
y = "is"
z = "Para"
print(x, y, z)
x = "Para "
y = "is "
z = "Para"
print(x + y + z)
x = 122
y = 185
print(x + y)
x = 5
y = "Yusuf"
print(x, y)
x = "Para"
def myfunc():
print("Para is " + x)
myfunc()
x = " no muscle"
def myfunc():
x = "no gain"
print("No pain " + x)
myfunc()
print("No pain" + x)
x
myfunc()
def myfunc():
global x
x = "No pain"
myfunc()
print("No gain " + x)
|
# !pip install pyarrow
from datasets import list_datasets, load_dataset
from datasets import Features, Sequence, Value, ClassLabel
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
from rich.console import Console
from tqdm.auto import tqdm
cs = Console()
# # 一、 数据加载
file_dict = {
"train": "../input/emotions-dataset-for-nlp/train.txt",
"validation": "../input/emotions-dataset-for-nlp/val.txt",
"test": "../input/emotions-dataset-for-nlp/test.txt",
}
class_names = ["sadness", "joy", "love", "anger", "fear", "surprise"]
str2idx_map = dict(zip(class_names, range(len(class_names))))
# 装换数据结构
for k, f in tqdm(file_dict.items()):
df = pd.read_csv(f, sep=";", header=None)
df.loc[:, 1] = df.loc[:, 1].map(str2idx_map)
out = "../working/" + f.split("/")[-1]
df.to_csv(out, sep=";", header=None, index=False)
all_datasets = list_datasets()
cs.print(f"There are {len(all_datasets)} datasets currently available on the Hub")
# 加载emotion数据
file_dict = {
"train": "../working/train.txt",
"validation": "../working/val.txt",
"test": "../working/test.txt",
}
emotion_features = Features(
{"text": Value("string"), "label": ClassLabel(names=class_names)}
)
emotions = load_dataset(
"csv",
data_files=file_dict,
sep=";",
names=["text", "label"],
features=emotion_features,
)
train_ds = emotions["train"]
cs.print("train_ds[0] => ", train_ds[0])
cs.print("train_ds.features => ", train_ds.features)
cs.print("train_ds[:3] => ", train_ds[:3])
emotions.set_format(type="pandas")
df = emotions["train"][:]
df.head()
def label_int2str(r):
return emotions["train"].features["label"].int2str(r)
df["label_name"] = df["label"].map(label_int2str)
df.head()
# # 一、Looking at the Class Distribution
df["label_name"].value_counts(ascending=True).plot.barh()
plt.title("Frequency of Classes")
plt.show()
df["Words Per Tweet"] = df["text"].str.split().apply(len)
df.boxplot(
"Words Per Tweet", by="label_name", grid=False, showfliers=False, color="black"
)
plt.xlabel("")
plt.show()
# # 二、 From Text to Tokens
from transformers import AutoTokenizer
tokenizer = AutoTokenizer.from_pretrained("distilbert-base-uncased")
text = "Tokenizing text is a core task of NLP"
encoded_text = tokenizer(text)
# ['[CLS]', 'token', '##izing', 'text', 'is', 'a', 'core', 'task', 'of', 'nl', '##p', '[SEP]']
# 1- [CLS] -> 开始; [SEP] -> 结束
# 2- 都变小写了
# 3- tokenizing -> 'token', '##izing'; nlp -> 'nl', '##p' ##指明前面不是空白的
tokens = tokenizer.convert_ids_to_tokens(encoded_text.input_ids)
# [CLS] tokenizing text is a core task of nlp [SEP]
print(tokenizer.convert_tokens_to_string(tokens))
# (30522, 512)
tokenizer.vocab_size, tokenizer.model_max_length
# ## 2- Tokenizing the Whole DataSet
# [PAD]: 0, [UNK]: 100, [CLS]: 101, [SEP]: 102, [MASK]: 103
# --------------------------------
def tokenize_batch(batch):
return tokenizer(batch["text"], padding=True, truncation=True)
emotions.set_format()
cs.print(tokenize_batch(emotions["train"][:2]))
emotions_encoded = emotions.map(tokenize_batch, batched=True, batch_size=None)
# ['text', 'label', 'input_ids', 'attention_mask']
emotions_encoded["train"].column_names
# # 三、 Training a Text Classifier
# ## 1- **Feature extraction**
# -----------------------------------
from transformers import AutoModel
import torch
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
model = AutoModel.from_pretrained("distilbert-base-uncased").to(device)
# sample test
text = "This is a test"
ipt = tokenizer(text, return_tensors="pt")
ipts = {k: v.to(device) for k, v in ipt.items()}
with torch.no_grad():
out = model(**ipts)
# tokenizer.convert_ids_to_tokens(tokenizer(text).input_ids)
# 返回 [batch_size, n_tokens, hidden_dim] = (1, 6, 768)
# ['[CLS]', 'this', 'is', 'a', 'test', '[SEP]'] 对于分类仅仅需要拿取第一个就行
print(out.last_hidden_state[:, 0].shape)
def extract_hidden_states(batch):
ipts = {
k: v.to(device) for k, v in batch.items() if k in tokenizer.model_input_names
}
with torch.no_grad():
out = model(**ipts).last_hidden_state
return {"hidden_state": out[:, 0].cpu().numpy()}
emotions_encoded = emotions.map(tokenize_batch, batched=True, batch_size=1000)
emotions_encoded.set_format("torch", columns=["input_ids", "attention_mask", "label"])
emotions_hidden = emotions_encoded.map(extract_hidden_states, batched=True)
emotions_hidden["train"].column_names
# 1-1 Creating a feature matrix
# --------------------------
x_tr = np.array(emotions_hidden["train"]["hidden_state"])
x_val = np.array(emotions_hidden["validation"]["hidden_state"])
y_tr = np.array(emotions_hidden["train"]["label"])
y_val = np.array(emotions_hidden["validation"]["label"])
# 1-2 Visualizing the training set
# --------------------------
from umap import UMAP
from sklearn.preprocessing import MinMaxScaler
x_sc = MinMaxScaler().fit_transform(x_tr)
mapper = UMAP(n_components=2, metric="cosine").fit(x_sc)
df_emb = pd.DataFrame(mapper.embedding_, columns=["X", "Y"])
df_emb["label"] = y_tr
# plot
fig, axes = plt.subplots(2, 3, figsize=(7, 5))
axes = axes.flatten()
cmaps = ["Greys", "Blues", "Oranges", "Reds", "Purples", "Greens"]
labels = emotions["train"].features["label"].names
for idx, (label, cmap) in enumerate(zip(labels, cmaps)):
df_emb_sub = df_emb.query(f"label == {idx}")
axes[idx].hexbin(
df_emb_sub["X"], df_emb_sub["Y"], cmap=cmap, gridsize=20, linewidths=(0,)
)
axes[idx].set_title(label)
axes[idx].set_xticks([]), axes[idx].set_yticks([])
plt.tight_layout()
plt.show()
# 1-3 Training a simple classifier
# --------------------------
from sklearn.linear_model import LogisticRegression
from sklearn.dummy import DummyClassifier
from sklearn.metrics import ConfusionMatrixDisplay, confusion_matrix
lr_clf = LogisticRegression(max_iter=3000)
cs.print(lr_clf.fit(x_tr, y_tr))
cs.print(lr_clf.score(x_val, y_val))
dummy_clf = DummyClassifier(strategy="most_frequent")
dummy_clf.fit(x_tr, y_tr)
dummy_clf.score(x_val, y_val)
def plot_confusion_matrix(y_pred, y_true, labels):
cm = confusion_matrix(y_true, y_pred, normalize="true")
fig, ax = plt.subplots(figsize=(6, 6))
disp = ConfusionMatrixDisplay(cm, display_labels=labels)
disp.plot(cmap="Blues", values_format=".2f", ax=ax, colorbar=False)
plt.title("Normalized confusion matrix")
plt.show()
y_pred = lr_clf.predict(x_val)
plot_confusion_matrix(y_pred, y_val, labels)
# ## 2- **Fine-Tuning Transformers**
# -----------------------------------
# from huggingface_hub import notebook_login
# notebook_login()
from transformers import AutoModelForSequenceClassification
from sklearn.metrics import accuracy_score, f1_score
import torch
from huggingface_hub import login
from transformers import Trainer, TrainingArguments
from datasets import list_datasets, load_dataset
from transformers import AutoTokenizer
from torch.nn.functional import cross_entropy
model_ckpt = "distilbert-base-uncased"
tokenizer = AutoTokenizer.from_pretrained(model_ckpt)
def tokenize_batch(batch):
return tokenizer(batch["text"], padding=True, truncation=True)
emotions.set_format()
emotions_encoded = emotions.map(tokenize_batch, batched=True, batch_size=None)
# 2-1 加载模型
# AutoModelForSequenceClassification has a classification head on top of the pretrained model outputs
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
model = AutoModelForSequenceClassification.from_pretrained(model_ckpt, num_labels=6).to(
device
)
# 2-2 Defining the performance metrics
def compute_metrics(pred):
labels = pred.label_ids
preds = pred.predictions.argmax(-1)
f1 = f1_score(labels, preds, average="weighted")
acc = accuracy_score(labels, preds)
return {"accuracy": acc, "f1": f1}
# ### 2-3 train & metrics
# help(TrainingArguments)
batch_size = 64
logging_step = len(emotions_encoded["train"]) // batch_size
model_name = f"{model_ckpt}-finetuned-emotion"
training_args = TrainingArguments(
output_dir=model_name,
num_train_epochs=2,
learning_rate=2e-5,
per_device_train_batch_size=batch_size,
per_device_eval_batch_size=batch_size,
weight_decay=0.01, # Adam
evaluation_strategy="epoch",
disable_tqdm=False,
logging_steps=logging_step,
push_to_hub=False,
log_level="error",
)
trainer = Trainer(
model=model,
args=training_args,
compute_metrics=compute_metrics,
train_dataset=emotions_encoded["train"],
eval_dataset=emotions_encoded["validation"],
)
trainer.train()
from sklearn.metrics import ConfusionMatrixDisplay, confusion_matrix
def plot_confusion_matrix(y_pred, y_true, labels):
cm = confusion_matrix(y_true, y_pred, normalize="true")
fig, ax = plt.subplots(figsize=(6, 6))
disp = ConfusionMatrixDisplay(cm, display_labels=labels)
disp.plot(cmap="Blues", values_format=".2f", ax=ax, colorbar=False)
plt.title("Normalized confusion matrix")
plt.show()
preds_output = trainer.predict(emotions_encoded["validation"])
cs.print(preds_output.metrics)
y_pred = np.argmax(preds_output.predictions, axis=1)
y_val = np.array(emotions["validation"]["label"])
labels = emotions["train"].features["label"].names
plot_confusion_matrix(y_pred, y_val, labels)
# ### 2-4 error analysis
def forward_pass_with_label(batch):
ipts = {
k: v.to(device) for k, v in batch.items() if k in tokenizer.model_input_names
}
with torch.no_grad():
output = model(**ipts)
pred_label = torch.argmax(output.logits, axis=-1)
loss = cross_entropy(output.logits, batch["label"].to(device), reduction="none")
return {"loss": loss.cpu().numpy(), "predicted_label": pred_label.cpu().numpy()}
emotions_encoded.set_format("torch", columns=["input_ids", "attention_mask", "label"])
# 计算损失
emotions_encoded["validation"] = emotions_encoded["validation"].map(
forward_pass_with_label, batched=True, batch_size=16
)
# DataFrame -> texts, losses, predicted/true labels
emotions_encoded.set_format("pandas")
cols = ["text", "label", "predicted_label", "loss"]
df_test = emotions_encoded["validation"][:][cols]
df_test["label"] = df_test["label"].map(label_int2str)
df_test["predicted_label"] = df_test["predicted_label"].map(label_int2str)
df_test.sort_values("loss", ascending=False).head(10)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.preprocessing import MinMaxScaler, StandardScaler
from sklearn.model_selection import train_test_split
from sklearn.cluster import KMeans
from sklearn.metrics import silhouette_score
from kneed import KneeLocator
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Workflow
# * 1- Data Importing
# * 2- Exploratory Data Analysis
# * 3- Elbow Method
# * 4- Modelling KMeans
# * 5- Conslusion
df = pd.read_csv(
"/kaggle/input/student-behaviour-dataset/StudentBehaviouDatasetForAnalysis.csv"
)
df.head(10)
df.tail(10)
df.sample(10)
df.shape
columns = df.columns.values
columns
df.info()
# * There is no....
p = df.gender.value_counts(normalize=True)
ax = sns.countplot(x="gender", data=df)
patches = ax.patches
percentage = [p[0] * 100, p[1] * 100]
for i in range(len(patches)):
x = patches[i].get_x() + patches[i].get_width() / 2
y = patches[i].get_height() + 0.05
ax.annotate("{:.1f}%".format(percentage[i]), (x, y), ha="center")
plt.ylabel("Numbers")
plt.xlabel("Gender")
plt.grid(alpha=0.7)
plt.show()
# * There are more about 63 % of male and 36 % of females.
#
df.NationalITy.value_counts(normalize=True)
for column in columns:
print("************ " + column + ("************"))
print(df[column].unique())
print("\n\n")
# ### Duplicated Values
df.duplicated().value_counts()
# * There are 2 duplicated values
df.duplicated
df.drop_duplicates(inplace=True, keep="first")
df.shape
# * Two duplicated observations were removed and kept the first one.
# ### Check for nan values
df.isna().sum()
# * There are no null values
df.duplicated().value_counts()
# * There are no duplicated rows
fig, ax = plt.subplots(1, 1, figsize=(10, 5))
df.NationalITy.value_counts().plot(kind="bar")
plt.grid(alpha=0.5)
plt.show()
# * KW and Jordan is the top ones
fig, ax = plt.subplots(1, 1, figsize=(10, 5))
df.PlaceofBirth.value_counts().plot(kind="bar")
plt.grid(alpha=0.5)
plt.show()
stg = df.StageID.value_counts()
stg
import math
p = [
df.StageID.value_counts()[0] * 100 / sum(df.StageID.value_counts()),
df.StageID.value_counts()[1] * 100 / sum(df.StageID.value_counts()),
df.StageID.value_counts()[2] * 100 / sum(df.StageID.value_counts()),
]
ax = plt.subplot(projection="polar")
patches = ax.patches
ax.barh("Middle", math.radians(360 * 2 / stg[0]))
ax.barh("Lower", math.radians(360 * 2 / stg[1]))
ax.barh("High", math.radians(360 * 2 / stg[2]))
ax.set_theta_zero_location("N") # top
ax.set_theta_direction(1) # sets the direction of the rotation
ax.set_rlabel_position(0)
ax.set_thetagrids([0, 96, 192, 288], labels=[0, 20, 40, 60])
p = df.gender.value_counts(normalize=True)
ax = sns.countplot(x="StageID", data=df)
plt.ylabel("Numbers")
plt.xlabel("School Level")
plt.grid(alpha=0.7)
plt.show()
sns.countplot(x=df.GradeID)
plt.xlabel("Grade Level")
plt.ylabel("Number")
plt.show()
sns.countplot(data=df, x="SectionID")
fig = plt.figure(figsize=(10, 6))
sns.countplot(data=df, x="Topic")
num_f = [c for c in columns[9:13]]
fig, ax = plt.subplots(4, 1, figsize=(5, 20))
for i, c in enumerate(num_f):
sns.histplot(data=df, x=c, ax=ax[i], hue="gender", kde=True)
num_f = [c for c in columns[9:13]]
fig, ax = plt.subplots(4, 1, figsize=(5, 20))
for i, c in enumerate(num_f):
sns.histplot(
data=df, x=c, ax=ax[i], hue="gender", kde=True, cumulative=True, cbar=True
)
n_df = df[num_f]
n_df.describe().T
num_f = [c for c in columns[9:13]]
fig, ax = plt.subplots(4, 1, figsize=(5, 20))
for i, c in enumerate(num_f):
sns.boxplot(data=df, x=c, ax=ax[i], hue="gender")
fig = plt.figure(figsize=(10, 6))
sns.pairplot(data=df, kind="kde")
sns.scatterplot(data=df, x=num_f[1], y=num_f[2], hue="Class")
plt.show()
n_df
sns.scatterplot(data=df, x=num_f[1], y=num_f[2], hue="Semester")
plt.show()
sns.scatterplot(data=df, x=num_f[1], y=num_f[2], hue="GradeID")
plt.show()
sns.scatterplot(data=df, x=num_f[1], y=num_f[2], hue="ParentschoolSatisfaction")
plt.show()
# * Low Level Students's parents are not satisfied with the school while High Level Students's parents are satisfied.
sns.catplot(data=df, x="raisedhands", y="Class", kind="violin", inner=None)
sns.catplot(data=df, x="raisedhands", y="Class", kind="point")
plt.grid(alpha=0.6)
plt.show()
sns.catplot(
data=df, x="raisedhands", y="ParentschoolSatisfaction", kind="violin", inner=None
)
sns.catplot(data=df, x="raisedhands", y="ParentschoolSatisfaction", kind="point")
plt.grid(alpha=0.6)
plt.show()
sns.catplot(
data=df,
x="AnnouncementsView",
y="ParentschoolSatisfaction",
kind="violin",
inner=None,
)
sns.catplot(data=df, x="AnnouncementsView", y="ParentschoolSatisfaction", kind="point")
plt.grid(alpha=0.6)
plt.show()
sns.catplot(data=df, x="AnnouncementsView", y="Class", kind="violin", inner=None)
sns.catplot(data=df, x="AnnouncementsView", y="Class", kind="point")
plt.grid(alpha=0.6)
plt.show()
sns.catplot(data=df, x="Discussion", y="Class", kind="violin", inner=None)
sns.catplot(data=df, x="Discussion", y="Class", kind="point")
plt.grid(alpha=0.6)
plt.show()
#
# High Level Students:
# 1 Higher Level of raised hands.
# 2 More Announcement Views.
# 3 More Discussin Involvement.
# 4 Parents are satisfied.
# 5 Most resources visits.
#
#
# Low Level Students:
# 1 Lowest Number of raised hands.
# 2 Lowest Announcement Views.
# 3 Lowest Discussin Involvement.
# 4 Usatisfied Parents
# 5 Least resources visits.
#
sns.catplot(data=df, x="AnnouncementsView", y="gender", kind="violin", inner=None)
sns.catplot(data=df, x="AnnouncementsView", y="gender", kind="point")
plt.grid(alpha=0.6)
plt.show()
sns.catplot(data=df, x="Discussion", y="gender", kind="violin", inner=None)
sns.catplot(data=df, x="Discussion", y="gender", kind="point")
plt.grid(alpha=0.6)
plt.show()
sns.catplot(data=df, x="raisedhands", y="gender", kind="violin", inner=None, color=".9")
sns.swarmplot(data=df, x="raisedhands", y="gender")
sns.catplot(data=df, x="raisedhands", y="gender", kind="point")
plt.grid(alpha=0.6)
plt.show()
g = sns.PairGrid(data=df, hue="Class")
g.map_diag(sns.kdeplot)
g.map_upper(sns.scatterplot)
g.map_lower(sns.kdeplot)
g.add_legend()
sns.catplot(
data=df, x="VisITedResources", y="gender", kind="violin", inner=None, color="0.9"
)
sns.swarmplot(data=df, x="VisITedResources", y="gender")
sns.catplot(data=df, x="VisITedResources", y="gender", kind="point")
plt.grid(alpha=0.6)
plt.show()
# * From the above graphs , it is clear that
# Females:
# have more views of announcements,have most visits of resources,most invlovement in discussion and most hands are raised.
#
# * Conclusion:
# It is concluded that most of the High level students are females.
# # Modelling
# ## KMeans Clustering
# * KMeans is suitable for smaller datasets.
# * It is centroid based unsupervised machine learning algorithm.
# * It tries to minimize either the intracluster or intercluster distance (silhouette score).
# * It requires feature scaling/Standardization/Normalization
# # Feature Scaling
# ## Standardization
df_standardized = StandardScaler().fit_transform(n_df.iloc[:, :2])
df_standardized_frame = pd.DataFrame(
df_standardized, columns=["raisedhands", "visitedresources"]
)
df_standardized_frame
# ### Determining The best number of clusters using Elbow Method
def elbow_method(data, start=1, end=10):
score = []
for i in range(start, end):
classifier = KMeans(n_clusters=i, init="k-means++")
classifier.fit(data)
score.append(classifier.inertia_)
return score
# #### Preparing the data for modelling
# First we would find clusters based on **VisITedResources** and **raisedhands**
# score = elbow_method(n_df[['VisITedResources','raisedhands']])
score = elbow_method(df_standardized_frame)
sns.lineplot(x=np.arange(1, 10), y=score)
plt.style.use("ggplot")
plt.xlabel("Number of Clusters")
plt.ylabel("Distortion")
plt.grid(alpha=0.6)
plt.show()
# ### Locate the Knee
kneedl = KneeLocator(
x=np.arange(1, 10), y=score, curve="convex", direction="decreasing", S=1
)
plt.style.use("ggplot")
kneedl.plot_knee()
# * The Knee Point is 3...
# * So we need to make 3 clusters
clustering = KMeans(n_clusters=3)
clustering.fit(df_standardized_frame)
centroids = clustering.cluster_centers_
labels = clustering.labels_
sns.scatterplot(
x="raisedhands",
y="visitedresources",
data=df_standardized_frame,
c=labels,
)
plt.scatter(x=centroids[:, 0], y=centroids[:, 1], s=100)
plt.show()
# ## Score
silhouette = silhouette_score(df_standardized_frame, labels, metric="euclidean")
print("Silhouette Score is:\t" + str(round(silhouette, 2)) + " %")
scores = {}
for i in range(2, 10):
kmeans = KMeans(n_clusters=i)
kmeans.fit(df_standardized_frame)
scores[i] = silhouette_score(
df_standardized_frame, kmeans.labels_, metric="euclidean"
)
scores
number_clusters = list(scores.keys())
scores_clusters = scores.values()
sns.lineplot(x=number_clusters, y=scores_clusters)
plt.scatter(x=kneedl.knee, y=scores[kneedl.knee], s=100, c="r")
# * So the best number of Clusters is 3
# # Conclusion
# * There are three clusters:
# * 1- Smallest number of both raisedhands and visitedresources.
# * 2- Largest number of both raisedhands and visitedresources.
# * 3- Moderate Number of both raisedhands and visitedresources.
# # Clustering Based on Announcement Views and Discussion
n_df
sns.catplot(data=df, x="AnnouncementsView", y="gender")
sns.catplot(data=df, x="Discussion", y="gender", kind="violin", inner=None)
sns.pointplot(data=df, x="Discussion", y="gender", color="yellow")
# * The level of females in discussion in somehow higher than that of the males.
# * This hypothesis is clear from the above violin plot and can be further described below:
df.groupby("gender")[
["AnnouncementsView", "Discussion"]
].describe().T.style.background_gradient(cmap="Reds_r")
# * From the above analysis it is clear that females have more contribution than the males.
# * They have the higher values than males in both the announcementsview and discussion
sns.catplot(data=df, x="gender", kind="count", col="Class", col_wrap=3)
plt.show()
# * The above hypothesis has been proved ie there are more females belonging to the High Class than males. Therefore , we can easily say that females contributed more than males in announcementsview , discussion, raisedhands, and visitedresources.
# * Therefore, It implies that females are more talented than males but not necessay at all.
# * It also implies that the number of females belonging to rich families is higher than that of the males.
# # KMeans Clustering
# ### Standardization
second_data = n_df.iloc[:, 2:4]
second_data_scaled = StandardScaler().fit_transform(second_data)
second_data_scaled_frame = pd.DataFrame(
data=second_data_scaled, columns=["AnnouncementsView", "Discussion"]
)
second_data_scaled_frame
# ### Elbow Method
score = elbow_method(second_data_scaled_frame, 1, 10)
sns.lineplot(x=np.arange(1, 10), y=score)
plt.style.use("ggplot")
plt.xlabel("Number of Clusters")
plt.ylabel("Distortion")
plt.grid(alpha=0.6)
plt.show()
# ### Locate the Knee
kneedl = KneeLocator(
x=np.arange(1, 10), y=score, curve="convex", direction="decreasing", S=1
)
plt.style.use("ggplot")
kneedl.plot_knee()
kneedl.knee
# * The Knee Point is 4...
# * So we need to make 4 clusters
classifier = KMeans(n_clusters=kneedl.knee, init="k-means++")
classifier.fit(second_data_scaled_frame)
centroids = classifier.cluster_centers_
labels = classifier.labels_
sns.scatterplot(
data=second_data_scaled_frame,
x="AnnouncementsView",
y="Discussion",
c=labels,
hue=labels,
)
plt.scatter(x=centroids[:, 0], y=centroids[:, 1], c="yellow", s=100, edgecolors="red")
# ## Score
silhouette = silhouette_score(second_data_scaled_frame, labels, metric="euclidean")
print("Silhouette Score is:\t" + str(round(silhouette, 2)) + " %")
scores = {}
for i in range(2, 10):
kmeans = KMeans(n_clusters=i)
kmeans.fit(second_data_scaled_frame)
scores[i] = silhouette_score(
second_data_scaled_frame, kmeans.labels_, metric="euclidean"
)
number_clusters = list(scores.keys())
scores_clusters = scores.values()
sns.lineplot(x=number_clusters, y=scores_clusters)
plt.scatter(x=kneedl.knee, y=scores[kneedl.knee], s=100, c="r")
|
import os
import cv2
import timm
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.model_selection import train_test_split
from sklearn.metrics import roc_auc_score
import torch
from torch.utils.data import TensorDataset, DataLoader, Dataset, random_split
import torch.nn as nn
import torch.nn.functional as F
import torchvision
import torchvision.transforms as transforms
import torch.optim as optim
from torch.optim import lr_scheduler
import time
from PIL import Image
train_on_gpu = True
from torch.utils.data.sampler import SubsetRandomSampler
from torch.optim.lr_scheduler import StepLR, ReduceLROnPlateau, CosineAnnealingLR
from torchsummary import summary
import copy
torch.manual_seed(0)
# # Deep Learning Classification Flow
# 1. Dataset, DataLoader 정의
# - Data augmentation 정의
# - 데이터 셋의 특성 파악을 위한 visual inspection
# 2. Model (Encoder) 정의
# - 본인이 직접 디자인
# - 유명한 모델의 구조를 이용하여 scratch부터 학습
# - ImageNet 등의 대규모 이미지 데이터 셋으로 학습된 weight 가져와서 fine-tuning
# 3. Training & validation function
# - Optimizer, loss function, learning rate, learning rate scheduler 등등
# 4. Testing & performance evaluation
# - Generalization performance 측정, 어떤 evaluation metrics 를 사용할 것인지?
# - AUROC, AUPRC, F1-score, sensitivity, specificity, PPV, NPV, etc.
# 5. XAI
# - Explainable AI
# - 모델의 학습 과정이 black box라 불릴 정도로 내부는 복잡하고 알 수 없음
# - 결과를 통해 간접적으로 모델을 해석하려는 시도
# - 대표적으로 LIME, SHAP, Grad-CAM 등이 있음
# - 여기에서는 Grad-CAM 실습
path2labels = "/kaggle/input/histopathologic-cancer-detection/train_labels.csv"
labels_df = pd.read_csv(path2labels)
labels_df.head()
sns.set_theme()
sns.countplot(data=labels_df, x="label")
plt.xticks(range(2), ["Normal", "Tumor"], fontsize=10)
plt.xlabel("Labels", fontsize=12)
plt.ylabel("Count", fontsize=12)
plt.show()
fig = plt.figure(figsize=(10, 10))
path2data = "/kaggle/input/histopathologic-cancer-detection/train"
train_imgs = os.listdir(path2data)
for idx, img in enumerate(np.random.choice(train_imgs, 20)):
ax = fig.add_subplot(4, 20 // 4, idx + 1)
im = Image.open(path2data + "/" + img)
plt.imshow(im)
plt.xticks([])
plt.yticks([])
lab = labels_df.loc[labels_df["id"] == img.split(".")[0], "label"].values[0]
ax.set_title(f'Label: {["Tumor" if lab == 1 else "Normal"][0]}')
# # Data Preparation
# - Dataset, Dataloader 정의
# - Data augmentation 정의
class cancer_dataset(Dataset):
def __init__(self, data_dir, transform=None, data_type="train"):
# path to images
path2data = os.path.join(data_dir, data_type)
# list of images in directory
filenames = os.listdir(path2data)
# get full path to images
self.full_filenames = [os.path.join(path2data, f) for f in filenames]
# get labels
path2labels = os.path.join(data_dir, "train_labels.csv")
labels_df = pd.read_csv(path2labels)
# seg dataframe index to id
labels_df.set_index("id", inplace=True)
# obtain labels from df
self.labels = [labels_df.loc[filename[:-4]].values[0] for filename in filenames]
# For image data augmentions
if transform is not None:
self.transform = transform
else:
self.transform = None
def __len__(self):
# return size of dataset
return len(self.full_filenames)
def __getitem__(self, idx):
# open image, apply transforms and return with label
img = Image.open(self.full_filenames[idx]) # PIL image
if self.transform is not None:
img = self.transform(img)
return img, self.labels[idx].squeeze(-1)
data_transformer = transforms.Compose(
[transforms.ToTensor(), transforms.Resize((224, 224))]
)
data_dir = "/kaggle/input/histopathologic-cancer-detection"
img_dataset = cancer_dataset(data_dir, transform=data_transformer, data_type="train")
# # Splitting the Dataset
# - Train - Validation - Test 스플릿
# - Train - Test 가 주어졌으므로 우리는 Train을 valid만 스플릿
# - 8:2 비율로 진행 / 일반적으로 6:2:2
len_dataset = len(img_dataset)
len_train = int(0.8 * len_dataset)
len_val = len_dataset - len_train
train_ds, val_ds = random_split(img_dataset, [len_train, len_val])
print(f"train dataset length: {len(train_ds)}")
print(f"validation dataset length: {len(val_ds)}")
# # Image Transformation
# We will define some transformers for image augmentation
# transformer for trainging dataset
train_transf = transforms.Compose(
[
transforms.ToTensor(),
transforms.Resize((224, 224)),
transforms.RandomHorizontalFlip(p=0.5),
transforms.RandomVerticalFlip(p=0.5),
transforms.RandomRotation(45),
# transforms.RandomResizedCrop(96, scale=(0.8, 1.0), ratio=(1.0, 1.0)),
# transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5)),
]
)
# No augmentation for validation dataset
val_transf = transforms.Compose(
[
transforms.ToTensor(),
transforms.Resize((224, 224)),
# transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5)),
]
)
# Overwrite the transforms functions
train_ds.transform = train_transf
val_ds.transform = val_transf
# Dataloaders
train_dl = DataLoader(
train_ds, batch_size=256, pin_memory=True, num_workers=2, shuffle=True
)
val_dl = DataLoader(
val_ds, batch_size=256, pin_memory=True, num_workers=2, shuffle=False
)
# # Define Classifier
# ## Simple CNN model
# This model has:
# - Four CNN layers with poolin layer for each layer.
# - Two fully connected layers, with a dropout layer
# - activation function is log_softmax
timm.list_models()
model = timm.create_model("resnet18", pretrained=True, num_classes=1)
# # Loss function
# We will use **NLLLoss**
loss_func = nn.BCEWithLogitsLoss()
# # Optimiser
# We will use **Adam**
opt = optim.Adam(model.parameters(), lr=3e-4)
lr_scheduler = ReduceLROnPlateau(opt, mode="min", factor=0.5, patience=20, verbose=0)
# # Training Model
# ## Helper functions
# Function to get the learning rate
def get_lr(opt):
for param_group in opt.param_groups:
return param_group["lr"]
# Function to compute the loss value per batch of data
def loss_batch(loss_func, output, target, opt=None):
loss = loss_func(output, target) # get loss
pred = output.argmax(dim=1, keepdim=True) # Get Output Class
metric_b = pred.eq(target.view_as(pred)).sum().item() # get performance metric
if opt is not None:
opt.zero_grad()
loss.backward()
opt.step()
return loss.item(), metric_b
# Compute the loss value & performance metric for the entire dataset (epoch)
def loss_epoch(model, loss_func, dataset_dl, opt=None, device="cpu"):
run_loss = 0.0
t_metric = 0.0
len_data = len(dataset_dl.dataset)
# internal loop over dataset
for xb, yb in tqdm(dataset_dl, leave=False):
# move batch to device
xb = xb.to(device)
yb = yb.to(device).unsqueeze(-1).float()
output = model(xb) # get model output
loss_b, metric_b = loss_batch(loss_func, output, yb, opt) # get loss per batch
run_loss += loss_b # update running loss
if metric_b is not None: # update running metric
t_metric += metric_b
loss = run_loss / float(len_data) # average loss value
metric = t_metric / float(len_data) # average metric value
return loss, metric
# ## Training function
from tqdm.notebook import trange, tqdm
def train_val(model, params, verbose=False):
# Get parameters
epochs = params["epochs"]
opt = params["optimiser"]
loss_func = params["f_loss"]
train_dl = params["train"]
val_dl = params["val"]
lr_scheduler = params["lr_change"]
weight_path = params["weight_path"]
# history of loss and metric values in each epoch
loss_history = {"train": [], "val": []}
metric_history = {"train": [], "val": []}
# a deep copy of weights for the best model
best_model_wts = copy.deepcopy(model.state_dict())
best_loss = float("inf") # init loss
# Train loop
for epoch in tqdm(range(epochs), leave=False):
# get lr
current_lr = get_lr(opt)
if verbose:
print(f"Epoch {epoch +1}/{epochs}, current lr={current_lr}")
# train model
model.train()
train_loss, train_metric = loss_epoch(
model, loss_func, train_dl, opt, device=params["device"]
)
loss_history["train"].append(train_loss)
metric_history["train"].append(train_metric)
# evaluate model
model.eval()
with torch.no_grad():
val_loss, val_metric = loss_epoch(
model, loss_func, val_dl, opt=None, device=params["device"]
)
# store best model
if val_loss < best_loss:
best_loss = val_loss
best_model_wts = copy.deepcopy(model.state_dict())
# save weights in a local file
torch.save(model.state_dict(), weight_path)
if verbose:
print("Saved best model weights")
loss_history["val"].append(val_loss)
metric_history["val"].append(val_metric)
# lr schedule
lr_scheduler.step(val_loss)
if current_lr != get_lr(opt):
if verbose:
print("Loading best model weights")
model.load_state_dict(best_model_wts)
if verbose:
print(
f"train loss: {train_loss:.6f}, dev loss: {val_loss:.6f}, accuracy: {100*val_metric:.2f}"
)
print("-" * 20)
model.load_state_dict(best_model_wts)
return model, loss_history, metric_history
# ## Train
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
model.to(device)
params_train = {
"train": train_dl,
"val": val_dl,
"epochs": 1,
"optimiser": opt,
"lr_change": lr_scheduler,
"f_loss": loss_func,
"device": device,
"weight_path": "weights.pt",
}
# Train model
model, loss_hist, metric_hist = train_val(model, params_train, verbose=True)
# ## Loss & Metrics visual
#
import seaborn as sns
sns.set(style="whitegrid")
epochs = params_train["epochs"]
fig, ax = plt.subplots(1, 2, figsize=(12, 5))
sns.lineplot(
x=[*range(1, epochs + 1)],
y=loss_hist["train"],
ax=ax[0],
label='loss_hist["train"]',
)
sns.lineplot(
x=[*range(1, epochs + 1)], y=loss_hist["val"], ax=ax[0], label='loss_hist["val"]'
)
sns.lineplot(
x=[*range(1, epochs + 1)],
y=metric_hist["train"],
ax=ax[1],
label='metric_hist["train"]',
)
sns.lineplot(
x=[*range(1, epochs + 1)],
y=metric_hist["val"],
ax=ax[1],
label='metric_hist["val"]',
)
plt.title("Convergence History")
# # Inference
# We will use our trained model to classify the test dataset
class cancerdata_test(Dataset):
def __init__(self, data_dir, transform, data_type="train"):
path2data = os.path.join(data_dir, data_type)
filenames = os.listdir(path2data)
self.full_filenames = [os.path.join(path2data, f) for f in filenames]
# labels are in a csv file named train_labels.csv
csv_filename = "sample_submission.csv"
path2csvLabels = os.path.join(data_dir, csv_filename)
labels_df = pd.read_csv(path2csvLabels)
# set data frame index to id
labels_df.set_index("id", inplace=True)
# obtain labels from data frame
self.labels = [labels_df.loc[filename[:-4]].values[0] for filename in filenames]
self.transform = transform
def __len__(self):
# return size of dataset
return len(self.full_filenames)
def __getitem__(self, idx):
# open image, apply transforms and return with label
image = Image.open(self.full_filenames[idx]) # PIL image
image = self.transform(image)
return image, self.labels[idx]
# load any model weights for the model
model.load_state_dict(torch.load("weights.pt"))
path2sub = "/kaggle/input/histopathologic-cancer-detection/sample_submission.csv"
labels_df = pd.read_csv(path2sub)
data_dir = "/kaggle/input/histopathologic-cancer-detection/"
data_transformer = transforms.Compose(
[transforms.ToTensor(), transforms.Resize((46, 46))]
)
img_dataset_test = cancerdata_test(data_dir, data_transformer, data_type="test")
print(len(img_dataset_test), "samples found")
def inference(model, dataset, device, num_classes=1):
len_data = len(dataset)
y_out = torch.zeros(len_data, num_classes) # initialize output tensor on CPU
y_gt = np.zeros((len_data), dtype="uint8") # initialize ground truth on CPU
model = model.to(device) # move model to device
model.eval()
with torch.no_grad():
for i in tqdm(range(len_data)):
x, y = dataset[i]
y_gt[i] = y
y_out[i] = model(x.unsqueeze(0).to(device))
return y_out.numpy(), y_gt
y_test_out, _ = inference(model, img_dataset_test, device)
y_test_pred = np.argmax(y_test_out, axis=1)
test_ids = [
name.split("/")[-1].split(".")[0] for name in img_dataset_test.full_filenames
]
test_preds = pd.DataFrame({"img": test_ids, "preds": y_test_pred})
submission = pd.merge(labels_df, test_preds, left_on="id", right_on="img")
submission = submission[["id", "preds"]]
submission.columns = ["id", "label"]
submission.head()
submission.to_csv("submission.csv", index=False)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import matplotlib.pyplot as plt
# DATASET_FILEPATH = '/kaggle/input/la-liga-player-status-season-2022-2023/lalaiga 2 all player final - lalaiga 2 all player.csv'
df = pd.read_csv(
"/kaggle/input/la-liga-player-status-season-2022-2023/lalaiga 2 all player final - lalaiga 2 all player.csv"
)
df
df_h = df.head(10)
df_h
# # make figure for plot
fig, ax = plt.subplots(figsize=(10, 5))
# axes
ax = fig.add_axes([0, 0, 1, 1])
ax.set_title("Palyer vs Goal chart")
ax.set_xlabel("Player")
ax.set_ylabel("Goals")
df_h
# # Plot
fig, ax = plt.subplots(figsize=(15, 5))
# axes
ax = fig.add_axes([0, 0, 1, 1])
ax.set_title("Palyer vs Goal chart")
ax.set_xlabel("Player")
ax.set_ylabel("Goals")
bars = ax.bar(df_h["Player Name"], df_h["Goals"]) # creating PLot
plt.show()
# # Now as A ascending Order
df_h = df_h.sort_values("Goals", ascending=False) # sorting the value
df_h
colors = [
"#ff0a54",
"#ff477e",
"#ff5c8a",
"#ff7096",
"#ff85a1",
"#ff99ac",
"#fbb1bd",
"#f9bec7",
"#f7cad0",
"#fae0e4",
]
fig, ax = plt.subplots(figsize=(15, 5))
# axes
ax = fig.add_axes([0, 0, 1, 1])
ax.tick_params(axis="x", labelsize=11, colors="Blue") # set x axis font size and colors
ax.tick_params(axis="y", labelsize=11, colors="Blue")
# set x ,Y label and title
ax.set_title("Palyer vs Goal chart", color="Green", fontsize="30")
ax.set_xlabel("Player", color="Green", fontsize="30")
ax.set_ylabel("Goals", color="Green", fontsize="30")
bars = ax.bar(df_h["Player Name"], df_h["Goals"], color=colors)
plt.show()
# **More Deciorartive**
colors = [
"#ff0a54",
"#ff477e",
"#ff5c8a",
"#ff7096",
"#ff85a1",
"#ff99ac",
"#fbb1bd",
"#f9bec7",
"#f7cad0",
"#fae0e4",
]
fig, ax = plt.subplots(figsize=(15, 5))
# axes
ax = fig.add_axes([0, 0, 1, 1])
ax.tick_params(
axis="x", labelsize=11, colors="#ffffff"
) # set x axis font size and colors
ax.tick_params(axis="y", labelsize=11, colors="#ffffff")
# set x ,Y label and title
ax.set_title("Palyer vs Goal chart", color="#eff7f6", fontsize="30")
ax.set_xlabel("Player", color="#eff7f6", fontsize="30")
ax.set_ylabel("Goals", color="#eff7f6", fontsize="30")
ax.set_ylim(0, 20)
bars = ax.bar(df_h["Player Name"], df_h["Goals"], color=colors)
fig.patch.set_facecolor("#11001c") # background color
for i, bar in enumerate(bars): # itereate every time
ax.text(
bar.get_x() + bar.get_width() / 2,
bar.get_height() + 0.25,
df_h["Goals"].iloc[i],
ha="center",
va="bottom",
fontsize="20",
) # ha,va-Horizontal and vertical allignment
plt.show()
# # Player VS Yellow Card Chart
df
df_y = (
df.groupby(by="Team-name").sum().reset_index()
) # make yeloow card number and all athe team name as a group
df_y
df_YC = df_y[["Team-name", "YC"]] # separate those 2 column which we need
df_YC
# # Making Plot
fig, ax2 = plt.subplots(figsize=(25, 10))
ax2 = fig.add_axes(
[
0,
0,
1,
1,
]
)
ax2.tick_params(
axis="x", labelsize=18, colors="#ffffff"
) # set x axis font size and colors
ax2.tick_params(axis="y", labelsize=18, colors="#ffffff")
# set x ,Y label and title
ax2.set_title("Team vs Yellow Card", color="#eff7f6", fontsize="30")
ax2.set_xlabel("Team", color="#eff7f6", fontsize="30")
ax2.set_ylabel("Yellow Card", color="#eff7f6", fontsize="30")
# ax2.set_ylim(0,20)
bars_2 = ax2.bar(df_YC["Team-name"], df_YC["YC"])
ax2.set_xticks(range(len(df_YC["Team-name"])))
ax2.set_xticklabels(
df_YC["Team-name"],
va="center",
ha="center",
rotation=90,
fontsize=18,
color="#ffffff",
)
fig.patch.set_facecolor("#11001c") # background color
for i, bar in enumerate(bars_2): # itereate every time
ax2.text(
bar.get_x() + bar.get_width() / 2,
bar.get_height() + 1.50,
df_YC["YC"].iloc[i],
ha="center",
va="center",
fontsize="20",
) # ha,va-Horizontal and vertical allignment
plt.show()
# # Ascending Order
fig, ax2 = plt.subplots(figsize=(25, 10))
ax2 = fig.add_axes(
[
0,
0,
1,
1,
]
)
ax2.tick_params(
axis="x", labelsize=18, colors="#ffffff"
) # set x axis font size and colors
ax2.tick_params(axis="y", labelsize=18, colors="#ffffff")
# set x ,Y label and title
ax2.set_title("Team vs Yellow Card", color="#eff7f6", fontsize="30")
ax2.set_xlabel("Team", color="#eff7f6", fontsize="30")
ax2.set_ylabel("Yellow Card", color="#eff7f6", fontsize="30")
df_YC = df_YC.sort_values("YC", ascending=True)
# colors combination
"""'colors=['#ff5f5f' ,'#ffa500','#00ffff', '#ff69b4','#00ff7f', '#d8bfd8','#00ced1','#dda0dd','#1e90ff','#87cefa','#4169e1', '#f08080','#ff6347','#32cd32',
'#9932cc', '#ff8c00',
'#ff1493', '#008b8b',
'#8b0000','#9400d3']"""
colors = [
"#ff0000", # Red
"#ff4d4d", # Coral Red
"#ff8080", # Salomon Pink
"#ff9999", # Pink
"#ffb3b3", # Misty Rose
"#ffc6c6", # Cameo Pink
"#ffcccc", # Light Pink
"#ff9999", # Salmon
"#ff6666", # Coral
"#ff3333", # Dark Red
"#e60000", # Ruby Red
"#cc0000", # Fire Brick
"#b30000", # Maroon
"#990000", # Crimson
"#800000", # Dark Red
"#660000", # Blood Red
"#4d0000", # Brown Red
"#330000", # Chocolate
"#1a0000", # Black Rose
]
# ax2.set_ylim(0,20)
bars_2 = ax2.bar(df_YC["Team-name"], df_YC["YC"], color=colors)
ax2.set_xticks(range(len(df_YC["Team-name"])))
ax2.set_xticklabels(
df_YC["Team-name"],
va="center",
ha="center",
rotation="90",
fontsize=18,
color="#ffffff",
)
fig.patch.set_facecolor("#11001c") # background color
for i, bar in enumerate(bars_2): # itereate every time
ax2.text(
bar.get_x() + bar.get_width() / 2,
bar.get_height() + 1.50,
df_YC["YC"].iloc[i],
ha="center",
va="center",
fontsize="20",
) # ha,va-Horizontal and vertical allignment
plt.show()
fig, ax2 = plt.subplots(figsize=(25, 10))
ax2 = fig.add_axes(
[
0,
0,
1,
1,
]
)
ax2.tick_params(
axis="x", labelsize=18, colors="#ffdd00"
) # set x axis font size and colors
ax2.tick_params(axis="y", labelsize=18, colors="#ffdd00")
# set x ,Y label and title
ax2.set_title("Team vs Yellow Card", color="#eff7f6", fontsize="30")
ax2.set_xlabel("Team Name", color="#eff7f6", fontsize="30")
ax2.set_ylabel("Yellow Card", color="#eff7f6", fontsize="30")
df_YC = df_YC.sort_values("YC", ascending=True)
# colors combination
colors = [
"#02026f",
"#0d1b7c",
"#1a338a",
"#264a98",
"#3252a2",
"#4060ad",
"#4c69b7",
"#5862a3",
"#656cbb",
"#7275c6",
"#7e7ecf",
"#8b88d8",
"#9791e2",
"#a39bea",
"#b0a5f3",
"#bcaefc",
"#c9b8ff",
"#d6c2ff",
"#e2ccff",
"#efd6ff",
]
# ax2.set_ylim(0,20)
bars_2 = ax2.bar(df_YC["Team-name"], df_YC["YC"], color=colors)
ax2.set_xticks(range(len(df_YC["Team-name"])))
ax2.set_xticklabels(
df_YC["Team-name"],
va="center",
ha="center",
rotation="90",
fontsize=25,
color="#ffdd00",
)
fig.patch.set_facecolor("#03045e") # background color
for i, bar in enumerate(bars_2): # itereate every time
ax2.text(
bar.get_x() + bar.get_width() / 2,
bar.get_height() + 1.50,
df_YC["YC"].iloc[i],
ha="center",
va="center",
fontsize="20",
color="#ff7b00",
) # ha,va-Horizontal and vertical allignment
plt.show()
|
# ## Introduction
# Kaggle is launching a COVID-19 forecasting challenge to help answer a subset of the National Academies of Sciences, Engineering, and Medicine’s (NASEM) and the World Health Organization (WHO) questions on COVID-19. In this challenge, we will be predicting the daily number of confirmed COVID19 cases in various locations across the world, as well as the number of resulting fatalities, for future dates. In this notebook, we will use COVID19 Global Forecasting (Week 5) dataset including the train, test and submission csv files. First, we will perform data analysis to identify the factors that impact the transmission rate of COVID-19. Afterwards, we will analyze the the effect of COVID-19 in India. Afterwards, we will use XGBoost and Random Forest regressor as ensemble learning models as well as to predict the daily number of confirmed COVID19 cases as well as the number of resulting fatalities in various locations across the world.
# ## Modeling Goal
# I decided early on to not approach this as I usually build forecasting models. Reason is we are modeling a physical phenomenon where people get infected, then infect others for some time, then either recover or die. I therefore studied compartmental models used in epidemiology, SIR and the like. These models rely on two time series: cases and recoveries/deaths. If we have accurate values for both then we can fit these models and get reasonably accurate predictions.
# Issue is we don't have these series.
# For cases we have a proxy, confirmed cases. This is a proxy in many ways:
# * It depends on the testing policy of each geography. Some test a lot, and confirmed cases are close to all cases.
# * A large fraction of sick people are asymptomatic, hence are easily missed by testing.
# * Testing does not happen when people get infected or contagious, it often happens with a delay.For all these reasons the confirmed case nubers we get is a distorted view of actual cases.
# For fatalities the numbers aren't accurate either;
# * In some geos we only get deaths test at hospital, in other geos it includes fatalities from nursing homes.
# * We don't have recoveries data.The latter can be fixed by grabbing recovery data from other online source. This has been done by some top competitors, I wish I had done it.
# Despite all these caveat, I assumed that we still have some form of SIR model at play with the two series we have at hand: fatalities depend on cases detected some while ago. That led to my first model.
# 
# # 1. import library and package
# manipulation data
import pandas as pd
import numpy as np
# visualiation data
import matplotlib.pyplot as plt
import seaborn as sns
import matplotlib
import plotly.graph_objects as go
import plotly.express as px
import plotly.graph_objects as go
from plotly.offline import init_notebook_mode, iplot
# default theme
sns.set(
context="notebook",
style="darkgrid",
palette="Spectral",
font="sans-serif",
font_scale=1,
rc=None,
)
matplotlib.rcParams["figure.figsize"] = [8, 8]
matplotlib.rcParams.update({"font.size": 15})
matplotlib.rcParams["font.family"] = "sans-serif"
# dataprep library
from dataprep.eda import *
from dataprep.datasets import load_dataset
from dataprep.eda import create_report
from sklearn.ensemble import RandomForestRegressor
from sklearn.model_selection import GridSearchCV, KFold
from sklearn import ensemble
from sklearn.preprocessing import OrdinalEncoder
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from sklearn.pipeline import Pipeline
from sklearn import metrics
# ## load data
df = pd.read_csv("../input/covid19-global-forecasting-week-5/train.csv")
test = pd.read_csv("../input/covid19-global-forecasting-week-5/test.csv")
sub = pd.read_csv("../input/covid19-global-forecasting-week-5/submission.csv")
df
# # 2. data analysis
df.info()
df.shape
df.dtypes.value_counts().plot.pie(
explode=[0.1, 0.1, 0.1], autopct="%1.1f%%", shadow=True
)
plt.title("data type")
# 1. our data had (969640 Rows, 9 Columns)
# 2. like we see :
# * more then 55% our data is **object** type
# * 33% is integer
# * 11% float
df.describe(include="all")
# what we can see from the describtion :
# * most of Country_Region case are in US with 895440
# * most of Province_State case are in Texas with 71400
# * most of County cases are in Washington with 8680
# * the most case are at the date of 2020-05-20 with 6926
# * the moste Target are Fatalities with 484820
# # 3. finding missing values
# ### A.train data
missing = df.isnull().sum()
missing_pourcent = df.isnull().sum() / df.shape[0] * 100
dic = {"mising": missing, "missing_pourcent %": missing_pourcent}
frame = pd.DataFrame(dic)
frame
# ### B. test data
missing = test.isnull().sum()
missing_pourcent = test.isnull().sum() / df.shape[0] * 100
dic = {"mising": missing, "missing_pourcent %": missing_pourcent}
frame = pd.DataFrame(dic)
frame
# ### C. submission
sub
# # Data Visualization
df.hist(figsize=(15, 15), edgecolor="black")
# ## TargetValue
plot(df.TargetValue)
fig = px.pie(df, values="TargetValue", names="Target")
fig.update_traces(textposition="inside")
fig.update_layout(uniformtext_minsize=12, uniformtext_mode="hide")
fig.show()
fig = px.pie(df, values="TargetValue", names="Country_Region")
fig.update_traces(textposition="inside")
fig.update_layout(uniformtext_minsize=12, uniformtext_mode="hide")
fig.show()
# ## A. County
plot(df.County)
plt.figure(figsize=(30, 9))
county_plot = df.County.value_counts().head(100)
sns.barplot(county_plot.index, county_plot)
plt.xticks(rotation=90)
plt.title("County count")
# ## B. Province_State
plot(df.Province_State)
plt.figure(figsize=(30, 9))
Province_State_plot = df.Province_State.value_counts().head(100)
sns.barplot(Province_State_plot.index, Province_State_plot)
plt.xticks(rotation=90)
plt.title("Province State count")
# ## C. Country_Region
plot(df.Country_Region)
plt.figure(figsize=(30, 9))
Country_Region_plot = df.Country_Region.value_counts().head(30)
sns.barplot(Country_Region_plot.index, Country_Region_plot)
plt.xticks(rotation=90)
plt.title("Country Region count")
confirmed = df[df["Target"] == "ConfirmedCases"]
fig = px.treemap(
confirmed, path=["Country_Region"], values="TargetValue", width=900, height=600
)
fig.update_traces(textposition="middle center", textfont_size=15)
fig.update_layout(
title={
"text": "Total Share of Worldwide COVID19 Confirmed Cases",
"y": 0.92,
"x": 0.5,
"xanchor": "center",
"yanchor": "top",
}
)
fig.show()
dead = df[df["Target"] == "Fatalities"]
fig = px.treemap(
dead, path=["Country_Region"], values="TargetValue", width=900, height=600
)
fig.update_traces(textposition="middle center", textfont_size=15)
fig.update_layout(
title={
"text": "Total Share of Worldwide COVID19 Fatalities",
"y": 0.92,
"x": 0.5,
"xanchor": "center",
"yanchor": "top",
}
)
fig.show()
fig = px.treemap(
df,
path=["Country_Region"],
values="TargetValue",
color="Population",
hover_data=["Country_Region"],
color_continuous_scale="matter",
title="Current share of Worldwide COVID19 Confirmed Cases",
)
fig.show()
df.Population.value_counts()
df.columns
# ## D. Target
df.Target.value_counts()
df.Target.value_counts().plot.pie(explode=[0.1, 0.1], autopct="%1.1f%%", shadow=True)
# ## Date
last_date = df.Date.max()
df_countries = df[df["Date"] == last_date]
df_countries = df_countries.groupby("Country_Region", as_index=False)[
"TargetValue"
].sum()
df_countries = df_countries.nlargest(10, "TargetValue")
df_trend = df.groupby(["Date", "Country_Region"], as_index=False)["TargetValue"].sum()
df_trend = df_trend.merge(df_countries, on="Country_Region")
df_trend.rename(
columns={"Country_Region": "Country", "TargetValue_x": "Cases"}, inplace=True
)
px.line(
df_trend,
x="Date",
y="Cases",
color="Country",
title="COVID19 Total Cases growth for top 10 worst affected countries",
)
# # 4. Data Preprocessing
# We would drop some features Who have many Null values and not that much important.
df = df.drop(["County", "Province_State", "Country_Region", "Target"], axis=1)
test = test.drop(["County", "Province_State", "Country_Region", "Target"], axis=1)
df
# we gonna cheech if we had i Null values
df.isnull().sum()
# 1. first we gonna create features
# 2. then we gonna train_dev_split
def create_features(df):
df["day"] = df["Date"].dt.day
df["month"] = df["Date"].dt.month
df["dayofweek"] = df["Date"].dt.dayofweek
df["dayofyear"] = df["Date"].dt.dayofyear
df["quarter"] = df["Date"].dt.quarter
df["weekofyear"] = df["Date"].dt.weekofyear
return df
def train_dev_split(df, days):
# Last days data as dev set
date = df["Date"].max() - dt.timedelta(days=days)
return df[df["Date"] <= date], df[df["Date"] > date]
test_date_min = test["Date"].min()
test_date_max = test["Date"].max()
def avoid_data_leakage(df, date=test_date_min):
return df[df["Date"] < date]
def to_integer(dt_time):
return 10000 * dt_time.year + 100 * dt_time.month + dt_time.day
df["Date"] = pd.to_datetime(df["Date"])
test["Date"] = pd.to_datetime(test["Date"])
test["Date"] = test["Date"].dt.strftime("%Y%m%d")
df["Date"] = df["Date"].dt.strftime("%Y%m%d").astype(int)
# # split data
predictors = df.drop(["TargetValue", "Id"], axis=1)
target = df["TargetValue"]
X_train, X_test, y_train, y_test = train_test_split(
predictors, target, test_size=0.22, random_state=0
)
# # RandomForestRegressor
model = RandomForestRegressor(n_jobs=-1)
estimators = 100
model.set_params(n_estimators=estimators)
scores = []
pipeline = Pipeline([("scaler2", StandardScaler()), ("RandomForestRegressor: ", model)])
pipeline.fit(X_train, y_train)
prediction = pipeline.predict(X_test)
pipeline.fit(X_train, y_train)
scores.append(pipeline.score(X_test, y_test))
plt.figure(figsize=(8, 6))
plt.plot(y_test, y_test, color="deeppink")
plt.scatter(y_test, prediction, color="dodgerblue")
plt.xlabel("Actual Target Value", fontsize=15)
plt.ylabel("Predicted Target Value", fontsize=15)
plt.title("Random Forest Regressor (R2 Score= 0.95)", fontsize=14)
plt.show()
X_test
# drop the ForecastId fro test data
test.drop(["ForecastId"], axis=1, inplace=True)
test.index.name = "Id"
test
y_pred2 = pipeline.predict(X_test)
y_pred2
predictions = pipeline.predict(test)
pred_list = [int(x) for x in predictions]
output = pd.DataFrame({"Id": test.index, "TargetValue": pred_list})
print(output)
output
# # XGBoost Regressor
import xgboost as xgb
xgbr = xgb.XGBRegressor(
n_estimators=800,
learning_rate=0.01,
gamma=0,
subsample=0.7,
colsample_bytree=0.7,
max_depth=10,
min_child_weight=0,
objective="reg:squarederror",
nthread=-1,
scale_pos_weight=1,
seed=27,
reg_alpha=0.00006,
n_jobs=-1,
)
xgbr.fit(X_train, y_train)
prediction_xgbr = xgbr.predict(X_test)
print(
"RMSE_XGBoost Regression=",
np.sqrt(metrics.mean_squared_error(y_test, prediction_xgbr)),
)
print("R2 Score_XGBoost Regression=", metrics.r2_score(y_test, prediction_xgbr))
plt.figure(figsize=(8, 6))
plt.scatter(x=y_test, y=prediction_xgbr, color="dodgerblue")
plt.plot(y_test, y_test, color="deeppink")
plt.xlabel("Actual Target Value", fontsize=15)
plt.ylabel("Predicted Target Value", fontsize=15)
plt.title("XGBoost Regressor (R2 Score= 0.89)", fontsize=14)
plt.show()
# # Submission
a = output.groupby(["Id"])["TargetValue"].quantile(q=0.05).reset_index()
b = output.groupby(["Id"])["TargetValue"].quantile(q=0.5).reset_index()
c = output.groupby(["Id"])["TargetValue"].quantile(q=0.95).reset_index()
a.columns = ["Id", "q0.05"]
b.columns = ["Id", "q0.5"]
c.columns = ["Id", "q0.95"]
a = pd.concat([a, b["q0.5"], c["q0.95"]], 1)
a["q0.05"] = a["q0.05"].clip(0, 10000)
a["q0.5"] = a["q0.5"].clip(0, 10000)
a["q0.95"] = a["q0.95"].clip(0, 10000)
a["Id"] = a["Id"] + 1
a
sub = pd.melt(a, id_vars=["Id"], value_vars=["q0.05", "q0.5", "q0.95"])
sub["variable"] = sub["variable"].str.replace("q", "", regex=False)
sub["ForecastId_Quantile"] = sub["Id"].astype(str) + "_" + sub["variable"]
sub["TargetValue"] = sub["value"]
sub = sub[["ForecastId_Quantile", "TargetValue"]]
sub.reset_index(drop=True, inplace=True)
sub.head()
sub.to_csv("submission.csv", index=False)
|
# # Aim:
# ## To perform Ecommerce Customer Churn Analysis and Prediction
# ## Churn: The rate at which customers stop doing business with a company over a given period of time
# # Dataset Overview
# ## Import libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
# ## Read the Dataset
df = pd.read_excel(
"/kaggle/input/ecommerce-customer-churn-analysis-and-prediction/E Commerce Dataset.xlsx",
sheet_name="E Comm",
)
df.head(8)
df.info()
df.describe()
# ## Unique values of the Object Columns
for col in df.columns:
if df[col].dtype == "object":
print(f"Unique values in column {col}: {df[col].unique()}")
# # Exploratory Analysis
# ## Gender vs Churn
grouped = df.groupby(["Gender", "Churn"]).count()["CustomerID"]
grouped.unstack().plot(kind="bar", stacked=False)
plt.xlabel("Gender")
plt.ylabel("Count")
plt.title("Churn by Gender")
plt.show()
# ## MaritalStatus vs Churn
grouped = df.groupby(["MaritalStatus", "Churn"]).count()["CustomerID"]
grouped.unstack().plot(kind="bar", stacked=False)
plt.xlabel("MaritalStatus")
plt.ylabel("Count")
plt.title("Churn by MaritalStatus")
plt.show()
# ## PreferredLoginDevice vs Churn
grouped = df.groupby(["PreferredLoginDevice", "Churn"]).count()["CustomerID"]
grouped.unstack().plot(kind="bar", stacked=False)
plt.xlabel("Preferred Login Device")
plt.ylabel("Count")
plt.title("Churn by Preferred Login Device")
plt.show()
# ## PreferredPaymentMode vs Churn
grouped = df.groupby(["PreferredPaymentMode", "Churn"]).count()["CustomerID"]
grouped.unstack().plot(kind="bar", stacked=False)
plt.xlabel("Preferred Payment Method")
plt.ylabel("Count")
plt.title("Churn by Preferred Payment Method")
plt.show()
# ## Tenure vs Churn
grouped = df.groupby(["Tenure", "Churn"]).count()["CustomerID"]
grouped.unstack().plot(kind="bar", stacked=False)
plt.xlabel("Tenure")
plt.ylabel("Count")
plt.title("Churn by Tenure")
plt.show()
# # Data Cleaning
# ## Number of null values in each column
print(df.isnull().sum())
# ## Use mode to fill the null Values
# ### The mode is the value that appears most often in a set of data values.
df = df.fillna(df.mode().iloc[0])
print(df.isnull().sum())
# # Feature Selection
# ## Correlation Matrix
plt.figure(figsize=(15, 10))
sns.heatmap(df.corr(), cmap="coolwarm")
plt.show()
df = df.drop("CustomerID", axis=1)
# ## Chi-Square Test
# ### Test for independence between categorical variables.
# ### If p > alpha: not significant result, independent.
# ### If p < alpha: significant result, dependent.
from scipy.stats import chi2_contingency
cat_features = df.select_dtypes(include=["object"]).columns
for feature in cat_features:
cross_tab = pd.crosstab(df[feature], df["Churn"])
res = chi2_contingency(cross_tab)
print(f"{feature}: p-value = {res.p}")
alpha = 0.05
if res.p > alpha:
print(f"{feature} is NOT significantly related to churn")
else:
print(f"{feature} is significantly related to churn")
df = pd.get_dummies(df, columns=df.select_dtypes(["object"]).columns)
df.head(5)
# # Machine Learning
# ## Split test and train
from sklearn.model_selection import train_test_split
X = df.drop("Churn", axis=1)
y = df["Churn"]
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=0)
y_train.value_counts()
# ## Preprocessing
# ### StandardScaler() function to standardize the data values into a standard format.
from sklearn.preprocessing import StandardScaler
sc = StandardScaler()
X_train = sc.fit_transform(X_train)
X_test = sc.transform(X_test)
# # Models - Classification Algorithms
# ## Logistic Regression
from sklearn.linear_model import LogisticRegression
lr_clf = LogisticRegression(random_state=0)
lr_clf.fit(X_train, y_train)
print("Accuracy on training set:", lr_clf.score(X_train, y_train) * 100)
print("Accuracy on test set:", lr_clf.score(X_test, y_test) * 100)
# ## Naive Bayes
from sklearn.naive_bayes import GaussianNB
nb_clf = GaussianNB()
nb_clf.fit(X_train, y_train)
print("Accuracy on training set:", nb_clf.score(X_train, y_train) * 100)
print("Accuracy on test set:", nb_clf.score(X_test, y_test) * 100)
# ## Random Forest
from sklearn.ensemble import RandomForestRegressor
rf_clf = RandomForestRegressor(n_estimators=10, random_state=0)
rf_clf.fit(X_train, y_train)
print("Accuracy on training set:", rf_clf.score(X_train, y_train) * 100)
print("Accuracy on test set:", rf_clf.score(X_test, y_test) * 100)
# ## Decision Tree
from sklearn.tree import DecisionTreeClassifier
dec_clf = DecisionTreeClassifier()
dec_clf.fit(X_train, y_train)
print("Accuracy on training set:", dec_clf.score(X_train, y_train) * 100)
print("Accuracy on test set:", dec_clf.score(X_test, y_test) * 100)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Importing Libraries
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.model_selection import train_test_split
from sklearn.metrics import (
log_loss,
roc_auc_score,
precision_score,
f1_score,
recall_score,
roc_curve,
auc,
)
from sklearn.metrics import (
classification_report,
confusion_matrix,
accuracy_score,
fbeta_score,
matthews_corrcoef,
)
from sklearn import metrics
from sklearn.model_selection import StratifiedKFold
# machine learning algorithms
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import (
RandomForestClassifier,
VotingClassifier,
AdaBoostClassifier,
GradientBoostingClassifier,
RandomForestClassifier,
ExtraTreesClassifier,
)
from sklearn.neural_network import MLPClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.linear_model import SGDClassifier
from sklearn.svm import SVC
from scipy import stats
import warnings
warnings.filterwarnings("ignore")
df = pd.read_csv("/kaggle/input/heart-disease-statlog/Heart_disease_statlog.csv")
df.info()
df.describe()
df.columns
df.columns = [
"age",
"sex",
"chest_pain_type",
"resting_blood_pressure",
"cholesterol",
"fasting_blood_sugar",
"rest_ecg",
"max_heart_rate_achieved",
"exercise_induced_angina",
"st_depression",
"st_slope",
"ca",
"thalassemia",
"target",
]
df.head()
df["chest_pain_type"][df["chest_pain_type"] == 0] = "typical angina "
df["chest_pain_type"][df["chest_pain_type"] == 1] = "atypical angina"
df["chest_pain_type"][df["chest_pain_type"] == 2] = "non-angina pain"
df["chest_pain_type"][df["chest_pain_type"] == 3] = "asymptomatic"
df["rest_ecg"][df["rest_ecg"] == 0] = "normal"
df["rest_ecg"][df["rest_ecg"] == 1] = "Abnormality in ST-T wave"
df["rest_ecg"][df["rest_ecg"] == 2] = "left ventricular hypertrophy"
df["st_slope"][df["st_slope"] == 0] = "upsloping"
df["st_slope"][df["st_slope"] == 1] = "flat"
df["st_slope"][df["st_slope"] == 2] = "downsloping"
df["thalassemia"][df["thalassemia"] == 0] = "null"
df["thalassemia"][df["thalassemia"] == 1] = "fixed defect"
df["thalassemia"][df["thalassemia"] == 2] = "normal blood flow"
df["thalassemia"][df["thalassemia"] == 3] = "reversible defect"
df["sex"] = df.sex.apply(lambda x: "male" if x == 1 else "female")
df.head()
df["rest_ecg"].value_counts()
df["chest_pain_type"].value_counts()
df["thalassemia"].value_counts()
df.isna().sum()
df["target"].value_counts().plot.pie(
x="Heart disease",
y="no.of patients",
autopct="%1.0f%%",
labels=["Normal", "Heart Disease"],
startangle=60,
colors=sns.color_palette("crest"),
)
plt.bar(df["sex"], df["target"], color="green")
plt.bar(df["chest_pain_type"], df["target"], color="red")
plt.bar(df["rest_ecg"], df["target"], color="yellow")
plt.pie(df["ca"])
plt.show()
sns.pairplot(df, hue="target", palette="mako")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Chat GPT after fine-tuning GPT2 with command following data set
# https://github.com/tatsu-lab/stanford_alpaca
# alpaca data.json
# https://github.com/minimaxir/gpt-2-simple
import gpt_2_simple as gpt2
import os
import requests
model_name = "124M"
if not os.path.isdir(os.path.join("models", model_name)):
print(f"Downloading {model_name} model...")
gpt2.download_gpt2(
model_name=model_name
) # model is saved into current directory under /models/124M/
file_name1 = "/kaggle/input/alpaca-data/alpaca-data-en.txt"
sess = gpt2.start_tf_sess()
gpt2.finetune(
sess, file_name1, model_name=model_name, steps=1000
) # steps is max number of training steps 1000
gpt2.generate(sess)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Building of a spam detection's system based on SVM, LogisticRegession and Naive base, Machine learning algorithms
# ## Install independencies
from sklearn.naive_bayes import MultinomialNB
from sklearn.svm import LinearSVC
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import VotingClassifier
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.metrics import accuracy_score, precision_score, recall_score, f1_score
from sklearn.model_selection import train_test_split
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
# ## Importing the data
data_email = pd.read_csv("/kaggle/input/email-data/mail_data.csv")
data_email.head()
# ## Structure of the dataset
data_email.describe(include="all")
# Eliminate duplicates
data_email.drop_duplicates()
# Counts the number of spams and hams
spam_output = data_email["Category"].to_numpy()
numberOfham = 0
numberOfSpam = 0
for i in spam_output:
if i == "ham":
numberOfham += 1
else:
numberOfSpam += 1
counts = [numberOfham, numberOfSpam]
# create arrayofvalues
spam_output = data_email["Category"].to_numpy()
# create a bar chart
labels = ["genuines messages", "spams"]
plt.bar(labels, counts)
plt.xlabel("messages")
plt.ylabel("Number of messages")
plt.title("Graphic of messages")
plt.show()
# ## Arranging the category to boolean type
data_email.loc[data_email["Category"] == "spam", "Category"] = 0
data_email.loc[data_email["Category"] == "ham", "Category"] = 1
# ## Separating the data as texts and labels
# input
X = data_email["Message"]
# output
y = data_email["Category"]
# ## Splitting the data into training data and test data for anticipate the accuracy
x_train, x_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=3)
# We consider the train data for 80 percent and 20 percent for the test.
# To ensure that the same splitted data will be reproducible as though the dataset is trained many times, we use the random_state parameter equalized to 3 (as a key for the algorithm which generates the randoms numbers).
# We consider the train data for 80 percent and 20 percent for the test
# ## Checking of the result obtained
print(X.shape)
print(x_train.shape)
print(x_test.shape)
# ## Feature Extraction (convert text into numeric)
# - TfidfVectorizer is a common technique used in spam detection because it can convert textual data into numerical features that can be used in machine learning models based on natural language processing algorithms (NLPA).
# - Tfdif stands for "Term Frequency-Inverse Document Frequecy", which is a technique used to measure the importance of each word in a text corpus.
# - a text corpus is an input for a NLPA
# ### Initilization of a TFIDF
feature_extraction = TfidfVectorizer(min_df=1, stop_words="english", lowercase=True)
# - min_df means that for inclunding anyword, it is must be appear at least one time in the text;
# - the stop_words sets the word (commonly used in a language) that will be removed during the processing
# - lowercase sets all words in lowercase before the process.
x_train_features = feature_extraction.fit_transform(x_train)
# ### Feature Space
# Before talking about the feature space. Let us explain what is the feature vector:
# - A feature vector is a numerical vector that represents the features of an example in a dataset. Each feature represents a specific piece of information about the example, which can be used to train a machine learning model. For example, in the case of the textual data, each feature may represent the number of occurences of a specific word in the text, or TF-IDF score of that word.
# - Thus, the feature space is the set of all features vectors for all examples in a dataset. It is an abstract mathematical space that is defined by choice of features and the way they are transformed or normalized. In machine we usually train models on datasets that have been preprocessed into an appropriate feature space. This allows machine learning algorithms to process the data in a way that is more conducive to detecting relastionships and patterns.
x_test_features = feature_extraction.transform(x_test)
# ### Set the output values into int
# ensure that all output has int type
y_train = y_train.astype("int")
y_test = y_test.astype("int")
# ### Initialize Model for predictions
# Initialize the three models
nb_model = MultinomialNB()
svm_model = LinearSVC()
lr_model = LogisticRegression()
ensemble_model = VotingClassifier(
estimators=[("nb", nb_model), ("svm", svm_model), ("lr", lr_model)], voting="hard"
)
ensemble_model.fit(x_train_features, y_train)
# ## Evaluating the model
# compute evaluation metrics
predictions = ensemble_model.predict(x_test_features)
acc_score = accuracy_score(y_test, predictions)
prec_score = precision_score(y_test, predictions, pos_label=0)
rec_score = recall_score(y_test, predictions, pos_label=0)
f1 = f1_score(y_test, predictions, pos_label=0)
print("Accuracy score:", acc_score)
print("Precision score:", prec_score)
print("Recall score:", rec_score)
print("F1 score:", f1)
# ## Building a Predictive System
input_text = input("Enter your mail here: ")
input_vector = feature_extraction.transform([input_text])
prediction = ensemble_model.predict(input_vector)[0]
if prediction == 0:
print("This is a spam message")
else:
print("This is not a spam message")
|
#
# # Group Members : Vansh Patwari (123), Aadi Vora (129) , Siddharth Yadav (132)
# # Topic Name : Fake News Detection
# --------------------------------------------------------------------------------------------------
# # A brief information about the dataset we are using (For Fake news Detection)
# 1. id : unique id for a news article.
# 2. title : The title of a news article.
# 3. author : Author of the news article.
# 4. content : The text of the article , could be incomplete.
# 5. label : A label that marks whether the news article is real or fake.
# # Importing Dependencies
#
import numpy as np
import pandas as pd
import re
from nltk.corpus import stopwords
from nltk.stem.porter import PorterStemmer
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import accuracy_score, confusion_matrix
import pickle
import matplotlib.pyplot as plt
from sklearn.preprocessing import LabelEncoder
from sklearn.cluster import KMeans
import nltk
nltk.download("stopwords")
# # Prining the stopwords present in english
print(stopwords.words("english"))
# # Data Pre-Processing
# loading the dataset
news_dataset = pd.read_csv("/kaggle/input/fake-news/train.csv")
news_dataset.head()
# printing the null values present
news_dataset.isnull().sum()
# replacing the null vlaues with empty string
news_dataset = news_dataset.fillna("")
# Merging the author name and news title
news_dataset["content"] = news_dataset["author"] + "" + news_dataset["title"]
print(news_dataset["content"])
# Separating the data and label
X = news_dataset.drop(columns="label", axis=1)
Y = news_dataset["label"]
print(X)
print(Y)
# How many fake and real articles?
print(news_dataset.groupby(["content"])["label"].count())
# news_dataset.groupby(['content'])['label'].count().plot(kind="bar")
# plt.show()
# # Stemming :
# Stemming is the process of reducing a word to its Root word
# example :
# actor,actress,acting ---> act
port_stem = PorterStemmer()
def stemming(content):
stemmed_content = re.sub("[^a-zA-Z]", " ", content)
stemmed_content = stemmed_content.lower()
stemmed_content = stemmed_content.split()
stemmed_content = [
port_stem.stem(word)
for word in stemmed_content
if not word in stopwords.words("english")
]
stemmed_content = " ".join(stemmed_content)
return stemmed_content
news_dataset["content"] = news_dataset["content"].apply(stemming)
print(news_dataset["content"])
# Separating the data and label
X = news_dataset["content"].values
Y = news_dataset["label"].values
print(X)
print(Y)
Y.shape
# # What does TfidfVectorizer means ?
# Tf stands for Term Frequency
# idf stands for inverse document frequency
# Converting the textual data to numerical data
from sklearn.feature_extraction.text import TfidfVectorizer
vectorizer = TfidfVectorizer()
vectorizer.fit(X)
X = vectorizer.transform(X)
# # Coverted the content into a unique value of vector so that computer can understand
print(X)
# # Splitting the dataset to trainning and text data
from sklearn.model_selection import train_test_split
X_train, X_test, Y_train, Y_test = train_test_split(
X, Y, test_size=0.2, stratify=Y, random_state=2
)
# # Trainning the model : Logistic Regression
model = LogisticRegression()
model.fit(X_train, Y_train)
# # Evalution of the data
# # Accuracy Score
# accuarcy score on the trainning data
X_train_prediction = model.predict(X_train)
trainning_data_accuracy = accuracy_score(X_train_prediction, Y_train)
print("Accuracy of the trainning data :", trainning_data_accuracy)
# accuarcy score on the test data
X_test_prediction = model.predict(X_test)
test_data_accuracy = accuracy_score(X_test_prediction, Y_test)
print("Accuracy of the test data :", test_data_accuracy)
# # Creating a confusion matrix as the parameter for logistic regression
confusion_matrix(Y_test, X_test_prediction, labels=[1, 0])
# # Building A Predictive System
X_new = X_test[1000]
prediction = model.predict(X_new)
print(prediction)
if prediction[0] == 0:
print("The news is REAL")
else:
print("The news is FAKE")
# For checking whether the predicted answer is right or not
print(Y_test[100])
# # Saving the model to make a web application in flask
# Saving the model
filename = "finalized_model.pkl"
pickle.dump(model, open(filename, "wb"))
# save vectorizer
filename = "vectorizer.pkl"
pickle.dump(vectorizer, open(filename, "wb"))
loaded_model = pickle.load(open("finalized_model.pkl", "rb"))
vector_model = pickle.load(open("vectorizer.pkl", "rb"))
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import numpy as np
import pandas as pd
import re
from sklearn.model_selection import train_test_split
from sklearn.utils import shuffle
import tensorflow as tf
from tensorflow.keras.utils import to_categorical
import matplotlib.pyplot as plt
import warnings
warnings.filterwarnings("ignore")
# Reading Data
wp_df = pd.read_csv("../input/tmnist-alphabet-94-characters/94_character_TMNIST.csv")
wp_df.head()
wp_df
# Displaying info regarding the columns and datatypes
wp_df.info()
# Checking if there are null values in the dataframe
wp_df.isnull().sum()
wp_df.describe()
# checking count of the dataset
wp_df.count()
# Grouping All labels
all_ = list(wp_df["labels"].unique())
# Regex Pattern to check alphabets, digits and special symbols
pattern_uc = re.compile(r"[A-Z]")
pattern_lc = re.compile(r"[a-z]")
pattern_numbers = re.compile(r"[0-9]")
pattern_symbols = re.compile(r"[\W]|[\_\,]")
# Extracting Pattern
lower_case = pattern_lc.findall(str(all_))
Upper_case = pattern_uc.findall(str(all_))
Numbers_ = pattern_numbers.findall(str(all_))
Symbols_ = list(set(pattern_symbols.findall(str(all_))))
Symbols_.pop(27)
# Creating Gropus
group = 1
for list_ in (lower_case, Upper_case, Numbers_, Symbols_):
wp_df.loc[wp_df["labels"].isin(list_), "group"] = str(group)
group += 1
# defining X and y for training
X = wp_df.iloc[:, 2:-1].astype("float32")
y = wp_df[["labels"]]
# preping the data to create dictionary
labels = y["labels"].unique()
values = [num for num in range(len(wp_df["labels"].unique()))]
label_dict = dict(zip(labels, values)) # Creating Dictionary
label_dict_inv = dict(zip(values, labels))
# Mapping
y["labels"].replace(label_dict, inplace=True) # Maping Values
# Checking the mappings
print(label_dict)
print("That's all 94 characters.")
print(wp_df["labels"].unique())
# creating test train split of 80% and 20%
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
# preping the inputs for training
Length, Height = 28, 28
NCl = y_train.nunique()[0] # Unique targets
# N of images 28x28
X_train = np.reshape(X_train.values, (X_train.shape[0], Length, Height))
X_test = np.reshape(X_test.values, (X_test.shape[0], Length, Height))
# Target into Categorical Values
y_train = to_categorical(y_train, NCl, dtype="int")
y_test = to_categorical(y_test, NCl, dtype="int")
print(f"X:Train, Test data shape:{X_train.shape},{X_test.shape}")
print(f"Y:Train, Test data shape:{y_train.shape},{y_test.shape}")
# displaying the charecters of our dataset
random = shuffle(X_train[:500]) # Randomly shuffle
fig, ax = plt.subplots(3, 4, figsize=(10, 10))
axes = ax.flatten()
for i in range(12):
img = np.reshape(random[i], (28, 28)) # reshaping it for displaying
axes[i].imshow(img, cmap="Greys")
img_final = np.reshape(
img, (1, 28, 28, 1)
) # reshapng it for passing into model for prediction
axes[i].grid()
# importing libraries for building neural netwrok
from tensorflow.keras.models import Sequential, load_model
from tensorflow.keras.layers import (
Dense,
Conv2D,
MaxPool2D,
Flatten,
Dropout,
BatchNormalization,
)
from tensorflow.keras.optimizers import SGD, Adam, RMSprop
from tensorflow.keras.callbacks import EarlyStopping
RGB = 1 # In this case only one instead of 3 because we dont have Color images
X_train = X_train.reshape(X_train.shape[0], X_train.shape[1], X_train.shape[2], RGB)
X_test = X_test.reshape(X_test.shape[0], X_test.shape[1], X_test.shape[2], RGB)
# normalizing the image data
X_train = X_train / 255
X_test = X_test / 255
print(f"Train, Test shapes: {X_train.shape},{X_test.shape}")
model = Sequential()
# 4 Conv with Maxpool and Dropout [25%] -> Flatten - > Dense -> Dense -> output
model.add(
Conv2D(
filters=32,
kernel_size=(3, 3),
input_shape=(Length, Height, RGB),
padding="same",
)
)
model.add(BatchNormalization())
model.add(tf.keras.layers.Activation("relu"))
model.add(MaxPool2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Conv2D(filters=64, kernel_size=(3, 3), padding="same"))
model.add(BatchNormalization())
model.add(tf.keras.layers.Activation("relu"))
model.add(MaxPool2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Conv2D(filters=128, kernel_size=(3, 3), padding="same"))
model.add(BatchNormalization())
model.add(tf.keras.layers.Activation("relu"))
model.add(MaxPool2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Conv2D(filters=256, kernel_size=(3, 3), padding="same"))
model.add(BatchNormalization())
model.add(tf.keras.layers.Activation("relu"))
model.add(MaxPool2D(pool_size=(2, 2)))
model.add(Dropout(0.25))
model.add(Flatten())
model.add(Dense(350))
model.add(BatchNormalization())
model.add(tf.keras.layers.Activation("relu"))
model.add(Dropout(0.25))
model.add(Dense(NCl, activation="softmax"))
model.summary()
# defining parameters for training
optimizer = Adam(learning_rate=0.01)
callback = EarlyStopping(monitor="loss", patience=5)
Batch_ = 64
Epochs_ = 20
model.compile(
loss="categorical_crossentropy", optimizer=optimizer, metrics=["accuracy"]
)
# # **Training the Model:**
# Training
history = model.fit(
X_train,
y_train,
validation_data=(X_test, y_test),
batch_size=Batch_,
epochs=Epochs_,
verbose=1,
)
# Evaluating model accuracy on test data
score = model.evaluate(X_test, y_test, batch_size=Batch_, verbose=0)
print(f"Test Accuracy:{round(score[1],4)*100}%")
# Function for Plotting
def Plott(data):
fig, ax = plt.subplots(1, 2, figsize=(20, 7))
# summarize history for accuracy
ax[0].plot(data.history["accuracy"])
ax[0].plot(data.history["val_accuracy"])
ax[0].set_title("model accuracy")
ax[0].legend(["train", "test"], loc="upper left")
# summarize history for loss
ax[1].plot(data.history["loss"], label=["loss"])
ax[1].plot(data.history["val_loss"], label=["val_loss"])
ax[1].set_title("model loss")
ax[1].legend(["train", "test"], loc="upper left")
plt.show()
Plott(history)
# predicting the charecters using trained model
fig, axes = plt.subplots(3, 3, figsize=(8, 9))
axes = axes.flatten()
for i, ax in enumerate(axes):
img = np.reshape(X_test[i], (28, 28)) # reshaping it for displaying
ax.imshow(img, cmap="Greys")
img_final = np.reshape(
img, (1, 28, 28, 1)
) # reshapng it for passing into model for prediction
pred = label_dict_inv[np.argmax(model.predict(img_final))]
ax.set_title("Prediction: " + pred)
ax.grid()
|
# # About
# The initial analysis performed by the community members revealed that the feature calc has the highest impact on predicting the existence of kidney stones. In this analysis, we aim to investigate the relationship between the residual values of predictions made solely based on calc and the other features in the dataset.
# Specifically, we are interested in exploring whether incorporating other features can improve the accuracy of predictions made based on calc.
# In this notebook we created a predictor based on the prior calc -> target probability, and analyzed the relation between the residuals and the other variables in the dataset.
# # Setuping and importing libraries
import numpy as np
import pandas as pd
import os
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.preprocessing import MinMaxScaler
from sklearn.feature_selection import mutual_info_classif
from sklearn.metrics import roc_auc_score
from sklearn.model_selection import train_test_split
import pycaret.regression as pc
from sklearn.model_selection import KFold
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
df = pd.read_csv("/kaggle/input/playground-series-s3e12/train.csv").drop("id", axis=1)
palette = sns.color_palette("gray")
sns.set_palette(palette)
seed = 42
# # Data profiling
# We will start this study profiling the dataset for those who are not familiar with it.
# We can highlight some information regarding the provided data:
# The dataset is relatively small, containing 414 samples and 6 features.
# - There are no missing values
# - The target dataset is slightly unbalanced (66% = 0, 44% = 1)
# - Gravity, Osmo and Cond distributions are left skewed
# - Calc and PH distributions are high skewed
# - There are few Gravity and PH outliers in the training set
df.describe(percentiles=[0.05, 0.5, 0.95])
df.isna().sum()
fig = plt.figure(figsize=(6, 4))
ax = sns.countplot(
x=df["target"],
linewidth=1,
edgecolor="black",
alpha=0.8,
palette=[palette[0], palette[-1]],
)
for i in ax.containers:
ax.bar_label(
i,
)
fig.suptitle("Target variable distribution\n", fontsize=24)
ax.set_ylabel("")
plt.yticks([])
sns.despine()
plt.show()
scaler = MinMaxScaler()
df_scaled = pd.DataFrame(scaler.fit_transform(df), columns=df.columns)
fig = plt.figure(figsize=(6, 4))
fig.suptitle("Features Boxplot", fontsize=24)
ax = sns.boxplot(data=df_scaled.drop("target", axis=1))
plt.yticks([])
sns.despine()
plt.show()
# # Exploratory data analysis
corr = df.corr()
mask = np.triu(np.ones_like(corr, dtype=bool))
fig = plt.figure(figsize=(8, 6))
fig.suptitle("Features Correlation Map", fontsize=24)
sns.heatmap(corr, mask=mask, annot=True, cmap="gray")
plt.show()
# - Some features are highly correlated which is generally undesirable, highly correlated features tend to be redundant.
# - Calc has a fair correlation with the target.
mutual_info = mutual_info_classif(df.drop("target", axis=1), df["target"])
fig = plt.figure(figsize=(6, 4))
ax = sns.barplot(x=df.drop("target", axis=1).columns, y=mutual_info, color="grey")
for i in ax.containers:
ax.bar_label(i, fmt="%.2f")
fig.suptitle("Features mutual information gain\n", fontsize=24)
plt.yticks([])
sns.despine()
plt.show()
# Mutual information is a metric to measure how much a given feature can reduce the entropy of the target variable samples, this concept is widely used in tree based models to select the tree splitting features.
# - Once again calc leads the potential predictive power indicator.
fig, ax = plt.subplots(nrows=3, ncols=2, figsize=(10, 10))
ax = ax.flatten()
for i, col in enumerate(["gravity", "ph", "osmo", "cond", "urea", "calc"]):
sns.kdeplot(
data=df,
x=col,
ax=ax[i],
fill=True,
legend=False,
hue="target",
palette=[palette[0], palette[-2]],
)
ax[i].set_title(f"{col} Distribution")
ax[i].set_xlabel(None)
ax[i].set_ylabel(None)
fig.suptitle("Distribution of Features per Class\n", fontsize=24)
fig.legend(["Crystal (1)", "No Crystal (0)"])
plt.tight_layout()
plt.show()
# - The calc distribution is different among the targets which in theory can provide a good predictive power.
# - For the other features (except for PH) the target classes distributions are fairly separable what can somehow help classifying the samples.
# # Modeling I
# To evaluate the effectiveness of calc in the task of predicting the target we created a simple statistical model that works as following:
# 1. Split the calc values into 0.1 intervals
# 2. Calculate the target probability for each interval
# 3. Apply the rolling mean function to smooth the results and reduce variance and overfitting.
# 4. Calculate the target probability of unseen samples based on the prior probability observed in the training set.
def calc_predictor_prob(df, target):
df_copy = df.copy()
df_copy["target"] = target
df_copy["calc_bin"] = df_copy["calc"].round(1)
intervals = df_copy["calc_bin"].nunique()
calc_prob = df_copy.groupby("calc_bin").agg(prob=("target", "mean"))
calc_prob["smoothed_prob"] = calc_prob["prob"].rolling(7).mean()
calc_prob["smoothed_prob"] = calc_prob["smoothed_prob"].fillna(method="bfill")
probability_dict = calc_prob["smoothed_prob"].to_dict()
return probability_dict
X = df.drop("target", axis=1)
y = df["target"]
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=seed
)
probability_table = calc_predictor_prob(X_train, y_train)
X_train["calc_predictor_prob"] = X_train["calc"].round(1).replace(probability_table)
X_train["calc_residue"] = X_train["calc_predictor_prob"] - y_train
X_test["calc_predictor_prob"] = X_test["calc"].round(1).replace(probability_table)
X_test["calc_residue"] = X_test["calc_predictor_prob"] - y_test
fig = plt.figure(figsize=(6, 4))
ax = sns.lineplot(data=X_train, x="calc", y="calc_predictor_prob")
fig.suptitle("Calc positive class probability (smoothed)\n", fontsize=24)
ax.set_ylabel("probability")
ax.set_xlabel("calc")
ax.grid(True)
plt.show()
# The calc -> probability curves resembles the log function, the higher the value of calc the higher the kidney stone probability.
cv = KFold(n_splits=10, shuffle=True, random_state=seed)
roc_auc_list = []
for train_index, test_index in cv.split(X_train):
X_tr, X_val = X_train.iloc[train_index], X_train.iloc[test_index]
y_tr, y_val = y_train.iloc[train_index], y_train.iloc[test_index]
calc_prob_table = calc_predictor_prob(X_tr, y_tr)
y_pred = X_val["calc"].round(1).replace(calc_prob_table)
roc_auc = roc_auc_score(y_val, y_pred)
roc_auc_list.append(roc_auc)
print(f"Mean ROC AUC: {np.mean(roc_auc_list):.4f}")
# The model was evaluated in a 10-fold cross validation schema and archieved the average score of **0.77 AUC**.
# # Exploring residue
# Now we will analyze the residue of the calc prediction, trying to answer the question:
# **Can the other features fix previous model error?**
fig, ax = plt.subplots(nrows=3, ncols=2, figsize=(10, 10))
ax = ax.flatten()
for i, col in enumerate(["gravity", "ph", "osmo", "cond", "urea", "calc"]):
sns.regplot(data=X_train, x=col, y="calc_residue", ax=ax[i])
ax[i].set_title(f"{col} residue")
ax[i].set_ylabel("")
ax[i].set_xlabel("")
ax[i].grid(False)
fig.suptitle("Feature vs residue correlation\n", fontsize=24)
plt.tight_layout()
plt.show()
# The regression plots are not promising, there is no linear correlation between the features and the residual, which suggests that it is very unlikely that the prediction errors can be corrected by the variables.
# ### Residue mean
# The residue of the predictions made from the single variable 'calc' has an average of approximately 17% of the target mean. This indicates that the model based solely on 'calc' is consistently overestimating the target value by 17%, which suggests that 17% of the target outcome cannot be explained by the model.
# CHATGPT:
# > A zero-mean residual is generally desirable in machine learning. A zero-mean residual indicates that the model's predictions are, on average, just as accurate as the actual values, and that any errors are symmetrically distributed around the actual values. This suggests that the model has managed to capture the underlying patterns in the data and has generalized the problem well. In contrast, if the residuals have a non-zero mean, it indicates that the model is systematically over- or under-predicting the target variable, which suggests that there is bias in the model.
X_test["calc_residue"].mean() / y_test.mean()
# # Fitting the residue
# As expected all the regression machine learning models performed very poorly in the problem of fitting the calc residue using the other features as input.
# Such result indicates that it is improbable that the other variables are capable of adding any significant predictive power to the proposed model.
# However, it does not imply that the other features are entirely useless. It suggests that once an inference is made using solely calc, it is unlikely that any other variable will provide significant improvement to it.
# **Remembering: residue fitting is the core principle of boosting models such as LightGBM, CatBoost and XGBoost.**
reg_setup = pc.setup(
X_train[["gravity", "ph", "osmo", "cond", "urea", "calc_residue"]],
target="calc_residue",
session_id=123,
train_size=0.999,
)
best_model = pc.compare_models(fold=10, round=4)
|
# # Vesuvis Data Preparation
# # Installs
# # Imports
from pathlib import Path
import numpy as np
import pandas as pd
import PIL.Image as Image
from tqdm.auto import tqdm
# # Paths & Settings
KAGGLE_DIR = Path("/") / "kaggle"
INPUT_DIR = KAGGLE_DIR / "input"
COMPETITION_DATA_DIR = INPUT_DIR / "vesuvius-challenge-ink-detection"
DOWNSAMPLING = 1.0
NUM_Z_SLICES = 8
# # Prepare DataFrame
def create_df_from_mask_paths(stage, downsampling):
mask_paths = sorted(COMPETITION_DATA_DIR.glob(f"{stage}/*/mask.png"))
df = pd.DataFrame({"mask_png": mask_paths})
df["mask_png"] = df["mask_png"].astype(str)
df["stage"] = df["mask_png"].str.split("/").str[-3]
df["fragment_id"] = df["mask_png"].str.split("/").str[-2]
df["mask_npy"] = df["mask_png"].str.replace(
stage, f"{stage}_{downsampling}", regex=False
)
df["mask_npy"] = df["mask_npy"].str.replace("input", "working", regex=False)
df["mask_npy"] = df["mask_npy"].str.replace("png", "npy", regex=False)
if stage == "train":
df["label_png"] = df["mask_png"].str.replace("mask", "inklabels", regex=False)
df["label_npy"] = df["mask_npy"].str.replace("mask", "inklabels", regex=False)
df["volumes_dir"] = df["mask_png"].str.replace(
"mask.png", "surface_volume", regex=False
)
df["volume_npy"] = df["mask_npy"].str.replace("mask", "volume", regex=False)
return df
train_df = create_df_from_mask_paths("train", DOWNSAMPLING)
train_df["label_npy"].values[0]
train_df
# # Convert Data to NumPy
# ## Based on https://www.kaggle.com/code/jpposma/vesuvius-challenge-ink-detection-tutorial
def load_image(path):
return Image.open(path)
def resize_image(image, downsampling):
size = int(image.size[0] * downsampling), int(image.size[1] * downsampling)
return image.resize(size)
def load_and_resize_image(path, downsampling):
image = load_image(path)
return resize_image(image, downsampling)
def load_label_npy(path, downsampling):
label = load_and_resize_image(path, downsampling)
return np.array(label) > 0
def load_mask_npy(path, downsampling):
mask = load_and_resize_image(path, downsampling).convert("1")
return np.array(mask)
def load_z_slice_npy(path, downsampling):
z_slice = load_and_resize_image(path, downsampling)
return np.array(z_slice, dtype=np.float32) / 65535.0
def load_volume_npy(volumes_dir, num_z_slices, downsampling):
mid = 65 // 2
start = mid - num_z_slices // 2
end = mid + num_z_slices // 2
z_slices_paths = sorted(Path(volumes_dir).glob("*.tif"))[start:end]
batch_size = num_z_slices // 4
paths_batches = [
z_slices_paths[i : i + batch_size]
for i in range(0, len(z_slices_paths), batch_size)
]
volumes = []
for paths_batch in tqdm(
paths_batches, leave=False, desc="Processing batches", position=1
):
z_slices = [
load_z_slice_npy(path, downsampling)
for path in tqdm(
paths_batch, leave=False, desc="Processing paths", position=2
)
]
volumes.append(np.stack(z_slices, axis=0))
del z_slices
# break
volume = np.concatenate(volumes, axis=0)
return volume
def save_data_as_npy(df, train=True):
for row in tqdm(
df.itertuples(), total=len(df), desc="Processing fragments", position=0
):
mask_npy = load_mask_npy(row.mask_png, DOWNSAMPLING)
volume_npy = load_volume_npy(row.volumes_dir, NUM_Z_SLICES, DOWNSAMPLING)
Path(row.mask_npy).parent.mkdir(exist_ok=True, parents=True)
np.save(row.mask_npy, mask_npy)
np.save(row.volume_npy, volume_npy)
if train:
label_npy = load_label_npy(row.label_png, DOWNSAMPLING)
np.save(row.label_npy, label_npy)
tqdm.write(f"Created {row.volume_npy} with shape {volume_npy.shape}")
save_data_as_npy(train_df)
# # Fix paths
train_df["label_npy"] = train_df["label_npy"].str.replace(
"working", "input/vesuvis-data-preparation", regex=False
)
train_df["mask_npy"] = train_df["mask_npy"].str.replace(
"working", "input/vesuvis-data-preparation", regex=False
)
train_df["volume_npy"] = train_df["volume_npy"].str.replace(
"working", "input/vesuvis-data-preparation", regex=False
)
train_df.to_csv(f"data_{DOWNSAMPLING}.csv")
|
import pandas as pd
import re
path = "/kaggle/input/ir-project-dataset/"
path_out = "/kaggle/working/"
class PatentParser:
def __init__(self, file_path):
self.file_path = file_path
def extract_terms(self, text):
terms = {}
for match in re.finditer(
r"(\b(?:\w+\s){0,6}\w+) \((\d+)\)|(\b(?:\w+\s){0,6}\w+) (\d+)", text
):
term1, number1, term2, number2 = match.groups()
number = number1 if number1 else number2
term = term1 if term1 else term2
if number not in terms:
terms[number] = []
terms[number].append(term)
return terms
def find_longest_common_substring(self, strings):
reversed_strings = [" ".join(s.split()[::-1]) for s in strings]
common_substrings = []
for i in range(len(reversed_strings[0].split())):
substrings = [s.split()[: i + 1] for s in reversed_strings]
if all(substrings[0] == s for s in substrings[1:]):
common_substrings.append(" ".join(substrings[0][::-1]))
else:
break
return max(common_substrings, key=len, default="")
def parse_patent_file(self):
with open(self.file_path, "r") as file:
content = file.read()
patent_sections = content.split("\n\n")
parsed_patents = []
for i, patent_section in enumerate(patent_sections):
sections = patent_section.split("_____d:\n")
patent_lines = sections[0].split("\n")
claim_lines = []
for line in reversed(patent_lines):
if line.startswith("_"):
identifiers = line
title = patent_lines[patent_lines.index(line) - 1]
break
claim_lines.append(line)
claim = "\n".join(reversed(claim_lines))
claim_contains_references = "R" if re.search(r"\w+ \(\d+\)", claim) else "N"
# There's just 4 patents over
# around 2000 that don't behave as expected,
# I'll just ignore those.
if len(sections) != 2:
print("ERROR at", i)
print(identifiers)
continue
claim_code_list_terms = self.extract_terms(claim.lower())
claim_code_term = {}
for code, list_terms in claim_code_list_terms.items():
maybe_term = self.find_longest_common_substring(list_terms)
if maybe_term != "":
claim_code_term[code] = maybe_term
body = sections[1].split("_____c:")[0].strip()
body_contains_references = (
"R" if re.search(r"(\w+ \(\d+\)|\w+ \d+)", body) else "N"
)
body_code_list_terms = self.extract_terms(body.lower())
body_code_term = {}
for code, list_terms in body_code_list_terms.items():
maybe_term = self.find_longest_common_substring(list_terms)
if maybe_term != "":
body_code_term[code] = maybe_term
parsed_patents.append(
{
"title": title,
"identifiers": identifiers,
"claim": claim,
"claim_contains_references": claim_contains_references,
"claim_code_list_terms": claim_code_list_terms,
"claim_code_term": claim_code_term,
"body": body,
"body_contains_references": body_contains_references,
"body_code_list_terms": body_code_list_terms,
"body_code_term": body_code_term,
}
)
if i % 100 == 0:
print("#", i)
df = pd.DataFrame(parsed_patents)
return df
parser = PatentParser(path + "H04N.txt")
patents_df = parser.parse_patent_file()
patents_df
patents_df.to_csv(path_out + "patents_dataframe.csv", index=False)
# import pandas as pd
# import re
# import json
# from ast import literal_eval
# patents_df = pd.read_csv(path+'patents_dataframe.csv')
# patents_df['claim_code_list_terms'] = patents_df['claim_code_list_terms'].apply(literal_eval)
# patents_df['claim_code_term'] = patents_df['claim_code_term'].apply(literal_eval)
# patents_df['body_code_list_terms'] = patents_df['body_code_list_terms'].apply(literal_eval)
# patents_df['body_code_term'] = patents_df['body_code_term'].apply(literal_eval)
def merge_dicts_by_numerical_reference(df, column_name):
merged_dict = {}
for index, row in df.iterrows():
# print(index)
cell_dict = row[column_name]
# print(type(cell_dict))
# print(cell_dict)
for key, value in cell_dict.items():
if key in merged_dict:
merged_dict[key].append((value, index))
else:
merged_dict[key] = [(value, index)]
return merged_dict
merged_patents = merge_dicts_by_numerical_reference(patents_df, "body_code_term")
# merged_patents
print("Number of numerical references:")
len(merged_patents)
merged_patents_more_than_1 = {k: v for k, v in merged_patents.items() if len(v) > 1}
# merged_patents_more_than_1
print("Number of numerical references that appear more than once:")
len(merged_patents_more_than_1)
# The take-away here is that numerical references don't reference
# the same technical term in different patents
# (unfortunately,
# otherwise we could have applied again the find_longest_common_substring
# function on the merged dictionaries to again get better technical terms).
# But let's try to see what happens if we make this assumption anyway.
# In this case we can find the longest common substring for every numerical reference
# to see if from some of this references we can get a better technical term.
def find_longest_common_substring(strings):
reversed_strings = [" ".join(s.split()[::-1]) for s in strings]
common_substrings = []
for i in range(len(reversed_strings[0].split())):
substrings = [s.split()[: i + 1] for s in reversed_strings]
if all(substrings[0] == s for s in substrings[1:]):
common_substrings.append(" ".join(substrings[0][::-1]))
else:
break
return max(common_substrings, key=len, default="")
def merge_dicts_longest_common_substring(input_dict):
result_dict = {}
for key, tuples_list in input_dict.items():
terms = [t[0] for t in tuples_list]
longest_common_substring = find_longest_common_substring(terms)
if longest_common_substring != "":
indexes = [t[1] for t in tuples_list if longest_common_substring in t[0]]
result_dict[key] = (longest_common_substring, indexes)
return result_dict
merged_patents_largest_chunk = merge_dicts_longest_common_substring(merged_patents)
# merged_patents_largest_chunk
# How many are there?
len(merged_patents_largest_chunk)
# Let's find how many of there numerical references
# map to a term that appears in more than one patent:
numer_references_more_than_1 = {
k: v for k, v in merged_patents_largest_chunk.items() if len(v[1]) > 1
}
len(numer_references_more_than_1)
# Now we change point of view,
# we consider the candidate technical term as the key
# and the list of patents as the value.
def merge_dicts_by_term(df, column_name):
merged_dict = {}
for index, row in df.iterrows():
cell_dict = row[column_name]
for key, value in cell_dict.items():
if value in merged_dict:
merged_dict[value].append(index)
else:
merged_dict[value] = [index]
return merged_dict
merged_patents_by_term = merge_dicts_by_term(patents_df, "body_code_term")
# merged_patents_by_term
# How many terms are there?
len(merged_patents_by_term)
# How many of these terms appear in more than one patent?
terms_more_than_1 = {k: v for k, v in merged_patents_by_term.items() if len(v) > 1}
# terms_more_than_1
# Let's sort the dictionary by the number of patents in which the term appears:
terms_more_than_1_sorted = {
k: v
for k, v in sorted(
terms_more_than_1.items(), key=lambda item: len(item[1]), reverse=True
)
}
for term in list(terms_more_than_1_sorted.keys())[:30]:
print(term)
# Let's try filtering these terms with some heuristics
# A sample of terms
import random
random.seed(2023)
print("Sample of terms before filtering")
for key in random.sample(list(merged_patents_by_term.keys()), 50):
print(key)
list_terms = list(merged_patents_by_term.keys())
print(len(list_terms))
def filter_terms(terms, stop_words):
filtered_terms = []
for term in terms:
# Remove leading/trailing whitespace
term = term.strip()
# Remove trailing 'a', 'an', 'the'
term = re.sub(r"\s+(a|an|the)$", "", term, flags=re.IGNORECASE)
# Check if the term is a stop word, contains only digits, or contains a number followed by a letter
if (
term.lower() in stop_words
or term.isdigit()
or re.search(r"\b\d+\b", term)
or re.search(r"\b\d+[a-zA-Z]\b", term)
or re.search(r"\bstep\b", term.lower())
or re.search(r"\bsteps\b", term.lower())
or re.search(r"\bmethod\b", term.lower())
or re.search(r"\bmethods\b", term.lower())
or re.search(r"\bequation\b", term.lower())
or re.search(r"\bequations\b", term.lower())
or re.search(r"\boperation\b", term.lower())
or re.search(r"\boperations\b", term.lower())
or re.search(r"\bformula\b", term.lower())
or re.search(r"\bformulas\b", term.lower())
or re.search(r"\bfig\b", term.lower())
or re.search(r"\bfigs\b", term.lower())
or re.search(r"\bfigure\b", term.lower())
or re.search(r"\bimage\b", term.lower())
or re.search(r"\bimages\b", term.lower())
or re.search(r"\bfigures\b", term.lower())
or re.search(r"\btable\b", term.lower())
or re.search(r"\btables\b", term.lower())
or re.search(r"\bgraph\b", term.lower())
or re.search(r"\bgraphs\b", term.lower())
or re.search(r"\bdiagram\b", term.lower())
or re.search(r"\bdiagrams\b", term.lower())
or re.search(r"\bblock\b", term.lower())
or re.search(r"\bblocks\b", term.lower())
):
continue
# Remove some words and everything before them
term = re.sub(r".*\binclude\b\s*", "", term, flags=re.IGNORECASE)
term = re.sub(r".*\bincludes\b\s*", "", term, flags=re.IGNORECASE)
term = re.sub(r".*\bincluded\b\s*", "", term, flags=re.IGNORECASE)
term = re.sub(r".*\bcomprise\b\s*", "", term, flags=re.IGNORECASE)
term = re.sub(r".*\bcomprises\b\s*", "", term, flags=re.IGNORECASE)
term = re.sub(r".*\bcomprised\b\s*", "", term, flags=re.IGNORECASE)
term = re.sub(r".*\bselect\b\s*", "", term, flags=re.IGNORECASE)
term = re.sub(r".*\bselects\b\s*", "", term, flags=re.IGNORECASE)
term = re.sub(r".*\bsaid\b\s*", "", term, flags=re.IGNORECASE)
term = re.sub(r".*\bclause\b\s*", "", term, flags=re.IGNORECASE)
term = re.sub(r".*\bclauses\b\s*", "", term, flags=re.IGNORECASE)
term = re.sub(r".*\bsection\b\s*", "", term, flags=re.IGNORECASE)
term = re.sub(r".*\bsections\b\s*", "", term, flags=re.IGNORECASE)
term = re.sub(r".*\bpart\b\s*", "", term, flags=re.IGNORECASE)
term = re.sub(r".*\bparts\b\s*", "", term, flags=re.IGNORECASE)
term = re.sub(r".*\bparagraph\b\s*", "", term, flags=re.IGNORECASE)
term = re.sub(r".*\bparagraphs\b\s*", "", term, flags=re.IGNORECASE)
term = re.sub(r".*\bitem\b\s*", "", term, flags=re.IGNORECASE)
term = re.sub(r".*\bitems\b\s*", "", term, flags=re.IGNORECASE)
# Remove stop words from the term
term = " ".join(
[word for word in term.split() if word.lower() not in stop_words]
)
# Keep the filtered term
filtered_terms.append(term)
return filtered_terms
from spacy.lang.en.stop_words import STOP_WORDS
my_stop_words = [
"first",
"second",
"third",
"fourth",
"fifth",
"sixth",
"seventh",
"eighth",
"ninth",
"tenth",
"eleventh",
"twelfth",
"thirteenth",
"fourteenth",
"fifteenth",
"sixteenth",
"seventeenth",
"eighteenth",
"nineteenth",
"twentieth",
"thirtieth",
"fortieth",
"fiftieth",
"sixtieth",
"one",
"two",
"three",
"four",
"five",
"six",
"seven",
"eight",
"nine",
"ten",
"eleven",
"twelve",
"thirteen",
"fourteen",
"fifteen",
"sixteen",
"seventeen",
"eighteen",
"nineteen",
"twenty",
"thirty",
"forty",
"fifty",
"sixty",
"primary",
"secondary",
"tertiary",
"quaternary",
"quinary",
"senary",
"septenary",
]
STOP_WORDS = STOP_WORDS.union(my_stop_words)
filtered_terms = filter_terms(list_terms, STOP_WORDS)
print("List")
print(len(filtered_terms))
print("Set")
print(len(set(filtered_terms)))
print("-" * 64)
print("Sample of terms after filtering")
for term in random.sample(list(set(filtered_terms)), 30):
print(term)
|
from IPython.display import display, HTML
display(HTML("<style>.container { width:100% !important; }</style>"))
data_path = "/kaggle/input/segment-job-reachability-data"
file_full_user_count = "file_full_user_count"
file_delta_user_count = "file_delta_user_count"
job_version = "job_version"
epsilon = 1
import os
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from matplotlib.pyplot import figure
from tqdm.notebook import tqdm
os.path.exists(data_path)
# # EDA on segment jobs data
def extract_describe(df):
describe = df.describe()
mean = describe.loc["mean"]
std = describe.loc["std"]
maxVal = describe.loc["max"]
minVal = describe.loc["min"]
return mean, std, minVal, maxVal
def print_minimal_description(df, fieldname):
mean, std, minV, maxV = extract_describe(df[fieldname].dropna())
print(
"{} \t Mean: {:0.2f} \t\t Std: {:0.2f} \t\t Min: {:0.2f} \t\t Max: {:0.2f}".format(
fieldname, mean, std, minV, maxV
)
)
def plot_hist(df, fieldname):
figure(figsize=(16, 2), dpi=80)
plt.hist(df[fieldname], bins=30)
plt.title("Histogram for {}".format(fieldname))
plt.show()
def double_plot_value(df, fieldname1, fieldname2):
values1 = df[fieldname1]
values2 = df[fieldname2]
figure(figsize=(16, 2), dpi=80)
plt.plot(range(len(values1)), values1, color="red", marker=".", markersize=1)
plt.plot(range(len(values2)), values2, color="orange", marker=".", markersize=1)
plt.title("Plotting for {} + {}".format(fieldname1, fieldname2))
plt.show()
def plot_value(df, fieldname):
values = df[fieldname]
figure(figsize=(16, 2), dpi=80)
plt.plot(range(len(values)), values, color="red", marker=".", markersize=1)
plt.title("Plotting for {}".format(fieldname))
plt.show()
def plot_value_sorted(df, fieldname):
values = df[fieldname].sort_values(ascending=False)
figure(figsize=(16, 2), dpi=80)
plt.plot(range(len(values)), values, color="red", marker=".", markersize=1)
plt.title("Plotting sorted for {}".format(fieldname))
plt.show()
def calculate_metric(filename):
df = pd.read_csv(os.path.join(data_path, filename), index_col=False)
print("Report for segment: ", filename.split(".")[0])
print("Len: ", len(df))
metric_df = pd.DataFrame()
metric_df["full"] = df["file_full_user_count"]
metric_df["delta"] = df["file_delta_user_count"]
metric_df["full_rolling_20"] = df["file_full_user_count"].rolling(20).mean()
metric_df["delta_rolling_20"] = df["file_delta_user_count"].rolling(20).mean()
metric_df["fcr"] = metric_df["full"] / (metric_df["full"].shift(1) + 1)
metric_df["dcr"] = metric_df["delta"] / (metric_df["delta"].shift(1) + 1)
metric_df["f_diff_to_avg"] = (metric_df["full"] - metric_df["full"].mean()).abs()
metric_df["d_diff_to_avg"] = (metric_df["delta"] - metric_df["delta"].mean()).abs()
metric_df["f_diff_to_prev"] = metric_df["full"] - metric_df["full"].shift(1)
metric_df["d_diff_to_prev"] = metric_df["delta"] - metric_df["delta"].shift(1)
metric_df["f_diff_rolling_20"] = (
metric_df["full"] - metric_df["full"].rolling(20).mean()
).abs()
metric_df["d_diff_rolling_20"] = (
metric_df["delta"] - metric_df["delta"].rolling(20).mean()
).abs()
# remove first value of fcr and dcr
metric_df["fcr"] = metric_df["fcr"].loc[2:]
metric_df["dcr"] = metric_df["dcr"].loc[2:]
print_minimal_description(metric_df, "full")
print_minimal_description(metric_df, "delta")
print_minimal_description(metric_df, "fcr")
print_minimal_description(metric_df, "dcr")
double_plot_value(metric_df, "full", "full_rolling_20")
double_plot_value(metric_df, "delta", "delta_rolling_20")
plot_value(metric_df, "f_diff_rolling_20")
plot_value(metric_df, "d_diff_rolling_20")
plot_value_sorted(metric_df, "f_diff_to_avg")
plot_value_sorted(metric_df, "d_diff_to_avg")
plot_value_sorted(metric_df, "f_diff_to_prev")
plot_value_sorted(metric_df, "d_diff_to_prev")
print("\n\n\n")
listdir = os.listdir(data_path)
for filedir in listdir:
calculate_metric(filedir)
# # Testing abnormal detection algo & mechanism
def get_metric(c_full, c_delta, p_full, p_delta):
FCR = c_full / (p_full + epsilon)
DCR = c_delta / (p_delta + epsilon)
return {
"full": c_full,
"delta": c_delta,
"fcr": FCR,
"dcr": DCR,
}
class DataContainer:
def __init__(self, name):
self.name = name
self.metric_names = [
"full",
"delta",
"fcr",
"dcr",
]
self.df = pd.DataFrame(columns=self.metric_names)
self.logs = []
def add(self, props, logs=None):
self.df = self.df.append(props, ignore_index=True)
if logs is not None:
self.logs.append(logs)
def get_data(self, offset):
if len(self.df) >= offset:
return self.df.iloc[-offset], True
else:
return 0, False
def get_mean(self, bound=None):
if bound is None:
df = {}
for field in self.df.columns:
df[field] = self.df[field].astype("float").describe()["mean"]
return df, True
else:
if len(self.df) >= bound:
df = {}
for field in self.df.columns:
df[field] = self.df[field].astype("float")[-bound:].mean()
return df, True
else:
return {}, False
def get_std(self, bound=None):
if bound is None:
df = {}
for field in self.df.columns:
df[field] = self.df[field].astype("float").describe()["std"]
return df, True
else:
if len(self.df) >= bound:
df = {}
for field in self.df.columns:
df[field] = self.df[field].astype("float")[-bound:].std()
return df, True
else:
return {}, False
def get_distribution(self, offset):
if len(self.df) >= offset:
df = self.df.iloc[-offset]
return {
"delta": df["delta"],
"other": df["full"] - df["delta"],
}, True
else:
return {}, False
def show(self):
print("Total number of record: ", len(self.df))
print("Total number of logs: ", len(self.logs))
self.plot()
def plot(self):
print("Ta-daaa!!!")
pass
class AnomalyDetector:
def __init__(self, name):
self.name = name
self.valid_data = DataContainer("valid_data_container")
self.anomaly_data = DataContainer("anomaly_data_container")
self.threshold = {
"full": 0.25,
"delta": 0.25,
"dcr": 0.25,
"lcr": 0.25,
}
def detect(self, props, threshold):
full = props["full"]
delta = props["delta"]
fcr = props["fcr"]
dcr = props["dcr"]
violate_rules = []
metrics = {}
# Logic rule
# Directly compare
def directly_compare_detection(data, name, fields):
for field in fields:
real = abs((props[field] - data[field]) / (props[field] + epsilon))
expected = threshold[field]
if real > expected:
violate_rules.append(
"[Anomaly][{}][{}][Group method:{}] Difference in {} is greater then threshold, {} > {}".format(
name,
field,
2,
field,
real,
expected,
)
)
metrics["directly_compare_{}_{}".format(name, field)] = real
last_valid_data, ok = self.valid_data.get_data(1)
if ok:
directly_compare_detection(
last_valid_data,
"last",
["full", "delta", "leave", "dcr", "lcr", "d_prop", "l_prop"],
)
same_day_last_week_data, ok = self.valid_data.get_data(7)
if ok:
directly_compare_detection(
same_day_last_week_data,
"same day last week",
["delta", "leave", "dcr", "lcr", "d_prop", "l_prop"],
)
# All_partner: unavailable
# Quantity-based detection
def quantity_based_detection(props, mean, std, name, fields):
for field in fields:
z_score = (props[field] - mean[field]) / (std[field] + 0.01)
if z_score < -threshold["z_score"] or z_score > threshold["z_score"]:
violate_rules.append(
"[Anomaly][{}][{}][Group method:{}] Value of Z_score in {} outside the range: {} for [{},{}]".format(
name,
field,
3,
field,
z_score,
-1.65,
1.65,
)
)
metrics["z_score_{}_{}".format(name, field)] = z_score
all_mean, ok = self.valid_data.get_mean()
all_std, ok = self.valid_data.get_std()
if ok:
quantity_based_detection(
props,
all_mean,
all_std,
"all mean",
["delta", "leave", "dcr", "lcr", "d_prop", "l_prop"],
)
rolling_mean, ok = self.valid_data.get_mean(7)
rolling_std, ok = self.valid_data.get_std(7)
if ok:
quantity_based_detection(
props,
rolling_mean,
rolling_std,
"7 day mean",
["delta", "leave", "dcr", "lcr", "d_prop", "l_prop"],
)
# distributed-based detection
# not available, since have no distribution in this kind of work. Try distribution of delta in full
def distribution_base_detection(
expected_distribution, actually_distribution, name
):
## Kullback leibler convergence
kl = 0.0
is_ok = True
for field in actually_distribution.keys():
if actually_distribution[field] == 0.0:
is_ok = False
break
if is_ok:
for field in expected_distribution.keys():
kl = kl + expected_distribution[field] * np.log(
expected_distribution[field] / actually_distribution[field]
)
if kl > threshold["kl"]:
violate_rules.append(
"[Anomaly][{}][Group method:{}] Value of KL greater then threshold: {} > {}".format(
name,
4,
kl,
threshold["kl"],
)
)
metrics["kl_{}".format(name)] = kl
else:
metrics["kl_{}".format(name)] = 0.0
## Chi-Squared test
actually_count = {
"delta": delta,
"other": full - delta,
}
expected_count = {
"delta": full * expected_distribution["delta"],
"other": full * expected_distribution["other"],
}
if full > 30:
chi_squared = 0.0
for field in actually_count.keys():
chi_squared = (
chi_squared
+ ((actually_count[field] - expected_count[field]) ** 2)
/ expected_count[field]
)
if chi_squared > threshold["chi2"]:
violate_rules.append(
"[Anomaly][{}][Group method:{}] Value of Chi2 greater then threshold: {} > {}".format(
name,
4,
chi_squared,
threshold["chi2"],
)
)
metrics["chi2_{}".format(name)] = chi_squared
distribution = {
"delta": delta / full,
"other": (full - delta) / full,
}
last_distribution, ok = self.valid_data.get_distribution(1)
if ok:
distribution_base_detection(distribution, last_distribution, "last")
same_day_last_week_distribution, ok = self.valid_data.get_distribution(7)
if ok:
distribution_base_detection(
distribution, same_day_last_week_distribution, "same_day_last_week"
)
return metrics, violate_rules
def add(self, props, index):
prv_data, ok = self.valid_data.get_data(1)
violate_rules = []
if ok:
props = get_metric(
props["file_full_user_count"],
props["file_delta_user_count"],
prv_data["full"],
prv_data["delta"],
prv_data["leave"],
)
else:
props = get_metric(
props["file_full_user_count"], props["file_delta_user_count"], 0, 0, 0
)
metric, rule = self.detect(props, self.threshold)
violate_rules = violate_rules + rule
self.valid_data.add(props)
print("Record {}:".format(index))
for field in props:
print("{}\t{}".format(field, props[field]))
for field in metric:
print("{}\t{}".format(field, metric[field]))
print("What wrongs:\n", "\n ".join(violate_rules))
def show(self):
print("Total number of anomaly data")
self.anomaly_data.show()
print("Total number of valid data")
self.valid_data.show()
filename = "New_Query_{}.csv".format(12904)
df = pd.read_csv(os.path.join(data_path, filename), index_col=False)
detector = AnomalyDetector("anom_detector")
for i in tqdm(range(len(df)), "Detecting in process..."):
raw = df.iloc[i]
detector.add(raw, i)
detector.show()
def visualize_zscore(df):
cmax = len(df) + 2
threshold = 3.0
duration = 10
min_denom = 20
new_df = pd.DataFrame()
new_df["full"] = df["file_full_user_count"]
new_df["delta"] = df["file_delta_user_count"]
new_df["delta_diff"] = new_df["delta"].diff()
new_df["diff_current"] = new_df["full"] - new_df["delta"]
new_df["leave"] = new_df["full"].shift(periods=1) - new_df["diff_current"]
new_df["delta_diff_1"] = new_df["delta"] - new_df["delta"].shift(periods=1)
new_df["leave_diff_1"] = new_df["leave"] - new_df["leave"].shift(periods=1)
new_df["delta_change_rate"] = new_df["delta"] / new_df["delta"].shift(periods=1)
new_df["leave_change_rate"] = new_df["leave"] / new_df["leave"].shift(periods=1)
new_df["z_delta_diff_all"] = (
new_df["delta_diff_1"]
- new_df["delta_diff_1"].rolling(window=cmax, min_periods=1).mean().shift(1)
) / new_df["delta_diff_1"].rolling(window=cmax, min_periods=1).std().shift(1)
new_df["z_leave_diff_all"] = (
new_df["leave_diff_1"]
- new_df["leave_diff_1"].rolling(window=cmax, min_periods=1).mean().shift(1)
) / new_df["leave_diff_1"].rolling(window=cmax, min_periods=1).std().shift(1)
new_df["delta_prop"] = new_df["delta"] / (new_df["full"] + 30)
new_df["leave_prop"] = new_df["leave"] / (new_df["full"] + 30)
new_df["full_diff_1"] = new_df["full"] - new_df["full"].shift(periods=1)
new_df["full_diff_7"] = new_df["full"] - new_df["full"].shift(periods=7)
new_df["delta_diff_7"] = new_df["delta"] - new_df["delta"].shift(periods=7)
new_df["leave_diff_7"] = new_df["leave"] - new_df["leave"].shift(periods=7)
new_df["rolling_mean_7_shift_1"] = (
new_df["delta"].rolling(window=7, min_periods=1, closed="both").mean().shift(1)
)
new_df["rolling_std_7_shift_1"] = (
new_df["delta"].rolling(window=7, min_periods=1, closed="both").std().shift(1)
)
new_df["z_score_delta_7"] = (
new_df["delta"]
- new_df["delta"].rolling(window=7, min_periods=1).mean().shift(1)
) / (new_df["delta"].rolling(window=7, min_periods=1).std().shift(1) + 0.01)
new_df["z_score_leave_7"] = (
new_df["leave"]
- new_df["leave"].rolling(window=7, min_periods=1).mean().shift(1)
) / (new_df["leave"].rolling(window=7, min_periods=1).std().shift(1) + 0.01)
anomaly_point_info = []
for i in range(len(new_df)):
# Use rules that was accept in TDD
def calc_anon():
if abs(new_df["delta_diff"].iloc[i]) >= new_df["delta"].iloc[i - 1] * 0.3:
anomaly_point_info.append(
{
"pos": i,
"field": "delta_diff",
"value": new_df["delta"].iloc[i],
"info": {
"delta": new_df["delta"].iloc[i],
"prev_delta": new_df["delta"].iloc[i - 1],
},
}
)
if new_df["delta"].iloc[i] < 1 and new_df["full"].iloc[i] >= 1:
anomaly_point_info.append(
{"pos": i, "field": "delta", "value": new_df["delta"].iloc[i]}
)
if abs(new_df["z_delta_diff_all"].iloc[i]) > threshold:
if abs(new_df["delta_diff_1"].iloc[i]) > duration:
anomaly_point_info.append(
{
"pos": i,
"field": "z_delta_diff_all",
"value": new_df["z_delta_diff_all"].iloc[i],
"info": {
"delta": new_df["delta"].iloc[i],
"prev_delta": new_df["delta"].shift(periods=1).iloc[i],
"delta_diff": new_df["delta_diff_1"].iloc[i],
"mean": new_df["delta_diff_1"]
.rolling(window=cmax, min_periods=2)
.mean()
.shift(1)
.iloc[i],
"std": new_df["delta_diff_1"]
.rolling(window=cmax, min_periods=2)
.std()
.shift(1)
.iloc[i],
},
}
)
if abs(new_df["z_leave_diff_all"].iloc[i]) > threshold:
if abs(new_df["leave_diff_1"].iloc[i]) > duration:
anomaly_point_info.append(
{
"pos": i,
"field": "z_leave_diff_all",
"value": new_df["z_leave_diff_all"].iloc[i],
"info": {
"leave": new_df["leave"].iloc[i],
"prev_leave": new_df["leave"].shift(periods=1).iloc[i],
"leave_diff": new_df["leave_diff_1"].iloc[i],
"mean": new_df["leave_diff_1"]
.rolling(window=cmax, min_periods=2)
.mean()
.shift(1)
.iloc[i],
"std": new_df["leave_diff_1"]
.rolling(window=cmax, min_periods=2)
.std()
.shift(1)
.iloc[i],
},
}
)
if abs(new_df["z_score_delta_7"].iloc[i]) > threshold:
if (
abs(
new_df["delta"].iloc[i]
- new_df["delta"]
.rolling(window=7, min_periods=2, closed="both")
.mean()
.shift(1)
.iloc[i]
)
> duration
):
anomaly_point_info.append(
{
"pos": i,
"field": "z_score_delta_7",
"value": new_df["z_score_delta_7"].iloc[i],
"info": {
"delta": new_df["delta"].iloc[i],
"mean": new_df["delta"]
.rolling(window=7, min_periods=2, closed="both")
.mean()
.shift(1)
.iloc[i],
"std": new_df["delta"]
.rolling(window=7, min_periods=2, closed="both")
.std()
.shift(1)
.iloc[i],
},
}
)
if abs(new_df["z_score_leave_7"].iloc[i]) > threshold:
if (
abs(
new_df["leave"].iloc[i]
- new_df["leave"]
.rolling(window=7, min_periods=2, closed="both")
.mean()
.shift(1)
.iloc[i]
)
> duration
):
anomaly_point_info.append(
{
"pos": i,
"field": "z_score_leave_7",
"value": new_df["z_score_leave_7"].iloc[i],
"info": {
"leave": new_df["leave"].iloc[i],
"mean": new_df["leave"]
.rolling(window=7, min_periods=2, closed="both")
.mean()
.shift(1)
.iloc[i],
"std": new_df["leave"]
.rolling(window=7, min_periods=2, closed="both")
.std()
.shift(1)
.iloc[i],
},
}
)
# calc_anon()
plot_fields = [
"full",
"delta",
"leave",
"delta_change_rate",
"leave_change_rate",
"delta_prop",
"leave_prop",
]
f, ax = plt.subplots(len(plot_fields), 1)
f.set_figwidth(20)
f.set_figheight(18)
f.tight_layout()
for i, field in enumerate(plot_fields):
ax[i].bar(np.arange(len(new_df)), new_df[field])
ax[i].axhline(y=0, color="black", linewidth=0.8, alpha=0.5, ls="--")
ax[i].set_ylim(bottom=min(0.0, new_df[field].min()))
ax[i].set_xlim(left=-1.0, right=len(new_df) + 1)
ax[i].axhline(y=0, color="black", linewidth=0.8, alpha=0.5, ls="--")
ax[i].set_title(field)
for anom_point in anomaly_point_info:
ax[i].axvspan(
anom_point["pos"] - 0.5, anom_point["pos"] + 0.5, color="red", alpha=0.5
)
"""import seaborn as sns
plt.figure(figsize = (8, 6))
sns.displot(data=new_df, x="delta_diff", kind="kde")"""
return anomaly_point_info
infos = visualize_zscore(df)
for i, info in enumerate(infos):
print("Anomaly {}:".format(i))
for key in info.keys():
print("{}\t{}".format(key, info[key]))
print()
visualize_zscore(df)
visualize_zscore(pd.read_csv(os.path.join(data_path, "New_Query_15649.csv")))
# ## Using other anomaly detection method
from statsmodels.tsa.seasonal import seasonal_decompose
from matplotlib.pyplot import figure
def visualize_custom(
df,
min_denom=20,
window_size=7,
sigma_mul=4,
rolling=7,
method="decompose",
shift_amount=2,
verbose=0,
):
cmax = len(df) + 2
new_df = pd.DataFrame()
new_df["full"] = df["file_full_user_count"]
new_df["delta"] = df["file_delta_user_count"]
new_df["delta_diff"] = new_df["delta"].diff()
new_df["diff_current"] = new_df["full"] - new_df["delta"]
new_df["leave"] = new_df["full"].shift(periods=1) - new_df["diff_current"]
new_df["delta_change_rate"] = new_df["delta"] / (
new_df["delta"].shift(periods=1) + min_denom
)
new_df["leave_change_rate"] = new_df["leave"] / (
new_df["leave"].shift(periods=1) + min_denom
)
new_df["delta_prop"] = new_df["delta"] / (new_df["full"] + min_denom)
new_df["leave_prop"] = new_df["leave"] / (new_df["full"] + min_denom)
plot_fields = [
"full",
"delta",
"leave",
"delta_change_rate",
"leave_change_rate",
"delta_prop",
"leave_prop",
]
main_metrics = [
"full",
"delta",
"leave",
]
anomaly_point_info = {}
for metric in main_metrics:
if method == "decompose":
shifted = len(new_df[metric]) - len(new_df[metric].dropna())
result = seasonal_decompose(
new_df[metric].dropna(),
model="additive",
period=window_size,
two_sided=False,
)
new_df[metric + "_trend"] = result.trend
new_df[metric + "_seasonal"] = result.seasonal
new_df[metric + "_random"] = new_df[metric] - result.trend - result.seasonal
"""
f, ax = plt.subplots(4, 1)
f.set_figwidth(20)
f.set_figheight(6)
f.tight_layout()
Graph for trend
"""
figure(figsize=(16, 1), dpi=80)
plt.plot(np.arange(len(new_df)), new_df[metric + "_trend"])
plt.title(metric + "_trend")
plt.xlim(left=-1.0, right=len(new_df) + 1)
plt.show()
"""
Graph for seasonal
"""
figure(figsize=(16, 1), dpi=80)
plt.plot(np.arange(len(new_df)), new_df[metric + "_seasonal"])
plt.title(metric + "_seasonal")
plt.xlim(left=-1.0, right=len(new_df) + 1)
plt.show()
"""
End of graph
"""
new_df[metric + "_upper"] = (
new_df[metric + "_trend"]
+ new_df[metric + "_seasonal"]
+ sigma_mul * new_df[metric + "_random"].rolling(window_size).std()
)
new_df[metric + "_lower"] = (
new_df[metric + "_trend"]
+ new_df[metric + "_seasonal"]
- sigma_mul * new_df[metric + "_random"].rolling(window_size).std()
)
new_df[metric + "_lower"] = new_df[metric + "_lower"]
new_df[metric + "_upper"] = new_df[metric + "_upper"]
elif method == "median":
new_df[metric + "_upper"] = (
new_df[metric].rolling(rolling).median()
+ sigma_mul * new_df[metric].rolling(rolling).std()
)
new_df[metric + "_lower"] = (
new_df[metric].rolling(rolling).median()
- sigma_mul * new_df[metric].rolling(rolling).std()
)
elif method == "mean":
new_df[metric + "_upper"] = (
new_df[metric].rolling(rolling).mean()
+ sigma_mul * new_df[metric].rolling(rolling).std()
)
new_df[metric + "_lower"] = (
new_df[metric].rolling(rolling).mean()
- sigma_mul * new_df[metric].rolling(rolling).std()
)
for metric in main_metrics:
for i in range(rolling, len(new_df)):
if (
new_df[metric].iloc[i] < new_df[metric + "_lower"].shift().iloc[i]
or new_df[metric].iloc[i] > new_df[metric + "_upper"].shift().iloc[i]
):
if i not in anomaly_point_info.keys():
anomaly_point_info[i] = []
anomaly_point_info[i].append(
"Value of {} is out of bound. {} not in [{}, {}]. Last value: {}".format(
metric,
new_df[metric].iloc[i],
new_df[metric + "_lower"].shift().iloc[i],
new_df[metric + "_upper"].shift().iloc[i],
new_df[metric].iloc[i - 1],
)
)
for metric in main_metrics:
figure(figsize=(16, 1), dpi=80)
plt.plot(np.arange(len(new_df)), new_df[metric])
plt.fill_between(
np.arange(len(new_df)),
new_df[metric + "_upper"].shift(),
new_df[metric + "_lower"].shift(),
color="#C9C9C9",
)
plt.title(metric)
plt.xlim(left=-1.0, right=len(new_df) + 1)
for key in anomaly_point_info.keys():
plt.axvspan(key - 0.45, key + 0.45, color="red", alpha=0.25)
plt.show()
return anomaly_point_info
visualize_custom(
pd.read_csv(os.path.join(data_path, "New_Query_15649.csv")),
sigma_mul=4,
window_size=7,
rolling=7,
method="median",
shift_amount=1,
)
visualize_custom(
pd.read_csv(os.path.join(data_path, "New_Query_12904.csv")),
sigma_mul=4,
window_size=7,
rolling=7,
method="median",
shift_amount=1,
)
visualize_custom(
pd.read_csv(os.path.join(data_path, "New_Query_10678.csv")),
sigma_mul=4,
window_size=7,
method="decompose",
shift_amount=1,
)
|
# Khai báo thư viện
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
sns.set()
data = pd.read_csv(
"../input/pima-indians-diabetes-database/diabetes.csv"
) # reading the data
data.head()
data.describe()
# Biểu diễn dữ liệu
plt.figure(figsize=(20, 25))
plotnumber = 1
for column in data:
if plotnumber <= 9:
ax = plt.subplot(3, 3, plotnumber)
sns.distplot(data[column])
plt.xlabel(column, fontsize=15)
plotnumber += 1
plt.show()
# Một vài cột có giá trị bằng 0. Điều này không bình thường.
# Thay thế các giá trị bằng 0 bằng giá trị trung bình của cột tương ứng.
data["BMI"] = data["BMI"].replace(0, data["BMI"].mean())
data["BloodPressure"] = data["BloodPressure"].replace(0, data["BloodPressure"].mean())
data["Glucose"] = data["Glucose"].replace(0, data["Glucose"].mean())
data["Insulin"] = data["Insulin"].replace(0, data["Insulin"].mean())
data["SkinThickness"] = data["SkinThickness"].replace(0, data["SkinThickness"].mean())
# Biểu diễn lần nữa
plt.figure(figsize=(20, 25))
plotnumber = 1
for column in data:
if plotnumber <= 9:
ax = plt.subplot(3, 3, plotnumber)
sns.distplot(data[column])
plt.xlabel(column, fontsize=15)
plotnumber += 1
plt.show()
# Bây giờ chúng ta đã xử lý các giá trị bằng 0 và dữ liệu trông tốt hơn. Tuy nhiên, vẫn có sự xuất hiện của các điểm dữ liệu ngoại lệ (outliers) trong một số cột.
fig, ax = plt.subplots(figsize=(15, 10))
sns.boxplot(data=data, width=0.5, ax=ax, fliersize=3)
plt.show()
# Xóa dữ liệu ngoại lệ
outlier = data["Pregnancies"].quantile(0.98)
# Xóa bỏ 2% dữ liệu lớn nhất từ cột số lượng thai sản.
data = data[data["Pregnancies"] < outlier]
outlier = data["BMI"].quantile(0.99)
# Xóa bỏ 1% dữ liệu lớn nhất từ cột BMI
data = data[data["BMI"] < outlier]
outlier = data["SkinThickness"].quantile(0.99)
# Xóa bỏ 1% dữ liệu lớn nhất từ cột SkinThickness
data = data[data["SkinThickness"] < outlier]
outlier = data["Insulin"].quantile(0.95)
# Xóa bỏ 5% dữ liệu lớn nhất từ cột Insulin
data = data[data["Insulin"] < outlier]
outlier = data["DiabetesPedigreeFunction"].quantile(0.99)
# Xóa bỏ 1% dữ liệu lớn nhất từ cột DiabetesPedigreeFunction
data = data[data["DiabetesPedigreeFunction"] < outlier]
outlier = data["Age"].quantile(0.99)
# Xóa bỏ 1% dữ liệu lớn nhất từ cột Age
data = data[data["Age"] < outlier]
# Biểu diễn lần nữa
plt.figure(figsize=(20, 25))
plotnumber = 1
for column in data:
if plotnumber <= 9:
ax = plt.subplot(3, 3, plotnumber)
sns.distplot(data[column])
plt.xlabel(column, fontsize=15)
plotnumber += 1
plt.show()
plt.figure(figsize=(16, 8))
corr = data.corr()
mask = np.triu(np.ones_like(corr, dtype=bool))
sns.heatmap(corr, mask=mask, annot=True, fmt=".2g", linewidths=1)
plt.show()
|
# # Επιβλεπόμενη Μάθηση: Παλινδρόμηση
# #### Ονοματεπώνυμα:
# Ηλία Παναγιώτης, Κόρσακ Σεβαστιανός
# # Dataset: Auto MPG
# #### Ιστορικά στοιχεία:
# Αυτό το dataset πάρθηκε απο το StatLib library το οποίο βρίσκεται στο Carnegie Mellon University. Το dataset χρησιμοποιήθηκε το 1983 στην έκθεση της Αμερικανικής Στατιστικής Εταιρείας (American Statistical Association). Επίσης χρησιμοποιήθηκε το 1993 απο τον Ross Quinlan ο οποίος συνδύασε Instance-Based και Model-Based Learning, καθώς και σε διαδικασίες για το 10ο Διεθνές Συνέδριο Μηχανικής Μάθησης από τον Morgan Kaufmann στο Πανεπιστήμιο της Μασαχουσέτης στο Amherst.
# #### Περιγραφή:
# To dataset που θα χρησιμοποιήσουμε εμέις είναι μια ελαφρώς τροποποιημένη έκδοση του dataset του StatLib library. Στόχος μας είναι να προβλέψουμε την τιμή του χαρακτηριστικού "mpg". Οκτώ απο τα δείγματα του αρχικού dataset αφαιρέθηκαν επειδή οι τιμές τους για την ετικέτα "mpg" δεν υπήρχαν(αυτό έκανε και ο Ross Quinlan to 1993).
# Τα δεδομένα αφορούν την κατανάλωση καυσίμου στην πόλη σε μίλια ανά γαλόνι(miles per gallon-mpg), την τιμή του οποίου θέλουμε να προβλέψουμε από 3 κατηγορικά και 4 συνεχή χαρακτηριστικά. Το dataset αποτελείται από 398 δείγματα, τις ετικέτες/τιμές τους, και 8 χαρακτηριστικά, τα οποία είναι τα εξης:
# 1. mpg: συνεχής (ετικέτα/τιμή)
# 2. cylinders: κατηγορική
# 3. displacement: συνεχής
# 4. horsepower: συνεχής
# 5. weight: συνεχής
# 6. acceleration: συνεχής
# 7. model year: κατηγορική
# 8. origin: κατηγορική
# 9. car name: string (μοναδικό για κάθε δείγμα)
# ### Προεπεξεργασία των δεδομένων
# Αρχικά εισάγουμε τις βασικές βιβλιοθήκες που θα χρησιμοποιήσουμε.
import pandas as pd
import scipy as sc
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
# Διαβάζουμε το dataset μας, και βλέπουμε την μορφή του.
dataset = pd.read_csv("autompg.csv", header=None)
col_names = [
"mpg",
"cylinders",
"displacement",
"horsepower",
"weight",
"acceleration",
"model year",
"origin",
"car name",
]
dataset.columns = col_names
dataset
# Παρατηρούμε ότι όπως είπαμε τα δεδομένα μας έχουν διαστάσεις $398 \times 9$ όπως αναφέραμε και πιο πάνω. Επίσης πριν ξεκινήσουμε την ανάλυση μας καλό θα ήταν να δούμε τους τύπους των δεδομένων της κάθε στήλης.
print(dataset.dtypes)
# Παρατηρούμε ότι όλες οι στήλες έχουν τύπους δεδομένων που συμφωνούν με το τι βλέπουμε στο πίνακα πιο πάνω, εκτός από την στήλη με το χαρακτηριστικό horsepower (υπποδύναμη) του οποίου τα δεδομένα είναι αριθμητικά, όμως έχει σαν τύπο δεδομένων το object. Πάμε λοιπόν να δούμε τι συμβαίνει εδώ.
dataset["horsepower"].unique()
# Παρατηρούμε ότι έχουμε τον χαρακτήρα '?' ανάμεσα στις μοναδικές αναπαραστάσεις της στήλης του horsepower. Συνεπώς έχουμε missing values σε αυτό το χαρακτηριστικό, οι οποίες έχουν αυτή την αναπαράσταση στο dataset μας, και για αυτό τον λόγο η στήλη 'horsepower' έχει ως τύπο δεδομένων το object.
dataset.replace("?", np.NaN, inplace=True)
dataset["horsepower"] = dataset["horsepower"].astype("float")
print(dataset.dtypes)
dataset["horsepower"].isnull().sum()
# Αντικαταστήσαμε λοιπόν τον χαρακτήρα '?' με NaN και κάναμε την στήλη του 'horsepower' να έχει τύπο δεδομένων float. Με αυτό τον τρόπο βλέπουμε ότι στο dataset μας 6 δείγματα έχουν missing value στο χαρακτηριστικό 'horsepower'. \
# \
# Υπάρχουν διάφοροι τρόποι να χειριστούμε τα missing values. Μπορούμε να αντικαταστήσουμε τα missing values π.χ με το median ή το mean της στήλης. Έμεις απλά θα αγνοήσουμε πλήρως τα 6 δείγματα που περιέχουν τα missing values.
dataset = dataset.dropna(axis=0)
print(np.shape(dataset))
# Πριν προχωρήσουμε ας ελέγξουμε αν υπάρχουν missing values σε άλλα χαρακτηριστικά.
dataset.isnull().sum()
# Όπως βλέπουμε παραπάνω δεν υπάρχουν άλλα missing values. Τώρα είμαστε σε θέση να προχωρήσουμε παρακάτω. Πάμε λοιπόν να δούμε κάποια βασικά στατιστικά για το κάθε αριθμητικό χαρακτηριστικό.
dataset.describe()
# Όπως είπαμε παραπάνω, στόχος μας είναι να κάνουμε ακριβής προβλέψεις για την τιμή του mpg. Ας δούμε ξεχωριστά λοιπόν τα στατιστικά αυτής της στήλης.
dataset["mpg"].describe()
# Παρατηρούμε ότι το mpg έχει μέση τιμή 23.44 και τυπική απόκλιση 7.80. Επίσης παίρνει τιμές στο διάστημα $[9,46.6]$.
sns.histplot(dataset["mpg"])
# Από το παραπάνω ιστόγραμμα παρατηρούμε ότι η κατανομή του mpg έχει ουρά προς τα δεξία (skewed to the right), δηλαδή έχουμε περισσότερα αυτοκίνητα με μικρό mpg από ότι με μεγάλο.
# Ας δούμε και τα ιστογράμματα των κατανομών των υπόλοιπων χαρακτηριστικών.
sns.histplot(dataset["cylinders"])
sns.histplot(dataset["displacement"])
sns.histplot(dataset["horsepower"])
sns.histplot(dataset["weight"])
sns.histplot(dataset["acceleration"])
sns.histplot(dataset["model year"])
sns.histplot(dataset["origin"])
# Από τα παραπάνω ιστογράμματα παρατηρούμε ότι το χαρακτηριστικό 'cylinders' είναι διακριτό και παίρνει τις τιμές 3 και 5 σπάνια και συνήθως τις τιμες 4,6 και 8. Το χαρακτηριστικό 'dispacement' είναι 'skewed to the right', όπως και τα χαρακτηριστικά 'horsepower' και 'weight'. Το χαρακτηριστικό 'acceleration' ακολουθεί μια κανονική κατανομή. Τα χαρακτηριστικά 'model year' και 'origin' είναι κατηγορικά. Το χαρακτηριστικό 'model year' παίρνει διακριτές ακέραιες τιμές από το 70 μέχρι το 82 που αντιπροσωπεύουν την χρονολογία κατασκευής του κάθε δείγματος (αυτοκινήτου), ενώ το χαρακτηριστικό 'origin' παίρνει τις κατηγορικές τιμές 1,2 και 3 που αντιπροσωπεύουν την Ήπειρο στην οποία κατασκευάστηκε το κάθε δείγμα. Το 1 αντιστοιχεί στην Αμερική, το 2 στην Ευρώπη και το 3 στην Ασία. Αυτό το χαρακτηριστικό είναι μη διατεταγμένο, συνεπώς θα το μετατρέψουμε σε 3 binary χαρακτηριστικά από τα οποία μόνο ένα είναι ενεργό κάθε φορά, αν παραμείνει μέσα στο μοντέλο μας μετά την επιλογή χαρακτηριστικών που θα κάνουμε στην συνέχεια.
# Πάμε τώρα να δούμε πως συσχετίζεται η ετικέτα/τιμή με τα υπόλοιπα χαρακτηριστικά με την χρήση ενός πίνακα συσχέτισης.
corrmatrix = dataset.corr()
sns.heatmap(corrmatrix, square=True)
# Από τον παραπάνω πίνακα συαχέτισης παρατηρούμε ότι τα χαρακτηριστικά 'cylinders', 'displacement', 'horsepower' και 'weight' έχουν μεγάλη σε απόλυτη τιμή αρνητική συσχέτιση με την 'mpg'. Τα υπόλοιπα χαρακτηριστικά έχουν μέτρια συσχέτιση με την 'mpg'.
# ## Επιλογή Χαρακτηριστικών
# #### Recursive Feature Elimination
# Για να κάνουμε επιλογή χαρακτηριστικών θα χρησιμοποιήσουμε την μέθοδο περιτύλιξης (Wrapper Method) Recursive Feature Elimination
from sklearn.feature_selection import RFE
from sklearn.svm import SVR
features_list = [
"cylinders",
"displacement",
"horsepower",
"weight",
"acceleration",
"model year",
"origin",
]
X = dataset[features_list]
y = dataset["mpg"]
est = SVR(kernel="linear")
rfe = RFE(est, n_features_to_select=5, step=1)
fit = rfe.fit(X, y)
print("Num Features: %s" % (fit.n_features_))
print("Selected Features: %s" % (fit.support_))
print("Feature Ranking: %s" % (fit.ranking_))
dataset = pd.get_dummies(dataset, columns=["origin"])
dataset = dataset.drop(["car name"], axis=1)
col_names = dataset.columns
dataset
# Παραπάνω βλέπουμε την μορφή που πήρε το dataset μας μετά απο αυτόν τον μετασχηματισμό, και την αφαίρεση του χαρακτηριστικού 'car name' το οποίο δεν μας πρόσφερει κάτι ουσιαστικό στην ανάλυση μας. Τα χαρακτηριστικά ΄displacement΄ και ΄weight΄, τα οποία βάση της επιλογής χαρακτηριστικών που κάναμε είναι τα πιο ασθενή, δεν θα τα αφαιρέσουμε τελικά από το μοντέλο μας γιατί κάνοντας δοκιμές χωρίς αυτά παίρναμε χειρότερα αποτελέσματα από αυτά που παίρνουμε με αυτά στο μοντέλο μας
# ## Εκπαίδευση Μοντέλων
# Πριν ξεκινήσουμε την εκπαίδευση των μοντέλων μας πρέπει να χωρίσουμε το dataset μας σε training και test sets, όπως επίσης και να ορίσουμε μια συνάρτηση η οποία θα μας βοηθήσει να παρουσιάζουμε πιο ωραία τις μετρικές απόδοσης του κάθε μοντέλου.
from sklearn.preprocessing import StandardScaler
from sklearn.linear_model import LinearRegression
from sklearn.linear_model import Lasso
from sklearn.linear_model import ElasticNet
from sklearn import neighbors
from sklearn.neural_network import MLPRegressor
from sklearn.experimental import enable_hist_gradient_boosting
from sklearn.ensemble import ExtraTreesRegressor
from sklearn.ensemble import HistGradientBoostingRegressor
from sklearn.model_selection import train_test_split
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import mean_squared_error, r2_score
from sklearn.decomposition import PCA
from imblearn.pipeline import Pipeline
features_list = [
"cylinders",
"displacement",
"horsepower",
"weight",
"acceleration",
"model year",
"origin_1",
"origin_2",
"origin_3",
]
X = dataset[features_list]
y = dataset["mpg"]
# split into train and test sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=7)
print("Train", X_train.shape, y_train.shape)
print("Test", X_test.shape, y_test.shape)
# αρχικοποιούμε τους μετασχηματιστές χωρίς υπερ-παραμέτρους
scaler = StandardScaler()
pca = PCA()
n_components = [1, 2, 3, 4, 5, 6, 7, 8, 9]
def compute_metrics(model, x_tr, y_tr, x_ts, y_ts):
train_predict = model.predict(x_tr)
test_predict = model.predict(x_ts)
# Υπολογισμός Ορθότητας (Accuracy)
train_accuracy = r2_score(y_tr, train_predict)
test_accuracy = r2_score(y_ts, test_predict)
# Υπολογισμός Μέσου Τετραγωνικού Σφάλματος
train_MSE = mean_squared_error(y_tr, train_predict)
test_MSE = mean_squared_error(y_ts, test_predict)
print("Ορθότητα στο σύνολο δεδομένων εκπαίδευσης: {:.2%}".format(train_accuracy))
print("Ορθότητα στο σύνολο δεδομένων ελέγχου: {:.2%}\n".format(test_accuracy))
print("Μέσο Τετραγωνικό Σφάλμα στα δείγματα εκπαίδευσης: {:.4f}".format(train_MSE))
print("Μέσο Τετραγωνικό Σφάλμα στα δείγματα ελέγχου: {:.4f}".format(test_MSE))
# #### Γραμμική παλινδρόμηση
lr = LinearRegression()
pipe_lr = Pipeline(
steps=[("scaler", scaler), ("pca", pca), ("linear regressor", lr)], memory="tmp"
)
estimator_lr = GridSearchCV(
pipe_lr,
dict(
pca__n_components=n_components,
),
cv=5,
scoring="r2",
n_jobs=-1,
)
import time
start_time = time.time()
estimator_lr.fit(X_train, y_train)
print("Γραμμικό Μοντέλο:\n")
print("Πρόβλεψη της mpg:\n")
compute_metrics(estimator_lr, X_train, y_train, X_test, y_test)
print("Συνολικός χρόνος fit και predict: %s seconds" % (time.time() - start_time))
print(estimator_lr.best_estimator_)
print(estimator_lr.best_params_)
# #### Παλινδρόμηση Lasso
lasso = Lasso()
alphas = np.arange(0.01, 1, 0.01)
pipe_lasso = Pipeline(
steps=[("scaler", scaler), ("pca", pca), ("lasso", lasso)], memory="tmp"
)
estimator_lasso = GridSearchCV(
pipe_lasso,
dict(pca__n_components=n_components, lasso__alpha=alphas),
cv=5,
scoring="r2",
n_jobs=-1,
)
import time
start_time = time.time()
estimator_lasso.fit(X_train, y_train)
print("Γραμμικό Μοντέλο:\n")
print("Πρόβλεψη της mpg:\n")
compute_metrics(estimator_lasso, X_train, y_train, X_test, y_test)
print("Συνολικός χρόνος fit και predict: %s seconds" % (time.time() - start_time))
print(estimator_lasso.best_estimator_)
print(estimator_lasso.best_params_)
alphas_prog = np.arange(0.001, 0.02, 0.001)
estimator_lasso_prog = GridSearchCV(
pipe_lasso,
dict(pca__n_components=n_components, lasso__alpha=alphas_prog),
cv=5,
scoring="r2",
n_jobs=-1,
)
import time
start_time = time.time()
estimator_lasso_prog.fit(X_train, y_train)
print("Γραμμικό Μοντέλο:\n")
print("Πρόβλεψη της mpg:\n")
compute_metrics(estimator_lasso_prog, X_train, y_train, X_test, y_test)
print("Συνολικός χρόνος fit και predict: %s seconds" % (time.time() - start_time))
print(estimator_lasso_prog.best_estimator_)
print(estimator_lasso_prog.best_params_)
# #### Elastic-Net
en = ElasticNet()
l1 = np.arange(0.01, 1, 0.01)
pipe_en = Pipeline(
steps=[("scaler", scaler), ("pca", pca), ("ElasticNet", en)], memory="tmp"
)
estimator_en = GridSearchCV(
pipe_en,
dict(
pca__n_components=n_components,
ElasticNet__alpha=alphas_prog,
ElasticNet__l1_ratio=l1,
),
cv=5,
scoring="r2",
n_jobs=-1,
)
import time
start_time = time.time()
estimator_en.fit(X_train, y_train)
print("Γραμμικό Μοντέλο:\n")
print("Πρόβλεψη της mpg:\n")
compute_metrics(estimator_en, X_train, y_train, X_test, y_test)
print("Συνολικός χρόνος fit και predict: %s seconds" % (time.time() - start_time))
print(estimator_en.best_estimator_)
print(estimator_en.best_params_)
# #### Nearest Neighbors Regression
kNN = neighbors.KNeighborsRegressor()
k = [3, 5, 7, 9]
weights = ["uniform", "distance"]
pipe_kNN = Pipeline(
steps=[("scaler", scaler), ("pca", pca), ("kNN", kNN)], memory="tmp"
)
estimator_kNN = GridSearchCV(
pipe_kNN,
dict(pca__n_components=n_components, kNN__n_neighbors=k, kNN__weights=weights),
cv=5,
scoring="r2",
n_jobs=-1,
)
import time
start_time = time.time()
estimator_kNN.fit(X_train, y_train)
print("Γραμμικό Μοντέλο:\n")
print("Πρόβλεψη της mpg:\n")
compute_metrics(estimator_kNN, X_train, y_train, X_test, y_test)
print("Συνολικός χρόνος fit και predict: %s seconds" % (time.time() - start_time))
print(estimator_kNN.best_estimator_)
print(estimator_kNN.best_params_)
# #### Multi-layer Perceptron Regression
MLP = MLPRegressor(random_state=7, max_iter=2000)
hidden_layer_size = np.arange(50, 500, 50)
learning_rate_init = np.arange(0.001, 0.01, 0.001)
pipe_MLP = Pipeline(
steps=[("scaler", scaler), ("pca", pca), ("MLP", MLP)], memory="tmp"
)
estimator_MLP = GridSearchCV(
pipe_MLP,
dict(
pca__n_components=n_components,
MLP__hidden_layer_sizes=hidden_layer_size,
MLP__learning_rate_init=learning_rate_init,
),
cv=5,
scoring="r2",
n_jobs=-1,
)
import time
start_time = time.time()
estimator_MLP.fit(X_train, y_train)
print("Γραμμικό Μοντέλο:\n")
print("Πρόβλεψη της mpg:\n")
compute_metrics(estimator_MLP, X_train, y_train, X_test, y_test)
print("Συνολικός χρόνος fit και predict: %s seconds" % (time.time() - start_time))
print(estimator_MLP.best_estimator_)
print(estimator_MLP.best_params_)
# #### ExtraTreesRegressor
etr = ExtraTreesRegressor(random_state=7)
n_estimators = np.arange(10, 200, 10)
pipe_etr = Pipeline(
steps=[("scaler", scaler), ("pca", pca), ("etr", etr)], memory="tmp"
)
estimator_etr = GridSearchCV(
pipe_etr,
dict(pca__n_components=n_components, etr__n_estimators=n_estimators),
cv=5,
scoring="r2",
n_jobs=-1,
)
import time
start_time = time.time()
estimator_etr.fit(X_train, y_train)
print("Γραμμικό Μοντέλο:\n")
print("Πρόβλεψη της mpg:\n")
compute_metrics(estimator_etr, X_train, y_train, X_test, y_test)
print("Συνολικός χρόνος fit και predict: %s seconds" % (time.time() - start_time))
print(estimator_etr.best_estimator_)
print(estimator_etr.best_params_)
# #### HistGradientBoostingRegressor
hgbr = HistGradientBoostingRegressor(random_state=7)
loss = ["least_squares", "least_absolute_deviation", "poisson"]
learning_rate = np.arange(0.1, 1, 0.1)
max_leaf_nodes = np.arange(5, 50, 5)
pipe_hgbr = Pipeline(
steps=[("scaler", scaler), ("pca", pca), ("hgbr", hgbr)], memory="tmp"
)
estimator_hgbr = GridSearchCV(
pipe_hgbr,
dict(
pca__n_components=n_components,
hgbr__loss=loss,
hgbr__max_leaf_nodes=max_leaf_nodes,
hgbr__learning_rate=learning_rate,
),
cv=5,
scoring="r2",
n_jobs=-1,
)
import time
start_time = time.time()
estimator_hgbr.fit(X_train, y_train)
print("Γραμμικό Μοντέλο:\n")
print("Πρόβλεψη της mpg:\n")
compute_metrics(estimator_hgbr, X_train, y_train, X_test, y_test)
print("Συνολικός χρόνος fit και predict: %s seconds" % (time.time() - start_time))
print(estimator_hgbr.best_estimator_)
print(estimator_hgbr.best_params_)
learning_rate_prog = np.arange(0.01, 0.11, 0.01)
max_leaf_nodes_prog = np.arange(2, 9, 1)
estimator_hgbr_prog = GridSearchCV(
pipe_hgbr,
dict(
pca__n_components=n_components,
hgbr__loss=loss,
hgbr__max_leaf_nodes=max_leaf_nodes_prog,
hgbr__learning_rate=learning_rate_prog,
),
cv=5,
scoring="r2",
n_jobs=-1,
)
import time
start_time = time.time()
estimator_hgbr_prog.fit(X_train, y_train)
print("Γραμμικό Μοντέλο:\n")
print("Πρόβλεψη της mpg:\n")
compute_metrics(estimator_hgbr_prog, X_train, y_train, X_test, y_test)
print("Συνολικός χρόνος fit και predict: %s seconds" % (time.time() - start_time))
print(estimator_hgbr_prog.best_estimator_)
print(estimator_hgbr_prog.best_params_)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Standard Deviation:
import numpy
speed = [86, 87, 88, 86, 87, 85, 86]
x = numpy.std(speed)
print(x)
# Data distribution:
import numpy
x = numpy.random.uniform(0.0, 5.0, 250)
print(x)
import numpy
import matplotlib.pyplot as plt
x = numpy.random.uniform(0.0, 5.0, 250)
plt.hist(x, 5)
plt.show()
# Normal data distribution:
import numpy
import matplotlib.pyplot as plt
x = numpy.random.normal(5.0, 1.0, 100000)
plt.hist(x, 100)
plt.show()
|
import pandas as pd
import numpy as np
import matplotlib as mpl
import matplotlib.pyplot as plt
import seaborn as sns
import os
plt.style.use("ggplot")
pd.set_option("display.max_columns", 100)
os.chdir("/kaggle/input/airbnb-cleaned-europe-dataset")
df = pd.read_csv("Aemf1.csv")
df
df.info()
# # EDA
# Heat map
# optimizng range for color scale
min = df.corr().min().min()
max = df.corr()[df.corr() != 1].max().max()
# thresholding selected correlations
df_corr = df.corr()[np.absolute(df.corr()) > 0.3]
# Mask for selecting only bottom triangle
mask = np.triu(df_corr)
with plt.style.context("default"):
sns.heatmap(df_corr, vmin=min, vmax=max, mask=mask)
# raw data
sns.pairplot(
df[
[
"City Center (km)",
"Metro Distance (km)",
"Attraction Index",
"Restraunt Index",
"Price",
]
][df["Price"] < 2000],
kind="hist",
corner=True,
)
# rescaled data
df_trial = pd.DataFrame()
df_trial["City Center (km)"] = np.log(df["City Center (km)"])
df_trial["Metro Distance (km)"] = np.log(df["Metro Distance (km)"])
df_trial["Attraction Index"] = np.log(df["Attraction Index"])
df_trial["Restraunt Index"] = np.log(df["Restraunt Index"])
df_trial["Price"] = np.log(df["Price"])
sns.pairplot(df_trial[df_trial["Price"] < 2000], kind="hist", corner=True)
fig, ax = plt.subplots(1)
fig.set_size_inches(8, 4)
sns.boxplot(data=df, x="City", y="Price", showfliers=False)
ax.set_ylim([0, 1300])
plt.show()
sns.boxplot(data=df, x="Superhost", y="Price", showfliers=False)
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(8, 3))
plt.sca(ax1)
sns.boxplot(data=df, x="Room Type", y="Price", showfliers=False)
plt.sca(ax2)
sns.boxplot(data=df, x="Shared Room", y="Price", showfliers=False)
plt.tight_layout()
sns.boxplot(data=df, x="Day", y="Price", showfliers=False)
sns.regplot(
data=df[df["Price"] < 2000],
x="Cleanliness Rating",
y="Price",
scatter=True,
scatter_kws={"alpha": 0.05},
line_kws={"color": "black"},
)
sns.regplot(
data=df[df["Price"] < 2000],
x="Guest Satisfaction",
y="Price",
scatter_kws={"alpha": 0.05},
line_kws={"color": "black"},
)
plt.show()
sns.boxplot(
data=df, x="Person Capacity", y="Price", showfliers=False
) # ,scatter_kws={'alpha':0.05},line_kws={"color": "black"})
plt.ylim([0, 1000])
plt.show()
# # Cleaning Data
from sklearn.compose import ColumnTransformer
from sklearn.preprocessing import FunctionTransformer
from sklearn.preprocessing import OneHotEncoder
from sklearn.preprocessing import StandardScaler, MinMaxScaler
cat_encoder = OneHotEncoder()
df = df.drop(columns=["Normalised Attraction Index", "Normalised Restraunt Index"])
names = df.columns.values
names = names.remove("Day", "Room Type", "City")
names = names.append(
"Day_0",
"Day_1",
"Type_0",
"Type_1",
"Type_2",
"City_0",
"City_1",
"City_2",
"City_3",
"City_4",
"City_5",
"City_6",
"City_7",
"City_8",
)
preprocessing = ColumnTransformer(
[
(
"log",
FunctionTransformer(np.log),
[
"City Center (km)",
"Metro Distance (km)",
"Attraction Index",
"Restraunt Index",
"Price",
],
),
("cat", cat_encoder, ["Day", "Room Type", "City"]),
(
"standardScale",
StandardScaler(),
[
"City Center (km)",
"Metro Distance (km)",
"Attraction Index",
"Restraunt Index",
"Guest Satisfaction",
],
),
(
"minmaxScale",
MinMaxScaler(),
["Person Capacity", "Cleanliness Rating", "Bedrooms"],
),
]
)
df_prepared = preprocessing.fit_transform(df)
pd.DataFrame(df_prepared).describe()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv(
"/kaggle/input/credit-risk-dataset/credit_risk_dataset.csv", encoding="latin"
)
df.info()
# ## Checking Memory Usage for each column
df.memory_usage(deep=True)
# ## We could see person_home_ownership, loan_intent, loan_grade , cb_person_default_on_file is categorical column but it seems that its holding more memory.
# # Confirming categorical column date : If its appropriate to convert it into category
# ## If categorical columns is either
# Binary (Yes / No)
# Nominal ( Groups with no rank or order between them )
# Ordinal Groups that are ranked in a specific order.
# ## It will be better if the column type is category. Let's explore and confirm.
cat_cols = [col for col in df.columns if df[col].dtype == "O"]
cat_cols
for col in cat_cols:
print(df[col].value_counts())
print("-------------")
# ## Code above confirms that columns of type object hoilding correct cateogircal data.
# ## It will be better to convert it into category type and then we will compare the memory consumption, if there is any gain
# # Convert categorical column data type to Cateogry
for col in cat_cols:
df[col] = df[col].astype("category")
df.memory_usage(deep=True)
# # Comparing memory consumption
# ## Memory Consumption when data type is Object
#
#
# Column Name
# Object Memory Consumption
# Category Memory Consumption
#
#
#
#
# person_home_ownership
# 2038740
# 33001
#
#
# loan_intent
# 2184571
# 33158
#
#
# loan_grade
# 1889698
# 33287
#
#
# cb_person_default_on_file
# 1889698
# 32805
#
#
# ## As we could rightly see there is a good optimization
# # Best way to Define Column Types
# ## Once we have a confirmation, It should be better if we define data type at the time of reading dataframe itself
#
df = pd.read_csv(
"/kaggle/input/credit-risk-dataset/credit_risk_dataset.csv",
dtype={
"person_home_ownership": "category",
"loan_intent": "category",
"loan_grade": "category",
"cb_person_default_on_file": "category",
},
encoding="latin",
)
df.memory_usage(deep=True)
|
# # **IMPORTING LIBRARIES**
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import seaborn as sns
from sklearn.preprocessing import OneHotEncoder
from sklearn.compose import ColumnTransformer
from sklearn.preprocessing import LabelEncoder
from sklearn.model_selection import train_test_split
from sklearn.impute import KNNImputer
from sklearn.linear_model import LogisticRegression
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.neighbors import KNeighborsClassifier
from sklearn.svm import SVC
from sklearn.metrics import confusion_matrix, accuracy_score
# # **IMPORTING DATASET**
dataset = pd.read_csv(
"../input/stroke-prediction-dataset/healthcare-dataset-stroke-data.csv"
)
dataset
dataset.describe()
dataset.info()
# # **Checking for missing value**
dataset.isnull().sum()
# # **DEALING WITH CATEGORICAL DATA**
dataset = pd.get_dummies(data=dataset, drop_first=True)
# # **SPLITTING DATA INTO X AND Y**
y = dataset.iloc[:, 6].values
dataset = dataset.drop(["stroke"], axis=1)
x = dataset.iloc[:, 1:].values
# # **DEALING WITH MISSING VALUE**
imputer = KNNImputer(missing_values=np.nan, n_neighbors=3)
x = imputer.fit_transform(x)
# # **SPLITTING DATA INTO TRAINING AND TEST SET**
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.20, random_state=1
)
# # **TESTING LOGISTIC REGRESSION MODEL**
lr = LogisticRegression()
lr.fit(x_train, y_train)
y_pred_lr = lr.predict(x_test)
# # **TESTING DECISION TREE CLASSIFIER**
dtc = DecisionTreeClassifier(criterion="gini", max_depth=10, random_state=1)
dtc.fit(x_train, y_train)
y_pred_dtc = dtc.predict(x_test)
# # **TESTING RANDOM FOREST CLASSIFIER**
rfc = RandomForestClassifier(
n_estimators=10, criterion="gini", max_depth=10, random_state=1
)
rfc.fit(x_train, y_train)
y_pred_rfc = rfc.predict(x_test)
# # **TESTING SUPPORT VECTOR CLASSIFIER**
svc = SVC(random_state=1)
svc.fit(x_train, y_train)
y_pred_svc = svc.predict(x_test)
# # **TESTING K NEAREST NEIGHBORS**
knn = KNeighborsClassifier(n_neighbors=5, metric="minkowski")
knn.fit(x_train, y_train)
y_pred_knn = knn.predict(x_test)
# # **CHECKING THE ACCURACY OF DIFFERENT MODELS**
# **LOGISTIC REGRESSION**
cm_lr = confusion_matrix(y_test, y_pred_lr)
print(cm_lr)
accuracy_score(y_test, y_pred_lr)
# **DECISION TREE CLASSIFIER**
cm_dtc = confusion_matrix(y_test, y_pred_dtc)
print(cm_dtc)
accuracy_score(y_test, y_pred_dtc)
# **RANDOM FOREST CLASSIFIER**
cm_rfc = confusion_matrix(y_test, y_pred_rfc)
print(cm_rfc)
accuracy_score(y_test, y_pred_rfc)
# **SUPPORT VECTOR CLASSIFIER**
cm_svc = confusion_matrix(y_test, y_pred_svc)
print(cm_svc)
accuracy_score(y_test, y_pred_svc)
# **K NEAREST NEIGHBORS**
cm_knn = confusion_matrix(y_test, y_pred_knn)
print(cm_knn)
accuracy_score(y_test, y_pred_knn)
|
import numpy as np
import pandas as pd
import cv2 as cv
import matplotlib.pyplot as plt
from matplotlib.colors import Colormap
import seaborn as sns
import os
from sklearn.utils import shuffle
from sklearn.model_selection import train_test_split
# Color palette
cmap = ["#fcfbf9", "#fed766", "#009fb7", "#0e6eb5", "#2f3982"]
# ## Data Processing
# ### Training Data
# Set path of the train directory
training_dir = "/kaggle/input/brain-tumor-mri-dataset/Training/"
# Each folder contains 4 more folders. Every folder has images of
# the corresponding folder's name (i.e. label of image is name of folder)
train_paths = []
train_labels = []
for label in os.listdir(training_dir):
for image in os.listdir(training_dir + label):
train_paths.append(training_dir + label + "/" + image)
train_labels.append(label)
# Now we can shuffle data because our train_paths and train_labels
# have similar type of images in sequence
train_paths, train_labels = shuffle(train_paths, train_labels)
# Collect data for visualization
df = pd.DataFrame({"Image": train_paths, "Category": train_labels})
cat = df["Category"].value_counts().index.tolist()
cat = [i.title() for i in cat]
count = df["Category"].value_counts().tolist()
# Create a function to customize autopct parameter of plt.pie()
def make_autopct(values):
def my_autopct(pct):
# The pct is percentage value that matplotlib supplies for every wedge
total = sum(values)
val = int(round(pct * total / 100.0))
return f"{pct:.2f}% ({val})"
return my_autopct
# Let's visualize the distribution of instances in our training data
sns.set(style="whitegrid")
plt.figure(figsize=(6, 8))
plt.pie(count, labels=cat, shadow=True, autopct=make_autopct(count), colors=cmap[1:5])
plt.title("Types of Tumour in Train set")
plt.show()
# Split our training data for training and validation as X_train and X_val
# along with labels as y_train, y_val
X_train, X_val, y_train, y_val = train_test_split(
train_paths, train_labels, test_size=0.2, train_size=0.8
)
# Know the number of instances in our split datsets
print(f"Instances in X_train: {len(X_train)}")
print(f"Instances in X_val: {len(X_val)}")
print(f"Instances in y_train: {len(y_train)}")
print(f"Instances in y_val: {len(y_val)}")
# ### Test Data
# Set path of test directory
test_dir = "/kaggle/input/brain-tumor-mri-dataset/Testing/"
# Each folder contains 4 more folders. Every folder has images of
# the corresponding folder's name (i.e. label of image is name of folder)
test_paths = []
test_labels = []
for label in os.listdir(test_dir):
for image in os.listdir(test_dir + label):
test_paths.append(test_dir + label + "/" + image)
test_labels.append(label)
# Now we can shuffle data because our test_paths and test_labels
# have similar type of images in sequence
train_paths, train_labels = shuffle(train_paths, train_labels)
# Collect data for visualization
df_test = pd.DataFrame({"Image": test_paths, "Category": test_labels})
cat_test = df_test["Category"].value_counts().index.tolist()
cat_test = [i.title() for i in cat_test]
count_test = df_test["Category"].value_counts().tolist()
# Let's visualize the distribution of instances in our training data
sns.set(style="whitegrid")
plt.figure(figsize=(6, 8))
plt.pie(
count_test,
labels=cat_test,
shadow=True,
autopct=make_autopct(count_test),
colors=cmap[:4],
)
plt.title("Types of Tumour in Test set")
plt.show()
# ## View Images in Dataset
def plot_images(n_rows, n_cols, paths_img, paths_label):
"""
Plots a figure containing all images in paths_img
along with paths_label
Parameters:
n_rows: number of rows to plot in figure
n_cols: number of cols to plot in figure
paths_img: a list of path of image
paths_label: a list of labels for image plotted
(consistent with paths_img)
"""
plt.figure(figsize=(n_cols * 2.25, n_rows * 2.25))
for row in range(n_rows):
for col in range(n_cols):
index = n_cols * row + col
img = cv.imread(paths_img[index])
plt.subplot(n_rows, n_cols, index + 1)
plt.imshow(img, cmap="binary", interpolation="nearest")
plt.axis("off")
plt.title(paths_label[index].title(), fontsize=12, pad=8)
plt.subplots_adjust(wspace=0.25, hspace=0.25)
plt.show()
# View images from our training set
plot_images(3, 5, X_train, y_train)
|
# ## A Quick First Analysis
# To start, let's have a look at the most lines delivered by each charater in Deep Space Nine.
# > From the processed dataset, generate counts of all lines spoken by each charater.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
all_series_lines = pd.read_json("../input/start-trek-scripts/all_series_lines.json")
episodes = all_series_lines["TNG"].keys()
total_line_count = 0
dirty_line_count = 0
for i, ep in enumerate(episodes):
if all_series_lines["TNG"][ep] is not np.NaN:
for member in list(all_series_lines["TNG"][ep].keys()):
for line in all_series_lines["TNG"][ep][member]:
total_line_count += 1
if line is not np.NaN and len(line) > 0 and line[-1].isupper():
dirty_line_count += 1
print(f"Total lines: {total_line_count}")
print(f"Dirty lines: {dirty_line_count}")
print(f"Percent dirty: {round((dirty_line_count / total_line_count)*100, 3)}%")
# # Processing the data
# ## Import raw data.
all_scripts_raw = pd.read_json("../input/start-trek-scripts/all_scripts_raw.json")
episodes = all_scripts_raw["TNG"].keys()
series_lines = all_scripts_raw["TNG"]
# ## Clean data.
import re
def clean_episode(episode_text):
# Remove text at the beginning of each episode
cleaned = re.sub(
"(The Next Generation Transcripts(.|\n)*?)([A-Z]+:+)", r"\3", episode_text
)
# cleaned = re.sub("((\n)*?)([A-Z]+:+)", r"\3", cleaned)
# Remove text at the end of each episode
cleaned = re.sub("<Back(.|\n)*", "", cleaned)
# Remove instances of " [OC]", denoting when a character is speaking off-camera.
cleaned = cleaned.replace(" [OC]", "")
# Remove any descriptions of the scene, which are surrounded by [] and () parentheses.
cleaned = re.sub(" ?[\(\[](.|\n)*?[\)\]]", "", cleaned)
# Remove non-breaking space characters
cleaned = cleaned.replace("\u00a0", "")
# Remove multiple instances of '\n'. Could be cleaner but I'm tired right now.
cleaned = cleaned.replace("\n\n", "\n")
cleaned = cleaned.replace("\n \n", "\n")
cleaned = cleaned.replace("\n\n", "\n")
cleaned = cleaned.replace("\n\n", "\n")
# Remove multiple instances of ':'.
cleaned = cleaned.replace("::", ":")
# Remove lines starting with "Captain's log", since they do not include data on which character spoke them.
cleaned = re.sub(
"((Captain's log|Ship's log|First [Oo]fficer's log)(.|\n)*?)([A-Z]+:)",
r"\4",
cleaned,
)
# Remove newlines that are in the middle of a spoken line
cleaned = re.sub("(?<! )\n(?!([A-Z]*?:))", " ", cleaned)
return cleaned
cleaned_episode_example = clean_episode(series_lines[1])
print("★★★★★ ORIGINAL ★★★★★")
print(series_lines[1][0:850])
print("\n★★★★★ CLEANED ★★★★★")
print(cleaned_episode_two[0:301])
for i, ep in enumerate(episodes):
series_lines[i] = clean_episode(series_lines[i])
# ## Group lines by character.
def group_by_character(episode_text):
lines_by_character = {}
split_lines = episode_text.split("\n")
for line in split_lines:
name = re.search("([A-Z]+)(?=:+)", line)
words = re.search("(?<=:)(.*)", line)
if name is not None:
name = name.group(0).strip()
words = words.group(0).strip()
if name in lines_by_character.keys():
lines_by_character[name].append(words)
else:
lines_by_character[name] = [words]
return lines_by_character
series_lines_by_character = []
for i, ep in enumerate(episodes):
series_lines_by_character.append(group_by_character(series_lines[i]))
total_word_counts = {}
total_line_counts = {}
for i, ep in enumerate(episodes):
if series_lines_by_character[i] is not np.NaN:
for member in list(series_lines_by_character[i].keys()):
total_words_by_member_in_ep = sum(
[len(line.split()) for line in series_lines_by_character[i][member]]
)
total_lines_by_member_in_ep = len(series_lines_by_character[i][member])
if member in total_word_counts.keys():
total_word_counts[member] = (
total_word_counts[member] + total_words_by_member_in_ep
)
total_line_counts[member] = (
total_line_counts[member] + total_lines_by_member_in_ep
)
else:
total_word_counts[member] = total_words_by_member_in_ep
total_line_counts[member] = total_lines_by_member_in_ep
words_df = pd.DataFrame(
list(total_word_counts.items()), columns=["Character", "No. of Words"]
)
most_words = words_df.sort_values(by="No. of Words", ascending=False).head(25)
most_words.plot.bar(x="Character", y="No. of Words")
plt.show()
lines_df = pd.DataFrame(
list(total_line_counts.items()), columns=["Character", "No. of Lines"]
)
most_lines = lines_df.sort_values(by="No. of Lines", ascending=False).head(25)
most_lines.plot.bar(x="Character", y="No. of Lines")
plt.show()
|
# **Car Price Prediction using Linear Regression**
# Linear Regression is a statistical method to model a linear relationship between input variables (X) and output variables (Y). Mathematically, linear regression can be described using the line equation:
# 𝑌=𝑎+𝑏𝑋
#
# where:
# 𝑌 is the output variable (target),
# 𝑋 is the input variable (feature),
# 𝑎 is the intercept (constant)
# and 𝑏 is the regression coefficient (gradient).
# **1. First we need to import the library**
import warnings
warnings.filterwarnings("ignore")
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
# **2. then we need to read the dataset**
df = pd.read_csv("/kaggle/input/car-price-prediction/CarPrice_Assignment.csv")
df.head()
# **3. finding the shape of our dataset**
df.shape
# **4. get some information about our dataset**
df.info()
# **5. checking for null values using missingno module**
df.isna().sum()
# Now we can draw following conclusion about our dataset.
# Our dataset total 26 feature. Amoung them 25 are independent and 1(Price) is dependent feature.
# Our dataset contains total of 256 rows
# Dataset contains both categorical and numerical feature
# Dataset has no missing values.
# **6. looking at descriptive statistic parameters for the dataset**
df.describe()
# **7. working with numerical features by Creating a list to store all numerical variable**
numerical_feature = [
feature for feature in df.columns if df[feature].dtypes not in ["O", "object"]
]
print("Number of Numerical Variable ", len(numerical_feature))
df[numerical_feature].head()
# 8. gathering information about our target feature
plt.figure(figsize=(20, 8))
plt.subplot(1, 2, 1)
plt.title("Car Price Distribution Plot")
sns.distplot(df["price"])
plt.subplot(1, 2, 2)
plt.title("Car Price Spread")
sns.boxplot(y=df["price"])
plt.show()
# Inference :
# The plot seemed to be right-skewed, meaning that the most prices in the dataset are low(Below 15,000).
# There is a significant difference between the mean and the median of the price distribution.
# The data points are far spread out from the mean, which indicates a high variance in the car prices.
# (85% of the prices are below 18,500, whereas the remaining 15% are between 18,500 and 45,400.)
# **9. finding the relationship between independent and dependent numerical features**
for feature in numerical_feature:
if feature not in ["car_ID"]:
plt.scatter(y=df["price"], x=df[feature])
plt.ylabel("Car Price")
plt.xlabel(feature)
plt.show()
# **9. finding the relationship between each feature with others.**
# We will now look at the correlation between them and plot with the help of a heatmap.
plt.figure(figsize=(18, 7))
sns.heatmap(
df[numerical_feature].corr(),
annot=True,
fmt="0.2f",
annot_kws={"size": 15},
linewidth=2,
linecolor="orange",
)
plt.show()
# correlation of numerical variables
# wheelbase have positive correlation with price of 58%.
# car length and car width have positive correlation with price of 68% and 76%.
# curbweight have positive correlation with price of 84%.
# enginesize have positive correlation with price of 87%.
# boreratio have positive correlation with price of 55%.
# horsepower have positive correlation with price of 81%.
# citympg and highwaympg have negative correlation with price of 69% and 70%.
# **10. finding the distribution for each of these numerical features and also check for outliers.**
index = 1
for feature in numerical_feature:
if feature not in ["car_ID"]:
plt.figure(figsize=(30, 8))
# first plot
plt.subplot(index, 2, 1)
plt.title(feature + "Distribution Plot")
sns.distplot(df[feature])
# second plot
plt.subplot(index, 2, 2)
plt.title(feature + "Box Plot")
sns.boxplot(y=df[feature])
plt.show()
index = index + 1
# **11. working with categorical values, by Creating a list to store all Categorical variable**
categorical_feature = [
feature for feature in df.columns if df[feature].dtypes in ["O", "object"]
]
print("Number of Categorical Variable ", len(categorical_feature))
df[categorical_feature].head()
# **12. Get counter plots for our categorical features**
plt.figure(figsize=(16, 20))
plotnumber = 1
for i in range(1, len(categorical_feature)):
if plotnumber <= 10:
ax = plt.subplot(5, 2, plotnumber)
sns.countplot(x=categorical_feature[i], data=df, ax=ax)
plt.title(f"\n{categorical_feature[i]} \n", fontsize=20)
plotnumber += 1
plt.tight_layout()
plt.show()
# **13. Box plot for each categorical features**
plt.figure(figsize=(16, 25))
plotnumber = 1
for i in range(1, len(categorical_feature)):
if plotnumber <= 10:
ax = plt.subplot(5, 2, plotnumber)
sns.boxplot(x=categorical_feature[i], y=df["price"], data=df, ax=ax)
plt.title(f"\n{categorical_feature[i]} \n", fontsize=20)
plotnumber += 1
plt.tight_layout()
plt.show()
# convertible, sedan and hardtop cars are costliers than others.
# hardtop cars are very less manufactured but their price is also higher.
# very few cars have engine at rear but if they have their price is higher.
# ohc is the most type of engine used in cars and ohcv is used only in costly cars.
# mostly cars have 4 no of cylinders.
# if a car have higher no of cylinders then its price is also higher.
# mpfi and 2bbl are the most commonly fuel system used in cars.
# idi and spdi type of fuel system is used only in costly cars.
# **14. Univeriant Analysis**
categorical_feature
df["CarName"].count()
df["CarName"].unique()
print(df["CarName"].value_counts())
# **15. extracting the car company name and use it instead of car name, by Splitting company name from CarName column**
CompanyName = df["CarName"].apply(lambda x: x.split(" ")[0])
df.insert(3, "CompanyName", CompanyName)
df.drop(["CarName"], axis=1, inplace=True)
df.head()
# **16. Fixing invalid values There seems to be some spelling error in the CompanyName column.**
# maxda = mazda
# Nissan = nissan
# porsche = porcshce
# toyota = toyouta
# vokswagen = volkswagen = vw
df["CompanyName"] = df["CompanyName"].str.lower()
def replace_name(a, b):
df["CompanyName"].replace(a, b, inplace=True)
replace_name("maxda", "mazda")
replace_name("porcshce", "porsche")
replace_name("toyouta", "toyota")
replace_name("vokswagen", "volkswagen")
replace_name("vw", "volkswagen")
df["CompanyName"].unique()
# **17. Checking for duplicates**
df.loc[df.duplicated()]
plt.figure(figsize=(25, 6))
plt.subplot(1, 3, 1)
plt1 = df["CompanyName"].value_counts().plot(kind="bar")
plt.title("Car Company Histogram")
plt1.set(xlabel="Car", ylabel="Frequency of car")
plt.subplot(1, 3, 2)
plt1 = df["fueltype"].value_counts().plot(kind="bar")
plt.title("Fuel Type Histogram")
plt1.set(xlabel="Fuel Type", ylabel="Frequency of fuel type")
plt.subplot(1, 3, 3)
plt1 = df["carbody"].value_counts().plot(kind="bar")
plt.title("Car Type Histogram")
plt1.set(xlabel="Car Type", ylabel="Frequency of Car type")
plt.show()
# Inference :
# Toyota seemed to be favored car company.
# mercury is the less used car.
# gas fueled cars are more than diesel.
# sedan is the top car type prefered.
# **18. analysing other features**
cat_columns = [
"aspiration",
"doornumber",
"drivewheel",
"enginelocation",
"enginetype",
"cylindernumber",
"fuelsystem",
]
for feature in cat_columns:
plt.figure(figsize=(20, 8))
plt.subplot(1, 2, 1)
plt.title(feature.title() + "Histogram")
sns.countplot(df[feature], palette=("Blues_d"))
plt.subplot(1, 2, 2)
plt.title(feature.title() + "vs Price")
sns.boxplot(x=df[feature], y=df["price"], palette=("PuBuGn"))
plt.show()
sns.pairplot(df)
plt.show()
# **19. Data Preprocessing**
df.head()
# **20. Dropping the index from the dataset**
df.drop(columns=["car_ID"], axis=1, inplace=True)
# **21. encoding ordinal categorical columns**
df["doornumber"] = df["doornumber"].map({"two": 2, "four": 4})
df["cylindernumber"] = df["cylindernumber"].map(
{"two": 2, "three": 3, "four": 4, "five": 5, "six": 6, "eight": 8, "twelve": 12}
)
# **22. creating features and label variable**
X = df.drop(columns="price", axis=1)
y = df["price"]
# **23. encoding categorical columns**
X = pd.get_dummies(X, drop_first=True)
X.head()
X.shape
# **24. checking for multicollinearity using `VIF` and `correlation matrix`**
from statsmodels.stats.outliers_influence import variance_inflation_factor
vif = pd.DataFrame()
vif["VIF"] = [variance_inflation_factor(X, i) for i in range(X.shape[1])]
vif["Features"] = X.columns
vif
# **25. Dropping curbweight because of high VIF value. (shows that curbweight has high multicollinearity.)**
# The higher the VIF, the higher the possibility that multicollinearity exists, and further research is required. When VIF is higher than 10, there is significant multicollinearity that needs to be corrected.
X = X.drop(["CompanyName_subaru", "enginelocation_rear", "enginetype_ohcf"], axis=1)
# **26. Splitting data into traing and testing set**
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=42
)
print("Traing Data Shape of x and y respectively: ", X_train.shape, y_train.shape)
print("Testing Data Shape of x and y respectively: ", X_test.shape, y_test.shape)
# **27. Model Building**
from sklearn.linear_model import LinearRegression
lr_model = LinearRegression()
lr_model.fit(X_train, y_train)
y_pred = lr_model.predict(X_test)
# **28. Model Evaluation**
from sklearn.metrics import mean_squared_error
from sklearn.metrics import r2_score
mse = mean_squared_error(y_pred, y_test)
r2_score = r2_score(y_pred, y_test)
lr_model.score(X_test, y_test)
mse
rmse = np.sqrt(mse)
rmse
r2_score
# **Car Price Prediction using Multiple Linear Regression**
# Multiple Linear Regression is a statistical method used to model the relationship between two or more input variables (X) and output variables (Y) which are linear. In multiple linear regression, the output variable (Y) is considered to be influenced by several input variables (X) with different regression coefficients for each input variable.
# **1. First we need to import the library**
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn import preprocessing
from sklearn.model_selection import train_test_split
from sklearn.metrics import r2_score
from sklearn.metrics import mean_absolute_error, mean_squared_error
from sklearn.ensemble import RandomForestRegressor
import math
carprice_estimated = pd.read_csv(
"/kaggle/input/car-price-prediction/CarPrice_Assignment.csv"
)
carprice_estimated.head()
carprice_estimated.tail()
carprice_estimated.shape
carprice_estimated.columns
carprice_estimated.info()
carprice_estimated.isnull().sum()
carprice_estimated.describe().T
carprice_estimated = carprice_estimated.drop(["car_ID", "symboling"], axis=1)
corr = carprice_estimated.corr()
corr
plt.figure(figsize=(15, 5))
sns.heatmap(corr, annot=True)
sns.scatterplot(x="carlength", y="wheelbase", data=carprice_estimated)
sns.scatterplot(x="carwidth", y="wheelbase", data=carprice_estimated)
sns.scatterplot(x="curbweight", y="carlength", data=carprice_estimated)
sns.scatterplot(x="carlength", y="carwidth", data=carprice_estimated)
sns.scatterplot(x="carlength", y="carwidth", hue="price", data=carprice_estimated)
sns.scatterplot(x="price", y="enginesize", data=carprice_estimated)
carprice_estimated["CarName"].unique()
carprice_estimated["CarName"] = carprice_estimated["CarName"].str.split(
" ", expand=True
)[0]
carprice_estimated["CarName"].unique()
carprice_estimated["CarName"] = carprice_estimated["CarName"].replace(
{
"maxda": "mazda",
"nissan": "Nissan",
"porcshce": "porsche",
"toyouta": "toyota",
"vokswagen": "volkswagen",
"vw": "volkswagen",
}
)
carprice_estimated["CarName"].nunique()
carprice_estimated["CarName"].value_counts()
plt.figure(figsize=(10, 10))
ax = sns.countplot(x=carprice_estimated["CarName"])
ax.bar_label(ax.containers[0])
plt.xticks(rotation=90)
plt.figure(figsize=(6, 6))
sns.distplot(carprice_estimated["price"], hist=True)
carprice_estimated["fueltype"].value_counts()
plt.figure(figsize=(3, 3))
sns.countplot(x="fueltype", data=carprice_estimated)
plt.figure(figsize=(3, 3))
colors = ["blue", "yellow"]
carprice_estimated.fueltype.value_counts().plot.pie(autopct="%1.1f%%", colors=colors)
plt.show()
carprice_estimated["doornumber"].value_counts()
plt.figure(figsize=(3, 3))
sns.countplot(x="doornumber", data=carprice_estimated)
plt.figure(figsize=(3, 3))
colors = ["red", "yellow"]
carprice_estimated.doornumber.value_counts().plot.pie(autopct="%1.1f%%", colors=colors)
plt.show()
sns.pairplot(carprice_estimated)
plt.figure(figsize=(25, 5))
sns.countplot(x="horsepower", data=carprice_estimated)
plt.figure(figsize=(20, 5))
sns.countplot(x="peakrpm", data=carprice_estimated)
label_encoder = preprocessing.LabelEncoder()
carprice_estimated["fueltype"] = label_encoder.fit_transform(
carprice_estimated["fueltype"]
)
carprice_estimated["doornumber"] = label_encoder.fit_transform(
carprice_estimated["doornumber"]
)
carprice_estimated["aspiration"] = label_encoder.fit_transform(
carprice_estimated["aspiration"]
)
carprice_estimated["enginelocation"] = label_encoder.fit_transform(
carprice_estimated["enginelocation"]
)
carprice_estimated["drivewheel"] = label_encoder.fit_transform(
carprice_estimated["drivewheel"]
)
carprice_estimated["CarName"] = pd.Categorical(carprice_estimated["CarName"])
dfDummiess = pd.get_dummies(carprice_estimated["CarName"], prefix="CarName")
dfDummiess
carprice_estimated = pd.concat([carprice_estimated, dfDummiess], axis=1)
carprice_estimated = carprice_estimated.drop(["CarName"], axis=1)
carprice_estimated["enginetype"] = pd.Categorical(carprice_estimated["enginetype"])
dfDummiess = pd.get_dummies(carprice_estimated["enginetype"], prefix="enginetype")
dfDummiess
carprice_estimated = pd.concat([carprice_estimated, dfDummiess], axis=1)
carprice_estimated = carprice_estimated.drop(["enginetype"], axis=1)
carprice_estimated["cylindernumber"] = pd.Categorical(
carprice_estimated["cylindernumber"]
)
dfDummiess = pd.get_dummies(
carprice_estimated["cylindernumber"], prefix="cylindernumber"
)
dfDummiess
carprice_estimated = pd.concat([carprice_estimated, dfDummiess], axis=1)
carprice_estimated = carprice_estimated.drop(["cylindernumber"], axis=1)
carprice_estimated["carbody"] = pd.Categorical(carprice_estimated["carbody"])
dfDummiess = pd.get_dummies(carprice_estimated["carbody"], prefix="carbody")
dfDummiess
carprice_estimated = pd.concat([carprice_estimated, dfDummiess], axis=1)
carprice_estimated = carprice_estimated.drop(["carbody"], axis=1)
carprice_estimated["fuelsystem"] = pd.Categorical(carprice_estimated["fuelsystem"])
dfDummiess = pd.get_dummies(carprice_estimated["fuelsystem"], prefix="fuelsystem")
dfDummiess
carprice_estimated = pd.concat([carprice_estimated, dfDummiess], axis=1)
carprice_estimated = carprice_estimated.drop(["fuelsystem"], axis=1)
carprice_estimated.head()
x = carprice_estimated.drop(["price"], axis=1)
y = carprice_estimated["price"]
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.2, random_state=41
)
from sklearn.ensemble import RandomForestRegressor
rf_reg = RandomForestRegressor(n_estimators=10, random_state=0)
model = rf_reg.fit(x_train, y_train)
y_pred = rf_reg.predict(x_test)
print("R2 score:", r2_score(y_test, y_pred))
MAE = mean_absolute_error(y_test, y_pred)
print("Mean Absolute Error:", MAE)
MSE = mean_squared_error(y_test, y_pred)
print("Mean Squared Error:", MSE)
RMSE = math.sqrt(MSE)
print("Root Mean Square Error:", RMSE)
|
# ## **IMGW Hydro & Meteo Archive - automatic download of archived data**
# ### **Introductory information**
# The notebook contains a script for automatically downloading archival hydrological and meteorological data provided by the Institute of Meteorology and Water Management - National Research Institute (IMGW-PIB). This entity provides meteorological and hydrological services on the territory of the Republic of Poland. Before working with this script, read the full information on the source of the data and the rules for its use:
# * https://danepubliczne.imgw.pl/datastore
# * https://www.kaggle.com/datasets/krystianadammolenda/imgw-hydro-and-meteo-archive-poland
# 
# > Source: https://www.nasa.gov/mission_pages/noaa-n/climate/climate_weather.html
# ## **Here we go!**
import requests, zipfile
import pandas as pd
import numpy as np
from io import BytesIO
# ---
# ### **Declaration of variables**
# In the `select_year` and `select_month` variables, set the appropriate time range for which you want to retrieve data.
# In the variables `select_categories` and `select_id_datatype` declare the type of data you want to retrieve.
select_years = ["2020", "2021"]
select_month = ["07", "08", "09"]
select_categories = ["Hydro", "Meteo"]
select_id_datatype = ["B00020S", "B00604S"]
# Meaning of 'Hydro' parameter codes:
# `B00020S` Water level (operational)
# `B00050S` Water flow rate (operational)
# `B00014A` Water level (observer)
# `B00101A` Water temperature (observer)
# Meaning of 'Meteo' parameter codes:
# `B00300S` Air temperature (official)
# `B00305A` Ground temperature (sensor)
# `B00202A` Wind direction (sensor)
# `B00702A` Average wind speed (sensor)
# `B00703A` Maximum speed (sensor)
# `B00608S` Rainfall total 10 minute sensor
# `B00604S` Daily precipitation total
# `B00606S` Hourly precipitation total
# `B00802A` Relative humidity (sensor)
# `B00714A` Largest wind gust in a 10-minute period from a synoptic station
# `B00910A` Stock of water in snow (observer)
# ### **Data download script**
# The script is written in such a way that it is not necessary to manually download each archive from https://danepubliczne.imgw.pl/datastore.
# The data will be stored under a variable that is the ID of the parameter.
for id_datatype in select_id_datatype:
globals()[id_datatype] = pd.DataFrame()
for category in select_categories:
for year in select_years:
for month in select_month:
print(f"\nSearching resources: {category}-{year}-{month}:")
try:
url = f"https://danepubliczne.imgw.pl/datastore/getfiledown/Arch/Telemetria/{category}/{year}/{category}_{year}-{month}.zip"
request = requests.get(url)
archive = zipfile.ZipFile(BytesIO(request.content))
except:
url = f"https://danepubliczne.imgw.pl/datastore/getfiledown/Arch/Telemetria/{category}/{year}/{category}_{year}-{month}.ZIP"
request = requests.get(url)
archive = zipfile.ZipFile(BytesIO(request.content))
for file in archive.namelist():
for id_datatype in select_id_datatype:
if file.split("_")[0] == id_datatype:
print(f" -- File found: {file}")
df = pd.read_csv(
archive.open(file),
sep=";",
decimal=",",
header=None,
low_memory=False,
usecols=[0, 1, 2, 3],
)
globals()[id_datatype] = pd.concat(
[globals()[id_datatype], df], ignore_index=True, sort=False
)
archive.close()
for id_datatype in select_id_datatype:
globals()[id_datatype].columns = [
"Measurement point ID",
"Measurement parameter ID",
"Measurement time stamp",
"Measured value",
]
print(f"\nDownload completed!")
print(f"Access to the data is under the variables: {select_id_datatype}")
# ### **Preview of downloaded data**
B00020S
B00020S.to_csv("Examples_of_data_downloaded_by_the_script_1.csv")
B00604S
B00604S.to_csv("Examples_of_data_downloaded_by_the_script_2.csv")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Financial Prediction in Retail with LSTM
# In this project, I will be performing financial income prediction / forecasting project with using the time series approach on the customer's records from a retail industry firm's database. Financial prediction is the practice of time series approach with single or multiple time series input. I will also add more notebooks about financial prediction with different algorithms and approaches. One notebook already published for the seasonality test ( https://www.kaggle.com/code/shedai/sales-volume-seasonality ) with a single time series approach, more notebooks are coming.
# - Single Time Series Approach: Sales volume is one of the mostly predicted feature in the time series. In this approach, project aims to create a time series from daily, weekly or monthly sales data. Most of the time, there is only one time series with 2 features : time (dates) and sales volume.
# - Multiple Parameter Time Series Approach: This type of approaches have multiple inputs, where all inputs are temporal (time dependent series) or we do feature engineering to create multiple inputs for the prediction model.
# For this notebook, we will be using the Long-Short Term Memory (LSTM) technique from Keras / Tensorflow library. ( you can check the wikipedia link for further details : https://en.wikipedia.org/wiki/Long_short-term_memory
# Note: You can also check a very similar notebook about CatBoost prediction, I have created a few days ago: https://www.kaggle.com/code/shedai/financial-prediction-in-retail-with-catboost/edit
# Another notebook for the same dataset and problem with Fast Fourier Transform (FFT) approach is available: https://www.kaggle.com/code/shedai/sales-prediction-with-fft
#
# TABLE OF CONTENTS
#
# * [1. IMPORTING LIBRARIES](#1)
#
# * [2. LOADING DATA](#2)
#
# * [3. Creating Time Series](#3)
#
# * [4. Data Visualization and Analysis](#4)
#
# * [5. Prediction / Forecasting with FFT](#5)
#
# * [6. Evaluation](#6)
#
# * [7. Conclusion](#7)
#
# * [8. END](#8)
# # 1. Importing Libraries
import warnings
warnings.filterwarnings("ignore")
import logging
logging.disable(logging.CRITICAL)
#
# # 2. Loading Data
#
# Now, we can load the data set and convert it to time series. The conversion requires the grouping dataset with the sum of sales for each day and type conversion to datetime format in pandas.
df = pd.read_csv("/kaggle/input/retail-data-set/file_out2.csv")
df = df.groupby("Date").sum()
df["Date"] = df.index
df = df[["Date", "TotalSales"]]
df["Date"] = pd.to_datetime(df["Date"], format="%Y-%m-%d")
df.head()
#
# # 3. Creating Time Series
# Now we will try to use the forecasting with LSTM. LSTM gets multiple parameters and one of the crucial point is, we need to normalize the data. Also the Keras library has a special shape requirement for the input layer of the neural network. We are starting to convert the data for the most suitable form for the LSTM input layer.
# Lets start with the target column first:
data = df[["TotalSales"]]
# Convert the dataframe to a numpy array
dataset = data.values
training_data_len = int(np.ceil(len(dataset) * 0.95))
dataset
# Now, we can normalize the data with min-max normalizer (scaler) from sklearn framework.
# Scale the data
from sklearn.preprocessing import MinMaxScaler
scaler = MinMaxScaler(feature_range=(0, 1))
scaled_data = scaler.fit_transform(dataset)
scaled_data
# Finally we split the train and test (validation) data sets and reshape the data set for LSTM input layer:
# Create the training data set
# Create the scaled training data set
train_data = scaled_data[0 : int(training_data_len), :]
# Split the data into x_train and y_train data sets
x_train = []
y_train = []
# we use 60 days ahead for forecasting range
for i in range(60, len(train_data)):
x_train.append(train_data[i - 60 : i, 0])
y_train.append(train_data[i, 0])
if i <= 61:
print(x_train)
print(y_train)
print()
# Convert the x_train and y_train to numpy arrays
x_train, y_train = np.array(x_train), np.array(y_train)
# Reshape the data
x_train = np.reshape(x_train, (x_train.shape[0], x_train.shape[1], 1))
# x_train.shape
#
# # 4. Data Visualization
# We can simply visualize the data set in time series with dataframe plot function.
data.plot()
#
# # 5. Prediction and Forecasting with LSTM
# Step by step, we will create neural network (deep learning) model with 1 input and model will have a shape like below:
# input(60) x LSTM(128) x LSTM(64) x Dense(25) x Output(1)
# the time series will run with 1 input and 1 output as the forecast. The LSTM and Dense layers will try to model the time series.
from keras.models import Sequential
from keras.layers import Dense, LSTM
# Build the LSTM model
model = Sequential()
model.add(LSTM(128, return_sequences=True, input_shape=(x_train.shape[1], 1)))
model.add(LSTM(64, return_sequences=False))
model.add(Dense(25))
model.add(Dense(1))
# Compile the model
model.compile(optimizer="adam", loss="mean_squared_error")
# Train the model
model.fit(x_train, y_train, batch_size=1, epochs=1)
# We can now create a testing scenario and evaluate the success of the model:
# Create the testing data set
# Create a new array containing scaled values from index 1543 to 2002
test_data = scaled_data[training_data_len - 60 :, :]
# Create the data sets x_test and y_test
x_test = []
y_test = dataset[training_data_len:, :]
for i in range(60, len(test_data)):
x_test.append(test_data[i - 60 : i, 0])
# Convert the data to a numpy array
x_test = np.array(x_test)
# Reshape the data
x_test = np.reshape(x_test, (x_test.shape[0], x_test.shape[1], 1))
# Get the models predicted price values
predictions = model.predict(x_test)
predictions = scaler.inverse_transform(predictions)
# Get the root mean squared error (RMSE)
rmse = np.sqrt(np.mean(((predictions - y_test) ** 2)))
rmse
# Finally, lets visualize the train / test sets and the prediction on test set for the visual evaluation of succes.
# # 6. Evaluation
# **What about future?** We can also use the whole data set for training and predict the future with 10% of data set date range again.
import matplotlib.pyplot as plt
# Plot the data
train = data[:training_data_len]
valid = data[training_data_len:]
valid["Predictions"] = predictions
# Visualize the data
plt.figure(figsize=(16, 6))
plt.title("Model")
plt.xlabel("Date", fontsize=18)
plt.ylabel("TotalSales", fontsize=18)
plt.plot(train["TotalSales"])
plt.plot(valid[["TotalSales", "Predictions"]])
plt.legend(["Train", "Val", "Predictions"], loc="upper left")
plt.show()
|
import pandas as pd
inpath = "/kaggle/input/fathomnet-2023-first-glance/"
train = pd.read_csv(inpath + "train_with_labels.csv")
test = pd.read_csv(inpath + "eval_images.csv")
# Find the most common values in a column categories
most_common = train["categories"].mode()[0].replace(".0", "")
print(most_common)
test["id"] = test["file_name"].str[:-4]
test["categories"] = most_common
test["osd"] = 0.1
test[["id", "categories", "osd"]]
test[["id", "categories", "osd"]].to_csv("submission.csv", index=False)
|
from transformers import ConvNextFeatureExtractor, ConvNextForImageClassification
import torch
from datasets import load_dataset
import torch
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
import numpy as np
import os
import matplotlib.pyplot as plt
from torch.utils.data import DataLoader
import torchvision
from torchvision import datasets
import torchvision.transforms as transforms
from torchview import draw_graph
from sklearn.metrics import accuracy_score, classification_report, roc_auc_score
from tqdm import tqdm_notebook as tqdm
import time
import warnings
warnings.simplefilter("ignore")
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
device
training_path = "/kaggle/input/brain-tumor-mri-images-44c/"
IMAGE_SIZE = (224, 224)
batch_size = 32
learning_rate = 1e-4
epochs = 15
def images_transforms(phase):
if phase == "training":
data_transformation = transforms.Compose(
[
transforms.Resize(IMAGE_SIZE),
transforms.RandomRotation(degrees=(-25, 20)),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225]),
]
)
else:
data_transformation = transforms.Compose(
[
transforms.Resize(IMAGE_SIZE),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225]),
]
)
return data_transformation
trainset = datasets.ImageFolder(training_path, transform=images_transforms("training"))
len(trainset)
num_classes = len(trainset.classes)
classes = trainset.classes
classes
trainset, valset = torch.utils.data.random_split(trainset, [3584, 895])
len(trainset), len(valset)
train_loader = DataLoader(trainset, batch_size=batch_size, shuffle=True)
val_loader = DataLoader(valset, batch_size=batch_size, shuffle=True)
def imshow(img):
plt.figure(figsize=(20, 20))
img = img / 2 + 0.5 # unnormalize
npimg = img.numpy()
plt.imshow(np.transpose(npimg, (1, 2, 0)))
plt.show()
examples = iter(train_loader)
images, labels = next(examples)
imshow(torchvision.utils.make_grid(images))
model = ConvNextForImageClassification.from_pretrained("facebook/convnext-tiny-224")
model = model.convnext
model = model.to(device)
model_graph = draw_graph(
model, input_size=(2, 3, 224, 224), device=device, expand_nested=True
)
model_graph.visual_graph
sample = torch.rand((2, 3, 224, 224)).to(device)
model(sample)[1].shape
class ConvNextTumorClassifier(nn.Module):
def __init__(self, base_model, base_model_output_shape, num_classes):
super().__init__()
self.base_model = base_model
self.fc = nn.Sequential(
nn.Linear(base_model_output_shape, 1024),
nn.ReLU(),
nn.Linear(1024, num_classes),
)
def forward(self, x):
x = self.base_model(x)[1]
out = self.fc(x)
return out
myModel = ConvNextTumorClassifier(model, 768, num_classes)
myModel = myModel.to(device)
criterion = nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(model.parameters(), lr=learning_rate)
model_graph = draw_graph(
myModel, input_size=(1, 3, 224, 224), device=device, expand_nested=True
)
model_graph.visual_graph.format = "png"
model_graph.visual_graph.graph_attr.update(dpi="1000")
model_graph.visual_graph.render("model_graph")
model_graph.visual_graph
def train(
model,
train_loader,
criterion,
optimizer,
val_loader,
epochs=25,
model_save_path="best_model.pt",
):
train_losses = []
val_losses = []
train_auc = []
val_auc = []
train_auc_epoch = []
val_auc_epoch = []
best_acc = 0.0
min_loss = np.Inf
since = time.time()
y_actual = []
y_pred = []
for e in range(epochs):
y_actual = []
y_pred = []
train_loss = 0.0
val_loss = 0.0
# Train the model
model.train()
for i, (images, labels) in enumerate(
tqdm(train_loader, total=int(len(train_loader)))
):
images = images.to(device).float()
labels = labels.to(device).long()
# Forward pass
outputs = model(images.to(device))
loss = criterion(outputs, labels)
# Backward and optimize
optimizer.zero_grad()
loss.backward()
optimizer.step()
# Loss and accuracy
train_loss += loss.item()
_, predictes = torch.max(outputs, 1)
y_actual += list(labels.data.cpu().numpy().flatten())
y_pred += list(predictes.detach().cpu().numpy().flatten())
train_auc.append(accuracy_score(y_actual, y_pred))
# Evaluate the model
model.eval()
for i, (images, labels) in enumerate(
tqdm(val_loader, total=int(len(val_loader)))
):
images = images.to(device).float()
labels = labels.to(device).long()
# Forward pass
outputs = model(images.to(device))
loss = criterion(outputs, labels)
# Loss and accuracy
val_loss += loss.item()
_, predictes = torch.max(outputs, 1)
y_actual += list(labels.data.cpu().numpy().flatten())
y_pred += list(predictes.detach().cpu().numpy().flatten())
val_auc.append(accuracy_score(y_actual, y_pred))
# Average losses and accuracies
train_loss = train_loss / len(train_loader)
val_loss = val_loss / len(val_loader)
train_losses.append(train_loss)
val_losses.append(val_loss)
training_auc = train_auc[-1]
validation_auc = val_auc[-1]
train_auc_epoch.append(training_auc)
val_auc_epoch.append(validation_auc)
# Updating best validation accuracy
if best_acc < validation_auc:
best_acc = validation_auc
# Saving best model
if min_loss >= val_loss:
torch.save(model.state_dict(), model_save_path)
min_loss = val_loss
print(
"EPOCH {}/{} Train loss: {:.6f},Validation loss: {:.6f}, Train AUC: {:.4f} Validation AUC: {:.4f}\n ".format(
e + 1, epochs, train_loss, val_loss, training_auc, validation_auc
)
)
print("-" * 10)
time_elapsed = time.time() - since
print(
"Training completed in {:.0f}m {:.0f}s".format(
time_elapsed // 60, time_elapsed % 60
)
)
print("Best validation accuracy: {:4f}".format(best_acc))
return train_losses, val_losses, train_auc, val_auc, train_auc_epoch, val_auc_epoch
def test(model, testloader, target_labels):
with torch.no_grad():
n_correct = 0
n_samples = 0
y_pred = []
y_actual = []
for i, (images, labels) in enumerate(testloader):
images = images.to(device)
labels = labels.to(device)
outputs = model(images)
y_actual += list(np.array(labels.detach().to("cpu")).flatten())
# value ,index
_, predictes = torch.max(outputs, 1)
y_pred += list(np.array(predictes.detach().to("cpu")).flatten())
# number of samples in current batch
n_samples += labels.shape[0]
n_correct += (predictes == labels).sum().item()
y_actual = np.array(y_actual).flatten()
y_pred = np.array(y_pred).flatten()
acc = classification_report(y_actual, y_pred, target_names=target_labels)
accuracy = accuracy_score(y_actual, y_pred)
print(f"{acc}")
print(f"Accuracy Score : {accuracy}")
train_losses, val_losses, train_auc, val_auc, train_auc_epoch, val_auc_epoch = train(
myModel,
train_loader,
criterion,
optimizer,
val_loader,
epochs,
model_save_path="tumor_model.pt",
)
test(myModel, train_loader, target_labels=classes)
test(myModel, val_loader, target_labels=classes)
plt.plot(range(1, epochs + 1), train_losses, label="Training")
plt.plot(range(1, epochs + 1), val_losses, label="Validation")
plt.title("Loss VS Epochs")
plt.xlabel("Epochs")
plt.ylabel("Loss")
plt.legend()
plt.plot(range(1, epochs + 1), train_auc, label="Training")
plt.plot(range(1, epochs + 1), val_auc, label="Validation")
plt.title("Accuracy VS Epochs")
plt.xlabel("Epochs")
plt.ylabel("Accuracy")
plt.legend()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.preprocessing import LabelEncoder
from sklearn.model_selection import train_test_split
from xgboost import XGBRegressor
from sklearn import metrics
import warnings
warnings.filterwarnings("ignore")
df = pd.read_csv("/kaggle/input/big-mart-sales/Train-Set.csv")
df
df["ProductID"].unique()
df["ProductID"].value_counts()
# number of data points and number of features
df.shape
# getting some information about the dataset
df.info()
df.isnull().sum()
# Categorical Features
# Item Identifier
# Item Fat content
# Item type
# outlet identifier
# outlet size
# outlet location type
# outlet type
# Handling Missing values ---imputation
# average value for numeric data
# mode value for
for i in df.columns:
df[i].unique()
print(i, df[i].unique())
print("--" * 50)
# Handling missing values-- imputation
# average/mean for numeric values
# mode for categorical values
# mean value of "item weight" column
df["Weight"].mean().round(2)
# filling the missing values in "item weight" column with mean values
df["Weight"].fillna(df["Weight"].mean(), inplace=True)
df.head()
df.isnull().sum()
df["Weight"].nunique()
df["OutletType"].value_counts()
df["OutletSize"].value_counts()
# Replacing the missing values in "Outlet Size" with mode
mode_of_OutletSize = df.pivot_table(
values="OutletSize", columns="OutletType", aggfunc=(lambda x: x.mode()[0])
)
print(mode_of_OutletSize)
missing_values = df["OutletSize"].isnull()
print(missing_values)
df.loc[missing_values, "OutletSize"] = df.loc[missing_values, "OutletType"].apply(
lambda x: mode_of_OutletSize
)
df["OutletSize"]
df.isnull().sum()
# Data Analysis
# statistical measures about the data
df.describe().round(2)
# Numerival features
sns.set()
# item weight distribution
plt.figure(figsize=(6, 6))
sns.distplot(df["Weight"])
plt.show()
# item Visibility distribution
plt.figure(figsize=(6, 6))
sns.distplot(df["ProductVisibility"])
plt.show()
# item MRP distribution
plt.figure(figsize=(6, 6))
sns.distplot(df["MRP"])
plt.show()
df.head(2)
# Outlet Sales distribution
plt.figure(figsize=(6, 6))
sns.distplot(df["OutletSales"])
plt.show()
# Outlet Establishment year column
plt.figure(figsize=(6, 6))
sns.countplot(x="EstablishmentYear", data=df)
plt.show()
# Ffat Content column
plt.figure(figsize=(6, 6))
sns.countplot(x="FatContent", data=df)
plt.show()
# Product Type column
plt.figure(figsize=(25, 6))
sns.countplot(x="ProductType", data=df)
plt.show()
df.head()
df["OutletSize"].value_counts()
df["OutletSize"] = df["OutletSize"].astype("category")
# Outlet Size column
plt.figure(figsize=(10, 6))
sns.countplot(x="OutletSize", data=df)
plt.show()
df.head(2)
# Outlet Establishment year column
plt.figure(figsize=(10, 6))
sns.countplot(x="OutletType", data=df)
plt.show()
# Outlet Establishment year column
plt.figure(figsize=(6, 6))
sns.countplot(x="LocationType", data=df)
plt.show()
# Data Preprocessing
df["FatContent"].value_counts()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Python Booleans - Mantıksal Operatörler
# Mantıksal operatörler iki değerden oluşur. True(doğru) - False(yanlış)
# # Boolean Values
# Programlamada genellikle bir ifadenin Doğru mu yoksa Yanlış mı olduğunu bilmeniz gerekir.
# Python'da herhangi bir ifadeyi değerlendirebilir ve True veya False olmak üzere iki yanıttan birini alabilirsiniz.
# İki değeri karşılaştırdığınızda, ifade değerlendirilir ve Python, Boole yanıtını döndürür:
# is not her iki değişken de aynı nesne değilse true döndürür
print(20 > 8)
print(20 > 8)
print(20 == 8)
# * if ifadesinde bir koşul çalıştırdığınızda, Python True veya False değerini döndürür koşulun doğru veya yanlış olmasına bağlı olarak mesaj yazdıralım
a = 50
b = 60
if b == a:
print("b ile a eşittir")
else:
print("b ile a eşit değildir")
# # Değerleri ve Değişkenleri Değerlendirme
# bool() işlevi, herhangi bir değeri değerlendirmenize ve karşılığında True veya False vermenize izin verir,
# Örneğin: Bir diziyi ve bir sayıyı değerlendirin:
print(bool("mandalina"))
print(bool(12))
# # Çoğu Değer Doğrudur
# Bir tür içeriğe sahipse, hemen hemen her değer True olarak değerlendirilir.
# Boş diziler dışında tüm diziler True'dur.
# 0 dışında herhangi bir sayı True'dur.
# Boş olanlar dışında tüm liste, demet, küme ve sözlük True'dur.
bool("gizem")
bool(987)
bool(["rachel", "joey", "phoebe"])
# # Bazı Değerler Yanlıştır
# Aslında, (), [], {}, "", 0 sayısı ve Yok değeri gibi boş değerler dışında False olarak değerlendirilen çok fazla değer yoktur. Ve elbette False değeri False olarak değerlendirilir.
# Aşağıdaki örnekler False olarak çıktı verecektir
bool(False)
bool(None)
bool(())
bool({})
bool(0)
bool("")
bool([])
# # Fonksiyonlar bir Boole Döndürebilir
# Bir Boole Değeri döndüren fonksiyonlar oluşturabilirsiniz
def myFunction():
return True
print(myFunction())
# Bir işlevin Boole yanıtına göre kod çalıştırabilirsiniz:
# Örnek "tatlı" Yazdır işlev True döndürürse, aksi takdirde "tuzlu" yazdıralım
def myFunction():
return True
if myFunction():
print("tatlı")
else:
print("tuzlu")
# Python ayrıca, bir nesnenin belirli bir veri türünde olup olmadığını belirlemek için kullanılabilen isinstance() fonksiyonu gibi bir boolean değeri döndüren birçok yerleşik işleve sahiptir:
# Örnek Bir nesnenin tamsayı olup olmadığını kontrol edin
x = 65
print(isinstance(x, int))
y = "meyve"
print(isinstance(y, str))
print(10 > 20)
print(10 == 10)
print(10 < 20)
print(bool(777))
print(bool("klon"))
# # Python Operatörleri
# Operatörler, değişkenler ve değerler üzerinde işlem yapmak için kullanılır.
# Aşağıdaki örnekte, iki değeri bir araya getirmek için + operatörünü kullanıyoruz:
print(77 + 7)
# # python operatörleri ikiye ayrılır.
# # 1 ) Python aritmetik operatörleri
# Aritmetik opetaörler, yaygın matematiksel işlemleri gerçekleştirmek için sayısal değerlerle birlikte kullanılır
# Name Example Try it
# + Addition x + y
# - Subtraction x - y
# * Multiplication x * y
# / Division x / y
# % Modulus x % y
# ** Exponentiation x ** y
# // Floor division x // y
a = 55
b = 5
print(a + b) # toplama
print(a - b) # çıkarma
print(a * b) # çarpma
print(a / b) # bölme
print(a % b) # modüler
print(a**b) # üst alma
print(a // b) # taban fonksiyonu // sonuca en yakın tamsayıya yuvarlar
# # 2) Python Atama Operatörleri
# Atama işleçleri, değişkenlere değer atamak için kullanılır:
# eşittir
x = 7
x
# artı eşittir
x = 7
x += 5
print(x)
# eksi eşittir
x = 7
x -= 5
print(x)
# çarpı eşittir
x = 7
x *= 5
print(x)
x = 7
x /= 5
print(x)
# bölümden kalan sayıyı verir
x = 7
x %= 5
print(x)
# kaç defa bölündüğü
x = 7
x //= 5
print(x)
# sayının üssü alınır
x = 7
x **= 5
print(x)
# # Python Karşılaştırma Operatörleri
# Karşılaştırma işleçleri iki değeri karşılaştırmak için kullanılır:
# == Eşit mi
x = 7
y = 17
print(x == y)
# 17, 7'ye eşit olmadığı için False döndürür
# != Eşit değil
x = 17
y = 7
print(x != y)
# 17, 7'ye eşit olmadığı için True döndürür
# > büyüktür
x = 17
y = 7
print(x > y)
# < küçüktür
x = 17
y = 7
print(x < y)
# >= Büyük eşittir
x = 17
y = 7
print(x >= y)
# küçük eşittir
x = 17
y = 7
print(x <= y)
# # Python Mantıksal Operatörler
# Mantıksal işleçler, koşullu ifadeleri birleştirmek için kullanılır: "and, or, not"
# And , her iki ifade de doğruysa true döndürür;
x = 3
print(x > 3 and x < 9)
# * Or , ifadelerden biri doğruysa true döndürür;
x = 7
print(x > 5 or x < 10)
# * Not , sonucu tersine çevirmek için kullanılır. sonuç doğruysa false döndürür
x = 9
print(not (x > 1 and x < 50))
# # Python Kimlik Operatörleri
# Kimlik fonksiyonları, nesneleri eşit olup olmadıklarını değil, aslında aynı nesne olup olmadıklarını ve aynı bellek konumuna sahip olup olmadıklarını karşılaştırmak için kullanılır
# is
# her iki değişken de aynı nesneye true döndürür
x = ["elma", "armut"]
y = ["elma", "armut"]
z = x
print(x is z)
# True değerini döndürür çünkü z, x ile aynı nesnedir
print(x is y)
# aynı içeriğe sahip olsalar bile x, y ile aynı nesne olmadığı için False döndürür
print(x == y)
# "is" ve "==" arasındaki farkı göstermek için: x eşittir y olduğu için bu karşılaştırma True değerini döndürür
# * is not her iki değişken de aynı nesne değilse true döndürür
x = ["elma", "armut"]
y = ["elma", "armut"]
z = x
print(x is not z)
print(x is not y)
print(x != y)
|
import numpy as np
import pandas as pd
import math
from sklearn.linear_model import LinearRegression
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Above shows the basics, what libraries are used and how data is pulled
# ----------------------------------------------------------------------
# Below shows the first 5 entris of the dataset
# ----------------------------------------------------------------------
data = pd.read_csv("/kaggle/input/world-happiness/2019.csv")
print(data.head())
print(data.shape)
# Below I did 3 things:
# 1. categorized the data into the input feature X and actual outcome y as training data.
# 2. fit the training set into the built-in sklearn's linear regression model.
# 3. printed the coefficient of determination metric of the model's prediction.
X = data[
[
"GDP per capita",
"Social support",
"Healthy life expectancy",
"Freedom to make life choices",
"Generosity",
"Perceptions of corruption",
]
]
y = data["Score"]
print(f"Shape of X is {X.shape}")
print(f"Shape of y is {y.shape}")
linear_model = LinearRegression()
linear_model.fit(X, y)
predict = linear_model.predict(X)
print("Prediction on training set (first 5):", predict[:5])
print("Actual target Value (first 5):", y.values[:5])
goodness = linear_model.score(X, y)
print("Coefficient of Determination:", goodness)
# After verification, Scikit-learn's LinearRegression uses the Ordinary Least Squares (OLS) method to estimate the model parameters. OLS is a closed-form solution, which means it directly computes the optimal model parameters without requiring an iterative process like gradient descent.
# Therefore, my plan from here is to learn about the meaning of some suggested metrics for sklearn's linear regression.
from sklearn.metrics import mean_squared_error, mean_absolute_error, r2_score
mse = mean_squared_error(y, predict)
rmse = np.sqrt(mse)
mae = mean_absolute_error(y, predict)
r2 = r2_score(y, predict)
print(f"Mean Squrared Error: ", mse)
print(f"Root Mean Squrared Error: ", rmse)
print(f"Mean Absolute Error: ", mae)
print(f"R-squared: ", r2)
# **Mean Squared Error**: A smaller MSE indicates a better fit. However, since the errors are squared, it can be challenging to interpret the MSE in the same units as the target variable.
# **Root Mean Squared Error**: RMSE is more interpretable than MSE and is widely used in regression problems. A smaller RMSE indicates better model performance.
# **Mean Absolute Error**: MAE is less sensitive to large errors compared to MSE or RMSE, making it more robust to outliers.
# **R-squared**: Coefficient of determination or the "goodness-of-fit" of the model
# **Summary of different metrics:**
# * High MSE or RMSE indicates the model making large errors in predictions
# * High MAE indicates the model making consistent errors across the board.
# * Comparing RMSE and MAE:
# * If RMSE is significantly GREATER than MAE, that indicates the model has a few large errors (require more outlier-robust techniques)
#
# **Review of the first 5 samples from the dataset:**
# Prediction 1: 7.00548205 | Actual: 7.769 | Error: 0.76351795
# Prediction 2: 7.09306376 | Actual: 7.6 | Error: 0.50693624
# Prediction 3: 7.17731778 | Actual: 7.554 | Error: 0.37668222
# Prediction 4: 6.94501295 | Actual: 7.494 | Error: 0.54898705
# Prediction 5: 6.92351457 | Actual: 7.488 | Error: 0.56448543
#
# **Evaluation:**
# Overall, the performance of this model seems decent, with an R-squared of 0.7792, indicating that it captures a significant portion(77.92%) of the variance in the target variable. However, there's room for improvement. The errors for each prediction vary between 0.37 and 0.76 points in the first 5 samples, but the happiness score ranges between 0-10 so it's fair to suggest that the errors are rather large in comparison and can be disruptive for meaningful interpretation of the study.
import matplotlib.pyplot as plt
plt.scatter(y, predict)
plt.xlabel("Actual score")
plt.ylabel("Predicted score")
plt.title("Prediction vs Training data")
plt.show()
# Above shows a simple scatter plot that the dots should form a straight line when x[j] = y[j] which forms a perfect fit model. It's unrealistic to make a perfect fit model however, because it would be an overfitting one. Below, I'll create multiple scatter plots for each input feature within X against the y to see if there's any nonlinearity in the relationship.
#
# by dataframe convention (m, n) where m = number of sample and n = the n th item among the input features
n = X.shape[1] # which should be 6, but I prefer not to hard code it:)
n_cols = 2
n_rows = round(n / n_cols)
# this is just to make a easy-to-read visualization
plt.figure(figsize=(10, 12))
for index, feature in enumerate(X.columns):
plt.subplot(n_rows, n_cols, index + 1)
plt.scatter(X[feature], y)
plt.xlabel(feature)
plt.ylabel("Score")
plt.title(f"Score vs {feature}")
# tight_layout() automates optimal spacing for my subplot
plt.tight_layout()
plt.show()
|
# #Implement CNN for classifying an apple's image kept in any 1 of 4 quadrants.
# Import required Libraries
import tensorflow as tf
from tensorflow.keras.preprocessing.image import ImageDataGenerator
from tensorflow.keras.applications import VGG16
from tensorflow.keras.layers import Dense, Flatten
from tensorflow.keras.models import Model
# Define Path
train_dir = "/kaggle/input/apples-in-four-quadrants-classification-cnn/custom-generated-apple-dataset/train"
test_dir = "/kaggle/input/apples-in-four-quadrants-classification-cnn/custom-generated-apple-dataset/test"
# Number of samples processed before the model is trained
batch_size = 32
# Image Size for VGG16 model
image_size = (224, 224)
# Use ImageDataGenerator to augment the training data with random transformations
train_datagen = ImageDataGenerator(
rescale=1.0 / 255,
rotation_range=20,
width_shift_range=0.2,
height_shift_range=0.2,
shear_range=0.2,
zoom_range=0.2,
fill_mode="nearest",
)
test_datagen = ImageDataGenerator(rescale=1.0 / 255)
# Load the training data and apply data augmentation
train_data = train_datagen.flow_from_directory(
train_dir, target_size=image_size, batch_size=batch_size, class_mode="categorical"
)
# Load the testing data (no data augmentation)
test_data = test_datagen.flow_from_directory(
test_dir, target_size=image_size, batch_size=batch_size, class_mode="categorical"
)
# Load the VGG16 model pre-trained on ImageNet (without the top layer)
base_model = VGG16(weights="imagenet", include_top=False, input_shape=image_size + (3,))
# Freeze the pre-trained layers of the VGG16 model
for layer in base_model.layers:
layer.trainable = False
# Add a custom top layer to the model for 4 class classification
x = Flatten()(base_model.output)
x = Dense(128, activation="relu")(x)
predictions = Dense(4, activation="softmax")(x)
# Build the final model by combining the pre-trained VGG16 base model with the custom top layer
model = Model(inputs=base_model.input, outputs=predictions)
# Compile the model with categorical crossentropy loss and Adam optimizer
model.compile(loss="categorical_crossentropy", optimizer="adam", metrics=["accuracy"])
# Model Trained
history = model.fit(train_data, epochs=20, validation_data=test_data, workers=4)
# Evaluate the trained model on the testing data
test_loss, test_acc = model.evaluate(test_data)
print("Test accuracy:", test_acc)
# Plot the training and validation accuracy as a line
import matplotlib.pyplot as plt
training_accuracy = history.history["accuracy"]
validation_accuracy = history.history["val_accuracy"]
epochs = range(1, len(training_accuracy) + 1)
plt.plot(epochs, training_accuracy, "bo-", label="Training accuracy")
plt.plot(epochs, validation_accuracy, "b-", label="Validation accuracy")
plt.title("Training and validation accuracy")
plt.xlabel("Epochs")
plt.ylabel("Accuracy")
plt.legend()
plt.show()
# Model Testing on unseen data
# Classes of Apples in quadrants
train_data.class_indices
import numpy as np
from tensorflow.keras.preprocessing.image import load_img, img_to_array
img_path = "/kaggle/input/apples-in-four-quadrants-classification-cnn/custom-generated-apple-dataset/unseen-data.png"
img = load_img(img_path, target_size=(224, 224))
x = img_to_array(img)
x = np.expand_dims(x, axis=0)
x = x / 255.0
# Use the model to make a prediction
preds = model.predict(x)
print(preds.argmax(axis=-1))
|
# ## Imports
import os
import numpy as np
import pandas as pd
import seaborn as sns
# ## Data Collection
filepath = "/kaggle/input/enem-2010-2021-pedreiras-maranhao/enem-pedreiras.csv"
df = pd.read_csv(filepath)
# ## Data wrangling
df.info()
# ## Data Visualization
sns.set_theme()
sns.histplot(data=df, x="NU_NOTA_MT", kde=True, hue="TP_SEXO")
sns.lineplot(data=df, x="NU_ANO", y="NU_NOTA_MT", hue="TP_SEXO")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Introduction
# This capstone project is proposed by google, as a conclusion of the course Google Data Analystics Certification. This case study is about Bellafast, a high-tech manufacturer with the main products focused on women. This small company, have the potential to become market leader. However, this project will focus on the company's products., the level of aggregation of your consumers and define a new marketing strategy for the company.
# # Ask
# My approach for this project is to divide into two tasks:
# First identify in the information provided by the company, how its consumers use the products.
# Second, perform a customer-oriented analysis.
# To begin, we will focus on the following questions:
# How many consumers do we have? How many use their services? By observing the data, we can initially identify some trends?
# Do your subscriptions take into account the needs of consumers? What other options can you offer? And how can we implement this information in Bellabeat marketing strategy?
# # Prepare
# **Dataset**: FitBit Fitness Tracker Data. This Kaggle data set contains personal fitness tracker from thirty fitbit users. Thirty eligible Fitbit users consented to the submission of personal tracker data, including minute-level output for physical activity, heart rate, and sleep monitoring. It includes information about daily activity, steps, and heart rate that can be used to explore users’ habits.
# To determine the credibility and integrity of the date we will use **ROCCC** system:
# This sample only contains 30 individuals, and since it was through research, it does not show accurate results that may lead to dishonest answers. However, it is possible to identify the lack of some information, such as water intake e 8 user data in relation to weight This data was collected in 2016, which means it is currently outdated and may not represent the current trends in smart device usage.
# # Process
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import plotly.express as px
import plotly.graph_objects as go
data = pd.read_csv(
"/kaggle/input/fitbit/Fitabase Data 4.12.16-5.12.16/dailyActivity_merged.csv"
)
data.head()
data.isnull().sum()
data.info()
# Changing type of date
data["ActivityDate"] = pd.to_datetime(data["ActivityDate"], format="%m/%d/%Y")
data["ActivityDate"].head()
# Create new column for dar of the week
data["DayWeek"] = data["ActivityDate"].dt.day_name()
data["DayWeek"].head()
data.info()
# # Analyse
# TotalSteps vs Calories
figure = px.scatter(
data_frame=data,
x="TotalSteps",
y="Calories",
size="TotalDistance",
title="Carlories Burned for Total Distance",
)
figure.update_layout(
title={
"text": "Carlories Burned for Total Distance",
"y": 0.95,
"x": 0.5,
"xanchor": "center",
"yanchor": "top",
},
title_font=dict(size=24),
)
figure.show()
# You can see that there is a linear relationship between the total number of steps and the number of calories burned in a day.
# Relation Between Sleep and Time in Bed
data_sleepday = pd.read_csv(
"/kaggle/input/fitbit/Fitabase Data 4.12.16-5.12.16/sleepDay_merged.csv"
)
data_sleepday.info()
data_sleepday.isnull().sum()
# Relation Between Sleep and Time in Bed
figure_sleep = px.scatter(
data_frame=data_sleepday,
x="TotalTimeInBed",
y="TotalMinutesAsleep",
title="Relation Between Sleep and Time in Bed",
)
figure_sleep.update_layout(
title={
"text": "Relation Between Sleep and Time in Bed",
"y": 0.95,
"x": 0.5,
"xanchor": "center",
"yanchor": "top",
},
title_font=dict(size=24),
)
figure_sleep.show()
# We can observe that many people have poor quality of sleep.
# Weekly Activity Summary
fig = go.Figure()
fig.add_trace(
go.Bar(
x=data["DayWeek"],
y=data["VeryActiveMinutes"],
name="Very Active",
marker_color="red",
)
)
fig.add_trace(
go.Bar(
x=data["DayWeek"],
y=data["FairlyActiveMinutes"],
name="Fairly Active",
marker_color="blue",
)
)
fig.add_trace(
go.Bar(
x=data["DayWeek"],
y=data["LightlyActiveMinutes"],
name="Lightly Active",
marker_color="pink",
)
)
fig.update_layout(
barmode="group",
xaxis_tickangle=-45,
title={
"text": "Weekly Activity Summary",
"x": 0.5,
"y": 0.95,
"xanchor": "center",
"yanchor": "top",
},
)
fig.show()
# We can observe more activity in the middle of the week, and a significant decrease from Friday and continue on weekends and Monday.
#
# Total Active Minutes
VeryActiveMinutes = data["VeryActiveMinutes"].sum()
FairlyActiveMinutes = data["FairlyActiveMinutes"].sum()
LightlyActiveMinutes = data["LightlyActiveMinutes"].sum()
SedentaryMinutes = data["SedentaryMinutes"].sum()
slices = [
VeryActiveMinutes,
FairlyActiveMinutes,
LightlyActiveMinutes,
SedentaryMinutes,
]
labels = [
"Very active minutes",
"Fairly active minutes",
"Lightly active minutes",
"Sedentary minutes",
]
colours = ["purple", "yellow", "orange", "red"]
explode = [0, 0, 0, 0.1]
plt.style.use("default")
plt.pie(
slices,
labels=labels,
colors=colours,
wedgeprops={"edgecolor": "black"},
explode=explode,
autopct="%1.1f%%",
)
plt.title("Percentage of Total Active Minutes")
plt.tight_layout()
plt.show()
|
import numpy as np
import cv2
from pydicom import dcmread
import pandas as pd
import matplotlib.pyplot as plt
from IPython.display import clear_output
import os
from matplotlib.patches import Rectangle
import torch
from torch import nn
from torch.utils import data
from torchvision import transforms
import torchvision
train_folder = "/kaggle/input/train-data/train"
def load_file(path):
return np.load(path).astype(np.float32)
# transform = transforms.Compose([
# transforms.ToTensor(),
# transforms.Normalize(0.49, 0.248),
# transforms.RandomAffine(degrees=(-5, 5), translate=(0, 0.05), scale=(0.9, 1.1)),
# transforms.RandomResizedCrop((224, 224), scale=(0.35, 1))
# ])
transform = transforms.ToTensor()
train_dataset = torchvision.datasets.DatasetFolder(
train_folder, loader=load_file, extensions="npy", transform=transform
)
# val_dataset = torchvision.datasets.DatasetFolder(val_folder, loader=load_file, extensions="npy", transform=transform)
class CNN(nn.Module):
def __init__(self):
super(CNN, self).__init__()
self.conv1 = nn.LazyConv2d(out_channels=128, kernel_size=[5, 5])
self.relu1 = nn.LeakyReLU()
self.maxpool1 = nn.MaxPool2d(2, 2)
self.conv2 = nn.LazyConv2d(out_channels=64, kernel_size=[3, 3])
self.relu2 = nn.LeakyReLU()
self.maxpool2 = nn.MaxPool2d(2, 2)
self.conv3 = nn.LazyConv2d(out_channels=32, kernel_size=[2, 2])
self.relu3 = nn.LeakyReLU()
self.maxpool3 = nn.MaxPool2d(2, 2)
self.dropout = nn.Dropout(0.2)
self.fc1 = nn.LazyLinear(out_features=4)
self.relu4 = nn.LeakyReLU()
self.fc2 = nn.LazyLinear(out_features=64)
self.softmax = nn.Softmax(dim=1)
def forward(self, x):
x = self.conv1(x)
x = self.relu1(x)
x = self.maxpool1(x)
x = self.conv2(x)
x = self.relu2(x)
x = self.maxpool2(x)
x = self.conv3(x)
x = self.relu3(x)
x = self.maxpool3(x)
x = torch.flatten(x, 1)
x = self.dropout(x)
x = self.fc1(x)
x = self.relu4(x)
x = self.fc2(x)
x = self.softmax(x)
return x
class Model:
def __init__(
self,
train=False,
filename="trained_model",
train_dataset=train_dataset,
epochs=10,
batch_size=64,
learning_rate=0.01,
save=True,
):
self.device = torch.device("cuda")
self.filename = filename + ".pt"
self.save = save
self.batch_size = batch_size
self.learning_rate = learning_rate
self.epochs = epochs
self.train_dataset = train_dataset
self.train_loader = torch.utils.data.DataLoader(
self.train_dataset, batch_size=self.batch_size, shuffle=True, num_workers=2
)
if train:
cnn = CNN()
self.trained_model = self._fit(model=cnn, train_loader=self.train_loader)
else:
self.trained_model = torch.load(self.filename)
def _fit(self, model, train_loader):
model = model.to(self.device)
loss_function = nn.NLLLoss() # Initialize loss function
optimizer = torch.optim.SGD(
params=model.parameters(), lr=self.learning_rate
) # initialize our SGD optimizer
print(
f" Starting training with Epochs = {self.epochs}, Batch size = {self.batch_size}, Learning rate = {self.learning_rate} "
)
for epoch in range(self.epochs):
running_loss, train_accuracy = self._train_one_epoch(
model, train_loader, loss_function, optimizer
)
print(
f"EPOCH: {epoch + 1} Training loss: {running_loss} Training accuracy: {train_accuracy} "
)
if self.save:
torch.save(model.state_dict(), self.filename)
return model
def _val_one_epoch(self, model, val_loader, loss_function):
model.eval()
epoch_val_loss = 0
correct, num = 0, 0
for inputs, labels in val_loader:
inputs = inputs.to(self.device)
labels = labels.to(self.device)
with torch.no_grad():
pred = model(inputs)
loss = loss_function(pred, labels)
epoch_val_loss += loss.item()
correct += (pred.argmax(1) == labels).type(torch.float).sum().item()
num += labels.size(0)
validation_accuracy = 100 * correct / num
return validation_accuracy
def _train_one_epoch(self, model, train_loader, loss_function, optimizer):
running_loss = 0
correct, num = 0, 0
model.train(True)
for inputs, labels in train_loader:
inputs = inputs.to(self.device)
labels = labels.to(self.device)
# Zero your gradients for every batch!
optimizer.zero_grad()
# Make predictions for this batch
pred = model(inputs)
# Compute the loss and its gradients
loss = loss_function(pred, labels)
loss.backward()
# Adjust learning weights
optimizer.step()
correct += (pred.argmax(1) == labels).type(torch.float).sum().item()
num += labels.size(0)
# Gather data and report
running_loss += loss.item()
training_accuracy = 100 * correct / num
return running_loss, training_accuracy
def predict(self):
pass
for lr in [0.001]:
filename = f"trained_model_learningrate_{lr}"
model = Model(train=True, learning_rate=lr, filename=filename)
|
# # 1. Import Dependencies and load dataset #
# ## 1.1 Import dependencies ##
import os
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
import seaborn as sns
import missingno as msno
from sklearn import tree
from sklearn.ensemble import RandomForestClassifier
from sklearn.preprocessing import LabelEncoder
from sklearn.ensemble import HistGradientBoostingClassifier
from sklearn.model_selection import cross_val_score
# ## 1.2 Load dataset ##
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
train = pd.read_csv("/kaggle/input/spaceship-titanic/train.csv")
test = pd.read_csv("/kaggle/input/spaceship-titanic/test.csv")
# # 2. Target Analysis #
# The analysis will be done to describe the following items:
# * the features,
# * data distribution and its relation to the target variable
# * missing values
# * possible correlations
# ## 2.1 Data Description and Visualization ##
# * **PassengerId** - A unique Id for each passenger. Each id takes the form of gggg_pp (gggg = group passenger; pp = number within group). People in group are often family members, but not always.
# * **HomePlanet** - The planet the passenger departed from (permanent residence)
# * **CryoSleep** - Indicates whether the passenger elected to be put into suspended animation for the duration of the voyage. These passenger are confined in their cabins.
# * **Cabin** - The cabin number where the passenger is staying in the form of deck/num/side, where side can be either P (Port) or S (Starboard)
# * **Destination** - Debarking planet
# * **Age** - The age of passenger
# * **VIP** - Whether the passenger has paid for special VIP service during the voyage
# * **RoomService, FoodCourt, ShoppingMall, Spa, VRDeck** - Amount the passenger has billed at each of the Spaceship Titanic's many luxury amenities
# * **Name** - The first and last name of passengers
# * **Transported** - Whether the passenger was transported to another dimension.
# Display first 5 rows of the training dataset
train.head()
train.info()
# Convert CryoSleep and VIP values from objects to boolean for later analysis
train[["CryoSleep", "VIP"]] = train[["CryoSleep", "VIP"]].astype("boolean")
# Describe training dataset of all numerical features and transpose
train.describe(include=float).applymap(lambda x: f"{x:0.2f}").T
# Describe training dataset including the categorical and boolean features
train.describe(include=[object, bool])
# ### Target Variable ###
# The following plot shows that the dataset is balanced in corresponding to the target variable **"Transported"**
train.Transported.value_counts().plot(
kind="bar", title="Distribution of target variable", xlabel="Transported"
)
plt.show()
# ### Colorcoded Pairplots of Numerical Data ###
# The following plot shows the numerical columns as a whole with pairplots, and colorcoded determines the Transportation status.
sns.pairplot(
train.drop(["CryoSleep", "VIP"], axis=1),
hue="Transported",
kind="scatter",
diag_kind="kde",
plot_kws={"alpha": 0.7},
)
plt.show()
# The columns represents the expenses on amenities are strongly skewed to the right, so in 'low or 0 expenses area', there are many amounts counted. Since the datacolumns represents in positive values, one can do is log-transformed to get better overview of possible differences in the distribution of expenses of "Transported" and "Non-Transported" passengers.
# Log transform and plot a pairplots
log_train = train.copy()
to_log_cols = ["RoomService", "FoodCourt", "ShoppingMall", "Spa", "VRDeck"]
log_train[to_log_cols] = log_train[to_log_cols].apply(
lambda col: np.log(col + 1), raw=True, axis=1
)
sns.pairplot(
log_train.drop(["CryoSleep", "VIP"], axis=1),
hue="Transported",
kind="scatter",
diag_kind="kde",
plot_kws={"alpha": 0.7},
)
plt.show()
# From the first overview of the diagonal elements, the passengers who were transported have a higher probability mass in their KDE-plots on lower expenses. Vice versa for passengers at higher expenses, therefore there are possibly relationship between the target variable and the amount of money people spent. Hypothetically, passengers with higher budget rather less likely to be transported. The following analysis observes the connection between VIP status and value of transported passengers
# The scatterplots is not adequate to provide details in the relationship between expense-categories. Therefore, univariative visualizations is introduced
# ## 2.2 Univariate Visualizations
# Total expense = sum of all expenses
train["TotalExp"] = train[
["RoomService", "FoodCourt", "ShoppingMall", "Spa", "VRDeck"]
].sum(1)
# ### 2.2.1 Numerical features ###
# Age plot
sns.kdeplot(data=train, x="Age", hue="Transported", fill=True)
plt.show()
# The two age distributions look similar, besides the peak at young ages for transported passengers. This could be meaning that children has higher possibility of being transported
# all expense calculated and rounded
all_exp = (
train[["RoomService", "FoodCourt", "ShoppingMall", "Spa", "VRDeck"]]
.sum()
.sort_values(ascending=False)
)
all_exp.name = "Share of the individual expense categories"
np.round(all_exp / all_exp.sum(), decimals=2)
# log transformation
num_features = [
"Total Expenses",
"RoomService",
"FoodCourt",
"ShoppingMall",
"Spa",
"VRDeck",
]
log_train["Total Expenses"] = np.log(train.TotalExp + 1)
plt.figure(figsize=(25, 25))
for i, feat in enumerate(num_features):
plt.subplot(4, 2, i + 1)
sns.kdeplot(data=log_train, x=feat, hue="Transported", common_norm=False, fill=True)
plt.xlabel("log " + feat, fontsize=12)
plt.title("Distribution of log " + feat, fontsize=16)
# ### 2.2.3 Categorical Features ###
cat_feat = ["HomePlanet", "CryoSleep", "Destination", "VIP"]
plt.figure(figsize=(25, 15))
for i, feat in enumerate(cat_feat):
plt.subplot(2, 2, i + 1)
p1 = (
train.groupby(feat)["Transported"]
.value_counts()
.unstack()
.plot(kind="bar", stacked=True, rot=0, ax=plt.gca())
)
nrows = train.dropna(subset=None).shape[0]
y_upper = train[feat].value_counts().to_numpy().max() * 1.1
plt.ylim((0, y_upper))
plt.title("Distribution of " + feat, fontsize=18)
plt.xlabel("")
plt.xticks(fontsize=16)
plt.subplots_adjust(wspace=0.3)
# In the case of homeplanet, higher percentage of transported passengers come from the Europe and similarly observed from people in CryoSleep which tend to more likely to be transported.
# In the VIP passengers, the transported passengers were found quite rare, and more observed in the non-VIP passengers
# ### 2.3 NaN-values ###
print(f"Number of rows: {len(train)}")
print(f"Number of rows with >= 1 NaN-value: {(train.isna().any(1).sum())}")
print(
f"\nPercentage of 'full'-rows: {100-(train.isna().any(1).sum()/len(train))*100:.2f}%"
)
train.isna().sum().sort_values().plot(kind="barh")
plt.show()
# We can confirm that from the PassengerID are filled with 100% data (there is no NaN value).
# Accordingly, all column shows couple amounts of NaN values
# To check the position of NaN-values inside the columns:
msno.matrix(train)
plt.show()
# ### 2.4 Possible correlations of NaN values ###
plt.figure(figsize=(10, 8))
mask = np.triu(np.ones_like(train.corr(), dtype=bool))
cmap = sns.diverging_palette(230, 20, as_cmap=True)
sns.heatmap(
train.corr(),
vmin=-1,
vmax=1,
annot=True,
cmap=cmap,
mask=mask,
square=True,
linewidth=0.5,
)
plt.show()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
nRowsRead = None # specify 'None' if want to read whole file
# Apply_Rate_2019.csv has 1200890 rows in reality, but we are only loading/previewing the first 1000 rows
data = pd.read_csv(
"/kaggle/input/clickthrough-rate-prediction/ad.csv", delimiter=",", nrows=nRowsRead
)
data.dataframeName = "Apply_Rate_2019.csv"
nRow, nCol = data.shape
print(f"There are {nRow} rows and {nCol} columns")
data.head(5)
data = data[
[
"Daily Time Spent on Site",
"Age",
"Area Income",
"Daily Internet Usage",
"Ad Topic Line",
"City",
"Gender",
"Clicked on Ad",
]
]
label = "Clicked on Ad"
data.head(5)
cat_features = ["Ad Topic Line", "City", "Gender"]
continue_var = [
"Daily Time Spent on Site",
"Age",
"Area Income",
"Daily Internet Usage",
]
fill_mean = lambda x: x.fillna(x.mean())
for col in continue_var:
data[col] = data[col].fillna(data[col].mean())
data[col] = data[col].astype("float64")
data = data.fillna(
"-1",
)
from sklearn.preprocessing import LabelEncoder, MinMaxScaler
mms = MinMaxScaler(feature_range=(0, 1))
data[continue_var] = mms.fit_transform(data[continue_var])
train = data
from pyffm import PyFFM
training_params = {
"epochs": 5,
"reg_lambda": 0.002,
"sigmoid": True,
"parallel": True,
"early_stop": True,
}
model = PyFFM(model="ffm", training_params=training_params)
train_data = train.sample(frac=0.75)
predict_data = train.drop(train_data.index)
model.train(train_data, label_name=label)
test = predict_data
preds = model.predict(predict_data.drop(columns=label))
test = test[label]
preds = list(preds)
test = list(test)
test
from sklearn.metrics import log_loss, roc_auc_score, accuracy_score
from sklearn.metrics import f1_score
from sklearn.metrics import cohen_kappa_score
from sklearn.metrics import roc_auc_score, roc_curve
from sklearn.metrics import precision_score, recall_score
print("test LogLoss", round(log_loss(preds, test), 4))
print("test roc_auc_score", round(roc_auc_score(preds, test), 4))
print("test f1 score", round(f1_score(preds, test), 4))
print("test precision score", round(precision_score(preds, test), 4))
print("test recall score", round(recall_score(preds, test), 4))
print("test accuracy score", round(accuracy_score(preds, test), 4))
|
# # Simple Linear Regression
# In this notebook, a simple linear regression model is built to predict the salary using an years of experience.
# 1. Reading and Understanding the Data
# 2. Training the model
# 3. Residual Analysis
# 4. Predicting and evaluating the model on the test set
# ## Step 1: Reading and Understanding the Data
# import warnings
import warnings
warnings.filterwarnings("ignore")
# import important libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import statsmodels
import statsmodels.api as sm
import sklearn
from sklearn.model_selection import train_test_split
from sklearn.metrics import r2_score
# read the data
data = pd.read_csv(
"/kaggle/input/salary-dataset-simple-linear-regression/Salary_dataset.csv"
)
data.head()
# drop the first column as it is redundant
data.drop("Unnamed: 0", axis=1, inplace=True)
data.head()
# shape of the data
data.shape
# There are 30 rows and 2 columns present in the data.
# info of the data
data.info()
# statistical summary of the dataset
data.describe()
# missing values in the data
data.isnull().sum()
# There are no missing values in the data.
# Visualise the data using regression plot
sns.regplot(x="YearsExperience", y="Salary", data=data)
plt.show()
# From the above regression plot, it is clear that Experience and Salary are linearly correlated.
# ## Step 2: Performing Simple Linear Regression
# The equation of simple linear regression is:
# y = mx + c
# where,
# y is the dependent (outcome or response) variable
# m is the slope of the line
# x is the independent (predictor) variable
# c is the y-intercept of the line
#
# Here, the independent variable is 'YearsExperience' and dependent variable is 'Salary'.
# ### Steps of model building:
# - Create X and y
# - create train and test sets
# - Train the model on training set (i.e. learn the coefficients)
# - Evaluate the model on the test set
# create X and y
X = data["YearsExperience"]
y = data["Salary"]
# train-test split
X_train, X_test, y_train, y_test = train_test_split(
X, y, train_size=0.67, random_state=20
)
print("The shape of X_train is ", X_train.shape)
print("The shape of X_test is ", X_test.shape)
print("The shape of y_train is ", y_train.shape)
print("The shape of y_test is ", y_test.shape)
# The statsmodels uses the 'Ordinary Least Squares' to fit the line. By default, the statsmodels library doesn't include the intercept. It only includes the coefficient of the predictor variables. We need to add the y-intercept explicitely in the statsmodels so we use the command 'add_constant' to fit an intercept. If we don't fit an intercept, the statmodels will fit a line passing through the origin.
# training the model using statsmodels
X_train_sm = sm.add_constant(X_train)
X_train_sm.head()
# fitting the model
lr = sm.OLS(y_train, X_train_sm) # This creates a Linear Regression Object
lr_model = lr.fit()
lr_model.params
# summary of the model
lr_model.summary()
# The p-value is 0. It means that, the coefficients are significant. The relationship between the number of years of experience and salary is not obtained purely by chance, there is a real statistically significant relationship.
# The p-value below 0.001 is considered as very low.
# The R-squared is 0.953. It means that, the 95% of the variance in the data is explained by the model.
# F-statistic tells us whether the overall model fit is significant or not. This parameter is examined because many a time it happens that even though all of betas are significant, but the overall model fit might happen just by chance. If the P(F-statistic) is less than 0.05 then the overall model fit is significant. The p-value of the F-statistic is 1.94e-13, which is practically a zero value. This shows that the model fit is significant since it is less than 0.05.
# make predictions on the training set
y_train_pred = lr_model.predict(X_train_sm)
# plot the model
plt.scatter(X_train, y_train)
plt.plot(X_train, y_train_pred, color="r")
plt.show()
# ## Step 3: Residual Analysis
# residual
res = y_train - y_train_pred
# plot the residuals
sns.distplot(res)
plt.title("Residual Plot")
plt.show()
# The center of the normal distribution is around zero. The shape is also looks like a normal distribution.
# look for patterns in residuals (we should not able to identify any patterns)
plt.scatter(X_train, res)
plt.show()
# The residuals don't follow any patterns. So, the asssumptions of Simple Linear Regression are validated.
# ## Step 4: Predictions and Evaluation on the Test Set
# add a constant/intercept to the test set
X_test_sm = sm.add_constant(X_test)
# make prediction on test set
y_test_pred = lr_model.predict(X_test_sm)
# evaluate the model on the training and test set
print(
"The R-squared for the training set is",
round(r2_score(y_true=y_train, y_pred=y_train_pred), 2),
)
print(
"The R-squared for the test set is",
round(r2_score(y_true=y_test, y_pred=y_test_pred), 2),
)
# plot the model with the test set
plt.scatter(X_train, y_train)
plt.scatter(X_test, y_test)
plt.plot(X_test, y_test_pred, color="g")
plt.show()
|
# # Imports
import numpy as np #
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list the files in the input directory
import os
print(os.listdir("../input"))
# Any results you write to the current directory are saved as output.
import glob
import tensorflow as tf
from sklearn.model_selection import train_test_split
from collections import Counter
from sklearn.model_selection import train_test_split
from collections import Counter
import cv2
from concurrent import futures
import threading
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.preprocessing import LabelEncoder
import datetime
# # Data import
# getting the total number of images in the training set
base_dir = "../input"
train_dir = os.path.join(base_dir, "train", "train")
type1_dir = os.path.join(base_dir, "Type_1")
type2_dir = os.path.join(base_dir, "Type_2")
type3_dir = os.path.join(base_dir, "Type_3")
type1_files = glob.glob(type1_dir + "/*.jpg")
type2_files = glob.glob(type2_dir + "/*.jpg")
type3_files = glob.glob(type3_dir + "/*.jpg")
added_type1_files = glob.glob(
os.path.join(base_dir, "additional_Type_1_v2", "Type_1") + "/*.jpg"
)
added_type2_files = glob.glob(
os.path.join(base_dir, "additional_Type_2_v2", "Type_2") + "/*.jpg"
)
added_type3_files = glob.glob(
os.path.join(base_dir, "additional_Type_3_v2", "Type_3") + "/*.jpg"
)
type1_files = type1_files + added_type1_files
type2_files = type2_files + added_type2_files
type3_files = type3_files + added_type3_files
print("Number of images in a train set of type 1: ", len(type1_files))
print("Number of images in a train set of type 2: ", len(type2_files))
print("Number of images in a train set of type 3: ", len(type3_files))
print(
"Total number of images in a train set: ",
sum([len(type1_files), len(type2_files), len(type3_files)]),
)
# Building a dataframe mapping images and Cancer type
files_df = pd.DataFrame(
{
"filename": type1_files + type2_files + type3_files,
"label": ["Type_1"] * len(type1_files)
+ ["Type_2"] * len(type2_files)
+ ["Type_3"] * len(type3_files),
}
)
files_df
# Shuffle data
random_state = 42
files_df = files_df.sample(frac=1, random_state=random_state)
files_df
# # Data exploration
files_df.describe()
# Check for duplicates
len(files_df[files_df.duplicated()])
# Get count of each type
type_count = pd.DataFrame(files_df["label"].value_counts()).rename(
columns={"label": "Num_Values"}
)
type_count
# Display barplot of type count
plt.figure(figsize=(15, 6))
sns.barplot(x=type_count["Num_Values"], y=type_count.index.to_list())
plt.title("Cervical Cancer Type Distribution")
plt.grid(True)
plt.show()
# Display sample images of types
for label in ("Type_1", "Type_2", "Type_3"):
filepaths = files_df[files_df["label"] == label]["filename"].values[:5]
fig = plt.figure(figsize=(15, 6))
for i, path in enumerate(filepaths):
img = cv2.imread(path)
img = cv2.cvtColor(img, cv2.COLOR_RGB2BGR)
img = cv2.resize(img, (224, 224))
fig.add_subplot(1, 5, i + 1)
plt.imshow(img)
plt.subplots_adjust(hspace=0.5)
plt.axis(False)
plt.title(label)
# # Data propocessing
# Split training,val and test set : 70:15:15
train_files, test_files, train_labels, test_labels = train_test_split(
files_df["filename"].values,
files_df["label"].values,
test_size=0.3,
random_state=random_state,
)
test_files, val_files, test_labels, val_labels = train_test_split(
test_files, test_labels, test_size=0.5, random_state=random_state
)
print("Number of images in train set: ", train_files.shape)
print("Number of images in validation set: ", val_files.shape)
print("Number of images in test set: ", test_files.shape, "\n")
print(
"Train:",
Counter(train_labels),
"\nVal:",
Counter(val_labels),
"\nTest:",
Counter(test_labels),
)
def load_images(files, labels):
features = []
correct_labels = []
bad_images = 0
for i in range(len(files)):
try:
img = cv2.imread(files[i])
resized_img = cv2.resize(img, (160, 160))
features.append(np.array(resized_img))
correct_labels.append(labels[i])
except Exception as e:
bad_images += 1
print("Encoutered bad image")
print("Bad images ecountered:", bad_images)
return np.array(features), np.array(correct_labels)
# Load training and evaluation data
train_features, train_labels = load_images(train_files, train_labels)
print("Train images loaded")
val_features, val_labels = load_images(val_files, val_labels)
print("Validation images loaded")
test_features, test_labels = load_images(test_files, test_labels)
print("test images loaded")
# check lengths of training and evaluation sets
len(train_features), len(train_labels), len(val_features), len(val_labels), len(
test_features
), len(test_labels)
BATCH_SIZE = 32
NUM_CLASSES = 3
EPOCHS = 20
INPUT_SHAPE = (160, 160, 3)
# encode train+val sets text categories with labels
le = LabelEncoder()
le.fit(train_labels)
train_labels_enc = le.transform(train_labels)
val_labels_enc = le.transform(val_labels)
train_labels_1hotenc = tf.keras.utils.to_categorical(
train_labels_enc, num_classes=NUM_CLASSES
)
val_labels_1hotenc = tf.keras.utils.to_categorical(
val_labels_enc, num_classes=NUM_CLASSES
)
print(train_labels[:6], train_labels_enc[:6])
print(train_labels[:6], train_labels_1hotenc[:6])
le = LabelEncoder()
le.fit(test_labels)
test_labels_enc = le.transform(test_labels)
test_labels_1hotenc = tf.keras.utils.to_categorical(
test_labels_enc, num_classes=NUM_CLASSES
)
print(test_labels[:6], test_labels_enc[:6])
print(test_labels[:6], test_labels_1hotenc[:6])
# # Data augmentation
data_augmentation = tf.keras.Sequential(
[
tf.keras.layers.RandomFlip("horizontal"),
tf.keras.layers.RandomRotation(0.2),
]
)
plt.figure(figsize=(10, 10))
first_image = train_features[0]
for i in range(9):
ax = plt.subplot(3, 3, i + 1)
augmented_image = data_augmentation(tf.expand_dims(first_image, 0))
plt.imshow(augmented_image[0] / 255)
plt.axis("off")
# # MobileNet V2 pre-trained
# ##Transfer learning
base_model = tf.keras.applications.MobileNetV2(
include_top=False, weights="imagenet", input_shape=INPUT_SHAPE
)
base_model.trainable = False
inputs = tf.keras.Input(shape=(160, 160, 3))
x = data_augmentation(inputs)
# rescale pixel values
preprocess_input = tf.keras.applications.mobilenet_v2.preprocess_input
x = preprocess_input(x)
x = base_model(x, training=False)
x = tf.keras.layers.GlobalAveragePooling2D()(x)
x = tf.keras.layers.Dropout(0.2)(x)
outputs = tf.keras.layers.Dense(3, activation="softmax")(x)
model = tf.keras.Model(inputs, outputs)
# compile the model
learning_rate = 1e-4
model.compile(
optimizer=tf.keras.optimizers.Adam(learning_rate=learning_rate),
loss="categorical_crossentropy",
metrics=["accuracy"],
)
model.summary()
history = model.fit(
x=train_features,
y=train_labels_1hotenc,
batch_size=BATCH_SIZE,
epochs=EPOCHS,
validation_data=(val_features, val_labels_1hotenc),
verbose=1,
)
def learning_performance_chart(title="Learning Perfomance", history=history):
# plots a chart showing the change in accuracy and loss function over epochs
f, (ax1, ax2) = plt.subplots(1, 2, figsize=(12, 4))
t = f.suptitle(title, fontsize=12)
f.subplots_adjust(top=0.85, wspace=0.3)
max_epoch = len(history.history["accuracy"]) + 1
epoch_list = list(range(1, max_epoch))
ax1.plot(epoch_list, history.history["accuracy"], label="Train Accuracy")
ax1.plot(epoch_list, history.history["val_accuracy"], label="Validation Accuracy")
ax1.set_xticks(np.arange(1, max_epoch, 5))
ax1.set_ylabel("Accuracy Value")
ax1.set_xlabel("Epoch")
ax1.set_title("Accuracy")
l1 = ax1.legend(loc="best")
ax2.plot(epoch_list, history.history["loss"], label="Train Loss")
ax2.plot(epoch_list, history.history["val_loss"], label="Validation Loss")
ax2.set_xticks(np.arange(1, max_epoch, 5))
ax2.set_ylabel("Loss Value")
ax2.set_xlabel("Epoch")
ax2.set_title("Loss")
l2 = ax2.legend(loc="best")
learning_performance_chart(title="MobileNetV2 baseline performance", history=history)
print("MobileNetV2 performance on the test set:")
results = model.evaluate(test_features, test_labels_1hotenc, verbose=1)
# keep track of the models performance
performance_df = pd.DataFrame(columns=["model", "test set accuracy"])
def record_performance(df, model_name, test_accuracy):
return df.append(
{"model": model_name, "test set accuracy": test_accuracy}, ignore_index=True
)
performance_df = record_performance(performance_df, "MobileNetV2 base", results[1])
performance_df
# # Fine tuning
tuning_model_1 = base_model
tuning_model_1.trainable = True
# Let's take a look to see how many layers are in the base model
print("Number of layers in the base model: ", len(tuning_model_1.layers))
# ## Fine tune at 120 layer
# Fine-tune from this layer onwards
fine_tune_at = 120
# Freeze all the layers before the `fine_tune_at` layer
for layer in tuning_model_1.layers[:fine_tune_at]:
layer.trainable = False
# Compile the model
# Use smaller learning rate
model.compile(
loss="categorical_crossentropy",
optimizer=tf.keras.optimizers.Adam(learning_rate=learning_rate / 10),
metrics=["accuracy"],
)
len(model.trainable_variables)
# Train the model some more
fine_tune_epochs = 20
total_epochs = EPOCHS + fine_tune_epochs
history_fine = model.fit(
x=train_features,
y=train_labels_1hotenc,
batch_size=BATCH_SIZE,
epochs=total_epochs,
initial_epoch=history.epoch[-1],
validation_data=(val_features, val_labels_1hotenc),
verbose=1,
)
learning_performance_chart(
title="MobileNetV2 fine tuned baseline performance", history=history_fine
)
print("fine tuned MobileNetV2 performance on the test set:")
results = model.evaluate(test_features, test_labels_1hotenc, verbose=1)
performance_df = record_performance(
performance_df, "MobileNetV2 fine tune top 38 layers", results[1]
)
performance_df
# ## Fine tune at 100 layer
tuning_model_2 = base_model
tuning_model_2.trainable = True
# Fine-tune from this layer onwards
fine_tune_at = 100
# Freeze all the layers before the `fine_tune_at` layer
for layer in tuning_model_2.layers[:fine_tune_at]:
layer.trainable = False
# Compile the model
# Use smaller learning rate
model.compile(
loss="categorical_crossentropy",
optimizer=tf.keras.optimizers.Adam(learning_rate=learning_rate / 10),
metrics=["accuracy"],
)
len(model.trainable_variables)
# Train the model some more
fine_tune_epochs = 20
total_epochs = EPOCHS + fine_tune_epochs
history_fine = model.fit(
x=train_features,
y=train_labels_1hotenc,
batch_size=BATCH_SIZE,
epochs=total_epochs,
initial_epoch=history.epoch[-1],
validation_data=(val_features, val_labels_1hotenc),
verbose=1,
)
learning_performance_chart(
title="MobileNetV2 fine tuned baseline performance", history=history_fine
)
print("fine tuned MobileNetV2 performance on the test set:")
results = model.evaluate(test_features, test_labels_1hotenc, verbose=1)
performance_df = record_performance(
performance_df, "MobileNetV2 fine tune top 56 layers", results[1]
)
performance_df
# The last model showed the best results on the test set with accuracy at ~65%
# # Testing
# ## Stage 1 test
test_dir = os.path.join("../input/test/test")
test_files = glob.glob(test_dir + "/*.jpg")
print("Number of images in a test set:", len(test_files))
# sort test files in correct order for submission
from tkinter import Tcl
test_files = Tcl().call("lsort", "-dict", test_files)
test_files[:5]
# Load images
features = []
bad_images = 0
for i in range(len(test_files)):
try:
img = cv2.imread(test_files[i])
resized_img = cv2.resize(img, (160, 160))
features.append(np.array(resized_img))
except Exception as e:
bad_images += 1
print("Encoutered bad image")
print("Bad images ecountered:", bad_images)
test_images = np.array(features)
test_images.shape
predictions = model.predict(test_images)
predictions[:5]
test_submissions = pd.DataFrame(
{
"image_name": test_files,
"Type_1": predictions[:, 0],
"Type_2": predictions[:, 1],
"Type_3": predictions[:, 2],
}
)
test_submissions.to_csv("submission.csv", index=False)
test_submissions.head()
|
# ## Introduction
# Trying out some EDA for the first time by myself. Initially, I didn't have a plan. I just happened to stumble across the fact that US Unemployment rates were the highest during the early stages of the Covid-19 Pandemic so I decided to do some Feature Engineering and EDA for the data from those months. I've put explainations for Python code wherever I can :)
# ## General Analysis
# Let's import the necessary libraries and check the contents of the csv file.
import numpy as np # linear algebra
import pandas as pd # data processing
import matplotlib.pyplot as plt
import seaborn as sns
df = pd.read_csv(
"/kaggle/input/unemployment-in-america-per-us-state/Unemployment in America Per US State.csv"
)
df
df.info()
# Instead of using `df.isnull().sum()` after `df.info()`, **we can see that there are 29892 entries and each column has a non-null count of 29892 so we can infer that there are no null values in this dataset even without using `df.isnull.sum()`.**
# One issue with this DataFrame is that all the civilian statistics are stored with object datatype. We need to convert these to a numeric datatype so that we can work with them. Let's create a function that does exactly this.
def objToInt(col):
if df.dtypes[i] != "int64" and df.dtypes[i] != "float64":
df[col] = df[col].str.replace(",", "").astype(int)
# Notice that the function is defined such that its parameter is **something that is callable from the DataFrame `df`.** In this case, `col` refers to the column names in `df` as seen by the changing of value and datatype of `df[col]`.
# We have 5 columns with `object` datatype. Instead of calling this function 5 times separately, we can create a list containing the names of the columns in `df` and execute the function inside a `for` loop.
# Let's create a list containing all the columns and call the function for all columns of object datatype except `State/ Area`. Since that is the second column in the dataset and the first column is already `int`, we will call the function from the third column onwards.
columns = df.columns
for i in range(2, len(columns)):
objToInt(columns[i])
df.head()
# You will see me use the above 2 steps frequently as you read through this notebook :)
# Let's combine the Month and Year to create a new feature having `datetime` datatype
df["Date"] = pd.to_datetime(df["Year"] * 100 + df["Month"], format="%Y%m")
df.drop(columns=["Year", "Month"], inplace=True)
df.head()
# ## Trends of US Unemployment rate over the years
# Let's take a look at the Percent of the entire Labor Force of the US that is unemployed in each month. We do this by adding the Total Unemployment for each state and divide that by the sum of the Total Civilian Labor Force across all states.
k = pd.DataFrame(df.groupby("Date")[["Total Unemployment in State/Area"]].sum())
m = pd.DataFrame(df.groupby("Date")[["Total Civilian Labor Force in State/Area"]].sum())
k.reset_index(inplace=True)
m.reset_index(inplace=True)
k["Total Unemployment in State/Area"] *= 100
monthly_stats = pd.DataFrame()
monthly_stats["Date"] = df["Date"].unique()
monthly_stats["% Unemployment in US"] = (
k["Total Unemployment in State/Area"]
/ m["Total Civilian Labor Force in State/Area"]
)
monthly_stats
# Let's create a lineplot to see the total Unemployment rate of the US over the years to see if there is any trend.
sns.set(style="whitegrid")
sns.lineplot(x="Date", y="% Unemployment in US", data=monthly_stats)
# We can see that there are big spikes at around 1982-83, 2009-10 and 2020. Let's plot 10 months having the all-time highest US Civilian Unemployment % in a graph.
top = monthly_stats.sort_values(by="% Unemployment in US", ascending=False)[:10]
bottom = monthly_stats.sort_values(by="% Unemployment in US", ascending=False)[-9:]
top
fig, ax = plt.subplots(figsize=(12, 6))
fig = sns.barplot(x="Date", y="% Unemployment in US", data=top, ax=ax)
x_dates = top["Date"].dt.strftime("%Y-%m")
ax.set_xticklabels(labels=x_dates, rotation=45, ha="right")
# ## Covid-19 EDA and Feature Engineering
# It's no surprise that the unemployment rates were among the highest from April 2020 till July 2020. The initial spread of Covid-19 lead to widespread lockdowns and mass layoffs as many businesses found it difficult to conduct operations. Let's see which states were affected the most during this time period.
# Let's create a new dataframe `covid_data` which will have the monthly unemployment rates from April till July 2020 for each State/Area. I plan to do this by initializing a DataFrame and then adding each month's column by defining a function and calling it iteratively.
# We initialize the dataframe and we create our first column which contains the names of all states. The parameter `date` will represent the `Date` value in `df` for which we wish to get the unemployment rates for each of the states/areas. We store these rates in a DataFrame newMonth after which we reset the index and drop the old index. We will then add it to `covid_data` under the name `colname`, which is also another parameter we need to specify while callin the function.
covid_data = pd.DataFrame()
covid_data["States"] = df["State/Area"].unique()
def addMonth(date, colname):
newMonth = pd.DataFrame(
df.loc[
df["Date"] == date, "Percent (%) of Labor Force Unemployed in State/Area"
]
)
newMonth.reset_index(inplace=True, drop=True)
covid_data[colname] = newMonth
# Cool! Now that we have defined a function it means that we just need to call it four times right? Or is there a faster way?
# I created two lists `date` and `months`. These contain column names from `df` and `colname` respectively. I did this so that I can run functions with parameters belonging to these lists inside a `for` loop to save lines of code. Does this sound familiar? ;)
date = ["2020-04-01", "2020-05-01", "2020-06-01", "2020-07-01"]
months = ["April", "May", "June", "July"]
for i in range(4):
addMonth(date[i], months[i])
covid_data.head()
# In the above code, instead of calling the `addMonth` function for each month separately, I ran it inside a `for` loop that calls the function for each value in `months` and `date`. This calls the function repeatedly without any problem.
# **To add context before moving forward, every time I use the phrase "Unemployed Civilians", I am referring to Civilians in the Labor Force who are Unemployed.**
# We got the Unemployment rates for each state for these four months. Let's plot some graphs now.
def monthPlot(month):
plt.figure(figsize=(8, 4))
fig = sns.barplot(
y="States",
x=month,
data=covid_data,
order=covid_data.sort_values(by=month, ascending=False).States.head(10),
orient="h",
palette=("Spectral"),
)
fig.set(
xlabel=str(
"% of working population unemployed in the month of " + month + " 2020"
)
)
for i in range(4):
monthPlot(months[i])
# We can see that the states with the highest Unemployment rates in the given months had a sizeable fraction (>10% atleast) of civilians who were unemployed. Let's see how much these top States contributed towards the total number of unemployed civilians in the country in the given months.
# The function below will compute:
# * Total sum of unemployed civilians from each state in a given month in `temp` (i.e., the total number of unemployed civilians in the US in a given month)
# * Names of 10 states with highest unemployed rates in a given month in `sort`
# * Total sum of unemployed civilians from each of the 10 states with the highest unemployment rates in a given month in `unemp_top`
# * Percentage of unemployed civilians in the US who are from the 10 states with the highest unemployment rates in a given month in `var`
# We will also plot pie charts to represent the data visually
def fracTop(date, month):
temp = int(df.loc[df["Date"] == date][["Total Unemployment in State/Area"]].sum())
sort = list(covid_data.sort_values(by=month, ascending=False).States.head(10))
unemp_top = int(
df.loc[(df["Date"] == date) & (df["State/Area"].isin(sort))][
["Total Unemployment in State/Area"]
].sum()
)
var = unemp_top * 100 / temp
# Plotting a pie chart
explode = [0, 0.1]
keys = ["Remaining States", "Top 10 States with highest Unemployment Rate"]
data = [100 - var, var]
color = sns.color_palette("dark")
plt.pie(data, labels=keys, colors=color, explode=explode, autopct="%.02f%%")
plt.title(
str(
"Share of Unemployed Civilians in the US for the month of "
+ month
+ " 2020:"
)
)
plt.show()
print(
"The top 10 states make up "
+ "{:.2f}".format(var)
+ " percent of all unemployed civilians in the US Labor Force in the month of "
+ month
+ " 2020\n\n\n\n"
)
# Using our two nifty lists, we can call the function inside a `for` loop again :)
for i in range(len(date)):
fracTop(date[i], months[i])
|
import pandas as pd
import re
path = "/kaggle/input/ir-project-dataset/"
path_out = "/kaggle/working/"
class PatentParser:
def __init__(self, file_path):
self.file_path = file_path
def extract_terms(self, text):
terms = {}
for match in re.finditer(
r"(\b(?:\w+\s){0,6}\w+) \((\d+)\)|(\b(?:\w+\s){0,6}\w+) (\d+)", text
):
term1, number1, term2, number2 = match.groups()
number = number1 if number1 else number2
term = term1 if term1 else term2
if number not in terms:
terms[number] = []
terms[number].append(term)
return terms
def find_longest_common_substring(self, strings):
reversed_strings = [" ".join(s.split()[::-1]) for s in strings]
common_substrings = []
for i in range(len(reversed_strings[0].split())):
substrings = [s.split()[: i + 1] for s in reversed_strings]
if all(substrings[0] == s for s in substrings[1:]):
common_substrings.append(" ".join(substrings[0][::-1]))
else:
break
return max(common_substrings, key=len, default="")
def parse_patent_file(self):
with open(self.file_path, "r") as file:
content = file.read()
patent_sections = content.split("\n\n")
parsed_patents = []
for i, patent_section in enumerate(patent_sections):
sections = patent_section.split("_____d:\n")
patent_lines = sections[0].split("\n")
claim_lines = []
for line in reversed(patent_lines):
if line.startswith("_"):
identifiers = line
title = patent_lines[patent_lines.index(line) - 1]
break
claim_lines.append(line)
claim = "\n".join(reversed(claim_lines))
claim_contains_references = "R" if re.search(r"\w+ \(\d+\)", claim) else "N"
# There's just 4 patents over
# around 2000 that don't behave as expected,
# I'll just ignore those.
if len(sections) != 2:
print("ERROR at", i)
print(identifiers)
continue
claim_code_list_terms = self.extract_terms(claim.lower())
claim_code_term = {}
for code, list_terms in claim_code_list_terms.items():
maybe_term = self.find_longest_common_substring(list_terms)
if maybe_term != "":
claim_code_term[code] = maybe_term
body = sections[1].split("_____c:")[0].strip()
body_contains_references = (
"R" if re.search(r"(\w+ \(\d+\)|\w+ \d+)", body) else "N"
)
body_code_list_terms = self.extract_terms(body.lower())
body_code_term = {}
for code, list_terms in body_code_list_terms.items():
maybe_term = self.find_longest_common_substring(list_terms)
if maybe_term != "":
body_code_term[code] = maybe_term
parsed_patents.append(
{
"title": title,
"identifiers": identifiers,
"claim": claim,
"claim_contains_references": claim_contains_references,
"claim_code_list_terms": claim_code_list_terms,
"claim_code_term": claim_code_term,
"body": body,
"body_contains_references": body_contains_references,
"body_code_list_terms": body_code_list_terms,
"body_code_term": body_code_term,
}
)
if i % 100 == 0:
print("#", i)
df = pd.DataFrame(parsed_patents)
return df
parser = PatentParser(path + "H04N.txt")
patents_df = parser.parse_patent_file()
patents_df
patents_df.to_csv(path_out + "patents_dataframe.csv", index=False)
|
import pandas as pd
import numpy as np
from sklearn.preprocessing import MinMaxScaler
from sklearn.model_selection import GridSearchCV
from sklearn.ensemble import RandomForestClassifier
from xgboost import XGBClassifier, cv
import plotly.graph_objects as go
train_df = pd.read_csv("../input/mobile-price-classification/train.csv")
print(train_df.isnull().sum(axis=0))
print(train_df.info())
train = train_df.iloc[:, :-1]
label = train_df.iloc[:, -1]
train.head()
corr = train.corr()
fig = go.Figure()
fig.add_trace(go.Heatmap(z=corr.values, x=corr.index.values, y=corr.columns.values))
fig.show()
mc = MinMaxScaler()
mc.fit(train)
train = mc.transform(train)
rf = RandomForestClassifier()
rf_param_grid = {
"n_estimators": [5, 6, 7, 8, 9, 10, 11, 12],
"max_depth": [5, 6, 7, 8, 9, 10, 11, 12],
}
rf_grid = GridSearchCV(
estimator=rf,
param_grid=rf_param_grid,
scoring="accuracy",
cv=4,
return_train_score=True,
)
rf_grid.fit(train, label)
rf_grid_df = pd.DataFrame(rf_grid.cv_results_)
rf_grid_df.loc[rf_grid_df["rank_test_score"] == 1,]
from tensorflow.keras.models import Model, Sequential
from tensorflow.keras import layers, Input
from tensorflow.keras import losses, optimizers
from keras.utils.np_utils import to_categorical
from keras.callbacks import (
EarlyStopping,
LearningRateScheduler,
ReduceLROnPlateau,
ModelCheckpoint,
)
import matplotlib.pyplot as plt
train_np = np.array(train)
label_np = np.array(label)
input_shape = train.shape[1]
label_np = to_categorical(label_np)
label_np
input_tensor = Input(shape=(20,))
x = layers.Dense(128, activation="relu")(input_tensor)
x = layers.BatchNormalization()(x)
x = layers.Dense(64, activation="relu")(x)
x = layers.BatchNormalization()(x)
x = layers.Dense(64, activation="relu")(x)
x = layers.BatchNormalization()(x)
x = layers.Dense(64, activation="relu")(x)
x = layers.BatchNormalization()(x)
x = layers.Dense(64, activation="relu")(x)
x = layers.BatchNormalization()(x)
x = layers.Dense(32, activation="relu")(x)
x = layers.BatchNormalization()(x)
x = layers.Dense(16, activation="relu")(x)
x = layers.BatchNormalization()(x)
x = layers.Dense(8, activation="relu")(x)
x = layers.BatchNormalization()(x)
x = layers.Dense(4, activation="relu")(x)
output_tensor = layers.Dense(4, activation="softmax")(x)
model = Model(input_tensor, output_tensor)
model.compile(
optimizer=optimizers.Adam(),
loss=losses.categorical_crossentropy,
metrics=["accuracy"],
)
callback_list = [
EarlyStopping(monitor="accuracy", patience=2),
ReduceLROnPlateau(monitor="loss", factor=0.1, patience=1),
]
history = model.fit(
train_np, label_np, callbacks=callback_list, batch_size=8, epochs=100
)
acc = history.history["accuracy"]
# val_acc = history.history['val_acc']
loss = history.history["loss"]
# val_loss = history.history['val_loss']
epochs = range(1, len(acc) + 1)
plt.figure()
plt.plot(epochs, acc, "bo", label="Train Acc")
# plt.plot(epochs, val_acc, 'b', label='Validation Acc')
plt.title("Training and validation Acc")
plt.legend()
plt.figure()
plt.plot(epochs, loss, "bo", label="Train Loss")
# plt.plot(epochs, val_loss, 'b', label='Validation Loss')
plt.title("Training and validation Loss")
plt.legend()
plt.show()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
data = pd.read_csv(
"/kaggle/input/leetcode-indian-user-ratings/leetcode_indian_userrating.csv"
)
data
data.info()
data["country"].unique()
## In our analysis, username, country, Unnamed: 0 columns are not needed
data = data.drop(["username", "country", "Unnamed: 0"], axis=1)
data
## So, now we will found global_rating count between different buckets
## of rating.
data.describe()
## So, according to above table,
## for rating column, minimum is 1471.29, maximum is 2841.96.
## Therefore, we create the bucket size of 100.
def result1(x):
min1 = (x // 100) * 100
max1 = ((x // 100) + 1) * 100
return f"{min1}-{max1}"
data["bucket"] = data["rating"].apply(lambda x: result1(x))
data
## Now, we group by bucket column and take count of records of global id.
df = (
data.groupby("bucket")
.agg(total_praticipants_present=("global rank", "count"))
.reset_index()
)
df
list(df["total_praticipants_present"])
list(df["bucket"])
## Now, we will create a bar chart between rating and participants
x = list(df["bucket"])
y = list(df["total_praticipants_present"])
plt.bar(x, y)
plt.title("Rating vs Number of Indian participants present")
plt.xlabel("Rating")
plt.ylabel("Number of participants present")
plt.xticks(rotation=90)
plt.show()
plt.savefig("Leetcode_India_Users_data_analysis.png")
# ### According to this analysis, we can say that most participants from India present in [1500-1600) rating range. and least were present in more than or equal to 2400 rating.
# ## Now, we will do analysis for unitedstates participants rating
us_data = pd.read_csv(
"/kaggle/input/leetcode-indian-user-ratings/leetcode_unitedstates_userrating.csv"
)
us_data
us_data["country"].unique()
## So, same as above, columns country, username, Unnamed: 0 are not needed
us_data = us_data.drop(["country", "username", "Unnamed: 0"], axis=1)
us_data
us_data.info()
## it means, all entries are non-null.
us_data.describe()
def result_us(x):
min1 = (x // 100) * 100
max1 = ((x // 100) + 1) * 100
return f"{min1}-{max1}"
us_data["bucket"] = us_data["rating"].apply(lambda x: result_us(x))
us_data
## Now, we group by bucket column
df_us = (
us_data.groupby("bucket")
.agg(total_participants_count=("global rank", "count"))
.reset_index()
)
df_us
## Now, we bill draw a bar grpah.
x = list(df_us["bucket"])
y = list(df_us["total_participants_count"])
plt.bar(x, y)
plt.xlabel("Rating Range")
plt.ylabel("total participants present")
plt.title("Rating vs Total US Participants present")
plt.xticks(rotation=90)
plt.show()
plt.savefig("Leetcode_US_Users_data_analysis.png")
# ## Above graph clearly shows, most participants were present in rating range '1500-1600'.
## Here, some users are also present after rating range 2700. So, we will make a seperate bar graph for them
x = list(df_us["bucket"])[13:]
y = list(df_us["total_participants_count"])[13:]
plt.bar(x, y)
plt.xlabel("Rating Range >=2700")
plt.ylabel("Number of participants present")
plt.title("Rating Range vs Number of US participants present")
plt.xticks(rotation=90)
plt.show()
|
import pandas as pd
df = pd.read_csv("/kaggle/input/super-conductivity/Super-Conductivity.csv")
df.shape
df.head()
df.isnull().sum()
df.isna().sum()
df.describe()
# # Data Analysis and Visualization
import seaborn as sns
import matplotlib.pyplot as plt
g = sns.catplot(x="critical_temp", data=df, kind="count")
for ax in g.axes.flat:
for p in ax.patches:
ax.annotate(
p.get_height(),
xy=(p.get_x() + p.get_width() / 2.0, p.get_height()),
xytext=(0, 10),
textcoords="offset points",
ha="center",
va="center",
)
plt.title("Fig 1: Distribution of Critical Temperature", y=-0.2, fontsize=12)
plt.show()
plt.figure(figsize=(10, 10))
sns.heatmap(
df.corr(),
cbar=True,
square=True,
annot=True,
fmt=".3f",
annot_kws={"size": 10},
cmap="rocket",
linewidths=0.2,
linecolor="orange",
)
plt.title("Fig 2: Correlation Matrix ", y=-0.3, fontsize=12)
plt.show()
# # Data Preprocessing
data = df.drop(columns="critical_temp")
target = df["critical_temp"]
target = target.apply(lambda x: 0 if x < 5 else (1 if x < 7 else 2))
# # Splitting The Data
from sklearn.model_selection import train_test_split as t_t_s
from sklearn.metrics import accuracy_score
x_test, x_train, y_test, y_train = t_t_s(df, target, test_size=0.2, random_state=2)
# # Implementatng Perceprtron
from sklearn.linear_model import Perceptron
p = Perceptron(penalty="l2", random_state=4)
p.fit(x_train, y_train)
train_pred = p.predict(x_train)
test_pred = p.predict(x_test)
plt.figure(figsize=(7, 2))
plt.barh(
y=["Testing", "Training"],
width=[accuracy_score(test_pred, y_test), accuracy_score(train_pred, y_train)],
color=["green", "blue"],
)
plt.xlim(0, 1)
plt.xlabel("Accuracy")
plt.text(
accuracy_score(train_pred, y_train) + 0.01,
1,
f"{accuracy_score(train_pred, y_train):.2f}",
va="center",
)
plt.text(
accuracy_score(test_pred, y_test) + 0.01,
0,
f"{accuracy_score(test_pred,y_test):.2f}",
va="center",
)
plt.title("Fig 3: Training and Testing Accuracy Using Perceptron", y=-0.5)
plt.show()
from sklearn.tree import DecisionTreeClassifier
D_Tr = DecisionTreeClassifier(criterion="entropy", max_depth=3)
D_Tr.fit(x_train, y_train)
D_Tr_train_pred = D_Tr.predict(x_train)
D_Tr_test_pred = D_Tr.predict(x_test)
plt.figure(figsize=(7, 2))
plt.barh(
y=["Testing", "Training"],
width=[
accuracy_score(D_Tr_test_pred, y_test),
accuracy_score(D_Tr_train_pred, y_train),
],
color=["purple", "blue"],
)
plt.xlim(0, 1)
plt.xlabel("Accuracy")
plt.text(
accuracy_score(D_Tr_test_pred, y_test) + 0.01,
0,
f"{accuracy_score(D_Tr_test_pred, y_test):.2f}",
va="center",
)
plt.text(
accuracy_score(D_Tr_train_pred, y_train) + 0.01,
1,
f"{accuracy_score(D_Tr_train_pred, y_train):.2f}",
va="center",
)
plt.title(
"Fig 4: Training and Testing Accuracy Using Entropy in Decistion Tree", y=-0.5
)
# plt.show()
from sklearn.tree import plot_tree
plt.figure(figsize=(17, 10))
plot_tree(D_Tr, fontsize="10", rounded="True", filled=True)
plt.title("Fig 4: Decision Tree", y=0, fontsize="20")
plt.show()
# # Implementing SVM Model
from sklearn.svm import SVC
svc = SVC(kernel="rbf")
svc.fit(x_train, y_train)
svc_train_pred = svc1.predict(x_train)
svc_test_pred = svc1.predict(x_test)
plt.figure(figsize=(7, 2))
plt.barh(
y=["Testing", "Training"],
width=[
accuracy_score(y_test, svc_test_pred),
accuracy_score(y_train, svc_train_pred),
],
color=["green", "blue"],
)
plt.xlim(0, 1)
plt.xlabel("Accuracy")
plt.text(
accuracy_score(y_train, svc_train_pred) + 0.01,
1,
f"{accuracy_score(y_train,svc_train_pred):.2f}",
va="center",
)
plt.text(
accuracy_score(y_test, svc_test_pred) + 0.01,
0,
f"{accuracy_score(y_test, svc_test_pred):.2f}",
va="center",
)
plt.title("Fig 5: Training and Testing Accuracy Using SVM", y=-0.5)
plt.show()
# # Implementing SVM model with Pipeline
from sklearn.preprocessing import StandardScaler
from sklearn.decomposition import PCA
from sklearn.pipeline import Pipeline
pipe = Pipeline([("sc", StandardScaler()), ("pca", PCA()), ("svm", svc)])
pipe.fit(x_train, y_train)
pipeline_train_prediction = pipe.predict(x_train)
pipeline_test_prediction = pipe.predict(x_test)
plt.figure(figsize=(7, 2))
plt.barh(
y=["Testing", "Training"],
width=[
accuracy_score(pipeline_test_prediction, y_test),
accuracy_score(pipeline_train_prediction, y_train),
],
color=["purple", "blue"],
)
plt.xlim(0, 1)
plt.xlabel("Accuracy")
plt.text(
accuracy_score(pipeline_train_prediction, y_train) + 0.01,
1,
f"{accuracy_score(pipeline_train_prediction,y_train):.2f}",
va="center",
)
plt.text(
accuracy_score(pipeline_test_prediction, y_test) + 0.01,
0,
f"{accuracy_score(pipeline_test_prediction,y_test):.2f}",
va="center",
)
plt.title("Fig 6: Training and Testing Accuracy Using SVM with Pipeline", y=-0.5)
plt.show()
# # Ratio of the Data Model Test Accuracy
fig = plt.figure(figsize=(10, 10))
labels = "Perceptron", "Decision Tree", "SVM", "SVM with Pipeline"
sizes = [
accuracy_score(test_pred, y_test),
accuracy_score(D_Tr_test_pred, y_test),
accuracy_score(y_test, svc_test_pred),
accuracy_score(pipeline_test_prediction, y_test),
]
plt.pie(sizes, labels=labels, autopct="%.2f%%", startangle=0)
plt.axis("equal")
plt.title(
"\n Fig 7: Distribution of Testing Accuracy of Super-Conductivity Dataset using Different Models",
y=-0.1,
fontsize="18",
)
plt.show()
|
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import gc
from sklearn.model_selection import StratifiedKFold
from sklearn.metrics import roc_auc_score
import catboost as cb
import tensorflow as tf
import tensorflow_addons as tfa
from tensorflow.keras.utils import plot_model
from tensorflow.keras.models import Model
from tensorflow.keras.callbacks import EarlyStopping
from tensorflow.keras.callbacks import ReduceLROnPlateau
from tensorflow.keras.layers import Dense, Dropout, Input
seed = 228
np.random.seed(seed)
tf.random.set_seed(seed)
import warnings
warnings.filterwarnings("ignore")
train = pd.read_csv("/kaggle/input/playground-series-s3e12/train.csv")
test = pd.read_csv("/kaggle/input/playground-series-s3e12/test.csv")
sub = pd.read_csv("/kaggle/input/playground-series-s3e12/sample_submission.csv")
# concat dataset
df = pd.concat([train, test], axis=0)
df.head()
# ### About the dataset
df.shape
df.describe()
# ## Visualization
# pie plot for target variable
plt.figure(figsize=(8, 8))
plt.pie(
df["target"].value_counts(),
labels=df["target"].value_counts().index,
autopct="%.2f%%",
)
hfont = {"fontname": "serif", "weight": "bold"}
plt.title("distribution of target variable", size=15, **hfont)
plt.show()
# numerical data distribution plot
for i in df.columns:
if i != "target":
plt.figure(figsize=(15, 6))
sns.histplot(df[i], kde=True, bins=30, palette="hls")
plt.xticks(rotation=90)
plt.show()
# box plot
for i in df.columns:
if i != "target":
plt.figure(figsize=(15, 6))
sns.boxplot(x=df[i], data=df, palette="hls")
plt.xticks(rotation=90)
plt.show()
# violin plot
for i in df.columns:
if i != "target":
plt.figure(figsize=(15, 6))
sns.violinplot(x=df[i], data=df, palette="hls")
plt.xticks(rotation=90)
plt.show()
plt.figure(figsize=(15, 6))
sns.pairplot(data=df, palette="hls")
plt.show()
plt.figure(figsize=(20, 10))
df_corr = df.corr()
matrix = np.triu(df.corr())
sns.heatmap(df_corr, annot=True, linewidth=0.8, mask=matrix, cmap="rocket")
plt.show()
# ## Modeling
# feature importance function
def feature_imp_plot(df):
df["avg_imp"] = df[df.columns[1:]].mean(axis=1)
df.sort_values("avg_imp", ascending=False, inplace=True)
fig = plt.figure(figsize=(15, 0.40 * len(df)))
plt.title(
"Feature importance",
size=20,
y=1.0,
fontname="Calibri",
fontweight="bold",
color="#444444",
)
a = sns.barplot(
data=df,
x="avg_imp",
y="feature",
palette="Blues_d",
linestyle="-",
linewidth=1,
edgecolor="black",
)
plt.xlabel("")
plt.xticks([])
plt.ylabel("")
plt.yticks(size=11, color="#444444")
for i in ["right", "top", "bottom"]:
a.spines[i].set_visible(False)
for i in ["left"]:
a.spines[i].set_linewidth(0.5)
plt.show()
def norm_0to1(preds):
return (preds - np.min(preds)) / (np.max(preds) - np.min(preds))
def preds_plot(preds):
plt.figure(figsize=(12, 8))
plt.title(
"Distribution of Prediction",
size=20,
y=1.0,
fontname="Calibri",
fontweight="bold",
color="#444444",
)
a = sns.histplot(preds, color="#72bfd6", bins=100)
plt.xlabel("")
plt.xticks(fontname="Calibri", size=12)
plt.ylabel("")
plt.yticks([])
for s in ["right", "top", "left"]:
a.spines[s].set_visible(False)
plt.show()
# data split
X = train.drop("target", axis=1)
y = train["target"]
# ## Catboost
train_cb = train.copy()
# removed two patients data because,they are outliers
# after reomving them model performing well
train_cb.drop(
train[(train["gravity"] > 1.035) & (train["target"] == 1)].index, inplace=True
) # 1 patient
train_cb.drop(train[(train["ph"] > 7.9)].index, inplace=True) # 1 patient
X_cb = train_cb.drop("target", axis=1)
y_cb = train_cb["target"]
seed = 228
FOLDS = 10
cb_params = {
"depth": 3,
"learning_rate": 0.15,
"rsm": 0.5,
"subsample": 0.631,
"l2_leaf_reg": 69,
"min_data_in_leaf": 20,
"random_strength": 0.775,
"random_seed": 228,
"use_best_model": True,
"task_type": "CPU",
"bootstrap_type": "Bernoulli",
"grow_policy": "SymmetricTree",
"loss_function": "Logloss",
"eval_metric": "AUC",
}
df = pd.DataFrame({"feature": X.columns})
predictions, scores = np.zeros(len(test)), []
k = StratifiedKFold(n_splits=FOLDS, random_state=seed, shuffle=True)
for fold, (train_idx, val_idx) in enumerate(k.split(X_cb, y_cb)):
cb_train = cb.Pool(data=X_cb.iloc[train_idx], label=y_cb.iloc[train_idx])
cb_valid = cb.Pool(data=X_cb.iloc[val_idx], label=y_cb.iloc[val_idx])
model = cb.train(
params=cb_params,
dtrain=cb_train,
num_boost_round=10000,
evals=cb_valid,
early_stopping_rounds=500,
verbose=False,
)
df["fold_" + str(fold + 1)] = model.get_feature_importance()
val_preds = model.predict(cb_valid)
val_score = roc_auc_score(y_cb.iloc[val_idx], val_preds)
scores.append(val_score)
predictions += model.predict(test) / FOLDS
print(f"- FOLD {fold+1} AUC: {round(val_score, 4)} -")
del cb_train, cb_valid, val_preds, val_score, model
gc.collect()
print("*" * 45)
print(f"Mean AUC: {round(np.mean(scores), 4)}")
predictions = norm_0to1(predictions)
cb_preds = predictions.copy()
feature_imp_plot(df)
preds_plot(predictions)
# ## NN
# scaling
from sklearn.preprocessing import StandardScaler
sc = StandardScaler()
X = sc.fit_transform(X)
test = sc.transform(test)
X = X
y = y
test = test
def my_model():
x_input = Input(shape=(X.shape[-1]), name="input")
x1 = Dense(256, activation="relu")(x_input)
d1 = Dropout(0.1)(x1)
x2 = Dense(128, activation="relu")(d1)
d2 = Dropout(0.1)(x2)
x3 = Dense(64, activation="relu")(d2)
d3 = Dropout(0.1)(x3)
output = Dense(1, activation="sigmoid", name="output")(d3)
model = Model(x_input, output, name="nn_model")
return model
VERBOSE = False
BATCH_SIZE = 32
predictions, scores = [], []
lr = ReduceLROnPlateau(monitor="val_auc", factor=0.5, patience=5, verbose=VERBOSE)
es = EarlyStopping(
monitor="val_auc",
patience=15,
verbose=VERBOSE,
mode="max",
restore_best_weights=True,
)
k = StratifiedKFold(n_splits=FOLDS, random_state=seed, shuffle=True)
for fold, (train_idx, val_idx) in enumerate(k.split(X, y)):
X_train, X_val = X[train_idx], X[val_idx]
y_train, y_val = y[train_idx], y[val_idx]
model = my_model()
model.compile(
optimizer="adam",
loss=tfa.losses.SigmoidFocalCrossEntropy(alpha=0.1, gamma=3),
metrics="AUC",
)
model.fit(
X_train,
y_train,
validation_data=(X_val, y_val),
epochs=300,
verbose=VERBOSE,
batch_size=BATCH_SIZE,
class_weight={0: 1, 1: 10},
callbacks=[lr, es],
)
y_pred = norm_0to1(model.predict(X_val))
val_score = roc_auc_score(y_val, y_pred)
scores.append(val_score)
predictions.append(model.predict(test))
print(f"- FOLD {fold+1} AUC: {round(val_score, 4)} -")
print(f"Mean AUC: {round(np.mean(scores), 4)}")
predictions = norm_0to1(np.squeeze(np.mean(predictions, axis=0)))
nn_preds = predictions.copy()
preds_plot(predictions)
# ## Submission
sub["target"] = (cb_preds * 0.7 + nn_preds * 0.3) * 0.4
sub.to_csv("submission_.csv", index=False)
preds_plot(sub["target"])
sub.head()
|
# # Sales Analysis
# **Introduction**
# Here, in this sales analysis, we focus on the data that represent the sales figure of various items in an electronics store company.
# The company has shops in multiple cities. It sells items that range from AAA batteries to Iphone and charging cables to washers and dryers.
# Due to recent tough financial period, the company has decided to scale back from certain locations and has decided to renegotiate some of the contracts with suppliers and transporters.
# Our objective here is to analyze data and to suggest best ways to cut cost and where more investment should be directed to get better results.
# This Project will be divided into 6 phases\
# ASK - Where we ask business questions\
# PREPARE - Where we get the data for the analysis\
# PROCESS - Where we get the data arranged for the analysis\
# ANALYZE - Where we analyze the data\
# SHARE - Where we share the data after analysis\
# ACT - Where we recommend course of action ahead
# # ASK Phase
# Here we divide the objective into three distinct tasks.
# - To find out which month did the store recorded most sales
# - To find out which city has generated most sales and which city has generated least sales
# - To find out which products were sold most and which products were sold least
# *Looking at the data, we can find that the data has one single specific identifier that can cause an issue. That is that the dataset has a column called Purchase_Address. Although this address can not be linked to any ohter identifier such as a phone number, social security number, or name, it must be specified that such data points needed to be collected more securely and the exact addresses apart from the City, State and Zip codes must be masked or completely removed whatsoever.*
# The data that we have here is available in 7 columns.
# Index\
# Order_ID\
# Product\
# Quantity_Ordered\
# Price_Each\
# Order_Date_Time\
# Purchase_Address
# **Data_Credibility :**\
# The dataset has almost all the data that we might need to make the conclusions for the questions asked above.\
# But the performance of a business is determined by many facotrs other than the factors mentioned above. Alse the data is dynamic, it will change every second and it is not necessarily true that all the assumptions and conclusions made on the data available will be true all the time.
# # PREPARE Phase
# ## Merging Files into one
import warnings
warnings.filterwarnings("ignore")
import os
os.listdir(
"/kaggle/input/sales-data-analysis"
) # Checking if files have been successfully uploaded
files = [file for file in os.listdir("/kaggle/input/sales-data-analysis")]
files.sort()
print(files)
import pandas as pd
all_month = pd.DataFrame() # Creating our main dataframe
for file in files:
df = pd.read_csv(
"/kaggle/input/sales-data-analysis/" + file,
delimiter=",",
encoding="unicode_escape",
)
all_month = pd.concat([all_month, df]) # Combining our files into one
print(all_month)
# # PROCESS Phase
# ## Here we try to get the data for analysis in the format that supports our analysis requirements
# Now for first task we need a new special column for month of the order\
# We would also need all the values in that month column to be integers since month numbers are integers\
# Considering our objective, we would need certain data in the file in certain type.\
# For example\
# Price in numerical value\
# Quantity in integer
# all_month['Month'] = all_month['Order_Date_Time'].str[0:2]
# running the line above gives us an error that there are NaN values in our databases,
# which means our data might have blank spaces to remove this first we would have to
# find out how many rows have the issue of NaN
nan_df = all_month[all_month.isna().any(axis=1)]
print(nan_df)
all_month = all_month.dropna(how="all") # Dropping the rows with NaN values
print(all_month)
# all_month['Month'] = all_month['Order_Date_Time'].str[0:2]
# Now that we have removed the NaN values, we get another error.
# For some reason, month column is having error that it has 'or' in it so let's get that too
# In the code below, we simply reject the values that have value 'or' in our supposed to be month column.
all_month["Month"] = all_month["Order_Date_Time"].str[0:2]
all_month = all_month[all_month["Month"] != "Or"]
print(all_month)
# Now that we have extracted the month column, we want that column as an integer since month numbers are integers, for that we use following code.
all_month["Month"] = all_month["Month"].astype("int32")
all_month.head()
all_month.dtypes
# Below, we apply the conversion principle that we applied to month column to other columns Quantity ordered cannot be a string, it has to be a number and Price need to be a numeric value.
all_month["Quantity_Ordered"] = pd.to_numeric(all_month["Quantity_Ordered"])
all_month["Price_Each"] = pd.to_numeric(all_month["Price_Each"])
all_month.dtypes
# Now that we have price and quantity column ready for processing, we can get a new sales column to get the idea of total sales.
all_month["sales"] = all_month["Quantity_Ordered"] * all_month["Price_Each"]
all_month.head()
# # ANALYZE Phase
# Now we try to get the data grouped and plotted so we can analyze it
for_month = all_month.groupby("Month") # Grouping data for first objective task
print(for_month.sum())
months = [month for month, something in for_month]
sales = for_month.sum()["sales"]
import matplotlib.pyplot as plt
plt.bar(months, sales)
plt.title("Month_Sales_Data")
plt.xlabel("Months")
plt.ylabel("Sales_In_That_Month")
plt.xticks(months)
plt.ticklabel_format(style="plain", axis="y")
plt.show()
# ## Conclusion 1
# Here we find out that the we have most sales in December and least sales in January.
# To find out the city-wise sales figure,we first would have to figure out how to extract the city and state information int one column.
def get_city(x):
city = x.split(",")[1] # Function to get the city out of Purchase Address
return city
def get_state(x):
state = x.split(",")[2].split(" ")[
1
] # Function to get the state out of Purchase Address
return state
all_month["City"] = all_month["Purchase_Address"].apply(
lambda x: (get_city(x)) + " " + (get_state(x))
)
all_month.head()
# Then we group by city name and plot it against sales just the way we did with the month-wise sales data
for_city = all_month.groupby("City") # Grouping data for second objective task
for_city.sum()
Cities = [city for city, something in for_city]
Sales_data = for_city.sum()["sales"]
plt.bar(Cities, Sales_data)
plt.title("City_Sales_Data")
plt.xlabel("Cities")
plt.ylabel("City_Wise_Sales")
plt.xticks(Cities, rotation="vertical")
plt.ticklabel_format(style="plain", axis="y")
plt.show()
# ## Conclusion 2
# Here we find out that we have most sales from San-Fransisco and least sales from Portland.
# Finally, to get the insight on last question, we group by products
for_product = all_month.groupby("Product")
for_product.sum()
# Above, we see that along with quantity ordered and sales,Price and month also got grouped since all these values are integers/floating points. But we have to ignore it and focus only on quantity ordered and sales.
product_list = [product for product, something in for_product]
Quantity_sold = for_product.sum()["Quantity_Ordered"]
plt.bar(product_list, Quantity_sold)
plt.title("Product_Quantity_Sold_Data")
plt.xlabel("Products")
plt.ylabel("Quantity Ordered")
plt.xticks(product_list, rotation="vertical")
plt.ticklabel_format(style="plain", axis="y")
plt.show()
# From the above plot, we can conclude that maximum orders are either for the AA and AAA batteries or involve these items, But this in no way can be used to determine product and sales relation. For that we need a pie chart that will put the Products against their share of sales amount.
# To plot a pie chart, we first need our products in str format and sales data separated and put against each product in int format. Below we do that.
type(product_list[0]) # Checking for format of products
product_sales = for_product.sum()["sales"] # Selecting sales from grouped series
# plt.bar(product_list,product_sales)
# plt.xticks(product_list,rotation = 'vertical')
# plt.ticklabel_format(style = 'plain',axis = 'y')
# plt.show()
# for_product.sum().values[0][3] # Checking attempts to extract the sales figure from grouped series
sales_product = [] # List where we will store the sales figure values
for i in range(len(product_list)):
sales_product.append(int(for_product.sum().values[i][3]))
sales_product # Checking if we got the list containing sales figure values
plt.pie(
sales_product, labels=product_list, radius=2.5
) # plotting the pie chart we need
font = {"family": "normal", "weight": "regular", "size": 20} # adjusting font
plt.rc("font", **font) # implementing font details
plt.title("Product_Wise Sales Representation", loc="right")
plt.show()
|
# İş Problemi
# Özellikleri belirtildiğinde kişilerin diyabet hastası olup olmadıklarını tahmin
# edebilecek bir makine öğrenmesi modeli geliştirilmesi istenmektedir. Modeli
# geliştirmeden önce gerekli olan veri analizi ve özellik mühendisliği adımlarını
# gerçekleştirmeniz beklenmektedir.
# Veri Seti Hikayesi
# Veri seti ABD'deki Ulusal Diyabet-Sindirim-Böbrek Hastalıkları Enstitüleri'nde tutulan büyük veri setinin parçasıdır. ABD'deki
# Arizona Eyaleti'nin en büyük 5. şehri olan Phoenix şehrinde yaşayan 21 yaş ve üzerinde olan Pima Indian kadınları üzerinde
# yapılan diyabet araştırması için kullanılan verilerdir.
# Hedef değişken "outcome" olarak belirtilmiş olup; 1 diyabet test sonucunun pozitif oluşunu, 0 ise negatif oluşunu belirtmektedir.
# 9 Değişken 768 Gözlem 24 KB
# Pregnancies: Hamilelik sayısı
# Glucose: Oral glikoz tolerans testinde 2 saatlik plazma glikoz konsantrasyonu
# Blood Pressure: Kan Basıncı (Küçük tansiyon) (mm Hg)
# SkinThickness: Cilt Kalınlığı
# Insulin: 2 saatlik serum insülini (mu U/ml)
# DiabetesPedigreeFunction: Fonksiyon (Oral glikoz tolerans testinde 2 saatlik plazma glikoz konsantrasyonu)
# BMI: Vücut kitle endeksi
# Age: Yaş (yıl)
# Outcome: Hastalığa sahip (1) ya da değil (0)
# Proje Görevleri
# Görev 1 : Keşifçi Veri Analizi
# Adım 1: Genel resmi inceleyiniz.
# Adım 2: Numerik ve kategorik değişkenleri yakalayınız.
# Adım 3: Numerik ve kategorik değişkenlerin analizini yapınız.
# Adım 4: Hedef değişken analizi yapınız. (Kategorik değişkenlere göre hedef değişkenin ortalaması, hedef değişkene göre
# numerik değişkenlerin ortalaması)
# Adım 5: Aykırı gözlem analizi yapınız.
# Adım 6: Eksik gözlem analizi yapınız.
# Adım 7: Korelasyon analizi yapınız.
# Görev 2 : Feature Engineering
# Adım 1: Eksik ve aykırı değerler için gerekli işlemleri yapınız. Veri setinde eksik gözlem bulunmamakta ama Glikoz, Insulin vb.
# değişkenlerde 0 değeri içeren gözlem birimleri eksik değeri ifade ediyor olabilir. Örneğin; bir kişinin glikoz veya insulin değeri 0
# olamayacaktır. Bu durumu dikkate alarak sıfır değerlerini ilgili değerlerde NaN olarak atama yapıp sonrasında eksik
# değerlere işlemleri uygulayabilirsiniz.
# Adım 2: Yeni değişkenler oluşturunuz.
# Adım 3: Encoding işlemlerini gerçekleştiriniz.
# Adım 4: Numerik değişkenler için standartlaştırma yapınız.
# Adım 5: Model oluşturunuz.
import numpy as np
import pandas as pd
import seaborn as sns
from matplotlib import pyplot as plt
# !pip install missingno
import missingno as msno
from datetime import date
from sklearn.metrics import accuracy_score
from sklearn.model_selection import train_test_split
from sklearn.neighbors import LocalOutlierFactor
from sklearn.preprocessing import (
MinMaxScaler,
LabelEncoder,
StandardScaler,
RobustScaler,
)
pd.set_option("display.max_columns", None)
pd.set_option("display.max_rows", None)
pd.set_option("display.float_format", lambda x: "%.3f" % x)
pd.set_option("display.width", 500)
def load():
data = pd.read_csv("/kaggle/input/pima-indians-diabetes-database/diabetes.csv")
return data
df = load()
df.head()
# Görev 1 : Keşifçi Veri Analizi
# Adım 1: Genel resmi inceleyiniz.
def check_df(dataframe):
print("############## Shape #############")
print(dataframe.shape)
print("############## Type #############")
print(dataframe.dtypes)
print("############## Head #############")
print(dataframe.head())
print("############## Tail #############")
print(dataframe.tail())
print("############## NA #############")
print(dataframe.isnull().sum())
print("############## Quantiles #############")
print(dataframe.describe([0, 0.05, 0.5, 0.95, 0.99, 1]).T)
check_df(df)
# Adım 2: Numerik ve kategorik değişkenleri yakalayınız.
# Adım 3: Numerik ve kategorik değişkenlerin analizini yapınız.
def grab_col_names(dataframe, cat_th=10, car_th=20):
"""
Veri setindeki kategorik, numerik ve kategorik fakat kardinal değişkenlerin isimlerini verir.
Not: Kategorik değişkenlerin içerisine numerik görünümlü kategorik değişkenler de dahildir.
Parameters
------
dataframe: dataframe
Değişken isimleri alınmak istenilen dataframe
cat_th: int, optional
numerik fakat kategorik olan değişkenler için sınıf eşik değeri
car_th: int, optinal
kategorik fakat kardinal değişkenler için sınıf eşik değeri
Returns
------
cat_cols: list
Kategorik değişken listesi
num_cols: list
Numerik değişken listesi
cat_but_car: list
Kategorik görünümlü kardinal değişken listesi
Examples
------
import seaborn as sns
df = sns.load_dataset("iris")
print(grab_col_names(df))
Notes
------
cat_cols + num_cols + cat_but_car = toplam değişken sayısı
num_but_cat cat_cols'un içerisinde.
Return olan 3 liste toplamı toplam değişken sayısına eşittir: cat_cols + num_cols + cat_but_car = değişken sayısı
"""
# cat_cols, cat_but_car
cat_cols = [col for col in dataframe.columns if dataframe[col].dtypes == "O"]
num_but_cat = [
col
for col in dataframe.columns
if dataframe[col].nunique() < cat_th
and dataframe[ # ilgili değişkendeki eşsiz değer sayısana bak. eğer belirlediğim thrsholdan küçükse ve
col
].dtypes
!= "O"
] # değişken tipi object değilse numeric ama kategorikleri yakala.
cat_but_car = [
col
for col in dataframe.columns
if dataframe[col].nunique() > car_th
and dataframe[ # ilgili değişkendeki eşsiz değer sayısana bak. eğer belirlediğim thrsholdan büyükse ve
col
].dtypes
== "O"
] # değişken tipi object ise kategorik ama kardinalsin yani çok fazla bilgi taşıyor.
cat_cols = (
cat_cols + num_but_cat
) # cat_cols listesi baştan oluştırıyoruz.cat_cols + num_but_cat değişeknleri al ve
cat_cols = [
col for col in cat_cols if col not in cat_but_car
] # cat_cols a kategorik ama kardinal değişkenlerde olmayanları da seç
# num_cols
num_cols = [
col for col in dataframe.columns if dataframe[col].dtypes != "O"
] # tipi objecten farklı olanları getir.
num_cols = [
col for col in num_cols if col not in num_but_cat
] # num görünen ve kategorik olanları çıkar. bunları ekleme.
print(f"Observations: {dataframe.shape[0]}")
print(f"Variables: {dataframe.shape[1]}")
print(f"cat_cols: {len(cat_cols)}")
print(f"num_cols: {len(num_cols)}")
print(f"cat_but_car: {len(cat_but_car)}")
print(f"num_but_cat: {len(num_but_cat)}")
return cat_cols, num_cols, cat_but_car
cat_cols, num_cols, cat_but_car = grab_col_names(df)
# Adım 4: Hedef değişken analizi yapınız. (Kategorik değişkenlere göre hedef değişkenin ortalaması, hedef değişkene göre numerik değişkenlerin ortalaması)
# kategorik değişenler
def cat_summary(dataframe, col_name, plot=False):
print(
pd.DataFrame(
{
col_name: dataframe[col_name].value_counts(),
"Ratio": 100 * dataframe[col_name].value_counts() / len(dataframe),
}
)
)
print("##########################################")
if plot:
sns.countplot(x=dataframe[col_name], data=dataframe)
plt.show(block=True)
for col in cat_cols:
cat_summary(df, col)
# numerik değişkenler
def num_summary(dataframe, numerical_col, plot=False):
quantiles = [0.05, 0.10, 0.20, 0.30, 0.40, 0.50, 0.60, 0.70, 0.80, 0.90, 0.95, 0.99]
print(dataframe[numerical_col].describe(quantiles).T)
if plot:
dataframe[numerical_col].hist(bins=20)
plt.xlabel(numerical_col)
plt.title(numerical_col)
plt.show(block=True)
for col in num_cols:
num_summary(df, col, plot=True)
# numerik değişkenlerin hedef değişkene göre analizi
def target_summary_with_num(dataframe, target, numerical_col):
print(dataframe.groupby(target).agg({numerical_col: "mean"}), end="\n\n\n")
for col in num_cols:
target_summary_with_num(df, "Outcome", col)
# Adım 5: Aykırı gözlem analizi yapınız.
for col in num_cols:
sns.boxplot(x=df[col])
plt.show(block=True)
def outlier_thresholds(dataframe, col_name, q1=0.25, q3=0.75):
quartile1 = dataframe[col_name].quantile(q1)
quartile3 = dataframe[col_name].quantile(q3)
interquantile_range = quartile3 - quartile1
up_limit = quartile3 + 1.5 * interquantile_range
low_limit = quartile1 - 1.5 * interquantile_range
return low_limit, up_limit
def check_outlier(dataframe, col_name):
low_limit, up_limit = outlier_thresholds(dataframe, col_name)
if dataframe[
(dataframe[col_name] > up_limit) | (dataframe[col_name] < low_limit)
].any(axis=None):
return (
True # if koşulunda aykırı gzlem varsa true dönecek. yoksa else e geçecek.
)
else:
return False
for col in num_cols:
print(col, check_outlier(df, col))
# Adım 6: Eksik gözlem analizi yapınız.
# eksik gozlem var mı yok mu sorgusu - False
df.isnull().values.any()
# Adım 7: Korelasyon analizi yapınız.
df.corr()
corr_matrix = df.corr()
sns.clustermap(corr_matrix, annot=True, fmt=".2f")
plt.show(block=True)
# Görev 2 : Feature Engineering
# Adım 1: Eksik ve aykırı değerler için gerekli işlemleri yapınız. Veri setinde eksik gözlem bulunmamakta ama Glikoz, Insulin vb.
# değişkenlerde 0 değeri içeren gözlem birimleri eksik değeri ifade ediyor olabilir. Örneğin; bir kişinin glikoz veya insulin değeri 0
# olamayacaktır. Bu durumu dikkate alarak sıfır değerlerini ilgili değerlerde NaN olarak atama yapıp sonrasında eksik
# değerlere işlemleri uygulayabilirsiniz.
na_values = [
col
for col in df.columns
if (df[col].min() == 0 and col not in ["Pregnancies", "Outcome"])
]
df[na_values] = df[na_values].replace(0, np.NaN)
df.isnull().sum()
def missing_values_table(
dataframe, na_name=False
): # na_name eksik değerlerin barındığı değişkenlerin ismini ve verisetine oranlarını verir.
na_columns = [col for col in dataframe.columns if dataframe[col].isnull().sum() > 0]
n_miss = dataframe[na_columns].isnull().sum().sort_values(ascending=False)
ratio = (
dataframe[na_columns].isnull().sum() / dataframe.shape[0] * 100
).sort_values(ascending=False)
missing_df = pd.concat(
[n_miss, np.round(ratio, 2)], axis=1, keys=["n_miss", "ratio"]
)
print(missing_df, end="\n")
if na_name:
return na_columns
missing_values_table(df, True)
# eksik değerlerin birbirleriyle ilişkisi?
msno.heatmap(df)
plt.show(block=True)
# eksik değerleri KNN ile giderme
cat_cols, num_cols, cat_but_car = grab_col_names(df)
dff = pd.get_dummies(df[cat_cols + num_cols], drop_first=True)
dff.head()
scaler = MinMaxScaler() # değerleri 1-0 a dönüştürerek standartlaştrma işlemi
dff = pd.DataFrame(scaler.fit_transform(dff), columns=dff.columns)
from sklearn.impute import KNNImputer
imputer = KNNImputer(n_neighbors=5)
dff = pd.DataFrame(imputer.fit_transform(dff), columns=dff.columns)
dff.head()
dff = pd.DataFrame(scaler.inverse_transform(dff), columns=dff.columns)
# kontroller (eksik değerleri KNN ile doldurmuştuk. eski yeni halini İnsulin-Glikoz ile karşılaştırılır.)
df["Insulin_imputed_knn"] = dff[["Insulin"]]
df.loc[df["Insulin"].isnull(), ["Insulin", "Insulin_imputed_knn"]]
df.head()
df["Glucose_imputed_knn"] = dff[["Glucose"]]
df.loc[df["Glucose"].isnull(), ["Glucose", "Glucose_imputed_knn"]]
# eksik değrelerin yerini imputed_knn ler alsın.
df["Insulin"] = dff["Insulin"]
df["Glucose"] = dff["Glucose"]
df["SkinThickness"] = dff["SkinThickness"]
df["BloodPressure"] = dff["BloodPressure"]
df["BMI"] = dff["BMI"]
df = df.drop(["Insulin_imputed_knn", "Glucose_imputed_knn"], axis=1)
df.head()
df.columns
# AYKIRI DEGERLER
# aykırı değerleri thresholdlar ile değiştir.
def replace_with_thresholds(dataframe, variable):
low_limit, up_limit = outlier_thresholds(dataframe, variable)
dataframe.loc[(dataframe[variable] < low_limit), variable] = low_limit
dataframe.loc[(dataframe[variable] > up_limit), variable] = up_limit
for col in num_cols:
replace_with_thresholds(df, col)
# tekrar kontrol ettiğimizde False.
for col in num_cols:
print(col, check_outlier(df, col))
# Adım 2: Yeni değişkenler oluşturunuz.
df.loc[(df["Glucose"] < 70), "N_Glucose"] = "Hipoglisemi"
df.loc[(df["Glucose"] >= 70) & (df["Glucose"] < 100), "N_Glucose"] = "Normal"
df.loc[(df["Glucose"] >= 100) & (df["Glucose"] < 126), "N_Glucose"] = "Hidden_Diabetes"
df.loc[(df["Glucose"] >= 126), "N_Glucose"] = "Diabetes"
df.loc[(df["BloodPressure"] < 70), "N_BloodPressure"] = "Optimal"
df.loc[
(df["BloodPressure"] >= 70) & (df["BloodPressure"] < 90), "N_BloodPressure"
] = "Normal"
df.loc[(df["BloodPressure"] >= 90), "N_BloodPressure"] = "High"
hr = 22.5 * 0.10
df["HOMA-IR"] = (df["Glucose"] * df["Insulin"]) % hr
df.loc[(df["BMI"] < 25), "N_BMI"] = "Normal"
df.loc[(df["BMI"] >= 25) & (df["BMI"] < 30), "N_BMI"] = "Overweight"
df.loc[(df["BMI"] >= 30) & (df["BMI"] < 35), "N_BMI"] = "Obese"
df.loc[(df["BMI"] >= 35) & (df["BMI"] < 40), "N_BMI"] = "Advanced_Obese"
df.loc[(df["BMI"] >= 40), "N_BMI"] = "Morbid_Obese"
df.loc[(df["Age"] >= 18) & (df["Age"] < 30), "N_Age"] = "Young_Female"
df.loc[(df["Age"] >= 30) & (df["Age"] < 45), "N_Age"] = "Mature_Female"
df.loc[(df["Age"] >= 45) & (df["Age"] < 65), "N_Age"] = "Senior_Female"
df.loc[(df["Age"] >= 65), "N_Age"] = "Elder_Female"
df.head(10)
# Adım 3: Encoding işlemlerini gerçekleştiriniz.
cat_cols, num_cols, cat_but_car = grab_col_names(df)
binary_cols = [
col
for col in df.columns
if df[col].dtype not in [int, float] and df[col].nunique() == 2
]
cat_cols = [
col for col in cat_cols if col not in binary_cols and col not in ["OUTCOME"]
]
def one_hot_encoder(dataframe, categorical_cols, drop_first=True):
dataframe = pd.get_dummies(
dataframe, columns=categorical_cols, drop_first=drop_first
)
return dataframe
ohe_cols = [col for col in df.columns if 10 >= df[col].nunique() > 2]
df = one_hot_encoder(df, ohe_cols)
df.head()
# Adım 4: Numerik değişkenler için standartlaştırma yapınız.
scaler = StandardScaler()
df[num_cols] = scaler.fit_transform(df[num_cols])
# Adım 5: Model oluşturunuz.
# Random Forest Classifier
y = df["Outcome"]
X = df.drop(["Outcome"], axis=1)
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.30, random_state=17
)
from sklearn.ensemble import RandomForestClassifier
rf_model = RandomForestClassifier(random_state=46).fit(X_train, y_train)
y_pred = rf_model.predict(X_test)
accuracy_score(y_pred, y_test)
# 0.8095238095238095 doğruluk oranı / başarı oranı testi.
# bağımsız değişkenlerdeki benzer özelliklere sahip gelen kişilerin diyabet olup olmama olasılığı %81 başarı oranı ile doğru tahmin edebilirim.
# KNN
from sklearn.metrics import classification_report, roc_auc_score
from sklearn.model_selection import GridSearchCV, cross_validate
from sklearn.neighbors import KNeighborsClassifier
from sklearn.preprocessing import StandardScaler
knn_model = KNeighborsClassifier().fit(X, y)
random_user = X.sample(1, random_state=45)
knn_model.predict(random_user)
# Confusion matrix için y_pred:
y_pred = knn_model.predict(X)
# AUC için y_prob: (y_prob olasılık degerleri üzerinden)
y_prob = knn_model.predict_proba(X)[:, 1]
print(classification_report(y, y_pred))
# AUC (ROC Eğrisi)
roc_auc_score(y, y_prob)
# acc 0.84 # 0.84 başarılı sınıflandırma oranı. %16sinde tahmin başarısız.
# f1 0.77
# auc 0.92
# 3 metrik üzerinden genel başarımızı görüyoruz.
# Presicion %78 (1 sınfına yönelik tahminlrin başarısı), recall %76 (gerçekte 1 olanları 1 olarak tahmın etme), f1 harmonık ortalama
# CV ile 5 katlı çapraz doğrulama:
cv_results = cross_validate(
knn_model, X, y, cv=5, scoring=["accuracy", "f1", "roc_auc"]
) # knnmodel, blı ve bsız degıskenler, cv=kac katlı, scorıng=kullanılmak istenen skorlar
cv_results["test_accuracy"].mean() # 0.78
cv_results["test_f1"].mean() # 0.68
cv_results["test_roc_auc"].mean() # 0.84
knn_model.get_params()
# 5. Hyperparameter Optimization
# amaç komşuluk sayısını değiştirerek en optimum komşuluk sayısının ne olacağını bulmak.
knn_model = KNeighborsClassifier()
knn_model.get_params()
knn_params = {"n_neighbors": range(2, 50)}
knn_gs_best = GridSearchCV(knn_model, knn_params, cv=5, n_jobs=-1, verbose=1).fit(X, y)
knn_gs_best.best_params_
# {'n_neighbors': 13} 13 komşuluk sayısı en iyisi,
# final modeli bununla kurarsam daha başarılı bir sonuç elde edebileceğim
# 6. Final Model
knn_final = knn_model.set_params(**knn_gs_best.best_params_).fit(X, y)
cv_results = cross_validate(
knn_final, X, y, cv=5, scoring=["accuracy", "f1", "roc_auc"]
)
cv_results["test_accuracy"].mean() # %79
cv_results["test_f1"].mean() # %68
cv_results["test_roc_auc"].mean() # %85
random_user = X.sample(
1
) # random kullanıcı seçildiğinde bu kişinin diyabet hastası olup olmadığı
knn_final.predict(random_user) # array([0] diyabet hastası değil)
# Logistic Regression
from sklearn.preprocessing import RobustScaler
from sklearn.linear_model import LogisticRegression
from sklearn.metrics import (
accuracy_score,
roc_auc_score,
confusion_matrix,
classification_report,
plot_roc_curve,
)
from sklearn.model_selection import train_test_split, cross_validate
log_model = LogisticRegression().fit(X, y)
log_model.intercept_
log_model.coef_
# tahmin
y_pred = log_model.predict(X) # tahmin edilen değerlerin ilk 10 tanesi
y_pred[0:10]
y[0:10] # gerçek değerlerin ilk 10 tanesi
# karmaşıklık matrisi
def plot_confusion_matrix(y, y_pred):
acc = round(accuracy_score(y, y_pred), 2)
cm = confusion_matrix(y, y_pred)
sns.heatmap(cm, annot=True, fmt=".0f")
plt.xlabel("y_pred")
plt.ylabel("y")
plt.title("Accuracy Score: {0}".format(acc), size=10)
plt.show(block=True)
plot_confusion_matrix(y, y_pred)
print(classification_report(y, y_pred))
# model doğrulamadan önceki oranlar
# Accuracy: 0.80
# Precision: 0.74
# Recall: 0.67
# F1-score: 0.70
# Model Validation: Holdout
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.20, random_state=17
)
log_model = LogisticRegression().fit(X_train, y_train)
y_pred = log_model.predict(X_test)
y_prob = log_model.predict_proba(X_test)[:, 1]
print(classification_report(y_test, y_pred))
# model doğrulamadan önceki oranlar
# Accuracy: 0.80
# Precision: 0.74
# Recall: 0.67
# F1-score: 0.70
# model doğruladıktan sonraki oranlar
# Accuracy: 0.81
# Precision: 0.80
# Recall: 0.63
# F1-score: 0.71
plot_roc_curve(log_model, X_test, y_test)
plt.title("ROC Curve")
plt.plot([0, 1], [0, 1], "r--")
plt.show(block=True)
# AUC değeri %89
# Model Validation: 10-Fold Cross Validation
y = df["Outcome"]
X = df.drop(["Outcome"], axis=1)
log_model = LogisticRegression().fit(X, y)
cv_results = cross_validate(
log_model, X, y, cv=5, scoring=["accuracy", "precision", "recall", "f1", "roc_auc"]
)
cv_results["test_accuracy"].mean()
# Accuracy: 0.7826 (ortalama acc. değeri) (doğru sınıflandırma oranı %78
cv_results["test_precision"].mean()
# Precision: 0.7196
cv_results["test_recall"].mean()
# Recall: 0.6234 # gerçekte 1 olanları tahmin etme başarımız %59
cv_results["test_f1"].mean()
# F1-score: 0.6673
cv_results["test_roc_auc"].mean()
# AUC: 0.8567
# Prediction for A New Observation
X.columns
random_user = X.sample(1, random_state=45) # dışarıdan bir tane kullanıcı gelsin
random_user
log_model.predict(
random_user
) # kullanıcının bilgisini verdiğimde bağımsız değişkeni verecek.
# array([1], dtype=int64) 1 döndü yani bu kişi diyabettir
|
# ### Build your own Neural Network from scratch using only Numpy
# Deep Learning has become a popular topic in recent times. It involves emulating the neural structure of the human brain through a network of nodes called a **Neural Network**. While our brain's neurons have physical components like nucleus, dendrites, and synapses, Neural Network neurons are interconnected and have weights and biases assigned to them.
# A neural network typically consists of an input layer, an output layer, and one or more hidden layers. In conventional neural networks, all nodes in these layers are interconnected to form a dense network. However, there are cases where certain nodes in the network are not connected to others, which are referred to as **Sparse Neural Networks**. InceptionNet models for image classification use Sparse Neural Networks. The following figure illustrates the structure of a neural network.
# 
# The neurons will be activated or fired, which is the input will be passed through the neuron to the next layer, this process is called as **Feedforward**. Each neuron inside the neural network will have a linear function using weights and biases like the following equation. The input will be transformed using the function below and produce a new output to the next layer.
# 
# You can see at the end there will be something called as the ***Activation function***. An activation function is a function which decides wheather the neuron needs to be activated or not. Some people say, a neuron without an activation function is just a linear regression model. There are several activation functions called Sigmoid, Softmax, Tanh, ReLu and many more. We'll see about activation functions in detail later.
# let's start to build a neural network from scratch. We'll get into code without much further ado. Let's start by importing numpy library for linear algebra functions.
#
import numpy as np # linear algebra
# **Numpy** is a python library which can be used to implement linear algebra functions. So for creating a neural network we need the base for building a nueral network, neurons. We'll create a class neuron to implement the weights and biases. And a function called as feedforward, which implements the process of feedforward equation as shown in the below figure.
# 
class Neuron:
def __init__(self, weights, bias):
self.weights = weights
self.bias = bias
def feedforward(self, x):
return np.dot(weights, x) + bias
# The above code provides the structure of a neuron. Now we'll verify it by initializing values to weights and biases, and implementing the feedforward network.
weights = np.array([0, 1])
bias = 1
n = Neuron(weights, bias)
n.feedforward([1, 1])
# Now we'll see the implementation of sigmoid and ReLu activation functions. The equation of sigmoid, softmax and ReLu activation functions are given below.
# 
# 
# 
# 
# We are gonna define two functions sigmoid, softmax and ReLu containing the above equations.
def sigmoid(input):
return 1 / (1 + np.exp(input))
def relu(input):
return max(0, input)
def softmax(input):
return np.exp(a) / np.sum(np.exp(a))
|
# # AFL Data Scrape from afltables.com
import pandas as pd
import requests
from bs4 import BeautifulSoup
import re
from datetime import datetime
from dateutil.relativedelta import relativedelta
import numpy as np
# ## Getting all unique player urls from teams
# How teams appear in team url
team_url_list = [
"adelaide",
"brisbanel",
"carlton",
"collingwood",
"essendon",
"fremantle",
"geelong",
"goldcoast",
"gws",
"hawthorn",
"melbourne",
"kangaroos",
"padelaide",
"richmond",
"stkilda",
"swans",
"westcoast",
"bullldogs",
]
player_url_set = set()
for team in team_url_list:
print(team)
page = requests.get("https://afltables.com/afl/stats/teams/" + team + ".html")
soup = BeautifulSoup(page.text)
tables = soup.findAll("table")
print("team tables count:", len(tables))
print()
if not len(tables):
raise Exception("Missing Table")
for row in tables[0].tbody.findAll("tr"):
try:
href = row.find("a").get("href")
except:
href = None
print("Error with row:")
print(row)
print()
if not href:
raise Exception("Missing href")
player_url_set.add(href)
print(len(player_url_set))
player_url_set
# ## Getting all player data from each player url
players_data_list = []
for i, player_url in enumerate(player_url_set):
print(i, player_url)
tables_list = pd.read_html("https://afltables.com/afl/stats" + player_url[2:])
try:
season_tables = [
table for table in tables_list if isinstance(table.columns, pd.MultiIndex)
]
except Exception as e:
print(e)
print()
if not season_tables:
print("season table missing")
print(len(season_tables))
players_data_list.append((player_url, season_tables))
len(players_data_list)
# ## Creating player data
player_df = pd.DataFrame(
columns=[
"name",
"afl_tables_url",
"team",
"year",
"game_num",
"opponent",
"Rd",
"R",
"#",
"KI",
"MK",
"HB",
"DI",
"GL",
"BH",
"HO",
"TK",
"RB",
"IF",
"CL",
"CG",
"FF",
"FA",
"BR",
"CP",
"UP",
"CM",
"MI",
"1%",
"BO",
"GA",
"%P",
]
)
player_df
def func(i, data):
player_url, player_table_list = data[0], data[1]
print(i, player_url)
player_name = player_url.split("/")[-1][:-5].replace("_", " ")
year_set = {
int(season.columns[0][0].split("-")[1].strip()) > 2011
for season in player_table_list
}
print(year_set)
if not all(year_set):
return
for season in player_table_list:
labels = [d.strip() for d in season.columns[0][0].split("-")]
if len(labels) != 2:
raise Exception(f"Error with {player_url}, {season.columns[0]}")
team_name, year = labels
for i, row in season.iterrows():
created_row = [player_name, player_url, team_name, year] + list(row.values)
player_df.loc[len(player_df)] = created_row
for i, data in enumerate(players_data_list):
func(i, data)
player_df[
[
"KI",
"MK",
"HB",
"DI",
"GL",
"BH",
"HO",
"TK",
"RB",
"IF",
"CL",
"CG",
"FF",
"FA",
"BR",
"CP",
"UP",
"CM",
"MI",
"1%",
"BO",
"GA",
"%P",
]
] = player_df[
[
"KI",
"MK",
"HB",
"DI",
"GL",
"BH",
"HO",
"TK",
"RB",
"IF",
"CL",
"CG",
"FF",
"FA",
"BR",
"CP",
"UP",
"CM",
"MI",
"1%",
"BO",
"GA",
"%P",
]
].fillna(
0
)
# ### Separating off aggregate data from each season into different dataset
player_agg_df = player_df[player_df["opponent"] == "Totals"]
player_df = player_df.drop(player_agg_df.index)
player_df.info()
# ## Creating a player bio table
players_bio_list = []
for i, player_url in enumerate(player_df["afl_tables_url"].unique()):
print(i, player_url)
page = requests.get("https://afltables.com/afl/stats" + player_url[2:])
soup = BeautifulSoup(page.text)
name = soup.find("h1").text
height_and_weight = soup.text.split("\n")[8]
birthday_debut = soup.text.split("\n")[7]
print("name", name, "h and w", height_and_weight)
players_bio_list.append((player_url, name, height_and_weight, birthday_debut))
player_table_data = []
for player in players_bio_list:
try:
url = player[0]
name = player[1]
height = re.split(" |:", player[2])[1]
weight = re.split(" |:", player[2])[4]
dob = re.split(":| |\(", player[3])[9]
debut_list = [re.split(":| |\(", player[3])[12]]
if re.split(":| |\(", player[3])[13] != "Last":
debut_list.append(re.split(":| |\(", player[3])[13])
last_list = [re.split(":| |\(", player[3])[15]]
try:
last_list.append(re.split(":| |\(", player[3])[16])
except:
pass
else:
last_list = [re.split(":| |\(", player[3])[14]]
try:
last_list.append(re.split(":| |\(", player[3])[15])
except:
pass
last = " ".join(last_list)
debut = " ".join(debut_list)
player_table_data.append((url, name, height, weight, dob, debut, last))
except:
print(player)
break
player_data_df = pd.DataFrame(
player_table_data,
columns=["afl_tables_url", "name", "height", "weight", "dob", "debut", "last"],
)
player_data_df["height"] = player_data_df["height"].astype(int)
player_data_df["weight"] = player_data_df["weight"].astype(int)
player_data_df["dob"] = pd.to_datetime(player_data_df["dob"], format="%d-%b-%Y")
player_data_df.info()
player_data_df["debut_year"] = (
player_data_df["debut"]
.apply(
lambda x: 0
if len(re.findall(r"\d+(?=y)", x)) == 0
else re.findall(r"\d+(?=y)", x)[0]
)
.astype(int)
)
player_data_df["debut_day"] = (
player_data_df["debut"]
.apply(
lambda x: 0
if len(re.findall(r"y (\d+)d", x)) == 0
else re.findall(r"y (\d+)d", x)[0].strip()
)
.astype(int)
)
player_data_df["last_year"] = (
player_data_df["last"]
.apply(
lambda x: 0
if len(re.findall(r"\d+(?=y)", x)) == 0
else re.findall(r"\d+(?=y)", x)[0]
)
.astype(int)
)
player_data_df["last_day"] = (
player_data_df["last"]
.apply(
lambda x: 0
if len(re.findall(r"y (\d+)d", x)) == 0
else re.findall(r"y (\d+)d", x)[0].strip()
)
.astype(int)
)
today = datetime.today()
def get_date(row, column):
return (
today - relativedelta(years=row[f"{column}_year"], days=row[f"{column}_day"])
).date()
player_data_df["debut_date"] = player_data_df.apply(
lambda x: get_date(x, "debut"), axis=1
)
player_data_df["last_date"] = player_data_df.apply(
lambda x: get_date(x, "last"), axis=1
)
player_data_df = player_data_df.drop(
["debut", "last", "debut1", "debut_year", "debut_day", "last_year", "last_day"],
axis=1,
)
# ## Creating a games stats table
game_df = pd.DataFrame(
columns=[
"year",
"round",
"date_time",
"venue",
"home_team",
"away_team",
"attendance",
"home_team_quarter_score",
"away_team_quarter_score",
"home_team_final_score",
"away_team_final_score",
]
)
game_df
round_list = [
"Round 1",
"Round 2",
"Round 3",
"Round 4",
"Round 5",
"Round 6",
"Round 7",
"Round 8",
"Round 9",
"Round 10",
"Round 11",
"Round 12",
"Round 13",
"Round 14",
"Round 15",
"Round 16",
"Round 17",
"Round 18",
"Round 19",
"Round 20",
"Round 21",
"Round 22",
"Round 23",
"Qualifying Final",
"Elimination Final",
"Semi Final",
"Preliminary Final",
"Grand Final",
]
year_list = sorted(list(player_df["year"].unique()))
for year in year_list:
game_tables = pd.read_html(f"https://afltables.com/afl/seas/{year}.html")
for game_table in game_tables:
try:
if len(game_table) == 1:
if game_table[0][0] in round_list:
rnd_num = game_table[0][0]
else:
continue
elif len(game_table) == 2:
home_team = game_table[0][0]
away_team = game_table[0][1]
home_team_quarter_scores = game_table[1][0]
away_team_quarter_scores = game_table[1][1]
home_team_final_score = game_table[2][0]
away_team_final_score = game_table[2][1]
d = re.split(" Att: | Venue: ", game_table[3][0])
if len(d) == 3:
date_time = d[0]
attendance = d[1]
venue = d[2]
elif len(d) == 2:
date_time = d[0]
attendance = 0
venue = d[1]
else:
raise Exception(f"Unexpected data {d}")
created_row = (
year,
rnd_num,
date_time,
venue,
home_team,
away_team,
attendance,
home_team_quarter_scores,
away_team_quarter_scores,
home_team_final_score,
away_team_final_score,
)
game_df.loc[len(game_df)] = created_row
else:
continue
except Exception as e:
print(f"{year}, {game_table}")
raise Exception(e)
game_df = game_df[~game_df["home_team_final_score"].isna()]
game_df[
["day_of_week", "date", "AEST_time", "AEST_am_pm", "local_time", "local_am_pm"]
] = game_df["date_time"].str.split(" ", expand=True)
game_df = game_df.drop("date_time", axis=1)
game_df["date"] = pd.to_datetime(game_df["date"], format="%d-%b-%Y")
game_df["local_time"] = game_df["local_time"].apply(lambda x: x[1:] if x else x)
game_df["local_am_pm"] = game_df["local_am_pm"].apply(lambda x: x[:-1] if x else x)
game_df["AEST"] = pd.to_datetime(
game_df["AEST_time"] + " " + game_df["AEST_am_pm"], format="%I:%M %p"
).dt.time
game_df["local"] = pd.to_datetime(
game_df["local_time"] + " " + game_df["local_am_pm"], format="%I:%M %p"
).dt.time
game_df = game_df.drop(["AEST_time", "AEST_am_pm", "local_time", "local_am_pm"], axis=1)
game_df["local"] = game_df["local"].fillna(game_df["AEST"])
game_df["home_team_quarter_score"].str.split(" ")
game_df[["home_Q1", "home_Q2", "home_Q3", "home_Q4", "home_extra_time"]] = game_df[
"home_team_quarter_score"
].str.split(re.compile(r"[ |\xa0]+"), regex=True, expand=True)
game_df[["away_Q1", "away_Q2", "away_Q3", "away_Q4", "away_extra_time"]] = game_df[
"away_team_quarter_score"
].str.split(re.compile(r"[ |\xa0]+"), regex=True, expand=True)
game_df = game_df.drop(["home_team_quarter_score", "away_team_quarter_score"], axis=1)
game_df["round"] = game_df["round"].replace(
{
"Qualifying Final": "QF",
"Elimination Final": "EF",
"Semi Final": "SF",
"Preliminary Final": "PF",
"Grand Final": "GF",
}
)
game_df["round"] = game_df["round"].str.replace("Round ", "")
player_data_df
player_df
game_df
# ## Creating player position and draft data
combined_df = player_df.drop("name", axis=1).merge(player_data_df, on="afl_tables_url")
combined_df["team"].unique()
footywire_map = {
"Brisbane Lions": "brisbane-lions",
"Port Adelaide": "port-adelaide-power",
"St Kilda": "st-kilda-saints",
"Geelong": "geelong-cats",
"Fremantle": "fremantle-dockers",
"Hawthorn": "hawthorn-hawks",
"Gold Coast": "gold-coast-suns",
"Essendon": "essendon-bombers",
"Carlton": "carlton-blues",
"Adelaide": "adelaide-crows",
"North Melbourne": "kangaroos",
"West Coast": "west-coast-eagles",
"Collingwood": "collingwood-magpies",
"Greater Western Sydney": "greater-western-sydney-giants",
"Sydney": "sydney-swans",
"Melbourne": "melbourne-demons",
"Western Bulldogs": "western-bulldogs",
"Richmond": "richmond-tigers",
}
footywire_name_map = {
"jack": "jackson",
"ollie": "oliver",
"zach": "zachary",
"josh": "joshua",
"lachie": "lachlan",
"dom": "dominic",
"tom": "thomas",
"cam": "cameron",
"nick": "nicholas",
"tim": "timothy",
"will": "william",
"mitch": "mitchell",
"matt": "matthew",
"brad": "bradley",
"alex": "alexis",
"sam": "samuel",
"timmy": "tim",
"harry": "harrison",
"matthew": "matt",
"brett": "bret",
"zac": "zachary",
"dan": "daniel",
"ed": "edward",
"steve": "stephen",
"bobby": "robert",
"robbie": "robert",
"ned": "edward",
"pat": "patrick",
"nic": "nicholas",
"junior": "willie",
"nik": "nicholas",
}
url_map = {
"pp-fremantle-dockers--michael-frederick": "pp-fremantle-dockers--minairo-frederick",
"pp-greater-western-sydney-giants--robert-hill": "pp-greater-western-sydney-giants--ian-hill",
"pp-adelaide-crows--elliott-himmelberg": "pp-adelaide-crows--elliot-himmelberg",
"pp-greater-western-sydney-giants--zachary-langdon": "pp-west-coast-eagles--zac-giles-langdon",
"pp-essendon-bombers--nicholas-cox": "pp-essendon-bombers--nikolas-cox",
"pp-hawthorn-hawks--angus-dewar": "pp-west-coast-eagles--angus-litherland",
"pp-west-coast-eagles--zachary-langdon": "pp-west-coast-eagles--zac-giles-langdon",
"pp-essendon-bombers--cory-dellolio": "pp-essendon-bombers--corey-dell-olio",
"pp-st-kilda-saints--mitchell-o-wens": "pp-st-kilda-saints--mitchito-owens",
"pp-essendon-bombers--massimo-dambrosio": "pp-essendon-bombers--massimo-d-ambrosio",
"pp-carlton-blues--joshua-deluca": "pp-carlton-blues--josh-deluca-cardillo",
"pp-kangaroos--lachlan-hosie": "pp-kangaroos--lachie-hosie",
"pp-fremantle-dockers--joshua-deluca": "pp-carlton-blues--josh-deluca-cardillo",
"pp-collingwood-magpies--robert-hill": "pp-collingwood-magpies--ian-hill",
"pp-fremantle-dockers--jason-carter": "pp-fremantle-dockers--jason-carter-1",
"pp-geelong-cats--oliver-dempsey": "pp-geelong-cats--ollie-dempsey",
"pp-west-coast-eagles--angus-dewar": "pp-west-coast-eagles--angus-litherland",
}
def get_footywire_url(team, player_name):
return (
f"https://www.footywire.com/afl/footy/pp-{footywire_map[team]}--{player_name}"
)
extra_player_data_list = []
count = 0
for afl_tables_url, name, team in list(
combined_df[["afl_tables_url", "name", "team"]].value_counts().index
):
count += 1
if count % 100 == 0:
print(count)
flag = None
player_name = name.lower().replace(" ", "-")
while True:
url_string = get_footywire_url(team, player_name)
try:
page = requests.get(url_string)
except:
print(afl_tables_url, name, team, url_string)
soup = BeautifulSoup(page.text)
if not soup.find(id="playerProfileData"):
if not flag:
player_name = "-".join(
[footywire_name_map.get(n, n) for n in name.lower().split(" ")]
)
flag = 1
elif flag == 1:
flag = 2
if name.lower().split(" ")[-1][0] != "o":
flag = 4
break
name = (
" ".join(name.lower().split(" ")[:-1])
+ " "
+ name.lower().split(" ")[-1][0]
+ " "
+ name.lower().split(" ")[-1][1:]
)
player_name = "-".join([n for n in name.lower().split(" ")])
elif flag == 2:
player_name = "-".join(
[footywire_name_map.get(n, n) for n in name.lower().split(" ")]
)
flag = 3
else:
flag = 4
break
else:
break
if flag == 4:
player_name = "-".join(
[footywire_name_map.get(n, n) for n in name.lower().split(" ")]
)
url_string = get_footywire_url(team, player_name)
url_string = (
"/".join(url_string.split("/")[:-1])
+ "/"
+ url_map.get(url_string.split("/")[-1], url_string.split("/")[-1])
)
try:
page = requests.get(url_string)
except:
print(afl_tables_url, name, team, url_string)
soup = BeautifulSoup(page.text)
if not soup.find(id="playerProfileData"):
print(afl_tables_url, name, team, url_string)
continue
try:
if soup.find(id="playerProfileTeamDiv"):
team_data = (
soup.find(id="playerProfileTeamDiv")
.find("b")
.text.replace("\xa0", "")
.replace("\n", "")
)
else:
team_data = None
if soup.find(id="playerProfileData1"):
data_1 = (
soup.find(id="playerProfileData1")
.text.replace("\xa0", "")
.replace("\n", " ")
)
else:
data_1 = None
if soup.find(id="playerProfileData2"):
data_2 = (
soup.find(id="playerProfileData2")
.text.replace("\xa0", "")
.replace("\n", " ")
)
else:
data_2 = None
if soup.find(id="playerProfileDraftInfo"):
draft_data = (
soup.find(id="playerProfileDraftInfo")
.text.replace("\xa0", "")
.replace("\n", " ")
)
else:
draft_data = None
except AttributeError:
print(afl_tables_url, name, team, url_string)
continue
extra_player_data_list.append(
(afl_tables_url, team, team_data, data_1, data_2, draft_data)
)
extra_df = pd.DataFrame(
extra_player_data_list,
columns=["afl_tables_url", "team", "team_data", "data_1", "data_2", "draft_data"],
)
extra_df[["misc", "origin"]] = extra_df["data_1"].str.split("Origin: ", expand=True)
extra_df[["misc2", "position"]] = extra_df["data_2"].str.split(
"Position: ", expand=True
)
extra_df["draft_round"] = (
extra_df["draft_data"].str.extract(r"Round\s(\d+)\s*,", expand=False).astype(float)
)
extra_df["draft_pick_number"] = (
extra_df["draft_data"].str.extract(r"#(\d+)* ", expand=False).astype(float)
)
extra_df["draft_year"] = extra_df["draft_data"].str.extract(
r"Last Drafted:.*(\d{4})", expand=False
)
extra_df["extra_draft_data"] = extra_df["draft_data"].str.extract(
r"Last Drafted:.*\d{4}.*?(.*)", expand=True
)
extra_df[["draft_type", "draft_team"]] = extra_df["extra_draft_data"].str.split(
" by ", expand=True
)
extra_df = extra_df.drop(
[
"team_data",
"data_1",
"data_2",
"draft_data",
"misc",
"misc2",
"extra_draft_data",
],
axis=1,
)
extra_df
temp_df = pd.DataFrame(
list(
extra_df[
[
"afl_tables_url",
"origin",
"draft_round",
"draft_pick_number",
"draft_year",
"draft_type",
"draft_team",
]
]
.value_counts()
.index
),
columns=[
"afl_tables_url",
"origin",
"draft_round",
"draft_pick_number",
"draft_year",
"draft_type",
"draft_team",
],
)
temp_df2 = pd.DataFrame(
list(extra_df[["afl_tables_url", "team", "position"]].value_counts().index),
columns=["afl_tables_url", "team", "position"],
)
player_df_final = player_df.merge(temp_df2, how="left", on=["afl_tables_url", "team"])
player_data_df_final = player_data_df.merge(temp_df, how="left", on="afl_tables_url")
# ## Exporting
player_df.to_csv("afltables_stats")
player_agg_df.to_csv("afltables_agg_stats")
player_data_df.to_csv("afltables_player")
game_df.to_csv("afltables_game")
extra_df.to_csv("extra_bio_info")
player_data_df_final.to_csv("afltables_player_final")
player_df_final.to_csv("afltables_stats_final")
# ## Final dfs
game_df
player_data_df_final
player_df_final
player_data_df
player_df
|
# # PART 1: Loading/preprocessing images and creating additional datasets
## Importing the necessary packages
import numpy as np
import pandas as pd
import seaborn as sns
import os
import cv2
import csv
from skimage import feature
import matplotlib.pyplot as plt
from sklearn import preprocessing
# I use this to print the plots in the notebook
# I use this to give a white background to the images in my dark theme notebook
from matplotlib import style
style.use("dark_background")
sns.set(style="whitegrid")
# I downloaded the dataset from [here](https://github.com/chandrikadeb7/Face-Mask-Detection), removing some instances manually to have a total of 1600 images of faces without a mask and 600 images of people with mask.
# With the following code it is possible to load the dataset, resize/binarise the images, and then create an additional image repository using Histogram of Gradients (HOG).
################################ 1. DEFINE CLASS TO EXTRACT HOG FEATURES ################################
class HOG:
def __init__(
self,
orientations=9,
pixelsPerCell=(8, 8),
cellsPerBlock=(3, 3),
transform=False,
):
# store the number of orientations, pixels per cell,
# cells per block, and whether or not power law
# compression should be applied
self.orienations = orientations
self.pixelsPerCell = pixelsPerCell
self.cellsPerBlock = cellsPerBlock
self.transform = transform
def describe(self, image):
# compute HOG for the image
hist = feature.hog(
image,
orientations=self.orienations,
pixels_per_cell=self.pixelsPerCell,
cells_per_block=self.cellsPerBlock,
transform_sqrt=self.transform,
)
## return the HOG features
return hist
hog = HOG(orientations=18, pixelsPerCell=(10, 10), cellsPerBlock=(1, 1), transform=True)
######## 2. LOAD IMAGES (FOR ORIGINAL REPO), BINARISE (FOR PREPROCESSED REPO) & EXTRACT HOG FEATURES (FOR FEATURE REPO) ########
bin_thresh = 180 # Threshold for binarisation
resize_factor = 100 # applies for both height and width
path = "../input/cmm536-cw-p1-model-sol/data"
datarepo = [] # List to append the images as 2D numpy arrays
originalrepo = [] # Create a repo for flattened pixels
binarisedrepo = [] # Create a list to append the binarised pixels
hogrepo = [] # Create a list to append the HOG features
target = [] # List to append the target/class/label
print("\nLoading images...")
for root, dirs, files in os.walk(path):
for file in files:
with open(os.path.join(root, file), "r") as auto:
img = cv2.imread(root + "/" + file, 0)
img = cv2.resize(img, (resize_factor, resize_factor))
datarepo.append(img)
originalrepo.append(img.flatten())
_, img_bin = cv2.threshold(img, bin_thresh, 255, 0)
# Append the flattened image to the pixel repo
binarisedrepo.append(img_bin.flatten())
# Extract HOG and append to HOG repo
hogfeatures = hog.describe(img)
hogrepo.append(hogfeatures)
# Append the folder where the image is to the target list
target.append(root.replace(path, "").replace("\\", ""))
# Convert the repo lists into numpy arrays
originalrepo = np.array(originalrepo)
binarisedrepo = np.array(binarisedrepo)
hogrepo = np.array(hogrepo)
########################## 3. CALCULATE THE DISTRIBUTION AND SHOW REPOS ##########################
print("\nCalculating class distribution...")
histo = [["Class", "Number of Samples"]]
for i, label1 in enumerate(sorted(list(set(target)))):
cont = 0
for j, label2 in enumerate(target):
if label1 == label2:
cont += 1
histo.append([label1, cont])
histo.append(["Total Samples", len(target)])
## Save the histogram as a .csv file
with open("./classdistribution.csv", "w", newline="", encoding="utf-8") as csvfile:
filewriter = csv.writer(csvfile, delimiter=",")
for i, hist in enumerate(histo):
filewriter.writerow(hist)
## Load as a panda
histo_panda = pd.DataFrame.from_records(histo[1:-1], columns=histo[0])
print(histo_panda)
print("Total images: " + str(len(target)))
## Create a histogram using seaborn
sns_plot = sns.barplot(y="Class", x="Number of Samples", data=histo_panda)
## Save the image
sns_plot.figure.set_size_inches(10, 6)
sns_plot.figure.savefig(
"barchart.jpg", orientation="landscape", dpi=600, transparent=True
)
print("\nShowing class distribution bar chart...")
plt.show()
print("Size of target: ", len(target))
print("Size of original repository: ", originalrepo.shape)
print("Example of the original repository: ")
print(originalrepo)
print("Size of binarised data structure: ", binarisedrepo.shape)
print("Example of the binarised repository: ")
print(binarisedrepo)
print("Size of HOG features data structure: ", hogrepo.shape)
print("Example of the HOG repository: ")
print(hogrepo)
################################ 4. SAVE THE DATASETS AS CSV FILES ################################
print("\nSaving datasets as .csv files...")
with open("./original.csv", "w", newline="", encoding="utf-8") as csvfile:
filewriter = csv.writer(csvfile, delimiter=",")
for i, pix in enumerate(originalrepo):
row = np.concatenate((pix, [target[i]]))
filewriter.writerow(row)
with open("./hog.csv", "w", newline="", encoding="utf-8") as csvfile:
filewriter = csv.writer(csvfile, delimiter=",")
for i, h in enumerate(hogrepo):
row = np.concatenate((h, [target[i]]))
filewriter.writerow(row)
with open("./binarised.csv", "w", newline="", encoding="utf-8") as csvfile:
filewriter = csv.writer(csvfile, delimiter=",")
for i, l in enumerate(binarisedrepo):
row = np.concatenate((l, [target[i]]))
filewriter.writerow(row)
print("\nData saved into .csv files!")
# In the next cell, I implemented Random OverSampling (ROS, more info about this algorithm [here](https://imbalanced-learn.readthedocs.io/en/stable/over_sampling.html)) to perform augmentation of the HOG dataset to balance the classes to 1600 each. This new dataset has 3200 samples in total.
########################## 1. CALCULATE NEW DATASET USING RANDOM OVERSAMPLING (ROS) ##########################
from imblearn.over_sampling import RandomOverSampler
ros = RandomOverSampler(random_state=0)
hogrepo_ros, target_ros = ros.fit_resample(hogrepo, target)
################################ 2. CALCULATE THE DISTRIBUTION AFTER ROS ################################
print("\nCalculating class distribution (after ROS)...")
histo = [["Class", "Number of Samples"]]
for i, label1 in enumerate(sorted(list(set(target_ros)))):
cont = 0
for j, label2 in enumerate(target_ros):
if label1 == label2:
cont += 1
histo.append([label1, cont])
histo.append(["Total Samples", len(target_ros)])
## Save the histogram as a .csv file
with open("./classdistribution_ros.csv", "w", newline="", encoding="utf-8") as csvfile:
filewriter = csv.writer(csvfile, delimiter=",")
for i, hist in enumerate(histo):
filewriter.writerow(hist)
## Convert histo into a panda dataframe
histo_panda = pd.DataFrame.from_records(histo[1:-1], columns=histo[0])
print(histo_panda)
print("Total images: " + str(len(target_ros)))
## Create a histogram using seaborn
sns_plot = sns.barplot(y="Class", x="Number of Samples", data=histo_panda)
## Save the image
sns_plot.figure.set_size_inches(10, 6)
sns_plot.figure.savefig(
"barchart_ros.jpg", orientation="landscape", dpi=600, transparent=True
)
print("\nShowing class distribution bar chart (after ros)...")
plt.show()
print("Size of ROS augmented target: ", len(target_ros))
print("Size of ROS augmented repository: ", hogrepo_ros.shape)
print("Example of the ROS augmented repository: ")
print(hogrepo_ros)
################################ 3. SAVE THE ROS DATASET AS CSV FILE ################################
print("\nSaving dataset as .csv file...")
with open("./hog_ros.csv", "w", newline="", encoding="utf-8") as csvfile:
filewriter = csv.writer(csvfile, delimiter=",")
for i, h in enumerate(hogrepo_ros):
row = np.concatenate((h, [target_ros[i]]))
filewriter.writerow(row)
print("\nData saved!")
# Finally, the following cell performs the classical class decomposition with *kmeans* to create a pixel repository with the decomposed classes. The size of the final repository remains at 2200 samples but now it has 3 classes in total.
################################ 1. CALCULATE K USING STANDARD BALANCING EQ ################################
def standardbalancingequation(target):
"""This function follows the standard balancing equation to calculate k for each class"""
import math
print("\nCalculating k values...")
## Obtain the number of classes in label list and sort
labelsIndexesUnique = list(set(target))
labelsIndexesUnique.sort()
## For each class, count the number of instances and calculate ki
k = []
for label in labelsIndexesUnique:
k.append(target.count(label))
avgInst = sum(k) / len(k)
k = [math.floor((ki / avgInst) + 1) for ki in k]
print("Values of k for each class: ", k)
return k
k_kmeans = standardbalancingequation(target)
print("Number of classes after class decomposition: ", sum(k_kmeans))
################################ 2. CLASS DECOMPOSITION USING KMEANS ################################
def CDKmeans(data, target, k):
print("\nClass decomposition by applying k-means...")
from sklearn.cluster import KMeans
target_cd = [""] * len(target)
IndexesUnique = list(set(target))
IndexesUnique.sort()
for i, label in enumerate(IndexesUnique):
print("Number of clusters for class " + str(label) + ": " + str(k[i]))
## Split the dataset
data_tocluster = []
data_tocluster_index = []
for j, dat in enumerate(data):
if target[j] == label:
data_tocluster.append(dat)
data_tocluster_index.append(j)
if 1 < k[i] <= len(data_tocluster):
## Apply k-means to the list
kmeans = KMeans(n_clusters=k[i], random_state=0).fit(data_tocluster)
for n, m in enumerate(kmeans.labels_):
target_cd[data_tocluster_index[n]] = str(label) + "_c" + str(m)
else:
for m in data_tocluster_index:
target_cd[m] = str(label) + "_c0"
return target_cd
target_cd = CDKmeans(originalrepo, target, k_kmeans)
################################ 3. CALCULATE THE DISTRIBUTION AFTER CD ################################
print("\nCalculating class distribution (after class decomposition)...")
histo = [["Class", "Number of Samples"]]
for i, label1 in enumerate(sorted(list(set(target_cd)))):
cont = 0
for j, label2 in enumerate(target_cd):
if label1 == label2:
cont += 1
histo.append([label1, cont])
histo.append(["Total Samples", len(target_cd)])
## Save the histogram as a .csv file
with open("./classdistribution_cd.csv", "w", newline="", encoding="utf-8") as csvfile:
filewriter = csv.writer(csvfile, delimiter=",")
for i, hist in enumerate(histo):
filewriter.writerow(hist)
## Convert histo into a panda dataframe
histo_panda = pd.DataFrame.from_records(histo[1:-1], columns=histo[0])
print(histo_panda)
print("Total images: " + str(len(target_cd)))
## Create a histogram using seaborn
sns_plot = sns.barplot(y="Class", x="Number of Samples", data=histo_panda)
## Save the image
sns_plot.figure.set_size_inches(10, 6)
sns_plot.figure.savefig(
"barchart_cd.jpg", orientation="landscape", dpi=600, transparent=True
)
print("\nShowing class distribution bar chart (after cd)...")
plt.show()
################################ 4. SAVE THE CD DATASET AS CSV FILE ################################
print("\nSaving dataset as .csv file...")
with open("./original_cd.csv", "w", newline="", encoding="utf-8") as csvfile:
filewriter = csv.writer(csvfile, delimiter=",")
for i, pix in enumerate(originalrepo):
row = np.concatenate((pix, [target_cd[i]]))
filewriter.writerow(row)
print("\nData saved!")
|
#
# # Superstore Analysis🛒
#
# <p style="padding:15px;
# background-color:#a2c77d;
# margin:0;
# border-radius: 15px 15px;
# overflow:hidden;
# font-weight:350;font-size:15px;max-width:100%;text-align:justify">The notebook includes visualization and analysis of data from an online superstore. Our goal is to gain insights into the performance of the store and identify opportunities for improvement. We need to identify patterns in the data in terms of customer segments, product categories, sales etc. We will use various data visualization tools to explore the data and draw meaningful conclusions. Through this project, we hope to provide valuable insights that can help the store optimize its operations and drive growth.
# # Knowing your data
#
#
#
# 📌The dataset has 21 columns whose descriptions are given below. Each row represents a product in an order placed by the customer. Don't confuse each row for a seperate order. We will treat Orders, Customers and Products as our main entities and perform our analysis accordingly.
# 📌The defined entities have attributes associated to them like Order ID, Order date, Customer Name, Customer ID, Category(of product), Sub-category(of product) etc.
#
# 📌Our analysis will mostly revolve around these attributes to get a better understanding of the data
# Metadata
# Row ID => Unique ID for each row.
# Order ID => Unique Order ID for each Customer.
# Order Date => Order Date of the product.
# Ship Date => Shipping Date of the Product.
# Ship Mode=> Shipping Mode specified by the Customer.
# Customer ID => Unique ID to identify each Customer.
# Customer Name => Name of the Customer.
# Segment => The segment where the Customer belongs.
# Country => Country of residence of the Customer.
# City => City of residence of of the Customer.
# State => State of residence of the Customer.
# Postal Code => Postal Code of every Customer.
# Region => Region where the Customer belong.
# Product ID => Unique ID of the Product.
# Category => Category of the product ordered.
# Sub-Category => Sub-Category of the product ordered.
# Product Name => Name of the Product
# Sales => Sales of the Product.
# Quantity => Quantity of the Product.
# Discount => Discount provided.
# Profit => Profit/Loss incurred.
# # Installations and imports
import pandas as pd
import matplotlib.pyplot as plt
from warnings import filterwarnings
filterwarnings("ignore")
import seaborn as sns
from xplotter.insights import *
# show all columns in functions like head()
pd.set_option("display.max_columns", None)
# to reset option use:
# pd.reset_option('max_columns')
sns.set_style("white")
sns.despine()
# # 👨🍳Preprocessing
df = pd.read_csv(
"/kaggle/input/superstore-dataset-final/Sample - Superstore.csv",
encoding="windows-1252",
parse_dates=True,
)
# Renaming columns with better naming conventions
dict = {
"Row ID": "Row_ID",
"Order ID": "Order_ID",
"Ship Date": "Ship_Date",
"Ship Mode": "Ship_Mode",
"Order Date": "Order_Date",
"Customer ID": "Customer_ID",
"Customer Name": "Customer_Name",
"Postal Code": "Postal_Code",
"Product ID": "Product_ID",
"Sub-Category": "Sub_category",
"Product Name": "Product_Name",
}
df.rename(columns=dict, inplace=True)
# Converting date columns to Timestamp type for easy processing
df["Ship_Date"] = pd.to_datetime(df.Ship_Date)
df["Order_Date"] = pd.to_datetime(df.Order_Date)
type(df["Ship_Date"][0]) # to check whether date parsing worked
# feature engineering: extracting time series features from pre-existing features
df["Order_date_year"] = df["Order_Date"].apply(lambda x: x.year)
df["Order_date_month"] = df["Order_Date"].apply(lambda x: x.month)
df["Order_date_month_name"] = df["Order_Date"].apply(lambda x: x.strftime("%b"))
df["Order_date_year_month"] = df["Order_Date"].apply(lambda x: x.strftime("%Y%m"))
df["Order_date"] = df["Order_Date"].apply(lambda x: x.strftime("%Y%m%d"))
df["Order_date_day"] = df["Order_Date"].apply(lambda x: x.day)
df["Order_date_dayofweek"] = df["Order_Date"].apply(lambda x: x.dayofweek)
df["Order_date_dayofweek_name"] = df["Order_Date"].apply(lambda x: x.strftime("%a"))
df["Order_date_year_month"].astype(str).astype(int)
# checking the updates
df.head(1)
# # 📊General Analysis
print("Shipping modes: ", end="")
print(df["Ship_Mode"].unique())
print("Countries: ", end="")
print(df["Country"].unique())
print("Number of states: " + str(df["State"].unique().shape[0]))
print("Region: ", end="")
print(df["Region"].unique())
print("Number of cities: " + str(df["City"].unique().shape[0]))
print("Shipping modes: ", end="")
print(df["Ship_Mode"].unique())
print("Categories: ", end="")
print(df["Category"].unique())
print("Number of Sub categories: " + str(df["Sub_category"].unique().shape[0]))
print("Number of Products: " + str(df["Product_ID"].unique().shape[0]))
# ### General Analysis
# <div style="float:left;margin-left:20px; max-width:85%; text-align:justify;
# padding:15px;
# background-color: #E5E4E2;
# margin:0;
# border-radius: 13px 13px;
# overflow:hidden;">📌Dataset has four Shipping modes: 'Second Class' 'Standard Class' 'First Class' 'Same Day'
#
# 📌It covers data from 531 cities, 49 states of United States divided into 4 regions: 'South' 'West' 'Central' 'East'
#
# 📌4 shipping modes are available: 'Second Class' 'Standard Class' 'First Class' 'Same Day'
#
# 📌1862 unique products have been divided into 4 catergories: 'Furniture' 'Office Supplies' 'Technology' and 17 sub categories
#
df1 = df.groupby(["State", "City"]).Row_ID.count()
df1 = df1.reset_index().sort_values(ascending=False, by="Row_ID").head(10)
df1
# top 10 cities with respect to sales
# Top 5 dates with most entries
df2 = df["Order_Date"].value_counts()
df2.sort_values(ascending=False).head(5)
# total discount offered in each category
df2 = df.groupby(["Category"]).Discount.sum()
df2.sort_values(ascending=False)
df3 = df.groupby(["Category"]).Profit.sum()
df3.sort_values(ascending=False)
# Technology is the most profitable sector followed by Office Supplies and Furniture
# choosing color palettes for plots
sns.choose_colorbrewer_palette(data_type="qualitative", as_cmap=False)
# data_type : {‘sequential’, ‘diverging’, ‘qualitative’}
# print(sns.color_palette("pastel6").as_hex())
# # 👥Customer segment analysis
fig = plt.figure(figsize=(13, 13))
grid = plt.GridSpec(2, 2, wspace=0, hspace=0.3, figure=fig)
plt.subplot(grid[0, :])
segment_shipmode = (
df.groupby(["Segment", "Ship_Mode", "Order_ID"]).Row_ID.count().reset_index()
)
segment_shipmode = (
segment_shipmode.groupby(["Segment", "Ship_Mode"]).Order_ID.count().reset_index()
)
segment_shipmode
sns.barplot(
data=segment_shipmode,
x=segment_shipmode.Segment,
y=segment_shipmode.Order_ID,
hue=segment_shipmode.Ship_Mode,
palette="Paired",
)
plt.title("Ship mode distribution in each segment", fontsize=17, color="dimgrey")
sns.despine()
plt.subplot(grid[1, 0])
# creating a seperate dateset to use for visualization
# you can print df_segment to get a better understanding of how the plot is made.
df_segment = df.groupby(["Customer_ID", "Segment"]).Row_ID.count().reset_index()
df_segment = df_segment.Segment.value_counts().reset_index()
plt.pie(
df_segment["Segment"],
labels=df_segment["index"],
textprops={"fontsize": 14, "fontweight": "medium"},
autopct="%1.1f%%",
pctdistance=0.7,
shadow=True,
colors=sns.color_palette("Paired"),
)
plt.title("Customers per segment", fontsize=17, color="dimgrey")
# Consumer Segment has the most customers followed by Corporate and Home Office
plt.subplot(grid[1, 1])
df_segment_sales = df.groupby(["Segment"]).Sales.sum().reset_index()
plt.pie(
df_segment_sales["Sales"],
labels=df_segment_sales["Segment"],
textprops={"fontsize": 14, "fontweight": "medium"},
autopct="%1.1f%%",
pctdistance=0.7,
shadow=True,
colors=sns.color_palette("Paired"),
)
plt.title("Sales per segment", fontsize=17, color="dimgrey")
plt.show()
# ### Customer segment insights
# <div style="float:left;margin-left:20px; max-width:85%; text-align:justify;
# padding:15px;
# background-color: #E5E4E2;
# margin:0;
# border-radius: 13px 13px;
# overflow:hidden;">Customers have been divided into 3 segments based on the type of account or the place where the order is being delivered. The segments are: Consumer, Corporate, Home Office
#
# 📌The Ship mode distribution for each segment is plotted. We can see that for all ths segments Standard class has the highest occurence followed by second class, first class and same day respectively.
#
# 📌Number of customers and total sales for a segment have been plotted. We see a similar pattern in both the cases. Consumer Segment has the highest number of customers and sales followed by Corporate and Home office respectively.
#
# # 🌍Geographical (state-wise) analysis
def plot(title, x, y, xrot, xfont, yrot, yfont):
plt.title(title, fontsize=16, color="dimgrey")
sns.barplot(x=x, y=y, palette="YlGnBu")
plt.xticks(rotation=xrot, fontsize=xfont)
plt.yticks(rotation=yrot, fontsize=yfont)
sns.despine()
plt.figure(figsize=(28, 6))
df_state_sales = df.groupby(["State"], as_index=False).Sales.sum()
df_state_sales = df_state_sales.sort_values(ascending=False, by="Sales").head(10)
# Top 10 states having highest sales
plt.subplot(1, 3, 1)
plt.xticks(rotation=90, fontsize=13)
plt.yticks(rotation=90, fontsize=10)
plt.title("Top 10 State wise Sales", fontsize=17, color="dimgrey")
sns.barplot(x=df_state_sales.State, y=df_state_sales.Sales, palette="rocket")
df_state_profit = df.groupby(["State"], as_index=False)[["Sales", "Profit"]].sum()
# series becomes a dataframe by adding as_index=False
df_state_profit = df_state_profit.sort_values(ascending=False, by="Sales").head(10)
df_state_profit.reset_index()
# df_state_profit
plt.subplot(1, 3, 2)
plt.xticks(rotation=90, fontsize=13)
plt.yticks(rotation=90, fontsize=10)
plt.title("Aggregate Sales and Profit of top 10 States", fontsize=17, color="dimgrey")
plt.bar(
df_state_profit["State"],
df_state_profit["Sales"],
width=0.5,
align="edge",
color="palegreen",
label="Sales",
)
plt.bar(
df_state_profit["State"],
df_state_profit["Profit"],
width=0.5,
color="turquoise",
label="Profit",
)
plt.legend()
plt.subplot(1, 3, 3)
df_state_orders = df.groupby(["Order_ID", "State"]).Row_ID.count().reset_index()
df_state_orders = df_state_orders["State"].value_counts().reset_index().head(10)
plt.xticks(rotation=90, fontsize=13)
plt.yticks(rotation=90, fontsize=10)
plt.title("Number of orders per state", fontsize=17, color="dimgrey")
sns.barplot(x=df_state_orders["index"], y=df_state_orders.State, palette="viridis")
plt.show()
# ### 🌍Geographical (State-wise) insights
# <div style="float:left;margin-left:20px; max-width:85%; text-align:justify;
# padding:15px;
# background-color:#E5E4E2;
# margin:0;
# border-radius: 13px 13px;
# overflow:hidden;">
# 📌California and New York lead in state wise sales, profits and number of orders. Yet they have a lower profit percentage compared to states like washington which generate lower sales yet the profits are on par with the top states .
# 📌Texas being the third in terms of sales still generates no profits and deals with a heavy loss.
#
df_category_offerings = (
df.groupby(["Category", "Product_ID"]).Row_ID.count().reset_index()
)
df_category_offerings = df_category_offerings["Category"].value_counts().reset_index()
df_offerings = df.groupby(["Sub_category", "Product_ID"]).Row_ID.count().reset_index()
df_offerings = df_offerings["Sub_category"].value_counts().reset_index()
# df_offerings = df_offerings.sort_values(ascending=False, by='Product_ID')
# Binders offer the most variety folowed by Paper and Furnishings
plt.figure(figsize=(13, 13))
df_ship_mode = df.groupby(["Order_ID", "Ship_Mode"]).Row_ID.count().reset_index()
df_ship_mode = df_ship_mode["Ship_Mode"].value_counts().reset_index()
df_region = df.groupby(["Order_ID", "Region"]).Row_ID.count().reset_index()
df_region = df_region["Region"].value_counts().reset_index()
plt.subplot(2, 2, 1)
plt.pie(
df_ship_mode["Ship_Mode"],
labels=df_ship_mode["index"],
textprops={"fontsize": 14, "fontweight": "medium"},
autopct="%1.1f%%",
pctdistance=0.8,
shadow=True,
colors=sns.color_palette("Set2"),
)
plt.title("Ship mode wise orders", fontsize=18, color="dimgrey")
plt.subplot(2, 2, 2)
plt.pie(
df_region["Region"],
labels=df_region["index"],
textprops={"fontsize": 14, "fontweight": "medium"},
explode=[0, 0, 0, 0.1],
autopct="%1.1f%%",
pctdistance=0.7,
shadow=True,
colors=sns.color_palette("Paired"),
)
plt.title("Region wise orders", fontsize=18, color="dimgrey")
plt.subplot(2, 2, 3)
plt.pie(
df_offerings["Sub_category"],
labels=df_offerings["index"],
textprops={"fontsize": 14, "fontweight": "medium"},
autopct="%1.1f%%",
pctdistance=0.9,
shadow=True,
colors=sns.color_palette("crest"),
)
# fontweight: 'ultralight', 'light', 'normal', 'regular', 'book', 'medium', 'roman', 'semibold', 'demibold', 'demi', 'bold', 'heavy', 'extra bold', 'black'
plt.title("% of Products in a sub category", fontsize=18, color="dimgrey")
plt.subplot(2, 2, 4)
plt.pie(
df_category_offerings["Category"],
labels=df_category_offerings["index"],
textprops={"fontsize": 14, "fontweight": "roman"},
autopct="%1.1f%%",
pctdistance=0.7,
shadow=True,
colors=sns.color_palette("flare"),
)
plt.title("% of Products in a category", fontsize=18, color="dimgrey")
plt.show()
# <div style="float:left;margin-left:20px; max-width:85%; text-align:justify;
# padding:15px;
# background-color:#E5E4E2;
# margin:0;
# border-radius: 13px 13px;
# overflow:hidden;">
# 📌Standard class contributes for about 60% of the total orders. First class(19.2%) and Second class(15.7%) also have a bunch of customers. 5.3% of orders were same say orders.
# 📌West and East are the top regions(thanks to states like California and Texas) while the south region that contributes the least sales still makes up 16.4%. This reveals that the sales have uniform geographical distribution.
# 📌58.2% of the products from the product catalogue belong to office supplies. We also know that technoloogy contributes the highest profits.This suggests that technology products are more profitable compared to office supplies. Furniture has the least amount of products and also generates the least amount of profit
#
# # ⏱️Time series analysis
fig = plt.figure(figsize=(18, 20))
# Axis definition
grid = plt.GridSpec(3, 2, wspace=0.2, hspace=0.3, figure=fig)
plt.subplot(grid[0, :])
odym_orders = (
df.groupby(["Order_date_year_month", "Order_ID"]).Row_ID.count().reset_index()
)
odym_orders = odym_orders.Order_date_year_month.value_counts().reset_index()
odym_orders = odym_orders.sort_values(by="index")
odym_profit = df.groupby(["Order_date_year_month"]).Profit.sum().reset_index()
sns.lineplot(
data=odym_orders,
x=odym_orders["index"],
y=odym_orders.Order_date_year_month,
color="#a1c9f4",
linewidth=4,
)
plt.xticks(rotation=45)
plt.xlabel("Year and Month", fontsize=14, color="dimgrey")
plt.ylabel("Number of orders", fontsize=14, color="dimgrey")
plt.title("Orders by year and month", fontsize=16, color="dimgrey")
plt.subplot(grid[1, :])
odym_sales = df.groupby(["Order_date_year_month"]).Sales.sum().reset_index()
odym_profit = df.groupby(["Order_date_year_month"]).Profit.sum().reset_index()
sns.lineplot(
data=odym_sales,
x="Order_date_year_month",
y="Sales",
label="Sales",
color="#8de5a1",
linewidth=4,
)
sns.lineplot(
data=odym_profit,
x="Order_date_year_month",
y="Profit",
label="Profit",
color="#ff9f9b",
linewidth=4,
)
plt.xticks(rotation=45)
plt.legend()
plt.xlabel("Year and Month", fontsize=14, color="dimgrey")
plt.ylabel("Sales and Profit", fontsize=14, color="dimgrey")
plt.title("Sales and Profit by year and month", fontsize=16, color="dimgrey")
plt.subplot(grid[2, 0])
day_orders = (
df.groupby(["Order_date_dayofweek", "Order_ID"]).Row_ID.count().reset_index()
)
day_orders = day_orders.Order_date_dayofweek.value_counts().reset_index()
day_orders
sns.barplot(data=day_orders, x="index", y="Order_date_dayofweek", palette="magma")
for i in day_orders["index"]:
plt.text(
x=day_orders["index"][i] - 0.3,
y=day_orders["Order_date_dayofweek"][i] - 0.3,
s=str(
round(
(day_orders["Order_date_dayofweek"][i])
/ (day_orders["Order_date_dayofweek"].sum())
* 100,
2,
)
)
+ "%",
size=12,
)
plt.xticks(
[r + 0.1 for r in range(len(day_orders["index"]))],
["Monday", "Tuesday", "Wednesday", "Thursday", "Friday", "Saturday", "Sunday"],
rotation=90,
)
plt.xlabel("Days of the week", fontsize=14, color="dimgrey")
plt.ylabel("Number of orders", fontsize=14, color="dimgrey")
plt.title("Orders by day of week", fontsize=16, color="dimgrey")
plt.subplot(grid[2, 1])
odm_orders = df.groupby(["Order_date_month", "Order_ID"]).Row_ID.count().reset_index()
odm_orders = odm_orders.Order_date_month.value_counts().reset_index()
odm_orders = odm_orders.sort_values(by="index")
sns.barplot(
data=odm_orders,
x="index",
y="Order_date_month",
palette="crest",
hue="Order_date_month",
dodge=False,
)
plt.legend([], [], frameon=False)
for i in odm_orders["index"]:
plt.text(
x=odm_orders["index"][i - 1] - 1.3,
y=odm_orders["Order_date_month"][i - 1] - 0.3,
s=str(odm_orders["Order_date_month"][i - 1]),
size=12,
)
plt.xticks(
[r + 0.1 for r in range(len(odm_orders["index"]))],
[
"Jan",
"Feb",
"Mar",
"Apr",
"May",
"Jun",
"Jul",
"Aug",
"Sep",
"Oct",
"Nov",
"Dec",
],
rotation=90,
)
plt.xlabel("Month", fontsize=14, color="dimgrey")
plt.ylabel("Number of orders", fontsize=14, color="dimgrey")
sns.despine()
plt.title("Orders by Month", fontsize=16, color="dimgrey")
# # Categorical analysis 📱🪑📎
import plotly.express as px
from plotly.offline import init_notebook_mode, iplot
init_notebook_mode(connected=True)
product_data = (
df.groupby(["Category", "Sub_category"])
.agg({"Profit": "sum"})
.sort_values("Profit")
.reset_index()
)
product_data = product_data[product_data.Profit > 0].sort_values(
"Profit", ascending=False
)
product_data["Value"] = "Profit"
product_data
px.sunburst(product_data, path=["Value", "Category", "Sub_category"], values="Profit")
# Profits of each Sub category in a category
fig = plt.figure(figsize=(13, 13))
grid = plt.GridSpec(2, 2, wspace=0.3, hspace=0.4, figure=fig)
subcat_sales = df.groupby(["Sub_category"]).Sales.sum().reset_index()
subcat_profit = df.groupby(["Sub_category"]).Profit.sum().reset_index()
subcat_sales["Profit"] = subcat_profit["Profit"]
subcat_sales = subcat_sales.sort_values(ascending=False, by="Sales")
subcat_sales["Ratio"] = subcat_sales["Profit"] / subcat_sales["Sales"]
cat_sales = df.groupby(["Category"]).Sales.sum().reset_index()
cat_profit = df.groupby(["Category"]).Profit.sum().reset_index()
cat_sales["Profit"] = cat_profit["Profit"]
cat_sales = cat_sales.sort_values(ascending=False, by="Sales")
cat_sales["Ratio"] = cat_sales["Profit"] / cat_sales["Sales"]
plt.subplot(grid[0, 0])
plt.xticks(rotation=90, fontsize=13)
plt.title("Aggregate Sales and Profit of sub categories", fontsize=17, color="dimgrey")
plt.bar(
subcat_sales["Sub_category"],
subcat_sales["Sales"],
width=0.5,
align="edge",
color="palegreen",
label="Sales",
)
plt.bar(
subcat_sales["Sub_category"],
subcat_sales["Profit"],
width=0.5,
color="turquoise",
label="Profit",
)
sns.despine()
plt.legend()
plt.subplot(grid[0, 1])
plt.xticks(rotation=90, fontsize=13)
plt.title("Sale : Profit ratio of sub categories", fontsize=17, color="dimgrey")
sns.barplot(
x=subcat_sales["Sub_category"],
y=subcat_sales["Ratio"],
color="turquoise",
hue=subcat_sales["Ratio"],
dodge=False,
)
plt.legend([], [], frameon=False)
sns.despine()
plt.subplot(grid[1, :])
plt.xticks(rotation=90, fontsize=13)
plt.title("Profit from categories", fontsize=17, color="dimgrey")
sns.barplot(
x=cat_sales["Profit"],
y=cat_sales["Category"],
color="turquoise",
hue=cat_sales["Ratio"],
dodge=False,
)
plt.legend([], [], frameon=False)
sns.despine()
# <div style="float:left;margin-left:20px; max-width:85%; text-align:justify;
# padding:15px;
# background-color:#E5E4E2;
# margin:0;
# border-radius: 13px 13px;
# overflow:hidden;">Products have been distributerd into 3 Categories and 17 sub_categories. A new attribute profit ratio has been created by dividing profit by sales. It gives us better insights into the profitability of a category. It represents profit generated for every dollar of sale.
#
# 📌For example, Phones are the most sold category but rank very low in the terms of profitability.
#
# 📌Labels, Envelopes, Paper, Copiers and fasteners have the top 5 profit ratio.
# 📌Furniture has the lowest profitability. Technology and office supplies have higher profitability comparatively.
#
fig = plt.figure(figsize=(18, 20))
df_sales = df.groupby(["Sub_category"], as_index=False).Sales.sum()
df_sales = df_sales.sort_values(ascending=False, by="Sales")
# Phones followed by Chairs and Storage generated the highest revenue
df["Sales_per_unit"] = df["Sales"] / df["Quantity"] # generated a new column
df_sales_mean = df.groupby(["Sub_category"], as_index=False).Sales_per_unit.mean()
df_sales_mean = df_sales_mean.sort_values(ascending=False, by="Sales_per_unit")
df_profit = df.groupby(["Sub_category"], as_index=False).Profit.sum()
df_profit = df_profit.sort_values(ascending=False, by="Profit")
# Copiers follwed by phones and Accessories have generated the most profit
df["Profit_per_unit"] = df["Profit"] / df["Quantity"]
df_profit_mean = df.groupby(["Sub_category"], as_index=False).Profit_per_unit.mean()
df_profit_mean = df_profit_mean.sort_values(ascending=False, by="Profit_per_unit")
# Copiers generate the most profit per unit
df_quantity = df.groupby(["Sub_category"], as_index=False)["Quantity"].sum()
df_quantity = df_quantity.sort_values(ascending=False, by="Quantity")
# Binders are the most sold sub category
df_offerings = df.groupby(["Sub_category", "Product_ID"]).Row_ID.count().reset_index()
df_offerings = df_offerings["Sub_category"].value_counts().reset_index()
# Binders offer the most variety folowed by Paper and Furnishings
# Axis definition
grid = plt.GridSpec(3, 2, wspace=0.2, hspace=0.6, figure=fig)
# plots
plt.subplot(grid[0, 0])
plot(
"Total sales of Sub-categories",
df_sales.Sub_category,
df_sales.Sales,
90,
13,
0,
10,
)
plt.subplot(grid[0, 1])
plot(
"Average cost of sub category",
df_sales_mean.Sub_category,
df_sales_mean.Sales_per_unit,
90,
13,
0,
10,
)
plt.subplot(grid[1, 0])
plot(
"Profit generated by each sub-category",
df_profit.Sub_category,
df_profit.Profit,
90,
13,
0,
10,
)
plt.subplot(grid[1, 1])
plot(
"Average profit per product belonging to a sub category",
df_profit_mean.Sub_category,
df_profit_mean.Profit_per_unit,
90,
13,
0,
10,
)
plt.subplot(grid[2, 0])
plot(
"Qunatity of products sold in each sub category",
df_quantity.Sub_category,
df_quantity.Quantity,
90,
13,
0,
10,
)
plt.subplot(grid[2, 1])
plot(
"Product Offerings in each sub category",
df_offerings["index"],
df_offerings.Sub_category,
90,
13,
0,
10,
)
plt.xlabel("Sub Category")
plt.ylabel("Products")
plt.show()
# <div style="float:left;margin-left:20px; max-width:85%; text-align:justify;
# padding:15px;
# background-color:#E5E4E2;
# margin:0;
# border-radius: 13px 13px;
# overflow:hidden;">
# 📌Copiers in general are the highest profit generating products. They have the least variety in offerings and are rarely sold in bulk.
# 📌Phones and chairs are the highest selling products. Binders and papers are mostly ordered in bulk. Paper has the highest variety (offerings) in products.
# 📌Tables are mostly sold at a lost and are the least profitable products.
#
plt.figure(figsize=(18, 6))
plt.subplot(1, 2, 1)
# sns.histplot(data=df, x="Quantity", kde=True)
sns.histplot(
data=df, x=df["Quantity"], hue="Category", palette="hls", element="step", binwidth=1
)
plt.title("Quantity distribution of Categories", fontsize=16, color="dimgrey")
plt.subplot(1, 2, 2)
sns.histplot(
data=df,
x=df["Order_date_month"],
hue="Category",
palette="Accent",
element="step",
binwidth=1,
)
plt.title("Category wise monthly order distribution", fontsize=16, color="dimgrey")
sns.despine()
plt.figure(figsize=(20, 6))
week_segment = (
df.groupby(["Order_date_dayofweek", "Segment", "Order_ID"])
.Row_ID.count()
.reset_index()
)
week_segment = (
week_segment.groupby(["Order_date_dayofweek", "Segment"])
.Order_ID.count()
.reset_index()
)
week_segment_sum = week_segment.groupby(["Order_date_dayofweek"]).Order_ID.sum()
week_segment_sum
week_segment["sum"] = [
920,
920,
920,
558,
558,
558,
182,
182,
182,
746,
746,
746,
916,
916,
916,
837,
837,
837,
850,
850,
850,
]
week_segment["percent"] = week_segment["Order_ID"] / week_segment["sum"] * 100
week_segment
plt.subplot(1, 2, 2)
sns.barplot(
data=week_segment,
x="Order_date_dayofweek",
y="percent",
hue="Segment",
palette="Pastel1",
)
plt.title(
"% of orders belonging to a segment on days of the week",
fontsize=16,
color="dimgrey",
)
plt.ylabel("Percent of total orders", fontsize=13, color="dimgrey")
plt.xlabel("Day", fontsize=13, color="dimgrey")
plt.xticks(
[r + 0.1 for r in range(len(day_orders["index"]))],
["Monday", "Tuesday", "Wednesday", "Thursday", "Friday", "Saturday", "Sunday"],
rotation=90,
)
sns.despine()
plt.subplot(1, 2, 1)
sns.barplot(
data=week_segment,
x="Order_date_dayofweek",
y="Order_ID",
hue="Segment",
palette="Pastel1",
)
plt.title(
"Orders belonging to a segment on days of the week", fontsize=16, color="dimgrey"
)
plt.ylabel("Number of orders", fontsize=13, color="dimgrey")
plt.xlabel("Day", fontsize=13, color="dimgrey")
plt.xticks(
[r + 0.1 for r in range(len(day_orders["index"]))],
["Monday", "Tuesday", "Wednesday", "Thursday", "Friday", "Saturday", "Sunday"],
rotation=90,
)
sns.despine()
plt.figure(figsize=(20, 6))
seg_cat = df.groupby(["Segment", "Category"]).Profit.sum().reset_index()
seg_cat["seg_cat"] = seg_cat.Segment + seg_cat.Category
plt.subplot(1, 2, 1)
sns.barplot(data=seg_cat, x="Segment", y="Profit", hue="Category", palette="Pastel1")
plt.title(
"Category wise profits of each Costumer segment", fontsize=16, color="dimgrey"
)
seg_cat1 = df.groupby(["Segment", "Category"]).Sales.sum().reset_index()
seg_cat1["seg_cat"] = seg_cat1.Segment + seg_cat.Category
plt.subplot(1, 2, 2)
sns.barplot(data=seg_cat1, x="Segment", y="Sales", hue="Category", palette="Pastel1")
plt.title("Category wise Sales of each Costumer segment", fontsize=16, color="dimgrey")
# # 🍬Miscellaneous
# products ordered in bulk
bulk = df.groupby(["Sub_category"]).Quantity.mean().reset_index()
sns.barplot(data=bulk, x=bulk.Sub_category, y=bulk.Quantity)
plt.xticks(rotation=90, fontsize=13)
plt.show()
# average sales on days of the month
# The start of the month sees higher sales compared to end. Sales also peak in the middle
order_day = df.groupby(["Order_date_day"]).Sales.sum().reset_index()
order_day
sns.barplot(
data=order_day,
x="Order_date_day",
y="Sales",
palette="crest",
hue="Sales",
dodge=False,
)
plt.legend([], [], frameon=False)
# most of the sales lie below 1000
# distibution of sales after removing outliers
# a highly positively skewed graph
df_clean = df.loc[(df.Sales <= 1000)]
sns.histplot(data=df_clean, x="Sales", palette="Accent", element="step")
sns.regplot(x=df_clean["Sales"], y=df_clean["Profit"])
df.loc[df["Profit"] == df["Profit"].max()]
# most profitable sale ever
df.loc[df["Sales"] == df["Sales"].max()]
# largest sale ever
|
# installing the needed upgrade optimum performance
# installing the needed upgrade optimum performance
# installing the needed upgrade optimum performance
# installing Panel
import pandas as pd
import numpy as np
import panel as pn
pn.extension("tabulator")
import hvplot.pandas
# this will help in making the Python code more structured automatically (good coding practice)
# %load_ext nb_black
# Libraries to help with reading and manipulating data
import numpy as np
import pandas as pd
# Libraries to help with data visualization
import matplotlib.pyplot as plt
import seaborn as sns
# split the data into train and test
from sklearn.model_selection import train_test_split
# to build linear regression_model
from sklearn.linear_model import LinearRegression
# to check model performance
from sklearn.metrics import mean_absolute_error, mean_squared_error, r2_score
# to build linear regression_model using statsmodels
import statsmodels.api as sm
# to compute VIF
from statsmodels.stats.outliers_influence import variance_inflation_factor
# # Command to tell Python to actually display the graphs
pd.set_option(
"display.float_format", lambda x: "%.2f" % x
) # To supress numerical display in scientific notations
# Removes the limit for the number of displayed columns
pd.set_option("display.max_columns", None)
# Sets the limit for the number of displayed rows
pd.set_option("display.max_rows", 200)
import warnings
warnings.filterwarnings("ignore")
path = (
"https://raw.githubusercontent.com/hayfordosmandata/DataBank/main/Bank_Churn1.csv"
)
churn_data = pd.read_csv(path)
# copying data to another variable to avoid any changes to original data
data = churn_data.copy()
import panel as pn
import seaborn as sb
# Create a Tabulator widget with pagination
table = pn.widgets.Tabulator(data.head(10000), pagination="remote")
# Display the widget in a Panel
panel = pn.Column("## First 20 Rows of Insurance Premium Dataset with Pages", table)
panel.servable()
# checking the shape of the data
print(f"There are {data.shape[0]} rows and {data.shape[1]} columns.")
# Checking the data types of the variables/columns for the dataset
data.info()
# Dropping the irrelevant columns
data.drop(["RowNumber", "CustomerId", "Surname"], axis=1, inplace=True)
import pandas as pd
import panel as pn
import hvplot.pandas
# Get the list of numerical columns
numeric_cols = data.select_dtypes(include=["float", "int"]).columns.tolist()
# Create the dropdown menu with numerical columns
dropdown = pn.widgets.Select(options=numeric_cols, name="Select a column")
# Define a function to update the plot based on the dropdown selection
def update_plot(event):
plot = data.hvplot.box(y=event.obj.value, title="Box plot of " + event.obj.value)
return plot
# Create the initial plot with the first numerical column
initial_col = numeric_cols[0]
plot = data.hvplot.box(y=initial_col, title="Box plot of " + initial_col, color="red")
# Combine the dropdown and the plot in a panel layout
dropdown.param.watch(update_plot, "value")
layout = pn.Row(dropdown, plot)
# Create a layout and serve the app
layout.servable()
import pandas as pd
import panel as pn
import hvplot.pandas
import holoviews as hv
# Define the available columns for the dropdowns
columns = list(data.columns)
# Define the available plot types for the dropdown
plot_types = {
"Displot": "hist",
"Boxplot": "box",
"Line Chart": "line",
"Scatter Plot": "scatter",
}
# Define the plots
def get_plots(column_x, column_y, plot_type):
if plot_type == "hist":
plot = data.hvplot.hist(column_x, title=f"{column_x} Distribution", bins=20)
elif plot_type == "box":
plot = data.hvplot.box(y=column_y, by="sex", title=f"{column_y} Distribution")
elif plot_type == "line":
plot = data.hvplot.line(column_x, column_y, title=f"{column_y} by {column_x}")
elif plot_type == "scatter":
plot = data.hvplot.scatter(
column_x,
column_y,
c="sex",
cmap="viridis",
title=f"{column_y} vs {column_x}",
)
return plot
# Define the dropdown widgets
dropdown_x = pn.widgets.Select(name="Horizontal Variables", options=columns)
dropdown_y = pn.widgets.Select(name="Vertical Variables", options=columns)
dropdown_plot_type = pn.widgets.Select(name="Plot Type", options=plot_types)
def update_plots(*events):
# Get the current values of the dropdown widgets
column_x = dropdown_x.value
column_y = dropdown_y.value
plot_type = dropdown_plot_type.value
# Generate the plot based on the selected values
plot = get_plots(column_x, column_y, plot_type)
# Update the panel with the new plot
plot_panel[1] = pn.pane.HoloViews(plot)
# Bind the dropdowns to the update function
dropdown_x.param.watch(update_plots, "value")
dropdown_y.param.watch(update_plots, "value")
dropdown_plot_type.param.watch(update_plots, "value")
# Create a layout for the plots and dropdowns
plot_panel = pn.Column(
pn.Row(dropdown_x, dropdown_y, dropdown_plot_type),
pn.pane.HoloViews(),
sizing_mode="stretch_width",
)
# Serve the dashboard
update_plots(None)
plot_panel.servable()
import pandas as pd
import panel as pn
import hvplot.pandas
import holoviews as hv
# Load data (not shown)
# data = pd.read_csv('my_data.csv')
# Define the available columns for the dropdowns
columns = list(data.columns)
# Define the available plot types for the dropdown
plot_types = {
"Histogram": "hist",
"Boxplot": "box",
"Line Chart": "line",
"Scatter Plot": "scatter",
"Violin Plot": "violin",
"Displot": "kde",
"Summary Statistics Table": "table",
"Heatmap": "heatmap",
}
# Define the plots
def get_plots(column_x, column_y, plot_type):
if plot_type == "hist":
plot = data.hvplot.hist(column_x, title=f"{column_x} Distribution", bins=20)
elif plot_type == "box":
plot = data.hvplot.box(
y=column_y, title=f"{column_y} Distribution", groupby=None
)
elif plot_type == "line":
plot = data.hvplot.line(column_x, column_y, title=f"{column_y} by {column_x}")
elif plot_type == "scatter":
plot = data.hvplot.scatter(
column_x,
column_y,
c="color",
cmap="viridis",
title=f"{column_y} vs {column_x}",
)
elif plot_type == "violin":
plot = data.hvplot.violin(
column_x, column_y, title=f"{column_y} by {column_x}", groupby=None
)
elif plot_type == "kde":
plot = data.hvplot.kde(column_x, title=f"{column_x} Distribution")
elif plot_type == "table":
plot = hv.Table(data[column_x].describe())
elif plot_type == "heatmap":
plot = hv.HeatMap(data.corr())
return plot
# Define the dropdown widgets
dropdown_x = pn.widgets.Select(name="Horizontal Variables", options=columns)
dropdown_y = pn.widgets.Select(name="Vertical Variables", options=columns)
dropdown_plot_type = pn.widgets.Select(name="Plot Type", options=plot_types)
def update_plots(*events):
# Get the current values of the dropdown widgets
column_x = dropdown_x.value
column_y = dropdown_y.value
plot_type = dropdown_plot_type.value
# Generate the plot based on the selected values
plot = get_plots(column_x, column_y, plot_type)
# Update the panel with the new plot
plot_panel[1] = pn.pane.HoloViews(plot)
# Bind the dropdowns to the update function
dropdown_x.param.watch(update_plots, "value")
dropdown_y.param.watch(update_plots, "value")
dropdown_plot_type.param.watch(update_plots, "value")
# Create a layout for the plots and dropdowns
plot_panel = pn.Column(
pn.Row(dropdown_x, dropdown_y, dropdown_plot_type),
pn.pane.HoloViews(),
sizing_mode="stretch_width",
)
# Serve the dashboard
update_plots(None)
plot_panel.servable()
# And we’ve written some code that smooths a time series and plots it using Matplotlib with outliers highlighted:
import matplotlib as mpl
mpl.use("agg")
from matplotlib.figure import Figure
def mpl_plot(avg, highlight):
fig = Figure()
ax = fig.add_subplot()
avg.plot(ax=ax)
if len(highlight):
highlight.plot(style="o", ax=ax)
return fig
def find_outliers(variable="age", window=30, sigma=10, view_fn=mpl_plot):
avg = data[variable].rolling(window=window).mean()
residual = data[variable] - avg
std = residual.rolling(window=window).std()
outliers = np.abs(residual) > std * sigma
return view_fn(avg, avg[outliers])
import param
import hvplot.pandas
import matplotlib as mpl
mpl.use("agg")
from matplotlib.figure import Figure
def mpl_plot(avg, highlight):
fig = Figure()
ax = fig.add_subplot()
avg.plot(ax=ax, kind="box", color="red")
if len(highlight):
highlight.plot(style="o", ax=ax)
return fig
def find_outliers(variable="CreditScore", window=30, sigma=10, view_fn=mpl_plot):
avg = data[variable].rolling(window=window).mean()
residual = data[variable] - avg
std = residual.rolling(window=window).std()
outliers = np.abs(residual) > std * sigma
return view_fn(avg, avg[outliers])
def hvplot(avg, highlight):
return avg.hvplot(height=200) * highlight.hvplot.scatter(
color="orange", padding=0.1
)
class Select_Dropdown_Variable(param.Parameterized):
variable = param.Selector(objects=list(data.columns))
window = param.Integer(default=10, bounds=(1, 20))
sigma = param.Number(default=10, bounds=(0, 20))
def view(self):
return find_outliers(self.variable, self.window, self.sigma, view_fn=hvplot)
obj = Select_Dropdown_Variable()
pn.Row(obj.param, obj.view)
columns = list(data.columns[0:-3])
x = pn.widgets.Select(value="CreditScore", options=columns, name="Horizontal Variables")
y = pn.widgets.Select(value="Gender", options=columns, name="Vertical Variables")
pn.Row(
pn.Column("## Select Feature Option", x, y),
pn.bind(data.hvplot.scatter, x, y, by="Gender"),
)
import pandas as pd
import panel as pn
import hvplot.pandas
import seaborn as sns
# Define the plots and tables
plots = {
"Histogram": data.hvplot.hist(y="CreditScore", title="Age Distribution", bins=20),
"Box Plot": data.hvplot.box(y="Balance", by="Exited", title="Balance Distribution"),
"Line Chart": data.hvplot.line(x="Age", y="Balance", title="Balance by Age"),
"Scatter Plot": data.hvplot.scatter(
x="CreditScore",
y="Age",
c="Exited",
cmap="viridis",
title="Exited by CreditScore and Age",
),
}
tables = {
"Summary Statistics 1": data[["CreditScore", "Balance"]]
.describe()
.T.hvplot.table(title="Summary Statistics 1"),
"Summary Statistics 2": data[["Age", "Tenure", "NumOfProducts", "EstimatedSalary"]]
.describe()
.T.hvplot.table(title="Summary Statistics 2"),
}
# Create the dropdown to select which plot or table to display
plot_dropdown = pn.widgets.Select(options=list(plots.keys()), name="Select a Plot")
table_dropdown = pn.widgets.Select(options=list(tables.keys()), name="Select a Table")
# Define a function to update the display based on the dropdown selection
def update_display(event):
if event.obj == plot_dropdown:
display_widget.object = plots[event.new]
elif event.obj == table_dropdown:
display_widget.object = tables[event.new]
# Create the initial display with the first plot
initial_plot = list(plots.keys())[0]
display_widget = pn.panel(plots[initial_plot])
# Combine the dropdown and the display in a panel layout
plot_dropdown.param.watch(update_display, "value")
table_dropdown.param.watch(update_display, "value")
dropdown_layout = pn.Row(plot_dropdown, table_dropdown)
layout = pn.Column(dropdown_layout, display_widget)
# Serve the dashboard
layout.servable()
import pandas as pd
import panel as pn
import hvplot.pandas
# Define the dropdown menu options
numeric_cols = data.select_dtypes(include=["float", "int"]).columns.tolist()
dropdown_scatter = pn.widgets.Select(
options=numeric_cols, name="Select a column for X-axis"
)
dropdown_box = pn.widgets.Select(options=numeric_cols, name="Select a column")
# Define a function to update the scatter plot based on the dropdown selection
def update_scatter(event):
plot = data.hvplot.scatter(
x=event.obj.value, y="Age", c="Exited", cmap="viridis", title="Scatter Plot"
)
return plot
# Define a function to update the box plot based on the dropdown selection
def update_box(event):
plot = data.hvplot.box(y=event.obj.value, by="Exited", title="Box Plot")
return plot
# Create the initial plots with the first numerical column
initial_col_scatter = numeric_cols[0]
initial_col_box = numeric_cols[0]
# Create Scatter and Boxplots
scatter_plot = data.hvplot.scatter(
x=initial_col_scatter, y="Age", c="Exited", cmap="viridis", title="Scatter Plot"
)
box_plot = data.hvplot.box(y=initial_col_box, by="Exited", title="Box Plot")
# Create Numerical Statistics
table1 = (
data[["CreditScore", "Balance"]]
.describe()
.T.hvplot.table(title="Summary Statistics 1")
)
table2 = (
data[["Age", "Tenure", "NumOfProducts", "EstimatedSalary"]]
.describe()
.T.hvplot.table(title="Summary Statistics 2")
)
# Create Correlation Heatmap
plt.figure(figsize=(30, 10))
heat_map = sns.heatmap(
data[
[
"CreditScore",
"Age",
"Tenure",
"Balance",
"NumOfProducts",
"HasCrCard",
"EstimatedSalary",
"Exited",
]
].corr(),
annot=True,
cmap="coolwarm",
vmin=-1,
vmax=1,
)
# Creat KDE and Bar Charts
kde_plot = data.hvplot.kde(y="CreditScore", by="Exited", title="KDE Plot")
bar_plot = data.hvplot.bar(y="Balance", x="Geography", stacked=True, title="Bar Plot")
# Define the Bar and Stack Chart functions
def create_pie_chart(column):
return data[column].value_counts().hvplot.bar(title=column, legend="right")
def create_bar_chart(column):
return data[column].value_counts().hvplot.bar(title=column, legend="right")
def create_stacked_bar_chart(column):
return (
data.groupby([column, "Exited"])
.size()
.unstack()
.hvplot.bar(stacked=True, title=column, legend="right")
)
# Creat Histograms and line charts
line_plot = data.hvplot.line(x="Age", y="Balance", groupby="Exited", title="Line Plot")
hist_plot = data.hvplot.hist(y="CreditScore", by="Gender", title="Histogram")
# Create the charts
pie_chart1 = create_pie_chart("Geography")
bar_chart1 = create_bar_chart("Gender")
stacked_bar_chart1 = create_stacked_bar_chart("Tenure")
pie_chart2 = create_pie_chart("HasCrCard")
bar_chart2 = create_bar_chart("IsActiveMember")
stacked_bar_chart2 = create_stacked_bar_chart("NumOfProducts")
# Create Vilin Charts and Heatmape
violin_plot = data.hvplot.violin(y="Age", by="Exited", title="Violin Plot")
heatmap_plot = data.hvplot.heatmap(
x="Geography", y="Age", C="Exited", reduce_function="mean", title="Heatmap"
)
# Combine the dropdowns and plots in a panel layout
dropdown_scatter.param.watch(update_scatter, "value")
dropdown_box.param.watch(update_box, "value")
scatter_box_layout = pn.Column(
pn.Row(dropdown_scatter, dropdown_box), pn.Row(scatter_plot, box_plot)
)
kde_bar_layout = pn.Column(kde_plot, bar_plot)
tables_layout = pn.Row(table1, table2)
violin_heatmap_layout = pn.Column(violin_plot, heatmap_plot)
line_hist_layout = pn.Column(line_plot, hist_plot)
# Create a tabbed layout for the dashboard
dashboard_tabs = pn.Tabs(
("Scatter & Box", scatter_box_layout),
("KDE & Bar", kde_bar_layout),
("Violin & Heatmap", violin_heatmap_layout),
("Line & Histogram", line_hist_layout),
("Summary Tables", tables_layout),
("Geography", pn.Row(pie_chart1)),
("Gender", pn.Row(bar_chart1)),
("Tenure", pn.Row(stacked_bar_chart1)),
("HasCrCard", pn.Row(pie_chart2)),
("IsActiveMember", pn.Row(bar_chart2)),
("NumOfProducts", pn.Row(stacked_bar_chart2)),
)
# Serve the dashboard
dashboard_tabs.servable()
import pandas as pd
import panel as pn
import hvplot.pandas
# Get the list of numerical columns
numeric_cols = data.select_dtypes(include=["float", "int"]).columns.tolist()
# Create the dropdown menu with numerical columns
dropdown = pn.widgets.Select(options=numeric_cols, name="Select a column")
# Define a function to update the plot based on the dropdown selection
def update_plot(event):
plot = data.hvplot.box(y=event.obj.value, title="Box plot of " + event.obj.value)
return plot
# Create the initial plot with the first numerical column
initial_col = numeric_cols[0]
plot = data.hvplot.box(y=initial_col, title="Box plot of " + initial_col, color="red")
# Combine the dropdown and the plot in a panel layout
dropdown.param.watch(update_plot, "value")
layout = pn.Row(dropdown, plot)
# Create a layout and serve the app
layout.servable()
import holoviews as hv
import numpy as np
import pandas as pd
import param
import panel as pn
# Define a function to create the boxplot
def create_boxplot(data, variable):
return data.hvplot.box(y=variable)
# Define the RoomOccupancy class with the dropdown and plot
class RoomOccupancy(param.Parameterized):
variable = param.ObjectSelector(default=data.columns[0], objects=list(data.columns))
@param.depends("variable")
def view(self):
return create_boxplot(data, self.variable)
# Define the dropdown widget to select the variable
variable_selector = hv.streams.Stream.define("Variable", variable=data.columns[0])(
variable=data.columns[0]
)
variable_menu = pn.widgets.Select(options=list(data.columns), value=data.columns[0])
# Define a function to update the plot based on the dropdown value
def update_boxplot(variable):
return create_boxplot(data, variable)
# Combine the widgets and the plot using a layout
layout = pn.Row(
pn.Column(variable_menu),
pn.Column(hv.DynamicMap(update_boxplot, streams=[variable_selector])),
)
# Create a layout and serve the app
layout.servable()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import heapq
import matplotlib
import matplotlib.pyplot as plt
from PIL import Image
import random
import math
from sklearn.neural_network import MLPClassifier
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
test_csv = pd.read_csv("/kaggle/input/mnist-in-csv/mnist_test.csv")
train_csv = pd.read_csv("/kaggle/input/mnist-in-csv/mnist_train.csv")
X_test = test_csv.drop(columns=["label"]).to_numpy()
X_train = train_csv.drop(columns=["label"]).to_numpy()
y_test = test_csv["label"].copy().to_numpy()
y_train = train_csv["label"].copy().to_numpy()
class NeuralNetwork:
def __init__(self, shape, weights=[], biases=[], lr=1e-4):
layers = []
self.lr = lr
for n in shape:
layers.append(np.random.uniform(-0.1, 0.1, n))
if not weights:
for n in range(len(shape) - 1):
layer_weights = np.array(
[
np.random.uniform(-0.1, 0.1, shape[n])
for _ in range(shape[n + 1])
]
)
weights.append(layer_weights)
if not biases:
for n in range(1, len(shape)):
layer_bias = np.random.uniform(-0.1, 0.1, shape[n])
biases.append(layer_bias)
self.layers = layers
self.weights = weights
self.biases = biases
def forward_prop(self, input_data):
z_list = []
self.layers[0] = input_data
for i in range(1, len(self.layers)):
layer_weights = self.weights[i - 1]
z = (layer_weights @ self.layers[i - 1]) + self.biases[i - 1]
z_list.append(z)
a = np.array([self.activate(num) for num in z])
self.layers[i] = a
return z_list
def activate(self, x):
return 1 / (1 + np.exp(-x))
def back_prop(self, label, z_list):
error_vector = 0.5 * (self.layers[-1] - label) ** 2
for i in range(len(self.layers) - 2, -1, -1):
weights = self.weights[i]
new_error = weights.T @ error_vector
for j in range(len(weights)):
self.biases[i][j] -= (
self.lr * error_vector[j] * (1 - z_list[i][j]) * z_list[i][j]
)
for k in range(len(weights[j])):
weights[j][k] -= (
self.lr
* error_vector[j]
* (1 - z_list[i][j])
* z_list[i][j]
* self.layers[i][k]
)
error_vector = np.copy(new_error)
def train(self, epochs, examples, training_labels):
examples = (examples - np.min(examples)) / (np.max(examples) - np.min(examples))
clean_labels = np.zeros((training_labels.size, training_labels.max() + 1))
clean_labels[np.arange(training_labels.size), training_labels] = 1
training_labels = clean_labels
for _ in range(epochs):
for example, label in zip(examples, training_labels):
z_list = self.forward_prop(example)
self.back_prop(label, z_list)
print("done with epoch")
def predict(self, features):
features = (features - np.min(features)) / (np.max(features) - np.min(features))
self.forward_prop(features)
return self.layers[-1]
nn = NeuralNetwork([784, 16, 16, 10])
nn.train(2, X_train[:5000], y_train[:5000])
prediction1 = nn.predict(X_test[1])
print(f"prediction1={prediction1} label={y_test[1]}")
prediction2 = nn.predict(X_test[3])
print(f"prediction2={prediction2} label={y_test[3]}")
# model = MLPClassifier(alpha=1e-6, hidden_layer_sizes=(75, 50, 25))
# model.fit(X_train, y_train)
# model.score(X_test, y_test)
# prediction = model.predict([X_test[157]])[0]
# test1 = np.array(Image.open("/kaggle/input/seventest/Capture (2).png"))
# test1_pred = model.predict(test1.reshape(1, 784))[0]
# test2 = np.array(Image.open("/kaggle/input/test-2/newimg.jpeg"))
# test2_pred = model.predict(test2.reshape(1, 784))[0]
# fig, (ax1, ax2, ax3) = plt.subplots(1, 3)
# ax1.imshow(X_test[157].reshape(28, 28))
# ax1.set_title(f"prediction={prediction}")
# ax2.imshow(test1.reshape(28, 28))
# ax2.set_title(f"prediction={test1_pred}")
# ax3.imshow(test2.reshape(28, 28))
# ax3.set_title(f"prediction={test2_pred}")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Yerleşik Veri Türleri
# Programlamada veri tipi önemli bir kavramdır.
# Değişkenler farklı türde verileri depolayabilir ve farklı türler farklı şeyler yapabilir.
# Python, bu kategorilerde varsayılan olarak yerleşik olarak aşağıdaki veri türlerine sahiptir:
# Text Type: str Numeric Types: int, float, complex Sequence Types: list, tuple, range Mapping Type: dict Set Types: set, frozenset Boolean Type: bool Binary Types: bytes, bytearray, memoryview None Type: NoneType
# Veri Türünü Öğrenme
# type() işlevini kullanarak herhangi bir nesnenin veri türünü öğrenebilirsiniz.
x = 2
print(type(x))
# Setting the Data Type
# Example Data Ty pe Try it x = "Hello World" str x = 20 int x = 20.5 float x = 1j complex x = ["apple", "banana", "cherry"] list x = ("apple", "banana", "cherry") tuple x = range(6) range x = {"name" : "John", "age" : 36} dict x = {"apple", "banana", "cherry"} set x = frozenset({"apple", "banana", "cherry"}) frozenset x = True bool x = b"Hello" bytes x = bytearray(5) bytearray x = memoryview(bytes(5)) memoryview x = None NoneType
# Belirli Veri Türünü Ayarlama
# If you want to specify the data type, you can use the following constructor functions:
# Example Data Type Try it x = str("Hello World") str x = int(20) int x = float(20.5) float x = complex(1j) complex x = list(("apple", "banana", "cherry")) list x = tuple(("apple", "banana", "cherry")) tuple x = range(6) range x = dict(name="John", age=36) dict x = set(("apple", "banana", "cherry")) set x = frozenset(("apple", "banana", "cherry")) frozenset x = bool(5) bool x = bytes(5) bytes x = bytearray(5) bytearray x = memoryview(bytes(5)) memoryview
# Python Numbers
# Python'da üç sayısal tür vardır:
# int float complex Sayısal tipteki değişkenler, onlara bir değer atadığınızda oluşturulur:
x = 2 # int
y = 3.8 # float
z = 3j # complex
# Python'da herhangi bir nesnenin türünü doğrulamak için type() işlevini kullanırız.
print(type(x))
print(type(y))
print(type(z))
# Int - Integer
# Int veya tamsayı, pozitif veya negatif, ondalık basamak içermeyen, sınırsız uzunlukta bir tam sayıdır.
# integers
x = 2
y = 38656222554887711
z = -3155522
print(type(x))
print(type(y))
print(type(z))
# Float
# Bir veya daha fazla ondalık basamak içeren pozitif veya negatif bir sayıdır.
x = 2.10
y = 3.8
z = -31.59
print(type(x))
print(type(y))
print(type(z))
# Float, 10'un kuvvetini belirtmek için "e" harfi bulunan bilimsel sayılar da olabilir.
# floats
x = 35e3
y = 12e4
z = -87.7e100
print(type(x))
print(type(y))
print(type(z))
# Complex
# Karmaşık sayılar sanal kısım olarak "j" ile yazılır:
x = 2 + 5j
y = 5j
z = -5j
print(type(x))
print(type(y))
# Tip Dönüşümü
# int(), float() ve Complex() yöntemleriyle bir türden diğerine dönüştürebilirsiniz:
x = 2 # int
y = 3.8 # float
z = 3j # complex
# convert from int to float:
a = float(x)
# convert from float to int:
b = int(y)
# convert from int to complex:
c = complex(x)
print(a)
print(b)
print(c)
print(type(a))
print(type(b))
print(type(c))
# Not: Karmaşık sayıları başka bir sayı türüne dönüştüremezsiniz.
# Rastgele Sayılar
# Python'un rasgele bir sayı yapmak için bir random() işlevi yoktur, ancak Python'un rasgele sayılar yapmak için kullanılabilecek random adlı yerleşik bir modülü vardır:
import random
print(random.randrange(1, 10))
# Bir Değişken Türü Oluşturma
# Bir değişkene bir tür belirtmek istediğiniz zamanlar olabilir. Bu döküm ile yapılabilir. Python, nesne yönelimli bir dildir ve bu nedenle, ilkel türleri de dahil olmak üzere veri türlerini tanımlamak için sınıfları kullanır.
# Bir Değişken Türü Belirtin
# Bir değişkene bir tür belirtmek istediğiniz zamanlar olabilir. Python, nesne yönelimli bir dildir
# int() - bir tamsayı hazır bilgisinden, bir değişken değişmez bilgisinden (tüm ondalık sayıları kaldırarak) veya bir dize değişmez bilgisinden (dizgenin bir tam sayıyı temsil etmesi koşuluyla) bir tamsayı oluşturur float() - bir tamsayı hazır bilgisinden, bir değişken sabit değerden veya bir dize değişmez bilgisinden bir kayan sayı oluşturur (dizenin bir kayan nokta veya bir tamsayıyı temsil etmesi koşuluyla) str() - diziler, tamsayı sabit değerleri ve değişken sabit değerler dahil olmak üzere çok çeşitli veri türlerinden bir dize oluşturur
# integers
x = int(2) # x will be 1
y = int(3.8) # y will be 2
z = int("3") # z will be 3
print(x)
print(y)
print(z)
# floats
x = float(1) # x will be 1.0
y = float(3.8) # y will be 3.8
z = float("3") # z will be 3.0
w = float("4.2") # w will be 4.2
print(x)
print(y)
print(z)
print(w)
# strings
x = str("s1") # x will be 's1'
y = str(2) # y will be '2'
z = str(3.0) # z will be '3.0'
print(x)
print(y)
print(z)
# Strings
# Python'daki dizeler, tek tırnak işaretleri veya çift tırnak işaretleri içine alınır.
# "merhaba", "merhaba" ile aynıdır.
# print() işleviyle bir dize hazır bilgisini görüntüleyebilirsiniz:
print("merhaba")
print("merhaba")
# Dizeyi bir Değişkene Atama
# Bir değişkene bir dize atamak, değişken adının ardından eşittir işareti ve dize ile yapılır:
a = "merhaba"
print(a)
# Çok Satırlı Dizeler
# Üç tırnak kullanarak bir değişkene çok satırlı bir dize atayabilirsiniz:
a = """Låt det vara själva smärtan,
elit bir yapı att bilda
men de hände vid en sådan tidpunkt
med några stora förlossningar och smärta.
tryck"""
print(a)
# Sringsler Dizilerdir
# Diğer birçok popüler programlama dili gibi, Python'daki dizeler de unicode karakterleri temsil eden bayt dizileridir.
# Bununla birlikte, Python'un bir karakter veri türü yoktur, tek bir karakter yalnızca 1 uzunluğunda bir dizedir.
# Dizenin öğelerine erişmek için köşeli parantezler kullanılabilir.
# 1 konumundaki karakteri alın (ilk karakterin 0 konumunda olduğunu unutmayın):
#
a = "merhaba insanlar!"
print(a[1])
# Bir Dizide Döngü Yapmak
# Dizeler dizi olduğundan, bir dizideki karakterler arasında bir for döngüsü ile döngü yapabiliriz.
# "aslan" kelimesindeki harfler arasında dolaşın:
for x in "lion":
print(x)
# String Length
# Bir dizenin uzunluğunu almak için len() işlevini kullanın.
# len() işlevi, bir dizenin uzunluğunu döndürür:
a = "merhaba insanlar!"
print(len(a))
# Dizeyi Kontrol Et
# Bir dizgede belirli bir ifadenin veya karakterin olup olmadığını kontrol etmek için in anahtar kelimesini kullanabiliriz.
# Aşağıdaki metinde "free" olup olmadığını kontrol edin:
txt = "If you're not going to enjoy life, why exist?"
print("exist" in txt)
# Bir if ifadesinde kullanın:
txt = "If you're not going to enjoy life, why exist?"
if "exist" in txt:
print("Yes, 'exist' is present.")
# OLMADIĞINI kontrol edin
# Belirli bir kelime öbeğinin veya karakterin bir dizgede OLMADIĞINI kontrol etmek için not in anahtar kelimesini kullanabiliriz.
# Aşağıdaki metinde "pahalı" ifadesinin OLMADIĞINI kontrol edin
txt = "If you're not going to enjoy life, why exist?"
print("free" not in txt)
# Use it in an if statement:
txt = "If you're not going to enjoy life, why exist?"
if "expensive" not in txt:
print("No, 'free' is NOT present.")
|
# ***Notbook still underwork***
# # **Import Dependncies**
import numpy as np
import pandas as pd
import os
import seaborn as sns
from matplotlib import pyplot as plt
plt.style.use("ggplot")
from math import ceil
from datashader.utils import lnglat_to_meters as webm
import holoviews as hv
import geoviews as gv
import datashader as ds
from colorcet import fire, rainbow, bgy, bjy, bkr, kb, kr
from datashader.colors import colormap_select, Greys9
from holoviews.streams import RangeXY
from holoviews.operation.datashader import datashade, dynspread, rasterize
from bokeh.io import push_notebook, show, output_notebook
output_notebook()
hv.extension("bokeh")
from datashader import transfer_functions as tf
from functools import partial
from datashader.utils import export_image
from IPython.core.display import HTML, display
from colorcet import fire, rainbow, bgy, bjy, bkr, kb, kr
# # **the data path**
path = "/kaggle/input/brazilian-ecommerce/"
# # **EDA**
olist_customer = pd.read_csv(path + "olist_customers_dataset.csv")
olist_geolocation = pd.read_csv(path + "olist_geolocation_dataset.csv")
olist_order_items = pd.read_csv(path + "olist_order_items_dataset.csv")
olist_order_payments = pd.read_csv(path + "olist_order_payments_dataset.csv")
olist_order_reviews = pd.read_csv(path + "olist_order_reviews_dataset.csv")
olist_orders = pd.read_csv(path + "olist_orders_dataset.csv")
olist_products = pd.read_csv(path + "olist_products_dataset.csv")
olist_sellers = pd.read_csv(path + "olist_sellers_dataset.csv")
# **Function to return the shape of the dataset**
def shape_of_dataset(df, dataset_name="df"):
print(f"{dataset_name} dataset has {df.shape[0]} nrows and {df.shape[1]} ncolumns")
return df.shape[0], df.shape[1]
customer_r, customer_c = shape_of_dataset(olist_customer, "Olist Customer")
geolocation_r, geolocation_c = shape_of_dataset(olist_geolocation, "Olist Geolocation")
order_items_r, order_items_c = shape_of_dataset(olist_order_items, "Olist Order Items")
order_payments_r, order_payments_c = shape_of_dataset(
olist_order_payments, "Olist Order Payments"
)
order_reviews_r, order_reviews_c = shape_of_dataset(
olist_order_reviews, "Olist Order Reviews"
)
orders_r, orders_c = shape_of_dataset(olist_orders, "Olist Orders")
products_r, products_c = shape_of_dataset(olist_products, "Olist Products")
sellers_r, sellers_c = shape_of_dataset(olist_sellers, "Olist Sellers")
# **Function to return how many null values in the dataset**
def count_null_values(df, dataset_name):
num_of_total_null_values = sum(df.isnull().sum().values)
print(f"{dataset_name} dataset has {num_of_total_null_values} null values")
return num_of_total_null_values
customer_null = count_null_values(olist_customer, "Olist Customer")
geolocation_null = count_null_values(olist_geolocation, "Olist Geolocation")
order_items_null = count_null_values(olist_order_items, "Olist Order Items")
order_payments_null = count_null_values(olist_order_payments, "Olist Order Payments")
order_reviews_null = count_null_values(olist_order_reviews, "Olist Order Reviews")
orders_null = count_null_values(olist_orders, "Olist Orders")
products_null = count_null_values(olist_products, "Olist Products")
sellers_null = count_null_values(olist_sellers, "Olist Products")
# **Olist Order Reviews, Orders and Products datasets have null values**
# **Function to return columns with null values in the dataset**
def detect_null_columns(df, dataset_name):
col = []
s = df.isnull().sum()
for x in range(len(s)):
if s[x] > 0:
col.append(s.index[x])
tot_cols = len(col)
if tot_cols == 0:
print(f"{dataset_name} dataset has no null columns")
else:
print(f"{dataset_name} dataset has {tot_cols} null columns and they are:")
for x in col:
print(x, end=",")
print()
return col, len(col)
total_customer_null_cols, customer_null_cols = detect_null_columns(
olist_customer, "Olist Customer"
)
total_geolocation_null_cols, geolocation_null_cols = detect_null_columns(
olist_geolocation, "Olist Geolocation"
)
total_order_items_null_cols, order_items_null_cols = detect_null_columns(
olist_order_items, "Olist Order Items"
)
total_order_payments_null_cols, order_payments_null_cols = detect_null_columns(
olist_order_payments, "Olist Order Payments"
)
total_order_reviews_null_cols, order_reviews_null_cols = detect_null_columns(
olist_order_reviews, "Olist Order Reviews"
)
total_orders_null_cols, orders_null_cols = detect_null_columns(
olist_orders, "Olist Orders"
)
total_products_null_cols, products_null_cols = detect_null_columns(
olist_products, "Olist Products"
)
total_sellers_null_cols, sellers_null_cols = detect_null_columns(
olist_sellers, "Olist Products"
)
# ### **Create df to compare between the datasets**
detailed_db = pd.DataFrame(
{
"dataset": [],
"nrows": [],
"ncols": [],
"null_amount": [],
"names_of_null_cols": [],
"num_null_cols": [],
}
)
def fill_db_dataset(
dataset_name, nrows, ncols, null_amount, name_null_cols, num_null_cols
):
detailed_db.loc[len(detailed_db.index)] = [
dataset_name,
nrows,
ncols,
null_amount,
", ".join(name_null_cols),
int(num_null_cols),
]
fill_db_dataset(
"Olist Customer",
customer_r,
customer_c,
customer_null,
total_customer_null_cols,
customer_null_cols,
)
fill_db_dataset(
"Olist Geolocation",
geolocation_r,
geolocation_c,
geolocation_null,
total_geolocation_null_cols,
geolocation_null_cols,
)
fill_db_dataset(
"Olist Order Items",
order_items_r,
order_items_c,
order_items_null,
total_order_items_null_cols,
order_items_null_cols,
)
fill_db_dataset(
"Olist Order Payments",
order_payments_r,
order_payments_c,
order_payments_null,
total_order_payments_null_cols,
order_payments_null_cols,
)
fill_db_dataset(
"Olist Order Reviews",
order_reviews_r,
order_reviews_c,
order_reviews_null,
total_order_reviews_null_cols,
order_reviews_null_cols,
)
fill_db_dataset(
"Olist Orders",
orders_r,
orders_c,
orders_null,
total_orders_null_cols,
orders_null_cols,
)
fill_db_dataset(
"Olist Products",
products_r,
products_c,
products_null,
total_products_null_cols,
products_null_cols,
)
fill_db_dataset(
"Olist Sellers",
sellers_r,
sellers_c,
sellers_null,
total_sellers_null_cols,
sellers_null_cols,
)
detailed_db
olist_orders.head()
olist_orders.dtypes
olist_orders.info()
olist_orders.describe(include="all")
olist_orders.nunique()
# ## **Function to countplot any column**
def count_plot(
x,
df,
title,
xlabel,
ylabel,
width,
height,
order=None,
rotation=False,
palette="winter",
hue=None,
):
ncount = len(df)
plt.figure(figsize=(width, height))
ax = sns.countplot(x=x, palette=palette, order=order, hue=hue)
plt.title(title, fontsize=20)
if rotation:
plt.xticks(rotation="vertical")
plt.xlabel(xlabel, fontsize=15)
plt.ylabel(ylabel, fontsize=15)
ax.yaxis.set_label_position("left")
for p in ax.patches:
x = p.get_bbox().get_points()[:, 0]
y = p.get_bbox().get_points()[1, 1]
ax.annotate(
"{:.1f}%".format(100.0 * y / ncount),
(x.mean(), y),
ha="center",
va="bottom",
) # set the alignment of the text
plt.show()
def bar_plot(
x,
y,
df,
title,
xlabel,
ylabel,
width,
height,
order=None,
rotation=False,
palette="winter",
hue=None,
):
ncount = len(df)
plt.figure(figsize=(width, height))
ax = sns.barplot(x=x, y=y, palette=palette, order=order, hue=hue)
plt.title(title, fontsize=20)
if rotation:
plt.xticks(rotation="vertical")
plt.xlabel(xlabel, fontsize=15)
plt.ylabel(ylabel, fontsize=15)
ax.yaxis.set_label_position("left")
for p in ax.patches:
x = p.get_bbox().get_points()[:, 0]
y = p.get_bbox().get_points()[1, 1]
ax.annotate(
"{:.1f}%".format(100.0 * y / ncount),
(x.mean(), y),
ha="center",
va="bottom",
) # set the alignment of the text
plt.show()
olist_orders.order_status.value_counts()
x = olist_orders["order_status"]
order = olist_orders["order_status"].value_counts().index
count_plot(
x,
olist_orders,
"Order Status",
"Status",
"Frequency",
12,
8,
order=order,
rotation=True,
)
# #### **97% of the orders are delivered**
# ### **Convert the datatype of all the time columns to datetime**
olist_orders["orders_delivered_carrier_date"] = pd.to_datetime(
olist_orders.order_delivered_carrier_date
)
olist_orders["order_estimated_delivery_date"] = pd.to_datetime(
olist_orders.order_estimated_delivery_date
)
olist_orders["order_delivered_customer_date"] = pd.to_datetime(
olist_orders.order_delivered_customer_date
)
olist_orders["order_delivered_carrier_date"] = pd.to_datetime(
olist_orders.order_delivered_carrier_date
)
olist_orders["order_purchase_timestamp"] = pd.to_datetime(
olist_orders.order_delivered_carrier_date
)
olist_orders.dtypes
olist_customer.head()
olist_orders_customers = pd.merge(olist_orders, olist_customer, on="customer_id")
olist_orders_customers.head()
olist_orders_customers.shape
olist_orders_customers.info()
olist_orders_customers.nunique()
olist_orders_customers["order_purchase_year"] = olist_orders_customers[
"order_purchase_timestamp"
].dt.strftime("%Y")
olist_orders_customers["order_purchase_month"] = olist_orders_customers[
"order_purchase_timestamp"
].dt.strftime("%-m")
olist_orders_customers["order_purchase_month_name"] = olist_orders_customers[
"order_purchase_timestamp"
].dt.strftime("%h")
olist_orders_customers["order_purchase_year_month"] = olist_orders_customers[
"order_purchase_year"
] + olist_orders_customers["order_purchase_timestamp"].dt.strftime("%m")
olist_orders_customers["order_purchase_day"] = olist_orders_customers[
"order_purchase_timestamp"
].dt.strftime("%-d")
olist_orders_customers["order_purchase_date"] = (
olist_orders_customers["order_purchase_year"]
+ olist_orders_customers["order_purchase_month"]
+ olist_orders_customers["order_purchase_day"]
)
olist_orders_customers["order_purchase_dayofweek"] = olist_orders_customers[
"order_purchase_timestamp"
].dt.strftime("%w")
olist_orders_customers["order_purchase_dayofweek_name"] = olist_orders_customers[
"order_purchase_timestamp"
].dt.strftime("%a")
olist_orders_customers["order_purchase_hour"] = olist_orders_customers[
"order_purchase_timestamp"
].dt.strftime("%H")
olist_orders_customers["order_purchase_time_day"] = olist_orders_customers[
"order_purchase_timestamp"
].dt.strftime("%p")
olist_orders_customers.head()
olist_orders_customers.dtypes
olist_orders_customers.isnull().sum()
olist_orders_customers["order_purchase_year"].value_counts()
x = olist_orders_customers["order_purchase_year"]
order = olist_orders_customers["order_purchase_year"].value_counts().index
count_plot(
x,
olist_orders_customers,
"Order Purchase year count",
"Year",
"num of orders",
12,
8,
order=order,
palette="spring",
)
# #### **There is a clear increase in online sales over years**
olist_orders_customers["order_purchase_month_name"].value_counts()
order = [
"Jun",
"Feb",
"Mar",
"Apr",
"May",
"Jun",
"Jul",
"Aug",
"Sep",
"Oct",
"Nov",
"Dec",
]
x = olist_orders_customers["order_purchase_month_name"]
count_plot(
x,
olist_orders_customers,
"Order Purchase per month",
"Month",
"num of orders",
12,
8,
order=order,
palette="summer",
)
olist_orders_customers["order_purchase_day"].value_counts()
x = olist_orders_customers["order_purchase_day"]
order = [str(i) for i in range(1, 32)]
count_plot(
x, olist_orders_customers, "Orders Per day", "day", "count", 14, 8, order=order
)
olist_orders_customers["order_purchase_dayofweek_name"].value_counts()
# I think there is an error in the dayofweek column
# as the Sun is mentioned only 37 times
order = ["Sat", "Sun", "Mon", "Tue", "Wed", "Thu", "Fri"]
x = olist_orders_customers["order_purchase_dayofweek_name"]
count_plot(
x,
olist_orders_customers,
"Order Purchase per day",
"Day",
"num of orders",
12,
8,
order=order,
palette="pastel",
)
olist_orders_customers["order_purchase_hour"].value_counts()
x = olist_orders_customers["order_purchase_hour"]
order = [str(i).zfill(2) for i in range(24)]
count_plot(
x,
olist_orders_customers,
"Order Purchase per hour",
"Hour",
"num of orders",
12,
8,
order=order,
palette="deep",
)
# ***brazilian's customers are prefered to buy 1:00PM and 2:00PM and they tend to buy more at afternoons.***
olist_orders_customers["order_purchase_time_day"].value_counts()
x = olist_orders_customers["order_purchase_time_day"]
count_plot(
x,
olist_orders_customers,
"Order Purchase time day",
"time day",
"num of orders",
8,
6,
order=["AM", "PM"],
)
# ***PM time day are the prefered time for brazilian's customers***
order = [
"Jan",
"Feb",
"Mar",
"Apr",
"May",
"Jun",
"Jul",
"Aug",
"Sep",
"Oct",
"Nov",
"Dec",
]
x = olist_orders_customers["order_purchase_month_name"]
hue = olist_orders_customers["order_purchase_year"]
count_plot(
x,
olist_orders_customers,
"Total orders comparison between years (JAN to DEC)",
"Month",
"Order Frequency",
15,
8,
order=order,
palette="Dark2",
hue=hue,
)
# ***It is clear that there is a significant increase in sales from 2017 (JAN to AUG) to 2018 (JAN to AUG)***
x = olist_orders_customers["order_purchase_month_name"]
hue = olist_orders_customers["order_purchase_time_day"]
order = [
"Jan",
"Feb",
"Mar",
"Apr",
"May",
"Jun",
"Jul",
"Aug",
"Sep",
"Oct",
"Nov",
"Dec",
]
count_plot(
x,
olist_orders_customers,
"Total orders time days across the months",
"Month",
"Order Frequency",
15,
8,
order=order,
palette="summer",
hue=hue,
)
x = olist_orders_customers["order_purchase_year"]
hue = olist_orders_customers["order_purchase_time_day"]
order = ["2016", "2017", "2018"]
count_plot(
x,
olist_orders_customers,
"Total orders time days across the years",
"year",
"Order Frequency",
12,
8,
order=order,
palette="dark",
hue=hue,
)
time_of_day = []
for time in olist_orders_customers["order_purchase_hour"]:
try:
time = int(time)
if time >= 6 and time < 12:
time_of_day.append("Morning")
elif time >= 12 and time < 17:
time_of_day.append("Afternoon")
elif time >= 17 and time <= 20:
time_of_day.append("Evening")
else:
time_of_day.append("Night")
except:
time_of_day.append("Unknown")
olist_orders_customers["classification_time_purchase"] = time_of_day
olist_orders_customers["classification_time_purchase"].value_counts()
x = olist_orders_customers["classification_time_purchase"]
order = x.value_counts().index
count_plot(
x,
olist_orders_customers,
"Total orders by time of the day",
"Time of day",
"Frequency",
13,
9,
order=order,
palette="dark",
)
# **Most of the orders are in afternoon period**
olist_orders_customers["order_purchase_year_month"].value_counts()
x = olist_orders_customers["order_purchase_year_month"].value_counts().index
y = olist_orders_customers["order_purchase_year_month"].value_counts().values
order = sorted(x)
bar_plot(
x,
y,
olist_orders_customers,
"Evolution of total orders in Brazilian Ecommerce",
"year_month",
"count",
12,
8,
rotation=True,
palette="flare",
order=order,
)
# ***we can see clear that customers are more prone to buy things online than before.***
olist_orders_customers["customer_state"].value_counts()
count_plot(
olist_orders_customers["customer_state"],
olist_orders_customers,
"Products Sold in states",
"State",
"Frequency",
13,
9,
order=olist_orders_customers["customer_state"].value_counts().index,
palette="bright",
)
# ## **A huge sales in SP state**
top_20_cities = olist_orders_customers["customer_city"].value_counts().head(20)
x = top_20_cities.index
y = top_20_cities.values
bar_plot(
x,
y,
olist_orders_customers,
"Top 20 Brazilian cities with most orders",
"City",
"Count",
12,
8,
rotation=True,
palette="flare",
)
olist_order_items.head()
olist_order_items.shape
olist_order_items.dtypes
olist_order_items.info()
olist_order_items.describe()
olist_cust_orders_items = pd.merge(
olist_orders_customers, olist_order_items, on="order_id"
)
olist_cust_orders_items.head()
prices_over_year_month = (
olist_cust_orders_items.groupby("order_purchase_year_month")["price"]
.agg("sum")
.reset_index()
)
prices_over_year_month
prices_over_year_month["price_ratio"] = (
prices_over_year_month["price"] / prices_over_year_month["price"].sum()
)
prices_over_year_month
x = prices_over_year_month["order_purchase_year_month"]
y = prices_over_year_month["price"]
plt.figure(figsize=(12, 8))
plt.plot(x, y, marker=".", color="purple", markersize=10, markeredgecolor="blue")
ax = sns.barplot(x=x, y=y, palette="flare")
plt.xticks(rotation="vertical")
c = 0
for p in ax.patches:
x = p.get_bbox().get_points()[:, 0]
y = p.get_bbox().get_points()[1, 1]
ax.annotate(
"{:.1f}%".format(prices_over_year_month["price_ratio"][c] * 100),
(x.mean(), y),
ha="center",
va="bottom",
) # set the alignment of the text
c += 1
plt.show()
avg_freight_value = (
olist_cust_orders_items.groupby("order_purchase_year_month")["freight_value"]
.agg("sum")
.reset_index()
)
avg_freight_value
avg_fr_val = []
for i in range(len(avg_freight_value)):
filter_df = olist_cust_orders_items[
olist_cust_orders_items["order_purchase_year_month"]
== avg_freight_value["order_purchase_year_month"][i]
]
avg_fr_val.append(ceil(filter_df["freight_value"].mean()))
avg_freight_value["avg_freight_value"] = avg_fr_val
avg_freight_value
x = avg_freight_value["order_purchase_year_month"]
y = avg_freight_value["avg_freight_value"]
plt.figure(figsize=(12, 8))
plt.plot(x, y, marker=".", color="purple", markersize=10, markeredgecolor="blue")
ax = sns.barplot(x=x, y=y, palette="bright")
plt.xticks(rotation="vertical")
plt.title("Evoloution of Average Freight value")
c = 0
for p in ax.patches:
x = p.get_bbox().get_points()[:, 0]
y = p.get_bbox().get_points()[1, 1]
ax.annotate(
"{:}".format(avg_freight_value["avg_freight_value"][c]),
(x.mean(), y),
ha="center",
va="bottom",
) # set the alignment of the text
c += 1
plt.show()
state_price = (
olist_cust_orders_items.groupby("customer_state")["price"]
.agg("mean")
.sort_values(ascending=False)
.reset_index()
)
state_price
x = state_price["customer_state"]
y = state_price["price"]
plt.figure(figsize=(12, 8))
ax = sns.barplot(x=x, y=y, palette="flare")
plt.xticks(rotation="vertical")
plt.title("Evoloution of Average Freight value")
c = 0
for p in ax.patches:
x = p.get_bbox().get_points()[:, 0]
y = p.get_bbox().get_points()[1, 1]
ax.annotate(
"{:}".format(ceil(state_price["price"][c])),
(x.mean(), y),
ha="center",
va="bottom",
) # set the alignment of the text
c += 1
plt.show()
olist_geolocation.head()
olist_geolocation.shape
olist_geolocation.nunique()
olist_geolocation.info()
olist_geolocation["geolocation_zip_code_prefix"] = olist_geolocation[
"geolocation_zip_code_prefix"
].astype("str")
olist_geolocation.dtypes
olist_geolocation["geolocation_zip_code_prefix_1_digits"] = olist_geolocation[
"geolocation_zip_code_prefix"
].str[0:1]
olist_geolocation["geolocation_zip_code_prefix_2_digits"] = olist_geolocation[
"geolocation_zip_code_prefix"
].str[0:2]
olist_geolocation["geolocation_zip_code_prefix_3_digits"] = olist_geolocation[
"geolocation_zip_code_prefix"
].str[0:3]
olist_geolocation["geolocation_zip_code_prefix_4_digits"] = olist_geolocation[
"geolocation_zip_code_prefix"
].str[0:4]
olist_geolocation.nunique()
olist_geolocation["geolocation_zip_code_prefix"].value_counts().to_frame().describe()
olist_geolocation = olist_geolocation[olist_geolocation.geolocation_lat <= 5.27438888]
# it’s most Western spot is at 73 deg, 58′ 58.19″W Long.
olist_geolocation = olist_geolocation[olist_geolocation.geolocation_lng >= -73.98283055]
# It’s most southern spot is at 33 deg, 45′ 04.21″ S Latitude.
olist_geolocation = olist_geolocation[olist_geolocation.geolocation_lat >= -33.75116944]
# It’s most Eastern spot is 34 deg, 47′ 35.33″ W Long.
olist_geolocation = olist_geolocation[olist_geolocation.geolocation_lng <= -34.79314722]
x, y = webm(olist_geolocation.geolocation_lng, olist_geolocation.geolocation_lat)
x
y
olist_geolocation["x"] = pd.Series(x)
olist_geolocation["y"] = pd.Series(y)
olist_geolocation.head()
olist_geolocation["geolocation_zip_code_prefix"] = olist_geolocation[
"geolocation_zip_code_prefix"
].astype(int)
olist_geolocation["geolocation_zip_code_prefix_1_digits"] = olist_geolocation[
"geolocation_zip_code_prefix_1_digits"
].astype(int)
olist_geolocation["geolocation_zip_code_prefix_2_digits"] = olist_geolocation[
"geolocation_zip_code_prefix_2_digits"
].astype(int)
olist_geolocation["geolocation_zip_code_prefix_3_digits"] = olist_geolocation[
"geolocation_zip_code_prefix_3_digits"
].astype(int)
olist_geolocation["geolocation_zip_code_prefix_4_digits"] = olist_geolocation[
"geolocation_zip_code_prefix_4_digits"
].astype(int)
olist_geolocation.dtypes
brazil = olist_geolocation
agg_name = "geolocation_zip_code_prefix"
brazil[agg_name].describe().to_frame()
T = 0.05
PX = 1
def plot_map(data, label, agg_data, agg_name, cmap):
url = "http://server.arcgisonline.com/ArcGIS/rest/services/Canvas/World_Dark_Gray_Base/MapServer/tile/{Z}/{Y}/{X}.png"
geomap = gv.WMTS(url)
points = hv.Points(gv.Dataset(data, kdims=["x", "y"], vdims=[agg_name]))
agg = datashade(points, element_type=gv.Image, aggregator=agg_data, cmap=cmap)
zip_codes = dynspread(agg, threshold=T, max_px=PX)
hover = hv.util.Dynamic(
rasterize(points, aggregator=agg_data, width=50, height=25, streams=[RangeXY]),
operation=hv.QuadMesh,
)
hover = hover.options(cmap=cmap)
img = geomap * zip_codes * hover
img = img.relabel(label)
return img
plot_map(brazil, "Zip Codes in Brazil", ds.min(agg_name), agg_name, cmap=rainbow)
# ***Python Packages used to make this plot:
# 1 - GeoViews is a Python library that makes it easy to explore and visualize geographical, meteorological, and oceanographic datasets.
# 2 - holoviews: annotate your data and it will visuliaze itself.
# 3 - datashader: Data visualization toolchain based on aggregating into a grid.***
background = "black"
cm = partial(colormap_select, reverse=(background != "black"))
export = partial(export_image, background=background, export_path="export")
display(HTML("<style>.container { width:100% !important; }</style>"))
W = 700
def create_map(data, cmap, data_agg, export_name="img"):
pad = (data.x.max() - data.x.min()) / 50
x_range, y_range = (
(data.x.min() - pad, data.x.max() + pad),
(data.y.min() - pad, data.y.max() + pad),
)
ratio = (y_range[1] - y_range[0]) / (x_range[1] - x_range[0])
plot_width = int(W)
plot_height = int(plot_width * ratio)
if ratio > 1.5:
plot_height = 550
plot_width = int(plot_height / ratio)
cvs = ds.Canvas(
plot_width=plot_width, plot_height=plot_height, x_range=x_range, y_range=y_range
)
agg = cvs.points(data, "x", "y", data_agg)
img = tf.shade(agg, cmap=cmap, how="eq_hist")
return export(img, export_name)
create_map(brazil, rainbow, ds.mean(agg_name), "brazil_zip_codes")
top_20_geo_states = olist_geolocation["geolocation_state"].value_counts().head(20)
top_20_geo_states
plt.figure(figsize=(10, 8))
x = top_20_geo_states.index
y = top_20_geo_states.values
sns.barplot(x=x, y=y, palette="dark")
plt.xticks(rotation="vertical")
plt.title("Geolocation States", fontsize=20)
plt.xlabel("State", fontsize=15)
plt.ylabel("Frequency", fontsize=15)
plt.show()
sp = olist_geolocation[olist_geolocation["geolocation_state"] == "SP"]
agg_name = "geolocation_zip_code_prefix"
sp[agg_name].describe().to_frame()
plot_map(sp, "Zip Codes in Sao Paulo State", ds.min(agg_name), agg_name, cmap=rainbow)
create_map(sp, rainbow, ds.mean(agg_name), "sp_zip_codes")
mg = olist_geolocation[olist_geolocation["geolocation_state"] == "MG"]
agg_name = "geolocation_zip_code_prefix"
mg[agg_name].describe().to_frame()
plot_map(
mg, "Zip Codes in Minas Gerais State", ds.min(agg_name), agg_name, cmap=rainbow
)
create_map(mg, rainbow, ds.mean(agg_name), "sp_zip_codes")
rj = olist_geolocation[olist_geolocation["geolocation_state"] == "RJ"]
agg_name = "geolocation_zip_code_prefix"
mg[agg_name].describe().to_frame()
plot_map(rj, "Zip Codes in RJ State", ds.min(agg_name), agg_name, cmap=rainbow)
create_map(rj, rainbow, ds.mean(agg_name), "rj_zip_codes")
top_20_geo_cities = olist_geolocation["geolocation_city"].value_counts().head(20)
top_20_geo_cities
plt.figure(figsize=(10, 8))
x = top_20_geo_cities.index
y = top_20_geo_cities.values
sns.barplot(x=x, y=y, palette="flare")
plt.xticks(rotation="vertical")
plt.title("Geolocation Cites", fontsize=20)
plt.xlabel("City", fontsize=15)
plt.ylabel("Frequency", fontsize=15)
plt.show()
saopaulo = olist_geolocation[olist_geolocation["geolocation_city"] == "sao paulo"]
agg_name = "geolocation_zip_code_prefix"
saopaulo[agg_name].describe().to_frame()
plot_map(
saopaulo, "Zip Codes in Sao Paulo City", ds.min(agg_name), agg_name, cmap=rainbow
)
create_map(saopaulo, rainbow, ds.mean(agg_name), "sp_zip_codes")
rio_de_janeiro = olist_geolocation[
olist_geolocation["geolocation_city"] == "rio de janeiro"
]
agg_name = "geolocation_zip_code_prefix"
rio_de_janeiro[agg_name].describe().to_frame()
plot_map(
rio_de_janeiro,
"Zip Codes in Rio De Janeiro City",
ds.min(agg_name),
agg_name,
cmap=rainbow,
)
create_map(rio_de_janeiro, rainbow, ds.mean(agg_name), "sp_zip_codes")
belo_horizonte = olist_geolocation[
olist_geolocation["geolocation_city"] == "belo horizonte"
]
agg_name = "geolocation_zip_code_prefix"
belo_horizonte[agg_name].describe().to_frame()
plot_map(
belo_horizonte,
"Zip Codes in Belo Horizonte City",
ds.min(agg_name),
agg_name,
cmap=rainbow,
)
create_map(belo_horizonte, rainbow, ds.mean(agg_name), "sp_zip_codes")
olist_geolocation["geolocation_zip_code_prefix_1_digits"].value_counts()
olist_geolocation["geolocation_zip_code_prefix"].head()
geo_zipCode_1dig = olist_geolocation[
olist_geolocation["geolocation_zip_code_prefix_1_digits"] == 2
]
create_map(geo_zipCode_1dig, cm(Greys9), ds.count(), "zip_code_2")
geo_zipCode_2dig = olist_geolocation[
olist_geolocation["geolocation_zip_code_prefix_2_digits"] == 22
]
create_map(geo_zipCode_2dig, cm(Greys9), ds.count(), "zip_code_22")
df = olist_geolocation[olist_geolocation["geolocation_zip_code_prefix_3_digits"] == 220]
create_map(df, cm(Greys9), ds.count(), "zip_code_220")
olist_customer.dtypes
olist_customer["customer_zip_code_prefix"] = olist_customer[
"customer_zip_code_prefix"
].astype("str")
olist_customer["customer_zip_code_prefix_3_digits"] = olist_customer[
"customer_zip_code_prefix"
].str[0:3]
olist_customer["customer_zip_code_prefix_3_digits"] = olist_customer[
"customer_zip_code_prefix_3_digits"
].astype("int32")
brazil_geo = olist_geolocation.set_index("geolocation_zip_code_prefix_3_digits").copy()
brazil_geo.head()
orders_df = olist_orders.merge(olist_order_items, on="order_id")
orders_df = orders_df.merge(olist_customer, on="customer_id")
orders_df = orders_df.merge(olist_order_reviews, on="order_id")
orders_df.head()
orders_df.shape
gp = orders_df.groupby("customer_zip_code_prefix_3_digits")["price"].sum().to_frame()
gp.head()
revenue = brazil_geo.join(gp)
revenue.head()
revenue.shape
agg_name = "revenue"
revenue[agg_name] = revenue.price / 1000
revenue.head()
plot_map(
revenue, "Orders Revenue (thousands R$)", ds.mean(agg_name), agg_name, cmap=fire
)
create_map(revenue, fire, ds.mean(agg_name), "revenue_brazil")
gp = orders_df.groupby("order_id").agg(
{"price": "sum", "customer_zip_code_prefix_3_digits": "max"}
)
gp.head()
gp = gp.groupby("customer_zip_code_prefix_3_digits")["price"].mean().to_frame()
gp.head()
avg_ticket = brazil_geo.join(gp)
avg_ticket.head()
agg_name = "avg_ticket"
avg_ticket[agg_name] = avg_ticket.price
avg_ticket.head()
plot_map(
avg_ticket, "Orders Average Ticket (R$)", ds.mean(agg_name), agg_name, cmap=bgy
)
create_map(avg_ticket, bgy, ds.mean("avg_ticket"), "avg_ticket_brazil")
gp = orders_df.groupby("order_id").agg(
{"price": "sum", "freight_value": "sum", "customer_zip_code_prefix_3_digits": "max"}
)
gp.head()
agg_name = "freight_ratio"
gp[agg_name] = gp.freight_value / gp.price
gp.head()
gp = gp.groupby("customer_zip_code_prefix_3_digits")[agg_name].mean().to_frame()
gp.head()
freight_ratio = brazil_geo.join(gp)
freight_ratio.head()
plot_map(
freight_ratio, "Orders Average Freight Ratio", ds.mean(agg_name), agg_name, cmap=bgy
)
create_map(freight_ratio, bgy, ds.mean("freight_ratio"), "freight_ratio_brazil")
orders_df["order_delivered_customer_date"] = pd.to_datetime(
orders_df["order_delivered_customer_date"]
)
orders_df["order_estimated_delivery_date"] = pd.to_datetime(
orders_df["order_estimated_delivery_date"]
)
orders_df["order_delivered_carrier_date"] = pd.to_datetime(
orders_df["order_delivered_carrier_date"]
)
orders_df["actual_delivery_time"] = (
orders_df["order_delivered_customer_date"]
- orders_df["order_delivered_carrier_date"]
)
orders_df["actual_delivery_time"] = orders_df["actual_delivery_time"].dt.days
orders_df[
[
"order_delivered_customer_date",
"order_delivered_carrier_date",
"actual_delivery_time",
]
].head()
gp = (
orders_df.groupby("customer_zip_code_prefix_3_digits")["actual_delivery_time"]
.mean()
.to_frame()
)
gp.head()
delivery_time = brazil_geo.join(gp)
delivery_time.head()
agg_name = "avg_delivery_time"
delivery_time[agg_name] = delivery_time["actual_delivery_time"]
delivery_time.head()
plot_map(
delivery_time,
"Orders Average Delivery Time (days)",
ds.mean(agg_name),
agg_name,
cmap=bjy,
)
create_map(delivery_time, bjy, ds.mean(agg_name), "avg_delivery_time_brazil")
pr = olist_geolocation[olist_geolocation["geolocation_state"] == "PR"]
pr.head()
pr.shape
pr = pr.set_index("geolocation_zip_code_prefix_3_digits")
pr.head()
gp = (
orders_df.groupby("customer_zip_code_prefix_3_digits")["actual_delivery_time"]
.mean()
.to_frame()
)
gp.head()
pr_delivery_time = pr.join(gp)
pr_delivery_time.head()
pr_delivery_time.shape
agg_name = "avg_delivery_time"
pr_delivery_time[agg_name] = pr_delivery_time["actual_delivery_time"]
pr.head()
plot_map(
pr_delivery_time,
"Orders Average Delivery Time in Parana State (days)",
ds.mean(agg_name),
agg_name,
cmap=bjy,
)
create_map(pr_delivery_time, bjy, ds.mean(agg_name), "avg_delivery_time_pr")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
import matplotlib.ticker as mtick
import re
import time
sns.set()
action_movie = pd.read_csv(
"/kaggle/input/imdb-movies-dataset-based-on-genre/action.csv"
)
crime_movie = pd.read_csv("/kaggle/input/imdb-movies-dataset-based-on-genre/crime.csv")
adventure_movie = pd.read_csv(
"/kaggle/input/imdb-movies-dataset-based-on-genre/adventure.csv"
)
thriller_movie = pd.read_csv(
"/kaggle/input/imdb-movies-dataset-based-on-genre/thriller.csv"
)
family_movie = pd.read_csv(
"/kaggle/input/imdb-movies-dataset-based-on-genre/family.csv"
)
mystery_movie = pd.read_csv(
"/kaggle/input/imdb-movies-dataset-based-on-genre/mystery.csv"
)
scifi_movie = pd.read_csv("/kaggle/input/imdb-movies-dataset-based-on-genre/scifi.csv")
history_movie = pd.read_csv(
"/kaggle/input/imdb-movies-dataset-based-on-genre/history.csv"
)
sports_movie = pd.read_csv(
"/kaggle/input/imdb-movies-dataset-based-on-genre/sports.csv"
)
animation_movie = pd.read_csv(
"/kaggle/input/imdb-movies-dataset-based-on-genre/animation.csv"
)
war_movie = pd.read_csv("/kaggle/input/imdb-movies-dataset-based-on-genre/war.csv")
biography_movie = pd.read_csv(
"/kaggle/input/imdb-movies-dataset-based-on-genre/biography.csv"
)
horror_movie = pd.read_csv(
"/kaggle/input/imdb-movies-dataset-based-on-genre/horror.csv"
)
fantasy_movie = pd.read_csv(
"/kaggle/input/imdb-movies-dataset-based-on-genre/fantasy.csv"
)
romance_movie = pd.read_csv(
"/kaggle/input/imdb-movies-dataset-based-on-genre/romance.csv"
)
film_noir_movie = pd.read_csv(
"/kaggle/input/imdb-movies-dataset-based-on-genre/film-noir.csv"
)
# Add genre column to each dataframe
action_movie["genre"] = "Action"
crime_movie["genre"] = "Crime"
adventure_movie["genre"] = "Adventure"
thriller_movie["genre"] = "Thriller"
family_movie["genre"] = "Family"
mystery_movie["genre"] = "Mystery"
scifi_movie["genre"] = "Sci-Fi"
history_movie["genre"] = "History"
sports_movie["genre"] = "Sports"
animation_movie["genre"] = "Animation"
war_movie["genre"] = "War"
biography_movie["genre"] = "Biography"
horror_movie["genre"] = "Horror"
fantasy_movie["genre"] = "Fantasy"
romance_movie["genre"] = "Romance"
film_noir_movie["genre"] = "Film-Noir"
# Concatenate all dataframes
df = pd.concat(
[
action_movie,
crime_movie,
adventure_movie,
thriller_movie,
family_movie,
mystery_movie,
scifi_movie,
history_movie,
sports_movie,
animation_movie,
war_movie,
biography_movie,
horror_movie,
fantasy_movie,
romance_movie,
film_noir_movie,
]
)
# Reset index
df = df.reset_index(drop=True)
# Preview the dataframe
df
# # Make new DF and clear it
# Drop unwanted values in year columns
unwanted_values = [
"I",
"II",
"V",
"III",
"VII",
"IV",
"XXIII",
"IX",
"XV",
"VI",
"X",
"XIV",
"XIX",
"XXIX",
"XXI",
"VIII",
"XI",
"XVIII",
"XII",
"XIII",
"LXXI",
"XVI",
"XX",
"XXXIII",
"XXXII",
"XXXVI",
"XVII",
"LXIV",
"LXII",
"LXVIII",
"XL",
"XXXIV",
"XXXI",
"XLV",
"XLIV",
"XXIV",
"XXVII",
"LX",
"XXV",
"XXXIX",
"2029",
"XXVIII",
"XXX",
"LXXII",
"1909",
"XXXVIII",
"XXII",
"LVI",
"LVII" "XLI",
"LII",
"XXXVII",
"LIX",
"LVIII",
"LXX",
"XLIII",
"XLIX",
"LXXIV",
"XXVI",
"C",
"XLI",
"LVII",
"LV",
"XLVI",
"LXXVII",
"XXXV",
"LIV",
"LI",
"LXXXII",
"XCIX",
"LXIII",
]
new_df = df[~df["year"].isin(unwanted_values)]
# # Clear year column
# fill the NaN with 0
new_df["year"] = new_df["year"].fillna(0).astype(int)
# replace 0 with np.nan
new_df["year"] = new_df["year"].replace(0, np.nan)
# drop rows with missing values in 'year' column
new_df = new_df.dropna(subset=["year"])
# Make into int
new_df["year"] = new_df["year"].astype(int)
new_df = new_df.drop(
["movie_id", "movie_id", "director_id", "star_id", "description"], axis=1
)
new_df
# # Clear director and Star columns from \n
new_df["director"] = new_df["director"].str.replace("\n", "")
new_df["star"] = new_df["star"].str.replace("\n", "")
# Drop Nan in runtime columns
new_df = new_df.dropna(subset=["runtime"])
# # Clear Runtime Column to be in time format
# Change string type in runtime to be int
new_df["runtime"] = new_df["runtime"].str.replace("min", "")
new_df["runtime"] = new_df["runtime"].str.replace(",", "").astype(int)
# Make it into datetime format
new_df["runtime"] = pd.to_timedelta(new_df["runtime"], unit="m")
new_df
# # Get movie with the highest rating for each year
new_df = new_df.sort_values(by="year", ascending=False)
# create an empty DataFrame with the same columns as new_df
highest_rated_movies = pd.DataFrame(columns=new_df.columns)
# doing for loop for each year
for year in new_df["year"].unique():
if year not in [2025, 2024, 2023]: # exclude years 2025, 2024, and 2023
df_year = new_df[new_df["year"] == year] # filter by year
if not df_year.empty: # check if the resulting DataFrame is not empty
df_year_sorted = df_year.sort_values(
by="rating", ascending=False
) # sort by rating in descending order
highest_rated_movie = df_year_sorted.iloc[
0
] # get the row with the highest rating
highest_rated_movies = highest_rated_movies.append(
highest_rated_movie
) # add the row to the new DataFrame
highest_rated_movies.head()
# # Get movie with the highest votes for each year
# create an empty DataFrame with the same columns as new_df
highest_voted_movies = pd.DataFrame(columns=new_df.columns)
# iterate through each unique year in the DataFrame
for year in new_df["year"].unique():
if year not in [2025, 2024, 2023]: # exclude years 2025, 2024, and 2023
df_year = new_df[new_df["year"] == year] # filter by year
if not df_year.empty: # check if the resulting DataFrame is not empty
df_year_sorted = df_year.sort_values(
by="votes", ascending=False
) # sort by votes in descending order
highest_voted_movie = df_year_sorted.iloc[
0
] # get the row with the highest votes
highest_voted_movies = highest_voted_movies.append(
highest_voted_movie
) # add the row to the new DataFrame
# highest_voted_movies = highest_voted_movies.drop(['movie_id', 'movie_id', 'director_id', 'star_id'], axis = 1)
highest_voted_movies.head(10)
### To get only votes based in one year
# df_2022 = new_df[new_df['year'] == 2022] # filter by year 2022
# df_2022_sorted = df_2022.sort_values(by='votes', ascending=False) # sort by votes in descending order
# highest_voted_movie_2022 = df_2022_sorted.iloc[0] # get the row with the highest votes for year 2022
highest_voted_movies.columns
# # Visualize relationship between votes, gross $ and rating
# create a function to format the y-axis tick labels for 'gross(in $)'
def gross_formatter(x, pos):
return "${:.1f}M".format(x * 1e-6)
# create the pair plot
g = sns.pairplot(new_df[["votes", "gross(in $)", "rating"]])
# set the y-axis tick label formatter for 'gross(in $)'
g.axes[1, 0].yaxis.set_major_formatter(mtick.FuncFormatter(gross_formatter))
# display the plot
plt.show()
# # Get genre that is popular in year
# create the new DataFrame with columns 'year', 'popular_genre', and 'number of release'
popular_genres = pd.DataFrame(columns=["year", "popular_genre", "number of release"])
# iterate through each unique year in the DataFrame
for year in new_df["year"].unique():
if year not in [2025, 2024, 2023]: # exclude years 2025, 2024, and 2023
df_year = new_df[new_df["year"] == year] # filter by year
if not df_year.empty: # check if the resulting DataFrame is not empty
# count the occurrences of each genre in the year
genre_counts = df_year["genre"].value_counts()
# get the most frequent genre
popular_genre = genre_counts.index[0]
# get the count of the popular genre
count = genre_counts.iloc[0]
# add the year, popular_genre, and count to the new DataFrame
popular_genres = popular_genres.append(
{
"year": year,
"popular_genre": popular_genre,
"number of release": count,
},
ignore_index=True,
)
popular_genres.head(23)
### Check by year manual
# df_year = new_df[new_df['year'] == 2022] # filter by year 2022
# genre_counts = df_year['genre'].value_counts().sort_values(ascending=False)
# genre_counts
# # Get movie with the gross revenue for each year
# create an empty DataFrame with the same columns as new_df
highest_revenue_movies = pd.DataFrame(columns=new_df.columns)
# iterate through each unique year in the DataFrame
for year in new_df["year"].unique():
if year not in [2025, 2024, 2023]: # exclude years 2025, 2024, and 2023
df_year = new_df[new_df["year"] == year] # filter by year
if not df_year.empty: # check if the resulting DataFrame is not empty
df_year_sorted = df_year.sort_values(
by="gross(in $)", ascending=False
) # sort by votes in descending order
highest_revenue_movie = df_year_sorted.iloc[
0
] # get the row with the highest votes
highest_revenue_movies = highest_revenue_movies.append(
highest_revenue_movie
) # add the row to the new DataFrame
# Drop columns that is unnecessary
highest_revenue_movies = highest_revenue_movies.sort_values(by="year", ascending=False)
highest_revenue_movies.head(23)
# sort the DataFrame by gross revenue in descending order and select the top 23 rows
top_grossing = highest_revenue_movies.sort_values("year", ascending=False).head(23)
# create the bar chart using seaborn
fig, ax = plt.subplots(figsize=(10, 6))
sns.barplot(x="year", y="gross(in $)", data=top_grossing, ax=ax)
# format y-axis tick labels in millions
ax.yaxis.set_major_formatter(mtick.FuncFormatter(lambda x, pos: f"{x/1e8:.0f}"))
# set the title and axis labels
ax.set_title("Top Movie High Revenue by Year")
ax.set_xlabel("Year")
ax.set_ylabel("Gross Revenue (in million $)")
# rotate the x-axis labels
plt.xticks(rotation=90)
# display the plot
plt.show()
certificate_counts_high_revenue = (
highest_revenue_movies["certificate"].value_counts().sort_values(ascending=False)
)
certificate_counts_high_revenue_df = (
certificate_counts_high_revenue.to_frame().reset_index()
)
certificate_counts_high_revenue_df.columns = ["certificate", "count"]
plt.figure(figsize=(10, 8))
sns.barplot(x="count", y="certificate", data=certificate_counts_high_revenue_df)
plt.title("Top Rated Certificates for High Revenue Movies")
plt.xlabel("Number of rated")
plt.ylabel("Rated")
plt.show()
plot = sns.boxplot(data=highest_revenue_movies, x="certificate", y="gross(in $)")
# get x axis to rotate 90 degrees
plot.set_xticklabels(plot.get_xticklabels(), rotation=90)
# set y-axis ticks to range from 0 to 1000 million
y_fmt = mtick.StrMethodFormatter("${x:,.0f}")
plot.yaxis.set_major_formatter(y_fmt)
plot.set_ylim([0, 10e8])
# set y-axis label
plt.title("Rating Categories that Generate the Highest Revenues")
plt.xlabel("Rated")
plt.ylabel("Gross Revenue")
plot = sns.boxplot(data=highest_revenue_movies, x="genre", y="gross(in $)")
# get x axis to rotate 90 degrees
plot.set_xticklabels(plot.get_xticklabels(), rotation=90)
# set y-axis ticks to range from 0 to 1000 million
y_fmt = mtick.StrMethodFormatter("${x:,.0f}")
plot.yaxis.set_major_formatter(y_fmt)
plot.set_ylim([0, 10e8])
# set y-axis label
plt.title("Genre Movie that Generate the Highest Revenues")
plt.xlabel("Genre Movie")
plt.ylabel("Gross Revenue")
### To get only revenue based in one year
# df_2022 = new_df[new_df['year'] == 2022] # filter by year 2022
# df_2022_sorted = df_2022.sort_values(by='gross(in $)', ascending=False) # sort by votes in descending order
# highest_voted_movie_2022 = df_2022_sorted.iloc[0:10] # get the row with the highest votes for year 2022
# highest_voted_movie_2022
# # Get average duration for each year
# group the DataFrame by 'year'
grouped = new_df.groupby("year")
# calculate the average runtime for each year
avg_runtime_by_year = grouped["runtime"].mean()
# convert the resulting Series to a DataFrame
df_avg_runtime_by_year = avg_runtime_by_year.reset_index()
df_avg_runtime_by_year.columns = ["year", "avg_runtime"]
# sort the DataFrame in descending order by 'year'
df_avg_runtime_by_year = df_avg_runtime_by_year.sort_values("year", ascending=False)
# exclude years 2025, 2024, and 2023
df_avg_runtime_by_year_excl = df_avg_runtime_by_year[
~df_avg_runtime_by_year["year"].isin([2025, 2024, 2023])
]
# select the data to plot
from1990_to_2000 = df_avg_runtime_by_year_excl.sort_values(
"year", ascending=False
).head(23)
# create the bar chart using seaborn
fig, ax = plt.subplots(figsize=(10, 6))
sns.barplot(
x="year",
y=from1990_to_2000["avg_runtime"].dt.total_seconds() / 3600,
data=from1990_to_2000,
ax=ax,
)
# set the title and axis labels
ax.set_title("Average Movie Runtime by Year")
ax.set_xlabel("Year")
ax.set_ylabel("Average Runtime (in hours)")
# rotate the x-axis labels
plt.xticks(rotation=90)
# display the plot
plt.show()
# # Get Director with the highest make movie
new_df.columns
director_counts = new_df["director"].value_counts().sort_values(ascending=False)
director_counts_df = director_counts.to_frame().reset_index()
director_counts_df.columns = ["director", "count"]
plt.figure(figsize=(10, 8))
sns.barplot(x="count", y="director", data=director_counts_df.head(10))
plt.title("Top 10 Directors with the Most Movie Releases")
plt.xlabel("Number of Movies")
plt.ylabel("Director")
plt.show()
# # See relationship between runtime and Gross Revenue
# convert runtime to minutes
new_df["runtime_mins"] = new_df["runtime"].dt.total_seconds() / 60
# set the style of the plots
sns.set_style("darkgrid")
# create a scatter plot using seaborn
sns.scatterplot(x="runtime_mins", y="gross(in $)", data=new_df)
# set the x and y labels
plt.xlabel("Runtime (mins)")
plt.ylabel("Gross (in $)")
# set the title of the plot
plt.title("Relationship between Runtime and Gross (in $)")
# set the limits of the x-axis to show only the values between the 20th and 75th percentile
xlim_min = new_df["runtime_mins"].quantile(0.1)
xlim_max = new_df["runtime_mins"].quantile(0.99)
plt.xlim(xlim_min, xlim_max)
# set the y-axis tick formatter to display values in millions
formatter = mtick.FuncFormatter(lambda x, pos: "{:.1f}M".format(x / 1000000))
plt.gca().yaxis.set_major_formatter(formatter)
# show the plot
plt.show()
|
# **Droping null values**
import pandas as pd
d = {
"student": ["Aman", "Yojit", None, "Mahendra", None],
"marks": [23, 21, 25, 24, None],
"age": [11, None, 13, 11, None],
}
d
a = pd.DataFrame(d)
a
a.dropna()
a.dropna(how="all") # drops those rows where all the values are null
a.dropna(how="all", inplace=True)
a
# **concept of axis**
a
a.dropna(axis=1)
# **Subset**
a.dropna(axis=0, subset=["marks"])
a.dropna(axis=1, subset=[1])
# **Videogames Datasheet **
b = pd.read_csv("/kaggle/input/videogamesales/vgsales.csv")
b
b.info()
b["Year"].dropna()
# **Fillna() - Handling null values by some relevant data**
d
a = pd.DataFrame(d)
a
a.dropna(how="all", inplace=True)
a
a["age"].fillna(a["age"].mean(), inplace=True)
a
|
# ## İş Problemi
# Şirketi terk edecek müşterileri tahmin edebilecek bir makine öğrenmesi modeli geliştirilmesi beklenmektedir.
# ## Veri Seti Hikayesi
# Telco müşteri kaybı verileri, üçüncü çeyrekte Kaliforniya'daki 7043 müşteriye ev telefonu ve İnternet hizmetleri sağlayan hayali bir telekom şirketi hakkında bilgi içerir. Hangi müşterilerin hizmetlerinden ayrıldığını, kaldığını veya hizmete kaydolduğunu gösterir.
import pandas as pd
import seaborn as sns
import numpy as np
import matplotlib.pyplot as plt
from sklearn.ensemble import RandomForestClassifier, GradientBoostingClassifier
from sklearn.linear_model import LogisticRegression
from sklearn.neighbors import KNeighborsClassifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.model_selection import GridSearchCV, cross_validate
from sklearn.preprocessing import StandardScaler, LabelEncoder
from xgboost import XGBClassifier
from lightgbm import LGBMClassifier
from catboost import CatBoostClassifier
import warnings
warnings.simplefilter(action="ignore")
pd.set_option("display.max_columns", None)
pd.set_option("display.float_format", lambda x: "%.3f" % x)
pd.set_option("display.width", 500)
# ## EDA
df_ = pd.read_csv(
"/kaggle/input/telco-customer-churn/WA_Fn-UseC_-Telco-Customer-Churn.csv"
)
df = df_.copy()
# Numerik ve Kategorik değişkenleri yakalayalım.
def grab_col_names(dataframe, cat_th=10, car_th=20):
cat_cols = [col for col in dataframe.columns if dataframe[col].dtypes == "O"]
num_but_cat = [
col
for col in dataframe.columns
if dataframe[col].dtypes != "O" and dataframe[col].nunique() < cat_th
]
cat_but_car = [
col
for col in dataframe.columns
if dataframe[col].dtypes == "O" and dataframe[col].nunique() > car_th
]
cat_cols = cat_cols + num_but_cat
cat_cols = [col for col in cat_cols if col not in cat_but_car]
num_cols = [col for col in dataframe.columns if dataframe[col].dtypes != "O"]
num_cols = [col for col in num_cols if col not in num_but_cat]
print(f"Observations: {dataframe.shape[0]}")
print(f"Variables: {dataframe.shape[1]}")
print(f"cat_cols: {len(cat_cols)}")
print(f"num_cols: {len(num_cols)}")
print(f"cat_but_car: {len(cat_but_car)}")
print(f"num_but_cat: {len(num_but_cat)}")
return cat_cols, num_cols, cat_but_car
cat_cols, num_cols, cat_but_car = grab_col_names(df)
# Gerekli düzenlemeleri yapalım.
df["TotalCharges"] = pd.to_numeric(df["TotalCharges"], errors="coerce")
df["Churn"] = df["Churn"].apply(lambda x: 1 if x == "Yes" else 0)
def cat_summary(dataframe, col_name, plot=False):
print(
pd.DataFrame(
{
col_name: dataframe[col_name].value_counts(),
"Ratio": 100 * dataframe[col_name].value_counts() / len(dataframe),
}
)
)
print("######################")
if plot:
sns.countplot(x=dataframe[col_name], data=dataframe)
plt.show()
for col in cat_cols:
cat_summary(df, col)
def num_summary(dataframe, col_name, plot=False):
quantiles = [0.05, 0.1, 0.20, 0.30, 0.40, 0.50, 0.60, 0.70, 0.80, 0.90, 0.95, 0.99]
print(dataframe[col_name].describe(quantiles).T)
print("######################")
if plot:
dataframe[col_name].hist(bins=20)
plt.xlabel(col_name)
plt.title(col_name)
plt.show()
for col in num_cols:
num_summary(df, col)
# Kategorik değişkenler ile hedef değişken incelemesini yapalım.
def target_summary_with_cat(dataframe, target, categorical_col):
print(
pd.DataFrame(
{
"TARGET_MEAN": dataframe.groupby(categorical_col)[target].mean(),
"Count": dataframe[categorical_col].value_counts(),
"Ratio": 100
* dataframe[categorical_col].value_counts()
/ len(dataframe),
}
),
end="\n\n\n",
)
for col in cat_cols:
target_summary_with_cat(df, "Churn", col)
# Aykırı gözlem var mı inceleyelim.
def outlier_tesholds(dataframe, variable, q1=0.25, q3=0.75):
quartile1 = dataframe[variable].quantile(q1)
quartile3 = dataframe[variable].quantile(q3)
iqr = quartile3 - quartile1
low_limit = quartile1 - 1.5 * iqr
up_limit = quartile3 + 1.5 * iqr
return low_limit, up_limit
def check_outlier(dataframe, variable):
low_limit, up_limit = outlier_tesholds(dataframe, variable)
if dataframe[
(dataframe[variable] < low_limit) | (dataframe[variable] > up_limit)
].any(axis=None):
return True
else:
return False
for col in num_cols:
print(check_outlier(df, col))
# Eksik gözlem var mı inceleyelim.
def missing_values_table(dataframe, na_name=False):
na_cols = [col for col in dataframe.columns if dataframe[col].isnull().sum() > 0]
n_miss = dataframe[na_cols].isnull().sum().sort_values(ascending=False)
ratio = (dataframe[na_cols].isnull().sum() / dataframe.shape[0] * 100).sort_values(
ascending=False
)
missing_df = pd.concat(
[n_miss, np.round(ratio, 2)], axis=1, keys=["n_miss", "ratio"]
)
print(missing_df, end="\n")
if na_name:
return na_cols
missing_values_table(df, na_name=True)
# ## Feature Engineering
# Eksik ve aykırı gözlemler için gerekli işlemleri yapalım.
df["TotalCharges"].fillna(df["TotalCharges"].median(), inplace=True)
# Yeni değişkenler oluşturalım.
df.loc[(df["tenure"] >= 0) & (df["tenure"] <= 12), "NEW_TENURE_YEAR"] = "0-1 Year"
df.loc[(df["tenure"] > 12) & (df["tenure"] <= 24), "NEW_TENURE_YEAR"] = "1-2 Year"
df.loc[(df["tenure"] > 24) & (df["tenure"] <= 36), "NEW_TENURE_YEAR"] = "2-3 Year"
df.loc[(df["tenure"] > 36) & (df["tenure"] <= 48), "NEW_TENURE_YEAR"] = "3-4 Year"
df.loc[(df["tenure"] > 48) & (df["tenure"] <= 60), "NEW_TENURE_YEAR"] = "4-5 Year"
df.loc[(df["tenure"] > 60) & (df["tenure"] <= 72), "NEW_TENURE_YEAR"] = "5-6 Year"
# Kontratı 1 veya 2 yıllık müşterileri Engaged olarak belirtme
df["NEW_Engaged"] = df["Contract"].apply(
lambda x: 1 if x in ["One year", "Two year"] else 0
)
# Herhangi bir destek, yedek veya koruma almayan kişiler
df["NEW_noProt"] = df.apply(
lambda x: 1
if (x["OnlineBackup"] != "Yes")
or (x["DeviceProtection"] != "Yes")
or (x["TechSupport"] != "Yes")
else 0,
axis=1,
)
# Aylık sözleşmesi bulunan ve genç olan müşteriler
df["NEW_Young_Not_Engaged"] = df.apply(
lambda x: 1 if (x["NEW_Engaged"] == 0) and (x["SeniorCitizen"] == 0) else 0, axis=1
)
# Kişinin toplam aldığı servis sayısı
df["NEW_TotalServices"] = (
df[
[
"PhoneService",
"InternetService",
"OnlineSecurity",
"OnlineBackup",
"DeviceProtection",
"TechSupport",
"StreamingTV",
"StreamingMovies",
]
]
== "Yes"
).sum(axis=1)
# Herhangi bir streaming hizmeti alan kişiler
df["NEW_FLAG_ANY_STREAMING"] = df.apply(
lambda x: 1
if (x["StreamingTV"] == "Yes") or (x["StreamingMovies"] == "Yes")
else 0,
axis=1,
)
# Kişi otomatik ödeme yapıyor mu?
df["NEW_FLAG_AutoPayment"] = df["PaymentMethod"].apply(
lambda x: 1 if x in ["Bank transfer (automatic)", "Credit card (automatic)"] else 0
)
# ortalama aylık ödeme
df["NEW_AVG_Charges"] = df["TotalCharges"] / (df["tenure"] + 1)
# Güncel Fiyatın ortalama fiyata göre artışı
df["NEW_Increase"] = df["NEW_AVG_Charges"] / df["MonthlyCharges"]
# Servis başına ücret
df["NEW_AVG_Service_Fee"] = df["MonthlyCharges"] / (df["NEW_TotalServices"] + 1)
# Encoding işlemlerini gerçekleştirelim.
cat_cols, num_cols, cat_but_car = grab_col_names(df)
def label_encoder(dataframe, binary_cols):
le = LabelEncoder()
dataframe[binary_cols] = le.fit_transform(dataframe[binary_cols])
return dataframe
binary_cols = [
col for col in df.columns if df[col].dtype == "O" and df[col].nunique() == 2
]
for col in binary_cols:
df = label_encoder(df, col)
def one_hot_encoder(dataframe, cat_cols, drop_first=True):
dataframe = pd.get_dummies(dataframe, columns=cat_cols, drop_first=drop_first)
return dataframe
cat_cols = [col for col in cat_cols if col not in binary_cols and col not in ["Churn"]]
df = one_hot_encoder(df, cat_cols)
df.head()
# Numerik değişkenler için standartlaştırma yapalım.
ss = StandardScaler()
df[num_cols] = ss.fit_transform(df[num_cols])
df.head()
# ## Modelleme
X = df.drop(["Churn", "customerID"], axis=1)
y = df["Churn"]
def base_models(X, y):
print("Base Models....")
models = [
("LR", LogisticRegression()),
("KNN", KNeighborsClassifier()),
("CART", DecisionTreeClassifier()),
("RF", RandomForestClassifier()),
("GBM", GradientBoostingClassifier()),
("XGBoost", XGBClassifier(eval_metric="logloss")),
("LightGBM", LGBMClassifier()),
("CatBoost", CatBoostClassifier(verbose=False)),
]
for name, model in models:
cv_results = cross_validate(
model,
X,
y,
cv=10,
scoring=["accuracy", "f1", "roc_auc", "precision", "recall"],
)
print(f"########## {name} ##########")
print(f"Accuracy: {round(cv_results['test_accuracy'].mean(), 4)}")
print(f"Auc: {round(cv_results['test_roc_auc'].mean(), 4)}")
print(f"Recall: {round(cv_results['test_recall'].mean(), 4)}")
print(f"Precision: {round(cv_results['test_precision'].mean(), 4)}")
print(f"F1: {round(cv_results['test_f1'].mean(), 4)}")
base_models(X, y)
# Modeller ile hiperparametre optimizasyonu gerçekleştirelim ve yeni hiparparametreler ile modeli tekrar kuralım.
lr_params = {"C": [0.001, 0.01, 0.1, 1, 10, 100, 1000]}
lightgbm_params = {
"learning_rate": [0.01, 0.1],
"n_estimators": [300, 500],
"colsample_bytree": [0.7, 1],
}
gbm_params = {
"learning_rate": [0.01, 0.1],
"max_depth": [3, 8, 10],
"n_estimators": [100, 500, 1000],
"subsample": [1, 0.5, 0.7],
}
catboost_params = {
"iterations": [200, 500],
"learning_rate": [0.01, 0.1],
"depth": [3, 6],
}
classifiers = [
("LR", LogisticRegression(), lr_params),
("GBM", GradientBoostingClassifier(), gbm_params),
("LightGBM", LGBMClassifier(), lightgbm_params),
("CatBoost", CatBoostClassifier(), catboost_params),
]
def hyperparameter_optimization(X, y, cv=3, scoring="accuracy"):
print("Hyperparameter Optimization....")
best_models = {}
for name, classifier, params in classifiers:
print(f"########## {name} ##########")
cv_results = cross_validate(classifier, X, y, cv=cv, scoring=scoring)
print(f"{scoring} (Before): {round(cv_results['test_score'].mean(), 4)}")
gs_best = GridSearchCV(classifier, params, cv=cv, n_jobs=-1, verbose=False).fit(
X, y
)
final_model = classifier.set_params(**gs_best.best_params_)
cv_results = cross_validate(final_model, X, y, cv=cv, scoring=scoring)
print(f"{scoring} (After): {round(cv_results['test_score'].mean(), 4)}")
print(f"{name} best params: {gs_best.best_params_}", end="\n\n")
best_models[name] = final_model
return best_models
best_models = hyperparameter_optimization(X, y)
|
# # Few examples on how to use the dataset
# 1. Determine whether a point (x, y) belongs to the corridor geometry
# 2. Find the closest point to the corridor
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import pickle
import matplotlib.pyplot as plt
# Plotting purposes
from shapely.geometry import Polygon
def plot_shape(shapes):
if type(shapes) == Polygon:
shapes = [shapes]
for shape in shapes:
for interior in shape.interiors:
plt.plot(*interior.xy)
plt.plot(*shape.exterior.xy)
# # 1. Determine whether a point (x, y) belongs to the corridor geometry
# Load one file from the dataset
file = "/kaggle/input/indoor-location-navigation-scaled-geojson/scaled_geojson/5cdbc652853bc856e89a8694/F1/shapely_geometry.pkl"
with open(file, "rb") as f:
corridor = pickle.load(f)
# Create a couple of points
from shapely.geometry import Point
p1 = Point(65, 70)
p2 = Point(150, 80)
p3 = Point(125, 117)
# Check if the points belong to the polygon
print(p1.within(corridor))
print(p2.within(corridor))
print(p3.within(corridor))
# Visually ...
fig, ax = plt.subplots(figsize=(12, 8), dpi=100)
for p in [p1, p2, p3]:
plt.scatter(p.x, p.y)
ax.annotate(p.within(corridor), (p.x, p.y))
plot_shape(corridor)
# # 2. Find the closest point to the corridor
# Predicting a point outside the corridor can't be less wrong than predicting the closest point within the geometry, isn't it ?
from shapely.ops import nearest_points
from shapely.geometry import Point
# Create a point
p = Point(150, 70)
# Find the nearest point (only first attribute matters)
nearest_p, _ = nearest_points(corridor, p)
nearest_p.xy
# Visually ...
fig, ax = plt.subplots(figsize=(12, 8), dpi=100)
ax.scatter(p.x, p.y, label="p")
ax.scatter(nearest_p.x, nearest_p.y, label="nearest_p")
ax.plot([p.x, nearest_p.x], [p.y, nearest_p.y], "--")
ax.legend()
plot_shape(corridor)
|
# # **Importing Data**
import numpy as np
x_train = []
y_train = []
x_eval = []
y_eval = []
path = "../input/glaucoma-detection/Fundus_Train_Val_Data/Fundus_Scanes_Sorted/Train/Glaucoma_Positive"
path2 = "../input/glaucoma-detection/Fundus_Train_Val_Data/Fundus_Scanes_Sorted/Train/Glaucoma_Negative"
pathEval = "../input/glaucoma-detection/Fundus_Train_Val_Data/Fundus_Scanes_Sorted/Validation/Glaucoma_Positive"
pathEval2 = "../input/glaucoma-detection/Fundus_Train_Val_Data/Fundus_Scanes_Sorted/Validation/Glaucoma_Negative"
from PIL import Image
width = 3000
height = 2000
left = (width - 1500) / 2
top = (height - 1500) / 2
right = (width + 1500) / 2
bottom = (height + 1500) / 2
import os
for filename in os.listdir(path):
if filename.endswith("jpg") or filename.endswith(".jpg"):
img = Image.open(os.path.join(path, filename))
img = img.crop((left, top, right, bottom))
img = img.convert("L")
img = img.resize((500, 500))
img = np.array(img)
x_train.append(img)
y_train.append(1)
for filename in os.listdir(path2):
if filename.endswith("jpg") or filename.endswith(".jpg"):
img = Image.open(os.path.join(path2, filename))
img = img.crop((left, top, right, bottom))
img = img.convert("L")
img = img.resize((500, 500))
img = np.array(img)
x_train.append(img)
y_train.append(0)
for filename in os.listdir(pathEval):
if filename.endswith("jpg") or filename.endswith(".jpg"):
img = Image.open(os.path.join(pathEval, filename))
img = img.crop((left, top, right, bottom))
img = img.convert("L")
img = img.resize((500, 500))
img = np.array(img)
x_eval.append(img)
y_eval.append(0)
for filename in os.listdir(pathEval2):
if filename.endswith("jpg") or filename.endswith(".jpg"):
img = Image.open(os.path.join(pathEval2, filename))
img = img.crop((left, top, right, bottom))
img = img.convert("L")
img = img.resize((500, 500))
img = np.array(img)
x_eval.append(img)
y_eval.append(0)
x_train = np.array(x_train)
y_train = np.array(y_train)
x_eval = np.array(x_eval)
y_eval = np.array(y_eval)
import matplotlib.pyplot as plt
plt.imshow(x_train[6])
# # *Shuffle Arrays*
idx = np.random.permutation(len(x_train))
# Shuffle both arrays using the same permutation
x_trainShuff = x_train[idx]
y_trainShuff = y_train[idx]
idx = np.random.permutation(len(x_eval))
# Shuffle both arrays using the same permutation
x_evalShuff = x_eval[idx]
y_evalShuff = y_eval[idx]
# # The Model
from tensorflow import keras
model = keras.Sequential(
[
keras.layers.Conv2D(64, (3, 3), input_shape=(500, 500, 1), activation="relu"),
keras.layers.MaxPooling2D(2, 2),
keras.layers.Conv2D(64, (3, 3), activation="relu"),
keras.layers.MaxPooling2D(2, 2),
keras.layers.Conv2D(64, (3, 3), activation="relu"),
keras.layers.MaxPooling2D(2, 2),
keras.layers.Conv2D(64, (3, 3), activation="relu"),
keras.layers.MaxPooling2D(2, 2),
keras.layers.Conv2D(64, (3, 3), activation="relu"),
keras.layers.MaxPooling2D(2, 2),
keras.layers.Conv2D(64, (3, 3), activation="relu"),
keras.layers.MaxPooling2D(2, 2),
keras.layers.Flatten(),
keras.layers.Dense(128, activation="relu"),
keras.layers.Dropout(0.5),
keras.layers.Dense(128, activation="relu"),
keras.layers.Dense(128, activation="relu"),
keras.layers.Dense(2, activation="softmax"),
]
)
model.compile(
optimizer="Adam", loss="SparseCategoricalCrossentropy", metrics=["accuracy"]
)
# ### Note: I had to reduce the number of epochs because it took longer than expected. When i coded this on my colab notebook with GPU activated, the number of epochs was 30, And the accuracy was about 74%.
# 
model.fit(x_trainShuff, y_trainShuff, epochs=1, batch_size=16)
model.evaluate(x_evalShuff, y_evalShuff)
weights = model.layers[0].get_weights()[0]
# Normalize the weights to [0, 1] for visualization
weights = (weights - weights.min()) / (weights.max() - weights.min())
# Plot the filters
n_filters = weights.shape[3]
for i in range(n_filters):
plt.subplot(8, 8, i + 1)
plt.imshow(weights[:, :, 0, i], cmap="gray")
plt.axis("off")
plt.show()
from keras.utils import plot_model
plot_model(model)
|
import numpy as np
import keras
from keras import layers
from keras.datasets import mnist
from keras.models import Model
from keras.layers import Input, Dense, Conv2D, MaxPooling2D, UpSampling2D
import os
import matplotlib.pyplot as plt
from keras import regularizers
from enum import Enum
class Logger:
RESET = "\033[0m"
RED = "\033[31m"
GREEN = "\033[32m"
def info(self, message: str):
print(f"{self.GREEN}[INFO]{self.RESET} {message}")
def error(self, message: str):
print(f"{self.RED}[ERROR]{self.RESET} {message}")
logger = Logger()
class ModelType(Enum):
FIRST = 1
SECOND = 2
THIRD = 3
class GenType(Enum):
FIRST = 1
SECOND = 2
THIRD = 3
class DiscrType(Enum):
FIRST = 1
SECOND = 2
THIRD = 3
def load_mnist_data(
path_to_file: str,
) -> ((np.ndarray, np.ndarray), (np.ndarray, np.ndarray)):
with np.load(path_to_file, allow_pickle=True) as mnist:
x_train, y_train = mnist["x_train"], mnist["y_train"]
x_test, y_test = mnist["x_test"], mnist["y_test"]
logger.info("Data loaded")
return (x_train, y_train), (x_test, y_test)
def reshape(x_train: np.ndarray, x_test: np.ndarray) -> (np.ndarray, np.ndarray):
x_train = x_train.astype("float32") / 255.0
x_test = x_test.astype("float32") / 255.0
x_train = x_train.reshape((len(x_train), np.prod(x_train.shape[1:])))
x_test = x_test.reshape((len(x_test), np.prod(x_test.shape[1:])))
logger.info("Data converted")
return x_train, x_test
def reshape_list(data: list) -> list:
for i in data:
i = i.astype("float32") / 255.0
i = i.reshape((len(i), np.prod(i.shape[1:])))
logger.info("Data converted")
return data
def create_encoder_model(
encoded: keras.engine.keras_tensor.KerasTensor,
input_img: keras.engine.keras_tensor.KerasTensor,
) -> keras.engine.functional.Functional:
# This model maps an input to its encoded representation
return keras.Model(input_img, encoded)
def create_decoder_model(
encoding_dim: int, autoencoder: keras.engine.functional.Functional
):
# This is our encoded (32-dimensional) input
encoded_input = keras.Input(shape=(encoding_dim,))
# Retrieve the last layer of the autoencoder model
decoder_layer = autoencoder.layers[-1]
# Create the decoder model
return keras.Model(encoded_input, decoder_layer(encoded_input))
def create_full_connected_model(
IMG_SIZE: int,
encoding_dim: int,
input_img: keras.engine.keras_tensor.KerasTensor,
model_type: ModelType,
) -> (
keras.engine.functional.Functional,
keras.engine.keras_tensor.KerasTensor,
keras.engine.keras_tensor.KerasTensor,
):
def first_model() -> (
keras.engine.functional.Functional,
keras.engine.keras_tensor.KerasTensor,
keras.engine.keras_tensor.KerasTensor,
):
# "encoded" is the encoded representation of the input
encoded = layers.Dense(encoding_dim, activation="relu")(input_img)
logger.info("Encoded layer created")
# "decoded" is the lossy reconstruction of the input
decoded = layers.Dense(IMG_SIZE, activation="sigmoid")(encoded)
logger.info("Decoded layer created")
# This model maps an input to its reconstruction
autoencoder = keras.Model(input_img, decoded)
logger.info("Autoencoder layer created")
return autoencoder, encoded, decoded
def second_model() -> (
keras.engine.functional.Functional,
keras.engine.keras_tensor.KerasTensor,
keras.engine.keras_tensor.KerasTensor,
):
encoded = layers.Dense(
encoding_dim, activation="relu", activity_regularizer=regularizers.l1(10e-5)
)(input_img)
logger.info("Encoded layer created")
decoded = layers.Dense(IMG_SIZE, activation="sigmoid")(encoded)
logger.info("Decoded layer created")
autoencoder = keras.Model(input_img, decoded)
logger.info("Autoencoder layer created")
return autoencoder, encoded, decoded
def third_model() -> (
keras.engine.functional.Functional,
keras.engine.keras_tensor.KerasTensor,
keras.engine.keras_tensor.KerasTensor,
):
encoded = layers.Dense(128, activation="relu")(input_img)
encoded = layers.Dense(64, activation="relu")(encoded)
encoded = layers.Dense(32, activation="relu")(encoded)
logger.info("Encoded layer created")
decoded = layers.Dense(64, activation="relu")(encoded)
decoded = layers.Dense(128, activation="relu")(decoded)
decoded = layers.Dense(784, activation="sigmoid")(decoded)
logger.info("Decoded layer created")
autoencoder = keras.Model(input_img, decoded)
logger.info("Autoencoder layer created")
return autoencoder, encoded, decoded
get_model_type = {
ModelType.FIRST: first_model,
ModelType.SECOND: second_model,
ModelType.THIRD: third_model,
}
return get_model_type.get(model_type)()
def get_prediction(
x_test: np.ndarray,
encoder: keras.engine.keras_tensor.KerasTensor,
decoder: keras.engine.keras_tensor.KerasTensor,
) -> np.ndarray:
encoded_imgs = encoder.predict(x_test)
return decoder.predict(encoded_imgs)
def display_result(
digits_amount: int, x_test: np.ndarray, decoded_imgs: np.ndarray
) -> None:
plt.figure(figsize=(20, 4))
for i in range(digits_amount):
# Display original
ax = plt.subplot(2, digits_amount, i + 1)
plt.imshow(x_test[i].reshape(28, 28))
plt.gray()
ax.get_xaxis().set_visible(False)
ax.get_yaxis().set_visible(False)
# Display reconstruction
ax = plt.subplot(2, digits_amount, i + 1 + digits_amount)
plt.imshow(decoded_imgs[i].reshape(28, 28))
plt.gray()
ax.get_xaxis().set_visible(False)
ax.get_yaxis().set_visible(False)
plt.show()
def simple_generator(orig_data: str) -> None:
encoding_dim = 32
(x_train, _), (x_test, _) = load_mnist_data(orig_data)
x_train, x_test = reshape(x_train, x_test)
input_img = keras.Input(shape=(784,))
autoencoder, encoded, decoded = create_full_connected_model(
784, encoding_dim, input_img, ModelType.FIRST
)
encoder = create_encoder_model(encoded, input_img)
decoder = create_decoder_model(encoding_dim, autoencoder)
autoencoder.compile(optimizer="adam", loss="binary_crossentropy")
autoencoder.fit(
x_train,
x_train,
epochs=50,
batch_size=256,
shuffle=True,
validation_data=(x_test, x_test),
)
decoded_imgs = get_prediction(x_test, encoder, decoder)
display_result(10, x_test, decoded_imgs)
def simple_generator_v2(orig_data: str) -> None:
encoding_dim = 32
(x_train, _), (x_test, _) = load_mnist_data(orig_data)
x_train, x_test = reshape(x_train, x_test)
input_img = keras.Input(shape=(784,))
autoencoder, encoded, decoded = create_full_connected_model(
784, encoding_dim, input_img, ModelType.SECOND
)
encoder = create_encoder_model(encoded, input_img)
decoder = create_decoder_model(encoding_dim, autoencoder)
autoencoder.compile(optimizer="adam", loss="binary_crossentropy")
autoencoder.fit(
x_train,
x_train,
epochs=50,
batch_size=256,
shuffle=True,
validation_data=(x_test, x_test),
)
decoded_imgs = get_prediction(x_test, encoder, decoder)
display_result(10, x_test, decoded_imgs)
def get_image_generator(curr_model_type: ModelType) -> object:
if curr_model_type == ModelType.FIRST:
return simple_generator
elif 1:
return simple_generator_v2
def get_generator(g_type: GenType) -> keras.engine.sequential.Sequential:
def first_type() -> keras.engine.sequential.Sequential:
return keras.Sequential(
[
layers.Dense(256, input_dim=100, activation="relu"),
layers.BatchNormalization(),
layers.Dense(512, activation="relu"),
layers.BatchNormalization(),
layers.Dense(28 * 28, activation="sigmoid"),
layers.Reshape((28, 28, 1)),
],
name="generator",
)
def second_type() -> keras.engine.sequential.Sequential:
return keras.Sequential(
[
layers.Dense(256, input_dim=100, activation="relu"),
layers.BatchNormalization(),
layers.Dense(512, activation="relu"),
layers.BatchNormalization(),
layers.Dense(1024, activation="relu"),
layers.BatchNormalization(),
layers.Dense(784, activation="sigmoid"),
layers.Reshape((28, 28, 1)),
],
name="generator_v2",
)
def third_type() -> keras.engine.sequential.Sequential:
return keras.Sequential(
[
layers.Dense(128, input_dim=100, activation="relu"),
layers.BatchNormalization(),
layers.Dense(256, activation="relu"),
layers.BatchNormalization(),
layers.Dense(512, activation="relu"),
layers.BatchNormalization(),
layers.Dense(1024, activation="relu"),
layers.BatchNormalization(),
layers.Dense(28 * 28, activation="sigmoid"),
layers.Reshape((28, 28, 1)),
],
name="generator_v3",
)
get_type = {
GenType.FIRST: first_type,
GenType.SECOND: second_type,
GenType.THIRD: third_type,
}
generator = get_type.get(g_type)()
logger.info("Generator created")
return generator
def get_discriminator(d_type: DiscrType) -> keras.engine.sequential.Sequential:
def first_type() -> keras.engine.sequential.Sequential:
return keras.Sequential(
[
layers.Flatten(input_shape=(28, 28, 1)),
layers.Dense(512, activation="relu"),
layers.Dense(256, activation="relu"),
layers.Dense(1, activation="sigmoid"),
],
name="discriminator",
)
def second_type() -> keras.engine.sequential.Sequential:
return keras.Sequential(
[
layers.Flatten(input_shape=(28, 28, 1)),
layers.Dense(512, activation="relu"),
layers.BatchNormalization(),
layers.Dense(256, activation="relu"),
layers.BatchNormalization(),
layers.Dense(128, activation="relu"),
layers.BatchNormalization(),
layers.Dense(1, activation="sigmoid"),
],
name="discriminator_v2",
)
def third_type() -> keras.engine.sequential.Sequential:
return keras.Sequential(
[
layers.Flatten(input_shape=(28, 28, 1)),
layers.Dense(512, activation="relu"),
layers.BatchNormalization(),
layers.Dense(256, activation="relu"),
layers.BatchNormalization(),
layers.Dense(128, activation="relu"),
layers.BatchNormalization(),
layers.Dense(64, activation="relu"),
layers.BatchNormalization(),
layers.Dense(1, activation="sigmoid"),
],
name="discriminator_v3",
)
get_type = {
DiscrType.FIRST: first_type,
DiscrType.SECOND: second_type,
DiscrType.THIRD: third_type,
}
discriminator = get_type.get(d_type)()
logger.info("Discriminator created")
return discriminator
def get_model(
generator: keras.engine.sequential.Sequential,
discriminator: keras.engine.sequential.Sequential,
) -> keras.engine.sequential.Sequential:
gan = keras.Sequential(
[
generator,
discriminator,
],
name="gan",
)
logger.info("Model created")
discriminator.compile(
optimizer=keras.optimizers.Adam(learning_rate=0.0002),
loss=keras.losses.BinaryCrossentropy(from_logits=True),
)
discriminator.trainable = False
logger.info("Discriminator compiled")
gan.compile(
optimizer=keras.optimizers.Adam(learning_rate=0.0002),
loss=keras.losses.BinaryCrossentropy(from_logits=True),
)
logger.info("Model compiled")
return gan
def train_model(
model: keras.engine.sequential.Sequential,
generator: keras.engine.sequential.Sequential,
discriminator: keras.engine.sequential.Sequential,
orig_data: str,
) -> None:
batch_size = 128
(x_train, _), (x_test, _) = load_mnist_data(orig_data)
x_train = (x_train.astype(np.float32) - 127.5) / 127.5
x_train = np.expand_dims(x_train, axis=3)
# Create labels for real and fake images
real_labels = np.ones((batch_size, 1))
fake_labels = np.zeros((batch_size, 1))
# Train the GAN for the specified number of epochs
epochs = 150
for epoch in range(epochs):
# ---------------------
# Train Discriminator
# ---------------------
# Select a random batch of images
idx = np.random.randint(0, x_train.shape[0], batch_size)
imgs = x_train[idx]
# Generate a batch of fake images
noise = np.random.normal(0, 1, (batch_size, 100))
gen_imgs = generator.predict(noise)
# Train the discriminator on real and fake images
d_loss_real = discriminator.train_on_batch(imgs, real_labels)
d_loss_fake = discriminator.train_on_batch(gen_imgs, fake_labels)
d_loss = 0.5 * np.add(d_loss_real, d_loss_fake)
print(f"Epoch: {epoch}, Discriminator loss: {d_loss}")
# ---------------------
# Train Generator
# ---------------------
# Generate a batch of fake images
noise = np.random.normal(0, 1, (batch_size, 100))
# Train the generator to fool the discriminator
g_loss = model.train_on_batch(noise, real_labels)
print(f"Epoch: {epoch}, Generator loss: {g_loss}")
# 1. Реалізувати згортковий автокодувальних для генерації нових
# зображень набору даних MNIST (використовувати документацію
# https://blog.keras.io/building-autoencoders-in-keras.html).
simple_egnerator = get_image_generator(ModelType.FIRST)
simple_egnerator("/kaggle/input/mnist-from-link/mnist.npz")
simple_generator_v2 = get_image_generator(ModelType.SECOND)
simple_generator_v2("/kaggle/input/mnist-from-link/mnist.npz")
# 2. Реалізувати GAN мережу для генерації нових зображень набору даних MNIST (використовувати приклад https://www.kaggle.com/avk256/gan-cifar10/ ).
generator = get_generator(GenType.FIRST)
discriminator = get_discriminator(DiscrType.FIRST)
model = get_model(generator, discriminator)
train_model(model, generator, discriminator, "/kaggle/input/mnist-from-link/mnist.npz")
# 3. Провести обчислювальні експерименти з розробленими нейронними мережами прикладу. Змінювати кількість шарів, кількість нейронів в шарах, види функцій активації, розмірність матриць згортки та вибірки, початкові ініціалізатори тощо
generator = get_generator(GenType.SECOND)
discriminator = get_discriminator(DiscrType.SECOND)
model = get_model(generator, discriminator)
train_model(model, generator, discriminator, "/kaggle/input/mnist-from-link/mnist.npz")
generator = get_generator(GenType.THIRD)
discriminator = get_discriminator(DiscrType.THIRD)
model = get_model(generator, discriminator)
train_model(model, generator, discriminator, "/kaggle/input/mnist-from-link/mnist.npz")
generator = get_generator(GenType.FIRST)
discriminator = get_discriminator(DiscrType.SECOND)
model = get_model(generator, discriminator)
train_model(model, generator, discriminator, "/kaggle/input/mnist-from-link/mnist.npz")
generator = get_generator(GenType.FIRST)
discriminator = get_discriminator(DiscrType.THIRD)
model = get_model(generator, discriminator)
train_model(model, generator, discriminator, "/kaggle/input/mnist-from-link/mnist.npz")
generator = get_generator(GenType.SECOND)
discriminator = get_discriminator(DiscrType.FIRST)
model = get_model(generator, discriminator)
train_model(model, generator, discriminator, "/kaggle/input/mnist-from-link/mnist.npz")
generator = get_generator(GenType.SECOND)
discriminator = get_discriminator(DiscrType.THIRD)
model = get_model(generator, discriminator)
train_model(model, generator, discriminator, "/kaggle/input/mnist-from-link/mnist.npz")
generator = get_generator(GenType.THIRD)
discriminator = get_discriminator(DiscrType.FIRST)
model = get_model(generator, discriminator)
train_model(model, generator, discriminator, "/kaggle/input/mnist-from-link/mnist.npz")
generator = get_generator(GenType.THIRD)
discriminator = get_discriminator(DiscrType.SECOND)
model = get_model(generator, discriminator)
train_model(model, generator, discriminator, "/kaggle/input/mnist-from-link/mnist.npz")
|
mode = "DL"
temp_bs = 256 # [8,64,256,32]
temp_lr = 4 # [3,4,5]
temp_model = "nume" # ['sae','wnd','nume']
DATASET = "UNSW" # PILIH SALAH SATU (CIC, UNSW, NSL)
# !pip install -U tensorflow_decision_forests
from git import Repo
Repo.clone_from("https://github.com/vianhandika/dataset.git", "dataset")
# !gdown https://drive.google.com/uc?id=1daV2etOwxZ70womJ-K32UPGCD7l-Q_Hk ### CSE Train
# !gdown https://drive.google.com/uc?id=1-gqGtSle3ZSa3zzm88aghlO4wZjTs31Q ### CSE Test
# !gdown https://drive.google.com/uc?id=1mFRojhoHN9UKDWWi4tKmBm4QmRMbqaMW ### UNSW Train
# !gdown https://drive.google.com/uc?id=1-L57aVAAIjGglTBMF3HRUSIqLCOTqtXf ### UNSW Test
# !gdown https://drive.google.com/uc?id=1K0bjOKawP9uVc5HXuOPU17pBvvyjUzkP ### NSL Train
# !gdown https://drive.google.com/uc?id=1LlnVKvVBlMnKXhh-qKvW_S2ufv37ZH-4 ### NSL Test
# mode = 'DL'
# temp_bs = 32 #[8,64,256,32]
# temp_lr = 3 #[3,4,5]
# temp_model = 'wnd' #['sae','wnd','nume']
# DATASET = 'CIC' # PILIH SALAH SATU (CIC, UNSW, NSL)
import os
import math
import shutil
import timeit
import warnings
import itertools
import numpy as np
import pandas as pd
import seaborn as sns
import tensorflow as tf
import matplotlib.pyplot as plt
warnings.filterwarnings("ignore")
from gc import freeze
from tqdm import tqdm
from tensorflow import keras
from tensorflow.keras import layers
from sklearn.preprocessing import LabelEncoder, MinMaxScaler, OneHotEncoder
from category_encoders import TargetEncoder
from sklearn.metrics import confusion_matrix, f1_score
from sklearn.model_selection import train_test_split
from tensorflow.keras.callbacks import ModelCheckpoint, EarlyStopping
from git import Repo
def mkdir(folder):
try:
os.mkdir(folder)
except:
pass
mkdir("/kaggle/working/result")
UNSEEN_VALUE = "9999999"
def classweights(label):
return {k: max(np.bincount(label)) / v for k, v in enumerate(np.bincount(label))}
def get_split(data, label, val=0.2, rand=10, encoder=[], X_test=None, y_test=None):
print("--- Get Split")
random_state = 10
X_train, X_valid, y_train, y_valid = train_test_split(
data, label, test_size=val, random_state=rand, stratify=label
)
X_train_back = X_train.copy()
try:
print("For Testing with shape:", X_test.shape)
X_train, y_train = data, label
X_valid, y_valid = X_test, y_test
except:
pass
## Encoding
# if not encoder:
# print('Apply Label Encoder')
# enc = LabelEncoder() #
# for col in X_train.columns:
# if str(X_train[col].dtypes)=='object':
# enc.fit(X_train[col].tolist()+[UNSEEN_VALUE])
# X_valid[col][~X_valid[col].isin(enc.classes_.tolist())] = UNSEEN_VALUE
# X_train[col] = enc.transform(X_train[col].values)
# X_valid[col] = enc.transform(X_valid[col].values)
# if not encoder:
# print('Apply Target Encoder')
# enc = TargetEncoder() #
# for col in X_train.columns:
# if str(X_train[col].dtypes)=='object':
# enc.fit(X_train[col].tolist()+[UNSEEN_VALUE],y_train.tolist()+[9999])
# X_valid[col][~X_valid[col].isin(X_train[col].unique())] = UNSEEN_VALUE
# X_train[col] = enc.transform(X_train[col].values)
# X_valid[col] = enc.transform(X_valid[col].values)
if not encoder:
print("Apply One Hot Encoder")
COLOM = [col for col in X_train.columns if str(X_train[col].dtypes) == "object"]
enc = OneHotEncoder(categories="auto", handle_unknown="ignore")
enc.fit(X_train[COLOM].values)
X_T = enc.transform(X_train[COLOM].values).toarray()
X_V = enc.transform(X_valid[COLOM].values).toarray()
X_T = pd.DataFrame(X_T, columns=enc.get_feature_names_out(COLOM))
X_V = pd.DataFrame(X_V, columns=enc.get_feature_names_out(COLOM))
X_train = pd.concat(
[X_T, X_train.drop(COLOM, 1).reset_index(drop=True)], axis=1
)
X_valid = pd.concat(
[X_V, X_valid.drop(COLOM, 1).reset_index(drop=True)], axis=1
)
else:
for col in encoder:
X_valid[col][
~X_valid[col].isin(np.unique(X_train_back[col]))
] = UNSEEN_VALUE
## Fillna
X_train = X_train.fillna(X_train.median())
X_valid = X_valid.fillna(X_valid.median())
## Normalization
scaler = MinMaxScaler()
X_train[X_train.drop(encoder, 1).columns] = scaler.fit_transform(
X_train.drop(encoder, 1).values
)
X_valid[X_valid.drop(encoder, 1).columns] = scaler.transform(
X_valid.drop(encoder, 1).values
)
return X_train, X_valid, y_train, y_valid
def pddataframe(dataframe, label=None, batch_size=1000) -> tf.data.Dataset:
# Make sure that missing values for string columns are not represented as
# float(NaN).
for col in dataframe.columns:
if dataframe[col].dtype in [str, object]:
dataframe[col] = dataframe[col].fillna("")
if label is not None:
features_dataframe = dataframe.drop(label, 1)
numtarget = len(y.value_counts())
output = (
dict(features_dataframe),
tf.keras.utils.to_categorical(dataframe[label].values, numtarget),
)
# output = (dict(features_dataframe), features_dataframe[NUMERIC_FEATURE_NAMES].values)
tf_dataset = tf.data.Dataset.from_tensor_slices(output)
# The batch size does not impact the training of TF-DF.
if batch_size is not None:
tf_dataset = tf_dataset.batch(batch_size)
# Seems to provide a small (measured as ~4% on a 32k rows dataset) speed-up.
tf_dataset = tf_dataset.prefetch(tf.data.AUTOTUNE)
return tf_dataset
def set_callbacks(filepath):
checkpoint = ModelCheckpoint(
filepath=filepath + "_weight.h5",
monitor="val_loss", # nilai yang dimonitor adalah val_loss
verbose=1, # print informasi setiap 1 epoch
save_best_only=True, # perolehan model terbaik saja yang di simpan
save_weights_only=True, # menyimpan weights saja supaya filesizenya tidak terlalu besar
mode="min",
) # model yang terbaik adalah yang memiliki nilai minimal dari nilai yang dimonitor
early_stop = EarlyStopping(
monitor="val_loss",
verbose=1,
patience=50, # pelatihan akan stop otomatis jika tidak ada improve setelah validasi terbaik dalam jangka 5 epoch
mode="min",
)
return [checkpoint, early_stop]
## LOAD DATASET
# DATASET = 'CIC' # PILIH SALAH SATU (CIC, UNSW, NSL)
if DATASET == "CIC":
df_train = pd.read_csv("../dataset/CIC_2018_multiclass_train_v1.csv")
df_test = pd.read_csv("../dataset/CIC_2018_multiclass_test_v1.csv")
# Feature engineering
common_port = [8080, 80, 21, 53, 443, 3389, 445, 22, 500, 0]
df_train["Dst Port"] = list(
map(lambda x: 10000 if x not in common_port else x, df_train["Dst Port"])
)
df_test["Dst Port"] = list(
map(lambda x: 10000 if x not in common_port else x, df_test["Dst Port"])
)
df_train["label"] = df_train["Label"]
df_test["label"] = df_test["Label"]
catcol = [
"Dst Port",
"Protocol",
"Fwd PSH Flags",
"Bwd PSH Flags",
"Fwd URG Flags",
"Bwd URG Flags",
"CWE Flag Count",
"ECE Flag Cnt",
"URG Flag Cnt",
"ACK Flag Cnt",
"PSH Flag Cnt",
"RST Flag Cnt",
"FIN Flag Cnt",
"SYN Flag Cnt",
]
dropcol = ["Timestamp", "Label"]
labelcol = "label"
df_train = df_train.drop(dropcol, 1)
df_test = df_test.drop(dropcol, 1)
df_train[catcol] = df_train[catcol].astype(str)
df_test[catcol] = df_test[catcol].astype(str)
featcol = df_train.drop(labelcol, 1).columns
labels = sorted(df_train[labelcol].value_counts().index.tolist())
df_train[labelcol] = df_train[labelcol].apply(lambda x: labels.index(x))
df_test[labelcol] = df_test[labelcol].apply(lambda x: labels.index(x))
elif DATASET == "UNSW":
df_train = pd.read_csv("../dataset/UNSW_NB15_multiclass_test_v1.csv")
df_test = pd.read_csv("../dataset/UNSW_NB15_multiclass_train_v1.csv")
catcol = ["proto", "service", "state"]
dropcol = ["id"]
labelcol = "label"
df_train = df_train.drop(dropcol, 1)
df_test = df_test.drop(dropcol, 1)
df_train[catcol] = df_train[catcol].astype(str)
df_test[catcol] = df_test[catcol].astype(str)
featcol = df_train.drop(labelcol, 1).columns
labels = sorted(df_train[labelcol].value_counts().index.tolist())
df_train[labelcol] = df_train[labelcol].apply(lambda x: labels.index(x))
df_test[labelcol] = df_test[labelcol].apply(lambda x: labels.index(x))
elif DATASET == "NSL":
df_train = pd.read_csv("../dataset/NSL_KDD_multiclass_train_v1.csv")
df_test = pd.read_csv("../dataset/NSL_KDD_multiclass_test_v1.csv")
catcol = ["protocol_type", "service", "flag"]
dropcol = []
labelcol = "label"
df_train = df_train.drop(dropcol, 1)
df_test = df_test.drop(dropcol, 1)
df_train[catcol] = df_train[catcol].astype(str)
df_test[catcol] = df_test[catcol].astype(str)
featcol = df_train.drop(labelcol, 1).columns
labels = sorted(df_train[labelcol].value_counts().index.tolist())
df_train[labelcol] = df_train[labelcol].apply(lambda x: labels.index(x))
df_test[labelcol] = df_test[labelcol].apply(lambda x: labels.index(x))
else:
raise "SALAH DATA"
X_train, X_valid, y_train, y_valid = get_split(
df_train.drop(labelcol, 1), df_train[labelcol], val=0.2, rand=10, encoder=catcol
)
X_train_enc, X_valid_enc, _, __ = get_split(
df_train.drop(labelcol, 1), df_train[labelcol], val=0.2, rand=10, encoder=[]
)
X, X_test, y, y_test = get_split(
df_train.drop(labelcol, 1),
df_train[labelcol],
val=None,
rand=None,
encoder=catcol,
X_test=df_test.drop(labelcol, 1),
y_test=df_test[labelcol],
)
X_enc, X_test_enc, _, __ = get_split(
df_train.drop(labelcol, 1),
df_train[labelcol],
val=None,
rand=None,
encoder=[],
X_test=df_test.drop(labelcol, 1),
y_test=df_test[labelcol],
)
NUMERIC_FEATURE_NAMES = [col for col in featcol if col not in catcol]
CATEGORICAL_FEATURE_NAMES = [col for col in catcol]
TARGET_COLUMN_NAME = labelcol
import random
from tensorflow.keras import *
from tensorflow import keras
class DistanceLayer(layers.Layer):
"""
This layer is responsible for computing the distance between the anchor
embedding and the positive embedding, and the anchor embedding and the
negative embedding.
"""
def __init__(self, **kwargs):
super().__init__(**kwargs)
def call(self, anchor, positive, negative):
ap_distance = tf.reduce_sum(tf.square(anchor - positive), -1)
an_distance = tf.reduce_sum(tf.square(anchor - negative), -1)
return (ap_distance, an_distance)
anchor_input = layers.Input(name="anchor", shape=X_train_enc.shape[1:])
positive_input = layers.Input(name="positive", shape=X_train_enc.shape[1:])
negative_input = layers.Input(name="negative", shape=X_train_enc.shape[1:])
embedding = keras.Sequential(
[
layers.Dense(512, activation="relu", name="layer1"),
layers.Dropout(0.5),
layers.Dense(256, activation="relu", name="layer2"),
layers.Dropout(0.5),
layers.Dense(256, name="layer3"),
],
name="Embedding",
)
distances = DistanceLayer()(
embedding(anchor_input),
embedding(positive_input),
embedding(negative_input),
)
siamese_network = models.Model(
inputs=[anchor_input, positive_input, negative_input],
outputs=distances,
name="siamese",
)
class SiameseModel(Model):
"""The Siamese Network model with a custom training and testing loops.
Computes the triplet loss using the three embeddings produced by the
Siamese Network.
The triplet loss is defined as:
L(A, P, N) = max(‖f(A) - f(P)‖² - ‖f(A) - f(N)‖² + margin, 0)
"""
def __init__(self, siamese_network, margin=0.5):
super().__init__()
self.siamese_network = siamese_network
self.margin = margin
self.loss_tracker = metrics.Mean(name="loss")
def call(self, inputs):
return self.siamese_network(inputs)
def train_step(self, data):
# GradientTape is a context manager that records every operation that
# you do inside. We are using it here to compute the loss so we can get
# the gradients and apply them using the optimizer specified in
# `compile()`.
with tf.GradientTape() as tape:
loss = self._compute_loss(data)
# Storing the gradients of the loss function with respect to the
# weights/parameters.
gradients = tape.gradient(loss, self.siamese_network.trainable_weights)
# Applying the gradients on the model using the specified optimizer
self.optimizer.apply_gradients(
zip(gradients, self.siamese_network.trainable_weights)
)
# Let's update and return the training loss metric.
self.loss_tracker.update_state(loss)
return {"loss": self.loss_tracker.result()}
def test_step(self, data):
loss = self._compute_loss(data)
# Let's update and return the loss metric.
self.loss_tracker.update_state(loss)
return {"loss": self.loss_tracker.result()}
def _compute_loss(self, data):
# The output of the network is a tuple containing the distances
# between the anchor and the positive example, and the anchor and
# the negative example.
ap_distance, an_distance = self.siamese_network(data)
# Computing the Triplet Loss by subtracting both distances and
# making sure we don't get a negative value.
loss = ap_distance - an_distance
loss = tf.maximum(loss + self.margin, 0.0)
return loss
@property
def metrics(self):
# We need to list our metrics here so the `reset_states()` can be
# called automatically.
return [self.loss_tracker]
class DataGenerator(keras.utils.Sequence):
"Generates data for Keras"
def __init__(self, Dict, Xv, yv, batch_size=64, shuffle=False):
"Initialization"
self.batch_size = batch_size
self.Dict = Dict
self.Xv = Xv
self.yv = yv
self.shuffle = shuffle
self.on_epoch_end()
def __len__(self):
"Denotes the number of batches per epoch"
return int(np.floor(len(self.Xv) / self.batch_size))
def __getitem__(self, index):
"Generate one batch of data"
# Generate indexes of the batch
indexes = self.indexes[index * self.batch_size : (index + 1) * self.batch_size]
# Find list of IDs
list_IDs_temp = [np.arange(len(self.yv))[k] for k in indexes]
# Generate data
X = self.__data_generation(list_IDs_temp)
return X
def on_epoch_end(self):
"Updates indexes after each epoch"
self.indexes = np.arange(len(self.yv))
if self.shuffle == True:
np.random.shuffle(self.indexes)
def __data_generation(self, list_IDs_temp):
"Generates data containing batch_size samples" # X : (n_samples, *dim, n_channels)
# Initialization
anchor, positive, negative = [], [], []
# Generate data
for i in list_IDs_temp:
anchor += [self.Xv.values[self.Dict.iloc[i]["a"]]]
positive += [self.Xv.values[self.Dict.iloc[i]["p"]]]
negative += [self.Xv.values[self.Dict.iloc[i]["n"]]]
return [np.array(anchor), np.array(positive), np.array(negative)]
import random
from sklearn.decomposition import PCA
def getdict(X_enc, y):
pca = PCA(n_components=1)
X_pca = pca.fit_transform(X_enc)[:, 0]
X_pca = pd.Series(X_pca)
yv = y.reset_index(drop=True)
Dict = pd.DataFrame(
{
"y": yv,
"pca": X_pca.apply(lambda x: 0 if x < 0 else 1),
"a": np.arange(yv.shape[0]),
}
)
dicty = [
{j: Dict[Dict["y"] == j][Dict["pca"] != 0] for j in yv.value_counts().index},
{j: Dict[Dict["y"] == j][Dict["pca"] != 1] for j in yv.value_counts().index},
]
print("create p")
Dict["p"] = Dict.apply(
lambda x: dicty[x["pca"]][x["y"]].sample(1, random_state=x["a"]).index[0],
axis=1,
)
print("create n")
Dict["n"] = Dict.apply(
lambda x: random.choices(
[
dicty[x["pca"]][j].sample(1, random_state=x["a"]).index[0]
for j in yv.value_counts().index
if j != x["y"]
]
)[0],
axis=1,
)
return Dict
Dict_train = getdict(X_train_enc, y_train)
Dict_valid = getdict(X_valid_enc, y_valid)
Dict_test = getdict(X_test_enc, y_test)
print(X_train_enc.shape, X_valid_enc.shape, y_train.shape, y_valid.shape)
train_dataset = DataGenerator(
Dict_train,
X_train_enc.reset_index(drop=True),
y_train.reset_index(drop=True),
batch_size=64,
shuffle=True,
)
val_dataset = DataGenerator(
Dict_valid,
X_valid_enc.reset_index(drop=True),
y_valid.reset_index(drop=True),
batch_size=64,
)
callbacks = [
keras.callbacks.ModelCheckpoint(
"save_at_{epoch}.keras",
monitor="val_loss",
save_best_only=True,
save_weights_only=True,
mode="min",
),
]
siamese_model = SiameseModel(siamese_network)
siamese_model.compile(optimizer=optimizers.Adam(0.0001))
siamese_model.fit(
train_dataset, epochs=50, validation_data=val_dataset, callbacks=callbacks
)
train_dataset = DataGenerator(
Dict_train,
X_train_enc.reset_index(drop=True),
y_train.reset_index(drop=True),
batch_size=len(y_train),
)
test_dataset = DataGenerator(
Dict_test,
X_test_enc.reset_index(drop=True),
y_test.reset_index(drop=True),
batch_size=len(y_test),
)
trainx = embedding.predict(train_dataset[0][0])
pred = embedding.predict(test_dataset[0][0])
from sklearn.neighbors import KNeighborsClassifier
from sklearn.ensemble import RandomForestClassifier
clf = RandomForestClassifier()
clf.fit(trainx, y_train.reset_index(drop=True))
ypred = clf.predict(pred)
print(f1_score(ypred, y_test.reset_index(drop=True).values, average="macro"))
print((ypred == y_test.reset_index(drop=True).values).sum())
from sklearn.neighbors import KNeighborsClassifier
from sklearn.ensemble import RandomForestClassifier
clf = RandomForestClassifier()
clf.fit(X_train_enc.values, y_train.reset_index(drop=True))
ypred = clf.predict(X_test_enc.values)
print(f1_score(ypred, y_test.reset_index(drop=True).values, average="macro"))
print((ypred == y_test.reset_index(drop=True).values).sum())
print()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
from PIL import Image
from fastai.vision.all import *
from fastai.vision.widgets import *
dest = Path("/kaggle/input/lego-brick-images/LEGO brick images v1")
path = "/kaggle/input/lego-brick-images/LEGO brick images v1/3003 Brick 2x2/0001.png"
im = Image.open(path)
im.resize((200, 200))
im
dls = DataBlock(
blocks=(ImageBlock, CategoryBlock),
get_items=get_image_files,
splitter=RandomSplitter(valid_pct=0.1, seed=23),
get_y=parent_label,
item_tfms=[Resize(100, method="squish")],
).dataloaders(dest, bs=32)
dls.show_batch(max_n=6)
learn = vision_learner(dls, resnet18, metrics=error_rate)
learn.fine_tune(15)
brick, _, probs = learn.predict(
PILImage.create(
"/kaggle/input/lego-brick-images/LEGO brick images v1/3003 Brick 2x2/0003.png"
)
)
print(f"This is a: {brick}.")
print(f"Probability: {probs[3]:.3f}")
interp = ClassificationInterpretation.from_learner(learn)
interp.plot_confusion_matrix(figsize=(20, 10))
interp.plot_top_losses(12, nrows=4, figsize=(20, 20))
cleaner = ImageClassifierCleaner(learn)
cleaner
learn.export("lego-bricks-model.pkl")
|
import pandas as pd # IMPORTING PANDAS
import matplotlib.pyplot as plt # IMPORTING PYPLOT
import numpy as np # IMPORTING NUMPY
import re # IMPORTING REGEX
nt = pd.read_csv(
"../input/netflix-shows/netflix_titles.csv"
) # READING THE CSV FILE AS DATAFRAME
print(nt) # PRINTING THE DATAFRAME
print(type(nt)) # PRINTING TYPE OF DATAFRAME TO CONFIRM
print(nt.columns) # PRINTING COLUMNS
nt.set_index("show_id", inplace=True) # SETTING "show_id" AS INDEX
print(nt) # PRINTING DATAFRAME
nt.shape # CHECKING SHAPE OF THE DATAFRAME
nt.columns # PRINTING COLUMNS
nt.type.unique() # SEEING UNIQUE VALUES IN TYPE COLUMN
# BAR GRAPH FOR NUMBER OF MOVIES/TV SHOWS
plt.figure(figsize=(7, 5))
plt.bar(["TV Shows", "Movies"], [sum(nt.type == "TV Show"), sum(nt.type == "Movie")])
plt.title("Bar plot for number of movies/TV Shows")
plt.xlabel("MEDIA TYPE")
plt.ylabel("NUMBER")
plt.show()
n = 0
for media in nt.title.unique():
n += 1
print(n) # PRINTING NUMBER OF UNIQUE MEDIA
n = 0
for d in nt.director.unique():
n += 1
print(n) # PRINTING NUMBER OF UNIQUE directors
nt["director"].fillna("None", inplace=True) # REMOVING THE NaN VALUES from DIRECTOR
direc_dict = {} # CREATING DICTIONARY FOR UNIQUE DIRECTORS AND COUNT OF MOVIES/TV SHOWS
for d in list(nt.director):
if d in direc_dict:
direc_dict[d] += 1
else:
direc_dict[d] = 1
# print(direc_dict)
# REMOVING NaN
del direc_dict["None"]
# PRINTING DIRECTORS FOR MORE THAN 7 MOVIES
for key, value in direc_dict.items():
if value > 7:
print(key)
# NUMBER OF COUNTRIES PRESENT
pd.DataFrame(nt.country.unique()).count()
coun_dict = {} # CREATING DICTIONARY FOR UNIQUE DIRECTORS AND COUNT OF MOVIES/TV SHOWS
for c in list(nt.country):
if c in coun_dict:
coun_dict[c] += 1
else:
coun_dict[c] = 1
# PRINTING COUNTRIES FOR MORE THAN 30 MOVIES
for key, value in coun_dict.items():
if value > 30:
print(key)
# PRINTING COUNTRY WITH MAXIMUM NUMBER OF MOVIES
max_val = max(coun_dict.values())
ls = []
for key, value in coun_dict.items():
if value == max_val:
ls.append(key)
print(ls, max_val)
# DATE_ADDED VARIABLE
nt.date_added
# CREATING BINS FOR HISTOGRAM
bins = [i for i in range(2010, 2021)]
# print(bins)
nt["date_added"].fillna("0000", inplace=True) # Removing NA values
# Retreiving year from dates
l = []
for date in nt.date_added:
temp = date[-4::]
l.append(int(temp))
# HISTOGRAM FOR Number of movies added in respective year
plt.figure(figsize=(7, 5))
plt.title("Number of movies added in respective year")
plt.xlabel("Years")
plt.ylabel("Number of movies")
plt.xticks(bins)
plt.hist(l, bins, align="right", rwidth=0.8, color="coral")
plt.show()
# RELEASE YEAR
nt.release_year
# NUMBER OF MOVIES RELEASED IN SPECIFIC YEAR
plt.figure(figsize=(7, 5))
plt.title("Number of movies released in respective year")
plt.xlabel("Years")
plt.ylabel("Number of movies")
plt.xticks(bins)
plt.hist(nt.release_year, bins, align="right", rwidth=0.8, color="teal")
plt.show()
# RATING
nt.rating
# REMOVING NAN AND MAKING UNIQUE RATING LIST
nt.rating.fillna("None", inplace=True)
unique_rating_list = nt.rating.unique() # UNIQUE RATING
# unique_rating_list.fillna('None', inplace = True) # Making Nan None
# unique_rating_list = [x for x in unique_rating_list if x is not 'None'] # Removing None by list comprehension
# print(unique_rating_list) # Printing Unique Ratings
rate_dict = {} # CREATING DICTIONARY FOR UNIQUE RATINGS AND COUNT OF MOVIES/TV SHOWS
for r in list(nt.rating):
if r in rate_dict:
rate_dict[r] += 1
else:
rate_dict[r] = 1
# BAR CHART FOR NUMBER OF MOVIES/SHOWS WITH THEIR RATINGS
plt.figure(figsize=(12, 7))
plt.title("Number of Movies/Shows with their respective ratings")
plt.xlabel("Ratings")
plt.ylabel("Count of Shows")
plt.bar(
rate_dict.keys(), rate_dict.values(), color="#4042e4"
) # PLOTTING BAR CHART FOR NUMBER OF RATINGS
plt.show()
# DURATION VARIABLE
nt.duration
# UNIQUE DURATION
unique_duration = list(nt.duration.unique())
# print(unique_duration)
# DEMARCATING TV SERIES AND MOVIES SEPERATELY
season_TVSeries = []
minutes_Movies = []
for el in unique_duration:
if el[-7::] == "Seasons" or el[-6::] == "Season":
season_TVSeries.append(el)
else:
minutes_Movies.append(el)
# print(season_TVSeries)
# print(minutes_Movies)
# FOR SEASONS
bins = [x for x in range(1, 16)]
# print(bins)
list_num_seasons = [int(re.sub(" .*", "", x)) for x in season_TVSeries]
# print(list_num_seasons)
# BAR CHART FOR NUMBER OF UNIQUE SEASONS IN SHOWS
plt.figure(figsize=(7, 5))
plt.title("Number of Unique Seasons in shows")
plt.ylabel("Shows")
plt.tick_params(axis="x", which="both", bottom=False, top=False) # REMOVING TICKS
plt.xticks(bins, " ") # REMOVING X LABEL VALUES
plt.scatter(bins, list_num_seasons, color="#4062e4")
plt.show()
# FOR MOVIES
bins = [x for x in range(0, 321, 40)]
list_time_movies = [int(re.sub(" .*", "", x)) for x in minutes_Movies]
# print(bins)
# print(list_time_movies)
# BAR CHART FOR NUMBER OF RATINGS
plt.figure(figsize=(7, 5))
plt.title("Number of Movies with their respective duration")
plt.xlabel("Number of Movies")
plt.ylabel("Duration")
plt.hist(
list_time_movies, bins, color="#657482", orientation="horizontal", align="mid"
) # PLOTTING
# BAR CHART FOR NUMBER OF RATINGS
plt.grid(True, axis="both", which="major")
plt.show()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
# for dirname, _, filenames in os.walk('/kaggle/input'):
# for filename in filenames:
# print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# @title Import libraries we use for this notebook
import os
import time
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from PIL import Image
import matplotlib.image as mpimg
import random
import pathlib
import tensorflow as tf
import tensorflow_hub as hub
from tensorflow.keras.preprocessing.image import ImageDataGenerator
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Conv2D, MaxPool2D, Dense, Activation
from tensorflow.keras.layers import Flatten
from tensorflow.keras.optimizers import Adam
from tensorflow.keras import layers
# Download helper functions script
# Import helper functions we're going to use in this notebook
from helper_functions import (
create_tensorboard_callback,
plot_loss_curves,
unzip_data,
walk_through_dir,
)
# ## Checking images
for dirpath, dirnames, filenames in os.walk(
"/kaggle/input/car-model-variants-and-images-dataset/images_512x512/images_512x512/AUDI"
):
print(
f"There are {len(dirnames)} directories and {len(filenames)} images in {dirpath}"
)
# ## Make Audi train and test datasets
import os
import random
import shutil
# Source and destination directories
src_dir = "/kaggle/input/car-model-variants-and-images-dataset/images_512x512/images_512x512/AUDI"
dest_dir = "/kaggle/working/AUDI"
# Create the train and test directories
os.makedirs(os.path.join(dest_dir, "train"), exist_ok=True)
os.makedirs(os.path.join(dest_dir, "test"), exist_ok=True)
# Loop through the directories and move the images to train or test sets
for dirpath, dirnames, filenames in os.walk(src_dir):
if len(filenames) == 0: # skip directories with no images
continue
# Create the corresponding directories in the destination folder
relative_dir = os.path.relpath(dirpath, src_dir)
train_dir = os.path.join(dest_dir, "train", relative_dir)
test_dir = os.path.join(dest_dir, "test", relative_dir)
os.makedirs(train_dir, exist_ok=True)
os.makedirs(test_dir, exist_ok=True)
# Split the images into train and test sets
train_size = int(0.8 * len(filenames))
test_size = len(filenames) - train_size
random.shuffle(filenames)
train_files = filenames[:train_size]
test_files = filenames[train_size:]
# Move the images to the train or test directories
for filename in train_files:
src_path = os.path.join(dirpath, filename)
dest_path = os.path.join(train_dir, filename)
shutil.copy(src_path, dest_path)
for filename in test_files:
src_path = os.path.join(dirpath, filename)
dest_path = os.path.join(test_dir, filename)
shutil.copy(src_path, dest_path)
# Setup data inputs
from tensorflow.keras.preprocessing.image import ImageDataGenerator
IMAGE_SHAPE = (512, 512)
BATCH_SIZE = 32
train_dir = "/kaggle/working/AUDI/train"
test_dir = "/kaggle/working/AUDI/test"
train_datagen = ImageDataGenerator(rescale=1 / 255.0)
test_datagen = ImageDataGenerator(rescale=1 / 255.0)
print("Train images:")
train_data = train_datagen.flow_from_directory(
train_dir, target_size=IMAGE_SHAPE, batch_size=BATCH_SIZE, class_mode="categorical"
)
print("Test images:")
test_data = train_datagen.flow_from_directory(
test_dir, target_size=IMAGE_SHAPE, batch_size=BATCH_SIZE, class_mode="categorical"
)
import datetime
def create_tensorboard_callback(dir_name, experiment_name):
log_dir = (
dir_name
+ "/"
+ experiment_name
+ "/"
+ datetime.datetime.now().strftime("%Y%m%d-%H%M%S")
)
tensorboard_callback = tf.keras.callbacks.TensorBoard(log_dir=log_dir)
print(f"Saving TensorBoard log files to: {log_dir}")
return tensorboard_callback
# Let's compare the following two models
resnet_url = "https://tfhub.dev/google/imagenet/resnet_v2_50/feature_vector/5"
effnet_url = "https://tfhub.dev/tensorflow/efficientnet/b0/feature-vector/1"
# Let's make a create_model() function to create a model from a URL
def create_model(model_url, num_classes):
"""
Takes a TensorFlow Hub URL and creates a Keras Sequential model with it.
Args:
model_url (str): A TensorFlow Hub feature extraction URL.
num_classes (int): Number of output neurons in the output layer,
should be equal to number of target classes, default 10.
Returns:
An uncompiled Keras Sequential model with model_url as feature extractor
layer and Dense output layer with num_classes output neurons.
"""
# Download the pretrained model and save it as a Keras layer
feature_extraction_layer = hub.KerasLayer(
model_url,
trainable=False, # freeze the already learned patterns
name="feature_extraction_layer",
input_shape=IMAGE_SHAPE + (3,),
)
# Create our model
model = tf.keras.Sequential(
[
feature_extraction_layer,
layers.Dense(num_classes, activation="softmax", name="output_layer"),
]
)
return model
# Create Resnet model
resnet_model = create_model(resnet_url, num_classes=train_data.num_classes)
resnet_model.summary()
# Compile our Resnet model
resnet_model.compile(
loss=tf.keras.losses.CategoricalCrossentropy(),
optimizer=Adam(),
metrics=["accuracy"],
)
# ## GPU required to run.
# > ⚠️ Fit models only with GPU runtime. Do not run this on CPU :)
# Let's fit our ResNet model to the data
history_resnet = resnet_model.fit(
train_data,
epochs=5,
steps_per_epoch=len(train_data),
validation_data=test_data,
validation_steps=len(test_data),
callbacks=[
create_tensorboard_callback(
dir_name="tensorboard", experiment_name="resnet50v2"
)
],
)
plot_loss_curves(history_resnet)
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.