script
stringlengths 113
767k
|
|---|
import numpy as np
import pandas as pd
from pandas_profiling import ProfileReport
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# # 1. Load Data & Check Information
df_net = pd.read_csv("../input/netflix-shows/netflix_titles.csv")
# dfqwefdsafqwe
df_net.head()
ProfileReport(df_net)
|
import pandas as pd
import numpy as np
import os
import matplotlib.pyplot as plt
from keras import models
from keras.utils import to_categorical, np_utils
from tensorflow import convert_to_tensor
from tensorflow.image import grayscale_to_rgb
from tensorflow.data import Dataset
from tensorflow.keras.layers import Flatten, Dense, GlobalAvgPool2D, GlobalMaxPool2D
from tensorflow.keras.callbacks import Callback, EarlyStopping, ReduceLROnPlateau
from tensorflow.keras import optimizers
from tensorflow.keras.utils import plot_model
# import tensorflow as tf
# tf.__version__
# Define the input path and show all files
path = "/kaggle/input/challenges-in-representation-learning-facial-expression-recognition-challenge/"
os.listdir(path)
# Load the image data with labels.
data = pd.read_csv(path + "icml_face_data.csv")
data.head()
# Overview
data[" Usage"].value_counts()
emotions = {
0: "Angry",
1: "Disgust",
2: "Fear",
3: "Happy",
4: "Sad",
5: "Surprise",
6: "Neutral",
}
def prepare_data(data):
"""Prepare data for modeling
input: data frame with labels und pixel data
output: image and label array"""
image_array = np.zeros(shape=(len(data), 48, 48))
image_label = np.array(list(map(int, data["emotion"])))
for i, row in enumerate(data.index):
image = np.fromstring(data.loc[row, " pixels"], dtype=int, sep=" ")
image = np.reshape(image, (48, 48))
image_array[i] = image
return image_array, image_label
# Define training, validation and test data:
train_image_array, train_image_label = prepare_data(data[data[" Usage"] == "Training"])
val_image_array, val_image_label = prepare_data(data[data[" Usage"] == "PrivateTest"])
test_image_array, test_image_label = prepare_data(data[data[" Usage"] == "PublicTest"])
# Reshape and scale the images:
train_images = train_image_array.reshape((train_image_array.shape[0], 48, 48, 1))
train_images = train_images.astype("float32") / 255
val_images = val_image_array.reshape((val_image_array.shape[0], 48, 48, 1))
val_images = val_images.astype("float32") / 255
test_images = test_image_array.reshape((test_image_array.shape[0], 48, 48, 1))
test_images = test_images.astype("float32") / 255
# As the pretrained model expects rgb images, we convert our grayscale images with a single channel to pseudo-rgb images with 3 channels
train_images_rgb = grayscale_to_rgb(convert_to_tensor(train_images))
val_images_rgb = grayscale_to_rgb(convert_to_tensor(val_images))
test_images_rgb = grayscale_to_rgb(convert_to_tensor(test_images))
# Data Augmentation using ImageDataGenerator
# sources:
# https://www.tensorflow.org/api_docs/python/tf/keras/preprocessing/image/ImageDataGenerator
# https://pyimagesearch.com/2019/07/08/keras-imagedatagenerator-and-data-augmentation/
from tensorflow.keras.preprocessing.image import ImageDataGenerator
train_rgb_datagen = ImageDataGenerator(
rotation_range=0.15,
width_shift_range=0.15,
height_shift_range=0.15,
shear_range=0.15,
zoom_range=0.15,
horizontal_flip=True,
zca_whitening=False,
)
train_rgb_datagen.fit(train_images_rgb)
# Encoding of the target value:
train_labels = to_categorical(train_image_label)
val_labels = to_categorical(val_image_label)
test_labels = to_categorical(test_image_label)
def plot_examples(label=0):
fig, axs = plt.subplots(1, 5, figsize=(25, 12))
fig.subplots_adjust(hspace=0.2, wspace=0.2)
axs = axs.ravel()
for i in range(5):
idx = data[data["emotion"] == label].index[i]
axs[i].imshow(train_images[idx][:, :, 0], cmap="gray")
axs[i].set_title(emotions[train_labels[idx].argmax()])
axs[i].set_xticklabels([])
axs[i].set_yticklabels([])
plot_examples(label=0)
plot_examples(label=1)
plot_examples(label=2)
plot_examples(label=3)
plot_examples(label=4)
plot_examples(label=5)
plot_examples(label=6)
# In case we may want to save some examples:
from PIL import Image
def save_all_emotions(channels=1, imgno=0):
for i in range(7):
idx = data[data["emotion"] == i].index[imgno]
emotion = emotions[train_labels[idx].argmax()]
img = train_images[idx]
if channels == 1:
img = img.squeeze()
else:
img = grayscale_to_rgb(
convert_to_tensor(img)
).numpy() # convert to tensor, then to 3ch, back to numpy
img_shape = img.shape
# print(f'img shape: {img_shape[0]},{img_shape[1]}, type: {type(img)}') #(48,48)
img = img * 255
img = img.astype(np.uint8)
suf = "_%d_%d_%d" % (img_shape[0], img_shape[1], channels)
os.makedirs("examples" + suf, exist_ok=True)
fname = os.path.join("examples" + suf, emotion + suf + ".png")
Image.fromarray(img).save(fname)
print(f"saved: {fname}")
save_all_emotions(channels=3, imgno=0)
def plot_compare_distributions(array1, array2, title1="", title2=""):
df_array1 = pd.DataFrame()
df_array2 = pd.DataFrame()
df_array1["emotion"] = array1.argmax(axis=1)
df_array2["emotion"] = array2.argmax(axis=1)
fig, axs = plt.subplots(1, 2, figsize=(12, 6), sharey=False)
x = emotions.values()
y = df_array1["emotion"].value_counts()
keys_missed = list(set(emotions.keys()).difference(set(y.keys())))
for key_missed in keys_missed:
y[key_missed] = 0
axs[0].bar(x, y.sort_index(), color="orange")
axs[0].set_title(title1)
axs[0].grid()
y = df_array2["emotion"].value_counts()
keys_missed = list(set(emotions.keys()).difference(set(y.keys())))
for key_missed in keys_missed:
y[key_missed] = 0
axs[1].bar(x, y.sort_index())
axs[1].set_title(title2)
axs[1].grid()
plt.show()
plot_compare_distributions(
train_labels, val_labels, title1="train labels", title2="val labels"
)
# Calculate the class weights of the label distribution:
class_weight = dict(
zip(
range(0, 7),
(
(
(
data[data[" Usage"] == "Training"]["emotion"].value_counts()
).sort_index()
)
/ len(data[data[" Usage"] == "Training"]["emotion"])
).tolist(),
)
)
class_weight
# ## General defintions and helper functions
# Define callbacks
early_stopping = EarlyStopping(
monitor="val_accuracy",
min_delta=0.00008,
patience=11,
verbose=1,
restore_best_weights=True,
)
lr_scheduler = ReduceLROnPlateau(
monitor="val_accuracy",
min_delta=0.0001,
factor=0.25,
patience=4,
min_lr=1e-7,
verbose=1,
)
callbacks = [
early_stopping,
lr_scheduler,
]
# General shape parameters
IMG_SIZE = 48
NUM_CLASSES = 7
BATCH_SIZE = 64
# A plotting function to visualize training progress
def render_history(history, suf=""):
fig, (ax1, ax2) = plt.subplots(1, 2)
plt.subplots_adjust(left=0.1, bottom=0.1, right=0.95, top=0.9, wspace=0.4)
ax1.set_title("Losses")
ax1.plot(history.history["loss"], label="loss")
ax1.plot(history.history["val_loss"], label="val_loss")
ax1.set_xlabel("epochs")
ax1.set_ylabel("value of the loss function")
ax1.legend()
ax2.set_title("Accuracies")
ax2.plot(history.history["accuracy"], label="accuracy")
ax2.plot(history.history["val_accuracy"], label="val_accuracy")
ax2.set_xlabel("epochs")
ax2.set_ylabel("value of accuracy")
ax2.legend()
plt.show()
suf = "" if suf == "" else "_" + suf
fig.savefig("loss_and_acc" + suf + ".png")
# ## Model construction
from tensorflow.keras.applications import MobileNet
from tensorflow.keras.models import Model
# By specifying the include_top=False argument, we load a network that
# doesn't include the classification layers at the top, which is ideal for feature extraction.
base_net = MobileNet(
input_shape=(IMG_SIZE, IMG_SIZE, 3), include_top=False, weights="imagenet"
)
# plot_model(base_net, show_shapes=True, show_layer_names=True, expand_nested=True, dpi=50, to_file='mobilenet_full.png')
# For these small images, mobilenet is a very large model. Observing that there is nothing left to convolve further, we take the model only until the 12.block
base_model = Model(
inputs=base_net.input,
outputs=base_net.get_layer("conv_pw_12_relu").output,
name="mobilenet_trunc",
)
# this is the same as:
# base_model = Model(inputs = base_net.input,outputs = base_net.layers[-7].output)
# plot_model(base_model, show_shapes=True, show_layer_names=True, expand_nested=True, dpi=50, to_file='mobilenet_truncated.png')
# from: https://www.tensorflow.org/tutorials/images/transfer_learning
from tensorflow.keras import Sequential, layers
from tensorflow.keras import Input, Model
# from tensor
# base_model.trainable = False
# This model expects pixel values in [-1, 1], but at this point, the pixel values in your images are in [0, 255].
# To rescale them, use the preprocessing method included with the model.
# preprocess_input = tf.keras.applications.mobilenet_v2.preprocess_input
# Add a classification head: To generate predictions from the block of features,
# average over the spatial 2x2 spatial locations, using a tf.keras.layers.GlobalAveragePooling2D layer
# to convert the features to a single 1280-element vector per image.
global_average_layer = GlobalAvgPool2D()
# feature_batch_average = global_average_layer(feature_batch)
# print(feature_batch_average.shape)
# Apply a tf.keras.layers.Dense layer to convert these features into a single prediction per image.
# You don't need an activation function here because this prediction will be treated as a logit,
# or a raw prediction value. Positive numbers predict class 1, negative numbers predict class 0.
prediction_layer = Dense(NUM_CLASSES, activation="softmax", name="pred")
# prediction_batch = prediction_layer(feature_batch_average)
# print(prediction_batch.shape)
# Build a model by chaining together the data augmentation, rescaling, base_model and feature extractor layers
# using the Keras Functional API. As previously mentioned, use training=False as our model contains a BatchNormalization layer.
inputs_raw = Input(shape=(IMG_SIZE, IMG_SIZE, 3))
# inputs_pp = preprocess_input(inputs_aug)
# x = base_model(inputs_pp, training=False)
x = base_model(inputs_raw, training=False)
x = global_average_layer(x)
# x = tf.keras.layers.Dropout(0.2)(x)
outputs = prediction_layer(x)
model = Model(inputs=inputs_raw, outputs=outputs)
model.summary()
plot_model(
model,
show_shapes=True,
show_layer_names=True,
expand_nested=True,
dpi=50,
to_file="MobileNet12blocks_structure.png",
)
# Train the classification head:
# base_model.trainable = True #if we included the model layers, but not the model itself, this doesn't have any effect
for layer in base_model.layers[:]:
layer.trainable = False
# for layer in base_model.layers[81:]:
# layer.trainable = True
optims = {
"sgd": optimizers.SGD(lr=0.1, momentum=0.9, decay=0.01),
"adam": optimizers.Adam(0.01),
"nadam": optimizers.Nadam(
learning_rate=0.1, beta_1=0.9, beta_2=0.999, epsilon=1e-07
),
}
model.compile(
loss="categorical_crossentropy", optimizer=optims["adam"], metrics=["accuracy"]
)
model.summary()
initial_epochs = 5
# total_epochs = initial_epochs + 5
history = model.fit_generator(
train_rgb_datagen.flow(train_images_rgb, train_labels, batch_size=BATCH_SIZE),
validation_data=(val_images_rgb, val_labels),
class_weight=class_weight,
steps_per_epoch=len(train_images) / BATCH_SIZE,
# initial_epoch = history.epoch[-1],
# epochs = total_epochs,
epochs=initial_epochs,
callbacks=callbacks,
use_multiprocessing=True,
)
# ### Fine-tuning
iterative_finetuning = False
# #### First iteration: partial fine-tuning of the base_model
if iterative_finetuning:
# fine-tune the top layers (blocks 7-12):
# Let's take a look to see how many layers are in the base model
print("Number of layers in the base model: ", len(base_model.layers))
# base_model.trainable = True #if we included the model layers, but not the model itself, this doesn't have any effect
for layer in base_model.layers:
layer.trainable = False
for layer in base_model.layers[-37:]: # blocks 7-12
layer.trainable = True
optims = {
"sgd": optimizers.SGD(lr=0.01, momentum=0.9, decay=0.01),
"adam": optimizers.Adam(0.001),
"nadam": optimizers.Nadam(
learning_rate=0.01, beta_1=0.9, beta_2=0.999, epsilon=1e-07
),
}
model.compile(
loss="categorical_crossentropy", optimizer=optims["adam"], metrics=["accuracy"]
)
model.summary()
if iterative_finetuning:
fine_tune_epochs = 40
total_epochs = history.epoch[-1] + fine_tune_epochs
history = model.fit_generator(
train_rgb_datagen.flow(train_images_rgb, train_labels, batch_size=BATCH_SIZE),
validation_data=(val_images_rgb, val_labels),
class_weight=class_weight,
steps_per_epoch=len(train_images) / BATCH_SIZE,
initial_epoch=history.epoch[-1],
epochs=total_epochs,
callbacks=callbacks,
use_multiprocessing=True,
)
if iterative_finetuning:
test_loss, test_acc = model.evaluate(test_images_rgb, test_labels) # , test_labels
print("test caccuracy:", test_acc)
if iterative_finetuning:
render_history(history, "mobilenet12blocks_wdgenaug_finetuning1")
# #### Second Iteration (or the main iteration, if iterative_finetuning was set to False): fine-tuning of the entire base_model
if iterative_finetuning:
ftsuf = "ft_2"
else:
ftsuf = "ft_atonce"
# fine-tune all layers
# Let's take a look to see how many layers are in the base model
print("Number of layers in the base model: ", len(base_model.layers))
# base_model.trainable = True #if we included the model layers, but not the model itself, this doesn't have any effect
for layer in base_model.layers:
layer.trainable = False
for layer in base_model.layers[:]:
layer.trainable = True
optims = {
"sgd": optimizers.SGD(lr=0.01, momentum=0.9, decay=0.01),
"adam": optimizers.Adam(0.0001),
"nadam": optimizers.Nadam(
learning_rate=0.01, beta_1=0.9, beta_2=0.999, epsilon=1e-07
),
}
model.compile(
loss="categorical_crossentropy", optimizer=optims["adam"], metrics=["accuracy"]
)
model.summary()
fine_tune_epochs = 100
total_epochs = history.epoch[-1] + fine_tune_epochs
history = model.fit_generator(
train_rgb_datagen.flow(train_images_rgb, train_labels, batch_size=BATCH_SIZE),
validation_data=(val_images_rgb, val_labels),
class_weight=class_weight,
steps_per_epoch=len(train_images) / BATCH_SIZE,
initial_epoch=history.epoch[-1],
epochs=total_epochs,
callbacks=callbacks,
use_multiprocessing=True,
)
test_loss, test_acc = model.evaluate(test_images_rgb, test_labels) # , test_labels
print("test caccuracy:", test_acc)
render_history(history, "mobilenet12blocks_wdgenaug_" + ftsuf)
pred_test_labels = model.predict(test_images_rgb)
model_yaml = model.to_yaml()
with open(
"MobileNet12blocks_wdgenaug_onrawdata_valacc_" + ftsuf + ".yaml", "w"
) as yaml_file:
yaml_file.write(model_yaml)
model.save("MobileNet12blocks_wdgenaug_onrawdata_valacc_" + ftsuf + ".h5")
# ### Analyze the predictions made for the test data
def plot_image_and_emotion(
test_image_array, test_image_label, pred_test_labels, image_number
):
"""Function to plot the image and compare the prediction results with the label"""
fig, axs = plt.subplots(1, 2, figsize=(12, 6), sharey=False)
bar_label = emotions.values()
axs[0].imshow(test_image_array[image_number], "gray")
axs[0].set_title(emotions[test_image_label[image_number]])
axs[1].bar(bar_label, pred_test_labels[image_number], color="orange", alpha=0.7)
axs[1].grid()
plt.show()
import ipywidgets as widgets
@widgets.interact
def f(x=106):
# print(x)
plot_image_and_emotion(test_image_array, test_image_label, pred_test_labels, x)
# ### Make inference for a single image from scratch:
def predict_emotion_of_image(
test_image_array, test_image_label, pred_test_labels, image_number
):
input_arr = test_image_array[image_number] / 255
input_arr = input_arr.reshape((48, 48, 1))
input_arr_rgb = grayscale_to_rgb(convert_to_tensor(input_arr))
predictions = model.predict(np.array([input_arr_rgb]))
predictions_f = [
"%s:%5.2f" % (emotions[i], p * 100) for i, p in enumerate(predictions[0])
]
label = emotions[test_image_label[image_number]]
return f"Label: {label}\nPredictions: {predictions_f}"
import ipywidgets as widgets
@widgets.interact
def f(x=106):
result = predict_emotion_of_image(
test_image_array, test_image_label, pred_test_labels, x
)
print(result)
# ## Compare the distribution of labels and predicted labels
def plot_compare_distributions(array1, array2, title1="", title2=""):
df_array1 = pd.DataFrame()
df_array2 = pd.DataFrame()
df_array1["emotion"] = array1.argmax(axis=1)
df_array2["emotion"] = array2.argmax(axis=1)
fig, axs = plt.subplots(1, 2, figsize=(12, 6), sharey=False)
x = emotions.values()
y = df_array1["emotion"].value_counts()
keys_missed = list(set(emotions.keys()).difference(set(y.keys())))
for key_missed in keys_missed:
y[key_missed] = 0
axs[0].bar(x, y.sort_index(), color="orange")
axs[0].set_title(title1)
axs[0].grid()
y = df_array2["emotion"].value_counts()
keys_missed = list(set(emotions.keys()).difference(set(y.keys())))
for key_missed in keys_missed:
y[key_missed] = 0
axs[1].bar(x, y.sort_index())
axs[1].set_title(title2)
axs[1].grid()
plt.show()
plot_compare_distributions(
test_labels, pred_test_labels, title1="test labels", title2="predict labels"
)
df_compare = pd.DataFrame()
df_compare["real"] = test_labels.argmax(axis=1)
df_compare["pred"] = pred_test_labels.argmax(axis=1)
df_compare["wrong"] = np.where(df_compare["real"] != df_compare["pred"], 1, 0)
from sklearn.metrics import confusion_matrix
from mlxtend.plotting import plot_confusion_matrix
conf_mat = confusion_matrix(test_labels.argmax(axis=1), pred_test_labels.argmax(axis=1))
fig, ax = plot_confusion_matrix(
conf_mat=conf_mat,
show_normed=True,
show_absolute=False,
class_names=emotions.values(),
figsize=(8, 8),
)
fig.show()
|
# #### EEMT 5400 IT for E-Commerce Applications
# ##### HW4 Max score: (1+1+1)+(1+1+2+2)+(1+2)+2
# You will use two different datasets in this homework and you can find their csv files in the below hyperlinks.
# 1. Car Seat:
# https://raw.githubusercontent.com/selva86/datasets/master/Carseats.csv
# 2. Bank Personal Loan:
# https://raw.githubusercontent.com/ChaithrikaRao/DataChime/master/Bank_Personal_Loan_Modelling.csv
# #### Q1.
# a) Perform PCA for both datasets. Create the scree plots (eigenvalues).
# b) Suggest the optimum number of compenents for each dataset with explanation.
# c) Save the PCAs as carseat_pca and ploan_pca respectively.
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.decomposition import PCA
from sklearn.preprocessing import StandardScaler
carseat_df = pd.read_csv(
"https://raw.githubusercontent.com/selva86/datasets/master/Carseats.csv"
)
ploan_df = pd.read_csv(
"https://raw.githubusercontent.com/ChaithrikaRao/DataChime/master/Bank_Personal_Loan_Modelling.csv"
)
scaler = StandardScaler()
numeric_carseat = carseat_df.select_dtypes(include=[np.number])
carseat_std = scaler.fit_transform(numeric_carseat)
numeric_ploan = ploan_df.select_dtypes(include=[np.number])
ploan_std = scaler.fit_transform(numeric_ploan)
pca_carseat = PCA()
pca_ploan = PCA()
carseat_pca_result = pca_carseat.fit(carseat_std)
ploan_pca_result = pca_ploan.fit(ploan_std)
def scree_plot(pca_result, title):
plt.figure()
plt.plot(np.cumsum(pca_result.explained_variance_ratio_))
plt.xlabel("Number of Components")
plt.ylabel("Cumulative Explained Variance")
plt.title(title)
plt.show()
scree_plot(carseat_pca_result, "Car Seat Dataset")
scree_plot(ploan_pca_result, "Bank Personal Loan Dataset")
# (b)The optimal number of components can be determined by looking at the point where the cumulative explained variance "elbows" or starts to level off.
# For the Car Seat dataset, it appears to be around 3 components.
# For the Bank Personal Loan dataset, it appears to be around 4 components.
carseat_pca = PCA(n_components=3)
ploan_pca = PCA(n_components=4)
carseat_pca_result = carseat_pca.fit_transform(carseat_std)
ploan_pca_result = ploan_pca.fit_transform(ploan_std)
# #### Q2. (Car Seat Dataset)
# a) Convert the non-numeric variables to numeric by using get_dummies() method in pandas. Use it in this question.
# b) Use the scikit learn variance filter to reduce the dimension of the dataset. Try different threshold and suggest the best one.
# c) Some columns may have high correlation. For each set of highly correlated variables, keep one variable only and remove the rest of highly correlated columns. (Tips: You can find the correlations among columns by using .corr() method of pandas dataframe. Reference: https://pandas.pydata.org/docs/reference/api/pandas.DataFrame.corr.html)
# d) Perform linear regression to predict the Sales with datasets from part b and part c respectively and compare the result
#
carseat_dummies = pd.get_dummies(carseat_df, drop_first=True)
from sklearn.feature_selection import VarianceThreshold
def filter_by_variance(data, threshold):
var_filter = VarianceThreshold(threshold=threshold)
return pd.DataFrame(
var_filter.fit_transform(data), columns=data.columns[var_filter.get_support()]
)
carseat_filtered_001 = filter_by_variance(carseat_dummies, 0.01)
carseat_filtered_01 = filter_by_variance(carseat_dummies, 0.1)
carseat_filtered_1 = filter_by_variance(carseat_dummies, 1)
print(f"0.01 threshold: {carseat_filtered_001.shape[1]} columns")
print(f"0.1 threshold: {carseat_filtered_01.shape[1]} columns")
print(f"1 threshold: {carseat_filtered_1.shape[1]} columns")
def remove_high_corr(data, threshold):
corr_matrix = data.corr().abs()
upper = corr_matrix.where(np.triu(np.ones(corr_matrix.shape), k=1).astype(np.bool))
to_drop = [column for column in upper.columns if any(upper[column] > threshold)]
return data.drop(columns=to_drop)
carseat_no_high_corr = remove_high_corr(carseat_filtered_01, 0.8)
def linear_regression_score(X, y):
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=42
)
lr = LinearRegression()
lr.fit(X_train, y_train)
y_pred = lr.predict(X_test)
return r2_score(y_test, y_pred)
X_filtered = carseat_filtered_01.drop(columns=["Sales"])
y_filtered = carseat_filtered_01["Sales"]
X_no_high_corr = carseat_no_high_corr.drop(columns=["Sales"])
y_no_high_corr = carseat_no_high_corr["Sales"]
filtered_score = linear_regression_score(X_filtered, y_filtered)
no_high_corr_score = linear_regression_score(X_no_high_corr, y_no_high_corr)
print(f"Filtered dataset R-squared: {filtered_score}")
print(f"No high correlation dataset R-squared: {no_high_corr_score}")
# #### Q3. (Bank Personal Loan Dataset)
# a) Find the variable which has the highest correlations with CCAvg
# b) Perform polynomial regression to predict CCAvg with the variable identified in part a.
# ##### Tips:
# step 1 - convert the dataset to polynomial using PolynomialFeatures from scikit learn (https://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.PolynomialFeatures.html)
# step 2 - Perform linear regression using scikit learn
ploan_df = pd.read_csv(
"https://raw.githubusercontent.com/ChaithrikaRao/DataChime/master/Bank_Personal_Loan_Modelling.csv"
)
correlations = ploan_df.corr().abs()
highest_corr = correlations["CCAvg"].sort_values(ascending=False).index[1]
print(f"The variable with the highest correlation with CCAvg is {highest_corr}")
X = ploan_df[[highest_corr]]
y = ploan_df["CCAvg"]
poly = PolynomialFeatures(degree=2)
X_poly = poly.fit_transform(X)
X_train, X_test, y_train, y_test = train_test_split(
X_poly, y, test_size=0.3, random_state=42
)
lr = LinearRegression()
lr.fit(X_train, y_train)
y_pred = lr.predict(X_test)
poly_r_squared = r2_score(y_test, y_pred)
print(f"Polynomial regression R-squared: {poly_r_squared}")
# #### Q4. (Bank Personal Loan Dataset)
# Perform linear regression with all variables in the dataset and compare the result with the model in question 3 using R-Squared value.
X_all = ploan_df.drop(columns=["ID", "CCAvg"])
y_all = ploan_df["CCAvg"]
X_train, X_test, y_train, y_test = train_test_split(
X_all, y_all, test_size=0.3, random_state=42
)
lr_all = LinearRegression()
lr_all.fit(X_train, y_train)
y_pred_all = lr_all.predict(X_test)
all_r_squared = r2_score(y_test, y_pred_all)
print(f"All variables linear regression R-squared: {all_r_squared}")
print(f"Polynomial regression R-squared: {poly_r_squared}")
print(f"All variables linear regression R-squared: {all_r_squared}")
|
# 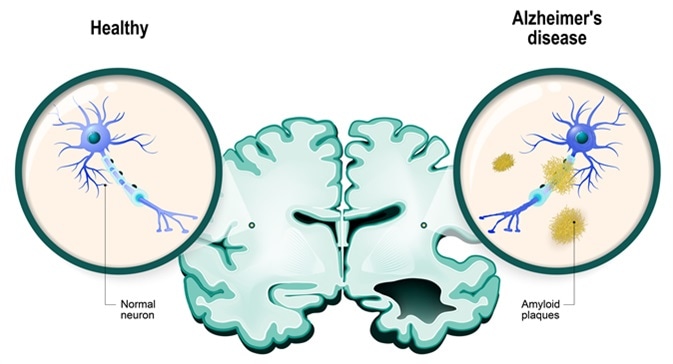
# **Alzheimer's disease** is the most common type of dementia. It is a progressive disease beginning with mild memory loss and possibly leading to loss of the ability to carry on a conversation and respond to the environment. Alzheimer's disease involves parts of the brain that control thought, memory, and language.
# # **Importing libraries**
import pandas as pd
import numpy as np
import os
import matplotlib.pyplot as plt
import warnings
from tensorflow.keras.applications.vgg19 import preprocess_input
from tensorflow.keras.preprocessing import image, image_dataset_from_directory
from tensorflow.keras.preprocessing.image import ImageDataGenerator as IDG
from imblearn.over_sampling import SMOTE
from tensorflow.keras.models import Sequential
from tensorflow import keras
import tensorflow
import tensorflow as tf
from tensorflow.keras.models import Model
from tensorflow.keras.applications.vgg16 import VGG16
from tensorflow.keras.layers import Input, Lambda, Dense, Flatten, Dropout
from tensorflow.keras.models import Model
from sklearn.model_selection import train_test_split
import seaborn as sns
import pathlib
from tensorflow.keras.utils import plot_model
from keras.callbacks import ModelCheckpoint, ReduceLROnPlateau, EarlyStopping
from sklearn.metrics import classification_report, confusion_matrix
# # **Identify dataset**
train_ds = tf.keras.preprocessing.image_dataset_from_directory(
"/kaggle/input/adni-extracted-axial/Axial",
validation_split=0.2,
subset="training",
seed=1337,
image_size=[180, 180],
batch_size=16,
)
val_ds = tf.keras.preprocessing.image_dataset_from_directory(
"/kaggle/input/adni-extracted-axial/Axial",
validation_split=0.2,
subset="validation",
seed=1337,
image_size=[180, 180],
batch_size=16,
)
# number and names of Classes
classnames = train_ds.class_names
len(classnames), train_ds.class_names
# # **Data Visualization**
plt.figure(figsize=(10, 10))
for images, labels in train_ds.take(1):
for i in range(9):
ax = plt.subplot(3, 3, i + 1)
plt.imshow(images[i].numpy().astype("uint8"))
plt.title(train_ds.class_names[labels[i]])
plt.axis("off")
# Number of images in each class
NUM_IMAGES = []
for label in classnames:
dir_name = "/kaggle/input/adni-extracted-axial/Axial" + "/" + label
NUM_IMAGES.append(len([name for name in os.listdir(dir_name)]))
NUM_IMAGES, classnames
# Rename class names
class_names = ["Alzheimer Disease", "Cognitively Impaired", "Cognitively Normal"]
train_ds.class_names = class_names
val_ds.class_names = class_names
NUM_CLASSES = len(class_names)
NUM_CLASSES
# Before Oversampling
# Visualization of each class with pie chart
import matplotlib.pyplot as plt
fig1, ax1 = plt.subplots()
ax1.pie(NUM_IMAGES, autopct="%1.1f%%", labels=train_ds.class_names)
plt.legend(title="Three Classes:", bbox_to_anchor=(0.75, 1.15))
# Performing Image Augmentation to have more data samples
IMG_SIZE = 180
IMAGE_SIZE = [180, 180]
DIM = (IMG_SIZE, IMG_SIZE)
ZOOM = [0.99, 1.01]
BRIGHT_RANGE = [0.8, 1.2]
HORZ_FLIP = True
FILL_MODE = "constant"
DATA_FORMAT = "channels_last"
WORK_DIR = "/kaggle/input/adni-extracted-axial/Axial"
work_dr = IDG(
rescale=1.0 / 255,
brightness_range=BRIGHT_RANGE,
zoom_range=ZOOM,
data_format=DATA_FORMAT,
fill_mode=FILL_MODE,
horizontal_flip=HORZ_FLIP,
)
train_data_gen = work_dr.flow_from_directory(
directory=WORK_DIR, target_size=DIM, batch_size=8000, shuffle=False
)
train_data, train_labels = train_data_gen.next()
# # **Oversampling technique**
# 
# Shape of data before oversampling
print(train_data.shape, train_labels.shape)
# Performing over-sampling of the data, since the classes are imbalanced
# After oversampling using SMOTE
sm = SMOTE(random_state=42)
train_data, train_labels = sm.fit_resample(
train_data.reshape(-1, IMG_SIZE * IMG_SIZE * 3), train_labels
)
train_data = train_data.reshape(-1, IMG_SIZE, IMG_SIZE, 3)
print(train_data.shape, train_labels.shape)
# Show pie plot for dataset (after oversampling)
# Visualization of each class with pie chart
images_after = [2590, 2590, 2590]
import matplotlib.pyplot as plt
fig1, ax1 = plt.subplots()
ax1.pie(images_after, autopct="%1.1f%%", labels=train_ds.class_names)
plt.legend(title="Three Classes:", bbox_to_anchor=(0.75, 1.15))
# # **Spliting data**
train_data, test_data, train_labels, test_labels = train_test_split(
train_data, train_labels, test_size=0.2, random_state=42
)
train_data, val_data, train_labels, val_labels = train_test_split(
train_data, train_labels, test_size=0.2, random_state=42
)
# # **Building the model**
# -------VGG16--------
vgg = VGG16(input_shape=(180, 180, 3), weights="imagenet", include_top=False)
for layer in vgg.layers:
layer.trainable = False
x = Flatten()(vgg.output)
prediction = Dense(3, activation="softmax")(x)
modelvgg = Model(inputs=vgg.input, outputs=prediction)
# Plotting layers as an image
plot_model(modelvgg, to_file="alzahimer.png", show_shapes=True)
# Optimizing model
modelvgg.compile(
optimizer="adam", loss="categorical_crossentropy", metrics=["accuracy"]
)
# Callbacks
checkpoint = ModelCheckpoint(
filepath="best_weights.hdf5", save_best_only=True, save_weights_only=True
)
lr_reduce = ReduceLROnPlateau(
monitor="val_loss", factor=0.3, patience=2, verbose=2, mode="max"
)
early_stop = EarlyStopping(monitor="val_loss", min_delta=0.1, patience=1, mode="min")
# # **Training model using my data**
# Fitting the model
hist = modelvgg.fit(
train_data,
train_labels,
epochs=10,
validation_data=(val_data, val_labels),
callbacks=[checkpoint, lr_reduce],
)
# Plotting accuracy and loss of the model
fig, ax = plt.subplots(1, 2, figsize=(20, 3))
ax = ax.ravel()
for i, met in enumerate(["accuracy", "loss"]):
ax[i].plot(hist.history[met])
ax[i].plot(hist.history["val_" + met])
ax[i].set_title("Model {}".format(met))
ax[i].set_xlabel("epochs")
ax[i].set_ylabel(met)
ax[i].legend(["train", "val"])
# Evaluation using test data
test_scores = modelvgg.evaluate(test_data, test_labels)
print("Testing Accuracy: %.2f%%" % (test_scores[1] * 100))
pred_labels = modelvgg.predict(test_data)
# # **Confusion matrix**
pred_ls = np.argmax(pred_labels, axis=1)
test_ls = np.argmax(test_labels, axis=1)
conf_arr = confusion_matrix(test_ls, pred_ls)
plt.figure(figsize=(8, 6), dpi=80, facecolor="w", edgecolor="k")
ax = sns.heatmap(
conf_arr,
cmap="Greens",
annot=True,
fmt="d",
xticklabels=classnames,
yticklabels=classnames,
)
plt.title("Alzheimer's Disease Diagnosis")
plt.xlabel("Prediction")
plt.ylabel("Truth")
plt.show(ax)
# # **Classification report**
print(classification_report(test_ls, pred_ls, target_names=classnames))
# # **Save the model for a Mobile app as tflite**
export_dir = "/kaggle/working/"
tf.saved_model.save(modelvgg, export_dir)
tflite_model_name = "alzheimerfinaly.tflite"
# Convert the model.
converter = tf.lite.TFLiteConverter.from_saved_model(export_dir)
converter.optimizations = [tf.lite.Optimize.OPTIMIZE_FOR_SIZE]
tflite_model = converter.convert()
tflite_model_file = pathlib.Path(tflite_model_name)
tflite_model_file.write_bytes(tflite_model)
# # **Save the model for a Web app as hdf5**
tf.keras.models.save_model(modelvgg, "Alzheimer_finaly.hdf5")
|
# # Tracking COVID-19 from New York City wastewater
# **TABLE OF CONTENTS**
# * [1. Introduction](#chapter_1)
# * [2. Data exploration](#chapter_2)
# * [3. Analysis](#chapter_3)
# * [4. Baseline model](#chapter_4)
# ## 1. Introduction
# The **New York City OpenData Project** (*link:* __[project home page](https://opendata.cityofnewyork.us)__) has hundreds of open, New York-related datasets available for everyone to use. On the website, all datasets are labeled by different city functions (business, government, education etc.).
# While browsing through different subcategories, I came across data by the Department of Environmental Protection (DEP). One dataset they had made available in public concerned the SARS-CoV-2 gene level concentrations measured in NYC wastewater (*link:* __[dataset page](https://data.cityofnewyork.us/Health/SARS-CoV-2-concentrations-measured-in-NYC-Wastewat/f7dc-2q9f)__). As one can guess, SARS-CoV-2 is the virus causing COVID-19.
# Since I had earlier used the NYC data on COVID (*link:* __[dataset page](https://data.cityofnewyork.us/Health/COVID-19-Daily-Counts-of-Cases-Hospitalizations-an/rc75-m7u3)__) in several notebooks since the pandemic, I decided to create a new notebook combining these two datasets.
# This notebook is a brief exploratory analysis on the relationship between the COVID-causing virus concentrations in NYC wastewater and actual COVID-19 cases detected in New York. Are these two related, and if so, how? Let's find out.
# *All data sources are read directly from the OpenData Project website, so potential errors are caused by temporary issues (update etc.) in the website data online availability.*
# **April 13th, 2023
# Jari Peltola**
# ******
# ## 2. Data Exploration
# import modules
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
from sklearn.ensemble import RandomForestRegressor
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import MinMaxScaler
from sklearn.metrics import r2_score, mean_absolute_error, mean_squared_error
from sklearn.model_selection import cross_val_score
# set column and row display
pd.set_option("display.max_columns", None)
pd.set_option("display.max_rows", None)
# disable warnings
pd.options.mode.chained_assignment = None
# wastewater dataset url
url = "https://data.cityofnewyork.us/api/views/f7dc-2q9f/rows.csv?accessType=DOWNLOAD"
# read the data from url
data = pd.read_csv(url, low_memory=False)
# drop rows with NaN values
data = data.dropna()
# reset index
data.reset_index(drop=True, inplace=True)
data.head()
data.shape
# A large metropolis area such as New York City has several wastewater resource recovery facilities (WRRF). Each of these facilities takes care of a specific area in the city's wastewater system (*link:* __[list of NYC wastewater treatment facilities](https://www.nyc.gov/site/dep/water/wastewater-treatment-plants.page)__). The 14 facilities serve city population ranging from less than one hundred thousand (Rockaway) up to one million people (Newtown Creek, Wards Island).
# We start by adding some additional data from the webpage linked above. For each facility, the added data includes receiving waterbody, drainage acres as well as verbal description of the drainage area. This is done to get a better comprehension of where exactly in the city each facility is located. Another possible solution would be to create some kind of map of all WRRF facilities in the area, but since we are actually dealing with water with ever-evolving drainage instead of buildings, this approach would not in my opinion work too well.
# It's good to keep the original data intact, so first we make a list **wrrf_list** consisting of all unique WRRF names, create a new dataframe **df_location** and add the facility names as a column **WRRF Name**.
# create list of all pickup locations
wrrf_list = data["WRRF Name"].unique()
# create new dataframe with empty column
df_location = pd.DataFrame(columns=["WRRF Name"])
# add column values from list
df_location["WRRF Name"] = np.array(wrrf_list)
df_location.head()
df_location["WRRF Name"].unique()
# Next we add three more columns including the additional data.
# list of receiving waterbodies
Receiving_Waterbody = [
"Jamaica Bay",
"Jamaica Bay",
"East River",
"Jamaica Bay",
"Upper East River",
"Jamaica Bay",
"Lower East River",
"Hudson River",
"Upper New York Bay",
"Lower New York Bay",
"Upper East River",
"Upper East River",
"Kill Van Kull",
"Upper East River",
]
# list of drainage acres
Drainage_Acres = [
5907,
15087,
15656,
6259,
16860,
25313,
3200,
6030,
12947,
10779,
16664,
12056,
9665,
15203,
]
# list of drainage areas
Drainage_Area = [
"Eastern section of Brooklyn, near Jamaica Bay",
"South and central Brooklyn",
"South and eastern midtown sections of Manhattan, northeast section of Brooklyn and western section of Queens",
"Rockaway Peninsula",
"Northeast section of Queens",
"Southern section of Queens",
"Northwest section of Brooklyn and Governors Island",
"West side of Manhattan above Bank Street",
"Western section of Brooklyn",
"Southern section of Staten Island",
"Eastern section of the Bronx",
"Western section of the Bronx and upper east side of Manhattan",
"Northern section of Staten Island",
"Northeast section of Queens",
]
# add new columns
df_location["Receiving_Waterbody"] = np.array(Receiving_Waterbody)
df_location["Drainage_Acres"] = np.array(Drainage_Acres)
df_location["Drainage_Area"] = np.array(Drainage_Area)
df_location.head()
# Now we may merge the **df_location** dataframe with our original data. For this we can use the common column **WRRF Name**. For clarity we keep calling also the merged results **data**.
# merge dataframes
data = pd.merge(data, df_location, on="WRRF Name", how="left")
data.dtypes
data.head()
# The date data we have is not in datetime format, so we will take care of that next, along with renaming some columns and dropping others.
# column to datetime
data["Test date"] = pd.to_datetime(data["Test date"])
# rename columns by their index location
data = data.rename(columns={data.columns[1]: "TestDate"})
data = data.rename(columns={data.columns[4]: "SARS_CoV2_Concentration"})
data = data.rename(columns={data.columns[5]: "SARS_CoV2_Concentration_PerCap"})
data = data.rename(columns={data.columns[7]: "Population"})
# drop columns by their index location
data = data.drop(data.columns[[0, 3, 6]], axis=1)
data.head()
# Let's take a look at the COVID concentration when some specific WRRF facilities and their respective waterbodies are concerned. For this example, we choose the **Coney Island** and **Jamaica Bay** facilities and plot the result.
# select facility data to two dataframes
df_coney = data.loc[data["WRRF Name"] == "Coney Island"]
df_jamaica = data.loc[data["WRRF Name"] == "Jamaica Bay"]
# set figure size
plt.figure(figsize=(10, 8))
# set parameters
plt.plot(
df_coney.TestDate,
df_coney.SARS_CoV2_Concentration,
label="Coney Island",
linewidth=3,
)
plt.plot(
df_jamaica.TestDate,
df_jamaica.SARS_CoV2_Concentration,
color="red",
label="Jamaica Bay",
linewidth=3,
)
# add title and axis labels
plt.title(
"COVID virus concentration (Coney Island, Jamaica Bay)", weight="bold", fontsize=16
)
plt.xlabel("Date", weight="bold", fontsize=14)
plt.ylabel("Concentration", weight="bold", fontsize=14)
# add legend
plt.legend()
plt.show()
# We can see that the COVID virus concentration in the two selected wastewater facilites is for the most time pretty similar. The big exception is the New Year period 2021-2022, when Coney Island recorded a significant concentration spike, but in Jamaica Bay the change was much more moderate.
# Next we bring in the New York City COVID-19 dataset and upload it as dataframe **covid_cases**.
# COVID-19 dataset url
url = "https://data.cityofnewyork.us/api/views/rc75-m7u3/rows.csv?accessType=DOWNLOAD"
# read the data from url
covid_cases = pd.read_csv(url, low_memory=False)
# drop rows with NaN values
covid_cases = covid_cases.dropna()
covid_cases.head()
covid_cases.dtypes
# One thing we can see is that the COVID-19 data includes both overall data and more specific figures on the city's five boroughs (the Bronx, Brooklyn, Manhattan, Queens, Staten Island).
# In order to merge the data, we rename and reformat the **date_of_interest** column to fit our existing data, since that's the common column we will use.
# rename column
covid_cases = covid_cases.rename(columns={covid_cases.columns[0]: "TestDate"})
# change format to datetime
covid_cases["TestDate"] = pd.to_datetime(covid_cases["TestDate"])
# merge dataframes
data = pd.merge(data, covid_cases, on="TestDate", how="left")
data.head()
data.shape
# What we don't know is the workload of different facilities when it comes to wastewater treatment. Let's see that next.
# check percentages
wrrf_perc = data["WRRF Name"].value_counts(normalize=True) * 100
wrrf_perc
# The percentages tell us that there is no dominant facility when it comes to wastewater treatment in New York City: it's all one big puzzle with relatively equal pieces.
# Now we are ready to find out more about the potential relationship between COVID virus concentration in wastewater and actual COVID cases.
# ******
# ## 3. Analysis
# First it would be good to know more about boroughs and their different wastewater facilities. Mainly it would be useful to find out if there are differences between wastewater treatment facilities and their measured COVID virus concentration levels when a particular borough is concerned.
# Let's take Brooklyn for example. Based on the verbal descriptions of different wastewater facilities, Brooklyn area is mostly served by the **26th Ward**, **Coney Island** and **Owl's Head** facilities. Next we select those three locations with all necessary columns to dataframe **brooklyn_data**.
# select Brooklyn data
brooklyn_data = data.loc[
data["WRRF Name"].isin(["26th Ward", "Coney Island", "Owls Head"])
]
# select columns
brooklyn_data = brooklyn_data.iloc[:, np.r_[0:8, 29:40]]
brooklyn_data.shape
brooklyn_data.head()
# To access the data more conveniently, we take a step back and create separate dataframes for the three facilities before plotting the result.
# create dataframes
df_26th = brooklyn_data.loc[brooklyn_data["WRRF Name"] == "26th Ward"]
df_coney = brooklyn_data.loc[brooklyn_data["WRRF Name"] == "Coney Island"]
df_owls = brooklyn_data.loc[brooklyn_data["WRRF Name"] == "Owls Head"]
# set figure size
plt.figure(figsize=(10, 8))
# set parameters
plt.plot(df_26th.TestDate, df_26th.SARS_CoV2_Concentration, label="26th", linewidth=3)
plt.plot(
df_coney.TestDate,
df_coney.SARS_CoV2_Concentration,
color="red",
label="Coney Island",
linewidth=3,
)
plt.plot(
df_owls.TestDate,
df_owls.SARS_CoV2_Concentration,
color="green",
label="Owls Head",
linewidth=3,
)
# add title and axis labels
plt.title(
"Virus concentration (26th Ward, Coney Island, Owls Head)",
weight="bold",
fontsize=16,
)
plt.xlabel("Date", weight="bold", fontsize=14)
plt.ylabel("Concentration", weight="bold", fontsize=14)
# add legend
plt.legend()
plt.show()
# There are some differences in concentration intensity, but all in all the figures pretty much follow the same pattern. Also, one must remember that for example the Coney Island facility serves population about twice as large as the Owl's Head facility. Then again, this makes the data from beginning of year 2023 even more intriguing, since Owl's Head had much larger COVID concentration then compared to other two Brooklyn facilities.
# Next we plot the third Brooklyn facility (26th Ward) data and compare it to the 7-day average of hospitalized COVID-19 patients in the area. It is notable that in the plot the concentration is multiplied by 0.01 to make a better visual fit with patient data. This change is made merely for plotting purposes and does not alter the actual values in our data.
plt.figure(figsize=(10, 8))
plt.plot(
df_26th.TestDate,
df_26th.SARS_CoV2_Concentration * 0.01,
label="CoV-2 concentration",
linewidth=3,
)
plt.plot(
df_26th.TestDate,
df_26th.BK_HOSPITALIZED_COUNT_7DAY_AVG,
color="red",
label="Hospitalizations avg",
linewidth=3,
)
# add title and axis labels
plt.title(
"Virus concentration in 26th Ward and COVID hospitalizations",
weight="bold",
fontsize=16,
)
plt.xlabel("Date", weight="bold", fontsize=14)
plt.ylabel("Count", weight="bold", fontsize=14)
# add legend
plt.legend()
# display plot
plt.show()
# The two lines definitely share a similar pattern, which in the end is not that surprising considering they actually describe two different viewpoints to the same phenomenon (COVID-19 pandemic/endemic).
# Next we take a closer look at the first 18 months of the COVID-19 pandemic and narrow our date selection accordingly. Also, we change our COVID-19 measurement to daily hospitalized patients instead of a weekly average.
# mask dataframe
start_date = "2020-01-01"
end_date = "2021-12-31"
# wear a mask
mask = (df_26th["TestDate"] >= start_date) & (df_26th["TestDate"] < end_date)
df_26th_mask = df_26th.loc[mask]
plt.figure(figsize=(10, 8))
plt.plot(
df_26th_mask.TestDate,
df_26th_mask.SARS_CoV2_Concentration * 0.01,
label="CoV-2 concentration",
linewidth=3,
)
plt.plot(
df_26th_mask.TestDate,
df_26th_mask.BK_HOSPITALIZED_COUNT,
color="red",
label="Hospitalizations",
linewidth=3,
)
plt.title(
"26th Ward virus concentration and daily hospitalizations",
weight="bold",
fontsize=16,
)
plt.xlabel("Date", weight="bold", fontsize=14)
plt.ylabel("Count", weight="bold", fontsize=14)
plt.legend()
plt.show()
# Now the similarities are even more clear. Taking a look at the late 2020/early 2021 situation, it seems that **some sort of threshold of daily COVID virus concentration difference in wastewater might actually become a valid predictor for future hospitalizations**, if one omitted the viewpoint of a ML model constructor.
# In this notebook we will not go that far, but next a simple baseline model is created as a sort of first step toward that goal.
# ******
# ## 4. Baseline model
# The regressive baseline model we will produce is all about comparing relative (per capita) COVID virus concentration in wasterwater with different features presented in COVID-19 patient data.
# For our baseline model we will use wastewater data from all five boroughs and their wastewater facilities, meaning we must return to our original **data**.
data.head(2)
data.shape
# As we need only some of the numerical data here, next we select the approriate columns to a new dataframe **df_model**.
# select columns by index
df_model = data.iloc[:, np.r_[3, 8:18]]
df_model.head()
df_model.dtypes
# To ensure better compatibility between different data features, the dataframe is next scaled with MinMaxScaler. This is not a necessary step, but as the COVID virus concentration per capita values are relatively large compared to COVID patient data, by scaling all data we may get a bit better results in actual baseline modeling.
# scale data
scaler = MinMaxScaler()
scaler.fit(df_model)
scaled = scaler.fit_transform(df_model)
scaled_df = pd.DataFrame(scaled, columns=df_model.columns)
scaled_df.head()
# plot scatterplot of COVID-19 case count and virus concentration level
scaled_df.plot(kind="scatter", x="SARS_CoV2_Concentration_PerCap", y="CASE_COUNT")
plt.show()
# Some outliers excluded, the data we have is relatively well concentrated and should therefore fit regressive model pretty well. Just to make sure, we drop potential NaN rows before proceeding.
# drop NaN rows
data = scaled_df.dropna()
# set random seed
np.random.seed(42)
# select features
X = data.drop("SARS_CoV2_Concentration_PerCap", axis=1)
y = data["SARS_CoV2_Concentration_PerCap"]
# separate data into training and validation sets
X_train, X_test, y_train, y_test = train_test_split(
X, y, train_size=0.8, test_size=0.2, random_state=1
)
# define model
model = RandomForestRegressor()
# fit model
model.fit(X_train, y_train)
# Now we can make predictions with the model based on our data.
y_preds = model.predict(X_test)
print("Regression model metrics on test set")
print(f"R2: {r2_score(y_test, y_preds)}")
print(f"MAE: {mean_absolute_error(y_test, y_preds)}")
print(f"MSE: {mean_squared_error(y_test, y_preds)}")
# The worst R2 score is basically minus infinity and the perfect score 1, with zero predicting the mean value every time. Taking this into account, the score of about 0.5 (the exact result changes every time the notebook is run as the online datasets are constantly updated) for a test set score is quite typical when using only a simple baseline fit.
# Also, as seen below, the score on train dataset is significantly better than the one of test dataset. As we had less than 500 datapoints i.e. rows overall for the baseline model to learn from, this is in the end not a big surprise.
print("Train score:")
print(model.score(X_train, y_train))
print("Test score:")
print(model.score(X_test, y_test))
# We can also use the cross-valuation score to run the same baseline model several times and take the average R2 score of the process. In this case we will run the model five times (cv=5).
np.random.seed(42)
# create regressor
rf_reg = RandomForestRegressor(n_estimators=100)
# create five models for cross-valuation
# setting "scoring=None" uses default scoring parameter
# cross_val_score function's default scoring parameter is R2
cv_r2 = cross_val_score(rf_reg, X_train, y_train, cv=5, scoring=None)
# print average R2 score of five baseline regressor models
np.mean(cv_r2)
|
# ## Project 4
# We're going to start with the dataset from Project 1.
# This time the goal is to compare data wrangling runtime by either using **Pandas** or **Polar**.
data_dir = "/kaggle/input/project-4-dataset/data-p1"
sampled = False
path_suffix = "" if not sampled else "_sampled"
from time import time
import pandas as pd
import numpy as np
import polars as pl
from polars import col
def print_time(t):
"""Function that converts time period in seconds into %m:%s:%ms expression.
Args:
t (int): time period in seconds
Returns:
s (string): time period formatted
"""
ms = t * 1000
m, ms = divmod(ms, 60000)
s, ms = divmod(ms, 1000)
return "%dm:%ds:%dms" % (m, s, ms)
# ## Load data
# #### Pandas
start = time()
pandas_data = pd.read_csv(f"{data_dir}/transactions_data{path_suffix}.csv")
print("\nProcessing took {}".format(print_time(time() - start)))
start = time()
pandas_data["date"] = pd.to_datetime(pandas_data["date"])
print("\nProcessing took {}".format(print_time(time() - start)))
## Create sales column
start = time()
pandas_data = (
pandas_data.groupby(
[
pandas_data.date.dt.date,
"id",
"item_id",
"dept_id",
"cat_id",
"store_id",
"state_id",
]
)
.agg("count")
.rename(columns={"date": "sales"})
.reset_index()
.assign(date=lambda df: pd.to_datetime(df.date))
)
print("\nProcessing took {}".format(print_time(time() - start)))
## Convert data types
start = time()
pandas_data = pandas_data.assign(
id=pandas_data.id.astype("category"),
item_id=pandas_data.item_id.astype("category"),
dept_id=pandas_data.dept_id.astype("category"),
cat_id=pandas_data.cat_id.astype("category"),
store_id=pandas_data.store_id.astype("category"),
state_id=pandas_data.state_id.astype("category"),
)
print("\nProcessing took {}".format(print_time(time() - start)))
## Filling with zeros
start = time()
pandas_data = pandas_data.set_index(["date", "id"])
min_date, max_date = (
pandas_data.index.get_level_values("date").min(),
pandas_data.index.get_level_values("date").max(),
)
dates_to_select = pd.date_range(min_date, max_date, freq="1D")
ids = pandas_data.index.get_level_values("id").unique()
index_to_select = pd.MultiIndex.from_product(
[dates_to_select, ids], names=["date", "id"]
)
def fill_category_nans(df, col_name, level_start, level_end):
return np.where(
df[col_name].isna(),
df.index.get_level_values("id")
.str.split("_")
.str[level_start:level_end]
.str.join("_"),
df[col_name],
)
pandas_data = (
pandas_data.reindex(index_to_select)
.fillna({"sales": 0})
.assign(
sales=lambda df: df.sales.astype("int"),
item_id=lambda df: fill_category_nans(df, "item_id", 0, 3),
dept_id=lambda df: fill_category_nans(df, "dept_id", 0, 2),
cat_id=lambda df: fill_category_nans(df, "cat_id", 0, 1),
store_id=lambda df: fill_category_nans(df, "store_id", 3, 5),
state_id=lambda df: fill_category_nans(df, "state_id", 3, 4),
)
.assign(
item_id=lambda df: df.item_id.astype("category"),
dept_id=lambda df: df.dept_id.astype("category"),
cat_id=lambda df: df.cat_id.astype("category"),
store_id=lambda df: df.store_id.astype("category"),
state_id=lambda df: df.state_id.astype("category"),
)
)
print("\nProcessing took {}".format(print_time(time() - start)))
# #### Polars
start = time()
transactions_pl = pl.read_csv(f"{data_dir}/transactions_data{path_suffix}.csv")
print("\nProcessing took {}".format(print_time(time() - start)))
start = time()
transactions_pl = transactions_pl.with_columns(
pl.col("date").str.strptime(pl.Date, fmt="%Y-%m-%d %H:%M:%S", strict=False)
)
print("\nProcessing took {}".format(print_time(time() - start)))
## Create sales column
start = time()
polars_data = (
transactions_pl.lazy()
.with_column(pl.lit(1).alias("sales"))
.groupby(["date", "id", "item_id", "dept_id", "cat_id", "store_id", "state_id"])
.agg(pl.col("sales").sum())
.collect()
)
print("\nProcessing took {}".format(print_time(time() - start)))
## Convert data types
start = time()
polars_data = (
polars_data.lazy()
.with_columns(
[
pl.col("id").cast(pl.Categorical),
pl.col("item_id").cast(pl.Categorical),
pl.col("dept_id").cast(pl.Categorical),
pl.col("cat_id").cast(pl.Categorical),
pl.col("store_id").cast(pl.Categorical),
pl.col("state_id").cast(pl.Categorical),
]
)
.collect()
)
print("\nProcessing took {}".format(print_time(time() - start)))
## Filling with zeros
start = time()
min_date, max_date = (
polars_data.with_columns(pl.col("date")).min()["date"][0],
polars_data.with_columns(pl.col("date")).max()["date"][0],
)
dates_to_select = pl.date_range(min_date, max_date, "1d")
# df with all combinations of daily dates and ids
date_id_df = pl.DataFrame({"date": dates_to_select}).join(
polars_data.select(pl.col("id").unique()), how="cross"
)
# join with original df
polars_data = polars_data.join(date_id_df, on=["date", "id"], how="outer").sort(
"id", "date"
)
# create tmp columns to assemble strings from item_id to fill columns for cells with null values
polars_data = (
polars_data.lazy()
.with_columns(
[
col("id"),
*[
col("id").apply(lambda s, i=i: s.split("_")[i]).alias(col_name)
for i, col_name in enumerate(["1", "2", "3", "4", "5", "6"])
],
]
)
.drop(["item_id", "dept_id", "cat_id", "store_id", "state_id"])
.collect()
)
# concat string components
item_id = polars_data.select(
pl.concat_str(
[
pl.col("1"),
pl.col("2"),
pl.col("3"),
],
separator="_",
).alias("item_id")
)
dept_id = polars_data.select(
pl.concat_str(
[
pl.col("1"),
pl.col("2"),
],
separator="_",
).alias("dept_id")
)
cat_id = polars_data.select(
pl.concat_str(
[
pl.col("1"),
],
separator="_",
).alias("cat_id")
)
store_id = polars_data.select(
pl.concat_str(
[
pl.col("4"),
pl.col("5"),
],
separator="_",
).alias("store_id")
)
state_id = polars_data.select(
pl.concat_str(
[
pl.col("4"),
],
separator="_",
).alias("state_id")
)
# fill sales columns with null values with 0
polars_data = (
polars_data.lazy()
.with_column(
pl.col("sales").fill_null(0),
)
.collect()
)
# recreate other columns with the string components
polars_data = (
pl.concat(
[polars_data, item_id, dept_id, cat_id, store_id, state_id], how="horizontal"
)
.drop(["1", "2", "3", "4", "5", "6"])
.sort("date", "id")
)
print("\nProcessing took {}".format(print_time(time() - start)))
# #### Comparison
polars_data.sort("sales", descending=True).head()
len(polars_data)
pandas_data.reset_index(drop=True).sort_values(by=["sales"], ascending=False).head()
len(pandas_data)
|
# This notebook reveals my solution for __RFM Analysis Task__ offered by Renat Alimbekov.
# This task is part of the __Task Series__ for Data Analysts/Scientists
# __Task Series__ - is a rubric where Alimbekov challenges his followers to solve tasks and share their solutions.
# So here I am :)
# Original solution can be found at - https://alimbekov.com/rfm-python/
# The task is to perform RFM Analysis.
# * __olist_orders_dataset.csv__ and __olist_order_payments_dataset.csv__ should be used
# * order_delivered_carrier_date - should be used in this task
# * Since the dataset is not actual by 2021, thus we should assume that we were asked to perform RFM analysis the day after the last record
# # Importing the modules
import pandas as pd
import numpy as np
import squarify
import matplotlib.pyplot as plt
import seaborn as sns
plt.style.use("ggplot")
import warnings
warnings.filterwarnings("ignore")
# # Loading the data
orders = pd.read_csv("../input/brazilian-ecommerce/olist_orders_dataset.csv")
payments = pd.read_csv("../input/brazilian-ecommerce/olist_order_payments_dataset.csv")
# # Dataframes join
orders["order_delivered_carrier_date"] = pd.to_datetime(
orders["order_delivered_carrier_date"]
) # datetime conversion
payments = payments.set_index("order_id") # preparation before the join
orders = orders.set_index("order_id") # preparation before the join
joined = orders.join(payments) # join on order_id
joined.isna().sum().sort_values(ascending=False)
joined.nunique().sort_values(ascending=False)
# It seems like we have missing values. And unfortunately order_delivered_carrier_date also has missing values. Thus, they should be dropped
last_date = joined["order_delivered_carrier_date"].max() + pd.to_timedelta(1, "D")
RFM = (
joined.dropna(subset=["order_delivered_carrier_date"])
.reset_index()
.groupby("customer_id")
.agg(
Recency=("order_delivered_carrier_date", lambda x: (last_date - x.max()).days),
Frequency=("order_id", "size"),
Monetary=("payment_value", "sum"),
)
)
# Sanity check - do we have NaN values or not?
RFM.isna().sum()
RFM.describe([0.01, 0.05, 0.1, 0.25, 0.5, 0.75, 0.9, 0.95, 0.99]).T
# So, here we can see that we have some outliers in Freqency and Monetary groups. Thus, they should be dropped and be analyzed separately
# # Recency
plt.figure(figsize=(12, 6))
sns.boxplot(x="Recency", data=RFM)
plt.title("Boxplot of Recency")
# # Frequency
RFM["Frequency"].value_counts(normalize=True) * 100
# I guess here we should select only frequency values that are greater than 5, because by doing this we only drop 0.11% of records
RFM["Frequency"].apply(
lambda x: "less or equal to 5" if x <= 5 else "greater than 5"
).value_counts(normalize=True) * 100
RFM = RFM[RFM["Frequency"] <= 5]
# # Monetary
RFM["Monetary"].describe([0.25, 0.5, 0.75, 0.9, 0.95, 0.99])
# Here, it seems like 95% percentile should be used to drop the outliers
plt.figure(figsize=(12, 6))
plt.title("Distribution of Monetary < 95%")
sns.distplot(RFM[RFM["Monetary"] < 447].Monetary)
RFM = RFM[RFM["Monetary"] < 447]
# # RFM groups
# I have used quantiles for assigning scores for Recency and Monetary.
# * groups are 0-33, 33-66, 66-100 quantiles
# For Frequency I have decided to group them by hand
# * score=1 if the frequency value is 1
# * otherwise, the score will be 2
RFM["R_score"] = pd.qcut(RFM["Recency"], 3, labels=[1, 2, 3]).astype(str)
RFM["M_score"] = pd.qcut(RFM["Monetary"], 3, labels=[1, 2, 3]).astype(str)
RFM["F_score"] = RFM["Frequency"].apply(lambda x: "1" if x == 1 else "2")
RFM["RFM_score"] = RFM["R_score"] + RFM["F_score"] + RFM["M_score"]
# 1. CORE - '123' - most recent, frequent, revenue generating - core customers that should be considered as most valuable clients
# 2. GONE - '311', '312', '313' - gone, one-timers - those clients are probably gone;
# 3. ROOKIE - '111', '112', '113' - just have joined - new clients that have joined recently
# 4. WHALES - '323', '213', '223 - most revenue generating - whales that generate revenue
# 5. LOYAL - '221', '222', '321', '322' - loyal users
# 6. REGULAR - '121', '122', '211', '212', - average users - just regular customers that don't stand out
#
def segment(x):
if x == "123":
return "Core"
elif x in ["311", "312", "313"]:
return "Gone"
elif x in ["111", "112", "113"]:
return "Rookies"
elif x in ["323", "213", "223"]:
return "Whales"
elif x in ["221", "222", "321", "322"]:
return "Loyal"
else:
return "Regular"
RFM["segments"] = RFM["RFM_score"].apply(segment)
RFM["segments"].value_counts(normalize=True) * 100
segmentwise = RFM.groupby("segments").agg(
RecencyMean=("Recency", "mean"),
FrequencyMean=("Frequency", "mean"),
MonetaryMean=("Monetary", "mean"),
GroupSize=("Recency", "size"),
)
segmentwise
font = {"family": "normal", "weight": "normal", "size": 18}
plt.rc("font", **font)
fig = plt.gcf()
ax = fig.add_subplot()
fig.set_size_inches(16, 16)
squarify.plot(
sizes=segmentwise["GroupSize"],
label=segmentwise.index,
color=["gold", "teal", "steelblue", "limegreen", "darkorange", "coral"],
alpha=0.8,
)
plt.title("RFM Segments", fontsize=18, fontweight="bold")
plt.axis("off")
plt.show()
# # Cohort Analysis
#
from operator import attrgetter
joined["order_purchase_timestamp"] = pd.to_datetime(joined["order_purchase_timestamp"])
joined["order_months"] = joined["order_purchase_timestamp"].dt.to_period("M")
joined["cohorts"] = joined.groupby("customer_id")["order_months"].transform("min")
cohorts_data = (
joined.reset_index()
.groupby(["cohorts", "order_months"])
.agg(
ClientsCount=("customer_id", "nunique"),
Revenue=("payment_value", "sum"),
Orders=("order_id", "count"),
)
.reset_index()
)
cohorts_data["periods"] = (cohorts_data.order_months - cohorts_data.cohorts).apply(
attrgetter("n")
) # periods for which the client have stayed
cohorts_data.head()
# Since, majority of our clients are not recurring ones, we can't perform proper cohort analysis on retention and other possible metrics.
# Fortunately, we can analyze dynamics of the bussiness and maybe will be even able to identify some relatively good cohorts that might be used as a prototype (e.g. by marketers).
font = {"family": "normal", "weight": "normal", "size": 12}
plt.rc("font", **font)
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(24, 6)) # for 2 parrallel plots
cohorts_data.set_index("cohorts").Revenue.plot(ax=ax1)
ax1.set_title("Cohort-wise revenue")
cohorts_data.set_index("cohorts").ClientsCount.plot(ax=ax2, c="b")
ax2.set_title("Cohort-wise clients counts")
# The figure above reveals the dynamics of Revenue and Number of Clients per cohort.
# On the left side we can see Revenue plot and on the right side can see ClientsCount plot.
# Overall, we can come to the next conclusions:
# * dynamics of two graphs are almost identical. Thus, it seems like the Average Order Amount was the same almost for each cohort. It could mean that the only way to get more revenue is to get more clients. Also, we know that we have 97% of non-recurring clients, thus maybe resolving this issue and stimulating customers to comeback would also result in revenue increase
# * I suspect that we don't have the full data for the last several months, because we can see abnormal drop. Thus, these last months shouldn't be taken into considerations
# * Cohort of November-2017 looks like out of trend, since this cohort showed outstanding results. It can be due to Black Friday sales that often happen at Novembers, or maybe during the November of 2017 some experimental marketing campaigns were performed that lead to good results. Thus, this cohort should be investigated by the company in order to identify the reason behind such an outstanding result, and take it into account
ig, (ax1, ax2) = plt.subplots(1, 2, figsize=(24, 6))
(cohorts_data["Revenue"] / cohorts_data["Orders"]).plot(ax=ax1)
ax1.set_title("Average Order Amount per cohort")
sns.boxplot((cohorts_data["Revenue"] / cohorts_data["Orders"]), ax=ax2)
ax2.set_title("Boxplot of the Average Order Amount")
|
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
from sklearn.preprocessing import LabelEncoder
from sklearn.ensemble import RandomForestRegressor
from sklearn.metrics import r2_score
from sklearn.model_selection import train_test_split, GridSearchCV, cross_val_score
# from bayes_opt import BayesianOptimization
import xgboost as xgb
import warnings
warnings.filterwarnings("ignore")
# ## 1. Data Preprocessing
train = pd.read_csv(
"/kaggle/input/house-prices-advanced-regression-techniques/train.csv"
)
test = pd.read_csv("/kaggle/input/house-prices-advanced-regression-techniques/test.csv")
# combining training set and testing set
df = train.drop(["SalePrice"], axis=1).append(test, ignore_index=True)
# check data types and missing values
df.info()
# 80 predictors in the data frame.
# Data types are string, integer, and float.
# Variables like 'Alley','FireplaceQu','PoolQC','Fence','MiscFeature' contains limited information (too much missing values). But note that some NA values represent "no such equipment" not missing values. (see data description)
# ### 1.1 Missing value analysis
# extract columns contain any null values
missing_values = df.columns[df.isna().any()].tolist()
df[missing_values].info()
# check number of missing values
df[missing_values].isnull().sum()
# Some columns contain limited information (only 198 records in Alley), but note that some of them represent "no such equipment/facility" not missing values (see data description).
# It is true that some predictors there contains a mixing of missing values and "no equipment/facility". "BsmtQual", "BsmtCond" and "BsmtExposure" supposed to be same counts and same number of NAs if no missing values inside.
# To fix this problem I will assume all of the missing values in **some predictors** as "no equipment/facility" if NA is explained in the description file.
# ### 1.2 Missing values imputation (part 1)
df["Alley"] = df["Alley"].fillna("No Access")
df["MasVnrType"] = df["MasVnrType"].fillna("None")
df["BsmtQual"] = df["BsmtQual"].fillna("No Basement")
df["FireplaceQu"] = df["FireplaceQu"].fillna("No Fireplace")
df["GarageType"] = df["GarageType"].fillna("No Garage")
df["PoolQC"] = df["PoolQC"].fillna("No Pool")
df["Fence"] = df["Fence"].fillna("No Fence")
df["MiscFeature"] = df["MiscFeature"].fillna("None")
# the following predictors are linked to the above predictors, should apply a further analysis
# df['BsmtCond'].fillna('No Basement')
# df['BsmtExposure'].fillna('No Basement')
# df['BsmtFinType1'].fillna('No Basement')
# df['BsmtFinType2'].fillna('No Basement')
# df['GarageFinish'].fillna('No Garage')
#
# Now consider other variables (in the comment), there are relationship between variables like 'BsmQual' and 'BsmtCond': only if the value of 'BsmQual' is "no equipment", another should be considered "no equipment" as well.
# check the remaining missing values
missing_values = df.columns[df.isna().any()].tolist()
df[missing_values].isnull().sum()
# clean categorical variables
# fill missing values in 'BsmtCond' column where 'BamtQual' column is 'No Basement'
df.loc[df["BsmtQual"] == "No Basement", "BsmtCond"] = df.loc[
df["BsmtQual"] == "No Basement", "BsmtCond"
].fillna("No Basement")
df.loc[df["BsmtQual"] == "No Basement", "BsmtExposure"] = df.loc[
df["BsmtQual"] == "No Basement", "BsmtExposure"
].fillna("No Basement")
df.loc[df["BsmtQual"] == "No Basement", "BsmtFinType1"] = df.loc[
df["BsmtQual"] == "No Basement", "BsmtFinType1"
].fillna("No Basement")
df.loc[df["BsmtQual"] == "No Basement", "BsmtFinType2"] = df.loc[
df["BsmtQual"] == "No Basement", "BsmtFinType2"
].fillna("No Basement")
df.loc[df["GarageType"] == "No Garage", "GarageFinish"] = df.loc[
df["GarageType"] == "No Garage", "GarageFinish"
].fillna("No Garage")
df.loc[df["GarageType"] == "No Garage", "GarageQual"] = df.loc[
df["GarageType"] == "No Garage", "GarageQual"
].fillna("No Garage")
df.loc[df["GarageType"] == "No Garage", "GarageCond"] = df.loc[
df["GarageType"] == "No Garage", "GarageCond"
].fillna("No Garage")
# clean numerical variables
# fill missing values in 'MasVnrArea' columns where 'MasVnrType' column is 'None'
df.loc[df["MasVnrType"] == "None", "MasVnrArea"] = df.loc[
df["MasVnrType"] == "None", "MasVnrArea"
].fillna(0)
df.loc[df["GarageType"] == "No Garage", "GarageYrBlt"] = df.loc[
df["GarageType"] == "No Garage", "GarageYrBlt"
].fillna(0)
missing_values = df.columns[df.isna().any()].tolist()
df[missing_values].isnull().sum()
# Now most of them only contains one or two missing values.
# Now let's check 'BsmtFinSF1', 'BsmtFinSF2', and 'TotalBsmtSF', since there are only one missing value
df.loc[df["BsmtFinSF1"].isna()].iloc[:, 30:39]
# They are from the same observation index = 2120, since BsmtQual and other variables are labels as "No basement", the Nan values can be labeled as '0'
df.loc[2120] = df.loc[2120].fillna(0)
# check na values in a row
df.loc[2120].isnull().sum()
missing_values = df.columns[df.isna().any()].tolist()
df[missing_values].isnull().sum()
df[missing_values].info()
# ### 1.3 Missing value imputation (part 2)
# Now that most of them only contains 1 or 2 missing values, I will check the distribution of the variables to see what imputation method is more reasonable.
# categorical variables
v1 = [
"MSZoning",
"Utilities",
"Exterior1st",
"Exterior2nd",
"BsmtCond",
"BsmtExposure",
"BsmtFinType2",
"Electrical",
"KitchenQual",
"Functional",
"GarageFinish",
"GarageQual",
"GarageCond",
"SaleType",
]
# quantitative variables
v2 = [x for x in missing_values if x not in v1]
v2.remove("LotFrontage")
for var in v1:
sns.countplot(x=df[var])
plt.show()
for var in v2:
plt.hist(df[var])
plt.title(var)
plt.show()
# Impute categorical variables using **mode**
#
# Impute quantitative variables using **median**
for var in v1:
df[var].fillna(df[var].mode()[0], inplace=True)
for var in v2:
df[var].fillna(df[var].median(), inplace=True)
# Check the missing values one more time, supposed to only have one variable "LotFrontage"
missing_values = df.columns[df.isna().any()].tolist()
df[missing_values].info()
# ### 1.4 Missing value imputation (step 3)
# Missing imputation using Random Forest(RF)
# Encoding
df = df.apply(
lambda x: pd.Series(
LabelEncoder().fit_transform(x[x.notnull()]), index=x[x.notnull()].index
)
)
df.info()
l_known = df[df.LotFrontage.notnull()]
l_unknown = df[df.LotFrontage.isnull()]
l_ytrain = l_known.iloc[:, 3]
l_xtrain = l_known.drop(columns=l_known.columns[3])
l_xtest = l_unknown.drop(columns=l_unknown.columns[3])
rf = RandomForestRegressor()
rf.fit(l_xtrain, l_ytrain)
pred = rf.predict(l_xtest)
df.loc[(df.LotFrontage.isnull()), "LotFrontage"] = pred
df.info()
# ## 2. Modelling
# ### 2.1 train test split
df = df.drop(["Id"], axis=1)
X = df.iloc[0 : train.shape[0], :]
y = train["SalePrice"]
X_test = df.iloc[train.shape[0] :, :]
X_train, X_val, y_train, y_val = train_test_split(
X, y, random_state=123, train_size=0.8
)
# ### 2.2 Hyperparameter tuning
# Benchmark
model = xgb.XGBRegressor()
model.fit(X_train, y_train)
pred = model.predict(X_val)
r2_score(y_val, pred)
params = {
"max_depth": [3, 5, 7],
"gamma": [0.0, 0.3, 0.5, 0.7],
}
model = xgb.XGBRegressor(learning_rate=0.1)
grid_search = GridSearchCV(estimator=model, param_grid=params, cv=5, n_jobs=-1)
grid_search.fit(X_train, y_train)
# Print the best hyperparameters
grid_search.best_params_, grid_search.best_score_
params = {
"reg_alpha": [x / 10 for x in range(1, 11)],
"reg_lambda": [x / 10 for x in range(1, 11)],
}
model = xgb.XGBRegressor(max_depth=3, learning_rate=0.1)
grid_search = GridSearchCV(estimator=model, param_grid=params, cv=5, n_jobs=-1)
grid_search.fit(X_train, y_train)
grid_search.best_params_, grid_search.best_score_
params = {"learning_rate": [x / 10 for x in range(1, 21)]}
model = xgb.XGBRegressor(max_depth=3, reg_alpha=0.8, reg_lambda=0.5)
grid_search = GridSearchCV(estimator=model, param_grid=params, cv=5, n_jobs=-1)
grid_search.fit(X_train, y_train)
grid_search.best_params_, grid_search.best_score_
# ### 2.3 Model fitting & Validating accuracy
model = xgb.XGBRegressor(max_depth=3, learning_rate=0.1, reg_alpha=0.8, reg_lambda=0.5)
model.fit(X_train, y_train)
pred = model.predict(X_val)
r2_score(y_val, pred)
pred = model.predict(X_test)
output = pd.DataFrame({"Id": test["Id"], "SalePrice": pred})
output.head(5)
output.to_csv("submission.csv", index=False)
|
# Note: This notebook was referece for my self-training from https://www.kaggle.com/mathchi/ab-test-for-real-data/ by [Mehmet A.](https://www.kaggle.com/mathchi)
# Since the original dataset is private, I faked one for running it through. Some row of the data was copied data from originally showed. Others was kind of randomly generated.
import os # accessing directory structure
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# # Data Science Analyze and Present A/B Test Results
# AA company recently introduced a new bidding type, “average bidding”, as an alternative to its exisiting bidding
# type, called “maximum bidding”. One of our clients, --------.com, has decided to test this new feature
# and wants to conduct an A/B test to understand if average bidding brings more conversions than maximum
# bidding.
# In this A/B test, --------.com randomly splits its audience into two equally sized groups, e.g. the test
# and the control group. AA company ad campaign with “maximum bidding” is served to “control group” and
# another campaign with “average bidding” is served to the “test group”.
# The A/B test has run for 1 month and --------.com now expects you to analyze and present the results
# of this A/B test.
# ##### You should answer the following questions in your presentation:
# 1. How would you define the hypothesis of this A/B test?
# 2. Can we conclude statistically significant results?
# 3. Which statistical test did you use, and why?
# 4. Based on your answer to Question 2, what would be your recommendation to client?
# ##### Hints:
# 1. Your presentation should last about 15 minutes, and should be presented in English.
# 2. The ultimate success metric for HotelsForYou.com is Number of Purchases. Therefore, you should focus on Purchase metrics for statistical testing.
# 3. Explain the concept of statistical testing for a non-technical audience.
# 4. The customer journey for this campaign is:
# 1. User sees an ad (Impression)
# 2. User clicks on the website link on the ad (Website Click)
# 3. User makes a search on the website (Search)
# 4. User views details of a product (View Content)
# 5. User adds the product to the cart (Add to Cart)
# 6. User purchases the product (Purchase)
# 5. Use visualizations to compare test and control group metrics, such as Website Click Through Rate, Cost per Action, and Conversion Rates in addition to Purchase numbers.
# 6. If you see trends, anomalies or other patterns, discuss these in your presentation.
# 7. You can make assumptions if needed.
# ## 1. Import libraries
import pandas as pd
import numpy as np
import seaborn as sns
import matplotlib.pyplot as plt
from scipy import stats
from scipy.stats import shapiro
from sklearn.neighbors import LocalOutlierFactor
from scipy.stats import levene
from sklearn.impute import KNNImputer
pd.set_option("display.max_columns", None)
pd.set_option("display.max_rows", None)
# ## 2. Functions
def checkReject(p_value, alpha=0.05):
if p_value > alpha:
print(f"p_value is {p_value} > alpha {alpha}, cannot reject null hypoposis")
elif p_value < alpha:
print("p_value is {p_value} < alpha {alpha}, reject null hypoposis")
# ## 3. Data Preparation
control = pd.read_excel(
"/kaggle/input/ab-case-study/AB_Case_Study.xlsx",
sheet_name="Control Group",
index_col=0,
)
test = pd.read_excel(
"/kaggle/input/ab-case-study/AB_Case_Study.xlsx",
sheet_name="Test Group",
index_col=0,
)
control.head()
test.head()
control.info()
# Categorical Columns
categorical = [
col
for col in control.columns
if (control[col].dtype == "object") | (control[col].dtype == "bool")
]
# Numerical Variables
numerical = [col for col in control.columns if col not in categorical]
numerical
kat_control = control[categorical]
num_control = control[numerical]
kat_control.head()
# If we look at the 12th line, we can see the NaN value.
num_control.head(15)
# Fill the control dataframe values NaN value with KNNImputer.
# Fill the control dataframe values NaN value with KNNImputer.
imputer = KNNImputer(n_neighbors=8)
num_control = pd.DataFrame(
imputer.fit_transform(num_control), columns=num_control.columns
)
num_control.iloc[11:12, :]
# Let's combine categorical and numerical variables with the concept.
control = pd.concat([kat_control, num_control], axis=1)
control.head(3)
# data type conversions from float64 to int64.
# Data Type Conversions
control = control.astype(
{
"# of Impressions": "int64",
"Reach": "int64",
"# of Website Clicks": "int64",
"# of Searches": "int64",
"# of View Content": "int64",
"# of Add to Cart": "int64",
"# of Purchase": "int64",
}
)
test = test.astype(
{
"# of Impressions": "int64",
"Reach": "int64",
"# of Website Clicks": "int64",
"# of Searches": "int64",
"# of View Content": "int64",
"# of Add to Cart": "int64",
"# of Purchase": "int64",
}
)
print(control.shape)
print(test.shape)
print(control.info())
print(test.info())
# ## 4. Preparing Data for Hypothesis Testing (A/B Test)
# Let's create a feature with the help of functions and create a feature for visualization and prepare it.
# add columns "Website Click Through Rate" "Number of Action", "Cost per Action"
control["Website Click Through Rate"] = (
control["# of Website Clicks"] / control["# of Impressions"] * 100
)
test["Website Click Through Rate"] = (
test["# of Website Clicks"] / test["# of Impressions"] * 100
)
control["Number of Action"] = (
control["# of Impressions"]
+ control["# of Website Clicks"]
+ control["# of Searches"]
+ control["# of View Content"]
+ control["# of Add to Cart"]
+ control["# of Purchase"]
)
test["Number of Action"] = (
test["# of Impressions"]
+ test["# of Website Clicks"]
+ test["# of Searches"]
+ test["# of View Content"]
+ test["# of Add to Cart"]
+ test["# of Purchase"]
)
control["Cost per Action"] = control["Spend [USD]"] / control["Number of Action"]
test["Cost per Action"] = test["Spend [USD]"] / test["Number of Action"]
control["Conversion Rate"] = (
control["Number of Action"] / control["# of Website Clicks"] * 100
)
test["Conversion Rate"] = test["Number of Action"] / test["# of Website Clicks"] * 100
control_nop = pd.DataFrame(control["# of Purchase"])
test_nop = pd.DataFrame(test["# of Purchase"])
Group_A = control.loc[:, "# of Purchase":"Conversion Rate"].drop(
columns="Number of Action"
)
Group_A["Group"] = "A (MaximumBidding)"
Group_B = test.loc[:, "# of Purchase":"Conversion Rate"].drop(
columns="Number of Action"
)
Group_B["Group"] = "B (AverageBidding)"
AB = pd.concat([Group_A, Group_B])
AB.head(3)
# Let's look at the numbers A and B.
AB["Group"].value_counts()
# ### 4.1. Website Click Through Rate (CTR)
# - It is a rate that shows how often the users who saw the ad CLICK the ad.
# - Number of Ad Clicks / Number of Ad Displays
# - Example: 5 clicks, CTR at 100 impressions = 5%
# - ##### Use visualizations to compare test and control group metrics, such as Website Click Through Rate, Cost per Action, and Conversion Rates in addition to Purchase numbers.
cols = [
"Website Click Through Rate",
"Cost per Action",
"Conversion Rate",
"# of Purchase",
]
fig, axarr = plt.subplots(2, 2, figsize=(18, 9))
for i in [0, 1]:
for j in [0, 1]:
for c in cols:
sns.boxplot(y=c, x="Group", hue="Group", data=AB, ax=axarr[i][j])
cols.remove(c)
break
# Let's create our df for AB test.
df_AB = pd.concat([control_nop, test_nop], axis=1)
df_AB.columns = ["A", "B"]
df_AB.head(3)
# ## 5. Two Independent Sample T Test
# ### 5.1. Assumption Check
# #### 5.1.1. Normality Assumption (**Shapiro Test**)
# * **H0**: Normal distribution assumption is provided.
# * **H1**: ... not provided.
p_value = shapiro(df_AB.A)[1]
checkReject(p_value)
p_value = shapiro(df_AB.B)[1]
checkReject(p_value)
# ### 5.1.2. Variance Homogeneity Assumption (**Levene Test**)
# * **H0** : Variances are homogeneous.
# * **H1** : Variances are not homogeneous.
# **H0** : Varyanslar homojendir.
# **H1** : Varyanslar homojen değildir.
p_value = stats.levene(df_AB.A, df_AB.B)[1]
checkReject(p_value)
# **Comment:** Since the pvalue = 0.36> 0.05, the H0 hypothesis, that is, **the variances were not statistically rejected as homogeneous.**
# ### 5.1.3. Nonparametric Independent Two-Sample T Test (Mann–Whitney U test)
# - Since the normality of the distribution of the data set in which Average Bidding (Test Group) was measured was rejected, the [NonParametric](https://machinelearningmastery.com/nonparametric-statistical-significance-tests-in-python/) Independent Two-Sample T Test was applied for the Hypothesis Test.
# **Hypothesis:**
# * **H0** : When it comes to Maximum Bidding and Average Bidding, there is no statistically significant difference between the purchasing amounts of the two groups. ($\mu_1 = \mu_2$)
# * **H1** : ... there is a difference ($\mu_1 \neq \mu_2$)
p_value = stats.mannwhitneyu(df_AB.A, df_AB.B)[1]
checkReject(p_value)
|
# The main goal of this notebook is provide step by step data analysis, data preprocessing and implement various machine learning tasks. The goal is not just to build a model which gives better results but also to learn various analysis and modeling techniques in the process of building the best model.
# import the required packages
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import seaborn as sns
import matplotlib.pyplot as plt
sns.set(color_codes=True)
import os
# load train and test data
train_data = pd.read_csv("../input/home-data-for-ml-course/train.csv")
test_data = pd.read_csv("../input/home-data-for-ml-course/test.csv")
train_data.head()
print(
"In training data, there are ",
train_data.shape[0],
" records and ",
train_data.shape[1],
" columns",
)
print(
"In test data, there are ",
test_data.shape[0],
" records and ",
test_data.shape[1],
" columns",
)
# Main data analysis steps:
# remove duplicates,outliers
# remove irrelelvant observations
# Major tasks of data preprocessing
# 1. Handling misssing values
# 2. Handling categorical variables
# 3. Handling outliers
train_data.describe()
# Remove unnnecessary fields
# The column id doesnt provide any additional informaion so we can drop this column
train_data = train_data.drop(["Id"], axis=1)
test_data = test_data.drop(["Id"], axis=1)
y = train_data["SalePrice"]
del train_data["SalePrice"]
# Check if there are any duplicate records
sum(train_data.duplicated())
# # **Handling Missing Data**
train_missing_cols = list(train_data.columns[train_data.isnull().any()])
test_missing_cols = list(test_data.columns[test_data.isnull().any()])
print(
"There are ",
len(train_missing_cols),
" columns with missing values in training data",
)
print("There are ", len(test_missing_cols), " columns with missing values in test data")
train_data.info()
test_data.info()
# percentage of missing values in train data
train_mis_cols = pd.Series(train_data.isnull().sum()) * 100 / train_data.shape[0]
train_mis_cols = train_mis_cols[train_mis_cols > 0]
train_mis_cols
# percentage of missing values in test data
test_mis_cols = pd.Series(test_data.isnull().sum()) * 100 / test_data.shape[0]
test_mis_cols = test_mis_cols[test_mis_cols > 0]
test_mis_cols
# from the above results notice that Alley,FireplaceQu,PoolQC,Fence,MiscFeature have more than 50% missing data. So we can ignore this columns for training the model.
# Theoretically if there are more than 30% of missing values then those columns are said to be ignored
train_data = train_data.drop(
["Alley", "FireplaceQu", "PoolQC", "Fence", "MiscFeature"], axis=1
)
test_data = test_data.drop(
["Alley", "FireplaceQu", "PoolQC", "Fence", "MiscFeature"], axis=1
)
# There will be a relation between lot frontage and lot area. Lets get the square root of lot area and check if it has any correlation with the lot frontage
test_data["SqrtLotArea"] = np.sqrt(test_data["LotArea"])
train_data["SqrtLotArea"] = np.sqrt(train_data["LotArea"])
print(test_data["LotFrontage"].corr(test_data["SqrtLotArea"]))
print(train_data["LotFrontage"].corr(train_data["SqrtLotArea"]))
# From the above results we can see that there is a linear relation between square root of lot area and lot frontage. So lets replace missin vlues of lot frontage with lot area.
test_data.LotFrontage[test_data["LotFrontage"].isnull()] = test_data.SqrtLotArea[
test_data["LotFrontage"].isnull()
]
train_data.LotFrontage[train_data["LotFrontage"].isnull()] = train_data.SqrtLotArea[
train_data["LotFrontage"].isnull()
]
del test_data["SqrtLotArea"]
del train_data["SqrtLotArea"]
def get_counts(data, column):
return data[column].value_counts()
def impute_values(data, column, value):
data.loc[data[column].isnull(), column] = value
print(get_counts(test_data, "MasVnrType"), len(test_data))
print(get_counts(train_data, "MasVnrType"), len(train_data))
# As most the values are none lets replace missing values with None
impute_values(test_data, "MasVnrType", "None")
impute_values(train_data, "MasVnrType", "None")
print(get_counts(test_data, "MasVnrArea"), len(test_data))
print(get_counts(train_data, "MasVnrArea"), len(train_data))
impute_values(test_data, "MasVnrArea", 0.0)
impute_values(train_data, "MasVnrArea", 0.0)
print(get_counts(train_data, "Electrical"), len(train_data))
impute_values(test_data, "Electrical", "SBrkr")
impute_values(train_data, "Electrical", "SBrkr")
def weightedAvg(data, col):
tmp = get_counts(data, col)
return sum(tmp.index * tmp.values) / sum(tmp.values)
impute_values(train_data, "GarageYrBlt", round(weightedAvg(train_data, "GarageYrBlt")))
impute_values(test_data, "GarageYrBlt", round(weightedAvg(test_data, "GarageYrBlt")))
get_counts(train_data, "GarageType")
impute_values(train_data, "GarageType", "Attchd")
impute_values(test_data, "GarageType", "Attchd")
train_data = train_data.replace(
{"ExterQual": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
test_data = test_data.replace(
{"ExterQual": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
train_data = train_data.replace(
{"ExterCond": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
test_data = test_data.replace(
{"ExterCond": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
train_data = train_data.replace(
{"BsmtQual": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
test_data = test_data.replace(
{"BsmtQual": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
train_data = train_data.replace(
{"BsmtCond": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
test_data = test_data.replace(
{"BsmtCond": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
train_data = train_data.replace(
{"BsmtExposure": {"Gd": 4, "Av": 3, "Mn": 2, "No": 1, np.NaN: 0}}
)
test_data = test_data.replace(
{"BsmtExposure": {"Gd": 4, "Av": 3, "Mn": 2, "No": 1, np.NaN: 0}}
)
train_data = train_data.replace(
{
"BsmtFinType1": {
"GLQ": 6,
"ALQ": 5,
"BLQ": 4,
"Rec": 3,
"LwQ": 2,
"Unf": 1,
np.NaN: 0,
}
}
)
test_data = test_data.replace(
{
"BsmtFinType1": {
"GLQ": 6,
"ALQ": 5,
"BLQ": 4,
"Rec": 3,
"LwQ": 2,
"Unf": 1,
np.NaN: 0,
}
}
)
train_data = train_data.replace(
{
"BsmtFinType2": {
"GLQ": 6,
"ALQ": 5,
"BLQ": 4,
"Rec": 3,
"LwQ": 2,
"Unf": 1,
np.NaN: 0,
}
}
)
test_data = test_data.replace(
{
"BsmtFinType2": {
"GLQ": 6,
"ALQ": 5,
"BLQ": 4,
"Rec": 3,
"LwQ": 2,
"Unf": 1,
np.NaN: 0,
}
}
)
train_data = train_data.replace(
{"HeatingQC": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
test_data = test_data.replace(
{"HeatingQC": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
train_data = train_data.replace(
{"KitchenQual": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
test_data = test_data.replace(
{"KitchenQual": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
train_data = train_data.replace(
{
"Functional": {
"Typ": 8,
"Min1": 7,
"Min2": 6,
"Mod": 5,
"Maj1": 4,
"Maj2": 3,
"Sev": 2,
"Sal": 1,
np.NaN: 0,
}
}
)
test_data = test_data.replace(
{
"Functional": {
"Typ": 8,
"Min1": 7,
"Min2": 6,
"Mod": 5,
"Maj1": 4,
"Maj2": 3,
"Sev": 2,
"Sal": 1,
np.NaN: 0,
}
}
)
train_data = train_data.replace(
{"FireplaceQu": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
test_data = test_data.replace(
{"FireplaceQu": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
train_data = train_data.replace(
{"GarageFinish": {"Fin": 3, "RFn": 2, "Unf": 1, np.NaN: 0}}
)
test_data = test_data.replace(
{"GarageFinish": {"Fin": 3, "RFn": 2, "Unf": 1, np.NaN: 0}}
)
train_data = train_data.replace(
{"GarageQual": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
test_data = test_data.replace(
{"GarageQual": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
train_data = train_data.replace(
{"GarageCond": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
test_data = test_data.replace(
{"GarageCond": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
train_data = train_data.replace({"PavedDrive": {"Y": 3, "P": 2, "N": 1, np.NaN: 0}})
test_data = test_data.replace({"PavedDrive": {"Y": 3, "P": 2, "N": 1, np.NaN: 0}})
train_data = train_data.replace(
{"PoolQC": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
test_data = test_data.replace(
{"PoolQC": {"Ex": 5, "Gd": 4, "TA": 3, "Fa": 2, "Po": 1, np.NaN: 0}}
)
train_data = train_data.replace(
{"Fence": {"GdPrv": 4, "MnPrv": 3, "GdWo": 2, "MnWw": 1, np.NaN: 0}}
)
test_data = test_data.replace(
{"Fence": {"GdPrv": 4, "MnPrv": 3, "GdWo": 2, "MnWw": 1, np.NaN: 0}}
)
print(pd.crosstab(test_data.MSSubClass, test_data.MSZoning))
print(test_data[test_data["MSZoning"].isnull() == True])
test_data.loc[
(test_data["MSZoning"].isnull()) & (test_data["MSSubClass"] == 20), "MSZoning"
] = "RL"
test_data.loc[
(test_data["MSZoning"].isnull()) & (test_data["MSSubClass"] == 30), "MSZoning"
] = "RM"
test_data.loc[
(test_data["MSZoning"].isnull()) & (test_data["MSSubClass"] == 70), "MSZoning"
] = "RM"
print(get_counts(test_data, "Utilities"), len(test_data))
impute_values(test_data, "Utilities", "AllPub")
print(pd.crosstab(test_data.Exterior1st, test_data.Exterior2nd))
impute_values(test_data, "Exterior1st", "VinylSd")
impute_values(test_data, "Exterior2nd", "VinylSd")
print(pd.crosstab(test_data.BsmtFinSF1, test_data.BsmtFinSF2))
impute_values(test_data, "BsmtFinSF1", 0.0)
impute_values(test_data, "BsmtFinSF2", 0.0)
print(get_counts(test_data, "BsmtUnfSF"), len(test_data))
impute_values(test_data, "BsmtUnfSF", round(weightedAvg(test_data, "BsmtUnfSF")))
test_data = test_data.drop(["TotalBsmtSF"], axis=1)
train_data = train_data.drop(["TotalBsmtSF"], axis=1)
print(get_counts(test_data, "BsmtFullBath"), len(test_data))
impute_values(test_data, "BsmtFullBath", round(weightedAvg(test_data, "BsmtFullBath")))
print(get_counts(test_data, "BsmtHalfBath"), len(test_data))
impute_values(test_data, "BsmtHalfBath", 0.0)
print(get_counts(test_data, "KitchenQual"), len(test_data))
impute_values(test_data, "KitchenQual", "TA")
print(get_counts(test_data, "Functional"), len(test_data))
impute_values(test_data, "Functional", "Typ")
print(get_counts(test_data, "GarageCars"), len(test_data))
impute_values(test_data, "GarageCars", 2.0)
print(get_counts(test_data, "GarageArea"), len(test_data))
impute_values(test_data, "GarageArea", round(weightedAvg(test_data, "GarageArea")))
print(get_counts(test_data, "SaleType"), len(test_data))
impute_values(test_data, "SaleType", "WD")
s = pd.Series(train_data.isnull().sum()) * 100 / train_data.shape[0]
s = s[s > 0]
print(s)
s = pd.Series(test_data.isnull().sum()) * 100 / test_data.shape[0]
s = s[s > 0]
print(s)
# There are no more missing values.
# check correlation between all columns and ignore few columns if they are higly correlated with other columns.
corr_table = train_data.corr(method="pearson")
corr_table[(corr_table > 0.7) | (corr_table < -0.7)]
# from the above results notice that external quality and over quality are highly correlated.
# from the above results notice that year built and garage year built are highly correlated.
# so lets drop garage year built and external quality
del train_data["ExterQual"]
del train_data["GarageYrBlt"]
del test_data["ExterQual"]
del test_data["GarageYrBlt"]
train_data.head()
# # **Handle categorical data**
tmp = train_data.columns.to_series().groupby(train_data.dtypes).groups
print({k.name: v for k, v in tmp.items()})
# feature list for one-hot encoding
oneHotCol = [
"MSSubClass",
"MSZoning",
"Street",
"LotShape",
"LandContour",
"Utilities",
"LotConfig",
"LandSlope",
"Neighborhood",
"Condition1",
"Condition2",
"BldgType",
"HouseStyle",
"RoofStyle",
"RoofMatl",
"Exterior1st",
"Exterior2nd",
"MasVnrType",
"Foundation",
"Heating",
"CentralAir",
"Electrical",
"Functional",
"GarageType",
"SaleType",
"SaleCondition",
]
# process each column in the list
for cols in oneHotCol:
train_data = pd.concat(
(train_data, pd.get_dummies(train_data[cols], prefix=cols)), axis=1
)
del train_data[cols]
test_data = pd.concat(
(test_data, pd.get_dummies(test_data[cols], prefix=cols)), axis=1
)
del test_data[cols]
# # **Normalize data**
# min-max scaling
train_data = (train_data - train_data.min()) / (train_data.max() - train_data.min())
test_data = (test_data - test_data.min()) / (test_data.max() - test_data.min())
print(train_data.shape)
print(test_data.shape)
# Notice that there are different number of columns in train and test data. Lets just consider the columns which are present in both
for col in train_data.columns:
if col not in test_data.columns:
del train_data[col]
for col in test_data.columns:
if col not in train_data.columns:
del test_data[col]
X = train_data
X_test = test_data
print(get_counts(test_data, "Utilities_AllPub"), len(test_data))
impute_values(test_data, "Utilities_AllPub", 1)
# # **Simple Linear Regression Model**
from sklearn import linear_model
lm = linear_model.LinearRegression()
lm.fit(X, y)
results = lm.predict(X_test)
np.mean((np.log(y.values) - np.log(lm.predict(X))) ** 2)
# # **Ridge regression**
from sklearn import linear_model
ridge = linear_model.Ridge(alpha=1.0)
ridge.fit(X, y)
results = ridge.predict(X_test)
np.mean((np.log(y.values) - np.log(ridge.predict(X))) ** 2)
# # Lasso Regression
las = linear_model.Lasso(alpha=1.0)
las.fit(X, y)
results = las.predict(X_test)
np.mean((np.log(y.values) - np.log(las.predict(X))) ** 2)
las_weight = las.coef_
las_weight = pd.DataFrame({"feature": X.columns, "weight": las_weight})
print(las_weight[las_weight["weight"] == 0])
# delete all the above columns
X_new = X.drop(
[
"MSSubClass_40",
"MSSubClass_90",
"MSZoning_RL",
"LotConfig_Inside",
"LandSlope_Mod",
"Neighborhood_Sawyer",
"Condition1_RRNn",
"BldgType_1Fam",
"RoofStyle_Hip",
"RoofStyle_Shed",
"MasVnrType_Stone",
"CentralAir_Y",
"Electrical_SBrkr",
"Functional_7",
"SaleType_Oth",
],
axis=1,
)
# delete all the above columns
X_test_new = X_test.drop(
[
"MSSubClass_40",
"MSSubClass_90",
"MSZoning_RL",
"LotConfig_Inside",
"LandSlope_Mod",
"Neighborhood_Sawyer",
"Condition1_RRNn",
"BldgType_1Fam",
"RoofStyle_Hip",
"RoofStyle_Shed",
"MasVnrType_Stone",
"CentralAir_Y",
"Electrical_SBrkr",
"Functional_7",
"SaleType_Oth",
],
axis=1,
)
lm = linear_model.LinearRegression()
lm.fit(X_new, y)
results = lm.predict(X_test_new)
np.mean((np.log(y.values) - np.log(lm.predict(X_new))) ** 2)
ridge = linear_model.Ridge(alpha=1.0)
ridge.fit(X_new, y)
results = ridge.predict(X_test_new)
np.mean((np.log(y.values) - np.log(ridge.predict(X_new))) ** 2)
las = linear_model.Lasso(alpha=2.0)
las.fit(X_new, y)
results = las.predict(X_test_new)
np.mean((np.log(y.values) - np.log(las.predict(X_new))) ** 2)
# # **Gridsearchcv for selecting parameters for ridge regression**
from sklearn.model_selection import GridSearchCV
parameters = {
"alpha": [0.001, 0.01, 0.1, 1, 10, 15, 20, 30, 50, 100, 200, 300, 500, 1000]
}
model = linear_model.Ridge()
Ridge_reg = GridSearchCV(model, parameters, scoring="neg_mean_squared_error", cv=5)
Ridge_reg.fit(X_new, y)
print(Ridge_reg.best_estimator_)
best_model = Ridge_reg.best_estimator_
best_model.fit(X_new, y)
results = best_model.predict(X_test_new)
np.mean((np.log(y.values) - np.log(best_model.predict(X_new))) ** 2)
sample_data = pd.read_csv("../input/home-data-for-ml-course/sample_submission.csv")
sample_data["SalePrice"] = results
sample_data.to_csv("submission.csv", index=False)
sample_data
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input/"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Load data
train_dataset_path = "/kaggle/input/jog-description-and-salary-in-indonesia/train.csv"
test_dataset_path = "/kaggle/input/jog-description-and-salary-in-indonesia/test.csv"
train_dataset_raw = pd.read_csv(train_dataset_path, sep="|")
test_dataset_raw = pd.read_csv(test_dataset_path, sep="|")
all_dataset_path = "/kaggle/input/jog-description-and-salary-in-indonesia/all.csv"
all_dataset_raw = pd.read_csv(all_dataset_path, sep="|")
all_dataset_raw.shape
all_dataset_raw.head()
# Return info on dataset
all_dataset_raw.info()
# Display number missing values per column
all_dataset_raw.isna().sum()
target_column = "salary"
train_data = train_dataset_raw.copy()
test_data = test_dataset_raw.copy()
all_data = all_dataset_raw.copy()
all_data
all_data.loc[:, ["salary_currency"]].groupby(by="salary_currency").size()
all_dataset_raw.loc[all_dataset_raw["salary"].isna(), :].index
# Outlier remover
all_data_low = all_data[target_column].quantile(0.01)
all_data_high = all_data[target_column].quantile(0.99)
all_data[target_column] = all_data[target_column][
(all_data_low < all_data[target_column]) & (all_data_high > all_data[target_column])
]
all_data[target_column]
# Null remover
all_data = all_dataset_raw.drop(
index=all_dataset_raw.loc[all_dataset_raw["salary"].isna(), :].index
)
all_data = all_data.reset_index().drop(columns=["id", "index"])
# Check the count for each category in the "gender" columnall_data
# Check the count for each category in the "career_level" column
all_data["career_level"].value_counts()
# Create a countplot to visualize the count of each category in the gender column.
sns.countplot(data=all_data, x="career_level")
plt.title("Number of Workers per level")
plt.ylabel("Number of Workers")
plt.show()
# Check the mean annual mileage for the different driving experience groups
all_data.groupby(by="career_level")["salary"].mean()
|
a = 2
print(a)
type(a)
b = 3.4
print(b)
type(b)
c = "abc"
print(c)
type(c)
# **Variable with number**
# **interger , floating , complex numbar**
d = 3 + 4j
print(d)
type(d)
# **Working with numerical variable**
Gross_profit = 30
Revenue = 100
Gross_profit_margin = (Gross_profit / Revenue) * 100
print(Gross_profit_margin)
type(Gross_profit_margin)
RevA = 8
Revenue = Revenue + RevA
Revenue
# **Variable with String**
address = "sdjhdghdff 1/fvfdjgssff fsdh"
print(address)
type(address)
# **Variable with boolean**
a = 5
b = 10
a < b
# **multipal assignment variable**
a, b, c = "sdgfgh", 23, 23.4
print(a)
print(b)
print(c)
|
# # Setup
import os
import gc
import time
import warnings
gc.enable()
warnings.filterwarnings("ignore")
import numpy as np
import pandas as pd
pd.set_option("display.max_columns", None)
pd.set_option("display.precision", 4)
import matplotlib.pyplot as plt
import seaborn as sns
SEED = 23
os.environ["PYTHONHASHSEED"] = str(SEED)
np.random.seed(SEED)
from sklearn.model_selection import KFold, StratifiedKFold
from sklearn.ensemble import ExtraTreesRegressor, ExtraTreesClassifier
from sklearn.metrics import mean_squared_log_error, roc_auc_score
DATA_DIR = "/kaggle/input/playground-series-s3e11"
train = pd.read_csv(f"{DATA_DIR}/train.csv")
test = pd.read_csv(f"{DATA_DIR}/test.csv")
sample_sub = pd.read_csv(f"{DATA_DIR}/sample_submission.csv")
original = pd.read_csv("/kaggle/input/media-campaign-cost-prediction/train_dataset.csv")
# # Overview
train.info()
train.sample()
original.sample()
original.shape, train.shape, test.shape, sample_sub.shape
original.isna().sum().sum(), train.isna().sum().sum(), test.isna().sum().sum()
train.drop("id", axis=1, inplace=True)
test.drop("id", axis=1, inplace=True)
TARGET = "cost"
features = list(test.columns)
# # Feature and Target distributions
plt.figure(figsize=(10, 4))
plt.suptitle("Train vs Original target distribution", fontsize=14)
sns.kdeplot(x=TARGET, data=train, cut=0, label="train")
sns.kdeplot(x=TARGET, data=original, cut=0, label="original")
plt.legend(fontsize="x-small")
plt.show()
train.nunique()
continuous_features = [
"store_sales(in millions)",
"gross_weight",
"units_per_case",
"store_sqft",
]
discrete_features = [
"unit_sales(in millions)",
"total_children",
"num_children_at_home",
"avg_cars_at home(approx).1",
]
categorical_features = [
"recyclable_package",
"low_fat",
"coffee_bar",
"video_store",
"salad_bar",
"prepared_food",
"florist",
]
fig, ax = plt.subplots(2, 2, figsize=(9, 6))
plt.suptitle(
"Train vs Test vs Original distribution - CONTINUOUS FEATURES", fontsize=14
)
for i, f in enumerate(continuous_features):
row, col = i // 2, i % 2
ax[row][col].set_title(f"{f}")
sns.kdeplot(x=f, data=train, cut=0, ax=ax[row][col], label="train")
sns.kdeplot(x=f, data=test, cut=0, ax=ax[row][col], label="test")
sns.kdeplot(x=f, data=original, cut=0, ax=ax[row][col], label="original")
ax[row][col].legend(fontsize="small")
plt.tight_layout()
plt.show()
print("Train vs Test vs Original distribution - DISCRETE NUMERICAL FEATURES\n")
for f in discrete_features:
counts_df = pd.DataFrame()
counts_df["original"] = np.round(
original[f].value_counts(normalize=True).sort_index(), 4
)
counts_df["train"] = np.round(train[f].value_counts(normalize=True).sort_index(), 4)
counts_df["test"] = np.round(test[f].value_counts(normalize=True).sort_index(), 4)
counts_df.index = sorted(train[f].unique())
print(f"{f} - unique values: {sorted(original[f].unique())}\n")
display(counts_df)
print()
print("Train vs Test vs Original distribution - CATEGORICAL FEATURES\n")
for f in categorical_features:
counts_df = pd.DataFrame()
counts_df["original"] = np.round(
original[f].value_counts(normalize=True).sort_index(), 4
)
counts_df["train"] = np.round(train[f].value_counts(normalize=True).sort_index(), 4)
counts_df["test"] = np.round(test[f].value_counts(normalize=True).sort_index(), 4)
counts_df.index = sorted(train[f].unique())
print(f"{f} - unique values: {sorted(original[f].unique())}\n")
display(counts_df)
print()
# ### Fixing datatypes
train[categorical_features] = train[categorical_features].astype("int8")
test[categorical_features] = test[categorical_features].astype("int8")
original[categorical_features] = original[categorical_features].astype("int8")
train[discrete_features] = train[discrete_features].astype("int8")
test[discrete_features] = test[discrete_features].astype("int8")
original[discrete_features] = original[discrete_features].astype("int8")
train[["store_sqft", "units_per_case"]] = train[
["store_sqft", "units_per_case"]
].astype("int")
test[["store_sqft", "units_per_case"]] = test[["store_sqft", "units_per_case"]].astype(
"int"
)
original[["store_sqft", "units_per_case"]] = original[
["store_sqft", "units_per_case"]
].astype("int")
# # Adverserial Validation
def adversarial_validation(first_dataset, second_dataset, model, features):
scores = []
oof_preds = {}
first_dataset["set"] = 0
second_dataset["set"] = 1
composite = pd.concat([first_dataset, second_dataset], axis=0, ignore_index=True)
X, y = composite[features], composite["set"]
cv = StratifiedKFold(n_splits=5, shuffle=True, random_state=SEED)
for fold, (train_idx, val_idx) in enumerate(cv.split(X, y)):
X_train, y_train = X.loc[train_idx], y.iloc[train_idx]
X_val, y_val = X.loc[val_idx], y.iloc[val_idx]
model.fit(X_train, y_train)
val_preds = model.predict_proba(X_val)[:, 1]
oof_preds.update(dict(zip(val_idx, val_preds)))
auc = roc_auc_score(y_val, val_preds)
scores.append(auc)
print(f"Fold #{fold}: AUC = {auc:.5f}")
_ = gc.collect()
oof_preds = pd.Series(oof_preds).sort_index()
print(f"OOF AUC = {roc_auc_score(y, oof_preds):.5f}\n")
model = ExtraTreesClassifier(
n_estimators=150, max_depth=7, n_jobs=-1, random_state=SEED
)
print("Original vs Train")
adversarial_validation(original, train, model, features)
print("Original vs Test")
adversarial_validation(original, test, model, features)
print("Train vs Test")
adversarial_validation(train, test, model, features)
print("Original + Train vs Test")
composite = pd.concat([original, train], axis=0, ignore_index=True)
adversarial_validation(composite, test, model, features)
gc.collect()
# # Baseline
def eval_metric(y_true, y_pred):
return mean_squared_log_error(y_true, y_pred, squared=False)
def cross_val_predict(model, features, num_folds=5, seed=SEED, extended=False):
scores = []
test_preds = {}
oof_preds = {}
X, y = train[features], train[TARGET]
X_test = test[features]
cv = KFold(n_splits=num_folds, shuffle=True, random_state=seed)
for fold, (train_idx, val_idx) in enumerate(cv.split(X, y)):
X_train, y_train = X.loc[train_idx], y.iloc[train_idx]
X_val, y_val = X.loc[val_idx], y.iloc[val_idx]
if extended: # original data added only to training folds
X_train = pd.concat(
[X_train, original[features]], axis=0, ignore_index=True
)
y_train = pd.concat([y_train, original[TARGET]], axis=0, ignore_index=True)
model.fit(X_train, y_train)
val_preds = model.predict(X_val)
oof_preds.update(dict(zip(val_idx, val_preds)))
test_preds[f"fold{fold}"] = model.predict(X_test)
score = eval_metric(y_val, val_preds)
scores.append(score)
print(f"Fold #{fold}: {score:.5f}", end=" | ")
_ = gc.collect()
test_preds = pd.DataFrame.from_dict(test_preds)
test_preds["mean"] = test_preds.mean(axis=1) # mean of fold-wise predictions
oof_preds = pd.Series(oof_preds).sort_index()
print(f"OOF score: {eval_metric(y, oof_preds):.5f}\n")
return oof_preds, test_preds
model = ExtraTreesRegressor(
n_estimators=200, max_depth=12, n_jobs=-1, random_state=SEED
)
# **Training data only:**
_, test_preds_trn = cross_val_predict(model, features)
# **Training data extended using original dataset:**
_, test_preds_ext = cross_val_predict(model, features, extended=True)
# # Submission files
def create_submission_files(test_preds, model_name, notebook="00"):
for col in test_preds.columns:
sub = sample_sub.copy()
sub[TARGET] = test_preds[col]
sub.to_csv(f"{notebook}_{model_name}_{col}.csv", index=False)
create_submission_files(test_preds_trn, "baseline_trn")
create_submission_files(test_preds_ext, "baseline_ext")
|
# # Electricity DayAhead Prices 2022
# This dataset provides hourly day ahead electricity prices for France and interconnections, sourced from the ENTSO-E Transparency Platform, which is a reputable market data provider for European electricity markets. It is valuable resource for businesses, investors, researchers, and energy consumers interested in analyzing and understanding the dynamics of the electricity market with a high level of granularity.
# The dataset includes historical and forecasted day ahead (DAH) electricity prices for France and interconnections (Germany, Italy, Spain, UK, Belgium). The data is presented at an hourly granularity, covering a specific timeframe, and includes information such as hourly electricity prices in Euros per MWh (Megawatt-hour), date, time.
# The dataset is presented in a structured format, such as CSV or Excel, making it easy to manipulate and analyze using various data analysis tools and techniques. It is ideal for conducting research, building predictive models, or gaining insights into the day ahead electricity prices for France and interconnections at an hourly level.
# **Use Case Examples**
# - Developing hourly price forecasting models for France using machine learning algorithms (Deep Learning, Regressions, Random Forests)
# - Analyzing the impact of cross-border electricity trading on hourly prices (EDA)
# - Studying historical hourly trends and patterns in electricity prices for France (Time Series Analysis, LSTM)
# - Building energy pricing models for business planning and strategy with hourly granularity
# - Conducting research on hourly energy market dynamics and trends
# **Dataset Features**
# - Hourly day ahead electricity prices for France
# - Interconnections prices (Italy, Belgium, Germany, Spain, UK)
# - Hourly granularity with date and time information (datetime format)
# - Structured format (e.g., CSV or Excel) for easy data manipulation
import pandas as pd
import matplotlib.pyplot as plt
# # 1. Load the Data
data = pd.read_csv(
"/kaggle/input/electricity-dayahead-prices-entsoe/electricity_dah_prices.csv",
index_col="date", # set date day as index column
parse_dates=True, # use as datetime format
)
# # 2. Quick EDA
data.columns
data.head(10)
data.info()
countries = ["france", "italy", "belgium", "spain", "uk", "germany"]
for country in countries:
data.groupby("date")[country].mean().plot(legend=country)
plt.show()
data[countries].corr()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # 1 Data
# # 1.1 Introduction
# The data preprocessing part is inspired by [SUPER ONE](https://www.kaggle.com/ailobster)'s notebook [Python Titanic Predictions Top 3%](https://www.kaggle.com/code/ailobster/python-titanic-predictions-top-3/notebook), which provides a comprehensive analysis, visualization, and explanation of feature engineering. It is strongly recommended to read this excellent resource.
# In my notebook, I will simply keep the data preprocessing part as simple as possible and focus more on the model.
# ## 1.2 Initialization
import pandas as pd
import numpy as np
import re
from sklearn.ensemble import RandomForestRegressor
# Load datasets
raw_train = pd.read_csv("/kaggle/input/titanic/train.csv")
raw_test = pd.read_csv("/kaggle/input/titanic/test.csv")
# Merge train and test data
all_data = raw_train.merge(raw_test, how="outer")
# ## 1.3 Feature processing
# Generate Ticket_num
all_data["Ticket_num"] = all_data["Ticket"].map(lambda x: re.sub("\D", "", x))
all_data["Ticket_num"] = pd.to_numeric(all_data["Ticket_num"])
# Fill Embarked na with the mode
all_data["Embarked"] = all_data["Embarked"].fillna("S")
# Random forest regressor to fill na of Age
age_df = all_data[["Age", "Pclass", "Sex", "Parch", "SibSp"]]
age_df = pd.get_dummies(age_df)
known_age = age_df[age_df.Age.notnull()].values
unknown_age = age_df[age_df.Age.isnull()].values
y = known_age[:, 0]
X = known_age[:, 1:]
rfr = RandomForestRegressor(random_state=0, n_estimators=100, n_jobs=-1)
rfr.fit(X, y)
predictedAges = rfr.predict(unknown_age[:, 1::])
all_data.loc[(all_data.Age.isnull()), "Age"] = predictedAges
# Fill Fare
fare = all_data.loc[
(all_data["Embarked"] == "S") & (all_data["Pclass"] == 3), "Fare"
].median()
all_data["Fare"] = all_data["Fare"].fillna(fare)
# ## 1.4 Feature engineering
# Extract the titles from Name
titles = set()
for name in all_data["Name"]:
titles.add(name.split(",")[1].split(".")[0].strip())
Title_Dictionary = {
"Capt": "Officer",
"Col": "Officer",
"Major": "Officer",
"Jonkheer": "Master",
"Don": "Royalty",
"Sir": "Royalty",
"Dr": "Officer",
"Rev": "Officer",
"the Countess": "Royalty",
"Mme": "Mrs",
"Mlle": "Miss",
"Ms": "Mrs",
"Mr": "Mr",
"Mrs": "Mrs",
"Miss": "Miss",
"Master": "Master",
"Lady": "Royalty",
"Dona": "Royalty",
}
# Surname frequency
all_data["Surname"] = all_data["Name"].map(lambda name: name.split(",")[0].strip())
all_data["FamilyGroup"] = all_data["Surname"].map(all_data["Surname"].value_counts())
Female_Child_Group = all_data.loc[
(all_data["FamilyGroup"] >= 2)
& ((all_data["Age"] <= 16) | (all_data["Sex"] == "female"))
]
Female_Child_Group = Female_Child_Group.groupby("Surname")["Survived"].mean()
Dead_List = set(Female_Child_Group[Female_Child_Group.apply(lambda x: x == 0)].index)
Male_Adult_Group = all_data.loc[
(all_data["FamilyGroup"] >= 2)
& (all_data["Age"] > 16)
& (all_data["Sex"] == "male")
]
Male_Adult_List = Male_Adult_Group.groupby("Surname")["Survived"].mean()
Survived_List = set(Male_Adult_List[Male_Adult_List.apply(lambda x: x == 1)].index)
all_data.loc[
(all_data["Survived"].isnull())
& (all_data["Surname"].apply(lambda x: x in Dead_List)),
["Sex", "Age", "Title"],
] = ["male", 28.0, "Mr"]
all_data.loc[
(all_data["Survived"].isnull())
& (all_data["Surname"].apply(lambda x: x in Survived_List)),
["Sex", "Age", "Title"],
] = ["female", 5.0, "Miss"]
# FamilySize
all_data["FamilySize"] = all_data["SibSp"] + all_data["Parch"] + 1
# FamilyLabel
def Fam_label(s):
if (s >= 2) & (s <= 4):
return 2
elif ((s > 4) & (s <= 7)) | (s == 1):
return 1
elif s > 7:
return 0
all_data["FamilyLabel"] = all_data["FamilySize"].apply(Fam_label)
# Deck
all_data["Cabin"] = all_data["Cabin"].fillna("Unknown")
all_data["Deck"] = all_data["Cabin"].str.get(0)
# TicketGroup
Ticket_Count = dict(all_data["Ticket"].value_counts())
all_data["TicketGroup"] = all_data["Ticket"].map(Ticket_Count)
def Ticket_Label(s):
if (s >= 2) & (s <= 4):
return 2
elif ((s > 4) & (s <= 8)) | (s == 1):
return 1
elif s > 8:
return 0
all_data["TicketGroup"] = all_data["TicketGroup"].apply(Ticket_Label)
# ## 1.5 Processed Data
# Select columns
all_data = all_data[
[
"Survived",
"Pclass",
"Sex",
"Age",
"Fare",
"Embarked",
"Title",
"FamilyLabel",
"Deck",
"TicketGroup",
]
]
# Split into train and test sets
all_data = all_data[
[
"Survived",
"Pclass",
"Sex",
"Age",
"Fare",
"Embarked",
"Title",
"FamilyLabel",
"Deck",
"TicketGroup",
]
]
all_data = pd.get_dummies(all_data)
processed_train = all_data[all_data["Survived"].notnull()]
processed_test = all_data[all_data["Survived"].isnull()].drop("Survived", axis=1)
# # 2 Model
# ## 2.1 Introduction
# We are going to use [LibFM](http://libfm.org/) as the our model. [PyFM](https://github.com/coreylynch/pyFM) is a Python implementation of LibFM.
# - Factorization Machines, a general approach that mimics most factorization models through feature engineering, combine the generality of feature engineering with the effectiveness of factorization models in estimating interactions between categorical variables of large domains.
# - The implementation uses stochastic gradient descent with adaptive regularization as a learning method. The adaptive regularization adjusts the regularization automatically while training the model parameters.
# ## 2.2 Initialization
# Install PyFM
import pandas as pd
import numpy as np
from scipy.sparse import csr_matrix
from pyfm import pylibfm
from sklearn.preprocessing import MinMaxScaler
from sklearn.feature_selection import SelectKBest
# Get X, y based on the data
X = processed_train.drop(["Survived"], axis=1)
y = processed_train["Survived"]
# Scale the data
scaler = MinMaxScaler()
X = scaler.fit_transform(X)
# Select important features
selector = SelectKBest(k=20)
X = selector.fit_transform(X, y)
# Compress sparse matrix: necessary step for FM
X_array = csr_matrix(X)
np.random.seed(524)
# Fit the FM model
fm_model = pylibfm.FM(
num_factors=20, # After fine-tuning, initial guesses: [5, 10, 20, 50, 100]
num_iter=200, # After fine-tuning, initial guesses: [10, 50, 100, 150, 200, 250]
verbose=True,
task="classification",
initial_learning_rate=0.001, # After fine-tuning, intial guesses: [0.1, 0.01, 0.001]
learning_rate_schedule="optimal",
validation_size=0.1,
k0=True,
k1=True,
init_stdev=0.01,
seed=524,
)
fm_model.fit(X_array, y)
# It is worth mentioning that the threshold for the last step (x > 0.5) is also fine-tuned (just try different thresholds (0.4 ~ 0.7) and submit each result).
# Submission
X_sub = scaler.transform(processed_test)
X_sub = selector.transform(X_sub)
X_sub_array = csr_matrix(X_sub)
y_sub_soft = fm_model.predict(X_sub_array)
y_sub_hard = [int(1) if x > 0.5 else int(0) for x in y_sub_soft] # Threshold
final_data = raw_test.copy()
final_data["Survived"] = y_sub_hard
submission = final_data[["PassengerId", "Survived"]]
submission_data = pd.DataFrame(submission)
submission_data = submission_data.reset_index(drop=True)
submission_data.to_csv("submission.csv", index=False)
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.linear_model import Perceptron
from sklearn.metrics import accuracy_score
from sklearn.tree import DecisionTreeClassifier
from sklearn.svm import SVC
from sklearn.linear_model import LinearRegression
from sklearn.linear_model import LogisticRegression
from keras.models import Sequential
from keras.layers import Dense
from sklearn.neural_network import MLPClassifier
dataset = pd.DataFrame(
{
"Age": [79, 69, 73, 75, 81, 77, 80, 85],
"Weight": [310, 240, 258, 300, 200, 217, 241, 264],
}
)
target = np.array([1, 1, 1, 1, -1, -1, -1, -1])
dataset
PLA = Perceptron()
PLA.fit(dataset, target)
y_pred = PLA.predict(dataset)
accuracy = (y_pred == target).mean()
print("Accuracy:", accuracy)
plt.scatter(dataset["Age"], dataset["Weight"], c=target)
plt.plot([70, 90], [300, 200], "-r")
plt.xlabel("Age")
plt.ylabel("Weight")
plt.show()
Tree = DecisionTreeClassifier(max_depth=1)
Tree.fit(dataset, target)
y_pred = Tree.predict(dataset)
Accuracy = accuracy_score(target, y_pred)
print("Accuracy:", Accuracy)
plt.scatter(dataset["Age"], dataset["Weight"], c=target)
xx, yy = np.meshgrid(range(70, 90), range(200, 300))
Z = Tree.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
plt.contour(xx, yy, Z, colors="red", alpha=0.5)
plt.xlabel("Age")
plt.ylabel("Weight")
plt.show()
svm = SVC(kernel="linear")
svm.fit(dataset, target)
y_pred = svm.predict(dataset)
Accuracy = accuracy_score(target, y_pred)
print("Accuracy:", Accuracy)
plt.scatter(dataset["Age"], dataset["Weight"], c=target)
xx, yy = np.meshgrid(range(70, 90), range(200, 300))
Z = svm.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
plt.contour(xx, yy, Z, colors="red", alpha=0.5)
plt.xlabel("Age")
plt.ylabel("Weight")
plt.show()
regression = LinearRegression().fit(dataset, target)
y_pred = np.where(regression.predict(dataset) > 0, 1, -1)
Accuracy = np.mean(y_pred == target)
print("Accuracy:", Accuracy)
plt.scatter(dataset["Age"], dataset["Weight"], c=target)
plt.plot(dataset["Age"], regression.predict(dataset), color="red")
plt.xlabel("Age")
plt.ylabel("Weight")
plt.show()
logisticregression = LogisticRegression()
logisticregression.fit(dataset, target)
y_pred = logisticregression.predict(dataset)
plt.scatter(dataset["Age"], dataset["Weight"], c=target)
xx, yy = np.meshgrid(np.linspace(69, 90, num=100), np.linspace(200, 300, num=100))
Z = logisticregression.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
plt.contour(xx, yy, Z, colors="red", alpha=0.1)
plt.xlabel("Age")
plt.ylabel("Weight")
plt.show()
model = Sequential()
model.add(Dense(1, input_dim=2, activation="sigmoid"))
model.compile(loss="binary_crossentropy", optimizer="adam", metrics=["accuracy"])
model.fit(dataset, target, epochs=1000, batch_size=8, verbose=0)
loss, accuracy = model.evaluate(dataset, target, verbose=0)
print("Accuracy:", accuracy)
clf = MLPClassifier(
hidden_layer_sizes=(10,),
activation="logistic",
solver="sgd",
max_iter=1000,
random_state=0,
)
clf.fit(dataset, target)
plt.scatter(dataset["Age"], dataset["Weight"], c=target)
xx, yy = np.meshgrid(np.linspace(70, 86, 300), np.linspace(200, 311, 300))
Z = clf.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
plt.contour(xx, yy, Z, colors="red", alpha=0.5)
plt.xlabel("Age")
plt.ylabel("Weight")
plt.show()
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
train_csv = pd.read_csv("/kaggle/input/playground-series-s3e12/train.csv")
test_csv = pd.read_csv("/kaggle/input/playground-series-s3e12/test.csv")
train_csv.head()
train_csv.shape
train_csv.describe()
import seaborn as sns
fig, axes = plt.subplots(2, 3, figsize=(12, 5))
feature_names = ["gravity", "ph", "osmo", "cond", "urea", "calc"]
ax = axes.flatten()
for i, feature in enumerate(feature_names):
axes = ax[i]
sns.kdeplot(data=train_csv, x=feature, hue="target", ax=axes)
plt.tight_layout()
sns.scatterplot(data=train_csv, x="calc", y="gravity", hue="target")
# **There is good amount of overlapping , therefore , chances of overfitting is quite high .**
corr_mat = train_csv.corr()["target"].sort_values(ascending=True)
corr_mat
def heatmap(dataset, label=None):
corr = dataset.corr()
plt.figure(figsize=(10, 6), dpi=300)
mask = np.zeros_like(corr)
# print(mask)
mask[np.triu_indices_from(mask)] = True
sns.heatmap(corr, mask=mask, annot=True, annot_kws={"size": 12}, cmap="viridis")
plt.yticks(fontsize=10)
plt.xticks(fontsize=10)
plt.title(f"{label} Dataset Correlation Matrix\n", fontsize=20, weight="bold")
plt.show()
heatmap(train_csv, label="Train")
# OSMO IS LOOKING TO BE A GREAT FEATURE
# # DATA PROCESSING
training_features = ["cond", "calc"]
target_feature = ["target"]
x_train = train_csv[training_features].values
y_train = train_csv[target_feature].values
x_train.shape
y_train.shape
from sklearn.pipeline import Pipeline
from sklearn.preprocessing import StandardScaler
from sklearn.linear_model import LogisticRegression
num_pipeline = Pipeline([("std_scaler", StandardScaler())])
x_train = num_pipeline.fit_transform(x_train)
x_train.shape
x_train[0]
# # MODELLING
log_reg = LogisticRegression().fit(x_train, y_train.ravel())
proba_predictions = log_reg.predict_proba(x_train)[:, 1]
from sklearn.metrics import roc_auc_score
score = roc_auc_score(y_train.ravel(), proba_predictions)
score
# TESTING THE MODEL
from sklearn.model_selection import StratifiedKFold
from sklearn.base import clone
def test_the_model(model):
results = []
scores = []
skfolds = StratifiedKFold(n_splits=20, random_state=42, shuffle=True)
for train_index, test_index in skfolds.split(x_train, y_train.ravel()):
clone_clf = clone(model)
y = y_train.ravel()
X_train_folds = x_train[train_index]
y_train_folds = y[train_index]
X_test_fold = x_train[test_index]
y_test_fold = y[test_index]
clone_clf.fit(X_train_folds, y_train_folds)
y_pred = clone_clf.predict_proba(X_test_fold)[:, 1]
scores.append(roc_auc_score(y_test_fold, y_pred))
return scores
log_regressor = LogisticRegression()
scores = test_the_model(log_regressor)
val_score = np.mean(np.array(scores))
val_score
import xgboost as xgb
xgb_reg = xgb.XGBClassifier().fit(x_train, y_train.ravel())
proba_predictions = xgb_reg.predict_proba(x_train)[:, 1]
proba_predictions
score = roc_auc_score(y_train.ravel(), proba_predictions)
score
xgb_regressor = xgb.XGBClassifier()
val_scores = test_the_model(xgb_regressor)
val_score = np.mean(val_scores)
val_score
# FINETUNING THE XGB PARAMETERS
regressor = xgb.XGBClassifier(
objective="binary:logistic",
eval_metric="auc",
tree_method="exact",
n_jobs=-1,
max_depth=2,
eta=0.01,
n_estimators=100,
).fit(x_train, y_train.ravel())
y_pred = regressor.predict_proba(x_train)[:, 1]
score = roc_auc_score(y_train.ravel(), y_pred)
print("Train set Score : ", score)
xgb_reg = xgb.XGBClassifier(
objective="binary:logistic",
eval_metric="auc",
tree_method="exact",
n_jobs=-1,
max_depth=2,
eta=0.01,
n_estimators=100,
)
val_score = np.mean(test_the_model(xgb_reg))
val_score
# # SUBMISSION
test_values = test_csv[training_features].values
test_val = num_pipeline.fit_transform(test_values)
test_predictions = regressor.predict_proba(test_val)[:, 1]
test_predictions.shape
submission = pd.DataFrame({"id": test_csv["id"].values, "target": test_predictions})
submission.head()
submission.to_csv("submission.csv", index=False)
test_predictions
|
import pandas as pd
import re
import numpy as np
sla = pd.read_excel(
r"../input/shopee-code-league-20/_DA_Logistics/SLA_matrix.xlsx", engine="openpyxl"
)
orders = pd.read_csv(
r"../input/shopee-code-league-20/_DA_Logistics/delivery_orders_march.csv"
)
sla
# 看起來很奇怪,不過從表中,大概可以猜出是一個對照表,而且index是出發地(from),column是目的地(to),然後表裡面是運送時間
# 他的columns看起來是自動建立的,應該是要第一個row拿來做column才對
# row 6-8看起來像是其他資訊,而且只有第一個column有東西,應該是註解之類的,不過要把它祥細看一下
# 看起來column:'Unnamed: 1'才是我要的index
# row:0才是我要的column
for i in sla.iloc[:, 0]:
print(i)
# 確實row 6-8只是註解而已
# 看起來column:'Unnamed: 1'才是我要的index
sla = sla.set_index("Unnamed: 1")
# row:0才是我要的column
sla.columns = sla.iloc[0]
# 設定表格index和column的name,這樣比較看得懂
sla.index.name = "from"
sla.columns.name = "to"
display(sla)
# 把多的內容拿掉
sla = sla.iloc[1:5, 1:5]
display(sla)
# 把表格中的string轉換成int
sla = sla.applymap(lambda x: int(re.match("^(\d+) working days$", x).group(1)))
display(sla)
display(sla.dtypes)
# 把sla的index和column內容都變小寫
sla.index = sla.index.map(lambda x: x.lower())
sla.columns = sla.columns.map(lambda x: x.lower())
display(sla)
# 因為沒有後面重複的地點,所以只用最後一個字
sla.index = sla.index.map(lambda x: x.split()[-1])
sla.columns = sla.columns.map(lambda x: x.split()[-1])
display(sla)
# 到這邊sla整理完成了
orders.head()
# 先整理時間格式
# 以下是範的應對關係
# 1583137548 (Converted to 2020-03-02 4:25:48 PM Local Time)
print(pd.to_datetime(1583137548, unit="s") + pd.Timedelta(hours=8))
# 1583733540 (Converted to 2020-03-09 1:59:00 PM Local Time)
print(pd.to_datetime(1583733540, unit="s") + pd.Timedelta(hours=8))
# 確定這樣轉換是對的,套用到資料上
# 因為時區都是用同一個時區,所以這邊就直接暴力加8小時,不另外設定時區
orders["pick"] = pd.to_datetime(orders["pick"], unit="s") + pd.Timedelta(hours=8)
orders["1st_deliver_attempt"] = pd.to_datetime(
orders["1st_deliver_attempt"], unit="s"
) + pd.Timedelta(hours=8)
orders["2nd_deliver_attempt"] = pd.to_datetime(
orders["2nd_deliver_attempt"], unit="s"
) + pd.Timedelta(hours=8)
display(orders.head())
# 因為題目只看日期,不看時間,把詳細時間丟掉
orders[["pick", "1st_deliver_attempt", "2nd_deliver_attempt"]] = orders[
["pick", "1st_deliver_attempt", "2nd_deliver_attempt"]
].applymap(lambda x: x.date())
orders[["pick", "1st_deliver_attempt", "2nd_deliver_attempt"]] = orders[
["pick", "1st_deliver_attempt", "2nd_deliver_attempt"]
].astype(
"datetime64"
) # 把datatype變回pd datetime object
display(orders.head())
# 整理一下欄位名稱,這樣比較好打
orders.columns = ["orderid", "pick", "st", "nd", "to", "from"]
display(orders.head())
# 整理地址,變成全小寫
orders["to"] = orders["to"].map(lambda x: x.lower())
orders["from"] = orders["from"].map(lambda x: x.lower())
display(orders.head())
# 這邊可以證明地址的最後一個字都是我們sla裡面有的地點
t = orders["to"].map(lambda x: x.split()[-1])
f = orders["from"].map(lambda x: x.split()[-1])
print(t.isin(sla.columns).all()) # sla的columns是to
print(f.isin(sla.index).all()) # sla的index是from
# 只留下要的地名
orders["to"] = orders["to"].map(lambda x: x.split()[-1])
orders["from"] = orders["from"].map(lambda x: x.split()[-1])
display(orders.head())
# 計算時間距離
# 製作一張表,利用查詢可以把所有日期都映射到int,因為假日的部分不計,所以數字不會增加,
# 這邊看不懂沒關係,直接往下拉,看display出來是甚麼東西就會懂了
# 要先知道製作的區間,所以找到整個dataset的日期最大值及最小值,才有辦法知道起訖日期
max_date = orders[["pick", "st", "nd"]].max().max()
min_date = orders[["pick", "st", "nd"]].min().min()
all_dates = pd.date_range(min_date, max_date) # 包含最後一個日期
# 找出星期日
sunday = all_dates[all_dates.weekday == 6]
# 登記國定假日
holiday = pd.DatetimeIndex(["2020-03-08", "2020-03-25", "2020-03-30", "2020-03-31"])
# 2020-03-08 (Sunday);
# 2020-03-25 (Wednesday);
# 2020-03-30 (Monday);
# 2020-03-31 (Tuesday)
# 找出區間內所有不用上班的日子
day_off = sunday.union(holiday)
# 開始製作表
date_delta = pd.Series(index=all_dates, dtype=int)
# 先從0開始計算
delta_value = 0
for date in all_dates:
if not date in day_off:
# 只有工作天的日子才會+1
delta_value += 1
# 紀錄這天的delta value
date_delta[date] = delta_value
display(date_delta)
# 計算工作日的方法
def calaulate_working_days(start, end):
# 因為nd有可能有nat所以要另外處理
if pd.isnull(end):
return np.nan
return date_delta[end] - date_delta[start]
# 下面這個計算時間的方法,理論上可行,可是跑超久,跑不出來,所以我就放棄了
# holiday = pd.DatetimeIndex(['2020-03-08','2020-03-25','2020-03-30','2020-03-31'])
# def calaulate_working_days(start,end):
# if pd.isnull(end):
# return np.nan
# all_days = pd.date_range(start,end)
# all_days = all_days.difference(all_days[[0]])
# all_days = all_days[all_days.weekday != 6]
# all_days = all_days.difference(holiday)
# return all_days.size
# orders['st_delta'] = orders.apply(lambda df: calaulate_working_days(*df[['pick','st']]),axis=1)
# orders['nd_delta'] = orders.apply(lambda df: calaulate_working_days(*df[['st','nd']]),axis=1)
# 把實際日期抽換成delta_value
# 用replace比用apply花較少時間,所以用replace
orders["pick_value"] = orders["pick"].replace(date_delta)
orders["st_value"] = orders["st"].replace(date_delta)
orders["nd_value"] = orders["nd"].replace(date_delta)
# 利用delta_value相減求得兩個事件之間實際距離幾個工作天
orders["st_delta"] = orders["st_value"] - orders["pick_value"] # 從pick到first_attempt
orders["nd_delta"] = (
orders["nd_value"] - orders["st_value"]
) # 從first_attempt到second_attempt
display(orders.head())
# 因為要用replace減少運算時間,所以sla要改成能被replace使用的樣子
stack_sla = sla.stack()
display(stack_sla)
stack_sla.index = stack_sla.index.map(lambda x: ",".join(x))
display(stack_sla)
# 新增一個欄位,能夠利用stack_sla,做replace
orders["route"] = orders["from"] + "," + orders["to"]
# 用replace取代成送貨時間
orders["transport"] = orders["route"].replace(stack_sla)
display(orders.head())
# 依照定義,開始記錄哪些有late
orders["st_late"] = 0
orders["nd_late"] = 0
orders["is_late"] = 0
# first_attempt_time > sla_matrix ---> st_late
orders.loc[orders["st_delta"] > orders["transport"], "st_late"] = 1
# second_attempt - first_attempt > 3 ---> nd_late
orders.loc[orders["nd_delta"] > 3, "nd_late"] = 1
# first_attempt_time > sla_matrix ---> is_late
orders.loc[orders["st_delta"] > orders["transport"], "is_late"] = 1
# second_attempt - first_attempt > 3 ---> is_late
orders.loc[orders["nd_delta"] > 3, "is_late"] = 1
display(orders.query("(st_late==0)&(nd_late==1)").head(20))
display(orders.query("(st_late==1)&(nd_late==0)").head(20))
display(orders.query("(st_late==1)&(nd_late==1)").head(20))
display(orders.query("(st_late==0)&(nd_late==0)").head(20))
orders[["st_late", "nd_late", "orderid"]].groupby(["st_late", "nd_late"]).agg("count")
answer = orders[["orderid", "is_late"]]
print(answer["is_late"].value_counts())
answer
|
# # Introduction
# Recommender systems are a big part of our lives, recommending products and movies that we want to buy or watch. Recommender systems have been around for decades but have recently come into the spotlight.
# In this notebook, We will discuss three types of recommender system: **(1)Association rule learning** (ARL), **(2)content-based** and **(3)collaborative filtering** approaches. In this notebook, we will explain how to build a recommender system with these three methods.
# The functions of a recommender system are to suggest things to the user based on a variety of criteria. These systems forecast the most probable product that customers would buy, and it is interesting to them. Netflix, Amazon, and other businesses employ recommendation algorithms to assist their clients in locating appropriate items or movies.
# Content:
#
# 1. [**Association Rule Learning - ARL**](#1)
# * 1.1 [Data Preprocessing](#2)
# * 1.1.2 [Business Problem](#3)
# * 1.1.3 [Dataset Story](#4)
# * 1.1.4 [Variables](#5)
# * 1.1.5 [Libraries](#6)
# * 1.1.6 [Load and Check Data](#7)
# * 1.1.7 [Outlier Observations](#8)
# * 1.2 [Preparing ARL Data Structure (Invoice-Product Matrix)](#9)
# * 1.3 [Association Rules](#10)
# * 1.4 [Making Product Suggestions to Users at the Shopping Cart Stage](#11)
#
# 1. [**Content-Based Filtering**](#12)
# * 2.1 [Generating TF-IDF Matrix](#13)
# * 2.1.1 [Libraries](#14)
# * 2.1.2 [CountVectorizer](#15)
# * 2.1.3 [tf-idf](#16)
# * 2.2 [Creating Cosine Similarity Matrix](#17)
# * 2.3 [Making Suggestions Based on Similarities](#18)
# * 2.4 [Functionalize All Code of Content-Based Filtering](#19)
#
# 1. [**Collaborative Filtering**](#20)
# * 3.1 [Item-Based Collaborative Filtering](#21)
# * 3.1.1 [Data Preprocessing](#22)
# * 3.1.2 [Creating the User Movie Df](#23)
# * 3.1.3 [Making Item-Based Movie Suggestions](#24)
# * 3.2 [User-Based Collaborative Filtering](#25)
# * 3.2.1 [Data Preprocessing](#26)
# * 3.2.2 [Determining The Movies Watched By The User To Make A Suggestion](#27)
# * 3.2.3 [Accessing Data and Ids of Other Users Watching the Same Movies](#28)
# * 3.2.4 [Identifying Users with the Most Similar Behaviors to the User to Suggest](#29)
# * 3.2.5 [Calculating the Weighted Average Recommendation Score](#30)
# * 3.3 [Model-Based Collaborative Filtering - Matrix Factorization](#31)
# * 3.3.1 [Data Preprocessing](#32)
# * 3.3.2 [Modelling](#33)
# * 3.3.3 [Model Tuning](#34)
# * 3.3.4 [Final Model and Prediction](#35)
#
# 1. [References](#36)
#
#
# # 1. Association Rule Learning
# Association rule learning is a rule-based machine learning approach for finding significant connections between variables in large databases. It is designed to identify strong rules that have been identified in databases using various measures of interest.
# Our aim is to suggest products to users in the product purchasing process by applying association analysis to the online retail II dataset.
# 1. Data Preprocessing
# 2. Preparing ARL Data Structure (Invoice-Product Matrix))
# 3. Association Rules
# 4. Making Product Suggestions to Users at the Shopping Cart Stage
# ## 1.1 Data Preprocessing
# ### 1.1.2 Business Problem
# To suggest products to customers who have reached the basket stage.
# ### 1.1.3 Dataset Story
# * The data set named Online Retail II is a UK-based online sale.
# * Store's sales between 01/12/2009 - 09/12/2011.
# * The product catalog of this company includes souvenirs. promotion can be considered as products.
# * There is also information that most of its customers are wholesalers..
# ### 1.1.4 Variables
# * **InvoiceNo**: Invoice number. The unique number of each transaction, namely the invoice. Aborted operation if it starts with C.
# * **StockCode**: Product code. Unique number for each product.
# * **Description**: Product name
# * **Quantity**: Number of products. It expresses how many of the products on the invoices have been sold.
# * **InvoiceDate**: Invoice date and time.
# * **UnitPrice**: Product price (in GBP)
# * **CustomerID**: Unique customer number
# * **Country**: The country where the customer lives.
# ### 1.1.5 LIBRARIES
### installlation required
#!pip install mlxtend
# libraries
import pandas as pd
pd.set_option("display.max_columns", None)
# pd.set_option('display.max_rows', None)
pd.set_option("display.width", 500)
# çıktının tek bir satırda olmasını sağlar.
pd.set_option("display.expand_frame_repr", False)
from mlxtend.frequent_patterns import apriori, association_rules
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
#
# ### 1.1.6 Load and Check Data
df = pd.read_csv(
"/kaggle/input/online-retail-ii-data-set-from-ml-repository/Year 2010-2011.csv",
encoding="unicode_escape",
)
df.head()
def check_df(dataframe):
print("################ Shape ####################")
print(dataframe.shape)
print("############### Columns ###################")
print(dataframe.columns)
print("############### Types #####################")
print(dataframe.dtypes)
print("############### Head ######################")
print(dataframe.head())
print("############### Tail ######################")
print(dataframe.tail())
print("############### Describe ###################")
print(dataframe.describe().T)
check_df(df)
#
# ### 1.1.7 Outlier Observations
def outlier_thresholds(dataframe, variable):
quartile1 = dataframe[variable].quantile(0.01)
quartile3 = dataframe[variable].quantile(0.99)
interquantile_range = quartile3 - quartile1
up_limit = quartile3 + 1.5 * interquantile_range
low_limit = quartile1 - 1.5 * interquantile_range
return low_limit, up_limit
def replace_with_thresholds(dataframe, variable):
low_limit, up_limit = outlier_thresholds(dataframe, variable)
dataframe.loc[(dataframe[variable] < low_limit), variable] = low_limit
dataframe.loc[(dataframe[variable] > up_limit), variable] = up_limit
def retail_data_prep(dataframe):
dataframe.dropna(inplace=True)
dataframe = dataframe[~dataframe["Invoice"].str.contains("C", na=False)]
dataframe = dataframe[dataframe["Quantity"] > 0]
dataframe = dataframe[dataframe["Price"] > 0]
replace_with_thresholds(dataframe, "Quantity")
replace_with_thresholds(dataframe, "Price")
return dataframe
df = retail_data_prep(df)
#
# ## 1.2 Preparing ARL Data Structure (Invoice-Product Matrix)
# * In this section, we will create a matrix of invoice and products as in the example below.
# EXAMPLE:
# Description NINE DRAWER OFFICE TIDY SET 2 TEA TOWELS I LOVE LONDON SPACEBOY BABY GIFT SET
# Invoice
# 536370 0 1 0
# 536852 1 0 1
# 536974 0 0 0
# 537065 1 0 0
# 537463 0 0 1
# * We will only work on Germany, let's filter.
df_de = df[df["Country"] == "Germany"]
df_de.head(10)
df_de.groupby(["Invoice", "Description"]).agg({"Quantity": "sum"}).head(10)
# * The ouput indicates **Invoice 536527** has these above items.
# * We want to see the description (products) in the columns.
# * And we want to see whether there are products at the intersections of the matrix or not. We can do this with unstack function, we could have used pivot funct instead of it.
df_de.groupby(["Invoice", "Description"]).agg({"Quantity": "sum"}).unstack().iloc[
0:5, 0:5
]
# * If there is a product in the columns, we expect 1, if not 0.
# * Firstly, we will assign 0 to NaN values.
df_de.groupby(["Invoice", "Description"]).agg({"Quantity": "sum"}).unstack().fillna(
0
).iloc[0:5, 0:5]
# * Seconly, If there is a product in the columns, we will convert it to 1.
# applymap will itinirate all cells in the dataframe. apply would only itinirate in row or columns
df_de.groupby(["Invoice", "Description"]).agg({"Quantity": "sum"}).unstack().fillna(
0
).applymap(lambda x: 1 if x > 0 else 0).iloc[0:5, 0:5]
# * Let's turn all these codes into a single function (named create_invoice_product_df).
def create_invoice_product_df(dataframe, id=False):
if id:
return (
dataframe.groupby(["Invoice", "StockCode"])["Quantity"]
.sum()
.unstack()
.fillna(0)
.applymap(lambda x: 1 if x > 0 else 0)
)
else:
return (
dataframe.groupby(["Invoice", "Description"])["Quantity"]
.sum()
.unstack()
.fillna(0)
.applymap(lambda x: 1 if x > 0 else 0)
)
de_inv_pro_df = create_invoice_product_df(df_de)
de_inv_pro_df.head()
# * It doesn't look good, let's make id = True in **create_invoice_product_df** for better looking matrix
def create_invoice_product_df(dataframe, id=True):
if id:
return (
dataframe.groupby(["Invoice", "StockCode"])["Quantity"]
.sum()
.unstack()
.fillna(0)
.applymap(lambda x: 1 if x > 0 else 0)
)
else:
return (
dataframe.groupby(["Invoice", "Description"])["Quantity"]
.sum()
.unstack()
.fillna(0)
.applymap(lambda x: 1 if x > 0 else 0)
)
de_inv_pro_df = create_invoice_product_df(df_de)
de_inv_pro_df.head()
de_inv_pro_df = create_invoice_product_df(df_de, id=True)
# * Let's define a function for checking StockCode number
def check_id(dataframe, stock_code):
product_name = (
dataframe[dataframe["StockCode"] == stock_code][["Description"]]
.values[0]
.tolist()
)
print(product_name)
# * Let's check first stockcode's name **10002**
check_id(df_de, "10002")
#
# ## 1.3. Association Rules
# * We will subtract the probabilities of all possible products being together.
# minumum support value 0.01, we don't want to get below 0.01
# In real life scenarios, this minimum support value is very low.
frequent_itemsets = apriori(de_inv_pro_df, min_support=0.01, use_colnames=True)
frequent_itemsets.sort_values("support", ascending=False).head(10)
rules = association_rules(frequent_itemsets, metric="support", min_threshold=0.01)
rules.sort_values("support", ascending=False).head()
rules.sort_values("lift", ascending=False).head(10)
# * **antecedent support:** probability of the first product
# * **consequent support:** probability of the second product and others
# * **support:** probability of two products (or more) appearing together
# * **confidence:** when product x is bought, the probability of purchasing product y
# * **lift:** when x is taken, the probability of getting y increases by this much (lift)
# ## 1.4 Making Product Suggestions to Users at the Shopping Cart Stage
# Example Product Id: 22492
product_id = "22492"
check_id(df, product_id)
sorted_rules = rules.sort_values("lift", ascending=False)
recommendation_list = []
for i, product in enumerate(sorted_rules["antecedents"]):
for j in list(product):
if j == product_id:
recommendation_list.append(list(sorted_rules.iloc[i]["consequents"])[0])
recommendation_list[0:3]
check_id(df, "21915")
def arl_recommender(rules_df, product_id, rec_count=1):
sorted_rules = rules_df.sort_values("lift", ascending=False)
recommendation_list = []
for i, product in sorted_rules["antecedents"].items():
for j in list(product):
if j == product_id:
recommendation_list.append(list(sorted_rules.iloc[i]["consequents"]))
recommendation_list = list(
{item for item_list in recommendation_list for item in item_list}
)
return recommendation_list[:rec_count]
check_id(df, "23049")
arl_recommender(rules, "22492", 1)
# If want to see two product suggection
arl_recommender(rules, "22492", 2)
# #### Some Notes
# * For example, if I had 10,000 products, I wouldn't be interested in all of them. In this case it should be done at category level
# * When the person adds a product to the cart, what I will suggest should already be clear.
# * I know what to suggest with product X, but if the person has already bought this product, it is necessary to make a correction accordingly. There must be an intermediate control mechanism. At the database level, the userid should be checked. If the person has not bought that product after checking, it is necessary to recommend that product. This cannot be done. There is a fine line you should consider
# # 2. Content-Based Filtering
# * Represent texts mathematically (vectoring texts)
# * Count Vector (word count)
# * TF-IDF
# In this content-based section, we will go through these steps below.
# 1. Creating the TF-IDF Matrix
# 2. Creating the Cosine Similarity Matrix
# 3. Making Suggestions Based on Similarities
# 4. Preparation of the Working Script
# ### Euclidean Distance
# $ d(p, q) = \sqrt \sum_{i = 1}^\infty x_i$
# Ürün içeriklerinin benzerlikleri üzerinden tavsiyeler geliştirilir.
# ### Cosine Similarity
import pandas as pd
pd.set_option("display.max_columns", None)
pd.set_option("display.width", 500)
pd.set_option("display.expand_frame_repr", False)
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.metrics.pairwise import cosine_similarity
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Euclidean Distance
# It finds the distance between two vectors.
# Cosine Similarity
# A metric focused on the similarity of two vectors.
# ## 2.1 Generating TF-IDF Matrix
# Recommendation System Based on Movie Overviews
# ### 2.1.1 Libraries
import pandas as pd
pd.set_option("display.max_columns", None)
pd.set_option("display.width", 500)
pd.set_option("display.expand_frame_repr", False)
from sklearn.feature_extraction.text import TfidfVectorizer
from sklearn.metrics.pairwise import cosine_similarity
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
df2 = pd.read_csv(
"/kaggle/input/movies-metadata/movies_metadata.csv",
sep=";",
encoding="unicode_escape",
low_memory=False,
)
df2.head()
df2["overview"].head()
#
# #### 2.1.2 CountVectorizer
# In the Count operation, the number of times each word occurs in each document is counted. For instace; let's look at the example below. We have four sentences. We will convert all words to the matrix. If the word is in it, it will count 1 or more, othewise 0.
from sklearn.feature_extraction.text import CountVectorizer
corpus = [
"This is the first document.",
"This document is the second document.",
"And this is the third one.",
"Is this the first document?",
]
vectorizer = CountVectorizer()
X = vectorizer.fit_transform(corpus)
vectorizer.get_feature_names()
X.toarray()
#
# #### 2.1.3 tf-idf
# There are normalized numeric representations.
# * **STEP 1:** TF(t) = (Frequency of occurrence of a t term in the relevant document) / (Total number of terms in the document)(term frequency)
# * **Step 2:** IDF(t) = 1 + log_e(Total number of documents + 1) / (number of documents with t term + 1) (inverse document frequency)
# * **Step 3:** TF-IDF = TF(t) * IDF(t)
# * **Step 4:** L2 normalization to TF-IDF values
from sklearn.feature_extraction.text import TfidfVectorizer
vectorizer = TfidfVectorizer(analyzer="word")
X = vectorizer.fit_transform(corpus)
vectorizer.get_feature_names()
X.toarray()
df2["overview"].head()
tfidf = TfidfVectorizer(stop_words="english")
df2["overview"] = df2["overview"].fillna("")
tfidf_matrix = tfidf.fit_transform(df2["overview"])
tfidf_matrix.shape
#
# ## 2.2 Creating Cosine Similarity Matrix
cosine_sim = cosine_similarity(tfidf_matrix, tfidf_matrix)
cosine_sim.shape
cosine_sim[1]
#
# ## 2.3 Making Suggestions Based on Similarities
indices = pd.Series(df2.index, index=df2["title"])
indices = indices[~indices.index.duplicated(keep="last")]
indices.shape
indices[:10]
# showing the index number of the films
indices["The American President"]
movie_index = indices["The American President"]
# Cosine similarities of The American President movie and other movies.
cosine_sim[movie_index]
similarity_scores = pd.DataFrame(cosine_sim[movie_index], columns=["score"])
movie_indices = similarity_scores.sort_values("score", ascending=False)[1:11].index
df2["title"].iloc[movie_indices]
#
# ## 2.4 Functionalize All Code of Content-Based Filtering
def content_based_recommender(title, cosine_sim, dataframe):
# generates index
indices = pd.Series(dataframe.index, index=dataframe["title"])
indices = indices[~indices.index.duplicated(keep="last")]
# capturing the index of the title
movie_index = indices[title]
# Calculating similarity scores by title
similarity_scores = pd.DataFrame(cosine_sim[movie_index], columns=["score"])
# the top 10 movies except itself(the movie we chose)
movie_indices = similarity_scores.sort_values("score", ascending=False)[1:11].index
return dataframe["title"].iloc[movie_indices]
# Example
content_based_recommender("The Matrix", cosine_sim, df2)
# Example
content_based_recommender("The Godfather", cosine_sim, df2)
## Cosine Similarity Function, if you want to use it again.
# def calculate_cosine_sim(dataframe):
# tfidf = TfidfVectorizer(stop_words='english')
# dataframe['overview'] = dataframe['overview'].fillna('')
# tfidf_matrix = tfidf.fit_transform(dataframe['overview'])
# cosine_sim = cosine_similarity(tfidf_matrix, tfidf_matrix)
# return cosine_sim
# cosine_sim = calculate_cosine_sim(df)
# content_based_recommender('The Dark Knight Rises', cosine_sim, df)
#
# # 3. Collaborative Filtering
# * Item-Based Collaborative Filtering
# * User-Based Collaborative Filtering
# * Model-Based Collaborative Filtering
# ## 3.1 Item-Based Collaborative Filtering
# * Suggestions are made on item similarity.
# * For instance; there are movies that show the same liking structure as The Lord of The Rings movie.
# ### 3.1.2 Data Preprocessing
import pandas as pd
pd.set_option("display.max_columns", 20)
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
movie = pd.read_csv("/kaggle/input/movielens-20m-dataset/movie.csv")
rating = pd.read_csv("/kaggle/input/movielens-20m-dataset/rating.csv")
df3 = movie.merge(rating, how="left", on="movieId")
df3.head()
movie = pd.read_csv("/kaggle/input/movielens-20m-dataset/movie.csv")
rating = pd.read_csv("/kaggle/input/movielens-20m-dataset/rating.csv")
df3 = movie.merge(rating, how="left", on="movieId")
df3.head()
df3.shape
# * We have 20000797 rows, 6 columns, we don't want to use all of them. let's take some sample from this data.
# * You can make the dataset smaller with this below code. It takes sample from orginal data. Frac is in ration of percentage you desire.
# * df = df.sample(frac =.50)
# ### 3.1.2 Creating the User Movie Df
df3.shape
# movie counts
df3["title"].nunique()
# rating count of each movies
df3["title"].value_counts().head()
rating_counts = pd.DataFrame(df3["title"].value_counts())
# we don't want all ratings, therefore we add trashold. It will not bring under 10000
rare_movies = rating_counts[rating_counts["title"] <= 10000].index
# taking out rare movies from dataframe
common_movies = df3[~df3["title"].isin(rare_movies)]
# all ratings
common_movies.shape
# we have 462 movies now
common_movies["title"].nunique()
# let's pivot it
user_movie_df = common_movies.pivot_table(
index=["userId"], columns=["title"], values="rating"
)
user_movie_df.shape
user_movie_df.head(10)
user_movie_df.columns
# columns count and title count are equal
len(user_movie_df.columns)
common_movies["title"].nunique()
#
# ### 3.1.3 Making Item-Based Movie Suggestions
movie_name = "Matrix, The (1999)"
movie_name = user_movie_df[movie_name]
user_movie_df.corrwith(movie_name).sort_values(ascending=False).head(10)
# Another Example
movie_name = "12 Angry Men (1957)"
movie_name = user_movie_df[movie_name]
user_movie_df.corrwith(movie_name).sort_values(ascending=False).head(10)
# random selection of movies
movie_name = pd.Series(user_movie_df.columns).sample(1).values[0]
movie_name = user_movie_df[movie_name]
user_movie_df.corrwith(movie_name).sort_values(ascending=False).head(10)
# Let's put all the codes in a single script
# script of all codes
def create_user_movie_df():
movie = pd.read_csv("/kaggle/input/movielens-20m-dataset/movie.csv")
rating = pd.read_csv("/kaggle/input/movielens-20m-dataset/rating.csv")
df = movie.merge(rating, how="left", on="movieId")
comment_counts = pd.DataFrame(df["title"].value_counts())
rare_movies = comment_counts[comment_counts["title"] <= 1000].index
common_movies = df[~df["title"].isin(rare_movies)]
user_movie_df = common_movies.pivot_table(
index=["userId"], columns=["title"], values="rating"
)
return user_movie_df
# user_movie_df = create_user_movie_df()
def item_based_recommender(movie_name, user_movie_df):
movie_name = user_movie_df[movie_name]
return user_movie_df.corrwith(movie_name).sort_values(ascending=False).head(10)
item_based_recommender("Fight Club (1999)", user_movie_df)
movie_name = pd.Series(user_movie_df.columns).sample(1).values[0]
item_based_recommender(movie_name, user_movie_df)
# this function helps to find the movie names
def check_film(keyword, user_movie_df):
return [col for col in user_movie_df.columns if keyword in col]
check_film("Str", user_movie_df)
item_based_recommender("Forrest Gump (1994)", user_movie_df)
#
# ## 3.2. User-Based Collaborative Filtering
# Suggestions are made based on user similarities.
# * Step 1: Preparing the Data Set
# * Step 2: Determining the Movies Watched by the User to Suggest
# * Step 3: Accessing Data and Ids of Other Users Watching the Same Movies
# * Step 4: Identifying Users with the Most Similar Behaviors to the User to Suggest
# * Step 5: Calculating the Weighted Average Recommendation Score
# ### 3.2.1. Data Preprocessing
# We have already defined user movie matrix above as named **user_movie_df**. We will use this data, instead of making all the data process again.
user_movie_df
# Let's define a random user
random_user = int(pd.Series(user_movie_df.index).sample(1, random_state=35).values)
# the output random user id
random_user
#
# ### 3.2.2. Determining The Movies Watched By The User To Make A Suggestion
# we selected the random_user's movie here
random_user_df = user_movie_df[user_movie_df.index == random_user]
random_user_df
# Taking out all NaN
movies_watched = random_user_df.columns[random_user_df.notna().any()].tolist()
# if you want to see all the movies that watched by random user, execute this
movies_watched
# Let's check if random user watch 2001: A Space Odyssey (1968)
user_movie_df.loc[
user_movie_df.index == random_user, user_movie_df.columns == "Aladdin (1992)"
]
# how many movies he watched
len(movies_watched)
#
# ### 3.2.3. Accessing Data and Ids of Other Users Watching the Same Movies
# James' movies
movies_watched_df = user_movie_df[movies_watched]
movies_watched_df.head()
# Number of people who watched at least one movie in common with James. 137658 people watched at least on movie, common movies count 191
movies_watched_df.shape
# As we can see above, there are lots of people watched at least one movie in common, but we need to put a threshold here. Common movies count 50
# user_movie_count indicates how many movies each user watched
# notnull gives us binary output (1 or 0) if we don't do that, the ratings count, like 3.0 + 4.0 = 7.0
user_movie_count = movies_watched_df.T.notnull().sum()
# moving user_movie_count in the columns
user_movie_count = user_movie_count.reset_index()
user_movie_count.columns = ["userId", "movie_count"]
user_movie_count.head()
# 40 treshold in common movies
user_movie_count[user_movie_count["movie_count"] > 40].sort_values(
"movie_count", ascending=False
)
# how many people watch the same movies with James (he watched 50 movies)
# there is one person watching the same movies
user_movie_count[user_movie_count["movie_count"] == 50].count()
# let's bring users id watching the same movies
users_same_movies = user_movie_count[user_movie_count["movie_count"] > 40]["userId"]
users_same_movies.head()
users_same_movies.count()
#
# ### 3.2.4. Identifying Users with the Most Similar Behaviors to the User to Suggest
# * We will perform 3 steps:
# * 1. We will aggregate data from James and other users.
# * 2. We will create the correlation df.
# * 3. We will find the most similar finders (Top Users)
final_df = pd.concat(
[
movies_watched_df[movies_watched_df.index.isin(users_same_movies)],
random_user_df[movies_watched],
]
)
final_df.head()
final_df.shape
# We set all user in the columns, but it doesn't look good,therefore, we will make them tidy
final_df.T.corr()
# making above matrix tidy
corr_df = final_df.T.corr().unstack().sort_values().drop_duplicates()
corr_df = pd.DataFrame(corr_df, columns=["corr"])
corr_df.index.names = ["user_id_1", "user_id_2"]
corr_df = corr_df.reset_index()
corr_df.head()
# Users with 65 percent or more correlation with James
top_users = corr_df[(corr_df["user_id_1"] == random_user) & (corr_df["corr"] >= 0.65)][
["user_id_2", "corr"]
].reset_index(drop=True)
top_users = top_users.sort_values(by="corr", ascending=False)
top_users.rename(columns={"user_id_2": "userId"}, inplace=True)
top_users
# lets merge our new table with ratings
rating = pd.read_csv("/kaggle/input/movielens-20m-dataset/rating.csv")
top_users_ratings = top_users.merge(
rating[["userId", "movieId", "rating"]], how="inner"
)
# taking out James from the table
top_users_ratings = top_users_ratings[top_users_ratings["userId"] != random_user]
# We have a problem here. There are two levels in data. One is corr, other one is rating. Someone has high corr but rating is 1.0 with James, another person has low corr, but high rating. Which one should we consider?
top_users_ratings.head()
# * We have a problem here. There are two levels in data. One is corr, other one is rating. Someone has high corr but rating is 1.0 with James, another person has low corr, but high rating. Which one should we consider?
# * I need to make a weighting based on rating and correlation.
# ### 3.2.5. Calculating the Weighted Average Recommendation Score
# * We will create a single score by simultaneously considering the impact of the users most similar to James (correlation) and the rating.
# Calculation of weighted_rating
top_users_ratings["weighted_rating"] = (
top_users_ratings["corr"] * top_users_ratings["rating"]
)
top_users_ratings.head()
# * There is one more problem this above table. There are many ratings given to a movie. We will use groupby for this problem.
recommendation_df = top_users_ratings.groupby("movieId").agg(
{"weighted_rating": "mean"}
)
recommendation_df = recommendation_df.reset_index()
recommendation_df.head()
# there are 8071 movies
recommendation_df[["movieId"]].nunique()
# There are 8071 movies, we can't recommend all these movies. we need a trashold. We do not want to recommend **weighted_score** below **3.5**
movies_to_be_recommend = recommendation_df[
recommendation_df["weighted_rating"] > 3.5
].sort_values("weighted_rating", ascending=False)
movies_to_be_recommend
# list of movies to recommend james
movie = pd.read_csv("/kaggle/input/movielens-20m-dataset/movie.csv")
movies_to_be_recommend.merge(movie[["movieId", "title"]])
# def user_based_recommender():
# import pickle
# import pandas as pd
# # user_movie_df = pickle.load(open('user_movie_df.pkl', 'rb'))
# random_user = int(pd.Series(user_movie_df.index).sample(1, random_state=45).values)
# random_user_df = user_movie_df[user_movie_df.index == random_user]
# movies_watched = random_user_df.columns[random_user_df.notna().any()].tolist()
# movies_watched_df = user_movie_df[movies_watched]
# user_movie_count = movies_watched_df.T.notnull().sum()
# user_movie_count = user_movie_count.reset_index()
# user_movie_count.columns = ["userId", "movie_count"]
# users_same_movies = user_movie_count[user_movie_count["movie_count"] > 20]["userId"]
#
# final_df = pd.concat([movies_watched_df[movies_watched_df.index.isin(users_same_movies)],
# random_user_df[movies_watched]])
#
# corr_df = final_df.T.corr().unstack().sort_values().drop_duplicates()
# corr_df = pd.DataFrame(corr_df, columns=["corr"])
# corr_df.index.names = ['user_id_1', 'user_id_2']
# corr_df = corr_df.reset_index()
#
# top_users = corr_df[(corr_df["user_id_1"] == random_user) & (corr_df["corr"] >= 0.65)][
# ["user_id_2", "corr"]].reset_index(drop=True)
#
# top_users = top_users.sort_values(by='corr', ascending=False)
# top_users.rename(columns={"user_id_2": "userId"}, inplace=True)
# rating = pd.read_csv('/kaggle/input/movielens-20m-dataset/rating.csv')
# top_users_ratings = top_users.merge(rating[["userId", "movieId", "rating"]], how='inner')
# top_users_ratings['weighted_rating'] = top_users_ratings['corr'] * top_users_ratings['rating']
# top_users_ratings = top_users_ratings[top_users_ratings["userId"] != random_user]
#
# recommendation_df = top_users_ratings.groupby('movieId').agg({"weighted_rating": "mean"})
# recommendation_df = recommendation_df.reset_index()
#
# movies_to_be_recommend = recommendation_df[recommendation_df["weighted_rating"] > 3.7].sort_values("weighted_rating", ascending=False)
# movie = pd.read_csv('/kaggle/input/movielens-20m-dataset/movie.csv')
# return movies_to_be_recommend.merge(movie[["movieId", "title"]])
# user_based_recommender()
#
# ## 3.3 Matrix Factorization
# * To fill in the blanks, the weights of the latent features that are assumed to exist for users and movles are found over the existing data and predictions are made for non-existent observations with these weights.
# * Matrix factorization assume; There are some latent factors when users like a movie. These hidden factors are also present in movies.
# 1. Decomposes the user-item matrix into 2 less dimensional matrices.
# 2. It assumes that the transition from two matrices to the user-item matrix occurs with latent factors. We will assume the latent variables.
# 3. The weights of the latent factors are found on the filled observations.
# 4. Empty observations are filled with the weights found.
# * The reason why James watches the movie; the genre of the film, the director or actors of the film, the duration of the film, the language in which the film was shot. While you are liking the movie, there are some factors that you are not aware of. These are called latent factors, features in machine learning.
# **add FORMULA OF MATRIX FAC.**
# * It's an optimization, it's actually a gradient descent work.
# Let's make it more clear.
# * It is assumed that the rating matrix is formed by the product of two factor matrices (dot product).
# * Factor matrices? User latent factors, movie latent factors, are actually two separate matrices.
# * Latent factors? Or latent features? Latent factors or variables.
# * Users and movies are considered to have scores for latent features.
# * These weights (scores) are first found on the existing data and then the empty sections are filled according to these weights.
# #### What are these factors in this data?
# Comedy, horror, adventure, action, thriller, youth, having a specific actor, director.
# ADD TABLES | PICTURE
# $ r_{11} = p_{11} * q_{11} + p_{12} * q_{21} $
# * All p and q are found iteratively over the existing values and then used.
# * Initially, random p and q values and the values in the rating matrix are tried to be estimated.
# * In each iteration, erroneous estimations are arranged and the values in the rating matrix are tried to be approached.
# * For example, if 5 is called 3 in one iteration, the next one is called 4, then 5 is called.
# * Thus, p and q values are filled as a result of a certain iteration.
# * Estimation is made for null observations based on the available p and q.
# #### Some Notes
# * Matrix Factorization vs SVD is not the same
# * SVD (Singular Value Decomposition) is a size reduction method.
# * ALS --> Spark ALS for big data. Only difference ALS make some changes on p and q values.
# ### 3.3.1 Data Preprocessing
# pip install surprise
import pandas as pd
from surprise import Reader, SVD, Dataset, accuracy
from surprise.model_selection import GridSearchCV, train_test_split, cross_validate
pd.set_option("display.max_columns", None)
movie = pd.read_csv("/kaggle/input/movielens-20m-dataset/movie.csv")
rating = pd.read_csv("/kaggle/input/movielens-20m-dataset/rating.csv")
df = movie.merge(rating, how="left", on="movieId")
df.head()
# We reduce the dataset to these four movies in terms of both followability and performance..
movie_ids = [130219, 356, 4422, 541]
movies = [
"The Dark Knight (2011)",
"Cries and Whispers (Viskningar och rop) (1972)",
"Forrest Gump (1994)",
"Blade Runner (1982)",
]
sample_df = df[df.movieId.isin(movie_ids)]
sample_df.shape
sample_df.head()
# creating the user movie dataframe
user_movie_df = sample_df.pivot_table(
index=["userId"], columns=["title"], values="rating"
)
user_movie_df.head()
# The surprise library requires between which numbers it will be. We give 1-5 range.
reader = Reader(rating_scale=(1, 5))
# The data we created in accordance with the data structure of the surprise library
data = Dataset.load_from_df(sample_df[["userId", "movieId", "rating"]], reader)
type(data)
#
# ### 3.3.2 Modelling
trainset, testset = train_test_split(data, test_size=0.25)
svd_model = SVD()
svd_model.fit(trainset)
predictions = svd_model.test(testset)
# predictions
accuracy.rmse(predictions)
cross_validate(svd_model, data, measures=["RMSE", "MAE"], cv=5, verbose=True)
user_movie_df.head()
# Let's guess blade runner( 541) for userid 1
svd_model.predict(uid=1.0, iid=541, verbose=True)
# Let's guess Whispers (356) for userid 1. We guessed for the movie she didn't watc
svd_model.predict(uid=1.0, iid=356, verbose=True)
#
# ### 3.3.3 Model Tuning
# There is the problem of how long I will do the process of changing the values. Should I do this replacement 10 times, like 100 times? So this is a hyperparameter for me that needs to be optimized by the user. How many times will I update the weight? With this question, what will my learning speed be? There is a learning rate that represents the speed of these updates.
# here it is our parameters for the model, epoch and learning rate
param_grid = {"n_epochs": [5, 10], "lr_all": [0.002, 0.005]}
gs = GridSearchCV(
SVD, param_grid, measures=["rmse", "mae"], cv=3, n_jobs=-1, joblib_verbose=True
)
gs.fit(data)
gs.best_score["rmse"]
gs.best_params["rmse"]
#
# ### 3.3.4 Final Model and Prediction
svd_model = SVD(**gs.best_params["rmse"])
data = data.build_full_trainset()
svd_model.fit(data)
user_movie_df.head()
# Let's guess blade runner(541) for userid 1
svd_model.predict(uid=1.0, iid=541, verbose=True)
# Another example Cries and Whispers (356) for user id1.
svd_model.predict(uid=1.0, iid=356, verbose=True)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
df = pd.read_csv("/kaggle/input/customer-shopping-dataset/customer_shopping_data.csv")
# # **You can find the whole Jupyter Notebook along with a web application for this notebook in the GitHub link [here](https://github.com/Ishrak30/Customer-Spending-Prediction-using-ML-Techniqiues)**
# # **Understanding Data**
df.head(10)
df.describe()
df[["day", "month", "year"]] = df["invoice_date"].str.split("/", expand=True)
print(df.shape)
print(df.columns)
print(df.dtypes)
df.isnull().sum()
categorical_feature = ["gender", "category", "payment_method", "shopping_mall"]
numerical_feature = ["age", "quantity", "month", "year"]
dropping = ["customer_id", "invoice_no", "day", "invoice_date"]
result = ["price"]
df.drop(dropping, axis=1, inplace=True)
df.columns
dt = df[categorical_feature]
for col in dt:
print("Features: ", col, "--", dt[col].unique())
# # **Exploratory Data Analysis**
import seaborn as sns
import plotly.express as px
import matplotlib.pyplot as plt
from matplotlib import style
# ## **Categorical Feature Plot**
# fig, ax = plt.subplots(figsize=(15, 5))
fig = px.histogram(
df,
x="category",
y="price",
color="gender",
labels={"category": "Category", "gender": "Gender"},
)
fig.show()
fig = px.histogram(
df,
x="year",
y="quantity",
color="gender",
labels={"year": "Years", "gender": "Gender", "sum of quantity": "Quantity"},
)
fig.show()
fig = px.histogram(
df,
x="gender",
y="price",
color="gender",
labels={"category": "Category", "gender": "Gender"},
)
fig.show()
# df['shopping_mall'].unique()
fig = px.histogram(
df,
x="shopping_mall",
y="price",
color="gender",
labels={"shopping_mall": "Mall Name", "gender": "Gender"},
)
fig.show()
df_mall_cat = (
df.groupby(["shopping_mall", "category"])["price"]
.sum()
.unstack("category")
.plot(kind="bar", figsize=(15, 8))
)
plt.xlabel("Price")
plt.title("Price for each item in different mall")
plt.ylabel("Mall")
plt.xticks(rotation=45)
plt.show()
# ## Numerical Feature Plot
# how many quantity is bought each year by gender
fig = px.histogram(
df,
x="year",
y="quantity",
color="gender",
labels={"year": "Year", "gender": "Gender"},
)
fig.show()
# payment method by gender every category
fig = px.box(
df,
x="category",
y="payment_method",
color="gender",
labels={
"category": "Categories",
"gender": "Gender",
"payment_method": "Payment Methods",
},
)
fig.show()
# # **Feature Encoding**
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
for col in categorical_feature:
df[col] = le.fit_transform(df[col])
df.head()
# # **Correlation Matrix**
##Correlation Matrix using Seaborn
# corr = df.corr()
# #mask for upper triangle
# mask = np.triu(np.ones_like(corr, dtype=bool))
# #matplotlib figure
# fig, ax = plt.subplots(figsize=(11,9))
# #colormap
# cmap = sns.color_palette("coolwarm", as_cmap=True)
# #cmap = sns.diverging_palette(230, 20, as_cmap=True)
# #plot
# dataplot = sns.heatmap(corr, mask=mask, cmap=cmap, vmax=.3, center=0,
# square=True, linewidths=.5, cbar_kws={"shrink": .5})
##Correlation Matrix using Plotly
import plotly.graph_objects as go
import plotly.figure_factory as ff
df_corr = df.corr() # Generate correlation matrix
fig = go.Figure()
fig.add_trace(
go.Heatmap(
x=df_corr.columns,
y=df_corr.index,
z=np.array(df_corr),
colorscale="teal",
text=np.array(df_corr),
texttemplate="%{text}",
)
)
fig.show()
# # Models
# split data
from sklearn.model_selection import train_test_split
# data modelling
from sklearn.linear_model import LinearRegression
from sklearn.metrics import (
confusion_matrix,
accuracy_score,
roc_curve,
classification_report,
)
from sklearn import metrics
y = df["price"]
X = df.drop("price", axis=1)
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.20, random_state=0
)
# from sklearn.preprocessing import StandardScaler
# sc_std = StandardScaler()
# X_train = sc_std.fit_transform(X_train)
# X_test = sc_std.transform(X_test)
X_train
# ## Working on Models
# ### Linear Regression
m1 = "Linear Regression"
lr = LinearRegression()
model = lr.fit(X_train, y_train)
plt.scatter(lr.predict(X_train), y_train)
plt.xlabel("Predicted value of Y")
plt.ylabel("Real value of Y")
plt.show()
from sklearn.metrics import mean_squared_error
pred = lr.predict(X_test)
rms = mean_squared_error(y_test, pred, squared=False)
print(rms)
print("Training R2 Score - ", lr.score(X_train, y_train) * 100)
print("Test R2 Score - ", lr.score(X_test, y_test) * 100)
# ### Random Forest
from sklearn.ensemble import RandomForestRegressor
m2 = "Random Forest"
rf = RandomForestRegressor(n_estimators=250, n_jobs=-1)
rf.fit(X_train, y_train)
plt.scatter(rf.predict(X_train), y_train)
plt.xlabel("Predicted value of Y")
plt.ylabel("Real value of Y")
plt.show()
print("Training R2 Score - ", rf.score(X_train, y_train) * 100)
print("Test R2 Score - ", rf.score(X_test, y_test) * 100)
# ### Decision Tree
from sklearn.tree import DecisionTreeRegressor
m3 = "Decision Tree"
dtm = DecisionTreeRegressor(max_depth=5, min_samples_split=6, max_leaf_nodes=10)
dtm.fit(X_train, y_train)
plt.scatter(dtm.predict(X_train), y_train)
plt.xlabel("Predicted value of Y")
plt.ylabel("Real value of Y")
plt.show()
print("R2 on train dataset = ", dtm.score(X_train, y_train) * 100)
print("R2 on test dataset = ", dtm.score(X_test, y_test) * 100)
# ### Lasso Regression
from sklearn.linear_model import Lasso, LassoCV
ls_cv = LassoCV(alphas=None, cv=10, max_iter=100000)
ls_cv.fit(X_train, y_train)
alpha = ls_cv.alpha_
alpha
ls = Lasso(alpha=ls_cv.alpha_)
ls.fit(X_train, y_train)
plt.scatter(ls.predict(X_train), y_train)
plt.xlabel("Predicted value of Y")
plt.ylabel("Real value of Y")
plt.show()
print("Train score: ", ls.score(X_train, y_train) * 100)
print("Test score: ", ls.score(X_test, y_test) * 100)
# ### Ridge Regression
from sklearn.linear_model import Ridge, RidgeCV
alphas = np.random.uniform(0, 10, 50)
r_cv = RidgeCV(alphas=alphas, cv=10)
r_cv.fit(X_train, y_train)
alpha = r_cv.alpha_
alpha
ri = Ridge(alpha=r_cv.alpha_)
ri.fit(X_train, y_train)
plt.scatter(ri.predict(X_train), y_train)
plt.xlabel("Predicted value of Y")
plt.ylabel("Real value of Y")
plt.show()
print("Train score: ", ri.score(X_train, y_train) * 100)
print("Test score: ", ri.score(X_test, y_test) * 100)
# # Here I will be using Decision tree for output since it has the best score and does not seem to be overfitting or underfitting.
# Export pickle file for webapp
# import pickle
# filename = 'webapp-pickle-dtm'
# pickle.dump(dtm,open(filename,'wb'))
# model = pickle.load(open(filename, 'rb'))
# user_input=[[0,53,0,4,0,4,10,2021]]
# user_input=np.array(user_input)
# prediction = model.predict(user_input)
# prediction
|
# Progetto Big Data & Analytics a.a. 2022/2023
# Kaggle Competition : " UW-Madison GI Tract Image Segmentation"
# Prof : Roberto Pirrone , Studente : Luca La Barbera
# Corso di Laurea Magistrale in Ingegneria Informatica - Università degli Studi di Palermo.
# Presentazione della Competition
# Descrizione Generale
# In questa competizione, si dovrà creare un modello per segmentare automaticamente lo stomaco e l'intestino nelle scansioni MRI.
# Le scansioni MRI provengono da pazienti oncologici reali che hanno effettuato 1-5 scansioni MRI in giorni diversi durante il loro trattamento con radiazioni. Dovrete basare il vostro algoritmo su un set di dati di queste scansioni per trovare soluzioni creative di deep learning che aiutino i pazienti oncologici a ricevere cure migliori.
# Descrizione del Dataset
# In questa competizione stiamo segmentando le cellule degli organi nelle immagini. Le annotazioni di addestramento sono fornite come maschere codificate in RLE e le immagini sono in formato PNG a 16 bit in scala di grigi.
# Ogni caso in questa competizione è rappresentato da più serie di fette di scansione (ogni serie è identificata dal giorno in cui è stata effettuata la scansione). Alcuni casi sono divisi per tempo (i primi giorni sono in train, i giorni successivi sono in test), mentre altri sono divisi per caso - l'intero caso è in train o in test. L'obiettivo di questa competizione è quello di essere in grado di generalizzare sia a casi parzialmente che interamente non visti.
# Si noti che, in questo caso, il set di test è interamente inedito. Si tratta di circa 50 casi, con un numero variabile di giorni e fette, come nel set di addestramento.
# Attenzione : Si noti che i nomi dei file delle immagini includono 4 numeri (es. 276_276_1.63_1.63.png). Questi quattro numeri sono la larghezza/altezza della fetta (numeri interi in pixel) e la distanza tra i pixel di larghezza/altezza (punti fluttuanti in mm). I primi due definiscono la risoluzione della diapositiva. Gli ultimi due registrano la dimensione fisica di ciascun pixel.
# Inizio del Codice
# Import delle librerie utili
# In Questa sezione vengono importate tutte le librerie utili per il data mining e l'alaborazione delle immagini come : Pandas, Numpy, PIL.Image.
# Si è scelto di implementare il modello utilizzando il frameword Tensorflow
# Importo le Librerie Utili
import numpy as np
import pandas as pd
pd.options.plotting.backend = "plotly"
import random
from glob import glob
import os, shutil
from tqdm import tqdm
tqdm.pandas()
import time
import copy
import joblib
from collections import defaultdict
import gc
from IPython import display as ipd
# visualizzazione
import matplotlib.pyplot as plt
from matplotlib.patches import Rectangle
# Sci-Kit Learn import
from sklearn.model_selection import StratifiedKFold, KFold, StratifiedGroupKFold
import tensorflow as tf
from tensorflow import keras
import PIL
from PIL import Image
import cv2
import warnings
warnings.filterwarnings("ignore")
import matplotlib.pyplot as plt
# For descriptive error messages
os.environ["CUDA_LAUNCH_BLOCKING"] = "1"
# Raccolta dei dati
# In questa sezione avviene la raccolta dei dati e la creazione dei dataframe, impostandone i campi in maniera tale da potere essere disponibili per l'elaborazione .
# Vengono implementate due funzioni di utilità per ricavare i dati a partire dai nomi dei file e delle cartelle dei dataset che vengono forniti dalla competition Kaggle :
# 1. funzione wordMap(x, s, e ) : questa funzione permette di mappare all'interno di una stringa una sottostringa passando :
# a. x : la stringa originale
# b. s : la stringa che la precede
# c. e : la stringa che la segue
# Ritorna la sottostringa desiderata
# 2. funzione pathMatch(dir, path) questa funzione permette di effettuare il matching di un file data la cartella e il pattern del FileName
# Prende in Input :
# a. dir : la cartella dentro cui cercare il file da matchare
# b. path : il path da matchare
# Ritorna il nome del file
# Elaborazione del Dataframe a partire dal file .csv
# La funzione che si occupa di creare il Dataframe è la "getProcessedDF(path, mode="train")".
# Questa funzione prend in input il path del file .csv e la "mode" ovvero la modalità con cui processare il file .
# Vengono individuate due elaborazioni differenti in base alla modalità :
# 1. La modalità "train" : il dataset viene elaborato in maniera tale da ottenere un dataframe con i campi utili per l'elaborazine sucessiva sulle immagini
# 2. La modalità diversa da train -> ovvero test set mode : il dataframe viene elaborato per la essere dato in submit per la competition.
import fnmatch
# metodo che permette di mappare all'interno di una stringa una sottostringa passando
# x : la stringa originale, s : la stringa che la precede , e : la stringa che la segue
def wordMap(x, s, e):
start = x.index(s) + len(s)
end = x.index(e, start)
subw = x[start:end]
return subw
# metodo che permette di effettuare il matching di un file data la cartella e il pattern del FileName
def pathMatch(dir, path):
files = os.scandir(dir)
for file in files:
if fnmatch.fnmatch(file.name, path):
return dir + "/" + file.name
# path delle immagini e del csv
images_path = "uw-madison-gi-tract-image-segmentation/train/"
train_csv_path = "uw-madison-gi-tract-image-segmentation/train.csv"
# definizione dei campi del dataframe , funzione che processa il file .csv e restituisce un df strutturato per l'elaborazione
def getProcessedDF(path, mode="train"):
data = pd.read_csv(path)
df = data.pivot_table(
values="segmentation", index="id", columns="class", aggfunc="max"
).reset_index()
df2 = data.drop_duplicates(subset=["id"])
df3 = pd.DataFrame(columns=["id", "large_bowel", "small_bowel", "stomach"])
df3["id"] = df2[~df2["id"].isin(df["id"])].id.values
df = pd.concat([df, df3])
df.reset_index(inplace=True, drop=True)
if mode == "train":
df["id"] = df.id.map(lambda x: x)
df["case"] = df.id.map(lambda x: int(x.split("_")[0].replace("case", "")))
df["day"] = df.id.map(lambda x: wordMap(x, "_day", "_"))
df["slice"] = df.id.map(lambda x: x.split("_")[-1])
df["image_path"] = df.id.map(
lambda x: images_path
+ wordMap(x, "", "_")
+ "/"
+ wordMap(x, "", "_")
+ "_day"
+ wordMap(x, "_day", "_")
+ "/scans/slice_"
+ x.split("_")[-1]
+ "|"
)
df["image_path"] = df.image_path.map(
lambda x: pathMatch(
wordMap(x, "", "/slice_"), wordMap(x, "scans/", "|") + "*.png"
)
)
df["width"] = df["image_path"].apply(lambda x: int(x[:-4].rsplit("_", 4)[1]))
df["height"] = df["image_path"].apply(lambda x: int(x[:-4].rsplit("_", 4)[2]))
df["empty"] = df.id
for i in range(len(df.id)):
if str(df["large_bowel"].iloc[i]) == "nan":
df["large_bowel"].iloc[i] = ""
if str(df["small_bowel"].iloc[i]) == "nan":
df["small_bowel"].iloc[i] = ""
if str(df["stomach"].iloc[i]) == "nan":
df["stomach"].iloc[i] = ""
df["empty"].iloc[i] = (
df["large_bowel"].iloc[i] == ""
and df["small_bowel"].iloc[i] == ""
and df["stomach"].iloc[i] == ""
)
df["count"] = np.sum(df.iloc[:, 1:4] != "", axis=1).values
else:
df["id"] = df.id.map(lambda x: x)
df["case"] = df.id.map(lambda x: int(x.split("_")[0].replace("case", "")))
df["day"] = df.id.map(lambda x: wordMap(x, "_day", "_"))
df["slice"] = df.id.map(lambda x: x.split("_")[-1])
df["image_path"] = df.id.map(
lambda x: images_path
+ wordMap(x, "", "_")
+ "/"
+ wordMap(x, "", "_")
+ "_day"
+ wordMap(x, "_day", "_")
+ "/scans/slice_"
+ x.split("_")[-1]
+ "|"
)
df["image_path"] = df.image_path.map(
lambda x: pathMatch(
wordMap(x, "", "/slice_"), wordMap(x, "scans/", "|") + "*.png"
)
)
df["width"] = df["image_path"].apply(lambda x: int(x[:-4].rsplit("_", 4)[1]))
df["height"] = df["image_path"].apply(lambda x: int(x[:-4].rsplit("_", 4)[2]))
df.sort_values(by=["case", "day", "slice"], ignore_index=True, inplace=True)
return df
df = getProcessedDF(path=train_csv_path, mode="train")
df
# Rimozione errori
fault1 = "case7_day0"
fault2 = "case81_day30"
df = df[~df["id"].str.contains(fault1) & ~df["id"].str.contains(fault2)].reset_index(
drop=True
)
df.to_csv("prova.csv")
gc.collect()
# Distribuzione delle classi
# Plotting di un Istogramma per evidenziare quanti casi presentano la maschera di segmentazione e quanti no.
df["empty"].value_counts().plot.bar(color=["blue", "red"])
df.info()
# RLE Function
# In questa sezione vengono implelemtate le due funzioni per la codifica e la decodifica RLE
# Codifica RLE: Run-length encoding
# L'algoritmo di RLE cerca nei dati da comprimere una serie di elementi uguali (in un'immagine bitmap, essa corrisponde ad una campitura piatta), e la sostituisce con un solo elemento, quindi un carattere speciale e infine il numero di volte che esso va ripetuto. Per esempio supponiamo di avere un'immagine dove la prima riga è formata da cento pixel neri, il RLE memorizzerà il primo pixel nero poi metterà il carattere speciale e in seguito memorizzerà il numero 100. Così invece di occupare cento locazioni la prima riga ne occuperà solo 3. Il carattere speciale è definito diversamente da ogni implementazione dell'algoritmo, e serve a distinguere un elemento normale da uno compresso.
# Questo algoritmo funziona bene in presenza di immagini con pochi colori molto uniformi, ovvero in serie di dati che abbiano molte ripetizioni al loro interno. Attualmente è utilizzato solo in alcune immagini bitmap; per esempio le bitmap utilizzate sui sistemi Microsoft possono essere compresse con RLE.
#
def rle_decode(mask_rle, height, width):
mask_rle = [int(k) for k in mask_rle.split()]
start_index = mask_rle[::2]
length = mask_rle[1::2]
start_index = [x - 1 for x in start_index]
end_index = [x + y for x, y in zip(start_index, length)]
img = np.zeros(height * width)
for i in range(len(start_index)):
img[start_index[i] : end_index[i]] = 255
return img.reshape((height, width)).astype(np.uint8)
# Run-length encoding
def rle_encode(img):
"""
img: numpy array, 1 - mask, 0 - background
Returns run length as string formated
"""
pixels = img.flatten()
pixels = np.concatenate([[0], pixels, [0]])
runs = np.where(pixels[1:] != pixels[:-1])[0] + 1
runs[1::2] -= runs[::2]
return " ".join(str(x) for x in runs)
# Funzioni di utilità per il calcolo di maschere e per la gestione delle immagini
# Visualizzazione delle Immagini
# In questa sezione si mostrano a campione delle immagini
# importiamo una immagine di classe "large_bowel" con segmentazione non nulla
example_row = df[df["empty"] == False]
example_row = example_row[example_row["height"] > 300].iloc[0, :]
print(example_row)
gc.collect()
path_img = example_row.image_path
path_img
# definizione di un metoto per plottare una immagine e le sue info
# display an image
# + image size
# + values of 100th row
import matplotlib.pyplot as plt
def print_img_info(img):
img_arr = np.asarray(img)
print(img_arr.shape)
print(img_arr[100])
plt.imshow(img, cmap="bone")
img = Image.open(example_row.image_path)
print_img_info(img)
mask_large_bowel = example_row.large_bowel
mask_small_bowel = example_row.small_bowel
mask_stomach = example_row.stomach
mask = rle_decode(mask_stomach, width=example_row.width, height=example_row.height)
print(mask.shape)
print(mask)
plt.imshow(mask)
mask = rle_decode(mask_small_bowel, width=example_row.width, height=example_row.height)
print(mask.shape)
print(mask)
plt.imshow(mask)
mask = rle_decode(mask_large_bowel, width=example_row.width, height=example_row.height)
print(mask.shape)
print(mask)
plt.imshow(mask)
gc.collect()
num = 5
segmentation_df_example = df[df["empty"] == False].sample(num)
import matplotlib.colors as col
fig, ax = plt.subplots(num, 5, figsize=(18, 8 * num))
colors = ["yellow", "green", "red"]
cmap1 = col.ListedColormap(colors[0])
cmap2 = col.ListedColormap(colors[1])
cmap3 = col.ListedColormap(colors[2])
for i in range(num):
record = segmentation_df_example.iloc[i, :]
img = tf.keras.preprocessing.image.load_img(
record.image_path, color_mode="grayscale"
)
img = tf.keras.preprocessing.image.img_to_array(img)
ax[i, 0].imshow(img[:, :, 0], cmap="bone")
ax[i, 0].set_title(record.id)
large_bowel = rle_decode(
record.large_bowel, width=record.width, height=record.height
)
small_bowel = rle_decode(
record.small_bowel, width=record.width, height=record.height
)
stomach = rle_decode(record.stomach, width=record.width, height=record.height)
ax[i, 1].imshow(large_bowel, cmap="bone")
ax[i, 1].set_title("Large Bowel Mask")
ax[i, 2].imshow(small_bowel, cmap="bone")
ax[i, 2].set_title("Small Bowel Mask")
ax[i, 3].imshow(stomach, cmap="bone")
ax[i, 3].set_title("Stomach Mask")
ax[i, 4].imshow(img[:, :, 0], cmap="bone")
if record.large_bowel != "":
ax[i, 4].imshow(
np.ma.masked_where(large_bowel == False, large_bowel), cmap=cmap1, alpha=1
)
if record.small_bowel != "":
ax[i, 4].imshow(
np.ma.masked_where(small_bowel == False, small_bowel), cmap=cmap2, alpha=1
)
if record.stomach != "":
ax[i, 4].imshow(
np.ma.masked_where(stomach == False, stomach), cmap=cmap3, alpha=1
)
ax[i, 4].set_title("IMG + MASKs")
gc.collect()
# Preprocessing
# In questa sezione vengono valutate ed effettuate tutte quelle operazioni di preprocessing utili per l'ottimizazione del calcolo del modello come il Resizing, Nomalizzione dei valori dei Pixel delle Immagini e codifica a 8 bit ("uint8").
# Al termine verranno scelti quelli che sono i parametri e i metodi che restituiscono risultati migliori per la pre-elaborazione.
# Funzioni di Preprocessing
# In questa sezione vengono applicate delle funzioni di preprocessing su un campione, successivamente viene costruita una pipeline di preprocessing e applicata a 5 campioni per validare il processo di preprocessing e applicarlo alla pipeline per il modello
def preprocess(img_path, width, height):
img = Image.open(img_path).resize((width, height))
img = (img - np.min(img)) * (1 / (np.max(img) - np.min(img)))
img = np.expand_dims(img, axis=-1)
return img
sample1 = segmentation_df_example.iloc[0, :]
img_s = Image.open(sample1.image_path)
print_img_info(img_s)
gc.collect()
img_s = preprocess(sample1.image_path, 128, 128)
print_img_info(img_s)
gc.collect()
# Preprocessing su Più esempi diversi
import matplotlib.colors as col
const_resize = (128, 128)
fig, ax = plt.subplots(num, 5, figsize=(18, 8 * num))
colors = ["yellow", "green", "red"]
cmap1 = col.ListedColormap(colors[0])
cmap2 = col.ListedColormap(colors[1])
cmap3 = col.ListedColormap(colors[2])
for i in range(num):
record = segmentation_df_example.iloc[i, :]
img = preprocess(record.image_path, 128, 128)
ax[i, 0].imshow(img, cmap="bone")
ax[i, 0].set_title(record.id)
large_bowel = rle_decode(
record.large_bowel, width=record.width, height=record.height
)
small_bowel = rle_decode(
record.small_bowel, width=record.width, height=record.height
)
stomach = rle_decode(record.stomach, width=record.width, height=record.height)
large_bowel = np.array(Image.fromarray(large_bowel).resize(const_resize))
large_bowel = (mask == 255).astype(np.float32)
small_bowel = np.array(Image.fromarray(small_bowel).resize(const_resize))
small_bowel = (small_bowel == 255).astype(np.float32)
stomach = np.array(Image.fromarray(stomach).resize(const_resize))
stomach = (stomach == 255).astype(np.float32)
ax[i, 1].imshow(large_bowel, cmap="bone")
ax[i, 1].set_title("Large Bowel Mask")
ax[i, 2].imshow(small_bowel, cmap="bone")
ax[i, 2].set_title("Small Bowel Mask")
ax[i, 3].imshow(stomach, cmap="bone")
ax[i, 3].set_title("Stomach Mask")
ax[i, 4].imshow(img, cmap="bone")
if record.large_bowel != "":
ax[i, 4].imshow(
np.ma.masked_where(large_bowel == False, large_bowel), cmap=cmap1, alpha=1
)
if record.small_bowel != "":
ax[i, 4].imshow(
np.ma.masked_where(small_bowel == False, small_bowel), cmap=cmap2, alpha=1
)
if record.stomach != "":
ax[i, 4].imshow(
np.ma.masked_where(stomach == False, stomach), cmap=cmap3, alpha=1
)
ax[i, 4].set_title("IMG + MASKs")
gc.collect()
# Generazione dei dataset Train e Test
# Sezione nella quale vengono creati i set di test e validation
# Definizione dei parametri per il modello
# Dopo vari valori, questi scelti per i parametri hanno restituito i migliori risultati in fase di addestramento.
# In Particolare per quanto riguarda la BATCH_SIZE si è scelto di avere dei batch di 64 poichè questo valore permette di avere la giusta probabilita che il batch singolo riesca ad intercettare la giusta distribuzione di probabilità dei dati.
# Scegliere un batch size troppo piccolo aumenta la probabilità di incappare in batch sfortunati che sembrano provenire da una distribuzione di probabilità diversa.
#
BATCH_SIZE = 64
EPOCHS = 40
n_spilts = 5
fold_selected = 2
IMG_SIZE = 128
classes = ["large_bowel", "small_bowel", "stomach"]
# Creazione del train-set e del validation-set
# Data Generator
# La Classe DataGenerator è una classe di utilità per la generazione dei dataset di immagini.
# Una volta caricare le immagini vengono pre elaborate e aggiunte ad un array X di immagini di input per il modello.
# Vengono anche create le maschere per le immagini e aggiunte all'array delle labels y .
#
from tensorflow import keras
from keras.utils import Sequence
from PIL import Image
import numpy as np
import tensorflow as tf
import cv2
class DataGenerator(Sequence):
def __init__(
self, df, batch_size=BATCH_SIZE, width=IMG_SIZE, height=IMG_SIZE, mode="train"
):
self.df = df
self.batch_size = batch_size
self.width = width
self.height = height
self.indexes = np.arange(len(df))
self.mode = mode
def __len__(self):
return int(len(self.df) // self.batch_size)
def __load_image(self, img_path):
img = preprocess(img_path, width=self.width, height=self.height)
return img.astype(np.float32)
def __getitem__(self, index):
X = np.empty((self.batch_size, self.height, self.width, 3))
y = np.empty((self.batch_size, self.height, self.width, 3))
indexes = self.indexes[index * self.batch_size : (index + 1) * self.batch_size]
for i, img_path in enumerate(self.df["image_path"].iloc[indexes]):
# Load the image
image = self.__load_image(img_path)
w = self.df["width"].iloc[indexes[i]]
h = self.df["height"].iloc[indexes[i]]
X[i,] = image
if self.mode == "train":
for k, j in enumerate(["stomach", "large_bowel", "small_bowel"]):
rle = self.df[j].iloc[indexes[i]]
mask = rle_decode(rle, height=h, width=w)
mask = np.array(
Image.fromarray(mask).resize((self.height, self.width))
)
mask = (mask == 255).astype(np.float32)
y[i, :, :, k] = mask
if self.mode == "train":
return (X, y)
return X
# Definizione delle Metriche e Loss
# Dice Coeff : L'indice è conosciuto con diversi altri nomi, in particolare indice di Sørensen-Dice, indice di Sørensen e coefficiente di Dice. Altre varianti includono il "coefficiente di somiglianza" o "indice", come il coefficiente di somiglianza di Dice (DSC). Le grafie alternative comuni per Sørensen sono Sorenson, Soerenson e Sörenson, e tutte e tre possono essere viste anche con la desinenza -sen.
# Link wikipedia per la formula : Dice Coeff Wiki
#
import keras.backend as K
from keras.losses import binary_crossentropy
import tensorflow as tf
def iou_coef(y_true, y_pred, smooth=1):
intersection = K.sum(K.abs(y_true * y_pred), axis=[1, 2, 3])
union = K.sum(y_true, [1, 2, 3]) + K.sum(y_pred, [1, 2, 3]) - intersection
iou = K.mean((intersection + smooth) / (union + smooth), axis=0)
return iou
def dice_coef(y_true, y_pred, threshold=0.5):
y_true_f = K.flatten(y_true)
y_pred_f = K.flatten(y_pred)
y_pred_f = K.cast(K.greater_equal(y_pred_f, threshold), "float32")
intersection = K.sum(y_true_f * y_pred_f)
dice_coef = (2 * intersection + 1) / (K.sum(y_true_f) + K.sum(y_pred_f) + 1)
return dice_coef
def dice_coef_loss(y_true, y_pred, epsilon=1e-5):
return 1 - (2.0 * K.sum(y_true * y_pred) + epsilon) / (
K.sum(y_true) + K.sum(y_pred) + epsilon
)
def bce_dice_loss(y_true, y_pred):
return binary_crossentropy(tf.cast(y_true, tf.float32), y_pred) + dice_coef_loss(
tf.cast(y_true, tf.float32), y_pred
)
# Train Set e Validation Set
# Si è scelto di dividere il dataframe in test set e validation set seguendo le percentuali 75% - 25%.
# Infatti queste hanno permesso di ottenere i migliori risultati in fase di addestramento e validazione
#
from sklearn.model_selection import train_test_split
train_set, val_set = train_test_split(df, test_size=0.25, shuffle=True, random_state=42)
train_set.info()
val_set.info()
train_generator = DataGenerator(train_set)
val_generator = DataGenerator(val_set)
# Costruzione del Modello
# Come modello di segmentazione viene scelta U-Net, un modello di segmentazione in Keras.
# U-Net è un metodo di segmentazione semantica originariamente proposto per la segmentazione delle immagini mediche. È uno dei primi modelli di segmentazione deep learning e l'architettura U-Net è utilizzata anche in molte varianti GAN, come il generatore Pix2Pix.
# Struttura di U-Net
# MODELLO CON SEGMENTATION MODELS
import segmentation_models as sm
sm.set_framework("tf.keras")
sm.framework()
from segmentation_models import Unet
from segmentation_models.utils import set_trainable
model = Unet(
"resnet34",
input_shape=(IMG_SIZE, IMG_SIZE, 3),
classes=3,
activation="sigmoid",
encoder_weights="imagenet",
)
model.compile(optimizer="adam", loss=dice_coef_loss, metrics=[dice_coef])
# Model summary
model.summary()
gc.collect()
# callbacks
from tensorflow.keras.callbacks import (
EarlyStopping,
LearningRateScheduler,
ReduceLROnPlateau,
ModelCheckpoint,
)
checkpoint = ModelCheckpoint(
"UNET_model",
monitor="val_loss",
verbose=1,
save_best_only=True,
save_weights_only=False,
mode="auto",
)
# Stop early if no improvement seen
early_stopping = EarlyStopping(
patience=5,
min_delta=0.0001,
restore_best_weights=True,
)
# Reduce learning rate on plateau
lr_plateau = keras.callbacks.ReduceLROnPlateau(
monitor="val_loss",
factor=0.1,
patience=5,
verbose=0,
min_delta=0.0001,
)
model.save("final_model")
# Train model
history = model.fit(
train_generator,
validation_data=val_generator,
callbacks=[checkpoint, early_stopping],
use_multiprocessing=False,
workers=4,
epochs=EPOCHS,
)
# Risultati Addestramento
# In Questa sezione vengono riportati i risultati dell'addestramento.
# Al termine dell'addesramento il modello avente con gli iperparametri precedentemente specificati ha raggiunto i seguenti risultati :
# loss: 0.0925 - dice_coef: 0.9077 - val_loss: 0.1262 - val_dice_coef: 0.8739 (Epoca 39/40)
hist_df = pd.DataFrame(history.history)
hist_df.to_csv("history.csv")
# PLOT TRAINING
plt.figure(figsize=(15, 5))
plt.subplot(1, 3, 1)
plt.plot(range(history.epoch[-1] + 1), history.history["loss"], label="Train_Loss")
plt.plot(range(history.epoch[-1] + 1), history.history["val_loss"], label="Val_loss")
plt.title("LOSS")
plt.xlabel("Epoch")
plt.ylabel("loss")
plt.legend()
plt.subplot(1, 3, 2)
plt.plot(
range(history.epoch[-1] + 1), history.history["dice_coef"], label="Train_dice_coef"
)
plt.plot(
range(history.epoch[-1] + 1),
history.history["val_dice_coef"],
label="Val_dice_coef",
)
plt.title("DICE")
plt.xlabel("Epoch")
plt.ylabel("dice_coef")
plt.legend()
plt.show()
# Mostro alcune Predizioni
import matplotlib.colors as col
pred_batches = DataGenerator(val_set.iloc[203:208, :], batch_size=1)
preds = model.predict_generator(pred_batches, verbose=1)
Threshold = 0.5
# Visualizing
fig, ax = plt.subplots(num, 3, figsize=(18, 8 * num))
colors = ["yellow", "green", "red"]
labels = ["Large Bowel", "Small Bowel", "Stomach"]
cmap1 = col.ListedColormap(colors[0])
cmap2 = col.ListedColormap(colors[1])
cmap3 = col.ListedColormap(colors[2])
for i in range(5):
images, mask = pred_batches[i]
sample_img = images[0, :, :, 0]
mask1 = mask[0, :, :, 0]
mask2 = mask[0, :, :, 1]
mask3 = mask[0, :, :, 2]
pre = preds[i]
predict1 = pre[:, :, 0]
predict2 = pre[:, :, 1]
predict3 = pre[:, :, 2]
predict1 = (predict1 > Threshold).astype(np.float32)
predict2 = (predict2 > Threshold).astype(np.float32)
predict3 = (predict3 > Threshold).astype(np.float32)
ax[i, 0].imshow(sample_img, cmap="bone")
ax[i, 0].set_title("Image", fontsize=12, y=1.01)
# --------------------------
ax[i, 1].imshow(sample_img, cmap="bone")
ax[i, 1].set_title("Mask", fontsize=12, y=1.01)
l0 = ax[i, 1].imshow(sample_img, cmap="bone")
l1 = ax[i, 1].imshow(np.ma.masked_where(mask1 == False, mask1), cmap=cmap1, alpha=1)
l2 = ax[i, 1].imshow(np.ma.masked_where(mask2 == False, mask2), cmap=cmap2, alpha=1)
l3 = ax[i, 1].imshow(np.ma.masked_where(mask3 == False, mask3), cmap=cmap3, alpha=1)
# --------------------------
ax[i, 2].set_title("Predict", fontsize=12, y=1.01)
l0 = ax[i, 2].imshow(sample_img, cmap="bone")
l1 = ax[i, 2].imshow(
np.ma.masked_where(predict1 == False, predict1), cmap=cmap1, alpha=1
)
l2 = ax[i, 2].imshow(
np.ma.masked_where(predict2 == False, predict2), cmap=cmap2, alpha=1
)
l3 = ax[i, 2].imshow(
np.ma.masked_where(predict3 == False, predict3), cmap=cmap3, alpha=1
)
# Test Set -- con la submission viene rilasciato il testset reale ---
test_dir = "/kaggle/input/uw-madison-gi-tract-image-segmentation/sample_submission.csv"
test_df = getProcessedDF(path=test_dir, mode="test")
if len(test_df) == 0:
test_df = df.iloc[: 10 * 64 * 3, :]
test_df = test_df.drop(
columns=["large_bowel", "small_bowel", "stomach", "empty", "count"]
)
test_df.head()
from keras.models import load_model
custom_objects = custom_objects = {
"dice_coef": dice_coef,
"dice_coef_loss": dice_coef_loss,
}
model_path = "/kaggle/working/UNET_model"
model = load_model(model_path, custom_objects=custom_objects)
from PIL import Image
pred_batches = DataGenerator(test_df, batch_size=BATCH_SIZE, mode="test")
num_batches = int(len(test_df) / BATCH_SIZE)
submission = pd.read_csv(
"/kaggle/input/uw-madison-gi-tract-image-segmentation/sample_submission.csv"
)
for i in range(num_batches):
# Predizioni
preds = model.predict(pred_batches[i], verbose=0)
# Codifica RLE
for j in range(BATCH_SIZE):
for k in range(3):
classes = {0: "large_bowel", 1: "small_bowel", 2: "stomach"}
pred_img = cv2.resize(
preds[j, :, :, k],
(
test_df.loc[i * BATCH_SIZE + j, "width"],
test_df.loc[i * BATCH_SIZE + j, "height"],
),
interpolation=cv2.INTER_NEAREST,
)
pred_img = (pred_img > 0.5).astype(dtype="uint8") # classify
submission.loc[3 * (i * BATCH_SIZE + j) + k, "predicted"] = rle_encode(
pred_img
)
submission.loc[3 * (i * BATCH_SIZE + j) + k, "id"] = test_df.loc[
i * BATCH_SIZE + j, "id"
]
submission.loc[3 * (i * BATCH_SIZE + j) + k, "class"] = classes.get(k)
submission.to_csv("submission.csv", index=False)
submission
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the "../input/" directory.
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Any results you write to the current directory are saved as output.
# Use this as path to input the database file "conn = sqlite3.connect('/kaggle/input/ipldatabase/database.sqlite')"
import sqlite3
import matplotlib.pyplot as plt
import seaborn as sns
# path = "../input/" #Insert path here
# database = path + 'database.sqlite'
# from subprocess import check_output
conn = sqlite3.connect("/kaggle/input/ipldatabase/database.sqlite")
# to check the table names in the database
tables = pd.read_sql_query(
"""SELECT *
FROM sqlite_master
WHERE type='table';""",
conn,
)
tables
countries = pd.read_sql_query(
"""SELECT *
FROM Country;""",
conn,
)
countries
teams = pd.read_sql(
"""SELECT *
FROM Team
""",
conn,
)
teams
# Joining tables Team_id and Player_match with match to get the team performance
team_table = pd.read_sql_query(
"""Select m.Match_id,Team_1,Team_2,Team_Name,Match_winner,Win_type,Toss_Decide from Match m
join Player_Match pm
on pm.match_id = m.match_id
join team t
on t.team_id = pm.team_id""",
conn,
)
team_table.head(20)
# connecting table Player_Match, Player adn Rolee to ccheck the roles of players and their skills only for country =1 i.e India
role = pd.read_sql_query(
"""Select pm.Role_Id,Role_Desc,p.Player_Id,Player_Name,Country_Name,Batting_hand,Bowling_skill
from Rolee r join Player_match pm
on r.Role_Id = pm.Role_Id
join Player p
on p.Player_Id = pm.Player_Id
where Country_Name=1
order by DOB desc""",
conn,
)
role.head(20)
seasons = pd.read_sql_query("""Select * from Season order by Season_Year""", conn)
seasons
ball_by_ball = pd.read_sql_query(
"""Select bs.Match_Id,Runs_Scored,Team_batting,Team_Bowling
from Ball_by_Ball bb join Batsman_Scored bs
on bs.Match_Id = bb.Match_Id""",
conn,
)
ball_by_ball.head()
## Using Group By and having
venue_city = pd.read_sql_query(
"""Select Venue_Id, Country_Name, count(City_Name) as 'Number of Cities'
from Venue v join City ct on ct.City_Id = v.City_Id
join Country cy
on cy.Country_Id= ct.Country_Id
group by Country_Name
having Country_Name= "India" or Country_Name='U.A.E'
""",
conn,
)
venue_city
metric = pd.read_sql_query(
"""SELECT 'Matches' As Dimension , COUNT(*) As 'Measure'
FROM Match
UNION ALL
SELECT 'Extra_Runs' As Dimension , SUM(Extra_Runs.Extra_Runs) As 'Measure'
FROM Extra_Runs
UNION ALL
SELECT 'Batsman_runs' As Dimension , SUM(B.Runs_Scored) As 'Value'
FROM Batsman_Scored B
UNION ALL
SELECT 'Wickets' As Dimension , COUNT(*) As 'Measure'
FROM Wicket_Taken
UNION ALL
SELECT 'Sixes' As Dimension , COUNT(*) As 'Measure'
FROM Batsman_Scored
WHERE Batsman_Scored.Runs_Scored = 6
UNION ALL
SELECT 'Fours' As Dimension , COUNT(*) As 'Measure'
FROM Batsman_Scored
WHERE Batsman_Scored.Runs_Scored = 4
UNION ALL
SELECT 'Singles' As Dimension , COUNT(*) As 'Measure'
FROM Batsman_Scored
WHERE Batsman_Scored.Runs_Scored = 1""",
conn,
)
metric
captain = pd.read_sql(
"""SELECT C.Player_Name , COUNT(*) As 'Matches_captained'
FROM Player_Match A JOIN Rolee B
ON A.Role_Id = B.Role_Id
JOIN Player C
ON A.Player_Id = C.Player_Id
WHERE A.Role_Id = 4
GROUP BY C.Player_Name
ORDER BY Matches_captained DESC;""",
conn,
)
captain
# labels = captain["Player_Name"]
plt.figure(figsize=(12, 6))
y = captain["Matches_captained"]
x = captain["Player_Name"]
plt.bar(x, y, align="center", color="g")
plt.show()
|
# ## March Machine Learning Mania 2021 - NCAAM
# 
# **What to predict**
# **Stage 1** - You should submit predicted probabilities for every possible matchup in the past 5 NCAA® tournaments (2015-2019).
# **Stage 2** - You should submit predicted probabilities for every possible matchup before the 2021 tournament begins.
import numpy as np
import pandas as pd
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# ## Data Section 1 - The Basics
# ### Teams.csv
# This file identifies the different college teams present in the dataset. Each school is uniquely identified by a 4 digit id number. You will not see games present for all teams in all seasons, because the games listing is only for matchups where both teams are Division-I teams. There are 357 teams currently in Division-I, and an overall total of 371 teams in our team listing (each year, some teams might start being Division-I programs, and others might stop being Division-I programs). This year there are four teams that are new to Division I: Bellarmine (TeamID=1469), North Alabama (TeamID=1469), Tarleton State (TeamID=1470), and UC_San Diego (TeamID=1471) and so you will not see any historical data for these teams prior to the current season. In addition, some teams opted not to play during the 2021 season due to the impact of COVID-19 and will not have any games listed.
# * **TeamID** - a 4 digit id number, from 1000-1999, uniquely identifying each NCAA® men's team. A school's TeamID does not change from one year to the next, so for instance the Duke men's TeamID is 1181 for all seasons. To avoid possible confusion between the men's data and the women's data, all of the men's team ID's range from 1000-1999, whereas all of the women's team ID's range from 3000-3999.
# * **TeamName** - a compact spelling of the team's college name, 16 characters or fewer. There are no commas or double-quotes in the team names, but you will see some characters that are not letters or spaces, e.g., Texas A&M, St Mary's CA, TAM C. Christi, and Bethune-Cookman.
# * **FirstD1Season** - the first season in our dataset that the school was a Division-I school. For instance, FL Gulf Coast (famously) was not a Division-I school until the 2008 season, despite their two wins just five years later in the 2013 NCAA® tourney. Of course, many schools were Division-I far earlier than 1985, but since we don't have any data included prior to 1985, all such teams are listed with a FirstD1Season of 1985.
# * **LastD1Season** - the last season in our dataset that the school was a Division-I school. For any teams that are currently Division-I, they will be listed with LastD1Season=2021, and you can confirm there are 357 such teams.
#
teams = pd.read_csv("/kaggle/input/ncaam-march-mania-2021/MTeams.csv")
teams.head()
teams.shape
# ## MSeasons.csv
# This file identifies the different seasons included in the historical data, along with certain season-level properties.
# * **Season** - indicates the year in which the tournament was played. Remember that the current season counts as 2021.
# * **DayZero** - tells you the date corresponding to DayNum=0 during that season. All game dates have been aligned upon a common scale so that (each year) the Monday championship game of the men's tournament is on DayNum=154. Working backward, the national semifinals are always on DayNum=152, the "play-in" games are on days 135, Selection Sunday is on day 132, the final day of the regular season is also day 132, and so on. All game data includes the day number in order to make it easier to perform date calculations. If you need to know the exact date a game was played on, you can combine the game's "DayNum" with the season's "DayZero". For instance, since day zero during the 2011-2012 season was 10/31/2011, if we know that the earliest regular season games that year were played on DayNum=7, they were therefore played on 11/07/2011.
#
# * **RegionW, RegionX, Region Y, Region Z** - by our contests' convention, each of the four regions in the final tournament is assigned a letter of W, X, Y, or Z. Whichever region's name comes first alphabetically, that region will be Region W. And whichever Region plays against Region W in the national semifinals, that will be Region X. For the other two regions, whichever region's name comes first alphabetically, that region will be Region Y, and the other will be Region Z. This allows us to identify the regions and brackets in a standardized way in other files, even if the region names change from year to year. For instance, during the 2012 tournament, the four regions were East, Midwest, South, and West. Being the first alphabetically, East becomes W. Since the East regional champion (Ohio State) played against the Midwest regional champion (Kansas) in the national semifinals, that makes Midwest be region X. For the other two (South and West), since South comes first alphabetically, that makes South Y and therefore West is Z. So for that season, the W/X/Y/Z are East,Midwest,South,West. And so for instance, Ohio State, the #2 seed in the East, is listed in the MNCAATourneySeeds file that year with a seed of W02, meaning they were the #2 seed in the W region (the East region). We will not know the final W/X/Y/Z designations until Selection Sunday, because the national semifinal pairings in the Final Four will depend upon the overall ranks of the four #1 seeds.
#
seasons = pd.read_csv("/kaggle/input/ncaam-march-mania-2021/MSeasons.csv")
seasons.head()
seasons.head()
# ## MNCAATourneySeeds.csv
# this file identifies the seeds for all teams in each NCAA® tournament, for all seasons of historical data. Thus, there are between 64-68 rows for each year, depending on whether there were any play-in games and how many there were. In recent years the structure has settled at 68 total teams, with four "play-in" games leading to the final field of 64 teams entering Round 1 on Thursday of the first week (by definition, that is DayNum=136 each season). We will not know the seeds of the respective tournament teams, or even exactly which 68 teams it will be, until Selection Sunday on March 14, 2021 (DayNum=132).
# * **Season** - the year that the tournament was played in
# * **Seed** - this is a 3/4-character identifier of the seed, where the first character is either W, X, Y, or Z (identifying the region the team was in) and the next two digits (either 01, 02, ..., 15, or 16) tell you the seed within the region. For play-in teams, there is a fourth character (a or b) to further distinguish the seeds, since teams that face each other in the play-in games will have seeds with the same first three characters. The "a" and "b" are assigned based on which Team ID is lower numerically. As an example of the format of the seed, the first record in the file is seed W01 from 1985, which means we are looking at the #1 seed in the W region (which we can see from the "MSeasons.csv" file was the East region).
# * **TeamID** - this identifies the id number of the team, as specified in the MTeams.csv file
#
MNCAATourneySeeds = pd.read_csv("../input/ncaam-march-mania-2021/MNCAATourneySeeds.csv")
MNCAATourneySeeds.head()
MNCAATourneySeeds.shape
# ## MRegularSeasonCompactResults.csv
# This file identifies the game-by-game results for many seasons of historical data, starting with the 1985 season (the first year the NCAA® had a 64-team tournament). For each season, the file includes all games played from DayNum 0 through 132. It is important to realize that the "Regular Season" games are simply defined to be all games played on DayNum=132 or earlier (DayNum=132 is Selection Sunday, and there are always a few conference tournament finals actually played early in the day on Selection Sunday itself). Thus a game played on or before Selection Sunday will show up here whether it was a pre-season tournament, a non-conference game, a regular conference game, a conference tournament game, or whatever.
# Season - this is the year of the associated entry in MSeasons.csv (the year in which the final tournament occurs). For example, during the 2016 season, there were regular season games played between November 2015 and March 2016, and all of those games will show up with a Season of 2016.
# DayNum - this integer always ranges from 0 to 132, and tells you what day the game was played on. It represents an offset from the "DayZero" date in the "MSeasons.csv" file. For example, the first game in the file was DayNum=20. Combined with the fact from the "MSeasons.csv" file that day zero was 10/29/1984 that year, this means the first game was played 20 days later, or 11/18/1984. There are no teams that ever played more than one game on a given date, so you can use this fact if you need a unique key (combining Season and DayNum and WTeamID). In order to accomplish this uniqueness, we had to adjust one game's date. In March 2008, the SEC postseason tournament had to reschedule one game (Georgia-Kentucky) to a subsequent day because of a tornado, so Georgia had to actually play two games on the same day. In order to enforce this uniqueness, we moved the game date for the Georgia-Kentucky game back to its original scheduled date.
# WTeamID - this identifies the id number of the team that won the game, as listed in the "MTeams.csv" file. No matter whether the game was won by the home team or visiting team, or if it was a neutral-site game, the "WTeamID" always identifies the winning team.
# WScore - this identifies the number of points scored by the winning team.
# LTeamID - this identifies the id number of the team that lost the game.
# LScore - this identifies the number of points scored by the losing team. Thus you can be confident that WScore will be greater than LScore for all games listed.
# WLoc - this identifies the "location" of the winning team. If the winning team was the home team, this value will be "H". If the winning team was the visiting team, this value will be "A". If it was played on a neutral court, then this value will be "N". Sometimes it is unclear whether the site should be considered neutral, since it is near one team's home court, or even on their court during a tournament, but for this determination we have simply used the Kenneth Massey data in its current state, where the "@" sign is either listed with the winning team, the losing team, or neither team. If you would like to investigate this factor more closely, we invite you to explore Data Section 3, which provides the city that each game was played in, irrespective of whether it was considered to be a neutral site.
# NumOT - this indicates the number of overtime periods in the game, an integer 0 or higher.
#
MRegularSeasonCompactResults = pd.read_csv(
"../input/ncaam-march-mania-2021/MRegularSeasonCompactResults.csv"
)
MRegularSeasonCompactResults.head()
MRegularSeasonCompactResults.shape
# ## MNCAATourneyCompactResults.csv
# This file identifies the game-by-game NCAA® tournament results for all seasons of historical data. The data is formatted exactly like the MRegularSeasonCompactResults data. All games will show up as neutral site (so WLoc is always N). Note that this tournament game data also includes the play-in games (which always occurred on day 134/135) for those years that had play-in games. Thus each season you will see between 63 and 67 games listed, depending on how many play-in games there were.
# Because of the consistent structure of the NCAA® tournament schedule, you can actually tell what round a game was, depending on the exact DayNum. Thus:
# * **DayNum=134 or 135 (Tue/Wed)** - play-in games to get the tournament field down to the final 64 teams
# * **DayNum=136 or 137 (Thu/Fri)** - Round 1, to bring the tournament field from 64 teams to 32 teams
# * **DayNum=138 or 139 (Sat/Sun)** - Round 2, to bring the tournament field from 32 teams to 16 teams
# * **DayNum=143 or 144 (Thu/Fri)** - Round 3, otherwise known as "Sweet Sixteen", to bring the tournament field from 16 teams to 8 teams
# * **DayNum=145 or 146 (Sat/Sun)** - Round 4, otherwise known as "Elite Eight" or "regional finals", to bring the tournament field from 8 teams to 4 teams
# * **DayNum=152 (Sat)** - Round 5, otherwise known as "Final Four" or "national semifinals", to bring the tournament field from 4 teams to 2 teams
# * **DayNum=154 (Mon)** - Round 6, otherwise known as "national final" or "national championship", to bring the tournament field from 2 teams to 1 champion team
MNCAATourneyCompactResults = pd.read_csv(
"../input/ncaam-march-mania-2021/MNCAATourneyCompactResults.csv"
)
MNCAATourneyCompactResults.head()
MNCAATourneyCompactResults.shape
# ## MSampleSubmissionStage1.csv
# This file illustrates the submission file format for Stage 1. It is the simplest possible submission: a 50% winning percentage is predicted for each possible matchup.
# A submission file lists every possible matchup between tournament teams for one or more years. During Stage 1, you are asked to make predictions for all possible matchups from the past five NCAA® tournaments (seasons 2015, 2016, 2017, 2018, 2019). In Stage 2, you will be asked to make predictions for all possible matchups from the current NCAA® tournament (season 2021).
# When there are 68 teams in the tournament, there are 68*67/2=2,278 predictions to make for that year, so a Stage 1 submission file will have 2,278*5=11,390 data rows.
# * **ID** - this is a 14-character string of the format SSSS_XXXX_YYYY, where SSSS is the four digit season number, XXXX is the four-digit TeamID of the lower-ID team, and YYYY is the four-digit TeamID of the higher-ID team.
# * **Pred** - this contains the predicted winning percentage for the first team identified in the ID field, the one represented above by XXXX.
# Example #1: You want to make a prediction for Duke (TeamID=1181) against Arizona (TeamID=1112) in the 2017 tournament, with Duke given a 53% chance to win and Arizona given a 47% chance to win. In this case, Arizona has the lower numerical ID so they would be listed first, and the winning percentage would be expressed from Arizona's perspective (47%):
# 2017_1112_1181,0.47
# Example #2: You want to make a prediction for Duke (TeamID=1181) against North Carolina (TeamID=1314) in the 2018 tournament, with Duke given a 51.6% chance to win and North Carolina given a 48.4% chance to win. In this case, Duke has the lower numerical ID so they would be listed first, and the winning percentage would be expressed from Duke's perspective (51.6%):
# 2018_1181_1314,0.516
# Also note that a single prediction row serves as a prediction for each of the two teams' winning chances. So for instance, in Example #1, the submission row of "2017_1112_1181,0.47" specifically gives a 47% chance for Arizona to win, and doesn't explicitly mention Duke's 53% chance to win. However, our evaluation utility will automatically infer the winning percentage in the other direction, so a 47% prediction for Arizona to win also means a 53% prediction for Duke to win. And similarly, because the submission row in Example #2 gives Duke a 51.6% chance to beat North Carolina, we will automatically figure out that this also means North Carolina has a 48.4% chance to beat Duke.
MSampleSubmissionStage1 = pd.read_csv(
"../input/ncaam-march-mania-2021/MSampleSubmissionStage1.csv"
)
MSampleSubmissionStage1.head()
MSampleSubmissionStage1.head()
|
# (Case Study - 1) Analysis Books Scraping
# For this datasets use this url : https://www.kaggle.com/datasets/repl4y/books-scraping
import pandas as pd
import numpy as np
import matplotlib as mp
import matplotlib.pyplot as plt
import random as rd
df = pd.read_csv("Books_scrapingV3.csv")
# 1.Observe Column in Top and Bottom
df.shape
df.head(5)
df.tail()
df.describe().T
# 2. Check Datatype of Each Column
df.info()
# 3. Check Null In The Dataset
df.isnull().head()
df.isnull().sum()
# 4. Hight And Lowset Price of Book
df.columns
# Highest price
df[df["price"] == df.price.max()]
# Lowest price
df[df["price"] == df.price.min()]
# 5. Count The Genres of Books
# Top 5 Genre of Books
df["Genre"].value_counts()
# 6. Top 5 Genres of Books
df["Genre"].value_counts().head()
# 7. What is the average price of items in each genre?
l = df["Genre"].unique()
l2 = []
for x in l:
l2.append(int(df[df["Genre"] == x]["price"].mean()))
print(
f""" {x}
average price : {int(df[df["Genre"] == x]["price"].mean())}
"""
)
y = np.array(l[:5])
x = np.array(l2[:5])
font1 = {"color": "blue", "size": 10}
# marker|line|color
plt.plot(y, x, "v--r")
plt.title("Explore the distribution of average prices of first five items in Genres")
plt.xlabel("First five Genre", fontdict=font1)
plt.ylabel("Average value in dollars", fontdict=font1)
plt.grid()
plt.show()
# 8. Genre most Popular ?
df.columns
df["Genre"].value_counts().head(1)
# 9. What is the average star rating for each genre?
df.columns
l = df["Genre"].unique()
for x in l:
print(
f""" {x}
average star : {int(df[df["Genre"] == x]["Star"].mean())}
"""
)
# 11. How many unique UPC codes are there in the dataset?
len(df["UPC"].unique())
# 10. What is the total revenue generated by each genre, taking into account the price, tax, and number of items sold?
l = df["Genre"].unique()
for x in l:
total = df[df["Genre"] == x]["price"].sum()
print(
f"""{x}:
*Total revenue : {int(total)}
"""
)
# 12.Which item has the most reviews, and what is its star rating ?
len(df[df["Star"] == 5])
# 13.How many items have a price above a certain threshold '25DH'?
df.columns
len(df[df["price"] > 25])
# 14. Which genre has the most items in stock
df[df["Stock"] == df["Stock"].max()]["Genre"]
# Make a random number reviews ....
new_Column = (
df["Number of reviews"]
.apply(lambda x: int(rd.randrange(0, 350)))
.rename("Number of reviews RD")
)
df2 = pd.concat([df, new_Column], axis=1)
df2.drop("Number of reviews", inplace=True, axis=1)
# 15. Determine the top 5 products with the highest number of reviews.
df2.columns
df2[["Title", "price", "Star", "Number of reviews RD"]].nlargest(
5, "Number of reviews RD"
)
# 17 . Find out the average star rating of products with a price higher than $25.
df2[df2["price"] == df2["price"]]["Star"].mean()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Python Booleans - Mantıksal Operatörler
# Mantıksal operatörler iki değerden oluşur. True - False True: doğru False: Yanlış
# # Boolean Values
# Programlamada genellikle bir ifadenin Doğru mu yoksa Yanlış mı olduğunu bilmeniz gerekir.
# Python'da herhangi bir ifadeyi değerlendirebilir ve True veya False olmak üzere iki yanıttan birini alabilirsiniz.
# İki değeri karşılaştırdığınızda, ifade değerlendirilir ve Python, Boole yanıtını döndürür:
print(3 > 2)
print(5 == 4)
print(21 < 18)
# if ifadesinde bir koşul çalıştırdığınızda, Python True veya False değerini döndürür:
# Koşulun Doğru veya Yanlış olmasına bağlı olarak bir mesaj yazdıralım.
y = 50
x = 23
if x > y:
print("x y'den büyüktür")
else:
print("x y'den küçüktür")
# # Değerleri ve Değişkenleri Değerlendirme
# bool() işlevi, herhangi bir değeri değerlendirmenize ve karşılığında True veya False vermenize izin verir,
# **Örneğin:** Bir diziyi ve bir sayıyı değerlendirin:
#
print(bool("Nike"))
print(bool(8))
# iki değişkeni değerlendirme
a = "Nike"
b = 8
print(bool(a))
print(bool(b))
# # Çoğu Değer Doğrudur
# Bir tür içeriğe sahipse, hemen hemen her değer True olarak değerlendirilir.
# Boş diziler dışında tüm diziler True'dur.
# 0 dışında herhangi bir sayı True'dur.
# Boş olanlar dışında tüm liste, demet, küme ve sözlük True'dur.
# Aşağıdakiler True olarak çıktı verir
bool("ulas")
bool(21)
bool(["samsung", "lg", "apple"])
# # Bazı Değerler Yanlış
# Aslında, (), [], {}, "", 0 sayısı ve Yok değeri gibi boş değerler dışında False olarak değerlendirilen çok fazla değer yoktur. Ve elbette False değeri False olarak değerlendirilir.
# Aşağıdaki örnekler False olarak çıktı verecektir
bool(False)
bool(None)
bool(0)
bool("")
bool(())
bool([])
bool({})
# # Fonksiyonlar bir Boole Döndürebilir
# Bir Boole Değeri döndüren fonksiyonlar oluşturabilirsiniz:
def Fonksiyon():
return True
print(Fonksiyon())
# Bir işlevin Boole yanıtına göre kod çalıştırabilirsiniz:
# **Örnek** "Doğru" Yazdır işlev True döndürürse, aksi takdirde "Yanlış" yazdırın:
def Fonksiyon():
return True
if Fonksiyon():
print("Doğru")
else:
print("Yanlış")
# Python ayrıca, bir nesnenin belirli bir veri türünde olup olmadığını belirlemek için kullanılabilen isinstance() fonksiyonu gibi bir boolean değeri döndüren birçok yerleşik işleve sahiptir:
# **Örnek** Bir sayının tamsayı olup olmadığını kontrol edin:
sayi = 1232
print(isinstance(sayi, int))
a = "Müzik Dinlemeyi Severim"
print(isinstance(a, str))
# Aşağıdaki boolean hangi değeri çıktı verir.
#
print(28 > 11)
print(2 == 54)
print(7 < 2)
print(bool("ulas"))
print(bool(0))
# # Python Operatörleri
# Operatörler, değişkenler ve değerler üzerinde işlem yapmak için kullanılır.
# Aşağıdaki örnekte, iki değeri bir araya getirmek için + operatörünü kullanıyoruz:
print(7 + 9)
# Python, operatörleri aşağıdaki gruplara ayırır:
# * Arithmetic operators
# * Assignment operators
# * Comparison operators
# * Logical operators
# * Identity operators
# * Membership operators
# * Bitwise operators
# # Python Arithmetic Operators
# Aritmetik opetaörler, yaygın matematiksel işlemleri gerçekleştirmek için sayısal değerlerle birlikte kullanılır:
# Name ExampleTry it
# + Toplama x + y
# - Çıkarma x - y
# * Çarpma x * y
# / Bölme x / y
# % Modüller x % y
# ** Üst Alma x ** y
# // taban fonksiyonu x // y
# toplama (addition)
a = 2
b = 8
print(a + b)
# çıkarma (Subtraction)
c = 30
d = 21
print(c - d)
# Çarpma (Multiplication)
x = 8
y = 8
print(x * y)
# Bölme (Division)
a = 100
b = 10
print(a / b)
# modüller (modulus)
e = 8
f = 5
print(e % f)
# üst alma (Exponentiation)
u = 4
l = 6
print(x**y) # 4*4*4*4*4*4
# taban fonksiyonu (Floor division)
a = 20
b = 3
print(a // b)
# taban fonksiyonu // sonucu en yakın tam sayıya yuvarlar
# # Python Atama Operatörleri
# Atama işleçleri, değişkenlere değer atamak için kullanılır:
# = (EŞİTTİR)
a = 12
a
# += (ARTI EŞİTTİR)
a = 4
a += 5
print(a)
# -= (EKSİ EŞİTTİR)
z = 10
z -= 2
print(z)
# *= (ÇARPI EŞİTTİR)
w = 7
w *= 9
print(w)
# /= (BÖLME EŞİTTİR)
Z = 10
Z /= 4
print(Z)
# %= (bölümden kalan sayıyı verir)
e = 32
e %= 3
print(e)
# //= (kaç defa bölündüğü)
q = 212
q //= 4
print(q)
# **= (sayının üssünü alır)
f = 4
f **= 4
print(f)
# # Python Karşılaştırma Operatörleri
# Karşılaştırma işleçleri iki değeri karşılaştırmak için kullanılır:
# == Eşit mi (Equal)
a = 10
b = 8
print(a == b)
# 10, 8'e eşit olmadığı için 'False' döndürür
# != Eşit değil (Not equal)
w = 4
x = 1
print(w != x)
# 4, 1'e eşit olmadığı için 'True' döndürür
# > büyüktür (Greater than)
t = 99
y = 3
print(t > y)
# 99, 3'ten büyük olduğu için 'True' döndürür
# < küçüktür (Less than)
a = 44
c = 22
print(a < c)
# 44, 22'ten küçük olmadığı için False döndürür
# >= Büyük eşittir (reater than or equal to)
p = 99
o = 3
print(p >= o)
# 99, 3'ten büyük veya Eşit olduğu için 'True' döndürür
# küçük eşittir (Less than or equal to )
a = 9
b = 6
print(a <= b)
# # Python Mantıksal Operatörler
# Mantıksal işleçler, koşullu ifadeleri birleştirmek için kullanılır: "and, or, not"
# and, Her iki ifade de doğruysa True döndürür
a = 56
print(a > 8 and a < 70)
# a 8'den büyük ve 70 den küçük olduğu için TRUE döndürür
# or İfadelerden biri doğruysa True döndürür
b = 9
print(b > 2 or b < 10)
# b, koşullardan biri doğru olduğu için True döndürür (9, 2'ten büyüktür, ancak 9, 10'dan küçük değildir)
# not Sonucu tersine çevirin, sonuç doğruysa False döndürür
w = 9
print(not (w > 1 and w < 10))
# sonucu tersine çevirmek için kullanılır. Norlamde true çıktısı verecekti
# # Python Kimlik Operatörleri
# Kimlik fonksiyonları, nesneleri eşit olup olmadıklarını değil, aslında aynı nesne olup olmadıklarını ve aynı bellek konumuna sahip olup olmadıklarını karşılaştırmak için kullanılır:
# is
# Her iki değişken de aynı nesneyse True döndürür
a = ["Samsung", "Apple"]
b = ["Samsung", "Apple"]
c = a
print(a is c)
# True değerini döndürür çünkü a, c ile aynı nesnedir
print(a is b)
# aynı içeriğe sahip olsalar bile a, b ile aynı nesne olmadığı için False döndürür
print(a == b)
# "is" ve "==" arasındaki farkı göstermek için: a eşittir b olduğu için bu karşılaştırma True değerini döndürür
# **is not Her iki değişken de aynı nesne değilse True döndürür**
# is not
# Her iki değişken de aynı nesne değilse True döndürür
z = ["Samsung", "Apple"]
x = ["Samsung", "Apple"]
c = z
print(z is not c)
# z, c ile aynı nesne olduğu için False döndürür
print(z is not x)
# True döndürür çünkü z, aynı içeriğe sahip olsalar bile x ile aynı nesne değildir
print(z != x)
# "is not" ve "!=" arasındaki farkı göstermek için: z eşittir x olduğu için bu karşılaştırma False döndürür
|
# # Intorduction
# Setup
import os
import cv2
import numpy as np
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
import matplotlib.pyplot as plt
# Avoid OMM Error
physical_devices = tf.config.experimental.list_physical_devices("GPU")
if len(physical_devices) > 0:
tf.config.experimental.set_memory_growth(physical_devices[0], True)
# check the GPU
physical_devices
# define the images directory
data_dir = "/kaggle/input/shai-level-2-training-2023"
print(os.listdir(data_dir))
# let`s make EDA for the dataset
data_splits = os.listdir(data_dir)[3:]
for data_kind in data_splits:
classes = os.listdir(os.path.join(data_dir, data_kind))
print(f"*We Process the {data_kind}")
for class_ in classes:
path = os.path.join(os.path.join(data_dir, data_kind), class_)
if os.path.isdir(path):
print(f" we have a {len(os.listdir(path))} images from class {class_}")
print("")
# # Genrate a Dataset and Visualize
batch_size = 32
img_size = (224, 224)
directory = "/kaggle/input/shai-level-2-training-2023/train"
# Define training dataset with 80% of the data
train_ds = tf.keras.preprocessing.image_dataset_from_directory(
directory,
validation_split=0.3,
subset="training",
seed=123,
image_size=img_size,
batch_size=batch_size,
)
# Define validation dataset with the remaining 20% of the data
val_ds = tf.keras.preprocessing.image_dataset_from_directory(
directory,
validation_split=0.3,
subset="validation",
seed=123,
image_size=img_size,
batch_size=batch_size,
)
class_names = list(train_ds.class_names)
class_names
# plot Visualize
plt.figure(figsize=(10, 10))
for image, label in train_ds.take(1):
for i in range(9):
plt.subplot(3, 3, i + 1)
plt.imshow(image[i].numpy().astype("uint8"))
plt.title(train_ds.class_names[label[i]])
plt.axis("off")
# # Configure the dataset for performance
#
# Configure dataset for performance
AUTOTUNE = tf.data.AUTOTUNE
train_ds = train_ds.cache().prefetch(buffer_size=AUTOTUNE)
val_ds = val_ds.cache().prefetch(buffer_size=AUTOTUNE)
# # Build a Model
NUM_CLASSES = 3
IMAGE_SIZE = (224, 224)
INPUT_SHAPE = IMAGE_SIZE + (3,)
# Preprocessing input layers
preprocess_input = tf.keras.applications.vgg19.preprocess_input
# Base Model `VGG19`
base_model = tf.keras.applications.VGG19(
input_shape=INPUT_SHAPE, include_top=False, weights="imagenet"
)
# Unfreeze some layers of the base model for fine-tuning
for layer in base_model.layers[:-4]:
layer.trainable = False
# Feature extractor layers
global_average_layer = tf.keras.layers.GlobalAveragePooling2D()
dense_layer_1 = tf.keras.layers.Dense(1024, activation="relu")
dropout_layer_1 = tf.keras.layers.Dropout(0.5)
dense_layer_2 = tf.keras.layers.Dense(512, activation="relu")
dropout_layer_2 = tf.keras.layers.Dropout(0.4)
prediction_layer = tf.keras.layers.Dense(NUM_CLASSES, activation="softmax")
# Build the model
inputs = tf.keras.Input(shape=INPUT_SHAPE)
x = preprocess_input(inputs)
x = base_model(x, training=True)
x = global_average_layer(x)
x = dense_layer_1(x)
x = dropout_layer_1(x)
x = dense_layer_2(x)
x = dropout_layer_2(x)
outputs = prediction_layer(x)
Mido = tf.keras.Model(inputs, outputs, name="Mido")
# Compile the model
Mido.compile(
optimizer=tf.keras.optimizers.Adam(learning_rate=0.0001),
loss="sparse_categorical_crossentropy",
metrics=["accuracy"],
)
# Print model summary
Mido.summary()
# plot the model
keras.utils.plot_model(Mido, show_shapes=True)
# # Define Callback
#
# Define Callback
from tensorflow.keras.callbacks import ModelCheckpoint
# Define the checkpoint filepath and callback
checkpoint_filepath = "best_model_weights.h5"
model_checkpoint_callback = ModelCheckpoint(
filepath=checkpoint_filepath,
save_weights_only=True,
monitor="val_accuracy",
mode="max",
save_best_only=True,
)
# Train the model with the callback
num_epochs = 150
history = Mido.fit(
train_ds,
epochs=num_epochs,
validation_data=val_ds,
callbacks=[model_checkpoint_callback],
)
# # Model Evaluation
# list all data in history
print(history.history.keys())
print("")
plt.figure(figsize=(10, 10))
plt.subplot(2, 1, 1)
# summarize history for accuracy
plt.plot(history.history["accuracy"])
plt.plot(history.history["val_accuracy"])
plt.title("model accuracy")
plt.ylabel("accuracy")
plt.legend(["train", "test"], loc="upper left")
# summarize history for loss
plt.subplot(2, 1, 2)
plt.plot(history.history["loss"])
plt.plot(history.history["val_loss"])
plt.title("model loss")
plt.ylabel("loss")
plt.xlabel("epoch")
plt.legend(["train", "test"], loc="upper left")
plt.show()
# Load the best model weights from the checkpoint
Mido.load_weights(checkpoint_filepath)
# Evaluate the best model
test_loss, test_accuracy = Mido.evaluate(val_ds)
print(f"Test Loss: {test_loss:.2f}")
print(f"Test Accuracy: {test_accuracy*100:.2f}%")
# # Test the Model and make Sample_submission
# make a prediction
import csv
# Path to the directory containing the images
images_dir = "/kaggle/input/shai-level-2-training-2023/test"
# List to hold the predictions
predictions = []
# Loop over the images in the directory
for image_file in os.listdir(images_dir):
# Load the image
image_path = os.path.join(images_dir, image_file)
image = cv2.imread(image_path)
image = cv2.resize(image, IMAGE_SIZE)
# Make a prediction on the image
prediction = Mido.predict(np.expand_dims(image, axis=0))
predicted_class = class_names[np.argmax(prediction)]
# Append the prediction to the list
predictions.append((image_file, predicted_class))
# Save the predictions to a CSV file
with open("submission.csv", "w", newline="") as csvfile:
writer = csv.writer(csvfile)
writer.writerow(["Image", "Label"])
for prediction in predictions:
writer.writerow(prediction)
import pandas as pd
pred = pd.read_csv("/kaggle/working/submission.csv")
pred.head()
pred.Label.value_counts()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# In this notebook, we'll be analyzing the National Football League (NFL) data. This dataset contains all regular season games from 2009-2016. It contains 356,768 rows and 100 columns.
# Well, let's get started!
# Libraries we will need:
import pandas as pd
import numpy as np
nfl_play = pd.read_csv(
"../input/nflplaybyplay2009to2016/NFL Play by Play 2009-2017 (v4).csv"
)
np.random.seed(0)
# Now, let's take a lok at sample data
nfl_play.sample(10)
# Above, we can clearly see, that the dataset has some missing values. Les's find them.
mis_values = nfl_play.isnull().sum()
mis_values
# We have a fairly large percent of missing value, let's clarify how large...
tot_cells = np.product(nfl_play.shape)
tot_mis = mis_values.sum()
# missing percent
(tot_mis / tot_cells) * 100
# As predicted, we have a whole 24.9% of missing value.
# # Dropping Missing Value
# Theoretically, 30% is the maximum missing values are allowed, beyond which we might want to drop the variable from analysis. But practically this varies. At times we get variables with ~50% of missing values but still, the customer insists to have it for analysis.h When the data goes missing on 60–70 percent of the variable, dropping the variable should be considered.
#
# Let's drop rows with at least one missing value.
nfl_play.dropna()
# Now, let's do the same with columns.We'll remove those that have at least one missing value.
col_with_na_dropped = nfl_play.dropna(axis=1)
col_with_na_dropped.head()
# let's see hom much data did we lose at this point.
print("Columns original dataset: %d \n" % nfl_play.shape[1])
print("Collumns with na's removed: %d" % col_with_na_dropped.shape[1])
# All Nan's have been excluded from data.
# # Filling in Missing Values
# Now,instead of removing, we'll try filling in the missing values.
#
subset_nfl = nfl_play.loc[:, "EPA":"Season"].head()
subset_nfl
# Time to replace Nan's with some value, in our case we'll replace them with 0
# all Nan's replaced with 0
subset_nfl.fillna(0)
# replace all NA's the value that comes directly after it in the same column,then replace all the remaining na's with 0
subset_nfl.fillna(method="bfill", axis=0).fillna(0)
# # Normalization and Scaling
# Scaling is a method used to normalize the range of independent variables or features of data. In data processing, it is also known as data normalization. We use it during the data preprocessing step.
# Libraries we"ll use:
import pandas as pd
import numpy as np
# for Box-Cox Transf
from scipy import stats
# for min_max scal
from mlxtend.preprocessing import minmax_scaling
# plotting
import seaborn as sns
import matplotlib.pyplot as plt
# set seed for reproducibility
np.random.seed(0)
# we'll generate 1000 data points randomly drawn from an exponential distribution
orig_data = np.random.exponential(size=1000)
# mix-max scale the data between 0 and 1
scaled_data = minmax_scaling(orig_data, columns=[0])
# plot both together to compare
fig, ax = plt.subplots(1, 2)
sns.distplot(orig_data, ax=ax[0])
ax[0].set_title("Original Nfl Data")
sns.distplot(scaled_data, ax=ax[1])
ax[1].set_title("Scaled data")
# The goal of normalization is to change the values of numeric columns in the dataset to a common scale, without distorting differences in the ranges of values. For machine learning, every dataset does not require normalization. It is required only when features have different ranges.
# We'll normalize the exponerial data with boxcox
normal_data = stats.boxcox(orig_data)
# plot both together to compare
fig, ax = plt.subplots(1, 2)
sns.distplot(orig_data, ax=ax[0])
ax[0].set_title("Original Nfl Data")
sns.distplot(normal_data[0], ax=ax[1])
ax[1].set_title("Normalized Nfl data")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# ## Importing the dataset
data = pd.read_csv("/kaggle/input/country-gdp/countries.csv")
data
# ## Analysis of dataset
data.info()
data.isnull().sum()
## there are no null records in given dataset
## there is no need of Rank Column
data.drop(["Rank"], axis=1, inplace=True)
data
## there is no need of ID Column
data.drop(["ID"], axis=1, inplace=True)
data
data["Continent"].unique()
# #### Task 1: Create a bar graph that shows number of Country in each continent
#
data_group_continent = (
data.groupby("Continent")
.agg(Number_of_countries=("Country", "count"))
.reset_index()
)
data_group_continent
x = data_group_continent["Continent"]
y = data_group_continent["Number_of_countries"]
plt.bar(x, y)
plt.xlabel("Continent")
plt.ylabel("Number of countries")
plt.title("Continent vs Number of Countries")
plt.xticks(rotation=45)
plt.show()
## It means, according to this dataset Africa has highest number of countries
# #### Task2: Find highest population country in each continent
data_task2 = (
data.groupby("Continent").agg(Highly_Populated=("Population", "max")).reset_index()
)
data_task2
merged_df = pd.merge(data_task2, data, on="Continent")
merged_df
data_task2 = merged_df[merged_df["Highly_Populated"] == merged_df["Population"]]
data_task2
x = data_task2.Continent
y = data_task2.Highly_Populated
plt.bar(x, y)
plt.xlabel("Continent")
plt.ylabel("Highest Population")
plt.xticks(rotation=45)
plt.show()
## it means, Asis has Highest population country "India"
# #### Task3: Now, find Continent which has highest GDP_per_capita and their respective country
data.head()
data_task3 = (
data.groupby("Continent")
.agg(Max_GDP_per_capita=("GDP_per_capita", "max"))
.reset_index()
)
data_task3
merged_df = pd.merge(data_task3, data, on="Continent")
merged_df
data_task3 = merged_df[merged_df["Max_GDP_per_capita"] == merged_df["GDP_per_capita"]]
data_task3
x = data_task3.Continent
y = data_task3.Max_GDP_per_capita
plt.bar(x, y)
plt.title("Continent vs Max GDP per capita")
plt.xlabel("Continent")
plt.ylabel("Max_GDP_per_capita")
plt.xticks(rotation=45)
plt.show()
## So, Continent Europe - Monaco has highest GDP per capita
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import os
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Observations
# Valid - Only event annotations where the series is marked true should be considered as unambiguous.
# This could be important, what happens if we include/exclude these?
# Task
# Your objective is to detect the start and stop of each freezing episode and the occurrence in these series of three types of freezing of gait events: Start Hesitation, Turn, and Walking.
# Will probably start with train/tdcsfog (lab data) + tdcsfog_metadata to get a view of the patient + motion that leads to events
# for dirname, _, filenames in os.walk('/kaggle/input/tlvmc-parkinsons-freezing-gait-prediction/train/tdcsfog/'):
# for filename in filenames:
# train_df = pd.read_csv(os.path.join(dirname, filename))
# Merge all of the tdcsfog data into 1 file and send it in to the model
combined_df = pd.DataFrame()
for dirname, _, filenames in os.walk(
"/kaggle/input/tlvmc-parkinsons-freezing-gait-prediction/train/tdcsfog/"
):
for filename in filenames:
# Add a column with the id from the file
curr_df = pd.read_csv(os.path.join(dirname, filename))
curr_df["Id"] = filename.split(".")[0]
combined_df = pd.concat([combined_df, curr_df], axis=0)
# Combine the Id column with the time column so our output is in the correct format
combined_df["Id"] = combined_df["Id"] + "_" + combined_df["Time"].astype(str)
combined_df.set_index("Id", inplace=True)
combined_df
from sklearn.tree import DecisionTreeClassifier
from sklearn.model_selection import train_test_split
x = combined_df[["AccV", "AccML", "AccAP"]]
y = combined_df[["StartHesitation", "Turn", "Walking"]]
x_train, x_test, y_train, y_test = train_test_split(
x, y, test_size=0.2, random_state=12
)
decision_tree = DecisionTreeClassifier()
decision_tree.fit(x_train, y_train)
y_pred = decision_tree.predict(x_test)
y_df = pd.DataFrame(y_pred)
y_df
# acc_decision_tree = round(decision_tree.score(x_test, y_test) * 100, 2)
# acc_decision_tree
# Prepare our test data for predictions
combined_test_df = pd.DataFrame()
for dirname, _, filenames in os.walk(
"/kaggle/input/tlvmc-parkinsons-freezing-gait-prediction/test/tdcsfog"
):
for filename in filenames:
# Add a column with the id from the file
curr_df = pd.read_csv(os.path.join(dirname, filename))
curr_df["Id"] = filename.split(".")[0]
combined_test_df = pd.concat([combined_test_df, curr_df], axis=0)
for dirname, _, filenames in os.walk(
"/kaggle/input/tlvmc-parkinsons-freezing-gait-prediction/test/defog"
):
for filename in filenames:
# Add a column with the id from the file
curr_df = pd.read_csv(os.path.join(dirname, filename))
curr_df["Id"] = filename.split(".")[0]
combined_test_df = pd.concat([combined_test_df, curr_df], axis=0)
# Combine the Id column with the time column so our output is in the correct format
combined_test_df["Id"] = (
combined_test_df["Id"] + "_" + combined_test_df["Time"].astype(str)
)
combined_test_df.set_index("Id", inplace=True)
combined_test_df = combined_test_df.drop(["Time"], axis=1)
combined_test_df
# Get predictions
y_pred = decision_tree.predict(combined_test_df)
y_df = pd.DataFrame(y_pred)
# Join our predictions with our ids, and reformat data for submission
submission = combined_test_df.reset_index("Id")
submission = submission.join(y_df)
submission = submission.rename(columns={0: "StartHesitation", 1: "Turn", 2: "Walking"})
submission = submission[["Id", "StartHesitation", "Turn", "Walking"]]
submission
submission.to_csv("/kaggle/working/submission.csv", index=False)
|
# install external libraries
# standard libraries
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import os, warnings
warnings.simplefilter("ignore")
import time
# sentence transformer library
from sentence_transformers import SentenceTransformer
# FAISS library (Facebook AI Similarity Search)
import faiss
# Torch library
import torch
# ### Load Data
# load data
file = "/kaggle/input/ca-independent-medical-review/Independent_Medical_Review__IMR__Determinations__Trend.csv"
df = pd.read_csv(file)
# filter the data
"""on gender=female & year >= 2010 & treatment = OB/GYN Procedures """
df = df.loc[
(df["Report Year"] >= 2010)
& (df["Patient Gender"] == "Female")
& (df["Treatment Category"] == "OB/GYN Procedures")
]
# drop NULLs & reset index
df.dropna(inplace=True)
df.reset_index(drop=True, inplace=True)
# custom function - extract sentences from column = Findings
def extract(txt):
if txt.startswith("Nature"):
split_txt = txt.split(" Findings")
re_split_txt = split_txt[0].split(":")
return re_split_txt[1].strip()
else:
split_txt = txt.split(" Findings")[0]
return split_txt.strip()
# apply the custom function to column = Findings
df["Findings"] = df["Findings"].apply(lambda x: extract(x))
# view
df.head()
# ### Load Model
# 
# load a pretrained model
model = SentenceTransformer("multi-qa-MiniLM-L6-cos-v1")
# ### Pre-process Data
# pre-process 'Findings' text data
patient_findings = df["Findings"].tolist()
# embed patient findings
patient_findings_embds = model.encode(patient_findings)
# ### Create Index using FAISS.
# using FAISS to create an index that allows for fast similarity search.
# Create an index using FAISS
index = faiss.IndexFlatL2(patient_findings_embds.shape[1])
index.add(patient_findings_embds)
faiss.write_index(index, "index_patient_findings")
index = faiss.read_index("index_patient_findings")
# ### Perform semantic search
# Using the index-ed data to perform semantic search. This will be achieved by inputting a query sentence and comparing it to the sentences in our index using search function. The custom-function will return the sentences that are most semantically similar to the query.
# custom function - semantic search
def search(query):
query_vector = model.encode([query])
k = 5
top_k = index.search(query_vector, k) # top3 only
return [patient_findings[_id] for _id in top_k[1].tolist()[0]]
# results
qry = "fibroids"
results = search(qry)
print("\n\n======================\n\n")
print("Query:", qry)
print("\nTop 5 most similar sentences in patient's findings:\n")
for r in enumerate(results, start=1):
print(f"{r[0]}). {r[1]}")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
import seaborn as sns
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Introduction:
# In this project, we will use big data technologies to process, analyze and leverage consumer data to implement algorithms for RFM model analysis based on big data technologies. Our team will use various big data analytics tools and techniques, such as Pandas library and Seaborn visualization library, to extract meaningful insights from the data, build RFM models, visualize the analysis results, and help companies make better decisions.
# Group No:14
# Team Member:
# * Wang Weiran 22412190
# * Chen Dingyuan 22407944
# * Li Zhenyou 22415106
# # Part A. Data Collection
# *** Dataset source: https://www.kaggle.com/datasets/tunguz/online-retail-ii**
online_retail_II = pd.read_csv("/kaggle/input/comp7095/online_retail_II.csv")
online_retail_II.head()
# # Part B. Data Clean & Preprocessing
# * According to the official explanation of this dataset, the 'Price' column refers to the unit price, so it was renamed 'UnitPrice'.
# * In order to make it easier to call the data of the column "Customer ID", it is renamed to "CustomerID".
# Copy the data to a new dataframe and perform data processing
dataset = online_retail_II.copy()
# According to the official explanation of this dataset, the 'Price' column refers to the unit price, so it was renamed 'UnitPrice'
dataset.rename(columns={"Price": "UnitPrice"}, inplace=True)
dataset.rename(columns={"Customer ID": "CustomerID"}, inplace=True)
dataset.head()
dataset.describe()
# **Missing value handling**
# Total up the number of NaN values in each row of the DataFrame.
dataset.isna().sum()
# Calculate the percentage of missing values
online_retail_II.isna().sum() / len(online_retail_II)
# Due to the sufficient amount of data, the missing values are deleted
dataset.dropna(how="any")
# Recheck the number of NaN values in each row of the DataFrame.
dataset.isna().sum()
# Print out the information about the DataFrame.
dataset.info()
# **Data set attribute resolution and outlier handling**
# From the "Invoice" attribute, we can see that there are 21 items per invioce on average
invoice_no = dataset.Invoice.unique().shape[0]
product_per_invoice = len(dataset) / invoice_no
product_per_invoice
# From the "Stockcode" attribute, we can see that there are 4070 stock codes in total
dataset.StockCode.unique().shape[0]
# From the "Description" attribute, we can see that there are 4224 products
dataset.Description.unique().shape[0]
# View the data distribution of the "Quantity" attribute
dataset.Quantity.describe()
# View the data distribution of the "UnitPrice" attribute
dataset.UnitPrice.describe()
# Check if there is data with unit price less than 0
dataset[dataset.UnitPrice < 0]
# Since the unit price of a product cannot be negative,
# the data with unit price less than 0 is deleted
dataset = dataset[dataset["UnitPrice"] > 0]
# From the "CustomerID" attribute, we can see that there are 4372 customers
dataset.CustomerID.unique().shape[0]
# From the "Country" attribute, we can see that there are 38 countrys
dataset.Country.unique().shape[0]
dataset.Country.value_counts(1)
# Create a new column named "TotalPrice", whose value is obtained by multiplying the "UnitPrice" and "Quantity"
dataset["TotalPrice"] = dataset["Quantity"] * dataset["UnitPrice"]
dataset.head()
# Extract the "Year" , "Month" and "Day" from the "InvoiceDate" attribute to a new column
dataset["Year"] = pd.to_datetime(dataset.InvoiceDate).dt.date.map(lambda x: x.year)
dataset["Month"] = pd.to_datetime(dataset.InvoiceDate).dt.date.map(lambda x: x.month)
dataset["Day"] = pd.to_datetime(dataset.InvoiceDate).dt.date.map(lambda x: x.day)
dataset.head()
# **Understand the properties in the Dataframe and get the following information:**
# 1. From the "Invoice" attribute, we can see that there are 21 items per invioce on average
# 2. From the "Stockcode" attribute, we can see that there are 4070 stock codes in total
# 3. From the "Description" attribute, we can see that there are 4224 products
# 4. From the "Quantity" attribute:
# * Negative values may refer to the number of returned goods
# * Two percent are return orders (Using "online_retail_II[online_retail_II.Quantity<0].count().Price/len(online_retail_II)" could get the result equal to 0.0196047... ~ 2%)
# 5. From the "UnitPrice" property, we can see that the highest unit price is 38970 and the lowest unit price is -11062.06, etc.
# 6. From the "CustomerID" attribute, we can see that there are 4372 customers
# 7. From the "Country" attribute, we can see that there are 38 countrys
# * 91 percent of the data is from the UK
# 8. From the "Stockcode" and "Description" attribute, we can see that these two attributes are not in one-to-one correspondence
# 9. From the "TotalPrice" attribute, we can see the total price of the invoice
# # Part C. Data Processing & Storage
# Copy the pre-processed data to a new dataframe, and perform the next operation in the new dataframe
dataset1 = dataset.copy()
# Observe the correlation between the variables in the table:
# Calculate the correlation between attributes
correlation = dataset1.corr()
sns.heatmap(correlation)
# # Part D. Data Analytics & Visualization
# Since this project aims to promote the purchase of goods by customers, it is necessary to exclude all return order information.
# In order to prevent the presence of Outlier in the "Quantity" column, all data values exceeding "Q3+1.5*IQR" are replaced with average values.
# The specific operation is as follows:
# Further analysis can be performed for return orders to provide reference for merchants,
# which will not be explored in this project
dataset_return = dataset1[dataset1.Quantity < 0]
dataset_return.head(10)
# Since this project aims to promote the purchase of goods by customers,
# it is necessary to exclude all return order information.
dataset2 = dataset1[dataset1.Quantity > 0]
# In order to prevent the presence of Outlier in the "Quantity" column,
# all data values exceeding "Q3+1.5*IQR" are replaced with average values.
IQR = dataset2.Quantity.describe()["75%"] - dataset2.Quantity.describe()["25%"]
# After caculate, Threshold = 23.5 , After rounding it, Threshold = 24
Threshold = dataset2.Quantity.describe()["75%"] + 1.5 * IQR
# The "Quantity" columns‘s mean value is 9.552234,
# and after rounding it, the mean value should be 10
dataset3 = dataset2.copy()
dataset3.loc[dataset2["Quantity"] > 24, "Quantity"] = 10
# **This project aims to increase user purchase rates, so the number of items sold per month is calculated to give the company advice on what time of the year it should increase its promotions to customers.**
# Display a bar chart of monthly sales using countplot()
sns.countplot(x="Month", data=dataset3)
# From the chart above we can conclude that there are more merchandise sales taught from October to December, a time when there are important holidays in the UK such as Halloween, Thanksgiving and Christmas, driving consumer shopping enthusiasm, so the company should increase the shopping offers during this time to achieve the goal of increasing customer purchase rates.
# The following chart shows the total price of sales for each month, which confirms the conclusion drawn above that the public's willingness to shop is strongest from October to December, and that companies should prepare their hardware and market well in advance for the peak shopping period during this time.
# Shows the total price of sales for each month
price_monthly = pd.DataFrame(
dataset2.groupby(["Month"])["TotalPrice"].sum()
).reset_index()
sns.relplot(x="Month", y="TotalPrice", kind="line", data=price_monthly)
plt.xlabel("Month", fontsize=15)
plt.ylabel("TotalPrice", fontsize=15)
plt.show()
# # Part E. Data Analysis Using RFM Models
# **We found that RFM model is the analysis model and means used by most companies to stratify the importance of customers. Therefore, in order to further utilize the dataset effectively and improve the purchase rate of customers, we will use RFM model to analyze the customers of the dataset and filter the customer information into different priority customer groups to facilitate the company to prioritize the needs of important customers or to develop corresponding marketing plans for different groups, hoping to achieve the goal of reducing the number of customers who need to conduct marketing, reducing marketing costs and improving the efficiency of marketing.**
# RFM is a method used for analyzing customer value and segmenting customers which is commonly used in database marketing and direct marketing.
# RFM stands for the three dimensions:
# * Recency – How recently did the customer purchase?
# * Frequency – How often do they purchase?
# * Monetary Value – How much do they spend?
dataset4 = dataset3.copy()
dataset4.head()
pd.to_datetime(dataset4.InvoiceDate).dt.date.max()
# **R(Recency)-value calculation**
# Set the nearest time in the dataset as the current time
Current_Date = pd.to_datetime(dataset4.InvoiceDate).dt.date.max()
# R value is equal to the difference between the time of the data and the current time
R_value = pd.to_datetime(dataset4.groupby(["CustomerID"])["InvoiceDate"].max()).dt.date
R_value = Current_Date - R_value
# Converts the R_value array to a time difference of type timedelta64 and rounds it down to the nearest integer.
R_value = R_value.astype("timedelta64[D]").astype(int)
# Rename the data column named "InvoiceDate" in the R_value array to "recency"
R_value = pd.DataFrame(R_value).rename(columns={"InvoiceDate": "recency"})
plt.figure(figsize=(10, 5))
plt.hist(R_value["recency"], bins=20)
plt.xlabel("Recency", fontsize=15)
plt.show()
# **F(Frequency)-value calculation**
# Group the dataset4 with the keywords 'CustomerID' and 'Invoice', and count the number of 'InvoiceDate' columns in each group
F_value = (
dataset4.groupby(["CustomerID", "Invoice"])["InvoiceDate"].count().sort_values()
)
# Rename the data column named "InvoiceDate" in the F_value array to "frequency"
F_value = (
pd.DataFrame(F_value).reset_index().rename(columns={"InvoiceDate": "frequency"})
)
F_value = F_value.groupby(["CustomerID"])["Invoice"].count().sort_values()
F_value = pd.DataFrame(F_value).reset_index().rename(columns={"Invoice": "frequency"})
plt.figure(figsize=(10, 5))
plt.hist(F_value[F_value.frequency < 100].frequency, bins=100)
plt.xlabel("Frequency", fontsize=15)
plt.show()
# dataset4.head()
# **M(Monetary)-value calculation**
# Group the dataset4 with the keywords 'CustomerID' and 'TotalPrice', and sum the value of 'TotalPrice' columns in each group
M_value = dataset4.groupby(["CustomerID"])["TotalPrice"].sum().sort_values()
M_value = pd.DataFrame(M_value).reset_index()
plt.figure(figsize=(10, 5))
plt.hist(M_value[M_value.TotalPrice < 6000]["TotalPrice"], bins=50)
plt.xlabel("Monetary", fontsize=15)
plt.show()
# **Customer priority classification**
RFM = R_value.copy()
# Rename the data column named "recency" in the RFM array to "R",
# and columns are added for "F" (frequency) and "M" (monetary value) using data from Dataframe F_value and M_value
RFM = RFM.rename(columns={"recency": "R"})
RFM["F"] = F_value.set_index("CustomerID")["frequency"]
RFM["M"] = M_value.set_index("CustomerID")["TotalPrice"]
# Define the bin used to group customers for all three metrics
bins_R = [0, 100, 200, 300, 400, 500]
bins_F = [0, 4, 8, 13, 25, 500]
bins_M = [0, 200, 1000, 5000, 10000, 300000]
# Divided into 5 levels from 1 to 5
labels_sequential = [1, 2, 3, 4, 5]
labels_Inverted = [5, 4, 3, 2, 1]
RFM["R_grade"] = pd.cut(RFM["R"], bins_R, labels=labels_Inverted, right=False)
RFM["F_grade"] = pd.cut(RFM["F"], bins_F, labels=labels_sequential, right=False)
RFM["M_grade"] = pd.cut(RFM["M"], bins_M, labels=labels_sequential, right=False)
RFM.head(10)
from numpy import *
mean(list(map(int, RFM["R_grade"]))) # result = 4.15
mean(list(map(int, RFM["F_grade"]))) # result = 1.55
mean(list(map(int, RFM["M_grade"]))) # result = 2.32
RFM["R_level"] = RFM["R_grade"].apply(lambda x: 1 if x > 4.15 else 0)
RFM["F_level"] = RFM["F_grade"].apply(lambda x: 1 if x > 1.55 else 0)
RFM["M_level"] = RFM["M_grade"].apply(lambda x: 1 if x > 2.32 else 0)
RFM["RFM_CODE"] = (
RFM["R_level"].astype("str")
+ RFM["F_level"].astype("str")
+ RFM["M_level"].astype("str")
)
def Classification(x):
if x == "111":
return "Key Value Customers"
elif x == "101":
return "Key Development Customers"
elif x == "011":
return "Key Maintain Customers"
elif x == "001":
return "Important Retention Customers"
elif x == "110":
return "General Value Customers"
elif x == "100":
return "General Development Customers"
elif x == "010":
return "General Maintain Customers"
elif x == "000":
return "General Retention Customers"
RFM["type"] = RFM["RFM_CODE"].apply(Classification)
RFM["type"].value_counts()
RFM.head(10)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Loading data into training set and test set
training_data = pd.read_csv("/kaggle/input/cap-4611-spring-21-assignment-1/train.csv")
test_data = pd.read_csv("/kaggle/input/cap-4611-spring-21-assignment-1/test.csv")
training_data
# # Check for Missing Data
# We check for null values and find that there are none in the data.
# We also check for values of zero but we cannot tell if it is a real zero value or a placeholder so we ignore it.
#
print(training_data.isnull().sum().sum())
print(test_data.isnull().sum().sum())
print(training_data.count(0).sum().sum())
print(test_data.count(0).sum().sum())
# # Check for Outliers
# We can remove features where the standard deviation is zero (which means every value is the same) since it doesn't help us with classification.
# By computing the zscore, we can find rows which contain values multiple standard deviations away from the mean and drop these rows as they are outliers.
# However, I found that not doing anything about the outliers gives the best results so I elected not to do anything. I commented out the code below that I initially used to remove features.
from scipy.stats import zscore
# remove features that do not help with classification (when standard deviation is zero)
# training_data = training_data.loc[:, training_data.std() > 0]
# test_data = test_data.loc[:, test_data.std() > 0]
# compute zscore for each value
# training_data_zscores = training_data.apply(zscore)
# print(training_data_zscores[training_data_zscores > 4].count())
# print(training_data_zscores[training_data_zscores < -4].count())
# find rows containing outliers
# rows_containing_outlier = training_data[((training_data_zscores > 1000) | (training_data_zscores < -1000)).any(1)]
# drop these rows
# training_data = training_data.drop(rows_containing_outlier['id'].to_list(), axis=0)
# training_data
# # Normalization or Standardization?
# I ended up getting good results without normalizing or standardizing the data so I decided not to do it.
# # Splitting the Training Data
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
# Set seed for reproducibility
SEED = 73
y = training_data["Bankrupt"]
X = training_data.drop(columns=["id", "Bankrupt"])
test_data = test_data.drop(columns=["id"])
# Split the data in 70% train and 30% test
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.05, random_state=SEED
)
# # Decision Tree
# I print the best scores I managed to get with my decision tree model.
from sklearn.tree import DecisionTreeClassifier
from sklearn.model_selection import RandomizedSearchCV, GridSearchCV
from scipy.stats import randint
from sklearn.metrics import accuracy_score, roc_auc_score, f1_score
np.random.seed(SEED)
dt = DecisionTreeClassifier(criterion="gini", max_depth=2000, random_state=SEED)
dt.fit(X_train, y_train)
prediction = [x[1] for x in dt.predict_proba(X_test)]
acc = accuracy_score(y_test, prediction)
roc_acc = roc_auc_score(y_test, prediction)
f1_acc = f1_score(y_test, prediction)
print("Accuracy score = ", acc)
print("ROC_ACC score = ", roc_acc)
print("F1_acc score = ", f1_acc)
# # Random Forest Model
# I print the best scores I could get with my random forest model. The ROC_ACC was pretty good so I submitted that. For both the random forest model and the decision tree I tried random values for hyper parameters until I got decent results.
from sklearn.ensemble import RandomForestClassifier
rf = RandomForestClassifier(
n_estimators=600, max_depth=150, criterion="entropy", random_state=SEED
)
rf.fit(X_train, y_train)
prediction1 = rf.predict(X_test)
prediction2 = [x[1] for x in rf.predict_proba(X_test)]
acc1 = accuracy_score(y_test, prediction1)
f1_acc1 = f1_score(y_test, prediction1)
roc_acc1 = roc_auc_score(y_test, prediction2)
print("Accuracy score = ", acc1)
print("ROC_ACC score = ", roc_acc1)
print("F1_acc score = ", f1_acc1)
prediction = [x[1] for x in rf.predict_proba(test_data)]
output = pd.DataFrame({"Bankrupt": prediction})
output.to_csv("MySubmission.csv", index=True, index_label="id")
print(output)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
import numpy as np
from sklearn.tree import DecisionTreeRegressor
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import (
BaggingRegressor,
RandomForestRegressor,
RandomForestClassifier,
)
from sklearn.linear_model import LinearRegression
from sklearn.model_selection import train_test_split
from sklearn.metrics import mean_squared_error, accuracy_score
# from sklearn.externals.six import StringIO
from sklearn import tree
from six import StringIO
from sklearn.model_selection import KFold, cross_val_score
from sklearn import metrics
# visualization
import seaborn as sns
from IPython.display import Image
# import pydotplus
from sklearn.tree import export_graphviz
df = pd.read_csv("../input/arketing-campaign/marketing_campaign.csv", delimiter=";")
df.head()
df_cleaned = df.copy()
del df_cleaned["Z_CostContact"]
del df_cleaned["Z_Revenue"]
# create account age
df_cleaned["Dt_Customer"] = pd.to_datetime(df_cleaned["Dt_Customer"])
df_cleaned["Dt_Customer_year_month"] = df_cleaned["Dt_Customer"].dt.to_period("M")
df_cleaned["account_age"] = (
pd.to_datetime("2014-12").year - df_cleaned["Dt_Customer_year_month"].dt.year
) * 12 + (
pd.to_datetime("2014-12").month - df_cleaned["Dt_Customer_year_month"].dt.month
)
del df_cleaned["Dt_Customer_year_month"]
del df_cleaned["Dt_Customer"]
df_cleaned.head()
# create customer age
df_cleaned["Age"] = 2014 - df_cleaned["Year_Birth"]
del df_cleaned["Year_Birth"]
df_cleaned.head()
df_cleaned.describe()
# Fillna in Income by using avg value
avg_income = np.mean(df_cleaned.Income)
df_cleaned["Income"] = df_cleaned["Income"].fillna(avg_income, axis=0)
df_cleaned.describe()
df_cleaned.corr()
# from sklearn.compose import ColumnTransformer
from sklearn.preprocessing import OneHotEncoder
# Create Dummy Variables
enc_edu = OneHotEncoder(handle_unknown="ignore")
enc_edu_df = pd.DataFrame(enc_edu.fit_transform(df_cleaned[["Education"]]).toarray())
df_cleaned = df_cleaned.join(enc_edu_df)
df_cleaned.head()
df_cleaned.rename(
columns={0: "2n Cycle", 1: "Basic", 2: "Graduation", 3: "Master", 4: "PhD"},
inplace=True,
)
enc_marital = OneHotEncoder(handle_unknown="ignore")
enc_marital_df = pd.DataFrame(
enc_edu.fit_transform(df_cleaned[["Marital_Status"]]).toarray()
)
df_cleaned = df_cleaned.join(enc_marital_df)
df_cleaned.rename(
columns={
0: "Absurd",
1: "Alone",
2: "Divorced",
3: "Married",
4: "Single",
5: "Together",
6: "Widow",
7: "YOLO",
},
inplace=True,
)
df_cleaned.head()
del df_cleaned["Marital_Status"]
del df_cleaned["Education"]
df_cleaned["total_Mnt"] = (
df_cleaned["MntWines"]
+ df_cleaned["MntFruits"]
+ df_cleaned["MntMeatProducts"]
+ df_cleaned["MntFishProducts"]
+ df_cleaned["MntSweetProducts"]
+ df_cleaned["MntGoldProds"]
)
df_cleaned["MntWines_pct"] = df_cleaned["MntWines"] / df_cleaned["total_Mnt"]
df_cleaned["MntFruits_pct"] = df_cleaned["MntFruits"] / df_cleaned["total_Mnt"]
df_cleaned["MntMeatProducts_pct"] = (
df_cleaned["MntMeatProducts"] / df_cleaned["total_Mnt"]
)
df_cleaned["MntFishProducts_pct"] = (
df_cleaned["MntFishProducts"] / df_cleaned["total_Mnt"]
)
df_cleaned["MntSweetProducts_pct"] = (
df_cleaned["MntSweetProducts"] / df_cleaned["total_Mnt"]
)
df_cleaned["MntGoldProds_pct"] = df_cleaned["MntGoldProds"] / df_cleaned["total_Mnt"]
df_cleaned[
[
"MntWines_pct",
"MntFruits_pct",
"MntMeatProducts_pct",
"MntFishProducts_pct",
"MntSweetProducts_pct",
"MntGoldProds_pct",
]
].describe()
# Create AcceptedCmps to count number of offers each customer accepted
df_cleaned["AcceptedCmps"] = df_cleaned[
["AcceptedCmp1", "AcceptedCmp2", "AcceptedCmp3", "AcceptedCmp4", "AcceptedCmp5"]
].sum(axis=1)
df_cleaned_for_DS = df_cleaned.copy()
# Start to Create Random Forest
# Balanced dataset by oversampling
count_response_0, count_response_1 = df_cleaned_for_DS.Response.value_counts()
print(count_response_0)
print(count_response_1)
df_cleaned_for_DS_0 = df_cleaned_for_DS[df_cleaned_for_DS["Response"] == 0]
df_cleaned_for_DS_1 = df_cleaned_for_DS[df_cleaned_for_DS["Response"] == 1]
df_cleaned_for_DS_1_over = df_cleaned_for_DS_1.sample(n=count_response_0, replace=True)
df_cleaned_for_DS_1_over.shape
df_cleaned_for_DS_resampling = pd.concat(
[df_cleaned_for_DS_1_over, df_cleaned_for_DS_0], axis=0
)
# split the train and test dataset
X = df_cleaned_for_DS_resampling[feature]
y = df_cleaned_for_DS_resampling.Response
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.4, random_state=123
)
feature = [
"2n Cycle",
"Basic",
"Graduation",
"Master",
"PhD",
"Absurd",
"Alone",
"Divorced",
"Married",
"Single",
"Together",
"Widow",
"YOLO",
"Income",
"Kidhome",
"Teenhome",
"Recency",
"MntWines_pct",
"MntFruits_pct",
"MntMeatProducts_pct",
"MntFishProducts_pct",
"MntSweetProducts_pct",
"MntGoldProds_pct",
"NumDealsPurchases",
"NumWebPurchases",
"NumCatalogPurchases",
"NumStorePurchases",
"NumWebVisitsMonth",
"AcceptedCmp1",
"AcceptedCmp2",
"AcceptedCmp3",
"AcceptedCmp4",
"AcceptedCmp5",
"AcceptedCmps",
"Complain",
"account_age",
"Age",
]
from sklearn.model_selection import GridSearchCV
rfc = RandomForestClassifier(n_estimators=30, random_state=1)
max_depth_range = range(1, 16)
param_grid = dict(max_depth=max_depth_range)
grid = GridSearchCV(rfc, param_grid, cv=10, scoring="accuracy")
grid.fit(X_train, y_train)
grid.cv_results_
grid_mean_scores = grid.cv_results_["mean_test_score"]
grid_test_mean_scores = metrics
# plot the results
sns.mpl.pyplot.plot(max_depth_range, grid_mean_scores)
sns.mpl.pyplot.xlabel("max_depth")
sns.mpl.pyplot.ylabel("Cross-Validated Mean Train Set Accuracy")
best_rfc = RandomForestClassifier(n_estimators=50, random_state=1, max_depth=12)
best_rfc.fit(X_train, y_train)
rfc_pred = best_rfc.predict(X_test)
accuracy_train = metrics.accuracy_score(y_train, best_rfc.predict(X_train))
accuracy_test = metrics.accuracy_score(y_test, rfc_pred)
print("Accuracy of Random Forest train is: ", accuracy_train)
print("Accuracy of Random Forest test is: ", accuracy_test)
# compute feature importances
pd.DataFrame(
{"feature": feature, "importance": best_rfc.feature_importances_}
).sort_values(by="importance", ascending=False)
DecisionTree_cross_val = DecisionTreeClassifier(max_depth=10, random_state=10)
DecisionTree_cross_val.fit(X_train, y_train)
cross_val_scores = cross_val_score(DecisionTree_cross_val, X_train, y_train, cv=10)
print("10-fold accuracies:\n", cross_val_scores)
print("10-fold mean accuracy:\n", cross_val_scores.mean())
y_test_pred_rcf = best_rfc.predict(X_test)
print(
"Classification Report:\n", metrics.classification_report(y_test, y_test_pred_rcf)
)
pred_probs_rcf = best_rfc.predict_proba(X_test)
pred_probs_rcf
fpr, tpr, thresholds = metrics.roc_curve(y_test, pred_probs_rcf[:, 1])
sns.mpl.pyplot.plot(fpr, tpr, label="DS")
sns.mpl.pyplot.xlim([0, 1])
sns.mpl.pyplot.ylim([0, 1.05])
sns.mpl.pyplot.legend(loc="lower right")
sns.mpl.pyplot.xlabel("False Positive Rate (1 - Specificity)")
sns.mpl.pyplot.ylabel("True Positive Rate (Sensitivity)")
print("Test AUC: ", metrics.roc_auc_score(y_test, best_rfc.predict(X_test)))
|
# importing modules
import os
import tarfile
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
from sklearn.model_selection import train_test_split
from sklearn.model_selection import cross_val_score
from sklearn.model_selection import KFold
from sklearn import linear_model
from sklearn.linear_model import LinearRegression, LogisticRegression
from sklearn.linear_model import Lasso, LassoLars, BayesianRidge
from sklearn.linear_model import Ridge, ElasticNet, HuberRegressor
from sklearn.svm import SVR, NuSVR, LinearSVR
from xgboost import XGBRegressor
from sklearn.neighbors import KNeighborsRegressor
from sklearn.gaussian_process import GaussianProcessRegressor
from sklearn.naive_bayes import GaussianNB
from sklearn.ensemble import RandomForestRegressor
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import mean_squared_error, r2_score
# importing the data
car_prices = pd.read_csv("/kaggle/input/sports-car-prices-dataset/Sport car price.csv")
car_prices
# types of data
car_prices.dtypes
# checking for NaN in the df
car_prices.isnull().sum().sort_values(ascending=False)
# show NaN values
car_prices[car_prices.isna().any(axis=1)]
car_prices[car_prices["Car Model"] == "C_Two"]
# assign engine size to electric for C_Two
car_prices.loc[car_prices["Car Model"] == "C_Two", "Engine Size (L)"] = car_prices.loc[
car_prices["Car Model"] == "C_Two", "Engine Size (L)"
].fillna("Electric")
car_prices[car_prices["Car Model"] == "C_Two"]
car_prices[car_prices["Car Model"] == "Model S Plaid"]
# assign engine size to electric for Tesla Model S Plaid
car_prices.loc[
car_prices["Car Model"] == "Model S Plaid", "Engine Size (L)"
] = car_prices.loc[
car_prices["Car Model"] == "Model S Plaid", "Engine Size (L)"
].fillna(
"Electric"
)
car_prices[car_prices.isna().any(axis=1)]
# assign engine size to electric for Tesla Roadster
car_prices.loc[
car_prices["Car Model"] == "Roadster", "Engine Size (L)"
] = car_prices.loc[car_prices["Car Model"] == "Roadster", "Engine Size (L)"].fillna(
"Electric"
)
car_prices[car_prices["Car Model"] == "Roadster"]
car_prices[car_prices.isna().any(axis=1)]
# the same for other electric cars
car_prices.loc[
car_prices["Car Model"] == "Taycan Turbo S", "Engine Size (L)"
] = car_prices.loc[
car_prices["Car Model"] == "Taycan Turbo S", "Engine Size (L)"
].fillna(
"Electric"
)
car_prices.loc[car_prices["Car Model"] == "Evija", "Engine Size (L)"] = car_prices.loc[
car_prices["Car Model"] == "Evija", "Engine Size (L)"
].fillna("Electric")
car_prices.loc[car_prices["Car Model"] == "Taycan", "Engine Size (L)"] = car_prices.loc[
car_prices["Car Model"] == "Taycan", "Engine Size (L)"
].fillna("Electric")
car_prices[car_prices.isna().any(axis=1)]
car_prices[car_prices["Car Model"] == "Model S Plaid"]
# changing the engine size, horsepower, torque and 0-60 MPH acceleration time for Tesla Model S
car_prices.loc[
car_prices["Car Model"] == "Model S Plaid", "Engine Size (L)"
] = "Electric"
car_prices.loc[car_prices["Car Model"] == "Model S Plaid", "Horsepower"] = 1020
car_prices.loc[car_prices["Car Model"] == "Model S Plaid", "Torque (lb-ft)"] = 1050
car_prices.loc[
car_prices["Car Model"] == "Model S Plaid", "0-60 MPH Time (seconds)"
] = 1.98
car_prices[car_prices["Car Model"] == "Model S Plaid"]
car_prices[car_prices.isna().any(axis=1)]
car_prices[car_prices["Car Model"] == "Roadster"]
# changing the engine size, horsepower, torque and 0-60 MPH acceleration time for Tesla Roadster
car_prices.loc[car_prices["Car Model"] == "Roadster", "Engine Size (L)"] = "Electric"
car_prices.loc[car_prices["Car Model"] == "Roadster", "Horsepower"] = 1020
car_prices.loc[car_prices["Car Model"] == "Roadster", "Torque (lb-ft)"] = 1050
car_prices.loc[car_prices["Car Model"] == "Roadster", "0-60 MPH Time (seconds)"] = 1.9
car_prices[car_prices["Car Model"] == "Roadster"]
car_prices[
(car_prices["Car Model"] == "GranTurismo")
& (car_prices["Engine Size (L)"] == "Electric")
]
car_prices.loc[
(car_prices["Car Model"] == "GranTurismo")
& (car_prices["Engine Size (L)"] == "Electric"),
"Year",
] = 2023
car_prices.loc[
(car_prices["Car Model"] == "GranTurismo")
& (car_prices["Engine Size (L)"] == "Electric"),
"Torque (lb-ft)",
] = 1350
car_prices[
(car_prices["Car Model"] == "GranTurismo")
& (car_prices["Engine Size (L)"] == "Electric")
]
car_prices.loc[car_prices["Horsepower"] == "1,500", "Horsepower"] = 1500
car_prices[car_prices["Car Model"] == "Chiron"]
car_prices.loc[car_prices["Car Model"] == "Chiron", "Horsepower"] = 1500
car_prices.loc[car_prices["Car Model"] == "Chiron", "Torque (lb-ft)"] = 1180
car_prices[car_prices["Car Model"] == "Chiron"]
# checking engine size distribution
car_prices["Engine Size (L)"].value_counts().sort_index()
car_prices[car_prices.isna().any(axis=1)]
car_prices
# car makes hist
fig, ax = plt.subplots(1, 1)
make_hist = ax.hist(car_prices["Car Make"], bins=100)
for tick in ax.get_xticklabels():
tick.set_rotation(90)
# checking for distribution of the data in the df
"""also, we need to check if there are any kind of artificially
unique values which may be a result of typo"""
for i in list(car_prices):
print(car_prices[i].value_counts(), "\n\n")
## horsepower distribution
car_prices["Horsepower"].value_counts()
car_prices.loc[
~car_prices["Horsepower"].astype(str).str.isdigit(), "Horsepower"
].tolist()
car_prices[car_prices.isna().any(axis=1)]
car_prices[car_prices["Horsepower"] == "1,500"]
car_prices[car_prices["Car Make"] == "Acura"]
car_prices.loc[car_prices["Car Model"] == "NSX", "0-60 MPH Time (seconds)"] = 2.9
car_prices[car_prices["Car Make"] == "Alfa Romeo"]
columns = list(car_prices)
columns
# rows with object in engine size
car_prices[car_prices["Engine Size (L)"] == "1.5 + Electric"]
car_prices[car_prices["Car Model"] == "i8"]
# changing i8 engine size
car_prices.loc[car_prices["Car Model"] == "i8", "Engine Size (L)"] = 1.5
car_prices.loc[car_prices["Car Model"] == "i8", "Torque (lb-ft)"] = 570
car_prices[car_prices["Car Model"] == "i8"]
# adding powertrain column
car_prices["Powertrain"] = car_prices["Car Model"].apply(
lambda x: "Hybrid" if x == "i8" else "Gas"
)
# assinging electric cars to electric powertrain
car_prices.loc[
(car_prices["Car Make"] == "Tesla") | (car_prices["Car Make"] == "Rimac"),
"Powertrain",
] = "Electric"
# data cleaning (engine size)
car_prices.loc[
car_prices["Engine Size (L)"] == "2.0 (Electric)", "Powertrain"
] = "Electric"
car_prices.loc[
car_prices["Engine Size (L)"] == "2.0 (Electric)", "Engine Size (L)"
] = 0.0
car_prices.loc[car_prices["Engine Size (L)"] == "4.0 (Hybrid)", "Powertrain"] = "Hybrid"
car_prices.loc[car_prices["Engine Size (L)"] == "4.0 (Hybrid)", "Engine Size (L)"] = 4.0
car_prices.loc[car_prices["Engine Size (L)"] == "Hybrid", "Powertrain"] = "Hybrid"
car_prices.loc[car_prices["Engine Size (L)"] == "Hybrid", "Engine Size (L)"] = 4.0
car_prices.loc[
car_prices["Engine Size (L)"] == "Electric (93 kWh)", "Powertrain"
] = "Electric"
car_prices.loc[
car_prices["Engine Size (L)"] == "Electric (93 kWh)", "Engine Size (L)"
] = 0
car_prices.loc[
car_prices["Engine Size (L)"] == "Electric Motor", "Powertrain"
] = "Electric"
car_prices.loc[car_prices["Engine Size (L)"] == "Electric Motor", "Engine Size (L)"] = 0
car_prices.loc[car_prices["Engine Size (L)"] == "Electric", "Engine Size (L)"] = 0
car_prices[car_prices["Engine Size (L)"] == "Electric (93 kWh)"]
car_prices.loc[
car_prices["Engine Size (L)"] == "Electric (93 kWh)", "Engine Size (L)"
] = 0
car_prices.loc[car_prices["Engine Size (L)"] == "Hybrid (4.0)", "Powertrain"] = "Hybrid"
car_prices.loc[car_prices["Engine Size (L)"] == "Hybrid (4.0)", "Engine Size (L)"] = 4.0
car_prices.dtypes
# changing engine size to float
car_prices = car_prices.astype({"Engine Size (L)": "float64"})
# removing obj characters from the price column
car_prices["Price (in USD)"] = car_prices["Price (in USD)"].str.replace("'", "")
car_prices["Price (in USD)"] = car_prices["Price (in USD)"].str.replace(",", "")
# changing price column to int
car_prices = car_prices.astype({"Price (in USD)": "int64"})
# changing other columns to int/float
car_prices = car_prices.astype(
{"Horsepower": "int64", "Torque (lb-ft)": "int64", "0-60 MPH Time (seconds)": float}
)
# dtypes changed successfully
car_prices.dtypes
# distribution of the numerical data
car_prices.hist(bins=50, figsize=(20, 20))
# showing year below 2010
car_prices[car_prices["Year"] < 2010]
# removing skewed values (year)
del_idx = car_prices.index[car_prices["Year"] < 2010].tolist()
car_prices = car_prices.drop(del_idx)
# reseting index count
car_prices.reset_index(drop=True, inplace=True)
car_prices
# heavily skewed price distribution
car_prices["Price (in USD)"].hist(bins=50, figsize=(20, 20))
# changing price distribution to log scale
np.log10(car_prices["Price (in USD)"]).hist(bins=50, figsize=(20, 20))
car_prices["Price (in USD)"] = np.log10(car_prices["Price (in USD)"])
# adding columns 1 hp price and 1 lb-ft price
car_prices["1 hp price"] = car_prices["Horsepower"] / car_prices["Price (in USD)"]
car_prices["1 lb-ft price"] = (
car_prices["Torque (lb-ft)"] / car_prices["Price (in USD)"]
)
car_prices
# powertrain distribution
car_prices["Powertrain"].hist(bins=50)
# scaling and encoding the categorical data
cat_cols = car_prices.select_dtypes(include=["object"])
num_cols = car_prices.select_dtypes(include=["float64", "int64"])
from sklearn.preprocessing import OrdinalEncoder
ordinal_encoder = OrdinalEncoder()
car_prices_cat_cols = pd.DataFrame(
ordinal_encoder.fit_transform(cat_cols), columns=cat_cols.columns
)
car_prices_encoded = pd.concat([car_prices_cat_cols, num_cols], axis=1)
car_prices_encoded
# separating the data to X and y
X = car_prices_encoded.drop("Price (in USD)", axis=1)
y = car_prices_encoded["Price (in USD)"].copy()
# splitting the data
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=13
)
print(X_train.shape, y_train.shape, X_test.shape, y_test.shape)
# checking the main models to see the performance
n_folds = 5
def cvs(model):
kf = KFold(n_folds, shuffle=True, random_state=42).get_n_splits(num_cols)
return cross_val_score(model, X_train, y_train, cv=kf)
def display_scores(scores):
print(Model.__name__)
print("Scores:", scores)
print("Mean:", scores.mean())
print("Standard deviation:", scores.std(), "\n\n\n")
for Model in [
LinearRegression,
Ridge,
Lasso,
LassoLars,
BayesianRidge,
ElasticNet,
HuberRegressor,
SVR,
NuSVR,
LinearSVR,
XGBRegressor,
KNeighborsRegressor,
RandomForestRegressor,
]:
if Model == LogisticRegression:
cvs_scores = cvs(LogisticRegression(solver="lbfgs"))
display_scores(cvs_scores)
else:
cvs_scores = cvs(Model())
display_scores(cvs_scores)
# choosing the best models to find optimal parameters to increase scoring
# Ridge parameters
ridge_reg = Ridge()
ridge_reg.fit(X_train, y_train)
ridge_reg.fit(X_train, y_train).get_params()
# GridSearchCV to find parameter set to increase the performance
param_grid = [
{
"alpha": [1e-2, 1e-1, 1, 10],
"max_iter": [1000, 10000],
"solver": ["auto", "svd", "cholesky", "lsqr", "sparse_cg"],
"tol": [1e-8, 1e-7, 1e-6, 1e-5, 1e-4, 1e-3, 1e-2],
}
]
ridge_reg = Ridge()
grid_search = GridSearchCV(ridge_reg, param_grid, cv=5)
grid_search.fit(X_train, y_train)
print(
"Best parameters: ",
grid_search.best_params_,
"Best score: ",
grid_search.best_score_,
)
# BayesianRidge parameters
br_reg = BayesianRidge()
br_reg.fit(X_train, y_train)
br_reg.fit(X_train, y_train).get_params()
# GridSearchCV to find parameter set to increase the performance
param_grid = [
{
"alpha_1": [1e-8, 1e-7, 1e-6, 1e-5, 1e-4],
"alpha_2": [1e-8, 1e-7, 1e-6, 1e-5, 1e-4],
"lambda_1": [1e-8, 1e-7, 1e-6, 1e-5, 1e-4],
"lambda_2": [1e-8, 1e-7, 1e-6, 1e-5, 1e-4],
"tol": [1e-8, 1e-7, 1e-6, 1e-5, 1e-4, 1e-3, 1e-2],
}
]
br_reg = BayesianRidge()
grid_search = GridSearchCV(br_reg, param_grid, cv=5)
grid_search.fit(X_train, y_train)
print(
"Best parameters: ",
grid_search.best_params_,
"Best score: ",
grid_search.best_score_,
)
# KNeighborsRegressor parameters
knn_reg = KNeighborsRegressor()
knn_reg.fit(X_train, y_train)
knn_reg.fit(X_train, y_train).get_params()
# GridSearchCV to find parameter set to increase the performance
param_grid = [
{
"n_neighbors": [3, 8, 10, 15, 25],
"weights": ["uniform", "distance"],
"algorithm": ["auto", "ball_tree", "kd_tree", "brute"],
"leaf_size": [20, 40, 60, 100],
"p": [1, 2, 5, 10],
}
]
knn_reg = KNeighborsRegressor()
grid_search = GridSearchCV(knn_reg, param_grid, cv=5)
grid_search.fit(X_train, y_train)
print(
"Best parameters: ",
grid_search.best_params_,
"Best score: ",
grid_search.best_score_,
)
# RandomForestRegressor parameters
rf_reg = RandomForestRegressor()
rf_reg.fit(X_train, y_train)
rf_reg.fit(X_train, y_train).get_params()
# GridSearchCV to find parameter set to increase the performance
param_grid = [
{
"n_estimators": [100],
"random_state": [42],
"criterion": ["squared_error", "absolute_error", "friedman_mse", "poisson"],
"bootstrap": [False, True],
"max_depth": [
10,
20,
],
"max_features": [1, None],
"min_samples_split": [2, 10],
"max_leaf_nodes": [2, None],
"min_impurity_decrease": [0, 1],
"min_samples_leaf": [1, 3],
"min_weight_fraction_leaf": [0.0, 0.25],
}
]
rf_reg = RandomForestRegressor()
grid_search = GridSearchCV(rf_reg, param_grid, cv=5)
grid_search.fit(X_train, y_train)
print(
"Best parameters: ",
grid_search.best_params_,
"Best score: ",
grid_search.best_score_,
)
# lst = [i for i in range(100)]
param_grid = [
{
"n_estimators": [100],
"random_state": [65],
"criterion": ["friedman_mse"],
"bootstrap": [False],
"max_depth": [20],
"max_features": [None],
"min_samples_split": [2],
"max_leaf_nodes": [None],
"min_impurity_decrease": [0],
"min_samples_leaf": [1],
"min_weight_fraction_leaf": [0.0],
}
]
rf_reg = RandomForestRegressor()
grid_search = GridSearchCV(rf_reg, param_grid, cv=5)
grid_search.fit(X_train, y_train)
print(
"Best parameters: ",
grid_search.best_params_,
"Best score: ",
grid_search.best_score_,
)
# Function to get RMSE and R2 results for selected models
def display_results(X_train, y_train, X_test, y_test, reg):
reg.fit(X_train, y_train)
y_pred = reg.predict(X_test)
print("RMSE: ", np.sqrt(mean_squared_error(y_test, y_pred)))
print("R2: ", r2_score(y_test, y_pred))
# Running the test set with the best paratemeters using Linear Regression
lin_reg = LinearRegression()
display_results(X_train, y_train, X_test, y_test, lin_reg)
# Running the test set with the best paratemeters using Ridge Regression
ridge_reg = Ridge(alpha=1, max_iter=1000, solver="sparse_cg", tol=1e-06)
display_results(X_train, y_train, X_test, y_test, ridge_reg)
# Running the test set with the best paratemeters using BayesianRidge Regression
br_reg = BayesianRidge(
alpha_1=1e-08, alpha_2=0.0001, lambda_1=0.0001, lambda_2=0.0001, tol=0.01
)
display_results(X_train, y_train, X_test, y_test, br_reg)
# Running the test set with the best paratemeters using XGB Regression
xgb_reg = XGBRegressor()
display_results(X_train, y_train, X_test, y_test, xgb_reg)
# Running the test set with the best paratemeters using KNN Regression
knn_reg = KNeighborsRegressor(leaf_size=100, n_neighbors=8, p=1, weights="distance")
display_results(X_train, y_train, X_test, y_test, knn_reg)
# Running the test set with the best paratemeters using Random Forest Regression
rf_reg = RandomForestRegressor(
bootstrap=False, criterion="friedman_mse", max_depth=20, max_features=None
)
display_results(X_train, y_train, X_test, y_test, rf_reg)
|
import tensorflow as tf
from tensorflow.keras.preprocessing.image import ImageDataGenerator
from tensorflow.keras.models import Sequential
from tensorflow.keras.layers import Conv2D, MaxPooling2D, Flatten, Dense, Dropout
from tensorflow.keras.callbacks import EarlyStopping, ModelCheckpoint
import matplotlib.pyplot as plt
train_data_dir = "/kaggle/input/soybean-seedsclassification-dataset"
train_datagen = ImageDataGenerator(
rescale=1.0 / 255, shear_range=0.2, zoom_range=0.2, horizontal_flip=True
)
train_generator = train_datagen.flow_from_directory(
train_data_dir,
target_size=(150, 150),
batch_size=32,
class_mode="categorical",
shuffle=True,
)
model = Sequential()
model.add(Conv2D(32, (3, 3), activation="relu", input_shape=(150, 150, 3)))
model.add(MaxPooling2D((2, 2)))
model.add(Conv2D(64, (3, 3), activation="relu"))
model.add(MaxPooling2D((2, 2)))
model.add(Conv2D(128, (3, 3), activation="relu"))
model.add(MaxPooling2D((2, 2)))
model.add(Conv2D(128, (3, 3), activation="relu"))
model.add(MaxPooling2D((2, 2)))
model.add(Flatten())
model.add(Dense(512, activation="relu"))
model.add(Dropout(0.5))
model.add(Dense(5, activation="softmax"))
model.compile(loss="categorical_crossentropy", optimizer="adam", metrics=["accuracy"])
# Train the model
history = model.fit(train_generator, steps_per_epoch=100, epochs=30, verbose=1)
# Save the model to a different directory
model.save("/kaggle/working/my_model.h5")
test_data_dir = "/kaggle/input/soybean-seedsclassification-dataset"
test_datagen = ImageDataGenerator(rescale=1.0 / 255)
test_generator = test_datagen.flow_from_directory(
test_data_dir,
target_size=(150, 150),
batch_size=32,
class_mode="categorical",
shuffle=False,
)
scores = model.evaluate(test_generator, verbose=1)
print("Test loss:", scores[0])
print("Test accuracy:", scores[1])
test_data_dir = "/kaggle/input/soybean-seedsclassification-dataset"
test_datagen = ImageDataGenerator(rescale=1.0 / 255)
test_generator = test_datagen.flow_from_directory(
test_data_dir,
target_size=(150, 150),
batch_size=16,
class_mode="categorical",
shuffle=False,
)
scores = model.evaluate(test_generator, verbose=1)
print("Test loss:", scores[0])
print("Test accuracy:", scores[1])
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
data = pd.read_csv(
"/kaggle/input/heart-failure-clinical-data/heart_failure_clinical_records_dataset.csv"
)
data.head()
train = data.iloc[:, :-1]
y = data.iloc[:, -1]
from sklearn.preprocessing import StandardScaler
sc_X = StandardScaler()
train = sc_X.fit_transform(train)
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
train, y, test_size=0.20, random_state=42
)
from sklearn.ensemble import RandomForestClassifier
clf = RandomForestClassifier(max_depth=15, random_state=0)
clf.fit(X_train, y_train)
pred = clf.predict(X_test)
pred
from sklearn.metrics import accuracy_score
acc_rf = accuracy_score(y_test, pred)
acc_rf
from xgboost import XGBClassifier
model = XGBClassifier(learning_rate=0.04)
model.fit(X_train, y_train)
y_pred_xg = model.predict(X_test)
y_pred_xg
acc_rf = accuracy_score(y_test, y_pred_xg)
acc_rf
|
# TASK:
# As a part of our project for the data modelling and visualisation class at Singapore Management University, we were given the Sales Data of a fictional company, TGL, which has multiple branches.
# We utilised Tableau to visualise the performance of our assigned branch, branch 2. We measured its performance over time and compared it against other branches. We even measured the performance of the products it sells over time for a complete performance analysis.
# The visualisation is catered towards the senior management of the company and we were asked to fulfil 3 tasks:
# a) measure the performance of the branch in 2018-20 and 2021-22
# b) identify the product which TGL should focus its resources on
# c)Identify the best performing branch over the last 5 years.
# Finally,We utilised python to extract the saved images of the tableau visualizations into Kaggle
# I would like to thank my professor and my groupmates for their help in completing this project
# DESIGN CONSIDERATIONS:
#
# our design choices were influenced by the expertise and understanding of our audience, TGL’s Senior Management team. Our data is represented with tables with numerical information for those familiar with accounting terms. And for those unfamiliar with accounting, we used highlight tables which change the intensity of the colors based on the relative values of the table to facilitate comparison. This combination of visual and numerical presentation accounts for differences in skill levels across our audience and helps them in achieving a comparable visual understanding. To further ensure that the data is easily accessible, we utilized the following visual tools to translate numbers into accessible visual data: Bar charts, Line graphs, and Pie charts.
# For the Branch performance, we utilized two contrasting colors at different intensities based on the period to separate sales value and net income. We differentiated based on color and intensity so that the audience can easily make an association between the color groups and the time groups. While measuring the product and inter-branch performance, we utilized distinct colors for each product/branch to make it easier to distinguish between them. By creating visualizations and using unique colors, we can derive various insights from the same data while also making it accessible to a variety of users.
from IPython.display import Image
Image(filename="/kaggle/input/branch-performance/Branch 2 performance.png")
# Using the Branch 2 Sales and Net Income table, the senior management will be able to understand the total dollar sales revenue and net income that the branch has generated over the last five years. The bar chart below represents the same data visually. The management can compare the heights of the bars to compare the branch's performance over time.
# The highest year-over-year net income growth for branch 2 occurred between 2018 and 2019, where it experienced a 267.42% growth in net income. Branch 2 also experienced similar patterns relating to its sales revenue, with the highest year-on year sales growth of 128.77% occurring between 2018 and 2019.
# During the timespan between 2018 and 2020, Branch 2 has increased its net income from 1.24 million to 6.45 million, and increased dollar sales from 5.45 million to 17.73 million USD. This implies an overall growth of 420% in net income and a 223% growth in Sales.
# Between 2021 and 2022, Branch 2 has increased its net income from 14.12 million to 16.9 million USD, and increased dollar sales from 34.39 million to 43.38 million USD. This implies an overall growth of 19.73% in Net income and a 26.13% growth in sales.
# When compared to the different time periods, we observe that the 2018-2020 period experiences a larger year-on-year growth than the 2021-2022 period. But it is important to consider that despite the lower growth, the 2021-2022 period brings in a larger number of sales and net income when measured in USD.
from IPython.display import Image
Image(filename="/kaggle/input/branch-performance/Inter branch performance.png")
# From the Branch Net income table, the line charts, and pie charts, the senior management will be able to draw insights in the same fashion as the product performance section. However, instead of products, they will compare the performance of different branches.
# Between 2018 and 2022, all branches have an increase in their net income, with the highest percentage and dollar value increase coming from Branch 2 with a 1,266.6% increase from 2018 to 2022 and representing an SGD 15,664,417 increase over its original income. In terms of the magnitude of increase, Branch 2 is the best performer out of the 5 branches. Branches 1, 3, 4 and 5 had percentage increases of 27.2%, 81.4%, 45.3% and 7.2% respectively.
# Ideally, we would want to identify the best performer with a steady upwards trend from 2018 to 2022. Therefore, with a visual comparison, we can compare the year-on-year increase of each branch. We note that the branches show a general increasing trend except for Branch 1 and Branch 5 which experienced a slight decrease in net income between 2020 and 2021. We also note that out of the 5 branches, Branch 2 showed the steepest increase in net income every year from 2018 to 2020. Branch 2 grew from being the 2nd smallest contributor to TGL’s net income to the largest contributor to its net income over the same period.
# Thus, based on the above data and analysis, Branch 2 is the best performer between 2018 to 2022.
from IPython.display import Image
Image(filename="/kaggle/input/branch-performance/Product performance.png")
|
# # Predicting Airbnb Prices
# # Importing Data
import pandas as pd
import numpy as np
import matplotlib as mpl
import matplotlib.pyplot as plt
import seaborn as sns
import os
from sklearn import set_config
plt.style.use("ggplot")
pd.set_option("display.max_columns", 100)
# set_config(transform_output="pandas") #doesn't work here :(
os.chdir("/kaggle/input/airbnb-cleaned-europe-dataset")
df = pd.read_csv("Aemf1.csv")
df
df.info()
# # Selecting price range for model
# Using a predictive model to predict the price of outliers may not be appropriate. The existing price distribution appears to contain high price outliers.
sns.histplot(np.log(df["Price"]))
df.sort_values("Price", ascending=False).head()
# To identify outliers, the price distribution from the most expensive case, where the city is Amsterdam and the room capacity is > 6, is used.
from scipy.stats import zscore
df_expensive = df[(df["City"] == "Amsterdam") & (df["Person Capacity"] == 6.0)]
mean_val = np.mean(df_expensive["Price"])
std_dev = np.std(df_expensive["Price"])
threshold = 3
lower_bound = mean_val - threshold * std_dev
upper_bound = mean_val + threshold * std_dev
df_no_outliers = df[df["Price"] < upper_bound] # &(z_scores>-4)]
print("With outliers exclusded..")
print(f"New max price: ${upper_bound:.2f}")
print(f"New min price: ${lower_bound:.2f}")
sns.histplot(np.log(df_no_outliers["Price"]))
plt.show()
# Only prices under \\\$6,500 will be considered by this model, and no outliers will be included in our analyses. In application, this model should not be used to predict the price of an airbnb if it is suspected that its price will exceed \\\$6,500/night.
df = df[df["Price"] < 6500].copy()
df.info()
# # EDA
# ## Heat map
# Correlations between variables are visualized in the heat map below.
# - Cleaner rooms are rated higher.
# - Attractions and restraunts are found in the same locations.
# - City centers are further from the metro.
# Heat map
# optimizng range for color scale
min = df.corr().min().min()
max = df.corr()[df.corr() != 1].max().max()
# thresholding selected correlations
df_corr = df.corr()[np.absolute(df.corr()) > 0.3]
# Mask for selecting only bottom triangle
mask = np.triu(df_corr)
with plt.style.context("default"):
sns.heatmap(df_corr, vmin=min, vmax=max, mask=mask)
# ## Pair plots
# Distributions and relationships are plotted for selected parameters below:
# raw data
sns.pairplot(
df[
[
"City Center (km)",
"Metro Distance (km)",
"Attraction Index",
"Restraunt Index",
"Price",
]
],
kind="hist",
corner=True,
)
# We see that these features are better represented in log space:
# rescaled data
df_trial = pd.DataFrame()
df_trial["City Center (km)"] = np.log(df["City Center (km)"])
df_trial["Metro Distance (km)"] = np.log(df["Metro Distance (km)"])
df_trial["Attraction Index"] = np.log(df["Attraction Index"])
df_trial["Restraunt Index"] = np.log(df["Restraunt Index"])
df_trial["Price"] = np.log(df["Price"])
sns.pairplot(df_trial, kind="hist", corner=True)
# ## RBF Definition
# We replace features with an apparent radial price distribution with an radial basis function.
# Metro, city center and restraunt index appear to have radial price distributions
from sklearn.metrics.pairwise import rbf_kernel
df["rbf_metro"] = rbf_kernel(df_trial[["Metro Distance (km)"]], [[-0.5]], gamma=1)
df["rbf_city"] = rbf_kernel(df_trial[["City Center (km)"]], [[0.3]], gamma=0.5)
df["rbf_res"] = rbf_kernel(df_trial[["Restraunt Index"]], [[6.25]], gamma=0.5)
# visualizing metro rbf function
fig, ax1 = plt.subplots(1)
plt.bar(df_trial["Metro Distance (km)"], df["Price"])
plt.xlabel("Log Metro Distance (km)")
plt.ylabel("Price")
ax2 = ax1.twinx()
ax2.scatter(df_trial["Metro Distance (km)"], df["rbf_metro"], color="k", s=0.5)
ax2.set_ylim([0, 1])
ax2.set_ylabel("Price rbf")
plt.show()
# visualizing city rbf function
fig, ax1 = plt.subplots(1)
plt.bar(df_trial["City Center (km)"], df["Price"])
plt.xlabel("Log City Center (km)")
plt.ylabel("Price")
ax2 = ax1.twinx()
ax2.scatter(df_trial["City Center (km)"], df["rbf_city"], color="k", s=0.5)
ax2.set_ylim([0, 1])
ax2.set_ylabel("City rbf")
plt.show()
# visualizing city rbf function
fig, ax1 = plt.subplots(1)
plt.bar(df_trial["Restraunt Index"], df["Price"])
plt.xlabel("Log Restraunt Index (km)")
plt.ylabel("Price")
ax2 = ax1.twinx()
ax2.scatter(df_trial["Restraunt Index"], df["rbf_res"], color="k", s=0.5)
ax2.set_ylim([0, 1])
ax2.set_ylabel("Restraunt rbf")
plt.show()
# ## Visualizing categorical data
fig, ax = plt.subplots(1)
fig.set_size_inches(8, 4)
sns.boxplot(data=df, x="City", y="Price", showfliers=False)
ax.set_ylim([0, 1300])
plt.show()
sns.boxplot(data=df, x="Superhost", y="Price", showfliers=False)
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(8, 3))
plt.sca(ax1)
sns.boxplot(data=df, x="Room Type", y="Price", showfliers=False)
plt.sca(ax2)
sns.boxplot(data=df, x="Shared Room", y="Price", showfliers=False)
plt.tight_layout()
sns.boxplot(data=df, x="Day", y="Price", showfliers=False)
sns.boxplot(data=df, x="Person Capacity", y="Price", showfliers=False)
plt.ylim([0, 1000])
plt.show()
# ## Visualizing consumer rankings
# sns.regplot(data = df[df["Price"]<2000], x='Cleanliness Rating', y='Price',scatter=True,scatter_kws={'alpha':0.05},line_kws={"color": "black"});
sns.jointplot(
x=df["Cleanliness Rating"],
y=np.log(df["Price"]),
kind="reg",
scatter_kws={"alpha": 0.05},
line_kws={"color": "black"},
)
plt.show()
# sns.regplot(data = df[df['Price']<2000], x='Guest Satisfaction', y='Price',scatter_kws={'alpha':0.05},line_kws={"color": "black"})
sns.jointplot(
x=df["Guest Satisfaction"],
y=np.log(df["Price"]),
kind="reg",
scatter_kws={"alpha": 0.05},
line_kws={"color": "black"},
)
plt.show()
# # Cleaning Data
# Three preprocessing transformations are defined for selecting different subsets of input features.
from sklearn.compose import ColumnTransformer
from sklearn.preprocessing import FunctionTransformer
from sklearn.preprocessing import OneHotEncoder
from sklearn.preprocessing import MinMaxScaler
from sklearn.base import BaseEstimator, TransformerMixin
from sklearn.pipeline import make_pipeline
from sklearn.model_selection import train_test_split
# defining functions for column transformer
cat_encoder = OneHotEncoder()
log_pipeline = make_pipeline(
FunctionTransformer(np.log), MinMaxScaler() # , inverse_func=np.exp),
)
def day_2_num(X):
return X == "Weekend"
def day_pipeline():
return make_pipeline(FunctionTransformer(day_2_num))
day_pipe = day_pipeline()
# defining standard column transformer
preprocessing = ColumnTransformer(
[
("day", day_pipe, ["Day"]),
(
"drop",
"drop",
[
"Normalised Attraction Index",
"Normalised Restraunt Index",
"rbf_metro",
"rbf_city",
"rbf_res",
],
),
(
"pass",
"passthrough",
["Private Room", "Shared Room", "Superhost", "Business", "Multiple Rooms"],
),
(
"maxscale",
MinMaxScaler(),
["Cleanliness Rating", "Bedrooms", "Guest Satisfaction"],
),
(
"log",
log_pipeline,
[
"Attraction Index",
"City Center (km)",
"Metro Distance (km)",
"Restraunt Index",
],
),
("cat", cat_encoder, ["Room Type", "City"]),
]
)
# column transformer with rbf functions instead of metro, city, and restraunts
preprocessing_rbf = ColumnTransformer(
[
("day", day_pipe, ["Day"]),
(
"drop",
"drop",
[
"Normalised Attraction Index",
"Normalised Restraunt Index",
"Metro Distance (km)",
"City Center (km)",
"Restraunt Index",
],
),
(
"pass",
"passthrough",
["Private Room", "Shared Room", "Superhost", "Business", "Multiple Rooms"],
),
(
"maxscale",
MinMaxScaler(),
["Cleanliness Rating", "Bedrooms", "Guest Satisfaction"],
),
("log", log_pipeline, ["Attraction Index"]),
("pass2", "passthrough", ["rbf_metro", "rbf_city", "rbf_res"]),
("cat", cat_encoder, ["Room Type", "City"]),
]
)
# column transformer with given normalized features
preprocessing_norm = ColumnTransformer(
[
("day", day_pipe, ["Day"]),
(
"drop",
"drop",
["Attraction Index", "Restraunt Index", "rbf_metro", "rbf_city", "rbf_res"],
),
(
"pass",
"passthrough",
["Private Room", "Shared Room", "Superhost", "Business", "Multiple Rooms"],
),
(
"maxscale",
MinMaxScaler(),
["Cleanliness Rating", "Bedrooms", "Guest Satisfaction"],
),
("pass2", "passthrough", ["Normalised Attraction Index"]),
("log", log_pipeline, ["City Center (km)", "Metro Distance (km)"]),
("pass3", "passthrough", ["Normalised Restraunt Index"]),
("cat", cat_encoder, ["Room Type", "City"]),
]
)
# naming output columns
names = pd.Series(
[
"Weekend",
"Private Room",
"Shared Room",
"Superhost",
"Business",
"Multiple Rooms",
"Cleanliness Rating",
"Bedrooms",
"Guest Satisfaction",
"Attraction Index",
"City Center",
"Metro Distance",
"Restraunt Index",
"Private room",
"Entire home/apt",
"Shared room",
"Amsterdam",
"Athens",
"Barcelona",
"Berlin",
"Budapest",
"Lisbon",
"Paris",
"Rome",
"Vienna",
]
)
# transforming data
df_processed = pd.DataFrame(preprocessing.fit_transform(df), columns=names)
# splitting data into test and train sets
y = df["Price"].copy()
X = df.drop(columns={"Price"})
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.2, random_state=10
)
# # Predicing Price
# importing packages
from sklearn.linear_model import LinearRegression, Ridge, ElasticNet
from sklearn.neighbors import KNeighborsRegressor
from sklearn.svm import SVR
from sklearn.ensemble import (
RandomForestRegressor,
ExtraTreesRegressor,
AdaBoostRegressor,
GradientBoostingRegressor,
)
from sklearn.metrics import mean_squared_error
import xgboost as xgb
from sklearn.decomposition import PCA
# ## Sampling regression models
# Common regression models were evaluated with six variations:
# 1. No rerescaling or feature engineering
# 2. Price target transformed to log-space
# 3. Dimensionality rediction via PCA
# 4. Feature engineering via replacement with RBF
# 5. Provided normalized resraunt and attraction indexes used
# 5. Combining variations 2-4
# defining mean absolute percentage error
def mape(y_true, y_pred):
return np.mean(np.abs((y_true - y_pred) / y_true)) * 100
# unscaled y
np.random.seed(10)
models = [
Ridge(),
ElasticNet(),
SVR(),
KNeighborsRegressor(),
RandomForestRegressor(),
ExtraTreesRegressor(),
AdaBoostRegressor(),
GradientBoostingRegressor(),
xgb.XGBRegressor(),
]
model_names = [
"Ridge",
"ElasticNet",
"SVR",
"K-nearest neighbors",
"Random Forest",
"Extra Trees",
"Adaptive Boosting",
"Gradient Boosting",
"XGBoost",
]
mse = []
mape_err = []
for model in models:
sample_pipe = make_pipeline(preprocessing, model)
sample_pipe.fit(X_train, y_train)
y_pred = sample_pipe.predict(X_test)
mse.append(mean_squared_error(y_test, y_pred))
mape_err.append(mape(y_test, y_pred))
# scaled y
scaled_mse = []
scaled_mape = []
for model in models:
scaled_pipe = make_pipeline(preprocessing, model)
scaled_pipe.fit(X_train, np.log(y_train))
y_pred = scaled_pipe.predict(X_test)
y_pred = np.exp(y_pred)
scaled_mse.append(mean_squared_error(y_test, y_pred))
scaled_mape.append(mape(y_test, y_pred))
# with dimensionality reduction
pca = PCA(n_components=0.95)
pca_mse = []
pca_mape = []
for model in models:
pca_pipe = make_pipeline(preprocessing, pca, model)
pca_pipe.fit(X_train, y_train)
y_pred = pca_pipe.predict(X_test)
pca_mse.append(mean_squared_error(y_test, y_pred))
pca_mape.append(mape(y_test, y_pred))
# with rbf
rbf_mse = []
rbf_mape = []
for model in models:
rbf_pipe = make_pipeline(preprocessing_rbf, model)
rbf_pipe.fit(X_train, y_train)
y_pred = rbf_pipe.predict(X_test)
rbf_mse.append(mean_squared_error(y_test, y_pred))
rbf_mape.append(mape(y_test, y_pred))
# with normalized features
norm_mse = []
norm_mape = []
for model in models:
norm_pipe = make_pipeline(preprocessing_norm, model)
norm_pipe.fit(X_train, y_train)
y_pred = norm_pipe.predict(X_test)
norm_mse.append(mean_squared_error(y_test, y_pred))
norm_mape.append(mape(y_test, y_pred))
# combination
combo_error = []
combo_mape = []
for model in models:
combo_pipe = make_pipeline(preprocessing_rbf, pca, model)
combo_pipe.fit(X_train, np.log(y_train))
y_pred = combo_pipe.predict(X_test)
y_pred = np.exp(y_pred)
combo_mse.append(mean_squared_error(y_test, y_pred))
combo_mape.append(mape(y_test, y_pred))
mse_results = pd.DataFrame(
[mse, scaled_mse, pca_mse, rbf_mse, norm_mse, combo_mse],
index=[
"Unscaled",
"Scaled",
"Reduced Dimensions",
"RBF Features",
"Normalized Features",
"Combination",
],
columns=model_names,
).T
mape_results = pd.DataFrame(
[mape_err, scaled_mape, pca_mape, rbf_mape, norm_mape, combo_mape],
index=[
"Unscaled",
"Scaled",
"Reduced Dimensions",
"RBF Features",
"Normalized Features",
"Combination",
],
columns=model_names,
).T
fig, (ax1, ax2) = plt.subplots(2)
plt.sca(ax1)
mse_results.plot(kind="barh")
# plt.axvline((sum(error)+sum(scaled_error)+sum(pca_error))/(len(error)+len(scaled_error)+len(pca_error)), color='k', linestyle='--')
plt.title("Regressor RMSE")
plt.xlim([0, 100000])
plt.sca(ax2)
mape_results.plot(kind="barh")
plt.legend(loc=4, facecolor="white")
plt.show()
sample_results.style.format("{:.0f}")
# The top performing classifier was the extra trees regressor with a scaled price target.
# ## Optimizing the extra tree regressor
from sklearn.model_selection import RandomizedSearchCV, GridSearchCV
from scipy.stats import loguniform, expon, uniform, randint
from skopt import BayesSearchCV
from skopt.space import Real, Categorical, Integer
from skopt.plots import plot_objective
# ### GridSearchCV
# grid search
tree_pipe = make_pipeline(
preprocessing, ExtraTreesRegressor(n_jobs=-1, random_state=10)
)
n_estimators = np.arange(20, 200, 5)
max_features = ["sqrt", "log2"]
max_depth = np.arange(1, 20, 2)
tree_params = {
"extratreesregressor__n_estimators": n_estimators,
"extratreesregressor__max_features": max_features,
"extratreesregressor__max_depth": max_depth,
}
tree_search = GridSearchCV(
tree_pipe, tree_params, scoring="neg_root_mean_squared_error", cv=5, n_jobs=-1
)
tree_search.fit(X_train, np.log(y_train))
tree_search.best_score_, tree_search.best_params_
# comparison of best gridsearch hyperparameter set to default set
baseline = error[5]
improvement = (baseline - tree_search.best_score_ ^ 2) / baseline * 100
# 100% improvement with max_depth = 17, max_features='sqrt',and n_estimators=85
print("Regressor performance improved by %.0f%%" % improvement)
# formats results of gridsearch into a dataframe
cv_results = pd.DataFrame(rnd_search.cv_results_)
cv_results = cv_results[
[
"param_randomforestregressor__n_estimators",
"param_randomforestregressor__max_features",
"param_randomforestregressor__max_depth",
"mean_test_score",
]
]
cv_results = cv_results.rename(
columns={
"param_randomforestregressor__n_estimators": "n_estimators",
"param_randomforestregressor__max_features": "max_features",
"param_randomforestregressor__max_depth": "max_depth",
"mean_test_score": "rmse",
}
)
cv_results["rmse"] = cv_results["rmse"] * -1
cv_results["max_features"] = pd.get_dummies(
cv_results["max_features"], drop_first=True
) # 1 = sqrt, 0=log2
# plots gridsearch results
with plt.style.context("default"):
fig = plt.figure(figsize=(8, 8))
ax = fig.add_subplot(111, projection="3d")
sc = ax.scatter(
cv_results["n_estimators"],
cv_results["max_depth"],
cv_results["max_features"],
c=cv_results["rmse"],
cmap="coolwarm",
)
ax.set_xlabel("n_estimators")
ax.set_ylabel("max_depth")
ax.set_zlabel("max_features")
cbar = fig.colorbar(sc, shrink=0.1)
cbar.set_label("RMSE")
plt.show()
# ### BayesSearchCV
rf_pipeline = make_pipeline(
preprocessing, RandomForestRegressor(random_state=10, n_jobs=-1)
)
rf_search = {
"randomforestregressor__n_estimators": Integer(70, 90),
"randomforestregressor__max_depth": Integer(15, 25),
#'randomforestregressor__min_weight_fraction_leaf': Real(0,.1,'uniform'),
#'randomforestregressor__max_features':Categorical(['sqrt','log2']),
#'randomforestregressor__max_leaf_nodes': Integer(2,10),
#'randomforestregressor__bootstrap':Categorical([True,False])
}
opt = BayesSearchCV(
rf_pipeline, [(rf_search, 50)], scoring="neg_root_mean_squared_error", cv=3
)
opt.fit(X_train, y_train)
print("val. score: %s" % opt.best_score_)
print("test score: %s" % opt.score(X_test, y_test))
print("best params: %s" % str(opt.best_params_))
rnd_best_pipe = make_pipeline(
preprocessing, RandomForestRegressor(n_estimators=132)
).fit(X_train, y_train)
y_pred = rnd_best_pipe.predict(X_test)
mean_squared_error(y_test, y_pred)
_ = plot_objective(
opt.optimizer_results_[0],
# dimensions=['bootstrap','max_features','max_leaf_nodes','min_leaf_weight_fraction','n_estimators'],
n_minimum_search=int(1e8),
)
plt.show()
|
import numpy as np
import pandas as pd
train_labels = pd.read_csv("../input/bms-molecular-translation/train_labels.csv")
# # All training label strings start with "InChI=1S/"
train_labels["first9"] = [train_label[:9] for train_label in train_labels["InChI"]]
train_labels["drop9"] = [train_label[9:] for train_label in train_labels["InChI"]]
train_labels.value_counts("first9")
train_labels["InChI"].values[0]
train_labels["drop9"].values[0]
|
# # Google Landmark Recognition Challenge 2020
# Simplified image similarity ranking and re-ranking implementation with:
# * EfficientNetB0 backbone for global feature similarity search
# * DELF module for local feature reranking
# Reference papers:
# * 2020 Recognition challenge winner: https://arxiv.org/abs/2010.01650
# * 2019 Recognition challend 2nd place: https://arxiv.org/abs/1906.03990
# Importing libraries
import os
import cv2
import shutil
import numpy as np
import pandas as pd
from scipy import spatial
from tqdm import tqdm
import tensorflow as tf
from tensorflow.keras import layers
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.model_selection import train_test_split
# Directories and file paths
TRAIN_DIR = "../input/landmark-recognition-2020/train"
TRAIN_CSV = "../input/landmark-recognition-2020/train.csv"
train_df = pd.read_csv(TRAIN_CSV)
TRAIN_PATHS = [
os.path.join(TRAIN_DIR, f"{img[0]}/{img[1]}/{img[2]}/{img}.jpg")
for img in train_df["id"]
]
train_df["path"] = TRAIN_PATHS
train_df
# Subsetting
train_df_grouped = pd.DataFrame(train_df.landmark_id.value_counts())
train_df_grouped.reset_index(inplace=True)
train_df_grouped.columns = ["landmark_id", "count"]
# Selected landmarks based on inclass frequency
selected_landmarks = train_df_grouped[
(train_df_grouped["count"] <= 155) & (train_df_grouped["count"] >= 150)
]
train_df_sub = train_df[train_df["landmark_id"].isin(selected_landmarks["landmark_id"])]
new_id = []
current_id = 0
previous_id = int(train_df_sub.head(1)["landmark_id"])
for landmark_id in train_df_sub["landmark_id"]:
if landmark_id == previous_id:
new_id.append(current_id)
else:
current_id += 1
new_id.append(current_id)
previous_id = landmark_id
train_df_sub["new_id"] = new_id
NUM_CLASSES = train_df_sub["landmark_id"].nunique()
print(f"Unique classes found: {NUM_CLASSES}")
train_df_sub
# Training and validation splits
# 90/10 stratified split for training and validation
X_train, X_val, y_train, y_val = train_test_split(
train_df_sub[["id", "path"]],
train_df_sub["new_id"],
train_size=0.9,
random_state=123,
shuffle=True,
stratify=train_df_sub["new_id"],
)
# Held-out test set for inference
# Further 95/5 split -> 5% of original training set left for test set
X_train, X_test, y_train, y_test = train_test_split(
X_train, y_train, train_size=0.95, random_state=123, shuffle=True, stratify=y_train
)
assert X_train.shape[0] + X_val.shape[0] + X_test.shape[0] == train_df_sub.shape[0]
print(f"Training data shape: {X_train.shape}")
print(f"Training label shape: {y_train.shape}")
print(f"Validation data shape: {X_val.shape}")
print(f"Validation label shape: {y_val.shape}")
print(f"Test data shape: {X_test.shape}")
print(f"Test label shape: {y_test.shape}")
print(f"Unique classes on y_train: {y_train.nunique()}")
print(f"Unique classes on y_val: {y_val.nunique()}")
print(f"Unique classes on y_test: {y_test.nunique()}")
# Classes distribution on training, validation and test sets
plt.figure(figsize=(10, 3))
ax = sns.histplot(y_train, bins=75, kde=True)
ax.set_title("Distribution of Landmarks on training set")
plt.tight_layout()
plt.figure(figsize=(10, 3))
ax = sns.histplot(y_val, bins=75, kde=True)
ax.set_title("Distribution of Landmarks on validation set")
plt.tight_layout()
plt.figure(figsize=(10, 3))
ax = sns.histplot(y_test, bins=75, kde=True)
ax.set_title("Distribution of Landmarks on test set")
plt.tight_layout()
plt.show()
# Creating image directories for classes subset
NEW_BASE_DIR = "/kaggle/working"
# Training set directory
for file, path, landmark in tqdm(zip(X_train["id"], X_train["path"], y_train)):
dir = f"{NEW_BASE_DIR}/train_sub/{str(landmark)}"
os.makedirs(dir, exist_ok=True)
fname = f"{file}.jpg"
shutil.copyfile(src=path, dst=f"{dir}/{fname}")
# Validation set directory
for file, path, landmark in tqdm(zip(X_val["id"], X_val["path"], y_val)):
dir = f"{NEW_BASE_DIR}/val_sub/{str(landmark)}"
os.makedirs(dir, exist_ok=True)
fname = f"{file}.jpg"
shutil.copyfile(src=path, dst=f"{dir}/{fname}")
# Training set directory
for file, path, landmark in tqdm(zip(X_test["id"], X_test["path"], y_test)):
dir = f"{NEW_BASE_DIR}/test_sub/{str(landmark)}"
os.makedirs(dir, exist_ok=True)
fname = f"{file}.jpg"
shutil.copyfile(src=path, dst=f"{dir}/{fname}")
# Creating tensorflow tf.data.Dataset
from tensorflow.keras.utils import image_dataset_from_directory
IMG_SIZE = 224
BATCH_SIZE = 16
print("Building training dataset...")
# Training tf.data.Dataset
train_ds = image_dataset_from_directory(
f"{NEW_BASE_DIR}/train_sub",
label_mode="int",
shuffle=True,
image_size=(IMG_SIZE, IMG_SIZE),
batch_size=BATCH_SIZE,
)
print("Building validation dataset...")
# Validation tf.data.Dataset
val_ds = image_dataset_from_directory(
f"{NEW_BASE_DIR}/val_sub",
label_mode="int",
shuffle=True,
image_size=(IMG_SIZE, IMG_SIZE),
batch_size=BATCH_SIZE,
)
print("Building test dataset...")
# Test tf.data.Dataset
test_ds = image_dataset_from_directory(
f"{NEW_BASE_DIR}/test_sub",
label_mode="int",
shuffle=True,
image_size=(IMG_SIZE, IMG_SIZE),
batch_size=BATCH_SIZE,
)
# Visualizing a random batch from training dataset
for data_batch, labels_batch in train_ds.take(1):
ncols = 4
nrows = int(data_batch.shape[0] / ncols)
fig, ax = plt.subplots(
nrows=nrows, ncols=ncols, figsize=(10, 11), sharex=True, sharey=True
)
img_counter = 0
for image, label in zip(data_batch, labels_batch):
axi = ax.flat[img_counter]
axi.imshow(image / 255.0)
label = label.numpy()
# axi.set_title(np.where(label == 1)[0])
axi.set_title(label)
img_counter += 1
plt.show()
####### ALTERNATIVE CODE FOR UNBATCHED DATASET #######
# ncols = 4
# nrows = 4
# fig, ax = plt.subplots(nrows = nrows, ncols = ncols, figsize=(10, 11),
# sharex = True, sharey = True)
# img_counter = 0
# for image, label in train_ds.take(16):
# axi = ax.flat[img_counter]
# axi.imshow(image[0]/255.)
# label = label.numpy()
# # axi.set_title(np.where(label == 1)[0])
# axi.set_title(label)
# img_counter += 1
# plt.show()
# Defining a data augmentation stage
img_augmentation = tf.keras.Sequential(
# [layers.RandomFlip("horizontal"),
[
layers.RandomTranslation(height_factor=0.1, width_factor=0.1),
layers.RandomRotation(0.02),
layers.RandomZoom(0.2),
],
name="img_augmentation",
)
# Displaying variations of a randomly augmented training image
plt.figure(figsize=(9, 9))
for image, label in train_ds.take(1):
for i in range(9):
ax = plt.subplot(3, 3, i + 1)
augmented_image = img_augmentation(image, training=True)
plt.imshow(augmented_image[15].numpy().astype("uint8"))
plt.axis("off")
####### ALTERNATIVE CODE FOR UNBATCHED DATASET #######
# # Displaying variations of a randomly augmented training image
# plt.figure(figsize=(9, 9))
# for image, label in train_ds.take(16):
# for i in range(9):
# ax = plt.subplot(3, 3, i + 1)
# augmented_image = img_augmentation(image[0], training = True)
# plt.imshow(augmented_image.numpy().astype("uint8"))
# plt.axis("off")
# Model
from tensorflow import keras
from tensorflow.keras import layers
from tensorflow.keras.applications import EfficientNetB0
MODELS_DIR = f"{NEW_BASE_DIR}/models"
os.makedirs(MODELS_DIR, exist_ok=True)
# Model instantiator
def build_model(num_classes=None):
inputs = keras.Input(shape=(IMG_SIZE, IMG_SIZE, 3))
x = img_augmentation(inputs)
# EfficientNetB0 backbone
model = EfficientNetB0(
input_tensor=x,
weights="imagenet",
include_top=False,
drop_connect_rate=DROP_CONNECT_RATE,
)
# Freeze pretrained weights
model.trainable = False
# Rebuild top
x = layers.GlobalAveragePooling2D(name="avg_pool")(model.output)
x = layers.BatchNormalization()(x)
x = layers.Dropout(TOP_DROPOUT_RATE, name="top_dropout")(x)
# Embedding
embedding = layers.Dense(512, name="embedding_512")(x)
outputs = layers.Dense(num_classes, activation="softmax", name="softmax")(embedding)
# Compile
model = tf.keras.Model(inputs, outputs, name="EfficientNetB0")
optimizer = tf.keras.optimizers.Adam(learning_rate=ADAM_LR)
model.compile(
optimizer=optimizer,
loss="sparse_categorical_crossentropy",
metrics=["accuracy"],
)
return model
# Training history visualization
def plot_hist(hist):
plt.plot(hist.history["accuracy"])
plt.plot(hist.history["val_accuracy"])
plt.title("model accuracy")
plt.ylabel("accuracy")
plt.xlabel("epoch")
plt.legend(["train", "validation"], loc="upper left")
plt.show()
# Instantiating model
# Hyperparameters
DROP_CONNECT_RATE = 0.2 # Dropout rate for stochastic depth on EfficientNet
TOP_DROPOUT_RATE = 0.2 # Top dropout
INIT_LR = 5e-3 # Initial learning rate
EPOCHS = 20
# Adam optimizer learning rate schedule
ADAM_LR = tf.keras.optimizers.schedules.ExponentialDecay(
INIT_LR, decay_steps=100, decay_rate=0.96, staircase=True
)
model = build_model(num_classes=NUM_CLASSES)
# Training embedding layer
model_file_path = os.path.join(MODELS_DIR, "EfficientNetB0_softmax.keras")
callbacks = [
keras.callbacks.ModelCheckpoint(
model_file_path, save_best_only=True, monitor="val_accuracy"
),
keras.callbacks.EarlyStopping(patience=2, monitor="val_accuracy"),
]
hist = model.fit(
train_ds,
epochs=EPOCHS,
validation_data=val_ds,
shuffle="batch",
callbacks=callbacks,
)
plot_hist(hist)
# Evaluating best model
model = keras.models.load_model(model_file_path)
print("Predictions on validation set...")
print(f"Validation accuracy: {model.evaluate(val_ds)[1]*100:.2f} %")
print("Predictions on test set...")
print(f"Test accuracy: {model.evaluate(test_ds)[1]*100:.2f} %")
# ## Cosine Similarity
# Pairwise query: key search for similarity candidates. In the following example:
# * Query images: validation set
# * Key images: training set
# Auxiliar functions
# Load image
def get_image(path, resize=False, reshape=False, target_size=None):
img = cv2.imread(path)
img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)
if resize:
img = cv2.resize(img, dsize=(target_size, target_size))
if reshape:
img = tf.reshape(img, [1, target_size, target_size, 3])
return img
# Get landmark samples
def get_landmark(landmark_id, samples=16):
nrows = samples // 4
random_imgs = np.random.choice(
train_df_sub[train_df_sub["new_id"] == landmark_id].index,
samples,
replace=False,
)
plt.figure(figsize=(12, 10))
for i, img in enumerate(train_df_sub.loc[random_imgs, :].values):
ax = plt.subplot(nrows, 4, i + 1)
plt.imshow(get_image(img[2]))
plt.title(f"{img[0]}")
plt.suptitle(
f"Samples of landmark {landmark_id}", fontsize=14, y=0.94, weight="bold"
)
plt.axis("off")
# Get image embeddings
def get_embeddings(model, image_paths, input_size, as_df=True):
embeddings = {}
embeddings["images_paths"] = []
embeddings["embedded_images"] = []
target_dir = os.path.split(os.path.split(image_paths[0])[0])[0]
print(f"Retrieving embeddings for {target_dir} with {model.name}...")
for image_path in tqdm(image_paths):
embeddings["images_paths"].append(image_path)
embedded_image = model.predict(
get_image(image_path, resize=True, reshape=True, target_size=input_size)
)
embeddings["embedded_images"].append(embedded_image)
if as_df:
embeddings = pd.DataFrame(embeddings)
return embeddings
# Get similarities between query key pair
def get_similarities(query, key):
"""
Get cosine similarity matrix between query and key pairs
Arguments:
query, key: embedded images
"""
query_array = np.stack(query.tolist()).reshape(query.shape[0], query[0].shape[1])
key_array = np.stack(key.tolist()).reshape(key.shape[0], key[0].shape[1])
# Initializing similarity matrix
similarity = np.zeros((query_array.shape[0], key_array.shape[0]))
# Getting pairwise similarities
print(
f"Getting pairwise {query_array.shape[0]} query: {key_array.shape[0]} key similarities..."
)
for query_index in tqdm(range(query_array.shape[0])):
similarity[query_index] = (
1
- spatial.distance.cdist(
query_array[np.newaxis, query_index, :], key_array, "cosine"
)[0]
)
return similarity
# Plot top ranked images
def plot_similar(similar_imgs, img_paths):
"""
Plot top N similar samples from similarity index
"""
plt.figure(figsize=(18, 6))
nrows = similar_imgs.shape[0] // 5
for i, img in enumerate(similar_imgs):
ax = plt.subplot(nrows, 5, i + 1)
plt.imshow(get_image(img_paths[img]))
plt.title(f"Landmark id: {os.path.split(os.path.split(img_paths[img])[0])[1]}")
plt.axis("off")
# Aggregate query and top similar plots
def query_top(image_index, top_n=5, figsize=(6, 6), reranked=None):
"""
Plot top N similar samples against queried image
If reranked, provide reranked dataframe with ['top_similar'] index reordered by reranked confidence
"""
image_id = os.path.split(val_embeddings["images_paths"][image_index])[1]
query_landmark_id = os.path.split(
os.path.split(val_embeddings["images_paths"][image_index])[0]
)[1]
if type(reranked) == pd.core.frame.DataFrame:
similar_n = reranked["top_similar"][:top_n]
else:
similar_n = np.argsort(val_train_similarity[image_index])[::-1][:top_n]
print(f"Queried image: {image_id}")
plt.figure(figsize=figsize)
plt.imshow(get_image(val_embeddings["images_paths"][image_index]))
plt.title(f"Landmark id: {query_landmark_id}")
plt.axis("off")
plot_similar(similar_n, train_embeddings["images_paths"])
# Embedding models
embedding_layer = "embedding_512"
embedding_model = tf.keras.Model(
inputs=model.input,
outputs=model.get_layer(embedding_layer).output,
name="EfficientNetB0_embed512",
)
# Retrieving embeddings
train_img_paths = train_ds.file_paths
val_img_paths = val_ds.file_paths
train_embeddings = get_embeddings(
model=embedding_model, image_paths=train_img_paths, input_size=IMG_SIZE
)
val_embeddings = get_embeddings(
model=embedding_model, image_paths=val_img_paths, input_size=IMG_SIZE
)
train_embeddings.head()
val_embeddings.head()
val_train_similarity = get_similarities(
val_embeddings["embedded_images"], train_embeddings["embedded_images"]
)
val_train_similarity.shape
# Calculating confidence score per submission
def confidence_top(
query=None, key=None, similarity=None, query_image_index=None, top=5
):
"""
Arguments:
query_image_index = index of query image on similarity matrix query axis
Return confidence scores for top N predictions
"""
query_paths = query["images_paths"]
key_paths = key["images_paths"]
similar_n = np.argsort(similarity[query_image_index])[::-1][:top]
confidence_df = {}
confidence_df["top_similar"] = []
for similar in similar_n:
confidence_df["top_similar"].append(similar)
confidence_df["image_paths"] = []
for similar in similar_n:
similar_image_path = key_paths[similar]
confidence_df["image_paths"].append(similar_image_path)
confidence_df["prediction"] = []
for similar in similar_n:
similar_image_path = key_paths[similar]
y = int(os.path.split(os.path.split(similar_image_path)[0])[1])
confidence_df["prediction"].append(y)
confidence_df["cos_similarity"] = []
for similar in similar_n:
confidence_df["cos_similarity"].append(similarity[query_image_index][similar])
return pd.DataFrame(confidence_df)
query_image_index = 0
query_top(query_image_index)
confidence_df = confidence_top(
query=val_embeddings,
key=train_embeddings,
similarity=val_train_similarity,
query_image_index=query_image_index,
top=top_n,
)
confidence_df
query_image_index = 4
query_top(query_image_index)
confidence_df = confidence_top(
query=val_embeddings,
key=train_embeddings,
similarity=val_train_similarity,
query_image_index=query_image_index,
top=5,
)
confidence_df
query_image_index = 65
query_top(query_image_index)
confidence_df = confidence_top(
query=val_embeddings,
key=train_embeddings,
similarity=val_train_similarity,
query_image_index=query_image_index,
top=5,
)
confidence_df
query_image_index = 13
query_top(query_image_index)
confidence_df = confidence_top(
query=val_embeddings,
key=train_embeddings,
similarity=val_train_similarity,
query_image_index=query_image_index,
top=top_n,
)
confidence_df
# ## Object oclusion example
# Object oclusion is only one of the examples of how a local feature reranking method improves query performance
query_image_index = 887
top_n = 10
query_top(query_image_index, top_n)
confidence_df = confidence_top(
query=val_embeddings,
key=train_embeddings,
similarity=val_train_similarity,
query_image_index=query_image_index,
top=top_n,
)
confidence_df
# ## DELF module
# Local features search
# References:
# * Large-Scale Image Retrieval with Attentive Deep Local Features: https://arxiv.org/abs/1612.06321
# * DELF on Tensorflow Hub: https://github.com/tensorflow/models/tree/master/research/delf
#
DELF_IMG_SIZE = 600
image_1 = get_image(
val_embeddings["images_paths"][887], resize=True, target_size=DELF_IMG_SIZE
)
plt.figure(figsize=(6, 6))
plt.imshow(image_1)
plt.axis("off")
plt.show()
image_2 = get_image(
train_embeddings["images_paths"][similar_n[5]],
resize=True,
target_size=DELF_IMG_SIZE,
)
plt.figure(figsize=(6, 6))
plt.imshow(image_2)
plt.axis("off")
plt.show()
from absl import logging
from PIL import Image, ImageOps
from scipy.spatial import cKDTree
from skimage.feature import plot_matches
from skimage.measure import ransac
from skimage.transform import AffineTransform
from six import BytesIO
import tensorflow_hub as hub
from six.moves.urllib.request import urlopen
delf = hub.load("https://tfhub.dev/google/delf/1").signatures["default"]
# DELF module
def run_delf(image):
"""
Apply DELF module to the input image
Arguments:
image: np.array resized image
"""
float_image = tf.image.convert_image_dtype(image, tf.float32)
return delf(
image=float_image,
score_threshold=tf.constant(100.0),
image_scales=tf.constant([0.25, 0.3536, 0.5, 0.7071, 1.0, 1.4142, 2.0]),
max_feature_num=tf.constant(1000),
)
def match_images(image1, image2, result1, result2, verbose=True):
distance_threshold = 0.8
# Read features.
num_features_1 = result1["locations"].shape[0]
num_features_2 = result2["locations"].shape[0]
if verbose:
print("Loaded image 1's %d features" % num_features_1)
print("Loaded image 2's %d features" % num_features_2)
# Find nearest-neighbor matches using a KD tree.
d1_tree = cKDTree(result1["descriptors"])
_, indices = d1_tree.query(
result2["descriptors"], distance_upper_bound=distance_threshold
)
# Select feature locations for putative matches.
locations_2_to_use = np.array(
[
result2["locations"][i,]
for i in range(num_features_2)
if indices[i] != num_features_1
]
)
locations_1_to_use = np.array(
[
result1["locations"][indices[i],]
for i in range(num_features_2)
if indices[i] != num_features_1
]
)
# Perform geometric verification using RANSAC.
_, inliers = ransac(
(locations_1_to_use, locations_2_to_use),
AffineTransform,
min_samples=3,
residual_threshold=20,
max_trials=1000,
)
if verbose:
print("Found %d inliers" % sum(inliers))
# Visualize correspondences.
_, ax = plt.subplots(figsize=(9, 9))
inlier_idxs = np.nonzero(inliers)[0]
plot_matches(
ax,
image1,
image2,
locations_1_to_use,
locations_2_to_use,
np.column_stack((inlier_idxs, inlier_idxs)),
matches_color="b",
)
ax.axis("off")
ax.set_title(f"DELF correspondences: Found {sum(inliers)} inliers")
delf_result1 = run_delf(image_1)
delf_result2 = run_delf(image_2)
match_images(image_1, image_2, delf_result1, delf_result2)
for image_index in similar_n[:6]:
key_image = get_image(
train_embeddings["images_paths"][image_index],
resize=True,
target_size=DELF_IMG_SIZE,
)
try:
delf_key_image_result = run_delf(key_image)
match_images(
image_1, key_image, delf_result1, delf_key_image_result, verbose=False
)
except:
print("No inliers found")
# ## Reranking
# Reranking using DELF local features descriptor
def delf_rerank(
query=None, key=None, query_image_index=None, confidence_df=None, re_sort=True
):
distance_threshold = 0.8
query_paths = query["images_paths"]
key_paths = key["images_paths"]
query_image = get_image(
query_paths[query_image_index], resize=True, target_size=DELF_IMG_SIZE
)
delf_result_query = run_delf(query_image)
# Read query features
num_features_query = delf_result_query["locations"].shape[0]
inliers_list = []
print(
f"Retrieving local features for top {len(confidence_df['image_paths'])} key images..."
)
for image_path in tqdm(confidence_df["image_paths"]):
key_image = get_image(image_path, resize=True, target_size=DELF_IMG_SIZE)
delf_result_key = run_delf(key_image)
# Read key features
num_features_key = delf_result_key["locations"].shape[0]
# Find nearest-neighbor matches using a KD tree.
d1_tree = cKDTree(delf_result_query["descriptors"])
_, indices = d1_tree.query(
delf_result_key["descriptors"], distance_upper_bound=distance_threshold
)
# Select feature locations for putative matches.
locations_k_to_use = np.array(
[
delf_result_key["locations"][i,]
for i in range(num_features_key)
if indices[i] != num_features_query
]
)
locations_q_to_use = np.array(
[
delf_result_query["locations"][indices[i],]
for i in range(num_features_key)
if indices[i] != num_features_query
]
)
# Perform geometric verification using RANSAC.
try:
_, inliers = ransac(
(locations_q_to_use, locations_k_to_use),
AffineTransform,
min_samples=3,
residual_threshold=20,
max_trials=1000,
)
except:
inliers = [0]
# Handling 0 inliers
try:
total_inliers = sum(inliers)
inliers_list.append(total_inliers)
except:
inliers_list.append(1) # Appending inlier = 1 to avoid null confidence
confidence_df["inliers"] = inliers_list
original_confidence = confidence_df["inliers"]
reranked_confidence = np.sqrt(original_confidence) * confidence_df["cos_similarity"]
confidence_df["reranked_conf"] = reranked_confidence
if re_sort:
confidence_df.sort_values("reranked_conf", ascending=False, inplace=True)
return confidence_df
reranked_df = delf_rerank(
query=val_embeddings,
key=train_embeddings,
query_image_index=query_image_index,
confidence_df=confidence_df,
re_sort=True,
)
reranked_df
query_image_index = 887
top_n = 10
query_top(query_image_index, top_n, reranked=reranked_df)
# ## Reranking examples
query_image_index = 11
top_n = 10
query_top(query_image_index, top_n)
confidence_df = confidence_top(
query=val_embeddings,
key=train_embeddings,
similarity=val_train_similarity,
query_image_index=query_image_index,
top=top_n,
)
confidence_df
reranked_df = delf_rerank(
query=val_embeddings,
key=train_embeddings,
query_image_index=query_image_index,
confidence_df=confidence_df,
re_sort=True,
)
reranked_df
query_image_index = 11
top_n = 10
query_top(query_image_index, top_n, reranked=reranked_df)
query_image_index = 395
top_n = 10
query_top(query_image_index, top_n)
confidence_df = confidence_top(
query=val_embeddings,
key=train_embeddings,
similarity=val_train_similarity,
query_image_index=query_image_index,
top=top_n,
)
confidence_df
reranked_df = delf_rerank(
query=val_embeddings,
key=train_embeddings,
query_image_index=query_image_index,
confidence_df=confidence_df,
re_sort=True,
)
reranked_df
query_image_index = 395
top_n = 10
query_top(query_image_index, top_n, reranked=reranked_df)
query_image_index = 40
top_n = 10
query_top(query_image_index, top_n)
confidence_df = confidence_top(
query=val_embeddings,
key=train_embeddings,
similarity=val_train_similarity,
query_image_index=query_image_index,
top=top_n,
)
confidence_df
reranked_df = delf_rerank(
query=val_embeddings,
key=train_embeddings,
query_image_index=query_image_index,
confidence_df=confidence_df,
re_sort=True,
)
reranked_df
query_image_index = 40
top_n = 10
query_top(query_image_index, top_n, reranked=reranked_df)
# ## Under-represented query image
# Effect of querying images not well represented on the key set
query_image_index = 822
top_n = 10
query_top(query_image_index, top_n)
# Let's investigate what the model has seen for landmark 36 during training...
get_landmark(36)
|
# # Linear Regression Explained
# This uses the data and code from the medium article [Linear Regression from Scratch](https://link.medium.com/dJlTSvMUfeb)
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
# # Set up functions to do the work for us. Explanations before each line of code.
# variables to store mean and standard deviation for each feature
mu = []
std = []
def load_data(filename):
df = pd.read_csv(filename, sep=",", index_col=False)
df.columns = ["housesize", "rooms", "price"]
data = np.array(df, dtype=float)
plot_data(data[:, :2], data[:, -1])
normalize(data)
return data[:, :2], data[:, -1]
def plot_data(x, y):
plt.xlabel("house size")
plt.ylabel("price")
plt.plot(x[:, 0], y, "bo")
plt.show()
def normalize(data):
for i in range(0, data.shape[1] - 1):
mu.append(np.mean(data[:, i]))
std.append(np.std(data[:, i]))
data[:, i] = (data[:, i] - np.mean(data[:, i])) / np.std(data[:, i])
def h(x, theta):
return np.matmul(x, theta)
def cost_function(x, y, theta):
return ((h(x, theta) - y).T @ (h(x, theta) - y)) / (2 * y.shape[0])
def gradient_descent(x, y, theta, learning_rate=0.1, num_epochs=10):
m = x.shape[0]
J_all = []
for _ in range(num_epochs):
h_x = h(x, theta)
cost_ = (1 / m) * (x.T @ (h_x - y))
theta = theta - (learning_rate) * cost_
J_all.append(cost_function(x, y, theta))
return theta, J_all
def plot_cost(J_all, num_epochs):
plt.xlabel("Epochs")
plt.ylabel("Cost")
plt.plot(num_epochs, J_all, "m", linewidth="5")
plt.show()
def test(theta, x):
x[0] = (x[0] - mu[0]) / std[0]
x[1] = (x[1] - mu[1]) / std[1]
y = theta[0] + theta[1] * x[0] + theta[2] * x[1]
print("Price of house: ", y)
x, y = load_data(
"../input/mediumlinear-regression/linear_regression/house_price_data.txt"
)
y = np.reshape(y, (46, 1))
x = np.hstack((np.ones((x.shape[0], 1)), x))
theta = np.zeros((x.shape[1], 1))
learning_rate = 0.1
num_epochs = 50
theta, J_all = gradient_descent(x, y, theta, learning_rate, num_epochs)
J = cost_function(x, y, theta)
print("Cost: ", J)
print("Parameters: ", theta)
# for testing and plotting cost
n_epochs = []
jplot = []
count = 0
for i in J_all:
jplot.append(i[0][0])
n_epochs.append(count)
count += 1
jplot = np.array(jplot)
n_epochs = np.array(n_epochs)
plot_cost(jplot, n_epochs)
test(theta, [1600, 3])
|
# ## Instalação e Importação de Pacotes
# Instalando o DuckDB:
# Importando os pacotes que serão utilizados e configurando o Pandas para mostrar até 200 linhas e 200 colunas, bem como tentar exibir valores decimais com 4 casas depois da vírgula (evitando notação científica):
import pandas as pd
import plotly.express as px
import duckdb
pd.set_option("display.max_columns", 200)
pd.set_option("display.max_rows", 200)
pd.set_option("display.float_format", lambda x: "%0.4f" % x)
# ## Objetivo
# Esse notebook tem como objetivo mostrar formas de trabalhar com arquivos que, quando importados diretamente via Pandas, necessitariam de mais memória RAM do que temos disponível.
# O intuito principal é de realizar análise exploratória diretamente com as ferramentas que serão mostradas, mas pode também servir para simplesmente reduzir o tamanho (quantidade de linhas e colunas) de uma tabela, descartando o que não for útil, de forma que seja possível trabalhar no Pandas sem "estourar" a memória.
# ## Introdução
# O comportamento padrão do Pandas exige que, antes de aplicar qualquer tipo de alteração ou consulta numa tabela, ela seja trazida "completamente" para a memória - exceto quando usamos *chunks*, mas daí continuamos não tendo todo o dado, certo?
# Com o DuckDB podemos utilizar o conceito de *lazy evaluation*, que permite que criemos de forma antecipada as etapas com as transformações e filtros, trazendo para a memória apenas a tabela que seria o resultado dessas etapas.
# Para facilitar o entendimento, podemos pensar no funcionamento de um banco de dados: quando passamos uma query SQL nosso retorno é apenas a parte da tabela que corresponde à query, não sendo necessário importar a tabela na sua totalidade.
# ## DuckDB
# Apesar de ser um banco de dados - daqueles que ficam salvos em um arquivo, "offline", similar ao SQLite - o DuckDB conta também com uma integração muito simples e poderosa com Pandas, permitindo inclusive que façamos queries em SQL diretamente em DataFrames Pandas, passando apenas a variável à qual esse DataFrame está atribuído no "FROM" da query.
# Nesse exemplo vamos "enganar" o DuckDB e, ao invés de criar uma conexão com um arquivo de banco de dados, vamos criar uma conexão "em branco", sem passar nenhum caminho ou arquivo nos parâmetros da função .connect(), para usufruir somente das queries SQL nas nossas tabelas.
con = duckdb.connect()
# Feito isso, basta usar a função .sql() nessa conexão, passando a query em forma de texto e o caminho para nosso arquivo no "FROM" dessa query.
# Nesse exemplo iremos retornar as primeiras vinte linhas do arquivo de microdados do ENEM de 2019.
con.sql(
"""
SELECT *
FROM '/kaggle/input/microdados-enem-2019-2020-e-2021-parquet/enem_2019.parquet'
LIMIT 20
"""
)
# Podemos notar que o retorno não está no formato de tabela do Pandas - o que é justamente o que buscamos, visto que dessa forma estamos usando apenas uma fração da memória que o Pandas ocuparia.
# Para transformar em um DataFrame Pandas (após termos certeza que a tabela não vai estourar a memória 😂) basta adicionar .df() após o comando .sql().
con.sql(
"""
SELECT *
FROM '/kaggle/input/microdados-enem-2019-2020-e-2021-parquet/enem_2019.parquet'
LIMIT 20
"""
).df()
# Não sei você, mas esse caminho no "FROM" me parece ocupar muito espaço, então vou usar a função .read_parquet() do DuckDB e atribuir a uma variável para substituir esse caminho.
# Vou aproveitar para fazer para os outros 2 arquivos também.
# Note que dessa forma continuamos usufruindo de *lazy evaluation* e não estamos trazendo essas tabelas para a memória, apenas atribuindo o caminho a uma variável.
enem_2019 = duckdb.read_parquet(
"/kaggle/input/microdados-enem-2019-2020-e-2021-parquet/enem_2019.parquet"
)
enem_2020 = duckdb.read_parquet(
"/kaggle/input/microdados-enem-2019-2020-e-2021-parquet/enem_2020.parquet"
)
enem_2021 = duckdb.read_parquet(
"/kaggle/input/microdados-enem-2019-2020-e-2021-parquet/enem_2021.parquet"
)
con.sql(
"""
SELECT *
FROM enem_2019
LIMIT 20
"""
).df()
# Ainda não acredita que o DuckDB seja "isso tudo"? Que tal contar o total de registros em cada arquivo? No Pandas precisaríamos das tabelas inteiras em memória para fazer essa contagem. Com o DuckDB é instantâneo e não ocupa praticamente nada de memória - é literalmente uma tabela com 1 linha e 1 coluna.
con.sql(
"""
SELECT COUNT(*) AS QTDE_REGISTROS
FROM enem_2019
"""
).df()
con.sql(
"""
SELECT COUNT(*) AS QTDE_REGISTROS
FROM enem_2020
"""
).df()
con.sql(
"""
SELECT COUNT(*) AS QTDE_REGISTROS
FROM enem_2021
"""
).df()
# Bora contar a quantidade de registros por "município da prova" e ordenar do maior pro menor.
con.sql(
"""
SELECT NO_MUNICIPIO_PROVA, COUNT(*) AS QTDE_REGISTROS
FROM enem_2019
GROUP BY 1
ORDER BY 2 DESC
"""
).df()
# Bom, 1.692 municípios é muita coisa pra caber num gráfico. Vamos filtrar só os 50 primeiros e plotar num gráfico de barras.
con.sql(
"""
SELECT NO_MUNICIPIO_PROVA, COUNT(*) AS QTDE_REGISTROS
FROM enem_2019
GROUP BY 1
ORDER BY 2 DESC
LIMIT 50
"""
).df().plot(backend="plotly", kind="bar", x="NO_MUNICIPIO_PROVA", y="QTDE_REGISTROS")
# Vamos então comparar os municípios com maior participação entre os 3 anos.
# Dessa vez, atribuindo o DataFrame resultante da query a uma variável.
df_compara_qtde_municipio = con.sql(
"""
SELECT *
FROM
(SELECT
NO_MUNICIPIO_PROVA,
COUNT(*) AS QTDE_REGISTROS,
2019 AS ANO_REF,
ROW_NUMBER() OVER (PARTITION BY "ANO_REF" ORDER BY "QTDE_REGISTROS" DESC) AS 'NUM_LINHA'
FROM enem_2019
GROUP BY 1
UNION
SELECT
NO_MUNICIPIO_PROVA,
COUNT(*) AS QTDE_REGISTROS,
2020 AS ANO_REF,
ROW_NUMBER() OVER (PARTITION BY "ANO_REF" ORDER BY "QTDE_REGISTROS" DESC) AS 'NUM_LINHA'
FROM enem_2020
GROUP BY 1
UNION
SELECT
NO_MUNICIPIO_PROVA,
COUNT(*) AS QTDE_REGISTROS,
2021 AS ANO_REF,
ROW_NUMBER() OVER (PARTITION BY "ANO_REF" ORDER BY "QTDE_REGISTROS" DESC) AS 'NUM_LINHA'
FROM enem_2021
GROUP BY 1) t1
WHERE t1.NUM_LINHA <= 50
ORDER BY t1.ANO_REF ASC, t1.NUM_LINHA ASC
"""
).df()
df_compara_qtde_municipio.head()
# Vamos usar um gráfico de *treemap*, que ordena cada uma das categorias (total, ano referência, município) da esquerda para a direita e de cima para baixo, de acordo com o(s) valor(es) passados.
# Podemos perceber que na categoria ano referência, o ano de 2020 (em azul) está mais à esquerda, indicando que o valor total de 2020 é superior ao de 2019 e 2021.
# O gráfico é interativo: passando o mouse ele mostra o valor de cada bloco/grupo e clicando ele "dá zoom".
fig = px.treemap(
df_compara_qtde_municipio,
path=[px.Constant("Total"), "ANO_REF", "NO_MUNICIPIO_PROVA"],
values="QTDE_REGISTROS",
)
fig.update_traces(root_color="lightgrey")
fig.update_layout(margin=dict(t=25, l=15, r=15, b=15))
fig.show()
# "Ah, mas eu não curto SQL... Prefiro pegar uma amostra de cada arquivo e jogar no meu bom e velho Pandas".
# Então vamos usar o "USING SAMPLE" do DuckDB. Ele aceita tanto um percentual quanto uma quantidade de linhas para fazer a amostragem. Nesse exemplo vamos usar um percentual e mostrar a quantidade de registros que a query retornaria.
#
con.sql(
"""
SELECT COUNT(*)
FROM enem_2019
USING SAMPLE 10%;
"""
)
# Para salvar o resultado como um arquivo .parquet, basta adicionar .write_parquet() após o .sql(), passando o nome do arquivo e o caminho, se necessário.
con.sql(
"""
SELECT COUNT(*)
FROM enem_2019
USING SAMPLE 10%;
"""
).write_parquet("enem_2019_amostra_10.parquet")
|
# # Diffusion Source Images View & Prompts
import os
import cv2
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import pyarrow.parquet as pq
paths = []
for dirname, _, filenames in os.walk("/kaggle/input/"):
for filename in filenames:
if filename[-4:] == ".png":
paths += [(os.path.join(dirname, filename))]
table = pq.read_table("/kaggle/input/diffusiondb-metadata/metadata.parquet")
print(table.to_pydict().keys())
df = pd.DataFrame.from_dict(table.to_pydict())
df2 = df[["image_name", "prompt"]]
mapping = dict(zip(df2["image_name"], df2["prompt"]))
# prpids=df2['image_name'].tolist()
# # Images
selected_images = np.random.choice(paths, size=30, replace=False)
fig, axes = plt.subplots(nrows=10, ncols=3, figsize=(14, 50))
for i, ax in enumerate(axes.flatten()):
if i < len(selected_images):
image = cv2.imread(selected_images[i])
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
ax.imshow(image)
ax.axis("off")
plt.show()
# # Images and Prompts
for i in range(len(selected_images)):
idi = selected_images[i].split("/")[-1]
prp = mapping.get(idi, "")
image = cv2.imread(selected_images[i])
image = cv2.cvtColor(image, cv2.COLOR_BGR2RGB)
plt.imshow(image)
plt.axis("off")
plt.show()
print(prp)
print()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import seaborn as sns
import matplotlib.pyplot as plt
from scipy import stats
from scipy.stats import randint
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import roc_auc_score
from sklearn.model_selection import train_test_split
from sklearn.metrics import f1_score
from sklearn.model_selection import RandomizedSearchCV
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# # Import train & test data
# Loaded and prepped the files- dropped the id columns off both, trimmed the white space.
train = pd.read_csv("../input/cap4611-assignment-1-data/train.csv").drop(columns="id")
test = pd.read_csv("../input/cap4611-assignment-1-data/test.csv").drop(columns="id")
# Cleaning whitespace from the column names
train.columns = train.columns.str.strip()
test.columns = test.columns.str.strip()
# # Interpreting our training data & Standardization vs Normalization
# Above we can see that each column in our training data contains 3,410 non-null values. This shows that there aren't any clearly missing values, as our Range Index is 3,410 entries. Perhaps there are some hidden missing values? I'll get to this later.
# There is some weird indentation/spacing going on with some of the columns, fixed that using the strip function
# Tree based models do not require any standardization or normalization. If we were to scale our model up or down, and do so consistently, it will not change the outcome of our trees.
train.info()
train.head()
train.describe()
# # Outliers!
# A simple way to detect outliers is to plot our data with some box plots, which would be great, but in our case we're given quite a few columns which could get ugly. Instead, I have decided to loop all columns in the training data and handle the outliers based on the interquartile range. Initially I wanted to replace each value outside the IQR with the bounds of the IQR. I then decided to use the mean of the column as a way of preserving the mean.
# replace all values outside the column IQR with a value 3 standard deviations away from the column mean
for col in train.columns:
if (
col == "id"
or col == "Bankrupt"
or col == "one if total liabilities exceeds total assets zero otherwise"
or col == "one if net income was negative for the last two year zero otherwise"
):
continue
val = train[col].mean()
Q1 = train[col].quantile(0.25)
Q3 = train[col].quantile(0.75)
IQR = Q3 - Q1
max = Q3 + (1.5 * IQR)
min = Q1 - (1.5 * IQR)
for x in [col]:
# train.loc[train[x] < min, x] = (val)
# train.loc[train[x] > max, x] = (val)
train.loc[train[x] < min, x] = val - 3 * train[col].std()
train.loc[train[x] > max, x] = val + 3 * train[col].std()
train.describe()
# # Decision Tree Testing
# First step is to drop the target feature, "Bankrupt" in this case, and then I will split the training data up into a validation set, this way I can get a more accurate model while also keeping my models from overfitting.
#
X = train.drop("Bankrupt", axis=1)
Y = train["Bankrupt"]
print("Dropped the target feature")
from sklearn import metrics
X_train, X_test, y_train, y_test = train_test_split(X, Y)
decision_tree = DecisionTreeClassifier(max_depth=5, criterion="gini")
decision_tree.fit(X_train, y_train)
y_pred = decision_tree.predict(X_test)
y_pred_proba = decision_tree.predict_proba(X_test)[:, 1]
accuracy = metrics.accuracy_score(y_test, y_pred)
score = metrics.f1_score(y_test, y_pred)
ROC_AUC = roc_auc_score(y_test, y_pred_proba)
print(ROC_AUC)
print(score)
print(accuracy)
# # Best Scores: Decision Tree
# ROC_AUC - 0.8893119105833042
# F1 - 0.34920634920634924
# Accuracy - 0.9519343493552169
# # Random Forest Testing
# I wasn't very impressed with the Decision Tree scores, so why not attempt a bunch of decision trees at once- i.e. a Random Forest!!!
# As you'll see below I tried my best to find the best parameters possible. I took screenshots of the outputs, notes on what worked best, and then once I got a rough idea of the parameters that returned the best results I went manual for the sake of time.
# To find the best set of Parameters I built a parameter grid(space?) and used RandomSearchCV. Went with RSCV over GSCV due to time. I found that smaller parameters responded better to scoring- I started a bit larger as I was afraid of building Random Stumps.
# 1. n_estimators: Number of trees to build before taking the best model.
# 2. max_features: Maximum # of features it will try in a tree
# 3. max_depth: Maximum # of levels in the tree, not wanting to build a stump- not wanting to build a redwood
# 4. min_samples_split: number of samples required to split the node
# 5. min_samples_leaf: number of samples required to be at the leaf node
X_train, X_test, Y_train, Y_test = train_test_split(X, Y, test_size=0.35)
n_estimators = [100, 125, 150, 200, 400]
max_features = ["auto", "sqrt", "log2"]
max_depth = [3, 4, 6, 8, 10]
max_depth.append(None)
min_samples_split = [4, 5, 6, 8, 10]
min_samples_leaf = [4, 6, 8, 10, 12]
bootstrap = [True]
random_grid = {
"n_estimators": n_estimators,
"max_features": max_features,
"min_samples_split": min_samples_split,
"max_depth": max_depth,
"min_samples_leaf": min_samples_leaf,
"bootstrap": bootstrap,
}
# random_forest = RandomForestClassifier(bootstrap=True, criterion='gini', max_depth=20, min_samples_leaf=2, min_samples_split=10, n_estimators=75, max_features='auto')
# random_forest = RandomForestClassifier(max_depth=6, n_estimators=1000, min_samples_leaf=5, min_samples_split=12)
random_forest = RandomForestClassifier(
max_depth=8, n_estimators=1500, min_samples_leaf=4, min_samples_split=8
)
# rf_random = RandomizedSearchCV(random_forest, random_grid, n_iter = 500, cv=5, random_state = 42, verbose=2)
# model = rf_random.fit(X_train, Y_train)
# best_random = rf_random.best_estimator_
random_forest.fit(X_train, Y_train)
y_pred = random_forest.predict(X_test)
y_pred_proba = random_forest.predict_proba(X_test)[:, 1]
accuracy = random_forest.score(X_test, Y_test)
score = f1_score(Y_test, y_pred)
ROC_AUC = roc_auc_score(Y_test, y_pred_proba)
print(accuracy)
print(score)
print(ROC_AUC)
# # Best Scores: Random Forest
# ROC_AUC - 0.977725674091442
# F1 - 0.3870967741935483
# Accuracy - 0.9745677523120225
# The below were used to print the parameters & score of the RSCV models I built
# from pprint import pprint
# pprint(best_random.get_params())
# pprint(random_forest.get_params())
# score = model.score(X_test, Y_test)
# y_pred = model.predict(X_test)
# y_pred_proba = model.predict_proba(X_test)[:,1]
# score = f1_score(Y_test, y_pred)
# ROC_AUC = roc_auc_score(Y_test, y_pred_proba)
# # accuracy = model.score(Y_test, y_pred_proba)
# # print(accuracy)
# print(score)
# print(ROC_AUC)
rf_proba = random_forest.predict_proba(test)[:, 1]
# prediction = model.predict_proba(test)[:, 1]
# dt = decision_tree.predict_proba(test)[:, 1]
submission = pd.DataFrame({"Bankrupt": rf_proba})
submission = submission.rename_axis("id")
print(submission)
submission.to_csv("submission.csv")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import random
import string
import numpy as np
import tensorflow as tf
from tensorflow import keras
from tensorflow.keras import layers
df = pd.read_csv("/kaggle/input/wikipedia-movie-plots/wiki_movie_plots_deduped.csv")
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
chunk_size = 2
num_chunks = 4
hashed_buckets = 1024
vocab = list(string.ascii_lowercase) + list(" ().,!?\"'-:;")
def split_text(text):
split = [(text[i : i + chunk_size]) for i in range(0, len(text), chunk_size)]
return split
def encode_text(text):
x = layers.experimental.preprocessing.StringLookup(vocabulary=vocab)(
list(text.lower())
)
x = layers.experimental.preprocessing.CategoryEncoding(max_tokens=len(vocab) + 1)(x)
return x
def decode_text(tensor):
x = tf.argmax(tensor, axis=1)
x = layers.experimental.preprocessing.StringLookup(
vocabulary=["[?]"] + vocab, invert=True
)(x)
x = "".join([tf.compat.as_str_any(tensor.numpy()) for tensor in x])
return x
inp = x = "This is a test! Let's see how it goes."
x = encode_text(x)
x = decode_text(x)
print(x)
def gen():
while True:
batch_inputs = []
batch_outputs = []
while len(batch_inputs) < 64:
text = df["Plot"].sample().values[0].lower()
if len(text) <= num_chunks * chunk_size + 1:
continue
segment_start = random.randint(0, len(text) - num_chunks * chunk_size - 1)
text_segment = text[segment_start : segment_start + num_chunks * chunk_size]
answer = text[
segment_start
+ num_chunks * chunk_size : segment_start
+ num_chunks * chunk_size
+ 1
]
batch_inputs.append(split_text(text_segment))
batch_outputs.append(encode_text(answer)[0])
yield np.array(batch_inputs), np.array(batch_outputs)
def calc_freq():
letters = {"[?]": 0}
total = 0
for letter in vocab:
letters[letter] = 0
for i in range(1000):
text = df["Plot"].sample().values[0].lower()
for letter in text:
total += 1
try:
letters[letter] += 1
except:
letters["[?]"] += 1
for letter in letters:
letters[letter] /= total
return letters
frequencies = calc_freq()
print(frequencies)
inputs = x = keras.Input(shape=(num_chunks,), dtype="string")
x = layers.experimental.preprocessing.Hashing(num_bins=hashed_buckets)(x)
x = layers.Lambda(
lambda x: keras.backend.one_hot(keras.backend.cast(x, "int64"), hashed_buckets)
)(x)
x = layers.Flatten()(x)
x = layers.Dense(1024, activation="relu")(x)
x = layers.Dense(512, activation="relu")(x)
x = layers.Dense(128, activation="relu")(x)
outputs = x = layers.Dense(len(vocab) + 1, activation="softmax")(x)
model = keras.Model(inputs=inputs, outputs=outputs, name="text_predict_model")
model.summary()
model.compile(
loss=keras.losses.CategoricalCrossentropy(),
optimizer=keras.optimizers.Adam(0.01),
metrics=["categorical_crossentropy", "categorical_accuracy"],
)
model.fit(gen(), epochs=50, steps_per_epoch=100)
def predict(text):
prediction_raw = model.predict(
np.array([split_text(text[-num_chunks * chunk_size :])])
)
print(tf.argmax(prediction_raw, axis=1))
# print(layers.experimental.preprocessing.StringLookup(vocabulary=['[?]']+vocab, invert=True)(prediction_raw))
print(decode_text(prediction_raw))
prediction = np.array(list(prediction_raw[0][1:]) + [0])
possibilities = ["[?]"] + vocab
data = [(possibilities[i], prediction[i]) for i in range(len(prediction))]
data.sort(reverse=True, key=lambda letter: letter[1])
print(data)
print(len(prediction), len(possibilities))
print(random.choices(possibilities, prediction))
def generate_next(text, temperature):
prediction_raw = model.predict(
np.array([split_text(text[-num_chunks * chunk_size :])])
)
prediction = np.array(list(prediction_raw[0][1:]) + [0])
possibilities = ["[?]"] + vocab
return random.choices(possibilities, prediction ** (1 / temperature))[0]
text = "This is a story about "
for i in range(100):
text += generate_next(text, 0.35)
print(text)
|
import numpy as np
import pandas as pd
from typing import Sequence, Tuple
from collections import defaultdict
import matplotlib.pyplot as plt
kaggle = False
if kaggle:
root = "/kaggle/input/amp-parkinsons-disease-progression-prediction"
else:
root = "data/"
# load dataset
train_proteins = pd.read_csv(f"{root}/train_proteins.csv")
train_peptides = pd.read_csv(f"{root}/train_peptides.csv")
train_clinical = pd.read_csv(f"{root}/train_clinical_data.csv")
supplemental_data = pd.read_csv(f"{root}/supplemental_clinical_data.csv")
t = set(train_clinical.patient_id.unique())
s = set(supplemental_data.patient_id.unique())
df = pd.concat([train_clinical, supplemental_data])
df.head()
gr = (
df[["visit_month", "updrs_1", "updrs_2", "updrs_3", "updrs_4"]]
.groupby("visit_month")
.median()
.dropna()
)
gr
gr.plot()
# # Data interpolation
# Idea
# - Find a way to predict N months into the future based on uprds (in submissin these will be predictions)
# Goal:
# - Be able to predict uprds for a visit_id with no protein/peptide data for a patient that has had some data in the past (inference iteration).
# You have to store a lit of "seen patient Ids"
# TODO:
# - model data per uprds to predict it's behavior. As we see from above it's not always growing, there might be some seasonality trends to it ie. (uprds_3_
# - Train a time series model per uprds (ML or DL with RNN)
# - Add model to existing submission notebooks to better predict uprds for a visit_id for a patient that has already been there in the past but curently does not have any peptide/protein data associated with this visit.
# Resources:
# - https://towardsdatascience.com/temporal-loops-intro-to-recurrent-neural-networks-for-time-series-forecasting-in-python-b0398963dc1f
# - https://www.analyticsvidhya.com/blog/2021/07/stock-market-forecasting-using-time-series-analysis-with-arima-model/
# - https://www.kaggle.com/code/humamfauzi/multiple-stock-prediction-using-single-nn/notebook
# - https://stats.stackexchange.com/questions/479211/how-to-cluster-non-aligned-time-series-with-different-length
all_months = {"updrs_1": {}, "updrs_2": {}, "updrs_3": {}, "updrs_4": {}}
for patient in df["patient_id"].unique(): # [:10]:
x = df[df["patient_id"] == patient]
uprds = x[["visit_month", "updrs_1", "updrs_2", "updrs_3", "updrs_4"]]
for ind, (vm, u1, u2, u3, u4) in uprds.iterrows():
pass
break
u1
# all_months = dict()
# for patient in df['patient_id'].unique():#[:10]:
# x = df[df['patient_id']==patient]
# uprds = x[['visit_month', 'updrs_1', 'updrs_2', 'updrs_3', 'updrs_4']]
# for vm, u1, u2, u3, u4 in uprds.iterrows():
# #months = len(x.visit_month.values.tolist())#[-1]
# #all_months.append(months)
# break
# # for uprds in ['updrs_1', 'updrs_2', 'updrs_3', 'updrs_4']:
# # uprds_vals = x[uprds].values
# # plt.plot(months, uprds_vals, marker='x', label=uprds)
# # break
# # # for i in x['updrs'].unique():
# # # tmp_uprds = x[x['updrs']==i]
# # # #tmp_uprds = tmp_uprds[tmp_uprds['visit_month'].isin([0, 6, 12, 24])]
# # # plt.plot(tmp_uprds['visit_month'], tmp_uprds['rating'], marker='x', label=i)
# # #assert tmp_uprds.shape[0]==4
# # plt.legend()
# # plt.show()
# # plt.show()
# plt.hist(all_months, bins = 10)
# plt.title("number of visits per patient")
# plt.xlabel("number of visits")
# plt.ylabel("count")
|
# First step to do pricing of a options derivative is Binomial Model.
# - Short call derivative and buy $\Delta$ units stock to evaluate the derivative price
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
#
def binomial_model(s0, s1u, s1d, K, r):
u, d = s1u / s0, s1d / s0
d1u, d1d = np.max([s1u - K, 0]), np.max([s1d - K, 0])
delta = (d1u - d1d) / (s1u - s1d)
p, q = (1 + r - d) / (u - d), (u - 1 - r) / (u - d)
d0 = (p * d1u / (1 + r)) + (q * d1d / (1 + r))
print(f"No of stocks and Derivative price is : {delta, d0}")
return delta, d0
binomial_model(50, 65, 40, 55, 0.08)
# We could take advantage of this model to get the price of two step binomial model.
# - Apply BM to get from step 2 to 1
# - Do the same from step 1 to 2.
def binomial_model1(u, d, d1u, d1d, r):
# u, d = s1u/s0, s1d/s0
# d1u, d1d = np.max([s1u-K,0]), np.max([s1d-K,0])
# delta = (d1u - d1d)/(s1u - s1d)
p, q = (1 + r - d) / (u - d), (u - 1 - r) / (u - d)
d0 = (p * d1u / (1 + r)) + (q * d1d / (1 + r))
# print(f'Derivative price is : {d0}')
return d0
d2pp, d2pm, d2mm = 25, 10, 5
u, d, r = 1.2, 0.9, 0.06
d1p, d1m = binomial_model1(u, d, d2pp, d2pm, r), binomial_model1(u, d, d2pm, d2mm, r)
binomial_model1(u, d, d1p, d1m, r)
# derivative price at 10 times step.
der_price = [np.sort(np.random.choice(range(1, 50), size=11, replace=False))[::-1]]
while len(der_price[-1]) != 1:
temp = []
p = der_price[-1]
k = len(p)
for i in range(k - 1):
d1p, d1m = p[i], p[i + 1]
temp.append(binomial_model1(u, d, d1p, d1m, r))
der_price.append(temp)
print(
f"For a given price at 10 timestep : {der_price[0]},\nDerivative price is {der_price[-1][-1]}"
)
# One can calculate above price using binomial coefficient. Due to round off error we shall see that the values do not match.
from scipy.special import binom
coeff = np.array([binom(10, i) for i in range(11)])
p, q = (1 + r - d) / (u - d), (u - 1 - r) / (u - d)
a = np.array([(p**i) * (q ** (10 - i)) for i in range(11)])
b = coeff * a
print(f"Derivative price is {b.dot(der_price[0])/((1+r)**10)}")
def f(k):
coeff = np.array([binom(k, i) for i in range(k + 1)])
# p, q = (1+r-d)/(u-d), (u-1-r)/(u-d)
a = np.array([(p**i) * (q ** (k - i)) for i in range(k + 1)])
b = coeff * a
# print(coeff,a,b)
print(f"Derivative price is {b.dot(der_price[10-k])/((1+r)**k)}")
for i in range(11):
f(i)
|
# # Cricket Umpire Mediapipe Images
# Mediapipe pose detection
import cv2
import os
import math
import random
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import mediapipe as mp
mp_pose = mp.solutions.pose
mp_drawing = mp.solutions.drawing_utils
mp_drawing_styles = mp.solutions.drawing_styles
paths0 = []
for dirname, _, filenames in os.walk(
"/kaggle/input/cricket-umpires-action-classification/umpire/train"
):
for filename in filenames:
if filename[-4:] == ".jpg":
paths0 += [(os.path.join(dirname, filename))]
print(paths0[0:3])
paths = random.sample(paths0, 200)
labels2 = []
paths2 = []
for i, path in enumerate(paths):
if i % 250 == 0:
print("i=", i)
file = path.split("/")[-1]
label = path.split("/")[-2]
image = cv2.imread(path)
with mp_pose.Pose(
static_image_mode=True,
# max_num_pose=2,
min_detection_confidence=0.1,
) as poses:
try:
results = pose.process(cv2.flip(image, 1))
if results.multi_pose_landmarks:
image_hight, image_width, _ = image.shape
annotated_image = cv2.flip(image.copy(), 1)
for pose_landmarks in results.multi_hand_landmarks:
mp_drawing.draw_landmarks(
annotated_image,
pose_landmarks,
mp_pose.POSE_CONNECTIONS,
mp_drawing_styles.get_default_pose_landmarks_style(),
mp_drawing_styles.get_default_pose_connections_style(),
)
anno_img = cv2.flip(annotated_image, 1)
cv2.imwrite(file, anno_img)
paths2 += [file]
labels2 += [label]
except:
continue
data = pd.DataFrame(columns=["path", "label"])
data["path"] = paths2
data["label"] = labels2
data.to_csv("data.csv", index=False)
display(data)
selected_num = random.sample(range(len(data)), 9)
fig, axes = plt.subplots(3, 3, figsize=(10, 10))
for i, ax in enumerate(axes.flat):
j = selected_num[i]
img_path = data.iloc[j, 0]
label = data.iloc[j, 1]
img = plt.imread(img_path)
ax.imshow(img)
ax.axis("off")
ax.set_title(label)
plt.tight_layout()
plt.show()
|
# # RSNA Pneumonia Detection Challenge
# building an algorithm that automatically detects potential pneumonia cases using Pytorch Lightning
# **About Challenge:** The competition challenges us to create an algorithm that can detect lung opacities on chest radiographs to aid in the accurate diagnosis of pneumonia, which is responsible for many deaths of children under 5 and emergency room visits in the US. Diagnosing pneumonia is difficult because other lung conditions and factors can cause increased opacity on the radiograph. CXRs are commonly used but their interpretation can be complicated and time-consuming, making an automated solution helpful in improving the efficiency and reach of diagnostic services. The Radiological Society of North America (RSNA®) has collaborated with Kaggle's machine learning community, the US National Institutes of Health, The Society of Thoracic Radiology, and MD.ai to create a dataset for this challenge.
# **About Dataset:** This is an update to a two-stage machine learning challenge to develop an algorithm that detects pneumonia in medical images by automatically locating lung opacities on chest radiographs. The dataset contains training and test images in DICOM format, along with training labels that include bounding boxes for pneumonia and a binary target column indicating whether evidence of pneumonia is present. Competitors are expected to predict bounding boxes for areas of the lung where pneumonia is present. The file format for predictions should include confidence, x-min, y-min, width, and height values, and there should be only one predicted row per image.
# The dataset for this challenge consists of chest radiographs in DICOM format. The training set contains a total of 26,684 images, which includes both the images from the stage 1 train set and the new stage 2 train set. The stage 2 test set contains 3,640 new, unseen images.
# ## Importing Necessary Libraries and reading files
from pathlib import Path
import pydicom
import cv2
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from tqdm.notebook import tqdm
# reading labels data
labels = pd.read_csv(
"/kaggle/input/rsna-pneumonia-detection-challenge/stage_2_train_labels.csv"
)
labels.head()
# defining the root path to importing images and files and save path to save the models and images
ROOT_PATH = Path(
"/kaggle/input/rsna-pneumonia-detection-challenge/stage_2_train_images/"
)
SAVE_PATH = Path("/kaggle/working/")
# Now, we are going to generate a 3x3 grid of subplots, where each subplot displays a chest X-ray image from the training set along with its associated label. Where label 0 indicating no Pneumonia and 1 indicating there is some evidence of Pneuomia.
fig, axis = plt.subplots(3, 3, figsize=(9, 9))
c = 0
for i in range(3):
for j in range(3):
patient_id = labels.patientId.iloc[c]
dcm_path = ROOT_PATH / patient_id
dcm_path = dcm_path.with_suffix(".dcm")
dcm = pydicom.read_file(dcm_path).pixel_array
label = labels["Target"].iloc[c]
axis[i][j].imshow(dcm, cmap="bone")
axis[i][j].set_title(label)
c += 1
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import plotly.express as px
# # 特徵
# * session_id - user_id
# * index - the index of the event for the session:一個user會有多個index
# * elapsed_time - how much time has passed (in milliseconds) between the start of the session and when the event was recorded - Time series
# * event_name - the name of the event type
# * name - the event name
# * level - what level of the game the event occurred in: 0~22
# * page - the page number of the event (刪除)
# * room_coor_x - the coordinates of the click in reference to the in-game room
# * room_coor_y - the coordinates of the click in reference to the in-game room
# * screen_coor_x - the coordinates of the click in reference to the player’s screen
# * screen_coor_y - the coordinates of the click in reference to the player’s screen
# * hover_duration - how long (in milliseconds) the hover happened for (刪除)
# * text - the text the player sees during this event (刪除)
# * fqid - the fully-qualified ID of the event (31%遺缺,刪除)
# * room_fqid - the fully-qualified ID of the room the event took place in
# * text_fqid - the fully-qualified ID of the (等同fqid+room_fqid,63%遺缺,刪除)
# * fullscreen - whether the player is in fullscreen mode(刪除)
# * hq - whether the game is in high-quality(刪除)
# * music - whether the game music is on or off(刪除)
# * level_group - which group of levels - and group of questions - this row belongs to (0-4, 5-12, 13-22)
X_train = pd.read_csv(
"/kaggle/input/predict-student-performance-from-game-play/train.csv",
usecols=[
"session_id", # user_id
"index",
"elapsed_time",
"event_name", # event type
"name", # the event name
"level",
"room_coor_x",
"room_coor_y",
"screen_coor_x",
"screen_coor_y",
"room_fqid",
"level_group",
],
nrows=1000000,
) # 共2000多萬筆資料,只使用100萬筆
y_train = pd.read_csv(
"/kaggle/input/predict-student-performance-from-game-play/train_labels.csv"
)
X_train.shape, y_train.shape
data = X_train.copy()
target = y_train.copy()
data.elapsed_time = data.elapsed_time / 60000 # 從millsecond變分鐘
data.shape, target.shape
### 因為選100萬筆資料近來,但最後一個人的資料可能不完整被我截掉,因此也扣除最後一個user的資料,最後剩999961筆資料
del_uncompleted_data = data.loc[data.session_id == data.session_id.to_list()[-1]].index[
0
] # 999961
data = data[:del_uncompleted_data]
data.shape
data.to_csv(
"/kaggle/working/predict-student-performance-from-game-play_smalltrain_data",
index=False,
)
data.head(3)
import re
# target的session_id包含題目q1~q8,不好與data比對,因此新增一個user_id
target["user_id"] = target.session_id.str.split("_", expand=True)[0]
target["question"] = target.session_id.str.split("_", expand=True)[1]
target["question"] = target["question"].apply(lambda x: re.sub("\D", "", x))
target["question"] = pd.to_numeric(target["question"]) # 轉為數值型別
target["user_id"] = pd.to_numeric(target["user_id"]) # 轉為數值型別
target.head(3)
target.question.unique()
data.session_id.nunique(), target.user_id.nunique() # target擁有的是2000萬筆資料的23562個玩家 但目前只取出50萬筆有885個玩家
# 發現每個question的排序和data相同
data.session_id.unique()[:5] == target.user_id[:5]
# 使target變成只有885個user
target = pd.DataFrame(target.groupby("question").apply(lambda x: x[:885]))
target.index = np.arange(len(target.index))
target.to_csv(
"/kaggle/working/predict-student-performance-from-game-play_smalltarget",
index=False,
)
target.head(3)
data.shape, target.shape
data.session_id.nunique(), target.user_id.nunique()
import gc
gc.collect()
# # EDA
data.info()
data.isnull().sum()
# # 先拿第一位的資料來分析:
# 先選一個user來看他的行為
temp_user_id = data.session_id[0]
temp_user_data = data.loc[data.session_id == temp_user_id]
temp_user_data
# temp_user回答問題的資料
target.loc[target.user_id == temp_user_id].sort_values(by="question", ascending=True)
temp_user_data.info()
# 三種level_group經常出現的位置有所不同
fig = px.bar(temp_user_data, x="room_fqid", color="level_group")
fig.show()
# elapsed_time vs. index
px.scatter(
temp_user_data,
x="index",
y="elapsed_time",
color=temp_user_data.level_group,
hover_name=temp_user_data.event_name,
)
# 有兩個回答問題的checkpoint:
# 1. index 164直接跳175
# 2. index 470直接跳512
# # 全部人的資料:
# # 1.是否有人未完成0~22個level?
data.session_id.nunique()
# 有人重複出現在checkpoint:在0~4多出8個,5~12多出6個,13~22多出4個,共18個
data[data.event_name == "checkpoint"].level.value_counts() - 885
# 取出重複出現的user
checkpoint_df = data[data.event_name == "checkpoint"]
len(
checkpoint_df[
checkpoint_df.duplicated(
subset=["session_id", "event_name", "level_group"], keep="last"
)
]
) # 剛好18個,因此其他checkpoint沒有重複的玩家,都有完成3個checkpoint
dup_checkpoint_df = checkpoint_df[
checkpoint_df.duplicated(
subset=["session_id", "event_name", "level_group"], keep=False
)
]
dup_checkpoint_df.groupby(["session_id"])["level_group"].value_counts()
# 8位玩家在checkpoint的資料有重複
# 畫出第一位重複玩的elapse_time vs. index
px.scatter(
data[data.session_id == 20100110332615344],
x="index",
y="elapsed_time",
color=data[data.session_id == 20100110332615344].level_group,
hover_name=data[data.session_id == 20100110332615344].event_name,
)
# 玩了兩次,而且在第二次到5~12關結束後沒有再繼續到13關
# 有人玩超過一次,且第二次玩不一定會玩到結束
# 建立一個所以有玩家的重要資訊
user_info_df = pd.DataFrame(
{
"user_id": data.session_id.unique(),
"playtime": data.groupby("session_id")["elapsed_time"].agg("max"),
"event_nb": data.groupby("session_id")["index"].agg("max"),
"score": target.groupby("user_id")["correct"].agg("sum"),
}
).reset_index(drop=True)
user_info_df
user_info_df[user_info_df.user_id.isin(dup_checkpoint_df.session_id)]
# 比較重複玩和未重複玩的playtime, event_nb, score 是否有顯著差異
my_dict = {
"play_again": user_info_df[
user_info_df.user_id.isin(dup_checkpoint_df.session_id)
].playtime,
"not_play_again": user_info_df[
(~user_info_df.user_id.isin(dup_checkpoint_df.session_id))
& (user_info_df.playtime < user_info_df.playtime.quantile(0.5))
].playtime,
}
fig, ax = plt.subplots()
ax.boxplot(my_dict.values())
ax.set_xticklabels(my_dict.keys())
ax.set_ylabel("playtime")
fig.show()
from scipy import stats
leveneTestRes = stats.levene(
user_info_df[user_info_df.user_id.isin(dup_checkpoint_df.session_id)].playtime,
user_info_df.playtime < user_info_df.playtime.quantile(0.5),
center="mean",
) # levene法齐性检验
print(f"{leveneTestRes.pvalue:.5f}") # 變異數不同質->welch’s t-test
tind = stats.ttest_ind(
a=user_info_df[user_info_df.user_id.isin(dup_checkpoint_df.session_id)].playtime,
b=user_info_df.playtime < user_info_df.playtime.quantile(0.5),
equal_var=False,
)
print(tind.pvalue) # 顯著不同
my_dict = {
"play_again": user_info_df[
user_info_df.user_id.isin(dup_checkpoint_df.session_id)
].score,
"not_play_again": user_info_df[
(~user_info_df.user_id.isin(dup_checkpoint_df.session_id))
].score,
}
fig, ax = plt.subplots()
ax.boxplot(my_dict.values())
ax.set_xticklabels(my_dict.keys())
ax.set_ylabel("score")
fig.show()
leveneTestRes = stats.levene(
user_info_df[user_info_df.user_id.isin(dup_checkpoint_df.session_id)].score,
user_info_df[(~user_info_df.user_id.isin(dup_checkpoint_df.session_id))].score,
center="mean",
) # levene法齐性检验
tind = stats.ttest_ind(
a=user_info_df[user_info_df.user_id.isin(dup_checkpoint_df.session_id)].score,
b=user_info_df[(~user_info_df.user_id.isin(dup_checkpoint_df.session_id))].score,
equal_var=True,
)
print(f"{leveneTestRes.pvalue:.5f}") # 變異數同質
print(tind.pvalue) # 有顯著不同
my_dict = {
"play_again": user_info_df[
user_info_df.user_id.isin(dup_checkpoint_df.session_id)
].event_nb,
"not_play_again": user_info_df[
(~user_info_df.user_id.isin(dup_checkpoint_df["index"]))
& (user_info_df.event_nb < user_info_df.event_nb.quantile(0.5))
].event_nb,
}
fig, ax = plt.subplots()
ax.boxplot(my_dict.values())
ax.set_xticklabels(my_dict.keys())
ax.set_ylabel("event_nb")
fig.show()
leveneTestRes = stats.levene(
user_info_df[user_info_df.user_id.isin(dup_checkpoint_df.session_id)].event_nb,
user_info_df[
(~user_info_df.user_id.isin(dup_checkpoint_df["index"]))
& (user_info_df.event_nb < user_info_df.event_nb.quantile(0.5))
].event_nb,
center="mean",
) # levene法齐性检验
tind = stats.ttest_ind(
a=user_info_df[user_info_df.user_id.isin(dup_checkpoint_df.session_id)].event_nb,
b=user_info_df[
(~user_info_df.user_id.isin(dup_checkpoint_df["index"]))
& (user_info_df.event_nb < user_info_df.event_nb.quantile(0.5))
].event_nb,
equal_var=False,
)
print(f"{leveneTestRes.pvalue:.5f}") # 變異數不同質
print(tind.pvalue) # 有顯著不同
# 重複玩的與未重複玩的playtime、score、event_nb皆有顯著差異。
# 重複玩的score比未重複玩的要來得低。
# 完兩次的人score越高 event_nb越低
user_info_df[user_info_df.user_id.isin(dup_checkpoint_df.session_id)].plot.scatter(
x="score", y="event_nb"
)
plt.xticks(np.arange(1, 19))
plt.show()
# # 2. 遊戲時間
# 多數人在半小時完成遊戲、發生約1000多次事件、平均得分為12
user_info_df.loc[:, ["playtime", "event_nb", "score"]].describe()
# playtime
px.box(user_info_df.playtime)
long_time_player = (
user_info_df[user_info_df.playtime > 91].sort_values(by="playtime").tail()
)
long_time_player
short_time_player = (
user_info_df[user_info_df.playtime < 91].sort_values(by="playtime").head()
)
short_time_player
# long_time
plt.figure(figsize=(12, 6))
plt.subplot(2, 2, 1)
plt.plot(
data[data.session_id == long_time_player.user_id.iloc[-1]]["index"],
data[data.session_id == long_time_player.user_id.iloc[-1]].elapsed_time,
)
plt.xlabel("index")
plt.ylabel("elapsed_time")
plt.subplot(2, 2, 2)
plt.plot(
data[data.session_id == long_time_player.user_id.iloc[-2]]["index"],
data[data.session_id == long_time_player.user_id.iloc[-2]].elapsed_time,
)
plt.xlabel("index")
plt.ylabel("elapsed_time")
# short_time
plt.subplot(2, 2, 3)
plt.plot(
data[data.session_id == short_time_player.user_id.iloc[0]]["index"],
data[data.session_id == short_time_player.user_id.iloc[0]].elapsed_time,
)
plt.xlabel("index")
plt.ylabel("elapsed_time")
plt.subplot(2, 2, 4)
plt.plot(
data[data.session_id == short_time_player.user_id.iloc[1]]["index"],
data[data.session_id == short_time_player.user_id.iloc[1]].elapsed_time,
)
plt.xlabel("index")
plt.ylabel("elapsed_time")
plt.show()
# 那些遊戲時間比較久的玩家,是因為在遊戲中中斷動作,持續一段時間後才又回到遊戲。因此event不一定會比遊戲時間短的玩家要多。
# 刪除離群值後 發現得分越高 使用的時間越低
playtime_nb_vs_score = (
user_info_df[user_info_df.playtime < 91]
.groupby("score")["playtime"]
.agg("mean")
.reset_index()
)
playtime_nb_vs_score.plot.scatter(x="score", y="playtime")
plt.xticks(np.arange(1, 19))
plt.show()
# # 3. event
# event_nb
px.box(user_info_df.event_nb)
more_event = (
user_info_df[user_info_df.event_nb > 1819].sort_values(by="event_nb").tail()
)
more_event
little_event = (
user_info_df[user_info_df.event_nb < 1819].sort_values(by="event_nb").head()
)
little_event
# navigate_click 多出許多
px.bar(
pd.DataFrame(
{
"more_event": data[
data.session_id == more_event.user_id.iloc[-1]
].event_name.value_counts(),
"little_event": data[
data.session_id == little_event.user_id.iloc[0]
].event_name.value_counts(),
}
)
)
# 發生較多事件的玩家,在每一關點及次數都較多
plt.figure(figsize=(8, 3))
plt.subplot(1, 2, 1)
plt.plot(
data[data.session_id == more_event.user_id.iloc[-1]]["index"],
data[data.session_id == more_event.user_id.iloc[-1]].level,
)
plt.title("more_event")
plt.xlabel("index")
plt.ylabel("elapsed_time")
plt.subplot(1, 2, 2)
plt.plot(
data[data.session_id == little_event.user_id.iloc[0]]["index"],
data[data.session_id == little_event.user_id.iloc[0]].level,
)
plt.title("little_event")
plt.xlabel("index")
plt.ylabel("elapsed_time")
# 刪除離群值後 ,發現動作越多 得分越低
event_nb_vs_score = (
user_info_df[user_info_df.event_nb < 1819]
.groupby("score")
.agg({"event_nb": "mean"})
.reset_index()
)
event_nb_vs_score.plot.scatter(x="score", y="event_nb")
plt.xticks(np.arange(1, 19))
plt.show()
# playtime vs. event: 集中在左下角,玩的時間越短,發生的事件也越少
px.scatter(
user_info_df[(user_info_df.event_nb < 1819) & (user_info_df.playtime < 91)],
x="event_nb",
y="playtime",
)
# # 4. 得分
# 多數人能能答對13題
px.bar(user_info_df["score"].value_counts())
# 常錯的題目
ans_correct_or_not = pd.DataFrame()
Q = dict(target.groupby("question")["correct"].value_counts()).keys()
res = list(zip(*Q))
Q = res[0]
A = res[1]
ans_correct_or_not["question"] = Q
ans_correct_or_not["question"] = ans_correct_or_not["question"].apply(lambda x: str(x))
ans_correct_or_not["ans"] = A
ans_correct_or_not["count"] = dict(
target.groupby("question")["correct"].value_counts()
).values()
ans_correct_or_not.head()
# 10 13 15 錯的比對的多
fig = px.bar(ans_correct_or_not, x="question", y="count", color="ans", text_auto=True)
fig.show()
# # 5. level、level_group
# 所有玩家都有回答完所有問題
target.question.value_counts().sort_index()
# 根據所有玩家在0~22關所花費的總時間,推測各關卡的難度
def calculate_range(x): # 全距
return x.max() - x.min()
# 需先刪除離群值
elapsed_time_groupby_level = (
data.loc[data.elapsed_time < 91, ["session_id", "elapsed_time", "level"]]
.groupby(["level", "session_id"])["elapsed_time"]
.apply(lambda x: calculate_range(x))
)
elapsed_time_groupby_level
# 第6、18、明顯大家平均花的時間較多
px.bar(elapsed_time_groupby_level.groupby("level").mean(), y="elapsed_time")
# level_group
elapsed_time_groupby_level_group = (
data.loc[data.elapsed_time < 91, ["session_id", "elapsed_time", "level_group"]]
.groupby(["level_group", "session_id"])["elapsed_time"]
.apply(lambda x: calculate_range(x))
)
elapsed_time_groupby_level_group
# 越後面的關卡,大家所花的平均時間越多
px.bar(
elapsed_time_groupby_level_group.groupby("level_group").mean().sort_values(),
y="elapsed_time",
)
gc.collect()
# # 6. checkpoint的停留時間
level4_checkpoint = data[
(data.event_name == "checkpoint") & (data.level == 4)
].drop_duplicates(subset="session_id", keep="first")
level5 = data.iloc[level4_checkpoint.index + 1]
level4_checkpoint_elapsed_time = level5["elapsed_time"].reset_index(
drop=True
) - level4_checkpoint.elapsed_time.reset_index(drop=True)
# level4_checkpoint_elapsed_time
level12_checkpoint = data[
(data.event_name == "checkpoint") & (data.level == 12)
].drop_duplicates(subset="session_id", keep="first")
level13 = data.iloc[level12_checkpoint.index + 1]
level12_checkpoint_elapsed_time = level13["elapsed_time"].reset_index(
drop=True
) - level12_checkpoint.elapsed_time.reset_index(drop=True)
# level12_checkpoint_elapsed_time
ans_time = level4_checkpoint_elapsed_time + level12_checkpoint_elapsed_time
px.box(ans_time)
user_info_df["ans_time"] = ans_time
user_info_df.head()
short_ans_time = (
user_info_df[user_info_df.ans_time < 12.9].sort_values(by="ans_time").head()
)
short_ans_time
# 觀察離群值
long_ans_time = (
user_info_df[user_info_df.ans_time > 12.9].sort_values(by="ans_time").tail()
)
long_ans_time
# 正常作答回答
px.line(
data[data.session_id == 20100213263066652],
x="index",
y="elapsed_time",
color="level_group",
)
# 回答時間過長,但點擊數量沒有比較多,回答時間過長的原因不是在嘗試其他答案,而是中途離開遊戲。
px.line(
data[data.session_id == 20100117123192490],
x="index",
y="elapsed_time",
color="level_group",
)
# 刪除ans_time的離群值後,發現答對題數越多者,回答時間也越短
px.scatter(
user_info_df[user_info_df.ans_time < 12.9]
.groupby("score")
.agg({"ans_time": "mean"})
)
gc.collect()
playtime_vs_anstime = user_info_df[
(user_info_df.ans_time < user_info_df.ans_time.quantile(0.75))
& (user_info_df.playtime < user_info_df.playtime.quantile(0.75))
]
plt.scatter(playtime_vs_anstime.ans_time, playtime_vs_anstime.playtime)
plt.xlabel("ans_time")
plt.ylabel("play_time")
plt.legend()
plt.show()
|
# Problem statement
# Predict on building satety during an earthquake
# Import libraries
import numpy as np
import pandas as pd
import os
import matplotlib.pyplot as plt
import seaborn as sns
# Get files
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Read files
train = pd.read_csv(
"/kaggle/input/predict-the-building-safety-under-the-earthquake/train.csv"
)
test = pd.read_csv(
"/kaggle/input/predict-the-building-safety-under-the-earthquake/test.csv"
)
submission = pd.read_csv(
"/kaggle/input/predict-the-building-safety-under-the-earthquake/sample_submission.csv"
)
train
train.describe()
train.info()
train.isna().sum()
test
test.isna().sum()
submission
# Analyse train
train["Max drift mm"] = round(train["Max drift mm"], 2)
sns.displot(train["Max drift mm"], kde=True)
train["Max drift mm"].value_counts()
is_multi = train["Max drift mm"].value_counts() > 1
filtered = train[train["Max drift mm"].isin(is_multi[is_multi].index)]
train = filtered
train
sns.displot(train["Max drift mm"], kde=True)
sns.boxplot(data=train, x="Number of floors", y="Max drift mm")
sns.boxplot(data=train, x="Floor height m", y="Max drift mm")
sns.boxplot(data=train, x="Distance to fault km", y="Max drift mm")
corr = train.corr()
sns.heatmap(corr)
corr
# Split dataset into training and validation sets
from sklearn.model_selection import train_test_split
train_pd, val_pd = train_test_split(train, test_size=0.1, random_state=42)
train_pd.shape, val_pd.shape, test.shape
# Tensorflow
import tensorflow as tf
import tensorflow_decision_forests as tfdf
print(tf.__version__)
# Name of the label column.
label = "Max drift mm"
train_ds = tfdf.keras.pd_dataframe_to_tf_dataset(
train_pd, fix_feature_names=False, label=label, task=tfdf.keras.Task.REGRESSION
)
val_ds = tfdf.keras.pd_dataframe_to_tf_dataset(
val_pd, fix_feature_names=False, label=label, task=tfdf.keras.Task.REGRESSION
)
test_ds = tfdf.keras.pd_dataframe_to_tf_dataset(
test, fix_feature_names=False, task=tfdf.keras.Task.REGRESSION
)
# Configure the model.
argument = tfdf.keras.AdvancedArguments(fail_on_non_keras_compatible_feature_name=False)
model = tfdf.keras.RandomForestModel(
task=tfdf.keras.Task.REGRESSION, advanced_arguments=argument
)
# Train the model.
model.fit(train_ds)
model.summary()
# Evaluate the model on the test dataset.
model.compile(metrics=["mse"])
evaluation = model.evaluate(val_ds, return_dict=True)
print(evaluation)
print()
print(f"MSE: {evaluation['mse']}")
print(f"RMSE: {np.sqrt(evaluation['mse'])}")
# Make predictions
predictions = model.predict(test_ds)
predictions = np.round_(predictions, decimals=2)
predictions = predictions.flatten()
predictions
# Prepare submission
submission["Max drift mm"] = predictions
submission.to_csv("submission.csv", index=False)
submission = pd.read_csv("submission.csv")
submission
|
# Semantic segmentation chalenges expose us to a lot of metrics. So I have decided to make a list of as many
# as I can and try to explain each one.
# Let's go.
# # Semantic segmentation targets
# In semantic segmentation tasks, we predict a mask, i.e. where the object of interest is present.
# To make things simple, let's assume we only have one type of object to predict thus it is a binary task.
# We can thus assign the following mapping:
#
#
# 1. **0** for the **background**
# 2. **1** for objecs of interest. Here in this competition for **glomuerli**.
# ## Accuracy
# This isn't the most clever one but it is for sure the simplest so we should start with it.
# How does it work?
# We check all the image pixels and give a score of 1 if the pixel is correctly predicted, 0 othrewise.
# Thus, this can mathmeatically be represented by the indicator function.
# To illustrate the accuracy metric, let's see an example. An image with half background, and half object of interest.
#
cv2.cvtColor(255 * accuracy_mask, 1)
import numpy as np
import matplotlib.pylab as plt
import cv2
accuracy_mask = np.zeros((256, 256), dtype=np.uint8)
accuracy_mask[: 256 // 2, :] = 1
plt.imshow(cv2.cvtColor(255 * accuracy_mask, cv2.COLOR_GRAY2RGB))
|
# Tabular Playground Series(feb)
# Table Playground Series are beginner friendly monthly competitions organised by kaggle.
#
# In this competition we have to make a regrssion model based on categorical and continous features provided
# This notebook is beginner friendly guide for creating supercool EDA and making baseline model.
#
# **Feel free to ask any question and if you find any mistake tell me in the comments**
# ## Importing Libraries 📗.
import os
import gc
import sys
import time
import tqdm
import random
import numpy as np
import pandas as pd
import seaborn as sns
import datatable as dt
import matplotlib.pyplot as plt
plt.style.use("fivethirtyeight")
import plotly.express as px
import plotly.graph_objs as go
import plotly.figure_factory as ff
from sklearn.decomposition import PCA
from sklearn.impute import SimpleImputer
from sklearn.model_selection import train_test_split
from sklearn.manifold import TSNE
from colorama import Fore, Back, Style
y_ = Fore.YELLOW
r_ = Fore.RED
g_ = Fore.GREEN
b_ = Fore.BLUE
m_ = Fore.MAGENTA
c_ = Fore.CYAN
sr_ = Style.RESET_ALL
import warnings
warnings.filterwarnings("ignore")
import lightgbm as lgb
import torch
import torch.nn as nn
import torch.optim as optim
import torch.nn.functional as F
from torch.utils.data import TensorDataset, DataLoader, Dataset
from sklearn.metrics import roc_auc_score, mean_squared_error
from sklearn.compose import ColumnTransformer
from sklearn.model_selection import KFold, StratifiedKFold
from sklearn.pipeline import FeatureUnion
from sklearn.decomposition import PCA, TruncatedSVD
from sklearn.preprocessing import (
StandardScaler,
PowerTransformer,
QuantileTransformer,
LabelEncoder,
OneHotEncoder,
OrdinalEncoder,
RobustScaler,
)
# 1. Given Data 💽
# we are provided with 3 files.
# 1. sample_submission.csv
# 2. test.csv
# 3. train.csv
# Train data contains 10 categorical featues and 14 continous features
# we have to make prediction on test data and make submission using sample_submission's format
# 2. Metric 📐
# Here metric used is very simple which is root mean squared error.
# rmse = root(1/n (yit-yip)^2)
# Here yit is true target values
# and yip is predicted target values.
# 3. Loading Data 💽
# We will use pandas to load data
#
folder_path = "../input/tabular-playground-series-feb-2021"
train_data = pd.read_csv(f"{folder_path}/train.csv")
test_data = pd.read_csv(f"{folder_path}/test.csv")
sample = pd.read_csv(f"{folder_path}/sample_submission.csv")
print(
"{0}Number of rows in train data: {1}{2}\n{0}Number of columns in train data: {1}{3}".format(
y_, r_, train_data.shape[0], train_data.shape[1]
)
)
print(
"{0}Number of rows in test data: {1}{2}\n{0}Number of columns in test data: {1}{3}".format(
m_, r_, test_data.shape[0], test_data.shape[1]
)
)
print(
"{0}Number of rows in sample : {1}{2}\n{0}Number of columns in sample : {1}{3}".format(
c_, r_, sample.shape[0], sample.shape[1]
)
)
train_data.head()
test_data.head()
sample.head()
# 4. Exploratory Data Analysis 📊📈📉👀
cont_features = [f"cont{i}" for i in range(14)]
cat_features = [f"cat{i}" for i in range(10)]
all_features = cont_features + cat_features
# 4.1 Checking for Null Values
#
train_data.isnull().sum().sum()
# 4.2 Distribution of cont0
plt.style.use("fivethirtyeight")
def distribution1(feature, color1, color2, df=train_data):
plt.figure(figsize=(15, 7))
plt.subplot(121)
dist = sns.distplot(df[feature], color=color1)
a = dist.patches
xy = [
(a[i].get_x() + a[i].get_width() / 2, a[i].get_height())
for i in range(1, len(a) - 1)
if (
a[i].get_height() > a[i - 1].get_height()
and a[i].get_height() > a[i + 1].get_height()
)
]
for i, j in xy:
dist.annotate(
s=f"{i:.3f}",
xy=(i, j),
xycoords="data",
ha="center",
va="center",
fontsize=11,
color="black",
xytext=(0, 7),
textcoords="offset points",
)
qnt = df[feature].quantile([0.25, 0.5, 0.75]).reset_index(level=0).to_numpy()
plt.subplot(122)
box = sns.boxplot(df[feature], color=color2)
for i, j in qnt:
box.annotate(str(j)[:4], xy=(j - 0.05, -0.01), horizontalalignment="center")
print(
"{}Max value of {} is: {} {:.2f} \n{}Min value of {} is: {} {:.2f}\n{}Mean of {} is: {}{:.2f}\n{}Standard Deviation of {} is:{}{:.2f}".format(
y_,
feature,
r_,
df[feature].max(),
g_,
feature,
r_,
df[feature].min(),
b_,
feature,
r_,
df[feature].mean(),
m_,
feature,
r_,
df[feature].std(),
)
)
distribution1("cont0", "yellow", "red")
# 4.3 Distribution of targets
distribution1("target", "red", "blue")
# Target ranges from 0 to 10.31
# but distribution really starts from target values >4
# We can see 2 peaks in target distribution
# One may assume that data is taken from 2 different distribution and then combined.
# so we are seeing two peaks
# But judging from TPS(jan) actually target is from same distribution but some values of targets
# belong to test data so we will most probably see the distribution of predictions with one peak around value 7
# 4.3 Distribution of all the continous features.
plt.figure(figsize=(20, 15))
colors = [
"#8ECAE6",
"#219EBC",
"#023047",
"#023047",
"#023047",
"#0E402D",
"#023047",
"#023047",
"#F77F00",
"#D62828",
"#4285F4",
"#EA4335",
"#FBBC05",
"#34A853",
]
for i, feature in enumerate(cont_features):
plt.subplot(2, 7, i + 1)
sns.distplot(train_data[feature], color=colors[i])
# It is clear that all the features have different disbtributions and none of them looks like normal distribution
# cont1 is very interesting as there are gaps in cont1 as if it is partly continous and partly categorical
# Maybe we should look into correlations between this features to get better insight
# 4.4 Correlation Matrix
corr = train_data[all_features + ["target"]].corr()
fig = px.imshow(corr)
fig.show()
# We see that there is some correlation between featues cont5 to cont12 but they are not very high.
#
# One interesting thing is no features have any correlation with target
# 4.5 Pairplot of features
def plot_grid(data, color1, color2, color3):
f = sns.PairGrid(data)
plt.figure(figsize=(10, 10))
f.map_upper(plt.scatter, color=color1)
f.map_lower(sns.kdeplot, color=color2)
# f.map_diag(sns.histplot,color = c3 );
f.map_diag(sns.kdeplot, lw=3, legend=False, color=color3)
plot_grid(
train_data.loc[:1000, all_features + ["target"]], "#EA4335", "#FBBC05", "#34A853"
)
# Pair plot gives very good idea about relationship between the features
# all the features have a square relationship means for every value in one feature
# there are enough values in entire range of other feature which makes this data little challanging
# 4.7 Let's play with PCA
# 4.7.1 How good are 4 components of pca at seprating data
def pca_plot1(features, n_components, target, nrows=10**4):
pca = PCA(n_components=n_components)
train_d = train_data.sample(n=nrows).fillna(train_data.mean())
train_g_pca = pca.fit_transform(train_d[features])
total_var = pca.explained_variance_ratio_.sum() * 100
labels = {str(i): f"PC {i+1}" for i in range(n_components)}
fig = px.scatter_matrix(
train_g_pca,
dimensions=range(n_components),
labels=labels,
title=f"Total explained variance ratio{total_var:.2f}%",
color=train_d[target].values,
)
fig.update_traces(diagonal_visible=True, opacity=0.5)
fig.show()
pca_plot1(cont_features, 4, "target")
# 4.7.2 How good are 3 components of pca at seprating data (3d plot)
def pca_plot_3d(features, target, nrows=10**4):
pca = PCA(n_components=3)
train_d = train_data.sample(n=nrows).fillna(train_data.mean())
train_g_pca = pca.fit_transform(train_d[features])
total_var = pca.explained_variance_ratio_.sum() * 100
fig = px.scatter_3d(
train_g_pca,
x=0,
y=1,
z=2,
title=f"Total explained variance ratio{total_var:.2f}%",
color=train_d[target].values,
labels={"0": "PC 1", "1": "PC 2", "2": "PC 3"},
)
fig.show()
pca_plot_3d(cont_features, "target")
# 4.7.3 Ploting explained variance
def plot_exp_var(features, nrows=10**4):
pca = PCA()
train_d = train_data.sample(n=nrows).fillna(train_data.mean())
pca.fit(train_d[features])
exp_var_cumul = np.cumsum(pca.explained_variance_ratio_)
fig = px.area(
x=range(1, exp_var_cumul.shape[0] + 1),
y=exp_var_cumul,
labels={"x": "# Components", "y": "Explained Variance"},
)
fig.show()
plot_exp_var(cont_features)
# 4.8 Countplot of all categorical fearues
plt.style.use("ggplot")
plt.figure(figsize=(25, 20))
for i, feature in enumerate(cat_features):
plt.subplot(2, 5, i + 1)
sns.countplot(train_data[feature])
# It is clear that distribution of categories are evry skewed
# It would be good to remove some of the cat features like
# cat4,cat0, cat2, cat6, cat7,
# The reason for this is that almost all rows will have same values for these columns
# so it will not provide any usefull information to model.
# 4.9 cont0 distribution based on cat9
# As cat9 has most categories let us see distribution of one feature based on cat9.
def distribution3(feature, category, df=train_data):
plt.subplots(figsize=(15, 7))
sns.histplot(train_data, x=feature, hue=category)
distribution3("cont0", "cat9")
# 4.10 cont0 box-plot based on cat9
# I think box-plot will give use better idea than histplot
def boxploting1(feature, category, df=train_data, figure_size=(15, 7)):
plt.subplots(figsize=figure_size)
sns.boxplot(
x=feature, y=category, data=df, whis=[0, 100], width=0.6, palette="vlag"
)
boxploting1("cont0", "cat9")
# 4.11 Boxenplot for cont1 and cat8
#
def boxploting2(feature, category, df=train_data, figure_size=(15, 7)):
plt.subplots(figsize=figure_size)
sns.boxenplot(y=feature, x=category, color="pink", scale="linear", data=df)
boxploting2("cont0", "cat8", figure_size=(10, 7))
# 4.12 Trivariate histogram with 2 category
def distribution3(feature, category1, category2, df=train_data, figure_size=(15, 15)):
sns.set_theme(style="dark")
sns.displot(
data=df,
x=feature,
y=category1,
col=category2,
log_scale=(True, False),
col_wrap=4,
height=4,
aspect=0.7,
)
distribution3("cont5", "cat8", "cat9")
# 4.13 swarmplot of target
def swarmplot(feature, category1, category2, df=train_data, figure_size=(15, 7)):
sns.set_theme(style="whitegrid", palette="muted")
plt.figure(figsize=figure_size)
ax = sns.swarmplot(data=df, x=feature, y=category1, hue=category2)
ax.set(ylabel="")
swarmplot("target", "cat8", "cat9", df=train_data.sample(n=10000))
# 4.13 Trend lines and templates plotly
# Now I will use plotly for plotting
def scatterplot1(feature1, feature2, category, df=train_data):
fig = px.scatter(
train_data,
x=feature1,
y=feature2,
color=category,
marginal_y="violin",
marginal_x="box",
trendline="ols",
template="simple_white",
)
fig.show()
scatterplot1("cont6", "cont7", "cat1")
# 4.14 Error bar
def errorbar(feature1, feature2, feature3, category, df=train_data):
df["e"] = df[feature3] / 100
fig = px.scatter(
df, x=feature1, y=feature2, color=category, error_x="e", error_y="e"
)
fig.show()
errorbar("cont1", "cont2", "cont3", "cat9", df=train_data.sample(n=1000))
# 4.15 parellel categories
def parallelcat(feature, df=train_data):
fig = px.parallel_categories(
df, color=feature, color_continuous_scale=px.colors.sequential.Inferno
)
fig.show()
parallelcat("cont2", df=train_data.sample(n=1000))
#
# 4.16 Parallel lines
def parallellines(feature, df=train_data):
fig = px.parallel_coordinates(
df,
color=feature,
color_continuous_scale=px.colors.diverging.Tealrose,
color_continuous_midpoint=2,
)
fig.show()
parallellines("target", df=train_data.sample(n=1000))
# 4.17 sunburst Chart
def sunburst(category1, category2, df=train_data):
fig = px.sunburst(df, path=[category1, category2], color="target")
fig.show()
sunburst("cat8", "cat9", df=train_data.sample(n=1000))
# 5. Data Preprocessing
# I am using StratifiedKFold for regression by dividing target into bins
#
folder_path = "../input/tabular-playground-series-feb-2021"
train_data = pd.read_csv(f"{folder_path}/train.csv")
test_data = pd.read_csv(f"{folder_path}/test.csv")
sample = pd.read_csv(f"{folder_path}/sample_submission.csv")
cont_features = [f"cont{i}" for i in range(14)]
cat_features = [f"cat{i}" for i in range(10)]
# cat_features = ['cat1','cat3','cat5','cat8','cat9']
all_features = cat_features + cont_features
target_feature = "target"
num_bins = int(1 + np.log2(len(train_data)))
train_data.loc[:, "bins"] = pd.cut(
train_data["target"].to_numpy(), bins=num_bins, labels=False
)
bins = train_data["bins"].to_numpy()
for feat in cat_features:
le = LabelEncoder()
train_data.loc[:, feat] = le.fit_transform(train_data[feat].fillna("-1"))
test_data.loc[:, feat] = le.transform(test_data[feat].fillna("-1"))
qt = QuantileTransformer()
train_data.loc[:, cont_features] = qt.fit_transform(train_data.loc[:, cont_features])
test_data.loc[:, cont_features] = qt.transform(test_data.loc[:, cont_features])
emb_c = {
cat: int(train_data[cat].nunique())
for cat in cat_features
if int(train_data[cat].nunique()) > 2
}
emb_cols = emb_c.keys()
embedding_sizes = [(c, min(50, (c + 1) // 2)) for _, c in emb_c.items()]
cont_features = cont_features + [cat for cat in cat_features if cat not in emb_cols]
cat_features = emb_cols
# target = train_data[target_feature].to_numpy()
# train_data = train_data[all_features].to_numpy()
# test_data = test_data[all_features].to_numpy()
train_data.shape, test_data.shape
# 6. Pytorch Model
def rmse_score(y_true, y_pred):
return np.sqrt(mean_squared_error(y_true, y_pred))
seed = 2021
def seed_everything(seed=42):
random.seed(seed)
os.environ["PYTHONHASHSEED"] = str(seed)
np.random.seed(seed)
torch.manual_seed(seed)
torch.cuda.manual_seed(seed)
torch.backends.cudnn.deterministic = True
seed_everything(seed=seed)
config = {
"epochs": 15,
"train_batch_size": 1024,
"valid_batch_size": 1024,
"test_batch_size": 1024,
"nfolds": 5,
"learning_rate": 0.001,
"input_size": len(all_features),
"cont_size": len(cont_features),
"hidden_sizes": [128, 64, 32, 16],
"output_size": 1,
}
class TPSDataset(Dataset):
def __init__(self, df, cat_features, cont_features):
self.cat_data = df.loc[:, cat_features].to_numpy()
self.cont_data = df.loc[:, cont_features].to_numpy()
self.target = df.loc[:, target_feature].to_numpy()
def __getitem__(self, idx):
input1 = torch.tensor(self.cat_data[idx], dtype=torch.long)
input2 = torch.tensor(self.cont_data[idx], dtype=torch.long)
target = torch.tensor(self.target[idx], dtype=torch.float)
return input1, input2, target
def __len__(self):
return len(self.target)
class Model(nn.Module):
def __init__(self, cont_size, output_size, hidden_sizes, embedding_sizes):
super().__init__()
self.embeddings = nn.ModuleList(
[nn.Embedding(cat, size) for (cat, size) in embedding_sizes]
)
self.emb_size = sum(e.embedding_dim for e in self.embeddings)
self.cont_size = cont_size
self.layer1 = self.batch_linear_drop(
self.emb_size + self.cont_size, hidden_sizes[0], 0.1, activation=nn.ReLU
)
self.layer2 = self.batch_linear_drop(
hidden_sizes[0], hidden_sizes[1], 0.1, activation=nn.ReLU
)
self.layer3 = self.batch_linear_drop(
hidden_sizes[1], hidden_sizes[2], 0.1, activation=nn.ReLU
)
self.layer4 = self.batch_linear_drop(
hidden_sizes[2], hidden_sizes[3], 0.1, activation=nn.ReLU
)
self.layer5 = self.batch_linear(hidden_sizes[3], output_size)
def batch_linear_drop(self, inp, out, drop, activation=None):
if activation:
return nn.Sequential(
nn.BatchNorm1d(inp), nn.Dropout(drop), nn.Linear(inp, out), activation()
)
else:
return nn.Sequential(
nn.BatchNorm1d(inp), nn.Dropout(drop), nn.Linear(inp, out)
)
def batch_linear(self, inp, out, activation=None):
if activation:
return nn.Sequential(nn.BatchNorm1d(inp), nn.Linear(inp, out), activation())
else:
return nn.Sequential(nn.BatchNorm1d(inp), nn.Linear(inp, out))
def forward(self, input1, input2):
x = [e(input1[:, i]) for i, e in enumerate(self.embeddings)]
x = torch.cat(x, 1)
x = torch.cat([x, input2], 1)
x = self.layer1(x)
x = self.layer2(x)
x = self.layer3(x)
x = self.layer4(x)
x = self.layer5(x)
return x
def run(plot_losses=True, verbose=True):
def loss_fn(outputs, targets):
loss = nn.MSELoss()(outputs, targets)
return loss
def train_loop(train_loader, model, loss_fn, device, optimizer, lr_scheduler=None):
model.train()
total_loss = 0
for i, (inputs1, inputs2, targets) in enumerate(train_loader):
inputs1 = inputs1.to(device)
inputs2 = inputs2.to(device)
targets = targets.to(device)
optimizer.zero_grad()
outputs = model(inputs1, inputs2)
loss = loss_fn(outputs, targets)
loss.backward()
total_loss += loss.item()
optimizer.step()
total_loss /= len(train_loader)
return total_loss
def valid_loop(valid_loader, model, loss_fn, device):
model.eval()
total_loss = 0
predictions = list()
for i, (inputs1, inputs2, targets) in enumerate(valid_loader):
inputs1 = inputs1.to(device)
inputs2 = inputs2.to(device)
targets = targets.to(device)
outputs = model(inputs1, inputs2)
loss = loss_fn(outputs, targets)
predictions.extend(outputs.detach().cpu().numpy())
total_loss += loss.item()
total_loss /= len(valid_loader)
return total_loss, np.array(predictions)
fold_train_losses = list()
fold_valid_losses = list()
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
print(f"{device} is used")
# kfold = KFold(n_splits=config['nfolds'])
kfold = StratifiedKFold(n_splits=config["nfolds"])
for k, (train_idx, valid_idx) in enumerate(kfold.split(X=train_data, y=bins)):
x_train, x_valid = train_data.iloc[train_idx, :], train_data.iloc[valid_idx, :]
input_size = config["cont_size"]
hiddens_sizes = config["hidden_sizes"]
output_size = 1
model = Model(input_size, output_size, hiddens_sizes, embedding_sizes)
model.to(device)
train_ds = TPSDataset(x_train, cat_features, cont_features)
train_dl = DataLoader(
train_ds,
batch_size=config["train_batch_size"],
shuffle=True,
num_workers=4,
pin_memory=True,
)
valid_ds = TPSDataset(x_valid, cat_features, cont_features)
valid_dl = DataLoader(
valid_ds,
batch_size=config["valid_batch_size"],
shuffle=False,
num_workers=4,
pin_memory=True,
)
optimizer = optim.Adam(model.parameters(), lr=config["learning_rate"])
lr_scheduler = optim.lr_scheduler.ReduceLROnPlateau(
optimizer, mode="min", factor=0.8, patience=2, verbose=True
)
# lr_scheduler = optim.lr_scheduler.StepLR(optimizer,step_size=10,gamma=0.8)
# lr_scheduler = None
print(f"Fold {k}")
best_loss = 999
train_losses = list()
valid_losses = list()
start = time.time()
for i in range(config["epochs"]):
train_loss = train_loop(
train_dl, model, loss_fn, device, optimizer, lr_scheduler=lr_scheduler
)
valid_loss, predictions = valid_loop(valid_dl, model, loss_fn, device)
if lr_scheduler:
lr_scheduler.step(valid_loss)
train_losses.append(train_loss)
valid_losses.append(valid_loss)
end = time.time()
epoch_time = end - start
start = end
score = rmse_score(x_valid[target_feature], predictions)
if verbose:
print(
f"epoch:{i} Training loss:{train_loss} | Validation loss:{valid_loss}| Score: {score} | epoch time {epoch_time:.2f}s "
)
if valid_loss <= best_loss:
if verbose:
print(
f"{g_}Validation loss Decreased from {best_loss} to {valid_loss}{sr_}"
)
best_loss = valid_loss
torch.save(model.state_dict(), f"model{k}.bin")
fold_train_losses.append(train_losses)
fold_valid_losses.append(valid_losses)
# break
if plot_losses == True:
plt.figure(figsize=(20, 14))
for i, (t, v) in enumerate(zip(fold_train_losses, fold_valid_losses)):
plt.subplot(2, 5, i + 1)
plt.title(f"Fold {i}")
plt.plot(t, label="train_loss")
plt.plot(v, label="valid_loss")
plt.legend()
plt.show()
run()
# Inference
class TPSDataset(Dataset):
def __init__(self, df, cat_features, cont_features):
self.cat_data = df.loc[:, cat_features].to_numpy()
self.cont_data = df.loc[:, cont_features].to_numpy()
def __getitem__(self, idx):
input1 = torch.tensor(self.cat_data[idx], dtype=torch.long)
input2 = torch.tensor(self.cont_data[idx], dtype=torch.long)
return input1, input2
def __len__(self):
return len(self.cat_data)
models = list()
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
input_size = config["cont_size"]
hiddens_sizes = config["hidden_sizes"]
output_size = 1
for i in range(config["nfolds"]):
model = Model(input_size, output_size, hiddens_sizes, embedding_sizes)
model.load_state_dict(torch.load(f"./model{i}.bin", map_location=device))
model.to(device)
model.eval()
models.append(model)
def inference(test):
all_prediction = np.zeros((test.shape[0], 1))
test_ds = TPSDataset(test, cat_features, cont_features)
test_dl = DataLoader(
test_ds,
batch_size=config["test_batch_size"],
shuffle=False,
num_workers=4,
pin_memory=True,
)
for model in models:
prediction = list()
for inputs1, inputs2 in test_dl:
inputs1 = inputs1.to(device)
inputs2 = inputs2.to(device)
outputs = model(inputs1, inputs2)
prediction.extend(outputs.detach().cpu().numpy())
all_prediction += prediction
return all_prediction / config["nfolds"]
predictions = inference(test_data)
sample.target = predictions
sample.to_csv("submission.csv", index=False)
sample.head()
plt.figure(figsize=(15, 7))
plt.subplot(131)
sns.distplot(sample.target)
plt.title("Distribution of test target")
plt.subplot(132)
sns.distplot(train_data[target_feature])
plt.title("Distribution of train target")
plt.subplot(133)
sns.distplot(sample.target, label="test")
sns.distplot(train_data[target_feature], label="train")
plt.legend()
plt.title("Distribution of train-test target")
|
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
sns.set_theme(style="whitegrid")
# Reading the csv
ds_salaries = pd.read_csv("/kaggle/input/data-science-job-salaries/ds_salaries.csv")
ds_salaries.head(10)
# Check data
ds_salaries.info()
# Check for missing values
ds_salaries.isnull().values.any()
# ## Analyzing data
# How many Data Analyst jobs were there in 2020 located in the US?
da_jobs_us_2020 = ds_salaries[ds_salaries["job_title"].str.contains("Data Analyst")]
da_jobs_us_2020 = da_jobs_us_2020.query(
"(work_year == 2020) & (company_location == 'US')"
)
da_jobs_us_2020
# The average salary of data analyst jobs in the US in 2020
average = da_jobs_us_2020["salary_in_usd"].mean()
print(average)
# How many Data Analyst jobs were there in 2021 around the world?
da_jobs_2021 = ds_salaries[ds_salaries["job_title"].str.contains("Data Analyst")]
da_jobs_2021 = da_jobs_2021.query("work_year == 2021")
da_jobs_2021["job_title"].count()
# The average salary of a data analyst in 2021
da_jobs_2021["salary_in_usd"].mean()
# Comparing companies
ds_salaries["company_size"].value_counts()
# company size for each country
company_size_pivot = pd.pivot_table(
ds_salaries,
index="company_location",
columns="company_size",
aggfunc={"company_size": ["count"]},
)
company_size_pivot
# Counting how many companies are large, small, medium
sns.catplot(data=ds_salaries, x="company_size", kind="count")
plt.title("Companies size")
plt.show()
# Salary payed per company size
sns.stripplot(
data=ds_salaries, x="company_size", y="salary_in_usd", order=["L", "M", "S"]
)
plt.title("Salary payed per company size")
plt.show()
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(15, 8), sharex="row")
non_us_companies = ds_salaries.query("company_location != 'US'")
us_companies = ds_salaries.query("company_location == 'US'")
count_of_companies_US = us_companies["company_size"].value_counts()
count_of_companies_non_US = non_us_companies["company_size"].value_counts()
sns.barplot(
non_us_companies,
x=count_of_companies_non_US.index,
y=count_of_companies_non_US.values,
ax=ax1,
)
ax1.set_title("Companies oversea")
ax1.set_xlabel("Company size")
sns.barplot(
us_companies, x=count_of_companies_US.index, y=count_of_companies_US.values, ax=ax2
)
ax2.set_title("Companies from US")
ax2.set_xlabel("Company size")
plt.show()
fig, (ax1, ax2) = plt.subplots(
1,
2,
figsize=(14, 6),
sharey=True,
)
sns.stripplot(
non_us_companies, x="company_size", y="salary_in_usd", ax=ax1, order=["L", "M", "S"]
)
ax1.set_title("Salary payed oversea")
sns.stripplot(
us_companies, x="company_size", y="salary_in_usd", ax=ax2, order=["L", "M", "S"]
)
ax2.set_title("Salary payed in US")
plt.show()
# comparing remote radio
print("Overseas")
print(non_us_companies.groupby(["work_year"])["remote_ratio"].value_counts())
print("United States")
print(us_companies.groupby(["work_year"])["remote_ratio"].value_counts())
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(15, 7))
remote_not_us = non_us_companies["remote_ratio"].value_counts()
remote_us = us_companies["remote_ratio"].value_counts()
sns.barplot(x=remote_us.index, y=remote_us.values, ax=ax1)
ax1.set_title("Remote jobs in US")
sns.barplot(x=remote_not_us.index, y=remote_not_us.values, ax=ax2)
ax2.set_title("Remote jobs outside US")
plt.show()
# experience level depending on the company size in the US
us_companies.groupby(["company_size"])["experience_level"].value_counts()
sns.catplot(
us_companies,
x="company_size",
kind="count",
hue="experience_level",
order=["L", "M", "S"],
)
plt.title("Experience level in US companies", fontsize=14, loc="left")
plt.show()
non_us_companies.groupby(["company_size"])["experience_level"].value_counts()
sns.catplot(
non_us_companies,
x="company_size",
kind="count",
hue="experience_level",
order=["L", "M", "S"],
)
plt.title("Experience level in companies overseas", fontsize=14, loc="left")
plt.show()
jobs_pivot = pd.pivot_table(
ds_salaries,
values="salary_in_usd",
index=["job_title"],
aggfunc={"salary_in_usd": ["mean"]},
)
jobs_pivot
jobs_pivot.plot.barh(
figsize=(8, 16), title="Average Salary of jobs", xlabel="Average Salary"
)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
kidney_train = pd.read_csv("../input/playground-series-s3e12/train.csv")
kidney_test = pd.read_csv("../input/playground-series-s3e12/test.csv")
print(kidney_train.info(), kidney_test.info())
kidney_train.head
kidney_train.columns
# from sklearn.model_selection import train_test_split
# X_train,X_test,y_train,y_test = train_test_split(X,y,random_state = True,test_size = 0.3)
X_train = kidney_train[["id", "gravity", "ph", "osmo", "cond", "urea", "calc"]]
X_test = kidney_test[["id", "gravity", "ph", "osmo", "cond", "urea", "calc"]]
y_train = kidney_train.target
from sklearn.ensemble import RandomForestClassifier
model = RandomForestClassifier(max_depth=53)
model.fit(X_train, y_train)
pred = model.predict(X_test)
print(pred)
kidney_test["target"] = pred
kidney_test.columns
kidney_test.drop(
["gravity", "ph", "osmo", "cond", "urea", "calc"], axis=1, inplace=True
)
kidney_test.to_csv("submission.csv", header=True, index=False)
# accuracy_score(y_test,pred)
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from scipy import stats
data = pd.read_csv("../input/diamonds/diamonds.csv")
print(data.info())
print("No Null Value")
data.head()
data.drop("Unnamed: 0", axis=1, inplace=True)
# **Description of data:**
# price price in US dollars (\$326--\$18,823)
# carat weight of the diamond (0.2--5.01)
# cut quality of the cut (Fair, Good, Very Good, Premium, Ideal)
# color diamond colour, from J (worst) to D (best)
# clarity a measurement of how clear the diamond is (I1 (worst), SI2, SI1, VS2, VS1, VVS2, VVS1, IF (best))
# x length in mm (0--10.74)
# y width in mm (0--58.9)
# z depth in mm (0--31.8)
# depth total depth percentage = z / mean(x, y) = 2 * z / (x + y) (43--79)
# table width of top of diamond relative to widest point (43--95)
data.describe()
# ## Let's Find Outlier:
# ### Option showfliers=Fasle does not show the outlier from boxplot
fig, [[ax1, ax2], [ax3, ax4]] = plt.subplots(nrows=2, ncols=2, figsize=(10, 14))
ax1.boxplot(data["x"])
ax1.set_title("x Outlier")
ax2.boxplot(data["y"])
ax2.set_title("y Outlier")
ax3.boxplot(data["z"])
ax3.set_title("z Outlier")
ax4.boxplot(data["price"])
ax4.set_title("price Outlier")
print("cut quality of the cut Fair, Good, Very Good, Premium, Ideal: ")
print(data[data["cut"] == "Fair"]["price"].mean())
print(data[data["cut"] == "Ideal"]["price"].mean())
print(data[data["cut"] == "Good"]["price"].mean())
print(data[data["cut"] == "Premium"]["price"].mean())
print(data[data["cut"] == "Very Good"]["price"].mean())
print("color diamond colour, from J (worst) to D (best):")
print(data[(data["cut"] == "Fair") & (data["color"] == "D")]["price"].mean())
print(data[(data["cut"] == "Ideal") & (data["color"] == "D")]["price"].mean())
print(data[(data["cut"] == "Good") & (data["color"] == "D")]["price"].mean())
print(data[(data["cut"] == "Very Good") & (data["color"] == "D")]["price"].mean())
print(data[(data["cut"] == "Premium") & (data["color"] == "D")]["price"].mean())
print("carat weight of the diamond (0.2--5.01):")
print(data[(data["cut"] == "Fair")]["carat"].mean())
print(data[(data["cut"] == "Ideal")]["carat"].mean())
print(data[(data["cut"] == "Good")]["carat"].mean())
print(data[(data["cut"] == "Very Good")]["carat"].mean())
print(data[(data["cut"] == "Premium")]["carat"].mean())
# ## **Lets Remove Outlier:**
# ### befor Remove let's talk About Mean And Trid_Mean
# ### Mean is sensetive to outlier although trid-mean delete 10 element from start and end data then calculate the mean because of outlier and more trustable than mean
print(stats.trim_mean(data["price"].sort_values(), 0.2))
print(data["price"].mean())
print("---------- see the different between trim_mean and mean--------------")
# ## Now Lets Define Function To Remove Outlier:
data.describe()
data["x"].plot(kind="hist", bins=100)
print(len(data[data["x"] < 4]))
print(len(data[data["x"] > 7.6]))
print("-------------------(remove x)----------------------")
def remove_x(x):
if x < 4:
return np.nan
elif x > 7.6:
return np.nan
else:
return x
data["x"] = data["x"].apply(remove_x)
data["y"].plot(kind="hist", bins=1000, figsize=(10, 10))
print(len(data[data["y"] < 4.7]))
print(len(data[data["y"] > 8]))
print("------------------(remove y)-----------------------")
def remove_x(x):
if x < 4.7:
return np.nan
elif x > 8:
return np.nan
else:
return x
data["y"] = data["y"].apply(remove_x)
data["z"].plot(kind="hist", bins=100, figsize=(10, 10))
print(len(data[data["z"] < 2]))
print(len(data[data["z"] > 5]))
print("------------------(remove z)-----------------------")
def remove_x(x):
if x < 2:
return np.nan
elif x > 5:
return np.nan
else:
return x
data["z"] = data["z"].apply(remove_x)
data["price"].plot(kind="hist", bins=100)
print(len(data[data["price"] < 1000]))
print(len(data[data["price"] > 10100]))
def remove_x(x):
if x < 1000:
return np.nan
elif x > 10100:
return np.nan
else:
return x
data["price"] = data["price"].apply(remove_x)
data["table"].plot(kind="hist", bins=100)
print(len(data[data["table"] < 56]))
print(len(data[data["table"] > 62]))
def remove_x(x):
if x < 56:
return np.nan
elif x > 62:
return np.nan
else:
return x
data["table"] = data["table"].apply(remove_x)
data["table"].plot(kind="box")
print(len(data[data["depth"] < 59]))
print(len(data[data["depth"] > 64]))
def remove_x(x):
if x < 59:
return np.nan
elif x > 64:
return np.nan
else:
return x
data["depth"] = data["depth"].apply(remove_x)
data["depth"].plot(kind="box")
data.dropna(inplace=True)
data["price"].plot(kind="box")
fig, [[ax1, ax2], [ax3, ax4]] = plt.subplots(nrows=2, ncols=2, figsize=(10, 14))
ax1.boxplot(data["x"])
ax1.set_title("x Outlier")
ax2.boxplot(data["y"])
ax2.set_title("y Outlier")
ax3.boxplot(data["z"])
ax3.set_title("z Outlier")
ax4.boxplot(data["price"])
ax4.set_title("price Outlier")
data.reset_index(drop=True, inplace=True)
y = data["price"]
x = data.drop("price", axis=1)
x_dummie = pd.get_dummies(x)
from sklearn.model_selection import train_test_split
from sklearn.feature_selection import SelectKBest, chi2
best_f = SelectKBest(chi2, k=24)
best_f.fit(x_dummie, y)
x_feat = best_f.transform(x_dummie)
print(best_f.get_support())
dfscores = pd.DataFrame(best_f.scores_)
dfcolumns = pd.DataFrame(x_dummie.columns)
featureScores = pd.concat([dfcolumns, dfscores], axis=1)
featureScores.columns = ["feature", "score"]
print(featureScores.nlargest(10, "score"))
print(x_feat.shape)
print(y.shape)
x_train, x_test, y_train, y_test = train_test_split(x_feat, y, test_size=0.3)
import xgboost as xgb
model = xgb.XGBRFRegressor()
model.fit(x_train, y_train)
print(model.score(x_train, y_train))
y_predict = model.predict(x_test)
from sklearn.metrics import r2_score
print(r2_score(y_predict, y_test))
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
train = pd.read_csv(
"/kaggle/input/competitive-data-science-predict-future-sales/sales_train.csv",
parse_dates=["date"],
infer_datetime_format=True,
dayfirst=True,
)
test = pd.read_csv(
"/kaggle/input/competitive-data-science-predict-future-sales/test.csv"
)
shops = pd.read_csv(
"/kaggle/input/competitive-data-science-predict-future-sales/shops.csv"
)
items = pd.read_csv(
"/kaggle/input/competitive-data-science-predict-future-sales/items.csv"
)
item_categories = pd.read_csv(
"/kaggle/input/competitive-data-science-predict-future-sales/item_categories.csv"
)
train = train.drop_duplicates()
train = train.loc[
train["item_id"] != 6066
] # Removing the outlier whose price is 307980.0
train = train.loc[train["item_price"] > 0] # Removing the outlier whose price is -1
# making a new column day
train["day"] = train["date"].dt.day
# making a new column month
train["month"] = train["date"].dt.month
# making a new column year
train["year"] = train["date"].dt.year
# making a new column week
train["week"] = train["date"].dt.week
# checking the new columns
train.columns
train["revenue"] = train["item_price"] * train["item_cnt_day"]
# converting the data into monthly sales data
# making a dataset with only monthly sales data
data = (
train.groupby(
[train["date"].apply(lambda x: x.strftime("%Y-%m")), "item_id", "shop_id"]
)
.sum()
.reset_index()
)
# specifying the important attributes which we want to add to the data
data = data[["date", "item_id", "shop_id", "item_cnt_day"]]
# at last we can select the specific attributes from the dataset which are important
data = data.pivot_table(
index=["item_id", "shop_id"], columns="date", values="item_cnt_day", fill_value=0
).reset_index()
# looking at the newly prepared datset
data.shape
# let's merge the monthly sales data prepared to the test data set
test = pd.merge(test, data, on=["item_id", "shop_id"], how="left")
# filling the empty values found in the dataset
test.fillna(0, inplace=True)
# checking the dataset
test.head()
train.head()
# now let's create the actual training data
X_train = test.drop(["2015-10", "item_id", "shop_id"], axis=1)
y_train = test["2015-10"]
# deleting the first column so that it can predict the future sales data
X_test = test.drop(["2013-01", "item_id", "shop_id"], axis=1)
from sklearn.model_selection import train_test_split
X_train, X_valid, y_train, y_valid = train_test_split(
X_train, y_train, test_size=0.2, random_state=0
)
model_scores = []
from sklearn.linear_model import LinearRegression
lr = LinearRegression()
lr.fit(X_train, y_train)
y_pred = lr.predict(X_test)
# Create the submission file and submit
preds = pd.DataFrame(y_pred, columns=["item_cnt_month"])
preds.to_csv("submission.csv", index_label="ID")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
from sklearn.tree import DecisionTreeClassifier
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
from sklearn import tree
from sklearn.metrics import roc_auc_score
# Plotting libraries
pd.plotting.register_matplotlib_converters()
import matplotlib.pyplot as plt
import seaborn as sns
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
pd.set_option("display.max_rows", 500)
pd.set_option("display.max_columns", 500)
pd.set_option("display.width", 1000)
# read in all of the data
train_data = pd.read_csv("/kaggle/input/assignment1cap/train.csv").drop(columns=["id"])
test_data = pd.read_csv("/kaggle/input/testdata/test.csv").drop(columns=["id"])
train_data.drop(train_data.tail(1).index, inplace=True)
train_data.describe(include="all")
# check if the data is all there.
test_data.describe(include="all")
# it looks like all of the data is there, 3410 across the board.
plt.subplots(figsize=(40, 30))
sns.heatmap(train_data.corr(), cmap="RdYlGn", linewidths=0.5)
# There's no real need to standardize or normalization the data because most of it is already in ratio's etc. There also aren't significant enough outliers to the extent that I think we can ignore them.
def H(dataset, target):
N = len(dataset)
levels = np.unique(dataset[target])
probs = [len(dataset.loc[dataset[target] == c]) / N for c in levels]
return entropy(probs)
def rem(dataset, feature, target):
N = len(dataset)
levels = np.unique(dataset[feature])
ans = 0
for c in levels:
dataset_c = dataset.loc[dataset[feature] == c]
ans += len(dataset_c) / N * H(dataset_c, target)
return ans
def information_gain(dataset, feature, target):
return H(dataset, target) - rem(dataset, feature, target)
def entropy(xs):
# This calculation is a truer representation of the general equation for calculating entropy; the sum operator becomes a for loop
return np.sum([-x * np.log2(x) for x in xs])
for x in features:
print(f"Feature: {x}")
print(f" H({target}, D): {H(train_data, target)}")
print(f" rem({x}, D): {rem(train_data, x, target)}")
print(f" IG({x}, D): {information_gain(train_data, x, target)}")
print()
ig_values = {x: information_gain(train_data, x, target) for x in features}
high_IG = {k: v for (k, v) in ig_values.items() if v > 0.20}
high_IG
# get a dictionary with only the highest IG values
max(ig_values, key=ig_values.get)
# Getting the node with the highest information gain to start building the decision tree.
high_ig_DF = train_data[high_IG]
high_ig_DF
features = train_data.columns[:-1]
target = train_data.columns[0]
X = train_data.loc[:, features]
Y = train_data.loc[:, target]
clf = tree.DecisionTreeClassifier()
clf = clf.fit(X, Y)
clf.predict(test_data)
roc_auc_score(Y, clf.predict_proba(X)[:, 1])
X_train, X_test, y_train, y_test = train_test_split(
X, Y, test_size=0.2, stratify=Y, random_state=1
)
dt = DecisionTreeClassifier(max_depth=2, random_state=1)
dt.fit(X_train, y_train)
y_pred = dt.predict(X_test)
accuracy_score(y_test, y_pred)
|
import random
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import seaborn as sns
import missingno
import matplotlib.pyplot as plt
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# ## Data preparation
# load all data available
train_data = pd.read_csv("/kaggle/input/spaceship-titanic/train.csv")
test_data = pd.read_csv("/kaggle/input/spaceship-titanic/test.csv")
sample_submission = pd.read_csv("/kaggle/input/spaceship-titanic/sample_submission.csv")
print(
"train_data",
train_data.shape,
"test_data",
test_data.shape,
"sample_submission",
sample_submission.shape,
)
# connect together train and test data to process all columns in a same way
data = pd.concat([train_data, test_data])
data
data.info()
missingno.matrix(data)
# function to fill NaN in series wth random non NaN value
def fill_with_random(series: pd.Series):
series2 = series.copy()
series2 = series2.apply(
lambda x: random.choice(series2.dropna().values) if x != x or x is None else x
) # x!=x is
return series2
# ---
# **Passenger ID**
# split passenger id to its group id and place in a group
data["group_id"] = data["PassengerId"].apply(lambda x: x.split("_")[0]).astype(int)
data["num_in_group"] = data["PassengerId"].apply(lambda x: x.split("_")[1]).astype(int)
print("Groups total:", data["group_id"].nunique())
print("Persons in group:")
sns.histplot(data["num_in_group"])
# ---
# **Home Planet**
# fill homw planet with random values in a same percentage, as existing data
print(data["HomePlanet"].value_counts(dropna=False), data["HomePlanet"].shape)
data["HomePlanet"] = fill_with_random(data["HomePlanet"])
sns.histplot(data["HomePlanet"])
# **CryoSleep**
# ---
# fill CryoSleep NaN with random data
print(data["CryoSleep"].value_counts(dropna=False))
data["CryoSleep"] = fill_with_random(data["CryoSleep"])
sns.histplot(data["CryoSleep"])
# ---
# **Cabin**
print(data["Cabin"].value_counts(dropna=False))
def split_cabin_code(x: str, n):
try:
split = x.split("/")
except:
return None
return split[n]
# extract specific featires from cabin description
data["cabin_deck"] = data["Cabin"].apply(lambda x: split_cabin_code(x, 0))
data["cabin_num"] = data["Cabin"].apply(lambda x: split_cabin_code(x, 1))
data["cabin_side"] = data["Cabin"].apply(lambda x: split_cabin_code(x, 2))
# fill NaN randomly
data["cabin_deck"] = fill_with_random(data["cabin_deck"])
data["cabin_num"] = fill_with_random(data["cabin_num"]).astype(int)
data["cabin_side"] = fill_with_random(data["cabin_side"])
fig, axs = plt.subplots(1, 3, figsize=(15, 5))
sns.histplot(data["cabin_deck"], ax=axs[0])
sns.histplot(data["cabin_num"], ax=axs[1])
sns.histplot(data["cabin_side"], ax=axs[2])
# ---
# **Destination**
# fill NaN randomly for destination point
print(data["Destination"].value_counts(dropna=False))
data["Destination"] = fill_with_random(data["Destination"])
sns.histplot(data["Destination"])
# ---
# **Age**
# fill age randomly
print(data["Age"].value_counts(dropna=False))
data["Age"] = fill_with_random(data["Age"])
sns.histplot(data["Age"])
# ---
# **VIP**
print(data["VIP"].value_counts(dropna=False))
# check percentage of VIP on different decks and cabin sides
deck_to_vip = data.pivot_table(index="cabin_deck", values="VIP", aggfunc="mean")
sns.heatmap(deck_to_vip)
plt.show()
# fill VIP status randomly
for deck in data["cabin_deck"].unique():
# fill subset of passengers in deck/side
data.loc[(data["cabin_deck"] == deck), "VIP"] = fill_with_random(
data.loc[(data["cabin_deck"] == deck)]["VIP"]
)
data["VIP"] = data["VIP"].astype(bool)
# ---
# **RoomService, FoodCourt, ShoppingMall, Spa, VRDeck**
# `RoomService`, `FoodCourt`, `ShoppingMall`, `Spa`, `VRDeck` - Amount the passenger has billed at each of the Spaceship Titanic's many luxury amenities.
fig, axs = plt.subplots(1, 3, figsize=(15, 5))
# check some correlations between bills and different possibly affecting factors
deck_to_bill = data.pivot_table(
index="cabin_deck",
values=["RoomService", "FoodCourt", "ShoppingMall", "Spa", "VRDeck"],
aggfunc="mean",
)
sns.heatmap(deck_to_bill, annot=True, fmt=".0f", ax=axs[0])
num_in_group_to_bill = data.pivot_table(
index="num_in_group",
values=["RoomService", "FoodCourt", "ShoppingMall", "Spa", "VRDeck"],
aggfunc="mean",
)
sns.heatmap(num_in_group_to_bill, annot=True, fmt=".0f", ax=axs[1])
age_to_bill = data.pivot_table(
index="Age",
values=["RoomService", "FoodCourt", "ShoppingMall", "Spa", "VRDeck"],
aggfunc="median",
)
sns.heatmap(age_to_bill, ax=axs[2])
plt.show()
# calculate total bill
data["total_bill"] = data[
["RoomService", "FoodCourt", "ShoppingMall", "Spa", "VRDeck"]
].apply(lambda x: x.sum(), axis=1)
sns.histplot(data["total_bill"])
# there is no specific bill trend visible, so fill all NaN with zeros
data = data.fillna(
{"RoomService": 0, "FoodCourt": 0, "ShoppingMall": 0, "Spa": 0, "VRDeck": 0}
)
# ---
# **Name**
def split_full_name(x: str, n):
try:
split = x.split(" ")
except:
return None
return split[n]
# extract name and family name
data["first_name"] = data["Name"].apply(lambda x: split_full_name(x, 0))
data["last_name"] = data["Name"].apply(lambda x: split_full_name(x, 1))
# fill gaps with
data["first_name"] = fill_with_random(data["first_name"])
data["last_name"] = fill_with_random(data["last_name"])
# **Target**
data["Transported"].value_counts()
# **Finalize**
data.set_index("PassengerId", inplace=True)
data.info()
# ## Feature engineering
# **OHE**
# encode home planet
data = data.join(pd.get_dummies(data["HomePlanet"], prefix="home", drop_first=True))
data.drop(columns="HomePlanet", inplace=True)
# encode destination
data = data.join(
pd.get_dummies(data["Destination"], prefix="destination", drop_first=True)
)
data.drop(columns="Destination", inplace=True)
# encode cabin size
data["cabin_side"] = data["cabin_side"].map({"S": 1, "P": 0})
# ---
# **Add custom features**
data
# get deck cabin amount
deck_cabins = data.groupby("cabin_deck")["cabin_num"].max()
deck_cabins.name = "deck_cabins"
ids = data.index
data = data.merge(deck_cabins.reset_index(), how="left", on="cabin_deck")
data.index = ids
data["cabin_location"] = data["cabin_num"] / data["deck_cabins"]
data
group_sizes = data["group_id"].value_counts().reset_index()
group_sizes.columns = ["group_id", "group_size"]
data = (
data.reset_index()
.merge(group_sizes, how="left", on="group_id")
.set_index("PassengerId")
)
sns.histplot(data["group_size"])
# rename T deck to A deck
data["cabin_deck"] = (
data["cabin_deck"]
.map({"T": 0, "A": 1, "B": 2, "C": 3, "D": 4, "E": 5, "F": 6, "G": 7})
.astype(int)
)
# OHE decks
# data['cabin_deck'] = data['cabin_deck'].map({'T': 'A'})
# data = data.join(pd.get_dummies(data['cabin_deck'], prefix='cabin_deck', drop_first=True))
# data.drop(columns='cabin_deck', inplace=True)
# group aggregated features
group_aggregates = data.groupby("group_id")[["Age", "total_bill"]].agg(
{
"Age": ["median", "max", "min"],
"total_bill": ["sum", "mean", "median", "max", "min"],
}
)
group_aggregates.columns = (
"group_"
+ group_aggregates.columns.get_level_values(0)
+ "_"
+ group_aggregates.columns.get_level_values(1)
)
# data = data.reset_index().merge(group_aggregates, how='left', on='group_id').set_index('PassengerId')
# sex by name
# here we train
from nltk.corpus import names
import nltk
def gender_features(word):
return {"last_letter": word[-1]}
# preparing a list of examples and corresponding class labels.
labeled_names = [(name, "male") for name in names.words("male.txt")] + [
(name, "female") for name in names.words("female.txt")
]
random.shuffle(labeled_names)
# we use the feature extractor to process the names data.
featuresets = [(gender_features(n), gender) for (n, gender) in labeled_names]
# Divide the resulting list of feature
# sets into a training set and a test set.
train_set, test_set = featuresets[500:], featuresets[:500]
# The training set is used to
# train a new "naive Bayes" classifier.
classifier = nltk.NaiveBayesClassifier.train(featuresets)
def classify_name(x):
return classifier.prob_classify(gender_features(x)).prob("male")
# define gender
data["is_male"] = data["first_name"].apply(classify_name)
# ---
# **Drop columns**
data.drop(columns="Cabin", inplace=True)
data.drop(columns="Name", inplace=True)
data.drop(columns="last_name", inplace=True)
data.drop(columns="first_name", inplace=True)
data.drop(columns="is_male", inplace=True)
data.drop(columns="cabin_num", inplace=True)
data.drop(columns="deck_cabins", inplace=True)
# data.drop(columns='VIP', inplace=True)
# data.drop(columns='home_Europa', inplace=True)
# data.drop(columns='home_Mars', inplace=True)
# data.drop(columns='destination_PSO J318.5-22', inplace=True)
# data.drop(columns='destination_TRAPPIST-1e', inplace=True)
data
# ## Feature scaling and prepare train data
from sklearn.preprocessing import StandardScaler
# scale data
scaler = StandardScaler()
scaled_data = pd.DataFrame(
scaler.fit_transform(data.drop(columns="Transported")),
index=data.index,
columns=data.drop(columns="Transported").columns,
)
scaled_data["Transported"] = data["Transported"]
# prepare data for train, validation and submission
x = data.drop(columns="Transported") # scaled_data
y = data["Transported"] # scaled_data
# train data
train_x = x[~y.isna()]
train_y = y[~y.isna()].astype(int)
# submission data
test_x = x[y.isna()]
print("train_x", train_x.shape, ", train_y", train_y.shape, ", test_x", test_x.shape)
# ## Feature selection
from sklearn.feature_selection import RFECV
from sklearn.model_selection import StratifiedKFold
from sklearn.linear_model import LogisticRegression
min_features_to_select = 1 # Minimum number of features to consider
clf = LogisticRegression(solver="liblinear")
cv = StratifiedKFold(5)
# recursive feature elimination
rfecv = RFECV(
estimator=clf,
step=1,
cv=cv,
scoring="accuracy",
min_features_to_select=min_features_to_select,
n_jobs=2,
)
rfecv.fit(train_x, train_y)
print(f"Total number of features: {train_x.shape[1]}")
print(f"Optimal number of features: {rfecv.n_features_}")
# cut train and submission data to optimal number of features
features = rfecv.get_feature_names_out()
print(features)
"""train_x = train_x[features]
test_x = test_x[features]"""
# ## Feature importance
from sklearn.ensemble import RandomForestClassifier
model = RandomForestClassifier()
model.fit(train_x, train_y)
importances = pd.Series(model.feature_importances_, train_x.columns)
fig, ax = plt.subplots(figsize=(15, 5))
importances.plot.bar(ax=ax)
ax.set_title("Feature importances")
ax.set_ylabel("Importance")
fig.tight_layout()
# ## Train model
from sklearn.model_selection import cross_val_score
from sklearn.linear_model import LogisticRegression
model = LogisticRegression(solver="liblinear")
print(
"Cross val score:",
cross_val_score(model, train_x, train_y, cv=5, scoring="accuracy").mean(),
)
model.fit(train_x, train_y)
# - baseline Cross val score: 0.78454 => Public 0.79424
# ## Submission
preds = model.predict(test_x)
preds = pd.DataFrame(preds, index=test_x.index).astype(bool)
preds.columns = ["Transported"]
preds
submission = sample_submission.drop(columns="Transported").merge(
preds.reset_index(), how="left", on="PassengerId"
)
submission.to_csv("submission.csv", index=False)
|
# Data Preprocessing
#
import os
import cv2
import numpy as np
import pandas as pd
from keras.utils import np_utils
from keras.datasets import mnist
from sklearn.utils import shuffle
from matplotlib import pyplot as plt
from sklearn.model_selection import train_test_split
data_root = (
"/kaggle/input/az-handwritten-alphabets-in-csv-format/A_Z Handwritten Data.csv"
)
dataset = pd.read_csv(data_root).astype("float32")
dataset.rename(columns={"0": "label"}, inplace=True)
letter_x = dataset.drop("label", axis=1)
letter_y = dataset["label"]
(digit_train_x, digit_train_y), (digit_test_x, digit_test_y) = mnist.load_data()
letter_x = letter_x.values
print(letter_x.shape, letter_y.shape)
print(digit_train_x.shape, digit_train_y.shape)
print(digit_test_x.shape, digit_test_y.shape)
digit_data = np.concatenate((digit_train_x, digit_test_x))
digit_target = np.concatenate((digit_train_y, digit_test_y))
print(digit_data.shape, digit_target.shape)
digit_target += 26
data = []
for flatten in letter_x:
image = np.reshape(flatten, (28, 28, 1))
data.append(image)
letter_data = np.array(data, dtype=np.float32)
letter_target = letter_y
digit_data = np.reshape(
digit_data, (digit_data.shape[0], digit_data.shape[1], digit_data.shape[2], 1)
)
print(letter_data.shape, letter_target.shape)
print(digit_data.shape, digit_target.shape)
shuffled_data = shuffle(letter_data)
rows, cols = 10, 10
plt.figure(figsize=(20, 20))
for i in range(rows * cols):
plt.subplot(cols, rows, i + 1)
plt.imshow(shuffled_data[i].reshape(28, 28), interpolation="nearest", cmap="gray")
plt.show()
shuffled_data = shuffle(digit_data)
rows, cols = 10, 10
plt.figure(figsize=(20, 20))
for i in range(rows * cols):
plt.subplot(cols, rows, i + 1)
plt.imshow(shuffled_data[i].reshape(28, 28), interpolation="nearest", cmap="gray")
plt.show()
data = np.concatenate((digit_data, letter_data))
target = np.concatenate((digit_target, letter_target))
print(data.shape, target.shape)
shuffled_data = shuffle(data)
rows, cols = 10, 10
plt.figure(figsize=(20, 20))
for i in range(rows * cols):
plt.subplot(cols, rows, i + 1)
plt.imshow(shuffled_data[i].reshape(28, 28), interpolation="nearest", cmap="gray")
plt.show()
train_data, test_data, train_labels, test_labels = train_test_split(
data, target, test_size=0.2
)
print(train_data.shape, train_labels.shape)
print(test_data.shape, test_labels.shape)
train_data = train_data / 255.0
test_data = test_data / 255.0
train_labels = np_utils.to_categorical(train_labels)
test_labels = np_utils.to_categorical(test_labels)
train_data = np.reshape(
train_data, (train_data.shape[0], train_data.shape[1], train_data.shape[2], 1)
)
test_data = np.reshape(
test_data, (test_data.shape[0], test_data.shape[1], test_data.shape[2], 1)
)
print(train_data.shape, test_data.shape)
print(train_labels.shape, test_labels.shape)
train_label_counts = [0 for i in range(36)]
test_label_counts = [0 for i in range(36)]
for i in range(train_data.shape[0]):
train_label_counts[np.argmax(train_labels[i])] += 1
for i in range(test_data.shape[0]):
test_label_counts[np.argmax(test_labels[i])] += 1
frequency = [train_label_counts, test_label_counts]
fig = plt.figure(figsize=(8, 6))
ax = fig.add_axes([0, 0, 1, 1])
x = [
"A",
"B",
"C",
"D",
"E",
"F",
"G",
"H",
"I",
"J",
"K",
"L",
"M",
"N",
"O",
"P",
"Q",
"R",
"S",
"T",
"U",
"V",
"W",
"X",
"Y",
"Z",
"0",
"1",
"2",
"3",
"4",
"5",
"6",
"7",
"8",
"9",
]
plt.xticks(range(len(frequency[0])), x)
plt.title("train vs. test data distribution")
plt.xlabel("character")
plt.ylabel("frequency")
ax.bar(np.arange(len(frequency[0])), frequency[0], color="b", width=0.35)
ax.bar(np.arange(len(frequency[1])) + 0.35, frequency[1], color="r", width=0.35)
ax.legend(labels=["train", "test"])
np.save("train_data.npy", train_data)
np.save("train_labels.npy", train_labels)
np.save("test_data.npy", test_data)
np.save("test_labels.npy", test_labels)
# CNN Architecture
#
import numpy as np
import visualkeras as vk # pip install visualkeras
import pandas as pd
import seaborn as sn
from keras.models import Sequential
from matplotlib import pyplot as plt
from keras.callbacks import ModelCheckpoint
from sklearn.metrics import confusion_matrix
from keras.preprocessing.image import ImageDataGenerator
from keras.layers import (
Conv2D,
Activation,
MaxPooling2D,
Flatten,
Dense,
Dropout,
BatchNormalization,
)
train_data = np.load("train_data.npy")
test_data = np.load("test_data.npy")
train_labels = np.load("train_labels.npy")
test_labels = np.load("test_labels.npy")
model = Sequential()
model.add(Conv2D(32, (5, 5), input_shape=(28, 28, 1), activation="relu"))
model.add(BatchNormalization())
model.add(Conv2D(32, (5, 5), activation="relu"))
model.add(BatchNormalization())
model.add(MaxPooling2D(2, 2))
model.add(Dropout(0.25))
model.add(BatchNormalization())
model.add(Flatten())
model.add(Dense(256, activation="relu"))
model.add(Dense(36, activation="softmax"))
model.compile(loss="categorical_crossentropy", optimizer="adam", metrics=["accuracy"])
model.summary()
vk.layered_view(model)
best_loss_checkpoint = ModelCheckpoint(
filepath="/kaggle/input/models/models/best_loss_model",
monitor="loss",
save_best_only=True,
save_weights_only=True,
mode="min",
)
best_val_loss_checkpoint = ModelCheckpoint(
filepath="/kaggle/input/models/models/best_val_loss_model",
monitor="val_loss",
save_best_only=True,
save_weights_only=True,
mode="min",
)
print(train_data.shape, test_data.shape)
print(train_labels.shape, test_labels.shape)
history = model.fit(
train_data,
train_labels,
validation_data=(test_data, test_labels),
epochs=10,
batch_size=200,
callbacks=[best_loss_checkpoint, best_val_loss_checkpoint],
)
plt.plot(history.history["loss"], "b", label="loss")
plt.plot(history.history["val_loss"], "r", label="val_loss")
plt.xlabel("epoch")
plt.ylabel("frequency")
plt.legend()
plt.show()
plt.plot(history.history["accuracy"], "b", label="accuracy")
plt.plot(history.history["val_accuracy"], "r", label="val_accuracy")
plt.xlabel("epoch")
plt.ylabel("frequency")
plt.legend()
plt.show()
model.load_weights("/kaggle/input/models/models/best_val_loss_model.h5")
loss, acc = model.evaluate(test_data, test_labels)
print(loss, acc)
predictions = model.predict(test_data)
confusion = confusion_matrix(
np.argmax(test_labels, axis=1), np.argmax(predictions, axis=1)
)
print(confusion)
labels = [
"A",
"B",
"C",
"D",
"E",
"F",
"G",
"H",
"I",
"J",
"K",
"L",
"M",
"N",
"O",
"P",
"Q",
"R",
"S",
"T",
"U",
"V",
"W",
"X",
"Y",
"Z",
"0",
"1",
"2",
"3",
"4",
"5",
"6",
"7",
"8",
"9",
]
df_cm = pd.DataFrame(confusion, columns=np.unique(labels), index=np.unique(labels))
df_cm.index.name = "actual"
df_cm.columns.name = "predicted"
plt.figure(figsize=(20, 15))
sn.set(font_scale=1.4)
sn.heatmap(df_cm, cmap="Blues", annot=True, annot_kws={"size": 15}, fmt="d")
# Application
import cv2
import numpy as np
from keras.models import Sequential
from keras.layers import (
Conv2D,
Activation,
MaxPooling2D,
Flatten,
Dense,
Dropout,
BatchNormalization,
)
def clear_whiteboard(display):
wb_x1, wb_x2, wb_y1, wb_y2 = (
whiteboard_region["x"][0],
whiteboard_region["x"][1],
whiteboard_region["y"][0],
whiteboard_region["y"][1],
)
display[wb_y1 - 10 : wb_y2 + 12, wb_x1 - 10 : wb_x2 + 12] = (255, 255, 255)
def setup_display():
title = np.zeros((80, 950, 3), dtype=np.uint8)
board = np.zeros((600, 650, 3), dtype=np.uint8)
panel = np.zeros((600, 300, 3), dtype=np.uint8)
board[5:590, 8:645] = (255, 255, 255)
board = cv2.rectangle(board, (8, 5), (645, 590), (255, 0, 0), 3)
panel = cv2.rectangle(panel, (1, 4), (290, 590), (0, 255, 192), 2)
panel = cv2.rectangle(panel, (22, 340), (268, 560), (255, 255, 255), 1)
panel = cv2.rectangle(panel, (22, 65), (268, 280), (255, 255, 255), 1)
cv2.line(panel, (145, 340), (145, 560), (255, 255, 255), 1)
cv2.line(panel, (22, 380), (268, 380), (255, 255, 255), 1)
cv2.putText(
title,
" " + window_name,
(10, 50),
cv2.FONT_HERSHEY_SIMPLEX,
1.2,
(255, 255, 255),
2,
)
cv2.putText(
panel, "Action: ", (23, 40), cv2.FONT_HERSHEY_SIMPLEX, 0.6, (255, 255, 255), 1
)
cv2.putText(
panel,
"Best Predictions",
(52, 320),
cv2.FONT_HERSHEY_SIMPLEX,
0.7,
(255, 255, 255),
1,
)
cv2.putText(
panel,
"Prediction",
(42, 362),
cv2.FONT_HERSHEY_SIMPLEX,
0.5,
(255, 255, 255),
1,
)
cv2.putText(
panel, "Accuracy", (168, 362), cv2.FONT_HERSHEY_SIMPLEX, 0.5, (255, 255, 255), 1
)
cv2.putText(
panel,
actions[0],
(95, 40),
cv2.FONT_HERSHEY_SIMPLEX,
0.6,
action_colors[actions[0]],
1,
)
display = np.concatenate((board, panel), axis=1)
display = np.concatenate((title, display), axis=0)
return display
def setup_panel(display):
action_region_pt1, action_region_pt2 = status_regions["action"]
preview_region_pt1, preview_region_pt2 = status_regions["preview"]
label_region_pt1, label_region_pt2 = status_regions["labels"]
acc_region_pt1, acc_region_pt2 = status_regions["accs"]
display[
action_region_pt1[1] : action_region_pt2[1],
action_region_pt1[0] : action_region_pt2[0],
] = (0, 0, 0)
display[
preview_region_pt1[1] : preview_region_pt2[1],
preview_region_pt1[0] : preview_region_pt2[0],
] = (0, 0, 0)
display[
label_region_pt1[1] : label_region_pt2[1],
label_region_pt1[0] : label_region_pt2[0],
] = (0, 0, 0)
display[
acc_region_pt1[1] : acc_region_pt2[1], acc_region_pt1[0] : acc_region_pt2[0]
] = (0, 0, 0)
if crop_preview is not None:
display[
preview_region_pt1[1] : preview_region_pt2[1],
preview_region_pt1[0] : preview_region_pt2[0],
] = cv2.resize(crop_preview, (crop_preview_h, crop_preview_w))
if best_predictions:
labels = list(best_predictions.keys())
accs = list(best_predictions.values())
prediction_status_cordinate = [
((725, 505), (830, 505), (0, 0, 255)),
((725, 562), (830, 562), (0, 255, 0)),
((725, 619), (830, 619), (255, 0, 0)),
]
for i in range(len(labels)):
label_cordinate, acc_cordinate, color = prediction_status_cordinate[i]
cv2.putText(
display,
labels[i],
label_cordinate,
cv2.FONT_HERSHEY_SIMPLEX,
0.8,
color,
2,
)
cv2.putText(
display,
str(accs[i]),
acc_cordinate,
cv2.FONT_HERSHEY_SIMPLEX,
0.8,
color,
2,
)
for i in range(len(labels), 3):
label_cordinate, acc_cordinate, color = prediction_status_cordinate[i]
cv2.putText(
display, "_", label_cordinate, cv2.FONT_HERSHEY_SIMPLEX, 0.8, color, 2
)
cv2.putText(
display, "_", acc_cordinate, cv2.FONT_HERSHEY_SIMPLEX, 0.8, color, 2
)
cv2.putText(
display,
current_action,
(745, 120),
cv2.FONT_HERSHEY_SIMPLEX,
0.6,
action_colors[current_action],
1,
)
def arrange_crop_rectangle_cordinates(cor1, cor2):
if cor1 is None or cor2 is None:
return
result = ()
if cor1[1] < cor2[1]:
if cor1[0] > cor2[0]:
result = ((cor2[0], cor1[1]), (cor1[0], cor2[1]))
else:
result = (cor1, cor2)
else:
if cor2[0] > cor1[0]:
result = ((cor1[0], cor2[1]), (cor2[0], cor1[1]))
else:
result = (cor2, cor1)
return result
def mouse_click_event(event, x, y, flags, params):
if current_action is actions[1]:
whiteboard_draw(event, x, y)
elif current_action is actions[2]:
character_crop(event, x, y)
def whiteboard_draw(event, x, y):
global left_button_down, right_button_down
wb_x1, wb_x2, wb_y1, wb_y2 = (
whiteboard_region["x"][0],
whiteboard_region["x"][1],
whiteboard_region["y"][0],
whiteboard_region["y"][1],
)
if event is cv2.EVENT_LBUTTONUP:
left_button_down = False
elif event is cv2.EVENT_RBUTTONUP:
right_button_down = False
elif wb_x1 <= x <= wb_x2 and wb_y1 <= y <= wb_y2:
color = (0, 0, 0)
if event in [
cv2.EVENT_LBUTTONDOWN,
cv2.EVENT_LBUTTONUP,
cv2.EVENT_RBUTTONDOWN,
cv2.EVENT_RBUTTONUP,
cv2.EVENT_MOUSEMOVE,
]:
if event is cv2.EVENT_LBUTTONDOWN:
color = (0, 0, 0)
left_button_down = True
elif left_button_down and event is cv2.EVENT_MOUSEMOVE:
color = (0, 0, 0)
elif event is cv2.EVENT_RBUTTONDOWN:
color = (255, 255, 255)
right_button_down = True
elif right_button_down and event is cv2.EVENT_MOUSEMOVE:
color = (255, 255, 255)
else:
return
cv2.circle(display, (x, y), 10, color, -1)
cv2.imshow(window_name, display)
def character_crop(event, x, y):
global bound_rect_cordinates, lbd_cordinate, lbu_cordinate, crop_preview, display, best_predictions
wb_x1, wb_x2, wb_y1, wb_y2 = (
whiteboard_region["x"][0],
whiteboard_region["x"][1],
whiteboard_region["y"][0],
whiteboard_region["y"][1],
)
if wb_x1 <= x <= wb_x2 and wb_y1 <= y <= wb_y2:
if event is cv2.EVENT_LBUTTONDOWN:
lbd_cordinate = (x, y)
elif event is cv2.EVENT_LBUTTONUP:
lbu_cordinate = (x, y)
if lbd_cordinate is not None and lbu_cordinate is not None:
bound_rect_cordinates = arrange_crop_rectangle_cordinates(
lbd_cordinate, lbu_cordinate
)
elif lbd_cordinate is not None:
if event is cv2.EVENT_MOUSEMOVE:
mouse_move_cordinate = (x, y)
mouse_move_rect_cordinates = arrange_crop_rectangle_cordinates(
lbd_cordinate, mouse_move_cordinate
)
top_cordinate, bottom_cordinate = (
mouse_move_rect_cordinates[0],
mouse_move_rect_cordinates[1],
)
display_copy = display.copy()
cropped_region = display_copy[
top_cordinate[1] : bottom_cordinate[1],
top_cordinate[0] : bottom_cordinate[0],
]
filled_rect = np.zeros((cropped_region.shape[:]))
filled_rect[:, :, :] = (0, 255, 0)
filled_rect = filled_rect.astype(np.uint8)
cropped_rect = cv2.addWeighted(
cropped_region, 0.3, filled_rect, 0.5, 1.0
)
if cropped_rect is not None:
display_copy[
top_cordinate[1] : bottom_cordinate[1],
top_cordinate[0] : bottom_cordinate[0],
] = cropped_rect
cv2.imwrite("captured/filled.jpg", display_copy)
cv2.imshow(window_name, display_copy)
if bound_rect_cordinates is not None:
top_cordinate, bottom_cordinate = (
bound_rect_cordinates[0],
bound_rect_cordinates[1],
)
crop_preview = display[
top_cordinate[1] : bottom_cordinate[1],
top_cordinate[0] : bottom_cordinate[0],
].copy()
crop_preview = np.invert(crop_preview)
best_predictions = predict(model, crop_preview)
display_copy = display.copy()
bound_rect_cordinates = lbd_cordinate = lbu_cordinate = None
setup_panel(display)
cv2.imshow(window_name, display)
elif event is cv2.EVENT_LBUTTONUP:
lbd_cordinate = lbu_cordinate = None
cv2.imshow(window_name, display)
def load_model(path):
model = Sequential()
model.add(Conv2D(32, (5, 5), input_shape=(28, 28, 1), activation="relu"))
model.add(BatchNormalization())
model.add(Conv2D(32, (5, 5), activation="relu"))
model.add(BatchNormalization())
model.add(MaxPooling2D(2, 2))
model.add(Dropout(0.25))
model.add(BatchNormalization())
model.add(Flatten())
model.add(Dense(256, activation="relu"))
model.add(Dense(36, activation="softmax"))
model.compile(
loss="categorical_crossentropy", optimizer="adam", metrics=["accuracy"]
)
model.load_weights(path)
return model
def predict(model, image):
labels = [
"A",
"B",
"C",
"D",
"E",
"F",
"G",
"H",
"I",
"J",
"K",
"L",
"M",
"N",
"O",
"P",
"Q",
"R",
"S",
"T",
"U",
"V",
"W",
"X",
"Y",
"Z",
"0",
"1",
"2",
"3",
"4",
"5",
"6",
"7",
"8",
"9",
]
image = cv2.cvtColor(image, cv2.COLOR_BGR2GRAY)
image = cv2.resize(image, (28, 28))
image = image / 255.0
image = np.reshape(image, (1, image.shape[0], image.shape[1], 1))
prediction = model.predict(image)
best_predictions = dict()
for i in range(3):
max_i = np.argmax(prediction[0])
acc = round(prediction[0][max_i], 1)
if acc > 0:
label = labels[max_i]
best_predictions[label] = acc
prediction[0][max_i] = 0
else:
break
return best_predictions
left_button_down = False
right_button_down = False
bound_rect_cordinates = lbd_cordinate = lbu_cordinate = None
whiteboard_region = {"x": (20, 632), "y": (98, 656)}
window_name = "Live Cropped Character Recognition"
best_predictions = dict()
crop_preview_h, crop_preview_w = 238, 206
crop_preview = None
actions = ["N/A", "DRAW", "CROP"]
action_colors = {
actions[0]: (0, 0, 255),
actions[1]: (0, 255, 0),
actions[2]: (0, 255, 192),
}
current_action = actions[0]
status_regions = {
"action": ((736, 97), (828, 131)),
"preview": ((676, 150), (914, 356)),
"labels": ((678, 468), (790, 632)),
"accs": ((801, 468), (913, 632)),
}
model = load_model("/kaggle/input/models/models/best_val_loss_model.h5")
display = setup_display()
cv2.imshow(window_name, display)
cv2.setMouseCallback(window_name, mouse_click_event)
pre_action = None
while True:
k = cv2.waitKey(1)
if k == ord("d") or k == ord("c"):
if k == ord("d"):
current_action = actions[1]
elif k == ord("c"):
current_action = actions[2]
if pre_action is not current_action:
setup_panel(display)
cv2.imshow(window_name, display)
pre_action = current_action
elif k == ord("e"):
clear_whiteboard(display)
cv2.imshow(window_name, display)
elif k == 27:
break
cv2.destroyAllWindows()
|
from pathlib import Path
import json
import numpy as np
import pandas as pd
import torch
import torch.nn as nn
from torch.utils.data import Dataset, DataLoader
import torchinfo
from tqdm import tqdm
import onnx
import onnxruntime
import onnx_tf
import tensorflow as tf
import tflite_runtime.interpreter as tflite
INPUT_DIR = Path("/kaggle/input/")
ASL_DIR = INPUT_DIR / "asl-signs"
DATASET_DIR = INPUT_DIR / "asl-dataset"
device = (
"cuda"
if torch.cuda.is_available()
else "mps"
if torch.backends.mps.is_available()
else "cpu"
)
data = np.load(INPUT_DIR / "asl-dataset" / "data.npz")
tensor = torch.tensor(data[data.files[0]])
tensor
with (INPUT_DIR / "asl-dataset" / "landmarks.json").open() as f:
landmarks = json.load(f)
points = torch.cat(
[torch.tensor(value).unfold(0, 3, 1) for value in landmarks.values()]
)
view = tensor[:, points]
view
vectors = torch.stack(
(view[:, :, 1] - view[:, :, 0], view[:, :, 2] - view[:, :, 1]), dim=2
).float()
angles = torch.div(
vectors.prod(dim=2).sum(dim=2), vectors.square().sum(dim=3).sqrt().prod(dim=2)
).acos()
torch.cat((tensor.flatten(1), angles), 1).size()
signs_df = pd.read_csv(DATASET_DIR / "train.csv")
signs_df
train_df = signs_df.sample(frac=0.8)
test_df = signs_df.drop(train_df.index).sample(frac=1)
train_df.shape, test_df.shape
class ASLDataset(Dataset):
def __init__(self, dataset_df, agg, preload=None):
files = np.load(DATASET_DIR / "data.npz")
if len(preload or []) != len(dataset_df):
preload = None
self.items = preload or [
torch.Tensor(files[str(i)]).to(device)
for i in tqdm(
dataset_df.sequence_id, desc="Loading data", total=len(dataset_df)
)
]
self.labels = torch.Tensor(dataset_df.label.values).long().to(device)
self.agg = agg
def __len__(self):
return len(self.items)
def __getitem__(self, index):
return self.agg(self.items[index]).float(), self.labels[index]
ROWS_PER_FRAME = 543 # number of landmarks per frame
def load_relevant_data_subset(pq_path):
data_columns = ["x", "y", "z"]
data = pd.read_parquet(pq_path, columns=data_columns)
n_frames = int(len(data) / ROWS_PER_FRAME)
data = data.values.reshape(n_frames, ROWS_PER_FRAME, len(data_columns))
return data.astype(np.float32)
POINTS = torch.cat(
[torch.tensor(value).unfold(0, 3, 1) for value in landmarks.values()]
)
INDICES = np.load(DATASET_DIR / "indices.npy")
class Preprocess(nn.Module):
def __init__(self, agg):
super().__init__()
self.agg = agg
def forward(self, x):
x = x[:, INDICES]
x = self.agg(x)
return x
class MLPModel(nn.Module):
def __init__(self):
super().__init__()
self.sequential = nn.Sequential(
nn.Flatten(),
nn.Linear(534 * 2, 1536),
nn.BatchNorm1d(1536),
nn.Dropout(0.3),
nn.ReLU(),
nn.Linear(1536, 2048),
nn.BatchNorm1d(2048),
nn.Dropout(0.2),
nn.ReLU(),
nn.Linear(2048, 1024),
nn.BatchNorm1d(1024),
nn.Dropout(0.2),
nn.ReLU(),
nn.Linear(1024, 250),
)
def forward(self, x):
# x = self.agg(x)
x = self.sequential(x)
return x
@staticmethod
def agg(x):
# Find "angles"
view = x[:, POINTS]
vectors = torch.stack(
(view[..., 1, :] - view[..., 0, :], view[..., 2, :] - view[..., 1, :]),
dim=-2,
).float()
angles = torch.div(
vectors.prod(dim=-2).sum(dim=-1),
vectors.square().sum(dim=-1).sqrt().prod(dim=-1),
) # .acos()
# Coordinate normalisation
coord_counts = (~x.isnan()).sum(dim=(0, 1))
coord_no_nan = x.clone()
coord_no_nan[coord_no_nan.isnan()] = 0
coord_mean = coord_no_nan.sum(dim=(0, 1)) / coord_counts
normed = x - coord_mean
# normed[normed.isnan()] = 0
# normed = nn.functional.normalize(normed, dim=-1)
# Coords + Angles
tensor = torch.cat((normed.flatten(-2), angles), 1)
# Mean
counts = (~tensor.isnan()).sum(dim=0)
no_nan = tensor.clone()
no_nan[no_nan.isnan()] = 0
mean = no_nan.sum(dim=0) / counts
# Standard Deviation
diff = tensor - mean
diff[diff.isnan()] = 0
correction = 1
std = (diff.square().sum(dim=0) / (counts - correction)).float().sqrt()
out = torch.cat((mean, std))
out[out.isnan()] = 0
return out
MLPModel.agg(tensor)
preprocess = Preprocess(MLPModel.agg)
preprocess(
torch.Tensor(
load_relevant_data_subset(
"/kaggle/input/asl-signs/train_landmark_files/16069/100015657.parquet"
)
)
)
if "train_preload" not in locals():
train_preload = None
if "test_preload" not in locals():
test_preload = None
train_dataset = ASLDataset(train_df, MLPModel.agg, train_preload)
train_preload = train_dataset.items
test_dataset = ASLDataset(test_df, MLPModel.agg, test_preload)
test_preload = test_dataset.items
len(train_dataset), len(test_dataset)
train_dataloader = DataLoader(train_dataset, batch_size=128, shuffle=True)
test_dataloader = DataLoader(test_dataset, batch_size=128, shuffle=True)
model = MLPModel().to(device)
learning_rate = 1e-3
weight_decay = 1e-2
loss_fn = nn.CrossEntropyLoss()
optimizer = torch.optim.AdamW(
model.parameters(), lr=learning_rate, weight_decay=weight_decay
)
scheduler = torch.optim.lr_scheduler.CosineAnnealingWarmRestarts(
optimizer, T_0=8, eta_min=1e-4
)
def train_val_loop(epoch, train_dataloader, val_dataloader, model, loss_fn, optimizer):
total_batches = len(train_dataloader)
train_size, train_batches = 0, 0
train_loss, train_correct = 0, 0
with tqdm(desc=f"Epoch {epoch}", total=total_batches) as bar:
for batch, (X, y) in enumerate(train_dataloader):
# Compute prediction and loss
pred = model(X)
loss = loss_fn(pred, y)
train_loss += loss.item()
train_correct += (pred.argmax(1) == y).type(torch.float).sum().item()
train_size += len(y)
train_batches += 1
# Backpropagation
optimizer.zero_grad()
loss.backward()
optimizer.step()
scheduler.step(epoch + batch / total_batches)
bar.update()
if batch % 10 == 0:
bar.set_postfix(
accuracy=train_correct / train_size,
loss=train_loss / train_batches,
lr=scheduler.get_last_lr(),
)
bar.set_postfix(
accuracy=train_correct / train_size, loss=train_loss / train_batches
)
with torch.no_grad():
val_size, val_batches = 0, 0
val_loss, val_correct = 0, 0
for batch, (X, y) in enumerate(val_dataloader):
pred = model(X)
val_loss += loss_fn(pred, y).item()
val_correct += (pred.argmax(1) == y).type(torch.float).sum().item()
val_size += len(y)
val_batches += 1
if batch % 10 == 0 or batch + 1 == len(val_dataloader):
bar.set_postfix(
accuracy=train_correct / train_size,
loss=train_loss / train_batches,
val_accuracy=val_correct / val_size,
val_loss=val_loss / val_batches,
)
return (
train_correct / train_size,
train_loss / train_batches,
val_correct / val_size,
val_loss / val_batches,
)
best_loss = float("inf")
saved_state = model.state_dict()
epochs = 64
for epoch in range(epochs):
acc, loss, v_acc, v_loss = train_val_loop(
epoch, train_dataloader, test_dataloader, model, loss_fn, optimizer
)
if v_loss < best_loss:
best_loss = v_loss
saved_state = model.state_dict()
torch.save(saved_state, "model_weights.pth")
torchinfo.summary(model, (1, 534 * 2))
# ### PyTorch → ONNX
eval_model = MLPModel()
eval_model.load_state_dict(saved_state)
preprocess.eval()
preprocess_sample = torch.rand((23, 543, 3)).to(
device
) # 23 is an arbitrary number of frames, 543 is the number of rows/landmarks, 3 is the x, y, z columns
onnx_preprocess_path = "preprocess.onnx"
torch.onnx.export(
preprocess,
preprocess_sample,
onnx_preprocess_path,
opset_version=12,
input_names=["inputs"],
output_names=["outputs"],
dynamic_axes={"inputs": {0: "frames"}},
)
eval_model.eval()
model_sample = torch.rand((1, 534 * 2)).to(device) # 1 is the batch size
onnx_model_path = "model.onnx"
torch.onnx.export(
eval_model,
model_sample,
onnx_model_path,
opset_version=12,
input_names=["inputs"],
output_names=["outputs"],
dynamic_axes={"inputs": {0: "batch_size"}},
)
# Will raise an exception if checks fail
onnx_preprocess = onnx.load(onnx_preprocess_path)
onnx.checker.check_model(onnx_preprocess)
onnx_model = onnx.load(onnx_model_path)
onnx.checker.check_model(onnx_model)
# ### ONNX → Tensorflow
tf_preprocess_path = "tf_preprocess"
tf_preprocess = onnx_tf.backend.prepare(onnx_preprocess)
tf_preprocess.export_graph(tf_preprocess_path)
tf_model_path = "tf_model"
tf_model = onnx_tf.backend.prepare(onnx_model)
tf_model.export_graph(tf_model_path)
class InferenceModel(tf.Module):
def __init__(self):
super().__init__()
self.preprocess = tf.saved_model.load(tf_preprocess_path)
self.model = tf.saved_model.load(tf_model_path)
self.preprocess.trainable = False
self.model.trainable = False
@tf.function(
input_signature=[
tf.TensorSpec(shape=[None, 543, 3], dtype=tf.float32, name="inputs")
]
)
def call(self, x):
outputs = {}
preprocessed = self.preprocess(**{"inputs": x})["outputs"]
pred = self.model(**{"inputs": tf.expand_dims(preprocessed, 0)})["outputs"][
0, :
]
# pred = tf.nn.softmax(pred)
return {"outputs": pred}
tf_inference = InferenceModel()
tf_inference_path = "tf_inference"
tf.saved_model.save(
tf_inference, tf_inference_path, signatures={"serving_default": tf_inference.call}
)
# ### Tensorflow → TFLite
model_converter = tf.lite.TFLiteConverter.from_saved_model(
tf_inference_path
) # path to the SavedModel directory
tflite_model = model_converter.convert()
# Save the model.
with open("model.tflite", "wb") as f:
f.write(tflite_model)
# The submission file (submission.zip) is created by compressing the TFLite model.
# ## Evaluating the TFLite model
import tflite_runtime.interpreter as tflite
interpreter = tflite.Interpreter("model.tflite")
found_signatures = list(interpreter.get_signature_list().keys())
# if REQUIRED_SIGNATURE not in found_signatures:
# raise KernelEvalException('Required input signature not found.')
frames = load_relevant_data_subset(
"/kaggle/input/asl-signs/train_landmark_files/16069/100015657.parquet"
)
prediction_fn = interpreter.get_signature_runner("serving_default")
output = prediction_fn(inputs=frames)
sign = np.argmax(output["outputs"])
print(sign, output["outputs"].shape)
interpreter.get_input_details()
interpreter.get_output_details()
interpreter.get_signature_list()
tests = []
for index, row in signs_df.iloc[:50].iterrows():
frames = load_relevant_data_subset(ASL_DIR / row.path)
interpreter = tflite.Interpreter("model.tflite")
prediction_fn = interpreter.get_signature_runner("serving_default")
output = prediction_fn(inputs=frames)
# output = tf_inference.call(frames)
# output = model(preprocess(torch.Tensor(frames)).unsqueeze(0))
sign = np.argmax(output["outputs"])
# sign = torch.argmax(output)
tests.append((sign, row.label))
tests
|
# # MNIST Baseline
# In this notebook, we create a baseline model to predict labels on the MNIST data set.
# ## Import packages
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.neighbors import KNeighborsClassifier
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import GridSearchCV
from sklearn.neighbors import KNeighborsClassifier
from sklearn.metrics import (
accuracy_score,
f1_score,
recall_score,
precision_score,
classification_report,
)
from random import shuffle
import random
random.seed(123)
# ## Define constants
IMAGE_SIZE = (28, 28)
# ## Load data
# We load both the train set and test set as Pandas data frames.
train_data = pd.read_csv("/kaggle/input/digit-recognizer/train.csv")
test_data = pd.read_csv("/kaggle/input/digit-recognizer/test.csv")
train_features = train_data.drop("label", axis=1)
train_label = train_data["label"]
# ## Data normalization
# Normalizing the images array to be in the range of 0-1 by dividing them by the max possible value.
train_features = train_features / 255.0
test_data = test_data / 255.0
# ## Fit KNN model
clf = KNeighborsClassifier(n_neighbors=1)
clf.fit(train_features, train_label)
# ## Evaluate KNN model
test_label_prediction = clf.predict(test_data)
submission = pd.DataFrame({"Label": test_label_prediction})
submission.index += 1
submission
submission.to_csv("submission.csv", index=True, index_label="ImageId")
# ## Split the traindata into two subsets and normalize the features of samples¶
train_data.shape
# Check for possible test sizes for 100-fold cross validation
folds = 100
possible_test_sizes = [
round(1 - x / train_data.shape[0], ndigits=4) for x in range(30000, 420000, folds)
]
# Print only positive test_size value
test_size = list(filter(lambda x: (x >= 0), possible_test_sizes))
print(test_size)
# Chose 0.2143 as the test size
print("\n\033[1mI choose 0.2857 as the test size")
# Split the dataset into the train_val set and testing set
# train_features = train_data.drop('label', axis=1)
# train_label = train_data['label']
X_train, X_test, y_train, y_test = train_test_split(
train_features, train_label, test_size=0.2857, random_state=0
)
print(
f"samples: {train_data.shape[0]}, train_val: {X_train.shape[0]}, test: {X_test.shape[0]}"
)
# Normalize features
normalizer = StandardScaler()
X_train = normalizer.fit_transform(X_train)
X_test = normalizer.transform(X_test)
# ## Train the KNN model and select the hyperparameter with cross-validation¶
k_range = range(1, 10)
param_grid = dict(n_neighbors=k_range)
print(param_grid)
clf_knn = KNeighborsClassifier(n_neighbors=1)
grid = GridSearchCV(clf_knn, param_grid, cv=2, scoring="accuracy", n_jobs=-1)
grid.fit(X_train, y_train)
print(grid.best_score_)
print(grid.best_params_)
# ## Evaluate model
clf_knn = KNeighborsClassifier(n_neighbors=grid.best_params_["n_neighbors"])
clf_knn.fit(X_train, y_train)
# Evaluate the model on the testing set
knn_predict = clf_knn.predict(X_test)
# Report prediction
print(classification_report(y_test, knn_predict))
knn_acc = accuracy_score(knn_predict, y_test)
print(
"\n\033[31;1;48;5;226mK-Nearest Neighbour Accuracy: \033[1m {:.2f}%".format(
knn_acc * 100
)
)
knn_predict_new = clf.predict(test_data)
submission = pd.DataFrame({"Label": knn_predict_new})
submission.index += 1
submission
submission.to_csv("submission.csv", index=True, index_label="ImageId")
|
# # House Prices: Advanced Regression Techniques
# _[Link to kaggle](https://www.kaggle.com/c/house-prices-advanced-regression-techniques)_
# **Author: Piotr Cichacki**
# ## Goal of the data analysis: predict the sales prices for each house
# ### Loading necessary libraries
# Data manipulation
import numpy as np
import pandas as pd
# Data visualization
import seaborn as sns
import matplotlib.pyplot as plt
plt.style.use("ggplot")
plt.rcParams["figure.figsize"] = (10, 6)
import warnings
warnings.filterwarnings("ignore")
# ### Read train and test datasets
train = pd.read_csv("../input/house-prices-advanced-regression-techniques/train.csv")
test = pd.read_csv("../input/house-prices-advanced-regression-techniques/test.csv")
# ### First look on our data
print("Train data shape: ", train.shape)
train.head(5)
train.info()
# ### Exploring house prices - our target variable
print(train["SalePrice"].describe())
plt.figure(figsize=(15, 4))
plt.title("House prices", fontdict={"fontsize": 20})
plt.boxplot(train.SalePrice, vert=False)
plt.show()
from scipy.stats import norm
from scipy import stats
fig, axs = plt.subplots(1, 2)
fig.set_size_inches(15, 6)
axs[0].set_title("House prices", fontdict={"fontsize": 20})
axs[0].hist(train["SalePrice"], bins=40, density=True)
mu, sigma = norm.fit(train["SalePrice"])
x = np.linspace(min(train["SalePrice"]), max(train["SalePrice"]))
axs[0].plot(x, norm.pdf(x, mu, sigma), linewidth=3, label="Normal distribution")
axs[1] = stats.probplot(train["SalePrice"], plot=plt)
axs[0].set_xlabel("Price")
axs[0].set_ylabel("Density")
print(f"Skewness: {train.SalePrice.skew()}")
print(f"Kurtosis: {train.SalePrice.kurtosis()}")
# SalePrice is positively skewed, so we will use log transformation to achieve more normal distribution and adjust the relative scales in the data so that they can be captured better by a linear model.
train["SalePrice"] = np.log(train["SalePrice"])
from scipy.stats import norm
from scipy import stats
fig, axs = plt.subplots(1, 2)
fig.set_size_inches(15, 6)
axs[0].set_title("House prices", fontdict={"fontsize": 20})
axs[0].hist(train["SalePrice"], bins=40, density=True)
mu, sigma = norm.fit(train["SalePrice"])
x = np.linspace(min(train["SalePrice"]), max(train["SalePrice"]))
axs[0].plot(x, norm.pdf(x, mu, sigma), linewidth=3, label="Normal distribution")
axs[1] = stats.probplot(train["SalePrice"], plot=plt)
axs[0].set_xlabel("Price")
axs[0].set_ylabel("Density")
print(f"Skewness: {train.SalePrice.skew()}")
print(f"Kurtosis: {train.SalePrice.kurtosis()}")
# Looks better.
# ### Dealing with missing data
sns.heatmap(train.isnull(), cmap="inferno", cbar=False)
plt.show()
total = train.isnull().sum().sort_values(ascending=False)
percent = total * 100 / train.shape[0]
missing_data = pd.concat([total, percent], axis=1, keys=["Total", "Percent"])
missing_data[missing_data.Total > 0]
# Firstly let's take a look on variables which have more than 10% missing data.
# **LotFrontage** - linear feet of street connected to property.
# Not very high correlation so I will delete this column from dataset.
lotfrontage = train.loc[:, ["SalePrice", "LotFrontage"]]
lotfrontage[lotfrontage["LotFrontage"].notnull()].corr()
train = train.drop(["LotFrontage"], axis=1)
# **Alley** - type of alley access to property. 'na' values for properties with no alley access.
train["Alley"].fillna("No alley access", inplace=True)
alley = train.loc[:, ["SalePrice", "Alley"]]
sns.boxplot(
alley["Alley"], alley["SalePrice"], order=["No alley access", "Pave", "Grvl"]
)
# **PoolQC** - pool quality. 'na' values for properties with no pool.
# Significant differences in prices between properties with pool.
train["PoolQC"].fillna("No pool", inplace=True)
pool = train.loc[:, ["SalePrice", "PoolQC"]]
sns.boxplot(pool["PoolQC"], pool["SalePrice"], order=["No pool", "Fa", "Gd", "Ex"])
# **MiscFeature** - miscellaneaous features not covered in other categories. 'na' values for none miscellaneous feature.
# Not significant correlation.
train["MiscFeature"].fillna("None", inplace=True)
misc = train.loc[:, ["SalePrice", "MiscFeature"]]
sns.boxplot(misc["MiscFeature"], misc["SalePrice"])
# **Fence** - fence quality. 'na' values for properties with no fence.
# Not significant correlation.
train["Fence"].fillna("No fence", inplace=True)
fence = train[["SalePrice", "Fence"]]
sns.boxplot(fence["Fence"], fence["SalePrice"])
# **FireplaceQu** - fireplace quality. 'na' values for properties with no fireplace.
# Significant correlation.
train["FireplaceQu"].fillna("No fireplace", inplace=True)
fireplace = train.loc[:, ["SalePrice", "FireplaceQu"]]
sns.boxplot(
fireplace["FireplaceQu"],
fireplace["SalePrice"],
order=["No fireplace", "Po", "Fa", "TA", "Gd", "Ex"],
)
# Now let's take a look on variables **"Garage..."** with missing values
# All of them have the same number of missing data and they refer to the same set of observations -> properties with no garage.
train["GarageCond"].fillna("No garage", inplace=True)
train["GarageType"].fillna("No garage", inplace=True)
train["GarageFinish"].fillna("No garage", inplace=True)
train["GarageQual"].fillna("No garage", inplace=True)
train = train.drop(["GarageYrBlt"], axis=1)
# Similar situation with **"Bsmt..."** variables
# Observations with missing values are the properties with no basement.
train["BsmtExposure"].fillna("No basement", inplace=True)
train["BsmtFinType1"].fillna("No basement", inplace=True)
train["BsmtFinType2"].fillna("No basement", inplace=True)
train["BsmtCond"].fillna("No basement", inplace=True)
train["BsmtQual"].fillna("No basement", inplace=True)
# Now I have 1 observation with missing information about **electrical** system and 8 observation with missing information about **masonry veneer type and area**.
# I will just delete them, since masonry veneer area is important variable (high correlation coefficient).
train = train.drop(train.loc[train["MasVnrType"].isnull()].index)
train = train.drop(train.loc[train["Electrical"].isnull()].index)
print("No more missing data: ", train.isnull().sum().max() == 0)
print(
"Number of observations after deleting observations with missing data: ",
train.shape[0],
)
# MSSubClass feature identifies the type of dweeling involved in the sale. Since it is categorical variable, I will change data type to string.
train["MSSubClass"] = train["MSSubClass"].astype(str)
# I will drop column with ID since it is meaningless in our model.
train = train.drop(["Id"], axis=1)
# ### Finding dependencies between our target variable and independent continouos variables with correlation matrix and some plots.
plt.figure(figsize=(3, 10))
plt.title(
"Correlation between SalePrice and dependent variables", fontdict={"fontsize": 20}
)
corr_matrix = train.corr().round(decimals=4)
corr_SalePrice = corr_matrix["SalePrice"].sort_values(ascending=False)
corr_df = pd.DataFrame(
corr_SalePrice.values, columns=["HousePrice"], index=corr_SalePrice.index
)
sns.heatmap(corr_df, annot=True, cmap="inferno")
plt.show()
print("Most meaningful dependent variables: \n", corr_SalePrice[corr_SalePrice > 0.4])
plt.title("Relationship between OverallQual and SalePrice", fontdict={"fontsize": 20})
sns.boxplot(x="OverallQual", y="SalePrice", data=train)
plt.xlabel("Rates the overall material and finish of the house")
plt.xticks(
np.arange(0, 10),
[
"Very Poor",
"Poor",
"Fair",
"Below average",
"Average",
"Above average",
"Good",
"Very good",
"Excellent",
"Very excellent",
],
rotation=50,
)
plt.show()
# Conclusion: Overall material and finish of the house quality seem to be strongly related with house prices.
fig, axs = plt.subplots(2, 2)
fig.set_size_inches(15, 15)
axs[0, 0].set_title("Relationship between GrLivArea and SalePrice")
axs[0, 0].set_xlabel("Above ground living area square feat")
axs[0, 0].set_ylabel("Sale price")
axs[0, 0].plot(train["GrLivArea"], train["SalePrice"], linestyle="none", marker="o")
axs[1, 0].set_title("Distribution of GrLivArea")
axs[1, 0].set_xlabel("Above ground living area square feat")
axs[1, 0].set_ylabel("Density")
axs[1, 0].hist(train["GrLivArea"], bins=40, density=True)
axs[0, 1].set_title(
"Relationship between GrLivArea and SalePrice with log transformation"
)
axs[0, 1].set_xlabel("Above ground living area square feat")
axs[0, 1].set_ylabel("Sale price")
axs[0, 1].plot(
np.log(train["GrLivArea"]), train["SalePrice"], linestyle="none", marker="o"
)
axs[1, 1].set_title("Distribution of GrLivArea with log transformation")
axs[1, 1].set_xlabel("Above ground living area square feat")
axs[1, 1].set_ylabel("Density")
axs[1, 1].hist(np.log(train["GrLivArea"]), bins=40, density=True)
plt.show()
# Conclusion: Above ground living area seem to be linearly and positive related with house prices. Looks better with log transformation.
train["GrLivArea"] = np.log(train["GrLivArea"])
plt.title("Relationship between GarageCars and SalePrice", fontdict={"fontsize": 20})
sns.boxplot(x="GarageCars", y="SalePrice", data=train)
plt.xlabel("Size of garage in car capacity")
plt.show()
plt.title("Relationship between GarageArea and SalePrice", fontdict={"fontsize": 20})
plt.xlabel("Garage area")
plt.ylabel("Sale price")
plt.plot(train.GarageArea, train.SalePrice, linestyle="none", marker="o")
plt.show()
plt.title("Relationship between TotalBsmtSF and SalePrice", fontdict={"fontsize": 20})
plt.xlabel("Total square feet of basement area")
plt.ylabel("Sale price")
plt.plot(train.TotalBsmtSF, train.SalePrice, linestyle="none", marker="o")
plt.show()
# Conclusion: Basement area also seem to be linearly and positive related with house prices. But there is one evident outlier.
train = train[~(train.TotalBsmtSF > 4000)]
fig, axs = plt.subplots(2, 2)
fig.set_size_inches(15, 15)
axs[0, 0].set_title("Relationship between 1stFlrSF and SalePrice")
axs[0, 0].set_xlabel("1stFlrSF")
axs[0, 0].set_ylabel("Sale price")
axs[0, 0].plot(train["1stFlrSF"], train["SalePrice"], linestyle="none", marker="o")
axs[1, 0].set_title("Distribution of 1stFlrSF")
axs[1, 0].set_xlabel("1stFlrSF")
axs[1, 0].set_ylabel("Density")
axs[1, 0].hist(train["1stFlrSF"], bins=40, density=True)
axs[0, 1].set_title(
"Relationship between 1stFlrSF and SalePrice with log transformation"
)
axs[0, 1].set_xlabel("1stFlrSF")
axs[0, 1].set_ylabel("Sale price")
axs[0, 1].plot(
np.log(train["1stFlrSF"]), train["SalePrice"], linestyle="none", marker="o"
)
axs[1, 1].set_title("Distribution of 1stFlrSF with log transformation")
axs[1, 1].set_xlabel("1stFlrSF")
axs[1, 1].set_ylabel("Density")
axs[1, 1].hist(np.log(train["1stFlrSF"]), bins=40, density=True)
plt.show()
# Seems fine but with log transformation looks better.
train["1stFlrSF"] = np.log(train["1stFlrSF"])
plt.title("Relationship between FullBath and SalePrice", fontdict={"fontsize": 20})
sns.boxplot(x="FullBath", y="SalePrice", data=train)
plt.xlabel("Full bathrooms above grade")
plt.show()
plt.title("Relationship between TotRmsAbvGrd and SalePrice", fontdict={"fontsize": 20})
sns.boxplot(x="TotRmsAbvGrd", y="SalePrice", data=train)
plt.xlabel("Total rooms above grade (does not include bathrooms)")
plt.show()
# Instead of considering separately total rooms above grad and full bathrooms above grade, I will add them.
train["TotRmsAbvGrd + FullBath"] = train.FullBath + train.TotRmsAbvGrd
test["TotRmsAbvGrd + FullBath"] = test.FullBath + test.TotRmsAbvGrd
plt.title(
"Relationship between TotRmsAbvGrd + FullBath and SalePrice",
fontdict={"fontsize": 20},
)
sns.boxplot(x="TotRmsAbvGrd + FullBath", y="SalePrice", data=train)
plt.xlabel("Total rooms and bathrooms above grade")
plt.show()
plt.figure(figsize=(14, 8))
plt.title("Relationship between YearBuilt and SalePrice", fontdict={"fontsize": 20})
sns.boxplot(x="YearBuilt", y="SalePrice", data=train)
plt.xlabel("Original construction date")
plt.show()
plt.title("Relationship between MasVnrArea and SalePrice", fontdict={"fontsize": 20})
plt.xlabel("Masonry veneer area in square feet")
plt.ylabel("Sale price")
plt.plot(train["MasVnrArea"], train.SalePrice, linestyle="none", marker="o")
plt.show()
# ### Preparing test dataset
test.info()
testID = test["Id"]
total = test.isnull().sum().sort_values(ascending=False)
percent = total * 100 / test.shape[0]
missing_data = pd.concat([total, percent], axis=1, keys=["Total", "Percent"])
missing_data[missing_data.Total > 0]
test = test.drop(["LotFrontage", "GarageYrBlt", "Id"], axis=1)
test["Alley"].fillna("No alley access", inplace=True)
test["PoolQC"].fillna("No pool", inplace=True)
test["MiscFeature"].fillna("None", inplace=True)
test["Fence"].fillna("No fence", inplace=True)
test["FireplaceQu"].fillna("No fireplace", inplace=True)
test["GarageCond"].fillna("No garage", inplace=True)
test["GarageType"].fillna("No garage", inplace=True)
test["GarageFinish"].fillna("No garage", inplace=True)
test["GarageQual"].fillna("No garage", inplace=True)
test["GarageCars"].fillna(
test.loc[test["GarageCars"].notnull(), "GarageCars"].mean(), inplace=True
)
test["GarageArea"].fillna(
test.loc[test["GarageArea"].notnull(), "GarageArea"].mean(), inplace=True
)
test["BsmtExposure"].fillna("No basement", inplace=True)
test["BsmtFinType1"].fillna("No basement", inplace=True)
test["BsmtFinType2"].fillna("No basement", inplace=True)
test["BsmtCond"].fillna("No basement", inplace=True)
test["BsmtQual"].fillna("No basement", inplace=True)
test["BsmtFullBath"].fillna(0.0, inplace=True)
test["BsmtHalfBath"].fillna(0.0, inplace=True)
test["BsmtFinSF1"].fillna(
test.loc[test["BsmtFinSF1"].notnull(), "BsmtFinSF1"].mean(), inplace=True
)
test["BsmtFinSF2"].fillna(
test.loc[test["BsmtFinSF2"].notnull(), "BsmtFinSF2"].mean(), inplace=True
)
test["BsmtUnfSF"].fillna(
test.loc[test["BsmtUnfSF"].notnull(), "BsmtUnfSF"].mean(), inplace=True
)
test["TotalBsmtSF"].fillna(
test.loc[test["TotalBsmtSF"].notnull(), "TotalBsmtSF"].mean(), inplace=True
)
test["MasVnrType"].fillna("None", inplace=True)
test["MasVnrArea"].fillna(
test.loc[test["MasVnrArea"].notnull(), "MasVnrArea"].mean(), inplace=True
)
test["KitchenQual"].fillna("TA", inplace=True)
test["Functional"].fillna("Typ", inplace=True)
test["SaleType"].fillna("Other", inplace=True)
test["MSZoning"].value_counts()
test["MSZoning"].fillna("RL", inplace=True)
test["Utilities"].value_counts()
test["Utilities"].fillna("AllPub", inplace=True)
test["Exterior1st"].value_counts()
test["Exterior1st"].fillna("VinylSd", inplace=True)
test["Exterior2nd"].value_counts()
test["Exterior2nd"].fillna("VinylSd", inplace=True)
test["MSSubClass"] = test["MSSubClass"].astype(str)
test["GrLivArea"] = np.log(test["GrLivArea"])
test["1stFlrSF"] = np.log(test["1stFlrSF"])
# ### Dummies variables
train.shape
test["SalePrice"] = -1
data = pd.concat([train, test], axis=0)
data.shape
data = pd.get_dummies(data)
train = data.loc[data["SalePrice"] > -1, :]
test = data.loc[data["SalePrice"] == -1, :]
train.shape
test = test.drop(["SalePrice"], axis=1)
test.shape
# ## Building models
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(
train.drop(["SalePrice"], axis=1), train["SalePrice"], random_state=0
)
# ### Linear regression models
from sklearn.linear_model import LinearRegression
lr = LinearRegression().fit(X_train, y_train)
print("Training set score: ", lr.score(X_train, y_train))
print("Test set score: ", lr.score(X_test, y_test))
from sklearn.linear_model import Ridge
ridge = Ridge(alpha=10).fit(X_train, y_train)
print("Training set score: ", ridge.score(X_train, y_train))
print("Test set score: ", ridge.score(X_test, y_test))
print("Number of features used: ", np.sum(ridge.coef_ != 0))
from sklearn.linear_model import Lasso
lasso = Lasso(alpha=0.001, max_iter=100000).fit(X_train, y_train)
print("Training set score: ", lasso.score(X_train, y_train))
print("Test set score: ", lasso.score(X_test, y_test))
print("Number of features used: ", np.sum(lasso.coef_ != 0))
# ### Decision Tree models
from sklearn.ensemble import RandomForestRegressor
forest = RandomForestRegressor(random_state=0, n_estimators=1000).fit(X_train, y_train)
print("Training set score: ", forest.score(X_train, y_train))
print("Test set score: ", forest.score(X_test, y_test))
# ### Saving answers to file
submission = pd.DataFrame()
submission["Id"] = testID
submission["SalePrice"] = np.exp(lasso.predict(test))
submission.to_csv("submission_lasso.csv", index=False)
submission["SalePrice"] = np.exp(ridge.predict(test))
submission.to_csv("submission_ridge.csv", index=False)
|
import io
import os
import cv2
import csv
import time
import copy
import math
import torch
import shutil
import logging
import argparse
import numpy as np
import torchvision
import numpy as np
import pandas as pd
import seaborn as sb
import torch.nn as nn
from PIL import Image
from tqdm import tqdm
import torch.optim as optim
from sklearn import datasets
import matplotlib.pyplot as plt
from tqdm.notebook import trange
from statistics import mean, stdev
from torchvision.utils import make_grid
import torch.utils.model_zoo as model_zoo
from torch.utils.data import Dataset, DataLoader
from torchvision import datasets, models, transforms
from sklearn.model_selection import (
train_test_split,
StratifiedKFold,
StratifiedShuffleSplit,
KFold,
)
class SAM(torch.optim.Optimizer):
def __init__(self, params, base_optimizer, rho=0.05, adaptive=False, **kwargs):
assert rho >= 0.0, f"Invalid rho, should be non-negative: {rho}"
defaults = dict(rho=rho, adaptive=adaptive, **kwargs)
super(SAM, self).__init__(params, defaults)
self.base_optimizer = base_optimizer(self.param_groups, **kwargs)
self.param_groups = self.base_optimizer.param_groups
self.defaults.update(self.base_optimizer.defaults)
@torch.no_grad()
def first_step(self, zero_grad=False):
grad_norm = self._grad_norm()
for group in self.param_groups:
scale = group["rho"] / (grad_norm + 1e-12)
for p in group["params"]:
if p.grad is None:
continue
self.state[p]["old_p"] = p.data.clone()
e_w = (
(torch.pow(p, 2) if group["adaptive"] else 1.0)
* p.grad
* scale.to(p)
)
p.add_(e_w) # climb to the local maximum "w + e(w)"
if zero_grad:
self.zero_grad()
@torch.no_grad()
def second_step(self, zero_grad=False):
for group in self.param_groups:
for p in group["params"]:
if p.grad is None:
continue
p.data = self.state[p]["old_p"] # get back to "w" from "w + e(w)"
self.base_optimizer.step() # do the actual "sharpness-aware" update
if zero_grad:
self.zero_grad()
@torch.no_grad()
def step(self, closure=None):
assert (
closure is not None
), "Sharpness Aware Minimization requires closure, but it was not provided"
closure = torch.enable_grad()(
closure
) # the closure should do a full forward-backward pass
self.first_step(zero_grad=True)
closure()
self.second_step()
def _grad_norm(self):
shared_device = self.param_groups[0]["params"][
0
].device # put everything on the same device, in case of model parallelism
norm = torch.norm(
torch.stack(
[
((torch.abs(p) if group["adaptive"] else 1.0) * p.grad)
.norm(p=2)
.to(shared_device)
for group in self.param_groups
for p in group["params"]
if p.grad is not None
]
),
p=2,
)
return norm
def load_state_dict(self, state_dict):
super().load_state_dict(state_dict)
self.base_optimizer.param_groups = self.param_groups
print("PyTorch Version: ", torch.__version__)
print("Torchvision Version: ", torchvision.__version__)
num_classes = 100
batch_size = 64
num_epochs = 20
model_choice = "Resnet34"
learning_rate = 0.01
SGD_momentum = 0.9
SGD_weight_decay = 4e-4
feature_extract = False
transform_train = transforms.Compose(
[
transforms.Resize((224, 224)),
transforms.RandomHorizontalFlip(0.1),
transforms.RandomRotation(20),
transforms.ToTensor(),
transforms.RandomAdjustSharpness(sharpness_factor=2, p=0.1),
transforms.ColorJitter(brightness=0.1, contrast=0.1, saturation=0.1),
transforms.Normalize((0.4914, 0.4822, 0.4465), (0.247, 0.243, 0.261)),
transforms.RandomErasing(p=0.75, scale=(0.02, 0.1), value=1.0, inplace=False),
]
)
transform_test = transforms.Compose(
[
transforms.ToTensor(),
transforms.Resize((224, 224)),
transforms.Normalize((0.4914, 0.4822, 0.4465), (0.247, 0.243, 0.261)),
]
)
""" Training Dataset & Dataloaders
"""
train_set = torchvision.datasets.CIFAR100(
root="./data", train=True, download=True, transform=transform_train
)
train_loader = torch.utils.data.DataLoader(
train_set, batch_size=batch_size, shuffle=True, num_workers=1
)
""" Validation Dataset & Dataoaders
"""
validation_set = torchvision.datasets.CIFAR100(
root="./data", train=False, download=True, transform=transform_test
)
validation_loader = torch.utils.data.DataLoader(
validation_set, batch_size=batch_size, shuffle=False, num_workers=1
)
dataloaders_dict = {}
dataloaders_dict["Train"] = train_loader
dataloaders_dict["Validation"] = validation_loader
# helper function to show a batch of training instances
def show_batch(data):
for images, labels in data:
fig, ax = plt.subplots(figsize=(30, 30))
ax.set_xticks([])
ax.set_yticks([])
ax.imshow(make_grid(images, nrow=8).permute(1, 2, 0))
break
# show_batch(train_loader)
def Learning_Rate(optimizer):
for param_group in optimizer.param_groups:
return param_group["lr"]
def train_model(model, dataloaders, criterion, optimizer, scheduler, num_epochs=10):
since = time.time()
train_acc_history = []
val_acc_history = []
best_acc = 0.0
for epoch in trange(
num_epochs,
desc=f"Model: {model_choice}, Number of Epochs: {num_epochs}, Batch Size: {batch_size}, Learning Rate: {(Learning_Rate(optimizer)):.9f} ",
):
print("Epoch {}/{}".format(epoch + 1, num_epochs))
print("-" * 10)
# Each epoch has a training and validation phase
for phase in ["Train", "Validation"]:
if phase == "Train":
model.train() # Set model to training mode
else:
model.eval() # Set model to evaluate mode
running_loss = 0.0
running_corrects = 0
# Iterate over data.
for inputs, labels in dataloaders[phase]:
inputs = inputs.to(device)
labels = labels.to(device)
optimizer.zero_grad()
with torch.set_grad_enabled(phase == "Train"):
outputs = model_ft(inputs.to(device))
loss = criterion(outputs, labels)
_, preds = torch.max(outputs, 1)
if phase == "Train":
loss.backward()
def closure():
outputs = model_ft(inputs)
_, preds = torch.max(outputs, 1)
loss = criterion(outputs, labels)
loss.backward()
return loss
optimizer.step(closure)
running_loss += loss.item() * inputs.size(0)
running_corrects += torch.sum(preds == labels.data)
epoch_loss = running_loss / len(dataloaders[phase].dataset)
epoch_acc = running_corrects.double() / len(dataloaders[phase].dataset)
print(
f"{phase} Loss: {epoch_loss:.9f}, Accuracy: {(epoch_acc * 100):.9f}%, Learning Rate: {(Learning_Rate(optimizer)):.9f}"
)
if phase == "Validation" and epoch_acc > best_acc:
best_acc = epoch_acc
# best_model_wts = copy.deepcopy(model.state_dict())
torch.save(model_ft.state_dict(), "./Best_Checkpoint.pth")
if phase == "Train":
train_acc_history.append(epoch_acc.item() * 100)
if phase == "Validation":
val_acc_history.append(epoch_acc.item() * 100)
torch.save(model_ft.state_dict(), "./Last_Checkpoint.pth")
scheduler.step()
print()
time_elapsed = time.time() - since
print(
"Training completed in {:.0f}m {:.0f}s".format(
time_elapsed // 60, time_elapsed % 60
)
)
print("Best Validation Accuracy: {:9f}".format(best_acc * 100))
model.load_state_dict(torch.load("./Best_Checkpoint.pth"))
return model, val_acc_history, train_acc_history
def initialize_model(model_name, num_classes, feature_extract, use_pretrained=True):
model_ft = None
if model_name == "Resnet34":
"""Resnet34"""
model_ft = models.resnet34(pretrained=use_pretrained)
set_parameter_requires_grad(model_ft, feature_extract)
num_ftrs = model_ft.fc.in_features
model_ft.fc = nn.Sequential(nn.Dropout(0.1), nn.Linear(num_ftrs, num_classes))
elif model_name == "Resnet18":
"""Resnet18"""
model_ft = models.resnet18(pretrained=use_pretrained)
set_parameter_requires_grad(model_ft, feature_extract)
num_ftrs = model_ft.fc.in_features
torch.nn.init.xavier_uniform_(model_ft.fc.weight)
model_ft.fc = nn.Sequential(nn.Dropout(0.1), nn.Linear(num_ftrs, num_classes))
elif model_name == "Resnet101":
"""Resnet101"""
model_ft = models.resnet101(pretrained=use_pretrained)
set_parameter_requires_grad(model_ft, feature_extract)
num_ftrs = model_ft.fc.in_features
model_ft.fc = nn.Sequential(nn.Dropout(0.1), nn.Linear(num_ftrs, num_classes))
elif model_name == "Resnext101":
"""Resnext101"""
model_ft = models.resnext101_32x8d(pretrained=use_pretrained)
set_parameter_requires_grad(model_ft, feature_extract)
num_ftrs = model_ft.fc.in_features
model_ft.fc = nn.Sequential(nn.Dropout(0.1), nn.Linear(num_ftrs, num_classes))
elif model_name == "Resnet152":
"""Resnet152"""
model_ft = models.resnet152(models.ResNet152_Weights.DEFAULT)
set_parameter_requires_grad(model_ft, feature_extract)
num_ftrs = model_ft.fc.in_features
model_ft.fc = nn.Sequential(nn.Dropout(0.1), nn.Linear(num_ftrs, num_classes))
elif model_name == "Alexnet":
"""Alexnet"""
model_ft = models.alexnet(pretrained=use_pretrained)
set_parameter_requires_grad(model_ft, feature_extract)
num_ftrs = model_ft.classifier[6].in_features
model_ft.classifier[6] = nn.Linear(num_ftrs, num_classes)
elif model_name == "VGG11":
"""VGG11"""
model_ft = models.vgg11_bn(pretrained=use_pretrained)
set_parameter_requires_grad(model_ft, feature_extract)
num_ftrs = model_ft.classifier[6].in_features
model_ft.classifier[6] = nn.Linear(num_ftrs, num_classes)
elif model_name == "Squeezenet":
"""Squeezenet"""
model_ft = models.squeezenet1_0(pretrained=use_pretrained)
set_parameter_requires_grad(model_ft, feature_extract)
model_ft.classifier[1] = nn.Conv2d(
512, num_classes, kernel_size=(1, 1), stride=(1, 1)
)
model_ft.num_classes = num_classes
model_ft.classifier[1] = nn.Conv2d(
512, num_classes, kernel_size=(1, 1), stride=(1, 1)
)
elif model_name == "Densenet121":
"""Densenet121"""
model_ft = models.densenet121(pretrained=use_pretrained)
set_parameter_requires_grad(model_ft, feature_extract)
num_ftrs = model_ft.classifier.in_features
model_ft.classifier = nn.Linear(num_ftrs, num_classes)
else:
print("Invalid model name, exiting...")
exit()
return model_ft
def set_parameter_requires_grad(model, feature_extracting):
if feature_extracting:
for param in model.parameters():
param.requires_grad = False
model_ft = initialize_model(
model_choice, num_classes, feature_extract, use_pretrained=True
)
params_to_update = model_ft.parameters()
# print("Params to learn:")
if feature_extract:
params_to_update = []
for name, param in model_ft.named_parameters():
if param.requires_grad == True:
params_to_update.append(param)
pass
# print("\t",name)
else:
for name, param in model_ft.named_parameters():
if param.requires_grad == True:
pass
# print("\t",name)
device = torch.device("cuda:0")
if torch.cuda.device_count() > 1:
print("Using", torch.cuda.device_count(), "GPUs")
model_ft = nn.DataParallel(model_ft)
model_ft.to(device)
criterion = nn.CrossEntropyLoss().to(device)
# optimizer = optim.SGD(params_to_update, lr = learning_rate, momentum = SGD_momentum, weight_decay = SGD_weight_decay)
base_optimizer = torch.optim.SGD
optimizer = SAM(
model_ft.parameters(),
base_optimizer,
lr=learning_rate,
momentum=SGD_momentum,
weight_decay=SGD_weight_decay,
)
# optimizer = optim.Adam(params_to_update, lr=1e-3)
# scheduler = torch.optim.lr_scheduler.ReduceLROnPlateau(optimizer, mode = 'min', factor = 0.001, patience = 5, threshold = 0.0001, threshold_mode='abs')
scheduler = torch.optim.lr_scheduler.CosineAnnealingLR(optimizer, T_max=num_epochs)
model_ft, validation_accuracy_history, training_accuracy_history = train_model(
model_ft, dataloaders_dict, criterion, optimizer, scheduler, num_epochs=num_epochs
)
plt.figure(figsize=[8, 6])
plt.plot(training_accuracy_history, "black", linewidth=2.0)
plt.plot(validation_accuracy_history, "blue", linewidth=2.0)
plt.legend(["Training Accuracy", "Validation Accuracy"], fontsize=14)
plt.xlabel("Epochs", fontsize=10)
plt.ylabel("Accuracy", fontsize=10)
plt.title("Accuracy Curves", fontsize=12)
inference_data_loader = torch.utils.data.DataLoader(
validation_set, batch_size=1, shuffle=False, num_workers=2
)
images, labels = next(iter(inference_data_loader))
labels = labels.to(device)
images = images.to(device)
model_ft = model_ft.to(device)
start = time.time()
outputs = model_ft(images)
end = time.time()
infrence_time = end - start
print(f"The inference time is: {infrence_time}")
predictions = []
with torch.no_grad():
for data in validation_loader:
images, labels = data
outputs = model_ft(images)
_, predicted = torch.max(outputs.data, 1)
predictions.append(predicted)
predictions_transformed = [x.item() for x in torch.cat(predictions)]
with open("submission.csv", "w", encoding="utf-8", newline="") as out:
writer = csv.writer(out)
writer.writerow(["ID", "Label"])
for ID, Label in enumerate(predictions_transformed):
writer.writerow([ID, Label])
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import matplotlib.pyplot as plt
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Use this for multiple df outputs from same cell
from IPython.core.interactiveshell import InteractiveShell
InteractiveShell.ast_node_interactivity = "all"
# fillter warnings
# not recommanded for beginners
import warnings
warnings.filterwarnings("ignore")
df_train = pd.read_csv("../input/dapprojekt23/train.csv")
df_test = pd.read_csv("../input/dapprojekt23/test.csv")
# # Preparation of dataset
# Based on my first mandatory notebook, I will perform operations to have meaningful dataset without missing observations and negative values.
mask = df_train.loc[:, "total"] < 0
df_train.loc[mask, "total"] = df_train.loc[mask, "total"] + 65536
mask = df_test.loc[:, "total"] < 0
df_test.loc[mask, "total"] = df_test.loc[mask, "total"] + 65536
# replacing negative values in feature total - train
df_train.replace({"total": 0}, np.nan, inplace=True)
df_train["total"].interpolate(method="pchip", inplace=True)
df_train = df_train.astype({"total": int})
# replacing negative values in feature total - test
df_test.replace({"total": 0}, np.nan, inplace=True)
df_test["total"].interpolate(method="pchip", inplace=True)
df_test = df_test.astype({"total": int})
# dropping monotonic feature id
df_train.drop(columns=["id"], inplace=True)
def process_missing(df, train=False):
grouped = df.groupby("machine_name")
missing_ones = pd.DataFrame()
for name, group in grouped:
group = group.sort_values(by=["day"])
group["day_diff"] = group["day"].diff()
diff_greater_than_one = group[group["day_diff"] > 1]
for index, row in diff_greater_than_one.iterrows():
for i in range(int(row["day"]) - int(row["day_diff"]) + 1, int(row["day"])):
if train:
new_row = {
"day": i,
"broken": np.nan,
"total": np.nan,
"label": row["label"],
"machine_name": row["machine_name"],
}
else:
new_row = {
"day": i,
"broken": np.nan,
"total": np.nan,
"machine_name": row["machine_name"],
}
missing_ones = missing_ones.append(new_row, ignore_index=True)
df = df.append(missing_ones, ignore_index=True)
df.sort_values(by=["machine_name", "day"], ignore_index=True, inplace=True)
df["total"].interpolate(method="pchip", inplace=True)
df["broken"].interpolate(method="pad", inplace=True)
# negative values set to zero, because of interpolation
df.loc[df["total"] < df["broken"], "broken"] = 0
if train:
df = df.astype({"day": int, "broken": int, "total": int, "label": int})
else:
df = df.astype({"day": int, "broken": int, "total": int})
return df
df_train = process_missing(df_train, train=True)
# Also, I will remove records after the occurence of an anomaly.
def dataset_after_removal(df):
# remove records after the last anomaly for each group
def remove_after_anomaly(group):
last_anomaly_index = group["label"][::-1].idxmax()
return group.loc[:last_anomaly_index]
grouped = df.groupby("machine_name")
df = grouped.apply(remove_after_anomaly)
df = df.reset_index(drop=True)
return df
df_train = dataset_after_removal(df_train)
# # 1. Cross-validation function 1
# > Write a function that receives data as input, does a 5-fold cross-validation split by sample, and tests the following models (with default hyper-parameters): GaussianNB, LogisticRegression, RandomForestClassifier, ExtraTreesClassifier, and XGBClassifier.
import wittgenstein as lw
from xgboost import XGBClassifier
from sklearn.metrics import f1_score
from sklearn.ensemble import RandomForestClassifier
from sklearn.linear_model import LogisticRegression
from sklearn.naive_bayes import GaussianNB
from sklearn.ensemble import ExtraTreesClassifier
from tabulate import tabulate
from sklearn.model_selection import KFold
def construct_model(name):
switch = {
"GaussianNB": GaussianNB(),
"LogisticRegression": LogisticRegression(),
"RandomForestClassifier": RandomForestClassifier(),
"XGBClassifier": XGBClassifier(),
"ExtraTreesClassifier": ExtraTreesClassifier(),
"RIPPER": lw.RIPPER(),
}
return switch.get(name, "Invalid input")
def train_by_samples(X, Y, name):
folds_macro = []
kf = KFold(n_splits=5)
for train_index, test_index in kf.split(X):
X_train, X_test = X.iloc[train_index], X.iloc[test_index]
y_train, y_test = Y.iloc[train_index], Y.iloc[test_index]
model = construct_model(name)
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
score = f1_score(y_test, y_pred, average="macro")
folds_macro.append(score)
return [
name,
folds_macro[0],
folds_macro[1],
folds_macro[2],
folds_macro[3],
folds_macro[4],
sum(folds_macro) / 5,
]
import time
def cross_validation_function1(df):
df_copied = df.copy(deep=True)
df_copied = df_copied.loc[df_copied.loc[:, "day"] > 365, :]
Y = pd.DataFrame(df_copied["label"].tolist())
df_copied.drop(columns=["machine_name", "label"], inplace=True)
X = pd.DataFrame(df_copied)
table = []
table.append(train_by_samples(X, Y, "GaussianNB"))
table.append(train_by_samples(X, Y, "LogisticRegression"))
table.append(train_by_samples(X, Y, "RandomForestClassifier"))
table.append(train_by_samples(X, Y, "XGBClassifier"))
table.append(train_by_samples(X, Y, "ExtraTreesClassifier"))
# table.append(train_by_samples(X, Y, "RIPPER"))
print(
tabulate(
table,
headers=[
"Algorithm",
"Fold 1",
"Fold 2",
"Fold 3",
"Fold 4",
"Fold 5",
"Average",
],
)
)
# # 2. Run the cross-validation function 1
# > Use the entire dataset. Submit the best model to the Kaggle leaderboard. Write the score obtained on the Public Leaderboard.
# Based on the printout below, we can see that model GaussianNB performs the best if the average f1-macro is measured.
# One possible drawback is that samples in folds are not grouped by machine.
cross_validation_function1(df_train)
def train(model, df):
df_copied = df.copy(deep=True)
df_copied = df_copied.loc[df_copied.loc[:, "day"] > 365, :]
Y = pd.DataFrame(df_copied["label"].tolist())
df_copied.drop(columns=["machine_name", "label"], inplace=True)
X = pd.DataFrame(df_copied)
model.fit(X, Y)
print(f"\tTrain f1 macro: {f1_score(Y, model.predict(X), average='macro')}")
return model
def test(df, model):
df_copied = df.copy(deep=True)
df_copied.drop(columns=["machine_name", "id"], inplace=True)
X_test = pd.DataFrame(df_copied)
Y_pred = model.predict(X_test)
return Y_pred
model = train(GaussianNB(), df_train)
Y_pred = test(df_test, model)
submission = df_test.loc[:, df_test.columns.isin(("id",))]
submission = submission.rename(columns={"id": "Id"})
submission.loc[:, "Predicted"] = Y_pred
submission.to_csv("submission.csv", index=None)
submission.head()
# **Leaderboard score: 0.54203**
# # 3. Cross-validation function 2
# > Create a second cross-validation function that receives data as input, does 5-fold cross-validation split by machine (all samples from one machine are in the same fold), and tests the same models as before.
from sklearn.model_selection import GroupKFold
# During the split, GroupKFold ensures that samples from the same group are kept together in the same fold.
def train_by_machines(X, Y, name):
folds_macro = []
gkf = GroupKFold(n_splits=5)
groups = X["machine_name"]
folds_macro = []
for train_index, test_index in gkf.split(X, Y, groups):
X_train, X_test = X.iloc[train_index], X.iloc[test_index]
y_train, y_test = Y.iloc[train_index], Y.iloc[test_index]
model = construct_model(name)
model.fit(X_train.drop("machine_name", axis=1), y_train)
y_pred = model.predict(X_test.drop("machine_name", axis=1))
score = f1_score(y_test, y_pred, average="macro")
folds_macro.append(score)
return [
name,
folds_macro[0],
folds_macro[1],
folds_macro[2],
folds_macro[3],
folds_macro[4],
sum(folds_macro) / 5,
]
def cross_validation_function2(df):
df_copied = df.copy(deep=True)
df_copied = df_copied.loc[df_copied.loc[:, "day"] > 365, :]
Y = pd.DataFrame(df_copied["label"].tolist())
df_copied.drop(columns=["label"], inplace=True)
X = pd.DataFrame(df_copied)
table = []
table.append(train_by_machines(X, Y, "GaussianNB"))
table.append(train_by_machines(X, Y, "LogisticRegression"))
table.append(train_by_machines(X, Y, "RandomForestClassifier"))
table.append(train_by_machines(X, Y, "XGBClassifier"))
table.append(train_by_machines(X, Y, "ExtraTreesClassifier"))
# table.append(train_by_machines(X, Y, "RIPPER"))
print(
tabulate(
table,
headers=[
"Algorithm",
"Fold 1",
"Fold 2",
"Fold 3",
"Fold 4",
"Fold 5",
"Average",
],
)
)
# # 4. Run the cross-validation function 2
# > Use the entire dataset. Submit the best model to the Kaggle leaderboard. Write the score obtained on the Public Leaderboard.
# Based on the printout below, we can see that also GaussianNB performs the best if the average f1-macro is measured.
# The connection between those those functions will be questioned later.
cross_validation_function2(df_train)
model = train(GaussianNB(), df_train)
Y_pred = test(df_test, model)
submission = df_test.loc[:, df_test.columns.isin(("id",))]
submission = submission.rename(columns={"id": "Id"})
submission.loc[:, "Predicted"] = Y_pred
submission.to_csv("submission.csv", index=None)
submission.head()
# **Leaderboard score: 0.54203**
# # 5. New features
# > Create 20 new features. You are free to create whatever features you believe will be good predictors.
# Hints: Simple math operations, probability and statistics, rolling and expanding windows, entropies, …
# In this chapter, we will perform some operations to get new features.
# I used the following techniques:
# * rolling mean and sum windows of size 7 (could be optimized) for features broken, total and broken_ratio
# * expanding mean and sum windows for features broken, total and broken_ratio
# * z-score for features broken, total and broken_ratio
# * min-max score for features broken, total and broken_ratio
for df in [df_train, df_test]:
grouped = df.groupby("machine_name")
# Broken ratio
df["broken_ratio"] = grouped["broken"].transform(lambda x: x / x.sum())
# Rolling broken mean
df["rolling_broken_mean"] = grouped["broken"].transform(
lambda x: x.rolling(window=7).mean()
)
df["rolling_broken_mean"] = df["rolling_broken_mean"].fillna(method="bfill")
# Rolling total mean
df["rolling_total_mean"] = grouped["total"].transform(
lambda x: x.rolling(window=7).mean()
)
df["rolling_total_mean"] = df["rolling_total_mean"].fillna(method="bfill")
# Rolling broken sum
df["rolling_broken_sum"] = grouped["broken"].transform(
lambda x: x.rolling(window=7).sum()
)
df["rolling_broken_sum"] = df["rolling_broken_sum"].fillna(method="bfill")
# Rolling total sum
df["rolling_total_sum"] = grouped["total"].transform(
lambda x: x.rolling(window=7).sum()
)
df["rolling_total_sum"] = df["rolling_total_sum"].fillna(method="bfill")
# Rolling broken ratio mean
df["rolling_broken_ratio_mean"] = grouped["broken_ratio"].transform(
lambda x: x.rolling(window=7).mean()
)
df["rolling_broken_ratio_mean"] = df["rolling_broken_ratio_mean"].fillna(
method="bfill"
)
# Expanding broken mean
df["expanding_broken_mean"] = grouped["broken"].transform(
lambda x: x.expanding().mean()
)
# Expanding total mean
df["expanding_total_mean"] = grouped["total"].transform(
lambda x: x.expanding().mean()
)
# Expanding broken sum
df["expanding_broken_sum"] = grouped["broken"].transform(
lambda x: x.expanding().sum()
)
# Expanding total sum
df["expanding_total_sum"] = grouped["total"].transform(
lambda x: x.expanding().sum()
)
# Expanding broken ratio mean
df["expanding_broken_ratio_mean"] = grouped["broken_ratio"].transform(
lambda x: x.expanding().mean()
)
# Broken entropy
alpha = 1e-9
p_broken = grouped["broken"].transform(lambda x: (x + alpha) / (x.sum() + alpha))
df["broken_entropy"] = -p_broken * np.log2(p_broken)
# Total entropy
p_total = grouped["total"].transform(lambda x: (x + alpha) / (x.sum() + alpha))
df["total_entropy"] = -p_total * np.log2(p_total)
# Broken ratio entropy
p_broken_ratio = grouped["broken_ratio"].transform(
lambda x: (x + alpha) / (x.sum() + alpha)
)
df["broken_ratio_entropy"] = -p_broken_ratio * np.log2(p_broken_ratio)
# Broken z-score
df["broken_zscore"] = grouped["broken"].transform(
lambda x: (x - x.mean()) / x.std()
)
# Total z-score
df["total_zscore"] = grouped["total"].transform(lambda x: (x - x.mean()) / x.std())
# Broken ratio z-score
broken_ratio = df["broken"] / df["total"]
df["broken_ratio_zscore"] = grouped["broken_ratio"].transform(
lambda x: (x - x.mean()) / x.std()
)
# Broken min-max
df["broken_minmax"] = grouped["broken"].transform(
lambda x: (x - x.min()) / (x.max() - x.min())
)
# Total min-max
df["total_minmax"] = grouped["total"].transform(
lambda x: (x - x.min()) / (x.max() - x.min())
)
# Broken ratio min-max
broken_ratio = df["broken"] / df["total"]
df["broken_ratio_minmax"] = grouped["broken_ratio"].transform(
lambda x: (x - x.min()) / (x.max() - x.min())
)
# # 6. Run cross-validation functions
# > Run cross-validation function 1 and 2. Use features created in the previous step. Submit the best model from function 1 and the best model from function 2.
cross_validation_function1(df_train)
cross_validation_function2(df_train)
# Based on the printout above, best model from both functions is XGBClassifier.
model = train(XGBClassifier(), df_train)
Y_pred = test(df_test, model)
submission = df_test.loc[:, df_test.columns.isin(("id",))]
submission = submission.rename(columns={"id": "Id"})
submission.loc[:, "Predicted"] = Y_pred
submission.to_csv("submission.csv", index=None)
submission.head()
# **Leaderboard score: 0.67785**
# # 7. Filter method
# > Determine which model has the best results based on the printout of the previous step and use only that model in this step. Use mutual_info_classif, f_classif, or chi2 (from the sklearn library) to rank the features. Take the first 2 best-ranked features and calculate the f1_score of the selected model using 5-fold cross-validation. Then add the next 2 ranked features and repeat the process. Keep adding the next 2 features until all features are included in the dataset. Draw a graph showing the f1_score on the y-axis and the number of features used in the algorithm on the x-axis. What is the optimal choice of the number of features?
# As I said before, best model is XGBClassifier and it will be used in research below. I used mutual_info_classif to rank the features.
from sklearn.feature_selection import mutual_info_classif, f_classif, chi2
df_copied = df_train.copy(deep=True)
df_copied = df_copied.loc[df_copied.loc[:, "day"] > 365, :]
y = pd.DataFrame(df_copied["label"].tolist())
df_copied.drop(columns=["machine_name", "label"], inplace=True)
X = pd.DataFrame(df_copied)
feature_scores = mutual_info_classif(X, y)
sorted_indices = feature_scores.argsort()[::-1]
ranked_features = X.columns[sorted_indices]
ranked_features = ranked_features.to_list()
print(f"Ranked features:\n{ranked_features}")
f1_measures = []
for num in range(2, len(ranked_features) + 2, 2):
features = ranked_features[:num]
print(f"Iteration {num // 2} with features: {features}\n")
features.append("machine_name")
df_copied = df_train.copy(deep=True)
df_copied = df_copied.loc[df_copied.loc[:, "day"] > 365, :]
X = pd.DataFrame(df_copied[features])
table = []
table.append(train_by_machines(X, y, "XGBClassifier"))
print(
tabulate(
table,
headers=[
"Algorithm",
"Fold 1",
"Fold 2",
"Fold 3",
"Fold 4",
"Fold 5",
"Average",
],
)
)
print("\n\n")
f1_measures.append(table[0][-1])
print(f1_measures)
plt.plot(range(1, len(f1_measures) + 1), f1_measures)
plt.title("The dependency of f1 measures on the number of features")
plt.xlabel("Iteration (number_of_features = iteration * 2)")
plt.ylabel("F1 score")
plt.show()
# Based on the plot above, some conclusions can be made. Local optimum can be considered in 10th iteration, but the global maximum is in iteration 11 with 22 features. To be more precise and sure about conclusion, additional ranking needs to be made. (not in the interest in this notebook)
# # 8. Run the cross-validation functions
# > Use features selected in the previous step. Comment on the changes in the evaluation metrics. Submit the best model to the Kaggle leaderboard. Write the score obtained on the Public Leaderboard. Based on the results determine which cross-validation function is better and why. Is the Public Leaderboard score improved? Does cross-validation on the training set linearly correlate with Public Leaderboard scores?
# As I mentioned above, highest f1-macro is in iteration 11 with 22 features, so I will do rest of the experiments with that number.
features = ranked_features[:22]
features.append("machine_name")
features.append("label")
features_test = ranked_features[:22]
features_test.append("id")
features_test.append("machine_name")
cross_validation_function1(df_train[features])
cross_validation_function2(df_train[features])
# Based on the previous cross-validation functions, XGBClassifier is the best model in both of them. Conclusion about which function is better is not straight-forward, but we can observe that models perform better when we use function2 which uses KFold (grouping by machines).
model = train(XGBClassifier(), df_train[features])
Y_pred = test(df_test[features_test], model)
submission = df_test.loc[:, df_test.columns.isin(("id",))]
submission = submission.rename(columns={"id": "Id"})
submission.loc[:, "Predicted"] = Y_pred
submission.to_csv("submission.csv", index=None)
submission.head()
# **Leaderboard score: 0.64488**
# The Leaderboard score has not improved if we use less features. There has to be some connection between which makes previous model better.
# To test linear correlation, I will try to submit second (RandomForestClassifier) and third (ExtraTreesClassifier) best model and observe the results.
model = train(RandomForestClassifier(), df_train[features])
Y_pred = test(df_test[features_test], model)
submission = df_test.loc[:, df_test.columns.isin(("id",))]
submission = submission.rename(columns={"id": "Id"})
submission.loc[:, "Predicted"] = Y_pred
submission.to_csv("submission.csv", index=None)
submission.head()
# **Leaderboard score RandomForestClassifier : 0.66305**
model = train(ExtraTreesClassifier(), df_train[features])
Y_pred = test(df_test[features_test], model)
submission = df_test.loc[:, df_test.columns.isin(("id",))]
submission = submission.rename(columns={"id": "Id"})
submission.loc[:, "Predicted"] = Y_pred
submission.to_csv("submission.csv", index=None)
submission.head()
# **Leaderboard score ExtraTreesClassifier : 0.66704**
# As we can see, the hypothesis about linear correlation between results from cross-val functions and Public Leaderboard is not so true. The third model ExtraTreesClassifier performs the best. One problem with mentioned functions is that we use average f1-score to decide which model is the best (could be weighted average based on number of samples). The other problem is that training is performed on subset of training dataset.
# # 9.Explainability with SHAP
# > Choose the best ensemble algorithm based on the printout of the previous step and train it on the whole train set. Use the TreeExplainer from the SHAP library (import shap) on the trained model. Extract shap_values from the resulting explainer. Select one machine with an anomaly that lasts for at least 5 days. Isolate the samples with anomaly and 5 samples before it. Display a single force_plot (from the shap library) for each of the isolated samples. Comment on the results. Which features have the greatest impact on the decision of the algorithm? Did the algorithm accurately predict those samples? Plot signals from that machine with the function from the previous exercise.
# In the research below, used model is XGBClassifier and dataset has 22 features.
model = train(XGBClassifier(), df_train[features])
import shap
df_copied = df_train[features].copy(deep=True)
y = pd.DataFrame(df_copied["label"].tolist())
df_copied.drop(columns=["machine_name", "label"], inplace=True)
X = pd.DataFrame(df_copied)
explainer = shap.TreeExplainer(model)
shap_values = explainer.shap_values(X)
def visualize_machine(machine_df, machine_name):
day = machine_df.loc[:, "day"].to_numpy()
broken = machine_df.loc[:, "broken"].to_numpy()
total = machine_df.loc[:, "total"].to_numpy()
label = machine_df.loc[:, "label"].to_numpy()
print(f"Number of anomalous days: {label.sum()}")
fig, ax = plt.subplots(2, 1, figsize=(15, 7))
fig.suptitle(machine_name)
ax[0].plot(day, broken, color="red")
ax[0].grid()
ax[0].set_ylabel("Broken")
ax[1].plot(day, total)
ax[1].grid()
ax[1].set_ylabel("Total")
tmp = label.nonzero()[0]
interval = (tmp[0], tmp[-1])
ax[0].axvspan(day[interval[0]], day[interval[1]], color="g", alpha=0.4)
ax[1].axvspan(day[interval[0]], day[interval[1]], color="g", alpha=0.4)
plt.show()
# Plots below show the continous signal for machine BS13597 with 12 anomalous days and extracted signal which show only those 12 days with 5 days before anomaly.
machine_df = df_train.loc[df_train.loc[:, "machine_name"] == "BS13597", :]
visualize_machine(machine_df, machine_name="BS13597")
mask = machine_df.loc[:, "label"] == 1
index = machine_df.loc[mask].index[0] - 5
# Extracted signal
extracted = machine_df.loc[index:]
visualize_machine(extracted, machine_name="Extracted signal BS13597")
extracted = extracted[features]
extracted.drop(columns=["machine_name", "label"], inplace=True)
# To explain which features contribute the most to the decision, we used shap values and force plot.
# Red features push the output to the higher values -> they say that output should be labeled as 1 (anomaly).
# Blue features say the opposite, that predicted class should be 0.
# Based on the force plots we can observe some interesting outcomes:
# * for every sample in extracted signal,non-anomalous days have negative output and anomalous days have positive -> predictions are correct
# * the greatest impact on predicting class 0 (non-anomalous) are: rolling_broken_mean, broken_min_max, broken_ration_min_max
# * the greatest impact on predicting class 1 (anomalous) are: rolling_broken_sum, expanding_total_sum, expanding_broken_sum
for idx in extracted.index.to_list():
shap.force_plot(
explainer.expected_value, shap_values[idx], X.iloc[idx], matplotlib=True
)
# # 10. RIPPER optimization
# > Use 33% of the data as a test data set and 67% as a training data set. Train RIPPER (from the wittgenstein library), calculate the f1_score on a test set and print the rules by which the RIPPER algorithm makes a decision. Comment on the rules.
# Optimize the RIPPER algorithm hyper-parameters with grid search: k (the number of RIPPERk optimization iterations), prune_size (the proportion of the training set to be used for pruning), and dl_allowance (description length).
# Calculate the f1_score on the test set and print the rules based on which the RIPPER algorithm makes a decision.
# Set the value of the parameter max_rules (Maximum number of rules) to three and max_rule_conds (maximum number of conditions per rule) to two. Are there any differences in the rules?
# What is the accuracy of the RIPPER algorithm compared to other tested algorithms? What are the advantages and disadvantages of the RIPPER algorithm?
# To reduce runtime of the notebook, I will use 18 features.
from sklearn.model_selection import train_test_split
features = ranked_features[:18]
y = df_train["label"]
X = df_train[features]
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.33, random_state=42
)
ripper_clf = lw.RIPPER()
ripper_clf.fit(X_train, y_train)
y_pred = ripper_clf.predict(X_test)
score = f1_score(y_test, y_pred, average="macro")
print(f"F1 macro score on test set: {score}")
print("Rules:\n------------------------------------------------------\n")
print(ripper_clf.ruleset_)
from sklearn.model_selection import GridSearchCV
def train_ripper_grid_search(rules, conds, X_train, y_train, X_test, y_test):
ripper_clf = lw.RIPPER(max_rules=rules, max_rule_conds=conds)
param_grid = {
"k": [1, 2, 3],
"prune_size": [0.1, 0.2, 0.3],
"dl_allowance": [1.0, 1.5, 2.0],
}
grid_search = GridSearchCV(
ripper_clf, param_grid=param_grid, scoring="f1_macro", cv=5
)
grid_search.fit(X_train, y_train)
best_clf = grid_search.best_estimator_
y_pred = best_clf.predict(X_test)
score = f1_score(y_test, y_pred, average="macro")
print(f"F1 macro score on test set: {score}")
print("Rules:\n------------------------------------------------------\n")
print(best_clf.ruleset_)
train_ripper_grid_search(None, None, X_train, y_train, X_test, y_test)
train_ripper_grid_search(
rules=3, conds=2, X_train=X_train, y_train=y_train, X_test=X_test, y_test=y_test
)
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from sklearn.linear_model import Lasso
from sklearn.model_selection import train_test_split
from scipy import integrate
# ## Homework
# * Get the trainning part of weather dataset at https://www.kaggle.com/c/australian-weather-prediction, a goal is to predict RainTomorrow label using binary logistic regression.
# * Explore the variables. Think what to do with categorical variables and missing values.
# * Explore data collinearity.
# * Think about data normalization.
# * Think to compose new features.
# * Using a standard sklearn.linear_model.LogisticRegression model build the classification model.
# * Try to improve the model varying model hyperparameters.
# * Hint: look at the tutorial https://www.kaggle.com/prashant111/logistic-regression-classifier-tutorial for some ideas. Think broader!
# * Make a prediction on a test set with your best model and submit to kaggle competition together with notebook file.
data = "../input/australian-weather-prediction/weatherAUS_train.csv"
data_train = pd.read_csv(data)
data_train.head(10)
df = data_train
df.columns
len(df["Location"].unique()), df["Location"].unique()
len(df["WindGustDir"].unique()), df["WindGustDir"].unique()
len(df["WindGustDir"].unique()), df["WindGustDir"].unique()
# so must be changed 'WindDir9am' 'WindDir3pm'
len(df["RainToday"].unique()), df["RainToday"].unique()
[df[i][:5] for i in df.columns]
np.shape(data_train)
for i in range(len(df.columns)):
# if there 17 and less informative columns
print(
23 - i,
") ",
"~%.2f%% of data is corrupted "
% (
(df.notna().sum(axis=1) < len(df.columns) - i).sum(axis=0)
/ df.shape[0]
* 100
),
)
# where and how much data is corrupted
for i in df.keys():
print(i, (df[i].notna() == False).sum(axis=0))
dfq = df.drop(df[(df.notna().sum(axis=1) < 17)].index).copy()
np.shape(dfq)
dfq.head(10)
dfq["RainToday"] = pd.factorize(dfq["RainToday"])[0]
dfq.head(5)
dir_wind = {
0: 0,
"N": 1,
"NNE": 2,
"NE": 3,
"ENE": 4,
"E": 5,
"ESE": 6,
"SE": 7,
"SSE": 8,
"S": 9,
"SSW": 10,
"SW": 11,
"WSW": 12,
"W": 13,
"WNW": 14,
"NW": 15,
"NNW": 16,
}
dir_wind.get("N")
dir_wind.get(0)
dfq["WindGustDir"] = dfq["WindGustDir"].fillna(value=0)
dfq["WindDir9am"] = dfq["WindDir9am"].fillna(value=0)
dfq["WindDir3pm"] = dfq["WindDir3pm"].fillna(value=0)
dfq["WindGustDirNum"] = dfq["WindGustDir"].apply(lambda x: dir_wind.get(x))
dfq["WindDir9amNum"] = dfq["WindDir9am"].apply(lambda x: dir_wind.get(x))
dfq["WindDir3pmNum"] = dfq["WindDir3pm"].apply(lambda x: dir_wind.get(x))
dfq.tail(10)
# 6 types of cities depending on their geography
city_dict = {
"Uluru": 1,
"AliceSprings": 1,
"Katherine": 2,
"Darwin": 2,
"Cairns": 2,
"Townsville": 2,
"PearceRAAF": 3,
"PerthAirport": 3,
"Perth": 3,
"Witchcliffe": 3,
"Walpole": 3,
"Albany": 3,
"SalmonGums": 3,
"Launceston": 4,
"Hobart": 4,
"Woomera": 5,
"Mildura": 5,
"Cobar": 5,
"Moree": 5,
"Sydney": 6,
"Sale": 6,
"Richmond": 6,
"Wollongong": 6,
"Canberra": 6,
"Nuriootpa": 6,
"Dartmoor": 6,
"Williamtown": 6,
"WaggaWagga": 6,
"GoldCoast": 6,
"Watsonia": 6,
"SydneyAirport": 6,
"Albury": 6,
"MountGinini": 6,
"Nhil": 6,
"BadgerysCreek": 6,
"MelbourneAirport": 6,
"Brisbane": 6,
"MountGambier": 6,
"Bendigo": 6,
"Newcastle": 6,
"Tuggeranong": 6,
"NorahHead": 6,
"CoffsHarbour": 6,
"Penrith": 6,
"Portland": 6,
"NorfolkIsland": 6,
"Melbourne": 6,
"Ballarat": 6,
"Adelaide": 6,
}
# Seasons -- winter and summer
# season_dict = {1 : 0, 2 : 0, 3 : 0, 4 : 0, 5 : 1, 6 : 1, 7 : 1, 8 : 1, 9 : 1, 10 : 1, 11 : 0, 12 : 0}
season_dict = {
1: 2,
2: 2,
3: 3,
4: 3,
5: 3,
6: 4,
7: 4,
8: 4,
9: 1,
10: 1,
11: 1,
12: 2,
}
# 1 - spring (in Australia)
# 2 - summer
# 3 - autumn
# 4 - winter
len(city_dict)
dfq["LocationNum"] = dfq["Location"].apply(lambda x: city_dict.get(x))
dfq["Month"] = dfq["Date"].apply(lambda x: int(x[5:7]))
dfq["Season"] = dfq["Date"].apply(lambda x: season_dict.get(int(x[5:7])))
dfq.head(10)
dfq["Evaporation"].sum() / len(dfq["Evaporation"].dropna()), dfq["Evaporation"].mean()
dfq_prep = dfq.drop(
labels=["Date", "Location", "WindGustDir", "WindDir9am", "WindDir3pm", "Month"],
axis=1,
)
sample = dfq_prep.groupby(["LocationNum", "Season"]).median()
# sample.loc([[3,1, 'MinTemp']])
sample
(sample.notna().sum(axis=1) < len(sample.columns)).sum(axis=0)
for i in range(1, 7): # LocationNum
for j in range(1, 5): # Season
for k in dfq_prep.columns[:-7]: # float numbers
# print(dfq_prep.loc[(i, j), k].head(1))
# dfq_prep.loc[(i, j), k] = dfq_prep.loc[(i, j), k].fillna(sample.loc[(i, j), k])
dfq_prep.loc[
(dfq_prep["LocationNum"] == i) & (dfq_prep["Season"] == j), k
] = dfq_prep[(dfq_prep["LocationNum"] == i) & (dfq_prep["Season"] == j)][
k
].fillna(
sample.loc[(i, j), k]
)
for k in dfq_prep.columns[-7:-2]: # integer numbers
# dfq_prep.loc[(i, j), k] = dfq_prep.loc[(i, j), k].fillna(int(round(sample.loc[(i, j), k])))
dfq_prep.loc[
(dfq_prep["LocationNum"] == i) & (dfq_prep["Season"] == j), k
] = dfq_prep[(dfq_prep["LocationNum"] == i) & (dfq_prep["Season"] == j)][
k
].fillna(
int(round(sample.loc[(i, j), k]))
)
dfq_prep.head()
# the data is ready
print(
"~%.2f%% of data is corrupted "
% (
(dfq_prep.notna().sum(axis=1) < len(dfq_prep.columns)).sum(axis=0)
/ dfq_prep.shape[0]
* 100
)
)
(dfq_prep.notna().sum(axis=1) < len(dfq_prep.columns)).sum(axis=0)
a = np.asarray(dfq_prep.corr())
corr_matrix = a - (np.indices((23, 23))[0] == np.indices((23, 23))[1]).astype(
np.float32
)
print(np.round(np.max(corr_matrix, axis=0), decimals=2))
print(np.round(np.min(corr_matrix, axis=0), decimals=2))
dfq_prep.corr().round(decimals=2)
from statsmodels.stats.outliers_influence import variance_inflation_factor
def calc_vif(X):
# Calculating VIF
vif = pd.DataFrame()
vif["variables"] = X.columns
vif["VIF"] = [variance_inflation_factor(X.values, i) for i in range(X.shape[1])]
return vif
X = dfq_prep.drop(
["Evaporation", "Sunshine", "Pressure9am", "Temp3pm", "Temp9am"], axis=1
)
calc_vif(X)
def standartization(features):
return (features - features.mean(axis=0)) / features.std(axis=0)
dfq_std = standartization(dfq_prep.drop(["RainTomorrow", "RainToday"], axis=1)[::])
dfq_std["RainTomorrow"] = dfq_prep["RainTomorrow"]
dfq_std["RainToday"] = dfq_prep["RainToday"]
dfq_std.head()
# Divide data into test and train
df_in = dfq_std.drop(["RainTomorrow"], axis=1)
X = np.vstack([df_in[df_in.columns[i]] for i in range(len(df_in.columns))]).T
X_train, X_test, y_train, y_test = train_test_split(
X, np.array(dfq_prep["RainTomorrow"]), test_size=0.3, random_state=42
)
X_train, y_train, np.shape(X_train), np.shape(y_train)
from sklearn.linear_model import LogisticRegression
model = LogisticRegression(multi_class="multinomial", solver="lbfgs")
model.fit(X_train, y_train)
y_train_pred = model.predict(X_train)
y_test_pred = model.predict(X_test)
from sklearn.metrics import classification_report
print("Train metrics")
print(classification_report(y_train, y_train_pred))
print("Test metrics")
print(classification_report(y_test, y_test_pred))
data_test = "../input/australian-weather-prediction/weatherAUS_test.csv"
data_test = pd.read_csv(data_test)
data_test.head(10)
data_test["RainToday"] = pd.factorize(data_test["RainToday"])[0]
data_test["WindGustDir"] = data_test["WindGustDir"].fillna(value=0)
data_test["WindDir9am"] = data_test["WindDir9am"].fillna(value=0)
data_test["WindDir3pm"] = data_test["WindDir3pm"].fillna(value=0)
data_test["WindGustDirNum"] = data_test["WindGustDir"].apply(lambda x: dir_wind.get(x))
data_test["WindDir9amNum"] = data_test["WindDir9am"].apply(lambda x: dir_wind.get(x))
data_test["WindDir3pmNum"] = data_test["WindDir3pm"].apply(lambda x: dir_wind.get(x))
data_test["LocationNum"] = data_test["Location"].apply(lambda x: city_dict.get(x))
data_test["Season"] = data_test["Date"].apply(lambda x: season_dict.get(int(x[5:7])))
data_prep = data_test.drop(
labels=["Date", "Location", "WindGustDir", "WindDir9am", "WindDir3pm"], axis=1
)
sample_test = data_prep.groupby(["LocationNum", "Season"]).mean()
for i in range(1, 7): # LocationNum
for j in range(1, 5): # Season
for k in data_prep.columns[:-7]: # float numbers
data_prep.loc[
(data_prep["LocationNum"] == i) & (data_prep["Season"] == j), k
] = data_prep[(data_prep["LocationNum"] == i) & (data_prep["Season"] == j)][
k
].fillna(
sample_test.loc[(i, j), k]
)
for k in data_prep.columns[-7:-2]: # integer numbers
data_prep.loc[
(data_prep["LocationNum"] == i) & (data_prep["Season"] == j), k
] = data_prep[(data_prep["LocationNum"] == i) & (data_prep["Season"] == j)][
k
].fillna(
int(round(sample_test.loc[(i, j), k]))
)
data_std = standartization(data_prep.drop(["RainToday"], axis=1)[::])
data_std["RainToday"] = data_prep["RainToday"]
data_in = data_std.drop(["Id"], axis=1)
X_real = np.vstack([data_in[data_in.columns[i]] for i in range(len(data_in.columns))]).T
y_pred = model.predict(X_real)
y_pred
data_real = "../input/australian-weather-prediction/weatherAUS_test_sample_solution.csv"
data_real = pd.read_csv(data_real)
data_real["RainTomorrow"] = y_pred
data_real.head()
data_real["RainTomorrow"].to_csv("weatherAUS_test_solution.csv")
|
# # Library
import os
import random
import numpy as np
import pandas as pd
from PIL import Image
from tqdm.notebook import tqdm
from scipy import spatial
from sklearn.model_selection import train_test_split
import torch
from torch import nn
from torch.utils.data import Dataset, DataLoader
from torch.optim.lr_scheduler import CosineAnnealingLR
from torchvision import transforms
import timm
from timm.utils import AverageMeter
import sys
sys.path.append("../input/sentence-transformers-222/sentence-transformers")
from sentence_transformers import SentenceTransformer
import warnings
warnings.filterwarnings("ignore")
# # Config
class CFG:
model_name = "vit_base_patch16_224"
model_path = "/kaggle/input/self-train-model/vit-b-self.pth"
input_size = 224
batch_size = 64
num_epochs = 3
lr = 1e-4
seed = 42
def seed_everything(seed):
os.environ["PYTHONHASHSEED"] = str(seed)
random.seed(seed)
np.random.seed(seed)
torch.manual_seed(seed)
if torch.cuda.is_available():
torch.cuda.manual_seed(seed)
torch.backends.cudnn.deterministic = True
seed_everything(CFG.seed)
# # Dataset
class DiffusionDataset(Dataset):
def __init__(self, df, transform):
self.df = df
self.transform = transform
def __len__(self):
return len(self.df)
def __getitem__(self, idx):
row = self.df.iloc[idx]
image = Image.open(row["filepath"])
image = self.transform(image)
prompt = row["prompt"]
return image, prompt
class DiffusionCollator:
def __init__(self):
self.st_model = SentenceTransformer(
"/kaggle/input/sentence-transformers-222/all-MiniLM-L6-v2", device="cpu"
)
def __call__(self, batch):
images, prompts = zip(*batch)
images = torch.stack(images)
prompt_embeddings = self.st_model.encode(
prompts, show_progress_bar=False, convert_to_tensor=True
)
return images, prompt_embeddings
def get_dataloaders(trn_df, val_df, input_size, batch_size):
transform = transforms.Compose(
[
transforms.Resize(input_size),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225]),
]
)
trn_dataset = DiffusionDataset(trn_df, transform)
val_dataset = DiffusionDataset(val_df, transform)
collator = DiffusionCollator()
dataloaders = {}
dataloaders["train"] = DataLoader(
dataset=trn_dataset,
shuffle=True,
batch_size=batch_size,
pin_memory=True,
num_workers=2,
drop_last=True,
collate_fn=collator,
)
dataloaders["val"] = DataLoader(
dataset=val_dataset,
shuffle=False,
batch_size=batch_size,
pin_memory=True,
num_workers=2,
drop_last=False,
collate_fn=collator,
)
return dataloaders
# # Train
def cosine_similarity(y_trues, y_preds):
return np.mean(
[
1 - spatial.distance.cosine(y_true, y_pred)
for y_true, y_pred in zip(y_trues, y_preds)
]
)
def train(trn_df, val_df, model_name, input_size, batch_size, num_epochs, lr):
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
dataloaders = get_dataloaders(trn_df, val_df, input_size, batch_size)
model = timm.create_model(model_name, pretrained=True, num_classes=384)
model.set_grad_checkpointing()
model.to(device)
optimizer = torch.optim.AdamW(model.parameters(), lr=lr)
ttl_iters = num_epochs * len(dataloaders["train"])
scheduler = CosineAnnealingLR(optimizer, T_max=ttl_iters, eta_min=1e-6)
criterion = nn.CosineEmbeddingLoss()
best_score = -1.0
for epoch in range(num_epochs):
train_meters = {
"loss": AverageMeter(),
"cos": AverageMeter(),
}
model.train()
for X, y in tqdm(dataloaders["train"], leave=False):
X, y = X.to(device), y.to(device)
optimizer.zero_grad()
X_out = model(X)
target = torch.ones(X.size(0)).to(device)
loss = criterion(X_out, y, target)
loss.backward()
optimizer.step()
scheduler.step()
trn_loss = loss.item()
trn_cos = cosine_similarity(
X_out.detach().cpu().numpy(), y.detach().cpu().numpy()
)
train_meters["loss"].update(trn_loss, n=X.size(0))
train_meters["cos"].update(trn_cos, n=X.size(0))
print(
"Epoch {:d} / trn/loss={:.4f}, trn/cos={:.4f}".format(
epoch + 1, train_meters["loss"].avg, train_meters["cos"].avg
)
)
val_meters = {
"loss": AverageMeter(),
"cos": AverageMeter(),
}
model.eval()
for X, y in tqdm(dataloaders["val"], leave=False):
X, y = X.to(device), y.to(device)
with torch.no_grad():
X_out = model(X)
target = torch.ones(X.size(0)).to(device)
loss = criterion(X_out, y, target)
val_loss = loss.item()
val_cos = cosine_similarity(
X_out.detach().cpu().numpy(), y.detach().cpu().numpy()
)
val_meters["loss"].update(val_loss, n=X.size(0))
val_meters["cos"].update(val_cos, n=X.size(0))
print(
"Epoch {:d} / val/loss={:.4f}, val/cos={:.4f}".format(
epoch + 1, val_meters["loss"].avg, val_meters["cos"].avg
)
)
if val_meters["cos"].avg > best_score:
best_score = val_meters["cos"].avg
torch.save(model.state_dict(), f"{model_name}.pth")
# df = pd.read_csv('/kaggle/input/diffusiondb-data-cleansing/diffusiondb.csv')
# trn_df, val_df = train_test_split(df, test_size=0.1, random_state=CFG.seed)
# train(trn_df, val_df, CFG.model_name, CFG.input_size, CFG.batch_size, CFG.num_epochs, CFG.lr)
# # Prediction
class DiffusionTestDataset(Dataset):
def __init__(self, images, transform):
self.images = images
self.transform = transform
def __len__(self):
return len(self.images)
def __getitem__(self, idx):
image = Image.open(self.images[idx])
image = self.transform(image)
return image
def predict(images, model_path, model_name, input_size, batch_size):
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
transform = transforms.Compose(
[
transforms.Resize(input_size),
transforms.RandomHorizontalFlip(p=0.5),
# transforms.RandomRotation(degrees=10),
# transforms.RandomVerticalFlip(p=0.5),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225]),
]
)
dataset = DiffusionTestDataset(images, transform)
dataloader = DataLoader(
dataset=dataset,
shuffle=False,
batch_size=1,
pin_memory=True,
num_workers=0,
drop_last=False,
)
model = timm.create_model(model_name, pretrained=False, num_classes=384)
# 加载训练好的模型
state_dict = torch.load(model_path)
model.load_state_dict(state_dict)
model.to(device)
model.eval()
tta_preds = None
for _ in range(2):
preds = []
for X in tqdm(dataloader, leave=False):
X = X.to(device)
with torch.no_grad():
X_out = model(X)
preds.append(X_out.cpu().numpy())
if tta_preds is None:
tta_preds = np.vstack(preds).flatten()
else:
tta_preds += np.vstack(preds).flatten()
return tta_preds / 2
from pathlib import Path
images = list(Path("./input/stable-diffusion-image-to-prompts/images").glob("*.png"))
imgIds = [i.stem for i in images]
EMBEDDING_LENGTH = 384
imgId_eId = [
"_".join(map(str, i))
for i in zip(
np.repeat(imgIds, EMBEDDING_LENGTH),
np.tile(range(EMBEDDING_LENGTH), len(imgIds)),
)
]
prompt_embeddings = predict(
images, CFG.model_path, CFG.model_name, CFG.input_size, CFG.batch_size
)
submission = pd.DataFrame(
index=imgId_eId, data=prompt_embeddings, columns=["val"]
).rename_axis("imgId_eId")
submission.to_csv("submission.csv")
|
# # 🛳 Titanic 3D modeling
# Hey sailor ! 🧜
# In this notebook we try a different an intuitive approach on the titanic dataset.
# We will model the cabins in 3D and then apply a quick kNN algorithm on this new space.
# 
# You can find the titanic decks plan here: https://www.encyclopedia-titanica.org/titanic-deckplans/. You only have to create an account (for free) to see it.
# Our modeling will be an approximation of it, feel free to fork the code and improve it !
# Libraries
import numpy as np
import pandas as pd
import plotly.graph_objects as go
from sklearn.neighbors import NearestNeighbors
from sklearn.preprocessing import StandardScaler
# Get train and test sets
X_train = pd.read_csv("/kaggle/input/titanic/train.csv")
y_train = X_train[["Survived"]]
X_test = pd.read_csv("/kaggle/input/titanic/test.csv")
# # NaN cabins filling
# In the train dataset, we only have 20% of the cabin provided... It's a bad news for our project, we can't fill each passenger in his respective cabin.
# In order to handle this, for each cabinless passenger, we'll find his nearest neighbor who has one, and allocate him to the closest available cabin.
# In order to find the nearest neighbor, we'll use the `['Pclass', 'Fare', 'Embarked', 'Sex']` space and the euclidean distance.
# We have to be careful to not use irrelevant features here, for example, `PassengerId` doesn't contain any useful information about the proximity of passengers, while the `Fare` or `Pclass` features seem interesting (expensive tickets and class might be a good start for finfing similar cabins).
# We note that we use the `Sex` feature here, it may be surprising: indeed we'll have women closer to women and vice-versa. It's a small cheat, because women are more likely to survive than men.
# There are obviously some ways to improve this cabin-filling algorithm, for example the number of children/parents could be mapped, as the potential same names, in order to have family in the same cabins.
# Concatening dataframes for complete cabin filling
X_tot = pd.concat([X_train, X_test])
X_tot.reset_index(drop=True, inplace=True)
# Columns / dimensions selected for the NearestNeighbors cabins filling
X_tot_for_neighbors = X_tot[["Pclass", "Fare"]].copy()
# Label mapping in order to have numeric values
X_tot_for_neighbors["Sex"] = X_tot["Sex"].map({"male": 0, "female": 1})
X_tot_for_neighbors["Embarked"] = X_tot["Embarked"].map({"S": 0, "C": 1, "Q": 2})
# Quick NaN filling with mean
X_tot_for_neighbors = X_tot_for_neighbors.fillna(X_tot_for_neighbors.mean())
# StandardScaler preprocessing
standard_scaler = StandardScaler()
X_tot_for_neighbors = standard_scaler.fit_transform(X_tot_for_neighbors)
# 100 NearestNeighbors fitting
nns = NearestNeighbors(n_neighbors=100, metric="euclidean").fit(X_tot_for_neighbors)
distances, indices = nns.kneighbors(X_tot_for_neighbors) # both of shape (1309, 100)
# We have two arrays returned, `distances` and `indices`, both of shape (1309, 100) (number of total passenger in X_train and X_test, number of neighbors)
# - `distances`: for each row $i$ (each passenger), the ascending euclidean distance of each $k$-neighbor as columns.
# - `indices`: for each row $i$ (each passenger), the index of the corresponding $k$-neighbor as columns
# In our modeling, the cabin will be simple: An association of a deck letter (A, B, C, D, E, F or G) and a number between $1$ and the maximum number of cabins for the deck.
# In the dataset, some cabins need to be reshaped in order to fit with our logic. These reformating takes us away from the real titanic but it's not so important, we don't change too much of them.
# Replacing special cabins (like "CXX CXX CXX" or "F GXXX") by the first cabin in the string
for idx, row in X_tot.iterrows():
cabin = row["Cabin"]
# If the cabin is NaN
if not isinstance(cabin, str):
continue
# Is a space is in the cabin string
if " " in cabin and "F" in cabin:
X_tot.loc[idx, "Cabin"] = row["Cabin"].split(" ")[1]
elif " " in cabin:
X_tot.loc[idx, "Cabin"] = row["Cabin"].split(" ")[0]
# Replacing T cabin by A1 cabin
X_tot.loc[X_tot["Cabin"] == "T", "Cabin"] = "A1"
# Replacing cabins with only one letter by the first cabin of the deck
X_tot.loc[X_tot["Cabin"] == "D", "Cabin"] = "D1"
X_tot.loc[X_tot["Cabin"] == "F", "Cabin"] = "F1"
# Maximum number of cabins per decks (close but not exact).
max_nb_cabins_decks = {
"A": 42,
"B": 120, # 123 in reality
"C": 310,
"D": 280, # 285 in reality
"E": 580, # 583 in reality
"F": 684,
"G": 362,
}
# Now let's dive into the cabin-filling algorithm !
# During a loop, if 0 passenger are allocated (for all of them, none of the 1-nearest-neighbors have a cabin), we'll re-run the loop, looking for the 2-nearest-neighbor cabin. If it's the same for the 2-NN, we do the same for $k=3$, etc. Then once one has found its cabin, we reset to $k=0$.
# We'll run the loops until each cabin has been filled.
# Closest cabin getter
def get_closest_cabin(cabin, X_tot):
"""
Returns the closest available cabin (with the cabin number above or below)
"""
# Deck letter
deck = cabin[0]
# Number of cabin
c_digit = int("".join([l for l in cabin if l.isdigit()]))
# Loop over alternatives number to add to the cabin one (+1, -1, +2, ...)
for k in [
n * -1 if idx % 2 == 1 else n
for idx, n in enumerate(np.repeat(np.arange(1, max_nb_cabins_decks[deck]), 2))
]:
# Clipping the cabin num w.r.t the maximum one for this deck
nb_cabin = np.clip(c_digit + k, 1, max_nb_cabins_decks[deck])
new_cabin = deck + str(nb_cabin)
# Checking if the cabin is available
if len(X_tot[X_tot["Cabin"] == new_cabin]) == 0:
break
return new_cabin
# ------- Cabin filling algorithm
already_filled = []
# Number of NaN cabin at the end of the loop (useful to check if no passenger has been allocated to any cabin during the entire loop)
end_loop_nb_nan_cabins = [0] # Initialize at 0 but not important
# If no passenger allocated during the loop, allocate passengers to the (k+1)-nearest-neighbor
add_to_k = 0
# Running loops until all passengers are allocated to a cabin
while X_tot["Cabin"].isna().sum() > 0:
for i in range(distances.shape[0]):
# If the passenger is already allocated to a cabin
if i in already_filled:
continue
# If the cabin is not NaN we don't have to fill it
if isinstance(X_tot.iloc[i]["Cabin"], str):
already_filled.append(i)
continue
# If the nearest neighbor is the instance itself (most of the case it is but sometimes not, if two instances are the same the second can be at first index)
if indices[i, 0] == i:
j = 1
else:
j = 0
# Select the distance and idx of nearest neighbor (ideally the first neighbor)
dist_nn = distances[i, j + add_to_k]
idx_nn = indices[i, j + add_to_k]
# Getting the corresponding cabin
cabin_nn = X_tot.iloc[idx_nn]["Cabin"]
# We'll run the loop a lot of time until each cabin has been filled by its nearest neighbor
if not isinstance(cabin_nn, str):
continue
# Closest cabin
new_cabin = get_closest_cabin(cabin_nn, X_tot)
# Run it to monitor the passengers allocation
print(f"Index {i}: allocating {X_tot.loc[i, 'Name']} to cabin {new_cabin}")
# New passenger allocated
already_filled.append(i)
X_tot.at[i, "Cabin"] = new_cabin
# compute current number of NaNs cabins and get the one of previous loop
current_loop_nb_nans_cabins = X_tot["Cabin"].isna().sum()
last_loop_nb_nan_cabins = end_loop_nb_nan_cabins.pop()
# If there is currently no position 1-nearest-neighbor with a cabin available (different to nan)
if current_loop_nb_nans_cabins == last_loop_nb_nan_cabins:
add_to_k += 1
else:
add_to_k = 0
end_loop_nb_nan_cabins.append(current_loop_nb_nans_cabins)
print(end_loop_nb_nan_cabins, add_to_k)
# - Ok ! Each passenger has now a cabin. Let's check the number of cabins filled per deck.
# Compute the number of cabins filled per decks
cabins_summary = pd.Series(
{
deck: X_tot[X_tot["Cabin"].str.contains(deck)].count()[0]
for deck in X_tot["Cabin"].apply(lambda x: x[0]).unique()
}
).sort_values()
# Assert each passenger has a cabin
assert cabins_summary.sum() == X_tot.shape[0]
cabins_summary.to_frame("Number of cabins")
# # 3D Modeling
# We model each deck and then concatenate them.
# The functions below are actually not intuitive at all and are likely to be badly coded so can be skipped.
# --------- Functions required for the modeling
marker_symbol = "circle"
def get_cabin_color(passengers, survived_mean):
"""
Get color, marker symbol, opacity and status for each passenger
"""
if survived_mean == 1:
return "green", marker_symbol, 0.7, "Survived"
elif survived_mean > 0 and survived_mean < 1:
return "orange", marker_symbol, 0.7, "Some survived"
elif survived_mean == 0 and isinstance(passengers, list):
return "black", marker_symbol, 0.3, "RIP"
elif np.isnan(survived_mean) and isinstance(passengers, list):
return "gold", marker_symbol, 1, "Test set"
else:
return "lightgrey", marker_symbol, 0.5, "Empty cabin"
def round_side(x, y, sides_range_cabin_n_deck, where, p=1):
"""
Function in order to cuvre a set of points (in order to get a "ship shape")
"""
return (
-np.abs(y[-sides_range_cabin_n_deck:]) ** p
+ np.max(y[-sides_range_cabin_n_deck:]) ** p
+ where
)
# Arbitrary max and min x, not so important
max_x = 5000
min_x = -5000
# --------- Function for coordinates creation of each deck
def create_deck(
deck_letter,
nb_cabins,
z,
first_range_nb_cabins=8,
big_cabin_corner=False,
back_curvature_exponent=3,
front_curvature_exponent=4,
):
# --------- Cabins coordinates
# x
x = (
[0] * first_range_nb_cabins
+ np.sort(
np.repeat(
np.arange(1, (nb_cabins - first_range_nb_cabins * 2 + 1) / 4).astype(
int
),
4,
)
).tolist()
+ [int((nb_cabins - first_range_nb_cabins * 2 + 1) / 4) + 1]
* first_range_nb_cabins
)
# y
y = []
for i in range(1, int(first_range_nb_cabins / 2) + 1):
y.append(i)
y.append(-i)
if big_cabin_corner:
y[-2:] += np.sign(y[-2:]) * 2
y += [3 if i % 2 == 0 else -3 for i in range(nb_cabins - first_range_nb_cabins * 2)]
k = 0
for i in range(first_range_nb_cabins, nb_cabins - first_range_nb_cabins):
if k % 4 == 2 or k % 4 == 3:
y[i] += np.sign(y[i])
k += 1
if deck_letter != "A":
for i in range(1, int(first_range_nb_cabins / 2) + 1):
y.append(i)
y.append(-i)
# Deck specificities
if deck_letter == "A":
x[-2:] = [8, 8]
y.extend([np.max(y), -np.max(y)] * 2)
# Create dataframe for the deck
deck = {f"{deck_letter}1": {"x": x[0], "y": y[0], "z": z}}
deck.update(
{
f"{deck_letter}{i + 2 if (i%2==1 and i >= 1) else i}": {
"x": x[i],
"y": y[i],
"z": z,
}
for i in range(1, nb_cabins)
}
)
deck = pd.DataFrame(deck).T
# First x axis spacing
lin_x = np.linspace(0, max_x, len(deck["x"].unique())).astype(int)
new_x = pd.Series(
[lin_x[i] for i in range(len(deck["x"].unique()))], index=deck["x"].unique()
)
deck["x"] = deck["x"].map(new_x)
# Round the sides points
to_round = first_range_nb_cabins
if deck_letter != "A":
deck["x"].iloc[-to_round:] = round_side(
x, y, to_round, where=deck["x"].iloc[-to_round], p=back_curvature_exponent
)
# deck['y'].iloc[-to_round:] = deck['y'].iloc[-to_round:] + np.sign(deck['y'].iloc[-to_round:])
deck["x"].iloc[:to_round] = -round_side(
x, y, to_round, where=deck["x"].iloc[to_round], p=front_curvature_exponent
)
deck["x"].iloc[: to_round - 2] -= (
deck.iloc[to_round - 3]["x"] - 4 * deck.iloc[to_round - 2]["x"]
)
# deck['y'].iloc[:to_round-2] = deck['y'].iloc[:to_round-2] + np.sign(deck['y'].iloc[:to_round-2])
# Spacing x-axis between the two curvatures
lin_x = np.linspace(
deck[deck["y"] == 2]["x"].min() + 2,
deck[deck["y"] == 2]["x"].max() - 2,
deck[deck["y"] == 4]["x"].count() + 2,
)[1:-1].astype(int)
new_x = pd.Series(lin_x, index=deck[deck["y"] == 4]["x"].astype(int))
other_x = pd.Series(deck["x"].where(~deck["x"].isin(new_x.index)).dropna())
other_x.index = other_x.values
map_x = pd.concat([new_x, other_x]).drop_duplicates()
deck["x"] = deck["x"].map(map_x)
# Filling deck with passengers from dataset
deck["passengers"] = pd.Series(dtype=object)
for cabin in deck.index:
cabin_subset = X_tot[X_tot["Cabin"] == cabin]
if len(cabin_subset["Name"]) > 0:
deck.at[cabin, "passengers"] = cabin_subset["Name"].tolist()
deck.at[cabin, "survived_mean"] = cabin_subset["Survived"].mean()
# Color and marker symbol
deck["color"] = deck.apply(
lambda x: get_cabin_color(x["passengers"], x["survived_mean"])[0], axis=1
)
deck["marker_symbol"] = deck.apply(
lambda x: get_cabin_color(x["passengers"], x["survived_mean"])[1], axis=1
)
deck["marker_opacity"] = deck.apply(
lambda x: get_cabin_color(x["passengers"], x["survived_mean"])[2], axis=1
)
deck["status"] = deck.apply(
lambda x: get_cabin_color(x["passengers"], x["survived_mean"])[3], axis=1
)
return deck
# Creating each decks
deck_A = create_deck(
deck_letter="A", nb_cabins=40, z=6, first_range_nb_cabins=4, big_cabin_corner=True
)
deck_B = create_deck(deck_letter="B", z=5, nb_cabins=92, front_curvature_exponent=7)
deck_C = create_deck(deck_letter="C", z=4, nb_cabins=144, front_curvature_exponent=7.2)
deck_D = create_deck(deck_letter="D", z=3, nb_cabins=80, front_curvature_exponent=7)
deck_E = create_deck(deck_letter="E", z=2, nb_cabins=460, front_curvature_exponent=7.3)
deck_F = create_deck(deck_letter="F", z=1, nb_cabins=272, front_curvature_exponent=7.3)
deck_G = create_deck(deck_letter="G", z=0, nb_cabins=296, front_curvature_exponent=7.3)
# Concatenate them
decks = pd.concat([deck_A, deck_B, deck_C, deck_D, deck_E, deck_F, deck_G])
# Visualization
fig = go.Figure()
# 3D points
# No survived peaople
decks_tmp = decks[decks["color"] == "black"]
fig.add_trace(
go.Scatter3d(
x=decks_tmp["x"],
y=decks_tmp["y"],
z=decks_tmp["z"],
mode="markers",
marker_color=decks_tmp["color"],
marker_symbol=decks_tmp["marker_symbol"],
marker_size=4,
name="Not Survived",
hovertemplate="x: %{x}" + "<br>y: %{y}<br>" + "%{text}",
text=[
f'Cabin: {cabin}<br>People: {decks_tmp.loc[cabin, "passengers"]}<br>{decks_tmp.loc[cabin, "status"]}'
for cabin in decks_tmp.index
],
)
)
# Survived
decks_tmp = decks[decks["color"] == "green"]
fig.add_trace(
go.Scatter3d(
x=decks_tmp["x"],
y=decks_tmp["y"],
z=decks_tmp["z"],
mode="markers",
marker_color=decks_tmp["color"],
marker_symbol=decks_tmp["marker_symbol"],
marker_size=4,
name="Survived",
hovertemplate="x: %{x}" + "<br>y: %{y}<br>" + "%{text}",
text=[
f'Cabin: {cabin}<br>People: {decks_tmp.loc[cabin, "passengers"]}<br>{decks_tmp.loc[cabin, "status"]}'
for cabin in decks_tmp.index
],
)
)
# Empty cabins
decks_tmp = decks[decks["color"] == "lightgrey"]
fig.add_trace(
go.Scatter3d(
x=decks_tmp["x"],
y=decks_tmp["y"],
z=decks_tmp["z"],
mode="markers",
marker_color=decks_tmp["color"],
marker_symbol=decks_tmp["marker_symbol"],
marker_size=4,
name="Empty cabins",
hovertemplate="x: %{x}" + "<br>y: %{y}<br>" + "%{text}",
text=[
f'Cabin: {cabin}<br>People: {decks_tmp.loc[cabin, "passengers"]}<br>{decks_tmp.loc[cabin, "status"]}'
for cabin in decks_tmp.index
],
)
)
# Test set
decks_tmp = decks[decks["color"] == "gold"]
fig.add_trace(
go.Scatter3d(
x=decks_tmp["x"],
y=decks_tmp["y"],
z=decks_tmp["z"],
mode="markers",
marker_color=decks_tmp["color"],
marker_symbol=decks_tmp["marker_symbol"],
marker_size=4,
name="Test set",
hovertemplate="x: %{x}" + "<br>y: %{y}<br>" + "%{text}",
text=[
f'Cabin: {cabin}<br>People: {decks_tmp.loc[cabin, "passengers"]}<br>{decks_tmp.loc[cabin, "status"]}'
for cabin in decks_tmp.index
],
)
)
# Waves
x = np.linspace(decks["x"].min(), decks["x"].max(), 100)
y = np.linspace(decks["y"].min(), decks["y"].max(), 100)
x, y = np.meshgrid(x, y)
z = (np.sin(abs(x) + abs(y) ** 0.2) * np.cos(abs(y) - abs(x) ** 0.4)).T - 1
fig.add_trace(
go.Surface(z=z, x=x, y=y, colorscale="ice", showlegend=False, hoverinfo="none")
)
# Layout
fig.update_layout(
scene=dict(
xaxis=dict(title="x axis", autorange="reversed"),
yaxis=dict(title="y axis"),
zaxis=dict(
ticktext=["A", "B", "C", "D", "E", "F", "G"],
tickvals=[6, 5, 4, 3, 2, 1, 0],
title="Decks",
ticks="outside",
),
),
legend=dict(orientation="h", x=0.3, y=1.1),
title=dict(text="Titanic 3D Modeling", x=0.5)
# paper_bgcolor='rgba(0,0,0,0)',
# plot_bgcolor='rgba(0,0,0,0)'
)
# Let only z-axis visible
fig.update_scenes(xaxis_visible=False, yaxis_visible=False, zaxis_visible=True)
fig.update_coloraxes(showscale=False)
fig.show()
# # Classification in this new space
# Now let's classify the passengers from the test set: we'll use again a $k$-NN, but in our new 3D space.
# For each passenger to be classified, we get the $k=5$ closest cabins (with (x, y, z) vectors euclidean distance), and average the survived feature values of the 5 neighbors.. Then for the prediction we have $0$ if the survived average value is below $0.5$ and $1$ in the other case.
# We note that we obviously don't expect our algorithm to have a strong predictive power (not enough right cabins provided in the dataset and even too strong dimensionnality reduction here actually), so we don't try to get the optimal $k$, it's not so important.
# Let's normalize the coords
decks[["x", "y", "z"]] = (
decks[["x", "y", "z"]] - decks[["x", "y", "z"]].mean()
) / decks[["x", "y", "z"]].std()
# Initialization
k = 5
y_pred = pd.Series(index=X_test["PassengerId"], dtype=int)
for i, row in X_test.iterrows():
name = row["Name"]
# Get the cabin corresponding to the name
cabin = decks["passengers"].explode().loc[lambda x: x == name].index[0]
# Get the corresponding coords
coords = decks.loc[cabin, ["x", "y", "z"]]
# Combpute and sort the euclidean distance with all other passengers
eucl_dist_sorted = (
np.sqrt(((decks[["x", "y", "z"]] - coords) ** 2).sum(1))
.sort_values()
.to_frame("dist")[1:]
)
# Get the prediction for this passenger depending on its k neighbors cabin survived mean
eucl_dist_sorted["survived_mean"] = decks["survived_mean"]
survived_mean = 0 if eucl_dist_sorted.iloc[:k].mean()["survived_mean"] <= 0.5 else 1
decks.loc[cabin, "survived_mean"] = survived_mean
y_pred.loc[row["PassengerId"]] = survived_mean
# Reformat and submit
y_pred = y_pred.to_frame("Survived").reset_index()
y_pred.to_csv("submission.csv", index=False)
|
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
# ## Contents
# - [Introduction](#introduction)
# * [Problem Statement](#problem-statement)
# - [Exploratory Data Analysis](#eda)
# - [Feature Engineering](#feature-engineering)
# - [Model Building](#model-building)
# * [Linear Regression](#linear-regression)
# * [Gradient Bososted Tree](#gradient-boosted-trees)
# - [Model Analysis](#model-analysis)
# - [Conclusion](#conclusion)
# ## Introduction
# Elo, one of Brazil's largest payment brands, is partnered with many merchants to offer promotions and discounts to their cardholders. Elo aimed to reduce marketting that is irrelevant to members and offer them custom-tailored promotions, thereby providing an enjoyable experience and beneficial service. To that end, Elo launched a Kaggle competition, enlisting the Kaggle community's help, to produce a machine learning model that can find signal between trasaction data and loyalty. Such a model will help Elo gauge customer loyalty and how promotional strategies affect it.
# **The data provided is simulated and fictitious. It does not contain real customer data.*
# ### Problem Statement
# Build a machine learning model that can effectively predict customer loyalty scores using trasaction data.
# ## Exploratory Data Analysis
# The data contained five main tables consisting **train data**, **test data**, **historical transactions**, **new merchant transactions** and **merchants**.
# **train data** table contained card_ids, loyalty scores and 3 arbitrary features provided by ELO. The aribtrary features were not very useful as they did not provide much signal in predicting loyalty scores.
# **test data** table contained the same arbitrary features as **train data** and card_id but did not contain loyalty scores.
# **historical transactions** contained details on purchases made by all the cardholders in the dataset. Details included purchase amount, merchant category, etc. Every card_id had atleaset 3 months of records.
# **new merchant transactions** contained transaction data from new merchants the cardholder had not yet purchased from in the historical transaction data. Every card_id had upto two months of new merchant data after a reference date (which differed for each card_id).
# **merchants** contained details on merchants seen in **historical transactions** and **new merchant transactions**
# The data was heavily anonymized. Non-numerical data such as city of purchase and merchant category were reassigned with arbitrary value so it is difficult to connect real world knowledge and find insights.
#
# load data
# train data
traindf = pd.read_csv("../input/elo-merchant-category-recommendation/train.csv")
# given test data with no loyaltly score
giventestdf = pd.read_csv("../input/elo-merchant-category-recommendation/test.csv")
giventestdf["card_id"].nunique()
# historical transaction data
histtransdf = pd.read_csv(
"../input/elo-merchant-category-recommendation/historical_transactions.csv"
)
# new merchant transactional data
newtransdf = pd.read_csv(
"../input/elo-merchant-category-recommendation/new_merchant_transactions.csv"
)
# merchant data
merchdf = pd.read_csv("../input/elo-merchant-category-recommendation/merchants.csv")
# training dataset at glance
traindf.head()
traindf.card_id.nunique()
histtransdf.head()
histtransdf["card_id"].nunique()
histtransdf.shape
newtransdf.card_id.nunique()
merchdf["merchant_id"].nunique()
# There are 201,917 card ids in the training data set.
# There are 123,623 card ids in the test data. This data cannot be used to train the model as it does not have loyalty scores (the response variable)
# Hisotrical transaction data has 325,540 card ids with 29,112,361 transactions. Train and test card ids are both included in this historical data.
# New merchant transactional data set had 290,001 card ids.
# There were 334,633 different merchants in the merchant data.
# plotting loyalty score distribution
traindf["target"].hist()
plt.xlabel("Loyalty Score")
plt.ylabel("Count")
plt.title("Loyalty Score Distribution")
# Loyalty scores are normally distriubted ranging from -10 to 10. There are some outliers at -33.
# ## Feature Engineering
# `NDR` - (New Dollar Ratio) Calculated by dividing amount of dollars
# concatenating all transaction data and adding indicator of new merchant
histtransdf["new"] = False
newtransdf["new"] = True
alltransdf = pd.concat([histtransdf, newtransdf])
alltransdf["purchase_date"] = pd.to_datetime(alltransdf["purchase_date"])
trx.authorized_flag = trx.authorized_flag.apply(lambda x: True if x == "Y" else False)
trx.city_id = trx.city_id.apply(str)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
x = 1
print(type(x))
x = 5
y = 1.4
z = 5j
print(type(x))
print(type(y))
print(type(z))
x = 121
y = 121111111
z = -2122121212121
print(type(x))
print(type(y))
print(type(z))
x = 3.2
y = 5.0
z = -89.59
print(type(x))
print(type(y))
print(type(z))
x = 78e1
y = 55e4
z = -64.7e177
print(type(x))
print(type(y))
print(type(z))
x = 4 + 7j
y = 8j
z = -4j
print(type(x))
print(type(y))
x = 6
y = 4.8
z = 8j
a = float(x)
b = int(y)
c = complex(x)
print(a)
print(b)
print(c)
print(type(a))
print(type(b))
print(type(c))
import random
print(random.randrange(1, 44))
x = int(4)
y = int(7.6)
z = int("7")
print(x)
print(y)
print(z)
x = float(8)
y = float(1.4)
z = float("8")
w = float("7.5")
print(x)
print(y)
print(z)
print(w)
x = str("s4")
y = str(4)
z = str(6.0)
print(x)
print(y)
print(z)
print("Hi")
print("Hi")
a = "Hi"
print(a)
a = """hayaller içinde gün
görmeye bak saklı kalan
güne can vermeye bak."""
print(a)
a = """aşktan yana gülmedim
kara bahtım ben
ölmedim kime geriye
dönmedi."""
print(a)
a = "Hi, people!"
print(a[1])
for x in "muz":
print(x)
a = "Hi, people!"
print(len(a))
txt = "hayat çok güzel!"
print("güzel" in txt)
txt = "hayat çok güzel!"
if "güzel" in txt:
print("Evet, 'güzel' şuan.")
txt = "hayat çok pahalı!"
print("pahalı" not in txt)
txt = "hayat çok pahalı!"
if "pahalı" not in txt:
print("Hayır, 'pahalı' şuan değil.")
|
import os
import tensorflow as tf
from tensorflow.keras import Model, callbacks
from tensorflow.keras.applications.densenet import DenseNet201
from tensorflow.keras.applications.mobilenet_v2 import MobileNetV2, preprocess_input
from tensorflow.keras.layers import Dense, Dropout, GlobalAveragePooling2D, Input
from tensorflow.keras.metrics import TopKCategoricalAccuracy
from tensorflow.keras.preprocessing.image import ImageDataGenerator
from tensorflow.keras.optimizers import Adam
try: # detect TPUs
tpu = tf.distribute.cluster_resolver.TPUClusterResolver.connect() # TPU detection
strategy = tf.distribute.TPUStrategy(tpu)
except ValueError: # detect GPUs
strategy = tf.distribute.MirroredStrategy() # for GPU or multi-GPU machines
# strategy = tf.distribute.get_strategy() # default strategy that works on CPU and single GPU
# strategy = tf.distribute.experimental.MultiWorkerMirroredStrategy() # for clusters of multi-GPU machines
print("Number of accelerators: ", strategy.num_replicas_in_sync)
data_dir = "../input/new-plant-diseases-dataset/New Plant Diseases Dataset(Augmented)/New Plant Diseases Dataset(Augmented)"
train_dir = data_dir + "/train"
test_dir = data_dir + "/valid"
diseases = os.listdir(train_dir)
print("Total disease classes are: {}".format(len(diseases)))
train_datagen_aug = ImageDataGenerator(
rescale=1.0 / 255,
shear_range=0.2,
zoom_range=0.2,
fill_mode="nearest",
rotation_range=20,
width_shift_range=0.2,
height_shift_range=0.2,
horizontal_flip=True,
validation_split=0.2,
) # set validation split
test_datagen_aug = ImageDataGenerator(
rescale=1.0 / 255,
shear_range=0.2,
zoom_range=0.2,
rotation_range=20,
horizontal_flip=True,
)
training_set_aug = train_datagen_aug.flow_from_directory(
directory=train_dir,
target_size=(224, 224), # As we choose 64*64 for our convolution model
batch_size=64,
class_mode="categorical",
subset="training",
)
validation_set_aug = train_datagen_aug.flow_from_directory(
directory=train_dir,
target_size=(224, 224), # As we choose 64*64 for our convolution model
batch_size=64,
class_mode="categorical",
subset="validation",
shuffle=False,
)
label_map = training_set_aug.class_indices
print("Target Classes Mapping Dict:\n")
print(label_map)
label_map = validation_set_aug.class_indices
print("Target Classes Mapping Dict:\n")
print(label_map)
test_set_aug = test_datagen_aug.flow_from_directory(
directory=test_dir,
target_size=(224, 224), # As we choose 64*64 for our convolution model
batch_size=64,
class_mode="categorical",
) # for 2 class binary
label_map = test_set_aug.class_indices
print("Target Classes Mapping Dict:\n")
print(label_map)
with strategy.scope():
image_input = Input(shape=(224, 224, 3))
base_model = tf.keras.applications.InceptionV3(
include_top=False, input_shape=(224, 224, 3)
)
# base_model = MobileNetV2(weights='imagenet', include_top=False, input_shape=(224,224,3))
num_classes = 38
x = base_model(image_input, training=False)
base_model.trainable = True
for layer in base_model.layers[:-10]:
layer.trainable = False
# x = Dense(256,activation = "relu")(x)
# x = Dropout(0.2)(x)
# x = Dense(128,activation = "relu")(x)
x = GlobalAveragePooling2D()(x)
x = Dropout(0.2)(x)
output = Dense(num_classes, activation="softmax")(x)
#
model = Model(inputs=image_input, outputs=output)
print(model.summary())
optimizer = Adam(learning_rate=0.01)
model.compile(
optimizer=optimizer,
loss="categorical_crossentropy",
metrics=[
"accuracy",
"top_k_categorical_accuracy",
TopKCategoricalAccuracy(k=1, name="top1"),
],
)
early_stopping_cb = callbacks.EarlyStopping(monitor="val_loss", patience=3)
reduce_lr = callbacks.ReduceLROnPlateau(
monitor="val_loss", factor=0.2, patience=2, verbose=1, min_lr=1e-7
)
history = model.fit(
training_set_aug,
epochs=30,
verbose=1,
callbacks=[early_stopping_cb, reduce_lr],
validation_data=validation_set_aug,
)
model.evaluate(test_set_aug)
import matplotlib.pyplot as plt
# Plotting
hist = history.history
def show_plt(type):
if type == 1:
plt.plot(hist["accuracy"], label="accuracy")
plt.plot(hist["val_accuracy"], label="val_accuracy")
plt.ylabel("Aaccuracy")
plt.xlabel("Epochs #")
plt.legend()
plt.show()
else:
plt.plot(hist["loss"], label="loss")
plt.plot(hist["val_loss"], label="val_loss")
plt.ylabel("Losss")
plt.xlabel("Epochs #")
plt.legend()
plt.show()
show_plt(1)
show_plt(0)
|
import matplotlib.pyplot as plt
from keras.models import Sequential
from keras.layers import Dense, Flatten, Conv2D, MaxPool2D, Dropout
from keras.utils import np_utils
import pandas as pd
from sklearn.model_selection import train_test_split
from keras.layers.normalization import BatchNormalization
import numpy as np
data = pd.read_csv("../input/digit-recognizer/train.csv")
data.head()
teste = pd.read_csv("../input/digit-recognizer/test.csv")
teste.head()
x_train = data.drop("label", axis=1)
y_train = data["label"]
x_train = x_train.values.reshape(x_train.shape[0], 28, 28, 1)
x_train = x_train.astype("float32")
teste = teste.values.reshape(teste.shape[0], 28, 28, 1)
teste = teste.astype("float32")
x_train /= 255
teste /= 255
y_train = np_utils.to_categorical(y_train, 10)
x_treino, x_teste, y_treino, y_teste = train_test_split(x_train, y_train, test_size=0.2)
classificador = Sequential()
# pré processamento da rede neural
classificador.add(
Conv2D(32, kernel_size=(3, 3), input_shape=(28, 28, 1), activation="relu")
)
classificador.add(BatchNormalization())
classificador.add(MaxPool2D(pool_size=(2, 2)))
# classificador.add(Flatten()) - em redes com mais de uma camada convolucional o flatten é aplicado apenas na ultima camada
classificador.add(Conv2D(32, kernel_size=(3, 3), activation="relu"))
classificador.add(BatchNormalization())
classificador.add(MaxPool2D(pool_size=(2, 2)))
classificador.add(Flatten())
# processamento da rede neural.
classificador.add(Dense(units=128, activation="relu"))
classificador.add(Dropout(0.2))
classificador.add(Dense(units=128, activation="relu"))
classificador.add(Dropout(0.2))
classificador.add(Dense(units=10, activation="softmax"))
# compile da rede
classificador.compile(
loss="categorical_crossentropy", optimizer="adam", metrics=["accuracy"]
)
# fit
classificador.fit(
x_treino, y_treino, epochs=35, batch_size=128, validation_data=(x_teste, y_teste)
)
predicao = classificador.predict(teste)
predicao.shape
predicao = np.argmax(predicao, axis=1)
predicao = pd.Series(predicao)
sub = pd.concat([pd.Series(range(1, 28001)), predicao], axis=1)
sub.columns = ["ImageId", "Label"]
sub
# sub.to_csv("submissao_dig5.csv", index=False)
|
# # Importing the libraries
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.model_selection import train_test_split, StratifiedKFold
from sklearn.metrics import roc_auc_score
from lightgbm import LGBMClassifier
from lightgbm.callback import early_stopping, log_evaluation
from catboost import CatBoostClassifier
color_pallete = sns.color_palette()
plt.style.use("fivethirtyeight")
color_pallete
#
# # Reading Data
# Data paths
orig_data_path = "/kaggle/input/kidney-stone-prediction-based-on-urine-analysis/kindey stone urine analysis.csv"
train_path = "/kaggle/input/playground-series-s3e12/train.csv"
test_path = "/kaggle/input/playground-series-s3e12/test.csv"
sub_file_path = "/kaggle/input/playground-series-s3e12/sample_submission.csv"
# Original Data
orig = pd.read_csv(orig_data_path)
# Training Data
train = pd.read_csv(train_path).drop("id", axis=1)
# Testing Data
test = pd.read_csv(test_path)
# Sample Submission file
sub_file = pd.read_csv(sub_file_path)
# Copying the datasets to new variables
df_train = train.copy()
df_test = test.copy()
df_train.head()
df_test.head()
orig.info()
df_train.info()
df_test.info()
#
# ``` The datasets donot have any missing values ```
# # Data Exploration & Visualization
df_train.describe().T
## Visualizations
# Distributions of Features
fig, axes = plt.subplots(
2,
3,
figsize=(12, 8),
)
for ax, col in zip(axes.flatten(), df_train.columns):
sns.histplot(x=df_train[col], kde=True, ax=ax)
sns.histplot(x=df_test[col], kde=True, ax=ax)
sns.histplot(x=orig[col], kde=True, ax=ax)
ax.legend(labels=["Train", "Test", "Orignial"])
fig.tight_layout()
# * The feature distributions of both the training and testing data looks almost similar
# * Also the feature distribution fo original data set is similar to the traing and test dataset but the "ranges" of features ar different
# Ratio of positive and negative target classes
fig, ((ax1, ax2), (ax3, ax4)) = plt.subplots(2, 2, figsize=(10, 8))
sns.countplot(data=orig, x="target", ax=ax1)
ax1.set_title("Original Data Kidney Stone")
ax2.pie(
orig["target"].value_counts(normalize=True),
explode=[0.01, 0],
labels=[0, 1],
shadow=True,
autopct="%.0f%%",
)
ax2.legend(loc="upper right", bbox_to_anchor=[1.5, 1])
sns.countplot(data=df_train, x="target", ax=ax3)
ax3.set_title("Train Data Kidney Stone")
ax4.pie(
df_train["target"].value_counts(normalize=True),
explode=[0.01, 0],
labels=[0, 1],
shadow=True,
autopct="%.0f%%",
)
ax4.legend(loc="upper right", bbox_to_anchor=[1.5, 1])
fig.tight_layout()
plt.show()
#
# ***
# ``` "target" feature in both orignal and train dataset is slghtly imabalAnced ```
# ***
# # Correlation Heatmap
# Heatmap For correlation
columns = ["gravity", "ph", "osmo", "cond", "urea", "calc"]
fig, axs = plt.subplots(1, 2, figsize=(15, 6))
# Create a mask
mask_train = np.triu(np.ones_like(df_train[columns].corr(), dtype=bool))
mask_test = np.triu(np.ones_like(df_test[columns].corr(), dtype=bool))
# Create a custom divergin palette
cmap = sns.diverging_palette(100, 7, s=75, l=40, n=5, center="light", as_cmap=True)
plt.figure(figsize=(10, 6))
sns.heatmap(
df_train[columns].corr(),
mask=mask_train,
center=0,
annot=True,
fmt=".2f",
cmap=cmap,
ax=axs[0],
).set_title("Correlation in Train Data")
sns.heatmap(
df_test[columns].corr(),
mask=mask_test,
center=0,
annot=True,
fmt=".2f",
cmap=cmap,
ax=axs[1],
).set_title("Correlation in Test Data")
#
# ***
# ``` Correlation between features on both Train and Test dataset is similar ```
# ***
#
# Merging Orginal dataset with df_train dataset
df_full = pd.concat([orig, df_train])
df_full.info()
# Checking for Duplicated values
df_full[df_full.duplicated()]
#
# # Outliers Detection and Removal
# Using boxplots to check for outliers
plt.style.use("bmh")
fig, axs = plt.subplots(2, 3, figsize=(12, 10))
axs[0, 0].boxplot(df_full["gravity"], 0, "gd")
axs[0, 0].set_title("gravity")
axs[0, 1].boxplot(df_full["ph"], 0, "gd")
axs[0, 1].set_title("ph")
axs[0, 2].boxplot(df_full["osmo"])
axs[0, 2].set_title("osmo")
axs[1, 0].boxplot(df_full["calc"])
axs[1, 0].set_title("calc")
axs[1, 1].boxplot(df_full["urea"])
axs[1, 1].set_title("urea")
axs[1, 2].boxplot(df_full["cond"])
axs[1, 2].set_title("cond")
fig.tight_layout()
#
# ***
# ``` From the above boxplots we can see that "gravity" & "ph" have some outliers ```
# ***
# Removing Outliers using quartile
columns = ["gravity", "ph", "osmo", "cond", "urea", "calc"]
# Function to remove the outliers
def remove_outliers(df, column):
Q1 = np.quantile(df[col], 0.25)
Q3 = np.quantile(df[col], 0.75)
IQR = Q3 - Q1
lower_limit = Q1 - 1.5 * IQR
upper_limit = Q3 + 1.5 * IQR
df.drop(df[df[col] > upper_limit].index, inplace=True)
return df
# Removing Outliers
# for col in columns:
# df_full = remove_outliers(df_full, col)
df_full.head()
# ## Featuer Engineering
# df_full["calc_to_ph_ratio"] = df_full["calc"] / df_full["ph"]
# df_full.head()
#
# # Model Training Using LightGBM
#
# Selecting features and target variables
# Using train dataset only
X = df_train.drop("target", axis=1)
y = df_train.target
# Using both train and original dataset
# X = df_full.drop("target", axis=1)
# y = df_full.target
# Splitting the data into training and testing set
X_train, X_val, y_train, y_val = train_test_split(
X, y, test_size=0.10, random_state=1234
)
print("X_train: {}, X_test: {} ".format(X_train.shape, X_val.shape))
print("y_train: {}, y_test: {} ".format(y_train.shape, y_val.shape))
# Simple function to df_train the model
def train_model(X, y, model, parameters):
clf = model(**parameters)
# clf = model()
clf.fit(X, y)
return clf
# Parameters for LGBMClassifier
lgbm_params = {
"n_estimators": 50,
"learning_rate": 0.019,
"max_depth": 3,
"num_leaves": 15,
"colsample_bytree": 0.6,
"min_child_samples": 15,
}
# Training the model
lgbm_model = train_model(X_train, y_train, LGBMClassifier, lgbm_params)
# Predictions
train_preds = lgbm_model.predict_proba(X_train.values)[:, 1]
val_preds = lgbm_model.predict_proba(X_val.values)[:, 1]
print("Train ==> roc_auc_score = {:.4f}".format(roc_auc_score(y_train, train_preds)))
print("Test ===> roc_auc_score = {:.4f}".format(roc_auc_score(y_val, val_preds)))
#
# Plotting Feature Importances
fi = lgbm_model.feature_importances_
col_names = X.columns
# Function to plot the feature importances
def plot_feature_imps(features_importances, feature_names):
fi_df = pd.DataFrame(
{"feature_names": feature_names, "feature_importance": fi}
).sort_values("feature_importance", ascending=False)
plt.figure(figsize=(12, 7))
sns.barplot(data=fi_df, x="feature_names", y="feature_importance")
plt.xlabel("Features", fontdict={"fontsize": 15})
plt.ylabel("Feature Importance", fontdict={"fontsize": 15})
# Plotting the feature importances
plot_feature_imps(fi, col_names)
#
# ``` It seems that feature "calc" has a very high feature importance ```
# # Cross Validation
# StrarifiedKFold cross validation
skf = StratifiedKFold(n_splits=10, shuffle=True, random_state=42)
lgbm_clfs = dict()
cv_roc_auc_scores = []
for fold_id, (train_idx, val_idx) in enumerate(skf.split(X, y)):
X_train, X_val = X.iloc[train_idx], X.iloc[val_idx]
y_train, y_val = y.iloc[train_idx], y.iloc[val_idx]
clf = LGBMClassifier(**lgbm_params)
clf.fit(X_train, y_train)
lgbm_clfs["clf_" + str(fold_id)] = clf
preds = clf.predict_proba(X_val)[:, 1]
score = roc_auc_score(y_val, preds)
cv_roc_auc_scores.append(score)
print(">> Training Fold: {} ==> roc_auc_score: {:.2f}".format(fold_id, score))
print("\n")
# print(" roc_auc_score: {:.2f}\n".format(score))
avg_roc_auc = np.mean(cv_roc_auc_scores)
print("Avg Roc_Auc_Score across 10 folds: {:.2f}".format(avg_roc_auc))
#
# # Prediction on Test dataset
# Predicting probabilities for test dataset
test_preds = []
for clf in lgbm_clfs.values():
preds = clf.predict_proba(df_test[X.columns])[:, 1]
test_preds.append(preds)
# Combining the predictions
combined_preds = np.array(test_preds).sum(0) / 10
# Top 10 predictions
combined_preds[:10]
#
# # Creating a Submission File
sub_file["id"] = df_test["id"]
sub_file["target"] = combined_preds
# Saving the submission file
sub_file.to_csv("submission.csv", index=False)
|
#
# $$$$$$$$$$$$$$$$$$$$$$$$$$$$$$' `$$$$$$$$$$$$$$$$$$$$$$$$$$$$$$
# $$$$$$$$$$$$$$$$$$$$$$$$$$$$' `$$$$$$$$$$$$$$$$$$$$$$$$$$$$
# $$$'`$$$$$$$$$$$$$'`$$$$$$! !$$$$$$'`$$$$$$$$$$$$$'`$$$
# $$$$ $$$$$$$$$$$ $$$$$$$ $$$$$$$ $$$$$$$$$$$ $$$$
# $$$$. `$' \' \$` $$$$$$$! !$$$$$$$ '$/ `/ `$' .$$$$
# $$$$$. !\ i i .$$$$$$$$ $$$$$$$$. i i /! .$$$$$
# $$$$$$ `--`--.$$$$$$$$$ $$$$$$$$$.--'--' $$$$$$
# $$$$$$L `$$$$$^^$$ $$^^$$$$$' J$$$$$$
# $$$$$$$. .' ""~ $$$ $. .$ $$$ ~"" `. .$$$$$$$
# $$$$$$$$. ; .e$$$$$! $$. .$$ !$$$$$e, ; .$$$$$$$$
# $$$$$$$$$ `.$$$$$$$$$$$$ $$$. .$$$ $$$$$$$$$$$$.' $$$$$$$$$
# $$$$$$$$ .$$$$$$$$$$$$$! $$`$$$$$$$$'$$ !$$$$$$$$$$$$$. $$$$$$$$
# $JT&yd$ $$$$$$$$$$$$$$$$. $ $$ $ .$$$$$$$$$$$$$$$$ $by&TL$
# $ $$ $
# $. $$ .$
# `$ $'
# `$$$$$$$$'
#
# Face recognition using Grid Search
# by Alin Cijov
#
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.svm import SVC
from sklearn.datasets import fetch_lfw_people
from sklearn.decomposition import PCA as RandomizedPCA
from sklearn.pipeline import make_pipeline
from sklearn.model_selection import train_test_split
from sklearn.model_selection import GridSearchCV
from sklearn.metrics import classification_report
from sklearn.metrics import confusion_matrix
sns.set(rc={"figure.figsize": (11.7, 8.27)})
# # Dataset
class lfwPeopleDataset:
def __init__(self, min_faces_per_person):
self.faces = fetch_lfw_people(min_faces_per_person=min_faces_per_person)
def draw_sample(self):
fig, ax = plt.subplots(3, 5)
for i, axi in enumerate(ax.flat):
axi.imshow(self.faces.images[i], cmap="bone")
axi.set(
xticks=[],
yticks=[],
xlabel=self.faces.target_names[self.faces.target[i]],
)
def get_features_labels(self):
return self.faces.data, self.faces.target, self.faces.target_names
lfw_ds = lfwPeopleDataset(60)
lfw_ds.draw_sample()
features, labels, target_names = lfw_ds.get_features_labels()
X_train, X_test, y_train, y_test = train_test_split(features, labels, random_state=42)
# # Model
class GridSearch:
def __init__(self):
pca = RandomizedPCA(n_components=150, whiten=True, random_state=42)
svc = SVC(kernel="rbf", class_weight="balanced")
self.model = make_pipeline(pca, svc)
param_grid = {
"svc__C": [1, 5, 10, 50],
"svc__gamma": [0.0001, 0.0005, 0.001, 0.005],
}
self.grid = GridSearchCV(self.model, param_grid)
def fit(self, X, y):
print(self.grid.best_params_)
self.model = self.grid.best_estimator_
def predict(self, X):
return self.model.predict(X)
def plot_sample(self, target_names, y_hat):
fig, ax = plt.subplots(4, 6)
for i, axi in enumerate(ax.flat):
axi.imshow(X_test[i].reshape(62, 47), cmap="bone")
axi.set(xticks=[], yticks=[])
axi.set_ylabel(
target_names[y_hat[i]].split()[-1],
color="black" if y_hat[i] == y_test[i] else "red",
)
fig.suptitle("Predicted Names; Incorrect Labels in Red", size=14)
def report(self, target_names, y, y_hat):
print(classification_report(y, y_hat, target_names=target_names))
def heatmap(self, target_names, y, y_hat):
mat = confusion_matrix(y, y_hat)
sns.heatmap(
mat.T,
square=True,
annot=True,
fmt="d",
cbar=False,
xticklabels=target_names,
yticklabels=target_names,
)
plt.xlabel("true label")
plt.ylabel("predicted label")
grid = GridSearch()
grid.fit(X_train, y_train)
# # Analysis
y_hat = grid.predict(X_test)
grid.plot_sample(target_names, y_hat)
grid.report(target_names, y_test, y_hat)
grid.heatmap(target_names, y_test, y_hat)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) zwill list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 5GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import torch
import pandas as pd
import torch.optim as optim
import numpy as np
torch.manual_seed(1)
device = torch.device("cpu")
# 데이터
xy_data = pd.read_csv("../input/mlregression-cabbage-price/train_cabbage_price.csv")
x_test = pd.read_csv("../input/mlregression-cabbage-price/test_cabbage_price.csv")
submit = pd.read_csv("../input/mlregression-cabbage-price/sample_submit.csv")
xy_data
xy_np = xy_data.to_numpy()
xy_np[:, 0] %= 10000
x_train = xy_np[:, 0:-1]
y_train = xy_np[:, -1]
from sklearn.neighbors import KNeighborsRegressor
regressor = KNeighborsRegressor(n_neighbors=3, weights="distance")
regressor.fit(x_train, y_train)
x_test
x_test_np = x_test.to_numpy()
x_test_np[:, 0] %= 10000
x_test_np.shape
x_train.shape
predict = regressor.predict(x_test_np)
predict
submit
for i in range(len(predict)):
submit["Expected"][i] = predict[i]
submit = submit.astype(np.int32)
submit.to_csv("submit.csv", mode="w", header=True, index=False)
submit
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
train = pd.read_csv("/kaggle/input/classificationcopy/data_inlf_train.csv")
test = pd.read_csv("/kaggle/input/classificationcopy/data_inlf_test.csv")
train.head
train.shape, test.shape
train.columns
# You have two datasets. One for training, another one for testing. Training data have these columns:
# kidslt6: kids at home under six
# age: years old
# educ: years of education
# huseduc: husband years of education
# husage: husband years old
# huswage : husband wage in dollars
# motheduc: mother years of education
# fatheduc: father years of education
# **inlf: 1 = in labor force, 0 = not in labor force (target)**
# The goal is to properly predict whether a person is in labor force or not. Testing data don't include (of course) a target, you have to predict it.
train.isnull().sum()
# ## Splitting data
X = train.drop(["inlf"], axis=1)
y = train["inlf"]
# ## Modeling
from sklearn.tree import DecisionTreeClassifier
clf = DecisionTreeClassifier()
clf.fit(X, y)
predictions = clf.predict(X)
predictions[:10]
# ## Metric - Accuracy
from sklearn import metrics
train_acc = metrics.accuracy_score(y, predictions)
train_acc
# ## Prepare Submission
predictions = clf.predict(test)
predictions[:10]
submission = pd.DataFrame(np.array(predictions), columns=["Expected"])
submission.head()
submission["id"] = submission.index
submission.head()
submission[["id", "Expected"]].to_csv("submission_clf.csv", index=False)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# ## TV Shows and Movies listed on Netflix
# - This dataset consists of tv shows and movies available on Netflix as of 2019. The dataset is collected from Flixable which is a third-party Netflix search engine.
# - In 2018, they released an interesting report which shows that the number of TV shows on Netflix has nearly tripled since 2010. The streaming service’s number of movies has decreased by more than 2,000 titles since 2010, while its number of TV shows has nearly tripled. It will be interesting to explore what all other insights can be obtained from the same dataset.
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.filterwarnings("ignore")
plt.style.use("fivethirtyeight")
sns.set_style("darkgrid")
import plotly
import plotly.graph_objs as go
import plotly.express as px
from plotly.offline import download_plotlyjs, plot, init_notebook_mode, iplot
df = pd.read_csv("/kaggle/input/netflix-shows/netflix_titles.csv")
df.head()
df.tail()
df.shape
df.info()
# Check if there is any duplicated records
df.duplicated().any()
# Number of unique data for each column
df.nunique()
# missing values
df.isnull().sum()
# - we can observe **11** of the datatype are **object** and **1 is integer**.
plt.figure(figsize=(17, 10))
sns.heatmap(df.isnull(), yticklabels=False, cbar=False, cmap="viridis")
df["country"].value_counts()
# ### Comparison of Tv Shows and Movies
movies_count = df[df.type == "Movie"]
tvshows_count = df[df["type"] == "TV Show"]
plt.figure(figsize=(10, 8))
sns.countplot(x="type", data=df, palette="viridis")
# ### Movies and TV Shows Content Comparison
colors = ["#809fff", "#66ff66"]
trace = go.Pie(
labels=["Movies", "Tv Show"],
values=[movies_count.type.count(), tvshows_count.type.count()],
hoverinfo="label+percent",
textinfo="label+percent",
marker=dict(colors=colors, line=dict(color="#2d2d2d", width=2)),
)
fig = go.Figure(data=[trace])
iplot(fig)
year_wise_content = df.release_year.value_counts().index[:20]
year_wise_content
plt.figure(figsize=(12, 10))
sns.countplot(data=df, y="release_year", order=year_wise_content, palette="viridis")
# ### Indian Content On Netflix
netflix_india = df[df.country == "India"]
netflix_india.head()
plt.figure(figsize=(12, 10))
sns.set_style("dark")
sns.countplot(
data=netflix_india,
y="release_year",
order=netflix_india.release_year.value_counts().index[:20],
palette="viridis",
)
# ## Let's see the Trends of the country which invented Netflix (USA)
US = df[df["country"] == "United States"]
US.head()
plt.figure(figsize=(8, 6))
sns.countplot(x="type", data=US)
# Get current axis on current figure
ax = plt.gca()
# ylim max value to be set
y_max = US["type"].value_counts().max()
ax.set_ylim([0, 2000])
# Iterate through the list of axes' patches
for p in ax.patches:
ax.text(
p.get_x() + p.get_width() / 2.0,
p.get_height(),
"%d" % int(p.get_height()),
fontsize=12,
color="black",
ha="center",
va="bottom",
)
plt.title("Comparison of Total TV Shows & Movies", size="15")
plt.show()
plt.figure(figsize=(8, 6))
US_5 = US[US.release_year >= 2016]
g = sns.catplot(
data=US_5,
kind="count",
x="release_year",
hue="type",
ci="sd",
palette="dark",
alpha=0.6,
height=6,
)
ax = plt.gca()
# ylim max value to be set
y_max = US_5["type"].value_counts().max()
ax.set_ylim([0, 350])
# Iterate through the list of axes' patches
for p in ax.patches:
ax.text(
p.get_x() + p.get_width() / 2.0,
p.get_height(),
"%d" % int(p.get_height()),
fontsize=12,
color="black",
ha="center",
va="bottom",
)
plt.title("Last 5 years trends in Netflix by USA", size="15")
plt.show()
movies = df[df.type == "Movie"]
shows = df[df.type == "TV Show"]
plt.figure(figsize=(9, 7))
top_contributors = (
movies.groupby(["country"])["show_id"]
.count()
.reset_index(name="count")
.sort_values(by="count", ascending=False)
)
sns.barplot(x="country", y="count", data=top_contributors.head(5))
# Get current axis on current figure
ax = plt.gca()
# ylim max value to be set
y_max = top_contributors["country"].value_counts().max()
ax.set_ylim([0, 2000])
# Iterate through the list of axes' patches
for p in ax.patches:
ax.text(
p.get_x() + p.get_width() / 2.0,
p.get_height(),
"%d" % int(p.get_height()),
fontsize=12,
color="black",
ha="center",
va="bottom",
)
plt.title("Contribution by country in Movies", size="15")
plt.show()
|
# Import statements
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
from scipy import stats
from sklearn.preprocessing import MinMaxScaler
from sklearn import tree
from sklearn.model_selection import cross_val_score
from sklearn.ensemble import RandomForestClassifier
from sklearn import metrics
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# Working with the data files, splitting into train and test sets and dropping any row or columns I needed in order for the input into the models to work out.
# Just separating my training and testing data with easy variable names for me to remember
trainData = pd.read_csv("/kaggle/input/homework1/train.csv")
trainData.drop("id", axis=1, inplace=True)
# Until I dropped these for both train and test data, my tree and forest were not liking my inputs
trainDataInput = trainData.drop("Bankrupt", axis=1)
trainDataOutput = trainData["Bankrupt"]
testData = pd.read_csv("/kaggle/input/homework1/test.csv")
testData.drop("id", axis=1, inplace=True)
# Describing the data so I see columns, rows, etc.
trainData.describe()
# I can find any missing values by finding the sum of the isna function in pandas. If I iterate through my loop of columns and check for any missing values and only iterate numMissing if I have any. Then print out the value of numMissing will tell me how many missing values I have.
# In stats there is a function to give me zscores so I can just check if the absolute value of a z score is larger than 3. This allows me to find any outliers I may have.
# Looking for missing values
# The .sum() returns the sum of my isna() function, anytime the return is 1 that says there is a missing value
numMissing = 0
for i in trainData.columns:
if trainData[i].isna().sum() != 0:
numMissing = numMissing + 1
# Looking for outliers in my axis 1 since that's where they would be shown
# Using .all() gave me everything iterated at once
trainOutlier = trainData[(np.abs(stats.zscore(trainData)) > 3).all(axis=1)]
print("Missing Values: " + str(numMissing))
print("Outliers: " + str(len(trainOutlier)))
# Since I have zero missing values and zero outliers I don't have the need to implement anything to handle them
# # Explaining some of my choices with the models
# So first off I tried both the tree and the random forest with criterion = 'gini' but the accuracies I had with criterion='entropy' was far worse.
# I also tried to normalize my data but the scores I got were lower for that as well. Since standardization doesn't work well with non-Gaussian distributions I decided against using it. The predictions from my raw data actually worked out quite well compared to my normalized data anyways.
# I will try to change any other parameters if I need, but these seemed to be the basic ones and I wanted to start simple.
# Note, Most of the other parameters seem to go further than I need, in a different course I had taken I very rarely had more than 3 or 4 parameters on more complicated models. So it didn't make sense to add any extra inputs since this one passed the benchmark.
# Normalizing data
trainNorm = MinMaxScaler().fit(trainDataInput)
trainDataNorm = trainNorm.transform(trainDataInput)
# Again I didn't actually use this for my predictions
# But I wanted to keep this code in case I decide to use it at a later time
# Actually making the tree and random forest
myTree = tree.DecisionTreeClassifier(criterion="entropy")
myTree.fit(trainDataInput, trainDataOutput)
myForest = RandomForestClassifier(max_depth=6, criterion="entropy")
myForest.fit(trainDataInput, trainDataOutput)
# Separating the outcome to find the column with the actual predictions
myTreeProba = myTree.predict_proba(testData)
myTreePredictions = myTreeProba[:, 1]
myForestProba = myForest.predict_proba(testData)
myForestPredictions = myForestProba[:, 1]
# Sending predictions to a csv so I can see them
myTreeOutput = pd.DataFrame({"Bankrupt": myTreePredictions})
myTreeOutput.to_csv("treePredictions.csv", index=True, index_label="id")
myForestOutput = pd.DataFrame({"Bankrupt": myForestPredictions})
myForestOutput.to_csv("forestPredictions.csv", index=True, index_label="id")
# RocAuc Score
treeRocAucScore = metrics.roc_auc_score(trainDataOutput, myTree.predict(trainDataInput))
RFSRocAucScore = metrics.roc_auc_score(
trainDataOutput, myForest.predict(trainDataInput)
)
# F1 Score
treeF1Score = metrics.f1_score(trainDataOutput, myTree.predict(trainDataInput))
RFSF1Score = metrics.f1_score(trainDataOutput, myForest.predict(trainDataInput))
# Accuracy Score
treeAccScore = metrics.accuracy_score(trainDataOutput, myTree.predict(trainDataInput))
RFSAccScore = metrics.accuracy_score(trainDataOutput, myForest.predict(trainDataInput))
# Printing everything
print("ROC Auc Scores")
print("Tree Model: " + str(treeRocAucScore) + " RFS Model: " + str(RFSRocAucScore))
print("F1 Scores")
print("Tree Model: " + str(treeF1Score) + " RFS Model: " + str(RFSF1Score))
print("Accuracy Scores")
print("Tree Model: " + str(treeAccScore) + " RFS Model: " + str(RFSAccScore))
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# import necessary libraries
import numpy as np
import pandas as pd
dataset = pd.read_csv("/kaggle/input/data-set-new/IRIS.csv")
dataset.head()
x = dataset.iloc[:, :-1]
y = dataset.iloc[:, -1]
print(x)
print(y)
# Splitting dataset into training set & test set
from sklearn.model_selection import train_test_split
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.2, random_state=0)
from sklearn.preprocessing import StandardScaler
sc = StandardScaler()
x_train = sc.fit_transform(x_train)
x_test = sc.fit_transform(x_test)
from sklearn.decomposition import PCA
pca = PCA()
x_train = pca.fit_transform(x_train)
x_test = pca.fit_transform(x_test)
print(x_train)
explained_variance = pca.explained_variance_ratio_
from sklearn.decomposition import PCA
pca = PCA(n_components=1)
x_train = pca.fit_transform(x_train)
x_test = pca.fit_transform(x_test)
print(x_train)
from sklearn.ensemble import RandomForestClassifier
classifier = RandomForestClassifier(max_depth=2, random_state=0)
classifier.fit(x_train, y_train)
# Predicting test set results
y_pred = classifier.predict(x_test)
print(y_train)
print(y_pred)
from sklearn.metrics import confusion_matrix
from sklearn.metrics import accuracy_score
cm = confusion_matrix(y_test, y_pred)
print(cm)
print(accuracy_score(y_test, y_pred))
|
# #### EEMT 5400 IT for E-Commerce Applications
# ##### HW4 Max score: (1+1+1)+(1+1+2+2)+(1+2)+2
# You will use two different datasets in this homework and you can find their csv files in the below hyperlinks.
# 1. Car Seat:
# https://raw.githubusercontent.com/selva86/datasets/master/Carseats.csv
# 2. Bank Personal Loan:
# https://raw.githubusercontent.com/ChaithrikaRao/DataChime/master/Bank_Personal_Loan_Modelling.csv
# #### Q1.
# a) Perform PCA for both datasets. Create the scree plots (eigenvalues).
# b) Suggest the optimum number of compenents for each dataset with explanation.
# c) Save the PCAs as carseat_pca and ploan_pca respectively.
# ### (a)
# import data
import pandas as pd
import numpy as np
import sklearn
from sklearn import preprocessing
CarSeat = pd.read_csv(
"https://raw.githubusercontent.com/selva86/datasets/master/Carseats.csv"
)
BPL = pd.read_csv(
"https://raw.githubusercontent.com/ChaithrikaRao/DataChime/master/Bank_Personal_Loan_Modelling.csv"
)
# Check the datatype of columns
CarSeat.dtypes
BPL.dtypes
# For non-numerical data,we delete it
CS_numeric = CarSeat.select_dtypes(include=["number"])
BPL_numeric = BPL.select_dtypes(include=["number"])
# Min-Max Scaler
min_max_scaler = preprocessing.MinMaxScaler()
min_max_scaler.fit(CS_numeric)
CarSeat_min_max_scaled = min_max_scaler.transform(CS_numeric)
min_max_scaler.fit(BPL_numeric)
BPL_min_max_scaled = min_max_scaler.transform(BPL_numeric)
# PCA
from sklearn.decomposition import PCA
pca = PCA(n_components=5)
# Missing Values input
from sklearn.impute import SimpleImputer
mean_imputer = SimpleImputer()
CarSeat_min_max_scaled = pd.DataFrame(
CarSeat_min_max_scaled, columns=CS_numeric.columns
)
mean_imputer.fit_transform(CarSeat_min_max_scaled)
CS_min_max_scaled_imputed = pd.DataFrame(
CarSeat_min_max_scaled, columns=CarSeat_min_max_scaled.columns
)
# Missing Values input
BPL_min_max_scaled = pd.DataFrame(BPL_min_max_scaled, columns=BPL_numeric.columns)
mean_imputer.fit_transform(BPL_min_max_scaled)
BPL_min_max_scaled_imputed = pd.DataFrame(
BPL_min_max_scaled, columns=BPL_min_max_scaled.columns
)
pca.fit(CS_min_max_scaled_imputed)
print(pca.explained_variance_ratio_)
print(pca.explained_variance_)
pca.fit(BPL_min_max_scaled_imputed)
print(pca.explained_variance_ratio_)
print(pca.explained_variance_)
# Scree plot for BPL
import matplotlib.pyplot as plt
principalComponents = pca.fit_transform(CarSeat_min_max_scaled)
pca.explained_variance_ratio_
importance = pca.explained_variance_ratio_
plt.scatter(range(1, 6), importance)
plt.plot(range(1, 6), importance)
plt.title("Scree Plot of CarSeat")
plt.xlabel("Factors")
plt.ylabel("Eigenvalue")
plt.grid()
plt.show()
# Scree plot for BPL
principalComponents = pca.fit_transform(BPL_min_max_scaled)
pca.explained_variance_ratio_
importance = pca.explained_variance_ratio_
plt.scatter(range(1, 6), importance)
plt.plot(range(1, 6), importance)
plt.title("Scree Plot for BPL")
plt.xlabel("Factors")
plt.ylabel("Eigenvalue")
plt.grid()
plt.show()
# ### (b) Suggest the optimum number of compenents for each dataset with explanation.
# We apply 95% as the benchmark
for i in range(9):
pca = PCA(n_components=i)
pca.fit(CS_min_max_scaled_imputed)
pca.explained_variance_ratio_
importance = pca.explained_variance_ratio_
if sum(importance) >= 0.95:
print("The optimum number of components for CarSeat data is:", i)
break
# We apply 95% as the benchmark
for i in range(15):
pca = PCA(n_components=i)
pca.fit(BPL_min_max_scaled_imputed)
pca.explained_variance_ratio_
importance = pca.explained_variance_ratio_
if sum(importance) >= 0.95:
print("The optimum number of components for BPL data is:", i)
break
# ### (c) Save the PCAs as carseat_pca and ploan_pca respectively.
pca = PCA(n_components=7)
pca.fit(CS_min_max_scaled_imputed)
PCA_components_CS = pd.DataFrame(
pca.components_,
columns=CS_min_max_scaled_imputed.columns,
index=["PC {}".format(i + 1) for i in range(pca.n_components_)],
).transpose()
PCA_components_CS.to_csv("CarSeat_PCA.csv")
pca = PCA(n_components=9)
pca.fit(BPL_min_max_scaled_imputed)
PCA_components = pd.DataFrame(
pca.components_,
columns=BPL_min_max_scaled_imputed.columns,
index=["PC {}".format(i + 1) for i in range(pca.n_components_)],
).transpose()
PCA_components.to_csv("BPL_PCA.csv")
# #### Q2. (Car Seat Dataset)
# a) Convert the non-numeric variables to numeric by using get_dummies() method in pandas. Use it in this question.
# b) Use the scikit learn variance filter to reduce the dimension of the dataset. Try different threshold and suggest the best one.
# c) Some columns may have high correlation. For each set of highly correlated variables, keep one variable only and remove the rest of highly correlated columns. (Tips: You can find the correlations among columns by using .corr() method of pandas dataframe. Reference: https://pandas.pydata.org/docs/reference/api/pandas.DataFrame.corr.html)
# d) Perform linear regression to predict the Sales with datasets from part b and part c respectively and compare the result
# ### a) Convert the non-numeric variables to numeric by using get_dummies() method in pandas. Use it in this question.
CS_one_hot = pd.get_dummies(CarSeat)
CS_one_hot.head(5)
# ### b) Use the scikit learn variance filter to reduce the dimension of the dataset. Try different threshold and suggest the best one.
#
from sklearn.feature_selection import VarianceThreshold
threshold_num = [0, 0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9, 1.0]
output = []
for i in threshold_num:
zero_var_filter = VarianceThreshold(threshold=i)
zero_var_filter.fit(CS_one_hot)
# Indicate the columns with variance larger than one
a = zero_var_filter.get_support()
output.append(a)
output
# As we can see, from threshold = 0.3,the output doesn't change. So we choose threshold = 0.3.
b_df = CS_one_hot[
[
"Sales",
"CompPrice",
"Income",
"Advertising",
"Population",
"Price",
"Age",
"Education",
]
]
#
# ### c) Some columns may have high correlation. For each set of highly correlated variables, keep one variable only and remove the rest of highly correlated columns. (Tips: You can find the correlations among columns by using .corr() method of pandas dataframe.
Corr = CS_one_hot.corr()
Corr
c_df = CS_one_hot[
[
"Sales",
"CompPrice",
"Income",
"Advertising",
"Population",
"Age",
"Education",
"ShelveLoc_Bad",
"Urban_Yes",
]
]
# ### d) Perform linear regression to predict the Sales with datasets from part b and part c respectively and compare the result
# Linear regression with (b) dataset
# Fit the data into linear regression model
from sklearn.linear_model import LinearRegression
lr = LinearRegression()
data_by = b_df[["Sales"]]
data_bx = b_df.drop(columns=["Sales"])
lr.fit(X=data_bx, y=data_by)
# Return the coefficients in pandas series
pd.Series(lr.coef_[0], index=data_bx.columns)
# Return the R-squared
lr.score(X=data_bx, y=data_by)
# Linear regression with (c) dataset
# Fit the data into linear regression model
data_cy = c_df[["Sales"]]
data_cx = c_df.drop(columns=["Sales"])
lr.fit(X=data_cx, y=data_cy)
# Return the coefficients in pandas series
pd.Series(lr.coef_[0], index=data_cx.columns)
# Return the R-squared
lr.score(X=data_cx, y=data_cy)
# # The result of b is better than c.
# #### Q3. (Bank Personal Loan Dataset)(a) Find the variable which has the highest correlations with CCAvg
import seaborn as sn
corr_matrix = BPL.corr()
plt.figure(figsize=(10, 8), dpi=100)
sn.heatmap(corr_matrix, annot=True)
plt.show()
# From the graph we find that the Income factor has the highest correlation with CCAvg.
# ### b) Perform polynomial regression to predict CCAvg with the variable identified in part a.
# ##### Tips:
# step 1 - convert the dataset to polynomial using PolynomialFeatures from scikit learn (https://scikit-learn.org/stable/modules/generated/sklearn.preprocessing.PolynomialFeatures.html)
# step 2 - Perform linear regression using scikit learn
BPL[["Income", "CCAvg"]]
from sklearn.preprocessing import PolynomialFeatures
X = BPL[["Income"]]
y = BPL[["CCAvg"]]
poly = PolynomialFeatures(4)
# Transform income to the power of two dimension
X2 = poly.fit_transform(X)
X2.shape
# Only Linear Regression with simple data
lin_reg = LinearRegression()
lin_reg.fit(X, y)
y_predict = lin_reg.predict(X)
plt.scatter(X, y)
plt.plot(X, y_predict, color="r")
plt.show()
# R-sqaure of simple linear regression
lin_reg.score(X, y)
# RE-apply linear regression
lin_reg2 = LinearRegression()
lin_reg2.fit(X2, BPL[["CCAvg"]])
y_predict2 = lin_reg2.predict(X2)
plt.scatter(X, y)
plt.plot(X, y_predict2, color="r")
plt.show()
# R-sqaure of polynomial linear regression
lin_reg2.score(X2, y)
|
import pandas as pd
import numpy as np
from glob import glob
from collections import defaultdict
from tqdm import tqdm
import time
import os
import copy
import gc
from PIL import Image
# visualization
import cv2
import matplotlib.pyplot as plt
from scipy import spatial
# Sklearn
# PyTorch
import torch
import torch.nn as nn
import torch.optim as optim
from torch.optim import lr_scheduler
from torch.utils.data import Dataset, DataLoader
from torch.cuda import amp
from torchvision.ops import sigmoid_focal_loss
from sklearn.metrics import confusion_matrix
# import timm
# Albumentations for augmentations
import albumentations as A
from albumentations.pytorch import ToTensorV2
import sys
sys.path.append("../input/sentence-transformers-222/sentence-transformers")
from sentence_transformers import SentenceTransformer
from glob import glob
from diffusers.pipelines.stable_diffusion.pipeline_stable_diffusion_img2img import *
# import vit_pytorch
# For colored terminal text
from colorama import Fore, Back, Style
c_ = Fore.GREEN
sr_ = Style.RESET_ALL
import warnings
warnings.filterwarnings("ignore")
class CFG:
seed = 2307
debug = False # set debug=False for Full Training
comment = ""
folds = 10
train_bs = 12
valid_bs = 12
epochs = 3
lr = 12e-6
scheduler = "CosineAnnealingLR"
min_lr = 1e-7
T_max = int(30000 / train_bs * epochs) + 50
T_0 = 25
warmup_epochs = 1
wd = 1e-2
img_size = (512, 512)
n_accumulate = 1
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")
def set_seed(seed=42):
np.random.seed(seed)
# random.seed(seed)
torch.manual_seed(seed)
torch.cuda.manual_seed(seed)
# When running on the CuDNN backend, two further options must be set
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = False
# Set a fixed value for the hash seed
os.environ["PYTHONHASHSEED"] = str(seed)
print("> SEEDING DONE")
set_seed(CFG.seed)
df = pd.read_csv("/kaggle/input/sd2gpt2/gpt_generated_prompts.csv")
df
def load_img(path):
img = Image.open(path)
img = img.resize(CFG.img_size)
return img
class DiffusionDataset(Dataset):
def __init__(self, df, transforms):
self.df = df
self.transforms = transforms
def __len__(self):
return len(self.df)
def __getitem__(self, idx):
row = self.df.iloc[idx]
image = load_img(
f"/kaggle/input/sd2gpt2/gpt_generated_images/gpt_generated_images/{idx}.png"
)
image = preprocess(image).squeeze()
return (
torch.tensor(image),
f"/kaggle/input/sd2gpt2/gpt_generated_images/gpt_generated_images/{idx}.png",
)
data_transforms = {
"train": A.Compose(
[
A.Sharpen(),
A.Flip(),
A.ShiftScaleRotate(shift_limit=0.2, scale_limit=0.5, rotate_limit=35),
A.Cutout(num_holes=10, max_h_size=16, max_w_size=16),
A.ColorJitter(
brightness=0.15, contrast=[0.8, 1.2], saturation=[0.7, 1.3], hue=0.05
),
A.Normalize(),
],
p=1.0,
),
"valid": A.Compose([A.Normalize()], p=1.0),
}
def prepare_loaders(df, debug=False):
valid_dataset = DiffusionDataset(df, transforms=data_transforms["valid"])
valid_loader = DataLoader(
valid_dataset,
batch_size=CFG.valid_bs if not debug else 3,
num_workers=4,
shuffle=False,
pin_memory=True,
)
return valid_loader
valid_loader = prepare_loaders(df=df, debug=False)
batch = next(iter(valid_loader))
img, p = batch
print(img.shape, len(p))
class revDiff(nn.Module):
def __init__(self):
super().__init__()
self.base = StableDiffusionImg2ImgPipeline.from_pretrained(
"stabilityai/stable-diffusion-2",
use_auth_token="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",
).vae.encoder
# self.encode = StableDiffusionImg2ImgPipeline.from_pretrained("stabilityai/stable-diffusion-2",use_auth_token="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")._encode_prompt
def forward(self, x, p):
x = self.base(x)
# p = self.encode(p,"cpu",1,True)
return x # , p.reshape(x.shape[0],2,77,1024)
m = revDiff()
# m(torch.rand(1,3,512,512),"have fun")[1].shape
@torch.no_grad()
def run_test(model, dataloader):
pbar = tqdm(enumerate(dataloader), total=len(dataloader), desc="Valid ")
preds = []
ids = []
ps = []
for step, (images, labels) in pbar:
images = images.to(CFG.device, dtype=torch.float)
pred = model(images, p).cpu().numpy()
for idx, i in enumerate(labels):
i = i.split("/")[-1].split(".")[0]
np.save(i, pred[idx])
# ps.extend(p.cpu().numpy().tolist())
# df_dict = {'image_id':ids , "preds" : preds }
return 0
# df = df.sample(24).reset_index(drop =True)
test_loader = prepare_loaders(df=df, debug=CFG.debug)
model = revDiff().to(CFG.device)
model.eval()
#
preds = run_test(model, test_loader)
|
# ## Table of Contents
# 1. [Introduction](#Introduction)
# 2. [Import Libraries and Read Dataset](#Import_Libraries_and_Read_Dataset)
# 3. [Data Exploration](#Data_Exploration)
# 4. [Data Cleaning](#Data_Cleaning)
# 5. [Data Visualization](#Data_Visualization)
# 6. [Data Preprocessing](#Data_Preprocessing)
# 7. [Modeling](#Modeling)
# * [Imbalanced Data](#Imbalanced_Data)
# * [Synthetic Minority Oversampling Technique (SMOTE)](#SMOTE)
# # 1. Introduction
# #### Churn occurs when a customer stops using a company's products or services. When a customer churns, there are usually early indicators or metrics that can be uncovered through churn analysis.
# #### This chart provides you with a handy reference for understanding the reasons for churn and how they relate to each other ["Telecom Churn Management Handbook" by Rob Mattison].
# 
# # 2. Import Libraries and Read Dataset
# for data preprocessing & manipulation
import numpy as np
import pandas as pd
# Visualization Libraries
import matplotlib.pyplot as plt
from pylab import rcParams # Customize Matplotlib plots using rcParams
import seaborn as sns
# Prepare Data for Machine Learning Classifier
from sklearn.preprocessing import LabelEncoder
from sklearn.preprocessing import StandardScaler
# Preparing Data for ML Classifier
from sklearn.preprocessing import LabelEncoder
from sklearn.preprocessing import StandardScaler
# Modeling
from sklearn.model_selection import train_test_split
from sklearn.linear_model import LogisticRegression
from sklearn.ensemble import RandomForestClassifier
from sklearn.svm import SVC
from sklearn.ensemble import AdaBoostClassifier
from xgboost.sklearn import XGBClassifier
from sklearn.metrics import (
precision_score,
recall_score,
confusion_matrix,
classification_report,
)
# Load Our Data
df = pd.read_csv(
"/kaggle/input/telco-customer-churn/WA_Fn-UseC_-Telco-Customer-Churn.csv"
)
df.head()
# # 3. Data Exploration
df.shape
df.columns
df.info()
# check what kind of catagorical data we have
cat_cols = [i for i in df.columns if df[i].dtypes == "object"]
for col in cat_cols:
print(col, ":", df[col].unique())
# Summarize our dataset
print("Rows: ", df.shape[0])
print("Columns: ", df.shape[1])
print("\nFeatures: \n", df.columns.tolist())
print("\nMissing Values: ", df.isnull().sum().values.sum(), "\n", df.isnull().sum())
print("\nUnique Values: \n", df.nunique())
# # 4. Data Cleaning
# set index: "customerID"
df = df.set_index(["customerID"])
# Converting "TotalCharges" to a numerical data type
print(df["TotalCharges"].dtypes)
df.TotalCharges = pd.to_numeric(df.TotalCharges, errors="coerce")
print(df["TotalCharges"].dtypes)
# Removing missing values
df.dropna(inplace=True)
# Double Check that nulls have been removed
print("\nMissing Values: ", df.isnull().sum().values.sum())
# Get summary stats on our numeric columns
df.describe()
# I check if the data is imbalanced or not
g = sns.countplot(x="Churn", data=df, palette="muted")
g.set_ylabel("Customers", fontsize=14)
g.set_xlabel("Churn", fontsize=14)
df.Churn.value_counts(normalize=True)
# # 5. Data Visualization
# Keep a copy in case we need to look at the original dataset in the future
df_copy = df.copy()
df_copy.drop(["tenure", "MonthlyCharges", "TotalCharges"], axis=1, inplace=True)
df_copy.head()
# Create a new dataset called summary so that we can summarize our churn data
summary = pd.concat(
[pd.crosstab(df_copy[x], df_copy["Churn"]) for x in df_copy.columns[:-1]],
keys=df_copy.columns[:-1],
)
summary
# Data to plot
sizes = df["Churn"].value_counts()
labels = df["Churn"].value_counts().index
colors = ["lightgreen", "red"]
explode = (0.05, 0)
rcParams["figure.figsize"] = 7, 7
# Plot
plt.pie(
sizes,
labels=labels,
colors=colors,
explode=explode,
autopct="%1.1f%%",
shadow=False,
startangle=90,
)
plt.title("Customer Churn Breakdown")
plt.show()
# Creat a Volin Plot showing how monthly charges relate to Churn
g = sns.factorplot(
x="Churn", y="MonthlyCharges", data=df, kind="violin", palette="Pastel1"
)
# **Note:** We can see that Churned customers tend to be higher paying customers
# Let's look at Tenure
g = sns.factorplot(x="Churn", y="tenure", data=df, kind="violin", palette="Pastel1")
# Correlation plot doesn't end up being too informative
def plot_corr(df, size=10):
"""
Function plots a graphical corelation matrix for each pair of columns in the dataframe.
Input:
df: pandas dataframe
size: vertical and horizontal size of the plot
"""
corr = df.corr()
fig, ax = plt.subplots(figsize=(size, size))
ax.legend()
cax = ax.matshow(corr)
fig.colorbar(cax)
plt.xticks(range(len(corr.columns)), corr.columns)
plt.yticks(range(len(corr.columns)), corr.columns)
plot_corr(df)
# # 6. Data Preprocessing
df.head()
cat_cols = [col for col in df.columns if df[col].dtypes == "object"]
print("Categorical Columns are: ", cat_cols)
# for col in cat_cols:
# print(col, ":", df[col].unique())
# Binary Columns with 2 Values
bin_cols = df.nunique()[df.nunique() == 2].keys().tolist()
# Columns more than 2 Values
multi_cols = [x for x in cat_cols if x not in bin_cols]
num_cols = [col for col in df.columns if col not in cat_cols]
print("Numerical Columns are: ", num_cols)
# Label Encoding Binary Columns
le = LabelEncoder()
for i in bin_cols:
df[i] = le.fit_transform(df[i])
# Duplicationg Columns for Multi-Value Columns
df = pd.get_dummies(data=df, columns=multi_cols)
df.head()
# # Scaling Numerical Columns
# std = StandardScaler()
# # Scale data
# scaled = std.fit_transform(df[num_cols])
# scaled = pd.DataFrame(scaled, columns = num_cols)
# # Dropping OriginalValues Merging Scaled Values for Numerical Columns
# df_og = df.copy()
# df = df.drop(columns = num_cols, axis =1)
# df = df.merge(scaled, left_index = True, right_index = True, how = "left")
df.head()
# Get Correlation of "Churn" with other variables
plt.figure(figsize=(15, 8))
df.corr()["Churn"].sort_values(ascending=False).plot(kind="bar")
# # 7. Modeling
# ## Imbalanced Data
# X, y
X = df.drop(["Churn"], axis=1)
y = df["Churn"].values
# Split it to a 80:20 Ratio Train:Test
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, stratify=y)
pd.DataFrame(y_train).iloc[:, 0].value_counts()
# Def results of a model
def model_results(model, X_train, y_train, X_test, y_test):
result = {}
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
# accuracy_train
result["Accuracy_Train"] = model.score(X_train, y_train)
# accuracy_test
result["Accuracy_Test"] = model.score(X_test, y_test)
# precision
result["Precision"] = precision_score(y_test, y_pred)
# recall
result["Recall"] = recall_score(y_test, y_pred)
# confusion_matrix
# print('\nConfusion Matrix:')
# print(confusion_matrix(y_test, y_pred))
# # classification_report
# print('\nClassification Report:')
# print(classification_report(y_test, y_pred))
print("\n")
return result
models = [
("Logistic Regression", LogisticRegression()),
("Random Forest", RandomForestClassifier()),
# ('Support Vector Machine (SVM)', SVC(kernel='linear')),
("Ada Boost", AdaBoostClassifier()),
("XG Boost", XGBClassifier()),
]
results = pd.DataFrame()
for name, model in models:
print(name, "...")
result = model_results(model, X_train, y_train, X_test, y_test)
d = pd.DataFrame(list(result.items()), columns=["KPI", name]).set_index("KPI")
results.index = list(result.keys())
results = pd.concat([results, d], axis=1)
results
# ## Synthetic Minority Oversampling Technique (SMOTE)
from imblearn.over_sampling import SMOTE
X_smote, y_smote = SMOTE().fit_resample(X, y)
pd.DataFrame(y_smote).iloc[:, 0].value_counts()
results_oversampling = pd.DataFrame()
for name, model in models:
print(name, "...")
result = model_results(model, X_smote, y_smote, X_test, y_test)
d = pd.DataFrame(list(result.items()), columns=["KPI", name]).set_index("KPI")
results_oversampling.index = list(result.keys())
results_oversampling = pd.concat([results_oversampling, d], axis=1)
results_oversampling
|
# # Tabular Data Classification and Baseline with EDA
# **Table of Contents:**
# 1. [Load Data and Inspect Top Level Features](#load)
# 2. [Exploratory Data Analysis (EDA)](#eda)
# 3. [Data Preparation and Preprocessing](#data-preprocessing)
# 4. [Model Training and Evaluation](#model-training)
# - 4.1. [Basic Analysis using Random Forest](#random-forest)
# - 4.2 [CatBoost Classification model](#catboost)
# 5. [Test set predictions](#test-predictions)
import gc
import matplotlib.pyplot as plt
import numpy as np
import os
import pandas as pd
import seaborn as sns
from catboost import CatBoostClassifier, cv, Pool
from collections import defaultdict
from imblearn.over_sampling import SMOTE
from IPython.display import Image
from pydotplus import graph_from_dot_data
from sklearn.decomposition import PCA
from sklearn.tree import DecisionTreeClassifier, export_graphviz
from sklearn.ensemble import (
RandomForestClassifier,
GradientBoostingClassifier,
BaggingClassifier,
)
from sklearn.preprocessing import LabelEncoder, StandardScaler, OneHotEncoder
from sklearn.model_selection import (
train_test_split,
KFold,
cross_val_score,
cross_val_predict,
)
from sklearn.model_selection import cross_validate, StratifiedKFold
from sklearn.metrics import (
accuracy_score,
f1_score,
precision_score,
roc_curve,
recall_score,
confusion_matrix,
classification_report,
auc,
precision_recall_curve,
)
from sklearn.linear_model import LinearRegression, Lasso, Ridge
from sklearn.pipeline import Pipeline
from sklearn.linear_model import LogisticRegression, Perceptron, RidgeClassifier
from sklearn.naive_bayes import GaussianNB
from sklearn.neural_network import MLPClassifier
from sklearn.svm import SVC
# ---
# ## 1. Load Data and inspect top level features
data_dir = "/kaggle/input/tabular-playground-series-mar-2021/"
train_df = pd.read_csv(os.path.join(data_dir, "train.csv"))
test_df = pd.read_csv(os.path.join(data_dir, "test.csv"))
train_df.head()
train_df.info()
train_df.isna().sum().sum()
# Good, we have absolutely no null or missing values at all!
# ---
# ## 2. Exploratory Data Analysis (EDA)
# Since we've got a range of both categorical and numerical features we should briefly explore these for insights.
# ### 2.1 Analysis of Categorical Features
def custom_countplot(data_df, col_name, ax=None, annotate=True):
"""Plot seaborn countplot for selected dataframe col"""
c_plot = sns.countplot(x=col_name, data=data_df, ax=ax)
if annotate:
for g in c_plot.patches:
c_plot.annotate(
f"{g.get_height()}",
(g.get_x() + g.get_width() / 3, g.get_height() + 60),
)
# We can plot all of our categorical columns using this basic helper function:
cat_cols = [x for x in train_df.columns.values if x.startswith("cat")]
n = len(cat_cols)
print(f"Number of categorical columns: {n}")
n = len(cat_cols)
fig, axs = plt.subplots(5, 4, figsize=(18, 10))
axs = axs.flatten()
# iterate through each col and plot
for i, col_name in enumerate(cat_cols):
custom_countplot(train_df, col_name, ax=axs[i], annotate=False)
axs[i].set_xlabel(f"{col_name}", weight="bold")
axs[i].set_ylabel("Count", weight="bold")
# only apply y label to left-most plots
if i not in [0, 4, 8, 12, 16]:
axs[i].set_ylabel("")
plt.tight_layout()
plt.show()
# Lets also look at our target output:
train_df["target"].value_counts().plot.bar()
plt.show()
train_df["target"].value_counts(normalize=True)
# So we've got a slight imbalance of data for our outputs. We could consider correcting this through the use of various imbalanced data techniques.
num_cols = [x for x in train_df.columns.values if x.startswith("cont")]
num_n = len(num_cols)
print(f"Number of numerical columns: {num_n}")
def target_boxplot(
y_val_col, x_val_col, data_df, figsize=(9, 6), ax=None, name="Boxplot"
):
"""Custom boxplot function - plot a chosen value against target x col"""
if not ax:
fig, ax = plt.subplots(figsize=figsize)
b_plot = sns.boxplot(x=x_val_col, y=y_val_col, data=data_df, ax=ax)
medians = data_df.groupby(x_val_col)[y_val_col].median()
vert_offset = data_df[y_val_col].median() * 0.05
for xtick in b_plot.get_xticks():
b_plot.text(
xtick,
medians[xtick] + vert_offset,
medians[xtick],
horizontalalignment="center",
size="small",
color="w",
weight="semibold",
)
if not ax:
plt.title(f"{name}", weight="bold")
plt.show()
num_n = len(num_cols)
fig, axs = plt.subplots(3, 4, figsize=(16, 9))
axs = axs.flatten()
# iterate through each col and plot
for i, col_name in enumerate(num_cols):
target_boxplot(col_name, "target", train_df, name=f"{col_name}", ax=axs[i])
axs[i].set_xlabel(f"{col_name}", weight="bold")
axs[i].set_ylabel("Value", weight="bold")
# only apply y label to left-most plots
if i not in [
0,
5,
]:
axs[i].set_ylabel("")
plt.tight_layout()
plt.show()
sns.pairplot(train_df.loc[:20000, num_cols], height=3, plot_kws={"alpha": 0.2})
plt.show()
# find the correlation between our variables
corr = train_df.loc[:, num_cols].corr()
plt.figure(figsize=(12, 8))
sns.heatmap(corr, annot=True)
plt.show()
# We have a few fairly strong correlations between our variables, which might be worth exploring further and seeing if we can reduce any unnecessary redundancy. For this it will be worth experimenting with some dimensionality reduction and/or feature augmentation techniques.
# Despite this, we dont have a huge number of numerical features, and so dont want to throw any useful information about our dependent output variable away if we can avoid it. Therefore, we will keep all features as they are during preprocessing for this notebook.
# ---
# ## 3. Preprocessing
# We need to suitably encode our categorical variables, and standardise (if required) our numerical features. In addition, we can also add some additional features as part of feature engineering if we're feeling curious.
# From a simplistic categorical encoding perspective, we can either encode our categorical features with appropriate integer labels, or perform one-hot encoding. The latter method will produce a much larger dataset, and hence introduced more complexity, but with tabular models this is often at the benefit of improved performance. This is not a simple rule however, and results will vary from problem to problem - thus, it is worth exploring both approaches and seeing which works best for your application.
class DataProcessor(object):
def __init__(self):
self.encoder = None
self.standard_scaler = None
self.num_cols = None
self.cat_cols = None
def preprocess(
self, data_df, train=True, one_hot_encode=False, add_pca_feats=False
):
"""Preprocess train / test as required"""
# if training, fit our transformers
if train:
self.train_ids = data_df.loc[:, "id"]
train_cats = data_df.loc[:, data_df.dtypes == object]
self.cat_cols = train_cats.columns
# if selected, one hot encode our cat features
if one_hot_encode:
self.encoder = OneHotEncoder(handle_unknown="ignore")
oh_enc = self.encoder.fit_transform(train_cats).toarray()
train_cats_enc = pd.DataFrame(
oh_enc, columns=self.encoder.get_feature_names()
)
self.final_cat_cols = list(train_cats_enc.columns)
# otherwise just encode our cat feats with ints
else:
# encode all of our categorical variables
self.encoder = defaultdict(LabelEncoder)
train_cats_enc = train_cats.apply(
lambda x: self.encoder[x.name].fit_transform(x)
)
self.final_cat_cols = list(self.cat_cols)
# standardise all numerical columns
train_num = data_df.loc[:, data_df.dtypes != object].drop(
columns=["target", "id"]
)
self.num_cols = train_num.columns
self.standard_scaler = StandardScaler()
train_num_std = self.standard_scaler.fit_transform(train_num)
# add pca reduced num feats if selected, else just combine num + cat feats
if add_pca_feats:
pca_feats = self._return_num_pca(train_num_std)
self.final_num_feats = list(self.num_cols) + list(self.pca_cols)
X = pd.DataFrame(
np.hstack((train_cats_enc, train_num_std, pca_feats)),
columns=list(self.final_cat_cols)
+ list(self.num_cols)
+ list(self.pca_cols),
)
else:
self.final_num_feats = list(self.num_cols)
X = pd.DataFrame(
np.hstack((train_cats_enc, train_num_std)),
columns=list(self.final_cat_cols) + list(self.num_cols),
)
# otherwise, treat as test data
else:
# transform categorical and numerical data
self.test_ids = data_df.loc[:, "id"]
cat_data = data_df.loc[:, self.cat_cols]
if one_hot_encode:
oh_enc = self.encoder.transform(cat_data).toarray()
cats_enc = pd.DataFrame(
oh_enc, columns=self.encoder.get_feature_names()
)
else:
cats_enc = cat_data.apply(lambda x: self.encoder[x.name].transform(x))
# transform test numerical data
num_data = data_df.loc[:, self.num_cols]
num_std = self.standard_scaler.transform(num_data)
if add_pca_feats:
pca_feats = self._return_num_pca(num_std, train=False)
X = pd.DataFrame(
np.hstack((cats_enc, num_std, pca_feats)),
columns=list(self.final_cat_cols)
+ list(self.num_cols)
+ list(self.pca_cols),
)
else:
X = pd.DataFrame(
np.hstack((cats_enc, num_std)),
columns=list(self.final_cat_cols) + list(self.num_cols),
)
return X
def _return_num_pca(self, num_df, n_components=0.85, train=True):
"""return dim reduced numerical features using PCA"""
if train:
self.pca = PCA(n_components=n_components)
num_rd = self.pca.fit_transform(num_df)
# create new col names for our reduced features
self.pca_cols = [f"pca_{x}" for x in range(num_rd.shape[1])]
else:
num_rd = self.pca.transform(num_df)
return pd.DataFrame(num_rd, columns=self.pca_cols)
# Lets transform our data into a form suitable for training various models. This includes encoding our categorical variables, and standardising our numerical variables.
# We can either encode our categorical feature values, or one-hot encode them. Our preprocessing function supports whichever we want, through simply setting the one_hot_encode argument as true (one-hot encoding) or false (simple numerical encoding). We obtain a larger number of feature columns if we one-hot encode, and therefore introduce more complexity. However, many models perform better with one-hot encoding, so it is worth trying both techniques for our range of models.
# We'll be using mainly tree-based methods in this notebook, and as such one-hot encoding and simple encoding of features does not actually make any noticeable difference (as demonstrated through years of empirical research and comparisons). Therefore, we'll keep our dataset simpler and just use categorical encoding.
data_proc = DataProcessor()
# advanced preprocessing- include pca feats + one hot encoding
X_train_full = data_proc.preprocess(train_df, one_hot_encode=True, add_pca_feats=False)
y_train_full = train_df.loc[:, "target"]
X_test = data_proc.preprocess(
test_df, train=False, one_hot_encode=True, add_pca_feats=False
)
print(
f"X_train_full: {X_train_full.shape} \\ny_train_full: {y_train_full.shape}, \nX_test: {X_test.shape}"
)
# Lets obtain a further split containing a validation and training split for model training, optimising and evaluation purposes:
X_train, X_val, y_train, y_val = train_test_split(
X_train_full, y_train_full, test_size=0.2, random_state=12, stratify=y_train_full
)
print(
f"X_train_full: {X_train.shape} \ny_train_full: {y_train.shape} \nX_val: {X_val.shape}, \ny_val: {y_val.shape}"
)
# ---
# ## 4. Exploring our dataset using different models
# ### 4.1 Random Forest Analysis
def show_tree_graph(tree_model, feature_names):
"""Output a decision tree to notebook"""
draw_data = export_graphviz(
tree_model,
filled=True,
rounded=True,
feature_names=feature_names,
out_file=None,
rotate=True,
class_names=True,
)
graph = graph_from_dot_data(draw_data)
return Image(graph.create_png())
rf_clf = RandomForestClassifier(n_estimators=100, max_depth=3)
rf_clf.fit(X_train, y_train)
show_tree_graph(rf_clf.estimators_[0], list(X_train.columns))
# Lets train our random forest again, but this time not limiting the depth of our trees:
rf_clf = RandomForestClassifier(n_estimators=100, n_jobs=-1)
# The great thing with random forests is the ease of being able to see the relative importance of our features used for making predictions:
def feature_importances(rf_model, dataframe):
"""Return dataframe of feat importances from random forest model"""
return pd.DataFrame(
{"columns": dataframe.columns, "importance": rf_model.feature_importances_}
).sort_values("importance", ascending=False)
importances = feature_importances(rf_clf, X_train)
TOP_N = 45
plt.figure(figsize=(14, 6))
sns.barplot(x="columns", y="importance", data=importances[:TOP_N])
plt.ylabel("Feature Importances", weight="bold")
plt.xlabel("Features", weight="bold")
plt.title("Random Forest Feature Importances", weight="bold")
plt.xticks(rotation=90)
plt.show()
print(importances[:TOP_N])
# Lets now make some predictions on our validation set to get a rough idea of the performance of our model:
val_preds = rf_clf.predict(X_val)
val_acc = accuracy_score(val_preds, y_val)
print(f"Random Forest accuracy on validation set: {val_acc}\n")
print(classification_report(val_preds, y_val))
# These metrics are hard to appreciate from the values alone, however they do highlight a severe limitation of our model. Lets plot a confusion matrix, which will help illustrate what this is.
def plot_confusion_matrix(true_y, pred_y, title="Confusion Matrix", figsize=(8, 6)):
"""Custom function for plotting a confusion matrix for predicted results"""
conf_matrix = confusion_matrix(true_y, pred_y)
conf_df = pd.DataFrame(
conf_matrix, columns=np.unique(true_y), index=np.unique(true_y)
)
conf_df.index.name = "Actual"
conf_df.columns.name = "Predicted"
plt.figure(figsize=figsize)
plt.title(title)
sns.set(font_scale=1.4)
sns.heatmap(conf_df, cmap="Blues", annot=True, annot_kws={"size": 16}, fmt="g")
plt.show()
return
plot_confusion_matrix(y_val, val_preds)
def plot_roc_curve(y_train, y_train_probs, y_val, y_val_probs, figsize=(8, 8)):
"""Helper function to plot the ROC AUC from given labels"""
# obtain true positive and false positive rates for roc_auc
fpr, tpr, thresholds = roc_curve(y_train, y_train_probs[:, 1], pos_label=1)
roc_auc = auc(fpr, tpr)
# obtain true positive and false positive rates for roc_auc
val_fpr, val_tpr, val_thresholds = roc_curve(y_val, y_val_probs[:, 1], pos_label=1)
val_roc_auc = auc(val_fpr, val_tpr)
plt.figure(figsize=figsize)
plt.plot(fpr, tpr, label=f"Train ROC AUC = {roc_auc}", color="blue")
plt.plot(val_fpr, val_tpr, label=f"Val ROC AUC = {val_roc_auc}", color="red")
plt.plot(
[0, 1], [0, 1], label="Random Guessing", linestyle=":", color="grey", alpha=0.6
)
plt.plot(
[0, 0, 1],
[0, 1, 1],
label="Perfect Performance",
linestyle="--",
color="black",
alpha=0.6,
)
plt.xlabel("False Positive Rate")
plt.ylabel("True Positive Rate")
plt.title("Receiver Operating Characteristic", weight="bold")
plt.legend(loc="best")
plt.show()
# obtain prediction probabilities for trg and val
y_val_probs = rf_clf.predict_proba(X_val)
y_trg_probs = rf_clf.predict_proba(X_train)
# plot our ROC curve
plot_roc_curve(y_train, y_trg_probs, y_val, y_val_probs)
# The AUC is the metric we're trying to maximise for this competition, and therefore we should seek to obtain a model that scores particularly well in this regard.
# Just for interest, we can also inspect the precision-recall curve for our models:
def plot_prec_rec_curve(y_train, y_train_probs, y_val, y_val_probs, figsize=(14, 6)):
"""Helper function to plot the ROC AUC from given labels"""
# obtain true positive and false positive rates for roc_auc
prec, rec, thresholds = precision_recall_curve(
y_train, y_train_probs[:, 1], pos_label=1
)
prec_rec_auc = auc(rec, prec)
# obtain true positive and false positive rates for roc_auc
val_prec, val_rec, val_thresholds = precision_recall_curve(
y_val, y_val_probs[:, 1], pos_label=1
)
val_prec_rec_auc = auc(val_rec, val_prec)
plt.figure(figsize=figsize)
plt.plot(
prec, rec, label=f"Train Precision-Recall AUC = {prec_rec_auc}", color="blue"
)
plt.plot(
val_prec,
val_rec,
label=f"Val Precision-Recall AUC = {val_prec_rec_auc}",
color="red",
)
plt.plot(
[0, 0, 1],
[0, 1, 1],
label="Perfect Performance",
linestyle="--",
color="black",
alpha=0.6,
)
plt.xlabel("Recall")
plt.ylabel("Precision")
plt.title("Precision-Recall Curve", weight="bold")
plt.legend(loc="best")
plt.show()
# plot our precision recall curve
plot_prec_rec_curve(y_train, y_trg_probs, y_val, y_val_probs)
#
# ### 4.2 CatBoost Classifier
# Since training can take a long time on CPU, we want to ensure we select GPU as the task type for our catboost model:
cb_learn_rate = 0.006
n_iterations = 15000
early_stop_rounds = 400
cb_params = {
"iterations": n_iterations,
"learning_rate": cb_learn_rate,
"task_type": "GPU",
"random_seed": 13,
"verbose": 500,
}
# cb_params = {'iterations' : n_iterations, 'learning_rate' : cb_learn_rate,
# 'random_seed' : 13, 'verbose' : 500}
cb_clf = CatBoostClassifier(**cb_params)
# A nice feature of CatBoost is the option of adding an interactive plot of training, which allows us to analyse the performance in real time:
cb_clf.fit(
X_train,
y_train,
eval_set=(X_val, y_val),
use_best_model=True,
plot=True,
early_stopping_rounds=early_stop_rounds,
)
# Lets make some predictions on the validation set and compare to our previous random forest model:
val_preds = cb_clf.predict(X_val)
val_acc = accuracy_score(val_preds, y_val)
print(f"CatBoost accuracy on validation set: {val_acc}\n")
print(classification_report(val_preds, y_val))
plot_confusion_matrix(y_val, val_preds)
# Finally, lets look at the ROC AUC for the CatBoost model:
# obtain prediction probabilities for trg and val
y_val_probs = cb_clf.predict_proba(X_val)
y_trg_probs = cb_clf.predict_proba(X_train)
# plot our ROC curve
plot_roc_curve(y_train, y_trg_probs, y_val, y_val_probs)
# ---
# ## 5. Test set predictions
# With some basic models under our belt, lets make some predictions on the test set and submit these:
test_preds = cb_clf.predict(X_test)
submission_df = pd.read_csv(os.path.join(data_dir, "sample_submission.csv"))
submission_df["target"] = test_preds
submission_df.to_csv("submission.csv", index=False)
|
# # 1 Dataset
import pandas as pd
import matplotlib.pyplot as plt
import re
import nltk
data = pd.read_csv("/kaggle/input/nlp-ulta-skincare-reviews/Ulta Skincare Reviews.csv")
data.head()
data.info()
data.isnull().sum()
data.fillna("Unknown", inplace=True)
data.isnull().sum()
# # 2 Auto Labeling Sentiment Using VaderSentiment
from vaderSentiment.vaderSentiment import SentimentIntensityAnalyzer
vds = SentimentIntensityAnalyzer()
text = "I am good for today."
vds.polarity_scores(text)
# doc = data['Review_Text']
# vds.polarity_scores(doc)
data["score"] = data["Review_Text"].apply(lambda review: vds.polarity_scores(review))
data.head()
data["compound"] = data["score"].apply(lambda score_dict: score_dict["compound"])
data.head()
data["sentiment"] = data["compound"].apply(lambda c: "pos" if c >= 0 else "neg")
data.head()
data["Product"].value_counts()
data["Brand"].value_counts()
data["sentiment"].value_counts()
data.loc[data["sentiment"] == "neg", "sentiment"] = 0
data.loc[data["sentiment"] == "pos", "sentiment"] = 1
data["sentiment"].value_counts()
data = data.astype({"sentiment": int})
data["sentiment"].value_counts().plot.pie(
figsize=(6, 6),
title="Distribution of reviews per sentiment",
labels=["", ""],
autopct="%1.1f%%",
)
labels = ["Positive", "Negative"]
plt.legend(labels, loc=3)
plt.gca().set_aspect("equal")
# # 3 Data Pre-processing
# ## 3.1 Cleaning Data
# Cleaning data
def remove(tweet):
# remove mention
tweet = re.sub("@[A-Za-z0-9_]+", "", tweet)
# remove stock market tickers like $GE
tweet = re.sub(r"\$\w*", "", tweet)
# remove old style retweet text "RT"
tweet = re.sub(r"^RT[\s]+", "", tweet)
tweet = re.sub(r"^rt[\s]+", "", tweet)
# remove hyperlinks
tweet = re.sub(r"https?:\/\/.*[\r\n]*", "", tweet)
tweet = re.sub(r"^https[\s]+", "", tweet)
# remove hashtags
# only removing the hash # sign from the word
tweet = re.sub(r"#", "", tweet)
tweet = re.sub(r"%", "", tweet)
# remove coma
tweet = re.sub(r",", "", tweet)
# remove angka
tweet = re.sub("[0-9]+", "", tweet)
tweet = re.sub(r":", "", tweet)
# remove space
tweet = tweet.strip()
# remove double space
tweet = re.sub("\s+", " ", tweet)
return tweet
data["cleaning_data"] = data["Review_Text"].apply(lambda x: remove(x.lower()))
data.drop_duplicates(subset="cleaning_data", keep="first", inplace=True)
data
# ## 3.2 Remove Stopwords
from nltk.corpus import stopwords
stopword = set(stopwords.words("english"))
def clean_stopwords(text):
text = " ".join(word for word in text.split() if word not in stopword)
return text
data["tweet_sw"] = data["cleaning_data"].apply(clean_stopwords)
# ## 3.3 Stemming
from nltk.stem import PorterStemmer
ps = PorterStemmer()
def porterstemmer(text):
text = " ".join(ps.stem(word) for word in text.split() if word in text)
return text
data["tweet_stem"] = data["tweet_sw"].apply(porterstemmer)
# ## 3.4 Lemmatization
import spacy
nlp = spacy.load("en_core_web_sm")
def lemmatization(text):
doc = nlp(text)
tokens = [
token.lemma_.lower()
for token in doc
if not token.is_stop and not token.is_punct
]
return " ".join(tokens)
data["tweet_clean"] = data["tweet_stem"].apply(lemmatization)
# ## 3.5 Data Final
data
data.to_excel("skincaredata.xlsx")
# # 4 TF - IDF
from sklearn.feature_extraction.text import TfidfVectorizer
vectorizer = TfidfVectorizer()
vectorizer.fit(data["tweet_clean"])
vector = vectorizer.transform(data["tweet_clean"])
print(vector)
# # 5 Split Dataset
from sklearn.model_selection import train_test_split
X = data["tweet_clean"]
y = data["sentiment"]
X_train, X_test, y_train, y_test = train_test_split(vector, y, test_size=0.2)
# # 6 SMOTE
# Class Imbalance Check
plt.pie(
y_train.value_counts(),
labels=["Negative Tweet", "Positive Tweet"],
autopct="%0.1f%%",
)
plt.axis("equal")
plt.title("Data Training before SMOTE")
plt.show()
from imblearn.over_sampling import SMOTE
sm = SMOTE(random_state=42)
X_train_sm, y_train_sm = sm.fit_resample(X_train, y_train)
# Class Imbalance Check
plt.pie(
y_train_sm.value_counts(),
labels=["Negative Tweet", "Positive Tweet"],
autopct="%0.1f%%",
)
plt.axis("equal")
plt.title("Data Training after SMOTE")
plt.show()
from collections import Counter
print("Original dataset shape %s" % Counter(y_train))
print("Resampled dataset shape %s" % Counter(y_train_sm))
# # 6 Model
from sklearn.linear_model import LogisticRegression
import sklearn.metrics as metrics
from sklearn.metrics import classification_report
from sklearn.metrics import (
accuracy_score,
confusion_matrix,
precision_score,
recall_score,
)
# ## 6.1 Model + SMOTE
# create logistic regression model + SMOTE
logreg = LogisticRegression()
# train model on vectorised training data
model = logreg.fit(X_train_sm, y_train_sm)
# ## 6.2 Model
# create logistic regression model
logreg1 = LogisticRegression()
# train model on vectorised training data
model1 = logreg.fit(X_train, y_train)
# # 7 Evaluation Model
# ## 7.1 Evaluation Model + SMOTE
y_preds_training = logreg.fit(X_train_sm, y_train_sm).predict(X_train)
y_preds_testing = logreg.fit(X_train_sm, y_train_sm).predict(X_test)
accuracy_training = accuracy_score(y_train, y_preds_training)
accuracy_testing = accuracy_score(y_test, y_preds_testing)
print("Accuracy: ", accuracy_training * 100, "%")
print("Accuracy: ", accuracy_testing * 100, "%")
print("Confusion Matrix training : ")
print(confusion_matrix(y_train, y_preds_training))
print("Confusion Matrix testing : ")
print(confusion_matrix(y_test, y_preds_testing))
print("Classification report testing: ")
print(classification_report(y_test, y_preds_testing))
print("Classification report training : ")
print(classification_report(y_train, y_preds_training))
# ## 7.2 Evaluation Model
y_preds_training1 = logreg1.fit(X_train, y_train).predict(X_train)
y_preds_testing1 = logreg1.fit(X_train, y_train).predict(X_test)
accuracy_training1 = accuracy_score(y_train, y_preds_training1)
accuracy_testing1 = accuracy_score(y_test, y_preds_testing1)
print("Accuracy training: ", accuracy_training1 * 100, "%")
print("Accuracy testing: ", accuracy_testing1 * 100, "%")
print("Confusion Matrix training : ")
print(confusion_matrix(y_train, y_preds_training1))
print("Confusion Matrix testing : ")
print(confusion_matrix(y_test, y_preds_testing1))
print("Classification report testing: ")
print(classification_report(y_test, y_preds_testing1))
print("Classification report training : ")
print(classification_report(y_train, y_preds_training1))
# # 8 SHAP
# ## 8 SHAP with SMOTE
import shap
explainer = shap.Explainer(
model, X_train_sm, feature_names=vectorizer.get_feature_names()
)
shap_values = explainer(X_test)
shap.plots.beeswarm(shap_values, max_display=25)
# ## 8.2 SHAP without SMOTE
explainer1 = shap.Explainer(
model, X_train, feature_names=vectorizer.get_feature_names()
)
shap_values1 = explainer1(X_test)
shap.plots.beeswarm(shap_values1, max_display=25)
|
import pandas as pd
import numpy as np
from gensim.parsing import (
strip_tags,
strip_numeric,
strip_multiple_whitespaces,
stem_text,
strip_punctuation,
remove_stopwords,
)
from gensim.parsing import preprocess_string
from gensim import parsing
import re
from rouge import Rouge
rouge = Rouge()
from collections import Counter
df_train = pd.read_csv(
"/kaggle/input/newspaper-text-summarization-cnn-dailymail/cnn_dailymail/train.csv"
)
df_train
number_doc = 30000
abstract = []
introduction = []
cnt = 0
for index, row in df_train.iterrows():
cnt += 1
abstract.append(row["highlights"])
introduction.append(row["article"])
if cnt == number_doc:
break
abstract = np.array(abstract)
introduction = np.array(introduction)
abstract[43]
introduction[43]
import nltk
from nltk.corpus import stopwords
from nltk.tokenize import sent_tokenize, word_tokenize
nltk.download("punkt")
nltk.download("stopwords")
nltk.download("wordnet")
import string
# tokens = word_tokenize(text.lower())
# stop_words = set(stopwords.words('english'))
# filtered_tokens = [token for token in tokens if token.lower() not in stop_words]
def get_Sentences(introduction):
sentences = sent_tokenize(introduction.lower())
sentences = [s for s in sentences if len(s.strip()) > 1]
return sentences
def preprocessing(sentences):
# remove_emails = lambda s: re.sub(r'^[a-zA-Z0-9+_.-]+@[a-zA-Z0-9.-]+$', '', s)
# remove_single_char = lambda s: re.sub(r'\s+\w{1}\s+', '', s)
CLEAN_FILTERS = [
# remove_emails,
strip_tags,
strip_numeric,
strip_multiple_whitespaces,
]
processed_words = preprocess_string(sentences, CLEAN_FILTERS)
text = " ".join(processed_words)
tokens = word_tokenize(text.lower())
stop_words = set(stopwords.words("english"))
stop_words.add("\x1a")
punctuations = set(string.punctuation)
filtered_tokens = [token for token in tokens if token.lower() not in stop_words]
return filtered_tokens
def get_word_freq(tokens):
word_freq = Counter(tokens)
# print(word_freq)
total_words = sum(word_freq.values())
# print(total_words)
return word_freq, total_words
def get_word_pd(word_freq, total_words):
word_pd = {word: count / total_words for word, count in word_freq.items()}
# print(word_pd)
return word_pd
# Calculate the sentence scores based on KL divergence
# doc * log (doc/sentences)
def KL(sentences, word_pd):
sentence_scores = {}
for i, sentence in enumerate(sentences):
sentence_words = preprocessing(sentence.lower())
sentence_word_freq = Counter(sentence_words)
sent_ps = {
word: count / len(sentence_words)
for word, count in sentence_word_freq.items()
}
# check kl divergence value for each sentence and store it in an array
score = 0
for word in sentence_words:
if word in word_pd and word in sent_ps:
score += word_pd[word] * np.log(word_pd[word] / sent_ps[word])
sentence_scores[i] = score
return sentence_scores
def get_Summary(sentence_scores, no_sentences):
# all_sentences = sorted(sentence_scores, key=sentence_scores.get, reverse=True)
# print(all_sentences)
# Sort the sentences in descending order of scores and select the top summary_size sentences
summary_sentences = sorted(sentence_scores, key=sentence_scores.get, reverse=True)[
:no_sentences
]
summary_sentences = sorted(summary_sentences)
# print(summary_sentences)
summary = " ".join([sentences[i] for i in summary_sentences])
return summary
def get_rouge_score(summary, abstract):
scores = rouge.get_scores(summary, abstract)
return scores
def preprocessing_summaries(sentences):
# remove_emails = lambda s: re.sub(r'^[a-zA-Z0-9+_.-]+@[a-zA-Z0-9.-]+$', '', s)
# remove_single_char = lambda s: re.sub(r'\s+\w{1}\s+', '', s)
CLEAN_FILTERS = [
# remove_emails,
strip_tags,
strip_numeric,
strip_multiple_whitespaces,
]
processed_words = preprocess_string(sentences, CLEAN_FILTERS)
text = " ".join(processed_words)
tokens = word_tokenize(text.lower())
stop_words = set(stopwords.words("english"))
stop_words.add("\x1a")
punctuations = set(string.punctuation)
filtered_tokens = " ".join(
token for token in tokens if token.lower() not in stop_words
)
return filtered_tokens
import warnings
warnings.filterwarnings("ignore")
columns = [
"summary",
"gold_summary",
"ROUGE-1 Precision",
"ROUGE-1 Recall",
"ROUGE-1 F1",
"ROUGE-L Precision",
"ROUGE-L Recall",
"ROUGE-L F1",
]
df = pd.DataFrame(columns=columns)
for i in range(len(abstract)):
sentences = get_Sentences(introduction[i])
tokens = preprocessing(introduction[i])
word_freq, total_words = get_word_freq(tokens)
word_pd = get_word_pd(word_freq, total_words)
sentence_scores = KL(sentences, word_pd)
summary = get_Summary(sentence_scores, 6)
# print(summary)
gold_summary = abstract[i].lower()
# print(i)
scores = get_rouge_score(
preprocessing_summaries(summary), preprocessing_summaries(gold_summary)
)
all_scores = {
"summary": summary,
"gold_summary": gold_summary,
"ROUGE-1 Precision": scores[0]["rouge-1"]["p"],
"ROUGE-1 Recall": scores[0]["rouge-1"]["r"],
"ROUGE-1 F1": scores[0]["rouge-1"]["f"],
"ROUGE-L Precision": scores[0]["rouge-l"]["p"],
"ROUGE-L Recall": scores[0]["rouge-l"]["r"],
"ROUGE-L F1": scores[0]["rouge-l"]["f"],
}
new_row = pd.DataFrame(all_scores, index=[0])
df = df.append(new_row, ignore_index=True)
df
sorted_df = df.sort_values(by=["ROUGE-1 F1"], ascending=False)
sorted_df.reset_index(drop=True)
top_5_rows = sorted_df.head(5)
top_5_rows
avg_precision = df["ROUGE-1 Precision"].mean()
avg_recall = df["ROUGE-1 Recall"].mean()
avg_f1 = df["ROUGE-1 F1"].mean()
avg_l_precision = df["ROUGE-L Precision"].mean()
avg_l_recall = df["ROUGE-L Recall"].mean()
avg_l_f1 = df["ROUGE-L F1"].mean()
# Print the results
print("Average ROUGE-1 Precision: ", avg_precision)
print("Average ROUGE-1 Recall: ", avg_recall)
print("Average ROUGE-1 F1: ", avg_f1)
print("Average ROUGE-L Precision: ", avg_l_precision)
print("Average ROUGE-L Recall: ", avg_l_recall)
print("Average ROUGE-L F1: ", avg_l_f1)
df["summary"][17285]
df["gold_summary"][17285]
introduction[17285]
|
# #### Import required libraries
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
from sklearn.model_selection import train_test_split
from sklearn.metrics import (
confusion_matrix,
classification_report,
ConfusionMatrixDisplay,
roc_curve,
precision_recall_curve,
auc,
RocCurveDisplay,
PrecisionRecallDisplay,
average_precision_score,
)
from sklearn.utils.class_weight import compute_class_weight
import tensorflow as tf
from tensorflow.keras import Model
from tensorflow.keras.layers import (
LayerNormalization,
MultiHeadAttention,
Dropout,
LayerNormalization,
Conv1D,
Dense,
GlobalAveragePooling1D,
Input,
Embedding,
)
from tensorflow.keras.metrics import BinaryAccuracy
from tensorflow.keras.optimizers import Adam
from tensorflow.keras.losses import BinaryCrossentropy
from tensorflow.keras.callbacks import EarlyStopping, ModelCheckpoint
# #### Define variables
window_size = 256
# #### Read data
# These data were created by me and are somewhat random. Data is for representation purpose only.
sequence_data = pd.read_csv(
"/kaggle/input/introduction-to-sequence-data-analytics-vidhya/sequence_data.csv"
)
sequence_labels = pd.read_csv(
"/kaggle/input/introduction-to-sequence-data-analytics-vidhya/sequence_labels.csv"
)
values = pd.read_csv(
"/kaggle/input/introduction-to-sequence-data-analytics-vidhya/possible_values.csv"
)
sequence_data
sequence_labels
values
# #### Preprocessing
values = (
values
# Ensure that index is numbered 0, 1, etc
.reset_index(drop=True)
# Use index as column (label encoding)
.reset_index()
# Rename column 'index' to 'label'
.rename(columns={"index": "value_label"})
)
# Labels should start with 1.
# 0 is reserved for missing values which will be masked.
values["value_label"] += 1
values
# Include value labels in sequence data
# Left join values with sequence data on value column
sequence_data = pd.merge(left=sequence_data, right=values, on="value", how="left")
sequence_data
# Drop redundant columns
sequence_data = sequence_data.drop(columns=["value"])
sequence_data
# Perform 'windowing'
for i in range(1, window_size):
sequence_data[f"step_{i}"] = (
sequence_data
# Make groups of ID
.groupby(by=["ID"])["value_label"]
# Shift value labels by i steps
.shift(i)
# Populate missing values with 0
.fillna(0).astype(int)
)
sequence_data
# Rename column 'value_label' as 'step_0' for consistent nomenclature
sequence_data = sequence_data.rename(columns={"value_label": "step_0"})
sequence_data
# Select the last observation for each ID
sequence_data = sequence_data.groupby(by=["ID"]).tail(1).reset_index(drop=True)
sequence_data
# Include outcomes in sequence data
# Left join sequence labels with sequence data on ID column
sequence_data = pd.merge(
left=sequence_data, right=sequence_labels, on=["ID"], how="left"
)
sequence_data
# Drop redundant column 'timestamp'
sequence_data = sequence_data.drop(columns=["timestamp"])
sequence_data
# Order column names
input_cols = [f"step_{i}" for i in range(window_size)]
input_cols.reverse()
input_cols[:10]
# Order columns as desired
sequence_data = sequence_data[["ID"] + input_cols + ["outcome"]]
sequence_data
# Split into train and validation
# (TRAINING - VALIDATION - TESTING is ideal)
X_train, X_valid, y_train, y_valid = train_test_split(
sequence_data.drop(columns=["outcome"]),
sequence_data["outcome"],
test_size=300,
random_state=0,
)
X_train
# #### Build and train model
def transformer_encoder(inputs, head_size, num_heads, ff_dim, dropout=0):
# Normalization and Attention
x = LayerNormalization(epsilon=1e-6)(inputs)
x = MultiHeadAttention(key_dim=head_size, num_heads=num_heads, dropout=dropout)(
x, x
)
x = Dropout(dropout)(x)
res = x + inputs
# Feed Forward Part
x = LayerNormalization(epsilon=1e-6)(res)
x = Conv1D(filters=ff_dim, kernel_size=1, activation="relu")(x)
x = Dropout(dropout)(x)
x = Conv1D(filters=inputs.shape[-1], kernel_size=1)(x)
return x + res
def build_model(
input_shape,
head_size,
num_heads,
ff_dim,
num_transformer_blocks,
mlp_units,
dropout=0,
mlp_dropout=0,
):
inputs = Input(shape=input_shape)
embeddings = Embedding(
input_dim=values.shape[0]
+ 1, # Number of distinct values + 1 (for '0' label encoding)
output_dim=64, # Desired embedding size
trainable=True,
mask_zero=True, # Mask 0s (padding)
)(inputs)
x = embeddings
for _ in range(num_transformer_blocks):
x = transformer_encoder(x, head_size, num_heads, ff_dim, dropout)
x = GlobalAveragePooling1D(data_format="channels_first")(x)
for dim in mlp_units:
x = Dense(dim, activation="relu")(x)
x = Dropout(mlp_dropout)(x)
outputs = Dense(1, activation="sigmoid")(x)
return Model(inputs, outputs)
# Create model architecture
model = build_model(
input_shape=(window_size,),
head_size=256,
num_heads=4,
ff_dim=4,
num_transformer_blocks=4,
mlp_units=[128],
mlp_dropout=0.4,
dropout=0.25,
)
# Compile the model with loss, metrics, optimizer, etc
model.compile(
loss=BinaryCrossentropy(),
optimizer=Adam(learning_rate=1e-4),
metrics=[BinaryAccuracy(threshold=0.5, name="binary_accuracy")],
)
# Overview of model architecture
model.summary()
# Since the number of examples per class (Damaged / Healthy) are unequal, we have 'class imbalance'
# Use class weights to offset this problem.
class_weights = compute_class_weight(class_weight="balanced", classes=[0, 1], y=y_train)
class_weights = dict(zip([0, 1], class_weights))
# Using early stopping to exit training if validation loss is not decreasing even after certain epochs (patience)
earlystopping = EarlyStopping(monitor="val_loss", verbose=True, patience=50)
# Save the best model with lower loss
checkpointer = ModelCheckpoint(
filepath="model_weights.hdf5", verbose=True, save_best_only=True
)
callbacks = [earlystopping, checkpointer]
history = model.fit(
X_train[input_cols].values,
y_train.values,
validation_data=(X_valid[input_cols].values, y_valid.values),
epochs=10,
batch_size=64,
class_weight=class_weights,
callbacks=callbacks,
)
# Save model
model.save("model")
# #### Evaluate model
# Overview of history
history.history
# Plot learning curves
fig, axs = plt.subplots(1, 2, figsize=(15, 3))
train_loss = history.history["loss"]
valid_loss = history.history["val_loss"]
train_acc = history.history["binary_accuracy"]
valid_acc = history.history["val_binary_accuracy"]
axs[0].plot(train_loss)
axs[0].plot(valid_loss)
axs[0].legend(["train", "valid"])
axs[0].set_title("Loss")
axs[1].plot(train_acc)
axs[1].plot(valid_acc)
axs[1].legend(["train", "valid"])
axs[1].set_title("Accuracy")
plt.tight_layout()
# Make prediction on test data
# We don't have test data so we will use validation data for demo.
# In real world applications, validation data should not be a substitute for testing data.
test_pred_proba = model.predict(X_valid[input_cols].values)
test_pred = tf.where(test_pred_proba > 0.5, 1, 0) # classification_threshold,
test_actual = y_valid.values
# Plot confusion matrix
cm = confusion_matrix(y_true=test_actual, y_pred=test_pred, labels=[0, 1])
cm = ConfusionMatrixDisplay(
confusion_matrix=cm,
display_labels=["Healthy", "Damaged"],
)
cm.plot(cmap="Blues")
# Plot classification report
print(
classification_report(
test_actual, test_pred, labels=[0, 1], target_names=["Healthy", "Damaged"]
)
)
# Plot ROC Curves for both classes
fig, axs = plt.subplots(1, 2, figsize=(15, 5))
fpr, tpr, _ = roc_curve(test_actual, test_pred_proba)
plot_obj = RocCurveDisplay(
fpr=fpr, tpr=tpr, roc_auc=auc(fpr, tpr), estimator_name=["Damaged"]
)
plot_obj.plot(color="darkorange", ax=axs[0])
axs[0].set_title("ROC Curve")
axs[0].plot([0, 1], [0, 1], color="navy", lw=2, linestyle="--")
fpr, tpr, _ = roc_curve(1 - test_actual, 1 - test_pred_proba)
plot_obj = RocCurveDisplay(
fpr=fpr, tpr=tpr, roc_auc=auc(fpr, tpr), estimator_name=["Healthy"]
)
plot_obj.plot(color="darkorange", ax=axs[1])
axs[1].set_title("ROC Curve")
axs[1].plot([0, 1], [0, 1], color="navy", lw=2, linestyle="--")
# Plot PR Curves for both classes
fig, axs = plt.subplots(1, 2, figsize=(15, 5))
precision, recall, thresholds = precision_recall_curve(test_actual, test_pred_proba)
pr_display = PrecisionRecallDisplay(
precision=precision,
recall=recall,
average_precision=average_precision_score(test_actual, test_pred_proba),
estimator_name=["Damaged"],
)
pr_display.plot(color="darkorange", ax=axs[0])
axs[0].set_title("PR Curve")
axs[0].set_xlabel("Recall")
axs[0].set_ylabel("Precision")
axs[0].legend(loc="upper right")
precision, recall, thresholds = precision_recall_curve(
1 - test_actual, 1 - test_pred_proba
)
pr_display = PrecisionRecallDisplay(
precision=precision,
recall=recall,
average_precision=average_precision_score(1 - test_actual, 1 - test_pred_proba),
estimator_name=["Healthy"],
)
pr_display.plot(color="darkorange", ax=axs[1])
axs[1].set_title("PR Curve")
axs[1].set_xlabel("Recall")
axs[1].set_ylabel("Precision")
axs[1].legend(loc="upper right")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Load in the datasets
street = pd.read_csv("/kaggle/input/london-police-records/london-street.csv")
search = pd.read_csv("/kaggle/input/london-police-records/london-stop-and-search.csv")
outcomes = pd.read_csv("/kaggle/input/london-police-records/london-outcomes.csv")
# Get look at the dataset
street.head()
street.info()
# Find proportion of missing data in street dataset
street.isnull().sum() / len(street)
# Drop Context Column from dataset
street.drop(columns=["Context"])
# Checking dataset too see if we need too keep Crime ID column
search.head()
# Checking dataset too see if any matches for Crime ID column
outcomes.info() # It looks like later we might be able to join datasets on Crime ID so we will leave it alone for now
# Quick look at Longitude
street.Longitude.describe()
# In terms of looking at specific locations of events happening it does not make sense to impute a mean or median to the column. We will just have to get rid of missing value rows in the column. Same with the Latitude column
# Drop
new_street = street[street.notna()]
new_street.isnull().sum()
new_street.head()
new_street["Month"] = new_street["Month"].astype(str)
new_street["Year"] = new_street.Month.str[0:3]
new_street.head()
|
import pandas as pd
import numpy as np
from scipy.stats import chi2_contingency
import matplotlib.pyplot as plt
import seaborn as sns
df = pd.read_csv("../input/mushroom-classification/mushrooms.csv")
df.head()
# # EDA
plt.subplots(4, 6, figsize=(20, 10))
a = 1
for i, s in enumerate(df.columns):
plt.subplot(4, 6, a)
sns.countplot(x=df[s], hue=df["class"])
a += 1
#
# # Cramer V
#
def cramerV(var_1, var2):
cont_tab = np.array(pd.crosstab(var1, var2))
chi2 = chi2_contingency(cont_tab)[0]
sample = np.sum(cont_tab)
minimum = min(cont_tab.shape) - 1
return chi2 / (sample * minimum)
from sklearn.preprocessing import LabelEncoder
le = LabelEncoder()
df_coded = pd.DataFrame()
for i in df.columns:
df_coded[i] = le.fit_transform(df[i])
# # Feature Engineering
df = pd.get_dummies(df, drop_first=True)
y = df["class_p"]
X = df.drop("class_p", axis=1)
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y)
#
# # Logistic Regression
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import cross_val_score
from sklearn.metrics import classification_report, confusion_matrix
lr = LogisticRegression()
lr.fit(X_train, y_train)
print(
"The cross validation sample score is",
cross_val_score(lr, X_train, y_train, cv=5).mean(),
)
pred = lr.predict(X_test)
print(classification_report(y_test, pred))
sns.heatmap(confusion_matrix(y_test, pred), annot=True)
# # Decision Tree Classifier
from sklearn.tree import DecisionTreeClassifier
from sklearn.model_selection import cross_val_score
from sklearn.metrics import classification_report, confusion_matrix
dtc = DecisionTreeClassifier()
dtc.fit(X_train, y_train)
print(
"The cross validation sample score is",
cross_val_score(lr, X_train, y_train, cv=5).mean(),
)
pred = dtc.predict(X_test)
print(classification_report(y_test, pred))
sns.heatmap(confusion_matrix(y_test, pred), annot=True)
# # Random Forest Classifer
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import cross_val_score
from sklearn.metrics import classification_report, confusion_matrix
rf = RandomForestClassifier()
rf.fit(X_train, y_train)
print(
"The cross validation sample score is",
cross_val_score(lr, X_train, y_train, cv=5).mean(),
)
pred = rf.predict(X_test)
print(classification_report(y_test, pred))
sns.heatmap(confusion_matrix(y_test, pred), annot=True)
pd.DataFrame(rf.feature_importances_, index=X_train.columns).sort_values(
0, ascending=False
).head(10).plot(kind="barh")
|
#
#
import numpy as np
import pandas as pd
import torch
import torchvision
import torch.nn as nn
import torch.optim as optim
from torchvision import datasets, models, transforms
from torch.utils.data.sampler import SubsetRandomSampler
import matplotlib.pyplot as plt
import time
import copy
from random import shuffle
import tqdm.notebook as tqdm
import sklearn
from sklearn.metrics import accuracy_score, cohen_kappa_score
from sklearn.metrics import classification_report
from PIL import Image
import cv2
import os
import shutil
df = pd.read_csv("./covid-chestxray-dataset/metadata.csv")
selected_df = df[df.finding == "Pneumonia/Viral/COVID-19"]
selected_df = selected_df[(selected_df.view == "AP") | (selected_df.view == "PA")]
selected_df.head(2)
images = selected_df.filename.values.tolist()
os.makedirs("./COVID19-DATASET/train/covid19")
os.makedirs("./COVID19-DATASET/train/normal")
COVID_PATH = "./COVID19-DATASET/train/covid19"
NORMAL_PATH = "./COVID19-DATASET/train/normal"
for image in images:
shutil.copy(
os.path.join("./covid-chestxray-dataset/images", image),
os.path.join(COVID_PATH, image),
)
for image in os.listdir("../input/chest-xray-pneumonia/chest_xray/train/NORMAL")[:300]:
shutil.copy(
os.path.join("../input/chest-xray-pneumonia/chest_xray/train/NORMAL", image),
os.path.join(NORMAL_PATH, image),
)
DATA_PATH = "./COVID19-DATASET/train"
class_names = os.listdir(DATA_PATH)
image_count = {}
for i in class_names:
image_count[i] = len(os.listdir(os.path.join(DATA_PATH, i)))
# Plotting Distribution of Each Classes
fig1, ax1 = plt.subplots()
ax1.pie(
image_count.values(),
labels=image_count.keys(),
shadow=True,
autopct="%1.1f%%",
startangle=90,
)
plt.show()
# I have taken correct propositions of data from each classes while staging my data to avoid overfitted results. Medical Image Datasets will always be like this, we don't have enough data on victim rather we have so many healthy ones. That is what we called is a "Skewed Dataset", obviously we developed methods to approach those problems (like K-Fold Cross Validation) that will explain that in my next notebook.
# Lets view some images to know what we are dealing with here. Feel free the run the cell to view images at random from the repository.
fig = plt.figure(figsize=(16, 5))
fig.suptitle("COVID19 Positive", size=22)
img_paths = os.listdir(COVID_PATH)
shuffle(img_paths)
for i, image in enumerate(img_paths[:4]):
img = cv2.imread(os.path.join(COVID_PATH, image))
plt.subplot(1, 4, i + 1, frameon=False)
plt.imshow(img)
fig.show()
fig = plt.figure(figsize=(16, 5))
fig.suptitle("COVID19 Negative - Healthy", size=22)
img_paths = os.listdir(NORMAL_PATH)
shuffle(img_paths)
for i, image in enumerate(img_paths[:4]):
img = cv2.imread(os.path.join(NORMAL_PATH, image))
plt.subplot(1, 4, i + 1, frameon=False)
plt.imshow(img)
fig.show()
# Have you seen how similar they look!!. Yeah right, sometimes even for a medical expert. It is hard to diagnose with Xray images. That's why we can help them with our model.
# ## Transforms
# Data Transforms or Augmentation is the synthesis of new data using the available with some little manipulations and image processing. Augmentation will help generalizing our model, avoids Over-fitting to the training data. Since we have relatively little amounts of data for training and validation we will synthesis some extra data through Image Transforms.
# torchvision from PyTorch provides,various tools that we can use to perform various tasks for Computer Vision with ease of use.
# Few most used tools include,
# - [transforms](https://pytorch.org/docs/stable/torchvision/transforms.html)- Image Data Augementation
# - [datasets](https://pytorch.org/docs/stable/torchvision/datasets.html)- Dataset loading and handling
# - [models](https://pytorch.org/docs/stable/torchvision/models.html)- Deep Learning Pre-Defined SOTA Models
# These tools will be so handy for us, so that we can concentrate on optimizing our results better.
# Statistics Based on ImageNet Data for Normalisation
mean_nums = [0.485, 0.456, 0.406]
std_nums = [0.229, 0.224, 0.225]
data_transforms = {
"train": transforms.Compose(
[
transforms.Resize((150, 150)), # Resizes all images into same dimension
transforms.RandomRotation(10), # Rotates the images upto Max of 10 Degrees
transforms.RandomHorizontalFlip(
p=0.4
), # Performs Horizantal Flip over images
transforms.ToTensor(), # Coverts into Tensors
transforms.Normalize(mean=mean_nums, std=std_nums),
]
), # Normalizes
"val": transforms.Compose(
[
transforms.Resize((150, 150)),
transforms.CenterCrop(
150
), # Performs Crop at Center and resizes it to 150x150
transforms.ToTensor(),
transforms.Normalize(mean=mean_nums, std=std_nums),
]
),
}
# ## Train and Validation Data Split
# We split our dataset into train and validation sets for training and validating our model with seperate datasets.
def load_split_train_test(datadir, valid_size=0.3):
train_data = datasets.ImageFolder(
datadir, transform=data_transforms["train"]
) # Picks up Image Paths from its respective folders and label them
test_data = datasets.ImageFolder(datadir, transform=data_transforms["val"])
num_train = len(train_data)
indices = list(range(num_train))
split = int(np.floor(valid_size * num_train))
np.random.shuffle(indices)
train_idx, test_idx = indices[split:], indices[:split]
dataset_size = {"train": len(train_idx), "val": len(test_idx)}
train_sampler = SubsetRandomSampler(
train_idx
) # Sampler for splitting train and val images
test_sampler = SubsetRandomSampler(test_idx)
trainloader = torch.utils.data.DataLoader(
train_data, sampler=train_sampler, batch_size=8
) # DataLoader provides data from traininng and validation in batches
testloader = torch.utils.data.DataLoader(
test_data, sampler=test_sampler, batch_size=8
)
return trainloader, testloader, dataset_size
trainloader, valloader, dataset_size = load_split_train_test(DATA_PATH, 0.2)
dataloaders = {"train": trainloader, "val": valloader}
data_sizes = {x: len(dataloaders[x].sampler) for x in ["train", "val"]}
class_names = trainloader.dataset.classes
print(class_names)
# Like I said, `datasets.ImageFolder` takes the images from each folder named after the class name and automatically labels them. Then `data.DataLoader` loads those labelled images and tracks of the Train Data(Image) and Label(Class Name). Those are the X and Y value which we take for training.
def imshow(inp, size=(30, 30), title=None):
"""Imshow for Tensor."""
inp = inp.numpy().transpose((1, 2, 0))
mean = mean_nums
std = std_nums
inp = std * inp + mean
inp = np.clip(inp, 0, 1)
plt.figure(figsize=size)
plt.imshow(inp)
if title is not None:
plt.title(title, size=30)
plt.pause(0.001) # pause a bit so that plots are updated
# Get a batch of training data
inputs, classes = next(iter(dataloaders["train"]))
# Make a grid from batch
out = torchvision.utils.make_grid(inputs)
imshow(out, title=[class_names[x] for x in classes])
# Ofcourse, we don't want to sit for hours to see our model training. That's why we have GPU with us, now we are going to play game using GPU but this game is *The Imitation Game*
# PyTorch has `device` object to load the data into the either of two hardware [CPU or CUDA(GPU)]
if torch.cuda.is_available():
device = torch.device("cuda:0")
print("Training on GPU... Ready for HyperJump...")
else:
device = torch.device("cpu")
print("Training on CPU... May the force be with you...")
torch.cuda.empty_cache()
# ## Dense-net 121
def CNN_Model(pretrained=True):
model = models.densenet121(
pretrained=pretrained
) # Returns Defined Densenet model with weights trained on ImageNet
num_ftrs = (
model.classifier.in_features
) # Get the number of features output from CNN layer
model.classifier = nn.Linear(
num_ftrs, len(class_names)
) # Overwrites the Classifier layer with custom defined layer for transfer learning
model = model.to(device) # Transfer the Model to GPU if available
return model
model = CNN_Model(pretrained=True)
# specify loss function (categorical cross-entropy loss)
criterion = nn.CrossEntropyLoss()
# Specify optimizer which performs Gradient Descent
optimizer = optim.Adam(model.parameters(), lr=1e-3)
# Decay LR by a factor of 0.1 every 7 epochs
exp_lr_scheduler = optim.lr_scheduler.StepLR(
optimizer, step_size=7, gamma=0.1
) # Learning Scheduler
# Since we haven't froze the CNN layer parameters untrainable, we are going to train a huge number of parameters.
pytorch_total_params = sum(p.numel() for p in model.parameters() if p.requires_grad)
print("Number of trainable parameters: \n{}".format(pytorch_total_params))
# ### Training
def train_model(model, criterion, optimizer, scheduler, num_epochs=10):
since = time.time()
best_model_wts = copy.deepcopy(model.state_dict())
best_loss = np.inf
for epoch in range(num_epochs):
print("Epoch {}/{}".format(epoch + 1, num_epochs))
print("-" * 10)
# Each epoch has a training and validation phase
for phase in ["train", "val"]:
if phase == "train":
model.train() # Set model to training mode
else:
model.eval() # Set model to evaluate mode
current_loss = 0.0
current_corrects = 0
current_kappa = 0
val_kappa = list()
for inputs, labels in tqdm.tqdm(
dataloaders[phase], desc=phase, leave=False
):
inputs = inputs.to(device)
labels = labels.to(device)
# We need to zero the gradients in the Cache.
optimizer.zero_grad()
# Time to carry out the forward training poss
# We only need to log the loss stats if we are in training phase
with torch.set_grad_enabled(phase == "train"):
outputs = model(inputs)
_, preds = torch.max(outputs, 1)
loss = criterion(outputs, labels)
# backward + optimize only if in training phase
if phase == "train":
loss.backward()
optimizer.step()
if phase == "train":
scheduler.step()
# We want variables to hold the loss statistics
current_loss += loss.item() * inputs.size(0)
current_corrects += torch.sum(preds == labels.data)
val_kappa.append(
cohen_kappa_score(preds.cpu().numpy(), labels.data.cpu().numpy())
)
epoch_loss = current_loss / data_sizes[phase]
epoch_acc = current_corrects.double() / data_sizes[phase]
if phase == "val":
epoch_kappa = np.mean(val_kappa)
print(
"{} Loss: {:.4f} | {} Accuracy: {:.4f} | Kappa Score: {:.4f}".format(
phase, epoch_loss, phase, epoch_acc, epoch_kappa
)
)
else:
print(
"{} Loss: {:.4f} | {} Accuracy: {:.4f}".format(
phase, epoch_loss, phase, epoch_acc
)
)
# EARLY STOPPING
if phase == "val" and epoch_loss < best_loss:
print(
"Val loss Decreased from {:.4f} to {:.4f} \nSaving Weights... ".format(
best_loss, epoch_loss
)
)
best_loss = epoch_loss
best_model_wts = copy.deepcopy(model.state_dict())
print()
time_since = time.time() - since
print(
"Training complete in {:.0f}m {:.0f}s".format(time_since // 60, time_since % 60)
)
print("Best val loss: {:.4f}".format(best_loss))
# Now we'll load in the best model weights and return it
model.load_state_dict(best_model_wts)
return model
# Let's define a function which would give image visualisations along with the predicted label to see if the model is really outputing relevant answers.
def visualize_model(model, num_images=6):
was_training = model.training
model.eval()
images_handeled = 0
ax = plt.figure()
with torch.no_grad():
for i, (inputs, labels) in enumerate(dataloaders["val"]):
inputs = inputs.to(device)
labels = labels.to(device)
outputs = model(inputs)
_, preds = torch.max(outputs, 1)
for j in range(inputs.size()[0]):
images_handeled += 1
ax = plt.subplot(num_images // 2, 2, images_handeled)
ax.axis("off")
ax.set_title(
"Actual: {} predicted: {}".format(
class_names[labels[j].item()], class_names[preds[j]]
)
)
imshow(inputs.cpu().data[j], (5, 5))
if images_handeled == num_images:
model.train(mode=was_training)
return
model.train(mode=was_training)
# I am have run with setting of 10 Epochs due to Kaggle, feel free to tweak up the no.of epochs to get even better model
base_model = train_model(model, criterion, optimizer, exp_lr_scheduler, num_epochs=10)
visualize_model(base_model)
plt.show()
|
import os # import the data file
import numpy as np # calculations
import pandas as pd # dataframes
pd.set_option("max_columns", None) # to show all the columns
import matplotlib.pyplot as plt # visualization
import seaborn as sns # visualization
from sklearn.model_selection import train_test_split # train test split
from sklearn.model_selection import GridSearchCV # parameter optimization
from imblearn.over_sampling import SMOTE # oversampling
from imblearn.under_sampling import RandomUnderSampler
from sklearn.linear_model import LogisticRegression # logistic regression
from sklearn.neighbors import KNeighborsClassifier # knn
from sklearn.ensemble import RandomForestClassifier # random forest
import xgboost as xgb # xgboost
from sklearn.metrics import classification_report # classification report
# upload the file
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
data = pd.read_csv(
"/kaggle/input/telco-customer-churn/WA_Fn-UseC_-Telco-Customer-Churn.csv"
)
# check the number of columns, rows, missing values and data types in the dataframe
data.info()
data.head()
# remove unwanted columns
data = data.drop("customerID", axis=1)
# Convert TotalCharges to numeric
data["TotalCharges"] = pd.to_numeric(data["TotalCharges"], errors="coerce")
data.isnull().sum()
# remove missing values
data = data.dropna()
# renaming the senior citizen category
data["SeniorCitizen"] = data["SeniorCitizen"].replace({0: "No", 1: "Yes"})
data.head()
# # Exploratory Analysis
# response variable
churn_count = data["Churn"].value_counts()
plt.pie(churn_count, labels=churn_count.index, autopct="%1.1f%%")
plt.title("Customer Churn")
# From the above plot it is visible that the proportions of churned and non-churned customers are very different from each other. Only a 1/4th of customer have churned whereas 3/4rd of customers have not churned.
# categorical variables
cat_col = [
"gender",
"SeniorCitizen",
"Partner",
"Dependents",
"PhoneService",
"MultipleLines",
"InternetService",
"OnlineSecurity",
"OnlineBackup",
"TechSupport",
"DeviceProtection",
"StreamingTV",
"StreamingMovies",
"Contract",
"PaperlessBilling",
"PaymentMethod",
]
color_palette = [
["#8B8378", "#FAEBD7"],
["#458B74", "#7FFFD4"],
["#8B2323", "#FF4040"],
["#53868B", "#98F5FF"],
["#458B00", "#76EE00"],
["#68228B", "#BF3EFF"],
["#698B69", "#C1FFC1"],
["#8B6914", "#FFC125"],
["#8B3A62", "#FF6EB4"],
["#8B864E", "#F0E68C"],
["#698B22", "#C0FF3E"],
["#8B4789", "#DA70D6"],
["#548B54", "#98FB98"],
["#668B8B", "#BBFFFF"],
["#8B668B", "#FFBBFF"],
["#2E8B57", "#54FF9F"],
]
fig, axes = plt.subplots(nrows=4, ncols=4, figsize=(20, 20))
for k, ax in zip(range(16), axes.flatten()):
sns.countplot(
data=data, x=f"{cat_col[k]}", hue="Churn", palette=color_palette[k], ax=ax
)
ax.set_title(f"Churn vs {cat_col[k]}")
if k == 15:
ax.set_xticklabels(ax.get_xticklabels(), rotation=25, ha="right")
else:
pass
# 1. Gender seems to have no impact over the churn rate.
# 2. Non-senior citizens have a more possibility to churn than the senior citizens.
# 3. Having a partner has reduced the churning rate.
# 4. When there are dependents there's a high possibility of churning.
# 5. Customers who have a phone service have churned most.
# 6. There's no effect from having multiple lines to churn rates.
# 7. From the different internet services, fiber optic service have high churning rates.
# 8. Customers who haven't got addition facilities such as online security, online backup, tech support, device protection, streaming TV, streaming movies have the high chance of churning than the others.
# 9. With the contract type, customers who have short term contracts like month-to-month contract has the lowest probability of renewing their contracts.
# 10. Paperless billing seems to contribute more to the churning than the other methods.
# 11. Out of the four payment methods electronic-check method has the highest churning rate.
# quantitative variables
qnt_col = ["tenure", "MonthlyCharges", "TotalCharges"]
fig, axes = plt.subplots(nrows=1, ncols=3, figsize=(20, 8))
for k, ax in zip(range(3), axes.flatten()):
sns.boxplot(
data=data, x="Churn", y=f"{qnt_col[k]}", palette=color_palette[1], ax=ax
)
ax.set_title(f"Churn vs {qnt_col[k]}")
# 1. Tenure is the length of time a customer stays with a company. So it is obvious that the tenure is high in non-churning customers. This depicts from the above chart as well as the average tenure is hugh in non-churning category.
# 2. The customers who have high monthly charges have churned more.
# 3. The situation is different when it comes to the total charges. Average total charge is high in non-churning customers.
# # Data Pre-processing
data["Churn"] = data["Churn"].replace({"No": 0, "Yes": 1})
y = data["Churn"]
x = data.drop("Churn", axis=1)
x = pd.get_dummies(x, drop_first=True)
x.head()
# split train and test set
x_train, x_test, y_train, y_test = train_test_split(x, y, test_size=0.3, random_state=0)
# oversampling
sm = SMOTE(random_state=0)
x_sm, y_sm = sm.fit_resample(x_train, y_train)
# undersampling
rus = RandomUnderSampler(random_state=0)
x_rus, y_rus = rus.fit_resample(x_train, y_train)
# # Logistic Model
acc_summary = pd.DataFrame(columns=["Model", "Resampling Technique", "Accuracy"])
# Imbalanced Sample
model_lr_im = LogisticRegression(random_state=0)
model_lr_im.fit(x_train, y_train)
y_pred_lr_im = model_lr_im.predict(x_test)
cr_lr_im = classification_report(y_pred_lr_im, y_test)
print(cr_lr_im)
# OverSampling
model_lr_os = LogisticRegression(random_state=0)
model_lr_os.fit(x_sm, y_sm)
y_pred_lr_os = model_lr_os.predict(x_test)
cr_lr_os = classification_report(y_pred_lr_os, y_test)
print(cr_lr_os)
# UnderSampling
model_lr_us = LogisticRegression(random_state=0)
model_lr_us.fit(x_rus, y_rus)
y_pred_lr_us = model_lr_us.predict(x_test)
cr_lr_us = classification_report(y_pred_lr_us, y_test)
print(cr_lr_us)
row1 = pd.Series(["Logistic", "Imbalanced", 0.80], index=acc_summary.columns)
row2 = pd.Series(["Logistic", "Over Sampling", 0.76], index=acc_summary.columns)
row3 = pd.Series(["Logistic", "Under Sampling", 0.75], index=acc_summary.columns)
acc_summary = acc_summary.append([row1, row2, row3], ignore_index=True)
# # KNN
# Imbalanced Sample
knn = KNeighborsClassifier()
param_grid = {"n_neighbors": range(1, 20)}
grid = GridSearchCV(knn, param_grid, cv=5)
grid.fit(x_train, y_train)
print("Best n_neighbors:", grid.best_params_["n_neighbors"])
knn = KNeighborsClassifier(n_neighbors=10)
model_knn_im = knn.fit(x_train, y_train)
y_pred_knn_im = model_knn_im.predict(x_test)
cr_knn_im = classification_report(y_pred_knn_im, y_test)
print(cr_knn_im)
# OverSampling
knn = KNeighborsClassifier()
param_grid = {"n_neighbors": range(1, 20)}
grid = GridSearchCV(knn, param_grid, cv=5)
grid.fit(x_sm, y_sm)
print("Best n_neighbors:", grid.best_params_["n_neighbors"])
knn = KNeighborsClassifier(n_neighbors=1)
model_knn_os = knn.fit(x_sm, y_sm)
y_pred_knn_os = model_knn_os.predict(x_test)
cr_knn_os = classification_report(y_pred_knn_os, y_test)
print(cr_knn_os)
# UnderSampling
knn = KNeighborsClassifier()
param_grid = {"n_neighbors": range(1, 20)}
grid = GridSearchCV(knn, param_grid, cv=5)
grid.fit(x_rus, y_rus)
print("Best n_neighbors:", grid.best_params_["n_neighbors"])
knn = KNeighborsClassifier(n_neighbors=19)
model_knn_us = knn.fit(x_rus, y_rus)
y_pred_knn_us = model_knn_us.predict(x_test)
cr_knn_us = classification_report(y_pred_knn_us, y_test)
print(cr_knn_us)
row4 = pd.Series(["KNN", "Imbalanced", 0.78], index=acc_summary.columns)
row5 = pd.Series(["KNN", "Over Sampling", 0.69], index=acc_summary.columns)
row6 = pd.Series(["KNN", "Under Sampling", 0.71], index=acc_summary.columns)
acc_summary = acc_summary.append([row4, row5, row6], ignore_index=True)
# # Random Forest
# Imbalanced Sample
rf = RandomForestClassifier()
param_grid = {
"n_estimators": [100, 200, 300],
"max_depth": [10, 20, 30],
"min_samples_split": [2, 4, 6],
"min_samples_leaf": [1, 2, 3],
"bootstrap": [True, False],
}
grid_search = GridSearchCV(
estimator=rf, param_grid=param_grid, cv=5, n_jobs=-1, verbose=False
)
grid_search.fit(x_train, y_train)
print("Best parameters:", grid_search.best_params_)
rf = RandomForestClassifier(
n_estimators=300,
bootstrap=True,
max_depth=10,
min_samples_leaf=3,
min_samples_split=6,
)
model_rf_im = rf.fit(x_train, y_train)
y_pred_rf_im = model_rf_im.predict(x_test)
cr_rf_im = classification_report(y_pred_rf_im, y_test)
print(cr_rf_im)
# OverSampling
rf = RandomForestClassifier()
param_grid = {
"n_estimators": [100, 200, 300],
"max_depth": [10, 20, 30],
"min_samples_split": [2, 4, 6],
"min_samples_leaf": [1, 2, 3],
"bootstrap": [True, False],
}
grid_search = GridSearchCV(
estimator=rf, param_grid=param_grid, cv=5, n_jobs=-1, verbose=False
)
grid_search.fit(x_sm, y_sm)
print("Best parameters:", grid_search.best_params_)
rf = RandomForestClassifier(
n_estimators=300,
bootstrap=True,
max_depth=30,
min_samples_leaf=1,
min_samples_split=4,
)
model_rf_os = rf.fit(x_sm, y_sm)
y_pred_rf_os = model_rf_os.predict(x_test)
cr_rf_os = classification_report(y_pred_rf_os, y_test)
print(cr_rf_os)
# UnderSampling
rf = RandomForestClassifier()
param_grid = {
"n_estimators": [100, 200, 300],
"max_depth": [10, 20, 30],
"min_samples_split": [2, 4, 6],
"min_samples_leaf": [1, 2, 3],
"bootstrap": [True, False],
}
grid_search = GridSearchCV(
estimator=rf, param_grid=param_grid, cv=5, n_jobs=-1, verbose=False
)
grid_search.fit(x_rus, y_rus)
print("Best parameters:", grid_search.best_params_)
rf = RandomForestClassifier(
n_estimators=200,
bootstrap=True,
max_depth=20,
min_samples_leaf=3,
min_samples_split=4,
)
model_rf_us = rf.fit(x_rus, y_rus)
y_pred_rf_us = model_rf_us.predict(x_test)
cr_rf_us = classification_report(y_pred_rf_us, y_test)
print(cr_rf_us)
row7 = pd.Series(["Random Forest", "Imbalanced", 0.80], index=acc_summary.columns)
row8 = pd.Series(["Random Forest", "Over Sampling", 0.77], index=acc_summary.columns)
row9 = pd.Series(["Random Forest", "Under Sampling", 0.75], index=acc_summary.columns)
acc_summary = acc_summary.append([row7, row8, row9], ignore_index=True)
# # XgBoost
# Imbalanced Sample
xb = xgb.XGBClassifier()
param_grid = {
"learning_rate": [0.01, 0.1, 0.5],
"max_depth": [3, 5, 7],
"n_estimators": [100, 500, 1000],
"subsample": [0.5, 1],
"colsample_bytree": [0.5, 1],
}
grid_search = GridSearchCV(estimator=xb, param_grid=param_grid, cv=5, verbose=False)
grid_search.fit(x_train, y_train)
print("Best parameters: ", grid_search.best_params_)
xg = xgb.XGBClassifier(
learning_rate=0.1, max_depth=3, n_estimators=100, subsample=0.5, colsample_bytree=1
)
model_xg_im = xg.fit(x_train, y_train)
y_pred_xg_im = model_xg_im.predict(x_test)
cr_xg_im = classification_report(y_pred_xg_im, y_test)
print(cr_xg_im)
# OverSampling
xb = xgb.XGBClassifier()
param_grid = {
"learning_rate": [0.01, 0.1, 0.5],
"max_depth": [3, 5, 7],
"n_estimators": [100, 500, 1000],
"subsample": [0.5, 1],
"colsample_bytree": [0.5, 1],
}
grid_search = GridSearchCV(estimator=xb, param_grid=param_grid, cv=5, verbose=False)
grid_search.fit(x_sm, y_sm)
print("Best parameters: ", grid_search.best_params_)
xg = xgb.XGBClassifier(
learning_rate=0.1, max_depth=7, n_estimators=100, subsample=1, colsample_bytree=0.5
)
model_xg_os = xg.fit(x_sm, y_sm)
y_pred_xg_os = model_xg_os.predict(x_test)
cr_xg_os = classification_report(y_pred_xg_os, y_test)
print(cr_xg_os)
# UnderSampling
xb = xgb.XGBClassifier()
param_grid = {
"learning_rate": [0.01, 0.1, 0.5],
"max_depth": [3, 5, 7],
"n_estimators": [100, 500, 1000],
"subsample": [0.5, 1],
"colsample_bytree": [0.5, 1],
}
grid_search = GridSearchCV(estimator=xb, param_grid=param_grid, cv=5, verbose=False)
grid_search.fit(x_rus, y_rus)
print("Best parameters: ", grid_search.best_params_)
xg = xgb.XGBClassifier(
learning_rate=0.01,
max_depth=3,
n_estimators=500,
subsample=0.5,
colsample_bytree=0.5,
)
model_xg_us = xg.fit(x_rus, y_rus)
y_pred_xg_us = model_xg_us.predict(x_test)
cr_xg_us = classification_report(y_pred_xg_us, y_test)
print(cr_xg_us)
row10 = pd.Series(["XgBoost", "Imbalanced", 0.80], index=acc_summary.columns)
row11 = pd.Series(["XgBoost", "Over Sampling", 0.78], index=acc_summary.columns)
row12 = pd.Series(["XgBoost", "Under Sampling", 0.75], index=acc_summary.columns)
acc_summary = acc_summary.append([row10, row11, row12], ignore_index=True)
# # Model Summary
acc_summary
|
from huggingface_hub import login
login(token="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")
import torch
from transformers import (
AutoTokenizer,
AutoModelForSeq2SeqLM,
Seq2SeqTrainingArguments,
Seq2SeqTrainer,
T5Tokenizer,
)
from datasets import load_dataset
raw_datasets = load_dataset("cfilt/iitb-english-hindi")
src_lang = "en"
target_lang = "hi"
model_checkpoint = "google/mt5-small"
tokenizer = T5Tokenizer.from_pretrained(model_checkpoint)
def preprocess_function(examples):
inputs = [example[src_lang] for example in examples["translation"]]
targets = [example[target_lang] for example in examples["translation"]]
model_inputs = tokenizer(
inputs, text_target=targets, max_length=128, truncation=True, padding=True
)
return model_inputs
tokenized_dataset = raw_datasets.map(preprocess_function, batched=True)
# from datasets import load_from_disk
# tokenized_dataset = load_from_disk("tokenized_dataset")
tokenized_dataset
# tokenized_dataset.save_to_disk("tokenized_dataset")
del raw_datasets
model = AutoModelForSeq2SeqLM.from_pretrained(model_checkpoint)
from transformers import DataCollatorForSeq2Seq
data_collator = DataCollatorForSeq2Seq(tokenizer=tokenizer, model=model_checkpoint)
import evaluate, numpy as np
metric = evaluate.load("sacrebleu")
def postprocess_text(preds, labels):
preds = [pred.strip() for pred in preds]
labels = [[label.strip()] for label in labels]
return preds, labels
def compute_metrics(eval_preds):
preds, labels = eval_preds
if isinstance(preds, tuple):
preds = preds[0]
decoded_preds = tokenizer.batch_decode(preds, skip_special_tokens=True)
labels = np.where(labels != -100, labels, tokenizer.pad_token_id)
decoded_labels = tokenizer.batch_decode(labels, skip_special_tokens=True)
decoded_preds, decoded_labels = postprocess_text(decoded_preds, decoded_labels)
result = metric.compute(predictions=decoded_preds, references=decoded_labels)
result = {"bleu": result["score"]}
prediction_lens = [
np.count_nonzero(pred != tokenizer.pad_token_id) for pred in preds
]
result["gen_len"] = np.mean(prediction_lens)
result = {k: round(v, 4) for k, v in result.items()}
return result
training_args = Seq2SeqTrainingArguments(
output_dir="mt5-en-to-hi",
evaluation_strategy="epoch",
learning_rate=2e-5,
per_device_train_batch_size=16,
per_device_eval_batch_size=16,
weight_decay=0.01,
save_total_limit=3,
num_train_epochs=4,
predict_with_generate=True,
fp16=False,
report_to="none",
push_to_hub=True,
)
trainer = Seq2SeqTrainer(
model=model,
args=training_args,
train_dataset=tokenized_dataset["train"].shuffle(seed=42).select(range(500000)),
eval_dataset=tokenized_dataset["validation"],
tokenizer=tokenizer,
data_collator=data_collator,
compute_metrics=compute_metrics,
)
trainer.train()
def translate(text):
inputs = tokenizer(text, return_tensors="pt").input_ids.to("cuda")
outputs = model.generate(
inputs, max_new_tokens=40, do_sample=True, top_k=30, top_p=0.95
)
outputs = tokenizer.decode(outputs[0], skip_special_tokens=True)
return outputs
text = "How are you?"
print(translate(text))
trainer.push_to_hub()
|
key = "AIzaSyCLx1cVxGxez6FsHD0uE671_B2W7q7q8XE"
import requests
import json, os
import urllib.request
from shapely.geometry import Point, Polygon
from matplotlib import pyplot as plt
import shapely
import pickle
import random
import numpy as np
import pandas as pd
from PIL import Image
from tqdm import tqdm
# output data directory
data_dir = "./all_images"
if not os.path.exists(data_dir):
os.mkdir(data_dir)
indPolyGrid = pickle.load(open("/kaggle/input/geodataindia/indPolyGrid.pkl", "rb"))
for i in indPolyGrid.values():
plt.plot(i[:, 1], i[:, 0])
print(len(indPolyGrid))
search_grids = indPolyGrid.keys()
# search_grids = list(range(50,60))
print(
"Searching grids : {}".format(
"All" if search_grids == indPolyGrid.keys() else search_grids
)
)
base = "https://maps.googleapis.com/maps/api/streetview"
ext = "?size=600x300&location={}&fov=100&heading={}&radius={}&pitch=10&key={}"
print(
"Seacrching Grids: {}".format(
"All" if search_grids == indPolyGrid.keys() else search_grids
)
)
latitudes = []
longitudes = []
paths = []
grids = []
for grid, coor in indPolyGrid.items():
poly = Polygon(np.flip(coor))
minx, miny, maxx, maxy = poly.bounds
count = 0
trials = 0
loc_list = set()
if grid in search_grids:
loc_list = set(os.listdir(data_dir))
print("=" * 15 + f" Searching grid {grid} " + "=" * 15)
while count < 20 and trials < 10:
point = Point(random.uniform(minx, maxx), random.uniform(miny, maxy))
location = str(point.y) + "," + str(point.x)
if (poly.contains(point)) and (location not in loc_list):
meta_url = base + "/metadata" + ext.format(location, 0, 1000, key)
r = requests.get(meta_url).json()
trials += 1
print("Trial: {}, count: {}".format(trials, count))
if r["status"] == "OK" and poly.contains(
Point(r["location"]["lng"], r["location"]["lat"])
):
location = (
str(r["location"]["lat"]) + "," + str(r["location"]["lng"])
)
if location not in loc_list:
print("Valid location found: {}".format(location))
loc_list.add(location)
for heading in [0, 90, 180]:
img_url = base + ext.format(location, heading, 10000, key)
urllib.request.urlretrieve(
img_url,
data_dir
+ "/{}_{}_{}_{}.jpg".format(
grid, location, heading, r["date"]
),
)
paths.append(
"/{}_{}_{}_{}.jpg".format(
grid, location, heading, r["date"]
)
)
latitudes.append(r["location"]["lat"])
longitudes.append(r["location"]["lng"])
grids.append(grid)
print(img_url)
count += 1
trials = 0
else:
print("Failed trial {} location exist".format(trials))
print("Location : {}".format(location))
else:
print("Failed trial {} status or contains".format(trials))
print("Location {}".format(location))
print(loc_list)
print(
"=" * 15 + f" Final locations for grid {grid}: {len(loc_list)} " + "=" * 15
)
df = pd.DataFrame()
df["latitudes"] = latitudes
df["longitudes"] = longitudes
df["paths"] = paths
df["class"] = grids
df.to_csv("all.csv", index=None)
df
df = pd.read_csv("/kaggle/working/all.csv")
# random sampling from df
samples = df.sample(n=6)
samples.reset_index(drop=True, inplace=True)
for i in range(6):
plt.figure(figsize=(20, 20))
plt.subplot(6, 1, i + 1)
im = Image.open(data_dir + samples["paths"][i])
plt.xlabel(
f"lat: {samples['latitudes'][i]:.2f}, long: {samples['longitudes'][i]:.2f}, grid: {samples['class'][i]}"
)
plt.imshow(im)
plt.show()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
import seaborn as sns
import matplotlib.pyplot as plt
import plotly.express as px
import missingno as msno
from plotly.subplots import make_subplots
import plotly.graph_objects as go
import warnings
warnings.filterwarnings("ignore")
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# Loading data
esport = pd.read_csv(
"/kaggle/input/esport-earnings/ESport_Earnings.csv",
error_bad_lines=False,
engine="python",
sep=",",
encoding="latin-1",
)
esport.head()
# checking the shape
esport.shape
# Checking for nulls
esport.isnull().sum()
# duplicates
esport.duplicated().sum()
esport.columns
# summary statistics
esport.describe()
# we have invalid years
# so will drop these rows are not 2006 and above on release year column
esport = esport.drop(esport[esport["Releaseyear"] < 2006].index)
esport.shape
esport.describe()
# viewing genre in the data
esport["Genre"].value_counts()
# # EDA
# ### Performance of different games in esports
# Which game has the highest total prize money?
highest_prize_money = (
esport.groupby("GameName")["TotalMoney"].sum().sort_values(ascending=False).head(1)
)
print("The game with the highest total prize money is:", highest_prize_money.index[0])
# How does the prize money vary across different game genres?
genre_prize_money = (
esport.groupby("Genre")["TotalMoney"].sum().sort_values(ascending=False)
)
print("Total prize money by genre:\n", genre_prize_money)
# Which game has the highest number of players and tournaments?
most_players = (
esport.groupby("GameName")["PlayerNo"].sum().sort_values(ascending=False).head(1)
)
most_tournaments = (
esport.groupby("GameName")["TournamentNo"]
.sum()
.sort_values(ascending=False)
.head(1)
)
print("The game with the most players is:", most_players.index[0])
print("The game with the most tournaments is:", most_tournaments.index[0])
sns.set_style("whitegrid")
sns.set(rc={"figure.figsize": (11.7, 8.27)})
# Create a horizontal bar chart of total prize money by game genre with bars in descending order
genre_order = (
esport.groupby(["Genre"])["TotalMoney"].sum().sort_values(ascending=False).index
)
sns.barplot(
x="TotalMoney", y="Genre", data=esport, estimator=sum, ci=None, order=genre_order
)
plt.title("Total Prize Money by Game Genre")
plt.xlabel("Total Prize Money")
plt.ylabel("Game Genre")
plt.show()
# How does the release year of a game affect its total prize money and popularity in esports?
year_prize_money = (
esport.groupby("Releaseyear")["TotalMoney"].sum().sort_values(ascending=False)
)
print(year_prize_money.head(15))
# How does the release year of a game affect its total prize money and popularity in esports?
year_players = esport.groupby("Releaseyear")["PlayerNo"].sum()
print(year_players.head(15))
# How does the release year of a game affect its total prize money and popularity in esports?
year_tournaments = esport.groupby("Releaseyear")["TournamentNo"].sum()
print(year_tournaments.head(15))
sns.set_style("whitegrid")
sns.set(rc={"figure.figsize": (11.7, 8.27)})
# Create a scatter plot of release year vs total prize money
sns.scatterplot(x="Releaseyear", y="TotalMoney", data=esport)
plt.title("Release Year vs Total Prize Money")
plt.xlabel("Release Year")
plt.ylabel("Total Prize Money")
plt.show()
# ### Top countries in esports and analyze their earnings
# Which country has the highest earnings in esports?
highest_earnings = (
esport.groupby("Top_Country")["Top_Country_Earnings"]
.sum()
.sort_values(ascending=False)
.head(1)
)
print("The country with the highest earnings in esports is:", highest_earnings.index[0])
sns.set_style("whitegrid")
sns.set(rc={"figure.figsize": (11.7, 8.27)})
# Create a horizontal bar chart of total earnings by country with bars in descending order
country_order = (
esport.groupby(["Top_Country"])["Top_Country_Earnings"]
.sum()
.sort_values(ascending=False)
.index
)
sns.barplot(
x="Top_Country_Earnings",
y="Top_Country",
data=esport,
estimator=sum,
ci=None,
order=country_order,
)
plt.title("Total Earnings by Country")
plt.xlabel("Total Earnings")
plt.ylabel("Country")
plt.show()
# How does the earnings of the top countries vary across different game genres?
genre_earnings = (
esport.groupby(["Genre", "Top_Country"])["Top_Country_Earnings"]
.sum()
.sort_values(ascending=False)
)
print("Total earnings by genre and country:\n", genre_earnings)
# How does the number of players and tournaments affect the earnings of the top countries?
country_players = (
esport.groupby("Top_Country")["PlayerNo"].sum().sort_values(ascending=False)
)
print(country_players.head(10))
sns.set_style("whitegrid")
sns.set(rc={"figure.figsize": (11.7, 8.27)})
# Create a horizontal bar chart of total earnings by country with bars in descending order
country_order = (
esport.groupby(["Top_Country"])["PlayerNo"].sum().sort_values(ascending=False).index
)
sns.barplot(
x="PlayerNo",
y="Top_Country",
data=esport,
estimator=sum,
ci=None,
order=country_order,
)
plt.title("PlayerNo by Country")
plt.xlabel("PlayerNo")
plt.ylabel("Country")
plt.show()
# How does the number of players and tournaments affect the earnings of the top countries?
country_tournaments = (
esport.groupby("Top_Country")["TournamentNo"].sum().sort_values(ascending=False)
)
print(country_tournaments.head(10))
sns.set_style("whitegrid")
sns.set(rc={"figure.figsize": (11.7, 8.27)})
# Create a horizontal bar chart of total earnings by country with bars in descending order
country_order = (
esport.groupby(["Top_Country"])["TournamentNo"]
.sum()
.sort_values(ascending=False)
.index
)
sns.barplot(
x="TournamentNo",
y="Top_Country",
data=esport,
estimator=sum,
ci=None,
order=country_order,
)
plt.title("TournamentNo by Country")
plt.xlabel("TournamentNo")
plt.ylabel("Country")
plt.show()
# ### Relationship between player and tournament numbers and total prize money.
# Is there a correlation between the number of players and the total prize money of a tournament?
sns.scatterplot(x="PlayerNo", y="TotalMoney", data=esport)
plt.show()
# Is there a correlation between the number of tournaments and the total prize money of a game?
sns.scatterplot(x="TournamentNo", y="TotalMoney", data=esport)
plt.show()
# How does the number of players and tournaments affect the prize money distribution across different game genres?
genre_players = esport.groupby("Genre")["PlayerNo"].sum().sort_values(ascending=False)
print(genre_players.head(10))
sns.set_style("whitegrid")
sns.set(rc={"figure.figsize": (11.7, 8.27)})
# Create a horizontal bar chart of total earnings by country with bars in descending order
country_order = (
esport.groupby(["Genre"])["PlayerNo"].sum().sort_values(ascending=False).index
)
sns.barplot(
x="PlayerNo", y="Genre", data=esport, estimator=sum, ci=None, order=country_order
)
plt.title("PlayerNo by Country")
plt.xlabel("PlayerNo")
plt.ylabel("Genre")
plt.show()
# How does the number of players and tournaments affect the prize money distribution across different game genres?
genre_tournaments = (
esport.groupby("Genre")["TournamentNo"].sum().sort_values(ascending=False)
)
print(genre_tournaments.head(10))
sns.set_style("whitegrid")
sns.set(rc={"figure.figsize": (11.7, 8.27)})
# Create a horizontal bar chart of total earnings by country with bars in descending order
country_order = (
esport.groupby(["Genre"])["TournamentNo"].sum().sort_values(ascending=False).index
)
sns.barplot(
x="TournamentNo",
y="Genre",
data=esport,
estimator=sum,
ci=None,
order=country_order,
)
plt.title("TournamentNo by Country")
plt.xlabel("TournamentNo")
plt.ylabel("Genre")
plt.show()
# How does the number of players and tournaments affect the prize money distribution across different game genres?
genre_total_money = (
esport.groupby("Genre")["TotalMoney"].sum().sort_values(ascending=False)
)
print(genre_total_money.head(10))
sns.set_style("whitegrid")
sns.set(rc={"figure.figsize": (11.7, 8.27)})
# Create a horizontal bar chart of total earnings by country with bars in descending order
country_order = (
esport.groupby(["Genre"])["TotalMoney"].sum().sort_values(ascending=False).index
)
sns.barplot(
x="TotalMoney", y="Genre", data=esport, estimator=sum, ci=None, order=country_order
)
plt.title("TotalMoney by Country")
plt.xlabel("TotalMoney")
plt.ylabel("Genre")
plt.show()
# Which player has earned the most money in esports?
highest_earnings_player = esport.groupby("IdNo")["TotalMoney"].sum()
print(highest_earnings_player.head())
sns.set_style("whitegrid")
sns.set(rc={"figure.figsize": (11.7, 8.27)})
# Create a scatter plot matrix of selected variables with title and legend
selected_vars = esport[
["IdNo", "TotalMoney", "Genre", "Top_Country", "Top_Country_Earnings"]
]
scatter_plot = sns.pairplot(selected_vars, hue="Genre")
scatter_plot.fig.suptitle(
"Relationships between 'IdNo', 'TotalMoney', 'Genre', 'Top_Country', and 'Top_Country_Earnings'"
)
scatter_plot._legend.set_title("Game Genre")
plt.show()
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import numpy as np
import matplotlib.image as mpimg
import matplotlib.pyplot as plt
import tensorflow as tf
plt.figure(figsize=(12, 5))
sp = plt.subplot(2, 5, 1)
sp.axis("Off")
img = mpimg.imread("/kaggle/input/messy-vs-clean-room/images/train/clean/30.png")
plt.imshow(img)
sp = plt.subplot(2, 5, 2)
sp.axis("Off")
img = mpimg.imread("/kaggle/input/messy-vs-clean-room/images/train/clean/24.png")
plt.imshow(img)
sp = plt.subplot(2, 5, 3)
sp.axis("Off")
img = mpimg.imread("/kaggle/input/messy-vs-clean-room/images/train/clean/32.png")
plt.imshow(img)
sp = plt.subplot(2, 5, 4)
sp.axis("Off")
img = mpimg.imread("/kaggle/input/messy-vs-clean-room/images/train/clean/21.png")
plt.imshow(img)
sp = plt.subplot(2, 5, 5)
sp.axis("Off")
img = mpimg.imread("/kaggle/input/messy-vs-clean-room/images/train/clean/89.png")
plt.imshow(img)
sp = plt.subplot(2, 5, 6)
sp.axis("Off")
img = mpimg.imread("/kaggle/input/messy-vs-clean-room/images/train/messy/2.png")
plt.imshow(img)
sp = plt.subplot(2, 5, 7)
sp.axis("Off")
img = mpimg.imread("/kaggle/input/messy-vs-clean-room/images/train/messy/89.png")
plt.imshow(img)
from tensorflow.keras.preprocessing.image import ImageDataGenerator
# train and test directories
train_dir = "/kaggle/input/messy-vs-clean-room/images/train/"
validation_dir = "/kaggle/input/messy-vs-clean-room/images/val/"
# All images will be rescaled by 1./255.
train_datagen = ImageDataGenerator(rescale=1.0 / 255.0)
test_datagen = ImageDataGenerator(rescale=1.0 / 255.0)
# --------------------
# Flow training images in batches of 20 using train_datagen generator & test_datagen
# --------------------
train_generator = train_datagen.flow_from_directory(
train_dir, batch_size=20, class_mode="binary", target_size=(150, 150)
)
validation_generator = test_datagen.flow_from_directory(
validation_dir, batch_size=20, class_mode="binary", target_size=(150, 150)
)
# Building the model.
#
from tensorflow.keras.layers import Dense, Conv2D, MaxPooling2D, Flatten, Dropout
model = tf.keras.models.Sequential(
[
# input shape is the desired size of the image 150x150 with 3 bytes color
Conv2D(16, (3, 3), activation="relu", input_shape=(150, 150, 3)),
MaxPooling2D(pool_size=(2, 2)),
Conv2D(32, (3, 3), activation="relu"),
MaxPooling2D(pool_size=(2, 2)),
Conv2D(64, (3, 3), activation="relu"),
MaxPooling2D(2, 2),
Flatten(),
Dense(512, activation="relu"),
Dropout(rate=0.2),
Dense(200, activation="relu"),
Dropout(0.1),
Dense(1, activation="sigmoid")
# output : clean(0or1) , messy (1or0)
]
)
# Summary of the model.**
model.summary()
from tensorflow import keras
keras.utils.plot_model(model)
from tensorflow.keras.optimizers import RMSprop
from tensorflow.keras.callbacks import EarlyStopping
monitor = EarlyStopping(
monitor="val_loss",
min_delta=1e-3,
patience=5,
verbose=1,
mode="auto",
restore_best_weights=True,
)
model.compile(optimizer="adam", loss="binary_crossentropy", metrics=["acc"])
history = model.fit(
train_generator,
validation_data=validation_generator,
steps_per_epoch=100,
epochs=5,
validation_steps=50,
verbose=2,
callbacks=[monitor],
)
# test path: /kaggle/input/messy-vs-clean-room/images/images/test/
test2_datagen = ImageDataGenerator(rescale=1.0 / 255.0)
test_dir = "/kaggle/input/messy-vs-clean-room/images/images/val/"
test_generator = test2_datagen.flow_from_directory(
test_dir, batch_size=6, class_mode=None, target_size=(150, 150), shuffle=False
)
y_prob = model.predict_generator(test_generator, callbacks=[monitor])
y_prob
y_pred = ["R" if probs > 0.5 else "O" for probs in y_prob]
y_pred
for dirname, _, filenames in os.walk(
"/kaggle/input/messy-vs-clean-room/images/images/val/"
):
images = []
for filename in filenames:
img_path = os.path.join(dirname, filename)
# print(img_path)
images.append(mpimg.imread(img_path))
plt.figure(figsize=(20, 10))
columns = 5
for i, image in enumerate(images):
plt.subplot(len(images) / columns + 1, columns, i + 1)
plt.imshow(image)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import pandas as pd
data = pd.read_csv(
"/kaggle/input/entity-annotated-corpus/ner_dataset.csv", encoding="unicode_escape"
)
data.head()
from itertools import chain
def get_dict_map(data, token_or_tag):
tok2idx = {}
idx2tok = {}
if token_or_tag == "token":
vocab = list(set(data["Word"].to_list()))
else:
vocab = list(set(data["Tag"].to_list()))
idx2tok = {idx: tok for idx, tok in enumerate(vocab)}
tok2idx = {tok: idx for idx, tok in enumerate(vocab)}
return tok2idx, idx2tok
token2idx, idx2token = get_dict_map(data, "token")
tag2idx, idx2tag = get_dict_map(data, "tag")
data["Word_idx"] = data["Word"].map(token2idx)
data["Tag_idx"] = data["Tag"].map(tag2idx)
data.head()
# Fill na
data_fillna = data.fillna(method="ffill", axis=0)
# Groupby and collect columns
data_group = data_fillna.groupby(["Sentence #"], as_index=False)[
"Word", "POS", "Tag", "Word_idx", "Tag_idx"
].agg(lambda x: list(x))
# Visualise data
data_group.head()
from sklearn.model_selection import train_test_split
from tensorflow.keras.preprocessing.sequence import pad_sequences
from tensorflow.keras.utils import to_categorical
import numpy as np
import tensorflow
from tensorflow.keras import Sequential, Model, Input
from tensorflow.keras.layers import (
LSTM,
Embedding,
Dense,
TimeDistributed,
Dropout,
Bidirectional,
)
from tensorflow.keras.utils import plot_model
import os
from PIL import Image
import matplotlib.pyplot as plt
import numpy as np
import torch
from torch import nn
from torch.utils.data import DataLoader
from torchvision import transforms
import torch.optim as optim
import time
from tqdm.auto import tqdm
from torch.utils.data import ConcatDataset, DataLoader, Subset, Dataset
from torchvision.datasets import DatasetFolder, VisionDataset
def get_pad_train_test_val(data_group, data):
# get max token and tag length
n_token = len(list(set(data["Word"].to_list())))
n_tag = len(list(set(data["Tag"].to_list())))
# Pad tokens (X var)
tokens = data_group["Word_idx"].tolist()
maxlen = max([len(s) for s in tokens])
pad_tokens = pad_sequences(
tokens, maxlen=maxlen, dtype="int32", padding="post", value=n_token - 1
)
# Pad Tags (y var) and convert it into one hot encoding
tags = data_group["Tag_idx"].tolist()
pad_tags = pad_sequences(
tags, maxlen=maxlen, dtype="int32", padding="post", value=tag2idx["O"]
)
n_tags = len(tag2idx)
pad_tags = [to_categorical(i, num_classes=n_tags) for i in pad_tags]
# Split train, test and validation set
tokens_, test_tokens, tags_, test_tags = train_test_split(
pad_tokens, pad_tags, test_size=0.1, train_size=0.9, random_state=2023
)
train_tokens, val_tokens, train_tags, val_tags = train_test_split(
tokens_, tags_, test_size=0.25, train_size=0.75, random_state=2023
)
print(
"train_tokens length:",
len(train_tokens),
"\ntrain_tokens length:",
len(train_tokens),
"\ntest_tokens length:",
len(test_tokens),
"\ntest_tags:",
len(test_tags),
"\nval_tokens:",
len(val_tokens),
"\nval_tags:",
len(val_tags),
)
return train_tokens, val_tokens, test_tokens, train_tags, val_tags, test_tags
(
train_tokens,
val_tokens,
test_tokens,
train_tags,
val_tags,
test_tags,
) = get_pad_train_test_val(data_group, data)
train_tokens, val_tokens, test_tokens, train_tags, val_tags, test_tags = (
torch.Tensor(i).long()
for i in [train_tokens, val_tokens, test_tokens, train_tags, val_tags, test_tags]
)
train_tokens[0]
from numpy.random import seed
seed(1)
tensorflow.random.set_seed(2)
input_dim = len(list(set(data["Word"].to_list()))) + 1
output_dim = 64
input_length = max([len(s) for s in data_group["Word_idx"].tolist()])
n_tags = len(tag2idx)
print(
"input_dim: ",
input_dim,
"\noutput_dim: ",
output_dim,
"\ninput_length: ",
input_length,
"\nn_tags: ",
n_tags,
)
# train_tokens[0],train_tags[0]
# torch.from_numpy (train_tokens[0]),torch.from_numpy (train_tags[0])
def get_bilstm_lstm_model():
model = Sequential()
# Add Embedding layer
model.add(
Embedding(input_dim=input_dim, output_dim=output_dim, input_length=input_length)
)
# Add bidirectional LSTM
model.add(
Bidirectional(
LSTM(
units=output_dim,
return_sequences=True,
dropout=0.2,
recurrent_dropout=0.2,
),
merge_mode="concat",
)
)
# Add LSTM
model.add(
LSTM(
units=output_dim, return_sequences=True, dropout=0.5, recurrent_dropout=0.5
)
)
# Add timeDistributed Layer
model.add(TimeDistributed(Dense(n_tags, activation="relu")))
# Optimiser
# adam = k.optimizers.Adam(lr=0.0005, beta_1=0.9, beta_2=0.999)
# Compile model
model.compile(
loss="categorical_crossentropy", optimizer="adam", metrics=["accuracy"]
)
model.summary()
return model
config = {
"batch_size": 64,
"lr": 0.0005,
"epochs": 2,
"weight_decay": 1e-5,
"save_path": "./models/model.ckpt",
"patience": 1000,
"hidden_size_LSTM_1": 384,
"dropout_1": 0.2,
"hidden_size_LSTM_2": 384,
"dropout_2": 0.5,
}
device = "cuda" if torch.cuda.is_available() else "cpu"
_save_name = f"{time.asctime( time.localtime(time.time()) )}"
class NerDataset(Dataset):
def __init__(self, data, labels):
super().__init__()
self.data = data
self.labels = labels
def __len__(self):
return self.data.shape[0]
def __getitem__(self, idx):
_data = self.data[idx]
# _data=self.tfm(_data.numpy())
_labels = self.labels[idx]
return _data, _labels
# train_set = NerDataset(train_tokens,train_tags)
# train_loader = DataLoader(train_set, batch_size=config['batch_size'], shuffle=True, num_workers=4, pin_memory=True)
# valid_set = NerDataset(test_tokens,test_tags)
# valid_loader = DataLoader(valid_set, batch_size=config['batch_size'], shuffle=True, num_workers=4, pin_memory=True)
train_set = NerDataset(train_tokens, train_tags)
train_loader = DataLoader(
train_set,
batch_size=config["batch_size"],
shuffle=True,
num_workers=1,
pin_memory=True,
)
valid_set = NerDataset(test_tokens, test_tags)
valid_loader = DataLoader(
valid_set,
batch_size=config["batch_size"],
shuffle=True,
num_workers=1,
pin_memory=True,
)
results = pd.DataFrame()
class NerNet(nn.Module):
def __init__(self):
super().__init__()
self.embedding = nn.Embedding(input_dim, output_dim)
self.LSTM_1 = nn.LSTM(
input_size=output_dim,
hidden_size=config["hidden_size_LSTM_1"],
num_layers=1,
bidirectional=True,
batch_first=True,
)
self.dropout_1 = nn.Dropout(config["dropout_1"])
self.LSTM_2 = nn.LSTM(
input_size=768,
hidden_size=config["hidden_size_LSTM_2"],
num_layers=1,
bidirectional=False,
batch_first=True,
)
self.dropout_2 = nn.Dropout(config["dropout_2"])
self.relu = nn.ReLU()
self.linear = nn.Linear(384, n_tags)
def forward(self, x):
# print(x.shape)
x = self.embedding(x)
# print('embed:',end="")
# print(x.shape,x.dtype)
# x=x.float()
x, (hn, cn) = self.LSTM_1(x)
# print('LSTM_1:',end="")
# print(x.shape,x.dtype)
x = self.dropout_1(x)
# print('dropout_1:',end="")
# print(x.shape,x.dtype)
x, (hn, cn) = self.LSTM_2(x)
# print('LSTM_2:',end="")
# print(x.shape,x.dtype)
x = self.dropout_2(x)
# print('dropout_2:',end="")
# print(x.shape,x.dtype)
x = self.linear(x)
# print('linear:',end="")
# print(x.shape,x.dtype)
return x
# for batch in tqdm(train_loader):
# x,y=batch
# print(x)
# print(y)
# print(x.shape)
# print(y.shape)
# break
model = NerNet().to(device)
loss_fn = nn.CrossEntropyLoss()
optimizer = torch.optim.Adam(
model.parameters(), lr=config["lr"], weight_decay=config["weight_decay"]
)
train_loss_ls = []
train_acc_ls = []
valid_loss_ls = []
valid_acc_ls = []
batch_train_loss_ls = []
batch_train_acc_ls = []
batch_valid_loss_ls = []
batch_valid_acc_ls = []
def train(model, optimizer, loss_fn, train_ds, test_ds, epochs, device=device):
# Initialize trackers, these are not parameters and should not be changed
stale = 0
best_acc = 0
train_loss_ls = []
train_acc_ls = []
valid_loss_ls = []
valid_acc_ls = []
for epoch in range(epochs):
# ---------- Training ----------
# Make sure the model is in train mode before training.
model.train()
# These are used to record information in training.
train_loss = []
train_accs = []
for batch in tqdm(train_loader):
# A batch consists of image data and corresponding labels.
imgs, labels = batch
# imgs, labels = imgs[0], labels[0]
# imgs = imgs.half()
# print(imgs.shape,labels.shape)
# print(imgs)
# print(labels)
# print(imgs.shape)
# print(labels.shape)
# Forward the data. (Make sure data and model are on the same device.)
logits = model(imgs.to(device))
# Calculate the cross-entropy loss.
# We don't need to apply softmax before computing cross-entropy as it is done automatically.
loss = loss_fn(logits, labels.float().to(device))
# Gradients stored in the parameters in the previous step should be cleared out first.
optimizer.zero_grad()
# Compute the gradients for parameters.
loss.backward()
# Clip the gradient norms for stable training.
grad_norm = nn.utils.clip_grad_norm_(model.parameters(), max_norm=10)
# Update the parameters with computed gradients.
optimizer.step()
# Compute the accuracy for current batch.
# print(logits)
# print(labels)
# print(logits.shape)
# print(labels.shape)
# print(logits.argmax(dim=-1))
acc = (labels[0][logits.argmax(dim=-1)] == labels.to(device)).float().mean()
# Record the loss and accuracy.
train_loss.append(loss.item())
train_accs.append(acc)
batch_train_loss_ls.append(train_loss[-1])
batch_train_acc_ls.append(train_accs[-1])
train_loss = sum(train_loss) / len(train_loss)
train_acc = sum(train_accs) / len(train_accs)
# Print the information.
print(
f"[ Train | {epoch + 1:03d}/{epochs:03d} ] loss = {train_loss:.5f}, acc = {train_acc:.5f}"
)
train_loss_ls.append(train_loss)
train_acc_ls.append(train_acc)
# ---------- Validation ----------
# Make sure the model is in eval mode so that some modules like dropout are disabled and work normally.
model.eval()
# These are used to record information in validation.
valid_loss = []
valid_accs = []
# Iterate the validation set by batches.
for batch in tqdm(valid_loader):
# A batch consists of image data and corresponding labels.
imgs, labels = batch
# imgs = imgs.half()
# We don't need gradient in validation.
# Using torch.no_grad() accelerates the forward process.
with torch.no_grad():
logits = model(imgs.to(device))
# We can still compute the loss (but not the gradient).
loss = loss_fn(logits, labels.float().to(device))
# Compute the accuracy for current batch.
# acc = (logits.argmax(dim=-1) == labels.to(device)).float().mean()
acc = (labels[0][logits.argmax(dim=-1)] == labels.to(device)).float().mean()
# Record the loss and accuracy.
valid_loss.append(loss.item())
valid_accs.append(acc)
# break
batch_valid_loss_ls.append(valid_loss[-1])
batch_valid_acc_ls.append(valid_accs[-1])
# The average loss and accuracy for entire validation set is the average of the recorded values.
valid_loss = sum(valid_loss) / len(valid_loss)
valid_acc = sum(valid_accs) / len(valid_accs)
# Print the information.
print(
f"[ Valid | {epoch + 1:03d}/{epochs:03d} ] loss = {valid_loss:.5f}, acc = {valid_acc:.5f}"
)
valid_loss_ls.append(valid_loss)
valid_acc_ls.append(valid_acc)
# update logs
if valid_acc > best_acc:
with open(f"./{_save_name}_log.txt", "a"):
print(
f"[ Valid | {epoch + 1:03d}/{epochs:03d} ] loss = {valid_loss:.5f}, acc = {valid_acc:.5f} -> best"
)
else:
with open(f"./{_save_name}_log.txt", "a"):
print(
f"[ Valid | {epoch + 1:03d}/{epochs:03d} ] loss = {valid_loss:.5f}, acc = {valid_acc:.5f}"
)
# save models
if valid_acc > best_acc:
print(f"Best model found at epoch {epoch}, saving model")
torch.save(
model.state_dict(), f"{_save_name}_best.ckpt"
) # only save best to prevent output memory exceed error
best_acc = valid_acc
stale = 0
else:
stale += 1
if stale > config["patience"]:
print(
f"No improvment {config['patience']} consecutive epochs, early stopping"
)
break
train(
model,
optimizer,
loss_fn,
train_set,
valid_set,
epochs=config["epochs"],
device=device,
)
import matplotlib.pyplot as plt
def plot_lists(train, valid, legend_labels):
# 创建一个新的图形
fig = plt.figure()
# 将数据添加到图形中
for x, y, label in zip(train, valid, legend_labels):
plt.plot(x, y, label=label)
# 添加图例
plt.legend()
# 显示图像
plt.show()
plot_lists(train_loss_ls, valid_loss_ls, ["train_loss", "valid_loss"])
plot_lists(train_acc_ls, valid_acc_ls, ["train_loss", "valid_loss"])
train_loss_ls
# def train_model(X, y, model):
# loss = list()
# for i in range(3):
# # fit model for one epoch on this sequence
# hist = model.fit(X, y, batch_size=1000, verbose=1, epochs=1, validation_split=0.2)
# loss.append(hist.history['loss'][0])
# return loss
# results = pd.DataFrame()
# model_bilstm_lstm = get_bilstm_lstm_model()
# plot_model(model_bilstm_lstm)
# results['with_add_lstm'] = train_model(train_tokens, np.array(train_tags), model_bilstm_lstm)
results["with_add_lstm"]
import spacy
from spacy import displacy
nlp = spacy.load("en_core_web_sm")
text = nlp(
"Jim bought 300 shares of Acme Corp. in 2006. And producing an annotated block of text that highlights the names of entities: [Jim]Person bought 300 shares of [Acme Corp.]Organization in [2006]Time. In this example, a person name consisting of one token, a two-token company name and a temporal expression have been detected and classified.State-of-the-art NER systems for English produce near-human performance. For example, the best system entering MUC-7 scored 93.39% of F-measure while human annotators scored 97.60% and 96.95%.[1][2]"
)
displacy.render(text, style="ent", jupyter=True)
|
# # HR Analytics: Job Change of Data Scientists
# 
# ## 1. Moduls to Use
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
import seaborn as sns
import warnings
warnings.simplefilter(action="ignore", category=FutureWarning)
# # 2. Import Datasets
path_train = "../input/hr-analytics-job-change-of-data-scientists/aug_train.csv"
path_test = "../input/hr-analytics-job-change-of-data-scientists/aug_test.csv"
path_submission = (
"../input/hr-analytics-job-change-of-data-scientists/sample_submission.csv"
)
df_train = pd.read_csv(path_train)
df_test = pd.read_csv(path_test)
# First 5 rows - Train
df_train.head()
# First 5 rows - Test
df_test.head()
# # 3. Exploratory Data Analysis
# ## 3.1 General analysis
df_train.dtypes
# Columns (features) - Train
list(df_train.columns)
# Columns (features) - Test
list(df_test.columns)
# Shape of the DataFrame - Train
df_train.shape
# Shape of the DataFrame - Test
df_test.shape
# Number of NaN per column
def percentage_nulls(df):
number_nulls = pd.DataFrame(df.isnull().sum(), columns=["Total"])
number_nulls["% nulls"] = round((number_nulls["Total"] / df.shape[0]) * 100, 1)
return number_nulls
# NaN for Train
percentage_nulls(df_train)
# NaN for Test
percentage_nulls(df_test)
# As seen in some columns there is a considerable amount of NaN.
# The % are similar in master datasets (train and test)
# ## 3.2 Feature *'city'* - City code
# City - Train
city_train = df_train["city"]
city_train.value_counts()
# City - Test
city_test = df_test["city"]
city_test.value_counts()
# it is observed that the cities are coded with a number and they are 123 different citys for Train and 108 for test
# ## 3.3 Feature *'city_ development _index'* - Developement index of the city (scaled)
# For more information: https://en.wikipedia.org/wiki/City_development_index
# Distribution city development index - Train
sns.displot(data=df_train, x="city_development_index", height=6, color="lightblue")
# Distribution city development index - Train
sns.displot(data=df_test, x="city_development_index", height=6, color="coral")
# ## 3.4 Feature *'gender'* - Gender of candidate
#
gender_train = df_train["gender"]
gender_test = df_test["gender"]
def percentage(df):
number = pd.DataFrame(df.value_counts())
number.columns = ["Total"]
number["%"] = round((number["Total"] / df.notnull().sum()) * 100, 1)
return number
percentage(gender_train)
percentage(gender_test)
# As you can see the% are similar in both datasets
fig, ax = plt.subplots(nrows=1, ncols=2, figsize=(10, 6))
sns.countplot(x="gender", data=df_train, palette="Set1", ax=ax[0]).set_title("Train")
sns.countplot(x="gender", data=df_test, palette="Set1", ax=ax[1]).set_title("Test")
fig.tight_layout()
fig.show()
# ## 3.5 Feature *'relevent_experience'* - Relevant experience of candidate
#
relevent_experience_train = df_train["relevent_experience"]
relevent_experience_test = df_test["relevent_experience"]
percentage(relevent_experience_train)
percentage(relevent_experience_test)
# As you can see the% are similar in both datasets
fig, ax = plt.subplots(nrows=1, ncols=2, figsize=(10, 6))
sns.countplot(
x="relevent_experience", data=df_train, palette="Set2", ax=ax[0]
).set_title("Train")
sns.countplot(
x="relevent_experience", data=df_test, palette="Set2", ax=ax[1]
).set_title("Test")
fig.tight_layout()
fig.show()
# ## 3.6 Feature *'enrolled_university'* - Type of University course enrolled if any
#
enrolled_university_train = df_train["enrolled_university"]
enrolled_university_test = df_test["enrolled_university"]
percentage(enrolled_university_train)
percentage(enrolled_university_test)
order_enrolled_university = percentage(enrolled_university_train).index
# As you can see the% are similar in both datasets
fig, ax = plt.subplots(nrows=1, ncols=2, figsize=(10, 6))
sns.countplot(
x="enrolled_university",
data=df_train,
palette="Set3",
ax=ax[0],
order=order_enrolled_university,
).set_title("Train")
sns.countplot(
x="enrolled_university",
data=df_test,
palette="Set3",
ax=ax[1],
order=order_enrolled_university,
).set_title("Test")
fig.tight_layout()
fig.show()
# ## 3.7 Feature *'education_level'* - Education level of candidate
#
education_level_train = df_train["education_level"]
education_level_test = df_test["education_level"]
percentage(education_level_train)
percentage(education_level_test)
# As you can see the% are similar in both datasets
order_education_level = percentage(education_level_train).index
fig, ax = plt.subplots(nrows=1, ncols=2, figsize=(12, 8))
sns.countplot(
x="education_level",
data=df_train,
palette="Set1",
ax=ax[0],
order=order_education_level,
).set_title("Train")
sns.countplot(
x="education_level",
data=df_test,
palette="Set1",
ax=ax[1],
order=order_education_level,
).set_title("Test")
fig.tight_layout()
fig.show()
# ## 3.8 Feature *'major_discipline'* - Education major discipline of candidate
#
major_discipline_train = df_train["major_discipline"]
major_discipline_test = df_test["major_discipline"]
percentage(major_discipline_train)
percentage(major_discipline_test)
# As you can see the% are similar in both datasets
order_major_discipline = percentage(major_discipline_train).index
fig, ax = plt.subplots(nrows=1, ncols=2, figsize=(14, 10))
sns.countplot(
x="major_discipline",
data=df_train,
palette="Set2",
ax=ax[0],
order=order_major_discipline,
).set_title("Train")
sns.countplot(
x="major_discipline",
data=df_test,
palette="Set2",
ax=ax[1],
order=order_major_discipline,
).set_title("Test")
fig.tight_layout()
fig.show()
# ## 3.9 Feature *'experience'* - Candidate total experience in years
#
experience_train = df_train["experience"]
experience_test = df_test["experience"]
percentage(experience_train)
percentage(experience_test)
order_experience = percentage(experience_train).index
fig, ax = plt.subplots(nrows=1, ncols=2, figsize=(14, 12))
sns.countplot(
x="experience", data=df_train, palette="Set3", ax=ax[0], order=order_experience
).set_title("Train")
sns.countplot(
x="experience", data=df_test, palette="Set3", ax=ax[1], order=order_experience
).set_title("Test")
fig.tight_layout()
fig.show()
# ## 3.10 Feature *'company_size'* - No of employees in current employer's company
company_size_train = df_train["company_size"]
company_size_test = df_test["company_size"]
percentage(company_size_train)
percentage(company_size_test)
# As you can see the% are similar in both datasets
order_company_size = percentage(company_size_train).index
fig, ax = plt.subplots(nrows=1, ncols=2, figsize=(14, 12))
sns.countplot(
x="company_size", data=df_train, palette="Set1", ax=ax[0], order=order_company_size
).set_title("Train")
sns.countplot(
x="company_size", data=df_test, palette="Set1", ax=ax[1], order=order_company_size
).set_title("Test")
fig.tight_layout()
fig.show()
# ## 3.11 Feature *'company_type'* - Type of current employer
company_type_train = df_train["company_type"]
company_type_test = df_test["company_type"]
percentage(company_type_train)
percentage(company_type_test)
# As you can see the% are similar in both datasets
order_company_type = percentage(company_type_train).index
fig, ax = plt.subplots(nrows=1, ncols=2, figsize=(14, 12))
sns.countplot(
x="company_type", data=df_train, palette="Set2", ax=ax[0], order=order_company_type
).set_title("Train")
sns.countplot(
x="company_type", data=df_test, palette="Set2", ax=ax[1], order=order_company_type
).set_title("Test")
fig.tight_layout()
fig.show()
# ## 3.12 Feature *'lastnewjob'* - Difference in years between previous job and current job
last_new_job_train = df_train["last_new_job"]
last_new_job_test = df_test["last_new_job"]
percentage(last_new_job_train)
percentage(last_new_job_test)
# As you can see the% are similar in both datasets
order_last_new_job = percentage(last_new_job_train).index
fig, ax = plt.subplots(nrows=1, ncols=2, figsize=(10, 8))
sns.countplot(
x="last_new_job", data=df_train, palette="Set3", ax=ax[0], order=order_last_new_job
).set_title("Train")
sns.countplot(
x="last_new_job", data=df_test, palette="Set3", ax=ax[1], order=order_last_new_job
).set_title("Test")
fig.tight_layout()
fig.show()
# ## 3.13 Feature *'training_hours'* - Training hours completed
# Distribution of training hours - Train
sns.displot(data=df_train, x="training_hours", height=6, color="lightblue")
# Distribution of training hours - Test
sns.displot(data=df_train, x="training_hours", height=6, color="coral")
# ## 3.14 *'Target'*
# 0 - Not looking for job change
# 1 - Looking for a job change
target = df_train["target"]
percentage(target)
sns.countplot(x="target", data=df_train, palette="Set1").set_title("Train")
# ### Target by city development index
# Distribution
sns.displot(
data=df_train, x="city_development_index", hue="target", kind="kde", height=6
)
# ### Target by gender
sns.catplot(
x="target", hue="gender", data=df_train, palette="Set1", kind="count", height=6
)
# ### Target by relevant experience
sns.catplot(
x="target",
hue="relevent_experience",
data=df_train,
palette="Set2",
kind="count",
height=6,
)
# ### Target by enrolled university
sns.catplot(
x="target",
hue="enrolled_university",
data=df_train,
palette="Set3",
kind="count",
height=6,
)
# ### Target by education level
sns.catplot(
x="target",
hue="education_level",
data=df_train,
palette="Set1",
kind="count",
height=8,
)
# ### Target by major discipline
sns.catplot(
x="target",
hue="major_discipline",
data=df_train,
palette="Set2",
kind="count",
height=8,
)
# ### Target by major experience
sns.catplot(x="target", hue="experience", data=df_train, kind="count", height=16)
# ### Target by training hours
# Distribution
sns.displot(data=df_train, x="training_hours", hue="target", kind="kde", height=6)
# ## 4. Data Preprocessing
# ### 4.1 Unnecessary columns
# Remember the train dataset
df_train.head()
df_train.columns
# I consider that these columns are not useful to use as features,therefore I delete them:
# 'enrollee_id'
# I remove the column 'enrollee_id' as it has no use
df_train = df_train.drop(["enrollee_id"], axis=1)
df_train.head()
# Select the target
target = df_train["target"]
# Drop the target
df_train = df_train.drop("target", axis=1)
df_train
# ## 4.2 Deal with NaN values
# As seen previously, some columns have a significant amount of NaN. We see what % is for the columns that remained
percentage_nulls(df_train)
# View the dtype for every column
df_train.dtypes
# Select columns name with categorical data
cat_columns = df_train.columns[df_train.dtypes == "object"]
cat_columns
# ### 4.2.1 Simple Imputation - Mode
# As a first approach I am going to make a simple imputation through mode
df_train_impute_mode = df_train.copy()
for columna in cat_columns:
df_train_impute_mode[columna].fillna(
df_train_impute_mode[columna].mode()[0], inplace=True
)
percentage_nulls(df_train_impute_mode)
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
import matplotlib.pyplot as plt
import seaborn as sns
sns.set(style="ticks")
flatui = ["#9b59b6", "#3498db", "#95a5a6", "#e74c3c", "#34495e", "#2ecc71"]
flatui = sns.color_palette(flatui)
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import LabelEncoder
from sklearn.metrics import accuracy_score
data = pd.read_csv("../input/iris-csv/IRIS.csv")
data.head()
data.shape
data.info()
print(data.columns)
data.species = LabelEncoder().fit_transform(data.species)
data.species.unique()
data
a_sepal_length = sns.swarmplot(x="species", y="sepal_length", data=data)
a_sepal_width = sns.swarmplot(x="species", y="sepal_width", data=data)
a_petal_length = sns.swarmplot(x="species", y="petal_length", data=data)
a_petal_width = sns.swarmplot(x="species", y="petal_width", data=data)
corr = data.corr()
g = sns.heatmap(
corr,
vmax=0.10,
center=0,
square=False,
linewidths=0.5,
cbar_kws={"shrink": 0.50},
annot=True,
fmt=".2f",
cmap="coolwarm",
)
sns.despine()
g.figure.set_size_inches(14, 10)
plt.show()
correlation = data.corr()
print(correlation["species"].sort_values(ascending=False), "\n")
delete_pl = data.drop(["petal_length"], axis=1)
delete_pl
delete_pl
# data = data.drop(columns = ['petal_width'])
delete_pl
# train test split
train, test = train_test_split(delete_pl, test_size=0.2, random_state=0)
train_X = train.drop(columns=["species"], axis=1)
train_Y = train["species"]
test_X = test.drop(columns=["species"], axis=1)
test_Y = test["species"]
train_X
train_Y
test_X
test_Y
model = LogisticRegression()
model.fit(train_X, train_Y)
predict = model.predict(test_X)
print("\nAccuracy score on test data: \n")
print(accuracy_score(test_Y, predict))
|
import os, sys, shutil, random
root_address = "/kaggle/input/diabetic-retinopathy-224x224-gaussian-filtered"
import imagehash
import numpy as np
import pandas as pd
import seaborn as sns
import tensorflow as tf
import matplotlib.pyplot as plt
from PIL import Image
from sklearn.model_selection import train_test_split
plt.style.use(
"https://github.com/dhaitz/matplotlib-stylesheets/raw/master/pitayasmoothie-dark.mplstyle"
)
# # Part I: Data handling
data_address = f"{root_address}/train.csv"
data = pd.read_csv(data_address)
data.head()
# ## Data overview
# We know that our dataset is comprised of images. First we have to investigate the distribution of data labels.
sns.countplot(data=data, x="diagnosis")
# The values in `diagnosis` which indicate the category of data, are unevenly distributed. This leads to a highly biased neural network.
# We can merge `Mild`, `Moderate`, `Proliferate_DR` and `Severe` to one category, beside `No_DR` which is technicaly true: The network will predict whether there is signs of retinopathy in the patience or not. The stage of the disease is out of the context.
# copy data
data2 = data.copy()
# select and merge every category other than `No_DR`
data2["diagnosis"] = data2["diagnosis"] > 0
data2["diagnosis"] = data2["diagnosis"].astype("int32")
# plot the distribution of categories
sns.countplot(data=data2, x="diagnosis")
# As it's shown above, now there is an almost evenly divided distribution.
# ## Data split
# Now data must be splitted into `train`, `test` and `val` sections.
# First we split data on 'train.csv' file. We will then know each filename belongs to what subdirectory.
# test and validation fraction
test_frac, val_frac = 0.25, 0.15
# separate `val` data from the `whole`
_, val = train_test_split(data2, test_size=val_frac, stratify=data2["diagnosis"])
# separate `test` and `train` from the remaining
train, test = train_test_split(
_,
test_size=test_frac / (1 - val_frac), # to have the same frac on the `whole`
stratify=_["diagnosis"],
)
# verify
print(f"{train.shape}, {test.shape}, {val.shape}\n")
# Here we verify the percentage of each subdirectory again.
# Make a new empty columns
data2["status"] = None
# fill values based on the `status`
for name, source in zip(["train", "test", "val"], [train, test, val]):
for j in source.index:
data2.at[j, "status"] = name
# check the results
print(data2.head(), "\n")
# plot the distribution
fig = plt.subplots(figsize=(6, 6))
sns.histplot(data=data2, x="status", hue="diagnosis", multiple="stack", stat="percent")
# ## Data packing
# A little bit of bash scripting to build subdirectories and copy the images into them. The desired directory structure is:
# ```
# data
# |-- test
# |-- DR
# |-- No_DR
# |-- train
# |-- DR
# |-- No_DR
# |-- val
# |-- DR
# |-- No_DR
# ```
# Merge all images into one temprory folder.
# here we copy all of the images into one folder. Files then will be copied to
# related folders according to `data2`
for cat in ["Mild", "Moderate", "No_DR", "Proliferate_DR", "Severe"]:
os.system(
f"cp -R {root_address}/gaussian_filtered_images/gaussian_filtered_images/{cat}/* ./Images"
)
# Copy the data from the temproray folder into the related subdirectories.
# loop over the splits
for name, dataframe in zip(["train", "test", "val"], [train, test, val]):
# loop over the categories
for filename, diag_code in zip(dataframe["id_code"], dataframe["diagnosis"]):
if diag_code:
os.system(f"cp ./Images/{filename}.png ./data/{name}/DR/")
else:
os.system(f"cp ./Images/{filename}.png ./data/{name}/No_DR/")
print(f"✅ {name}")
dup_dict = {}
for split in ["train", "test", "val"]:
for im_class in ["DR", "No_DR"]:
for image_name in os.listdir(f"./data/{split}/{im_class}"):
image = Image.open(f"./data/{split}/{im_class}/{image_name}")
image_hash = str(imagehash.phash(image))
if image_hash not in dup_dict.keys():
dup_dict[image_hash] = set()
dup_dict[image_hash].add(f"./data/{split}/{im_class}/{image_name}")
dup_dict2 = {}
num_same = 0
for key in dup_dict:
if len(dup_dict[key]) > 1:
dup_dict2[key] = dup_dict[key]
num_same += len(dup_dict[key])
del dup_dict
print(f"There are {num_same} almost similar images.")
num_to_show = 10
figure = plt.figure(figsize=(9, 45))
for i, im_hash in enumerate(random.sample([*dup_dict2.keys()], num_to_show)):
images = list(dup_dict2[im_hash])
im1 = np.asarray(Image.open(f"{images[0]}"))
im2 = np.asarray(Image.open(f"{images[1]}"))
plt.subplot(num_to_show, 2, 2 * i + 1)
plt.title(f"{images[0]}")
plt.imshow(im1)
plt.subplot(num_to_show, 2, 2 * i + 2)
plt.title(f"{images[1]}")
plt.imshow(im2)
train_data = tf.keras.preprocessing.image.ImageDataGenerator(
rescale=1.0 / 255.0
).flow_from_directory("./data/train", target_size=(224, 224), shuffle=True)
test_data = tf.keras.preprocessing.image.ImageDataGenerator(
rescale=1.0 / 255.0
).flow_from_directory("./data/test", target_size=(224, 224), shuffle=True)
val_data = tf.keras.preprocessing.image.ImageDataGenerator(
rescale=1.0 / 255.0
).flow_from_directory("./data/val", target_size=(224, 224), shuffle=True)
test_data.n
fig, ax = plt.subplots(3, 3, figsize=(18, 18), sharey="col")
for i, name, data_gen in [
*zip(range(3), ["train", "test", "validation"], [train_data, test_data, val_data])
]:
for j in range(3):
n_batch, n_image = np.random.randint(32), np.random.randint(data_gen.n // 32)
ax[i][j].imshow(train_data[n_image][0][n_batch])
label = np.argmax(train_data[n_image][1][n_batch])
ax[i][j].set_title(f'{["DR", "No_DR"][label]} - ({name})')
ax[i][j].axis("off")
# # Part II: Model fitting
base_model = tf.keras.applications.MobileNetV2(
include_top=False, input_shape=train_data.image_shape
)
base_model.trainable = False
input = tf.keras.layers.Input(shape=train_data.image_shape)
preprocess = tf.keras.applications.mobilenet_v2.preprocess_input
global_average_pooling = tf.keras.layers.GlobalAveragePooling2D()
dense0 = tf.keras.layers.Dense(100, activation="leaky_relu")
dense1 = tf.keras.layers.Dense(50, activation="leaky_relu")
predict = tf.keras.layers.Dense(2, activation="sigmoid")
x = preprocess(input)
x = base_model(x)
x = global_average_pooling(x)
x = dense0(x)
x = tf.keras.layers.Dropout(0.3)(x)
x = dense1(x)
model = tf.keras.Model(input, predict(x))
model.summary()
primary_learning_rate = 5e-3
model.compile(
optimizer=tf.keras.optimizers.Adam(learning_rate=primary_learning_rate),
loss=tf.keras.losses.BinaryCrossentropy(from_logits=True),
metrics=["accuracy", tf.keras.metrics.Precision(), tf.keras.metrics.Recall()],
)
model.evaluate(test_data)
history = model.fit(train_data, epochs=50, validation_data=val_data)
# make only the pre-trained model trainable
base_model.trainable = True
# select some of the layers to be untrainable
# we need the previous knowledge of the model (the pre-trained weights and bias)
for layer in base_model.layers[:150]:
layer.trainable = False
fine_learning_rate = 1e-5
model.compile(
optimizer=tf.keras.optimizers.Adam(learning_rate=fine_learning_rate),
loss=tf.keras.losses.BinaryCrossentropy(from_logits=True),
metrics=["accuracy", tf.keras.metrics.Precision(), tf.keras.metrics.Recall()],
)
model.summary()
history_fine = model.fit(train_data, epochs=30, validation_data=val_data)
model.evaluate(test_data)
fig, ax = plt.subplots(1, 2, figsize=(12, 6))
epochs = range(1, len(history.history["loss"]) + len(history_fine.history["loss"]) + 1)
ax[0].plot(
epochs, history.history["loss"] + history_fine.history["loss"], label="training"
)
ax[0].plot(
epochs,
history.history["val_loss"] + history_fine.history["val_loss"],
label="validation",
)
ax[0].set_title("Loss")
ax[0].legend()
ax[1].plot(
epochs,
history.history["accuracy"] + history_fine.history["accuracy"],
label="training",
)
ax[1].plot(
epochs,
history.history["val_accuracy"] + history_fine.history["val_accuracy"],
label="validation",
)
ax[1].set_title("Accuracy")
ax[1].legend()
input_layer = tf.keras.layers.Input(shape=(224, 224, 3))
max_pool = tf.keras.layers.MaxPooling2D(pool_size=(3, 3))
conv2d1 = tf.keras.layers.Conv2D(8, kernel_size=(3, 3), activation="relu")
batch_norm1 = tf.keras.layers.BatchNormalization()
conv2d2 = tf.keras.layers.Conv2D(8, kernel_size=(3, 3), activation="relu")
batch_norm2 = tf.keras.layers.BatchNormalization()
conv2d3 = tf.keras.layers.Conv2D(8, kernel_size=(3, 3), activation="relu")
batch_norm3 = tf.keras.layers.BatchNormalization()
conv2d4 = tf.keras.layers.Conv2D(8, kernel_size=(3, 3), activation="relu")
batch_norm4 = tf.keras.layers.BatchNormalization()
flatten = tf.keras.layers.Flatten()
dense1 = tf.keras.layers.Dense(50, activation="relu")
dropout1 = tf.keras.layers.Dropout(0.2)
predict = tf.keras.layers.Dense(2, activation="sigmoid")
x = input_layer
x = conv2d1(x)
x = max_pool(x)
x = batch_norm1(x)
x = conv2d2(x)
x = max_pool(x)
x = batch_norm2(x)
x = conv2d3(x)
x = max_pool(x)
x = batch_norm3(x)
x = flatten(x)
x = dense1(x)
x = dropout1(x)
model = tf.keras.Model(input_layer, predict(x))
model.summary()
model.compile(
loss=tf.keras.losses.BinaryCrossentropy(),
metrics="accuracy",
optimizer=tf.keras.optimizers.Adam(learning_rate=1e-4),
)
model_mobilenet_history = model.fit(train_data, epochs=30, validation_data=val_data)
model.save("mobilenet")
model.evaluate(test_data)
import matplotlib.pyplot as plt
def plot_curves(history):
loss = history.history["loss"]
val_loss = history.history["val_loss"]
accuracy = history.history["accuracy"]
val_accuracy = history.history["val_accuracy"]
epochs = range(len(history.history["loss"]))
# plot loss
plt.plot(epochs, loss, label="training_loss")
plt.plot(epochs, val_loss, label="val_loss")
plt.title("loss")
plt.xlabel("epochs")
plt.legend()
# plot accuracy
plt.figure()
plt.plot(epochs, accuracy, label="training_accuracy")
plt.plot(epochs, val_accuracy, label="val_accuracy")
plt.title("accuracy")
plt.xlabel("epochs")
plt.legend()
import tensorflow as tf
import cv2
import numpy as np
import matplotlib.pyplot as plt
def predict_class(path):
img = cv2.imread(path)
RGBImg = cv2.cvtColor(img, cv2.COLOR_BGR2RGB)
RGBImg = cv2.resize(RGBImg, (224, 224))
plt.imshow(RGBImg)
image = np.array(RGBImg) / 255.0
new_model = tf.keras.models.load_model("mobilenet")
predict = new_model.predict(np.array([image]))
per = np.argmax(predict, axis=1)
if per == 1:
print("No DR")
else:
print("DR")
predict_class(
"/kaggle/input/diabetic-retinopathy-224x224-gaussian-filtered/gaussian_filtered_images/gaussian_filtered_images/Mild/0024cdab0c1e.png"
)
# Necessary utility modules and libraries
import os
import shutil
import pathlib
import random
import datetime
# Plotting libraries
import numpy as np
import matplotlib.pyplot as plt
import matplotlib.image as mpimg
import seaborn as sns
import pandas as pd
# Libraries for building the model
import tensorflow as tf
import tensorflow_hub as hub
from tensorflow.keras.preprocessing import image_dataset_from_directory
from tensorflow.keras.preprocessing.image import ImageDataGenerator
from tensorflow.keras.layers import (
Input,
Dense,
Conv2D,
Flatten,
MaxPool2D,
Dropout,
GlobalAveragePooling2D,
BatchNormalization,
GlobalMaxPooling2D,
)
from tensorflow.keras.applications import (
DenseNet121,
ResNet50,
MobileNetV2,
InceptionV3,
EfficientNetB0,
)
from tensorflow.keras.models import Sequential
from tensorflow.keras import backend
from sklearn.model_selection import StratifiedKFold, KFold
# !pip install -U tensorflow==2.8.0
root_dir = "/kaggle/input/diabetic-retinopathy-224x224-gaussian-filtered/gaussian_filtered_images/gaussian_filtered_images"
classes = os.listdir(root_dir)
classes.pop(3)
classes
# Walk through gaussian_filtered_images directory and list names of files
for dirpath, dirnames, filenames in os.walk(root_dir):
print(
f"There are {len(dirnames)} directories and {len(filenames)} images in {dirpath.split('/')[-1]}"
)
# View random images in the dataset
def view_random_images(root_dir=root_dir, classes=classes):
class_paths = [root_dir + "/" + image_class for image_class in classes]
# print(class_paths)
images_path = []
labels = []
for i in range(len(class_paths)):
random_images = random.sample(os.listdir(class_paths[i]), 10)
random_images_path = [class_paths[i] + "/" + img for img in random_images]
for j in random_images_path:
images_path.append(j)
labels.append(classes[i])
images_path
plt.figure(figsize=(17, 10))
plt.suptitle("Image Dataset", fontsize=20)
for i in range(1, 51):
plt.subplot(5, 10, i)
img = mpimg.imread(images_path[i - 1])
plt.imshow(img, aspect="auto")
plt.title(labels[i - 1])
plt.axis(False)
# Observing the images
view_random_images()
# 2nd time
view_random_images()
# ## Applying K-Fold cross validation on dataset
train_csv = pd.read_csv(
"/kaggle/input/diabetic-retinopathy-224x224-gaussian-filtered/train.csv"
)
train_csv
train_csv["diagnosis"].value_counts()
train_df = {}
test_df = {}
for i in range(5):
df = train_csv[train_csv["diagnosis"] == i]["id_code"].to_list()
for j in random.sample(df, int(0.8 * len(df))):
train_df[j] = i
for j in df:
if j not in train_df.keys():
test_df[j] = i
train_df = pd.DataFrame(train_df.items(), columns=["id_code", "diagnosis"]).sample(
frac=1, random_state=42
)
test_df = pd.DataFrame(test_df.items(), columns=["id_code", "diagnosis"]).sample(
frac=1, random_state=42
)
train_df
def mapping(df):
class_code = {
0: "No_DR",
1: "Mild",
2: "Moderate",
3: "Severe",
4: "Proliferate_DR",
}
df["label"] = list(map(class_code.get, df["diagnosis"]))
df["path"] = [i[1]["label"] + "/" + i[1]["id_code"] + ".png" for i in df.iterrows()]
return df
mapping(train_df), mapping(test_df)
len(train_df), len(test_df)
# Initializing the input size
IMG_SHAPE = (224, 224)
N_SPLIT = 5
EPOCHS = 60
# Defining euclidean distance function to be used as a evaluation metrics
def euclideanDist(img1, img2):
return backend.sqrt(backend.sum((img1 - img2) ** 2))
# Function to perform k-fold validation on test model
def validation_k_fold(model_test, k=5, epochs=EPOCHS, n_splits=N_SPLIT, lr=0.001):
kfold = StratifiedKFold(n_splits=N_SPLIT, shuffle=True, random_state=42)
train_datagen = ImageDataGenerator(rescale=1.0 / 255)
validation_datagen = ImageDataGenerator(rescale=1.0 / 255)
test_datagen = ImageDataGenerator(rescale=1.0 / 255)
train_y = train_df["label"]
train_x = train_df["path"]
# Variable for keeping the count of the splits we're executing
j = 0
es = tf.keras.callbacks.EarlyStopping(monitor="loss", patience=5)
for train_idx, val_idx in list(kfold.split(train_x, train_y)):
x_train_df = train_df.iloc[train_idx]
x_valid_df = train_df.iloc[val_idx]
j += 1
train_data = train_datagen.flow_from_dataframe(
dataframe=x_train_df,
directory=root_dir,
x_col="path",
y_col="label",
class_mode="categorical",
target_size=IMG_SHAPE,
)
valid_data = validation_datagen.flow_from_dataframe(
dataframe=x_valid_df,
directory=root_dir,
x_col="path",
y_col="label",
class_mode="categorical",
target_size=IMG_SHAPE,
)
test_data = test_datagen.flow_from_dataframe(
dataframe=test_df,
directory=root_dir,
x_col="path",
y_col="label",
class_mode="categorical",
target_size=IMG_SHAPE,
)
# Initializing the early stopping callback
es = tf.keras.callbacks.EarlyStopping(monitor="loss", patience=5)
# Compile the model
model_test.compile(
loss="categorical_crossentropy",
optimizer=tf.keras.optimizers.Adamax(learning_rate=lr),
metrics=[euclideanDist, "accuracy", tf.keras.metrics.CosineSimilarity()],
)
history = model_test.fit_generator(
train_data,
validation_data=valid_data,
epochs=epochs,
validation_steps=len(valid_data),
callbacks=[es],
)
# Evaluate the model
result = model_test.evaluate(test_data)
model_test_result = {
"test_loss": result[0],
"test_accuracy": result[1],
}
return [history, model_test_result]
# Function to plot the performance metrics
def plot_result(hist):
plt.figure(figsize=(10, 7))
plt.suptitle(f"Performance Metrics", fontsize=20)
# Actual and validation losses
plt.subplot(2, 2, 1)
plt.plot(hist.history["loss"], label="train")
plt.plot(hist.history["val_loss"], label="validation")
plt.title("Train and validation loss curve")
plt.legend()
# Actual and validation accuracy
plt.subplot(2, 2, 2)
plt.plot(hist.history["accuracy"], label="train")
plt.plot(hist.history["val_accuracy"], label="validation")
plt.title("Training and validation accuracy curve")
plt.legend()
# ## Modelling (base Models)
# We'll use the following ImageNet models for training the images and observe the variations of the accuracy of the predicitions as predicted by the models:
# * AlexNet
# * DenseNet121
# * ResNet50
# * EfficientNetB0
# ## 1. AlexNet
# Basic CNN model for AlexNet
model_alexnet = tf.keras.Sequential(
[
Conv2D(
input_shape=IMG_SHAPE + (3,),
filters=96,
kernel_size=11,
strides=4,
activation="relu",
),
MaxPool2D(pool_size=3, strides=2),
Conv2D(
filters=256, kernel_size=5, strides=1, padding="valid", activation="relu"
),
MaxPool2D(pool_size=3, strides=2),
Conv2D(
filters=384, kernel_size=3, strides=1, padding="same", activation="relu"
),
Conv2D(
filters=384, kernel_size=3, strides=1, padding="same", activation="relu"
),
Conv2D(
filters=256, kernel_size=3, strides=1, padding="same", activation="relu"
),
MaxPool2D(pool_size=3, strides=2),
Flatten(),
Dense(len(classes), activation="softmax"),
],
name="model_AlexNet",
)
# Summary of AlexNet model
# from tensorflow.keras.utils import plot_model
# plot_model(model_alexnet)
model_alexnet.summary()
model_alexnet_history, model_alexnet_result = validation_k_fold(model_alexnet, lr=0.001)
# Evaluation metrics for alexnet
model_alexnet_result
# Performance metrics for AlexNet
plot_result(model_alexnet_history)
# ## 2. DenseNet
# Basic architecture of DenseNet
model_densenet = DenseNet121(
weights="imagenet", include_top=False, input_shape=(224, 224, 3)
)
x = model_densenet.output
x = GlobalMaxPooling2D()(x)
x = BatchNormalization()(x)
x = Dropout(0.5)(x)
# x= Dense(256, activation="relu")(x)
# x= Dense(128, activation="relu")(x)
x = BatchNormalization()(x)
x = Dropout(0.5)(x)
output = Dense(len(classes), activation="softmax")(x) # FC-layer
model_denseNet = tf.keras.Model(inputs=model_densenet.input, outputs=output)
len(model_denseNet.layers)
# for layer in model_denseNet.layers[:-50]:
# layer.trainable=False
# Summary of the denseNet model
model_denseNet.summary()
model_denseNet_history, model_denseNet_result = validation_k_fold(
model_denseNet, lr=0.001, epochs=50
)
# Evaluation metrics for denseNet model
model_denseNet_result
# Visualizing the evaluation metrics
plot_result(model_denseNet_history)
# ## 3. ResNet50
# Basic architecture model of ResNet50
model_resnet50 = ResNet50(
weights="imagenet", include_top=False, input_shape=(224, 224, 3)
)
x = model_resnet50.output
x = GlobalAveragePooling2D()(x)
x = BatchNormalization()(x)
x = Dropout(0.5)(x)
x = BatchNormalization()(x)
x = Dropout(0.5)(x)
output = Dense(len(classes), activation="softmax")(x) # FC-layer
model_resNet = tf.keras.Model(inputs=model_resnet50.input, outputs=output)
# Summary of the ResNet50 model
model_resNet.summary()
model_resNet_history, model_resNet_result = validation_k_fold(
model_resNet, lr=0.0001, epochs=50
)
# Evaluation metrics for resNet
model_resNet_result
plot_result(model_resNet_history)
# ## 4. EfficientNetB0
# Basic architecture model of MobileNetV2
model_efficientnet = EfficientNetB0(
weights="imagenet", include_top=False, input_shape=(224, 224, 3)
)
x = model_efficientnet.output
x = GlobalMaxPooling2D()(x)
x = BatchNormalization()(x)
x = Dropout(0.5)(x)
x = BatchNormalization()(x)
x = Dropout(0.5)(x)
output = Dense(len(classes), activation="softmax")(x) # FC-layer
model_efficientNet = tf.keras.Model(inputs=model_efficientnet.input, outputs=output)
# Summary of mobilenetv2 model
model_efficientNet.summary()
model_efficientnet_history, model_efficientnet_result = validation_k_fold(
model_efficientNet, lr=0.0001
)
# Evaluate metrics for MobileNetV2
model_efficientnet_result
plot_result(model_efficientnet_history)
# ## Visualizing the predictions of all the base models
def pred_vals(model_history):
d = {}
d["acc"] = np.mean(model_history.history["accuracy"])
d["val_acc"] = np.mean(model_history.history["val_accuracy"])
d["loss"] = np.mean(model_history.history["loss"])
d["val_loss"] = np.mean(model_history.history["val_loss"])
return d
df_result = pd.DataFrame(
{
"AlexNet": pred_vals(model_alexnet_history),
"DenseNet": pred_vals(model_denseNet_history),
"ResNet50": pred_vals(model_resNet_history),
"EfficientNetB0": pred_vals(model_efficientnet_history),
"MobileNet": pred_vals(model_mobilenet_history),
}
).transpose()
df_result = df_result.reset_index().rename(columns={"index": "model"})
df_result
# plt.style.use('seaborn')
# Plotting models and their accuracy performance
df_result.plot(x="model", y=["acc", "val_acc"], kind="bar", figsize=(15, 7))
plt.xticks(rotation=0)
plt.title("Respective Models and their training and validation accuracy")
|
import numpy as np # linear algebra
import pandas as pd # data processing, CSV file I/O (e.g. pd.read_csv)
# Input data files are available in the read-only "../input/" directory
# For example, running this (by clicking run or pressing Shift+Enter) will list all files under the input directory
import os
for dirname, _, filenames in os.walk("/kaggle/input"):
for filename in filenames:
print(os.path.join(dirname, filename))
# You can write up to 20GB to the current directory (/kaggle/working/) that gets preserved as output when you create a version using "Save & Run All"
# You can also write temporary files to /kaggle/temp/, but they won't be saved outside of the current session
# ## Introduction
# ##### Based on the presented data, it is necessary to build a linear classification model and, based on this model, try to accurately predict the presence of a kidney stone.
# ## Import libraries
import numpy as np
import pandas as pd
import seaborn as sns
import matplotlib.pyplot as plt
import scipy.stats as st
import warnings
warnings.filterwarnings("ignore")
plt.style.use("seaborn")
plt.rcParams["figure.figsize"] = (10, 8)
pd.options.display.max_rows = 2000
pd.options.display.max_columns = 500
# ## Load data
train = pd.read_csv("/kaggle/input/playground-series-s3e12/train.csv")
test = pd.read_csv("/kaggle/input/playground-series-s3e12/test.csv")
# ## Preliminary analyse
train.info()
train.sample(5)
train.iloc[:, 1:-1].describe()
# ## Exploratory data analysis
# ### Plot some statistics graphics for each features our data
sns.set_theme()
data_1 = train.iloc[:, 1:-1]
for i in range(6):
plt.subplot(3, 2, i + 1)
col = data_1.iloc[:, i]
sns.barplot(data=train, x="target", y=col)
plt.tight_layout()
plt.show()
sns.set_theme()
data_1 = train.iloc[:, 1:-1]
for i in range(6):
plt.subplot(3, 2, i + 1)
col = data_1.iloc[:, i]
sns.violinplot(data=train, x="target", y=col)
plt.tight_layout()
plt.show()
sns.set_theme()
data_1 = train.iloc[:, 1:-1]
for i in range(6):
plt.subplot(3, 2, i + 1)
sns.kdeplot(data=data_1, x=data_1.columns[i])
plt.tight_layout()
plt.show()
sns.set_theme()
data_1 = train.iloc[:, 1:-1]
for i in range(6):
plt.subplot(3, 2, i + 1)
sns.boxplot(data=data_1, x=data_1.columns[i])
plt.tight_layout()
plt.show()
sns.set_theme()
data_1 = train.iloc[:, 1:-1]
for i in range(6):
plt.subplot(3, 2, i + 1)
sns.boxenplot(data=data_1, x=data_1.columns[i])
plt.tight_layout()
plt.show()
# ### Pairplot features of data with use scatter kind
sns.set_theme()
sns.pairplot(train.iloc[:, 1:], hue="target", kind="scatter")
# ### With use kde kind
sns.set_theme()
sns.pairplot(train.iloc[:, 1:], kind="kde", hue="target")
# ### Pairplot with reg kind
sns.set_theme()
sns.pairplot(train.iloc[:, 1:], kind="reg", diag_kind="kde", hue="target")
# ### Let's build a correlation map of features
sns.set_theme()
sns.heatmap(
train.iloc[:, 1:-1].corr(),
cmap="PiYG",
center=0,
annot_kws={"size": 8},
fmt=".1f",
linewidths=0.1,
square=True,
annot=True,
)
# ### These graphs show that there is a correlation between the signs. It can be seen that there is no clear division between the features of the ranking class. We can say in advance that for training here the methods of bagging or boosting are better suited.
# ### Let's check each feature for a normal distribution
def checking_normality_func(feature: str):
plt.subplot(2, 3, 1)
sns.histplot(data=train, x=train.loc[train["target"] == 1, feature], kde=True)
plt.title("Class 1")
plt.subplot(2, 3, 2)
st.probplot(train.loc[train["target"] == 1, feature], plot=plt)
plt.title("Class 1")
plt.subplot(2, 3, 3)
sns.ecdfplot(data=train, x=train.loc[train["target"] == 1, feature])
plt.title("Class 1")
plt.subplot(2, 3, 4)
sns.histplot(data=train, x=train.loc[train["target"] == 0, feature], kde=True)
plt.title("Class 0")
plt.subplot(2, 3, 5)
st.probplot(train.loc[train["target"] == 0, feature], plot=plt)
plt.title("Class 0")
plt.subplot(2, 3, 6)
sns.ecdfplot(data=train, x=train.loc[train["target"] == 0, feature])
plt.title("Class 0")
plt.suptitle(
f"Distribution Plot and Checking the Distribution for Normality \n ${feature}$"
)
plt.tight_layout()
plt.show()
for i in train.columns[1:-1]:
print("*" * 100)
print("*" * 100)
sns.set_theme()
checking_normality_func(i)
# ## Preprocessing data
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
X = train.iloc[:, 1:-1]
_ = scaler.fit(X)
X = scaler.transform(X)
X.shape
Y = np.array(train.iloc[:, -1:])
Y.shape
A = test.iloc[:, 1:]
_ = scaler.fit(A)
A = scaler.transform(A)
A.shape
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, Y, test_size=0.2)
print(f"X_train: {X_train.shape}, y_train: {y_train.shape}")
print("-" * 50)
print(f"X_test: {X_test.shape}, y_test: {y_test.shape}")
# ## Build model
from lazypredict.Supervised import LazyClassifier
clf = LazyClassifier(predictions=True)
models, predictions = clf.fit(X_train, X_test, y_train, y_test)
models
|
# # Extreme Fine Tuning of LGBM using Incremental training
# In my efforts to push leaderboard i stumbled across a small trick to improve predictions in 4th to 5th decimal using same parameters and a single model, essentially it is a trick to improve prediction of your best parameter, squeezing more out of them!!. Trick is executed in following steps:
# * Find the best parameters for your LGBM, manually or using optimization methods of your choice.
# * train the model to the best RMSE you can get in one training round using high early stopping.
# * train the model for 1 or 2 rounds with reduced learning rate.
# * once the first few rounds are over, start reducing regularization params by a factor at each incremental training iteration, you will start observing improvements in 5th decimal place... which is enough to get 5th decimal improvement on your models leaderboard score.
# At the top of leaderboard this make a huge difference, i pushed my rank from `39` at **0.84202** to my best `6th place`(17th Feb 2021) with **0.84193**
# Lets check out.
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import seaborn as sns
from sklearn.model_selection import KFold, GridSearchCV, train_test_split
from sklearn.metrics import mean_squared_error
from sklearn.preprocessing import LabelEncoder
from lightgbm import LGBMRegressor
import optuna
from functools import partial
import warnings
warnings.filterwarnings("ignore")
train = pd.read_csv("../input/tabular-playground-series-feb-2021/train.csv")
test = pd.read_csv("../input/tabular-playground-series-feb-2021/test.csv")
X_train = train.drop(["id", "target"], axis=1)
y_train = train.target
X_test = test.drop(["id"], axis=1)
cat_cols = [feature for feature in train.columns if "cat" in feature]
def label_encoder(df):
for feature in cat_cols:
le = LabelEncoder()
le.fit(df[feature])
df[feature] = le.transform(df[feature])
return df
X_train = label_encoder(X_train)
X_test = label_encoder(X_test)
split = KFold(n_splits=5, random_state=2)
def objective(trial, X, y, name="xgb"):
params = {
"max_depth": trial.suggest_int("max_depth", 5, 50),
"n_estimators": 200000,
#'boosting':trial.suggest_categorical('boosting', ['gbdt', 'dart', 'goss']),
"subsample": trial.suggest_uniform("subsample", 0.2, 1.0),
"colsample_bytree": trial.suggest_uniform("colsample_bytree", 0.2, 1.0),
"learning_rate": trial.suggest_uniform("learning_rate", 0.007, 0.02),
"reg_lambda": trial.suggest_uniform("reg_lambda", 0.01, 50),
"reg_alpha": trial.suggest_uniform("reg_alpha", 0.01, 50),
"min_child_samples": trial.suggest_int("min_child_samples", 5, 100),
"num_leaves": trial.suggest_int("num_leaves", 10, 200),
"n_jobs": -1,
"metric": "rmse",
"max_bin": trial.suggest_int("max_bin", 300, 1000),
"cat_smooth": trial.suggest_int("cat_smooth", 5, 100),
"cat_l2": trial.suggest_loguniform("cat_l2", 1e-3, 100),
}
model = LGBMRegressor(**params)
X_train, X_val, y_train, y_val = train_test_split(
X, y, test_size=0.2, random_state=0
)
model.fit(
X_train,
y_train,
eval_set=[(X_val, y_val)],
eval_metric=["rmse"],
early_stopping_rounds=250,
categorical_feature=[0, 1, 2, 3, 4, 5, 6, 7, 8, 9],
# callbacks=[optuna.integration.LightGBMPruningCallback(trial, metric='rmse')],
verbose=0,
)
train_score = np.round(
np.sqrt(mean_squared_error(y_train, model.predict(X_train))), 5
)
test_score = np.round(np.sqrt(mean_squared_error(y_val, model.predict(X_val))), 5)
print(f"TRAIN RMSE : {train_score} || TEST RMSE : {test_score}")
return test_score
optimize = partial(objective, X=X_train, y=y_train)
study_lgbm = optuna.create_study(direction="minimize")
# study_lgbm.optimize(optimize, n_trials=300)
# i have commented out the trials so as to cut short the notebook execution time.
# From the above optuna trials the best parameters i could find were the following ones!
lgbm_params = {
"max_depth": 16,
"subsample": 0.8032697250789377,
"colsample_bytree": 0.21067140508531404,
"learning_rate": 0.009867383057779643,
"reg_lambda": 10.987474846877767,
"reg_alpha": 17.335285595031994,
"min_child_samples": 31,
"num_leaves": 66,
"max_bin": 522,
"cat_smooth": 81,
"cat_l2": 0.029690334194270022,
"metric": "rmse",
"n_jobs": -1,
"n_estimators": 20000,
}
preds_list_base = []
preds_list_final_iteration = []
preds_list_all = []
for train_idx, val_idx in split.split(X_train):
X_tr = X_train.iloc[train_idx]
X_val = X_train.iloc[val_idx]
y_tr = y_train.iloc[train_idx]
y_val = y_train.iloc[val_idx]
Model = LGBMRegressor(**lgbm_params).fit(
X_tr,
y_tr,
eval_set=[(X_val, y_val)],
eval_metric=["rmse"],
early_stopping_rounds=250,
categorical_feature=[0, 1, 2, 3, 4, 5, 6, 7, 8, 9],
# callbacks=[optuna.integration.LightGBMPruningCallback(trial, metric='rmse')],
verbose=0,
)
preds_list_base.append(Model.predict(X_test))
preds_list_all.append(Model.predict(X_test))
print(
f"RMSE for Base model is {np.sqrt(mean_squared_error(y_val, Model.predict(X_val)))}"
)
first_rmse = np.sqrt(mean_squared_error(y_val, Model.predict(X_val)))
params = lgbm_params.copy()
for i in range(1, 8):
if i > 2:
# reducing regularizing params if
params["reg_lambda"] *= 0.9
params["reg_alpha"] *= 0.9
params["num_leaves"] += 40
params["learning_rate"] = 0.003
Model = LGBMRegressor(**params).fit(
X_tr,
y_tr,
eval_set=[(X_val, y_val)],
eval_metric=["rmse"],
early_stopping_rounds=200,
categorical_feature=[0, 1, 2, 3, 4, 5, 6, 7, 8, 9],
# callbacks=[optuna.integration.LightGBMPruningCallback(trial, metric='rmse')],
verbose=0,
init_model=Model,
)
preds_list_all.append(Model.predict(X_test))
print(
f"RMSE for Incremental trial {i} model is {np.sqrt(mean_squared_error(y_val, Model.predict(X_val)))}"
)
last_rmse = np.sqrt(mean_squared_error(y_val, Model.predict(X_val)))
print("", end="\n\n")
print(f"Improvement of : {first_rmse - last_rmse}")
print("-" * 100)
preds_list_final_iteration.append(Model.predict(X_test))
# Great!! we can see that we have observed some further improvement in all the folds. Lets point out few findings:
# * The first few iterations are just using very low learning_rate.. after the 2nd iteration we can see that there are iterations with very good improvement, observed by reducing regularization.
# * There are also iterations where loss increased at later iterations slightly compared to previous iteration, showing that we have reached the limit in few iterations before the max iteration.
# * If you try setting verbose=1, you will observe that these improvements are observed only in first few trees created... after that loss starts to increase, LGBM keeps the best model. But reducing regularization does improve loss for first few trees!!!!
# I have 3 different sets of predictions, one for only the base model and one for all the predictions done and last one for only final iteration.
# * `y_preds_base` : **0.84196 - 0.84199** (keeps jumping between these)
# * `y_preds_all` : **0.84195 - 0.84196**
# * `y_preds_final_iteration` : **0.84193**
y_preds_base = np.array(preds_list_base).mean(axis=0)
y_preds_base
y_preds_all = np.array(preds_list_all).mean(axis=0)
y_preds_all
y_preds_final_iteration = np.array(preds_list_final_iteration).mean(axis=0)
y_preds_final_iteration
submission = pd.DataFrame({"id": test.id, "target": y_preds_final_iteration})
submission.to_csv("submission.csv", index=False)
pd.read_csv("submission.csv")
# ### Finding the right regularization reducing factors using optuna
# you may even try reducing or increasing few params and find the best mix of factors using optuna, it may even be possible to improve results more than achieved above, an example of the technique is shown below...
# creating a pre trained model to use in objective.
X_tr, X_val, y_tr, y_val = train_test_split(
X_train, y_train, test_size=0.2, random_state=0
)
lgbm = LGBMRegressor(**lgbm_params).fit(
X_tr,
y_tr,
eval_set=[(X_val, y_val)],
eval_metric=["rmse"],
early_stopping_rounds=250,
categorical_feature=[0, 1, 2, 3, 4, 5, 6, 7, 8, 9],
verbose=0,
)
def objective(trial, model, X, y, iterations=5):
X_train, X_val, y_train, y_val = train_test_split(
X, y, test_size=0.2, random_state=0
)
f1 = trial.suggest_uniform("f1", 0.1, 1.0)
f2 = trial.suggest_uniform("f2", 0.1, 3)
f3 = trial.suggest_int("f3", 20, 100)
f4 = trial.suggest_int("f4", 20, 50)
f5 = trial.suggest_int("f5", 1, 5)
lr_factor = trial.suggest_uniform("lr_factor", 0.1, 0.7)
params = lgbm_params.copy()
print(
f"RMSE for base model is {np.sqrt(mean_squared_error(y_val, Model.predict(X_val)))}"
)
for i in range(1, iterations):
if i > 2:
params["reg_lambda"] *= f1
params["reg_alpha"] += f2
params["num_leaves"] += f3
params["min_child_samples"] -= f4
params["cat_smooth"] -= f5
params["learning_rate"] *= lr_factor
# params['max_depth'] += f5
params["learning_rate"] = (
params["learning_rate"] if params["learning_rate"] > 0.0009 else 0.0009
)
# need to stop learning rate to reduce to a very insignificant value, hence we use this threshold
Model = model(**params).fit(
X_train,
y_train,
eval_set=[(X_val, y_val)],
eval_metric=["rmse"],
early_stopping_rounds=200,
categorical_feature=[0, 1, 2, 3, 4, 5, 6, 7, 8, 9],
verbose=1000,
init_model=Model if i > 1 else lgbm,
) # we will use pre trained model for first iteration
print(
f"RMSE for {i}th model is {np.sqrt(mean_squared_error(y_val, Model.predict(X_val)))}"
)
RMSE = mean_squared_error(y_val, Model.predict(X_val), squared=False)
return RMSE
study = optuna.create_study(direction="minimize")
optimize = partial(objective, X=X_train, y=y_train, model=LGBMRegressor)
# study.optimize(optimize, n_trials=100)
|
Subsets and Splits
No community queries yet
The top public SQL queries from the community will appear here once available.